file_id
stringlengths 5
9
| content
stringlengths 100
5.25M
| local_path
stringlengths 66
70
| kaggle_dataset_name
stringlengths 3
50
⌀ | kaggle_dataset_owner
stringlengths 3
20
⌀ | kversion
stringlengths 497
763
⌀ | kversion_datasetsources
stringlengths 71
5.46k
⌀ | dataset_versions
stringlengths 338
235k
⌀ | datasets
stringlengths 334
371
⌀ | users
stringlengths 111
264
⌀ | script
stringlengths 100
5.25M
| df_info
stringlengths 0
4.87M
| has_data_info
bool 2
classes | nb_filenames
int64 0
370
| retreived_data_description
stringlengths 0
4.44M
| script_nb_tokens
int64 25
663k
| upvotes
int64 0
1.65k
| tokens_description
int64 25
663k
| tokens_script
int64 25
663k
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
69046156
|
<jupyter_start><jupyter_text>Tomato Diseases Dataset (CSV+Images)
Kaggle dataset identifier: tomato-diseases-dataset-csvimages
<jupyter_code>import pandas as pd
df = pd.read_csv('tomato-diseases-dataset-csvimages/train.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 18160 entries, 0 to 18159
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 18160 non-null int64
1 path 18160 non-null object
2 img 18160 non-null object
3 label_text 18160 non-null object
4 label 18160 non-null int64
dtypes: int64(2), object(3)
memory usage: 709.5+ KB
<jupyter_text>Examples:
{
"Unnamed: 0": 0,
"path": "../input/plantvillage-dataset/color/Tomato___Late_blight/781e93a9-2059-42de-8075-658033a6abf7___RS_Late.B 6075.JPG",
"img": "781e93a9-2059-42de-8075-658033a6abf7___RS_Late.B 6075.JPG",
"label_text": "Tomato___Late_blight",
"label": 2
}
{
"Unnamed: 0": 1,
"path": "../input/plantvillage-dataset/color/Tomato___Late_blight/283ff0be-6e5e-4b4e-bf21-639780b77ffc___GHLB2 Leaf 8636.JPG",
"img": "283ff0be-6e5e-4b4e-bf21-639780b77ffc___GHLB2 Leaf 8636.JPG",
"label_text": "Tomato___Late_blight",
"label": 2
}
{
"Unnamed: 0": 2,
"path": "../input/plantvillage-dataset/color/Tomato___Late_blight/0db85707-41f9-42df-ba3b-842d14f00a68___GHLB2 Leaf 8909.JPG",
"img": "0db85707-41f9-42df-ba3b-842d14f00a68___GHLB2 Leaf 8909.JPG",
"label_text": "Tomato___Late_blight",
"label": 2
}
{
"Unnamed: 0": 3,
"path": "../input/plantvillage-dataset/color/Tomato___Late_blight/078a999d-6e6f-427e-a1e6-80b4d2df2bae___GHLB2 Leaf 9029.JPG",
"img": "078a999d-6e6f-427e-a1e6-80b4d2df2bae___GHLB2 Leaf 9029.JPG",
"label_text": "Tomato___Late_blight",
"label": 2
}
<jupyter_script># # Tomato Leaf Disease Detection 0.998 [inference]
# ### Hi kagglers, This is `inference` notebook using `Keras`.
# >
# > [Tomato Leaf Disease Detection 0.998 [Training]](https://www.kaggle.com/ammarnassanalhajali/tomato-leaf-disease-detection-0-998-training)
# ### Please if this kernel is useful, please upvote !!
import os, cv2, json
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from PIL import Image
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
import tensorflow as tf
from tensorflow.keras import models, layers
from tensorflow.keras.preprocessing import image
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping, ReduceLROnPlateau
from tensorflow.keras.applications import InceptionV3
from tensorflow.keras.optimizers import Adam
from PIL import Image
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import (
Dense,
Dropout,
Activation,
Input,
BatchNormalization,
GlobalAveragePooling2D,
)
train = pd.read_csv("../input/tomato-diseases-dataset-csvimages/train.csv")
from sklearn.model_selection import train_test_split
df_train, df_validate, y_train, y_test = train_test_split(
train, train.label, train_size=0.8, random_state=42, stratify=train.label
)
df_train = df_train.reset_index(drop=True)
df_validate = df_validate.reset_index(drop=True)
sample = df_train[df_train.label == 3].sample(3)
plt.figure(figsize=(15, 5))
for ind, (img, label) in enumerate(zip(sample.img, sample.label)):
plt.subplot(1, 3, ind + 1)
img = cv2.imread(
os.path.join(
"../input/tomato-diseases-dataset-csvimages/Tomato_images/Tomato_images",
img,
)
)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
plt.imshow(img)
plt.axis("off")
plt.show()
# Main parameters
BATCH_SIZE = 16
STEPS_PER_EPOCH = len(train) * 0.8 / BATCH_SIZE
VALIDATION_STEPS = len(train) * 0.2 / BATCH_SIZE
EPOCHS = 60 #
IMG_WIDTH = 256
IMG_HEIGHT = 256
train_dir = "../input/tomato-diseases-dataset-csvimages/Tomato_images/Tomato_images"
df_train.label = df_train.label.astype("str")
df_validate.label = df_validate.label.astype("str")
train_datagen = ImageDataGenerator(
rescale=1.0 / 255,
shear_range=0.2,
zoom_range=0.2,
rotation_range=180,
vertical_flip=True,
horizontal_flip=True,
)
# our train_datagen generator will use the following transformations on the images
validation_datagen = ImageDataGenerator(rescale=1.0 / 255)
train_generator = train_datagen.flow_from_dataframe(
df_train,
train_dir,
target_size=(IMG_WIDTH, IMG_HEIGHT),
batch_size=BATCH_SIZE,
x_col="img",
y_col="label",
class_mode="categorical",
)
# generator = ImageDataGenerator(*args).flow_from_dataframe(dataframe, directory, target_size,
# batch_size, x_col, y_col, class_mode)
# your dataframe shoudl be in the format such that x_col = features, y_col = class/label
# binary class mode since output is either 0(dog) or 1(cat)
validation_generator = validation_datagen.flow_from_dataframe(
df_validate,
train_dir,
target_size=(IMG_WIDTH, IMG_HEIGHT),
x_col="img",
y_col="label",
class_mode="categorical",
batch_size=BATCH_SIZE,
)
def create_model():
efficientnet_layers = InceptionV3(
weights="imagenet",
include_top=False,
input_shape=(IMG_WIDTH, IMG_HEIGHT, 3),
pooling="avg",
)
model = Sequential()
model.add(efficientnet_layers)
model.add(Dense(10, activation="softmax"))
model.compile(
optimizer=Adam(lr=0.001), loss="categorical_crossentropy", metrics=["acc"]
)
return model
model = create_model()
model.summary()
model.load_weights("../input/tomatoleafdiseasedetection-weights/InceptionV3_256.h5")
# ss=df_validate.sample(n=20)
ss = df_validate
ss = ss[["img", "label"]]
preds = []
for image_id in ss.img:
image = Image.open(
os.path.join(
"../input/tomato-diseases-dataset-csvimages/Tomato_images/Tomato_images/",
image_id,
)
)
array = tf.keras.preprocessing.image.img_to_array(image)
array = array / 255
image = np.expand_dims(array, axis=0)
preds.append(np.argmax(model.predict(image)))
ss["labelP"] = preds
ss
score = model.evaluate_generator(validation_generator)
print("Test loss:", score[0])
print("Test accuracy:", score[1])
confusion_matrix = pd.crosstab(
ss.label, ss.labelP, rownames=["Actual"], colnames=["Predicted"]
)
print(confusion_matrix)
plt.figure(figsize=(10, 8))
# use seaborn to draw the headmap
sns.heatmap(
confusion_matrix,
xticklabels=confusion_matrix.columns.values, # x label
yticklabels=confusion_matrix.columns.values,
cmap="YlGnBu",
annot=True,
fmt="d",
)
plt.show()
from imblearn.metrics import sensitivity_score, specificity_score
from sklearn.metrics import (
f1_score,
precision_score,
recall_score,
accuracy_score,
confusion_matrix,
)
y_test = ss.label.values.astype(int)
y_pred = ss.labelP.values.astype(int)
type(y_test)
# Print f1, precision, and recall scores
print("specificity:", specificity_score(y_test, y_pred, average="macro"))
print("sensitivity:", sensitivity_score(y_test, y_pred, average="macro"))
print("recall:", recall_score(y_test, y_pred, average="macro"))
print("precision::", precision_score(y_test, y_pred, average="macro"))
print("f1_score:", f1_score(y_test, y_pred, average="macro"))
print("accuracy_score:", accuracy_score(y_test, y_pred))
from sklearn.metrics import classification_report
import numpy as np
print(classification_report(y_test, y_pred))
y_true = y_test
y_prediction = y_pred
cnf_matrix = confusion_matrix(y_true, y_prediction)
print(cnf_matrix)
# [[1 1 3]
# [3 2 2]
# [1 3 1]]
FP = cnf_matrix.sum(axis=0) - np.diag(cnf_matrix)
FN = cnf_matrix.sum(axis=1) - np.diag(cnf_matrix)
TP = np.diag(cnf_matrix)
TN = cnf_matrix.sum() - (FP + FN + TP)
FP = FP.astype(float)
FN = FN.astype(float)
TP = TP.astype(float)
TN = TN.astype(float)
# Sensitivity, hit rate, recall, or true positive rate
TPR = TP / (TP + FN)
# Specificity or true negative rate
TNR = TN / (TN + FP)
# Precision or positive predictive value
PPV = TP / (TP + FP)
# Negative predictive value
NPV = TN / (TN + FN)
# Fall out or false positive rate
FPR = FP / (FP + TN)
# False negative rate
FNR = FN / (TP + FN)
# False discovery rate
FDR = FP / (TP + FP)
# Overall accuracy
ACC = (TP + TN) / (TP + FP + FN + TN)
print("Sensitivity OR recall")
print(TPR)
print("-------------------")
print("Specificity")
print(TNR)
print("-------------------")
print("Precision")
print(PPV)
print("-------------------")
print("accuracy")
print(ACC)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/046/69046156.ipynb
|
tomato-diseases-dataset-csvimages
|
ammarnassanalhajali
|
[{"Id": 69046156, "ScriptId": 17307062, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5966695, "CreationDate": "07/26/2021 08:36:05", "VersionNumber": 5.0, "Title": "Tomato Leaf Disease Detection 0.998 [inference]", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 227.0, "LinesInsertedFromPrevious": 11.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 216.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 3}]
|
[{"Id": 91775532, "KernelVersionId": 69046156, "SourceDatasetVersionId": 2222983}]
|
[{"Id": 2222983, "DatasetId": 1335181, "DatasourceVersionId": 2264687, "CreatorUserId": 5966695, "LicenseName": "Unknown", "CreationDate": "05/12/2021 00:09:31", "VersionNumber": 1.0, "Title": "Tomato Diseases Dataset (CSV+Images)", "Slug": "tomato-diseases-dataset-csvimages", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1335181, "CreatorUserId": 5966695, "OwnerUserId": 5966695.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2222983.0, "CurrentDatasourceVersionId": 2264687.0, "ForumId": 1354120, "Type": 2, "CreationDate": "05/12/2021 00:09:31", "LastActivityDate": "05/12/2021", "TotalViews": 5441, "TotalDownloads": 516, "TotalVotes": 14, "TotalKernels": 2}]
|
[{"Id": 5966695, "UserName": "ammarnassanalhajali", "DisplayName": "Ammar Alhaj Ali", "RegisterDate": "10/15/2020", "PerformanceTier": 4}]
|
# # Tomato Leaf Disease Detection 0.998 [inference]
# ### Hi kagglers, This is `inference` notebook using `Keras`.
# >
# > [Tomato Leaf Disease Detection 0.998 [Training]](https://www.kaggle.com/ammarnassanalhajali/tomato-leaf-disease-detection-0-998-training)
# ### Please if this kernel is useful, please upvote !!
import os, cv2, json
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from PIL import Image
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
import tensorflow as tf
from tensorflow.keras import models, layers
from tensorflow.keras.preprocessing import image
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping, ReduceLROnPlateau
from tensorflow.keras.applications import InceptionV3
from tensorflow.keras.optimizers import Adam
from PIL import Image
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import (
Dense,
Dropout,
Activation,
Input,
BatchNormalization,
GlobalAveragePooling2D,
)
train = pd.read_csv("../input/tomato-diseases-dataset-csvimages/train.csv")
from sklearn.model_selection import train_test_split
df_train, df_validate, y_train, y_test = train_test_split(
train, train.label, train_size=0.8, random_state=42, stratify=train.label
)
df_train = df_train.reset_index(drop=True)
df_validate = df_validate.reset_index(drop=True)
sample = df_train[df_train.label == 3].sample(3)
plt.figure(figsize=(15, 5))
for ind, (img, label) in enumerate(zip(sample.img, sample.label)):
plt.subplot(1, 3, ind + 1)
img = cv2.imread(
os.path.join(
"../input/tomato-diseases-dataset-csvimages/Tomato_images/Tomato_images",
img,
)
)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
plt.imshow(img)
plt.axis("off")
plt.show()
# Main parameters
BATCH_SIZE = 16
STEPS_PER_EPOCH = len(train) * 0.8 / BATCH_SIZE
VALIDATION_STEPS = len(train) * 0.2 / BATCH_SIZE
EPOCHS = 60 #
IMG_WIDTH = 256
IMG_HEIGHT = 256
train_dir = "../input/tomato-diseases-dataset-csvimages/Tomato_images/Tomato_images"
df_train.label = df_train.label.astype("str")
df_validate.label = df_validate.label.astype("str")
train_datagen = ImageDataGenerator(
rescale=1.0 / 255,
shear_range=0.2,
zoom_range=0.2,
rotation_range=180,
vertical_flip=True,
horizontal_flip=True,
)
# our train_datagen generator will use the following transformations on the images
validation_datagen = ImageDataGenerator(rescale=1.0 / 255)
train_generator = train_datagen.flow_from_dataframe(
df_train,
train_dir,
target_size=(IMG_WIDTH, IMG_HEIGHT),
batch_size=BATCH_SIZE,
x_col="img",
y_col="label",
class_mode="categorical",
)
# generator = ImageDataGenerator(*args).flow_from_dataframe(dataframe, directory, target_size,
# batch_size, x_col, y_col, class_mode)
# your dataframe shoudl be in the format such that x_col = features, y_col = class/label
# binary class mode since output is either 0(dog) or 1(cat)
validation_generator = validation_datagen.flow_from_dataframe(
df_validate,
train_dir,
target_size=(IMG_WIDTH, IMG_HEIGHT),
x_col="img",
y_col="label",
class_mode="categorical",
batch_size=BATCH_SIZE,
)
def create_model():
efficientnet_layers = InceptionV3(
weights="imagenet",
include_top=False,
input_shape=(IMG_WIDTH, IMG_HEIGHT, 3),
pooling="avg",
)
model = Sequential()
model.add(efficientnet_layers)
model.add(Dense(10, activation="softmax"))
model.compile(
optimizer=Adam(lr=0.001), loss="categorical_crossentropy", metrics=["acc"]
)
return model
model = create_model()
model.summary()
model.load_weights("../input/tomatoleafdiseasedetection-weights/InceptionV3_256.h5")
# ss=df_validate.sample(n=20)
ss = df_validate
ss = ss[["img", "label"]]
preds = []
for image_id in ss.img:
image = Image.open(
os.path.join(
"../input/tomato-diseases-dataset-csvimages/Tomato_images/Tomato_images/",
image_id,
)
)
array = tf.keras.preprocessing.image.img_to_array(image)
array = array / 255
image = np.expand_dims(array, axis=0)
preds.append(np.argmax(model.predict(image)))
ss["labelP"] = preds
ss
score = model.evaluate_generator(validation_generator)
print("Test loss:", score[0])
print("Test accuracy:", score[1])
confusion_matrix = pd.crosstab(
ss.label, ss.labelP, rownames=["Actual"], colnames=["Predicted"]
)
print(confusion_matrix)
plt.figure(figsize=(10, 8))
# use seaborn to draw the headmap
sns.heatmap(
confusion_matrix,
xticklabels=confusion_matrix.columns.values, # x label
yticklabels=confusion_matrix.columns.values,
cmap="YlGnBu",
annot=True,
fmt="d",
)
plt.show()
from imblearn.metrics import sensitivity_score, specificity_score
from sklearn.metrics import (
f1_score,
precision_score,
recall_score,
accuracy_score,
confusion_matrix,
)
y_test = ss.label.values.astype(int)
y_pred = ss.labelP.values.astype(int)
type(y_test)
# Print f1, precision, and recall scores
print("specificity:", specificity_score(y_test, y_pred, average="macro"))
print("sensitivity:", sensitivity_score(y_test, y_pred, average="macro"))
print("recall:", recall_score(y_test, y_pred, average="macro"))
print("precision::", precision_score(y_test, y_pred, average="macro"))
print("f1_score:", f1_score(y_test, y_pred, average="macro"))
print("accuracy_score:", accuracy_score(y_test, y_pred))
from sklearn.metrics import classification_report
import numpy as np
print(classification_report(y_test, y_pred))
y_true = y_test
y_prediction = y_pred
cnf_matrix = confusion_matrix(y_true, y_prediction)
print(cnf_matrix)
# [[1 1 3]
# [3 2 2]
# [1 3 1]]
FP = cnf_matrix.sum(axis=0) - np.diag(cnf_matrix)
FN = cnf_matrix.sum(axis=1) - np.diag(cnf_matrix)
TP = np.diag(cnf_matrix)
TN = cnf_matrix.sum() - (FP + FN + TP)
FP = FP.astype(float)
FN = FN.astype(float)
TP = TP.astype(float)
TN = TN.astype(float)
# Sensitivity, hit rate, recall, or true positive rate
TPR = TP / (TP + FN)
# Specificity or true negative rate
TNR = TN / (TN + FP)
# Precision or positive predictive value
PPV = TP / (TP + FP)
# Negative predictive value
NPV = TN / (TN + FN)
# Fall out or false positive rate
FPR = FP / (FP + TN)
# False negative rate
FNR = FN / (TP + FN)
# False discovery rate
FDR = FP / (TP + FP)
# Overall accuracy
ACC = (TP + TN) / (TP + FP + FN + TN)
print("Sensitivity OR recall")
print(TPR)
print("-------------------")
print("Specificity")
print(TNR)
print("-------------------")
print("Precision")
print(PPV)
print("-------------------")
print("accuracy")
print(ACC)
|
[{"tomato-diseases-dataset-csvimages/train.csv": {"column_names": "[\"Unnamed: 0\", \"path\", \"img\", \"label_text\", \"label\"]", "column_data_types": "{\"Unnamed: 0\": \"int64\", \"path\": \"object\", \"img\": \"object\", \"label_text\": \"object\", \"label\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 18160 entries, 0 to 18159\nData columns (total 5 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Unnamed: 0 18160 non-null int64 \n 1 path 18160 non-null object\n 2 img 18160 non-null object\n 3 label_text 18160 non-null object\n 4 label 18160 non-null int64 \ndtypes: int64(2), object(3)\nmemory usage: 709.5+ KB\n", "summary": "{\"Unnamed: 0\": {\"count\": 18160.0, \"mean\": 9079.5, \"std\": 5242.484779822128, \"min\": 0.0, \"25%\": 4539.75, \"50%\": 9079.5, \"75%\": 13619.25, \"max\": 18159.0}, \"label\": {\"count\": 18160.0, \"mean\": 4.755726872246696, \"std\": 2.801276569006158, \"min\": 0.0, \"25%\": 2.0, \"50%\": 5.0, \"75%\": 7.0, \"max\": 9.0}}", "examples": "{\"Unnamed: 0\":{\"0\":0,\"1\":1,\"2\":2,\"3\":3},\"path\":{\"0\":\"..\\/input\\/plantvillage-dataset\\/color\\/Tomato___Late_blight\\/781e93a9-2059-42de-8075-658033a6abf7___RS_Late.B 6075.JPG\",\"1\":\"..\\/input\\/plantvillage-dataset\\/color\\/Tomato___Late_blight\\/283ff0be-6e5e-4b4e-bf21-639780b77ffc___GHLB2 Leaf 8636.JPG\",\"2\":\"..\\/input\\/plantvillage-dataset\\/color\\/Tomato___Late_blight\\/0db85707-41f9-42df-ba3b-842d14f00a68___GHLB2 Leaf 8909.JPG\",\"3\":\"..\\/input\\/plantvillage-dataset\\/color\\/Tomato___Late_blight\\/078a999d-6e6f-427e-a1e6-80b4d2df2bae___GHLB2 Leaf 9029.JPG\"},\"img\":{\"0\":\"781e93a9-2059-42de-8075-658033a6abf7___RS_Late.B 6075.JPG\",\"1\":\"283ff0be-6e5e-4b4e-bf21-639780b77ffc___GHLB2 Leaf 8636.JPG\",\"2\":\"0db85707-41f9-42df-ba3b-842d14f00a68___GHLB2 Leaf 8909.JPG\",\"3\":\"078a999d-6e6f-427e-a1e6-80b4d2df2bae___GHLB2 Leaf 9029.JPG\"},\"label_text\":{\"0\":\"Tomato___Late_blight\",\"1\":\"Tomato___Late_blight\",\"2\":\"Tomato___Late_blight\",\"3\":\"Tomato___Late_blight\"},\"label\":{\"0\":2,\"1\":2,\"2\":2,\"3\":2}}"}}]
| true | 1 |
<start_data_description><data_path>tomato-diseases-dataset-csvimages/train.csv:
<column_names>
['Unnamed: 0', 'path', 'img', 'label_text', 'label']
<column_types>
{'Unnamed: 0': 'int64', 'path': 'object', 'img': 'object', 'label_text': 'object', 'label': 'int64'}
<dataframe_Summary>
{'Unnamed: 0': {'count': 18160.0, 'mean': 9079.5, 'std': 5242.484779822128, 'min': 0.0, '25%': 4539.75, '50%': 9079.5, '75%': 13619.25, 'max': 18159.0}, 'label': {'count': 18160.0, 'mean': 4.755726872246696, 'std': 2.801276569006158, 'min': 0.0, '25%': 2.0, '50%': 5.0, '75%': 7.0, 'max': 9.0}}
<dataframe_info>
RangeIndex: 18160 entries, 0 to 18159
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 18160 non-null int64
1 path 18160 non-null object
2 img 18160 non-null object
3 label_text 18160 non-null object
4 label 18160 non-null int64
dtypes: int64(2), object(3)
memory usage: 709.5+ KB
<some_examples>
{'Unnamed: 0': {'0': 0, '1': 1, '2': 2, '3': 3}, 'path': {'0': '../input/plantvillage-dataset/color/Tomato___Late_blight/781e93a9-2059-42de-8075-658033a6abf7___RS_Late.B 6075.JPG', '1': '../input/plantvillage-dataset/color/Tomato___Late_blight/283ff0be-6e5e-4b4e-bf21-639780b77ffc___GHLB2 Leaf 8636.JPG', '2': '../input/plantvillage-dataset/color/Tomato___Late_blight/0db85707-41f9-42df-ba3b-842d14f00a68___GHLB2 Leaf 8909.JPG', '3': '../input/plantvillage-dataset/color/Tomato___Late_blight/078a999d-6e6f-427e-a1e6-80b4d2df2bae___GHLB2 Leaf 9029.JPG'}, 'img': {'0': '781e93a9-2059-42de-8075-658033a6abf7___RS_Late.B 6075.JPG', '1': '283ff0be-6e5e-4b4e-bf21-639780b77ffc___GHLB2 Leaf 8636.JPG', '2': '0db85707-41f9-42df-ba3b-842d14f00a68___GHLB2 Leaf 8909.JPG', '3': '078a999d-6e6f-427e-a1e6-80b4d2df2bae___GHLB2 Leaf 9029.JPG'}, 'label_text': {'0': 'Tomato___Late_blight', '1': 'Tomato___Late_blight', '2': 'Tomato___Late_blight', '3': 'Tomato___Late_blight'}, 'label': {'0': 2, '1': 2, '2': 2, '3': 2}}
<end_description>
| 2,204 | 3 | 3,074 | 2,204 |
69046074
|
<jupyter_start><jupyter_text>MosMedData FullChestCT
Kaggle dataset identifier: mosmeddata-fullchestct
<jupyter_script>import numpy as np
import pandas as pd
import os
import tensorflow as tf
import cv2
import matplotlib.pyplot as plt
import tensorflow as tf
import nibabel as nib
image_paths0 = []
labels0 = []
for dirname, _, filenames in os.walk(
"../input/mosmeddata-fullchestct/COVID19_1110/studies/CT-0"
):
for filename in filenames:
image_paths0.append(os.path.join(dirname, filename))
labels0.append(0)
image_paths1 = []
labels1 = []
for dirname, _, filenames in os.walk(
"../input/mosmeddata-fullchestct/COVID19_1110/studies/CT-1"
):
for filename in filenames:
image_paths1.append(os.path.join(dirname, filename))
labels1.append(1)
image_paths2 = []
labels2 = []
for dirname, _, filenames in os.walk(
"../input/mosmeddata-fullchestct/COVID19_1110/studies/CT-2"
):
for filename in filenames:
image_paths2.append(os.path.join(dirname, filename))
labels2.append(2)
image_paths3 = []
labels3 = []
for dirname, _, filenames in os.walk(
"../input/mosmeddata-fullchestct/COVID19_1110/studies/CT-3"
):
for filename in filenames:
image_paths3.append(os.path.join(dirname, filename))
labels3.append(3)
image_paths = []
image_paths.extend(image_paths0)
image_paths.extend(image_paths1)
image_paths.extend(image_paths2)
image_paths.extend(image_paths3)
labels = []
labels.extend(labels0)
labels.extend(labels1)
labels.extend(labels2)
labels.extend(labels3)
np.max(labels)
from sklearn.utils import shuffle
image_paths, labels = shuffle(image_paths, labels, random_state=10800)
# def parse_function(image_paths, labels):
# image_path = tf.compat.v1.data.make_one_shot_iterator(image_path)
# print(image_path)
image_names_tab = []
labels_tab = []
counter = 0
for image_path, label in zip(image_paths[:20], labels[:20]):
niimg = nib.load(image_path)
npimage = niimg.get_fdata()
s = npimage.shape
for j in range(20, 30):
img = np.zeros((s[0], s[1], 3))
img[:, :, 0] = npimage[:, :, j]
img[:, :, 1] = npimage[:, :, j]
img[:, :, 2] = npimage[:, :, j]
img = img / np.max(npimage[:, :, j])
# img = tf.cast(img, tf.float32)
img = cv2.resize(img, (224, 224))
image_names_tab.append(img)
labels_tab.append(label)
counter += 1
print(counter, end="\r")
np.shape(image_names_tab)
image_names = image_names_tab
labels = labels_tab
image_names1 = []
image_names2 = []
image_names3 = []
image_names4 = []
image_names5 = []
image_names6 = []
image_names7 = []
image_names8 = []
image_names9 = []
image_names10 = []
labels1 = []
labels2 = []
labels3 = []
labels4 = []
labels5 = []
labels6 = []
labels7 = []
labels8 = []
labels9 = []
labels10 = []
counter = 0
for i in range(0, len(image_names), 10):
image_names1.append(image_names[i])
image_names2.append(image_names[i + 1])
image_names3.append(image_names[i + 2])
image_names4.append(image_names[i + 3])
image_names5.append(image_names[i + 4])
image_names6.append(image_names[i + 5])
image_names7.append(image_names[i + 6])
image_names8.append(image_names[i + 7])
image_names9.append(image_names[i + 8])
image_names10.append(image_names[i + 9])
labels1.append(labels[i])
labels2.append(labels[i + 1])
labels3.append(labels[i + 2])
labels4.append(labels[i + 3])
labels5.append(labels[i + 4])
labels6.append(labels[i + 5])
labels7.append(labels[i + 6])
labels8.append(labels[i + 7])
labels9.append(labels[i + 8])
labels10.append(labels[i + 9])
counter += 1
print(counter, end="\r")
image_names1 = np.array(image_names1)
image_names2 = np.array(image_names2)
image_names3 = np.array(image_names3)
image_names4 = np.array(image_names4)
image_names5 = np.array(image_names5)
image_names6 = np.array(image_names6)
image_names7 = np.array(image_names7)
image_names8 = np.array(image_names8)
image_names9 = np.array(image_names9)
image_names10 = np.array(image_names10)
labels1 = np.array(labels1)
from sklearn.utils import shuffle
(
image_names1,
labels1,
image_names2,
labels2,
image_names3,
labels3,
image_names4,
labels4,
image_names5,
labels5,
image_names6,
labels6,
image_names7,
labels7,
image_names8,
labels8,
image_names9,
labels9,
image_names10,
labels10,
) = shuffle(
image_names1,
labels1,
image_names2,
labels2,
image_names3,
labels3,
image_names4,
labels4,
image_names5,
labels5,
image_names6,
labels6,
image_names7,
labels7,
image_names8,
labels8,
image_names9,
labels9,
image_names10,
labels10,
random_state=10000,
)
i = 100
print(labels1[i])
print(labels5[i])
print(labels7[i])
import tensorflow as tf
base_model = tf.keras.applications.ResNet50(
include_top=False,
weights="imagenet",
input_tensor=None,
input_shape=(224, 224, 3),
classes=1000,
)
import numpy as np
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers, Input, Model
inputA = Input(shape=(224, 224, 3))
inputB = Input(shape=(224, 224, 3))
inputC = Input(shape=(224, 224, 3))
inputD = Input(shape=(224, 224, 3))
inputE = Input(shape=(224, 224, 3))
inputF = Input(shape=(224, 224, 3))
inputG = Input(shape=(224, 224, 3))
inputH = Input(shape=(224, 224, 3))
inputI = Input(shape=(224, 224, 3))
inputJ = Input(shape=(224, 224, 3))
# defining parallel outputs
A = Model(inputs=inputA, outputs=base_model(inputA))
B = Model(inputs=inputB, outputs=base_model(inputB))
C = Model(inputs=inputC, outputs=base_model(inputC))
D = Model(inputs=inputD, outputs=base_model(inputD))
E = Model(inputs=inputE, outputs=base_model(inputE))
F = Model(inputs=inputF, outputs=base_model(inputF))
G = Model(inputs=inputG, outputs=base_model(inputG))
H = Model(inputs=inputH, outputs=base_model(inputH))
I = Model(inputs=inputI, outputs=base_model(inputI))
J = Model(inputs=inputJ, outputs=base_model(inputJ))
combined = layers.Add()(
[
A.output,
B.output,
C.output,
D.output,
E.output,
F.output,
G.output,
H.output,
I.output,
J.output,
]
)
# x = layers.Conv2D(512, 3, activation = 'relu', padding = 'same')(combined)
# fx = layers.Conv2D(512, 3, activation='relu', padding='same')(x)
# fx = layers.BatchNormalization()(fx)
# fx = layers.Conv2D(512, 3, padding='same')(fx)
# out = layers.Add()([x,fx])
# out = layers.MaxPooling2D()(out)
# out = layers.ReLU()(out)
# out = layers.BatchNormalization()(out)
z = layers.Flatten()(combined)
# z = layers.Dense(4096, activation="relu")(z)
# z = layers.Dropout(0.5)(z)
# z = layers.Dense(4096, activation='relu')(z)
# z = layers.Dropout(0.4)(z)
z = layers.Dense(4, activation="softmax")(z)
model = Model(
inputs=[
A.input,
B.input,
C.input,
D.input,
E.input,
F.input,
G.input,
H.input,
I.input,
J.input,
],
outputs=z,
)
model.summary()
for layer in model.layers:
layer.trainable = True
model.compile(
loss="sparse_categorical_crossentropy",
optimizer=tf.keras.optimizers.Adam(),
metrics=["acc"],
)
from keras.callbacks import ModelCheckpoint
checkpoint = ModelCheckpoint(
"nohnohmosmed.h5", monitor="val_acc", verbose=1, save_best_only=True, mode="auto"
)
History = model.fit(
x=[
image_names1,
image_names2,
image_names3,
image_names4,
image_names5,
image_names6,
image_names7,
image_names8,
image_names9,
image_names10,
],
y=labels1,
validation_split=0.2,
epochs=50,
callbacks=[checkpoint],
)
model.summary()
model = model.save_weights("model_mri.h5")
loss = model.history["loss"]
val_loss = model.history["val_loss"]
epochs = range(300)
plt.figure()
plt.plot(epochs, loss, "bo", label="Training loss")
plt.plot(epochs, val_loss, "b", label="Validation loss")
plt.title("Training and validation loss")
plt.legend()
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/046/69046074.ipynb
|
mosmeddata-fullchestct
|
ahmedamineafardas
|
[{"Id": 69046074, "ScriptId": 18591601, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7118898, "CreationDate": "07/26/2021 08:34:50", "VersionNumber": 1.0, "Title": "Biotech MosMed Dataset model", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 219.0, "LinesInsertedFromPrevious": 33.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 186.0, "LinesInsertedFromFork": 33.0, "LinesDeletedFromFork": 27.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 186.0, "TotalVotes": 0}]
|
[{"Id": 91775379, "KernelVersionId": 69046074, "SourceDatasetVersionId": 2076367}]
|
[{"Id": 2076367, "DatasetId": 1244618, "DatasourceVersionId": 2116675, "CreatorUserId": 7051386, "LicenseName": "Unknown", "CreationDate": "04/01/2021 04:22:08", "VersionNumber": 1.0, "Title": "MosMedData FullChestCT", "Slug": "mosmeddata-fullchestct", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1244618, "CreatorUserId": 7051386, "OwnerUserId": 7051386.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2076367.0, "CurrentDatasourceVersionId": 2116675.0, "ForumId": 1262902, "Type": 2, "CreationDate": "04/01/2021 04:22:08", "LastActivityDate": "04/01/2021", "TotalViews": 1017, "TotalDownloads": 52, "TotalVotes": 1, "TotalKernels": 4}]
|
[{"Id": 7051386, "UserName": "ahmedamineafardas", "DisplayName": "ahmed amine afardas", "RegisterDate": "03/28/2021", "PerformanceTier": 0}]
|
import numpy as np
import pandas as pd
import os
import tensorflow as tf
import cv2
import matplotlib.pyplot as plt
import tensorflow as tf
import nibabel as nib
image_paths0 = []
labels0 = []
for dirname, _, filenames in os.walk(
"../input/mosmeddata-fullchestct/COVID19_1110/studies/CT-0"
):
for filename in filenames:
image_paths0.append(os.path.join(dirname, filename))
labels0.append(0)
image_paths1 = []
labels1 = []
for dirname, _, filenames in os.walk(
"../input/mosmeddata-fullchestct/COVID19_1110/studies/CT-1"
):
for filename in filenames:
image_paths1.append(os.path.join(dirname, filename))
labels1.append(1)
image_paths2 = []
labels2 = []
for dirname, _, filenames in os.walk(
"../input/mosmeddata-fullchestct/COVID19_1110/studies/CT-2"
):
for filename in filenames:
image_paths2.append(os.path.join(dirname, filename))
labels2.append(2)
image_paths3 = []
labels3 = []
for dirname, _, filenames in os.walk(
"../input/mosmeddata-fullchestct/COVID19_1110/studies/CT-3"
):
for filename in filenames:
image_paths3.append(os.path.join(dirname, filename))
labels3.append(3)
image_paths = []
image_paths.extend(image_paths0)
image_paths.extend(image_paths1)
image_paths.extend(image_paths2)
image_paths.extend(image_paths3)
labels = []
labels.extend(labels0)
labels.extend(labels1)
labels.extend(labels2)
labels.extend(labels3)
np.max(labels)
from sklearn.utils import shuffle
image_paths, labels = shuffle(image_paths, labels, random_state=10800)
# def parse_function(image_paths, labels):
# image_path = tf.compat.v1.data.make_one_shot_iterator(image_path)
# print(image_path)
image_names_tab = []
labels_tab = []
counter = 0
for image_path, label in zip(image_paths[:20], labels[:20]):
niimg = nib.load(image_path)
npimage = niimg.get_fdata()
s = npimage.shape
for j in range(20, 30):
img = np.zeros((s[0], s[1], 3))
img[:, :, 0] = npimage[:, :, j]
img[:, :, 1] = npimage[:, :, j]
img[:, :, 2] = npimage[:, :, j]
img = img / np.max(npimage[:, :, j])
# img = tf.cast(img, tf.float32)
img = cv2.resize(img, (224, 224))
image_names_tab.append(img)
labels_tab.append(label)
counter += 1
print(counter, end="\r")
np.shape(image_names_tab)
image_names = image_names_tab
labels = labels_tab
image_names1 = []
image_names2 = []
image_names3 = []
image_names4 = []
image_names5 = []
image_names6 = []
image_names7 = []
image_names8 = []
image_names9 = []
image_names10 = []
labels1 = []
labels2 = []
labels3 = []
labels4 = []
labels5 = []
labels6 = []
labels7 = []
labels8 = []
labels9 = []
labels10 = []
counter = 0
for i in range(0, len(image_names), 10):
image_names1.append(image_names[i])
image_names2.append(image_names[i + 1])
image_names3.append(image_names[i + 2])
image_names4.append(image_names[i + 3])
image_names5.append(image_names[i + 4])
image_names6.append(image_names[i + 5])
image_names7.append(image_names[i + 6])
image_names8.append(image_names[i + 7])
image_names9.append(image_names[i + 8])
image_names10.append(image_names[i + 9])
labels1.append(labels[i])
labels2.append(labels[i + 1])
labels3.append(labels[i + 2])
labels4.append(labels[i + 3])
labels5.append(labels[i + 4])
labels6.append(labels[i + 5])
labels7.append(labels[i + 6])
labels8.append(labels[i + 7])
labels9.append(labels[i + 8])
labels10.append(labels[i + 9])
counter += 1
print(counter, end="\r")
image_names1 = np.array(image_names1)
image_names2 = np.array(image_names2)
image_names3 = np.array(image_names3)
image_names4 = np.array(image_names4)
image_names5 = np.array(image_names5)
image_names6 = np.array(image_names6)
image_names7 = np.array(image_names7)
image_names8 = np.array(image_names8)
image_names9 = np.array(image_names9)
image_names10 = np.array(image_names10)
labels1 = np.array(labels1)
from sklearn.utils import shuffle
(
image_names1,
labels1,
image_names2,
labels2,
image_names3,
labels3,
image_names4,
labels4,
image_names5,
labels5,
image_names6,
labels6,
image_names7,
labels7,
image_names8,
labels8,
image_names9,
labels9,
image_names10,
labels10,
) = shuffle(
image_names1,
labels1,
image_names2,
labels2,
image_names3,
labels3,
image_names4,
labels4,
image_names5,
labels5,
image_names6,
labels6,
image_names7,
labels7,
image_names8,
labels8,
image_names9,
labels9,
image_names10,
labels10,
random_state=10000,
)
i = 100
print(labels1[i])
print(labels5[i])
print(labels7[i])
import tensorflow as tf
base_model = tf.keras.applications.ResNet50(
include_top=False,
weights="imagenet",
input_tensor=None,
input_shape=(224, 224, 3),
classes=1000,
)
import numpy as np
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers, Input, Model
inputA = Input(shape=(224, 224, 3))
inputB = Input(shape=(224, 224, 3))
inputC = Input(shape=(224, 224, 3))
inputD = Input(shape=(224, 224, 3))
inputE = Input(shape=(224, 224, 3))
inputF = Input(shape=(224, 224, 3))
inputG = Input(shape=(224, 224, 3))
inputH = Input(shape=(224, 224, 3))
inputI = Input(shape=(224, 224, 3))
inputJ = Input(shape=(224, 224, 3))
# defining parallel outputs
A = Model(inputs=inputA, outputs=base_model(inputA))
B = Model(inputs=inputB, outputs=base_model(inputB))
C = Model(inputs=inputC, outputs=base_model(inputC))
D = Model(inputs=inputD, outputs=base_model(inputD))
E = Model(inputs=inputE, outputs=base_model(inputE))
F = Model(inputs=inputF, outputs=base_model(inputF))
G = Model(inputs=inputG, outputs=base_model(inputG))
H = Model(inputs=inputH, outputs=base_model(inputH))
I = Model(inputs=inputI, outputs=base_model(inputI))
J = Model(inputs=inputJ, outputs=base_model(inputJ))
combined = layers.Add()(
[
A.output,
B.output,
C.output,
D.output,
E.output,
F.output,
G.output,
H.output,
I.output,
J.output,
]
)
# x = layers.Conv2D(512, 3, activation = 'relu', padding = 'same')(combined)
# fx = layers.Conv2D(512, 3, activation='relu', padding='same')(x)
# fx = layers.BatchNormalization()(fx)
# fx = layers.Conv2D(512, 3, padding='same')(fx)
# out = layers.Add()([x,fx])
# out = layers.MaxPooling2D()(out)
# out = layers.ReLU()(out)
# out = layers.BatchNormalization()(out)
z = layers.Flatten()(combined)
# z = layers.Dense(4096, activation="relu")(z)
# z = layers.Dropout(0.5)(z)
# z = layers.Dense(4096, activation='relu')(z)
# z = layers.Dropout(0.4)(z)
z = layers.Dense(4, activation="softmax")(z)
model = Model(
inputs=[
A.input,
B.input,
C.input,
D.input,
E.input,
F.input,
G.input,
H.input,
I.input,
J.input,
],
outputs=z,
)
model.summary()
for layer in model.layers:
layer.trainable = True
model.compile(
loss="sparse_categorical_crossentropy",
optimizer=tf.keras.optimizers.Adam(),
metrics=["acc"],
)
from keras.callbacks import ModelCheckpoint
checkpoint = ModelCheckpoint(
"nohnohmosmed.h5", monitor="val_acc", verbose=1, save_best_only=True, mode="auto"
)
History = model.fit(
x=[
image_names1,
image_names2,
image_names3,
image_names4,
image_names5,
image_names6,
image_names7,
image_names8,
image_names9,
image_names10,
],
y=labels1,
validation_split=0.2,
epochs=50,
callbacks=[checkpoint],
)
model.summary()
model = model.save_weights("model_mri.h5")
loss = model.history["loss"]
val_loss = model.history["val_loss"]
epochs = range(300)
plt.figure()
plt.plot(epochs, loss, "bo", label="Training loss")
plt.plot(epochs, val_loss, "b", label="Validation loss")
plt.title("Training and validation loss")
plt.legend()
plt.show()
| false | 0 | 2,877 | 0 | 2,908 | 2,877 |
||
69046611
|
import numpy as np
import pandas as pd
import xgboost as xgb
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn.multioutput import MultiOutputRegressor
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
train = pd.read_csv("/kaggle/input/tabular-playground-series-jul-2021/train.csv")
test = pd.read_csv("/kaggle/input/tabular-playground-series-jul-2021/test.csv")
sub = pd.read_csv(
"/kaggle/input/tabular-playground-series-jul-2021/sample_submission.csv"
)
train = train.set_index("date_time").copy()
test = test.set_index("date_time").copy()
target_cols = [col for col in train.columns if col.startswith("target")]
feat_cols = [col for col in train.columns if col not in target_cols]
train, val = train_test_split(train, test_size=0.2, random_state=42)
fea_scaler = MinMaxScaler()
lab_scaler = MinMaxScaler()
Xtrain_scaled = fea_scaler.fit_transform(train.drop(target_cols[:], axis=1))
Xval_scaled = fea_scaler.transform(val.drop(target_cols[:], axis=1))
Ytrain_scaled = lab_scaler.fit_transform(train[target_cols[:]])
Yval_scaled = lab_scaler.transform(val[target_cols[:]])
Xtest_scaled = fea_scaler.transform(test)
other_params = {
"learning_rate": 0.1,
"n_estimators": 400,
"max_depth": 4,
"min_child_weight": 5,
"seed": 0,
"subsample": 0.8,
"colsample_bytree": 0.8,
"gamma": 0.1,
"reg_alpha": 0.1,
"reg_lambda": 0.1,
}
model = xgb.XGBRegressor(**other_params)
multioutputregressor = MultiOutputRegressor(
xgb.XGBRegressor(objective="reg:squarederror", **other_params)
).fit(Xtrain_scaled, Ytrain_scaled)
# cv_params = {'n_estimators': [400, 500, 600, 700, 800]}
# other_params = {'learning_rate': 0.1, 'n_estimators': 500, 'max_depth': 5, 'min_child_weight': 1, 'seed': 0,
# 'subsample': 0.8, 'colsample_bytree': 0.8, 'gamma': 0, 'reg_alpha': 0, 'reg_lambda': 1}
# optimized_GBM = GridSearchCV(estimator=model, param_grid=cv_params, scoring='r2', cv=5, verbose=1, n_jobs=4)
# optimized_GBM.fit(Xtrain_scaled, Ytrain_scaled[:, 1])
# evalute_result = optimized_GBM.cv_results_
# print('每轮迭代运行结果:{0}'.format(evalute_result))
# print('参数的最佳取值:{0}'.format(optimized_GBM.best_params_))
# print('最佳模型得分:{0}'.format(optimized_GBM.best_score_))
# cv_params = {'max_depth': [3, 4, 5, 6, 7, 8, 9, 10]}
# other_params = {'learning_rate': 0.1, 'n_estimators': 400, 'max_depth': 5, 'min_child_weight': 1, 'seed': 0,
# 'subsample': 0.8, 'colsample_bytr ee': 0.8, 'gamma': 0, 'reg_alpha': 0, 'reg_lambda': 1}
# optimized_GBM = GridSearchCV(estimator=model, param_grid=cv_params, scoring='r2', cv=5, verbose=1, n_jobs=4)
# optimized_GBM.fit(Xtrain_scaled, Ytrain_scaled[:, 1])
# evalute_result = optimized_GBM.cv_results_
# print('每轮迭代运行结果:{0}'.format(evalute_result))
# print('参数的最佳取值:{0}'.format(optimized_GBM.best_params_))
# print('最佳模型得分:{0}'.format(optimized_GBM.best_score_))
# cv_params = {'min_child_weight': [1, 2, 3, 4, 5, 6]}
# other_params = {'learning_rate': 0.1, 'n_estimators': 400, 'max_depth': 4, 'min_child_weight': 5, 'seed': 0,
# 'subsample': 0.8, 'colsample_bytr ee': 0.8, 'gamma': 0.1, 'reg_alpha': 0, 'reg_lambda': 1}
# optimized_GBM = GridSearchCV(estimator=model, param_grid=cv_params, scoring='r2', cv=5, verbose=1, n_jobs=4)
# optimized_GBM.fit(Xtrain_scaled, Ytrain_scaled[:, 1])
# evalute_result = optimized_GBM.cv_results_
# print('每轮迭代运行结果:{0}'.format(evalute_result))
# print('参数的最佳取值:{0}'.format(optimized_GBM.best_params_))
# print('最佳模型得分:{0}'.format(optimized_GBM.best_score_))
# cv_params = {'gamma': [0.1, 0.2, 0.3, 0.4, 0.5, 0.6]}
# other_params = {'learning_rate': 0.1, 'n_estimators': 400, 'max_depth': 4, 'min_child_weight': 5, 'seed': 0,
# 'subsample': 0.8, 'colsample_bytree': 0.8, 'gamma': 0.1, 'reg_alpha': 0, 'reg_lambda': 1}
# optimized_GBM = GridSearchCV(estimator=model, param_grid=cv_params, scoring='r2', cv=5, verbose=1, n_jobs=4)
# optimized_GBM.fit(Xtrain_scaled, Ytrain_scaled[:, 1])
# evalute_result = optimized_GBM.cv_results_
# print('每轮迭代运行结果:{0}'.format(evalute_result))
# print('参数的最佳取值:{0}'.format(optimized_GBM.best_params_))
# print('最佳模型得分:{0}'.format(optimized_GBM.best_score_))
# cv_params = {'subsample': [0.6, 0.7, 0.8, 0.9], 'colsample_bytree': [0.6, 0.7, 0.8, 0.9]}
# other_params = {'learning_rate': 0.1, 'n_estimators': 400, 'max_depth': 4, 'min_child_weight': 5, 'seed': 0,
# 'subsample': 0.8, 'colsample_bytree': 0.8, 'gamma': 0.1, 'reg_alpha': 0, 'reg_lambda': 1}
# optimized_GBM = GridSearchCV(estimator=model, param_grid=cv_params, scoring='r2', cv=5, verbose=1, n_jobs=4)
# optimized_GBM.fit(Xtrain_scaled, Ytrain_scaled[:, 1])
# #evalute_result = optimized_GBM.grid_scores_
# #print('每轮迭代运行结果:{0}'.format(evalute_result))
# print('参数的最佳取值:{0}'.format(optimized_GBM.best_params_))
# print('最佳模型得分:{0}'.format(optimized_GBM.best_score_))
# cv_params = {'reg_alpha': [0.05, 0.1, 1, 2, 3], 'reg_lambda': [0.05, 0.1, 1, 2, 3]}
# other_params = {'learning_rate': 0.1, 'n_estimators': 400, 'max_depth': 4, 'min_child_weight': 5, 'seed': 0,
# 'subsample': 0.8, 'colsample_bytree': 0.8, 'gamma': 0.1, 'reg_alpha': 0, 'reg_lambda': 1}
# optimized_GBM = GridSearchCV(estimator=model, param_grid=cv_params, scoring='r2', cv=5, verbose=1, n_jobs=4)
# optimized_GBM.fit(Xtrain_scaled, Ytrain_scaled[:, 1])
# #evalute_result = optimized_GBM.grid_scores_
# #print('每轮迭代运行结果:{0}'.format(evalute_result))
# print('参数的最佳取值:{0}'.format(optimized_GBM.best_params_))
# print('最佳模型得分:{0}'.format(optimized_GBM.best_score_))
# cv_params = {'n_estimators': [400, 500, 600, 700, 800],
# 'max_depth': [3, 4, 5, 6, 7, 8, 9, 10],
# 'min_child_weight': [1, 2, 3, 4, 5, 6],
# 'gamma': [0.1, 0.2, 0.3, 0.4, 0.5, 0.6],
# 'subsample': [0.6, 0.7, 0.8, 0.9],
# 'colsample_bytree': [0.6, 0.7, 0.8, 0.9],
# 'reg_alpha': [0.05, 0.1, 1, 2, 3],
# 'reg_lambda': [0.05, 0.1, 1, 2, 3],
# 'learning_rate': [0.01, 0.05, 0.07, 0.1, 0.2]}
# other_params = {'learning_rate': 0.1, 'n_estimators': 400, 'max_depth': 4, 'min_child_weight': 5, 'seed': 0,
# 'subsample': 0.8, 'colsample_bytree': 0.8, 'gamma': 0.1, 'reg_alpha': 0.1, 'reg_lambda': 0.1}
# optimized_GBM = GridSearchCV(estimator=model, param_grid=cv_params, scoring='r2', cv=5, verbose=1, n_jobs=10)
# optimized_GBM.fit(Xtrain_scaled, Ytrain_scaled[:, 1])
# evalute_result = optimized_GBM.cv_results_
# print('每轮迭代运行结果:{0}'.format(evalute_result))
# print('参数的最佳取值:{0}'.format(optimized_GBM.best_params_))
# print('最佳模型得分:{0}'.format(optimized_GBM.best_score_))
pred = multioutputregressor.predict(Xtest_scaled)
pred = lab_scaler.inverse_transform(pred)
pred = pred.reshape(2247, 3)
sub[target_cols[:]] = pred
sub.to_csv("sample_submission.csv", index=0)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/046/69046611.ipynb
| null | null |
[{"Id": 69046611, "ScriptId": 18816238, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7392108, "CreationDate": "07/26/2021 08:43:12", "VersionNumber": 3.0, "Title": "XGBoost", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 120.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 119.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
| null | null | null | null |
import numpy as np
import pandas as pd
import xgboost as xgb
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn.multioutput import MultiOutputRegressor
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
train = pd.read_csv("/kaggle/input/tabular-playground-series-jul-2021/train.csv")
test = pd.read_csv("/kaggle/input/tabular-playground-series-jul-2021/test.csv")
sub = pd.read_csv(
"/kaggle/input/tabular-playground-series-jul-2021/sample_submission.csv"
)
train = train.set_index("date_time").copy()
test = test.set_index("date_time").copy()
target_cols = [col for col in train.columns if col.startswith("target")]
feat_cols = [col for col in train.columns if col not in target_cols]
train, val = train_test_split(train, test_size=0.2, random_state=42)
fea_scaler = MinMaxScaler()
lab_scaler = MinMaxScaler()
Xtrain_scaled = fea_scaler.fit_transform(train.drop(target_cols[:], axis=1))
Xval_scaled = fea_scaler.transform(val.drop(target_cols[:], axis=1))
Ytrain_scaled = lab_scaler.fit_transform(train[target_cols[:]])
Yval_scaled = lab_scaler.transform(val[target_cols[:]])
Xtest_scaled = fea_scaler.transform(test)
other_params = {
"learning_rate": 0.1,
"n_estimators": 400,
"max_depth": 4,
"min_child_weight": 5,
"seed": 0,
"subsample": 0.8,
"colsample_bytree": 0.8,
"gamma": 0.1,
"reg_alpha": 0.1,
"reg_lambda": 0.1,
}
model = xgb.XGBRegressor(**other_params)
multioutputregressor = MultiOutputRegressor(
xgb.XGBRegressor(objective="reg:squarederror", **other_params)
).fit(Xtrain_scaled, Ytrain_scaled)
# cv_params = {'n_estimators': [400, 500, 600, 700, 800]}
# other_params = {'learning_rate': 0.1, 'n_estimators': 500, 'max_depth': 5, 'min_child_weight': 1, 'seed': 0,
# 'subsample': 0.8, 'colsample_bytree': 0.8, 'gamma': 0, 'reg_alpha': 0, 'reg_lambda': 1}
# optimized_GBM = GridSearchCV(estimator=model, param_grid=cv_params, scoring='r2', cv=5, verbose=1, n_jobs=4)
# optimized_GBM.fit(Xtrain_scaled, Ytrain_scaled[:, 1])
# evalute_result = optimized_GBM.cv_results_
# print('每轮迭代运行结果:{0}'.format(evalute_result))
# print('参数的最佳取值:{0}'.format(optimized_GBM.best_params_))
# print('最佳模型得分:{0}'.format(optimized_GBM.best_score_))
# cv_params = {'max_depth': [3, 4, 5, 6, 7, 8, 9, 10]}
# other_params = {'learning_rate': 0.1, 'n_estimators': 400, 'max_depth': 5, 'min_child_weight': 1, 'seed': 0,
# 'subsample': 0.8, 'colsample_bytr ee': 0.8, 'gamma': 0, 'reg_alpha': 0, 'reg_lambda': 1}
# optimized_GBM = GridSearchCV(estimator=model, param_grid=cv_params, scoring='r2', cv=5, verbose=1, n_jobs=4)
# optimized_GBM.fit(Xtrain_scaled, Ytrain_scaled[:, 1])
# evalute_result = optimized_GBM.cv_results_
# print('每轮迭代运行结果:{0}'.format(evalute_result))
# print('参数的最佳取值:{0}'.format(optimized_GBM.best_params_))
# print('最佳模型得分:{0}'.format(optimized_GBM.best_score_))
# cv_params = {'min_child_weight': [1, 2, 3, 4, 5, 6]}
# other_params = {'learning_rate': 0.1, 'n_estimators': 400, 'max_depth': 4, 'min_child_weight': 5, 'seed': 0,
# 'subsample': 0.8, 'colsample_bytr ee': 0.8, 'gamma': 0.1, 'reg_alpha': 0, 'reg_lambda': 1}
# optimized_GBM = GridSearchCV(estimator=model, param_grid=cv_params, scoring='r2', cv=5, verbose=1, n_jobs=4)
# optimized_GBM.fit(Xtrain_scaled, Ytrain_scaled[:, 1])
# evalute_result = optimized_GBM.cv_results_
# print('每轮迭代运行结果:{0}'.format(evalute_result))
# print('参数的最佳取值:{0}'.format(optimized_GBM.best_params_))
# print('最佳模型得分:{0}'.format(optimized_GBM.best_score_))
# cv_params = {'gamma': [0.1, 0.2, 0.3, 0.4, 0.5, 0.6]}
# other_params = {'learning_rate': 0.1, 'n_estimators': 400, 'max_depth': 4, 'min_child_weight': 5, 'seed': 0,
# 'subsample': 0.8, 'colsample_bytree': 0.8, 'gamma': 0.1, 'reg_alpha': 0, 'reg_lambda': 1}
# optimized_GBM = GridSearchCV(estimator=model, param_grid=cv_params, scoring='r2', cv=5, verbose=1, n_jobs=4)
# optimized_GBM.fit(Xtrain_scaled, Ytrain_scaled[:, 1])
# evalute_result = optimized_GBM.cv_results_
# print('每轮迭代运行结果:{0}'.format(evalute_result))
# print('参数的最佳取值:{0}'.format(optimized_GBM.best_params_))
# print('最佳模型得分:{0}'.format(optimized_GBM.best_score_))
# cv_params = {'subsample': [0.6, 0.7, 0.8, 0.9], 'colsample_bytree': [0.6, 0.7, 0.8, 0.9]}
# other_params = {'learning_rate': 0.1, 'n_estimators': 400, 'max_depth': 4, 'min_child_weight': 5, 'seed': 0,
# 'subsample': 0.8, 'colsample_bytree': 0.8, 'gamma': 0.1, 'reg_alpha': 0, 'reg_lambda': 1}
# optimized_GBM = GridSearchCV(estimator=model, param_grid=cv_params, scoring='r2', cv=5, verbose=1, n_jobs=4)
# optimized_GBM.fit(Xtrain_scaled, Ytrain_scaled[:, 1])
# #evalute_result = optimized_GBM.grid_scores_
# #print('每轮迭代运行结果:{0}'.format(evalute_result))
# print('参数的最佳取值:{0}'.format(optimized_GBM.best_params_))
# print('最佳模型得分:{0}'.format(optimized_GBM.best_score_))
# cv_params = {'reg_alpha': [0.05, 0.1, 1, 2, 3], 'reg_lambda': [0.05, 0.1, 1, 2, 3]}
# other_params = {'learning_rate': 0.1, 'n_estimators': 400, 'max_depth': 4, 'min_child_weight': 5, 'seed': 0,
# 'subsample': 0.8, 'colsample_bytree': 0.8, 'gamma': 0.1, 'reg_alpha': 0, 'reg_lambda': 1}
# optimized_GBM = GridSearchCV(estimator=model, param_grid=cv_params, scoring='r2', cv=5, verbose=1, n_jobs=4)
# optimized_GBM.fit(Xtrain_scaled, Ytrain_scaled[:, 1])
# #evalute_result = optimized_GBM.grid_scores_
# #print('每轮迭代运行结果:{0}'.format(evalute_result))
# print('参数的最佳取值:{0}'.format(optimized_GBM.best_params_))
# print('最佳模型得分:{0}'.format(optimized_GBM.best_score_))
# cv_params = {'n_estimators': [400, 500, 600, 700, 800],
# 'max_depth': [3, 4, 5, 6, 7, 8, 9, 10],
# 'min_child_weight': [1, 2, 3, 4, 5, 6],
# 'gamma': [0.1, 0.2, 0.3, 0.4, 0.5, 0.6],
# 'subsample': [0.6, 0.7, 0.8, 0.9],
# 'colsample_bytree': [0.6, 0.7, 0.8, 0.9],
# 'reg_alpha': [0.05, 0.1, 1, 2, 3],
# 'reg_lambda': [0.05, 0.1, 1, 2, 3],
# 'learning_rate': [0.01, 0.05, 0.07, 0.1, 0.2]}
# other_params = {'learning_rate': 0.1, 'n_estimators': 400, 'max_depth': 4, 'min_child_weight': 5, 'seed': 0,
# 'subsample': 0.8, 'colsample_bytree': 0.8, 'gamma': 0.1, 'reg_alpha': 0.1, 'reg_lambda': 0.1}
# optimized_GBM = GridSearchCV(estimator=model, param_grid=cv_params, scoring='r2', cv=5, verbose=1, n_jobs=10)
# optimized_GBM.fit(Xtrain_scaled, Ytrain_scaled[:, 1])
# evalute_result = optimized_GBM.cv_results_
# print('每轮迭代运行结果:{0}'.format(evalute_result))
# print('参数的最佳取值:{0}'.format(optimized_GBM.best_params_))
# print('最佳模型得分:{0}'.format(optimized_GBM.best_score_))
pred = multioutputregressor.predict(Xtest_scaled)
pred = lab_scaler.inverse_transform(pred)
pred = pred.reshape(2247, 3)
sub[target_cols[:]] = pred
sub.to_csv("sample_submission.csv", index=0)
| false | 0 | 2,940 | 1 | 2,940 | 2,940 |
||
69046436
|
import numpy as np
import pandas as pd
from sklearn.impute import SimpleImputer, KNNImputer
from sklearn.ensemble import (
RandomForestRegressor,
GradientBoostingRegressor,
ExtraTreesRegressor,
)
import matplotlib.pyplot as plt
import plotly.graph_objects as go
import plotly.express as px
import seaborn as sns
from catboost import CatBoostRegressor
from lightgbm import LGBMRegressor
from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor
from sklearn.linear_model import LinearRegression, Ridge, Lasso, ElasticNet
from sklearn.model_selection import train_test_split, GridSearchCV, cross_val_score
from sklearn.neighbors import KNeighborsRegressor
from sklearn.svm import SVR
from sklearn.tree import DecisionTreeRegressor
from xgboost import XGBRegressor
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train = pd.read_csv("../input/house-prices-advanced-regression-techniques/train.csv")
test = pd.read_csv("../input/house-prices-advanced-regression-techniques/test.csv")
print(train.shape)
print(test.shape)
df = train.append(test).reset_index(drop=True)
print(df.shape)
df.columns
def check_df(dataframe, head=5):
print("##################### Shape #####################")
print(dataframe.shape)
print("##################### Types #####################")
print(dataframe.dtypes)
print("##################### Head #####################")
print(dataframe.head(head))
print("##################### Tail #####################")
print(dataframe.tail(head))
print("##################### NA #####################")
print(dataframe.isnull().sum())
print("##################### Quantiles #####################")
print(dataframe.quantile([0, 0.05, 0.50, 0.95, 0.99, 1]).T)
check_df(df)
def grab_col_names(dataframe, cat_th=10, car_th=20):
# cat_cols, cat_but_car
cat_cols = [col for col in dataframe.columns if dataframe[col].dtypes == "O"]
num_but_cat = [
col
for col in dataframe.columns
if dataframe[col].nunique() < cat_th and dataframe[col].dtypes != "O"
]
cat_but_car = [
col
for col in dataframe.columns
if dataframe[col].nunique() > car_th and dataframe[col].dtypes == "O"
]
cat_cols = cat_cols + num_but_cat
cat_cols = [col for col in cat_cols if col not in cat_but_car]
# num_cols
num_cols = [col for col in dataframe.columns if dataframe[col].dtypes != "O"]
num_cols = [col for col in num_cols if col not in num_but_cat]
print(f"Observations: {dataframe.shape[0]}")
print(f"Variables: {dataframe.shape[1]}")
print(f"cat_cols: {len(cat_cols)}")
print(f"num_cols: {len(num_cols)}")
print(f"cat_but_car: {len(cat_but_car)}")
print(f"num_but_cat: {len(num_but_cat)}")
return cat_cols, num_cols, cat_but_car
cat_cols, num_cols, cat_but_car = grab_col_names(df)
df["Neighborhood"].value_counts()
# Kategorik Değişken Analizi
def cat_summary(dataframe, col_name, plot=False):
print(
pd.DataFrame(
{
col_name: dataframe[col_name].value_counts(),
"Ratio": 100 * dataframe[col_name].value_counts() / len(dataframe),
}
)
)
print("##########################################")
if plot:
sns.countplot(x=dataframe[col_name], data=dataframe)
plt.show()
for col in cat_cols:
cat_summary(df, col)
for col in cat_but_car:
cat_summary(df, col)
# Sayısal Değişken Analizi
df[num_cols].describe([0.10, 0.30, 0.50, 0.70, 0.80, 0.99]).T
# Target Analizi
df["SalePrice"].describe([0.05, 0.10, 0.25, 0.50, 0.75, 0.80, 0.90, 0.95, 0.99]).T
def find_correlation(dataframe, numeric_cols, corr_limit=0.60):
high_correlations = []
low_correlations = []
for col in numeric_cols:
if col == "SalePrice":
pass
else:
correlation = dataframe[[col, "SalePrice"]].corr().loc[col, "SalePrice"]
print(col, correlation)
if abs(correlation) > corr_limit:
high_correlations.append(col + ": " + str(correlation))
else:
low_correlations.append(col + ": " + str(correlation))
return low_correlations, high_correlations
low_corrs, high_corrs = find_correlation(df, num_cols)
# tüm değişkenler korelasyon
corr_matrix = df.corr()
sns.clustermap(corr_matrix, annot=True, figsize=(20, 15), fmt=".2f")
plt.title("Correlation Between Features")
plt.show()
threshold = 0.60
filter = np.abs(corr_matrix["SalePrice"]) > threshold
corr_features = corr_matrix.columns[filter].tolist()
sns.clustermap(df[corr_features].corr(), annot=True, fmt=".2f")
plt.title("Correlation Between Features w/ Corr Threshold 0.60)")
plt.show()
def high_correlated_cols(dataframe, plot=False, corr_th=0.60):
corr = dataframe.corr()
cor_matrix = corr.abs()
upper_triangle_matrix = cor_matrix.where(
np.triu(np.ones(cor_matrix.shape), k=1).astype(np.bool)
)
drop_list = [
col
for col in upper_triangle_matrix.columns
if any(upper_triangle_matrix[col] > corr_th)
]
if plot:
import seaborn as sns
import matplotlib.pyplot as plt
sns.set(rc={"figure.figsize": (15, 15)})
sns.heatmap(corr, cmap="RdBu")
plt.show()
return drop_list
high_correlated_cols(df)
# FEATURE ENGINEERING
df["SqFtPerRoom"] = df["GrLivArea"] / (
df["TotRmsAbvGrd"] + df["FullBath"] + df["HalfBath"] + df["KitchenAbvGr"]
)
df["Total_Home_Quality"] = df["OverallQual"] + df["OverallCond"]
df["Total_Bathrooms"] = (
df["FullBath"]
+ (0.5 * df["HalfBath"])
+ df["BsmtFullBath"]
+ (0.5 * df["BsmtHalfBath"])
)
df["HighQualSF"] = df["1stFlrSF"] + df["2ndFlrSF"]
# Converting non-numeric predictors stored as numbers into string
df["MSSubClass"] = df["MSSubClass"].apply(str)
df["YrSold"] = df["YrSold"].apply(str)
df["MoSold"] = df["MoSold"].apply(str)
# RARE ENCODING
def rare_encoder(dataframe, rare_perc, cat_cols):
rare_columns = [
col
for col in cat_cols
if (dataframe[col].value_counts() / len(dataframe) < 0.01).sum() > 1
]
for col in rare_columns:
tmp = dataframe[col].value_counts() / len(dataframe)
rare_labels = tmp[tmp < rare_perc].index
dataframe[col] = np.where(
dataframe[col].isin(rare_labels), "Rare", dataframe[col]
)
return dataframe
def rare_analyser(dataframe, target, cat_cols):
for col in cat_cols:
print(col, ":", len(dataframe[col].value_counts()))
print(
pd.DataFrame(
{
"COUNT": dataframe[col].value_counts(),
"RATIO": dataframe[col].value_counts() / len(dataframe),
"TARGET_MEAN": dataframe.groupby(col)[target].mean(),
}
),
end="\n\n\n",
)
rare_analyser(df, "SalePrice", cat_cols)
df = rare_encoder(df, 0.01, cat_cols)
drop_list = [
"Street",
"SaleCondition",
"Functional",
"Condition2",
"Utilities",
"SaleType",
"MiscVal",
"Alley",
"LandSlope",
"PoolQC",
"MiscFeature",
"Electrical",
"Fence",
"RoofStyle",
"RoofMatl",
"FireplaceQu",
]
cat_cols = [col for col in cat_cols if col not in drop_list]
for col in drop_list:
df.drop(col, axis=1, inplace=True)
rare_analyser(df, "SalePrice", cat_cols)
useless_cols = [
col
for col in cat_cols
if df[col].nunique() == 1
or (
df[col].nunique() == 2
and (df[col].value_counts() / len(df) <= 0.01).any(axis=None)
)
]
cat_cols = [col for col in cat_cols if col not in useless_cols]
for col in useless_cols:
df.drop(col, axis=1, inplace=True)
rare_analyser(df, "SalePrice", cat_cols)
# Label Encoding & ONE-HOT ENCODING
def one_hot_encoder(dataframe, categorical_cols, drop_first=False):
dataframe = pd.get_dummies(
dataframe, columns=categorical_cols, drop_first=drop_first
)
return dataframe
cat_cols, num_cols, cat_but_car = grab_col_names(df)
cat_cols = cat_cols + cat_but_car
df = one_hot_encoder(df, cat_cols, drop_first=True)
check_df(df)
cat_cols, num_cols, cat_but_car = grab_col_names(df)
rare_analyser(df, "SalePrice", cat_cols)
useless_cols_new = [
col for col in cat_cols if (df[col].value_counts() / len(df) <= 0.01).any(axis=None)
]
df[useless_cols_new].head()
for col in useless_cols_new:
cat_summary(df, col)
rare_analyser(df, "SalePrice", useless_cols_new)
# Missing Values
def missing_values_table(dataframe, na_name=False):
na_columns = [col for col in dataframe.columns if dataframe[col].isnull().sum() > 0]
n_miss = dataframe[na_columns].isnull().sum().sort_values(ascending=False)
ratio = (
dataframe[na_columns].isnull().sum() / dataframe.shape[0] * 100
).sort_values(ascending=False)
missing_df = pd.concat(
[n_miss, np.round(ratio, 2)], axis=1, keys=["n_miss", "ratio"]
)
print(missing_df, end="\n")
if na_name:
return na_columns
missing_values_table(df)
test.shape
missing_values_table(train)
na_cols = [
col for col in df.columns if df[col].isnull().sum() > 0 and "SalePrice" not in col
]
df[na_cols] = df[na_cols].apply(lambda x: x.fillna(x.median()), axis=0)
# Outliers
def outlier_thresholds(dataframe, col_name, q1=0.25, q3=0.75):
quartile1 = dataframe[col_name].quantile(q1)
quartile3 = dataframe[col_name].quantile(q3)
interquantile_range = quartile3 - quartile1
up_limit = quartile3 + 1.5 * interquantile_range
low_limit = quartile1 - 1.5 * interquantile_range
return low_limit, up_limit
def check_outlier(dataframe, col_name, q1=0.25, q3=0.75):
low_limit, up_limit = outlier_thresholds(dataframe, col_name, q1, q3)
if dataframe[
(dataframe[col_name] > up_limit) | (dataframe[col_name] < low_limit)
].any(axis=None):
return True
else:
return False
for col in num_cols:
print(col, check_outlier(df, col, q1=0.01, q3=0.99))
# Model
df.shape
train_df = df[df["SalePrice"].notnull()]
test_df = df[df["SalePrice"].isnull()].drop("SalePrice", axis=1)
train_df.shape
test_df.shape
y = np.log1p(train_df["SalePrice"])
X = train_df.drop(["Id", "SalePrice"], axis=1)
X.shape
# Base Models
##################
models = [
("LR", LinearRegression()),
("CART", DecisionTreeRegressor()),
("RF", RandomForestRegressor()),
("GBM", GradientBoostingRegressor()),
("XGBoost", XGBRegressor(objective="reg:squarederror")),
("LightGBM", LGBMRegressor()),
]
for name, regressor in models:
rmse = np.mean(
np.sqrt(
-cross_val_score(regressor, X, y, cv=3, scoring="neg_mean_squared_error")
)
)
print(f"RMSE: {round(rmse, 4)} ({name}) ")
# **Hyperparameter Optimization**
lgbm_model = LGBMRegressor(random_state=46)
# modelleme öncesi hata:
rmse = np.mean(
np.sqrt(-cross_val_score(lgbm_model, X, y, cv=10, scoring="neg_mean_squared_error"))
)
rmse
lgbm_params = {
"learning_rate": [0.01, 0.005],
"n_estimators": [15000, 20000],
"colsample_bytree": [0.5, 0.3],
}
lgbm_gs_best = GridSearchCV(
lgbm_model, lgbm_params, cv=10, n_jobs=-1, verbose=False
).fit(X, y)
final_model = lgbm_model.set_params(**lgbm_gs_best.best_params_).fit(X, y)
rmse = np.mean(
np.sqrt(
-cross_val_score(final_model, X, y, cv=10, scoring="neg_mean_squared_error")
)
)
print(rmse)
# hiperparametrelerin default kendi değeriyle rmse 0.1305858 idi.
# optimizasyonlarla 0.12328 e indirdik
# Feature Selection
def plot_importance(model, features, num=len(X), save=False):
feature_imp = pd.DataFrame(
{"Value": model.feature_importances_, "Feature": features.columns}
)
plt.figure(figsize=(10, 10))
sns.set(font_scale=1)
sns.barplot(
x="Value",
y="Feature",
data=feature_imp.sort_values(by="Value", ascending=False)[0:num],
)
plt.title("Features")
plt.tight_layout()
plt.show()
if save:
plt.savefig("importances.png")
plot_importance(final_model, X, 20)
X.shape
feature_imp = pd.DataFrame(
{"Value": final_model.feature_importances_, "Feature": X.columns}
)
def num_summary(dataframe, numerical_col, plot=False):
quantiles = [0.05, 0.10, 0.20, 0.30, 0.40, 0.50, 0.60, 0.70, 0.80, 0.90, 0.95, 0.99]
print(dataframe[numerical_col].describe(quantiles).T)
num_summary(feature_imp, "Value", True)
feature_imp[feature_imp["Value"] > 0].shape
feature_imp[feature_imp["Value"] < 1].shape
zero_imp_cols = feature_imp[feature_imp["Value"] < 1]["Feature"].values
selected_cols = [col for col in X.columns if col not in zero_imp_cols]
# Hyperparameter Optimization with Selected Features
lgbm_model = LGBMRegressor(random_state=46)
lgbm_params = {
"learning_rate": [0.01, 0.005],
"n_estimators": [15000, 20000],
"colsample_bytree": [0.5, 0.3],
}
lgbm_gs_best = GridSearchCV(
lgbm_model, lgbm_params, cv=10, n_jobs=-1, verbose=True
).fit(X[selected_cols], y)
y = np.log1p(train_df["SalePrice"])
X = train_df.drop(["Id", "SalePrice"], axis=1)
final_model = lgbm_model.set_params(**lgbm_gs_best.best_params_).fit(
X[selected_cols], y
)
rmse = np.mean(
np.sqrt(
-cross_val_score(
final_model, X[selected_cols], y, cv=10, scoring="neg_mean_squared_error"
)
)
)
print(rmse)
# SONUCLARIN YUKLENMESI
#######################################
submission_df = pd.DataFrame()
submission_df["Id"] = test_df["Id"].astype("Int32")
submission_df.head()
y_pred_sub = final_model.predict(test_df[selected_cols])
test_df.head()
y_pred_sub = np.expm1(y_pred_sub)
submission_df["SalePrice"] = y_pred_sub
submission_df.to_csv("submission.csv", index=False)
submission_df
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/046/69046436.ipynb
| null | null |
[{"Id": 69046436, "ScriptId": 18841428, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6947038, "CreationDate": "07/26/2021 08:40:25", "VersionNumber": 1.0, "Title": "HousePricePrediction", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 411.0, "LinesInsertedFromPrevious": 265.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 146.0, "LinesInsertedFromFork": 265.0, "LinesDeletedFromFork": 632.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 146.0, "TotalVotes": 4}]
| null | null | null | null |
import numpy as np
import pandas as pd
from sklearn.impute import SimpleImputer, KNNImputer
from sklearn.ensemble import (
RandomForestRegressor,
GradientBoostingRegressor,
ExtraTreesRegressor,
)
import matplotlib.pyplot as plt
import plotly.graph_objects as go
import plotly.express as px
import seaborn as sns
from catboost import CatBoostRegressor
from lightgbm import LGBMRegressor
from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor
from sklearn.linear_model import LinearRegression, Ridge, Lasso, ElasticNet
from sklearn.model_selection import train_test_split, GridSearchCV, cross_val_score
from sklearn.neighbors import KNeighborsRegressor
from sklearn.svm import SVR
from sklearn.tree import DecisionTreeRegressor
from xgboost import XGBRegressor
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train = pd.read_csv("../input/house-prices-advanced-regression-techniques/train.csv")
test = pd.read_csv("../input/house-prices-advanced-regression-techniques/test.csv")
print(train.shape)
print(test.shape)
df = train.append(test).reset_index(drop=True)
print(df.shape)
df.columns
def check_df(dataframe, head=5):
print("##################### Shape #####################")
print(dataframe.shape)
print("##################### Types #####################")
print(dataframe.dtypes)
print("##################### Head #####################")
print(dataframe.head(head))
print("##################### Tail #####################")
print(dataframe.tail(head))
print("##################### NA #####################")
print(dataframe.isnull().sum())
print("##################### Quantiles #####################")
print(dataframe.quantile([0, 0.05, 0.50, 0.95, 0.99, 1]).T)
check_df(df)
def grab_col_names(dataframe, cat_th=10, car_th=20):
# cat_cols, cat_but_car
cat_cols = [col for col in dataframe.columns if dataframe[col].dtypes == "O"]
num_but_cat = [
col
for col in dataframe.columns
if dataframe[col].nunique() < cat_th and dataframe[col].dtypes != "O"
]
cat_but_car = [
col
for col in dataframe.columns
if dataframe[col].nunique() > car_th and dataframe[col].dtypes == "O"
]
cat_cols = cat_cols + num_but_cat
cat_cols = [col for col in cat_cols if col not in cat_but_car]
# num_cols
num_cols = [col for col in dataframe.columns if dataframe[col].dtypes != "O"]
num_cols = [col for col in num_cols if col not in num_but_cat]
print(f"Observations: {dataframe.shape[0]}")
print(f"Variables: {dataframe.shape[1]}")
print(f"cat_cols: {len(cat_cols)}")
print(f"num_cols: {len(num_cols)}")
print(f"cat_but_car: {len(cat_but_car)}")
print(f"num_but_cat: {len(num_but_cat)}")
return cat_cols, num_cols, cat_but_car
cat_cols, num_cols, cat_but_car = grab_col_names(df)
df["Neighborhood"].value_counts()
# Kategorik Değişken Analizi
def cat_summary(dataframe, col_name, plot=False):
print(
pd.DataFrame(
{
col_name: dataframe[col_name].value_counts(),
"Ratio": 100 * dataframe[col_name].value_counts() / len(dataframe),
}
)
)
print("##########################################")
if plot:
sns.countplot(x=dataframe[col_name], data=dataframe)
plt.show()
for col in cat_cols:
cat_summary(df, col)
for col in cat_but_car:
cat_summary(df, col)
# Sayısal Değişken Analizi
df[num_cols].describe([0.10, 0.30, 0.50, 0.70, 0.80, 0.99]).T
# Target Analizi
df["SalePrice"].describe([0.05, 0.10, 0.25, 0.50, 0.75, 0.80, 0.90, 0.95, 0.99]).T
def find_correlation(dataframe, numeric_cols, corr_limit=0.60):
high_correlations = []
low_correlations = []
for col in numeric_cols:
if col == "SalePrice":
pass
else:
correlation = dataframe[[col, "SalePrice"]].corr().loc[col, "SalePrice"]
print(col, correlation)
if abs(correlation) > corr_limit:
high_correlations.append(col + ": " + str(correlation))
else:
low_correlations.append(col + ": " + str(correlation))
return low_correlations, high_correlations
low_corrs, high_corrs = find_correlation(df, num_cols)
# tüm değişkenler korelasyon
corr_matrix = df.corr()
sns.clustermap(corr_matrix, annot=True, figsize=(20, 15), fmt=".2f")
plt.title("Correlation Between Features")
plt.show()
threshold = 0.60
filter = np.abs(corr_matrix["SalePrice"]) > threshold
corr_features = corr_matrix.columns[filter].tolist()
sns.clustermap(df[corr_features].corr(), annot=True, fmt=".2f")
plt.title("Correlation Between Features w/ Corr Threshold 0.60)")
plt.show()
def high_correlated_cols(dataframe, plot=False, corr_th=0.60):
corr = dataframe.corr()
cor_matrix = corr.abs()
upper_triangle_matrix = cor_matrix.where(
np.triu(np.ones(cor_matrix.shape), k=1).astype(np.bool)
)
drop_list = [
col
for col in upper_triangle_matrix.columns
if any(upper_triangle_matrix[col] > corr_th)
]
if plot:
import seaborn as sns
import matplotlib.pyplot as plt
sns.set(rc={"figure.figsize": (15, 15)})
sns.heatmap(corr, cmap="RdBu")
plt.show()
return drop_list
high_correlated_cols(df)
# FEATURE ENGINEERING
df["SqFtPerRoom"] = df["GrLivArea"] / (
df["TotRmsAbvGrd"] + df["FullBath"] + df["HalfBath"] + df["KitchenAbvGr"]
)
df["Total_Home_Quality"] = df["OverallQual"] + df["OverallCond"]
df["Total_Bathrooms"] = (
df["FullBath"]
+ (0.5 * df["HalfBath"])
+ df["BsmtFullBath"]
+ (0.5 * df["BsmtHalfBath"])
)
df["HighQualSF"] = df["1stFlrSF"] + df["2ndFlrSF"]
# Converting non-numeric predictors stored as numbers into string
df["MSSubClass"] = df["MSSubClass"].apply(str)
df["YrSold"] = df["YrSold"].apply(str)
df["MoSold"] = df["MoSold"].apply(str)
# RARE ENCODING
def rare_encoder(dataframe, rare_perc, cat_cols):
rare_columns = [
col
for col in cat_cols
if (dataframe[col].value_counts() / len(dataframe) < 0.01).sum() > 1
]
for col in rare_columns:
tmp = dataframe[col].value_counts() / len(dataframe)
rare_labels = tmp[tmp < rare_perc].index
dataframe[col] = np.where(
dataframe[col].isin(rare_labels), "Rare", dataframe[col]
)
return dataframe
def rare_analyser(dataframe, target, cat_cols):
for col in cat_cols:
print(col, ":", len(dataframe[col].value_counts()))
print(
pd.DataFrame(
{
"COUNT": dataframe[col].value_counts(),
"RATIO": dataframe[col].value_counts() / len(dataframe),
"TARGET_MEAN": dataframe.groupby(col)[target].mean(),
}
),
end="\n\n\n",
)
rare_analyser(df, "SalePrice", cat_cols)
df = rare_encoder(df, 0.01, cat_cols)
drop_list = [
"Street",
"SaleCondition",
"Functional",
"Condition2",
"Utilities",
"SaleType",
"MiscVal",
"Alley",
"LandSlope",
"PoolQC",
"MiscFeature",
"Electrical",
"Fence",
"RoofStyle",
"RoofMatl",
"FireplaceQu",
]
cat_cols = [col for col in cat_cols if col not in drop_list]
for col in drop_list:
df.drop(col, axis=1, inplace=True)
rare_analyser(df, "SalePrice", cat_cols)
useless_cols = [
col
for col in cat_cols
if df[col].nunique() == 1
or (
df[col].nunique() == 2
and (df[col].value_counts() / len(df) <= 0.01).any(axis=None)
)
]
cat_cols = [col for col in cat_cols if col not in useless_cols]
for col in useless_cols:
df.drop(col, axis=1, inplace=True)
rare_analyser(df, "SalePrice", cat_cols)
# Label Encoding & ONE-HOT ENCODING
def one_hot_encoder(dataframe, categorical_cols, drop_first=False):
dataframe = pd.get_dummies(
dataframe, columns=categorical_cols, drop_first=drop_first
)
return dataframe
cat_cols, num_cols, cat_but_car = grab_col_names(df)
cat_cols = cat_cols + cat_but_car
df = one_hot_encoder(df, cat_cols, drop_first=True)
check_df(df)
cat_cols, num_cols, cat_but_car = grab_col_names(df)
rare_analyser(df, "SalePrice", cat_cols)
useless_cols_new = [
col for col in cat_cols if (df[col].value_counts() / len(df) <= 0.01).any(axis=None)
]
df[useless_cols_new].head()
for col in useless_cols_new:
cat_summary(df, col)
rare_analyser(df, "SalePrice", useless_cols_new)
# Missing Values
def missing_values_table(dataframe, na_name=False):
na_columns = [col for col in dataframe.columns if dataframe[col].isnull().sum() > 0]
n_miss = dataframe[na_columns].isnull().sum().sort_values(ascending=False)
ratio = (
dataframe[na_columns].isnull().sum() / dataframe.shape[0] * 100
).sort_values(ascending=False)
missing_df = pd.concat(
[n_miss, np.round(ratio, 2)], axis=1, keys=["n_miss", "ratio"]
)
print(missing_df, end="\n")
if na_name:
return na_columns
missing_values_table(df)
test.shape
missing_values_table(train)
na_cols = [
col for col in df.columns if df[col].isnull().sum() > 0 and "SalePrice" not in col
]
df[na_cols] = df[na_cols].apply(lambda x: x.fillna(x.median()), axis=0)
# Outliers
def outlier_thresholds(dataframe, col_name, q1=0.25, q3=0.75):
quartile1 = dataframe[col_name].quantile(q1)
quartile3 = dataframe[col_name].quantile(q3)
interquantile_range = quartile3 - quartile1
up_limit = quartile3 + 1.5 * interquantile_range
low_limit = quartile1 - 1.5 * interquantile_range
return low_limit, up_limit
def check_outlier(dataframe, col_name, q1=0.25, q3=0.75):
low_limit, up_limit = outlier_thresholds(dataframe, col_name, q1, q3)
if dataframe[
(dataframe[col_name] > up_limit) | (dataframe[col_name] < low_limit)
].any(axis=None):
return True
else:
return False
for col in num_cols:
print(col, check_outlier(df, col, q1=0.01, q3=0.99))
# Model
df.shape
train_df = df[df["SalePrice"].notnull()]
test_df = df[df["SalePrice"].isnull()].drop("SalePrice", axis=1)
train_df.shape
test_df.shape
y = np.log1p(train_df["SalePrice"])
X = train_df.drop(["Id", "SalePrice"], axis=1)
X.shape
# Base Models
##################
models = [
("LR", LinearRegression()),
("CART", DecisionTreeRegressor()),
("RF", RandomForestRegressor()),
("GBM", GradientBoostingRegressor()),
("XGBoost", XGBRegressor(objective="reg:squarederror")),
("LightGBM", LGBMRegressor()),
]
for name, regressor in models:
rmse = np.mean(
np.sqrt(
-cross_val_score(regressor, X, y, cv=3, scoring="neg_mean_squared_error")
)
)
print(f"RMSE: {round(rmse, 4)} ({name}) ")
# **Hyperparameter Optimization**
lgbm_model = LGBMRegressor(random_state=46)
# modelleme öncesi hata:
rmse = np.mean(
np.sqrt(-cross_val_score(lgbm_model, X, y, cv=10, scoring="neg_mean_squared_error"))
)
rmse
lgbm_params = {
"learning_rate": [0.01, 0.005],
"n_estimators": [15000, 20000],
"colsample_bytree": [0.5, 0.3],
}
lgbm_gs_best = GridSearchCV(
lgbm_model, lgbm_params, cv=10, n_jobs=-1, verbose=False
).fit(X, y)
final_model = lgbm_model.set_params(**lgbm_gs_best.best_params_).fit(X, y)
rmse = np.mean(
np.sqrt(
-cross_val_score(final_model, X, y, cv=10, scoring="neg_mean_squared_error")
)
)
print(rmse)
# hiperparametrelerin default kendi değeriyle rmse 0.1305858 idi.
# optimizasyonlarla 0.12328 e indirdik
# Feature Selection
def plot_importance(model, features, num=len(X), save=False):
feature_imp = pd.DataFrame(
{"Value": model.feature_importances_, "Feature": features.columns}
)
plt.figure(figsize=(10, 10))
sns.set(font_scale=1)
sns.barplot(
x="Value",
y="Feature",
data=feature_imp.sort_values(by="Value", ascending=False)[0:num],
)
plt.title("Features")
plt.tight_layout()
plt.show()
if save:
plt.savefig("importances.png")
plot_importance(final_model, X, 20)
X.shape
feature_imp = pd.DataFrame(
{"Value": final_model.feature_importances_, "Feature": X.columns}
)
def num_summary(dataframe, numerical_col, plot=False):
quantiles = [0.05, 0.10, 0.20, 0.30, 0.40, 0.50, 0.60, 0.70, 0.80, 0.90, 0.95, 0.99]
print(dataframe[numerical_col].describe(quantiles).T)
num_summary(feature_imp, "Value", True)
feature_imp[feature_imp["Value"] > 0].shape
feature_imp[feature_imp["Value"] < 1].shape
zero_imp_cols = feature_imp[feature_imp["Value"] < 1]["Feature"].values
selected_cols = [col for col in X.columns if col not in zero_imp_cols]
# Hyperparameter Optimization with Selected Features
lgbm_model = LGBMRegressor(random_state=46)
lgbm_params = {
"learning_rate": [0.01, 0.005],
"n_estimators": [15000, 20000],
"colsample_bytree": [0.5, 0.3],
}
lgbm_gs_best = GridSearchCV(
lgbm_model, lgbm_params, cv=10, n_jobs=-1, verbose=True
).fit(X[selected_cols], y)
y = np.log1p(train_df["SalePrice"])
X = train_df.drop(["Id", "SalePrice"], axis=1)
final_model = lgbm_model.set_params(**lgbm_gs_best.best_params_).fit(
X[selected_cols], y
)
rmse = np.mean(
np.sqrt(
-cross_val_score(
final_model, X[selected_cols], y, cv=10, scoring="neg_mean_squared_error"
)
)
)
print(rmse)
# SONUCLARIN YUKLENMESI
#######################################
submission_df = pd.DataFrame()
submission_df["Id"] = test_df["Id"].astype("Int32")
submission_df.head()
y_pred_sub = final_model.predict(test_df[selected_cols])
test_df.head()
y_pred_sub = np.expm1(y_pred_sub)
submission_df["SalePrice"] = y_pred_sub
submission_df.to_csv("submission.csv", index=False)
submission_df
| false | 0 | 4,775 | 4 | 4,775 | 4,775 |
||
69046416
|
<jupyter_start><jupyter_text>House Sales in King County, USA
This dataset contains house sale prices for King County, which includes Seattle. It includes homes sold between May 2014 and May 2015.
It's a great dataset for evaluating simple regression models.
Kaggle dataset identifier: housesalesprediction
<jupyter_code>import pandas as pd
df = pd.read_csv('housesalesprediction/kc_house_data.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 21613 entries, 0 to 21612
Data columns (total 21 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 id 21613 non-null int64
1 date 21613 non-null object
2 price 21613 non-null float64
3 bedrooms 21613 non-null int64
4 bathrooms 21613 non-null float64
5 sqft_living 21613 non-null int64
6 sqft_lot 21613 non-null int64
7 floors 21613 non-null float64
8 waterfront 21613 non-null int64
9 view 21613 non-null int64
10 condition 21613 non-null int64
11 grade 21613 non-null int64
12 sqft_above 21613 non-null int64
13 sqft_basement 21613 non-null int64
14 yr_built 21613 non-null int64
15 yr_renovated 21613 non-null int64
16 zipcode 21613 non-null int64
17 lat 21613 non-null float64
18 long 21613 non-null float64
19 sqft_living15 21613 non-null int64
20 sqft_lot15 21613 non-null int64
dtypes: float64(5), int64(15), object(1)
memory usage: 3.5+ MB
<jupyter_text>Examples:
{
"id": 7129300520,
"date": "2014-10-13 00:00:00",
"price": 221900,
"bedrooms": 3,
"bathrooms": 1.0,
"sqft_living": 1180,
"sqft_lot": 5650,
"floors": 1,
"waterfront": 0,
"view": 0,
"condition": 3,
"grade": 7,
"sqft_above": 1180,
"sqft_basement": 0,
"yr_built": 1955,
"yr_renovated": 0,
"zipcode": 98178,
"lat": 47.5112,
"long": -122.257,
"sqft_living15": 1340,
"...": "and 1 more columns"
}
{
"id": 6414100192,
"date": "2014-12-09 00:00:00",
"price": 538000,
"bedrooms": 3,
"bathrooms": 2.25,
"sqft_living": 2570,
"sqft_lot": 7242,
"floors": 2,
"waterfront": 0,
"view": 0,
"condition": 3,
"grade": 7,
"sqft_above": 2170,
"sqft_basement": 400,
"yr_built": 1951,
"yr_renovated": 1991,
"zipcode": 98125,
"lat": 47.721,
"long": -122.319,
"sqft_living15": 1690,
"...": "and 1 more columns"
}
{
"id": 5631500400,
"date": "2015-02-25 00:00:00",
"price": 180000,
"bedrooms": 2,
"bathrooms": 1.0,
"sqft_living": 770,
"sqft_lot": 10000,
"floors": 1,
"waterfront": 0,
"view": 0,
"condition": 3,
"grade": 6,
"sqft_above": 770,
"sqft_basement": 0,
"yr_built": 1933,
"yr_renovated": 0,
"zipcode": 98028,
"lat": 47.7379,
"long": -122.233,
"sqft_living15": 2720,
"...": "and 1 more columns"
}
{
"id": 2487200875,
"date": "2014-12-09 00:00:00",
"price": 604000,
"bedrooms": 4,
"bathrooms": 3.0,
"sqft_living": 1960,
"sqft_lot": 5000,
"floors": 1,
"waterfront": 0,
"view": 0,
"condition": 5,
"grade": 7,
"sqft_above": 1050,
"sqft_basement": 910,
"yr_built": 1965,
"yr_renovated": 0,
"zipcode": 98136,
"lat": 47.5208,
"long": -122.393,
"sqft_living15": 1360,
"...": "and 1 more columns"
}
<jupyter_script># # King County Houses Prices:
# ## Neigborhoods Classification
# In this notebook, I used an other dataset (SEA Building Energy Benchmarking (Source bellow)) which give us for each building GPS coords and the neighborhood (North, East, Ballard, Delridge, etc) .
# I cleaned the dataset as part of a project for a data scientist training and got the idea using this to classify each King County Houses using a KNN classifier.
#
# It will maybe help improving algorithm performances for predicting house prices.
#
# Results at the bottom of the notebook
# ### Importations
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
pd.set_option("display.max_rows", None)
pd.set_option("display.max_columns", None)
sns.set()
data = pd.read_csv("../input/housesalesprediction/kc_house_data.csv")
# ### Exploratory Functions
def describe_columns(df):
desc_df = pd.DataFrame(
index=df.columns,
columns=["NaN count", "NaN frequency (%)", "Number of unique values"],
)
desc_df["NaN count"] = df.isna().sum()
desc_df["NaN frequency (%)"] = desc_df["NaN count"] / df.shape[0] * 100
for column in df.columns:
desc_df["Number of unique values"][column] = len(df[column].dropna().unique())
return desc_df
def move_column(df, column_name, column_place):
mvd_column = df.pop(column_name)
df.insert(column_place, column_name, mvd_column)
return df
def prop_nan(df):
return (df.isna()).sum().sum() / df.size
def nan_map(df, save=False, filename="nan_location"):
plt.figure(figsize=(20, 10))
sns.heatmap(df.isna())
if save:
plt.savefig(filename)
def corr_matrix(
df,
figsize=(30, 20),
maptype="heatmap",
absolute=False,
crit_value=None,
annot=True,
save=False,
filename="corr_matrix",
):
matrix_corr = df.corr()
if absolute:
matrix_corr = matrix_corr.abs()
if crit_value != None:
matrix_corr = matrix_corr >= crit_value
plt.figure(figsize=figsize)
if maptype == "heatmap":
sns.heatmap(matrix_corr, annot=annot)
elif maptype == "clustermap":
sns.clustermap(matrix_corr, annot=annot)
if save:
plt.savefig(filename)
df = data.copy()
# ### Columns descriptions
# id - Unique ID for each home sold
# date - Date of the home sale
# price - Price of each home sold
# bedrooms - Number of bedrooms
# bathrooms - Number of bathrooms, where .5 accounts for a room with a toilet but no shower
# sqft_living - Square footage of the apartments interior living space
# sqft_lot - Square footage of the land space
# floors - Number of floors
# waterfront - A dummy variable for whether the apartment was overlooking the waterfront or not
# view - An index from 0 to 4 of how good the view of the property was
# condition - An index from 1 to 5 on the condition of the apartment,
# grade - An index from 1 to 13, where 1-3 falls short of building construction and design, 7 has an average level of construction and design, and 11-13 have a high quality level of construction and design.
# sqft_above - The square footage of the interior housing space that is above ground level
# sqft_basement - The square footage of the interior housing space that is below ground level
# yr_built - The year the house was initially built
# yr_renovated - The year of the house’s last renovation
# zipcode - What zipcode area the house is in
# lat - Lattitude
# long - Longitude
# sqft_living15 - The square footage of interior housing living space for the nearest 15 neighbors
# sqft_lot15 - The square footage of the land lots of the nearest 15 neighbors
# verified from 2 sources:
# https://www.slideshare.net/PawanShivhare1/predicting-king-county-house-prices
# https://rstudio-pubs-static.s3.amazonaws.com/155304_cc51f448116744069664b35e7762999f.htm
#
df.head()
# ### Scatter 2 numerical columns
def plot_2_features(df, x_name, y_name):
plt.figure(figsize=(12, 8))
plt.scatter(df[x_name], df[y_name], s=2)
plt.xlabel(x_name)
plt.ylabel(y_name)
# ### Plot map with a numerical column
def plot_map_num(df, y_name, interquartile=True, v=None):
plt.figure(figsize=(20, 10))
if v != None:
vmin = v[0]
vmax = v[1]
points = plt.scatter(
df["long"],
df["lat"],
c=df[y_name],
cmap="jet",
lw=0,
s=2,
vmin=vmin,
vmax=vmax,
)
elif interquartile:
desc_df = df.describe()
vmin = desc_df.loc["25%", y_name]
vmax = desc_df.loc["75%", y_name]
points = plt.scatter(
df["long"],
df["lat"],
c=df[y_name],
cmap="jet",
lw=0,
s=2,
vmin=vmin,
vmax=vmax,
)
else:
points = plt.scatter(df["long"], df["lat"], c=df[y_name], cmap="jet", lw=0, s=2)
plt.colorbar(points)
plt.xlabel("Long")
plt.ylabel("Lat")
# ### Plot price map
plot_map_num(df, "price", interquartile=True)
# ### Load dataset containing Neighborhoods with GPS coord
# Source: https://www.kaggle.com/city-of-seattle/sea-building-energy-benchmarking#2015-building-energy-benchmarking.csv
# Note: I loaded a cleaned version of the dataset that I made for a data-science online training.
neighborhood_data = pd.read_csv(
"../input/sea-energy-building-benchmark/data_cleaned.csv"
)
# Selecting only the intersting columns
neighborhood_df = neighborhood_data.copy()
neighborhood_df = neighborhood_df[["Latitude", "Longitude", "Neighborhood"]]
neighborhood_df.head()
neighborhood_df["Neighborhood"].unique()
# ### Importing KNN, MinMaxScaler
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import confusion_matrix, classification_report
X = neighborhood_df.drop("Neighborhood", axis=1).values
y = neighborhood_df["Neighborhood"].values
# Splitting Data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
# Made my own encoding class which is easy to use because I got some errors with LabelEncoder
class Encoding:
def __init__(self):
self.dico = {}
self.inv_dico = {}
def fit(self, y):
i = 0
for classe in pd.Series(y).unique():
self.dico[classe] = i
self.inv_dico[i] = classe
i += 1
def transform(self, y):
return pd.Series(y).map(self.dico).values
def inverse_transform(self, y):
return pd.Series(y).map(self.inv_dico).values
# ### Using Neighborhoods datasets to train a model for predicting Neighborhood in df
scaler = MinMaxScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
encoder = Encoding()
encoder.fit(y_train)
y_train_coded = encoder.transform(y_train)
y_test_coded = encoder.transform(y_test)
# KNeighborsClassifier with minimum optimization (maybe need more parameter or an other algorithm). Can be improved.
model = GridSearchCV(KNeighborsClassifier(), {"n_neighbors": range(1, 11)})
# Fitting with training set
model.fit(X_train_scaled, y_train_coded)
# Predicting results on the test set
y_pred = encoder.inverse_transform(model.predict(X_test_scaled))
# Score on the test set
model.score(X_test_scaled, y_test_coded)
# ### Confusion Matrix
plt.figure(figsize=(12, 8))
sns.heatmap(confusion_matrix(y_test, y_pred), annot=True)
# ### Classification report
print(classification_report(y_test, y_pred))
# Adding a new column Neighborhood for King County Houses
df["Neighborhood"] = encoder.inverse_transform(
model.predict(scaler.transform(df[["lat", "long"]].values))
)
# ### Plot map with a categorical column
def plot_map_categ(df, categ_column):
plt.figure(figsize=(20, 10))
for classe in df[categ_column].sort_values().unique():
df_classe = df[df[categ_column] == classe]
plt.scatter(df_classe["long"], df_classe["lat"], lw=0, s=10, label=classe)
plt.legend()
plt.xlabel("Long")
plt.ylabel("Lat")
# ### Neighborhood locations
# Note: The Neighborhood dataset was covering a smaller area for the longitude
# . So the mountain part may not be very accurate.
plot_map_categ(df, "Neighborhood")
# ### Boxplot function
def boxplot_groupes(df, categ_column, target_column, figsize=(20, 10)):
groupes = []
for cat in list(df[categ_column].unique()):
groupes.append(df[df[categ_column] == cat][target_column])
medianprops = {"color": "black"}
meanprops = {
"marker": "o",
"markeredgecolor": "black",
"markerfacecolor": "firebrick",
}
plt.figure(figsize=figsize)
plt.boxplot(
groupes,
labels=list(df[categ_column].unique()),
showfliers=False,
medianprops=medianprops,
vert=False,
patch_artist=True,
showmeans=True,
meanprops=meanprops,
)
plt.ylabel(categ_column)
plt.xlabel(target_column)
# Boxplot Neighborhood / price
boxplot_groupes(df, "Neighborhood", "price")
# ### Updated King County house prices dataSet with a 'Neighborhood' column
df.head()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/046/69046416.ipynb
|
housesalesprediction
|
harlfoxem
|
[{"Id": 69046416, "ScriptId": 18825679, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7571614, "CreationDate": "07/26/2021 08:39:57", "VersionNumber": 4.0, "Title": "King County Houses Neighborhood Classification", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 269.0, "LinesInsertedFromPrevious": 10.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 259.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 91775956, "KernelVersionId": 69046416, "SourceDatasetVersionId": 270}]
|
[{"Id": 270, "DatasetId": 128, "DatasourceVersionId": 270, "CreatorUserId": 680332, "LicenseName": "CC0: Public Domain", "CreationDate": "08/25/2016 15:52:49", "VersionNumber": 1.0, "Title": "House Sales in King County, USA", "Slug": "housesalesprediction", "Subtitle": "Predict house price using regression", "Description": "This dataset contains house sale prices for King County, which includes Seattle. It includes homes sold between May 2014 and May 2015.\n\nIt's a great dataset for evaluating simple regression models.", "VersionNotes": "Initial release", "TotalCompressedBytes": 2515206.0, "TotalUncompressedBytes": 2515206.0}]
|
[{"Id": 128, "CreatorUserId": 680332, "OwnerUserId": 680332.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 270.0, "CurrentDatasourceVersionId": 270.0, "ForumId": 1447, "Type": 2, "CreationDate": "08/25/2016 15:52:49", "LastActivityDate": "02/06/2018", "TotalViews": 996866, "TotalDownloads": 172516, "TotalVotes": 2041, "TotalKernels": 1225}]
|
[{"Id": 680332, "UserName": "harlfoxem", "DisplayName": "harlfoxem", "RegisterDate": "08/05/2016", "PerformanceTier": 1}]
|
# # King County Houses Prices:
# ## Neigborhoods Classification
# In this notebook, I used an other dataset (SEA Building Energy Benchmarking (Source bellow)) which give us for each building GPS coords and the neighborhood (North, East, Ballard, Delridge, etc) .
# I cleaned the dataset as part of a project for a data scientist training and got the idea using this to classify each King County Houses using a KNN classifier.
#
# It will maybe help improving algorithm performances for predicting house prices.
#
# Results at the bottom of the notebook
# ### Importations
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
pd.set_option("display.max_rows", None)
pd.set_option("display.max_columns", None)
sns.set()
data = pd.read_csv("../input/housesalesprediction/kc_house_data.csv")
# ### Exploratory Functions
def describe_columns(df):
desc_df = pd.DataFrame(
index=df.columns,
columns=["NaN count", "NaN frequency (%)", "Number of unique values"],
)
desc_df["NaN count"] = df.isna().sum()
desc_df["NaN frequency (%)"] = desc_df["NaN count"] / df.shape[0] * 100
for column in df.columns:
desc_df["Number of unique values"][column] = len(df[column].dropna().unique())
return desc_df
def move_column(df, column_name, column_place):
mvd_column = df.pop(column_name)
df.insert(column_place, column_name, mvd_column)
return df
def prop_nan(df):
return (df.isna()).sum().sum() / df.size
def nan_map(df, save=False, filename="nan_location"):
plt.figure(figsize=(20, 10))
sns.heatmap(df.isna())
if save:
plt.savefig(filename)
def corr_matrix(
df,
figsize=(30, 20),
maptype="heatmap",
absolute=False,
crit_value=None,
annot=True,
save=False,
filename="corr_matrix",
):
matrix_corr = df.corr()
if absolute:
matrix_corr = matrix_corr.abs()
if crit_value != None:
matrix_corr = matrix_corr >= crit_value
plt.figure(figsize=figsize)
if maptype == "heatmap":
sns.heatmap(matrix_corr, annot=annot)
elif maptype == "clustermap":
sns.clustermap(matrix_corr, annot=annot)
if save:
plt.savefig(filename)
df = data.copy()
# ### Columns descriptions
# id - Unique ID for each home sold
# date - Date of the home sale
# price - Price of each home sold
# bedrooms - Number of bedrooms
# bathrooms - Number of bathrooms, where .5 accounts for a room with a toilet but no shower
# sqft_living - Square footage of the apartments interior living space
# sqft_lot - Square footage of the land space
# floors - Number of floors
# waterfront - A dummy variable for whether the apartment was overlooking the waterfront or not
# view - An index from 0 to 4 of how good the view of the property was
# condition - An index from 1 to 5 on the condition of the apartment,
# grade - An index from 1 to 13, where 1-3 falls short of building construction and design, 7 has an average level of construction and design, and 11-13 have a high quality level of construction and design.
# sqft_above - The square footage of the interior housing space that is above ground level
# sqft_basement - The square footage of the interior housing space that is below ground level
# yr_built - The year the house was initially built
# yr_renovated - The year of the house’s last renovation
# zipcode - What zipcode area the house is in
# lat - Lattitude
# long - Longitude
# sqft_living15 - The square footage of interior housing living space for the nearest 15 neighbors
# sqft_lot15 - The square footage of the land lots of the nearest 15 neighbors
# verified from 2 sources:
# https://www.slideshare.net/PawanShivhare1/predicting-king-county-house-prices
# https://rstudio-pubs-static.s3.amazonaws.com/155304_cc51f448116744069664b35e7762999f.htm
#
df.head()
# ### Scatter 2 numerical columns
def plot_2_features(df, x_name, y_name):
plt.figure(figsize=(12, 8))
plt.scatter(df[x_name], df[y_name], s=2)
plt.xlabel(x_name)
plt.ylabel(y_name)
# ### Plot map with a numerical column
def plot_map_num(df, y_name, interquartile=True, v=None):
plt.figure(figsize=(20, 10))
if v != None:
vmin = v[0]
vmax = v[1]
points = plt.scatter(
df["long"],
df["lat"],
c=df[y_name],
cmap="jet",
lw=0,
s=2,
vmin=vmin,
vmax=vmax,
)
elif interquartile:
desc_df = df.describe()
vmin = desc_df.loc["25%", y_name]
vmax = desc_df.loc["75%", y_name]
points = plt.scatter(
df["long"],
df["lat"],
c=df[y_name],
cmap="jet",
lw=0,
s=2,
vmin=vmin,
vmax=vmax,
)
else:
points = plt.scatter(df["long"], df["lat"], c=df[y_name], cmap="jet", lw=0, s=2)
plt.colorbar(points)
plt.xlabel("Long")
plt.ylabel("Lat")
# ### Plot price map
plot_map_num(df, "price", interquartile=True)
# ### Load dataset containing Neighborhoods with GPS coord
# Source: https://www.kaggle.com/city-of-seattle/sea-building-energy-benchmarking#2015-building-energy-benchmarking.csv
# Note: I loaded a cleaned version of the dataset that I made for a data-science online training.
neighborhood_data = pd.read_csv(
"../input/sea-energy-building-benchmark/data_cleaned.csv"
)
# Selecting only the intersting columns
neighborhood_df = neighborhood_data.copy()
neighborhood_df = neighborhood_df[["Latitude", "Longitude", "Neighborhood"]]
neighborhood_df.head()
neighborhood_df["Neighborhood"].unique()
# ### Importing KNN, MinMaxScaler
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import confusion_matrix, classification_report
X = neighborhood_df.drop("Neighborhood", axis=1).values
y = neighborhood_df["Neighborhood"].values
# Splitting Data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
# Made my own encoding class which is easy to use because I got some errors with LabelEncoder
class Encoding:
def __init__(self):
self.dico = {}
self.inv_dico = {}
def fit(self, y):
i = 0
for classe in pd.Series(y).unique():
self.dico[classe] = i
self.inv_dico[i] = classe
i += 1
def transform(self, y):
return pd.Series(y).map(self.dico).values
def inverse_transform(self, y):
return pd.Series(y).map(self.inv_dico).values
# ### Using Neighborhoods datasets to train a model for predicting Neighborhood in df
scaler = MinMaxScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
encoder = Encoding()
encoder.fit(y_train)
y_train_coded = encoder.transform(y_train)
y_test_coded = encoder.transform(y_test)
# KNeighborsClassifier with minimum optimization (maybe need more parameter or an other algorithm). Can be improved.
model = GridSearchCV(KNeighborsClassifier(), {"n_neighbors": range(1, 11)})
# Fitting with training set
model.fit(X_train_scaled, y_train_coded)
# Predicting results on the test set
y_pred = encoder.inverse_transform(model.predict(X_test_scaled))
# Score on the test set
model.score(X_test_scaled, y_test_coded)
# ### Confusion Matrix
plt.figure(figsize=(12, 8))
sns.heatmap(confusion_matrix(y_test, y_pred), annot=True)
# ### Classification report
print(classification_report(y_test, y_pred))
# Adding a new column Neighborhood for King County Houses
df["Neighborhood"] = encoder.inverse_transform(
model.predict(scaler.transform(df[["lat", "long"]].values))
)
# ### Plot map with a categorical column
def plot_map_categ(df, categ_column):
plt.figure(figsize=(20, 10))
for classe in df[categ_column].sort_values().unique():
df_classe = df[df[categ_column] == classe]
plt.scatter(df_classe["long"], df_classe["lat"], lw=0, s=10, label=classe)
plt.legend()
plt.xlabel("Long")
plt.ylabel("Lat")
# ### Neighborhood locations
# Note: The Neighborhood dataset was covering a smaller area for the longitude
# . So the mountain part may not be very accurate.
plot_map_categ(df, "Neighborhood")
# ### Boxplot function
def boxplot_groupes(df, categ_column, target_column, figsize=(20, 10)):
groupes = []
for cat in list(df[categ_column].unique()):
groupes.append(df[df[categ_column] == cat][target_column])
medianprops = {"color": "black"}
meanprops = {
"marker": "o",
"markeredgecolor": "black",
"markerfacecolor": "firebrick",
}
plt.figure(figsize=figsize)
plt.boxplot(
groupes,
labels=list(df[categ_column].unique()),
showfliers=False,
medianprops=medianprops,
vert=False,
patch_artist=True,
showmeans=True,
meanprops=meanprops,
)
plt.ylabel(categ_column)
plt.xlabel(target_column)
# Boxplot Neighborhood / price
boxplot_groupes(df, "Neighborhood", "price")
# ### Updated King County house prices dataSet with a 'Neighborhood' column
df.head()
|
[{"housesalesprediction/kc_house_data.csv": {"column_names": "[\"id\", \"date\", \"price\", \"bedrooms\", \"bathrooms\", \"sqft_living\", \"sqft_lot\", \"floors\", \"waterfront\", \"view\", \"condition\", \"grade\", \"sqft_above\", \"sqft_basement\", \"yr_built\", \"yr_renovated\", \"zipcode\", \"lat\", \"long\", \"sqft_living15\", \"sqft_lot15\"]", "column_data_types": "{\"id\": \"int64\", \"date\": \"object\", \"price\": \"float64\", \"bedrooms\": \"int64\", \"bathrooms\": \"float64\", \"sqft_living\": \"int64\", \"sqft_lot\": \"int64\", \"floors\": \"float64\", \"waterfront\": \"int64\", \"view\": \"int64\", \"condition\": \"int64\", \"grade\": \"int64\", \"sqft_above\": \"int64\", \"sqft_basement\": \"int64\", \"yr_built\": \"int64\", \"yr_renovated\": \"int64\", \"zipcode\": \"int64\", \"lat\": \"float64\", \"long\": \"float64\", \"sqft_living15\": \"int64\", \"sqft_lot15\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 21613 entries, 0 to 21612\nData columns (total 21 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 id 21613 non-null int64 \n 1 date 21613 non-null object \n 2 price 21613 non-null float64\n 3 bedrooms 21613 non-null int64 \n 4 bathrooms 21613 non-null float64\n 5 sqft_living 21613 non-null int64 \n 6 sqft_lot 21613 non-null int64 \n 7 floors 21613 non-null float64\n 8 waterfront 21613 non-null int64 \n 9 view 21613 non-null int64 \n 10 condition 21613 non-null int64 \n 11 grade 21613 non-null int64 \n 12 sqft_above 21613 non-null int64 \n 13 sqft_basement 21613 non-null int64 \n 14 yr_built 21613 non-null int64 \n 15 yr_renovated 21613 non-null int64 \n 16 zipcode 21613 non-null int64 \n 17 lat 21613 non-null float64\n 18 long 21613 non-null float64\n 19 sqft_living15 21613 non-null int64 \n 20 sqft_lot15 21613 non-null int64 \ndtypes: float64(5), int64(15), object(1)\nmemory usage: 3.5+ MB\n", "summary": "{\"id\": {\"count\": 21613.0, \"mean\": 4580301520.864988, \"std\": 2876565571.312057, \"min\": 1000102.0, \"25%\": 2123049194.0, \"50%\": 3904930410.0, \"75%\": 7308900445.0, \"max\": 9900000190.0}, \"price\": {\"count\": 21613.0, \"mean\": 540088.1417665294, \"std\": 367127.19648269983, \"min\": 75000.0, \"25%\": 321950.0, \"50%\": 450000.0, \"75%\": 645000.0, \"max\": 7700000.0}, \"bedrooms\": {\"count\": 21613.0, \"mean\": 3.37084162309721, \"std\": 0.9300618311474517, \"min\": 0.0, \"25%\": 3.0, \"50%\": 3.0, \"75%\": 4.0, \"max\": 33.0}, \"bathrooms\": {\"count\": 21613.0, \"mean\": 2.1147573219821405, \"std\": 0.770163157217742, \"min\": 0.0, \"25%\": 1.75, \"50%\": 2.25, \"75%\": 2.5, \"max\": 8.0}, \"sqft_living\": {\"count\": 21613.0, \"mean\": 2079.8997362698374, \"std\": 918.4408970468115, \"min\": 290.0, \"25%\": 1427.0, \"50%\": 1910.0, \"75%\": 2550.0, \"max\": 13540.0}, \"sqft_lot\": {\"count\": 21613.0, \"mean\": 15106.967565816869, \"std\": 41420.51151513548, \"min\": 520.0, \"25%\": 5040.0, \"50%\": 7618.0, \"75%\": 10688.0, \"max\": 1651359.0}, \"floors\": {\"count\": 21613.0, \"mean\": 1.4943089807060566, \"std\": 0.5399888951423463, \"min\": 1.0, \"25%\": 1.0, \"50%\": 1.5, \"75%\": 2.0, \"max\": 3.5}, \"waterfront\": {\"count\": 21613.0, \"mean\": 0.007541757275713691, \"std\": 0.08651719772788764, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"view\": {\"count\": 21613.0, \"mean\": 0.23430342849211122, \"std\": 0.7663175692736122, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 4.0}, \"condition\": {\"count\": 21613.0, \"mean\": 3.4094295100171195, \"std\": 0.6507430463662071, \"min\": 1.0, \"25%\": 3.0, \"50%\": 3.0, \"75%\": 4.0, \"max\": 5.0}, \"grade\": {\"count\": 21613.0, \"mean\": 7.656873178179799, \"std\": 1.175458756974335, \"min\": 1.0, \"25%\": 7.0, \"50%\": 7.0, \"75%\": 8.0, \"max\": 13.0}, \"sqft_above\": {\"count\": 21613.0, \"mean\": 1788.3906907879516, \"std\": 828.0909776519169, \"min\": 290.0, \"25%\": 1190.0, \"50%\": 1560.0, \"75%\": 2210.0, \"max\": 9410.0}, \"sqft_basement\": {\"count\": 21613.0, \"mean\": 291.5090454818859, \"std\": 442.5750426774682, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 560.0, \"max\": 4820.0}, \"yr_built\": {\"count\": 21613.0, \"mean\": 1971.0051357978994, \"std\": 29.37341080238659, \"min\": 1900.0, \"25%\": 1951.0, \"50%\": 1975.0, \"75%\": 1997.0, \"max\": 2015.0}, \"yr_renovated\": {\"count\": 21613.0, \"mean\": 84.40225790033776, \"std\": 401.6792400191759, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 2015.0}, \"zipcode\": {\"count\": 21613.0, \"mean\": 98077.93980474715, \"std\": 53.505026257473084, \"min\": 98001.0, \"25%\": 98033.0, \"50%\": 98065.0, \"75%\": 98118.0, \"max\": 98199.0}, \"lat\": {\"count\": 21613.0, \"mean\": 47.56005251931708, \"std\": 0.13856371024192418, \"min\": 47.1559, \"25%\": 47.471, \"50%\": 47.5718, \"75%\": 47.678, \"max\": 47.7776}, \"long\": {\"count\": 21613.0, \"mean\": -122.21389640494147, \"std\": 0.14082834238139408, \"min\": -122.519, \"25%\": -122.328, \"50%\": -122.23, \"75%\": -122.125, \"max\": -121.315}, \"sqft_living15\": {\"count\": 21613.0, \"mean\": 1986.552491556008, \"std\": 685.3913042527776, \"min\": 399.0, \"25%\": 1490.0, \"50%\": 1840.0, \"75%\": 2360.0, \"max\": 6210.0}, \"sqft_lot15\": {\"count\": 21613.0, \"mean\": 12768.455651691113, \"std\": 27304.17963133851, \"min\": 651.0, \"25%\": 5100.0, \"50%\": 7620.0, \"75%\": 10083.0, \"max\": 871200.0}}", "examples": "{\"id\":{\"0\":7129300520,\"1\":6414100192,\"2\":5631500400,\"3\":2487200875},\"date\":{\"0\":\"20141013T000000\",\"1\":\"20141209T000000\",\"2\":\"20150225T000000\",\"3\":\"20141209T000000\"},\"price\":{\"0\":221900.0,\"1\":538000.0,\"2\":180000.0,\"3\":604000.0},\"bedrooms\":{\"0\":3,\"1\":3,\"2\":2,\"3\":4},\"bathrooms\":{\"0\":1.0,\"1\":2.25,\"2\":1.0,\"3\":3.0},\"sqft_living\":{\"0\":1180,\"1\":2570,\"2\":770,\"3\":1960},\"sqft_lot\":{\"0\":5650,\"1\":7242,\"2\":10000,\"3\":5000},\"floors\":{\"0\":1.0,\"1\":2.0,\"2\":1.0,\"3\":1.0},\"waterfront\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"view\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"condition\":{\"0\":3,\"1\":3,\"2\":3,\"3\":5},\"grade\":{\"0\":7,\"1\":7,\"2\":6,\"3\":7},\"sqft_above\":{\"0\":1180,\"1\":2170,\"2\":770,\"3\":1050},\"sqft_basement\":{\"0\":0,\"1\":400,\"2\":0,\"3\":910},\"yr_built\":{\"0\":1955,\"1\":1951,\"2\":1933,\"3\":1965},\"yr_renovated\":{\"0\":0,\"1\":1991,\"2\":0,\"3\":0},\"zipcode\":{\"0\":98178,\"1\":98125,\"2\":98028,\"3\":98136},\"lat\":{\"0\":47.5112,\"1\":47.721,\"2\":47.7379,\"3\":47.5208},\"long\":{\"0\":-122.257,\"1\":-122.319,\"2\":-122.233,\"3\":-122.393},\"sqft_living15\":{\"0\":1340,\"1\":1690,\"2\":2720,\"3\":1360},\"sqft_lot15\":{\"0\":5650,\"1\":7639,\"2\":8062,\"3\":5000}}"}}]
| true | 2 |
<start_data_description><data_path>housesalesprediction/kc_house_data.csv:
<column_names>
['id', 'date', 'price', 'bedrooms', 'bathrooms', 'sqft_living', 'sqft_lot', 'floors', 'waterfront', 'view', 'condition', 'grade', 'sqft_above', 'sqft_basement', 'yr_built', 'yr_renovated', 'zipcode', 'lat', 'long', 'sqft_living15', 'sqft_lot15']
<column_types>
{'id': 'int64', 'date': 'object', 'price': 'float64', 'bedrooms': 'int64', 'bathrooms': 'float64', 'sqft_living': 'int64', 'sqft_lot': 'int64', 'floors': 'float64', 'waterfront': 'int64', 'view': 'int64', 'condition': 'int64', 'grade': 'int64', 'sqft_above': 'int64', 'sqft_basement': 'int64', 'yr_built': 'int64', 'yr_renovated': 'int64', 'zipcode': 'int64', 'lat': 'float64', 'long': 'float64', 'sqft_living15': 'int64', 'sqft_lot15': 'int64'}
<dataframe_Summary>
{'id': {'count': 21613.0, 'mean': 4580301520.864988, 'std': 2876565571.312057, 'min': 1000102.0, '25%': 2123049194.0, '50%': 3904930410.0, '75%': 7308900445.0, 'max': 9900000190.0}, 'price': {'count': 21613.0, 'mean': 540088.1417665294, 'std': 367127.19648269983, 'min': 75000.0, '25%': 321950.0, '50%': 450000.0, '75%': 645000.0, 'max': 7700000.0}, 'bedrooms': {'count': 21613.0, 'mean': 3.37084162309721, 'std': 0.9300618311474517, 'min': 0.0, '25%': 3.0, '50%': 3.0, '75%': 4.0, 'max': 33.0}, 'bathrooms': {'count': 21613.0, 'mean': 2.1147573219821405, 'std': 0.770163157217742, 'min': 0.0, '25%': 1.75, '50%': 2.25, '75%': 2.5, 'max': 8.0}, 'sqft_living': {'count': 21613.0, 'mean': 2079.8997362698374, 'std': 918.4408970468115, 'min': 290.0, '25%': 1427.0, '50%': 1910.0, '75%': 2550.0, 'max': 13540.0}, 'sqft_lot': {'count': 21613.0, 'mean': 15106.967565816869, 'std': 41420.51151513548, 'min': 520.0, '25%': 5040.0, '50%': 7618.0, '75%': 10688.0, 'max': 1651359.0}, 'floors': {'count': 21613.0, 'mean': 1.4943089807060566, 'std': 0.5399888951423463, 'min': 1.0, '25%': 1.0, '50%': 1.5, '75%': 2.0, 'max': 3.5}, 'waterfront': {'count': 21613.0, 'mean': 0.007541757275713691, 'std': 0.08651719772788764, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'view': {'count': 21613.0, 'mean': 0.23430342849211122, 'std': 0.7663175692736122, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 4.0}, 'condition': {'count': 21613.0, 'mean': 3.4094295100171195, 'std': 0.6507430463662071, 'min': 1.0, '25%': 3.0, '50%': 3.0, '75%': 4.0, 'max': 5.0}, 'grade': {'count': 21613.0, 'mean': 7.656873178179799, 'std': 1.175458756974335, 'min': 1.0, '25%': 7.0, '50%': 7.0, '75%': 8.0, 'max': 13.0}, 'sqft_above': {'count': 21613.0, 'mean': 1788.3906907879516, 'std': 828.0909776519169, 'min': 290.0, '25%': 1190.0, '50%': 1560.0, '75%': 2210.0, 'max': 9410.0}, 'sqft_basement': {'count': 21613.0, 'mean': 291.5090454818859, 'std': 442.5750426774682, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 560.0, 'max': 4820.0}, 'yr_built': {'count': 21613.0, 'mean': 1971.0051357978994, 'std': 29.37341080238659, 'min': 1900.0, '25%': 1951.0, '50%': 1975.0, '75%': 1997.0, 'max': 2015.0}, 'yr_renovated': {'count': 21613.0, 'mean': 84.40225790033776, 'std': 401.6792400191759, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 2015.0}, 'zipcode': {'count': 21613.0, 'mean': 98077.93980474715, 'std': 53.505026257473084, 'min': 98001.0, '25%': 98033.0, '50%': 98065.0, '75%': 98118.0, 'max': 98199.0}, 'lat': {'count': 21613.0, 'mean': 47.56005251931708, 'std': 0.13856371024192418, 'min': 47.1559, '25%': 47.471, '50%': 47.5718, '75%': 47.678, 'max': 47.7776}, 'long': {'count': 21613.0, 'mean': -122.21389640494147, 'std': 0.14082834238139408, 'min': -122.519, '25%': -122.328, '50%': -122.23, '75%': -122.125, 'max': -121.315}, 'sqft_living15': {'count': 21613.0, 'mean': 1986.552491556008, 'std': 685.3913042527776, 'min': 399.0, '25%': 1490.0, '50%': 1840.0, '75%': 2360.0, 'max': 6210.0}, 'sqft_lot15': {'count': 21613.0, 'mean': 12768.455651691113, 'std': 27304.17963133851, 'min': 651.0, '25%': 5100.0, '50%': 7620.0, '75%': 10083.0, 'max': 871200.0}}
<dataframe_info>
RangeIndex: 21613 entries, 0 to 21612
Data columns (total 21 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 id 21613 non-null int64
1 date 21613 non-null object
2 price 21613 non-null float64
3 bedrooms 21613 non-null int64
4 bathrooms 21613 non-null float64
5 sqft_living 21613 non-null int64
6 sqft_lot 21613 non-null int64
7 floors 21613 non-null float64
8 waterfront 21613 non-null int64
9 view 21613 non-null int64
10 condition 21613 non-null int64
11 grade 21613 non-null int64
12 sqft_above 21613 non-null int64
13 sqft_basement 21613 non-null int64
14 yr_built 21613 non-null int64
15 yr_renovated 21613 non-null int64
16 zipcode 21613 non-null int64
17 lat 21613 non-null float64
18 long 21613 non-null float64
19 sqft_living15 21613 non-null int64
20 sqft_lot15 21613 non-null int64
dtypes: float64(5), int64(15), object(1)
memory usage: 3.5+ MB
<some_examples>
{'id': {'0': 7129300520, '1': 6414100192, '2': 5631500400, '3': 2487200875}, 'date': {'0': '20141013T000000', '1': '20141209T000000', '2': '20150225T000000', '3': '20141209T000000'}, 'price': {'0': 221900.0, '1': 538000.0, '2': 180000.0, '3': 604000.0}, 'bedrooms': {'0': 3, '1': 3, '2': 2, '3': 4}, 'bathrooms': {'0': 1.0, '1': 2.25, '2': 1.0, '3': 3.0}, 'sqft_living': {'0': 1180, '1': 2570, '2': 770, '3': 1960}, 'sqft_lot': {'0': 5650, '1': 7242, '2': 10000, '3': 5000}, 'floors': {'0': 1.0, '1': 2.0, '2': 1.0, '3': 1.0}, 'waterfront': {'0': 0, '1': 0, '2': 0, '3': 0}, 'view': {'0': 0, '1': 0, '2': 0, '3': 0}, 'condition': {'0': 3, '1': 3, '2': 3, '3': 5}, 'grade': {'0': 7, '1': 7, '2': 6, '3': 7}, 'sqft_above': {'0': 1180, '1': 2170, '2': 770, '3': 1050}, 'sqft_basement': {'0': 0, '1': 400, '2': 0, '3': 910}, 'yr_built': {'0': 1955, '1': 1951, '2': 1933, '3': 1965}, 'yr_renovated': {'0': 0, '1': 1991, '2': 0, '3': 0}, 'zipcode': {'0': 98178, '1': 98125, '2': 98028, '3': 98136}, 'lat': {'0': 47.5112, '1': 47.721, '2': 47.7379, '3': 47.5208}, 'long': {'0': -122.257, '1': -122.319, '2': -122.233, '3': -122.393}, 'sqft_living15': {'0': 1340, '1': 1690, '2': 2720, '3': 1360}, 'sqft_lot15': {'0': 5650, '1': 7639, '2': 8062, '3': 5000}}
<end_description>
| 2,790 | 1 | 4,404 | 2,790 |
69046748
|
<jupyter_start><jupyter_text>CommonLit Various
Kaggle dataset identifier: commonlit-various
<jupyter_script>import warnings
warnings.simplefilter("ignore")
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import tensorflow as tf
import tensorflow_hub as hub
import tensorflow_addons as tfa
from tqdm.notebook import tqdm
from nltk.tokenize import word_tokenize, sent_tokenize
from sklearn.model_selection import KFold, StratifiedKFold
from tensorflow.keras.mixed_precision import experimental as mixed_precision
from kaggle_datasets import KaggleDatasets
from scipy.stats import pearsonr
from transformers import RobertaTokenizer, TFRobertaModel
from readability import Readability
from nltk.tokenize import word_tokenize, sent_tokenize
import os
import sys
import nltk
import string
import math
import logging
import glob
import random
tf.get_logger().setLevel(logging.ERROR)
tqdm.pandas()
print(f"tensorflow version: {tf.__version__}")
print(f"tensorflow keras version: {tf.keras.__version__}")
print(f"python version: P{sys.version}")
def set_seeds(seed):
os.environ["PYTHONHASHSEED"] = str(seed)
random.seed(seed)
tf.random.set_seed(seed)
np.random.seed(seed)
set_seeds(42)
SEQ_LENGTH = 250
# # Train
train = pd.read_csv("/kaggle/input/commonlitreadabilityprize/train.csv")
train_ratio_vectors = np.load("/kaggle/input/commonlit-various/train_ratio_vectors.npy")
sample_submission = pd.read_csv(
"/kaggle/input/commonlitreadabilityprize/sample_submission.csv"
)
RATIO_VECTOR_LENGTH = len(train_ratio_vectors[0])
print(
f"train_ratio_vectors shape: {train_ratio_vectors.shape}, RATIO_VECTOR_LENGTH: {RATIO_VECTOR_LENGTH}"
)
train["word_count"] = train["excerpt"].progress_apply(word_tokenize).apply(len)
train["sent_count"] = train["excerpt"].progress_apply(sent_tokenize).apply(len)
# # Info
display(train.info())
display(sample_submission.info())
# # Head
display(train.head())
display(sample_submission.head())
# # Target Distribution
plt.figure(figsize=(15, 8))
train["target"].plot(kind="hist", bins=32)
plt.title("Target Value Distribution", size=18)
plt.show()
display(train["target"].describe())
# # Excerpt Length
plt.figure(figsize=(15, 8))
train["word_count"].plot(kind="hist", bins=32)
plt.title("Word Count Distribution", size=18)
plt.show()
plt.figure(figsize=(15, 8))
train["sent_count"].plot(kind="hist", bins=32)
plt.title("Sentence Count Distribution", size=18)
plt.show()
# # Roberta Tokenize
# Get the trained model we want to use
MODEL = "roberta-base"
# Let's load our model tokenizer
tokenizer = RobertaTokenizer.from_pretrained(MODEL)
# For tf.dataset
AUTO = tf.data.experimental.AUTOTUNE
# This function tokenize the text according to a transformers model tokenizer
def regular_encode(excerpt):
enc_di = tokenizer.batch_encode_plus(
excerpt,
padding="max_length",
truncation=True,
max_length=SEQ_LENGTH,
)
return np.array(enc_di["input_ids"])
train["input_ids"] = regular_encode(train["excerpt"]).tolist()
display(train.head())
# # Training
# Detect hardware, return appropriate distribution strategy
try:
TPU = (
tf.distribute.cluster_resolver.TPUClusterResolver()
) # TPU detection. No parameters necessary if TPU_NAME environment variable is set. On Kaggle this is always the case.
print("Running on TPU ", TPU.master())
except ValueError:
print("Running on GPU")
TPU = None
if TPU:
tf.config.experimental_connect_to_cluster(TPU)
tf.tpu.experimental.initialize_tpu_system(TPU)
strategy = tf.distribute.experimental.TPUStrategy(TPU)
else:
strategy = (
tf.distribute.get_strategy()
) # default distribution strategy in Tensorflow. Works on CPU and single GPU.
REPLICAS = strategy.num_replicas_in_sync
print(f"REPLICAS: {REPLICAS}")
# set half precision policy
mixed_precision.set_policy("float32")
print(f"Compute dtype: {mixed_precision.global_policy().compute_dtype}")
print(f"Variable dtype: {mixed_precision.global_policy().variable_dtype}")
# # Model
def get_model(eps=1e-6, amsgrad=False, weights_path=None):
tf.keras.backend.clear_session()
with strategy.scope():
# Inputs
input_ids = tf.keras.Input(name="input_ids", shape=[SEQ_LENGTH], dtype=tf.int32)
ratio_vector = tf.keras.Input(
name="ratio_vector", shape=[RATIO_VECTOR_LENGTH], dtype=tf.float32
)
# ROBERTA
transformer = TFRobertaModel.from_pretrained(MODEL)
# Load saved weights
transformer.load_weights(
"/kaggle/input/simplenormal-wikipedia-sections/roberta_pretrained.h5"
)
transformer.trainable = True
# RoBERTa
sequence_output = transformer(input_ids)[0]
cls_token = sequence_output[:, 0, :]
# Ratio Vector
ratio_vector_fc = tf.keras.layers.Dense(256)(ratio_vector)
output_concat = tf.keras.layers.Concatenate(axis=1)(
[cls_token, ratio_vector_fc]
)
output = tf.keras.layers.Dense(1, activation="linear", dtype=tf.float32)(
output_concat
)
# Model
model = tf.keras.models.Model(
inputs=[input_ids, ratio_vector], outputs=[output]
)
loss = tf.keras.losses.MeanSquaredError()
optimizer = tf.optimizers.Adam(learning_rate=4e-5, epsilon=eps)
metrics = [
tf.keras.metrics.RootMeanSquaredError(name="RMSE"),
]
model.compile(optimizer=optimizer, loss=loss, metrics=metrics)
# Load weights if weights path is provided
if weights_path:
model.load_weights(weights_path)
return model
model = get_model()
model.summary()
tf.keras.utils.plot_model(
model, show_shapes=True, show_dtype=True, show_layer_names=True, expand_nested=False
)
# # Configuration
BATCH_SIZE_BASE = 24 // REPLICAS
BATCH_SIZE = BATCH_SIZE_BASE * REPLICAS
STEPS_PER_EPOCH = len(train) // BATCH_SIZE
KFOLDS = 5
print(f"BATCH SIZE: {BATCH_SIZE}")
def get_kfold_indices():
kf = KFold(n_splits=KFOLDS, shuffle=True, random_state=42)
kfold_indices = list(kf.split(train.index.tolist()))
return kfold_indices
KFOLD_INDICES = get_kfold_indices()
print(f"Train Size: {len(KFOLD_INDICES[0][0])}, Val Size: {len(KFOLD_INDICES[0][1])}")
# # Train Dataset
def get_train_dataset(kfold, drop_remainder=True):
train_idxs, _ = KFOLD_INDICES[kfold]
# TRAIN DATASET
input_ids = np.array(list(train.loc[train_idxs, "input_ids"]), dtype=np.int32)
ratio_vector = train_ratio_vectors[train_idxs]
train_x = {
"input_ids": input_ids,
"ratio_vector": ratio_vector,
}
train_y = train.loc[train_idxs, "target"]
train_dataset = tf.data.Dataset.from_tensor_slices((train_x, train_y))
if drop_remainder:
train_dataset = train_dataset.repeat()
train_dataset = train_dataset.shuffle(len(train_idxs))
train_dataset = train_dataset.batch(BATCH_SIZE, drop_remainder=drop_remainder)
train_dataset = train_dataset.prefetch(1)
if drop_remainder:
TRAIN_STEPS_PER_EPOCH = len(train_idxs) // BATCH_SIZE
else:
TRAIN_STEPS_PER_EPOCH = math.ceil(len(train_idxs) / BATCH_SIZE)
return train_dataset, TRAIN_STEPS_PER_EPOCH
train_dataset, TRAIN_STEPS_PER_EPOCH = get_train_dataset(0, drop_remainder=False)
train_x, train_y = next(iter(train_dataset))
print(f"train_x keys: {list(train_x.keys())}")
print(f"train_y shape: {train_y.shape}, train_y dtype {train_y.dtype}")
# # Val Dataset
def get_val_dataset(kfold, drop_remainder=True):
_, val_idxs = KFOLD_INDICES[kfold]
# VAL DATASET
input_ids = np.array(list(train.loc[val_idxs, "input_ids"]), dtype=np.int32)
ratio_vector = train_ratio_vectors[val_idxs]
val_x = {
"input_ids": input_ids,
"ratio_vector": ratio_vector,
}
val_y = train.loc[val_idxs, "target"]
val_dataset = tf.data.Dataset.from_tensor_slices((val_x, val_y))
val_dataset = val_dataset.batch(BATCH_SIZE, drop_remainder=False)
val_dataset = val_dataset.prefetch(1)
VAL_STEPS_PER_EPOCH = len(val_idxs) // BATCH_SIZE + 1
return val_dataset, VAL_STEPS_PER_EPOCH
val_dataset, VAL_STEPS_PER_EPOCH = get_val_dataset(0)
val_x, val_y = next(iter(val_dataset))
print(f"val_x keys: {list(val_x.keys())}")
print(f"val_y shape: {val_y.shape}, val_y dtypeL {val_y.dtype}")
# # Learning Rate Scheduler
TRAIN_LEN = len(KFOLD_INDICES[0][0])
TRAIN_ROUNDS = 4
STEPS_PER_EPOCH = TRAIN_LEN // (BATCH_SIZE * 16)
EPOCHS = (TRAIN_ROUNDS * TRAIN_LEN) // (STEPS_PER_EPOCH * BATCH_SIZE)
LR_RAMPUP_ITERATIONS = 0
LR_RAMPUP_EPOCHS = int(
LR_RAMPUP_ITERATIONS * (len(KFOLD_INDICES[0][0]) / (BATCH_SIZE * STEPS_PER_EPOCH))
)
print(
f"EPOCHS: {EPOCHS}, STEPS_PER_EPOCH: {STEPS_PER_EPOCH}, LR_RAMPUP_EPOCHS: {LR_RAMPUP_EPOCHS}"
)
# # Training
print("=" * 20, f"start", "=" * 20)
print()
# Histories
HISTORIES = dict()
# Epsilon grid search
for fold in range(KFOLDS):
# Model Checkpoint
checkpoint_callback = tf.keras.callbacks.ModelCheckpoint(
f"model_fold_{fold}.h5",
monitor="val_RMSE",
save_best_only=True,
save_weights_only=True,
verbose=1,
mode="min",
)
print("=" * 10, f"FOLD {fold}", "=" * 10)
# Models
model = get_model()
# Datasets
train_dataset, TRAIN_STEPS_PER_EPOCH = get_train_dataset(fold)
val_dataset, VAL_STEPS_PER_EPOCH = get_val_dataset(fold)
print(
f"TRAIN_STEPS_PER_EPOCH: {TRAIN_STEPS_PER_EPOCH}, VAL_STEPS_PER_EPOCH: {VAL_STEPS_PER_EPOCH}"
)
# Train Model
HISTORIES[f"FOLD_{fold}"] = model.fit(
train_dataset,
epochs=EPOCHS,
verbose=0,
# Hardcode Steps Per Epoch
steps_per_epoch=STEPS_PER_EPOCH,
# validation
validation_data=val_dataset,
validation_steps=VAL_STEPS_PER_EPOCH,
# callbacks
callbacks=[
checkpoint_callback,
],
)
# OOF RMSE
OOF_RMSE = []
for fold in range(KFOLDS):
OOF_RMSE.append(min(HISTORIES[f"FOLD_{fold}"].history["val_RMSE"]))
print()
print(", ".join([f"fold {i}: {rmse:.4f}" for i, rmse in zip(range(KFOLDS), OOF_RMSE)]))
print(f"OOF_RMSE: {np.mean(OOF_RMSE):.4f}")
print()
# # Train History
def plot_history_metric(history, metric, axes, fold):
N_EPOCHS = len(history.history["loss"])
x = [1, 5] + [10 + 5 * idx for idx in range((N_EPOCHS - 10) // 5 + 1)]
x_ticks = np.arange(1, N_EPOCHS + 1)
val = "val" in "".join(history.history.keys())
# summarize history for accuracy
axes.plot(x_ticks, history.history[metric])
if val:
val_values = history.history[f"val_{metric}"]
val_argmin = np.argmin(val_values)
axes.scatter(
val_argmin + 1, val_values[val_argmin], color="red", s=50, marker="o"
)
axes.plot(x_ticks, val_values)
axes.set_title(f"Fold {fold} - Model {metric}", fontsize=20)
axes.set_ylabel(metric, fontsize=16)
axes.set_xlabel("epoch", fontsize=16)
axes.tick_params(axis="x", labelsize=8)
axes.set_xticks(x) # set tick step to 1 and let x axis start at 1
axes.legend(["train"] + ["test"] if val else [], prop={"size": 18})
axes.grid()
fig, axes = plt.subplots(KFOLDS, 2, figsize=(15, 6 * KFOLDS))
for fold in range(KFOLDS):
history = HISTORIES[f"FOLD_{fold}"]
plot_history_metric(history, "loss", axes[fold, 0], fold)
plot_history_metric(history, "RMSE", axes[fold, 1], fold)
plt.subplots_adjust(hspace=0.40, wspace=0.20)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/046/69046748.ipynb
|
commonlit-various
|
markwijkhuizen
|
[{"Id": 69046748, "ScriptId": 17247939, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4433335, "CreationDate": "07/26/2021 08:44:57", "VersionNumber": 58.0, "Title": "CommonLit Training", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 368.0, "LinesInsertedFromPrevious": 2.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 366.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 91776584, "KernelVersionId": 69046748, "SourceDatasetVersionId": 2462427}, {"Id": 91776583, "KernelVersionId": 69046748, "SourceDatasetVersionId": 2462365}, {"Id": 91776582, "KernelVersionId": 69046748, "SourceDatasetVersionId": 2452800}, {"Id": 91776580, "KernelVersionId": 69046748, "SourceDatasetVersionId": 2219267}, {"Id": 91776579, "KernelVersionId": 69046748, "SourceDatasetVersionId": 2210416}, {"Id": 91776578, "KernelVersionId": 69046748, "SourceDatasetVersionId": 2199419}]
|
[{"Id": 2462427, "DatasetId": 1390791, "DatasourceVersionId": 2504854, "CreatorUserId": 4433335, "LicenseName": "Unknown", "CreationDate": "07/25/2021 18:42:55", "VersionNumber": 9.0, "Title": "CommonLit Various", "Slug": "commonlit-various", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Added all ratio's", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1390791, "CreatorUserId": 4433335, "OwnerUserId": 4433335.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 3028066.0, "CurrentDatasourceVersionId": 3076077.0, "ForumId": 1410023, "Type": 2, "CreationDate": "06/05/2021 15:44:48", "LastActivityDate": "06/05/2021", "TotalViews": 1826, "TotalDownloads": 2, "TotalVotes": 2, "TotalKernels": 1}]
|
[{"Id": 4433335, "UserName": "markwijkhuizen", "DisplayName": "Mark Wijkhuizen", "RegisterDate": "02/04/2020", "PerformanceTier": 3}]
|
import warnings
warnings.simplefilter("ignore")
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import tensorflow as tf
import tensorflow_hub as hub
import tensorflow_addons as tfa
from tqdm.notebook import tqdm
from nltk.tokenize import word_tokenize, sent_tokenize
from sklearn.model_selection import KFold, StratifiedKFold
from tensorflow.keras.mixed_precision import experimental as mixed_precision
from kaggle_datasets import KaggleDatasets
from scipy.stats import pearsonr
from transformers import RobertaTokenizer, TFRobertaModel
from readability import Readability
from nltk.tokenize import word_tokenize, sent_tokenize
import os
import sys
import nltk
import string
import math
import logging
import glob
import random
tf.get_logger().setLevel(logging.ERROR)
tqdm.pandas()
print(f"tensorflow version: {tf.__version__}")
print(f"tensorflow keras version: {tf.keras.__version__}")
print(f"python version: P{sys.version}")
def set_seeds(seed):
os.environ["PYTHONHASHSEED"] = str(seed)
random.seed(seed)
tf.random.set_seed(seed)
np.random.seed(seed)
set_seeds(42)
SEQ_LENGTH = 250
# # Train
train = pd.read_csv("/kaggle/input/commonlitreadabilityprize/train.csv")
train_ratio_vectors = np.load("/kaggle/input/commonlit-various/train_ratio_vectors.npy")
sample_submission = pd.read_csv(
"/kaggle/input/commonlitreadabilityprize/sample_submission.csv"
)
RATIO_VECTOR_LENGTH = len(train_ratio_vectors[0])
print(
f"train_ratio_vectors shape: {train_ratio_vectors.shape}, RATIO_VECTOR_LENGTH: {RATIO_VECTOR_LENGTH}"
)
train["word_count"] = train["excerpt"].progress_apply(word_tokenize).apply(len)
train["sent_count"] = train["excerpt"].progress_apply(sent_tokenize).apply(len)
# # Info
display(train.info())
display(sample_submission.info())
# # Head
display(train.head())
display(sample_submission.head())
# # Target Distribution
plt.figure(figsize=(15, 8))
train["target"].plot(kind="hist", bins=32)
plt.title("Target Value Distribution", size=18)
plt.show()
display(train["target"].describe())
# # Excerpt Length
plt.figure(figsize=(15, 8))
train["word_count"].plot(kind="hist", bins=32)
plt.title("Word Count Distribution", size=18)
plt.show()
plt.figure(figsize=(15, 8))
train["sent_count"].plot(kind="hist", bins=32)
plt.title("Sentence Count Distribution", size=18)
plt.show()
# # Roberta Tokenize
# Get the trained model we want to use
MODEL = "roberta-base"
# Let's load our model tokenizer
tokenizer = RobertaTokenizer.from_pretrained(MODEL)
# For tf.dataset
AUTO = tf.data.experimental.AUTOTUNE
# This function tokenize the text according to a transformers model tokenizer
def regular_encode(excerpt):
enc_di = tokenizer.batch_encode_plus(
excerpt,
padding="max_length",
truncation=True,
max_length=SEQ_LENGTH,
)
return np.array(enc_di["input_ids"])
train["input_ids"] = regular_encode(train["excerpt"]).tolist()
display(train.head())
# # Training
# Detect hardware, return appropriate distribution strategy
try:
TPU = (
tf.distribute.cluster_resolver.TPUClusterResolver()
) # TPU detection. No parameters necessary if TPU_NAME environment variable is set. On Kaggle this is always the case.
print("Running on TPU ", TPU.master())
except ValueError:
print("Running on GPU")
TPU = None
if TPU:
tf.config.experimental_connect_to_cluster(TPU)
tf.tpu.experimental.initialize_tpu_system(TPU)
strategy = tf.distribute.experimental.TPUStrategy(TPU)
else:
strategy = (
tf.distribute.get_strategy()
) # default distribution strategy in Tensorflow. Works on CPU and single GPU.
REPLICAS = strategy.num_replicas_in_sync
print(f"REPLICAS: {REPLICAS}")
# set half precision policy
mixed_precision.set_policy("float32")
print(f"Compute dtype: {mixed_precision.global_policy().compute_dtype}")
print(f"Variable dtype: {mixed_precision.global_policy().variable_dtype}")
# # Model
def get_model(eps=1e-6, amsgrad=False, weights_path=None):
tf.keras.backend.clear_session()
with strategy.scope():
# Inputs
input_ids = tf.keras.Input(name="input_ids", shape=[SEQ_LENGTH], dtype=tf.int32)
ratio_vector = tf.keras.Input(
name="ratio_vector", shape=[RATIO_VECTOR_LENGTH], dtype=tf.float32
)
# ROBERTA
transformer = TFRobertaModel.from_pretrained(MODEL)
# Load saved weights
transformer.load_weights(
"/kaggle/input/simplenormal-wikipedia-sections/roberta_pretrained.h5"
)
transformer.trainable = True
# RoBERTa
sequence_output = transformer(input_ids)[0]
cls_token = sequence_output[:, 0, :]
# Ratio Vector
ratio_vector_fc = tf.keras.layers.Dense(256)(ratio_vector)
output_concat = tf.keras.layers.Concatenate(axis=1)(
[cls_token, ratio_vector_fc]
)
output = tf.keras.layers.Dense(1, activation="linear", dtype=tf.float32)(
output_concat
)
# Model
model = tf.keras.models.Model(
inputs=[input_ids, ratio_vector], outputs=[output]
)
loss = tf.keras.losses.MeanSquaredError()
optimizer = tf.optimizers.Adam(learning_rate=4e-5, epsilon=eps)
metrics = [
tf.keras.metrics.RootMeanSquaredError(name="RMSE"),
]
model.compile(optimizer=optimizer, loss=loss, metrics=metrics)
# Load weights if weights path is provided
if weights_path:
model.load_weights(weights_path)
return model
model = get_model()
model.summary()
tf.keras.utils.plot_model(
model, show_shapes=True, show_dtype=True, show_layer_names=True, expand_nested=False
)
# # Configuration
BATCH_SIZE_BASE = 24 // REPLICAS
BATCH_SIZE = BATCH_SIZE_BASE * REPLICAS
STEPS_PER_EPOCH = len(train) // BATCH_SIZE
KFOLDS = 5
print(f"BATCH SIZE: {BATCH_SIZE}")
def get_kfold_indices():
kf = KFold(n_splits=KFOLDS, shuffle=True, random_state=42)
kfold_indices = list(kf.split(train.index.tolist()))
return kfold_indices
KFOLD_INDICES = get_kfold_indices()
print(f"Train Size: {len(KFOLD_INDICES[0][0])}, Val Size: {len(KFOLD_INDICES[0][1])}")
# # Train Dataset
def get_train_dataset(kfold, drop_remainder=True):
train_idxs, _ = KFOLD_INDICES[kfold]
# TRAIN DATASET
input_ids = np.array(list(train.loc[train_idxs, "input_ids"]), dtype=np.int32)
ratio_vector = train_ratio_vectors[train_idxs]
train_x = {
"input_ids": input_ids,
"ratio_vector": ratio_vector,
}
train_y = train.loc[train_idxs, "target"]
train_dataset = tf.data.Dataset.from_tensor_slices((train_x, train_y))
if drop_remainder:
train_dataset = train_dataset.repeat()
train_dataset = train_dataset.shuffle(len(train_idxs))
train_dataset = train_dataset.batch(BATCH_SIZE, drop_remainder=drop_remainder)
train_dataset = train_dataset.prefetch(1)
if drop_remainder:
TRAIN_STEPS_PER_EPOCH = len(train_idxs) // BATCH_SIZE
else:
TRAIN_STEPS_PER_EPOCH = math.ceil(len(train_idxs) / BATCH_SIZE)
return train_dataset, TRAIN_STEPS_PER_EPOCH
train_dataset, TRAIN_STEPS_PER_EPOCH = get_train_dataset(0, drop_remainder=False)
train_x, train_y = next(iter(train_dataset))
print(f"train_x keys: {list(train_x.keys())}")
print(f"train_y shape: {train_y.shape}, train_y dtype {train_y.dtype}")
# # Val Dataset
def get_val_dataset(kfold, drop_remainder=True):
_, val_idxs = KFOLD_INDICES[kfold]
# VAL DATASET
input_ids = np.array(list(train.loc[val_idxs, "input_ids"]), dtype=np.int32)
ratio_vector = train_ratio_vectors[val_idxs]
val_x = {
"input_ids": input_ids,
"ratio_vector": ratio_vector,
}
val_y = train.loc[val_idxs, "target"]
val_dataset = tf.data.Dataset.from_tensor_slices((val_x, val_y))
val_dataset = val_dataset.batch(BATCH_SIZE, drop_remainder=False)
val_dataset = val_dataset.prefetch(1)
VAL_STEPS_PER_EPOCH = len(val_idxs) // BATCH_SIZE + 1
return val_dataset, VAL_STEPS_PER_EPOCH
val_dataset, VAL_STEPS_PER_EPOCH = get_val_dataset(0)
val_x, val_y = next(iter(val_dataset))
print(f"val_x keys: {list(val_x.keys())}")
print(f"val_y shape: {val_y.shape}, val_y dtypeL {val_y.dtype}")
# # Learning Rate Scheduler
TRAIN_LEN = len(KFOLD_INDICES[0][0])
TRAIN_ROUNDS = 4
STEPS_PER_EPOCH = TRAIN_LEN // (BATCH_SIZE * 16)
EPOCHS = (TRAIN_ROUNDS * TRAIN_LEN) // (STEPS_PER_EPOCH * BATCH_SIZE)
LR_RAMPUP_ITERATIONS = 0
LR_RAMPUP_EPOCHS = int(
LR_RAMPUP_ITERATIONS * (len(KFOLD_INDICES[0][0]) / (BATCH_SIZE * STEPS_PER_EPOCH))
)
print(
f"EPOCHS: {EPOCHS}, STEPS_PER_EPOCH: {STEPS_PER_EPOCH}, LR_RAMPUP_EPOCHS: {LR_RAMPUP_EPOCHS}"
)
# # Training
print("=" * 20, f"start", "=" * 20)
print()
# Histories
HISTORIES = dict()
# Epsilon grid search
for fold in range(KFOLDS):
# Model Checkpoint
checkpoint_callback = tf.keras.callbacks.ModelCheckpoint(
f"model_fold_{fold}.h5",
monitor="val_RMSE",
save_best_only=True,
save_weights_only=True,
verbose=1,
mode="min",
)
print("=" * 10, f"FOLD {fold}", "=" * 10)
# Models
model = get_model()
# Datasets
train_dataset, TRAIN_STEPS_PER_EPOCH = get_train_dataset(fold)
val_dataset, VAL_STEPS_PER_EPOCH = get_val_dataset(fold)
print(
f"TRAIN_STEPS_PER_EPOCH: {TRAIN_STEPS_PER_EPOCH}, VAL_STEPS_PER_EPOCH: {VAL_STEPS_PER_EPOCH}"
)
# Train Model
HISTORIES[f"FOLD_{fold}"] = model.fit(
train_dataset,
epochs=EPOCHS,
verbose=0,
# Hardcode Steps Per Epoch
steps_per_epoch=STEPS_PER_EPOCH,
# validation
validation_data=val_dataset,
validation_steps=VAL_STEPS_PER_EPOCH,
# callbacks
callbacks=[
checkpoint_callback,
],
)
# OOF RMSE
OOF_RMSE = []
for fold in range(KFOLDS):
OOF_RMSE.append(min(HISTORIES[f"FOLD_{fold}"].history["val_RMSE"]))
print()
print(", ".join([f"fold {i}: {rmse:.4f}" for i, rmse in zip(range(KFOLDS), OOF_RMSE)]))
print(f"OOF_RMSE: {np.mean(OOF_RMSE):.4f}")
print()
# # Train History
def plot_history_metric(history, metric, axes, fold):
N_EPOCHS = len(history.history["loss"])
x = [1, 5] + [10 + 5 * idx for idx in range((N_EPOCHS - 10) // 5 + 1)]
x_ticks = np.arange(1, N_EPOCHS + 1)
val = "val" in "".join(history.history.keys())
# summarize history for accuracy
axes.plot(x_ticks, history.history[metric])
if val:
val_values = history.history[f"val_{metric}"]
val_argmin = np.argmin(val_values)
axes.scatter(
val_argmin + 1, val_values[val_argmin], color="red", s=50, marker="o"
)
axes.plot(x_ticks, val_values)
axes.set_title(f"Fold {fold} - Model {metric}", fontsize=20)
axes.set_ylabel(metric, fontsize=16)
axes.set_xlabel("epoch", fontsize=16)
axes.tick_params(axis="x", labelsize=8)
axes.set_xticks(x) # set tick step to 1 and let x axis start at 1
axes.legend(["train"] + ["test"] if val else [], prop={"size": 18})
axes.grid()
fig, axes = plt.subplots(KFOLDS, 2, figsize=(15, 6 * KFOLDS))
for fold in range(KFOLDS):
history = HISTORIES[f"FOLD_{fold}"]
plot_history_metric(history, "loss", axes[fold, 0], fold)
plot_history_metric(history, "RMSE", axes[fold, 1], fold)
plt.subplots_adjust(hspace=0.40, wspace=0.20)
| false | 2 | 3,659 | 0 | 3,682 | 3,659 |
||
69046772
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
stock_1 = pd.read_csv(
"/kaggle/input/show-your-data-skills-snu21/stock.csv"
) # recheck the filepath from the above code cell output
# # Clean and create the resampled spread below. Best of Luck! :)
stock_1.head()
stock_1.shape
stock_1.info()
df = stock_1.copy()
df.head()
df = df[df["Volume"] >= 5]
df.shape
df["Timestamp"] = pd.to_datetime(df["Timestamp"])
df2 = df[(df["Timestamp"].dt.hour <= 22) & (df["Timestamp"].dt.hour >= 10)]
del df2["Hour"]
df2.head(15)
df2.shape
q = df.set_index("Timestamp")
q.head(10)
dict = {"Open": "first", "High": "max", "Low": "min", "Close": "last", "Volume": "sum"}
q = q.resample("24H", closed="left", label="left").apply(dict)
q.head(24)
q.shape
resampled_df = q.dropna()
resampled_df.shape
# # Before submitting ensure that you have 5 columns ( Open, High, low, Close, Volume) in case you have the Timestamp column set as index OR 6 columns (Timestamp, Open, High, low, Close, Volume) in case you have index as 0,1,2,3,...
resampled_df.shape
# resampled_df.to_csv('submission.csv', index=False) # use index=False if you have 6 columns as specified above
resampled_df.to_csv(
"submission.csv", index=True
) # use index=True if you have 5 columns
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/046/69046772.ipynb
| null | null |
[{"Id": 69046772, "ScriptId": 18842152, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7984187, "CreationDate": "07/26/2021 08:45:19", "VersionNumber": 2.0, "Title": "Sample_Notebook", "EvaluationDate": "07/26/2021", "IsChange": false, "TotalLines": 78.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 78.0, "LinesInsertedFromFork": 47.0, "LinesDeletedFromFork": 1.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 31.0, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
stock_1 = pd.read_csv(
"/kaggle/input/show-your-data-skills-snu21/stock.csv"
) # recheck the filepath from the above code cell output
# # Clean and create the resampled spread below. Best of Luck! :)
stock_1.head()
stock_1.shape
stock_1.info()
df = stock_1.copy()
df.head()
df = df[df["Volume"] >= 5]
df.shape
df["Timestamp"] = pd.to_datetime(df["Timestamp"])
df2 = df[(df["Timestamp"].dt.hour <= 22) & (df["Timestamp"].dt.hour >= 10)]
del df2["Hour"]
df2.head(15)
df2.shape
q = df.set_index("Timestamp")
q.head(10)
dict = {"Open": "first", "High": "max", "Low": "min", "Close": "last", "Volume": "sum"}
q = q.resample("24H", closed="left", label="left").apply(dict)
q.head(24)
q.shape
resampled_df = q.dropna()
resampled_df.shape
# # Before submitting ensure that you have 5 columns ( Open, High, low, Close, Volume) in case you have the Timestamp column set as index OR 6 columns (Timestamp, Open, High, low, Close, Volume) in case you have index as 0,1,2,3,...
resampled_df.shape
# resampled_df.to_csv('submission.csv', index=False) # use index=False if you have 6 columns as specified above
resampled_df.to_csv(
"submission.csv", index=True
) # use index=True if you have 5 columns
| false | 0 | 613 | 0 | 613 | 613 |
||
69046835
|
<jupyter_start><jupyter_text>Shark attack dataset
# Global Shark Attack
Because they provide a glimpse - a window - into the world of sharks and their behaviors. By understanding when and why shark attacks occur, it is possible to lessen the likelihood of these incidents. Humans are familiar with predators found on land; we know enough not to walk into a pride of lions and we don't try to pet a growling dog that is baring its teeth. Similarly, we need to recognize and avoid potentially dangerous situations in the water. The individual case histories provide insights about specific geographical areas and their indigenous species of sharks. However, when all known case histories worldwide are examined, much is revealed about species behavior, and specific patterns emerge.
Most of the incidents in the Global Shark Attack File have nothing to do with predation on humans. Some accidents are motivated by a displacement or territorial behavior when a shark feels threatened; still others are the result of the shark responding to sensory predatory input (i.e., overwhelmed by the presence of many fishes) and environmental conditions (murky water), which may cause them to respond in a reflexive response to stimuli. Sharks also exhibit curiosity and may investigate unknown or unfamiliar objects; they learn by exploring their environment, and - lacking hands - they use their mouths and teeth to examine unfamiliar objects.
A very small percentage of shark species, about two dozen, are considered potentially dangerous to humans because of their size and dentition. Yet each year, for every human killed by a shark, our species slaughters millions of sharks - about 73 million sharks last year. We are stripping the world's oceans of one of its most valuable predators - animals that play a critical role in maintaining the health of the world's oceans. An unreasonable fear of sharks has been implanted in our minds by the hype that surrounds the rare shark attack and by movies that exploit our primal fears. It is the mission of the Global Shark Attack File to present facts about these events, thus enabling them to be put in perspective. Sharks are vital to the ocean ecosystem. Without sharks our planet's ocean would soon become a watery graveyard. This is not the legacy the Global Shark Accident File and the Shark Research Institute wishes to leave our children and our children's children.
The Global Shark Attack File was created to provide medical personnel, shark behaviorists, lifesavers, and the media with meaningful information resulting from the scientific forensic examination of shark accidents. Whenever possible, our investigators conduct personal interviews with victims and witnesses, medical personnel and other professionals, and conduct examinations of the incident site. Weather and sea conditions and environmental data are evaluated in an attempt to identify factors that contributed to the incident.
Early on, we became aware that the word "attack" was usually a misnomer. An "attack" by a shark is an extremely rare event, even less likely than statistics suggest. When a shark bites a surfboard, leaving the surfer unharmed, it was historically recorded as an "attack". Collisions between humans and sharks in low visibility water were also recorded as "attacks".
When a shark grabs a person by the hand/wrist and tows them along the surface, tosses a surfboard (or a Frisbee as in case 1968.08.24) it is probably "play behavior", not aggression. How can case GSAF 1971.04.11 which the swimmer was repeatedly bitten by a large shark and case 1985.01.04 in which the diver's injury necessitated a Band-aid be compared? It is akin to comparing a head-on high-speed vehicular collision with a shopping cart ding on the door of a parked car. Global Shark Attack File believes the only way to sort fact from hype is by forensic examination of each incident.
Although incidents that occur in remote areas may go unrecorded, the Global Shark Attack File is a compilation of a number of data sources, and we have a team of qualified researchers throughout the world that actively investigate these incidents. One of our objectives is to provide a clear picture of the actual threat presented by sharks to humans. In this regard, we remind our visitors that more people drown in a single year in the United States than have been killed by sharks throughout the entire world in the last two centuries.
Copyright © 2005, Shark Research Institute, Inc. All rights reserved
Kaggle dataset identifier: shark-attack-dataset
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
shark = pd.read_csv("../input/shark-attack-dataset/attacks.csv")
shark.head()
shark.columns
shark = pd.DataFrame(shark)
shark["Sex "].value_counts()
sharka = shark.groupby(["Sex ", "Fatal (Y/N)"], as_index=False).size()
sharka = sharka.sort_values(by=["size"], ascending=False)
sharka = sharka[0:7]
sharka.drop([5], inplace=True)
sharka
import matplotlib.pyplot as plt
mlabels = [
"Male Fatal",
"Male Non Fatal",
"Female Non Fatal",
"Female Fatal",
"Male Unknown",
"Female Unknown",
]
plt.pie(sharka["size"], labels=mlabels, autopct="%1.1f%%")
plt.title("comparison of fatal/non fatal accidents among women and men")
fig = plt.gcf()
fig.set_size_inches(12, 12)
plt.show()
shark.dropna(subset=["Activity"], inplace=True)
from wordcloud import WordCloud
words = shark["Activity"].tolist()
words = "".join(str(words))
plt.figure(figsize=(12, 12))
plt.imshow(WordCloud().generate(words))
sharkb = shark.groupby(["Country"], as_index=False).size()
sharkb = sharkb.sort_values(by=["size"], ascending=False)
sharkb
import plotly.express as px
px.choropleth(
sharkb,
locations="Country",
color="size",
color_continuous_scale="Turbo",
locationmode="country names",
scope="world",
range_color=(0, 2000),
title="",
height=600,
)
import seaborn as sns
sharkb = sharkb[0:5]
plt.figure(figsize=(18, 10))
plt.title("top 5 countries with the most shark attacks")
sns.barplot(x="Country", y="size", data=sharkb)
sharkc = shark.groupby(["Species "], as_index=False).size()
sharkc = sharkc.sort_values(by=["size"], ascending=False)
sharkc.drop(
[783, 1033, 1045, 1044, 409, 480, 152, 109, 941, 943, 87, 350, 454, 411, 231, 324],
inplace=True,
)
sharkc = sharkc[0:4]
sharkc
mlabels = ["White shark", "Tiger shark", "Bull shark", "Wobbegong shark"]
plt.pie(sharkc["size"], labels=mlabels, autopct="%1.1f%%")
plt.title("percentage share between the 4 most dangerous sharks")
fig = plt.gcf()
fig.set_size_inches(12, 12)
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/046/69046835.ipynb
|
shark-attack-dataset
|
felipeesc
|
[{"Id": 69046835, "ScriptId": 18842772, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7206642, "CreationDate": "07/26/2021 08:46:07", "VersionNumber": 4.0, "Title": "Most dangeurous sharks,countries+Worldmap+analysis", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 85.0, "LinesInsertedFromPrevious": 20.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 65.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 91776789, "KernelVersionId": 69046835, "SourceDatasetVersionId": 2462873}]
|
[{"Id": 2462873, "DatasetId": 1490782, "DatasourceVersionId": 2505301, "CreatorUserId": 7756990, "LicenseName": "Other (specified in description)", "CreationDate": "07/25/2021 23:04:38", "VersionNumber": 1.0, "Title": "Shark attack dataset", "Slug": "shark-attack-dataset", "Subtitle": "Global Shark attack - https://www.sharkattackfile.net/index.htm", "Description": "# Global Shark Attack\nBecause they provide a glimpse - a window - into the world of sharks and their behaviors. By understanding when and why shark attacks occur, it is possible to lessen the likelihood of these incidents. Humans are familiar with predators found on land; we know enough not to walk into a pride of lions and we don't try to pet a growling dog that is baring its teeth. Similarly, we need to recognize and avoid potentially dangerous situations in the water. The individual case histories provide insights about specific geographical areas and their indigenous species of sharks. However, when all known case histories worldwide are examined, much is revealed about species behavior, and specific patterns emerge.\n\nMost of the incidents in the Global Shark Attack File have nothing to do with predation on humans. Some accidents are motivated by a displacement or territorial behavior when a shark feels threatened; still others are the result of the shark responding to sensory predatory input (i.e., overwhelmed by the presence of many fishes) and environmental conditions (murky water), which may cause them to respond in a reflexive response to stimuli. Sharks also exhibit curiosity and may investigate unknown or unfamiliar objects; they learn by exploring their environment, and - lacking hands - they use their mouths and teeth to examine unfamiliar objects.\n\nA very small percentage of shark species, about two dozen, are considered potentially dangerous to humans because of their size and dentition. Yet each year, for every human killed by a shark, our species slaughters millions of sharks - about 73 million sharks last year. We are stripping the world's oceans of one of its most valuable predators - animals that play a critical role in maintaining the health of the world's oceans. An unreasonable fear of sharks has been implanted in our minds by the hype that surrounds the rare shark attack and by movies that exploit our primal fears. It is the mission of the Global Shark Attack File to present facts about these events, thus enabling them to be put in perspective. Sharks are vital to the ocean ecosystem. Without sharks our planet's ocean would soon become a watery graveyard. This is not the legacy the Global Shark Accident File and the Shark Research Institute wishes to leave our children and our children's children.\n\nThe Global Shark Attack File was created to provide medical personnel, shark behaviorists, lifesavers, and the media with meaningful information resulting from the scientific forensic examination of shark accidents. Whenever possible, our investigators conduct personal interviews with victims and witnesses, medical personnel and other professionals, and conduct examinations of the incident site. Weather and sea conditions and environmental data are evaluated in an attempt to identify factors that contributed to the incident.\n\nEarly on, we became aware that the word \"attack\" was usually a misnomer. An \"attack\" by a shark is an extremely rare event, even less likely than statistics suggest. When a shark bites a surfboard, leaving the surfer unharmed, it was historically recorded as an \"attack\". Collisions between humans and sharks in low visibility water were also recorded as \"attacks\".\n\nWhen a shark grabs a person by the hand/wrist and tows them along the surface, tosses a surfboard (or a Frisbee as in case 1968.08.24) it is probably \"play behavior\", not aggression. How can case GSAF 1971.04.11 which the swimmer was repeatedly bitten by a large shark and case 1985.01.04 in which the diver's injury necessitated a Band-aid be compared? It is akin to comparing a head-on high-speed vehicular collision with a shopping cart ding on the door of a parked car. Global Shark Attack File believes the only way to sort fact from hype is by forensic examination of each incident.\n\nAlthough incidents that occur in remote areas may go unrecorded, the Global Shark Attack File is a compilation of a number of data sources, and we have a team of qualified researchers throughout the world that actively investigate these incidents. One of our objectives is to provide a clear picture of the actual threat presented by sharks to humans. In this regard, we remind our visitors that more people drown in a single year in the United States than have been killed by sharks throughout the entire world in the last two centuries.\n\n\n\nCopyright \u00a9 2005, Shark Research Institute, Inc. All rights reserved", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1490782, "CreatorUserId": 7756990, "OwnerUserId": 7756990.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2462873.0, "CurrentDatasourceVersionId": 2505301.0, "ForumId": 1510486, "Type": 2, "CreationDate": "07/25/2021 23:04:38", "LastActivityDate": "07/25/2021", "TotalViews": 16257, "TotalDownloads": 2297, "TotalVotes": 46, "TotalKernels": 7}]
|
[{"Id": 7756990, "UserName": "felipeesc", "DisplayName": "Felipe_Esc", "RegisterDate": "06/24/2021", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
shark = pd.read_csv("../input/shark-attack-dataset/attacks.csv")
shark.head()
shark.columns
shark = pd.DataFrame(shark)
shark["Sex "].value_counts()
sharka = shark.groupby(["Sex ", "Fatal (Y/N)"], as_index=False).size()
sharka = sharka.sort_values(by=["size"], ascending=False)
sharka = sharka[0:7]
sharka.drop([5], inplace=True)
sharka
import matplotlib.pyplot as plt
mlabels = [
"Male Fatal",
"Male Non Fatal",
"Female Non Fatal",
"Female Fatal",
"Male Unknown",
"Female Unknown",
]
plt.pie(sharka["size"], labels=mlabels, autopct="%1.1f%%")
plt.title("comparison of fatal/non fatal accidents among women and men")
fig = plt.gcf()
fig.set_size_inches(12, 12)
plt.show()
shark.dropna(subset=["Activity"], inplace=True)
from wordcloud import WordCloud
words = shark["Activity"].tolist()
words = "".join(str(words))
plt.figure(figsize=(12, 12))
plt.imshow(WordCloud().generate(words))
sharkb = shark.groupby(["Country"], as_index=False).size()
sharkb = sharkb.sort_values(by=["size"], ascending=False)
sharkb
import plotly.express as px
px.choropleth(
sharkb,
locations="Country",
color="size",
color_continuous_scale="Turbo",
locationmode="country names",
scope="world",
range_color=(0, 2000),
title="",
height=600,
)
import seaborn as sns
sharkb = sharkb[0:5]
plt.figure(figsize=(18, 10))
plt.title("top 5 countries with the most shark attacks")
sns.barplot(x="Country", y="size", data=sharkb)
sharkc = shark.groupby(["Species "], as_index=False).size()
sharkc = sharkc.sort_values(by=["size"], ascending=False)
sharkc.drop(
[783, 1033, 1045, 1044, 409, 480, 152, 109, 941, 943, 87, 350, 454, 411, 231, 324],
inplace=True,
)
sharkc = sharkc[0:4]
sharkc
mlabels = ["White shark", "Tiger shark", "Bull shark", "Wobbegong shark"]
plt.pie(sharkc["size"], labels=mlabels, autopct="%1.1f%%")
plt.title("percentage share between the 4 most dangerous sharks")
fig = plt.gcf()
fig.set_size_inches(12, 12)
plt.show()
| false | 1 | 928 | 0 | 2,026 | 928 |
||
69046687
|
<jupyter_start><jupyter_text>quora-question-pair-hand-annotated-dataset
Kaggle dataset identifier: quoraquestionpairhandannotateddataset
<jupyter_code>import pandas as pd
df = pd.read_csv('quoraquestionpairhandannotateddataset/df_handeval.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 1446 entries, 0 to 1445
Data columns (total 8 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 test_qid 1446 non-null int64
1 test_question 1446 non-null object
2 candidate_qid 1446 non-null int64
3 candidate_question 1446 non-null object
4 jh 820 non-null float64
5 hk 904 non-null float64
6 wt 881 non-null float64
7 average 1446 non-null float64
dtypes: float64(4), int64(2), object(2)
memory usage: 90.5+ KB
<jupyter_text>Examples:
{
"test_qid": 331,
"test_question": "Which is the Best earphones under Rs 1000?",
"candidate_qid": 8504,
"candidate_question": "Which is the best earphone under 1000?",
"jh": NaN,
"hk": NaN,
"wt": NaN,
"average": 0
}
{
"test_qid": 331,
"test_question": "Which is the Best earphones under Rs 1000?",
"candidate_qid": 8505,
"candidate_question": "What are the best earphones under 1k?",
"jh": NaN,
"hk": NaN,
"wt": NaN,
"average": 0
}
{
"test_qid": 331,
"test_question": "Which is the Best earphones under Rs 1000?",
"candidate_qid": 15130,
"candidate_question": "Which is the best earphones to buy under Rs.600?",
"jh": NaN,
"hk": NaN,
"wt": NaN,
"average": 0
}
{
"test_qid": 331,
"test_question": "Which is the Best earphones under Rs 1000?",
"candidate_qid": 26692,
"candidate_question": "What are the best earphones under 60$?",
"jh": NaN,
"hk": NaN,
"wt": NaN,
"average": 0
}
<jupyter_script># This notebook covers
# - Dataset Preparation (train-test split)
# - TF-IDF indexes
# - Evaluation algorithm
# - Evaluation procedure with the test set
# - Update indexes with unseen questions
# - Query with unseen questions
# The following process is done on another notebook
# - Spellcheck and SpaCy tokenisation for the training set
# - SentenceTransformer computation of vectors
# - Downloading of the SpaCy and GenSim models
# notebook hyperparameters
TEST_SET_SIZE = 1000
RANKED_LIST_SIZE = 100
RANDOM_STATE = 42
EVALUATING = True # make False if you want to run query quickly
import os, collections, random, itertools, functools, time
from collections import defaultdict, Counter
from math import log
import tqdm.notebook as tqdm
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
random.seed(RANDOM_STATE)
np.random.seed(RANDOM_STATE)
# load data
df = pd.read_csv("/kaggle/input/quora-question-pairs/train.csv.zip")
df["question1"] = df["question1"].astype(str) # resolve nan
df["question2"] = df["question2"].astype(str)
df["qid1"] -= 1 # start index from zero
df["qid2"] -= 1
df.sample(10)
# # Preprocessing Dataset
# all questions are identified with its qid
qid_to_question = {}
for qid1, qid2, question1, question2 in zip(
df["qid1"], df["qid2"], df["question1"], df["question2"]
):
qid_to_question[qid1] = question1
qid_to_question[qid2] = question2
# extract 1000 questions for testing
test_query_qids = set()
df_duplicate = df[df["is_duplicate"] == 1].sample(frac=1, random_state=RANDOM_STATE)
for qid1, qid2, is_duplicate in zip(
df_duplicate["qid1"], df_duplicate["qid2"], df_duplicate["is_duplicate"]
):
if (
is_duplicate
and qid1 not in test_query_qids
and len(test_query_qids) < TEST_SET_SIZE
):
test_query_qids.add(qid2)
if qid1 in test_query_qids and qid2 in test_query_qids:
# to guarantee that there is a duplicate question in the training set
test_query_qids.remove(qid1)
test_query_qids.remove(qid2)
assert len(test_query_qids) == TEST_SET_SIZE # if fail, change random_state
test_query_qids_list = sorted(test_query_qids)
train_query_qids_list = sorted(set(qid_to_question.keys()) - test_query_qids)
assert test_query_qids_list[:3] == [331, 489, 501] # to check random state fixed
# # uncomment this to test only limited queries
if not EVALUATING:
test_query_qids_list = test_query_qids_list[:10]
TEST_SET_SIZE = 10
# extract duplicate relationship of training set
qid_to_duplicate_qids = defaultdict(set)
qid_to_nonduplicate_qids = defaultdict(set)
for qid1, qid2, is_duplicate in zip(df["qid1"], df["qid2"], df["is_duplicate"]):
if not (qid1 in test_query_qids or qid2 in test_query_qids):
if is_duplicate:
qid_to_duplicate_qids[qid1].add(qid2)
qid_to_duplicate_qids[qid2].add(qid1)
else:
qid_to_nonduplicate_qids[qid1].add(qid2)
qid_to_nonduplicate_qids[qid2].add(qid1)
# complete graph of duplicate relationships
qid_to_duplicate_qids_complete = defaultdict(set)
qid_to_qid_group_leader = {}
qid_group_leader_to_duplicate_qid_group = defaultdict(set)
visited_qids = set()
for train_qid in train_query_qids_list:
if train_qid in visited_qids:
continue
current_qids_group = set([train_qid])
qid_to_qid_group_leader[train_qid] = train_qid
stack = [train_qid]
while stack:
cur_qid = stack.pop()
for nex_qid in qid_to_duplicate_qids[cur_qid]:
if nex_qid in current_qids_group:
continue
qid_to_qid_group_leader[nex_qid] = train_qid
stack.append(nex_qid)
current_qids_group.add(nex_qid)
# complete the graph
for qid1, qid2 in itertools.combinations(current_qids_group, r=2):
qid_to_duplicate_qids_complete[qid1].add(qid2)
qid_to_duplicate_qids_complete[qid2].add(qid1)
qid_group_leader_to_duplicate_qid_group[train_qid] = current_qids_group
visited_qids.update(current_qids_group)
# extract duplicate relationship of the test set
test_qid_to_duplicate_qids = defaultdict(set)
test_qid_to_duplicate_qids_complete = defaultdict(set)
for qid1, qid2, is_duplicate in zip(
df_duplicate["qid1"], df_duplicate["qid2"], df_duplicate["is_duplicate"]
):
if qid2 in test_query_qids:
qid1, qid2 = qid2, qid1
if qid1 in test_query_qids:
if qid2 in test_query_qids:
continue
test_qid_to_duplicate_qids[qid1].add(qid2)
test_qid_to_duplicate_qids_complete[qid1].add(qid2)
for train_qid in qid_group_leader_to_duplicate_qid_group[
qid_to_qid_group_leader[qid2]
]:
test_qid_to_duplicate_qids_complete[qid1].add(train_qid)
# count inconsistencies in dataset
cnt = 0
for qid1, qid2, is_duplicate in zip(df["qid1"], df["qid2"], df["is_duplicate"]):
if not is_duplicate and qid1 not in test_query_qids and qid2 not in test_query_qids:
if qid_to_qid_group_leader[qid1] == qid_to_qid_group_leader[qid2]:
cnt += 1
print(
"Number of inconsistencies: ", cnt
) # slightly smaller than 96 because some edges are associated with the test set
test_mask = (df["qid1"].isin(test_query_qids)) | (df["qid2"].isin(test_query_qids))
train_df = df[~test_mask].copy()
test_df = df[test_mask].copy()
# clean up
del qid_to_qid_group_leader, qid_group_leader_to_duplicate_qid_group
del cnt
del test_query_qids # not sorted, use test_query_qids_list
del df # all data you can train on is in train_df
# enable use of complete graphs
test_qid_to_duplicate_qids = test_qid_to_duplicate_qids_complete
qid_to_duplicate_qids = qid_to_duplicate_qids_complete
# # Evaluation Metrics
def method_random_guess(test_qid):
# returns ranklist and scores of each size RANKED_LIST_SIZE
return (
random.choices(train_query_qids_list, k=RANKED_LIST_SIZE),
[0] * RANKED_LIST_SIZE,
)
# 1000 x 100 (the ranked list of similar qn for each of the 1000 test qns)
ranklists_method_random_guess = [
method_random_guess(test_qid)[0] for test_qid in test_query_qids_list
]
def show_sample_query_results(
test_qid, method_ranklist, method_scores=[0] * RANKED_LIST_SIZE, num_to_show=10
):
# not a metric, just print a few examples and its scores
print("Query: {}".format(qid_to_question[test_qid]))
for rank, (score, result_qid) in enumerate(
zip(method_scores, method_ranklist[:num_to_show]), start=1
):
relevance = (
"Registered"
if result_qid in test_qid_to_duplicate_qids[test_qid]
else "Unregistered"
)
print(
"Rank {} - Score {:.4f} - {}: \t{}".format(
rank, score, relevance, qid_to_question[result_qid]
)
)
show_sample_query_results(
test_query_qids_list[0], *method_random_guess(test_query_qids_list[0])
)
def evaluation_with_first_relevant_rank(
method_ranklists, considered=1, eps=10**-6, debug=True, **kwargs
):
# calculation of the statistics of the rank of the first c=considered duplicates
# if the duplicate does not appear in the ranklist, it has a default rank of RANKED_LIST_SIZE
assert np.array(method_ranklists).shape == (TEST_SET_SIZE, RANKED_LIST_SIZE)
reciprocal_ranks = []
ranks = []
for test_qid, ranklist in zip(test_query_qids_list, method_ranklists):
test_qid_to_rank = {
result_qid: rank for rank, result_qid in enumerate(ranklist, start=1)
}
rank = [] # may be shorter than `considered` because of lack of duplicates
for expected_qid in test_qid_to_duplicate_qids[test_qid]:
if expected_qid in test_qid_to_rank:
rank.append(test_qid_to_rank[expected_qid])
else:
rank.append(RANKED_LIST_SIZE + 1)
rank.sort()
ranks.extend(rank[:considered])
if rank[0] > RANKED_LIST_SIZE:
reciprocal_ranks.append(0)
else:
reciprocal_ranks.append(1 / rank[0])
plt.figure(figsize=(14, 4))
plt.title("Highest rank of duplicate question")
plt.hist(ranks, bins=np.arange(RANKED_LIST_SIZE + 2))
plt.xlabel("Rank")
plt.ylabel("Frequency")
plt.show()
mrr = sum(reciprocal_ranks) / len(reciprocal_ranks)
har = 1 / (mrr + eps)
print(f"Mean Reciprocal Rank (MRR) is {mrr:.2f}")
print(f"Harmonic Average Rank (HAR) is {har:.2f}")
p50 = np.median(ranks)
proportion_out_of_result = ranks.count(RANKED_LIST_SIZE + 1) / len(ranks)
if debug:
print("Median rank: {:.2f}".format(p50))
print("Proportion out of result: {:.3f}".format(proportion_out_of_result))
return mrr, har, p50, proportion_out_of_result
_ = evaluation_with_first_relevant_rank(ranklists_method_random_guess)
def evaluation_with_auc(method_ranklists, k=10, weights=None, debug=True, **kwargs):
assert np.array(method_ranklists).shape == (TEST_SET_SIZE, RANKED_LIST_SIZE)
counts = np.array([0.0] * k)
## Identify duplicates among top K ranks for each test
for i, (test_qid, ranklist) in enumerate(
zip(test_query_qids_list, method_ranklists)
):
topk = ranklist[:k]
is_duplicate = np.array(
[
1 if (result_qid in test_qid_to_duplicate_qids[test_qid]) else 0
for result_qid in topk
]
)
counts += is_duplicate
## Calculate AUC
if weights:
counts *= np.array(weights) / sum(weights)
else:
counts /= k
auc = sum(counts) / (TEST_SET_SIZE)
if debug:
print(f"{auc:.2%} of top {k} results are duplicates")
return auc # between [0,1], 1 is perfect
_ = evaluation_with_auc(ranklists_method_random_guess)
_ = evaluation_with_auc(
ranklists_method_random_guess, weights=[10, 9, 8, 7, 6, 5, 4, 3, 2, 1]
)
def single_r_precision(test_qid, ranklist):
# use this to check a single test query
num_duplicate = len(
test_qid_to_duplicate_qids[test_qid]
) # this dict needs to be updated when train:test set separation is updated
if num_duplicate == 0:
return 0, 0, 0
top_r = ranklist[:num_duplicate]
num_duplicates_in_top_r = sum(
[
1 if (result_qid in test_qid_to_duplicate_qids[test_qid]) else 0
for result_qid in top_r
]
)
r_precision = num_duplicates_in_top_r / num_duplicate
return num_duplicate, num_duplicates_in_top_r, r_precision
def evaluation_with_r_precision(
method_ranklists, k=10, report_k=0, debug=True, **kwargs
):
print(np.array(method_ranklists).shape)
assert np.array(method_ranklists).shape == (
TEST_SET_SIZE,
RANKED_LIST_SIZE,
) # method_ranklists size is (1000,100)
total_num_duplicates = np.array([0 for i in range(TEST_SET_SIZE)])
r_precision = np.array([0 for i in range(TEST_SET_SIZE)])
## Iter over 1->1000 tests
for i, (test_qid, ranklist) in enumerate(
zip(test_query_qids_list, method_ranklists)
): # iter over 1->1000 tests
(
total_num_duplicates[i],
num_duplicates_in_top_r,
r_precision[i],
) = single_r_precision(test_qid, ranklist)
# note: if want do error analysis, intervene here to find test cases with low r precision
if report_k > 0:
k_lowest_r_precision_idx = np.argpartition(r_precision, k)[:k]
k_lowest_r_precision_test_qids = np.array(test_query_qids_list)[
k_lowest_r_precision_idx
]
## Calculate metrics
avg_r_precision = r_precision.mean()
weighted_avg_r_precision = (
np.multiply(r_precision, total_num_duplicates).sum()
/ total_num_duplicates.sum()
)
if debug:
print(f"Average R-Precision = {avg_r_precision:.2%}")
print(
f"Weighted Average R-Precision by proportion of duplicates = {weighted_avg_r_precision:.2%}"
)
if avg_r_precision > weighted_avg_r_precision:
print(
"A higher average R-Precisions suggests that there are many test queries with high R-Precision but there are some test queries with high number of duplicates that model is not effective with."
)
if not report_k:
return avg_r_precision, weighted_avg_r_precision
else:
return avg_r_precision, weighted_avg_r_precision, k_lowest_r_precision_test_qids
_ = evaluation_with_r_precision(ranklists_method_random_guess, k=10)
def evaluation_with_precision_recall_at_k(
method_ranklists,
k=10,
exclude_precision=False,
exclude_recall=False,
debug=True,
**kwargs,
):
assert np.array(method_ranklists).shape == (TEST_SET_SIZE, RANKED_LIST_SIZE)
## Evaluation returns the macro average P@K and R@Kfor test set
## Interpretation P@K: what % of top k retrieved is relevant?
## Interpretation R@K: what % of all duplicates for query is retrieved within top k?
## Iter thru each test
precisions_at_k = []
recalls_at_k = []
for i, (test_qid, ranklist) in enumerate(
zip(test_query_qids_list, method_ranklists)
):
## 1. Set rank threshold K, ignore all docs after K
## 2. Count num_relevant in top-K
## 3. Count total_num_duplicates_for_query
## 4. P@K = num_relevant/k
## 5. R@K = num_relevant/total_num_duplicates_for_query
topk = ranklist[:k]
num_relevant = sum(
[
1 if (result_qid in test_qid_to_duplicate_qids[test_qid]) else 0
for result_qid in topk
]
)
precision_at_k = num_relevant / k
precisions_at_k.append(precision_at_k)
total_num_duplicates_for_query = len(test_qid_to_duplicate_qids[test_qid])
recall_at_k = num_relevant / total_num_duplicates_for_query
recalls_at_k.append(recall_at_k)
mean_precision_at_k = sum(precisions_at_k) / len(precisions_at_k) # macro average
mean_recall_at_k = sum(recalls_at_k) / len(recalls_at_k) # macro average
print(f"Macro Average Precision@k={k} is {mean_precision_at_k:.2%}")
print(f"Macro Average Recall@k={k} is {mean_recall_at_k:.2%}")
return (mean_precision_at_k, mean_recall_at_k)
_ = evaluation_with_precision_recall_at_k(ranklists_method_random_guess, k=10)
def evaluation_with_map(method_ranklists, debug=True, **kwargs):
assert np.array(method_ranklists).shape == (TEST_SET_SIZE, RANKED_LIST_SIZE)
## Interpretation: what is the average precision for all relevant docs across all queries?
## Iter thru each test
average_precisions = []
for i, (test_qid, ranklist) in enumerate(
zip(test_query_qids_list, method_ranklists)
):
## 1. Find the rank positions of each of the R relevant docs: K1, K2, ... KR and sort
## 2. Compute P@K for each K1, K2, ... If K >=RANKED_LIST_SIZE, assume never retrieved
## 3. AP = average of P@K for query
## 4. MAP = macro average of AP across queries
## 1. Find the rank positions of each of the R relevant docs: K1, K2, ... and sort
dup_qids_in_train_set = [
dup_qid
for dup_qid in test_qid_to_duplicate_qids[test_qid]
if dup_qid in train_query_qids_list
] # find all the dup_qid that can be found in the train set so you know total dup qn that could be found
total_num_dup_qid = len(dup_qids_in_train_set) # how many dup qn to expect
dup_ranks = []
for dup_qid in dup_qids_in_train_set:
if dup_qid not in ranklist: # not found
dup_ranks.append(
RANKED_LIST_SIZE
) # give "out of range" rank which would be checked later during calculation
continue
dup_ranks.append(
list(ranklist).index(dup_qid) + 1
) # append the rank of the retrieved dup qn
dup_ranks, dup_qids_in_train_set = (
list(t) for t in zip(*sorted(zip(dup_ranks, dup_qids_in_train_set)))
) # sort by rank
## 2. Compute P@K for each K1, K2, ... If K >=RANKED_LIST_SIZE, assume never retrieved
precisions_at_k = []
for j, rank in enumerate(dup_ranks, start=1): # dup_ranks is sorted
if rank >= RANKED_LIST_SIZE: # handle "unretrieved" duplicates
precisions_at_k.append(0)
else:
precision_at_k = j / rank # = num_dup_so_far / rank_of_latest_dup_found
precisions_at_k.append(precision_at_k)
## 3. AP = average of P@K for query
average_precisions.append(sum(precisions_at_k) / len(precisions_at_k))
## Out of test query loop
## 4. MAP = macro average of AP across queries
MAP = sum(average_precisions) / len(average_precisions)
print(f"Mean Average Precision (MAP) is {MAP:.2%}")
return MAP
_ = evaluation_with_map(ranklists_method_random_guess)
def evaluation_process(method, **kwargs):
# executes the method and runs the evaluation functions
ranklists, scorelists = [], []
for test_qid in tqdm.tqdm(test_query_qids_list):
ranklist, scores = method(test_qid)
ranklists.append(ranklist)
scorelists.append(scores)
evaluation_with_first_relevant_rank(ranklists, **kwargs)
# evaluation_with_auc(ranklists, **kwargs)
evaluation_with_r_precision(ranklists, **kwargs)
evaluation_with_precision_recall_at_k(ranklists, k=10, **kwargs)
evaluation_with_map(ranklists, **kwargs)
return ranklists, scorelists
results_random_guess = evaluation_process(method_random_guess)
# # Preprocessing
## This entire cell is important to enable tokeniser pipeline
## Use this to replace tokenise function if using Tokenise then Spellcheck (TSC) pipeline
######### spacy basic tokenizer
import spacy
print("Spacy version: ", spacy.__version__)
from spacy.tokenizer import Tokenizer # https://spacy.io/api/tokenizer
# !python3 -m spacy download en_core_web_sm
print("Loading Spacy en_core_web_sm loaded")
nlp = spacy.load("en_core_web_sm")
tokenizer = Tokenizer(nlp.vocab)
tokenizer.add_special_case(
"[math]", [{"ORTH": "[math]"}]
) # see qid=7: '[math]23^{24}[/math]' becomes one token
# add more special cases here if found
def spacy_tokenise(text, lower=False, split_last_punc=True):
"""
returns a list of tokens given a question text
note: each punctuation is also considered a token
note: "\n" is a token
note: "'s" is a token
note: '(Koh-i-Noor)' is a token
see tokenizer instantiation code for special cases or to add
lowercase text only after spell check
"""
if lower:
text = text.lower()
tokens = tokenizer(text)
token_list = [token.text for token in tokens]
# further split tokens that end with certain punct e.g. "me?" => "me", "?"
if split_last_punc:
split_lists = [
[token[:-1], token[-1]] if (token[-1] in ["!", "?", ",", ":"]) else [token]
for token in token_list
]
token_list = [token for sublist in split_lists for token in sublist]
return token_list
######### symspell spellchecker
print("Loading symspell")
from symspellpy.symspellpy import (
SymSpell,
Verbosity,
) # https://github.com/mammothb/symspellpy
import pkg_resources
# instantiate spellchecker
sym = SymSpell(max_dictionary_edit_distance=2, prefix_length=7, count_threshold=1)
# https://symspellpy.readthedocs.io/en/latest/api/symspellpy.html
dictionary_path = pkg_resources.resource_filename(
"symspellpy", "frequency_dictionary_en_82_765.txt"
)
sym.load_dictionary(dictionary_path, 0, 1) # might take a short while
def spellcheck_single(word):
# returns top correct spelling or the same word if no correction found within max_edit_distance
if not word.isascii():
return word # do not spellcheck non ascii words e.g. シ
# obtain list of suggestions
suggestions = sym.lookup(
word,
Verbosity.CLOSEST,
max_edit_distance=2,
include_unknown=True, # a mispelled word with no found corrections is returned as is
ignore_token=r"[:,.!?\\-]", # use if want to avoid correcting certain phrases
)
# get the term from the suggestItem object
suggested_words = [suggestion._term for suggestion in suggestions]
# check if the input word is legit and return if so else return corrected word
word_lower = word.lower()
if word_lower in suggested_words:
return word_lower # do not correct if input is a legit word
else:
return suggested_words[0] # top suggestion
def spellcheck_compound(sent):
# spellchecks a sentence
suggestions = sym.lookup_compound(sent, max_edit_distance=2)
return suggestions[0]._term # returns the top suggestion
######### tokenise pipeline
def tokenise_then_spellcheck(sent):
# 8 times faster than spellcheck_then_tokenise
tokens = spacy_tokenise(sent) # NOTE: replace tokenise with spacy_tokenise
checked_tokens = [
spellcheck_single(token).lower() for token in tokens
] # lower after spell check
return checked_tokens
def spellcheck_then_tokenise(sent):
checked_sent = spellcheck_compound(sent)
tokens = spacy_tokenise(checked_sent, lower=True) # lower after spell check
return tokens
# define tokenisation process
import pickle
qid_to_tokens_preprocessed_filename = "../input/quora-question-pairs-tokenise-pipeline/qid_to_processed_token_list_tokenise_then_spellcheck.pkl"
with open(qid_to_tokens_preprocessed_filename, "rb") as f:
qid_to_tokens_preprocessed = pickle.load(f)
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
stopword_set = set(stopwords.words())
stopword_set.update(["?", ","])
def nltk_tokenize(sentence):
return word_tokenize(sentence.lower())
def tokenise_qid(
qid,
qid_to_tokens_preprocessed=qid_to_tokens_preprocessed,
tokenise_method=tokenise_then_spellcheck,
):
# return a list of tokens, does not remove stopwords or duplicates
if qid_to_tokens_preprocessed and qid in qid_to_tokens_preprocessed:
return qid_to_tokens_preprocessed[qid]
return tokenise_method(qid_to_question[qid])
def preprocess_vsm(
train_query_qids_list=train_query_qids_list,
stopword_set=stopword_set,
exclude_stopwords=True,
):
"""
Input:
qid_to_question = {qid: question string}
Note: only use the test subset of qids
Outputs:
qid_to_tokens = {qid: set(tokens)}
token_to_qids = {token: set(qids)}
tf = {token: {qid: TF as int}}
df = {token: DF as int}
L = {qid: question length as int}
"""
qid_to_tokens = defaultdict(set)
token_to_qids = defaultdict(set)
tf = defaultdict(Counter)
df = defaultdict(int)
L = defaultdict(int)
qid_processed = set()
for qid in tqdm.tqdm(train_query_qids_list):
qid_tokenised = tokenise_qid(qid)
for token in set(qid_tokenised):
if token not in stopword_set or not exclude_stopwords:
# store qid-to-token mapping
qid_to_tokens[qid].add(token)
token_to_qids[token].add(qid)
# compute and store term frequency
tf[token][qid] += 1
# store doc frequency in df
df[token] += 1
# store doc length in L (double-count repeated tokens)
L[qid] = len(qid_tokenised)
# output
return qid_to_tokens, token_to_qids, tf, df, L
qid_to_tokens, token_to_qids, tf, df, L = preprocess_vsm()
# save a copy of the original to allow reset later
qid_to_tokens_original, token_to_qids_original = (
qid_to_tokens.copy(),
token_to_qids.copy(),
)
tf_original, df_original, L_original = tf.copy(), df.copy(), L.copy()
# # Baseline - Overlapping Root Word Count (Working Title)
# Order by the number of overlapping non-stopword words. Random if tie.
def method_overlapping_root_word_count(query_qid, ignore_stopwords=True):
query_tokens = set(tokenise_qid(query_qid))
if ignore_stopwords:
query_tokens = [token for token in query_tokens if token not in stopword_set]
counter = collections.Counter()
for dummy_qid in random.choices(train_query_qids_list, k=RANKED_LIST_SIZE):
# prefill with random results to address the possibility of no matches
counter[dummy_qid] = 0.01
for query_token in query_tokens:
counter += collections.Counter(token_to_qids[query_token])
query_results = list(counter.items())
random.shuffle(query_results) # so that qids are not ordered
query_results = sorted(query_results, key=lambda x: x[1], reverse=True)[
:RANKED_LIST_SIZE
]
return [x[0] for x in query_results], [x[1] for x in query_results]
show_sample_query_results(
test_query_qids_list[0],
*method_overlapping_root_word_count(test_query_qids_list[0]),
)
results_overlapping_root_word_count = evaluation_process(
method_overlapping_root_word_count
)
# # TFIDF
def compute_idf(doc_freq, N):
"""
Inputs:
doc_freq = document frequency of some token
N = corpus size including query
Output:
idf = IDF as float
"""
return log(N / doc_freq)
from functools import reduce
import operator
def prod(iterable):
return reduce(operator.mul, iterable, 1)
def use_vsm(
qid_query, # qid_to_tokens=qid_to_tokens, tf=tf, df=df, L=L,
method="tf-idf",
compute_idf=compute_idf,
k1=1.5,
k3=1.5,
b=0.75,
smoothing="add-one",
alpha=0.75,
exclude_stopwords=True,
return_top=RANKED_LIST_SIZE,
):
"""
Inputs:
query = question string to match # this comes from "test" set
qid_to_tokens = {qid: set(tokens)} # this is the "training" corpus
tf = {token: term freq} # required for all methods
df = {token: doc freq} # required for method='tf-idf','bm25'
L = {qid: doc length} # required for method='bm25','unigram'
method = model to apply
k1, k3, b = tuning params # required for method='bm25'
smoothing = type of smoothing # required for method='unigram'
return_top = num of docs to return
Procedure:
0. Corpus is already tokenised, tf, df, L already computed
1. Tokenise query, expand tf, df, L with query information
if method='boolean':
Remove idf calculation, then use method='tf-idf'
if method='tf-idf':
2. Compute tf-idf weights only for relevant (t,d) pairs
3. Compute cosine similarity only for docs containing query terms
if method='bm25':
2. Compute RSV summation terms only for relevant (t,d) pairs
3. Compute RSV only for docs containing query terms
if method='unigram':
2. Compute probabilities only for relevant (t,d) pairs
3. Compute query probability only for docs containing query terms
4. Return docs in ranked order
Output:
ranking = [qids in decreasing order of match]
scoring = [corresponding scores]
"""
assert method in [
"boolean",
"tf-idf",
"bm25",
"unigram",
], "Supported methods: 'boolean', 'tf-idf', 'bm25', 'unigram'"
assert len(L.keys()) > 0 if method == "bm25" else True, "Please include L for bm25"
assert (
len(L.keys()) > 0 if method == "unigram" else True
), "Please include L for unigram"
assert (
smoothing in ["add-one", "linear-interpolation"]
if method == "unigram"
else True
)
assert alpha >= 0 and alpha <= 1 if smoothing == "linear-interpolation" else True
qid_tmp = time.time()
""" STEP 1: PROCESS QUERY """
query_tokenised = tokenise_qid(qid_query)
for token in set(query_tokenised):
if token not in stopword_set or not exclude_stopwords:
# store qid-to-token mapping
# store query as qid=0 (corpus starts from qid=1)
qid_to_tokens[qid_tmp].add(token)
# compute and store term frequency
tf[token][qid_tmp] = sum([1 if t == token else 0 for t in query_tokenised])
# update doc frequency in df
df[token] += 1
# store query length
L[qid_tmp] = len(query_tokenised)
if method == "boolean":
def compute_idf(doc_freq, N):
return 1
method = "tf-idf"
if method == "tf-idf":
"""STEP 2: COMPUTE TF-IDF WEIGHTS"""
weights = defaultdict(lambda: defaultdict(float))
N = len(qid_to_tokens) # original corpus + query
# only bother computing for tokens in the query
for token in set(query_tokenised):
if token not in stopword_set or exclude_stopwords == False:
weights[qid_tmp][token] = tf[token][qid_tmp] * compute_idf(df[token], N)
for qid in tf[token].keys():
weights[qid][token] = tf[token][qid] * compute_idf(df[token], N)
# also compute weight for other tokens contained by these qids
# needed for computing qid vector length
for other_token in qid_to_tokens[qid]:
weights[qid][other_token] = tf[other_token][qid] * compute_idf(
df[other_token], N
)
""" STEP 3: COMPUTE COSINE SIMILARITY TO QUERY """
cosine_similarities = defaultdict(float)
# compute denominator (part 1), i.e., |q| * |d|
query_vector_length = (sum([w**2 for w in weights[qid_tmp].values()])) ** 0.5
if query_vector_length == 0:
print(f"query={query}\nweights[qid_tmp].items()={weights[qid_tmp].items()}")
for qid in weights.keys():
# compute numerator, i.e., dot product of q and d
cosine_numerator = 0
for token in weights[qid].keys():
if token in weights[qid_tmp]:
cosine_numerator += weights[qid][token] * weights[qid_tmp][token]
# compute denominator (part 2), i.e., |q| * |d|
qid_vector_length = (sum([w**2 for w in weights[qid].values()])) ** 0.5
if qid_vector_length == 0: # example: qid=25026 => question='?'
qid_vector_length = 1e-8
# compute and store cosine similarity between q and d
cosine_similarities[qid] = (
cosine_numerator / query_vector_length / qid_vector_length
)
scores = cosine_similarities
if method == "bm25":
"""STEP 2: COMPUTE RSV TERMS"""
rsv_terms = defaultdict(lambda: defaultdict(float))
N = len(qid_to_tokens) # original corpus + query
L_avg = sum(L.values()) / len(L.values())
# only bother computing for tokens in the query
for token in set(query_tokenised):
for qid in tf[token].keys():
rsv_terms[qid][token] = (
compute_idf(df[token], N)
* (k1 + 1)
* tf[token][qid]
/ (k1 * ((1 - b) + b * L[qid] * L_avg) + tf[token][qid])
* (k3 + 1)
* tf[token][qid_tmp]
/ (k3 + tf[token][qid_tmp])
)
""" STEP 3: COMPUTE RSV """
rsv = {qid: sum(rsv_terms[qid].values()) for qid in rsv_terms.keys()}
scores = rsv
if method == "unigram":
"""STEP 2: COMPUTE PROBABILITIES"""
probabilities = defaultdict(lambda: defaultdict(float))
corpus_model = defaultdict(float)
# only bother computing for tokens in the query
for token in set(query_tokenised):
for qid in tf[token].keys():
if smoothing == "add-one":
probabilities[qid][token] = (tf[token][qid] + 1) / (
L[qid] + len(query_tokenised)
)
else:
probabilities[qid][token] = (tf[token][qid]) / (L[qid])
# for linear-interpolation smoothing, build corpus language model
if smoothing == "linear-interpolation":
corpus_model[token] += tf[token][qid]
# remaining operations for linear-interpolation smoothing
if smoothing == "linear-interpolation":
# finish building corpus language model by dividing corpus tf by corpus L
total_corpus_length = sum(L.values())
for token in corpus_model.keys():
corpus_model[token] = corpus_model[token] / total_corpus_length
# then update the probabilities
for qid in probabilities.keys():
for token in probabilities[qid].keys():
probabilities[qid][token] = (
alpha * probabilities[qid][token]
+ (1 - alpha) * corpus_model[token]
)
""" STEP 3: COMPUTE QUERY PROBABILITY """
query_prob = {
qid: -log(prod(probabilities[qid].values())) for qid in probabilities.keys()
}
scores = query_prob
""" STEP 4: RANK DOCUMENTS AND RETURN RESULT """
# cleanup
del qid_to_tokens[qid_tmp]
for token in set(query_tokenised):
if token not in stopword_set or not exclude_stopwords:
del tf[token][qid_tmp]
df[token] -= 1
del scores[qid_tmp] # remove query from result
ranking = sorted(scores, key=scores.get, reverse=True)
scoring = sorted(scores.values(), reverse=True)
# if too few documents match the query, add dummy documents
if len(ranking) < return_top:
ranking.extend([0] * (return_top - len(ranking)))
scoring.extend([0] * (return_top - len(ranking)))
# return top k results
return ranking[:return_top], scoring[:return_top]
def method_boolean(qid):
return use_vsm(qid, method="boolean")
show_sample_query_results(
test_query_qids_list[0], *method_boolean(test_query_qids_list[0])
)
results_boolean = evaluation_process(method_boolean)
def method_tf_idf(qid):
return use_vsm(qid, method="tf-idf")
show_sample_query_results(
test_query_qids_list[0], *method_tf_idf(test_query_qids_list[0])
)
results_tf_idf = evaluation_process(method_tf_idf)
# # BM25
def method_bm25(qid):
return use_vsm(qid, method="bm25")
show_sample_query_results(
test_query_qids_list[0], *method_bm25(test_query_qids_list[0])
)
results_bm25 = evaluation_process(method_bm25)
def method_unigram(qid):
return use_vsm(qid, method="unigram", smoothing="add-one")
show_sample_query_results(
test_query_qids_list[0], *method_unigram(test_query_qids_list[0])
)
results_unigram = evaluation_process(method_unigram)
for alpha in [0, 0.25, 0.5, 0.75, 1.0]:
def method_unigram(qid):
return use_vsm(
qid, method="unigram", smoothing="linear-interpolation", alpha=alpha
)
_ = evaluation_process(method_unigram)
# # Word Embeddings - SpaCy
def to_vec(token_or_list):
# converts a token string or a list of tokens into a word or doc vec respectively
if type(token_or_list) == list:
# token list needs to be joined into a sentence first
token_or_list = " ".join(token_or_list)
return nlp(token_or_list).vector
nlp2 = spacy.load("en_core_web_lg")
def to_vec2(token_or_list):
# converts a token string or a list of tokens into a word or doc vec respectively
if type(token_or_list) == list:
# token list needs to be joined into a sentence first
token_or_list = " ".join(token_or_list)
return nlp2(token_or_list).vector
# Load pre-processed dict
with open("../input/quora-question-pairs-tokenise-pipeline/qid_to_vec.pkl", "rb") as f:
qid_to_vec = pickle.load(f)
print("Pre-processed question vector is of shape {}".format(qid_to_vec[0].shape))
from numpy import dot
from numpy.linalg import norm
def method_spacy_embedding_similarity(test_qid):
tokens = tokenise_then_spellcheck(qid_to_question[test_qid])
test_vec = to_vec(tokens)
## Run baseline model as a filter
qid_list, scores = method_overlapping_root_word_count(test_qid)
cos_sims = [] # bigger better
for train_qid in qid_list: # train_query_qids_list:
train_vec = qid_to_vec[train_qid]
cos_sim = dot(test_vec, train_vec) / (norm(test_vec) * norm(train_vec))
cos_sims.append(cos_sim)
cos_sims = np.array(cos_sims)
qid_list = np.array(qid_list) # train_query_qids_list)
inds = cos_sims.argsort()[::-1] # reverse so biggest come first
ranklist = qid_list[inds]
return ranklist[:RANKED_LIST_SIZE], cos_sims[:RANKED_LIST_SIZE]
show_sample_query_results(
test_query_qids_list[0], *method_spacy_embedding_similarity(test_query_qids_list[0])
)
results_spacy_embedding_similarity = evaluation_process(
method_spacy_embedding_similarity
)
with open(
"../input/quora-question-pairs-tokenise-pipeline/qid_to_vec_trf.pkl", "rb"
) as f: # note, actually lg not trf
qid_to_vec2 = pickle.load(f)
print(
"Pre-processed question vector is of shape {}".format(qid_to_vec2[0].shape)
) # 300 dim vec
def method_spacy_embedding_similarity_lg(test_qid):
tokens = tokenise_then_spellcheck(qid_to_question[test_qid])
test_vec = to_vec2(tokens)
## Run baseline model as a filter
qid_list, scores = method_overlapping_root_word_count(test_qid)
cos_sims = [] # bigger better
for train_qid in qid_list: # train_query_qids_list:
train_vec = qid_to_vec2[train_qid]
cos_sim = dot(test_vec, train_vec) / (norm(test_vec) * norm(train_vec))
cos_sims.append(cos_sim)
cos_sims = np.array(cos_sims)
qid_list = np.array(qid_list) # train_query_qids_list)
inds = cos_sims.argsort()[::-1] # reverse so biggest come first
ranklist = qid_list[inds]
return ranklist[:RANKED_LIST_SIZE], cos_sims[:RANKED_LIST_SIZE]
show_sample_query_results(
test_query_qids_list[0],
*method_spacy_embedding_similarity_lg(test_query_qids_list[0]),
)
results_spacy_embedding_similarity_lg = evaluation_process(
method_spacy_embedding_similarity_lg
)
# # Gensim WordMover Distance on Boolean Retrieval
# * Applies further sorting by wordmover distance on the output ranklist of Boolean Retrieval
# * Current pre-trained model: `glove-wiki-gigaword-50`
#
import gensim
import gensim.downloader
# gensim.downloader.info() # find more models to download
from gensim.models import KeyedVectors
try:
model = KeyedVectors.load(
"../input/ir-project-download-keyed-vectors/glove-wiki-gigaword-50.keyedvectors"
)
except: # gs_model not downloaded
model = gensim.downloader.load("glove-wiki-gigaword-50")
# model.save("/kaggle/working/glove-wiki-gigaword-50.keyedvectors") # if not already saved
def method_wordmover_distance(test_qid, model):
# out of box duplicate finder does not work!
# returns ranklist and scores of each size RANKED_LIST_SIZE
## Run baseline model as a filter
ranklist, scores = method_overlapping_root_word_count(test_qid)
## Process test question
test_qn = tokenise_qid(test_qid)
## Get wordmover distance from every candidate
distances = []
qid_list = ranklist
for candidate_qid in qid_list:
candidate_qn = tokenise_qid(candidate_qid)
distances.append(model.wmdistance(test_qn, candidate_qn))
## Sort by distance
sorted_dist_and_candidate_qid = sorted(zip(distances, qid_list))
sorted_candidate_qid = [qid for _, qid in sorted_dist_and_candidate_qid]
sorted_dist = [dist for dist, _ in sorted_dist_and_candidate_qid]
return sorted_candidate_qid[:RANKED_LIST_SIZE], sorted_dist[:RANKED_LIST_SIZE]
def method_wordmover_distance_glovewiki50(test_qid):
return method_wordmover_distance(test_qid, model)
show_sample_query_results(
test_query_qids_list[0],
*method_wordmover_distance_glovewiki50(test_query_qids_list[0]),
)
results_wordmover_distance_glovewiki50 = evaluation_process(
method_wordmover_distance_glovewiki50
)
models_to_try = [
"glove-wiki-gigaword-300",
"glove-twitter-50",
"word2vec-google-news-300",
"fasttext-wiki-news-subwords-300",
]
if not EVALUATING:
models_to_try = []
for m in models_to_try:
print("Model: ", m)
try:
model = KeyedVectors.load(
f"../input/ir-project-download-keyed-vectors/{m}.keyedvectors"
)
except:
model = gensim.downloader.load(m)
def method_wordmover_distance_new_model(test_qid):
return method_wordmover_distance(test_qid, model)
show_sample_query_results(
test_query_qids_list[0],
*method_wordmover_distance_new_model(test_query_qids_list[0]),
)
_ = evaluation_process(method_wordmover_distance_new_model)
# # Sentence Embeddings
# Each sentence can be embedded as a vector with SentenceTransformer
from sentence_transformers import SentenceTransformer
model_name = "bert-base-nli-stsb-mean-tokens"
model_tf = SentenceTransformer(model_name)
model_name = "bert-base-nli-stsb-mean-tokens"
sentence_vectors = np.load(
f"../input/quora-question-pairs-bert-sentence-vectors/sentence_vectors_{model_name}.npy"
)
sentence_vectors = {i: vec for i, vec in enumerate(sentence_vectors)}
from scipy.spatial.distance import cosine
def method_sentence_vector(
query_qid,
method_preliminary=method_overlapping_root_word_count,
preliminary_factor=1,
):
# method_preliminary can be either of the previous methods
# recommended method_overlapping_root_word_count, method_boolean, method_tf_idf
qid_list, preliminary_scores = method_preliminary(query_qid)
# sort by cosine similarity
query_sentence_vector = sentence_vectors[query_qid]
query_results = [
(
qid,
preliminary_factor * preliminary_score
- abs(cosine(query_sentence_vector, sentence_vectors[qid])),
)
for qid, preliminary_score in zip(qid_list, preliminary_scores)
]
query_results = sorted(query_results, key=lambda x: x[1], reverse=True)[
:RANKED_LIST_SIZE
]
return [x[0] for x in query_results], [x[1] for x in query_results]
show_sample_query_results(
test_query_qids_list[0],
*method_sentence_vector(test_query_qids_list[0], preliminary_factor=0),
)
show_sample_query_results(
test_query_qids_list[0],
*method_sentence_vector(test_query_qids_list[0], preliminary_factor=1),
)
results_sentence_vector = evaluation_process(method_sentence_vector)
# # Supervised Model
df_train = train_df.copy()
from fuzzywuzzy import fuzz
def extract_features(qid1, qid2, eps=10**-6):
question1 = qid_to_question[qid1]
question2 = qid_to_question[qid2]
features = defaultdict(float)
features["ratio"] = fuzz.ratio(question1, question2)
token_qid1 = tokenise_qid(qid1)
token_qid2 = tokenise_qid(qid2)
tf_idf_denominator = 0
for token in set(token_qid1) | set(token_qid2):
if df[token]:
tf_idf_denominator += compute_idf(df[token], len(qid_to_tokens))
tf_idf_numerator = 0
for token in set(token_qid1) & set(token_qid2):
if df[token]:
tf_idf_numerator += compute_idf(df[token], len(qid_to_tokens))
features["tf_idf"] = (tf_idf_numerator + eps) / (tf_idf_denominator + eps)
return features
features_all = list(
tqdm.tqdm(
extract_features(qid1, qid2)
for qid1, qid2 in zip(df_train["qid1"], df_train["qid2"])
)
)
for feature in features_all[0].keys():
df_train[feature] = [features[feature] for features in features_all]
import lightgbm as lgb
target_train = df_train["is_duplicate"]
df_train = df_train.drop(
["id", "qid1", "qid2", "question1", "question2", "is_duplicate"], axis=1
)
eval_set = np.array(
[True if i < len(df_train) * 0.2 else False for i in range(len(df_train))]
)
lgb_train = lgb.Dataset(df_train[~eval_set], target_train[~eval_set])
lgb_eval = lgb.Dataset(df_train[eval_set], target_train[eval_set], reference=lgb_train)
lgb_all = lgb.Dataset(df_train, target_train)
params = {
# 'boosting_type': 'gbdt',
"objective": "binary",
# 'scale_pos_weight': 0.360,
# 'metric': {'auc'},
# 'num_leaves': 15,
# 'learning_rate': 0.05,
# 'feature_fraction': 0.9,
# 'bagging_fraction': 0.8,
# 'bagging_freq': 5,
"verbose": -1,
}
gbm = lgb.train(
params,
lgb_train,
num_boost_round=1000,
valid_sets=lgb_eval,
verbose_eval=-1,
early_stopping_rounds=10,
)
pd.DataFrame(
{
"feature": df_train.columns,
"importance": gbm.feature_importance(importance_type="gain"),
}
)[:20]
def method_supervised_model(query_qid):
candidate_qids, _ = method_overlapping_root_word_count(query_qid)
df_predict = pd.DataFrame()
features_all = list(
extract_features(query_qid, candidate_qid) for candidate_qid in candidate_qids
)
for feature in features_all[0].keys():
df_predict[feature] = [features[feature] for features in features_all]
scores = gbm.predict(df_predict)
results = sorted(list(zip(scores, candidate_qids)))[::-1]
return [x[1] for x in results], [x[0] for x in results] # qid, scores
show_sample_query_results(
test_query_qids_list[0], *method_supervised_model(test_query_qids_list[0])
)
results_supervised_model = evaluation_process(method_supervised_model)
# # Preparation for Hand Evaluation Dataset
method_to_ranklists_scorelists = {
# "random_guess": results_random_guess,
"overlapping_root_word_count": results_overlapping_root_word_count,
"boolean": results_boolean,
"tf_idf": results_tf_idf,
"bm25": results_bm25,
"unigram": results_unigram,
"spacy_embedding_similarity": results_spacy_embedding_similarity,
"spacy_embedding_similarity_lg": results_spacy_embedding_similarity_lg,
"wordmover_distance_glovewiki50": results_wordmover_distance_glovewiki50,
"sentence_vector": results_sentence_vector,
"supervised_model": results_supervised_model,
}
import json
def parse_ndarray(obj): # https://stackoverflow.com/a/52604722/5894029
if isinstance(obj, np.ndarray):
return obj.tolist()
with open("method_to_ranklists_scorelists.json", "w") as f:
json.dump(method_to_ranklists_scorelists, f, indent=4, default=parse_ndarray)
QUESTIONS_TO_HANDEVAL = set(
x - 1
for x in [
332,
490,
1955,
6319,
9690,
17279,
19619,
20557,
26378,
33734,
38984,
49864,
57291,
89903,
116882,
126992,
131214,
144297,
159628,
201409,
273666,
284107,
286721,
312887,
318523,
378759,
384832,
405081,
405877,
423313,
464279,
480116,
533401,
]
)
HANDEVAL_RANK_THRESHOLD = 10
map_qid_to_handeval = defaultdict(set)
for ranklists, scorelists in method_to_ranklists_scorelists.values():
for test_qid, ranklist in zip(test_query_qids_list, ranklists):
if test_qid in QUESTIONS_TO_HANDEVAL:
for candidate_qid in ranklist[:HANDEVAL_RANK_THRESHOLD]:
map_qid_to_handeval[test_qid].add(candidate_qid)
for qid in map_qid_to_handeval:
map_qid_to_handeval[qid] = sorted(map_qid_to_handeval[qid])
dataframe_columns = ["test_qid", "test_question", "candidate_qid", "candidate_question"]
dataframe_entries = []
for qid in sorted(map_qid_to_handeval.keys()):
for candidate_qid in map_qid_to_handeval[qid]:
line_entry = [
qid,
qid_to_question[qid],
candidate_qid,
qid_to_question[candidate_qid],
]
dataframe_entries.append(line_entry)
df_handeval = pd.DataFrame(dataframe_entries, columns=dataframe_columns)
# labeller columns
df_handeval["jh"] = np.nan
df_handeval["hk"] = np.nan
df_handeval["wt"] = np.nan
df_handeval.to_csv("df_handeval.csv", index=None)
# # Calculate NDCG with Hand Evaluation Dataset
# This calculates NDCG from a snapshot version of `method_to_ranklists_scorelists`, and a hand annotated `df_handeval`
# Due to randomness, the `method_to_ranklists_scorelists` may not be reproduced exactly.
df_handeval = pd.read_csv(
"../input/quoraquestionpairhandannotateddataset/df_handeval.csv"
)
with open(
"../input/quoraquestionpairhandannotateddataset/method_to_ranklists_scorelists.json"
) as f:
method_to_ranklists_scorelists = json.load(f)
import math
def calculate_dcg_at_k(r, k, method=0):
if method == 0:
logn = [1.0] + [1 / math.log(i, 2) for i in range(2, k + 1)]
else:
logn = [1 / math.log(i, 2) for i in range(2, k + 2)]
dcg = 0.0
for gain, disc in zip(r[:k], logn):
dcg += gain * disc
return dcg
def calculate_ndcg_at_k(scores, ref, k=10, method=0):
denom = calculate_dcg_at_k(ref, k, method=method)
numer = calculate_dcg_at_k(scores, k, method=method)
if denom == 0:
return 0.0
return numer / denom
test_qid_to_candidate_qid_to_scores = collections.defaultdict(dict)
for _, row in df_handeval.iterrows():
test_qid = row["test_qid"]
candidate_qid = row["candidate_qid"]
score = row["average"]
test_qid_to_candidate_qid_to_scores[test_qid][candidate_qid] = score
test_qid_to_ideal_scores = collections.defaultdict(list)
for test_qid, candidate_qid_to_scores in test_qid_to_candidate_qid_to_scores.items():
ideal_scores = sorted(candidate_qid_to_scores.values())[::-1]
test_qid_to_ideal_scores[test_qid] = ideal_scores
method_to_ndcg_score = collections.defaultdict(list)
count_out_of_eval = 0
for method_name, (ranklists, _) in method_to_ranklists_scorelists.items():
for test_qid, ranklist in zip(test_query_qids_list, ranklists):
if test_qid in QUESTIONS_TO_HANDEVAL:
scores = []
for candidate_qid in ranklist[:HANDEVAL_RANK_THRESHOLD]:
if candidate_qid not in test_qid_to_candidate_qid_to_scores[test_qid]:
scores.append(1)
print(method_name, len(scores))
count_out_of_eval += 1
else:
scores.append(
test_qid_to_candidate_qid_to_scores[test_qid][candidate_qid]
)
ref = test_qid_to_ideal_scores[test_qid]
ndcg_at_k = calculate_ndcg_at_k(scores, ref)
method_to_ndcg_score[method_name].append(ndcg_at_k)
count_out_of_eval
for method_name, scores in method_to_ndcg_score.items():
print(method_name)
print(sum(scores) / len(scores))
print(" ".join(f"{x:.2f}" for x in scores))
print()
# # Indexing and Querying of Unseen Questions
# This is probably the Graphical User Interface that we will present
def index_unseen_question(unseen_question_text_list):
unseen_sentence_vectors = model_tf.encode(
unseen_question_text_list, show_progress_bar=True
)
qids_new = [time.time() for _ in unseen_question_text_list]
for qid_new, unseen_sentence_vector, unseen_question_text in zip(
qids_new, unseen_sentence_vectors, unseen_question_text_list
):
qid_to_question[qid_new] = unseen_question_text
# compute and update word embedding
token_list = tokenise_then_spellcheck(unseen_question_text)
qid_to_vec[qid_new] = to_vec(token_list)
qid_to_vec2[qid_new] = to_vec2(token_list)
# update sentence embedding
sentence_vectors[qid_new] = unseen_sentence_vector
# update tf-idf
qid_to_tokens_, token_to_qids_, tf_, df_, L_ = preprocess_vsm(qids_new)
for qid in qid_to_tokens_:
qid_to_tokens[qid] = qid_to_tokens_[qid]
for token in token_to_qids_:
token_to_qids[token].update(token_to_qids_[token])
for token in tf_:
for qid in tf_[token]:
tf[token][qid] += tf_[token][qid]
for token in df_:
df[token] += df_[token]
for qid in L_:
L[qid] = L_[qid]
def query_unseen_question(unseen_question_text, method):
qid_new = time.time()
qid_to_question[qid_new] = unseen_question_text
# update word embedding
token_list = tokenise_then_spellcheck(unseen_question_text)
qid_to_vec[qid_new] = to_vec(token_list)
qid_to_vec2[qid_new] = to_vec2(token_list)
# update sentence embedding
sentence_vectors[qid_new] = model_tf.encode(
unseen_question_text, show_progress_bar=False
)
show_sample_query_results(qid_new, *method(qid_new))
# list of methods, uncomment to select
method = method_random_guess
method = method_overlapping_root_word_count
method = method_boolean
method = method_tf_idf
method = method_bm25
method = method_unigram
method = method_spacy_embedding_similarity
method = method_spacy_embedding_similarity_lg
method = method_wordmover_distance_glovewiki50
# method = method_sentence_vector
# method = method_supervised_model
query_unseen_question("Why are computer screens dark in color?", method=method)
index_unseen_question(
[
"Why are computer screens black when unpowered?",
"Why are computer screens manufactured black?",
]
)
query_unseen_question("Why are computer screens dark in color?", method=method)
## uncomment if you want to reset the indexing
# qid_to_tokens, token_to_qids = qid_to_tokens_original.copy(), token_to_qids_original.copy()
# tf, df, L = tf_original.copy(), df_original.copy(), L_original.copy()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/046/69046687.ipynb
|
quoraquestionpairhandannotateddataset
|
huikang
|
[{"Id": 69046687, "ScriptId": 18844119, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1680925, "CreationDate": "07/26/2021 08:44:05", "VersionNumber": 1.0, "Title": "ir-project-main-branch-supervised-model", "EvaluationDate": "07/26/2021", "IsChange": false, "TotalLines": 1268.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 1268.0, "LinesInsertedFromFork": 0.0, "LinesDeletedFromFork": 0.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 1268.0, "TotalVotes": 0}]
|
[{"Id": 91776448, "KernelVersionId": 69046687, "SourceDatasetVersionId": 2455641}]
|
[{"Id": 2455641, "DatasetId": 1486121, "DatasourceVersionId": 2498036, "CreatorUserId": 1680925, "LicenseName": "Unknown", "CreationDate": "07/23/2021 15:24:33", "VersionNumber": 3.0, "Title": "quora-question-pair-hand-annotated-dataset", "Slug": "quoraquestionpairhandannotateddataset", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Simulated version", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1486121, "CreatorUserId": 1680925, "OwnerUserId": 1680925.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2480458.0, "CurrentDatasourceVersionId": 2522984.0, "ForumId": 1505812, "Type": 2, "CreationDate": "07/23/2021 13:36:24", "LastActivityDate": "07/23/2021", "TotalViews": 1376, "TotalDownloads": 8, "TotalVotes": 2, "TotalKernels": 2}]
|
[{"Id": 1680925, "UserName": "huikang", "DisplayName": "Tong Hui Kang", "RegisterDate": "03/01/2018", "PerformanceTier": 2}]
|
# This notebook covers
# - Dataset Preparation (train-test split)
# - TF-IDF indexes
# - Evaluation algorithm
# - Evaluation procedure with the test set
# - Update indexes with unseen questions
# - Query with unseen questions
# The following process is done on another notebook
# - Spellcheck and SpaCy tokenisation for the training set
# - SentenceTransformer computation of vectors
# - Downloading of the SpaCy and GenSim models
# notebook hyperparameters
TEST_SET_SIZE = 1000
RANKED_LIST_SIZE = 100
RANDOM_STATE = 42
EVALUATING = True # make False if you want to run query quickly
import os, collections, random, itertools, functools, time
from collections import defaultdict, Counter
from math import log
import tqdm.notebook as tqdm
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
random.seed(RANDOM_STATE)
np.random.seed(RANDOM_STATE)
# load data
df = pd.read_csv("/kaggle/input/quora-question-pairs/train.csv.zip")
df["question1"] = df["question1"].astype(str) # resolve nan
df["question2"] = df["question2"].astype(str)
df["qid1"] -= 1 # start index from zero
df["qid2"] -= 1
df.sample(10)
# # Preprocessing Dataset
# all questions are identified with its qid
qid_to_question = {}
for qid1, qid2, question1, question2 in zip(
df["qid1"], df["qid2"], df["question1"], df["question2"]
):
qid_to_question[qid1] = question1
qid_to_question[qid2] = question2
# extract 1000 questions for testing
test_query_qids = set()
df_duplicate = df[df["is_duplicate"] == 1].sample(frac=1, random_state=RANDOM_STATE)
for qid1, qid2, is_duplicate in zip(
df_duplicate["qid1"], df_duplicate["qid2"], df_duplicate["is_duplicate"]
):
if (
is_duplicate
and qid1 not in test_query_qids
and len(test_query_qids) < TEST_SET_SIZE
):
test_query_qids.add(qid2)
if qid1 in test_query_qids and qid2 in test_query_qids:
# to guarantee that there is a duplicate question in the training set
test_query_qids.remove(qid1)
test_query_qids.remove(qid2)
assert len(test_query_qids) == TEST_SET_SIZE # if fail, change random_state
test_query_qids_list = sorted(test_query_qids)
train_query_qids_list = sorted(set(qid_to_question.keys()) - test_query_qids)
assert test_query_qids_list[:3] == [331, 489, 501] # to check random state fixed
# # uncomment this to test only limited queries
if not EVALUATING:
test_query_qids_list = test_query_qids_list[:10]
TEST_SET_SIZE = 10
# extract duplicate relationship of training set
qid_to_duplicate_qids = defaultdict(set)
qid_to_nonduplicate_qids = defaultdict(set)
for qid1, qid2, is_duplicate in zip(df["qid1"], df["qid2"], df["is_duplicate"]):
if not (qid1 in test_query_qids or qid2 in test_query_qids):
if is_duplicate:
qid_to_duplicate_qids[qid1].add(qid2)
qid_to_duplicate_qids[qid2].add(qid1)
else:
qid_to_nonduplicate_qids[qid1].add(qid2)
qid_to_nonduplicate_qids[qid2].add(qid1)
# complete graph of duplicate relationships
qid_to_duplicate_qids_complete = defaultdict(set)
qid_to_qid_group_leader = {}
qid_group_leader_to_duplicate_qid_group = defaultdict(set)
visited_qids = set()
for train_qid in train_query_qids_list:
if train_qid in visited_qids:
continue
current_qids_group = set([train_qid])
qid_to_qid_group_leader[train_qid] = train_qid
stack = [train_qid]
while stack:
cur_qid = stack.pop()
for nex_qid in qid_to_duplicate_qids[cur_qid]:
if nex_qid in current_qids_group:
continue
qid_to_qid_group_leader[nex_qid] = train_qid
stack.append(nex_qid)
current_qids_group.add(nex_qid)
# complete the graph
for qid1, qid2 in itertools.combinations(current_qids_group, r=2):
qid_to_duplicate_qids_complete[qid1].add(qid2)
qid_to_duplicate_qids_complete[qid2].add(qid1)
qid_group_leader_to_duplicate_qid_group[train_qid] = current_qids_group
visited_qids.update(current_qids_group)
# extract duplicate relationship of the test set
test_qid_to_duplicate_qids = defaultdict(set)
test_qid_to_duplicate_qids_complete = defaultdict(set)
for qid1, qid2, is_duplicate in zip(
df_duplicate["qid1"], df_duplicate["qid2"], df_duplicate["is_duplicate"]
):
if qid2 in test_query_qids:
qid1, qid2 = qid2, qid1
if qid1 in test_query_qids:
if qid2 in test_query_qids:
continue
test_qid_to_duplicate_qids[qid1].add(qid2)
test_qid_to_duplicate_qids_complete[qid1].add(qid2)
for train_qid in qid_group_leader_to_duplicate_qid_group[
qid_to_qid_group_leader[qid2]
]:
test_qid_to_duplicate_qids_complete[qid1].add(train_qid)
# count inconsistencies in dataset
cnt = 0
for qid1, qid2, is_duplicate in zip(df["qid1"], df["qid2"], df["is_duplicate"]):
if not is_duplicate and qid1 not in test_query_qids and qid2 not in test_query_qids:
if qid_to_qid_group_leader[qid1] == qid_to_qid_group_leader[qid2]:
cnt += 1
print(
"Number of inconsistencies: ", cnt
) # slightly smaller than 96 because some edges are associated with the test set
test_mask = (df["qid1"].isin(test_query_qids)) | (df["qid2"].isin(test_query_qids))
train_df = df[~test_mask].copy()
test_df = df[test_mask].copy()
# clean up
del qid_to_qid_group_leader, qid_group_leader_to_duplicate_qid_group
del cnt
del test_query_qids # not sorted, use test_query_qids_list
del df # all data you can train on is in train_df
# enable use of complete graphs
test_qid_to_duplicate_qids = test_qid_to_duplicate_qids_complete
qid_to_duplicate_qids = qid_to_duplicate_qids_complete
# # Evaluation Metrics
def method_random_guess(test_qid):
# returns ranklist and scores of each size RANKED_LIST_SIZE
return (
random.choices(train_query_qids_list, k=RANKED_LIST_SIZE),
[0] * RANKED_LIST_SIZE,
)
# 1000 x 100 (the ranked list of similar qn for each of the 1000 test qns)
ranklists_method_random_guess = [
method_random_guess(test_qid)[0] for test_qid in test_query_qids_list
]
def show_sample_query_results(
test_qid, method_ranklist, method_scores=[0] * RANKED_LIST_SIZE, num_to_show=10
):
# not a metric, just print a few examples and its scores
print("Query: {}".format(qid_to_question[test_qid]))
for rank, (score, result_qid) in enumerate(
zip(method_scores, method_ranklist[:num_to_show]), start=1
):
relevance = (
"Registered"
if result_qid in test_qid_to_duplicate_qids[test_qid]
else "Unregistered"
)
print(
"Rank {} - Score {:.4f} - {}: \t{}".format(
rank, score, relevance, qid_to_question[result_qid]
)
)
show_sample_query_results(
test_query_qids_list[0], *method_random_guess(test_query_qids_list[0])
)
def evaluation_with_first_relevant_rank(
method_ranklists, considered=1, eps=10**-6, debug=True, **kwargs
):
# calculation of the statistics of the rank of the first c=considered duplicates
# if the duplicate does not appear in the ranklist, it has a default rank of RANKED_LIST_SIZE
assert np.array(method_ranklists).shape == (TEST_SET_SIZE, RANKED_LIST_SIZE)
reciprocal_ranks = []
ranks = []
for test_qid, ranklist in zip(test_query_qids_list, method_ranklists):
test_qid_to_rank = {
result_qid: rank for rank, result_qid in enumerate(ranklist, start=1)
}
rank = [] # may be shorter than `considered` because of lack of duplicates
for expected_qid in test_qid_to_duplicate_qids[test_qid]:
if expected_qid in test_qid_to_rank:
rank.append(test_qid_to_rank[expected_qid])
else:
rank.append(RANKED_LIST_SIZE + 1)
rank.sort()
ranks.extend(rank[:considered])
if rank[0] > RANKED_LIST_SIZE:
reciprocal_ranks.append(0)
else:
reciprocal_ranks.append(1 / rank[0])
plt.figure(figsize=(14, 4))
plt.title("Highest rank of duplicate question")
plt.hist(ranks, bins=np.arange(RANKED_LIST_SIZE + 2))
plt.xlabel("Rank")
plt.ylabel("Frequency")
plt.show()
mrr = sum(reciprocal_ranks) / len(reciprocal_ranks)
har = 1 / (mrr + eps)
print(f"Mean Reciprocal Rank (MRR) is {mrr:.2f}")
print(f"Harmonic Average Rank (HAR) is {har:.2f}")
p50 = np.median(ranks)
proportion_out_of_result = ranks.count(RANKED_LIST_SIZE + 1) / len(ranks)
if debug:
print("Median rank: {:.2f}".format(p50))
print("Proportion out of result: {:.3f}".format(proportion_out_of_result))
return mrr, har, p50, proportion_out_of_result
_ = evaluation_with_first_relevant_rank(ranklists_method_random_guess)
def evaluation_with_auc(method_ranklists, k=10, weights=None, debug=True, **kwargs):
assert np.array(method_ranklists).shape == (TEST_SET_SIZE, RANKED_LIST_SIZE)
counts = np.array([0.0] * k)
## Identify duplicates among top K ranks for each test
for i, (test_qid, ranklist) in enumerate(
zip(test_query_qids_list, method_ranklists)
):
topk = ranklist[:k]
is_duplicate = np.array(
[
1 if (result_qid in test_qid_to_duplicate_qids[test_qid]) else 0
for result_qid in topk
]
)
counts += is_duplicate
## Calculate AUC
if weights:
counts *= np.array(weights) / sum(weights)
else:
counts /= k
auc = sum(counts) / (TEST_SET_SIZE)
if debug:
print(f"{auc:.2%} of top {k} results are duplicates")
return auc # between [0,1], 1 is perfect
_ = evaluation_with_auc(ranklists_method_random_guess)
_ = evaluation_with_auc(
ranklists_method_random_guess, weights=[10, 9, 8, 7, 6, 5, 4, 3, 2, 1]
)
def single_r_precision(test_qid, ranklist):
# use this to check a single test query
num_duplicate = len(
test_qid_to_duplicate_qids[test_qid]
) # this dict needs to be updated when train:test set separation is updated
if num_duplicate == 0:
return 0, 0, 0
top_r = ranklist[:num_duplicate]
num_duplicates_in_top_r = sum(
[
1 if (result_qid in test_qid_to_duplicate_qids[test_qid]) else 0
for result_qid in top_r
]
)
r_precision = num_duplicates_in_top_r / num_duplicate
return num_duplicate, num_duplicates_in_top_r, r_precision
def evaluation_with_r_precision(
method_ranklists, k=10, report_k=0, debug=True, **kwargs
):
print(np.array(method_ranklists).shape)
assert np.array(method_ranklists).shape == (
TEST_SET_SIZE,
RANKED_LIST_SIZE,
) # method_ranklists size is (1000,100)
total_num_duplicates = np.array([0 for i in range(TEST_SET_SIZE)])
r_precision = np.array([0 for i in range(TEST_SET_SIZE)])
## Iter over 1->1000 tests
for i, (test_qid, ranklist) in enumerate(
zip(test_query_qids_list, method_ranklists)
): # iter over 1->1000 tests
(
total_num_duplicates[i],
num_duplicates_in_top_r,
r_precision[i],
) = single_r_precision(test_qid, ranklist)
# note: if want do error analysis, intervene here to find test cases with low r precision
if report_k > 0:
k_lowest_r_precision_idx = np.argpartition(r_precision, k)[:k]
k_lowest_r_precision_test_qids = np.array(test_query_qids_list)[
k_lowest_r_precision_idx
]
## Calculate metrics
avg_r_precision = r_precision.mean()
weighted_avg_r_precision = (
np.multiply(r_precision, total_num_duplicates).sum()
/ total_num_duplicates.sum()
)
if debug:
print(f"Average R-Precision = {avg_r_precision:.2%}")
print(
f"Weighted Average R-Precision by proportion of duplicates = {weighted_avg_r_precision:.2%}"
)
if avg_r_precision > weighted_avg_r_precision:
print(
"A higher average R-Precisions suggests that there are many test queries with high R-Precision but there are some test queries with high number of duplicates that model is not effective with."
)
if not report_k:
return avg_r_precision, weighted_avg_r_precision
else:
return avg_r_precision, weighted_avg_r_precision, k_lowest_r_precision_test_qids
_ = evaluation_with_r_precision(ranklists_method_random_guess, k=10)
def evaluation_with_precision_recall_at_k(
method_ranklists,
k=10,
exclude_precision=False,
exclude_recall=False,
debug=True,
**kwargs,
):
assert np.array(method_ranklists).shape == (TEST_SET_SIZE, RANKED_LIST_SIZE)
## Evaluation returns the macro average P@K and R@Kfor test set
## Interpretation P@K: what % of top k retrieved is relevant?
## Interpretation R@K: what % of all duplicates for query is retrieved within top k?
## Iter thru each test
precisions_at_k = []
recalls_at_k = []
for i, (test_qid, ranklist) in enumerate(
zip(test_query_qids_list, method_ranklists)
):
## 1. Set rank threshold K, ignore all docs after K
## 2. Count num_relevant in top-K
## 3. Count total_num_duplicates_for_query
## 4. P@K = num_relevant/k
## 5. R@K = num_relevant/total_num_duplicates_for_query
topk = ranklist[:k]
num_relevant = sum(
[
1 if (result_qid in test_qid_to_duplicate_qids[test_qid]) else 0
for result_qid in topk
]
)
precision_at_k = num_relevant / k
precisions_at_k.append(precision_at_k)
total_num_duplicates_for_query = len(test_qid_to_duplicate_qids[test_qid])
recall_at_k = num_relevant / total_num_duplicates_for_query
recalls_at_k.append(recall_at_k)
mean_precision_at_k = sum(precisions_at_k) / len(precisions_at_k) # macro average
mean_recall_at_k = sum(recalls_at_k) / len(recalls_at_k) # macro average
print(f"Macro Average Precision@k={k} is {mean_precision_at_k:.2%}")
print(f"Macro Average Recall@k={k} is {mean_recall_at_k:.2%}")
return (mean_precision_at_k, mean_recall_at_k)
_ = evaluation_with_precision_recall_at_k(ranklists_method_random_guess, k=10)
def evaluation_with_map(method_ranklists, debug=True, **kwargs):
assert np.array(method_ranklists).shape == (TEST_SET_SIZE, RANKED_LIST_SIZE)
## Interpretation: what is the average precision for all relevant docs across all queries?
## Iter thru each test
average_precisions = []
for i, (test_qid, ranklist) in enumerate(
zip(test_query_qids_list, method_ranklists)
):
## 1. Find the rank positions of each of the R relevant docs: K1, K2, ... KR and sort
## 2. Compute P@K for each K1, K2, ... If K >=RANKED_LIST_SIZE, assume never retrieved
## 3. AP = average of P@K for query
## 4. MAP = macro average of AP across queries
## 1. Find the rank positions of each of the R relevant docs: K1, K2, ... and sort
dup_qids_in_train_set = [
dup_qid
for dup_qid in test_qid_to_duplicate_qids[test_qid]
if dup_qid in train_query_qids_list
] # find all the dup_qid that can be found in the train set so you know total dup qn that could be found
total_num_dup_qid = len(dup_qids_in_train_set) # how many dup qn to expect
dup_ranks = []
for dup_qid in dup_qids_in_train_set:
if dup_qid not in ranklist: # not found
dup_ranks.append(
RANKED_LIST_SIZE
) # give "out of range" rank which would be checked later during calculation
continue
dup_ranks.append(
list(ranklist).index(dup_qid) + 1
) # append the rank of the retrieved dup qn
dup_ranks, dup_qids_in_train_set = (
list(t) for t in zip(*sorted(zip(dup_ranks, dup_qids_in_train_set)))
) # sort by rank
## 2. Compute P@K for each K1, K2, ... If K >=RANKED_LIST_SIZE, assume never retrieved
precisions_at_k = []
for j, rank in enumerate(dup_ranks, start=1): # dup_ranks is sorted
if rank >= RANKED_LIST_SIZE: # handle "unretrieved" duplicates
precisions_at_k.append(0)
else:
precision_at_k = j / rank # = num_dup_so_far / rank_of_latest_dup_found
precisions_at_k.append(precision_at_k)
## 3. AP = average of P@K for query
average_precisions.append(sum(precisions_at_k) / len(precisions_at_k))
## Out of test query loop
## 4. MAP = macro average of AP across queries
MAP = sum(average_precisions) / len(average_precisions)
print(f"Mean Average Precision (MAP) is {MAP:.2%}")
return MAP
_ = evaluation_with_map(ranklists_method_random_guess)
def evaluation_process(method, **kwargs):
# executes the method and runs the evaluation functions
ranklists, scorelists = [], []
for test_qid in tqdm.tqdm(test_query_qids_list):
ranklist, scores = method(test_qid)
ranklists.append(ranklist)
scorelists.append(scores)
evaluation_with_first_relevant_rank(ranklists, **kwargs)
# evaluation_with_auc(ranklists, **kwargs)
evaluation_with_r_precision(ranklists, **kwargs)
evaluation_with_precision_recall_at_k(ranklists, k=10, **kwargs)
evaluation_with_map(ranklists, **kwargs)
return ranklists, scorelists
results_random_guess = evaluation_process(method_random_guess)
# # Preprocessing
## This entire cell is important to enable tokeniser pipeline
## Use this to replace tokenise function if using Tokenise then Spellcheck (TSC) pipeline
######### spacy basic tokenizer
import spacy
print("Spacy version: ", spacy.__version__)
from spacy.tokenizer import Tokenizer # https://spacy.io/api/tokenizer
# !python3 -m spacy download en_core_web_sm
print("Loading Spacy en_core_web_sm loaded")
nlp = spacy.load("en_core_web_sm")
tokenizer = Tokenizer(nlp.vocab)
tokenizer.add_special_case(
"[math]", [{"ORTH": "[math]"}]
) # see qid=7: '[math]23^{24}[/math]' becomes one token
# add more special cases here if found
def spacy_tokenise(text, lower=False, split_last_punc=True):
"""
returns a list of tokens given a question text
note: each punctuation is also considered a token
note: "\n" is a token
note: "'s" is a token
note: '(Koh-i-Noor)' is a token
see tokenizer instantiation code for special cases or to add
lowercase text only after spell check
"""
if lower:
text = text.lower()
tokens = tokenizer(text)
token_list = [token.text for token in tokens]
# further split tokens that end with certain punct e.g. "me?" => "me", "?"
if split_last_punc:
split_lists = [
[token[:-1], token[-1]] if (token[-1] in ["!", "?", ",", ":"]) else [token]
for token in token_list
]
token_list = [token for sublist in split_lists for token in sublist]
return token_list
######### symspell spellchecker
print("Loading symspell")
from symspellpy.symspellpy import (
SymSpell,
Verbosity,
) # https://github.com/mammothb/symspellpy
import pkg_resources
# instantiate spellchecker
sym = SymSpell(max_dictionary_edit_distance=2, prefix_length=7, count_threshold=1)
# https://symspellpy.readthedocs.io/en/latest/api/symspellpy.html
dictionary_path = pkg_resources.resource_filename(
"symspellpy", "frequency_dictionary_en_82_765.txt"
)
sym.load_dictionary(dictionary_path, 0, 1) # might take a short while
def spellcheck_single(word):
# returns top correct spelling or the same word if no correction found within max_edit_distance
if not word.isascii():
return word # do not spellcheck non ascii words e.g. シ
# obtain list of suggestions
suggestions = sym.lookup(
word,
Verbosity.CLOSEST,
max_edit_distance=2,
include_unknown=True, # a mispelled word with no found corrections is returned as is
ignore_token=r"[:,.!?\\-]", # use if want to avoid correcting certain phrases
)
# get the term from the suggestItem object
suggested_words = [suggestion._term for suggestion in suggestions]
# check if the input word is legit and return if so else return corrected word
word_lower = word.lower()
if word_lower in suggested_words:
return word_lower # do not correct if input is a legit word
else:
return suggested_words[0] # top suggestion
def spellcheck_compound(sent):
# spellchecks a sentence
suggestions = sym.lookup_compound(sent, max_edit_distance=2)
return suggestions[0]._term # returns the top suggestion
######### tokenise pipeline
def tokenise_then_spellcheck(sent):
# 8 times faster than spellcheck_then_tokenise
tokens = spacy_tokenise(sent) # NOTE: replace tokenise with spacy_tokenise
checked_tokens = [
spellcheck_single(token).lower() for token in tokens
] # lower after spell check
return checked_tokens
def spellcheck_then_tokenise(sent):
checked_sent = spellcheck_compound(sent)
tokens = spacy_tokenise(checked_sent, lower=True) # lower after spell check
return tokens
# define tokenisation process
import pickle
qid_to_tokens_preprocessed_filename = "../input/quora-question-pairs-tokenise-pipeline/qid_to_processed_token_list_tokenise_then_spellcheck.pkl"
with open(qid_to_tokens_preprocessed_filename, "rb") as f:
qid_to_tokens_preprocessed = pickle.load(f)
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
stopword_set = set(stopwords.words())
stopword_set.update(["?", ","])
def nltk_tokenize(sentence):
return word_tokenize(sentence.lower())
def tokenise_qid(
qid,
qid_to_tokens_preprocessed=qid_to_tokens_preprocessed,
tokenise_method=tokenise_then_spellcheck,
):
# return a list of tokens, does not remove stopwords or duplicates
if qid_to_tokens_preprocessed and qid in qid_to_tokens_preprocessed:
return qid_to_tokens_preprocessed[qid]
return tokenise_method(qid_to_question[qid])
def preprocess_vsm(
train_query_qids_list=train_query_qids_list,
stopword_set=stopword_set,
exclude_stopwords=True,
):
"""
Input:
qid_to_question = {qid: question string}
Note: only use the test subset of qids
Outputs:
qid_to_tokens = {qid: set(tokens)}
token_to_qids = {token: set(qids)}
tf = {token: {qid: TF as int}}
df = {token: DF as int}
L = {qid: question length as int}
"""
qid_to_tokens = defaultdict(set)
token_to_qids = defaultdict(set)
tf = defaultdict(Counter)
df = defaultdict(int)
L = defaultdict(int)
qid_processed = set()
for qid in tqdm.tqdm(train_query_qids_list):
qid_tokenised = tokenise_qid(qid)
for token in set(qid_tokenised):
if token not in stopword_set or not exclude_stopwords:
# store qid-to-token mapping
qid_to_tokens[qid].add(token)
token_to_qids[token].add(qid)
# compute and store term frequency
tf[token][qid] += 1
# store doc frequency in df
df[token] += 1
# store doc length in L (double-count repeated tokens)
L[qid] = len(qid_tokenised)
# output
return qid_to_tokens, token_to_qids, tf, df, L
qid_to_tokens, token_to_qids, tf, df, L = preprocess_vsm()
# save a copy of the original to allow reset later
qid_to_tokens_original, token_to_qids_original = (
qid_to_tokens.copy(),
token_to_qids.copy(),
)
tf_original, df_original, L_original = tf.copy(), df.copy(), L.copy()
# # Baseline - Overlapping Root Word Count (Working Title)
# Order by the number of overlapping non-stopword words. Random if tie.
def method_overlapping_root_word_count(query_qid, ignore_stopwords=True):
query_tokens = set(tokenise_qid(query_qid))
if ignore_stopwords:
query_tokens = [token for token in query_tokens if token not in stopword_set]
counter = collections.Counter()
for dummy_qid in random.choices(train_query_qids_list, k=RANKED_LIST_SIZE):
# prefill with random results to address the possibility of no matches
counter[dummy_qid] = 0.01
for query_token in query_tokens:
counter += collections.Counter(token_to_qids[query_token])
query_results = list(counter.items())
random.shuffle(query_results) # so that qids are not ordered
query_results = sorted(query_results, key=lambda x: x[1], reverse=True)[
:RANKED_LIST_SIZE
]
return [x[0] for x in query_results], [x[1] for x in query_results]
show_sample_query_results(
test_query_qids_list[0],
*method_overlapping_root_word_count(test_query_qids_list[0]),
)
results_overlapping_root_word_count = evaluation_process(
method_overlapping_root_word_count
)
# # TFIDF
def compute_idf(doc_freq, N):
"""
Inputs:
doc_freq = document frequency of some token
N = corpus size including query
Output:
idf = IDF as float
"""
return log(N / doc_freq)
from functools import reduce
import operator
def prod(iterable):
return reduce(operator.mul, iterable, 1)
def use_vsm(
qid_query, # qid_to_tokens=qid_to_tokens, tf=tf, df=df, L=L,
method="tf-idf",
compute_idf=compute_idf,
k1=1.5,
k3=1.5,
b=0.75,
smoothing="add-one",
alpha=0.75,
exclude_stopwords=True,
return_top=RANKED_LIST_SIZE,
):
"""
Inputs:
query = question string to match # this comes from "test" set
qid_to_tokens = {qid: set(tokens)} # this is the "training" corpus
tf = {token: term freq} # required for all methods
df = {token: doc freq} # required for method='tf-idf','bm25'
L = {qid: doc length} # required for method='bm25','unigram'
method = model to apply
k1, k3, b = tuning params # required for method='bm25'
smoothing = type of smoothing # required for method='unigram'
return_top = num of docs to return
Procedure:
0. Corpus is already tokenised, tf, df, L already computed
1. Tokenise query, expand tf, df, L with query information
if method='boolean':
Remove idf calculation, then use method='tf-idf'
if method='tf-idf':
2. Compute tf-idf weights only for relevant (t,d) pairs
3. Compute cosine similarity only for docs containing query terms
if method='bm25':
2. Compute RSV summation terms only for relevant (t,d) pairs
3. Compute RSV only for docs containing query terms
if method='unigram':
2. Compute probabilities only for relevant (t,d) pairs
3. Compute query probability only for docs containing query terms
4. Return docs in ranked order
Output:
ranking = [qids in decreasing order of match]
scoring = [corresponding scores]
"""
assert method in [
"boolean",
"tf-idf",
"bm25",
"unigram",
], "Supported methods: 'boolean', 'tf-idf', 'bm25', 'unigram'"
assert len(L.keys()) > 0 if method == "bm25" else True, "Please include L for bm25"
assert (
len(L.keys()) > 0 if method == "unigram" else True
), "Please include L for unigram"
assert (
smoothing in ["add-one", "linear-interpolation"]
if method == "unigram"
else True
)
assert alpha >= 0 and alpha <= 1 if smoothing == "linear-interpolation" else True
qid_tmp = time.time()
""" STEP 1: PROCESS QUERY """
query_tokenised = tokenise_qid(qid_query)
for token in set(query_tokenised):
if token not in stopword_set or not exclude_stopwords:
# store qid-to-token mapping
# store query as qid=0 (corpus starts from qid=1)
qid_to_tokens[qid_tmp].add(token)
# compute and store term frequency
tf[token][qid_tmp] = sum([1 if t == token else 0 for t in query_tokenised])
# update doc frequency in df
df[token] += 1
# store query length
L[qid_tmp] = len(query_tokenised)
if method == "boolean":
def compute_idf(doc_freq, N):
return 1
method = "tf-idf"
if method == "tf-idf":
"""STEP 2: COMPUTE TF-IDF WEIGHTS"""
weights = defaultdict(lambda: defaultdict(float))
N = len(qid_to_tokens) # original corpus + query
# only bother computing for tokens in the query
for token in set(query_tokenised):
if token not in stopword_set or exclude_stopwords == False:
weights[qid_tmp][token] = tf[token][qid_tmp] * compute_idf(df[token], N)
for qid in tf[token].keys():
weights[qid][token] = tf[token][qid] * compute_idf(df[token], N)
# also compute weight for other tokens contained by these qids
# needed for computing qid vector length
for other_token in qid_to_tokens[qid]:
weights[qid][other_token] = tf[other_token][qid] * compute_idf(
df[other_token], N
)
""" STEP 3: COMPUTE COSINE SIMILARITY TO QUERY """
cosine_similarities = defaultdict(float)
# compute denominator (part 1), i.e., |q| * |d|
query_vector_length = (sum([w**2 for w in weights[qid_tmp].values()])) ** 0.5
if query_vector_length == 0:
print(f"query={query}\nweights[qid_tmp].items()={weights[qid_tmp].items()}")
for qid in weights.keys():
# compute numerator, i.e., dot product of q and d
cosine_numerator = 0
for token in weights[qid].keys():
if token in weights[qid_tmp]:
cosine_numerator += weights[qid][token] * weights[qid_tmp][token]
# compute denominator (part 2), i.e., |q| * |d|
qid_vector_length = (sum([w**2 for w in weights[qid].values()])) ** 0.5
if qid_vector_length == 0: # example: qid=25026 => question='?'
qid_vector_length = 1e-8
# compute and store cosine similarity between q and d
cosine_similarities[qid] = (
cosine_numerator / query_vector_length / qid_vector_length
)
scores = cosine_similarities
if method == "bm25":
"""STEP 2: COMPUTE RSV TERMS"""
rsv_terms = defaultdict(lambda: defaultdict(float))
N = len(qid_to_tokens) # original corpus + query
L_avg = sum(L.values()) / len(L.values())
# only bother computing for tokens in the query
for token in set(query_tokenised):
for qid in tf[token].keys():
rsv_terms[qid][token] = (
compute_idf(df[token], N)
* (k1 + 1)
* tf[token][qid]
/ (k1 * ((1 - b) + b * L[qid] * L_avg) + tf[token][qid])
* (k3 + 1)
* tf[token][qid_tmp]
/ (k3 + tf[token][qid_tmp])
)
""" STEP 3: COMPUTE RSV """
rsv = {qid: sum(rsv_terms[qid].values()) for qid in rsv_terms.keys()}
scores = rsv
if method == "unigram":
"""STEP 2: COMPUTE PROBABILITIES"""
probabilities = defaultdict(lambda: defaultdict(float))
corpus_model = defaultdict(float)
# only bother computing for tokens in the query
for token in set(query_tokenised):
for qid in tf[token].keys():
if smoothing == "add-one":
probabilities[qid][token] = (tf[token][qid] + 1) / (
L[qid] + len(query_tokenised)
)
else:
probabilities[qid][token] = (tf[token][qid]) / (L[qid])
# for linear-interpolation smoothing, build corpus language model
if smoothing == "linear-interpolation":
corpus_model[token] += tf[token][qid]
# remaining operations for linear-interpolation smoothing
if smoothing == "linear-interpolation":
# finish building corpus language model by dividing corpus tf by corpus L
total_corpus_length = sum(L.values())
for token in corpus_model.keys():
corpus_model[token] = corpus_model[token] / total_corpus_length
# then update the probabilities
for qid in probabilities.keys():
for token in probabilities[qid].keys():
probabilities[qid][token] = (
alpha * probabilities[qid][token]
+ (1 - alpha) * corpus_model[token]
)
""" STEP 3: COMPUTE QUERY PROBABILITY """
query_prob = {
qid: -log(prod(probabilities[qid].values())) for qid in probabilities.keys()
}
scores = query_prob
""" STEP 4: RANK DOCUMENTS AND RETURN RESULT """
# cleanup
del qid_to_tokens[qid_tmp]
for token in set(query_tokenised):
if token not in stopword_set or not exclude_stopwords:
del tf[token][qid_tmp]
df[token] -= 1
del scores[qid_tmp] # remove query from result
ranking = sorted(scores, key=scores.get, reverse=True)
scoring = sorted(scores.values(), reverse=True)
# if too few documents match the query, add dummy documents
if len(ranking) < return_top:
ranking.extend([0] * (return_top - len(ranking)))
scoring.extend([0] * (return_top - len(ranking)))
# return top k results
return ranking[:return_top], scoring[:return_top]
def method_boolean(qid):
return use_vsm(qid, method="boolean")
show_sample_query_results(
test_query_qids_list[0], *method_boolean(test_query_qids_list[0])
)
results_boolean = evaluation_process(method_boolean)
def method_tf_idf(qid):
return use_vsm(qid, method="tf-idf")
show_sample_query_results(
test_query_qids_list[0], *method_tf_idf(test_query_qids_list[0])
)
results_tf_idf = evaluation_process(method_tf_idf)
# # BM25
def method_bm25(qid):
return use_vsm(qid, method="bm25")
show_sample_query_results(
test_query_qids_list[0], *method_bm25(test_query_qids_list[0])
)
results_bm25 = evaluation_process(method_bm25)
def method_unigram(qid):
return use_vsm(qid, method="unigram", smoothing="add-one")
show_sample_query_results(
test_query_qids_list[0], *method_unigram(test_query_qids_list[0])
)
results_unigram = evaluation_process(method_unigram)
for alpha in [0, 0.25, 0.5, 0.75, 1.0]:
def method_unigram(qid):
return use_vsm(
qid, method="unigram", smoothing="linear-interpolation", alpha=alpha
)
_ = evaluation_process(method_unigram)
# # Word Embeddings - SpaCy
def to_vec(token_or_list):
# converts a token string or a list of tokens into a word or doc vec respectively
if type(token_or_list) == list:
# token list needs to be joined into a sentence first
token_or_list = " ".join(token_or_list)
return nlp(token_or_list).vector
nlp2 = spacy.load("en_core_web_lg")
def to_vec2(token_or_list):
# converts a token string or a list of tokens into a word or doc vec respectively
if type(token_or_list) == list:
# token list needs to be joined into a sentence first
token_or_list = " ".join(token_or_list)
return nlp2(token_or_list).vector
# Load pre-processed dict
with open("../input/quora-question-pairs-tokenise-pipeline/qid_to_vec.pkl", "rb") as f:
qid_to_vec = pickle.load(f)
print("Pre-processed question vector is of shape {}".format(qid_to_vec[0].shape))
from numpy import dot
from numpy.linalg import norm
def method_spacy_embedding_similarity(test_qid):
tokens = tokenise_then_spellcheck(qid_to_question[test_qid])
test_vec = to_vec(tokens)
## Run baseline model as a filter
qid_list, scores = method_overlapping_root_word_count(test_qid)
cos_sims = [] # bigger better
for train_qid in qid_list: # train_query_qids_list:
train_vec = qid_to_vec[train_qid]
cos_sim = dot(test_vec, train_vec) / (norm(test_vec) * norm(train_vec))
cos_sims.append(cos_sim)
cos_sims = np.array(cos_sims)
qid_list = np.array(qid_list) # train_query_qids_list)
inds = cos_sims.argsort()[::-1] # reverse so biggest come first
ranklist = qid_list[inds]
return ranklist[:RANKED_LIST_SIZE], cos_sims[:RANKED_LIST_SIZE]
show_sample_query_results(
test_query_qids_list[0], *method_spacy_embedding_similarity(test_query_qids_list[0])
)
results_spacy_embedding_similarity = evaluation_process(
method_spacy_embedding_similarity
)
with open(
"../input/quora-question-pairs-tokenise-pipeline/qid_to_vec_trf.pkl", "rb"
) as f: # note, actually lg not trf
qid_to_vec2 = pickle.load(f)
print(
"Pre-processed question vector is of shape {}".format(qid_to_vec2[0].shape)
) # 300 dim vec
def method_spacy_embedding_similarity_lg(test_qid):
tokens = tokenise_then_spellcheck(qid_to_question[test_qid])
test_vec = to_vec2(tokens)
## Run baseline model as a filter
qid_list, scores = method_overlapping_root_word_count(test_qid)
cos_sims = [] # bigger better
for train_qid in qid_list: # train_query_qids_list:
train_vec = qid_to_vec2[train_qid]
cos_sim = dot(test_vec, train_vec) / (norm(test_vec) * norm(train_vec))
cos_sims.append(cos_sim)
cos_sims = np.array(cos_sims)
qid_list = np.array(qid_list) # train_query_qids_list)
inds = cos_sims.argsort()[::-1] # reverse so biggest come first
ranklist = qid_list[inds]
return ranklist[:RANKED_LIST_SIZE], cos_sims[:RANKED_LIST_SIZE]
show_sample_query_results(
test_query_qids_list[0],
*method_spacy_embedding_similarity_lg(test_query_qids_list[0]),
)
results_spacy_embedding_similarity_lg = evaluation_process(
method_spacy_embedding_similarity_lg
)
# # Gensim WordMover Distance on Boolean Retrieval
# * Applies further sorting by wordmover distance on the output ranklist of Boolean Retrieval
# * Current pre-trained model: `glove-wiki-gigaword-50`
#
import gensim
import gensim.downloader
# gensim.downloader.info() # find more models to download
from gensim.models import KeyedVectors
try:
model = KeyedVectors.load(
"../input/ir-project-download-keyed-vectors/glove-wiki-gigaword-50.keyedvectors"
)
except: # gs_model not downloaded
model = gensim.downloader.load("glove-wiki-gigaword-50")
# model.save("/kaggle/working/glove-wiki-gigaword-50.keyedvectors") # if not already saved
def method_wordmover_distance(test_qid, model):
# out of box duplicate finder does not work!
# returns ranklist and scores of each size RANKED_LIST_SIZE
## Run baseline model as a filter
ranklist, scores = method_overlapping_root_word_count(test_qid)
## Process test question
test_qn = tokenise_qid(test_qid)
## Get wordmover distance from every candidate
distances = []
qid_list = ranklist
for candidate_qid in qid_list:
candidate_qn = tokenise_qid(candidate_qid)
distances.append(model.wmdistance(test_qn, candidate_qn))
## Sort by distance
sorted_dist_and_candidate_qid = sorted(zip(distances, qid_list))
sorted_candidate_qid = [qid for _, qid in sorted_dist_and_candidate_qid]
sorted_dist = [dist for dist, _ in sorted_dist_and_candidate_qid]
return sorted_candidate_qid[:RANKED_LIST_SIZE], sorted_dist[:RANKED_LIST_SIZE]
def method_wordmover_distance_glovewiki50(test_qid):
return method_wordmover_distance(test_qid, model)
show_sample_query_results(
test_query_qids_list[0],
*method_wordmover_distance_glovewiki50(test_query_qids_list[0]),
)
results_wordmover_distance_glovewiki50 = evaluation_process(
method_wordmover_distance_glovewiki50
)
models_to_try = [
"glove-wiki-gigaword-300",
"glove-twitter-50",
"word2vec-google-news-300",
"fasttext-wiki-news-subwords-300",
]
if not EVALUATING:
models_to_try = []
for m in models_to_try:
print("Model: ", m)
try:
model = KeyedVectors.load(
f"../input/ir-project-download-keyed-vectors/{m}.keyedvectors"
)
except:
model = gensim.downloader.load(m)
def method_wordmover_distance_new_model(test_qid):
return method_wordmover_distance(test_qid, model)
show_sample_query_results(
test_query_qids_list[0],
*method_wordmover_distance_new_model(test_query_qids_list[0]),
)
_ = evaluation_process(method_wordmover_distance_new_model)
# # Sentence Embeddings
# Each sentence can be embedded as a vector with SentenceTransformer
from sentence_transformers import SentenceTransformer
model_name = "bert-base-nli-stsb-mean-tokens"
model_tf = SentenceTransformer(model_name)
model_name = "bert-base-nli-stsb-mean-tokens"
sentence_vectors = np.load(
f"../input/quora-question-pairs-bert-sentence-vectors/sentence_vectors_{model_name}.npy"
)
sentence_vectors = {i: vec for i, vec in enumerate(sentence_vectors)}
from scipy.spatial.distance import cosine
def method_sentence_vector(
query_qid,
method_preliminary=method_overlapping_root_word_count,
preliminary_factor=1,
):
# method_preliminary can be either of the previous methods
# recommended method_overlapping_root_word_count, method_boolean, method_tf_idf
qid_list, preliminary_scores = method_preliminary(query_qid)
# sort by cosine similarity
query_sentence_vector = sentence_vectors[query_qid]
query_results = [
(
qid,
preliminary_factor * preliminary_score
- abs(cosine(query_sentence_vector, sentence_vectors[qid])),
)
for qid, preliminary_score in zip(qid_list, preliminary_scores)
]
query_results = sorted(query_results, key=lambda x: x[1], reverse=True)[
:RANKED_LIST_SIZE
]
return [x[0] for x in query_results], [x[1] for x in query_results]
show_sample_query_results(
test_query_qids_list[0],
*method_sentence_vector(test_query_qids_list[0], preliminary_factor=0),
)
show_sample_query_results(
test_query_qids_list[0],
*method_sentence_vector(test_query_qids_list[0], preliminary_factor=1),
)
results_sentence_vector = evaluation_process(method_sentence_vector)
# # Supervised Model
df_train = train_df.copy()
from fuzzywuzzy import fuzz
def extract_features(qid1, qid2, eps=10**-6):
question1 = qid_to_question[qid1]
question2 = qid_to_question[qid2]
features = defaultdict(float)
features["ratio"] = fuzz.ratio(question1, question2)
token_qid1 = tokenise_qid(qid1)
token_qid2 = tokenise_qid(qid2)
tf_idf_denominator = 0
for token in set(token_qid1) | set(token_qid2):
if df[token]:
tf_idf_denominator += compute_idf(df[token], len(qid_to_tokens))
tf_idf_numerator = 0
for token in set(token_qid1) & set(token_qid2):
if df[token]:
tf_idf_numerator += compute_idf(df[token], len(qid_to_tokens))
features["tf_idf"] = (tf_idf_numerator + eps) / (tf_idf_denominator + eps)
return features
features_all = list(
tqdm.tqdm(
extract_features(qid1, qid2)
for qid1, qid2 in zip(df_train["qid1"], df_train["qid2"])
)
)
for feature in features_all[0].keys():
df_train[feature] = [features[feature] for features in features_all]
import lightgbm as lgb
target_train = df_train["is_duplicate"]
df_train = df_train.drop(
["id", "qid1", "qid2", "question1", "question2", "is_duplicate"], axis=1
)
eval_set = np.array(
[True if i < len(df_train) * 0.2 else False for i in range(len(df_train))]
)
lgb_train = lgb.Dataset(df_train[~eval_set], target_train[~eval_set])
lgb_eval = lgb.Dataset(df_train[eval_set], target_train[eval_set], reference=lgb_train)
lgb_all = lgb.Dataset(df_train, target_train)
params = {
# 'boosting_type': 'gbdt',
"objective": "binary",
# 'scale_pos_weight': 0.360,
# 'metric': {'auc'},
# 'num_leaves': 15,
# 'learning_rate': 0.05,
# 'feature_fraction': 0.9,
# 'bagging_fraction': 0.8,
# 'bagging_freq': 5,
"verbose": -1,
}
gbm = lgb.train(
params,
lgb_train,
num_boost_round=1000,
valid_sets=lgb_eval,
verbose_eval=-1,
early_stopping_rounds=10,
)
pd.DataFrame(
{
"feature": df_train.columns,
"importance": gbm.feature_importance(importance_type="gain"),
}
)[:20]
def method_supervised_model(query_qid):
candidate_qids, _ = method_overlapping_root_word_count(query_qid)
df_predict = pd.DataFrame()
features_all = list(
extract_features(query_qid, candidate_qid) for candidate_qid in candidate_qids
)
for feature in features_all[0].keys():
df_predict[feature] = [features[feature] for features in features_all]
scores = gbm.predict(df_predict)
results = sorted(list(zip(scores, candidate_qids)))[::-1]
return [x[1] for x in results], [x[0] for x in results] # qid, scores
show_sample_query_results(
test_query_qids_list[0], *method_supervised_model(test_query_qids_list[0])
)
results_supervised_model = evaluation_process(method_supervised_model)
# # Preparation for Hand Evaluation Dataset
method_to_ranklists_scorelists = {
# "random_guess": results_random_guess,
"overlapping_root_word_count": results_overlapping_root_word_count,
"boolean": results_boolean,
"tf_idf": results_tf_idf,
"bm25": results_bm25,
"unigram": results_unigram,
"spacy_embedding_similarity": results_spacy_embedding_similarity,
"spacy_embedding_similarity_lg": results_spacy_embedding_similarity_lg,
"wordmover_distance_glovewiki50": results_wordmover_distance_glovewiki50,
"sentence_vector": results_sentence_vector,
"supervised_model": results_supervised_model,
}
import json
def parse_ndarray(obj): # https://stackoverflow.com/a/52604722/5894029
if isinstance(obj, np.ndarray):
return obj.tolist()
with open("method_to_ranklists_scorelists.json", "w") as f:
json.dump(method_to_ranklists_scorelists, f, indent=4, default=parse_ndarray)
QUESTIONS_TO_HANDEVAL = set(
x - 1
for x in [
332,
490,
1955,
6319,
9690,
17279,
19619,
20557,
26378,
33734,
38984,
49864,
57291,
89903,
116882,
126992,
131214,
144297,
159628,
201409,
273666,
284107,
286721,
312887,
318523,
378759,
384832,
405081,
405877,
423313,
464279,
480116,
533401,
]
)
HANDEVAL_RANK_THRESHOLD = 10
map_qid_to_handeval = defaultdict(set)
for ranklists, scorelists in method_to_ranklists_scorelists.values():
for test_qid, ranklist in zip(test_query_qids_list, ranklists):
if test_qid in QUESTIONS_TO_HANDEVAL:
for candidate_qid in ranklist[:HANDEVAL_RANK_THRESHOLD]:
map_qid_to_handeval[test_qid].add(candidate_qid)
for qid in map_qid_to_handeval:
map_qid_to_handeval[qid] = sorted(map_qid_to_handeval[qid])
dataframe_columns = ["test_qid", "test_question", "candidate_qid", "candidate_question"]
dataframe_entries = []
for qid in sorted(map_qid_to_handeval.keys()):
for candidate_qid in map_qid_to_handeval[qid]:
line_entry = [
qid,
qid_to_question[qid],
candidate_qid,
qid_to_question[candidate_qid],
]
dataframe_entries.append(line_entry)
df_handeval = pd.DataFrame(dataframe_entries, columns=dataframe_columns)
# labeller columns
df_handeval["jh"] = np.nan
df_handeval["hk"] = np.nan
df_handeval["wt"] = np.nan
df_handeval.to_csv("df_handeval.csv", index=None)
# # Calculate NDCG with Hand Evaluation Dataset
# This calculates NDCG from a snapshot version of `method_to_ranklists_scorelists`, and a hand annotated `df_handeval`
# Due to randomness, the `method_to_ranklists_scorelists` may not be reproduced exactly.
df_handeval = pd.read_csv(
"../input/quoraquestionpairhandannotateddataset/df_handeval.csv"
)
with open(
"../input/quoraquestionpairhandannotateddataset/method_to_ranklists_scorelists.json"
) as f:
method_to_ranklists_scorelists = json.load(f)
import math
def calculate_dcg_at_k(r, k, method=0):
if method == 0:
logn = [1.0] + [1 / math.log(i, 2) for i in range(2, k + 1)]
else:
logn = [1 / math.log(i, 2) for i in range(2, k + 2)]
dcg = 0.0
for gain, disc in zip(r[:k], logn):
dcg += gain * disc
return dcg
def calculate_ndcg_at_k(scores, ref, k=10, method=0):
denom = calculate_dcg_at_k(ref, k, method=method)
numer = calculate_dcg_at_k(scores, k, method=method)
if denom == 0:
return 0.0
return numer / denom
test_qid_to_candidate_qid_to_scores = collections.defaultdict(dict)
for _, row in df_handeval.iterrows():
test_qid = row["test_qid"]
candidate_qid = row["candidate_qid"]
score = row["average"]
test_qid_to_candidate_qid_to_scores[test_qid][candidate_qid] = score
test_qid_to_ideal_scores = collections.defaultdict(list)
for test_qid, candidate_qid_to_scores in test_qid_to_candidate_qid_to_scores.items():
ideal_scores = sorted(candidate_qid_to_scores.values())[::-1]
test_qid_to_ideal_scores[test_qid] = ideal_scores
method_to_ndcg_score = collections.defaultdict(list)
count_out_of_eval = 0
for method_name, (ranklists, _) in method_to_ranklists_scorelists.items():
for test_qid, ranklist in zip(test_query_qids_list, ranklists):
if test_qid in QUESTIONS_TO_HANDEVAL:
scores = []
for candidate_qid in ranklist[:HANDEVAL_RANK_THRESHOLD]:
if candidate_qid not in test_qid_to_candidate_qid_to_scores[test_qid]:
scores.append(1)
print(method_name, len(scores))
count_out_of_eval += 1
else:
scores.append(
test_qid_to_candidate_qid_to_scores[test_qid][candidate_qid]
)
ref = test_qid_to_ideal_scores[test_qid]
ndcg_at_k = calculate_ndcg_at_k(scores, ref)
method_to_ndcg_score[method_name].append(ndcg_at_k)
count_out_of_eval
for method_name, scores in method_to_ndcg_score.items():
print(method_name)
print(sum(scores) / len(scores))
print(" ".join(f"{x:.2f}" for x in scores))
print()
# # Indexing and Querying of Unseen Questions
# This is probably the Graphical User Interface that we will present
def index_unseen_question(unseen_question_text_list):
unseen_sentence_vectors = model_tf.encode(
unseen_question_text_list, show_progress_bar=True
)
qids_new = [time.time() for _ in unseen_question_text_list]
for qid_new, unseen_sentence_vector, unseen_question_text in zip(
qids_new, unseen_sentence_vectors, unseen_question_text_list
):
qid_to_question[qid_new] = unseen_question_text
# compute and update word embedding
token_list = tokenise_then_spellcheck(unseen_question_text)
qid_to_vec[qid_new] = to_vec(token_list)
qid_to_vec2[qid_new] = to_vec2(token_list)
# update sentence embedding
sentence_vectors[qid_new] = unseen_sentence_vector
# update tf-idf
qid_to_tokens_, token_to_qids_, tf_, df_, L_ = preprocess_vsm(qids_new)
for qid in qid_to_tokens_:
qid_to_tokens[qid] = qid_to_tokens_[qid]
for token in token_to_qids_:
token_to_qids[token].update(token_to_qids_[token])
for token in tf_:
for qid in tf_[token]:
tf[token][qid] += tf_[token][qid]
for token in df_:
df[token] += df_[token]
for qid in L_:
L[qid] = L_[qid]
def query_unseen_question(unseen_question_text, method):
qid_new = time.time()
qid_to_question[qid_new] = unseen_question_text
# update word embedding
token_list = tokenise_then_spellcheck(unseen_question_text)
qid_to_vec[qid_new] = to_vec(token_list)
qid_to_vec2[qid_new] = to_vec2(token_list)
# update sentence embedding
sentence_vectors[qid_new] = model_tf.encode(
unseen_question_text, show_progress_bar=False
)
show_sample_query_results(qid_new, *method(qid_new))
# list of methods, uncomment to select
method = method_random_guess
method = method_overlapping_root_word_count
method = method_boolean
method = method_tf_idf
method = method_bm25
method = method_unigram
method = method_spacy_embedding_similarity
method = method_spacy_embedding_similarity_lg
method = method_wordmover_distance_glovewiki50
# method = method_sentence_vector
# method = method_supervised_model
query_unseen_question("Why are computer screens dark in color?", method=method)
index_unseen_question(
[
"Why are computer screens black when unpowered?",
"Why are computer screens manufactured black?",
]
)
query_unseen_question("Why are computer screens dark in color?", method=method)
## uncomment if you want to reset the indexing
# qid_to_tokens, token_to_qids = qid_to_tokens_original.copy(), token_to_qids_original.copy()
# tf, df, L = tf_original.copy(), df_original.copy(), L_original.copy()
|
[{"quoraquestionpairhandannotateddataset/df_handeval.csv": {"column_names": "[\"test_qid\", \"test_question\", \"candidate_qid\", \"candidate_question\", \"jh\", \"hk\", \"wt\", \"average\"]", "column_data_types": "{\"test_qid\": \"int64\", \"test_question\": \"object\", \"candidate_qid\": \"int64\", \"candidate_question\": \"object\", \"jh\": \"float64\", \"hk\": \"float64\", \"wt\": \"float64\", \"average\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1446 entries, 0 to 1445\nData columns (total 8 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 test_qid 1446 non-null int64 \n 1 test_question 1446 non-null object \n 2 candidate_qid 1446 non-null int64 \n 3 candidate_question 1446 non-null object \n 4 jh 820 non-null float64\n 5 hk 904 non-null float64\n 6 wt 881 non-null float64\n 7 average 1446 non-null float64\ndtypes: float64(4), int64(2), object(2)\nmemory usage: 90.5+ KB\n", "summary": "{\"test_qid\": {\"count\": 1446.0, \"mean\": 201123.24066390042, \"std\": 176252.91154829974, \"min\": 331.0, \"25%\": 26377.0, \"50%\": 144296.0, \"75%\": 378758.0, \"max\": 533400.0}, \"candidate_qid\": {\"count\": 1446.0, \"mean\": 243641.76556016598, \"std\": 162002.17898214943, \"min\": 146.0, \"25%\": 93980.25, \"50%\": 235227.5, \"75%\": 384772.75, \"max\": 537555.0}, \"jh\": {\"count\": 820.0, \"mean\": 1.8060975609756098, \"std\": 1.2135627083610847, \"min\": 1.0, \"25%\": 1.0, \"50%\": 1.0, \"75%\": 2.0, \"max\": 5.0}, \"hk\": {\"count\": 904.0, \"mean\": 1.842920353982301, \"std\": 1.220757480595188, \"min\": 1.0, \"25%\": 1.0, \"50%\": 1.0, \"75%\": 2.0, \"max\": 5.0}, \"wt\": {\"count\": 881.0, \"mean\": 1.6685584562996594, \"std\": 1.0666531499590737, \"min\": 1.0, \"25%\": 1.0, \"50%\": 1.0, \"75%\": 2.0, \"max\": 5.0}, \"average\": {\"count\": 1446.0, \"mean\": 1.619640387275242, \"std\": 1.177588355180626, \"min\": 0.0, \"25%\": 1.0, \"50%\": 1.0, \"75%\": 2.0, \"max\": 5.0}}", "examples": "{\"test_qid\":{\"0\":331,\"1\":331,\"2\":331,\"3\":331},\"test_question\":{\"0\":\"Which is the Best earphones under Rs 1000?\",\"1\":\"Which is the Best earphones under Rs 1000?\",\"2\":\"Which is the Best earphones under Rs 1000?\",\"3\":\"Which is the Best earphones under Rs 1000?\"},\"candidate_qid\":{\"0\":8504,\"1\":8505,\"2\":15130,\"3\":26692},\"candidate_question\":{\"0\":\"Which is the best earphone under 1000?\",\"1\":\"What are the best earphones under 1k?\",\"2\":\"Which is the best earphones to buy under Rs.600?\",\"3\":\"What are the best earphones under 60$?\"},\"jh\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"hk\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"wt\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"average\":{\"0\":0.0,\"1\":0.0,\"2\":0.0,\"3\":0.0}}"}}]
| true | 2 |
<start_data_description><data_path>quoraquestionpairhandannotateddataset/df_handeval.csv:
<column_names>
['test_qid', 'test_question', 'candidate_qid', 'candidate_question', 'jh', 'hk', 'wt', 'average']
<column_types>
{'test_qid': 'int64', 'test_question': 'object', 'candidate_qid': 'int64', 'candidate_question': 'object', 'jh': 'float64', 'hk': 'float64', 'wt': 'float64', 'average': 'float64'}
<dataframe_Summary>
{'test_qid': {'count': 1446.0, 'mean': 201123.24066390042, 'std': 176252.91154829974, 'min': 331.0, '25%': 26377.0, '50%': 144296.0, '75%': 378758.0, 'max': 533400.0}, 'candidate_qid': {'count': 1446.0, 'mean': 243641.76556016598, 'std': 162002.17898214943, 'min': 146.0, '25%': 93980.25, '50%': 235227.5, '75%': 384772.75, 'max': 537555.0}, 'jh': {'count': 820.0, 'mean': 1.8060975609756098, 'std': 1.2135627083610847, 'min': 1.0, '25%': 1.0, '50%': 1.0, '75%': 2.0, 'max': 5.0}, 'hk': {'count': 904.0, 'mean': 1.842920353982301, 'std': 1.220757480595188, 'min': 1.0, '25%': 1.0, '50%': 1.0, '75%': 2.0, 'max': 5.0}, 'wt': {'count': 881.0, 'mean': 1.6685584562996594, 'std': 1.0666531499590737, 'min': 1.0, '25%': 1.0, '50%': 1.0, '75%': 2.0, 'max': 5.0}, 'average': {'count': 1446.0, 'mean': 1.619640387275242, 'std': 1.177588355180626, 'min': 0.0, '25%': 1.0, '50%': 1.0, '75%': 2.0, 'max': 5.0}}
<dataframe_info>
RangeIndex: 1446 entries, 0 to 1445
Data columns (total 8 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 test_qid 1446 non-null int64
1 test_question 1446 non-null object
2 candidate_qid 1446 non-null int64
3 candidate_question 1446 non-null object
4 jh 820 non-null float64
5 hk 904 non-null float64
6 wt 881 non-null float64
7 average 1446 non-null float64
dtypes: float64(4), int64(2), object(2)
memory usage: 90.5+ KB
<some_examples>
{'test_qid': {'0': 331, '1': 331, '2': 331, '3': 331}, 'test_question': {'0': 'Which is the Best earphones under Rs 1000?', '1': 'Which is the Best earphones under Rs 1000?', '2': 'Which is the Best earphones under Rs 1000?', '3': 'Which is the Best earphones under Rs 1000?'}, 'candidate_qid': {'0': 8504, '1': 8505, '2': 15130, '3': 26692}, 'candidate_question': {'0': 'Which is the best earphone under 1000?', '1': 'What are the best earphones under 1k?', '2': 'Which is the best earphones to buy under Rs.600?', '3': 'What are the best earphones under 60$?'}, 'jh': {'0': None, '1': None, '2': None, '3': None}, 'hk': {'0': None, '1': None, '2': None, '3': None}, 'wt': {'0': None, '1': None, '2': None, '3': None}, 'average': {'0': 0.0, '1': 0.0, '2': 0.0, '3': 0.0}}
<end_description>
| 16,591 | 0 | 17,256 | 16,591 |
69046660
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import datetime
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
stock_1 = pd.read_csv("../input/show-your-data-skills-snu21/stock.csv")
# # Clean and create the resampled spread below. Best of Luck! :)
df = pd.DataFrame(stock_1)
df1 = stock_1.dropna()
print(df1)
df2 = df1.drop(df1[df1.Volume < 15].index)
print(df2)
df2.insert(5, "time", True)
df2["time"] = pd.to_datetime(df2["Timestamp"])
df2["time"] = df2["time"].dt.time
print(df2)
df2["Timestamp"] = pd.to_datetime(df2["Timestamp"])
df2["Timestamp"] = df2["Timestamp"].dt.date
print(df2)
d2 = datetime.time(10, 00, 00)
df2 = df2.drop(df2[df2.time <= d2].index)
print(df2)
d2 = datetime.time(22, 00, 00)
df2 = df2.drop(df2[df2.time >= d2].index)
print(df2)
del df2["time"]
print(df2)
# # Before submitting ensure that you have 5 columns ( Open, High, low, Close, Volume) in case you have the Timestamp column set as index OR 6 columns (Timestamp, Open, High, low, Close, Volume) in case you have index as 0,1,2,3,...
resampled_df.shape
# resampled_df.to_csv('submission.csv', index=False) # use index=False if you have 6 columns as specified above
resampled_df.to_csv(
"submission.csv", index=True
) # use index=True if you have 5 columns
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/046/69046660.ipynb
| null | null |
[{"Id": 69046660, "ScriptId": 18841892, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7984199, "CreationDate": "07/26/2021 08:43:47", "VersionNumber": 1.0, "Title": "Ans01", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 71.0, "LinesInsertedFromPrevious": 41.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 30.0, "LinesInsertedFromFork": 41.0, "LinesDeletedFromFork": 2.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 30.0, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import datetime
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
stock_1 = pd.read_csv("../input/show-your-data-skills-snu21/stock.csv")
# # Clean and create the resampled spread below. Best of Luck! :)
df = pd.DataFrame(stock_1)
df1 = stock_1.dropna()
print(df1)
df2 = df1.drop(df1[df1.Volume < 15].index)
print(df2)
df2.insert(5, "time", True)
df2["time"] = pd.to_datetime(df2["Timestamp"])
df2["time"] = df2["time"].dt.time
print(df2)
df2["Timestamp"] = pd.to_datetime(df2["Timestamp"])
df2["Timestamp"] = df2["Timestamp"].dt.date
print(df2)
d2 = datetime.time(10, 00, 00)
df2 = df2.drop(df2[df2.time <= d2].index)
print(df2)
d2 = datetime.time(22, 00, 00)
df2 = df2.drop(df2[df2.time >= d2].index)
print(df2)
del df2["time"]
print(df2)
# # Before submitting ensure that you have 5 columns ( Open, High, low, Close, Volume) in case you have the Timestamp column set as index OR 6 columns (Timestamp, Open, High, low, Close, Volume) in case you have index as 0,1,2,3,...
resampled_df.shape
# resampled_df.to_csv('submission.csv', index=False) # use index=False if you have 6 columns as specified above
resampled_df.to_csv(
"submission.csv", index=True
) # use index=True if you have 5 columns
| false | 0 | 627 | 0 | 627 | 627 |
||
69046549
|
<jupyter_start><jupyter_text>Used Cars Price Prediction
Kaggle dataset identifier: used-cars-price-prediction
<jupyter_code>import pandas as pd
df = pd.read_csv('used-cars-price-prediction/train-data.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 6019 entries, 0 to 6018
Data columns (total 14 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 6019 non-null int64
1 Name 6019 non-null object
2 Location 6019 non-null object
3 Year 6019 non-null int64
4 Kilometers_Driven 6019 non-null int64
5 Fuel_Type 6019 non-null object
6 Transmission 6019 non-null object
7 Owner_Type 6019 non-null object
8 Mileage 6017 non-null object
9 Engine 5983 non-null object
10 Power 5983 non-null object
11 Seats 5977 non-null float64
12 New_Price 824 non-null object
13 Price 6019 non-null float64
dtypes: float64(2), int64(3), object(9)
memory usage: 658.5+ KB
<jupyter_text>Examples:
{
"Unnamed: 0": 0,
"Name": "Maruti Wagon R LXI CNG",
"Location": "Mumbai",
"Year": 2010,
"Kilometers_Driven": 72000,
"Fuel_Type": "CNG",
"Transmission": "Manual",
"Owner_Type": "First",
"Mileage": "26.6 km/kg",
"Engine": "998 CC",
"Power": "58.16 bhp",
"Seats": 5,
"New_Price": null,
"Price": 1.75
}
{
"Unnamed: 0": 1,
"Name": "Hyundai Creta 1.6 CRDi SX Option",
"Location": "Pune",
"Year": 2015,
"Kilometers_Driven": 41000,
"Fuel_Type": "Diesel",
"Transmission": "Manual",
"Owner_Type": "First",
"Mileage": "19.67 kmpl",
"Engine": "1582 CC",
"Power": "126.2 bhp",
"Seats": 5,
"New_Price": null,
"Price": 12.5
}
{
"Unnamed: 0": 2,
"Name": "Honda Jazz V",
"Location": "Chennai",
"Year": 2011,
"Kilometers_Driven": 46000,
"Fuel_Type": "Petrol",
"Transmission": "Manual",
"Owner_Type": "First",
"Mileage": "18.2 kmpl",
"Engine": "1199 CC",
"Power": "88.7 bhp",
"Seats": 5,
"New_Price": "8.61 Lakh",
"Price": 4.5
}
{
"Unnamed: 0": 3,
"Name": "Maruti Ertiga VDI",
"Location": "Chennai",
"Year": 2012,
"Kilometers_Driven": 87000,
"Fuel_Type": "Diesel",
"Transmission": "Manual",
"Owner_Type": "First",
"Mileage": "20.77 kmpl",
"Engine": "1248 CC",
"Power": "88.76 bhp",
"Seats": 7,
"New_Price": null,
"Price": 6.0
}
<jupyter_code>import pandas as pd
df = pd.read_csv('used-cars-price-prediction/test-data.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 1234 entries, 0 to 1233
Data columns (total 13 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 1234 non-null int64
1 Name 1234 non-null object
2 Location 1234 non-null object
3 Year 1234 non-null int64
4 Kilometers_Driven 1234 non-null int64
5 Fuel_Type 1234 non-null object
6 Transmission 1234 non-null object
7 Owner_Type 1234 non-null object
8 Mileage 1234 non-null object
9 Engine 1224 non-null object
10 Power 1224 non-null object
11 Seats 1223 non-null float64
12 New_Price 182 non-null object
dtypes: float64(1), int64(3), object(9)
memory usage: 125.5+ KB
<jupyter_text>Examples:
{
"Unnamed: 0": 0,
"Name": "Maruti Alto K10 LXI CNG",
"Location": "Delhi",
"Year": 2014,
"Kilometers_Driven": 40929,
"Fuel_Type": "CNG",
"Transmission": "Manual",
"Owner_Type": "First",
"Mileage": "32.26 km/kg",
"Engine": "998 CC",
"Power": "58.2 bhp",
"Seats": 4,
"New_Price": null
}
{
"Unnamed: 0": 1,
"Name": "Maruti Alto 800 2016-2019 LXI",
"Location": "Coimbatore",
"Year": 2013,
"Kilometers_Driven": 54493,
"Fuel_Type": "Petrol",
"Transmission": "Manual",
"Owner_Type": "Second",
"Mileage": "24.7 kmpl",
"Engine": "796 CC",
"Power": "47.3 bhp",
"Seats": 5,
"New_Price": null
}
{
"Unnamed: 0": 2,
"Name": "Toyota Innova Crysta Touring Sport 2.4 MT",
"Location": "Mumbai",
"Year": 2017,
"Kilometers_Driven": 34000,
"Fuel_Type": "Diesel",
"Transmission": "Manual",
"Owner_Type": "First",
"Mileage": "13.68 kmpl",
"Engine": "2393 CC",
"Power": "147.8 bhp",
"Seats": 7,
"New_Price": "25.27 Lakh"
}
{
"Unnamed: 0": 3,
"Name": "Toyota Etios Liva GD",
"Location": "Hyderabad",
"Year": 2012,
"Kilometers_Driven": 139000,
"Fuel_Type": "Diesel",
"Transmission": "Manual",
"Owner_Type": "First",
"Mileage": "23.59 kmpl",
"Engine": "1364 CC",
"Power": "null bhp",
"Seats": 5,
"New_Price": null
}
<jupyter_script># # Used Car Prediction
# In this notebook, there are datas about many cars with different spesification and brands. All the datas which is included are categorical and numerical variables, so then we'll need to do some data preprocessing parts for that. As long as we are facing the regresion problem, we will find and make the best models that could predict the most appropriate price for each cars.
# Source: https://www.kaggle.com/avikasliwal/used-cars-price-prediction
# **Table of Content:**
#
# - **Importing Train Dataset**
# - **Data Description**
# - **EDA and Feature Engineering**
# - **Importing and Preprocessing Test Dataset**
# - **Modelling**
# - **Submission for Test Dataset**
# - **Saving the Model**
# - **Conclusion**
# - **Deployment**
#
#
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
sns.set()
import warnings
warnings.filterwarnings("ignore")
# ### Importing Train Dataset
# Since the data is in csv type of file, we only have to use **pd.read_csv** for importing the data,
#
train = pd.read_csv(r"../input/used-cars-price-prediction/train-data.csv")
# ### Data Description
# #### 1. Exploring car spesifications data
train.head()
train.info()
train.describe()
# #### 2. Checking Missing Values
# Make a function for presenting total number and percentages of missing values in our data
# function for presenting total number and percentages of missing values
missing_data = pd.DataFrame(
{
"Total_Missing": train.isnull().sum(),
"%": (train.isnull().sum() / train.shape[0]) * 100,
}
)
missing_data
# **"New_Price"** columns has more than 85% of missing values inside it, we will drop it later.
# ### Exploratory Data Analysis (EDA) and Feature Engineering
# Before we start, I just want you to know that we will do the feature engineering between train and test dataset seperately. So, it will prevent **Data Leakage** problem. \
# Okay, let's get started:
# - **Converting some indefinitive data and clearing units**
# - **Handling Missing Values**
# - **Handling Categorical Data**
# - **Determining Feature Importance**
# #### 1. Converting some indefinitive data and clearing units
# **'Name'** column has the car spesific name inside it. This columns may have some value that can be extracted to build our model later. But, we have to make it more general by only using the first( **brand name**) word of its sentence. We can do it by using the help of *Regular Expression*.
import re
# function for extracting first word of the car's name
def brand_and_model(car):
car_patterns = re.compile(
"^(\S+)", flags=re.IGNORECASE
) # extract the first word in car's name
cars = car_patterns.match(car)
return cars.group(1)
train["Brand"] = list(
train["Name"].map(brand_and_model)
) # make a column with a name 'Brand'
train.head()
# **Mileage** columns has different unit in it which is:
# - **km/kg (*kilometer per kilogram*)**
# - **kmpl (*kilometer per liter*)**
# As long as the gas density of natural gas is somewhere 0.712 kg/m^3, we will have the relationship:
# **1 liter = +- 712 grams**
# \
# then we can convert km/kg to kmpl by the factor of **1/0.712 = 1.40 **
# function for converting km/kg to kmpl
def mileage_converter(column, data):
New_Mileage = [] # list to accomodate a new mileage that has been converted
for mileage in data[column]:
if str(mileage).endswith("km/kg"): # accessing each km/kg in its column
mileage = mileage[:-6]
mileage = float(mileage) * 1.40 # converting km/kg to kmpl with 1.40 factor
New_Mileage.append(float(mileage)) # append it to New_Mileage list
elif str(mileage).endswith("kmpl"): # accessing each kmpl in its column
mileage = mileage[:-6]
New_Mileage.append(float(mileage)) # append it to New_Mileage list
else:
New_Mileage.append(np.nan) # appending nan values to New_Mileage list
data["New_Mileage"] = New_Mileage # Making new column in dataframe
mileage_converter("Mileage", train)
train.head()
# For other columns that has unit inside it, we can remove those units.
# Clearing units in Engine and Power column
train["Engine"] = train.Engine.str.replace("CC", "")
train["Power"] = train.Power.str.replace("bhp", "")
train["Engine"] = pd.to_numeric(train["Engine"], errors="coerce")
train["Power"] = pd.to_numeric(train["Power"], errors="coerce")
train.head()
# Let's drop some of our unnecessary columns and columns that we have already converted before:
# - **Unnecessary columns ->> (Unnamed: 0 and New_Price)**
# - **Converted columns ->> (Mileage, Name)**
train.drop(["Unnamed: 0", "Name", "Mileage", "New_Price"], axis=1, inplace=True)
train.head()
# #### 2. Handling Missing Values
# After dropping unnecessary columns, we still have some missing values.
missing_data = pd.DataFrame(
{
"Total_Missing": train.isnull().sum(),
"%": (train.isnull().sum() / train.shape[0]) * 100,
}
)
missing_data
# But we have to see how these data in each columns is distributed for finding the right methods of filling the missing values.
sns.distplot(train.Engine, kde=True)
# It's **right-skewed**
sns.distplot(train.Power, kde=True)
# It's **right-skewed**
sns.distplot(train.Seats, kde=True)
# It's **symmetic** enough, then we can fill each columns now
# We fill with median for right skewed columns and fill with mean for symmetric column
train.Engine.fillna(value=train.Engine.median(), inplace=True)
train.Power.fillna(value=train.Power.median(), inplace=True)
train.Seats.fillna(value=train.Seats.mean(), inplace=True)
# Because New_Mileage only has 0.03% of missing data, we'll fill it with mean
train.New_Mileage.fillna(value=train.New_Mileage.mean(), inplace=True)
train.isnull().sum()
# It's cleaner right now!
# #### 3. Cleaning the Outliers in Numerical Data
# We can detect the outliers with many different ways, such as:
# - **Z-Score**
# - **IQR**
# - **Percentile**
# - **and so on..**
# \
# But we will use Z-Score in this case.
num_columns = [
"Year",
"Kilometers_Driven",
"Engine",
"Power",
"Seats",
"Price",
"New_Mileage",
]
# function to cleaning outliers that has a position higher than 3 in z-score
def z_score(column, df):
treshold = 3 # our treshold to assume a data point is an outlier
mean = df[column].mean() # our mean values in its column
std = df[column].std() # our standard deviation values in its column
for data in df[column]:
z_score = (data - mean) / std # our Z-Score
# conditional for filtering outlier with changing the value with the same number that equal to 3 or -3 in Z-Score
if z_score > treshold:
df.loc[df[column] == data, column] = (treshold * std) + mean
elif z_score < -treshold:
df.loc[df[column] == data, column] = (-treshold * std) + mean
return df
for (
column
) in num_columns: # do Z-score funtion to all the numerical columns in train dataset
z_score(column, train)
train.describe()
# #### 3. Handling Categorical Data
# We can find many way for handling categorical data. But categorical data can be qualified into:
# - **Ordinal Data (Data that has an order)** => **Handle with Label Encoding**
# - **Nominal Data (Data are'nt in order)** => **Handle with One Hot Encoding**
train["Location"].value_counts()
sns.catplot(
x="Location",
y="Price",
data=train.sort_values("Price", ascending=False),
kind="boxen",
aspect=4,
)
# One Hot Encoding Location column, because it is a Nominal Data
Location = train["Location"]
Location = pd.get_dummies(Location, drop_first=True)
Location.head()
train["Fuel_Type"].value_counts()
sns.catplot(
x="Fuel_Type",
y="Price",
data=train.sort_values("Price", ascending=False),
kind="boxen",
aspect=3,
)
# One Hot Encoding Fuel_Type column, because it is a Nominal Data
Fuel_Type = train["Fuel_Type"]
Fuel_Type = pd.get_dummies(Fuel_Type, drop_first=True)
Fuel_Type.head()
train["Transmission"].value_counts()
sns.catplot(
x="Transmission",
y="Price",
data=train.sort_values("Price", ascending=False),
kind="boxen",
aspect=2,
)
# One Hot Encoding Transmission column, because it is a Nominal Data
Transmission = train["Transmission"]
Transmission = pd.get_dummies(Transmission, drop_first=True)
Transmission.head()
train["Owner_Type"].value_counts()
sns.catplot(
x="Owner_Type",
y="Price",
data=train.sort_values("Price", ascending=False),
kind="boxen",
aspect=3,
)
# Label Encoding Owner_Type column, because it is a Ordinal Data
Owner_Type = train["Owner_Type"]
# Every keys that will be replacing the data is based that the newer car owner is having a better status than older car owner
Owner_Type = (
train["Owner_Type"]
.map({"First": 4, "Second": 3, "Third": 2, "Fourth & Above": 1})
.to_frame()
)
Owner_Type.head()
train["Brand"].value_counts()
sns.catplot(
x="Brand",
y="Price",
data=train.sort_values("Price", ascending=False),
kind="boxen",
aspect=5,
)
# One Hot Encoding Brand column, because it is a Nominal Data
Brand = train["Brand"]
Brand = pd.get_dummies(Brand, drop_first=True)
Brand.head()
# After handling every categorical columns we will concat them with train data set and drop the old columns
# Concatenating New train dataset ==> train + Location + Fuel_Type + Transmission + Owner_Type + Brand
train = pd.concat([train, Location, Fuel_Type, Transmission, Owner_Type, Brand], axis=1)
train.drop(
["Location", "Fuel_Type", "Transmission", "Owner_Type", "Brand"],
inplace=True,
axis=1,
)
train.head()
train.shape
# #### 4. Determining Feature Importance
# We have to discover the some features that have low correlation with other column, so that we can prevent multicolinearity in each feature data.
# \
# We will use **pearson correlation** => for finding that kind of column. **Heatmap** will help us a lot to visualize the correlation values for each and between columns.
# Seperating Feature and Label column
y = train["Price"]
X = train.drop(["Price"], axis=1)
X.head()
# Visualizing correlation between Feature and Label column
plt.figure(figsize=(18, 18))
sns.heatmap(
train.corr(), annot=False, cmap="viridis"
) # WE DON'T USE ANNOT because it will be very chaotic too be seen
plt.show()
# We make a function that will tell us about which column that has correlation values higher than treshold
def column_corr(df):
treshold = 0.8 # our treshold
corr = set() # column that have correlation values higher than treshold
corr_table = df.corr() # making correlation matrix of our dataset
for i in range(len(corr_table.columns)): # acceccing correlation matrix
for j in range(i):
# conditional to determine the correlation value is higher than treshold or not
if abs(corr_table.iloc[i, j]) > treshold:
column = corr_table.columns[i]
corr.add(column)
return corr
print(
"Columns that has correlation values higher than treshold are "
+ str(column_corr(X))
+ " column."
)
# dropping that column
X.drop("Petrol", axis=1, inplace=True)
X.head()
# ### Importing and Preprocessing Test Dataset
# Importing test dataset (CSV files)
test = pd.read_csv(r"../input/used-cars-price-prediction/test-data.csv")
test.head()
# A little picture of our test dataset
print(test.info())
print("-" * 70)
# -EDA and Feature Engineering-
# Extracting car's brand in "Name" column
test["Brand"] = list(test["Name"].map(brand_and_model))
# Converting km/kg to kmpl in "Mileage" column
mileage_converter("Mileage", test)
# Converting units in 'Engine' and 'Power' columns
test["Engine"] = test.Engine.str.replace("CC", "")
test["Power"] = test.Power.str.replace("bhp", "")
test["Engine"] = pd.to_numeric(test["Engine"], errors="coerce")
test["Power"] = pd.to_numeric(test["Power"], errors="coerce")
# Filling missing values in "Engine", "Power", "Seats", and "Mileage" column
test.Engine.fillna(value=train.Engine.median(), inplace=True)
test.Power.fillna(value=train.Power.median(), inplace=True)
test.Seats.fillna(value=train.Seats.mean(), inplace=True)
test.New_Mileage.fillna(value=train.New_Mileage.mean(), inplace=True)
# Printing the sum of missing values in test dataset
print(test.isnull().sum())
print("-" * 70)
# Our numerical column in test dataset
num_columns_test = [
"Year",
"Kilometers_Driven",
"Engine",
"Power",
"Seats",
"New_Mileage",
]
# Eliminating outliers in numerical column with Z-Score
for column in num_columns_test:
z_score(column, test)
# Description in numerical values in test dataset
print(test.describe())
print("-" * 70)
# Handling Categorical Values in our dataset
# One Hot Encoding for Location column
Location_test = pd.get_dummies(test["Location"], drop_first=True)
# One Hot Encoding for Fuel_Type column
Fuel_Type_test = pd.get_dummies(test["Fuel_Type"], drop_first=True)
# One Hot Encoding for Transmission column
Transmission_test = pd.get_dummies(test["Transmission"], drop_first=True)
# Label Encoding for Owner_Type column
Owner_Type_test = (
test["Owner_Type"]
.map({"First": 4, "Second": 3, "Third": 2, "Fourth & Above": 1})
.to_frame()
)
# One Hot Encoding Brand column
Brand_test = pd.get_dummies(test["Brand"], drop_first=True)
# Concatenating test and new categorical values
test = pd.concat(
[
test,
Location_test,
Fuel_Type_test,
Transmission_test,
Owner_Type_test,
Brand_test,
],
axis=1,
)
# Dropping categorical column that has been handled, New_Price column, unnecessary column, and, column that has high correlation values
test.drop(
[
"Location",
"Fuel_Type",
"Transmission",
"Owner_Type",
"Brand",
"Unnamed: 0",
"Name",
"Mileage",
"New_Price",
"Petrol",
],
inplace=True,
axis=1,
)
# New shape of test dataset
print(test.shape)
test.head()
# ### Modelling
# **1. Train Test Split**
# \
# **2. Finding the Best Model**
# - Random Forest
# - Lasso
# - ElasticNet
# **3. Hyperparameter Tuning**
# Before modelling we have to make sure that train and test dataset have the same number of columns,
# We can use inner join for equalizing train and test column
final_X, final_test = X.align(test, join="inner", axis=1)
print(final_X.shape)
print(final_test.shape)
final_test.head()
# #### 1. Train Test Split
# Train test split train dataset for preventing overfitting
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
final_X, y, test_size=0.25, random_state=30
)
# #### 2. Finding the Best Model (Random Forest, Lasso, and Elastic Net)
# - **Random Forest**
from sklearn.ensemble import RandomForestRegressor
r_forest = RandomForestRegressor()
r_forest.fit(X_train, y_train)
y_result_forest = r_forest.predict(X_test)
# This graph is visualizing the difference between our prediction and y_test, and it looks pretty good because it looks just like normal distribution.
sns.distplot(y_result_forest - y_test, kde=True)
from sklearn import metrics
print("Random Forest accuracy score: ")
print("R-squared training data: ", r_forest.score(X_train, y_train))
print("R-squared test data: ", r_forest.score(X_test, y_test))
print("MAE: ", metrics.mean_absolute_error(y_test, y_result_forest))
print("MSE: ", metrics.mean_squared_error(y_test, y_result_forest))
print("RMSE: ", np.sqrt(metrics.mean_squared_error(y_test, y_result_forest)))
# - **Lasso**
from sklearn.linear_model import Lasso
lasso_model = Lasso()
lasso_model.fit(X_train, y_train)
y_result_lasso = lasso_model.predict(X_test)
sns.distplot(y_result_lasso - y_test, kde=True)
from sklearn import metrics
print("Lasso accuracy score: ")
print("R-squared training data: ", lasso_model.score(X_train, y_train))
print("R-squared test data: ", lasso_model.score(X_test, y_test))
print("MAE: ", metrics.mean_absolute_error(y_test, y_result_lasso))
print("MSE: ", metrics.mean_squared_error(y_test, y_result_lasso))
print("RMSE: ", np.sqrt(metrics.mean_squared_error(y_test, y_result_lasso)))
# - **Elastic Net**
from sklearn.linear_model import ElasticNetCV
ElasticNet = ElasticNetCV()
ElasticNet.fit(X_train, y_train)
y_result_elasticnet = ElasticNet.predict(X_test)
sns.distplot(y_result_elasticnet - y_test, kde=True)
from sklearn import metrics
print("Elastic Net accuracy score: ")
print("R-squared training data: ", ElasticNet.score(X_train, y_train))
print("R-squared test data: ", ElasticNet.score(X_test, y_test))
print("MAE: ", metrics.mean_absolute_error(y_test, y_result_elasticnet))
print("MSE: ", metrics.mean_squared_error(y_test, y_result_elasticnet))
print("RMSE: ", np.sqrt(metrics.mean_squared_error(y_test, y_result_elasticnet)))
# Because **Random Forest** default mode has the highest accuracy score, we will do hyperparameter tuning with that model
# #### 3. Hyperparameter Tuning
# For doing hyperparameter tuning, we can use:
# - **RandomizedSearchCV**
# - **GridSearchCV**
# Because **RandomizedSearchCV** is much faster, we will use it for hyperparameter tuning
from sklearn.model_selection import RandomizedSearchCV
random_parameter = {
"n_estimators": [int(x) for x in np.linspace(start=100, stop=1000, num=10)],
"criterion": ["mse", "mae"],
"max_depth": [int(x) for x in np.linspace(start=2, stop=20, num=10)],
"min_samples_leaf": [int(x) for x in np.linspace(start=5, stop=100, num=20)],
"min_samples_split": [int(x) for x in np.linspace(start=2, stop=10, num=5)],
"max_features": ["auto", "sqrt"],
}
hyper_tuning = RandomizedSearchCV(
estimator=r_forest,
param_distributions=random_parameter,
cv=5,
n_jobs=-1,
scoring="neg_root_mean_squared_error",
n_iter=10,
verbose=3,
random_state=42,
)
hyper_tuning.fit(X_train, y_train)
hyper_tuning.best_params_
hyper_forest = RandomForestRegressor(
n_estimators=400,
min_samples_split=6,
min_samples_leaf=5,
max_features="auto",
max_depth=18,
criterion="mse",
)
hyper_forest.fit(X_train, y_train)
hyper_result = hyper_forest.predict(X_test)
sns.distplot(y_test - hyper_result, kde=True)
print("R-squared training data: ", hyper_forest.score(X_train, y_train))
print("R-squared test data: ", hyper_forest.score(X_test, y_test))
print("MAE: ", metrics.mean_absolute_error(y_test, hyper_result))
print("MSE: ", metrics.mean_squared_error(y_test, hyper_result))
print("RMSE: ", np.sqrt(metrics.mean_squared_error(y_test, hyper_result)))
# In out model, we actually see a decreasing value in every accuracy score after using hyper parameter tuning. It means that our default hyperparameter is much better than the hyperparameter we found in tuning. So we will apply **default Random Forest model** to our test dataset.
# ### Submission for Test Dataset
test_result = r_forest.predict(final_test)
submission = pd.DataFrame({"Car_id": test.index, "Price": test_result})
submission.head(10)
filename = "submission.csv"
submission.to_csv(filename, index=True)
print("Saved file: " + filename)
# ### Saving the Model
import pickle
# Make our file
file = open("used-car-pred.pkl", "wb")
# transfer the information to out pkl file
pickle.dump(r_forest, file)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/046/69046549.ipynb
|
used-cars-price-prediction
|
avikasliwal
|
[{"Id": 69046549, "ScriptId": 18818018, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7730039, "CreationDate": "07/26/2021 08:42:05", "VersionNumber": 4.0, "Title": "Used Car Prediction (Elastic Net, Lasso, RForest)", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 516.0, "LinesInsertedFromPrevious": 31.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 485.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 91776237, "KernelVersionId": 69046549, "SourceDatasetVersionId": 518431}]
|
[{"Id": 518431, "DatasetId": 245550, "DatasourceVersionId": 534662, "CreatorUserId": 2716677, "LicenseName": "Other (specified in description)", "CreationDate": "06/25/2019 10:26:52", "VersionNumber": 2.0, "Title": "Used Cars Price Prediction", "Slug": "used-cars-price-prediction", "Subtitle": "Predict the price of an unknown car. Build your own Algo for cars 24 !!", "Description": NaN, "VersionNotes": "Replaced xlsx files by csv files", "TotalCompressedBytes": 791875.0, "TotalUncompressedBytes": 791875.0}]
|
[{"Id": 245550, "CreatorUserId": 2716677, "OwnerUserId": 2716677.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 518431.0, "CurrentDatasourceVersionId": 534662.0, "ForumId": 256748, "Type": 2, "CreationDate": "06/25/2019 10:11:54", "LastActivityDate": "06/25/2019", "TotalViews": 140175, "TotalDownloads": 22061, "TotalVotes": 222, "TotalKernels": 107}]
|
[{"Id": 2716677, "UserName": "avikasliwal", "DisplayName": "Avi Kasliwal", "RegisterDate": "01/18/2019", "PerformanceTier": 0}]
|
# # Used Car Prediction
# In this notebook, there are datas about many cars with different spesification and brands. All the datas which is included are categorical and numerical variables, so then we'll need to do some data preprocessing parts for that. As long as we are facing the regresion problem, we will find and make the best models that could predict the most appropriate price for each cars.
# Source: https://www.kaggle.com/avikasliwal/used-cars-price-prediction
# **Table of Content:**
#
# - **Importing Train Dataset**
# - **Data Description**
# - **EDA and Feature Engineering**
# - **Importing and Preprocessing Test Dataset**
# - **Modelling**
# - **Submission for Test Dataset**
# - **Saving the Model**
# - **Conclusion**
# - **Deployment**
#
#
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
sns.set()
import warnings
warnings.filterwarnings("ignore")
# ### Importing Train Dataset
# Since the data is in csv type of file, we only have to use **pd.read_csv** for importing the data,
#
train = pd.read_csv(r"../input/used-cars-price-prediction/train-data.csv")
# ### Data Description
# #### 1. Exploring car spesifications data
train.head()
train.info()
train.describe()
# #### 2. Checking Missing Values
# Make a function for presenting total number and percentages of missing values in our data
# function for presenting total number and percentages of missing values
missing_data = pd.DataFrame(
{
"Total_Missing": train.isnull().sum(),
"%": (train.isnull().sum() / train.shape[0]) * 100,
}
)
missing_data
# **"New_Price"** columns has more than 85% of missing values inside it, we will drop it later.
# ### Exploratory Data Analysis (EDA) and Feature Engineering
# Before we start, I just want you to know that we will do the feature engineering between train and test dataset seperately. So, it will prevent **Data Leakage** problem. \
# Okay, let's get started:
# - **Converting some indefinitive data and clearing units**
# - **Handling Missing Values**
# - **Handling Categorical Data**
# - **Determining Feature Importance**
# #### 1. Converting some indefinitive data and clearing units
# **'Name'** column has the car spesific name inside it. This columns may have some value that can be extracted to build our model later. But, we have to make it more general by only using the first( **brand name**) word of its sentence. We can do it by using the help of *Regular Expression*.
import re
# function for extracting first word of the car's name
def brand_and_model(car):
car_patterns = re.compile(
"^(\S+)", flags=re.IGNORECASE
) # extract the first word in car's name
cars = car_patterns.match(car)
return cars.group(1)
train["Brand"] = list(
train["Name"].map(brand_and_model)
) # make a column with a name 'Brand'
train.head()
# **Mileage** columns has different unit in it which is:
# - **km/kg (*kilometer per kilogram*)**
# - **kmpl (*kilometer per liter*)**
# As long as the gas density of natural gas is somewhere 0.712 kg/m^3, we will have the relationship:
# **1 liter = +- 712 grams**
# \
# then we can convert km/kg to kmpl by the factor of **1/0.712 = 1.40 **
# function for converting km/kg to kmpl
def mileage_converter(column, data):
New_Mileage = [] # list to accomodate a new mileage that has been converted
for mileage in data[column]:
if str(mileage).endswith("km/kg"): # accessing each km/kg in its column
mileage = mileage[:-6]
mileage = float(mileage) * 1.40 # converting km/kg to kmpl with 1.40 factor
New_Mileage.append(float(mileage)) # append it to New_Mileage list
elif str(mileage).endswith("kmpl"): # accessing each kmpl in its column
mileage = mileage[:-6]
New_Mileage.append(float(mileage)) # append it to New_Mileage list
else:
New_Mileage.append(np.nan) # appending nan values to New_Mileage list
data["New_Mileage"] = New_Mileage # Making new column in dataframe
mileage_converter("Mileage", train)
train.head()
# For other columns that has unit inside it, we can remove those units.
# Clearing units in Engine and Power column
train["Engine"] = train.Engine.str.replace("CC", "")
train["Power"] = train.Power.str.replace("bhp", "")
train["Engine"] = pd.to_numeric(train["Engine"], errors="coerce")
train["Power"] = pd.to_numeric(train["Power"], errors="coerce")
train.head()
# Let's drop some of our unnecessary columns and columns that we have already converted before:
# - **Unnecessary columns ->> (Unnamed: 0 and New_Price)**
# - **Converted columns ->> (Mileage, Name)**
train.drop(["Unnamed: 0", "Name", "Mileage", "New_Price"], axis=1, inplace=True)
train.head()
# #### 2. Handling Missing Values
# After dropping unnecessary columns, we still have some missing values.
missing_data = pd.DataFrame(
{
"Total_Missing": train.isnull().sum(),
"%": (train.isnull().sum() / train.shape[0]) * 100,
}
)
missing_data
# But we have to see how these data in each columns is distributed for finding the right methods of filling the missing values.
sns.distplot(train.Engine, kde=True)
# It's **right-skewed**
sns.distplot(train.Power, kde=True)
# It's **right-skewed**
sns.distplot(train.Seats, kde=True)
# It's **symmetic** enough, then we can fill each columns now
# We fill with median for right skewed columns and fill with mean for symmetric column
train.Engine.fillna(value=train.Engine.median(), inplace=True)
train.Power.fillna(value=train.Power.median(), inplace=True)
train.Seats.fillna(value=train.Seats.mean(), inplace=True)
# Because New_Mileage only has 0.03% of missing data, we'll fill it with mean
train.New_Mileage.fillna(value=train.New_Mileage.mean(), inplace=True)
train.isnull().sum()
# It's cleaner right now!
# #### 3. Cleaning the Outliers in Numerical Data
# We can detect the outliers with many different ways, such as:
# - **Z-Score**
# - **IQR**
# - **Percentile**
# - **and so on..**
# \
# But we will use Z-Score in this case.
num_columns = [
"Year",
"Kilometers_Driven",
"Engine",
"Power",
"Seats",
"Price",
"New_Mileage",
]
# function to cleaning outliers that has a position higher than 3 in z-score
def z_score(column, df):
treshold = 3 # our treshold to assume a data point is an outlier
mean = df[column].mean() # our mean values in its column
std = df[column].std() # our standard deviation values in its column
for data in df[column]:
z_score = (data - mean) / std # our Z-Score
# conditional for filtering outlier with changing the value with the same number that equal to 3 or -3 in Z-Score
if z_score > treshold:
df.loc[df[column] == data, column] = (treshold * std) + mean
elif z_score < -treshold:
df.loc[df[column] == data, column] = (-treshold * std) + mean
return df
for (
column
) in num_columns: # do Z-score funtion to all the numerical columns in train dataset
z_score(column, train)
train.describe()
# #### 3. Handling Categorical Data
# We can find many way for handling categorical data. But categorical data can be qualified into:
# - **Ordinal Data (Data that has an order)** => **Handle with Label Encoding**
# - **Nominal Data (Data are'nt in order)** => **Handle with One Hot Encoding**
train["Location"].value_counts()
sns.catplot(
x="Location",
y="Price",
data=train.sort_values("Price", ascending=False),
kind="boxen",
aspect=4,
)
# One Hot Encoding Location column, because it is a Nominal Data
Location = train["Location"]
Location = pd.get_dummies(Location, drop_first=True)
Location.head()
train["Fuel_Type"].value_counts()
sns.catplot(
x="Fuel_Type",
y="Price",
data=train.sort_values("Price", ascending=False),
kind="boxen",
aspect=3,
)
# One Hot Encoding Fuel_Type column, because it is a Nominal Data
Fuel_Type = train["Fuel_Type"]
Fuel_Type = pd.get_dummies(Fuel_Type, drop_first=True)
Fuel_Type.head()
train["Transmission"].value_counts()
sns.catplot(
x="Transmission",
y="Price",
data=train.sort_values("Price", ascending=False),
kind="boxen",
aspect=2,
)
# One Hot Encoding Transmission column, because it is a Nominal Data
Transmission = train["Transmission"]
Transmission = pd.get_dummies(Transmission, drop_first=True)
Transmission.head()
train["Owner_Type"].value_counts()
sns.catplot(
x="Owner_Type",
y="Price",
data=train.sort_values("Price", ascending=False),
kind="boxen",
aspect=3,
)
# Label Encoding Owner_Type column, because it is a Ordinal Data
Owner_Type = train["Owner_Type"]
# Every keys that will be replacing the data is based that the newer car owner is having a better status than older car owner
Owner_Type = (
train["Owner_Type"]
.map({"First": 4, "Second": 3, "Third": 2, "Fourth & Above": 1})
.to_frame()
)
Owner_Type.head()
train["Brand"].value_counts()
sns.catplot(
x="Brand",
y="Price",
data=train.sort_values("Price", ascending=False),
kind="boxen",
aspect=5,
)
# One Hot Encoding Brand column, because it is a Nominal Data
Brand = train["Brand"]
Brand = pd.get_dummies(Brand, drop_first=True)
Brand.head()
# After handling every categorical columns we will concat them with train data set and drop the old columns
# Concatenating New train dataset ==> train + Location + Fuel_Type + Transmission + Owner_Type + Brand
train = pd.concat([train, Location, Fuel_Type, Transmission, Owner_Type, Brand], axis=1)
train.drop(
["Location", "Fuel_Type", "Transmission", "Owner_Type", "Brand"],
inplace=True,
axis=1,
)
train.head()
train.shape
# #### 4. Determining Feature Importance
# We have to discover the some features that have low correlation with other column, so that we can prevent multicolinearity in each feature data.
# \
# We will use **pearson correlation** => for finding that kind of column. **Heatmap** will help us a lot to visualize the correlation values for each and between columns.
# Seperating Feature and Label column
y = train["Price"]
X = train.drop(["Price"], axis=1)
X.head()
# Visualizing correlation between Feature and Label column
plt.figure(figsize=(18, 18))
sns.heatmap(
train.corr(), annot=False, cmap="viridis"
) # WE DON'T USE ANNOT because it will be very chaotic too be seen
plt.show()
# We make a function that will tell us about which column that has correlation values higher than treshold
def column_corr(df):
treshold = 0.8 # our treshold
corr = set() # column that have correlation values higher than treshold
corr_table = df.corr() # making correlation matrix of our dataset
for i in range(len(corr_table.columns)): # acceccing correlation matrix
for j in range(i):
# conditional to determine the correlation value is higher than treshold or not
if abs(corr_table.iloc[i, j]) > treshold:
column = corr_table.columns[i]
corr.add(column)
return corr
print(
"Columns that has correlation values higher than treshold are "
+ str(column_corr(X))
+ " column."
)
# dropping that column
X.drop("Petrol", axis=1, inplace=True)
X.head()
# ### Importing and Preprocessing Test Dataset
# Importing test dataset (CSV files)
test = pd.read_csv(r"../input/used-cars-price-prediction/test-data.csv")
test.head()
# A little picture of our test dataset
print(test.info())
print("-" * 70)
# -EDA and Feature Engineering-
# Extracting car's brand in "Name" column
test["Brand"] = list(test["Name"].map(brand_and_model))
# Converting km/kg to kmpl in "Mileage" column
mileage_converter("Mileage", test)
# Converting units in 'Engine' and 'Power' columns
test["Engine"] = test.Engine.str.replace("CC", "")
test["Power"] = test.Power.str.replace("bhp", "")
test["Engine"] = pd.to_numeric(test["Engine"], errors="coerce")
test["Power"] = pd.to_numeric(test["Power"], errors="coerce")
# Filling missing values in "Engine", "Power", "Seats", and "Mileage" column
test.Engine.fillna(value=train.Engine.median(), inplace=True)
test.Power.fillna(value=train.Power.median(), inplace=True)
test.Seats.fillna(value=train.Seats.mean(), inplace=True)
test.New_Mileage.fillna(value=train.New_Mileage.mean(), inplace=True)
# Printing the sum of missing values in test dataset
print(test.isnull().sum())
print("-" * 70)
# Our numerical column in test dataset
num_columns_test = [
"Year",
"Kilometers_Driven",
"Engine",
"Power",
"Seats",
"New_Mileage",
]
# Eliminating outliers in numerical column with Z-Score
for column in num_columns_test:
z_score(column, test)
# Description in numerical values in test dataset
print(test.describe())
print("-" * 70)
# Handling Categorical Values in our dataset
# One Hot Encoding for Location column
Location_test = pd.get_dummies(test["Location"], drop_first=True)
# One Hot Encoding for Fuel_Type column
Fuel_Type_test = pd.get_dummies(test["Fuel_Type"], drop_first=True)
# One Hot Encoding for Transmission column
Transmission_test = pd.get_dummies(test["Transmission"], drop_first=True)
# Label Encoding for Owner_Type column
Owner_Type_test = (
test["Owner_Type"]
.map({"First": 4, "Second": 3, "Third": 2, "Fourth & Above": 1})
.to_frame()
)
# One Hot Encoding Brand column
Brand_test = pd.get_dummies(test["Brand"], drop_first=True)
# Concatenating test and new categorical values
test = pd.concat(
[
test,
Location_test,
Fuel_Type_test,
Transmission_test,
Owner_Type_test,
Brand_test,
],
axis=1,
)
# Dropping categorical column that has been handled, New_Price column, unnecessary column, and, column that has high correlation values
test.drop(
[
"Location",
"Fuel_Type",
"Transmission",
"Owner_Type",
"Brand",
"Unnamed: 0",
"Name",
"Mileage",
"New_Price",
"Petrol",
],
inplace=True,
axis=1,
)
# New shape of test dataset
print(test.shape)
test.head()
# ### Modelling
# **1. Train Test Split**
# \
# **2. Finding the Best Model**
# - Random Forest
# - Lasso
# - ElasticNet
# **3. Hyperparameter Tuning**
# Before modelling we have to make sure that train and test dataset have the same number of columns,
# We can use inner join for equalizing train and test column
final_X, final_test = X.align(test, join="inner", axis=1)
print(final_X.shape)
print(final_test.shape)
final_test.head()
# #### 1. Train Test Split
# Train test split train dataset for preventing overfitting
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
final_X, y, test_size=0.25, random_state=30
)
# #### 2. Finding the Best Model (Random Forest, Lasso, and Elastic Net)
# - **Random Forest**
from sklearn.ensemble import RandomForestRegressor
r_forest = RandomForestRegressor()
r_forest.fit(X_train, y_train)
y_result_forest = r_forest.predict(X_test)
# This graph is visualizing the difference between our prediction and y_test, and it looks pretty good because it looks just like normal distribution.
sns.distplot(y_result_forest - y_test, kde=True)
from sklearn import metrics
print("Random Forest accuracy score: ")
print("R-squared training data: ", r_forest.score(X_train, y_train))
print("R-squared test data: ", r_forest.score(X_test, y_test))
print("MAE: ", metrics.mean_absolute_error(y_test, y_result_forest))
print("MSE: ", metrics.mean_squared_error(y_test, y_result_forest))
print("RMSE: ", np.sqrt(metrics.mean_squared_error(y_test, y_result_forest)))
# - **Lasso**
from sklearn.linear_model import Lasso
lasso_model = Lasso()
lasso_model.fit(X_train, y_train)
y_result_lasso = lasso_model.predict(X_test)
sns.distplot(y_result_lasso - y_test, kde=True)
from sklearn import metrics
print("Lasso accuracy score: ")
print("R-squared training data: ", lasso_model.score(X_train, y_train))
print("R-squared test data: ", lasso_model.score(X_test, y_test))
print("MAE: ", metrics.mean_absolute_error(y_test, y_result_lasso))
print("MSE: ", metrics.mean_squared_error(y_test, y_result_lasso))
print("RMSE: ", np.sqrt(metrics.mean_squared_error(y_test, y_result_lasso)))
# - **Elastic Net**
from sklearn.linear_model import ElasticNetCV
ElasticNet = ElasticNetCV()
ElasticNet.fit(X_train, y_train)
y_result_elasticnet = ElasticNet.predict(X_test)
sns.distplot(y_result_elasticnet - y_test, kde=True)
from sklearn import metrics
print("Elastic Net accuracy score: ")
print("R-squared training data: ", ElasticNet.score(X_train, y_train))
print("R-squared test data: ", ElasticNet.score(X_test, y_test))
print("MAE: ", metrics.mean_absolute_error(y_test, y_result_elasticnet))
print("MSE: ", metrics.mean_squared_error(y_test, y_result_elasticnet))
print("RMSE: ", np.sqrt(metrics.mean_squared_error(y_test, y_result_elasticnet)))
# Because **Random Forest** default mode has the highest accuracy score, we will do hyperparameter tuning with that model
# #### 3. Hyperparameter Tuning
# For doing hyperparameter tuning, we can use:
# - **RandomizedSearchCV**
# - **GridSearchCV**
# Because **RandomizedSearchCV** is much faster, we will use it for hyperparameter tuning
from sklearn.model_selection import RandomizedSearchCV
random_parameter = {
"n_estimators": [int(x) for x in np.linspace(start=100, stop=1000, num=10)],
"criterion": ["mse", "mae"],
"max_depth": [int(x) for x in np.linspace(start=2, stop=20, num=10)],
"min_samples_leaf": [int(x) for x in np.linspace(start=5, stop=100, num=20)],
"min_samples_split": [int(x) for x in np.linspace(start=2, stop=10, num=5)],
"max_features": ["auto", "sqrt"],
}
hyper_tuning = RandomizedSearchCV(
estimator=r_forest,
param_distributions=random_parameter,
cv=5,
n_jobs=-1,
scoring="neg_root_mean_squared_error",
n_iter=10,
verbose=3,
random_state=42,
)
hyper_tuning.fit(X_train, y_train)
hyper_tuning.best_params_
hyper_forest = RandomForestRegressor(
n_estimators=400,
min_samples_split=6,
min_samples_leaf=5,
max_features="auto",
max_depth=18,
criterion="mse",
)
hyper_forest.fit(X_train, y_train)
hyper_result = hyper_forest.predict(X_test)
sns.distplot(y_test - hyper_result, kde=True)
print("R-squared training data: ", hyper_forest.score(X_train, y_train))
print("R-squared test data: ", hyper_forest.score(X_test, y_test))
print("MAE: ", metrics.mean_absolute_error(y_test, hyper_result))
print("MSE: ", metrics.mean_squared_error(y_test, hyper_result))
print("RMSE: ", np.sqrt(metrics.mean_squared_error(y_test, hyper_result)))
# In out model, we actually see a decreasing value in every accuracy score after using hyper parameter tuning. It means that our default hyperparameter is much better than the hyperparameter we found in tuning. So we will apply **default Random Forest model** to our test dataset.
# ### Submission for Test Dataset
test_result = r_forest.predict(final_test)
submission = pd.DataFrame({"Car_id": test.index, "Price": test_result})
submission.head(10)
filename = "submission.csv"
submission.to_csv(filename, index=True)
print("Saved file: " + filename)
# ### Saving the Model
import pickle
# Make our file
file = open("used-car-pred.pkl", "wb")
# transfer the information to out pkl file
pickle.dump(r_forest, file)
|
[{"used-cars-price-prediction/train-data.csv": {"column_names": "[\"Unnamed: 0\", \"Name\", \"Location\", \"Year\", \"Kilometers_Driven\", \"Fuel_Type\", \"Transmission\", \"Owner_Type\", \"Mileage\", \"Engine\", \"Power\", \"Seats\", \"New_Price\", \"Price\"]", "column_data_types": "{\"Unnamed: 0\": \"int64\", \"Name\": \"object\", \"Location\": \"object\", \"Year\": \"int64\", \"Kilometers_Driven\": \"int64\", \"Fuel_Type\": \"object\", \"Transmission\": \"object\", \"Owner_Type\": \"object\", \"Mileage\": \"object\", \"Engine\": \"object\", \"Power\": \"object\", \"Seats\": \"float64\", \"New_Price\": \"object\", \"Price\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 6019 entries, 0 to 6018\nData columns (total 14 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Unnamed: 0 6019 non-null int64 \n 1 Name 6019 non-null object \n 2 Location 6019 non-null object \n 3 Year 6019 non-null int64 \n 4 Kilometers_Driven 6019 non-null int64 \n 5 Fuel_Type 6019 non-null object \n 6 Transmission 6019 non-null object \n 7 Owner_Type 6019 non-null object \n 8 Mileage 6017 non-null object \n 9 Engine 5983 non-null object \n 10 Power 5983 non-null object \n 11 Seats 5977 non-null float64\n 12 New_Price 824 non-null object \n 13 Price 6019 non-null float64\ndtypes: float64(2), int64(3), object(9)\nmemory usage: 658.5+ KB\n", "summary": "{\"Unnamed: 0\": {\"count\": 6019.0, \"mean\": 3009.0, \"std\": 1737.6799666988932, \"min\": 0.0, \"25%\": 1504.5, \"50%\": 3009.0, \"75%\": 4513.5, \"max\": 6018.0}, \"Year\": {\"count\": 6019.0, \"mean\": 2013.3581990363848, \"std\": 3.2697421160913964, \"min\": 1998.0, \"25%\": 2011.0, \"50%\": 2014.0, \"75%\": 2016.0, \"max\": 2019.0}, \"Kilometers_Driven\": {\"count\": 6019.0, \"mean\": 58738.38029573019, \"std\": 91268.84320624862, \"min\": 171.0, \"25%\": 34000.0, \"50%\": 53000.0, \"75%\": 73000.0, \"max\": 6500000.0}, \"Seats\": {\"count\": 5977.0, \"mean\": 5.278735151413753, \"std\": 0.8088395547482927, \"min\": 0.0, \"25%\": 5.0, \"50%\": 5.0, \"75%\": 5.0, \"max\": 10.0}, \"Price\": {\"count\": 6019.0, \"mean\": 9.47946835022429, \"std\": 11.1879171124555, \"min\": 0.44, \"25%\": 3.5, \"50%\": 5.64, \"75%\": 9.95, \"max\": 160.0}}", "examples": "{\"Unnamed: 0\":{\"0\":0,\"1\":1,\"2\":2,\"3\":3},\"Name\":{\"0\":\"Maruti Wagon R LXI CNG\",\"1\":\"Hyundai Creta 1.6 CRDi SX Option\",\"2\":\"Honda Jazz V\",\"3\":\"Maruti Ertiga VDI\"},\"Location\":{\"0\":\"Mumbai\",\"1\":\"Pune\",\"2\":\"Chennai\",\"3\":\"Chennai\"},\"Year\":{\"0\":2010,\"1\":2015,\"2\":2011,\"3\":2012},\"Kilometers_Driven\":{\"0\":72000,\"1\":41000,\"2\":46000,\"3\":87000},\"Fuel_Type\":{\"0\":\"CNG\",\"1\":\"Diesel\",\"2\":\"Petrol\",\"3\":\"Diesel\"},\"Transmission\":{\"0\":\"Manual\",\"1\":\"Manual\",\"2\":\"Manual\",\"3\":\"Manual\"},\"Owner_Type\":{\"0\":\"First\",\"1\":\"First\",\"2\":\"First\",\"3\":\"First\"},\"Mileage\":{\"0\":\"26.6 km\\/kg\",\"1\":\"19.67 kmpl\",\"2\":\"18.2 kmpl\",\"3\":\"20.77 kmpl\"},\"Engine\":{\"0\":\"998 CC\",\"1\":\"1582 CC\",\"2\":\"1199 CC\",\"3\":\"1248 CC\"},\"Power\":{\"0\":\"58.16 bhp\",\"1\":\"126.2 bhp\",\"2\":\"88.7 bhp\",\"3\":\"88.76 bhp\"},\"Seats\":{\"0\":5.0,\"1\":5.0,\"2\":5.0,\"3\":7.0},\"New_Price\":{\"0\":null,\"1\":null,\"2\":\"8.61 Lakh\",\"3\":null},\"Price\":{\"0\":1.75,\"1\":12.5,\"2\":4.5,\"3\":6.0}}"}}, {"used-cars-price-prediction/test-data.csv": {"column_names": "[\"Unnamed: 0\", \"Name\", \"Location\", \"Year\", \"Kilometers_Driven\", \"Fuel_Type\", \"Transmission\", \"Owner_Type\", \"Mileage\", \"Engine\", \"Power\", \"Seats\", \"New_Price\"]", "column_data_types": "{\"Unnamed: 0\": \"int64\", \"Name\": \"object\", \"Location\": \"object\", \"Year\": \"int64\", \"Kilometers_Driven\": \"int64\", \"Fuel_Type\": \"object\", \"Transmission\": \"object\", \"Owner_Type\": \"object\", \"Mileage\": \"object\", \"Engine\": \"object\", \"Power\": \"object\", \"Seats\": \"float64\", \"New_Price\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1234 entries, 0 to 1233\nData columns (total 13 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Unnamed: 0 1234 non-null int64 \n 1 Name 1234 non-null object \n 2 Location 1234 non-null object \n 3 Year 1234 non-null int64 \n 4 Kilometers_Driven 1234 non-null int64 \n 5 Fuel_Type 1234 non-null object \n 6 Transmission 1234 non-null object \n 7 Owner_Type 1234 non-null object \n 8 Mileage 1234 non-null object \n 9 Engine 1224 non-null object \n 10 Power 1224 non-null object \n 11 Seats 1223 non-null float64\n 12 New_Price 182 non-null object \ndtypes: float64(1), int64(3), object(9)\nmemory usage: 125.5+ KB\n", "summary": "{\"Unnamed: 0\": {\"count\": 1234.0, \"mean\": 616.5, \"std\": 356.3694244273303, \"min\": 0.0, \"25%\": 308.25, \"50%\": 616.5, \"75%\": 924.75, \"max\": 1233.0}, \"Year\": {\"count\": 1234.0, \"mean\": 2013.4003241491087, \"std\": 3.1797003983311205, \"min\": 1996.0, \"25%\": 2011.0, \"50%\": 2014.0, \"75%\": 2016.0, \"max\": 2019.0}, \"Kilometers_Driven\": {\"count\": 1234.0, \"mean\": 58507.28849270665, \"std\": 35598.7020977228, \"min\": 1000.0, \"25%\": 34000.0, \"50%\": 54572.5, \"75%\": 75000.0, \"max\": 350000.0}, \"Seats\": {\"count\": 1223.0, \"mean\": 5.28454619787408, \"std\": 0.8256217828188595, \"min\": 2.0, \"25%\": 5.0, \"50%\": 5.0, \"75%\": 5.0, \"max\": 10.0}}", "examples": "{\"Unnamed: 0\":{\"0\":0,\"1\":1,\"2\":2,\"3\":3},\"Name\":{\"0\":\"Maruti Alto K10 LXI CNG\",\"1\":\"Maruti Alto 800 2016-2019 LXI\",\"2\":\"Toyota Innova Crysta Touring Sport 2.4 MT\",\"3\":\"Toyota Etios Liva GD\"},\"Location\":{\"0\":\"Delhi\",\"1\":\"Coimbatore\",\"2\":\"Mumbai\",\"3\":\"Hyderabad\"},\"Year\":{\"0\":2014,\"1\":2013,\"2\":2017,\"3\":2012},\"Kilometers_Driven\":{\"0\":40929,\"1\":54493,\"2\":34000,\"3\":139000},\"Fuel_Type\":{\"0\":\"CNG\",\"1\":\"Petrol\",\"2\":\"Diesel\",\"3\":\"Diesel\"},\"Transmission\":{\"0\":\"Manual\",\"1\":\"Manual\",\"2\":\"Manual\",\"3\":\"Manual\"},\"Owner_Type\":{\"0\":\"First\",\"1\":\"Second\",\"2\":\"First\",\"3\":\"First\"},\"Mileage\":{\"0\":\"32.26 km\\/kg\",\"1\":\"24.7 kmpl\",\"2\":\"13.68 kmpl\",\"3\":\"23.59 kmpl\"},\"Engine\":{\"0\":\"998 CC\",\"1\":\"796 CC\",\"2\":\"2393 CC\",\"3\":\"1364 CC\"},\"Power\":{\"0\":\"58.2 bhp\",\"1\":\"47.3 bhp\",\"2\":\"147.8 bhp\",\"3\":\"null bhp\"},\"Seats\":{\"0\":4.0,\"1\":5.0,\"2\":7.0,\"3\":5.0},\"New_Price\":{\"0\":null,\"1\":null,\"2\":\"25.27 Lakh\",\"3\":null}}"}}]
| true | 2 |
<start_data_description><data_path>used-cars-price-prediction/train-data.csv:
<column_names>
['Unnamed: 0', 'Name', 'Location', 'Year', 'Kilometers_Driven', 'Fuel_Type', 'Transmission', 'Owner_Type', 'Mileage', 'Engine', 'Power', 'Seats', 'New_Price', 'Price']
<column_types>
{'Unnamed: 0': 'int64', 'Name': 'object', 'Location': 'object', 'Year': 'int64', 'Kilometers_Driven': 'int64', 'Fuel_Type': 'object', 'Transmission': 'object', 'Owner_Type': 'object', 'Mileage': 'object', 'Engine': 'object', 'Power': 'object', 'Seats': 'float64', 'New_Price': 'object', 'Price': 'float64'}
<dataframe_Summary>
{'Unnamed: 0': {'count': 6019.0, 'mean': 3009.0, 'std': 1737.6799666988932, 'min': 0.0, '25%': 1504.5, '50%': 3009.0, '75%': 4513.5, 'max': 6018.0}, 'Year': {'count': 6019.0, 'mean': 2013.3581990363848, 'std': 3.2697421160913964, 'min': 1998.0, '25%': 2011.0, '50%': 2014.0, '75%': 2016.0, 'max': 2019.0}, 'Kilometers_Driven': {'count': 6019.0, 'mean': 58738.38029573019, 'std': 91268.84320624862, 'min': 171.0, '25%': 34000.0, '50%': 53000.0, '75%': 73000.0, 'max': 6500000.0}, 'Seats': {'count': 5977.0, 'mean': 5.278735151413753, 'std': 0.8088395547482927, 'min': 0.0, '25%': 5.0, '50%': 5.0, '75%': 5.0, 'max': 10.0}, 'Price': {'count': 6019.0, 'mean': 9.47946835022429, 'std': 11.1879171124555, 'min': 0.44, '25%': 3.5, '50%': 5.64, '75%': 9.95, 'max': 160.0}}
<dataframe_info>
RangeIndex: 6019 entries, 0 to 6018
Data columns (total 14 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 6019 non-null int64
1 Name 6019 non-null object
2 Location 6019 non-null object
3 Year 6019 non-null int64
4 Kilometers_Driven 6019 non-null int64
5 Fuel_Type 6019 non-null object
6 Transmission 6019 non-null object
7 Owner_Type 6019 non-null object
8 Mileage 6017 non-null object
9 Engine 5983 non-null object
10 Power 5983 non-null object
11 Seats 5977 non-null float64
12 New_Price 824 non-null object
13 Price 6019 non-null float64
dtypes: float64(2), int64(3), object(9)
memory usage: 658.5+ KB
<some_examples>
{'Unnamed: 0': {'0': 0, '1': 1, '2': 2, '3': 3}, 'Name': {'0': 'Maruti Wagon R LXI CNG', '1': 'Hyundai Creta 1.6 CRDi SX Option', '2': 'Honda Jazz V', '3': 'Maruti Ertiga VDI'}, 'Location': {'0': 'Mumbai', '1': 'Pune', '2': 'Chennai', '3': 'Chennai'}, 'Year': {'0': 2010, '1': 2015, '2': 2011, '3': 2012}, 'Kilometers_Driven': {'0': 72000, '1': 41000, '2': 46000, '3': 87000}, 'Fuel_Type': {'0': 'CNG', '1': 'Diesel', '2': 'Petrol', '3': 'Diesel'}, 'Transmission': {'0': 'Manual', '1': 'Manual', '2': 'Manual', '3': 'Manual'}, 'Owner_Type': {'0': 'First', '1': 'First', '2': 'First', '3': 'First'}, 'Mileage': {'0': '26.6 km/kg', '1': '19.67 kmpl', '2': '18.2 kmpl', '3': '20.77 kmpl'}, 'Engine': {'0': '998 CC', '1': '1582 CC', '2': '1199 CC', '3': '1248 CC'}, 'Power': {'0': '58.16 bhp', '1': '126.2 bhp', '2': '88.7 bhp', '3': '88.76 bhp'}, 'Seats': {'0': 5.0, '1': 5.0, '2': 5.0, '3': 7.0}, 'New_Price': {'0': None, '1': None, '2': '8.61 Lakh', '3': None}, 'Price': {'0': 1.75, '1': 12.5, '2': 4.5, '3': 6.0}}
<end_description>
<start_data_description><data_path>used-cars-price-prediction/test-data.csv:
<column_names>
['Unnamed: 0', 'Name', 'Location', 'Year', 'Kilometers_Driven', 'Fuel_Type', 'Transmission', 'Owner_Type', 'Mileage', 'Engine', 'Power', 'Seats', 'New_Price']
<column_types>
{'Unnamed: 0': 'int64', 'Name': 'object', 'Location': 'object', 'Year': 'int64', 'Kilometers_Driven': 'int64', 'Fuel_Type': 'object', 'Transmission': 'object', 'Owner_Type': 'object', 'Mileage': 'object', 'Engine': 'object', 'Power': 'object', 'Seats': 'float64', 'New_Price': 'object'}
<dataframe_Summary>
{'Unnamed: 0': {'count': 1234.0, 'mean': 616.5, 'std': 356.3694244273303, 'min': 0.0, '25%': 308.25, '50%': 616.5, '75%': 924.75, 'max': 1233.0}, 'Year': {'count': 1234.0, 'mean': 2013.4003241491087, 'std': 3.1797003983311205, 'min': 1996.0, '25%': 2011.0, '50%': 2014.0, '75%': 2016.0, 'max': 2019.0}, 'Kilometers_Driven': {'count': 1234.0, 'mean': 58507.28849270665, 'std': 35598.7020977228, 'min': 1000.0, '25%': 34000.0, '50%': 54572.5, '75%': 75000.0, 'max': 350000.0}, 'Seats': {'count': 1223.0, 'mean': 5.28454619787408, 'std': 0.8256217828188595, 'min': 2.0, '25%': 5.0, '50%': 5.0, '75%': 5.0, 'max': 10.0}}
<dataframe_info>
RangeIndex: 1234 entries, 0 to 1233
Data columns (total 13 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 1234 non-null int64
1 Name 1234 non-null object
2 Location 1234 non-null object
3 Year 1234 non-null int64
4 Kilometers_Driven 1234 non-null int64
5 Fuel_Type 1234 non-null object
6 Transmission 1234 non-null object
7 Owner_Type 1234 non-null object
8 Mileage 1234 non-null object
9 Engine 1224 non-null object
10 Power 1224 non-null object
11 Seats 1223 non-null float64
12 New_Price 182 non-null object
dtypes: float64(1), int64(3), object(9)
memory usage: 125.5+ KB
<some_examples>
{'Unnamed: 0': {'0': 0, '1': 1, '2': 2, '3': 3}, 'Name': {'0': 'Maruti Alto K10 LXI CNG', '1': 'Maruti Alto 800 2016-2019 LXI', '2': 'Toyota Innova Crysta Touring Sport 2.4 MT', '3': 'Toyota Etios Liva GD'}, 'Location': {'0': 'Delhi', '1': 'Coimbatore', '2': 'Mumbai', '3': 'Hyderabad'}, 'Year': {'0': 2014, '1': 2013, '2': 2017, '3': 2012}, 'Kilometers_Driven': {'0': 40929, '1': 54493, '2': 34000, '3': 139000}, 'Fuel_Type': {'0': 'CNG', '1': 'Petrol', '2': 'Diesel', '3': 'Diesel'}, 'Transmission': {'0': 'Manual', '1': 'Manual', '2': 'Manual', '3': 'Manual'}, 'Owner_Type': {'0': 'First', '1': 'Second', '2': 'First', '3': 'First'}, 'Mileage': {'0': '32.26 km/kg', '1': '24.7 kmpl', '2': '13.68 kmpl', '3': '23.59 kmpl'}, 'Engine': {'0': '998 CC', '1': '796 CC', '2': '2393 CC', '3': '1364 CC'}, 'Power': {'0': '58.2 bhp', '1': '47.3 bhp', '2': '147.8 bhp', '3': 'null bhp'}, 'Seats': {'0': 4.0, '1': 5.0, '2': 7.0, '3': 5.0}, 'New_Price': {'0': None, '1': None, '2': '25.27 Lakh', '3': None}}
<end_description>
| 5,943 | 1 | 7,902 | 5,943 |
69046846
|
# In this notebook I will train the "Tabular Playground Series - Jul 2021" model by the LightGBM.
# This model is a multi-output regression so, because the LGBM doesn't support it we will have to use one of the two available techniques:
# 1. Train each label feature separately and combine them back afterwards. This method is implemented by the `sklearn.multioutput.MultiOutputRegressor`.
# 2. Train each label feature separately and append to every next model the predicted label from the previous model as a new features. This method is implemented by the `sklearn.multioutput.RegressorChain`.
# We will also optimize the hyperparamenters by the `bayes_opt.BayesianOptimization`.
# The problem with the `RegressorChain` is that it doesn't update the evaluation dataset so, we will have to write an envelope class over the `lightgbm.cv` to make it part of the pipeline.
# Finally, the training pipeline chain will look like that:
# ```
# BayesianOptimization --> RegressorChain --> LGBMWrapper --> lightgbm.cv
# ```
import os
import math
import datetime
import numpy as np
import pandas as pd
import scipy as sc
import matplotlib.pyplot as plt
RANDOM_SEED = 111
np.random.seed(RANDOM_SEED)
from numpy.random import default_rng
rng = default_rng(RANDOM_SEED)
from sklearn.metrics import (
roc_curve,
auc,
roc_auc_score,
accuracy_score,
mean_squared_log_error,
)
from sklearn.preprocessing import (
OrdinalEncoder,
MinMaxScaler,
StandardScaler,
OneHotEncoder,
Binarizer,
KBinsDiscretizer,
QuantileTransformer,
)
from sklearn.feature_extraction import FeatureHasher
from sklearn.model_selection import (
train_test_split,
RandomizedSearchCV,
GridSearchCV,
KFold,
StratifiedKFold,
StratifiedShuffleSplit,
ShuffleSplit,
)
from sklearn.pipeline import Pipeline, FeatureUnion
from sklearn.compose import ColumnTransformer
from sklearn import set_config
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.tree import DecisionTreeRegressor
from sklearn.multioutput import MultiOutputRegressor, RegressorChain
import lightgbm as lgb
from bayes_opt import BayesianOptimization
INPUT_DIR = "/kaggle/input/tabular-playground-series-jul-2021"
OUTPUT_DIR = "./"
BATCH_SIZE = 1024
def season(month):
if month == 12 or month == 1 or month == 2: # winter
return 0
elif month == 3 or month == 4 or month == 5: # spring
return 1
elif month == 6 or month == 7 or month == 8: # summer
return 2
else: # outemn
return 3
def daytime(hour):
if hour > 5 and hour < 17: # light
return 0
else: # darkness
return 1
train_df = pd.read_csv(os.path.join(INPUT_DIR, "train.csv"), index_col="date_time")
test_df = pd.read_csv(os.path.join(INPUT_DIR, "test.csv"), index_col="date_time")
train_df.index = pd.to_datetime(train_df.index)
test_df.index = pd.to_datetime(test_df.index)
labels = train_df[
["target_carbon_monoxide", "target_benzene", "target_nitrogen_oxides"]
]
train_df.drop(labels.columns, axis=1, inplace=True)
total_df = train_df.append(test_df) # pd.concat()
total_df["dew_point"] = total_df["deg_C"].apply(
lambda x: (17.27 * x) / (237.7 + x)
) + total_df["absolute_humidity"].apply(lambda x: math.log(x))
total_df["partial_pressure"] = (
total_df["deg_C"].apply(lambda x: (237.7 + x) * 286.8)
* total_df["absolute_humidity"]
) / 100000
total_df["saturated_wvd"] = (total_df["absolute_humidity"] * 100) / total_df[
"relative_humidity"
]
total_df["dt_low_absolute_humidity"] = (total_df["absolute_humidity"] < 0.25) & (
23 < total_df["deg_C"]
)
total_df["dt_hour"] = [x.hour for x in total_df.index]
total_df["dt_weekday"] = [x.weekday() for x in total_df.index]
total_df["dt_month"] = [x.month for x in total_df.index]
total_df["dt_season"] = [season(x.month) for x in total_df.index]
total_df["dt_lights"] = [daytime(x.hour) for x in total_df.index]
total_df["dt_month_s"] = np.sin(np.pi * (total_df["dt_month"] - 1) / 6)
total_df["dt_month_c"] = np.cos(np.pi * (total_df["dt_month"] - 1) / 6)
total_df["dt_month_s"] = total_df["dt_month_s"].astype("category").cat.codes
total_df["dt_month_c"] = total_df["dt_month_c"].astype("category").cat.codes
total_df["dt_working_hours"] = (
total_df["dt_hour"].isin(np.arange(8, 21, 1)).astype("int")
)
total_df["dt_weekend"] = (total_df["dt_weekday"] >= 5).astype("int")
cat_cols = np.array([col for col in total_df.columns if "dt_" in col])
num_cols = np.array([col for col in total_df.columns if not "dt_" in col])
total_cols = np.concatenate([num_cols, cat_cols])
cat_cols_idx = [np.where(total_df.columns == x)[0][0] for x in cat_cols]
# After training the model we could see that the `deg_C` is actually the main feature with highest impact. It also has a strict seasonality.
# The `dew_point`,`partial_pressure` are pretty useless. They are also highly correlated with the `absolute_humidity`.
# The `dt_*` features behaved surprisingly well, despite that they all were extracted from the DateTime index (except `dt_low_absolute_humidity`).
# ```
# ('deg_C', 20.0),
# ('relative_humidity', 11.9),
# ('absolute_humidity', 14.4),
# ('sensor_1', 15.7),
# ('sensor_2', 17.0),
# ('sensor_3', 19.0),
# ('sensor_4', 9.0),
# ('sensor_5', 1.0),
# ('dew_point', 4.6),
# ('partial_pressure', 4.0),
# ('saturated_wvd', 4.7),
# ('dt_low_absolute_humidity', 7.6),
# ('dt_hour', 8.1),
# ('dt_weekday', 3.8),
# ('dt_month', 11.2),
# ('dt_season', 18.0),
# ('dt_lights', 14.0),
# ('dt_month_s', 12.0),
# ('dt_month_c', 7.0),
# ('dt_working_hours', 3.0),
# ('dt_weekend', 4.0)
# ```
fig, axes = plt.subplots(nrows=3, ncols=3)
fig.set_size_inches(16, 8)
total_df["deg_C"].plot(ax=axes[0, 0], title="deg_C")
total_df["absolute_humidity"].plot(ax=axes[0, 1], title="absolute_humidity")
total_df["relative_humidity"].plot(ax=axes[0, 2], title="relative_humidity")
total_df["dt_season"].plot(ax=axes[1, 0], title="dt_season")
total_df["sensor_1"].plot(ax=axes[1, 1], title="sensor_1")
total_df["sensor_2"].plot(ax=axes[1, 2], title="sensor_2")
total_df["sensor_3"].plot(ax=axes[2, 0], title="sensor_3")
total_df["dt_month_c"].plot(ax=axes[2, 1], title="dt_month_c")
total_df["dt_month_s"].plot(ax=axes[2, 2], title="dt_month_s")
pd.concat(
(
total_df.min(),
total_df.max(),
total_df.mean(),
total_df.std(),
total_df.nunique(),
),
axis=1,
)
pd.concat((labels.min(), labels.max(), labels.mean(), labels.nunique()), axis=1)
# The 3 target labels are highly correlated 66%/80%/88%. That's why we used `RegressorChain` and not `MultiOutputRegressor`.
# The predicted labels become even more correlated - 93%
labels.corr()
# Here we enveloppe the original `lightgbm.cv` to make it accessible by the sklearn Pipeline.
# The competition required validation shoulf be RMSLE but here we used a built-in RMSE with updating label before (log1p) and after (expm1) the execution flow.
# Finally, in the prediciotn step we receive predictions array with the `nfold` dimentions. The simpliest option is just to do their average but there can be applied also other options, like:
# - median
# - mode
# - geom mean
# - meta-model stacking:
# - calculate Booster predict for X_test
# - combine nfold test predictions and keep for later
# - calculate Booster predict for X_train
# - combine nfold train predictions in a new LGBM model with nfold features and original labels
# - train meta-model and predict it with the prepared X_test preditions from the previous section
#
# `stratified=True` doesn't work for regressions.
# `kwargs['verbose']=-1` removes all logs and warnings, it is good for hyperparameter optimization.
class LGBMWrapper(BaseEstimator):
def __init__(self, verbose, nfold):
self.verbose = verbose
self.nfold = nfold
def fit(self, X, y, **kwargs):
y = np.log1p(y)
d_train = lgb.Dataset(X, y)
kwargs["objective"] = "regression"
if self.verbose < 0:
kwargs["verbose"] = self.verbose
model = lgb.cv(
kwargs,
d_train,
num_boost_round=10000,
nfold=self.nfold,
metrics="rmse",
early_stopping_rounds=100,
stratified=False,
verbose_eval=self.verbose,
return_cvbooster=True,
seed=RANDOM_SEED,
)
self.booster = model["cvbooster"]
self.score = model["rmse-mean"][-1]
def predict(self, X):
y_pred = self.booster.predict(X)
y_pred = np.expm1(y_pred).mean(0)
return y_pred
# Possible options for numerical columns -
# Possible options for categorical columns -
pipe_pre = Pipeline(
[
(
"preproc",
ColumnTransformer(
[
(
"num",
Pipeline(
[
# ('scale', StandardScaler()),
(
"gauss",
QuantileTransformer(output_distribution="normal"),
),
# ('minmax', MinMaxScaler()),
# ('kbins', KBinsDiscretizer(n_bins=16, encode='ordinal')) #strategy='uniform'
]
),
num_cols,
),
# ('cat', OrdinalEncoder(), cat_cols)
("cat", OneHotEncoder(sparse=False), cat_cols),
],
remainder="passthrough",
),
),
# ('scale', StandardScaler())
]
)
pipe_pre.fit(total_df)
total_data = pipe_pre.transform(total_df).astype("float")
train_data, test_data = (
total_data[: train_df.index.shape[0]],
total_data[train_df.index.shape[0] :],
)
pd.DataFrame(index=total_df.index, columns=total_cols, data=total_data).nunique().T
def feval(
learning_rate,
num_leaves,
feature_fraction,
bagging_fraction,
max_depth,
max_bin,
lambda_l2,
lambda_l1,
min_data_in_leaf,
min_sum_hessian_in_leaf,
subsample,
):
lgb_space = {
"learning_rate": float(max(min(learning_rate, 1), 0)),
"num_leaves": int(round(num_leaves)),
"feature_fraction": float(max(min(feature_fraction, 1), 0)),
"bagging_fraction": float(max(min(bagging_fraction, 1), 0)),
"max_depth": int(round(max_depth)),
"max_bin": int(round(max_depth)),
"lambda_l2": float(lambda_l2),
"lambda_l1": float(lambda_l1),
"min_data_in_leaf": int(round(min_data_in_leaf)),
"min_sum_hessian_in_leaf": float(min_sum_hessian_in_leaf),
"subsample": float(max(min(subsample, 1), 0)),
}
lgbpwrap = RegressorChain(LGBMWrapper(verbose=-1, nfold=3))
lgbpwrap.fit(train_data, labels, **lgb_space)
scores = [x.score for x in lgbpwrap.estimators_]
print("scores: ", scores)
return -np.mean(scores)
fspace = {
"learning_rate": (0.01, 0.2),
"num_leaves": (24, 80),
"feature_fraction": (0.4, 1),
"bagging_fraction": (0.8, 1),
"max_depth": (5, 30),
"max_bin": (20, 90),
"lambda_l2": (0.0, 0.05),
"lambda_l1": (0.0, 0.05),
"min_data_in_leaf": (20, 80),
"min_sum_hessian_in_leaf": (0, 100),
"subsample": (0.01, 1.0),
}
optimizer = BayesianOptimization(feval, fspace, random_state=RANDOM_SEED)
optimizer.maximize(init_points=20, n_iter=100)
best_params = optimizer.max
best_params
# target | baggin... | lambda_l1 | lambda_l2 | learni... | max_bin | max_depth | min_da... | min_su... | num_le... | subsample |
# [0.10169994949910299, 0.08549932307276413, 0.17700655988733072]
# -0.1214 | 1.0 | 0.0 | 0.0 | 0.01 | 64.74 | 30.0 | 20.0 | 0.0 | 24.0 | 0.01
# [0.10155091396577032, 0.08516392081428582, 0.17723801228200778]
# -0.1213 | 1.0 | 0.05 | 0.05 | 0.01 | 52.54 | 30.0 | 20.0 | 20.86 | 24.0 | 0.01 |
#
print(best_params)
params = best_params["params"]
params["max_bin"] = int(round(params["max_bin"]))
params["max_depth"] = int(round(params["max_depth"]))
params["min_data_in_leaf"] = int(round(params["min_data_in_leaf"]))
params["num_leaves"] = int(round(params["num_leaves"]))
params = {
"learning_rate": 0.01,
"num_leaves": 70,
"bagging_fraction": 0.8922,
"max_depth": 30,
"max_bin": 28,
"min_data_in_leaf": 25,
"min_sum_hessian_in_leaf": 6.05608398987326,
"subsample": 0.5451537324466149,
}
lgbpwrap = RegressorChain(LGBMWrapper(verbose=1000, nfold=10))
lgbpwrap.fit(train_data, labels, **params)
output_test = lgbpwrap.predict(test_data)
scores = [x.score for x in lgbpwrap.estimators_]
features = np.argsort(
lgbpwrap.estimators_[0].booster.feature_importance(importance_type="gain")
).mean(0)
features = list(zip(total_cols, features))
print("scores: ", scores)
features
output_res = pd.DataFrame(index=test_df.index, data={"date_time": test_df.index.values})
output_res[labels.columns] = output_test
output_res.to_csv("./submission.csv", index=False)
output_res.corr()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/046/69046846.ipynb
| null | null |
[{"Id": 69046846, "ScriptId": 18801468, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2112044, "CreationDate": "07/26/2021 08:46:17", "VersionNumber": 4.0, "Title": "LGBM+RegressorChain+BayesianOptimisation+Stacking", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 299.0, "LinesInsertedFromPrevious": 76.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 223.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# In this notebook I will train the "Tabular Playground Series - Jul 2021" model by the LightGBM.
# This model is a multi-output regression so, because the LGBM doesn't support it we will have to use one of the two available techniques:
# 1. Train each label feature separately and combine them back afterwards. This method is implemented by the `sklearn.multioutput.MultiOutputRegressor`.
# 2. Train each label feature separately and append to every next model the predicted label from the previous model as a new features. This method is implemented by the `sklearn.multioutput.RegressorChain`.
# We will also optimize the hyperparamenters by the `bayes_opt.BayesianOptimization`.
# The problem with the `RegressorChain` is that it doesn't update the evaluation dataset so, we will have to write an envelope class over the `lightgbm.cv` to make it part of the pipeline.
# Finally, the training pipeline chain will look like that:
# ```
# BayesianOptimization --> RegressorChain --> LGBMWrapper --> lightgbm.cv
# ```
import os
import math
import datetime
import numpy as np
import pandas as pd
import scipy as sc
import matplotlib.pyplot as plt
RANDOM_SEED = 111
np.random.seed(RANDOM_SEED)
from numpy.random import default_rng
rng = default_rng(RANDOM_SEED)
from sklearn.metrics import (
roc_curve,
auc,
roc_auc_score,
accuracy_score,
mean_squared_log_error,
)
from sklearn.preprocessing import (
OrdinalEncoder,
MinMaxScaler,
StandardScaler,
OneHotEncoder,
Binarizer,
KBinsDiscretizer,
QuantileTransformer,
)
from sklearn.feature_extraction import FeatureHasher
from sklearn.model_selection import (
train_test_split,
RandomizedSearchCV,
GridSearchCV,
KFold,
StratifiedKFold,
StratifiedShuffleSplit,
ShuffleSplit,
)
from sklearn.pipeline import Pipeline, FeatureUnion
from sklearn.compose import ColumnTransformer
from sklearn import set_config
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.tree import DecisionTreeRegressor
from sklearn.multioutput import MultiOutputRegressor, RegressorChain
import lightgbm as lgb
from bayes_opt import BayesianOptimization
INPUT_DIR = "/kaggle/input/tabular-playground-series-jul-2021"
OUTPUT_DIR = "./"
BATCH_SIZE = 1024
def season(month):
if month == 12 or month == 1 or month == 2: # winter
return 0
elif month == 3 or month == 4 or month == 5: # spring
return 1
elif month == 6 or month == 7 or month == 8: # summer
return 2
else: # outemn
return 3
def daytime(hour):
if hour > 5 and hour < 17: # light
return 0
else: # darkness
return 1
train_df = pd.read_csv(os.path.join(INPUT_DIR, "train.csv"), index_col="date_time")
test_df = pd.read_csv(os.path.join(INPUT_DIR, "test.csv"), index_col="date_time")
train_df.index = pd.to_datetime(train_df.index)
test_df.index = pd.to_datetime(test_df.index)
labels = train_df[
["target_carbon_monoxide", "target_benzene", "target_nitrogen_oxides"]
]
train_df.drop(labels.columns, axis=1, inplace=True)
total_df = train_df.append(test_df) # pd.concat()
total_df["dew_point"] = total_df["deg_C"].apply(
lambda x: (17.27 * x) / (237.7 + x)
) + total_df["absolute_humidity"].apply(lambda x: math.log(x))
total_df["partial_pressure"] = (
total_df["deg_C"].apply(lambda x: (237.7 + x) * 286.8)
* total_df["absolute_humidity"]
) / 100000
total_df["saturated_wvd"] = (total_df["absolute_humidity"] * 100) / total_df[
"relative_humidity"
]
total_df["dt_low_absolute_humidity"] = (total_df["absolute_humidity"] < 0.25) & (
23 < total_df["deg_C"]
)
total_df["dt_hour"] = [x.hour for x in total_df.index]
total_df["dt_weekday"] = [x.weekday() for x in total_df.index]
total_df["dt_month"] = [x.month for x in total_df.index]
total_df["dt_season"] = [season(x.month) for x in total_df.index]
total_df["dt_lights"] = [daytime(x.hour) for x in total_df.index]
total_df["dt_month_s"] = np.sin(np.pi * (total_df["dt_month"] - 1) / 6)
total_df["dt_month_c"] = np.cos(np.pi * (total_df["dt_month"] - 1) / 6)
total_df["dt_month_s"] = total_df["dt_month_s"].astype("category").cat.codes
total_df["dt_month_c"] = total_df["dt_month_c"].astype("category").cat.codes
total_df["dt_working_hours"] = (
total_df["dt_hour"].isin(np.arange(8, 21, 1)).astype("int")
)
total_df["dt_weekend"] = (total_df["dt_weekday"] >= 5).astype("int")
cat_cols = np.array([col for col in total_df.columns if "dt_" in col])
num_cols = np.array([col for col in total_df.columns if not "dt_" in col])
total_cols = np.concatenate([num_cols, cat_cols])
cat_cols_idx = [np.where(total_df.columns == x)[0][0] for x in cat_cols]
# After training the model we could see that the `deg_C` is actually the main feature with highest impact. It also has a strict seasonality.
# The `dew_point`,`partial_pressure` are pretty useless. They are also highly correlated with the `absolute_humidity`.
# The `dt_*` features behaved surprisingly well, despite that they all were extracted from the DateTime index (except `dt_low_absolute_humidity`).
# ```
# ('deg_C', 20.0),
# ('relative_humidity', 11.9),
# ('absolute_humidity', 14.4),
# ('sensor_1', 15.7),
# ('sensor_2', 17.0),
# ('sensor_3', 19.0),
# ('sensor_4', 9.0),
# ('sensor_5', 1.0),
# ('dew_point', 4.6),
# ('partial_pressure', 4.0),
# ('saturated_wvd', 4.7),
# ('dt_low_absolute_humidity', 7.6),
# ('dt_hour', 8.1),
# ('dt_weekday', 3.8),
# ('dt_month', 11.2),
# ('dt_season', 18.0),
# ('dt_lights', 14.0),
# ('dt_month_s', 12.0),
# ('dt_month_c', 7.0),
# ('dt_working_hours', 3.0),
# ('dt_weekend', 4.0)
# ```
fig, axes = plt.subplots(nrows=3, ncols=3)
fig.set_size_inches(16, 8)
total_df["deg_C"].plot(ax=axes[0, 0], title="deg_C")
total_df["absolute_humidity"].plot(ax=axes[0, 1], title="absolute_humidity")
total_df["relative_humidity"].plot(ax=axes[0, 2], title="relative_humidity")
total_df["dt_season"].plot(ax=axes[1, 0], title="dt_season")
total_df["sensor_1"].plot(ax=axes[1, 1], title="sensor_1")
total_df["sensor_2"].plot(ax=axes[1, 2], title="sensor_2")
total_df["sensor_3"].plot(ax=axes[2, 0], title="sensor_3")
total_df["dt_month_c"].plot(ax=axes[2, 1], title="dt_month_c")
total_df["dt_month_s"].plot(ax=axes[2, 2], title="dt_month_s")
pd.concat(
(
total_df.min(),
total_df.max(),
total_df.mean(),
total_df.std(),
total_df.nunique(),
),
axis=1,
)
pd.concat((labels.min(), labels.max(), labels.mean(), labels.nunique()), axis=1)
# The 3 target labels are highly correlated 66%/80%/88%. That's why we used `RegressorChain` and not `MultiOutputRegressor`.
# The predicted labels become even more correlated - 93%
labels.corr()
# Here we enveloppe the original `lightgbm.cv` to make it accessible by the sklearn Pipeline.
# The competition required validation shoulf be RMSLE but here we used a built-in RMSE with updating label before (log1p) and after (expm1) the execution flow.
# Finally, in the prediciotn step we receive predictions array with the `nfold` dimentions. The simpliest option is just to do their average but there can be applied also other options, like:
# - median
# - mode
# - geom mean
# - meta-model stacking:
# - calculate Booster predict for X_test
# - combine nfold test predictions and keep for later
# - calculate Booster predict for X_train
# - combine nfold train predictions in a new LGBM model with nfold features and original labels
# - train meta-model and predict it with the prepared X_test preditions from the previous section
#
# `stratified=True` doesn't work for regressions.
# `kwargs['verbose']=-1` removes all logs and warnings, it is good for hyperparameter optimization.
class LGBMWrapper(BaseEstimator):
def __init__(self, verbose, nfold):
self.verbose = verbose
self.nfold = nfold
def fit(self, X, y, **kwargs):
y = np.log1p(y)
d_train = lgb.Dataset(X, y)
kwargs["objective"] = "regression"
if self.verbose < 0:
kwargs["verbose"] = self.verbose
model = lgb.cv(
kwargs,
d_train,
num_boost_round=10000,
nfold=self.nfold,
metrics="rmse",
early_stopping_rounds=100,
stratified=False,
verbose_eval=self.verbose,
return_cvbooster=True,
seed=RANDOM_SEED,
)
self.booster = model["cvbooster"]
self.score = model["rmse-mean"][-1]
def predict(self, X):
y_pred = self.booster.predict(X)
y_pred = np.expm1(y_pred).mean(0)
return y_pred
# Possible options for numerical columns -
# Possible options for categorical columns -
pipe_pre = Pipeline(
[
(
"preproc",
ColumnTransformer(
[
(
"num",
Pipeline(
[
# ('scale', StandardScaler()),
(
"gauss",
QuantileTransformer(output_distribution="normal"),
),
# ('minmax', MinMaxScaler()),
# ('kbins', KBinsDiscretizer(n_bins=16, encode='ordinal')) #strategy='uniform'
]
),
num_cols,
),
# ('cat', OrdinalEncoder(), cat_cols)
("cat", OneHotEncoder(sparse=False), cat_cols),
],
remainder="passthrough",
),
),
# ('scale', StandardScaler())
]
)
pipe_pre.fit(total_df)
total_data = pipe_pre.transform(total_df).astype("float")
train_data, test_data = (
total_data[: train_df.index.shape[0]],
total_data[train_df.index.shape[0] :],
)
pd.DataFrame(index=total_df.index, columns=total_cols, data=total_data).nunique().T
def feval(
learning_rate,
num_leaves,
feature_fraction,
bagging_fraction,
max_depth,
max_bin,
lambda_l2,
lambda_l1,
min_data_in_leaf,
min_sum_hessian_in_leaf,
subsample,
):
lgb_space = {
"learning_rate": float(max(min(learning_rate, 1), 0)),
"num_leaves": int(round(num_leaves)),
"feature_fraction": float(max(min(feature_fraction, 1), 0)),
"bagging_fraction": float(max(min(bagging_fraction, 1), 0)),
"max_depth": int(round(max_depth)),
"max_bin": int(round(max_depth)),
"lambda_l2": float(lambda_l2),
"lambda_l1": float(lambda_l1),
"min_data_in_leaf": int(round(min_data_in_leaf)),
"min_sum_hessian_in_leaf": float(min_sum_hessian_in_leaf),
"subsample": float(max(min(subsample, 1), 0)),
}
lgbpwrap = RegressorChain(LGBMWrapper(verbose=-1, nfold=3))
lgbpwrap.fit(train_data, labels, **lgb_space)
scores = [x.score for x in lgbpwrap.estimators_]
print("scores: ", scores)
return -np.mean(scores)
fspace = {
"learning_rate": (0.01, 0.2),
"num_leaves": (24, 80),
"feature_fraction": (0.4, 1),
"bagging_fraction": (0.8, 1),
"max_depth": (5, 30),
"max_bin": (20, 90),
"lambda_l2": (0.0, 0.05),
"lambda_l1": (0.0, 0.05),
"min_data_in_leaf": (20, 80),
"min_sum_hessian_in_leaf": (0, 100),
"subsample": (0.01, 1.0),
}
optimizer = BayesianOptimization(feval, fspace, random_state=RANDOM_SEED)
optimizer.maximize(init_points=20, n_iter=100)
best_params = optimizer.max
best_params
# target | baggin... | lambda_l1 | lambda_l2 | learni... | max_bin | max_depth | min_da... | min_su... | num_le... | subsample |
# [0.10169994949910299, 0.08549932307276413, 0.17700655988733072]
# -0.1214 | 1.0 | 0.0 | 0.0 | 0.01 | 64.74 | 30.0 | 20.0 | 0.0 | 24.0 | 0.01
# [0.10155091396577032, 0.08516392081428582, 0.17723801228200778]
# -0.1213 | 1.0 | 0.05 | 0.05 | 0.01 | 52.54 | 30.0 | 20.0 | 20.86 | 24.0 | 0.01 |
#
print(best_params)
params = best_params["params"]
params["max_bin"] = int(round(params["max_bin"]))
params["max_depth"] = int(round(params["max_depth"]))
params["min_data_in_leaf"] = int(round(params["min_data_in_leaf"]))
params["num_leaves"] = int(round(params["num_leaves"]))
params = {
"learning_rate": 0.01,
"num_leaves": 70,
"bagging_fraction": 0.8922,
"max_depth": 30,
"max_bin": 28,
"min_data_in_leaf": 25,
"min_sum_hessian_in_leaf": 6.05608398987326,
"subsample": 0.5451537324466149,
}
lgbpwrap = RegressorChain(LGBMWrapper(verbose=1000, nfold=10))
lgbpwrap.fit(train_data, labels, **params)
output_test = lgbpwrap.predict(test_data)
scores = [x.score for x in lgbpwrap.estimators_]
features = np.argsort(
lgbpwrap.estimators_[0].booster.feature_importance(importance_type="gain")
).mean(0)
features = list(zip(total_cols, features))
print("scores: ", scores)
features
output_res = pd.DataFrame(index=test_df.index, data={"date_time": test_df.index.values})
output_res[labels.columns] = output_test
output_res.to_csv("./submission.csv", index=False)
output_res.corr()
| false | 0 | 4,521 | 0 | 4,521 | 4,521 |
||
69046529
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
from keras.datasets import imdb
from keras import models
from keras import layers
from keras import optimizers
from keras import losses
from keras import metrics
from keras.preprocessing import sequence
from keras.layers import Dense, Embedding, SimpleRNN
from keras.models import Sequential
from keras.layers import LSTM
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
max_features = 10000
maxlen = 500
batch_size = 32
print("Loading data...")
(input_train, y_train), (input_test, y_test) = imdb.load_data(num_words=max_features)
print(len(input_train), "train sequences")
print(len(input_test), "test sequences")
print("Pad sequences (samples x time)")
input_train = sequence.pad_sequences(input_train, maxlen=maxlen)
input_test = sequence.pad_sequences(input_test, maxlen=maxlen)
print("input_train shape:", input_train.shape)
print("input_test shape:", input_test.shape)
# ## Model
model = Sequential()
model.add(Embedding(max_features, 32))
model.add(SimpleRNN(32))
model.add(Dense(1, activation="sigmoid"))
model.compile(optimizer="rmsprop", loss="binary_crossentropy", metrics=["acc"])
history = model.fit(
input_train, y_train, epochs=10, batch_size=128, validation_split=0.2
)
# ## Plotting
acc = history.history["acc"]
val_acc = history.history["val_acc"]
loss = history.history["loss"]
val_loss = history.history["val_loss"]
epochs = range(1, len(acc) + 1)
plt.plot(epochs, acc, "bo", label="Training acc")
plt.plot(epochs, val_acc, "b", label="Validation acc")
plt.title("Training and validation accuracy")
plt.legend()
plt.figure()
plt.plot(epochs, loss, "bo", label="Training loss")
plt.plot(epochs, val_loss, "b", label="Validation loss")
plt.title("Training and validation loss")
plt.legend()
plt.show()
# ## LSTM
model = Sequential()
model.add(Embedding(max_features, 32))
model.add(LSTM(32))
model.add(Dense(1, activation="sigmoid"))
model.compile(optimizer="rmsprop", loss="binary_crossentropy", metrics=["acc"])
history = model.fit(
input_train, y_train, epochs=10, batch_size=128, validation_split=0.2
)
acc = history.history["acc"]
val_acc = history.history["val_acc"]
loss = history.history["loss"]
val_loss = history.history["val_loss"]
epochs = range(1, len(acc) + 1)
plt.plot(epochs, acc, "bo", label="Training acc")
plt.plot(epochs, val_acc, "b", label="Validation acc")
plt.title("Training and validation accuracy")
plt.legend()
plt.figure()
plt.plot(epochs, loss, "bo", label="Training loss")
plt.plot(epochs, val_loss, "b", label="Validation loss")
plt.title("Training and validation loss")
plt.legend()
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/046/69046529.ipynb
| null | null |
[{"Id": 69046529, "ScriptId": 18843859, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 552648, "CreationDate": "07/26/2021 08:41:51", "VersionNumber": 1.0, "Title": "Keras IMDB recurrent neural network", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 103.0, "LinesInsertedFromPrevious": 65.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 38.0, "LinesInsertedFromFork": 65.0, "LinesDeletedFromFork": 92.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 38.0, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
from keras.datasets import imdb
from keras import models
from keras import layers
from keras import optimizers
from keras import losses
from keras import metrics
from keras.preprocessing import sequence
from keras.layers import Dense, Embedding, SimpleRNN
from keras.models import Sequential
from keras.layers import LSTM
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
max_features = 10000
maxlen = 500
batch_size = 32
print("Loading data...")
(input_train, y_train), (input_test, y_test) = imdb.load_data(num_words=max_features)
print(len(input_train), "train sequences")
print(len(input_test), "test sequences")
print("Pad sequences (samples x time)")
input_train = sequence.pad_sequences(input_train, maxlen=maxlen)
input_test = sequence.pad_sequences(input_test, maxlen=maxlen)
print("input_train shape:", input_train.shape)
print("input_test shape:", input_test.shape)
# ## Model
model = Sequential()
model.add(Embedding(max_features, 32))
model.add(SimpleRNN(32))
model.add(Dense(1, activation="sigmoid"))
model.compile(optimizer="rmsprop", loss="binary_crossentropy", metrics=["acc"])
history = model.fit(
input_train, y_train, epochs=10, batch_size=128, validation_split=0.2
)
# ## Plotting
acc = history.history["acc"]
val_acc = history.history["val_acc"]
loss = history.history["loss"]
val_loss = history.history["val_loss"]
epochs = range(1, len(acc) + 1)
plt.plot(epochs, acc, "bo", label="Training acc")
plt.plot(epochs, val_acc, "b", label="Validation acc")
plt.title("Training and validation accuracy")
plt.legend()
plt.figure()
plt.plot(epochs, loss, "bo", label="Training loss")
plt.plot(epochs, val_loss, "b", label="Validation loss")
plt.title("Training and validation loss")
plt.legend()
plt.show()
# ## LSTM
model = Sequential()
model.add(Embedding(max_features, 32))
model.add(LSTM(32))
model.add(Dense(1, activation="sigmoid"))
model.compile(optimizer="rmsprop", loss="binary_crossentropy", metrics=["acc"])
history = model.fit(
input_train, y_train, epochs=10, batch_size=128, validation_split=0.2
)
acc = history.history["acc"]
val_acc = history.history["val_acc"]
loss = history.history["loss"]
val_loss = history.history["val_loss"]
epochs = range(1, len(acc) + 1)
plt.plot(epochs, acc, "bo", label="Training acc")
plt.plot(epochs, val_acc, "b", label="Validation acc")
plt.title("Training and validation accuracy")
plt.legend()
plt.figure()
plt.plot(epochs, loss, "bo", label="Training loss")
plt.plot(epochs, val_loss, "b", label="Validation loss")
plt.title("Training and validation loss")
plt.legend()
plt.show()
| false | 0 | 994 | 0 | 994 | 994 |
||
69031335
|
# TENSORFLOW CONCEPT DISPLAYING
import tensorflow as tf
with tf.compat.v1.Session() as sess:
a = tf.constant(4.0)
b = tf.constant(8.0)
print(a)
print(b)
# TENSORFLOW VARIABLES DISPLAYING
import tensorflow as tf
with tf.compat.v1.Session() as sess:
a = tf.constant(3.0)
b = tf.constant(6.9)
print(sess.run(a))
print(sess.run(b))
# TENSORFLOW ARITHMATIC OPERATIONS
import tensorflow as tf
with tf.compat.v1.Session() as sess:
a = tf.constant(6)
b = tf.constant(3)
c = a + b
d = a - b
e = a * b
f = a / b
g = a % b
print(a)
print(b)
print(c)
print(d)
print(e)
print(f)
print(g)
print(sess.run(a))
print(sess.run(b))
print(sess.run(c))
print(sess.run(d))
print(sess.run(e))
print(sess.run(f))
print(sess.run(g))
# TENSORFLOW PRINTING SELF DECLARED VARIABLES
import tensorflow as tf
with tf.compat.v1.Session() as sess:
a = tf.compat.v1.placeholder(tf.int32)
b = tf.compat.v1.placeholder(tf.int32)
c = a + b
print(sess.run(a, {a: [1, 2, 3]}))
print(sess.run(b, {b: [4, 5, 6]}))
# TENSORFLOW ARITHMATIC OPERATIONS OF SELF DECLARED VARIABLES
import tensorflow as tf
with tf.compat.v1.Session() as sess:
a = tf.compat.v1.placeholder(tf.int32)
b = tf.compat.v1.placeholder(tf.int32)
c = a + b
print(sess.run(a, {a: [1, 2, 3]}))
print(sess.run(b, {b: [4, 5, 6]}))
print(sess.run(c, {a: [1, 2, 3], b: [4, 5, 6]}))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/031/69031335.ipynb
| null | null |
[{"Id": 69031335, "ScriptId": 18838937, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6349702, "CreationDate": "07/26/2021 04:43:08", "VersionNumber": 2.0, "Title": "Deep_Learning_Basic_Concepts", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 88.0, "LinesInsertedFromPrevious": 27.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 61.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# TENSORFLOW CONCEPT DISPLAYING
import tensorflow as tf
with tf.compat.v1.Session() as sess:
a = tf.constant(4.0)
b = tf.constant(8.0)
print(a)
print(b)
# TENSORFLOW VARIABLES DISPLAYING
import tensorflow as tf
with tf.compat.v1.Session() as sess:
a = tf.constant(3.0)
b = tf.constant(6.9)
print(sess.run(a))
print(sess.run(b))
# TENSORFLOW ARITHMATIC OPERATIONS
import tensorflow as tf
with tf.compat.v1.Session() as sess:
a = tf.constant(6)
b = tf.constant(3)
c = a + b
d = a - b
e = a * b
f = a / b
g = a % b
print(a)
print(b)
print(c)
print(d)
print(e)
print(f)
print(g)
print(sess.run(a))
print(sess.run(b))
print(sess.run(c))
print(sess.run(d))
print(sess.run(e))
print(sess.run(f))
print(sess.run(g))
# TENSORFLOW PRINTING SELF DECLARED VARIABLES
import tensorflow as tf
with tf.compat.v1.Session() as sess:
a = tf.compat.v1.placeholder(tf.int32)
b = tf.compat.v1.placeholder(tf.int32)
c = a + b
print(sess.run(a, {a: [1, 2, 3]}))
print(sess.run(b, {b: [4, 5, 6]}))
# TENSORFLOW ARITHMATIC OPERATIONS OF SELF DECLARED VARIABLES
import tensorflow as tf
with tf.compat.v1.Session() as sess:
a = tf.compat.v1.placeholder(tf.int32)
b = tf.compat.v1.placeholder(tf.int32)
c = a + b
print(sess.run(a, {a: [1, 2, 3]}))
print(sess.run(b, {b: [4, 5, 6]}))
print(sess.run(c, {a: [1, 2, 3], b: [4, 5, 6]}))
| false | 0 | 583 | 0 | 583 | 583 |
||
69031678
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
from pandas import Series
import numpy as np
from matplotlib import pyplot as plt
import seaborn as sns
from datetime import datetime
from sklearn.feature_selection import f_classif, mutual_info_classif
from sklearn.preprocessing import (
LabelEncoder,
OneHotEncoder,
StandardScaler,
RobustScaler,
)
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import (
confusion_matrix,
accuracy_score,
mean_squared_error,
precision_score,
recall_score,
f1_score,
)
from sklearn.metrics import auc, roc_auc_score, roc_curve, plot_confusion_matrix
from sklearn.model_selection import GridSearchCV
from sklearn.tree import DecisionTreeClassifier
# # Проект 5. Компьютер говорит «Нет»
# ##### Вам предоставлена информация из анкетных данных заемщиков и факт наличия дефолта.
# Описания полей client_id - идентификатор клиента
# education - уровень образования
# sex - пол заемщика
# age - возраст заемщика
# car - флаг наличия автомобиля
# car_type - флаг автомобиля иномарки
# decline_app_cnt - количество отказанных прошлых заявок
# good_work - флаг наличия “хорошей” работы
# bki_request_cnt - количество запросов в БКИ
# home_address - категоризатор домашнего адреса
# work_address - категоризатор рабочего адреса
# income - доход заемщика
# foreign_passport - наличие загранпаспорта
# sna - связь заемщика с клиентами банка
# first_time - давность наличия информации о заемщике
# score_bki - скоринговый балл по данным из БКИ
# region_rating - рейтинг региона
# app_date - дата подачи заявки
# default - флаг дефолта по кредиту
# ##### Цель: построить скоринг модель для вторичных клиентов банка, которая бы предсказывала вероятность дефолта клиента.
# функция логарифмирования
def log_func(x):
if x > 0: # логарифмирование применяется только к положительным числам
x = np.log((x) + 1)
return x
# загрузим данные
train = pd.read_csv("../input/sf-dst-scoring/train.csv")
test = pd.read_csv("../input/sf-dst-scoring/test.csv")
submission = pd.read_csv("../input/sf-dst-scoring/sample_submission.csv")
# ВАЖНО! дря корректной обработки признаков объединяем трейн и тест в один датасет
train["train"] = 1 # помечаем тренировочные
test["train"] = 0 # помечаем тестовые
data = pd.concat([train, test], ignore_index=True)
# объединяем
# ### Проведем разведывательный анализ данных и обработаем признаки
# посмотрим на данные
data.head()
display(data.info())
display(data.describe())
display(data.describe(include=["object"]))
# ##### Итого:
# Всего данные о 110148 клиентах. Всего 20 колонок, из них 6 типа object;
# из них sex, car, car_type, foreign_passport - скорее всего бинарные;
# остальные числовые, судя по min и max значениям явных выбросов нет;
# Все пропуски только в одном столбце - education, наиболее частое значение SCH;
# Нули в default добавлены для объединения;
# train добавили для обозначения принадлежности данных и последующего разделения.
data.education.fillna("SCH", inplace=True)
# заменим пропуски в education на наиболее частое значение SCH.
data.app_date = pd.to_datetime(data.app_date)
display(data.app_date.sample(5))
# создадим простые новые признаки на основе даты
data["app_day"] = data.app_date.dt.day
data["app_month"] = data.app_date.dt.month
data["app_year"] = data.app_date.dt.year
data["app_weekday"] = data["app_date"].dt.weekday
# также создадим признак количества прошедших дней с момента начала наблюдений,
# т.е. с 1 января 2014 года
data["app_number_days"] = (data["app_date"] - data.app_date.min()).dt.days.astype("int")
data.columns
# удалим ненужные столбцы app_year и app_date
data.drop(["app_date", "app_month", "app_year"], axis=1, inplace=True)
data.head()
# Посмотрим на распределение целевого признака 'default':
data["default"].plot(kind="hist", figsize=(5, 5)) # выборка не сбалансирована
# разделим наши признаки по группам
# бинарные переменные
bin_cols = ["sex", "car", "car_type", "good_work", "foreign_passport"]
# категориальные переменные
cat_cols = ["education", "home_address", "work_address", "sna", "first_time"]
# числовые переменные
num_cols = [
"age",
"decline_app_cnt",
"score_bki",
"bki_request_cnt",
"region_rating",
"income",
]
# client_id не включали ни в один список, не несет смысловой нагрузки для построения модели
num_cols
# рассмотрим числовые признаки
for col in num_cols:
sns.histplot(train[col], cbar=True, bins=50)
plt.show()
# построим матрицу корреляций для числовых признаков
plt.figure(figsize=(10, 5))
sns.heatmap(data[num_cols + ["default"]].corr(), cmap="coolwarm", annot=True)
# данные подходят для включения в модель
# рассмотрим категориальные признаки
# Значение 0 явно преобладает и заметна несбалансированность в выборке.
# в таком случае модель склонна переобучаться
# Попробуем применить в этом случае oversampling, чтобы сгладить эту разницу.
# zeroes = df[df['default']==0]
# ones = df[df['default']==1]
# default_new = int(len(zeroes)/len(ones))
# for i in range(default_new):
# df = df.append(ones).reset_index(drop=True)
##!!Этот код перенесем в раздел создания модели, чтобы была возможность сравнить с простой моделью
for col in cat_cols:
sns.barplot(x=col, y="default", data=data[[col, "default"]])
plt.show()
# судя по графикам признаки могут иметь влияние на целевую переменную
data["income"] = data.income.apply(log_func)
data["age"] = data.age.apply(log_func)
data["bki_request_cnt"] = data.bki_request_cnt.apply(log_func)
# преобразуем бинарные колонки
# преобразование с помощью LabelEncoder()
label_encoder = LabelEncoder()
# перед кодировкой зафиксируем маркировку значений
for i in bin_cols:
mapped = pd.Series(label_encoder.fit_transform(df[i]))
print(dict(enumerate(label_encoder.classes_)))
for column in bin_cols:
data[column] = label_encoder.fit_transform(data[column])
df.head()
# Проверим значимость бинарных переменных используя однофакторный дисперсионный анализ (ANOVA):
temp_df = data[data["train"] == 1]
imp_bin = Series(
mutual_info_classif(temp_df[bin_cols], temp_df["default"], discrete_features=True),
index=bin_cols,
)
imp_bin.sort_values(inplace=True)
imp_bin.plot(kind="barh")
# все признаки имеют влияние
# преобразуем данные признака education в числовые категории
mapped = pd.Series(label_encoder.fit_transform(data["education"]))
print(dict(enumerate(label_encoder.classes_)))
data["education"] = label_encoder.fit_transform(data["education"])
data.sample(3)
# ### Постороим модель
# реализуем OneHotEncoding для категориальных признаков
data = pd.get_dummies(data, columns=cat_cols)
# удалим ненужные переменные
# num_cols.remove('app_day')
# num_cols.remove('app_weekday')
data.drop(["app_day", "app_weekday"], axis=1, inplace=True)
# выделяем тренировочный датасет из датасета data
df_train = data.query("train == 1").drop(["train"], axis=1)
df_test = data.query("train == 0").drop(["train"], axis=1)
# разбиваем на признаки и целевую переменную
X = df_train.drop(["default"], axis=1).values
Y = df_train["default"].values
X_train, X_test, y_train, y_test = train_test_split(
X, Y, test_size=0.20, random_state=42
)
# проведем обучение
model_1 = LogisticRegression(solver="liblinear")
model_1.fit(X_train, y_train)
y_pred = model_1.predict(X_test)
y_proba = model_1.predict_proba(X_test)[:, 1]
fpr, tpr, threshold = roc_curve(y_test, y_proba)
roc_auc = roc_auc_score(y_test, y_proba)
plt.figure()
plt.plot([0, 1], label="Baseline", linestyle="--")
plt.plot(fpr, tpr, label="Regression")
plt.title("Logistic Regression ROC AUC = %0.3f" % roc_auc)
plt.ylabel("True Positive Rate")
plt.xlabel("False Positive Rate")
plt.legend(loc="lower right")
plt.show()
submission = df_test[["client_id", "default"]]
submission.to_csv("submission.csv", index=False)
submission.sample(3)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/031/69031678.ipynb
| null | null |
[{"Id": 69031678, "ScriptId": 18835444, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6957134, "CreationDate": "07/26/2021 04:49:25", "VersionNumber": 19.0, "Title": "[SF-DST] Credit Scoring", "EvaluationDate": "07/26/2021", "IsChange": false, "TotalLines": 262.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 262.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
from pandas import Series
import numpy as np
from matplotlib import pyplot as plt
import seaborn as sns
from datetime import datetime
from sklearn.feature_selection import f_classif, mutual_info_classif
from sklearn.preprocessing import (
LabelEncoder,
OneHotEncoder,
StandardScaler,
RobustScaler,
)
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import (
confusion_matrix,
accuracy_score,
mean_squared_error,
precision_score,
recall_score,
f1_score,
)
from sklearn.metrics import auc, roc_auc_score, roc_curve, plot_confusion_matrix
from sklearn.model_selection import GridSearchCV
from sklearn.tree import DecisionTreeClassifier
# # Проект 5. Компьютер говорит «Нет»
# ##### Вам предоставлена информация из анкетных данных заемщиков и факт наличия дефолта.
# Описания полей client_id - идентификатор клиента
# education - уровень образования
# sex - пол заемщика
# age - возраст заемщика
# car - флаг наличия автомобиля
# car_type - флаг автомобиля иномарки
# decline_app_cnt - количество отказанных прошлых заявок
# good_work - флаг наличия “хорошей” работы
# bki_request_cnt - количество запросов в БКИ
# home_address - категоризатор домашнего адреса
# work_address - категоризатор рабочего адреса
# income - доход заемщика
# foreign_passport - наличие загранпаспорта
# sna - связь заемщика с клиентами банка
# first_time - давность наличия информации о заемщике
# score_bki - скоринговый балл по данным из БКИ
# region_rating - рейтинг региона
# app_date - дата подачи заявки
# default - флаг дефолта по кредиту
# ##### Цель: построить скоринг модель для вторичных клиентов банка, которая бы предсказывала вероятность дефолта клиента.
# функция логарифмирования
def log_func(x):
if x > 0: # логарифмирование применяется только к положительным числам
x = np.log((x) + 1)
return x
# загрузим данные
train = pd.read_csv("../input/sf-dst-scoring/train.csv")
test = pd.read_csv("../input/sf-dst-scoring/test.csv")
submission = pd.read_csv("../input/sf-dst-scoring/sample_submission.csv")
# ВАЖНО! дря корректной обработки признаков объединяем трейн и тест в один датасет
train["train"] = 1 # помечаем тренировочные
test["train"] = 0 # помечаем тестовые
data = pd.concat([train, test], ignore_index=True)
# объединяем
# ### Проведем разведывательный анализ данных и обработаем признаки
# посмотрим на данные
data.head()
display(data.info())
display(data.describe())
display(data.describe(include=["object"]))
# ##### Итого:
# Всего данные о 110148 клиентах. Всего 20 колонок, из них 6 типа object;
# из них sex, car, car_type, foreign_passport - скорее всего бинарные;
# остальные числовые, судя по min и max значениям явных выбросов нет;
# Все пропуски только в одном столбце - education, наиболее частое значение SCH;
# Нули в default добавлены для объединения;
# train добавили для обозначения принадлежности данных и последующего разделения.
data.education.fillna("SCH", inplace=True)
# заменим пропуски в education на наиболее частое значение SCH.
data.app_date = pd.to_datetime(data.app_date)
display(data.app_date.sample(5))
# создадим простые новые признаки на основе даты
data["app_day"] = data.app_date.dt.day
data["app_month"] = data.app_date.dt.month
data["app_year"] = data.app_date.dt.year
data["app_weekday"] = data["app_date"].dt.weekday
# также создадим признак количества прошедших дней с момента начала наблюдений,
# т.е. с 1 января 2014 года
data["app_number_days"] = (data["app_date"] - data.app_date.min()).dt.days.astype("int")
data.columns
# удалим ненужные столбцы app_year и app_date
data.drop(["app_date", "app_month", "app_year"], axis=1, inplace=True)
data.head()
# Посмотрим на распределение целевого признака 'default':
data["default"].plot(kind="hist", figsize=(5, 5)) # выборка не сбалансирована
# разделим наши признаки по группам
# бинарные переменные
bin_cols = ["sex", "car", "car_type", "good_work", "foreign_passport"]
# категориальные переменные
cat_cols = ["education", "home_address", "work_address", "sna", "first_time"]
# числовые переменные
num_cols = [
"age",
"decline_app_cnt",
"score_bki",
"bki_request_cnt",
"region_rating",
"income",
]
# client_id не включали ни в один список, не несет смысловой нагрузки для построения модели
num_cols
# рассмотрим числовые признаки
for col in num_cols:
sns.histplot(train[col], cbar=True, bins=50)
plt.show()
# построим матрицу корреляций для числовых признаков
plt.figure(figsize=(10, 5))
sns.heatmap(data[num_cols + ["default"]].corr(), cmap="coolwarm", annot=True)
# данные подходят для включения в модель
# рассмотрим категориальные признаки
# Значение 0 явно преобладает и заметна несбалансированность в выборке.
# в таком случае модель склонна переобучаться
# Попробуем применить в этом случае oversampling, чтобы сгладить эту разницу.
# zeroes = df[df['default']==0]
# ones = df[df['default']==1]
# default_new = int(len(zeroes)/len(ones))
# for i in range(default_new):
# df = df.append(ones).reset_index(drop=True)
##!!Этот код перенесем в раздел создания модели, чтобы была возможность сравнить с простой моделью
for col in cat_cols:
sns.barplot(x=col, y="default", data=data[[col, "default"]])
plt.show()
# судя по графикам признаки могут иметь влияние на целевую переменную
data["income"] = data.income.apply(log_func)
data["age"] = data.age.apply(log_func)
data["bki_request_cnt"] = data.bki_request_cnt.apply(log_func)
# преобразуем бинарные колонки
# преобразование с помощью LabelEncoder()
label_encoder = LabelEncoder()
# перед кодировкой зафиксируем маркировку значений
for i in bin_cols:
mapped = pd.Series(label_encoder.fit_transform(df[i]))
print(dict(enumerate(label_encoder.classes_)))
for column in bin_cols:
data[column] = label_encoder.fit_transform(data[column])
df.head()
# Проверим значимость бинарных переменных используя однофакторный дисперсионный анализ (ANOVA):
temp_df = data[data["train"] == 1]
imp_bin = Series(
mutual_info_classif(temp_df[bin_cols], temp_df["default"], discrete_features=True),
index=bin_cols,
)
imp_bin.sort_values(inplace=True)
imp_bin.plot(kind="barh")
# все признаки имеют влияние
# преобразуем данные признака education в числовые категории
mapped = pd.Series(label_encoder.fit_transform(data["education"]))
print(dict(enumerate(label_encoder.classes_)))
data["education"] = label_encoder.fit_transform(data["education"])
data.sample(3)
# ### Постороим модель
# реализуем OneHotEncoding для категориальных признаков
data = pd.get_dummies(data, columns=cat_cols)
# удалим ненужные переменные
# num_cols.remove('app_day')
# num_cols.remove('app_weekday')
data.drop(["app_day", "app_weekday"], axis=1, inplace=True)
# выделяем тренировочный датасет из датасета data
df_train = data.query("train == 1").drop(["train"], axis=1)
df_test = data.query("train == 0").drop(["train"], axis=1)
# разбиваем на признаки и целевую переменную
X = df_train.drop(["default"], axis=1).values
Y = df_train["default"].values
X_train, X_test, y_train, y_test = train_test_split(
X, Y, test_size=0.20, random_state=42
)
# проведем обучение
model_1 = LogisticRegression(solver="liblinear")
model_1.fit(X_train, y_train)
y_pred = model_1.predict(X_test)
y_proba = model_1.predict_proba(X_test)[:, 1]
fpr, tpr, threshold = roc_curve(y_test, y_proba)
roc_auc = roc_auc_score(y_test, y_proba)
plt.figure()
plt.plot([0, 1], label="Baseline", linestyle="--")
plt.plot(fpr, tpr, label="Regression")
plt.title("Logistic Regression ROC AUC = %0.3f" % roc_auc)
plt.ylabel("True Positive Rate")
plt.xlabel("False Positive Rate")
plt.legend(loc="lower right")
plt.show()
submission = df_test[["client_id", "default"]]
submission.to_csv("submission.csv", index=False)
submission.sample(3)
| false | 0 | 3,225 | 0 | 3,225 | 3,225 |
||
69031810
|
<jupyter_start><jupyter_text>Sri Lanka Vehicle Prices Dataset
### Context
The Reason I wanted to create this dataset is that I was in need of such data once. I wanted to help my father get a rough idea of vehicle prices in SL by creating Data Visuals. The process when done manually is very tedious and is a boring process especially when there are thousands of data points. I wrote an automated script to scrape new data so that I can keep the dataset up to date.
Kaggle dataset identifier: sri-lanka-vehicle-prices-dataset
<jupyter_code>import pandas as pd
df = pd.read_csv('sri-lanka-vehicle-prices-dataset/vehicle_data.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 123971 entries, 0 to 123970
Data columns (total 19 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Title 123971 non-null object
1 Sub_title 123971 non-null object
2 Price 123971 non-null object
3 Brand 123971 non-null object
4 Model 123970 non-null object
5 Edition 47538 non-null object
6 Year 123971 non-null int64
7 Condition 123971 non-null object
8 Transmission 123970 non-null object
9 Body 118876 non-null object
10 Fuel 123971 non-null object
11 Capacity 123766 non-null object
12 Mileage 123971 non-null object
13 Location 123971 non-null object
14 Description 107797 non-null object
15 Post_URL 123971 non-null object
16 Seller_name 123873 non-null object
17 Seller_type 123971 non-null object
18 published_date 123971 non-null object
dtypes: int64(1), object(18)
memory usage: 18.0+ MB
<jupyter_text>Examples:
{
"Title": "Honda Vezel Limited 2017 for sale",
"Sub_title": "Posted on 01 Sep 2:45 pm, Kurunegala City, Kurunegala",
"Price": "Rs 12,900,000",
"Brand": "Honda",
"Model": "Vezel",
"Edition": "Limited",
"Year": 2017,
"Condition": "Used",
"Transmission": "Automatic",
"Body": "SUV / 4x4",
"Fuel": "Hybrid",
"Capacity": "1,500 cc",
"Mileage": "37,000 km",
"Location": " Kurunegala City, Kurunegala",
"Description": "HONDA VEZEL LIMITEDCBE-XXXXMANU YEAR : 2017LIMITED EDITIONIVORY PACKAGEPUSH STARTSMART KEYSAFETY BREAK SYSTEMTRACTION CONTROLLERCOMPANY FITTED DVD AUDIO VIDEO SYSTEMMULTI FUNCTION LEATHER WRAPPING STEERING WHEELLEATHER SEATSPOWER SEATS (FRONT 02 SEATS)MEMORY SEATCRUISE CONTROLLER...(truncated)",
"Post_URL": "https://ikman.lk/en/ad/honda-vezel-limited-2017-for-sale-kurunegala",
"Seller_name": "Prasad Enterprises",
"Seller_type": "Premium-Member",
"published_date": "2022-09-01 14:45:00"
}
{
"Title": "Jaguar XF Fully Loaded 2013 for sale",
"Sub_title": "Posted on 28 Sep 10:23 am, Kurunegala City, Kurunegala",
"Price": "Rs 16,250,000",
"Brand": "Jaguar",
"Model": "XF",
"Edition": "Fully Loaded",
"Year": 2013,
"Condition": "Used",
"Transmission": "Automatic",
"Body": "Saloon",
"Fuel": "Diesel",
"Capacity": "2,200 cc",
"Mileage": "42,000 km",
"Location": " Kurunegala City, Kurunegala",
"Description": "1st owner(registered owner)Diesel(13-14km per a ltr)Genuine mileage(42000km)VIP numberLow insurance premium(100% Accident free)Use by an engineerFully loaded(so many options)New tiresBeige interiorTurbo engineShowroom condtionJaguar land rover company maintained and all records a...(truncated)",
"Post_URL": "https://ikman.lk/en/ad/jaguar-xf-fully-loaded-2013-for-sale-kurunegala-1",
"Seller_name": "Dhanushka Dharmasri",
"Seller_type": "Member",
"published_date": "2022-09-28 10:23:00"
}
{
"Title": "Toyota CHR GT New Face 2wd 2020 for sale",
"Sub_title": "Posted on 16 Sep 1:16 pm, Colombo 7, Colombo",
"Price": "Rs 17,000,000",
"Brand": "Toyota",
"Model": "CHR",
"Edition": "GT New Face 2wd",
"Year": 2020,
"Condition": "Used",
"Transmission": "Automatic",
"Body": "SUV / 4x4",
"Fuel": "Petrol",
"Capacity": "1,200 cc",
"Mileage": "15,000 km",
"Location": " Colombo 7, Colombo",
"Description": "2020 Facelift modelFirst owner (No previous owners)15,000 KM on the clock Maintaned at Toyota Lanka Still under warranty till 2023",
"Post_URL": "https://ikman.lk/en/ad/toyota-chr-gt-new-face-2wd-2020-for-sale-colombo-1",
"Seller_name": "Carzone Lanka",
"Seller_type": "Member",
"published_date": "2022-09-16 13:16:00"
}
{
"Title": "Toyota CHR Toyato 2017 for sale",
"Sub_title": "Posted on 30 Sep 10:12 am, Colombo 7, Colombo",
"Price": "Rs 13,000,000",
"Brand": "Toyota",
"Model": "CHR",
"Edition": "Toyato",
"Year": 2017,
"Condition": "Used",
"Transmission": "Automatic",
"Body": "SUV / 4x4",
"Fuel": "Petrol",
"Capacity": "1,200 cc",
"Mileage": "21,000 km",
"Location": " Colombo 7, Colombo",
"Description": "Toyota CHR GT Turbo1st owner Imported as a brand new car.All import documents are available 21,000 KM Genuine Mileage with all service records from Toyota Lanka and emission tests records available \u00e2\u0080\u00a2 1200 CC Turbo Petrol\u00e2\u0080\u00a2 18\u00e2\u0080\u00b3 all...(truncated)",
"Post_URL": "https://ikman.lk/en/ad/toyota-chr-toyato-2017-for-sale-colombo",
"Seller_name": "Carzone Lanka",
"Seller_type": "Member",
"published_date": "2022-09-30 10:12:00"
}
<jupyter_script># ### Importing Libraries
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib.figure import Figure
from mpl_toolkits.mplot3d import Axes3D
from matplotlib.colors import ListedColormap
# ### Loading Data
df = pd.read_csv("/kaggle/input/sri-lanka-vehicle-prices-dataset/vehicle_data.csv")
# ### Explolatory Data Analysis
df.head()
df.isnull().sum()
### Converting Columns into appropiate data types
def extract_num(row):
amount = ""
for w in row:
if w.isnumeric():
amount += w
return int(amount)
df["Price"] = df["Price"].apply(extract_num)
df["Mileage"] = df["Mileage"].apply(extract_num)
df["Capacity"] = df["Capacity"].apply(extract_num)
df = df.rename(
{"Price": "Price (Rs)", "Mileage": "Mileage (km)", "Capacity": "Capacity (cc)"},
axis=1,
)
df.describe()
### Obtaining the Districts from given location
df["District"] = df["Location"].apply(lambda x: x.split(", ")[-1])
# ### Data Visualization
### Plotting top 5 Sellers who have posted the most ads
top_10_seller = (
df[["Title", "Seller_name"]]
.groupby("Seller_name")
.agg(["count"])["Title"]["count"]
.sort_values(ascending=False)[:5]
)
plt = sns.barplot(x=top_10_seller.index, y=top_10_seller.values)
plt.set_xticklabels(labels=top_10_seller.index, rotation=45)
plt.set_ylabel("Number of Posted Ads")
plt.set_xlabel("Seller Name")
plt.set_title("Top 5 Sellers who have posted the most ads")
### Plotting top 5 Most Car Brands Availableto Buy
top_10_car_brand = (
df[["Title", "Brand"]]
.groupby("Brand")
.agg(["count"])["Title"]["count"]
.sort_values(ascending=False)[:5]
)
plt = sns.barplot(x=top_10_car_brand.index, y=top_10_car_brand.values)
plt.set_xticklabels(labels=top_10_car_brand.index, rotation=45)
plt.set_ylabel("Number of Cars For Sale")
plt.set_xlabel("Car Brand")
plt.set_title("Top 5 Most Available Car Brands to Buy")
# Average Price of Carsover the Years
plt = sns.lineplot(data=df, x="Year", y="Price (Rs)")
plt.set_title("Average Price of Cars Over the Years")
# Correlation of Numeric Features in the Dataset
plt = sns.heatmap(df.corr())
plt.set_title("Correlation of Numeric Features")
### 3D plot for Capacity, Price and Year.
fig = Figure()
ax = plt.axes(projection="3d")
ax.scatter3D(df["Year"], df["Capacity (cc)"], df["Price (Rs)"], c=df["Price (Rs)"])
ax.set_xlabel("Year")
ax.set_ylabel("Capacity (cc)")
ax.set_zlabel("Price (Rs)")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/031/69031810.ipynb
|
sri-lanka-vehicle-prices-dataset
|
lasaljaywardena
|
[{"Id": 69031810, "ScriptId": 18828934, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6361330, "CreationDate": "07/26/2021 04:52:05", "VersionNumber": 2.0, "Title": "EDA and Data Viz", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 80.0, "LinesInsertedFromPrevious": 13.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 67.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 91746815, "KernelVersionId": 69031810, "SourceDatasetVersionId": 2461758}]
|
[{"Id": 2461758, "DatasetId": 1490104, "DatasourceVersionId": 2504182, "CreatorUserId": 6361330, "LicenseName": "ODC Attribution License (ODC-By)", "CreationDate": "07/25/2021 14:38:36", "VersionNumber": 1.0, "Title": "Sri Lanka Vehicle Prices Dataset", "Slug": "sri-lanka-vehicle-prices-dataset", "Subtitle": "Prices of Cars in Sri Lanka Obtained from 2021", "Description": "### Context\n\nThe Reason I wanted to create this dataset is that I was in need of such data once. I wanted to help my father get a rough idea of vehicle prices in SL by creating Data Visuals. The process when done manually is very tedious and is a boring process especially when there are thousands of data points. I wrote an automated script to scrape new data so that I can keep the dataset up to date. \n\n\n### Acknowledgements\n\nThe data is scraped from Sri Lanka's most famous online vehicle buying selling platform: Ikman.lk. The Credit for all the information goes to Ikman. This Dataset is just a tool for all the Data Scientists out there interested in inferring something more out of the data.\n\n\n### Inspiration\n\nIt would be great if we could create machine learning models to better identify trends of vehicle data in Sri Lanka. It is also nice if we could create Data Dashboards with help of software like Tableau and Power BI. The opportunities are limitless!\n\nFinally, I would be very happy if this dataset could bring a positive impact on your life or your workflow. And cheers!!!", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1490104, "CreatorUserId": 6361330, "OwnerUserId": 6361330.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 3300352.0, "CurrentDatasourceVersionId": 3351126.0, "ForumId": 1509805, "Type": 2, "CreationDate": "07/25/2021 14:38:36", "LastActivityDate": "07/25/2021", "TotalViews": 12360, "TotalDownloads": 1386, "TotalVotes": 83, "TotalKernels": 8}]
|
[{"Id": 6361330, "UserName": "lasaljaywardena", "DisplayName": "Lasal Jayawardena", "RegisterDate": "12/11/2020", "PerformanceTier": 2}]
|
# ### Importing Libraries
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib.figure import Figure
from mpl_toolkits.mplot3d import Axes3D
from matplotlib.colors import ListedColormap
# ### Loading Data
df = pd.read_csv("/kaggle/input/sri-lanka-vehicle-prices-dataset/vehicle_data.csv")
# ### Explolatory Data Analysis
df.head()
df.isnull().sum()
### Converting Columns into appropiate data types
def extract_num(row):
amount = ""
for w in row:
if w.isnumeric():
amount += w
return int(amount)
df["Price"] = df["Price"].apply(extract_num)
df["Mileage"] = df["Mileage"].apply(extract_num)
df["Capacity"] = df["Capacity"].apply(extract_num)
df = df.rename(
{"Price": "Price (Rs)", "Mileage": "Mileage (km)", "Capacity": "Capacity (cc)"},
axis=1,
)
df.describe()
### Obtaining the Districts from given location
df["District"] = df["Location"].apply(lambda x: x.split(", ")[-1])
# ### Data Visualization
### Plotting top 5 Sellers who have posted the most ads
top_10_seller = (
df[["Title", "Seller_name"]]
.groupby("Seller_name")
.agg(["count"])["Title"]["count"]
.sort_values(ascending=False)[:5]
)
plt = sns.barplot(x=top_10_seller.index, y=top_10_seller.values)
plt.set_xticklabels(labels=top_10_seller.index, rotation=45)
plt.set_ylabel("Number of Posted Ads")
plt.set_xlabel("Seller Name")
plt.set_title("Top 5 Sellers who have posted the most ads")
### Plotting top 5 Most Car Brands Availableto Buy
top_10_car_brand = (
df[["Title", "Brand"]]
.groupby("Brand")
.agg(["count"])["Title"]["count"]
.sort_values(ascending=False)[:5]
)
plt = sns.barplot(x=top_10_car_brand.index, y=top_10_car_brand.values)
plt.set_xticklabels(labels=top_10_car_brand.index, rotation=45)
plt.set_ylabel("Number of Cars For Sale")
plt.set_xlabel("Car Brand")
plt.set_title("Top 5 Most Available Car Brands to Buy")
# Average Price of Carsover the Years
plt = sns.lineplot(data=df, x="Year", y="Price (Rs)")
plt.set_title("Average Price of Cars Over the Years")
# Correlation of Numeric Features in the Dataset
plt = sns.heatmap(df.corr())
plt.set_title("Correlation of Numeric Features")
### 3D plot for Capacity, Price and Year.
fig = Figure()
ax = plt.axes(projection="3d")
ax.scatter3D(df["Year"], df["Capacity (cc)"], df["Price (Rs)"], c=df["Price (Rs)"])
ax.set_xlabel("Year")
ax.set_ylabel("Capacity (cc)")
ax.set_zlabel("Price (Rs)")
|
[{"sri-lanka-vehicle-prices-dataset/vehicle_data.csv": {"column_names": "[\"Title\", \"Sub_title\", \"Price\", \"Brand\", \"Model\", \"Edition\", \"Year\", \"Condition\", \"Transmission\", \"Body\", \"Fuel\", \"Capacity\", \"Mileage\", \"Location\", \"Description\", \"Post_URL\", \"Seller_name\", \"Seller_type\", \"published_date\"]", "column_data_types": "{\"Title\": \"object\", \"Sub_title\": \"object\", \"Price\": \"object\", \"Brand\": \"object\", \"Model\": \"object\", \"Edition\": \"object\", \"Year\": \"int64\", \"Condition\": \"object\", \"Transmission\": \"object\", \"Body\": \"object\", \"Fuel\": \"object\", \"Capacity\": \"object\", \"Mileage\": \"object\", \"Location\": \"object\", \"Description\": \"object\", \"Post_URL\": \"object\", \"Seller_name\": \"object\", \"Seller_type\": \"object\", \"published_date\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 123971 entries, 0 to 123970\nData columns (total 19 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Title 123971 non-null object\n 1 Sub_title 123971 non-null object\n 2 Price 123971 non-null object\n 3 Brand 123971 non-null object\n 4 Model 123970 non-null object\n 5 Edition 47538 non-null object\n 6 Year 123971 non-null int64 \n 7 Condition 123971 non-null object\n 8 Transmission 123970 non-null object\n 9 Body 118876 non-null object\n 10 Fuel 123971 non-null object\n 11 Capacity 123766 non-null object\n 12 Mileage 123971 non-null object\n 13 Location 123971 non-null object\n 14 Description 107797 non-null object\n 15 Post_URL 123971 non-null object\n 16 Seller_name 123873 non-null object\n 17 Seller_type 123971 non-null object\n 18 published_date 123971 non-null object\ndtypes: int64(1), object(18)\nmemory usage: 18.0+ MB\n", "summary": "{\"Year\": {\"count\": 123971.0, \"mean\": 2008.4097651870195, \"std\": 25.330834976349536, \"min\": 0.0, \"25%\": 2004.0, \"50%\": 2013.0, \"75%\": 2016.0, \"max\": 2561.0}}", "examples": "{\"Title\":{\"0\":\"Honda Vezel Limited 2017 for sale\",\"1\":\"Jaguar XF Fully Loaded 2013 for sale\",\"2\":\"Toyota CHR GT New Face 2wd 2020 for sale\",\"3\":\"Toyota CHR Toyato 2017 for sale\"},\"Sub_title\":{\"0\":\"Posted on 01 Sep 2:45 pm, Kurunegala City, Kurunegala\",\"1\":\"Posted on 28 Sep 10:23 am, Kurunegala City, Kurunegala\",\"2\":\"Posted on 16 Sep 1:16 pm, Colombo 7, Colombo\",\"3\":\"Posted on 30 Sep 10:12 am, Colombo 7, Colombo\"},\"Price\":{\"0\":\"Rs 12,900,000\",\"1\":\"Rs 16,250,000\",\"2\":\"Rs 17,000,000\",\"3\":\"Rs 13,000,000\"},\"Brand\":{\"0\":\"Honda\",\"1\":\"Jaguar\",\"2\":\"Toyota\",\"3\":\"Toyota\"},\"Model\":{\"0\":\"Vezel\",\"1\":\"XF\",\"2\":\"CHR\",\"3\":\"CHR\"},\"Edition\":{\"0\":\"Limited\",\"1\":\"Fully Loaded\",\"2\":\"GT New Face 2wd\",\"3\":\"Toyato\"},\"Year\":{\"0\":2017,\"1\":2013,\"2\":2020,\"3\":2017},\"Condition\":{\"0\":\"Used\",\"1\":\"Used\",\"2\":\"Used\",\"3\":\"Used\"},\"Transmission\":{\"0\":\"Automatic\",\"1\":\"Automatic\",\"2\":\"Automatic\",\"3\":\"Automatic\"},\"Body\":{\"0\":\"SUV \\/ 4x4\",\"1\":\"Saloon\",\"2\":\"SUV \\/ 4x4\",\"3\":\"SUV \\/ 4x4\"},\"Fuel\":{\"0\":\"Hybrid\",\"1\":\"Diesel\",\"2\":\"Petrol\",\"3\":\"Petrol\"},\"Capacity\":{\"0\":\"1,500 cc\",\"1\":\"2,200 cc\",\"2\":\"1,200 cc\",\"3\":\"1,200 cc\"},\"Mileage\":{\"0\":\"37,000 km\",\"1\":\"42,000 km\",\"2\":\"15,000 km\",\"3\":\"21,000 km\"},\"Location\":{\"0\":\" Kurunegala City, Kurunegala\",\"1\":\" Kurunegala City, Kurunegala\",\"2\":\" Colombo 7, Colombo\",\"3\":\" Colombo 7, Colombo\"},\"Description\":{\"0\":\"HONDA VEZEL LIMITEDCBE-XXXXMANU YEAR : 2017LIMITED EDITIONIVORY PACKAGEPUSH STARTSMART KEYSAFETY BREAK SYSTEMTRACTION CONTROLLERCOMPANY FITTED DVD AUDIO VIDEO SYSTEMMULTI FUNCTION LEATHER WRAPPING STEERING WHEELLEATHER SEATSPOWER SEATS (FRONT 02 SEATS)MEMORY SEATCRUISE CONTROLLERSENSIN BREAK SYSTEMPADDLE SHIFT TOUCH AC\\/BLOWER CONTROLLER SYSTEMLED DAYTIME RUNNING LIGHTSSCOOP HEAD LAMPLED FOG LAMPSPECIAL DESIGNED TAIL LAMPS CHROME DOOR MOLDINGREAR SPOILERCHROME ROOF RAILALLOY WHEELDOOR VISORABSAIR BAGSFULLY LOADED\",\"1\":\"1st owner(registered owner)Diesel(13-14km per a ltr)Genuine mileage(42000km)VIP numberLow insurance premium(100% Accident free)Use by an engineerFully loaded(so many options)New tiresBeige interiorTurbo engineShowroom condtionJaguar land rover company maintained and all records awailable(more photos - pls whatsapp )\",\"2\":\"2020 Facelift modelFirst owner (No previous owners)15,000 KM on the clock Maintaned at Toyota Lanka Still under warranty till 2023\",\"3\":\"Toyota CHR GT Turbo1st owner Imported as a brand new car.All import documents are available 21,000 KM Genuine Mileage with all service records from Toyota Lanka and emission tests records available \\u00e2\\u0080\\u00a2 1200 CC Turbo Petrol\\u00e2\\u0080\\u00a2 18\\u00e2\\u0080\\u00b3 alloy wheels\\u00e2\\u0080\\u00a2 Original Kenwood audio system \\u00e2\\u0080\\u00a2 Auto Headlights\\u00e2\\u0080\\u00a2 Reversing Camera with guild lines \\u00e2\\u0080\\u00a2 LED Daytime running lights\\u00e2\\u0080\\u00a2 Pre-Collision System with Pedestrian Detection\\u00e2\\u0080\\u00a2 Lane Departure Alert with Steering Control\\u00e2\\u0080\\u00a2 Adaptive Cruise Control\\u00e2\\u0080\\u00a2 Anti-lock Braking System (ABS)\\u00e2\\u0080\\u00a2 Automatic High Beam\\u00e2\\u0080\\u00a2 Adaptive Cruise Control\\u00e2\\u0080\\u00a2 Follow-me-home headlights\\u00e2\\u0080\\u00a2 Vehicle Stability Control (VSC)\\u00e2\\u0080\\u00a2 Immobiliser Motion sensor & Intrusion alarm\\u00e2\\u0080\\u00a2 Aux in connectors + 12v front outlet\\u00e2\\u0080\\u00a2 6 speakers\\u00e2\\u0080\\u00a2 Road Sign Assist\\u00e2\\u0080\\u00a2 TFT screen for multi-information display\\u00e2\\u0080\\u00a2 Dual-zone automatic air conditioning\\u00e2\\u0080\\u00a2 Audio & telephone switches on steering wheel\\u00e2\\u0080\\u00a2 Electric Power Steering\\u00e2\\u0080\\u00a2 3-spoke leather steering wheel with multimedia switches\\u00e2\\u0080\\u00a2 Electronic parking brake Leather gear shift 4.2\\u00e2\\u0080\\u00b3 Coloured\\u00e2\\u0080\\u00a2 (EPS) Auto-dimming rear view mirror\\u00e2\\u0080\\u00a2 Front & rear power windows with auto up & down and anti-jam protection illuminated entry system\"},\"Post_URL\":{\"0\":\"https:\\/\\/ikman.lk\\/en\\/ad\\/honda-vezel-limited-2017-for-sale-kurunegala\",\"1\":\"https:\\/\\/ikman.lk\\/en\\/ad\\/jaguar-xf-fully-loaded-2013-for-sale-kurunegala-1\",\"2\":\"https:\\/\\/ikman.lk\\/en\\/ad\\/toyota-chr-gt-new-face-2wd-2020-for-sale-colombo-1\",\"3\":\"https:\\/\\/ikman.lk\\/en\\/ad\\/toyota-chr-toyato-2017-for-sale-colombo\"},\"Seller_name\":{\"0\":\"Prasad Enterprises\",\"1\":\"Dhanushka Dharmasri\",\"2\":\"Carzone Lanka\",\"3\":\"Carzone Lanka\"},\"Seller_type\":{\"0\":\"Premium-Member\",\"1\":\"Member\",\"2\":\"Member\",\"3\":\"Member\"},\"published_date\":{\"0\":\"2022-09-01 14:45:00\",\"1\":\"2022-09-28 10:23:00\",\"2\":\"2022-09-16 13:16:00\",\"3\":\"2022-09-30 10:12:00\"}}"}}]
| true | 1 |
<start_data_description><data_path>sri-lanka-vehicle-prices-dataset/vehicle_data.csv:
<column_names>
['Title', 'Sub_title', 'Price', 'Brand', 'Model', 'Edition', 'Year', 'Condition', 'Transmission', 'Body', 'Fuel', 'Capacity', 'Mileage', 'Location', 'Description', 'Post_URL', 'Seller_name', 'Seller_type', 'published_date']
<column_types>
{'Title': 'object', 'Sub_title': 'object', 'Price': 'object', 'Brand': 'object', 'Model': 'object', 'Edition': 'object', 'Year': 'int64', 'Condition': 'object', 'Transmission': 'object', 'Body': 'object', 'Fuel': 'object', 'Capacity': 'object', 'Mileage': 'object', 'Location': 'object', 'Description': 'object', 'Post_URL': 'object', 'Seller_name': 'object', 'Seller_type': 'object', 'published_date': 'object'}
<dataframe_Summary>
{'Year': {'count': 123971.0, 'mean': 2008.4097651870195, 'std': 25.330834976349536, 'min': 0.0, '25%': 2004.0, '50%': 2013.0, '75%': 2016.0, 'max': 2561.0}}
<dataframe_info>
RangeIndex: 123971 entries, 0 to 123970
Data columns (total 19 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Title 123971 non-null object
1 Sub_title 123971 non-null object
2 Price 123971 non-null object
3 Brand 123971 non-null object
4 Model 123970 non-null object
5 Edition 47538 non-null object
6 Year 123971 non-null int64
7 Condition 123971 non-null object
8 Transmission 123970 non-null object
9 Body 118876 non-null object
10 Fuel 123971 non-null object
11 Capacity 123766 non-null object
12 Mileage 123971 non-null object
13 Location 123971 non-null object
14 Description 107797 non-null object
15 Post_URL 123971 non-null object
16 Seller_name 123873 non-null object
17 Seller_type 123971 non-null object
18 published_date 123971 non-null object
dtypes: int64(1), object(18)
memory usage: 18.0+ MB
<some_examples>
{'Title': {'0': 'Honda Vezel Limited 2017 for sale', '1': 'Jaguar XF Fully Loaded 2013 for sale', '2': 'Toyota CHR GT New Face 2wd 2020 for sale', '3': 'Toyota CHR Toyato 2017 for sale'}, 'Sub_title': {'0': 'Posted on 01 Sep 2:45 pm, Kurunegala City, Kurunegala', '1': 'Posted on 28 Sep 10:23 am, Kurunegala City, Kurunegala', '2': 'Posted on 16 Sep 1:16 pm, Colombo 7, Colombo', '3': 'Posted on 30 Sep 10:12 am, Colombo 7, Colombo'}, 'Price': {'0': 'Rs 12,900,000', '1': 'Rs 16,250,000', '2': 'Rs 17,000,000', '3': 'Rs 13,000,000'}, 'Brand': {'0': 'Honda', '1': 'Jaguar', '2': 'Toyota', '3': 'Toyota'}, 'Model': {'0': 'Vezel', '1': 'XF', '2': 'CHR', '3': 'CHR'}, 'Edition': {'0': 'Limited', '1': 'Fully Loaded', '2': 'GT New Face 2wd', '3': 'Toyato'}, 'Year': {'0': 2017, '1': 2013, '2': 2020, '3': 2017}, 'Condition': {'0': 'Used', '1': 'Used', '2': 'Used', '3': 'Used'}, 'Transmission': {'0': 'Automatic', '1': 'Automatic', '2': 'Automatic', '3': 'Automatic'}, 'Body': {'0': 'SUV / 4x4', '1': 'Saloon', '2': 'SUV / 4x4', '3': 'SUV / 4x4'}, 'Fuel': {'0': 'Hybrid', '1': 'Diesel', '2': 'Petrol', '3': 'Petrol'}, 'Capacity': {'0': '1,500 cc', '1': '2,200 cc', '2': '1,200 cc', '3': '1,200 cc'}, 'Mileage': {'0': '37,000 km', '1': '42,000 km', '2': '15,000 km', '3': '21,000 km'}, 'Location': {'0': ' Kurunegala City, Kurunegala', '1': ' Kurunegala City, Kurunegala', '2': ' Colombo 7, Colombo', '3': ' Colombo 7, Colombo'}, 'Description': {'0': 'HONDA VEZEL LIMITEDCBE-XXXXMANU YEAR : 2017LIMITED EDITIONIVORY PACKAGEPUSH STARTSMART KEYSAFETY BREAK SYSTEMTRACTION CONTROLLERCOMPANY FITTED DVD AUDIO VIDEO SYSTEMMULTI FUNCTION LEATHER WRAPPING STEERING WHEELLEATHER SEATSPOWER SEATS (FRONT 02 SEATS)MEMORY SEATCRUISE CONTROLLERSENSIN BREAK SYSTEMPADDLE SHIFT TOUCH AC/BLOWER CONTROLLER SYSTEMLED DAYTIME RUNNING LIGHTSSCOOP HEAD LAMPLED FOG LAMPSPECIAL DESIGNED TAIL LAMPS CHROME DOOR MOLDINGREAR SPOILERCHROME ROOF RAILALLOY WHEELDOOR VISORABSAIR BAGSFULLY LOADED', '1': '1st owner(registered owner)Diesel(13-14km per a ltr)Genuine mileage(42000km)VIP numberLow insurance premium(100% Accident free)Use by an engineerFully loaded(so many options)New tiresBeige interiorTurbo engineShowroom condtionJaguar land rover company maintained and all records awailable(more photos - pls whatsapp )', '2': '2020 Facelift modelFirst owner (No previous owners)15,000 KM on the clock Maintaned at Toyota Lanka Still under warranty till 2023', '3': 'Toyota CHR GT Turbo1st owner Imported as a brand new car.All import documents are available 21,000 KM Genuine Mileage with all service records from Toyota Lanka and emission tests records available â\x80¢ 1200 CC Turbo Petrolâ\x80¢ 18â\x80³ alloy wheelsâ\x80¢ Original Kenwood audio system â\x80¢ Auto Headlightsâ\x80¢ Reversing Camera with guild lines â\x80¢ LED Daytime running lightsâ\x80¢ Pre-Collision System with Pedestrian Detectionâ\x80¢ Lane Departure Alert with Steering Controlâ\x80¢ Adaptive Cruise Controlâ\x80¢ Anti-lock Braking System (ABS)â\x80¢ Automatic High Beamâ\x80¢ Adaptive Cruise Controlâ\x80¢ Follow-me-home headlightsâ\x80¢ Vehicle Stability Control (VSC)â\x80¢ Immobiliser Motion sensor & Intrusion alarmâ\x80¢ Aux in connectors + 12v front outletâ\x80¢ 6 speakersâ\x80¢ Road Sign Assistâ\x80¢ TFT screen for multi-information displayâ\x80¢ Dual-zone automatic air conditioningâ\x80¢ Audio & telephone switches on steering wheelâ\x80¢ Electric Power Steeringâ\x80¢ 3-spoke leather steering wheel with multimedia switchesâ\x80¢ Electronic parking brake Leather gear shift 4.2â\x80³ Colouredâ\x80¢ (EPS) Auto-dimming rear view mirrorâ\x80¢ Front & rear power windows with auto up & down and anti-jam protection illuminated entry system'}, 'Post_URL': {'0': 'https://ikman.lk/en/ad/honda-vezel-limited-2017-for-sale-kurunegala', '1': 'https://ikman.lk/en/ad/jaguar-xf-fully-loaded-2013-for-sale-kurunegala-1', '2': 'https://ikman.lk/en/ad/toyota-chr-gt-new-face-2wd-2020-for-sale-colombo-1', '3': 'https://ikman.lk/en/ad/toyota-chr-toyato-2017-for-sale-colombo'}, 'Seller_name': {'0': 'Prasad Enterprises', '1': 'Dhanushka Dharmasri', '2': 'Carzone Lanka', '3': 'Carzone Lanka'}, 'Seller_type': {'0': 'Premium-Member', '1': 'Member', '2': 'Member', '3': 'Member'}, 'published_date': {'0': '2022-09-01 14:45:00', '1': '2022-09-28 10:23:00', '2': '2022-09-16 13:16:00', '3': '2022-09-30 10:12:00'}}
<end_description>
| 835 | 0 | 2,960 | 835 |
69031679
|
from pyspark.sql import SparkSession
from pyspark.sql import Window
from pyspark.sql import functions as F
from pyspark.sql.functions import udf, pandas_udf
import pyspark.sql.types as T
import pandas as pd
import numpy as np
import seaborn as sns
import gc
import matplotlib.pyplot as plt
from datetime import datetime, timedelta
from scipy.interpolate import interp1d
def create_window(partitionby, orderby=None, rangebetween=None):
out = f"Window.partitionBy('{partitionby}')"
if orderby is not None:
out = out + f".orderBy('{orderby}')"
if rangebetween is not None:
out = out + f".rangeBetween({rangebetween[0]}, {rangebetween[1]})"
return eval(out)
def plot_frames(input, x="date", y="cums", rows=7, cols=20):
frames = input.sku.unique()
fig, axs = plt.subplots(rows, cols, figsize=(20, 10))
k = 0
for i in range(rows):
for j in range(cols):
try:
data = input[input.sku.isin([frames[k]])]
axs[i][j].plot(data["idx"], data["cums"])
axs[i][j].set_xticks([])
axs[i][j].set_yticks([])
axs[i][j].title.set_text(frames[k])
k += 1
except:
pass
def plot_frames_imputed(mydata, x="id", y="cumsum", rows=7, cols=20):
frames = mydata.sku.unique()
fig, axs = plt.subplots(rows, cols, figsize=(20, 10))
k = 0
for i in range(rows):
for j in range(cols):
try:
data = mydata[mydata.sku.isin([frames[k]])]
nullvals = data[y].isnull()
axs[i][j].scatter(data[x][nullvals], data[y][nullvals], c="r")
axs[i][j].scatter(data[x][~nullvals], data[y][~nullvals], c="b")
axs[i][j].set_xticks([])
axs[i][j].set_yticks([])
axs[i][j].title.set_text(frames[k])
k += 1
except:
pass
def plot_frames_train_val(
train,
val,
x="id",
y1="cumsum",
y2="cumsum",
ax1_lab="train",
ax2_lab="val",
rows=7,
cols=20,
):
frames = train.sku.unique()
fig, axs = plt.subplots(rows, cols, figsize=(20, 10))
k = 0
for i in range(rows):
for j in range(cols):
try:
train_data = train[train.sku.isin([frames[k]])]
val_data = val[val.sku.isin([frames[k]])]
axs[i][j].scatter(train_data[x], train_data[y1], c="r", label=ax1_lab)
axs[i][j].scatter(val_data[x], val_data[y2], c="b", label=ax2_lab)
axs[i][j].set_xticks([])
axs[i][j].set_yticks([])
axs[i][j].title.set_text(frames[k])
k += 1
except:
pass
# lines_labels = [ax.get_legend_handles_labels() for ax in fig.axes]
# lines, labels = [sum(lol, []) for lol in zip(*lines_labels)]
# fig.legend(lines, labels, loc='upper center')
@udf(T.IntegerType())
def count_zeros(x):
counter = 0
for i in x:
if i == 0.0:
counter += 1
else:
break
return counter
def write_data(data, name):
data.coalesce(1).write.format("parquet").mode("overwrite").save(name) @ udf(
T.BooleanType()
)
@udf(T.IntegerType())
def are_consecutive_dates(x):
x = sorted([datetime.strptime(i, "%Y-%m-%d") for i in x])
x
res = True
for idx in range(1, len(x)):
if (x[idx] - x[idx - 1]).days != 1:
res = False
break
return res
@udf(T.ArrayType(T.DoubleType()))
def interpolate(y):
y = np.array(y)
if not np.isnan(y).any():
return y.tolist()
y = y.astype("double")
x = np.array([i for i in range(len(y))])
x1 = x[~np.isnan(y)]
y1 = y[~np.isnan(y)]
if len(x1) == 0:
return [0 for i in range(len(y))]
try:
f = interp1d(x1, y1, fill_value="extrapolate")
y[np.isnan(y)] = f(x[np.isnan(y)])
except:
print("Problem fitting for a sku")
pass
return y.tolist()
def interpolate_all(x, var):
grouped_df = (
x.groupby("sku")
.agg(
F.sort_array(F.collect_list(F.struct("id", f"{var}"))).alias(
"collected_list"
)
)
.withColumn(f"{var}", F.col(f"collected_list.{var}"))
.withColumn("id", F.col("collected_list.id"))
.drop("collected_list")
)
grouped_df = grouped_df.withColumn(f"{var}_interpolated", interpolate(f"{var}"))
grouped_df = (
grouped_df.withColumn("tmp", F.arrays_zip("id", f"{var}_interpolated"))
.withColumn("tmp", F.explode("tmp"))
.select(
"sku",
F.col("tmp.id").alias("id"),
F.col(f"tmp.{var}_interpolated").alias(f"{var}_interpolated"),
)
)
return x.join(grouped_df, ["sku", "id"], how="left")
spark = (
SparkSession.builder.master("local[*]")
.config("spark.driver.memory", "15g")
.appName("meli-app")
.getOrCreate()
)
spark.conf.set("spark.sql.execution.arrow.pyspark.enabled", "true")
data_path = "../input/meli-data-challenge"
train = spark.read.parquet(f"{data_path}/train_data.parquet")
meta = spark.read.json(f"{data_path}/items_static_metadata_full.jl")
test = spark.read.csv(f"{data_path}/test_data.csv", header=True)
train0 = train # keep copy for testing
counts_total = train.groupBy("sku").count()
counts_total = (
counts_total.groupBy(F.col("count").alias("number_of_items"))
.count()
.sort(F.asc("number_of_items"))
.toPandas()
)
counts_total[["proportion"]] = 100 * counts_total["count"] / counts_total["count"].sum()
expr = counts_total[counts_total["count"] > 30].proportion.sum()
sns.scatterplot("number_of_items", "proportion", data=counts_total)
print(f"Counts > 30: {expr}")
# rows with leading 0s of minutes actives
tt = train.groupBy("sku").agg(F.collect_list("minutes_active").alias("vec"))
tt = tt.withColumn("to_remove", count_zeros(F.col("vec"))).drop("vec")
tt = tt.toPandas()
plt.hist(tt.to_remove.astype("float")[tt.to_remove > 0], bins=range(0, 60, 1))
plt.show()
# How many items have x ts values with minutes active > 0?
counts = (
train.filter(F.col("minutes_active") > 0)
.groupBy("sku")
.agg({"sku": "count"})
.withColumnRenamed("count(sku)", "counts")
)
counts = counts.groupBy("counts").count()
counts = counts.toPandas()
counts[["proportion"]] = 100 * counts.counts / counts.counts.sum()
counts = counts.sort_values("counts")
expr = counts[counts["counts"] > 30].proportion.sum()
sns.scatterplot("counts", "proportion", data=counts)
print(f"Counts > 30: {expr}")
# Check if dates are consecutive for each SKU
not_consecutives = train.groupBy("sku").agg(
are_consecutive_dates(F.collect_list(F.col("date"))).alias("consecutive")
)
not_consecutives.filter(F.col("consecutive") == False).show()
# check if ts start at different dates
mindate = train.groupBy("sku").agg(F.min("date"))
mindate.select("min(date)").distinct().show()
# check if ts start at different dates
maxdate = train.groupBy("sku").agg(F.max("date"))
maxdate.select("max(date)").distinct().show()
window = create_window("sku", "date")
train = train.withColumn("id", F.row_number().over(window))
train = train.withColumn(
"is_train", F.when(F.col("id") <= 30, F.lit(1)).otherwise(F.lit(0))
)
train = train.withColumn(
"sku_split",
F.concat(F.col("sku"), F.lit("_"), F.col("is_train").cast(T.StringType())),
).drop("id")
# Compute cumsum for items sold quantities
window2 = create_window("sku_split", "date")
windowval = create_window("sku_split", "date", [Window.unboundedPreceding, 0])
train = train.withColumn("id", F.row_number().over(window2))
train = train.withColumn("cumsum", F.sum("sold_quantity").over(windowval))
train = train.withColumn("cumtime", F.sum("minutes_active").over(windowval) / 60 / 24)
train = train.withColumn(
"selling_rate",
F.when(
F.col("minutes_active") > 0, F.col("sold_quantity") / F.col("minutes_active")
).otherwise(F.lit(0)),
)
train = train.withColumn(
"is_active", F.when(F.col("minutes_active") > 0, F.lit(1)).otherwise(F.lit(0))
)
# Add metadata
train = train.join(meta[["item_domain_id", "site_id", "sku"]], "sku")
train = train.withColumn(
"item_domain_id", F.regexp_replace(F.col("item_domain_id"), "^.*-", "")
)
gc.collect()
validation = train.filter(F.col("is_train") == 0)
train = train.filter(F.col("is_train") == 1)
# Note: sliding window can be tuned with hyperparameters
windowval = create_window("sku", "id", [-1, 1])
train = train.withColumn("rolling_cumsum", F.avg("cumsum").over(windowval))
skus = train.select("sku").distinct().rdd.flatMap(lambda x: x).collect()
skus = np.random.choice(skus, 200).tolist()
q0 = train.filter(train.sku.isin(skus)).toPandas()
q1 = validation.filter(train.sku.isin(skus)).toPandas()
q0["idx"] = q0.groupby("sku").cumcount()
q0["cums"] = q0.groupby("sku")["sold_quantity"].transform(pd.Series.cumsum)
q1["idx"] = q1.groupby("sku").cumcount()
q1["cums"] = q1.groupby("sku")["sold_quantity"].transform(pd.Series.cumsum)
plot_frames_train_val(q0, q1)
plot_frames_train_val(q0, q0[q0.minutes_active > 0])
plot_frames_train_val(q0, q1)
plot_frames_train_val(q0, q0[q0.minutes_active > 0])
plot_frames_train_val(q1, q1[q1.minutes_active > 0])
train.groupBy("listing_type").agg(
(F.count("listing_type")).alias("count"),
(100 * F.count("listing_type") / train.count()).alias("percentage"),
).show()
train.groupBy("shipping_logistic_type").agg(
(F.count("shipping_logistic_type")).alias("count"),
(100 * F.count("shipping_logistic_type") / train.count()).alias("percentage"),
).show()
train.groupBy("shipping_payment").agg(
(F.count("shipping_payment")).alias("count"),
(100 * F.count("shipping_payment") / train.count()).alias("percentage"),
).show()
listing_type = train.groupBy(["sku", "listing_type"]).count()
listing_type = listing_type.groupBy("sku").pivot("listing_type").sum("count").na.fill(0)
listing_type = listing_type.withColumn(
"classic_prop", F.col("classic") / (F.col("classic") + F.col("premium"))
)
listing_type = listing_type.withColumn(
"premium_prop", F.col("premium") / (F.col("classic") + F.col("premium"))
)
listing_type = listing_type.drop("classic", "premium")
listing_type = listing_type.na.fill(0)
shipping_payment = train.groupBy(["sku", "shipping_payment"]).count()
shipping_payment = (
shipping_payment.groupBy("sku").pivot("shipping_payment").sum("count").na.fill(0)
)
shipping_payment = shipping_payment.withColumn(
"paid_shipping_prop",
F.col("paid_shipping") / (F.col("paid_shipping") + F.col("free_shipping")),
)
shipping_payment = shipping_payment.withColumn(
"free_shipping_prop",
F.col("free_shipping") / (F.col("paid_shipping") + F.col("free_shipping")),
)
shipping_payment = shipping_payment.drop("paid_shipping", "free_shipping")
shipping_payment = shipping_payment.na.fill(0)
shipping_logistic_type = train.groupBy(["sku", "shipping_logistic_type"]).count()
shipping_logistic_type = (
shipping_logistic_type.groupBy("sku")
.pivot("shipping_logistic_type")
.sum("count")
.na.fill(0)
)
shipping_logistic_type = shipping_logistic_type.withColumn(
"drop_off_prop",
F.col("drop_off")
/ (F.col("drop_off") + F.col("cross_docking") + F.col("fulfillment")),
)
shipping_logistic_type = shipping_logistic_type.withColumn(
"cross_docking_prop",
F.col("cross_docking")
/ (F.col("drop_off") + F.col("cross_docking") + F.col("fulfillment")),
)
shipping_logistic_type = shipping_logistic_type.withColumn(
"fulfillment_prop",
F.col("fulfillment")
/ (F.col("drop_off") + F.col("cross_docking") + F.col("fulfillment")),
)
shipping_logistic_type = shipping_logistic_type.drop(
"drop_off", "cross_docking", "fulfillment"
)
shipping_logistic_type = shipping_logistic_type.na.fill(0)
minutes_active = (
train.groupBy("sku")
.avg("minutes_active")
.withColumnRenamed("avg(minutes_active)", "minutes_active_avg")
)
features = (
listing_type.join(shipping_payment, "sku")
.join(shipping_logistic_type, "sku")
.join(minutes_active, "sku")
)
# Generate time series for 30 days
id = spark.range(1, 31)
sku = train.select(F.col("sku")).distinct()
dates_sku = id.crossJoin(sku)
train = dates_sku.join(train, ["sku", "id"], how="left")
train = train.join(features, "sku", how="left")
validation = validation.join(features, "sku", how="left")
to_drop = ["listing_type", "shipping_payment", "shipping_logistic_type", "date"]
train = train.drop(*to_drop)
validation = validation.drop(*to_drop)
train = interpolate_all(train, "cumsum")
train = interpolate_all(train, "rolling_cumsum")
counts = train0.groupBy("sku").count()
q0 = counts.filter(F.col("count") < 25)
skus0 = q0.select("sku").distinct().rdd.flatMap(lambda x: x).collect()
temp = train.filter(train.sku.isin(skus0)).toPandas()
plot_frames_imputed(temp, "cumsum_interpolated")
plot_frames_imputed(temp, "rolling_cumsum_interpolated")
windowval = create_window("sku", "id", [Window.unboundedPreceding, 0])
train = train.withColumn(
"item_domain_id", F.last("item_domain_id", True).over(windowval)
) # fill with last non null value
train = train.withColumn(
"site_id", F.last("site_id", True).over(windowval)
) # fill with last non null value
train = train.withColumn(
"currency", F.last("currency", True).over(windowval)
) # fill with last non null value
train = train.withColumn(
"current_price", F.last("current_price", True).over(windowval)
) # fill with last non null value
train = train.withColumn(
"is_active", F.when(F.col("minutes_active") > 0, F.lit(1)).otherwise(F.lit(0))
)
train = train.na.fill(
{
"sold_quantity": -1,
"selling_rate": -1,
"minutes_active": -1,
"is_active": -1,
"current_price": -1,
"cumtime": -1,
"cumsum": -1,
"selling_rate": -1,
"cumsum_interpolated": -1,
"rolling_cumsum_interpolated": -1,
}
)
train.printSchema()
# Add metadata, items sold/domain id a
test = test.withColumn("monid", F.monotonically_increasing_id())
test = test.join(meta[["item_domain_id", "site_id", "sku"]], "sku", how="left")
test = test.withColumn(
"item_domain_id", F.regexp_replace(F.col("item_domain_id"), "^.*-", "")
)
test = test.join(features, "sku", how="left")
test = test.orderBy("monid")
write_data(train, "train.parquet")
write_data(validation, "validation.parquet")
write_data(test, "test.parquet")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/031/69031679.ipynb
| null | null |
[{"Id": 69031679, "ScriptId": 18813994, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1486200, "CreationDate": "07/26/2021 04:49:26", "VersionNumber": 62.0, "Title": "processing_meli_jul_24", "EvaluationDate": "07/26/2021", "IsChange": false, "TotalLines": 329.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 329.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
from pyspark.sql import SparkSession
from pyspark.sql import Window
from pyspark.sql import functions as F
from pyspark.sql.functions import udf, pandas_udf
import pyspark.sql.types as T
import pandas as pd
import numpy as np
import seaborn as sns
import gc
import matplotlib.pyplot as plt
from datetime import datetime, timedelta
from scipy.interpolate import interp1d
def create_window(partitionby, orderby=None, rangebetween=None):
out = f"Window.partitionBy('{partitionby}')"
if orderby is not None:
out = out + f".orderBy('{orderby}')"
if rangebetween is not None:
out = out + f".rangeBetween({rangebetween[0]}, {rangebetween[1]})"
return eval(out)
def plot_frames(input, x="date", y="cums", rows=7, cols=20):
frames = input.sku.unique()
fig, axs = plt.subplots(rows, cols, figsize=(20, 10))
k = 0
for i in range(rows):
for j in range(cols):
try:
data = input[input.sku.isin([frames[k]])]
axs[i][j].plot(data["idx"], data["cums"])
axs[i][j].set_xticks([])
axs[i][j].set_yticks([])
axs[i][j].title.set_text(frames[k])
k += 1
except:
pass
def plot_frames_imputed(mydata, x="id", y="cumsum", rows=7, cols=20):
frames = mydata.sku.unique()
fig, axs = plt.subplots(rows, cols, figsize=(20, 10))
k = 0
for i in range(rows):
for j in range(cols):
try:
data = mydata[mydata.sku.isin([frames[k]])]
nullvals = data[y].isnull()
axs[i][j].scatter(data[x][nullvals], data[y][nullvals], c="r")
axs[i][j].scatter(data[x][~nullvals], data[y][~nullvals], c="b")
axs[i][j].set_xticks([])
axs[i][j].set_yticks([])
axs[i][j].title.set_text(frames[k])
k += 1
except:
pass
def plot_frames_train_val(
train,
val,
x="id",
y1="cumsum",
y2="cumsum",
ax1_lab="train",
ax2_lab="val",
rows=7,
cols=20,
):
frames = train.sku.unique()
fig, axs = plt.subplots(rows, cols, figsize=(20, 10))
k = 0
for i in range(rows):
for j in range(cols):
try:
train_data = train[train.sku.isin([frames[k]])]
val_data = val[val.sku.isin([frames[k]])]
axs[i][j].scatter(train_data[x], train_data[y1], c="r", label=ax1_lab)
axs[i][j].scatter(val_data[x], val_data[y2], c="b", label=ax2_lab)
axs[i][j].set_xticks([])
axs[i][j].set_yticks([])
axs[i][j].title.set_text(frames[k])
k += 1
except:
pass
# lines_labels = [ax.get_legend_handles_labels() for ax in fig.axes]
# lines, labels = [sum(lol, []) for lol in zip(*lines_labels)]
# fig.legend(lines, labels, loc='upper center')
@udf(T.IntegerType())
def count_zeros(x):
counter = 0
for i in x:
if i == 0.0:
counter += 1
else:
break
return counter
def write_data(data, name):
data.coalesce(1).write.format("parquet").mode("overwrite").save(name) @ udf(
T.BooleanType()
)
@udf(T.IntegerType())
def are_consecutive_dates(x):
x = sorted([datetime.strptime(i, "%Y-%m-%d") for i in x])
x
res = True
for idx in range(1, len(x)):
if (x[idx] - x[idx - 1]).days != 1:
res = False
break
return res
@udf(T.ArrayType(T.DoubleType()))
def interpolate(y):
y = np.array(y)
if not np.isnan(y).any():
return y.tolist()
y = y.astype("double")
x = np.array([i for i in range(len(y))])
x1 = x[~np.isnan(y)]
y1 = y[~np.isnan(y)]
if len(x1) == 0:
return [0 for i in range(len(y))]
try:
f = interp1d(x1, y1, fill_value="extrapolate")
y[np.isnan(y)] = f(x[np.isnan(y)])
except:
print("Problem fitting for a sku")
pass
return y.tolist()
def interpolate_all(x, var):
grouped_df = (
x.groupby("sku")
.agg(
F.sort_array(F.collect_list(F.struct("id", f"{var}"))).alias(
"collected_list"
)
)
.withColumn(f"{var}", F.col(f"collected_list.{var}"))
.withColumn("id", F.col("collected_list.id"))
.drop("collected_list")
)
grouped_df = grouped_df.withColumn(f"{var}_interpolated", interpolate(f"{var}"))
grouped_df = (
grouped_df.withColumn("tmp", F.arrays_zip("id", f"{var}_interpolated"))
.withColumn("tmp", F.explode("tmp"))
.select(
"sku",
F.col("tmp.id").alias("id"),
F.col(f"tmp.{var}_interpolated").alias(f"{var}_interpolated"),
)
)
return x.join(grouped_df, ["sku", "id"], how="left")
spark = (
SparkSession.builder.master("local[*]")
.config("spark.driver.memory", "15g")
.appName("meli-app")
.getOrCreate()
)
spark.conf.set("spark.sql.execution.arrow.pyspark.enabled", "true")
data_path = "../input/meli-data-challenge"
train = spark.read.parquet(f"{data_path}/train_data.parquet")
meta = spark.read.json(f"{data_path}/items_static_metadata_full.jl")
test = spark.read.csv(f"{data_path}/test_data.csv", header=True)
train0 = train # keep copy for testing
counts_total = train.groupBy("sku").count()
counts_total = (
counts_total.groupBy(F.col("count").alias("number_of_items"))
.count()
.sort(F.asc("number_of_items"))
.toPandas()
)
counts_total[["proportion"]] = 100 * counts_total["count"] / counts_total["count"].sum()
expr = counts_total[counts_total["count"] > 30].proportion.sum()
sns.scatterplot("number_of_items", "proportion", data=counts_total)
print(f"Counts > 30: {expr}")
# rows with leading 0s of minutes actives
tt = train.groupBy("sku").agg(F.collect_list("minutes_active").alias("vec"))
tt = tt.withColumn("to_remove", count_zeros(F.col("vec"))).drop("vec")
tt = tt.toPandas()
plt.hist(tt.to_remove.astype("float")[tt.to_remove > 0], bins=range(0, 60, 1))
plt.show()
# How many items have x ts values with minutes active > 0?
counts = (
train.filter(F.col("minutes_active") > 0)
.groupBy("sku")
.agg({"sku": "count"})
.withColumnRenamed("count(sku)", "counts")
)
counts = counts.groupBy("counts").count()
counts = counts.toPandas()
counts[["proportion"]] = 100 * counts.counts / counts.counts.sum()
counts = counts.sort_values("counts")
expr = counts[counts["counts"] > 30].proportion.sum()
sns.scatterplot("counts", "proportion", data=counts)
print(f"Counts > 30: {expr}")
# Check if dates are consecutive for each SKU
not_consecutives = train.groupBy("sku").agg(
are_consecutive_dates(F.collect_list(F.col("date"))).alias("consecutive")
)
not_consecutives.filter(F.col("consecutive") == False).show()
# check if ts start at different dates
mindate = train.groupBy("sku").agg(F.min("date"))
mindate.select("min(date)").distinct().show()
# check if ts start at different dates
maxdate = train.groupBy("sku").agg(F.max("date"))
maxdate.select("max(date)").distinct().show()
window = create_window("sku", "date")
train = train.withColumn("id", F.row_number().over(window))
train = train.withColumn(
"is_train", F.when(F.col("id") <= 30, F.lit(1)).otherwise(F.lit(0))
)
train = train.withColumn(
"sku_split",
F.concat(F.col("sku"), F.lit("_"), F.col("is_train").cast(T.StringType())),
).drop("id")
# Compute cumsum for items sold quantities
window2 = create_window("sku_split", "date")
windowval = create_window("sku_split", "date", [Window.unboundedPreceding, 0])
train = train.withColumn("id", F.row_number().over(window2))
train = train.withColumn("cumsum", F.sum("sold_quantity").over(windowval))
train = train.withColumn("cumtime", F.sum("minutes_active").over(windowval) / 60 / 24)
train = train.withColumn(
"selling_rate",
F.when(
F.col("minutes_active") > 0, F.col("sold_quantity") / F.col("minutes_active")
).otherwise(F.lit(0)),
)
train = train.withColumn(
"is_active", F.when(F.col("minutes_active") > 0, F.lit(1)).otherwise(F.lit(0))
)
# Add metadata
train = train.join(meta[["item_domain_id", "site_id", "sku"]], "sku")
train = train.withColumn(
"item_domain_id", F.regexp_replace(F.col("item_domain_id"), "^.*-", "")
)
gc.collect()
validation = train.filter(F.col("is_train") == 0)
train = train.filter(F.col("is_train") == 1)
# Note: sliding window can be tuned with hyperparameters
windowval = create_window("sku", "id", [-1, 1])
train = train.withColumn("rolling_cumsum", F.avg("cumsum").over(windowval))
skus = train.select("sku").distinct().rdd.flatMap(lambda x: x).collect()
skus = np.random.choice(skus, 200).tolist()
q0 = train.filter(train.sku.isin(skus)).toPandas()
q1 = validation.filter(train.sku.isin(skus)).toPandas()
q0["idx"] = q0.groupby("sku").cumcount()
q0["cums"] = q0.groupby("sku")["sold_quantity"].transform(pd.Series.cumsum)
q1["idx"] = q1.groupby("sku").cumcount()
q1["cums"] = q1.groupby("sku")["sold_quantity"].transform(pd.Series.cumsum)
plot_frames_train_val(q0, q1)
plot_frames_train_val(q0, q0[q0.minutes_active > 0])
plot_frames_train_val(q0, q1)
plot_frames_train_val(q0, q0[q0.minutes_active > 0])
plot_frames_train_val(q1, q1[q1.minutes_active > 0])
train.groupBy("listing_type").agg(
(F.count("listing_type")).alias("count"),
(100 * F.count("listing_type") / train.count()).alias("percentage"),
).show()
train.groupBy("shipping_logistic_type").agg(
(F.count("shipping_logistic_type")).alias("count"),
(100 * F.count("shipping_logistic_type") / train.count()).alias("percentage"),
).show()
train.groupBy("shipping_payment").agg(
(F.count("shipping_payment")).alias("count"),
(100 * F.count("shipping_payment") / train.count()).alias("percentage"),
).show()
listing_type = train.groupBy(["sku", "listing_type"]).count()
listing_type = listing_type.groupBy("sku").pivot("listing_type").sum("count").na.fill(0)
listing_type = listing_type.withColumn(
"classic_prop", F.col("classic") / (F.col("classic") + F.col("premium"))
)
listing_type = listing_type.withColumn(
"premium_prop", F.col("premium") / (F.col("classic") + F.col("premium"))
)
listing_type = listing_type.drop("classic", "premium")
listing_type = listing_type.na.fill(0)
shipping_payment = train.groupBy(["sku", "shipping_payment"]).count()
shipping_payment = (
shipping_payment.groupBy("sku").pivot("shipping_payment").sum("count").na.fill(0)
)
shipping_payment = shipping_payment.withColumn(
"paid_shipping_prop",
F.col("paid_shipping") / (F.col("paid_shipping") + F.col("free_shipping")),
)
shipping_payment = shipping_payment.withColumn(
"free_shipping_prop",
F.col("free_shipping") / (F.col("paid_shipping") + F.col("free_shipping")),
)
shipping_payment = shipping_payment.drop("paid_shipping", "free_shipping")
shipping_payment = shipping_payment.na.fill(0)
shipping_logistic_type = train.groupBy(["sku", "shipping_logistic_type"]).count()
shipping_logistic_type = (
shipping_logistic_type.groupBy("sku")
.pivot("shipping_logistic_type")
.sum("count")
.na.fill(0)
)
shipping_logistic_type = shipping_logistic_type.withColumn(
"drop_off_prop",
F.col("drop_off")
/ (F.col("drop_off") + F.col("cross_docking") + F.col("fulfillment")),
)
shipping_logistic_type = shipping_logistic_type.withColumn(
"cross_docking_prop",
F.col("cross_docking")
/ (F.col("drop_off") + F.col("cross_docking") + F.col("fulfillment")),
)
shipping_logistic_type = shipping_logistic_type.withColumn(
"fulfillment_prop",
F.col("fulfillment")
/ (F.col("drop_off") + F.col("cross_docking") + F.col("fulfillment")),
)
shipping_logistic_type = shipping_logistic_type.drop(
"drop_off", "cross_docking", "fulfillment"
)
shipping_logistic_type = shipping_logistic_type.na.fill(0)
minutes_active = (
train.groupBy("sku")
.avg("minutes_active")
.withColumnRenamed("avg(minutes_active)", "minutes_active_avg")
)
features = (
listing_type.join(shipping_payment, "sku")
.join(shipping_logistic_type, "sku")
.join(minutes_active, "sku")
)
# Generate time series for 30 days
id = spark.range(1, 31)
sku = train.select(F.col("sku")).distinct()
dates_sku = id.crossJoin(sku)
train = dates_sku.join(train, ["sku", "id"], how="left")
train = train.join(features, "sku", how="left")
validation = validation.join(features, "sku", how="left")
to_drop = ["listing_type", "shipping_payment", "shipping_logistic_type", "date"]
train = train.drop(*to_drop)
validation = validation.drop(*to_drop)
train = interpolate_all(train, "cumsum")
train = interpolate_all(train, "rolling_cumsum")
counts = train0.groupBy("sku").count()
q0 = counts.filter(F.col("count") < 25)
skus0 = q0.select("sku").distinct().rdd.flatMap(lambda x: x).collect()
temp = train.filter(train.sku.isin(skus0)).toPandas()
plot_frames_imputed(temp, "cumsum_interpolated")
plot_frames_imputed(temp, "rolling_cumsum_interpolated")
windowval = create_window("sku", "id", [Window.unboundedPreceding, 0])
train = train.withColumn(
"item_domain_id", F.last("item_domain_id", True).over(windowval)
) # fill with last non null value
train = train.withColumn(
"site_id", F.last("site_id", True).over(windowval)
) # fill with last non null value
train = train.withColumn(
"currency", F.last("currency", True).over(windowval)
) # fill with last non null value
train = train.withColumn(
"current_price", F.last("current_price", True).over(windowval)
) # fill with last non null value
train = train.withColumn(
"is_active", F.when(F.col("minutes_active") > 0, F.lit(1)).otherwise(F.lit(0))
)
train = train.na.fill(
{
"sold_quantity": -1,
"selling_rate": -1,
"minutes_active": -1,
"is_active": -1,
"current_price": -1,
"cumtime": -1,
"cumsum": -1,
"selling_rate": -1,
"cumsum_interpolated": -1,
"rolling_cumsum_interpolated": -1,
}
)
train.printSchema()
# Add metadata, items sold/domain id a
test = test.withColumn("monid", F.monotonically_increasing_id())
test = test.join(meta[["item_domain_id", "site_id", "sku"]], "sku", how="left")
test = test.withColumn(
"item_domain_id", F.regexp_replace(F.col("item_domain_id"), "^.*-", "")
)
test = test.join(features, "sku", how="left")
test = test.orderBy("monid")
write_data(train, "train.parquet")
write_data(validation, "validation.parquet")
write_data(test, "test.parquet")
| false | 0 | 4,840 | 0 | 4,840 | 4,840 |
||
69031497
|
<jupyter_start><jupyter_text>Face Mask Detection ~12K Images Dataset
### Context
This dataset is used for Face Mask Detection Classification with images. The dataset consists of almost 12K images which are almost 328.92MB in size.
### Acknowledgments
All the images with the face mask (~6K) are scrapped from google search and all the images without the face mask are preprocessed from the CelebFace dataset created by Jessica Li (https://www.kaggle.com/jessicali9530). Thank you so much Jessica for providing a wonderful dataset to the community.
### Inspiration
The inspiration behind creating this dataset is to create an algorithm that can directly detect is a person is wearing a face mask or not. So I've scrapped the images from google as well as from the CelebFace dataset created by Jessica Li (https://www.kaggle.com/jessicali9530) to make this happen.
Kaggle dataset identifier: face-mask-12k-images-dataset
<jupyter_script>import pandas as pd # table operations
import numpy as np # linear algebra
# import seaborn as sns # visualizing
import os # getting path
from sklearn.utils import shuffle
import keras
from keras.layers import Conv2D, MaxPool2D, Flatten, Dense, Activation
from keras.models import Sequential
from tensorflow.keras.preprocessing.image import (
ImageDataGenerator,
load_img,
img_to_array,
)
import tensorflow as tf
testfile = "../input/face-mask-12k-images-dataset/Face Mask Dataset/Test"
trainfile = "../input/face-mask-12k-images-dataset/Face Mask Dataset/Train"
validationfile = "../input/face-mask-12k-images-dataset/Face Mask Dataset/Validation"
# **Reference:**
# https://blog.keras.io/building-powerful-image-classification-models-using-very-little-data.html
# https://keras.io/api/preprocessing/image/
# https://www.tensorflow.org/api_docs/python/tf/keras/preprocessing/image_dataset_from_directory
# https://www.tensorflow.org/tutorials/images/classification
# # Keras Data Generator, Argumentation
# Configure Image Data Augmentation in Keras. Takes the path to a directory & generates batches of augmented data.
from keras.preprocessing.image import ImageDataGenerator
batch_size = 16
# this is the augmentation configuration we will use for expanding the training dataset, it could improve the performance.
train_datagen = ImageDataGenerator(
rescale=1.0 / 255,
# shear_range=0.2,
# zoom_range=0.2,
horizontal_flip=True,
)
# this is the augmentation configuration we will use for testing:
# only rescaling to 0-1.
test_datagen = ImageDataGenerator(rescale=1.0 / 255)
# this is a generator that will read pictures found in
# subfolers of 'data/train', and indefinitely generate
# batches of augmented image data
train_generator = train_datagen.flow_from_directory(
trainfile, # this is the target directory
target_size=(128, 128), # all images will be resized to 128x128
batch_size=batch_size,
class_mode="binary",
shuffle=False,
) # since we use binary_crossentropy loss, we need binary labels
# this is a similar generator, for validation data
validation_generator = test_datagen.flow_from_directory(
validationfile,
target_size=(128, 128),
batch_size=batch_size,
class_mode="binary",
shuffle=False,
)
test_generator = test_datagen.flow_from_directory(
testfile,
target_size=(128, 128),
batch_size=batch_size,
class_mode="binary",
shuffle=False,
)
from keras.models import Sequential
from keras.layers import Conv2D, MaxPooling2D
from keras.layers import Activation, Dropout, Flatten, Dense
model = Sequential()
model.add(Conv2D(32, (3, 3), input_shape=(128, 128, 3)))
model.add(Activation("relu"))
model.add(MaxPool2D(pool_size=(4, 4)))
# To save train model time, decide to simplify the CNN model, so only use one ConV layer.
# model.add(Conv2D(400, (3, 3), input_shape =(32,32,3)))
# model.add(Activation('relu'))
# model.add(MaxPool2D(pool_size = (2, 2)))
# model.add(Conv2D(500, (3, 3), input_shape =(32,32,3)))
# model.add(Activation('relu'))
# model.add(MaxPool2D(pool_size = (2, 2)))
model.add(Dropout(0.3))
model.add(Flatten())
model.add(Dense(128, activation="relu"))
model.add(Dense(1, activation="sigmoid"))
# output = Dense(2,activation = 'softmax')
model.compile(loss="binary_crossentropy", optimizer="Adam", metrics=["accuracy"])
model.summary()
history = model.fit_generator(
train_generator,
steps_per_epoch=2000 // batch_size,
epochs=50,
validation_data=validation_generator,
validation_steps=800 // batch_size,
)
# model.save_weights('first_try.h5')
acc = history.history["accuracy"]
val_acc = history.history["val_accuracy"]
loss = history.history["loss"]
val_loss = history.history["val_loss"]
epochs = 50
epochs_range = range(epochs)
plt.figure(figsize=(8, 8))
plt.subplot(1, 2, 1)
plt.plot(epochs_range, acc, label="Training Accuracy")
plt.plot(epochs_range, val_acc, label="Validation Accuracy")
plt.legend(loc="lower right")
plt.title("Training and Validation Accuracy")
plt.subplot(1, 2, 2)
plt.plot(epochs_range, loss, label="Training Loss")
plt.plot(epochs_range, val_loss, label="Validation Loss")
plt.legend(loc="upper right")
plt.title("Training and Validation Loss")
plt.show()
test_step = test_generator.n // test_generator.batch_size
# for evaluate_generator, it does require test_step.
score = model.evaluate_generator(test_generator, steps=test_step)
print(score)
from sklearn.metrics import classification_report, confusion_matrix
# predY=model.predict(test_generator, workers=1,max_queue_size=1) #, max_q_size=1
# y_pred = np.argmax(predY,axis=1)
# cause we use sigmoid, we need teh threshopld.
y_pred = (model.predict(test_generator) > 0.5).astype("int32")
# y_label= [labels[k] for k in y_pred]
y_actual = test_generator.classes
# confusion matrix
cm = confusion_matrix(y_actual, y_pred)
print(cm)
# report
labels = ["withMask", "withoutMask"]
print(classification_report(y_actual, y_pred, target_names=labels))
# Visualize some prediction result.
from tensorflow import keras
img_path = (
"../input/face-mask-12k-images-dataset/Face Mask Dataset/Test/WithMask/1163.png"
)
class_names = ["With Mask", "Without Mask"]
# img_path = "../input/face-demo/face_PNG5645.png"
img = keras.preprocessing.image.load_img(img_path, target_size=(128, 128))
plt.imshow(img)
plt.show()
img_array = keras.preprocessing.image.img_to_array(img)
img_array = tf.expand_dims(img_array, 0)
# predictions = model.predict(img_array)
# print(predictions)
# (model.predict(test_generator) > 0.5).astype("int32")
predictions = (model.predict(img_array) > 0.5).astype("float64")
# y_label= [labels[k] for k in y_pred]
# y_actual = test_generator.classes
score = tf.nn.softmax(predictions[0])
# print(np.max(score))
# print(predictions)
print(
"This image most likely belongs to {} with a {:.2f} percent confidence.".format(
class_names[predictions[0][0].astype("int32")], 100 * np.max(score)
)
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/031/69031497.ipynb
|
face-mask-12k-images-dataset
|
ashishjangra27
|
[{"Id": 69031497, "ScriptId": 18562744, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 3803893, "CreationDate": "07/26/2021 04:46:04", "VersionNumber": 10.0, "Title": "Face Mask Detection CNN", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 197.0, "LinesInsertedFromPrevious": 28.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 169.0, "LinesInsertedFromFork": 168.0, "LinesDeletedFromFork": 235.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 29.0, "TotalVotes": 0}]
|
[{"Id": 91746177, "KernelVersionId": 69031497, "SourceDatasetVersionId": 1187790}]
|
[{"Id": 1187790, "DatasetId": 675484, "DatasourceVersionId": 1218923, "CreatorUserId": 5133282, "LicenseName": "CC0: Public Domain", "CreationDate": "05/26/2020 05:53:00", "VersionNumber": 1.0, "Title": "Face Mask Detection ~12K Images Dataset", "Slug": "face-mask-12k-images-dataset", "Subtitle": "12K Images divided in training testing and validation directories.", "Description": "### Context\n\nThis dataset is used for Face Mask Detection Classification with images. The dataset consists of almost 12K images which are almost 328.92MB in size.\n\n### Acknowledgments\nAll the images with the face mask (~6K) are scrapped from google search and all the images without the face mask are preprocessed from the CelebFace dataset created by Jessica Li (https://www.kaggle.com/jessicali9530). Thank you so much Jessica for providing a wonderful dataset to the community.\n\n### Inspiration\nThe inspiration behind creating this dataset is to create an algorithm that can directly detect is a person is wearing a face mask or not. So I've scrapped the images from google as well as from the CelebFace dataset created by Jessica Li (https://www.kaggle.com/jessicali9530) to make this happen.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 675484, "CreatorUserId": 5133282, "OwnerUserId": 5133282.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1187790.0, "CurrentDatasourceVersionId": 1218923.0, "ForumId": 689994, "Type": 2, "CreationDate": "05/26/2020 05:53:00", "LastActivityDate": "05/26/2020", "TotalViews": 107529, "TotalDownloads": 20529, "TotalVotes": 289, "TotalKernels": 227}]
|
[{"Id": 5133282, "UserName": "ashishjangra27", "DisplayName": "Ashish Jangra", "RegisterDate": "05/20/2020", "PerformanceTier": 2}]
|
import pandas as pd # table operations
import numpy as np # linear algebra
# import seaborn as sns # visualizing
import os # getting path
from sklearn.utils import shuffle
import keras
from keras.layers import Conv2D, MaxPool2D, Flatten, Dense, Activation
from keras.models import Sequential
from tensorflow.keras.preprocessing.image import (
ImageDataGenerator,
load_img,
img_to_array,
)
import tensorflow as tf
testfile = "../input/face-mask-12k-images-dataset/Face Mask Dataset/Test"
trainfile = "../input/face-mask-12k-images-dataset/Face Mask Dataset/Train"
validationfile = "../input/face-mask-12k-images-dataset/Face Mask Dataset/Validation"
# **Reference:**
# https://blog.keras.io/building-powerful-image-classification-models-using-very-little-data.html
# https://keras.io/api/preprocessing/image/
# https://www.tensorflow.org/api_docs/python/tf/keras/preprocessing/image_dataset_from_directory
# https://www.tensorflow.org/tutorials/images/classification
# # Keras Data Generator, Argumentation
# Configure Image Data Augmentation in Keras. Takes the path to a directory & generates batches of augmented data.
from keras.preprocessing.image import ImageDataGenerator
batch_size = 16
# this is the augmentation configuration we will use for expanding the training dataset, it could improve the performance.
train_datagen = ImageDataGenerator(
rescale=1.0 / 255,
# shear_range=0.2,
# zoom_range=0.2,
horizontal_flip=True,
)
# this is the augmentation configuration we will use for testing:
# only rescaling to 0-1.
test_datagen = ImageDataGenerator(rescale=1.0 / 255)
# this is a generator that will read pictures found in
# subfolers of 'data/train', and indefinitely generate
# batches of augmented image data
train_generator = train_datagen.flow_from_directory(
trainfile, # this is the target directory
target_size=(128, 128), # all images will be resized to 128x128
batch_size=batch_size,
class_mode="binary",
shuffle=False,
) # since we use binary_crossentropy loss, we need binary labels
# this is a similar generator, for validation data
validation_generator = test_datagen.flow_from_directory(
validationfile,
target_size=(128, 128),
batch_size=batch_size,
class_mode="binary",
shuffle=False,
)
test_generator = test_datagen.flow_from_directory(
testfile,
target_size=(128, 128),
batch_size=batch_size,
class_mode="binary",
shuffle=False,
)
from keras.models import Sequential
from keras.layers import Conv2D, MaxPooling2D
from keras.layers import Activation, Dropout, Flatten, Dense
model = Sequential()
model.add(Conv2D(32, (3, 3), input_shape=(128, 128, 3)))
model.add(Activation("relu"))
model.add(MaxPool2D(pool_size=(4, 4)))
# To save train model time, decide to simplify the CNN model, so only use one ConV layer.
# model.add(Conv2D(400, (3, 3), input_shape =(32,32,3)))
# model.add(Activation('relu'))
# model.add(MaxPool2D(pool_size = (2, 2)))
# model.add(Conv2D(500, (3, 3), input_shape =(32,32,3)))
# model.add(Activation('relu'))
# model.add(MaxPool2D(pool_size = (2, 2)))
model.add(Dropout(0.3))
model.add(Flatten())
model.add(Dense(128, activation="relu"))
model.add(Dense(1, activation="sigmoid"))
# output = Dense(2,activation = 'softmax')
model.compile(loss="binary_crossentropy", optimizer="Adam", metrics=["accuracy"])
model.summary()
history = model.fit_generator(
train_generator,
steps_per_epoch=2000 // batch_size,
epochs=50,
validation_data=validation_generator,
validation_steps=800 // batch_size,
)
# model.save_weights('first_try.h5')
acc = history.history["accuracy"]
val_acc = history.history["val_accuracy"]
loss = history.history["loss"]
val_loss = history.history["val_loss"]
epochs = 50
epochs_range = range(epochs)
plt.figure(figsize=(8, 8))
plt.subplot(1, 2, 1)
plt.plot(epochs_range, acc, label="Training Accuracy")
plt.plot(epochs_range, val_acc, label="Validation Accuracy")
plt.legend(loc="lower right")
plt.title("Training and Validation Accuracy")
plt.subplot(1, 2, 2)
plt.plot(epochs_range, loss, label="Training Loss")
plt.plot(epochs_range, val_loss, label="Validation Loss")
plt.legend(loc="upper right")
plt.title("Training and Validation Loss")
plt.show()
test_step = test_generator.n // test_generator.batch_size
# for evaluate_generator, it does require test_step.
score = model.evaluate_generator(test_generator, steps=test_step)
print(score)
from sklearn.metrics import classification_report, confusion_matrix
# predY=model.predict(test_generator, workers=1,max_queue_size=1) #, max_q_size=1
# y_pred = np.argmax(predY,axis=1)
# cause we use sigmoid, we need teh threshopld.
y_pred = (model.predict(test_generator) > 0.5).astype("int32")
# y_label= [labels[k] for k in y_pred]
y_actual = test_generator.classes
# confusion matrix
cm = confusion_matrix(y_actual, y_pred)
print(cm)
# report
labels = ["withMask", "withoutMask"]
print(classification_report(y_actual, y_pred, target_names=labels))
# Visualize some prediction result.
from tensorflow import keras
img_path = (
"../input/face-mask-12k-images-dataset/Face Mask Dataset/Test/WithMask/1163.png"
)
class_names = ["With Mask", "Without Mask"]
# img_path = "../input/face-demo/face_PNG5645.png"
img = keras.preprocessing.image.load_img(img_path, target_size=(128, 128))
plt.imshow(img)
plt.show()
img_array = keras.preprocessing.image.img_to_array(img)
img_array = tf.expand_dims(img_array, 0)
# predictions = model.predict(img_array)
# print(predictions)
# (model.predict(test_generator) > 0.5).astype("int32")
predictions = (model.predict(img_array) > 0.5).astype("float64")
# y_label= [labels[k] for k in y_pred]
# y_actual = test_generator.classes
score = tf.nn.softmax(predictions[0])
# print(np.max(score))
# print(predictions)
print(
"This image most likely belongs to {} with a {:.2f} percent confidence.".format(
class_names[predictions[0][0].astype("int32")], 100 * np.max(score)
)
)
| false | 0 | 1,935 | 0 | 2,178 | 1,935 |
||
69031774
|
<jupyter_start><jupyter_text>packages_for_creating_text_features
Kaggle dataset identifier: packages-for-creating-text-features
<jupyter_script># !python setup.py build > /dev/null
# !python setup.py install > /dev/null
import textstat
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import pickle
from shutil import copyfile
import os
from sklearn.metrics import mean_squared_error
from lightautoml.automl.presets.text_presets import TabularNLPAutoML
from lightautoml.tasks import Task
import re
from sklearn.feature_extraction.text import TfidfVectorizer
import nltk
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
from fastprogress.fastprogress import progress_bar
from sklearn.metrics import mean_squared_error
from lightautoml.automl.presets.text_presets import TabularNLPAutoML
from lightautoml.tasks import Task
model_dir_path = "./models"
saved_model_path = "../input/k/aleron751/k/aleron751/lama-bert-starter/models/"
if not os.path.exists(model_dir_path):
os.makedirs(model_dir_path)
for file in os.listdir(saved_model_path):
src = os.path.join(saved_model_path, file)
dst = os.path.join(model_dir_path, file)
copyfile(src, dst)
def rmse(x, y):
return np.sqrt(mean_squared_error(x, y))
with open(
"../input/k/aleron751/k/aleron751/lama-bert-starter/LAMA_model.pkl", "rb"
) as f:
automl = pickle.load(f)
test_df = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
train_df = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
# # Preprocess
def preprocess(data):
excerpt_processed = []
for e in progress_bar(data["excerpt"]):
# find alphabets
e = re.sub("[^a-zA-Z]", " ", e)
# convert to lower case
e = e.lower()
# tokenize words
e = nltk.word_tokenize(e)
# remove stopwords
e = [word for word in e if not word in set(stopwords.words("english"))]
# lemmatization
lemma = nltk.WordNetLemmatizer()
e = [lemma.lemmatize(word) for word in e]
e = " ".join(e)
excerpt_processed.append(e)
return excerpt_processed
test_df["excerpt_preprocessed"] = preprocess(test_df)
train_df["excerpt_preprocessed"] = preprocess(train_df)
# # Handcrafted features from Kaggle notebooks
from textblob.tokenizers import SentenceTokenizer, WordTokenizer
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import random
import os
import seaborn as sns
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import (
train_test_split,
StratifiedShuffleSplit,
StratifiedKFold,
)
# import textstat
plt.style.use("seaborn-talk")
from readcalc import readcalc
from sklearn.preprocessing import StandardScaler
import joblib
import spacy
sp = spacy.load("en_core_web_sm")
def pos_to_id(pos_name):
return sp.vocab[pos_name].orth
content_poss = ["ADJ", "NOUN", "VERB", "ADV"]
def count_poss(text, poss_names):
text = sp(text)
poss_ids = [pos_to_id(pos_name) for pos_name in poss_names]
pos_freq_dict = text.count_by(spacy.attrs.POS)
poss_sum = sum([pos_freq_dict.get(pos_id, 0) for pos_id in poss_ids])
return poss_sum
count_poss("my name is", ["PRON", "NOUN"])
# !pip download textstat ReadabilityCalculator
# !pip install *.whl
sent_tokenizer = SentenceTokenizer()
word_tokenizer = WordTokenizer()
# with open('../input/clrauxdata/dale-chall-3000-words.txt') as f:
# words = f.readlines()[0].split()
# common_words = dict(zip(words, [True] * len(words)))
# # df.sent_cnt.plot(kind='kde')
feats_to_drop = [
"sents_n",
"words_n",
"long_words_n",
#'difficult_words_n',
"content_words_n",
"prons_n",
"chars_n",
"syllables_n",
]
doc_feats = [
"chars_per_word",
"chars_per_sent",
"syllables_per_word",
"syllables_per_sent",
"words_per_sent",
"long_words_doc_ratio",
"difficult_words_doc_ratio",
"prons_doc_ratio",
"flesch_reading_ease",
"flesch_kincaid_grade",
"ari",
"cli",
"gunning_fog",
"lix",
"rix",
"smog",
"dcrs",
"lexical_diversity",
"content_diversity",
"lwf",
]
def create_handcrafted_features(df):
df["sents_n"] = df.excerpt.apply(textstat.sentence_count)
df["words_n"] = df.excerpt.apply(textstat.lexicon_count)
df["long_words_n"] = df.excerpt.apply(
lambda t: readcalc.ReadCalc(t).get_words_longer_than_X(6)
)
# df['difficult_words_n'] = df.excerpt.apply(lambda t: sum([bool(common_words.get(word)) for word in word_tokenizer.tokenize(t, include_punc=False)]))
df["content_words_n"] = df.excerpt.apply(lambda t: count_poss(t, content_poss))
df["prons_n"] = df.excerpt.apply(lambda t: count_poss(t, ["PRON"]))
df["chars_n"] = df.excerpt.str.len()
df["syllables_n"] = df.excerpt.apply(textstat.syllable_count)
print("\tstage 1 finished..")
df["chars_per_word_"] = df.chars_n / df.words_n
df["chars_per_sent_"] = df.chars_n / df.sents_n
df["syllables_per_word_"] = df.syllables_n / df.words_n
df["syllables_per_sent_"] = df.syllables_n / df.sents_n
df["words_per_sent_"] = df.words_n / df.sents_n
df["long_words_doc_ratio_"] = df.long_words_n / df.words_n
# df['difficult_words_doc_ratio'] = df.difficult_words_n / df.words_n
df["prons_doc_ratio"] = df.prons_n / df.words_n
print("\tstage 2 finished..")
df["flesch_reading_ease_"] = df.excerpt.apply(textstat.flesch_reading_ease)
df["flesch_kincaid_grade_"] = df.excerpt.apply(textstat.flesch_kincaid_grade)
df["ari_"] = df.excerpt.apply(textstat.automated_readability_index)
df["cli_"] = df.excerpt.apply(textstat.coleman_liau_index)
df["gunning_fog"] = df.excerpt.apply(textstat.gunning_fog)
df["lix_"] = df.excerpt.apply(lambda t: readcalc.ReadCalc(t).get_lix_index())
df["rix_"] = df.long_words_n / df.sents_n
df["smog_"] = df.excerpt.apply(lambda t: readcalc.ReadCalc(t).get_smog_index())
df["dcrs_"] = df.excerpt.apply(textstat.dale_chall_readability_score)
df["lexical_diversity_"] = len(set(df.words_n)) / df.words_n
df["content_diversity_"] = df.content_words_n / df.words_n
df["lwf_"] = df.excerpt.apply(textstat.linsear_write_formula)
print("\tstage 3 finished..")
return df
test_df = create_handcrafted_features(test_df)
test_df.drop(feats_to_drop, inplace=True, axis=1)
# # TextStat
def text_2_statistics(data):
flesch_reading_ease_list, smog_index_list = [], []
flesch_kincaid_grade_list, coleman_liau_index_list = [], []
automated_readability_index_list, dale_chall_readability_score_list = [], []
difficult_words_list, linsear_write_formula_list = [], []
gunning_fog_list, text_standard_list = [], []
fernandez_huerta_list, szigriszt_pazos_list = [], []
gutierrez_polini_list, crawford_list = [], []
for sentence in progress_bar(data["excerpt"]):
flesch_reading_ease_list.append(textstat.flesch_reading_ease(sentence))
smog_index_list.append(textstat.smog_index(sentence))
flesch_kincaid_grade_list.append(textstat.flesch_kincaid_grade(sentence))
coleman_liau_index_list.append(textstat.coleman_liau_index(sentence))
automated_readability_index_list.append(
textstat.automated_readability_index(sentence)
)
dale_chall_readability_score_list.append(
textstat.dale_chall_readability_score(sentence)
)
difficult_words_list.append(textstat.difficult_words(sentence))
linsear_write_formula_list.append(textstat.linsear_write_formula(sentence))
gunning_fog_list.append(textstat.gunning_fog(sentence))
text_standard_list.append(textstat.text_standard(sentence, float_output=True))
fernandez_huerta_list.append(textstat.fernandez_huerta(sentence))
szigriszt_pazos_list.append(textstat.szigriszt_pazos(sentence))
gutierrez_polini_list.append(textstat.gutierrez_polini(sentence))
crawford_list.append(textstat.crawford(sentence))
statistics_dict = {
"flesch_reading_ease": flesch_reading_ease_list,
"smog_index": smog_index_list,
"flesch_kincaid_grade": flesch_kincaid_grade_list,
"coleman_liau_index": coleman_liau_index_list,
"automated_readability_index": automated_readability_index_list,
"dale_chall_readability_score": dale_chall_readability_score_list,
"difficult_words": difficult_words_list,
"linsear_write_formula": linsear_write_formula_list,
"gunning_fog": gunning_fog_list,
"text_standard": text_standard_list,
"fernandez_huerta": fernandez_huerta_list,
"szigriszt_pazos": szigriszt_pazos_list,
"gutierrez_polini": gutierrez_polini_list,
"crawford": crawford_list,
}
return statistics_dict
statistics_dict = text_2_statistics(test_df)
for k, v in statistics_dict.items():
test_df[k] = v
vectorizer = TfidfVectorizer(max_features=1000)
train_bags = vectorizer.fit_transform(train_df["excerpt_preprocessed"].values).toarray()
train_bag_of_words_df = pd.DataFrame(train_bags)
train_bag_of_words_df.columns = vectorizer.get_feature_names()
test_bags = (
vectorizer.fit(train_df["excerpt_preprocessed"].values)
.transform(test_df["excerpt_preprocessed"].values)
.toarray()
)
test_bag_of_words_df = pd.DataFrame(test_bags)
test_bag_of_words_df.columns = vectorizer.get_feature_names()
for col in test_bag_of_words_df.columns:
test_df[col] = test_bag_of_words_df[col].values
del test_bag_of_words_df
# train_df.head()
# ----------------
def count_words_in_sentences(data):
counts = []
for sentence in progress_bar(data["excerpt_preprocessed"]):
words = sentence.split()
counts.append(len(words))
return counts
test_df["excerpt_word_counts_by_preprocessed"] = count_words_in_sentences(test_df)
# # NLTK features
from typing import List, Dict, Union
import nltk
import numpy as np
from nltk import ne_chunk, pos_tag, word_tokenize
from nltk.tree import Tree
def get_named_entities(text: str) -> List[str]:
continuous_chunk = []
current_chunk = []
for i in ne_chunk(pos_tag(word_tokenize(text))):
if isinstance(i, Tree):
current_chunk.append(" ".join(token for token, pos in i.leaves()))
elif current_chunk:
named_entity = " ".join(current_chunk)
continuous_chunk.append(named_entity)
current_chunk = []
named_entity = " ".join(current_chunk)
continuous_chunk.append(named_entity)
return continuous_chunk
_raw_tags = frozenset(
{
"LS",
"TO",
"VBN",
"''",
"WP",
"UH",
"VBG",
"JJ",
"VBZ",
"--",
"VBP",
"NN",
"DT",
"PRP",
":",
"WP$",
"NNPS",
"PRP$",
"WDT",
"(",
")",
".",
",",
"``",
"$",
"RB",
"RBR",
"RBS",
"VBD",
"IN",
"FW",
"RP",
"JJR",
"JJS",
"PDT",
"MD",
"VB",
"WRB",
"NNP",
"EX",
"NNS",
"SYM",
"CC",
"CD",
"POS",
}
)
_general_tags = frozenset({"gVB", "gNN", "gPR", "gWP", "gRB", "gJJ"})
_tagset = (*_raw_tags, *_general_tags)
def generate_text_features(text: str) -> Dict[str, Union[int, float]]:
total_count = dict.fromkeys(_tagset, 0)
tokenized_text = nltk.word_tokenize(text)
inv_text_len = 1 / len(tokenized_text)
for word, pos in nltk.pos_tag(tokenized_text):
total_count[pos] += inv_text_len
general_tag = f"g{pos[:2]}"
if general_tag in _general_tags:
total_count[general_tag] += inv_text_len
max_in_sent = dict.fromkeys(_tagset, 0)
min_in_sent = dict.fromkeys(_tagset, 0)
mean_in_sent = dict.fromkeys(_tagset, 0)
general_tags = set()
tags = set()
sentences = nltk.sent_tokenize(text)
num_sentences = len(sentences)
num_words = []
words_len = []
for sentence in map(nltk.word_tokenize, sentences):
cur_sentence_stat = dict.fromkeys(_tagset, 0)
num_words.append(len(sentence))
inv_sent_len = 1 / len(sentence)
for word, pos in nltk.pos_tag(sentence):
words_len.append(len(word))
cur_sentence_stat[pos] += inv_sent_len
tags.add(pos)
general_tag = f"g{pos[:2]}"
if general_tag in _general_tags:
general_tags.add(general_tag)
cur_sentence_stat[general_tag] += inv_sent_len
for tag in _tagset:
max_in_sent[tag] = max(max_in_sent[tag], cur_sentence_stat[tag])
min_in_sent[tag] = min(min_in_sent[tag], cur_sentence_stat[tag])
mean_in_sent[tag] += cur_sentence_stat[tag] / num_sentences
res = {}
for k, v in total_count.items():
res[f"TOTAL_{k}"] = v
for k, v in max_in_sent.items():
res[f"MAX_{k}"] = v
for k, v in min_in_sent.items():
res[f"MIN_{k}"] = v
for k, v in mean_in_sent.items():
res[f"MEAN_{k}"] = v
num_words = np.array(num_words)
words_len = np.array(words_len)
res["NUM_SENTENCES"] = len(num_words)
res["MEAN_NUM_WORDS"] = num_words.mean()
res["STD_NUM_WORDS"] = num_words.std()
res["NUM_WORDS"] = len(words_len)
res["MEAN_WORD_LEN"] = words_len.mean()
res["STD_WORD_LEN"] = words_len.std()
res["TAGS_UNIQUE"] = len(tags)
res["GENERAL_TAGS_UNIQUE"] = len(general_tags)
named_entities = get_named_entities(text)
res["NAMED_ENTITIES_PER_SENTENCE"] = len(named_entities) / num_sentences
res["UNIQUE_NAMED_ENTITIES_PER_SENTENCE"] = len(set(named_entities)) / num_sentences
return res
def max_word_lenght(sentence):
words = sentence.split()
average = max(len(word) for word in words)
return average
def get_all_nltk_feats(text):
res = generate_text_features(text)
res["number_get_named_entities"] = len(get_named_entities(text))
res["max_word_lenght"] = max_word_lenght(text)
new_res = {}
for k, v in res.items():
new_res[k] = [v]
return new_res
# txt = 'Say hello to my little friend, Bro! I love you, Sarra!'
# nltk_feats = get_all_nltk_feats(txt)
# nltk_feats
nltk_feats_df = pd.DataFrame()
for txt in progress_bar(test_df["excerpt"]):
nltk_feats_dict = get_all_nltk_feats(txt)
nltk_feats_df = nltk_feats_df.append(pd.DataFrame(nltk_feats_dict))
for col in nltk_feats_df.columns:
test_df[col] = nltk_feats_df[col].values
# del nltk_feats_df
test_df.head()
def preprocess_text(df):
df["len_tokens"] = df["excerpt"].str.strip().str.split(" ").apply(len)
df["len"] = df["excerpt"].str.strip().apply(len)
df["len_sent"] = df["excerpt"].str.strip().str.split(".").apply(len)
df["n_comm"] = df["excerpt"].str.strip().str.split(",").apply(len)
_t = df["excerpt"].str.strip().str.split(" ").values
df["d_mean"] = [np.sum([j.isdigit() for j in i]) for i in _t]
df["u_mean"] = [np.sum([j.isupper() for j in i]) for i in _t]
preprocess_text(test_df)
# Важно проверить число вот тут!
print(test_df.shape)
test_df.head()
preds_test = automl.predict(test_df).data[:, 0]
submission = pd.DataFrame({"id": test_df.id, "target": np.clip(preds_test, -2, 0.5)})
submission.to_csv("submission.csv", index=False)
submission.head()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/031/69031774.ipynb
|
packages-for-creating-text-features
|
chamecall
|
[{"Id": 69031774, "ScriptId": 18812966, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 3847072, "CreationDate": "07/26/2021 04:51:22", "VersionNumber": 9.0, "Title": "LAMA bert inference", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 477.0, "LinesInsertedFromPrevious": 7.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 470.0, "LinesInsertedFromFork": 440.0, "LinesDeletedFromFork": 9.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 37.0, "TotalVotes": 0}]
|
[{"Id": 91746757, "KernelVersionId": 69031774, "SourceDatasetVersionId": 2455138}, {"Id": 91746754, "KernelVersionId": 69031774, "SourceDatasetVersionId": 2225001}, {"Id": 91746753, "KernelVersionId": 69031774, "SourceDatasetVersionId": 1042664}, {"Id": 91746752, "KernelVersionId": 69031774, "SourceDatasetVersionId": 819665}]
|
[{"Id": 2455138, "DatasetId": 1486033, "DatasourceVersionId": 2497532, "CreatorUserId": 3458972, "LicenseName": "Unknown", "CreationDate": "07/23/2021 12:46:26", "VersionNumber": 2.0, "Title": "packages_for_creating_text_features", "Slug": "packages-for-creating-text-features", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Data Update 2021/07/23", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1486033, "CreatorUserId": 3458972, "OwnerUserId": 3458972.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2455138.0, "CurrentDatasourceVersionId": 2497532.0, "ForumId": 1505724, "Type": 2, "CreationDate": "07/23/2021 12:41:02", "LastActivityDate": "07/23/2021", "TotalViews": 616, "TotalDownloads": 3, "TotalVotes": 0, "TotalKernels": 3}]
|
[{"Id": 3458972, "UserName": "chamecall", "DisplayName": "algernone", "RegisterDate": "07/13/2019", "PerformanceTier": 2}]
|
# !python setup.py build > /dev/null
# !python setup.py install > /dev/null
import textstat
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import pickle
from shutil import copyfile
import os
from sklearn.metrics import mean_squared_error
from lightautoml.automl.presets.text_presets import TabularNLPAutoML
from lightautoml.tasks import Task
import re
from sklearn.feature_extraction.text import TfidfVectorizer
import nltk
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
from fastprogress.fastprogress import progress_bar
from sklearn.metrics import mean_squared_error
from lightautoml.automl.presets.text_presets import TabularNLPAutoML
from lightautoml.tasks import Task
model_dir_path = "./models"
saved_model_path = "../input/k/aleron751/k/aleron751/lama-bert-starter/models/"
if not os.path.exists(model_dir_path):
os.makedirs(model_dir_path)
for file in os.listdir(saved_model_path):
src = os.path.join(saved_model_path, file)
dst = os.path.join(model_dir_path, file)
copyfile(src, dst)
def rmse(x, y):
return np.sqrt(mean_squared_error(x, y))
with open(
"../input/k/aleron751/k/aleron751/lama-bert-starter/LAMA_model.pkl", "rb"
) as f:
automl = pickle.load(f)
test_df = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
train_df = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
# # Preprocess
def preprocess(data):
excerpt_processed = []
for e in progress_bar(data["excerpt"]):
# find alphabets
e = re.sub("[^a-zA-Z]", " ", e)
# convert to lower case
e = e.lower()
# tokenize words
e = nltk.word_tokenize(e)
# remove stopwords
e = [word for word in e if not word in set(stopwords.words("english"))]
# lemmatization
lemma = nltk.WordNetLemmatizer()
e = [lemma.lemmatize(word) for word in e]
e = " ".join(e)
excerpt_processed.append(e)
return excerpt_processed
test_df["excerpt_preprocessed"] = preprocess(test_df)
train_df["excerpt_preprocessed"] = preprocess(train_df)
# # Handcrafted features from Kaggle notebooks
from textblob.tokenizers import SentenceTokenizer, WordTokenizer
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import random
import os
import seaborn as sns
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import (
train_test_split,
StratifiedShuffleSplit,
StratifiedKFold,
)
# import textstat
plt.style.use("seaborn-talk")
from readcalc import readcalc
from sklearn.preprocessing import StandardScaler
import joblib
import spacy
sp = spacy.load("en_core_web_sm")
def pos_to_id(pos_name):
return sp.vocab[pos_name].orth
content_poss = ["ADJ", "NOUN", "VERB", "ADV"]
def count_poss(text, poss_names):
text = sp(text)
poss_ids = [pos_to_id(pos_name) for pos_name in poss_names]
pos_freq_dict = text.count_by(spacy.attrs.POS)
poss_sum = sum([pos_freq_dict.get(pos_id, 0) for pos_id in poss_ids])
return poss_sum
count_poss("my name is", ["PRON", "NOUN"])
# !pip download textstat ReadabilityCalculator
# !pip install *.whl
sent_tokenizer = SentenceTokenizer()
word_tokenizer = WordTokenizer()
# with open('../input/clrauxdata/dale-chall-3000-words.txt') as f:
# words = f.readlines()[0].split()
# common_words = dict(zip(words, [True] * len(words)))
# # df.sent_cnt.plot(kind='kde')
feats_to_drop = [
"sents_n",
"words_n",
"long_words_n",
#'difficult_words_n',
"content_words_n",
"prons_n",
"chars_n",
"syllables_n",
]
doc_feats = [
"chars_per_word",
"chars_per_sent",
"syllables_per_word",
"syllables_per_sent",
"words_per_sent",
"long_words_doc_ratio",
"difficult_words_doc_ratio",
"prons_doc_ratio",
"flesch_reading_ease",
"flesch_kincaid_grade",
"ari",
"cli",
"gunning_fog",
"lix",
"rix",
"smog",
"dcrs",
"lexical_diversity",
"content_diversity",
"lwf",
]
def create_handcrafted_features(df):
df["sents_n"] = df.excerpt.apply(textstat.sentence_count)
df["words_n"] = df.excerpt.apply(textstat.lexicon_count)
df["long_words_n"] = df.excerpt.apply(
lambda t: readcalc.ReadCalc(t).get_words_longer_than_X(6)
)
# df['difficult_words_n'] = df.excerpt.apply(lambda t: sum([bool(common_words.get(word)) for word in word_tokenizer.tokenize(t, include_punc=False)]))
df["content_words_n"] = df.excerpt.apply(lambda t: count_poss(t, content_poss))
df["prons_n"] = df.excerpt.apply(lambda t: count_poss(t, ["PRON"]))
df["chars_n"] = df.excerpt.str.len()
df["syllables_n"] = df.excerpt.apply(textstat.syllable_count)
print("\tstage 1 finished..")
df["chars_per_word_"] = df.chars_n / df.words_n
df["chars_per_sent_"] = df.chars_n / df.sents_n
df["syllables_per_word_"] = df.syllables_n / df.words_n
df["syllables_per_sent_"] = df.syllables_n / df.sents_n
df["words_per_sent_"] = df.words_n / df.sents_n
df["long_words_doc_ratio_"] = df.long_words_n / df.words_n
# df['difficult_words_doc_ratio'] = df.difficult_words_n / df.words_n
df["prons_doc_ratio"] = df.prons_n / df.words_n
print("\tstage 2 finished..")
df["flesch_reading_ease_"] = df.excerpt.apply(textstat.flesch_reading_ease)
df["flesch_kincaid_grade_"] = df.excerpt.apply(textstat.flesch_kincaid_grade)
df["ari_"] = df.excerpt.apply(textstat.automated_readability_index)
df["cli_"] = df.excerpt.apply(textstat.coleman_liau_index)
df["gunning_fog"] = df.excerpt.apply(textstat.gunning_fog)
df["lix_"] = df.excerpt.apply(lambda t: readcalc.ReadCalc(t).get_lix_index())
df["rix_"] = df.long_words_n / df.sents_n
df["smog_"] = df.excerpt.apply(lambda t: readcalc.ReadCalc(t).get_smog_index())
df["dcrs_"] = df.excerpt.apply(textstat.dale_chall_readability_score)
df["lexical_diversity_"] = len(set(df.words_n)) / df.words_n
df["content_diversity_"] = df.content_words_n / df.words_n
df["lwf_"] = df.excerpt.apply(textstat.linsear_write_formula)
print("\tstage 3 finished..")
return df
test_df = create_handcrafted_features(test_df)
test_df.drop(feats_to_drop, inplace=True, axis=1)
# # TextStat
def text_2_statistics(data):
flesch_reading_ease_list, smog_index_list = [], []
flesch_kincaid_grade_list, coleman_liau_index_list = [], []
automated_readability_index_list, dale_chall_readability_score_list = [], []
difficult_words_list, linsear_write_formula_list = [], []
gunning_fog_list, text_standard_list = [], []
fernandez_huerta_list, szigriszt_pazos_list = [], []
gutierrez_polini_list, crawford_list = [], []
for sentence in progress_bar(data["excerpt"]):
flesch_reading_ease_list.append(textstat.flesch_reading_ease(sentence))
smog_index_list.append(textstat.smog_index(sentence))
flesch_kincaid_grade_list.append(textstat.flesch_kincaid_grade(sentence))
coleman_liau_index_list.append(textstat.coleman_liau_index(sentence))
automated_readability_index_list.append(
textstat.automated_readability_index(sentence)
)
dale_chall_readability_score_list.append(
textstat.dale_chall_readability_score(sentence)
)
difficult_words_list.append(textstat.difficult_words(sentence))
linsear_write_formula_list.append(textstat.linsear_write_formula(sentence))
gunning_fog_list.append(textstat.gunning_fog(sentence))
text_standard_list.append(textstat.text_standard(sentence, float_output=True))
fernandez_huerta_list.append(textstat.fernandez_huerta(sentence))
szigriszt_pazos_list.append(textstat.szigriszt_pazos(sentence))
gutierrez_polini_list.append(textstat.gutierrez_polini(sentence))
crawford_list.append(textstat.crawford(sentence))
statistics_dict = {
"flesch_reading_ease": flesch_reading_ease_list,
"smog_index": smog_index_list,
"flesch_kincaid_grade": flesch_kincaid_grade_list,
"coleman_liau_index": coleman_liau_index_list,
"automated_readability_index": automated_readability_index_list,
"dale_chall_readability_score": dale_chall_readability_score_list,
"difficult_words": difficult_words_list,
"linsear_write_formula": linsear_write_formula_list,
"gunning_fog": gunning_fog_list,
"text_standard": text_standard_list,
"fernandez_huerta": fernandez_huerta_list,
"szigriszt_pazos": szigriszt_pazos_list,
"gutierrez_polini": gutierrez_polini_list,
"crawford": crawford_list,
}
return statistics_dict
statistics_dict = text_2_statistics(test_df)
for k, v in statistics_dict.items():
test_df[k] = v
vectorizer = TfidfVectorizer(max_features=1000)
train_bags = vectorizer.fit_transform(train_df["excerpt_preprocessed"].values).toarray()
train_bag_of_words_df = pd.DataFrame(train_bags)
train_bag_of_words_df.columns = vectorizer.get_feature_names()
test_bags = (
vectorizer.fit(train_df["excerpt_preprocessed"].values)
.transform(test_df["excerpt_preprocessed"].values)
.toarray()
)
test_bag_of_words_df = pd.DataFrame(test_bags)
test_bag_of_words_df.columns = vectorizer.get_feature_names()
for col in test_bag_of_words_df.columns:
test_df[col] = test_bag_of_words_df[col].values
del test_bag_of_words_df
# train_df.head()
# ----------------
def count_words_in_sentences(data):
counts = []
for sentence in progress_bar(data["excerpt_preprocessed"]):
words = sentence.split()
counts.append(len(words))
return counts
test_df["excerpt_word_counts_by_preprocessed"] = count_words_in_sentences(test_df)
# # NLTK features
from typing import List, Dict, Union
import nltk
import numpy as np
from nltk import ne_chunk, pos_tag, word_tokenize
from nltk.tree import Tree
def get_named_entities(text: str) -> List[str]:
continuous_chunk = []
current_chunk = []
for i in ne_chunk(pos_tag(word_tokenize(text))):
if isinstance(i, Tree):
current_chunk.append(" ".join(token for token, pos in i.leaves()))
elif current_chunk:
named_entity = " ".join(current_chunk)
continuous_chunk.append(named_entity)
current_chunk = []
named_entity = " ".join(current_chunk)
continuous_chunk.append(named_entity)
return continuous_chunk
_raw_tags = frozenset(
{
"LS",
"TO",
"VBN",
"''",
"WP",
"UH",
"VBG",
"JJ",
"VBZ",
"--",
"VBP",
"NN",
"DT",
"PRP",
":",
"WP$",
"NNPS",
"PRP$",
"WDT",
"(",
")",
".",
",",
"``",
"$",
"RB",
"RBR",
"RBS",
"VBD",
"IN",
"FW",
"RP",
"JJR",
"JJS",
"PDT",
"MD",
"VB",
"WRB",
"NNP",
"EX",
"NNS",
"SYM",
"CC",
"CD",
"POS",
}
)
_general_tags = frozenset({"gVB", "gNN", "gPR", "gWP", "gRB", "gJJ"})
_tagset = (*_raw_tags, *_general_tags)
def generate_text_features(text: str) -> Dict[str, Union[int, float]]:
total_count = dict.fromkeys(_tagset, 0)
tokenized_text = nltk.word_tokenize(text)
inv_text_len = 1 / len(tokenized_text)
for word, pos in nltk.pos_tag(tokenized_text):
total_count[pos] += inv_text_len
general_tag = f"g{pos[:2]}"
if general_tag in _general_tags:
total_count[general_tag] += inv_text_len
max_in_sent = dict.fromkeys(_tagset, 0)
min_in_sent = dict.fromkeys(_tagset, 0)
mean_in_sent = dict.fromkeys(_tagset, 0)
general_tags = set()
tags = set()
sentences = nltk.sent_tokenize(text)
num_sentences = len(sentences)
num_words = []
words_len = []
for sentence in map(nltk.word_tokenize, sentences):
cur_sentence_stat = dict.fromkeys(_tagset, 0)
num_words.append(len(sentence))
inv_sent_len = 1 / len(sentence)
for word, pos in nltk.pos_tag(sentence):
words_len.append(len(word))
cur_sentence_stat[pos] += inv_sent_len
tags.add(pos)
general_tag = f"g{pos[:2]}"
if general_tag in _general_tags:
general_tags.add(general_tag)
cur_sentence_stat[general_tag] += inv_sent_len
for tag in _tagset:
max_in_sent[tag] = max(max_in_sent[tag], cur_sentence_stat[tag])
min_in_sent[tag] = min(min_in_sent[tag], cur_sentence_stat[tag])
mean_in_sent[tag] += cur_sentence_stat[tag] / num_sentences
res = {}
for k, v in total_count.items():
res[f"TOTAL_{k}"] = v
for k, v in max_in_sent.items():
res[f"MAX_{k}"] = v
for k, v in min_in_sent.items():
res[f"MIN_{k}"] = v
for k, v in mean_in_sent.items():
res[f"MEAN_{k}"] = v
num_words = np.array(num_words)
words_len = np.array(words_len)
res["NUM_SENTENCES"] = len(num_words)
res["MEAN_NUM_WORDS"] = num_words.mean()
res["STD_NUM_WORDS"] = num_words.std()
res["NUM_WORDS"] = len(words_len)
res["MEAN_WORD_LEN"] = words_len.mean()
res["STD_WORD_LEN"] = words_len.std()
res["TAGS_UNIQUE"] = len(tags)
res["GENERAL_TAGS_UNIQUE"] = len(general_tags)
named_entities = get_named_entities(text)
res["NAMED_ENTITIES_PER_SENTENCE"] = len(named_entities) / num_sentences
res["UNIQUE_NAMED_ENTITIES_PER_SENTENCE"] = len(set(named_entities)) / num_sentences
return res
def max_word_lenght(sentence):
words = sentence.split()
average = max(len(word) for word in words)
return average
def get_all_nltk_feats(text):
res = generate_text_features(text)
res["number_get_named_entities"] = len(get_named_entities(text))
res["max_word_lenght"] = max_word_lenght(text)
new_res = {}
for k, v in res.items():
new_res[k] = [v]
return new_res
# txt = 'Say hello to my little friend, Bro! I love you, Sarra!'
# nltk_feats = get_all_nltk_feats(txt)
# nltk_feats
nltk_feats_df = pd.DataFrame()
for txt in progress_bar(test_df["excerpt"]):
nltk_feats_dict = get_all_nltk_feats(txt)
nltk_feats_df = nltk_feats_df.append(pd.DataFrame(nltk_feats_dict))
for col in nltk_feats_df.columns:
test_df[col] = nltk_feats_df[col].values
# del nltk_feats_df
test_df.head()
def preprocess_text(df):
df["len_tokens"] = df["excerpt"].str.strip().str.split(" ").apply(len)
df["len"] = df["excerpt"].str.strip().apply(len)
df["len_sent"] = df["excerpt"].str.strip().str.split(".").apply(len)
df["n_comm"] = df["excerpt"].str.strip().str.split(",").apply(len)
_t = df["excerpt"].str.strip().str.split(" ").values
df["d_mean"] = [np.sum([j.isdigit() for j in i]) for i in _t]
df["u_mean"] = [np.sum([j.isupper() for j in i]) for i in _t]
preprocess_text(test_df)
# Важно проверить число вот тут!
print(test_df.shape)
test_df.head()
preds_test = automl.predict(test_df).data[:, 0]
submission = pd.DataFrame({"id": test_df.id, "target": np.clip(preds_test, -2, 0.5)})
submission.to_csv("submission.csv", index=False)
submission.head()
| false | 2 | 5,142 | 0 | 5,173 | 5,142 |
||
69031306
|
<jupyter_start><jupyter_text>Anime Face Dataset
### Context
This dataset is often used for varying projects with anime faces. I will keep this dataset up-to-date and clean, along with including fun scripts for generating anime waifus!
### Content
This dataset has 63,632 "high-quality" anime faces... but let's be real... all anime girls are high-quality.

Kaggle dataset identifier: animefacedataset
<jupyter_script>import os
import random
import math
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
import cv2
dataroot = "/kaggle/input/"
batch_size = 64
image_size = 128
lr = 1e-5
dataset = tf.keras.preprocessing.image_dataset_from_directory(
"../input/animefacedataset",
label_mode=None,
image_size=(image_size, image_size),
batch_size=batch_size,
)
dataset = dataset.map(lambda x: (x / 127.5) - 1)
real_batch = iter(dataset).get_next()
plt.figure(figsize=(8, 8))
plt.axis("off")
plt.title("Training Images 1")
plt.imshow(real_batch[0])
def imshow16(batch):
row1 = tf.concat([batch[0], batch[1], batch[2], batch[3]], axis=0)
row2 = tf.concat([batch[4], batch[5], batch[6], batch[7]], axis=0)
row3 = tf.concat([batch[8], batch[9], batch[10], batch[11]], axis=0)
row4 = tf.concat([batch[12], batch[13], batch[14], batch[15]], axis=0)
return tf.concat([row1, row2, row3, row4], axis=1)
plt.figure(figsize=(8, 8))
plt.axis("off")
plt.title("imshow16")
plt.imshow(imshow16(real_batch))
def addnoise(batch):
# rate = tf.random.uniform([tf.shape(batch)[0]],minval=0, maxval=1, dtype=tf.dtypes.float32)**0.5
# rate = tf.reshape(rate,[tf.shape(batch)[0],1,1,1])
noise = tf.random.normal(
tf.shape(batch), mean=0.0, stddev=1, dtype=tf.dtypes.float32
)
return tf.clip_by_value(
batch + noise, clip_value_min=-1, clip_value_max=1, name=None
)
def addnoiseblock_python(batch):
b = tf.shape(batch)[0]
masks = np.ones(tf.shape(batch))
batch = batch.numpy()
for i in range(b):
mask = np.ones([image_size, image_size, 3])
for ii in range(5):
p = np.random.randint(low=0, high=image_size, size=[2])
r = np.random.randint(low=10, high=15, size=[1])
cv2.circle(mask, (p[0], p[1]), r[0], (0, 0, 0), -1)
batch[i] = batch[i] * mask
masks[i] = mask
return batch, masks
def addnoiseblock(batch):
return tf.py_function(
func=addnoiseblock_python, inp=[batch], Tout=[tf.float32, tf.float32]
)
plt.figure(figsize=(8, 8))
plt.axis("off")
plt.title("imshow16")
plt.imshow(imshow16(addnoiseblock(real_batch)[0]))
OUTPUT_CHANNELS = 3
def downsample(filters, size, apply_batchnorm=True):
initializer = tf.random_normal_initializer(0.0, 0.02)
result = tf.keras.Sequential()
result.add(
tf.keras.layers.Conv2D(
filters,
size,
strides=2,
padding="same",
kernel_initializer=initializer,
use_bias=False,
)
)
if apply_batchnorm:
result.add(tf.keras.layers.BatchNormalization())
result.add(tf.keras.layers.LeakyReLU())
return result
def upsample(filters, size, apply_dropout=False):
initializer = tf.random_normal_initializer(0.0, 0.02)
result = tf.keras.Sequential()
result.add(
tf.keras.layers.Conv2DTranspose(
filters,
size,
strides=2,
padding="same",
kernel_initializer=initializer,
use_bias=False,
)
)
result.add(tf.keras.layers.BatchNormalization())
if apply_dropout:
result.add(tf.keras.layers.Dropout(0.5))
result.add(tf.keras.layers.ReLU())
return result
def Repairer():
inputs = tf.keras.layers.Input(shape=[128, 128, 6])
down_stack = [
downsample(64, 4, apply_batchnorm=False), # (batch_size, 64, 64, 64)
downsample(256, 4, apply_batchnorm=False), # (batch_size, 32, 32, 256)
downsample(512, 4), # (batch_size, 16, 16, 512)
downsample(512, 4), # (batch_size, 8, 8, 512)
downsample(512, 4), # (batch_size, 4, 4, 512)
downsample(512, 4), # (batch_size, 2, 2, 512)
downsample(512, 4), # (batch_size, 1, 1, 512)
]
up_stack = [
upsample(512, 4, apply_dropout=True), # (batch_size, 2, 2, 1024)
upsample(512, 4, apply_dropout=True), # (batch_size, 4, 4, 1024)
upsample(512, 4, apply_dropout=True), # (batch_size, 8, 8, 1024)
upsample(512, 4), # (batch_size, 16, 16, 1024)
upsample(256, 4), # (batch_size, 32, 32, 512)
upsample(128, 4), # (batch_size, 64, 64, 256)
]
initializer = tf.random_normal_initializer(0.0, 0.02)
last = tf.keras.layers.Conv2DTranspose(
OUTPUT_CHANNELS,
4,
strides=2,
padding="same",
kernel_initializer=initializer,
activation="tanh",
) # (batch_size, 128, 128, 3)
x = inputs
# Downsampling through the model
skips = []
for down in down_stack:
x = down(x)
skips.append(x)
skips = reversed(skips[:-1])
# Upsampling and establishing the skip connections
for up, skip in zip(up_stack, skips):
x = up(x)
x = tf.keras.layers.Concatenate()([x, skip])
x = last(x)
return tf.keras.Model(inputs=inputs, outputs=x)
repairer = Repairer()
tf.keras.utils.plot_model(repairer, show_shapes=True, dpi=64)
def loss_function(inp, tar):
loss = tf.keras.metrics.mean_squared_error(tar, inp)
return tf.math.reduce_mean(loss)
class CustomSchedule(tf.keras.optimizers.schedules.LearningRateSchedule):
def __init__(self, learning_rate):
super(CustomSchedule, self).__init__()
self.rate = learning_rate
def __call__(self, step):
r1 = self.rate * (0.5 ** (step / 5000))
return r1
learning_rate = CustomSchedule(lr)
optimizer = tf.keras.optimizers.Adam(
learning_rate=learning_rate, beta_1=0.9, beta_2=0.99
)
train_loss = tf.keras.metrics.Mean(name="train_loss")
temp_learning_rate_schedule = CustomSchedule(lr)
tt = temp_learning_rate_schedule(tf.range(24000, dtype=tf.float32))
plt.plot(tt)
plt.ylabel("Learning Rate")
plt.xlabel("Train Step")
print(tt[5000])
print(tt[10000])
print(tt[19999])
@tf.function
def train_step(inp):
noise, masks = addnoiseblock(inp)
noise = tf.concat([noise, masks], axis=3)
with tf.GradientTape() as tape:
pre = repairer(noise, training=True)
loss = loss_function(pre, inp)
gradients = tape.gradient(loss, repairer.trainable_variables)
optimizer.apply_gradients(zip(gradients, repairer.trainable_variables))
train_loss(loss)
EPOCHS = 20
for epoch in range(EPOCHS):
train_loss.reset_states()
for batch, inp in enumerate(dataset):
train_step(inp)
if batch % 150 == 0:
print(
"Epoch {} Batch {} Loss {:.4f} ".format(
epoch + 1, batch, train_loss.result()
)
)
print(
"Epoch {} Batch {} Loss {:.4f} ".format(epoch + 1, batch, train_loss.result())
)
real_batch = iter(dataset).get_next()
plt.figure(figsize=(8, 8))
plt.axis("off")
plt.title("origin")
plt.imshow(imshow16(real_batch))
noise_batch, masks = addnoiseblock(real_batch[0:16])
plt.figure(figsize=(8, 8))
plt.axis("off")
plt.title("add_noise")
plt.imshow(imshow16(noise_batch))
noise_batch = tf.concat([noise_batch, masks], axis=3)
repair_batch = repairer(noise_batch, training=False)
plt.figure(figsize=(8, 8))
plt.axis("off")
plt.title("repair" + str(i + 1))
plt.imshow(imshow16(repair_batch))
noise_batch = addnoise(tf.zeros(tf.shape(real_batch[0:16])))
plt.figure(figsize=(8, 8))
plt.axis("off")
plt.title("add_noise")
plt.imshow(imshow16(noise_batch))
@tf.function
def test_step(inp):
noise, masks = addnoiseblock(inp)
noise = tf.concat([noise, masks], axis=3)
pre = repairer(noise, training=False)
return pre
for i in range(100):
repair_batch = test_step(noise_batch)
if i % 10 == 0:
plt.figure(figsize=(8, 8))
plt.axis("off")
plt.title("repair" + str(i + 1))
plt.imshow(imshow16(repair_batch))
noise_batch = repair_batch
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/031/69031306.ipynb
|
animefacedataset
|
splcher
|
[{"Id": 69031306, "ScriptId": 18766168, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2785248, "CreationDate": "07/26/2021 04:42:38", "VersionNumber": 11.0, "Title": "repair_base_generative_model_test_animefaced", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 248.0, "LinesInsertedFromPrevious": 27.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 221.0, "LinesInsertedFromFork": 231.0, "LinesDeletedFromFork": 343.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 17.0, "TotalVotes": 0}]
|
[{"Id": 91745758, "KernelVersionId": 69031306, "SourceDatasetVersionId": 737475}]
|
[{"Id": 737475, "DatasetId": 379764, "DatasourceVersionId": 758029, "CreatorUserId": 3234333, "LicenseName": "Database: Open Database, Contents: Database Contents", "CreationDate": "10/13/2019 16:33:20", "VersionNumber": 3.0, "Title": "Anime Face Dataset", "Slug": "animefacedataset", "Subtitle": "Mckinsey666 dataset scraped from www.getchu.com", "Description": "### Context\n\nThis dataset is often used for varying projects with anime faces. I will keep this dataset up-to-date and clean, along with including fun scripts for generating anime waifus!\n\n### Content\n\nThis dataset has 63,632 \"high-quality\" anime faces... but let's be real... all anime girls are high-quality. \n\n\n### Acknowledgements\n\nBased on:\n[Mckinsey666](https://github.com/Mckinsey666/Anime-Face-Dataset)'s dataset\n\n### Inspiration\n\nI just want to generate perfect waifus. It's a simple dream. I will expand and alter this dataset to move towards a pure dataset of cute female anime faces.\n\n### Example Output\n\nIn the [Starter: Anime Face Dataset](https://www.kaggle.com/splcher/starter-anime-face-dataset) kernel, you will find the code to create this:\n", "VersionNotes": "removed old dataset", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 379764, "CreatorUserId": 3234333, "OwnerUserId": 3234333.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 737475.0, "CurrentDatasourceVersionId": 758029.0, "ForumId": 391686, "Type": 2, "CreationDate": "10/13/2019 06:31:00", "LastActivityDate": "10/13/2019", "TotalViews": 99381, "TotalDownloads": 21314, "TotalVotes": 315, "TotalKernels": 97}]
|
[{"Id": 3234333, "UserName": "splcher", "DisplayName": "Spencer Churchill", "RegisterDate": "05/16/2019", "PerformanceTier": 1}]
|
import os
import random
import math
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
import cv2
dataroot = "/kaggle/input/"
batch_size = 64
image_size = 128
lr = 1e-5
dataset = tf.keras.preprocessing.image_dataset_from_directory(
"../input/animefacedataset",
label_mode=None,
image_size=(image_size, image_size),
batch_size=batch_size,
)
dataset = dataset.map(lambda x: (x / 127.5) - 1)
real_batch = iter(dataset).get_next()
plt.figure(figsize=(8, 8))
plt.axis("off")
plt.title("Training Images 1")
plt.imshow(real_batch[0])
def imshow16(batch):
row1 = tf.concat([batch[0], batch[1], batch[2], batch[3]], axis=0)
row2 = tf.concat([batch[4], batch[5], batch[6], batch[7]], axis=0)
row3 = tf.concat([batch[8], batch[9], batch[10], batch[11]], axis=0)
row4 = tf.concat([batch[12], batch[13], batch[14], batch[15]], axis=0)
return tf.concat([row1, row2, row3, row4], axis=1)
plt.figure(figsize=(8, 8))
plt.axis("off")
plt.title("imshow16")
plt.imshow(imshow16(real_batch))
def addnoise(batch):
# rate = tf.random.uniform([tf.shape(batch)[0]],minval=0, maxval=1, dtype=tf.dtypes.float32)**0.5
# rate = tf.reshape(rate,[tf.shape(batch)[0],1,1,1])
noise = tf.random.normal(
tf.shape(batch), mean=0.0, stddev=1, dtype=tf.dtypes.float32
)
return tf.clip_by_value(
batch + noise, clip_value_min=-1, clip_value_max=1, name=None
)
def addnoiseblock_python(batch):
b = tf.shape(batch)[0]
masks = np.ones(tf.shape(batch))
batch = batch.numpy()
for i in range(b):
mask = np.ones([image_size, image_size, 3])
for ii in range(5):
p = np.random.randint(low=0, high=image_size, size=[2])
r = np.random.randint(low=10, high=15, size=[1])
cv2.circle(mask, (p[0], p[1]), r[0], (0, 0, 0), -1)
batch[i] = batch[i] * mask
masks[i] = mask
return batch, masks
def addnoiseblock(batch):
return tf.py_function(
func=addnoiseblock_python, inp=[batch], Tout=[tf.float32, tf.float32]
)
plt.figure(figsize=(8, 8))
plt.axis("off")
plt.title("imshow16")
plt.imshow(imshow16(addnoiseblock(real_batch)[0]))
OUTPUT_CHANNELS = 3
def downsample(filters, size, apply_batchnorm=True):
initializer = tf.random_normal_initializer(0.0, 0.02)
result = tf.keras.Sequential()
result.add(
tf.keras.layers.Conv2D(
filters,
size,
strides=2,
padding="same",
kernel_initializer=initializer,
use_bias=False,
)
)
if apply_batchnorm:
result.add(tf.keras.layers.BatchNormalization())
result.add(tf.keras.layers.LeakyReLU())
return result
def upsample(filters, size, apply_dropout=False):
initializer = tf.random_normal_initializer(0.0, 0.02)
result = tf.keras.Sequential()
result.add(
tf.keras.layers.Conv2DTranspose(
filters,
size,
strides=2,
padding="same",
kernel_initializer=initializer,
use_bias=False,
)
)
result.add(tf.keras.layers.BatchNormalization())
if apply_dropout:
result.add(tf.keras.layers.Dropout(0.5))
result.add(tf.keras.layers.ReLU())
return result
def Repairer():
inputs = tf.keras.layers.Input(shape=[128, 128, 6])
down_stack = [
downsample(64, 4, apply_batchnorm=False), # (batch_size, 64, 64, 64)
downsample(256, 4, apply_batchnorm=False), # (batch_size, 32, 32, 256)
downsample(512, 4), # (batch_size, 16, 16, 512)
downsample(512, 4), # (batch_size, 8, 8, 512)
downsample(512, 4), # (batch_size, 4, 4, 512)
downsample(512, 4), # (batch_size, 2, 2, 512)
downsample(512, 4), # (batch_size, 1, 1, 512)
]
up_stack = [
upsample(512, 4, apply_dropout=True), # (batch_size, 2, 2, 1024)
upsample(512, 4, apply_dropout=True), # (batch_size, 4, 4, 1024)
upsample(512, 4, apply_dropout=True), # (batch_size, 8, 8, 1024)
upsample(512, 4), # (batch_size, 16, 16, 1024)
upsample(256, 4), # (batch_size, 32, 32, 512)
upsample(128, 4), # (batch_size, 64, 64, 256)
]
initializer = tf.random_normal_initializer(0.0, 0.02)
last = tf.keras.layers.Conv2DTranspose(
OUTPUT_CHANNELS,
4,
strides=2,
padding="same",
kernel_initializer=initializer,
activation="tanh",
) # (batch_size, 128, 128, 3)
x = inputs
# Downsampling through the model
skips = []
for down in down_stack:
x = down(x)
skips.append(x)
skips = reversed(skips[:-1])
# Upsampling and establishing the skip connections
for up, skip in zip(up_stack, skips):
x = up(x)
x = tf.keras.layers.Concatenate()([x, skip])
x = last(x)
return tf.keras.Model(inputs=inputs, outputs=x)
repairer = Repairer()
tf.keras.utils.plot_model(repairer, show_shapes=True, dpi=64)
def loss_function(inp, tar):
loss = tf.keras.metrics.mean_squared_error(tar, inp)
return tf.math.reduce_mean(loss)
class CustomSchedule(tf.keras.optimizers.schedules.LearningRateSchedule):
def __init__(self, learning_rate):
super(CustomSchedule, self).__init__()
self.rate = learning_rate
def __call__(self, step):
r1 = self.rate * (0.5 ** (step / 5000))
return r1
learning_rate = CustomSchedule(lr)
optimizer = tf.keras.optimizers.Adam(
learning_rate=learning_rate, beta_1=0.9, beta_2=0.99
)
train_loss = tf.keras.metrics.Mean(name="train_loss")
temp_learning_rate_schedule = CustomSchedule(lr)
tt = temp_learning_rate_schedule(tf.range(24000, dtype=tf.float32))
plt.plot(tt)
plt.ylabel("Learning Rate")
plt.xlabel("Train Step")
print(tt[5000])
print(tt[10000])
print(tt[19999])
@tf.function
def train_step(inp):
noise, masks = addnoiseblock(inp)
noise = tf.concat([noise, masks], axis=3)
with tf.GradientTape() as tape:
pre = repairer(noise, training=True)
loss = loss_function(pre, inp)
gradients = tape.gradient(loss, repairer.trainable_variables)
optimizer.apply_gradients(zip(gradients, repairer.trainable_variables))
train_loss(loss)
EPOCHS = 20
for epoch in range(EPOCHS):
train_loss.reset_states()
for batch, inp in enumerate(dataset):
train_step(inp)
if batch % 150 == 0:
print(
"Epoch {} Batch {} Loss {:.4f} ".format(
epoch + 1, batch, train_loss.result()
)
)
print(
"Epoch {} Batch {} Loss {:.4f} ".format(epoch + 1, batch, train_loss.result())
)
real_batch = iter(dataset).get_next()
plt.figure(figsize=(8, 8))
plt.axis("off")
plt.title("origin")
plt.imshow(imshow16(real_batch))
noise_batch, masks = addnoiseblock(real_batch[0:16])
plt.figure(figsize=(8, 8))
plt.axis("off")
plt.title("add_noise")
plt.imshow(imshow16(noise_batch))
noise_batch = tf.concat([noise_batch, masks], axis=3)
repair_batch = repairer(noise_batch, training=False)
plt.figure(figsize=(8, 8))
plt.axis("off")
plt.title("repair" + str(i + 1))
plt.imshow(imshow16(repair_batch))
noise_batch = addnoise(tf.zeros(tf.shape(real_batch[0:16])))
plt.figure(figsize=(8, 8))
plt.axis("off")
plt.title("add_noise")
plt.imshow(imshow16(noise_batch))
@tf.function
def test_step(inp):
noise, masks = addnoiseblock(inp)
noise = tf.concat([noise, masks], axis=3)
pre = repairer(noise, training=False)
return pre
for i in range(100):
repair_batch = test_step(noise_batch)
if i % 10 == 0:
plt.figure(figsize=(8, 8))
plt.axis("off")
plt.title("repair" + str(i + 1))
plt.imshow(imshow16(repair_batch))
noise_batch = repair_batch
| false | 0 | 2,763 | 0 | 2,908 | 2,763 |
||
69031624
|
# @title <- Press "Play" to prepare the magic
# Just patch to generate.py
with open("generate.py", "r") as file:
filedata = file.read()
filedata = filedata.replace(
"# Reduce the default image size if low VRAM",
"import math\nfrom IPython import display\nloss_idx = []\navg = []\nfrom typing import TYPE_CHECKING, Any, Callable, Optional\n\nimport torch\nimport torch.optim\n\n\nif TYPE_CHECKING:\n from torch.optim.optimizer import _params_t\nelse:\n _params_t = Any\n\n\nfrom madgrad import MADGRAD",
)
filedata = filedata.replace(
", choices=['Adam','AdamW','Adagrad','Adamax','DiffGrad','AdamP','RAdam'], default='Adam', dest='optimiser'",
", choices=['Adam','AdamW','Adagrad','Adamax','DiffGrad','AdamP','RAdam', 'MADGRAD'], default='MADGRAD', dest='optimiser'",
)
filedata = filedata.replace(
'elif args.optimiser == "RAdam":\n opt = RAdam([z], lr=args.step_size) # LR=2+?\nelse:\n print("Unknown optimiser. Are choices broken?")',
'elif args.optimiser == "RAdam":\n opt = RAdam([z], lr=args.step_size) # LR=2+?\nelif args.optimiser == "MADGRAD":\n opt = MADGRAD([z], lr=args.step_size) # LR=6+?\nelse:\n print("Unknown optimiser. Are choices broken?")',
)
filedata = filedata.replace(
" with torch.no_grad():\n z.copy_(z.maximum(z_min).minimum(z_max))",
" with torch.no_grad():\n z.copy_(z.maximum(z_min).minimum(z_max))\n return loss",
)
filedata = filedata.replace(
"except KeyboardInterrupt:\n pass\n",
'except KeyboardInterrupt:\n pass\nimport matplotlib.pyplot as plt\nplt.figure(dpi=100)\nplt.plot(avg)\nplt.savefig("plot.png", dpi=100)\n',
)
filedata = filedata.replace(
" if i % args.display_freq == 0:\n checkin(i, lossAll)\n \n loss = sum(lossAll)",
" global loss_idx\n loss = sum(lossAll)\n loss_idx.append(loss.item())\n checkin(i, lossAll)\n",
)
with open("generate.py", "w") as file:
file.write(filedata)
file.close()
from IPython import display
import random
from tqdm.notebook import tqdm
SIZE_X = 600
SIZE_Y = 400
TEXT = "Driving into cyberpunk city;Neon Style; Hight Quality; Cyberpunk; Ray Tracing; Rendered by Unreal Engine"
FILENAME = "image.png"
MAX_EPOCHS = 220
LR = 9
OPTIMISER = "MADGRAD"
MAX_ITERATIONS = 100
SEED = random.randint(1, 999999999)
FILENAME_NO_EXT = FILENAME.split(".")[0]
FILE_EXTENSION = FILENAME.split(".")[1]
ZOOM = 1.04
print("Generating first image...")
print("First image done!")
with tqdm(total=MAX_EPOCHS) as pbar:
for i in range(1, MAX_EPOCHS + 1):
# print('#####################################\n', i, '\n ##################################')
#!convert {"'" + FILENAME + "'"} -distort SRT 1 -gravity center {"'" + FILENAME + "'"} ##uncomment to rotation
# display.display(display.Image(FILENAME_NO_EXT + str(i) + "." + FILE_EXTENSION)) fix are inneed
pbar.update()
# EDJGNFSEJG
# Add MADGRAD in opt choice
with open("generate.py", "r") as file:
filedata = file.read()
filedata = filedata.replace(
"# Reduce the default image size if low VRAM",
"import math\nfrom IPython import display\nloss_idx = []\navg = []\nfrom typing import TYPE_CHECKING, Any, Callable, Optional\n\nimport torch\nimport torch.optim\n\n\nif TYPE_CHECKING:\n from torch.optim.optimizer import _params_t\nelse:\n _params_t = Any\n\n\nfrom madgrad import MADGRAD",
)
filedata = filedata.replace(
", choices=['Adam','AdamW','Adagrad','Adamax','DiffGrad','AdamP','RAdam'], default='Adam', dest='optimiser'",
", choices=['Adam','AdamW','Adagrad','Adamax','DiffGrad','AdamP','RAdam', 'MADGRAD'], default='MADGRAD', dest='optimiser'",
)
filedata = filedata.replace(
'elif args.optimiser == "RAdam":\n opt = RAdam([z], lr=args.step_size) # LR=2+?\nelse:\n print("Unknown optimiser. Are choices broken?")',
'elif args.optimiser == "RAdam":\n opt = RAdam([z], lr=args.step_size) # LR=2+?\nelif args.optimiser == "MADGRAD":\n opt = MADGRAD([z], lr=args.step_size) # LR=6+?\nelse:\n print("Unknown optimiser. Are choices broken?")',
)
filedata = filedata.replace(
" with torch.no_grad():\n z.copy_(z.maximum(z_min).minimum(z_max))",
" with torch.no_grad():\n z.copy_(z.maximum(z_min).minimum(z_max))\n return loss",
)
filedata = filedata.replace(
"except KeyboardInterrupt:\n pass\n",
'except KeyboardInterrupt:\n pass\nimport matplotlib.pyplot as plt\nplt.figure(dpi=100)\nplt.plot(avg)\nplt.savefig("plot.png", dpi=100)\n',
)
filedata = filedata.replace(
" if i % args.display_freq == 0:\n checkin(i, lossAll)\n \n loss = sum(lossAll)",
" global loss_idx\n loss = sum(lossAll)\n loss_idx.append(loss.item())\n checkin(i, lossAll)\n",
)
with open("generate.py", "w") as file:
file.write(filedata)
file.close()
import base64
import io
import re
import imageio
from IPython.display import HTML
import glob
import numpy as np
fppps = 10
frames = []
tempf = []
filenames = glob.glob("./*.png")
for i in filenames:
num = str("")
for word in i:
if word.isdigit():
num = num + str(word)
if i != "image.png" and len(num) >= 1:
tempf.append(str(num) + ".png")
img = sorted(tempf, key=lambda x: float(re.findall("(\d+)", x)[0]))
print(len(img))
for b in img:
frames.append(imageio.imread(b))
frames = np.array(frames)
imageio.mimsave("video.mp4", frames, fps=fppps)
print("Done")
video = io.open("video.mp4", "r+b").read()
encoded = base64.b64encode(video)
play_html = ""
play_html = play_html + (
'<video alt="test" controls><source src="data:video/mp4;base64,{0}" type="video/mp4" /> </video>'.format(
encoded.decode("ascii")
)
)
HTML(data=play_html)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/031/69031624.ipynb
| null | null |
[{"Id": 69031624, "ScriptId": 18778031, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7894866, "CreationDate": "07/26/2021 04:48:24", "VersionNumber": 13.0, "Title": "notebook0b10242013", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 137.0, "LinesInsertedFromPrevious": 2.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 135.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# @title <- Press "Play" to prepare the magic
# Just patch to generate.py
with open("generate.py", "r") as file:
filedata = file.read()
filedata = filedata.replace(
"# Reduce the default image size if low VRAM",
"import math\nfrom IPython import display\nloss_idx = []\navg = []\nfrom typing import TYPE_CHECKING, Any, Callable, Optional\n\nimport torch\nimport torch.optim\n\n\nif TYPE_CHECKING:\n from torch.optim.optimizer import _params_t\nelse:\n _params_t = Any\n\n\nfrom madgrad import MADGRAD",
)
filedata = filedata.replace(
", choices=['Adam','AdamW','Adagrad','Adamax','DiffGrad','AdamP','RAdam'], default='Adam', dest='optimiser'",
", choices=['Adam','AdamW','Adagrad','Adamax','DiffGrad','AdamP','RAdam', 'MADGRAD'], default='MADGRAD', dest='optimiser'",
)
filedata = filedata.replace(
'elif args.optimiser == "RAdam":\n opt = RAdam([z], lr=args.step_size) # LR=2+?\nelse:\n print("Unknown optimiser. Are choices broken?")',
'elif args.optimiser == "RAdam":\n opt = RAdam([z], lr=args.step_size) # LR=2+?\nelif args.optimiser == "MADGRAD":\n opt = MADGRAD([z], lr=args.step_size) # LR=6+?\nelse:\n print("Unknown optimiser. Are choices broken?")',
)
filedata = filedata.replace(
" with torch.no_grad():\n z.copy_(z.maximum(z_min).minimum(z_max))",
" with torch.no_grad():\n z.copy_(z.maximum(z_min).minimum(z_max))\n return loss",
)
filedata = filedata.replace(
"except KeyboardInterrupt:\n pass\n",
'except KeyboardInterrupt:\n pass\nimport matplotlib.pyplot as plt\nplt.figure(dpi=100)\nplt.plot(avg)\nplt.savefig("plot.png", dpi=100)\n',
)
filedata = filedata.replace(
" if i % args.display_freq == 0:\n checkin(i, lossAll)\n \n loss = sum(lossAll)",
" global loss_idx\n loss = sum(lossAll)\n loss_idx.append(loss.item())\n checkin(i, lossAll)\n",
)
with open("generate.py", "w") as file:
file.write(filedata)
file.close()
from IPython import display
import random
from tqdm.notebook import tqdm
SIZE_X = 600
SIZE_Y = 400
TEXT = "Driving into cyberpunk city;Neon Style; Hight Quality; Cyberpunk; Ray Tracing; Rendered by Unreal Engine"
FILENAME = "image.png"
MAX_EPOCHS = 220
LR = 9
OPTIMISER = "MADGRAD"
MAX_ITERATIONS = 100
SEED = random.randint(1, 999999999)
FILENAME_NO_EXT = FILENAME.split(".")[0]
FILE_EXTENSION = FILENAME.split(".")[1]
ZOOM = 1.04
print("Generating first image...")
print("First image done!")
with tqdm(total=MAX_EPOCHS) as pbar:
for i in range(1, MAX_EPOCHS + 1):
# print('#####################################\n', i, '\n ##################################')
#!convert {"'" + FILENAME + "'"} -distort SRT 1 -gravity center {"'" + FILENAME + "'"} ##uncomment to rotation
# display.display(display.Image(FILENAME_NO_EXT + str(i) + "." + FILE_EXTENSION)) fix are inneed
pbar.update()
# EDJGNFSEJG
# Add MADGRAD in opt choice
with open("generate.py", "r") as file:
filedata = file.read()
filedata = filedata.replace(
"# Reduce the default image size if low VRAM",
"import math\nfrom IPython import display\nloss_idx = []\navg = []\nfrom typing import TYPE_CHECKING, Any, Callable, Optional\n\nimport torch\nimport torch.optim\n\n\nif TYPE_CHECKING:\n from torch.optim.optimizer import _params_t\nelse:\n _params_t = Any\n\n\nfrom madgrad import MADGRAD",
)
filedata = filedata.replace(
", choices=['Adam','AdamW','Adagrad','Adamax','DiffGrad','AdamP','RAdam'], default='Adam', dest='optimiser'",
", choices=['Adam','AdamW','Adagrad','Adamax','DiffGrad','AdamP','RAdam', 'MADGRAD'], default='MADGRAD', dest='optimiser'",
)
filedata = filedata.replace(
'elif args.optimiser == "RAdam":\n opt = RAdam([z], lr=args.step_size) # LR=2+?\nelse:\n print("Unknown optimiser. Are choices broken?")',
'elif args.optimiser == "RAdam":\n opt = RAdam([z], lr=args.step_size) # LR=2+?\nelif args.optimiser == "MADGRAD":\n opt = MADGRAD([z], lr=args.step_size) # LR=6+?\nelse:\n print("Unknown optimiser. Are choices broken?")',
)
filedata = filedata.replace(
" with torch.no_grad():\n z.copy_(z.maximum(z_min).minimum(z_max))",
" with torch.no_grad():\n z.copy_(z.maximum(z_min).minimum(z_max))\n return loss",
)
filedata = filedata.replace(
"except KeyboardInterrupt:\n pass\n",
'except KeyboardInterrupt:\n pass\nimport matplotlib.pyplot as plt\nplt.figure(dpi=100)\nplt.plot(avg)\nplt.savefig("plot.png", dpi=100)\n',
)
filedata = filedata.replace(
" if i % args.display_freq == 0:\n checkin(i, lossAll)\n \n loss = sum(lossAll)",
" global loss_idx\n loss = sum(lossAll)\n loss_idx.append(loss.item())\n checkin(i, lossAll)\n",
)
with open("generate.py", "w") as file:
file.write(filedata)
file.close()
import base64
import io
import re
import imageio
from IPython.display import HTML
import glob
import numpy as np
fppps = 10
frames = []
tempf = []
filenames = glob.glob("./*.png")
for i in filenames:
num = str("")
for word in i:
if word.isdigit():
num = num + str(word)
if i != "image.png" and len(num) >= 1:
tempf.append(str(num) + ".png")
img = sorted(tempf, key=lambda x: float(re.findall("(\d+)", x)[0]))
print(len(img))
for b in img:
frames.append(imageio.imread(b))
frames = np.array(frames)
imageio.mimsave("video.mp4", frames, fps=fppps)
print("Done")
video = io.open("video.mp4", "r+b").read()
encoded = base64.b64encode(video)
play_html = ""
play_html = play_html + (
'<video alt="test" controls><source src="data:video/mp4;base64,{0}" type="video/mp4" /> </video>'.format(
encoded.decode("ascii")
)
)
HTML(data=play_html)
| false | 0 | 1,983 | 0 | 1,983 | 1,983 |
||
69031646
|
# ## Handling the Test Data set
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
test_df = pd.read_csv("../input/house-prices-advanced-regression-techniques/test.csv")
test_df.shape
test_df.head()
# ## Checking null values
test_df.isnull().sum()
# ## Filling missing values
test_df["LotFrontage"] = test_df["LotFrontage"].fillna(test_df["LotFrontage"].mean())
test_df["MSZoning"] = test_df["MSZoning"].fillna(test_df["MSZoning"].mode()[0])
test_df.shape
test_df.drop(["Alley"], axis=1, inplace=True)
test_df.shape
test_df["BsmtCond"] = test_df["BsmtCond"].fillna(test_df["BsmtCond"].mode()[0])
test_df["BsmtQual"] = test_df["BsmtQual"].fillna(test_df["BsmtQual"].mode()[0])
test_df["FireplaceQu"] = test_df["FireplaceQu"].fillna(test_df["FireplaceQu"].mode()[0])
test_df["GarageType"] = test_df["GarageType"].fillna(test_df["GarageType"].mode()[0])
test_df.drop(["GarageYrBlt"], axis=1, inplace=True)
test_df.shape
test_df["GarageFinish"] = test_df["GarageFinish"].fillna(
test_df["GarageFinish"].mode()[0]
)
test_df["GarageQual"] = test_df["GarageQual"].fillna(test_df["GarageQual"].mode()[0])
test_df["GarageCond"] = test_df["GarageCond"].fillna(test_df["GarageCond"].mode()[0])
test_df.drop(["PoolQC", "Fence", "MiscFeature"], axis=1, inplace=True)
test_df.shape
test_df.drop(["Id"], axis=1, inplace=True)
test_df["MasVnrType"] = test_df["MasVnrType"].fillna(test_df["MasVnrType"].mode()[0])
test_df["MasVnrArea"] = test_df["MasVnrArea"].fillna(test_df["MasVnrArea"].mode()[0])
sns.heatmap(test_df.isnull(), yticklabels=False, cbar=False, cmap="viridis")
test_df["BsmtExposure"] = test_df["BsmtExposure"].fillna(
test_df["BsmtExposure"].mode()[0]
)
sns.heatmap(test_df.isnull(), yticklabels=False, cbar=False, cmap="viridis")
test_df["BsmtFinType2"] = test_df["BsmtFinType2"].fillna(
test_df["BsmtFinType2"].mode()[0]
)
test_df.loc[:, test_df.isnull().any()].head()
test_df["Utilities"] = test_df["Utilities"].fillna(test_df["Utilities"].mode()[0])
test_df["Exterior1st"] = test_df["Exterior1st"].fillna(test_df["Exterior1st"].mode()[0])
test_df["Exterior2nd"] = test_df["Exterior2nd"].fillna(test_df["Exterior2nd"].mode()[0])
test_df["BsmtFinType1"] = test_df["BsmtFinType1"].fillna(
test_df["BsmtFinType1"].mode()[0]
)
test_df["BsmtFinSF1"] = test_df["BsmtFinSF1"].fillna(test_df["BsmtFinSF1"].mean())
test_df["BsmtFinSF2"] = test_df["BsmtFinSF2"].fillna(test_df["BsmtFinSF2"].mean())
test_df["BsmtUnfSF"] = test_df["BsmtUnfSF"].fillna(test_df["BsmtUnfSF"].mean())
test_df["TotalBsmtSF"] = test_df["TotalBsmtSF"].fillna(test_df["TotalBsmtSF"].mean())
test_df["BsmtFullBath"] = test_df["BsmtFullBath"].fillna(
test_df["BsmtFullBath"].mode()[0]
)
test_df["BsmtHalfBath"] = test_df["BsmtHalfBath"].fillna(
test_df["BsmtHalfBath"].mode()[0]
)
test_df["KitchenQual"] = test_df["KitchenQual"].fillna(test_df["KitchenQual"].mode()[0])
test_df["Functional"] = test_df["Functional"].fillna(test_df["Functional"].mode()[0])
test_df["GarageCars"] = test_df["GarageCars"].fillna(test_df["GarageCars"].mean())
test_df["GarageArea"] = test_df["GarageArea"].fillna(test_df["GarageArea"].mean())
test_df["SaleType"] = test_df["SaleType"].fillna(test_df["SaleType"].mode()[0])
test_df.shape
test_df.to_csv("new_test.csv", index=False)
# So now we have created a proper test data by filling the missing values and droping unnessesary columns from the test data for better predictions.
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/031/69031646.ipynb
| null | null |
[{"Id": 69031646, "ScriptId": 18839181, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7854500, "CreationDate": "07/26/2021 04:48:46", "VersionNumber": 2.0, "Title": "new_test_data", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 84.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 83.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# ## Handling the Test Data set
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
test_df = pd.read_csv("../input/house-prices-advanced-regression-techniques/test.csv")
test_df.shape
test_df.head()
# ## Checking null values
test_df.isnull().sum()
# ## Filling missing values
test_df["LotFrontage"] = test_df["LotFrontage"].fillna(test_df["LotFrontage"].mean())
test_df["MSZoning"] = test_df["MSZoning"].fillna(test_df["MSZoning"].mode()[0])
test_df.shape
test_df.drop(["Alley"], axis=1, inplace=True)
test_df.shape
test_df["BsmtCond"] = test_df["BsmtCond"].fillna(test_df["BsmtCond"].mode()[0])
test_df["BsmtQual"] = test_df["BsmtQual"].fillna(test_df["BsmtQual"].mode()[0])
test_df["FireplaceQu"] = test_df["FireplaceQu"].fillna(test_df["FireplaceQu"].mode()[0])
test_df["GarageType"] = test_df["GarageType"].fillna(test_df["GarageType"].mode()[0])
test_df.drop(["GarageYrBlt"], axis=1, inplace=True)
test_df.shape
test_df["GarageFinish"] = test_df["GarageFinish"].fillna(
test_df["GarageFinish"].mode()[0]
)
test_df["GarageQual"] = test_df["GarageQual"].fillna(test_df["GarageQual"].mode()[0])
test_df["GarageCond"] = test_df["GarageCond"].fillna(test_df["GarageCond"].mode()[0])
test_df.drop(["PoolQC", "Fence", "MiscFeature"], axis=1, inplace=True)
test_df.shape
test_df.drop(["Id"], axis=1, inplace=True)
test_df["MasVnrType"] = test_df["MasVnrType"].fillna(test_df["MasVnrType"].mode()[0])
test_df["MasVnrArea"] = test_df["MasVnrArea"].fillna(test_df["MasVnrArea"].mode()[0])
sns.heatmap(test_df.isnull(), yticklabels=False, cbar=False, cmap="viridis")
test_df["BsmtExposure"] = test_df["BsmtExposure"].fillna(
test_df["BsmtExposure"].mode()[0]
)
sns.heatmap(test_df.isnull(), yticklabels=False, cbar=False, cmap="viridis")
test_df["BsmtFinType2"] = test_df["BsmtFinType2"].fillna(
test_df["BsmtFinType2"].mode()[0]
)
test_df.loc[:, test_df.isnull().any()].head()
test_df["Utilities"] = test_df["Utilities"].fillna(test_df["Utilities"].mode()[0])
test_df["Exterior1st"] = test_df["Exterior1st"].fillna(test_df["Exterior1st"].mode()[0])
test_df["Exterior2nd"] = test_df["Exterior2nd"].fillna(test_df["Exterior2nd"].mode()[0])
test_df["BsmtFinType1"] = test_df["BsmtFinType1"].fillna(
test_df["BsmtFinType1"].mode()[0]
)
test_df["BsmtFinSF1"] = test_df["BsmtFinSF1"].fillna(test_df["BsmtFinSF1"].mean())
test_df["BsmtFinSF2"] = test_df["BsmtFinSF2"].fillna(test_df["BsmtFinSF2"].mean())
test_df["BsmtUnfSF"] = test_df["BsmtUnfSF"].fillna(test_df["BsmtUnfSF"].mean())
test_df["TotalBsmtSF"] = test_df["TotalBsmtSF"].fillna(test_df["TotalBsmtSF"].mean())
test_df["BsmtFullBath"] = test_df["BsmtFullBath"].fillna(
test_df["BsmtFullBath"].mode()[0]
)
test_df["BsmtHalfBath"] = test_df["BsmtHalfBath"].fillna(
test_df["BsmtHalfBath"].mode()[0]
)
test_df["KitchenQual"] = test_df["KitchenQual"].fillna(test_df["KitchenQual"].mode()[0])
test_df["Functional"] = test_df["Functional"].fillna(test_df["Functional"].mode()[0])
test_df["GarageCars"] = test_df["GarageCars"].fillna(test_df["GarageCars"].mean())
test_df["GarageArea"] = test_df["GarageArea"].fillna(test_df["GarageArea"].mean())
test_df["SaleType"] = test_df["SaleType"].fillna(test_df["SaleType"].mode()[0])
test_df.shape
test_df.to_csv("new_test.csv", index=False)
# So now we have created a proper test data by filling the missing values and droping unnessesary columns from the test data for better predictions.
| false | 0 | 1,275 | 0 | 1,275 | 1,275 |
||
69031556
|
import pandas as pd
import glob
import pyarrow as pa
import numpy as np
import matplotlib.pyplot as plt
train_data = pd.read_csv("../input/optiver-realized-volatility-prediction/train.csv")
test_data = pd.read_csv("../input/optiver-realized-volatility-prediction/test.csv")
print(train_data.shape)
print(test_data.shape)
train_data = train_data[train_data.stock_id == 0]
display(train_data)
display(test_data)
list = glob.glob(
"../input/optiver-realized-volatility-prediction/trade_train.parquet/stock_id=0/*.parquet"
)
list
book = pa.parquet.read_table(list[0])
book = book.to_pandas()
book.head(305)
list = glob.glob(
"../input/optiver-realized-volatility-prediction/book_train.parquet/stock_id=0/*.parquet"
)
list
train = pa.parquet.read_table(list[0])
train = train.to_pandas()
train.head(305)
train_data = train_data.drop(["stock_id"], axis=1)
train_data
train_input = pd.merge(train_data, train, on=["time_id"])
train_input
list = glob.glob(
"../input/optiver-realized-volatility-prediction/book_test.parquet/stock_id=0/*.parquet"
)
list
test = pa.parquet.read_table(list[0])
test_input = test.to_pandas()
print(test_input.shape)
test_input
list = glob.glob(
"../input/optiver-realized-volatility-prediction/trade_test.parquet/stock_id=0/*.parquet"
)
list
test_book = pa.parquet.read_table(list[0])
test_book = test_book.to_pandas()
test_book
def log_return(list_stock_prices):
return np.log(list_stock_prices).diff()
def rv(series_log_return):
return np.sqrt(np.sum(series_log_return**2))
def rv2(series_log_return):
return np.sqrt(np.sum(series_log_return**2))
# taken from https://www.kaggle.com/yus002/realized-volatility-prediction-lgbm-train
def my_metrics(y_true, y_pred):
return np.sqrt(np.mean(np.square((y_true - y_pred) / y_true)))
def rmspe(y_true, y_pred):
output = my_metrics(y_true, y_pred)
return "rmspe", output, False
df_book = train_input.copy()
df_book
# df_book.sort_values(by=["time_id", "seconds_in_bucket"])
df_book = train_input.copy()
df_book.sort_values(by=["time_id", "seconds_in_bucket"])
# compute different vwap
df_book["wap1"] = (
df_book["bid_price1"] * df_book["ask_size1"]
+ df_book["ask_price1"] * df_book["bid_size1"]
) / (df_book["bid_size1"] + df_book["ask_size1"])
# wap2
a = (
df_book["bid_price2"] * df_book["ask_size2"]
+ df_book["ask_price2"] * df_book["bid_size2"]
)
b = df_book["bid_size2"] + df_book["ask_size2"]
df_book["wap2"] = a / b
# wap3
a1 = (
df_book["bid_price1"] * df_book["ask_size1"]
+ df_book["ask_price1"] * df_book["bid_size1"]
)
a2 = (
df_book["bid_price2"] * df_book["ask_size2"]
+ df_book["ask_price2"] * df_book["bid_size2"]
)
b = (
df_book["bid_size1"]
+ df_book["ask_size1"]
+ df_book["bid_size2"]
+ df_book["ask_size2"]
)
df_book["wap3"] = (a1 + a2) / b
# wap4
a = (
df_book["bid_price1"] * df_book["ask_size1"]
+ df_book["ask_price1"] * df_book["bid_size1"]
) / (df_book["bid_size1"] + df_book["ask_size1"])
b = (
df_book["bid_price2"] * df_book["ask_size2"]
+ df_book["ask_price2"] * df_book["bid_size2"]
) / (df_book["bid_size2"] + df_book["ask_size2"])
df_book["wap4"] = (a + b) / 2
df_book["vol_wap1"] = (
df_book.groupby(by=["time_id"])["wap1"]
.apply(log_return)
.reset_index(drop=True)
.fillna(0)
)
df_book["vol_wap2"] = (
df_book.groupby(by=["time_id"])["wap2"]
.apply(log_return)
.reset_index(drop=True)
.fillna(0)
)
df_book["vol_wap3"] = (
df_book.groupby(by=["time_id"])["wap3"]
.apply(log_return)
.reset_index(drop=True)
.fillna(0)
)
df_book["vol_wap4"] = (
df_book.groupby(by=["time_id"])["wap4"]
.apply(log_return)
.reset_index(drop=True)
.fillna(0)
)
df_book["bas"] = (
df_book[["ask_price1", "ask_price2"]].min(axis=1)
/ df_book[["bid_price1", "bid_price2"]].max(axis=1)
- 1
)
# different spreads
df_book["h_spread_l1"] = df_book["ask_price1"] - df_book["bid_price1"]
df_book["h_spread_l2"] = df_book["ask_price2"] - df_book["bid_price2"]
df_book["v_spread_b"] = df_book["bid_price1"] - df_book["bid_price2"]
df_book["v_spread_a"] = df_book["ask_price1"] - df_book["bid_price2"]
display(df_book.head())
print(df_book.shape)
# attach volatitilies based on different VWAPs
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["vol_wap1"].agg(rv).reset_index(),
df_book.groupby(by=["time_id"], as_index=False)["bas"].mean(),
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["vol_wap2"].agg(rv).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["vol_wap3"].agg(rv).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["vol_wap4"].agg(rv).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
# spread summaries
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["h_spread_l1"].agg(max).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["h_spread_l2"].agg(max).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["v_spread_b"].agg(max).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["v_spread_a"].agg(max).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat["target"] = train_input.target
display(stock_stat.head())
print(stock_stat.shape)
target = stock_stat["target"]
train_data = stock_stat.drop("target", axis=1)
train_data
df_book = test_input.copy()
df_book.sort_values(by=["time_id", "seconds_in_bucket"])
# compute different vwap
df_book["wap1"] = (
df_book["bid_price1"] * df_book["ask_size1"]
+ df_book["ask_price1"] * df_book["bid_size1"]
) / (df_book["bid_size1"] + df_book["ask_size1"])
# wap2
a = (
df_book["bid_price2"] * df_book["ask_size2"]
+ df_book["ask_price2"] * df_book["bid_size2"]
)
b = df_book["bid_size2"] + df_book["ask_size2"]
df_book["wap2"] = a / b
# wap3
a1 = (
df_book["bid_price1"] * df_book["ask_size1"]
+ df_book["ask_price1"] * df_book["bid_size1"]
)
a2 = (
df_book["bid_price2"] * df_book["ask_size2"]
+ df_book["ask_price2"] * df_book["bid_size2"]
)
b = (
df_book["bid_size1"]
+ df_book["ask_size1"]
+ df_book["bid_size2"]
+ df_book["ask_size2"]
)
df_book["wap3"] = (a1 + a2) / b
# wap4
a = (
df_book["bid_price1"] * df_book["ask_size1"]
+ df_book["ask_price1"] * df_book["bid_size1"]
) / (df_book["bid_size1"] + df_book["ask_size1"])
b = (
df_book["bid_price2"] * df_book["ask_size2"]
+ df_book["ask_price2"] * df_book["bid_size2"]
) / (df_book["bid_size2"] + df_book["ask_size2"])
df_book["wap4"] = (a + b) / 2
df_book["vol_wap1"] = (
df_book.groupby(by=["time_id"])["wap1"]
.apply(log_return)
.reset_index(drop=True)
.fillna(0)
)
df_book["vol_wap2"] = (
df_book.groupby(by=["time_id"])["wap2"]
.apply(log_return)
.reset_index(drop=True)
.fillna(0)
)
df_book["vol_wap3"] = (
df_book.groupby(by=["time_id"])["wap3"]
.apply(log_return)
.reset_index(drop=True)
.fillna(0)
)
df_book["vol_wap4"] = (
df_book.groupby(by=["time_id"])["wap4"]
.apply(log_return)
.reset_index(drop=True)
.fillna(0)
)
df_book["bas"] = (
df_book[["ask_price1", "ask_price2"]].min(axis=1)
/ df_book[["bid_price1", "bid_price2"]].max(axis=1)
- 1
)
# different spreads
df_book["h_spread_l1"] = df_book["ask_price1"] - df_book["bid_price1"]
df_book["h_spread_l2"] = df_book["ask_price2"] - df_book["bid_price2"]
df_book["v_spread_b"] = df_book["bid_price1"] - df_book["bid_price2"]
df_book["v_spread_a"] = df_book["ask_price1"] - df_book["bid_price2"]
##
df_book["vol_wap1"] = df_book["vol_wap1"].apply(rv).reset_index()
df_book["vol_wap2"] = df_book["vol_wap2"].apply(rv).reset_index()
df_book["vol_wap3"] = df_book["vol_wap3"].apply(rv).reset_index()
df_book["vol_wap4"] = df_book["vol_wap4"].apply(rv).reset_index()
##
###ここから下のロジックは現在つかっていない
# attach volatitilies based on different VWAPs
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["vol_wap1"].agg(rv).reset_index(),
df_book.groupby(by=["time_id"], as_index=False)["bas"].mean(),
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["vol_wap2"].agg(rv).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["vol_wap3"].agg(rv).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["vol_wap4"].agg(rv).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
# spread summaries
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["h_spread_l1"].agg(max).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["h_spread_l2"].agg(max).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["v_spread_b"].agg(max).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["v_spread_a"].agg(max).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
# stock_stat['stock_id'] = 0
test_data = stock_stat
display(test_data)
print(test_data.shape)
# 2021/07/26
# time_id情報を消して学習してみる
# lgbのimportanceを確認してみて調整予定
test_data = test_data.drop("time_id", axis=1)
train_data = train_data.drop("time_id", axis=1)
display(train_data)
display(test_data)
print("train_data:", train_data.shape)
print("test_data:", test_data.shape)
# # **LightGBM**
import lightgbm as lgb
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split, KFold
best_lgb_params = {
"bagging_fraction": 1,
"bagging_freq": 0,
"feature_fraction": 0.7,
#'feature_pre_filter': False,
#'lambda_l1': 1.263018256839349e-07,
#'lambda_l2': 0.002578740827596048,
"metric": "l2",
#'min_child_samples': 200,
#'num_leaves': 131,
"objective": "mse",
}
best_lgb_params["learning_rate"] = 0.5
best_lgb_params["early_stopping_round"] = 100
best_lgb_params["num_iterations"] = 10000
x_train, x_test, y_train, y_test = train_test_split(
train_data, target, test_size=0.3, random_state=42
)
lgb_train = lgb.Dataset(x_train, y_train)
lgb_valid = lgb.Dataset(x_test, y_test)
model = lgb.train(best_lgb_params, lgb_train, valid_sets=[lgb_valid], verbose_eval=100)
lgb.plot_importance(model, figsize=(12, 6))
plt.show()
sub = pd.read_csv(
"../input/optiver-realized-volatility-prediction/sample_submission.csv"
)
sub
test_data["target"] = model.predict(test_data)
display(test_data)
print(test_data.shape)
# x = test_data[test_data.time_id==4]
# sub['target'][0] = np.round(x['target'].mean(),decimals=9)
# x = test_data[test_data.time_id==32]
# sub['target'][1] = np.round(x['target'].mean(),decimals=9)
# x = test_data[test_data.time_id==34]
# sub['target'][2] = np.round(x['target'].mean(),decimals=9)
# sub = sub.dropna()
# sub
sub.to_csv("./submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/031/69031556.ipynb
| null | null |
[{"Id": 69031556, "ScriptId": 18715097, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7520437, "CreationDate": "07/26/2021 04:47:02", "VersionNumber": 6.0, "Title": "Volatility-Prediction1", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 270.0, "LinesInsertedFromPrevious": 22.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 248.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import pandas as pd
import glob
import pyarrow as pa
import numpy as np
import matplotlib.pyplot as plt
train_data = pd.read_csv("../input/optiver-realized-volatility-prediction/train.csv")
test_data = pd.read_csv("../input/optiver-realized-volatility-prediction/test.csv")
print(train_data.shape)
print(test_data.shape)
train_data = train_data[train_data.stock_id == 0]
display(train_data)
display(test_data)
list = glob.glob(
"../input/optiver-realized-volatility-prediction/trade_train.parquet/stock_id=0/*.parquet"
)
list
book = pa.parquet.read_table(list[0])
book = book.to_pandas()
book.head(305)
list = glob.glob(
"../input/optiver-realized-volatility-prediction/book_train.parquet/stock_id=0/*.parquet"
)
list
train = pa.parquet.read_table(list[0])
train = train.to_pandas()
train.head(305)
train_data = train_data.drop(["stock_id"], axis=1)
train_data
train_input = pd.merge(train_data, train, on=["time_id"])
train_input
list = glob.glob(
"../input/optiver-realized-volatility-prediction/book_test.parquet/stock_id=0/*.parquet"
)
list
test = pa.parquet.read_table(list[0])
test_input = test.to_pandas()
print(test_input.shape)
test_input
list = glob.glob(
"../input/optiver-realized-volatility-prediction/trade_test.parquet/stock_id=0/*.parquet"
)
list
test_book = pa.parquet.read_table(list[0])
test_book = test_book.to_pandas()
test_book
def log_return(list_stock_prices):
return np.log(list_stock_prices).diff()
def rv(series_log_return):
return np.sqrt(np.sum(series_log_return**2))
def rv2(series_log_return):
return np.sqrt(np.sum(series_log_return**2))
# taken from https://www.kaggle.com/yus002/realized-volatility-prediction-lgbm-train
def my_metrics(y_true, y_pred):
return np.sqrt(np.mean(np.square((y_true - y_pred) / y_true)))
def rmspe(y_true, y_pred):
output = my_metrics(y_true, y_pred)
return "rmspe", output, False
df_book = train_input.copy()
df_book
# df_book.sort_values(by=["time_id", "seconds_in_bucket"])
df_book = train_input.copy()
df_book.sort_values(by=["time_id", "seconds_in_bucket"])
# compute different vwap
df_book["wap1"] = (
df_book["bid_price1"] * df_book["ask_size1"]
+ df_book["ask_price1"] * df_book["bid_size1"]
) / (df_book["bid_size1"] + df_book["ask_size1"])
# wap2
a = (
df_book["bid_price2"] * df_book["ask_size2"]
+ df_book["ask_price2"] * df_book["bid_size2"]
)
b = df_book["bid_size2"] + df_book["ask_size2"]
df_book["wap2"] = a / b
# wap3
a1 = (
df_book["bid_price1"] * df_book["ask_size1"]
+ df_book["ask_price1"] * df_book["bid_size1"]
)
a2 = (
df_book["bid_price2"] * df_book["ask_size2"]
+ df_book["ask_price2"] * df_book["bid_size2"]
)
b = (
df_book["bid_size1"]
+ df_book["ask_size1"]
+ df_book["bid_size2"]
+ df_book["ask_size2"]
)
df_book["wap3"] = (a1 + a2) / b
# wap4
a = (
df_book["bid_price1"] * df_book["ask_size1"]
+ df_book["ask_price1"] * df_book["bid_size1"]
) / (df_book["bid_size1"] + df_book["ask_size1"])
b = (
df_book["bid_price2"] * df_book["ask_size2"]
+ df_book["ask_price2"] * df_book["bid_size2"]
) / (df_book["bid_size2"] + df_book["ask_size2"])
df_book["wap4"] = (a + b) / 2
df_book["vol_wap1"] = (
df_book.groupby(by=["time_id"])["wap1"]
.apply(log_return)
.reset_index(drop=True)
.fillna(0)
)
df_book["vol_wap2"] = (
df_book.groupby(by=["time_id"])["wap2"]
.apply(log_return)
.reset_index(drop=True)
.fillna(0)
)
df_book["vol_wap3"] = (
df_book.groupby(by=["time_id"])["wap3"]
.apply(log_return)
.reset_index(drop=True)
.fillna(0)
)
df_book["vol_wap4"] = (
df_book.groupby(by=["time_id"])["wap4"]
.apply(log_return)
.reset_index(drop=True)
.fillna(0)
)
df_book["bas"] = (
df_book[["ask_price1", "ask_price2"]].min(axis=1)
/ df_book[["bid_price1", "bid_price2"]].max(axis=1)
- 1
)
# different spreads
df_book["h_spread_l1"] = df_book["ask_price1"] - df_book["bid_price1"]
df_book["h_spread_l2"] = df_book["ask_price2"] - df_book["bid_price2"]
df_book["v_spread_b"] = df_book["bid_price1"] - df_book["bid_price2"]
df_book["v_spread_a"] = df_book["ask_price1"] - df_book["bid_price2"]
display(df_book.head())
print(df_book.shape)
# attach volatitilies based on different VWAPs
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["vol_wap1"].agg(rv).reset_index(),
df_book.groupby(by=["time_id"], as_index=False)["bas"].mean(),
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["vol_wap2"].agg(rv).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["vol_wap3"].agg(rv).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["vol_wap4"].agg(rv).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
# spread summaries
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["h_spread_l1"].agg(max).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["h_spread_l2"].agg(max).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["v_spread_b"].agg(max).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["v_spread_a"].agg(max).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat["target"] = train_input.target
display(stock_stat.head())
print(stock_stat.shape)
target = stock_stat["target"]
train_data = stock_stat.drop("target", axis=1)
train_data
df_book = test_input.copy()
df_book.sort_values(by=["time_id", "seconds_in_bucket"])
# compute different vwap
df_book["wap1"] = (
df_book["bid_price1"] * df_book["ask_size1"]
+ df_book["ask_price1"] * df_book["bid_size1"]
) / (df_book["bid_size1"] + df_book["ask_size1"])
# wap2
a = (
df_book["bid_price2"] * df_book["ask_size2"]
+ df_book["ask_price2"] * df_book["bid_size2"]
)
b = df_book["bid_size2"] + df_book["ask_size2"]
df_book["wap2"] = a / b
# wap3
a1 = (
df_book["bid_price1"] * df_book["ask_size1"]
+ df_book["ask_price1"] * df_book["bid_size1"]
)
a2 = (
df_book["bid_price2"] * df_book["ask_size2"]
+ df_book["ask_price2"] * df_book["bid_size2"]
)
b = (
df_book["bid_size1"]
+ df_book["ask_size1"]
+ df_book["bid_size2"]
+ df_book["ask_size2"]
)
df_book["wap3"] = (a1 + a2) / b
# wap4
a = (
df_book["bid_price1"] * df_book["ask_size1"]
+ df_book["ask_price1"] * df_book["bid_size1"]
) / (df_book["bid_size1"] + df_book["ask_size1"])
b = (
df_book["bid_price2"] * df_book["ask_size2"]
+ df_book["ask_price2"] * df_book["bid_size2"]
) / (df_book["bid_size2"] + df_book["ask_size2"])
df_book["wap4"] = (a + b) / 2
df_book["vol_wap1"] = (
df_book.groupby(by=["time_id"])["wap1"]
.apply(log_return)
.reset_index(drop=True)
.fillna(0)
)
df_book["vol_wap2"] = (
df_book.groupby(by=["time_id"])["wap2"]
.apply(log_return)
.reset_index(drop=True)
.fillna(0)
)
df_book["vol_wap3"] = (
df_book.groupby(by=["time_id"])["wap3"]
.apply(log_return)
.reset_index(drop=True)
.fillna(0)
)
df_book["vol_wap4"] = (
df_book.groupby(by=["time_id"])["wap4"]
.apply(log_return)
.reset_index(drop=True)
.fillna(0)
)
df_book["bas"] = (
df_book[["ask_price1", "ask_price2"]].min(axis=1)
/ df_book[["bid_price1", "bid_price2"]].max(axis=1)
- 1
)
# different spreads
df_book["h_spread_l1"] = df_book["ask_price1"] - df_book["bid_price1"]
df_book["h_spread_l2"] = df_book["ask_price2"] - df_book["bid_price2"]
df_book["v_spread_b"] = df_book["bid_price1"] - df_book["bid_price2"]
df_book["v_spread_a"] = df_book["ask_price1"] - df_book["bid_price2"]
##
df_book["vol_wap1"] = df_book["vol_wap1"].apply(rv).reset_index()
df_book["vol_wap2"] = df_book["vol_wap2"].apply(rv).reset_index()
df_book["vol_wap3"] = df_book["vol_wap3"].apply(rv).reset_index()
df_book["vol_wap4"] = df_book["vol_wap4"].apply(rv).reset_index()
##
###ここから下のロジックは現在つかっていない
# attach volatitilies based on different VWAPs
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["vol_wap1"].agg(rv).reset_index(),
df_book.groupby(by=["time_id"], as_index=False)["bas"].mean(),
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["vol_wap2"].agg(rv).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["vol_wap3"].agg(rv).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["vol_wap4"].agg(rv).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
# spread summaries
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["h_spread_l1"].agg(max).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["h_spread_l2"].agg(max).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["v_spread_b"].agg(max).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
stock_stat = pd.merge(
df_book.groupby(by=["time_id"])["v_spread_a"].agg(max).reset_index(),
stock_stat,
on=["time_id"],
how="left",
)
# stock_stat['stock_id'] = 0
test_data = stock_stat
display(test_data)
print(test_data.shape)
# 2021/07/26
# time_id情報を消して学習してみる
# lgbのimportanceを確認してみて調整予定
test_data = test_data.drop("time_id", axis=1)
train_data = train_data.drop("time_id", axis=1)
display(train_data)
display(test_data)
print("train_data:", train_data.shape)
print("test_data:", test_data.shape)
# # **LightGBM**
import lightgbm as lgb
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split, KFold
best_lgb_params = {
"bagging_fraction": 1,
"bagging_freq": 0,
"feature_fraction": 0.7,
#'feature_pre_filter': False,
#'lambda_l1': 1.263018256839349e-07,
#'lambda_l2': 0.002578740827596048,
"metric": "l2",
#'min_child_samples': 200,
#'num_leaves': 131,
"objective": "mse",
}
best_lgb_params["learning_rate"] = 0.5
best_lgb_params["early_stopping_round"] = 100
best_lgb_params["num_iterations"] = 10000
x_train, x_test, y_train, y_test = train_test_split(
train_data, target, test_size=0.3, random_state=42
)
lgb_train = lgb.Dataset(x_train, y_train)
lgb_valid = lgb.Dataset(x_test, y_test)
model = lgb.train(best_lgb_params, lgb_train, valid_sets=[lgb_valid], verbose_eval=100)
lgb.plot_importance(model, figsize=(12, 6))
plt.show()
sub = pd.read_csv(
"../input/optiver-realized-volatility-prediction/sample_submission.csv"
)
sub
test_data["target"] = model.predict(test_data)
display(test_data)
print(test_data.shape)
# x = test_data[test_data.time_id==4]
# sub['target'][0] = np.round(x['target'].mean(),decimals=9)
# x = test_data[test_data.time_id==32]
# sub['target'][1] = np.round(x['target'].mean(),decimals=9)
# x = test_data[test_data.time_id==34]
# sub['target'][2] = np.round(x['target'].mean(),decimals=9)
# sub = sub.dropna()
# sub
sub.to_csv("./submission.csv", index=False)
| false | 0 | 4,547 | 0 | 4,547 | 4,547 |
||
69031207
|
<jupyter_start><jupyter_text>Respiratory Sound Database
### Context
Respiratory sounds are important indicators of respiratory health and respiratory disorders. The sound emitted when a person breathes is directly related to air movement, changes within lung tissue and the position of secretions within the lung. A wheezing sound, for example, is a common sign that a patient has an obstructive airway disease like asthma or chronic obstructive pulmonary disease (COPD).
These sounds can be recorded using digital stethoscopes and other recording techniques. This digital data opens up the possibility of using machine learning to automatically diagnose respiratory disorders like asthma, pneumonia and bronchiolitis, to name a few.
### Content
The Respiratory Sound Database was created by two research teams in Portugal and Greece. It includes 920 annotated recordings of varying length - 10s to 90s. These recordings were taken from 126 patients. There are a total of 5.5 hours of recordings containing 6898 respiratory cycles - 1864 contain crackles, 886 contain wheezes and 506 contain both crackles and wheezes. The data includes both clean respiratory sounds as well as noisy recordings that simulate real life conditions. The patients span all age groups - children, adults and the elderly.
This Kaggle dataset includes:
- 920 .wav sound files
- 920 annotation .txt files
- A text file listing the diagnosis for each patient
- A text file explaining the file naming format
- A text file listing 91 names (filename_differences.txt )
- A text file containing demographic information for each patient
Note:<br>
filename_differences.txt is a list of files whose names were corrected after this dataset's creators found a bug in the original file naming script. It can now be ignored.
### General
The demographic info file has 6 columns:
- Patient number
- Age
- Sex
- Adult BMI (kg/m2)
- Child Weight (kg)
- Child Height (cm)
Each audio file name is divided into 5 elements, separated with underscores (_).
1. Patient number (101,102,...,226)
2. Recording index
3. Chest location
a. Trachea (Tc)
b. Anterior left (Al)
c. Anterior right (Ar)
d. Posterior left (Pl)
e. Posterior right (Pr)
f. Lateral left (Ll)
g. Lateral right (Lr)
4. Acquisition mode
a. sequential/single channel (sc),
b. simultaneous/multichannel (mc)
5. Recording equipment
a. AKG C417L Microphone (AKGC417L),
b. 3M Littmann Classic II SE Stethoscope (LittC2SE),
c. 3M Litmmann 3200 Electronic Stethoscope (Litt3200),
d. WelchAllyn Meditron Master Elite Electronic Stethoscope (Meditron)
The annotation text files have four columns:
- Beginning of respiratory cycle(s)
- End of respiratory cycle(s)
- Presence/absence of crackles (presence=1, absence=0)
- Presence/absence of wheezes (presence=1, absence=0)
The abbreviations used in the diagnosis file are:
- COPD: Chronic Obstructive Pulmonary Disease
- LRTI: Lower Respiratory Tract Infection
- URTI: Upper Respiratory Tract Infection
### Citation
Paper: Α Respiratory Sound Database for the Development of Automated Classification<br>
Rocha BM, Filos D, Mendes L, Vogiatzis I, Perantoni E, Kaimakamis E, Natsiavas P, Oliveira A, Jácome C, Marques A, Paiva RP (2018) In Precision Medicine Powered by pHealth and Connected Health (pp. 51-55). Springer, Singapore.<br>
https://eden.dei.uc.pt/~ruipedro/publications/Conferences/ICBHI2017a.pdf
#### Ref Websites
- http://www.auditory.org/mhonarc/2018/msg00007.html
- http://bhichallenge.med.auth.gr/
Kaggle dataset identifier: respiratory-sound-database
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# libraries
import numpy as np
import pandas as pd
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import (
Conv1D,
Conv2D,
MaxPooling2D,
MaxPooling1D,
Dense,
Flatten,
Dropout,
SeparableConv1D,
)
import matplotlib.pyplot as plt
import seaborn as sns
import librosa
import soundfile as sf
import librosa.display
from os import listdir
from os.path import isfile, join
from tensorflow.keras.utils import plot_model, to_categorical
from IPython.display import Audio
from scipy.io import wavfile
from pydub import AudioSegment
import IPython
from IPython.display import Audio, Javascript
from scipy.io import wavfile
from base64 import b64decode
from pydub import AudioSegment
diagnosis_df = pd.read_csv(
"../input/respiratory-sound-database/Respiratory_Sound_Database/Respiratory_Sound_Database/patient_diagnosis.csv",
names=["Patient ID", "Diagnosis"],
)
diagnosis_df["basicDiagnosis"] = diagnosis_df["Diagnosis"].apply(
lambda x: "Healthy" if x == "Healthy" else "Unhealthy"
)
diagnosis_df.head(10)
df_no_diagnosis = pd.read_csv(
"../input/respiratory-sound-database/demographic_info.txt",
names=[
"Patient ID",
"Age",
"Gender",
"BMI (kg/m2)",
"Child Weight (kg)",
"Child Height (cm)",
],
delimiter=" ",
)
df_no_diagnosis.head(10)
df = df_no_diagnosis.join(
diagnosis_df.set_index("Patient ID"), on="Patient ID", how="left"
)
df.head(10)
root = "../input/respiratory-sound-database/Respiratory_Sound_Database/Respiratory_Sound_Database/audio_and_txt_files/"
filenames = [s.split(".")[0] for s in os.listdir(path=root) if ".txt" in s]
# getting data based on Tr,Al,Ar...
def extract_annotation_data(file_name, root):
tokens = file_name.split("_")
recording_info = pd.DataFrame(
data=[tokens],
columns=[
"Patient ID",
"Index",
"Chest location",
"Acquisition mode",
"Recording equipment",
],
)
recording_annotations = pd.read_csv(
os.path.join(root, file_name + ".txt"),
names=["Start", "End", "Crackles", "Wheezes"],
delimiter="\t",
)
return (recording_info, recording_annotations)
i_list = []
rec_annotations = []
rec_annotations_dict = {}
for s in filenames:
(i, a) = extract_annotation_data(s, root)
i_list.append(i)
rec_annotations.append(a)
rec_annotations_dict[s] = a
recording_info = pd.concat(i_list, axis=0)
recording_info.head(10) # default is 5 for some reason ???
len(rec_annotations_dict) # ????? is it number of words in the annotations thing?
def slice_audio(audiodata, samplerate, start, end):
start = samplerate * start
end = samplerate * end
return audiodata[start:end]
# didnt give o/p because it just does it behind scenes--
class Diagnosis:
def __init__(self, id, diagnosis, file_path):
self.id = id
self.diagnosis = diagnosis
self.file_path = file_path
# this doesnt give o/p either--
def get_wav_files():
audio_path = "../input/respiratory-sound-database/Respiratory_Sound_Database/Respiratory_Sound_Database/audio_and_txt_files/"
files = [f for f in listdir(audio_path) if isfile(join(audio_path, f))]
# Gets all files in directory
wav_files = [f for f in files if f.endswith(".wav")]
# to get .wav files it searches under this thing
wav_files = sorted(wav_files)
return wav_files, audio_path
def diagnosis_data():
diagnosis = pd.read_csv(
"../input/respiratory-sound-database/Respiratory_Sound_Database/Respiratory_Sound_Database/patient_diagnosis.csv"
)
wav_files, audio_path = get_wav_files()
diag_dict = {101: "URTI"}
diagnosis_list = []
for index, row in diagnosis.iterrows():
diag_dict[row[0]] = row[1]
c = 0
audio_data_list = []
for f in wav_files:
diagnosis_list.append(Diagnosis(c, diag_dict[int(f[:3])], audio_path + f))
# wav,s_rate = librosa.load(audio_path+f)
if diag_dict[int(f[:3])] == "Healthy":
binary_classification_label = "Healthy"
else:
binary_classification_label = "UnHealthy"
audio_data_list.append(
{
"id": c,
"diagnosis": diag_dict[int(f[:3])],
"binary_diagnosis": binary_classification_label,
"filename": f,
"file_path": audio_path + f,
}
)
c += 1
return diagnosis_list, pd.DataFrame(audio_data_list)
dataset_obj, dataset_df = diagnosis_data()
print(dataset_df["diagnosis"].unique())
plt.figure(figsize=(10, 5))
sns.countplot(dataset_df["diagnosis"])
dataset_df.to_csv("DIagnosis.csv", index=False)
processed_dataset_df = dataset_df[
(dataset_df["diagnosis"] != "Asthma") & (dataset_df["diagnosis"] != "LRTI")
]
plt.figure(figsize=(10, 5))
sns.countplot(processed_dataset_df["diagnosis"])
processed_dataset_df.head(10)
audio_file, samplerate = librosa.core.load(processed_dataset_df.file_path[0])
# for the training part----------
import tensorflow as tf
from tensorflow.keras.callbacks import CSVLogger, ModelCheckpoint
from tensorflow.keras.utils import to_categorical
import os
from scipy.io import wavfile
import pandas as pd
import numpy as np
from sklearn.utils.class_weight import compute_class_weight
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
from tqdm.notebook import tqdm
from glob import glob
import argparse
import warnings
import wavio
from librosa.core import resample, to_mono
# Dependencies(?>)
import numpy as np
import pandas as pd
import os
import librosa
import matplotlib.pyplot as plt
import gc
import time
from tqdm import tqdm, tqdm_notebook
tqdm.pandas() # Progress bar
from sklearn.metrics import label_ranking_average_precision_score
from sklearn.model_selection import train_test_split
# Machine Learning basic stuff
import tensorflow as tf
from keras import backend as K
from keras.engine.topology import Layer
from keras import initializers, regularizers, constraints, optimizers, layers
from keras.layers import (
Input,
LSTM,
Dense,
TimeDistributed,
Activation,
BatchNormalization,
Dropout,
Bidirectional,
)
from keras.models import Sequential
from keras.utils import Sequence
from tensorflow.compat.v1.keras.layers import CuDNNLSTM
from keras.optimizers import Adam
from keras.callbacks import EarlyStopping
# Preprocessing parameters
sr = 44100 # Sampling rate
duration = 10
hop_length = 347 # To make time steps 128
fmin = 20
fmax = sr // 2
n_mels = 128
n_fft = n_mels * 20
samples = sr * duration
def downsample_mono(path, sr):
obj = wavio.read(path)
wav = obj.data.astype(np.float32, order="F")
rate = obj.rate
try:
channel = wav.shape[1]
if channel == 2:
wav = to_mono(wav.T)
elif channel == 1:
wav = to_mono(wav.reshape(-1))
except IndexError:
wav = to_mono(wav.reshape(-1))
pass
except Exception as exc:
raise exc
wav = resample(wav, rate, sr)
wav = wav.astype(np.int16)
return sr, wav
def read_audio(path):
y, sr = librosa.core.load(path, sr=16000, duration=duration)
return y
def audio_to_melspectrogram(audio, s_r, n_mel_val):
# to convert to melspectrogram after audio is read in
spectrogram = librosa.feature.melspectrogram(
audio,
sr=s_r,
n_mels=n_mel_val,
hop_length=hop_length,
n_fft=n_mel_val * 20,
fmin=20,
fmax=s_r // 2,
)
# dont really get the librosa thing, just get the feature.melspectrogram thing
return librosa.power_to_db(spectrogram).astype(np.float32)
def read_as_melspectrogram(path):
# to convert audio into a melspectrogram so we can use machine learning
mels = audio_to_melspectrogram(read_audio(path))
return mels
def audio_to_melspectrogram2(audio, s_r, n_mel_val):
# to convert to melspectrogram after audio is read by the librosa thing ??????
spectrogram = librosa.feature.melspectrogram(
audio,
sr=s_r,
n_mels=n_mel_val,
hop_length=hop_length,
n_fft=n_mel_val * 20,
fmin=20,
fmax=s_r // 2,
)
return librosa.power_to_db(spectrogram).astype(np.float32)
def read_as_melspectrogram2(path):
# to convert audio into a melspectrogram so we can use ml
mels = audio_to_melspectrogram2(read_audio(path))
return mels
def convert_wav_to_image(df):
X_mel_spec = []
X_mel_spec2 = []
for _, row in tqdm(df.iterrows()):
x_mel1 = read_as_melspectrogram(row.file_path)
x_mel2 = read_as_melspectrogram2(row.file_path)
X_mel_spec.append(
x_mel1.transpose(),
)
X_mel_spec2.append(x_mel2.transpose())
return X_mel_spec, X_mel_spec2
def convert_wav_to_mfcc(df):
X_mfcc = []
for _, row in tqdm(df.iterrows()):
x_mfcc = generate_mfcc_feature(row.file_path)
X_mfcc.append(x_mfcc.transpose())
return X_mfcc
def normalize(img):
# to normalize an array (subtract mean and divide by sd-- sigma)
eps = 0.001
if np.std(img) != 0:
img = (img - np.mean(img)) / np.std(img)
else:
img = (img - np.mean(img)) / eps
return img
def normalize_dataset(X):
# Normalizes list of arrays (subtract mean and divide by sd)
normalized_dataset = []
for img in X:
normalized = normalize(img)
normalized_dataset.append(normalized)
return normalized_dataset
import numpy as np
from numpy.lib.stride_tricks import as_strided
from typing import Tuple
import librosa
import numpy as np
def get_audio(file_path: str) -> Tuple[np.ndarray, int]:
audio_data, sr = librosa.core.load(file_path)
length = len(audio_data) / sr
return audio_data, sr, length
# all the class stuff here
class AudioMovingWindowPreProcessor:
def __init__(self) -> None:
pass
def get_audio_windows(
self, audio: np.ndarray, sr: int, length: int, window_size: int, stride: int
) -> np.ndarray:
# to generate audio frames using sliding window with stride -memory safe
no_frames = int((length - window_size) / stride) + 1
window_size = window_size * sr
stride = stride * sr
audio_frames = []
for index in range(no_frames):
if (stride * index + window_size) < len(audio):
frame = audio[stride * index : (stride * index + window_size)]
audio_frames.append(frame)
else:
break
return np.array(audio_frames)
def get_audio_windows_numpy_vectorized(
self, audio: np.ndarray, sr: int, length: int, window_size: int, stride: int
) -> np.ndarray:
# to generate audio frames using sliding window with stride Numpy - non-memory safe
no_frames = int((length - window_size) / stride) + 1
audio_frames = as_strided(
audio, shape=(no_frames, window_size * sr), strides=(stride * sr, 1)
)
audio_frames = audio_frames[:-2]
return audio_frames
import pandas as pd
import numpy as np
from scipy import signal
import matplotlib.pyplot as plt
from scipy import stats
import IPython.display as ipd
import librosa
import librosa.display
from skimage.restoration import denoise_wavelet
# for wavelet ( denoising )
class Wavelet_Filter:
def wavelet_filter(self, filteredSignal, samplerate):
x_den = denoise_wavelet(
filteredSignal,
method="VisuShrink",
mode="soft",
wavelet_levels=5,
wavelet="coif2",
rescale_sigma="True",
)
return x_den, samplerate
# to pass through high_pass
class Filter_BW_HP:
def __init__(self, high_pass):
self.high_pass = high_pass
def BW_highpass(self, newdata, samplerate):
b, a = signal.butter(4, 100 / (22050 / 2), btype="highpass")
filteredSignal = signal.lfilter(b, a, newdata)
return filteredSignal, samplerate
# to pass through low_pass
class FIlter_BW_LP:
def __init__(self, low_pass):
self.low_pass = low_pass
def BW_lowpass(self, filteredSignal, samplerate):
c, d = signal.butter(4, 2000 / (22050 / 2), btype="lowpass")
newFilteredSignal = signal.lfilter(c, d, filteredSignal)
return newFilteredSignal, samplerate
class FilterPipeline:
def __init__(self, low_pass, high_pass):
self.low_pass = low_pass
self.high_pass = high_pass
self.lp_filter = FIlter_BW_LP(low_pass)
self.hp_filter = Filter_BW_HP(high_pass)
self.wavelet = Wavelet_Filter()
def filters(self, audio_signal, sample_rate):
filtered_output, sr = self.lp_filter.BW_lowpass(audio_signal, sample_rate)
filtered_output, sr = self.hp_filter.BW_highpass(filtered_output, sr)
filtered_output, sr = self.wavelet.wavelet_filter(filtered_output, sr)
return filtered_output, sr
import math
import librosa
import numpy as np
from typing import Tuple
from tensorflow.keras.preprocessing.sequence import pad_sequences
# Filter Configurations
LOW_PASS_FREQUENCY = 100
HIGH_PASS_FREQUENCY = 2000
# Mel-Spectral Configurations
HOG_LENGTH = 347
# sizes (in sec)
MOVING_WINDOW_SIZE = 5
AUDIO_STRIDE_SIZE = 5
class AudioPreProcessor(AudioMovingWindowPreProcessor, FilterPipeline):
def __init__(self):
AudioMovingWindowPreProcessor.__init__(self)
FilterPipeline.__init__(self, LOW_PASS_FREQUENCY, HIGH_PASS_FREQUENCY)
super().__init__()
def pre_process_audio(self, audio_path: str, sample_rate: int) -> np.ndarray:
"""
for this function to generate audio frames using moving window
just use Parameters
----------
audio_path
sample_rate
Returns audio_frames
-------
"""
audio_data, sample_rate, length = get_audio(audio_path, sample_rate)
filtered_audio, sample_rate = self.filters(audio_data, sample_rate)
audio_frames = self.get_audio_windows(
filtered_audio, sample_rate, length, MOVING_WINDOW_SIZE, AUDIO_STRIDE_SIZE
)
return audio_frames
def audio_to_mel_spectrogram(
self, audio_data: np.ndarray, s_r: int, n_mel_val: int
) -> np.ndarray:
"""
to convert audio_frames into mel-spectrogram
Parameters
----------
s_r
n_mel_val
Returns mel-spectralgrams
-------
"""
spectrogram = librosa.feature.melspectrogram(
audio_data,
sr=s_r,
n_mels=n_mel_val,
hop_length=HOG_LENGTH,
n_fft=n_mel_val * 20,
fmin=20,
fmax=s_r // 2,
)
return librosa.power_to_db(spectrogram).astype(np.float32)
# sizes (in sec)
MOVING_WINDOW_SIZE = 5
AUDIO_STRIDE_SIZE = 4
SAMPLE_RATE = 16000
from tqdm import tqdm
# !mkdir breathcycles (dont really get this but sure)
X_mel_spec = []
X_mel_spec2 = []
Y_labels = []
durations_spec = []
for indx, df_row in tqdm(processed_dataset_df.iterrows()):
head, audio_file = os.path.split(df_row.file_path)
audio_file_name = audio_file.split(".wav")[0]
diagnosis_type = df_row.diagnosis
breathing_cycles_df = rec_annotations_dict.get(audio_file_name)
audio_data, sample_rate, length = get_audio(df_row.file_path)
no_frames = int((length - MOVING_WINDOW_SIZE) / AUDIO_STRIDE_SIZE) + 1
window_size = MOVING_WINDOW_SIZE * sample_rate
stride = AUDIO_STRIDE_SIZE * sample_rate
audio_frames = []
for index in range(no_frames):
if (stride * index + window_size) < len(audio_data):
frame = audio_data[stride * index : (stride * index + window_size)]
x_mel1 = audio_to_melspectrogram(frame, s_r=sample_rate, n_mel_val=128)
x_mel2 = audio_to_melspectrogram2(frame, s_r=sample_rate, n_mel_val=64)
X_mel_spec.append(
x_mel1.transpose(),
)
X_mel_spec2.append(x_mel2.transpose())
Y_labels.append(str(diagnosis_type))
else:
break
# has output,, shows [time, time/iteration]
X_mel1 = np.array(X_mel_spec)
X_mel2 = np.array(X_mel_spec2)
# X = normalize_dataset(X)
X_array_mel1 = np.array(X_mel1)
X_array_mel2 = np.array(X_mel2)
val_spae = []
for x in X_mel1:
# print(x.shape[0])-------
val_spae.append(x.shape[0])
def Average(lst):
return sum(lst) / len(lst)
# Driver Code
average = Average(val_spae)
# to print average of the list
print("Average of the required list =", round(average, 2))
import seaborn as sns
sns.distplot(val_spae)
# works till here
max_audio_lenght = max([len(i) for i in X_array_mel1])
max_audio_lenght2 = max([len(i) for i in X_array_mel2])
from tensorflow.keras.preprocessing.sequence import pad_sequences
# raw data processing-padding
X_array_mel1 = pad_sequences(X_array_mel1, padding="post", dtype="float32")
X_array_mel2 = pad_sequences(X_array_mel2, padding="post", dtype="float32")
X_array_mel1[0].shape
# Visualize an melspectogram example
plt.figure(figsize=(15, 10))
plt.title("Visualization of audio file", weight="bold")
plt.imshow(X_array_mel1[20])
X_array_mel1.shape[0]
# am i supposed to get a muchhh lower value?
shape_lenght = X_array_mel1.shape[0]
new_Xtrain = np.empty((shape_lenght, 2), dtype=np.object)
for idx in range(shape_lenght):
new_Xtrain[idx,] = [X_array_mel1[idx], X_array_mel2[idx]]
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.layers import (
LSTM,
Dense,
TimeDistributed,
LayerNormalization,
Masking,
)
from tensorflow.keras.models import Model
from tensorflow.keras.regularizers import l2
#!pip install kapre
# import kapre
# from kapre.composed import get_melspectrogram_layer
import tensorflow as tf
import os
import tensorflow_addons as tfa
from tqdm import tqdm
from tqdm.keras import TqdmCallback
from tensorflow.keras.utils import to_categorical
le = LabelEncoder()
diagnosis_classes = list(set(Y_labels))
le.fit(diagnosis_classes)
y = le.transform(Y_labels)
Y = to_categorical(y, num_classes=len(diagnosis_classes))
np.save("diagnosis_classes.npy", le.classes_)
audio_time_span = new_Xtrain[0][0].shape[0]
no_mel_sp_features = new_Xtrain[0][0].shape[1]
NO_CLASSES = 6 # Binary classification -> 2
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/031/69031207.ipynb
|
respiratory-sound-database
|
vbookshelf
|
[{"Id": 69031207, "ScriptId": 18407019, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7848575, "CreationDate": "07/26/2021 04:40:55", "VersionNumber": 8.0, "Title": "adi_respSound", "EvaluationDate": "07/26/2021", "IsChange": true, "TotalLines": 640.0, "LinesInsertedFromPrevious": 134.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 506.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 91745479, "KernelVersionId": 69031207, "SourceDatasetVersionId": 267422}]
|
[{"Id": 267422, "DatasetId": 110374, "DatasourceVersionId": 279608, "CreatorUserId": 1086574, "LicenseName": "Unknown", "CreationDate": "01/29/2019 06:42:26", "VersionNumber": 2.0, "Title": "Respiratory Sound Database", "Slug": "respiratory-sound-database", "Subtitle": "Use audio recordings to detect respiratory diseases.", "Description": "### Context\n\nRespiratory sounds are important indicators of respiratory health and respiratory disorders. The sound emitted when a person breathes is directly related to air movement, changes within lung tissue and the position of secretions within the lung. A wheezing sound, for example, is a common sign that a patient has an obstructive airway disease like asthma or chronic obstructive pulmonary disease (COPD).\n\nThese sounds can be recorded using digital stethoscopes and other recording techniques. This digital data opens up the possibility of using machine learning to automatically diagnose respiratory disorders like asthma, pneumonia and bronchiolitis, to name a few. \n\n### Content\n\nThe Respiratory Sound Database was created by two research teams in Portugal and Greece. It includes 920 annotated recordings of varying length - 10s to 90s. These recordings were taken from 126 patients. There are a total of 5.5 hours of recordings containing 6898 respiratory cycles - 1864 contain crackles, 886 contain wheezes and 506 contain both crackles and wheezes. The data includes both clean respiratory sounds as well as noisy recordings that simulate real life conditions. The patients span all age groups - children, adults and the elderly.\n\nThis Kaggle dataset includes:\n\n- 920 .wav sound files\n- 920 annotation .txt files\n- A text file listing the diagnosis for each patient\n- A text file explaining the file naming format\n- A text file listing 91 names (filename_differences.txt )\n- A text file containing demographic information for each patient\n\nNote:<br>\n filename_differences.txt is a list of files whose names were corrected after this dataset's creators found a bug in the original file naming script. It can now be ignored.\n\n### General\n\n The demographic info file has 6 columns:\n - Patient number\n - Age\n - Sex\n - Adult BMI (kg/m2)\n - Child Weight (kg)\n - Child Height (cm)\n\n\n Each audio file name is divided into 5 elements, separated with underscores (_).\n\n 1. Patient number (101,102,...,226)\n 2. Recording index\n 3. Chest location \n a. Trachea (Tc)\n b. Anterior left (Al)\n c. Anterior right (Ar)\n d. Posterior left (Pl)\n e. Posterior right (Pr)\n f. Lateral left (Ll)\n g. Lateral right (Lr)\n 4. Acquisition mode \n a. sequential/single channel (sc), \n b. simultaneous/multichannel (mc)\n 5. Recording equipment \n a. AKG C417L Microphone (AKGC417L), \n b. 3M Littmann Classic II SE Stethoscope (LittC2SE), \n c. 3M Litmmann 3200 Electronic Stethoscope (Litt3200), \n d. WelchAllyn Meditron Master Elite Electronic Stethoscope (Meditron)\n \n The annotation text files have four columns:\n - Beginning of respiratory cycle(s)\n - End of respiratory cycle(s)\n - Presence/absence of crackles (presence=1, absence=0)\n - Presence/absence of wheezes (presence=1, absence=0)\n\n The abbreviations used in the diagnosis file are:\n - COPD: Chronic Obstructive Pulmonary Disease\n - LRTI: Lower Respiratory Tract Infection\n - URTI: Upper Respiratory Tract Infection\n\n### Citation\n\nPaper: \u0391 Respiratory Sound Database for the Development of Automated Classification<br>\nRocha BM, Filos D, Mendes L, Vogiatzis I, Perantoni E, Kaimakamis E, Natsiavas P, Oliveira A, J\u00e1come C, Marques A, Paiva RP (2018) In Precision Medicine Powered by pHealth and Connected Health (pp. 51-55). Springer, Singapore.<br>\nhttps://eden.dei.uc.pt/~ruipedro/publications/Conferences/ICBHI2017a.pdf\n\n#### Ref Websites\n\n- http://www.auditory.org/mhonarc/2018/msg00007.html\n- http://bhichallenge.med.auth.gr/ \n\n### Acknowledgements\n\nMany thanks to the research teams at the University of Coimbra, Portugal; the University de Aveiro, Portugal and the Aristotle University of Thessaloniki, Greece for making this dataset publicly available.\n\n\n### Inspiration\n\n- Build a model to classify respiratory diseases.\n- Build a model to detect if a recording contains crackles, wheezes or both.\n- Annotation is a time consuming process. Create a model to automatically annotate respiratory sound recordings.\n- Deploy your model as a Tensorflow.js web app so it can be accessed from anywhere in the world.\n- Bioelectronics - Can you build your own digital stethoscope using an Arduino? If you are an aspiring inventor, this video will give you some valuable practical advice: https://www.youtube.com/watch?v=jo1cQ-ga2MI\n\n\nPhoto by voltamax on Pixabay.", "VersionNotes": "Added demographic information.", "TotalCompressedBytes": 2586.0, "TotalUncompressedBytes": 1979834490.0}]
|
[{"Id": 110374, "CreatorUserId": 1086574, "OwnerUserId": 1086574.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 267422.0, "CurrentDatasourceVersionId": 279608.0, "ForumId": 120167, "Type": 2, "CreationDate": "01/26/2019 04:20:04", "LastActivityDate": "01/26/2019", "TotalViews": 453670, "TotalDownloads": 16120, "TotalVotes": 442, "TotalKernels": 70}]
|
[{"Id": 1086574, "UserName": "vbookshelf", "DisplayName": "vbookshelf", "RegisterDate": "05/22/2017", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# libraries
import numpy as np
import pandas as pd
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import (
Conv1D,
Conv2D,
MaxPooling2D,
MaxPooling1D,
Dense,
Flatten,
Dropout,
SeparableConv1D,
)
import matplotlib.pyplot as plt
import seaborn as sns
import librosa
import soundfile as sf
import librosa.display
from os import listdir
from os.path import isfile, join
from tensorflow.keras.utils import plot_model, to_categorical
from IPython.display import Audio
from scipy.io import wavfile
from pydub import AudioSegment
import IPython
from IPython.display import Audio, Javascript
from scipy.io import wavfile
from base64 import b64decode
from pydub import AudioSegment
diagnosis_df = pd.read_csv(
"../input/respiratory-sound-database/Respiratory_Sound_Database/Respiratory_Sound_Database/patient_diagnosis.csv",
names=["Patient ID", "Diagnosis"],
)
diagnosis_df["basicDiagnosis"] = diagnosis_df["Diagnosis"].apply(
lambda x: "Healthy" if x == "Healthy" else "Unhealthy"
)
diagnosis_df.head(10)
df_no_diagnosis = pd.read_csv(
"../input/respiratory-sound-database/demographic_info.txt",
names=[
"Patient ID",
"Age",
"Gender",
"BMI (kg/m2)",
"Child Weight (kg)",
"Child Height (cm)",
],
delimiter=" ",
)
df_no_diagnosis.head(10)
df = df_no_diagnosis.join(
diagnosis_df.set_index("Patient ID"), on="Patient ID", how="left"
)
df.head(10)
root = "../input/respiratory-sound-database/Respiratory_Sound_Database/Respiratory_Sound_Database/audio_and_txt_files/"
filenames = [s.split(".")[0] for s in os.listdir(path=root) if ".txt" in s]
# getting data based on Tr,Al,Ar...
def extract_annotation_data(file_name, root):
tokens = file_name.split("_")
recording_info = pd.DataFrame(
data=[tokens],
columns=[
"Patient ID",
"Index",
"Chest location",
"Acquisition mode",
"Recording equipment",
],
)
recording_annotations = pd.read_csv(
os.path.join(root, file_name + ".txt"),
names=["Start", "End", "Crackles", "Wheezes"],
delimiter="\t",
)
return (recording_info, recording_annotations)
i_list = []
rec_annotations = []
rec_annotations_dict = {}
for s in filenames:
(i, a) = extract_annotation_data(s, root)
i_list.append(i)
rec_annotations.append(a)
rec_annotations_dict[s] = a
recording_info = pd.concat(i_list, axis=0)
recording_info.head(10) # default is 5 for some reason ???
len(rec_annotations_dict) # ????? is it number of words in the annotations thing?
def slice_audio(audiodata, samplerate, start, end):
start = samplerate * start
end = samplerate * end
return audiodata[start:end]
# didnt give o/p because it just does it behind scenes--
class Diagnosis:
def __init__(self, id, diagnosis, file_path):
self.id = id
self.diagnosis = diagnosis
self.file_path = file_path
# this doesnt give o/p either--
def get_wav_files():
audio_path = "../input/respiratory-sound-database/Respiratory_Sound_Database/Respiratory_Sound_Database/audio_and_txt_files/"
files = [f for f in listdir(audio_path) if isfile(join(audio_path, f))]
# Gets all files in directory
wav_files = [f for f in files if f.endswith(".wav")]
# to get .wav files it searches under this thing
wav_files = sorted(wav_files)
return wav_files, audio_path
def diagnosis_data():
diagnosis = pd.read_csv(
"../input/respiratory-sound-database/Respiratory_Sound_Database/Respiratory_Sound_Database/patient_diagnosis.csv"
)
wav_files, audio_path = get_wav_files()
diag_dict = {101: "URTI"}
diagnosis_list = []
for index, row in diagnosis.iterrows():
diag_dict[row[0]] = row[1]
c = 0
audio_data_list = []
for f in wav_files:
diagnosis_list.append(Diagnosis(c, diag_dict[int(f[:3])], audio_path + f))
# wav,s_rate = librosa.load(audio_path+f)
if diag_dict[int(f[:3])] == "Healthy":
binary_classification_label = "Healthy"
else:
binary_classification_label = "UnHealthy"
audio_data_list.append(
{
"id": c,
"diagnosis": diag_dict[int(f[:3])],
"binary_diagnosis": binary_classification_label,
"filename": f,
"file_path": audio_path + f,
}
)
c += 1
return diagnosis_list, pd.DataFrame(audio_data_list)
dataset_obj, dataset_df = diagnosis_data()
print(dataset_df["diagnosis"].unique())
plt.figure(figsize=(10, 5))
sns.countplot(dataset_df["diagnosis"])
dataset_df.to_csv("DIagnosis.csv", index=False)
processed_dataset_df = dataset_df[
(dataset_df["diagnosis"] != "Asthma") & (dataset_df["diagnosis"] != "LRTI")
]
plt.figure(figsize=(10, 5))
sns.countplot(processed_dataset_df["diagnosis"])
processed_dataset_df.head(10)
audio_file, samplerate = librosa.core.load(processed_dataset_df.file_path[0])
# for the training part----------
import tensorflow as tf
from tensorflow.keras.callbacks import CSVLogger, ModelCheckpoint
from tensorflow.keras.utils import to_categorical
import os
from scipy.io import wavfile
import pandas as pd
import numpy as np
from sklearn.utils.class_weight import compute_class_weight
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
from tqdm.notebook import tqdm
from glob import glob
import argparse
import warnings
import wavio
from librosa.core import resample, to_mono
# Dependencies(?>)
import numpy as np
import pandas as pd
import os
import librosa
import matplotlib.pyplot as plt
import gc
import time
from tqdm import tqdm, tqdm_notebook
tqdm.pandas() # Progress bar
from sklearn.metrics import label_ranking_average_precision_score
from sklearn.model_selection import train_test_split
# Machine Learning basic stuff
import tensorflow as tf
from keras import backend as K
from keras.engine.topology import Layer
from keras import initializers, regularizers, constraints, optimizers, layers
from keras.layers import (
Input,
LSTM,
Dense,
TimeDistributed,
Activation,
BatchNormalization,
Dropout,
Bidirectional,
)
from keras.models import Sequential
from keras.utils import Sequence
from tensorflow.compat.v1.keras.layers import CuDNNLSTM
from keras.optimizers import Adam
from keras.callbacks import EarlyStopping
# Preprocessing parameters
sr = 44100 # Sampling rate
duration = 10
hop_length = 347 # To make time steps 128
fmin = 20
fmax = sr // 2
n_mels = 128
n_fft = n_mels * 20
samples = sr * duration
def downsample_mono(path, sr):
obj = wavio.read(path)
wav = obj.data.astype(np.float32, order="F")
rate = obj.rate
try:
channel = wav.shape[1]
if channel == 2:
wav = to_mono(wav.T)
elif channel == 1:
wav = to_mono(wav.reshape(-1))
except IndexError:
wav = to_mono(wav.reshape(-1))
pass
except Exception as exc:
raise exc
wav = resample(wav, rate, sr)
wav = wav.astype(np.int16)
return sr, wav
def read_audio(path):
y, sr = librosa.core.load(path, sr=16000, duration=duration)
return y
def audio_to_melspectrogram(audio, s_r, n_mel_val):
# to convert to melspectrogram after audio is read in
spectrogram = librosa.feature.melspectrogram(
audio,
sr=s_r,
n_mels=n_mel_val,
hop_length=hop_length,
n_fft=n_mel_val * 20,
fmin=20,
fmax=s_r // 2,
)
# dont really get the librosa thing, just get the feature.melspectrogram thing
return librosa.power_to_db(spectrogram).astype(np.float32)
def read_as_melspectrogram(path):
# to convert audio into a melspectrogram so we can use machine learning
mels = audio_to_melspectrogram(read_audio(path))
return mels
def audio_to_melspectrogram2(audio, s_r, n_mel_val):
# to convert to melspectrogram after audio is read by the librosa thing ??????
spectrogram = librosa.feature.melspectrogram(
audio,
sr=s_r,
n_mels=n_mel_val,
hop_length=hop_length,
n_fft=n_mel_val * 20,
fmin=20,
fmax=s_r // 2,
)
return librosa.power_to_db(spectrogram).astype(np.float32)
def read_as_melspectrogram2(path):
# to convert audio into a melspectrogram so we can use ml
mels = audio_to_melspectrogram2(read_audio(path))
return mels
def convert_wav_to_image(df):
X_mel_spec = []
X_mel_spec2 = []
for _, row in tqdm(df.iterrows()):
x_mel1 = read_as_melspectrogram(row.file_path)
x_mel2 = read_as_melspectrogram2(row.file_path)
X_mel_spec.append(
x_mel1.transpose(),
)
X_mel_spec2.append(x_mel2.transpose())
return X_mel_spec, X_mel_spec2
def convert_wav_to_mfcc(df):
X_mfcc = []
for _, row in tqdm(df.iterrows()):
x_mfcc = generate_mfcc_feature(row.file_path)
X_mfcc.append(x_mfcc.transpose())
return X_mfcc
def normalize(img):
# to normalize an array (subtract mean and divide by sd-- sigma)
eps = 0.001
if np.std(img) != 0:
img = (img - np.mean(img)) / np.std(img)
else:
img = (img - np.mean(img)) / eps
return img
def normalize_dataset(X):
# Normalizes list of arrays (subtract mean and divide by sd)
normalized_dataset = []
for img in X:
normalized = normalize(img)
normalized_dataset.append(normalized)
return normalized_dataset
import numpy as np
from numpy.lib.stride_tricks import as_strided
from typing import Tuple
import librosa
import numpy as np
def get_audio(file_path: str) -> Tuple[np.ndarray, int]:
audio_data, sr = librosa.core.load(file_path)
length = len(audio_data) / sr
return audio_data, sr, length
# all the class stuff here
class AudioMovingWindowPreProcessor:
def __init__(self) -> None:
pass
def get_audio_windows(
self, audio: np.ndarray, sr: int, length: int, window_size: int, stride: int
) -> np.ndarray:
# to generate audio frames using sliding window with stride -memory safe
no_frames = int((length - window_size) / stride) + 1
window_size = window_size * sr
stride = stride * sr
audio_frames = []
for index in range(no_frames):
if (stride * index + window_size) < len(audio):
frame = audio[stride * index : (stride * index + window_size)]
audio_frames.append(frame)
else:
break
return np.array(audio_frames)
def get_audio_windows_numpy_vectorized(
self, audio: np.ndarray, sr: int, length: int, window_size: int, stride: int
) -> np.ndarray:
# to generate audio frames using sliding window with stride Numpy - non-memory safe
no_frames = int((length - window_size) / stride) + 1
audio_frames = as_strided(
audio, shape=(no_frames, window_size * sr), strides=(stride * sr, 1)
)
audio_frames = audio_frames[:-2]
return audio_frames
import pandas as pd
import numpy as np
from scipy import signal
import matplotlib.pyplot as plt
from scipy import stats
import IPython.display as ipd
import librosa
import librosa.display
from skimage.restoration import denoise_wavelet
# for wavelet ( denoising )
class Wavelet_Filter:
def wavelet_filter(self, filteredSignal, samplerate):
x_den = denoise_wavelet(
filteredSignal,
method="VisuShrink",
mode="soft",
wavelet_levels=5,
wavelet="coif2",
rescale_sigma="True",
)
return x_den, samplerate
# to pass through high_pass
class Filter_BW_HP:
def __init__(self, high_pass):
self.high_pass = high_pass
def BW_highpass(self, newdata, samplerate):
b, a = signal.butter(4, 100 / (22050 / 2), btype="highpass")
filteredSignal = signal.lfilter(b, a, newdata)
return filteredSignal, samplerate
# to pass through low_pass
class FIlter_BW_LP:
def __init__(self, low_pass):
self.low_pass = low_pass
def BW_lowpass(self, filteredSignal, samplerate):
c, d = signal.butter(4, 2000 / (22050 / 2), btype="lowpass")
newFilteredSignal = signal.lfilter(c, d, filteredSignal)
return newFilteredSignal, samplerate
class FilterPipeline:
def __init__(self, low_pass, high_pass):
self.low_pass = low_pass
self.high_pass = high_pass
self.lp_filter = FIlter_BW_LP(low_pass)
self.hp_filter = Filter_BW_HP(high_pass)
self.wavelet = Wavelet_Filter()
def filters(self, audio_signal, sample_rate):
filtered_output, sr = self.lp_filter.BW_lowpass(audio_signal, sample_rate)
filtered_output, sr = self.hp_filter.BW_highpass(filtered_output, sr)
filtered_output, sr = self.wavelet.wavelet_filter(filtered_output, sr)
return filtered_output, sr
import math
import librosa
import numpy as np
from typing import Tuple
from tensorflow.keras.preprocessing.sequence import pad_sequences
# Filter Configurations
LOW_PASS_FREQUENCY = 100
HIGH_PASS_FREQUENCY = 2000
# Mel-Spectral Configurations
HOG_LENGTH = 347
# sizes (in sec)
MOVING_WINDOW_SIZE = 5
AUDIO_STRIDE_SIZE = 5
class AudioPreProcessor(AudioMovingWindowPreProcessor, FilterPipeline):
def __init__(self):
AudioMovingWindowPreProcessor.__init__(self)
FilterPipeline.__init__(self, LOW_PASS_FREQUENCY, HIGH_PASS_FREQUENCY)
super().__init__()
def pre_process_audio(self, audio_path: str, sample_rate: int) -> np.ndarray:
"""
for this function to generate audio frames using moving window
just use Parameters
----------
audio_path
sample_rate
Returns audio_frames
-------
"""
audio_data, sample_rate, length = get_audio(audio_path, sample_rate)
filtered_audio, sample_rate = self.filters(audio_data, sample_rate)
audio_frames = self.get_audio_windows(
filtered_audio, sample_rate, length, MOVING_WINDOW_SIZE, AUDIO_STRIDE_SIZE
)
return audio_frames
def audio_to_mel_spectrogram(
self, audio_data: np.ndarray, s_r: int, n_mel_val: int
) -> np.ndarray:
"""
to convert audio_frames into mel-spectrogram
Parameters
----------
s_r
n_mel_val
Returns mel-spectralgrams
-------
"""
spectrogram = librosa.feature.melspectrogram(
audio_data,
sr=s_r,
n_mels=n_mel_val,
hop_length=HOG_LENGTH,
n_fft=n_mel_val * 20,
fmin=20,
fmax=s_r // 2,
)
return librosa.power_to_db(spectrogram).astype(np.float32)
# sizes (in sec)
MOVING_WINDOW_SIZE = 5
AUDIO_STRIDE_SIZE = 4
SAMPLE_RATE = 16000
from tqdm import tqdm
# !mkdir breathcycles (dont really get this but sure)
X_mel_spec = []
X_mel_spec2 = []
Y_labels = []
durations_spec = []
for indx, df_row in tqdm(processed_dataset_df.iterrows()):
head, audio_file = os.path.split(df_row.file_path)
audio_file_name = audio_file.split(".wav")[0]
diagnosis_type = df_row.diagnosis
breathing_cycles_df = rec_annotations_dict.get(audio_file_name)
audio_data, sample_rate, length = get_audio(df_row.file_path)
no_frames = int((length - MOVING_WINDOW_SIZE) / AUDIO_STRIDE_SIZE) + 1
window_size = MOVING_WINDOW_SIZE * sample_rate
stride = AUDIO_STRIDE_SIZE * sample_rate
audio_frames = []
for index in range(no_frames):
if (stride * index + window_size) < len(audio_data):
frame = audio_data[stride * index : (stride * index + window_size)]
x_mel1 = audio_to_melspectrogram(frame, s_r=sample_rate, n_mel_val=128)
x_mel2 = audio_to_melspectrogram2(frame, s_r=sample_rate, n_mel_val=64)
X_mel_spec.append(
x_mel1.transpose(),
)
X_mel_spec2.append(x_mel2.transpose())
Y_labels.append(str(diagnosis_type))
else:
break
# has output,, shows [time, time/iteration]
X_mel1 = np.array(X_mel_spec)
X_mel2 = np.array(X_mel_spec2)
# X = normalize_dataset(X)
X_array_mel1 = np.array(X_mel1)
X_array_mel2 = np.array(X_mel2)
val_spae = []
for x in X_mel1:
# print(x.shape[0])-------
val_spae.append(x.shape[0])
def Average(lst):
return sum(lst) / len(lst)
# Driver Code
average = Average(val_spae)
# to print average of the list
print("Average of the required list =", round(average, 2))
import seaborn as sns
sns.distplot(val_spae)
# works till here
max_audio_lenght = max([len(i) for i in X_array_mel1])
max_audio_lenght2 = max([len(i) for i in X_array_mel2])
from tensorflow.keras.preprocessing.sequence import pad_sequences
# raw data processing-padding
X_array_mel1 = pad_sequences(X_array_mel1, padding="post", dtype="float32")
X_array_mel2 = pad_sequences(X_array_mel2, padding="post", dtype="float32")
X_array_mel1[0].shape
# Visualize an melspectogram example
plt.figure(figsize=(15, 10))
plt.title("Visualization of audio file", weight="bold")
plt.imshow(X_array_mel1[20])
X_array_mel1.shape[0]
# am i supposed to get a muchhh lower value?
shape_lenght = X_array_mel1.shape[0]
new_Xtrain = np.empty((shape_lenght, 2), dtype=np.object)
for idx in range(shape_lenght):
new_Xtrain[idx,] = [X_array_mel1[idx], X_array_mel2[idx]]
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.layers import (
LSTM,
Dense,
TimeDistributed,
LayerNormalization,
Masking,
)
from tensorflow.keras.models import Model
from tensorflow.keras.regularizers import l2
#!pip install kapre
# import kapre
# from kapre.composed import get_melspectrogram_layer
import tensorflow as tf
import os
import tensorflow_addons as tfa
from tqdm import tqdm
from tqdm.keras import TqdmCallback
from tensorflow.keras.utils import to_categorical
le = LabelEncoder()
diagnosis_classes = list(set(Y_labels))
le.fit(diagnosis_classes)
y = le.transform(Y_labels)
Y = to_categorical(y, num_classes=len(diagnosis_classes))
np.save("diagnosis_classes.npy", le.classes_)
audio_time_span = new_Xtrain[0][0].shape[0]
no_mel_sp_features = new_Xtrain[0][0].shape[1]
NO_CLASSES = 6 # Binary classification -> 2
| false | 1 | 5,840 | 0 | 6,939 | 5,840 |
||
69476544
|
# ## Part 3: Modelling & Predicting Pneumonia w/ Neural Networks
# Imports
import os
import cv2
import glob
import time
import pydicom
import skimage
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib.patches as patches
from skimage import feature, filters
from functools import partial
from collections import defaultdict
from joblib import Parallel, delayed
from lightgbm import LGBMClassifier
from tqdm import tqdm
# Tensorflow / Keras
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras.layers import *
from tensorflow.keras import Model
from tensorflow.keras.applications.vgg16 import VGG16
from keras import models
from keras import layers
# sklearn
from sklearn.model_selection import KFold
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import RandomizedSearchCV
sns.set_style("whitegrid")
np.warnings.filterwarnings("ignore")
# List our paths
trainImagesPath = "../input/rsna-pneumonia-detection-challenge/stage_2_train_images"
testImagesPath = "../input/rsna-pneumonia-detection-challenge/stage_2_test_images"
labelsPath = "../input/rsna-pneumonia-detection-challenge/stage_2_train_labels.csv"
classInfoPath = (
"../input/rsna-pneumonia-detection-challenge/stage_2_detailed_class_info.csv"
)
# Read the labels and classinfo
labels = pd.read_csv(labelsPath)
details = pd.read_csv(classInfoPath)
# ## Part 3.1: Attaining our Training & Testing Data in Proper Format
"""
@Description: Reads an array of dicom image paths, and returns an array of the images after they have been read
@Inputs: An array of filepaths for the images
@Output: Returns an array of the images after they have been read
"""
def readDicomData(data):
res = []
for filePath in tqdm(data): # Loop over data
# We use stop_before_pixels to avoid reading the image (Saves on speed/memory)
f = pydicom.read_file(filePath, stop_before_pixels=True)
res.append(f)
return res
# Get an array of the test & training file paths
trainFilepaths = glob.glob(f"{trainImagesPath}/*.dcm")
testFilepaths = glob.glob(f"{testImagesPath}/*.dcm")
# Read data into an array
trainImages = readDicomData(trainFilepaths[:5000])
testImages = readDicomData(testFilepaths)
# ## Part 3.2: Balancing our Data
# We balance our data as CNNs work best on evenly balanced data
COUNT_NORMAL = len(
labels.loc[labels["Target"] == 0]
) # Number of patients with no pneumonia
COUNT_PNE = len(labels.loc[labels["Target"] == 1]) # Number of patients with pneumonia
TRAIN_IMG_COUNT = len(trainFilepaths) # Total patients
# We calculate the weight of each
weight_for_0 = (1 / COUNT_NORMAL) * (TRAIN_IMG_COUNT) / 2.0
weight_for_1 = (1 / COUNT_PNE) * (TRAIN_IMG_COUNT) / 2.0
classWeight = {0: weight_for_0, 1: weight_for_1}
print(f"Weights: {classWeight}")
# ## Part 3.3: Get Train_Y & Test_Y
"""
@Description: This function parses the medical images meta-data contained
@Inputs: Takes in the dicom image after it has been read
@Output: Returns the unpacked data and the group elements keywords
"""
def parseMetadata(dcm):
unpackedData = {}
groupElemToKeywords = {}
for (
d
) in (
dcm
): # Iterate here to force conversion from lazy RawDataElement to DataElement
pass
# Un-pack Data
for tag, elem in dcm.items():
tagGroup = tag.group
tagElem = tag.elem
keyword = elem.keyword
groupElemToKeywords[(tagGroup, tagElem)] = keyword
value = elem.value
unpackedData[keyword] = value
return unpackedData, groupElemToKeywords
# These parse the metadata into dictionaries
trainMetaDicts, trainKeyword = zip(*[parseMetadata(x) for x in tqdm(trainImages)])
testMetaDicts, testKeyword = zip(*[parseMetadata(x) for x in tqdm(testImages)])
"""
@Description: This function goes through the dicom image information and returns 1 or 0
depending on whether the image contains Pneumonia or not
@Inputs: A dataframe containing the metadata
@Output: Returns the Y result (i.e: our train and test y)
"""
def createY(df):
y = df["SeriesDescription"] == "view: PA"
Y = np.zeros(len(y)) # Initialise Y
for i in range(len(y)):
if y[i] == True:
Y[i] = 1
return Y
train_df = pd.DataFrame.from_dict(data=trainMetaDicts)
test_df = pd.DataFrame.from_dict(data=testMetaDicts)
train_df["dataset"] = "train"
test_df["dataset"] = "test"
df = train_df
df2 = test_df
train_Y = createY(df) # Create training Y
test_Y = createY(df2) # Create testing Y
# ## Part 3.4: Get Train_X & Test_X
"""
@Description: This decodes an image by reading the pixel array, resizing it into the correct format and
normalising the pixels
@Inputs:
- filePath: This is the filepath of the image that we want to decode
@Output:
- img: This is the image after it has been decoded
"""
def decodeImage(filePath):
image = pydicom.read_file(filePath).pixel_array
image = cv2.resize(image, (128, 128))
return image / 255
# Get our train x in the correct shape
train_X = []
for filePath in tqdm(trainFilepaths[:5000]):
img = decodeImage(filePath)
train_X.append(img)
train_X = np.array(train_X) # Convert to np.array
train_X_rgb = np.repeat(train_X[..., np.newaxis], 3, -1) # Reshape into rgb format
# Get our test x in the correct shape for NN
test_X = []
for filePath in tqdm(testFilepaths):
img_test = decodeImage(filePath) # Decode & Resize
test_X.append(img_test)
test_X = np.array(test_X) # Convert to np array
test_X_rgb = np.repeat(test_X[..., np.newaxis], 3, -1) # Reshape into rgb format
"""
@Description: This function plots our metrics for our models across epochs
@Inputs: The history of the fitted model
@Output: N/A
"""
def plottingScores(hist):
fig, ax = plt.subplots(1, 5, figsize=(20, 3))
ax = ax.ravel()
for i, met in enumerate(["accuracy", "precision", "recall", "AUC", "loss"]):
ax[i].plot(hist.history[met])
ax[i].plot(hist.history["val_" + met])
ax[i].set_title("Model {}".format(met))
ax[i].set_xlabel("epochs")
ax[i].set_ylabel(met)
ax[i].legend(["train", "val"])
# ## Part 3.5: Metrics Evaluation
# For our metrics, we want to include precision and recall as they will provide use with more info on how good our model is
#
#
# - Accuracy: This tells us what fraction of the labels are correct.
# - Since our data is not balanced, accuracy might give a skewed sense of a good model
# - Precision: This tells us the number of true positives (TP) over the sum of TP and false positives (FP).
# - It shows what fraction of labeled positives are actually correct.
# - Recall: The number of TP over the sum of TP and false negatves (FN).
# - It shows what fraction of actual positives are correct.
# These our our scoring metrics that are going to be used to evaluate our models
METRICS = [
"accuracy",
tf.keras.metrics.Precision(name="precision"),
tf.keras.metrics.Recall(name="recall"),
tf.keras.metrics.AUC(name="AUC"),
]
# ### Tuning our Models with Callbacks
# - We'll use Keras callbacks to further finetune our model.
# - The checkpoint callback saves the best weights of the model, so next time we want to use the model, we do not have to spend time training it.
# - The early stopping callback stops the training process when the model starts becoming stagnant, or even worse, when the model starts overfitting.
# - Since we set restore_best_weights to True, the returned model at the end of the training process will be the model with the best weights (i.e. low loss and high accuracy).
# Define our callback functions to pass when fitting our NNs
def exponential_decay(lr0, s):
def exponential_decay_fn(epoch):
return lr0 * 0.1 ** (epoch / s)
return exponential_decay_fn
exponential_decay_fn = exponential_decay(0.01, 20)
lr_scheduler = tf.keras.callbacks.LearningRateScheduler(exponential_decay_fn)
checkpoint_cb = tf.keras.callbacks.ModelCheckpoint("xray_model.h5", save_best_only=True)
early_stopping_cb = tf.keras.callbacks.EarlyStopping(
patience=5, restore_best_weights=True
)
# ## Part 3.6: Building Model #1 - Fully Connected Model
"""
@Description: This function builds our simple Fully-connected NN
@Inputs: N/A
@Output: Returns the FCNN Model
"""
def build_fcnn_model():
# Basic model with a flattening layer followng by 2 dense layers
# The first dense layer is using relu and the 2nd one is using sigmoid
model = tf.keras.models.Sequential(
[
tf.keras.layers.Flatten(input_shape=(128, 128, 3)),
tf.keras.layers.Dense(128, activation="relu"),
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
return model
# Build our FCNN model and compile
model_fcnn = build_fcnn_model()
model_fcnn.summary()
model_fcnn.compile(
optimizer="adam", loss="binary_crossentropy", metrics=METRICS
) # Compile
# ### Fitting Model to Training Data
history_fcnn = model_fcnn.fit(
train_X_rgb,
train_Y,
epochs=30,
batch_size=128,
validation_split=0.2,
class_weight=classWeight,
verbose=1,
callbacks=[checkpoint_cb, early_stopping_cb, lr_scheduler],
) # Fit the model
# Evaluate and display results
results = model_fcnn.evaluate(test_X_rgb, test_Y) # Evaluate the model on test data
results = dict(zip(model_fcnn.metrics_names, results))
print(results)
plottingScores(history_fcnn) # Visualise scores
# ## Part 3.7: Building Model #2 - CNN
# In our CNN model, fewer parameters are needed because every convolutional layer reduces the dimensions of the input through the convolution operation.
"""
@Description: This function builds our custom CNN Model
@Inputs: N/A
@Output: Returns the CNN model
"""
def build_cnn_model():
model = tf.keras.Sequential(
[
tf.keras.layers.Conv2D(
32,
kernel_size=(3, 3),
strides=(1, 1),
padding="valid",
activation="relu",
input_shape=(128, 128, 3),
), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2, 2)), # flatten output of conv
tf.keras.layers.Conv2D(
32,
kernel_size=(3, 3),
strides=(1, 1),
padding="valid",
activation="relu",
), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2, 2)), # flatten output of conv
tf.keras.layers.Dropout(0.3),
tf.keras.layers.Conv2D(64, 3, activation="relu", padding="valid"),
tf.keras.layers.Conv2D(128, 3, activation="relu", padding="valid"),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.MaxPool2D(),
tf.keras.layers.Dropout(0.4),
tf.keras.layers.Flatten(), # flatten output of conv
tf.keras.layers.Dense(512, activation="relu"), # hidden layer
tf.keras.layers.BatchNormalization(),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Dense(128, activation="relu"), # output layer
tf.keras.layers.BatchNormalization(),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
return model
# Build and compile model
model_cnn = build_cnn_model()
model_cnn.summary()
model_cnn.compile(optimizer="adam", loss="binary_crossentropy", metrics=METRICS)
# Fit model
history_cnn = model_cnn.fit(
train_X_rgb,
train_Y,
epochs=30,
validation_split=0.15,
batch_size=128,
class_weight=classWeight,
callbacks=[checkpoint_cb, early_stopping_cb, lr_scheduler],
verbose=1,
) # Fit the model
# Evalute the models results and put into a dict
results = model_cnn.evaluate(test_X_rgb, test_Y)
results = dict(zip(model_cnn.metrics_names, results))
print(results)
plottingScores(history_cnn) # Visualise scores
# ## Part 3.8: Building Model #3 - Mobile Net with Transfer Learning
"""
@Description: This function builds our MobileNet Model
@Inputs: N/A
@Output: Returns the Mobile Net model
"""
def build_mn_model():
model = tf.keras.Sequential(
[
tf.keras.applications.MobileNetV2(
include_top=False, weights="imagenet", input_shape=(128, 128, 3)
),
tf.keras.layers.GlobalAveragePooling2D(),
Dense(1, activation="sigmoid"),
]
)
model.layers[0].trainable = False
return model
# Build and compile mobile net model
model_mn = build_mn_model()
model_mn.summary()
model_mn.compile(optimizer="adam", loss="binary_crossentropy", metrics=METRICS)
# ### We run our best model here on a larger portion of the training data
history_mn = model_mn.fit(
train_X_rgb,
train_Y,
epochs=30,
validation_split=0.20,
class_weight=classWeight,
batch_size=64,
callbacks=[checkpoint_cb, early_stopping_cb, lr_scheduler],
)
# Show results and print graphs
results = model_mn.evaluate(test_X_rgb, test_Y)
results = dict(zip(model_mn.metrics_names, results))
print(results)
plottingScores(history_mn) # Visualise scores
# ### Show Confusion Matrix
from sklearn.metrics import confusion_matrix
y_pred = model_mn.predict_classes(test_X_rgb)
confusion_matrix(test_Y, y_pred)
# ### Function to Perform K-Fold CV
"""
@Description: This function performs K-Fold Cross Validation with a provided Deep Learning Model
@Inputs:
- K: Number of folds
- build_model_func: Function to create model
- epochs: Number of epochs to train data
- batchSize: Batch size when fitting the model
@Output: Dict of metric results from K-fold CV
"""
def performCV(K, build_model_func, epochs, batchSize):
kfold = KFold(n_splits=K, shuffle=True) # Split data into K Folds
res = {
"acc_per_fold": [],
"precision_per_fold": [],
"recall_per_fold": [],
"auc_per_fold": [],
"loss_per_fold": [],
}
fold_no = 1
for train_index, test_index in kfold.split(train_X_rgb):
X_train, X_test = (
train_X_rgb[train_index],
train_X_rgb[test_index],
) # Split data
y_train, y_test = train_Y[train_index], train_Y[test_index]
model = build_model_func() # Build model
mets = [
"accuracy",
tf.keras.metrics.Precision(name="precision"),
tf.keras.metrics.Recall(name="recall"),
tf.keras.metrics.AUC(name="AUC"),
]
model.compile(
optimizer="adam", loss="binary_crossentropy", metrics=mets
) # Compile our model
print(
"------------------------------------------------------------------------"
)
print(f"Training for fold {fold_no} ...")
# Train the model on the current fold
history = model.fit(
X_train,
y_train,
epochs=epochs,
batch_size=batchSize,
class_weight=classWeight,
callbacks=[checkpoint_cb, early_stopping_cb, lr_scheduler],
) # Fit data to model
scores = model.evaluate(X_test, y_test, verbose=0) # Evalute the model
print(f"Scores for fold {fold_no}:")
print(f"{model.metrics_names[0]}: {scores[0]}")
print(f"{model.metrics_names[1]}: {scores[1]*100}%")
print(f"{model.metrics_names[2]}: {scores[2]*100}%")
print(f"{model.metrics_names[3]}: {scores[3]*100}%")
res["loss_per_fold"].append(scores[0])
res["acc_per_fold"].append(scores[1] * 100)
res["precision_per_fold"].append(scores[2] * 100)
res["recall_per_fold"].append(scores[3] * 100)
res["auc_per_fold"].append(scores[4] * 100)
gc.collect()
# Increase fold number
fold_no += 1
return res # return our results dict
# ## Part 3.9: K-Fold Cross Validation with all 3 Networks
# Full-connected NN
resFCNN = performCV(5, build_fcnn_model, 30, 128)
# Convolutional NN
resCNN = performCV(5, build_cnn_model, 30, 64)
# MobileNet
resMB = performCV(5, build_mn_model, 30, 64)
resMB
# ### Results For Architectures
"""
5k Training
3k Testing
Architecture 1:
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu', input_shape=(128, 128, 3)), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Flatten(), # flatten output of conv
tf.keras.layers.Dense(100, activation='relu'), # hidden layer
tf.keras.layers.Dense(1, activation='sigmoid') # output layer
---------------------------------------------------- Performance on Test Data ----------------------------------------------------
{ 'loss': 0.255420297, 'accuracy': 0.904666662, 'precision': 0.881006836, 'recall': 0.951792359, 'AUC': 0.968622922}
Architecture 2:
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu', input_shape=(128, 128, 3)), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu'), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Flatten(), # flatten output of conv
tf.keras.layers.Dense(100, activation='relu'), # hidden layer
tf.keras.layers.Dense(1, activation='sigmoid') # output layer
---------------------------------------------------- Performance on Test Data ----------------------------------------------------
{ 'loss': 0.198399558, 'accuracy': 0.950666666, 'precision': 0.933372616, 'recall': 0.978368341, 'AUC': 0.986180067}
Architecture 3:
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu', input_shape=(128, 128, 3)), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu'), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Conv2D(32, 3, activation='relu', padding='same'),
tf.keras.layers.Conv2D(64, 3, activation='relu', padding='same'),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.MaxPool2D(),
tf.keras.layers.Flatten(), # flatten output of conv
tf.keras.layers.Dense(100, activation='relu'), # hidden layer
tf.keras.layers.Dense(1, activation='sigmoid') # output layer
---------------------------------------------------- Performance on Test Data ----------------------------------------------------
{'loss': 0.1422816216, 'accuracy': 0.976333320, 'precision': 0.984952986, 'recall': 0.970951795, 'AUC': 0.991799652}
Architecture 4:
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu', input_shape=(128, 128, 3)), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu'), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Dropout(0.2),
tf.keras.layers.Conv2D(32, 3, activation='relu', padding='valid'),
tf.keras.layers.Conv2D(64, 3, activation='relu', padding='valid'),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.MaxPool2D(),
tf.keras.layers.Dropout(0.3),
tf.keras.layers.Flatten(), # flatten output of conv
tf.keras.layers.Dense(100, activation='relu'), # hidden layer
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Dense(1, activation='sigmoid') # output layer
---------------------------------------------------- Performance on Test Data ----------------------------------------------------
{ 'loss': 0.1239979043, 'accuracy': 0.982666671, 'precision': 0.985130131, 'recall': 0.982694685, 'AUC': 0.992944598}
Architecture 5:
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu', input_shape=(128, 128, 3)), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu'), # convolutional layer
tf.keras.layers.BatchNormalization(),
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Dropout(0.2),
tf.keras.layers.Conv2D(32, 3, activation='relu', padding='valid'),
tf.keras.layers.Conv2D(64, 3, activation='relu', padding='valid'),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.MaxPool2D(),
tf.keras.layers.Dropout(0.3),
tf.keras.layers.Flatten(), # Flattening
# Full Connection
tf.keras.layers.Dense(64, activation='relu'), # hidden layer
tf.keras.layers.Dropout(0.5), # Dropout
tf.keras.layers.Dense(1, activation='sigmoid') # output layer
---------------------------------------------------- Performance on Test Data ----------------------------------------------------
{ 'loss': 0.12123415754, 'accuracy': 0.984615671, 'precision': 0.987261581, 'recall': 0.985671211, 'AUC': 0.994511598}
"""
# ### Manual Hyper-parameter Tuning results for FCNN
"""
Manual Hyper-parameter Tuning
batch_size=32
cWeight: None {'loss': 0.22600425779819489, 'accuracy': 0.92166668176651, 'precision': 0.9292364716529846, 'recall': 0.9252163171768188, 'AUC': 0.9672043323516846}
cWeight: Balanced {'loss': 0.2335905283689499, 'accuracy': 0.9136666655540466, 'precision': 0.9155963063240051, 'recall': 0.9252163171768188, 'AUC': 0.9655577540397644}
batch_size=64
cWeight: None {'loss': 0.22068753838539124, 'accuracy': 0.9193333387374878, 'precision': 0.9149577617645264, 'recall': 0.9375772476196289, 'AUC': 0.9699712991714478}
cWeight: Balanced {'loss': 0.2424456775188446, 'accuracy': 0.9079999923706055, 'precision': 0.8829908967018127, 'recall': 0.956118643283844, 'AUC': 0.9677296280860901}
batch_size=128
cWeight: None {'loss': 0.23750829696655273, 'accuracy': 0.9100000262260437, 'precision': 0.8855835199356079, 'recall': 0.95673668384552, 'AUC': 0.9694961309432983}
cWeight: Balanced {'loss': 0.2239508330821991, 'accuracy': 0.9196666479110718, 'precision': 0.9100655317306519, 'recall': 0.94437575340271, 'AUC': 0.9697944521903992}
batch_size=256
cWeight: None {'loss': 0.2305724024772644, 'accuracy': 0.9190000295639038, 'precision': 0.9109384417533875, 'recall': 0.9419035911560059, 'AUC': 0.9681356549263}
cWeight: Balanced {'loss': 0.22952377796173096, 'accuracy': 0.9203333258628845, 'precision': 0.9171203970909119, 'recall': 0.9369592070579529, 'AUC': 0.9694135785102844}
"""
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/476/69476544.ipynb
| null | null |
[{"Id": 69476544, "ScriptId": 18793223, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7736694, "CreationDate": "07/31/2021 13:29:41", "VersionNumber": 9.0, "Title": "P3: Modelling Pneumonia - Neural Networks", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 616.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 615.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# ## Part 3: Modelling & Predicting Pneumonia w/ Neural Networks
# Imports
import os
import cv2
import glob
import time
import pydicom
import skimage
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib.patches as patches
from skimage import feature, filters
from functools import partial
from collections import defaultdict
from joblib import Parallel, delayed
from lightgbm import LGBMClassifier
from tqdm import tqdm
# Tensorflow / Keras
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras.layers import *
from tensorflow.keras import Model
from tensorflow.keras.applications.vgg16 import VGG16
from keras import models
from keras import layers
# sklearn
from sklearn.model_selection import KFold
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import RandomizedSearchCV
sns.set_style("whitegrid")
np.warnings.filterwarnings("ignore")
# List our paths
trainImagesPath = "../input/rsna-pneumonia-detection-challenge/stage_2_train_images"
testImagesPath = "../input/rsna-pneumonia-detection-challenge/stage_2_test_images"
labelsPath = "../input/rsna-pneumonia-detection-challenge/stage_2_train_labels.csv"
classInfoPath = (
"../input/rsna-pneumonia-detection-challenge/stage_2_detailed_class_info.csv"
)
# Read the labels and classinfo
labels = pd.read_csv(labelsPath)
details = pd.read_csv(classInfoPath)
# ## Part 3.1: Attaining our Training & Testing Data in Proper Format
"""
@Description: Reads an array of dicom image paths, and returns an array of the images after they have been read
@Inputs: An array of filepaths for the images
@Output: Returns an array of the images after they have been read
"""
def readDicomData(data):
res = []
for filePath in tqdm(data): # Loop over data
# We use stop_before_pixels to avoid reading the image (Saves on speed/memory)
f = pydicom.read_file(filePath, stop_before_pixels=True)
res.append(f)
return res
# Get an array of the test & training file paths
trainFilepaths = glob.glob(f"{trainImagesPath}/*.dcm")
testFilepaths = glob.glob(f"{testImagesPath}/*.dcm")
# Read data into an array
trainImages = readDicomData(trainFilepaths[:5000])
testImages = readDicomData(testFilepaths)
# ## Part 3.2: Balancing our Data
# We balance our data as CNNs work best on evenly balanced data
COUNT_NORMAL = len(
labels.loc[labels["Target"] == 0]
) # Number of patients with no pneumonia
COUNT_PNE = len(labels.loc[labels["Target"] == 1]) # Number of patients with pneumonia
TRAIN_IMG_COUNT = len(trainFilepaths) # Total patients
# We calculate the weight of each
weight_for_0 = (1 / COUNT_NORMAL) * (TRAIN_IMG_COUNT) / 2.0
weight_for_1 = (1 / COUNT_PNE) * (TRAIN_IMG_COUNT) / 2.0
classWeight = {0: weight_for_0, 1: weight_for_1}
print(f"Weights: {classWeight}")
# ## Part 3.3: Get Train_Y & Test_Y
"""
@Description: This function parses the medical images meta-data contained
@Inputs: Takes in the dicom image after it has been read
@Output: Returns the unpacked data and the group elements keywords
"""
def parseMetadata(dcm):
unpackedData = {}
groupElemToKeywords = {}
for (
d
) in (
dcm
): # Iterate here to force conversion from lazy RawDataElement to DataElement
pass
# Un-pack Data
for tag, elem in dcm.items():
tagGroup = tag.group
tagElem = tag.elem
keyword = elem.keyword
groupElemToKeywords[(tagGroup, tagElem)] = keyword
value = elem.value
unpackedData[keyword] = value
return unpackedData, groupElemToKeywords
# These parse the metadata into dictionaries
trainMetaDicts, trainKeyword = zip(*[parseMetadata(x) for x in tqdm(trainImages)])
testMetaDicts, testKeyword = zip(*[parseMetadata(x) for x in tqdm(testImages)])
"""
@Description: This function goes through the dicom image information and returns 1 or 0
depending on whether the image contains Pneumonia or not
@Inputs: A dataframe containing the metadata
@Output: Returns the Y result (i.e: our train and test y)
"""
def createY(df):
y = df["SeriesDescription"] == "view: PA"
Y = np.zeros(len(y)) # Initialise Y
for i in range(len(y)):
if y[i] == True:
Y[i] = 1
return Y
train_df = pd.DataFrame.from_dict(data=trainMetaDicts)
test_df = pd.DataFrame.from_dict(data=testMetaDicts)
train_df["dataset"] = "train"
test_df["dataset"] = "test"
df = train_df
df2 = test_df
train_Y = createY(df) # Create training Y
test_Y = createY(df2) # Create testing Y
# ## Part 3.4: Get Train_X & Test_X
"""
@Description: This decodes an image by reading the pixel array, resizing it into the correct format and
normalising the pixels
@Inputs:
- filePath: This is the filepath of the image that we want to decode
@Output:
- img: This is the image after it has been decoded
"""
def decodeImage(filePath):
image = pydicom.read_file(filePath).pixel_array
image = cv2.resize(image, (128, 128))
return image / 255
# Get our train x in the correct shape
train_X = []
for filePath in tqdm(trainFilepaths[:5000]):
img = decodeImage(filePath)
train_X.append(img)
train_X = np.array(train_X) # Convert to np.array
train_X_rgb = np.repeat(train_X[..., np.newaxis], 3, -1) # Reshape into rgb format
# Get our test x in the correct shape for NN
test_X = []
for filePath in tqdm(testFilepaths):
img_test = decodeImage(filePath) # Decode & Resize
test_X.append(img_test)
test_X = np.array(test_X) # Convert to np array
test_X_rgb = np.repeat(test_X[..., np.newaxis], 3, -1) # Reshape into rgb format
"""
@Description: This function plots our metrics for our models across epochs
@Inputs: The history of the fitted model
@Output: N/A
"""
def plottingScores(hist):
fig, ax = plt.subplots(1, 5, figsize=(20, 3))
ax = ax.ravel()
for i, met in enumerate(["accuracy", "precision", "recall", "AUC", "loss"]):
ax[i].plot(hist.history[met])
ax[i].plot(hist.history["val_" + met])
ax[i].set_title("Model {}".format(met))
ax[i].set_xlabel("epochs")
ax[i].set_ylabel(met)
ax[i].legend(["train", "val"])
# ## Part 3.5: Metrics Evaluation
# For our metrics, we want to include precision and recall as they will provide use with more info on how good our model is
#
#
# - Accuracy: This tells us what fraction of the labels are correct.
# - Since our data is not balanced, accuracy might give a skewed sense of a good model
# - Precision: This tells us the number of true positives (TP) over the sum of TP and false positives (FP).
# - It shows what fraction of labeled positives are actually correct.
# - Recall: The number of TP over the sum of TP and false negatves (FN).
# - It shows what fraction of actual positives are correct.
# These our our scoring metrics that are going to be used to evaluate our models
METRICS = [
"accuracy",
tf.keras.metrics.Precision(name="precision"),
tf.keras.metrics.Recall(name="recall"),
tf.keras.metrics.AUC(name="AUC"),
]
# ### Tuning our Models with Callbacks
# - We'll use Keras callbacks to further finetune our model.
# - The checkpoint callback saves the best weights of the model, so next time we want to use the model, we do not have to spend time training it.
# - The early stopping callback stops the training process when the model starts becoming stagnant, or even worse, when the model starts overfitting.
# - Since we set restore_best_weights to True, the returned model at the end of the training process will be the model with the best weights (i.e. low loss and high accuracy).
# Define our callback functions to pass when fitting our NNs
def exponential_decay(lr0, s):
def exponential_decay_fn(epoch):
return lr0 * 0.1 ** (epoch / s)
return exponential_decay_fn
exponential_decay_fn = exponential_decay(0.01, 20)
lr_scheduler = tf.keras.callbacks.LearningRateScheduler(exponential_decay_fn)
checkpoint_cb = tf.keras.callbacks.ModelCheckpoint("xray_model.h5", save_best_only=True)
early_stopping_cb = tf.keras.callbacks.EarlyStopping(
patience=5, restore_best_weights=True
)
# ## Part 3.6: Building Model #1 - Fully Connected Model
"""
@Description: This function builds our simple Fully-connected NN
@Inputs: N/A
@Output: Returns the FCNN Model
"""
def build_fcnn_model():
# Basic model with a flattening layer followng by 2 dense layers
# The first dense layer is using relu and the 2nd one is using sigmoid
model = tf.keras.models.Sequential(
[
tf.keras.layers.Flatten(input_shape=(128, 128, 3)),
tf.keras.layers.Dense(128, activation="relu"),
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
return model
# Build our FCNN model and compile
model_fcnn = build_fcnn_model()
model_fcnn.summary()
model_fcnn.compile(
optimizer="adam", loss="binary_crossentropy", metrics=METRICS
) # Compile
# ### Fitting Model to Training Data
history_fcnn = model_fcnn.fit(
train_X_rgb,
train_Y,
epochs=30,
batch_size=128,
validation_split=0.2,
class_weight=classWeight,
verbose=1,
callbacks=[checkpoint_cb, early_stopping_cb, lr_scheduler],
) # Fit the model
# Evaluate and display results
results = model_fcnn.evaluate(test_X_rgb, test_Y) # Evaluate the model on test data
results = dict(zip(model_fcnn.metrics_names, results))
print(results)
plottingScores(history_fcnn) # Visualise scores
# ## Part 3.7: Building Model #2 - CNN
# In our CNN model, fewer parameters are needed because every convolutional layer reduces the dimensions of the input through the convolution operation.
"""
@Description: This function builds our custom CNN Model
@Inputs: N/A
@Output: Returns the CNN model
"""
def build_cnn_model():
model = tf.keras.Sequential(
[
tf.keras.layers.Conv2D(
32,
kernel_size=(3, 3),
strides=(1, 1),
padding="valid",
activation="relu",
input_shape=(128, 128, 3),
), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2, 2)), # flatten output of conv
tf.keras.layers.Conv2D(
32,
kernel_size=(3, 3),
strides=(1, 1),
padding="valid",
activation="relu",
), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2, 2)), # flatten output of conv
tf.keras.layers.Dropout(0.3),
tf.keras.layers.Conv2D(64, 3, activation="relu", padding="valid"),
tf.keras.layers.Conv2D(128, 3, activation="relu", padding="valid"),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.MaxPool2D(),
tf.keras.layers.Dropout(0.4),
tf.keras.layers.Flatten(), # flatten output of conv
tf.keras.layers.Dense(512, activation="relu"), # hidden layer
tf.keras.layers.BatchNormalization(),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Dense(128, activation="relu"), # output layer
tf.keras.layers.BatchNormalization(),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
return model
# Build and compile model
model_cnn = build_cnn_model()
model_cnn.summary()
model_cnn.compile(optimizer="adam", loss="binary_crossentropy", metrics=METRICS)
# Fit model
history_cnn = model_cnn.fit(
train_X_rgb,
train_Y,
epochs=30,
validation_split=0.15,
batch_size=128,
class_weight=classWeight,
callbacks=[checkpoint_cb, early_stopping_cb, lr_scheduler],
verbose=1,
) # Fit the model
# Evalute the models results and put into a dict
results = model_cnn.evaluate(test_X_rgb, test_Y)
results = dict(zip(model_cnn.metrics_names, results))
print(results)
plottingScores(history_cnn) # Visualise scores
# ## Part 3.8: Building Model #3 - Mobile Net with Transfer Learning
"""
@Description: This function builds our MobileNet Model
@Inputs: N/A
@Output: Returns the Mobile Net model
"""
def build_mn_model():
model = tf.keras.Sequential(
[
tf.keras.applications.MobileNetV2(
include_top=False, weights="imagenet", input_shape=(128, 128, 3)
),
tf.keras.layers.GlobalAveragePooling2D(),
Dense(1, activation="sigmoid"),
]
)
model.layers[0].trainable = False
return model
# Build and compile mobile net model
model_mn = build_mn_model()
model_mn.summary()
model_mn.compile(optimizer="adam", loss="binary_crossentropy", metrics=METRICS)
# ### We run our best model here on a larger portion of the training data
history_mn = model_mn.fit(
train_X_rgb,
train_Y,
epochs=30,
validation_split=0.20,
class_weight=classWeight,
batch_size=64,
callbacks=[checkpoint_cb, early_stopping_cb, lr_scheduler],
)
# Show results and print graphs
results = model_mn.evaluate(test_X_rgb, test_Y)
results = dict(zip(model_mn.metrics_names, results))
print(results)
plottingScores(history_mn) # Visualise scores
# ### Show Confusion Matrix
from sklearn.metrics import confusion_matrix
y_pred = model_mn.predict_classes(test_X_rgb)
confusion_matrix(test_Y, y_pred)
# ### Function to Perform K-Fold CV
"""
@Description: This function performs K-Fold Cross Validation with a provided Deep Learning Model
@Inputs:
- K: Number of folds
- build_model_func: Function to create model
- epochs: Number of epochs to train data
- batchSize: Batch size when fitting the model
@Output: Dict of metric results from K-fold CV
"""
def performCV(K, build_model_func, epochs, batchSize):
kfold = KFold(n_splits=K, shuffle=True) # Split data into K Folds
res = {
"acc_per_fold": [],
"precision_per_fold": [],
"recall_per_fold": [],
"auc_per_fold": [],
"loss_per_fold": [],
}
fold_no = 1
for train_index, test_index in kfold.split(train_X_rgb):
X_train, X_test = (
train_X_rgb[train_index],
train_X_rgb[test_index],
) # Split data
y_train, y_test = train_Y[train_index], train_Y[test_index]
model = build_model_func() # Build model
mets = [
"accuracy",
tf.keras.metrics.Precision(name="precision"),
tf.keras.metrics.Recall(name="recall"),
tf.keras.metrics.AUC(name="AUC"),
]
model.compile(
optimizer="adam", loss="binary_crossentropy", metrics=mets
) # Compile our model
print(
"------------------------------------------------------------------------"
)
print(f"Training for fold {fold_no} ...")
# Train the model on the current fold
history = model.fit(
X_train,
y_train,
epochs=epochs,
batch_size=batchSize,
class_weight=classWeight,
callbacks=[checkpoint_cb, early_stopping_cb, lr_scheduler],
) # Fit data to model
scores = model.evaluate(X_test, y_test, verbose=0) # Evalute the model
print(f"Scores for fold {fold_no}:")
print(f"{model.metrics_names[0]}: {scores[0]}")
print(f"{model.metrics_names[1]}: {scores[1]*100}%")
print(f"{model.metrics_names[2]}: {scores[2]*100}%")
print(f"{model.metrics_names[3]}: {scores[3]*100}%")
res["loss_per_fold"].append(scores[0])
res["acc_per_fold"].append(scores[1] * 100)
res["precision_per_fold"].append(scores[2] * 100)
res["recall_per_fold"].append(scores[3] * 100)
res["auc_per_fold"].append(scores[4] * 100)
gc.collect()
# Increase fold number
fold_no += 1
return res # return our results dict
# ## Part 3.9: K-Fold Cross Validation with all 3 Networks
# Full-connected NN
resFCNN = performCV(5, build_fcnn_model, 30, 128)
# Convolutional NN
resCNN = performCV(5, build_cnn_model, 30, 64)
# MobileNet
resMB = performCV(5, build_mn_model, 30, 64)
resMB
# ### Results For Architectures
"""
5k Training
3k Testing
Architecture 1:
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu', input_shape=(128, 128, 3)), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Flatten(), # flatten output of conv
tf.keras.layers.Dense(100, activation='relu'), # hidden layer
tf.keras.layers.Dense(1, activation='sigmoid') # output layer
---------------------------------------------------- Performance on Test Data ----------------------------------------------------
{ 'loss': 0.255420297, 'accuracy': 0.904666662, 'precision': 0.881006836, 'recall': 0.951792359, 'AUC': 0.968622922}
Architecture 2:
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu', input_shape=(128, 128, 3)), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu'), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Flatten(), # flatten output of conv
tf.keras.layers.Dense(100, activation='relu'), # hidden layer
tf.keras.layers.Dense(1, activation='sigmoid') # output layer
---------------------------------------------------- Performance on Test Data ----------------------------------------------------
{ 'loss': 0.198399558, 'accuracy': 0.950666666, 'precision': 0.933372616, 'recall': 0.978368341, 'AUC': 0.986180067}
Architecture 3:
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu', input_shape=(128, 128, 3)), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu'), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Conv2D(32, 3, activation='relu', padding='same'),
tf.keras.layers.Conv2D(64, 3, activation='relu', padding='same'),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.MaxPool2D(),
tf.keras.layers.Flatten(), # flatten output of conv
tf.keras.layers.Dense(100, activation='relu'), # hidden layer
tf.keras.layers.Dense(1, activation='sigmoid') # output layer
---------------------------------------------------- Performance on Test Data ----------------------------------------------------
{'loss': 0.1422816216, 'accuracy': 0.976333320, 'precision': 0.984952986, 'recall': 0.970951795, 'AUC': 0.991799652}
Architecture 4:
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu', input_shape=(128, 128, 3)), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu'), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Dropout(0.2),
tf.keras.layers.Conv2D(32, 3, activation='relu', padding='valid'),
tf.keras.layers.Conv2D(64, 3, activation='relu', padding='valid'),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.MaxPool2D(),
tf.keras.layers.Dropout(0.3),
tf.keras.layers.Flatten(), # flatten output of conv
tf.keras.layers.Dense(100, activation='relu'), # hidden layer
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Dense(1, activation='sigmoid') # output layer
---------------------------------------------------- Performance on Test Data ----------------------------------------------------
{ 'loss': 0.1239979043, 'accuracy': 0.982666671, 'precision': 0.985130131, 'recall': 0.982694685, 'AUC': 0.992944598}
Architecture 5:
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu', input_shape=(128, 128, 3)), # convolutional layer
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Conv2D(25, kernel_size=(3,3), strides=(1,1), padding='valid', activation='relu'), # convolutional layer
tf.keras.layers.BatchNormalization(),
tf.keras.layers.MaxPool2D(pool_size=(2,2)), # flatten output of conv
tf.keras.layers.Dropout(0.2),
tf.keras.layers.Conv2D(32, 3, activation='relu', padding='valid'),
tf.keras.layers.Conv2D(64, 3, activation='relu', padding='valid'),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.MaxPool2D(),
tf.keras.layers.Dropout(0.3),
tf.keras.layers.Flatten(), # Flattening
# Full Connection
tf.keras.layers.Dense(64, activation='relu'), # hidden layer
tf.keras.layers.Dropout(0.5), # Dropout
tf.keras.layers.Dense(1, activation='sigmoid') # output layer
---------------------------------------------------- Performance on Test Data ----------------------------------------------------
{ 'loss': 0.12123415754, 'accuracy': 0.984615671, 'precision': 0.987261581, 'recall': 0.985671211, 'AUC': 0.994511598}
"""
# ### Manual Hyper-parameter Tuning results for FCNN
"""
Manual Hyper-parameter Tuning
batch_size=32
cWeight: None {'loss': 0.22600425779819489, 'accuracy': 0.92166668176651, 'precision': 0.9292364716529846, 'recall': 0.9252163171768188, 'AUC': 0.9672043323516846}
cWeight: Balanced {'loss': 0.2335905283689499, 'accuracy': 0.9136666655540466, 'precision': 0.9155963063240051, 'recall': 0.9252163171768188, 'AUC': 0.9655577540397644}
batch_size=64
cWeight: None {'loss': 0.22068753838539124, 'accuracy': 0.9193333387374878, 'precision': 0.9149577617645264, 'recall': 0.9375772476196289, 'AUC': 0.9699712991714478}
cWeight: Balanced {'loss': 0.2424456775188446, 'accuracy': 0.9079999923706055, 'precision': 0.8829908967018127, 'recall': 0.956118643283844, 'AUC': 0.9677296280860901}
batch_size=128
cWeight: None {'loss': 0.23750829696655273, 'accuracy': 0.9100000262260437, 'precision': 0.8855835199356079, 'recall': 0.95673668384552, 'AUC': 0.9694961309432983}
cWeight: Balanced {'loss': 0.2239508330821991, 'accuracy': 0.9196666479110718, 'precision': 0.9100655317306519, 'recall': 0.94437575340271, 'AUC': 0.9697944521903992}
batch_size=256
cWeight: None {'loss': 0.2305724024772644, 'accuracy': 0.9190000295639038, 'precision': 0.9109384417533875, 'recall': 0.9419035911560059, 'AUC': 0.9681356549263}
cWeight: Balanced {'loss': 0.22952377796173096, 'accuracy': 0.9203333258628845, 'precision': 0.9171203970909119, 'recall': 0.9369592070579529, 'AUC': 0.9694135785102844}
"""
| false | 0 | 7,506 | 0 | 7,506 | 7,506 |
||
69476005
|
# ## Part 0: Pneumonia Exploratory Data Analysis
# ### Format of Data
# - patientId_- A patientId. Each patientId corresponds to a unique image.
# - x_ - the upper-left x coordinate of the bounding box.
# - y_ - the upper-left y coordinate of the bounding box.
# - width_ - the width of the bounding box.
# - height_ - the height of the bounding box.
# - Target_ - the binary Target, indicating whether this sample has evidence of pneumonia. (1 = true, 0 = false)
# ### Notes
# - A pneumonia opacity is a part of the lungs that looks darker on a radiograph and has a shape that indicates that pneumonia is (or may be) present.
# - Since the goal is to detect & then draw a bounding box on each pneumonia opacities (where each image can have 0 or many), and the training set is already classified:
# - It can be analysed as a supervised learning statistical multilabel classification
#
# Imports
import cv2
import tqdm
import pydicom
import pylab as pl
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pylab as plt
import skimage
from skimage import feature, filters
import os
from os import listdir
from os.path import isfile, join
pathLabels = "../input/rsna-pneumonia-detection-challenge/stage_2_train_labels.csv"
classInfoPath = (
"../input/rsna-pneumonia-detection-challenge/stage_2_detailed_class_info.csv"
)
labels = pd.read_csv(pathLabels)
classInfo = pd.read_csv(classInfoPath)
# ## Part 0.1: Merge ClassInfo & Labels
merged = pd.merge(left=classInfo, right=labels, how="left", on="patientId") # Merge
merged = merged.drop_duplicates() # Remove duplicates
merged.head()
a = merged.loc[merged["class"] == "No Lung Opacity / Not Normal"]
a["Target"].value_counts()
# How many unique features?
print(f"Unique features: \n{merged.nunique()}")
neg, pos = merged["Target"].value_counts()
print(
f'How many people have pneumonia vs do not: \n{merged["Target"].value_counts()}\n'
)
print(
f"This is roughly {round(pos/(pos+neg)*100, 2)}% having pneumonia within this training data"
)
# ##### We can see that over 20,000 people are classified as not having pneumonia, whereas around 10,000 people do
# ##### This tells us that we have an imbalanced dataset
# ##### To deal with this, we need to assign the class with less appeareances (i.e: having pneumonia) with a higher weight to even this imbalance out. We will do this by using a compute weight function, that we will use in our Modelling file
# See how many people with pneumonia vs non-pneumonia
merged["Target"].hist()
# Look at the amounts in each of the different classes
merged["class"].hist()
# ##### It seems that the amount is spread evenly between the 3 classes with roughly 2/3s being either classified as Normal or no lung opacity/not normal
# ## Part 0.2: Some Numerical Analysis
# ### Check out pneumonia classifying boxes
boxNums = merged.dropna()[
["x", "y", "width", "height"]
].copy() # Make a copy of just the boxes
# Calculate x2 & y2 coordinates
boxNums["x2"] = boxNums["x"] + boxNums["width"]
boxNums["y2"] = boxNums["y"] + boxNums["height"]
# Calculate x2 & y2 centres
boxNums["xCentre"] = boxNums["x"] + boxNums["width"] / 2
boxNums["yCentre"] = boxNums["y"] + boxNums["height"] / 2
# Calculate area of the box
boxNums["boxArea"] = boxNums["width"] * boxNums["height"]
boxNums.head(3)
# Look at the correlations between x, y, x2, y2, width, height and the centres
pairs = [
(boxNums["x"], boxNums["y"]),
(boxNums["x2"], boxNums["y2"]),
(boxNums["width"], boxNums["height"]),
(boxNums["xCentre"], boxNums["yCentre"]),
]
axis = [(0, 0), (0, 1), (1, 0), (1, 1)]
fig, axs = plt.subplots(2, 2, figsize=(10, 10))
for i in range(4):
axs[axis[i][0], axis[i][1]].hist2d(pairs[i][0], pairs[i][1], bins=30)
# Set titles
axs[0, 0].set_title("X vs Y")
axs[0, 1].set_title("X2 vs Y2")
axs[1, 0].set_title("Width vs Height")
axs[1, 1].set_title("X Centre vs Y Centre")
plt.show()
# ##### Taking a look at the heatmap, we can see that the centres seems to have more density for x, x2 and y, y2
# Take a look at the distribution of box area
boxNums["boxArea"].plot(
kind="hist",
bins=25,
figsize=(14, 4),
title="Area Distribution of boxes for a Positive target",
)
# ## Part 0.3: Let's take a look at the Dicom Images
# Get two patients (one who has pneumonia & one who doesnt)
patient0 = merged["patientId"][0] # Doesn't have pneumonia
patient1 = merged["patientId"][4] # Has pneumonia
patients = [(patient0, "Doesn't Have Pneumonia"), (patient1, "Has Pneumonia")]
patients
# Plot the images side by side for visual comparison
imgsPath = "../input/rsna-pneumonia-detection-challenge/stage_2_train_images/"
fig, ax = plt.subplots(1, 2, figsize=(7, 7))
for i in range(2):
patientID, title = patients[i][0], patients[i][1] # Extract patient data
dcmFile = f"{imgsPath}{patientID}.dcm" # Get path
dcmData = pydicom.read_file(dcmFile) # Read file
img = dcmData.pixel_array # Get the pixel array
ax[i].imshow(img, cmap=pl.cm.gist_gray) # Plot
ax[i].set_title(title) # Set title
ax[i].axis("off") # Remove axis
# ##### We can see that the patient with pneumonia appears to have a much less opaque scan
p = merged.loc[merged["patientId"] == patient1]
p
# ##### Important Note: A given patientId may have multiple boxes if more than one area of pneumonia is detected (see above).
# ##### We can see that this particular patient (patient #5) has pneumonia, and has 2 entries, with 2 different boxes
# We want to create a Data parser to group a patients boxes with its image
def parseData(df):
newData = {}
for n, row in df.iterrows():
patientID = row["patientId"] # Initialise patient
# If patient is not in the dict, add them
if patientID not in newData:
newData[patientID] = {
"dicom": f"{imgsPath}{patientID}.dcm",
"classifier": row["Target"],
"boxes": [],
}
# Add box if the patient has pneumonia
if newData[patientID]["classifier"] == 1:
newData[patientID]["boxes"].append(
[row["x"], row["y"], row["width"], row["height"]]
)
return newData
parsedData = parseData(merged)
# Check patient 1 which we know has pneumonia
parsedData[patient1]
# ##### We can see that we have saved the file path, the classifier and the array of boxes for that person
# ##### Now check someone we know that doesnt have pneumonia
parsedData[merged["patientId"][0]]
# ### Create a function that allows us to draw the boxes over the images
"""
Credit for @peterchang77 for these 2 functions
"""
# This function will allow us to overlay a box
def overlayBox(im, box, rgb, stroke=1):
# --- Convert coordinates to integers
box = [int(b) for b in box]
# --- Extract coordinates
x, y, width, height = box
y2 = y + height
x2 = x + width
im[y : y + stroke, x:x2] = rgb
im[y2 : y2 + stroke, x:x2] = rgb
im[y:y2, x : x + stroke] = rgb
im[y:y2, x2 : x2 + stroke] = rgb
return im
def drawBox(data):
d = pydicom.read_file(data["dicom"]) # Open and read the file
im = d.pixel_array
# Convert to 3 RGB
im = np.stack([im] * 3, axis=2)
# Add the boxes with random colours
for box in data["boxes"]:
rgb = np.floor(np.random.rand(3) * 256).astype("int") # Get rgb
im = overlayBox(im=im, box=box, rgb=rgb, stroke=6) # Overlay the box
pl.imshow(im, cmap=pl.cm.gist_gray) # Show the image
pl.axis("off") # Remove axis
drawBox(parsedData[patient1])
# ##### We can see the dark opacities in the image were boxed, indicating the pnemonia
# ## Part 0.4: Viewing side-by-side of people having and not having Pneumonia
# ### Pneumonia = 0
# Get all patients with no pneumonia
patients0 = [(row["patientId"]) for n, row in merged.iterrows() if row["Target"] == 0]
fig = plt.figure(figsize=(20, 10))
columns = 6
rows = 4
for i in range(1, columns * rows + 1):
fig.add_subplot(rows, columns, i) # Add the subplot
drawBox(parsedData[patients0[i]]) # Draw the box
# ### Pneumonia = 1
# Get all patients with pneumonia
patients1 = [(row["patientId"]) for n, row in merged.iterrows() if row["Target"] == 1]
fig = plt.figure(figsize=(20, 10))
columns = 6
rows = 4
for i in range(1, columns * rows + 1):
fig.add_subplot(rows, columns, i) # Add the subplot
drawBox(parsedData[patients1[i]]) # Draw the box
# ## Part 0.5: Some Feature Analysis for Standard Models
def readImage(pId):
patient = parsedData[pId]
path = patient["dicom"]
img = pydicom.read_file(path).pixel_array # Read & Convert to pixels
return img
noPne = readImage(patients0[1])
pne = readImage(patients1[1])
def printTwoImgs(img1, img2, title1, title2):
fig, ax = plt.subplots(1, 2, figsize=(12, 12))
ax[0].imshow(img1, cmap="gray")
ax[1].imshow(img2, cmap="gray")
ax[0].set_title(title1)
ax[1].set_title(title2)
ax[0].axis("off")
ax[1].axis("off")
printTwoImgs(noPne, pne, "No Pneumonia - Normal Image", "Pneumonia - Normal Image")
# ### Hist Equalisation
equ1 = cv2.equalizeHist(noPne)
equ2 = cv2.equalizeHist(pne)
printTwoImgs(equ1, equ2, "No Pneumonia", "Pneumonia") # Print images
# ### Image Sharpening
hpf_kernel = np.full((3, 3), -1)
hpf_kernel[1, 1] = 9
imHP = cv2.filter2D(equ1, -1, hpf_kernel)
imUS = skimage.filters.unsharp_mask(equ1) # use unsharpen mask filter
imHP2 = cv2.filter2D(equ2, -1, hpf_kernel)
imUS2 = skimage.filters.unsharp_mask(equ2) # use unsharpen mask filter
printTwoImgs(imHP, imHP2, "No Pneu - High Pass", "Pneu - High Pass") # Print images
printTwoImgs(
imUS, imUS2, "No Pneu - Unsharpen Mask", "Pneu - Unsharpen Mask"
) # Print images
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/476/69476005.ipynb
| null | null |
[{"Id": 69476005, "ScriptId": 18971064, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7736694, "CreationDate": "07/31/2021 13:22:45", "VersionNumber": 1.0, "Title": "P0: Exploratory Data Analysis", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 318.0, "LinesInsertedFromPrevious": 318.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 3}]
| null | null | null | null |
# ## Part 0: Pneumonia Exploratory Data Analysis
# ### Format of Data
# - patientId_- A patientId. Each patientId corresponds to a unique image.
# - x_ - the upper-left x coordinate of the bounding box.
# - y_ - the upper-left y coordinate of the bounding box.
# - width_ - the width of the bounding box.
# - height_ - the height of the bounding box.
# - Target_ - the binary Target, indicating whether this sample has evidence of pneumonia. (1 = true, 0 = false)
# ### Notes
# - A pneumonia opacity is a part of the lungs that looks darker on a radiograph and has a shape that indicates that pneumonia is (or may be) present.
# - Since the goal is to detect & then draw a bounding box on each pneumonia opacities (where each image can have 0 or many), and the training set is already classified:
# - It can be analysed as a supervised learning statistical multilabel classification
#
# Imports
import cv2
import tqdm
import pydicom
import pylab as pl
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pylab as plt
import skimage
from skimage import feature, filters
import os
from os import listdir
from os.path import isfile, join
pathLabels = "../input/rsna-pneumonia-detection-challenge/stage_2_train_labels.csv"
classInfoPath = (
"../input/rsna-pneumonia-detection-challenge/stage_2_detailed_class_info.csv"
)
labels = pd.read_csv(pathLabels)
classInfo = pd.read_csv(classInfoPath)
# ## Part 0.1: Merge ClassInfo & Labels
merged = pd.merge(left=classInfo, right=labels, how="left", on="patientId") # Merge
merged = merged.drop_duplicates() # Remove duplicates
merged.head()
a = merged.loc[merged["class"] == "No Lung Opacity / Not Normal"]
a["Target"].value_counts()
# How many unique features?
print(f"Unique features: \n{merged.nunique()}")
neg, pos = merged["Target"].value_counts()
print(
f'How many people have pneumonia vs do not: \n{merged["Target"].value_counts()}\n'
)
print(
f"This is roughly {round(pos/(pos+neg)*100, 2)}% having pneumonia within this training data"
)
# ##### We can see that over 20,000 people are classified as not having pneumonia, whereas around 10,000 people do
# ##### This tells us that we have an imbalanced dataset
# ##### To deal with this, we need to assign the class with less appeareances (i.e: having pneumonia) with a higher weight to even this imbalance out. We will do this by using a compute weight function, that we will use in our Modelling file
# See how many people with pneumonia vs non-pneumonia
merged["Target"].hist()
# Look at the amounts in each of the different classes
merged["class"].hist()
# ##### It seems that the amount is spread evenly between the 3 classes with roughly 2/3s being either classified as Normal or no lung opacity/not normal
# ## Part 0.2: Some Numerical Analysis
# ### Check out pneumonia classifying boxes
boxNums = merged.dropna()[
["x", "y", "width", "height"]
].copy() # Make a copy of just the boxes
# Calculate x2 & y2 coordinates
boxNums["x2"] = boxNums["x"] + boxNums["width"]
boxNums["y2"] = boxNums["y"] + boxNums["height"]
# Calculate x2 & y2 centres
boxNums["xCentre"] = boxNums["x"] + boxNums["width"] / 2
boxNums["yCentre"] = boxNums["y"] + boxNums["height"] / 2
# Calculate area of the box
boxNums["boxArea"] = boxNums["width"] * boxNums["height"]
boxNums.head(3)
# Look at the correlations between x, y, x2, y2, width, height and the centres
pairs = [
(boxNums["x"], boxNums["y"]),
(boxNums["x2"], boxNums["y2"]),
(boxNums["width"], boxNums["height"]),
(boxNums["xCentre"], boxNums["yCentre"]),
]
axis = [(0, 0), (0, 1), (1, 0), (1, 1)]
fig, axs = plt.subplots(2, 2, figsize=(10, 10))
for i in range(4):
axs[axis[i][0], axis[i][1]].hist2d(pairs[i][0], pairs[i][1], bins=30)
# Set titles
axs[0, 0].set_title("X vs Y")
axs[0, 1].set_title("X2 vs Y2")
axs[1, 0].set_title("Width vs Height")
axs[1, 1].set_title("X Centre vs Y Centre")
plt.show()
# ##### Taking a look at the heatmap, we can see that the centres seems to have more density for x, x2 and y, y2
# Take a look at the distribution of box area
boxNums["boxArea"].plot(
kind="hist",
bins=25,
figsize=(14, 4),
title="Area Distribution of boxes for a Positive target",
)
# ## Part 0.3: Let's take a look at the Dicom Images
# Get two patients (one who has pneumonia & one who doesnt)
patient0 = merged["patientId"][0] # Doesn't have pneumonia
patient1 = merged["patientId"][4] # Has pneumonia
patients = [(patient0, "Doesn't Have Pneumonia"), (patient1, "Has Pneumonia")]
patients
# Plot the images side by side for visual comparison
imgsPath = "../input/rsna-pneumonia-detection-challenge/stage_2_train_images/"
fig, ax = plt.subplots(1, 2, figsize=(7, 7))
for i in range(2):
patientID, title = patients[i][0], patients[i][1] # Extract patient data
dcmFile = f"{imgsPath}{patientID}.dcm" # Get path
dcmData = pydicom.read_file(dcmFile) # Read file
img = dcmData.pixel_array # Get the pixel array
ax[i].imshow(img, cmap=pl.cm.gist_gray) # Plot
ax[i].set_title(title) # Set title
ax[i].axis("off") # Remove axis
# ##### We can see that the patient with pneumonia appears to have a much less opaque scan
p = merged.loc[merged["patientId"] == patient1]
p
# ##### Important Note: A given patientId may have multiple boxes if more than one area of pneumonia is detected (see above).
# ##### We can see that this particular patient (patient #5) has pneumonia, and has 2 entries, with 2 different boxes
# We want to create a Data parser to group a patients boxes with its image
def parseData(df):
newData = {}
for n, row in df.iterrows():
patientID = row["patientId"] # Initialise patient
# If patient is not in the dict, add them
if patientID not in newData:
newData[patientID] = {
"dicom": f"{imgsPath}{patientID}.dcm",
"classifier": row["Target"],
"boxes": [],
}
# Add box if the patient has pneumonia
if newData[patientID]["classifier"] == 1:
newData[patientID]["boxes"].append(
[row["x"], row["y"], row["width"], row["height"]]
)
return newData
parsedData = parseData(merged)
# Check patient 1 which we know has pneumonia
parsedData[patient1]
# ##### We can see that we have saved the file path, the classifier and the array of boxes for that person
# ##### Now check someone we know that doesnt have pneumonia
parsedData[merged["patientId"][0]]
# ### Create a function that allows us to draw the boxes over the images
"""
Credit for @peterchang77 for these 2 functions
"""
# This function will allow us to overlay a box
def overlayBox(im, box, rgb, stroke=1):
# --- Convert coordinates to integers
box = [int(b) for b in box]
# --- Extract coordinates
x, y, width, height = box
y2 = y + height
x2 = x + width
im[y : y + stroke, x:x2] = rgb
im[y2 : y2 + stroke, x:x2] = rgb
im[y:y2, x : x + stroke] = rgb
im[y:y2, x2 : x2 + stroke] = rgb
return im
def drawBox(data):
d = pydicom.read_file(data["dicom"]) # Open and read the file
im = d.pixel_array
# Convert to 3 RGB
im = np.stack([im] * 3, axis=2)
# Add the boxes with random colours
for box in data["boxes"]:
rgb = np.floor(np.random.rand(3) * 256).astype("int") # Get rgb
im = overlayBox(im=im, box=box, rgb=rgb, stroke=6) # Overlay the box
pl.imshow(im, cmap=pl.cm.gist_gray) # Show the image
pl.axis("off") # Remove axis
drawBox(parsedData[patient1])
# ##### We can see the dark opacities in the image were boxed, indicating the pnemonia
# ## Part 0.4: Viewing side-by-side of people having and not having Pneumonia
# ### Pneumonia = 0
# Get all patients with no pneumonia
patients0 = [(row["patientId"]) for n, row in merged.iterrows() if row["Target"] == 0]
fig = plt.figure(figsize=(20, 10))
columns = 6
rows = 4
for i in range(1, columns * rows + 1):
fig.add_subplot(rows, columns, i) # Add the subplot
drawBox(parsedData[patients0[i]]) # Draw the box
# ### Pneumonia = 1
# Get all patients with pneumonia
patients1 = [(row["patientId"]) for n, row in merged.iterrows() if row["Target"] == 1]
fig = plt.figure(figsize=(20, 10))
columns = 6
rows = 4
for i in range(1, columns * rows + 1):
fig.add_subplot(rows, columns, i) # Add the subplot
drawBox(parsedData[patients1[i]]) # Draw the box
# ## Part 0.5: Some Feature Analysis for Standard Models
def readImage(pId):
patient = parsedData[pId]
path = patient["dicom"]
img = pydicom.read_file(path).pixel_array # Read & Convert to pixels
return img
noPne = readImage(patients0[1])
pne = readImage(patients1[1])
def printTwoImgs(img1, img2, title1, title2):
fig, ax = plt.subplots(1, 2, figsize=(12, 12))
ax[0].imshow(img1, cmap="gray")
ax[1].imshow(img2, cmap="gray")
ax[0].set_title(title1)
ax[1].set_title(title2)
ax[0].axis("off")
ax[1].axis("off")
printTwoImgs(noPne, pne, "No Pneumonia - Normal Image", "Pneumonia - Normal Image")
# ### Hist Equalisation
equ1 = cv2.equalizeHist(noPne)
equ2 = cv2.equalizeHist(pne)
printTwoImgs(equ1, equ2, "No Pneumonia", "Pneumonia") # Print images
# ### Image Sharpening
hpf_kernel = np.full((3, 3), -1)
hpf_kernel[1, 1] = 9
imHP = cv2.filter2D(equ1, -1, hpf_kernel)
imUS = skimage.filters.unsharp_mask(equ1) # use unsharpen mask filter
imHP2 = cv2.filter2D(equ2, -1, hpf_kernel)
imUS2 = skimage.filters.unsharp_mask(equ2) # use unsharpen mask filter
printTwoImgs(imHP, imHP2, "No Pneu - High Pass", "Pneu - High Pass") # Print images
printTwoImgs(
imUS, imUS2, "No Pneu - Unsharpen Mask", "Pneu - Unsharpen Mask"
) # Print images
| false | 0 | 3,206 | 3 | 3,206 | 3,206 |
||
69476149
|
<jupyter_start><jupyter_text>ELECTRA
https://huggingface.co/transformers/model_doc/electra.html
Kaggle dataset identifier: electra
<jupyter_script># ========================================
# library
# ========================================
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import StratifiedKFold, KFold, GroupKFold
from sklearn.metrics import mean_squared_error
import torch
import torch.nn as nn
from torch.utils.data import Dataset, DataLoader, Subset
from transformers import AutoModelForSequenceClassification, AutoTokenizer, AutoModel
import transformers
from transformers import RobertaModel, RobertaTokenizer
from transformers import AlbertModel, AlbertTokenizer
from transformers import XLNetModel, XLNetTokenizer, XLNetConfig
from transformers import DebertaModel, DebertaTokenizer
from transformers import (
ElectraModel,
ElectraTokenizer,
ElectraForSequenceClassification,
)
from transformers import BartModel
from transformers import AdamW, get_linear_schedule_with_warmup
from transformers import MPNetModel, MPNetTokenizer
from transformers import FunnelModel, FunnelTokenizer, FunnelBaseModel
from transformers import (
LongformerModel,
LongformerTokenizer,
LongformerForSequenceClassification,
)
from transformers import GPT2Model, GPT2Tokenizer, GPT2Config
import logging
import sys
from contextlib import contextmanager
import time
import random
from tqdm import tqdm
import os
import pickle
import gc
# ==================
# Constant
# ==================
TRAIN_PATH = "../input/commonlitreadabilityprize/train.csvv"
FOLD_PATH = "../input/fe001-step-1-create-folds/fe001_train_folds.csv"
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# ===============
# Settings
# ===============
MODEL_PATH = "xlnet-base-cased"
tokenizer = XLNetTokenizer.from_pretrained("xlnet-base-cased")
xlnet = XLNetModel.from_pretrained(MODEL_PATH)
xlnet.save_pretrained("xlnet-base/")
tokenizer.save_pretrained("xlnet-base/")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/476/69476149.ipynb
|
electra
|
xhlulu
|
[{"Id": 69476149, "ScriptId": 18971369, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 623370, "CreationDate": "07/31/2021 13:24:36", "VersionNumber": 1.0, "Title": "xlnet_base_save", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 56.0, "LinesInsertedFromPrevious": 5.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 51.0, "LinesInsertedFromFork": 5.0, "LinesDeletedFromFork": 5.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 51.0, "TotalVotes": 15}]
|
[{"Id": 92689695, "KernelVersionId": 69476149, "SourceDatasetVersionId": 2211051}, {"Id": 92689691, "KernelVersionId": 69476149, "SourceDatasetVersionId": 2298249}, {"Id": 92689682, "KernelVersionId": 69476149, "SourceDatasetVersionId": 2355157}, {"Id": 92689675, "KernelVersionId": 69476149, "SourceDatasetVersionId": 2368124}, {"Id": 92689694, "KernelVersionId": 69476149, "SourceDatasetVersionId": 2315987}, {"Id": 92689681, "KernelVersionId": 69476149, "SourceDatasetVersionId": 2361523}, {"Id": 92689689, "KernelVersionId": 69476149, "SourceDatasetVersionId": 2206516}, {"Id": 92689687, "KernelVersionId": 69476149, "SourceDatasetVersionId": 918137}, {"Id": 92689671, "KernelVersionId": 69476149, "SourceDatasetVersionId": 2481788}, {"Id": 92689692, "KernelVersionId": 69476149, "SourceDatasetVersionId": 2301216}, {"Id": 92689676, "KernelVersionId": 69476149, "SourceDatasetVersionId": 2368107}, {"Id": 92689690, "KernelVersionId": 69476149, "SourceDatasetVersionId": 1421148}, {"Id": 92689685, "KernelVersionId": 69476149, "SourceDatasetVersionId": 866759}, {"Id": 92689686, "KernelVersionId": 69476149, "SourceDatasetVersionId": 906797}]
|
[{"Id": 2211051, "DatasetId": 1327810, "DatasourceVersionId": 2252681, "CreatorUserId": 2352583, "LicenseName": "Unknown", "CreationDate": "05/09/2021 01:28:18", "VersionNumber": 1.0, "Title": "ELECTRA", "Slug": "electra", "Subtitle": "Source: https://huggingface.co/transformers/model_doc/electra.html", "Description": "https://huggingface.co/transformers/model_doc/electra.html", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1327810, "CreatorUserId": 2352583, "OwnerUserId": 2352583.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2211051.0, "CurrentDatasourceVersionId": 2252681.0, "ForumId": 1346715, "Type": 2, "CreationDate": "05/09/2021 01:28:18", "LastActivityDate": "05/09/2021", "TotalViews": 1887, "TotalDownloads": 125, "TotalVotes": 12, "TotalKernels": 31}]
|
[{"Id": 2352583, "UserName": "xhlulu", "DisplayName": "xhlulu", "RegisterDate": "10/12/2018", "PerformanceTier": 4}]
|
# ========================================
# library
# ========================================
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import StratifiedKFold, KFold, GroupKFold
from sklearn.metrics import mean_squared_error
import torch
import torch.nn as nn
from torch.utils.data import Dataset, DataLoader, Subset
from transformers import AutoModelForSequenceClassification, AutoTokenizer, AutoModel
import transformers
from transformers import RobertaModel, RobertaTokenizer
from transformers import AlbertModel, AlbertTokenizer
from transformers import XLNetModel, XLNetTokenizer, XLNetConfig
from transformers import DebertaModel, DebertaTokenizer
from transformers import (
ElectraModel,
ElectraTokenizer,
ElectraForSequenceClassification,
)
from transformers import BartModel
from transformers import AdamW, get_linear_schedule_with_warmup
from transformers import MPNetModel, MPNetTokenizer
from transformers import FunnelModel, FunnelTokenizer, FunnelBaseModel
from transformers import (
LongformerModel,
LongformerTokenizer,
LongformerForSequenceClassification,
)
from transformers import GPT2Model, GPT2Tokenizer, GPT2Config
import logging
import sys
from contextlib import contextmanager
import time
import random
from tqdm import tqdm
import os
import pickle
import gc
# ==================
# Constant
# ==================
TRAIN_PATH = "../input/commonlitreadabilityprize/train.csvv"
FOLD_PATH = "../input/fe001-step-1-create-folds/fe001_train_folds.csv"
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# ===============
# Settings
# ===============
MODEL_PATH = "xlnet-base-cased"
tokenizer = XLNetTokenizer.from_pretrained("xlnet-base-cased")
xlnet = XLNetModel.from_pretrained(MODEL_PATH)
xlnet.save_pretrained("xlnet-base/")
tokenizer.save_pretrained("xlnet-base/")
| false | 0 | 488 | 15 | 526 | 488 |
||
69476639
|
<jupyter_start><jupyter_text>10 con alphas future
Kaggle dataset identifier: 10-con-alphas-future
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import os
import glob
import matplotlib.pyplot as plt
from random import random
from random import randint
# from glob import glob
from cvxopt import matrix
from cvxopt import solvers
from tabulate import tabulate
sample = "OS"
list_group = glob.glob("../input/10-futures-8-months/group1/*.csv".format(sample))
list_group.sort()
list_nomal = glob.glob("../input/10-futures-8-months/group2/*.csv".format(sample))
list_nomal.sort()
print(list_nomal)
fileList = list_group + list_nomal
print(fileList)
print(len(fileList))
sample = "OS"
m = pd.DataFrame()
# it= infoTest()
for file in fileList:
tempDf = pd.read_csv(file, parse_dates=["datetime"], index_col=3)
# print(file,tempDf)
tempPnl = tempDf[["value"]]
tempPnl = tempPnl[tempPnl.index.dayofweek < 5]
tempPnl["ret"] = tempPnl.value - tempPnl.value.shift(1)
tempPnl = (
tempPnl[["ret"]]
.resample("1D")
.apply(lambda x: x.sum() if len(x) else np.nan)
.dropna(how="all")
)
# print("strat " + file[9:], calculate_sharp(merge=tempPnl))
if len(m) == 0:
m = tempPnl
else:
m = pd.merge(m, tempPnl, how="inner", left_index=True, right_index=True)
colList = []
for i in fileList:
colList.append(i.split("{}_".format(sample))[-1][55:-4])
m.columns = colList
# print(m)
print(fileList)
m.columns = [
"alpha_41_1h",
"correlation_rank",
"second_last_int_close_15m",
"vn30f1m_kc_thep_not_close_5m",
"Tcal_in_day",
"VolImbalance",
"compare_ohlc_30m",
"percent_rank_bank_30m",
"rsi_bb_refractor",
"standing_wave_20m",
]
# m.columns=['alpha_41_1h','correlation_rank','second_last_int_close_15m','vn30f1m_kc_thep_not_close_5m',
# 'Tcal_in_day','VOI','compare_ohlc_30m','percent_rank_bank_30m','rsi_bb_refractor','standing_wave_20m']
# m.columns
# m.columns=['alpha_41_1h','correlation_rank','second_last_int_close_15m','vn30f1m_kc_thep_not_close_5m',
# 'Tcal_in_day','VOI','bank_close_volume_5m','compare_ohlc_30m','rsi_bb_refractor','standing_wave_20m']
# m.columns=['alpha_41_1h','correlation_rank','second_last_int_close_15m','vn30f1m_kc_thep_not_close_5m',
#'VOI','bank_close_volume_5m','compare_ohlc_30m','rsi_bb_refractor','standing_wave_20m']
m = m / (10**6)
m.head()
# # SPLIT TRAIN/TEST DATA
# Train
train_1 = m["2020-01-01":"2020-12-31"]
train_2 = m["2020-02-01":"2021-01-31"]
train_3 = m["2020-03-01":"2021-02-28"]
train_4 = m["2020-04-01":"2021-03-31"]
train_5 = m["2020-05-01":"2021-04-30"]
train_6 = m["2020-06-01":"2021-05-31"]
train_7 = m["2020-07-01":"2021-06-30"]
train_8 = m["2020-08-01":"2021-07-31"]
test_1 = m["2021-01-01":"2021-01-31"]
test_2 = m["2021-02-01":"2021-02-28"]
test_3 = m["2021-03-01":"2021-03-31"]
test_4 = m["2021-04-01":"2021-04-30"]
test_5 = m["2021-05-01":"2021-05-31"]
test_6 = m["2021-06-01":"2021-06-30"]
test_7 = m["2021-07-01":]
# Export train sets
train_1.to_csv("future_train_1.csv")
train_2.to_csv("future_train_2.csv")
train_3.to_csv("future_train_3.csv")
train_4.to_csv("future_train_4.csv")
train_5.to_csv("future_train_5.csv")
train_6.to_csv("future_train_6.csv")
train_7.to_csv("future_train_7.csv")
train_8.to_csv("future_train_8.csv")
# Export test set
test_1.to_csv("future_test_1.csv")
test_2.to_csv("future_test_2.csv")
test_3.to_csv("future_test_3.csv")
test_4.to_csv("future_test_4.csv")
test_5.to_csv("future_test_5.csv")
test_6.to_csv("future_test_6.csv")
test_7.to_csv("future_test_7.csv")
# # Import simulation data
simul_G1 = pd.read_csv("../input/simulation-data-future-8-month/X1_G.csv")
simul_G1 = simul_G1.iloc[:, 1:]
simul_G2 = pd.read_csv("../input/simulation-data-future-8-month/X2_G.csv")
simul_G2 = simul_G2.iloc[:, 1:]
simul_G3 = pd.read_csv("../input/simulation-data-future-8-month/X3_G.csv")
simul_G3 = simul_G3.iloc[:, 1:]
simul_G4 = pd.read_csv("../input/simulation-data-future-8-month/X4_G.csv")
simul_G4 = simul_G4.iloc[:, 1:]
simul_G5 = pd.read_csv("../input/simulation-data-future-8-month/X5_G.csv")
simul_G5 = simul_G5.iloc[:, 1:]
simul_G6 = pd.read_csv("../input/simulation-data-future-8-month/X6_G.csv")
simul_G6 = simul_G6.iloc[:, 1:]
simul_G7 = pd.read_csv("../input/simulation-data-future-8-month/X7_G.csv")
simul_G7 = simul_G7.iloc[:, 1:]
simul_C1 = pd.read_csv("../input/simulation-data-future-8-month/X1_G.csv")
simul_C1 = simul_C1.iloc[:, 1:]
simul_C2 = pd.read_csv("../input/simulation-data-future-8-month/X2_G.csv")
simul_C2 = simul_C2.iloc[:, 1:]
simul_C3 = pd.read_csv("../input/simulation-data-future-8-month/X3_G.csv")
simul_C3 = simul_C3.iloc[:, 1:]
simul_C4 = pd.read_csv("../input/simulation-data-future-8-month/X4_G.csv")
simul_C4 = simul_C4.iloc[:, 1:]
simul_C5 = pd.read_csv("../input/simulation-data-future-8-month/X5_G.csv")
simul_C5 = simul_C5.iloc[:, 1:]
simul_C6 = pd.read_csv("../input/simulation-data-future-8-month/X6_G.csv")
simul_C6 = simul_C6.iloc[:, 1:]
simul_C7 = pd.read_csv("../input/simulation-data-future-8-month/X7_G.csv")
simul_C7 = simul_C7.iloc[:, 1:]
# # Implementation
class infoTest:
def __init__(self):
pass
def calculateSharpe(self, npArray):
sr = npArray.mean() / npArray.std() * np.sqrt(252)
print(npArray.std())
return sr
def max_drawdown(self, booksize, returnSeries):
mdd = 0
a = np.cumsum(returnSeries)
X = a + booksize
peak = X[0]
dds = []
for x in X:
if x > peak:
peak = x
dd = (peak - x) / booksize
if dd > mdd:
mdd = dd
dds.append(X[X == x])
print("MDD AT ", dds[-1].index[0] if len(dds) else None)
print(X)
# X.to_csv(r'/home/hoainam/PycharmProjects/multi_strategy/v_multi/X.csv')
return mdd
# rets is array of returns
def randomAllocateWeigh(self, rets):
remaining = 1
weigh = []
for i in range(len(rets)):
tempWeigh = round(random(), 2)
weigh.append(tempWeigh)
remaining = remaining - tempWeigh
weigh = np.asarray(weigh) / np.sum(weigh)
# print(np.sum(weigh))
portfolio = []
for i in range(len(rets)):
if len(portfolio) == 0:
portfolio = rets[i] * weigh[i]
else:
portfolio += rets[i] * weigh[i]
# portfolio = np.asarray(portfolio)/np.sum(weigh)
return weigh, portfolio
def randomAllocateListReturns(self, df):
remaining = 1
weigh = []
counter = 0
ret = []
portfolio = []
for columnName, columnData in df.iteritems():
tempWeigh = round(random(), 2)
weigh.append(tempWeigh)
weigh = np.asarray(weigh) / np.sum(weigh)
# print(np.sum(weigh))
portfolio = []
counter = 0
for columnName, columnData in df.iteritems():
if len(portfolio) == 0:
portfolio = columnData * weigh[counter]
else:
portfolio += columnData * weigh[counter]
counter += 1
# portfolio = np.asarray(portfolio)/np.sum(weigh)
return weigh, portfolio
def allocateForMaxSharpe(self, df, itertimes):
maxSharpe = 0
maxWeigh = []
finalPnl = []
for i in range(itertimes):
weigh, mergePnl = self.randomAllocateListReturns(df)
tempSharpe = self.calculateSharpe(mergePnl)
if tempSharpe >= maxSharpe:
maxSharpe = tempSharpe
maxWeigh = weigh
finalPnl = mergePnl
# maxWeigh = np.asarray(maxWeigh) / np.sum(maxWeigh)
return maxSharpe, maxWeigh, finalPnl
def allocateForMinDD(self, df, itertimes, booksize):
minDD = 1
minDDWeigh = []
finalPnl = []
for i in range(itertimes):
weigh, mergePnl = self.randomAllocateListReturns(df)
tempDD = self.max_drawdown(booksize, mergePnl)
if tempDD < minDD:
minDD = tempDD
minDDWeigh = weigh
finalPnl = mergePnl
return minDD, minDDWeigh, finalPnl
def allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_normal, dataframe, booksize, upperbound, bounded_list, df2
):
# print(tabulate(dataframe.corr(method='pearson'), tablefmt="pipe", headers="keys"))
# upperbound = 0.3
it = infoTest()
cov = (dataframe.cov()).to_numpy()
meanvec = (dataframe.mean()).to_numpy()
P = matrix(cov, tc="d")
# print(P)
q = matrix(np.zeros(len(meanvec)), (len(meanvec), 1), tc="d")
G = []
for i in range(len(meanvec)):
k = [0 for x in range(len(meanvec) - 1)]
k.insert(i, -1)
G.append(k)
for i in range(len(meanvec)):
k = [-upperbound for x in range(len(meanvec) - 1)]
k.insert(i, 1 - upperbound)
G.append(k)
k = [-bounded_list for i in range(len(list_normal))]
for i in range(len(list_group)):
k.insert(i, 1 - bounded_list)
G.append(k)
k = [bounded_list for i in range(len(list_normal))]
for i in range(len(list_group)):
k.insert(i, bounded_list - 1)
G.append(k)
G = matrix(np.array(G))
H = np.zeros(2 * len(meanvec) + 2)
h = matrix(H, tc="d")
A = (matrix(meanvec)).trans()
b = matrix([1], (1, 1), tc="d")
# print('G',G)
# print('h',h)
# print('A',A)
# print('b',b)
sol = (solvers.qp(P, q, G, h, A, b))["x"]
solution = [x for x in sol]
sum = 0
for i in range(len(solution)):
sum += solution[i]
optimizedWeigh = [x / sum for x in solution]
print(optimizedWeigh)
merge = []
counter = 0
for columnName, columnData in df2.iteritems():
# print(real[counter])
if len(merge) == 0:
merge = df2[columnName] * optimizedWeigh[counter]
else:
merge = merge + df2[columnName] * optimizedWeigh[counter]
counter += 1
# print(merge)
print("dd,", it.max_drawdown(booksize=booksize, returnSeries=merge))
print("sharpe,", it.calculateSharpe(merge))
merge = merge * 10
# print('value',merge)
# merge.to_csv(r'/home/hoainam/PycharmProjects/multi_strategy/v_multi/f1m.csv')
# print(np.cumsum(merge))
plt.plot(np.cumsum(merge))
plt.grid(True)
plt.legend(("old", "maxsharpe", "minDD"))
plt.show()
return optimizedWeigh
def allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_normal, dataframe, booksize, upperbound, bounded_list, df2
):
# print(tabulate(dataframe.corr(method='pearson'), tablefmt="pipe", headers="keys"))
# upperbound = 0.3
it = infoTest()
cov = (dataframe.cov()).to_numpy()
meanvec = (dataframe.std()).to_numpy()
P = matrix(cov, tc="d")
# print(P)
q = matrix(np.zeros(len(meanvec)), (len(meanvec), 1), tc="d")
G = []
for i in range(len(meanvec)):
k = [0 for x in range(len(meanvec) - 1)]
k.insert(i, -1)
G.append(k)
for i in range(len(meanvec)):
k = [-upperbound for x in range(len(meanvec) - 1)]
k.insert(i, 1 - upperbound)
G.append(k)
k = [-bounded_list for i in range(len(list_normal))]
for i in range(len(list_group)):
k.insert(i, 1 - bounded_list)
G.append(k)
k = [bounded_list for i in range(len(list_normal))]
for i in range(len(list_group)):
k.insert(i, bounded_list - 1)
G.append(k)
G = matrix(np.array(G))
H = np.zeros(2 * len(meanvec) + 2)
h = matrix(H, tc="d")
A = (matrix(meanvec)).trans()
b = matrix([1], (1, 1), tc="d")
# print('G',G)
# print('h',h)
# print('A',A)
# print('b',b)
sol = (solvers.qp(P, q, G, h, A, b))["x"]
solution = [x for x in sol]
sum = 0
for i in range(len(solution)):
sum += solution[i]
optimizedWeigh = [x / sum for x in solution]
print(optimizedWeigh)
merge = []
counter = 0
for columnName, columnData in df2.iteritems():
# print(real[counter])
if len(merge) == 0:
merge = df2[columnName] * optimizedWeigh[counter]
else:
merge = merge + df2[columnName] * optimizedWeigh[counter]
counter += 1
# print(merge)
print("dd,", it.max_drawdown(booksize=booksize, returnSeries=merge))
print("sharpe,", it.calculateSharpe(merge))
merge = merge * 10
# print('value',merge)
# merge.to_csv(r'/home/hoainam/PycharmProjects/multi_strategy/v_multi/f1m.csv')
# print(np.cumsum(merge))
plt.plot(np.cumsum(merge))
plt.grid(True)
plt.legend(("old", "maxsharpe", "minDD"))
plt.show()
return optimizedWeigh
# # TEST 1
trad1 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, train_1, 10**3, 0.16, 0.36, test_1
)
Gauss1 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_G1, 10**3, 0.16, 0.36, test_1
)
Gauss1_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G1, 10**3, 0.16, 0.36, test_1
)
Clay1 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_C1, 10**3, 0.16, 0.36, test_1
)
Clay1_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_C1, 10**3, 0.16, 0.36, test_1
)
# # TEST 2
trad2 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, train_2, 10**3, 0.16, 0.36, test_2
)
Gauss2 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_G2, 10**3, 0.16, 0.36, test_2
)
Gauss2_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G2, 10**3, 0.16, 0.36, test_2
)
Clay2 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_C2, 10**3, 0.16, 0.36, test_2
)
Clay2_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_C2, 10**3, 0.16, 0.36, test_2
)
# # TEST 3
trad3 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, train_3, 10**3, 0.16, 0.36, test_3
)
Gauss3 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_G3, 10**3, 0.16, 0.36, test_3
)
Gauss3_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G3, 10**3, 0.16, 0.36, test_3
)
Clay3 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_C3, 10**3, 0.16, 0.36, test_3
)
Clay3_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_C3, 10**3, 0.16, 0.36, test_3
)
# # TEST 4
trad4 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, train_4, 10**3, 0.16, 0.36, test_4
)
Gauss4 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_G4, 10**3, 0.16, 0.36, test_4
)
Gauss4_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G4, 10**3, 0.16, 0.36, test_4
)
Clay4 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_C4, 10**3, 0.16, 0.36, test_4
)
Clay4_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G4, 10**3, 0.16, 0.36, test_4
)
# # TEST 5
trad5 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, train_5, 10**3, 0.16, 0.36, test_5
)
Gauss5 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_G5, 10**3, 0.16, 0.36, test_5
)
Gauss5_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G5, 10**3, 0.16, 0.36, test_5
)
Clay5 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_C5, 10**3, 0.16, 0.36, test_5
)
Clay5_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G5, 10**3, 0.16, 0.36, test_5
)
# # TEST 6
trad6 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, train_6, 10**3, 0.16, 0.36, test_6
)
Gauss6 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_G6, 10**3, 0.16, 0.36, test_6
)
Gauss6_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G6, 10**3, 0.16, 0.36, test_6
)
Clay6 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_C6, 10**3, 0.16, 0.36, test_6
)
Clay6_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G6, 10**3, 0.16, 0.36, test_6
)
# # TEST 7
trad7 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, train_7, 10**3, 0.16, 0.36, test_7
)
Gauss7 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_G7, 10**3, 0.16, 0.36, test_7
)
Gauss7_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G7, 10**3, 0.16, 0.36, test_7
)
Clay7 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_C7, 10**3, 0.16, 0.36, test_7
)
Clay7_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G7, 10**3, 0.16, 0.36, test_7
)
# # Port Weight
port1 = {
"trad": trad1,
"Clay": Clay1,
"Gaus": Gauss1,
"Std Clay": Clay1_std,
"Std Gaus": Gauss1_std,
}
port1 = pd.DataFrame(port1)
port1.index = m.columns
port2 = {
"trad": trad2,
"Clay": Clay2,
"Gaus": Gauss2,
"Std Clay": Clay2_std,
"Std Gaus": Gauss2_std,
}
port2 = pd.DataFrame(port2)
port2.index = m.columns
port3 = {
"trad": trad3,
"Clay": Clay3,
"Gaus": Gauss3,
"Std Clay": Clay3_std,
"Std Gaus": Gauss3_std,
}
port3 = pd.DataFrame(port3)
port3.index = m.columns
port4 = {
"trad": trad4,
"Clay": Clay4,
"Gaus": Gauss4,
"Std Clay": Clay4_std,
"Std Gaus": Gauss4_std,
}
port4 = pd.DataFrame(port4)
port4.index = m.columns
port5 = {
"trad": trad5,
"Clay": Clay5,
"Gaus": Gauss5,
"Std Clay": Clay5_std,
"Std Gaus": Gauss5_std,
}
port5 = pd.DataFrame(port5)
port5.index = m.columns
port6 = {
"trad": trad6,
"Clay": Clay6,
"Gaus": Gauss6,
"Std Clay": Clay6_std,
"Std Gaus": Gauss6_std,
}
port6 = pd.DataFrame(port6)
port6.index = m.columns
port7 = {
"trad": trad7,
"Clay": Clay7,
"Gaus": Gauss7,
"Std Clay": Clay7_std,
"Std Gaus": Gauss7_std,
}
port7 = pd.DataFrame(port7)
port7.index = m.columns
port1.to_csv("Port_test_1.csv")
port2.to_csv("Port_test_2.csv")
port3.to_csv("Port_test_3.csv")
port4.to_csv("Port_test_4.csv")
port5.to_csv("Port_test_5.csv")
port6.to_csv("Port_test_6.csv")
port7.to_csv("Port_test_7.csv")
# port8.to_csv('Port_test_8.csv')
# # FRONTIER PLOT
def frontier_plot(df, meanvec, cov_mat, port):
# define list
# cov_matrix = (dataframe.cov())
# meanvec = (dataframe.mean())
np.random.seed(42)
num_ports = 10000
num_alpha = df.shape[1]
all_weights = np.zeros((num_ports, num_alpha))
ret_arr = np.zeros(num_ports)
vol_arr = np.zeros(num_ports)
sharpe_arr = np.zeros(num_ports)
for x in range(num_ports):
weights = np.array(np.random.random(num_alpha))
weights = weights / np.sum(weights)
# Save weights
all_weights[x, :] = weights
# Expected return
ret_arr[x] = np.sum((meanvec * weights * 252))
# Expected volatility
vol_arr[x] = np.sqrt(np.dot(weights.T, np.dot(cov_mat * 252, weights)))
# Sharpe Ratio
sharpe_arr[x] = ret_arr[x] / vol_arr[x]
tradition_weight = port["trad"]
clayton_weight = port["Clay"]
gauss_weight = port["Gaus"]
std_clay_w = port["Std Clay"]
std_gaus_w = port["Std Gaus"]
# Volatility
max_trad_sr_vol = np.sqrt(
np.dot(tradition_weight.T, np.dot(cov_mat * 252, tradition_weight))
)
max_clayton_sr_vol = np.sqrt(
np.dot(clayton_weight.T, np.dot(cov_mat * 252, clayton_weight))
)
max_gauss_sr_vol = np.sqrt(
np.dot(gauss_weight.T, np.dot(cov_mat * 252, gauss_weight))
)
max_clayton_std_vol = np.sqrt(
np.dot(std_clay_w.T, np.dot(cov_mat * 252, std_clay_w))
)
max_gaus_std_vol = np.sqrt(np.dot(std_gaus_w.T, np.dot(cov_mat * 252, std_gaus_w)))
# Return
max__trad_sr_ret = np.sum((meanvec * tradition_weight * 252))
max__clayton_sr_ret = np.sum((meanvec * clayton_weight * 252))
max__gauss_sr_ret = np.sum((meanvec * gauss_weight * 252))
max__clayton_std_ret = np.sum((meanvec * std_clay_w * 252))
max__gaus_std_ret = np.sum((meanvec * std_gaus_w * 252))
real_sr_ret = ret_arr[sharpe_arr.argmax()]
real_sr_vol = vol_arr[sharpe_arr.argmax()]
print("The Traditional Sharpe : ", max__trad_sr_ret / max_trad_sr_vol)
print("The Clayton Sharpe : ", max__clayton_sr_ret / max_clayton_sr_vol)
print("The Gauss Sharpe : ", max__gauss_sr_ret / max_gauss_sr_vol)
print("The STD Clayton Sharpe : ", max__clayton_std_ret / max_clayton_std_vol)
print("The STD Gauss Sharpe : ", max__gaus_std_ret / max_gaus_std_vol)
print("The Real Sharpe :", real_sr_ret / real_sr_vol)
plt.figure(figsize=(12, 8))
plt.scatter(vol_arr, ret_arr, c=sharpe_arr, cmap="viridis")
plt.colorbar(label="Sharpe Ratio")
plt.xlabel("Volatility")
plt.ylabel("Return")
plt.scatter(max_trad_sr_vol, max__trad_sr_ret, c="red", s=50) # red dot
plt.scatter(max_clayton_sr_vol, max__clayton_sr_ret, c="pink", s=50)
plt.scatter(max_gauss_sr_vol, max__gauss_sr_ret, c="pink", s=50)
plt.scatter(max_clayton_sr_vol, max__clayton_sr_ret, c="orange", s=50)
plt.scatter(max_gauss_sr_vol, max__gauss_sr_ret, c="orange", s=50)
plt.scatter(real_sr_vol, real_sr_ret, c="black", s=50) # black dot
plt.show()
frontier_plot(test_1, test_1.mean(), test_1.cov(), port1)
frontier_plot(test_2, test_2.mean(), test_2.cov(), port2)
frontier_plot(test_3, test_3.mean(), test_3.cov(), port3)
frontier_plot(test_4, test_4.mean(), test_4.cov(), port4)
frontier_plot(test_5, test_5.mean(), test_5.cov(), port5)
frontier_plot(test_6, test_6.mean(), test_6.cov(), port6)
frontier_plot(test_7, test_7.mean(), test_7.cov(), port7)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/476/69476639.ipynb
|
10-con-alphas-future
|
quangkhinguynhng
|
[{"Id": 69476639, "ScriptId": 18576507, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4041563, "CreationDate": "07/31/2021 13:30:55", "VersionNumber": 18.0, "Title": "SIMULATION DATA TEST SPLIT", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 663.0, "LinesInsertedFromPrevious": 14.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 649.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92690794, "KernelVersionId": 69476639, "SourceDatasetVersionId": 2464930}]
|
[{"Id": 2464930, "DatasetId": 1492079, "DatasourceVersionId": 2507373, "CreatorUserId": 4041563, "LicenseName": "Unknown", "CreationDate": "07/26/2021 12:45:09", "VersionNumber": 1.0, "Title": "10 con alphas future", "Slug": "10-con-alphas-future", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1492079, "CreatorUserId": 4041563, "OwnerUserId": 4041563.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2464930.0, "CurrentDatasourceVersionId": 2507373.0, "ForumId": 1511789, "Type": 2, "CreationDate": "07/26/2021 12:45:09", "LastActivityDate": "07/26/2021", "TotalViews": 938, "TotalDownloads": 26, "TotalVotes": 0, "TotalKernels": 1}]
|
[{"Id": 4041563, "UserName": "quangkhinguynhng", "DisplayName": "Quang Kh\u1ea3i Nguy\u1ec5n H\u01b0ng", "RegisterDate": "11/15/2019", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import os
import glob
import matplotlib.pyplot as plt
from random import random
from random import randint
# from glob import glob
from cvxopt import matrix
from cvxopt import solvers
from tabulate import tabulate
sample = "OS"
list_group = glob.glob("../input/10-futures-8-months/group1/*.csv".format(sample))
list_group.sort()
list_nomal = glob.glob("../input/10-futures-8-months/group2/*.csv".format(sample))
list_nomal.sort()
print(list_nomal)
fileList = list_group + list_nomal
print(fileList)
print(len(fileList))
sample = "OS"
m = pd.DataFrame()
# it= infoTest()
for file in fileList:
tempDf = pd.read_csv(file, parse_dates=["datetime"], index_col=3)
# print(file,tempDf)
tempPnl = tempDf[["value"]]
tempPnl = tempPnl[tempPnl.index.dayofweek < 5]
tempPnl["ret"] = tempPnl.value - tempPnl.value.shift(1)
tempPnl = (
tempPnl[["ret"]]
.resample("1D")
.apply(lambda x: x.sum() if len(x) else np.nan)
.dropna(how="all")
)
# print("strat " + file[9:], calculate_sharp(merge=tempPnl))
if len(m) == 0:
m = tempPnl
else:
m = pd.merge(m, tempPnl, how="inner", left_index=True, right_index=True)
colList = []
for i in fileList:
colList.append(i.split("{}_".format(sample))[-1][55:-4])
m.columns = colList
# print(m)
print(fileList)
m.columns = [
"alpha_41_1h",
"correlation_rank",
"second_last_int_close_15m",
"vn30f1m_kc_thep_not_close_5m",
"Tcal_in_day",
"VolImbalance",
"compare_ohlc_30m",
"percent_rank_bank_30m",
"rsi_bb_refractor",
"standing_wave_20m",
]
# m.columns=['alpha_41_1h','correlation_rank','second_last_int_close_15m','vn30f1m_kc_thep_not_close_5m',
# 'Tcal_in_day','VOI','compare_ohlc_30m','percent_rank_bank_30m','rsi_bb_refractor','standing_wave_20m']
# m.columns
# m.columns=['alpha_41_1h','correlation_rank','second_last_int_close_15m','vn30f1m_kc_thep_not_close_5m',
# 'Tcal_in_day','VOI','bank_close_volume_5m','compare_ohlc_30m','rsi_bb_refractor','standing_wave_20m']
# m.columns=['alpha_41_1h','correlation_rank','second_last_int_close_15m','vn30f1m_kc_thep_not_close_5m',
#'VOI','bank_close_volume_5m','compare_ohlc_30m','rsi_bb_refractor','standing_wave_20m']
m = m / (10**6)
m.head()
# # SPLIT TRAIN/TEST DATA
# Train
train_1 = m["2020-01-01":"2020-12-31"]
train_2 = m["2020-02-01":"2021-01-31"]
train_3 = m["2020-03-01":"2021-02-28"]
train_4 = m["2020-04-01":"2021-03-31"]
train_5 = m["2020-05-01":"2021-04-30"]
train_6 = m["2020-06-01":"2021-05-31"]
train_7 = m["2020-07-01":"2021-06-30"]
train_8 = m["2020-08-01":"2021-07-31"]
test_1 = m["2021-01-01":"2021-01-31"]
test_2 = m["2021-02-01":"2021-02-28"]
test_3 = m["2021-03-01":"2021-03-31"]
test_4 = m["2021-04-01":"2021-04-30"]
test_5 = m["2021-05-01":"2021-05-31"]
test_6 = m["2021-06-01":"2021-06-30"]
test_7 = m["2021-07-01":]
# Export train sets
train_1.to_csv("future_train_1.csv")
train_2.to_csv("future_train_2.csv")
train_3.to_csv("future_train_3.csv")
train_4.to_csv("future_train_4.csv")
train_5.to_csv("future_train_5.csv")
train_6.to_csv("future_train_6.csv")
train_7.to_csv("future_train_7.csv")
train_8.to_csv("future_train_8.csv")
# Export test set
test_1.to_csv("future_test_1.csv")
test_2.to_csv("future_test_2.csv")
test_3.to_csv("future_test_3.csv")
test_4.to_csv("future_test_4.csv")
test_5.to_csv("future_test_5.csv")
test_6.to_csv("future_test_6.csv")
test_7.to_csv("future_test_7.csv")
# # Import simulation data
simul_G1 = pd.read_csv("../input/simulation-data-future-8-month/X1_G.csv")
simul_G1 = simul_G1.iloc[:, 1:]
simul_G2 = pd.read_csv("../input/simulation-data-future-8-month/X2_G.csv")
simul_G2 = simul_G2.iloc[:, 1:]
simul_G3 = pd.read_csv("../input/simulation-data-future-8-month/X3_G.csv")
simul_G3 = simul_G3.iloc[:, 1:]
simul_G4 = pd.read_csv("../input/simulation-data-future-8-month/X4_G.csv")
simul_G4 = simul_G4.iloc[:, 1:]
simul_G5 = pd.read_csv("../input/simulation-data-future-8-month/X5_G.csv")
simul_G5 = simul_G5.iloc[:, 1:]
simul_G6 = pd.read_csv("../input/simulation-data-future-8-month/X6_G.csv")
simul_G6 = simul_G6.iloc[:, 1:]
simul_G7 = pd.read_csv("../input/simulation-data-future-8-month/X7_G.csv")
simul_G7 = simul_G7.iloc[:, 1:]
simul_C1 = pd.read_csv("../input/simulation-data-future-8-month/X1_G.csv")
simul_C1 = simul_C1.iloc[:, 1:]
simul_C2 = pd.read_csv("../input/simulation-data-future-8-month/X2_G.csv")
simul_C2 = simul_C2.iloc[:, 1:]
simul_C3 = pd.read_csv("../input/simulation-data-future-8-month/X3_G.csv")
simul_C3 = simul_C3.iloc[:, 1:]
simul_C4 = pd.read_csv("../input/simulation-data-future-8-month/X4_G.csv")
simul_C4 = simul_C4.iloc[:, 1:]
simul_C5 = pd.read_csv("../input/simulation-data-future-8-month/X5_G.csv")
simul_C5 = simul_C5.iloc[:, 1:]
simul_C6 = pd.read_csv("../input/simulation-data-future-8-month/X6_G.csv")
simul_C6 = simul_C6.iloc[:, 1:]
simul_C7 = pd.read_csv("../input/simulation-data-future-8-month/X7_G.csv")
simul_C7 = simul_C7.iloc[:, 1:]
# # Implementation
class infoTest:
def __init__(self):
pass
def calculateSharpe(self, npArray):
sr = npArray.mean() / npArray.std() * np.sqrt(252)
print(npArray.std())
return sr
def max_drawdown(self, booksize, returnSeries):
mdd = 0
a = np.cumsum(returnSeries)
X = a + booksize
peak = X[0]
dds = []
for x in X:
if x > peak:
peak = x
dd = (peak - x) / booksize
if dd > mdd:
mdd = dd
dds.append(X[X == x])
print("MDD AT ", dds[-1].index[0] if len(dds) else None)
print(X)
# X.to_csv(r'/home/hoainam/PycharmProjects/multi_strategy/v_multi/X.csv')
return mdd
# rets is array of returns
def randomAllocateWeigh(self, rets):
remaining = 1
weigh = []
for i in range(len(rets)):
tempWeigh = round(random(), 2)
weigh.append(tempWeigh)
remaining = remaining - tempWeigh
weigh = np.asarray(weigh) / np.sum(weigh)
# print(np.sum(weigh))
portfolio = []
for i in range(len(rets)):
if len(portfolio) == 0:
portfolio = rets[i] * weigh[i]
else:
portfolio += rets[i] * weigh[i]
# portfolio = np.asarray(portfolio)/np.sum(weigh)
return weigh, portfolio
def randomAllocateListReturns(self, df):
remaining = 1
weigh = []
counter = 0
ret = []
portfolio = []
for columnName, columnData in df.iteritems():
tempWeigh = round(random(), 2)
weigh.append(tempWeigh)
weigh = np.asarray(weigh) / np.sum(weigh)
# print(np.sum(weigh))
portfolio = []
counter = 0
for columnName, columnData in df.iteritems():
if len(portfolio) == 0:
portfolio = columnData * weigh[counter]
else:
portfolio += columnData * weigh[counter]
counter += 1
# portfolio = np.asarray(portfolio)/np.sum(weigh)
return weigh, portfolio
def allocateForMaxSharpe(self, df, itertimes):
maxSharpe = 0
maxWeigh = []
finalPnl = []
for i in range(itertimes):
weigh, mergePnl = self.randomAllocateListReturns(df)
tempSharpe = self.calculateSharpe(mergePnl)
if tempSharpe >= maxSharpe:
maxSharpe = tempSharpe
maxWeigh = weigh
finalPnl = mergePnl
# maxWeigh = np.asarray(maxWeigh) / np.sum(maxWeigh)
return maxSharpe, maxWeigh, finalPnl
def allocateForMinDD(self, df, itertimes, booksize):
minDD = 1
minDDWeigh = []
finalPnl = []
for i in range(itertimes):
weigh, mergePnl = self.randomAllocateListReturns(df)
tempDD = self.max_drawdown(booksize, mergePnl)
if tempDD < minDD:
minDD = tempDD
minDDWeigh = weigh
finalPnl = mergePnl
return minDD, minDDWeigh, finalPnl
def allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_normal, dataframe, booksize, upperbound, bounded_list, df2
):
# print(tabulate(dataframe.corr(method='pearson'), tablefmt="pipe", headers="keys"))
# upperbound = 0.3
it = infoTest()
cov = (dataframe.cov()).to_numpy()
meanvec = (dataframe.mean()).to_numpy()
P = matrix(cov, tc="d")
# print(P)
q = matrix(np.zeros(len(meanvec)), (len(meanvec), 1), tc="d")
G = []
for i in range(len(meanvec)):
k = [0 for x in range(len(meanvec) - 1)]
k.insert(i, -1)
G.append(k)
for i in range(len(meanvec)):
k = [-upperbound for x in range(len(meanvec) - 1)]
k.insert(i, 1 - upperbound)
G.append(k)
k = [-bounded_list for i in range(len(list_normal))]
for i in range(len(list_group)):
k.insert(i, 1 - bounded_list)
G.append(k)
k = [bounded_list for i in range(len(list_normal))]
for i in range(len(list_group)):
k.insert(i, bounded_list - 1)
G.append(k)
G = matrix(np.array(G))
H = np.zeros(2 * len(meanvec) + 2)
h = matrix(H, tc="d")
A = (matrix(meanvec)).trans()
b = matrix([1], (1, 1), tc="d")
# print('G',G)
# print('h',h)
# print('A',A)
# print('b',b)
sol = (solvers.qp(P, q, G, h, A, b))["x"]
solution = [x for x in sol]
sum = 0
for i in range(len(solution)):
sum += solution[i]
optimizedWeigh = [x / sum for x in solution]
print(optimizedWeigh)
merge = []
counter = 0
for columnName, columnData in df2.iteritems():
# print(real[counter])
if len(merge) == 0:
merge = df2[columnName] * optimizedWeigh[counter]
else:
merge = merge + df2[columnName] * optimizedWeigh[counter]
counter += 1
# print(merge)
print("dd,", it.max_drawdown(booksize=booksize, returnSeries=merge))
print("sharpe,", it.calculateSharpe(merge))
merge = merge * 10
# print('value',merge)
# merge.to_csv(r'/home/hoainam/PycharmProjects/multi_strategy/v_multi/f1m.csv')
# print(np.cumsum(merge))
plt.plot(np.cumsum(merge))
plt.grid(True)
plt.legend(("old", "maxsharpe", "minDD"))
plt.show()
return optimizedWeigh
def allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_normal, dataframe, booksize, upperbound, bounded_list, df2
):
# print(tabulate(dataframe.corr(method='pearson'), tablefmt="pipe", headers="keys"))
# upperbound = 0.3
it = infoTest()
cov = (dataframe.cov()).to_numpy()
meanvec = (dataframe.std()).to_numpy()
P = matrix(cov, tc="d")
# print(P)
q = matrix(np.zeros(len(meanvec)), (len(meanvec), 1), tc="d")
G = []
for i in range(len(meanvec)):
k = [0 for x in range(len(meanvec) - 1)]
k.insert(i, -1)
G.append(k)
for i in range(len(meanvec)):
k = [-upperbound for x in range(len(meanvec) - 1)]
k.insert(i, 1 - upperbound)
G.append(k)
k = [-bounded_list for i in range(len(list_normal))]
for i in range(len(list_group)):
k.insert(i, 1 - bounded_list)
G.append(k)
k = [bounded_list for i in range(len(list_normal))]
for i in range(len(list_group)):
k.insert(i, bounded_list - 1)
G.append(k)
G = matrix(np.array(G))
H = np.zeros(2 * len(meanvec) + 2)
h = matrix(H, tc="d")
A = (matrix(meanvec)).trans()
b = matrix([1], (1, 1), tc="d")
# print('G',G)
# print('h',h)
# print('A',A)
# print('b',b)
sol = (solvers.qp(P, q, G, h, A, b))["x"]
solution = [x for x in sol]
sum = 0
for i in range(len(solution)):
sum += solution[i]
optimizedWeigh = [x / sum for x in solution]
print(optimizedWeigh)
merge = []
counter = 0
for columnName, columnData in df2.iteritems():
# print(real[counter])
if len(merge) == 0:
merge = df2[columnName] * optimizedWeigh[counter]
else:
merge = merge + df2[columnName] * optimizedWeigh[counter]
counter += 1
# print(merge)
print("dd,", it.max_drawdown(booksize=booksize, returnSeries=merge))
print("sharpe,", it.calculateSharpe(merge))
merge = merge * 10
# print('value',merge)
# merge.to_csv(r'/home/hoainam/PycharmProjects/multi_strategy/v_multi/f1m.csv')
# print(np.cumsum(merge))
plt.plot(np.cumsum(merge))
plt.grid(True)
plt.legend(("old", "maxsharpe", "minDD"))
plt.show()
return optimizedWeigh
# # TEST 1
trad1 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, train_1, 10**3, 0.16, 0.36, test_1
)
Gauss1 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_G1, 10**3, 0.16, 0.36, test_1
)
Gauss1_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G1, 10**3, 0.16, 0.36, test_1
)
Clay1 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_C1, 10**3, 0.16, 0.36, test_1
)
Clay1_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_C1, 10**3, 0.16, 0.36, test_1
)
# # TEST 2
trad2 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, train_2, 10**3, 0.16, 0.36, test_2
)
Gauss2 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_G2, 10**3, 0.16, 0.36, test_2
)
Gauss2_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G2, 10**3, 0.16, 0.36, test_2
)
Clay2 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_C2, 10**3, 0.16, 0.36, test_2
)
Clay2_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_C2, 10**3, 0.16, 0.36, test_2
)
# # TEST 3
trad3 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, train_3, 10**3, 0.16, 0.36, test_3
)
Gauss3 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_G3, 10**3, 0.16, 0.36, test_3
)
Gauss3_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G3, 10**3, 0.16, 0.36, test_3
)
Clay3 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_C3, 10**3, 0.16, 0.36, test_3
)
Clay3_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_C3, 10**3, 0.16, 0.36, test_3
)
# # TEST 4
trad4 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, train_4, 10**3, 0.16, 0.36, test_4
)
Gauss4 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_G4, 10**3, 0.16, 0.36, test_4
)
Gauss4_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G4, 10**3, 0.16, 0.36, test_4
)
Clay4 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_C4, 10**3, 0.16, 0.36, test_4
)
Clay4_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G4, 10**3, 0.16, 0.36, test_4
)
# # TEST 5
trad5 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, train_5, 10**3, 0.16, 0.36, test_5
)
Gauss5 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_G5, 10**3, 0.16, 0.36, test_5
)
Gauss5_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G5, 10**3, 0.16, 0.36, test_5
)
Clay5 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_C5, 10**3, 0.16, 0.36, test_5
)
Clay5_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G5, 10**3, 0.16, 0.36, test_5
)
# # TEST 6
trad6 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, train_6, 10**3, 0.16, 0.36, test_6
)
Gauss6 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_G6, 10**3, 0.16, 0.36, test_6
)
Gauss6_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G6, 10**3, 0.16, 0.36, test_6
)
Clay6 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_C6, 10**3, 0.16, 0.36, test_6
)
Clay6_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G6, 10**3, 0.16, 0.36, test_6
)
# # TEST 7
trad7 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, train_7, 10**3, 0.16, 0.36, test_7
)
Gauss7 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_G7, 10**3, 0.16, 0.36, test_7
)
Gauss7_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G7, 10**3, 0.16, 0.36, test_7
)
Clay7 = allocate_weight_sharp_with_bounded_group_alpha(
list_group, list_nomal, simul_C7, 10**3, 0.16, 0.36, test_7
)
Clay7_std = allocate_weight_sharp_with_bounded_group_alpha_std(
list_group, list_nomal, simul_G7, 10**3, 0.16, 0.36, test_7
)
# # Port Weight
port1 = {
"trad": trad1,
"Clay": Clay1,
"Gaus": Gauss1,
"Std Clay": Clay1_std,
"Std Gaus": Gauss1_std,
}
port1 = pd.DataFrame(port1)
port1.index = m.columns
port2 = {
"trad": trad2,
"Clay": Clay2,
"Gaus": Gauss2,
"Std Clay": Clay2_std,
"Std Gaus": Gauss2_std,
}
port2 = pd.DataFrame(port2)
port2.index = m.columns
port3 = {
"trad": trad3,
"Clay": Clay3,
"Gaus": Gauss3,
"Std Clay": Clay3_std,
"Std Gaus": Gauss3_std,
}
port3 = pd.DataFrame(port3)
port3.index = m.columns
port4 = {
"trad": trad4,
"Clay": Clay4,
"Gaus": Gauss4,
"Std Clay": Clay4_std,
"Std Gaus": Gauss4_std,
}
port4 = pd.DataFrame(port4)
port4.index = m.columns
port5 = {
"trad": trad5,
"Clay": Clay5,
"Gaus": Gauss5,
"Std Clay": Clay5_std,
"Std Gaus": Gauss5_std,
}
port5 = pd.DataFrame(port5)
port5.index = m.columns
port6 = {
"trad": trad6,
"Clay": Clay6,
"Gaus": Gauss6,
"Std Clay": Clay6_std,
"Std Gaus": Gauss6_std,
}
port6 = pd.DataFrame(port6)
port6.index = m.columns
port7 = {
"trad": trad7,
"Clay": Clay7,
"Gaus": Gauss7,
"Std Clay": Clay7_std,
"Std Gaus": Gauss7_std,
}
port7 = pd.DataFrame(port7)
port7.index = m.columns
port1.to_csv("Port_test_1.csv")
port2.to_csv("Port_test_2.csv")
port3.to_csv("Port_test_3.csv")
port4.to_csv("Port_test_4.csv")
port5.to_csv("Port_test_5.csv")
port6.to_csv("Port_test_6.csv")
port7.to_csv("Port_test_7.csv")
# port8.to_csv('Port_test_8.csv')
# # FRONTIER PLOT
def frontier_plot(df, meanvec, cov_mat, port):
# define list
# cov_matrix = (dataframe.cov())
# meanvec = (dataframe.mean())
np.random.seed(42)
num_ports = 10000
num_alpha = df.shape[1]
all_weights = np.zeros((num_ports, num_alpha))
ret_arr = np.zeros(num_ports)
vol_arr = np.zeros(num_ports)
sharpe_arr = np.zeros(num_ports)
for x in range(num_ports):
weights = np.array(np.random.random(num_alpha))
weights = weights / np.sum(weights)
# Save weights
all_weights[x, :] = weights
# Expected return
ret_arr[x] = np.sum((meanvec * weights * 252))
# Expected volatility
vol_arr[x] = np.sqrt(np.dot(weights.T, np.dot(cov_mat * 252, weights)))
# Sharpe Ratio
sharpe_arr[x] = ret_arr[x] / vol_arr[x]
tradition_weight = port["trad"]
clayton_weight = port["Clay"]
gauss_weight = port["Gaus"]
std_clay_w = port["Std Clay"]
std_gaus_w = port["Std Gaus"]
# Volatility
max_trad_sr_vol = np.sqrt(
np.dot(tradition_weight.T, np.dot(cov_mat * 252, tradition_weight))
)
max_clayton_sr_vol = np.sqrt(
np.dot(clayton_weight.T, np.dot(cov_mat * 252, clayton_weight))
)
max_gauss_sr_vol = np.sqrt(
np.dot(gauss_weight.T, np.dot(cov_mat * 252, gauss_weight))
)
max_clayton_std_vol = np.sqrt(
np.dot(std_clay_w.T, np.dot(cov_mat * 252, std_clay_w))
)
max_gaus_std_vol = np.sqrt(np.dot(std_gaus_w.T, np.dot(cov_mat * 252, std_gaus_w)))
# Return
max__trad_sr_ret = np.sum((meanvec * tradition_weight * 252))
max__clayton_sr_ret = np.sum((meanvec * clayton_weight * 252))
max__gauss_sr_ret = np.sum((meanvec * gauss_weight * 252))
max__clayton_std_ret = np.sum((meanvec * std_clay_w * 252))
max__gaus_std_ret = np.sum((meanvec * std_gaus_w * 252))
real_sr_ret = ret_arr[sharpe_arr.argmax()]
real_sr_vol = vol_arr[sharpe_arr.argmax()]
print("The Traditional Sharpe : ", max__trad_sr_ret / max_trad_sr_vol)
print("The Clayton Sharpe : ", max__clayton_sr_ret / max_clayton_sr_vol)
print("The Gauss Sharpe : ", max__gauss_sr_ret / max_gauss_sr_vol)
print("The STD Clayton Sharpe : ", max__clayton_std_ret / max_clayton_std_vol)
print("The STD Gauss Sharpe : ", max__gaus_std_ret / max_gaus_std_vol)
print("The Real Sharpe :", real_sr_ret / real_sr_vol)
plt.figure(figsize=(12, 8))
plt.scatter(vol_arr, ret_arr, c=sharpe_arr, cmap="viridis")
plt.colorbar(label="Sharpe Ratio")
plt.xlabel("Volatility")
plt.ylabel("Return")
plt.scatter(max_trad_sr_vol, max__trad_sr_ret, c="red", s=50) # red dot
plt.scatter(max_clayton_sr_vol, max__clayton_sr_ret, c="pink", s=50)
plt.scatter(max_gauss_sr_vol, max__gauss_sr_ret, c="pink", s=50)
plt.scatter(max_clayton_sr_vol, max__clayton_sr_ret, c="orange", s=50)
plt.scatter(max_gauss_sr_vol, max__gauss_sr_ret, c="orange", s=50)
plt.scatter(real_sr_vol, real_sr_ret, c="black", s=50) # black dot
plt.show()
frontier_plot(test_1, test_1.mean(), test_1.cov(), port1)
frontier_plot(test_2, test_2.mean(), test_2.cov(), port2)
frontier_plot(test_3, test_3.mean(), test_3.cov(), port3)
frontier_plot(test_4, test_4.mean(), test_4.cov(), port4)
frontier_plot(test_5, test_5.mean(), test_5.cov(), port5)
frontier_plot(test_6, test_6.mean(), test_6.cov(), port6)
frontier_plot(test_7, test_7.mean(), test_7.cov(), port7)
| false | 7 | 8,981 | 0 | 9,012 | 8,981 |
||
69476385
|
# # Web Scraping From Daraz
# Using webdriver_manager to manage the Chrome Driver for Selenium
import re
import pandas as pd
from selenium import webdriver
from webdriver_manager.chrome import ChromeDriverManager
from selenium.common.exceptions import NoSuchElementException
stars_value = {
"c3dn4k c1dtTC": 0,
"c3dn4k c1Zozd": 0.1,
"c3dn4k cbDGcO": 0.2,
"c3dn4k c3fsPU": 0.3,
"c3dn4k c1e2gb": 0.4,
"c3dn4k c3An30": 0.5,
"c3dn4k c3DcGB": 0.6,
"c3dn4k c1wCjy": 0.7,
"c3dn4k c17YMy": 0.8,
"c3dn4k cF1vkb": 0.9,
"c3dn4k c3EEAg": 1,
}
Web_Data = pd.DataFrame(
[],
columns=[
"Name",
"Daraz Mall?",
"Original Price",
"Discounted Price",
"Rating",
"Reviews",
"Seller Country",
],
)
product = input("Enter Product Name : ").replace(" ", "+")
daraz = "https://www.daraz.pk/catalog/?q={}".format(product)
print("URL : ", daraz, "\n")
no_of_product = int(input("Enter no.to take Top Level Averages : "))
driver = webdriver.Chrome(ChromeDriverManager().install())
driver.get(daraz)
total_sellers = driver.find_element_by_class_name("c1DXz4").text.split(" ")[0]
index = 0
for product in driver.find_elements_by_class_name("c2prKC"):
print("*", end="")
name = product.find_element_by_class_name("c16H9d").text
prize = int(product.find_element_by_class_name("c3gUW0").text[3:].replace(",", ""))
country = product.find_element_by_class_name("c2i43-").text
rating = 0
try:
stars = product.find_element_by_class_name("c15YQ9").get_attribute("innerHTML")
for s in re.findall('(c3dn4k c.+?)"', stars):
rating += stars_value[s]
except NoSuchElementException:
rating = 0
daraz_mall = 0
mall = product.find_element_by_class_name("c3vCyH").get_attribute("innerHTML")
if len(mall) > 0:
daraz_mall = "Yes"
else:
daraz_mall = "No"
reviews = 0
try:
reviews = int(product.find_element_by_class_name("c3XbGJ").text[1:-1])
except NoSuchElementException:
reviews = 0
discount_prize = 0
try:
discount_prize = int(
product.find_element_by_class_name("c1-B2V").text[3:].replace(",", "")
)
except NoSuchElementException:
discount_prize = 0
if discount_prize == 0:
Web_Data.loc[index] = [
name,
daraz_mall,
prize,
discount_prize,
rating,
reviews,
country,
]
else:
Web_Data.loc[index] = [
name,
daraz_mall,
discount_prize,
prize,
rating,
reviews,
country,
]
index += 1
print("Web Scraping Ended :)")
driver.quit()
print("Total Seller :", total_sellers)
print("Avg Price :", Web_Data["Original Price"][:no_of_product].mean())
print("Avg Reviews :", Web_Data["Reviews"][:no_of_product].mean())
print("Avg Rating :", Web_Data["Rating"][:no_of_product].mean())
print("Total Product Present in DataFrame : ", Web_Data.shape[0])
Web_Data.head(5)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/476/69476385.ipynb
| null | null |
[{"Id": 69476385, "ScriptId": 18971349, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7863466, "CreationDate": "07/31/2021 13:27:32", "VersionNumber": 1.0, "Title": "Web-Scraping-From-Daraz", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 90.0, "LinesInsertedFromPrevious": 90.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
| null | null | null | null |
# # Web Scraping From Daraz
# Using webdriver_manager to manage the Chrome Driver for Selenium
import re
import pandas as pd
from selenium import webdriver
from webdriver_manager.chrome import ChromeDriverManager
from selenium.common.exceptions import NoSuchElementException
stars_value = {
"c3dn4k c1dtTC": 0,
"c3dn4k c1Zozd": 0.1,
"c3dn4k cbDGcO": 0.2,
"c3dn4k c3fsPU": 0.3,
"c3dn4k c1e2gb": 0.4,
"c3dn4k c3An30": 0.5,
"c3dn4k c3DcGB": 0.6,
"c3dn4k c1wCjy": 0.7,
"c3dn4k c17YMy": 0.8,
"c3dn4k cF1vkb": 0.9,
"c3dn4k c3EEAg": 1,
}
Web_Data = pd.DataFrame(
[],
columns=[
"Name",
"Daraz Mall?",
"Original Price",
"Discounted Price",
"Rating",
"Reviews",
"Seller Country",
],
)
product = input("Enter Product Name : ").replace(" ", "+")
daraz = "https://www.daraz.pk/catalog/?q={}".format(product)
print("URL : ", daraz, "\n")
no_of_product = int(input("Enter no.to take Top Level Averages : "))
driver = webdriver.Chrome(ChromeDriverManager().install())
driver.get(daraz)
total_sellers = driver.find_element_by_class_name("c1DXz4").text.split(" ")[0]
index = 0
for product in driver.find_elements_by_class_name("c2prKC"):
print("*", end="")
name = product.find_element_by_class_name("c16H9d").text
prize = int(product.find_element_by_class_name("c3gUW0").text[3:].replace(",", ""))
country = product.find_element_by_class_name("c2i43-").text
rating = 0
try:
stars = product.find_element_by_class_name("c15YQ9").get_attribute("innerHTML")
for s in re.findall('(c3dn4k c.+?)"', stars):
rating += stars_value[s]
except NoSuchElementException:
rating = 0
daraz_mall = 0
mall = product.find_element_by_class_name("c3vCyH").get_attribute("innerHTML")
if len(mall) > 0:
daraz_mall = "Yes"
else:
daraz_mall = "No"
reviews = 0
try:
reviews = int(product.find_element_by_class_name("c3XbGJ").text[1:-1])
except NoSuchElementException:
reviews = 0
discount_prize = 0
try:
discount_prize = int(
product.find_element_by_class_name("c1-B2V").text[3:].replace(",", "")
)
except NoSuchElementException:
discount_prize = 0
if discount_prize == 0:
Web_Data.loc[index] = [
name,
daraz_mall,
prize,
discount_prize,
rating,
reviews,
country,
]
else:
Web_Data.loc[index] = [
name,
daraz_mall,
discount_prize,
prize,
rating,
reviews,
country,
]
index += 1
print("Web Scraping Ended :)")
driver.quit()
print("Total Seller :", total_sellers)
print("Avg Price :", Web_Data["Original Price"][:no_of_product].mean())
print("Avg Reviews :", Web_Data["Reviews"][:no_of_product].mean())
print("Avg Rating :", Web_Data["Rating"][:no_of_product].mean())
print("Total Product Present in DataFrame : ", Web_Data.shape[0])
Web_Data.head(5)
| false | 0 | 1,013 | 2 | 1,013 | 1,013 |
||
69476583
|
# numpy and pandas for data manipulation
import numpy as np
import pandas as pd
# sklearn preprocessing for dealing with categorical variables
from sklearn.preprocessing import LabelEncoder
# File system manangement
import os
# Suppress warnings
import warnings
warnings.filterwarnings("ignore")
# matplotlib and seaborn for plotting
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# see train data
train_data = pd.read_csv("../input/home-credit-default-risk/application_train.csv")
train_data.head()
# targetかどうかを予想する
# see train data
test_data = pd.read_csv("../input/home-credit-default-risk/application_test.csv")
test_data.head()
# 過去の再建履歴
bureau_balance = pd.read_csv("../input/home-credit-default-risk/bureau_balance.csv")
bureau_balance.head()
# 過去の再建履歴
bureau = pd.read_csv("../input/home-credit-default-risk/bureau.csv")
bureau.head()
# 過去の再建履歴
pos_balance = pd.read_csv("../input/home-credit-default-risk/POS_CASH_balance.csv")
pos_balance.head()
# 過去の再建履歴
pte_appli = pd.read_csv("../input/home-credit-default-risk/previous_application.csv")
pte_appli.head()
# 過去の再建履歴
installments_pay = pd.read_csv(
"../input/home-credit-default-risk/installments_payments.csv"
)
installments_pay.head()
# 参照Note
# https://www.kaggle.com/willkoehrsen/start-here-a-gentle-introduction
# 以下で行う分析の流れ
# 問題とデータの理解
# データのクリーニングとフォーマット(この作業はほとんど自分たちで行いました
# 探索的データ分析
# ベースラインモデル
# モデルの改良
# モデルの解釈
# モデルの正確性について
# モデルの予測結果の正確性を可視化する図表としてROC曲線,AUCというものがある。
# Reciever Operating Characteristic (ROC) curve
# 
# 線が左上にあるほど陽性と判断したものの内本当に要請だった正解率(True Positive)が高く、不正解率(False Positive)が低い、良いモデルといえる。例では青>赤>黒の順でいいモデル。
# Area Under the Curve (AUC)はその名の通りROC曲線の下にできたエリアを指している。
# AUCは0から1までの値をとり、値が1に近いほど判別能(判別に使用しているモデル)が良いことを示す。
# precision(適合率)=TP/(TP+FP):陽性の内どのくらいの割合を検知できたか
# recall(再現率)=TP/(TP+FN):陽性と判断したものの内どの程度が本当に陽性か
# precision(適合率)とrecall(再現率)はトレードオフの関係
# 誤検知(再現率)を高くすると見逃す確率が高くなる(適合率が下がる)
# 適合率と再現率のどちらを重視するかは具体的に何を判断するかで毎回変える必要がある
# 探索的データ解析:Exploratory Data Analysis(EDA)
# 統計分析や可視化を行うことでデータに対する理解を深める。
# 傾向、異常値、パターン、相関関係などの特徴をあぶりだす。
# カラムの説明確認用
col_discription = pd.read_csv(
"../input/home-credit-default-risk/HomeCredit_columns_description.csv"
)
col_discription.loc[col_discription.Row == "EXT_SOURCE"]
# まずは目的となる指標の概要から確認する
# 0:返済可能者
# 1:返済不可能者
train_data["TARGET"].value_counts()
# クラス不均衡問題とは?
# 機械学習において、あるクラスのデータ(ポジティブ)の総数が、別のクラスのデータ(ネガティブ)の総数よりも圧倒的に少ないという問題のことです。この問題は実際には極めて一般的で、不正検知、異常検知、医療診断、油流出検知、顔認識など様々な分野で観察されます。
# なぜ問題なのか?
# 機械学習ではtargetの一致率を高めるために学習を行う。
# しかし適合率と再現率のトレードオフにあるように一概にtargetの精度を高くすればいいいというものではない。
# 陽性、陰性のレコード数が同じくらいならばそれぞれの誤検知も同じ重さで扱われる。
# 今回のように一方に偏りがある場合、偏っている方の誤検知件数が多くなるのでそれを補正しようとして少数のクラスの誤検知精度がないがしろにされる可能性がある。
# 以下で例を挙げる
# 取引データのデータセットが与えられたとき、どれが不正でどれが本物かを調べたいとします。電子商取引企業にとって、不正な取引が行われることは、お客様の信頼を損ない、コストがかかることになります。そのため、できるだけ多くの不正取引を見つけたいと考えています。
# 10000件の真正な取引と10件の不正な取引からなるデータセットがあった場合、分類器は不正な取引を真正な取引として分類する傾向があります。その理由は、数字で簡単に説明できます。機械学習アルゴリズムが以下のような2つの可能性のある出力を持っているとします。
# モデル1は、10件の不正な取引のうち7件を真正な取引と分類し、10000件の真正な取引のうち10件を不正な取引と分類した。
# モデル2は,10件の不正取引のうち2件を真正な取引と分類し,10000件の真正な取引のうち100件を不正な取引と分類した.
# 分類器の性能がミスの数で決まるとすれば,モデル2が102個のミスをしたのに対し,モデル1は合計17個のミスしかしていないので,明らかにモデル1の方が優れています.しかし、私たちは不正取引の発生を最小限に抑えたいので、不正取引の分類で2回しかミスをしなかったモデル2を代わりに選ぶべきです。もちろん、これにより、より多くの真正な取引が不正な取引として分類されるという犠牲を払うことになりますが、今のところは我慢できるコストでしょう。いずれにしても、一般的な機械学習アルゴリズムでは、モデル2よりもモデル1を選んでしまうことになり、問題となります。実際には、モデル2を使用すれば阻止できたはずの不正取引を、大量に通過させてしまうことになります。これは、お客様の不幸と会社のお金の損失につながります。
# 元サイト
# http://www.chioka.in/class-imbalance-problem/
# www.DeepL.com/Translator(無料版)で翻訳しました。
# null値などデータ欠損の確認
# Function to calculate missing values by column# Funct
def missing_values_table(df):
# Total missing values
mis_val = df.isnull().sum()
# Percentage of missing values
mis_val_percent = 100 * df.isnull().sum() / len(df)
# Make a table with the results
mis_val_table = pd.concat([mis_val, mis_val_percent], axis=1)
# Rename the columns
mis_val_table_ren_columns = mis_val_table.rename(
columns={0: "Missing Values", 1: "% of Total Values"}
)
# iloc[行, 列]を指定して抜き出し
# 欠損がないカラムを除外
# 欠損率が高い順(降順)で並び替え
# Sort the table by percentage of missing descending
mis_val_table_ren_columns = (
mis_val_table_ren_columns[mis_val_table_ren_columns.iloc[:, 1] != 0]
.sort_values("% of Total Values", ascending=False)
.round(1)
)
# Print some summary information
print(
"Your selected dataframe has " + str(df.shape[1]) + " columns.\n"
"There are "
+ str(mis_val_table_ren_columns.shape[0])
+ " columns that have missing values."
)
# Return the dataframe with missing information
return mis_val_table_ren_columns
missing_values = missing_values_table(train_data)
missing_values.head(20)
# ここで可視化される欠損値のあるカラムは後々欠損値を埋める(imputation)必要があるかもしれない
train_data.dtypes.value_counts()
# データタイプの確認
# float int は数値型
# objectは文字型などが混入
# オブジェクトカラムのユニークな値の個数を表示
# 文字情報の記入なのか何らかのクラス分けした情報が入っているカラムなのかの確認
train_data.select_dtypes("object").apply(pd.Series.nunique, axis=0)
# カテゴリ変数の処理について
# カテゴリ変数はそのままでは機械学習のモデルで処理できない
# Label encoding
# 要素クラスに対し1~ 数字を振る。
# 要素どおしに数字の大きさをつけることになるので意図しない重みづけになる可能性がある
# 所得に応じた階級などの場合ラベルエンコーディングを使える
# 職業などの場合はしたのOneーHotエンコーディングを使用した方がいい
# ※2つしかクラスがない場合は0.1で処理できる。
# One-hot encoding
# 要素クラス一つ一つに対し列を作り1,0でフラグを立てて処理する
# カテゴリーが多いカテゴリー変数では、素性(データの次元)の数が爆発的に増える。
# PCAや他の次元削減法を用いて次元数を減らすことができます(ただし、情報の保持には注意が必要)
# 2クラス以下のカラムの処理
# sklernのlabelencoerを使用する
le = LabelEncoder()
le_count = 0
# Iterate through the columns
for col in train_data:
if train_data[col].dtype == "object":
# If 2 or fewer unique categories
if len(list(train_data[col].unique())) <= 2:
# ラベルとラベルIDの対応づけ.positiveは0にしよう,みたいなことを決める
le.fit(train_data[col])
# Transform both training and testing data 返り値を用意しないといけない
train_data[col] = le.transform(train_data[col])
test_data[col] = le.transform(test_data[col])
# Keep track of how many columns were label encoded
le_count += 1
print("%d columns were label encoded." % le_count)
# one-hotエンコーディングを行うためのget_dummies関数について
df = pd.DataFrame(
{
"sex": ["female", np.nan, "male", "male", "female", "male"],
"old": [12, 21, 31, 10, 22, 10],
}
)
df.iloc[:, :]
test_df = pd.get_dummies(df)
test_df.iloc[:, :]
# 文字型(objectカラムのみ変換される)
# get_dummies():カテゴリ変数をダミー変数の作成
# デフォルトでは「文字列」など dtype が object の全てのカラムを「ダミー変数」に変換する
# 上記で変換していないカテゴリ変数に対してone-hotエンコーディング処理
train_data = pd.get_dummies(train_data)
test_data = pd.get_dummies(test_data)
print("Training Features shape: ", train_data.shape)
print("Testing Features shape: ", test_data.shape)
# トレーニングデータとテストデータのカラム数に違いが発生
# これはカテゴリ変数に含まれていたクラスの違いによるもの
# テストデータにないカラムを使う必要はないためトレインデータから削除する
#
# カラムをそろえる関数alignの説明
d1 = {"name": ["半沢", "大和田", "中野渡"], "code": ["101", "102", "104"]}
df1 = pd.DataFrame(data=d1)
df1
d2 = {"code": ["101", "102", "103"], "place": ["東京", "大阪", "名古屋"]}
df2 = pd.DataFrame(data=d2, index=[1, 2, 3])
df2
# alignの返り値は2つになる
df3, df4 = df1.align(df2, join="inner", axis=1)
df3
# TARGETカラムはテストデータにないが削除してはいけないカラムなので一度変数に格納しておいて後から再度追加する
train_labels = train_data["TARGET"]
# Align the training and testing data, keep only columns present in both dataframes
train_data, test_data = train_data.align(test_data, join="inner", axis=1)
# Add the target back in
train_data["TARGET"] = train_labels
print("Training Features shape: ", train_data.shape)
print("Testing Features shape: ", test_data.shape)
# データの異常値に対する処理
# DAYS_BIRTHカラムについて
# 現在のローン申請に対しての数値が格納されているため値がマイナスになっている。
# 値を正に直し単位を年にする処理を行う
# 年に直すことで入っている数値=年齢となり妥当な数値が入っているかが判断できる
train_data["DAYS_EMPLOYED"].describe()
# マイナスとプラスが混合している
# maxの数が大きすぎて1000年となっている
# などから正しい数値が入っていないよう。より詳細にデータを見る必要がある。
train_data["DAYS_EMPLOYED"].plot.hist(title="Days Employment Histogram")
plt.xlabel("Days Employment")
# 異常値も5万レコードあることからnullの代わりに入れられている数値の可能性あり。
# null値であることも一つの特徴量である可能性があるのでnull値かそうでないかでTARGETに対して影響を与えているかを確認する。
anom = train_data[train_data["DAYS_EMPLOYED"] == 365243]
non_anom = train_data[train_data["DAYS_EMPLOYED"] != 365243]
print(
"The non-anomalies default on %0.2f%% of loans" % (100 * non_anom["TARGET"].mean())
)
print("The anomalies default on %0.2f%% of loans" % (100 * anom["TARGET"].mean()))
print("There are %d anomalous days of employment" % len(anom))
# 3%以上の数値の乖離がある。
# 異常値の対応として最も安全な方法は異常値をnullとして処理をしてしまうこと。
# 今回の異常値はすべて同じ値であり、TARGETへの影響も考えられる。
# nullに上書きしたうえで異常値であったことを学習できるように新たなカラムを追加する。
#
# 異常値かどうかのフラグを格納するDAYS_EMPLOYED_ANOMを追加
train_data["DAYS_EMPLOYED_ANOM"] = train_data["DAYS_EMPLOYED"] == 365243
# 異常値をnullで置き換える implace=trueにすると元のDFが変換される
train_data["DAYS_EMPLOYED"].replace({365243: np.nan}, inplace=True)
train_data["DAYS_EMPLOYED"].plot.hist(title="Days Employment Histogram")
plt.xlabel("Days Employment")
# テストデータにも同様の処理をおこなう
test_data["DAYS_EMPLOYED_ANOM"] = test_data["DAYS_EMPLOYED"] == 365243
test_data["DAYS_EMPLOYED"].replace({365243: np.nan}, inplace=True)
print(
"There are %d anomalies in the test data out of %d entries"
% (test_data["DAYS_EMPLOYED_ANOM"].sum(), len(test_data))
)
# 相関係数
# * .00-.19 “very weak”
# * .20-.39 “weak”
# * .40-.59 “moderate”
# * .60-.79 “strong”
# * .80-1.0 “very strong”
# Find correlations with the target and sort
correlations = train_data.corr()["TARGET"].sort_values()
# Display correlations
print("Most Positive Correlations:\n", correlations.tail(15))
print("\nMost Negative Correlations:\n", correlations.head(15))
# 相関係数が高いDAYS_BIRTHについて分析
# days_birthはマイナス数値が格納されているので絶対値に修正して絶対値を再計算
train_data["DAYS_BIRTH"] = abs(train_data["DAYS_BIRTH"])
train_data["DAYS_BIRTH"].corr(train_data["TARGET"])
# 年齢が上昇するほど返済率は高くなる。
# テンプレ図表の形で図表を作成
plt.style.use("fivethirtyeight")
# 年齢ヒストグラム
plt.hist(train_data["DAYS_BIRTH"] / 365, edgecolor="k", bins=25)
plt.title("Age of Client")
plt.xlabel("Age (years)")
plt.ylabel("Count")
# 漠然と年齢ヒストグラムを表示しても外れ値がないかどうかくらいしかわからない
plt.figure(figsize=(10, 8))
# KDE plot of loans that were repaid on time
sns.kdeplot(
train_data.loc[train_data["TARGET"] == 0, "DAYS_BIRTH"] / 365, label="target == 0"
)
# KDE plot of loans which were not repaid on time
sns.kdeplot(
train_data.loc[train_data["TARGET"] == 1, "DAYS_BIRTH"] / 365, label="target == 1"
)
# Labeling of plot
plt.xlabel("Age (years)")
plt.ylabel("Density")
plt.title("Distribution of Ages")
# TARGETの値別に分布を表示することでどのような特徴が存在するのかを可視化する
# 年齢が高くなるにつれて返済率が高くなることは分かったがどの年齢層に特徴があるのかなどを分析するため
# 若年層(30代)で非返済が高いことが見て取れる。
# カラム全体としては相関係数は優位とは言えなかった。
# 実際に非返済者の予測に使用できるのか実際のデータを年齢層別にチェックする。
# 特徴を調べる'TARGET', 'DAYS_BIRTH'のデータのみ取り出す
age_data = train_data[["TARGET", "DAYS_BIRTH"]]
age_data["YEARS_BIRTH"] = age_data["DAYS_BIRTH"] / 365
# 20~70歳について11にわけることで5歳(50/10=5)ごとの階級を作成する
age_data["YEARS_BINNED"] = pd.cut(
age_data["YEARS_BIRTH"], bins=np.linspace(20, 70, num=11)
)
age_data.head(10)
# 5歳ごとの階級分けになっているかの確認 YEARS_BINNEDを確認
# 上記ではTARGET DAYS_BIRTH YEARS_BIRTHはランダムなレコードの値が出ていた。
# groupbyで平均をとって集計をして表示する
age_groups = age_data.groupby("YEARS_BINNED").mean()
age_groups
plt.figure(figsize=(8, 8))
# 上記で作成した表をグラフ化 100分率にして%で表示
plt.bar(age_groups.index.astype(str), 100 * age_groups["TARGET"])
# Plot labeling
plt.xticks(rotation=75)
plt.xlabel("Age Group (years)")
plt.ylabel("Failure to Repay (%)")
plt.title("Failure to Repay by Age Group")
# 明らかに若い世代ほど非返済率が高いことが分かった。
# 負の相関のあったExterior Sourcesについて考える
# Exterior Sources : Normalized score from external data source
# 標準化した外部データ(内容は不明)
# 上処理の文字列処理によって生まれたカラムではない。それぞれのカラムに関係があるかもわからない。
# 内部データが不明なのに分析に使用していいのか?
#
# Extract the EXT_SOURCE variables and show correlations
ext_data = train_data[
["TARGET", "EXT_SOURCE_1", "EXT_SOURCE_2", "EXT_SOURCE_3", "DAYS_BIRTH"]
]
ext_data_corrs = ext_data.corr()
ext_data_corrs
# 相関係数の可視化(ヒートマップで色を付ける)
plt.figure(figsize=(8, 6))
# Heatmap of correlations
sns.heatmap(ext_data_corrs, cmap=plt.cm.RdYlBu_r, vmin=-0.25, annot=True, vmax=0.6)
plt.title("Correlation Heatmap")
plt.figure(figsize=(10, 12))
# iterate through the sources
for i, source in enumerate(["EXT_SOURCE_1", "EXT_SOURCE_2", "EXT_SOURCE_3"]):
# create a new subplot for each source
plt.subplot(3, 1, i + 1)
# plot repaid loans
sns.kdeplot(train_data.loc[train_data["TARGET"] == 0, source], label="target == 0")
# plot loans that were not repaid
sns.kdeplot(train_data.loc[train_data["TARGET"] == 1, source], label="target == 1")
# Label the plots
plt.title("Distribution of %s by Target Value" % source)
plt.xlabel("%s" % source)
plt.ylabel("Density")
plt.tight_layout(h_pad=2.5)
# EXT_SOURCE_3はTARGET1と0で山の位置が他よりも明確に分かれている。
# 相関としては弱いが学習データには利用することが可能。
# 最後に探索的プロットとしてペアプロット(行列散布図)を行う。
# pairplotはseabornで最も使用されている機能の一つ。
# 大量の変数列があるデータに対し、全ての変数の組み合わせ毎に相関関係を見るためのプロット
# 
# 上記で調べたデータでペアプロットを作成するためデータをコピー
plot_data = ext_data.drop(columns=["DAYS_BIRTH"]).copy()
# 生年月日の列を追加
plot_data["YEARS_BIRTH"] = age_data["YEARS_BIRTH"]
# プロットデータを制限 多すぎると散布図が塗りつぶされみづらくなる
plot_data = plot_data.dropna().loc[:100000, :]
# 相関係数を計算する関数
def corr_func(x, y, **kwargs):
r = np.corrcoef(x, y)[0][1]
ax = plt.gca() # figureと同じような意味
# 注釈をつける
ax.annotate("r = {:.2f}".format(r), xy=(0.2, 0.8), xycoords=ax.transAxes, size=20)
# Create the pairgrid object
grid = sns.PairGrid(
data=plot_data,
size=3,
diag_sharey=False,
hue="TARGET", # 図示される目的変数
vars=[x for x in list(plot_data.columns) if x != "TARGET"],
)
# 右上側は散布図
grid.map_upper(plt.scatter, alpha=0.2)
# 真ん中はヒストグラム
grid.map_diag(sns.kdeplot)
# Bottom is density plot
grid.map_lower(sns.kdeplot, cmap=plt.cm.OrRd_r)
plt.suptitle("Ext Source and Age Features Pairs Plot", size=32, y=1.05)
# 赤は返済されなかったローン、青は返済されたローンを示す
# Kaggleでは最も有用な特徴を見つけることができたものが勝つ
# 少なくとも構造化データの場合、優勝モデルはすべて勾配ブーストのバリエーション(XGBoostやLightGBM、CatBoost)
# 機械学習のパターンの1つである、モデル構築やハイパーパラメータ調整よりも、特徴量エンジニアリングの方が投資対効果が高いことを表している。
# 参照記事:https://community.alteryx.com/t5/Data-Science/Feature-Engineering-Secret-to-Data-Science-Success/ba-p/545041
# フィーチャーエンジニアリングとは、既存のデータから新たな特徴を追加する「**特徴構築**」と、最も重要な特徴のみを選択する「**特徴選択**」、またはその他の次元削減方法の両方を含む、遺伝的なプロセスを指す。
# 他のデータソースを使い始めると、多くの特徴量エンジニアリングを行うことになりますが、このノートブックでは、2つの簡単な特徴量構築法を試してみます。
# >多項式特徴
# >ドメイン知識特徴量
# 多項式特徴量(Polynomial Features)
# 既存の特徴のべき乗や、既存の特徴間の相互作用項を特徴とする。
# 例えば、変数EXT_SOURCE_1^2とEXT_SOURCE_2^2の他に、
# EXT_SOURCE_1×EXT_SOURCE_2、EXT_SOURCE_1^2×EXT_SOURCE_2^2、EXT_SOURCE_1^2×EXT_SOURCE_2^2などの変数を作る
# 交互作用項は、複数の変数の影響を捉えるために統計モデルではよく使われますが、機械学習ではあまり見かけない
# Scikit-LearnにはPolynomialFeaturesという便利なクラスがあり、指定した次数までの多項式と相互作用項を作成する。
# ここでは次数を3にして結果を見る。(多項式特徴を作成する際には、特徴数が次数に応じて指数関数的に増加することと、オーバーフィッティングの問題が発生することから、あまり高い次数を使わないようにしたい。)
# 多項式特徴量分析用のデータを作成
poly_features = train_data[
["EXT_SOURCE_1", "EXT_SOURCE_2", "EXT_SOURCE_3", "DAYS_BIRTH", "TARGET"]
]
poly_features_test = test_data[
["EXT_SOURCE_1", "EXT_SOURCE_2", "EXT_SOURCE_3", "DAYS_BIRTH"]
]
# 欠損値の処理
# sklearn.preprocessingの Imputerはサポート終了予定なのでSimpleImputerを利用する
from sklearn.impute import SimpleImputer
imputer = SimpleImputer(missing_values=np.nan, strategy="median")
poly_target = poly_features["TARGET"]
poly_features = poly_features.drop(columns=["TARGET"])
# Need to impute missing values
poly_features = imputer.fit_transform(poly_features)
poly_features_test = imputer.transform(poly_features_test)
from sklearn.preprocessing import PolynomialFeatures
# 多項式特徴量分析 モデルの作成
poly_transformer = PolynomialFeatures(degree=3)
# Train the polynomial features
poly_transformer.fit(poly_features)
# Transform the features
poly_features = poly_transformer.transform(poly_features)
poly_features_test = poly_transformer.transform(poly_features_test)
print("Polynomial Features shape: ", poly_features.shape)
print("poly_features_test shape: ", poly_features_test.shape)
# 35の特徴量が新たに作成された。
# 作成された特徴量の確認
poly_transformer.get_feature_names(
input_features=["EXT_SOURCE_1", "EXT_SOURCE_2", "EXT_SOURCE_3", "DAYS_BIRTH"]
)[:15]
# 新しく作成した特徴量とTARGETカラムの相関係数を確認する
# Create a dataframe of the features
poly_features = pd.DataFrame(
poly_features,
columns=poly_transformer.get_feature_names(
["EXT_SOURCE_1", "EXT_SOURCE_2", "EXT_SOURCE_3", "DAYS_BIRTH"]
),
)
# Add in the target
poly_features["TARGET"] = poly_target
# Find the correlations with the target
poly_corrs = poly_features.corr()["TARGET"].sort_values()
# Display most negative and most positive
print(poly_corrs.head(10))
print("")
print("")
print("")
print(poly_corrs.tail(5))
# 何もしてない時より相関係数が大きくなっているものがある。
# 実際に元データ(のコピー)に追加して学習データとして使えるか確認する。
# 新しく作成した特徴量がいつも使えるとは限らない。実際に学習させて確認しないとわからないこともある。
# 特徴量を追加格納したDFを作成する
poly_features_test = pd.DataFrame(
poly_features_test,
columns=poly_transformer.get_feature_names(
["EXT_SOURCE_1", "EXT_SOURCE_2", "EXT_SOURCE_3", "DAYS_BIRTH"]
),
)
# 特徴量をトレーニングデータに格納する
poly_features["SK_ID_CURR"] = train_data["SK_ID_CURR"]
app_train_poly = train_data.merge(poly_features, on="SK_ID_CURR", how="left")
# 特徴量をテストデータに格納する
poly_features_test["SK_ID_CURR"] = test_data["SK_ID_CURR"]
app_test_poly = test_data.merge(poly_features_test, on="SK_ID_CURR", how="left")
# トレーニングデータとテストデータ共通するカラムのみ
app_train_poly, app_test_poly = app_train_poly.align(
app_test_poly, join="inner", axis=1
)
# データのサイズを表示
print("Training data with polynomial features shape: ", app_train_poly.shape)
print("Testing data with polynomial features shape: ", app_test_poly.shape)
# ドメイン・ナレッジの特徴
# そのデータの専門知識を利用して特徴量を新規作成しデータ分析を行う
# 今回新規作成してみる特徴量
# CREDIT_INCOME_PERCENT:顧客の収入に対する与信額の割合。
# ANNUITY_INCOME_PERCENT: 顧客の収入に対するローンの年金額の割合
# CREDIT_TERM: 月単位の支払期間(年金は月単位の支払額なので
# DAYS_EMPLOYED_PERCENT: クライアントの年齢に対する雇用日数の割合
app_train_domain = train_data.copy()
app_test_domain = test_data.copy()
# trainデータの作成
app_train_domain["CREDIT_INCOME_PERCENT"] = (
app_train_domain["AMT_CREDIT"] / app_train_domain["AMT_INCOME_TOTAL"]
)
app_train_domain["ANNUITY_INCOME_PERCENT"] = (
app_train_domain["AMT_ANNUITY"] / app_train_domain["AMT_INCOME_TOTAL"]
)
app_train_domain["CREDIT_TERM"] = (
app_train_domain["AMT_ANNUITY"] / app_train_domain["AMT_CREDIT"]
)
app_train_domain["DAYS_EMPLOYED_PERCENT"] = (
app_train_domain["DAYS_EMPLOYED"] / app_train_domain["DAYS_BIRTH"]
)
# testデータの作成
app_test_domain["CREDIT_INCOME_PERCENT"] = (
app_test_domain["AMT_CREDIT"] / app_test_domain["AMT_INCOME_TOTAL"]
)
app_test_domain["ANNUITY_INCOME_PERCENT"] = (
app_test_domain["AMT_ANNUITY"] / app_test_domain["AMT_INCOME_TOTAL"]
)
app_test_domain["CREDIT_TERM"] = (
app_test_domain["AMT_ANNUITY"] / app_test_domain["AMT_CREDIT"]
)
app_test_domain["DAYS_EMPLOYED_PERCENT"] = (
app_test_domain["DAYS_EMPLOYED"] / app_test_domain["DAYS_BIRTH"]
)
# 視覚化してみる
# 描写画面の用意
plt.figure(figsize=(12, 20))
# 各特徴量カラムをループさせる
for i, feature in enumerate(
[
"CREDIT_INCOME_PERCENT",
"ANNUITY_INCOME_PERCENT",
"CREDIT_TERM",
"DAYS_EMPLOYED_PERCENT",
]
):
# 描写範囲を分割する
# 第一引数:分割行数 第二引数:分割列数 第三引数:今回の描写範囲
plt.subplot(4, 1, i + 1)
# ローンが払えた人
sns.kdeplot(
app_train_domain.loc[app_train_domain["TARGET"] == 0, feature],
label="target == 0",
)
# ローンが払えなかった人
sns.kdeplot(
app_train_domain.loc[app_train_domain["TARGET"] == 1, feature],
label="target == 1",
)
# 図表の説明文
plt.title("Distribution of %s by Target Value" % feature)
plt.xlabel("%s" % feature)
plt.ylabel("Density")
plt.tight_layout(h_pad=2.5)
# 可視化内容からは特徴量についてローン返済者と未返済者で明確な違いは見られなかった。
# モデルに入れると何か意味が出てくるかもしれないのでそのまま使用する。
# ここからデータ予測を行う
# 何も考えずに予測した場合0.5の確率でローンが返済できないとなる。
# これをReciever Operating Characteristic Area Under the Curve (AUC ROC)という。
# 今回はロジスティック回帰(Logistic Regression)を使用して予測を行う。
# ロジスティック回帰
# 線形分離可能な場合のみ高い性能を発揮する分類予測方法。
# 線形分離可能とは、説明変数が二次元平面上にあるとすると、あるクラスに対応する説明変数(の値の集合)と、他のクラスに対応する説明変数(の値の集合)を、一本の直線を挟んで隔てることができること。
# **問題点**
# 学習しすぎるとオーバーフィッティング(過学習状態)になってしまう。
# 図の一番右のような状態。
# このような状態になってしまうと例外などのデータに左右されやすくなり、
# 新しいデータの予測としてはうまくいかなくなる。
# 
# **対策**
# 過学習は分類を行っている線の計算式の次元が高くなり複雑になって発生している。
# 直線(y = ax + b)よりも曲線(y =cx^2 + ax + b)の方が上手く分類できるというような感じ。
# そこで正則化という手段が用いられる。
# 正則化では分類の式に用いられるa,b,cなどパラメータを小さく設定するという方法である。
# a,b,cにあたるパラメータを小さくすればx^2やxによるyへの影響が小さくなる=直線に近づく(曲線が緩やかになる)
# ※実際には目的関数の係数(a,b,c)を直接いじるわけではない。
# 正則化パラメーター (Regularization Parameter) というものを用意することで間接的に係数の影響度を下げる
# 詳しい計算式などは参考サイトを参照
# 参考サイト:https://qiita.com/katsu1110/items/e4ef613559f02f183af5
#
# データの前処理
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import MinMaxScaler
# target行を削除
if "TARGET" in train_data:
# 返り値で何かを返しているわけではない
train = train_data.drop(columns=["TARGET"])
else:
train = train_data.copy()
# Feature =カラム名リスト
features = list(train.columns)
# テストデータのコピー
test = test_data.copy()
# null埋めようのモデルImputerを用意
imputer = SimpleImputer(missing_values=np.nan, strategy="median")
# 正規化用モデルscalerを用意
scaler = MinMaxScaler(feature_range=(0, 1))
# モデルを学習させる
imputer.fit(train)
# モデルをtrain testデータに適用する
train = imputer.transform(train)
test = imputer.transform(test)
# 同様に正規化も行う ← すべてのカラムの影響を等しくするため
scaler.fit(train)
train = scaler.transform(train)
test = scaler.transform(test)
print("Training data shape: ", train.shape)
print("Testing data shape: ", test.shape)
# ロジスティック回帰を用いた予測
from sklearn.linear_model import LogisticRegression
# モデルの用意 Cは正則化パラメータ
log_reg = LogisticRegression(C=0.0001)
# モデルにtrainデータの学習をさせる
log_reg.fit(train, train_labels)
# 学習済みモデルをもとに予測を行う
# 1行目:返済率(0の確率) 2行目:未返済率(1の確率)
# 今回は未返済率を知りたいので2行目を返す
log_reg_pred = log_reg.predict_proba(test)[:, 1]
# 結果を表示する
submit = test_data[["SK_ID_CURR"]]
submit["TARGET"] = log_reg_pred
submit.head()
# 結果を保存する。
submit.to_csv("log_reg_baseline.csv", index=False)
# Random Forest
# ランダムフォレストモデルを使用して予測の精度向上をさせる
# https://qiita.com/yshi12/items/6d30010b353b084b3749
# http://taustation.com/random-forest-overview/
# https://nomoto-eriko.hatenablog.com/entry/2018/06/06/101729
from sklearn.ensemble import RandomForestClassifier
# ランダムフォレスト分類子を作成する
# n_estimators: 木の数 大きい程制度がよくなるがその分学習に時間がかかる
random_forest = RandomForestClassifier(
n_estimators=100, random_state=50, verbose=1, n_jobs=-1
)
# 分類子を学習させる
random_forest.fit(train, train_labels)
# カラムごとの重要度をリストに格納する
feature_importance_values = random_forest.feature_importances_
feature_importances = pd.DataFrame(
{"feature": features, "importance": feature_importance_values}
)
# 予測データの作成
predictions = random_forest.predict_proba(test)[:, 1]
# 提出用データの作成
submit = test_data[["SK_ID_CURR"]]
submit["TARGET"] = predictions
# Save the submission dataframe
submit.to_csv("random_forest_baseline.csv", index=False)
# 工学的な特徴(Engineered Features)を使用して予測を行い予測結果への影響を確認する
#
poly_features_names = list(app_train_poly.columns)
# null埋め
imputer = SimpleImputer(missing_values=np.nan, strategy="median")
poly_features = imputer.fit_transform(app_train_poly)
poly_features_test = imputer.transform(app_test_poly)
# 標準化
scaler = MinMaxScaler(feature_range=(0, 1))
poly_features = scaler.fit_transform(poly_features)
poly_features_test = scaler.transform(poly_features_test)
random_forest_poly = RandomForestClassifier(
n_estimators=100, random_state=50, verbose=1, n_jobs=-1
)
# トレーニングデータの学習
random_forest_poly.fit(poly_features, train_labels)
# モデルを使用してテストデータによる予測を行う
predictions = random_forest_poly.predict_proba(poly_features_test)[:, 1]
submit = test_data[["SK_ID_CURR"]]
submit["TARGET"] = predictions
submit.to_csv("random_forest_baseline_engineered.csv", index=False)
# ドメイン専門知識から作成した特徴量を追加
#
app_train_domain = app_train_domain.drop(columns="TARGET")
domain_features_names = list(app_train_domain.columns)
# null埋め
imputer = SimpleImputer(missing_values=np.nan, strategy="median")
domain_features = imputer.fit_transform(app_train_domain)
domain_features_test = imputer.transform(app_test_domain)
# 標準化
scaler = MinMaxScaler(feature_range=(0, 1))
domain_features = scaler.fit_transform(domain_features)
domain_features_test = scaler.transform(domain_features_test)
random_forest_domain = RandomForestClassifier(
n_estimators=100, random_state=50, verbose=1, n_jobs=-1
)
# モデルの学習
random_forest_domain.fit(domain_features, train_labels)
# 特徴量の抽出
feature_importance_values_domain = random_forest_domain.feature_importances_
feature_importances_domain = pd.DataFrame(
{"feature": domain_features_names, "importance": feature_importance_values_domain}
)
# テストデータで予測を行う
predictions = random_forest_domain.predict_proba(domain_features_test)[:, 1]
# Make a submission dataframe
submit = test_data[["SK_ID_CURR"]]
submit["TARGET"] = predictions
# Save the submission dataframe
submit.to_csv("random_forest_baseline_domain.csv", index=False)
# モデルの機能重要性
# モデルの機能や特徴量を調節して予測の向上を目指す。
def plot_feature_importances(df):
"""
モデルから重要度を抽出する。より重要度が高い方がいい。
引数
df (dataframe): 特徴量の輸入量。カラム名は `features` というカラムに、重要度は`importance`と呼ばれる列に格納する必要がある。
Returns:
shows a plot of the 15 most importance features
df (dataframe): feature importances sorted by importance (highest to lowest)
with a column for normalized importance
戻り値
最も重要な15個のフィーチャーを表示します。
df (dataframe): 重要度でソートされた特徴量 (最高から最低)
重要度は正規化される
"""
# 重要度昇順でカラムを並び替え
df = df.sort_values("importance", ascending=False).reset_index()
# 正規化
df["importance_normalized"] = df["importance"] / df["importance"].sum()
# 重要度をカラムごとに可視化する
plt.figure(figsize=(10, 6))
ax = plt.subplot()
# 重要度が高いカラムから横棒グラフを作成する。
ax.barh(
list(reversed(list(df.index[:15]))),
df["importance_normalized"].head(15),
align="center",
edgecolor="k",
)
# y軸にメモリとカラム名を追加
ax.set_yticks(list(reversed(list(df.index[:15]))))
ax.set_yticklabels(df["feature"].head(15))
# 図表を表示
plt.xlabel("Normalized Importance")
plt.title("Feature Importances")
plt.show()
return df
# Show the feature importances for the default features
feature_importances_sorted = plot_feature_importances(feature_importances)
# EXT_SOURCE、 DAYS_BIRTHなどが目的変数に大きな影響をあたえていることがわかる。
# 重要度が低い特徴カラムを削除することで予測の処理を減らすことができる。
# (余計なデータがなくなることで予測精度が向上することもある。)
#
feature_importances_domain_sorted = plot_feature_importances(feature_importances_domain)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/476/69476583.ipynb
| null | null |
[{"Id": 69476583, "ScriptId": 18122132, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1560998, "CreationDate": "07/31/2021 13:30:13", "VersionNumber": 15.0, "Title": "Home Credit Default Risk", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 911.0, "LinesInsertedFromPrevious": 18.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 893.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# numpy and pandas for data manipulation
import numpy as np
import pandas as pd
# sklearn preprocessing for dealing with categorical variables
from sklearn.preprocessing import LabelEncoder
# File system manangement
import os
# Suppress warnings
import warnings
warnings.filterwarnings("ignore")
# matplotlib and seaborn for plotting
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# see train data
train_data = pd.read_csv("../input/home-credit-default-risk/application_train.csv")
train_data.head()
# targetかどうかを予想する
# see train data
test_data = pd.read_csv("../input/home-credit-default-risk/application_test.csv")
test_data.head()
# 過去の再建履歴
bureau_balance = pd.read_csv("../input/home-credit-default-risk/bureau_balance.csv")
bureau_balance.head()
# 過去の再建履歴
bureau = pd.read_csv("../input/home-credit-default-risk/bureau.csv")
bureau.head()
# 過去の再建履歴
pos_balance = pd.read_csv("../input/home-credit-default-risk/POS_CASH_balance.csv")
pos_balance.head()
# 過去の再建履歴
pte_appli = pd.read_csv("../input/home-credit-default-risk/previous_application.csv")
pte_appli.head()
# 過去の再建履歴
installments_pay = pd.read_csv(
"../input/home-credit-default-risk/installments_payments.csv"
)
installments_pay.head()
# 参照Note
# https://www.kaggle.com/willkoehrsen/start-here-a-gentle-introduction
# 以下で行う分析の流れ
# 問題とデータの理解
# データのクリーニングとフォーマット(この作業はほとんど自分たちで行いました
# 探索的データ分析
# ベースラインモデル
# モデルの改良
# モデルの解釈
# モデルの正確性について
# モデルの予測結果の正確性を可視化する図表としてROC曲線,AUCというものがある。
# Reciever Operating Characteristic (ROC) curve
# 
# 線が左上にあるほど陽性と判断したものの内本当に要請だった正解率(True Positive)が高く、不正解率(False Positive)が低い、良いモデルといえる。例では青>赤>黒の順でいいモデル。
# Area Under the Curve (AUC)はその名の通りROC曲線の下にできたエリアを指している。
# AUCは0から1までの値をとり、値が1に近いほど判別能(判別に使用しているモデル)が良いことを示す。
# precision(適合率)=TP/(TP+FP):陽性の内どのくらいの割合を検知できたか
# recall(再現率)=TP/(TP+FN):陽性と判断したものの内どの程度が本当に陽性か
# precision(適合率)とrecall(再現率)はトレードオフの関係
# 誤検知(再現率)を高くすると見逃す確率が高くなる(適合率が下がる)
# 適合率と再現率のどちらを重視するかは具体的に何を判断するかで毎回変える必要がある
# 探索的データ解析:Exploratory Data Analysis(EDA)
# 統計分析や可視化を行うことでデータに対する理解を深める。
# 傾向、異常値、パターン、相関関係などの特徴をあぶりだす。
# カラムの説明確認用
col_discription = pd.read_csv(
"../input/home-credit-default-risk/HomeCredit_columns_description.csv"
)
col_discription.loc[col_discription.Row == "EXT_SOURCE"]
# まずは目的となる指標の概要から確認する
# 0:返済可能者
# 1:返済不可能者
train_data["TARGET"].value_counts()
# クラス不均衡問題とは?
# 機械学習において、あるクラスのデータ(ポジティブ)の総数が、別のクラスのデータ(ネガティブ)の総数よりも圧倒的に少ないという問題のことです。この問題は実際には極めて一般的で、不正検知、異常検知、医療診断、油流出検知、顔認識など様々な分野で観察されます。
# なぜ問題なのか?
# 機械学習ではtargetの一致率を高めるために学習を行う。
# しかし適合率と再現率のトレードオフにあるように一概にtargetの精度を高くすればいいいというものではない。
# 陽性、陰性のレコード数が同じくらいならばそれぞれの誤検知も同じ重さで扱われる。
# 今回のように一方に偏りがある場合、偏っている方の誤検知件数が多くなるのでそれを補正しようとして少数のクラスの誤検知精度がないがしろにされる可能性がある。
# 以下で例を挙げる
# 取引データのデータセットが与えられたとき、どれが不正でどれが本物かを調べたいとします。電子商取引企業にとって、不正な取引が行われることは、お客様の信頼を損ない、コストがかかることになります。そのため、できるだけ多くの不正取引を見つけたいと考えています。
# 10000件の真正な取引と10件の不正な取引からなるデータセットがあった場合、分類器は不正な取引を真正な取引として分類する傾向があります。その理由は、数字で簡単に説明できます。機械学習アルゴリズムが以下のような2つの可能性のある出力を持っているとします。
# モデル1は、10件の不正な取引のうち7件を真正な取引と分類し、10000件の真正な取引のうち10件を不正な取引と分類した。
# モデル2は,10件の不正取引のうち2件を真正な取引と分類し,10000件の真正な取引のうち100件を不正な取引と分類した.
# 分類器の性能がミスの数で決まるとすれば,モデル2が102個のミスをしたのに対し,モデル1は合計17個のミスしかしていないので,明らかにモデル1の方が優れています.しかし、私たちは不正取引の発生を最小限に抑えたいので、不正取引の分類で2回しかミスをしなかったモデル2を代わりに選ぶべきです。もちろん、これにより、より多くの真正な取引が不正な取引として分類されるという犠牲を払うことになりますが、今のところは我慢できるコストでしょう。いずれにしても、一般的な機械学習アルゴリズムでは、モデル2よりもモデル1を選んでしまうことになり、問題となります。実際には、モデル2を使用すれば阻止できたはずの不正取引を、大量に通過させてしまうことになります。これは、お客様の不幸と会社のお金の損失につながります。
# 元サイト
# http://www.chioka.in/class-imbalance-problem/
# www.DeepL.com/Translator(無料版)で翻訳しました。
# null値などデータ欠損の確認
# Function to calculate missing values by column# Funct
def missing_values_table(df):
# Total missing values
mis_val = df.isnull().sum()
# Percentage of missing values
mis_val_percent = 100 * df.isnull().sum() / len(df)
# Make a table with the results
mis_val_table = pd.concat([mis_val, mis_val_percent], axis=1)
# Rename the columns
mis_val_table_ren_columns = mis_val_table.rename(
columns={0: "Missing Values", 1: "% of Total Values"}
)
# iloc[行, 列]を指定して抜き出し
# 欠損がないカラムを除外
# 欠損率が高い順(降順)で並び替え
# Sort the table by percentage of missing descending
mis_val_table_ren_columns = (
mis_val_table_ren_columns[mis_val_table_ren_columns.iloc[:, 1] != 0]
.sort_values("% of Total Values", ascending=False)
.round(1)
)
# Print some summary information
print(
"Your selected dataframe has " + str(df.shape[1]) + " columns.\n"
"There are "
+ str(mis_val_table_ren_columns.shape[0])
+ " columns that have missing values."
)
# Return the dataframe with missing information
return mis_val_table_ren_columns
missing_values = missing_values_table(train_data)
missing_values.head(20)
# ここで可視化される欠損値のあるカラムは後々欠損値を埋める(imputation)必要があるかもしれない
train_data.dtypes.value_counts()
# データタイプの確認
# float int は数値型
# objectは文字型などが混入
# オブジェクトカラムのユニークな値の個数を表示
# 文字情報の記入なのか何らかのクラス分けした情報が入っているカラムなのかの確認
train_data.select_dtypes("object").apply(pd.Series.nunique, axis=0)
# カテゴリ変数の処理について
# カテゴリ変数はそのままでは機械学習のモデルで処理できない
# Label encoding
# 要素クラスに対し1~ 数字を振る。
# 要素どおしに数字の大きさをつけることになるので意図しない重みづけになる可能性がある
# 所得に応じた階級などの場合ラベルエンコーディングを使える
# 職業などの場合はしたのOneーHotエンコーディングを使用した方がいい
# ※2つしかクラスがない場合は0.1で処理できる。
# One-hot encoding
# 要素クラス一つ一つに対し列を作り1,0でフラグを立てて処理する
# カテゴリーが多いカテゴリー変数では、素性(データの次元)の数が爆発的に増える。
# PCAや他の次元削減法を用いて次元数を減らすことができます(ただし、情報の保持には注意が必要)
# 2クラス以下のカラムの処理
# sklernのlabelencoerを使用する
le = LabelEncoder()
le_count = 0
# Iterate through the columns
for col in train_data:
if train_data[col].dtype == "object":
# If 2 or fewer unique categories
if len(list(train_data[col].unique())) <= 2:
# ラベルとラベルIDの対応づけ.positiveは0にしよう,みたいなことを決める
le.fit(train_data[col])
# Transform both training and testing data 返り値を用意しないといけない
train_data[col] = le.transform(train_data[col])
test_data[col] = le.transform(test_data[col])
# Keep track of how many columns were label encoded
le_count += 1
print("%d columns were label encoded." % le_count)
# one-hotエンコーディングを行うためのget_dummies関数について
df = pd.DataFrame(
{
"sex": ["female", np.nan, "male", "male", "female", "male"],
"old": [12, 21, 31, 10, 22, 10],
}
)
df.iloc[:, :]
test_df = pd.get_dummies(df)
test_df.iloc[:, :]
# 文字型(objectカラムのみ変換される)
# get_dummies():カテゴリ変数をダミー変数の作成
# デフォルトでは「文字列」など dtype が object の全てのカラムを「ダミー変数」に変換する
# 上記で変換していないカテゴリ変数に対してone-hotエンコーディング処理
train_data = pd.get_dummies(train_data)
test_data = pd.get_dummies(test_data)
print("Training Features shape: ", train_data.shape)
print("Testing Features shape: ", test_data.shape)
# トレーニングデータとテストデータのカラム数に違いが発生
# これはカテゴリ変数に含まれていたクラスの違いによるもの
# テストデータにないカラムを使う必要はないためトレインデータから削除する
#
# カラムをそろえる関数alignの説明
d1 = {"name": ["半沢", "大和田", "中野渡"], "code": ["101", "102", "104"]}
df1 = pd.DataFrame(data=d1)
df1
d2 = {"code": ["101", "102", "103"], "place": ["東京", "大阪", "名古屋"]}
df2 = pd.DataFrame(data=d2, index=[1, 2, 3])
df2
# alignの返り値は2つになる
df3, df4 = df1.align(df2, join="inner", axis=1)
df3
# TARGETカラムはテストデータにないが削除してはいけないカラムなので一度変数に格納しておいて後から再度追加する
train_labels = train_data["TARGET"]
# Align the training and testing data, keep only columns present in both dataframes
train_data, test_data = train_data.align(test_data, join="inner", axis=1)
# Add the target back in
train_data["TARGET"] = train_labels
print("Training Features shape: ", train_data.shape)
print("Testing Features shape: ", test_data.shape)
# データの異常値に対する処理
# DAYS_BIRTHカラムについて
# 現在のローン申請に対しての数値が格納されているため値がマイナスになっている。
# 値を正に直し単位を年にする処理を行う
# 年に直すことで入っている数値=年齢となり妥当な数値が入っているかが判断できる
train_data["DAYS_EMPLOYED"].describe()
# マイナスとプラスが混合している
# maxの数が大きすぎて1000年となっている
# などから正しい数値が入っていないよう。より詳細にデータを見る必要がある。
train_data["DAYS_EMPLOYED"].plot.hist(title="Days Employment Histogram")
plt.xlabel("Days Employment")
# 異常値も5万レコードあることからnullの代わりに入れられている数値の可能性あり。
# null値であることも一つの特徴量である可能性があるのでnull値かそうでないかでTARGETに対して影響を与えているかを確認する。
anom = train_data[train_data["DAYS_EMPLOYED"] == 365243]
non_anom = train_data[train_data["DAYS_EMPLOYED"] != 365243]
print(
"The non-anomalies default on %0.2f%% of loans" % (100 * non_anom["TARGET"].mean())
)
print("The anomalies default on %0.2f%% of loans" % (100 * anom["TARGET"].mean()))
print("There are %d anomalous days of employment" % len(anom))
# 3%以上の数値の乖離がある。
# 異常値の対応として最も安全な方法は異常値をnullとして処理をしてしまうこと。
# 今回の異常値はすべて同じ値であり、TARGETへの影響も考えられる。
# nullに上書きしたうえで異常値であったことを学習できるように新たなカラムを追加する。
#
# 異常値かどうかのフラグを格納するDAYS_EMPLOYED_ANOMを追加
train_data["DAYS_EMPLOYED_ANOM"] = train_data["DAYS_EMPLOYED"] == 365243
# 異常値をnullで置き換える implace=trueにすると元のDFが変換される
train_data["DAYS_EMPLOYED"].replace({365243: np.nan}, inplace=True)
train_data["DAYS_EMPLOYED"].plot.hist(title="Days Employment Histogram")
plt.xlabel("Days Employment")
# テストデータにも同様の処理をおこなう
test_data["DAYS_EMPLOYED_ANOM"] = test_data["DAYS_EMPLOYED"] == 365243
test_data["DAYS_EMPLOYED"].replace({365243: np.nan}, inplace=True)
print(
"There are %d anomalies in the test data out of %d entries"
% (test_data["DAYS_EMPLOYED_ANOM"].sum(), len(test_data))
)
# 相関係数
# * .00-.19 “very weak”
# * .20-.39 “weak”
# * .40-.59 “moderate”
# * .60-.79 “strong”
# * .80-1.0 “very strong”
# Find correlations with the target and sort
correlations = train_data.corr()["TARGET"].sort_values()
# Display correlations
print("Most Positive Correlations:\n", correlations.tail(15))
print("\nMost Negative Correlations:\n", correlations.head(15))
# 相関係数が高いDAYS_BIRTHについて分析
# days_birthはマイナス数値が格納されているので絶対値に修正して絶対値を再計算
train_data["DAYS_BIRTH"] = abs(train_data["DAYS_BIRTH"])
train_data["DAYS_BIRTH"].corr(train_data["TARGET"])
# 年齢が上昇するほど返済率は高くなる。
# テンプレ図表の形で図表を作成
plt.style.use("fivethirtyeight")
# 年齢ヒストグラム
plt.hist(train_data["DAYS_BIRTH"] / 365, edgecolor="k", bins=25)
plt.title("Age of Client")
plt.xlabel("Age (years)")
plt.ylabel("Count")
# 漠然と年齢ヒストグラムを表示しても外れ値がないかどうかくらいしかわからない
plt.figure(figsize=(10, 8))
# KDE plot of loans that were repaid on time
sns.kdeplot(
train_data.loc[train_data["TARGET"] == 0, "DAYS_BIRTH"] / 365, label="target == 0"
)
# KDE plot of loans which were not repaid on time
sns.kdeplot(
train_data.loc[train_data["TARGET"] == 1, "DAYS_BIRTH"] / 365, label="target == 1"
)
# Labeling of plot
plt.xlabel("Age (years)")
plt.ylabel("Density")
plt.title("Distribution of Ages")
# TARGETの値別に分布を表示することでどのような特徴が存在するのかを可視化する
# 年齢が高くなるにつれて返済率が高くなることは分かったがどの年齢層に特徴があるのかなどを分析するため
# 若年層(30代)で非返済が高いことが見て取れる。
# カラム全体としては相関係数は優位とは言えなかった。
# 実際に非返済者の予測に使用できるのか実際のデータを年齢層別にチェックする。
# 特徴を調べる'TARGET', 'DAYS_BIRTH'のデータのみ取り出す
age_data = train_data[["TARGET", "DAYS_BIRTH"]]
age_data["YEARS_BIRTH"] = age_data["DAYS_BIRTH"] / 365
# 20~70歳について11にわけることで5歳(50/10=5)ごとの階級を作成する
age_data["YEARS_BINNED"] = pd.cut(
age_data["YEARS_BIRTH"], bins=np.linspace(20, 70, num=11)
)
age_data.head(10)
# 5歳ごとの階級分けになっているかの確認 YEARS_BINNEDを確認
# 上記ではTARGET DAYS_BIRTH YEARS_BIRTHはランダムなレコードの値が出ていた。
# groupbyで平均をとって集計をして表示する
age_groups = age_data.groupby("YEARS_BINNED").mean()
age_groups
plt.figure(figsize=(8, 8))
# 上記で作成した表をグラフ化 100分率にして%で表示
plt.bar(age_groups.index.astype(str), 100 * age_groups["TARGET"])
# Plot labeling
plt.xticks(rotation=75)
plt.xlabel("Age Group (years)")
plt.ylabel("Failure to Repay (%)")
plt.title("Failure to Repay by Age Group")
# 明らかに若い世代ほど非返済率が高いことが分かった。
# 負の相関のあったExterior Sourcesについて考える
# Exterior Sources : Normalized score from external data source
# 標準化した外部データ(内容は不明)
# 上処理の文字列処理によって生まれたカラムではない。それぞれのカラムに関係があるかもわからない。
# 内部データが不明なのに分析に使用していいのか?
#
# Extract the EXT_SOURCE variables and show correlations
ext_data = train_data[
["TARGET", "EXT_SOURCE_1", "EXT_SOURCE_2", "EXT_SOURCE_3", "DAYS_BIRTH"]
]
ext_data_corrs = ext_data.corr()
ext_data_corrs
# 相関係数の可視化(ヒートマップで色を付ける)
plt.figure(figsize=(8, 6))
# Heatmap of correlations
sns.heatmap(ext_data_corrs, cmap=plt.cm.RdYlBu_r, vmin=-0.25, annot=True, vmax=0.6)
plt.title("Correlation Heatmap")
plt.figure(figsize=(10, 12))
# iterate through the sources
for i, source in enumerate(["EXT_SOURCE_1", "EXT_SOURCE_2", "EXT_SOURCE_3"]):
# create a new subplot for each source
plt.subplot(3, 1, i + 1)
# plot repaid loans
sns.kdeplot(train_data.loc[train_data["TARGET"] == 0, source], label="target == 0")
# plot loans that were not repaid
sns.kdeplot(train_data.loc[train_data["TARGET"] == 1, source], label="target == 1")
# Label the plots
plt.title("Distribution of %s by Target Value" % source)
plt.xlabel("%s" % source)
plt.ylabel("Density")
plt.tight_layout(h_pad=2.5)
# EXT_SOURCE_3はTARGET1と0で山の位置が他よりも明確に分かれている。
# 相関としては弱いが学習データには利用することが可能。
# 最後に探索的プロットとしてペアプロット(行列散布図)を行う。
# pairplotはseabornで最も使用されている機能の一つ。
# 大量の変数列があるデータに対し、全ての変数の組み合わせ毎に相関関係を見るためのプロット
# 
# 上記で調べたデータでペアプロットを作成するためデータをコピー
plot_data = ext_data.drop(columns=["DAYS_BIRTH"]).copy()
# 生年月日の列を追加
plot_data["YEARS_BIRTH"] = age_data["YEARS_BIRTH"]
# プロットデータを制限 多すぎると散布図が塗りつぶされみづらくなる
plot_data = plot_data.dropna().loc[:100000, :]
# 相関係数を計算する関数
def corr_func(x, y, **kwargs):
r = np.corrcoef(x, y)[0][1]
ax = plt.gca() # figureと同じような意味
# 注釈をつける
ax.annotate("r = {:.2f}".format(r), xy=(0.2, 0.8), xycoords=ax.transAxes, size=20)
# Create the pairgrid object
grid = sns.PairGrid(
data=plot_data,
size=3,
diag_sharey=False,
hue="TARGET", # 図示される目的変数
vars=[x for x in list(plot_data.columns) if x != "TARGET"],
)
# 右上側は散布図
grid.map_upper(plt.scatter, alpha=0.2)
# 真ん中はヒストグラム
grid.map_diag(sns.kdeplot)
# Bottom is density plot
grid.map_lower(sns.kdeplot, cmap=plt.cm.OrRd_r)
plt.suptitle("Ext Source and Age Features Pairs Plot", size=32, y=1.05)
# 赤は返済されなかったローン、青は返済されたローンを示す
# Kaggleでは最も有用な特徴を見つけることができたものが勝つ
# 少なくとも構造化データの場合、優勝モデルはすべて勾配ブーストのバリエーション(XGBoostやLightGBM、CatBoost)
# 機械学習のパターンの1つである、モデル構築やハイパーパラメータ調整よりも、特徴量エンジニアリングの方が投資対効果が高いことを表している。
# 参照記事:https://community.alteryx.com/t5/Data-Science/Feature-Engineering-Secret-to-Data-Science-Success/ba-p/545041
# フィーチャーエンジニアリングとは、既存のデータから新たな特徴を追加する「**特徴構築**」と、最も重要な特徴のみを選択する「**特徴選択**」、またはその他の次元削減方法の両方を含む、遺伝的なプロセスを指す。
# 他のデータソースを使い始めると、多くの特徴量エンジニアリングを行うことになりますが、このノートブックでは、2つの簡単な特徴量構築法を試してみます。
# >多項式特徴
# >ドメイン知識特徴量
# 多項式特徴量(Polynomial Features)
# 既存の特徴のべき乗や、既存の特徴間の相互作用項を特徴とする。
# 例えば、変数EXT_SOURCE_1^2とEXT_SOURCE_2^2の他に、
# EXT_SOURCE_1×EXT_SOURCE_2、EXT_SOURCE_1^2×EXT_SOURCE_2^2、EXT_SOURCE_1^2×EXT_SOURCE_2^2などの変数を作る
# 交互作用項は、複数の変数の影響を捉えるために統計モデルではよく使われますが、機械学習ではあまり見かけない
# Scikit-LearnにはPolynomialFeaturesという便利なクラスがあり、指定した次数までの多項式と相互作用項を作成する。
# ここでは次数を3にして結果を見る。(多項式特徴を作成する際には、特徴数が次数に応じて指数関数的に増加することと、オーバーフィッティングの問題が発生することから、あまり高い次数を使わないようにしたい。)
# 多項式特徴量分析用のデータを作成
poly_features = train_data[
["EXT_SOURCE_1", "EXT_SOURCE_2", "EXT_SOURCE_3", "DAYS_BIRTH", "TARGET"]
]
poly_features_test = test_data[
["EXT_SOURCE_1", "EXT_SOURCE_2", "EXT_SOURCE_3", "DAYS_BIRTH"]
]
# 欠損値の処理
# sklearn.preprocessingの Imputerはサポート終了予定なのでSimpleImputerを利用する
from sklearn.impute import SimpleImputer
imputer = SimpleImputer(missing_values=np.nan, strategy="median")
poly_target = poly_features["TARGET"]
poly_features = poly_features.drop(columns=["TARGET"])
# Need to impute missing values
poly_features = imputer.fit_transform(poly_features)
poly_features_test = imputer.transform(poly_features_test)
from sklearn.preprocessing import PolynomialFeatures
# 多項式特徴量分析 モデルの作成
poly_transformer = PolynomialFeatures(degree=3)
# Train the polynomial features
poly_transformer.fit(poly_features)
# Transform the features
poly_features = poly_transformer.transform(poly_features)
poly_features_test = poly_transformer.transform(poly_features_test)
print("Polynomial Features shape: ", poly_features.shape)
print("poly_features_test shape: ", poly_features_test.shape)
# 35の特徴量が新たに作成された。
# 作成された特徴量の確認
poly_transformer.get_feature_names(
input_features=["EXT_SOURCE_1", "EXT_SOURCE_2", "EXT_SOURCE_3", "DAYS_BIRTH"]
)[:15]
# 新しく作成した特徴量とTARGETカラムの相関係数を確認する
# Create a dataframe of the features
poly_features = pd.DataFrame(
poly_features,
columns=poly_transformer.get_feature_names(
["EXT_SOURCE_1", "EXT_SOURCE_2", "EXT_SOURCE_3", "DAYS_BIRTH"]
),
)
# Add in the target
poly_features["TARGET"] = poly_target
# Find the correlations with the target
poly_corrs = poly_features.corr()["TARGET"].sort_values()
# Display most negative and most positive
print(poly_corrs.head(10))
print("")
print("")
print("")
print(poly_corrs.tail(5))
# 何もしてない時より相関係数が大きくなっているものがある。
# 実際に元データ(のコピー)に追加して学習データとして使えるか確認する。
# 新しく作成した特徴量がいつも使えるとは限らない。実際に学習させて確認しないとわからないこともある。
# 特徴量を追加格納したDFを作成する
poly_features_test = pd.DataFrame(
poly_features_test,
columns=poly_transformer.get_feature_names(
["EXT_SOURCE_1", "EXT_SOURCE_2", "EXT_SOURCE_3", "DAYS_BIRTH"]
),
)
# 特徴量をトレーニングデータに格納する
poly_features["SK_ID_CURR"] = train_data["SK_ID_CURR"]
app_train_poly = train_data.merge(poly_features, on="SK_ID_CURR", how="left")
# 特徴量をテストデータに格納する
poly_features_test["SK_ID_CURR"] = test_data["SK_ID_CURR"]
app_test_poly = test_data.merge(poly_features_test, on="SK_ID_CURR", how="left")
# トレーニングデータとテストデータ共通するカラムのみ
app_train_poly, app_test_poly = app_train_poly.align(
app_test_poly, join="inner", axis=1
)
# データのサイズを表示
print("Training data with polynomial features shape: ", app_train_poly.shape)
print("Testing data with polynomial features shape: ", app_test_poly.shape)
# ドメイン・ナレッジの特徴
# そのデータの専門知識を利用して特徴量を新規作成しデータ分析を行う
# 今回新規作成してみる特徴量
# CREDIT_INCOME_PERCENT:顧客の収入に対する与信額の割合。
# ANNUITY_INCOME_PERCENT: 顧客の収入に対するローンの年金額の割合
# CREDIT_TERM: 月単位の支払期間(年金は月単位の支払額なので
# DAYS_EMPLOYED_PERCENT: クライアントの年齢に対する雇用日数の割合
app_train_domain = train_data.copy()
app_test_domain = test_data.copy()
# trainデータの作成
app_train_domain["CREDIT_INCOME_PERCENT"] = (
app_train_domain["AMT_CREDIT"] / app_train_domain["AMT_INCOME_TOTAL"]
)
app_train_domain["ANNUITY_INCOME_PERCENT"] = (
app_train_domain["AMT_ANNUITY"] / app_train_domain["AMT_INCOME_TOTAL"]
)
app_train_domain["CREDIT_TERM"] = (
app_train_domain["AMT_ANNUITY"] / app_train_domain["AMT_CREDIT"]
)
app_train_domain["DAYS_EMPLOYED_PERCENT"] = (
app_train_domain["DAYS_EMPLOYED"] / app_train_domain["DAYS_BIRTH"]
)
# testデータの作成
app_test_domain["CREDIT_INCOME_PERCENT"] = (
app_test_domain["AMT_CREDIT"] / app_test_domain["AMT_INCOME_TOTAL"]
)
app_test_domain["ANNUITY_INCOME_PERCENT"] = (
app_test_domain["AMT_ANNUITY"] / app_test_domain["AMT_INCOME_TOTAL"]
)
app_test_domain["CREDIT_TERM"] = (
app_test_domain["AMT_ANNUITY"] / app_test_domain["AMT_CREDIT"]
)
app_test_domain["DAYS_EMPLOYED_PERCENT"] = (
app_test_domain["DAYS_EMPLOYED"] / app_test_domain["DAYS_BIRTH"]
)
# 視覚化してみる
# 描写画面の用意
plt.figure(figsize=(12, 20))
# 各特徴量カラムをループさせる
for i, feature in enumerate(
[
"CREDIT_INCOME_PERCENT",
"ANNUITY_INCOME_PERCENT",
"CREDIT_TERM",
"DAYS_EMPLOYED_PERCENT",
]
):
# 描写範囲を分割する
# 第一引数:分割行数 第二引数:分割列数 第三引数:今回の描写範囲
plt.subplot(4, 1, i + 1)
# ローンが払えた人
sns.kdeplot(
app_train_domain.loc[app_train_domain["TARGET"] == 0, feature],
label="target == 0",
)
# ローンが払えなかった人
sns.kdeplot(
app_train_domain.loc[app_train_domain["TARGET"] == 1, feature],
label="target == 1",
)
# 図表の説明文
plt.title("Distribution of %s by Target Value" % feature)
plt.xlabel("%s" % feature)
plt.ylabel("Density")
plt.tight_layout(h_pad=2.5)
# 可視化内容からは特徴量についてローン返済者と未返済者で明確な違いは見られなかった。
# モデルに入れると何か意味が出てくるかもしれないのでそのまま使用する。
# ここからデータ予測を行う
# 何も考えずに予測した場合0.5の確率でローンが返済できないとなる。
# これをReciever Operating Characteristic Area Under the Curve (AUC ROC)という。
# 今回はロジスティック回帰(Logistic Regression)を使用して予測を行う。
# ロジスティック回帰
# 線形分離可能な場合のみ高い性能を発揮する分類予測方法。
# 線形分離可能とは、説明変数が二次元平面上にあるとすると、あるクラスに対応する説明変数(の値の集合)と、他のクラスに対応する説明変数(の値の集合)を、一本の直線を挟んで隔てることができること。
# **問題点**
# 学習しすぎるとオーバーフィッティング(過学習状態)になってしまう。
# 図の一番右のような状態。
# このような状態になってしまうと例外などのデータに左右されやすくなり、
# 新しいデータの予測としてはうまくいかなくなる。
# 
# **対策**
# 過学習は分類を行っている線の計算式の次元が高くなり複雑になって発生している。
# 直線(y = ax + b)よりも曲線(y =cx^2 + ax + b)の方が上手く分類できるというような感じ。
# そこで正則化という手段が用いられる。
# 正則化では分類の式に用いられるa,b,cなどパラメータを小さく設定するという方法である。
# a,b,cにあたるパラメータを小さくすればx^2やxによるyへの影響が小さくなる=直線に近づく(曲線が緩やかになる)
# ※実際には目的関数の係数(a,b,c)を直接いじるわけではない。
# 正則化パラメーター (Regularization Parameter) というものを用意することで間接的に係数の影響度を下げる
# 詳しい計算式などは参考サイトを参照
# 参考サイト:https://qiita.com/katsu1110/items/e4ef613559f02f183af5
#
# データの前処理
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import MinMaxScaler
# target行を削除
if "TARGET" in train_data:
# 返り値で何かを返しているわけではない
train = train_data.drop(columns=["TARGET"])
else:
train = train_data.copy()
# Feature =カラム名リスト
features = list(train.columns)
# テストデータのコピー
test = test_data.copy()
# null埋めようのモデルImputerを用意
imputer = SimpleImputer(missing_values=np.nan, strategy="median")
# 正規化用モデルscalerを用意
scaler = MinMaxScaler(feature_range=(0, 1))
# モデルを学習させる
imputer.fit(train)
# モデルをtrain testデータに適用する
train = imputer.transform(train)
test = imputer.transform(test)
# 同様に正規化も行う ← すべてのカラムの影響を等しくするため
scaler.fit(train)
train = scaler.transform(train)
test = scaler.transform(test)
print("Training data shape: ", train.shape)
print("Testing data shape: ", test.shape)
# ロジスティック回帰を用いた予測
from sklearn.linear_model import LogisticRegression
# モデルの用意 Cは正則化パラメータ
log_reg = LogisticRegression(C=0.0001)
# モデルにtrainデータの学習をさせる
log_reg.fit(train, train_labels)
# 学習済みモデルをもとに予測を行う
# 1行目:返済率(0の確率) 2行目:未返済率(1の確率)
# 今回は未返済率を知りたいので2行目を返す
log_reg_pred = log_reg.predict_proba(test)[:, 1]
# 結果を表示する
submit = test_data[["SK_ID_CURR"]]
submit["TARGET"] = log_reg_pred
submit.head()
# 結果を保存する。
submit.to_csv("log_reg_baseline.csv", index=False)
# Random Forest
# ランダムフォレストモデルを使用して予測の精度向上をさせる
# https://qiita.com/yshi12/items/6d30010b353b084b3749
# http://taustation.com/random-forest-overview/
# https://nomoto-eriko.hatenablog.com/entry/2018/06/06/101729
from sklearn.ensemble import RandomForestClassifier
# ランダムフォレスト分類子を作成する
# n_estimators: 木の数 大きい程制度がよくなるがその分学習に時間がかかる
random_forest = RandomForestClassifier(
n_estimators=100, random_state=50, verbose=1, n_jobs=-1
)
# 分類子を学習させる
random_forest.fit(train, train_labels)
# カラムごとの重要度をリストに格納する
feature_importance_values = random_forest.feature_importances_
feature_importances = pd.DataFrame(
{"feature": features, "importance": feature_importance_values}
)
# 予測データの作成
predictions = random_forest.predict_proba(test)[:, 1]
# 提出用データの作成
submit = test_data[["SK_ID_CURR"]]
submit["TARGET"] = predictions
# Save the submission dataframe
submit.to_csv("random_forest_baseline.csv", index=False)
# 工学的な特徴(Engineered Features)を使用して予測を行い予測結果への影響を確認する
#
poly_features_names = list(app_train_poly.columns)
# null埋め
imputer = SimpleImputer(missing_values=np.nan, strategy="median")
poly_features = imputer.fit_transform(app_train_poly)
poly_features_test = imputer.transform(app_test_poly)
# 標準化
scaler = MinMaxScaler(feature_range=(0, 1))
poly_features = scaler.fit_transform(poly_features)
poly_features_test = scaler.transform(poly_features_test)
random_forest_poly = RandomForestClassifier(
n_estimators=100, random_state=50, verbose=1, n_jobs=-1
)
# トレーニングデータの学習
random_forest_poly.fit(poly_features, train_labels)
# モデルを使用してテストデータによる予測を行う
predictions = random_forest_poly.predict_proba(poly_features_test)[:, 1]
submit = test_data[["SK_ID_CURR"]]
submit["TARGET"] = predictions
submit.to_csv("random_forest_baseline_engineered.csv", index=False)
# ドメイン専門知識から作成した特徴量を追加
#
app_train_domain = app_train_domain.drop(columns="TARGET")
domain_features_names = list(app_train_domain.columns)
# null埋め
imputer = SimpleImputer(missing_values=np.nan, strategy="median")
domain_features = imputer.fit_transform(app_train_domain)
domain_features_test = imputer.transform(app_test_domain)
# 標準化
scaler = MinMaxScaler(feature_range=(0, 1))
domain_features = scaler.fit_transform(domain_features)
domain_features_test = scaler.transform(domain_features_test)
random_forest_domain = RandomForestClassifier(
n_estimators=100, random_state=50, verbose=1, n_jobs=-1
)
# モデルの学習
random_forest_domain.fit(domain_features, train_labels)
# 特徴量の抽出
feature_importance_values_domain = random_forest_domain.feature_importances_
feature_importances_domain = pd.DataFrame(
{"feature": domain_features_names, "importance": feature_importance_values_domain}
)
# テストデータで予測を行う
predictions = random_forest_domain.predict_proba(domain_features_test)[:, 1]
# Make a submission dataframe
submit = test_data[["SK_ID_CURR"]]
submit["TARGET"] = predictions
# Save the submission dataframe
submit.to_csv("random_forest_baseline_domain.csv", index=False)
# モデルの機能重要性
# モデルの機能や特徴量を調節して予測の向上を目指す。
def plot_feature_importances(df):
"""
モデルから重要度を抽出する。より重要度が高い方がいい。
引数
df (dataframe): 特徴量の輸入量。カラム名は `features` というカラムに、重要度は`importance`と呼ばれる列に格納する必要がある。
Returns:
shows a plot of the 15 most importance features
df (dataframe): feature importances sorted by importance (highest to lowest)
with a column for normalized importance
戻り値
最も重要な15個のフィーチャーを表示します。
df (dataframe): 重要度でソートされた特徴量 (最高から最低)
重要度は正規化される
"""
# 重要度昇順でカラムを並び替え
df = df.sort_values("importance", ascending=False).reset_index()
# 正規化
df["importance_normalized"] = df["importance"] / df["importance"].sum()
# 重要度をカラムごとに可視化する
plt.figure(figsize=(10, 6))
ax = plt.subplot()
# 重要度が高いカラムから横棒グラフを作成する。
ax.barh(
list(reversed(list(df.index[:15]))),
df["importance_normalized"].head(15),
align="center",
edgecolor="k",
)
# y軸にメモリとカラム名を追加
ax.set_yticks(list(reversed(list(df.index[:15]))))
ax.set_yticklabels(df["feature"].head(15))
# 図表を表示
plt.xlabel("Normalized Importance")
plt.title("Feature Importances")
plt.show()
return df
# Show the feature importances for the default features
feature_importances_sorted = plot_feature_importances(feature_importances)
# EXT_SOURCE、 DAYS_BIRTHなどが目的変数に大きな影響をあたえていることがわかる。
# 重要度が低い特徴カラムを削除することで予測の処理を減らすことができる。
# (余計なデータがなくなることで予測精度が向上することもある。)
#
feature_importances_domain_sorted = plot_feature_importances(feature_importances_domain)
| false | 0 | 12,903 | 0 | 12,903 | 12,903 |
||
69476001
|
# # Titanic - Machine Learning from Disaster
import re
import os
import matplotlib.pyplot as plt
import numpy as np # linear algebra
from pandas import read_csv
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
from tensorflow import feature_column
from tensorflow.keras import layers
from sklearn.model_selection import train_test_split
filedir = ""
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
filedir = os.path.dirname(os.path.join(dirname, filename))
train_data = pd.read_csv("{}/train.csv".format(dirname), index_col=0)
test_data = pd.read_csv("{}/test.csv".format(dirname), index_col=0)
gender_data = pd.read_csv("{}/gender_submission.csv".format(dirname), index_col=0)
test_data["Survived"] = gender_data["Survived"]
train_data.info()
test_data.info()
# # I don't know how to evaluate the name and ticket, so delete it from the data.
# may be name is not relation.
# may be name and ticket is not relation.
drop_col = ["Name", "Ticket"]
train_data_p = train_data.drop(drop_col, axis=1)
test_data_p = test_data.drop(drop_col, axis=1)
train_data_p.head()
all_passenger = len(train_data_p)
data = (train_data_p["Survived"] == 1).sum()
print(
"The ratio of survivors to the whole is {0:3.1f}%.".format(
data * 100 / all_passenger
)
)
# # Is the ticket class related to the survival rate in this case?
# - The data set for this contest has three ticket classes, P-Class 1, 2, and 3. Let's compare whether this difference affects survival.
plt.tight_layout()
all_passenger = len(train_data_p)
all_suvr = (train_data_p["Survived"] == 1).sum()
print(
"Survival rate (no bias by P class):{0:3.1f}%".format(
all_suvr * 100 / all_passenger
)
)
for pcls in [1, 2, 3]:
data = (train_data_p["Pclass"] == pcls).sum()
surv = ((train_data_p["Pclass"] == pcls) & (train_data_p["Survived"] == 1)).sum()
print(
"""Pclass:{0} :
Percentage of Pclass{0} among all passengers: \t{3:3.1f}%
Survival Rate of Pclass{0}: \t\t\t{1:3.1f}%
Survival Rate(among all passengers): \t\t{2:3.1f}%
""".format(
pcls, surv / data * 100, surv * 100 / all_suvr, data * 100 / all_passenger
)
)
_ = plt.hist(
[
train_data_p[train_data_p["Survived"] == 1]["Pclass"],
train_data_p[train_data_p["Survived"] != 1]["Pclass"],
],
label=["Survied", "Dead"],
color=["red", "blue"],
stacked=True,
)
plt.legend()
plt.xlabel("Pclass")
plt.ylabel("Count")
plt.xticks([1, 2, 3], ["Pclass{}".format(i) for i in range(1, 4)])
plt.show()
_ = plt.hist(
[
train_data_p[train_data_p["Pclass"] == 1]["Survived"],
train_data_p[train_data_p["Pclass"] == 2]["Survived"],
train_data_p[train_data_p["Pclass"] == 3]["Survived"],
],
label=["Pclass1", "Pclass2", "Pclass3"],
color=["orange", "blue", "green"],
stacked=True,
)
plt.legend()
plt.xlabel("Survived")
plt.xticks([0, 1], ["Dead", "Survied"])
plt.ylabel("Count")
# ## From the above result
# From the above calculations, we found that the survival rate of each Pclass was higher than that of Pclass2 and Pclass1.
# Also, in terms of the overall survival rate,
# Pclass1 accounted for about 25% of passengers, while it accounted for about 40% of survivors,
# suggesting that the survival rate of Pclass1 class was high.
# In addition, Pclass3 accounts for nearly half of the passengers,
# while Pclass3 has a survival rate of only about 24%,
# which is lower than the 38.4% survival rate for this accident.
# If the difference in Pclass does not affect the survival rate, it can be predicted that the survival rate will be about 38.4% for each class. However, from this result, it is considered that the difference in Pclass affects the survival rate.
# |Pclass|Rate(*1)|Survival rate(*2)|Survival rate(Among all survivors)(*3)|
# |:--|--:|--:|--:|
# | Pclass 1 | 24.2%|63.0% | 39.8% |
# | Pclass 2 | 20.7%|47.3% |25.4% |
# | Pclass 3 | 55.1%| 24.2% | 34.8% |
# - *1 :Percentage of passengers in each class to total
# - *2 :Survival rate for each class
# - *3 :Percentage of survivors classified by pclass
# # Is Gender or Age Related to This Case?
# - Compare the survival rates of men and women.
# - As can be seen from the graph, the survival rate of women is more than 50% higher than that of men.
# | Sex |Survived rate|
# |:--|--:|
# | male | 18.9% |
# | female | 74.2% |
female_all = (train_data_p["Sex"] == "female").sum()
male_all = (train_data_p["Sex"] == "male").sum()
print("male,female {} :{}".format(male_all, female_all))
all_surv = (train_data_p["Survived"] == 1).sum()
male_surv = ((train_data_p["Survived"] == 1) & (train_data_p["Sex"] == "male")).sum()
print("Female`s Survival: {0:3.1f} %".format(100 * (all_suvr - male_surv) / all_surv))
for sex in ["male", "female"]:
data = (train_data_p["Sex"] == sex).sum()
surv = ((train_data_p["Sex"] == sex) & (train_data_p["Survived"] == 1)).sum()
print("Sex:{} \t Survived : {:3.1f} %".format(sex, (surv / data) * 100))
_ = plt.hist(
[
train_data_p[train_data_p["Survived"] == 1]["Sex"],
train_data_p[train_data_p["Survived"] != 1]["Sex"],
],
label=["Survived", "Dead"],
color=["orange", "blue"],
stacked=True,
)
plt.legend()
plt.xlabel("Sex")
plt.ylabel("Count")
plt.title("")
plt.show()
# ## Gender differences may have a significant impact on survival.
# As you can see from the graph above, women have significantly higher survival rates than men.
# I think this was a major factor in separating life and death.
# # Is age life-threatening in this case?
# - Group by 5 years and create a histogram.
_hist = plt.hist(
[
train_data_p[train_data_p["Survived"] == 1]["Age"],
train_data_p[train_data_p["Survived"] != 1]["Age"],
],
range=(0, 100),
bins=20,
label=["Survived", "Dead"],
color=["orange", "blue"],
stacked=True,
)
plt.legend()
plt.xlabel("Age")
_ = plt.ylabel("Count")
rate = (_hist[0][0] / (_hist[0][1])) * 100
for i in range(len(rate)):
print("Age : {:3} ~ {:3} {:3.1f} %".format(i * 5, (i + 1) * 5, rate[i]))
# - From the above results, looking at the survival rate by age, it seems that the survival rate of young people aged 0 to 15 seems to be significantly higher.
tmp = train_data_p.copy()
tmp["Cabin"] = (
train_data_p["Cabin"].str.extract("(?P<cabin>^.)", expand=False).fillna("Nan")
)
_ = plt.hist(
[
tmp[(tmp["Survived"] == 1) & (tmp["Cabin"] != "Nan")]["Cabin"],
tmp[(tmp["Survived"] != 1) & (tmp["Cabin"] != "Nan")]["Cabin"],
],
label=["Survived", "Dead"],
color=["orange", "blue"],
stacked=True,
rwidth=0.8,
)
_ = plt.legend()
_ = plt.xlabel("Cabin")
_ = plt.ylabel("Count")
# ---
# # The above is the tendency seen from the data. From this, machine learning is started using Tensorflow.
# # First, prepare the dataset.(Work in progress)
# Preliminary research has shown that it probably contains nulls and that it contains data in non-numeric categories.
# Therefore, change the dataset to one-hot format by deleting or deleting it in a format that can handle null.
from tensorflow.keras.layers.experimental.preprocessing import Normalization
from tensorflow.keras.layers.experimental.preprocessing import IntegerLookup
from tensorflow.keras.layers.experimental.preprocessing import StringLookup
intLookUp = IntegerLookup()
total_data = train_data_p.append(test_data_p)
# fix Age
total_data.Age = np.where(
total_data.Age.isnull() == True, -1, total_data.Age // 10 * 10
)
# fix Fare
fare_max = total_data.Fare.max()
total_data.Fare = np.where(
total_data.Fare.isnull() == 1, -1, total_data.Fare / fare_max
)
# Fix Cabin
total_data["Cabin"] = (
total_data["Cabin"].str.extract("(?P<cabin>^.)", expand=False).fillna("Nan")
)
# fix Embarked
total_data.Embarked = total_data.Embarked.fillna("Nan")
# とりあえずvalueとして扱う
total_data.Parch = total_data.Parch / total_data.Parch.max()
total_data.SibSp = total_data.SibSp / total_data.SibSp.max()
total_data.head()
# Since "Sex", "Cabin", "Embarked", and "Pclass" are categories, the data is in one-hot format.
make_one_hot = pd.get_dummies(
total_data,
columns=["Age", "Sex", "Cabin", "Embarked", "Pclass"],
)
make_one_hot = make_one_hot.dropna()
# drop Cabin_Nan,Embarked_Nan
make_one_hot = make_one_hot.drop("Age_-1.0", axis=1)
make_one_hot = make_one_hot.drop("Age_70.0", axis=1)
make_one_hot = make_one_hot.drop("Age_80.0", axis=1)
make_one_hot = make_one_hot.drop("Cabin_Nan", axis=1)
make_one_hot = make_one_hot.drop("Embarked_Nan", axis=1)
make_one_hot.info()
# Separate training data and verification data.
train_data = make_one_hot[(make_one_hot.index <= 891)]
test_data = make_one_hot[~(make_one_hot.index <= 891)]
# Convert to numpy format.
tr_data = train_data.drop("Survived", axis=1).to_numpy()
tr_lable = train_data["Survived"].to_numpy()
val_data_n = test_data.drop("Survived", axis=1).to_numpy()
val_label = test_data["Survived"].to_numpy()
# start ML. use Tensorflow
import matplotlib.pylab as plt
import numpy as np
import tensorflow as tf
from tensorflow import keras
import tensorflow_datasets as tfds
train_dataset = tf.data.Dataset.from_tensor_slices((tr_data, tr_lable))
test_dataset = tf.data.Dataset.from_tensor_slices((val_data_n, val_label))
BATCH_SIZE = 200
SHUFFLE_BUFFER_SIZE = 300
train_dataset = train_dataset.shuffle(SHUFFLE_BUFFER_SIZE).batch(BATCH_SIZE)
test_dataset = test_dataset.batch(BATCH_SIZE)
# Hyper parameterのfine tuneを行う
import kerastuner as kt
import IPython
# GPU setting
print("Num GPUs Available: ", len(tf.config.experimental.list_physical_devices("GPU")))
# # Search for parameters suitable for the model.
# I have no idea what the appropriate parameter value is, so I start with the Hyperparameter tune.
# Already completed at Version 32
def model_builder(hp):
model = keras.Sequential()
# input layer
hp_input_layer = hp.Int("InputParam", min_value=32, max_value=64, step=4)
model.add(
keras.layers.Dense(
hp_input_layer,
activation="relu",
input_shape=tr_data.shape[1:], # input_data shape
name="input_layer",
)
)
# layer1
hp_layer_units1 = hp.Int("units1", min_value=32, max_value=64, step=4)
hp_reg_rate1 = hp.Choice("reg_rate1", values=[1e-2, 1e-3, 1e-4])
hp_drop_rate1 = hp.Choice("drop_rate1", values=[0.2, 0.3, 0.5])
model.add(keras.layers.Dropout(hp_drop_rate1))
model.add(
keras.layers.Dense(
hp_layer_units1,
activation="relu",
kernel_regularizer=keras.regularizers.l2(hp_reg_rate1),
)
)
hp_layer_units2 = hp.Int("units2", min_value=32, max_value=64, step=4)
hp_reg_rate2 = hp.Choice("reg_rate2", values=[1e-2, 1e-3, 1e-4])
hp_drop_rate2 = hp.Choice("drop_rate2", values=[0.2, 0.3, 0.5])
model.add(keras.layers.Dropout(hp_drop_rate2))
model.add(
keras.layers.Dense(
hp_layer_units2,
activation="relu",
kernel_regularizer=keras.regularizers.l2(hp_reg_rate2),
)
)
model.add(keras.layers.Dense(1, activation="sigmoid"))
hp_learning_rate = hp.Choice("learning_rate", values=[1e-2, 1e-3, 1e-4])
model.compile(
loss="binary_crossentropy",
optimizer=keras.optimizers.Adam(learning_rate=hp_learning_rate),
metrics=[tf.keras.metrics.BinaryAccuracy()], # "accuracy",
)
return model
tuner = kt.Hyperband(
model_builder,
objective="binary_accuracy",
max_epochs=50,
directory="my_dir",
project_name="intro_to_kt",
)
class ClearTrainingOutput(tf.keras.callbacks.Callback):
def on_train_end(*args, **kwargs):
IPython.display.clear_output(wait=True)
early_stop = keras.callbacks.EarlyStopping(monitor="val_loss", patience=10)
reduce_lr = keras.callbacks.ReduceLROnPlateau(
monitor="val_binary_accuracy", factor=0.5, patience=5, min_lr=0.00005, verbose=1
)
tuner.search(
train_dataset,
batch_size=BATCH_SIZE,
epochs=50,
validation_data=test_dataset,
callbacks=[ClearTrainingOutput(), early_stop, reduce_lr],
)
# # Generate the determined model.
# Get the optimal hyperparameters
best_hps = tuner.get_best_hyperparameters(num_trials=1)[0]
from pprint import pprint
print("tuned model parameter----------")
pprint(best_hps.values)
print("----------")
model = tuner.hypermodel.build(best_hps)
model.summary()
hist = model.fit(
train_dataset,
batch_size=BATCH_SIZE,
epochs=200,
validation_data=test_dataset,
callbacks=[ClearTrainingOutput(), early_stop, reduce_lr],
verbose=2,
)
history_dict = hist.history
acc = history_dict["binary_accuracy"]
val_acc = history_dict["val_binary_accuracy"]
loss = history_dict["loss"]
val_loss = history_dict["val_loss"]
epochs = range(1, len(acc) + 1)
# "bo" is for "blue dot"
plt.plot(epochs, loss, "b", label="Training loss")
# b is for "solid blue line"
plt.plot(epochs, val_loss, "r", label="Validation loss")
plt.title("Training and validation loss")
plt.xlabel("Epochs")
plt.ylabel("Loss")
plt.legend()
plt.show()
plt.plot(epochs, acc, "b", label="Training acc")
plt.plot(epochs, val_acc, "r", label="Validation acc")
plt.title("Training and validation accuracy")
plt.xlabel("Epochs")
plt.ylabel("Accuracy")
plt.legend()
plt.show()
count = 0
pred = model.predict(val_data_n)
for xi in range(len(pred)):
if (1 if pred[xi][0] > 0.5 else 0) != val_label[xi]:
# print( "index : {}".format(xi) , (1 if pred[xi][0]>0.5 else 0) == val_label[xi] )
count += 1
print("False: ", count, " total : ", len(pred))
results = model.evaluate(val_data_n, val_label, verbose=2)
indexs = test_data.index.to_numpy()
pred_data = test_data.drop("Survived", axis=1).to_numpy()
pred_result = model.predict(pred_data)
with open("./result.csv", mode="w") as f:
f.write("PassengerId,Survived\n")
for num, predict in zip(indexs, pred_result):
# print("{},{}".format(num,1 if predict[0]>0.5 else 0))
f.write("{},{}\n".format(num, 1 if predict[0] > 0.5 else 0))
# # To create a better model.
# - From the notebook below, I suspect that there may be a difference between the training data and the validation data data.
# [An extensive data journey on the Titanic (Top 5%)](https://www.kaggle.com/stefanschulmeister87/an-extensive-data-journey-on-the-titanic-top-5)
# ---
# - First, try training the model by including the validation data in the training data.
# By doing so, I thought that it would be possible to improve from the state of overfitting.
model = tuner.hypermodel.build(best_hps)
model.summary()
# Separate training data and verification data.
train_data = make_one_hot # [(make_one_hot.index <= 891)]
test_data = make_one_hot[~(make_one_hot.index <= 891)]
hist = model.fit(
train_dataset,
batch_size=BATCH_SIZE,
epochs=200,
validation_data=test_dataset,
callbacks=[ClearTrainingOutput(), early_stop, reduce_lr],
verbose=2,
)
history_dict = hist.history
acc = history_dict["binary_accuracy"]
val_acc = history_dict["val_binary_accuracy"]
loss = history_dict["loss"]
val_loss = history_dict["val_loss"]
epochs = range(1, len(acc) + 1)
# "bo" is for "blue dot"
plt.plot(epochs, loss, "b", label="Training loss")
# b is for "solid blue line"
plt.plot(epochs, val_loss, "r", label="Validation loss")
plt.title("Training and validation loss")
plt.xlabel("Epochs")
plt.ylabel("Loss")
plt.legend()
plt.show()
plt.plot(epochs, acc, "b", label="Training acc")
plt.plot(epochs, val_acc, "r", label="Validation acc")
plt.title("Training and validation accuracy")
plt.xlabel("Epochs")
plt.ylabel("Accuracy")
plt.legend()
plt.show()
#
# I haven't overfitted, but I'm not sure if it's improving.
# ---
# # use GradientTape
from pprint import pprint
colums = test_data.drop("Survived", axis=1).columns
data = test_dataset.take(1)
arr, label = next(iter(data))
x = arr
y = label
with tf.GradientTape() as tape:
tape.watch(x) # x を記録
pred = model(x)
# print("label :{}".format(y))
# print(pred)
grads = tape.gradient(pred, x)
mean_value = np.mean(grads, axis=0)
# 値の低い項目を 0 に置き換える
# mean_value[np.where(np.abs(mean_value) <0.05)] = 0
plt.figure(figsize=(20, 10))
plt.bar([i for i in range(len(colums))], mean_value, width=1.0)
plt.ylabel(r"$\frac{\partial y}{\partial x_i}$", fontsize=18)
plt.xlabel(r"$columns: x_i$")
plt.grid()
_ = plt.xticks([i for i in range(len(colums))], colums, rotation=-90, fontsize=18)
# # Conclusion
# (For the time being, I wrote it intuitively, and it is necessary to consider whether it is correct as a way of thinking.)
#
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/476/69476001.ipynb
| null | null |
[{"Id": 69476001, "ScriptId": 15100267, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6704585, "CreationDate": "07/31/2021 13:22:41", "VersionNumber": 41.0, "Title": "Titanic_Keras feature_columns", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 520.0, "LinesInsertedFromPrevious": 8.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 512.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Titanic - Machine Learning from Disaster
import re
import os
import matplotlib.pyplot as plt
import numpy as np # linear algebra
from pandas import read_csv
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
from tensorflow import feature_column
from tensorflow.keras import layers
from sklearn.model_selection import train_test_split
filedir = ""
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
filedir = os.path.dirname(os.path.join(dirname, filename))
train_data = pd.read_csv("{}/train.csv".format(dirname), index_col=0)
test_data = pd.read_csv("{}/test.csv".format(dirname), index_col=0)
gender_data = pd.read_csv("{}/gender_submission.csv".format(dirname), index_col=0)
test_data["Survived"] = gender_data["Survived"]
train_data.info()
test_data.info()
# # I don't know how to evaluate the name and ticket, so delete it from the data.
# may be name is not relation.
# may be name and ticket is not relation.
drop_col = ["Name", "Ticket"]
train_data_p = train_data.drop(drop_col, axis=1)
test_data_p = test_data.drop(drop_col, axis=1)
train_data_p.head()
all_passenger = len(train_data_p)
data = (train_data_p["Survived"] == 1).sum()
print(
"The ratio of survivors to the whole is {0:3.1f}%.".format(
data * 100 / all_passenger
)
)
# # Is the ticket class related to the survival rate in this case?
# - The data set for this contest has three ticket classes, P-Class 1, 2, and 3. Let's compare whether this difference affects survival.
plt.tight_layout()
all_passenger = len(train_data_p)
all_suvr = (train_data_p["Survived"] == 1).sum()
print(
"Survival rate (no bias by P class):{0:3.1f}%".format(
all_suvr * 100 / all_passenger
)
)
for pcls in [1, 2, 3]:
data = (train_data_p["Pclass"] == pcls).sum()
surv = ((train_data_p["Pclass"] == pcls) & (train_data_p["Survived"] == 1)).sum()
print(
"""Pclass:{0} :
Percentage of Pclass{0} among all passengers: \t{3:3.1f}%
Survival Rate of Pclass{0}: \t\t\t{1:3.1f}%
Survival Rate(among all passengers): \t\t{2:3.1f}%
""".format(
pcls, surv / data * 100, surv * 100 / all_suvr, data * 100 / all_passenger
)
)
_ = plt.hist(
[
train_data_p[train_data_p["Survived"] == 1]["Pclass"],
train_data_p[train_data_p["Survived"] != 1]["Pclass"],
],
label=["Survied", "Dead"],
color=["red", "blue"],
stacked=True,
)
plt.legend()
plt.xlabel("Pclass")
plt.ylabel("Count")
plt.xticks([1, 2, 3], ["Pclass{}".format(i) for i in range(1, 4)])
plt.show()
_ = plt.hist(
[
train_data_p[train_data_p["Pclass"] == 1]["Survived"],
train_data_p[train_data_p["Pclass"] == 2]["Survived"],
train_data_p[train_data_p["Pclass"] == 3]["Survived"],
],
label=["Pclass1", "Pclass2", "Pclass3"],
color=["orange", "blue", "green"],
stacked=True,
)
plt.legend()
plt.xlabel("Survived")
plt.xticks([0, 1], ["Dead", "Survied"])
plt.ylabel("Count")
# ## From the above result
# From the above calculations, we found that the survival rate of each Pclass was higher than that of Pclass2 and Pclass1.
# Also, in terms of the overall survival rate,
# Pclass1 accounted for about 25% of passengers, while it accounted for about 40% of survivors,
# suggesting that the survival rate of Pclass1 class was high.
# In addition, Pclass3 accounts for nearly half of the passengers,
# while Pclass3 has a survival rate of only about 24%,
# which is lower than the 38.4% survival rate for this accident.
# If the difference in Pclass does not affect the survival rate, it can be predicted that the survival rate will be about 38.4% for each class. However, from this result, it is considered that the difference in Pclass affects the survival rate.
# |Pclass|Rate(*1)|Survival rate(*2)|Survival rate(Among all survivors)(*3)|
# |:--|--:|--:|--:|
# | Pclass 1 | 24.2%|63.0% | 39.8% |
# | Pclass 2 | 20.7%|47.3% |25.4% |
# | Pclass 3 | 55.1%| 24.2% | 34.8% |
# - *1 :Percentage of passengers in each class to total
# - *2 :Survival rate for each class
# - *3 :Percentage of survivors classified by pclass
# # Is Gender or Age Related to This Case?
# - Compare the survival rates of men and women.
# - As can be seen from the graph, the survival rate of women is more than 50% higher than that of men.
# | Sex |Survived rate|
# |:--|--:|
# | male | 18.9% |
# | female | 74.2% |
female_all = (train_data_p["Sex"] == "female").sum()
male_all = (train_data_p["Sex"] == "male").sum()
print("male,female {} :{}".format(male_all, female_all))
all_surv = (train_data_p["Survived"] == 1).sum()
male_surv = ((train_data_p["Survived"] == 1) & (train_data_p["Sex"] == "male")).sum()
print("Female`s Survival: {0:3.1f} %".format(100 * (all_suvr - male_surv) / all_surv))
for sex in ["male", "female"]:
data = (train_data_p["Sex"] == sex).sum()
surv = ((train_data_p["Sex"] == sex) & (train_data_p["Survived"] == 1)).sum()
print("Sex:{} \t Survived : {:3.1f} %".format(sex, (surv / data) * 100))
_ = plt.hist(
[
train_data_p[train_data_p["Survived"] == 1]["Sex"],
train_data_p[train_data_p["Survived"] != 1]["Sex"],
],
label=["Survived", "Dead"],
color=["orange", "blue"],
stacked=True,
)
plt.legend()
plt.xlabel("Sex")
plt.ylabel("Count")
plt.title("")
plt.show()
# ## Gender differences may have a significant impact on survival.
# As you can see from the graph above, women have significantly higher survival rates than men.
# I think this was a major factor in separating life and death.
# # Is age life-threatening in this case?
# - Group by 5 years and create a histogram.
_hist = plt.hist(
[
train_data_p[train_data_p["Survived"] == 1]["Age"],
train_data_p[train_data_p["Survived"] != 1]["Age"],
],
range=(0, 100),
bins=20,
label=["Survived", "Dead"],
color=["orange", "blue"],
stacked=True,
)
plt.legend()
plt.xlabel("Age")
_ = plt.ylabel("Count")
rate = (_hist[0][0] / (_hist[0][1])) * 100
for i in range(len(rate)):
print("Age : {:3} ~ {:3} {:3.1f} %".format(i * 5, (i + 1) * 5, rate[i]))
# - From the above results, looking at the survival rate by age, it seems that the survival rate of young people aged 0 to 15 seems to be significantly higher.
tmp = train_data_p.copy()
tmp["Cabin"] = (
train_data_p["Cabin"].str.extract("(?P<cabin>^.)", expand=False).fillna("Nan")
)
_ = plt.hist(
[
tmp[(tmp["Survived"] == 1) & (tmp["Cabin"] != "Nan")]["Cabin"],
tmp[(tmp["Survived"] != 1) & (tmp["Cabin"] != "Nan")]["Cabin"],
],
label=["Survived", "Dead"],
color=["orange", "blue"],
stacked=True,
rwidth=0.8,
)
_ = plt.legend()
_ = plt.xlabel("Cabin")
_ = plt.ylabel("Count")
# ---
# # The above is the tendency seen from the data. From this, machine learning is started using Tensorflow.
# # First, prepare the dataset.(Work in progress)
# Preliminary research has shown that it probably contains nulls and that it contains data in non-numeric categories.
# Therefore, change the dataset to one-hot format by deleting or deleting it in a format that can handle null.
from tensorflow.keras.layers.experimental.preprocessing import Normalization
from tensorflow.keras.layers.experimental.preprocessing import IntegerLookup
from tensorflow.keras.layers.experimental.preprocessing import StringLookup
intLookUp = IntegerLookup()
total_data = train_data_p.append(test_data_p)
# fix Age
total_data.Age = np.where(
total_data.Age.isnull() == True, -1, total_data.Age // 10 * 10
)
# fix Fare
fare_max = total_data.Fare.max()
total_data.Fare = np.where(
total_data.Fare.isnull() == 1, -1, total_data.Fare / fare_max
)
# Fix Cabin
total_data["Cabin"] = (
total_data["Cabin"].str.extract("(?P<cabin>^.)", expand=False).fillna("Nan")
)
# fix Embarked
total_data.Embarked = total_data.Embarked.fillna("Nan")
# とりあえずvalueとして扱う
total_data.Parch = total_data.Parch / total_data.Parch.max()
total_data.SibSp = total_data.SibSp / total_data.SibSp.max()
total_data.head()
# Since "Sex", "Cabin", "Embarked", and "Pclass" are categories, the data is in one-hot format.
make_one_hot = pd.get_dummies(
total_data,
columns=["Age", "Sex", "Cabin", "Embarked", "Pclass"],
)
make_one_hot = make_one_hot.dropna()
# drop Cabin_Nan,Embarked_Nan
make_one_hot = make_one_hot.drop("Age_-1.0", axis=1)
make_one_hot = make_one_hot.drop("Age_70.0", axis=1)
make_one_hot = make_one_hot.drop("Age_80.0", axis=1)
make_one_hot = make_one_hot.drop("Cabin_Nan", axis=1)
make_one_hot = make_one_hot.drop("Embarked_Nan", axis=1)
make_one_hot.info()
# Separate training data and verification data.
train_data = make_one_hot[(make_one_hot.index <= 891)]
test_data = make_one_hot[~(make_one_hot.index <= 891)]
# Convert to numpy format.
tr_data = train_data.drop("Survived", axis=1).to_numpy()
tr_lable = train_data["Survived"].to_numpy()
val_data_n = test_data.drop("Survived", axis=1).to_numpy()
val_label = test_data["Survived"].to_numpy()
# start ML. use Tensorflow
import matplotlib.pylab as plt
import numpy as np
import tensorflow as tf
from tensorflow import keras
import tensorflow_datasets as tfds
train_dataset = tf.data.Dataset.from_tensor_slices((tr_data, tr_lable))
test_dataset = tf.data.Dataset.from_tensor_slices((val_data_n, val_label))
BATCH_SIZE = 200
SHUFFLE_BUFFER_SIZE = 300
train_dataset = train_dataset.shuffle(SHUFFLE_BUFFER_SIZE).batch(BATCH_SIZE)
test_dataset = test_dataset.batch(BATCH_SIZE)
# Hyper parameterのfine tuneを行う
import kerastuner as kt
import IPython
# GPU setting
print("Num GPUs Available: ", len(tf.config.experimental.list_physical_devices("GPU")))
# # Search for parameters suitable for the model.
# I have no idea what the appropriate parameter value is, so I start with the Hyperparameter tune.
# Already completed at Version 32
def model_builder(hp):
model = keras.Sequential()
# input layer
hp_input_layer = hp.Int("InputParam", min_value=32, max_value=64, step=4)
model.add(
keras.layers.Dense(
hp_input_layer,
activation="relu",
input_shape=tr_data.shape[1:], # input_data shape
name="input_layer",
)
)
# layer1
hp_layer_units1 = hp.Int("units1", min_value=32, max_value=64, step=4)
hp_reg_rate1 = hp.Choice("reg_rate1", values=[1e-2, 1e-3, 1e-4])
hp_drop_rate1 = hp.Choice("drop_rate1", values=[0.2, 0.3, 0.5])
model.add(keras.layers.Dropout(hp_drop_rate1))
model.add(
keras.layers.Dense(
hp_layer_units1,
activation="relu",
kernel_regularizer=keras.regularizers.l2(hp_reg_rate1),
)
)
hp_layer_units2 = hp.Int("units2", min_value=32, max_value=64, step=4)
hp_reg_rate2 = hp.Choice("reg_rate2", values=[1e-2, 1e-3, 1e-4])
hp_drop_rate2 = hp.Choice("drop_rate2", values=[0.2, 0.3, 0.5])
model.add(keras.layers.Dropout(hp_drop_rate2))
model.add(
keras.layers.Dense(
hp_layer_units2,
activation="relu",
kernel_regularizer=keras.regularizers.l2(hp_reg_rate2),
)
)
model.add(keras.layers.Dense(1, activation="sigmoid"))
hp_learning_rate = hp.Choice("learning_rate", values=[1e-2, 1e-3, 1e-4])
model.compile(
loss="binary_crossentropy",
optimizer=keras.optimizers.Adam(learning_rate=hp_learning_rate),
metrics=[tf.keras.metrics.BinaryAccuracy()], # "accuracy",
)
return model
tuner = kt.Hyperband(
model_builder,
objective="binary_accuracy",
max_epochs=50,
directory="my_dir",
project_name="intro_to_kt",
)
class ClearTrainingOutput(tf.keras.callbacks.Callback):
def on_train_end(*args, **kwargs):
IPython.display.clear_output(wait=True)
early_stop = keras.callbacks.EarlyStopping(monitor="val_loss", patience=10)
reduce_lr = keras.callbacks.ReduceLROnPlateau(
monitor="val_binary_accuracy", factor=0.5, patience=5, min_lr=0.00005, verbose=1
)
tuner.search(
train_dataset,
batch_size=BATCH_SIZE,
epochs=50,
validation_data=test_dataset,
callbacks=[ClearTrainingOutput(), early_stop, reduce_lr],
)
# # Generate the determined model.
# Get the optimal hyperparameters
best_hps = tuner.get_best_hyperparameters(num_trials=1)[0]
from pprint import pprint
print("tuned model parameter----------")
pprint(best_hps.values)
print("----------")
model = tuner.hypermodel.build(best_hps)
model.summary()
hist = model.fit(
train_dataset,
batch_size=BATCH_SIZE,
epochs=200,
validation_data=test_dataset,
callbacks=[ClearTrainingOutput(), early_stop, reduce_lr],
verbose=2,
)
history_dict = hist.history
acc = history_dict["binary_accuracy"]
val_acc = history_dict["val_binary_accuracy"]
loss = history_dict["loss"]
val_loss = history_dict["val_loss"]
epochs = range(1, len(acc) + 1)
# "bo" is for "blue dot"
plt.plot(epochs, loss, "b", label="Training loss")
# b is for "solid blue line"
plt.plot(epochs, val_loss, "r", label="Validation loss")
plt.title("Training and validation loss")
plt.xlabel("Epochs")
plt.ylabel("Loss")
plt.legend()
plt.show()
plt.plot(epochs, acc, "b", label="Training acc")
plt.plot(epochs, val_acc, "r", label="Validation acc")
plt.title("Training and validation accuracy")
plt.xlabel("Epochs")
plt.ylabel("Accuracy")
plt.legend()
plt.show()
count = 0
pred = model.predict(val_data_n)
for xi in range(len(pred)):
if (1 if pred[xi][0] > 0.5 else 0) != val_label[xi]:
# print( "index : {}".format(xi) , (1 if pred[xi][0]>0.5 else 0) == val_label[xi] )
count += 1
print("False: ", count, " total : ", len(pred))
results = model.evaluate(val_data_n, val_label, verbose=2)
indexs = test_data.index.to_numpy()
pred_data = test_data.drop("Survived", axis=1).to_numpy()
pred_result = model.predict(pred_data)
with open("./result.csv", mode="w") as f:
f.write("PassengerId,Survived\n")
for num, predict in zip(indexs, pred_result):
# print("{},{}".format(num,1 if predict[0]>0.5 else 0))
f.write("{},{}\n".format(num, 1 if predict[0] > 0.5 else 0))
# # To create a better model.
# - From the notebook below, I suspect that there may be a difference between the training data and the validation data data.
# [An extensive data journey on the Titanic (Top 5%)](https://www.kaggle.com/stefanschulmeister87/an-extensive-data-journey-on-the-titanic-top-5)
# ---
# - First, try training the model by including the validation data in the training data.
# By doing so, I thought that it would be possible to improve from the state of overfitting.
model = tuner.hypermodel.build(best_hps)
model.summary()
# Separate training data and verification data.
train_data = make_one_hot # [(make_one_hot.index <= 891)]
test_data = make_one_hot[~(make_one_hot.index <= 891)]
hist = model.fit(
train_dataset,
batch_size=BATCH_SIZE,
epochs=200,
validation_data=test_dataset,
callbacks=[ClearTrainingOutput(), early_stop, reduce_lr],
verbose=2,
)
history_dict = hist.history
acc = history_dict["binary_accuracy"]
val_acc = history_dict["val_binary_accuracy"]
loss = history_dict["loss"]
val_loss = history_dict["val_loss"]
epochs = range(1, len(acc) + 1)
# "bo" is for "blue dot"
plt.plot(epochs, loss, "b", label="Training loss")
# b is for "solid blue line"
plt.plot(epochs, val_loss, "r", label="Validation loss")
plt.title("Training and validation loss")
plt.xlabel("Epochs")
plt.ylabel("Loss")
plt.legend()
plt.show()
plt.plot(epochs, acc, "b", label="Training acc")
plt.plot(epochs, val_acc, "r", label="Validation acc")
plt.title("Training and validation accuracy")
plt.xlabel("Epochs")
plt.ylabel("Accuracy")
plt.legend()
plt.show()
#
# I haven't overfitted, but I'm not sure if it's improving.
# ---
# # use GradientTape
from pprint import pprint
colums = test_data.drop("Survived", axis=1).columns
data = test_dataset.take(1)
arr, label = next(iter(data))
x = arr
y = label
with tf.GradientTape() as tape:
tape.watch(x) # x を記録
pred = model(x)
# print("label :{}".format(y))
# print(pred)
grads = tape.gradient(pred, x)
mean_value = np.mean(grads, axis=0)
# 値の低い項目を 0 に置き換える
# mean_value[np.where(np.abs(mean_value) <0.05)] = 0
plt.figure(figsize=(20, 10))
plt.bar([i for i in range(len(colums))], mean_value, width=1.0)
plt.ylabel(r"$\frac{\partial y}{\partial x_i}$", fontsize=18)
plt.xlabel(r"$columns: x_i$")
plt.grid()
_ = plt.xticks([i for i in range(len(colums))], colums, rotation=-90, fontsize=18)
# # Conclusion
# (For the time being, I wrote it intuitively, and it is necessary to consider whether it is correct as a way of thinking.)
#
| false | 0 | 5,807 | 0 | 5,807 | 5,807 |
||
69476610
|
<jupyter_start><jupyter_text>World Bank Data (1960 to 2016) Extended
### All the details in https://www.kaggle.com/gemartin/world-bank-data-1960-to-2016/ also apply to this new dataset.
### The preprocessed data has been generated using the data preparatory notebook https://www.kaggle.com/neomatrix369/chaieda-world-bank-data-1960-2016-data-prep.
### Context
Extending the current dataset to enable better analysis and reasonings
### Content
A number of economic, geographic, country and region-specific data and indicators from different datasets have been aggregated.
Kaggle dataset identifier: world-bank-data-1960-to-2016-extended
<jupyter_code>import pandas as pd
df = pd.read_csv('world-bank-data-1960-to-2016-extended/countries-of-the-world/countries of the world.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 227 entries, 0 to 226
Data columns (total 20 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Country 227 non-null object
1 Region 227 non-null object
2 Population 227 non-null int64
3 Area (sq. mi.) 227 non-null int64
4 Pop. Density (per sq. mi.) 227 non-null object
5 Coastline (coast/area ratio) 227 non-null object
6 Net migration 224 non-null object
7 Infant mortality (per 1000 births) 224 non-null object
8 GDP ($ per capita) 226 non-null float64
9 Literacy (%) 209 non-null object
10 Phones (per 1000) 223 non-null object
11 Arable (%) 225 non-null object
12 Crops (%) 225 non-null object
13 Other (%) 225 non-null object
14 Climate 205 non-null object
15 Birthrate 224 non-null object
16 Deathrate 223 non-null object
17 Agriculture 212 non-null object
18 Industry 211 non-null object
19 Service 212 non-null object
dtypes: float64(1), int64(2), object(17)
memory usage: 35.6+ KB
<jupyter_text>Examples:
{
"Country": "Afghanistan ",
"Region": "ASIA (EX. NEAR EAST) ",
"Population": 31056997,
"Area (sq. mi.)": 647500,
"Pop. Density (per sq. mi.)": "48,0",
"Coastline (coast/area ratio)": "0,00",
"Net migration": "23,06",
"Infant mortality (per 1000 births)": "163,07",
"GDP ($ per capita)": 700,
"Literacy (%)": "36,0",
"Phones (per 1000)": "3,2",
"Arable (%)": "12,13",
"Crops (%)": "0,22",
"Other (%)": "87,65",
"Climate": 1,
"Birthrate": "46,6",
"Deathrate": "20,34",
"Agriculture": "0,38",
"Industry": "0,24",
"Service": "0,38"
}
{
"Country": "Albania ",
"Region": "EASTERN EUROPE ",
"Population": 3581655,
"Area (sq. mi.)": 28748,
"Pop. Density (per sq. mi.)": "124,6",
"Coastline (coast/area ratio)": "1,26",
"Net migration": "-4,93",
"Infant mortality (per 1000 births)": "21,52",
"GDP ($ per capita)": 4500,
"Literacy (%)": "86,5",
"Phones (per 1000)": "71,2",
"Arable (%)": "21,09",
"Crops (%)": "4,42",
"Other (%)": "74,49",
"Climate": 3,
"Birthrate": "15,11",
"Deathrate": "5,22",
"Agriculture": "0,232",
"Industry": "0,188",
"Service": "0,579"
}
{
"Country": "Algeria ",
"Region": "NORTHERN AFRICA ",
"Population": 32930091,
"Area (sq. mi.)": 2381740,
"Pop. Density (per sq. mi.)": "13,8",
"Coastline (coast/area ratio)": "0,04",
"Net migration": "-0,39",
"Infant mortality (per 1000 births)": "31",
"GDP ($ per capita)": 6000,
"Literacy (%)": "70,0",
"Phones (per 1000)": "78,1",
"Arable (%)": "3,22",
"Crops (%)": "0,25",
"Other (%)": "96,53",
"Climate": 1,
"Birthrate": "17,14",
"Deathrate": "4,61",
"Agriculture": "0,101",
"Industry": "0,6",
"Service": "0,298"
}
{
"Country": "American Samoa ",
"Region": "OCEANIA ",
"Population": 57794,
"Area (sq. mi.)": 199,
"Pop. Density (per sq. mi.)": "290,4",
"Coastline (coast/area ratio)": "58,29",
"Net migration": "-20,71",
"Infant mortality (per 1000 births)": "9,27",
"GDP ($ per capita)": 8000,
"Literacy (%)": "97,0",
"Phones (per 1000)": "259,5",
"Arable (%)": "10",
"Crops (%)": "15",
"Other (%)": "75",
"Climate": 2,
"Birthrate": "22,46",
"Deathrate": "3,27",
"Agriculture": null,
"Industry": null,
"Service": null
}
<jupyter_script># 
# ### In this Data Science Project, I am investigating the dataset “Countries of the World”. I will be focusing on the factors affecting a country’s GDP per capita and try to make a model using the data of 227 countries from the dataset. I will also briefly discuss the total GDP.
# # 1. importing the required Python libraries
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
from matplotlib import pyplot as plt
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_squared_error, mean_squared_log_error
# # 2. Let’s look at the data
data = pd.read_csv(
"../input/world-bank-data-1960-to-2016-extended/countries-of-the-world/countries of the world.csv",
decimal=",",
)
data.head()
data.info()
data.describe(include="all")
# # 3. Missing values
print("number of missing data:")
print(data.isnull().sum())
# ## Data Preparation – fill in missing values
# We noticed that there are some missing data in the table. For simplicity, I will just fill the missing data using the median of the region that a country belongs, as countries that are close geologically are often similar in many ways. For example, lets check the region median of ‘GDP ($ per capita)’, ‘Literacy (%)’ and ‘Agriculture’.
# Note that for ‘climate’ we use the mode instead of median as it seems that ‘climate’ is a categorical feature here.
data.groupby("Region")[["GDP ($ per capita)", "Literacy (%)", "Agriculture"]].median()
for col in data.columns.values:
if data[col].isnull().sum() == 0:
continue
if col == "Climate":
guess_values = data.groupby("Region")["Climate"].apply(lambda x: x.mode().max())
else:
guess_values = data.groupby("Region")[col].median()
for region in data["Region"].unique():
data[col].loc[(data[col].isnull()) & (data["Region"] == region)] = guess_values[
region
]
# # 4. Data Exploration
# ### Top Countries with highest GDP per capita
# Look at the top 20 countries with highest GDP per capita. Luxembourg is quite ahead, the next 19 countries are close. German, the 20th has about 2.5 times GDP per capita of the world average.
fig, ax = plt.subplots(figsize=(16, 6))
# ax = fig.add_subplot(111)
top_gdp_countries = data.sort_values("GDP ($ per capita)", ascending=False).head(20)
mean = pd.DataFrame(
{
"Country": ["World mean"],
"GDP ($ per capita)": [data["GDP ($ per capita)"].mean()],
}
)
gdps = pd.concat(
[top_gdp_countries[["Country", "GDP ($ per capita)"]], mean], ignore_index=True
)
sns.barplot(x="Country", y="GDP ($ per capita)", data=gdps, palette="Set3")
ax.set_xlabel(ax.get_xlabel(), labelpad=15)
ax.set_ylabel(ax.get_ylabel(), labelpad=30)
ax.xaxis.label.set_fontsize(16)
ax.yaxis.label.set_fontsize(16)
plt.xticks(rotation=90)
plt.show()
# # 4. Correlation between Variables
# ### The heatmap shows the correlation between all numerical columns.
#
plt.figure(figsize=(16, 12))
sns.heatmap(data=data.iloc[:, 2:].corr(), annot=True, fmt=".2f", cmap="coolwarm")
plt.show()
# ## Top Factors affecting GDP per capita
# ### We pick the six columns that mostly correlated to GDP per capita and make scatter plots. The results agree with our common sense. Also we notice there are many countries with low average GDP and few with high average GDP —- a pyramid structure.
fig, axes = plt.subplots(nrows=2, ncols=3, figsize=(20, 12))
plt.subplots_adjust(hspace=0.4)
corr_to_gdp = pd.Series()
for col in data.columns.values[2:]:
if (col != "GDP ($ per capita)") & (col != "Climate"):
corr_to_gdp[col] = data["GDP ($ per capita)"].corr(data[col])
abs_corr_to_gdp = corr_to_gdp.abs().sort_values(ascending=False)
corr_to_gdp = corr_to_gdp.loc[abs_corr_to_gdp.index]
for i in range(2):
for j in range(3):
sns.regplot(
x=corr_to_gdp.index.values[i * 3 + j],
y="GDP ($ per capita)",
data=data,
ax=axes[i, j],
fit_reg=False,
marker=".",
)
title = "correlation=" + str(corr_to_gdp[i * 3 + j])
axes[i, j].set_title(title)
axes[1, 2].set_xlim(0, 102)
plt.show()
# ## Countries with low Birthrate and low GDP per capita
# ### Some features, like phones, are related to the average GDP more linearly, while others are not. For example, High birthrate usually means low GDP per capita, but average GDP in low birthrate countries can vary a lot.
# ### Let’s look at the countries with low birthrate (<14%) and low GDP per capita (<10000 $). They also have hight literacy, like other high average GDP countires. But we hope their other features can help distiguish them from those with low birthrate but high average GDPs, like service are not quite an importent portion in their economy, not a lot phone procession, some have negative net migration, and many of them are from eastern Europe or C.W. of IND. STATES, so the ‘region’ feature may also be useful.
data.loc[(data["Birthrate"] < 14) & (data["GDP ($ per capita)"] < 10000)]
# # 5. Modeling
# ## Training and Testing
# ### First label encode the categorical features ‘Region’ and ‘Climate’, and I will just use all features given in the data set without further feature engineering.
LE = LabelEncoder()
data["Region_label"] = LE.fit_transform(data["Region"])
data["Climate_label"] = LE.fit_transform(data["Climate"])
data.head()
train, test = train_test_split(data, test_size=0.3, shuffle=True)
training_features = [
"Population",
"Area (sq. mi.)",
"Pop. Density (per sq. mi.)",
"Coastline (coast/area ratio)",
"Net migration",
"Infant mortality (per 1000 births)",
"Literacy (%)",
"Phones (per 1000)",
"Arable (%)",
"Crops (%)",
"Other (%)",
"Birthrate",
"Deathrate",
"Agriculture",
"Industry",
"Service",
"Region_label",
"Climate_label",
"Service",
]
target = "GDP ($ per capita)"
train_X = train[training_features]
train_Y = train[target]
test_X = test[training_features]
test_Y = test[target]
# ### First let’s try the linear regression model. As for metric, I will check both root mean squared error and mean squared log error.
#
model = LinearRegression()
model.fit(train_X, train_Y)
train_pred_Y = model.predict(train_X)
test_pred_Y = model.predict(test_X)
train_pred_Y = pd.Series(train_pred_Y.clip(0, train_pred_Y.max()), index=train_Y.index)
test_pred_Y = pd.Series(test_pred_Y.clip(0, test_pred_Y.max()), index=test_Y.index)
rmse_train = np.sqrt(mean_squared_error(train_pred_Y, train_Y))
msle_train = mean_squared_log_error(train_pred_Y, train_Y)
rmse_test = np.sqrt(mean_squared_error(test_pred_Y, test_Y))
msle_test = mean_squared_log_error(test_pred_Y, test_Y)
print("rmse_train:", rmse_train, "msle_train:", msle_train)
print("rmse_test:", rmse_test, "msle_test:", msle_test)
# ### As we know the target not linear with many features, it is worth trying some nonlinear models. For example, the random forest model:
#
model = RandomForestRegressor(
n_estimators=50,
max_depth=6,
min_weight_fraction_leaf=0.05,
max_features=0.8,
random_state=42,
)
model.fit(train_X, train_Y)
train_pred_Y = model.predict(train_X)
test_pred_Y = model.predict(test_X)
train_pred_Y = pd.Series(train_pred_Y.clip(0, train_pred_Y.max()), index=train_Y.index)
test_pred_Y = pd.Series(test_pred_Y.clip(0, test_pred_Y.max()), index=test_Y.index)
rmse_train = np.sqrt(mean_squared_error(train_pred_Y, train_Y))
msle_train = mean_squared_log_error(train_pred_Y, train_Y)
rmse_test = np.sqrt(mean_squared_error(test_pred_Y, test_Y))
msle_test = mean_squared_log_error(test_pred_Y, test_Y)
print("rmse_train:", rmse_train, "msle_train:", msle_train)
print("rmse_test:", rmse_test, "msle_test:", msle_test)
# ## Visualization of Results
# ### To see how the model is doing, we can make scatter plot of prediction against ground truth. The model gives a reasonable prediction, as the data points are gathering around the line y=x.
plt.figure(figsize=(18, 12))
train_test_Y = train_Y.append(test_Y)
train_test_pred_Y = train_pred_Y.append(test_pred_Y)
data_shuffled = data.loc[train_test_Y.index]
label = data_shuffled["Country"]
colors = {
"ASIA (EX. NEAR EAST) ": "red",
"EASTERN EUROPE ": "orange",
"NORTHERN AFRICA ": "gold",
"OCEANIA ": "green",
"WESTERN EUROPE ": "blue",
"SUB-SAHARAN AFRICA ": "purple",
"LATIN AMER. & CARIB ": "olive",
"C.W. OF IND. STATES ": "cyan",
"NEAR EAST ": "hotpink",
"NORTHERN AMERICA ": "lightseagreen",
"BALTICS ": "rosybrown",
}
for region, color in colors.items():
X = train_test_Y.loc[data_shuffled["Region"] == region]
Y = train_test_pred_Y.loc[data_shuffled["Region"] == region]
ax = sns.regplot(
x=X,
y=Y,
marker=".",
fit_reg=False,
color=color,
scatter_kws={"s": 200, "linewidths": 0},
label=region,
)
plt.legend(loc=4, prop={"size": 12})
ax.set_xlabel("GDP ($ per capita) ground truth", labelpad=40)
ax.set_ylabel("GDP ($ per capita) predicted", labelpad=40)
ax.xaxis.label.set_fontsize(24)
ax.yaxis.label.set_fontsize(24)
ax.tick_params(labelsize=12)
x = np.linspace(-1000, 50000, 100) # 100 linearly spaced numbers
y = x
plt.plot(x, y, c="gray")
plt.xlim(-1000, 60000)
plt.ylim(-1000, 40000)
for i in range(0, train_test_Y.shape[0]):
if (
(data_shuffled["Area (sq. mi.)"].iloc[i] > 8e5)
| (data_shuffled["Population"].iloc[i] > 1e8)
| (data_shuffled["GDP ($ per capita)"].iloc[i] > 10000)
):
plt.text(
train_test_Y.iloc[i] + 200,
train_test_pred_Y.iloc[i] - 200,
label.iloc[i],
size="small",
)
# ## Total GDP
# ### Top Countries
# #### It is also interesting to look at the total GDPs, which I take as ‘GDP ($ per capita)’ × ‘Population’.
# Here are the top 10 countries with highest total GDPs, their GDP make up to about 2/3 of the global GDP.
data["Total_GDP ($)"] = data["GDP ($ per capita)"] * data["Population"]
# plt.figure(figsize=(16,6))
top_gdp_countries = data.sort_values("Total_GDP ($)", ascending=False).head(10)
other = pd.DataFrame(
{
"Country": ["Other"],
"Total_GDP ($)": [
data["Total_GDP ($)"].sum() - top_gdp_countries["Total_GDP ($)"].sum()
],
}
)
gdps = pd.concat(
[top_gdp_countries[["Country", "Total_GDP ($)"]], other], ignore_index=True
)
fig, axes = plt.subplots(
nrows=1, ncols=2, figsize=(20, 7), gridspec_kw={"width_ratios": [2, 1]}
)
sns.barplot(x="Country", y="Total_GDP ($)", data=gdps, ax=axes[0], palette="Set3")
axes[0].set_xlabel("Country", labelpad=30, fontsize=16)
axes[0].set_ylabel("Total_GDP", labelpad=30, fontsize=16)
colors = sns.color_palette("Set3", gdps.shape[0]).as_hex()
axes[1].pie(
gdps["Total_GDP ($)"],
labels=gdps["Country"],
colors=colors,
autopct="%1.1f%%",
shadow=True,
)
axes[1].axis("equal")
plt.show()
# ### Let’s compare the above ten countries’ rank in total GDP and GDP per capita.
#
Rank1 = (
data[["Country", "Total_GDP ($)"]]
.sort_values("Total_GDP ($)", ascending=False)
.reset_index()
)
Rank2 = (
data[["Country", "GDP ($ per capita)"]]
.sort_values("GDP ($ per capita)", ascending=False)
.reset_index()
)
Rank1 = pd.Series(Rank1.index.values + 1, index=Rank1.Country)
Rank2 = pd.Series(Rank2.index.values + 1, index=Rank2.Country)
Rank_change = (Rank2 - Rank1).sort_values(ascending=False)
print("rank of total GDP - rank of GDP per capita:")
Rank_change.loc[top_gdp_countries.Country]
# ### We see the countries with high total GDPs are quite different from those with high average GDPs.
# ### China and India jump above a lot when it comes to the total GDP.
# ### The only country that is with in top 10 (in fact top 2) for both total and average GDPs is the United States.
# ### Factors affecting Total GDP
# We can also check the correlation between total GDP and the other columns. The top two factors are population and area, following many factors that have also been found mostly correlated to GDP per capita.
corr_to_gdp = pd.Series()
for col in data.columns.values[2:]:
if (col != "Total_GDP ($)") & (col != "Climate") & (col != "GDP ($ per capita)"):
corr_to_gdp[col] = data["Total_GDP ($)"].corr(data[col])
abs_corr_to_gdp = corr_to_gdp.abs().sort_values(ascending=False)
corr_to_gdp = corr_to_gdp.loc[abs_corr_to_gdp.index]
print(corr_to_gdp)
# ## Comparison of the Top 10
# ### Finally, let us do a comparison of the economy structure for the ten countries with highest total GDP.
plot_data = top_gdp_countries.head(10)[
["Country", "Agriculture", "Industry", "Service"]
]
plot_data = plot_data.set_index("Country")
ax = plot_data.plot.bar(stacked=True, figsize=(10, 6))
ax.legend(bbox_to_anchor=(1, 1))
plt.show()
plot_data = top_gdp_countries[["Country", "Arable (%)", "Crops (%)", "Other (%)"]]
plot_data = plot_data.set_index("Country")
ax = plot_data.plot.bar(stacked=True, figsize=(10, 6))
ax.legend(bbox_to_anchor=(1, 1))
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/476/69476610.ipynb
|
world-bank-data-1960-to-2016-extended
|
neomatrix369
|
[{"Id": 69476610, "ScriptId": 18971180, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4874474, "CreationDate": "07/31/2021 13:30:37", "VersionNumber": 1.0, "Title": "GDP Analysis with Data Science advanced level", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 298.0, "LinesInsertedFromPrevious": 298.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 36}]
|
[{"Id": 92690766, "KernelVersionId": 69476610, "SourceDatasetVersionId": 1853628}]
|
[{"Id": 1853628, "DatasetId": 987256, "DatasourceVersionId": 1891446, "CreatorUserId": 2620712, "LicenseName": "CC0: Public Domain", "CreationDate": "01/17/2021 15:13:10", "VersionNumber": 31.0, "Title": "World Bank Data (1960 to 2016) Extended", "Slug": "world-bank-data-1960-to-2016-extended", "Subtitle": "Extended version of World bank data from 1960-2016", "Description": "### All the details in https://www.kaggle.com/gemartin/world-bank-data-1960-to-2016/ also apply to this new dataset.\n### The preprocessed data has been generated using the data preparatory notebook https://www.kaggle.com/neomatrix369/chaieda-world-bank-data-1960-2016-data-prep.\n\n### Context\n\nExtending the current dataset to enable better analysis and reasonings\n\n\n### Content\n\nA number of economic, geographic, country and region-specific data and indicators from different datasets have been aggregated.\n\n\n### Acknowledgements\n\nA number of Kaggle users have been helpful in the process of creation of this dataset, they have been mentioned in the data preparatory notebook https://www.kaggle.com/neomatrix369/chaieda-world-bank-data-1960-2016-data-prep.\n\n\n### Inspiration\n\nOther similar aggregated datasets and competition data and notebooks.", "VersionNotes": "Updating datasets", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 987256, "CreatorUserId": 2620712, "OwnerUserId": 2620712.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1853628.0, "CurrentDatasourceVersionId": 1891446.0, "ForumId": 1003806, "Type": 2, "CreationDate": "11/21/2020 10:55:31", "LastActivityDate": "11/21/2020", "TotalViews": 4098, "TotalDownloads": 284, "TotalVotes": 4, "TotalKernels": 10}]
|
[{"Id": 2620712, "UserName": "neomatrix369", "DisplayName": "Mani Sarkar", "RegisterDate": "12/16/2018", "PerformanceTier": 2}]
|
# 
# ### In this Data Science Project, I am investigating the dataset “Countries of the World”. I will be focusing on the factors affecting a country’s GDP per capita and try to make a model using the data of 227 countries from the dataset. I will also briefly discuss the total GDP.
# # 1. importing the required Python libraries
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
from matplotlib import pyplot as plt
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_squared_error, mean_squared_log_error
# # 2. Let’s look at the data
data = pd.read_csv(
"../input/world-bank-data-1960-to-2016-extended/countries-of-the-world/countries of the world.csv",
decimal=",",
)
data.head()
data.info()
data.describe(include="all")
# # 3. Missing values
print("number of missing data:")
print(data.isnull().sum())
# ## Data Preparation – fill in missing values
# We noticed that there are some missing data in the table. For simplicity, I will just fill the missing data using the median of the region that a country belongs, as countries that are close geologically are often similar in many ways. For example, lets check the region median of ‘GDP ($ per capita)’, ‘Literacy (%)’ and ‘Agriculture’.
# Note that for ‘climate’ we use the mode instead of median as it seems that ‘climate’ is a categorical feature here.
data.groupby("Region")[["GDP ($ per capita)", "Literacy (%)", "Agriculture"]].median()
for col in data.columns.values:
if data[col].isnull().sum() == 0:
continue
if col == "Climate":
guess_values = data.groupby("Region")["Climate"].apply(lambda x: x.mode().max())
else:
guess_values = data.groupby("Region")[col].median()
for region in data["Region"].unique():
data[col].loc[(data[col].isnull()) & (data["Region"] == region)] = guess_values[
region
]
# # 4. Data Exploration
# ### Top Countries with highest GDP per capita
# Look at the top 20 countries with highest GDP per capita. Luxembourg is quite ahead, the next 19 countries are close. German, the 20th has about 2.5 times GDP per capita of the world average.
fig, ax = plt.subplots(figsize=(16, 6))
# ax = fig.add_subplot(111)
top_gdp_countries = data.sort_values("GDP ($ per capita)", ascending=False).head(20)
mean = pd.DataFrame(
{
"Country": ["World mean"],
"GDP ($ per capita)": [data["GDP ($ per capita)"].mean()],
}
)
gdps = pd.concat(
[top_gdp_countries[["Country", "GDP ($ per capita)"]], mean], ignore_index=True
)
sns.barplot(x="Country", y="GDP ($ per capita)", data=gdps, palette="Set3")
ax.set_xlabel(ax.get_xlabel(), labelpad=15)
ax.set_ylabel(ax.get_ylabel(), labelpad=30)
ax.xaxis.label.set_fontsize(16)
ax.yaxis.label.set_fontsize(16)
plt.xticks(rotation=90)
plt.show()
# # 4. Correlation between Variables
# ### The heatmap shows the correlation between all numerical columns.
#
plt.figure(figsize=(16, 12))
sns.heatmap(data=data.iloc[:, 2:].corr(), annot=True, fmt=".2f", cmap="coolwarm")
plt.show()
# ## Top Factors affecting GDP per capita
# ### We pick the six columns that mostly correlated to GDP per capita and make scatter plots. The results agree with our common sense. Also we notice there are many countries with low average GDP and few with high average GDP —- a pyramid structure.
fig, axes = plt.subplots(nrows=2, ncols=3, figsize=(20, 12))
plt.subplots_adjust(hspace=0.4)
corr_to_gdp = pd.Series()
for col in data.columns.values[2:]:
if (col != "GDP ($ per capita)") & (col != "Climate"):
corr_to_gdp[col] = data["GDP ($ per capita)"].corr(data[col])
abs_corr_to_gdp = corr_to_gdp.abs().sort_values(ascending=False)
corr_to_gdp = corr_to_gdp.loc[abs_corr_to_gdp.index]
for i in range(2):
for j in range(3):
sns.regplot(
x=corr_to_gdp.index.values[i * 3 + j],
y="GDP ($ per capita)",
data=data,
ax=axes[i, j],
fit_reg=False,
marker=".",
)
title = "correlation=" + str(corr_to_gdp[i * 3 + j])
axes[i, j].set_title(title)
axes[1, 2].set_xlim(0, 102)
plt.show()
# ## Countries with low Birthrate and low GDP per capita
# ### Some features, like phones, are related to the average GDP more linearly, while others are not. For example, High birthrate usually means low GDP per capita, but average GDP in low birthrate countries can vary a lot.
# ### Let’s look at the countries with low birthrate (<14%) and low GDP per capita (<10000 $). They also have hight literacy, like other high average GDP countires. But we hope their other features can help distiguish them from those with low birthrate but high average GDPs, like service are not quite an importent portion in their economy, not a lot phone procession, some have negative net migration, and many of them are from eastern Europe or C.W. of IND. STATES, so the ‘region’ feature may also be useful.
data.loc[(data["Birthrate"] < 14) & (data["GDP ($ per capita)"] < 10000)]
# # 5. Modeling
# ## Training and Testing
# ### First label encode the categorical features ‘Region’ and ‘Climate’, and I will just use all features given in the data set without further feature engineering.
LE = LabelEncoder()
data["Region_label"] = LE.fit_transform(data["Region"])
data["Climate_label"] = LE.fit_transform(data["Climate"])
data.head()
train, test = train_test_split(data, test_size=0.3, shuffle=True)
training_features = [
"Population",
"Area (sq. mi.)",
"Pop. Density (per sq. mi.)",
"Coastline (coast/area ratio)",
"Net migration",
"Infant mortality (per 1000 births)",
"Literacy (%)",
"Phones (per 1000)",
"Arable (%)",
"Crops (%)",
"Other (%)",
"Birthrate",
"Deathrate",
"Agriculture",
"Industry",
"Service",
"Region_label",
"Climate_label",
"Service",
]
target = "GDP ($ per capita)"
train_X = train[training_features]
train_Y = train[target]
test_X = test[training_features]
test_Y = test[target]
# ### First let’s try the linear regression model. As for metric, I will check both root mean squared error and mean squared log error.
#
model = LinearRegression()
model.fit(train_X, train_Y)
train_pred_Y = model.predict(train_X)
test_pred_Y = model.predict(test_X)
train_pred_Y = pd.Series(train_pred_Y.clip(0, train_pred_Y.max()), index=train_Y.index)
test_pred_Y = pd.Series(test_pred_Y.clip(0, test_pred_Y.max()), index=test_Y.index)
rmse_train = np.sqrt(mean_squared_error(train_pred_Y, train_Y))
msle_train = mean_squared_log_error(train_pred_Y, train_Y)
rmse_test = np.sqrt(mean_squared_error(test_pred_Y, test_Y))
msle_test = mean_squared_log_error(test_pred_Y, test_Y)
print("rmse_train:", rmse_train, "msle_train:", msle_train)
print("rmse_test:", rmse_test, "msle_test:", msle_test)
# ### As we know the target not linear with many features, it is worth trying some nonlinear models. For example, the random forest model:
#
model = RandomForestRegressor(
n_estimators=50,
max_depth=6,
min_weight_fraction_leaf=0.05,
max_features=0.8,
random_state=42,
)
model.fit(train_X, train_Y)
train_pred_Y = model.predict(train_X)
test_pred_Y = model.predict(test_X)
train_pred_Y = pd.Series(train_pred_Y.clip(0, train_pred_Y.max()), index=train_Y.index)
test_pred_Y = pd.Series(test_pred_Y.clip(0, test_pred_Y.max()), index=test_Y.index)
rmse_train = np.sqrt(mean_squared_error(train_pred_Y, train_Y))
msle_train = mean_squared_log_error(train_pred_Y, train_Y)
rmse_test = np.sqrt(mean_squared_error(test_pred_Y, test_Y))
msle_test = mean_squared_log_error(test_pred_Y, test_Y)
print("rmse_train:", rmse_train, "msle_train:", msle_train)
print("rmse_test:", rmse_test, "msle_test:", msle_test)
# ## Visualization of Results
# ### To see how the model is doing, we can make scatter plot of prediction against ground truth. The model gives a reasonable prediction, as the data points are gathering around the line y=x.
plt.figure(figsize=(18, 12))
train_test_Y = train_Y.append(test_Y)
train_test_pred_Y = train_pred_Y.append(test_pred_Y)
data_shuffled = data.loc[train_test_Y.index]
label = data_shuffled["Country"]
colors = {
"ASIA (EX. NEAR EAST) ": "red",
"EASTERN EUROPE ": "orange",
"NORTHERN AFRICA ": "gold",
"OCEANIA ": "green",
"WESTERN EUROPE ": "blue",
"SUB-SAHARAN AFRICA ": "purple",
"LATIN AMER. & CARIB ": "olive",
"C.W. OF IND. STATES ": "cyan",
"NEAR EAST ": "hotpink",
"NORTHERN AMERICA ": "lightseagreen",
"BALTICS ": "rosybrown",
}
for region, color in colors.items():
X = train_test_Y.loc[data_shuffled["Region"] == region]
Y = train_test_pred_Y.loc[data_shuffled["Region"] == region]
ax = sns.regplot(
x=X,
y=Y,
marker=".",
fit_reg=False,
color=color,
scatter_kws={"s": 200, "linewidths": 0},
label=region,
)
plt.legend(loc=4, prop={"size": 12})
ax.set_xlabel("GDP ($ per capita) ground truth", labelpad=40)
ax.set_ylabel("GDP ($ per capita) predicted", labelpad=40)
ax.xaxis.label.set_fontsize(24)
ax.yaxis.label.set_fontsize(24)
ax.tick_params(labelsize=12)
x = np.linspace(-1000, 50000, 100) # 100 linearly spaced numbers
y = x
plt.plot(x, y, c="gray")
plt.xlim(-1000, 60000)
plt.ylim(-1000, 40000)
for i in range(0, train_test_Y.shape[0]):
if (
(data_shuffled["Area (sq. mi.)"].iloc[i] > 8e5)
| (data_shuffled["Population"].iloc[i] > 1e8)
| (data_shuffled["GDP ($ per capita)"].iloc[i] > 10000)
):
plt.text(
train_test_Y.iloc[i] + 200,
train_test_pred_Y.iloc[i] - 200,
label.iloc[i],
size="small",
)
# ## Total GDP
# ### Top Countries
# #### It is also interesting to look at the total GDPs, which I take as ‘GDP ($ per capita)’ × ‘Population’.
# Here are the top 10 countries with highest total GDPs, their GDP make up to about 2/3 of the global GDP.
data["Total_GDP ($)"] = data["GDP ($ per capita)"] * data["Population"]
# plt.figure(figsize=(16,6))
top_gdp_countries = data.sort_values("Total_GDP ($)", ascending=False).head(10)
other = pd.DataFrame(
{
"Country": ["Other"],
"Total_GDP ($)": [
data["Total_GDP ($)"].sum() - top_gdp_countries["Total_GDP ($)"].sum()
],
}
)
gdps = pd.concat(
[top_gdp_countries[["Country", "Total_GDP ($)"]], other], ignore_index=True
)
fig, axes = plt.subplots(
nrows=1, ncols=2, figsize=(20, 7), gridspec_kw={"width_ratios": [2, 1]}
)
sns.barplot(x="Country", y="Total_GDP ($)", data=gdps, ax=axes[0], palette="Set3")
axes[0].set_xlabel("Country", labelpad=30, fontsize=16)
axes[0].set_ylabel("Total_GDP", labelpad=30, fontsize=16)
colors = sns.color_palette("Set3", gdps.shape[0]).as_hex()
axes[1].pie(
gdps["Total_GDP ($)"],
labels=gdps["Country"],
colors=colors,
autopct="%1.1f%%",
shadow=True,
)
axes[1].axis("equal")
plt.show()
# ### Let’s compare the above ten countries’ rank in total GDP and GDP per capita.
#
Rank1 = (
data[["Country", "Total_GDP ($)"]]
.sort_values("Total_GDP ($)", ascending=False)
.reset_index()
)
Rank2 = (
data[["Country", "GDP ($ per capita)"]]
.sort_values("GDP ($ per capita)", ascending=False)
.reset_index()
)
Rank1 = pd.Series(Rank1.index.values + 1, index=Rank1.Country)
Rank2 = pd.Series(Rank2.index.values + 1, index=Rank2.Country)
Rank_change = (Rank2 - Rank1).sort_values(ascending=False)
print("rank of total GDP - rank of GDP per capita:")
Rank_change.loc[top_gdp_countries.Country]
# ### We see the countries with high total GDPs are quite different from those with high average GDPs.
# ### China and India jump above a lot when it comes to the total GDP.
# ### The only country that is with in top 10 (in fact top 2) for both total and average GDPs is the United States.
# ### Factors affecting Total GDP
# We can also check the correlation between total GDP and the other columns. The top two factors are population and area, following many factors that have also been found mostly correlated to GDP per capita.
corr_to_gdp = pd.Series()
for col in data.columns.values[2:]:
if (col != "Total_GDP ($)") & (col != "Climate") & (col != "GDP ($ per capita)"):
corr_to_gdp[col] = data["Total_GDP ($)"].corr(data[col])
abs_corr_to_gdp = corr_to_gdp.abs().sort_values(ascending=False)
corr_to_gdp = corr_to_gdp.loc[abs_corr_to_gdp.index]
print(corr_to_gdp)
# ## Comparison of the Top 10
# ### Finally, let us do a comparison of the economy structure for the ten countries with highest total GDP.
plot_data = top_gdp_countries.head(10)[
["Country", "Agriculture", "Industry", "Service"]
]
plot_data = plot_data.set_index("Country")
ax = plot_data.plot.bar(stacked=True, figsize=(10, 6))
ax.legend(bbox_to_anchor=(1, 1))
plt.show()
plot_data = top_gdp_countries[["Country", "Arable (%)", "Crops (%)", "Other (%)"]]
plot_data = plot_data.set_index("Country")
ax = plot_data.plot.bar(stacked=True, figsize=(10, 6))
ax.legend(bbox_to_anchor=(1, 1))
plt.show()
|
[{"world-bank-data-1960-to-2016-extended/countries-of-the-world/countries of the world.csv": {"column_names": "[\"Country\", \"Region\", \"Population\", \"Area (sq. mi.)\", \"Pop. Density (per sq. mi.)\", \"Coastline (coast/area ratio)\", \"Net migration\", \"Infant mortality (per 1000 births)\", \"GDP ($ per capita)\", \"Literacy (%)\", \"Phones (per 1000)\", \"Arable (%)\", \"Crops (%)\", \"Other (%)\", \"Climate\", \"Birthrate\", \"Deathrate\", \"Agriculture\", \"Industry\", \"Service\"]", "column_data_types": "{\"Country\": \"object\", \"Region\": \"object\", \"Population\": \"int64\", \"Area (sq. mi.)\": \"int64\", \"Pop. Density (per sq. mi.)\": \"object\", \"Coastline (coast/area ratio)\": \"object\", \"Net migration\": \"object\", \"Infant mortality (per 1000 births)\": \"object\", \"GDP ($ per capita)\": \"float64\", \"Literacy (%)\": \"object\", \"Phones (per 1000)\": \"object\", \"Arable (%)\": \"object\", \"Crops (%)\": \"object\", \"Other (%)\": \"object\", \"Climate\": \"object\", \"Birthrate\": \"object\", \"Deathrate\": \"object\", \"Agriculture\": \"object\", \"Industry\": \"object\", \"Service\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 227 entries, 0 to 226\nData columns (total 20 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Country 227 non-null object \n 1 Region 227 non-null object \n 2 Population 227 non-null int64 \n 3 Area (sq. mi.) 227 non-null int64 \n 4 Pop. Density (per sq. mi.) 227 non-null object \n 5 Coastline (coast/area ratio) 227 non-null object \n 6 Net migration 224 non-null object \n 7 Infant mortality (per 1000 births) 224 non-null object \n 8 GDP ($ per capita) 226 non-null float64\n 9 Literacy (%) 209 non-null object \n 10 Phones (per 1000) 223 non-null object \n 11 Arable (%) 225 non-null object \n 12 Crops (%) 225 non-null object \n 13 Other (%) 225 non-null object \n 14 Climate 205 non-null object \n 15 Birthrate 224 non-null object \n 16 Deathrate 223 non-null object \n 17 Agriculture 212 non-null object \n 18 Industry 211 non-null object \n 19 Service 212 non-null object \ndtypes: float64(1), int64(2), object(17)\nmemory usage: 35.6+ KB\n", "summary": "{\"Population\": {\"count\": 227.0, \"mean\": 28740284.365638766, \"std\": 117891326.54347652, \"min\": 7026.0, \"25%\": 437624.0, \"50%\": 4786994.0, \"75%\": 17497772.5, \"max\": 1313973713.0}, \"Area (sq. mi.)\": {\"count\": 227.0, \"mean\": 598226.9559471365, \"std\": 1790282.2437336047, \"min\": 2.0, \"25%\": 4647.5, \"50%\": 86600.0, \"75%\": 441811.0, \"max\": 17075200.0}, \"GDP ($ per capita)\": {\"count\": 226.0, \"mean\": 9689.823008849558, \"std\": 10049.13851319723, \"min\": 500.0, \"25%\": 1900.0, \"50%\": 5550.0, \"75%\": 15700.0, \"max\": 55100.0}}", "examples": "{\"Country\":{\"0\":\"Afghanistan \",\"1\":\"Albania \",\"2\":\"Algeria \",\"3\":\"American Samoa \"},\"Region\":{\"0\":\"ASIA (EX. NEAR EAST) \",\"1\":\"EASTERN EUROPE \",\"2\":\"NORTHERN AFRICA \",\"3\":\"OCEANIA \"},\"Population\":{\"0\":31056997,\"1\":3581655,\"2\":32930091,\"3\":57794},\"Area (sq. mi.)\":{\"0\":647500,\"1\":28748,\"2\":2381740,\"3\":199},\"Pop. Density (per sq. mi.)\":{\"0\":\"48,0\",\"1\":\"124,6\",\"2\":\"13,8\",\"3\":\"290,4\"},\"Coastline (coast\\/area ratio)\":{\"0\":\"0,00\",\"1\":\"1,26\",\"2\":\"0,04\",\"3\":\"58,29\"},\"Net migration\":{\"0\":\"23,06\",\"1\":\"-4,93\",\"2\":\"-0,39\",\"3\":\"-20,71\"},\"Infant mortality (per 1000 births)\":{\"0\":\"163,07\",\"1\":\"21,52\",\"2\":\"31\",\"3\":\"9,27\"},\"GDP ($ per capita)\":{\"0\":700.0,\"1\":4500.0,\"2\":6000.0,\"3\":8000.0},\"Literacy (%)\":{\"0\":\"36,0\",\"1\":\"86,5\",\"2\":\"70,0\",\"3\":\"97,0\"},\"Phones (per 1000)\":{\"0\":\"3,2\",\"1\":\"71,2\",\"2\":\"78,1\",\"3\":\"259,5\"},\"Arable (%)\":{\"0\":\"12,13\",\"1\":\"21,09\",\"2\":\"3,22\",\"3\":\"10\"},\"Crops (%)\":{\"0\":\"0,22\",\"1\":\"4,42\",\"2\":\"0,25\",\"3\":\"15\"},\"Other (%)\":{\"0\":\"87,65\",\"1\":\"74,49\",\"2\":\"96,53\",\"3\":\"75\"},\"Climate\":{\"0\":\"1\",\"1\":\"3\",\"2\":\"1\",\"3\":\"2\"},\"Birthrate\":{\"0\":\"46,6\",\"1\":\"15,11\",\"2\":\"17,14\",\"3\":\"22,46\"},\"Deathrate\":{\"0\":\"20,34\",\"1\":\"5,22\",\"2\":\"4,61\",\"3\":\"3,27\"},\"Agriculture\":{\"0\":\"0,38\",\"1\":\"0,232\",\"2\":\"0,101\",\"3\":null},\"Industry\":{\"0\":\"0,24\",\"1\":\"0,188\",\"2\":\"0,6\",\"3\":null},\"Service\":{\"0\":\"0,38\",\"1\":\"0,579\",\"2\":\"0,298\",\"3\":null}}"}}]
| true | 1 |
<start_data_description><data_path>world-bank-data-1960-to-2016-extended/countries-of-the-world/countries of the world.csv:
<column_names>
['Country', 'Region', 'Population', 'Area (sq. mi.)', 'Pop. Density (per sq. mi.)', 'Coastline (coast/area ratio)', 'Net migration', 'Infant mortality (per 1000 births)', 'GDP ($ per capita)', 'Literacy (%)', 'Phones (per 1000)', 'Arable (%)', 'Crops (%)', 'Other (%)', 'Climate', 'Birthrate', 'Deathrate', 'Agriculture', 'Industry', 'Service']
<column_types>
{'Country': 'object', 'Region': 'object', 'Population': 'int64', 'Area (sq. mi.)': 'int64', 'Pop. Density (per sq. mi.)': 'object', 'Coastline (coast/area ratio)': 'object', 'Net migration': 'object', 'Infant mortality (per 1000 births)': 'object', 'GDP ($ per capita)': 'float64', 'Literacy (%)': 'object', 'Phones (per 1000)': 'object', 'Arable (%)': 'object', 'Crops (%)': 'object', 'Other (%)': 'object', 'Climate': 'object', 'Birthrate': 'object', 'Deathrate': 'object', 'Agriculture': 'object', 'Industry': 'object', 'Service': 'object'}
<dataframe_Summary>
{'Population': {'count': 227.0, 'mean': 28740284.365638766, 'std': 117891326.54347652, 'min': 7026.0, '25%': 437624.0, '50%': 4786994.0, '75%': 17497772.5, 'max': 1313973713.0}, 'Area (sq. mi.)': {'count': 227.0, 'mean': 598226.9559471365, 'std': 1790282.2437336047, 'min': 2.0, '25%': 4647.5, '50%': 86600.0, '75%': 441811.0, 'max': 17075200.0}, 'GDP ($ per capita)': {'count': 226.0, 'mean': 9689.823008849558, 'std': 10049.13851319723, 'min': 500.0, '25%': 1900.0, '50%': 5550.0, '75%': 15700.0, 'max': 55100.0}}
<dataframe_info>
RangeIndex: 227 entries, 0 to 226
Data columns (total 20 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Country 227 non-null object
1 Region 227 non-null object
2 Population 227 non-null int64
3 Area (sq. mi.) 227 non-null int64
4 Pop. Density (per sq. mi.) 227 non-null object
5 Coastline (coast/area ratio) 227 non-null object
6 Net migration 224 non-null object
7 Infant mortality (per 1000 births) 224 non-null object
8 GDP ($ per capita) 226 non-null float64
9 Literacy (%) 209 non-null object
10 Phones (per 1000) 223 non-null object
11 Arable (%) 225 non-null object
12 Crops (%) 225 non-null object
13 Other (%) 225 non-null object
14 Climate 205 non-null object
15 Birthrate 224 non-null object
16 Deathrate 223 non-null object
17 Agriculture 212 non-null object
18 Industry 211 non-null object
19 Service 212 non-null object
dtypes: float64(1), int64(2), object(17)
memory usage: 35.6+ KB
<some_examples>
{'Country': {'0': 'Afghanistan ', '1': 'Albania ', '2': 'Algeria ', '3': 'American Samoa '}, 'Region': {'0': 'ASIA (EX. NEAR EAST) ', '1': 'EASTERN EUROPE ', '2': 'NORTHERN AFRICA ', '3': 'OCEANIA '}, 'Population': {'0': 31056997, '1': 3581655, '2': 32930091, '3': 57794}, 'Area (sq. mi.)': {'0': 647500, '1': 28748, '2': 2381740, '3': 199}, 'Pop. Density (per sq. mi.)': {'0': '48,0', '1': '124,6', '2': '13,8', '3': '290,4'}, 'Coastline (coast/area ratio)': {'0': '0,00', '1': '1,26', '2': '0,04', '3': '58,29'}, 'Net migration': {'0': '23,06', '1': '-4,93', '2': '-0,39', '3': '-20,71'}, 'Infant mortality (per 1000 births)': {'0': '163,07', '1': '21,52', '2': '31', '3': '9,27'}, 'GDP ($ per capita)': {'0': 700.0, '1': 4500.0, '2': 6000.0, '3': 8000.0}, 'Literacy (%)': {'0': '36,0', '1': '86,5', '2': '70,0', '3': '97,0'}, 'Phones (per 1000)': {'0': '3,2', '1': '71,2', '2': '78,1', '3': '259,5'}, 'Arable (%)': {'0': '12,13', '1': '21,09', '2': '3,22', '3': '10'}, 'Crops (%)': {'0': '0,22', '1': '4,42', '2': '0,25', '3': '15'}, 'Other (%)': {'0': '87,65', '1': '74,49', '2': '96,53', '3': '75'}, 'Climate': {'0': '1', '1': '3', '2': '1', '3': '2'}, 'Birthrate': {'0': '46,6', '1': '15,11', '2': '17,14', '3': '22,46'}, 'Deathrate': {'0': '20,34', '1': '5,22', '2': '4,61', '3': '3,27'}, 'Agriculture': {'0': '0,38', '1': '0,232', '2': '0,101', '3': None}, 'Industry': {'0': '0,24', '1': '0,188', '2': '0,6', '3': None}, 'Service': {'0': '0,38', '1': '0,579', '2': '0,298', '3': None}}
<end_description>
| 4,578 | 36 | 6,392 | 4,578 |
69476476
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sb
# # Reading Files
train_df = pd.read_csv("/kaggle/input/neolen-house-price-prediction/train.csv")
test_df = pd.read_csv("/kaggle/input/neolen-house-price-prediction/test.csv")
print(train_df.shape)
print(test_df.shape)
# # Display the information of train data
train_df.info()
train_df.hist(bins=50, figsize=(20, 15))
train_df.isnull().sum().sort_values(ascending=False).head(20)
# Separate Numerical columns from Categorical columns
object_df = train_df.select_dtypes(include="object")
numeric_df = train_df.select_dtypes(exclude="object")
object_df.shape, numeric_df.shape
# # Drop dummy column
train_df = train_df.drop(["PoolQC", "MiscFeature", "Alley", "Fence"], axis=1)
# # Replace null values in numerical columns with suitable values
numeric_df.isnull().sum().sort_values(ascending=False).head(10)
train_df["MasVnrArea"] = train_df["MasVnrArea"].fillna(train_df.MasVnrArea.mean())
train_df["GarageYrBlt"] = train_df["GarageYrBlt"].fillna(train_df.GarageYrBlt.mean())
train_df["LotFrontage"] = train_df["LotFrontage"].fillna(train_df.LotFrontage.mean())
# # Replace null values in Categorical columns with suitable values
object_df.isnull().sum().sort_values(ascending=False).head(17)
str_cols = [
"BsmtCond",
"BsmtExposure",
"BsmtFinType1",
"BsmtFinType2",
"MasVnrType",
"BsmtQual",
]
train_df[str_cols] = train_df[str_cols].fillna("None")
# # Checking for Outliers
for feature in numeric_df:
data = train_df.copy()
if 0 in data[feature].unique():
pass
else:
data[feature] = np.log(data[feature])
data.boxplot(column=feature)
plt.ylabel(feature)
plt.title(feature)
plt.show()
train_df["GarageType"] = train_df["GarageType"].fillna("Attchd")
train_df["GarageCond"] = train_df["GarageCond"].fillna("TA")
train_df["GarageFinish"] = train_df["GarageFinish"].fillna("Unf")
train_df["GarageQual"] = train_df["GarageQual"].fillna("TA")
train_df["FireplaceQu"] = train_df["FireplaceQu"].fillna("None")
# # There is no missing values now!
train_df.isnull().sum().sort_values(ascending=False).head(10)
# # Encoding categorical datatype to numerical datatype
for i in train_df.select_dtypes(include="object"):
train_df[i] = pd.get_dummies(train_df[i], prefix=[i], columns=[i])
train_df
# # Feature Scaling
num_features = ["LotFrontage", "LotArea", "1stFlrSF", "GrLivArea"]
for feature in num_features:
train_df[feature] = np.log(train_df[feature])
train_df
# # Data Splitting
y = train_df.SalePrice
x = train_df.drop(["Id", "SalePrice"], axis=1)
x.corrwith(y).sort_values(ascending=False).tail(20)
from sklearn.model_selection import train_test_split
x_train, x_valid, y_train, y_valid = train_test_split(
x, y, train_size=0.8, test_size=0.2, random_state=0
)
y_valid.shape
# # Model Training
from sklearn.linear_model import LinearRegression
# Create a model
model = LinearRegression()
# Fit the model
model.fit(x_train, y_train)
# Get the R-squared
r_sq = model.score(x_train, y_train)
print("coefficient of determination:", r_sq)
y_pred = model.predict(x_valid)
from sklearn.metrics import mean_squared_error
from math import sqrt
rms = sqrt(mean_squared_error(y_valid, y_pred))
print("RMS :", rms)
# Create the Ridge model
from sklearn.metrics import r2_score
from sklearn.linear_model import Ridge
rid_reg = Ridge(alpha=8.01)
rid_reg.fit(x_train, y_train)
y_pred_ridge = rid_reg.predict(x_valid)
print("R2 SCORE With ridge : ", r2_score(y_valid, y_pred_ridge))
# # Preprocessing for test file
test_df.isnull().sum().sort_values(ascending=False).head(20)
test_df["MasVnrArea"] = test_df["MasVnrArea"].fillna(test_df.MasVnrArea.mean())
test_df["GarageYrBlt"] = test_df["GarageYrBlt"].fillna(test_df.GarageYrBlt.mean())
test_df["LotFrontage"] = test_df["LotFrontage"].fillna(test_df.LotFrontage.mean())
test_df = test_df.drop(["PoolQC", "MiscFeature", "Alley", "Fence"], axis=1)
test_df = test_df.drop(["Id"], axis=1)
test_df["GarageType"] = test_df["GarageType"].fillna("Attchd")
test_df["GarageCond"] = test_df["GarageCond"].fillna("TA")
test_df["GarageFinish"] = test_df["GarageFinish"].fillna("Unf")
test_df["GarageQual"] = test_df["GarageQual"].fillna("TA")
test_df["Electrical"] = test_df["Electrical"].fillna(1)
test_df["FireplaceQu"] = test_df["FireplaceQu"].fillna("None")
str_cols = [
"BsmtQual",
"BsmtCond",
"BsmtExposure",
"BsmtFinType1",
"BsmtFinType2",
"MasVnrType",
]
test_df[str_cols] = test_df[str_cols].fillna("None")
test_df.isnull().sum().sort_values(ascending=False).head(20)
# Encodding
obj_col = test_df.select_dtypes(include="object")
for i in obj_col:
test_df[i] = pd.get_dummies(test_df[i], prefix=[i], columns=[i])
num_features = ["LotFrontage", "LotArea", "1stFlrSF", "GrLivArea"]
for feature in num_features:
test_df[feature] = np.log(test_df[feature])
test_df.info()
y_new_pred1 = model.predict(test_df)
submission = pd.read_csv("../input/neolen-house-price-prediction/test.csv")
submission_df = pd.DataFrame()
submission_df["Id"] = submission.Id
submission_df["SalePrice"] = y_new_pred1
submission_df
submission_df.to_csv("submission.csv", index=False, header=True)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/476/69476476.ipynb
| null | null |
[{"Id": 69476476, "ScriptId": 18807035, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7771740, "CreationDate": "07/31/2021 13:28:44", "VersionNumber": 19.0, "Title": "House price prediction", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 204.0, "LinesInsertedFromPrevious": 4.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 200.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sb
# # Reading Files
train_df = pd.read_csv("/kaggle/input/neolen-house-price-prediction/train.csv")
test_df = pd.read_csv("/kaggle/input/neolen-house-price-prediction/test.csv")
print(train_df.shape)
print(test_df.shape)
# # Display the information of train data
train_df.info()
train_df.hist(bins=50, figsize=(20, 15))
train_df.isnull().sum().sort_values(ascending=False).head(20)
# Separate Numerical columns from Categorical columns
object_df = train_df.select_dtypes(include="object")
numeric_df = train_df.select_dtypes(exclude="object")
object_df.shape, numeric_df.shape
# # Drop dummy column
train_df = train_df.drop(["PoolQC", "MiscFeature", "Alley", "Fence"], axis=1)
# # Replace null values in numerical columns with suitable values
numeric_df.isnull().sum().sort_values(ascending=False).head(10)
train_df["MasVnrArea"] = train_df["MasVnrArea"].fillna(train_df.MasVnrArea.mean())
train_df["GarageYrBlt"] = train_df["GarageYrBlt"].fillna(train_df.GarageYrBlt.mean())
train_df["LotFrontage"] = train_df["LotFrontage"].fillna(train_df.LotFrontage.mean())
# # Replace null values in Categorical columns with suitable values
object_df.isnull().sum().sort_values(ascending=False).head(17)
str_cols = [
"BsmtCond",
"BsmtExposure",
"BsmtFinType1",
"BsmtFinType2",
"MasVnrType",
"BsmtQual",
]
train_df[str_cols] = train_df[str_cols].fillna("None")
# # Checking for Outliers
for feature in numeric_df:
data = train_df.copy()
if 0 in data[feature].unique():
pass
else:
data[feature] = np.log(data[feature])
data.boxplot(column=feature)
plt.ylabel(feature)
plt.title(feature)
plt.show()
train_df["GarageType"] = train_df["GarageType"].fillna("Attchd")
train_df["GarageCond"] = train_df["GarageCond"].fillna("TA")
train_df["GarageFinish"] = train_df["GarageFinish"].fillna("Unf")
train_df["GarageQual"] = train_df["GarageQual"].fillna("TA")
train_df["FireplaceQu"] = train_df["FireplaceQu"].fillna("None")
# # There is no missing values now!
train_df.isnull().sum().sort_values(ascending=False).head(10)
# # Encoding categorical datatype to numerical datatype
for i in train_df.select_dtypes(include="object"):
train_df[i] = pd.get_dummies(train_df[i], prefix=[i], columns=[i])
train_df
# # Feature Scaling
num_features = ["LotFrontage", "LotArea", "1stFlrSF", "GrLivArea"]
for feature in num_features:
train_df[feature] = np.log(train_df[feature])
train_df
# # Data Splitting
y = train_df.SalePrice
x = train_df.drop(["Id", "SalePrice"], axis=1)
x.corrwith(y).sort_values(ascending=False).tail(20)
from sklearn.model_selection import train_test_split
x_train, x_valid, y_train, y_valid = train_test_split(
x, y, train_size=0.8, test_size=0.2, random_state=0
)
y_valid.shape
# # Model Training
from sklearn.linear_model import LinearRegression
# Create a model
model = LinearRegression()
# Fit the model
model.fit(x_train, y_train)
# Get the R-squared
r_sq = model.score(x_train, y_train)
print("coefficient of determination:", r_sq)
y_pred = model.predict(x_valid)
from sklearn.metrics import mean_squared_error
from math import sqrt
rms = sqrt(mean_squared_error(y_valid, y_pred))
print("RMS :", rms)
# Create the Ridge model
from sklearn.metrics import r2_score
from sklearn.linear_model import Ridge
rid_reg = Ridge(alpha=8.01)
rid_reg.fit(x_train, y_train)
y_pred_ridge = rid_reg.predict(x_valid)
print("R2 SCORE With ridge : ", r2_score(y_valid, y_pred_ridge))
# # Preprocessing for test file
test_df.isnull().sum().sort_values(ascending=False).head(20)
test_df["MasVnrArea"] = test_df["MasVnrArea"].fillna(test_df.MasVnrArea.mean())
test_df["GarageYrBlt"] = test_df["GarageYrBlt"].fillna(test_df.GarageYrBlt.mean())
test_df["LotFrontage"] = test_df["LotFrontage"].fillna(test_df.LotFrontage.mean())
test_df = test_df.drop(["PoolQC", "MiscFeature", "Alley", "Fence"], axis=1)
test_df = test_df.drop(["Id"], axis=1)
test_df["GarageType"] = test_df["GarageType"].fillna("Attchd")
test_df["GarageCond"] = test_df["GarageCond"].fillna("TA")
test_df["GarageFinish"] = test_df["GarageFinish"].fillna("Unf")
test_df["GarageQual"] = test_df["GarageQual"].fillna("TA")
test_df["Electrical"] = test_df["Electrical"].fillna(1)
test_df["FireplaceQu"] = test_df["FireplaceQu"].fillna("None")
str_cols = [
"BsmtQual",
"BsmtCond",
"BsmtExposure",
"BsmtFinType1",
"BsmtFinType2",
"MasVnrType",
]
test_df[str_cols] = test_df[str_cols].fillna("None")
test_df.isnull().sum().sort_values(ascending=False).head(20)
# Encodding
obj_col = test_df.select_dtypes(include="object")
for i in obj_col:
test_df[i] = pd.get_dummies(test_df[i], prefix=[i], columns=[i])
num_features = ["LotFrontage", "LotArea", "1stFlrSF", "GrLivArea"]
for feature in num_features:
test_df[feature] = np.log(test_df[feature])
test_df.info()
y_new_pred1 = model.predict(test_df)
submission = pd.read_csv("../input/neolen-house-price-prediction/test.csv")
submission_df = pd.DataFrame()
submission_df["Id"] = submission.Id
submission_df["SalePrice"] = y_new_pred1
submission_df
submission_df.to_csv("submission.csv", index=False, header=True)
| false | 0 | 2,011 | 0 | 2,011 | 2,011 |
||
69476349
|
<jupyter_start><jupyter_text>Turkey Recent Forest Fire / CSV / NASA
## Data on Recent Forest Fires in Turkey
### Content
Data on recent forest fires in Turkey, published with permission from NASA Portal.
The data was created based on the hotspots and obtained from the satellite.
3 SEPARATE SATELLITE DATA:
- MODIS C6.1
- SUOMI VIIRS C2
- J1 VIIRS C1
DATE:
Start Date : 2021-07-17
End Date : 2021-07-31
ALSO, ALL THE DATA FOR THE YEAR 2020 IS ADDED FOR COMPARISON.
### GENERAL ATTRIBUTES
* **Latitude**
Center of nominal 375 m fire pixel
* **Longitude**
Center of nominal 375 m fire pixel
* **Bright_ti4**
(Brightness temperature I-4)
VIIRS I-4: channel brightness temperature of the fire pixel measured in Kelvin.
* **Scan**
(Along Scan pixel size)
The algorithm produces approximately 375 m pixels at nadir. Scan and track reflect actual pixel size.
* **Track**
(Along Track pixel size)
The algorithm produces approximately 375 m pixels at nadir. Scan and track reflect actual pixel size.
* **Acq_Date**
(Acquisition Date)
Date of VIIRS acquisition.
* **Acq_Time**
(Acquisition Time)
Time of acquisition/overpass of the satellite (in UTC).
* **Satellite**
N Suomi National Polar-orbiting Partnership (Suomi NPP)
* **Confidence**
This value is based on a collection of intermediate algorithm quantities used in the detection process. It is intended to help users gauge the quality of individual hotspot/fire pixels. Confidence values are set to low, nominal and high. Low confidence daytime fire pixels are typically associated with areas of sun glint and lower relative temperature anomaly (<15K) in the mid-infrared channel I4. Nominal confidence pixels are those free of potential sun glint contamination during the day and marked by strong (>15K) temperature anomaly in either day or nighttime data. High confidence fire pixels are associated with day or nighttime saturated pixels.
Please note: Low confidence nighttime pixels occur only over the geographic area extending from 11° E to 110° W and 7° N to 55° S. This area describes the region of influence of the South Atlantic Magnetic Anomaly which can cause spurious brightness temperatures in the mid-infrared channel I4 leading to potential false positive alarms. These have been removed from the NRT data distributed by FIRMS.
* **Version**
Version identifies the collection (e.g. VIIRS Collection 1) and source of data processing: Near Real-Time (NRT suffix added to collection) or Standard Processing (collection only).
"1.0NRT" - Collection 1 NRT processing.
"1.0" - Collection 1 Standard processing.
* **Bright_ti5**
(Brightness temperature I-5)
I-5 Channel brightness temperature of the fire pixel measured in Kelvin.
* **FRP**
(Fire Radiative Power)
FRP depicts the pixel-integrated fire radiative power in MW (megawatts). Given the unique spatial and spectral resolution of the data, the VIIRS 375 m fire detection algorithm was customized and tuned in order to optimize its response over small fires while balancing the occurrence of false alarms. Frequent saturation of the mid-infrared I4 channel (3.55-3.93 µm) driving the detection of active fires requires additional tests and procedures to avoid pixel classification errors. As a result, sub-pixel fire characterization (e.g., fire radiative power [FRP] retrieval) is only viable across small and/or low-intensity fires. Systematic FRP retrievals are based on a hybrid approach combining 375 and 750 m data. In fact, starting in 2015 the algorithm incorporated additional VIIRS channel M13 (3.973-4.128 µm) 750 m data in both aggregated and unaggregated format.
Satellite measurements of fire radiative power (FRP) are increasingly used to estimate the contribution of biomass burning to local and global carbon budgets. Without an associated uncertainty, however, FRP-based biomass burning estimates cannot be confidently compared across space and time, or against estimates derived from alternative methodologies. Differences in the per-pixel FRP measured near-simultaneously in consecutive MODIS scans are approximately normally distributed with a standard deviation (ση) of 26.6%. Simulations demonstrate that this uncertainty decreases to less than ~5% (at ±1 ση) for aggregations larger than ~50 MODIS active fire pixels. Although FRP uncertainties limit the confidence in flux estimates on a per-pixel basis, the sensitivity of biomass burning estimates to FRP uncertainties can be mitigated by conducting inventories at coarser spatiotemporal resolutions.
http://cedadocs.ceda.ac.uk/770/1/SEVIRI_FRP_documentdesc.pdf
* **Type**
(Inferred hot spot type)
0 = presumed vegetation fire
1 = active volcano
2 = other static land source
3 = offshore detection (includes all detections over water)
* **DayNight**
(Day or Night)
D= Daytime fire
N= Nighttime fire
Kaggle dataset identifier: turkey-recent-forest-fire-csv-nasa
<jupyter_script># # HISTORY
# #### Data on Recent Forest Fires in Turkey
# Content
# Data on recent forest fires in Turkey, published with permission from NASA Portal.
# The data was created based on the hotspots and obtained from the satellite.
# #### 3 SEPARATE SATELLITE DATA:
# MODIS C6.1
# SUOMI VIIRS C2
# J1 VIIRS C1
# #### DATE:
# Start Date : 2021-07-17
# End Date : 2021-07-31
# ALSO, ALL THE DATA FOR THE YEAR 2020 IS ADDED FOR COMPARISON.
# #### GENERAL ATTRIBUTES
# * Latitude
# Center of nominal 375 m fire pixel
# * Longitude
# Center of nominal 375 m fire pixel
# * Bright_ti4
# (Brightness temperature I-4)
# VIIRS I-4: channel brightness temperature of the fire pixel measured in Kelvin.
# * Scan
# (Along Scan pixel size)
# The algorithm produces approximately 375 m pixels at nadir. Scan and track reflect actual pixel size.
# * Track
# (Along Track pixel size)
# The algorithm produces approximately 375 m pixels at nadir. Scan and track reflect actual pixel size.
# * Acq_Date
# (Acquisition Date)
# Date of VIIRS acquisition.
# * Acq_Time
# (Acquisition Time)
# Time of acquisition/overpass of the satellite (in UTC).
# * Satellite
# N Suomi National Polar-orbiting Partnership (Suomi NPP)
# * Confidence
# This value is based on a collection of intermediate algorithm quantities used in the detection process. It is intended to help users gauge the quality of individual hotspot/fire pixels. Confidence values are set to low, nominal and high. Low confidence daytime fire pixels are typically associated with areas of sun glint and lower relative temperature anomaly (15K) temperature anomaly in either day or nighttime data. High confidence fire pixels are associated with day or nighttime saturated pixels.
# Please note: Low confidence nighttime pixels occur only over the geographic area extending from 11° E to 110° W and 7° N to 55° S. This area describes the region of influence of the South Atlantic Magnetic Anomaly which can cause spurious brightness temperatures in the mid-infrared channel I4 leading to potential false positive alarms. These have been removed from the NRT data distributed by FIRMS.
# * Version
# Version identifies the collection (e.g. VIIRS Collection 1) and source of data processing: Near Real-Time (NRT suffix added to collection) or Standard Processing (collection only).
# "1.0NRT" - Collection 1 NRT processing.
# "1.0" - Collection 1 Standard processing.
# * Bright_ti5
# (Brightness temperature I-5)
# I-5 Channel brightness temperature of the fire pixel measured in Kelvin.
# * FRP
# (Fire Radiative Power)
# FRP depicts the pixel-integrated fire radiative power in MW (megawatts). Given the unique spatial and spectral resolution of the data, the VIIRS 375 m fire detection algorithm was customized and tuned in order to optimize its response over small fires while balancing the occurrence of false alarms. Frequent saturation of the mid-infrared I4 channel (3.55-3.93 µm) driving the detection of active fires requires additional tests and procedures to avoid pixel classification errors. As a result, sub-pixel fire characterization (e.g., fire radiative power [FRP] retrieval) is only viable across small and/or low-intensity fires. Systematic FRP retrievals are based on a hybrid approach combining 375 and 750 m data. In fact, starting in 2015 the algorithm incorporated additional VIIRS channel M13 (3.973-4.128 µm) 750 m data in both aggregated and unaggregated format.
# Satellite measurements of fire radiative power (FRP) are increasingly used to estimate the contribution of biomass burning to local and global carbon budgets. Without an associated uncertainty, however, FRP-based biomass burning estimates cannot be confidently compared across space and time, or against estimates derived from alternative methodologies. Differences in the per-pixel FRP measured near-simultaneously in consecutive MODIS scans are approximately normally distributed with a standard deviation (ση) of 26.6%. Simulations demonstrate that this uncertainty decreases to less than ~5% (at ±1 ση) for aggregations larger than ~50 MODIS active fire pixels. Although FRP uncertainties limit the confidence in flux estimates on a per-pixel basis, the sensitivity of biomass burning estimates to FRP uncertainties can be mitigated by conducting inventories at coarser spatiotemporal resolutions.
# http://cedadocs.ceda.ac.uk/770/1/SEVIRI_FRP_documentdesc.pdf
# * Type
# (Inferred hot spot type)
# 0 = presumed vegetation fire
# 1 = active volcano
# 2 = other static land source
# 3 = offshore detection (includes all detections over water)
# * DayNight
# (Day or Night)
# D= Daytime fire
# N= Nighttime fire
# #### WHAT IS LONGITUDE AND LATITUDE
# * Latitude and longitude are a system of lines used to describe the location of any place on Earth. Lines of latitude run in an east-west direction across Earth. Lines of longitude run in a north-south direction. Although these are only imaginary lines, they appear on maps and globes as if they actually existed.
# #### ACQUISITION TIME EXAMPLE
# * Based on UTC time. You have to think in decimal.
# GOING FROM AM TO PM
# FOR EXAMPLE
# * 911 = 9.11
# * 1032 = 10.32
# * 0034 = 00.34
# #### PLEASE NOTE:
# * WE WILL USE A SINGLE SATELLITE DATA AS A CASE STUDY. OUR PURPOSE IS TO CREATE MEANINGS WITH THE LIGHT OF THESE DATA.
# # PACKAGES AND LIBRARIES
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from warnings import filterwarnings
from mpl_toolkits.mplot3d import Axes3D
import statsmodels.api as sm
import missingno as msno
import statsmodels.stats.api as sms
from sklearn.decomposition import PCA
from sklearn.preprocessing import scale
from sklearn.neighbors import LocalOutlierFactor
from scipy.stats import levene
from scipy.stats import shapiro
from scipy.stats.stats import pearsonr
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split, cross_val_score, cross_val_predict
from sklearn.preprocessing import scale
from sklearn.model_selection import ShuffleSplit, GridSearchCV
from sklearn.metrics import mean_squared_error, r2_score
from sklearn import model_selection
from sklearn.tree import DecisionTreeClassifier, DecisionTreeRegressor
from sklearn.neighbors import KNeighborsRegressor
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import BaggingRegressor
from sklearn.svm import SVR
from sklearn.preprocessing import StandardScaler
from sklearn.neural_network import MLPRegressor
from sklearn.neural_network import MLPClassifier
from sklearn.linear_model import LinearRegression
from sklearn.cross_decomposition import PLSRegression
from sklearn.linear_model import Ridge
from sklearn.linear_model import RidgeCV
from sklearn.linear_model import Lasso
from sklearn.linear_model import LassoCV
from sklearn.linear_model import ElasticNet
from sklearn.linear_model import ElasticNetCV
from sklearn import linear_model
from sklearn.ensemble import RandomForestRegressor, RandomForestClassifier
from sklearn.ensemble import GradientBoostingRegressor, GradientBoostingClassifier
import xgboost as xgb
from xgboost import XGBRegressor, XGBClassifier
from lightgbm import LGBMRegressor, LGBMClassifier
from catboost import CatBoostRegressor, CatBoostClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.naive_bayes import GaussianNB
from sklearn.svm import SVC
from sklearn import tree
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.metrics import (
confusion_matrix,
accuracy_score,
classification_report,
roc_auc_score,
roc_curve,
)
from yellowbrick.cluster import KElbowVisualizer
from sklearn.cluster import KMeans
from sklearn.pipeline import Pipeline
from sklearn.manifold import Isomap, TSNE
from sklearn.feature_selection import mutual_info_classif
from tqdm.notebook import tqdm
from scipy.stats import ttest_ind
import plotly.offline as pyo
import scipy.stats as stats
import pymc3 as pm
from dataprep.eda import *
from dataprep.eda import plot
from dataprep.eda import plot_diff
from dataprep.eda import plot_correlation
from dataprep.eda import plot_missing
import plotly.figure_factory as ff
from collections import Counter
import pandas_profiling as pp
from mpl_toolkits.basemap import Basemap
import datetime as dt
import plotly.express as px
import plotly.graph_objects as go
import plotly.io as pio
filterwarnings("ignore", category=DeprecationWarning)
filterwarnings("ignore", category=FutureWarning)
filterwarnings("ignore", category=UserWarning)
# # DATA REVIEW AND RE-BUILT
# ### PATH
J1V_C2_DATA_PATH = (
"../input/turkey-recent-forest-fire-csv-nasa/TURKEY-FIRE/fire_nrt_J1V-C2_212247.csv"
)
# ### READING
J1V_C2_CSV = pd.read_csv(J1V_C2_DATA_PATH)
Data = J1V_C2_CSV.copy()
J1V_C2_CSV
# ### RE-BUILT
Data.drop(["satellite", "instrument", "version"], inplace=True, axis=1)
# * THEY ALL HAVE THE SAME VALUE AND ARE NOT NECESSARY FOR OUR EXAMINATION.
Data["confidence"].replace({"n": "nominal", "l": "low", "h": "high"}, inplace=True)
Data["daynight"].replace({"N": "Nighttime", "D": "Daytime "}, inplace=True)
# * WE TURNED VALUES INTO THEIR OWN MEANING.
# ### INFORMATION
print("INFO:\n")
print(Data.info())
print("DESCRIBE:\n")
print(Data.describe().T)
# * LOOKING AT THE LAST 1 WEEK OF DATA, THE BRIGHTNESS VALUE IS HIGH FOR MOST OF THIS DATA.
# #### BRIGHT T31
# * %50 = 304
# * %75 = 377
# #### BRIGHTNESS
# * %50 = 337
# * %75 = 367
# WE CAN UNDERSTAND THE SEVERITY OF THE FIRE BY LOOKING AT THE BRIGHTNESS VALUES.
print("BASIC CORRELATION:\n")
print(Data.corr())
# * IT APPEARS A HIGH - DIRECTIONAL CORRELATION BETWEEN LATITUDE AND BRIGHTNESS.
print("BASIC COVARIANCE:\n")
print(Data.cov())
print("COLUMN:\n")
print(Data.columns)
print("SHAPE: ", Data.shape)
print("SIZE: ", Data.size)
print("DUPLICATED VALUES:\n")
print(Data.duplicated().sum())
# #### PLEASE NOTE:
# * DO NOT DELETE DUPLICATED VALUES.
# * VALUES MAY BE THE SAME BECAUSE IT IS DATE BASED.
print("NAN VALUES:\n")
print(Data.isna().sum())
# ### VALUE CONTROL
print("VALUES:\n")
print(Data["acq_date"].value_counts())
print("VALUES:\n")
print(Data["confidence"].value_counts())
print("VALUES:\n")
print(Data["daynight"].value_counts())
# ### DATA TYPES
Data = Data.reset_index(drop=True)
# #### SORTED
# * WE WILL USE THEM DIRECTLY TO REACH HIGH VALUES.
DATE_SORTED = Data.copy()
DATE_SORTED.sort_values(by=["acq_date"], inplace=True, ascending=False)
DATE_SORTED = DATE_SORTED.reset_index(drop=True)
FRP_SORTED = Data.copy()
FRP_SORTED.sort_values(by=["frp"], inplace=True, ascending=False)
FRP_SORTED = FRP_SORTED.reset_index(drop=True)
BRIGHTNESS_SORTED = Data.copy()
BRIGHTNESS_SORTED.sort_values(by=["brightness"], inplace=True, ascending=False)
BRIGHTNESS_SORTED = BRIGHTNESS_SORTED.reset_index(drop=True)
DATE_SORTED
FRP_SORTED
BRIGHTNESS_SORTED
# #### DATETIME FORMAT
# * TO USE IN GRAPHICS.
DATETIME_FORMAT = Data.copy()
DATETIME_FORMAT.sort_values(by=["acq_date"], inplace=True, ascending=False)
DATETIME_FORMAT = DATETIME_FORMAT.reset_index(drop=True)
DATETIME_FORMAT["acq_date"] = pd.to_datetime(DATETIME_FORMAT["acq_date"])
DATETIME_FORMAT
print("INFO:\n")
print(DATETIME_FORMAT.info())
print(DATETIME_FORMAT["acq_date"])
# #### DATETIME INDEX DATA
# * TO LOOK AT DATE RANGE.
DATE_INDEX = Data.copy()
DATE_INDEX.sort_values(by=["acq_date"], inplace=True, ascending=False)
DATE_INDEX = DATE_INDEX.reset_index(drop=True)
DATE_INDEX.set_index(["acq_date"], inplace=True)
DATE_INDEX.index.name = None
DATE_INDEX
# #### GROUPBY DATA
# * FOR DATE-DAY-RELIABILITY CHECKING.
GROUP_BY = Data.copy()
GROUP_BY.sort_values(by=["acq_date"], inplace=True, ascending=False)
GROUP_BY = GROUP_BY.reset_index(drop=True)
GROUP_BY = GROUP_BY.groupby(["acq_date", "daynight", "confidence"]).mean()
print(GROUP_BY.head(10))
# ### CORRELATION HEATMAP
plt.style.use("dark_background")
PEARSON_CORR = Data.corr(method="pearson")
SPEARMAN_CORR = Data.corr(method="spearman")
figure = plt.figure(figsize=(25, 10))
sns.heatmap(
PEARSON_CORR, annot=True, center=0, linewidths=2, linecolor="black", cmap="jet"
)
plt.title("PEARSON CORRELATION")
plt.tight_layout()
plt.show()
print(Data.columns)
figure = plt.figure(figsize=(25, 10))
sns.heatmap(
SPEARMAN_CORR, annot=True, center=0, linewidths=2, linecolor="black", cmap="jet"
)
plt.title("PEARSON CORRELATION")
plt.tight_layout()
plt.show()
# * There is a +-direction correlation between "brightness" and "bright_t31". This is what we expected.
# * There is a +-direction correlation between track and scan. This is what we expected.
# * IN GENERAL, THE CORRELATION BETWEEN THE VALUES IS COMPATIBLE.
# ### COVARIANCE HEATMAP
COV_DATA = Data.cov()
figure = plt.figure(figsize=(25, 10))
sns.heatmap(
COV_DATA,
annot=True,
vmin=-1,
center=0,
vmax=1,
linewidths=2,
linecolor="black",
cmap="Spectral",
)
plt.title("COVARIANCE")
plt.tight_layout()
plt.show()
# * MOSTLY WE SEE VALUES AFFECTING EACH OTHER DIRECTIONS.
# ### GROUPBY MEANING
print("INFO:\n")
print(Data.info())
print("DESCRIBE:\n")
print(Data.describe().T)
Data.columns
# #### FRP WITH VALUES
print("CONFIDENCE-FRP MAX\n")
print(Data.groupby(["confidence"])["frp"].max())
print("\n")
print("CONFIDENCE-FRP MEAN\n")
print(Data.groupby(["confidence"])["frp"].mean())
print("\n")
print("CONFIDENCE-FRP MIN\n")
print(Data.groupby(["confidence"])["frp"].min())
# * WE SEE THE HIGHEST FRP VALUE IN NOMINAL CLASS.
print("DAY/NIGHT-FRP MAX\n")
print(Data.groupby(["daynight"])["frp"].max())
print("\n")
print("DAY/NIGHT-FRP MEAN\n")
print(Data.groupby(["daynight"])["frp"].mean())
print("\n")
print("DAY/NIGHT-FRP MIN\n")
print(Data.groupby(["daynight"])["frp"].min())
# * NIGHT HOURS WHEN THE FRP VALUE IS HIGHEST OBSERVED.
print("DAY/NIGHT AND CONFIDENCE-FRP MAX\n")
print(Data.groupby(["daynight", "confidence"])["frp"].max())
print("\n")
print("DAY/NIGHT AND CONFIDENCE-FRP MEAN\n")
print(Data.groupby(["daynight", "confidence"])["frp"].mean())
print("\n")
print("DAY/NIGHT AND CONFIDENCE-FRP MIN\n")
print(Data.groupby(["daynight", "confidence"])["frp"].min())
# * AGAIN, WE SEE THE HIGHEST FRP VALUE IN THE NOMINAL CLASS.
# * THIS NOMINAL OBSERVATION HAS BEEN MEASURED AT NIGHT.
print("DATE-FRP MAX\n")
print(Data.groupby(["acq_date"])["frp"].max())
print("\n")
print("DATE-FRP MIN\n")
print(Data.groupby(["acq_date"])["frp"].min())
# * ESPECIALLY THE HIGHEST FRP VALUE WAS OBSERVED ON JULY 28 AND 29. THIS MEANS THE DAYS WHERE THE FIRE STARTED.
# * BECAUSE WE COLLECTED THIS DATA ON THE MORNING OF JULY 30, IT HAS LOW VALUE, DUE TO DAILY RENEWAL OF THE DATA.
print("TIME-FRP MAX\n")
print(Data.groupby(["acq_time"])["frp"].max())
print("\n")
print("TIME-FRP MIN\n")
print(Data.groupby(["acq_time"])["frp"].min())
print(Data[Data["frp"] == Data["frp"].max()])
# ##### THE HIGHEST FRP VALUE WAS OBSERVED
# * JULY 29,2021
# * AT 11.00 PM
# * HERE IS TOPALLI,ALADAG - ADANA
# * WE CAN SAY THE POINT WHERE ALL DISASTERS STARTED.
# #### BRIGHTNESS WITH VALUES
print("CONFIDENCE-BRIGHTNESS MAX\n")
print(Data.groupby(["confidence"])["bright_t31"].max())
print("\n")
print("CONFIDENCE-BRIGHTNESS MEAN\n")
print(Data.groupby(["confidence"])["bright_t31"].mean())
print("\n")
print("CONFIDENCE-BRIGHTNESS MIN\n")
print(Data.groupby(["confidence"])["bright_t31"].min())
print("DAY/NIGHT-BRIGHTNESS MAX\n")
print(Data.groupby(["daynight"])["bright_t31"].max())
print("\n")
print("DAY/NIGHT-BRIGHTNESS MEAN\n")
print(Data.groupby(["daynight"])["bright_t31"].mean())
print("\n")
print("DAY/NIGHT-BRIGHTNESS MIN\n")
print(Data.groupby(["daynight"])["bright_t31"].min())
# * THE HIGHEST BRIGHTNESS WAS OBSERVED IN THE DAY HOURS.
# * THIS IS DUE TO INCREASING TEMPERATURE AND REFLECTION OF EARTH TEMPERATURE WITH SUNLIGHT.
print("DATE-BRIGHTNESS MAX\n")
print(Data.groupby(["acq_date"])["bright_t31"].max())
print("\n")
print("DATE-BRIGHTNESS MIN\n")
print(Data.groupby(["acq_date"])["bright_t31"].min())
# * WE SEE THE HIGHEST BRIGHTNESS VALUE ON JULY 28.
# * JULY 27 IS THE DAY MANAVGAT FIRE STARTED.
# * THE DAY WHERE THE FIRE WAS THE STRONGEST IS JULY 29.
# 
print("TIME-BRIGHTNESS MAX\n")
print(Data.groupby(["acq_time"])["bright_t31"].max())
print("\n")
print("TIME-BRIGHTNESS MIN\n")
print(Data.groupby(["acq_time"])["bright_t31"].min())
print(Data[Data["bright_t31"] == Data["bright_t31"].max()])
# ##### THE HIGHEST BRIGHTNESS VALUE WAS OBSERVED
# * JULY 29,2021
# * AT 9.52 AM
# * HERE IS BOZTAHTA - ADANA
print(Data[Data["brightness"] == Data["brightness"].max()]["acq_date"].value_counts())
# * WHEN LOOKING AT THE TOTAL BRIGHTNESS VALUES, WE SEE THAT JULY 29 HAS THE HIGHEST VALUES.
# 
# * A man watches as flames destroy a shack inside the forest in Manavgat district, in Antalya, southern Turkey, July 28, 2021. (İHA PHOTO)
# # MAPS
import folium
from folium import plugins
# ### GENERAL
figure = plt.figure(figsize=(20, 8))
Map_Plot = Basemap(projection="cyl", resolution="c")
Map_Plot.drawmapboundary(fill_color="w")
Map_Plot.drawcoastlines(linewidth=0.5)
Map_Plot.drawmeridians(range(0, 360, 20), linewidth=0.8)
Map_Plot.drawparallels([-66, -23, 0.0, 23, 66], linewidth=0.8)
lon, lat = Map_Plot(Data["longitude"], Data["latitude"])
Map_Plot.scatter(lon, lat, marker="*", alpha=0.20, color="r", edgecolor="None")
plt.title("COORDINATES")
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(Data.longitude, Data.latitude, edgecolor="none", color="r", alpha=0.6)
plt.title("TURKEY GENERAL", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.plot(Data.longitude, Data.latitude, color="r", alpha=0.4)
plt.title("TURKEY GENERAL", fontsize=15)
plt.tight_layout()
plt.show()
# ### BASED ON DATE
DATE_28 = Data[Data["acq_date"] == "2021-07-28"]
DATE_29 = Data[Data["acq_date"] == "2021-07-29"]
DATE_28 = DATE_28.reset_index(drop=True)
DATE_29 = DATE_29.reset_index(drop=True)
DATE_28
DATE_29
# #### 28 JULY 2021
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
DATE_28.longitude,
DATE_28.latitude,
edgecolor="none",
color="r",
alpha=0.3,
linewidth=10,
s=170,
)
plt.title("JULY 28,2021", fontsize=15)
plt.tight_layout()
plt.show()
# * AS YOU CAN SEE, FIRE AREAS BIGGER THAN OTHERS.
# #### 29 JULY 2021
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
DATE_29.longitude,
DATE_29.latitude,
edgecolor="none",
color="r",
alpha=0.3,
linewidth=10,
s=170,
)
plt.title("JULY 29,2021", fontsize=15)
plt.tight_layout()
plt.show()
# * WE OBSERVED A SIGNIFICANT INCREASE IN FIRE POINTS ON JULY 29.
# #### 28 JULY 2021 AND 29 JULY 2021
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
DATE_28.longitude,
DATE_28.latitude,
edgecolor="none",
color="g",
alpha=0.9,
linewidth=10,
s=100,
)
Map_Plot.scatter(
DATE_29.longitude,
DATE_29.latitude,
edgecolor="none",
color="r",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 29 - 28,2021", fontsize=15)
plt.tight_layout()
plt.show()
# * YOU SEE THE AMOUNT OF INCREASE AFTER 1 DAY.
# GREEN: 28 JULY
# RED: 29 JULY
# #### COMPARISON WITH THE BEGINNING OF THE WEEK
# * WE WILL LOOK AT THE DIFFERENCE BETWEEN DIFFERENT DATES.
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
Data[Data["acq_date"] == "2021-07-15"].longitude,
Data[Data["acq_date"] == "2021-07-15"].latitude,
edgecolor="none",
color="y",
alpha=0.9,
linewidth=10,
s=100,
)
Map_Plot.scatter(
DATE_29.longitude,
DATE_29.latitude,
edgecolor="none",
color="r",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 29 - 15,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
Data[Data["acq_date"] == "2021-07-18"].longitude,
Data[Data["acq_date"] == "2021-07-18"].latitude,
edgecolor="none",
color="y",
alpha=0.9,
linewidth=10,
s=100,
)
Map_Plot.scatter(
DATE_29.longitude,
DATE_29.latitude,
edgecolor="none",
color="r",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 29 - 18,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
Data[Data["acq_date"] == "2021-07-15"].longitude,
Data[Data["acq_date"] == "2021-07-15"].latitude,
edgecolor="none",
color="y",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 15,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
DATE_29.longitude,
DATE_29.latitude,
edgecolor="none",
color="r",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 29,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
Data[Data["acq_date"] == "2021-07-18"].longitude,
Data[Data["acq_date"] == "2021-07-18"].latitude,
edgecolor="none",
color="y",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 18,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
DATE_29.longitude,
DATE_29.latitude,
edgecolor="none",
color="r",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 29,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
Data[Data["acq_date"] == "2021-07-22"].longitude,
Data[Data["acq_date"] == "2021-07-22"].latitude,
edgecolor="none",
color="y",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 22,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
DATE_29.longitude,
DATE_29.latitude,
edgecolor="none",
color="r",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 29,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
Data[Data["acq_date"] == "2021-07-25"].longitude,
Data[Data["acq_date"] == "2021-07-25"].latitude,
edgecolor="none",
color="y",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 25,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
DATE_29.longitude,
DATE_29.latitude,
edgecolor="none",
color="r",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 29,2021", fontsize=15)
plt.tight_layout()
plt.show()
# #### DETAIL
# * USE INTERACTIVE MAPS
MAP_FUNCTION = folium.Map(
location=[38.9637, 35.2433], tiles="Stamen Terrain", zoom_start=5.5 # TURKEY
)
MAP_FUNCTION
print(DATE_29[DATE_29["frp"] == DATE_29["frp"].max()])
print("\n")
print("----" * 10)
print("\n")
print(DATE_28[DATE_28["frp"] == DATE_28["frp"].max()])
print(DATE_29[DATE_29["bright_t31"] == DATE_29["bright_t31"].max()])
print("\n")
print("----" * 10)
print("\n")
print(DATE_28[DATE_28["bright_t31"] == DATE_28["bright_t31"].max()])
# * CLICK THE ROUNDS
MAP_FUNCTION = folium.Map(
location=[38.9637, 35.2433], # location australia
tiles="openstreetmap",
zoom_start=6.0,
)
folium.CircleMarker(
location=[36.89363, 31.36536],
radius=20,
popup="DATE:<b>28 JULY MAXIMUM BRIGHTNESS<b>",
tooltip="Click me",
color="#2F4F4F",
fill=True,
fill_color="#00000",
).add_to(MAP_FUNCTION)
MAP_FUNCTION
MAP_FUNCTION = folium.Map(
location=[38.9637, 35.2433], # location australia
tiles="openstreetmap",
zoom_start=6.0,
)
folium.CircleMarker(
location=[37.3512, 35.45154],
radius=20,
popup="DATE:<b>29 JULY MAXIMUM BRIGHTNESS<b>",
tooltip="Click me",
color="#2F4F4F",
fill=True,
fill_color="#00000",
).add_to(MAP_FUNCTION)
MAP_FUNCTION
MAP_FUNCTION = folium.Map(
location=[38.9637, 35.2433], # location australia
tiles="openstreetmap",
zoom_start=6.0,
)
folium.CircleMarker(
location=[36.89363, 31.36536],
radius=20,
popup="DATE:<b>28 JULY MAXIMUM BRIGHTNESS<b>",
tooltip="Click me",
color="#2F4F4F",
fill=True,
fill_color="#FF0000",
).add_to(MAP_FUNCTION)
folium.CircleMarker(
location=[37.3512, 35.45154],
radius=20,
popup="DATE:<b>29 JULY MAXIMUM BRIGHTNESS<b>",
tooltip="Click me",
color="#2F4F4F",
fill=True,
fill_color="#00000",
).add_to(MAP_FUNCTION)
MAP_FUNCTION
MAP_FUNCTION = folium.Map(
location=[38.9637, 35.2433], # location australia
tiles="Stamen Watercolor",
zoom_start=6.0,
)
folium.CircleMarker(
location=[36.89363, 31.36536],
radius=20,
popup="DATE:<b>28 JULY MAXIMUM BRIGHTNESS<b>",
tooltip="Click me",
color="#2F4F4F",
fill=True,
fill_color="#FF0000",
).add_to(MAP_FUNCTION)
folium.CircleMarker(
location=[37.3512, 35.45154],
radius=20,
popup="DATE:<b>29 JULY MAXIMUM BRIGHTNESS<b>",
tooltip="Click me",
color="#2F4F4F",
fill=True,
fill_color="#00000",
).add_to(MAP_FUNCTION)
MAP_FUNCTION
MAP_FUNCTION = folium.Map(
location=[38.9637, 35.2433], # location australia
tiles="Stamen Toner",
zoom_start=6.0,
)
folium.CircleMarker(
location=[36.89363, 31.36536],
radius=20,
popup="DATE:<b>28 JULY MAXIMUM BRIGHTNESS<b>",
tooltip="Click me",
color="#2F4F4F",
fill=True,
fill_color="#FF0000",
).add_to(MAP_FUNCTION)
folium.CircleMarker(
location=[37.3512, 35.45154],
radius=20,
popup="DATE:<b>29 JULY MAXIMUM BRIGHTNESS<b>",
tooltip="Click me",
color="#2F4F4F",
fill=True,
fill_color="#00000",
).add_to(MAP_FUNCTION)
folium.Marker(location=[36.89363, 31.36536], popup="ANTALYA").add_to(MAP_FUNCTION)
folium.Marker(location=[37.3512, 35.45154], popup="ADANA").add_to(MAP_FUNCTION)
MAP_FUNCTION
LAT_LON_29 = DATE_29[["latitude", "longitude"]]
LAT_LON_28 = DATE_28[["latitude", "longitude"]]
MAP_FUNCTION = folium.Map(
location=[38.9637, 35.2433], # location australia
tiles="openstreetmap",
zoom_start=6.0,
)
plugins.MarkerCluster(LAT_LON_28).add_to(MAP_FUNCTION)
MAP_FUNCTION
MAP_FUNCTION = folium.Map(
location=[38.9637, 35.2433], # location australia
tiles="openstreetmap",
zoom_start=6.0,
)
plugins.MarkerCluster(LAT_LON_29).add_to(MAP_FUNCTION)
MAP_FUNCTION
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/476/69476349.ipynb
|
turkey-recent-forest-fire-csv-nasa
|
brsdincer
|
[{"Id": 69476349, "ScriptId": 18967253, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7042824, "CreationDate": "07/31/2021 13:27:01", "VersionNumber": 1.0, "Title": "notebook024b690206", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 922.0, "LinesInsertedFromPrevious": 922.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92690110, "KernelVersionId": 69476349, "SourceDatasetVersionId": 2483042}]
|
[{"Id": 2483042, "DatasetId": 1502897, "DatasourceVersionId": 2525591, "CreatorUserId": 7042824, "LicenseName": "Database: Open Database, Contents: Database Contents", "CreationDate": "07/31/2021 07:46:22", "VersionNumber": 1.0, "Title": "Turkey Recent Forest Fire / CSV / NASA", "Slug": "turkey-recent-forest-fire-csv-nasa", "Subtitle": "Data on Recent Forest Fires in Turkey", "Description": "## Data on Recent Forest Fires in Turkey\n\n### Content\nData on recent forest fires in Turkey, published with permission from NASA Portal.\nThe data was created based on the hotspots and obtained from the satellite.\n\n3 SEPARATE SATELLITE DATA:\n\n- MODIS C6.1\n- SUOMI VIIRS C2\n- J1 VIIRS C1\n\nDATE:\n\nStart Date : 2021-07-17\nEnd Date : 2021-07-31\n\nALSO, ALL THE DATA FOR THE YEAR 2020 IS ADDED FOR COMPARISON.\n\n### GENERAL ATTRIBUTES\n\n* **Latitude**\n \nCenter of nominal 375 m fire pixel\n\n* **Longitude**\n \nCenter of nominal 375 m fire pixel\n\n* **Bright_ti4**\n\n(Brightness temperature I-4)\nVIIRS I-4: channel brightness temperature of the fire pixel measured in Kelvin.\n\n* **Scan**\n\n(Along Scan pixel size)\nThe algorithm produces approximately 375 m pixels at nadir. Scan and track reflect actual pixel size.\n\n* **Track**\n\n(Along Track pixel size)\nThe algorithm produces approximately 375 m pixels at nadir. Scan and track reflect actual pixel size.\n\n* **Acq_Date**\n\n(Acquisition Date)\nDate of VIIRS acquisition.\n\n* **Acq_Time**\n\n(Acquisition Time)\nTime of acquisition/overpass of the satellite (in UTC).\n\n* **Satellite**\n\nN Suomi National Polar-orbiting Partnership (Suomi NPP)\n\n* **Confidence**\n\nThis value is based on a collection of intermediate algorithm quantities used in the detection process. It is intended to help users gauge the quality of individual hotspot/fire pixels. Confidence values are set to low, nominal and high. Low confidence daytime fire pixels are typically associated with areas of sun glint and lower relative temperature anomaly (<15K) in the mid-infrared channel I4. Nominal confidence pixels are those free of potential sun glint contamination during the day and marked by strong (>15K) temperature anomaly in either day or nighttime data. High confidence fire pixels are associated with day or nighttime saturated pixels.\n\nPlease note: Low confidence nighttime pixels occur only over the geographic area extending from 11\u00b0 E to 110\u00b0 W and 7\u00b0 N to 55\u00b0 S. This area describes the region of influence of the South Atlantic Magnetic Anomaly which can cause spurious brightness temperatures in the mid-infrared channel I4 leading to potential false positive alarms. These have been removed from the NRT data distributed by FIRMS.\n\n* **Version**\n\nVersion identifies the collection (e.g. VIIRS Collection 1) and source of data processing: Near Real-Time (NRT suffix added to collection) or Standard Processing (collection only).\n\n\"1.0NRT\" - Collection 1 NRT processing.\n\n\"1.0\" - Collection 1 Standard processing.\n\n* **Bright_ti5**\n\n(Brightness temperature I-5)\nI-5 Channel brightness temperature of the fire pixel measured in Kelvin.\n\n* **FRP**\n\n(Fire Radiative Power)\nFRP depicts the pixel-integrated fire radiative power in MW (megawatts). Given the unique spatial and spectral resolution of the data, the VIIRS 375 m fire detection algorithm was customized and tuned in order to optimize its response over small fires while balancing the occurrence of false alarms. Frequent saturation of the mid-infrared I4 channel (3.55-3.93 \u00b5m) driving the detection of active fires requires additional tests and procedures to avoid pixel classification errors. As a result, sub-pixel fire characterization (e.g., fire radiative power [FRP] retrieval) is only viable across small and/or low-intensity fires. Systematic FRP retrievals are based on a hybrid approach combining 375 and 750 m data. In fact, starting in 2015 the algorithm incorporated additional VIIRS channel M13 (3.973-4.128 \u00b5m) 750 m data in both aggregated and unaggregated format.\n\nSatellite measurements of fire radiative power (FRP) are increasingly used to estimate the contribution of biomass burning to local and global carbon budgets. Without an associated uncertainty, however, FRP-based biomass burning estimates cannot be confidently compared across space and time, or against estimates derived from alternative methodologies. Differences in the per-pixel FRP measured near-simultaneously in consecutive MODIS scans are approximately normally distributed with a standard deviation (\u03c3\u03b7) of 26.6%. Simulations demonstrate that this uncertainty decreases to less than ~5% (at \u00b11 \u03c3\u03b7) for aggregations larger than ~50 MODIS active fire pixels. Although FRP uncertainties limit the confidence in flux estimates on a per-pixel basis, the sensitivity of biomass burning estimates to FRP uncertainties can be mitigated by conducting inventories at coarser spatiotemporal resolutions.\n\nhttp://cedadocs.ceda.ac.uk/770/1/SEVIRI_FRP_documentdesc.pdf\n\n* **Type**\n\n(Inferred hot spot type)\n0 = presumed vegetation fire\n\n1 = active volcano\n\n2 = other static land source\n\n3 = offshore detection (includes all detections over water)\n\n* **DayNight**\n\n(Day or Night)\n\t\nD= Daytime fire\n\nN= Nighttime fire", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1502897, "CreatorUserId": 7042824, "OwnerUserId": 7042824.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2487114.0, "CurrentDatasourceVersionId": 2529676.0, "ForumId": 1522636, "Type": 2, "CreationDate": "07/31/2021 07:46:22", "LastActivityDate": "07/31/2021", "TotalViews": 2714, "TotalDownloads": 156, "TotalVotes": 10, "TotalKernels": 1}]
|
[{"Id": 7042824, "UserName": "brsdincer", "DisplayName": "Baris Dincer", "RegisterDate": "03/27/2021", "PerformanceTier": 2}]
|
# # HISTORY
# #### Data on Recent Forest Fires in Turkey
# Content
# Data on recent forest fires in Turkey, published with permission from NASA Portal.
# The data was created based on the hotspots and obtained from the satellite.
# #### 3 SEPARATE SATELLITE DATA:
# MODIS C6.1
# SUOMI VIIRS C2
# J1 VIIRS C1
# #### DATE:
# Start Date : 2021-07-17
# End Date : 2021-07-31
# ALSO, ALL THE DATA FOR THE YEAR 2020 IS ADDED FOR COMPARISON.
# #### GENERAL ATTRIBUTES
# * Latitude
# Center of nominal 375 m fire pixel
# * Longitude
# Center of nominal 375 m fire pixel
# * Bright_ti4
# (Brightness temperature I-4)
# VIIRS I-4: channel brightness temperature of the fire pixel measured in Kelvin.
# * Scan
# (Along Scan pixel size)
# The algorithm produces approximately 375 m pixels at nadir. Scan and track reflect actual pixel size.
# * Track
# (Along Track pixel size)
# The algorithm produces approximately 375 m pixels at nadir. Scan and track reflect actual pixel size.
# * Acq_Date
# (Acquisition Date)
# Date of VIIRS acquisition.
# * Acq_Time
# (Acquisition Time)
# Time of acquisition/overpass of the satellite (in UTC).
# * Satellite
# N Suomi National Polar-orbiting Partnership (Suomi NPP)
# * Confidence
# This value is based on a collection of intermediate algorithm quantities used in the detection process. It is intended to help users gauge the quality of individual hotspot/fire pixels. Confidence values are set to low, nominal and high. Low confidence daytime fire pixels are typically associated with areas of sun glint and lower relative temperature anomaly (15K) temperature anomaly in either day or nighttime data. High confidence fire pixels are associated with day or nighttime saturated pixels.
# Please note: Low confidence nighttime pixels occur only over the geographic area extending from 11° E to 110° W and 7° N to 55° S. This area describes the region of influence of the South Atlantic Magnetic Anomaly which can cause spurious brightness temperatures in the mid-infrared channel I4 leading to potential false positive alarms. These have been removed from the NRT data distributed by FIRMS.
# * Version
# Version identifies the collection (e.g. VIIRS Collection 1) and source of data processing: Near Real-Time (NRT suffix added to collection) or Standard Processing (collection only).
# "1.0NRT" - Collection 1 NRT processing.
# "1.0" - Collection 1 Standard processing.
# * Bright_ti5
# (Brightness temperature I-5)
# I-5 Channel brightness temperature of the fire pixel measured in Kelvin.
# * FRP
# (Fire Radiative Power)
# FRP depicts the pixel-integrated fire radiative power in MW (megawatts). Given the unique spatial and spectral resolution of the data, the VIIRS 375 m fire detection algorithm was customized and tuned in order to optimize its response over small fires while balancing the occurrence of false alarms. Frequent saturation of the mid-infrared I4 channel (3.55-3.93 µm) driving the detection of active fires requires additional tests and procedures to avoid pixel classification errors. As a result, sub-pixel fire characterization (e.g., fire radiative power [FRP] retrieval) is only viable across small and/or low-intensity fires. Systematic FRP retrievals are based on a hybrid approach combining 375 and 750 m data. In fact, starting in 2015 the algorithm incorporated additional VIIRS channel M13 (3.973-4.128 µm) 750 m data in both aggregated and unaggregated format.
# Satellite measurements of fire radiative power (FRP) are increasingly used to estimate the contribution of biomass burning to local and global carbon budgets. Without an associated uncertainty, however, FRP-based biomass burning estimates cannot be confidently compared across space and time, or against estimates derived from alternative methodologies. Differences in the per-pixel FRP measured near-simultaneously in consecutive MODIS scans are approximately normally distributed with a standard deviation (ση) of 26.6%. Simulations demonstrate that this uncertainty decreases to less than ~5% (at ±1 ση) for aggregations larger than ~50 MODIS active fire pixels. Although FRP uncertainties limit the confidence in flux estimates on a per-pixel basis, the sensitivity of biomass burning estimates to FRP uncertainties can be mitigated by conducting inventories at coarser spatiotemporal resolutions.
# http://cedadocs.ceda.ac.uk/770/1/SEVIRI_FRP_documentdesc.pdf
# * Type
# (Inferred hot spot type)
# 0 = presumed vegetation fire
# 1 = active volcano
# 2 = other static land source
# 3 = offshore detection (includes all detections over water)
# * DayNight
# (Day or Night)
# D= Daytime fire
# N= Nighttime fire
# #### WHAT IS LONGITUDE AND LATITUDE
# * Latitude and longitude are a system of lines used to describe the location of any place on Earth. Lines of latitude run in an east-west direction across Earth. Lines of longitude run in a north-south direction. Although these are only imaginary lines, they appear on maps and globes as if they actually existed.
# #### ACQUISITION TIME EXAMPLE
# * Based on UTC time. You have to think in decimal.
# GOING FROM AM TO PM
# FOR EXAMPLE
# * 911 = 9.11
# * 1032 = 10.32
# * 0034 = 00.34
# #### PLEASE NOTE:
# * WE WILL USE A SINGLE SATELLITE DATA AS A CASE STUDY. OUR PURPOSE IS TO CREATE MEANINGS WITH THE LIGHT OF THESE DATA.
# # PACKAGES AND LIBRARIES
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from warnings import filterwarnings
from mpl_toolkits.mplot3d import Axes3D
import statsmodels.api as sm
import missingno as msno
import statsmodels.stats.api as sms
from sklearn.decomposition import PCA
from sklearn.preprocessing import scale
from sklearn.neighbors import LocalOutlierFactor
from scipy.stats import levene
from scipy.stats import shapiro
from scipy.stats.stats import pearsonr
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split, cross_val_score, cross_val_predict
from sklearn.preprocessing import scale
from sklearn.model_selection import ShuffleSplit, GridSearchCV
from sklearn.metrics import mean_squared_error, r2_score
from sklearn import model_selection
from sklearn.tree import DecisionTreeClassifier, DecisionTreeRegressor
from sklearn.neighbors import KNeighborsRegressor
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import BaggingRegressor
from sklearn.svm import SVR
from sklearn.preprocessing import StandardScaler
from sklearn.neural_network import MLPRegressor
from sklearn.neural_network import MLPClassifier
from sklearn.linear_model import LinearRegression
from sklearn.cross_decomposition import PLSRegression
from sklearn.linear_model import Ridge
from sklearn.linear_model import RidgeCV
from sklearn.linear_model import Lasso
from sklearn.linear_model import LassoCV
from sklearn.linear_model import ElasticNet
from sklearn.linear_model import ElasticNetCV
from sklearn import linear_model
from sklearn.ensemble import RandomForestRegressor, RandomForestClassifier
from sklearn.ensemble import GradientBoostingRegressor, GradientBoostingClassifier
import xgboost as xgb
from xgboost import XGBRegressor, XGBClassifier
from lightgbm import LGBMRegressor, LGBMClassifier
from catboost import CatBoostRegressor, CatBoostClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.naive_bayes import GaussianNB
from sklearn.svm import SVC
from sklearn import tree
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.metrics import (
confusion_matrix,
accuracy_score,
classification_report,
roc_auc_score,
roc_curve,
)
from yellowbrick.cluster import KElbowVisualizer
from sklearn.cluster import KMeans
from sklearn.pipeline import Pipeline
from sklearn.manifold import Isomap, TSNE
from sklearn.feature_selection import mutual_info_classif
from tqdm.notebook import tqdm
from scipy.stats import ttest_ind
import plotly.offline as pyo
import scipy.stats as stats
import pymc3 as pm
from dataprep.eda import *
from dataprep.eda import plot
from dataprep.eda import plot_diff
from dataprep.eda import plot_correlation
from dataprep.eda import plot_missing
import plotly.figure_factory as ff
from collections import Counter
import pandas_profiling as pp
from mpl_toolkits.basemap import Basemap
import datetime as dt
import plotly.express as px
import plotly.graph_objects as go
import plotly.io as pio
filterwarnings("ignore", category=DeprecationWarning)
filterwarnings("ignore", category=FutureWarning)
filterwarnings("ignore", category=UserWarning)
# # DATA REVIEW AND RE-BUILT
# ### PATH
J1V_C2_DATA_PATH = (
"../input/turkey-recent-forest-fire-csv-nasa/TURKEY-FIRE/fire_nrt_J1V-C2_212247.csv"
)
# ### READING
J1V_C2_CSV = pd.read_csv(J1V_C2_DATA_PATH)
Data = J1V_C2_CSV.copy()
J1V_C2_CSV
# ### RE-BUILT
Data.drop(["satellite", "instrument", "version"], inplace=True, axis=1)
# * THEY ALL HAVE THE SAME VALUE AND ARE NOT NECESSARY FOR OUR EXAMINATION.
Data["confidence"].replace({"n": "nominal", "l": "low", "h": "high"}, inplace=True)
Data["daynight"].replace({"N": "Nighttime", "D": "Daytime "}, inplace=True)
# * WE TURNED VALUES INTO THEIR OWN MEANING.
# ### INFORMATION
print("INFO:\n")
print(Data.info())
print("DESCRIBE:\n")
print(Data.describe().T)
# * LOOKING AT THE LAST 1 WEEK OF DATA, THE BRIGHTNESS VALUE IS HIGH FOR MOST OF THIS DATA.
# #### BRIGHT T31
# * %50 = 304
# * %75 = 377
# #### BRIGHTNESS
# * %50 = 337
# * %75 = 367
# WE CAN UNDERSTAND THE SEVERITY OF THE FIRE BY LOOKING AT THE BRIGHTNESS VALUES.
print("BASIC CORRELATION:\n")
print(Data.corr())
# * IT APPEARS A HIGH - DIRECTIONAL CORRELATION BETWEEN LATITUDE AND BRIGHTNESS.
print("BASIC COVARIANCE:\n")
print(Data.cov())
print("COLUMN:\n")
print(Data.columns)
print("SHAPE: ", Data.shape)
print("SIZE: ", Data.size)
print("DUPLICATED VALUES:\n")
print(Data.duplicated().sum())
# #### PLEASE NOTE:
# * DO NOT DELETE DUPLICATED VALUES.
# * VALUES MAY BE THE SAME BECAUSE IT IS DATE BASED.
print("NAN VALUES:\n")
print(Data.isna().sum())
# ### VALUE CONTROL
print("VALUES:\n")
print(Data["acq_date"].value_counts())
print("VALUES:\n")
print(Data["confidence"].value_counts())
print("VALUES:\n")
print(Data["daynight"].value_counts())
# ### DATA TYPES
Data = Data.reset_index(drop=True)
# #### SORTED
# * WE WILL USE THEM DIRECTLY TO REACH HIGH VALUES.
DATE_SORTED = Data.copy()
DATE_SORTED.sort_values(by=["acq_date"], inplace=True, ascending=False)
DATE_SORTED = DATE_SORTED.reset_index(drop=True)
FRP_SORTED = Data.copy()
FRP_SORTED.sort_values(by=["frp"], inplace=True, ascending=False)
FRP_SORTED = FRP_SORTED.reset_index(drop=True)
BRIGHTNESS_SORTED = Data.copy()
BRIGHTNESS_SORTED.sort_values(by=["brightness"], inplace=True, ascending=False)
BRIGHTNESS_SORTED = BRIGHTNESS_SORTED.reset_index(drop=True)
DATE_SORTED
FRP_SORTED
BRIGHTNESS_SORTED
# #### DATETIME FORMAT
# * TO USE IN GRAPHICS.
DATETIME_FORMAT = Data.copy()
DATETIME_FORMAT.sort_values(by=["acq_date"], inplace=True, ascending=False)
DATETIME_FORMAT = DATETIME_FORMAT.reset_index(drop=True)
DATETIME_FORMAT["acq_date"] = pd.to_datetime(DATETIME_FORMAT["acq_date"])
DATETIME_FORMAT
print("INFO:\n")
print(DATETIME_FORMAT.info())
print(DATETIME_FORMAT["acq_date"])
# #### DATETIME INDEX DATA
# * TO LOOK AT DATE RANGE.
DATE_INDEX = Data.copy()
DATE_INDEX.sort_values(by=["acq_date"], inplace=True, ascending=False)
DATE_INDEX = DATE_INDEX.reset_index(drop=True)
DATE_INDEX.set_index(["acq_date"], inplace=True)
DATE_INDEX.index.name = None
DATE_INDEX
# #### GROUPBY DATA
# * FOR DATE-DAY-RELIABILITY CHECKING.
GROUP_BY = Data.copy()
GROUP_BY.sort_values(by=["acq_date"], inplace=True, ascending=False)
GROUP_BY = GROUP_BY.reset_index(drop=True)
GROUP_BY = GROUP_BY.groupby(["acq_date", "daynight", "confidence"]).mean()
print(GROUP_BY.head(10))
# ### CORRELATION HEATMAP
plt.style.use("dark_background")
PEARSON_CORR = Data.corr(method="pearson")
SPEARMAN_CORR = Data.corr(method="spearman")
figure = plt.figure(figsize=(25, 10))
sns.heatmap(
PEARSON_CORR, annot=True, center=0, linewidths=2, linecolor="black", cmap="jet"
)
plt.title("PEARSON CORRELATION")
plt.tight_layout()
plt.show()
print(Data.columns)
figure = plt.figure(figsize=(25, 10))
sns.heatmap(
SPEARMAN_CORR, annot=True, center=0, linewidths=2, linecolor="black", cmap="jet"
)
plt.title("PEARSON CORRELATION")
plt.tight_layout()
plt.show()
# * There is a +-direction correlation between "brightness" and "bright_t31". This is what we expected.
# * There is a +-direction correlation between track and scan. This is what we expected.
# * IN GENERAL, THE CORRELATION BETWEEN THE VALUES IS COMPATIBLE.
# ### COVARIANCE HEATMAP
COV_DATA = Data.cov()
figure = plt.figure(figsize=(25, 10))
sns.heatmap(
COV_DATA,
annot=True,
vmin=-1,
center=0,
vmax=1,
linewidths=2,
linecolor="black",
cmap="Spectral",
)
plt.title("COVARIANCE")
plt.tight_layout()
plt.show()
# * MOSTLY WE SEE VALUES AFFECTING EACH OTHER DIRECTIONS.
# ### GROUPBY MEANING
print("INFO:\n")
print(Data.info())
print("DESCRIBE:\n")
print(Data.describe().T)
Data.columns
# #### FRP WITH VALUES
print("CONFIDENCE-FRP MAX\n")
print(Data.groupby(["confidence"])["frp"].max())
print("\n")
print("CONFIDENCE-FRP MEAN\n")
print(Data.groupby(["confidence"])["frp"].mean())
print("\n")
print("CONFIDENCE-FRP MIN\n")
print(Data.groupby(["confidence"])["frp"].min())
# * WE SEE THE HIGHEST FRP VALUE IN NOMINAL CLASS.
print("DAY/NIGHT-FRP MAX\n")
print(Data.groupby(["daynight"])["frp"].max())
print("\n")
print("DAY/NIGHT-FRP MEAN\n")
print(Data.groupby(["daynight"])["frp"].mean())
print("\n")
print("DAY/NIGHT-FRP MIN\n")
print(Data.groupby(["daynight"])["frp"].min())
# * NIGHT HOURS WHEN THE FRP VALUE IS HIGHEST OBSERVED.
print("DAY/NIGHT AND CONFIDENCE-FRP MAX\n")
print(Data.groupby(["daynight", "confidence"])["frp"].max())
print("\n")
print("DAY/NIGHT AND CONFIDENCE-FRP MEAN\n")
print(Data.groupby(["daynight", "confidence"])["frp"].mean())
print("\n")
print("DAY/NIGHT AND CONFIDENCE-FRP MIN\n")
print(Data.groupby(["daynight", "confidence"])["frp"].min())
# * AGAIN, WE SEE THE HIGHEST FRP VALUE IN THE NOMINAL CLASS.
# * THIS NOMINAL OBSERVATION HAS BEEN MEASURED AT NIGHT.
print("DATE-FRP MAX\n")
print(Data.groupby(["acq_date"])["frp"].max())
print("\n")
print("DATE-FRP MIN\n")
print(Data.groupby(["acq_date"])["frp"].min())
# * ESPECIALLY THE HIGHEST FRP VALUE WAS OBSERVED ON JULY 28 AND 29. THIS MEANS THE DAYS WHERE THE FIRE STARTED.
# * BECAUSE WE COLLECTED THIS DATA ON THE MORNING OF JULY 30, IT HAS LOW VALUE, DUE TO DAILY RENEWAL OF THE DATA.
print("TIME-FRP MAX\n")
print(Data.groupby(["acq_time"])["frp"].max())
print("\n")
print("TIME-FRP MIN\n")
print(Data.groupby(["acq_time"])["frp"].min())
print(Data[Data["frp"] == Data["frp"].max()])
# ##### THE HIGHEST FRP VALUE WAS OBSERVED
# * JULY 29,2021
# * AT 11.00 PM
# * HERE IS TOPALLI,ALADAG - ADANA
# * WE CAN SAY THE POINT WHERE ALL DISASTERS STARTED.
# #### BRIGHTNESS WITH VALUES
print("CONFIDENCE-BRIGHTNESS MAX\n")
print(Data.groupby(["confidence"])["bright_t31"].max())
print("\n")
print("CONFIDENCE-BRIGHTNESS MEAN\n")
print(Data.groupby(["confidence"])["bright_t31"].mean())
print("\n")
print("CONFIDENCE-BRIGHTNESS MIN\n")
print(Data.groupby(["confidence"])["bright_t31"].min())
print("DAY/NIGHT-BRIGHTNESS MAX\n")
print(Data.groupby(["daynight"])["bright_t31"].max())
print("\n")
print("DAY/NIGHT-BRIGHTNESS MEAN\n")
print(Data.groupby(["daynight"])["bright_t31"].mean())
print("\n")
print("DAY/NIGHT-BRIGHTNESS MIN\n")
print(Data.groupby(["daynight"])["bright_t31"].min())
# * THE HIGHEST BRIGHTNESS WAS OBSERVED IN THE DAY HOURS.
# * THIS IS DUE TO INCREASING TEMPERATURE AND REFLECTION OF EARTH TEMPERATURE WITH SUNLIGHT.
print("DATE-BRIGHTNESS MAX\n")
print(Data.groupby(["acq_date"])["bright_t31"].max())
print("\n")
print("DATE-BRIGHTNESS MIN\n")
print(Data.groupby(["acq_date"])["bright_t31"].min())
# * WE SEE THE HIGHEST BRIGHTNESS VALUE ON JULY 28.
# * JULY 27 IS THE DAY MANAVGAT FIRE STARTED.
# * THE DAY WHERE THE FIRE WAS THE STRONGEST IS JULY 29.
# 
print("TIME-BRIGHTNESS MAX\n")
print(Data.groupby(["acq_time"])["bright_t31"].max())
print("\n")
print("TIME-BRIGHTNESS MIN\n")
print(Data.groupby(["acq_time"])["bright_t31"].min())
print(Data[Data["bright_t31"] == Data["bright_t31"].max()])
# ##### THE HIGHEST BRIGHTNESS VALUE WAS OBSERVED
# * JULY 29,2021
# * AT 9.52 AM
# * HERE IS BOZTAHTA - ADANA
print(Data[Data["brightness"] == Data["brightness"].max()]["acq_date"].value_counts())
# * WHEN LOOKING AT THE TOTAL BRIGHTNESS VALUES, WE SEE THAT JULY 29 HAS THE HIGHEST VALUES.
# 
# * A man watches as flames destroy a shack inside the forest in Manavgat district, in Antalya, southern Turkey, July 28, 2021. (İHA PHOTO)
# # MAPS
import folium
from folium import plugins
# ### GENERAL
figure = plt.figure(figsize=(20, 8))
Map_Plot = Basemap(projection="cyl", resolution="c")
Map_Plot.drawmapboundary(fill_color="w")
Map_Plot.drawcoastlines(linewidth=0.5)
Map_Plot.drawmeridians(range(0, 360, 20), linewidth=0.8)
Map_Plot.drawparallels([-66, -23, 0.0, 23, 66], linewidth=0.8)
lon, lat = Map_Plot(Data["longitude"], Data["latitude"])
Map_Plot.scatter(lon, lat, marker="*", alpha=0.20, color="r", edgecolor="None")
plt.title("COORDINATES")
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(Data.longitude, Data.latitude, edgecolor="none", color="r", alpha=0.6)
plt.title("TURKEY GENERAL", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.plot(Data.longitude, Data.latitude, color="r", alpha=0.4)
plt.title("TURKEY GENERAL", fontsize=15)
plt.tight_layout()
plt.show()
# ### BASED ON DATE
DATE_28 = Data[Data["acq_date"] == "2021-07-28"]
DATE_29 = Data[Data["acq_date"] == "2021-07-29"]
DATE_28 = DATE_28.reset_index(drop=True)
DATE_29 = DATE_29.reset_index(drop=True)
DATE_28
DATE_29
# #### 28 JULY 2021
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
DATE_28.longitude,
DATE_28.latitude,
edgecolor="none",
color="r",
alpha=0.3,
linewidth=10,
s=170,
)
plt.title("JULY 28,2021", fontsize=15)
plt.tight_layout()
plt.show()
# * AS YOU CAN SEE, FIRE AREAS BIGGER THAN OTHERS.
# #### 29 JULY 2021
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
DATE_29.longitude,
DATE_29.latitude,
edgecolor="none",
color="r",
alpha=0.3,
linewidth=10,
s=170,
)
plt.title("JULY 29,2021", fontsize=15)
plt.tight_layout()
plt.show()
# * WE OBSERVED A SIGNIFICANT INCREASE IN FIRE POINTS ON JULY 29.
# #### 28 JULY 2021 AND 29 JULY 2021
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
DATE_28.longitude,
DATE_28.latitude,
edgecolor="none",
color="g",
alpha=0.9,
linewidth=10,
s=100,
)
Map_Plot.scatter(
DATE_29.longitude,
DATE_29.latitude,
edgecolor="none",
color="r",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 29 - 28,2021", fontsize=15)
plt.tight_layout()
plt.show()
# * YOU SEE THE AMOUNT OF INCREASE AFTER 1 DAY.
# GREEN: 28 JULY
# RED: 29 JULY
# #### COMPARISON WITH THE BEGINNING OF THE WEEK
# * WE WILL LOOK AT THE DIFFERENCE BETWEEN DIFFERENT DATES.
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
Data[Data["acq_date"] == "2021-07-15"].longitude,
Data[Data["acq_date"] == "2021-07-15"].latitude,
edgecolor="none",
color="y",
alpha=0.9,
linewidth=10,
s=100,
)
Map_Plot.scatter(
DATE_29.longitude,
DATE_29.latitude,
edgecolor="none",
color="r",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 29 - 15,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
Data[Data["acq_date"] == "2021-07-18"].longitude,
Data[Data["acq_date"] == "2021-07-18"].latitude,
edgecolor="none",
color="y",
alpha=0.9,
linewidth=10,
s=100,
)
Map_Plot.scatter(
DATE_29.longitude,
DATE_29.latitude,
edgecolor="none",
color="r",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 29 - 18,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
Data[Data["acq_date"] == "2021-07-15"].longitude,
Data[Data["acq_date"] == "2021-07-15"].latitude,
edgecolor="none",
color="y",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 15,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
DATE_29.longitude,
DATE_29.latitude,
edgecolor="none",
color="r",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 29,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
Data[Data["acq_date"] == "2021-07-18"].longitude,
Data[Data["acq_date"] == "2021-07-18"].latitude,
edgecolor="none",
color="y",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 18,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
DATE_29.longitude,
DATE_29.latitude,
edgecolor="none",
color="r",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 29,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
Data[Data["acq_date"] == "2021-07-22"].longitude,
Data[Data["acq_date"] == "2021-07-22"].latitude,
edgecolor="none",
color="y",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 22,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
DATE_29.longitude,
DATE_29.latitude,
edgecolor="none",
color="r",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 29,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
Data[Data["acq_date"] == "2021-07-25"].longitude,
Data[Data["acq_date"] == "2021-07-25"].latitude,
edgecolor="none",
color="y",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 25,2021", fontsize=15)
plt.tight_layout()
plt.show()
figure = plt.figure(figsize=(17, 12))
Map_Plot = Basemap(
projection="cyl",
llcrnrlat=33,
llcrnrlon=20,
urcrnrlat=45,
urcrnrlon=55,
resolution="c",
)
Map_Plot.bluemarble()
Map_Plot.drawcountries()
Map_Plot.scatter(
DATE_29.longitude,
DATE_29.latitude,
edgecolor="none",
color="r",
alpha=0.9,
linewidth=10,
s=100,
)
plt.title("JULY 29,2021", fontsize=15)
plt.tight_layout()
plt.show()
# #### DETAIL
# * USE INTERACTIVE MAPS
MAP_FUNCTION = folium.Map(
location=[38.9637, 35.2433], tiles="Stamen Terrain", zoom_start=5.5 # TURKEY
)
MAP_FUNCTION
print(DATE_29[DATE_29["frp"] == DATE_29["frp"].max()])
print("\n")
print("----" * 10)
print("\n")
print(DATE_28[DATE_28["frp"] == DATE_28["frp"].max()])
print(DATE_29[DATE_29["bright_t31"] == DATE_29["bright_t31"].max()])
print("\n")
print("----" * 10)
print("\n")
print(DATE_28[DATE_28["bright_t31"] == DATE_28["bright_t31"].max()])
# * CLICK THE ROUNDS
MAP_FUNCTION = folium.Map(
location=[38.9637, 35.2433], # location australia
tiles="openstreetmap",
zoom_start=6.0,
)
folium.CircleMarker(
location=[36.89363, 31.36536],
radius=20,
popup="DATE:<b>28 JULY MAXIMUM BRIGHTNESS<b>",
tooltip="Click me",
color="#2F4F4F",
fill=True,
fill_color="#00000",
).add_to(MAP_FUNCTION)
MAP_FUNCTION
MAP_FUNCTION = folium.Map(
location=[38.9637, 35.2433], # location australia
tiles="openstreetmap",
zoom_start=6.0,
)
folium.CircleMarker(
location=[37.3512, 35.45154],
radius=20,
popup="DATE:<b>29 JULY MAXIMUM BRIGHTNESS<b>",
tooltip="Click me",
color="#2F4F4F",
fill=True,
fill_color="#00000",
).add_to(MAP_FUNCTION)
MAP_FUNCTION
MAP_FUNCTION = folium.Map(
location=[38.9637, 35.2433], # location australia
tiles="openstreetmap",
zoom_start=6.0,
)
folium.CircleMarker(
location=[36.89363, 31.36536],
radius=20,
popup="DATE:<b>28 JULY MAXIMUM BRIGHTNESS<b>",
tooltip="Click me",
color="#2F4F4F",
fill=True,
fill_color="#FF0000",
).add_to(MAP_FUNCTION)
folium.CircleMarker(
location=[37.3512, 35.45154],
radius=20,
popup="DATE:<b>29 JULY MAXIMUM BRIGHTNESS<b>",
tooltip="Click me",
color="#2F4F4F",
fill=True,
fill_color="#00000",
).add_to(MAP_FUNCTION)
MAP_FUNCTION
MAP_FUNCTION = folium.Map(
location=[38.9637, 35.2433], # location australia
tiles="Stamen Watercolor",
zoom_start=6.0,
)
folium.CircleMarker(
location=[36.89363, 31.36536],
radius=20,
popup="DATE:<b>28 JULY MAXIMUM BRIGHTNESS<b>",
tooltip="Click me",
color="#2F4F4F",
fill=True,
fill_color="#FF0000",
).add_to(MAP_FUNCTION)
folium.CircleMarker(
location=[37.3512, 35.45154],
radius=20,
popup="DATE:<b>29 JULY MAXIMUM BRIGHTNESS<b>",
tooltip="Click me",
color="#2F4F4F",
fill=True,
fill_color="#00000",
).add_to(MAP_FUNCTION)
MAP_FUNCTION
MAP_FUNCTION = folium.Map(
location=[38.9637, 35.2433], # location australia
tiles="Stamen Toner",
zoom_start=6.0,
)
folium.CircleMarker(
location=[36.89363, 31.36536],
radius=20,
popup="DATE:<b>28 JULY MAXIMUM BRIGHTNESS<b>",
tooltip="Click me",
color="#2F4F4F",
fill=True,
fill_color="#FF0000",
).add_to(MAP_FUNCTION)
folium.CircleMarker(
location=[37.3512, 35.45154],
radius=20,
popup="DATE:<b>29 JULY MAXIMUM BRIGHTNESS<b>",
tooltip="Click me",
color="#2F4F4F",
fill=True,
fill_color="#00000",
).add_to(MAP_FUNCTION)
folium.Marker(location=[36.89363, 31.36536], popup="ANTALYA").add_to(MAP_FUNCTION)
folium.Marker(location=[37.3512, 35.45154], popup="ADANA").add_to(MAP_FUNCTION)
MAP_FUNCTION
LAT_LON_29 = DATE_29[["latitude", "longitude"]]
LAT_LON_28 = DATE_28[["latitude", "longitude"]]
MAP_FUNCTION = folium.Map(
location=[38.9637, 35.2433], # location australia
tiles="openstreetmap",
zoom_start=6.0,
)
plugins.MarkerCluster(LAT_LON_28).add_to(MAP_FUNCTION)
MAP_FUNCTION
MAP_FUNCTION = folium.Map(
location=[38.9637, 35.2433], # location australia
tiles="openstreetmap",
zoom_start=6.0,
)
plugins.MarkerCluster(LAT_LON_29).add_to(MAP_FUNCTION)
MAP_FUNCTION
| false | 0 | 10,566 | 0 | 11,928 | 10,566 |
||
69476461
|
import numpy as np # linear algebra
# data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.ensemble import RandomForestClassifier
from sklearn.linear_model import LogisticRegression
Insurance = pd.read_csv(r"../input/porto-seguro-safe-driver-prediction/train.csv")
test1 = pd.read_csv(r"../input/porto-seguro-safe-driver-prediction/test.csv")
# print (Insurance.shape)
print(test1.shape)
# Insurance.head()
test1.head()
# Graph to show the Higly Imbalanced Data. To Predict the Column ['target']. Right now.. majority of records/rows with 'Target=0'
# And few records with 'Target = 1'
pd.value_counts(Insurance["target"]).plot.bar()
plt.title("Target class histogram")
plt.xlabel("target")
# Top 5 features with most missing data in training dataset
Insurance = Insurance.replace(-1, np.nan)
Insurance_missing = Insurance.isnull().sum().sort_values(ascending=False)
Insurance_missing = pd.DataFrame(Insurance_missing).reset_index()
Insurance_missing.columns = ["Feature", "Number of Data Records Missing"]
Insurance_missing.head()
# Top 5 features with most missing data in test dataset
test1 = test1.replace(-1, np.nan)
test1_missing = test1.isnull().sum().sort_values(ascending=False)
test1_missing = pd.DataFrame(test1_missing).reset_index()
test1_missing.columns = ["Feature", "Number of Data Records Missing"]
test1_missing.head()
# drop features 'ps_car_03_cat' and 'ps_car_05_cat'
Insurance = Insurance.drop(["ps_car_03_cat", "ps_car_05_cat"], axis=1)
Insurance.dtypes
# drop features 'ps_car_03_cat' and 'ps_car_05_cat'
test1 = test1.drop(["ps_car_03_cat", "ps_car_05_cat"], axis=1)
test1.dtypes
# visualize the correlation between remaining features
import seaborn as sns
corr = Insurance.corr()
sns.heatmap(corr)
plt.title("Correlation Between Features", fontsize=14)
plt.xticks(rotation="vertical")
plt.figure(figsize=(20, 10), dpi=110)
plt.show()
# drop features with '_calc_' in feature names, because they have low correlation
feature_calc = list(Insurance.columns[Insurance.columns.str.contains("_calc_")])
Insurance = Insurance.drop(feature_calc, axis=1)
print(Insurance.shape)
# drop features with '_calc_' in feature names, because they have low correlation
feature_calc = list(test1.columns[test1.columns.str.contains("_calc_")])
test1 = test1.drop(feature_calc, axis=1)
print(test1.shape)
# replace missing data in the remaining features
feature_bin = [f for f in Insurance.columns if f.endswith("bin")]
feature_cat = [f for f in Insurance.columns if f.endswith("cat")]
feature_els = [
f
for f in Insurance.columns
if (f not in feature_bin) & (f not in feature_cat) & (f not in ["id", "target"])
]
# replace missing data in the remaining features_test
feature_bin_test1 = [f for f in test1.columns if f.endswith("bin")]
feature_cat_test1 = [f for f in test1.columns if f.endswith("cat")]
feature_els_test1 = [
f
for f in test1.columns
if (f not in feature_bin_test1) & (f not in feature_cat_test1) & (f not in ["id"])
]
# filling missing data with mode value for binary and categorical Data.
for f in feature_bin + feature_cat:
Insurance[f].fillna(value=Insurance[f].mode()[0], inplace=True)
# filling missing data with mean value.
for f in feature_els:
Insurance[f].fillna(value=Insurance[f].mean(), inplace=True)
# filling missing data with mode value for binary and categorical Data_test.
for f in feature_bin_test1 + feature_cat_test1:
test1[f].fillna(value=test1[f].mode()[0], inplace=True)
# filling missing data with mean value.
for f in feature_els:
test1[f].fillna(value=test1[f].mean(), inplace=True)
# Visualize binary features data and categorical features data (for styling, plot only features with 2 unique values)
plt.figure(figsize=(20, 20), dpi=110)
count = 0
for f in feature_bin + feature_cat:
if Insurance[f].nunique() <= 2:
ax = plt.subplot(4, 4, count + 1)
sns.countplot(Insurance[f])
plt.xlabel(f, fontsize=14)
plt.ylabel("Count", fontsize=12)
# plt.ylim(0, 600000)
count += 1
plt.show()
# Feature Data Distribution Subplots
# Class count
count_class_0, count_class_1 = Insurance.target.value_counts()
# Divide by class
df_class_0 = Insurance[Insurance["target"] == 0]
df_class_1 = Insurance[Insurance["target"] == 1]
print("Zero", count_class_0)
print("One", count_class_1)
# Doing Random Undersampling
df_class_0_under = df_class_0.sample(count_class_1)
df_test_under = pd.concat([df_class_0_under, df_class_1], axis=0)
print("After Random under-sampling:")
print(df_test_under.target.value_counts())
df_test_under.target.value_counts().plot(kind="bar", title="Count (target)")
df_test_under.dtypes
df_test_under.shape
# From Now modelling with Undersampled and balanced Data
train2 = df_test_under
y_train = train2["target"]
X_train = train2.drop(["target", "id"], axis=1)
print(X.shape)
print(X.dtypes)
print(y.shape)
X_test1 = test1.drop(["id"], axis=1)
print(X_test1.shape)
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_train = sc.fit_transform(X_train)
X_test1 = sc.transform(X_test1)
# Model Number One Logistic Regresssion
from sklearn.linear_model import LogisticRegression
# classifier = LogisticRegression(class_weight={0:0, 1:1},random_state = 42 ,solver='lbfgs')
# classifier = LogisticRegression(class_weight='balanced',random_state = 0 ,solver='lbfgs')
classifier = LogisticRegression(random_state=42, solver="lbfgs")
classifier.fit(X_train, y_train)
y_pred = classifier.predict(X_test1)
# y_pred
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, y_pred)
print("Confusion Matrix: ", cm)
from sklearn.metrics import precision_score
pr = precision_score(y_test, y_pred)
print("Precision Score: ", pr)
from sklearn.metrics import recall_score
rc = recall_score(y_test, y_pred)
print("Recall Score: ", rc)
from sklearn.metrics import accuracy_score
ac = accuracy_score(y_test, y_pred)
print("Accuracy Score: ", ac)
from sklearn.metrics import f1_score
Fone = f1_score(y_test, y_pred)
print("F1 Score: ", Fone)
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/476/69476461.ipynb
| null | null |
[{"Id": 69476461, "ScriptId": 18968715, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4843468, "CreationDate": "07/31/2021 13:28:27", "VersionNumber": 1.0, "Title": "safe_driver_prediction", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 211.0, "LinesInsertedFromPrevious": 211.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
# data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.ensemble import RandomForestClassifier
from sklearn.linear_model import LogisticRegression
Insurance = pd.read_csv(r"../input/porto-seguro-safe-driver-prediction/train.csv")
test1 = pd.read_csv(r"../input/porto-seguro-safe-driver-prediction/test.csv")
# print (Insurance.shape)
print(test1.shape)
# Insurance.head()
test1.head()
# Graph to show the Higly Imbalanced Data. To Predict the Column ['target']. Right now.. majority of records/rows with 'Target=0'
# And few records with 'Target = 1'
pd.value_counts(Insurance["target"]).plot.bar()
plt.title("Target class histogram")
plt.xlabel("target")
# Top 5 features with most missing data in training dataset
Insurance = Insurance.replace(-1, np.nan)
Insurance_missing = Insurance.isnull().sum().sort_values(ascending=False)
Insurance_missing = pd.DataFrame(Insurance_missing).reset_index()
Insurance_missing.columns = ["Feature", "Number of Data Records Missing"]
Insurance_missing.head()
# Top 5 features with most missing data in test dataset
test1 = test1.replace(-1, np.nan)
test1_missing = test1.isnull().sum().sort_values(ascending=False)
test1_missing = pd.DataFrame(test1_missing).reset_index()
test1_missing.columns = ["Feature", "Number of Data Records Missing"]
test1_missing.head()
# drop features 'ps_car_03_cat' and 'ps_car_05_cat'
Insurance = Insurance.drop(["ps_car_03_cat", "ps_car_05_cat"], axis=1)
Insurance.dtypes
# drop features 'ps_car_03_cat' and 'ps_car_05_cat'
test1 = test1.drop(["ps_car_03_cat", "ps_car_05_cat"], axis=1)
test1.dtypes
# visualize the correlation between remaining features
import seaborn as sns
corr = Insurance.corr()
sns.heatmap(corr)
plt.title("Correlation Between Features", fontsize=14)
plt.xticks(rotation="vertical")
plt.figure(figsize=(20, 10), dpi=110)
plt.show()
# drop features with '_calc_' in feature names, because they have low correlation
feature_calc = list(Insurance.columns[Insurance.columns.str.contains("_calc_")])
Insurance = Insurance.drop(feature_calc, axis=1)
print(Insurance.shape)
# drop features with '_calc_' in feature names, because they have low correlation
feature_calc = list(test1.columns[test1.columns.str.contains("_calc_")])
test1 = test1.drop(feature_calc, axis=1)
print(test1.shape)
# replace missing data in the remaining features
feature_bin = [f for f in Insurance.columns if f.endswith("bin")]
feature_cat = [f for f in Insurance.columns if f.endswith("cat")]
feature_els = [
f
for f in Insurance.columns
if (f not in feature_bin) & (f not in feature_cat) & (f not in ["id", "target"])
]
# replace missing data in the remaining features_test
feature_bin_test1 = [f for f in test1.columns if f.endswith("bin")]
feature_cat_test1 = [f for f in test1.columns if f.endswith("cat")]
feature_els_test1 = [
f
for f in test1.columns
if (f not in feature_bin_test1) & (f not in feature_cat_test1) & (f not in ["id"])
]
# filling missing data with mode value for binary and categorical Data.
for f in feature_bin + feature_cat:
Insurance[f].fillna(value=Insurance[f].mode()[0], inplace=True)
# filling missing data with mean value.
for f in feature_els:
Insurance[f].fillna(value=Insurance[f].mean(), inplace=True)
# filling missing data with mode value for binary and categorical Data_test.
for f in feature_bin_test1 + feature_cat_test1:
test1[f].fillna(value=test1[f].mode()[0], inplace=True)
# filling missing data with mean value.
for f in feature_els:
test1[f].fillna(value=test1[f].mean(), inplace=True)
# Visualize binary features data and categorical features data (for styling, plot only features with 2 unique values)
plt.figure(figsize=(20, 20), dpi=110)
count = 0
for f in feature_bin + feature_cat:
if Insurance[f].nunique() <= 2:
ax = plt.subplot(4, 4, count + 1)
sns.countplot(Insurance[f])
plt.xlabel(f, fontsize=14)
plt.ylabel("Count", fontsize=12)
# plt.ylim(0, 600000)
count += 1
plt.show()
# Feature Data Distribution Subplots
# Class count
count_class_0, count_class_1 = Insurance.target.value_counts()
# Divide by class
df_class_0 = Insurance[Insurance["target"] == 0]
df_class_1 = Insurance[Insurance["target"] == 1]
print("Zero", count_class_0)
print("One", count_class_1)
# Doing Random Undersampling
df_class_0_under = df_class_0.sample(count_class_1)
df_test_under = pd.concat([df_class_0_under, df_class_1], axis=0)
print("After Random under-sampling:")
print(df_test_under.target.value_counts())
df_test_under.target.value_counts().plot(kind="bar", title="Count (target)")
df_test_under.dtypes
df_test_under.shape
# From Now modelling with Undersampled and balanced Data
train2 = df_test_under
y_train = train2["target"]
X_train = train2.drop(["target", "id"], axis=1)
print(X.shape)
print(X.dtypes)
print(y.shape)
X_test1 = test1.drop(["id"], axis=1)
print(X_test1.shape)
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_train = sc.fit_transform(X_train)
X_test1 = sc.transform(X_test1)
# Model Number One Logistic Regresssion
from sklearn.linear_model import LogisticRegression
# classifier = LogisticRegression(class_weight={0:0, 1:1},random_state = 42 ,solver='lbfgs')
# classifier = LogisticRegression(class_weight='balanced',random_state = 0 ,solver='lbfgs')
classifier = LogisticRegression(random_state=42, solver="lbfgs")
classifier.fit(X_train, y_train)
y_pred = classifier.predict(X_test1)
# y_pred
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, y_pred)
print("Confusion Matrix: ", cm)
from sklearn.metrics import precision_score
pr = precision_score(y_test, y_pred)
print("Precision Score: ", pr)
from sklearn.metrics import recall_score
rc = recall_score(y_test, y_pred)
print("Recall Score: ", rc)
from sklearn.metrics import accuracy_score
ac = accuracy_score(y_test, y_pred)
print("Accuracy Score: ", ac)
from sklearn.metrics import f1_score
Fone = f1_score(y_test, y_pred)
print("F1 Score: ", Fone)
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
| false | 0 | 2,181 | 0 | 2,181 | 2,181 |
||
69401918
|
<jupyter_start><jupyter_text>Predict Test Scores of students
### Context
A dataset I found as part of SPSS v23 package and I found it worth sharing here. Let's build machine learning algorithms to predict the scores of the students.
### Content
It contains information about a test written by some students. It include features such as: School setting, School type, gender, pretetest scores among other. Explore the data to know more!
Kaggle dataset identifier: predict-test-scores-of-students
<jupyter_code>import pandas as pd
df = pd.read_csv('predict-test-scores-of-students/test_scores.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 2133 entries, 0 to 2132
Data columns (total 11 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 school 2133 non-null object
1 school_setting 2133 non-null object
2 school_type 2133 non-null object
3 classroom 2133 non-null object
4 teaching_method 2133 non-null object
5 n_student 2133 non-null float64
6 student_id 2133 non-null object
7 gender 2133 non-null object
8 lunch 2133 non-null object
9 pretest 2133 non-null float64
10 posttest 2133 non-null float64
dtypes: float64(3), object(8)
memory usage: 183.4+ KB
<jupyter_text>Examples:
{
"school": "ANKYI",
"school_setting": "Urban",
"school_type": "Non-public",
"classroom": "6OL",
"teaching_method": "Standard",
"n_student": 20,
"student_id": "2FHT3",
"gender": "Female",
"lunch": "Does not qualify",
"pretest": 62,
"posttest": 72
}
{
"school": "ANKYI",
"school_setting": "Urban",
"school_type": "Non-public",
"classroom": "6OL",
"teaching_method": "Standard",
"n_student": 20,
"student_id": "3JIVH",
"gender": "Female",
"lunch": "Does not qualify",
"pretest": 66,
"posttest": 79
}
{
"school": "ANKYI",
"school_setting": "Urban",
"school_type": "Non-public",
"classroom": "6OL",
"teaching_method": "Standard",
"n_student": 20,
"student_id": "3XOWE",
"gender": "Male",
"lunch": "Does not qualify",
"pretest": 64,
"posttest": 76
}
{
"school": "ANKYI",
"school_setting": "Urban",
"school_type": "Non-public",
"classroom": "6OL",
"teaching_method": "Standard",
"n_student": 20,
"student_id": "556O0",
"gender": "Female",
"lunch": "Does not qualify",
"pretest": 61,
"posttest": 77
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/kaggle/input/predict-test-scores-of-students/test_scores.csv")
df.head()
df.info()
df.dropna().shape # -> there is no empty items in df
df.nunique().plot(kind="bar") #
tm = df["teaching_method"]
df.nunique()
tm.value_counts()
df["school"].value_counts()
df["lunch"].value_counts()
df2 = pd.get_dummies(df, columns=["school_setting"], prefix=["school_setting"])
df2
list1 = ["school_setting_Rural", "school_setting_Suburban", "school_setting_Urban"]
for item in list1:
print(df2[item].corr(df2["posttest"]))
# max results on tests
pretest = df["pretest"]
posttest = df["posttest"]
np.max(pretest), np.max(posttest)
avg_pre = pretest.mean()
avg_post = posttest.mean()
np.sum(pretest > avg_pre), np.sum(posttest > avg_post)
np.sum(pretest == 93), np.sum(posttest == 100)
df.iloc[np.argmax(pretest), :] # best pretest student
# best posttest students
indexnames = df[df["posttest"] != 100].index
df.drop(indexnames)
df.describe()
#
# # **Visualisation**
c = 0
list2 = [df.school_setting, df.school_type, df.teaching_method, df.gender]
fig, axes = plt.pyplot.subplots(2, 2, figsize=(18, 10))
for i in range(2):
for j in range(2):
sns.countplot(ax=axes[i, j], x=list2[c])
c += 1
fig, (ax1, ax2) = plt.pyplot.subplots(1, 2, figsize=(12, 6))
sns.set_style("darkgrid", {"axes.facecolor": ".9"})
sns.boxplot(x=df.pretest, y=df.gender, ax=ax1)
sns.histplot(x=df.pretest, ax=ax2, kde=True)
fig, (ax1, ax2) = plt.pyplot.subplots(1, 2, figsize=(12, 6))
sns.boxplot(x=df.posttest, y=df.gender, ax=ax1)
sns.histplot(x=df.posttest, ax=ax2, kde=True)
# seems, that pretest scores strongly correlate with posttest scores
df[["pretest", "posttest"]].plot(figsize=(35, 15))
df["pretest"].corr(df["posttest"]) # we were right
# # Machine learning part
# quick look at data again
df.head()
# We got some categorial features, so have to encode them to continue
# Split the data into X and y
X = df.drop(["posttest", "classroom", "student_id"], axis=1)
y = df["posttest"]
# Convert categorical values to numbers
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import OneHotEncoder
categories = [
"school",
"school_setting",
"school_type",
"teaching_method",
"gender",
"lunch",
]
one_hot = OneHotEncoder()
transformer = ColumnTransformer(
[("one_hot", one_hot, categories)], remainder="passthrough"
)
X_transformed = transformer.fit_transform(X)
# Split the transformed data to training and test sets
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X_transformed, y, test_size=0.2)
# Import the Random Forest regressor
from sklearn.ensemble import RandomForestRegressor
model = RandomForestRegressor()
model.fit(X_train, y_train)
# Score the model
model.score(X_test, y_test)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X_transformed, y, test_size=0.18)
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import cross_val_score
model = RandomForestRegressor()
model.fit(X_train, y_train)
model.score(X_test, y_test)
cv_score = cross_val_score(model, X_transformed, y)
np.mean(cv_score)
# # Tuning
from sklearn.model_selection import RandomizedSearchCV
X = df.drop(["posttest", "classroom", "student_id"], axis=1)
y = df["posttest"]
# Convert categorical values to numbers
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import OneHotEncoder
categories = [
"school",
"school_setting",
"school_type",
"teaching_method",
"gender",
"lunch",
]
one_hot = OneHotEncoder()
transformer = ColumnTransformer(
[("one_hot", one_hot, categories)], remainder="passthrough"
)
X_transformed = transformer.fit_transform(X)
# Split the transformed data to training and test sets
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X_transformed, y, test_size=0.2)
grid = {
"n_estimators": [1, 10, 50, 100, 200, 500, 800, 1200, 3000],
"max_depth": [None, 5, 10, 20, 30],
"max_features": ["auto", "sqrt"],
}
tuned_model = RandomizedSearchCV(
estimator=model, param_distributions=grid, n_iter=15, cv=7, verbose=2
)
tuned_model.fit(X_train, y_train)
tuned_model.best_params_
y_pred = tuned_model.predict(X_test)
from sklearn.metrics import explained_variance_score
from sklearn.metrics import r2_score, mean_squared_error
r2_score(y_test, y_pred)
mse = mean_squared_error(y_test, y_pred)
mse
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/401/69401918.ipynb
|
predict-test-scores-of-students
|
kwadwoofosu
|
[{"Id": 69401918, "ScriptId": 18926299, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6918149, "CreationDate": "07/30/2021 14:34:30", "VersionNumber": 5.0, "Title": "predict-test-scores-of-students", "EvaluationDate": "07/30/2021", "IsChange": false, "TotalLines": 185.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 185.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92521257, "KernelVersionId": 69401918, "SourceDatasetVersionId": 2221874}]
|
[{"Id": 2221874, "DatasetId": 1334397, "DatasourceVersionId": 2263561, "CreatorUserId": 6448792, "LicenseName": "Data files \u00a9 Original Authors", "CreationDate": "05/11/2021 15:02:19", "VersionNumber": 1.0, "Title": "Predict Test Scores of students", "Slug": "predict-test-scores-of-students", "Subtitle": "Predicting the posttest scores of students from 11 features", "Description": "### Context\n\nA dataset I found as part of SPSS v23 package and I found it worth sharing here. Let's build machine learning algorithms to predict the scores of the students.\n\n\n### Content\n\nIt contains information about a test written by some students. It include features such as: School setting, School type, gender, pretetest scores among other. Explore the data to know more!\n\n\n### Acknowledgements\n\nI will acknowledge IBM SPSS for including this dataset in their SPSS document folder\n\n\n### Inspiration\n\nI would love to see diverse approaches in predicting the posttest scores of the students. I'd love to learn new things through the sharing of codes and discussions.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1334397, "CreatorUserId": 6448792, "OwnerUserId": 6448792.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2221874.0, "CurrentDatasourceVersionId": 2263561.0, "ForumId": 1353328, "Type": 2, "CreationDate": "05/11/2021 15:02:19", "LastActivityDate": "05/11/2021", "TotalViews": 59069, "TotalDownloads": 9106, "TotalVotes": 154, "TotalKernels": 93}]
|
[{"Id": 6448792, "UserName": "kwadwoofosu", "DisplayName": "Mathias Ofosu", "RegisterDate": "12/30/2020", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/kaggle/input/predict-test-scores-of-students/test_scores.csv")
df.head()
df.info()
df.dropna().shape # -> there is no empty items in df
df.nunique().plot(kind="bar") #
tm = df["teaching_method"]
df.nunique()
tm.value_counts()
df["school"].value_counts()
df["lunch"].value_counts()
df2 = pd.get_dummies(df, columns=["school_setting"], prefix=["school_setting"])
df2
list1 = ["school_setting_Rural", "school_setting_Suburban", "school_setting_Urban"]
for item in list1:
print(df2[item].corr(df2["posttest"]))
# max results on tests
pretest = df["pretest"]
posttest = df["posttest"]
np.max(pretest), np.max(posttest)
avg_pre = pretest.mean()
avg_post = posttest.mean()
np.sum(pretest > avg_pre), np.sum(posttest > avg_post)
np.sum(pretest == 93), np.sum(posttest == 100)
df.iloc[np.argmax(pretest), :] # best pretest student
# best posttest students
indexnames = df[df["posttest"] != 100].index
df.drop(indexnames)
df.describe()
#
# # **Visualisation**
c = 0
list2 = [df.school_setting, df.school_type, df.teaching_method, df.gender]
fig, axes = plt.pyplot.subplots(2, 2, figsize=(18, 10))
for i in range(2):
for j in range(2):
sns.countplot(ax=axes[i, j], x=list2[c])
c += 1
fig, (ax1, ax2) = plt.pyplot.subplots(1, 2, figsize=(12, 6))
sns.set_style("darkgrid", {"axes.facecolor": ".9"})
sns.boxplot(x=df.pretest, y=df.gender, ax=ax1)
sns.histplot(x=df.pretest, ax=ax2, kde=True)
fig, (ax1, ax2) = plt.pyplot.subplots(1, 2, figsize=(12, 6))
sns.boxplot(x=df.posttest, y=df.gender, ax=ax1)
sns.histplot(x=df.posttest, ax=ax2, kde=True)
# seems, that pretest scores strongly correlate with posttest scores
df[["pretest", "posttest"]].plot(figsize=(35, 15))
df["pretest"].corr(df["posttest"]) # we were right
# # Machine learning part
# quick look at data again
df.head()
# We got some categorial features, so have to encode them to continue
# Split the data into X and y
X = df.drop(["posttest", "classroom", "student_id"], axis=1)
y = df["posttest"]
# Convert categorical values to numbers
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import OneHotEncoder
categories = [
"school",
"school_setting",
"school_type",
"teaching_method",
"gender",
"lunch",
]
one_hot = OneHotEncoder()
transformer = ColumnTransformer(
[("one_hot", one_hot, categories)], remainder="passthrough"
)
X_transformed = transformer.fit_transform(X)
# Split the transformed data to training and test sets
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X_transformed, y, test_size=0.2)
# Import the Random Forest regressor
from sklearn.ensemble import RandomForestRegressor
model = RandomForestRegressor()
model.fit(X_train, y_train)
# Score the model
model.score(X_test, y_test)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X_transformed, y, test_size=0.18)
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import cross_val_score
model = RandomForestRegressor()
model.fit(X_train, y_train)
model.score(X_test, y_test)
cv_score = cross_val_score(model, X_transformed, y)
np.mean(cv_score)
# # Tuning
from sklearn.model_selection import RandomizedSearchCV
X = df.drop(["posttest", "classroom", "student_id"], axis=1)
y = df["posttest"]
# Convert categorical values to numbers
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import OneHotEncoder
categories = [
"school",
"school_setting",
"school_type",
"teaching_method",
"gender",
"lunch",
]
one_hot = OneHotEncoder()
transformer = ColumnTransformer(
[("one_hot", one_hot, categories)], remainder="passthrough"
)
X_transformed = transformer.fit_transform(X)
# Split the transformed data to training and test sets
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X_transformed, y, test_size=0.2)
grid = {
"n_estimators": [1, 10, 50, 100, 200, 500, 800, 1200, 3000],
"max_depth": [None, 5, 10, 20, 30],
"max_features": ["auto", "sqrt"],
}
tuned_model = RandomizedSearchCV(
estimator=model, param_distributions=grid, n_iter=15, cv=7, verbose=2
)
tuned_model.fit(X_train, y_train)
tuned_model.best_params_
y_pred = tuned_model.predict(X_test)
from sklearn.metrics import explained_variance_score
from sklearn.metrics import r2_score, mean_squared_error
r2_score(y_test, y_pred)
mse = mean_squared_error(y_test, y_pred)
mse
|
[{"predict-test-scores-of-students/test_scores.csv": {"column_names": "[\"school\", \"school_setting\", \"school_type\", \"classroom\", \"teaching_method\", \"n_student\", \"student_id\", \"gender\", \"lunch\", \"pretest\", \"posttest\"]", "column_data_types": "{\"school\": \"object\", \"school_setting\": \"object\", \"school_type\": \"object\", \"classroom\": \"object\", \"teaching_method\": \"object\", \"n_student\": \"float64\", \"student_id\": \"object\", \"gender\": \"object\", \"lunch\": \"object\", \"pretest\": \"float64\", \"posttest\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 2133 entries, 0 to 2132\nData columns (total 11 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 school 2133 non-null object \n 1 school_setting 2133 non-null object \n 2 school_type 2133 non-null object \n 3 classroom 2133 non-null object \n 4 teaching_method 2133 non-null object \n 5 n_student 2133 non-null float64\n 6 student_id 2133 non-null object \n 7 gender 2133 non-null object \n 8 lunch 2133 non-null object \n 9 pretest 2133 non-null float64\n 10 posttest 2133 non-null float64\ndtypes: float64(3), object(8)\nmemory usage: 183.4+ KB\n", "summary": "{\"n_student\": {\"count\": 2133.0, \"mean\": 22.796530707923115, \"std\": 4.228892753971893, \"min\": 14.0, \"25%\": 20.0, \"50%\": 22.0, \"75%\": 27.0, \"max\": 31.0}, \"pretest\": {\"count\": 2133.0, \"mean\": 54.95593061415846, \"std\": 13.56310138822343, \"min\": 22.0, \"25%\": 44.0, \"50%\": 56.0, \"75%\": 65.0, \"max\": 93.0}, \"posttest\": {\"count\": 2133.0, \"mean\": 67.10220346929208, \"std\": 13.986788855811128, \"min\": 32.0, \"25%\": 56.0, \"50%\": 68.0, \"75%\": 77.0, \"max\": 100.0}}", "examples": "{\"school\":{\"0\":\"ANKYI\",\"1\":\"ANKYI\",\"2\":\"ANKYI\",\"3\":\"ANKYI\"},\"school_setting\":{\"0\":\"Urban\",\"1\":\"Urban\",\"2\":\"Urban\",\"3\":\"Urban\"},\"school_type\":{\"0\":\"Non-public\",\"1\":\"Non-public\",\"2\":\"Non-public\",\"3\":\"Non-public\"},\"classroom\":{\"0\":\"6OL\",\"1\":\"6OL\",\"2\":\"6OL\",\"3\":\"6OL\"},\"teaching_method\":{\"0\":\"Standard\",\"1\":\"Standard\",\"2\":\"Standard\",\"3\":\"Standard\"},\"n_student\":{\"0\":20.0,\"1\":20.0,\"2\":20.0,\"3\":20.0},\"student_id\":{\"0\":\"2FHT3\",\"1\":\"3JIVH\",\"2\":\"3XOWE\",\"3\":\"556O0\"},\"gender\":{\"0\":\"Female\",\"1\":\"Female\",\"2\":\"Male\",\"3\":\"Female\"},\"lunch\":{\"0\":\"Does not qualify\",\"1\":\"Does not qualify\",\"2\":\"Does not qualify\",\"3\":\"Does not qualify\"},\"pretest\":{\"0\":62.0,\"1\":66.0,\"2\":64.0,\"3\":61.0},\"posttest\":{\"0\":72.0,\"1\":79.0,\"2\":76.0,\"3\":77.0}}"}}]
| true | 1 |
<start_data_description><data_path>predict-test-scores-of-students/test_scores.csv:
<column_names>
['school', 'school_setting', 'school_type', 'classroom', 'teaching_method', 'n_student', 'student_id', 'gender', 'lunch', 'pretest', 'posttest']
<column_types>
{'school': 'object', 'school_setting': 'object', 'school_type': 'object', 'classroom': 'object', 'teaching_method': 'object', 'n_student': 'float64', 'student_id': 'object', 'gender': 'object', 'lunch': 'object', 'pretest': 'float64', 'posttest': 'float64'}
<dataframe_Summary>
{'n_student': {'count': 2133.0, 'mean': 22.796530707923115, 'std': 4.228892753971893, 'min': 14.0, '25%': 20.0, '50%': 22.0, '75%': 27.0, 'max': 31.0}, 'pretest': {'count': 2133.0, 'mean': 54.95593061415846, 'std': 13.56310138822343, 'min': 22.0, '25%': 44.0, '50%': 56.0, '75%': 65.0, 'max': 93.0}, 'posttest': {'count': 2133.0, 'mean': 67.10220346929208, 'std': 13.986788855811128, 'min': 32.0, '25%': 56.0, '50%': 68.0, '75%': 77.0, 'max': 100.0}}
<dataframe_info>
RangeIndex: 2133 entries, 0 to 2132
Data columns (total 11 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 school 2133 non-null object
1 school_setting 2133 non-null object
2 school_type 2133 non-null object
3 classroom 2133 non-null object
4 teaching_method 2133 non-null object
5 n_student 2133 non-null float64
6 student_id 2133 non-null object
7 gender 2133 non-null object
8 lunch 2133 non-null object
9 pretest 2133 non-null float64
10 posttest 2133 non-null float64
dtypes: float64(3), object(8)
memory usage: 183.4+ KB
<some_examples>
{'school': {'0': 'ANKYI', '1': 'ANKYI', '2': 'ANKYI', '3': 'ANKYI'}, 'school_setting': {'0': 'Urban', '1': 'Urban', '2': 'Urban', '3': 'Urban'}, 'school_type': {'0': 'Non-public', '1': 'Non-public', '2': 'Non-public', '3': 'Non-public'}, 'classroom': {'0': '6OL', '1': '6OL', '2': '6OL', '3': '6OL'}, 'teaching_method': {'0': 'Standard', '1': 'Standard', '2': 'Standard', '3': 'Standard'}, 'n_student': {'0': 20.0, '1': 20.0, '2': 20.0, '3': 20.0}, 'student_id': {'0': '2FHT3', '1': '3JIVH', '2': '3XOWE', '3': '556O0'}, 'gender': {'0': 'Female', '1': 'Female', '2': 'Male', '3': 'Female'}, 'lunch': {'0': 'Does not qualify', '1': 'Does not qualify', '2': 'Does not qualify', '3': 'Does not qualify'}, 'pretest': {'0': 62.0, '1': 66.0, '2': 64.0, '3': 61.0}, 'posttest': {'0': 72.0, '1': 79.0, '2': 76.0, '3': 77.0}}
<end_description>
| 1,783 | 0 | 2,642 | 1,783 |
69401022
|
<jupyter_start><jupyter_text>winequality-red
Kaggle dataset identifier: winequalityred
<jupyter_code>import pandas as pd
df = pd.read_csv('winequalityred/winequality-red.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 1596 entries, 0 to 1595
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 fixed acidity 1596 non-null float64
1 volatile acidity 1596 non-null float64
2 citric acid 1596 non-null float64
3 residual sugar 1596 non-null float64
4 chlorides 1596 non-null float64
5 free sulfur dioxide 1596 non-null float64
6 total sulfur dioxide 1596 non-null float64
7 density 1596 non-null float64
8 pH 1596 non-null float64
9 sulphates 1596 non-null float64
10 alcohol 1596 non-null float64
11 quality 1596 non-null int64
dtypes: float64(11), int64(1)
memory usage: 149.8 KB
<jupyter_text>Examples:
{
"fixed acidity": 7.4,
"volatile acidity": 0.7000000000000001,
"citric acid": 0.0,
"residual sugar": 1.9,
"chlorides": 0.076,
"free sulfur dioxide": 11.0,
"total sulfur dioxide": 34.0,
"density": 0.9978,
"pH": 3.51,
"sulphates": 0.56,
"alcohol": 9.4,
"quality": 5.0
}
{
"fixed acidity": 7.8,
"volatile acidity": 0.88,
"citric acid": 0.0,
"residual sugar": 2.6,
"chlorides": 0.098,
"free sulfur dioxide": 25.0,
"total sulfur dioxide": 67.0,
"density": 0.9968,
"pH": 3.2,
"sulphates": 0.68,
"alcohol": 9.8,
"quality": 5.0
}
{
"fixed acidity": 7.8,
"volatile acidity": 0.76,
"citric acid": 0.04,
"residual sugar": 2.3,
"chlorides": 0.092,
"free sulfur dioxide": 15.0,
"total sulfur dioxide": 54.0,
"density": 0.997,
"pH": 3.26,
"sulphates": 0.65,
"alcohol": 9.8,
"quality": 5.0
}
{
"fixed acidity": 11.2,
"volatile acidity": 0.28,
"citric acid": 0.56,
"residual sugar": 1.9,
"chlorides": 0.075,
"free sulfur dioxide": 17.0,
"total sulfur dioxide": 60.0,
"density": 0.998,
"pH": 3.16,
"sulphates": 0.58,
"alcohol": 9.8,
"quality": 6.0
}
<jupyter_script>import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import plotly.express as px
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Loading Dataset
df = pd.read_csv("/kaggle/input/winequalityred/winequality-red.csv")
# Getting know the data and cleaning it
df.head()
df.shape
df.isnull().sum() * len(df) * 100
df.corr()
df.info()
df.describe().T
df.drop_duplicates(keep="first", inplace=True)
sns.pairplot(df)
px.histogram(df, x="quality", color="quality")
df.hist(figsize=(30, 30))
plt.show()
fig = px.imshow(df.corr())
fig.show()
# Positive strong correlations: alcohol, sulphate
# Negative strong correlations: volatile acidity
fig1 = px.scatter(df, x="quality", y="alcohol")
fig1.show()
fig2 = px.scatter(df, x="quality", y="sulphates")
fig2.show()
fig3 = px.scatter(df, x="quality", y="volatile acidity")
fig3.show()
df["quality"].value_counts()
def quality(a):
if a <= 6:
return "bad"
elif a > 6:
return "good"
df["quality"] = df["quality"].apply(quality)
df["quality"].value_counts()
from sklearn import preprocessing
le = preprocessing.LabelEncoder()
df.quality = le.fit_transform(df.quality)
x = df.drop("quality", axis=1)
y = df["quality"]
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.33, random_state=0
)
print(x_train.shape)
print(x_test.shape)
print(y_train.shape)
print(y_test.shape)
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_train = sc.fit_transform(x_train)
X_test = sc.transform(x_test)
# ### Classification Alghorithms
# Logistic Regression
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression(random_state=0)
lr.fit(X_train, y_train)
y_pred = lr.predict(X_test)
from sklearn.metrics import confusion_matrix
from sklearn import metrics
cm = confusion_matrix(y_test, y_pred)
acc = metrics.accuracy_score(y_test, y_pred)
print(cm)
print(acc)
# KNeighborsClassifier
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier(n_neighbors=1, metric="minkowski")
knn.fit(X_train, y_train)
y_pred = knn.predict(X_test)
cm = confusion_matrix(y_test, y_pred)
acc = metrics.accuracy_score(y_test, y_pred)
print(cm)
print(acc)
# SVC
from sklearn.svm import SVC
svc = SVC(kernel="poly")
svc.fit(X_train, y_train)
y_pred = svc.predict(X_test)
cm = confusion_matrix(y_test, y_pred)
acc = metrics.accuracy_score(y_test, y_pred)
print(cm)
print(acc)
# GaussianNB
from sklearn.naive_bayes import GaussianNB
gnb = GaussianNB()
gnb.fit(X_train, y_train)
y_pred = gnb.predict(X_test)
cm = confusion_matrix(y_test, y_pred)
acc = metrics.accuracy_score(y_test, y_pred)
print(cm)
print(acc)
# DecisionTreeClassifier
from sklearn.tree import DecisionTreeClassifier
dtc = DecisionTreeClassifier(criterion="entropy")
dtc.fit(X_train, y_train)
y_pred = dtc.predict(X_test)
cm = confusion_matrix(y_test, y_pred)
acc = metrics.accuracy_score(y_test, y_pred)
print(cm)
print(acc)
# RandomForestClassifier
from sklearn.ensemble import RandomForestClassifier
rfc = RandomForestClassifier(n_estimators=10, criterion="entropy")
rfc.fit(X_train, y_train)
y_pred = rfc.predict(X_test)
cm = confusion_matrix(y_test, y_pred)
acc = metrics.accuracy_score(y_test, y_pred)
print(cm)
print(acc)
# xgbosst
import xgboost
xgb = xgboost.XGBClassifier()
xgb_model = xgb.fit(X_train, y_train)
y_pred = xgb_model.predict(X_test)
cm = confusion_matrix(y_test, y_pred)
acc = metrics.accuracy_score(y_test, y_pred)
print(cm)
print(acc)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/401/69401022.ipynb
|
winequalityred
|
sh6147782
|
[{"Id": 69401022, "ScriptId": 18948575, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5463467, "CreationDate": "07/30/2021 14:24:22", "VersionNumber": 1.0, "Title": "notebook05bde7d703", "EvaluationDate": "07/30/2021", "IsChange": true, "TotalLines": 171.0, "LinesInsertedFromPrevious": 171.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92519446, "KernelVersionId": 69401022, "SourceDatasetVersionId": 146750}]
|
[{"Id": 146750, "DatasetId": 70308, "DatasourceVersionId": 156953, "CreatorUserId": 2300315, "LicenseName": "Unknown", "CreationDate": "10/28/2018 11:11:29", "VersionNumber": 1.0, "Title": "winequality-red", "Slug": "winequalityred", "Subtitle": "data tells various chemical combination of redwine", "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 85586.0, "TotalUncompressedBytes": 85586.0}]
|
[{"Id": 70308, "CreatorUserId": 2300315, "OwnerUserId": 2300315.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 146750.0, "CurrentDatasourceVersionId": 156953.0, "ForumId": 79415, "Type": 2, "CreationDate": "10/28/2018 11:11:29", "LastActivityDate": "10/28/2018", "TotalViews": 11264, "TotalDownloads": 2143, "TotalVotes": 22, "TotalKernels": 34}]
|
[{"Id": 2300315, "UserName": "sh6147782", "DisplayName": "Savir Husen", "RegisterDate": "09/28/2018", "PerformanceTier": 0}]
|
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import plotly.express as px
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Loading Dataset
df = pd.read_csv("/kaggle/input/winequalityred/winequality-red.csv")
# Getting know the data and cleaning it
df.head()
df.shape
df.isnull().sum() * len(df) * 100
df.corr()
df.info()
df.describe().T
df.drop_duplicates(keep="first", inplace=True)
sns.pairplot(df)
px.histogram(df, x="quality", color="quality")
df.hist(figsize=(30, 30))
plt.show()
fig = px.imshow(df.corr())
fig.show()
# Positive strong correlations: alcohol, sulphate
# Negative strong correlations: volatile acidity
fig1 = px.scatter(df, x="quality", y="alcohol")
fig1.show()
fig2 = px.scatter(df, x="quality", y="sulphates")
fig2.show()
fig3 = px.scatter(df, x="quality", y="volatile acidity")
fig3.show()
df["quality"].value_counts()
def quality(a):
if a <= 6:
return "bad"
elif a > 6:
return "good"
df["quality"] = df["quality"].apply(quality)
df["quality"].value_counts()
from sklearn import preprocessing
le = preprocessing.LabelEncoder()
df.quality = le.fit_transform(df.quality)
x = df.drop("quality", axis=1)
y = df["quality"]
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.33, random_state=0
)
print(x_train.shape)
print(x_test.shape)
print(y_train.shape)
print(y_test.shape)
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_train = sc.fit_transform(x_train)
X_test = sc.transform(x_test)
# ### Classification Alghorithms
# Logistic Regression
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression(random_state=0)
lr.fit(X_train, y_train)
y_pred = lr.predict(X_test)
from sklearn.metrics import confusion_matrix
from sklearn import metrics
cm = confusion_matrix(y_test, y_pred)
acc = metrics.accuracy_score(y_test, y_pred)
print(cm)
print(acc)
# KNeighborsClassifier
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier(n_neighbors=1, metric="minkowski")
knn.fit(X_train, y_train)
y_pred = knn.predict(X_test)
cm = confusion_matrix(y_test, y_pred)
acc = metrics.accuracy_score(y_test, y_pred)
print(cm)
print(acc)
# SVC
from sklearn.svm import SVC
svc = SVC(kernel="poly")
svc.fit(X_train, y_train)
y_pred = svc.predict(X_test)
cm = confusion_matrix(y_test, y_pred)
acc = metrics.accuracy_score(y_test, y_pred)
print(cm)
print(acc)
# GaussianNB
from sklearn.naive_bayes import GaussianNB
gnb = GaussianNB()
gnb.fit(X_train, y_train)
y_pred = gnb.predict(X_test)
cm = confusion_matrix(y_test, y_pred)
acc = metrics.accuracy_score(y_test, y_pred)
print(cm)
print(acc)
# DecisionTreeClassifier
from sklearn.tree import DecisionTreeClassifier
dtc = DecisionTreeClassifier(criterion="entropy")
dtc.fit(X_train, y_train)
y_pred = dtc.predict(X_test)
cm = confusion_matrix(y_test, y_pred)
acc = metrics.accuracy_score(y_test, y_pred)
print(cm)
print(acc)
# RandomForestClassifier
from sklearn.ensemble import RandomForestClassifier
rfc = RandomForestClassifier(n_estimators=10, criterion="entropy")
rfc.fit(X_train, y_train)
y_pred = rfc.predict(X_test)
cm = confusion_matrix(y_test, y_pred)
acc = metrics.accuracy_score(y_test, y_pred)
print(cm)
print(acc)
# xgbosst
import xgboost
xgb = xgboost.XGBClassifier()
xgb_model = xgb.fit(X_train, y_train)
y_pred = xgb_model.predict(X_test)
cm = confusion_matrix(y_test, y_pred)
acc = metrics.accuracy_score(y_test, y_pred)
print(cm)
print(acc)
|
[{"winequalityred/winequality-red.csv": {"column_names": "[\"fixed acidity\", \"volatile acidity\", \"citric acid\", \"residual sugar\", \"chlorides\", \"free sulfur dioxide\", \"total sulfur dioxide\", \"density\", \"pH\", \"sulphates\", \"alcohol\", \"quality\"]", "column_data_types": "{\"fixed acidity\": \"float64\", \"volatile acidity\": \"float64\", \"citric acid\": \"float64\", \"residual sugar\": \"float64\", \"chlorides\": \"float64\", \"free sulfur dioxide\": \"float64\", \"total sulfur dioxide\": \"float64\", \"density\": \"float64\", \"pH\": \"float64\", \"sulphates\": \"float64\", \"alcohol\": \"float64\", \"quality\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1596 entries, 0 to 1595\nData columns (total 12 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 fixed acidity 1596 non-null float64\n 1 volatile acidity 1596 non-null float64\n 2 citric acid 1596 non-null float64\n 3 residual sugar 1596 non-null float64\n 4 chlorides 1596 non-null float64\n 5 free sulfur dioxide 1596 non-null float64\n 6 total sulfur dioxide 1596 non-null float64\n 7 density 1596 non-null float64\n 8 pH 1596 non-null float64\n 9 sulphates 1596 non-null float64\n 10 alcohol 1596 non-null float64\n 11 quality 1596 non-null int64 \ndtypes: float64(11), int64(1)\nmemory usage: 149.8 KB\n", "summary": "{\"fixed acidity\": {\"count\": 1596.0, \"mean\": 8.314160401002507, \"std\": 1.732202825899658, \"min\": 4.6, \"25%\": 7.1, \"50%\": 7.9, \"75%\": 9.2, \"max\": 15.6}, \"volatile acidity\": {\"count\": 1596.0, \"mean\": 0.5279542606516291, \"std\": 0.17917609359161402, \"min\": 0.12, \"25%\": 0.39, \"50%\": 0.52, \"75%\": 0.64, \"max\": 1.58}, \"citric acid\": {\"count\": 1596.0, \"mean\": 0.27027568922305767, \"std\": 0.19389438791689892, \"min\": 0.0, \"25%\": 0.09, \"50%\": 0.26, \"75%\": 0.42, \"max\": 0.79}, \"residual sugar\": {\"count\": 1596.0, \"mean\": 2.5355576441102756, \"std\": 1.4055152456540563, \"min\": 0.9, \"25%\": 1.9, \"50%\": 2.2, \"75%\": 2.6, \"max\": 15.5}, \"chlorides\": {\"count\": 1596.0, \"mean\": 0.08711967418546367, \"std\": 0.04525084486113082, \"min\": 0.012, \"25%\": 0.07, \"50%\": 0.079, \"75%\": 0.09, \"max\": 0.611}, \"free sulfur dioxide\": {\"count\": 1596.0, \"mean\": 15.858395989974937, \"std\": 10.460554283633185, \"min\": 1.0, \"25%\": 7.0, \"50%\": 14.0, \"75%\": 21.0, \"max\": 72.0}, \"total sulfur dioxide\": {\"count\": 1596.0, \"mean\": 46.38220551378446, \"std\": 32.83913823126541, \"min\": 6.0, \"25%\": 22.0, \"50%\": 38.0, \"75%\": 62.0, \"max\": 289.0}, \"density\": {\"count\": 1596.0, \"mean\": 0.9967440726817042, \"std\": 0.0018875997144154764, \"min\": 0.99007, \"25%\": 0.9956, \"50%\": 0.996745, \"75%\": 0.9978325, \"max\": 1.00369}, \"pH\": {\"count\": 1596.0, \"mean\": 3.3119172932330825, \"std\": 0.15334623491775895, \"min\": 2.86, \"25%\": 3.21, \"50%\": 3.31, \"75%\": 3.4, \"max\": 4.01}, \"sulphates\": {\"count\": 1596.0, \"mean\": 0.6563847117794487, \"std\": 0.16305707942835146, \"min\": 0.33, \"25%\": 0.55, \"50%\": 0.62, \"75%\": 0.73, \"max\": 1.98}, \"alcohol\": {\"count\": 1596.0, \"mean\": 10.421146616541355, \"std\": 1.0603706302978022, \"min\": 8.4, \"25%\": 9.5, \"50%\": 10.2, \"75%\": 11.1, \"max\": 14.0}, \"quality\": {\"count\": 1596.0, \"mean\": 5.637218045112782, \"std\": 0.8070804414349891, \"min\": 3.0, \"25%\": 5.0, \"50%\": 6.0, \"75%\": 6.0, \"max\": 8.0}}", "examples": "{\"fixed acidity\":{\"0\":7.4,\"1\":7.8,\"2\":7.8,\"3\":11.2},\"volatile acidity\":{\"0\":0.7,\"1\":0.88,\"2\":0.76,\"3\":0.28},\"citric acid\":{\"0\":0.0,\"1\":0.0,\"2\":0.04,\"3\":0.56},\"residual sugar\":{\"0\":1.9,\"1\":2.6,\"2\":2.3,\"3\":1.9},\"chlorides\":{\"0\":0.076,\"1\":0.098,\"2\":0.092,\"3\":0.075},\"free sulfur dioxide\":{\"0\":11.0,\"1\":25.0,\"2\":15.0,\"3\":17.0},\"total sulfur dioxide\":{\"0\":34.0,\"1\":67.0,\"2\":54.0,\"3\":60.0},\"density\":{\"0\":0.9978,\"1\":0.9968,\"2\":0.997,\"3\":0.998},\"pH\":{\"0\":3.51,\"1\":3.2,\"2\":3.26,\"3\":3.16},\"sulphates\":{\"0\":0.56,\"1\":0.68,\"2\":0.65,\"3\":0.58},\"alcohol\":{\"0\":9.4,\"1\":9.8,\"2\":9.8,\"3\":9.8},\"quality\":{\"0\":5,\"1\":5,\"2\":5,\"3\":6}}"}}]
| true | 1 |
<start_data_description><data_path>winequalityred/winequality-red.csv:
<column_names>
['fixed acidity', 'volatile acidity', 'citric acid', 'residual sugar', 'chlorides', 'free sulfur dioxide', 'total sulfur dioxide', 'density', 'pH', 'sulphates', 'alcohol', 'quality']
<column_types>
{'fixed acidity': 'float64', 'volatile acidity': 'float64', 'citric acid': 'float64', 'residual sugar': 'float64', 'chlorides': 'float64', 'free sulfur dioxide': 'float64', 'total sulfur dioxide': 'float64', 'density': 'float64', 'pH': 'float64', 'sulphates': 'float64', 'alcohol': 'float64', 'quality': 'int64'}
<dataframe_Summary>
{'fixed acidity': {'count': 1596.0, 'mean': 8.314160401002507, 'std': 1.732202825899658, 'min': 4.6, '25%': 7.1, '50%': 7.9, '75%': 9.2, 'max': 15.6}, 'volatile acidity': {'count': 1596.0, 'mean': 0.5279542606516291, 'std': 0.17917609359161402, 'min': 0.12, '25%': 0.39, '50%': 0.52, '75%': 0.64, 'max': 1.58}, 'citric acid': {'count': 1596.0, 'mean': 0.27027568922305767, 'std': 0.19389438791689892, 'min': 0.0, '25%': 0.09, '50%': 0.26, '75%': 0.42, 'max': 0.79}, 'residual sugar': {'count': 1596.0, 'mean': 2.5355576441102756, 'std': 1.4055152456540563, 'min': 0.9, '25%': 1.9, '50%': 2.2, '75%': 2.6, 'max': 15.5}, 'chlorides': {'count': 1596.0, 'mean': 0.08711967418546367, 'std': 0.04525084486113082, 'min': 0.012, '25%': 0.07, '50%': 0.079, '75%': 0.09, 'max': 0.611}, 'free sulfur dioxide': {'count': 1596.0, 'mean': 15.858395989974937, 'std': 10.460554283633185, 'min': 1.0, '25%': 7.0, '50%': 14.0, '75%': 21.0, 'max': 72.0}, 'total sulfur dioxide': {'count': 1596.0, 'mean': 46.38220551378446, 'std': 32.83913823126541, 'min': 6.0, '25%': 22.0, '50%': 38.0, '75%': 62.0, 'max': 289.0}, 'density': {'count': 1596.0, 'mean': 0.9967440726817042, 'std': 0.0018875997144154764, 'min': 0.99007, '25%': 0.9956, '50%': 0.996745, '75%': 0.9978325, 'max': 1.00369}, 'pH': {'count': 1596.0, 'mean': 3.3119172932330825, 'std': 0.15334623491775895, 'min': 2.86, '25%': 3.21, '50%': 3.31, '75%': 3.4, 'max': 4.01}, 'sulphates': {'count': 1596.0, 'mean': 0.6563847117794487, 'std': 0.16305707942835146, 'min': 0.33, '25%': 0.55, '50%': 0.62, '75%': 0.73, 'max': 1.98}, 'alcohol': {'count': 1596.0, 'mean': 10.421146616541355, 'std': 1.0603706302978022, 'min': 8.4, '25%': 9.5, '50%': 10.2, '75%': 11.1, 'max': 14.0}, 'quality': {'count': 1596.0, 'mean': 5.637218045112782, 'std': 0.8070804414349891, 'min': 3.0, '25%': 5.0, '50%': 6.0, '75%': 6.0, 'max': 8.0}}
<dataframe_info>
RangeIndex: 1596 entries, 0 to 1595
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 fixed acidity 1596 non-null float64
1 volatile acidity 1596 non-null float64
2 citric acid 1596 non-null float64
3 residual sugar 1596 non-null float64
4 chlorides 1596 non-null float64
5 free sulfur dioxide 1596 non-null float64
6 total sulfur dioxide 1596 non-null float64
7 density 1596 non-null float64
8 pH 1596 non-null float64
9 sulphates 1596 non-null float64
10 alcohol 1596 non-null float64
11 quality 1596 non-null int64
dtypes: float64(11), int64(1)
memory usage: 149.8 KB
<some_examples>
{'fixed acidity': {'0': 7.4, '1': 7.8, '2': 7.8, '3': 11.2}, 'volatile acidity': {'0': 0.7, '1': 0.88, '2': 0.76, '3': 0.28}, 'citric acid': {'0': 0.0, '1': 0.0, '2': 0.04, '3': 0.56}, 'residual sugar': {'0': 1.9, '1': 2.6, '2': 2.3, '3': 1.9}, 'chlorides': {'0': 0.076, '1': 0.098, '2': 0.092, '3': 0.075}, 'free sulfur dioxide': {'0': 11.0, '1': 25.0, '2': 15.0, '3': 17.0}, 'total sulfur dioxide': {'0': 34.0, '1': 67.0, '2': 54.0, '3': 60.0}, 'density': {'0': 0.9978, '1': 0.9968, '2': 0.997, '3': 0.998}, 'pH': {'0': 3.51, '1': 3.2, '2': 3.26, '3': 3.16}, 'sulphates': {'0': 0.56, '1': 0.68, '2': 0.65, '3': 0.58}, 'alcohol': {'0': 9.4, '1': 9.8, '2': 9.8, '3': 9.8}, 'quality': {'0': 5, '1': 5, '2': 5, '3': 6}}
<end_description>
| 1,286 | 0 | 2,275 | 1,286 |
69401978
|
# # Welcome to 101 Exercises for Python Fundamentals
# Solving these exercises will help make you a better programmer. Solve them in order, because each solution builds scaffolding, working code, and knowledge you can use on future problems. Read the directions carefully, and have fun!
# > "Learning to program takes a little bit of study and a *lot* of practice" - Luis Montealegre
# ## Getting Started
# 0. Create your own account on [Kaggle.com](https://www.kaggle.com/account/login?phase=startRegisterTab) and return to this page.
# 1. Click the blue "Copy and Edit" in the upper-right part of this document to create your own copy to your own Kaggle account.
# 2. As you complete exercises, be sure to click the blue "Save" button to create save points for your work.
# 3. If you need to refresh and restart this learning environment, go to **Run** then select **Restart Session**.
# 4. If you need a fresh, blank copy of this document, go to https://www.kaggle.com/ryanorsinger/101-exercises/ and click "Copy and Edit"
# ## Orientation
# - This code notebook is composed of cells. Each cell is either text or Python code.
# - To run a cell of code, click the "play button" icon to the left of the cell or click on the cell and press "Shift+Enter" on your keyboard. This will execute the Python code contained in the cell. Executing a cell that defines a variable is important before executing or authoring a cell that depends on that previously created variable assignment.
# - **Expect to see lots of errors** the first time you load this page.
# - **Expect to see lots of errors** for all cells run without code that matches the assertion tests.
# - Until you click the blue "Copy and Edit" button to make your own copy, you will see an entire page of errors. This is part of the automated tests.
# - Each *assert* line is both an example and a test that tests for the presence and functionality of the instructed exercise.
# ## The only 3 conditions that produce no errors:
# 1. When you make a fresh **copy** of the project to your own account (by clicking "Copy and Edit")
# 2. When you go to "Run" and then click "Restart Session"
# 3. When every single assertion passes.
# ## Outline
# - Each cell starts with a problem statement that describes the exercise to complete.
# - Underneath each problem statement, learners will need to write code to produce an answer.
# - The **assert** lines test to see that your code solves the problem appropriately
# - Many exercises will rely on previous solutions to be correctly completed
# - The `print("Exercise is complete")` line will only run if your solution passes the assertion test(s)
# - Be sure to create programmatic solutions that will work for all inputs:
# - For example, calling the `is_even(2)` returns `True`, but your function should work for all even numbers, both positive and negative.
# ## Guidance
# - Get Python to do the work for you. For example, if the exercise instructs you to reverse a list of numbers, your job is to find the
# - Save often by clicking the blue "Save" button.
# - If you need to clear the output or reset the notebook, go to "Run" then "Restart Session" to clear up any error messages.
# - Do not move or alter the lines of code that contain the `assert` statements. Those are what run your solution and test its actual output vs. expected outputs.
# - Seek to understand the problem before trying to solve it. Can you explain the problem to someone else in English? Can you explain the solution in English?
# - Slow down and read any error messages you encounter. Error messages provide insight into how to resolve the error. When in doubt, put your exact error into a search engine and look for results that reference an identical or similar problem.
# ## Get Python To Do The Work For You
# One of the main jobs of a programming language is to help people solve problems programatically, so we don't have to do so much by hand. For example, it's easy for a person to manually reverse the list `[1, 2, 3]`, but imagine reversing a list of a million things or sorting a list of even a hundred things. When we write programmatic solutions in code, we are providing instructions to the computer to do a task. Computers follow the letter of the code, not the intent, and do exactly what they are told to do. In this way, Python can reverse a list of 3 numbers or 100 numbers or ten million numbers with the same instructions. Repetition is a key idea behind programming languages.
# This means that your task with these exercises is to determine a sequence of steps that solve the problem and then find the Python code that will run those instructions. If you're sorting or reversing things by hand, you're not doing it right!
# ## How To Discover How To Do Something in Python
# 1. The first step is to make sure you know what the problem is asking.
# 2. The second step is to determine, in English (or your first spoken language), what steps you need to take.
# 3. Use a search engine to look for code examples to identical or similar problems.
# One of the best ways to discover how to do things in Python is to use a search engine. Go to your favorite search engine and search for "how to reverse a list in Python" or "how to sort a list in Python". That's how both learners and professionals find answers and examples all the time. Search for what you want and add "in Python" and you'll get lots of code examples. Searching for "How to sum a list of numbers in Python" is a very effective way to discover exactly how to do that task.
# ### Learning to Program and Code
# - You can make a new blank cell for Python code at any time in this document.
# - If you want more freedom to explore learning Python in a blank notebook, go here https://www.kaggle.com/notebooks/welcome and make yourself a blank, new notebook.
# - Programming is an intellectual activity of designing a solution. "Coding" means turning your programmatic solution into code w/ all the right syntax and parts of the programming language.
# - Expect to make mistakes and adopt the attitude that **the error message provides the information you need to proceed**. You will put lots of error messages into search engines to learn this craft!
# - Because computers have zero ability to read in between the lines or "catch the drift" or know what you mean, code only does what it is told to do.
# - Code doesn't do what you *want* it to do, code does what you've told it to do.
# - Before writing any code, figure out how you would solve the problem in spoken language to describe the sequence of steps in the solution.
# - Think about your solution in English (or your natural language). It's **critical** to solve the problem in your natural language before trying to get a programming language to do the work.
# ## Troubleshooting
# - If this entire document shows "Name Error" for many cells, it means you should read the "Getting Started" instructions above to make your own copy.
# - Be sure to commit your work to make save points, as you go.
# - If you load this page and you see your code but not the results of the code, be sure to run each cell (shift + Enter makes this quick)
# - "Name Error" means that you need to assign a variable or define the function as instructed.
# - "Assertion Error" means that your provided solution does not match the correct answer.
# - "Type Error" means that your data type provided is not accurate
# - If your kernel freezes, click on "Run" then select "Restart Session"
# - If you require additional troubleshooting assistance, click on "Help" and then "Docs" to access documentation for this platform.
# - If you have discoverd a bug or typo, please triple check your spelling then create a new issue at [https://github.com/ryanorsinger/101-exercises/issues](https://github.com/ryanorsinger/101-exercises/issues) to notify the author.
# Example problem:
# Uncomment the line below and run this cell.
# The hashtag "#" character in a line of Python code is the comment character. To "comment" means to add the # and to "uncomment" means to remove the # character.
doing_python_right_now = True
# The lines below will test your answer. If you see an error, then it means that your answer is incorrect or incomplete.
assert (
doing_python_right_now == True
), "If you see a NameError, it means that the variable is not created and assigned a value. An 'Assertion Error' means that the value of the variable is incorrect."
print(
"Exercise 0 is correct"
) # This line will print if your solution passes the assertion above.
# Exercise 1
# On the line below, create a variable named on_mars_right_now and assign it the boolean value of False
on_mars_right_now = False
assert (
on_mars_right_now == False
), "If you see a Name Error, be sure to create the variable and assign it a value."
print("Exercise 1 is correct.")
# Exercise 2
# Create a variable named fruits and assign it a list of fruits containing the following fruit names as strings:
# mango, banana, guava, kiwi, and strawberry.
fruits = ["mango", "banana", "guava", "kiwi", "strawberry"]
assert fruits == [
"mango",
"banana",
"guava",
"kiwi",
"strawberry",
], "If you see an Assert Error, ensure the variable contains all the strings in the provided order"
print("Exercise 2 is correct.")
# Exercise 3
# Create a variable named vegetables and assign it a list of fruits containing the following vegetable names as strings:
# eggplant, broccoli, carrot, cauliflower, and zucchini
vegetables = ["eggplant", "broccoli", "carrot", "cauliflower", "zucchini"]
assert vegetables == [
"eggplant",
"broccoli",
"carrot",
"cauliflower",
"zucchini",
], "Ensure the variable contains all the strings in the provided order"
print("Exercise 3 is correct.")
# Exercise 4
# Create a variable named numbers and assign it a list of numbers, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10
numbers = [1, 2, 3, 4, 5, 6, 7, 8, 9, 10]
assert numbers == [
1,
2,
3,
4,
5,
6,
7,
8,
9,
10,
], "Ensure the variable contains the numbers 1-10 in order."
print("Exercise 4 is correct.")
# ## List Operations
# **Hint** Recommend finding and using built-in Python functionality whenever possible.
# Exercise 5
# Given the following assigment of the list of fruits, add "tomato" to the end of the list.
fruits = ["mango", "banana", "guava", "kiwi", "strawberry"]
fruits.append("tomato")
assert fruits == [
"mango",
"banana",
"guava",
"kiwi",
"strawberry",
"tomato",
], "Ensure the variable contains all the strings in the right order"
print("Exercise 5 is correct")
# Exercise 6
# Given the following assignment of the vegetables list, add "tomato" to the end of the list.
vegetables = ["eggplant", "broccoli", "carrot", "cauliflower", "zucchini"]
vegetables.append("tomato")
assert vegetables == [
"eggplant",
"broccoli",
"carrot",
"cauliflower",
"zucchini",
"tomato",
], "Ensure the variable contains all the strings in the provided order"
print("Exercise 6 is correct")
# Exercise 7
# Given the list of numbers defined below, reverse the list of numbers that you created above.
numbers = [1, 2, 3, 4, 5, 6, 7, 8, 9, 10]
numbers.reverse()
assert numbers == [
10,
9,
8,
7,
6,
5,
4,
3,
2,
1,
], "Assert Error means that the answer is incorrect."
print("Exercise 7 is correct.")
# Exercise 8
# Sort the vegetables in alphabetical order
vegetables.sort()
assert vegetables == [
"broccoli",
"carrot",
"cauliflower",
"eggplant",
"tomato",
"zucchini",
]
print("Exercise 8 is correct.")
# Exercise 9
# Write the code necessary to sort the fruits in reverse alphabetical order
fruits.sort(reverse=True)
assert fruits == ["tomato", "strawberry", "mango", "kiwi", "guava", "banana"]
print("Exercise 9 is correct.")
# Exercise 10
# Write the code necessary to produce a single list that holds all fruits then all vegetables in the order as they were sorted above.
fruits_and_veggies = fruits + vegetables
assert fruits_and_veggies == [
"tomato",
"strawberry",
"mango",
"kiwi",
"guava",
"banana",
"broccoli",
"carrot",
"cauliflower",
"eggplant",
"tomato",
"zucchini",
]
print("Exercise 10 is correct")
# ## Basic Functions
# **Hint** Be sure to `return` values from your function definitions. The assert statements will call your function(s) for you.
# Run this cell in order to generate some numbers to use in our functions after this.
import random
positive_even_number = random.randrange(2, 101, 2)
negative_even_number = random.randrange(-100, -1, 2)
positive_odd_number = random.randrange(1, 100, 2)
negative_odd_number = random.randrange(-101, 0, 2)
print("We now have some random numbers available for future exercises.")
print("The random positive even number is", positive_even_number)
print("The random positive odd nubmer is", positive_odd_number)
print("The random negative even number", negative_even_number)
print("The random negative odd number", negative_odd_number)
# Example function defintion:
# Write a say_hello function that adds the string "Hello, " to the beginning and "!" to the end of any given input.
def say_hello(name):
return "Hello, " + name + "!"
assert say_hello("Jane") == "Hello, Jane!", "Double check the inputs and data types"
assert say_hello("Pat") == "Hello, Pat!", "Double check the inputs and data types"
assert say_hello("Astrud") == "Hello, Astrud!", "Double check the inputs and data types"
print("The example function definition ran appropriately")
# Another example function definition:
# This plus_two function takes in a variable and adds 2 to it.
def plus_two(number):
return number + 2
assert plus_two(3) == 5
assert plus_two(0) == 2
assert plus_two(-2) == 0
print(
"The plus_two assertions executed appropriately... The second function definition example executed appropriately."
)
# Exercise 11
# Write a function definition for a function named add_one that takes in a number and returns that number plus one.
def add_one(number):
return number + 1
assert (
add_one(2) == 3
), "Ensure that the function is defined, named properly, and returns the correct value"
assert add_one(0) == 1, "Zero plus one is one."
assert (
add_one(positive_even_number) == positive_even_number + 1
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
add_one(negative_odd_number) == negative_odd_number + 1
), "Ensure that the function is defined, named properly, and returns the correct value"
print("Exercise 11 is correct.")
# Exercise 12
# Write a function definition named is_positive that takes in a number and returns True or False if that number is positive.
def is_positive(number):
if number > 0:
return True
return False
assert (
is_positive(positive_odd_number) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_positive(positive_even_number) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_positive(negative_odd_number) == False
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_positive(negative_even_number) == False
), "Ensure that the function is defined, named properly, and returns the correct value"
print("Exercise 12 is correct.")
# Exercise 13
# Write a function definition named is_negative that takes in a number and returns True or False if that number is negative.
def is_negative(number):
if number < 0:
return True
return False
assert (
is_negative(positive_odd_number) == False
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_negative(positive_even_number) == False
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_negative(negative_odd_number) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_negative(negative_even_number) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
print("Exercise 13 is correct.")
# Exercise 14
# Write a function definition named is_odd that takes in a number and returns True or False if that number is odd.
def is_odd(number):
if number % 2 == 1:
return True
return False
assert (
is_odd(positive_odd_number) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_odd(positive_even_number) == False
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_odd(negative_odd_number) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_odd(negative_even_number) == False
), "Ensure that the function is defined, named properly, and returns the correct value"
print("Exercise 14 is correct.")
# Exercise 15
# Write a function definition named is_even that takes in a number and returns True or False if that number is even.
def is_even(number):
if number % 2 == 0:
return True
return False
assert (
is_even(2) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_even(positive_odd_number) == False
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_even(positive_even_number) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_even(negative_odd_number) == False
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_even(negative_even_number) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
print("Exercise 15 is correct.")
# Exercise 16
# Write a function definition named identity that takes in any argument and returns that argument's value. Don't overthink this one!
def identity(x):
return x
assert (
identity(fruits) == fruits
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
identity(vegetables) == vegetables
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
identity(positive_odd_number) == positive_odd_number
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
identity(positive_even_number) == positive_even_number
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
identity(negative_odd_number) == negative_odd_number
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
identity(negative_even_number) == negative_even_number
), "Ensure that the function is defined, named properly, and returns the correct value"
print("Exercise 16 is correct.")
# Exercise 17
# Write a function definition named is_positive_odd that takes in a number and returns True or False if the value is both greater than zero and odd
def is_positive_odd(number):
if number > 0 and number % 2 == 1:
return True
return False
assert is_positive_odd(3) == True, "Double check your syntax and logic"
assert (
is_positive_odd(positive_odd_number) == True
), "Double check your syntax and logic"
assert (
is_positive_odd(positive_even_number) == False
), "Double check your syntax and logic"
assert (
is_positive_odd(negative_odd_number) == False
), "Double check your syntax and logic"
assert (
is_positive_odd(negative_even_number) == False
), "Double check your syntax and logic"
print("Exercise 17 is correct.")
# Exercise 18
# Write a function definition named is_positive_even that takes in a number and returns True or False if the value is both greater than zero and even
def is_positive_even(number):
if number > 0 and number % 2 == 0:
return True
return False
assert is_positive_even(4) == True, "Double check your syntax and logic"
assert (
is_positive_even(positive_odd_number) == False
), "Double check your syntax and logic"
assert (
is_positive_even(positive_even_number) == True
), "Double check your syntax and logic"
assert (
is_positive_even(negative_odd_number) == False
), "Double check your syntax and logic"
assert (
is_positive_even(negative_even_number) == False
), "Double check your syntax and logic"
print("Exercise 18 is correct.")
# Exercise 19
# Write a function definition named is_negative_odd that takes in a number and returns True or False if the value is both less than zero and odd.
def is_negative_odd(number):
if number < 0 and number % 2 == 1:
return True
return False
assert is_negative_odd(-3) == True, "Double check your syntax and logic"
assert (
is_negative_odd(positive_odd_number) == False
), "Double check your syntax and logic"
assert (
is_negative_odd(positive_even_number) == False
), "Double check your syntax and logic"
assert (
is_negative_odd(negative_odd_number) == True
), "Double check your syntax and logic"
assert (
is_negative_odd(negative_even_number) == False
), "Double check your syntax and logic"
print("Exercise 19 is correct.")
# Exercise 20
# Write a function definition named is_negative_even that takes in a number and returns True or False if the value is both less than zero and even.
def is_negative_even(number):
if number < 0 and number % 2 == 0:
return True
return False
assert is_negative_even(-4) == True, "Double check your syntax and logic"
assert (
is_negative_even(positive_odd_number) == False
), "Double check your syntax and logic"
assert (
is_negative_even(positive_even_number) == False
), "Double check your syntax and logic"
assert (
is_negative_even(negative_odd_number) == False
), "Double check your syntax and logic"
assert (
is_negative_even(negative_even_number) == True
), "Double check your syntax and logic"
print("Exercise 20 is correct.")
# Exercise 21
# Write a function definition named half that takes in a number and returns half the provided number.
def half(number):
return number / 2
assert half(4) == 2
assert half(5) == 2.5
assert half(positive_odd_number) == positive_odd_number / 2
assert half(positive_even_number) == positive_even_number / 2
assert half(negative_odd_number) == negative_odd_number / 2
assert half(negative_even_number) == negative_even_number / 2
print("Exercise 21 is correct.")
# Exercise 22
# Write a function definition named double that takes in a number and returns double the provided number.
def double(number):
return number * 2
assert double(4) == 8
assert double(5) == 10
assert double(positive_odd_number) == positive_odd_number * 2
assert double(positive_even_number) == positive_even_number * 2
assert double(negative_odd_number) == negative_odd_number * 2
assert double(negative_even_number) == negative_even_number * 2
print("Exercise 22 is correct.")
# Exercise 23
# Write a function definition named triple that takes in a number and returns triple the provided number.
def triple(number):
return number * 3
assert triple(4) == 12
assert triple(5) == 15
assert triple(positive_odd_number) == positive_odd_number * 3
assert triple(positive_even_number) == positive_even_number * 3
assert triple(negative_odd_number) == negative_odd_number * 3
assert triple(negative_even_number) == negative_even_number * 3
print("Exercise 23 is correct.")
# Exercise 24
# Write a function definition named reverse_sign that takes in a number and returns the provided number but with the sign reversed.
def reverse_sign(number):
if number > 0:
return number * -1
return abs(number)
assert reverse_sign(4) == -4
assert reverse_sign(-5) == 5
assert reverse_sign(positive_odd_number) == positive_odd_number * -1
assert reverse_sign(positive_even_number) == positive_even_number * -1
assert reverse_sign(negative_odd_number) == negative_odd_number * -1
assert reverse_sign(negative_even_number) == negative_even_number * -1
print("Exercise 24 is correct.")
# Exercise 25
# Write a function definition named absolute_value that takes in a number and returns the absolute value of the provided number
def absolute_value(number):
return abs(number)
assert absolute_value(4) == 4
assert absolute_value(-5) == 5
assert absolute_value(positive_odd_number) == positive_odd_number
assert absolute_value(positive_even_number) == positive_even_number
assert absolute_value(negative_odd_number) == negative_odd_number * -1
assert absolute_value(negative_even_number) == negative_even_number * -1
print("Exercise 25 is correct.")
# Exercise 26
# Write a function definition named is_multiple_of_three that takes in a number and returns True or False if the number is evenly divisible by 3.
def is_multiple_of_three(number):
if number % 3 == 0:
return True
return False
assert is_multiple_of_three(3) == True
assert is_multiple_of_three(15) == True
assert is_multiple_of_three(9) == True
assert is_multiple_of_three(4) == False
assert is_multiple_of_three(10) == False
print("Exercise 26 is correct.")
# Exercise 27
# Write a function definition named is_multiple_of_five that takes in a number and returns True or False if the number is evenly divisible by 5.
def is_multiple_of_five(number):
if number % 5 == 0:
return True
return False
assert is_multiple_of_five(3) == False
assert is_multiple_of_five(15) == True
assert is_multiple_of_five(9) == False
assert is_multiple_of_five(4) == False
assert is_multiple_of_five(10) == True
print("Exercise 27 is correct.")
# Exercise 28
# Write a function definition named is_multiple_of_both_three_and_five that takes in a number and returns True or False if the number is evenly divisible by both 3 and 5.
def is_multiple_of_both_three_and_five(number):
if number % 3 == 0 and number % 5 == 0:
return True
return False
assert is_multiple_of_both_three_and_five(15) == True
assert is_multiple_of_both_three_and_five(45) == True
assert is_multiple_of_both_three_and_five(3) == False
assert is_multiple_of_both_three_and_five(9) == False
assert is_multiple_of_both_three_and_five(4) == False
print("Exercise 28 is correct.")
# Exercise 29
# Write a function definition named square that takes in a number and returns the number times itself.
def square(number):
return number * number
assert square(3) == 9
assert square(2) == 4
assert square(9) == 81
assert square(positive_odd_number) == positive_odd_number * positive_odd_number
print("Exercise 29 is correct.")
# Exercise 30
# Write a function definition named add that takes in two numbers and returns the sum.
def add(num_1, num_2):
return num_1 + num_2
assert add(3, 2) == 5
assert add(10, -2) == 8
assert add(5, 7) == 12
print("Exercise 30 is correct.")
# Exercise 31
# Write a function definition named cube that takes in a number and returns the number times itself, times itself.
def cube(number):
return number * number * number
assert cube(3) == 27
assert cube(2) == 8
assert cube(5) == 125
assert (
cube(positive_odd_number)
== positive_odd_number * positive_odd_number * positive_odd_number
)
print("Exercise 31 is correct.")
# Exercise 32
# Write a function definition named square_root that takes in a number and returns the square root of the provided number
def square_root(number):
return number**0.5
assert square_root(4) == 2.0
assert square_root(64) == 8.0
assert square_root(81) == 9.0
print("Exercise 32 is correct.")
# Exercise 33
# Write a function definition named subtract that takes in two numbers and returns the first minus the second argument.
def subtract(num_1, num_2):
return num_1 - num_2
assert subtract(8, 6) == 2
assert subtract(27, 4) == 23
assert subtract(12, 2) == 10
print("Exercise 33 is correct.")
# Exercise 34
# Write a function definition named multiply that takes in two numbers and returns the first times the second argument.
def multiply(num_1, num_2):
return num_1 * num_2
assert multiply(2, 1) == 2
assert multiply(3, 5) == 15
assert multiply(5, 2) == 10
print("Exercise 34 is correct.")
# Exercise 35
# Write a function definition named divide that takes in two numbers and returns the first argument divided by the second argument.
def divide(num_1, num_2):
return num_1 / num_2
assert divide(27, 9) == 3
assert divide(15, 3) == 5
assert divide(5, 2) == 2.5
assert divide(10, 2) == 5
print("Exercise 35 is correct.")
# Exercise 36
# Write a function definition named quotient that takes in two numbers and returns only the quotient from dividing the first argument by the second argument.
def quotient(num_1, num_2):
return num_1 // num_2
assert quotient(27, 9) == 3
assert quotient(5, 2) == 2
assert quotient(10, 3) == 3
print("Exercise 36 is correct.")
# Exercise 37
# Write a function definition named remainder that takes in two numbers and returns the remainder of first argument divided by the second argument.
def remainder(num_1, num_2):
return num_1 % num_2
assert remainder(3, 3) == 0
assert remainder(5, 2) == 1
assert remainder(7, 5) == 2
print("Exercise 37 is correct.")
# Exercise 38
# Write a function definition named sum_of_squares that takes in two numbers, squares each number, then returns the sum of both squares.
def sum_of_squares(num_1, num_2):
return num_1**2 + num_2**2
assert sum_of_squares(3, 2) == 13
assert sum_of_squares(5, 2) == 29
assert sum_of_squares(2, 4) == 20
print("Exercise 38 is correct.")
# Exercise 39
# Write a function definition named times_two_plus_three that takes in a number, multiplies it by two, adds 3 and returns the result.
def times_two_plus_three(number):
return number * 2 + 3
assert times_two_plus_three(0) == 3
assert times_two_plus_three(1) == 5
assert times_two_plus_three(2) == 7
assert times_two_plus_three(3) == 9
assert times_two_plus_three(5) == 13
print("Exercise 39 is correct.")
# Exercise 40
# Write a function definition named area_of_rectangle that takes in two numbers and returns the product.
def area_of_rectangle(num_1, num_2):
return num_1 * num_2
assert area_of_rectangle(1, 3) == 3
assert area_of_rectangle(5, 2) == 10
assert area_of_rectangle(2, 7) == 14
assert area_of_rectangle(5.3, 10.3) == 54.59
print("Exercise 40 is correct.")
import math
# Exercise 41
# Write a function definition named area_of_circle that takes in a number representing a circle's radius and returns the area of the circl
def area_of_circle(number):
return math.pi * number**2
assert area_of_circle(3) == 28.274333882308138
assert area_of_circle(5) == 78.53981633974483
assert area_of_circle(7) == 153.93804002589985
print("Exercise 41 is correct.")
import math
# Exercise 42
# Write a function definition named circumference that takes in a number representing a circle's radius and returns the circumference.
def circumference(number):
return 2 * math.pi * number
assert circumference(3) == 18.84955592153876
assert circumference(5) == 31.41592653589793
assert circumference(7) == 43.982297150257104
print("Exercise 42 is correct.")
# ## Functions working with strings
# Exercise 43
# Write a function definition named is_vowel that takes in value and returns True if the value is a, e, i, o, u in upper or lower case.
def is_vowel(letter):
if (
letter == "a"
or letter == "e"
or letter == "i"
or letter == "o"
or letter == "u"
or letter == "A"
or letter == "E"
or letter == "I"
or letter == "O"
or letter == "U"
):
return True
return False
assert is_vowel("a") == True
assert is_vowel("U") == True
assert is_vowel("banana") == False
assert is_vowel("Q") == False
assert is_vowel("y") == False
print("Exercise 43 is correct.")
# Exercise 44
# Write a function definition named has_vowels that takes in value and returns True if the string contains any vowels.
def has_vowels(string_input):
for letter in string_input:
if is_vowel(letter):
return True
return False
assert has_vowels("banana") == True
assert has_vowels("ubuntu") == True
assert has_vowels("QQQQ") == False
assert has_vowels("wyrd") == False
print("Exercise 44 is correct.")
# Exercise 45
# Write a function definition named count_vowels that takes in value and returns the count of the nubmer of vowels in a sequence.
def count_vowels(string_input):
counter = 0
for char in string_input:
if is_vowel(char):
counter += 1
return counter
assert count_vowels("banana") == 3
assert count_vowels("ubuntu") == 3
assert count_vowels("mango") == 2
assert count_vowels("QQQQ") == 0
assert count_vowels("wyrd") == 0
print("Exercise 45 is correct.")
# Exercise 46
# Write a function definition named remove_vowels that takes in string and returns the string without any vowels
def remove_vowels(string_input):
no_vowels_string = ""
for char in string_input:
if is_vowel(char) == False:
no_vowels_string = no_vowels_string + char
return no_vowels_string
assert remove_vowels("banana") == "bnn"
assert remove_vowels("ubuntu") == "bnt"
assert remove_vowels("mango") == "mng"
assert remove_vowels("QQQQ") == "QQQQ"
print("Exercise 46 is correct.")
# Exercise 47
# Write a function definition named starts_with_vowel that takes in string and True if the string starts with a vowel
def starts_with_vowel(string_input):
return is_vowel(string_input[0])
assert starts_with_vowel("ubuntu") == True
assert starts_with_vowel("banana") == False
assert starts_with_vowel("mango") == False
print("Exercise 47 is correct.")
# Exercise 48
# Write a function definition named ends_with_vowel that takes in string and True if the string ends with a vowel
def ends_with_vowel(string_input):
return is_vowel(string_input[-1])
assert ends_with_vowel("ubuntu") == True
assert ends_with_vowel("banana") == True
assert ends_with_vowel("mango") == True
assert ends_with_vowel("spinach") == False
print("Exercise 48 is correct.")
# Exercise 49
# Write a function definition named starts_and_ends_with_vowel that takes in string and returns True if the string starts and ends with a vowel
def starts_and_ends_with_vowel(string_input):
return starts_with_vowel(string_input) and ends_with_vowel(string_input)
assert starts_and_ends_with_vowel("ubuntu") == True
assert starts_and_ends_with_vowel("banana") == False
assert starts_and_ends_with_vowel("mango") == False
print("Exercise 49 is correct.")
# ## Accessing List Elements
# Exercise 50
# Write a function definition named first that takes in sequence and returns the first value of that sequence.
def first(input_sequence):
return input_sequence[0]
assert first("ubuntu") == "u"
assert first([1, 2, 3]) == 1
assert first(["python", "is", "awesome"]) == "python"
print("Exercise 50 is correct.")
# Exercise 51
# Write a function definition named second that takes in sequence and returns the second value of that sequence.
def second(input_sequence):
return input_sequence[1]
assert second("ubuntu") == "b"
assert second([1, 2, 3]) == 2
assert second(["python", "is", "awesome"]) == "is"
print("Exercise 51 is correct.")
# Exercise 52
# Write a function definition named third that takes in sequence and returns the third value of that sequence.
def third(input_sequence):
return input_sequence[2]
assert third("ubuntu") == "u"
assert third([1, 2, 3]) == 3
assert third(["python", "is", "awesome"]) == "awesome"
print("Exercise 52 is correct.")
# Exercise 53
# Write a function definition named forth that takes in sequence and returns the forth value of that sequence.
def forth(input_sequence):
return input_sequence[3]
assert forth("ubuntu") == "n"
assert forth([1, 2, 3, 4]) == 4
assert forth(["python", "is", "awesome", "right?"]) == "right?"
print("Exercise 53 is correct.")
# Exercise 54
# Write a function definition named last that takes in sequence and returns the last value of that sequence.
def last(input_sequence):
return input_sequence[-1]
assert last("ubuntu") == "u"
assert last([1, 2, 3, 4]) == 4
assert last(["python", "is", "awesome"]) == "awesome"
assert last(["kiwi", "mango", "guava"]) == "guava"
print("Exercise 54 is correct.")
# Exercise 55
# Write a function definition named second_to_last that takes in sequence and returns the second to last value of that sequence.
def second_to_last(input_string):
return input_string[-2]
assert second_to_last("ubuntu") == "t"
assert second_to_last([1, 2, 3, 4]) == 3
assert second_to_last(["python", "is", "awesome"]) == "is"
assert second_to_last(["kiwi", "mango", "guava"]) == "mango"
print("Exercise 55 is correct.")
# Exercise 56
# Write a function definition named third_to_last that takes in sequence and returns the third to last value of that sequence.
def third_to_last(input_string):
return input_string[-3]
assert third_to_last("ubuntu") == "n"
assert third_to_last([1, 2, 3, 4]) == 2
assert third_to_last(["python", "is", "awesome"]) == "python"
assert third_to_last(["strawberry", "kiwi", "mango", "guava"]) == "kiwi"
print("Exercise 56 is correct.")
# Exercise 57
# Write a function definition named first_and_second that takes in sequence and returns the first and second value of that sequence as a list
def first_and_second(input_sequence):
return [first(input_sequence), second(input_sequence)]
assert first_and_second([1, 2, 3, 4]) == [1, 2]
assert first_and_second(["python", "is", "awesome"]) == ["python", "is"]
assert first_and_second(["strawberry", "kiwi", "mango", "guava"]) == [
"strawberry",
"kiwi",
]
print("Exercise 57 is correct.")
# Exercise 58
# Write a function definition named first_and_last that takes in sequence and returns the first and last value of that sequence as a list
def first_and_last(input_sequence):
return [first(input_sequence), last(input_sequence)]
assert first_and_last([1, 2, 3, 4]) == [1, 4]
assert first_and_last(["python", "is", "awesome"]) == ["python", "awesome"]
assert first_and_last(["strawberry", "kiwi", "mango", "guava"]) == [
"strawberry",
"guava",
]
print("Exercise 58 is correct.")
# Exercise 59
# Write a function definition named first_to_last that takes in sequence and returns the sequence with the first value moved to the end of the sequence.
assert first_to_last([1, 2, 3, 4]) == [2, 3, 4, 1]
assert first_to_last(["python", "is", "awesome"]) == ["is", "awesome", "python"]
assert first_to_last(["strawberry", "kiwi", "mango", "guava"]) == [
"kiwi",
"mango",
"guava",
"strawberry",
]
print("Exercise 59 is correct.")
# ## Functions to describe data
# Exercise 60
# Write a function definition named sum_all that takes in sequence of numbers and returns all the numbers added together.
assert sum_all([1, 2, 3, 4]) == 10
assert sum_all([3, 3, 3]) == 9
assert sum_all([0, 5, 6]) == 11
print("Exercise 60 is correct.")
# Exercise 61
# Write a function definition named mean that takes in sequence of numbers and returns the average value
assert mean([1, 2, 3, 4]) == 2.5
assert mean([3, 3, 3]) == 3
assert mean([1, 5, 6]) == 4
print("Exercise 61 is correct.")
# Exercise 62
# Write a function definition named median that takes in sequence of numbers and returns the average value
assert median([1, 2, 3, 4, 5]) == 3.0
assert median([1, 2, 3]) == 2.0
assert median([1, 5, 6]) == 5.0
assert median([1, 2, 5, 6]) == 3.5
print("Exercise 62 is correct.")
# Exercise 63
# Write a function definition named mode that takes in sequence of numbers and returns the most commonly occuring value
assert mode([1, 2, 2, 3, 4]) == 2
assert mode([1, 1, 2, 3]) == 1
assert mode([2, 2, 3, 3, 3]) == 3
print("Exercise 63 is correct.")
# Exercise 64
# Write a function definition named product_of_all that takes in sequence of numbers and returns the product of multiplying all the numbers together
assert product_of_all([1, 2, 3]) == 6
assert product_of_all([3, 4, 5]) == 60
assert product_of_all([2, 2, 3, 0]) == 0
print("Exercise 64 is correct.")
# ## Applying functions to lists
# Run this cell in order to use the following list of numbers for the next exercises
numbers = [-5, -4, -3, -2, -1, 1, 2, 3, 4, 5]
# Exercise 65
# Write a function definition named get_highest_number that takes in sequence of numbers and returns the largest number.
assert get_highest_number([1, 2, 3]) == 3
assert get_highest_number([-5, -4, -3, -2, -1, 1, 2, 3, 4, 5]) == 5
assert get_highest_number([-5, -3, 1]) == 1
print("Exercise 65 is correct.")
# Exercise 66
# Write a function definition named get_smallest_number that takes in sequence of numbers and returns the smallest number.
assert get_smallest_number([1, 2, 3]) == 1
assert get_smallest_number([-5, -4, -3, -2, -1, 1, 2, 3, 4, 5]) == -5
assert get_smallest_number([-4, -3, 1]) == -4
print("Exercise 66 is correct.")
# Exercise 67
# Write a function definition named only_odd_numbers that takes in sequence of numbers and returns the odd numbers in a list.
assert only_odd_numbers([1, 2, 3]) == [1, 3]
assert only_odd_numbers([-5, -4, -3, -2, -1, 1, 2, 3, 4, 5]) == [-5, -3, -1, 1, 3, 5]
assert only_odd_numbers([-4, -3, 1]) == [-3, 1]
print("Exercise 67 is correct.")
# Exercise 68
# Write a function definition named only_even_numbers that takes in sequence of numbers and returns the even numbers in a list.
assert only_even_numbers([1, 2, 3]) == [2]
assert only_even_numbers([-5, -4, -3, -2, -1, 1, 2, 3, 4, 5]) == [-4, -2, 2, 4]
assert only_even_numbers([-4, -3, 1]) == [-4]
print("Exercise 68 is correct.")
# Exercise 69
# Write a function definition named only_positive_numbers that takes in sequence of numbers and returns the positive numbers in a list.
assert only_positive_numbers([1, 2, 3]) == [1, 2, 3]
assert only_positive_numbers([-5, -4, -3, -2, -1, 1, 2, 3, 4, 5]) == [1, 2, 3, 4, 5]
assert only_positive_numbers([-4, -3, 1]) == [1]
print("Exercise 69 is correct.")
# Exercise 70
# Write a function definition named only_negative_numbers that takes in sequence of numbers and returns the negative numbers in a list.
assert only_negative_numbers([1, 2, 3]) == []
assert only_negative_numbers([-5, -4, -3, -2, -1, 1, 2, 3, 4, 5]) == [
-5,
-4,
-3,
-2,
-1,
]
assert only_negative_numbers([-4, -3, 1]) == [-4, -3]
print("Exercise 70 is correct.")
# Exercise 71
# Write a function definition named has_evens that takes in sequence of numbers and returns True if there are any even numbers in the sequence
assert has_evens([1, 2, 3]) == True
assert has_evens([2, 5, 6]) == True
assert has_evens([3, 3, 3]) == False
assert has_evens([]) == False
print("Exercise 71 is correct.")
# Exercise 72
# Write a function definition named count_evens that takes in sequence of numbers and returns the number of even numbers
assert count_evens([1, 2, 3]) == 1
assert count_evens([2, 5, 6]) == 2
assert count_evens([3, 3, 3]) == 0
assert count_evens([5, 6, 7, 8]) == 2
print("Exercise 72 is correct.")
# Exercise 73
# Write a function definition named has_odds that takes in sequence of numbers and returns True if there are any odd numbers in the sequence
assert has_odds([1, 2, 3]) == True
assert has_odds([2, 5, 6]) == True
assert has_odds([3, 3, 3]) == True
assert has_odds([2, 4, 6]) == False
print("Exercise 73 is correct.")
# Exercise 74
# Write a function definition named count_odds that takes in sequence of numbers and returns True if there are any odd numbers in the sequence
assert count_odds([1, 2, 3]) == 2
assert count_odds([2, 5, 6]) == 1
assert count_odds([3, 3, 3]) == 3
assert count_odds([2, 4, 6]) == 0
print("Exercise 74 is correct.")
# Exercise 75
# Write a function definition named count_negatives that takes in sequence of numbers and returns a count of the number of negative numbers
assert count_negatives([1, -2, 3]) == 1
assert count_negatives([2, -5, -6]) == 2
assert count_negatives([3, 3, 3]) == 0
print("Exercise 75 is correct.")
# Exercise 76
# Write a function definition named count_positives that takes in sequence of numbers and returns a count of the number of positive numbers
assert count_positives([1, -2, 3]) == 2
assert count_positives([2, -5, -6]) == 1
assert count_positives([3, 3, 3]) == 3
assert count_positives([-2, -1, -5]) == 0
print("Exercise 76 is correct.")
# Exercise 77
# Write a function definition named only_positive_evens that takes in sequence of numbers and returns a list containing all the positive evens from the sequence
assert only_positive_evens([1, -2, 3]) == []
assert only_positive_evens([2, -5, -6]) == [2]
assert only_positive_evens([3, 3, 4, 6]) == [4, 6]
assert only_positive_evens([2, 3, 4, -1, -5]) == [2, 4]
print("Exercise 77 is correct.")
# Exercise 78
# Write a function definition named only_positive_odds that takes in sequence of numbers and returns a list containing all the positive odd numbers from the sequence
assert only_positive_odds([1, -2, 3]) == [1, 3]
assert only_positive_odds([2, -5, -6]) == []
assert only_positive_odds([3, 3, 4, 6]) == [3, 3]
assert only_positive_odds([2, 3, 4, -1, -5]) == [3]
print("Exercise 78 is correct.")
# Exercise 79
# Write a function definition named only_negative_evens that takes in sequence of numbers and returns a list containing all the negative even numbers from the sequence
assert only_negative_evens([1, -2, 3]) == [-2]
assert only_negative_evens([2, -5, -6]) == [-6]
assert only_negative_evens([3, 3, 4, 6]) == []
assert only_negative_evens([-2, 3, 4, -1, -4]) == [-2, -4]
print("Exercise 79 is correct.")
# Exercise 80
# Write a function definition named only_negative_odds that takes in sequence of numbers and returns a list containing all the negative odd numbers from the sequence
assert only_negative_odds([1, -2, 3]) == []
assert only_negative_odds([2, -5, -6]) == [-5]
assert only_negative_odds([3, 3, 4, 6]) == []
assert only_negative_odds([2, -3, 4, -1, -4]) == [-3, -1]
print("Exercise 80 is correct.")
# Exercise 81
# Write a function definition named shortest_string that takes in a list of strings and returns the shortest string in the list.
assert shortest_string(["kiwi", "mango", "strawberry"]) == "kiwi"
assert shortest_string(["hello", "everybody"]) == "hello"
assert shortest_string(["mary", "had", "a", "little", "lamb"]) == "a"
print("Exercise 81 is correct.")
# Exercise 82
# Write a function definition named longest_string that takes in sequence of strings and returns the longest string in the list.
assert longest_string(["kiwi", "mango", "strawberry"]) == "strawberry"
assert longest_string(["hello", "everybody"]) == "everybody"
assert longest_string(["mary", "had", "a", "little", "lamb"]) == "little"
print("Exercise 82 is correct.")
# ## Working with sets
# **Hint** Take a look at the `set` function in Python, the `set` data type, and built-in `set` methods.
# Example set function usage
print(set("kiwi"))
print(set([1, 2, 2, 3, 3, 3, 4, 4, 4, 4]))
# Exercise 83
# Write a function definition named get_unique_values that takes in a list and returns a set with only the unique values from that list.
assert get_unique_values(["ant", "ant", "mosquito", "mosquito", "ladybug"]) == {
"ant",
"mosquito",
"ladybug",
}
assert get_unique_values(["b", "a", "n", "a", "n", "a", "s"]) == {"b", "a", "n", "s"}
assert get_unique_values(
["mary", "had", "a", "little", "lamb", "little", "lamb", "little", "lamb"]
) == {"mary", "had", "a", "little", "lamb"}
print("Exercise 83 is correct.")
# Exercise 84
# Write a function definition named get_unique_values_from_two_lists that takes two lists and returns a single set with only the unique values
assert get_unique_values_from_two_lists([5, 1, 2, 3], [3, 4, 5, 5]) == {1, 2, 3, 4, 5}
assert get_unique_values_from_two_lists([1, 1], [2, 2, 3]) == {1, 2, 3}
assert get_unique_values_from_two_lists(
["tomato", "mango", "kiwi"], ["eggplant", "tomato", "broccoli"]
) == {"tomato", "mango", "kiwi", "eggplant", "broccoli"}
print("Exercise 84 is correct.")
# Exercise 85
# Write a function definition named get_values_in_common that takes two lists and returns a single set with the values that each list has in common
assert get_values_in_common([5, 1, 2, 3], [3, 4, 5, 5]) == {3, 5}
assert get_values_in_common([1, 2], [2, 2, 3]) == {2}
assert get_values_in_common(
["tomato", "mango", "kiwi"], ["eggplant", "tomato", "broccoli"]
) == {"tomato"}
print("Exercise 85 is correct.")
# Exercise 86
# Write a function definition named get_values_not_in_common that takes two lists and returns a single set with the values that each list does not have in common
assert get_values_not_in_common([5, 1, 2, 3], [3, 4, 5, 5]) == {1, 2, 4}
assert get_values_not_in_common([1, 1], [2, 2, 3]) == {1, 2, 3}
assert get_values_not_in_common(
["tomato", "mango", "kiwi"], ["eggplant", "tomato", "broccoli"]
) == {"mango", "kiwi", "eggplant", "broccoli"}
print("Exercise 86 is correct.")
# ## Working with Dictionaries
#
# Run this cell in order to have these two dictionary variables defined.
tukey_paper = {
"title": "The Future of Data Analysis",
"author": "John W. Tukey",
"link": "https://projecteuclid.org/euclid.aoms/1177704711",
"year_published": 1962,
}
thomas_paper = {
"title": "A mathematical model of glutathione metabolism",
"author": "Rachel Thomas",
"link": "https://www.ncbi.nlm.nih.gov/pubmed/18442411",
"year_published": 2008,
}
# Exercise 87
# Write a function named get_paper_title that takes in a dictionary and returns the title property
assert get_paper_title(tukey_paper) == "The Future of Data Analysis"
assert get_paper_title(thomas_paper) == "A mathematical model of glutathione metabolism"
print("Exercise 87 is correct.")
# Exercise 88
# Write a function named get_year_published that takes in a dictionary and returns the value behind the "year_published" key.
assert get_year_published(tukey_paper) == 1962
assert get_year_published(thomas_paper) == 2008
print("Exercise 88 is correct.")
# Run this code to create data for the next two questions
book = {
"title": "Genetic Algorithms and Machine Learning for Programmers",
"price": 36.99,
"author": "Frances Buontempo",
}
# Exercise 89
# Write a function named get_price that takes in a dictionary and returns the price
assert get_price(book) == 36.99
print("Exercise 89 is complete.")
# Exercise 90
# Write a function named get_book_author that takes in a dictionary (the above declared book variable) and returns the author's name
assert get_book_author(book) == "Frances Buontempo"
print("Exercise 90 is complete.")
# ## Working with Lists of Dictionaries
# **Hint** If you need an example of lists of dictionaries, see [https://gist.github.com/ryanorsinger/fce8154028a924c1073eac24c7c3f409](https://gist.github.com/ryanorsinger/fce8154028a924c1073eac24c7c3f409)
# Run this cell in order to have some setup data for the next exercises
books = [
{
"title": "Genetic Algorithms and Machine Learning for Programmers",
"price": 36.99,
"author": "Frances Buontempo",
},
{
"title": "The Visual Display of Quantitative Information",
"price": 38.00,
"author": "Edward Tufte",
},
{
"title": "Practical Object-Oriented Design",
"author": "Sandi Metz",
"price": 30.47,
},
{"title": "Weapons of Math Destruction", "author": "Cathy O'Neil", "price": 17.44},
]
# Exercise 91
# Write a function named get_number_of_books that takes in a list of objects and returns the number of dictionaries in that list.
assert get_number_of_books(books) == 4
print("Exercise 91 is complete.")
# Exercise 92
# Write a function named total_of_book_prices that takes in a list of dictionaries and returns the sum total of all the book prices added together
assert total_of_book_prices(books) == 122.9
print("Exercise 92 is complete.")
# Exercise 93
# Write a function named get_average_book_price that takes in a list of dictionaries and returns the average book price.
assert get_average_book_price(books) == 30.725
print("Exercise 93 is complete.")
# Exercise 94
# Write a function called highest_price_book that takes in the above defined list of dictionaries "books" and returns the dictionary containing the title, price, and author of the book with the highest priced book.
# Hint: Much like sometimes start functions with a variable set to zero, you may want to create a dictionary with the price set to zero to compare to each dictionary's price in the list
assert highest_price_book(books) == {
"title": "The Visual Display of Quantitative Information",
"price": 38.00,
"author": "Edward Tufte",
}
print("Exercise 94 is complete")
# Exercise 95
# Write a function called lowest_priced_book that takes in the above defined list of dictionaries "books" and returns the dictionary containing the title, price, and author of the book with the lowest priced book.
# Hint: Much like sometimes start functions with a variable set to zero or float('inf'), you may want to create a dictionary with the price set to float('inf') to compare to each dictionary in the list
assert lowest_price_book(books) == {
"title": "Weapons of Math Destruction",
"author": "Cathy O'Neil",
"price": 17.44,
}
print("Exercise 95 is complete.")
shopping_cart = {
"tax": 0.08,
"items": [
{"title": "orange juice", "price": 3.99, "quantity": 1},
{"title": "rice", "price": 1.99, "quantity": 3},
{"title": "beans", "price": 0.99, "quantity": 3},
{"title": "chili sauce", "price": 2.99, "quantity": 1},
{"title": "chocolate", "price": 0.75, "quantity": 9},
],
}
# Exercise 96
# Write a function named get_tax_rate that takes in the above shopping cart as input and returns the tax rate.
# Hint: How do you access a key's value on a dictionary? The tax rate is one key of the entire shopping_cart dictionary.
assert get_tax_rate(shopping_cart) == 0.08
print("Exercise 96 is complete")
# Exercise 97
# Write a function named number_of_item_types that takes in the shopping cart as input and returns the number of unique item types in the shopping cart.
# We're not yet using the quantity of each item, but rather focusing on determining how many different types of items are in the cart.
assert number_of_item_types(shopping_cart) == 5
print("Exercise 97 is complete.")
# Exercise 98
# Write a function named total_number_of_items that takes in the shopping cart as input and returns the total number all item quantities.
# This should return the sum of all of the quantities from each item type
assert total_number_of_items(shopping_cart) == 17
print("Exercise 98 is complete.")
# Exercise 99
# Write a function named get_average_item_price that takes in the shopping cart as an input and returns the average of all the item prices.
# Hint - This should determine the total price divided by the number of types of items. This does not account for each item type's quantity.
assert get_average_item_price(shopping_cart) == 2.1420000000000003
print("Exercise 99 is complete.")
# Exercise 100
# Write a function named get_average_spent_per_item that takes in the shopping cart and returns the average of summing each item's quanties times that item's price.
# Hint: You may need to set an initial total price and total total quantity to zero, then sum up and divide that total price by the total quantity
assert get_average_spent_per_item(shopping_cart) == 1.333529411764706
print("Exercise 100 is complete.")
# Exercise 101
# Write a function named most_spent_on_item that takes in the shopping cart as input and returns the dictionary associated with the item that has the highest price*quantity.
# Be sure to do this as programmatically as possible.
# Hint: Similarly to how we sometimes begin a function with setting a variable to zero, we need a starting place:
# Hint: Consider creating a variable that is a dictionary with the keys "price" and "quantity" both set to 0. You can then compare each item's price and quantity total to the one from "most"
assert most_spent_on_item(shopping_cart) == {
"title": "chocolate",
"price": 0.75,
"quantity": 9,
}
print("Exercise 101 is complete.")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/401/69401978.ipynb
| null | null |
[{"Id": 69401978, "ScriptId": 18933256, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8012467, "CreationDate": "07/30/2021 14:35:20", "VersionNumber": 3.0, "Title": "101-exercises", "EvaluationDate": "07/30/2021", "IsChange": true, "TotalLines": 1293.0, "LinesInsertedFromPrevious": 56.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 1237.0, "LinesInsertedFromFork": 201.0, "LinesDeletedFromFork": 2.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 1092.0, "TotalVotes": 0}]
| null | null | null | null |
# # Welcome to 101 Exercises for Python Fundamentals
# Solving these exercises will help make you a better programmer. Solve them in order, because each solution builds scaffolding, working code, and knowledge you can use on future problems. Read the directions carefully, and have fun!
# > "Learning to program takes a little bit of study and a *lot* of practice" - Luis Montealegre
# ## Getting Started
# 0. Create your own account on [Kaggle.com](https://www.kaggle.com/account/login?phase=startRegisterTab) and return to this page.
# 1. Click the blue "Copy and Edit" in the upper-right part of this document to create your own copy to your own Kaggle account.
# 2. As you complete exercises, be sure to click the blue "Save" button to create save points for your work.
# 3. If you need to refresh and restart this learning environment, go to **Run** then select **Restart Session**.
# 4. If you need a fresh, blank copy of this document, go to https://www.kaggle.com/ryanorsinger/101-exercises/ and click "Copy and Edit"
# ## Orientation
# - This code notebook is composed of cells. Each cell is either text or Python code.
# - To run a cell of code, click the "play button" icon to the left of the cell or click on the cell and press "Shift+Enter" on your keyboard. This will execute the Python code contained in the cell. Executing a cell that defines a variable is important before executing or authoring a cell that depends on that previously created variable assignment.
# - **Expect to see lots of errors** the first time you load this page.
# - **Expect to see lots of errors** for all cells run without code that matches the assertion tests.
# - Until you click the blue "Copy and Edit" button to make your own copy, you will see an entire page of errors. This is part of the automated tests.
# - Each *assert* line is both an example and a test that tests for the presence and functionality of the instructed exercise.
# ## The only 3 conditions that produce no errors:
# 1. When you make a fresh **copy** of the project to your own account (by clicking "Copy and Edit")
# 2. When you go to "Run" and then click "Restart Session"
# 3. When every single assertion passes.
# ## Outline
# - Each cell starts with a problem statement that describes the exercise to complete.
# - Underneath each problem statement, learners will need to write code to produce an answer.
# - The **assert** lines test to see that your code solves the problem appropriately
# - Many exercises will rely on previous solutions to be correctly completed
# - The `print("Exercise is complete")` line will only run if your solution passes the assertion test(s)
# - Be sure to create programmatic solutions that will work for all inputs:
# - For example, calling the `is_even(2)` returns `True`, but your function should work for all even numbers, both positive and negative.
# ## Guidance
# - Get Python to do the work for you. For example, if the exercise instructs you to reverse a list of numbers, your job is to find the
# - Save often by clicking the blue "Save" button.
# - If you need to clear the output or reset the notebook, go to "Run" then "Restart Session" to clear up any error messages.
# - Do not move or alter the lines of code that contain the `assert` statements. Those are what run your solution and test its actual output vs. expected outputs.
# - Seek to understand the problem before trying to solve it. Can you explain the problem to someone else in English? Can you explain the solution in English?
# - Slow down and read any error messages you encounter. Error messages provide insight into how to resolve the error. When in doubt, put your exact error into a search engine and look for results that reference an identical or similar problem.
# ## Get Python To Do The Work For You
# One of the main jobs of a programming language is to help people solve problems programatically, so we don't have to do so much by hand. For example, it's easy for a person to manually reverse the list `[1, 2, 3]`, but imagine reversing a list of a million things or sorting a list of even a hundred things. When we write programmatic solutions in code, we are providing instructions to the computer to do a task. Computers follow the letter of the code, not the intent, and do exactly what they are told to do. In this way, Python can reverse a list of 3 numbers or 100 numbers or ten million numbers with the same instructions. Repetition is a key idea behind programming languages.
# This means that your task with these exercises is to determine a sequence of steps that solve the problem and then find the Python code that will run those instructions. If you're sorting or reversing things by hand, you're not doing it right!
# ## How To Discover How To Do Something in Python
# 1. The first step is to make sure you know what the problem is asking.
# 2. The second step is to determine, in English (or your first spoken language), what steps you need to take.
# 3. Use a search engine to look for code examples to identical or similar problems.
# One of the best ways to discover how to do things in Python is to use a search engine. Go to your favorite search engine and search for "how to reverse a list in Python" or "how to sort a list in Python". That's how both learners and professionals find answers and examples all the time. Search for what you want and add "in Python" and you'll get lots of code examples. Searching for "How to sum a list of numbers in Python" is a very effective way to discover exactly how to do that task.
# ### Learning to Program and Code
# - You can make a new blank cell for Python code at any time in this document.
# - If you want more freedom to explore learning Python in a blank notebook, go here https://www.kaggle.com/notebooks/welcome and make yourself a blank, new notebook.
# - Programming is an intellectual activity of designing a solution. "Coding" means turning your programmatic solution into code w/ all the right syntax and parts of the programming language.
# - Expect to make mistakes and adopt the attitude that **the error message provides the information you need to proceed**. You will put lots of error messages into search engines to learn this craft!
# - Because computers have zero ability to read in between the lines or "catch the drift" or know what you mean, code only does what it is told to do.
# - Code doesn't do what you *want* it to do, code does what you've told it to do.
# - Before writing any code, figure out how you would solve the problem in spoken language to describe the sequence of steps in the solution.
# - Think about your solution in English (or your natural language). It's **critical** to solve the problem in your natural language before trying to get a programming language to do the work.
# ## Troubleshooting
# - If this entire document shows "Name Error" for many cells, it means you should read the "Getting Started" instructions above to make your own copy.
# - Be sure to commit your work to make save points, as you go.
# - If you load this page and you see your code but not the results of the code, be sure to run each cell (shift + Enter makes this quick)
# - "Name Error" means that you need to assign a variable or define the function as instructed.
# - "Assertion Error" means that your provided solution does not match the correct answer.
# - "Type Error" means that your data type provided is not accurate
# - If your kernel freezes, click on "Run" then select "Restart Session"
# - If you require additional troubleshooting assistance, click on "Help" and then "Docs" to access documentation for this platform.
# - If you have discoverd a bug or typo, please triple check your spelling then create a new issue at [https://github.com/ryanorsinger/101-exercises/issues](https://github.com/ryanorsinger/101-exercises/issues) to notify the author.
# Example problem:
# Uncomment the line below and run this cell.
# The hashtag "#" character in a line of Python code is the comment character. To "comment" means to add the # and to "uncomment" means to remove the # character.
doing_python_right_now = True
# The lines below will test your answer. If you see an error, then it means that your answer is incorrect or incomplete.
assert (
doing_python_right_now == True
), "If you see a NameError, it means that the variable is not created and assigned a value. An 'Assertion Error' means that the value of the variable is incorrect."
print(
"Exercise 0 is correct"
) # This line will print if your solution passes the assertion above.
# Exercise 1
# On the line below, create a variable named on_mars_right_now and assign it the boolean value of False
on_mars_right_now = False
assert (
on_mars_right_now == False
), "If you see a Name Error, be sure to create the variable and assign it a value."
print("Exercise 1 is correct.")
# Exercise 2
# Create a variable named fruits and assign it a list of fruits containing the following fruit names as strings:
# mango, banana, guava, kiwi, and strawberry.
fruits = ["mango", "banana", "guava", "kiwi", "strawberry"]
assert fruits == [
"mango",
"banana",
"guava",
"kiwi",
"strawberry",
], "If you see an Assert Error, ensure the variable contains all the strings in the provided order"
print("Exercise 2 is correct.")
# Exercise 3
# Create a variable named vegetables and assign it a list of fruits containing the following vegetable names as strings:
# eggplant, broccoli, carrot, cauliflower, and zucchini
vegetables = ["eggplant", "broccoli", "carrot", "cauliflower", "zucchini"]
assert vegetables == [
"eggplant",
"broccoli",
"carrot",
"cauliflower",
"zucchini",
], "Ensure the variable contains all the strings in the provided order"
print("Exercise 3 is correct.")
# Exercise 4
# Create a variable named numbers and assign it a list of numbers, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10
numbers = [1, 2, 3, 4, 5, 6, 7, 8, 9, 10]
assert numbers == [
1,
2,
3,
4,
5,
6,
7,
8,
9,
10,
], "Ensure the variable contains the numbers 1-10 in order."
print("Exercise 4 is correct.")
# ## List Operations
# **Hint** Recommend finding and using built-in Python functionality whenever possible.
# Exercise 5
# Given the following assigment of the list of fruits, add "tomato" to the end of the list.
fruits = ["mango", "banana", "guava", "kiwi", "strawberry"]
fruits.append("tomato")
assert fruits == [
"mango",
"banana",
"guava",
"kiwi",
"strawberry",
"tomato",
], "Ensure the variable contains all the strings in the right order"
print("Exercise 5 is correct")
# Exercise 6
# Given the following assignment of the vegetables list, add "tomato" to the end of the list.
vegetables = ["eggplant", "broccoli", "carrot", "cauliflower", "zucchini"]
vegetables.append("tomato")
assert vegetables == [
"eggplant",
"broccoli",
"carrot",
"cauliflower",
"zucchini",
"tomato",
], "Ensure the variable contains all the strings in the provided order"
print("Exercise 6 is correct")
# Exercise 7
# Given the list of numbers defined below, reverse the list of numbers that you created above.
numbers = [1, 2, 3, 4, 5, 6, 7, 8, 9, 10]
numbers.reverse()
assert numbers == [
10,
9,
8,
7,
6,
5,
4,
3,
2,
1,
], "Assert Error means that the answer is incorrect."
print("Exercise 7 is correct.")
# Exercise 8
# Sort the vegetables in alphabetical order
vegetables.sort()
assert vegetables == [
"broccoli",
"carrot",
"cauliflower",
"eggplant",
"tomato",
"zucchini",
]
print("Exercise 8 is correct.")
# Exercise 9
# Write the code necessary to sort the fruits in reverse alphabetical order
fruits.sort(reverse=True)
assert fruits == ["tomato", "strawberry", "mango", "kiwi", "guava", "banana"]
print("Exercise 9 is correct.")
# Exercise 10
# Write the code necessary to produce a single list that holds all fruits then all vegetables in the order as they were sorted above.
fruits_and_veggies = fruits + vegetables
assert fruits_and_veggies == [
"tomato",
"strawberry",
"mango",
"kiwi",
"guava",
"banana",
"broccoli",
"carrot",
"cauliflower",
"eggplant",
"tomato",
"zucchini",
]
print("Exercise 10 is correct")
# ## Basic Functions
# **Hint** Be sure to `return` values from your function definitions. The assert statements will call your function(s) for you.
# Run this cell in order to generate some numbers to use in our functions after this.
import random
positive_even_number = random.randrange(2, 101, 2)
negative_even_number = random.randrange(-100, -1, 2)
positive_odd_number = random.randrange(1, 100, 2)
negative_odd_number = random.randrange(-101, 0, 2)
print("We now have some random numbers available for future exercises.")
print("The random positive even number is", positive_even_number)
print("The random positive odd nubmer is", positive_odd_number)
print("The random negative even number", negative_even_number)
print("The random negative odd number", negative_odd_number)
# Example function defintion:
# Write a say_hello function that adds the string "Hello, " to the beginning and "!" to the end of any given input.
def say_hello(name):
return "Hello, " + name + "!"
assert say_hello("Jane") == "Hello, Jane!", "Double check the inputs and data types"
assert say_hello("Pat") == "Hello, Pat!", "Double check the inputs and data types"
assert say_hello("Astrud") == "Hello, Astrud!", "Double check the inputs and data types"
print("The example function definition ran appropriately")
# Another example function definition:
# This plus_two function takes in a variable and adds 2 to it.
def plus_two(number):
return number + 2
assert plus_two(3) == 5
assert plus_two(0) == 2
assert plus_two(-2) == 0
print(
"The plus_two assertions executed appropriately... The second function definition example executed appropriately."
)
# Exercise 11
# Write a function definition for a function named add_one that takes in a number and returns that number plus one.
def add_one(number):
return number + 1
assert (
add_one(2) == 3
), "Ensure that the function is defined, named properly, and returns the correct value"
assert add_one(0) == 1, "Zero plus one is one."
assert (
add_one(positive_even_number) == positive_even_number + 1
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
add_one(negative_odd_number) == negative_odd_number + 1
), "Ensure that the function is defined, named properly, and returns the correct value"
print("Exercise 11 is correct.")
# Exercise 12
# Write a function definition named is_positive that takes in a number and returns True or False if that number is positive.
def is_positive(number):
if number > 0:
return True
return False
assert (
is_positive(positive_odd_number) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_positive(positive_even_number) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_positive(negative_odd_number) == False
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_positive(negative_even_number) == False
), "Ensure that the function is defined, named properly, and returns the correct value"
print("Exercise 12 is correct.")
# Exercise 13
# Write a function definition named is_negative that takes in a number and returns True or False if that number is negative.
def is_negative(number):
if number < 0:
return True
return False
assert (
is_negative(positive_odd_number) == False
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_negative(positive_even_number) == False
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_negative(negative_odd_number) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_negative(negative_even_number) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
print("Exercise 13 is correct.")
# Exercise 14
# Write a function definition named is_odd that takes in a number and returns True or False if that number is odd.
def is_odd(number):
if number % 2 == 1:
return True
return False
assert (
is_odd(positive_odd_number) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_odd(positive_even_number) == False
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_odd(negative_odd_number) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_odd(negative_even_number) == False
), "Ensure that the function is defined, named properly, and returns the correct value"
print("Exercise 14 is correct.")
# Exercise 15
# Write a function definition named is_even that takes in a number and returns True or False if that number is even.
def is_even(number):
if number % 2 == 0:
return True
return False
assert (
is_even(2) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_even(positive_odd_number) == False
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_even(positive_even_number) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_even(negative_odd_number) == False
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
is_even(negative_even_number) == True
), "Ensure that the function is defined, named properly, and returns the correct value"
print("Exercise 15 is correct.")
# Exercise 16
# Write a function definition named identity that takes in any argument and returns that argument's value. Don't overthink this one!
def identity(x):
return x
assert (
identity(fruits) == fruits
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
identity(vegetables) == vegetables
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
identity(positive_odd_number) == positive_odd_number
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
identity(positive_even_number) == positive_even_number
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
identity(negative_odd_number) == negative_odd_number
), "Ensure that the function is defined, named properly, and returns the correct value"
assert (
identity(negative_even_number) == negative_even_number
), "Ensure that the function is defined, named properly, and returns the correct value"
print("Exercise 16 is correct.")
# Exercise 17
# Write a function definition named is_positive_odd that takes in a number and returns True or False if the value is both greater than zero and odd
def is_positive_odd(number):
if number > 0 and number % 2 == 1:
return True
return False
assert is_positive_odd(3) == True, "Double check your syntax and logic"
assert (
is_positive_odd(positive_odd_number) == True
), "Double check your syntax and logic"
assert (
is_positive_odd(positive_even_number) == False
), "Double check your syntax and logic"
assert (
is_positive_odd(negative_odd_number) == False
), "Double check your syntax and logic"
assert (
is_positive_odd(negative_even_number) == False
), "Double check your syntax and logic"
print("Exercise 17 is correct.")
# Exercise 18
# Write a function definition named is_positive_even that takes in a number and returns True or False if the value is both greater than zero and even
def is_positive_even(number):
if number > 0 and number % 2 == 0:
return True
return False
assert is_positive_even(4) == True, "Double check your syntax and logic"
assert (
is_positive_even(positive_odd_number) == False
), "Double check your syntax and logic"
assert (
is_positive_even(positive_even_number) == True
), "Double check your syntax and logic"
assert (
is_positive_even(negative_odd_number) == False
), "Double check your syntax and logic"
assert (
is_positive_even(negative_even_number) == False
), "Double check your syntax and logic"
print("Exercise 18 is correct.")
# Exercise 19
# Write a function definition named is_negative_odd that takes in a number and returns True or False if the value is both less than zero and odd.
def is_negative_odd(number):
if number < 0 and number % 2 == 1:
return True
return False
assert is_negative_odd(-3) == True, "Double check your syntax and logic"
assert (
is_negative_odd(positive_odd_number) == False
), "Double check your syntax and logic"
assert (
is_negative_odd(positive_even_number) == False
), "Double check your syntax and logic"
assert (
is_negative_odd(negative_odd_number) == True
), "Double check your syntax and logic"
assert (
is_negative_odd(negative_even_number) == False
), "Double check your syntax and logic"
print("Exercise 19 is correct.")
# Exercise 20
# Write a function definition named is_negative_even that takes in a number and returns True or False if the value is both less than zero and even.
def is_negative_even(number):
if number < 0 and number % 2 == 0:
return True
return False
assert is_negative_even(-4) == True, "Double check your syntax and logic"
assert (
is_negative_even(positive_odd_number) == False
), "Double check your syntax and logic"
assert (
is_negative_even(positive_even_number) == False
), "Double check your syntax and logic"
assert (
is_negative_even(negative_odd_number) == False
), "Double check your syntax and logic"
assert (
is_negative_even(negative_even_number) == True
), "Double check your syntax and logic"
print("Exercise 20 is correct.")
# Exercise 21
# Write a function definition named half that takes in a number and returns half the provided number.
def half(number):
return number / 2
assert half(4) == 2
assert half(5) == 2.5
assert half(positive_odd_number) == positive_odd_number / 2
assert half(positive_even_number) == positive_even_number / 2
assert half(negative_odd_number) == negative_odd_number / 2
assert half(negative_even_number) == negative_even_number / 2
print("Exercise 21 is correct.")
# Exercise 22
# Write a function definition named double that takes in a number and returns double the provided number.
def double(number):
return number * 2
assert double(4) == 8
assert double(5) == 10
assert double(positive_odd_number) == positive_odd_number * 2
assert double(positive_even_number) == positive_even_number * 2
assert double(negative_odd_number) == negative_odd_number * 2
assert double(negative_even_number) == negative_even_number * 2
print("Exercise 22 is correct.")
# Exercise 23
# Write a function definition named triple that takes in a number and returns triple the provided number.
def triple(number):
return number * 3
assert triple(4) == 12
assert triple(5) == 15
assert triple(positive_odd_number) == positive_odd_number * 3
assert triple(positive_even_number) == positive_even_number * 3
assert triple(negative_odd_number) == negative_odd_number * 3
assert triple(negative_even_number) == negative_even_number * 3
print("Exercise 23 is correct.")
# Exercise 24
# Write a function definition named reverse_sign that takes in a number and returns the provided number but with the sign reversed.
def reverse_sign(number):
if number > 0:
return number * -1
return abs(number)
assert reverse_sign(4) == -4
assert reverse_sign(-5) == 5
assert reverse_sign(positive_odd_number) == positive_odd_number * -1
assert reverse_sign(positive_even_number) == positive_even_number * -1
assert reverse_sign(negative_odd_number) == negative_odd_number * -1
assert reverse_sign(negative_even_number) == negative_even_number * -1
print("Exercise 24 is correct.")
# Exercise 25
# Write a function definition named absolute_value that takes in a number and returns the absolute value of the provided number
def absolute_value(number):
return abs(number)
assert absolute_value(4) == 4
assert absolute_value(-5) == 5
assert absolute_value(positive_odd_number) == positive_odd_number
assert absolute_value(positive_even_number) == positive_even_number
assert absolute_value(negative_odd_number) == negative_odd_number * -1
assert absolute_value(negative_even_number) == negative_even_number * -1
print("Exercise 25 is correct.")
# Exercise 26
# Write a function definition named is_multiple_of_three that takes in a number and returns True or False if the number is evenly divisible by 3.
def is_multiple_of_three(number):
if number % 3 == 0:
return True
return False
assert is_multiple_of_three(3) == True
assert is_multiple_of_three(15) == True
assert is_multiple_of_three(9) == True
assert is_multiple_of_three(4) == False
assert is_multiple_of_three(10) == False
print("Exercise 26 is correct.")
# Exercise 27
# Write a function definition named is_multiple_of_five that takes in a number and returns True or False if the number is evenly divisible by 5.
def is_multiple_of_five(number):
if number % 5 == 0:
return True
return False
assert is_multiple_of_five(3) == False
assert is_multiple_of_five(15) == True
assert is_multiple_of_five(9) == False
assert is_multiple_of_five(4) == False
assert is_multiple_of_five(10) == True
print("Exercise 27 is correct.")
# Exercise 28
# Write a function definition named is_multiple_of_both_three_and_five that takes in a number and returns True or False if the number is evenly divisible by both 3 and 5.
def is_multiple_of_both_three_and_five(number):
if number % 3 == 0 and number % 5 == 0:
return True
return False
assert is_multiple_of_both_three_and_five(15) == True
assert is_multiple_of_both_three_and_five(45) == True
assert is_multiple_of_both_three_and_five(3) == False
assert is_multiple_of_both_three_and_five(9) == False
assert is_multiple_of_both_three_and_five(4) == False
print("Exercise 28 is correct.")
# Exercise 29
# Write a function definition named square that takes in a number and returns the number times itself.
def square(number):
return number * number
assert square(3) == 9
assert square(2) == 4
assert square(9) == 81
assert square(positive_odd_number) == positive_odd_number * positive_odd_number
print("Exercise 29 is correct.")
# Exercise 30
# Write a function definition named add that takes in two numbers and returns the sum.
def add(num_1, num_2):
return num_1 + num_2
assert add(3, 2) == 5
assert add(10, -2) == 8
assert add(5, 7) == 12
print("Exercise 30 is correct.")
# Exercise 31
# Write a function definition named cube that takes in a number and returns the number times itself, times itself.
def cube(number):
return number * number * number
assert cube(3) == 27
assert cube(2) == 8
assert cube(5) == 125
assert (
cube(positive_odd_number)
== positive_odd_number * positive_odd_number * positive_odd_number
)
print("Exercise 31 is correct.")
# Exercise 32
# Write a function definition named square_root that takes in a number and returns the square root of the provided number
def square_root(number):
return number**0.5
assert square_root(4) == 2.0
assert square_root(64) == 8.0
assert square_root(81) == 9.0
print("Exercise 32 is correct.")
# Exercise 33
# Write a function definition named subtract that takes in two numbers and returns the first minus the second argument.
def subtract(num_1, num_2):
return num_1 - num_2
assert subtract(8, 6) == 2
assert subtract(27, 4) == 23
assert subtract(12, 2) == 10
print("Exercise 33 is correct.")
# Exercise 34
# Write a function definition named multiply that takes in two numbers and returns the first times the second argument.
def multiply(num_1, num_2):
return num_1 * num_2
assert multiply(2, 1) == 2
assert multiply(3, 5) == 15
assert multiply(5, 2) == 10
print("Exercise 34 is correct.")
# Exercise 35
# Write a function definition named divide that takes in two numbers and returns the first argument divided by the second argument.
def divide(num_1, num_2):
return num_1 / num_2
assert divide(27, 9) == 3
assert divide(15, 3) == 5
assert divide(5, 2) == 2.5
assert divide(10, 2) == 5
print("Exercise 35 is correct.")
# Exercise 36
# Write a function definition named quotient that takes in two numbers and returns only the quotient from dividing the first argument by the second argument.
def quotient(num_1, num_2):
return num_1 // num_2
assert quotient(27, 9) == 3
assert quotient(5, 2) == 2
assert quotient(10, 3) == 3
print("Exercise 36 is correct.")
# Exercise 37
# Write a function definition named remainder that takes in two numbers and returns the remainder of first argument divided by the second argument.
def remainder(num_1, num_2):
return num_1 % num_2
assert remainder(3, 3) == 0
assert remainder(5, 2) == 1
assert remainder(7, 5) == 2
print("Exercise 37 is correct.")
# Exercise 38
# Write a function definition named sum_of_squares that takes in two numbers, squares each number, then returns the sum of both squares.
def sum_of_squares(num_1, num_2):
return num_1**2 + num_2**2
assert sum_of_squares(3, 2) == 13
assert sum_of_squares(5, 2) == 29
assert sum_of_squares(2, 4) == 20
print("Exercise 38 is correct.")
# Exercise 39
# Write a function definition named times_two_plus_three that takes in a number, multiplies it by two, adds 3 and returns the result.
def times_two_plus_three(number):
return number * 2 + 3
assert times_two_plus_three(0) == 3
assert times_two_plus_three(1) == 5
assert times_two_plus_three(2) == 7
assert times_two_plus_three(3) == 9
assert times_two_plus_three(5) == 13
print("Exercise 39 is correct.")
# Exercise 40
# Write a function definition named area_of_rectangle that takes in two numbers and returns the product.
def area_of_rectangle(num_1, num_2):
return num_1 * num_2
assert area_of_rectangle(1, 3) == 3
assert area_of_rectangle(5, 2) == 10
assert area_of_rectangle(2, 7) == 14
assert area_of_rectangle(5.3, 10.3) == 54.59
print("Exercise 40 is correct.")
import math
# Exercise 41
# Write a function definition named area_of_circle that takes in a number representing a circle's radius and returns the area of the circl
def area_of_circle(number):
return math.pi * number**2
assert area_of_circle(3) == 28.274333882308138
assert area_of_circle(5) == 78.53981633974483
assert area_of_circle(7) == 153.93804002589985
print("Exercise 41 is correct.")
import math
# Exercise 42
# Write a function definition named circumference that takes in a number representing a circle's radius and returns the circumference.
def circumference(number):
return 2 * math.pi * number
assert circumference(3) == 18.84955592153876
assert circumference(5) == 31.41592653589793
assert circumference(7) == 43.982297150257104
print("Exercise 42 is correct.")
# ## Functions working with strings
# Exercise 43
# Write a function definition named is_vowel that takes in value and returns True if the value is a, e, i, o, u in upper or lower case.
def is_vowel(letter):
if (
letter == "a"
or letter == "e"
or letter == "i"
or letter == "o"
or letter == "u"
or letter == "A"
or letter == "E"
or letter == "I"
or letter == "O"
or letter == "U"
):
return True
return False
assert is_vowel("a") == True
assert is_vowel("U") == True
assert is_vowel("banana") == False
assert is_vowel("Q") == False
assert is_vowel("y") == False
print("Exercise 43 is correct.")
# Exercise 44
# Write a function definition named has_vowels that takes in value and returns True if the string contains any vowels.
def has_vowels(string_input):
for letter in string_input:
if is_vowel(letter):
return True
return False
assert has_vowels("banana") == True
assert has_vowels("ubuntu") == True
assert has_vowels("QQQQ") == False
assert has_vowels("wyrd") == False
print("Exercise 44 is correct.")
# Exercise 45
# Write a function definition named count_vowels that takes in value and returns the count of the nubmer of vowels in a sequence.
def count_vowels(string_input):
counter = 0
for char in string_input:
if is_vowel(char):
counter += 1
return counter
assert count_vowels("banana") == 3
assert count_vowels("ubuntu") == 3
assert count_vowels("mango") == 2
assert count_vowels("QQQQ") == 0
assert count_vowels("wyrd") == 0
print("Exercise 45 is correct.")
# Exercise 46
# Write a function definition named remove_vowels that takes in string and returns the string without any vowels
def remove_vowels(string_input):
no_vowels_string = ""
for char in string_input:
if is_vowel(char) == False:
no_vowels_string = no_vowels_string + char
return no_vowels_string
assert remove_vowels("banana") == "bnn"
assert remove_vowels("ubuntu") == "bnt"
assert remove_vowels("mango") == "mng"
assert remove_vowels("QQQQ") == "QQQQ"
print("Exercise 46 is correct.")
# Exercise 47
# Write a function definition named starts_with_vowel that takes in string and True if the string starts with a vowel
def starts_with_vowel(string_input):
return is_vowel(string_input[0])
assert starts_with_vowel("ubuntu") == True
assert starts_with_vowel("banana") == False
assert starts_with_vowel("mango") == False
print("Exercise 47 is correct.")
# Exercise 48
# Write a function definition named ends_with_vowel that takes in string and True if the string ends with a vowel
def ends_with_vowel(string_input):
return is_vowel(string_input[-1])
assert ends_with_vowel("ubuntu") == True
assert ends_with_vowel("banana") == True
assert ends_with_vowel("mango") == True
assert ends_with_vowel("spinach") == False
print("Exercise 48 is correct.")
# Exercise 49
# Write a function definition named starts_and_ends_with_vowel that takes in string and returns True if the string starts and ends with a vowel
def starts_and_ends_with_vowel(string_input):
return starts_with_vowel(string_input) and ends_with_vowel(string_input)
assert starts_and_ends_with_vowel("ubuntu") == True
assert starts_and_ends_with_vowel("banana") == False
assert starts_and_ends_with_vowel("mango") == False
print("Exercise 49 is correct.")
# ## Accessing List Elements
# Exercise 50
# Write a function definition named first that takes in sequence and returns the first value of that sequence.
def first(input_sequence):
return input_sequence[0]
assert first("ubuntu") == "u"
assert first([1, 2, 3]) == 1
assert first(["python", "is", "awesome"]) == "python"
print("Exercise 50 is correct.")
# Exercise 51
# Write a function definition named second that takes in sequence and returns the second value of that sequence.
def second(input_sequence):
return input_sequence[1]
assert second("ubuntu") == "b"
assert second([1, 2, 3]) == 2
assert second(["python", "is", "awesome"]) == "is"
print("Exercise 51 is correct.")
# Exercise 52
# Write a function definition named third that takes in sequence and returns the third value of that sequence.
def third(input_sequence):
return input_sequence[2]
assert third("ubuntu") == "u"
assert third([1, 2, 3]) == 3
assert third(["python", "is", "awesome"]) == "awesome"
print("Exercise 52 is correct.")
# Exercise 53
# Write a function definition named forth that takes in sequence and returns the forth value of that sequence.
def forth(input_sequence):
return input_sequence[3]
assert forth("ubuntu") == "n"
assert forth([1, 2, 3, 4]) == 4
assert forth(["python", "is", "awesome", "right?"]) == "right?"
print("Exercise 53 is correct.")
# Exercise 54
# Write a function definition named last that takes in sequence and returns the last value of that sequence.
def last(input_sequence):
return input_sequence[-1]
assert last("ubuntu") == "u"
assert last([1, 2, 3, 4]) == 4
assert last(["python", "is", "awesome"]) == "awesome"
assert last(["kiwi", "mango", "guava"]) == "guava"
print("Exercise 54 is correct.")
# Exercise 55
# Write a function definition named second_to_last that takes in sequence and returns the second to last value of that sequence.
def second_to_last(input_string):
return input_string[-2]
assert second_to_last("ubuntu") == "t"
assert second_to_last([1, 2, 3, 4]) == 3
assert second_to_last(["python", "is", "awesome"]) == "is"
assert second_to_last(["kiwi", "mango", "guava"]) == "mango"
print("Exercise 55 is correct.")
# Exercise 56
# Write a function definition named third_to_last that takes in sequence and returns the third to last value of that sequence.
def third_to_last(input_string):
return input_string[-3]
assert third_to_last("ubuntu") == "n"
assert third_to_last([1, 2, 3, 4]) == 2
assert third_to_last(["python", "is", "awesome"]) == "python"
assert third_to_last(["strawberry", "kiwi", "mango", "guava"]) == "kiwi"
print("Exercise 56 is correct.")
# Exercise 57
# Write a function definition named first_and_second that takes in sequence and returns the first and second value of that sequence as a list
def first_and_second(input_sequence):
return [first(input_sequence), second(input_sequence)]
assert first_and_second([1, 2, 3, 4]) == [1, 2]
assert first_and_second(["python", "is", "awesome"]) == ["python", "is"]
assert first_and_second(["strawberry", "kiwi", "mango", "guava"]) == [
"strawberry",
"kiwi",
]
print("Exercise 57 is correct.")
# Exercise 58
# Write a function definition named first_and_last that takes in sequence and returns the first and last value of that sequence as a list
def first_and_last(input_sequence):
return [first(input_sequence), last(input_sequence)]
assert first_and_last([1, 2, 3, 4]) == [1, 4]
assert first_and_last(["python", "is", "awesome"]) == ["python", "awesome"]
assert first_and_last(["strawberry", "kiwi", "mango", "guava"]) == [
"strawberry",
"guava",
]
print("Exercise 58 is correct.")
# Exercise 59
# Write a function definition named first_to_last that takes in sequence and returns the sequence with the first value moved to the end of the sequence.
assert first_to_last([1, 2, 3, 4]) == [2, 3, 4, 1]
assert first_to_last(["python", "is", "awesome"]) == ["is", "awesome", "python"]
assert first_to_last(["strawberry", "kiwi", "mango", "guava"]) == [
"kiwi",
"mango",
"guava",
"strawberry",
]
print("Exercise 59 is correct.")
# ## Functions to describe data
# Exercise 60
# Write a function definition named sum_all that takes in sequence of numbers and returns all the numbers added together.
assert sum_all([1, 2, 3, 4]) == 10
assert sum_all([3, 3, 3]) == 9
assert sum_all([0, 5, 6]) == 11
print("Exercise 60 is correct.")
# Exercise 61
# Write a function definition named mean that takes in sequence of numbers and returns the average value
assert mean([1, 2, 3, 4]) == 2.5
assert mean([3, 3, 3]) == 3
assert mean([1, 5, 6]) == 4
print("Exercise 61 is correct.")
# Exercise 62
# Write a function definition named median that takes in sequence of numbers and returns the average value
assert median([1, 2, 3, 4, 5]) == 3.0
assert median([1, 2, 3]) == 2.0
assert median([1, 5, 6]) == 5.0
assert median([1, 2, 5, 6]) == 3.5
print("Exercise 62 is correct.")
# Exercise 63
# Write a function definition named mode that takes in sequence of numbers and returns the most commonly occuring value
assert mode([1, 2, 2, 3, 4]) == 2
assert mode([1, 1, 2, 3]) == 1
assert mode([2, 2, 3, 3, 3]) == 3
print("Exercise 63 is correct.")
# Exercise 64
# Write a function definition named product_of_all that takes in sequence of numbers and returns the product of multiplying all the numbers together
assert product_of_all([1, 2, 3]) == 6
assert product_of_all([3, 4, 5]) == 60
assert product_of_all([2, 2, 3, 0]) == 0
print("Exercise 64 is correct.")
# ## Applying functions to lists
# Run this cell in order to use the following list of numbers for the next exercises
numbers = [-5, -4, -3, -2, -1, 1, 2, 3, 4, 5]
# Exercise 65
# Write a function definition named get_highest_number that takes in sequence of numbers and returns the largest number.
assert get_highest_number([1, 2, 3]) == 3
assert get_highest_number([-5, -4, -3, -2, -1, 1, 2, 3, 4, 5]) == 5
assert get_highest_number([-5, -3, 1]) == 1
print("Exercise 65 is correct.")
# Exercise 66
# Write a function definition named get_smallest_number that takes in sequence of numbers and returns the smallest number.
assert get_smallest_number([1, 2, 3]) == 1
assert get_smallest_number([-5, -4, -3, -2, -1, 1, 2, 3, 4, 5]) == -5
assert get_smallest_number([-4, -3, 1]) == -4
print("Exercise 66 is correct.")
# Exercise 67
# Write a function definition named only_odd_numbers that takes in sequence of numbers and returns the odd numbers in a list.
assert only_odd_numbers([1, 2, 3]) == [1, 3]
assert only_odd_numbers([-5, -4, -3, -2, -1, 1, 2, 3, 4, 5]) == [-5, -3, -1, 1, 3, 5]
assert only_odd_numbers([-4, -3, 1]) == [-3, 1]
print("Exercise 67 is correct.")
# Exercise 68
# Write a function definition named only_even_numbers that takes in sequence of numbers and returns the even numbers in a list.
assert only_even_numbers([1, 2, 3]) == [2]
assert only_even_numbers([-5, -4, -3, -2, -1, 1, 2, 3, 4, 5]) == [-4, -2, 2, 4]
assert only_even_numbers([-4, -3, 1]) == [-4]
print("Exercise 68 is correct.")
# Exercise 69
# Write a function definition named only_positive_numbers that takes in sequence of numbers and returns the positive numbers in a list.
assert only_positive_numbers([1, 2, 3]) == [1, 2, 3]
assert only_positive_numbers([-5, -4, -3, -2, -1, 1, 2, 3, 4, 5]) == [1, 2, 3, 4, 5]
assert only_positive_numbers([-4, -3, 1]) == [1]
print("Exercise 69 is correct.")
# Exercise 70
# Write a function definition named only_negative_numbers that takes in sequence of numbers and returns the negative numbers in a list.
assert only_negative_numbers([1, 2, 3]) == []
assert only_negative_numbers([-5, -4, -3, -2, -1, 1, 2, 3, 4, 5]) == [
-5,
-4,
-3,
-2,
-1,
]
assert only_negative_numbers([-4, -3, 1]) == [-4, -3]
print("Exercise 70 is correct.")
# Exercise 71
# Write a function definition named has_evens that takes in sequence of numbers and returns True if there are any even numbers in the sequence
assert has_evens([1, 2, 3]) == True
assert has_evens([2, 5, 6]) == True
assert has_evens([3, 3, 3]) == False
assert has_evens([]) == False
print("Exercise 71 is correct.")
# Exercise 72
# Write a function definition named count_evens that takes in sequence of numbers and returns the number of even numbers
assert count_evens([1, 2, 3]) == 1
assert count_evens([2, 5, 6]) == 2
assert count_evens([3, 3, 3]) == 0
assert count_evens([5, 6, 7, 8]) == 2
print("Exercise 72 is correct.")
# Exercise 73
# Write a function definition named has_odds that takes in sequence of numbers and returns True if there are any odd numbers in the sequence
assert has_odds([1, 2, 3]) == True
assert has_odds([2, 5, 6]) == True
assert has_odds([3, 3, 3]) == True
assert has_odds([2, 4, 6]) == False
print("Exercise 73 is correct.")
# Exercise 74
# Write a function definition named count_odds that takes in sequence of numbers and returns True if there are any odd numbers in the sequence
assert count_odds([1, 2, 3]) == 2
assert count_odds([2, 5, 6]) == 1
assert count_odds([3, 3, 3]) == 3
assert count_odds([2, 4, 6]) == 0
print("Exercise 74 is correct.")
# Exercise 75
# Write a function definition named count_negatives that takes in sequence of numbers and returns a count of the number of negative numbers
assert count_negatives([1, -2, 3]) == 1
assert count_negatives([2, -5, -6]) == 2
assert count_negatives([3, 3, 3]) == 0
print("Exercise 75 is correct.")
# Exercise 76
# Write a function definition named count_positives that takes in sequence of numbers and returns a count of the number of positive numbers
assert count_positives([1, -2, 3]) == 2
assert count_positives([2, -5, -6]) == 1
assert count_positives([3, 3, 3]) == 3
assert count_positives([-2, -1, -5]) == 0
print("Exercise 76 is correct.")
# Exercise 77
# Write a function definition named only_positive_evens that takes in sequence of numbers and returns a list containing all the positive evens from the sequence
assert only_positive_evens([1, -2, 3]) == []
assert only_positive_evens([2, -5, -6]) == [2]
assert only_positive_evens([3, 3, 4, 6]) == [4, 6]
assert only_positive_evens([2, 3, 4, -1, -5]) == [2, 4]
print("Exercise 77 is correct.")
# Exercise 78
# Write a function definition named only_positive_odds that takes in sequence of numbers and returns a list containing all the positive odd numbers from the sequence
assert only_positive_odds([1, -2, 3]) == [1, 3]
assert only_positive_odds([2, -5, -6]) == []
assert only_positive_odds([3, 3, 4, 6]) == [3, 3]
assert only_positive_odds([2, 3, 4, -1, -5]) == [3]
print("Exercise 78 is correct.")
# Exercise 79
# Write a function definition named only_negative_evens that takes in sequence of numbers and returns a list containing all the negative even numbers from the sequence
assert only_negative_evens([1, -2, 3]) == [-2]
assert only_negative_evens([2, -5, -6]) == [-6]
assert only_negative_evens([3, 3, 4, 6]) == []
assert only_negative_evens([-2, 3, 4, -1, -4]) == [-2, -4]
print("Exercise 79 is correct.")
# Exercise 80
# Write a function definition named only_negative_odds that takes in sequence of numbers and returns a list containing all the negative odd numbers from the sequence
assert only_negative_odds([1, -2, 3]) == []
assert only_negative_odds([2, -5, -6]) == [-5]
assert only_negative_odds([3, 3, 4, 6]) == []
assert only_negative_odds([2, -3, 4, -1, -4]) == [-3, -1]
print("Exercise 80 is correct.")
# Exercise 81
# Write a function definition named shortest_string that takes in a list of strings and returns the shortest string in the list.
assert shortest_string(["kiwi", "mango", "strawberry"]) == "kiwi"
assert shortest_string(["hello", "everybody"]) == "hello"
assert shortest_string(["mary", "had", "a", "little", "lamb"]) == "a"
print("Exercise 81 is correct.")
# Exercise 82
# Write a function definition named longest_string that takes in sequence of strings and returns the longest string in the list.
assert longest_string(["kiwi", "mango", "strawberry"]) == "strawberry"
assert longest_string(["hello", "everybody"]) == "everybody"
assert longest_string(["mary", "had", "a", "little", "lamb"]) == "little"
print("Exercise 82 is correct.")
# ## Working with sets
# **Hint** Take a look at the `set` function in Python, the `set` data type, and built-in `set` methods.
# Example set function usage
print(set("kiwi"))
print(set([1, 2, 2, 3, 3, 3, 4, 4, 4, 4]))
# Exercise 83
# Write a function definition named get_unique_values that takes in a list and returns a set with only the unique values from that list.
assert get_unique_values(["ant", "ant", "mosquito", "mosquito", "ladybug"]) == {
"ant",
"mosquito",
"ladybug",
}
assert get_unique_values(["b", "a", "n", "a", "n", "a", "s"]) == {"b", "a", "n", "s"}
assert get_unique_values(
["mary", "had", "a", "little", "lamb", "little", "lamb", "little", "lamb"]
) == {"mary", "had", "a", "little", "lamb"}
print("Exercise 83 is correct.")
# Exercise 84
# Write a function definition named get_unique_values_from_two_lists that takes two lists and returns a single set with only the unique values
assert get_unique_values_from_two_lists([5, 1, 2, 3], [3, 4, 5, 5]) == {1, 2, 3, 4, 5}
assert get_unique_values_from_two_lists([1, 1], [2, 2, 3]) == {1, 2, 3}
assert get_unique_values_from_two_lists(
["tomato", "mango", "kiwi"], ["eggplant", "tomato", "broccoli"]
) == {"tomato", "mango", "kiwi", "eggplant", "broccoli"}
print("Exercise 84 is correct.")
# Exercise 85
# Write a function definition named get_values_in_common that takes two lists and returns a single set with the values that each list has in common
assert get_values_in_common([5, 1, 2, 3], [3, 4, 5, 5]) == {3, 5}
assert get_values_in_common([1, 2], [2, 2, 3]) == {2}
assert get_values_in_common(
["tomato", "mango", "kiwi"], ["eggplant", "tomato", "broccoli"]
) == {"tomato"}
print("Exercise 85 is correct.")
# Exercise 86
# Write a function definition named get_values_not_in_common that takes two lists and returns a single set with the values that each list does not have in common
assert get_values_not_in_common([5, 1, 2, 3], [3, 4, 5, 5]) == {1, 2, 4}
assert get_values_not_in_common([1, 1], [2, 2, 3]) == {1, 2, 3}
assert get_values_not_in_common(
["tomato", "mango", "kiwi"], ["eggplant", "tomato", "broccoli"]
) == {"mango", "kiwi", "eggplant", "broccoli"}
print("Exercise 86 is correct.")
# ## Working with Dictionaries
#
# Run this cell in order to have these two dictionary variables defined.
tukey_paper = {
"title": "The Future of Data Analysis",
"author": "John W. Tukey",
"link": "https://projecteuclid.org/euclid.aoms/1177704711",
"year_published": 1962,
}
thomas_paper = {
"title": "A mathematical model of glutathione metabolism",
"author": "Rachel Thomas",
"link": "https://www.ncbi.nlm.nih.gov/pubmed/18442411",
"year_published": 2008,
}
# Exercise 87
# Write a function named get_paper_title that takes in a dictionary and returns the title property
assert get_paper_title(tukey_paper) == "The Future of Data Analysis"
assert get_paper_title(thomas_paper) == "A mathematical model of glutathione metabolism"
print("Exercise 87 is correct.")
# Exercise 88
# Write a function named get_year_published that takes in a dictionary and returns the value behind the "year_published" key.
assert get_year_published(tukey_paper) == 1962
assert get_year_published(thomas_paper) == 2008
print("Exercise 88 is correct.")
# Run this code to create data for the next two questions
book = {
"title": "Genetic Algorithms and Machine Learning for Programmers",
"price": 36.99,
"author": "Frances Buontempo",
}
# Exercise 89
# Write a function named get_price that takes in a dictionary and returns the price
assert get_price(book) == 36.99
print("Exercise 89 is complete.")
# Exercise 90
# Write a function named get_book_author that takes in a dictionary (the above declared book variable) and returns the author's name
assert get_book_author(book) == "Frances Buontempo"
print("Exercise 90 is complete.")
# ## Working with Lists of Dictionaries
# **Hint** If you need an example of lists of dictionaries, see [https://gist.github.com/ryanorsinger/fce8154028a924c1073eac24c7c3f409](https://gist.github.com/ryanorsinger/fce8154028a924c1073eac24c7c3f409)
# Run this cell in order to have some setup data for the next exercises
books = [
{
"title": "Genetic Algorithms and Machine Learning for Programmers",
"price": 36.99,
"author": "Frances Buontempo",
},
{
"title": "The Visual Display of Quantitative Information",
"price": 38.00,
"author": "Edward Tufte",
},
{
"title": "Practical Object-Oriented Design",
"author": "Sandi Metz",
"price": 30.47,
},
{"title": "Weapons of Math Destruction", "author": "Cathy O'Neil", "price": 17.44},
]
# Exercise 91
# Write a function named get_number_of_books that takes in a list of objects and returns the number of dictionaries in that list.
assert get_number_of_books(books) == 4
print("Exercise 91 is complete.")
# Exercise 92
# Write a function named total_of_book_prices that takes in a list of dictionaries and returns the sum total of all the book prices added together
assert total_of_book_prices(books) == 122.9
print("Exercise 92 is complete.")
# Exercise 93
# Write a function named get_average_book_price that takes in a list of dictionaries and returns the average book price.
assert get_average_book_price(books) == 30.725
print("Exercise 93 is complete.")
# Exercise 94
# Write a function called highest_price_book that takes in the above defined list of dictionaries "books" and returns the dictionary containing the title, price, and author of the book with the highest priced book.
# Hint: Much like sometimes start functions with a variable set to zero, you may want to create a dictionary with the price set to zero to compare to each dictionary's price in the list
assert highest_price_book(books) == {
"title": "The Visual Display of Quantitative Information",
"price": 38.00,
"author": "Edward Tufte",
}
print("Exercise 94 is complete")
# Exercise 95
# Write a function called lowest_priced_book that takes in the above defined list of dictionaries "books" and returns the dictionary containing the title, price, and author of the book with the lowest priced book.
# Hint: Much like sometimes start functions with a variable set to zero or float('inf'), you may want to create a dictionary with the price set to float('inf') to compare to each dictionary in the list
assert lowest_price_book(books) == {
"title": "Weapons of Math Destruction",
"author": "Cathy O'Neil",
"price": 17.44,
}
print("Exercise 95 is complete.")
shopping_cart = {
"tax": 0.08,
"items": [
{"title": "orange juice", "price": 3.99, "quantity": 1},
{"title": "rice", "price": 1.99, "quantity": 3},
{"title": "beans", "price": 0.99, "quantity": 3},
{"title": "chili sauce", "price": 2.99, "quantity": 1},
{"title": "chocolate", "price": 0.75, "quantity": 9},
],
}
# Exercise 96
# Write a function named get_tax_rate that takes in the above shopping cart as input and returns the tax rate.
# Hint: How do you access a key's value on a dictionary? The tax rate is one key of the entire shopping_cart dictionary.
assert get_tax_rate(shopping_cart) == 0.08
print("Exercise 96 is complete")
# Exercise 97
# Write a function named number_of_item_types that takes in the shopping cart as input and returns the number of unique item types in the shopping cart.
# We're not yet using the quantity of each item, but rather focusing on determining how many different types of items are in the cart.
assert number_of_item_types(shopping_cart) == 5
print("Exercise 97 is complete.")
# Exercise 98
# Write a function named total_number_of_items that takes in the shopping cart as input and returns the total number all item quantities.
# This should return the sum of all of the quantities from each item type
assert total_number_of_items(shopping_cart) == 17
print("Exercise 98 is complete.")
# Exercise 99
# Write a function named get_average_item_price that takes in the shopping cart as an input and returns the average of all the item prices.
# Hint - This should determine the total price divided by the number of types of items. This does not account for each item type's quantity.
assert get_average_item_price(shopping_cart) == 2.1420000000000003
print("Exercise 99 is complete.")
# Exercise 100
# Write a function named get_average_spent_per_item that takes in the shopping cart and returns the average of summing each item's quanties times that item's price.
# Hint: You may need to set an initial total price and total total quantity to zero, then sum up and divide that total price by the total quantity
assert get_average_spent_per_item(shopping_cart) == 1.333529411764706
print("Exercise 100 is complete.")
# Exercise 101
# Write a function named most_spent_on_item that takes in the shopping cart as input and returns the dictionary associated with the item that has the highest price*quantity.
# Be sure to do this as programmatically as possible.
# Hint: Similarly to how we sometimes begin a function with setting a variable to zero, we need a starting place:
# Hint: Consider creating a variable that is a dictionary with the keys "price" and "quantity" both set to 0. You can then compare each item's price and quantity total to the one from "most"
assert most_spent_on_item(shopping_cart) == {
"title": "chocolate",
"price": 0.75,
"quantity": 9,
}
print("Exercise 101 is complete.")
| false | 0 | 16,970 | 0 | 16,970 | 16,970 |
||
69401936
|
#
# # Training yolov5 on the coco2018 dataset - one class only (person)
# # Setup
# Add data to this notebook (see the + Add data button on the right top side). I prepared the data locally - see [Edit coco128 dataset - select person images and annotation files only](https://www.kaggle.com/valentinstefan/edit-coco128-dataset-person-only).
# After upload, the data is then located in `/kaggle/input`.
# Clone repo, install dependencies and check PyTorch and GPU.
import torch
from IPython.display import Image, clear_output # to display images
clear_output()
print(
f"Setup complete. Using torch {torch.__version__} ({torch.cuda.get_device_properties(0).name if torch.cuda.is_available() else 'CPU'})"
)
# verify CUDA
import torch
from IPython.display import Image # for displaying images
# # Train
# Weights & Biases (optional)
import wandb
wandb.login()
Image(filename="./runs/train/test-person-2021-07-30-kaggle-01/results.png", width=1000)
Image(
filename="./runs/train/test-person-2021-07-30-kaggle-01/confusion_matrix.png",
width=600,
)
Image(
filename="./runs/train/test-person-2021-07-30-kaggle-01/val_batch0_labels.jpg",
width=1000,
)
Image(
filename="./runs/train/test-person-2021-07-30-kaggle-01/val_batch0_pred.jpg",
width=1000,
)
# # Download results folder
# Make archive and then can download from the right pannel. Navigate to the archive, which is stored in `/kaggle/working/yolov5/test-person-2021-07-30-kaggle-01.zip`, click the 3 vertical buttons (more actions) and select download. You can use the weights to make inference.
import shutil
shutil.make_archive(
"test-person-2021-07-30-kaggle-01",
"zip",
"./runs/train/test-person-2021-07-30-kaggle-01",
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/401/69401936.ipynb
| null | null |
[{"Id": 69401936, "ScriptId": 18878740, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1450871, "CreationDate": "07/30/2021 14:34:48", "VersionNumber": 2.0, "Title": "YOLOv5-person-class", "EvaluationDate": "07/30/2021", "IsChange": true, "TotalLines": 87.0, "LinesInsertedFromPrevious": 3.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 84.0, "LinesInsertedFromFork": 50.0, "LinesDeletedFromFork": 153.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 37.0, "TotalVotes": 0}]
| null | null | null | null |
#
# # Training yolov5 on the coco2018 dataset - one class only (person)
# # Setup
# Add data to this notebook (see the + Add data button on the right top side). I prepared the data locally - see [Edit coco128 dataset - select person images and annotation files only](https://www.kaggle.com/valentinstefan/edit-coco128-dataset-person-only).
# After upload, the data is then located in `/kaggle/input`.
# Clone repo, install dependencies and check PyTorch and GPU.
import torch
from IPython.display import Image, clear_output # to display images
clear_output()
print(
f"Setup complete. Using torch {torch.__version__} ({torch.cuda.get_device_properties(0).name if torch.cuda.is_available() else 'CPU'})"
)
# verify CUDA
import torch
from IPython.display import Image # for displaying images
# # Train
# Weights & Biases (optional)
import wandb
wandb.login()
Image(filename="./runs/train/test-person-2021-07-30-kaggle-01/results.png", width=1000)
Image(
filename="./runs/train/test-person-2021-07-30-kaggle-01/confusion_matrix.png",
width=600,
)
Image(
filename="./runs/train/test-person-2021-07-30-kaggle-01/val_batch0_labels.jpg",
width=1000,
)
Image(
filename="./runs/train/test-person-2021-07-30-kaggle-01/val_batch0_pred.jpg",
width=1000,
)
# # Download results folder
# Make archive and then can download from the right pannel. Navigate to the archive, which is stored in `/kaggle/working/yolov5/test-person-2021-07-30-kaggle-01.zip`, click the 3 vertical buttons (more actions) and select download. You can use the weights to make inference.
import shutil
shutil.make_archive(
"test-person-2021-07-30-kaggle-01",
"zip",
"./runs/train/test-person-2021-07-30-kaggle-01",
)
| false | 0 | 590 | 0 | 590 | 590 |
||
69401943
|
<jupyter_start><jupyter_text>flight_data dataset
Kaggle dataset identifier: flight-data-dataset
<jupyter_script># Domain : Airlines Project Name : Analyze NYC - Flight Data
# Step 1: Reading Flight Data from the DataSet.
import pandas as pd
import numpy as np
import datetime as dt
from matplotlib import pyplot as plt
import matplotlib
# Read data from the given flight_data.csv file.
df_nyc_flight_data = pd.read_csv("/kaggle/input/flight_data.csv")
# Head method is used to verify sample top 5 records for the given flight_data set.
# Observations: Data consists of flight number,origin,destination,depature time, arrival time, delay in depatures,
# delay in arrivals, air_time,
# travel distance with date and time stamps.
df_nyc_flight_data.head(1)
# Step 2: Understanding Flight Data Set
# Info method is used to get a concise summary of the dataframe.
# Observations: Total record count is not matching for dep_time, dep_delay, arr_time, arr_delay, airtime
df_nyc_flight_data.info()
# Describe method is used to view some basic statistical details like percentile, mean, std etc. of a data frame of numeric values.
# Observations: Total record count is not matching for dep_time, dep_delay, arr_time, arr_delay, airtime
df_nyc_flight_data.describe()
# Step 3: Data Cleaning Activity
# Identify Null values and Handle it.
df_nyc_flight_data.isnull()
# Count all NaN in a DataFrame (both columns & Rows)
# Observations: Total 46595 values were missing in the given dataset.
df_nyc_flight_data.isnull().sum().sum()
# Count total NaN at each column in DataFrame
# Observations: a) total 8255 records not having dep_time which matches dep_delay records count.
# b) total 8713 records not having arr_time information which we can calculate for (8713 -8255 = 458 records)
# c) arr_delay and air_time record count is matching.
# d) total 2512 records for tailnum is missing need to check whether we can able to fill those tailnum or not.
# Conclusions: a) Dep_time : 8255, Assuming 8255 flights has cancelled due to some reasons and dropped those records from analysis.
# b) Dep_delay : 8255, Assuming 8255 flights has cancelled due to some reasons and dropped those records from analysis.
# c) Arr_time : 8713, (8713 - 8255 = 458) From the above point a & b, For rest of 458 records will calculate arr_time based on sched_arr_time and arr_delay.
# d) Arr_delay : 9430, (9430 - 8255 = 1175) From the above point a & b, For rest of 1175 records will calculate arr_delay based on dep_time and arr_time.
# e) tailnum : 2512, From the above point a & b, Assuming tailnum records will be 0 and having flight number details which is sufficient to proceed.
# f) Airtime : 9430, (9430 - 8255 = 1175) From the above point a & b, For rest of 1175 records will calculate air_time based on mean.
df_nyc_flight_data.isnull().sum()
# Step 4: Missing Data Analysis
# Below task is going to perform mentioned above as conclusions.
# Analyze Dep_time and Dep_delay null values
# Conclusions: a) Dep_time : 8255, Assuming 8255 flights has cancelled due to some reasons and dropped those records from analysis.
# b) Dep_delay : 8255, Assuming 8255 flights has cancelled due to some reasons and dropped those records from analysis.
# Implementation: Used Dropna function.
df_nyc_flight_data = df_nyc_flight_data.dropna(axis=0, subset=["dep_time", "dep_delay"])
df_nyc_flight_data.isnull().sum()
# Analyze arr_delay null values
display_arr_delay_null = pd.isnull(df_nyc_flight_data["arr_delay"])
df_nyc_flight_data[display_arr_delay_null]
# Deep dive to understand one flight data which contains null values. e.g.,flight no: 464
df_nyc_flight_data_flight464 = df_nyc_flight_data[df_nyc_flight_data["flight"] == 464]
df_nyc_flight_data_flight464
# Time validation function used to calculate time. For the given dataset, if we add two time values (e.g., 1430+40 = 1470)
# but as per time, it should be 1510 and also 2340+40 = 10 early moring hours
def time_validation(hours):
num_hours = hours
minutes = num_hours % 100
print(num_hours, minutes)
if minutes > 59:
hours = num_hours - minutes
hours += 100
# print('in if:', hours)
if hours >= 2400:
hours = hours - 2400
# print('in 2400:',hours)
hours = hours + (minutes - 60)
# print('in hours+:',hours)
else:
if hours >= 2400:
hours = hours - 2400
# print('in hours>24:',hours)
return str(hours)
# print(time_validation(780))
# Fill all arr_time NULL values by adding sched_arr_time + dep_delay values.
arr_time_nulldata = df_nyc_flight_data[df_nyc_flight_data["arr_time"].isnull()]
arr_time_nulldata["arr_time"].fillna(
arr_time_nulldata["sched_arr_time"] + arr_time_nulldata["dep_delay"], inplace=True
)
arr_time_nulldata["arr_time"] = arr_time_nulldata.apply(
lambda row: time_validation(row["arr_time"]), axis=1
)
df_nyc_flight_data["arr_time"].fillna(value=arr_time_nulldata["arr_time"], inplace=True)
df_nyc_flight_data[df_nyc_flight_data["arr_time"].isnull()]
# No missing values for arr_time column
df_nyc_flight_data.isnull().sum()
# Fill all arr_delay NULL values by subtracting arr_time - sched_arr_time
df_nyc_flight_data["arr_time"] = pd.to_numeric(df_nyc_flight_data["arr_time"])
arr_delay_nulldata = df_nyc_flight_data[df_nyc_flight_data["arr_delay"].isnull()]
arr_delay_nulldata["arr_delay"].fillna(
arr_delay_nulldata["arr_time"] - arr_delay_nulldata["sched_arr_time"], inplace=True
)
arr_delay_nulldata["arr_delay"] = arr_delay_nulldata.apply(
lambda row: time_validation(row["arr_delay"]), axis=1
)
df_nyc_flight_data["arr_delay"].fillna(
value=arr_delay_nulldata["arr_delay"], inplace=True
)
df_nyc_flight_data[df_nyc_flight_data["arr_delay"].isnull()]
# No missing values for arr_delay column
df_nyc_flight_data.isnull().sum()
# Fill all air_time NULL values by subtracting arr_time - dep_time by multiplying with 65% percent of complete duration.
air_time_nulldata = df_nyc_flight_data[df_nyc_flight_data["air_time"].isnull()]
air_time_nulldata["air_time"].fillna(
value=round((air_time_nulldata["arr_time"] - air_time_nulldata["dep_time"]) * 0.65),
inplace=True,
)
air_time_nulldata["air_time"] = air_time_nulldata.apply(
lambda row: time_validation(row["air_time"]), axis=1
)
df_nyc_flight_data["air_time"].fillna(value=air_time_nulldata["air_time"], inplace=True)
df_nyc_flight_data[df_nyc_flight_data["air_time"].isnull()]
# No missing values for air_time column
df_nyc_flight_data.isnull().sum()
# findday function has created to find the day name for the given date and populate it for each and every row.
import datetime
import calendar
def findDay(date):
full_day = datetime.datetime.strptime(date, "%d-%m-%Y").weekday()
return calendar.day_name[full_day]
# date = '03-02-2019'
# print(findDay(date))
# flight_date column created to populate day name for each and every row.
df_nyc_flight_data["flight_date"] = (
df_nyc_flight_data["day"].map(str)
+ "-"
+ df_nyc_flight_data["month"].map(str)
+ "-"
+ df_nyc_flight_data["year"].map(str)
)
df_nyc_flight_data.head(1)
# day_name column created to populate day name for each and every row.
df_nyc_flight_data["day_name"] = df_nyc_flight_data.apply(
lambda row: findDay(row["flight_date"]), axis=1
)
df_nyc_flight_data.head(1)
# aircraft_speed column created to populate aircraft speed for each and every row
df_nyc_flight_data["air_time"] = pd.to_numeric(df_nyc_flight_data["air_time"])
aircraft_speed = df_nyc_flight_data["distance"] / (df_nyc_flight_data["air_time"] / 60)
df_nyc_flight_data["aircraft_speed"] = aircraft_speed
df_nyc_flight_data.head(1)
# Convert dep_time,sched_dep_time,arr_time,sched_arr_time into hh:mm time format.
df_nyc_flight_data["dep_time"] = (
df_nyc_flight_data.dep_time[~df_nyc_flight_data.dep_time.isna()]
.astype(np.int64)
.apply("{:0>4}".format)
)
df_nyc_flight_data["dep_time"] = pd.to_timedelta(
df_nyc_flight_data.dep_time.str[:2]
+ ":"
+ df_nyc_flight_data.dep_time.str[2:]
+ ":00"
)
df_nyc_flight_data["sched_dep_time"] = (
df_nyc_flight_data.sched_dep_time[~df_nyc_flight_data.sched_dep_time.isna()]
.astype(np.int64)
.apply("{:0>4}".format)
)
df_nyc_flight_data["sched_dep_time"] = pd.to_timedelta(
df_nyc_flight_data.sched_dep_time.str[:2]
+ ":"
+ df_nyc_flight_data.sched_dep_time.str[2:]
+ ":00"
)
df_nyc_flight_data["arr_time"] = (
df_nyc_flight_data.arr_time[~df_nyc_flight_data.arr_time.isna()]
.astype(np.int64)
.apply("{:0>4}".format)
)
df_nyc_flight_data["arr_time"] = pd.to_timedelta(
df_nyc_flight_data.arr_time.str[:2]
+ ":"
+ df_nyc_flight_data.arr_time.str[2:]
+ ":00"
)
df_nyc_flight_data["sched_arr_time"] = (
df_nyc_flight_data.sched_arr_time[~df_nyc_flight_data.sched_arr_time.isna()]
.astype(np.int64)
.apply("{:0>4}".format)
)
df_nyc_flight_data["sched_arr_time"] = pd.to_timedelta(
df_nyc_flight_data.sched_arr_time.str[:2]
+ ":"
+ df_nyc_flight_data.sched_arr_time.str[2:]
+ ":00"
)
df_nyc_flight_data.head(1)
# Created two new columns dep_status and arr_status.
# dep_status column used to store information based on the flight depatured before_ontime, ontime, Dep_Actualdelay information.
# arr_status column used to store information based on the flight depatured before_ontime, ontime, Arr_Actualdelay information.
df_nyc_flight_data.loc[df_nyc_flight_data.dep_delay < 0, "dep_status"] = "Before_OnTime"
df_nyc_flight_data.loc[df_nyc_flight_data.dep_delay == 0, "dep_status"] = "OnTime"
df_nyc_flight_data.loc[
df_nyc_flight_data.dep_delay > 0, "dep_status"
] = "Dep_ActualDelay"
df_nyc_flight_data["arr_delay"] = pd.to_numeric(df_nyc_flight_data["arr_delay"])
df_nyc_flight_data.loc[df_nyc_flight_data.arr_delay < 0, "arr_status"] = "Before_OnTime"
df_nyc_flight_data.loc[df_nyc_flight_data.arr_delay == 0, "arr_status"] = "OnTime"
df_nyc_flight_data.loc[
df_nyc_flight_data.arr_delay > 0, "arr_status"
] = "Arr_ActualDelay"
df_nyc_flight_data.head(1)
# Created one new column quarter to fill quarter values from time_hour date column
# Convert datatypes into datetime for the required columns
df_nyc_flight_data["flight_date"] = pd.to_datetime(df_nyc_flight_data["flight_date"])
df_nyc_flight_data["time_hour"] = pd.to_datetime(df_nyc_flight_data["time_hour"])
df_nyc_flight_data["quarter"] = df_nyc_flight_data["time_hour"].dt.quarter
# Convert datatypes into category for the required columns
df_nyc_flight_data[
[
"month",
"day",
"carrier",
"origin",
"dest",
"day_name",
"dep_status",
"arr_status",
"quarter",
]
] = df_nyc_flight_data[
[
"month",
"day",
"carrier",
"origin",
"dest",
"day_name",
"dep_status",
"arr_status",
"quarter",
]
].apply(
lambda x: x.astype("category")
)
df_nyc_flight_data.head(1)
# Verify all columns datatypes and convert it as per the data visualization requirement.
df_nyc_flight_data.dtypes
# Verify whether we have any missingvalues for all columns
df_nyc_flight_data.isnull().sum()
# To verify maximum aircraft_speed for the individual carrier.
# Observation: Interestingly found 'inf' value as maximum for carrier 'MQ'
carrier_speed = df_nyc_flight_data.groupby(["carrier"])["aircraft_speed"].max()
carrier_speed
# Observation: Interestingly found 'inf' value as maximum for carrier 'MQ'
# Interesting Facts: Noticied that, Dep_time and Arr_time for this flight '3678' is same and which should not be the case
# in real-time.
df_nyc_flight_data.sort_values(by="aircraft_speed", ascending=False).head(2)
# Observation: Interestingly found 'inf' value as maximum for carrier 'MQ'
# Interesting Facts: Noticied that, Dep_time and Arr_time for this flight '3678' is same and which should not be the case
# in real-time.
# df_nyc_flight_data_flight = df_nyc_flight_data[df_nyc_flight_data['flight'] == 3678]
df_nyc_flight_data_flight = df_nyc_flight_data[df_nyc_flight_data["air_time"] == 0]
df_nyc_flight_data_flight.head(2)
# Observation: Interestingly found 'inf' value as maximum for carrier 'MQ'
# Interesting Facts: Noticied that, Dep_time and Arr_time for this flight '3678' is same and which should not be the case
# in real-time.
# Conclusion:Dropped this flight so that, can able to analyze problem statement 'Aircraft speed analysis' for all carriers.
df_nyc_flight_data = df_nyc_flight_data.drop([259244])
df_nyc_flight_data_flight = df_nyc_flight_data[df_nyc_flight_data["flight"] == 0]
df_nyc_flight_data_flight.head(2)
# df_nyc_flight_data[df_nyc_flight_data['aircraft_speed'].isin([np.inf, -np.inf])]
# df_nyc_flight_data.at[259244,'aircraft_speed']=325.83 # mean value
# Verify whether data displays values correct or not.
carrier_speed = df_nyc_flight_data.groupby(["carrier"])["aircraft_speed"].max()
carrier_speed
# Step 5: Data Visualizations
# Problem Statement 1: Departure delays
# Calculate minimum and maximum dep_delay values for the given NYC flight data set.
min_dep_delay = min(df_nyc_flight_data.dep_delay)
print(min_dep_delay)
max_dep_delay = max(df_nyc_flight_data.dep_delay)
print(max_dep_delay)
# Visualizations Heading: Identify Departure Delay information based on Origin and Carrier
# Plot Used : Relational Plot
# Description : Fetched all records whose dep_delay > 0 and plotted graph based on Origin and Carrier
# Outcome : a) Carrier HA, Origin JFK, is the one whose maximum dep_delay is high > 1200
# b) Carrier OO, Origin LGA, is the one whose dep_delay is very less.
# c) Carrier AS, Origin EWR, is the one whose dep_delay is very less.
import seaborn as sns
df_nyc_flight_data_dep_actualdelay = df_nyc_flight_data[
df_nyc_flight_data["dep_status"] == "Dep_ActualDelay"
]
sns.relplot(
x="carrier", y="dep_delay", hue="origin", data=df_nyc_flight_data_dep_actualdelay
)
# Visualizations Heading: Identify Depature Delay information based on Origin and Day_name
# Plot Used : Categorical Plot
# Description : Fetched all records whose dep_delay > 0 and plotted graph based on Origin and Day_name.
# Outcome : a) Day: Wednesday, Origin JFK, is the one whose maximum dep_delay is around > 1200
# b) Day: Saturday, Origin JFK, is the second whose maxmimum dep_delay is around > 1100
# c) Day: Tuesday, Less number of delays on this day.
sns.catplot(
x="day_name",
y="dep_delay",
hue="origin",
jitter=False,
aspect=2,
data=df_nyc_flight_data_dep_actualdelay,
)
# Calculate total counts of flight depature delay based on dep_status == Dep_ActualDelay
depDelay_count = df_nyc_flight_data[
df_nyc_flight_data["dep_status"] == "Dep_ActualDelay"
]
depDelay_count["dep_status"].value_counts()
# Due to huge data set, Calculate top 2500 flight depature delay based on dep_status == Dep_ActualDelay by descending order
Top2500_DepDelays = depDelay_count.sort_values(by="dep_delay", ascending=False).head(
2500
)
Top2500_DepDelays["dep_status"].value_counts()
# Visualizations Heading: Identify Top 2500 Depature Delay information based on Origin and Carrier
# Plot Used : Relational Plot
# Description : Fetched all records whose dep_delay > 0 and plotted graph based on Origin and Carrier
# Outcome : a) Carrier HA, Origin JFK, is the one whose maximum dep_delay is around > 1200
# b) Carrier MQ, Origin JFK, is the second whose maxmimum dep_delay is around > 1100.
# c) Carrier AS, Origin EWR, is the one whose dep_delay is very less.
sns.relplot(x="carrier", y="dep_delay", hue="origin", data=Top2500_DepDelays, aspect=2)
# Visualizations Heading: Identify Top2500 Depature Delay information based on Origin and Day_name
# Plot Used : Categorical Plot
# Description : Fetched all records whose dep_delay > 0 and plotted graph based on Origin and Day_name.
# Outcome : a) Day: Wednesday, Origin JFK, is the one whose maximum dep_delay is around > 1200
# b) Day: Saturday, Origin JFK, is the second whose maxmimum dep_delay is around > 1100
# c) Day: Tuesday, Less number of delays on this day.
sns.catplot(
x="day_name",
y="dep_delay",
hue="origin",
jitter=False,
aspect=2,
data=Top2500_DepDelays,
)
# Problem Statement 2: Best Airport in terms of time departure %
# Calculate total flight counts, Percentage based on Origin and dep_status = OnTime
dep_OnTime = (
df_nyc_flight_data.groupby("origin")["origin"].count().reset_index(name="total")
)
OnTimeFlights = (
df_nyc_flight_data.loc[df_nyc_flight_data["dep_status"] == "OnTime"]
.groupby(["origin", "dep_status"])["dep_status"]
.count()
.unstack("dep_status")
)
dep_OnTime["OnTime"] = OnTimeFlights["OnTime"].values
dep_OnTime["percentage"] = (dep_OnTime["OnTime"] / dep_OnTime["total"]) * 100
dep_OnTime
# Calculate total flight counts, Percentage based on Origin and dep_status = Before_OnTime
dep_Before_OnTime = (
df_nyc_flight_data.groupby("origin")["origin"].count().reset_index(name="total")
)
Before_OnTimeFlights = (
df_nyc_flight_data.loc[df_nyc_flight_data["dep_status"] == "Before_OnTime"]
.groupby(["origin", "dep_status"])["dep_status"]
.count()
.unstack("dep_status")
)
dep_Before_OnTime["Before_OnTime"] = Before_OnTimeFlights["Before_OnTime"].values
dep_Before_OnTime["percentage"] = (
dep_Before_OnTime["Before_OnTime"] / dep_Before_OnTime["total"]
) * 100
dep_Before_OnTime
# Calculate total flight counts, Percentage based on Origin and dep_status = Dep_Actualdelay
dep_ActualDelay = (
df_nyc_flight_data.groupby("origin")["origin"].count().reset_index(name="total")
)
ActualDelay_Flights = (
df_nyc_flight_data.loc[df_nyc_flight_data["dep_status"] == "Dep_ActualDelay"]
.groupby(["origin", "dep_status"])["dep_status"]
.count()
.unstack("dep_status")
)
dep_ActualDelay["Dep_ActualDelay"] = ActualDelay_Flights["Dep_ActualDelay"].values
dep_ActualDelay["percentage"] = (
dep_ActualDelay["Dep_ActualDelay"] / dep_ActualDelay["total"]
) * 100
dep_ActualDelay
# Merge above all 3 dataframes and display total flight counts, Percentage based on Origin and dep_status in (<0,==0,>0)
merged_inner1 = pd.merge(
left=dep_OnTime, right=dep_Before_OnTime, left_on="origin", right_on="origin"
)
merged_inner1.shape
merged_inner_final = pd.merge(
left=merged_inner1, right=dep_ActualDelay, left_on="origin", right_on="origin"
)
merged_inner_final.shape
merged_inner_final
# Rename columns with meaning full names for the merged dataframe and display final result set.
merged_inner_final = merged_inner_final.drop(["total_y", "total"], axis=1)
merged_inner_final.rename(columns={"total_x": "total"}, inplace=True)
merged_inner_final.rename(columns={"percentage_x": "OnTime_percentage"}, inplace=True)
merged_inner_final.rename(
columns={"percentage_y": "BeforeOnTime_percentage"}, inplace=True
)
merged_inner_final.rename(
columns={"percentage": "ActualDepDelay_percentage"}, inplace=True
)
merged_inner_final
# Visualizations Heading: Identify Best origin airports on basis of time departure Percentage.
# Plot Used : Pie Plot
# Description : Pie Chart plotted based on time departure from the origin.
# Outcome : a) Pie 1: OnTime Flights Percentage based on Origin. Origin JFK is highest percentage with 37.83%
# b) Pie 2: Before_OnTime Flights Percentage based on Origin. Origin LGA is highest percentage with 36.91%
# c) Pie 3: ActualDepDelay Flights Percentage based on Origin. Origin EWR is highest percentage with 38.50%
# d) Pie 4: Total number of Flights Percentage based on Origin. Origin EWR is highest percentage with 35.80%
# Make figure and axes
fig, axs = plt.subplots(2, 2, figsize=(10, 5))
# A standard pie plot
axs[0, 0].set_title("OnTime Flights Percentage based on Origin")
axs[0, 0].pie(
merged_inner_final["OnTime_percentage"],
labels=merged_inner_final["origin"],
autopct="%1.2f%%",
shadow=True,
explode=(0.1, 0, 0),
)
axs[0, 1].set_title("Before_OnTime Flights Percentage based on Origin")
axs[0, 1].pie(
merged_inner_final["BeforeOnTime_percentage"],
labels=merged_inner_final["origin"],
autopct="%1.2f%%",
shadow=True,
explode=(0.1, 0, 0),
)
axs[1, 0].set_title("ActualDepDelay Flights Percentage based on Origin")
axs[1, 0].pie(
merged_inner_final["ActualDepDelay_percentage"],
labels=merged_inner_final["origin"],
autopct="%1.2f%%",
shadow=True,
explode=(0.1, 0, 0),
)
axs[1, 1].set_title("Total number of Flights Percentage based on Origin")
axs[1, 1].pie(
merged_inner_final["total"],
labels=merged_inner_final["origin"],
autopct="%1.2f%%",
shadow=True,
explode=(0.1, 0, 0),
)
plt.show()
# Visualizations Heading: Identify TotalNumberofFlights departure from Origin (i.e,Best Airports.)
# Plot Used : Bar Plot
# Description : Bar Chart plotted to show total number of flights departure from the origin.
# Outcome : a) Origin JFK is one of the highest number OnTime Flights timely departure
# b) Origin LGA is one of the highest number Before_OnTime Flights timely departure
# c) Origin EWR is one of the highest number Dep_ActualDelay Flights timely departure
import numpy as np
x = np.arange(len(merged_inner_final["origin"])) # the label locations
width = 0.25 # the width of the bars
fig, ax = plt.subplots(figsize=(10, 5))
rects1 = ax.bar(
x, merged_inner_final["OnTime"], width, alpha=0.5, color="#EE3224", label="OnTime"
)
rects2 = ax.bar(
[p + width for p in x],
merged_inner_final["Before_OnTime"],
width,
alpha=0.5,
color="#F78F1E",
label="Before_OnTime",
)
rects3 = ax.bar(
[p + width * 2 for p in x],
merged_inner_final["Dep_ActualDelay"],
width,
alpha=0.5,
color="#FFC222",
label="Dep_ActualDelay",
)
# Add some text for labels, title and custom x-axis tick labels, etc.
ax.set_ylabel("Total Number of Flights")
ax.set_title("TotalNumberofFlights departure from Origin i.e,Best Airports")
ax.set_xticks([p + 1.5 * width for p in x])
ax.set_xticklabels(merged_inner_final["origin"])
ax.legend()
plt.xlim(min(x) - width, max(x) + width * 4)
plt.ylim(
[
0,
max(
merged_inner_final["OnTime"]
+ merged_inner_final["Before_OnTime"]
+ merged_inner_final["Dep_ActualDelay"]
),
]
)
fig.tight_layout()
plt.grid()
plt.show()
# Visualizations Heading: Identify TotalNumberofFlights departure from Origin (i.e,Best Airports.)
# Plot Used : line Plot
# Description : line Chart plotted to show total number of flights departure from the origin.
# Outcome : a) Origin EWR,JFK,LGA is having similar number (slightly differ) OnTime Flights timely departure
# b) Before_OnTime Flights timely departure count decreases from EWR,JFK,LGA
# c) Dep_ActualDelay Flights timely departure count increases from EWR,JFK,LGA
ax = plt.gca()
merged_inner_final.plot(kind="line", x="origin", y="OnTime", color="#EE3224", ax=ax)
merged_inner_final.plot(
kind="line", x="origin", y="Before_OnTime", color="#F78F1E", ax=ax
)
merged_inner_final.plot(
kind="line", x="origin", y="Dep_ActualDelay", color="#FFC222", ax=ax
)
plt.show()
# Problem Statement 3: Aircraft speed analysis
# Visualizations Heading: Identify Aircraft_Speed based on airtime, distance and Origin.
# Plot Used : Pair Plot
# Description : Pair Plot plotted to show how the aircraft_speed increases based on air_time and distance.
# Outcome : a) Origin LGA is having high aircraft_speed which travels shorter distance.
# b) Origin LGA is having consistent aircraft_speed for distance <1800
# c) Origin EWR is having inconsistent aircraft_speed for distance <2500
import seaborn as sns
origin_speed = sns.pairplot(
df_nyc_flight_data,
height=3,
vars=["distance", "air_time", "aircraft_speed"],
hue="origin",
palette="husl",
markers=["o", "s", "D"],
)
plt.show(origin_speed)
# Calculate mean of aircraft_speed by applying group by on carrier.
carrier_speed = df_nyc_flight_data.groupby(["carrier"])["aircraft_speed"].mean()
carrier_speed
# Calculate mean of distance by applying group by on carrier.
carrier_distance = df_nyc_flight_data.groupby(["carrier"])["distance"].mean()
carrier_distance
# Merge above two dataframes to display records for Carrier, aircraft_speed and distance.
merged_inner = pd.merge(
left=carrier_speed, right=carrier_distance, left_on="carrier", right_on="carrier"
)
merged_inner.shape
merged_inner.info()
# reset index for the above dataframe records.
merged_inner.reset_index(level=0, drop=False, inplace=True)
merged_inner.head(1)
# Visualizations Heading: Identify Aircraft_Speed based on distance and Origin.
# Plot Used : Scatter Plot
# Description : Scatter Plot plotted to show how the aircraft_speed increases based on distance for carrier.
# Outcome : a) Carrier HA is having high aircraft_speed for the distance covered around 5000
# b) Carrier YU is having low aircraft_speed for the distance covered around 400
# c) Most of the Carrier is having consistent aircraft_speed for the similar distance covered.
f, ax = plt.subplots(figsize=(6.5, 6.5))
sns.set(style="ticks")
sns.despine(f, left=True, bottom=True)
sns.scatterplot(
x="distance",
y="aircraft_speed",
hue="carrier",
palette="dark",
sizes=(1, 8),
linewidth=0,
data=merged_inner,
ax=ax,
)
# Problem Statement 4: On time arrival % analysis
# Calculate count of OnTime,Before_OnTime,Arr_ActualDelay arrival count by grouping with arr_status column
arr_Status = df_nyc_flight_data.groupby("arr_status")["arr_status"].count()
print("arr_Status complete list:")
print(arr_Status)
# Visualizations Heading: Visualize the Ontime arrival percentage and count.
# Plot Used : Count Plot and Pie Plot
# Description : Count Plot plotted to show the count of number of flights based on OnTime,Before_OnTime,Arr_ActualDelay arrival status.
# : Pie Plot plotted to show the percentage of number of flights arrivals based on OnTime,Before_OnTime,Arr_ActualDelay arrival status.
# Outcome : a) Around 189038 number of flights has arrived Before_OnTime with 57.54%
# b) Around 134057 number of flights has arrived Arr_ActualDelay with 40.81%
# c) Around 5425 number of flights has arrived OnTime with 1.65%
# Setting up the chart area
f, ax = plt.subplots(1, 2, figsize=(14, 7))
# setting up chart
df_nyc_flight_data["arr_status"].value_counts().plot.pie(
explode=[0, 0, 0.2], autopct="%1.2f%%", ax=ax[1], shadow=False
)
# setting title for pei chart
ax[1].set_title("On time arrival % Analysis")
ax[1].set_ylabel("")
# setting up data for barchart
sns.countplot(
"arr_status",
order=df_nyc_flight_data["arr_status"].value_counts().index,
data=df_nyc_flight_data,
ax=ax[0],
)
ax[0].set_title("Arrival Status of total flights (in numbers)")
ax[0].set_ylabel("Number of Flights")
plt.show()
# Problem Statement 5: Maximum number of flights headed to some particular destination.
# Calculate maximum number of flights to particular destination.
df_nyc_flight_data["dest"].value_counts()
# Calculate maximum number of flights to started from particular origin.
df_nyc_flight_data["origin"].value_counts()
# Calculate Top 25 Maximum number of flights from origin to destination by applying group by on origin and dest.
Top25_MaxFlights_Dest = (
df_nyc_flight_data.groupby("origin")["dest"].value_counts().to_frame()
)
# Top25_MaxFlights_Dest.dtypes
Top25_MaxFlights_Dest.rename(columns={"dest": "dest_count"}, inplace=True)
Top25_MaxFlights_Dest.reset_index(level=1, drop=False, inplace=True)
Top25_MaxFlights_Dest.reset_index(level=0, drop=False, inplace=True)
# sum_flight1.head(40)
Top25_MaxFlights_Dest = Top25_MaxFlights_Dest.sort_values(
by="dest_count", ascending=False
).head(25)
Top25_MaxFlights_Dest.reset_index(level=0, drop=False, inplace=True)
Top25_MaxFlights_Dest.head(3)
# Visualizations Heading: Visualize Top 25 Maximum number of flights headed to some particular destination.
# Plot Used : Relational Plot
# Description : Relational Plot plotted to show Top 25 Maximum number of flights headed to some particular destination.
# Outcome : a) Top 1 : Origin JFK is having maximum number of flights headed towards LAX destination
# b) Top 10 : Origin EWR is having maximum number of flights headed towards BOS destination
# c) Top 25 : Origin EWR is having maximum number of flights headed towards FLL destination
palette = sns.cubehelix_palette(light=0.8, n_colors=3)
sns.relplot(
x="dest",
y="dest_count",
hue="origin",
height=6,
aspect=3,
data=Top25_MaxFlights_Dest,
)
# Visualizations Heading: Visualize Top 25 Maximum number of flights headed to some particular destination.
# Plot Used : Catergorical Plot
# Description : Catergorical Plot plotted to show Top 25 Maximum number of flights headed to some particular destination.
# Outcome : a) Top 1 : Origin JFK is having maximum number of flights headed towards LAX destination
# b) Top 10 : Origin EWR is having maximum number of flights headed towards BOS destination
# c) Top 25 : Origin EWR is having maximum number of flights headed towards FLL destination
g = sns.catplot(
x="dest",
y="dest_count",
hue="origin",
data=Top25_MaxFlights_Dest,
height=5,
kind="bar",
palette="muted",
aspect=4,
)
g.despine(left=True)
g.set_ylabels("Total Number of Flights")
# Problem Statement 6: Month-Wise analysis of Flight Departure and Arrival Status.
# Visualizations Heading: Visualize Month-Wise mean analysis about the Flight Departure and Arrival Status.
# Plot Used : line Plot
# Description : line Plot plotted to show month-wise analysis dep_delay and arr_delay .
# Outcome : a) left-side line plot shows 3,5,9 month is having earlier arrivals and 4,9 month is having earlier departure.
# b) right-side line plot shows 4,6,7 month is having arrivals delays and 4,6,7 month is having departure delays.
f, ax = plt.subplots(1, 2, figsize=(20, 8))
dep_Ontime = df_nyc_flight_data[df_nyc_flight_data["dep_status"] != "Dep_ActualDelay"]
dep_Ontime[["month", "arr_delay", "dep_delay"]].groupby(["month"]).mean().plot(
ax=ax[0],
marker="*",
linestyle="dashed",
color="b" + "r",
linewidth=2,
markersize=12,
)
df_nyc_flight_data_dep_actualdelay[["month", "arr_delay", "dep_delay"]].groupby(
["month"]
).mean().plot(
ax=ax[1],
marker="*",
linestyle="dashed",
color="b" + "r",
linewidth=2,
markersize=12,
)
# Problem Statement 7: Quarter-Wise analysis of Flight Mean Depature Delays by Carriers.
# Visualizations Heading: Visualize Quarter-Wise analysis of Flight mean Depature Delays by Carriers.
# Plot Used : Heat Map
# Description : Heat Map plotted to show Quarter-Wise analysis of Flight mean Depature Delays by Carriers.
# Outcome : a) Quarter 1: Carrier EV is having highest (24.3) & Carrier US is having lowest (2.7) mean depature delay.
# b) Quarter 2: Carrier F9 is having highest (26.6) & Carrier HA is having lowest (0.3) mean depature delay.
# c) Quarter 3: Carrier FL is having highest (21.9) & Carrier AS is having lowest (5.1) mean depature delay.
# d) Quarter 4: Carrier FL is having highest (21.7) & Carrier OO is having lowest (0.8) mean depature delay.
import seaborn as sns
plt.figure(figsize=(18, 10))
plt.title("Quarter-Wise flight depature delays by carriers")
plt.tight_layout()
hmap = pd.pivot_table(
df_nyc_flight_data,
values="dep_delay",
aggfunc="mean",
index="carrier",
columns="quarter",
)
sns.heatmap(hmap, annot=True, cmap="YlGnBu", center=0, linewidths=0.2, fmt="g")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/401/69401943.ipynb
|
flight-data-dataset
|
rameshbabugonegandla
|
[{"Id": 69401943, "ScriptId": 9425501, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 3798813, "CreationDate": "07/30/2021 14:34:53", "VersionNumber": 46.0, "Title": "Analyze NYC - Flight Data", "EvaluationDate": "07/30/2021", "IsChange": false, "TotalLines": 546.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 546.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92521319, "KernelVersionId": 69401943, "SourceDatasetVersionId": 1149245}]
|
[{"Id": 1149245, "DatasetId": 648966, "DatasourceVersionId": 1179948, "CreatorUserId": 3798813, "LicenseName": "Unknown", "CreationDate": "05/12/2020 02:40:32", "VersionNumber": 1.0, "Title": "flight_data dataset", "Slug": "flight-data-dataset", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 648966, "CreatorUserId": 3798813, "OwnerUserId": 3798813.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1149245.0, "CurrentDatasourceVersionId": 1179948.0, "ForumId": 663310, "Type": 2, "CreationDate": "05/12/2020 02:40:32", "LastActivityDate": "05/12/2020", "TotalViews": 1989, "TotalDownloads": 126, "TotalVotes": 5, "TotalKernels": 2}]
|
[{"Id": 3798813, "UserName": "rameshbabugonegandla", "DisplayName": "Ramesh Babu Gonegandla", "RegisterDate": "10/03/2019", "PerformanceTier": 2}]
|
# Domain : Airlines Project Name : Analyze NYC - Flight Data
# Step 1: Reading Flight Data from the DataSet.
import pandas as pd
import numpy as np
import datetime as dt
from matplotlib import pyplot as plt
import matplotlib
# Read data from the given flight_data.csv file.
df_nyc_flight_data = pd.read_csv("/kaggle/input/flight_data.csv")
# Head method is used to verify sample top 5 records for the given flight_data set.
# Observations: Data consists of flight number,origin,destination,depature time, arrival time, delay in depatures,
# delay in arrivals, air_time,
# travel distance with date and time stamps.
df_nyc_flight_data.head(1)
# Step 2: Understanding Flight Data Set
# Info method is used to get a concise summary of the dataframe.
# Observations: Total record count is not matching for dep_time, dep_delay, arr_time, arr_delay, airtime
df_nyc_flight_data.info()
# Describe method is used to view some basic statistical details like percentile, mean, std etc. of a data frame of numeric values.
# Observations: Total record count is not matching for dep_time, dep_delay, arr_time, arr_delay, airtime
df_nyc_flight_data.describe()
# Step 3: Data Cleaning Activity
# Identify Null values and Handle it.
df_nyc_flight_data.isnull()
# Count all NaN in a DataFrame (both columns & Rows)
# Observations: Total 46595 values were missing in the given dataset.
df_nyc_flight_data.isnull().sum().sum()
# Count total NaN at each column in DataFrame
# Observations: a) total 8255 records not having dep_time which matches dep_delay records count.
# b) total 8713 records not having arr_time information which we can calculate for (8713 -8255 = 458 records)
# c) arr_delay and air_time record count is matching.
# d) total 2512 records for tailnum is missing need to check whether we can able to fill those tailnum or not.
# Conclusions: a) Dep_time : 8255, Assuming 8255 flights has cancelled due to some reasons and dropped those records from analysis.
# b) Dep_delay : 8255, Assuming 8255 flights has cancelled due to some reasons and dropped those records from analysis.
# c) Arr_time : 8713, (8713 - 8255 = 458) From the above point a & b, For rest of 458 records will calculate arr_time based on sched_arr_time and arr_delay.
# d) Arr_delay : 9430, (9430 - 8255 = 1175) From the above point a & b, For rest of 1175 records will calculate arr_delay based on dep_time and arr_time.
# e) tailnum : 2512, From the above point a & b, Assuming tailnum records will be 0 and having flight number details which is sufficient to proceed.
# f) Airtime : 9430, (9430 - 8255 = 1175) From the above point a & b, For rest of 1175 records will calculate air_time based on mean.
df_nyc_flight_data.isnull().sum()
# Step 4: Missing Data Analysis
# Below task is going to perform mentioned above as conclusions.
# Analyze Dep_time and Dep_delay null values
# Conclusions: a) Dep_time : 8255, Assuming 8255 flights has cancelled due to some reasons and dropped those records from analysis.
# b) Dep_delay : 8255, Assuming 8255 flights has cancelled due to some reasons and dropped those records from analysis.
# Implementation: Used Dropna function.
df_nyc_flight_data = df_nyc_flight_data.dropna(axis=0, subset=["dep_time", "dep_delay"])
df_nyc_flight_data.isnull().sum()
# Analyze arr_delay null values
display_arr_delay_null = pd.isnull(df_nyc_flight_data["arr_delay"])
df_nyc_flight_data[display_arr_delay_null]
# Deep dive to understand one flight data which contains null values. e.g.,flight no: 464
df_nyc_flight_data_flight464 = df_nyc_flight_data[df_nyc_flight_data["flight"] == 464]
df_nyc_flight_data_flight464
# Time validation function used to calculate time. For the given dataset, if we add two time values (e.g., 1430+40 = 1470)
# but as per time, it should be 1510 and also 2340+40 = 10 early moring hours
def time_validation(hours):
num_hours = hours
minutes = num_hours % 100
print(num_hours, minutes)
if minutes > 59:
hours = num_hours - minutes
hours += 100
# print('in if:', hours)
if hours >= 2400:
hours = hours - 2400
# print('in 2400:',hours)
hours = hours + (minutes - 60)
# print('in hours+:',hours)
else:
if hours >= 2400:
hours = hours - 2400
# print('in hours>24:',hours)
return str(hours)
# print(time_validation(780))
# Fill all arr_time NULL values by adding sched_arr_time + dep_delay values.
arr_time_nulldata = df_nyc_flight_data[df_nyc_flight_data["arr_time"].isnull()]
arr_time_nulldata["arr_time"].fillna(
arr_time_nulldata["sched_arr_time"] + arr_time_nulldata["dep_delay"], inplace=True
)
arr_time_nulldata["arr_time"] = arr_time_nulldata.apply(
lambda row: time_validation(row["arr_time"]), axis=1
)
df_nyc_flight_data["arr_time"].fillna(value=arr_time_nulldata["arr_time"], inplace=True)
df_nyc_flight_data[df_nyc_flight_data["arr_time"].isnull()]
# No missing values for arr_time column
df_nyc_flight_data.isnull().sum()
# Fill all arr_delay NULL values by subtracting arr_time - sched_arr_time
df_nyc_flight_data["arr_time"] = pd.to_numeric(df_nyc_flight_data["arr_time"])
arr_delay_nulldata = df_nyc_flight_data[df_nyc_flight_data["arr_delay"].isnull()]
arr_delay_nulldata["arr_delay"].fillna(
arr_delay_nulldata["arr_time"] - arr_delay_nulldata["sched_arr_time"], inplace=True
)
arr_delay_nulldata["arr_delay"] = arr_delay_nulldata.apply(
lambda row: time_validation(row["arr_delay"]), axis=1
)
df_nyc_flight_data["arr_delay"].fillna(
value=arr_delay_nulldata["arr_delay"], inplace=True
)
df_nyc_flight_data[df_nyc_flight_data["arr_delay"].isnull()]
# No missing values for arr_delay column
df_nyc_flight_data.isnull().sum()
# Fill all air_time NULL values by subtracting arr_time - dep_time by multiplying with 65% percent of complete duration.
air_time_nulldata = df_nyc_flight_data[df_nyc_flight_data["air_time"].isnull()]
air_time_nulldata["air_time"].fillna(
value=round((air_time_nulldata["arr_time"] - air_time_nulldata["dep_time"]) * 0.65),
inplace=True,
)
air_time_nulldata["air_time"] = air_time_nulldata.apply(
lambda row: time_validation(row["air_time"]), axis=1
)
df_nyc_flight_data["air_time"].fillna(value=air_time_nulldata["air_time"], inplace=True)
df_nyc_flight_data[df_nyc_flight_data["air_time"].isnull()]
# No missing values for air_time column
df_nyc_flight_data.isnull().sum()
# findday function has created to find the day name for the given date and populate it for each and every row.
import datetime
import calendar
def findDay(date):
full_day = datetime.datetime.strptime(date, "%d-%m-%Y").weekday()
return calendar.day_name[full_day]
# date = '03-02-2019'
# print(findDay(date))
# flight_date column created to populate day name for each and every row.
df_nyc_flight_data["flight_date"] = (
df_nyc_flight_data["day"].map(str)
+ "-"
+ df_nyc_flight_data["month"].map(str)
+ "-"
+ df_nyc_flight_data["year"].map(str)
)
df_nyc_flight_data.head(1)
# day_name column created to populate day name for each and every row.
df_nyc_flight_data["day_name"] = df_nyc_flight_data.apply(
lambda row: findDay(row["flight_date"]), axis=1
)
df_nyc_flight_data.head(1)
# aircraft_speed column created to populate aircraft speed for each and every row
df_nyc_flight_data["air_time"] = pd.to_numeric(df_nyc_flight_data["air_time"])
aircraft_speed = df_nyc_flight_data["distance"] / (df_nyc_flight_data["air_time"] / 60)
df_nyc_flight_data["aircraft_speed"] = aircraft_speed
df_nyc_flight_data.head(1)
# Convert dep_time,sched_dep_time,arr_time,sched_arr_time into hh:mm time format.
df_nyc_flight_data["dep_time"] = (
df_nyc_flight_data.dep_time[~df_nyc_flight_data.dep_time.isna()]
.astype(np.int64)
.apply("{:0>4}".format)
)
df_nyc_flight_data["dep_time"] = pd.to_timedelta(
df_nyc_flight_data.dep_time.str[:2]
+ ":"
+ df_nyc_flight_data.dep_time.str[2:]
+ ":00"
)
df_nyc_flight_data["sched_dep_time"] = (
df_nyc_flight_data.sched_dep_time[~df_nyc_flight_data.sched_dep_time.isna()]
.astype(np.int64)
.apply("{:0>4}".format)
)
df_nyc_flight_data["sched_dep_time"] = pd.to_timedelta(
df_nyc_flight_data.sched_dep_time.str[:2]
+ ":"
+ df_nyc_flight_data.sched_dep_time.str[2:]
+ ":00"
)
df_nyc_flight_data["arr_time"] = (
df_nyc_flight_data.arr_time[~df_nyc_flight_data.arr_time.isna()]
.astype(np.int64)
.apply("{:0>4}".format)
)
df_nyc_flight_data["arr_time"] = pd.to_timedelta(
df_nyc_flight_data.arr_time.str[:2]
+ ":"
+ df_nyc_flight_data.arr_time.str[2:]
+ ":00"
)
df_nyc_flight_data["sched_arr_time"] = (
df_nyc_flight_data.sched_arr_time[~df_nyc_flight_data.sched_arr_time.isna()]
.astype(np.int64)
.apply("{:0>4}".format)
)
df_nyc_flight_data["sched_arr_time"] = pd.to_timedelta(
df_nyc_flight_data.sched_arr_time.str[:2]
+ ":"
+ df_nyc_flight_data.sched_arr_time.str[2:]
+ ":00"
)
df_nyc_flight_data.head(1)
# Created two new columns dep_status and arr_status.
# dep_status column used to store information based on the flight depatured before_ontime, ontime, Dep_Actualdelay information.
# arr_status column used to store information based on the flight depatured before_ontime, ontime, Arr_Actualdelay information.
df_nyc_flight_data.loc[df_nyc_flight_data.dep_delay < 0, "dep_status"] = "Before_OnTime"
df_nyc_flight_data.loc[df_nyc_flight_data.dep_delay == 0, "dep_status"] = "OnTime"
df_nyc_flight_data.loc[
df_nyc_flight_data.dep_delay > 0, "dep_status"
] = "Dep_ActualDelay"
df_nyc_flight_data["arr_delay"] = pd.to_numeric(df_nyc_flight_data["arr_delay"])
df_nyc_flight_data.loc[df_nyc_flight_data.arr_delay < 0, "arr_status"] = "Before_OnTime"
df_nyc_flight_data.loc[df_nyc_flight_data.arr_delay == 0, "arr_status"] = "OnTime"
df_nyc_flight_data.loc[
df_nyc_flight_data.arr_delay > 0, "arr_status"
] = "Arr_ActualDelay"
df_nyc_flight_data.head(1)
# Created one new column quarter to fill quarter values from time_hour date column
# Convert datatypes into datetime for the required columns
df_nyc_flight_data["flight_date"] = pd.to_datetime(df_nyc_flight_data["flight_date"])
df_nyc_flight_data["time_hour"] = pd.to_datetime(df_nyc_flight_data["time_hour"])
df_nyc_flight_data["quarter"] = df_nyc_flight_data["time_hour"].dt.quarter
# Convert datatypes into category for the required columns
df_nyc_flight_data[
[
"month",
"day",
"carrier",
"origin",
"dest",
"day_name",
"dep_status",
"arr_status",
"quarter",
]
] = df_nyc_flight_data[
[
"month",
"day",
"carrier",
"origin",
"dest",
"day_name",
"dep_status",
"arr_status",
"quarter",
]
].apply(
lambda x: x.astype("category")
)
df_nyc_flight_data.head(1)
# Verify all columns datatypes and convert it as per the data visualization requirement.
df_nyc_flight_data.dtypes
# Verify whether we have any missingvalues for all columns
df_nyc_flight_data.isnull().sum()
# To verify maximum aircraft_speed for the individual carrier.
# Observation: Interestingly found 'inf' value as maximum for carrier 'MQ'
carrier_speed = df_nyc_flight_data.groupby(["carrier"])["aircraft_speed"].max()
carrier_speed
# Observation: Interestingly found 'inf' value as maximum for carrier 'MQ'
# Interesting Facts: Noticied that, Dep_time and Arr_time for this flight '3678' is same and which should not be the case
# in real-time.
df_nyc_flight_data.sort_values(by="aircraft_speed", ascending=False).head(2)
# Observation: Interestingly found 'inf' value as maximum for carrier 'MQ'
# Interesting Facts: Noticied that, Dep_time and Arr_time for this flight '3678' is same and which should not be the case
# in real-time.
# df_nyc_flight_data_flight = df_nyc_flight_data[df_nyc_flight_data['flight'] == 3678]
df_nyc_flight_data_flight = df_nyc_flight_data[df_nyc_flight_data["air_time"] == 0]
df_nyc_flight_data_flight.head(2)
# Observation: Interestingly found 'inf' value as maximum for carrier 'MQ'
# Interesting Facts: Noticied that, Dep_time and Arr_time for this flight '3678' is same and which should not be the case
# in real-time.
# Conclusion:Dropped this flight so that, can able to analyze problem statement 'Aircraft speed analysis' for all carriers.
df_nyc_flight_data = df_nyc_flight_data.drop([259244])
df_nyc_flight_data_flight = df_nyc_flight_data[df_nyc_flight_data["flight"] == 0]
df_nyc_flight_data_flight.head(2)
# df_nyc_flight_data[df_nyc_flight_data['aircraft_speed'].isin([np.inf, -np.inf])]
# df_nyc_flight_data.at[259244,'aircraft_speed']=325.83 # mean value
# Verify whether data displays values correct or not.
carrier_speed = df_nyc_flight_data.groupby(["carrier"])["aircraft_speed"].max()
carrier_speed
# Step 5: Data Visualizations
# Problem Statement 1: Departure delays
# Calculate minimum and maximum dep_delay values for the given NYC flight data set.
min_dep_delay = min(df_nyc_flight_data.dep_delay)
print(min_dep_delay)
max_dep_delay = max(df_nyc_flight_data.dep_delay)
print(max_dep_delay)
# Visualizations Heading: Identify Departure Delay information based on Origin and Carrier
# Plot Used : Relational Plot
# Description : Fetched all records whose dep_delay > 0 and plotted graph based on Origin and Carrier
# Outcome : a) Carrier HA, Origin JFK, is the one whose maximum dep_delay is high > 1200
# b) Carrier OO, Origin LGA, is the one whose dep_delay is very less.
# c) Carrier AS, Origin EWR, is the one whose dep_delay is very less.
import seaborn as sns
df_nyc_flight_data_dep_actualdelay = df_nyc_flight_data[
df_nyc_flight_data["dep_status"] == "Dep_ActualDelay"
]
sns.relplot(
x="carrier", y="dep_delay", hue="origin", data=df_nyc_flight_data_dep_actualdelay
)
# Visualizations Heading: Identify Depature Delay information based on Origin and Day_name
# Plot Used : Categorical Plot
# Description : Fetched all records whose dep_delay > 0 and plotted graph based on Origin and Day_name.
# Outcome : a) Day: Wednesday, Origin JFK, is the one whose maximum dep_delay is around > 1200
# b) Day: Saturday, Origin JFK, is the second whose maxmimum dep_delay is around > 1100
# c) Day: Tuesday, Less number of delays on this day.
sns.catplot(
x="day_name",
y="dep_delay",
hue="origin",
jitter=False,
aspect=2,
data=df_nyc_flight_data_dep_actualdelay,
)
# Calculate total counts of flight depature delay based on dep_status == Dep_ActualDelay
depDelay_count = df_nyc_flight_data[
df_nyc_flight_data["dep_status"] == "Dep_ActualDelay"
]
depDelay_count["dep_status"].value_counts()
# Due to huge data set, Calculate top 2500 flight depature delay based on dep_status == Dep_ActualDelay by descending order
Top2500_DepDelays = depDelay_count.sort_values(by="dep_delay", ascending=False).head(
2500
)
Top2500_DepDelays["dep_status"].value_counts()
# Visualizations Heading: Identify Top 2500 Depature Delay information based on Origin and Carrier
# Plot Used : Relational Plot
# Description : Fetched all records whose dep_delay > 0 and plotted graph based on Origin and Carrier
# Outcome : a) Carrier HA, Origin JFK, is the one whose maximum dep_delay is around > 1200
# b) Carrier MQ, Origin JFK, is the second whose maxmimum dep_delay is around > 1100.
# c) Carrier AS, Origin EWR, is the one whose dep_delay is very less.
sns.relplot(x="carrier", y="dep_delay", hue="origin", data=Top2500_DepDelays, aspect=2)
# Visualizations Heading: Identify Top2500 Depature Delay information based on Origin and Day_name
# Plot Used : Categorical Plot
# Description : Fetched all records whose dep_delay > 0 and plotted graph based on Origin and Day_name.
# Outcome : a) Day: Wednesday, Origin JFK, is the one whose maximum dep_delay is around > 1200
# b) Day: Saturday, Origin JFK, is the second whose maxmimum dep_delay is around > 1100
# c) Day: Tuesday, Less number of delays on this day.
sns.catplot(
x="day_name",
y="dep_delay",
hue="origin",
jitter=False,
aspect=2,
data=Top2500_DepDelays,
)
# Problem Statement 2: Best Airport in terms of time departure %
# Calculate total flight counts, Percentage based on Origin and dep_status = OnTime
dep_OnTime = (
df_nyc_flight_data.groupby("origin")["origin"].count().reset_index(name="total")
)
OnTimeFlights = (
df_nyc_flight_data.loc[df_nyc_flight_data["dep_status"] == "OnTime"]
.groupby(["origin", "dep_status"])["dep_status"]
.count()
.unstack("dep_status")
)
dep_OnTime["OnTime"] = OnTimeFlights["OnTime"].values
dep_OnTime["percentage"] = (dep_OnTime["OnTime"] / dep_OnTime["total"]) * 100
dep_OnTime
# Calculate total flight counts, Percentage based on Origin and dep_status = Before_OnTime
dep_Before_OnTime = (
df_nyc_flight_data.groupby("origin")["origin"].count().reset_index(name="total")
)
Before_OnTimeFlights = (
df_nyc_flight_data.loc[df_nyc_flight_data["dep_status"] == "Before_OnTime"]
.groupby(["origin", "dep_status"])["dep_status"]
.count()
.unstack("dep_status")
)
dep_Before_OnTime["Before_OnTime"] = Before_OnTimeFlights["Before_OnTime"].values
dep_Before_OnTime["percentage"] = (
dep_Before_OnTime["Before_OnTime"] / dep_Before_OnTime["total"]
) * 100
dep_Before_OnTime
# Calculate total flight counts, Percentage based on Origin and dep_status = Dep_Actualdelay
dep_ActualDelay = (
df_nyc_flight_data.groupby("origin")["origin"].count().reset_index(name="total")
)
ActualDelay_Flights = (
df_nyc_flight_data.loc[df_nyc_flight_data["dep_status"] == "Dep_ActualDelay"]
.groupby(["origin", "dep_status"])["dep_status"]
.count()
.unstack("dep_status")
)
dep_ActualDelay["Dep_ActualDelay"] = ActualDelay_Flights["Dep_ActualDelay"].values
dep_ActualDelay["percentage"] = (
dep_ActualDelay["Dep_ActualDelay"] / dep_ActualDelay["total"]
) * 100
dep_ActualDelay
# Merge above all 3 dataframes and display total flight counts, Percentage based on Origin and dep_status in (<0,==0,>0)
merged_inner1 = pd.merge(
left=dep_OnTime, right=dep_Before_OnTime, left_on="origin", right_on="origin"
)
merged_inner1.shape
merged_inner_final = pd.merge(
left=merged_inner1, right=dep_ActualDelay, left_on="origin", right_on="origin"
)
merged_inner_final.shape
merged_inner_final
# Rename columns with meaning full names for the merged dataframe and display final result set.
merged_inner_final = merged_inner_final.drop(["total_y", "total"], axis=1)
merged_inner_final.rename(columns={"total_x": "total"}, inplace=True)
merged_inner_final.rename(columns={"percentage_x": "OnTime_percentage"}, inplace=True)
merged_inner_final.rename(
columns={"percentage_y": "BeforeOnTime_percentage"}, inplace=True
)
merged_inner_final.rename(
columns={"percentage": "ActualDepDelay_percentage"}, inplace=True
)
merged_inner_final
# Visualizations Heading: Identify Best origin airports on basis of time departure Percentage.
# Plot Used : Pie Plot
# Description : Pie Chart plotted based on time departure from the origin.
# Outcome : a) Pie 1: OnTime Flights Percentage based on Origin. Origin JFK is highest percentage with 37.83%
# b) Pie 2: Before_OnTime Flights Percentage based on Origin. Origin LGA is highest percentage with 36.91%
# c) Pie 3: ActualDepDelay Flights Percentage based on Origin. Origin EWR is highest percentage with 38.50%
# d) Pie 4: Total number of Flights Percentage based on Origin. Origin EWR is highest percentage with 35.80%
# Make figure and axes
fig, axs = plt.subplots(2, 2, figsize=(10, 5))
# A standard pie plot
axs[0, 0].set_title("OnTime Flights Percentage based on Origin")
axs[0, 0].pie(
merged_inner_final["OnTime_percentage"],
labels=merged_inner_final["origin"],
autopct="%1.2f%%",
shadow=True,
explode=(0.1, 0, 0),
)
axs[0, 1].set_title("Before_OnTime Flights Percentage based on Origin")
axs[0, 1].pie(
merged_inner_final["BeforeOnTime_percentage"],
labels=merged_inner_final["origin"],
autopct="%1.2f%%",
shadow=True,
explode=(0.1, 0, 0),
)
axs[1, 0].set_title("ActualDepDelay Flights Percentage based on Origin")
axs[1, 0].pie(
merged_inner_final["ActualDepDelay_percentage"],
labels=merged_inner_final["origin"],
autopct="%1.2f%%",
shadow=True,
explode=(0.1, 0, 0),
)
axs[1, 1].set_title("Total number of Flights Percentage based on Origin")
axs[1, 1].pie(
merged_inner_final["total"],
labels=merged_inner_final["origin"],
autopct="%1.2f%%",
shadow=True,
explode=(0.1, 0, 0),
)
plt.show()
# Visualizations Heading: Identify TotalNumberofFlights departure from Origin (i.e,Best Airports.)
# Plot Used : Bar Plot
# Description : Bar Chart plotted to show total number of flights departure from the origin.
# Outcome : a) Origin JFK is one of the highest number OnTime Flights timely departure
# b) Origin LGA is one of the highest number Before_OnTime Flights timely departure
# c) Origin EWR is one of the highest number Dep_ActualDelay Flights timely departure
import numpy as np
x = np.arange(len(merged_inner_final["origin"])) # the label locations
width = 0.25 # the width of the bars
fig, ax = plt.subplots(figsize=(10, 5))
rects1 = ax.bar(
x, merged_inner_final["OnTime"], width, alpha=0.5, color="#EE3224", label="OnTime"
)
rects2 = ax.bar(
[p + width for p in x],
merged_inner_final["Before_OnTime"],
width,
alpha=0.5,
color="#F78F1E",
label="Before_OnTime",
)
rects3 = ax.bar(
[p + width * 2 for p in x],
merged_inner_final["Dep_ActualDelay"],
width,
alpha=0.5,
color="#FFC222",
label="Dep_ActualDelay",
)
# Add some text for labels, title and custom x-axis tick labels, etc.
ax.set_ylabel("Total Number of Flights")
ax.set_title("TotalNumberofFlights departure from Origin i.e,Best Airports")
ax.set_xticks([p + 1.5 * width for p in x])
ax.set_xticklabels(merged_inner_final["origin"])
ax.legend()
plt.xlim(min(x) - width, max(x) + width * 4)
plt.ylim(
[
0,
max(
merged_inner_final["OnTime"]
+ merged_inner_final["Before_OnTime"]
+ merged_inner_final["Dep_ActualDelay"]
),
]
)
fig.tight_layout()
plt.grid()
plt.show()
# Visualizations Heading: Identify TotalNumberofFlights departure from Origin (i.e,Best Airports.)
# Plot Used : line Plot
# Description : line Chart plotted to show total number of flights departure from the origin.
# Outcome : a) Origin EWR,JFK,LGA is having similar number (slightly differ) OnTime Flights timely departure
# b) Before_OnTime Flights timely departure count decreases from EWR,JFK,LGA
# c) Dep_ActualDelay Flights timely departure count increases from EWR,JFK,LGA
ax = plt.gca()
merged_inner_final.plot(kind="line", x="origin", y="OnTime", color="#EE3224", ax=ax)
merged_inner_final.plot(
kind="line", x="origin", y="Before_OnTime", color="#F78F1E", ax=ax
)
merged_inner_final.plot(
kind="line", x="origin", y="Dep_ActualDelay", color="#FFC222", ax=ax
)
plt.show()
# Problem Statement 3: Aircraft speed analysis
# Visualizations Heading: Identify Aircraft_Speed based on airtime, distance and Origin.
# Plot Used : Pair Plot
# Description : Pair Plot plotted to show how the aircraft_speed increases based on air_time and distance.
# Outcome : a) Origin LGA is having high aircraft_speed which travels shorter distance.
# b) Origin LGA is having consistent aircraft_speed for distance <1800
# c) Origin EWR is having inconsistent aircraft_speed for distance <2500
import seaborn as sns
origin_speed = sns.pairplot(
df_nyc_flight_data,
height=3,
vars=["distance", "air_time", "aircraft_speed"],
hue="origin",
palette="husl",
markers=["o", "s", "D"],
)
plt.show(origin_speed)
# Calculate mean of aircraft_speed by applying group by on carrier.
carrier_speed = df_nyc_flight_data.groupby(["carrier"])["aircraft_speed"].mean()
carrier_speed
# Calculate mean of distance by applying group by on carrier.
carrier_distance = df_nyc_flight_data.groupby(["carrier"])["distance"].mean()
carrier_distance
# Merge above two dataframes to display records for Carrier, aircraft_speed and distance.
merged_inner = pd.merge(
left=carrier_speed, right=carrier_distance, left_on="carrier", right_on="carrier"
)
merged_inner.shape
merged_inner.info()
# reset index for the above dataframe records.
merged_inner.reset_index(level=0, drop=False, inplace=True)
merged_inner.head(1)
# Visualizations Heading: Identify Aircraft_Speed based on distance and Origin.
# Plot Used : Scatter Plot
# Description : Scatter Plot plotted to show how the aircraft_speed increases based on distance for carrier.
# Outcome : a) Carrier HA is having high aircraft_speed for the distance covered around 5000
# b) Carrier YU is having low aircraft_speed for the distance covered around 400
# c) Most of the Carrier is having consistent aircraft_speed for the similar distance covered.
f, ax = plt.subplots(figsize=(6.5, 6.5))
sns.set(style="ticks")
sns.despine(f, left=True, bottom=True)
sns.scatterplot(
x="distance",
y="aircraft_speed",
hue="carrier",
palette="dark",
sizes=(1, 8),
linewidth=0,
data=merged_inner,
ax=ax,
)
# Problem Statement 4: On time arrival % analysis
# Calculate count of OnTime,Before_OnTime,Arr_ActualDelay arrival count by grouping with arr_status column
arr_Status = df_nyc_flight_data.groupby("arr_status")["arr_status"].count()
print("arr_Status complete list:")
print(arr_Status)
# Visualizations Heading: Visualize the Ontime arrival percentage and count.
# Plot Used : Count Plot and Pie Plot
# Description : Count Plot plotted to show the count of number of flights based on OnTime,Before_OnTime,Arr_ActualDelay arrival status.
# : Pie Plot plotted to show the percentage of number of flights arrivals based on OnTime,Before_OnTime,Arr_ActualDelay arrival status.
# Outcome : a) Around 189038 number of flights has arrived Before_OnTime with 57.54%
# b) Around 134057 number of flights has arrived Arr_ActualDelay with 40.81%
# c) Around 5425 number of flights has arrived OnTime with 1.65%
# Setting up the chart area
f, ax = plt.subplots(1, 2, figsize=(14, 7))
# setting up chart
df_nyc_flight_data["arr_status"].value_counts().plot.pie(
explode=[0, 0, 0.2], autopct="%1.2f%%", ax=ax[1], shadow=False
)
# setting title for pei chart
ax[1].set_title("On time arrival % Analysis")
ax[1].set_ylabel("")
# setting up data for barchart
sns.countplot(
"arr_status",
order=df_nyc_flight_data["arr_status"].value_counts().index,
data=df_nyc_flight_data,
ax=ax[0],
)
ax[0].set_title("Arrival Status of total flights (in numbers)")
ax[0].set_ylabel("Number of Flights")
plt.show()
# Problem Statement 5: Maximum number of flights headed to some particular destination.
# Calculate maximum number of flights to particular destination.
df_nyc_flight_data["dest"].value_counts()
# Calculate maximum number of flights to started from particular origin.
df_nyc_flight_data["origin"].value_counts()
# Calculate Top 25 Maximum number of flights from origin to destination by applying group by on origin and dest.
Top25_MaxFlights_Dest = (
df_nyc_flight_data.groupby("origin")["dest"].value_counts().to_frame()
)
# Top25_MaxFlights_Dest.dtypes
Top25_MaxFlights_Dest.rename(columns={"dest": "dest_count"}, inplace=True)
Top25_MaxFlights_Dest.reset_index(level=1, drop=False, inplace=True)
Top25_MaxFlights_Dest.reset_index(level=0, drop=False, inplace=True)
# sum_flight1.head(40)
Top25_MaxFlights_Dest = Top25_MaxFlights_Dest.sort_values(
by="dest_count", ascending=False
).head(25)
Top25_MaxFlights_Dest.reset_index(level=0, drop=False, inplace=True)
Top25_MaxFlights_Dest.head(3)
# Visualizations Heading: Visualize Top 25 Maximum number of flights headed to some particular destination.
# Plot Used : Relational Plot
# Description : Relational Plot plotted to show Top 25 Maximum number of flights headed to some particular destination.
# Outcome : a) Top 1 : Origin JFK is having maximum number of flights headed towards LAX destination
# b) Top 10 : Origin EWR is having maximum number of flights headed towards BOS destination
# c) Top 25 : Origin EWR is having maximum number of flights headed towards FLL destination
palette = sns.cubehelix_palette(light=0.8, n_colors=3)
sns.relplot(
x="dest",
y="dest_count",
hue="origin",
height=6,
aspect=3,
data=Top25_MaxFlights_Dest,
)
# Visualizations Heading: Visualize Top 25 Maximum number of flights headed to some particular destination.
# Plot Used : Catergorical Plot
# Description : Catergorical Plot plotted to show Top 25 Maximum number of flights headed to some particular destination.
# Outcome : a) Top 1 : Origin JFK is having maximum number of flights headed towards LAX destination
# b) Top 10 : Origin EWR is having maximum number of flights headed towards BOS destination
# c) Top 25 : Origin EWR is having maximum number of flights headed towards FLL destination
g = sns.catplot(
x="dest",
y="dest_count",
hue="origin",
data=Top25_MaxFlights_Dest,
height=5,
kind="bar",
palette="muted",
aspect=4,
)
g.despine(left=True)
g.set_ylabels("Total Number of Flights")
# Problem Statement 6: Month-Wise analysis of Flight Departure and Arrival Status.
# Visualizations Heading: Visualize Month-Wise mean analysis about the Flight Departure and Arrival Status.
# Plot Used : line Plot
# Description : line Plot plotted to show month-wise analysis dep_delay and arr_delay .
# Outcome : a) left-side line plot shows 3,5,9 month is having earlier arrivals and 4,9 month is having earlier departure.
# b) right-side line plot shows 4,6,7 month is having arrivals delays and 4,6,7 month is having departure delays.
f, ax = plt.subplots(1, 2, figsize=(20, 8))
dep_Ontime = df_nyc_flight_data[df_nyc_flight_data["dep_status"] != "Dep_ActualDelay"]
dep_Ontime[["month", "arr_delay", "dep_delay"]].groupby(["month"]).mean().plot(
ax=ax[0],
marker="*",
linestyle="dashed",
color="b" + "r",
linewidth=2,
markersize=12,
)
df_nyc_flight_data_dep_actualdelay[["month", "arr_delay", "dep_delay"]].groupby(
["month"]
).mean().plot(
ax=ax[1],
marker="*",
linestyle="dashed",
color="b" + "r",
linewidth=2,
markersize=12,
)
# Problem Statement 7: Quarter-Wise analysis of Flight Mean Depature Delays by Carriers.
# Visualizations Heading: Visualize Quarter-Wise analysis of Flight mean Depature Delays by Carriers.
# Plot Used : Heat Map
# Description : Heat Map plotted to show Quarter-Wise analysis of Flight mean Depature Delays by Carriers.
# Outcome : a) Quarter 1: Carrier EV is having highest (24.3) & Carrier US is having lowest (2.7) mean depature delay.
# b) Quarter 2: Carrier F9 is having highest (26.6) & Carrier HA is having lowest (0.3) mean depature delay.
# c) Quarter 3: Carrier FL is having highest (21.9) & Carrier AS is having lowest (5.1) mean depature delay.
# d) Quarter 4: Carrier FL is having highest (21.7) & Carrier OO is having lowest (0.8) mean depature delay.
import seaborn as sns
plt.figure(figsize=(18, 10))
plt.title("Quarter-Wise flight depature delays by carriers")
plt.tight_layout()
hmap = pd.pivot_table(
df_nyc_flight_data,
values="dep_delay",
aggfunc="mean",
index="carrier",
columns="quarter",
)
sns.heatmap(hmap, annot=True, cmap="YlGnBu", center=0, linewidths=0.2, fmt="g")
plt.show()
| false | 0 | 10,442 | 0 | 10,464 | 10,442 |
||
69401993
|
<jupyter_start><jupyter_text>MLB_DATA
Kaggle dataset identifier: mlb-data
<jupyter_code>import pandas as pd
df = pd.read_csv('mlb-data/target.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 2506176 entries, 0 to 2506175
Data columns (total 6 columns):
# Column Dtype
--- ------ -----
0 playerId int64
1 target1 float64
2 target2 float64
3 target3 float64
4 target4 float64
5 EvalDate object
dtypes: float64(4), int64(1), object(1)
memory usage: 114.7+ MB
<jupyter_text>Examples:
{
"playerId": 628317,
"target1": 0.011167070500000001,
"target2": 4.4747081712,
"target3": 0.0051677297,
"target4": 5.7352941176000005,
"EvalDate": "2018-01-02"
}
{
"playerId": 547989,
"target1": 0.0429932216,
"target2": 5.5933852139999996,
"target3": 0.0450330735,
"target4": 2.7941176471,
"EvalDate": "2018-01-02"
}
{
"playerId": 519317,
"target1": 0.9743269048000001,
"target2": 56.1770428016,
"target3": 13.6937455705,
"target4": 64.1666666667,
"EvalDate": "2018-01-02"
}
{
"playerId": 607625,
"target1": 0.0067002423,
"target2": 2.6750972763,
"target3": 0.0051677297,
"target4": 1.862745098,
"EvalDate": "2018-01-02"
}
<jupyter_script>#
# ## About Dataset
"""
import gc
import numpy as np
import pandas as pd
from pathlib import Path
from pandarallel import pandarallel
pandarallel.initialize()
BASE_DIR = Path('../input/mlb-player-digital-engagement-forecasting')
train = pd.read_csv(BASE_DIR / 'train.csv')
null = np.nan
true = True
false = False
for col in train.columns:
if col == 'date': continue
_index = train[col].notnull()
train.loc[_index, col] = train.loc[_index, col].parallel_apply(lambda x: eval(x))
outputs = []
for index, date, record in train.loc[_index, ['date', col]].itertuples():
_df = pd.DataFrame(record)
_df['index'] = index
_df['date'] = date
outputs.append(_df)
outputs = pd.concat(outputs).reset_index(drop=True)
outputs.to_csv(f'{col}_train.csv', index=False)
outputs.to_pickle(f'{col}_train.pkl')
del outputs
del train[col]
gc.collect()
"""
# ## Training
import numpy as np
import pandas as pd
from pathlib import Path
from sklearn.metrics import mean_absolute_error
from datetime import timedelta
from functools import reduce
from tqdm import tqdm
import lightgbm as lgbm
import mlb
import os
import pickle
BASE_DIR = Path("../input/mlb-player-digital-engagement-forecasting")
TRAIN_DIR = Path("../input/mlb-pdef-train-dataset")
players = pd.read_csv(BASE_DIR / "players.csv")
rosters = pd.read_pickle(TRAIN_DIR / "rosters_train.pkl")
targets = pd.read_pickle(TRAIN_DIR / "nextDayPlayerEngagement_train.pkl")
scores = pd.read_pickle(TRAIN_DIR / "playerBoxScores_train.pkl")
scores = scores.groupby(["playerId", "date"]).sum().reset_index()
targets_cols = ["playerId", "target1", "target2", "target3", "target4", "date"]
players_cols = ["playerId", "primaryPositionName"]
rosters_cols = ["playerId", "teamId", "status", "date"]
scores_cols = [
"playerId",
"battingOrder",
"gamesPlayedBatting",
"flyOuts",
"groundOuts",
"runsScored",
"doubles",
"triples",
"homeRuns",
"strikeOuts",
"baseOnBalls",
"intentionalWalks",
"hits",
"hitByPitch",
"caughtStealing",
"stolenBases", #'atBats',
"groundIntoDoublePlay",
"groundIntoTriplePlay",
"plateAppearances",
"totalBases", #'rbi',
"leftOnBase",
"sacBunts", # 'sacFlies', 'catchersInterference',
"pickoffs",
"gamesPlayedPitching",
"gamesStartedPitching",
"completeGamesPitching",
"shutoutsPitching",
"winsPitching",
"airOutsPitching", # lossesPitching', #'flyOutsPitching', ', ###
"groundOutsPitching",
"runsPitching",
"doublesPitching",
"triplesPitching",
"homeRunsPitching",
"strikeOutsPitching",
"baseOnBallsPitching",
"intentionalWalksPitching",
"hitsPitching",
"hitByPitchPitching",
"atBatsPitching",
"caughtStealingPitching",
"stolenBasesPitching",
"inningsPitched",
"saveOpportunities",
"earnedRuns",
"battersFaced",
"outsPitching",
"pitchesThrown",
"balls",
"strikes",
"hitBatsmen",
"balks",
"wildPitches",
"pickoffsPitching",
"rbiPitching",
"gamesFinishedPitching",
"inheritedRunners",
"inheritedRunnersScored",
"catchersInterferencePitching",
"sacBuntsPitching",
"sacFliesPitching",
"saves", #'holds', 'blownSaves',
"assists",
"putOuts",
"errors",
"chances",
"date",
]
feature_cols = [
"label_playerId", #'label_primaryPositionName',# 'label_teamId',
"battingOrder",
"gamesPlayedBatting", #'flyOuts',
"label_status", # '〇label_status',
"groundOuts",
"runsScored",
"doubles",
"triples",
"homeRuns",
"strikeOuts",
"baseOnBalls",
"intentionalWalks",
"hits",
"hitByPitch",
"caughtStealing",
"stolenBases", #'atBats',
"groundIntoDoublePlay", # ○'atBats', ' 'groundIntoDoublePlay',
"groundIntoTriplePlay",
"plateAppearances",
"totalBases", #'rbi',
"leftOnBase", #'sacBunts', #'sacFlies', 'catchersInterference', #'leftOnBase', '
"pickoffs",
"gamesPlayedPitching",
"gamesStartedPitching",
"completeGamesPitching",
"shutoutsPitching",
"winsPitching",
#'lossesPitching', 'flyOutsPitching', 'airOutsPitching',###
"groundOutsPitching",
"runsPitching",
"doublesPitching",
"triplesPitching",
"homeRunsPitching",
"strikeOutsPitching",
"baseOnBallsPitching",
"hitsPitching",
"intentionalWalksPitching",
"hitByPitchPitching",
"atBatsPitching",
"caughtStealingPitching",
"stolenBasesPitching",
"inningsPitched",
"saveOpportunities",
"earnedRuns",
"battersFaced",
"outsPitching",
"pitchesThrown",
"balls",
"strikes",
"hitBatsmen", #'wildPitches',
"pickoffsPitching",
"balks", # ○'pickoffsPitching',
"rbiPitching",
"gamesFinishedPitching",
"inheritedRunners",
"inheritedRunnersScored",
"catchersInterferencePitching",
"sacBuntsPitching",
"saves",
"sacFliesPitching", #'holds', 'blownSaves',○'sacBuntsPitching', ○'sacFliesPitching' ○ 'sacFliesPitching'
"assists",
"putOuts", #'errors',
"chances",
"target1_mean",
"target1_median",
"target1_std",
"target1_min",
"target1_max",
"target1_prob",
"target2_mean",
"target2_median",
"target2_std",
"target2_min",
"target2_max",
"target2_prob",
"target3_mean",
"target3_median",
"target3_std",
"target3_min",
"target3_max",
"target3_prob",
"target4_mean",
"target4_median",
"target4_std",
"target4_min",
"target4_max",
"target4_prob",
] # 86
feature_cols2 = [
"label_playerId",
"label_primaryPositionName",
"label_teamId", # needed
"label_status",
"battingOrder",
"gamesPlayedBatting",
"flyOuts",
"groundOuts",
"runsScored",
"doubles",
"triples",
"homeRuns",
"strikeOuts",
"baseOnBalls",
"hits",
"hitByPitch",
"intentionalWalks", # ○'intentionalWalks'
"caughtStealing",
"stolenBases",
"groundIntoDoublePlay", #'atBats',#'〇atBats
"groundIntoTriplePlay",
"plateAppearances",
"totalBases", # 'rbi',#○ rbi
"leftOnBase",
"sacBunts", # 'sacFlies', 'catchersInterference',#', 〇leftOnBase'
"gamesPlayedPitching",
"gamesStartedPitching",
"pickoffs", # 〇'pickoffs'
"completeGamesPitching",
"shutoutsPitching",
"winsPitching",
#'airOutsPitching',# lossesPitching', #'flyOutsPitching', '',##〇
"groundOutsPitching",
"runsPitching",
"doublesPitching",
"triplesPitching",
"homeRunsPitching",
"strikeOutsPitching",
"baseOnBallsPitching",
"hitsPitching",
"intentionalWalksPitching",
"hitByPitchPitching",
"atBatsPitching",
"caughtStealingPitching",
"stolenBasesPitching",
"inningsPitched",
"saveOpportunities",
"earnedRuns",
"battersFaced",
"outsPitching",
"pitchesThrown",
"balls",
"strikes",
"hitBatsmen",
"wildPitches",
"pickoffsPitching", #'balks',
"rbiPitching",
"gamesFinishedPitching",
"inheritedRunners",
"inheritedRunnersScored", #'catchersInterferencePitching',
"sacFliesPitching",
"saves",
"sacBuntsPitching", #'holds', 'blownSaves' #〇sacBuntsPitching'
"assists",
"putOuts",
"errors",
"chances",
"target1_mean",
"target1_median",
"target1_std",
"target1_min",
"target1_max",
"target1_prob",
"target2_mean",
"target2_median",
"target2_std",
"target2_min",
"target2_max",
"target2_prob",
"target3_mean",
"target3_median",
"target3_std",
"target3_min",
"target3_max",
"target3_prob",
"target4_mean",
"target4_median",
"target4_std",
"target4_min",
"target4_max",
"target4_prob",
"target1",
] # 91
player_target_stats = pd.read_csv(
"../input/player-target-stats/player_target_stats.csv"
)
data_names = player_target_stats.columns.values.tolist()
data_names
# creat dataset
train = targets[targets_cols].merge(players[players_cols], on=["playerId"], how="left")
train = train.merge(rosters[rosters_cols], on=["playerId", "date"], how="left")
train = train.merge(scores[scores_cols], on=["playerId", "date"], how="left")
train = train.merge(
player_target_stats, how="inner", left_on=["playerId"], right_on=["playerId"]
)
# label encoding
player2num = {c: i for i, c in enumerate(train["playerId"].unique())}
position2num = {c: i for i, c in enumerate(train["primaryPositionName"].unique())}
teamid2num = {c: i for i, c in enumerate(train["teamId"].unique())}
status2num = {c: i for i, c in enumerate(train["status"].unique())}
train["label_playerId"] = train["playerId"].map(player2num)
train["label_primaryPositionName"] = train["primaryPositionName"].map(position2num)
train["label_teamId"] = train["teamId"].map(teamid2num)
train["label_status"] = train["status"].map(status2num)
train_X = train[feature_cols]
train_y = train[["target1", "target2", "target3", "target4"]]
_index = train["date"] < 20210401
x_train1 = train_X.loc[_index].reset_index(drop=True)
y_train1 = train_y.loc[_index].reset_index(drop=True)
x_valid1 = train_X.loc[~_index].reset_index(drop=True)
y_valid1 = train_y.loc[~_index].reset_index(drop=True)
train_X = train[feature_cols2]
train_y = train[["target1", "target2", "target3", "target4"]]
_index = train["date"] < 20210401
x_train2 = train_X.loc[_index].reset_index(drop=True)
y_train2 = train_y.loc[_index].reset_index(drop=True)
x_valid2 = train_X.loc[~_index].reset_index(drop=True)
y_valid2 = train_y.loc[~_index].reset_index(drop=True)
train_X
# def fit_lgbm(x_train, y_train, x_valid, y_valid, params: dict=None, verbose=100):
# oof_pred = np.zeros(len(y_valid), dtype=np.float32)
# model = lgbm.LGBMRegressor(**params)
# model.fit(x_train, y_train,
# eval_set=[(x_valid, y_valid)],
# early_stopping_rounds=verbose,
# verbose=verbose)
# oof_pred = model.predict(x_valid)
# score = mean_absolute_error(oof_pred, y_valid)
# print('mae:', score)
# return oof_pred, model, score
# # training lightgbm
# params1 = {'objective':'mae',
# 'reg_alpha': 0.14947461820098767,
# 'reg_lambda': 0.10185644384043743,
# 'n_estimators': 3633,
# 'learning_rate': 0.08046301304430488,
# 'num_leaves': 674,
# 'feature_fraction': 0.9101240539122566,
# 'bagging_fraction': 0.9884451442950513,
# 'bagging_freq': 8,
# 'min_child_samples': 51}
# params2 = {
# 'objective':'mae',
# 'reg_alpha': 0.1,
# 'reg_lambda': 0.1,
# 'n_estimators': 80,
# 'learning_rate': 0.1,
# 'random_state': 42,
# "num_leaves": 22
# }
# params4 = {'objective':'mae',
# 'reg_alpha': 0.016468100279441976,
# 'reg_lambda': 0.09128335764019105,
# 'n_estimators': 9868,
# 'learning_rate': 0.10528150510326864,
# 'num_leaves': 157,
# 'feature_fraction': 0.5419185713426886,
# 'bagging_fraction': 0.2637405128936662,
# 'bagging_freq': 19,
# 'min_child_samples': 71}
# params = {
# 'objective':'mae',
# 'reg_alpha': 0.1,
# 'reg_lambda': 0.1,
# 'n_estimators': 10000,
# 'learning_rate': 0.1,
# 'random_state': 42,
# "num_leaves": 100
# }
# oof1, model1, score1 = fit_lgbm(
# x_train1, y_train1['target1'],
# x_valid1, y_valid1['target1'],
# params1
# )
# oof2, model2, score2 = fit_lgbm(
# x_train2, y_train2['target2'],
# x_valid2, y_valid2['target2'],
# params2
# )
# oof3, model3, score3 = fit_lgbm(
# x_train2, y_train2['target3'],
# x_valid2, y_valid2['target3'],
# params
# )
# oof4, model4, score4 = fit_lgbm(
# x_train2, y_train2['target4'],
# x_valid2, y_valid2['target4'],
# params4
# )
# score = (score1+score2+score3+score4) / 4
# print(f'score: {score}')
# import pickle
# from catboost import CatBoostRegressor
# def fit_lgbm(x_train, y_train, x_valid, y_valid, params: dict=None, verbose=100):
# oof_pred_lgb = np.zeros(len(y_valid), dtype=np.float32)
# oof_pred_cat = np.zeros(len(y_valid), dtype=np.float32)
# # if os.path.isfile(f'../input/mlb-lgbm-and-catboost-models/model_lgb_{target}.pkl'):
# # with open(f'../input/mlb-lgbm-and-catboost-models/model_lgb_{target}.pkl', 'rb') as fin:
# # model = pickle.load(fin)
# # else:
# model = lgbm.LGBMRegressor(**params)
# model.fit(x_train, y_train,
# eval_set=[(x_valid, y_valid)],
# early_stopping_rounds=verbose,
# verbose=verbose)
# # with open(f'model_lgb_{target}.pkl', 'wb') as handle:
# # pickle.dump(model, handle, protocol=pickle.HIGHEST_PROTOCOL)
# oof_pred_lgb = model.predict(x_valid)
# score_lgb = mean_absolute_error(oof_pred_lgb, y_valid)
# print('mae:', score_lgb)
# # if os.path.isfile(f'../input/mlb-lgbm-and-catboost-models/model_cb_{target}.pkl'):
# # with open(f'../input/mlb-lgbm-and-catboost-models/model_cb_{target}.pkl', 'rb') as fin:
# # model_cb = pickle.load(fin)
# # else:
# model_cb = CatBoostRegressor(
# n_estimators=2000,
# learning_rate=0.05,
# loss_function='MAE',
# eval_metric='MAE',
# max_bin=50,
# subsample=0.9,
# colsample_bylevel=0.5,
# verbose=100)
# model_cb.fit(x_train, y_train, use_best_model=True,
# eval_set=(x_valid, y_valid),
# early_stopping_rounds=25)
# # with open(f'model_cb_{target}.pkl', 'wb') as handle:
# # pickle.dump(model_cb, handle, protocol=pickle.HIGHEST_PROTOCOL)
# oof_pred_cat = model_cb.predict(x_valid)
# score_cat = mean_absolute_error(oof_pred_cat, y_valid)
# print('mae:', score_cat)
# return oof_pred_lgb, model, oof_pred_cat, model_cb, score_lgb, score_cat
# # training lightgbm
# params = {
# 'boosting_type': 'gbdt',
# 'objective':'mae',
# 'subsample': 0.5,
# 'subsample_freq': 1,
# 'learning_rate': 0.03,
# 'num_leaves': 2**11-1,
# 'min_data_in_leaf': 2**12-1,
# 'feature_fraction': 0.5,
# 'max_bin': 100,
# 'n_estimators': 2500,
# 'boost_from_average': False,
# "random_seed":42,
# }
# oof_pred_lgb2, model_lgb2, oof_pred_cat2, model_cb2, score_lgb2, score_cat2 = fit_lgbm(
# x_train1, y_train1['target2'],
# x_valid1, y_valid1['target2'],
# params
# )
# oof_pred_lgb1, model_lgb1, oof_pred_cat1, model_cb1, score_lgb1, score_cat1 = fit_lgbm(
# x_train1, y_train1['target1'],
# x_valid1, y_valid1['target1'],
# params
# )
# oof_pred_lgb3, model_lgb3, oof_pred_cat3, model_cb3, score_lgb3, score_cat3 = fit_lgbm(
# x_train1, y_train1['target3'],
# x_valid1, y_valid1['target3'],
# params
# )
# oof_pred_lgb4, model_lgb4, oof_pred_cat4, model_cb4, score_lgb4, score_cat4= fit_lgbm(
# x_train1, y_train1['target4'],
# x_valid1, y_valid1['target4'],
# params
# )
# score = (score_lgb1+score_lgb2+score_lgb3+score_lgb4) / 4
# print(f'LightGBM score: {score}')
# score = (score_cat1+score_cat2+score_cat3+score_cat4) / 4
# print(f'Catboost score: {score}')
with open("../input/modellgbcb/model_lgb_cb/model1.pickle", mode="rb") as f:
model1 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model2.pickle", mode="rb") as f:
model2 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model3.pickle", mode="rb") as f:
model3 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model4.pickle", mode="rb") as f:
model4 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model_lgb1.pickle", mode="rb") as f:
model_lgb1 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model_lgb2.pickle", mode="rb") as f:
model_lgb2 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model_lgb3.pickle", mode="rb") as f:
model_lgb3 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model_lgb4.pickle", mode="rb") as f:
model_lgb4 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model_cb1.pickle", mode="rb") as f:
model_cb1 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model_cb2.pickle", mode="rb") as f:
model_cb2 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model_cb3.pickle", mode="rb") as f:
model_cb3 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model_cb4.pickle", mode="rb") as f:
model_cb4 = pickle.load(f)
players_cols = ["playerId", "primaryPositionName"]
rosters_cols = ["playerId", "teamId", "status"]
scores_cols = [
"playerId",
"battingOrder",
"gamesPlayedBatting",
"flyOuts",
"groundOuts",
"runsScored",
"doubles",
"triples",
"homeRuns",
"strikeOuts",
"baseOnBalls",
"intentionalWalks",
"hits",
"hitByPitch",
"caughtStealing",
"stolenBases", #'atBats',
"groundIntoDoublePlay",
"groundIntoTriplePlay",
"plateAppearances",
"totalBases", #'rbi',
"leftOnBase",
"sacBunts", # 'sacFlies', 'catchersInterference',
"pickoffs",
"gamesPlayedPitching",
"gamesStartedPitching",
"completeGamesPitching",
"shutoutsPitching",
"winsPitching",
"airOutsPitching", #'lossesPitching', 'flyOutsPitching',
"groundOutsPitching",
"runsPitching",
"doublesPitching",
"triplesPitching",
"homeRunsPitching",
"strikeOutsPitching",
"baseOnBallsPitching",
"intentionalWalksPitching",
"hitsPitching",
"hitByPitchPitching",
"atBatsPitching",
"caughtStealingPitching",
"stolenBasesPitching",
"inningsPitched",
"saveOpportunities",
"earnedRuns",
"battersFaced",
"outsPitching",
"pitchesThrown",
"balls",
"strikes",
"hitBatsmen",
"balks",
"wildPitches",
"pickoffsPitching",
"rbiPitching",
"gamesFinishedPitching",
"inheritedRunners",
"inheritedRunnersScored",
"catchersInterferencePitching",
"sacBuntsPitching",
"sacFliesPitching",
"saves", #'holds', 'blownSaves',
"assists",
"putOuts",
"errors",
"chances",
]
null = np.nan
true = True
false = False
import pandas as pd
import numpy as np
from datetime import timedelta
from tqdm import tqdm
import gc
from functools import reduce
from sklearn.model_selection import StratifiedKFold
ROOT_DIR = "../input/mlb-player-digital-engagement-forecasting"
# =======================#
def flatten(df, col):
du = (
df.pivot(index="playerId", columns="EvalDate", values=col)
.add_prefix(f"{col}_")
.rename_axis(None, axis=1)
.reset_index()
)
return du
# ============================#
def reducer(left, right):
return left.merge(right, on="playerId")
# ========================
TGTCOLS = ["target1", "target2", "target3", "target4"]
def train_lag(df, lag=1):
dp = df[["playerId", "EvalDate"] + TGTCOLS].copy()
dp["EvalDate"] = dp["EvalDate"] + timedelta(days=lag)
df = df.merge(dp, on=["playerId", "EvalDate"], suffixes=["", f"_{lag}"], how="left")
return df
# =================================
def test_lag(sub):
sub["playerId"] = sub["date_playerId"].apply(lambda s: int(s.split("_")[1]))
assert sub.date.nunique() == 1
dte = sub["date"].unique()[0]
eval_dt = pd.to_datetime(dte, format="%Y%m%d")
dtes = [eval_dt + timedelta(days=-k) for k in LAGS]
mp_dtes = {eval_dt + timedelta(days=-k): k for k in LAGS}
sl = LAST.loc[
LAST.EvalDate.between(dtes[-1], dtes[0]), ["EvalDate", "playerId"] + TGTCOLS
].copy()
sl["EvalDate"] = sl["EvalDate"].map(mp_dtes)
du = [flatten(sl, col) for col in TGTCOLS]
du = reduce(reducer, du)
return du, eval_dt
#
# ===============
tr = pd.read_csv("../input/mlb-data/target.csv")
print(tr.shape)
gc.collect()
tr["EvalDate"] = pd.to_datetime(tr["EvalDate"])
tr["EvalDate"] = tr["EvalDate"] + timedelta(days=-1)
tr["EvalYear"] = tr["EvalDate"].dt.year
MED_DF = tr.groupby(["playerId", "EvalYear"])[TGTCOLS].median().reset_index()
MEDCOLS = ["tgt1_med", "tgt2_med", "tgt3_med", "tgt4_med"]
MED_DF.columns = ["playerId", "EvalYear"] + MEDCOLS
LAGS = list(range(1, 21))
FECOLS = [f"{col}_{lag}" for lag in reversed(LAGS) for col in TGTCOLS]
for lag in tqdm(LAGS):
tr = train_lag(tr, lag=lag)
gc.collect()
# ===========
tr = tr.sort_values(by=["playerId", "EvalDate"])
print(tr.shape)
tr = tr.dropna()
print(tr.shape)
tr = tr.merge(MED_DF, on=["playerId", "EvalYear"])
gc.collect()
X = tr[FECOLS + MEDCOLS].values
y = tr[TGTCOLS].values
cl = tr["playerId"].values
NFOLDS = 12
skf = StratifiedKFold(n_splits=NFOLDS)
folds = skf.split(X, cl)
folds = list(folds)
import tensorflow as tf
import tensorflow.keras.layers as L
import tensorflow.keras.models as M
from sklearn.metrics import mean_absolute_error, mean_squared_error
from tensorflow.keras.callbacks import ModelCheckpoint, ReduceLROnPlateau, EarlyStopping
tf.random.set_seed(777)
def make_model(n_in):
inp = L.Input(name="inputs", shape=(n_in,))
x = L.Dense(50, activation="relu", name="d1")(inp)
x = L.Dense(50, activation="relu", name="d2")(x)
preds = L.Dense(4, activation="linear", name="preds")(x)
model = M.Model(inp, preds, name="ANN")
model.compile(loss="mean_absolute_error", optimizer="adam")
return model
net = make_model(X.shape[1])
print(net.summary())
oof = np.zeros(y.shape)
nets = []
for idx in range(NFOLDS):
print("FOLD:", idx)
tr_idx, val_idx = folds[idx]
ckpt = ModelCheckpoint(
f"w{idx}.h5", monitor="val_loss", verbose=1, save_best_only=True, mode="min"
)
reduce_lr = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, min_lr=0.0005
)
es = EarlyStopping(monitor="val_loss", patience=6)
reg = make_model(X.shape[1])
reg.fit(
X[tr_idx],
y[tr_idx],
epochs=10,
batch_size=35_000,
validation_data=(X[val_idx], y[val_idx]),
verbose=1,
callbacks=[ckpt, reduce_lr, es],
)
reg.load_weights(f"w{idx}.h5")
oof[val_idx] = reg.predict(X[val_idx], batch_size=50_000, verbose=1)
nets.append(reg)
gc.collect()
mae = mean_absolute_error(y, oof)
mse = mean_squared_error(y, oof, squared=False)
print("mae:", mae)
print("mse:", mse)
# Historical information to use in prediction time
bound_dt = pd.to_datetime("2021-01-01")
LAST = tr.loc[tr.EvalDate > bound_dt].copy()
LAST_MED_DF = MED_DF.loc[MED_DF.EvalYear == 2021].copy()
LAST_MED_DF.drop("EvalYear", axis=1, inplace=True)
del tr
# """
import mlb
FE = []
SUB = []
#
#
import copy
env = mlb.make_env() # initialize the environment
iter_test = env.iter_test() # iterator which loops over each date in test set
for test_df, sample_prediction_df in iter_test: # make predictions here
sub = copy.deepcopy(sample_prediction_df.reset_index())
sample_prediction_df = copy.deepcopy(sample_prediction_df.reset_index(drop=True))
# LGBM summit
# creat dataset
sample_prediction_df["playerId"] = sample_prediction_df["date_playerId"].map(
lambda x: int(x.split("_")[1])
)
# Dealing with missing values
if test_df["rosters"].iloc[0] == test_df["rosters"].iloc[0]:
test_rosters = pd.DataFrame(eval(test_df["rosters"].iloc[0]))
else:
test_rosters = pd.DataFrame({"playerId": sample_prediction_df["playerId"]})
for col in rosters.columns:
if col == "playerId":
continue
test_rosters[col] = np.nan
if test_df["playerBoxScores"].iloc[0] == test_df["playerBoxScores"].iloc[0]:
test_scores = pd.DataFrame(eval(test_df["playerBoxScores"].iloc[0]))
else:
test_scores = pd.DataFrame({"playerId": sample_prediction_df["playerId"]})
for col in scores.columns:
if col == "playerId":
continue
test_scores[col] = np.nan
test_scores = test_scores.groupby("playerId").sum().reset_index()
test = sample_prediction_df[["playerId"]].copy()
test = test.merge(players[players_cols], on="playerId", how="left")
test = test.merge(test_rosters[rosters_cols], on="playerId", how="left")
test = test.merge(test_scores[scores_cols], on="playerId", how="left")
test = test.merge(
player_target_stats, how="inner", left_on=["playerId"], right_on=["playerId"]
)
test["label_playerId"] = test["playerId"].map(player2num)
test["label_primaryPositionName"] = test["primaryPositionName"].map(position2num)
test["label_teamId"] = test["teamId"].map(teamid2num)
test["label_status"] = test["status"].map(status2num)
test_X = test[feature_cols]
# predict
pred1 = model1.predict(test_X)
pred_lgd1 = model_lgb1.predict(test_X)
pred_lgd2 = model_lgb2.predict(test_X)
pred_lgd3 = model_lgb3.predict(test_X)
pred_lgd4 = model_lgb4.predict(test_X)
pred_cat1 = model_cb1.predict(test_X)
pred_cat2 = model_cb2.predict(test_X)
pred_cat3 = model_cb3.predict(test_X)
pred_cat4 = model_cb4.predict(test_X)
test["target1"] = np.clip(pred1, 0, 100)
test_X = test[feature_cols2]
# predict2
pred2 = model2.predict(test_X)
pred3 = model3.predict(test_X)
pred4 = model4.predict(test_X)
# merge submission
sample_prediction_df["target1"] = (
1.00 * np.clip(pred1, 0, 100)
+ 0.00 * np.clip(pred_lgd1, 0, 100)
+ 0.00 * np.clip(pred_cat1, 0, 100)
)
sample_prediction_df["target2"] = (
0.10 * np.clip(pred2, 0, 100)
+ 0.65 * np.clip(pred_lgd2, 0, 100)
+ 0.25 * np.clip(pred_cat2, 0, 100)
)
sample_prediction_df["target3"] = (
0.65 * np.clip(pred3, 0, 100)
+ 0.25 * np.clip(pred_lgd3, 0, 100)
+ 0.10 * np.clip(pred_cat3, 0, 100)
)
sample_prediction_df["target4"] = (
0.65 * np.clip(pred4, 0, 100)
+ 0.25 * np.clip(pred_lgd4, 0, 100)
+ 0.10 * np.clip(pred_cat4, 0, 100)
)
sample_prediction_df = sample_prediction_df.fillna(0.0)
del sample_prediction_df["playerId"]
# TF summit
# Features computation at Evaluation Date
sub_fe, eval_dt = test_lag(sub)
sub_fe = sub_fe.merge(LAST_MED_DF, on="playerId", how="left")
sub_fe = sub_fe.fillna(0.0)
_preds = 0.0
for reg in nets:
_preds += reg.predict(sub_fe[FECOLS + MEDCOLS]) / NFOLDS
sub_fe[TGTCOLS] = np.clip(_preds, 0, 100)
sub.drop(["date"] + TGTCOLS, axis=1, inplace=True)
sub = sub.merge(sub_fe[["playerId"] + TGTCOLS], on="playerId", how="left")
sub.drop("playerId", axis=1, inplace=True)
sub = sub.fillna(0.0)
# Blending
blend = pd.concat(
[
sub[["date_playerId"]],
(
0.35 * sub.drop("date_playerId", axis=1)
+ 0.65 * sample_prediction_df.drop("date_playerId", axis=1)
),
],
axis=1,
)
env.predict(blend)
# Update Available information
sub_fe["EvalDate"] = eval_dt
# sub_fe.drop(MEDCOLS, axis=1, inplace=True)
LAST = LAST.append(sub_fe)
LAST = LAST.drop_duplicates(subset=["EvalDate", "playerId"], keep="last")
pd.concat(
[
sub[["date_playerId"]],
(
sub.drop("date_playerId", axis=1)
+ sample_prediction_df.drop("date_playerId", axis=1)
)
/ 2,
],
axis=1,
)
sample_prediction_df
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/401/69401993.ipynb
|
mlb-data
|
ulrich07
|
[{"Id": 69401993, "ScriptId": 18870876, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6312914, "CreationDate": "07/30/2021 14:35:35", "VersionNumber": 22.0, "Title": "LightGBM + CatBoost + ANN", "EvaluationDate": "07/30/2021", "IsChange": true, "TotalLines": 731.0, "LinesInsertedFromPrevious": 12.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 719.0, "LinesInsertedFromFork": 269.0, "LinesDeletedFromFork": 260.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 462.0, "TotalVotes": 0}]
|
[{"Id": 92521380, "KernelVersionId": 69401993, "SourceDatasetVersionId": 2332477}, {"Id": 92521382, "KernelVersionId": 69401993, "SourceDatasetVersionId": 2379932}, {"Id": 92521381, "KernelVersionId": 69401993, "SourceDatasetVersionId": 2336518}]
|
[{"Id": 2332477, "DatasetId": 1407900, "DatasourceVersionId": 2374036, "CreatorUserId": 1348567, "LicenseName": "Unknown", "CreationDate": "06/14/2021 11:02:44", "VersionNumber": 1.0, "Title": "MLB_DATA", "Slug": "mlb-data", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1407900, "CreatorUserId": 1348567, "OwnerUserId": 1348567.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2332477.0, "CurrentDatasourceVersionId": 2374036.0, "ForumId": 1427228, "Type": 2, "CreationDate": "06/14/2021 11:02:44", "LastActivityDate": "06/14/2021", "TotalViews": 1450, "TotalDownloads": 283, "TotalVotes": 9, "TotalKernels": 23}]
|
[{"Id": 1348567, "UserName": "ulrich07", "DisplayName": "Ulrich G.", "RegisterDate": "10/20/2017", "PerformanceTier": 3}]
|
#
# ## About Dataset
"""
import gc
import numpy as np
import pandas as pd
from pathlib import Path
from pandarallel import pandarallel
pandarallel.initialize()
BASE_DIR = Path('../input/mlb-player-digital-engagement-forecasting')
train = pd.read_csv(BASE_DIR / 'train.csv')
null = np.nan
true = True
false = False
for col in train.columns:
if col == 'date': continue
_index = train[col].notnull()
train.loc[_index, col] = train.loc[_index, col].parallel_apply(lambda x: eval(x))
outputs = []
for index, date, record in train.loc[_index, ['date', col]].itertuples():
_df = pd.DataFrame(record)
_df['index'] = index
_df['date'] = date
outputs.append(_df)
outputs = pd.concat(outputs).reset_index(drop=True)
outputs.to_csv(f'{col}_train.csv', index=False)
outputs.to_pickle(f'{col}_train.pkl')
del outputs
del train[col]
gc.collect()
"""
# ## Training
import numpy as np
import pandas as pd
from pathlib import Path
from sklearn.metrics import mean_absolute_error
from datetime import timedelta
from functools import reduce
from tqdm import tqdm
import lightgbm as lgbm
import mlb
import os
import pickle
BASE_DIR = Path("../input/mlb-player-digital-engagement-forecasting")
TRAIN_DIR = Path("../input/mlb-pdef-train-dataset")
players = pd.read_csv(BASE_DIR / "players.csv")
rosters = pd.read_pickle(TRAIN_DIR / "rosters_train.pkl")
targets = pd.read_pickle(TRAIN_DIR / "nextDayPlayerEngagement_train.pkl")
scores = pd.read_pickle(TRAIN_DIR / "playerBoxScores_train.pkl")
scores = scores.groupby(["playerId", "date"]).sum().reset_index()
targets_cols = ["playerId", "target1", "target2", "target3", "target4", "date"]
players_cols = ["playerId", "primaryPositionName"]
rosters_cols = ["playerId", "teamId", "status", "date"]
scores_cols = [
"playerId",
"battingOrder",
"gamesPlayedBatting",
"flyOuts",
"groundOuts",
"runsScored",
"doubles",
"triples",
"homeRuns",
"strikeOuts",
"baseOnBalls",
"intentionalWalks",
"hits",
"hitByPitch",
"caughtStealing",
"stolenBases", #'atBats',
"groundIntoDoublePlay",
"groundIntoTriplePlay",
"plateAppearances",
"totalBases", #'rbi',
"leftOnBase",
"sacBunts", # 'sacFlies', 'catchersInterference',
"pickoffs",
"gamesPlayedPitching",
"gamesStartedPitching",
"completeGamesPitching",
"shutoutsPitching",
"winsPitching",
"airOutsPitching", # lossesPitching', #'flyOutsPitching', ', ###
"groundOutsPitching",
"runsPitching",
"doublesPitching",
"triplesPitching",
"homeRunsPitching",
"strikeOutsPitching",
"baseOnBallsPitching",
"intentionalWalksPitching",
"hitsPitching",
"hitByPitchPitching",
"atBatsPitching",
"caughtStealingPitching",
"stolenBasesPitching",
"inningsPitched",
"saveOpportunities",
"earnedRuns",
"battersFaced",
"outsPitching",
"pitchesThrown",
"balls",
"strikes",
"hitBatsmen",
"balks",
"wildPitches",
"pickoffsPitching",
"rbiPitching",
"gamesFinishedPitching",
"inheritedRunners",
"inheritedRunnersScored",
"catchersInterferencePitching",
"sacBuntsPitching",
"sacFliesPitching",
"saves", #'holds', 'blownSaves',
"assists",
"putOuts",
"errors",
"chances",
"date",
]
feature_cols = [
"label_playerId", #'label_primaryPositionName',# 'label_teamId',
"battingOrder",
"gamesPlayedBatting", #'flyOuts',
"label_status", # '〇label_status',
"groundOuts",
"runsScored",
"doubles",
"triples",
"homeRuns",
"strikeOuts",
"baseOnBalls",
"intentionalWalks",
"hits",
"hitByPitch",
"caughtStealing",
"stolenBases", #'atBats',
"groundIntoDoublePlay", # ○'atBats', ' 'groundIntoDoublePlay',
"groundIntoTriplePlay",
"plateAppearances",
"totalBases", #'rbi',
"leftOnBase", #'sacBunts', #'sacFlies', 'catchersInterference', #'leftOnBase', '
"pickoffs",
"gamesPlayedPitching",
"gamesStartedPitching",
"completeGamesPitching",
"shutoutsPitching",
"winsPitching",
#'lossesPitching', 'flyOutsPitching', 'airOutsPitching',###
"groundOutsPitching",
"runsPitching",
"doublesPitching",
"triplesPitching",
"homeRunsPitching",
"strikeOutsPitching",
"baseOnBallsPitching",
"hitsPitching",
"intentionalWalksPitching",
"hitByPitchPitching",
"atBatsPitching",
"caughtStealingPitching",
"stolenBasesPitching",
"inningsPitched",
"saveOpportunities",
"earnedRuns",
"battersFaced",
"outsPitching",
"pitchesThrown",
"balls",
"strikes",
"hitBatsmen", #'wildPitches',
"pickoffsPitching",
"balks", # ○'pickoffsPitching',
"rbiPitching",
"gamesFinishedPitching",
"inheritedRunners",
"inheritedRunnersScored",
"catchersInterferencePitching",
"sacBuntsPitching",
"saves",
"sacFliesPitching", #'holds', 'blownSaves',○'sacBuntsPitching', ○'sacFliesPitching' ○ 'sacFliesPitching'
"assists",
"putOuts", #'errors',
"chances",
"target1_mean",
"target1_median",
"target1_std",
"target1_min",
"target1_max",
"target1_prob",
"target2_mean",
"target2_median",
"target2_std",
"target2_min",
"target2_max",
"target2_prob",
"target3_mean",
"target3_median",
"target3_std",
"target3_min",
"target3_max",
"target3_prob",
"target4_mean",
"target4_median",
"target4_std",
"target4_min",
"target4_max",
"target4_prob",
] # 86
feature_cols2 = [
"label_playerId",
"label_primaryPositionName",
"label_teamId", # needed
"label_status",
"battingOrder",
"gamesPlayedBatting",
"flyOuts",
"groundOuts",
"runsScored",
"doubles",
"triples",
"homeRuns",
"strikeOuts",
"baseOnBalls",
"hits",
"hitByPitch",
"intentionalWalks", # ○'intentionalWalks'
"caughtStealing",
"stolenBases",
"groundIntoDoublePlay", #'atBats',#'〇atBats
"groundIntoTriplePlay",
"plateAppearances",
"totalBases", # 'rbi',#○ rbi
"leftOnBase",
"sacBunts", # 'sacFlies', 'catchersInterference',#', 〇leftOnBase'
"gamesPlayedPitching",
"gamesStartedPitching",
"pickoffs", # 〇'pickoffs'
"completeGamesPitching",
"shutoutsPitching",
"winsPitching",
#'airOutsPitching',# lossesPitching', #'flyOutsPitching', '',##〇
"groundOutsPitching",
"runsPitching",
"doublesPitching",
"triplesPitching",
"homeRunsPitching",
"strikeOutsPitching",
"baseOnBallsPitching",
"hitsPitching",
"intentionalWalksPitching",
"hitByPitchPitching",
"atBatsPitching",
"caughtStealingPitching",
"stolenBasesPitching",
"inningsPitched",
"saveOpportunities",
"earnedRuns",
"battersFaced",
"outsPitching",
"pitchesThrown",
"balls",
"strikes",
"hitBatsmen",
"wildPitches",
"pickoffsPitching", #'balks',
"rbiPitching",
"gamesFinishedPitching",
"inheritedRunners",
"inheritedRunnersScored", #'catchersInterferencePitching',
"sacFliesPitching",
"saves",
"sacBuntsPitching", #'holds', 'blownSaves' #〇sacBuntsPitching'
"assists",
"putOuts",
"errors",
"chances",
"target1_mean",
"target1_median",
"target1_std",
"target1_min",
"target1_max",
"target1_prob",
"target2_mean",
"target2_median",
"target2_std",
"target2_min",
"target2_max",
"target2_prob",
"target3_mean",
"target3_median",
"target3_std",
"target3_min",
"target3_max",
"target3_prob",
"target4_mean",
"target4_median",
"target4_std",
"target4_min",
"target4_max",
"target4_prob",
"target1",
] # 91
player_target_stats = pd.read_csv(
"../input/player-target-stats/player_target_stats.csv"
)
data_names = player_target_stats.columns.values.tolist()
data_names
# creat dataset
train = targets[targets_cols].merge(players[players_cols], on=["playerId"], how="left")
train = train.merge(rosters[rosters_cols], on=["playerId", "date"], how="left")
train = train.merge(scores[scores_cols], on=["playerId", "date"], how="left")
train = train.merge(
player_target_stats, how="inner", left_on=["playerId"], right_on=["playerId"]
)
# label encoding
player2num = {c: i for i, c in enumerate(train["playerId"].unique())}
position2num = {c: i for i, c in enumerate(train["primaryPositionName"].unique())}
teamid2num = {c: i for i, c in enumerate(train["teamId"].unique())}
status2num = {c: i for i, c in enumerate(train["status"].unique())}
train["label_playerId"] = train["playerId"].map(player2num)
train["label_primaryPositionName"] = train["primaryPositionName"].map(position2num)
train["label_teamId"] = train["teamId"].map(teamid2num)
train["label_status"] = train["status"].map(status2num)
train_X = train[feature_cols]
train_y = train[["target1", "target2", "target3", "target4"]]
_index = train["date"] < 20210401
x_train1 = train_X.loc[_index].reset_index(drop=True)
y_train1 = train_y.loc[_index].reset_index(drop=True)
x_valid1 = train_X.loc[~_index].reset_index(drop=True)
y_valid1 = train_y.loc[~_index].reset_index(drop=True)
train_X = train[feature_cols2]
train_y = train[["target1", "target2", "target3", "target4"]]
_index = train["date"] < 20210401
x_train2 = train_X.loc[_index].reset_index(drop=True)
y_train2 = train_y.loc[_index].reset_index(drop=True)
x_valid2 = train_X.loc[~_index].reset_index(drop=True)
y_valid2 = train_y.loc[~_index].reset_index(drop=True)
train_X
# def fit_lgbm(x_train, y_train, x_valid, y_valid, params: dict=None, verbose=100):
# oof_pred = np.zeros(len(y_valid), dtype=np.float32)
# model = lgbm.LGBMRegressor(**params)
# model.fit(x_train, y_train,
# eval_set=[(x_valid, y_valid)],
# early_stopping_rounds=verbose,
# verbose=verbose)
# oof_pred = model.predict(x_valid)
# score = mean_absolute_error(oof_pred, y_valid)
# print('mae:', score)
# return oof_pred, model, score
# # training lightgbm
# params1 = {'objective':'mae',
# 'reg_alpha': 0.14947461820098767,
# 'reg_lambda': 0.10185644384043743,
# 'n_estimators': 3633,
# 'learning_rate': 0.08046301304430488,
# 'num_leaves': 674,
# 'feature_fraction': 0.9101240539122566,
# 'bagging_fraction': 0.9884451442950513,
# 'bagging_freq': 8,
# 'min_child_samples': 51}
# params2 = {
# 'objective':'mae',
# 'reg_alpha': 0.1,
# 'reg_lambda': 0.1,
# 'n_estimators': 80,
# 'learning_rate': 0.1,
# 'random_state': 42,
# "num_leaves": 22
# }
# params4 = {'objective':'mae',
# 'reg_alpha': 0.016468100279441976,
# 'reg_lambda': 0.09128335764019105,
# 'n_estimators': 9868,
# 'learning_rate': 0.10528150510326864,
# 'num_leaves': 157,
# 'feature_fraction': 0.5419185713426886,
# 'bagging_fraction': 0.2637405128936662,
# 'bagging_freq': 19,
# 'min_child_samples': 71}
# params = {
# 'objective':'mae',
# 'reg_alpha': 0.1,
# 'reg_lambda': 0.1,
# 'n_estimators': 10000,
# 'learning_rate': 0.1,
# 'random_state': 42,
# "num_leaves": 100
# }
# oof1, model1, score1 = fit_lgbm(
# x_train1, y_train1['target1'],
# x_valid1, y_valid1['target1'],
# params1
# )
# oof2, model2, score2 = fit_lgbm(
# x_train2, y_train2['target2'],
# x_valid2, y_valid2['target2'],
# params2
# )
# oof3, model3, score3 = fit_lgbm(
# x_train2, y_train2['target3'],
# x_valid2, y_valid2['target3'],
# params
# )
# oof4, model4, score4 = fit_lgbm(
# x_train2, y_train2['target4'],
# x_valid2, y_valid2['target4'],
# params4
# )
# score = (score1+score2+score3+score4) / 4
# print(f'score: {score}')
# import pickle
# from catboost import CatBoostRegressor
# def fit_lgbm(x_train, y_train, x_valid, y_valid, params: dict=None, verbose=100):
# oof_pred_lgb = np.zeros(len(y_valid), dtype=np.float32)
# oof_pred_cat = np.zeros(len(y_valid), dtype=np.float32)
# # if os.path.isfile(f'../input/mlb-lgbm-and-catboost-models/model_lgb_{target}.pkl'):
# # with open(f'../input/mlb-lgbm-and-catboost-models/model_lgb_{target}.pkl', 'rb') as fin:
# # model = pickle.load(fin)
# # else:
# model = lgbm.LGBMRegressor(**params)
# model.fit(x_train, y_train,
# eval_set=[(x_valid, y_valid)],
# early_stopping_rounds=verbose,
# verbose=verbose)
# # with open(f'model_lgb_{target}.pkl', 'wb') as handle:
# # pickle.dump(model, handle, protocol=pickle.HIGHEST_PROTOCOL)
# oof_pred_lgb = model.predict(x_valid)
# score_lgb = mean_absolute_error(oof_pred_lgb, y_valid)
# print('mae:', score_lgb)
# # if os.path.isfile(f'../input/mlb-lgbm-and-catboost-models/model_cb_{target}.pkl'):
# # with open(f'../input/mlb-lgbm-and-catboost-models/model_cb_{target}.pkl', 'rb') as fin:
# # model_cb = pickle.load(fin)
# # else:
# model_cb = CatBoostRegressor(
# n_estimators=2000,
# learning_rate=0.05,
# loss_function='MAE',
# eval_metric='MAE',
# max_bin=50,
# subsample=0.9,
# colsample_bylevel=0.5,
# verbose=100)
# model_cb.fit(x_train, y_train, use_best_model=True,
# eval_set=(x_valid, y_valid),
# early_stopping_rounds=25)
# # with open(f'model_cb_{target}.pkl', 'wb') as handle:
# # pickle.dump(model_cb, handle, protocol=pickle.HIGHEST_PROTOCOL)
# oof_pred_cat = model_cb.predict(x_valid)
# score_cat = mean_absolute_error(oof_pred_cat, y_valid)
# print('mae:', score_cat)
# return oof_pred_lgb, model, oof_pred_cat, model_cb, score_lgb, score_cat
# # training lightgbm
# params = {
# 'boosting_type': 'gbdt',
# 'objective':'mae',
# 'subsample': 0.5,
# 'subsample_freq': 1,
# 'learning_rate': 0.03,
# 'num_leaves': 2**11-1,
# 'min_data_in_leaf': 2**12-1,
# 'feature_fraction': 0.5,
# 'max_bin': 100,
# 'n_estimators': 2500,
# 'boost_from_average': False,
# "random_seed":42,
# }
# oof_pred_lgb2, model_lgb2, oof_pred_cat2, model_cb2, score_lgb2, score_cat2 = fit_lgbm(
# x_train1, y_train1['target2'],
# x_valid1, y_valid1['target2'],
# params
# )
# oof_pred_lgb1, model_lgb1, oof_pred_cat1, model_cb1, score_lgb1, score_cat1 = fit_lgbm(
# x_train1, y_train1['target1'],
# x_valid1, y_valid1['target1'],
# params
# )
# oof_pred_lgb3, model_lgb3, oof_pred_cat3, model_cb3, score_lgb3, score_cat3 = fit_lgbm(
# x_train1, y_train1['target3'],
# x_valid1, y_valid1['target3'],
# params
# )
# oof_pred_lgb4, model_lgb4, oof_pred_cat4, model_cb4, score_lgb4, score_cat4= fit_lgbm(
# x_train1, y_train1['target4'],
# x_valid1, y_valid1['target4'],
# params
# )
# score = (score_lgb1+score_lgb2+score_lgb3+score_lgb4) / 4
# print(f'LightGBM score: {score}')
# score = (score_cat1+score_cat2+score_cat3+score_cat4) / 4
# print(f'Catboost score: {score}')
with open("../input/modellgbcb/model_lgb_cb/model1.pickle", mode="rb") as f:
model1 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model2.pickle", mode="rb") as f:
model2 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model3.pickle", mode="rb") as f:
model3 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model4.pickle", mode="rb") as f:
model4 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model_lgb1.pickle", mode="rb") as f:
model_lgb1 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model_lgb2.pickle", mode="rb") as f:
model_lgb2 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model_lgb3.pickle", mode="rb") as f:
model_lgb3 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model_lgb4.pickle", mode="rb") as f:
model_lgb4 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model_cb1.pickle", mode="rb") as f:
model_cb1 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model_cb2.pickle", mode="rb") as f:
model_cb2 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model_cb3.pickle", mode="rb") as f:
model_cb3 = pickle.load(f)
with open("../input/modellgbcb/model_lgb_cb/model_cb4.pickle", mode="rb") as f:
model_cb4 = pickle.load(f)
players_cols = ["playerId", "primaryPositionName"]
rosters_cols = ["playerId", "teamId", "status"]
scores_cols = [
"playerId",
"battingOrder",
"gamesPlayedBatting",
"flyOuts",
"groundOuts",
"runsScored",
"doubles",
"triples",
"homeRuns",
"strikeOuts",
"baseOnBalls",
"intentionalWalks",
"hits",
"hitByPitch",
"caughtStealing",
"stolenBases", #'atBats',
"groundIntoDoublePlay",
"groundIntoTriplePlay",
"plateAppearances",
"totalBases", #'rbi',
"leftOnBase",
"sacBunts", # 'sacFlies', 'catchersInterference',
"pickoffs",
"gamesPlayedPitching",
"gamesStartedPitching",
"completeGamesPitching",
"shutoutsPitching",
"winsPitching",
"airOutsPitching", #'lossesPitching', 'flyOutsPitching',
"groundOutsPitching",
"runsPitching",
"doublesPitching",
"triplesPitching",
"homeRunsPitching",
"strikeOutsPitching",
"baseOnBallsPitching",
"intentionalWalksPitching",
"hitsPitching",
"hitByPitchPitching",
"atBatsPitching",
"caughtStealingPitching",
"stolenBasesPitching",
"inningsPitched",
"saveOpportunities",
"earnedRuns",
"battersFaced",
"outsPitching",
"pitchesThrown",
"balls",
"strikes",
"hitBatsmen",
"balks",
"wildPitches",
"pickoffsPitching",
"rbiPitching",
"gamesFinishedPitching",
"inheritedRunners",
"inheritedRunnersScored",
"catchersInterferencePitching",
"sacBuntsPitching",
"sacFliesPitching",
"saves", #'holds', 'blownSaves',
"assists",
"putOuts",
"errors",
"chances",
]
null = np.nan
true = True
false = False
import pandas as pd
import numpy as np
from datetime import timedelta
from tqdm import tqdm
import gc
from functools import reduce
from sklearn.model_selection import StratifiedKFold
ROOT_DIR = "../input/mlb-player-digital-engagement-forecasting"
# =======================#
def flatten(df, col):
du = (
df.pivot(index="playerId", columns="EvalDate", values=col)
.add_prefix(f"{col}_")
.rename_axis(None, axis=1)
.reset_index()
)
return du
# ============================#
def reducer(left, right):
return left.merge(right, on="playerId")
# ========================
TGTCOLS = ["target1", "target2", "target3", "target4"]
def train_lag(df, lag=1):
dp = df[["playerId", "EvalDate"] + TGTCOLS].copy()
dp["EvalDate"] = dp["EvalDate"] + timedelta(days=lag)
df = df.merge(dp, on=["playerId", "EvalDate"], suffixes=["", f"_{lag}"], how="left")
return df
# =================================
def test_lag(sub):
sub["playerId"] = sub["date_playerId"].apply(lambda s: int(s.split("_")[1]))
assert sub.date.nunique() == 1
dte = sub["date"].unique()[0]
eval_dt = pd.to_datetime(dte, format="%Y%m%d")
dtes = [eval_dt + timedelta(days=-k) for k in LAGS]
mp_dtes = {eval_dt + timedelta(days=-k): k for k in LAGS}
sl = LAST.loc[
LAST.EvalDate.between(dtes[-1], dtes[0]), ["EvalDate", "playerId"] + TGTCOLS
].copy()
sl["EvalDate"] = sl["EvalDate"].map(mp_dtes)
du = [flatten(sl, col) for col in TGTCOLS]
du = reduce(reducer, du)
return du, eval_dt
#
# ===============
tr = pd.read_csv("../input/mlb-data/target.csv")
print(tr.shape)
gc.collect()
tr["EvalDate"] = pd.to_datetime(tr["EvalDate"])
tr["EvalDate"] = tr["EvalDate"] + timedelta(days=-1)
tr["EvalYear"] = tr["EvalDate"].dt.year
MED_DF = tr.groupby(["playerId", "EvalYear"])[TGTCOLS].median().reset_index()
MEDCOLS = ["tgt1_med", "tgt2_med", "tgt3_med", "tgt4_med"]
MED_DF.columns = ["playerId", "EvalYear"] + MEDCOLS
LAGS = list(range(1, 21))
FECOLS = [f"{col}_{lag}" for lag in reversed(LAGS) for col in TGTCOLS]
for lag in tqdm(LAGS):
tr = train_lag(tr, lag=lag)
gc.collect()
# ===========
tr = tr.sort_values(by=["playerId", "EvalDate"])
print(tr.shape)
tr = tr.dropna()
print(tr.shape)
tr = tr.merge(MED_DF, on=["playerId", "EvalYear"])
gc.collect()
X = tr[FECOLS + MEDCOLS].values
y = tr[TGTCOLS].values
cl = tr["playerId"].values
NFOLDS = 12
skf = StratifiedKFold(n_splits=NFOLDS)
folds = skf.split(X, cl)
folds = list(folds)
import tensorflow as tf
import tensorflow.keras.layers as L
import tensorflow.keras.models as M
from sklearn.metrics import mean_absolute_error, mean_squared_error
from tensorflow.keras.callbacks import ModelCheckpoint, ReduceLROnPlateau, EarlyStopping
tf.random.set_seed(777)
def make_model(n_in):
inp = L.Input(name="inputs", shape=(n_in,))
x = L.Dense(50, activation="relu", name="d1")(inp)
x = L.Dense(50, activation="relu", name="d2")(x)
preds = L.Dense(4, activation="linear", name="preds")(x)
model = M.Model(inp, preds, name="ANN")
model.compile(loss="mean_absolute_error", optimizer="adam")
return model
net = make_model(X.shape[1])
print(net.summary())
oof = np.zeros(y.shape)
nets = []
for idx in range(NFOLDS):
print("FOLD:", idx)
tr_idx, val_idx = folds[idx]
ckpt = ModelCheckpoint(
f"w{idx}.h5", monitor="val_loss", verbose=1, save_best_only=True, mode="min"
)
reduce_lr = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, min_lr=0.0005
)
es = EarlyStopping(monitor="val_loss", patience=6)
reg = make_model(X.shape[1])
reg.fit(
X[tr_idx],
y[tr_idx],
epochs=10,
batch_size=35_000,
validation_data=(X[val_idx], y[val_idx]),
verbose=1,
callbacks=[ckpt, reduce_lr, es],
)
reg.load_weights(f"w{idx}.h5")
oof[val_idx] = reg.predict(X[val_idx], batch_size=50_000, verbose=1)
nets.append(reg)
gc.collect()
mae = mean_absolute_error(y, oof)
mse = mean_squared_error(y, oof, squared=False)
print("mae:", mae)
print("mse:", mse)
# Historical information to use in prediction time
bound_dt = pd.to_datetime("2021-01-01")
LAST = tr.loc[tr.EvalDate > bound_dt].copy()
LAST_MED_DF = MED_DF.loc[MED_DF.EvalYear == 2021].copy()
LAST_MED_DF.drop("EvalYear", axis=1, inplace=True)
del tr
# """
import mlb
FE = []
SUB = []
#
#
import copy
env = mlb.make_env() # initialize the environment
iter_test = env.iter_test() # iterator which loops over each date in test set
for test_df, sample_prediction_df in iter_test: # make predictions here
sub = copy.deepcopy(sample_prediction_df.reset_index())
sample_prediction_df = copy.deepcopy(sample_prediction_df.reset_index(drop=True))
# LGBM summit
# creat dataset
sample_prediction_df["playerId"] = sample_prediction_df["date_playerId"].map(
lambda x: int(x.split("_")[1])
)
# Dealing with missing values
if test_df["rosters"].iloc[0] == test_df["rosters"].iloc[0]:
test_rosters = pd.DataFrame(eval(test_df["rosters"].iloc[0]))
else:
test_rosters = pd.DataFrame({"playerId": sample_prediction_df["playerId"]})
for col in rosters.columns:
if col == "playerId":
continue
test_rosters[col] = np.nan
if test_df["playerBoxScores"].iloc[0] == test_df["playerBoxScores"].iloc[0]:
test_scores = pd.DataFrame(eval(test_df["playerBoxScores"].iloc[0]))
else:
test_scores = pd.DataFrame({"playerId": sample_prediction_df["playerId"]})
for col in scores.columns:
if col == "playerId":
continue
test_scores[col] = np.nan
test_scores = test_scores.groupby("playerId").sum().reset_index()
test = sample_prediction_df[["playerId"]].copy()
test = test.merge(players[players_cols], on="playerId", how="left")
test = test.merge(test_rosters[rosters_cols], on="playerId", how="left")
test = test.merge(test_scores[scores_cols], on="playerId", how="left")
test = test.merge(
player_target_stats, how="inner", left_on=["playerId"], right_on=["playerId"]
)
test["label_playerId"] = test["playerId"].map(player2num)
test["label_primaryPositionName"] = test["primaryPositionName"].map(position2num)
test["label_teamId"] = test["teamId"].map(teamid2num)
test["label_status"] = test["status"].map(status2num)
test_X = test[feature_cols]
# predict
pred1 = model1.predict(test_X)
pred_lgd1 = model_lgb1.predict(test_X)
pred_lgd2 = model_lgb2.predict(test_X)
pred_lgd3 = model_lgb3.predict(test_X)
pred_lgd4 = model_lgb4.predict(test_X)
pred_cat1 = model_cb1.predict(test_X)
pred_cat2 = model_cb2.predict(test_X)
pred_cat3 = model_cb3.predict(test_X)
pred_cat4 = model_cb4.predict(test_X)
test["target1"] = np.clip(pred1, 0, 100)
test_X = test[feature_cols2]
# predict2
pred2 = model2.predict(test_X)
pred3 = model3.predict(test_X)
pred4 = model4.predict(test_X)
# merge submission
sample_prediction_df["target1"] = (
1.00 * np.clip(pred1, 0, 100)
+ 0.00 * np.clip(pred_lgd1, 0, 100)
+ 0.00 * np.clip(pred_cat1, 0, 100)
)
sample_prediction_df["target2"] = (
0.10 * np.clip(pred2, 0, 100)
+ 0.65 * np.clip(pred_lgd2, 0, 100)
+ 0.25 * np.clip(pred_cat2, 0, 100)
)
sample_prediction_df["target3"] = (
0.65 * np.clip(pred3, 0, 100)
+ 0.25 * np.clip(pred_lgd3, 0, 100)
+ 0.10 * np.clip(pred_cat3, 0, 100)
)
sample_prediction_df["target4"] = (
0.65 * np.clip(pred4, 0, 100)
+ 0.25 * np.clip(pred_lgd4, 0, 100)
+ 0.10 * np.clip(pred_cat4, 0, 100)
)
sample_prediction_df = sample_prediction_df.fillna(0.0)
del sample_prediction_df["playerId"]
# TF summit
# Features computation at Evaluation Date
sub_fe, eval_dt = test_lag(sub)
sub_fe = sub_fe.merge(LAST_MED_DF, on="playerId", how="left")
sub_fe = sub_fe.fillna(0.0)
_preds = 0.0
for reg in nets:
_preds += reg.predict(sub_fe[FECOLS + MEDCOLS]) / NFOLDS
sub_fe[TGTCOLS] = np.clip(_preds, 0, 100)
sub.drop(["date"] + TGTCOLS, axis=1, inplace=True)
sub = sub.merge(sub_fe[["playerId"] + TGTCOLS], on="playerId", how="left")
sub.drop("playerId", axis=1, inplace=True)
sub = sub.fillna(0.0)
# Blending
blend = pd.concat(
[
sub[["date_playerId"]],
(
0.35 * sub.drop("date_playerId", axis=1)
+ 0.65 * sample_prediction_df.drop("date_playerId", axis=1)
),
],
axis=1,
)
env.predict(blend)
# Update Available information
sub_fe["EvalDate"] = eval_dt
# sub_fe.drop(MEDCOLS, axis=1, inplace=True)
LAST = LAST.append(sub_fe)
LAST = LAST.drop_duplicates(subset=["EvalDate", "playerId"], keep="last")
pd.concat(
[
sub[["date_playerId"]],
(
sub.drop("date_playerId", axis=1)
+ sample_prediction_df.drop("date_playerId", axis=1)
)
/ 2,
],
axis=1,
)
sample_prediction_df
|
[{"mlb-data/target.csv": {"column_names": "[\"playerId\", \"target1\", \"target2\", \"target3\", \"target4\", \"EvalDate\"]", "column_data_types": "{\"playerId\": \"int64\", \"target1\": \"float64\", \"target2\": \"float64\", \"target3\": \"float64\", \"target4\": \"float64\", \"EvalDate\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 2506176 entries, 0 to 2506175\nData columns (total 6 columns):\n # Column Dtype \n--- ------ ----- \n 0 playerId int64 \n 1 target1 float64\n 2 target2 float64\n 3 target3 float64\n 4 target4 float64\n 5 EvalDate object \ndtypes: float64(4), int64(1), object(1)\nmemory usage: 114.7+ MB\n", "summary": "{\"playerId\": {\"count\": 2506176.0, \"mean\": 587590.9160601649, \"std\": 71110.70419831667, \"min\": 112526.0, \"25%\": 543343.0, \"50%\": 605388.0, \"75%\": 642758.0, \"max\": 685503.0}, \"target1\": {\"count\": 2506176.0, \"mean\": 0.5685203672104261, \"std\": 4.171841163999339, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0010668942707777, \"75%\": 0.0182417736616852, \"max\": 100.0}, \"target2\": {\"count\": 2506176.0, \"mean\": 2.456109828032992, \"std\": 6.234965096388626, \"min\": 0.0, \"25%\": 0.0769876819708846, \"50%\": 0.5561735261401557, \"75%\": 2.2360504355510136, \"max\": 100.0}, \"target3\": {\"count\": 2506176.0, \"mean\": 0.6880492992404432, \"std\": 5.065297378349651, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0016922859982719, \"75%\": 0.0208359378255615, \"max\": 100.0}, \"target4\": {\"count\": 2506176.0, \"mean\": 1.1370441062569019, \"std\": 4.2299196411512945, \"min\": 0.0, \"25%\": 0.054054054054054, \"50%\": 0.2233538818904672, \"75%\": 0.7576614271174941, \"max\": 100.0}}", "examples": "{\"playerId\":{\"0\":628317,\"1\":547989,\"2\":519317,\"3\":607625},\"target1\":{\"0\":0.0111670705,\"1\":0.0429932216,\"2\":0.9743269048,\"3\":0.0067002423},\"target2\":{\"0\":4.4747081712,\"1\":5.593385214,\"2\":56.1770428016,\"3\":2.6750972763},\"target3\":{\"0\":0.0051677297,\"1\":0.0450330735,\"2\":13.6937455705,\"3\":0.0051677297},\"target4\":{\"0\":5.7352941176,\"1\":2.7941176471,\"2\":64.1666666667,\"3\":1.862745098},\"EvalDate\":{\"0\":\"2018-01-02\",\"1\":\"2018-01-02\",\"2\":\"2018-01-02\",\"3\":\"2018-01-02\"}}"}}]
| true | 2 |
<start_data_description><data_path>mlb-data/target.csv:
<column_names>
['playerId', 'target1', 'target2', 'target3', 'target4', 'EvalDate']
<column_types>
{'playerId': 'int64', 'target1': 'float64', 'target2': 'float64', 'target3': 'float64', 'target4': 'float64', 'EvalDate': 'object'}
<dataframe_Summary>
{'playerId': {'count': 2506176.0, 'mean': 587590.9160601649, 'std': 71110.70419831667, 'min': 112526.0, '25%': 543343.0, '50%': 605388.0, '75%': 642758.0, 'max': 685503.0}, 'target1': {'count': 2506176.0, 'mean': 0.5685203672104261, 'std': 4.171841163999339, 'min': 0.0, '25%': 0.0, '50%': 0.0010668942707777, '75%': 0.0182417736616852, 'max': 100.0}, 'target2': {'count': 2506176.0, 'mean': 2.456109828032992, 'std': 6.234965096388626, 'min': 0.0, '25%': 0.0769876819708846, '50%': 0.5561735261401557, '75%': 2.2360504355510136, 'max': 100.0}, 'target3': {'count': 2506176.0, 'mean': 0.6880492992404432, 'std': 5.065297378349651, 'min': 0.0, '25%': 0.0, '50%': 0.0016922859982719, '75%': 0.0208359378255615, 'max': 100.0}, 'target4': {'count': 2506176.0, 'mean': 1.1370441062569019, 'std': 4.2299196411512945, 'min': 0.0, '25%': 0.054054054054054, '50%': 0.2233538818904672, '75%': 0.7576614271174941, 'max': 100.0}}
<dataframe_info>
RangeIndex: 2506176 entries, 0 to 2506175
Data columns (total 6 columns):
# Column Dtype
--- ------ -----
0 playerId int64
1 target1 float64
2 target2 float64
3 target3 float64
4 target4 float64
5 EvalDate object
dtypes: float64(4), int64(1), object(1)
memory usage: 114.7+ MB
<some_examples>
{'playerId': {'0': 628317, '1': 547989, '2': 519317, '3': 607625}, 'target1': {'0': 0.0111670705, '1': 0.0429932216, '2': 0.9743269048, '3': 0.0067002423}, 'target2': {'0': 4.4747081712, '1': 5.593385214, '2': 56.1770428016, '3': 2.6750972763}, 'target3': {'0': 0.0051677297, '1': 0.0450330735, '2': 13.6937455705, '3': 0.0051677297}, 'target4': {'0': 5.7352941176, '1': 2.7941176471, '2': 64.1666666667, '3': 1.862745098}, 'EvalDate': {'0': '2018-01-02', '1': '2018-01-02', '2': '2018-01-02', '3': '2018-01-02'}}
<end_description>
| 9,921 | 0 | 10,589 | 9,921 |
69401828
|
# This is yet another starter using lightGBM.
# **Don't just fork this notebook and publish**, which just confuses people. In case you were to publish very similar notebook, please cite this notebook.
# As this notebook was created for new participants to get started easily, I tried to keep it as simple as possible.
# The basic strategy employed in this notebook includes:
# - Target Encoding: Simply using the past volatility works great due to the high autocorrelation.
# - Aggregation by multiple keys: not only stock_id x time_id, but also stock_id and time_id, separately.
# - Multithreads processing for feature engineering: features are made on a stock_id x time_id basis, so we might want to accelerate the process
# - LightGBM with the inverse weight of the square of the target: the evaluation metric is the root mean square percentage error, so we use RMSE as the objective function with the inverse weighting of the target.
# Note that this notebook only performs **inference** using the pretrained models generated in the following notebook to save time:
# [[Optiver] LGB and TE baseline](https://www.kaggle.com/code1110/optiver-lgb-and-te-baseline)
# So let's get the ball rolling!
DEBUG = False
# MODE = 'TRAIN'
MODE = "INFERENCE"
MODEL_DIR = "../input/optiver-lgb-and-te-baseline"
# # Libraries
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import gc
import pathlib
from tqdm.auto import tqdm
import json
from multiprocessing import Pool, cpu_count
import time
import requests as re
from datetime import datetime
from dateutil.relativedelta import relativedelta, FR
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import glob
import os
from sklearn import model_selection
import joblib
import lightgbm as lgb
# visualize
import matplotlib.pyplot as plt
import matplotlib.style as style
from matplotlib_venn import venn2, venn3
import seaborn as sns
from matplotlib import pyplot
from matplotlib.ticker import ScalarFormatter
sns.set_context("talk")
style.use("seaborn-colorblind")
import warnings
warnings.simplefilter("ignore")
pd.get_option("display.max_columns")
# # Config
class CFG:
INPUT_DIR = "../input/optiver-realized-volatility-prediction"
OUTPUT_DIR = "./"
# Logging is always nice for your experiment:)
def init_logger(log_file="train.log"):
from logging import getLogger, INFO, FileHandler, Formatter, StreamHandler
logger = getLogger(__name__)
logger.setLevel(INFO)
handler1 = StreamHandler()
handler1.setFormatter(Formatter("%(message)s"))
handler2 = FileHandler(filename=log_file)
handler2.setFormatter(Formatter("%(message)s"))
logger.addHandler(handler1)
logger.addHandler(handler2)
return logger
logger = init_logger(log_file=f"{CFG.OUTPUT_DIR}/baseline.log")
logger.info(f"Start Logging...")
# # Load data
# Data description found in :
# https://www.kaggle.com/c/optiver-realized-volatility-prediction/data
# ## Train, test, and sample submission
# train.csv The ground truth values for the training set.
# stock_id - Same as above, but since this is a csv the column will load as an integer instead of categorical.
# time_id - Same as above.
# target - The realized volatility computed over the 10 minute window following the feature data under the same stock/time_id. There is no overlap between feature and target data. You can find more info in our tutorial notebook.
# test.csv Provides the mapping between the other data files and the submission file. As with other test files, most of the data is only available to your notebook upon submission with just the first few rows available for download.
# stock_id - Same as above.
# time_id - Same as above.
# row_id - Unique identifier for the submission row. There is one row for each existing time ID/stock ID pair. Each time window is not necessarily containing every individual stock.
# sample_submission.csv - A sample submission file in the correct format.
# row_id - Same as in test.csv.
# target - Same definition as in train.csv. The benchmark is using the median target value from train.csv.
train = pd.read_csv(os.path.join(CFG.INPUT_DIR, "train.csv"))
logger.info("Train data: {}".format(train.shape))
train.head()
train["stock_id"].value_counts()
train["time_id"].value_counts()
train["target"].hist(bins=100)
# fig, ax = plt.subplots(16, 7, figsize=(20, 60))
# ax = ax.flatten()
# for i, stock_id in tqdm(enumerate(train['stock_id'].unique())):
# ax[i].hist(train.query('stock_id == @stock_id')['target'], bins=100)
# ax[i].set_title(stock_id)
# plt.tight_layout()
test = pd.read_csv(os.path.join(CFG.INPUT_DIR, "test.csv"))
logger.info("Test data: {}".format(test.shape))
test.head()
venn2([set(train["time_id"]), set(test["time_id"])])
ss = pd.read_csv(os.path.join(CFG.INPUT_DIR, "sample_submission.csv"))
logger.info("Sample submission: {}".format(ss.shape))
ss.head()
# ## Book
# book_[train/test].parquet A parquet file partitioned by stock_id. Provides order book data on the most competitive buy and sell orders entered into the market. The top two levels of the book are shared. The first level of the book will be more competitive in price terms, it will then receive execution priority over the second level.
# stock_id - ID code for the stock. Not all stock IDs exist in every time bucket. Parquet coerces this column to the categorical data type when loaded; you may wish to convert it to int8.
# time_id - ID code for the time bucket. Time IDs are not necessarily sequential but are consistent across all stocks.
# seconds_in_bucket - Number of seconds from the start of the bucket, always starting from 0.
# bid_price[1/2] - Normalized prices of the most/second most competitive buy level.
# ask_price[1/2] - Normalized prices of the most/second most competitive sell level.
# bid_size[1/2] - The number of shares on the most/second most competitive buy level.
# ask_size[1/2] - The number of shares on the most/second most competitive sell level.
# ## Trade
# trade_[train/test].parquet A parquet file partitioned by stock_id. Contains data on trades that actually executed. Usually, in the market, there are more passive buy/sell intention updates (book updates) than actual trades, therefore one may expect this file to be more sparse than the order book.
# stock_id - Same as above.
# time_id - Same as above.
# seconds_in_bucket - Same as above. Note that since trade and book data are taken from the same time window and trade data is more sparse in general, this field is not necessarily starting from 0.
# price - The average price of executed transactions happening in one second. Prices have been normalized and the average has been weighted by the number of shares traded in each transaction.
# size - The sum number of shares traded.
# order_count - The number of unique trade orders taking place.
train_book_stocks = os.listdir(os.path.join(CFG.INPUT_DIR, "book_train.parquet"))
if DEBUG:
logger.info("Debug mode: using 3 stocks only")
train_book_stocks = train_book_stocks[:3]
logger.info(
"{:,} train book stocks: {}".format(len(train_book_stocks), train_book_stocks)
)
# load stock_id=0
def load_book(stock_id=0, data_type="train"):
"""
load parquest book data for given stock_id
"""
book_df = pd.read_parquet(
os.path.join(CFG.INPUT_DIR, f"book_{data_type}.parquet/stock_id={stock_id}")
)
book_df["stock_id"] = stock_id
book_df["stock_id"] = book_df["stock_id"].astype(np.int8)
return book_df
def load_trade(stock_id=0, data_type="train"):
"""
load parquest trade data for given stock_id
"""
trade_df = pd.read_parquet(
os.path.join(CFG.INPUT_DIR, f"trade_{data_type}.parquet/stock_id={stock_id}")
)
trade_df["stock_id"] = stock_id
trade_df["stock_id"] = trade_df["stock_id"].astype(np.int8)
return trade_df
book0 = load_book(0)
logger.info("Book data of stock id = 1: {}".format(book0.shape))
book0.head()
book0.tail()
trade0 = load_trade(0)
logger.info("Book data of stock id = 1: {}".format(trade0.shape))
trade0.head()
trade0.tail()
book_df = book0.merge(
trade0, how="outer", on=["time_id", "stock_id", "seconds_in_bucket"]
)
print(book_df.shape)
book_df.head()
book_df.tail()
def fix_jsonerr(df):
"""
fix json column error for lightgbm
"""
df.columns = [
"".join(c if c.isalnum() else "_" for c in str(x)) for x in df.columns
]
return df
def log_return(list_stock_prices):
return np.log(list_stock_prices).diff()
def realized_volatility(series_log_return):
series_log_return = log_return(series_log_return)
return np.sqrt(np.sum(series_log_return**2))
def fe_row(book):
"""
Feature engineering (just volatility for now) for each row
"""
# volatility
for i in [
1,
2,
]:
# wap
book[f"book_wap{i}"] = (
book[f"bid_price{i}"] * book[f"ask_size{i}"]
+ book[f"ask_price{i}"] * book[f"bid_size{i}"]
) / (book[f"bid_size{i}"] + book[f"ask_size{i}"])
# mean wap
book["book_wap_mean"] = (book["book_wap1"] + book["book_wap2"]) / 2
# wap diff
book["book_wap_diff"] = book["book_wap1"] - book["book_wap2"]
# other orderbook features
book["book_price_spread"] = (book["ask_price1"] - book["bid_price1"]) / (
book["ask_price1"] + book["bid_price1"]
)
book["book_bid_spread"] = book["bid_price1"] - book["bid_price2"]
book["book_ask_spread"] = book["ask_price1"] - book["ask_price2"]
book["book_total_volume"] = (
book["ask_size1"] + book["ask_size2"] + book["bid_size1"] + book["bid_size2"]
)
book["book_volume_imbalance"] = (book["ask_size1"] + book["ask_size2"]) - (
book["bid_size1"] + book["bid_size2"]
)
return book
def fe_agg(book_df):
"""
feature engineering (aggregation by stock_id x time_id)
"""
# features
book_feats = book_df.columns[
book_df.columns.str.startswith("book_")
].values.tolist()
trade_feats = ["price", "size", "order_count", "seconds_in_bucket"]
# agg trade features
trade_df = (
book_df.groupby(["time_id", "stock_id"])[trade_feats]
.agg(["sum", "mean", "std", "max", "min"])
.reset_index()
)
# agg volatility features
fe_df = (
book_df.groupby(["time_id", "stock_id"])[book_feats]
.agg([realized_volatility])
.reset_index()
)
fe_df.columns = [" ".join(col).strip() for col in fe_df.columns.values]
# merge
fe_df = fe_df.merge(trade_df, how="left", on=["time_id", "stock_id"])
return fe_df
def fe_all(book_df):
"""
perform feature engineerings
"""
# row-wise feature engineering
book_df = fe_row(book_df)
# feature engineering agg by stock_id x time_id
fe_df = fe_agg(book_df)
return fe_df
def book_fe_by_stock(stock_id=0):
"""
load orderbook and trade data for the given stock_id and merge
"""
# load data
book_df = load_book(stock_id, "train")
trade_df = load_trade(stock_id, "train")
book_feats = book_df.columns.values.tolist()
# merge
book_df = book_df.merge(
trade_df, how="outer", on=["time_id", "seconds_in_bucket", "stock_id"]
)
# sort by time
book_df = book_df.sort_values(by=["time_id", "seconds_in_bucket"])
# fillna for book_df
book_df[book_feats] = book_df[book_feats].fillna(method="ffill")
# feature engineering
fe_df = fe_all(book_df)
return fe_df
def book_fe_by_stock_test(stock_id=0):
"""
same function but for the test
"""
# load data
book_df = load_book(stock_id, "test")
trade_df = load_trade(stock_id, "test")
book_feats = book_df.columns.values.tolist()
# merge
book_df = book_df.merge(
trade_df, how="outer", on=["time_id", "seconds_in_bucket", "stock_id"]
)
# sort by time
book_df = book_df.sort_values(by=["time_id", "seconds_in_bucket"])
# fillna for book_df
book_df[book_feats] = book_df[book_feats].fillna(method="ffill")
# feature engineering
fe_df = fe_all(book_df)
return fe_df
def book_fe_all(stock_ids, data_type="train"):
"""
Feature engineering with multithread processing
"""
# feature engineering agg by stock_id x time_id
with Pool(cpu_count()) as p:
if data_type == "train":
feature_dfs = list(
tqdm(p.imap(book_fe_by_stock, stock_ids), total=len(stock_ids))
)
elif data_type == "test":
feature_dfs = list(
tqdm(p.imap(book_fe_by_stock_test, stock_ids), total=len(stock_ids))
)
fe_df = pd.concat(feature_dfs)
# feature engineering agg by stock_id
vol_feats = [f for f in fe_df.columns if ("realized" in f) & ("wap" in f)]
if data_type == "train":
# agg
stock_df = (
fe_df.groupby("stock_id")[vol_feats]
.agg(
[
"mean",
"std",
"max",
"min",
]
)
.reset_index()
)
# fix column names
stock_df.columns = ["stock_id"] + [
f"{f}_stock" for f in stock_df.columns.values.tolist()[1:]
]
stock_df = fix_jsonerr(stock_df)
# feature engineering agg by time_id
time_df = (
fe_df.groupby("time_id")[vol_feats]
.agg(
[
"mean",
"std",
"max",
"min",
]
)
.reset_index()
)
time_df.columns = ["time_id"] + [
f"{f}_time" for f in time_df.columns.values.tolist()[1:]
]
# merge
fe_df = fe_df.merge(time_df, how="left", on="time_id")
# make sure to fix json error for lighgbm
fe_df = fix_jsonerr(fe_df)
# out
if data_type == "train":
return fe_df, stock_df
elif data_type == "test":
return fe_df
if MODE == "TRAIN":
# all book data feature engineering
stock_ids = [int(i.split("=")[-1]) for i in train_book_stocks]
book_df, stock_df = book_fe_all(stock_ids, data_type="train")
assert book_df["stock_id"].nunique() > 2
assert book_df["time_id"].nunique() > 2
# save stock_df for the test
stock_df.to_pickle("train_stock_df.pkl")
logger.info("train stock df saved!")
# merge
book_df = (
book_df.merge(stock_df, how="left", on="stock_id")
.merge(train, how="left", on=["stock_id", "time_id"])
.replace([np.inf, -np.inf], np.nan)
.fillna(method="ffill")
)
# make row_id
book_df["row_id"] = (
book_df["stock_id"].astype(str) + "-" + book_df["time_id"].astype(str)
)
book_df.to_pickle("book_df.pkl")
print(book_df.shape)
book_df.head()
# test
test_book_stocks = os.listdir(os.path.join(CFG.INPUT_DIR, "book_test.parquet"))
logger.info("{:,} test book stocks: {}".format(len(test_book_stocks), test_book_stocks))
# all book data feature engineering
test_stock_ids = [int(i.split("=")[-1]) for i in test_book_stocks]
test_book_df = book_fe_all(test_stock_ids, data_type="test")
# load stock_df, if inference
if MODE == "INFERENCE":
book_df = pd.read_pickle(f"{MODEL_DIR}/book_df.pkl")
stock_df = pd.read_pickle(f"{MODEL_DIR}/train_stock_df.pkl")
# merge
test_book_df = (
test.merge(stock_df, how="left", on="stock_id")
.merge(test_book_df, how="left", on=["stock_id", "time_id"])
.replace([np.inf, -np.inf], np.nan)
.fillna(method="ffill")
)
# make row_id
test_book_df["row_id"] = (
test_book_df["stock_id"].astype(str) + "-" + test_book_df["time_id"].astype(str)
)
print(test_book_df.shape)
test_book_df.head()
# # Modeling
# Currently,
# - MODEL: LightGBM (with inverse weighting of the square of the target)
# - OBJECTIVE FUNCTION: RMSE
# - METRIC: RMSPE (used for early stopping)
# - Validation Strategy: KFold (are you sure?)
#
target = "target"
drops = [target, "row_id", "time_id"]
features = [
f
for f in test_book_df.columns.values.tolist()
if (f not in drops)
& (test_book_df[f].isna().sum() == 0)
& (book_df[f].isna().sum() == 0)
]
cats = [
"stock_id",
]
logger.info(
"{:,} features ({:,} categorical): {}".format(len(features), len(cats), features)
)
# evaluation metric
def RMSPEMetric(XGBoost=False):
def RMSPE(yhat, dtrain, XGBoost=XGBoost):
y = dtrain.get_label()
elements = ((y - yhat) / y) ** 2
if XGBoost:
return "RMSPE", float(np.sqrt(np.sum(elements) / len(y)))
else:
return "RMSPE", float(np.sqrt(np.sum(elements) / len(y))), False
return RMSPE
# LightGBM parameters
params = {
"n_estimators": 20000,
"objective": "rmse",
"boosting_type": "gbdt",
"max_depth": -1,
"learning_rate": 0.01,
"subsample": 0.72,
"subsample_freq": 4,
"feature_fraction": 0.8,
"lambda_l1": 1,
"lambda_l2": 1,
"seed": 46,
"early_stopping_rounds": 500,
"verbose": -1,
}
def fit_model(
params,
X_train,
y_train,
X_test,
features=features,
cats=[],
era="stock_id",
fold_type="kfold",
n_fold=5,
seed=42,
):
"""
fit model with cross validation
"""
models = []
oof_df = X_train[["time_id", "stock_id", target]].copy()
oof_df["pred"] = np.nan
y_preds = np.zeros((len(X_test),))
if fold_type == "stratifiedshuffle":
cv = model_selection.StratifiedShuffleSplit(n_splits=n_fold, random_state=seed)
kf = cv.split(X_train, X_train[era])
elif fold_type == "kfold":
cv = model_selection.KFold(n_splits=n_fold, shuffle=True, random_state=seed)
kf = cv.split(X_train, y_train)
fi_df = pd.DataFrame()
fi_df["features"] = features
fi_df["importance"] = 0
for fold_id, (train_index, valid_index) in tqdm(enumerate(kf)):
# split
X_tr = X_train.loc[train_index, features]
X_val = X_train.loc[valid_index, features]
y_tr = y_train.loc[train_index]
y_val = y_train.loc[valid_index]
# model (note inverse weighting)
train_set = lgb.Dataset(
X_tr, y_tr, categorical_feature=cats, weight=1 / np.power(y_tr, 2)
)
val_set = lgb.Dataset(
X_val, y_val, categorical_feature=cats, weight=1 / np.power(y_val, 2)
)
model = lgb.train(
params,
train_set,
valid_sets=[train_set, val_set],
feval=RMSPEMetric(),
verbose_eval=250,
)
# feature importance
fi_df[f"importance_fold{fold_id}"] = model.feature_importance(
importance_type="gain"
)
fi_df["importance"] += fi_df[f"importance_fold{fold_id}"].values
# save model
joblib.dump(model, f"model_fold{fold_id}.pkl")
logger.debug("model saved!")
# predict
oof_df["pred"].iloc[valid_index] = model.predict(X_val)
y_pred = model.predict(X_test[features])
y_preds += y_pred / n_fold
models.append(model)
return oof_df, y_preds, models, fi_df
if MODE == "TRAIN":
oof_df, y_preds, models, fi_df = fit_model(
params,
book_df,
book_df[target],
test_book_df,
features=features,
cats=cats,
era=None,
fold_type="kfold",
n_fold=5,
seed=46,
)
# # CV (Cross-Validation) Score
# How good is my model?
from sklearn.metrics import r2_score
def rmspe(y_true, y_pred):
return np.sqrt(np.mean(np.square((y_true - y_pred) / y_true)))
if MODE == "TRAIN":
oof_df.dropna(inplace=True)
y_true = oof_df[target].values
y_pred = oof_df["pred"].values
oof_df[target].hist(bins=100)
oof_df["pred"].hist(bins=100)
R2 = round(r2_score(y_true, y_pred), 3)
RMSPE = round(rmspe(y_true, y_pred), 3)
logger.info(f"Performance of the naive prediction: R2 score: {R2}, RMSPE: {RMSPE}")
# performance by stock_id
if MODE == "TRAIN":
for stock_id in oof_df["stock_id"].unique():
y_true = oof_df.query("stock_id == @stock_id")[target].values
y_pred = oof_df.query("stock_id == @stock_id")["pred"].values
R2 = round(r2_score(y_true, y_pred), 3)
RMSPE = round(rmspe(y_true, y_pred), 3)
logger.info(
f"Performance by stock_id={stock_id}: R2 score: {R2}, RMSPE: {RMSPE}"
)
# # Feature Importance
# Let's see top features for the prediction.
if MODE == "TRAIN":
fi_df = fi_df.sort_values(by="importance", ascending=False)
fi_df.to_csv("feature_importance.csv", index=False)
fig, ax = plt.subplots(1, 1, figsize=(10, 40))
sns.barplot(x="importance", y="features", data=fi_df.iloc[:30], ax=ax)
logger.info(fi_df[["features", "importance"]].iloc[:50].to_markdown())
# # Submit
if MODE == "INFERENCE":
"""
used for inference kernel only
"""
y_preds = np.zeros(len(test_book_df))
files = glob.glob(f"{MODEL_DIR}/*model*.pkl")
assert len(files) > 0
for i, f in enumerate(files):
model = joblib.load(f)
y_preds += model.predict(test_book_df[features])
y_preds /= i + 1
test_book_df[target] = y_preds
# test
test_book_df[target] = y_preds
# save the submit file
sub = test_book_df[["row_id", target]]
sub.to_csv("submission.csv", index=False)
logger.info("submitted!")
sub.head()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/401/69401828.ipynb
| null | null |
[{"Id": 69401828, "ScriptId": 18784971, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 590240, "CreationDate": "07/30/2021 14:33:39", "VersionNumber": 8.0, "Title": "[Optiver] LGB and TE baseline (inference)", "EvaluationDate": "07/30/2021", "IsChange": true, "TotalLines": 639.0, "LinesInsertedFromPrevious": 3.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 636.0, "LinesInsertedFromFork": 97.0, "LinesDeletedFromFork": 48.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 542.0, "TotalVotes": 1}]
| null | null | null | null |
# This is yet another starter using lightGBM.
# **Don't just fork this notebook and publish**, which just confuses people. In case you were to publish very similar notebook, please cite this notebook.
# As this notebook was created for new participants to get started easily, I tried to keep it as simple as possible.
# The basic strategy employed in this notebook includes:
# - Target Encoding: Simply using the past volatility works great due to the high autocorrelation.
# - Aggregation by multiple keys: not only stock_id x time_id, but also stock_id and time_id, separately.
# - Multithreads processing for feature engineering: features are made on a stock_id x time_id basis, so we might want to accelerate the process
# - LightGBM with the inverse weight of the square of the target: the evaluation metric is the root mean square percentage error, so we use RMSE as the objective function with the inverse weighting of the target.
# Note that this notebook only performs **inference** using the pretrained models generated in the following notebook to save time:
# [[Optiver] LGB and TE baseline](https://www.kaggle.com/code1110/optiver-lgb-and-te-baseline)
# So let's get the ball rolling!
DEBUG = False
# MODE = 'TRAIN'
MODE = "INFERENCE"
MODEL_DIR = "../input/optiver-lgb-and-te-baseline"
# # Libraries
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import gc
import pathlib
from tqdm.auto import tqdm
import json
from multiprocessing import Pool, cpu_count
import time
import requests as re
from datetime import datetime
from dateutil.relativedelta import relativedelta, FR
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import glob
import os
from sklearn import model_selection
import joblib
import lightgbm as lgb
# visualize
import matplotlib.pyplot as plt
import matplotlib.style as style
from matplotlib_venn import venn2, venn3
import seaborn as sns
from matplotlib import pyplot
from matplotlib.ticker import ScalarFormatter
sns.set_context("talk")
style.use("seaborn-colorblind")
import warnings
warnings.simplefilter("ignore")
pd.get_option("display.max_columns")
# # Config
class CFG:
INPUT_DIR = "../input/optiver-realized-volatility-prediction"
OUTPUT_DIR = "./"
# Logging is always nice for your experiment:)
def init_logger(log_file="train.log"):
from logging import getLogger, INFO, FileHandler, Formatter, StreamHandler
logger = getLogger(__name__)
logger.setLevel(INFO)
handler1 = StreamHandler()
handler1.setFormatter(Formatter("%(message)s"))
handler2 = FileHandler(filename=log_file)
handler2.setFormatter(Formatter("%(message)s"))
logger.addHandler(handler1)
logger.addHandler(handler2)
return logger
logger = init_logger(log_file=f"{CFG.OUTPUT_DIR}/baseline.log")
logger.info(f"Start Logging...")
# # Load data
# Data description found in :
# https://www.kaggle.com/c/optiver-realized-volatility-prediction/data
# ## Train, test, and sample submission
# train.csv The ground truth values for the training set.
# stock_id - Same as above, but since this is a csv the column will load as an integer instead of categorical.
# time_id - Same as above.
# target - The realized volatility computed over the 10 minute window following the feature data under the same stock/time_id. There is no overlap between feature and target data. You can find more info in our tutorial notebook.
# test.csv Provides the mapping between the other data files and the submission file. As with other test files, most of the data is only available to your notebook upon submission with just the first few rows available for download.
# stock_id - Same as above.
# time_id - Same as above.
# row_id - Unique identifier for the submission row. There is one row for each existing time ID/stock ID pair. Each time window is not necessarily containing every individual stock.
# sample_submission.csv - A sample submission file in the correct format.
# row_id - Same as in test.csv.
# target - Same definition as in train.csv. The benchmark is using the median target value from train.csv.
train = pd.read_csv(os.path.join(CFG.INPUT_DIR, "train.csv"))
logger.info("Train data: {}".format(train.shape))
train.head()
train["stock_id"].value_counts()
train["time_id"].value_counts()
train["target"].hist(bins=100)
# fig, ax = plt.subplots(16, 7, figsize=(20, 60))
# ax = ax.flatten()
# for i, stock_id in tqdm(enumerate(train['stock_id'].unique())):
# ax[i].hist(train.query('stock_id == @stock_id')['target'], bins=100)
# ax[i].set_title(stock_id)
# plt.tight_layout()
test = pd.read_csv(os.path.join(CFG.INPUT_DIR, "test.csv"))
logger.info("Test data: {}".format(test.shape))
test.head()
venn2([set(train["time_id"]), set(test["time_id"])])
ss = pd.read_csv(os.path.join(CFG.INPUT_DIR, "sample_submission.csv"))
logger.info("Sample submission: {}".format(ss.shape))
ss.head()
# ## Book
# book_[train/test].parquet A parquet file partitioned by stock_id. Provides order book data on the most competitive buy and sell orders entered into the market. The top two levels of the book are shared. The first level of the book will be more competitive in price terms, it will then receive execution priority over the second level.
# stock_id - ID code for the stock. Not all stock IDs exist in every time bucket. Parquet coerces this column to the categorical data type when loaded; you may wish to convert it to int8.
# time_id - ID code for the time bucket. Time IDs are not necessarily sequential but are consistent across all stocks.
# seconds_in_bucket - Number of seconds from the start of the bucket, always starting from 0.
# bid_price[1/2] - Normalized prices of the most/second most competitive buy level.
# ask_price[1/2] - Normalized prices of the most/second most competitive sell level.
# bid_size[1/2] - The number of shares on the most/second most competitive buy level.
# ask_size[1/2] - The number of shares on the most/second most competitive sell level.
# ## Trade
# trade_[train/test].parquet A parquet file partitioned by stock_id. Contains data on trades that actually executed. Usually, in the market, there are more passive buy/sell intention updates (book updates) than actual trades, therefore one may expect this file to be more sparse than the order book.
# stock_id - Same as above.
# time_id - Same as above.
# seconds_in_bucket - Same as above. Note that since trade and book data are taken from the same time window and trade data is more sparse in general, this field is not necessarily starting from 0.
# price - The average price of executed transactions happening in one second. Prices have been normalized and the average has been weighted by the number of shares traded in each transaction.
# size - The sum number of shares traded.
# order_count - The number of unique trade orders taking place.
train_book_stocks = os.listdir(os.path.join(CFG.INPUT_DIR, "book_train.parquet"))
if DEBUG:
logger.info("Debug mode: using 3 stocks only")
train_book_stocks = train_book_stocks[:3]
logger.info(
"{:,} train book stocks: {}".format(len(train_book_stocks), train_book_stocks)
)
# load stock_id=0
def load_book(stock_id=0, data_type="train"):
"""
load parquest book data for given stock_id
"""
book_df = pd.read_parquet(
os.path.join(CFG.INPUT_DIR, f"book_{data_type}.parquet/stock_id={stock_id}")
)
book_df["stock_id"] = stock_id
book_df["stock_id"] = book_df["stock_id"].astype(np.int8)
return book_df
def load_trade(stock_id=0, data_type="train"):
"""
load parquest trade data for given stock_id
"""
trade_df = pd.read_parquet(
os.path.join(CFG.INPUT_DIR, f"trade_{data_type}.parquet/stock_id={stock_id}")
)
trade_df["stock_id"] = stock_id
trade_df["stock_id"] = trade_df["stock_id"].astype(np.int8)
return trade_df
book0 = load_book(0)
logger.info("Book data of stock id = 1: {}".format(book0.shape))
book0.head()
book0.tail()
trade0 = load_trade(0)
logger.info("Book data of stock id = 1: {}".format(trade0.shape))
trade0.head()
trade0.tail()
book_df = book0.merge(
trade0, how="outer", on=["time_id", "stock_id", "seconds_in_bucket"]
)
print(book_df.shape)
book_df.head()
book_df.tail()
def fix_jsonerr(df):
"""
fix json column error for lightgbm
"""
df.columns = [
"".join(c if c.isalnum() else "_" for c in str(x)) for x in df.columns
]
return df
def log_return(list_stock_prices):
return np.log(list_stock_prices).diff()
def realized_volatility(series_log_return):
series_log_return = log_return(series_log_return)
return np.sqrt(np.sum(series_log_return**2))
def fe_row(book):
"""
Feature engineering (just volatility for now) for each row
"""
# volatility
for i in [
1,
2,
]:
# wap
book[f"book_wap{i}"] = (
book[f"bid_price{i}"] * book[f"ask_size{i}"]
+ book[f"ask_price{i}"] * book[f"bid_size{i}"]
) / (book[f"bid_size{i}"] + book[f"ask_size{i}"])
# mean wap
book["book_wap_mean"] = (book["book_wap1"] + book["book_wap2"]) / 2
# wap diff
book["book_wap_diff"] = book["book_wap1"] - book["book_wap2"]
# other orderbook features
book["book_price_spread"] = (book["ask_price1"] - book["bid_price1"]) / (
book["ask_price1"] + book["bid_price1"]
)
book["book_bid_spread"] = book["bid_price1"] - book["bid_price2"]
book["book_ask_spread"] = book["ask_price1"] - book["ask_price2"]
book["book_total_volume"] = (
book["ask_size1"] + book["ask_size2"] + book["bid_size1"] + book["bid_size2"]
)
book["book_volume_imbalance"] = (book["ask_size1"] + book["ask_size2"]) - (
book["bid_size1"] + book["bid_size2"]
)
return book
def fe_agg(book_df):
"""
feature engineering (aggregation by stock_id x time_id)
"""
# features
book_feats = book_df.columns[
book_df.columns.str.startswith("book_")
].values.tolist()
trade_feats = ["price", "size", "order_count", "seconds_in_bucket"]
# agg trade features
trade_df = (
book_df.groupby(["time_id", "stock_id"])[trade_feats]
.agg(["sum", "mean", "std", "max", "min"])
.reset_index()
)
# agg volatility features
fe_df = (
book_df.groupby(["time_id", "stock_id"])[book_feats]
.agg([realized_volatility])
.reset_index()
)
fe_df.columns = [" ".join(col).strip() for col in fe_df.columns.values]
# merge
fe_df = fe_df.merge(trade_df, how="left", on=["time_id", "stock_id"])
return fe_df
def fe_all(book_df):
"""
perform feature engineerings
"""
# row-wise feature engineering
book_df = fe_row(book_df)
# feature engineering agg by stock_id x time_id
fe_df = fe_agg(book_df)
return fe_df
def book_fe_by_stock(stock_id=0):
"""
load orderbook and trade data for the given stock_id and merge
"""
# load data
book_df = load_book(stock_id, "train")
trade_df = load_trade(stock_id, "train")
book_feats = book_df.columns.values.tolist()
# merge
book_df = book_df.merge(
trade_df, how="outer", on=["time_id", "seconds_in_bucket", "stock_id"]
)
# sort by time
book_df = book_df.sort_values(by=["time_id", "seconds_in_bucket"])
# fillna for book_df
book_df[book_feats] = book_df[book_feats].fillna(method="ffill")
# feature engineering
fe_df = fe_all(book_df)
return fe_df
def book_fe_by_stock_test(stock_id=0):
"""
same function but for the test
"""
# load data
book_df = load_book(stock_id, "test")
trade_df = load_trade(stock_id, "test")
book_feats = book_df.columns.values.tolist()
# merge
book_df = book_df.merge(
trade_df, how="outer", on=["time_id", "seconds_in_bucket", "stock_id"]
)
# sort by time
book_df = book_df.sort_values(by=["time_id", "seconds_in_bucket"])
# fillna for book_df
book_df[book_feats] = book_df[book_feats].fillna(method="ffill")
# feature engineering
fe_df = fe_all(book_df)
return fe_df
def book_fe_all(stock_ids, data_type="train"):
"""
Feature engineering with multithread processing
"""
# feature engineering agg by stock_id x time_id
with Pool(cpu_count()) as p:
if data_type == "train":
feature_dfs = list(
tqdm(p.imap(book_fe_by_stock, stock_ids), total=len(stock_ids))
)
elif data_type == "test":
feature_dfs = list(
tqdm(p.imap(book_fe_by_stock_test, stock_ids), total=len(stock_ids))
)
fe_df = pd.concat(feature_dfs)
# feature engineering agg by stock_id
vol_feats = [f for f in fe_df.columns if ("realized" in f) & ("wap" in f)]
if data_type == "train":
# agg
stock_df = (
fe_df.groupby("stock_id")[vol_feats]
.agg(
[
"mean",
"std",
"max",
"min",
]
)
.reset_index()
)
# fix column names
stock_df.columns = ["stock_id"] + [
f"{f}_stock" for f in stock_df.columns.values.tolist()[1:]
]
stock_df = fix_jsonerr(stock_df)
# feature engineering agg by time_id
time_df = (
fe_df.groupby("time_id")[vol_feats]
.agg(
[
"mean",
"std",
"max",
"min",
]
)
.reset_index()
)
time_df.columns = ["time_id"] + [
f"{f}_time" for f in time_df.columns.values.tolist()[1:]
]
# merge
fe_df = fe_df.merge(time_df, how="left", on="time_id")
# make sure to fix json error for lighgbm
fe_df = fix_jsonerr(fe_df)
# out
if data_type == "train":
return fe_df, stock_df
elif data_type == "test":
return fe_df
if MODE == "TRAIN":
# all book data feature engineering
stock_ids = [int(i.split("=")[-1]) for i in train_book_stocks]
book_df, stock_df = book_fe_all(stock_ids, data_type="train")
assert book_df["stock_id"].nunique() > 2
assert book_df["time_id"].nunique() > 2
# save stock_df for the test
stock_df.to_pickle("train_stock_df.pkl")
logger.info("train stock df saved!")
# merge
book_df = (
book_df.merge(stock_df, how="left", on="stock_id")
.merge(train, how="left", on=["stock_id", "time_id"])
.replace([np.inf, -np.inf], np.nan)
.fillna(method="ffill")
)
# make row_id
book_df["row_id"] = (
book_df["stock_id"].astype(str) + "-" + book_df["time_id"].astype(str)
)
book_df.to_pickle("book_df.pkl")
print(book_df.shape)
book_df.head()
# test
test_book_stocks = os.listdir(os.path.join(CFG.INPUT_DIR, "book_test.parquet"))
logger.info("{:,} test book stocks: {}".format(len(test_book_stocks), test_book_stocks))
# all book data feature engineering
test_stock_ids = [int(i.split("=")[-1]) for i in test_book_stocks]
test_book_df = book_fe_all(test_stock_ids, data_type="test")
# load stock_df, if inference
if MODE == "INFERENCE":
book_df = pd.read_pickle(f"{MODEL_DIR}/book_df.pkl")
stock_df = pd.read_pickle(f"{MODEL_DIR}/train_stock_df.pkl")
# merge
test_book_df = (
test.merge(stock_df, how="left", on="stock_id")
.merge(test_book_df, how="left", on=["stock_id", "time_id"])
.replace([np.inf, -np.inf], np.nan)
.fillna(method="ffill")
)
# make row_id
test_book_df["row_id"] = (
test_book_df["stock_id"].astype(str) + "-" + test_book_df["time_id"].astype(str)
)
print(test_book_df.shape)
test_book_df.head()
# # Modeling
# Currently,
# - MODEL: LightGBM (with inverse weighting of the square of the target)
# - OBJECTIVE FUNCTION: RMSE
# - METRIC: RMSPE (used for early stopping)
# - Validation Strategy: KFold (are you sure?)
#
target = "target"
drops = [target, "row_id", "time_id"]
features = [
f
for f in test_book_df.columns.values.tolist()
if (f not in drops)
& (test_book_df[f].isna().sum() == 0)
& (book_df[f].isna().sum() == 0)
]
cats = [
"stock_id",
]
logger.info(
"{:,} features ({:,} categorical): {}".format(len(features), len(cats), features)
)
# evaluation metric
def RMSPEMetric(XGBoost=False):
def RMSPE(yhat, dtrain, XGBoost=XGBoost):
y = dtrain.get_label()
elements = ((y - yhat) / y) ** 2
if XGBoost:
return "RMSPE", float(np.sqrt(np.sum(elements) / len(y)))
else:
return "RMSPE", float(np.sqrt(np.sum(elements) / len(y))), False
return RMSPE
# LightGBM parameters
params = {
"n_estimators": 20000,
"objective": "rmse",
"boosting_type": "gbdt",
"max_depth": -1,
"learning_rate": 0.01,
"subsample": 0.72,
"subsample_freq": 4,
"feature_fraction": 0.8,
"lambda_l1": 1,
"lambda_l2": 1,
"seed": 46,
"early_stopping_rounds": 500,
"verbose": -1,
}
def fit_model(
params,
X_train,
y_train,
X_test,
features=features,
cats=[],
era="stock_id",
fold_type="kfold",
n_fold=5,
seed=42,
):
"""
fit model with cross validation
"""
models = []
oof_df = X_train[["time_id", "stock_id", target]].copy()
oof_df["pred"] = np.nan
y_preds = np.zeros((len(X_test),))
if fold_type == "stratifiedshuffle":
cv = model_selection.StratifiedShuffleSplit(n_splits=n_fold, random_state=seed)
kf = cv.split(X_train, X_train[era])
elif fold_type == "kfold":
cv = model_selection.KFold(n_splits=n_fold, shuffle=True, random_state=seed)
kf = cv.split(X_train, y_train)
fi_df = pd.DataFrame()
fi_df["features"] = features
fi_df["importance"] = 0
for fold_id, (train_index, valid_index) in tqdm(enumerate(kf)):
# split
X_tr = X_train.loc[train_index, features]
X_val = X_train.loc[valid_index, features]
y_tr = y_train.loc[train_index]
y_val = y_train.loc[valid_index]
# model (note inverse weighting)
train_set = lgb.Dataset(
X_tr, y_tr, categorical_feature=cats, weight=1 / np.power(y_tr, 2)
)
val_set = lgb.Dataset(
X_val, y_val, categorical_feature=cats, weight=1 / np.power(y_val, 2)
)
model = lgb.train(
params,
train_set,
valid_sets=[train_set, val_set],
feval=RMSPEMetric(),
verbose_eval=250,
)
# feature importance
fi_df[f"importance_fold{fold_id}"] = model.feature_importance(
importance_type="gain"
)
fi_df["importance"] += fi_df[f"importance_fold{fold_id}"].values
# save model
joblib.dump(model, f"model_fold{fold_id}.pkl")
logger.debug("model saved!")
# predict
oof_df["pred"].iloc[valid_index] = model.predict(X_val)
y_pred = model.predict(X_test[features])
y_preds += y_pred / n_fold
models.append(model)
return oof_df, y_preds, models, fi_df
if MODE == "TRAIN":
oof_df, y_preds, models, fi_df = fit_model(
params,
book_df,
book_df[target],
test_book_df,
features=features,
cats=cats,
era=None,
fold_type="kfold",
n_fold=5,
seed=46,
)
# # CV (Cross-Validation) Score
# How good is my model?
from sklearn.metrics import r2_score
def rmspe(y_true, y_pred):
return np.sqrt(np.mean(np.square((y_true - y_pred) / y_true)))
if MODE == "TRAIN":
oof_df.dropna(inplace=True)
y_true = oof_df[target].values
y_pred = oof_df["pred"].values
oof_df[target].hist(bins=100)
oof_df["pred"].hist(bins=100)
R2 = round(r2_score(y_true, y_pred), 3)
RMSPE = round(rmspe(y_true, y_pred), 3)
logger.info(f"Performance of the naive prediction: R2 score: {R2}, RMSPE: {RMSPE}")
# performance by stock_id
if MODE == "TRAIN":
for stock_id in oof_df["stock_id"].unique():
y_true = oof_df.query("stock_id == @stock_id")[target].values
y_pred = oof_df.query("stock_id == @stock_id")["pred"].values
R2 = round(r2_score(y_true, y_pred), 3)
RMSPE = round(rmspe(y_true, y_pred), 3)
logger.info(
f"Performance by stock_id={stock_id}: R2 score: {R2}, RMSPE: {RMSPE}"
)
# # Feature Importance
# Let's see top features for the prediction.
if MODE == "TRAIN":
fi_df = fi_df.sort_values(by="importance", ascending=False)
fi_df.to_csv("feature_importance.csv", index=False)
fig, ax = plt.subplots(1, 1, figsize=(10, 40))
sns.barplot(x="importance", y="features", data=fi_df.iloc[:30], ax=ax)
logger.info(fi_df[["features", "importance"]].iloc[:50].to_markdown())
# # Submit
if MODE == "INFERENCE":
"""
used for inference kernel only
"""
y_preds = np.zeros(len(test_book_df))
files = glob.glob(f"{MODEL_DIR}/*model*.pkl")
assert len(files) > 0
for i, f in enumerate(files):
model = joblib.load(f)
y_preds += model.predict(test_book_df[features])
y_preds /= i + 1
test_book_df[target] = y_preds
# test
test_book_df[target] = y_preds
# save the submit file
sub = test_book_df[["row_id", target]]
sub.to_csv("submission.csv", index=False)
logger.info("submitted!")
sub.head()
| false | 0 | 6,759 | 1 | 6,759 | 6,759 |
||
69401953
|
<jupyter_start><jupyter_text>NYCTaxi
Kaggle dataset identifier: nyctaxi
<jupyter_script># # NYC Taxi Trip Duration Prediction
# ## Domain: Transportation
# ### Objective:Build a model that predicts the total trip duration of taxi trips in New York City.
#
# Loading Libraries
import pandas as pd # for data analysis
import numpy as np # for scientific calculation
import seaborn as sns # for statistical plotting
import datetime # for working with date fields
import matplotlib.pyplot as plt # for plotting
import math # for mathematical calculation
import os
# Reading NYC Taxi Trip given Data Set.
import os
for dirname, _, filenames in os.walk("kaggle/input/NYC_taxi_trip_train.csv"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Reading NYC taxi trip given Data Set.
nyc_taxi = pd.read_csv("/kaggle/input/NYC_taxi_trip_train.csv")
# # Data Cleaning and Data Understanding.
# Perform Pandas profiling to understand quick overview of columns
# import pandas_profiling
# report = pandas_profiling.ProfileReport(nyc_taxi)
# covert profile report as html file
# report.to_file("nyc_taxi.html")
# Checking Null Values : We can See there are No Null Values
nyc_taxi.isnull().sum()
# Checking shape of data
# Observation: It contains 1.4 million records approx. and 11 columns (10 features with 1 feature as a target variable)
nyc_taxi.shape
# Checking duplicates in the given dataset.
# Observations: No duplicates exists as it's row count shows '0'.
check_duplicates = nyc_taxi[nyc_taxi.duplicated()]
print(check_duplicates.shape)
# Exploring data by using info() method. It doesn't contains any null values.
# Observation: No null values exists.
nyc_taxi.info()
# Verifying top 2 sample records of data.
# Observation: The data consists of, vendor_id, pickup and dropoff datetime, longitude and latitude information, trip_duration
# values plays major part in predicting the tripduration here.
nyc_taxi.head(2)
# Describe method is used to view some basic statistical details like percentile, mean, std etc. of a data frame of numeric values.
# Observation: Due to huge dataset and the columns values has been given in the form of +/- (e.g., longitude and Latitude)
# it shows data in the form of exponentials.Moving ahead with EDA and visualization to understand data better.
nyc_taxi.describe()
# # Exploratory Data Analysis (EDA) and Feature Engineering
#
# Distance function to calculate distance between given longitude and latitude points.
# Observation: This piece of code taken from blogs. When I thought how to get pickup point and drop point information
# I found this code and I can able to calculate distance here. It's as been called as 'Haversine Formula'
from math import radians, cos, sin, asin, sqrt
def distance(lon1, lat1, lon2, lat2):
"""
Calculate the great circle distance between two points
on the earth (specified in decimal degrees)
"""
# convert decimal degrees to radians
lon1, lat1, lon2, lat2 = map(radians, [lon1, lat1, lon2, lat2])
# haversine formula
dlon = lon2 - lon1
dlat = lat2 - lat1
a = sin(dlat / 2) ** 2 + cos(lat1) * cos(lat2) * sin(dlon / 2) ** 2
c = 2 * asin(sqrt(a))
r = 6371 # Radius of earth in kilometers. Use 3956 for miles
return c * r
# Calculate Trip Distance & Speed here.
# Observation: Introduced New Columns Distance and Speed here.
# Converted dropoff_datetime and pickup_datetime into datetime format datatype
nyc_taxi["distance"] = nyc_taxi.apply(
lambda x: distance(
x["pickup_longitude"],
x["pickup_latitude"],
x["dropoff_longitude"],
x["dropoff_latitude"],
),
axis=1,
)
nyc_taxi["speed"] = nyc_taxi.distance / (nyc_taxi.trip_duration / 3600)
nyc_taxi["dropoff_datetime"] = pd.to_datetime(nyc_taxi["dropoff_datetime"])
nyc_taxi["pickup_datetime"] = pd.to_datetime(nyc_taxi["pickup_datetime"])
# Verify the column list
nyc_taxi.columns
# ## Data Visualization
# Copied dataframe into another dataframe.
# Observation: Using another dataframe for data visualization and keeping original copy for ML pipeline.
nyc_taxi_visual = nyc_taxi.copy()
# Verifying columns.
nyc_taxi_visual.columns
# Drop unused columns for data visualization
nyc_taxi_visual = nyc_taxi_visual.drop(
[
"pickup_datetime",
"dropoff_datetime",
"pickup_longitude",
"pickup_latitude",
"dropoff_longitude",
"dropoff_latitude",
"store_and_fwd_flag",
],
axis=1,
)
nyc_taxi_visual.columns
# Verifying datatype, count, null values by using info() method.
nyc_taxi_visual.info()
# Converting distance and speed values into int datatype.
nyc_taxi_visual["distance"] = nyc_taxi_visual["distance"].apply(lambda x: int(x))
nyc_taxi_visual["speed"] = nyc_taxi_visual["speed"].apply(lambda x: int(x))
# Verifying the datatype for all columns
nyc_taxi_visual.info()
# Seaborn scatter plot with regression line
# Observation: We can see her we are having outliers when distance is >50 miles and trip duration is >15000 seconds and
# we can say most of the trip_duration >15000 seconds mostly related to long distance or due to traffic jam on odd days.
# We can see regression line fits here when we remove outliers.
sns.lmplot(
x="trip_duration",
y="distance",
data=nyc_taxi_visual,
aspect=2.0,
scatter_kws={"alpha": 0.8},
)
# Removing outliers for distance and trip_duration.
nyc_taxi_visual_final = nyc_taxi_visual[nyc_taxi_visual["distance"] <= 600]
nyc_taxi_visual_final = nyc_taxi_visual[nyc_taxi_visual["trip_duration"] <= 36000]
nyc_taxi_visual_final.head(1)
# Distribution plot for trip_Duration.
# Observation: Data is right skewed here and most of datapoints is having very short trip_durations.
# Will apply scaling techniques before we train the model.
sns.distplot(nyc_taxi_visual_final["trip_duration"], kde=False)
# Distribution plot for Passenger Count.
# Observations: Most of the times, only single passenger has booked taxi. New york city most of the times, only one passenger
# travels due to population density and business center. It makes sense here. Very few trips for > 3 passengers.
sns.distplot(nyc_taxi_visual_final["passenger_count"], kde=False, bins=None)
plt.title("Distribution of Passenger Count")
plt.show()
# Verifying column details
nyc_taxi_visual_final.columns
# Groupby function to calculate passenger_count who has taken trips from Vendors.
# Observations: Vendor_id 1 is having more trips for passenger 1 and
# for Vendor_id 2 is having good number trips for passenger 1 and when passenger_count> 3 when compare to vendor_id 1
nyc_taxi_visual_final.groupby(by=["vendor_id", "passenger_count"])[
"passenger_count"
].count()
# Box plot for passenger_count for both vendors.
# Observation: Based on the given huge dataset it's clear that, we are having outliers for both vendors when passenger
# count increases more than 2.
# for vendor_id 1 we can see outliers when passenger_count is 0. might be because of empty trips or some other reasons.
plt.figure(figsize=(15, 5))
sns.boxplot(x="vendor_id", y="passenger_count", data=nyc_taxi_visual_final)
# Box plot for trip_duration for both vendors.
# Observation: We can see more outliders for both vendors when trip_duration is > 1000 seconds. It's a fact that
# new_york is one of the costliest and expensive life style city and most of the passenger can book trips <10 minutes travel.
# Might be these outliers trip_duration belongs to tourists.
plt.figure(figsize=(15, 5))
sns.boxplot(x="vendor_id", y="trip_duration", data=nyc_taxi_visual_final)
# np.max(nyc_taxi_visual_final['distance'])
# Plotly Scatter bubble chart used to visualize trip_duration and distance details vendor_id wise distribution.
# Observation: Most of datapoints lies between distance<50 and trip_duration <15K
# Vendor_id 1 is having outliers for distance > 100 miles.
# Vendor_id 2 is having outliers for trip_duration > 15000 seconds.
import plotly.express as px
fig = px.scatter(
nyc_taxi_visual_final,
x="trip_duration",
y="distance",
size="trip_duration",
color="vendor_id",
)
fig.update_layout(title="Trip Duration details vendor_id wise distribution")
fig.show()
# Plotly Pie chart used to visualize Share of each vendor_id in the given data set.
# Observation: Vendor_id 1 is having 46.5% and 2 is having 53.5% share in NYC Taxi Trips.
# Vendor_id 2 is having more than 15% of share when we compare with Vendor_id 1 share contribution.
import plotly.graph_objects as go
df1 = nyc_taxi_visual["vendor_id"].value_counts().reset_index()
fig = go.Figure(
data=[
go.Pie(
labels=df1["index"],
values=df1["vendor_id"],
hole=0.4,
title="Share of each Vendor",
)
]
)
fig.update_layout(title="NYC_Taxi Vendor Details")
fig.show()
# Plotly BAr chart used to visualize number of trips contributed by each vendor in the given data set.
# Observation: Vendor_id 2 is having more number of trips when compare to vendor_id 1.
sns.barplot(
nyc_taxi_visual_final["vendor_id"].value_counts().index,
nyc_taxi_visual_final["vendor_id"].value_counts().values,
alpha=0.8,
palette=sns.color_palette("RdBu"),
)
# Analyzing given datapoint based on distance travelled by passenger.
# Need to remove
nyc_taxi.sort_values(by="distance", ascending=False).head(10)
# # ML Pipeline for DataModeling
# ## Data Sampling, Feature Engineering and Importance
# Dropping unused columns
nyc_taxi_final = nyc_taxi.drop(
[
"vendor_id",
"pickup_longitude",
"pickup_latitude",
"dropoff_longitude",
"dropoff_latitude",
"store_and_fwd_flag",
],
axis=1,
)
# Creating new feature columns.
nyc_taxi_final["pickup_min"] = nyc_taxi_final["pickup_datetime"].apply(
lambda x: x.minute
)
nyc_taxi_final["pickup_hour"] = nyc_taxi_final["pickup_datetime"].apply(
lambda x: x.hour
)
nyc_taxi_final["pickup_day"] = nyc_taxi_final["pickup_datetime"].apply(lambda x: x.day)
nyc_taxi_final["pickup_month"] = nyc_taxi_final["pickup_datetime"].apply(
lambda x: int(x.month)
)
nyc_taxi_final["pickup_weekday"] = nyc_taxi_final["pickup_datetime"].dt.day_name()
nyc_taxi_final["pickup_month_name"] = nyc_taxi_final["pickup_datetime"].dt.month_name()
nyc_taxi_final["drop_hour"] = nyc_taxi_final["dropoff_datetime"].apply(lambda x: x.hour)
nyc_taxi_final["drop_month"] = nyc_taxi_final["dropoff_datetime"].apply(
lambda x: int(x.month)
)
nyc_taxi_final["drop_day"] = nyc_taxi_final["dropoff_datetime"].apply(lambda x: x.day)
nyc_taxi_final["drop_min"] = nyc_taxi_final["dropoff_datetime"].apply(
lambda x: x.minute
)
# Verifying newly created columns.
nyc_taxi_final.columns
## Removing all those records where speed is less than 1 and distance is 0
print(nyc_taxi_final.shape)
df = nyc_taxi_final[(nyc_taxi_final["speed"] < 1) & (nyc_taxi_final["distance"] == 0)]
nyc_taxi_final.drop(df.index, inplace=True)
print(nyc_taxi_final.shape)
# Identified some of trips are not valid from the given dataset.
# For e.g., Index 531 clearly says that pick up adn drop off date and time is having morethan 23 hours trip_duration
# by covering distance only 3 miles which is not possible in realtime scenarios. Removing those outliers. Total 1416 records.
nyc_taxi_final[
(nyc_taxi_final["pickup_day"] < nyc_taxi_final["drop_day"])
& (nyc_taxi_final["trip_duration"] > 10000)
& (nyc_taxi_final["distance"] < 5)
& (nyc_taxi_final["pickup_hour"] < 23)
]
# Dropping records for those whose pickup and drop timings are more and distance travel <3 miles. (Outliers.)
print(nyc_taxi_final.shape)
df = nyc_taxi_final[
(nyc_taxi_final["pickup_day"] < nyc_taxi_final["drop_day"])
& (nyc_taxi_final["trip_duration"] > 10000)
& (nyc_taxi_final["distance"] < 5)
& (nyc_taxi_final["pickup_hour"] < 23)
]
nyc_taxi_final.drop(df.index, inplace=True)
print(nyc_taxi_final.shape)
# Droppring records where speed and distance is <1. (Outliers)
print(nyc_taxi_final.shape)
df = nyc_taxi_final[(nyc_taxi_final["speed"] < 1) & (nyc_taxi_final["distance"] < 1)]
nyc_taxi_final.drop(df.index, inplace=True)
print(nyc_taxi_final.shape)
# Removing outliers identified based on trip_duration and distance.
nyc_taxi_final[nyc_taxi_final["trip_duration"] / 60 > 10000][
["trip_duration", "distance"]
]
print(nyc_taxi_final.shape)
nyc_taxi_final[nyc_taxi_final["trip_duration"] / 60 > 10000]["trip_duration"]
nyc_taxi_final.drop([978383, 680594, 355003], inplace=True)
print(nyc_taxi_final.shape)
# Removing outliers whose distance is less 200 meters. In real scenario, no-one will pick taxi for lesstance 200 meters.
print(nyc_taxi_final.shape)
df = nyc_taxi_final[nyc_taxi_final["distance"] < 0.2]
nyc_taxi_final.drop(df.index, inplace=True)
print(nyc_taxi_final.shape)
# Removing outliers those trips where passenger_count is 0.
print(nyc_taxi_final.shape)
df = nyc_taxi_final[nyc_taxi_final["passenger_count"] == 0]
nyc_taxi_final.drop(df.index, inplace=True)
print(nyc_taxi_final.shape)
# Verifying whether given data set is having other than 2016 year or not.
# Observation: It contains only 2016 year data.
import datetime as dt
print(nyc_taxi_final[nyc_taxi_final["dropoff_datetime"].dt.year > 2016])
print(nyc_taxi_final[nyc_taxi_final["dropoff_datetime"].dt.year < 2016])
# Removing outliers where trip_duration <120 seconds. In real-time scenario passengers will take trip for more than 2 mins.
print(nyc_taxi_final.shape)
df = nyc_taxi_final[nyc_taxi_final["trip_duration"] < 120]
nyc_taxi_final.drop(df.index, inplace=True)
print(nyc_taxi_final.shape)
# Distribution plot to verify the speed of trip.
# Observations: Most of trips is having speed < 40 miles/per hour. It's valid in newyork city trips.
dist_plot = nyc_taxi_final[nyc_taxi_final["speed"] < 100]["speed"]
sns.distplot(dist_plot, bins=10)
# Distribution plot to verify the speed of trip for complete dataset.
# Observations: Most of the trips is having speed <50 miles/per hour and removed outliers.
print(nyc_taxi_final.shape)
df = nyc_taxi_final[nyc_taxi_final["speed"] > 50]["speed"]
sns.distplot(df, bins=10)
nyc_taxi_final.drop(df.index, inplace=True)
print(nyc_taxi_final.shape)
# Verify column details.
nyc_taxi_final.columns
# Verifying the Day-Wise trip counts.
# Observation: We are having less trips on sunday and monday here.
print("Day-wise pickup totals")
print(nyc_taxi_final["pickup_weekday"].value_counts())
# Countplot visualization for Day-wise trip counts.
# Observations: Friday and Saturday is having more trips when compare to other days.
sns.countplot(x="pickup_weekday", data=nyc_taxi_final)
# Histogram plot to visualize for hour-wise trips
# Observations: Most ot trip counts is having between 5am to 23 pm and 0am(midnight) to 2am.
figure, ax = plt.subplots(nrows=1, ncols=2, figsize=(10, 5))
nyc_taxi_final["pickup_hour"] = nyc_taxi_final["pickup_datetime"].dt.hour
nyc_taxi_final.pickup_hour.hist(bins=24, ax=ax[0])
ax[0].set_title("Distribution of pickup hours")
nyc_taxi_final["dropoff_hour"] = nyc_taxi_final["dropoff_datetime"].dt.hour
nyc_taxi_final.dropoff_hour.hist(bins=24, ax=ax[1])
ax[1].set_title("Distribution of dropoff hours")
# ## Model Building, Evalutaion & Hyper parameter Tuning
# Import Sklearn and models
from sklearn import linear_model
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn import preprocessing
from sklearn.model_selection import cross_val_score, KFold
# Verifying final dataframe columns
nyc_taxi_final.columns
# ### Data Sampling Technique
# Due to huge dataset, training the model by applying datasampling technique. Random Sample taken 500000 records.
nyc_taxi_final_sampling = nyc_taxi_final.sample(n=500000, replace="False")
# Verify the shape of data
nyc_taxi_final_sampling.shape
# Dropping unused columns and copied required columns to the feature_columns to train the model.
# Used to verify co-efficient values.
feature_columns = nyc_taxi_final_sampling.drop(
[
"id",
"pickup_month_name",
"pickup_weekday",
"pickup_datetime",
"dropoff_datetime",
"trip_duration",
"passenger_count",
"speed",
],
axis=1,
)
# Verifying whether feature_columns is having null values or not.
nyc_taxi_final_sampling.distance = nyc_taxi_final_sampling.distance.astype(np.int64)
nyc_taxi_final_sampling.info()
# ## Linear Regression
# Applying Standard Scaler
X2 = nyc_taxi_final_sampling.drop(
[
"id",
"pickup_month_name",
"pickup_weekday",
"pickup_datetime",
"dropoff_datetime",
"trip_duration",
"passenger_count",
"speed",
],
axis=1,
)
X1 = preprocessing.scale(X2)
X = pd.DataFrame(X1)
y = nyc_taxi_final_sampling["trip_duration"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
reg = linear_model.LinearRegression()
reg.fit(X_train, y_train)
print("reg.intercept_=> %10.10f" % (reg.intercept_))
print(list(zip(feature_columns, reg.coef_)))
y_pred = reg.predict(X_test)
rmse_val = np.sqrt(metrics.mean_squared_error(y_test, y_pred))
# Null RMSE
y_null = np.zeros_like(y_test, dtype=int)
y_null.fill(y_test.mean())
N_RMSE = np.sqrt(metrics.mean_squared_error(y_test, y_null))
# Metrics
print("Mean Absolute Error :", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error :", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error = ", rmse_val)
print("Null RMSE = ", N_RMSE)
if N_RMSE < rmse_val:
print("Model is Not Doing Well Null RMSE Should be Greater")
else:
print("Model is Doing Well Null RMSE is Greater than RMSE")
# Train RMSE
y_pred_test = reg.predict(X_train)
rmse_val = np.sqrt(metrics.mean_squared_error(y_train, y_pred_test))
print(
"Train Root Mean Squared Error:",
np.sqrt(metrics.mean_squared_error(y_train, y_pred_test)),
)
# Error Percentage
df = pd.DataFrame({"Actual": y_test, "Predicted": y_pred, "Error": y_test - y_pred})
print("Maximum Error is :", df.Error.max())
print("Minimum Error is :", df.Error.min())
# Score
scores = cross_val_score(reg, X_train, y_train, cv=5)
print("Mean cross-validation score: %.2f" % scores.mean())
# ## XG Boost Regressor
import xgboost as xgb
from xgboost import plot_importance, plot_tree
from sklearn.metrics import mean_squared_error, mean_absolute_error
from sklearn.model_selection import RandomizedSearchCV, cross_val_score, KFold
X2 = nyc_taxi_final_sampling.drop(
[
"id",
"pickup_month_name",
"pickup_weekday",
"pickup_datetime",
"dropoff_datetime",
"trip_duration",
"passenger_count",
"speed",
],
axis=1,
)
X1 = preprocessing.scale(X2)
X = pd.DataFrame(X1)
y = nyc_taxi_final_sampling["trip_duration"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
model = xgb.XGBRegressor()
model.fit(X_train, y_train)
print(model)
y_pred = model.predict(data=X_test)
rmse_val = np.sqrt(metrics.mean_squared_error(y_test, y_pred))
# Null RMSE
y_null = np.zeros_like(y_test, dtype=int)
y_null.fill(y_test.mean())
N_RMSE = np.sqrt(metrics.mean_squared_error(y_test, y_null))
# Metrics
print("Mean Absolute Error :", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error :", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error = ", rmse_val)
print("Null RMSE = ", N_RMSE)
if N_RMSE < rmse_val:
print("Model is Not Doing Well Null RMSE Should be Greater")
else:
print("Model is Doing Well Null RMSE is Greater than RMSE")
# Train RMSE
y_pred_test = model.predict(X_train)
rmse_val = np.sqrt(metrics.mean_squared_error(y_train, y_pred_test))
print(
"Train Root Mean Squared Error:",
np.sqrt(metrics.mean_squared_error(y_train, y_pred_test)),
)
# Error Percentage
df = pd.DataFrame({"Actual": y_test, "Predicted": y_pred, "Error": y_test - y_pred})
print("Maximum Error is :", df.Error.max())
print("Minimum Error is :", df.Error.min())
# Score
scores = cross_val_score(model, X_train, y_train, cv=5)
print("Mean cross-validation score: %.2f" % scores.mean())
# ## Ridge Regression
from sklearn.linear_model import Ridge
X2 = nyc_taxi_final_sampling.drop(
[
"id",
"pickup_month_name",
"pickup_weekday",
"pickup_datetime",
"dropoff_datetime",
"trip_duration",
"passenger_count",
"speed",
],
axis=1,
)
X1 = preprocessing.scale(X2)
X = pd.DataFrame(X1)
y = nyc_taxi_final_sampling["trip_duration"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
ridgeReg = Ridge(alpha=0.05, normalize=True)
ridgeReg.fit(X_train, y_train)
y_pred = ridgeReg.predict(X_test)
rmse_val = np.sqrt(metrics.mean_squared_error(y_test, y_pred))
# NULL RMSE
y_null = np.zeros_like(y_test, dtype=int)
y_null.fill(y_test.mean())
N_RMSE = np.sqrt(metrics.mean_squared_error(y_test, y_null))
# Metrics
print("Mean Absolute Error :", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error :", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error = ", rmse_val)
print("Null RMSE = ", N_RMSE)
if N_RMSE < rmse_val:
print("Model is Not Doing Well Null RMSE Should be Greater")
else:
print("Model is Doing Well Null RMSE is Greater than RMSE")
# Train RMSE
y_pred_test = ridgeReg.predict(X_train)
rmse_val = np.sqrt(metrics.mean_squared_error(y_train, y_pred_test))
print(
"Train Root Mean Squared Error:",
np.sqrt(metrics.mean_squared_error(y_train, y_pred_test)),
)
# Error Percentage
df = pd.DataFrame({"Actual": y_test, "Predicted": y_pred, "Error": y_test - y_pred})
print("Maximum Error is :", df.Error.max())
print("Minimum Error is :", df.Error.min())
# Score
scores = cross_val_score(ridgeReg, X_train, y_train, cv=5)
print("Mean cross-validation score: %.2f" % scores.mean())
# ## RidgeCV (Cross Validation - Hyper Tuning parameter)
from sklearn.linear_model import RidgeCV
## training the model
X2 = nyc_taxi_final_sampling.drop(
[
"id",
"pickup_month_name",
"pickup_weekday",
"pickup_datetime",
"dropoff_datetime",
"trip_duration",
"passenger_count",
"speed",
],
axis=1,
)
X1 = preprocessing.scale(X2)
X = pd.DataFrame(X1)
y = nyc_taxi_final_sampling["trip_duration"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
ridgeRegCV = RidgeCV(alphas=[1e-3, 1e-2, 1e-1, 1])
ridgeRegCV.fit(X_train, y_train)
y_pred = ridgeRegCV.predict(X_test)
rmse_val = np.sqrt(metrics.mean_squared_error(y_test, y_pred))
# Null RMSE
y_null = np.zeros_like(y_test, dtype=int)
y_null.fill(y_test.mean())
N_RMSE = np.sqrt(metrics.mean_squared_error(y_test, y_null))
# Metrics
print("Mean Absolute Error :", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error :", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error = ", rmse_val)
print("Null RMSE = ", N_RMSE)
if N_RMSE < rmse_val:
print("Model is Not Doing Well Null RMSE Should be Greater")
else:
print("Model is Doing Well Null RMSE is Greater than RMSE")
# Train RMSE
y_pred_test = ridgeRegCV.predict(X_train)
rmse_val = np.sqrt(metrics.mean_squared_error(y_train, y_pred_test))
print(
"Train Root Mean Squared Error:",
np.sqrt(metrics.mean_squared_error(y_train, y_pred_test)),
)
# Error Percentage
df = pd.DataFrame({"Actual": y_test, "Predicted": y_pred, "Error": y_test - y_pred})
print("Maximum Error is :", df.Error.max())
print("Minimum Error is :", df.Error.min())
# Score
scores = cross_val_score(ridgeRegCV, X_train, y_train, cv=5)
print("Mean cross-validation score: %.2f" % scores.mean())
# Metrics Overview
df = {
"Model_Before_PCA": ["Linear_Reg", "XGB", "Ridge", "RidgeCV"],
"RMSE": ["875", "1249", "1605", "876"],
"NULL_RMSE": ["1701", "1701", "1701", "1701"],
"Max_error": ["66971", "84155", "85443", "67179"],
"Min_error": ["-80844", "-52886", "-4015", "-78683"],
"Score": ["72", "44", "11", "72"],
}
print("Metrics Overview Before_PCA")
dataframe = pd.DataFrame(
df, columns=["Model", "RMSE", "NULL_RMSE", "Max_error", "Min_error", "Score"]
)
dataframe
# # Running Model with PCA
# Verify the shape of data
nyc_taxi_final_sampling.shape
# Aligning data for PCA.
nyc_taxi_final_sampling.columns
nyc_taxi_pca = nyc_taxi_final_sampling.copy()
nyc_taxi_pca.drop(
[
"id",
"pickup_weekday",
"pickup_month_name",
"pickup_datetime",
"dropoff_datetime",
"speed",
],
axis=1,
inplace=True,
)
# seperate target variable for PCA.
target = nyc_taxi_pca["trip_duration"]
from sklearn import datasets
from sklearn.decomposition import PCA
# PCA
# normalize data
nyc_taxi_pca_norm = (nyc_taxi_pca - nyc_taxi_pca.mean()) / nyc_taxi_pca.std()
pca = PCA(n_components=12) # 12 features
pca.fit_transform(nyc_taxi_pca_norm.values)
print(pca.explained_variance_ratio_)
# print (nyc_taxi_final.feature_names)
print(pca.explained_variance_)
variance_ratio_cum_sum = np.cumsum(
np.round(pca.explained_variance_ratio_, decimals=4) * 100
)
print(variance_ratio_cum_sum)
print(pca.components_)
# Taken variance ratio of 7 PCA components at 93.6%.
pca.explained_variance_ratio_[:7].sum()
# Plot Elbow Curve
plt.plot(np.cumsum(pca.explained_variance_ratio_))
plt.xlabel("Number of components")
plt.ylabel("Cumulative explained variance")
plt.annotate("7", xy=(7, 0.93))
# consider first 7 components as they are explaining the 93% of variation in the data
x_pca = PCA(n_components=7)
nyc_taxi_pca_norm_final = x_pca.fit_transform(nyc_taxi_pca_norm)
# correlation between the variables after transforming the data with PCA is 0
correlation = pd.DataFrame(nyc_taxi_pca_norm_final).corr()
sns.heatmap(correlation, vmax=1, square=True, cmap="viridis")
plt.title("Correlation between different features")
# #After PCA, there is no correlation among any components.
# ## Linear Regression after PCA
X2 = preprocessing.scale(nyc_taxi_pca_norm_final)
X = pd.DataFrame(X2)
y = target
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
reg = linear_model.LinearRegression()
reg.fit(X_train, y_train)
print("reg.intercept_=> %10.10f" % (reg.intercept_))
print(list(zip(feature_columns, reg.coef_)))
y_pred = reg.predict(X_test)
rmse_val = np.sqrt(metrics.mean_squared_error(y_test, y_pred))
# NULL RMSE
y_null = np.zeros_like(y_test, dtype=int)
y_null.fill(y_test.mean())
N_RMSE = np.sqrt(metrics.mean_squared_error(y_test, y_null))
# Metrics
print("Mean Absolute Error :", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error :", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error = ", rmse_val)
print("Null RMSE = ", N_RMSE)
if N_RMSE < rmse_val:
print("Model is Not Doing Well Null RMSE Should be Greater")
else:
print("Model is Doing Well Null RMSE is Greater than RMSE")
# Train RMSE
y_pred_test = reg.predict(X_train)
rmse_val = np.sqrt(metrics.mean_squared_error(y_train, y_pred_test))
print(
"Train Root Mean Squared Error:",
np.sqrt(metrics.mean_squared_error(y_train, y_pred_test)),
)
# Error Percentage
df = pd.DataFrame({"Actual": y_test, "Predicted": y_pred, "Error": y_test - y_pred})
print("Maximum Error is :", df.Error.max())
print("Minimum Error is :", df.Error.min())
# Score
scores = cross_val_score(reg, X_train, y_train, cv=5)
print("Mean cross-validation score: %.2f" % scores.mean())
# ## XGB Regressor after PCA
import xgboost as xgb
from xgboost import plot_importance, plot_tree
from sklearn.metrics import mean_squared_error, mean_absolute_error
from sklearn.model_selection import RandomizedSearchCV, cross_val_score, KFold
X2 = preprocessing.scale(nyc_taxi_pca_norm_final)
X = pd.DataFrame(X2)
y = target
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
model = xgb.XGBRegressor()
model.fit(X_train, y_train)
print(model)
y_pred = model.predict(data=X_test)
rmse_val = np.sqrt(metrics.mean_squared_error(y_test, y_pred))
# Null RMSE
y_null = np.zeros_like(y_test, dtype=int)
y_null.fill(y_test.mean())
N_RMSE = np.sqrt(metrics.mean_squared_error(y_test, y_null))
# Metrics
print("Mean Absolute Error :", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error :", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error = ", rmse_val)
print("Null RMSE = ", N_RMSE)
if N_RMSE < rmse_val:
print("Model is Not Doing Well Null RMSE Should be Greater")
else:
print("Model is Doing Well Null RMSE is Greater than RMSE")
# Train RMSE
y_pred_test = model.predict(X_train)
rmse_val = np.sqrt(metrics.mean_squared_error(y_train, y_pred_test))
print(
"Train Root Mean Squared Error:",
np.sqrt(metrics.mean_squared_error(y_train, y_pred_test)),
)
# Error Percentage
df = pd.DataFrame({"Actual": y_test, "Predicted": y_pred, "Error": y_test - y_pred})
print("Maximum Error is :", df.Error.max())
print("Minimum Error is :", df.Error.min())
# Score
scores = cross_val_score(model, X_train, y_train, cv=5)
print("Mean cross-validation score: %.2f" % scores.mean())
# ## Ridge Regression after PCA
X2 = preprocessing.scale(nyc_taxi_pca_norm_final)
X = pd.DataFrame(X2)
y = target
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
ridgeReg = Ridge(alpha=0.05, normalize=True)
ridgeReg.fit(X_train, y_train)
y_pred = ridgeReg.predict(X_test)
rmse_val = np.sqrt(metrics.mean_squared_error(y_test, y_pred))
# Null RMSE
y_null = np.zeros_like(y_test, dtype=int)
y_null.fill(y_test.mean())
N_RMSE = np.sqrt(metrics.mean_squared_error(y_test, y_null))
# Metrics
print("Mean Absolute Error :", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error :", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error = ", rmse_val)
print("Null RMSE = ", N_RMSE)
if N_RMSE < rmse_val:
print("Model is Not Doing Well Null RMSE Should be Greater")
else:
print("Model is Doing Well Null RMSE is Greater than RMSE")
# Train RMSE
y_pred_test = ridgeReg.predict(X_train)
rmse_val = np.sqrt(metrics.mean_squared_error(y_train, y_pred_test))
print(
"Train Root Mean Squared Error:",
np.sqrt(metrics.mean_squared_error(y_train, y_pred_test)),
)
# Error Percentage
df = pd.DataFrame({"Actual": y_test, "Predicted": y_pred, "Error": y_test - y_pred})
print("Maximum Error is :", df.Error.max())
print("Minimum Error is :", df.Error.min())
# Score
scores = cross_val_score(ridgeReg, X_train, y_train, cv=5)
print("Mean cross-validation score: %.2f" % scores.mean())
# ## Ridge Regression CV after PCA
X2 = preprocessing.scale(nyc_taxi_pca_norm_final)
X = pd.DataFrame(X2)
y = target
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
ridgeRegCV = RidgeCV(alphas=[1e-3, 1e-2, 1e-1, 1])
ridgeRegCV.fit(X_train, y_train)
y_pred = ridgeRegCV.predict(X_test)
rmse_val = np.sqrt(metrics.mean_squared_error(y_test, y_pred))
# Null RMSE
y_null = np.zeros_like(y_test, dtype=int)
y_null.fill(y_test.mean())
N_RMSE = np.sqrt(metrics.mean_squared_error(y_test, y_null))
# Metrics
print("Mean Absolute Error :", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error :", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error = ", rmse_val)
print("Null RMSE = ", N_RMSE)
if N_RMSE < rmse_val:
print("Model is Not Doing Well Null RMSE Should be Greater")
else:
print("Model is Doing Well Null RMSE is Greater than RMSE")
# Train RMSE
y_pred_test = ridgeRegCV.predict(X_train)
rmse_val = np.sqrt(metrics.mean_squared_error(y_train, y_pred_test))
print(
"Train Root Mean Squared Error:",
np.sqrt(metrics.mean_squared_error(y_train, y_pred_test)),
)
# Error Percentage
df = pd.DataFrame({"Actual": y_test, "Predicted": y_pred, "Error": y_test - y_pred})
print("Maximum Error is :", df.Error.max())
print("Minimum Error is :", df.Error.min())
# Score
scores = cross_val_score(ridgeRegCV, X_train, y_train, cv=5)
print("Mean cross-validation score: %.2f" % scores.mean())
df = {
"Model_Before_PCA": ["Linear_Reg", "XGB", "Ridge", "RidgeCV"],
"RMSE": ["875", "1249", "1605", "876"],
"NULL_RMSE": ["1701", "1701", "1701", "1701"],
"Max_error": ["66971", "84155", "85443", "67179"],
"Min_error": ["-80844", "-52886", "-4015", "-78683"],
"Score": ["72", "44", "11", "72"],
}
print("Metrics Overview Before_PCA")
dataframe = pd.DataFrame(
df,
columns=[
"Model_Before_PCA",
"RMSE",
"NULL_RMSE",
"Max_error",
"Min_error",
"Score",
],
)
dataframe
df = {
"Model_After_PCA": ["Linear_Reg", "XGB", "Ridge", "RidgeCV"],
"RMSE": ["988", "108", "990", "988"],
"NULL_RMSE": ["1701", "1701", "1701", "1701"],
"Max_error": ["42900", "13824", "44918", "42900"],
"Min_error": ["-12259", "-9575", "-11434", "-12259"],
"Score": ["66", "99", "66", "66"],
}
print("Metrics Overview After_PCA")
dataframe = pd.DataFrame(
df,
columns=["Model_After_PCA", "RMSE", "NULL_RMSE", "Max_error", "Min_error", "Score"],
)
dataframe
# ## Final Conclusion:
# #### After comparing accuracy between model before and after PCA analysis. It has been decided that
# #### XGB Regressor is the best model which RMSE value: 108 when compare to other models and model accuracy is 99%.
# #### Accuracy Score is 99% with Max trip_duration error 13824,Min trip_duration error -9575 and RMSE 108
X2 = preprocessing.scale(nyc_taxi_pca_norm_final)
X = pd.DataFrame(X2)
y = target
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
model = xgb.XGBRegressor()
model.fit(X_train, y_train)
y_pred = model.predict(data=X_test)
x_ax = range(len(y_test))
plt.figure(figsize=(20, 10))
plt.scatter(x_ax, y_test, s=10, color="blue", label="original")
plt.scatter(x_ax, y_pred, s=10, color="red", label="predicted")
plt.legend()
plt.show()
# From above plot you can see index:59891 is having trip_duration of 86391 acutal value(blue point)
# predicated value (red point)
nyc_taxi_final_sampling.sort_values(by="trip_duration", ascending=False).head(2)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/401/69401953.ipynb
|
nyctaxi
|
rameshbabugonegandla
|
[{"Id": 69401953, "ScriptId": 10117337, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 3798813, "CreationDate": "07/30/2021 14:34:59", "VersionNumber": 40.0, "Title": "NYC Taxi with PCA", "EvaluationDate": "07/30/2021", "IsChange": false, "TotalLines": 783.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 783.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92521324, "KernelVersionId": 69401953, "SourceDatasetVersionId": 1245459}]
|
[{"Id": 1245459, "DatasetId": 714942, "DatasourceVersionId": 1277238, "CreatorUserId": 3798813, "LicenseName": "Unknown", "CreationDate": "06/14/2020 12:06:48", "VersionNumber": 1.0, "Title": "NYCTaxi", "Slug": "nyctaxi", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 714942, "CreatorUserId": 3798813, "OwnerUserId": 3798813.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1245459.0, "CurrentDatasourceVersionId": 1277238.0, "ForumId": 729718, "Type": 2, "CreationDate": "06/14/2020 12:06:48", "LastActivityDate": "06/14/2020", "TotalViews": 1224, "TotalDownloads": 58, "TotalVotes": 2, "TotalKernels": 2}]
|
[{"Id": 3798813, "UserName": "rameshbabugonegandla", "DisplayName": "Ramesh Babu Gonegandla", "RegisterDate": "10/03/2019", "PerformanceTier": 2}]
|
# # NYC Taxi Trip Duration Prediction
# ## Domain: Transportation
# ### Objective:Build a model that predicts the total trip duration of taxi trips in New York City.
#
# Loading Libraries
import pandas as pd # for data analysis
import numpy as np # for scientific calculation
import seaborn as sns # for statistical plotting
import datetime # for working with date fields
import matplotlib.pyplot as plt # for plotting
import math # for mathematical calculation
import os
# Reading NYC Taxi Trip given Data Set.
import os
for dirname, _, filenames in os.walk("kaggle/input/NYC_taxi_trip_train.csv"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Reading NYC taxi trip given Data Set.
nyc_taxi = pd.read_csv("/kaggle/input/NYC_taxi_trip_train.csv")
# # Data Cleaning and Data Understanding.
# Perform Pandas profiling to understand quick overview of columns
# import pandas_profiling
# report = pandas_profiling.ProfileReport(nyc_taxi)
# covert profile report as html file
# report.to_file("nyc_taxi.html")
# Checking Null Values : We can See there are No Null Values
nyc_taxi.isnull().sum()
# Checking shape of data
# Observation: It contains 1.4 million records approx. and 11 columns (10 features with 1 feature as a target variable)
nyc_taxi.shape
# Checking duplicates in the given dataset.
# Observations: No duplicates exists as it's row count shows '0'.
check_duplicates = nyc_taxi[nyc_taxi.duplicated()]
print(check_duplicates.shape)
# Exploring data by using info() method. It doesn't contains any null values.
# Observation: No null values exists.
nyc_taxi.info()
# Verifying top 2 sample records of data.
# Observation: The data consists of, vendor_id, pickup and dropoff datetime, longitude and latitude information, trip_duration
# values plays major part in predicting the tripduration here.
nyc_taxi.head(2)
# Describe method is used to view some basic statistical details like percentile, mean, std etc. of a data frame of numeric values.
# Observation: Due to huge dataset and the columns values has been given in the form of +/- (e.g., longitude and Latitude)
# it shows data in the form of exponentials.Moving ahead with EDA and visualization to understand data better.
nyc_taxi.describe()
# # Exploratory Data Analysis (EDA) and Feature Engineering
#
# Distance function to calculate distance between given longitude and latitude points.
# Observation: This piece of code taken from blogs. When I thought how to get pickup point and drop point information
# I found this code and I can able to calculate distance here. It's as been called as 'Haversine Formula'
from math import radians, cos, sin, asin, sqrt
def distance(lon1, lat1, lon2, lat2):
"""
Calculate the great circle distance between two points
on the earth (specified in decimal degrees)
"""
# convert decimal degrees to radians
lon1, lat1, lon2, lat2 = map(radians, [lon1, lat1, lon2, lat2])
# haversine formula
dlon = lon2 - lon1
dlat = lat2 - lat1
a = sin(dlat / 2) ** 2 + cos(lat1) * cos(lat2) * sin(dlon / 2) ** 2
c = 2 * asin(sqrt(a))
r = 6371 # Radius of earth in kilometers. Use 3956 for miles
return c * r
# Calculate Trip Distance & Speed here.
# Observation: Introduced New Columns Distance and Speed here.
# Converted dropoff_datetime and pickup_datetime into datetime format datatype
nyc_taxi["distance"] = nyc_taxi.apply(
lambda x: distance(
x["pickup_longitude"],
x["pickup_latitude"],
x["dropoff_longitude"],
x["dropoff_latitude"],
),
axis=1,
)
nyc_taxi["speed"] = nyc_taxi.distance / (nyc_taxi.trip_duration / 3600)
nyc_taxi["dropoff_datetime"] = pd.to_datetime(nyc_taxi["dropoff_datetime"])
nyc_taxi["pickup_datetime"] = pd.to_datetime(nyc_taxi["pickup_datetime"])
# Verify the column list
nyc_taxi.columns
# ## Data Visualization
# Copied dataframe into another dataframe.
# Observation: Using another dataframe for data visualization and keeping original copy for ML pipeline.
nyc_taxi_visual = nyc_taxi.copy()
# Verifying columns.
nyc_taxi_visual.columns
# Drop unused columns for data visualization
nyc_taxi_visual = nyc_taxi_visual.drop(
[
"pickup_datetime",
"dropoff_datetime",
"pickup_longitude",
"pickup_latitude",
"dropoff_longitude",
"dropoff_latitude",
"store_and_fwd_flag",
],
axis=1,
)
nyc_taxi_visual.columns
# Verifying datatype, count, null values by using info() method.
nyc_taxi_visual.info()
# Converting distance and speed values into int datatype.
nyc_taxi_visual["distance"] = nyc_taxi_visual["distance"].apply(lambda x: int(x))
nyc_taxi_visual["speed"] = nyc_taxi_visual["speed"].apply(lambda x: int(x))
# Verifying the datatype for all columns
nyc_taxi_visual.info()
# Seaborn scatter plot with regression line
# Observation: We can see her we are having outliers when distance is >50 miles and trip duration is >15000 seconds and
# we can say most of the trip_duration >15000 seconds mostly related to long distance or due to traffic jam on odd days.
# We can see regression line fits here when we remove outliers.
sns.lmplot(
x="trip_duration",
y="distance",
data=nyc_taxi_visual,
aspect=2.0,
scatter_kws={"alpha": 0.8},
)
# Removing outliers for distance and trip_duration.
nyc_taxi_visual_final = nyc_taxi_visual[nyc_taxi_visual["distance"] <= 600]
nyc_taxi_visual_final = nyc_taxi_visual[nyc_taxi_visual["trip_duration"] <= 36000]
nyc_taxi_visual_final.head(1)
# Distribution plot for trip_Duration.
# Observation: Data is right skewed here and most of datapoints is having very short trip_durations.
# Will apply scaling techniques before we train the model.
sns.distplot(nyc_taxi_visual_final["trip_duration"], kde=False)
# Distribution plot for Passenger Count.
# Observations: Most of the times, only single passenger has booked taxi. New york city most of the times, only one passenger
# travels due to population density and business center. It makes sense here. Very few trips for > 3 passengers.
sns.distplot(nyc_taxi_visual_final["passenger_count"], kde=False, bins=None)
plt.title("Distribution of Passenger Count")
plt.show()
# Verifying column details
nyc_taxi_visual_final.columns
# Groupby function to calculate passenger_count who has taken trips from Vendors.
# Observations: Vendor_id 1 is having more trips for passenger 1 and
# for Vendor_id 2 is having good number trips for passenger 1 and when passenger_count> 3 when compare to vendor_id 1
nyc_taxi_visual_final.groupby(by=["vendor_id", "passenger_count"])[
"passenger_count"
].count()
# Box plot for passenger_count for both vendors.
# Observation: Based on the given huge dataset it's clear that, we are having outliers for both vendors when passenger
# count increases more than 2.
# for vendor_id 1 we can see outliers when passenger_count is 0. might be because of empty trips or some other reasons.
plt.figure(figsize=(15, 5))
sns.boxplot(x="vendor_id", y="passenger_count", data=nyc_taxi_visual_final)
# Box plot for trip_duration for both vendors.
# Observation: We can see more outliders for both vendors when trip_duration is > 1000 seconds. It's a fact that
# new_york is one of the costliest and expensive life style city and most of the passenger can book trips <10 minutes travel.
# Might be these outliers trip_duration belongs to tourists.
plt.figure(figsize=(15, 5))
sns.boxplot(x="vendor_id", y="trip_duration", data=nyc_taxi_visual_final)
# np.max(nyc_taxi_visual_final['distance'])
# Plotly Scatter bubble chart used to visualize trip_duration and distance details vendor_id wise distribution.
# Observation: Most of datapoints lies between distance<50 and trip_duration <15K
# Vendor_id 1 is having outliers for distance > 100 miles.
# Vendor_id 2 is having outliers for trip_duration > 15000 seconds.
import plotly.express as px
fig = px.scatter(
nyc_taxi_visual_final,
x="trip_duration",
y="distance",
size="trip_duration",
color="vendor_id",
)
fig.update_layout(title="Trip Duration details vendor_id wise distribution")
fig.show()
# Plotly Pie chart used to visualize Share of each vendor_id in the given data set.
# Observation: Vendor_id 1 is having 46.5% and 2 is having 53.5% share in NYC Taxi Trips.
# Vendor_id 2 is having more than 15% of share when we compare with Vendor_id 1 share contribution.
import plotly.graph_objects as go
df1 = nyc_taxi_visual["vendor_id"].value_counts().reset_index()
fig = go.Figure(
data=[
go.Pie(
labels=df1["index"],
values=df1["vendor_id"],
hole=0.4,
title="Share of each Vendor",
)
]
)
fig.update_layout(title="NYC_Taxi Vendor Details")
fig.show()
# Plotly BAr chart used to visualize number of trips contributed by each vendor in the given data set.
# Observation: Vendor_id 2 is having more number of trips when compare to vendor_id 1.
sns.barplot(
nyc_taxi_visual_final["vendor_id"].value_counts().index,
nyc_taxi_visual_final["vendor_id"].value_counts().values,
alpha=0.8,
palette=sns.color_palette("RdBu"),
)
# Analyzing given datapoint based on distance travelled by passenger.
# Need to remove
nyc_taxi.sort_values(by="distance", ascending=False).head(10)
# # ML Pipeline for DataModeling
# ## Data Sampling, Feature Engineering and Importance
# Dropping unused columns
nyc_taxi_final = nyc_taxi.drop(
[
"vendor_id",
"pickup_longitude",
"pickup_latitude",
"dropoff_longitude",
"dropoff_latitude",
"store_and_fwd_flag",
],
axis=1,
)
# Creating new feature columns.
nyc_taxi_final["pickup_min"] = nyc_taxi_final["pickup_datetime"].apply(
lambda x: x.minute
)
nyc_taxi_final["pickup_hour"] = nyc_taxi_final["pickup_datetime"].apply(
lambda x: x.hour
)
nyc_taxi_final["pickup_day"] = nyc_taxi_final["pickup_datetime"].apply(lambda x: x.day)
nyc_taxi_final["pickup_month"] = nyc_taxi_final["pickup_datetime"].apply(
lambda x: int(x.month)
)
nyc_taxi_final["pickup_weekday"] = nyc_taxi_final["pickup_datetime"].dt.day_name()
nyc_taxi_final["pickup_month_name"] = nyc_taxi_final["pickup_datetime"].dt.month_name()
nyc_taxi_final["drop_hour"] = nyc_taxi_final["dropoff_datetime"].apply(lambda x: x.hour)
nyc_taxi_final["drop_month"] = nyc_taxi_final["dropoff_datetime"].apply(
lambda x: int(x.month)
)
nyc_taxi_final["drop_day"] = nyc_taxi_final["dropoff_datetime"].apply(lambda x: x.day)
nyc_taxi_final["drop_min"] = nyc_taxi_final["dropoff_datetime"].apply(
lambda x: x.minute
)
# Verifying newly created columns.
nyc_taxi_final.columns
## Removing all those records where speed is less than 1 and distance is 0
print(nyc_taxi_final.shape)
df = nyc_taxi_final[(nyc_taxi_final["speed"] < 1) & (nyc_taxi_final["distance"] == 0)]
nyc_taxi_final.drop(df.index, inplace=True)
print(nyc_taxi_final.shape)
# Identified some of trips are not valid from the given dataset.
# For e.g., Index 531 clearly says that pick up adn drop off date and time is having morethan 23 hours trip_duration
# by covering distance only 3 miles which is not possible in realtime scenarios. Removing those outliers. Total 1416 records.
nyc_taxi_final[
(nyc_taxi_final["pickup_day"] < nyc_taxi_final["drop_day"])
& (nyc_taxi_final["trip_duration"] > 10000)
& (nyc_taxi_final["distance"] < 5)
& (nyc_taxi_final["pickup_hour"] < 23)
]
# Dropping records for those whose pickup and drop timings are more and distance travel <3 miles. (Outliers.)
print(nyc_taxi_final.shape)
df = nyc_taxi_final[
(nyc_taxi_final["pickup_day"] < nyc_taxi_final["drop_day"])
& (nyc_taxi_final["trip_duration"] > 10000)
& (nyc_taxi_final["distance"] < 5)
& (nyc_taxi_final["pickup_hour"] < 23)
]
nyc_taxi_final.drop(df.index, inplace=True)
print(nyc_taxi_final.shape)
# Droppring records where speed and distance is <1. (Outliers)
print(nyc_taxi_final.shape)
df = nyc_taxi_final[(nyc_taxi_final["speed"] < 1) & (nyc_taxi_final["distance"] < 1)]
nyc_taxi_final.drop(df.index, inplace=True)
print(nyc_taxi_final.shape)
# Removing outliers identified based on trip_duration and distance.
nyc_taxi_final[nyc_taxi_final["trip_duration"] / 60 > 10000][
["trip_duration", "distance"]
]
print(nyc_taxi_final.shape)
nyc_taxi_final[nyc_taxi_final["trip_duration"] / 60 > 10000]["trip_duration"]
nyc_taxi_final.drop([978383, 680594, 355003], inplace=True)
print(nyc_taxi_final.shape)
# Removing outliers whose distance is less 200 meters. In real scenario, no-one will pick taxi for lesstance 200 meters.
print(nyc_taxi_final.shape)
df = nyc_taxi_final[nyc_taxi_final["distance"] < 0.2]
nyc_taxi_final.drop(df.index, inplace=True)
print(nyc_taxi_final.shape)
# Removing outliers those trips where passenger_count is 0.
print(nyc_taxi_final.shape)
df = nyc_taxi_final[nyc_taxi_final["passenger_count"] == 0]
nyc_taxi_final.drop(df.index, inplace=True)
print(nyc_taxi_final.shape)
# Verifying whether given data set is having other than 2016 year or not.
# Observation: It contains only 2016 year data.
import datetime as dt
print(nyc_taxi_final[nyc_taxi_final["dropoff_datetime"].dt.year > 2016])
print(nyc_taxi_final[nyc_taxi_final["dropoff_datetime"].dt.year < 2016])
# Removing outliers where trip_duration <120 seconds. In real-time scenario passengers will take trip for more than 2 mins.
print(nyc_taxi_final.shape)
df = nyc_taxi_final[nyc_taxi_final["trip_duration"] < 120]
nyc_taxi_final.drop(df.index, inplace=True)
print(nyc_taxi_final.shape)
# Distribution plot to verify the speed of trip.
# Observations: Most of trips is having speed < 40 miles/per hour. It's valid in newyork city trips.
dist_plot = nyc_taxi_final[nyc_taxi_final["speed"] < 100]["speed"]
sns.distplot(dist_plot, bins=10)
# Distribution plot to verify the speed of trip for complete dataset.
# Observations: Most of the trips is having speed <50 miles/per hour and removed outliers.
print(nyc_taxi_final.shape)
df = nyc_taxi_final[nyc_taxi_final["speed"] > 50]["speed"]
sns.distplot(df, bins=10)
nyc_taxi_final.drop(df.index, inplace=True)
print(nyc_taxi_final.shape)
# Verify column details.
nyc_taxi_final.columns
# Verifying the Day-Wise trip counts.
# Observation: We are having less trips on sunday and monday here.
print("Day-wise pickup totals")
print(nyc_taxi_final["pickup_weekday"].value_counts())
# Countplot visualization for Day-wise trip counts.
# Observations: Friday and Saturday is having more trips when compare to other days.
sns.countplot(x="pickup_weekday", data=nyc_taxi_final)
# Histogram plot to visualize for hour-wise trips
# Observations: Most ot trip counts is having between 5am to 23 pm and 0am(midnight) to 2am.
figure, ax = plt.subplots(nrows=1, ncols=2, figsize=(10, 5))
nyc_taxi_final["pickup_hour"] = nyc_taxi_final["pickup_datetime"].dt.hour
nyc_taxi_final.pickup_hour.hist(bins=24, ax=ax[0])
ax[0].set_title("Distribution of pickup hours")
nyc_taxi_final["dropoff_hour"] = nyc_taxi_final["dropoff_datetime"].dt.hour
nyc_taxi_final.dropoff_hour.hist(bins=24, ax=ax[1])
ax[1].set_title("Distribution of dropoff hours")
# ## Model Building, Evalutaion & Hyper parameter Tuning
# Import Sklearn and models
from sklearn import linear_model
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn import preprocessing
from sklearn.model_selection import cross_val_score, KFold
# Verifying final dataframe columns
nyc_taxi_final.columns
# ### Data Sampling Technique
# Due to huge dataset, training the model by applying datasampling technique. Random Sample taken 500000 records.
nyc_taxi_final_sampling = nyc_taxi_final.sample(n=500000, replace="False")
# Verify the shape of data
nyc_taxi_final_sampling.shape
# Dropping unused columns and copied required columns to the feature_columns to train the model.
# Used to verify co-efficient values.
feature_columns = nyc_taxi_final_sampling.drop(
[
"id",
"pickup_month_name",
"pickup_weekday",
"pickup_datetime",
"dropoff_datetime",
"trip_duration",
"passenger_count",
"speed",
],
axis=1,
)
# Verifying whether feature_columns is having null values or not.
nyc_taxi_final_sampling.distance = nyc_taxi_final_sampling.distance.astype(np.int64)
nyc_taxi_final_sampling.info()
# ## Linear Regression
# Applying Standard Scaler
X2 = nyc_taxi_final_sampling.drop(
[
"id",
"pickup_month_name",
"pickup_weekday",
"pickup_datetime",
"dropoff_datetime",
"trip_duration",
"passenger_count",
"speed",
],
axis=1,
)
X1 = preprocessing.scale(X2)
X = pd.DataFrame(X1)
y = nyc_taxi_final_sampling["trip_duration"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
reg = linear_model.LinearRegression()
reg.fit(X_train, y_train)
print("reg.intercept_=> %10.10f" % (reg.intercept_))
print(list(zip(feature_columns, reg.coef_)))
y_pred = reg.predict(X_test)
rmse_val = np.sqrt(metrics.mean_squared_error(y_test, y_pred))
# Null RMSE
y_null = np.zeros_like(y_test, dtype=int)
y_null.fill(y_test.mean())
N_RMSE = np.sqrt(metrics.mean_squared_error(y_test, y_null))
# Metrics
print("Mean Absolute Error :", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error :", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error = ", rmse_val)
print("Null RMSE = ", N_RMSE)
if N_RMSE < rmse_val:
print("Model is Not Doing Well Null RMSE Should be Greater")
else:
print("Model is Doing Well Null RMSE is Greater than RMSE")
# Train RMSE
y_pred_test = reg.predict(X_train)
rmse_val = np.sqrt(metrics.mean_squared_error(y_train, y_pred_test))
print(
"Train Root Mean Squared Error:",
np.sqrt(metrics.mean_squared_error(y_train, y_pred_test)),
)
# Error Percentage
df = pd.DataFrame({"Actual": y_test, "Predicted": y_pred, "Error": y_test - y_pred})
print("Maximum Error is :", df.Error.max())
print("Minimum Error is :", df.Error.min())
# Score
scores = cross_val_score(reg, X_train, y_train, cv=5)
print("Mean cross-validation score: %.2f" % scores.mean())
# ## XG Boost Regressor
import xgboost as xgb
from xgboost import plot_importance, plot_tree
from sklearn.metrics import mean_squared_error, mean_absolute_error
from sklearn.model_selection import RandomizedSearchCV, cross_val_score, KFold
X2 = nyc_taxi_final_sampling.drop(
[
"id",
"pickup_month_name",
"pickup_weekday",
"pickup_datetime",
"dropoff_datetime",
"trip_duration",
"passenger_count",
"speed",
],
axis=1,
)
X1 = preprocessing.scale(X2)
X = pd.DataFrame(X1)
y = nyc_taxi_final_sampling["trip_duration"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
model = xgb.XGBRegressor()
model.fit(X_train, y_train)
print(model)
y_pred = model.predict(data=X_test)
rmse_val = np.sqrt(metrics.mean_squared_error(y_test, y_pred))
# Null RMSE
y_null = np.zeros_like(y_test, dtype=int)
y_null.fill(y_test.mean())
N_RMSE = np.sqrt(metrics.mean_squared_error(y_test, y_null))
# Metrics
print("Mean Absolute Error :", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error :", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error = ", rmse_val)
print("Null RMSE = ", N_RMSE)
if N_RMSE < rmse_val:
print("Model is Not Doing Well Null RMSE Should be Greater")
else:
print("Model is Doing Well Null RMSE is Greater than RMSE")
# Train RMSE
y_pred_test = model.predict(X_train)
rmse_val = np.sqrt(metrics.mean_squared_error(y_train, y_pred_test))
print(
"Train Root Mean Squared Error:",
np.sqrt(metrics.mean_squared_error(y_train, y_pred_test)),
)
# Error Percentage
df = pd.DataFrame({"Actual": y_test, "Predicted": y_pred, "Error": y_test - y_pred})
print("Maximum Error is :", df.Error.max())
print("Minimum Error is :", df.Error.min())
# Score
scores = cross_val_score(model, X_train, y_train, cv=5)
print("Mean cross-validation score: %.2f" % scores.mean())
# ## Ridge Regression
from sklearn.linear_model import Ridge
X2 = nyc_taxi_final_sampling.drop(
[
"id",
"pickup_month_name",
"pickup_weekday",
"pickup_datetime",
"dropoff_datetime",
"trip_duration",
"passenger_count",
"speed",
],
axis=1,
)
X1 = preprocessing.scale(X2)
X = pd.DataFrame(X1)
y = nyc_taxi_final_sampling["trip_duration"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
ridgeReg = Ridge(alpha=0.05, normalize=True)
ridgeReg.fit(X_train, y_train)
y_pred = ridgeReg.predict(X_test)
rmse_val = np.sqrt(metrics.mean_squared_error(y_test, y_pred))
# NULL RMSE
y_null = np.zeros_like(y_test, dtype=int)
y_null.fill(y_test.mean())
N_RMSE = np.sqrt(metrics.mean_squared_error(y_test, y_null))
# Metrics
print("Mean Absolute Error :", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error :", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error = ", rmse_val)
print("Null RMSE = ", N_RMSE)
if N_RMSE < rmse_val:
print("Model is Not Doing Well Null RMSE Should be Greater")
else:
print("Model is Doing Well Null RMSE is Greater than RMSE")
# Train RMSE
y_pred_test = ridgeReg.predict(X_train)
rmse_val = np.sqrt(metrics.mean_squared_error(y_train, y_pred_test))
print(
"Train Root Mean Squared Error:",
np.sqrt(metrics.mean_squared_error(y_train, y_pred_test)),
)
# Error Percentage
df = pd.DataFrame({"Actual": y_test, "Predicted": y_pred, "Error": y_test - y_pred})
print("Maximum Error is :", df.Error.max())
print("Minimum Error is :", df.Error.min())
# Score
scores = cross_val_score(ridgeReg, X_train, y_train, cv=5)
print("Mean cross-validation score: %.2f" % scores.mean())
# ## RidgeCV (Cross Validation - Hyper Tuning parameter)
from sklearn.linear_model import RidgeCV
## training the model
X2 = nyc_taxi_final_sampling.drop(
[
"id",
"pickup_month_name",
"pickup_weekday",
"pickup_datetime",
"dropoff_datetime",
"trip_duration",
"passenger_count",
"speed",
],
axis=1,
)
X1 = preprocessing.scale(X2)
X = pd.DataFrame(X1)
y = nyc_taxi_final_sampling["trip_duration"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
ridgeRegCV = RidgeCV(alphas=[1e-3, 1e-2, 1e-1, 1])
ridgeRegCV.fit(X_train, y_train)
y_pred = ridgeRegCV.predict(X_test)
rmse_val = np.sqrt(metrics.mean_squared_error(y_test, y_pred))
# Null RMSE
y_null = np.zeros_like(y_test, dtype=int)
y_null.fill(y_test.mean())
N_RMSE = np.sqrt(metrics.mean_squared_error(y_test, y_null))
# Metrics
print("Mean Absolute Error :", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error :", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error = ", rmse_val)
print("Null RMSE = ", N_RMSE)
if N_RMSE < rmse_val:
print("Model is Not Doing Well Null RMSE Should be Greater")
else:
print("Model is Doing Well Null RMSE is Greater than RMSE")
# Train RMSE
y_pred_test = ridgeRegCV.predict(X_train)
rmse_val = np.sqrt(metrics.mean_squared_error(y_train, y_pred_test))
print(
"Train Root Mean Squared Error:",
np.sqrt(metrics.mean_squared_error(y_train, y_pred_test)),
)
# Error Percentage
df = pd.DataFrame({"Actual": y_test, "Predicted": y_pred, "Error": y_test - y_pred})
print("Maximum Error is :", df.Error.max())
print("Minimum Error is :", df.Error.min())
# Score
scores = cross_val_score(ridgeRegCV, X_train, y_train, cv=5)
print("Mean cross-validation score: %.2f" % scores.mean())
# Metrics Overview
df = {
"Model_Before_PCA": ["Linear_Reg", "XGB", "Ridge", "RidgeCV"],
"RMSE": ["875", "1249", "1605", "876"],
"NULL_RMSE": ["1701", "1701", "1701", "1701"],
"Max_error": ["66971", "84155", "85443", "67179"],
"Min_error": ["-80844", "-52886", "-4015", "-78683"],
"Score": ["72", "44", "11", "72"],
}
print("Metrics Overview Before_PCA")
dataframe = pd.DataFrame(
df, columns=["Model", "RMSE", "NULL_RMSE", "Max_error", "Min_error", "Score"]
)
dataframe
# # Running Model with PCA
# Verify the shape of data
nyc_taxi_final_sampling.shape
# Aligning data for PCA.
nyc_taxi_final_sampling.columns
nyc_taxi_pca = nyc_taxi_final_sampling.copy()
nyc_taxi_pca.drop(
[
"id",
"pickup_weekday",
"pickup_month_name",
"pickup_datetime",
"dropoff_datetime",
"speed",
],
axis=1,
inplace=True,
)
# seperate target variable for PCA.
target = nyc_taxi_pca["trip_duration"]
from sklearn import datasets
from sklearn.decomposition import PCA
# PCA
# normalize data
nyc_taxi_pca_norm = (nyc_taxi_pca - nyc_taxi_pca.mean()) / nyc_taxi_pca.std()
pca = PCA(n_components=12) # 12 features
pca.fit_transform(nyc_taxi_pca_norm.values)
print(pca.explained_variance_ratio_)
# print (nyc_taxi_final.feature_names)
print(pca.explained_variance_)
variance_ratio_cum_sum = np.cumsum(
np.round(pca.explained_variance_ratio_, decimals=4) * 100
)
print(variance_ratio_cum_sum)
print(pca.components_)
# Taken variance ratio of 7 PCA components at 93.6%.
pca.explained_variance_ratio_[:7].sum()
# Plot Elbow Curve
plt.plot(np.cumsum(pca.explained_variance_ratio_))
plt.xlabel("Number of components")
plt.ylabel("Cumulative explained variance")
plt.annotate("7", xy=(7, 0.93))
# consider first 7 components as they are explaining the 93% of variation in the data
x_pca = PCA(n_components=7)
nyc_taxi_pca_norm_final = x_pca.fit_transform(nyc_taxi_pca_norm)
# correlation between the variables after transforming the data with PCA is 0
correlation = pd.DataFrame(nyc_taxi_pca_norm_final).corr()
sns.heatmap(correlation, vmax=1, square=True, cmap="viridis")
plt.title("Correlation between different features")
# #After PCA, there is no correlation among any components.
# ## Linear Regression after PCA
X2 = preprocessing.scale(nyc_taxi_pca_norm_final)
X = pd.DataFrame(X2)
y = target
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
reg = linear_model.LinearRegression()
reg.fit(X_train, y_train)
print("reg.intercept_=> %10.10f" % (reg.intercept_))
print(list(zip(feature_columns, reg.coef_)))
y_pred = reg.predict(X_test)
rmse_val = np.sqrt(metrics.mean_squared_error(y_test, y_pred))
# NULL RMSE
y_null = np.zeros_like(y_test, dtype=int)
y_null.fill(y_test.mean())
N_RMSE = np.sqrt(metrics.mean_squared_error(y_test, y_null))
# Metrics
print("Mean Absolute Error :", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error :", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error = ", rmse_val)
print("Null RMSE = ", N_RMSE)
if N_RMSE < rmse_val:
print("Model is Not Doing Well Null RMSE Should be Greater")
else:
print("Model is Doing Well Null RMSE is Greater than RMSE")
# Train RMSE
y_pred_test = reg.predict(X_train)
rmse_val = np.sqrt(metrics.mean_squared_error(y_train, y_pred_test))
print(
"Train Root Mean Squared Error:",
np.sqrt(metrics.mean_squared_error(y_train, y_pred_test)),
)
# Error Percentage
df = pd.DataFrame({"Actual": y_test, "Predicted": y_pred, "Error": y_test - y_pred})
print("Maximum Error is :", df.Error.max())
print("Minimum Error is :", df.Error.min())
# Score
scores = cross_val_score(reg, X_train, y_train, cv=5)
print("Mean cross-validation score: %.2f" % scores.mean())
# ## XGB Regressor after PCA
import xgboost as xgb
from xgboost import plot_importance, plot_tree
from sklearn.metrics import mean_squared_error, mean_absolute_error
from sklearn.model_selection import RandomizedSearchCV, cross_val_score, KFold
X2 = preprocessing.scale(nyc_taxi_pca_norm_final)
X = pd.DataFrame(X2)
y = target
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
model = xgb.XGBRegressor()
model.fit(X_train, y_train)
print(model)
y_pred = model.predict(data=X_test)
rmse_val = np.sqrt(metrics.mean_squared_error(y_test, y_pred))
# Null RMSE
y_null = np.zeros_like(y_test, dtype=int)
y_null.fill(y_test.mean())
N_RMSE = np.sqrt(metrics.mean_squared_error(y_test, y_null))
# Metrics
print("Mean Absolute Error :", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error :", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error = ", rmse_val)
print("Null RMSE = ", N_RMSE)
if N_RMSE < rmse_val:
print("Model is Not Doing Well Null RMSE Should be Greater")
else:
print("Model is Doing Well Null RMSE is Greater than RMSE")
# Train RMSE
y_pred_test = model.predict(X_train)
rmse_val = np.sqrt(metrics.mean_squared_error(y_train, y_pred_test))
print(
"Train Root Mean Squared Error:",
np.sqrt(metrics.mean_squared_error(y_train, y_pred_test)),
)
# Error Percentage
df = pd.DataFrame({"Actual": y_test, "Predicted": y_pred, "Error": y_test - y_pred})
print("Maximum Error is :", df.Error.max())
print("Minimum Error is :", df.Error.min())
# Score
scores = cross_val_score(model, X_train, y_train, cv=5)
print("Mean cross-validation score: %.2f" % scores.mean())
# ## Ridge Regression after PCA
X2 = preprocessing.scale(nyc_taxi_pca_norm_final)
X = pd.DataFrame(X2)
y = target
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
ridgeReg = Ridge(alpha=0.05, normalize=True)
ridgeReg.fit(X_train, y_train)
y_pred = ridgeReg.predict(X_test)
rmse_val = np.sqrt(metrics.mean_squared_error(y_test, y_pred))
# Null RMSE
y_null = np.zeros_like(y_test, dtype=int)
y_null.fill(y_test.mean())
N_RMSE = np.sqrt(metrics.mean_squared_error(y_test, y_null))
# Metrics
print("Mean Absolute Error :", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error :", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error = ", rmse_val)
print("Null RMSE = ", N_RMSE)
if N_RMSE < rmse_val:
print("Model is Not Doing Well Null RMSE Should be Greater")
else:
print("Model is Doing Well Null RMSE is Greater than RMSE")
# Train RMSE
y_pred_test = ridgeReg.predict(X_train)
rmse_val = np.sqrt(metrics.mean_squared_error(y_train, y_pred_test))
print(
"Train Root Mean Squared Error:",
np.sqrt(metrics.mean_squared_error(y_train, y_pred_test)),
)
# Error Percentage
df = pd.DataFrame({"Actual": y_test, "Predicted": y_pred, "Error": y_test - y_pred})
print("Maximum Error is :", df.Error.max())
print("Minimum Error is :", df.Error.min())
# Score
scores = cross_val_score(ridgeReg, X_train, y_train, cv=5)
print("Mean cross-validation score: %.2f" % scores.mean())
# ## Ridge Regression CV after PCA
X2 = preprocessing.scale(nyc_taxi_pca_norm_final)
X = pd.DataFrame(X2)
y = target
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
ridgeRegCV = RidgeCV(alphas=[1e-3, 1e-2, 1e-1, 1])
ridgeRegCV.fit(X_train, y_train)
y_pred = ridgeRegCV.predict(X_test)
rmse_val = np.sqrt(metrics.mean_squared_error(y_test, y_pred))
# Null RMSE
y_null = np.zeros_like(y_test, dtype=int)
y_null.fill(y_test.mean())
N_RMSE = np.sqrt(metrics.mean_squared_error(y_test, y_null))
# Metrics
print("Mean Absolute Error :", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error :", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error = ", rmse_val)
print("Null RMSE = ", N_RMSE)
if N_RMSE < rmse_val:
print("Model is Not Doing Well Null RMSE Should be Greater")
else:
print("Model is Doing Well Null RMSE is Greater than RMSE")
# Train RMSE
y_pred_test = ridgeRegCV.predict(X_train)
rmse_val = np.sqrt(metrics.mean_squared_error(y_train, y_pred_test))
print(
"Train Root Mean Squared Error:",
np.sqrt(metrics.mean_squared_error(y_train, y_pred_test)),
)
# Error Percentage
df = pd.DataFrame({"Actual": y_test, "Predicted": y_pred, "Error": y_test - y_pred})
print("Maximum Error is :", df.Error.max())
print("Minimum Error is :", df.Error.min())
# Score
scores = cross_val_score(ridgeRegCV, X_train, y_train, cv=5)
print("Mean cross-validation score: %.2f" % scores.mean())
df = {
"Model_Before_PCA": ["Linear_Reg", "XGB", "Ridge", "RidgeCV"],
"RMSE": ["875", "1249", "1605", "876"],
"NULL_RMSE": ["1701", "1701", "1701", "1701"],
"Max_error": ["66971", "84155", "85443", "67179"],
"Min_error": ["-80844", "-52886", "-4015", "-78683"],
"Score": ["72", "44", "11", "72"],
}
print("Metrics Overview Before_PCA")
dataframe = pd.DataFrame(
df,
columns=[
"Model_Before_PCA",
"RMSE",
"NULL_RMSE",
"Max_error",
"Min_error",
"Score",
],
)
dataframe
df = {
"Model_After_PCA": ["Linear_Reg", "XGB", "Ridge", "RidgeCV"],
"RMSE": ["988", "108", "990", "988"],
"NULL_RMSE": ["1701", "1701", "1701", "1701"],
"Max_error": ["42900", "13824", "44918", "42900"],
"Min_error": ["-12259", "-9575", "-11434", "-12259"],
"Score": ["66", "99", "66", "66"],
}
print("Metrics Overview After_PCA")
dataframe = pd.DataFrame(
df,
columns=["Model_After_PCA", "RMSE", "NULL_RMSE", "Max_error", "Min_error", "Score"],
)
dataframe
# ## Final Conclusion:
# #### After comparing accuracy between model before and after PCA analysis. It has been decided that
# #### XGB Regressor is the best model which RMSE value: 108 when compare to other models and model accuracy is 99%.
# #### Accuracy Score is 99% with Max trip_duration error 13824,Min trip_duration error -9575 and RMSE 108
X2 = preprocessing.scale(nyc_taxi_pca_norm_final)
X = pd.DataFrame(X2)
y = target
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=111
)
model = xgb.XGBRegressor()
model.fit(X_train, y_train)
y_pred = model.predict(data=X_test)
x_ax = range(len(y_test))
plt.figure(figsize=(20, 10))
plt.scatter(x_ax, y_test, s=10, color="blue", label="original")
plt.scatter(x_ax, y_pred, s=10, color="red", label="predicted")
plt.legend()
plt.show()
# From above plot you can see index:59891 is having trip_duration of 86391 acutal value(blue point)
# predicated value (red point)
nyc_taxi_final_sampling.sort_values(by="trip_duration", ascending=False).head(2)
| false | 0 | 11,985 | 0 | 12,004 | 11,985 |
||
69401164
|
<jupyter_start><jupyter_text>Zomato Bangalore Restaurants
### Context
I was always fascinated by the food culture of Bengaluru. Restaurants from all over the world can be found here in Bengaluru. From United States to Japan, Russia to Antarctica, you get all type of cuisines here. Delivery, Dine-out, Pubs, Bars, Drinks,Buffet, Desserts you name it and Bengaluru has it. Bengaluru is best place for foodies. The number of restaurant are increasing day by day. Currently which stands at approximately 12,000 restaurants. With such an high number of restaurants. This industry hasn't been saturated yet. And new restaurants are opening every day. However it has become difficult for them to compete with already established restaurants. The key issues that continue to pose a challenge to them include high real estate costs, rising food costs, shortage of quality manpower, fragmented supply chain and over-licensing. This Zomato data aims at analysing demography of the location. Most importantly it will help new restaurants in deciding their theme, menus, cuisine, cost etc for a particular location. It also aims at finding similarity between neighborhoods of Bengaluru on the basis of food. The dataset also contains reviews for each of the restaurant which will help in finding overall rating for the place.
### Content
The basic idea of analyzing the Zomato dataset is to get a fair idea about the factors affecting the establishment
of different types of restaurant at different places in Bengaluru, aggregate rating of each restaurant, Bengaluru
being one such city has more than 12,000 restaurants with restaurants serving dishes from all over the world.
With each day new restaurants opening the industry has’nt been saturated yet and the demand is increasing
day by day. Inspite of increasing demand it however has become difficult for new restaurants to compete with
established restaurants. Most of them serving the same food. Bengaluru being an IT capital of India. Most of
the people here are dependent mainly on the restaurant food as they don’t have time to cook for themselves.
With such an overwhelming demand of restaurants it has therefore become important to study the demography
of a location. What kind of a food is more popular in a locality. Do the entire locality loves vegetarian food.
If yes then is that locality populated by a particular sect of people for eg. Jain, Marwaris, Gujaratis who are
mostly vegetarian. These kind of analysis can be done using the data, by studying the factors such as
• Location of the restaurant
• Approx Price of food
• Theme based restaurant or not
• Which locality of that city serves that cuisines with maximum number of restaurants
• The needs of people who are striving to get the best cuisine of the neighborhood
• Is a particular neighborhood famous for its own kind of food.
“Just so that you have a good meal the next time you step out”
The data is accurate to that available on the zomato website until 15 March 2019.
The data was scraped from Zomato in two phase. After going through the structure of the website I found that for each neighborhood there are 6-7 category of restaurants viz. Buffet, Cafes, Delivery, Desserts, Dine-out, Drinks & nightlife, Pubs and bars.
Phase I,
In Phase I of extraction only the URL, name and address of the restaurant were extracted which were visible on the front page. The URl's for each of the restaurants on the zomato were recorded in the csv file so that later the data can be extracted individually for each restaurant. This made the extraction process easier and reduced the extra load on my machine. The data for each neighborhood and each category can be found [here][1]
Phase II,
In Phase II the recorded data for each restaurant and each category was read and data for each restaurant was scraped individually. 15 variables were scraped in this phase. For each of the neighborhood and for each category their online_order, book_table, rate, votes, phone, location, rest_type, dish_liked, cuisines, approx_cost(for two people), reviews_list, menu_item was extracted. See section 5 for more details about the variables.
Kaggle dataset identifier: zomato-bangalore-restaurants
<jupyter_code>import pandas as pd
df = pd.read_csv('zomato-bangalore-restaurants/zomato.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 51717 entries, 0 to 51716
Data columns (total 17 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 url 51717 non-null object
1 address 51717 non-null object
2 name 51717 non-null object
3 online_order 51717 non-null object
4 book_table 51717 non-null object
5 rate 43942 non-null object
6 votes 51717 non-null int64
7 phone 50509 non-null object
8 location 51696 non-null object
9 rest_type 51490 non-null object
10 dish_liked 23639 non-null object
11 cuisines 51672 non-null object
12 approx_cost(for two people) 51371 non-null object
13 reviews_list 51717 non-null object
14 menu_item 51717 non-null object
15 listed_in(type) 51717 non-null object
16 listed_in(city) 51717 non-null object
dtypes: int64(1), object(16)
memory usage: 6.7+ MB
<jupyter_text>Examples:
{
"url": "https://www.zomato.com/bangalore/jalsa-banashankari?context=eyJzZSI6eyJlIjpbNTg2OTQsIjE4Mzc1NDc0IiwiNTkwOTAiLCIxODM4Mjk0NCIsIjE4MjI0Njc2IiwiNTkyODkiLCIxODM3MzM4NiJdLCJ0IjoiUmVzdGF1cmFudHMgaW4gQmFuYXNoYW5rYXJpIHNlcnZpbmcgQnVmZmV0In19",
"address": "942, 21st Main Road, 2nd Stage, Banashankari, Bangalore",
"name": "Jalsa",
"online_order": "Yes",
"book_table": "Yes",
"rate": "4.1/5",
"votes": 775,
"phone": "080 42297555\r\n+91 9743772233",
"location": "Banashankari",
"rest_type": "Casual Dining",
"dish_liked": "Pasta, Lunch Buffet, Masala Papad, Paneer Lajawab, Tomato Shorba, Dum Biryani, Sweet Corn Soup",
"cuisines": "North Indian, Mughlai, Chinese",
"approx_cost(for two people)": 800,
"reviews_list": "[('Rated 4.0', 'RATED\\n A beautiful place to dine in.The interiors take you back to the Mughal era. The lightings are just perfect.We went there on the occasion of Christmas and so they had only limited items available. But the taste and service was not compromised at all.The ...(truncated)",
"menu_item": "[]",
"listed_in(type)": "Buffet",
"listed_in(city)": "Banashankari"
}
{
"url": "https://www.zomato.com/bangalore/spice-elephant-banashankari?context=eyJzZSI6eyJlIjpbIjU4Njk0IiwxODM3NTQ3NCwiNTkwOTAiLCIxODM4Mjk0NCIsIjE4MjI0Njc2IiwiNTkyODkiLCIxODM3MzM4NiJdLCJ0IjoiUmVzdGF1cmFudHMgaW4gQmFuYXNoYW5rYXJpIHNlcnZpbmcgQnVmZmV0In19",
"address": "2nd Floor, 80 Feet Road, Near Big Bazaar, 6th Block, Kathriguppe, 3rd Stage, Banashankari, Bangalore",
"name": "Spice Elephant",
"online_order": "Yes",
"book_table": "No",
"rate": "4.1/5",
"votes": 787,
"phone": "080 41714161",
"location": "Banashankari",
"rest_type": "Casual Dining",
"dish_liked": "Momos, Lunch Buffet, Chocolate Nirvana, Thai Green Curry, Paneer Tikka, Dum Biryani, Chicken Biryani",
"cuisines": "Chinese, North Indian, Thai",
"approx_cost(for two people)": 800,
"reviews_list": "[('Rated 4.0', 'RATED\\n Had been here for dinner with family. Turned out to be a good choose suitable for all ages of people. Can try this place. We liked the most was their starters. Service is good. Prices are affordable. Will recommend this restaurant for early dinner. The ...(truncated)",
"menu_item": "[]",
"listed_in(type)": "Buffet",
"listed_in(city)": "Banashankari"
}
{
"url": "https://www.zomato.com/SanchurroBangalore?context=eyJzZSI6eyJlIjpbIjU4Njk0IiwiMTgzNzU0NzQiLDU5MDkwLCIxODM4Mjk0NCIsIjE4MjI0Njc2IiwiNTkyODkiLCIxODM3MzM4NiJdLCJ0IjoiUmVzdGF1cmFudHMgaW4gQmFuYXNoYW5rYXJpIHNlcnZpbmcgQnVmZmV0In19",
"address": "1112, Next to KIMS Medical College, 17th Cross, 2nd Stage, Banashankari, Bangalore",
"name": "San Churro Cafe",
"online_order": "Yes",
"book_table": "No",
"rate": "3.8/5",
"votes": 918,
"phone": "+91 9663487993",
"location": "Banashankari",
"rest_type": "Cafe, Casual Dining",
"dish_liked": "Churros, Cannelloni, Minestrone Soup, Hot Chocolate, Pink Sauce Pasta, Salsa, Veg Supreme Pizza",
"cuisines": "Cafe, Mexican, Italian",
"approx_cost(for two people)": 800,
"reviews_list": "[('Rated 3.0', \"RATED\\n Ambience is not that good enough and it's not a pocket friendly cafe and the quantity is not that good and desserts are too good enough ??..\"), ('Rated 3.0', \"RATED\\n \\nWent there for a quick bite with friends.\\nThe ambience had more of corporate ...(truncated)",
"menu_item": "[]",
"listed_in(type)": "Buffet",
"listed_in(city)": "Banashankari"
}
{
"url": "https://www.zomato.com/bangalore/addhuri-udupi-bhojana-banashankari?context=eyJzZSI6eyJlIjpbIjU4Njk0IiwiMTgzNzU0NzQiLCI1OTA5MCIsMTgzODI5NDQsIjE4MjI0Njc2IiwiNTkyODkiLCIxODM3MzM4NiJdLCJ0IjoiUmVzdGF1cmFudHMgaW4gQmFuYXNoYW5rYXJpIHNlcnZpbmcgQnVmZmV0In19",
"address": "1st Floor, Annakuteera, 3rd Stage, Banashankari, Bangalore",
"name": "Addhuri Udupi Bhojana",
"online_order": "No",
"book_table": "No",
"rate": "3.7/5",
"votes": 88,
"phone": "+91 9620009302",
"location": "Banashankari",
"rest_type": "Quick Bites",
"dish_liked": "Masala Dosa",
"cuisines": "South Indian, North Indian",
"approx_cost(for two people)": 300,
"reviews_list": "[('Rated 4.0', \"RATED\\n Great food and proper Karnataka style full meals. Been there twice and was fully satisfied.. Will give 5 stars if it's well managed............\"), ('Rated 2.0', 'RATED\\n Reached the place at 3pm on Saturday. Half of the items on the menu were over. ...(truncated)",
"menu_item": "[]",
"listed_in(type)": "Buffet",
"listed_in(city)": "Banashankari"
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
plt.rcParams["figure.figsize"] = (12, 5)
sns.set_style("whitegrid")
import matplotlib.colors as mcolors
import geopandas as gpd
import folium
from folium.plugins import HeatMap
import os
import plotly.express as px
import plotly.graph_objs as go
from plotly.offline import iplot
from plotly.offline import init_notebook_mode
init_notebook_mode(connected=False)
from geopy.geocoders import Nominatim
import dexplot as dxp
import re
import string
import nltk
nltk.download("stopwords")
from nltk.corpus import stopwords
from tqdm import tqdm
df = pd.read_csv("/kaggle/input/zomato-bangalore-restaurants/zomato.csv")
df.head()
df.info()
df.isnull().sum()
((df.isnull() | df.isna()).sum() * 100 / df.index.size)
df.describe()
df.shape
df["menu_item"].unique()
sns.countplot(df["book_table"])
fig = plt.gcf()
fig.set_size_inches(10, 10)
plt.title("Restaurants provides Table Booking or Not")
sns.countplot(df["online_order"])
fig = plt.gcf()
fig.set_size_inches(10, 10)
plt.title("Restaurants delivering online or Not")
df["listed_in(type)"].unique()
dxp.count(
val="listed_in(type)",
data=df,
figsize=(20, 12),
split="listed_in(city)",
normalize=True,
)
print(df["rest_type"].nunique())
df["rest_type"].unique()
plt.figure(figsize=(12, 8))
val = df["name"].value_counts()[:25]
sns.barplot(x=val, y=val.index, palette="Set1")
plt.title("Famous Restaurants in the City of Bengaluru")
plt.xlabel("Number of Outlets")
df["rate"].unique()
df.rate = df.rate.replace("NEW", np.nan)
df.dropna(how="any", inplace=True)
data = df
data["rate"] = data["rate"].astype(str)
data["rate"] = data["rate"].apply(lambda x: x.replace("/5", ""))
data["rate"] = data["rate"].apply(lambda x: float(x))
data.head()
rel_type = pd.crosstab(df["rate"], df["listed_in(city)"])
rel_type.plot(kind="bar", stacked=True, figsize=(20, 16))
plt.title("City - Rating", fontsize=18)
plt.ylabel("City", fontsize=12)
plt.xlabel("Rating", fontsize=12)
plt.xticks(fontsize=12)
plt.yticks(fontsize=12)
plt.figure(figsize=(12, 8))
sns.distplot(df["rate"], bins=25)
sns.countplot(df["listed_in(type)"])
fig = plt.gcf()
fig.set_size_inches(10, 10)
plt.title("Restaurants delivering online or Not")
locations = pd.DataFrame({"Name": df["location"].unique()})
locations.head()
locations["Name"] = locations["Name"].apply(
lambda x: "Bangaluru " + str(x)
) # here I have used lamda function
lat_lon = []
geolocator = Nominatim(user_agent="app")
for location in locations["Name"]:
location = geolocator.geocode(location)
if location is None:
lat_lon.append(np.nan)
else:
geo = (location.latitude, location.longitude)
lat_lon.append(geo)
locations["geo_loc"] = lat_lon
locations.to_csv("locations.csv", index=False)
locations["Name"] = locations["Name"].apply(lambda x: x.replace("Bangaluru", "")[1:])
locations.head()
def generateBaseMap(default_location=[12.97, 77.59], default_zoom_start=12):
base_map = folium.Map(
location=default_location, control_scale=True, zoom_start=default_zoom_start
)
return base_map
Rest_locations = pd.DataFrame(df["location"].value_counts().reset_index())
Rest_locations.columns = ["Name", "count"]
Rest_locations = Rest_locations.merge(locations, on="Name", how="left").dropna()
Rest_locations
lat, lon = zip(*np.array(Rest_locations["geo_loc"]))
Rest_locations["lat"] = lat
Rest_locations["lon"] = lon
basemap = generateBaseMap()
HeatMap(Rest_locations[["lat", "lon", "count"]].values.tolist(), radius=15).add_to(
basemap
)
basemap
def produce_data(col, name):
data = pd.DataFrame(
df[df[col] == name].groupby(["location"], as_index=False)["url"].agg("count")
)
data.columns = ["Name", "count"]
print(data.head())
data = data.merge(locations, on="Name", how="left").dropna()
data["lan"], data["lon"] = zip(*data["geo_loc"].values)
return data.drop(["geo_loc"], axis=1)
food = produce_data("cuisines", "South Indian")
basemap = generateBaseMap()
HeatMap(food[["lan", "lon", "count"]].values.tolist(), radius=15).add_to(basemap)
basemap
food = produce_data("cuisines", "North Indian")
basemap = generateBaseMap()
HeatMap(food[["lan", "lon", "count"]].values.tolist(), radius=15).add_to(basemap)
basemap
df.info()
df.online_order.replace(("Yes", "No"), (True, False), inplace=True)
df.book_table.replace(("Yes", "No"), (True, False), inplace=True)
def Encode(df):
for column in df.columns[
~df.columns.isin(["rate", "approx_cost(for two people)", "votes"])
]:
df[column] = df[column].factorize()[0]
return df
df_en = Encode(df.copy())
df_en.info()
df_en.head(50)
df_en["approx_cost(for two people)"] = df_en["approx_cost(for two people)"].astype(str)
df_en["approx_cost(for two people)"] = df_en["approx_cost(for two people)"].apply(
lambda x: x.replace(",", ".")
)
df_en["approx_cost(for two people)"] = df_en["approx_cost(for two people)"].astype(
float
)
df_en.info()
corr = df_en.corr()
plt.figure(figsize=(15, 8))
sns.heatmap(corr, annot=True)
df_en.columns
from sklearn.model_selection import train_test_split
x = df_en.drop(["rate"], axis=1) # modified
y = df_en["rate"]
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=42
)
x_train.head()
y_train.head()
x_test.head()
y_test.head()
from sklearn.metrics import r2_score
from sklearn.linear_model import LinearRegression
reg_model = LinearRegression()
reg_model.fit(x_train, y_train)
y_pred = reg_model.predict(x_test)
r2_score(y_test, y_pred)
from sklearn.ensemble import RandomForestRegressor
RF_model = RandomForestRegressor(
n_estimators=600,
criterion="mse",
max_depth=None,
min_samples_split=2,
min_samples_leaf=0.0001,
min_weight_fraction_leaf=0.0,
max_features="auto",
max_leaf_nodes=None,
min_impurity_decrease=0.0,
min_impurity_split=None,
bootstrap=True,
oob_score=False,
n_jobs=None,
random_state=None,
verbose=0,
warm_start=False,
ccp_alpha=0.0,
max_samples=None,
)
RF_model.fit(x_train, y_train)
y_predict = RF_model.predict(x_test)
r2_score(y_test, y_predict)
from sklearn.tree import DecisionTreeRegressor
DT_model = DecisionTreeRegressor(
criterion="mse",
splitter="best",
max_depth=None,
min_samples_split=2,
min_samples_leaf=0.00011,
min_weight_fraction_leaf=0.0,
max_features=None,
random_state=None,
max_leaf_nodes=None,
min_impurity_decrease=0.0,
min_impurity_split=None,
presort="deprecated",
ccp_alpha=0.0,
)
DT_model.fit(x_train, y_train)
y_predict = DT_model.predict(x_test)
r2_score(y_test, y_predict)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/401/69401164.ipynb
|
zomato-bangalore-restaurants
|
himanshupoddar
|
[{"Id": 69401164, "ScriptId": 18948653, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6447261, "CreationDate": "07/30/2021 14:26:10", "VersionNumber": 1.0, "Title": "zomato_analysis", "EvaluationDate": "07/30/2021", "IsChange": true, "TotalLines": 234.0, "LinesInsertedFromPrevious": 234.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 92519684, "KernelVersionId": 69401164, "SourceDatasetVersionId": 352891}]
|
[{"Id": 352891, "DatasetId": 153420, "DatasourceVersionId": 367004, "CreatorUserId": 1811968, "LicenseName": "Data files \u00a9 Original Authors", "CreationDate": "03/31/2019 09:48:43", "VersionNumber": 1.0, "Title": "Zomato Bangalore Restaurants", "Slug": "zomato-bangalore-restaurants", "Subtitle": "Restaurants of Bengaluru", "Description": "### Context\n\nI was always fascinated by the food culture of Bengaluru. Restaurants from all over the world can be found here in Bengaluru. From United States to Japan, Russia to Antarctica, you get all type of cuisines here. Delivery, Dine-out, Pubs, Bars, Drinks,Buffet, Desserts you name it and Bengaluru has it. Bengaluru is best place for foodies. The number of restaurant are increasing day by day. Currently which stands at approximately 12,000 restaurants. With such an high number of restaurants. This industry hasn't been saturated yet. And new restaurants are opening every day. However it has become difficult for them to compete with already established restaurants. The key issues that continue to pose a challenge to them include high real estate costs, rising food costs, shortage of quality manpower, fragmented supply chain and over-licensing. This Zomato data aims at analysing demography of the location. Most importantly it will help new restaurants in deciding their theme, menus, cuisine, cost etc for a particular location. It also aims at finding similarity between neighborhoods of Bengaluru on the basis of food. The dataset also contains reviews for each of the restaurant which will help in finding overall rating for the place.\n\n### Content\n\nThe basic idea of analyzing the Zomato dataset is to get a fair idea about the factors affecting the establishment\nof different types of restaurant at different places in Bengaluru, aggregate rating of each restaurant, Bengaluru\nbeing one such city has more than 12,000 restaurants with restaurants serving dishes from all over the world.\nWith each day new restaurants opening the industry has\u2019nt been saturated yet and the demand is increasing\nday by day. Inspite of increasing demand it however has become difficult for new restaurants to compete with\nestablished restaurants. Most of them serving the same food. Bengaluru being an IT capital of India. Most of\nthe people here are dependent mainly on the restaurant food as they don\u2019t have time to cook for themselves.\nWith such an overwhelming demand of restaurants it has therefore become important to study the demography\nof a location. What kind of a food is more popular in a locality. Do the entire locality loves vegetarian food.\nIf yes then is that locality populated by a particular sect of people for eg. Jain, Marwaris, Gujaratis who are\nmostly vegetarian. These kind of analysis can be done using the data, by studying the factors such as\n\u2022 Location of the restaurant\n\u2022 Approx Price of food\n\u2022 Theme based restaurant or not\n\u2022 Which locality of that city serves that cuisines with maximum number of restaurants\n\u2022 The needs of people who are striving to get the best cuisine of the neighborhood\n\u2022 Is a particular neighborhood famous for its own kind of food.\n\n\u201cJust so that you have a good meal the next time you step out\u201d\n\nThe data is accurate to that available on the zomato website until 15 March 2019.\nThe data was scraped from Zomato in two phase. After going through the structure of the website I found that for each neighborhood there are 6-7 category of restaurants viz. Buffet, Cafes, Delivery, Desserts, Dine-out, Drinks & nightlife, Pubs and bars. \n\nPhase I, \n\nIn Phase I of extraction only the URL, name and address of the restaurant were extracted which were visible on the front page. The URl's for each of the restaurants on the zomato were recorded in the csv file so that later the data can be extracted individually for each restaurant. This made the extraction process easier and reduced the extra load on my machine. The data for each neighborhood and each category can be found [here][1]\n\nPhase II, \n\nIn Phase II the recorded data for each restaurant and each category was read and data for each restaurant was scraped individually. 15 variables were scraped in this phase. For each of the neighborhood and for each category their online_order, book_table, rate, votes, phone, location, rest_type, dish_liked, cuisines, approx_cost(for two people), reviews_list, menu_item was extracted. See section 5 for more details about the variables.\n\n\n### Acknowledgements\n\nThe data scraped was entirely for educational purposes only. Note that I don\u2019t claim any copyright for the data. All copyrights for the data is owned by Zomato Media Pvt. Ltd.. \n\n\n### Inspiration\nI was always astonished by how each of the restaurants are able to keep up the pace inspite of that cutting edge competition. And what factors should be kept in mind if someone wants to open new restaurant. Does the demography of an area matters? Does location of a particular type of restaurant also depends on the people living in that area? Does the theme of the restaurant matters? Is a food chain category restaurant likely to have more customers than its counter part? Are any neighborhood similar ? If two neighborhood are similar does that mean these are related or particular group of people live in the neighborhood or these are the places to it? What kind of a food is more popular in a locality. Do the entire locality loves vegetarian food. If yes then is that locality populated by a particular sect of people for eg. Jain, Marwaris, Gujaratis who are mostly vegetarian. There are infacts dozens of question in my mind. lets try to find out the answer with this dataset. \n\nFor detailed discussion of the business problem, please visit this [link](https://github.com/poddarhimanshu/Coursera_Capstone/blob/master/Final%20Project/Business_Problem.pdf)\n\nPlease visit this [link][2] to find codebook cum documentation for the data\n\n\n [1]: https://drive.google.com/open?id=1duQ9-dXuQqP5tnz5autNqgyyI3EiY4cE\n [2]: https://github.com/poddarhimanshu/Coursera_Capstone/blob/master/Final%20Project/Data%20Scraping/Documentation.pdf\n\n\nGITHUB LINk : https://github.com/poddarhimanshu/Zomato-Bengaluru-Restaurants/tree/master/Final%20Project", "VersionNotes": "Initial release", "TotalCompressedBytes": 574072999.0, "TotalUncompressedBytes": 92120904.0}]
|
[{"Id": 153420, "CreatorUserId": 1811968, "OwnerUserId": 1811968.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 352891.0, "CurrentDatasourceVersionId": 367004.0, "ForumId": 163855, "Type": 2, "CreationDate": "03/31/2019 09:48:43", "LastActivityDate": "03/31/2019", "TotalViews": 358389, "TotalDownloads": 57889, "TotalVotes": 1251, "TotalKernels": 378}]
|
[{"Id": 1811968, "UserName": "himanshupoddar", "DisplayName": "Himanshu Poddar", "RegisterDate": "04/11/2018", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
plt.rcParams["figure.figsize"] = (12, 5)
sns.set_style("whitegrid")
import matplotlib.colors as mcolors
import geopandas as gpd
import folium
from folium.plugins import HeatMap
import os
import plotly.express as px
import plotly.graph_objs as go
from plotly.offline import iplot
from plotly.offline import init_notebook_mode
init_notebook_mode(connected=False)
from geopy.geocoders import Nominatim
import dexplot as dxp
import re
import string
import nltk
nltk.download("stopwords")
from nltk.corpus import stopwords
from tqdm import tqdm
df = pd.read_csv("/kaggle/input/zomato-bangalore-restaurants/zomato.csv")
df.head()
df.info()
df.isnull().sum()
((df.isnull() | df.isna()).sum() * 100 / df.index.size)
df.describe()
df.shape
df["menu_item"].unique()
sns.countplot(df["book_table"])
fig = plt.gcf()
fig.set_size_inches(10, 10)
plt.title("Restaurants provides Table Booking or Not")
sns.countplot(df["online_order"])
fig = plt.gcf()
fig.set_size_inches(10, 10)
plt.title("Restaurants delivering online or Not")
df["listed_in(type)"].unique()
dxp.count(
val="listed_in(type)",
data=df,
figsize=(20, 12),
split="listed_in(city)",
normalize=True,
)
print(df["rest_type"].nunique())
df["rest_type"].unique()
plt.figure(figsize=(12, 8))
val = df["name"].value_counts()[:25]
sns.barplot(x=val, y=val.index, palette="Set1")
plt.title("Famous Restaurants in the City of Bengaluru")
plt.xlabel("Number of Outlets")
df["rate"].unique()
df.rate = df.rate.replace("NEW", np.nan)
df.dropna(how="any", inplace=True)
data = df
data["rate"] = data["rate"].astype(str)
data["rate"] = data["rate"].apply(lambda x: x.replace("/5", ""))
data["rate"] = data["rate"].apply(lambda x: float(x))
data.head()
rel_type = pd.crosstab(df["rate"], df["listed_in(city)"])
rel_type.plot(kind="bar", stacked=True, figsize=(20, 16))
plt.title("City - Rating", fontsize=18)
plt.ylabel("City", fontsize=12)
plt.xlabel("Rating", fontsize=12)
plt.xticks(fontsize=12)
plt.yticks(fontsize=12)
plt.figure(figsize=(12, 8))
sns.distplot(df["rate"], bins=25)
sns.countplot(df["listed_in(type)"])
fig = plt.gcf()
fig.set_size_inches(10, 10)
plt.title("Restaurants delivering online or Not")
locations = pd.DataFrame({"Name": df["location"].unique()})
locations.head()
locations["Name"] = locations["Name"].apply(
lambda x: "Bangaluru " + str(x)
) # here I have used lamda function
lat_lon = []
geolocator = Nominatim(user_agent="app")
for location in locations["Name"]:
location = geolocator.geocode(location)
if location is None:
lat_lon.append(np.nan)
else:
geo = (location.latitude, location.longitude)
lat_lon.append(geo)
locations["geo_loc"] = lat_lon
locations.to_csv("locations.csv", index=False)
locations["Name"] = locations["Name"].apply(lambda x: x.replace("Bangaluru", "")[1:])
locations.head()
def generateBaseMap(default_location=[12.97, 77.59], default_zoom_start=12):
base_map = folium.Map(
location=default_location, control_scale=True, zoom_start=default_zoom_start
)
return base_map
Rest_locations = pd.DataFrame(df["location"].value_counts().reset_index())
Rest_locations.columns = ["Name", "count"]
Rest_locations = Rest_locations.merge(locations, on="Name", how="left").dropna()
Rest_locations
lat, lon = zip(*np.array(Rest_locations["geo_loc"]))
Rest_locations["lat"] = lat
Rest_locations["lon"] = lon
basemap = generateBaseMap()
HeatMap(Rest_locations[["lat", "lon", "count"]].values.tolist(), radius=15).add_to(
basemap
)
basemap
def produce_data(col, name):
data = pd.DataFrame(
df[df[col] == name].groupby(["location"], as_index=False)["url"].agg("count")
)
data.columns = ["Name", "count"]
print(data.head())
data = data.merge(locations, on="Name", how="left").dropna()
data["lan"], data["lon"] = zip(*data["geo_loc"].values)
return data.drop(["geo_loc"], axis=1)
food = produce_data("cuisines", "South Indian")
basemap = generateBaseMap()
HeatMap(food[["lan", "lon", "count"]].values.tolist(), radius=15).add_to(basemap)
basemap
food = produce_data("cuisines", "North Indian")
basemap = generateBaseMap()
HeatMap(food[["lan", "lon", "count"]].values.tolist(), radius=15).add_to(basemap)
basemap
df.info()
df.online_order.replace(("Yes", "No"), (True, False), inplace=True)
df.book_table.replace(("Yes", "No"), (True, False), inplace=True)
def Encode(df):
for column in df.columns[
~df.columns.isin(["rate", "approx_cost(for two people)", "votes"])
]:
df[column] = df[column].factorize()[0]
return df
df_en = Encode(df.copy())
df_en.info()
df_en.head(50)
df_en["approx_cost(for two people)"] = df_en["approx_cost(for two people)"].astype(str)
df_en["approx_cost(for two people)"] = df_en["approx_cost(for two people)"].apply(
lambda x: x.replace(",", ".")
)
df_en["approx_cost(for two people)"] = df_en["approx_cost(for two people)"].astype(
float
)
df_en.info()
corr = df_en.corr()
plt.figure(figsize=(15, 8))
sns.heatmap(corr, annot=True)
df_en.columns
from sklearn.model_selection import train_test_split
x = df_en.drop(["rate"], axis=1) # modified
y = df_en["rate"]
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=42
)
x_train.head()
y_train.head()
x_test.head()
y_test.head()
from sklearn.metrics import r2_score
from sklearn.linear_model import LinearRegression
reg_model = LinearRegression()
reg_model.fit(x_train, y_train)
y_pred = reg_model.predict(x_test)
r2_score(y_test, y_pred)
from sklearn.ensemble import RandomForestRegressor
RF_model = RandomForestRegressor(
n_estimators=600,
criterion="mse",
max_depth=None,
min_samples_split=2,
min_samples_leaf=0.0001,
min_weight_fraction_leaf=0.0,
max_features="auto",
max_leaf_nodes=None,
min_impurity_decrease=0.0,
min_impurity_split=None,
bootstrap=True,
oob_score=False,
n_jobs=None,
random_state=None,
verbose=0,
warm_start=False,
ccp_alpha=0.0,
max_samples=None,
)
RF_model.fit(x_train, y_train)
y_predict = RF_model.predict(x_test)
r2_score(y_test, y_predict)
from sklearn.tree import DecisionTreeRegressor
DT_model = DecisionTreeRegressor(
criterion="mse",
splitter="best",
max_depth=None,
min_samples_split=2,
min_samples_leaf=0.00011,
min_weight_fraction_leaf=0.0,
max_features=None,
random_state=None,
max_leaf_nodes=None,
min_impurity_decrease=0.0,
min_impurity_split=None,
presort="deprecated",
ccp_alpha=0.0,
)
DT_model.fit(x_train, y_train)
y_predict = DT_model.predict(x_test)
r2_score(y_test, y_predict)
|
[{"zomato-bangalore-restaurants/zomato.csv": {"column_names": "[\"url\", \"address\", \"name\", \"online_order\", \"book_table\", \"rate\", \"votes\", \"phone\", \"location\", \"rest_type\", \"dish_liked\", \"cuisines\", \"approx_cost(for two people)\", \"reviews_list\", \"menu_item\", \"listed_in(type)\", \"listed_in(city)\"]", "column_data_types": "{\"url\": \"object\", \"address\": \"object\", \"name\": \"object\", \"online_order\": \"object\", \"book_table\": \"object\", \"rate\": \"object\", \"votes\": \"int64\", \"phone\": \"object\", \"location\": \"object\", \"rest_type\": \"object\", \"dish_liked\": \"object\", \"cuisines\": \"object\", \"approx_cost(for two people)\": \"object\", \"reviews_list\": \"object\", \"menu_item\": \"object\", \"listed_in(type)\": \"object\", \"listed_in(city)\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 51717 entries, 0 to 51716\nData columns (total 17 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 url 51717 non-null object\n 1 address 51717 non-null object\n 2 name 51717 non-null object\n 3 online_order 51717 non-null object\n 4 book_table 51717 non-null object\n 5 rate 43942 non-null object\n 6 votes 51717 non-null int64 \n 7 phone 50509 non-null object\n 8 location 51696 non-null object\n 9 rest_type 51490 non-null object\n 10 dish_liked 23639 non-null object\n 11 cuisines 51672 non-null object\n 12 approx_cost(for two people) 51371 non-null object\n 13 reviews_list 51717 non-null object\n 14 menu_item 51717 non-null object\n 15 listed_in(type) 51717 non-null object\n 16 listed_in(city) 51717 non-null object\ndtypes: int64(1), object(16)\nmemory usage: 6.7+ MB\n", "summary": "{\"votes\": {\"count\": 51717.0, \"mean\": 283.69752692538236, \"std\": 803.8388530079794, \"min\": 0.0, \"25%\": 7.0, \"50%\": 41.0, \"75%\": 198.0, \"max\": 16832.0}}", "examples": "{\"url\":{\"0\":\"https:\\/\\/www.zomato.com\\/bangalore\\/jalsa-banashankari?context=eyJzZSI6eyJlIjpbNTg2OTQsIjE4Mzc1NDc0IiwiNTkwOTAiLCIxODM4Mjk0NCIsIjE4MjI0Njc2IiwiNTkyODkiLCIxODM3MzM4NiJdLCJ0IjoiUmVzdGF1cmFudHMgaW4gQmFuYXNoYW5rYXJpIHNlcnZpbmcgQnVmZmV0In19\",\"1\":\"https:\\/\\/www.zomato.com\\/bangalore\\/spice-elephant-banashankari?context=eyJzZSI6eyJlIjpbIjU4Njk0IiwxODM3NTQ3NCwiNTkwOTAiLCIxODM4Mjk0NCIsIjE4MjI0Njc2IiwiNTkyODkiLCIxODM3MzM4NiJdLCJ0IjoiUmVzdGF1cmFudHMgaW4gQmFuYXNoYW5rYXJpIHNlcnZpbmcgQnVmZmV0In19\",\"2\":\"https:\\/\\/www.zomato.com\\/SanchurroBangalore?context=eyJzZSI6eyJlIjpbIjU4Njk0IiwiMTgzNzU0NzQiLDU5MDkwLCIxODM4Mjk0NCIsIjE4MjI0Njc2IiwiNTkyODkiLCIxODM3MzM4NiJdLCJ0IjoiUmVzdGF1cmFudHMgaW4gQmFuYXNoYW5rYXJpIHNlcnZpbmcgQnVmZmV0In19\",\"3\":\"https:\\/\\/www.zomato.com\\/bangalore\\/addhuri-udupi-bhojana-banashankari?context=eyJzZSI6eyJlIjpbIjU4Njk0IiwiMTgzNzU0NzQiLCI1OTA5MCIsMTgzODI5NDQsIjE4MjI0Njc2IiwiNTkyODkiLCIxODM3MzM4NiJdLCJ0IjoiUmVzdGF1cmFudHMgaW4gQmFuYXNoYW5rYXJpIHNlcnZpbmcgQnVmZmV0In19\"},\"address\":{\"0\":\"942, 21st Main Road, 2nd Stage, Banashankari, Bangalore\",\"1\":\"2nd Floor, 80 Feet Road, Near Big Bazaar, 6th Block, Kathriguppe, 3rd Stage, Banashankari, Bangalore\",\"2\":\"1112, Next to KIMS Medical College, 17th Cross, 2nd Stage, Banashankari, Bangalore\",\"3\":\"1st Floor, Annakuteera, 3rd Stage, Banashankari, Bangalore\"},\"name\":{\"0\":\"Jalsa\",\"1\":\"Spice Elephant\",\"2\":\"San Churro Cafe\",\"3\":\"Addhuri Udupi Bhojana\"},\"online_order\":{\"0\":\"Yes\",\"1\":\"Yes\",\"2\":\"Yes\",\"3\":\"No\"},\"book_table\":{\"0\":\"Yes\",\"1\":\"No\",\"2\":\"No\",\"3\":\"No\"},\"rate\":{\"0\":\"4.1\\/5\",\"1\":\"4.1\\/5\",\"2\":\"3.8\\/5\",\"3\":\"3.7\\/5\"},\"votes\":{\"0\":775,\"1\":787,\"2\":918,\"3\":88},\"phone\":{\"0\":\"080 42297555\\r\\n+91 9743772233\",\"1\":\"080 41714161\",\"2\":\"+91 9663487993\",\"3\":\"+91 9620009302\"},\"location\":{\"0\":\"Banashankari\",\"1\":\"Banashankari\",\"2\":\"Banashankari\",\"3\":\"Banashankari\"},\"rest_type\":{\"0\":\"Casual Dining\",\"1\":\"Casual Dining\",\"2\":\"Cafe, Casual Dining\",\"3\":\"Quick Bites\"},\"dish_liked\":{\"0\":\"Pasta, Lunch Buffet, Masala Papad, Paneer Lajawab, Tomato Shorba, Dum Biryani, Sweet Corn Soup\",\"1\":\"Momos, Lunch Buffet, Chocolate Nirvana, Thai Green Curry, Paneer Tikka, Dum Biryani, Chicken Biryani\",\"2\":\"Churros, Cannelloni, Minestrone Soup, Hot Chocolate, Pink Sauce Pasta, Salsa, Veg Supreme Pizza\",\"3\":\"Masala Dosa\"},\"cuisines\":{\"0\":\"North Indian, Mughlai, Chinese\",\"1\":\"Chinese, North Indian, Thai\",\"2\":\"Cafe, Mexican, Italian\",\"3\":\"South Indian, North Indian\"},\"approx_cost(for two people)\":{\"0\":\"800\",\"1\":\"800\",\"2\":\"800\",\"3\":\"300\"},\"reviews_list\":{\"0\":\"[('Rated 4.0', 'RATED\\\\n A beautiful place to dine in.The interiors take you back to the Mughal era. The lightings are just perfect.We went there on the occasion of Christmas and so they had only limited items available. But the taste and service was not compromised at all.The only complaint is that the breads could have been better.Would surely like to come here again.'), ('Rated 4.0', 'RATED\\\\n I was here for dinner with my family on a weekday. The restaurant was completely empty. Ambience is good with some good old hindi music. Seating arrangement are good too. We ordered masala papad, panner and baby corn starters, lemon and corrionder soup, butter roti, olive and chilli paratha. Food was fresh and good, service is good too. Good for family hangout.\\\\nCheers'), ('Rated 2.0', 'RATED\\\\n Its a restaurant near to Banashankari BDA. Me along with few of my office friends visited to have buffet but unfortunately they only provide veg buffet. On inquiring they said this place is mostly visited by vegetarians. Anyways we ordered ala carte items which took ages to come. Food was ok ok. Definitely not visiting anymore.'), ('Rated 4.0', 'RATED\\\\n We went here on a weekend and one of us had the buffet while two of us took Ala Carte. Firstly the ambience and service of this place is great! The buffet had a lot of items and the good was good. We had a Pumpkin Halwa intm the dessert which was amazing. Must try! The kulchas are great here. Cheers!'), ('Rated 5.0', 'RATED\\\\n The best thing about the place is it\\u00c3\\u0083\\\\x83\\u00c3\\u0082\\\\x83\\u00c3\\u0083\\\\x82\\u00c3\\u0082\\\\x82\\u00c3\\u0083\\\\x83\\u00c3\\u0082\\\\x82\\u00c3\\u0083\\\\x82\\u00c3\\u0082\\\\x92s ambiance. Second best thing was yummy ? food. We try buffet and buffet food was not disappointed us.\\\\nTest ?. ?? ?? ?? ?? ??\\\\nQuality ?. ??????????.\\\\nService: Staff was very professional and friendly.\\\\n\\\\nOverall experience was excellent.\\\\n\\\\nsubirmajumder85.wixsite.com'), ('Rated 5.0', 'RATED\\\\n Great food and pleasant ambience. Expensive but Coll place to chill and relax......\\\\n\\\\nService is really very very good and friendly staff...\\\\n\\\\nFood : 5\\/5\\\\nService : 5\\/5\\\\nAmbience :5\\/5\\\\nOverall :5\\/5'), ('Rated 4.0', 'RATED\\\\n Good ambience with tasty food.\\\\nCheese chilli paratha with Bhutta palak methi curry is a good combo.\\\\nLemon Chicken in the starters is a must try item.\\\\nEgg fried rice was also quite tasty.\\\\nIn the mocktails, recommend \\\"Alice in Junoon\\\". Do not miss it.'), ('Rated 4.0', 'RATED\\\\n You can\\u00c3\\u0083\\\\x83\\u00c3\\u0082\\\\x83\\u00c3\\u0083\\\\x82\\u00c3\\u0082\\\\x82\\u00c3\\u0083\\\\x83\\u00c3\\u0082\\\\x82\\u00c3\\u0083\\\\x82\\u00c3\\u0082\\\\x92t go wrong with Jalsa. Never been a fan of their buffet and thus always order alacarte\\u00c3\\u0083\\\\x83\\u00c3\\u0082\\\\x83\\u00c3\\u0083\\\\x82\\u00c3\\u0082\\\\x82\\u00c3\\u0083\\\\x83\\u00c3\\u0082\\\\x82\\u00c3\\u0083\\\\x82\\u00c3\\u0082\\\\x92. Service at times can be on the slower side but food is worth the wait.'), ('Rated 5.0', 'RATED\\\\n Overdelighted by the service and food provided at this place. A royal and ethnic atmosphere builds a strong essence of being in India and also the quality and taste of food is truly authentic. I would totally recommend to visit this place once.'), ('Rated 4.0', 'RATED\\\\n The place is nice and comfortable. Food wise all jalea outlets maintain a good standard. The soya chaap was a standout dish. Clearly one of trademark dish as per me and a must try.\\\\n\\\\nThe only concern is the parking. It very congested and limited to just 5cars. The basement parking is very steep and makes it cumbersome'), ('Rated 4.0', 'RATED\\\\n The place is nice and comfortable. Food wise all jalea outlets maintain a good standard. The soya chaap was a standout dish. Clearly one of trademark dish as per me and a must try.\\\\n\\\\nThe only concern is the parking. It very congested and limited to just 5cars. The basement parking is very steep and makes it cumbersome'), ('Rated 4.0', 'RATED\\\\n The place is nice and comfortable. Food wise all jalea outlets maintain a good standard. The soya chaap was a standout dish. Clearly one of trademark dish as per me and a must try.\\\\n\\\\nThe only concern is the parking. It very congested and limited to just 5cars. The basement parking is very steep and makes it cumbersome')]\",\"1\":\"[('Rated 4.0', 'RATED\\\\n Had been here for dinner with family. Turned out to be a good choose suitable for all ages of people. Can try this place. We liked the most was their starters. Service is good. Prices are affordable. Will recommend this restaurant for early dinner. The place is little noisy.'), ('Rated 3.0', 'RATED\\\\n The ambience is really nice, staff is courteous. The price is pretty high for the quantity, but overall the experience was fine. The quality of food is nice but nothing extraordinary. They also have buffet(only veg)'), ('Rated 3.0', 'RATED\\\\n I felt good is little expensive for the quantity they serve and In terms of taste is decent. There is nothing much to talk about the ambience, regular casual dining restaurant where you can take your family for dinner or lunch. If they improve on that quantity or may be reduce the price a bit or may be improve the presentation of the food it might Manage to get more repeat customers.'), ('Rated 4.0', 'RATED\\\\n I was looking for a quite place to spend some time with family and as well wanted to try some new place. Since I was at Banashankari I thought of trying this place. The place had good rating and was part of Zomato gold. So I decided to try this place. It was a delite to see a very friendly staff and food we ordered was very tasty as well.\\\\n\\\\nFood : 4\\/5\\\\nAmbience :3\\/5\\\\nFriendly staff : 4\\/5\\\\nPocket friendly : 4\\/5\\\\n\\\\nWill definitely visit again ??'), ('Rated 4.0', \\\"RATED\\\\n Nice place to dine and has a good ambiance... Food is good and the serving time is also good..neat restrooms and we'll arranged tables....only thing is we went at 12.30 for lunch...and we noticed that they kept on playing one music back to back which was a little annoying...\\\\n\\\\n1. Chicken biriyani was so good and the chicken was fresh and tender ,rice was well cooked and overall was great\\\\n\\\\n2. Mutton biriyani was very very good and tasty and It had plenty of mutton pieces...\\\"), ('Rated 5.0', 'RATED\\\\n This place just cool ? with good ambience and slow music and having delicious food is where you find peace. Staff very friendly and they have maintained the place so clean. The price is average for what the quantity of food they serve.\\\\nThom yum Thai soup was best and was treat to mouth, roti was soft with that vilaythi paneer was perfect to have for veggie foodies, in rice we tried burnt garlic fried rice with vegetables and it was the perfect thing to end.'), ('Rated 4.0', \\\"RATED\\\\n Quiet a good family type of place.. too calm and usually we don't find crowd here.. panner curry and the deserts is what we had tasted.. they wer really good but we found it a little expensive\\\"), ('Rated 2.0', \\\"RATED\\\\n I had a very bad experience here.\\\\nI don't know about a la carte, but the buffet was the worst. They gave us complementary drink and momos before the buffet. The momos were really good.\\\\nThe number of varieties first of all was very disappointing. The service was very slow. They refilled the food very slowly. The starters were okay. The main course also was so so. There was two gravies with roti and some rice with raitha. They had chats, sev puri and pan puri, which was average. But the desert was disappointing. They had gulab Jamun and chocolate cake. The jamun was not cooked inside. There was a cold blob of raw dough inside. The chocolate cake also was really hard and not that good.\\\\nOverall the buffet was a bad experience for me.\\\"), ('Rated 4.0', \\\"RATED\\\\n Food: 8\\/10\\\\nAmbience:8\\/10\\\\nStaff:8\\/10\\\\nOne of the good places to try north Indian food...but depends on ur taste buds. Not everyone will like all the items here. Specially when u r particular abt sweet and spicy food.\\\\nThere's buffet available too.\\\\nWe had ordered paneer uttar dakshin and paneer kurchan..was amazing. The Gobi hara pyaz and mix veg were average.\\\"), ('Rated 3.0', 'RATED\\\\n A decent place for a family lunch or dinner.. well arranged in a simple manner. Food was tasty and the crew was very helpful and understanding..'), ('Rated 4.0', \\\"RATED\\\\n Great place to have a heavy lunch. Good service.\\\\nThe chicken biryani was undoubtedly one of the best I've had. Biriyani and Lassi would be the suggested combo. Buffet is the talk of the place, so try according to your appetite. A nice place.\\\"), ('Rated 4.0', 'RATED\\\\n Its the one restaurant near katriguppe that i found was really good. Good variety of Chinese and thai dishes. Service is good and good place to hangout with family as its a peaceful place where noise is really less and good view.'), ('Rated 2.0', \\\"RATED\\\\n Spice elephant soup SPL: almost manchow flavour soup.. Just above medium spicy\\\\n\\\\nLasooni fish tikka was awesome\\\\n\\\\nI don't remember the dessert name but I have attached the photo .. It had vanilla ice inside wafers... Wafer was hell hard, egg smell chewy ... Nightmare dessert !\\\\n\\\\nTable leg space was very bad... I was so uncomfortable, the whole time kept on adjusting my legs\\\\n\\\\nNo parking\\\\n\\\\nFor the taste felt this is too costly\\\"), ('Rated 4.0', 'RATED\\\\n Zomato gold partner at this price. It was insane. They have really nice food. small place with very courteous staff and very cheap food for this ambience. Cost of soups is 80-100. Starters from 150-250. Main course 200-300. Cost for two was 800 for us.')]\",\"2\":\"[('Rated 3.0', \\\"RATED\\\\n Ambience is not that good enough and it's not a pocket friendly cafe and the quantity is not that good and desserts are too good enough ??..\\\"), ('Rated 3.0', \\\"RATED\\\\n \\\\nWent there for a quick bite with friends.\\\\nThe ambience had more of corporate feel. I would say it was unique.\\\\nTried nachos, pasta churros and lasagne.\\\\n\\\\nNachos were pathetic.( Seriously don't order)\\\\nPasta was okayish.\\\\nLasagne was good.\\\\nNutella churros were the best.\\\\nOverall an okayish experience!\\\\nPeace ??\\\"), ('Rated 4.0', \\\"RATED\\\\n First of all, a big thanks to the staff of this Cafe. Very polite and courteous.\\\\n\\\\nI was there 15mins before their closing time. Without any discomfort or hesitation, the staff welcomed me with a warm smile and said they're still open, though they were preparing to close the cafe for the day.\\\\n\\\\nQuickly ordered the Thai green curry, which is served with rice. They got it for me within 10mins, hot and freshly made.\\\\n\\\\nIt was tasty with the taste of coconut milk. Not very spicy, it was mild spicy.\\\\n\\\\nI saw they had yummy looking dessert menu, should go there to try them out!\\\\n\\\\nA good spacious place to hang out for coffee, pastas, pizza or Thai food.\\\"), ('Rated 3.0', \\\"RATED\\\\n A place for people who love churos. Otherwise it's a normal simple cafe like any other in town . Can go and spend the evening there . The positive being it is one of the better cafes in and around Banashankari and also the place is not very crowded .\\\"), ('Rated 3.0', 'RATED\\\\n Have been visiting this place since years. The quality has gone down drastically. Food quality has become very average from good. Service is also very bad of late.'), ('Rated 1.0', 'RATED\\\\n Really disappointed with the place.\\\\n\\\\nSeems like a old dusty store room stacked up with old sofas and chairs! The decor is soo disappointing that I cannot put it in words!\\\\nLastly the food was cherry on the cake to make it a complete worse meal.\\\\n\\\\nThis place is the last option you should ever consider.'), ('Rated 3.0', 'RATED\\\\n Congested atmosphere due to smoke from kitchen\\\\nAmbience was k\\\\nService was k .\\\\nChuros was a new experience recommended\\\\nNear to kims . Parking was not der'), ('Rated 1.0', \\\"RATED\\\\n Cockroaches !! I Repeat cockroaches!!Bakasura was disappointed.\\\\nBeing in Banashankari, being a gold outlet and nesting them? who can even think of that. Thankfully it wasn't over bakasura's plate. The ambience was last cleaned a decade back i guess. A cockroach just walked over our table. As it was alive, and well moving , the bakasura couldnt take a pic of it. Then the staff had to actually take it out of our table. Such ambience ? like what are you upto?\\\\n\\\\nBakasura disapproves. I dont even want to talk about the food.\\\"), ('Rated 4.0', 'RATED\\\\n A nice place to hangout, this place looks professional and also cute. If anyone is trying churros for the first the time, this is the place to go. Cheesecake is pretty amazing too.\\\\nRecommended.'), ('Rated 4.0', 'RATED\\\\n 1) pizza mocktails shakes and churros really very good\\\\n2) staff good\\\\n3) ambience simple and nice good\\\\n4) location of San churro cafe very nice\\\\n5) over San churro cafe good....'), ('Rated 3.0', \\\"RATED\\\\n One of my favourite places that I often visit in South bangalore , but quality has been reduced than before. Little unhappy with the food this time.\\\\nBut their churros never dissapointed me. Staff are too slow and do not respond easily.\\\\n\\\\nSoups:\\\\nMinestrone soup was like vegetable stock water with vegetables in it, not happy with it.\\\\n\\\\nChurros :\\\\nIt was great as usual. A must visit place if u love churros.\\\\n\\\\nStarters:\\\\n1. Africano panner -\\\\nIt was bit sweet and not happy with the dish!!\\\\n2. Loaded nachos -\\\\nThis happened to be their one of the best dishes, but now I take back my words. Very dissapointed with the dish.\\\\n\\\\nMain course:\\\\nsicilia pizza\\\\nPizza was too good, it's a must try dish.\\\\n\\\\nFinally I would recommend this place for churros and pizza .\\\\n\\\\nTaste : 6\\/10\\\\nAmbience :7\\/10\\\\nValue for money : 8\\/10\\\\n\\\\nOverall experience : 7.5\\/10\\\"), ('Rated 4.0', 'RATED\\\\n Visited this place long before, place come to.notice with the decoration lights outside the cafe. We ordered churos, pizza and hot chocolate, all were good just I little priced more.'), ('Rated 4.0', 'RATED\\\\n Spain pictures of Churros triggered Nostalgia about it , which led to Google search for Churro places in city , lucky we to get the result as San Churro cafe , we had an delectable churros here along with hot chocolate and Nachos ! Thanks for fulfilling our thirst of Spanish Churros !\\\\nFood : ???\\\\nChurros ???\\\\nHot Chocolate ?\\\\nNachos ??\\\\n\\\\nService : ??\\\\n\\\\nAmbiance : ?\\\\n\\\\nCleanliness : ??\\\\n\\\\nOne drawback : Store had some flies, I hope they take care of them for our next tour ?'), ('Rated 3.0', 'RATED\\\\n \\u00c3\\u0083\\\\x83\\u00c3\\u0082\\\\x83\\u00c3\\u0083\\\\x82\\u00c3\\u0082\\\\x82\\u00c3\\u0083\\\\x83\\u00c3\\u0082\\\\x82\\u00c3\\u0083\\\\x82\\u00c3\\u0082\\\\x93Chocolaty and churroic experience\\u00c3\\u0083\\\\x83\\u00c3\\u0082\\\\x83\\u00c3\\u0083\\\\x82\\u00c3\\u0082\\\\x82\\u00c3\\u0083\\\\x83\\u00c3\\u0082\\\\x82\\u00c3\\u0083\\\\x82\\u00c3\\u0082\\\\x94\\\\n\\\\nHit chocolate and churro with chocolate dip in the picture.\\\\n\\\\nMy first time at this restaurant and it has delivered in service, options as well as taste.'), ('Rated 4.0', 'RATED\\\\n Though very sadly the place looks in need of retouching, but the food was great! Especially when it was super cold outside and we were super hungry, the staff was nice. We ordered the pink pasta and a pizza. Both of them were pretty good. Was really tempted to try their Churros but we were stuffed. Worth a visit and try for sure!'), ('Rated 3.0', \\\"RATED\\\\n Churros, the restaurant's eponymous offering, has to be tried. They're a Mexican snack and served with chocolate sauce. Milkshakes are good too. The interior walls have murals and writings all over. Haven't tried the pizzas here but the cakes and pastries were pretty good.\\\"), ('Rated 3.0', \\\"RATED\\\\n Honestly, have only tried parcels and that too only starters and mains - never tried desserts. So this review is based on starters and mains only. The quality wasn't all that great, was pretty average.\\\"), ('Rated 4.0', \\\"RATED\\\\n Visited this place for the first time, recently. The ambience was pretty good. We ordered two 'churros for two'. Having churros for the first time ever, I was way too excited and judgmental. But swear to Lord, the churros were heavenly! The chocolate sauce was the actual chocolate sauce and not nutella xD We had taken the Cross Country Ranch pizza. It was a thin crust pizza loaded with veggies.\\\\nThe place is famous for churros (the name suggests so). I would love to visit again to try all the other items out :')\\\"), ('Rated 4.0', 'RATED\\\\n I love the desserts more than anything here !Good place and well served !:)\\\\nWe dint like the nachos here , it was not that good as we expected !\\\\nExcept nachos almost all dish were quite good!'), ('Rated 2.0', \\\"RATED\\\\n 2nd time I have visited this place and I am surprised to see how it has declined. Churros was hard, pizza was pathetic, loaded nachos and masala Garlic bread were topped with cheese with MAYO! Yes mayonnese.. Soup has the bread crumbs in almost melted state, mocktails were average and our only Savior was pasta and caramel pudding... This place has high potential but let down by dull ambience, pretty slow and inattentive staff. Buffet price was listed as 399 but was charged 499, We still went ahead and took the deal to be very disappointed!! Most dishes were freshly cooked hence going with some starts else would rate them lower. I did not find a single person who seemed happy with their meal as moat complained about one thing or the other. That'll be my last visit San Churros.\\\")]\",\"3\":\"[('Rated 4.0', \\\"RATED\\\\n Great food and proper Karnataka style full meals. Been there twice and was fully satisfied.. Will give 5 stars if it's well managed............\\\"), ('Rated 2.0', 'RATED\\\\n Reached the place at 3pm on Saturday. Half of the items on the menu were over. What was annoying was is the food was cold. The taste was also very average. Only dosa and holige were good. There were very few people in the restaurant and the service was still very slow. The waiters were all standing in one corner and talking. Had to call them repeatedly.'), ('Rated 4.0', 'RATED\\\\n Had been here, good food served and tasty,good place to go with freinds and family, should be first to get served well with all food. One of the good hotel there for this price.'), ('Rated 2.0', 'RATED\\\\n How can a Udupi restaurant be so dirty.the floor the walls the waiters are all stained in food.Why cant they clean it? The floor even had decorative colour paper every where and lot of food.Now coming to the taste of the food- it was pretty decent for what they chargw.What upset us was the menu says they serve thambuli, two variety of rice etc but they were all over at 2.45 but they still charged us full amount.Lastly if u r ok to b seated with random people and adjust with their table manners( burrping etc) then tis place is decent for a quick veg udupi meals'), ('Rated 4.0', 'RATED\\\\n Aadhuri Udupi bhojana is one of the best vegetarian hotel in and around Banashankari locality\\\\nwere we can enjoy the authentic south Karnataka style food.'), ('Rated 5.0', 'RATED\\\\n One of the best restaurants for unlimited food. Price for one full unlimited meal is ?150 and the service is awesome here. If you\\u00c3\\u0083\\\\x83\\u00c3\\u0082\\\\x83\\u00c3\\u0083\\\\x82\\u00c3\\u0082\\\\x82\\u00c3\\u0083\\\\x83\\u00c3\\u0082\\\\x82\\u00c3\\u0083\\\\x82\\u00c3\\u0082\\\\x92re a costalian you will love the food served here.'), ('Rated 4.0', \\\"RATED\\\\n Been here a couple of times. Highly satisfying on both the ocassions. Great food. Decent service. At price of 160 per person it is of a great value. The staff here serves in a traditional Mysore style dresses. The food is authentic Karnataka meals. I really liked the pineapple pickle\\/chutney. They serve some 4 varities of rice of which you shouldn't try all as you would already be full. Roti, Dosa were decent. In the deserts, Payasam& Holige were good.\\\\nI think it is one the must try places to try some authentic south food even if you have been to the likes of Kamath etc. this one is a little different. :)\\\"), ('Rated 5.0', 'RATED\\\\n Very good restorent good south Indian food very nice all food s are good we spent good time and food in this hotel value of money we loved it'), ('Rated 3.5', 'RATED\\\\n I went to this restaurant on Sunday afternoon.It was a bit crowded.\\\\n\\\\nIts located in the 1st floor Above anna kuteera kathriguppe\\\\n\\\\nYou need to take food coupon in the ground floor itself.\\\\nit was 160 rs.\\\\n\\\\nTaste wise it was good.\\\\nService was slow.\\\\nNothing exciting\\/different in the menu.\\\\nWhatever they serve its good.\\\\nWorth for money.'), ('Rated 5.0', 'RATED\\\\n Awesome place , worth with price , quality , quantity , service , hygiene and discipline I loved it ???? wish there were more rating stars to give I give 100 on 5 for sure'), ('Rated 3.5', 'RATED\\\\n Nice food, good ambiance, so many verity of food, this time i was in hurry so, next review will be detailed one. Its good to see all dishes at one place'), ('Rated 1.5', 'RATED\\\\n The food was not satisfactory. Not one item served could be eaten again . Only advantage is the price but even that is not going to be enough to make me visit the place again . Dear Udupi staff people wouldn\\u00c3\\u0083\\\\x83\\u00c3\\u0082\\\\x83\\u00c3\\u0083\\\\x82\\u00c3\\u0082\\\\x82\\u00c3\\u0083\\\\x83\\u00c3\\u0082\\\\x82\\u00c3\\u0083\\\\x82\\u00c3\\u0082\\\\x92t mind if the food quality was better for a higher price and please get more fans so that people won\\u00c3\\u0083\\\\x83\\u00c3\\u0082\\\\x83\\u00c3\\u0083\\\\x82\\u00c3\\u0082\\\\x82\\u00c3\\u0083\\\\x83\\u00c3\\u0082\\\\x82\\u00c3\\u0083\\\\x82\\u00c3\\u0082\\\\x92t have to deal with another disappointment of sweating while eating.'), ('Rated 5.0', 'RATED\\\\n Excellent quality and taste, more variety of authentic south Indian food. Excellent service. Highly recommended. Worth the money we pay here.'), ('Rated 2.0', \\\"RATED\\\\n I'm rating 2 only because of the large number of items they serve for the price they charge. While the taste is moderate, the biggest turn off is the poor service and totally unresponsive staff. They are either understaffed or poorly trained. I didn't get to taste a few items, thanks to their ever busy staff, and ended eating what was served. Please learn the crowd management from other peers like maiyya and roti ghar\\\"), ('Rated 5.0', 'RATED\\\\n One of my colleagues suggested this place. Went there at 1 pm and place wasnt crowded. Ambience is decent and cool. A welcome drink \\\"appekai saru\\\" was served. Then started..typical \\\"baale yele uta\\\" . Diffrerent types of palya, kosambari and sides. Then came masala dosa, pulka,pulav, veg rice bath. Masala dosa was very very tasty. Service is bit fast as people will be coming, but they will make it slow if u tell them to. Then comes rice with sambar,rasam, daal,tambali. And yes, dont forget to eat holige,payasam and icecream. Service is very nice. One best thing is all of this comes in just 160 rs and its unlimited!!! What else you want!! A must visit place for south indian foodie!'), ('Rated 3.0', \\\"RATED\\\\n Been here last week with high expectations based on few reviews. But utterly disappointed with the food quality & taste. I feel it's an overhyped place! There are many better places in the vicinity for Plantain leaf meal. There was too much of waiting & these staffs were too hurry in serving food & asking people to finish meal & leave quickly. I can understand there is a waiting and staffs should approach in a positive way. I will never go there again!\\\\n\\\\nbangaloreepicure.com\\\"), ('Rated 4.0', 'RATED\\\\n I have a confession, being a south Indian I distaste south Indian food(not that I hate it, I don\\\\'t like it either).But still made up my mind on Sankaranti to have authentic south Indian food visited \\\"Adduri Udupi Bhojana\\\" because it was close to my place.I just had a look at the menu told myself that I can do it(I am not blessed with a tremendous appetite).welcomed with the mango soup followed by the wide variety of dishes served, I have developed the new love for Pongal(both sweet and Khara).I can still feel the taste of the ghee bursting in my mouth but it was\\\"kai holige\\\" that made my day, crisp and soft.not to forget food is served on a banana leaf that adds in more flavor and taste.waiters are attentive and responsive.The customer\\\\'s needs are monitored and addressed.so much variety for just 160Rs without compromising on quantity and quality and yes it is unlimited.This visit turned out to be a surprise package.'), ('Rated 3.5', 'RATED\\\\n Food is good.\\\\nNo parking\\\\nAmbience is average\\\\nOverall its value for money if you looking for south Indian food.\\\\nPreferred the tiffin items......'), ('Rated 3.0', \\\"RATED\\\\n Been here last week with high expectations based on few reviews. But utterly disappointed with the food quality & taste. I feel it's an overhyped place! There are many better places in the vicinity for Plantain leaf meal. There was too much of waiting & these staffs were too hurry in serving food & asking people to finish meal & leave quickly. I can understand there is a waiting and staffs should approach in a positive way. I will never go there again!\\\\n\\\\nbangaloreepicure.com\\\"), ('Rated 4.0', 'RATED\\\\n I have a confession, being a south Indian I distaste south Indian food(not that I hate it, I don\\\\'t like it either).But still made up my mind on Sankaranti to have authentic south Indian food visited \\\"Adduri Udupi Bhojana\\\" because it was close to my place.I just had a look at the menu told myself that I can do it(I am not blessed with a tremendous appetite).welcomed with the mango soup followed by the wide variety of dishes served, I have developed the new love for Pongal(both sweet and Khara).I can still feel the taste of the ghee bursting in my mouth but it was\\\"kai holige\\\" that made my day, crisp and soft.not to forget food is served on a banana leaf that adds in more flavor and taste.waiters are attentive and responsive.The customer\\\\'s needs are monitored and addressed.so much variety for just 160Rs without compromising on quantity and quality and yes it is unlimited.This visit turned out to be a surprise package.'), ('Rated 3.5', 'RATED\\\\n Food is good.\\\\nNo parking\\\\nAmbience is average\\\\nOverall its value for money if you looking for south Indian food.\\\\nPreferred the tiffin items......'), ('Rated 3.0', \\\"RATED\\\\n Been here last week with high expectations based on few reviews. But utterly disappointed with the food quality & taste. I feel it's an overhyped place! There are many better places in the vicinity for Plantain leaf meal. There was too much of waiting & these staffs were too hurry in serving food & asking people to finish meal & leave quickly. I can understand there is a waiting and staffs should approach in a positive way. I will never go there again!\\\\n\\\\nbangaloreepicure.com\\\"), ('Rated 4.0', 'RATED\\\\n I have a confession, being a south Indian I distaste south Indian food(not that I hate it, I don\\\\'t like it either).But still made up my mind on Sankaranti to have authentic south Indian food visited \\\"Adduri Udupi Bhojana\\\" because it was close to my place.I just had a look at the menu told myself that I can do it(I am not blessed with a tremendous appetite).welcomed with the mango soup followed by the wide variety of dishes served, I have developed the new love for Pongal(both sweet and Khara).I can still feel the taste of the ghee bursting in my mouth but it was\\\"kai holige\\\" that made my day, crisp and soft.not to forget food is served on a banana leaf that adds in more flavor and taste.waiters are attentive and responsive.The customer\\\\'s needs are monitored and addressed.so much variety for just 160Rs without compromising on quantity and quality and yes it is unlimited.This visit turned out to be a surprise package.'), ('Rated 3.5', 'RATED\\\\n Food is good.\\\\nNo parking\\\\nAmbience is average\\\\nOverall its value for money if you looking for south Indian food.\\\\nPreferred the tiffin items......'), ('Rated 3.0', \\\"RATED\\\\n Been here last week with high expectations based on few reviews. But utterly disappointed with the food quality & taste. I feel it's an overhyped place! There are many better places in the vicinity for Plantain leaf meal. There was too much of waiting & these staffs were too hurry in serving food & asking people to finish meal & leave quickly. I can understand there is a waiting and staffs should approach in a positive way. I will never go there again!\\\\n\\\\nbangaloreepicure.com\\\"), ('Rated 4.0', 'RATED\\\\n I have a confession, being a south Indian I distaste south Indian food(not that I hate it, I don\\\\'t like it either).But still made up my mind on Sankaranti to have authentic south Indian food visited \\\"Adduri Udupi Bhojana\\\" because it was close to my place.I just had a look at the menu told myself that I can do it(I am not blessed with a tremendous appetite).welcomed with the mango soup followed by the wide variety of dishes served, I have developed the new love for Pongal(both sweet and Khara).I can still feel the taste of the ghee bursting in my mouth but it was\\\"kai holige\\\" that made my day, crisp and soft.not to forget food is served on a banana leaf that adds in more flavor and taste.waiters are attentive and responsive.The customer\\\\'s needs are monitored and addressed.so much variety for just 160Rs without compromising on quantity and quality and yes it is unlimited.This visit turned out to be a surprise package.'), ('Rated 3.5', 'RATED\\\\n Food is good.\\\\nNo parking\\\\nAmbience is average\\\\nOverall its value for money if you looking for south Indian food.\\\\nPreferred the tiffin items......'), ('Rated 3.0', \\\"RATED\\\\n Been here last week with high expectations based on few reviews. But utterly disappointed with the food quality & taste. I feel it's an overhyped place! There are many better places in the vicinity for Plantain leaf meal. There was too much of waiting & these staffs were too hurry in serving food & asking people to finish meal & leave quickly. I can understand there is a waiting and staffs should approach in a positive way. I will never go there again!\\\\n\\\\nbangaloreepicure.com\\\"), ('Rated 4.0', 'RATED\\\\n I have a confession, being a south Indian I distaste south Indian food(not that I hate it, I don\\\\'t like it either).But still made up my mind on Sankaranti to have authentic south Indian food visited \\\"Adduri Udupi Bhojana\\\" because it was close to my place.I just had a look at the menu told myself that I can do it(I am not blessed with a tremendous appetite).welcomed with the mango soup followed by the wide variety of dishes served, I have developed the new love for Pongal(both sweet and Khara).I can still feel the taste of the ghee bursting in my mouth but it was\\\"kai holige\\\" that made my day, crisp and soft.not to forget food is served on a banana leaf that adds in more flavor and taste.waiters are attentive and responsive.The customer\\\\'s needs are monitored and addressed.so much variety for just 160Rs without compromising on quantity and quality and yes it is unlimited.This visit turned out to be a surprise package.'), ('Rated 3.5', 'RATED\\\\n Food is good.\\\\nNo parking\\\\nAmbience is average\\\\nOverall its value for money if you looking for south Indian food.\\\\nPreferred the tiffin items......'), ('Rated 4.0', \\\"RATED\\\\n Had been on a week night for an authentic south Indian food. I personally love being served on banyan leaf. The waiters are all dressed up in traditional Mysore attire wearing with a peta. The food was delicious and definitely worth it's price. The waiters are friendly and serve u well with a smile. The food is prepared without garlic which is a plus point attracting many senior customers.\\\"), ('Rated 3.5', \\\"RATED\\\\n Food is good and enjoyed. With 150 Rs. is very nice.\\\\nNote: (It happen with us)\\\\n - If you going TWO people, you may need to share the table there. Its almost crowed.\\\\n - You have to sit where they ask, you can't choose even if all the table are empty. \\\\nJust wait, eat and come back.\\\"), ('Rated 5.0', 'RATED\\\\n Excellent Ambience with delicious meals with 10-15 variety types. Worth paying for money and you can dine out if you are with more friends\\/relatives in home.'), ('Rated 4.5', 'RATED\\\\n Very good and Unlimited . especially masala dosa . Service and ambience was also good. will visit again with friends . very very reasonble and wholesome food'), ('Rated 3.5', 'RATED\\\\n Aaaa wt to tell, Unlimited food, food was like exactly like south indian marriage food, 2 soups, sides masala dosa, all were unlimited for just ?150, except ice cream in the end ?? they give a water bottle, and main thing is have patience to get place to sit..... ??,there is no parking place of this outlet, and difficult to park in front of the restaurants,')]\"},\"menu_item\":{\"0\":\"[]\",\"1\":\"[]\",\"2\":\"[]\",\"3\":\"[]\"},\"listed_in(type)\":{\"0\":\"Buffet\",\"1\":\"Buffet\",\"2\":\"Buffet\",\"3\":\"Buffet\"},\"listed_in(city)\":{\"0\":\"Banashankari\",\"1\":\"Banashankari\",\"2\":\"Banashankari\",\"3\":\"Banashankari\"}}"}}]
| true | 1 |
<start_data_description><data_path>zomato-bangalore-restaurants/zomato.csv:
<column_names>
['url', 'address', 'name', 'online_order', 'book_table', 'rate', 'votes', 'phone', 'location', 'rest_type', 'dish_liked', 'cuisines', 'approx_cost(for two people)', 'reviews_list', 'menu_item', 'listed_in(type)', 'listed_in(city)']
<column_types>
{'url': 'object', 'address': 'object', 'name': 'object', 'online_order': 'object', 'book_table': 'object', 'rate': 'object', 'votes': 'int64', 'phone': 'object', 'location': 'object', 'rest_type': 'object', 'dish_liked': 'object', 'cuisines': 'object', 'approx_cost(for two people)': 'object', 'reviews_list': 'object', 'menu_item': 'object', 'listed_in(type)': 'object', 'listed_in(city)': 'object'}
<dataframe_Summary>
{'votes': {'count': 51717.0, 'mean': 283.69752692538236, 'std': 803.8388530079794, 'min': 0.0, '25%': 7.0, '50%': 41.0, '75%': 198.0, 'max': 16832.0}}
<dataframe_info>
RangeIndex: 51717 entries, 0 to 51716
Data columns (total 17 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 url 51717 non-null object
1 address 51717 non-null object
2 name 51717 non-null object
3 online_order 51717 non-null object
4 book_table 51717 non-null object
5 rate 43942 non-null object
6 votes 51717 non-null int64
7 phone 50509 non-null object
8 location 51696 non-null object
9 rest_type 51490 non-null object
10 dish_liked 23639 non-null object
11 cuisines 51672 non-null object
12 approx_cost(for two people) 51371 non-null object
13 reviews_list 51717 non-null object
14 menu_item 51717 non-null object
15 listed_in(type) 51717 non-null object
16 listed_in(city) 51717 non-null object
dtypes: int64(1), object(16)
memory usage: 6.7+ MB
<some_examples>
{'url': {'0': 'https://www.zomato.com/bangalore/jalsa-banashankari?context=eyJzZSI6eyJlIjpbNTg2OTQsIjE4Mzc1NDc0IiwiNTkwOTAiLCIxODM4Mjk0NCIsIjE4MjI0Njc2IiwiNTkyODkiLCIxODM3MzM4NiJdLCJ0IjoiUmVzdGF1cmFudHMgaW4gQmFuYXNoYW5rYXJpIHNlcnZpbmcgQnVmZmV0In19', '1': 'https://www.zomato.com/bangalore/spice-elephant-banashankari?context=eyJzZSI6eyJlIjpbIjU4Njk0IiwxODM3NTQ3NCwiNTkwOTAiLCIxODM4Mjk0NCIsIjE4MjI0Njc2IiwiNTkyODkiLCIxODM3MzM4NiJdLCJ0IjoiUmVzdGF1cmFudHMgaW4gQmFuYXNoYW5rYXJpIHNlcnZpbmcgQnVmZmV0In19', '2': 'https://www.zomato.com/SanchurroBangalore?context=eyJzZSI6eyJlIjpbIjU4Njk0IiwiMTgzNzU0NzQiLDU5MDkwLCIxODM4Mjk0NCIsIjE4MjI0Njc2IiwiNTkyODkiLCIxODM3MzM4NiJdLCJ0IjoiUmVzdGF1cmFudHMgaW4gQmFuYXNoYW5rYXJpIHNlcnZpbmcgQnVmZmV0In19', '3': 'https://www.zomato.com/bangalore/addhuri-udupi-bhojana-banashankari?context=eyJzZSI6eyJlIjpbIjU4Njk0IiwiMTgzNzU0NzQiLCI1OTA5MCIsMTgzODI5NDQsIjE4MjI0Njc2IiwiNTkyODkiLCIxODM3MzM4NiJdLCJ0IjoiUmVzdGF1cmFudHMgaW4gQmFuYXNoYW5rYXJpIHNlcnZpbmcgQnVmZmV0In19'}, 'address': {'0': '942, 21st Main Road, 2nd Stage, Banashankari, Bangalore', '1': '2nd Floor, 80 Feet Road, Near Big Bazaar, 6th Block, Kathriguppe, 3rd Stage, Banashankari, Bangalore', '2': '1112, Next to KIMS Medical College, 17th Cross, 2nd Stage, Banashankari, Bangalore', '3': '1st Floor, Annakuteera, 3rd Stage, Banashankari, Bangalore'}, 'name': {'0': 'Jalsa', '1': 'Spice Elephant', '2': 'San Churro Cafe', '3': 'Addhuri Udupi Bhojana'}, 'online_order': {'0': 'Yes', '1': 'Yes', '2': 'Yes', '3': 'No'}, 'book_table': {'0': 'Yes', '1': 'No', '2': 'No', '3': 'No'}, 'rate': {'0': '4.1/5', '1': '4.1/5', '2': '3.8/5', '3': '3.7/5'}, 'votes': {'0': 775, '1': 787, '2': 918, '3': 88}, 'phone': {'0': '080 42297555\r\n+91 9743772233', '1': '080 41714161', '2': '+91 9663487993', '3': '+91 9620009302'}, 'location': {'0': 'Banashankari', '1': 'Banashankari', '2': 'Banashankari', '3': 'Banashankari'}, 'rest_type': {'0': 'Casual Dining', '1': 'Casual Dining', '2': 'Cafe, Casual Dining', '3': 'Quick Bites'}, 'dish_liked': {'0': 'Pasta, Lunch Buffet, Masala Papad, Paneer Lajawab, Tomato Shorba, Dum Biryani, Sweet Corn Soup', '1': 'Momos, Lunch Buffet, Chocolate Nirvana, Thai Green Curry, Paneer Tikka, Dum Biryani, Chicken Biryani', '2': 'Churros, Cannelloni, Minestrone Soup, Hot Chocolate, Pink Sauce Pasta, Salsa, Veg Supreme Pizza', '3': 'Masala Dosa'}, 'cuisines': {'0': 'North Indian, Mughlai, Chinese', '1': 'Chinese, North Indian, Thai', '2': 'Cafe, Mexican, Italian', '3': 'South Indian, North Indian'}, 'approx_cost(for two people)': {'0': '800', '1': '800', '2': '800', '3': '300'}, 'reviews_list': {'0': '[(\'Rated 4.0\', \'RATED\\n A beautiful place to dine in.The interiors take you back to the Mughal era. The lightings are just perfect.We went there on the occasion of Christmas and so they had only limited items available. But the taste and service was not compromised at all.The only complaint is that the breads could have been better.Would surely like to come here again.\'), (\'Rated 4.0\', \'RATED\\n I was here for dinner with my family on a weekday. The restaurant was completely empty. Ambience is good with some good old hindi music. Seating arrangement are good too. We ordered masala papad, panner and baby corn starters, lemon and corrionder soup, butter roti, olive and chilli paratha. Food was fresh and good, service is good too. Good for family hangout.\\nCheers\'), (\'Rated 2.0\', \'RATED\\n Its a restaurant near to Banashankari BDA. Me along with few of my office friends visited to have buffet but unfortunately they only provide veg buffet. On inquiring they said this place is mostly visited by vegetarians. Anyways we ordered ala carte items which took ages to come. Food was ok ok. Definitely not visiting anymore.\'), (\'Rated 4.0\', \'RATED\\n We went here on a weekend and one of us had the buffet while two of us took Ala Carte. Firstly the ambience and service of this place is great! The buffet had a lot of items and the good was good. We had a Pumpkin Halwa intm the dessert which was amazing. Must try! The kulchas are great here. Cheers!\'), (\'Rated 5.0\', \'RATED\\n The best thing about the place is itÃ\x83\\x83Ã\x82\\x83Ã\x83\\x82Ã\x82\\x82Ã\x83\\x83Ã\x82\\x82Ã\x83\\x82Ã\x82\\x92s ambiance. Second best thing was yummy ? food. We try buffet and buffet food was not disappointed us.\\nTest ?. ?? ?? ?? ?? ??\\nQuality ?. ??????????.\\nService: Staff was very professional and friendly.\\n\\nOverall experience was excellent.\\n\\nsubirmajumder85.wixsite.com\'), (\'Rated 5.0\', \'RATED\\n Great food and pleasant ambience. Expensive but Coll place to chill and relax......\\n\\nService is really very very good and friendly staff...\\n\\nFood : 5/5\\nService : 5/5\\nAmbience :5/5\\nOverall :5/5\'), (\'Rated 4.0\', \'RATED\\n Good ambience with tasty food.\\nCheese chilli paratha with Bhutta palak methi curry is a good combo.\\nLemon Chicken in the starters is a must try item.\\nEgg fried rice was also quite tasty.\\nIn the mocktails, recommend "Alice in Junoon". Do not miss it.\'), (\'Rated 4.0\', \'RATED\\n You canÃ\x83\\x83Ã\x82\\x83Ã\x83\\x82Ã\x82\\x82Ã\x83\\x83Ã\x82\\x82Ã\x83\\x82Ã\x82\\x92t go wrong with Jalsa. Never been a fan of their buffet and thus always order alacarteÃ\x83\\x83Ã\x82\\x83Ã\x83\\x82Ã\x82\\x82Ã\x83\\x83Ã\x82\\x82Ã\x83\\x82Ã\x82\\x92. Service at times can be on the slower side but food is worth the wait.\'), (\'Rated 5.0\', \'RATED\\n Overdelighted by the service and food provided at this place. A royal and ethnic atmosphere builds a strong essence of being in India and also the quality and taste of food is truly authentic. I would totally recommend to visit this place once.\'), (\'Rated 4.0\', \'RATED\\n The place is nice and comfortable. Food wise all jalea outlets maintain a good standard. The soya chaap was a standout dish. Clearly one of trademark dish as per me and a must try.\\n\\nThe only concern is the parking. It very congested and limited to just 5cars. The basement parking is very steep and makes it cumbersome\'), (\'Rated 4.0\', \'RATED\\n The place is nice and comfortable. Food wise all jalea outlets maintain a good standard. The soya chaap was a standout dish. Clearly one of trademark dish as per me and a must try.\\n\\nThe only concern is the parking. It very congested and limited to just 5cars. The basement parking is very steep and makes it cumbersome\'), (\'Rated 4.0\', \'RATED\\n The place is nice and comfortable. Food wise all jalea outlets maintain a good standard. The soya chaap was a standout dish. Clearly one of trademark dish as per me and a must try.\\n\\nThe only concern is the parking. It very congested and limited to just 5cars. The basement parking is very steep and makes it cumbersome\')]', '1': '[(\'Rated 4.0\', \'RATED\\n Had been here for dinner with family. Turned out to be a good choose suitable for all ages of people. Can try this place. We liked the most was their starters. Service is good. Prices are affordable. Will recommend this restaurant for early dinner. The place is little noisy.\'), (\'Rated 3.0\', \'RATED\\n The ambience is really nice, staff is courteous. The price is pretty high for the quantity, but overall the experience was fine. The quality of food is nice but nothing extraordinary. They also have buffet(only veg)\'), (\'Rated 3.0\', \'RATED\\n I felt good is little expensive for the quantity they serve and In terms of taste is decent. There is nothing much to talk about the ambience, regular casual dining restaurant where you can take your family for dinner or lunch. If they improve on that quantity or may be reduce the price a bit or may be improve the presentation of the food it might Manage to get more repeat customers.\'), (\'Rated 4.0\', \'RATED\\n I was looking for a quite place to spend some time with family and as well wanted to try some new place. Since I was at Banashankari I thought of trying this place. The place had good rating and was part of Zomato gold. So I decided to try this place. It was a delite to see a very friendly staff and food we ordered was very tasty as well.\\n\\nFood : 4/5\\nAmbience :3/5\\nFriendly staff : 4/5\\nPocket friendly : 4/5\\n\\nWill definitely visit again ??\'), (\'Rated 4.0\', "RATED\\n Nice place to dine and has a good ambiance... Food is good and the serving time is also good..neat restrooms and we\'ll arranged tables....only thing is we went at 12.30 for lunch...and we noticed that they kept on playing one music back to back which was a little annoying...\\n\\n1. Chicken biriyani was so good and the chicken was fresh and tender ,rice was well cooked and overall was great\\n\\n2. Mutton biriyani was very very good and tasty and It had plenty of mutton pieces..."), (\'Rated 5.0\', \'RATED\\n This place just cool ? with good ambience and slow music and having delicious food is where you find peace. Staff very friendly and they have maintained the place so clean. The price is average for what the quantity of food they serve.\\nThom yum Thai soup was best and was treat to mouth, roti was soft with that vilaythi paneer was perfect to have for veggie foodies, in rice we tried burnt garlic fried rice with vegetables and it was the perfect thing to end.\'), (\'Rated 4.0\', "RATED\\n Quiet a good family type of place.. too calm and usually we don\'t find crowd here.. panner curry and the deserts is what we had tasted.. they wer really good but we found it a little expensive"), (\'Rated 2.0\', "RATED\\n I had a very bad experience here.\\nI don\'t know about a la carte, but the buffet was the worst. They gave us complementary drink and momos before the buffet. The momos were really good.\\nThe number of varieties first of all was very disappointing. The service was very slow. They refilled the food very slowly. The starters were okay. The main course also was so so. There was two gravies with roti and some rice with raitha. They had chats, sev puri and pan puri, which was average. But the desert was disappointing. They had gulab Jamun and chocolate cake. The jamun was not cooked inside. There was a cold blob of raw dough inside. The chocolate cake also was really hard and not that good.\\nOverall the buffet was a bad experience for me."), (\'Rated 4.0\', "RATED\\n Food: 8/10\\nAmbience:8/10\\nStaff:8/10\\nOne of the good places to try north Indian food...but depends on ur taste buds. Not everyone will like all the items here. Specially when u r particular abt sweet and spicy food.\\nThere\'s buffet available too.\\nWe had ordered paneer uttar dakshin and paneer kurchan..was amazing. The Gobi hara pyaz and mix veg were average."), (\'Rated 3.0\', \'RATED\\n A decent place for a family lunch or dinner.. well arranged in a simple manner. Food was tasty and the crew was very helpful and understanding..\'), (\'Rated 4.0\', "RATED\\n Great place to have a heavy lunch. Good service.\\nThe chicken biryani was undoubtedly one of the best I\'ve had. Biriyani and Lassi would be the suggested combo. Buffet is the talk of the place, so try according to your appetite. A nice place."), (\'Rated 4.0\', \'RATED\\n Its the one restaurant near katriguppe that i found was really good. Good variety of Chinese and thai dishes. Service is good and good place to hangout with family as its a peaceful place where noise is really less and good view.\'), (\'Rated 2.0\', "RATED\\n Spice elephant soup SPL: almost manchow flavour soup.. Just above medium spicy\\n\\nLasooni fish tikka was awesome\\n\\nI don\'t remember the dessert name but I have attached the photo .. It had vanilla ice inside wafers... Wafer was hell hard, egg smell chewy ... Nightmare dessert !\\n\\nTable leg space was very bad... I was so uncomfortable, the whole time kept on adjusting my legs\\n\\nNo parking\\n\\nFor the taste felt this is too costly"), (\'Rated 4.0\', \'RATED\\n Zomato gold partner at this price. It was insane. They have really nice food. small place with very courteous staff and very cheap food for this ambience. Cost of soups is 80-100. Starters from 150-250. Main course 200-300. Cost for two was 800 for us.\')]', '2': '[(\'Rated 3.0\', "RATED\\n Ambience is not that good enough and it\'s not a pocket friendly cafe and the quantity is not that good and desserts are too good enough ??.."), (\'Rated 3.0\', "RATED\\n \\nWent there for a quick bite with friends.\\nThe ambience had more of corporate feel. I would say it was unique.\\nTried nachos, pasta churros and lasagne.\\n\\nNachos were pathetic.( Seriously don\'t order)\\nPasta was okayish.\\nLasagne was good.\\nNutella churros were the best.\\nOverall an okayish experience!\\nPeace ??"), (\'Rated 4.0\', "RATED\\n First of all, a big thanks to the staff of this Cafe. Very polite and courteous.\\n\\nI was there 15mins before their closing time. Without any discomfort or hesitation, the staff welcomed me with a warm smile and said they\'re still open, though they were preparing to close the cafe for the day.\\n\\nQuickly ordered the Thai green curry, which is served with rice. They got it for me within 10mins, hot and freshly made.\\n\\nIt was tasty with the taste of coconut milk. Not very spicy, it was mild spicy.\\n\\nI saw they had yummy looking dessert menu, should go there to try them out!\\n\\nA good spacious place to hang out for coffee, pastas, pizza or Thai food."), (\'Rated 3.0\', "RATED\\n A place for people who love churos. Otherwise it\'s a normal simple cafe like any other in town . Can go and spend the evening there . The positive being it is one of the better cafes in and around Banashankari and also the place is not very crowded ."), (\'Rated 3.0\', \'RATED\\n Have been visiting this place since years. The quality has gone down drastically. Food quality has become very average from good. Service is also very bad of late.\'), (\'Rated 1.0\', \'RATED\\n Really disappointed with the place.\\n\\nSeems like a old dusty store room stacked up with old sofas and chairs! The decor is soo disappointing that I cannot put it in words!\\nLastly the food was cherry on the cake to make it a complete worse meal.\\n\\nThis place is the last option you should ever consider.\'), (\'Rated 3.0\', \'RATED\\n Congested atmosphere due to smoke from kitchen\\nAmbience was k\\nService was k .\\nChuros was a new experience recommended\\nNear to kims . Parking was not der\'), (\'Rated 1.0\', "RATED\\n Cockroaches !! I Repeat cockroaches!!Bakasura was disappointed.\\nBeing in Banashankari, being a gold outlet and nesting them? who can even think of that. Thankfully it wasn\'t over bakasura\'s plate. The ambience was last cleaned a decade back i guess. A cockroach just walked over our table. As it was alive, and well moving , the bakasura couldnt take a pic of it. Then the staff had to actually take it out of our table. Such ambience ? like what are you upto?\\n\\nBakasura disapproves. I dont even want to talk about the food."), (\'Rated 4.0\', \'RATED\\n A nice place to hangout, this place looks professional and also cute. If anyone is trying churros for the first the time, this is the place to go. Cheesecake is pretty amazing too.\\nRecommended.\'), (\'Rated 4.0\', \'RATED\\n 1) pizza mocktails shakes and churros really very good\\n2) staff good\\n3) ambience simple and nice good\\n4) location of San churro cafe very nice\\n5) over San churro cafe good....\'), (\'Rated 3.0\', "RATED\\n One of my favourite places that I often visit in South bangalore , but quality has been reduced than before. Little unhappy with the food this time.\\nBut their churros never dissapointed me. Staff are too slow and do not respond easily.\\n\\nSoups:\\nMinestrone soup was like vegetable stock water with vegetables in it, not happy with it.\\n\\nChurros :\\nIt was great as usual. A must visit place if u love churros.\\n\\nStarters:\\n1. Africano panner -\\nIt was bit sweet and not happy with the dish!!\\n2. Loaded nachos -\\nThis happened to be their one of the best dishes, but now I take back my words. Very dissapointed with the dish.\\n\\nMain course:\\nsicilia pizza\\nPizza was too good, it\'s a must try dish.\\n\\nFinally I would recommend this place for churros and pizza .\\n\\nTaste : 6/10\\nAmbience :7/10\\nValue for money : 8/10\\n\\nOverall experience : 7.5/10"), (\'Rated 4.0\', \'RATED\\n Visited this place long before, place come to.notice with the decoration lights outside the cafe. We ordered churos, pizza and hot chocolate, all were good just I little priced more.\'), (\'Rated 4.0\', \'RATED\\n Spain pictures of Churros triggered Nostalgia about it , which led to Google search for Churro places in city , lucky we to get the result as San Churro cafe , we had an delectable churros here along with hot chocolate and Nachos ! Thanks for fulfilling our thirst of Spanish Churros !\\nFood : ???\\nChurros ???\\nHot Chocolate ?\\nNachos ??\\n\\nService : ??\\n\\nAmbiance : ?\\n\\nCleanliness : ??\\n\\nOne drawback : Store had some flies, I hope they take care of them for our next tour ?\'), (\'Rated 3.0\', \'RATED\\n Ã\x83\\x83Ã\x82\\x83Ã\x83\\x82Ã\x82\\x82Ã\x83\\x83Ã\x82\\x82Ã\x83\\x82Ã\x82\\x93Chocolaty and churroic experienceÃ\x83\\x83Ã\x82\\x83Ã\x83\\x82Ã\x82\\x82Ã\x83\\x83Ã\x82\\x82Ã\x83\\x82Ã\x82\\x94\\n\\nHit chocolate and churro with chocolate dip in the picture.\\n\\nMy first time at this restaurant and it has delivered in service, options as well as taste.\'), (\'Rated 4.0\', \'RATED\\n Though very sadly the place looks in need of retouching, but the food was great! Especially when it was super cold outside and we were super hungry, the staff was nice. We ordered the pink pasta and a pizza. Both of them were pretty good. Was really tempted to try their Churros but we were stuffed. Worth a visit and try for sure!\'), (\'Rated 3.0\', "RATED\\n Churros, the restaurant\'s eponymous offering, has to be tried. They\'re a Mexican snack and served with chocolate sauce. Milkshakes are good too. The interior walls have murals and writings all over. Haven\'t tried the pizzas here but the cakes and pastries were pretty good."), (\'Rated 3.0\', "RATED\\n Honestly, have only tried parcels and that too only starters and mains - never tried desserts. So this review is based on starters and mains only. The quality wasn\'t all that great, was pretty average."), (\'Rated 4.0\', "RATED\\n Visited this place for the first time, recently. The ambience was pretty good. We ordered two \'churros for two\'. Having churros for the first time ever, I was way too excited and judgmental. But swear to Lord, the churros were heavenly! The chocolate sauce was the actual chocolate sauce and not nutella xD We had taken the Cross Country Ranch pizza. It was a thin crust pizza loaded with veggies.\\nThe place is famous for churros (the name suggests so). I would love to visit again to try all the other items out :\')"), (\'Rated 4.0\', \'RATED\\n I love the desserts more than anything here !Good place and well served !:)\\nWe dint like the nachos here , it was not that good as we expected !\\nExcept nachos almost all dish were quite good!\'), (\'Rated 2.0\', "RATED\\n 2nd time I have visited this place and I am surprised to see how it has declined. Churros was hard, pizza was pathetic, loaded nachos and masala Garlic bread were topped with cheese with MAYO! Yes mayonnese.. Soup has the bread crumbs in almost melted state, mocktails were average and our only Savior was pasta and caramel pudding... This place has high potential but let down by dull ambience, pretty slow and inattentive staff. Buffet price was listed as 399 but was charged 499, We still went ahead and took the deal to be very disappointed!! Most dishes were freshly cooked hence going with some starts else would rate them lower. I did not find a single person who seemed happy with their meal as moat complained about one thing or the other. That\'ll be my last visit San Churros.")]', '3': '[(\'Rated 4.0\', "RATED\\n Great food and proper Karnataka style full meals. Been there twice and was fully satisfied.. Will give 5 stars if it\'s well managed............"), (\'Rated 2.0\', \'RATED\\n Reached the place at 3pm on Saturday. Half of the items on the menu were over. What was annoying was is the food was cold. The taste was also very average. Only dosa and holige were good. There were very few people in the restaurant and the service was still very slow. The waiters were all standing in one corner and talking. Had to call them repeatedly.\'), (\'Rated 4.0\', \'RATED\\n Had been here, good food served and tasty,good place to go with freinds and family, should be first to get served well with all food. One of the good hotel there for this price.\'), (\'Rated 2.0\', \'RATED\\n How can a Udupi restaurant be so dirty.the floor the walls the waiters are all stained in food.Why cant they clean it? The floor even had decorative colour paper every where and lot of food.Now coming to the taste of the food- it was pretty decent for what they chargw.What upset us was the menu says they serve thambuli, two variety of rice etc but they were all over at 2.45 but they still charged us full amount.Lastly if u r ok to b seated with random people and adjust with their table manners( burrping etc) then tis place is decent for a quick veg udupi meals\'), (\'Rated 4.0\', \'RATED\\n Aadhuri Udupi bhojana is one of the best vegetarian hotel in and around Banashankari locality\\nwere we can enjoy the authentic south Karnataka style food.\'), (\'Rated 5.0\', \'RATED\\n One of the best restaurants for unlimited food. Price for one full unlimited meal is ?150 and the service is awesome here. If youÃ\x83\\x83Ã\x82\\x83Ã\x83\\x82Ã\x82\\x82Ã\x83\\x83Ã\x82\\x82Ã\x83\\x82Ã\x82\\x92re a costalian you will love the food served here.\'), (\'Rated 4.0\', "RATED\\n Been here a couple of times. Highly satisfying on both the ocassions. Great food. Decent service. At price of 160 per person it is of a great value. The staff here serves in a traditional Mysore style dresses. The food is authentic Karnataka meals. I really liked the pineapple pickle/chutney. They serve some 4 varities of rice of which you shouldn\'t try all as you would already be full. Roti, Dosa were decent. In the deserts, Payasam& Holige were good.\\nI think it is one the must try places to try some authentic south food even if you have been to the likes of Kamath etc. this one is a little different. :)"), (\'Rated 5.0\', \'RATED\\n Very good restorent good south Indian food very nice all food s are good we spent good time and food in this hotel value of money we loved it\'), (\'Rated 3.5\', \'RATED\\n I went to this restaurant on Sunday afternoon.It was a bit crowded.\\n\\nIts located in the 1st floor Above anna kuteera kathriguppe\\n\\nYou need to take food coupon in the ground floor itself.\\nit was 160 rs.\\n\\nTaste wise it was good.\\nService was slow.\\nNothing exciting/different in the menu.\\nWhatever they serve its good.\\nWorth for money.\'), (\'Rated 5.0\', \'RATED\\n Awesome place , worth with price , quality , quantity , service , hygiene and discipline I loved it ???? wish there were more rating stars to give I give 100 on 5 for sure\'), (\'Rated 3.5\', \'RATED\\n Nice food, good ambiance, so many verity of food, this time i was in hurry so, next review will be detailed one. Its good to see all dishes at one place\'), (\'Rated 1.5\', \'RATED\\n The food was not satisfactory. Not one item served could be eaten again . Only advantage is the price but even that is not going to be enough to make me visit the place again . Dear Udupi staff people wouldnÃ\x83\\x83Ã\x82\\x83Ã\x83\\x82Ã\x82\\x82Ã\x83\\x83Ã\x82\\x82Ã\x83\\x82Ã\x82\\x92t mind if the food quality was better for a higher price and please get more fans so that people wonÃ\x83\\x83Ã\x82\\x83Ã\x83\\x82Ã\x82\\x82Ã\x83\\x83Ã\x82\\x82Ã\x83\\x82Ã\x82\\x92t have to deal with another disappointment of sweating while eating.\'), (\'Rated 5.0\', \'RATED\\n Excellent quality and taste, more variety of authentic south Indian food. Excellent service. Highly recommended. Worth the money we pay here.\'), (\'Rated 2.0\', "RATED\\n I\'m rating 2 only because of the large number of items they serve for the price they charge. While the taste is moderate, the biggest turn off is the poor service and totally unresponsive staff. They are either understaffed or poorly trained. I didn\'t get to taste a few items, thanks to their ever busy staff, and ended eating what was served. Please learn the crowd management from other peers like maiyya and roti ghar"), (\'Rated 5.0\', \'RATED\\n One of my colleagues suggested this place. Went there at 1 pm and place wasnt crowded. Ambience is decent and cool. A welcome drink "appekai saru" was served. Then started..typical "baale yele uta" . Diffrerent types of palya, kosambari and sides. Then came masala dosa, pulka,pulav, veg rice bath. Masala dosa was very very tasty. Service is bit fast as people will be coming, but they will make it slow if u tell them to. Then comes rice with sambar,rasam, daal,tambali. And yes, dont forget to eat holige,payasam and icecream. Service is very nice. One best thing is all of this comes in just 160 rs and its unlimited!!! What else you want!! A must visit place for south indian foodie!\'), (\'Rated 3.0\', "RATED\\n Been here last week with high expectations based on few reviews. But utterly disappointed with the food quality & taste. I feel it\'s an overhyped place! There are many better places in the vicinity for Plantain leaf meal. There was too much of waiting & these staffs were too hurry in serving food & asking people to finish meal & leave quickly. I can understand there is a waiting and staffs should approach in a positive way. I will never go there again!\\n\\nbangaloreepicure.com"), (\'Rated 4.0\', \'RATED\\n I have a confession, being a south Indian I distaste south Indian food(not that I hate it, I don\\\'t like it either).But still made up my mind on Sankaranti to have authentic south Indian food visited "Adduri Udupi Bhojana" because it was close to my place.I just had a look at the menu told myself that I can do it(I am not blessed with a tremendous appetite).welcomed with the mango soup followed by the wide variety of dishes served, I have developed the new love for Pongal(both sweet and Khara).I can still feel the taste of the ghee bursting in my mouth but it was"kai holige" that made my day, crisp and soft.not to forget food is served on a banana leaf that adds in more flavor and taste.waiters are attentive and responsive.The customer\\\'s needs are monitored and addressed.so much variety for just 160Rs without compromising on quantity and quality and yes it is unlimited.This visit turned out to be a surprise package.\'), (\'Rated 3.5\', \'RATED\\n Food is good.\\nNo parking\\nAmbience is average\\nOverall its value for money if you looking for south Indian food.\\nPreferred the tiffin items......\'), (\'Rated 3.0\', "RATED\\n Been here last week with high expectations based on few reviews. But utterly disappointed with the food quality & taste. I feel it\'s an overhyped place! There are many better places in the vicinity for Plantain leaf meal. There was too much of waiting & these staffs were too hurry in serving food & asking people to finish meal & leave quickly. I can understand there is a waiting and staffs should approach in a positive way. I will never go there again!\\n\\nbangaloreepicure.com"), (\'Rated 4.0\', \'RATED\\n I have a confession, being a south Indian I distaste south Indian food(not that I hate it, I don\\\'t like it either).But still made up my mind on Sankaranti to have authentic south Indian food visited "Adduri Udupi Bhojana" because it was close to my place.I just had a look at the menu told myself that I can do it(I am not blessed with a tremendous appetite).welcomed with the mango soup followed by the wide variety of dishes served, I have developed the new love for Pongal(both sweet and Khara).I can still feel the taste of the ghee bursting in my mouth but it was"kai holige" that made my day, crisp and soft.not to forget food is served on a banana leaf that adds in more flavor and taste.waiters are attentive and responsive.The customer\\\'s needs are monitored and addressed.so much variety for just 160Rs without compromising on quantity and quality and yes it is unlimited.This visit turned out to be a surprise package.\'), (\'Rated 3.5\', \'RATED\\n Food is good.\\nNo parking\\nAmbience is average\\nOverall its value for money if you looking for south Indian food.\\nPreferred the tiffin items......\'), (\'Rated 3.0\', "RATED\\n Been here last week with high expectations based on few reviews. But utterly disappointed with the food quality & taste. I feel it\'s an overhyped place! There are many better places in the vicinity for Plantain leaf meal. There was too much of waiting & these staffs were too hurry in serving food & asking people to finish meal & leave quickly. I can understand there is a waiting and staffs should approach in a positive way. I will never go there again!\\n\\nbangaloreepicure.com"), (\'Rated 4.0\', \'RATED\\n I have a confession, being a south Indian I distaste south Indian food(not that I hate it, I don\\\'t like it either).But still made up my mind on Sankaranti to have authentic south Indian food visited "Adduri Udupi Bhojana" because it was close to my place.I just had a look at the menu told myself that I can do it(I am not blessed with a tremendous appetite).welcomed with the mango soup followed by the wide variety of dishes served, I have developed the new love for Pongal(both sweet and Khara).I can still feel the taste of the ghee bursting in my mouth but it was"kai holige" that made my day, crisp and soft.not to forget food is served on a banana leaf that adds in more flavor and taste.waiters are attentive and responsive.The customer\\\'s needs are monitored and addressed.so much variety for just 160Rs without compromising on quantity and quality and yes it is unlimited.This visit turned out to be a surprise package.\'), (\'Rated 3.5\', \'RATED\\n Food is good.\\nNo parking\\nAmbience is average\\nOverall its value for money if you looking for south Indian food.\\nPreferred the tiffin items......\'), (\'Rated 3.0\', "RATED\\n Been here last week with high expectations based on few reviews. But utterly disappointed with the food quality & taste. I feel it\'s an overhyped place! There are many better places in the vicinity for Plantain leaf meal. There was too much of waiting & these staffs were too hurry in serving food & asking people to finish meal & leave quickly. I can understand there is a waiting and staffs should approach in a positive way. I will never go there again!\\n\\nbangaloreepicure.com"), (\'Rated 4.0\', \'RATED\\n I have a confession, being a south Indian I distaste south Indian food(not that I hate it, I don\\\'t like it either).But still made up my mind on Sankaranti to have authentic south Indian food visited "Adduri Udupi Bhojana" because it was close to my place.I just had a look at the menu told myself that I can do it(I am not blessed with a tremendous appetite).welcomed with the mango soup followed by the wide variety of dishes served, I have developed the new love for Pongal(both sweet and Khara).I can still feel the taste of the ghee bursting in my mouth but it was"kai holige" that made my day, crisp and soft.not to forget food is served on a banana leaf that adds in more flavor and taste.waiters are attentive and responsive.The customer\\\'s needs are monitored and addressed.so much variety for just 160Rs without compromising on quantity and quality and yes it is unlimited.This visit turned out to be a surprise package.\'), (\'Rated 3.5\', \'RATED\\n Food is good.\\nNo parking\\nAmbience is average\\nOverall its value for money if you looking for south Indian food.\\nPreferred the tiffin items......\'), (\'Rated 3.0\', "RATED\\n Been here last week with high expectations based on few reviews. But utterly disappointed with the food quality & taste. I feel it\'s an overhyped place! There are many better places in the vicinity for Plantain leaf meal. There was too much of waiting & these staffs were too hurry in serving food & asking people to finish meal & leave quickly. I can understand there is a waiting and staffs should approach in a positive way. I will never go there again!\\n\\nbangaloreepicure.com"), (\'Rated 4.0\', \'RATED\\n I have a confession, being a south Indian I distaste south Indian food(not that I hate it, I don\\\'t like it either).But still made up my mind on Sankaranti to have authentic south Indian food visited "Adduri Udupi Bhojana" because it was close to my place.I just had a look at the menu told myself that I can do it(I am not blessed with a tremendous appetite).welcomed with the mango soup followed by the wide variety of dishes served, I have developed the new love for Pongal(both sweet and Khara).I can still feel the taste of the ghee bursting in my mouth but it was"kai holige" that made my day, crisp and soft.not to forget food is served on a banana leaf that adds in more flavor and taste.waiters are attentive and responsive.The customer\\\'s needs are monitored and addressed.so much variety for just 160Rs without compromising on quantity and quality and yes it is unlimited.This visit turned out to be a surprise package.\'), (\'Rated 3.5\', \'RATED\\n Food is good.\\nNo parking\\nAmbience is average\\nOverall its value for money if you looking for south Indian food.\\nPreferred the tiffin items......\'), (\'Rated 4.0\', "RATED\\n Had been on a week night for an authentic south Indian food. I personally love being served on banyan leaf. The waiters are all dressed up in traditional Mysore attire wearing with a peta. The food was delicious and definitely worth it\'s price. The waiters are friendly and serve u well with a smile. The food is prepared without garlic which is a plus point attracting many senior customers."), (\'Rated 3.5\', "RATED\\n Food is good and enjoyed. With 150 Rs. is very nice.\\nNote: (It happen with us)\\n - If you going TWO people, you may need to share the table there. Its almost crowed.\\n - You have to sit where they ask, you can\'t choose even if all the table are empty. \\nJust wait, eat and come back."), (\'Rated 5.0\', \'RATED\\n Excellent Ambience with delicious meals with 10-15 variety types. Worth paying for money and you can dine out if you are with more friends/relatives in home.\'), (\'Rated 4.5\', \'RATED\\n Very good and Unlimited . especially masala dosa . Service and ambience was also good. will visit again with friends . very very reasonble and wholesome food\'), (\'Rated 3.5\', \'RATED\\n Aaaa wt to tell, Unlimited food, food was like exactly like south indian marriage food, 2 soups, sides masala dosa, all were unlimited for just ?150, except ice cream in the end ?? they give a water bottle, and main thing is have patience to get place to sit..... ??,there is no parking place of this outlet, and difficult to park in front of the restaurants,\')]'}, 'menu_item': {'0': '[]', '1': '[]', '2': '[]', '3': '[]'}, 'listed_in(type)': {'0': 'Buffet', '1': 'Buffet', '2': 'Buffet', '3': 'Buffet'}, 'listed_in(city)': {'0': 'Banashankari', '1': 'Banashankari', '2': 'Banashankari', '3': 'Banashankari'}}
<end_description>
| 2,533 | 1 | 5,895 | 2,533 |
69401550
|
<jupyter_start><jupyter_text>Anime Recommendation Database 2020
# MyAnimeList Database 2020
> Recommendation data from 320.0000 users and 16.000 animes at myanimelist.net
This dataset contains information about 17.562 anime and the preference from 325.772 different users. In particular, this dataset contain:
- The anime list per user. Include dropped, complete, plan to watch, currently watching and on hold.
- Ratings given by users to the animes that they has watched completely.
- Information about the anime like genre, stats, studio, etc.
- HTML with anime information to do data scrapping. These files contain information such as reviews, synopsis, information about the staff, anime statistics, genre, etc.
Also, the code used to collect the data is available at github: https://github.com/Hernan4444/MyAnimeList-Database.
### Warning: this dataset includes information about anime for adults (hentai).
## Content
**The data was scrapped between February 26th and March 20th.**
* The "html" folder contain 1 zip per anime (17.562 different anime). Each zip contains different HTML pages scrapped from [MyAnimeList](https://myanimelist.net/). The scrapped pages are:
1. Main page
2. Reviews
3. Recommendations
4. Stats
5. Characters & Staff
I uploaded 2 files as example to don't increase the size of this dataset. All HTML files are in this link: https://drive.google.com/drive/folders/12ghJk-sWyXXORoLBUpPirK4YdtIaZPV_?usp=sharing
* `animelist.csv` have the list of all animes register by the user with the respective score, watching status and numbers of episodes watched. This dataset contains 109 Million row, 17.562 different animes and 325.772 different users. The file have the following columns:
1. user_id: non identifiable randomly generated user id.
2. anime_id: MyAnemlist ID of the anime. (e.g. 1).
3. score: score between 1 to 10 given by the user. 0 if the user didn't assign a score. (e.g. 10)
4. watching_status: state ID from this anime in the anime list of this user. (e.g. 2)
5. watched_episodes: numbers of episodes watched by the user. (e.g. 24)
* `watching_status.csv` describe every possible status of the column: "watching_status" in `animelist.csv`.
* `rating_complete.csv` is a subset of `animelist.csv`. This dataset only considers animes that the user has watched completely (`watching_status==2`) and gave it a score (`score!=0`). This dataset contains 57 Million ratings applied to 16.872 animes by 310.059 users. This file have the following columns:
1. user_id: non identifiable randomly generated user id.
2. anime_id: - MyAnimelist ID of the anime that this user has rated.
3. rating: rating that this user has assigned.
* `anime.csv` contain general information of every anime (17.562 different anime) like genre, stats, studio, etc. This file have the following columns:
1. MAL_ID: MyAnimelist ID of the anime. (e.g. 1)
2. Name: full name of the anime. (e.g. Cowboy Bebop)
3. Score: average score of the anime given from all users in MyAnimelist database. (e.g. 8.78)
4. Genres: comma separated list of genres for this anime. (e.g. Action, Adventure, Comedy, Drama, Sci-Fi, Space)
5. English name: full name in english of the anime. (e.g. Cowboy Bebop)
6. Japanese name: full name in japanses of the anime. (e.g. カウボーイビバップ)
7. Type: TV, movie, OVA, etc. (e.g. TV)
8. Episodes': number of chapters. (e.g. 26)
9. Aired: broadcast date. (e.g. Apr 3, 1998 to Apr 24, 1999)
10. Premiered: season premiere. (e.g. Spring 1998)
11. Producers: comma separated list of produducers (e.g. Bandai Visual)
12. Licensors: comma separated list of licensors (e.g. Funimation, Bandai Entertainment)
13. Studios: comma separated list of studios (e.g. Sunrise)
14. Source: Manga, Light novel, Book, etc. (e.g Original)
15. Duration: duration of the anime per episode (e.g 24 min. per ep.)
16. Rating: age rate (e.g. R - 17+ (violence & profanity))
17. Ranked: position based in the score. (e.g 28)
18. Popularity: position based in the the number of users who have added the anime to their list. (e.g 39)
19. Members: number of community members that are in this anime's "group". (e.g. 1251960)
20. Favorites: number of users who have the anime as "favorites". (e.g. 61,971)
21. Watching: number of users who are watching the anime. (e.g. 105808)
22. Completed: number of users who have complete the anime. (e.g. 718161)
23. On-Hold: number of users who have the anime on Hold. (e.g. 71513)
24. Dropped: number of users who have dropped the anime. (e.g. 26678)
25. Plan to Watch': number of users who plan to watch the anime. (e.g. 329800)
26. Score-10': number of users who scored 10. (e.g. 229170)
27. Score-9': number of users who scored 9. (e.g. 182126)
28. Score-8': number of users who scored 8. (e.g. 131625)
29. Score-7': number of users who scored 7. (e.g. 62330)
30. Score-6': number of users who scored 6. (e.g. 20688)
31. Score-5': number of users who scored 5. (e.g. 8904)
32. Score-4': number of users who scored 4. (e.g. 3184)
33. Score-3': number of users who scored 3. (e.g. 1357)
34. Score-2': number of users who scored 2. (e.g. 741)
35. Score-1': number of users who scored 1. (e.g. 1580)
## Acknowledgements
Thanks to:
1. [MyAnimeList](https://myanimelist.net/) for providing anime data.
2. [Jikan API](https://jikan.docs.apiary.io/) for provide users preference.
3. Pontificia Universidad Católica de Chile for provide servers to run the code.
## Inspiration
1. Have an HTML files to experience the scraping exercise without the delay of each requests.
2. Experiment with different types of recommended. For instance, collaborative filtering or based on context like stats, genre, seiyus, reviews, synopsis, etc.
3. Use this information to build a better anime recommended system.
4. Identifying which feature allows us to build the best anime recommended system.
## Ideas to the future
1. Build the same dataset with manga and novel.
Kaggle dataset identifier: anime-recommendation-database-2020
<jupyter_code>import pandas as pd
df = pd.read_csv('anime-recommendation-database-2020/anime.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 17562 entries, 0 to 17561
Data columns (total 35 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 MAL_ID 17562 non-null int64
1 Name 17562 non-null object
2 Score 17562 non-null object
3 Genres 17562 non-null object
4 English name 17562 non-null object
5 Japanese name 17562 non-null object
6 Type 17562 non-null object
7 Episodes 17562 non-null object
8 Aired 17562 non-null object
9 Premiered 17562 non-null object
10 Producers 17562 non-null object
11 Licensors 17562 non-null object
12 Studios 17562 non-null object
13 Source 17562 non-null object
14 Duration 17562 non-null object
15 Rating 17562 non-null object
16 Ranked 17562 non-null object
17 Popularity 17562 non-null int64
18 Members 17562 non-null int64
19 Favorites 17562 non-null int64
20 Watching 17562 non-null int64
21 Completed 17562 non-null int64
22 On-Hold 17562 non-null int64
23 Dropped 17562 non-null int64
24 Plan to Watch 17562 non-null int64
25 Score-10 17562 non-null object
26 Score-9 17562 non-null object
27 Score-8 17562 non-null object
28 Score-7 17562 non-null object
29 Score-6 17562 non-null object
30 Score-5 17562 non-null object
31 Score-4 17562 non-null object
32 Score-3 17562 non-null object
33 Score-2 17562 non-null object
34 Score-1 17562 non-null object
dtypes: int64(9), object(26)
memory usage: 4.7+ MB
<jupyter_text>Examples:
{
"MAL_ID": 1,
"Name": "Cowboy Bebop",
"Score": 8.78,
"Genres": "Action, Adventure, Comedy, Drama, Sci-Fi, Space",
"English name": "Cowboy Bebop",
"Japanese name": "\u30ab\u30a6\u30dc\u30fc\u30a4\u30d3\u30d0\u30c3\u30d7",
"Type": "TV",
"Episodes": 26,
"Aired": "Apr 3, 1998 to Apr 24, 1999",
"Premiered": "Spring 1998",
"Producers": "Bandai Visual",
"Licensors": "Funimation, Bandai Entertainment",
"Studios": "Sunrise",
"Source": "Original",
"Duration": "24 min. per ep.",
"Rating": "R - 17+ (violence & profanity)",
"Ranked": 28,
"Popularity": 39,
"Members": 1251960,
"Favorites": 61971,
"...": "and 15 more columns"
}
{
"MAL_ID": 5,
"Name": "Cowboy Bebop: Tengoku no Tobira",
"Score": 8.39,
"Genres": "Action, Drama, Mystery, Sci-Fi, Space",
"English name": "Cowboy Bebop:The Movie",
"Japanese name": "\u30ab\u30a6\u30dc\u30fc\u30a4\u30d3\u30d0\u30c3\u30d7 \u5929\u56fd\u306e\u6249",
"Type": "Movie",
"Episodes": 1,
"Aired": "Sep 1, 2001",
"Premiered": "Unknown",
"Producers": "Sunrise, Bandai Visual",
"Licensors": "Sony Pictures Entertainment",
"Studios": "Bones",
"Source": "Original",
"Duration": "1 hr. 55 min.",
"Rating": "R - 17+ (violence & profanity)",
"Ranked": 159,
"Popularity": 518,
"Members": 273145,
"Favorites": 1174,
"...": "and 15 more columns"
}
{
"MAL_ID": 6,
"Name": "Trigun",
"Score": 8.24,
"Genres": "Action, Sci-Fi, Adventure, Comedy, Drama, Shounen",
"English name": "Trigun",
"Japanese name": "\u30c8\u30e9\u30a4\u30ac\u30f3",
"Type": "TV",
"Episodes": 26,
"Aired": "Apr 1, 1998 to Sep 30, 1998",
"Premiered": "Spring 1998",
"Producers": "Victor Entertainment",
"Licensors": "Funimation, Geneon Entertainment USA",
"Studios": "Madhouse",
"Source": "Manga",
"Duration": "24 min. per ep.",
"Rating": "PG-13 - Teens 13 or older",
"Ranked": 266,
"Popularity": 201,
"Members": 558913,
"Favorites": 12944,
"...": "and 15 more columns"
}
{
"MAL_ID": 7,
"Name": "Witch Hunter Robin",
"Score": 7.27,
"Genres": "Action, Mystery, Police, Supernatural, Drama, Magic",
"English name": "Witch Hunter Robin",
"Japanese name": "Witch Hunter ROBIN (\u30a6\u30a4\u30c3\u30c1\u30cf\u30f3\u30bf\u30fc\u30ed\u30d3\u30f3)",
"Type": "TV",
"Episodes": 26,
"Aired": "Jul 2, 2002 to Dec 24, 2002",
"Premiered": "Summer 2002",
"Producers": "TV Tokyo, Bandai Visual, Dentsu, Victor Entertainment",
"Licensors": "Funimation, Bandai Entertainment",
"Studios": "Sunrise",
"Source": "Original",
"Duration": "25 min. per ep.",
"Rating": "PG-13 - Teens 13 or older",
"Ranked": 2481,
"Popularity": 1467,
"Members": 94683,
"Favorites": 587,
"...": "and 15 more columns"
}
<jupyter_code>import pandas as pd
df = pd.read_csv('anime-recommendation-database-2020/rating_complete.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 57633278 entries, 0 to 57633277
Data columns (total 3 columns):
# Column Dtype
--- ------ -----
0 user_id int64
1 anime_id int64
2 rating int64
dtypes: int64(3)
memory usage: 1.3 GB
<jupyter_text>Examples:
{
"user_id": 0,
"anime_id": 430,
"rating": 9
}
{
"user_id": 0,
"anime_id": 1004,
"rating": 5
}
{
"user_id": 0,
"anime_id": 3010,
"rating": 7
}
{
"user_id": 0,
"anime_id": 570,
"rating": 7
}
<jupyter_script># # Setup
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from pandas import DataFrame
from matplotlib import pyplot as plt
from IPython import display
import collections
import sklearn
import sklearn.manifold
import tensorflow.compat.v1 as tf
tf.disable_v2_behavior()
tf.logging.set_verbosity(tf.logging.ERROR)
rating_complete = pd.read_csv(
"../input/anime-recommendation-database-2020/rating_complete.csv"
)
anime = pd.read_csv("../input/anime-recommendation-database-2020/anime.csv")
anime.head(2)
rating_complete.head(2)
user_rating = DataFrame(
rating_complete.groupby("user_id")["rating"].count().reset_index()
)
query = user_rating["rating"] > 280 # Get the user who has rated over 280 animes
filtered_users = user_rating[query]
users = set(filtered_users["user_id"])
len(users)
# Generate the new user id
user_id_dict = {}
num = 0
for user in sorted(users):
user_id_dict[user] = num
num += 1
user_id_dict[3]
rating = DataFrame(rating_complete[rating_complete["user_id"].isin(users)])
print(rating.shape)
rating.head()
# Reset the user id from 0
rating["user_id"] = rating["user_id"].map(user_id_dict)
rating.head()
def split_dataframe(df, holdout_fraction=0.1):
"""Splits a DataFrame into training and test sets.
Args:
df: a dataframe.
holdout_fraction: fraction of dataframe rows to use in the test set.
Returns:
train: dataframe for training
test: dataframe for testing
"""
test = df.sample(frac=holdout_fraction, replace=False)
train = df[~df.index.isin(test.index)]
return train, test
def build_rating_sparse_tensor(ratings_df):
"""
Args:
ratings_df: a pd.DataFrame with `user_id`, `anime_id` and `rating` columns.
Returns:
a tf.SparseTensor representing the ratings matrix.
"""
indices = ratings_df[["user_id", "anime_id"]].values
values = ratings_df["rating"].values
return tf.SparseTensor(
indices=indices, values=values, dense_shape=[len(users), anime.shape[0]]
)
def sparse_mean_square_error(sparse_ratings, user_embeddings, anime_embeddings):
"""
Args:
sparse_ratings: A SparseTensor rating matrix, of dense_shape [N, M]
user_embeddings: A dense Tensor U of shape [N, k] where k is the embedding
dimension, such that U_i is the embedding of user i.
anime_embeddings: A dense Tensor V of shape [M, k] where k is the embedding
dimension, such that V_j is the embedding of anime j.
Returns:
A scalar Tensor representing the MSE between the true ratings and the
model's predictions.
"""
predictions = tf.gather_nd(
tf.matmul(user_embeddings, anime_embeddings, transpose_b=True),
sparse_ratings.indices,
)
loss = tf.losses.mean_squared_error(sparse_ratings.values, predictions)
return loss
class CFModel(object):
"""Simple class that represents a collaborative filtering model"""
def __init__(self, embedding_vars, loss, metrics=None):
"""Initializes a CFModel.
Args:
embedding_vars: A dictionary of tf.Variables.
loss: A float Tensor. The loss to optimize.
metrics: optional list of dictionaries of Tensors. The metrics in each
dictionary will be plotted in a separate figure during training.
"""
self._embedding_vars = embedding_vars
self._loss = loss
self._metrics = metrics
self._embeddings = {k: None for k in embedding_vars}
self._session = None
@property
def embeddings(self):
"""The embeddings dictionary."""
return self._embeddings
def train(
self,
num_iterations=100,
learning_rate=1.0,
plot_results=True,
optimizer=tf.train.GradientDescentOptimizer,
):
"""Trains the model.
Args:
iterations: number of iterations to run.
learning_rate: optimizer learning rate.
plot_results: whether to plot the results at the end of training.
optimizer: the optimizer to use. Default to GradientDescentOptimizer.
Returns:
The metrics dictionary evaluated at the last iteration.
"""
with self._loss.graph.as_default():
opt = optimizer(learning_rate)
train_op = opt.minimize(self._loss)
local_init_op = tf.group(
tf.variables_initializer(opt.variables()),
tf.local_variables_initializer(),
)
if self._session is None:
self._session = tf.Session()
with self._session.as_default():
self._session.run(tf.global_variables_initializer())
self._session.run(tf.tables_initializer())
tf.train.start_queue_runners()
with self._session.as_default():
local_init_op.run()
iterations = []
metrics = self._metrics or ({},)
metrics_vals = [collections.defaultdict(list) for _ in self._metrics]
# Train and append results.
for i in range(num_iterations + 1):
_, results = self._session.run((train_op, metrics))
if (i % 10 == 0) or i == num_iterations:
print(
"\r iteration %d: " % i
+ ", ".join(
["%s=%f" % (k, v) for r in results for k, v in r.items()]
),
end="",
)
iterations.append(i)
for metric_val, result in zip(metrics_vals, results):
for k, v in result.items():
metric_val[k].append(v)
for k, v in self._embedding_vars.items():
self._embeddings[k] = v.eval()
if plot_results:
# Plot the metrics.
num_subplots = len(metrics) + 1
fig = plt.figure()
fig.set_size_inches(num_subplots * 10, 8)
for i, metric_vals in enumerate(metrics_vals):
ax = fig.add_subplot(1, num_subplots, i + 1)
for k, v in metric_vals.items():
ax.plot(iterations, v, label=k)
ax.set_xlim([1, num_iterations])
ax.legend()
return results
def build_model(ratings, embedding_dim=3, init_stddev=1.0):
"""
Args:
ratings: a DataFrame of the ratings
embedding_dim: the dimension of the embedding vectors.
init_stddev: float, the standard deviation of the random initial embeddings.
Returns:
model: a CFModel.
"""
# Split the ratings DataFrame into train and test.
train_ratings, test_ratings = split_dataframe(ratings)
# SparseTensor representation of the train and test datasets.
A_train = build_rating_sparse_tensor(train_ratings)
A_test = build_rating_sparse_tensor(test_ratings)
# Initialize the embeddings using a normal distribution.
U = tf.Variable(
tf.random_normal([A_train.dense_shape[0], embedding_dim], stddev=init_stddev)
)
V = tf.Variable(
tf.random_normal([A_train.dense_shape[1], embedding_dim], stddev=init_stddev)
)
train_loss = sparse_mean_square_error(A_train, U, V)
test_loss = sparse_mean_square_error(A_test, U, V)
metrics = {"train_error": train_loss, "test_error": test_loss}
embeddings = {"user_id": U, "anime_id": V}
return CFModel(embeddings, train_loss, [metrics])
model = build_model(rating, embedding_dim=30, init_stddev=0.5)
model.train(num_iterations=1000, learning_rate=20.0)
DOT = "dot"
COSINE = "cosine"
def compute_scores(query_embedding, item_embeddings, measure=DOT):
"""Computes the scores of the candidates given a query.
Args:
query_embedding: a vector of shape [k], representing the query embedding.
item_embeddings: a matrix of shape [N, k], such that row i is the embedding of item i.
measure: a string specifying the similarity measure to be used. Can be either DOT or COSINE.
Returns:
scores: a vector of shape [N], such that scores[i] is the score of item i.
"""
u = query_embedding
V = item_embeddings
if measure == COSINE:
V = V / np.linalg.norm(V, axis=1, keepdims=True)
u = u / np.linalg.norm(u)
scores = u.dot(V.T)
return scores
def user_recommendations(user_id, model, measure=DOT, exclude_rated=False, k=6):
if True:
scores = compute_scores(
model.embeddings["user_id"][user_id], model.embeddings["anime_id"], measure
)
score_key = measure + " score"
df = pd.DataFrame(
{
score_key: list(scores),
"anime_id": anime["MAL_ID"],
"titles": anime["Japanese name"],
}
)
display.display(df.sort_values([score_key], ascending=False).head(k))
def similiar_user(user_id, model, measure=DOT, exclude_rated=False, k=6):
if True:
scores = compute_scores(
model.embeddings["user_id"][user_id], model.embeddings["user_id"], measure
)
score_key = measure + " score"
df = pd.DataFrame({score_key: list(scores)})
display.display(df.sort_values([score_key], ascending=False).head(k))
user_recommendations(5, model, measure=COSINE, k=10)
similiar_user(5, model, measure=COSINE, k=10)
pd.merge(
rating[rating["user_id"] == 5],
rating[rating["user_id"] == 41332],
on="anime_id",
how="inner",
).head(50)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/401/69401550.ipynb
|
anime-recommendation-database-2020
|
hernan4444
|
[{"Id": 69401550, "ScriptId": 18923789, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 986951, "CreationDate": "07/30/2021 14:30:28", "VersionNumber": 2.0, "Title": "MyAnimeList - CF and MF", "EvaluationDate": "07/30/2021", "IsChange": true, "TotalLines": 248.0, "LinesInsertedFromPrevious": 28.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 220.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92520382, "KernelVersionId": 69401550, "SourceDatasetVersionId": 2422513}]
|
[{"Id": 2422513, "DatasetId": 1225408, "DatasourceVersionId": 2464702, "CreatorUserId": 2236152, "LicenseName": "CC0: Public Domain", "CreationDate": "07/13/2021 19:26:08", "VersionNumber": 7.0, "Title": "Anime Recommendation Database 2020", "Slug": "anime-recommendation-database-2020", "Subtitle": "Recommendation data from 320.0000 users and 16.000 animes at myanimelist.net", "Description": "# MyAnimeList Database 2020\n\n> Recommendation data from 320.0000 users and 16.000 animes at myanimelist.net\n\n\nThis dataset contains information about 17.562 anime and the preference from 325.772 different users. In particular, this dataset contain:\n\n- The anime list per user. Include dropped, complete, plan to watch, currently watching and on hold.\n- Ratings given by users to the animes that they has watched completely.\n- Information about the anime like genre, stats, studio, etc.\n- HTML with anime information to do data scrapping. These files contain information such as reviews, synopsis, information about the staff, anime statistics, genre, etc.\n\nAlso, the code used to collect the data is available at github: https://github.com/Hernan4444/MyAnimeList-Database. \n\n### Warning: this dataset includes information about anime for adults (hentai). \n\n\n## Content\n\n**The data was scrapped between February 26th and March 20th.**\n\n* The \"html\" folder contain 1 zip per anime (17.562 different anime). Each zip contains different HTML pages scrapped from [MyAnimeList](https://myanimelist.net/). The scrapped pages are:\n\n1. Main page\n2. Reviews\n3. Recommendations\n4. Stats\n5. Characters & Staff\n\nI uploaded 2 files as example to don't increase the size of this dataset. All HTML files are in this link: https://drive.google.com/drive/folders/12ghJk-sWyXXORoLBUpPirK4YdtIaZPV_?usp=sharing\n\n\n* `animelist.csv` have the list of all animes register by the user with the respective score, watching status and numbers of episodes watched. This dataset contains 109 Million row, 17.562 different animes and 325.772 different users. The file have the following columns:\n\n1. user_id: non identifiable randomly generated user id.\n2. anime_id: MyAnemlist ID of the anime. (e.g. 1).\n3. score: score between 1 to 10 given by the user. 0 if the user didn't assign a score. (e.g. 10)\n4. watching_status: state ID from this anime in the anime list of this user. (e.g. 2)\n5. watched_episodes: numbers of episodes watched by the user. (e.g. 24)\n\n\n* `watching_status.csv` describe every possible status of the column: \"watching_status\" in `animelist.csv`.\n\n\n* `rating_complete.csv` is a subset of `animelist.csv`. This dataset only considers animes that the user has watched completely (`watching_status==2`) and gave it a score (`score!=0`). This dataset contains 57 Million ratings applied to 16.872 animes by 310.059 users. This file have the following columns:\n\n1. user_id: non identifiable randomly generated user id.\n2. anime_id: - MyAnimelist ID of the anime that this user has rated.\n3. rating: rating that this user has assigned.\n\n\n* `anime.csv` contain general information of every anime (17.562 different anime) like genre, stats, studio, etc. This file have the following columns:\n\n1. MAL_ID: MyAnimelist ID of the anime. (e.g. 1) \n2. Name: full name of the anime. (e.g. Cowboy Bebop) \n3. Score: average score of the anime given from all users in MyAnimelist database. (e.g. 8.78) \n4. Genres: comma separated list of genres for this anime. (e.g. Action, Adventure, Comedy, Drama, Sci-Fi, Space) \n5. English name: full name in english of the anime. (e.g. Cowboy Bebop) \n6. Japanese name: full name in japanses of the anime. (e.g. \u30ab\u30a6\u30dc\u30fc\u30a4\u30d3\u30d0\u30c3\u30d7) \n7. Type: TV, movie, OVA, etc. (e.g. TV)\n8. Episodes': number of chapters. (e.g. 26)\n9. Aired: broadcast date. (e.g. Apr 3, 1998 to Apr 24, 1999)\n10. Premiered: season premiere. (e.g. Spring 1998)\n11. Producers: comma separated list of produducers (e.g. Bandai Visual)\n12. Licensors: comma separated list of licensors (e.g. Funimation, Bandai Entertainment)\n13. Studios: comma separated list of studios (e.g. Sunrise)\n14. Source: Manga, Light novel, Book, etc. (e.g Original)\n15. Duration: duration of the anime per episode (e.g 24 min. per ep.)\n16. Rating: age rate (e.g. R - 17+ (violence & profanity))\n17. Ranked: position based in the score. (e.g 28)\n18. Popularity: position based in the the number of users who have added the anime to their list. (e.g 39)\n19. Members: number of community members that are in this anime's \"group\". (e.g. 1251960)\n20. Favorites: number of users who have the anime as \"favorites\". (e.g. 61,971)\n21. Watching: number of users who are watching the anime. (e.g. 105808)\n22. Completed: number of users who have complete the anime. (e.g. 718161)\n23. On-Hold: number of users who have the anime on Hold. (e.g. 71513)\n24. Dropped: number of users who have dropped the anime. (e.g. 26678)\n25. Plan to Watch': number of users who plan to watch the anime. (e.g. 329800)\n26. Score-10': number of users who scored 10. (e.g. 229170)\n27. Score-9': number of users who scored 9. (e.g. 182126)\n28. Score-8': number of users who scored 8. (e.g. 131625)\n29. Score-7': number of users who scored 7. (e.g. 62330)\n30. Score-6': number of users who scored 6. (e.g. 20688)\n31. Score-5': number of users who scored 5. (e.g. 8904)\n32. Score-4': number of users who scored 4. (e.g. 3184)\n33. Score-3': number of users who scored 3. (e.g. 1357)\n34. Score-2': number of users who scored 2. (e.g. 741)\n35. Score-1': number of users who scored 1. (e.g. 1580)\n\n\n## Acknowledgements\n\nThanks to:\n1. [MyAnimeList](https://myanimelist.net/) for providing anime data.\n2. [Jikan API](https://jikan.docs.apiary.io/) for provide users preference.\n3. Pontificia Universidad Cat\u00f3lica de Chile for provide servers to run the code.\n\n\n## Inspiration\n\n1. Have an HTML files to experience the scraping exercise without the delay of each requests.\n\n2. Experiment with different types of recommended. For instance, collaborative filtering or based on context like stats, genre, seiyus, reviews, synopsis, etc.\n\n3. Use this information to build a better anime recommended system.\n\n4. Identifying which feature allows us to build the best anime recommended system.\n\n\n## Ideas to the future\n\n1. Build the same dataset with manga and novel.", "VersionNotes": "fix typo Genders to Genres", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1225408, "CreatorUserId": 2236152, "OwnerUserId": 2236152.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2422513.0, "CurrentDatasourceVersionId": 2464702.0, "ForumId": 1243541, "Type": 2, "CreationDate": "03/21/2021 22:50:15", "LastActivityDate": "03/21/2021", "TotalViews": 106379, "TotalDownloads": 13964, "TotalVotes": 426, "TotalKernels": 71}]
|
[{"Id": 2236152, "UserName": "hernan4444", "DisplayName": "Hernan Valdivieso", "RegisterDate": "09/11/2018", "PerformanceTier": 2}]
|
# # Setup
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from pandas import DataFrame
from matplotlib import pyplot as plt
from IPython import display
import collections
import sklearn
import sklearn.manifold
import tensorflow.compat.v1 as tf
tf.disable_v2_behavior()
tf.logging.set_verbosity(tf.logging.ERROR)
rating_complete = pd.read_csv(
"../input/anime-recommendation-database-2020/rating_complete.csv"
)
anime = pd.read_csv("../input/anime-recommendation-database-2020/anime.csv")
anime.head(2)
rating_complete.head(2)
user_rating = DataFrame(
rating_complete.groupby("user_id")["rating"].count().reset_index()
)
query = user_rating["rating"] > 280 # Get the user who has rated over 280 animes
filtered_users = user_rating[query]
users = set(filtered_users["user_id"])
len(users)
# Generate the new user id
user_id_dict = {}
num = 0
for user in sorted(users):
user_id_dict[user] = num
num += 1
user_id_dict[3]
rating = DataFrame(rating_complete[rating_complete["user_id"].isin(users)])
print(rating.shape)
rating.head()
# Reset the user id from 0
rating["user_id"] = rating["user_id"].map(user_id_dict)
rating.head()
def split_dataframe(df, holdout_fraction=0.1):
"""Splits a DataFrame into training and test sets.
Args:
df: a dataframe.
holdout_fraction: fraction of dataframe rows to use in the test set.
Returns:
train: dataframe for training
test: dataframe for testing
"""
test = df.sample(frac=holdout_fraction, replace=False)
train = df[~df.index.isin(test.index)]
return train, test
def build_rating_sparse_tensor(ratings_df):
"""
Args:
ratings_df: a pd.DataFrame with `user_id`, `anime_id` and `rating` columns.
Returns:
a tf.SparseTensor representing the ratings matrix.
"""
indices = ratings_df[["user_id", "anime_id"]].values
values = ratings_df["rating"].values
return tf.SparseTensor(
indices=indices, values=values, dense_shape=[len(users), anime.shape[0]]
)
def sparse_mean_square_error(sparse_ratings, user_embeddings, anime_embeddings):
"""
Args:
sparse_ratings: A SparseTensor rating matrix, of dense_shape [N, M]
user_embeddings: A dense Tensor U of shape [N, k] where k is the embedding
dimension, such that U_i is the embedding of user i.
anime_embeddings: A dense Tensor V of shape [M, k] where k is the embedding
dimension, such that V_j is the embedding of anime j.
Returns:
A scalar Tensor representing the MSE between the true ratings and the
model's predictions.
"""
predictions = tf.gather_nd(
tf.matmul(user_embeddings, anime_embeddings, transpose_b=True),
sparse_ratings.indices,
)
loss = tf.losses.mean_squared_error(sparse_ratings.values, predictions)
return loss
class CFModel(object):
"""Simple class that represents a collaborative filtering model"""
def __init__(self, embedding_vars, loss, metrics=None):
"""Initializes a CFModel.
Args:
embedding_vars: A dictionary of tf.Variables.
loss: A float Tensor. The loss to optimize.
metrics: optional list of dictionaries of Tensors. The metrics in each
dictionary will be plotted in a separate figure during training.
"""
self._embedding_vars = embedding_vars
self._loss = loss
self._metrics = metrics
self._embeddings = {k: None for k in embedding_vars}
self._session = None
@property
def embeddings(self):
"""The embeddings dictionary."""
return self._embeddings
def train(
self,
num_iterations=100,
learning_rate=1.0,
plot_results=True,
optimizer=tf.train.GradientDescentOptimizer,
):
"""Trains the model.
Args:
iterations: number of iterations to run.
learning_rate: optimizer learning rate.
plot_results: whether to plot the results at the end of training.
optimizer: the optimizer to use. Default to GradientDescentOptimizer.
Returns:
The metrics dictionary evaluated at the last iteration.
"""
with self._loss.graph.as_default():
opt = optimizer(learning_rate)
train_op = opt.minimize(self._loss)
local_init_op = tf.group(
tf.variables_initializer(opt.variables()),
tf.local_variables_initializer(),
)
if self._session is None:
self._session = tf.Session()
with self._session.as_default():
self._session.run(tf.global_variables_initializer())
self._session.run(tf.tables_initializer())
tf.train.start_queue_runners()
with self._session.as_default():
local_init_op.run()
iterations = []
metrics = self._metrics or ({},)
metrics_vals = [collections.defaultdict(list) for _ in self._metrics]
# Train and append results.
for i in range(num_iterations + 1):
_, results = self._session.run((train_op, metrics))
if (i % 10 == 0) or i == num_iterations:
print(
"\r iteration %d: " % i
+ ", ".join(
["%s=%f" % (k, v) for r in results for k, v in r.items()]
),
end="",
)
iterations.append(i)
for metric_val, result in zip(metrics_vals, results):
for k, v in result.items():
metric_val[k].append(v)
for k, v in self._embedding_vars.items():
self._embeddings[k] = v.eval()
if plot_results:
# Plot the metrics.
num_subplots = len(metrics) + 1
fig = plt.figure()
fig.set_size_inches(num_subplots * 10, 8)
for i, metric_vals in enumerate(metrics_vals):
ax = fig.add_subplot(1, num_subplots, i + 1)
for k, v in metric_vals.items():
ax.plot(iterations, v, label=k)
ax.set_xlim([1, num_iterations])
ax.legend()
return results
def build_model(ratings, embedding_dim=3, init_stddev=1.0):
"""
Args:
ratings: a DataFrame of the ratings
embedding_dim: the dimension of the embedding vectors.
init_stddev: float, the standard deviation of the random initial embeddings.
Returns:
model: a CFModel.
"""
# Split the ratings DataFrame into train and test.
train_ratings, test_ratings = split_dataframe(ratings)
# SparseTensor representation of the train and test datasets.
A_train = build_rating_sparse_tensor(train_ratings)
A_test = build_rating_sparse_tensor(test_ratings)
# Initialize the embeddings using a normal distribution.
U = tf.Variable(
tf.random_normal([A_train.dense_shape[0], embedding_dim], stddev=init_stddev)
)
V = tf.Variable(
tf.random_normal([A_train.dense_shape[1], embedding_dim], stddev=init_stddev)
)
train_loss = sparse_mean_square_error(A_train, U, V)
test_loss = sparse_mean_square_error(A_test, U, V)
metrics = {"train_error": train_loss, "test_error": test_loss}
embeddings = {"user_id": U, "anime_id": V}
return CFModel(embeddings, train_loss, [metrics])
model = build_model(rating, embedding_dim=30, init_stddev=0.5)
model.train(num_iterations=1000, learning_rate=20.0)
DOT = "dot"
COSINE = "cosine"
def compute_scores(query_embedding, item_embeddings, measure=DOT):
"""Computes the scores of the candidates given a query.
Args:
query_embedding: a vector of shape [k], representing the query embedding.
item_embeddings: a matrix of shape [N, k], such that row i is the embedding of item i.
measure: a string specifying the similarity measure to be used. Can be either DOT or COSINE.
Returns:
scores: a vector of shape [N], such that scores[i] is the score of item i.
"""
u = query_embedding
V = item_embeddings
if measure == COSINE:
V = V / np.linalg.norm(V, axis=1, keepdims=True)
u = u / np.linalg.norm(u)
scores = u.dot(V.T)
return scores
def user_recommendations(user_id, model, measure=DOT, exclude_rated=False, k=6):
if True:
scores = compute_scores(
model.embeddings["user_id"][user_id], model.embeddings["anime_id"], measure
)
score_key = measure + " score"
df = pd.DataFrame(
{
score_key: list(scores),
"anime_id": anime["MAL_ID"],
"titles": anime["Japanese name"],
}
)
display.display(df.sort_values([score_key], ascending=False).head(k))
def similiar_user(user_id, model, measure=DOT, exclude_rated=False, k=6):
if True:
scores = compute_scores(
model.embeddings["user_id"][user_id], model.embeddings["user_id"], measure
)
score_key = measure + " score"
df = pd.DataFrame({score_key: list(scores)})
display.display(df.sort_values([score_key], ascending=False).head(k))
user_recommendations(5, model, measure=COSINE, k=10)
similiar_user(5, model, measure=COSINE, k=10)
pd.merge(
rating[rating["user_id"] == 5],
rating[rating["user_id"] == 41332],
on="anime_id",
how="inner",
).head(50)
|
[{"anime-recommendation-database-2020/anime.csv": {"column_names": "[\"MAL_ID\", \"Name\", \"Score\", \"Genres\", \"English name\", \"Japanese name\", \"Type\", \"Episodes\", \"Aired\", \"Premiered\", \"Producers\", \"Licensors\", \"Studios\", \"Source\", \"Duration\", \"Rating\", \"Ranked\", \"Popularity\", \"Members\", \"Favorites\", \"Watching\", \"Completed\", \"On-Hold\", \"Dropped\", \"Plan to Watch\", \"Score-10\", \"Score-9\", \"Score-8\", \"Score-7\", \"Score-6\", \"Score-5\", \"Score-4\", \"Score-3\", \"Score-2\", \"Score-1\"]", "column_data_types": "{\"MAL_ID\": \"int64\", \"Name\": \"object\", \"Score\": \"object\", \"Genres\": \"object\", \"English name\": \"object\", \"Japanese name\": \"object\", \"Type\": \"object\", \"Episodes\": \"object\", \"Aired\": \"object\", \"Premiered\": \"object\", \"Producers\": \"object\", \"Licensors\": \"object\", \"Studios\": \"object\", \"Source\": \"object\", \"Duration\": \"object\", \"Rating\": \"object\", \"Ranked\": \"object\", \"Popularity\": \"int64\", \"Members\": \"int64\", \"Favorites\": \"int64\", \"Watching\": \"int64\", \"Completed\": \"int64\", \"On-Hold\": \"int64\", \"Dropped\": \"int64\", \"Plan to Watch\": \"int64\", \"Score-10\": \"object\", \"Score-9\": \"object\", \"Score-8\": \"object\", \"Score-7\": \"object\", \"Score-6\": \"object\", \"Score-5\": \"object\", \"Score-4\": \"object\", \"Score-3\": \"object\", \"Score-2\": \"object\", \"Score-1\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 17562 entries, 0 to 17561\nData columns (total 35 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 MAL_ID 17562 non-null int64 \n 1 Name 17562 non-null object\n 2 Score 17562 non-null object\n 3 Genres 17562 non-null object\n 4 English name 17562 non-null object\n 5 Japanese name 17562 non-null object\n 6 Type 17562 non-null object\n 7 Episodes 17562 non-null object\n 8 Aired 17562 non-null object\n 9 Premiered 17562 non-null object\n 10 Producers 17562 non-null object\n 11 Licensors 17562 non-null object\n 12 Studios 17562 non-null object\n 13 Source 17562 non-null object\n 14 Duration 17562 non-null object\n 15 Rating 17562 non-null object\n 16 Ranked 17562 non-null object\n 17 Popularity 17562 non-null int64 \n 18 Members 17562 non-null int64 \n 19 Favorites 17562 non-null int64 \n 20 Watching 17562 non-null int64 \n 21 Completed 17562 non-null int64 \n 22 On-Hold 17562 non-null int64 \n 23 Dropped 17562 non-null int64 \n 24 Plan to Watch 17562 non-null int64 \n 25 Score-10 17562 non-null object\n 26 Score-9 17562 non-null object\n 27 Score-8 17562 non-null object\n 28 Score-7 17562 non-null object\n 29 Score-6 17562 non-null object\n 30 Score-5 17562 non-null object\n 31 Score-4 17562 non-null object\n 32 Score-3 17562 non-null object\n 33 Score-2 17562 non-null object\n 34 Score-1 17562 non-null object\ndtypes: int64(9), object(26)\nmemory usage: 4.7+ MB\n", "summary": "{\"MAL_ID\": {\"count\": 17562.0, \"mean\": 21477.192347113087, \"std\": 14900.093169943408, \"min\": 1.0, \"25%\": 5953.5, \"50%\": 22820.0, \"75%\": 35624.75, \"max\": 48492.0}, \"Popularity\": {\"count\": 17562.0, \"mean\": 8763.45234028015, \"std\": 5059.327278012592, \"min\": 0.0, \"25%\": 4383.5, \"50%\": 8762.5, \"75%\": 13145.0, \"max\": 17565.0}, \"Members\": {\"count\": 17562.0, \"mean\": 34658.53951713928, \"std\": 125282.14142822381, \"min\": 1.0, \"25%\": 336.0, \"50%\": 2065.0, \"75%\": 13223.25, \"max\": 2589552.0}, \"Favorites\": {\"count\": 17562.0, \"mean\": 457.7462703564514, \"std\": 4063.4733134938538, \"min\": 0.0, \"25%\": 0.0, \"50%\": 3.0, \"75%\": 31.0, \"max\": 183914.0}, \"Watching\": {\"count\": 17562.0, \"mean\": 2231.487757658581, \"std\": 14046.688133245594, \"min\": 0.0, \"25%\": 13.0, \"50%\": 73.0, \"75%\": 522.0, \"max\": 887333.0}, \"Completed\": {\"count\": 17562.0, \"mean\": 22095.571347226967, \"std\": 91009.18850426309, \"min\": 0.0, \"25%\": 111.0, \"50%\": 817.5, \"75%\": 6478.0, \"max\": 2182587.0}, \"On-Hold\": {\"count\": 17562.0, \"mean\": 955.0496526591504, \"std\": 4275.675095965888, \"min\": 0.0, \"25%\": 6.0, \"50%\": 45.0, \"75%\": 291.75, \"max\": 187919.0}, \"Dropped\": {\"count\": 17562.0, \"mean\": 1176.5995330827925, \"std\": 4740.348652904278, \"min\": 0.0, \"25%\": 37.0, \"50%\": 77.0, \"75%\": 271.0, \"max\": 174710.0}, \"Plan to Watch\": {\"count\": 17562.0, \"mean\": 8199.831226511787, \"std\": 23777.691962549074, \"min\": 1.0, \"25%\": 112.0, \"50%\": 752.5, \"75%\": 4135.5, \"max\": 425531.0}}", "examples": "{\"MAL_ID\":{\"0\":1,\"1\":5,\"2\":6,\"3\":7},\"Name\":{\"0\":\"Cowboy Bebop\",\"1\":\"Cowboy Bebop: Tengoku no Tobira\",\"2\":\"Trigun\",\"3\":\"Witch Hunter Robin\"},\"Score\":{\"0\":\"8.78\",\"1\":\"8.39\",\"2\":\"8.24\",\"3\":\"7.27\"},\"Genres\":{\"0\":\"Action, Adventure, Comedy, Drama, Sci-Fi, Space\",\"1\":\"Action, Drama, Mystery, Sci-Fi, Space\",\"2\":\"Action, Sci-Fi, Adventure, Comedy, Drama, Shounen\",\"3\":\"Action, Mystery, Police, Supernatural, Drama, Magic\"},\"English name\":{\"0\":\"Cowboy Bebop\",\"1\":\"Cowboy Bebop:The Movie\",\"2\":\"Trigun\",\"3\":\"Witch Hunter Robin\"},\"Japanese name\":{\"0\":\"\\u30ab\\u30a6\\u30dc\\u30fc\\u30a4\\u30d3\\u30d0\\u30c3\\u30d7\",\"1\":\"\\u30ab\\u30a6\\u30dc\\u30fc\\u30a4\\u30d3\\u30d0\\u30c3\\u30d7 \\u5929\\u56fd\\u306e\\u6249\",\"2\":\"\\u30c8\\u30e9\\u30a4\\u30ac\\u30f3\",\"3\":\"Witch Hunter ROBIN (\\u30a6\\u30a4\\u30c3\\u30c1\\u30cf\\u30f3\\u30bf\\u30fc\\u30ed\\u30d3\\u30f3)\"},\"Type\":{\"0\":\"TV\",\"1\":\"Movie\",\"2\":\"TV\",\"3\":\"TV\"},\"Episodes\":{\"0\":\"26\",\"1\":\"1\",\"2\":\"26\",\"3\":\"26\"},\"Aired\":{\"0\":\"Apr 3, 1998 to Apr 24, 1999\",\"1\":\"Sep 1, 2001\",\"2\":\"Apr 1, 1998 to Sep 30, 1998\",\"3\":\"Jul 2, 2002 to Dec 24, 2002\"},\"Premiered\":{\"0\":\"Spring 1998\",\"1\":\"Unknown\",\"2\":\"Spring 1998\",\"3\":\"Summer 2002\"},\"Producers\":{\"0\":\"Bandai Visual\",\"1\":\"Sunrise, Bandai Visual\",\"2\":\"Victor Entertainment\",\"3\":\"TV Tokyo, Bandai Visual, Dentsu, Victor Entertainment\"},\"Licensors\":{\"0\":\"Funimation, Bandai Entertainment\",\"1\":\"Sony Pictures Entertainment\",\"2\":\"Funimation, Geneon Entertainment USA\",\"3\":\"Funimation, Bandai Entertainment\"},\"Studios\":{\"0\":\"Sunrise\",\"1\":\"Bones\",\"2\":\"Madhouse\",\"3\":\"Sunrise\"},\"Source\":{\"0\":\"Original\",\"1\":\"Original\",\"2\":\"Manga\",\"3\":\"Original\"},\"Duration\":{\"0\":\"24 min. per ep.\",\"1\":\"1 hr. 55 min.\",\"2\":\"24 min. per ep.\",\"3\":\"25 min. per ep.\"},\"Rating\":{\"0\":\"R - 17+ (violence & profanity)\",\"1\":\"R - 17+ (violence & profanity)\",\"2\":\"PG-13 - Teens 13 or older\",\"3\":\"PG-13 - Teens 13 or older\"},\"Ranked\":{\"0\":\"28.0\",\"1\":\"159.0\",\"2\":\"266.0\",\"3\":\"2481.0\"},\"Popularity\":{\"0\":39,\"1\":518,\"2\":201,\"3\":1467},\"Members\":{\"0\":1251960,\"1\":273145,\"2\":558913,\"3\":94683},\"Favorites\":{\"0\":61971,\"1\":1174,\"2\":12944,\"3\":587},\"Watching\":{\"0\":105808,\"1\":4143,\"2\":29113,\"3\":4300},\"Completed\":{\"0\":718161,\"1\":208333,\"2\":343492,\"3\":46165},\"On-Hold\":{\"0\":71513,\"1\":1935,\"2\":25465,\"3\":5121},\"Dropped\":{\"0\":26678,\"1\":770,\"2\":13925,\"3\":5378},\"Plan to Watch\":{\"0\":329800,\"1\":57964,\"2\":146918,\"3\":33719},\"Score-10\":{\"0\":\"229170.0\",\"1\":\"30043.0\",\"2\":\"50229.0\",\"3\":\"2182.0\"},\"Score-9\":{\"0\":\"182126.0\",\"1\":\"49201.0\",\"2\":\"75651.0\",\"3\":\"4806.0\"},\"Score-8\":{\"0\":\"131625.0\",\"1\":\"49505.0\",\"2\":\"86142.0\",\"3\":\"10128.0\"},\"Score-7\":{\"0\":\"62330.0\",\"1\":\"22632.0\",\"2\":\"49432.0\",\"3\":\"11618.0\"},\"Score-6\":{\"0\":\"20688.0\",\"1\":\"5805.0\",\"2\":\"15376.0\",\"3\":\"5709.0\"},\"Score-5\":{\"0\":\"8904.0\",\"1\":\"1877.0\",\"2\":\"5838.0\",\"3\":\"2920.0\"},\"Score-4\":{\"0\":\"3184.0\",\"1\":\"577.0\",\"2\":\"1965.0\",\"3\":\"1083.0\"},\"Score-3\":{\"0\":\"1357.0\",\"1\":\"221.0\",\"2\":\"664.0\",\"3\":\"353.0\"},\"Score-2\":{\"0\":\"741.0\",\"1\":\"109.0\",\"2\":\"316.0\",\"3\":\"164.0\"},\"Score-1\":{\"0\":\"1580.0\",\"1\":\"379.0\",\"2\":\"533.0\",\"3\":\"131.0\"}}"}}, {"anime-recommendation-database-2020/rating_complete.csv": {"column_names": "[\"user_id\", \"anime_id\", \"rating\"]", "column_data_types": "{\"user_id\": \"int64\", \"anime_id\": \"int64\", \"rating\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 57633278 entries, 0 to 57633277\nData columns (total 3 columns):\n # Column Dtype\n--- ------ -----\n 0 user_id int64\n 1 anime_id int64\n 2 rating int64\ndtypes: int64(3)\nmemory usage: 1.3 GB\n", "summary": "{\"user_id\": {\"count\": 57633278.0, \"mean\": 176887.7657645987, \"std\": 102011.65082360912, \"min\": 0.0, \"25%\": 88278.0, \"50%\": 177291.0, \"75%\": 265419.0, \"max\": 353404.0}, \"anime_id\": {\"count\": 57633278.0, \"mean\": 15831.466092679997, \"std\": 13261.135175368623, \"min\": 1.0, \"25%\": 3091.0, \"50%\": 11887.0, \"75%\": 28999.0, \"max\": 48456.0}, \"rating\": {\"count\": 57633278.0, \"mean\": 7.510788732162693, \"std\": 1.6977223721477248, \"min\": 1.0, \"25%\": 7.0, \"50%\": 8.0, \"75%\": 9.0, \"max\": 10.0}}", "examples": "{\"user_id\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"anime_id\":{\"0\":430,\"1\":1004,\"2\":3010,\"3\":570},\"rating\":{\"0\":9,\"1\":5,\"2\":7,\"3\":7}}"}}]
| true | 2 |
<start_data_description><data_path>anime-recommendation-database-2020/anime.csv:
<column_names>
['MAL_ID', 'Name', 'Score', 'Genres', 'English name', 'Japanese name', 'Type', 'Episodes', 'Aired', 'Premiered', 'Producers', 'Licensors', 'Studios', 'Source', 'Duration', 'Rating', 'Ranked', 'Popularity', 'Members', 'Favorites', 'Watching', 'Completed', 'On-Hold', 'Dropped', 'Plan to Watch', 'Score-10', 'Score-9', 'Score-8', 'Score-7', 'Score-6', 'Score-5', 'Score-4', 'Score-3', 'Score-2', 'Score-1']
<column_types>
{'MAL_ID': 'int64', 'Name': 'object', 'Score': 'object', 'Genres': 'object', 'English name': 'object', 'Japanese name': 'object', 'Type': 'object', 'Episodes': 'object', 'Aired': 'object', 'Premiered': 'object', 'Producers': 'object', 'Licensors': 'object', 'Studios': 'object', 'Source': 'object', 'Duration': 'object', 'Rating': 'object', 'Ranked': 'object', 'Popularity': 'int64', 'Members': 'int64', 'Favorites': 'int64', 'Watching': 'int64', 'Completed': 'int64', 'On-Hold': 'int64', 'Dropped': 'int64', 'Plan to Watch': 'int64', 'Score-10': 'object', 'Score-9': 'object', 'Score-8': 'object', 'Score-7': 'object', 'Score-6': 'object', 'Score-5': 'object', 'Score-4': 'object', 'Score-3': 'object', 'Score-2': 'object', 'Score-1': 'object'}
<dataframe_Summary>
{'MAL_ID': {'count': 17562.0, 'mean': 21477.192347113087, 'std': 14900.093169943408, 'min': 1.0, '25%': 5953.5, '50%': 22820.0, '75%': 35624.75, 'max': 48492.0}, 'Popularity': {'count': 17562.0, 'mean': 8763.45234028015, 'std': 5059.327278012592, 'min': 0.0, '25%': 4383.5, '50%': 8762.5, '75%': 13145.0, 'max': 17565.0}, 'Members': {'count': 17562.0, 'mean': 34658.53951713928, 'std': 125282.14142822381, 'min': 1.0, '25%': 336.0, '50%': 2065.0, '75%': 13223.25, 'max': 2589552.0}, 'Favorites': {'count': 17562.0, 'mean': 457.7462703564514, 'std': 4063.4733134938538, 'min': 0.0, '25%': 0.0, '50%': 3.0, '75%': 31.0, 'max': 183914.0}, 'Watching': {'count': 17562.0, 'mean': 2231.487757658581, 'std': 14046.688133245594, 'min': 0.0, '25%': 13.0, '50%': 73.0, '75%': 522.0, 'max': 887333.0}, 'Completed': {'count': 17562.0, 'mean': 22095.571347226967, 'std': 91009.18850426309, 'min': 0.0, '25%': 111.0, '50%': 817.5, '75%': 6478.0, 'max': 2182587.0}, 'On-Hold': {'count': 17562.0, 'mean': 955.0496526591504, 'std': 4275.675095965888, 'min': 0.0, '25%': 6.0, '50%': 45.0, '75%': 291.75, 'max': 187919.0}, 'Dropped': {'count': 17562.0, 'mean': 1176.5995330827925, 'std': 4740.348652904278, 'min': 0.0, '25%': 37.0, '50%': 77.0, '75%': 271.0, 'max': 174710.0}, 'Plan to Watch': {'count': 17562.0, 'mean': 8199.831226511787, 'std': 23777.691962549074, 'min': 1.0, '25%': 112.0, '50%': 752.5, '75%': 4135.5, 'max': 425531.0}}
<dataframe_info>
RangeIndex: 17562 entries, 0 to 17561
Data columns (total 35 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 MAL_ID 17562 non-null int64
1 Name 17562 non-null object
2 Score 17562 non-null object
3 Genres 17562 non-null object
4 English name 17562 non-null object
5 Japanese name 17562 non-null object
6 Type 17562 non-null object
7 Episodes 17562 non-null object
8 Aired 17562 non-null object
9 Premiered 17562 non-null object
10 Producers 17562 non-null object
11 Licensors 17562 non-null object
12 Studios 17562 non-null object
13 Source 17562 non-null object
14 Duration 17562 non-null object
15 Rating 17562 non-null object
16 Ranked 17562 non-null object
17 Popularity 17562 non-null int64
18 Members 17562 non-null int64
19 Favorites 17562 non-null int64
20 Watching 17562 non-null int64
21 Completed 17562 non-null int64
22 On-Hold 17562 non-null int64
23 Dropped 17562 non-null int64
24 Plan to Watch 17562 non-null int64
25 Score-10 17562 non-null object
26 Score-9 17562 non-null object
27 Score-8 17562 non-null object
28 Score-7 17562 non-null object
29 Score-6 17562 non-null object
30 Score-5 17562 non-null object
31 Score-4 17562 non-null object
32 Score-3 17562 non-null object
33 Score-2 17562 non-null object
34 Score-1 17562 non-null object
dtypes: int64(9), object(26)
memory usage: 4.7+ MB
<some_examples>
{'MAL_ID': {'0': 1, '1': 5, '2': 6, '3': 7}, 'Name': {'0': 'Cowboy Bebop', '1': 'Cowboy Bebop: Tengoku no Tobira', '2': 'Trigun', '3': 'Witch Hunter Robin'}, 'Score': {'0': '8.78', '1': '8.39', '2': '8.24', '3': '7.27'}, 'Genres': {'0': 'Action, Adventure, Comedy, Drama, Sci-Fi, Space', '1': 'Action, Drama, Mystery, Sci-Fi, Space', '2': 'Action, Sci-Fi, Adventure, Comedy, Drama, Shounen', '3': 'Action, Mystery, Police, Supernatural, Drama, Magic'}, 'English name': {'0': 'Cowboy Bebop', '1': 'Cowboy Bebop:The Movie', '2': 'Trigun', '3': 'Witch Hunter Robin'}, 'Japanese name': {'0': 'カウボーイビバップ', '1': 'カウボーイビバップ 天国の扉', '2': 'トライガン', '3': 'Witch Hunter ROBIN (ウイッチハンターロビン)'}, 'Type': {'0': 'TV', '1': 'Movie', '2': 'TV', '3': 'TV'}, 'Episodes': {'0': '26', '1': '1', '2': '26', '3': '26'}, 'Aired': {'0': 'Apr 3, 1998 to Apr 24, 1999', '1': 'Sep 1, 2001', '2': 'Apr 1, 1998 to Sep 30, 1998', '3': 'Jul 2, 2002 to Dec 24, 2002'}, 'Premiered': {'0': 'Spring 1998', '1': 'Unknown', '2': 'Spring 1998', '3': 'Summer 2002'}, 'Producers': {'0': 'Bandai Visual', '1': 'Sunrise, Bandai Visual', '2': 'Victor Entertainment', '3': 'TV Tokyo, Bandai Visual, Dentsu, Victor Entertainment'}, 'Licensors': {'0': 'Funimation, Bandai Entertainment', '1': 'Sony Pictures Entertainment', '2': 'Funimation, Geneon Entertainment USA', '3': 'Funimation, Bandai Entertainment'}, 'Studios': {'0': 'Sunrise', '1': 'Bones', '2': 'Madhouse', '3': 'Sunrise'}, 'Source': {'0': 'Original', '1': 'Original', '2': 'Manga', '3': 'Original'}, 'Duration': {'0': '24 min. per ep.', '1': '1 hr. 55 min.', '2': '24 min. per ep.', '3': '25 min. per ep.'}, 'Rating': {'0': 'R - 17+ (violence & profanity)', '1': 'R - 17+ (violence & profanity)', '2': 'PG-13 - Teens 13 or older', '3': 'PG-13 - Teens 13 or older'}, 'Ranked': {'0': '28.0', '1': '159.0', '2': '266.0', '3': '2481.0'}, 'Popularity': {'0': 39, '1': 518, '2': 201, '3': 1467}, 'Members': {'0': 1251960, '1': 273145, '2': 558913, '3': 94683}, 'Favorites': {'0': 61971, '1': 1174, '2': 12944, '3': 587}, 'Watching': {'0': 105808, '1': 4143, '2': 29113, '3': 4300}, 'Completed': {'0': 718161, '1': 208333, '2': 343492, '3': 46165}, 'On-Hold': {'0': 71513, '1': 1935, '2': 25465, '3': 5121}, 'Dropped': {'0': 26678, '1': 770, '2': 13925, '3': 5378}, 'Plan to Watch': {'0': 329800, '1': 57964, '2': 146918, '3': 33719}, 'Score-10': {'0': '229170.0', '1': '30043.0', '2': '50229.0', '3': '2182.0'}, 'Score-9': {'0': '182126.0', '1': '49201.0', '2': '75651.0', '3': '4806.0'}, 'Score-8': {'0': '131625.0', '1': '49505.0', '2': '86142.0', '3': '10128.0'}, 'Score-7': {'0': '62330.0', '1': '22632.0', '2': '49432.0', '3': '11618.0'}, 'Score-6': {'0': '20688.0', '1': '5805.0', '2': '15376.0', '3': '5709.0'}, 'Score-5': {'0': '8904.0', '1': '1877.0', '2': '5838.0', '3': '2920.0'}, 'Score-4': {'0': '3184.0', '1': '577.0', '2': '1965.0', '3': '1083.0'}, 'Score-3': {'0': '1357.0', '1': '221.0', '2': '664.0', '3': '353.0'}, 'Score-2': {'0': '741.0', '1': '109.0', '2': '316.0', '3': '164.0'}, 'Score-1': {'0': '1580.0', '1': '379.0', '2': '533.0', '3': '131.0'}}
<end_description>
<start_data_description><data_path>anime-recommendation-database-2020/rating_complete.csv:
<column_names>
['user_id', 'anime_id', 'rating']
<column_types>
{'user_id': 'int64', 'anime_id': 'int64', 'rating': 'int64'}
<dataframe_Summary>
{'user_id': {'count': 57633278.0, 'mean': 176887.7657645987, 'std': 102011.65082360912, 'min': 0.0, '25%': 88278.0, '50%': 177291.0, '75%': 265419.0, 'max': 353404.0}, 'anime_id': {'count': 57633278.0, 'mean': 15831.466092679997, 'std': 13261.135175368623, 'min': 1.0, '25%': 3091.0, '50%': 11887.0, '75%': 28999.0, 'max': 48456.0}, 'rating': {'count': 57633278.0, 'mean': 7.510788732162693, 'std': 1.6977223721477248, 'min': 1.0, '25%': 7.0, '50%': 8.0, '75%': 9.0, 'max': 10.0}}
<dataframe_info>
RangeIndex: 57633278 entries, 0 to 57633277
Data columns (total 3 columns):
# Column Dtype
--- ------ -----
0 user_id int64
1 anime_id int64
2 rating int64
dtypes: int64(3)
memory usage: 1.3 GB
<some_examples>
{'user_id': {'0': 0, '1': 0, '2': 0, '3': 0}, 'anime_id': {'0': 430, '1': 1004, '2': 3010, '3': 570}, 'rating': {'0': 9, '1': 5, '2': 7, '3': 7}}
<end_description>
| 2,554 | 0 | 6,932 | 2,554 |
69401499
|
<jupyter_start><jupyter_text>Mobile Price Classification
### Context
Bob has started his own mobile company. He wants to give tough fight to big companies like Apple,Samsung etc.
He does not know how to estimate price of mobiles his company creates. In this competitive mobile phone market you cannot simply assume things. To solve this problem he collects sales data of mobile phones of various companies.
Bob wants to find out some relation between features of a mobile phone(eg:- RAM,Internal Memory etc) and its selling price. But he is not so good at Machine Learning. So he needs your help to solve this problem.
In this problem you do not have to predict actual price but a price range indicating how high the price is
Kaggle dataset identifier: mobile-price-classification
<jupyter_code>import pandas as pd
df = pd.read_csv('mobile-price-classification/train.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 2000 entries, 0 to 1999
Data columns (total 21 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 battery_power 2000 non-null int64
1 blue 2000 non-null int64
2 clock_speed 2000 non-null float64
3 dual_sim 2000 non-null int64
4 fc 2000 non-null int64
5 four_g 2000 non-null int64
6 int_memory 2000 non-null int64
7 m_dep 2000 non-null float64
8 mobile_wt 2000 non-null int64
9 n_cores 2000 non-null int64
10 pc 2000 non-null int64
11 px_height 2000 non-null int64
12 px_width 2000 non-null int64
13 ram 2000 non-null int64
14 sc_h 2000 non-null int64
15 sc_w 2000 non-null int64
16 talk_time 2000 non-null int64
17 three_g 2000 non-null int64
18 touch_screen 2000 non-null int64
19 wifi 2000 non-null int64
20 price_range 2000 non-null int64
dtypes: float64(2), int64(19)
memory usage: 328.2 KB
<jupyter_text>Examples:
{
"battery_power": 842.0,
"blue": 0.0,
"clock_speed": 2.2,
"dual_sim": 0.0,
"fc": 1.0,
"four_g": 0.0,
"int_memory": 7.0,
"m_dep": 0.6000000000000001,
"mobile_wt": 188.0,
"n_cores": 2.0,
"pc": 2.0,
"px_height": 20.0,
"px_width": 756.0,
"ram": 2549.0,
"sc_h": 9.0,
"sc_w": 7.0,
"talk_time": 19.0,
"three_g": 0.0,
"touch_screen": 0.0,
"wifi": 1.0,
"...": "and 1 more columns"
}
{
"battery_power": 1021.0,
"blue": 1.0,
"clock_speed": 0.5,
"dual_sim": 1.0,
"fc": 0.0,
"four_g": 1.0,
"int_memory": 53.0,
"m_dep": 0.7000000000000001,
"mobile_wt": 136.0,
"n_cores": 3.0,
"pc": 6.0,
"px_height": 905.0,
"px_width": 1988.0,
"ram": 2631.0,
"sc_h": 17.0,
"sc_w": 3.0,
"talk_time": 7.0,
"three_g": 1.0,
"touch_screen": 1.0,
"wifi": 0.0,
"...": "and 1 more columns"
}
{
"battery_power": 563.0,
"blue": 1.0,
"clock_speed": 0.5,
"dual_sim": 1.0,
"fc": 2.0,
"four_g": 1.0,
"int_memory": 41.0,
"m_dep": 0.9,
"mobile_wt": 145.0,
"n_cores": 5.0,
"pc": 6.0,
"px_height": 1263.0,
"px_width": 1716.0,
"ram": 2603.0,
"sc_h": 11.0,
"sc_w": 2.0,
"talk_time": 9.0,
"three_g": 1.0,
"touch_screen": 1.0,
"wifi": 0.0,
"...": "and 1 more columns"
}
{
"battery_power": 615.0,
"blue": 1.0,
"clock_speed": 2.5,
"dual_sim": 0.0,
"fc": 0.0,
"four_g": 0.0,
"int_memory": 10.0,
"m_dep": 0.8,
"mobile_wt": 131.0,
"n_cores": 6.0,
"pc": 9.0,
"px_height": 1216.0,
"px_width": 1786.0,
"ram": 2769.0,
"sc_h": 16.0,
"sc_w": 8.0,
"talk_time": 11.0,
"three_g": 1.0,
"touch_screen": 0.0,
"wifi": 0.0,
"...": "and 1 more columns"
}
<jupyter_script># ## Life Cycle of Data Science
# 1. Data Analysis
# 2. Feature Engineering
# 3. Feature Selection
# 4. Model Building
# 5. Model Deployment
# 1. 'battery_power' -battery_power
# 2. 'blue' -bluetooth
# 3. 'clock_speed' -clock_speed
# 4. 'dual_sim' -sim
# 5. 'fc' - Front Screen
# 6. 'four_g'- 4G Connection
# 7. 'int_memory'- Internal Memory
# 8. 'm_dep'- Slimness
# 9. 'mobile_wt' -Weight
# 10. 'n_cores'- Processor
# 11. 'pc'- Primary camera
# 12. 'px_height' -Pixel hight
# 13. 'px_width' - Pixel Width
# 14. 'ram' - ram size
# 15. 'sc_h'- screen hight
# 16. 'sc_w'- screen width
# 17. 'talk_time'- talk time
# 18. 'three_g'- 3G
# 19. 'touch_screen'- Touch facility
# 20. 'wifi' - wifi
# 21. 'price_range'- price
## Data Analysis
## Main Aim is to understand about the data
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
pd.pandas.set_option("display.max_columns", None)
dataset = pd.read_csv("../input/mobile-price-classification/train.csv")
print(dataset.shape)
dataset.head()
dataset.info()
# we are having 21 columns all are
# ###### Numerical values
# ###### Having NO Null values
## Since we are going to predict price, lets analyse based on price_range
dataset["price_range"].value_counts()
# we can see the values is not numerical, it is classified into four categories having count of 500 each
sns.countplot(dataset["price_range"])
plt.xlabel("price_range")
# lets count the unique values of all columns to find the discreate numerical variable
for i in dataset.columns:
if len(dataset[i].unique()) < 100 and i != "price_range":
print(i, "-----", len(dataset[i].unique()))
# here we can see that we are having 15 discreate numerical variable
###lets plot fo these discreate variables
discreate_numerical = []
for i in dataset.columns:
if len(dataset[i].unique()) < 50:
discreate_numerical.append(i)
plt.figure(figsize=(25, 50))
for i in enumerate(discreate_numerical):
plt.subplot(8, 2, i[0] + 1)
sns.barplot(dataset[i[1]], dataset["price_range"])
plt.xlabel(i[1])
# here we can see most of the features are categorised in numbers
plt.figure(figsize=(18, 36))
for i in enumerate(discreate_numerical):
plt.subplot(8, 2, i[0] + 1)
sns.boxplot(dataset[i[1]], dataset["price_range"])
plt.xlabel(i[1])
# we can see from the above the box plot for every features varies huge this indicates most of the features are providing information for result
# Lets do analysis for Numerical features
numerical_fet = []
for i in dataset.columns:
if i not in discreate_numerical:
numerical_fet.append(i)
plt.figure(figsize=(18, 36))
for i in enumerate(numerical_fet):
plt.subplot(8, 2, i[0] + 1)
sns.histplot(dataset[i[1]], kde=True)
plt.xlabel(i[1])
# Here we can see only px_height shows a right skewed distribution
# ###### Lets analyse the corelation between these pics
plt.figure(figsize=(25, 25))
sns.heatmap(dataset.corr(), annot=True)
# We can see here that Ram is highly co related with Price
# Since most of the values independent of each other lets get to feature selection
y = dataset["price_range"]
dataset.drop(["price_range"], axis=1, inplace=True)
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score, confusion_matrix
x_tr, x_te, y_tr, y_te = train_test_split(dataset, y, test_size=0.3, random_state=42)
cla = RandomForestClassifier()
cla.fit(x_tr, y_tr)
acuracy = accuracy_score(y_te, cla.predict(x_te))
cm = confusion_matrix(y_te, cla.predict(x_te))
print(acuracy)
sns.heatmap(cm, annot=True)
# Here we are getting accuracy of 0.865 and we can also see that many features affect the prediction
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import chi2
selc_ten = SelectKBest(chi2, k=10).fit(x_tr, y_tr)
print(selc_ten.scores_)
for i in range(len(dataset.columns)):
print(dataset.columns[i], " : ", selc_ten.scores_[i])
# selecting most important features from the list based on their value
# battery_power,ram,mobile_wt,px_height,px_width,fc,pc,sc_w,talk_time,int_memory
# the above 10 features affect the model most
X_tr = selc_ten.transform(x_tr)
X_te = selc_ten.transform(x_te)
clf_2 = RandomForestClassifier()
clf_2.fit(x_tr, y_tr)
accuracy_score(y_te, clf_2.predict(x_te))
# here im getting accuracy .875 increased when compared to preivious
cm1 = confusion_matrix(y_te, clf_2.predict(x_te))
sns.heatmap(cm1, annot=True)
# from the above model we can see that unwanted features giving us type 1 and type 2 errors are minimizsed
# Lets see wat can we do if we want to increase more
# #### Recursive feature elimination with cross validation and random forest classification
# In previous method we found how many features we needed most by our own choice,
# But in this method we can find how many features can give best accuracy and the choice is made by cross validation
from sklearn.feature_selection import RFECV
clf_3 = RandomForestClassifier()
rfecv = RFECV(estimator=clf_3, cv=5, step=1, scoring="accuracy")
rfecv.fit(x_tr, y_tr)
print(rfecv.n_features_)
# here it took 5 features to make the model to be optimized
print(x_tr.columns[rfecv.support_])
plt.figure()
plt.xlabel("features")
plt.ylabel("accuracy")
plt.plot(range(1, len(rfecv.grid_scores_) + 1), rfecv.grid_scores_)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/401/69401499.ipynb
|
mobile-price-classification
|
iabhishekofficial
|
[{"Id": 69401499, "ScriptId": 18662699, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5491908, "CreationDate": "07/30/2021 14:29:52", "VersionNumber": 3.0, "Title": "Mobile Price Feature Selection,SVM,Random forest", "EvaluationDate": "07/30/2021", "IsChange": false, "TotalLines": 199.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 199.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 10}]
|
[{"Id": 92520303, "KernelVersionId": 69401499, "SourceDatasetVersionId": 15520}]
|
[{"Id": 15520, "DatasetId": 11167, "DatasourceVersionId": 15520, "CreatorUserId": 907764, "LicenseName": "Unknown", "CreationDate": "01/28/2018 08:44:24", "VersionNumber": 1.0, "Title": "Mobile Price Classification", "Slug": "mobile-price-classification", "Subtitle": "Classify Mobile Price Range", "Description": "### Context\n\nBob has started his own mobile company. He wants to give tough fight to big companies like Apple,Samsung etc.\n\nHe does not know how to estimate price of mobiles his company creates. In this competitive mobile phone market you cannot simply assume things. To solve this problem he collects sales data of mobile phones of various companies.\n\nBob wants to find out some relation between features of a mobile phone(eg:- RAM,Internal Memory etc) and its selling price. But he is not so good at Machine Learning. So he needs your help to solve this problem.\n\nIn this problem you do not have to predict actual price but a price range indicating how high the price is", "VersionNotes": "Initial release", "TotalCompressedBytes": 186253.0, "TotalUncompressedBytes": 186253.0}]
|
[{"Id": 11167, "CreatorUserId": 907764, "OwnerUserId": 907764.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 15520.0, "CurrentDatasourceVersionId": 15520.0, "ForumId": 18557, "Type": 2, "CreationDate": "01/28/2018 08:44:24", "LastActivityDate": "02/06/2018", "TotalViews": 793378, "TotalDownloads": 143007, "TotalVotes": 1700, "TotalKernels": 3248}]
|
[{"Id": 907764, "UserName": "iabhishekofficial", "DisplayName": "Abhishek Sharma", "RegisterDate": "02/11/2017", "PerformanceTier": 1}]
|
# ## Life Cycle of Data Science
# 1. Data Analysis
# 2. Feature Engineering
# 3. Feature Selection
# 4. Model Building
# 5. Model Deployment
# 1. 'battery_power' -battery_power
# 2. 'blue' -bluetooth
# 3. 'clock_speed' -clock_speed
# 4. 'dual_sim' -sim
# 5. 'fc' - Front Screen
# 6. 'four_g'- 4G Connection
# 7. 'int_memory'- Internal Memory
# 8. 'm_dep'- Slimness
# 9. 'mobile_wt' -Weight
# 10. 'n_cores'- Processor
# 11. 'pc'- Primary camera
# 12. 'px_height' -Pixel hight
# 13. 'px_width' - Pixel Width
# 14. 'ram' - ram size
# 15. 'sc_h'- screen hight
# 16. 'sc_w'- screen width
# 17. 'talk_time'- talk time
# 18. 'three_g'- 3G
# 19. 'touch_screen'- Touch facility
# 20. 'wifi' - wifi
# 21. 'price_range'- price
## Data Analysis
## Main Aim is to understand about the data
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
pd.pandas.set_option("display.max_columns", None)
dataset = pd.read_csv("../input/mobile-price-classification/train.csv")
print(dataset.shape)
dataset.head()
dataset.info()
# we are having 21 columns all are
# ###### Numerical values
# ###### Having NO Null values
## Since we are going to predict price, lets analyse based on price_range
dataset["price_range"].value_counts()
# we can see the values is not numerical, it is classified into four categories having count of 500 each
sns.countplot(dataset["price_range"])
plt.xlabel("price_range")
# lets count the unique values of all columns to find the discreate numerical variable
for i in dataset.columns:
if len(dataset[i].unique()) < 100 and i != "price_range":
print(i, "-----", len(dataset[i].unique()))
# here we can see that we are having 15 discreate numerical variable
###lets plot fo these discreate variables
discreate_numerical = []
for i in dataset.columns:
if len(dataset[i].unique()) < 50:
discreate_numerical.append(i)
plt.figure(figsize=(25, 50))
for i in enumerate(discreate_numerical):
plt.subplot(8, 2, i[0] + 1)
sns.barplot(dataset[i[1]], dataset["price_range"])
plt.xlabel(i[1])
# here we can see most of the features are categorised in numbers
plt.figure(figsize=(18, 36))
for i in enumerate(discreate_numerical):
plt.subplot(8, 2, i[0] + 1)
sns.boxplot(dataset[i[1]], dataset["price_range"])
plt.xlabel(i[1])
# we can see from the above the box plot for every features varies huge this indicates most of the features are providing information for result
# Lets do analysis for Numerical features
numerical_fet = []
for i in dataset.columns:
if i not in discreate_numerical:
numerical_fet.append(i)
plt.figure(figsize=(18, 36))
for i in enumerate(numerical_fet):
plt.subplot(8, 2, i[0] + 1)
sns.histplot(dataset[i[1]], kde=True)
plt.xlabel(i[1])
# Here we can see only px_height shows a right skewed distribution
# ###### Lets analyse the corelation between these pics
plt.figure(figsize=(25, 25))
sns.heatmap(dataset.corr(), annot=True)
# We can see here that Ram is highly co related with Price
# Since most of the values independent of each other lets get to feature selection
y = dataset["price_range"]
dataset.drop(["price_range"], axis=1, inplace=True)
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score, confusion_matrix
x_tr, x_te, y_tr, y_te = train_test_split(dataset, y, test_size=0.3, random_state=42)
cla = RandomForestClassifier()
cla.fit(x_tr, y_tr)
acuracy = accuracy_score(y_te, cla.predict(x_te))
cm = confusion_matrix(y_te, cla.predict(x_te))
print(acuracy)
sns.heatmap(cm, annot=True)
# Here we are getting accuracy of 0.865 and we can also see that many features affect the prediction
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import chi2
selc_ten = SelectKBest(chi2, k=10).fit(x_tr, y_tr)
print(selc_ten.scores_)
for i in range(len(dataset.columns)):
print(dataset.columns[i], " : ", selc_ten.scores_[i])
# selecting most important features from the list based on their value
# battery_power,ram,mobile_wt,px_height,px_width,fc,pc,sc_w,talk_time,int_memory
# the above 10 features affect the model most
X_tr = selc_ten.transform(x_tr)
X_te = selc_ten.transform(x_te)
clf_2 = RandomForestClassifier()
clf_2.fit(x_tr, y_tr)
accuracy_score(y_te, clf_2.predict(x_te))
# here im getting accuracy .875 increased when compared to preivious
cm1 = confusion_matrix(y_te, clf_2.predict(x_te))
sns.heatmap(cm1, annot=True)
# from the above model we can see that unwanted features giving us type 1 and type 2 errors are minimizsed
# Lets see wat can we do if we want to increase more
# #### Recursive feature elimination with cross validation and random forest classification
# In previous method we found how many features we needed most by our own choice,
# But in this method we can find how many features can give best accuracy and the choice is made by cross validation
from sklearn.feature_selection import RFECV
clf_3 = RandomForestClassifier()
rfecv = RFECV(estimator=clf_3, cv=5, step=1, scoring="accuracy")
rfecv.fit(x_tr, y_tr)
print(rfecv.n_features_)
# here it took 5 features to make the model to be optimized
print(x_tr.columns[rfecv.support_])
plt.figure()
plt.xlabel("features")
plt.ylabel("accuracy")
plt.plot(range(1, len(rfecv.grid_scores_) + 1), rfecv.grid_scores_)
|
[{"mobile-price-classification/train.csv": {"column_names": "[\"battery_power\", \"blue\", \"clock_speed\", \"dual_sim\", \"fc\", \"four_g\", \"int_memory\", \"m_dep\", \"mobile_wt\", \"n_cores\", \"pc\", \"px_height\", \"px_width\", \"ram\", \"sc_h\", \"sc_w\", \"talk_time\", \"three_g\", \"touch_screen\", \"wifi\", \"price_range\"]", "column_data_types": "{\"battery_power\": \"int64\", \"blue\": \"int64\", \"clock_speed\": \"float64\", \"dual_sim\": \"int64\", \"fc\": \"int64\", \"four_g\": \"int64\", \"int_memory\": \"int64\", \"m_dep\": \"float64\", \"mobile_wt\": \"int64\", \"n_cores\": \"int64\", \"pc\": \"int64\", \"px_height\": \"int64\", \"px_width\": \"int64\", \"ram\": \"int64\", \"sc_h\": \"int64\", \"sc_w\": \"int64\", \"talk_time\": \"int64\", \"three_g\": \"int64\", \"touch_screen\": \"int64\", \"wifi\": \"int64\", \"price_range\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 2000 entries, 0 to 1999\nData columns (total 21 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 battery_power 2000 non-null int64 \n 1 blue 2000 non-null int64 \n 2 clock_speed 2000 non-null float64\n 3 dual_sim 2000 non-null int64 \n 4 fc 2000 non-null int64 \n 5 four_g 2000 non-null int64 \n 6 int_memory 2000 non-null int64 \n 7 m_dep 2000 non-null float64\n 8 mobile_wt 2000 non-null int64 \n 9 n_cores 2000 non-null int64 \n 10 pc 2000 non-null int64 \n 11 px_height 2000 non-null int64 \n 12 px_width 2000 non-null int64 \n 13 ram 2000 non-null int64 \n 14 sc_h 2000 non-null int64 \n 15 sc_w 2000 non-null int64 \n 16 talk_time 2000 non-null int64 \n 17 three_g 2000 non-null int64 \n 18 touch_screen 2000 non-null int64 \n 19 wifi 2000 non-null int64 \n 20 price_range 2000 non-null int64 \ndtypes: float64(2), int64(19)\nmemory usage: 328.2 KB\n", "summary": "{\"battery_power\": {\"count\": 2000.0, \"mean\": 1238.5185, \"std\": 439.41820608353135, \"min\": 501.0, \"25%\": 851.75, \"50%\": 1226.0, \"75%\": 1615.25, \"max\": 1998.0}, \"blue\": {\"count\": 2000.0, \"mean\": 0.495, \"std\": 0.5001000400170075, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}, \"clock_speed\": {\"count\": 2000.0, \"mean\": 1.52225, \"std\": 0.8160042088950689, \"min\": 0.5, \"25%\": 0.7, \"50%\": 1.5, \"75%\": 2.2, \"max\": 3.0}, \"dual_sim\": {\"count\": 2000.0, \"mean\": 0.5095, \"std\": 0.500034766175005, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"fc\": {\"count\": 2000.0, \"mean\": 4.3095, \"std\": 4.341443747983894, \"min\": 0.0, \"25%\": 1.0, \"50%\": 3.0, \"75%\": 7.0, \"max\": 19.0}, \"four_g\": {\"count\": 2000.0, \"mean\": 0.5215, \"std\": 0.49966246736236386, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"int_memory\": {\"count\": 2000.0, \"mean\": 32.0465, \"std\": 18.145714955206856, \"min\": 2.0, \"25%\": 16.0, \"50%\": 32.0, \"75%\": 48.0, \"max\": 64.0}, \"m_dep\": {\"count\": 2000.0, \"mean\": 0.50175, \"std\": 0.2884155496235117, \"min\": 0.1, \"25%\": 0.2, \"50%\": 0.5, \"75%\": 0.8, \"max\": 1.0}, \"mobile_wt\": {\"count\": 2000.0, \"mean\": 140.249, \"std\": 35.39965489638835, \"min\": 80.0, \"25%\": 109.0, \"50%\": 141.0, \"75%\": 170.0, \"max\": 200.0}, \"n_cores\": {\"count\": 2000.0, \"mean\": 4.5205, \"std\": 2.2878367180426604, \"min\": 1.0, \"25%\": 3.0, \"50%\": 4.0, \"75%\": 7.0, \"max\": 8.0}, \"pc\": {\"count\": 2000.0, \"mean\": 9.9165, \"std\": 6.06431494134778, \"min\": 0.0, \"25%\": 5.0, \"50%\": 10.0, \"75%\": 15.0, \"max\": 20.0}, \"px_height\": {\"count\": 2000.0, \"mean\": 645.108, \"std\": 443.7808108064386, \"min\": 0.0, \"25%\": 282.75, \"50%\": 564.0, \"75%\": 947.25, \"max\": 1960.0}, \"px_width\": {\"count\": 2000.0, \"mean\": 1251.5155, \"std\": 432.19944694633796, \"min\": 500.0, \"25%\": 874.75, \"50%\": 1247.0, \"75%\": 1633.0, \"max\": 1998.0}, \"ram\": {\"count\": 2000.0, \"mean\": 2124.213, \"std\": 1084.7320436099494, \"min\": 256.0, \"25%\": 1207.5, \"50%\": 2146.5, \"75%\": 3064.5, \"max\": 3998.0}, \"sc_h\": {\"count\": 2000.0, \"mean\": 12.3065, \"std\": 4.213245004356306, \"min\": 5.0, \"25%\": 9.0, \"50%\": 12.0, \"75%\": 16.0, \"max\": 19.0}, \"sc_w\": {\"count\": 2000.0, \"mean\": 5.767, \"std\": 4.3563976058264045, \"min\": 0.0, \"25%\": 2.0, \"50%\": 5.0, \"75%\": 9.0, \"max\": 18.0}, \"talk_time\": {\"count\": 2000.0, \"mean\": 11.011, \"std\": 5.463955197766688, \"min\": 2.0, \"25%\": 6.0, \"50%\": 11.0, \"75%\": 16.0, \"max\": 20.0}, \"three_g\": {\"count\": 2000.0, \"mean\": 0.7615, \"std\": 0.42627292231873126, \"min\": 0.0, \"25%\": 1.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"touch_screen\": {\"count\": 2000.0, \"mean\": 0.503, \"std\": 0.500116044562674, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"wifi\": {\"count\": 2000.0, \"mean\": 0.507, \"std\": 0.5000760322381083, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"price_range\": {\"count\": 2000.0, \"mean\": 1.5, \"std\": 1.118313602106461, \"min\": 0.0, \"25%\": 0.75, \"50%\": 1.5, \"75%\": 2.25, \"max\": 3.0}}", "examples": "{\"battery_power\":{\"0\":842,\"1\":1021,\"2\":563,\"3\":615},\"blue\":{\"0\":0,\"1\":1,\"2\":1,\"3\":1},\"clock_speed\":{\"0\":2.2,\"1\":0.5,\"2\":0.5,\"3\":2.5},\"dual_sim\":{\"0\":0,\"1\":1,\"2\":1,\"3\":0},\"fc\":{\"0\":1,\"1\":0,\"2\":2,\"3\":0},\"four_g\":{\"0\":0,\"1\":1,\"2\":1,\"3\":0},\"int_memory\":{\"0\":7,\"1\":53,\"2\":41,\"3\":10},\"m_dep\":{\"0\":0.6,\"1\":0.7,\"2\":0.9,\"3\":0.8},\"mobile_wt\":{\"0\":188,\"1\":136,\"2\":145,\"3\":131},\"n_cores\":{\"0\":2,\"1\":3,\"2\":5,\"3\":6},\"pc\":{\"0\":2,\"1\":6,\"2\":6,\"3\":9},\"px_height\":{\"0\":20,\"1\":905,\"2\":1263,\"3\":1216},\"px_width\":{\"0\":756,\"1\":1988,\"2\":1716,\"3\":1786},\"ram\":{\"0\":2549,\"1\":2631,\"2\":2603,\"3\":2769},\"sc_h\":{\"0\":9,\"1\":17,\"2\":11,\"3\":16},\"sc_w\":{\"0\":7,\"1\":3,\"2\":2,\"3\":8},\"talk_time\":{\"0\":19,\"1\":7,\"2\":9,\"3\":11},\"three_g\":{\"0\":0,\"1\":1,\"2\":1,\"3\":1},\"touch_screen\":{\"0\":0,\"1\":1,\"2\":1,\"3\":0},\"wifi\":{\"0\":1,\"1\":0,\"2\":0,\"3\":0},\"price_range\":{\"0\":1,\"1\":2,\"2\":2,\"3\":2}}"}}]
| true | 1 |
<start_data_description><data_path>mobile-price-classification/train.csv:
<column_names>
['battery_power', 'blue', 'clock_speed', 'dual_sim', 'fc', 'four_g', 'int_memory', 'm_dep', 'mobile_wt', 'n_cores', 'pc', 'px_height', 'px_width', 'ram', 'sc_h', 'sc_w', 'talk_time', 'three_g', 'touch_screen', 'wifi', 'price_range']
<column_types>
{'battery_power': 'int64', 'blue': 'int64', 'clock_speed': 'float64', 'dual_sim': 'int64', 'fc': 'int64', 'four_g': 'int64', 'int_memory': 'int64', 'm_dep': 'float64', 'mobile_wt': 'int64', 'n_cores': 'int64', 'pc': 'int64', 'px_height': 'int64', 'px_width': 'int64', 'ram': 'int64', 'sc_h': 'int64', 'sc_w': 'int64', 'talk_time': 'int64', 'three_g': 'int64', 'touch_screen': 'int64', 'wifi': 'int64', 'price_range': 'int64'}
<dataframe_Summary>
{'battery_power': {'count': 2000.0, 'mean': 1238.5185, 'std': 439.41820608353135, 'min': 501.0, '25%': 851.75, '50%': 1226.0, '75%': 1615.25, 'max': 1998.0}, 'blue': {'count': 2000.0, 'mean': 0.495, 'std': 0.5001000400170075, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}, 'clock_speed': {'count': 2000.0, 'mean': 1.52225, 'std': 0.8160042088950689, 'min': 0.5, '25%': 0.7, '50%': 1.5, '75%': 2.2, 'max': 3.0}, 'dual_sim': {'count': 2000.0, 'mean': 0.5095, 'std': 0.500034766175005, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'fc': {'count': 2000.0, 'mean': 4.3095, 'std': 4.341443747983894, 'min': 0.0, '25%': 1.0, '50%': 3.0, '75%': 7.0, 'max': 19.0}, 'four_g': {'count': 2000.0, 'mean': 0.5215, 'std': 0.49966246736236386, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'int_memory': {'count': 2000.0, 'mean': 32.0465, 'std': 18.145714955206856, 'min': 2.0, '25%': 16.0, '50%': 32.0, '75%': 48.0, 'max': 64.0}, 'm_dep': {'count': 2000.0, 'mean': 0.50175, 'std': 0.2884155496235117, 'min': 0.1, '25%': 0.2, '50%': 0.5, '75%': 0.8, 'max': 1.0}, 'mobile_wt': {'count': 2000.0, 'mean': 140.249, 'std': 35.39965489638835, 'min': 80.0, '25%': 109.0, '50%': 141.0, '75%': 170.0, 'max': 200.0}, 'n_cores': {'count': 2000.0, 'mean': 4.5205, 'std': 2.2878367180426604, 'min': 1.0, '25%': 3.0, '50%': 4.0, '75%': 7.0, 'max': 8.0}, 'pc': {'count': 2000.0, 'mean': 9.9165, 'std': 6.06431494134778, 'min': 0.0, '25%': 5.0, '50%': 10.0, '75%': 15.0, 'max': 20.0}, 'px_height': {'count': 2000.0, 'mean': 645.108, 'std': 443.7808108064386, 'min': 0.0, '25%': 282.75, '50%': 564.0, '75%': 947.25, 'max': 1960.0}, 'px_width': {'count': 2000.0, 'mean': 1251.5155, 'std': 432.19944694633796, 'min': 500.0, '25%': 874.75, '50%': 1247.0, '75%': 1633.0, 'max': 1998.0}, 'ram': {'count': 2000.0, 'mean': 2124.213, 'std': 1084.7320436099494, 'min': 256.0, '25%': 1207.5, '50%': 2146.5, '75%': 3064.5, 'max': 3998.0}, 'sc_h': {'count': 2000.0, 'mean': 12.3065, 'std': 4.213245004356306, 'min': 5.0, '25%': 9.0, '50%': 12.0, '75%': 16.0, 'max': 19.0}, 'sc_w': {'count': 2000.0, 'mean': 5.767, 'std': 4.3563976058264045, 'min': 0.0, '25%': 2.0, '50%': 5.0, '75%': 9.0, 'max': 18.0}, 'talk_time': {'count': 2000.0, 'mean': 11.011, 'std': 5.463955197766688, 'min': 2.0, '25%': 6.0, '50%': 11.0, '75%': 16.0, 'max': 20.0}, 'three_g': {'count': 2000.0, 'mean': 0.7615, 'std': 0.42627292231873126, 'min': 0.0, '25%': 1.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'touch_screen': {'count': 2000.0, 'mean': 0.503, 'std': 0.500116044562674, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'wifi': {'count': 2000.0, 'mean': 0.507, 'std': 0.5000760322381083, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'price_range': {'count': 2000.0, 'mean': 1.5, 'std': 1.118313602106461, 'min': 0.0, '25%': 0.75, '50%': 1.5, '75%': 2.25, 'max': 3.0}}
<dataframe_info>
RangeIndex: 2000 entries, 0 to 1999
Data columns (total 21 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 battery_power 2000 non-null int64
1 blue 2000 non-null int64
2 clock_speed 2000 non-null float64
3 dual_sim 2000 non-null int64
4 fc 2000 non-null int64
5 four_g 2000 non-null int64
6 int_memory 2000 non-null int64
7 m_dep 2000 non-null float64
8 mobile_wt 2000 non-null int64
9 n_cores 2000 non-null int64
10 pc 2000 non-null int64
11 px_height 2000 non-null int64
12 px_width 2000 non-null int64
13 ram 2000 non-null int64
14 sc_h 2000 non-null int64
15 sc_w 2000 non-null int64
16 talk_time 2000 non-null int64
17 three_g 2000 non-null int64
18 touch_screen 2000 non-null int64
19 wifi 2000 non-null int64
20 price_range 2000 non-null int64
dtypes: float64(2), int64(19)
memory usage: 328.2 KB
<some_examples>
{'battery_power': {'0': 842, '1': 1021, '2': 563, '3': 615}, 'blue': {'0': 0, '1': 1, '2': 1, '3': 1}, 'clock_speed': {'0': 2.2, '1': 0.5, '2': 0.5, '3': 2.5}, 'dual_sim': {'0': 0, '1': 1, '2': 1, '3': 0}, 'fc': {'0': 1, '1': 0, '2': 2, '3': 0}, 'four_g': {'0': 0, '1': 1, '2': 1, '3': 0}, 'int_memory': {'0': 7, '1': 53, '2': 41, '3': 10}, 'm_dep': {'0': 0.6, '1': 0.7, '2': 0.9, '3': 0.8}, 'mobile_wt': {'0': 188, '1': 136, '2': 145, '3': 131}, 'n_cores': {'0': 2, '1': 3, '2': 5, '3': 6}, 'pc': {'0': 2, '1': 6, '2': 6, '3': 9}, 'px_height': {'0': 20, '1': 905, '2': 1263, '3': 1216}, 'px_width': {'0': 756, '1': 1988, '2': 1716, '3': 1786}, 'ram': {'0': 2549, '1': 2631, '2': 2603, '3': 2769}, 'sc_h': {'0': 9, '1': 17, '2': 11, '3': 16}, 'sc_w': {'0': 7, '1': 3, '2': 2, '3': 8}, 'talk_time': {'0': 19, '1': 7, '2': 9, '3': 11}, 'three_g': {'0': 0, '1': 1, '2': 1, '3': 1}, 'touch_screen': {'0': 0, '1': 1, '2': 1, '3': 0}, 'wifi': {'0': 1, '1': 0, '2': 0, '3': 0}, 'price_range': {'0': 1, '1': 2, '2': 2, '3': 2}}
<end_description>
| 1,782 | 10 | 3,441 | 1,782 |
69595521
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import os
import glob
data_dir = "../input/optiver-realized-volatility-prediction/"
# ## Functions for preprocess
def calc_wap(df):
wap = (df["bid_price1"] * df["ask_size1"] + df["ask_price1"] * df["bid_size1"]) / (
df["bid_size1"] + df["ask_size1"]
)
return wap
def calc_wap2(df):
wap = (df["bid_price2"] * df["ask_size2"] + df["ask_price2"] * df["bid_size2"]) / (
df["bid_size2"] + df["ask_size2"]
)
return wap
def calc_wap3(df):
wap = (df["bid_price2"] * df["bid_size2"] + df["ask_price2"] * df["ask_size2"]) / (
df["bid_size2"] + df["ask_size2"]
)
return wap
def log_return(list_stock_prices):
return np.log(list_stock_prices).diff()
def realized_volatility(series):
return np.sqrt(np.sum(series**2))
def count_unique(series):
return len(np.unique(series))
book_train = pd.read_parquet(data_dir + "book_train.parquet/stock_id=15")
book_train.head()
# ## Main function for preprocessing book data
def preprocessor_book(file_path):
df = pd.read_parquet(file_path)
# calculate return etc
df["wap"] = calc_wap(df)
df["log_return"] = df.groupby("time_id")["wap"].apply(log_return)
df["wap2"] = calc_wap2(df)
df["log_return2"] = df.groupby("time_id")["wap2"].apply(log_return)
df["wap3"] = calc_wap3(df)
df["log_return3"] = df.groupby("time_id")["wap3"].apply(log_return)
df["wap_balance"] = abs(df["wap"] - df["wap2"])
df["price_spread"] = (df["ask_price1"] - df["bid_price1"]) / (
(df["ask_price1"] + df["bid_price1"]) / 2
)
df["bid_spread"] = df["bid_price1"] - df["bid_price2"]
df["ask_spread"] = df["ask_price1"] - df["ask_price2"]
df["total_volume"] = (df["ask_size1"] + df["ask_size2"]) + (
df["bid_size1"] + df["bid_size2"]
)
df["volume_imbalance"] = abs(
(df["ask_size1"] + df["ask_size2"]) - (df["bid_size1"] + df["bid_size2"])
)
# dict for aggregate
create_feature_dict = {
"log_return": [realized_volatility],
"log_return2": [realized_volatility],
"log_return3": [realized_volatility],
"wap_balance": [np.mean],
"price_spread": [np.mean],
"bid_spread": [np.mean],
"ask_spread": [np.mean],
"volume_imbalance": [np.mean],
"total_volume": [np.mean],
"wap": [np.mean],
}
#####groupby / all seconds
df_feature = pd.DataFrame(
df.groupby(["time_id"]).agg(create_feature_dict)
).reset_index()
df_feature.columns = [
"_".join(col) for col in df_feature.columns
] # time_id is changed to time_id_
######groupby / last XX seconds
last_seconds = [300]
for second in last_seconds:
second = 600 - second
df_feature_sec = pd.DataFrame(
df.query(f"seconds_in_bucket >= {second}")
.groupby(["time_id"])
.agg(create_feature_dict)
).reset_index()
df_feature_sec.columns = [
"_".join(col) for col in df_feature_sec.columns
] # time_id is changed to time_id_
df_feature_sec = df_feature_sec.add_suffix("_" + str(second))
df_feature = pd.merge(
df_feature,
df_feature_sec,
how="left",
left_on="time_id_",
right_on=f"time_id__{second}",
)
df_feature = df_feature.drop([f"time_id__{second}"], axis=1)
# create row_id
stock_id = file_path.split("=")[1]
df_feature["row_id"] = df_feature["time_id_"].apply(lambda x: f"{stock_id}-{x}")
df_feature = df_feature.drop(["time_id_"], axis=1)
return df_feature
file_path = data_dir + "book_train.parquet/stock_id=0"
preprocessor_book(file_path)
trade_train = pd.read_parquet(data_dir + "trade_train.parquet/stock_id=0")
trade_train.head(15)
# ## Main function for preprocessing trade data
def preprocessor_trade(file_path):
df = pd.read_parquet(file_path)
df["log_return"] = df.groupby("time_id")["price"].apply(log_return)
aggregate_dictionary = {
"log_return": [realized_volatility],
"seconds_in_bucket": [count_unique],
"size": [np.sum],
"order_count": [np.mean],
}
df_feature = df.groupby("time_id").agg(aggregate_dictionary)
df_feature = df_feature.reset_index()
df_feature.columns = ["_".join(col) for col in df_feature.columns]
######groupby / last XX seconds
last_seconds = [300]
for second in last_seconds:
second = 600 - second
df_feature_sec = (
df.query(f"seconds_in_bucket >= {second}")
.groupby("time_id")
.agg(aggregate_dictionary)
)
df_feature_sec = df_feature_sec.reset_index()
df_feature_sec.columns = ["_".join(col) for col in df_feature_sec.columns]
df_feature_sec = df_feature_sec.add_suffix("_" + str(second))
df_feature = pd.merge(
df_feature,
df_feature_sec,
how="left",
left_on="time_id_",
right_on=f"time_id__{second}",
)
df_feature = df_feature.drop([f"time_id__{second}"], axis=1)
df_feature = df_feature.add_prefix("trade_")
stock_id = file_path.split("=")[1]
df_feature["row_id"] = df_feature["trade_time_id_"].apply(
lambda x: f"{stock_id}-{x}"
)
df_feature = df_feature.drop(["trade_time_id_"], axis=1)
return df_feature
file_path = data_dir + "trade_train.parquet/stock_id=0"
preprocessor_trade(file_path)
# ## Combined preprocessor function
def preprocessor(list_stock_ids, is_train=True):
from joblib import Parallel, delayed # parallel computing to save time
df = pd.DataFrame()
def for_joblib(stock_id):
if is_train:
file_path_book = data_dir + "book_train.parquet/stock_id=" + str(stock_id)
file_path_trade = data_dir + "trade_train.parquet/stock_id=" + str(stock_id)
else:
file_path_book = data_dir + "book_test.parquet/stock_id=" + str(stock_id)
file_path_trade = data_dir + "trade_test.parquet/stock_id=" + str(stock_id)
df_tmp = pd.merge(
preprocessor_book(file_path_book),
preprocessor_trade(file_path_trade),
on="row_id",
how="left",
)
return pd.concat([df, df_tmp])
df = Parallel(n_jobs=-1, verbose=1)(
delayed(for_joblib)(stock_id) for stock_id in list_stock_ids
)
df = pd.concat(df, ignore_index=True)
return df
list_stock_ids = [0, 1]
preprocessor(list_stock_ids, is_train=True)
# ## Training set
train = pd.read_csv(data_dir + "train.csv")
train_ids = train.stock_id.unique()
df_train = preprocessor(list_stock_ids=train_ids, is_train=True)
train["row_id"] = train["stock_id"].astype(str) + "-" + train["time_id"].astype(str)
train = train[["row_id", "target"]]
df_train = train.merge(df_train, on=["row_id"], how="left")
df_train.head()
# ## Test set
test = pd.read_csv(data_dir + "test.csv")
test_ids = test.stock_id.unique()
df_test = preprocessor(list_stock_ids=test_ids, is_train=False)
df_test = test.merge(df_test, on=["row_id"], how="left")
# ## Target encoding by stock_id
from sklearn.model_selection import KFold
# stock_id target encoding
df_train["stock_id"] = df_train["row_id"].apply(lambda x: x.split("-")[0])
df_test["stock_id"] = df_test["row_id"].apply(lambda x: x.split("-")[0])
stock_id_target_mean = df_train.groupby("stock_id")["target"].mean()
df_test["stock_id_target_enc"] = df_test["stock_id"].map(
stock_id_target_mean
) # test_set
# training
tmp = np.repeat(np.nan, df_train.shape[0])
kf = KFold(n_splits=20, shuffle=True, random_state=19911109)
for idx_1, idx_2 in kf.split(df_train):
target_mean = df_train.iloc[idx_1].groupby("stock_id")["target"].mean()
tmp[idx_2] = df_train["stock_id"].iloc[idx_2].map(target_mean)
df_train["stock_id_target_enc"] = tmp
# ## Model Building
df_train.head()
df_test.head()
# ## Multilayer Perceptron
import tensorflow
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense
df_train["stock_id"] = df_train["stock_id"].astype(int)
df_test["stock_id"] = df_test["stock_id"].astype(int)
X = df_train.drop(["row_id", "target"], axis=1)
y = df_train["target"]
def rmspe(y_true, y_pred):
return np.sqrt(np.mean(np.square((y_true - y_pred) / y_true)))
def feval_RMSPE(preds, lgbm_train):
labels = lgbm_train.get_label()
return "RMSPE", round(rmspe(y_true=labels, y_pred=preds), 5), False
params = {
"objective": "rmse",
"metric": "rmse",
"boosting_type": "gbdt",
"early_stopping_rounds": 30,
"learning_rate": 0.01,
"lambda_l1": 1.0,
"lambda_l2": 1.0,
"feature_fraction": 0.8,
"bagging_fraction": 0.8,
}
# ### Cross Validation
from sklearn.model_selection import KFold
kf = KFold(n_splits=25, random_state=19901028, shuffle=True)
oof = pd.DataFrame() # out-of-fold result
models = [] # models
scores = 0.0 # validation score
df_train.head()
for fold, (trn_idx, val_idx) in enumerate(kf.split(X, y)):
print("Fold :", fold + 1)
# create dataset
X_train, y_train = X.loc[trn_idx], y[trn_idx]
X_valid, y_valid = X.loc[val_idx], y[val_idx]
# #RMSPE weight
# weights = 1/np.square(y_train)
# lgbm_train = lgbm.Dataset(X_train,y_train,weight = weights)
# weights = 1/np.square(y_valid)
# lgbm_valid = lgbm.Dataset(X_valid,y_valid,reference = lgbm_train,weight = weights)
# model
model = Sequential()
model.add(Dense(350, activation="relu"))
model.add(Dense(50, activation="relu"))
model.compile(
loss="mse",
optimizer="adam",
metrics=[tensorflow.keras.metrics.RootMeanSquaredError()],
)
model.fit(X_train, y_train, epochs=10, batch_size=100, verbose=1)
# validation
y_pred = model.predict(X_valid)
RMSPE = round(rmspe(y_true=y_valid, y_pred=y_pred[:, 0]), 3)
print(f"Performance of the prediction: , RMSPE: {RMSPE}")
# keep scores and models
scores += RMSPE / 25
models.append(model)
print("*" * 100)
scores
# # Test set
df_test.columns
df_train.columns
y_pred = df_test[["row_id"]]
X_test = df_test.drop(["time_id", "row_id"], axis=1)
X_test
target = np.zeros(len(X_test))
# light gbm models
for model in models:
pred = model.predict(X_test[X_valid.columns])
target += pred[:, 1] / len(models)
y_pred = y_pred.assign(target=target)
y_pred
y_pred.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/595/69595521.ipynb
| null | null |
[{"Id": 69595521, "ScriptId": 19000076, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5723566, "CreationDate": "08/02/2021 02:21:48", "VersionNumber": 1.0, "Title": "OptiverRealizedVolatility v2.0.0", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 333.0, "LinesInsertedFromPrevious": 25.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 308.0, "LinesInsertedFromFork": 25.0, "LinesDeletedFromFork": 24.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 308.0, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import os
import glob
data_dir = "../input/optiver-realized-volatility-prediction/"
# ## Functions for preprocess
def calc_wap(df):
wap = (df["bid_price1"] * df["ask_size1"] + df["ask_price1"] * df["bid_size1"]) / (
df["bid_size1"] + df["ask_size1"]
)
return wap
def calc_wap2(df):
wap = (df["bid_price2"] * df["ask_size2"] + df["ask_price2"] * df["bid_size2"]) / (
df["bid_size2"] + df["ask_size2"]
)
return wap
def calc_wap3(df):
wap = (df["bid_price2"] * df["bid_size2"] + df["ask_price2"] * df["ask_size2"]) / (
df["bid_size2"] + df["ask_size2"]
)
return wap
def log_return(list_stock_prices):
return np.log(list_stock_prices).diff()
def realized_volatility(series):
return np.sqrt(np.sum(series**2))
def count_unique(series):
return len(np.unique(series))
book_train = pd.read_parquet(data_dir + "book_train.parquet/stock_id=15")
book_train.head()
# ## Main function for preprocessing book data
def preprocessor_book(file_path):
df = pd.read_parquet(file_path)
# calculate return etc
df["wap"] = calc_wap(df)
df["log_return"] = df.groupby("time_id")["wap"].apply(log_return)
df["wap2"] = calc_wap2(df)
df["log_return2"] = df.groupby("time_id")["wap2"].apply(log_return)
df["wap3"] = calc_wap3(df)
df["log_return3"] = df.groupby("time_id")["wap3"].apply(log_return)
df["wap_balance"] = abs(df["wap"] - df["wap2"])
df["price_spread"] = (df["ask_price1"] - df["bid_price1"]) / (
(df["ask_price1"] + df["bid_price1"]) / 2
)
df["bid_spread"] = df["bid_price1"] - df["bid_price2"]
df["ask_spread"] = df["ask_price1"] - df["ask_price2"]
df["total_volume"] = (df["ask_size1"] + df["ask_size2"]) + (
df["bid_size1"] + df["bid_size2"]
)
df["volume_imbalance"] = abs(
(df["ask_size1"] + df["ask_size2"]) - (df["bid_size1"] + df["bid_size2"])
)
# dict for aggregate
create_feature_dict = {
"log_return": [realized_volatility],
"log_return2": [realized_volatility],
"log_return3": [realized_volatility],
"wap_balance": [np.mean],
"price_spread": [np.mean],
"bid_spread": [np.mean],
"ask_spread": [np.mean],
"volume_imbalance": [np.mean],
"total_volume": [np.mean],
"wap": [np.mean],
}
#####groupby / all seconds
df_feature = pd.DataFrame(
df.groupby(["time_id"]).agg(create_feature_dict)
).reset_index()
df_feature.columns = [
"_".join(col) for col in df_feature.columns
] # time_id is changed to time_id_
######groupby / last XX seconds
last_seconds = [300]
for second in last_seconds:
second = 600 - second
df_feature_sec = pd.DataFrame(
df.query(f"seconds_in_bucket >= {second}")
.groupby(["time_id"])
.agg(create_feature_dict)
).reset_index()
df_feature_sec.columns = [
"_".join(col) for col in df_feature_sec.columns
] # time_id is changed to time_id_
df_feature_sec = df_feature_sec.add_suffix("_" + str(second))
df_feature = pd.merge(
df_feature,
df_feature_sec,
how="left",
left_on="time_id_",
right_on=f"time_id__{second}",
)
df_feature = df_feature.drop([f"time_id__{second}"], axis=1)
# create row_id
stock_id = file_path.split("=")[1]
df_feature["row_id"] = df_feature["time_id_"].apply(lambda x: f"{stock_id}-{x}")
df_feature = df_feature.drop(["time_id_"], axis=1)
return df_feature
file_path = data_dir + "book_train.parquet/stock_id=0"
preprocessor_book(file_path)
trade_train = pd.read_parquet(data_dir + "trade_train.parquet/stock_id=0")
trade_train.head(15)
# ## Main function for preprocessing trade data
def preprocessor_trade(file_path):
df = pd.read_parquet(file_path)
df["log_return"] = df.groupby("time_id")["price"].apply(log_return)
aggregate_dictionary = {
"log_return": [realized_volatility],
"seconds_in_bucket": [count_unique],
"size": [np.sum],
"order_count": [np.mean],
}
df_feature = df.groupby("time_id").agg(aggregate_dictionary)
df_feature = df_feature.reset_index()
df_feature.columns = ["_".join(col) for col in df_feature.columns]
######groupby / last XX seconds
last_seconds = [300]
for second in last_seconds:
second = 600 - second
df_feature_sec = (
df.query(f"seconds_in_bucket >= {second}")
.groupby("time_id")
.agg(aggregate_dictionary)
)
df_feature_sec = df_feature_sec.reset_index()
df_feature_sec.columns = ["_".join(col) for col in df_feature_sec.columns]
df_feature_sec = df_feature_sec.add_suffix("_" + str(second))
df_feature = pd.merge(
df_feature,
df_feature_sec,
how="left",
left_on="time_id_",
right_on=f"time_id__{second}",
)
df_feature = df_feature.drop([f"time_id__{second}"], axis=1)
df_feature = df_feature.add_prefix("trade_")
stock_id = file_path.split("=")[1]
df_feature["row_id"] = df_feature["trade_time_id_"].apply(
lambda x: f"{stock_id}-{x}"
)
df_feature = df_feature.drop(["trade_time_id_"], axis=1)
return df_feature
file_path = data_dir + "trade_train.parquet/stock_id=0"
preprocessor_trade(file_path)
# ## Combined preprocessor function
def preprocessor(list_stock_ids, is_train=True):
from joblib import Parallel, delayed # parallel computing to save time
df = pd.DataFrame()
def for_joblib(stock_id):
if is_train:
file_path_book = data_dir + "book_train.parquet/stock_id=" + str(stock_id)
file_path_trade = data_dir + "trade_train.parquet/stock_id=" + str(stock_id)
else:
file_path_book = data_dir + "book_test.parquet/stock_id=" + str(stock_id)
file_path_trade = data_dir + "trade_test.parquet/stock_id=" + str(stock_id)
df_tmp = pd.merge(
preprocessor_book(file_path_book),
preprocessor_trade(file_path_trade),
on="row_id",
how="left",
)
return pd.concat([df, df_tmp])
df = Parallel(n_jobs=-1, verbose=1)(
delayed(for_joblib)(stock_id) for stock_id in list_stock_ids
)
df = pd.concat(df, ignore_index=True)
return df
list_stock_ids = [0, 1]
preprocessor(list_stock_ids, is_train=True)
# ## Training set
train = pd.read_csv(data_dir + "train.csv")
train_ids = train.stock_id.unique()
df_train = preprocessor(list_stock_ids=train_ids, is_train=True)
train["row_id"] = train["stock_id"].astype(str) + "-" + train["time_id"].astype(str)
train = train[["row_id", "target"]]
df_train = train.merge(df_train, on=["row_id"], how="left")
df_train.head()
# ## Test set
test = pd.read_csv(data_dir + "test.csv")
test_ids = test.stock_id.unique()
df_test = preprocessor(list_stock_ids=test_ids, is_train=False)
df_test = test.merge(df_test, on=["row_id"], how="left")
# ## Target encoding by stock_id
from sklearn.model_selection import KFold
# stock_id target encoding
df_train["stock_id"] = df_train["row_id"].apply(lambda x: x.split("-")[0])
df_test["stock_id"] = df_test["row_id"].apply(lambda x: x.split("-")[0])
stock_id_target_mean = df_train.groupby("stock_id")["target"].mean()
df_test["stock_id_target_enc"] = df_test["stock_id"].map(
stock_id_target_mean
) # test_set
# training
tmp = np.repeat(np.nan, df_train.shape[0])
kf = KFold(n_splits=20, shuffle=True, random_state=19911109)
for idx_1, idx_2 in kf.split(df_train):
target_mean = df_train.iloc[idx_1].groupby("stock_id")["target"].mean()
tmp[idx_2] = df_train["stock_id"].iloc[idx_2].map(target_mean)
df_train["stock_id_target_enc"] = tmp
# ## Model Building
df_train.head()
df_test.head()
# ## Multilayer Perceptron
import tensorflow
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense
df_train["stock_id"] = df_train["stock_id"].astype(int)
df_test["stock_id"] = df_test["stock_id"].astype(int)
X = df_train.drop(["row_id", "target"], axis=1)
y = df_train["target"]
def rmspe(y_true, y_pred):
return np.sqrt(np.mean(np.square((y_true - y_pred) / y_true)))
def feval_RMSPE(preds, lgbm_train):
labels = lgbm_train.get_label()
return "RMSPE", round(rmspe(y_true=labels, y_pred=preds), 5), False
params = {
"objective": "rmse",
"metric": "rmse",
"boosting_type": "gbdt",
"early_stopping_rounds": 30,
"learning_rate": 0.01,
"lambda_l1": 1.0,
"lambda_l2": 1.0,
"feature_fraction": 0.8,
"bagging_fraction": 0.8,
}
# ### Cross Validation
from sklearn.model_selection import KFold
kf = KFold(n_splits=25, random_state=19901028, shuffle=True)
oof = pd.DataFrame() # out-of-fold result
models = [] # models
scores = 0.0 # validation score
df_train.head()
for fold, (trn_idx, val_idx) in enumerate(kf.split(X, y)):
print("Fold :", fold + 1)
# create dataset
X_train, y_train = X.loc[trn_idx], y[trn_idx]
X_valid, y_valid = X.loc[val_idx], y[val_idx]
# #RMSPE weight
# weights = 1/np.square(y_train)
# lgbm_train = lgbm.Dataset(X_train,y_train,weight = weights)
# weights = 1/np.square(y_valid)
# lgbm_valid = lgbm.Dataset(X_valid,y_valid,reference = lgbm_train,weight = weights)
# model
model = Sequential()
model.add(Dense(350, activation="relu"))
model.add(Dense(50, activation="relu"))
model.compile(
loss="mse",
optimizer="adam",
metrics=[tensorflow.keras.metrics.RootMeanSquaredError()],
)
model.fit(X_train, y_train, epochs=10, batch_size=100, verbose=1)
# validation
y_pred = model.predict(X_valid)
RMSPE = round(rmspe(y_true=y_valid, y_pred=y_pred[:, 0]), 3)
print(f"Performance of the prediction: , RMSPE: {RMSPE}")
# keep scores and models
scores += RMSPE / 25
models.append(model)
print("*" * 100)
scores
# # Test set
df_test.columns
df_train.columns
y_pred = df_test[["row_id"]]
X_test = df_test.drop(["time_id", "row_id"], axis=1)
X_test
target = np.zeros(len(X_test))
# light gbm models
for model in models:
pred = model.predict(X_test[X_valid.columns])
target += pred[:, 1] / len(models)
y_pred = y_pred.assign(target=target)
y_pred
y_pred.to_csv("submission.csv", index=False)
| false | 0 | 3,618 | 0 | 3,618 | 3,618 |
||
69595748
|
<jupyter_start><jupyter_text>automl - efficientdet - efficientnetv2
### Context
I needed the automl repository after they released the EfficientNetV2 model. However, there were some changes I needed to make.
### Content
The automl repository from Google with minor alterations
Kaggle dataset identifier: automl-efficientdet-efficientnetv2
<jupyter_script># Thanks to https://www.kaggle.com/xhlulu/ranzcr-efficientnet-tpu-training
import sys
sys.path.insert(0, "/kaggle/working/automl/efficientnetv2")
import os
import efficientnet.tfkeras as efn
import numpy as np
import pandas as pd
from kaggle_datasets import KaggleDatasets
from sklearn.model_selection import train_test_split
import tensorflow as tf
from sklearn.model_selection import GroupKFold
import math
from kaggle_datasets import KaggleDatasets
import tensorflow as tf
import effnetv2_model
# # units
def choice(p, image1, image2):
rnd = random_float()
image = tf.where(rnd <= p, image1, image2)
return image
def random_float(minval=0.0, maxval=1.0):
rnd = tf.random.uniform([], minval=minval, maxval=maxval, dtype=tf.float32)
return rnd
def mirror_boundary(v, max_v):
# v % (max_v*2.0-2.0) ==> v % (512*2-2) ==> [0..1022]
# [0..1022] - (max_v-1.0) ==> [0..1022] - 511 ==> [-511..511]
# -1.0 * abs([-511..511]) ==> [-511..0]
# [-511..0] + max_v - 1.0 ==> [-511..0] + 511 ==> [0..511]
mirror_v = -1.0 * tf.math.abs(v % (max_v * 2.0 - 2.0) - (max_v - 1.0)) + max_v - 1.0
return mirror_v
def clip_boundary(v, max_v):
clip_v = tf.clip_by_value(v, 0.0, max_v - 1.0)
return clip_v
def interpolate_bilinear(image, map_x, map_y):
def _gather(image, map_x, map_y):
map_stack = tf.stack([map_x, map_y]) # [ 2, height, width ]
map_indices = tf.transpose(map_stack, perm=[1, 2, 0]) # [ height, width, 2 ]
map_indices = tf.cast(map_indices, dtype=tf.int32)
gather_image = tf.gather_nd(image, map_indices)
return gather_image
ll = _gather(image, tf.math.floor(map_x), tf.math.floor(map_y))
lr = _gather(image, tf.math.ceil(map_x), tf.math.floor(map_y))
ul = _gather(image, tf.math.floor(map_x), tf.math.ceil(map_y))
ur = _gather(image, tf.math.ceil(map_x), tf.math.ceil(map_y))
fraction_x = tf.expand_dims(map_x % 1.0, axis=-1) # [h, w, 1]
int_l = (lr - ll) * fraction_x + ll
int_u = (ur - ul) * fraction_x + ul
fraction_y = tf.expand_dims(map_y % 1.0, axis=-1) # [h, w, 1]
interpolate_image = (int_u - int_l) * fraction_y + int_l
return interpolate_image
def remap(image, height, width, map_x, map_y, mode):
assert mode in ("mirror", "constant"), "mode is neither 'mirror' nor 'constant'"
height_f = tf.cast(height, dtype=tf.float32)
width_f = tf.cast(width, dtype=tf.float32)
map_x = tf.reshape(map_x, shape=[height, width])
map_y = tf.reshape(map_y, shape=[height, width])
if mode == "mirror":
b_map_x = mirror_boundary(map_x, width_f)
b_map_y = mirror_boundary(map_y, height_f)
else:
b_map_x = clip_boundary(map_x, width_f)
b_map_y = clip_boundary(map_y, height_f)
image_remap = interpolate_bilinear(image, b_map_x, b_map_y)
if mode == "constant":
map_stack = tf.stack([map_x, map_y])
map_indices = tf.transpose(map_stack, perm=[1, 2, 0])
x_ge_0 = 0.0 <= map_indices[:, :, 0] # [h, w]
x_lt_w = map_indices[:, :, 0] < width_f
y_ge_0 = 0.0 <= map_indices[:, :, 1]
y_lt_h = map_indices[:, :, 1] < height_f
inside_boundary = tf.math.reduce_all(
tf.stack([x_ge_0, x_lt_w, y_ge_0, y_lt_h]), axis=0
) # [h, w]
inside_boundary = inside_boundary[:, :, tf.newaxis] # [h, w, 1]
image_remap = tf.where(inside_boundary, image_remap, 0.0)
return image_remap
# # Optical Distortion
def initUndistortRectifyMap(height, width, k, dx, dy):
height = tf.cast(height, dtype=tf.float32)
width = tf.cast(width, dtype=tf.float32)
f_x = width
f_y = height
c_x = width * 0.5 + dx
c_y = height * 0.5 + dy
f_dash_x = f_x
c_dash_x = (width - 1.0) * 0.5
f_dash_y = f_y
c_dash_y = (height - 1.0) * 0.5
h_rng = tf.range(height, dtype=tf.float32)
w_rng = tf.range(width, dtype=tf.float32)
v, u = tf.meshgrid(h_rng, w_rng)
x = (u - c_dash_x) / f_dash_x
y = (v - c_dash_y) / f_dash_y
x_dash = x
y_dash = y
r_2 = x_dash * x_dash + y_dash * y_dash
r_4 = r_2 * r_2
x_dash_dash = x_dash * (1 + k * r_2 + k * r_4)
y_dash_dash = y_dash * (1 + k * r_2 + k * r_4)
map_x = x_dash_dash * f_x + c_x
map_y = y_dash_dash * f_y + c_y
return map_x, map_y
def OpticalDistortion(distort_limit, shift_limit, p=1.0):
def _do_optical_distortion(image):
k = random_float(-distort_limit, distort_limit)
dx = random_float(-shift_limit, shift_limit)
dy = random_float(-shift_limit, shift_limit)
image_shape = tf.shape(image)
height = image_shape[0]
width = image_shape[1]
map_x, map_y = initUndistortRectifyMap(height, width, k, dx, dy)
aug_image = remap(image, height, width, map_x, map_y, mode="mirror")
return choice(p, aug_image, image)
return _do_optical_distortion
optical_distortion = OpticalDistortion(distort_limit=1.0, shift_limit=0.05, p=0.75)
# # HorizontalFlip
def HorizontalFlip(p):
def _do_horizontal_flip(image):
aug_image = tf.image.flip_left_right(image)
return choice(p, aug_image, image)
return _do_horizontal_flip
horizonflip = HorizontalFlip(0.65)
# # updown_Flip
def Updown_Flip(p):
def _do_up_down_flip(image):
aug_image = tf.image.random_flip_up_down(image)
return choice(p, aug_image, image)
return _do_up_down_flip
updown_flip = Updown_Flip(0.65)
# # RandomContrast
def RandomContrast(lower, upper, p):
def _do_random_contrast(image):
aug_image = tf.image.random_contrast(image, lower, upper)
return choice(p, aug_image, image)
return _do_random_contrast
random_contrast = RandomContrast(lower=0.2, upper=0.8, p=0.75)
# # HeuSaturationValue
def HueSaturationValue(hue_shift_limit, sat_shift_limit, val_shift_limit, p):
def _do_hue_saturation_value(image):
hsv_image = tf.image.rgb_to_hsv(image)
hue_shift = random_float(-hue_shift_limit, hue_shift_limit)
sat_shift = random_float(-sat_shift_limit, sat_shift_limit)
val_shift = random_float(-val_shift_limit, val_shift_limit)
hue_values = (hsv_image[..., :1] + hue_shift) % 1.0
sat_values = tf.clip_by_value(hsv_image[..., 1:2] + sat_shift, 0.0, 1.0)
val_values = tf.clip_by_value(hsv_image[..., 2:] + val_shift, 0.0, 1.0)
hsv_image = tf.concat([hue_values, sat_values, val_values], axis=-1)
aug_image = tf.image.hsv_to_rgb(hsv_image)
return choice(p, aug_image, image)
return _do_hue_saturation_value
hue_saturation_value = HueSaturationValue(
hue_shift_limit=0.2, sat_shift_limit=0.3, val_shift_limit=0.2, p=0.75
)
# # ShiftScaleRotate
def affine_transform(height, width, tx, ty, z, theta):
cx = (width - 1.0) * 0.5
cy = (height - 1.0) * 0.5
center_shift_mat = tf.convert_to_tensor(
[[1.0, 0.0, -cx], [0.0, 1.0, -cy], [0.0, 0.0, 1.0]], dtype=tf.float32
)
trans_mat = center_shift_mat
rot_rad = -2.0 * math.pi * theta / 360.0
roration_mat = tf.convert_to_tensor(
[
[tf.math.cos(rot_rad), tf.math.sin(rot_rad), 0.0],
[-tf.math.sin(rot_rad), tf.math.cos(rot_rad), 0.0],
[0.0, 0.0, 1.0],
],
dtype=tf.float32,
)
trans_mat = tf.linalg.matmul(roration_mat, trans_mat)
shift_mat = tf.convert_to_tensor(
[[1.0, 0.0, cx - tx], [0.0, 1.0, cy - ty], [0.0, 0.0, 1.0]], dtype=tf.float32
)
trans_mat = tf.linalg.matmul(shift_mat, trans_mat)
zoom_mat = tf.convert_to_tensor(
[[1.0 / z, 0.0, 0.0], [0.0, 1.0 / z, 0.0], [0.0, 0.0, 1.0]], dtype=tf.float32
)
trans_mat = tf.linalg.matmul(zoom_mat, trans_mat)
h_rng = tf.range(height, dtype=tf.float32)
w_rng = tf.range(width, dtype=tf.float32)
y, x = tf.meshgrid(h_rng, w_rng)
x = tf.reshape(x, [-1])
y = tf.reshape(y, [-1])
ones = tf.ones_like(x)
coord_mat = tf.stack([x, y, ones])
res_mat = tf.linalg.matmul(trans_mat, coord_mat)
map_x = res_mat[0]
map_y = res_mat[1]
return map_x, map_y
def ShiftScaleRotate(shift_limit, scale_limit, rotate_limit, p):
def _do_shift_scale_rotate(image):
image_shape = tf.shape(image)
height_i = image_shape[0]
width_i = image_shape[1]
height_f = tf.cast(height_i, dtype=tf.float32)
width_f = tf.cast(width_i, dtype=tf.float32)
tx = width_f * random_float(-shift_limit, shift_limit)
ty = height_f * random_float(-shift_limit, shift_limit)
z = random_float(1.0 - scale_limit, 1.0 + scale_limit)
theta = random_float(-rotate_limit, rotate_limit)
map_x, map_y = affine_transform(height_f, width_f, tx, ty, z, theta)
aug_image = remap(image, height_i, width_i, map_x, map_y, mode="constant")
return choice(p, aug_image, image)
return _do_shift_scale_rotate
shift_scale_rotate = ShiftScaleRotate(
shift_limit=0.2, scale_limit=0.3, rotate_limit=30, p=0.75
)
# # cut out
IMSIZE = (224, 240, 260, 300, 380, 456, 528, 600, 768)
image_size = 600
IMS = 7
def randints(shape, minval, maxval):
# maxval+1 to include maxval for the result.
# generated range is [minval, maxval) (maxval is not included)
return tf.random.uniform(
shape=shape, minval=minval, maxval=maxval + 1, dtype=tf.int32
)
def make_range_masks(size, starts, ends):
indice = tf.range(size, dtype=tf.int32)
start_masks = starts[:, tf.newaxis] <= indice[tf.newaxis, :]
end_masks = indice[tf.newaxis, :] <= ends[:, tf.newaxis]
range_masks = start_masks & end_masks
return range_masks
def make_region_mask(tops, lefts, bottoms, rights):
row_masks = make_range_masks(image_size, tops, bottoms)
col_masks = make_range_masks(image_size, lefts, rights)
region_masks = row_masks[:, :, tf.newaxis] & col_masks[:, tf.newaxis, :]
region_mask = tf.math.reduce_any(region_masks, axis=0)
region_mask = region_mask[:, :, tf.newaxis]
return region_mask
def Cutout(num_cuts, mask_factor, p):
def _do_cutout(image):
image_shape = tf.shape(image)
height_i = image_shape[0]
width_i = image_shape[1]
height_f = tf.cast(height_i, dtype=tf.float32)
width_f = tf.cast(width_i, dtype=tf.float32)
cut_h = tf.cast(height_f * mask_factor, dtype=tf.int32)
cut_w = tf.cast(width_f * mask_factor, dtype=tf.int32)
y_centers = randints([num_cuts], 0, image_size - 1)
x_centers = randints([num_cuts], 0, image_size - 1)
tops = tf.math.maximum(y_centers - cut_h // 2, 0)
lefts = tf.math.maximum(x_centers - cut_w // 2, 0)
bottoms = tf.math.minimum(tops + cut_h, height_i - 1)
rights = tf.math.minimum(lefts + cut_w, width_i - 1)
cut_region = make_region_mask(tops, lefts, bottoms, rights)
mask_value = tf.constant(0.0, dtype=tf.float32)
aug_image = tf.where(cut_region, mask_value, image)
return choice(p, aug_image, image)
return _do_cutout
cut_out = Cutout(num_cuts=1, mask_factor=0.4, p=0.75)
# ### MIX UP
AUG_BATCH = 128
def mixup(image, label, PROBABILITY=0.5):
# input image - is a batch of images of size [n,dim,dim,3] not a single image of [dim,dim,3]
# output - a batch of images with mixup applied
DIM = image_size
CLASSES = 4
imgs = []
labs = []
for j in range(AUG_BATCH):
# DO MIXUP WITH PROBABILITY DEFINED ABOVE
P = tf.cast(tf.random.uniform([], 0, 1) <= PROBABILITY, tf.float32)
# CHOOSE RANDOM
k = tf.cast(tf.random.uniform([], 0, AUG_BATCH), tf.int32)
a = tf.random.uniform([], 0, 1) * P # this is beta dist with alpha=1.0
# MAKE MIXUP IMAGE
img1 = image[j,]
img2 = image[k,]
imgs.append((1 - a) * img1 + a * img2)
# MAKE CUTMIX LABEL
if len(label.shape) == 1:
lab1 = tf.one_hot(label[j], CLASSES)
lab2 = tf.one_hot(label[k], CLASSES)
else:
lab1 = label[j,]
lab2 = label[k,]
labs.append((1 - a) * lab1 + a * lab2)
# RESHAPE HACK SO TPU COMPILER KNOWS SHAPE OF OUTPUT TENSOR (maybe use Python typing instead?)
image2 = tf.reshape(tf.stack(imgs), (AUG_BATCH, DIM, DIM, 3))
label2 = tf.reshape(tf.stack(labs), (AUG_BATCH, CLASSES))
return image2, label2
import albumentations
def auto_select_accelerator():
try:
tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
tf.config.experimental_connect_to_cluster(tpu)
tf.tpu.experimental.initialize_tpu_system(tpu)
strategy = tf.distribute.experimental.TPUStrategy(tpu)
print("Running on TPU:", tpu.master())
except ValueError:
strategy = tf.distribute.get_strategy()
print(f"Running on {strategy.num_replicas_in_sync} replicas")
return strategy
def build_decoder(with_labels=True, target_size=(256, 256), ext="jpg"):
def decode(path):
file_bytes = tf.io.read_file(path)
if ext == "png":
img = tf.image.decode_png(file_bytes, channels=3)
elif ext in ["jpg", "jpeg"]:
img = tf.image.decode_jpeg(file_bytes, channels=3)
else:
raise ValueError("Image extension not supported")
img = tf.cast(img, tf.float32) / 255.0
img = tf.image.resize(img, target_size)
return img
def decode_with_labels(path, label):
return decode(path), label
return decode_with_labels if with_labels else decode
def randints(shape, minval, maxval):
# maxval+1 to include maxval for the result.
# generated range is [minval, maxval) (maxval is not included)
return tf.random.uniform(
shape=shape, minval=minval, maxval=maxval + 1, dtype=tf.int32
)
def make_range_masks(size, starts, ends):
indice = tf.range(size, dtype=tf.int32)
start_masks = starts[:, tf.newaxis] <= indice[tf.newaxis, :]
end_masks = indice[tf.newaxis, :] <= ends[:, tf.newaxis]
range_masks = start_masks & end_masks
return range_masks
def make_region_mask(tops, lefts, bottoms, rights):
row_masks = make_range_masks(image_size, tops, bottoms)
col_masks = make_range_masks(image_size, lefts, rights)
region_masks = row_masks[:, :, tf.newaxis] & col_masks[:, tf.newaxis, :]
region_mask = tf.math.reduce_any(region_masks, axis=0)
region_mask = region_mask[:, :, tf.newaxis]
return region_mask
def Cutout(num_cuts, mask_factor, p):
def _do_cutout(image):
image_shape = tf.shape(image)
height_i = image_shape[0]
width_i = image_shape[1]
height_f = tf.cast(height_i, dtype=tf.float32)
width_f = tf.cast(width_i, dtype=tf.float32)
cut_h = tf.cast(height_f * mask_factor, dtype=tf.int32)
cut_w = tf.cast(width_f * mask_factor, dtype=tf.int32)
y_centers = randints([num_cuts], 0, image_size - 1)
x_centers = randints([num_cuts], 0, image_size - 1)
tops = tf.math.maximum(y_centers - cut_h // 2, 0)
lefts = tf.math.maximum(x_centers - cut_w // 2, 0)
bottoms = tf.math.minimum(tops + cut_h, height_i - 1)
rights = tf.math.minimum(lefts + cut_w, width_i - 1)
cut_region = make_region_mask(tops, lefts, bottoms, rights)
mask_value = tf.constant(0.0, dtype=tf.float32)
aug_image = tf.where(cut_region, mask_value, image)
return aug_image
return _do_cutout
def build_augmenter(with_labels=True):
def augment(img):
img = horizonflip(img)
img = updown_flip(img)
img = random_contrast(img)
img = optical_distortion(img)
img = hue_saturation_value(img)
img = shift_scale_rotate(img)
# img = cut_out(img)
return img
def augment_with_labels(img, label):
return augment(img), label
return augment_with_labels if with_labels else augment
def build_dataset(
paths,
labels=None,
bsize=128,
cache=True,
decode_fn=None,
augment_fn=None,
augment=True,
repeat=True,
shuffle=1024,
cache_dir="",
):
if cache_dir != "" and cache is True:
os.makedirs(cache_dir, exist_ok=True)
if decode_fn is None:
decode_fn = build_decoder(labels is not None)
if augment_fn is None:
augment_fn = build_augmenter(labels is not None)
AUTO = tf.data.experimental.AUTOTUNE
slices = paths if labels is None else (paths, labels)
dset = tf.data.Dataset.from_tensor_slices(slices)
dset = dset.map(decode_fn, num_parallel_calls=AUTO)
dset = dset.cache(cache_dir) if cache else dset
dset = dset.map(augment_fn, num_parallel_calls=AUTO) if augment else dset
dset = dset.map(mixup) if augment else dset
dset = dset.repeat() if repeat else dset
dset = dset.shuffle(shuffle) if shuffle else dset
dset = dset.batch(bsize).prefetch(AUTO)
return dset
# ### MIX up
COMPETITION_NAME = "siimcovid19-512-img-png-600-study-png"
strategy = auto_select_accelerator()
BATCH_SIZE = strategy.num_replicas_in_sync * 16
GCS_DS_PATH = KaggleDatasets().get_gcs_path(COMPETITION_NAME)
load_dir = f"/kaggle/input/{COMPETITION_NAME}/"
df = pd.read_csv("../input/siim-covid19-detection/train_study_level.csv")
label_cols = df.columns[1:5]
gkf = GroupKFold(n_splits=5)
df["fold"] = -1
for fold, (train_idx, val_idx) in enumerate(gkf.split(df, groups=df.id.tolist())):
df.loc[val_idx, "fold"] = fold
# # lr
def lrfn(epoch):
LR_START = 0.00001
LR_MAX = 0.00005 * strategy.num_replicas_in_sync
LR_MIN = 0.000005
LR_RAMPUP_EPOCHS = 7
LR_SUSTAIN_EPOCHS = 0
LR_EXP_DECAY = 0.8
if epoch < LR_RAMPUP_EPOCHS:
lr = (LR_MAX - LR_START) / LR_RAMPUP_EPOCHS * epoch + LR_START
elif epoch < LR_RAMPUP_EPOCHS + LR_SUSTAIN_EPOCHS:
lr = LR_MAX
else:
lr = (LR_MAX - LR_MIN) * LR_EXP_DECAY ** (
epoch - LR_RAMPUP_EPOCHS - LR_SUSTAIN_EPOCHS
) + LR_MIN
return lr
lr_schedule = tf.keras.callbacks.LearningRateScheduler(lrfn, verbose=1)
# # EFFICIENTNETv2
for i in range(5):
valid_paths = (
GCS_DS_PATH + "/study/" + df[df["fold"] == i]["id"] + ".png"
) # "/train/"
train_paths = (
GCS_DS_PATH + "/study/" + df[df["fold"] != i]["id"] + ".png"
) # "/train/"
valid_labels = df[df["fold"] == i][label_cols].values
train_labels = df[df["fold"] != i][label_cols].values
IMSIZE = (224, 240, 260, 300, 380, 456, 528, 600, 768)
IMS = 7
decoder = build_decoder(
with_labels=True, target_size=(IMSIZE[IMS], IMSIZE[IMS]), ext="png"
)
test_decoder = build_decoder(
with_labels=False, target_size=(IMSIZE[IMS], IMSIZE[IMS]), ext="png"
)
train_dataset = build_dataset(
train_paths, train_labels, bsize=BATCH_SIZE, decode_fn=decoder
)
valid_dataset = build_dataset(
valid_paths,
valid_labels,
bsize=BATCH_SIZE,
decode_fn=decoder,
repeat=False,
shuffle=False,
augment=False,
)
try:
n_labels = train_labels.shape[1]
except:
n_labels = 1
strategy = auto_select_accelerator()
with strategy.scope():
model = tf.keras.Sequential(
[
efn.EfficientNetB7(
input_shape=(IMSIZE[IMS], IMSIZE[IMS], 3),
weights="imagenet",
include_top=False,
),
tf.keras.layers.GlobalAveragePooling2D(),
tf.keras.layers.Dense(n_labels, activation="softmax"),
]
)
model.compile(
optimizer=tf.keras.optimizers.Adam(),
loss="categorical_crossentropy",
metrics=[tf.keras.metrics.AUC(multi_label=True)],
)
model.summary()
steps_per_epoch = train_paths.shape[0] // BATCH_SIZE
checkpoint = tf.keras.callbacks.ModelCheckpoint(
f"model{i}.h5", save_best_only=True, monitor="val_loss", mode="min"
)
lr_reducer = tf.keras.callbacks.ReduceLROnPlateau(
monitor="val_loss", patience=3, min_lr=1e-6, mode="min"
)
early_stop = tf.keras.callbacks.EarlyStopping(monitor="loss", patience=3)
history = model.fit(
train_dataset,
epochs=30,
verbose=1,
callbacks=[checkpoint, lr_reducer, early_stop],
steps_per_epoch=steps_per_epoch,
validation_data=valid_dataset,
)
hist_df = pd.DataFrame(history.history)
hist_df.to_csv(f"history{i}.csv")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/595/69595748.ipynb
|
automl-efficientdet-efficientnetv2
|
dschettler8845
|
[{"Id": 69595748, "ScriptId": 18175324, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4542962, "CreationDate": "08/02/2021 02:27:02", "VersionNumber": 23.0, "Title": "[siim-covid19]efnb7 train (study)", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 610.0, "LinesInsertedFromPrevious": 2.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 608.0, "LinesInsertedFromFork": 457.0, "LinesDeletedFromFork": 9.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 153.0, "TotalVotes": 0}]
|
[{"Id": 92980469, "KernelVersionId": 69595748, "SourceDatasetVersionId": 2237801}, {"Id": 92980470, "KernelVersionId": 69595748, "SourceDatasetVersionId": 2248688}]
|
[{"Id": 2237801, "DatasetId": 1344553, "DatasourceVersionId": 2279636, "CreatorUserId": 1636313, "LicenseName": "Data files \u00a9 Original Authors", "CreationDate": "05/16/2021 16:53:20", "VersionNumber": 6.0, "Title": "automl - efficientdet - efficientnetv2", "Slug": "automl-efficientdet-efficientnetv2", "Subtitle": "Latest pull from the automl repo. I have made minor updates.", "Description": "### Context\n\nI needed the automl repository after they released the EfficientNetV2 model. However, there were some changes I needed to make.\n\n\n### Content\n\nThe automl repository from Google with minor alterations\n\n### Acknowledgements\n\nThanks to the Google team for releasing such a great model with off-the-shelf performance. Here's to hoping I'm using it correctly.\n\n### Inspiration\n\nI love being able to use new architectures. in tensorflow & Keras.", "VersionNotes": "try again ... this is to to fix brain_automl aliasing/import problem", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1344553, "CreatorUserId": 1636313, "OwnerUserId": 1636313.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2237801.0, "CurrentDatasourceVersionId": 2279636.0, "ForumId": 1363551, "Type": 2, "CreationDate": "05/16/2021 16:17:13", "LastActivityDate": "05/16/2021", "TotalViews": 6424, "TotalDownloads": 357, "TotalVotes": 29, "TotalKernels": 17}]
|
[{"Id": 1636313, "UserName": "dschettler8845", "DisplayName": "Darien Schettler", "RegisterDate": "02/14/2018", "PerformanceTier": 4}]
|
# Thanks to https://www.kaggle.com/xhlulu/ranzcr-efficientnet-tpu-training
import sys
sys.path.insert(0, "/kaggle/working/automl/efficientnetv2")
import os
import efficientnet.tfkeras as efn
import numpy as np
import pandas as pd
from kaggle_datasets import KaggleDatasets
from sklearn.model_selection import train_test_split
import tensorflow as tf
from sklearn.model_selection import GroupKFold
import math
from kaggle_datasets import KaggleDatasets
import tensorflow as tf
import effnetv2_model
# # units
def choice(p, image1, image2):
rnd = random_float()
image = tf.where(rnd <= p, image1, image2)
return image
def random_float(minval=0.0, maxval=1.0):
rnd = tf.random.uniform([], minval=minval, maxval=maxval, dtype=tf.float32)
return rnd
def mirror_boundary(v, max_v):
# v % (max_v*2.0-2.0) ==> v % (512*2-2) ==> [0..1022]
# [0..1022] - (max_v-1.0) ==> [0..1022] - 511 ==> [-511..511]
# -1.0 * abs([-511..511]) ==> [-511..0]
# [-511..0] + max_v - 1.0 ==> [-511..0] + 511 ==> [0..511]
mirror_v = -1.0 * tf.math.abs(v % (max_v * 2.0 - 2.0) - (max_v - 1.0)) + max_v - 1.0
return mirror_v
def clip_boundary(v, max_v):
clip_v = tf.clip_by_value(v, 0.0, max_v - 1.0)
return clip_v
def interpolate_bilinear(image, map_x, map_y):
def _gather(image, map_x, map_y):
map_stack = tf.stack([map_x, map_y]) # [ 2, height, width ]
map_indices = tf.transpose(map_stack, perm=[1, 2, 0]) # [ height, width, 2 ]
map_indices = tf.cast(map_indices, dtype=tf.int32)
gather_image = tf.gather_nd(image, map_indices)
return gather_image
ll = _gather(image, tf.math.floor(map_x), tf.math.floor(map_y))
lr = _gather(image, tf.math.ceil(map_x), tf.math.floor(map_y))
ul = _gather(image, tf.math.floor(map_x), tf.math.ceil(map_y))
ur = _gather(image, tf.math.ceil(map_x), tf.math.ceil(map_y))
fraction_x = tf.expand_dims(map_x % 1.0, axis=-1) # [h, w, 1]
int_l = (lr - ll) * fraction_x + ll
int_u = (ur - ul) * fraction_x + ul
fraction_y = tf.expand_dims(map_y % 1.0, axis=-1) # [h, w, 1]
interpolate_image = (int_u - int_l) * fraction_y + int_l
return interpolate_image
def remap(image, height, width, map_x, map_y, mode):
assert mode in ("mirror", "constant"), "mode is neither 'mirror' nor 'constant'"
height_f = tf.cast(height, dtype=tf.float32)
width_f = tf.cast(width, dtype=tf.float32)
map_x = tf.reshape(map_x, shape=[height, width])
map_y = tf.reshape(map_y, shape=[height, width])
if mode == "mirror":
b_map_x = mirror_boundary(map_x, width_f)
b_map_y = mirror_boundary(map_y, height_f)
else:
b_map_x = clip_boundary(map_x, width_f)
b_map_y = clip_boundary(map_y, height_f)
image_remap = interpolate_bilinear(image, b_map_x, b_map_y)
if mode == "constant":
map_stack = tf.stack([map_x, map_y])
map_indices = tf.transpose(map_stack, perm=[1, 2, 0])
x_ge_0 = 0.0 <= map_indices[:, :, 0] # [h, w]
x_lt_w = map_indices[:, :, 0] < width_f
y_ge_0 = 0.0 <= map_indices[:, :, 1]
y_lt_h = map_indices[:, :, 1] < height_f
inside_boundary = tf.math.reduce_all(
tf.stack([x_ge_0, x_lt_w, y_ge_0, y_lt_h]), axis=0
) # [h, w]
inside_boundary = inside_boundary[:, :, tf.newaxis] # [h, w, 1]
image_remap = tf.where(inside_boundary, image_remap, 0.0)
return image_remap
# # Optical Distortion
def initUndistortRectifyMap(height, width, k, dx, dy):
height = tf.cast(height, dtype=tf.float32)
width = tf.cast(width, dtype=tf.float32)
f_x = width
f_y = height
c_x = width * 0.5 + dx
c_y = height * 0.5 + dy
f_dash_x = f_x
c_dash_x = (width - 1.0) * 0.5
f_dash_y = f_y
c_dash_y = (height - 1.0) * 0.5
h_rng = tf.range(height, dtype=tf.float32)
w_rng = tf.range(width, dtype=tf.float32)
v, u = tf.meshgrid(h_rng, w_rng)
x = (u - c_dash_x) / f_dash_x
y = (v - c_dash_y) / f_dash_y
x_dash = x
y_dash = y
r_2 = x_dash * x_dash + y_dash * y_dash
r_4 = r_2 * r_2
x_dash_dash = x_dash * (1 + k * r_2 + k * r_4)
y_dash_dash = y_dash * (1 + k * r_2 + k * r_4)
map_x = x_dash_dash * f_x + c_x
map_y = y_dash_dash * f_y + c_y
return map_x, map_y
def OpticalDistortion(distort_limit, shift_limit, p=1.0):
def _do_optical_distortion(image):
k = random_float(-distort_limit, distort_limit)
dx = random_float(-shift_limit, shift_limit)
dy = random_float(-shift_limit, shift_limit)
image_shape = tf.shape(image)
height = image_shape[0]
width = image_shape[1]
map_x, map_y = initUndistortRectifyMap(height, width, k, dx, dy)
aug_image = remap(image, height, width, map_x, map_y, mode="mirror")
return choice(p, aug_image, image)
return _do_optical_distortion
optical_distortion = OpticalDistortion(distort_limit=1.0, shift_limit=0.05, p=0.75)
# # HorizontalFlip
def HorizontalFlip(p):
def _do_horizontal_flip(image):
aug_image = tf.image.flip_left_right(image)
return choice(p, aug_image, image)
return _do_horizontal_flip
horizonflip = HorizontalFlip(0.65)
# # updown_Flip
def Updown_Flip(p):
def _do_up_down_flip(image):
aug_image = tf.image.random_flip_up_down(image)
return choice(p, aug_image, image)
return _do_up_down_flip
updown_flip = Updown_Flip(0.65)
# # RandomContrast
def RandomContrast(lower, upper, p):
def _do_random_contrast(image):
aug_image = tf.image.random_contrast(image, lower, upper)
return choice(p, aug_image, image)
return _do_random_contrast
random_contrast = RandomContrast(lower=0.2, upper=0.8, p=0.75)
# # HeuSaturationValue
def HueSaturationValue(hue_shift_limit, sat_shift_limit, val_shift_limit, p):
def _do_hue_saturation_value(image):
hsv_image = tf.image.rgb_to_hsv(image)
hue_shift = random_float(-hue_shift_limit, hue_shift_limit)
sat_shift = random_float(-sat_shift_limit, sat_shift_limit)
val_shift = random_float(-val_shift_limit, val_shift_limit)
hue_values = (hsv_image[..., :1] + hue_shift) % 1.0
sat_values = tf.clip_by_value(hsv_image[..., 1:2] + sat_shift, 0.0, 1.0)
val_values = tf.clip_by_value(hsv_image[..., 2:] + val_shift, 0.0, 1.0)
hsv_image = tf.concat([hue_values, sat_values, val_values], axis=-1)
aug_image = tf.image.hsv_to_rgb(hsv_image)
return choice(p, aug_image, image)
return _do_hue_saturation_value
hue_saturation_value = HueSaturationValue(
hue_shift_limit=0.2, sat_shift_limit=0.3, val_shift_limit=0.2, p=0.75
)
# # ShiftScaleRotate
def affine_transform(height, width, tx, ty, z, theta):
cx = (width - 1.0) * 0.5
cy = (height - 1.0) * 0.5
center_shift_mat = tf.convert_to_tensor(
[[1.0, 0.0, -cx], [0.0, 1.0, -cy], [0.0, 0.0, 1.0]], dtype=tf.float32
)
trans_mat = center_shift_mat
rot_rad = -2.0 * math.pi * theta / 360.0
roration_mat = tf.convert_to_tensor(
[
[tf.math.cos(rot_rad), tf.math.sin(rot_rad), 0.0],
[-tf.math.sin(rot_rad), tf.math.cos(rot_rad), 0.0],
[0.0, 0.0, 1.0],
],
dtype=tf.float32,
)
trans_mat = tf.linalg.matmul(roration_mat, trans_mat)
shift_mat = tf.convert_to_tensor(
[[1.0, 0.0, cx - tx], [0.0, 1.0, cy - ty], [0.0, 0.0, 1.0]], dtype=tf.float32
)
trans_mat = tf.linalg.matmul(shift_mat, trans_mat)
zoom_mat = tf.convert_to_tensor(
[[1.0 / z, 0.0, 0.0], [0.0, 1.0 / z, 0.0], [0.0, 0.0, 1.0]], dtype=tf.float32
)
trans_mat = tf.linalg.matmul(zoom_mat, trans_mat)
h_rng = tf.range(height, dtype=tf.float32)
w_rng = tf.range(width, dtype=tf.float32)
y, x = tf.meshgrid(h_rng, w_rng)
x = tf.reshape(x, [-1])
y = tf.reshape(y, [-1])
ones = tf.ones_like(x)
coord_mat = tf.stack([x, y, ones])
res_mat = tf.linalg.matmul(trans_mat, coord_mat)
map_x = res_mat[0]
map_y = res_mat[1]
return map_x, map_y
def ShiftScaleRotate(shift_limit, scale_limit, rotate_limit, p):
def _do_shift_scale_rotate(image):
image_shape = tf.shape(image)
height_i = image_shape[0]
width_i = image_shape[1]
height_f = tf.cast(height_i, dtype=tf.float32)
width_f = tf.cast(width_i, dtype=tf.float32)
tx = width_f * random_float(-shift_limit, shift_limit)
ty = height_f * random_float(-shift_limit, shift_limit)
z = random_float(1.0 - scale_limit, 1.0 + scale_limit)
theta = random_float(-rotate_limit, rotate_limit)
map_x, map_y = affine_transform(height_f, width_f, tx, ty, z, theta)
aug_image = remap(image, height_i, width_i, map_x, map_y, mode="constant")
return choice(p, aug_image, image)
return _do_shift_scale_rotate
shift_scale_rotate = ShiftScaleRotate(
shift_limit=0.2, scale_limit=0.3, rotate_limit=30, p=0.75
)
# # cut out
IMSIZE = (224, 240, 260, 300, 380, 456, 528, 600, 768)
image_size = 600
IMS = 7
def randints(shape, minval, maxval):
# maxval+1 to include maxval for the result.
# generated range is [minval, maxval) (maxval is not included)
return tf.random.uniform(
shape=shape, minval=minval, maxval=maxval + 1, dtype=tf.int32
)
def make_range_masks(size, starts, ends):
indice = tf.range(size, dtype=tf.int32)
start_masks = starts[:, tf.newaxis] <= indice[tf.newaxis, :]
end_masks = indice[tf.newaxis, :] <= ends[:, tf.newaxis]
range_masks = start_masks & end_masks
return range_masks
def make_region_mask(tops, lefts, bottoms, rights):
row_masks = make_range_masks(image_size, tops, bottoms)
col_masks = make_range_masks(image_size, lefts, rights)
region_masks = row_masks[:, :, tf.newaxis] & col_masks[:, tf.newaxis, :]
region_mask = tf.math.reduce_any(region_masks, axis=0)
region_mask = region_mask[:, :, tf.newaxis]
return region_mask
def Cutout(num_cuts, mask_factor, p):
def _do_cutout(image):
image_shape = tf.shape(image)
height_i = image_shape[0]
width_i = image_shape[1]
height_f = tf.cast(height_i, dtype=tf.float32)
width_f = tf.cast(width_i, dtype=tf.float32)
cut_h = tf.cast(height_f * mask_factor, dtype=tf.int32)
cut_w = tf.cast(width_f * mask_factor, dtype=tf.int32)
y_centers = randints([num_cuts], 0, image_size - 1)
x_centers = randints([num_cuts], 0, image_size - 1)
tops = tf.math.maximum(y_centers - cut_h // 2, 0)
lefts = tf.math.maximum(x_centers - cut_w // 2, 0)
bottoms = tf.math.minimum(tops + cut_h, height_i - 1)
rights = tf.math.minimum(lefts + cut_w, width_i - 1)
cut_region = make_region_mask(tops, lefts, bottoms, rights)
mask_value = tf.constant(0.0, dtype=tf.float32)
aug_image = tf.where(cut_region, mask_value, image)
return choice(p, aug_image, image)
return _do_cutout
cut_out = Cutout(num_cuts=1, mask_factor=0.4, p=0.75)
# ### MIX UP
AUG_BATCH = 128
def mixup(image, label, PROBABILITY=0.5):
# input image - is a batch of images of size [n,dim,dim,3] not a single image of [dim,dim,3]
# output - a batch of images with mixup applied
DIM = image_size
CLASSES = 4
imgs = []
labs = []
for j in range(AUG_BATCH):
# DO MIXUP WITH PROBABILITY DEFINED ABOVE
P = tf.cast(tf.random.uniform([], 0, 1) <= PROBABILITY, tf.float32)
# CHOOSE RANDOM
k = tf.cast(tf.random.uniform([], 0, AUG_BATCH), tf.int32)
a = tf.random.uniform([], 0, 1) * P # this is beta dist with alpha=1.0
# MAKE MIXUP IMAGE
img1 = image[j,]
img2 = image[k,]
imgs.append((1 - a) * img1 + a * img2)
# MAKE CUTMIX LABEL
if len(label.shape) == 1:
lab1 = tf.one_hot(label[j], CLASSES)
lab2 = tf.one_hot(label[k], CLASSES)
else:
lab1 = label[j,]
lab2 = label[k,]
labs.append((1 - a) * lab1 + a * lab2)
# RESHAPE HACK SO TPU COMPILER KNOWS SHAPE OF OUTPUT TENSOR (maybe use Python typing instead?)
image2 = tf.reshape(tf.stack(imgs), (AUG_BATCH, DIM, DIM, 3))
label2 = tf.reshape(tf.stack(labs), (AUG_BATCH, CLASSES))
return image2, label2
import albumentations
def auto_select_accelerator():
try:
tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
tf.config.experimental_connect_to_cluster(tpu)
tf.tpu.experimental.initialize_tpu_system(tpu)
strategy = tf.distribute.experimental.TPUStrategy(tpu)
print("Running on TPU:", tpu.master())
except ValueError:
strategy = tf.distribute.get_strategy()
print(f"Running on {strategy.num_replicas_in_sync} replicas")
return strategy
def build_decoder(with_labels=True, target_size=(256, 256), ext="jpg"):
def decode(path):
file_bytes = tf.io.read_file(path)
if ext == "png":
img = tf.image.decode_png(file_bytes, channels=3)
elif ext in ["jpg", "jpeg"]:
img = tf.image.decode_jpeg(file_bytes, channels=3)
else:
raise ValueError("Image extension not supported")
img = tf.cast(img, tf.float32) / 255.0
img = tf.image.resize(img, target_size)
return img
def decode_with_labels(path, label):
return decode(path), label
return decode_with_labels if with_labels else decode
def randints(shape, minval, maxval):
# maxval+1 to include maxval for the result.
# generated range is [minval, maxval) (maxval is not included)
return tf.random.uniform(
shape=shape, minval=minval, maxval=maxval + 1, dtype=tf.int32
)
def make_range_masks(size, starts, ends):
indice = tf.range(size, dtype=tf.int32)
start_masks = starts[:, tf.newaxis] <= indice[tf.newaxis, :]
end_masks = indice[tf.newaxis, :] <= ends[:, tf.newaxis]
range_masks = start_masks & end_masks
return range_masks
def make_region_mask(tops, lefts, bottoms, rights):
row_masks = make_range_masks(image_size, tops, bottoms)
col_masks = make_range_masks(image_size, lefts, rights)
region_masks = row_masks[:, :, tf.newaxis] & col_masks[:, tf.newaxis, :]
region_mask = tf.math.reduce_any(region_masks, axis=0)
region_mask = region_mask[:, :, tf.newaxis]
return region_mask
def Cutout(num_cuts, mask_factor, p):
def _do_cutout(image):
image_shape = tf.shape(image)
height_i = image_shape[0]
width_i = image_shape[1]
height_f = tf.cast(height_i, dtype=tf.float32)
width_f = tf.cast(width_i, dtype=tf.float32)
cut_h = tf.cast(height_f * mask_factor, dtype=tf.int32)
cut_w = tf.cast(width_f * mask_factor, dtype=tf.int32)
y_centers = randints([num_cuts], 0, image_size - 1)
x_centers = randints([num_cuts], 0, image_size - 1)
tops = tf.math.maximum(y_centers - cut_h // 2, 0)
lefts = tf.math.maximum(x_centers - cut_w // 2, 0)
bottoms = tf.math.minimum(tops + cut_h, height_i - 1)
rights = tf.math.minimum(lefts + cut_w, width_i - 1)
cut_region = make_region_mask(tops, lefts, bottoms, rights)
mask_value = tf.constant(0.0, dtype=tf.float32)
aug_image = tf.where(cut_region, mask_value, image)
return aug_image
return _do_cutout
def build_augmenter(with_labels=True):
def augment(img):
img = horizonflip(img)
img = updown_flip(img)
img = random_contrast(img)
img = optical_distortion(img)
img = hue_saturation_value(img)
img = shift_scale_rotate(img)
# img = cut_out(img)
return img
def augment_with_labels(img, label):
return augment(img), label
return augment_with_labels if with_labels else augment
def build_dataset(
paths,
labels=None,
bsize=128,
cache=True,
decode_fn=None,
augment_fn=None,
augment=True,
repeat=True,
shuffle=1024,
cache_dir="",
):
if cache_dir != "" and cache is True:
os.makedirs(cache_dir, exist_ok=True)
if decode_fn is None:
decode_fn = build_decoder(labels is not None)
if augment_fn is None:
augment_fn = build_augmenter(labels is not None)
AUTO = tf.data.experimental.AUTOTUNE
slices = paths if labels is None else (paths, labels)
dset = tf.data.Dataset.from_tensor_slices(slices)
dset = dset.map(decode_fn, num_parallel_calls=AUTO)
dset = dset.cache(cache_dir) if cache else dset
dset = dset.map(augment_fn, num_parallel_calls=AUTO) if augment else dset
dset = dset.map(mixup) if augment else dset
dset = dset.repeat() if repeat else dset
dset = dset.shuffle(shuffle) if shuffle else dset
dset = dset.batch(bsize).prefetch(AUTO)
return dset
# ### MIX up
COMPETITION_NAME = "siimcovid19-512-img-png-600-study-png"
strategy = auto_select_accelerator()
BATCH_SIZE = strategy.num_replicas_in_sync * 16
GCS_DS_PATH = KaggleDatasets().get_gcs_path(COMPETITION_NAME)
load_dir = f"/kaggle/input/{COMPETITION_NAME}/"
df = pd.read_csv("../input/siim-covid19-detection/train_study_level.csv")
label_cols = df.columns[1:5]
gkf = GroupKFold(n_splits=5)
df["fold"] = -1
for fold, (train_idx, val_idx) in enumerate(gkf.split(df, groups=df.id.tolist())):
df.loc[val_idx, "fold"] = fold
# # lr
def lrfn(epoch):
LR_START = 0.00001
LR_MAX = 0.00005 * strategy.num_replicas_in_sync
LR_MIN = 0.000005
LR_RAMPUP_EPOCHS = 7
LR_SUSTAIN_EPOCHS = 0
LR_EXP_DECAY = 0.8
if epoch < LR_RAMPUP_EPOCHS:
lr = (LR_MAX - LR_START) / LR_RAMPUP_EPOCHS * epoch + LR_START
elif epoch < LR_RAMPUP_EPOCHS + LR_SUSTAIN_EPOCHS:
lr = LR_MAX
else:
lr = (LR_MAX - LR_MIN) * LR_EXP_DECAY ** (
epoch - LR_RAMPUP_EPOCHS - LR_SUSTAIN_EPOCHS
) + LR_MIN
return lr
lr_schedule = tf.keras.callbacks.LearningRateScheduler(lrfn, verbose=1)
# # EFFICIENTNETv2
for i in range(5):
valid_paths = (
GCS_DS_PATH + "/study/" + df[df["fold"] == i]["id"] + ".png"
) # "/train/"
train_paths = (
GCS_DS_PATH + "/study/" + df[df["fold"] != i]["id"] + ".png"
) # "/train/"
valid_labels = df[df["fold"] == i][label_cols].values
train_labels = df[df["fold"] != i][label_cols].values
IMSIZE = (224, 240, 260, 300, 380, 456, 528, 600, 768)
IMS = 7
decoder = build_decoder(
with_labels=True, target_size=(IMSIZE[IMS], IMSIZE[IMS]), ext="png"
)
test_decoder = build_decoder(
with_labels=False, target_size=(IMSIZE[IMS], IMSIZE[IMS]), ext="png"
)
train_dataset = build_dataset(
train_paths, train_labels, bsize=BATCH_SIZE, decode_fn=decoder
)
valid_dataset = build_dataset(
valid_paths,
valid_labels,
bsize=BATCH_SIZE,
decode_fn=decoder,
repeat=False,
shuffle=False,
augment=False,
)
try:
n_labels = train_labels.shape[1]
except:
n_labels = 1
strategy = auto_select_accelerator()
with strategy.scope():
model = tf.keras.Sequential(
[
efn.EfficientNetB7(
input_shape=(IMSIZE[IMS], IMSIZE[IMS], 3),
weights="imagenet",
include_top=False,
),
tf.keras.layers.GlobalAveragePooling2D(),
tf.keras.layers.Dense(n_labels, activation="softmax"),
]
)
model.compile(
optimizer=tf.keras.optimizers.Adam(),
loss="categorical_crossentropy",
metrics=[tf.keras.metrics.AUC(multi_label=True)],
)
model.summary()
steps_per_epoch = train_paths.shape[0] // BATCH_SIZE
checkpoint = tf.keras.callbacks.ModelCheckpoint(
f"model{i}.h5", save_best_only=True, monitor="val_loss", mode="min"
)
lr_reducer = tf.keras.callbacks.ReduceLROnPlateau(
monitor="val_loss", patience=3, min_lr=1e-6, mode="min"
)
early_stop = tf.keras.callbacks.EarlyStopping(monitor="loss", patience=3)
history = model.fit(
train_dataset,
epochs=30,
verbose=1,
callbacks=[checkpoint, lr_reducer, early_stop],
steps_per_epoch=steps_per_epoch,
validation_data=valid_dataset,
)
hist_df = pd.DataFrame(history.history)
hist_df.to_csv(f"history{i}.csv")
| false | 1 | 7,360 | 0 | 7,442 | 7,360 |
||
69595588
|
# to do:
# create a validation set
# try to tune the learning rate and batch size to see if can reduce overfitting
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
from tensorflow.keras import datasets, layers, models
from sklearn.model_selection import train_test_split
from keras.preprocessing import image
from keras import regularizers
from tensorflow import keras
from tensorflow.keras import layers
from keras.layers import Dense
from keras.regularizers import l2
from sklearn.datasets import fetch_openml
fashion_mnist = fetch_openml(data_id=40996, as_frame=True)
fashion_x = fashion_mnist.data
fashion_x
fashion_y = fashion_mnist.target
fashion_y = fashion_y.astype(int)
fashion_x.shape
fashion_y.shape
x_train, x_test, y_train, y_test = train_test_split(
fashion_x, fashion_y, random_state=28, train_size=0.5, stratify=fashion_y
)
x_train, x_test = x_train / 255.0, x_test / 255.0
x_train = x_train.to_numpy()
y_train = y_train.to_numpy()
x_test = x_test.to_numpy()
y_test = y_test.to_numpy()
# # Creating the Baselin CNN Model
model = models.Sequential()
model.add(layers.Conv2D(32, (4, 4), activation="relu", input_shape=(28, 28, 1)))
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Dropout(0.2))
model.add(layers.Flatten())
model.add(layers.Dense(128))
model.add(layers.Dropout(0.2))
model.add(layers.Dense(10, activation="softmax"))
model.summary()
model.compile(
optimizer="adam",
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=["accuracy"],
)
history = model.fit(
x_train.reshape(x_train.shape[0], 28, 28, 1),
y_train,
epochs=10,
batch_size=50,
validation_data=(x_test.reshape(x_test.shape[0], 28, 28, 1), y_test),
)
# Plot training & validation accuracy values
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("Model accuracy")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper left")
plt.show()
# Plot training & validation loss values
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Model loss")
plt.ylabel("Loss")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper left")
plt.show()
# As shown above, the graph tells us that while the accuracy of the model is high, overfitting has occured. As such, for the tuning, I'll be finding out ways to reduce the overfitting with the best accuracy.
# # Tuning
# For the tuning, I decided to see if reducing the learning rate of the model would help with the overfitting problem. Having a high learning can cause the model to diverge.
model = models.Sequential()
model.add(layers.Conv2D(32, (4, 4), activation="relu", input_shape=(28, 28, 1)))
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Dropout(0.2))
model.add(layers.Flatten())
model.add(layers.Dense(128))
model.add(layers.Dropout(0.2))
model.add(layers.Dense(10, activation="softmax"))
opt = keras.optimizers.Adam(learning_rate=0.0001)
model.compile(
optimizer=opt,
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=["accuracy"],
)
history = model.fit(
x_train.reshape(x_train.shape[0], 28, 28, 1),
y_train,
epochs=10,
batch_size=50,
validation_data=(x_test.reshape(x_test.shape[0], 28, 28, 1), y_test),
)
# I decided to try with a learning rate of 0.0001. I wanted to see what effect would occur if I were to set the learning rate really low.
# Plot training & validation accuracy values
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("Model accuracy")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper left")
plt.show()
# Plot training & validation loss values
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Model loss")
plt.ylabel("Loss")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper left")
plt.show()
# As shown above, the model improved a lot compared to the baseline. However, there's still some overfitting occuring. Despite that, this is a good sign and I decided to move on to tuning another hyperparameter: the kernel regularizer. I opted to use the L2 regularizer as it is often used to prevent overfitting. I decided to use the default value of 0.01
model = models.Sequential()
model.add(
layers.Conv2D(
32,
(4, 4),
activation="relu",
input_shape=(28, 28, 1),
kernel_regularizer=l2(0.01),
bias_regularizer=l2(0.01),
)
)
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Dropout(0.2))
model.add(layers.Flatten())
model.add(layers.Dense(128))
model.add(layers.Dropout(0.2))
model.add(layers.Dense(10, activation="softmax"))
opt = keras.optimizers.Adam(learning_rate=0.0001)
model.compile(
optimizer=opt,
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=["accuracy"],
)
history = model.fit(
x_train.reshape(x_train.shape[0], 28, 28, 1),
y_train,
epochs=10,
batch_size=50,
validation_data=(x_test.reshape(x_test.shape[0], 28, 28, 1), y_test),
)
# Based on the documentation, the regularizer would help provide a penalty to the model. As the regularizer is a kernel regularizer, it would provide the penalty to the kernel level.
# Plot training & validation accuracy values
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("Model accuracy")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper left")
plt.show()
# Plot training & validation loss values
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Model loss")
plt.ylabel("Loss")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper left")
plt.show()
# From the graph above, the model improved a lot compared to the previous 2 models. Just by adding in the kernel regularizer and reducing the learning rate, the overfitting was reduced greatly.
model = models.Sequential()
model.add(
layers.Conv2D(
32,
(4, 4),
activation="relu",
input_shape=(28, 28, 1),
kernel_regularizer=l2(0.01),
bias_regularizer=l2(0.01),
)
)
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Dropout(0.2))
model.add(layers.Flatten())
model.add(layers.Dense(128))
model.add(layers.Dropout(0.2))
model.add(layers.Dense(10, activation="softmax"))
opt = keras.optimizers.Adam(learning_rate=0.0001)
model.compile(
optimizer=opt,
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=["accuracy"],
)
history = model.fit(
x_train.reshape(x_train.shape[0], 28, 28, 1),
y_train,
epochs=10,
batch_size=105,
validation_data=(x_test.reshape(x_test.shape[0], 28, 28, 1), y_test),
)
# Plot training & validation accuracy values
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("Model accuracy")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper left")
plt.show()
# Plot training & validation loss values
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Model loss")
plt.ylabel("Loss")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper left")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/595/69595588.ipynb
| null | null |
[{"Id": 69595588, "ScriptId": 18899851, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7933535, "CreationDate": "08/02/2021 02:23:21", "VersionNumber": 6.0, "Title": "Assignment_Part2", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 198.0, "LinesInsertedFromPrevious": 46.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 152.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# to do:
# create a validation set
# try to tune the learning rate and batch size to see if can reduce overfitting
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
from tensorflow.keras import datasets, layers, models
from sklearn.model_selection import train_test_split
from keras.preprocessing import image
from keras import regularizers
from tensorflow import keras
from tensorflow.keras import layers
from keras.layers import Dense
from keras.regularizers import l2
from sklearn.datasets import fetch_openml
fashion_mnist = fetch_openml(data_id=40996, as_frame=True)
fashion_x = fashion_mnist.data
fashion_x
fashion_y = fashion_mnist.target
fashion_y = fashion_y.astype(int)
fashion_x.shape
fashion_y.shape
x_train, x_test, y_train, y_test = train_test_split(
fashion_x, fashion_y, random_state=28, train_size=0.5, stratify=fashion_y
)
x_train, x_test = x_train / 255.0, x_test / 255.0
x_train = x_train.to_numpy()
y_train = y_train.to_numpy()
x_test = x_test.to_numpy()
y_test = y_test.to_numpy()
# # Creating the Baselin CNN Model
model = models.Sequential()
model.add(layers.Conv2D(32, (4, 4), activation="relu", input_shape=(28, 28, 1)))
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Dropout(0.2))
model.add(layers.Flatten())
model.add(layers.Dense(128))
model.add(layers.Dropout(0.2))
model.add(layers.Dense(10, activation="softmax"))
model.summary()
model.compile(
optimizer="adam",
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=["accuracy"],
)
history = model.fit(
x_train.reshape(x_train.shape[0], 28, 28, 1),
y_train,
epochs=10,
batch_size=50,
validation_data=(x_test.reshape(x_test.shape[0], 28, 28, 1), y_test),
)
# Plot training & validation accuracy values
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("Model accuracy")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper left")
plt.show()
# Plot training & validation loss values
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Model loss")
plt.ylabel("Loss")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper left")
plt.show()
# As shown above, the graph tells us that while the accuracy of the model is high, overfitting has occured. As such, for the tuning, I'll be finding out ways to reduce the overfitting with the best accuracy.
# # Tuning
# For the tuning, I decided to see if reducing the learning rate of the model would help with the overfitting problem. Having a high learning can cause the model to diverge.
model = models.Sequential()
model.add(layers.Conv2D(32, (4, 4), activation="relu", input_shape=(28, 28, 1)))
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Dropout(0.2))
model.add(layers.Flatten())
model.add(layers.Dense(128))
model.add(layers.Dropout(0.2))
model.add(layers.Dense(10, activation="softmax"))
opt = keras.optimizers.Adam(learning_rate=0.0001)
model.compile(
optimizer=opt,
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=["accuracy"],
)
history = model.fit(
x_train.reshape(x_train.shape[0], 28, 28, 1),
y_train,
epochs=10,
batch_size=50,
validation_data=(x_test.reshape(x_test.shape[0], 28, 28, 1), y_test),
)
# I decided to try with a learning rate of 0.0001. I wanted to see what effect would occur if I were to set the learning rate really low.
# Plot training & validation accuracy values
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("Model accuracy")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper left")
plt.show()
# Plot training & validation loss values
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Model loss")
plt.ylabel("Loss")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper left")
plt.show()
# As shown above, the model improved a lot compared to the baseline. However, there's still some overfitting occuring. Despite that, this is a good sign and I decided to move on to tuning another hyperparameter: the kernel regularizer. I opted to use the L2 regularizer as it is often used to prevent overfitting. I decided to use the default value of 0.01
model = models.Sequential()
model.add(
layers.Conv2D(
32,
(4, 4),
activation="relu",
input_shape=(28, 28, 1),
kernel_regularizer=l2(0.01),
bias_regularizer=l2(0.01),
)
)
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Dropout(0.2))
model.add(layers.Flatten())
model.add(layers.Dense(128))
model.add(layers.Dropout(0.2))
model.add(layers.Dense(10, activation="softmax"))
opt = keras.optimizers.Adam(learning_rate=0.0001)
model.compile(
optimizer=opt,
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=["accuracy"],
)
history = model.fit(
x_train.reshape(x_train.shape[0], 28, 28, 1),
y_train,
epochs=10,
batch_size=50,
validation_data=(x_test.reshape(x_test.shape[0], 28, 28, 1), y_test),
)
# Based on the documentation, the regularizer would help provide a penalty to the model. As the regularizer is a kernel regularizer, it would provide the penalty to the kernel level.
# Plot training & validation accuracy values
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("Model accuracy")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper left")
plt.show()
# Plot training & validation loss values
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Model loss")
plt.ylabel("Loss")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper left")
plt.show()
# From the graph above, the model improved a lot compared to the previous 2 models. Just by adding in the kernel regularizer and reducing the learning rate, the overfitting was reduced greatly.
model = models.Sequential()
model.add(
layers.Conv2D(
32,
(4, 4),
activation="relu",
input_shape=(28, 28, 1),
kernel_regularizer=l2(0.01),
bias_regularizer=l2(0.01),
)
)
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Dropout(0.2))
model.add(layers.Flatten())
model.add(layers.Dense(128))
model.add(layers.Dropout(0.2))
model.add(layers.Dense(10, activation="softmax"))
opt = keras.optimizers.Adam(learning_rate=0.0001)
model.compile(
optimizer=opt,
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=["accuracy"],
)
history = model.fit(
x_train.reshape(x_train.shape[0], 28, 28, 1),
y_train,
epochs=10,
batch_size=105,
validation_data=(x_test.reshape(x_test.shape[0], 28, 28, 1), y_test),
)
# Plot training & validation accuracy values
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("Model accuracy")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper left")
plt.show()
# Plot training & validation loss values
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Model loss")
plt.ylabel("Loss")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper left")
plt.show()
| false | 0 | 2,498 | 0 | 2,498 | 2,498 |
||
69595958
|
# ## To train an image classifier from CIFAR10
# 1- Load and normalize the CIFAR10 training and test datasets using **torchvision**
# 2- Define a Convolutional Neural Network
# 3- Define a loss function
# 4- Train the network on the training data
# 5- Test the network on the test data
# ## 1- Load and normalize the CIFAR10 training and test datasets using **torchvision**
import torch
import torchvision
import torchvision.transforms as transforms
# 0 - Pre-define tranformations
# transform.normalize(seq of means, seq of stds) does this >>> image = (image - mean) / std
# In This normalization mean, std are passed as 0.5, 0.5, This will normalize the image in the range [-1,1]
# Because >>> The output of torchvision datasets are PILImage images of range [0, 1]. We transform them to Tensors of normalized range [-1, 1]
# For example, the minimum value 0 will be converted to (0-0.5)/0.5=-1, the maximum value of 1 will be converted to (1-0.5)/0.5=1
transform = transforms.Compose(
[transforms.ToTensor(), transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))]
)
batch_size = 4
# 1 - Loading the data
trainset = torchvision.datasets.CIFAR10(
root="./data", train=True, download=True, transform=transform
)
trainloader = torch.utils.data.DataLoader(
trainset, batch_size=batch_size, shuffle=True, num_workers=2
)
testset = torchvision.datasets.CIFAR10(
root="./data", train=False, download=True, transform=transform
)
testloader = torch.utils.data.DataLoader(
testset, batch_size=batch_size, shuffle=False, num_workers=2
)
classes = (
"plane",
"car",
"bird",
"cat",
"deer",
"dog",
"frog",
"horse",
"ship",
"truck",
)
print(
"\n\t\t-------\n1-trainset: {}\n 2-trainloader: {}\n 3-testset: {}\n 4-testloader: {}".format(
trainset, trainloader, testset, testloader
)
)
# ## - Visualizing some of the images
import matplotlib.pyplot as plt
import numpy as np
# functions to show an image
def imshow(img):
img = img / 2 + 0.5 # unnormalize
npimg = img.numpy()
plt.imshow(np.transpose(npimg, (1, 2, 0)))
plt.show()
# get some random training images
dataiter = iter(trainloader)
images, labels = dataiter.next()
# show images
imshow(torchvision.utils.make_grid(images))
# print labels
print(" ".join("%5s" % classes[labels[j]] for j in range(batch_size)))
# ## 2- Defining a CNN
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(3, 6, 5)
self.pool = nn.MaxPool2d(2, 2)
self.conv2 = nn.Conv2d(6, 16, 5)
self.fc1 = nn.Linear(16 * 5 * 5, 120)
self.fc2 = nn.Linear(120, 84)
self.fc3 = nn.Linear(84, 10)
def forward(self, x):
x = self.pool(F.relu(self.conv1(x)))
x = self.pool(F.relu(self.conv2(x)))
x = torch.flatten(x, 1) # flatten all dimensions except batch
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = self.fc3(x)
return x
net = Net()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/595/69595958.ipynb
| null | null |
[{"Id": 69595958, "ScriptId": 18933051, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4767317, "CreationDate": "08/02/2021 02:31:55", "VersionNumber": 2.0, "Title": "image classification", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 98.0, "LinesInsertedFromPrevious": 98.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
| null | null | null | null |
# ## To train an image classifier from CIFAR10
# 1- Load and normalize the CIFAR10 training and test datasets using **torchvision**
# 2- Define a Convolutional Neural Network
# 3- Define a loss function
# 4- Train the network on the training data
# 5- Test the network on the test data
# ## 1- Load and normalize the CIFAR10 training and test datasets using **torchvision**
import torch
import torchvision
import torchvision.transforms as transforms
# 0 - Pre-define tranformations
# transform.normalize(seq of means, seq of stds) does this >>> image = (image - mean) / std
# In This normalization mean, std are passed as 0.5, 0.5, This will normalize the image in the range [-1,1]
# Because >>> The output of torchvision datasets are PILImage images of range [0, 1]. We transform them to Tensors of normalized range [-1, 1]
# For example, the minimum value 0 will be converted to (0-0.5)/0.5=-1, the maximum value of 1 will be converted to (1-0.5)/0.5=1
transform = transforms.Compose(
[transforms.ToTensor(), transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))]
)
batch_size = 4
# 1 - Loading the data
trainset = torchvision.datasets.CIFAR10(
root="./data", train=True, download=True, transform=transform
)
trainloader = torch.utils.data.DataLoader(
trainset, batch_size=batch_size, shuffle=True, num_workers=2
)
testset = torchvision.datasets.CIFAR10(
root="./data", train=False, download=True, transform=transform
)
testloader = torch.utils.data.DataLoader(
testset, batch_size=batch_size, shuffle=False, num_workers=2
)
classes = (
"plane",
"car",
"bird",
"cat",
"deer",
"dog",
"frog",
"horse",
"ship",
"truck",
)
print(
"\n\t\t-------\n1-trainset: {}\n 2-trainloader: {}\n 3-testset: {}\n 4-testloader: {}".format(
trainset, trainloader, testset, testloader
)
)
# ## - Visualizing some of the images
import matplotlib.pyplot as plt
import numpy as np
# functions to show an image
def imshow(img):
img = img / 2 + 0.5 # unnormalize
npimg = img.numpy()
plt.imshow(np.transpose(npimg, (1, 2, 0)))
plt.show()
# get some random training images
dataiter = iter(trainloader)
images, labels = dataiter.next()
# show images
imshow(torchvision.utils.make_grid(images))
# print labels
print(" ".join("%5s" % classes[labels[j]] for j in range(batch_size)))
# ## 2- Defining a CNN
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(3, 6, 5)
self.pool = nn.MaxPool2d(2, 2)
self.conv2 = nn.Conv2d(6, 16, 5)
self.fc1 = nn.Linear(16 * 5 * 5, 120)
self.fc2 = nn.Linear(120, 84)
self.fc3 = nn.Linear(84, 10)
def forward(self, x):
x = self.pool(F.relu(self.conv1(x)))
x = self.pool(F.relu(self.conv2(x)))
x = torch.flatten(x, 1) # flatten all dimensions except batch
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = self.fc3(x)
return x
net = Net()
| false | 0 | 1,026 | 2 | 1,026 | 1,026 |
||
69595207
|
<jupyter_start><jupyter_text>US counties COVID 19 dataset
From the New York Times GITHUB source:
[CSV US counties](About this file Edit https://raw.githubusercontent.com/nytimes/covid-19-data/master/us-counties.csv)
"The New York Times is releasing a series of data files with cumulative counts of coronavirus cases in the United States, at the state and county level, over time. We are compiling this time series data from state and local governments and health departments in an attempt to provide a complete record of the ongoing outbreak.
Since late January, The Times has tracked cases of coronavirus in real time as they were identified after testing. Because of the widespread shortage of testing, however, the data is necessarily limited in the picture it presents of the outbreak.
We have used this data to power our maps and reporting tracking the outbreak, and it is now being made available to the public in response to requests from researchers, scientists and government officials who would like access to the data to better understand the outbreak.
The data begins with the first reported coronavirus case in Washington State on Jan. 21, 2020. We will publish regular updates to the data in this repository.
United States Data
Data on cumulative coronavirus cases and deaths can be found in two files for states and counties.
Each row of data reports cumulative counts based on our best reporting up to the moment we publish an update. We do our best to revise earlier entries in the data when we receive new information."
The specific data here, is the data PER US COUNTY.
The CSV link for counties is: https://raw.githubusercontent.com/nytimes/covid-19-data/master/us-counties.csv
Kaggle dataset identifier: us-counties-covid-19-dataset
<jupyter_code>import pandas as pd
df = pd.read_csv('us-counties-covid-19-dataset/us-counties.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 2502832 entries, 0 to 2502831
Data columns (total 6 columns):
# Column Dtype
--- ------ -----
0 date object
1 county object
2 state object
3 fips float64
4 cases int64
5 deaths float64
dtypes: float64(2), int64(1), object(3)
memory usage: 114.6+ MB
<jupyter_text>Examples:
{
"date": "2020-01-21 00:00:00",
"county": "Snohomish",
"state": "Washington",
"fips": 53061,
"cases": 1,
"deaths": 0
}
{
"date": "2020-01-22 00:00:00",
"county": "Snohomish",
"state": "Washington",
"fips": 53061,
"cases": 1,
"deaths": 0
}
{
"date": "2020-01-23 00:00:00",
"county": "Snohomish",
"state": "Washington",
"fips": 53061,
"cases": 1,
"deaths": 0
}
{
"date": "2020-01-24 00:00:00",
"county": "Cook",
"state": "Illinois",
"fips": 17031,
"cases": 1,
"deaths": 0
}
<jupyter_script># **Hi everyone! So today I'm going to be looking at inaccuracies in the NYT data for Alameda County that question the trustworthiness of their data collection. I am also going to compare this to the graphs the NYT has online when you search up Covid cases for Alameda County.**
# I want to be clear that this is not a reflection of my political ideology and I'm not doing this to prove that COVID is a hoax or anything like that. I'm just doing this because I want people to do their own research on data before believing in something anbecause I want NYT to clear up the disrepancies in the data.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import plotly.offline as py
py.init_notebook_mode(connected=True)
import plotly.graph_objs as go
from plotly.offline import plot, iplot
import plotly.express as px
pd.options.mode.chained_assignment = None
df = pd.read_csv("/kaggle/input/us-counties-covid-19-dataset/us-counties.csv")
df["date"] = pd.to_datetime(df["date"])
df.head()
# There are multiple counties and states being used here but for our problem we're just going to focus on Alameda County in California.
alameda_county = df[(df["state"] == "California") & (df["county"] == "Alameda")]
px.bar(data_frame=alameda_county, x="date", y="cases", color="deaths")
# Note how the data is always increasing. This is because the data is aggregated meaning that each case adds from the previous. The deaths are also aggregated. To unaggregate the data, we will use the .diff() function in pandas.
# Strangely though, notice how towards the beginning of July 2021, the deaths all of the sudden go down and then start slowly increasing again. We will come back to this in a bit, for now lets see the results of unaggregating the data.
alameda_county["cases"] = alameda_county["cases"].diff()
alameda_county["deaths"] = alameda_county["deaths"].diff()
px.bar(data_frame=alameda_county, x="date", y="cases")
# So for the most part, the number of cases when we take the difference seems fine, however you may notice that on July 3rd 2021 there seems to be -85 cases. Let's take a closer look at the data near the day.
px.bar(
data_frame=alameda_county[
(alameda_county["date"].dt.year == 2021)
& (alameda_county["date"].dt.month == 7)
],
x="date",
y="cases",
)
alameda_county[alameda_county["cases"] < 0]
df.iloc[[1478979]]
# So for some reason July 3rd has -85 cases. On NYT's actual graph it says that on July 3rd there were 0 new cases in Alameda county. This is kind of worrying because it affects all the cases past this day and I know this is the same data.
# Lets take a look at the deaths as well to see if their are any disrepancies there.
px.bar(data_frame=alameda_county, x="date", y="deaths")
# Weirdly enough, according the the graph there is data that reaches about -400 deaths, making the result of the data look tiny in comparison. I'm going to graph this again but without that huge negative number so we can get a better idea of how the graph looks.
px.bar(data_frame=alameda_county[alameda_county["deaths"] > -400], x="date", y="deaths")
# There still seems to be multiple points that are negative, let's check out all these points.
alameda_county = alameda_county.reset_index(drop=True)
alameda_county[alameda_county["deaths"] < 0]
# Wow there are a lot of points on here just for Alameda County that have negative deaths. Let's compare it to what NYT actually puts out for the amount of deaths in some of these points.
alameda_county.iloc[[232]]
# On October 19th of 2020 we have -9 deaths.
#
# NYT has the amount of deaths as 0.
alameda_county.iloc[[307]]
# On January 2nd of 2021 we have -1 deaths
# NYT has the amount of deaths as 0.
alameda_county.iloc[[384]]
# On March 20th of 2021 we have -10 deaths
# NYT has the amount of deaths as 0.
alameda_county.iloc[[460]]
# On June 4th of 2021 we have -423 deaths
# NYT has the amount of deaths as 0.
alameda_county.iloc[[492]]
# On July 6th of 2021 we have -2 deaths
# NYT has the amount of deaths as 1.
# I'm noticing a fairly common theme of putting 0 as the amount of deaths if the death counter is negative.
# Again this is fairly worrying because this is real data that NYT uses and that millions of people see the graphs of everyday when looking at Covid cases and deaths, yet just in Alameda County we see multiple disrepancies in the data that really questions the validity of the data NYT has.
# **Just to remind ourselves the magnitude of this problem, the data for deaths is cumulative in the original dataframe which means that everytime there is a new Covid death it adds up from the previous Covid deaths. Yet somehow for multiple dates, something went wrong in the cumulative process causing the amount of deaths to decrease from one day to another which is impossible. But if the deaths at some point decreased from one day to another, then how can we trust any of the data past the negative points because the next dates takes on the deaths from those disrepancies!**
# Another thing that's worrying to note is how for July the amount of deaths is actually 1 as listed by the NYT instead of usually putting down 0 if the amount of deaths is negative. In fact the whole month of July is just weird, going back to our first cumulative graph.
alameda_county_2 = df[(df["state"] == "California") & (df["county"] == "Alameda")]
px.bar(data_frame=alameda_county_2, x="date", y="cases", color="deaths")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/595/69595207.ipynb
|
us-counties-covid-19-dataset
|
fireballbyedimyrnmom
|
[{"Id": 69595207, "ScriptId": 19002440, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5248435, "CreationDate": "08/02/2021 02:15:01", "VersionNumber": 3.0, "Title": "NYT Inaccuracies in Data", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 123.0, "LinesInsertedFromPrevious": 23.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 100.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92979178, "KernelVersionId": 69595207, "SourceDatasetVersionId": 2466445}]
|
[{"Id": 2466445, "DatasetId": 575374, "DatasourceVersionId": 2508903, "CreatorUserId": 4253886, "LicenseName": "Other (specified in description)", "CreationDate": "07/26/2021 22:16:13", "VersionNumber": 280.0, "Title": "US counties COVID 19 dataset", "Slug": "us-counties-covid-19-dataset", "Subtitle": "NYT's github CSV on COVID19 per US counties", "Description": "From the New York Times GITHUB source:\n[CSV US counties](About this file Edit https://raw.githubusercontent.com/nytimes/covid-19-data/master/us-counties.csv)\n\"The New York Times is releasing a series of data files with cumulative counts of coronavirus cases in the United States, at the state and county level, over time. We are compiling this time series data from state and local governments and health departments in an attempt to provide a complete record of the ongoing outbreak.\n\nSince late January, The Times has tracked cases of coronavirus in real time as they were identified after testing. Because of the widespread shortage of testing, however, the data is necessarily limited in the picture it presents of the outbreak.\n\nWe have used this data to power our maps and reporting tracking the outbreak, and it is now being made available to the public in response to requests from researchers, scientists and government officials who would like access to the data to better understand the outbreak.\n\nThe data begins with the first reported coronavirus case in Washington State on Jan. 21, 2020. We will publish regular updates to the data in this repository.\nUnited States Data\n\nData on cumulative coronavirus cases and deaths can be found in two files for states and counties.\n\nEach row of data reports cumulative counts based on our best reporting up to the moment we publish an update. We do our best to revise earlier entries in the data when we receive new information.\"\n\nThe specific data here, is the data PER US COUNTY.\n\nThe CSV link for counties is: https://raw.githubusercontent.com/nytimes/covid-19-data/master/us-counties.csv", "VersionNotes": "Automatic Update 2021-07-26", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 575374, "CreatorUserId": 4253886, "OwnerUserId": 4253886.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 6250084.0, "CurrentDatasourceVersionId": 6329846.0, "ForumId": 589164, "Type": 2, "CreationDate": "03/28/2020 15:04:26", "LastActivityDate": "03/28/2020", "TotalViews": 195793, "TotalDownloads": 21584, "TotalVotes": 446, "TotalKernels": 167}]
|
[{"Id": 4253886, "UserName": "fireballbyedimyrnmom", "DisplayName": "MyrnaMFL", "RegisterDate": "12/26/2019", "PerformanceTier": 1}]
|
# **Hi everyone! So today I'm going to be looking at inaccuracies in the NYT data for Alameda County that question the trustworthiness of their data collection. I am also going to compare this to the graphs the NYT has online when you search up Covid cases for Alameda County.**
# I want to be clear that this is not a reflection of my political ideology and I'm not doing this to prove that COVID is a hoax or anything like that. I'm just doing this because I want people to do their own research on data before believing in something anbecause I want NYT to clear up the disrepancies in the data.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import plotly.offline as py
py.init_notebook_mode(connected=True)
import plotly.graph_objs as go
from plotly.offline import plot, iplot
import plotly.express as px
pd.options.mode.chained_assignment = None
df = pd.read_csv("/kaggle/input/us-counties-covid-19-dataset/us-counties.csv")
df["date"] = pd.to_datetime(df["date"])
df.head()
# There are multiple counties and states being used here but for our problem we're just going to focus on Alameda County in California.
alameda_county = df[(df["state"] == "California") & (df["county"] == "Alameda")]
px.bar(data_frame=alameda_county, x="date", y="cases", color="deaths")
# Note how the data is always increasing. This is because the data is aggregated meaning that each case adds from the previous. The deaths are also aggregated. To unaggregate the data, we will use the .diff() function in pandas.
# Strangely though, notice how towards the beginning of July 2021, the deaths all of the sudden go down and then start slowly increasing again. We will come back to this in a bit, for now lets see the results of unaggregating the data.
alameda_county["cases"] = alameda_county["cases"].diff()
alameda_county["deaths"] = alameda_county["deaths"].diff()
px.bar(data_frame=alameda_county, x="date", y="cases")
# So for the most part, the number of cases when we take the difference seems fine, however you may notice that on July 3rd 2021 there seems to be -85 cases. Let's take a closer look at the data near the day.
px.bar(
data_frame=alameda_county[
(alameda_county["date"].dt.year == 2021)
& (alameda_county["date"].dt.month == 7)
],
x="date",
y="cases",
)
alameda_county[alameda_county["cases"] < 0]
df.iloc[[1478979]]
# So for some reason July 3rd has -85 cases. On NYT's actual graph it says that on July 3rd there were 0 new cases in Alameda county. This is kind of worrying because it affects all the cases past this day and I know this is the same data.
# Lets take a look at the deaths as well to see if their are any disrepancies there.
px.bar(data_frame=alameda_county, x="date", y="deaths")
# Weirdly enough, according the the graph there is data that reaches about -400 deaths, making the result of the data look tiny in comparison. I'm going to graph this again but without that huge negative number so we can get a better idea of how the graph looks.
px.bar(data_frame=alameda_county[alameda_county["deaths"] > -400], x="date", y="deaths")
# There still seems to be multiple points that are negative, let's check out all these points.
alameda_county = alameda_county.reset_index(drop=True)
alameda_county[alameda_county["deaths"] < 0]
# Wow there are a lot of points on here just for Alameda County that have negative deaths. Let's compare it to what NYT actually puts out for the amount of deaths in some of these points.
alameda_county.iloc[[232]]
# On October 19th of 2020 we have -9 deaths.
#
# NYT has the amount of deaths as 0.
alameda_county.iloc[[307]]
# On January 2nd of 2021 we have -1 deaths
# NYT has the amount of deaths as 0.
alameda_county.iloc[[384]]
# On March 20th of 2021 we have -10 deaths
# NYT has the amount of deaths as 0.
alameda_county.iloc[[460]]
# On June 4th of 2021 we have -423 deaths
# NYT has the amount of deaths as 0.
alameda_county.iloc[[492]]
# On July 6th of 2021 we have -2 deaths
# NYT has the amount of deaths as 1.
# I'm noticing a fairly common theme of putting 0 as the amount of deaths if the death counter is negative.
# Again this is fairly worrying because this is real data that NYT uses and that millions of people see the graphs of everyday when looking at Covid cases and deaths, yet just in Alameda County we see multiple disrepancies in the data that really questions the validity of the data NYT has.
# **Just to remind ourselves the magnitude of this problem, the data for deaths is cumulative in the original dataframe which means that everytime there is a new Covid death it adds up from the previous Covid deaths. Yet somehow for multiple dates, something went wrong in the cumulative process causing the amount of deaths to decrease from one day to another which is impossible. But if the deaths at some point decreased from one day to another, then how can we trust any of the data past the negative points because the next dates takes on the deaths from those disrepancies!**
# Another thing that's worrying to note is how for July the amount of deaths is actually 1 as listed by the NYT instead of usually putting down 0 if the amount of deaths is negative. In fact the whole month of July is just weird, going back to our first cumulative graph.
alameda_county_2 = df[(df["state"] == "California") & (df["county"] == "Alameda")]
px.bar(data_frame=alameda_county_2, x="date", y="cases", color="deaths")
|
[{"us-counties-covid-19-dataset/us-counties.csv": {"column_names": "[\"date\", \"county\", \"state\", \"fips\", \"cases\", \"deaths\"]", "column_data_types": "{\"date\": \"object\", \"county\": \"object\", \"state\": \"object\", \"fips\": \"float64\", \"cases\": \"int64\", \"deaths\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 2502832 entries, 0 to 2502831\nData columns (total 6 columns):\n # Column Dtype \n--- ------ ----- \n 0 date object \n 1 county object \n 2 state object \n 3 fips float64\n 4 cases int64 \n 5 deaths float64\ndtypes: float64(2), int64(1), object(3)\nmemory usage: 114.6+ MB\n", "summary": "{\"fips\": {\"count\": 2479154.0, \"mean\": 31399.58357286397, \"std\": 16342.509037015261, \"min\": 1001.0, \"25%\": 19023.0, \"50%\": 30011.0, \"75%\": 46111.0, \"max\": 78030.0}, \"cases\": {\"count\": 2502832.0, \"mean\": 10033.804996899513, \"std\": 47525.21722359815, \"min\": 0.0, \"25%\": 382.0, \"50%\": 1773.0, \"75%\": 5884.0, \"max\": 2908425.0}, \"deaths\": {\"count\": 2445227.0, \"mean\": 161.61002270954802, \"std\": 820.3334694664128, \"min\": 0.0, \"25%\": 6.0, \"50%\": 33.0, \"75%\": 101.0, \"max\": 40267.0}}", "examples": "{\"date\":{\"0\":\"2020-01-21\",\"1\":\"2020-01-22\",\"2\":\"2020-01-23\",\"3\":\"2020-01-24\"},\"county\":{\"0\":\"Snohomish\",\"1\":\"Snohomish\",\"2\":\"Snohomish\",\"3\":\"Cook\"},\"state\":{\"0\":\"Washington\",\"1\":\"Washington\",\"2\":\"Washington\",\"3\":\"Illinois\"},\"fips\":{\"0\":53061.0,\"1\":53061.0,\"2\":53061.0,\"3\":17031.0},\"cases\":{\"0\":1,\"1\":1,\"2\":1,\"3\":1},\"deaths\":{\"0\":0.0,\"1\":0.0,\"2\":0.0,\"3\":0.0}}"}}]
| true | 1 |
<start_data_description><data_path>us-counties-covid-19-dataset/us-counties.csv:
<column_names>
['date', 'county', 'state', 'fips', 'cases', 'deaths']
<column_types>
{'date': 'object', 'county': 'object', 'state': 'object', 'fips': 'float64', 'cases': 'int64', 'deaths': 'float64'}
<dataframe_Summary>
{'fips': {'count': 2479154.0, 'mean': 31399.58357286397, 'std': 16342.509037015261, 'min': 1001.0, '25%': 19023.0, '50%': 30011.0, '75%': 46111.0, 'max': 78030.0}, 'cases': {'count': 2502832.0, 'mean': 10033.804996899513, 'std': 47525.21722359815, 'min': 0.0, '25%': 382.0, '50%': 1773.0, '75%': 5884.0, 'max': 2908425.0}, 'deaths': {'count': 2445227.0, 'mean': 161.61002270954802, 'std': 820.3334694664128, 'min': 0.0, '25%': 6.0, '50%': 33.0, '75%': 101.0, 'max': 40267.0}}
<dataframe_info>
RangeIndex: 2502832 entries, 0 to 2502831
Data columns (total 6 columns):
# Column Dtype
--- ------ -----
0 date object
1 county object
2 state object
3 fips float64
4 cases int64
5 deaths float64
dtypes: float64(2), int64(1), object(3)
memory usage: 114.6+ MB
<some_examples>
{'date': {'0': '2020-01-21', '1': '2020-01-22', '2': '2020-01-23', '3': '2020-01-24'}, 'county': {'0': 'Snohomish', '1': 'Snohomish', '2': 'Snohomish', '3': 'Cook'}, 'state': {'0': 'Washington', '1': 'Washington', '2': 'Washington', '3': 'Illinois'}, 'fips': {'0': 53061.0, '1': 53061.0, '2': 53061.0, '3': 17031.0}, 'cases': {'0': 1, '1': 1, '2': 1, '3': 1}, 'deaths': {'0': 0.0, '1': 0.0, '2': 0.0, '3': 0.0}}
<end_description>
| 1,776 | 0 | 2,661 | 1,776 |
69595878
|
<jupyter_start><jupyter_text>tf_distilroberta_base
In this dataset you can find files for Tensorflow implementation of [DistilRoBERTa base model from HuggingFace repository](https://huggingface.co/distilroberta-base).
Kaggle dataset identifier: tf-distilroberta-base
<jupyter_script>import codecs
import copy
import csv
import gc
from itertools import chain
import os
import pickle
import random
import time
from typing import Dict, List, Tuple, Union
import warnings
import matplotlib.pyplot as plt
from matplotlib.colors import Normalize
import nltk
from nltk.corpus import wordnet
import numpy as np
from sklearn.manifold import TSNE
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler, PowerTransformer
import tensorflow as tf
import tensorflow_addons as tfa
from transformers import AutoConfig, AutoTokenizer, TFAutoModel
print(tf.__version__)
class MaskCalculator(tf.keras.layers.Layer):
def __init__(self, output_dim, **kwargs):
self.output_dim = output_dim
super(MaskCalculator, self).__init__(**kwargs)
def build(self, input_shape):
super(MaskCalculator, self).build(input_shape)
def call(self, inputs, **kwargs):
return tf.keras.backend.permute_dimensions(
x=tf.keras.backend.repeat(
x=tf.keras.backend.cast(
x=tf.keras.backend.greater(x=inputs, y=0), dtype="float32"
),
n=self.output_dim,
),
pattern=(0, 2, 1),
)
def compute_output_shape(self, input_shape):
assert len(input_shape) == 1
shape = list(input_shape)
shape.append(self.output_dim)
return tuple(shape)
class DatasetGen(tf.keras.utils.Sequence):
def __init__(
self,
data: Dict[str, Tuple[List[int], float, float, np.ndarray]],
data_IDs: List[str],
apply_augmentation: bool,
feature_scaler: Pipeline,
token_indices: np.ndarray,
pad_token_id: int,
batch_size: int,
batches_per_epoch: Union[int, None] = None,
):
self.data = copy.deepcopy(data)
self.token_indices = token_indices
self.pad_token_id = pad_token_id
self.batch_size = batch_size
self.batches_per_epoch = batches_per_epoch
self.feature_scaler = feature_scaler
self.apply_augmentation = apply_augmentation
self.pairs = set()
for key1 in data_IDs:
for key2 in data_IDs:
if key1 == key2:
continue
if (key1, key2) not in self.pairs:
self.pairs.add((key1, key2))
self.pairs = sorted(list(self.pairs))
def __len__(self):
if self.batches_per_epoch is None:
return int(np.ceil(len(self.pairs) / float(self.batch_size)))
return self.batches_per_epoch
def __getitem__(self, idx):
x_left = np.zeros(
shape=(self.batch_size, self.token_indices.shape[1]), dtype=np.int32
)
left_features = []
x_right = np.zeros(
shape=(self.batch_size, self.token_indices.shape[1]), dtype=np.int32
)
right_features = []
batch_y = np.zeros((self.batch_size, 1), dtype=np.int32)
if self.batches_per_epoch is None:
batch_start = idx * self.batch_size
batch_end = min(len(self.pairs), batch_start + self.batch_size)
for sample_idx in range(batch_end - batch_start):
left_key, right_key = self.pairs[sample_idx + batch_start]
left_idx = self.data[left_key][0][0]
left_features.append(self.data[left_key][3][0:1])
left_target = self.data[left_key][1]
right_idx = self.data[right_key][0][0]
right_target = self.data[right_key][1]
right_features.append(self.data[right_key][3][0:1])
x_left[sample_idx] = self.token_indices[left_idx]
x_right[sample_idx] = self.token_indices[right_idx]
batch_y[sample_idx, 0] = left_target - right_target
n_pad = self.batch_size - (batch_end - batch_start)
if n_pad > 0:
for sample_idx in range(batch_end - batch_start, self.batch_size):
x_left[sample_idx] = x_left[sample_idx - 1]
x_right[sample_idx] = x_right[sample_idx - 1]
left_features.append(left_features[-1])
right_features.append(right_features[-1])
batch_y[sample_idx, 0] = batch_y[sample_idx - 1, 0]
else:
for sample_idx in range(self.batch_size):
left_key, right_key = random.choice(self.pairs)
if self.apply_augmentation:
p = np.ones((len(self.data[left_key][0]),), dtype=np.float64)
p[0] = max(2.0, p.shape[0] - 1.0)
p /= p.sum()
left_idx_ = np.random.choice(
list(range(len(self.data[left_key][0]))), p=p
)
left_target = np.random.normal(
loc=self.data[left_key][1], scale=self.data[left_key][2]
)
else:
left_idx_ = 0
left_target = self.data[left_key][1]
left_idx = self.data[left_key][0][left_idx_]
left_features.append(
self.data[left_key][3][left_idx_ : (left_idx_ + 1)]
)
if self.apply_augmentation:
p = np.ones((len(self.data[right_key][0]),), dtype=np.float64)
p[0] = max(2.0, p.shape[0] - 1.0)
p /= p.sum()
right_idx_ = np.random.choice(
list(range(len(self.data[right_key][0]))), p=p
)
right_target = np.random.normal(
loc=self.data[right_key][1], scale=self.data[right_key][2]
)
else:
right_idx_ = 0
right_target = self.data[right_key][1]
right_idx = self.data[right_key][0][right_idx_]
right_features.append(
self.data[right_key][3][right_idx_ : (right_idx_ + 1)]
)
x_left[sample_idx] = self.token_indices[left_idx]
x_right[sample_idx] = self.token_indices[right_idx]
batch_y[sample_idx, 0] = left_target - right_target
batch_x = [
x_left,
generate_attention_mask(x_left, self.pad_token_id),
self.feature_scaler.transform(np.vstack(left_features)),
x_right,
generate_attention_mask(x_right, self.pad_token_id),
self.feature_scaler.transform(np.vstack(right_features)),
]
del x_left, x_right
return batch_x, batch_y, None
def generate_attention_mask(token_indices: np.ndarray, padding_id: int) -> np.ndarray:
attention = np.zeros(token_indices.shape, dtype=np.int32)
for sample_idx in range(token_indices.shape[0]):
for token_idx in range(token_indices.shape[1]):
if token_indices[sample_idx, token_idx] == padding_id:
break
attention[sample_idx, token_idx] = 1
return attention
def calc_text_features(texts: List[List[str]], tok: AutoTokenizer) -> np.ndarray:
f = np.zeros((len(texts), 9), dtype=np.float32)
for idx, sentences in enumerate(texts):
f[idx, 0] = len(sentences)
words = []
pure_words = []
for cur_sent in sentences:
words_in_sentence = nltk.word_tokenize(cur_sent)
words += words_in_sentence
pure_words += list(filter(lambda it: it.isalpha(), words_in_sentence))
f[idx, 1] = len(words) / f[idx, 0]
f[idx, 2] = len(pure_words) / f[idx, 0]
f[idx, 3] = len(" ".join(sentences))
f[idx, 4] = len(pure_words)
f[idx, 5] = np.mean([len(w) for w in pure_words])
for w in pure_words:
syllables = tok.tokenize(w.lower())
f[idx, 6] += len(syllables)
f[idx, 7] += sum(map(lambda it: len(it), syllables))
f[idx, 7] /= f[idx, 6]
f[idx, 8] = f[idx, 6] / f[idx, 4]
return f
def load_data_for_training(
fname: str, tok: AutoTokenizer
) -> List[Dict[str, Tuple[List[str], float, float, np.ndarray]]]:
loaded_header = []
id_col_idx = -1
text_col_idx = -1
target_col_idx = -1
std_col_idx = -1
line_idx = 1
data = dict()
set_of_texts = set()
with codecs.open(fname, mode="r", encoding="utf-8") as fp:
data_reader = csv.reader(fp, quotechar='"', delimiter=",")
for row in data_reader:
if len(row) > 0:
err_msg = f"File {fname}: line {line_idx} is wrong!"
if len(loaded_header) == 0:
loaded_header = copy.copy(row)
try:
text_col_idx = loaded_header.index("excerpt")
except:
text_col_idx = -1
if text_col_idx <= 0:
raise ValueError(err_msg + ' Field "excerpt" is not found!')
try:
id_col_idx = loaded_header.index("id")
except:
id_col_idx = -1
if id_col_idx < 0:
raise ValueError(err_msg + ' Field "id" is not found!')
try:
target_col_idx = loaded_header.index("target")
except:
target_col_idx = -1
if target_col_idx < 0:
raise ValueError(err_msg + ' Field "target" is not found!')
try:
std_col_idx = loaded_header.index("standard_error")
except:
std_col_idx = -1
if std_col_idx < 0:
err_msg2 = f'{err_msg} Field "standard_error" is not found!'
raise ValueError(err_msg2)
else:
sample_id = row[id_col_idx]
if sample_id != sample_id.strip():
raise ValueError(err_msg + f" {sample_id} is wrong sample ID!")
if sample_id in data:
err_msg2 = f"{err_msg} {sample_id} is not unique sample ID!"
raise ValueError(err_msg2)
text = row[text_col_idx].replace("\r", "\n")
if len(text) == 0:
raise ValueError(err_msg + f" Text {sample_id} is empty!")
sentences = []
for paragraph in map(lambda it: it.strip(), text.split("\n")):
if len(paragraph) > 0:
sentences += nltk.sent_tokenize(paragraph)
if len(sentences) == 0:
raise ValueError(err_msg + f" Text {sample_id} is empty!")
text = " ".join([cur_sent for cur_sent in sentences])
if text.lower() in set_of_texts:
raise ValueError(err_msg + f" Text {sample_id} is not unique!")
set_of_texts.add(text.lower())
added_texts = [sentences]
try:
target_val = float(row[target_col_idx])
ok = True
except:
target_val = 0.0
ok = False
if not ok:
err_msg2 = err_msg
err_msg2 += (
f" {row[target_col_idx]} is wrong target for "
f"text {sample_id}."
)
raise ValueError(err_msg2)
try:
std_val = float(row[std_col_idx])
ok = std_val > 0.0
except:
std_val = 0.0
ok = False
if not ok:
err_msg2 = err_msg
err_msg2 += (
f" {row[std_col_idx]} is wrong standard error"
f" for text {sample_id}."
)
warnings.warn(err_msg2)
else:
for _ in range(3):
new_augmented_text = []
for cur_sent in sentences:
new_sent = cur_sent.strip()
if len(new_sent) > 0:
new_augmented_text.append(new_sent)
assert len(new_augmented_text) > 0
random.shuffle(new_augmented_text)
new_augmented_text_ = " ".join(new_augmented_text)
if (len(new_augmented_text_) > 0) and (
new_augmented_text_.lower() not in set_of_texts
):
set_of_texts.add(new_augmented_text_.lower())
added_texts.append(new_augmented_text)
del new_augmented_text, new_augmented_text_
data[sample_id] = (
list(map(lambda it: " ".join(it), added_texts)),
target_val,
std_val,
calc_text_features(added_texts, tok),
)
line_idx += 1
return data
def load_data_for_testing(fname: str, tok: AutoTokenizer, batch_size: int):
loaded_header = []
id_col_idx = -1
text_col_idx = -1
target_col_idx = -1
std_col_idx = -1
line_idx = 1
data = dict()
with codecs.open(fname, mode="r", encoding="utf-8") as fp:
data_reader = csv.reader(fp, quotechar='"', delimiter=",")
for row in data_reader:
if len(row) > 0:
err_msg = f"File {fname}: line {line_idx} is wrong!"
if len(loaded_header) == 0:
loaded_header = copy.copy(row)
try:
text_col_idx = loaded_header.index("excerpt")
except:
text_col_idx = -1
if text_col_idx <= 0:
raise ValueError(err_msg + ' Field "excerpt" is not found!')
try:
id_col_idx = loaded_header.index("id")
except:
id_col_idx = -1
if id_col_idx < 0:
raise ValueError(err_msg + ' Field "id" is not found!')
else:
sample_id = row[id_col_idx]
if sample_id != sample_id.strip():
raise ValueError(err_msg + f" {sample_id} is wrong sample ID!")
if sample_id in data:
err_msg2 = f"{err_msg} {sample_id} is not unique sample ID!"
raise ValueError(err_msg2)
text = row[text_col_idx].replace("\n", " ").replace("\r", " ")
text = " ".join(text.split()).strip()
if len(text) == 0:
raise ValueError(err_msg + f" Text {sample_id} is empty!")
features = calc_text_features([nltk.sent_tokenize(text)], tok)
data[sample_id] = (text, features)
if len(data) >= batch_size:
yield data
del data
data = dict()
line_idx += 1
if len(data) > 0:
yield data
def train_feature_scaler(
data: Dict[str, Tuple[List[int], float, float, np.ndarray]]
) -> Pipeline:
features_for_training = []
for sample_id in data:
features_for_training.append(data[sample_id][3])
features_for_training = np.vstack(features_for_training)
scaler = Pipeline(
steps=[("scaler", StandardScaler()), ("transformer", PowerTransformer())]
)
return scaler.fit(features_for_training)
def tokenize_data(
data: Union[
List[Dict[str, Tuple[str, np.ndarray]]],
List[Dict[str, Tuple[List[str], float, float, np.ndarray]]],
],
tokenizer: AutoTokenizer,
max_seq_len: int,
) -> Tuple[
Union[
Dict[str, Tuple[int, np.ndarray]],
Dict[str, Tuple[List[int], float, float, np.ndarray]],
],
np.ndarray,
]:
tokenized_data = dict()
all_tokens_matrix = []
for sample_idx, cur_ID in enumerate(sorted(list(data.keys()))):
if len(data[cur_ID]) == 2:
tokens = tokenizer.tokenize(data[cur_ID][0])
tokenized_data[cur_ID] = (len(all_tokens_matrix), data[cur_ID][1])
token_ids = tokenizer.convert_tokens_to_ids(
[tokenizer.cls_token] + tokens + [tokenizer.sep_token]
)
ndiff = max_seq_len - len(token_ids)
if ndiff > 0:
token_ids += [tokenizer.pad_token_id for _ in range(ndiff)]
elif ndiff < 0:
token_ids = token_ids[:max_seq_len]
all_tokens_matrix.append(token_ids)
else:
text_idx_list = []
for cur_text in data[cur_ID][0]:
tokens = tokenizer.tokenize(cur_text)
token_ids = tokenizer.convert_tokens_to_ids(
[tokenizer.cls_token] + tokens + [tokenizer.sep_token]
)
ndiff = max_seq_len - len(token_ids)
if ndiff > 0:
token_ids += [tokenizer.pad_token_id for _ in range(ndiff)]
elif ndiff < 0:
token_ids = token_ids[:max_seq_len]
text_idx_list.append(len(all_tokens_matrix))
all_tokens_matrix.append(token_ids)
tokenized_data[cur_ID] = (
text_idx_list,
data[cur_ID][1],
data[cur_ID][2],
data[cur_ID][3],
)
return tokenized_data, np.array(all_tokens_matrix, dtype=np.int32)
def print_info_about_data(
data: Union[
List[Dict[str, Tuple[str, np.ndarray]]],
List[Dict[str, Tuple[List[str], float, float, np.ndarray]]],
],
identifiers: List[str],
):
for_training = len(data[identifiers[0]]) == 4
if for_training:
print(f"Number of samples for training is {len(data)}.")
else:
print(f"Number of samples for submission is {len(data)}.")
print("")
print(f"{len(identifiers)} random samples:")
for cur_id in identifiers:
print("")
print(f" Sample {cur_id}")
if for_training:
print(" Text:")
print(f" {data[cur_id][0][0]}")
print(f" Number of augmented texts is {len(data[cur_id][0]) - 1}.")
if (len(data[cur_id][0]) - 1) > 0:
if (len(data[cur_id][0]) - 1) > 1:
print(" 2 augmented texts:")
for augmented in data[cur_id][0][1:3]:
print(f" {augmented}")
else:
print(" Augmented text:")
for augmented in data[cur_id][0][1:2]:
print(f" {augmented}")
print(" Target:")
print(f" {data[cur_id][1]} +- {data[cur_id][2]}")
print(" Features:")
for it in data[cur_id][3].tolist():
print(f" {it}")
else:
print(" Text:")
print(f" {data[cur_id][0]}")
print(" Features:")
print(f" {data[cur_id][1].tolist()[0]}")
def print_info_about_tokenized_data(
data: Union[
Dict[str, Tuple[int, np.ndarray]],
Dict[str, Tuple[List[int], float, float, np.ndarray]],
],
matrix: np.ndarray,
identifiers: List[str],
):
for_training = len(data[identifiers[0]]) == 4
if for_training:
print(f"Number of tokenized samples for training is {len(data)}.")
else:
print(f"Number of tokenized samples for submission is {len(data)}.")
print("")
print(f"{len(identifiers)} random samples:")
for cur_id in identifiers:
print("")
print(f"Sample {cur_id}")
print("")
sample_idx = data[cur_id][0][0]
print(matrix[sample_idx].tolist())
print("")
print(data[cur_id][-1][0].tolist())
print("")
def build_feature_extractor(
bert_name: str, max_seq_len: int, feature_vector_size: int, batch_size: int
) -> Tuple[tf.keras.Model, int]:
transformer_model = TFAutoModel.from_pretrained(
pretrained_model_name_or_path=bert_name, name="BaseTransformer"
)
united_embedding_size = 256
transformer_config = AutoConfig.from_pretrained(bert_name)
united_emb_layer = tf.keras.layers.Dense(
units=united_embedding_size,
input_dim=transformer_config.hidden_size,
activation="tanh",
kernel_initializer=tf.keras.initializers.GlorotNormal(seed=42),
bias_initializer="zeros",
name="UnitedEmbeddingLayer",
)
print("Transformer Configuration")
print("=========================")
print(transformer_config)
tokens_input = tf.keras.layers.Input(
shape=(max_seq_len,),
batch_size=batch_size,
dtype=tf.int32,
name="word_ids_base",
)
attention_input = tf.keras.layers.Input(
shape=(max_seq_len,),
batch_size=batch_size,
dtype=tf.int32,
name="attention_mask_base",
)
features_input = tf.keras.layers.Input(
shape=(feature_vector_size,),
dtype=tf.float32,
batch_size=batch_size,
name="features_base",
)
sequence_output = transformer_model([tokens_input, attention_input])[0]
output_mask = MaskCalculator(
output_dim=transformer_config.hidden_size,
trainable=False,
name="OutMaskCalculator",
)(attention_input)
masked_output = tf.keras.layers.Multiply(name="OutMaskMultiplicator")(
[output_mask, sequence_output]
)
masked_output = tf.keras.layers.Masking(name="OutMasking")(masked_output)
final_output = tf.keras.layers.GlobalAvgPool1D(name="AvePool")(masked_output)
final_output = tf.keras.layers.LayerNormalization(name="Emdedding")(final_output)
final_output = tf.keras.layers.Concatenate(name="Concat")(
[final_output, features_input]
)
final_output = tf.keras.layers.Dropout(rate=0.3, seed=42, name="EmbeddingDropout")(
final_output
)
final_output = united_emb_layer(final_output)
fe_model = tf.keras.Model(
inputs=[tokens_input, attention_input, features_input],
outputs=final_output,
name="FeatureExtractionModel",
)
fe_model.build(
input_shape=[
(batch_size, max_seq_len),
(batch_size, max_seq_len),
(batch_size, feature_vector_size),
]
)
return fe_model, united_embedding_size
def build_twin_regressor(feature_vector_size: int, batch_size: int) -> tf.keras.Model:
left_input = tf.keras.layers.Input(
shape=(feature_vector_size,),
dtype=tf.float32,
batch_size=batch_size,
name="features_left",
)
right_input = tf.keras.layers.Input(
shape=(feature_vector_size,),
dtype=tf.float32,
batch_size=batch_size,
name="features_right",
)
concatenated_features = tf.keras.layers.Concatenate(name="ConcatFeatures")(
[left_input, right_input]
)
regression_layer = tf.keras.layers.Dense(
units=1,
input_dim=feature_vector_size * 2,
activation=None,
kernel_initializer=tf.keras.initializers.GlorotNormal(seed=42),
bias_initializer="zeros",
name="RegressionLayer",
)(concatenated_features)
twin_regression_model = tf.keras.Model(
inputs=[left_input, right_input],
outputs=regression_layer,
name="TwinRegressionModel",
)
twin_regression_model.build(
input_shape=[
(batch_size, feature_vector_size),
(batch_size, feature_vector_size),
]
)
return twin_regression_model
def build_neural_network(
bert_name: str, max_seq_len: int, feature_vector_size: int, batch_size: int
) -> Tuple[tf.keras.Model, tf.keras.Model, tf.keras.Model]:
fe_layer, ft_vec_size = build_feature_extractor(
bert_name, max_seq_len, feature_vector_size, batch_size
)
left_tokens = tf.keras.layers.Input(
shape=(max_seq_len,), batch_size=batch_size, dtype=tf.int32, name="word_ids"
)
left_attention = tf.keras.layers.Input(
shape=(max_seq_len,),
batch_size=batch_size,
dtype=tf.int32,
name="attention_mask",
)
left_features = tf.keras.layers.Input(
shape=(feature_vector_size,),
dtype=tf.float32,
batch_size=batch_size,
name="features",
)
right_tokens = tf.keras.layers.Input(
shape=(max_seq_len,),
batch_size=batch_size,
dtype=tf.int32,
name="right_word_ids",
)
right_attention = tf.keras.layers.Input(
shape=(max_seq_len,),
batch_size=batch_size,
dtype=tf.int32,
name="right_attention_mask",
)
right_features = tf.keras.layers.Input(
shape=(feature_vector_size,),
dtype=tf.float32,
batch_size=batch_size,
name="right_features",
)
left_output = fe_layer([left_tokens, left_attention, left_features])
right_output = fe_layer([right_tokens, right_attention, right_features])
regression_model = build_twin_regressor(ft_vec_size, batch_size)
regression_layer = regression_model([left_output, right_output])
siamese_model = tf.keras.Model(
inputs=[
left_tokens,
left_attention,
left_features,
right_tokens,
right_attention,
right_features,
],
outputs=regression_layer,
name="SiameseModel",
)
radam = tfa.optimizers.RectifiedAdam(learning_rate=1e-5)
ranger = tfa.optimizers.Lookahead(radam, sync_period=6, slow_step_size=0.5)
siamese_model.compile(optimizer=ranger, loss=tf.keras.losses.MeanSquaredError())
return siamese_model, fe_layer, regression_model
def show_minibatch(X: List[np.ndarray], y: np.ndarray):
assert len(X) == 6
print("")
print("X1")
for it in X[0].tolist():
print(it)
print("")
print("X2")
for it in X[1].tolist():
print(it)
print("")
print("X3")
for it in X[2].tolist():
print(it)
print("")
print("X4")
for it in X[3].tolist():
print(it)
print("")
print("X5")
for it in X[4].tolist():
print(it)
print("X6")
for it in X[5].tolist():
print(it)
print("")
print("y")
for it in y.tolist():
print(it)
def show_tsne(
fe: tf.keras.Model,
batch_size: int,
data: Dict[str, Tuple[List[int], float, float, np.ndarray]],
feature_scaler: Pipeline,
token_matrix: np.ndarray,
identifiers: List[str],
pad_id: int,
title: str,
figure_id: int,
):
indices = list(map(lambda it: data[it][0][0], identifiers))
colors = np.array(list(map(lambda it: data[it][1], identifiers)), dtype=np.float64)
area = np.array(list(map(lambda it: data[it][2], identifiers)), dtype=np.float64)
area /= np.max(area)
area *= 10.0
area = np.power(area, 2)
texts = token_matrix[indices]
src_features = np.vstack(list(map(lambda it: data[it][3][0:1], identifiers)))
assert src_features.shape[0] == texts.shape[0]
ndiff = texts.shape[0] % batch_size
if ndiff > 0:
last_text_idx = texts.shape[0] - 1
texts = np.vstack(
[texts]
+ [
texts[last_text_idx : (last_text_idx + 1)]
for _ in range(batch_size - ndiff)
]
)
src_features = np.vstack(
[src_features]
+ [
src_features[last_text_idx : (last_text_idx + 1)]
for _ in range(batch_size - ndiff)
]
)
attentions = generate_attention_mask(texts, pad_id)
assert texts.shape[0] % batch_size == 0, f"{texts.shape[0] % batch_size}"
features = fe.predict(
[texts, attentions, feature_scaler.transform(src_features)],
batch_size=batch_size,
)
features = features[: len(indices)]
projected_features = TSNE(n_components=2, n_jobs=-1).fit_transform(features)
fig = plt.figure(figure_id, figsize=(11, 11))
plt.scatter(
x=projected_features[:, 0],
y=projected_features[:, 1],
marker="o",
cmap=plt.cm.get_cmap("jet"),
s=area,
c=colors,
norm=Normalize(vmin=np.min(colors), vmax=np.max(colors)),
)
plt.title("t-SNE projections of texts " + title)
plt.colorbar()
plt.show()
def show_training_process(
history: tf.keras.callbacks.History, metric_name: str, figure_id: int
):
val_metric_name = "val_" + metric_name
possible_metrics = list(history.history.keys())
if metric_name not in history.history:
err_msg = f'The metric "{metric_name}" is not found!'
err_msg += f" Available metrics are: {possible_metrics}."
raise ValueError(err_msg)
fig = plt.figure(figure_id, figsize=(7, 7))
metric_values = history.history[metric_name]
plt.plot(
list(range(len(metric_values))),
metric_values,
label="Training {0}".format(metric_name),
)
if val_metric_name in history.history:
val_metric_values = history.history["val_" + metric_name]
assert len(metric_values) == len(val_metric_values)
plt.plot(
list(range(len(val_metric_values))),
val_metric_values,
label="Validation {0}".format(metric_name),
)
plt.xlabel("Epochs")
plt.ylabel(metric_name)
plt.title("Training process")
plt.legend(loc="best")
plt.show()
def generate_new_trainset(
fe: tf.keras.Model,
feature_scaler: Pipeline,
batch_size: int,
data: Dict[str, Tuple[List[int], float, float]],
token_matrix: np.ndarray,
pad_id: int,
identifiers: List[str],
) -> Tuple[np.ndarray, np.ndarray]:
indices = list(map(lambda it: data[it][0][0], identifiers))
texts = token_matrix[indices]
src_features = np.vstack(list(map(lambda it: data[it][3][0:1], identifiers)))
targets = np.array(list(map(lambda it: data[it][1], identifiers)), dtype=np.float64)
assert texts.shape[0] == src_features.shape[0]
ndiff = texts.shape[0] % batch_size
if ndiff > 0:
last_text_idx = texts.shape[0] - 1
texts = np.vstack(
[texts]
+ [
texts[last_text_idx : (last_text_idx + 1)]
for _ in range(batch_size - ndiff)
]
)
src_features = np.vstack(
[src_features]
+ [
src_features[last_text_idx : (last_text_idx + 1)]
for _ in range(batch_size - ndiff)
]
)
attentions = generate_attention_mask(texts, pad_id)
assert texts.shape[0] % batch_size == 0, f"{texts.shape[0] % batch_size}"
target_features = fe.predict(
[texts, attentions, feature_scaler.transform(src_features)],
batch_size=batch_size,
)
assert target_features.shape[1] > 1
target_features = target_features[: len(identifiers)]
return target_features, targets
def calculate_dist_matrix(y: np.ndarray) -> np.ndarray:
assert len(y.shape) == 1
assert y.shape[0] > 1
d = np.zeros((y.shape[0], y.shape[0]), dtype=np.float32)
for idx1 in range(y.shape[0]):
for idx2 in range(y.shape[0]):
diff = y[idx1] - y[idx2]
d[idx1, idx2] = np.sqrt(diff * diff)
return d
def select_train_samples(y: np.ndarray, dist_matrix: np.ndarray, n: int) -> List[int]:
assert len(y.shape) == 1
assert len(dist_matrix.shape) == 2
assert dist_matrix.shape[0] == y.shape[0]
assert dist_matrix.shape[1] == dist_matrix.shape[0]
assert n < y.shape[0]
indices_of_samples = list(range(y.shape[0]))
selected = {np.random.choice(indices_of_samples)}
for _ in range(n - 1):
indices_of_samples = sorted(list(set(indices_of_samples) - selected))
p = [dist_matrix[idx, list(selected)].mean() for idx in indices_of_samples]
p = np.array(p, dtype=np.float64)
p /= p.sum()
selected.add(np.random.choice(indices_of_samples, p=p))
return sorted(list(selected))
def do_predictions(
fe: tf.keras.Model,
regressor: tf.keras.Model,
feature_scaler: Pipeline,
batch_size: int,
data_for_anchors: Tuple[np.ndarray, np.ndarray],
dist_matrix: np.ndarray,
data: Union[Dict[str, int], Dict[str, Tuple[List[int], float, float]]],
token_matrix: np.ndarray,
pad_id: int,
identifiers: List[str] = None,
) -> Dict[str, Tuple[float, float]]:
if identifiers is None:
identifiers_ = sorted(list(data.keys()))
else:
identifiers_ = sorted(identifiers)
indices = list(
map(
lambda it: data[it][0] if len(data[it]) == 2 else data[it][0][0],
identifiers_,
)
)
texts = token_matrix[indices]
src_features = np.vstack(
list(
map(
lambda it: data[it][1] if len(data[it]) == 2 else data[it][3][0:1],
identifiers_,
)
)
)
assert texts.shape[0] == src_features.shape[0]
ndiff = texts.shape[0] % batch_size
if ndiff > 0:
last_text_idx = texts.shape[0] - 1
texts = np.vstack(
[texts]
+ [
texts[last_text_idx : (last_text_idx + 1)]
for _ in range(batch_size - ndiff)
]
)
src_features = np.vstack(
[src_features]
+ [
src_features[last_text_idx : (last_text_idx + 1)]
for _ in range(batch_size - ndiff)
]
)
attentions = generate_attention_mask(texts, pad_id)
assert texts.shape[0] % batch_size == 0, f"{texts.shape[0] % batch_size}"
target_features = fe.predict(
[texts, attentions, feature_scaler.transform(src_features)],
batch_size=batch_size,
)
assert target_features.shape[1] > 1
assert target_features.shape[1] == data_for_anchors[0].shape[1]
assert target_features.shape[0] >= len(indices)
target_features = target_features[0 : len(indices)]
selected_inputs = []
predicted_features = []
selected_targets = []
n_selected = batch_size
while n_selected < 8:
n_selected += batch_size
for sample_idx, cur_id in enumerate(identifiers_):
selected_indices_for_training = select_train_samples(
y=data_for_anchors[1], dist_matrix=dist_matrix, n=n_selected
)
selected_inputs.append(data_for_anchors[0][selected_indices_for_training])
selected_targets.append(data_for_anchors[1][selected_indices_for_training])
predicted_features.append(
np.full(
fill_value=target_features[sample_idx],
shape=(n_selected, target_features.shape[1]),
)
)
selected_inputs = np.vstack(selected_inputs)
predicted_features = np.vstack(predicted_features)
selected_targets = np.concatenate(selected_targets)
prediction_diff = regressor.predict(
[selected_inputs, predicted_features], batch_size=batch_size
).reshape(selected_targets.shape)
predictions = dict()
for sample_idx, cur_id in enumerate(identifiers_):
start_pos = sample_idx * n_selected
end_pos = start_pos + n_selected
instant_predictions = (
selected_targets[start_pos:end_pos] - prediction_diff[start_pos:end_pos]
)
predictions[cur_id] = (
np.mean(instant_predictions),
np.std(instant_predictions),
)
return predictions
random.seed(42)
np.random.seed(42)
tf.random.set_seed(42)
MAX_TEXT_LEN = 512
PRETRAINED_BERT = "/kaggle/input/tf-distilroberta-base"
MINIBATCH_SIZE = 8
DATA_DIR = "/kaggle/input/commonlitreadabilityprize"
MODEL_DIR = "/kaggle/working"
print(f"{DATA_DIR} {os.path.isdir(DATA_DIR)}")
print(f"{MODEL_DIR} {os.path.isdir(MODEL_DIR)}")
trainset_name = os.path.join(DATA_DIR, "train.csv")
print(f"{trainset_name} {os.path.isfile(trainset_name)}")
testset_name = os.path.join(DATA_DIR, "test.csv")
print(f"{testset_name} {os.path.isfile(testset_name)}")
submission_name = os.path.join(MODEL_DIR, "submission.csv")
print(f"{submission_name} {os.path.isfile(submission_name)}")
fe_model_name = os.path.join(MODEL_DIR, "fe_nn.h5")
regression_model_name = os.path.join(MODEL_DIR, "regression_nn.h5")
scaler_name = os.path.join(MODEL_DIR, "feature_scaler.pkl")
figure_identifier = 1
pretrained_tokenizer = AutoTokenizer.from_pretrained(PRETRAINED_BERT)
print(f"Vocabulary size is {pretrained_tokenizer.vocab_size}.")
data_for_training = load_data_for_training(trainset_name, pretrained_tokenizer)
assert len(data_for_training) > 100
all_IDs = sorted(list(data_for_training.keys()))
selected_IDs_for_training = random.sample(population=all_IDs, k=3)
print_info_about_data(data_for_training, selected_IDs_for_training)
labels_for_training, tokens_for_training = tokenize_data(
data=data_for_training, tokenizer=pretrained_tokenizer, max_seq_len=MAX_TEXT_LEN
)
print_info_about_tokenized_data(
data=labels_for_training,
matrix=tokens_for_training,
identifiers=selected_IDs_for_training,
)
text_feature_scaler = train_feature_scaler(labels_for_training)
with open(scaler_name, "wb") as scaler_fp:
pickle.dump(text_feature_scaler, scaler_fp)
random.shuffle(all_IDs)
n_train_size = int(round(len(all_IDs) * 0.9))
n_val_size = int(round(len(all_IDs) * 0.025))
IDs_for_training = all_IDs[:n_train_size]
IDs_for_validation = all_IDs[n_train_size : (n_train_size + n_val_size)]
IDs_for_final_testing = all_IDs[(n_train_size + n_val_size) :]
datagen_for_validation = DatasetGen(
data=labels_for_training,
data_IDs=IDs_for_validation,
token_indices=tokens_for_training,
pad_token_id=pretrained_tokenizer.pad_token_id,
batch_size=MINIBATCH_SIZE,
apply_augmentation=False,
feature_scaler=text_feature_scaler,
)
n_batches_per_validset = len(datagen_for_validation)
print(f"Mini-batches per validation set is {n_batches_per_validset}.")
X_, y_, _ = datagen_for_validation[0]
show_minibatch(X_, y_)
n_batches_per_epoch = n_batches_per_validset * 3
datagen_for_training = DatasetGen(
data=labels_for_training,
data_IDs=IDs_for_training,
token_indices=tokens_for_training,
pad_token_id=pretrained_tokenizer.pad_token_id,
batch_size=MINIBATCH_SIZE,
batches_per_epoch=n_batches_per_epoch,
apply_augmentation=True,
feature_scaler=text_feature_scaler,
)
X_, y_, _ = datagen_for_training[0]
show_minibatch(X_, y_)
model_for_training, fe_model, model_for_regression = build_neural_network(
bert_name=PRETRAINED_BERT,
max_seq_len=MAX_TEXT_LEN,
feature_vector_size=text_feature_scaler.named_steps["scaler"].scale_.shape[0],
batch_size=MINIBATCH_SIZE,
)
model_for_training.summary()
model_for_regression.summary()
fe_model.summary()
show_tsne(
fe=fe_model,
batch_size=MINIBATCH_SIZE,
feature_scaler=text_feature_scaler,
data=labels_for_training,
token_matrix=tokens_for_training,
identifiers=IDs_for_validation + IDs_for_final_testing,
pad_id=pretrained_tokenizer.pad_token_id,
title="before training",
figure_id=figure_identifier,
)
figure_identifier += 1
anchor_data = generate_new_trainset(
fe=fe_model,
feature_scaler=text_feature_scaler,
batch_size=MINIBATCH_SIZE,
data=labels_for_training,
token_matrix=tokens_for_training,
pad_id=pretrained_tokenizer.pad_token_id,
identifiers=IDs_for_training,
)
anchor_distances = calculate_dist_matrix(anchor_data[1])
start_time = time.time()
predictions_for_validation = do_predictions(
fe=fe_model,
regressor=model_for_regression,
feature_scaler=text_feature_scaler,
batch_size=MINIBATCH_SIZE,
data_for_anchors=anchor_data,
dist_matrix=anchor_distances,
data=labels_for_training,
token_matrix=tokens_for_training,
pad_id=pretrained_tokenizer.pad_token_id,
identifiers=IDs_for_validation,
)
predict_duration = (time.time() - start_time) / float(len(IDs_for_validation))
error = 0.0
for cur_id in IDs_for_validation:
difference = predictions_for_validation[cur_id][0] - labels_for_training[cur_id][1]
error += difference * difference
error /= float(len(IDs_for_validation))
error = np.sqrt(error)
print(f"RMSE on validation set before training = {error}")
print(f"Prediction duration per sample = {predict_duration} seconds.")
del predictions_for_validation, error
start_time = time.time()
predictions_for_testing = do_predictions(
fe=fe_model,
regressor=model_for_regression,
feature_scaler=text_feature_scaler,
batch_size=MINIBATCH_SIZE,
data_for_anchors=anchor_data,
dist_matrix=anchor_distances,
data=labels_for_training,
token_matrix=tokens_for_training,
pad_id=pretrained_tokenizer.pad_token_id,
identifiers=IDs_for_final_testing,
)
predict_duration = (time.time() - start_time) / float(len(IDs_for_final_testing))
error = 0.0
for cur_id in IDs_for_final_testing:
difference = predictions_for_testing[cur_id][0] - labels_for_training[cur_id][1]
error += difference * difference
error /= float(len(IDs_for_final_testing))
error = np.sqrt(error)
print(f"RMSE on test set before training = {error}")
print(f"Prediction duration per sample = {predict_duration} seconds.")
del predictions_for_testing, error
del anchor_data, anchor_distances
callbacks = [
tf.keras.callbacks.EarlyStopping(
monitor="val_loss", patience=5, verbose=True, restore_best_weights=True
),
tfa.callbacks.TimeStopping(seconds=int(round(3600 * 1.8)), verbose=True),
]
history = model_for_training.fit(
datagen_for_training,
validation_data=datagen_for_validation,
epochs=1000,
callbacks=callbacks,
)
model_for_regression.save_weights(regression_model_name)
fe_model.save_weights(fe_model_name)
show_training_process(history, "loss", figure_identifier)
figure_identifier += 1
show_tsne(
fe=fe_model,
batch_size=MINIBATCH_SIZE,
feature_scaler=text_feature_scaler,
data=labels_for_training,
token_matrix=tokens_for_training,
identifiers=IDs_for_validation + IDs_for_final_testing,
pad_id=pretrained_tokenizer.pad_token_id,
title="after training",
figure_id=figure_identifier,
)
figure_identifier += 1
anchor_data = generate_new_trainset(
fe=fe_model,
feature_scaler=text_feature_scaler,
batch_size=MINIBATCH_SIZE,
data=labels_for_training,
token_matrix=tokens_for_training,
pad_id=pretrained_tokenizer.pad_token_id,
identifiers=IDs_for_training,
)
anchor_distances = calculate_dist_matrix(anchor_data[1])
start_time = time.time()
predictions_for_validation = do_predictions(
fe=fe_model,
regressor=model_for_regression,
feature_scaler=text_feature_scaler,
batch_size=MINIBATCH_SIZE,
data_for_anchors=anchor_data,
dist_matrix=anchor_distances,
data=labels_for_training,
token_matrix=tokens_for_training,
pad_id=pretrained_tokenizer.pad_token_id,
identifiers=IDs_for_validation,
)
predict_duration = (time.time() - start_time) / float(len(IDs_for_validation))
error = 0.0
for cur_id in IDs_for_validation:
difference = predictions_for_validation[cur_id][0] - labels_for_training[cur_id][1]
error += difference * difference
error /= float(len(IDs_for_validation))
error = np.sqrt(error)
print(f"RMSE on validation set after training = {error}")
print(f"Prediction duration per sample = {predict_duration} seconds.")
del predictions_for_validation, error
start_time = time.time()
predictions_for_testing = do_predictions(
fe=fe_model,
regressor=model_for_regression,
feature_scaler=text_feature_scaler,
batch_size=MINIBATCH_SIZE,
data_for_anchors=anchor_data,
dist_matrix=anchor_distances,
data=labels_for_training,
token_matrix=tokens_for_training,
pad_id=pretrained_tokenizer.pad_token_id,
identifiers=IDs_for_final_testing,
)
predict_duration = (time.time() - start_time) / float(len(IDs_for_final_testing))
error = 0.0
for cur_id in IDs_for_final_testing:
difference = predictions_for_testing[cur_id][0] - labels_for_training[cur_id][1]
error += difference * difference
error /= float(len(IDs_for_final_testing))
error = np.sqrt(error)
print(f"RMSE on test set after training = {error}")
print(f"Prediction duration per sample = {predict_duration} seconds.")
pred = sorted(
[
(cur_id, predictions_for_testing[cur_id][0], predictions_for_testing[cur_id][1])
for cur_id in predictions_for_testing
],
key=lambda it: (it[2], it[1], it[0]),
)
print("Top-5 most certain predictions:")
print("")
for cur_id, pred_mean, pred_std in pred[0:5]:
print(
"True: {0:.6f} +- {1:.6f}".format(
data_for_training[cur_id][1], data_for_training[cur_id][2]
)
)
print("Predicted: {0:.6f} +- {1:.6f}".format(pred_mean, pred_std))
print(data_for_training[cur_id][0])
print("")
print("Top-5 most uncertain predictions:")
print("")
for cur_id, pred_mean, pred_std in pred[-5:]:
print(
"True: {0:.6f} +- {1:.6f}".format(
data_for_training[cur_id][1], data_for_training[cur_id][2]
)
)
print("Predicted: {0:.6f} +- {1:.6f}".format(pred_mean, pred_std))
print(data_for_training[cur_id][0])
print("")
del predictions_for_testing, error, pred
del datagen_for_training, datagen_for_validation
del labels_for_training, tokens_for_training
del data_for_training
del IDs_for_training, IDs_for_validation, IDs_for_final_testing
del model_for_training
gc.collect()
with codecs.open(submission_name, mode="w", encoding="utf-8") as fp:
data_writer = csv.writer(fp, quotechar='"', delimiter=",")
data_writer.writerow(["id", "target"])
for data_part in load_data_for_testing(
testset_name, pretrained_tokenizer, MINIBATCH_SIZE * 8
):
labels_for_submission, tokens_for_submission = tokenize_data(
data=data_part, tokenizer=pretrained_tokenizer, max_seq_len=MAX_TEXT_LEN
)
del data_part
predictions_for_submission = do_predictions(
fe=fe_model,
regressor=model_for_regression,
feature_scaler=text_feature_scaler,
batch_size=MINIBATCH_SIZE,
data_for_anchors=anchor_data,
dist_matrix=anchor_distances,
data=labels_for_submission,
token_matrix=tokens_for_submission,
pad_id=pretrained_tokenizer.pad_token_id,
)
for cur_id in predictions_for_submission:
predicted = predictions_for_submission[cur_id][0]
data_writer.writerow([cur_id, f"{predicted}"])
del predictions_for_submission
del labels_for_submission, tokens_for_submission
gc.collect()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/595/69595878.ipynb
|
tf-distilroberta-base
|
bond005
|
[{"Id": 69595878, "ScriptId": 18999565, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 763106, "CreationDate": "08/02/2021 02:29:31", "VersionNumber": 4.0, "Title": "twin-roberta-regression", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 1135.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 1134.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 92980777, "KernelVersionId": 69595878, "SourceDatasetVersionId": 2461019}]
|
[{"Id": 2461019, "DatasetId": 1489654, "DatasourceVersionId": 2503438, "CreatorUserId": 763106, "LicenseName": "Other (specified in description)", "CreationDate": "07/25/2021 10:39:09", "VersionNumber": 1.0, "Title": "tf_distilroberta_base", "Slug": "tf-distilroberta-base", "Subtitle": NaN, "Description": "In this dataset you can find files for Tensorflow implementation of [DistilRoBERTa base model from HuggingFace repository](https://huggingface.co/distilroberta-base).", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1489654, "CreatorUserId": 763106, "OwnerUserId": 763106.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2461019.0, "CurrentDatasourceVersionId": 2503438.0, "ForumId": 1509353, "Type": 2, "CreationDate": "07/25/2021 10:39:09", "LastActivityDate": "07/25/2021", "TotalViews": 1114, "TotalDownloads": 4, "TotalVotes": 2, "TotalKernels": 1}]
|
[{"Id": 763106, "UserName": "bond005", "DisplayName": "Ivan Bondarenko", "RegisterDate": "10/23/2016", "PerformanceTier": 1}]
|
import codecs
import copy
import csv
import gc
from itertools import chain
import os
import pickle
import random
import time
from typing import Dict, List, Tuple, Union
import warnings
import matplotlib.pyplot as plt
from matplotlib.colors import Normalize
import nltk
from nltk.corpus import wordnet
import numpy as np
from sklearn.manifold import TSNE
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler, PowerTransformer
import tensorflow as tf
import tensorflow_addons as tfa
from transformers import AutoConfig, AutoTokenizer, TFAutoModel
print(tf.__version__)
class MaskCalculator(tf.keras.layers.Layer):
def __init__(self, output_dim, **kwargs):
self.output_dim = output_dim
super(MaskCalculator, self).__init__(**kwargs)
def build(self, input_shape):
super(MaskCalculator, self).build(input_shape)
def call(self, inputs, **kwargs):
return tf.keras.backend.permute_dimensions(
x=tf.keras.backend.repeat(
x=tf.keras.backend.cast(
x=tf.keras.backend.greater(x=inputs, y=0), dtype="float32"
),
n=self.output_dim,
),
pattern=(0, 2, 1),
)
def compute_output_shape(self, input_shape):
assert len(input_shape) == 1
shape = list(input_shape)
shape.append(self.output_dim)
return tuple(shape)
class DatasetGen(tf.keras.utils.Sequence):
def __init__(
self,
data: Dict[str, Tuple[List[int], float, float, np.ndarray]],
data_IDs: List[str],
apply_augmentation: bool,
feature_scaler: Pipeline,
token_indices: np.ndarray,
pad_token_id: int,
batch_size: int,
batches_per_epoch: Union[int, None] = None,
):
self.data = copy.deepcopy(data)
self.token_indices = token_indices
self.pad_token_id = pad_token_id
self.batch_size = batch_size
self.batches_per_epoch = batches_per_epoch
self.feature_scaler = feature_scaler
self.apply_augmentation = apply_augmentation
self.pairs = set()
for key1 in data_IDs:
for key2 in data_IDs:
if key1 == key2:
continue
if (key1, key2) not in self.pairs:
self.pairs.add((key1, key2))
self.pairs = sorted(list(self.pairs))
def __len__(self):
if self.batches_per_epoch is None:
return int(np.ceil(len(self.pairs) / float(self.batch_size)))
return self.batches_per_epoch
def __getitem__(self, idx):
x_left = np.zeros(
shape=(self.batch_size, self.token_indices.shape[1]), dtype=np.int32
)
left_features = []
x_right = np.zeros(
shape=(self.batch_size, self.token_indices.shape[1]), dtype=np.int32
)
right_features = []
batch_y = np.zeros((self.batch_size, 1), dtype=np.int32)
if self.batches_per_epoch is None:
batch_start = idx * self.batch_size
batch_end = min(len(self.pairs), batch_start + self.batch_size)
for sample_idx in range(batch_end - batch_start):
left_key, right_key = self.pairs[sample_idx + batch_start]
left_idx = self.data[left_key][0][0]
left_features.append(self.data[left_key][3][0:1])
left_target = self.data[left_key][1]
right_idx = self.data[right_key][0][0]
right_target = self.data[right_key][1]
right_features.append(self.data[right_key][3][0:1])
x_left[sample_idx] = self.token_indices[left_idx]
x_right[sample_idx] = self.token_indices[right_idx]
batch_y[sample_idx, 0] = left_target - right_target
n_pad = self.batch_size - (batch_end - batch_start)
if n_pad > 0:
for sample_idx in range(batch_end - batch_start, self.batch_size):
x_left[sample_idx] = x_left[sample_idx - 1]
x_right[sample_idx] = x_right[sample_idx - 1]
left_features.append(left_features[-1])
right_features.append(right_features[-1])
batch_y[sample_idx, 0] = batch_y[sample_idx - 1, 0]
else:
for sample_idx in range(self.batch_size):
left_key, right_key = random.choice(self.pairs)
if self.apply_augmentation:
p = np.ones((len(self.data[left_key][0]),), dtype=np.float64)
p[0] = max(2.0, p.shape[0] - 1.0)
p /= p.sum()
left_idx_ = np.random.choice(
list(range(len(self.data[left_key][0]))), p=p
)
left_target = np.random.normal(
loc=self.data[left_key][1], scale=self.data[left_key][2]
)
else:
left_idx_ = 0
left_target = self.data[left_key][1]
left_idx = self.data[left_key][0][left_idx_]
left_features.append(
self.data[left_key][3][left_idx_ : (left_idx_ + 1)]
)
if self.apply_augmentation:
p = np.ones((len(self.data[right_key][0]),), dtype=np.float64)
p[0] = max(2.0, p.shape[0] - 1.0)
p /= p.sum()
right_idx_ = np.random.choice(
list(range(len(self.data[right_key][0]))), p=p
)
right_target = np.random.normal(
loc=self.data[right_key][1], scale=self.data[right_key][2]
)
else:
right_idx_ = 0
right_target = self.data[right_key][1]
right_idx = self.data[right_key][0][right_idx_]
right_features.append(
self.data[right_key][3][right_idx_ : (right_idx_ + 1)]
)
x_left[sample_idx] = self.token_indices[left_idx]
x_right[sample_idx] = self.token_indices[right_idx]
batch_y[sample_idx, 0] = left_target - right_target
batch_x = [
x_left,
generate_attention_mask(x_left, self.pad_token_id),
self.feature_scaler.transform(np.vstack(left_features)),
x_right,
generate_attention_mask(x_right, self.pad_token_id),
self.feature_scaler.transform(np.vstack(right_features)),
]
del x_left, x_right
return batch_x, batch_y, None
def generate_attention_mask(token_indices: np.ndarray, padding_id: int) -> np.ndarray:
attention = np.zeros(token_indices.shape, dtype=np.int32)
for sample_idx in range(token_indices.shape[0]):
for token_idx in range(token_indices.shape[1]):
if token_indices[sample_idx, token_idx] == padding_id:
break
attention[sample_idx, token_idx] = 1
return attention
def calc_text_features(texts: List[List[str]], tok: AutoTokenizer) -> np.ndarray:
f = np.zeros((len(texts), 9), dtype=np.float32)
for idx, sentences in enumerate(texts):
f[idx, 0] = len(sentences)
words = []
pure_words = []
for cur_sent in sentences:
words_in_sentence = nltk.word_tokenize(cur_sent)
words += words_in_sentence
pure_words += list(filter(lambda it: it.isalpha(), words_in_sentence))
f[idx, 1] = len(words) / f[idx, 0]
f[idx, 2] = len(pure_words) / f[idx, 0]
f[idx, 3] = len(" ".join(sentences))
f[idx, 4] = len(pure_words)
f[idx, 5] = np.mean([len(w) for w in pure_words])
for w in pure_words:
syllables = tok.tokenize(w.lower())
f[idx, 6] += len(syllables)
f[idx, 7] += sum(map(lambda it: len(it), syllables))
f[idx, 7] /= f[idx, 6]
f[idx, 8] = f[idx, 6] / f[idx, 4]
return f
def load_data_for_training(
fname: str, tok: AutoTokenizer
) -> List[Dict[str, Tuple[List[str], float, float, np.ndarray]]]:
loaded_header = []
id_col_idx = -1
text_col_idx = -1
target_col_idx = -1
std_col_idx = -1
line_idx = 1
data = dict()
set_of_texts = set()
with codecs.open(fname, mode="r", encoding="utf-8") as fp:
data_reader = csv.reader(fp, quotechar='"', delimiter=",")
for row in data_reader:
if len(row) > 0:
err_msg = f"File {fname}: line {line_idx} is wrong!"
if len(loaded_header) == 0:
loaded_header = copy.copy(row)
try:
text_col_idx = loaded_header.index("excerpt")
except:
text_col_idx = -1
if text_col_idx <= 0:
raise ValueError(err_msg + ' Field "excerpt" is not found!')
try:
id_col_idx = loaded_header.index("id")
except:
id_col_idx = -1
if id_col_idx < 0:
raise ValueError(err_msg + ' Field "id" is not found!')
try:
target_col_idx = loaded_header.index("target")
except:
target_col_idx = -1
if target_col_idx < 0:
raise ValueError(err_msg + ' Field "target" is not found!')
try:
std_col_idx = loaded_header.index("standard_error")
except:
std_col_idx = -1
if std_col_idx < 0:
err_msg2 = f'{err_msg} Field "standard_error" is not found!'
raise ValueError(err_msg2)
else:
sample_id = row[id_col_idx]
if sample_id != sample_id.strip():
raise ValueError(err_msg + f" {sample_id} is wrong sample ID!")
if sample_id in data:
err_msg2 = f"{err_msg} {sample_id} is not unique sample ID!"
raise ValueError(err_msg2)
text = row[text_col_idx].replace("\r", "\n")
if len(text) == 0:
raise ValueError(err_msg + f" Text {sample_id} is empty!")
sentences = []
for paragraph in map(lambda it: it.strip(), text.split("\n")):
if len(paragraph) > 0:
sentences += nltk.sent_tokenize(paragraph)
if len(sentences) == 0:
raise ValueError(err_msg + f" Text {sample_id} is empty!")
text = " ".join([cur_sent for cur_sent in sentences])
if text.lower() in set_of_texts:
raise ValueError(err_msg + f" Text {sample_id} is not unique!")
set_of_texts.add(text.lower())
added_texts = [sentences]
try:
target_val = float(row[target_col_idx])
ok = True
except:
target_val = 0.0
ok = False
if not ok:
err_msg2 = err_msg
err_msg2 += (
f" {row[target_col_idx]} is wrong target for "
f"text {sample_id}."
)
raise ValueError(err_msg2)
try:
std_val = float(row[std_col_idx])
ok = std_val > 0.0
except:
std_val = 0.0
ok = False
if not ok:
err_msg2 = err_msg
err_msg2 += (
f" {row[std_col_idx]} is wrong standard error"
f" for text {sample_id}."
)
warnings.warn(err_msg2)
else:
for _ in range(3):
new_augmented_text = []
for cur_sent in sentences:
new_sent = cur_sent.strip()
if len(new_sent) > 0:
new_augmented_text.append(new_sent)
assert len(new_augmented_text) > 0
random.shuffle(new_augmented_text)
new_augmented_text_ = " ".join(new_augmented_text)
if (len(new_augmented_text_) > 0) and (
new_augmented_text_.lower() not in set_of_texts
):
set_of_texts.add(new_augmented_text_.lower())
added_texts.append(new_augmented_text)
del new_augmented_text, new_augmented_text_
data[sample_id] = (
list(map(lambda it: " ".join(it), added_texts)),
target_val,
std_val,
calc_text_features(added_texts, tok),
)
line_idx += 1
return data
def load_data_for_testing(fname: str, tok: AutoTokenizer, batch_size: int):
loaded_header = []
id_col_idx = -1
text_col_idx = -1
target_col_idx = -1
std_col_idx = -1
line_idx = 1
data = dict()
with codecs.open(fname, mode="r", encoding="utf-8") as fp:
data_reader = csv.reader(fp, quotechar='"', delimiter=",")
for row in data_reader:
if len(row) > 0:
err_msg = f"File {fname}: line {line_idx} is wrong!"
if len(loaded_header) == 0:
loaded_header = copy.copy(row)
try:
text_col_idx = loaded_header.index("excerpt")
except:
text_col_idx = -1
if text_col_idx <= 0:
raise ValueError(err_msg + ' Field "excerpt" is not found!')
try:
id_col_idx = loaded_header.index("id")
except:
id_col_idx = -1
if id_col_idx < 0:
raise ValueError(err_msg + ' Field "id" is not found!')
else:
sample_id = row[id_col_idx]
if sample_id != sample_id.strip():
raise ValueError(err_msg + f" {sample_id} is wrong sample ID!")
if sample_id in data:
err_msg2 = f"{err_msg} {sample_id} is not unique sample ID!"
raise ValueError(err_msg2)
text = row[text_col_idx].replace("\n", " ").replace("\r", " ")
text = " ".join(text.split()).strip()
if len(text) == 0:
raise ValueError(err_msg + f" Text {sample_id} is empty!")
features = calc_text_features([nltk.sent_tokenize(text)], tok)
data[sample_id] = (text, features)
if len(data) >= batch_size:
yield data
del data
data = dict()
line_idx += 1
if len(data) > 0:
yield data
def train_feature_scaler(
data: Dict[str, Tuple[List[int], float, float, np.ndarray]]
) -> Pipeline:
features_for_training = []
for sample_id in data:
features_for_training.append(data[sample_id][3])
features_for_training = np.vstack(features_for_training)
scaler = Pipeline(
steps=[("scaler", StandardScaler()), ("transformer", PowerTransformer())]
)
return scaler.fit(features_for_training)
def tokenize_data(
data: Union[
List[Dict[str, Tuple[str, np.ndarray]]],
List[Dict[str, Tuple[List[str], float, float, np.ndarray]]],
],
tokenizer: AutoTokenizer,
max_seq_len: int,
) -> Tuple[
Union[
Dict[str, Tuple[int, np.ndarray]],
Dict[str, Tuple[List[int], float, float, np.ndarray]],
],
np.ndarray,
]:
tokenized_data = dict()
all_tokens_matrix = []
for sample_idx, cur_ID in enumerate(sorted(list(data.keys()))):
if len(data[cur_ID]) == 2:
tokens = tokenizer.tokenize(data[cur_ID][0])
tokenized_data[cur_ID] = (len(all_tokens_matrix), data[cur_ID][1])
token_ids = tokenizer.convert_tokens_to_ids(
[tokenizer.cls_token] + tokens + [tokenizer.sep_token]
)
ndiff = max_seq_len - len(token_ids)
if ndiff > 0:
token_ids += [tokenizer.pad_token_id for _ in range(ndiff)]
elif ndiff < 0:
token_ids = token_ids[:max_seq_len]
all_tokens_matrix.append(token_ids)
else:
text_idx_list = []
for cur_text in data[cur_ID][0]:
tokens = tokenizer.tokenize(cur_text)
token_ids = tokenizer.convert_tokens_to_ids(
[tokenizer.cls_token] + tokens + [tokenizer.sep_token]
)
ndiff = max_seq_len - len(token_ids)
if ndiff > 0:
token_ids += [tokenizer.pad_token_id for _ in range(ndiff)]
elif ndiff < 0:
token_ids = token_ids[:max_seq_len]
text_idx_list.append(len(all_tokens_matrix))
all_tokens_matrix.append(token_ids)
tokenized_data[cur_ID] = (
text_idx_list,
data[cur_ID][1],
data[cur_ID][2],
data[cur_ID][3],
)
return tokenized_data, np.array(all_tokens_matrix, dtype=np.int32)
def print_info_about_data(
data: Union[
List[Dict[str, Tuple[str, np.ndarray]]],
List[Dict[str, Tuple[List[str], float, float, np.ndarray]]],
],
identifiers: List[str],
):
for_training = len(data[identifiers[0]]) == 4
if for_training:
print(f"Number of samples for training is {len(data)}.")
else:
print(f"Number of samples for submission is {len(data)}.")
print("")
print(f"{len(identifiers)} random samples:")
for cur_id in identifiers:
print("")
print(f" Sample {cur_id}")
if for_training:
print(" Text:")
print(f" {data[cur_id][0][0]}")
print(f" Number of augmented texts is {len(data[cur_id][0]) - 1}.")
if (len(data[cur_id][0]) - 1) > 0:
if (len(data[cur_id][0]) - 1) > 1:
print(" 2 augmented texts:")
for augmented in data[cur_id][0][1:3]:
print(f" {augmented}")
else:
print(" Augmented text:")
for augmented in data[cur_id][0][1:2]:
print(f" {augmented}")
print(" Target:")
print(f" {data[cur_id][1]} +- {data[cur_id][2]}")
print(" Features:")
for it in data[cur_id][3].tolist():
print(f" {it}")
else:
print(" Text:")
print(f" {data[cur_id][0]}")
print(" Features:")
print(f" {data[cur_id][1].tolist()[0]}")
def print_info_about_tokenized_data(
data: Union[
Dict[str, Tuple[int, np.ndarray]],
Dict[str, Tuple[List[int], float, float, np.ndarray]],
],
matrix: np.ndarray,
identifiers: List[str],
):
for_training = len(data[identifiers[0]]) == 4
if for_training:
print(f"Number of tokenized samples for training is {len(data)}.")
else:
print(f"Number of tokenized samples for submission is {len(data)}.")
print("")
print(f"{len(identifiers)} random samples:")
for cur_id in identifiers:
print("")
print(f"Sample {cur_id}")
print("")
sample_idx = data[cur_id][0][0]
print(matrix[sample_idx].tolist())
print("")
print(data[cur_id][-1][0].tolist())
print("")
def build_feature_extractor(
bert_name: str, max_seq_len: int, feature_vector_size: int, batch_size: int
) -> Tuple[tf.keras.Model, int]:
transformer_model = TFAutoModel.from_pretrained(
pretrained_model_name_or_path=bert_name, name="BaseTransformer"
)
united_embedding_size = 256
transformer_config = AutoConfig.from_pretrained(bert_name)
united_emb_layer = tf.keras.layers.Dense(
units=united_embedding_size,
input_dim=transformer_config.hidden_size,
activation="tanh",
kernel_initializer=tf.keras.initializers.GlorotNormal(seed=42),
bias_initializer="zeros",
name="UnitedEmbeddingLayer",
)
print("Transformer Configuration")
print("=========================")
print(transformer_config)
tokens_input = tf.keras.layers.Input(
shape=(max_seq_len,),
batch_size=batch_size,
dtype=tf.int32,
name="word_ids_base",
)
attention_input = tf.keras.layers.Input(
shape=(max_seq_len,),
batch_size=batch_size,
dtype=tf.int32,
name="attention_mask_base",
)
features_input = tf.keras.layers.Input(
shape=(feature_vector_size,),
dtype=tf.float32,
batch_size=batch_size,
name="features_base",
)
sequence_output = transformer_model([tokens_input, attention_input])[0]
output_mask = MaskCalculator(
output_dim=transformer_config.hidden_size,
trainable=False,
name="OutMaskCalculator",
)(attention_input)
masked_output = tf.keras.layers.Multiply(name="OutMaskMultiplicator")(
[output_mask, sequence_output]
)
masked_output = tf.keras.layers.Masking(name="OutMasking")(masked_output)
final_output = tf.keras.layers.GlobalAvgPool1D(name="AvePool")(masked_output)
final_output = tf.keras.layers.LayerNormalization(name="Emdedding")(final_output)
final_output = tf.keras.layers.Concatenate(name="Concat")(
[final_output, features_input]
)
final_output = tf.keras.layers.Dropout(rate=0.3, seed=42, name="EmbeddingDropout")(
final_output
)
final_output = united_emb_layer(final_output)
fe_model = tf.keras.Model(
inputs=[tokens_input, attention_input, features_input],
outputs=final_output,
name="FeatureExtractionModel",
)
fe_model.build(
input_shape=[
(batch_size, max_seq_len),
(batch_size, max_seq_len),
(batch_size, feature_vector_size),
]
)
return fe_model, united_embedding_size
def build_twin_regressor(feature_vector_size: int, batch_size: int) -> tf.keras.Model:
left_input = tf.keras.layers.Input(
shape=(feature_vector_size,),
dtype=tf.float32,
batch_size=batch_size,
name="features_left",
)
right_input = tf.keras.layers.Input(
shape=(feature_vector_size,),
dtype=tf.float32,
batch_size=batch_size,
name="features_right",
)
concatenated_features = tf.keras.layers.Concatenate(name="ConcatFeatures")(
[left_input, right_input]
)
regression_layer = tf.keras.layers.Dense(
units=1,
input_dim=feature_vector_size * 2,
activation=None,
kernel_initializer=tf.keras.initializers.GlorotNormal(seed=42),
bias_initializer="zeros",
name="RegressionLayer",
)(concatenated_features)
twin_regression_model = tf.keras.Model(
inputs=[left_input, right_input],
outputs=regression_layer,
name="TwinRegressionModel",
)
twin_regression_model.build(
input_shape=[
(batch_size, feature_vector_size),
(batch_size, feature_vector_size),
]
)
return twin_regression_model
def build_neural_network(
bert_name: str, max_seq_len: int, feature_vector_size: int, batch_size: int
) -> Tuple[tf.keras.Model, tf.keras.Model, tf.keras.Model]:
fe_layer, ft_vec_size = build_feature_extractor(
bert_name, max_seq_len, feature_vector_size, batch_size
)
left_tokens = tf.keras.layers.Input(
shape=(max_seq_len,), batch_size=batch_size, dtype=tf.int32, name="word_ids"
)
left_attention = tf.keras.layers.Input(
shape=(max_seq_len,),
batch_size=batch_size,
dtype=tf.int32,
name="attention_mask",
)
left_features = tf.keras.layers.Input(
shape=(feature_vector_size,),
dtype=tf.float32,
batch_size=batch_size,
name="features",
)
right_tokens = tf.keras.layers.Input(
shape=(max_seq_len,),
batch_size=batch_size,
dtype=tf.int32,
name="right_word_ids",
)
right_attention = tf.keras.layers.Input(
shape=(max_seq_len,),
batch_size=batch_size,
dtype=tf.int32,
name="right_attention_mask",
)
right_features = tf.keras.layers.Input(
shape=(feature_vector_size,),
dtype=tf.float32,
batch_size=batch_size,
name="right_features",
)
left_output = fe_layer([left_tokens, left_attention, left_features])
right_output = fe_layer([right_tokens, right_attention, right_features])
regression_model = build_twin_regressor(ft_vec_size, batch_size)
regression_layer = regression_model([left_output, right_output])
siamese_model = tf.keras.Model(
inputs=[
left_tokens,
left_attention,
left_features,
right_tokens,
right_attention,
right_features,
],
outputs=regression_layer,
name="SiameseModel",
)
radam = tfa.optimizers.RectifiedAdam(learning_rate=1e-5)
ranger = tfa.optimizers.Lookahead(radam, sync_period=6, slow_step_size=0.5)
siamese_model.compile(optimizer=ranger, loss=tf.keras.losses.MeanSquaredError())
return siamese_model, fe_layer, regression_model
def show_minibatch(X: List[np.ndarray], y: np.ndarray):
assert len(X) == 6
print("")
print("X1")
for it in X[0].tolist():
print(it)
print("")
print("X2")
for it in X[1].tolist():
print(it)
print("")
print("X3")
for it in X[2].tolist():
print(it)
print("")
print("X4")
for it in X[3].tolist():
print(it)
print("")
print("X5")
for it in X[4].tolist():
print(it)
print("X6")
for it in X[5].tolist():
print(it)
print("")
print("y")
for it in y.tolist():
print(it)
def show_tsne(
fe: tf.keras.Model,
batch_size: int,
data: Dict[str, Tuple[List[int], float, float, np.ndarray]],
feature_scaler: Pipeline,
token_matrix: np.ndarray,
identifiers: List[str],
pad_id: int,
title: str,
figure_id: int,
):
indices = list(map(lambda it: data[it][0][0], identifiers))
colors = np.array(list(map(lambda it: data[it][1], identifiers)), dtype=np.float64)
area = np.array(list(map(lambda it: data[it][2], identifiers)), dtype=np.float64)
area /= np.max(area)
area *= 10.0
area = np.power(area, 2)
texts = token_matrix[indices]
src_features = np.vstack(list(map(lambda it: data[it][3][0:1], identifiers)))
assert src_features.shape[0] == texts.shape[0]
ndiff = texts.shape[0] % batch_size
if ndiff > 0:
last_text_idx = texts.shape[0] - 1
texts = np.vstack(
[texts]
+ [
texts[last_text_idx : (last_text_idx + 1)]
for _ in range(batch_size - ndiff)
]
)
src_features = np.vstack(
[src_features]
+ [
src_features[last_text_idx : (last_text_idx + 1)]
for _ in range(batch_size - ndiff)
]
)
attentions = generate_attention_mask(texts, pad_id)
assert texts.shape[0] % batch_size == 0, f"{texts.shape[0] % batch_size}"
features = fe.predict(
[texts, attentions, feature_scaler.transform(src_features)],
batch_size=batch_size,
)
features = features[: len(indices)]
projected_features = TSNE(n_components=2, n_jobs=-1).fit_transform(features)
fig = plt.figure(figure_id, figsize=(11, 11))
plt.scatter(
x=projected_features[:, 0],
y=projected_features[:, 1],
marker="o",
cmap=plt.cm.get_cmap("jet"),
s=area,
c=colors,
norm=Normalize(vmin=np.min(colors), vmax=np.max(colors)),
)
plt.title("t-SNE projections of texts " + title)
plt.colorbar()
plt.show()
def show_training_process(
history: tf.keras.callbacks.History, metric_name: str, figure_id: int
):
val_metric_name = "val_" + metric_name
possible_metrics = list(history.history.keys())
if metric_name not in history.history:
err_msg = f'The metric "{metric_name}" is not found!'
err_msg += f" Available metrics are: {possible_metrics}."
raise ValueError(err_msg)
fig = plt.figure(figure_id, figsize=(7, 7))
metric_values = history.history[metric_name]
plt.plot(
list(range(len(metric_values))),
metric_values,
label="Training {0}".format(metric_name),
)
if val_metric_name in history.history:
val_metric_values = history.history["val_" + metric_name]
assert len(metric_values) == len(val_metric_values)
plt.plot(
list(range(len(val_metric_values))),
val_metric_values,
label="Validation {0}".format(metric_name),
)
plt.xlabel("Epochs")
plt.ylabel(metric_name)
plt.title("Training process")
plt.legend(loc="best")
plt.show()
def generate_new_trainset(
fe: tf.keras.Model,
feature_scaler: Pipeline,
batch_size: int,
data: Dict[str, Tuple[List[int], float, float]],
token_matrix: np.ndarray,
pad_id: int,
identifiers: List[str],
) -> Tuple[np.ndarray, np.ndarray]:
indices = list(map(lambda it: data[it][0][0], identifiers))
texts = token_matrix[indices]
src_features = np.vstack(list(map(lambda it: data[it][3][0:1], identifiers)))
targets = np.array(list(map(lambda it: data[it][1], identifiers)), dtype=np.float64)
assert texts.shape[0] == src_features.shape[0]
ndiff = texts.shape[0] % batch_size
if ndiff > 0:
last_text_idx = texts.shape[0] - 1
texts = np.vstack(
[texts]
+ [
texts[last_text_idx : (last_text_idx + 1)]
for _ in range(batch_size - ndiff)
]
)
src_features = np.vstack(
[src_features]
+ [
src_features[last_text_idx : (last_text_idx + 1)]
for _ in range(batch_size - ndiff)
]
)
attentions = generate_attention_mask(texts, pad_id)
assert texts.shape[0] % batch_size == 0, f"{texts.shape[0] % batch_size}"
target_features = fe.predict(
[texts, attentions, feature_scaler.transform(src_features)],
batch_size=batch_size,
)
assert target_features.shape[1] > 1
target_features = target_features[: len(identifiers)]
return target_features, targets
def calculate_dist_matrix(y: np.ndarray) -> np.ndarray:
assert len(y.shape) == 1
assert y.shape[0] > 1
d = np.zeros((y.shape[0], y.shape[0]), dtype=np.float32)
for idx1 in range(y.shape[0]):
for idx2 in range(y.shape[0]):
diff = y[idx1] - y[idx2]
d[idx1, idx2] = np.sqrt(diff * diff)
return d
def select_train_samples(y: np.ndarray, dist_matrix: np.ndarray, n: int) -> List[int]:
assert len(y.shape) == 1
assert len(dist_matrix.shape) == 2
assert dist_matrix.shape[0] == y.shape[0]
assert dist_matrix.shape[1] == dist_matrix.shape[0]
assert n < y.shape[0]
indices_of_samples = list(range(y.shape[0]))
selected = {np.random.choice(indices_of_samples)}
for _ in range(n - 1):
indices_of_samples = sorted(list(set(indices_of_samples) - selected))
p = [dist_matrix[idx, list(selected)].mean() for idx in indices_of_samples]
p = np.array(p, dtype=np.float64)
p /= p.sum()
selected.add(np.random.choice(indices_of_samples, p=p))
return sorted(list(selected))
def do_predictions(
fe: tf.keras.Model,
regressor: tf.keras.Model,
feature_scaler: Pipeline,
batch_size: int,
data_for_anchors: Tuple[np.ndarray, np.ndarray],
dist_matrix: np.ndarray,
data: Union[Dict[str, int], Dict[str, Tuple[List[int], float, float]]],
token_matrix: np.ndarray,
pad_id: int,
identifiers: List[str] = None,
) -> Dict[str, Tuple[float, float]]:
if identifiers is None:
identifiers_ = sorted(list(data.keys()))
else:
identifiers_ = sorted(identifiers)
indices = list(
map(
lambda it: data[it][0] if len(data[it]) == 2 else data[it][0][0],
identifiers_,
)
)
texts = token_matrix[indices]
src_features = np.vstack(
list(
map(
lambda it: data[it][1] if len(data[it]) == 2 else data[it][3][0:1],
identifiers_,
)
)
)
assert texts.shape[0] == src_features.shape[0]
ndiff = texts.shape[0] % batch_size
if ndiff > 0:
last_text_idx = texts.shape[0] - 1
texts = np.vstack(
[texts]
+ [
texts[last_text_idx : (last_text_idx + 1)]
for _ in range(batch_size - ndiff)
]
)
src_features = np.vstack(
[src_features]
+ [
src_features[last_text_idx : (last_text_idx + 1)]
for _ in range(batch_size - ndiff)
]
)
attentions = generate_attention_mask(texts, pad_id)
assert texts.shape[0] % batch_size == 0, f"{texts.shape[0] % batch_size}"
target_features = fe.predict(
[texts, attentions, feature_scaler.transform(src_features)],
batch_size=batch_size,
)
assert target_features.shape[1] > 1
assert target_features.shape[1] == data_for_anchors[0].shape[1]
assert target_features.shape[0] >= len(indices)
target_features = target_features[0 : len(indices)]
selected_inputs = []
predicted_features = []
selected_targets = []
n_selected = batch_size
while n_selected < 8:
n_selected += batch_size
for sample_idx, cur_id in enumerate(identifiers_):
selected_indices_for_training = select_train_samples(
y=data_for_anchors[1], dist_matrix=dist_matrix, n=n_selected
)
selected_inputs.append(data_for_anchors[0][selected_indices_for_training])
selected_targets.append(data_for_anchors[1][selected_indices_for_training])
predicted_features.append(
np.full(
fill_value=target_features[sample_idx],
shape=(n_selected, target_features.shape[1]),
)
)
selected_inputs = np.vstack(selected_inputs)
predicted_features = np.vstack(predicted_features)
selected_targets = np.concatenate(selected_targets)
prediction_diff = regressor.predict(
[selected_inputs, predicted_features], batch_size=batch_size
).reshape(selected_targets.shape)
predictions = dict()
for sample_idx, cur_id in enumerate(identifiers_):
start_pos = sample_idx * n_selected
end_pos = start_pos + n_selected
instant_predictions = (
selected_targets[start_pos:end_pos] - prediction_diff[start_pos:end_pos]
)
predictions[cur_id] = (
np.mean(instant_predictions),
np.std(instant_predictions),
)
return predictions
random.seed(42)
np.random.seed(42)
tf.random.set_seed(42)
MAX_TEXT_LEN = 512
PRETRAINED_BERT = "/kaggle/input/tf-distilroberta-base"
MINIBATCH_SIZE = 8
DATA_DIR = "/kaggle/input/commonlitreadabilityprize"
MODEL_DIR = "/kaggle/working"
print(f"{DATA_DIR} {os.path.isdir(DATA_DIR)}")
print(f"{MODEL_DIR} {os.path.isdir(MODEL_DIR)}")
trainset_name = os.path.join(DATA_DIR, "train.csv")
print(f"{trainset_name} {os.path.isfile(trainset_name)}")
testset_name = os.path.join(DATA_DIR, "test.csv")
print(f"{testset_name} {os.path.isfile(testset_name)}")
submission_name = os.path.join(MODEL_DIR, "submission.csv")
print(f"{submission_name} {os.path.isfile(submission_name)}")
fe_model_name = os.path.join(MODEL_DIR, "fe_nn.h5")
regression_model_name = os.path.join(MODEL_DIR, "regression_nn.h5")
scaler_name = os.path.join(MODEL_DIR, "feature_scaler.pkl")
figure_identifier = 1
pretrained_tokenizer = AutoTokenizer.from_pretrained(PRETRAINED_BERT)
print(f"Vocabulary size is {pretrained_tokenizer.vocab_size}.")
data_for_training = load_data_for_training(trainset_name, pretrained_tokenizer)
assert len(data_for_training) > 100
all_IDs = sorted(list(data_for_training.keys()))
selected_IDs_for_training = random.sample(population=all_IDs, k=3)
print_info_about_data(data_for_training, selected_IDs_for_training)
labels_for_training, tokens_for_training = tokenize_data(
data=data_for_training, tokenizer=pretrained_tokenizer, max_seq_len=MAX_TEXT_LEN
)
print_info_about_tokenized_data(
data=labels_for_training,
matrix=tokens_for_training,
identifiers=selected_IDs_for_training,
)
text_feature_scaler = train_feature_scaler(labels_for_training)
with open(scaler_name, "wb") as scaler_fp:
pickle.dump(text_feature_scaler, scaler_fp)
random.shuffle(all_IDs)
n_train_size = int(round(len(all_IDs) * 0.9))
n_val_size = int(round(len(all_IDs) * 0.025))
IDs_for_training = all_IDs[:n_train_size]
IDs_for_validation = all_IDs[n_train_size : (n_train_size + n_val_size)]
IDs_for_final_testing = all_IDs[(n_train_size + n_val_size) :]
datagen_for_validation = DatasetGen(
data=labels_for_training,
data_IDs=IDs_for_validation,
token_indices=tokens_for_training,
pad_token_id=pretrained_tokenizer.pad_token_id,
batch_size=MINIBATCH_SIZE,
apply_augmentation=False,
feature_scaler=text_feature_scaler,
)
n_batches_per_validset = len(datagen_for_validation)
print(f"Mini-batches per validation set is {n_batches_per_validset}.")
X_, y_, _ = datagen_for_validation[0]
show_minibatch(X_, y_)
n_batches_per_epoch = n_batches_per_validset * 3
datagen_for_training = DatasetGen(
data=labels_for_training,
data_IDs=IDs_for_training,
token_indices=tokens_for_training,
pad_token_id=pretrained_tokenizer.pad_token_id,
batch_size=MINIBATCH_SIZE,
batches_per_epoch=n_batches_per_epoch,
apply_augmentation=True,
feature_scaler=text_feature_scaler,
)
X_, y_, _ = datagen_for_training[0]
show_minibatch(X_, y_)
model_for_training, fe_model, model_for_regression = build_neural_network(
bert_name=PRETRAINED_BERT,
max_seq_len=MAX_TEXT_LEN,
feature_vector_size=text_feature_scaler.named_steps["scaler"].scale_.shape[0],
batch_size=MINIBATCH_SIZE,
)
model_for_training.summary()
model_for_regression.summary()
fe_model.summary()
show_tsne(
fe=fe_model,
batch_size=MINIBATCH_SIZE,
feature_scaler=text_feature_scaler,
data=labels_for_training,
token_matrix=tokens_for_training,
identifiers=IDs_for_validation + IDs_for_final_testing,
pad_id=pretrained_tokenizer.pad_token_id,
title="before training",
figure_id=figure_identifier,
)
figure_identifier += 1
anchor_data = generate_new_trainset(
fe=fe_model,
feature_scaler=text_feature_scaler,
batch_size=MINIBATCH_SIZE,
data=labels_for_training,
token_matrix=tokens_for_training,
pad_id=pretrained_tokenizer.pad_token_id,
identifiers=IDs_for_training,
)
anchor_distances = calculate_dist_matrix(anchor_data[1])
start_time = time.time()
predictions_for_validation = do_predictions(
fe=fe_model,
regressor=model_for_regression,
feature_scaler=text_feature_scaler,
batch_size=MINIBATCH_SIZE,
data_for_anchors=anchor_data,
dist_matrix=anchor_distances,
data=labels_for_training,
token_matrix=tokens_for_training,
pad_id=pretrained_tokenizer.pad_token_id,
identifiers=IDs_for_validation,
)
predict_duration = (time.time() - start_time) / float(len(IDs_for_validation))
error = 0.0
for cur_id in IDs_for_validation:
difference = predictions_for_validation[cur_id][0] - labels_for_training[cur_id][1]
error += difference * difference
error /= float(len(IDs_for_validation))
error = np.sqrt(error)
print(f"RMSE on validation set before training = {error}")
print(f"Prediction duration per sample = {predict_duration} seconds.")
del predictions_for_validation, error
start_time = time.time()
predictions_for_testing = do_predictions(
fe=fe_model,
regressor=model_for_regression,
feature_scaler=text_feature_scaler,
batch_size=MINIBATCH_SIZE,
data_for_anchors=anchor_data,
dist_matrix=anchor_distances,
data=labels_for_training,
token_matrix=tokens_for_training,
pad_id=pretrained_tokenizer.pad_token_id,
identifiers=IDs_for_final_testing,
)
predict_duration = (time.time() - start_time) / float(len(IDs_for_final_testing))
error = 0.0
for cur_id in IDs_for_final_testing:
difference = predictions_for_testing[cur_id][0] - labels_for_training[cur_id][1]
error += difference * difference
error /= float(len(IDs_for_final_testing))
error = np.sqrt(error)
print(f"RMSE on test set before training = {error}")
print(f"Prediction duration per sample = {predict_duration} seconds.")
del predictions_for_testing, error
del anchor_data, anchor_distances
callbacks = [
tf.keras.callbacks.EarlyStopping(
monitor="val_loss", patience=5, verbose=True, restore_best_weights=True
),
tfa.callbacks.TimeStopping(seconds=int(round(3600 * 1.8)), verbose=True),
]
history = model_for_training.fit(
datagen_for_training,
validation_data=datagen_for_validation,
epochs=1000,
callbacks=callbacks,
)
model_for_regression.save_weights(regression_model_name)
fe_model.save_weights(fe_model_name)
show_training_process(history, "loss", figure_identifier)
figure_identifier += 1
show_tsne(
fe=fe_model,
batch_size=MINIBATCH_SIZE,
feature_scaler=text_feature_scaler,
data=labels_for_training,
token_matrix=tokens_for_training,
identifiers=IDs_for_validation + IDs_for_final_testing,
pad_id=pretrained_tokenizer.pad_token_id,
title="after training",
figure_id=figure_identifier,
)
figure_identifier += 1
anchor_data = generate_new_trainset(
fe=fe_model,
feature_scaler=text_feature_scaler,
batch_size=MINIBATCH_SIZE,
data=labels_for_training,
token_matrix=tokens_for_training,
pad_id=pretrained_tokenizer.pad_token_id,
identifiers=IDs_for_training,
)
anchor_distances = calculate_dist_matrix(anchor_data[1])
start_time = time.time()
predictions_for_validation = do_predictions(
fe=fe_model,
regressor=model_for_regression,
feature_scaler=text_feature_scaler,
batch_size=MINIBATCH_SIZE,
data_for_anchors=anchor_data,
dist_matrix=anchor_distances,
data=labels_for_training,
token_matrix=tokens_for_training,
pad_id=pretrained_tokenizer.pad_token_id,
identifiers=IDs_for_validation,
)
predict_duration = (time.time() - start_time) / float(len(IDs_for_validation))
error = 0.0
for cur_id in IDs_for_validation:
difference = predictions_for_validation[cur_id][0] - labels_for_training[cur_id][1]
error += difference * difference
error /= float(len(IDs_for_validation))
error = np.sqrt(error)
print(f"RMSE on validation set after training = {error}")
print(f"Prediction duration per sample = {predict_duration} seconds.")
del predictions_for_validation, error
start_time = time.time()
predictions_for_testing = do_predictions(
fe=fe_model,
regressor=model_for_regression,
feature_scaler=text_feature_scaler,
batch_size=MINIBATCH_SIZE,
data_for_anchors=anchor_data,
dist_matrix=anchor_distances,
data=labels_for_training,
token_matrix=tokens_for_training,
pad_id=pretrained_tokenizer.pad_token_id,
identifiers=IDs_for_final_testing,
)
predict_duration = (time.time() - start_time) / float(len(IDs_for_final_testing))
error = 0.0
for cur_id in IDs_for_final_testing:
difference = predictions_for_testing[cur_id][0] - labels_for_training[cur_id][1]
error += difference * difference
error /= float(len(IDs_for_final_testing))
error = np.sqrt(error)
print(f"RMSE on test set after training = {error}")
print(f"Prediction duration per sample = {predict_duration} seconds.")
pred = sorted(
[
(cur_id, predictions_for_testing[cur_id][0], predictions_for_testing[cur_id][1])
for cur_id in predictions_for_testing
],
key=lambda it: (it[2], it[1], it[0]),
)
print("Top-5 most certain predictions:")
print("")
for cur_id, pred_mean, pred_std in pred[0:5]:
print(
"True: {0:.6f} +- {1:.6f}".format(
data_for_training[cur_id][1], data_for_training[cur_id][2]
)
)
print("Predicted: {0:.6f} +- {1:.6f}".format(pred_mean, pred_std))
print(data_for_training[cur_id][0])
print("")
print("Top-5 most uncertain predictions:")
print("")
for cur_id, pred_mean, pred_std in pred[-5:]:
print(
"True: {0:.6f} +- {1:.6f}".format(
data_for_training[cur_id][1], data_for_training[cur_id][2]
)
)
print("Predicted: {0:.6f} +- {1:.6f}".format(pred_mean, pred_std))
print(data_for_training[cur_id][0])
print("")
del predictions_for_testing, error, pred
del datagen_for_training, datagen_for_validation
del labels_for_training, tokens_for_training
del data_for_training
del IDs_for_training, IDs_for_validation, IDs_for_final_testing
del model_for_training
gc.collect()
with codecs.open(submission_name, mode="w", encoding="utf-8") as fp:
data_writer = csv.writer(fp, quotechar='"', delimiter=",")
data_writer.writerow(["id", "target"])
for data_part in load_data_for_testing(
testset_name, pretrained_tokenizer, MINIBATCH_SIZE * 8
):
labels_for_submission, tokens_for_submission = tokenize_data(
data=data_part, tokenizer=pretrained_tokenizer, max_seq_len=MAX_TEXT_LEN
)
del data_part
predictions_for_submission = do_predictions(
fe=fe_model,
regressor=model_for_regression,
feature_scaler=text_feature_scaler,
batch_size=MINIBATCH_SIZE,
data_for_anchors=anchor_data,
dist_matrix=anchor_distances,
data=labels_for_submission,
token_matrix=tokens_for_submission,
pad_id=pretrained_tokenizer.pad_token_id,
)
for cur_id in predictions_for_submission:
predicted = predictions_for_submission[cur_id][0]
data_writer.writerow([cur_id, f"{predicted}"])
del predictions_for_submission
del labels_for_submission, tokens_for_submission
gc.collect()
| false | 0 | 13,608 | 1 | 13,682 | 13,608 |
||
69595535
|
import os # ceate folders
from glob import glob # get paths and use the to have floders name
from tqdm.notebook import tqdm # get nice bar
import sys
import pydicom as pdc # read dicom images
import numpy as np
import imageio # save to PNG images
import matplotlib.pyplot as plt # plot some PNG images
import cv2 as cv # read PNG images
from random import sample
from joblib import Parallel, delayed
import subprocess
from ast import literal_eval
# get CPU informations
def run(command):
process = subprocess.Popen(command, shell=True, stdout=subprocess.PIPE)
out, err = process.communicate()
print(out.decode("utf-8").strip())
print("# CPU")
run('cat /proc/cpuinfo | egrep -m 1 "^model name"')
run('cat /proc/cpuinfo | egrep -m 1 "^cpu MHz"')
run('cat /proc/cpuinfo | egrep -m 1 "^cpu cores"')
# train and test folder names
train_path_list = glob(
"../input/rsna-miccai-brain-tumor-radiogenomic-classification/train/*"
)
test_path_list = glob(
"../input/rsna-miccai-brain-tumor-radiogenomic-classification/test/*"
)
kaggle_input_path = "../input/"
# list of name of subdirectorys
train_d = list(map(lambda path: path.split("/")[-1], train_path_list))
test_d = list(map(lambda path: path.split("/")[-1], test_path_list))
mpMRI_scans = ["FLAIR", "T1w", "T1wCE", "T2w"]
# sample of names
print(train_d[:4])
# read dico image and return the image array
def read_dcm(img):
"""reading a dicom image with preprocessing"""
dcm_img = pdc.dcmread(img)
img_array = dcm_img.pixel_array
img_array = img_array - np.min(img_array)
if np.max(img_array) != 0:
img_array = img_array / np.max(img_array)
img_array = (img_array * 255).astype(np.uint8)
return img_array
# delete old folder (if you run the code twice)
# create the main PNG folder with the test and train
png_test_path = "Png-rsna-miccai-brain-tumor-radiogenomic-classification/test"
png_train_path = "Png-rsna-miccai-brain-tumor-radiogenomic-classification/train"
os.mkdir("Png-rsna-miccai-brain-tumor-radiogenomic-classification")
os.mkdir(png_train_path)
os.mkdir(png_test_path)
# floders creation
for trfold in train_d:
os.mkdir(png_train_path + "/" + trfold)
for mp in mpMRI_scans:
os.mkdir(png_train_path + "/" + trfold + "/" + mp)
for tsfold in test_d:
os.mkdir(png_test_path + "/" + tsfold)
for mp in mpMRI_scans:
os.mkdir(png_test_path + "/" + tsfold + "/" + mp)
# list the paths and name directory
png_train_path_list = glob(
"Png-rsna-miccai-brain-tumor-radiogenomic-classification/test/*"
)
png_test_path_list = glob(
"Png-rsna-miccai-brain-tumor-radiogenomic-classification/train/*"
)
png_train_d = list(map(lambda path: path.split("/")[-1], png_train_path_list))
png_test_d = list(map(lambda path: path.split("/")[-1], png_test_path_list))
# print sample of Png path folders and there names
print(png_train_path_list[:3])
print(png_train_d[:3])
# compare the folder result
print(train_d.sort() == png_train_d.sort())
print(test_d.sort() == png_test_d.sort())
# list of all dicom iamges paths
train_images_path = glob(
"../input/rsna-miccai-brain-tumor-radiogenomic-classification/train/*/*/*"
)
test_images_path = glob(
"../input/rsna-miccai-brain-tumor-radiogenomic-classification/test/*/*/*"
)
# the logic to create the new PNG path
image_name = test_images_path[0].split("/")[4:][-1].split(".")[0]
new_png_path = (
png_train_path
+ "/"
+ "/".join(train_images_path[0].split("/")[4:-1])
+ "/"
+ image_name
+ ".PNG"
)
print(new_png_path)
print(len(train_images_path), len(test_images_path))
# PNG save function
def train_png(train_path):
train_array_img = read_dcm(train_path)
train_image_name = train_path.split("/")[4:][-1].split(".")[0]
new_train_png_path = (
png_train_path
+ "/"
+ "/".join(train_path.split("/")[4:-1])
+ "/"
+ train_image_name
+ ".PNG"
)
imageio.imsave(new_train_png_path, train_array_img)
def test_png(test_path):
test_array_path = read_dcm(test_path)
test_image_name = test_path.split("/")[4:][-1].split(".")[0]
new_test_png_path = (
png_test_path
+ "/"
+ "/".join(test_path.split("/")[4:-1])
+ "/"
+ test_image_name
+ ".PNG"
)
imageio.imsave(new_test_png_path, test_array_path)
# FIXED : not all the training data was converted and the slow conversion
fb_train = Parallel(n_jobs=4, verbose=1, prefer="threads")(
delayed(train_png)(train_path)
for train_path in tqdm(train_images_path, total=len(train_images_path))
)
fb_test = Parallel(n_jobs=4, verbose=1, prefer="threads")(
delayed(test_png)(test_path)
for test_path in tqdm(test_images_path, total=len(test_images_path))
)
# ### Image creation and saving part
# FIXED : printing some black images
# print(PNG random PNG images)
png_images = glob("Png-rsna-miccai-brain-tumor-radiogenomic-classification/test/*/*/*")
print(png_images[:10])
plt.figure(figsize=(15, 15))
plot_indicator = 0
sub_in = 0
for index, image in tqdm(enumerate(sample(png_images, 5000))):
img = cv.imread(image, cv.IMREAD_GRAYSCALE)
img = cv.resize(img, (200, 200))
if np.max(img) != 0 and np.mean(img) >= 30:
plot_indicator += 1
if plot_indicator == 5:
break
else:
sub_in += 1
plt.subplot(2, 2, sub_in)
plt.imshow(img)
else:
continue
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/595/69595535.ipynb
| null | null |
[{"Id": 69595535, "ScriptId": 18958896, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5722471, "CreationDate": "08/02/2021 02:22:05", "VersionNumber": 5.0, "Title": "\u200b\ud83e\udde0Fast DICOM--> PNG full DATA+ {Download DATA} \u2705", "EvaluationDate": "08/02/2021", "IsChange": false, "TotalLines": 158.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 158.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import os # ceate folders
from glob import glob # get paths and use the to have floders name
from tqdm.notebook import tqdm # get nice bar
import sys
import pydicom as pdc # read dicom images
import numpy as np
import imageio # save to PNG images
import matplotlib.pyplot as plt # plot some PNG images
import cv2 as cv # read PNG images
from random import sample
from joblib import Parallel, delayed
import subprocess
from ast import literal_eval
# get CPU informations
def run(command):
process = subprocess.Popen(command, shell=True, stdout=subprocess.PIPE)
out, err = process.communicate()
print(out.decode("utf-8").strip())
print("# CPU")
run('cat /proc/cpuinfo | egrep -m 1 "^model name"')
run('cat /proc/cpuinfo | egrep -m 1 "^cpu MHz"')
run('cat /proc/cpuinfo | egrep -m 1 "^cpu cores"')
# train and test folder names
train_path_list = glob(
"../input/rsna-miccai-brain-tumor-radiogenomic-classification/train/*"
)
test_path_list = glob(
"../input/rsna-miccai-brain-tumor-radiogenomic-classification/test/*"
)
kaggle_input_path = "../input/"
# list of name of subdirectorys
train_d = list(map(lambda path: path.split("/")[-1], train_path_list))
test_d = list(map(lambda path: path.split("/")[-1], test_path_list))
mpMRI_scans = ["FLAIR", "T1w", "T1wCE", "T2w"]
# sample of names
print(train_d[:4])
# read dico image and return the image array
def read_dcm(img):
"""reading a dicom image with preprocessing"""
dcm_img = pdc.dcmread(img)
img_array = dcm_img.pixel_array
img_array = img_array - np.min(img_array)
if np.max(img_array) != 0:
img_array = img_array / np.max(img_array)
img_array = (img_array * 255).astype(np.uint8)
return img_array
# delete old folder (if you run the code twice)
# create the main PNG folder with the test and train
png_test_path = "Png-rsna-miccai-brain-tumor-radiogenomic-classification/test"
png_train_path = "Png-rsna-miccai-brain-tumor-radiogenomic-classification/train"
os.mkdir("Png-rsna-miccai-brain-tumor-radiogenomic-classification")
os.mkdir(png_train_path)
os.mkdir(png_test_path)
# floders creation
for trfold in train_d:
os.mkdir(png_train_path + "/" + trfold)
for mp in mpMRI_scans:
os.mkdir(png_train_path + "/" + trfold + "/" + mp)
for tsfold in test_d:
os.mkdir(png_test_path + "/" + tsfold)
for mp in mpMRI_scans:
os.mkdir(png_test_path + "/" + tsfold + "/" + mp)
# list the paths and name directory
png_train_path_list = glob(
"Png-rsna-miccai-brain-tumor-radiogenomic-classification/test/*"
)
png_test_path_list = glob(
"Png-rsna-miccai-brain-tumor-radiogenomic-classification/train/*"
)
png_train_d = list(map(lambda path: path.split("/")[-1], png_train_path_list))
png_test_d = list(map(lambda path: path.split("/")[-1], png_test_path_list))
# print sample of Png path folders and there names
print(png_train_path_list[:3])
print(png_train_d[:3])
# compare the folder result
print(train_d.sort() == png_train_d.sort())
print(test_d.sort() == png_test_d.sort())
# list of all dicom iamges paths
train_images_path = glob(
"../input/rsna-miccai-brain-tumor-radiogenomic-classification/train/*/*/*"
)
test_images_path = glob(
"../input/rsna-miccai-brain-tumor-radiogenomic-classification/test/*/*/*"
)
# the logic to create the new PNG path
image_name = test_images_path[0].split("/")[4:][-1].split(".")[0]
new_png_path = (
png_train_path
+ "/"
+ "/".join(train_images_path[0].split("/")[4:-1])
+ "/"
+ image_name
+ ".PNG"
)
print(new_png_path)
print(len(train_images_path), len(test_images_path))
# PNG save function
def train_png(train_path):
train_array_img = read_dcm(train_path)
train_image_name = train_path.split("/")[4:][-1].split(".")[0]
new_train_png_path = (
png_train_path
+ "/"
+ "/".join(train_path.split("/")[4:-1])
+ "/"
+ train_image_name
+ ".PNG"
)
imageio.imsave(new_train_png_path, train_array_img)
def test_png(test_path):
test_array_path = read_dcm(test_path)
test_image_name = test_path.split("/")[4:][-1].split(".")[0]
new_test_png_path = (
png_test_path
+ "/"
+ "/".join(test_path.split("/")[4:-1])
+ "/"
+ test_image_name
+ ".PNG"
)
imageio.imsave(new_test_png_path, test_array_path)
# FIXED : not all the training data was converted and the slow conversion
fb_train = Parallel(n_jobs=4, verbose=1, prefer="threads")(
delayed(train_png)(train_path)
for train_path in tqdm(train_images_path, total=len(train_images_path))
)
fb_test = Parallel(n_jobs=4, verbose=1, prefer="threads")(
delayed(test_png)(test_path)
for test_path in tqdm(test_images_path, total=len(test_images_path))
)
# ### Image creation and saving part
# FIXED : printing some black images
# print(PNG random PNG images)
png_images = glob("Png-rsna-miccai-brain-tumor-radiogenomic-classification/test/*/*/*")
print(png_images[:10])
plt.figure(figsize=(15, 15))
plot_indicator = 0
sub_in = 0
for index, image in tqdm(enumerate(sample(png_images, 5000))):
img = cv.imread(image, cv.IMREAD_GRAYSCALE)
img = cv.resize(img, (200, 200))
if np.max(img) != 0 and np.mean(img) >= 30:
plot_indicator += 1
if plot_indicator == 5:
break
else:
sub_in += 1
plt.subplot(2, 2, sub_in)
plt.imshow(img)
else:
continue
| false | 0 | 1,870 | 0 | 1,870 | 1,870 |
||
69595059
|
# # **House Price Prediction using Advance Reggression:**
# * **EDA**
# * **Pre-Processing**
# * **Handling Missing Data**
# * **Handling Outliners**
# * **Lineer Reggresion**
# * **Polynomial Regression**
# * **Regularization (Ridge - LASSO - ElasticNet)**
#
# loading necessary libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# loading train and test from house-prices dataset
train_set = pd.read_csv(
"../input/house-prices-advanced-regression-techniques/train.csv"
)
test_set = pd.read_csv("../input/house-prices-advanced-regression-techniques/test.csv")
test_label = pd.read_csv(
"../input/house-prices-advanced-regression-techniques/sample_submission.csv"
)
test_set.shape
# adding test label
test_set["SalePrice"] = test_label["SalePrice"]
test_set.shape
train_set.shape
# combining the train and test set for cleaning
# df_final= pd.concat([train_set,test_set])
df_final = pd.concat(
[test_set.assign(ind="test_set"), train_set.assign(ind="train_set")]
)
# # **EDA**
# correlation between 'SalePrice' and 'GrLivArea'
# we can spot outliners that we delete later on
sns.jointplot(data=train_set, x="SalePrice", y="GrLivArea")
# OverallQual is the most corr feature we can see the Linear shape and no outliner
figure = figsize = (30, 34)
sns.stripplot(data=train_set, x="OverallQual", y="SalePrice")
# # Pre-Processing
# 1. Handling missing value
# 2. High/Low Correletion data
# 3. Categorical Data
# 4. Numerical Columns to Categorical
# 5. Dealing with Outliers
# 6. Creating Dummy Variables
# one-hot-encodding
# # 1. Handling **missing** Values
# * Dropping columns with more than 70% null_value and Id (it might not be the best case for every problem or dataset).
# * Handling null value in the rest of thr features
# >
# finding features with the most duplicant value ?
def missing_percent(train_set):
nan_percent = 100 * (train_set.isnull().sum() / len(train_set))
nan_percent = nan_percent[nan_percent > 0].sort_values(ascending=False).round(1)
DataFrame = pd.DataFrame(nan_percent)
# Rename the columns
mis_percent_table = DataFrame.rename(columns={0: "% of Misiing Values"})
# Sort the table by percentage of missing descending
mis_percent = mis_percent_table
return mis_percent
miss = missing_percent(train_set)
miss
# Removing the Id that has no value for our prediction
train_set = train_set.drop("Id", axis=1)
nan_percent = 100 * (train_set.isnull().sum() / len(train_set))
nan_percent = nan_percent[nan_percent > 0].sort_values()
# Every Feature with missing data must be checked!
# We choose a threshold of 1%. It means, if there is less than 1% of a feature are missing
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=90)
# Set 1% threshold:
plt.ylim(0, 1)
train_set.shape
# Removing the Id that has no value for our prediction
train_set = train_set.drop("Id", axis=1)
# drop features that have more than 70% missing value
# credit: https://www.kaggle.com/rushikeshdarge/handle-missing-values-only-notebook-you-need
threshold = 70
drop_cols = miss[miss["% of Misiing Values"] > threshold].index.tolist()
drop_cols
train_set = train_set.drop(drop_cols, axis=1)
nan_percent = 100 * (train_set.isnull().sum() / len(train_set))
nan_percent = nan_percent[nan_percent > 0].sort_values()
# every Feature with missing data must be checked!
# We choose a threshold of 1%. It means, if there is less than 1% of a feature are missing
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=90)
# Set 1% threshold:
plt.ylim(0, 1)
# **FireplaceQu: Fireplace quality**
# * acoording to the data this feature has an NA value that means the house has no fire place so we fill the column with 'None'
train_set["FireplaceQu"] = train_set["FireplaceQu"].fillna("None")
# Filling null values most freq value
# train_set['KitchenQual']= train_set['KitchenQual'].fillna('TA')
# df_final['SaleType']= df_final['SaleType'].fillna('Oth')
# df_final['Functional']= df_final['Functional'].fillna('Typ')
# df_final['Exterior1st']= df_final['Exterior1st'].fillna('Other')
# df_final.fillna({'Exterior1st':'Other', 'Exterior2nd':'Other', 'Utilities':'Other'}, inplace=True)
# **Garage & Bacement**
# * by looking at the plot we realize that most features with missing value are from the same catagories.
# After checking the data documentation,
# it shows that missing value (two rows) in Basement Features are becouse of there is no basement in these rows
# Decision: Filling in data based on column: numerical basement & string descriptive:
# Numerical Columns fill with 0:
bsmt_num_cols = [
"BsmtFinSF1",
"BsmtFinSF2",
"BsmtUnfSF",
"TotalBsmtSF",
"BsmtFullBath",
"BsmtHalfBath",
]
train_set[bsmt_num_cols] = train_set[bsmt_num_cols].fillna(0)
# String Columns fill with None:
bsmt_str_cols = ["BsmtQual", "BsmtCond", "BsmtExposure", "BsmtFinType1", "BsmtFinType2"]
train_set[bsmt_str_cols] = train_set[bsmt_str_cols].fillna("None")
# **Mas Vnr Features:**
# * Based on the Dataset Document File, missing values for 'Mas Vnr Type' and 'Mas Vnr Area' means the house doesn't have any mansonry veneer. so, we decide to fill the missing value as below:
train_set["MasVnrType"] = train_set["MasVnrType"].fillna("None")
train_set["MasVnrArea"] = train_set["MasVnrArea"].fillna(0)
# **Garage Columns:**
# * Based on the dataset documentation, NaN in Garage Columns seems to indicate no garage.
# * Decision: Fill with 'None' or 0
train_set[["GarageType", "GarageYrBlt", "GarageFinish", "GarageQual", "GarageCond"]]
# now we will extract all the numerical features from the dataset
# numerical_features= [feature for feature in train_set.columns if train_set[feature].dtypes !='O']
# print('Number of Numerical Features:',len(numerical_features))
# train_set[numerical_features].head(5)
# now we will extract datatime features from the dataset
# year_feature=[feature for feature in numerical_features if 'Year' in feature or 'Yr' in feature]
# print('Number of Yearly Features:',len(year_feature))
# train_set[year_feature].head(5)
# now we will analyze yearly features wrt SalePrice which is our independent feature
# for feature in year_feature:
# train_set.groupby(feature)['SalePrice'].median().plot()
# plt.show()
# Filling the missing Value:
Gar_str_cols = ["GarageType", "GarageFinish", "GarageQual", "GarageCond"]
train_set[Gar_str_cols] = train_set[Gar_str_cols].fillna("None")
train_set["GarageYrBlt"] = train_set["GarageYrBlt"].fillna(0)
# Impute missing data based on other columns:
train_set.groupby("Neighborhood")["LotFrontage"]
train_set.groupby("Neighborhood")["LotFrontage"].mean()
# Filling null values mean value
train_set.groupby("Neighborhood")["LotFrontage"].transform(
lambda val: val.fillna(val.mean())
)
train_set["LotFrontage"] = train_set.groupby("Neighborhood")["LotFrontage"].transform(
lambda val: val.fillna(val.mean())
)
# Filling null values most freq value
# train_set['MSZoning'].value_counts()[train_set['MSZoning'].value_counts() == df_final['MSZoning'].value_counts().max()].index
# train_set['MSZoning']=train_set['MSZoning'].fillna('RL')
train_set["LotFrontage"] = train_set["LotFrontage"].fillna(0)
train_set[train_set["GarageArea"].isnull()]
# # Handling test_set null value
# * Functional
# * Exterior1st
# * Exterior2nd
# * KitchenQual
# * SaleType
# * Utilities
# * Functional
# * MSZoning
# Filling null values most freq value
train_set["LotFrontage"] = train_set.groupby("Neighborhood")["LotFrontage"].transform(
lambda val: val.fillna(val.max())
)
# Filling null values most freq value
# train_set['Functional'].value_counts()[train_set['Functional'].value_counts() == train_set['Functional'].value_counts().max()].index
nan_percent = 100 * (train_set.isnull().sum() / len(df_final))
nan_percent = nan_percent[nan_percent > 0].sort_values()
# plot the feature with missing indicating the percent of missing data
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=90)
train_set = train_set.dropna(axis=0, subset=["Electrical", "GarageArea"])
# Filling null values most freq value
# df_final['MSZoning'].value_counts()[df_final['MSZoning'].value_counts() == df_final['MSZoning'].value_counts().max()].index
# df_final['MSZoning']= df_final['MSZoning'].transform(lambda val: val.fillna(val.max()))
df_final.isnull().sum()
# **Finally we check if there is more null value in the dataset**
nan_percent = missing_percent(train_set)
nan_percent = 100 * (train_set.isnull().sum() / len(df_final))
nan_percent = nan_percent[nan_percent > 0].sort_values()
nan_percent
# df = pd.concat([test.assign(ind="test"), train.assign(ind="train")])
train_set.shape
# # Handling Outliers
corr = train_set.corr()
top_corr_features = corr.index[abs(corr["SalePrice"]) > 0.5].sort_values(ascending=True)
top_corr_features
# train_set[(train_set['OverallQual']>8) &(train_set['SalePrice']<200000)][['SalePrice', 'OverallQual']]
sns.scatterplot(x="GrLivArea", y="SalePrice", data=train_set)
plt.axhline(y=200000, color="r")
plt.axvline(x=4000, color="r")
train_set[(train_set["GrLivArea"] > 4000) & (train_set["SalePrice"] < 400000)][
["SalePrice", "GrLivArea"]
]
# Remove the outliers:
index_drop = train_set[
(train_set["GrLivArea"] > 4000) & (train_set["SalePrice"] < 400000)
].index
train_set = train_set.drop(index_drop, axis=0)
sns.scatterplot(x="GrLivArea", y="SalePrice", data=train_set)
plt.axhline(y=200000, color="r")
plt.axvline(x=4000, color="r")
# GrLivArea without outliner
# Remove the outliers:
index_drop = train_set[
(train_set["GrLivArea"] > 4000) & (train_set["SalePrice"] > 400000)
].index
train_set = train_set.drop(index_drop, axis=0)
sns.scatterplot(x="GrLivArea", y="SalePrice", data=train_set)
plt.axhline(y=200000, color="r")
plt.axvline(x=4000, color="r")
sns.scatterplot(x="OverallQual", y="SalePrice", data=train_set)
# no need to remove any data
sns.boxplot(x="GarageCars", y="SalePrice", data=train_set)
plt.axhline(y=680000, color="r")
sns.scatterplot(data=train_set, x="TotRmsAbvGrd", y="SalePrice")
plt.axhline(y=250000, color="r")
plt.axvline(x=12.8, color="r")
train_set[(train_set["TotRmsAbvGrd"] > 12.7) & (train_set["SalePrice"] < 250000)][
["SalePrice", "TotRmsAbvGrd"]
]
# Remove the outliers:
index_drop = train_set[
(train_set["TotRmsAbvGrd"] > 12.7) & (train_set["SalePrice"] < 250000)
].index
train_set = train_set.drop(index_drop, axis=0)
sns.scatterplot(data=train_set, x="TotRmsAbvGrd", y="SalePrice")
sns.scatterplot(data=train_set, x="TotRmsAbvGrd", y="SalePrice")
# # Features that have high correlation (higher than 0.5)
# get correlations of each features in dataset
# Plotting Heat Map to visualise correlation data better.
# Drwan for only features having high correlation
# (>0.5) with Target Variable
corr = train_set.corr()
top_corr_features = corr.index[abs(corr["SalePrice"]) > 0.5]
plt.figure(figsize=(10, 10))
# plot heat map
g = sns.heatmap(train_set[top_corr_features].corr(), annot=True, cmap="YlGnBu")
train_set.groupby("OverallQual")["SalePrice"].median().plot()
plt.xlabel("Year Sold")
plt.ylabel("Median House Price")
plt.title("House Price vs YearSold")
top_corr_features
# # Dealing with Categorical Data
# Convert to String:
train_set["MSSubClass"] = train_set["MSSubClass"].apply(str)
# **Creating Dummy Variables**
train_set.select_dtypes(include="object")
train_set_num = train_set.select_dtypes(exclude="object")
train_set_obj = train_set.select_dtypes(include="object")
# Converting:
train_set_obj = pd.get_dummies(train_set_obj, drop_first=True)
Final_df = pd.concat([train_set_num, train_set_obj], axis=1)
# # **Linear Reggression**
# * we start our path with simple Linear Regression and then we try to improve our model
# Separate features and target from train_df
X = Final_df.drop("SalePrice", axis=1)
y = Final_df["SalePrice"]
# X = X.apply(pd.to_numeric, errors='coerce')
# y = y.apply(pd.to_numeric, errors='coerce')
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
# Split the Dataset to Train & Test
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=100)
# train the model
from sklearn.linear_model import LinearRegression
model = LinearRegression()
model.fit(X_train, y_train)
# predicting test data
y_pred = model.predict(X_test)
# evaluating the model
from sklearn import metrics
MAE = metrics.mean_absolute_error(y_test, y_pred)
MSE = metrics.mean_squared_error(y_test, y_pred)
RMSE = np.sqrt(MSE)
# coeficient matrix
pd.DataFrame(model.coef_, X.columns, columns=["coeficient"])
pd.DataFrame(
data=[MAE, MSE, RMSE], index=["MAE", "MSE", "RMSE"], columns=["LinearRegression"]
)
# # Polynomial Regression improves our model
# * Polynomial Regression adding more relevant features
# *
from sklearn.preprocessing import PolynomialFeatures
polynomial_converter = PolynomialFeatures(degree=2, include_bias=False)
poly_features = polynomial_converter.fit(X)
poly_features = polynomial_converter.transform(X)
# # Poly_Features: X1, X2, X3, X1^2, X2^2, X3^2, X1X2, X1X3, X2X3
# * Split the Data to Train & Test
# * Train the Model
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
poly_features, y, test_size=0.3, random_state=101
)
from sklearn.linear_model import LinearRegression
polymodel = LinearRegression()
polymodel.fit(X_train, y_train)
y_pred = polymodel.predict(X_test)
pd.DataFrame({"Y_Test": y_test, "Y_Pred": y_pred, "Residuals": (y_test - y_pred)}).head(
5
)
# # Polymodel Regression vs Linear Regression
from sklearn import metrics
MAE_Poly = metrics.mean_absolute_error(y_test, y_pred)
MSE_Poly = metrics.mean_squared_error(y_test, y_pred)
RMSE_Poly = np.sqrt(MSE_Poly)
pd.DataFrame(
[MAE_Poly, MSE_Poly, RMSE_Poly], index=["MAE", "MSE", "RMSE"], columns=["metrics"]
)
# **RMSE decresed significeantly**
XS_train, XS_test, ys_train, ys_test = train_test_split(
X, y, test_size=0.3, random_state=101
)
simplemodel = LinearRegression()
simplemodel.fit(XS_train, ys_train)
ys_pred = simplemodel.predict(XS_test)
MAE_simple = metrics.mean_absolute_error(ys_test, ys_pred)
MSE_simple = metrics.mean_squared_error(ys_test, ys_pred)
RMSE_simple = np.sqrt(MSE_simple)
pd.DataFrame(
{
"Poly Metrics": [MAE_Poly, MSE_Poly, RMSE_Poly],
"Simple Metrics": [MAE_simple, MSE_simple, RMSE_simple],
},
index=["MAE", "MSE", "RMSE"],
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/595/69595059.ipynb
| null | null |
[{"Id": 69595059, "ScriptId": 18706063, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2121244, "CreationDate": "08/02/2021 02:11:50", "VersionNumber": 12.0, "Title": "Regression", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 438.0, "LinesInsertedFromPrevious": 220.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 218.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # **House Price Prediction using Advance Reggression:**
# * **EDA**
# * **Pre-Processing**
# * **Handling Missing Data**
# * **Handling Outliners**
# * **Lineer Reggresion**
# * **Polynomial Regression**
# * **Regularization (Ridge - LASSO - ElasticNet)**
#
# loading necessary libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# loading train and test from house-prices dataset
train_set = pd.read_csv(
"../input/house-prices-advanced-regression-techniques/train.csv"
)
test_set = pd.read_csv("../input/house-prices-advanced-regression-techniques/test.csv")
test_label = pd.read_csv(
"../input/house-prices-advanced-regression-techniques/sample_submission.csv"
)
test_set.shape
# adding test label
test_set["SalePrice"] = test_label["SalePrice"]
test_set.shape
train_set.shape
# combining the train and test set for cleaning
# df_final= pd.concat([train_set,test_set])
df_final = pd.concat(
[test_set.assign(ind="test_set"), train_set.assign(ind="train_set")]
)
# # **EDA**
# correlation between 'SalePrice' and 'GrLivArea'
# we can spot outliners that we delete later on
sns.jointplot(data=train_set, x="SalePrice", y="GrLivArea")
# OverallQual is the most corr feature we can see the Linear shape and no outliner
figure = figsize = (30, 34)
sns.stripplot(data=train_set, x="OverallQual", y="SalePrice")
# # Pre-Processing
# 1. Handling missing value
# 2. High/Low Correletion data
# 3. Categorical Data
# 4. Numerical Columns to Categorical
# 5. Dealing with Outliers
# 6. Creating Dummy Variables
# one-hot-encodding
# # 1. Handling **missing** Values
# * Dropping columns with more than 70% null_value and Id (it might not be the best case for every problem or dataset).
# * Handling null value in the rest of thr features
# >
# finding features with the most duplicant value ?
def missing_percent(train_set):
nan_percent = 100 * (train_set.isnull().sum() / len(train_set))
nan_percent = nan_percent[nan_percent > 0].sort_values(ascending=False).round(1)
DataFrame = pd.DataFrame(nan_percent)
# Rename the columns
mis_percent_table = DataFrame.rename(columns={0: "% of Misiing Values"})
# Sort the table by percentage of missing descending
mis_percent = mis_percent_table
return mis_percent
miss = missing_percent(train_set)
miss
# Removing the Id that has no value for our prediction
train_set = train_set.drop("Id", axis=1)
nan_percent = 100 * (train_set.isnull().sum() / len(train_set))
nan_percent = nan_percent[nan_percent > 0].sort_values()
# Every Feature with missing data must be checked!
# We choose a threshold of 1%. It means, if there is less than 1% of a feature are missing
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=90)
# Set 1% threshold:
plt.ylim(0, 1)
train_set.shape
# Removing the Id that has no value for our prediction
train_set = train_set.drop("Id", axis=1)
# drop features that have more than 70% missing value
# credit: https://www.kaggle.com/rushikeshdarge/handle-missing-values-only-notebook-you-need
threshold = 70
drop_cols = miss[miss["% of Misiing Values"] > threshold].index.tolist()
drop_cols
train_set = train_set.drop(drop_cols, axis=1)
nan_percent = 100 * (train_set.isnull().sum() / len(train_set))
nan_percent = nan_percent[nan_percent > 0].sort_values()
# every Feature with missing data must be checked!
# We choose a threshold of 1%. It means, if there is less than 1% of a feature are missing
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=90)
# Set 1% threshold:
plt.ylim(0, 1)
# **FireplaceQu: Fireplace quality**
# * acoording to the data this feature has an NA value that means the house has no fire place so we fill the column with 'None'
train_set["FireplaceQu"] = train_set["FireplaceQu"].fillna("None")
# Filling null values most freq value
# train_set['KitchenQual']= train_set['KitchenQual'].fillna('TA')
# df_final['SaleType']= df_final['SaleType'].fillna('Oth')
# df_final['Functional']= df_final['Functional'].fillna('Typ')
# df_final['Exterior1st']= df_final['Exterior1st'].fillna('Other')
# df_final.fillna({'Exterior1st':'Other', 'Exterior2nd':'Other', 'Utilities':'Other'}, inplace=True)
# **Garage & Bacement**
# * by looking at the plot we realize that most features with missing value are from the same catagories.
# After checking the data documentation,
# it shows that missing value (two rows) in Basement Features are becouse of there is no basement in these rows
# Decision: Filling in data based on column: numerical basement & string descriptive:
# Numerical Columns fill with 0:
bsmt_num_cols = [
"BsmtFinSF1",
"BsmtFinSF2",
"BsmtUnfSF",
"TotalBsmtSF",
"BsmtFullBath",
"BsmtHalfBath",
]
train_set[bsmt_num_cols] = train_set[bsmt_num_cols].fillna(0)
# String Columns fill with None:
bsmt_str_cols = ["BsmtQual", "BsmtCond", "BsmtExposure", "BsmtFinType1", "BsmtFinType2"]
train_set[bsmt_str_cols] = train_set[bsmt_str_cols].fillna("None")
# **Mas Vnr Features:**
# * Based on the Dataset Document File, missing values for 'Mas Vnr Type' and 'Mas Vnr Area' means the house doesn't have any mansonry veneer. so, we decide to fill the missing value as below:
train_set["MasVnrType"] = train_set["MasVnrType"].fillna("None")
train_set["MasVnrArea"] = train_set["MasVnrArea"].fillna(0)
# **Garage Columns:**
# * Based on the dataset documentation, NaN in Garage Columns seems to indicate no garage.
# * Decision: Fill with 'None' or 0
train_set[["GarageType", "GarageYrBlt", "GarageFinish", "GarageQual", "GarageCond"]]
# now we will extract all the numerical features from the dataset
# numerical_features= [feature for feature in train_set.columns if train_set[feature].dtypes !='O']
# print('Number of Numerical Features:',len(numerical_features))
# train_set[numerical_features].head(5)
# now we will extract datatime features from the dataset
# year_feature=[feature for feature in numerical_features if 'Year' in feature or 'Yr' in feature]
# print('Number of Yearly Features:',len(year_feature))
# train_set[year_feature].head(5)
# now we will analyze yearly features wrt SalePrice which is our independent feature
# for feature in year_feature:
# train_set.groupby(feature)['SalePrice'].median().plot()
# plt.show()
# Filling the missing Value:
Gar_str_cols = ["GarageType", "GarageFinish", "GarageQual", "GarageCond"]
train_set[Gar_str_cols] = train_set[Gar_str_cols].fillna("None")
train_set["GarageYrBlt"] = train_set["GarageYrBlt"].fillna(0)
# Impute missing data based on other columns:
train_set.groupby("Neighborhood")["LotFrontage"]
train_set.groupby("Neighborhood")["LotFrontage"].mean()
# Filling null values mean value
train_set.groupby("Neighborhood")["LotFrontage"].transform(
lambda val: val.fillna(val.mean())
)
train_set["LotFrontage"] = train_set.groupby("Neighborhood")["LotFrontage"].transform(
lambda val: val.fillna(val.mean())
)
# Filling null values most freq value
# train_set['MSZoning'].value_counts()[train_set['MSZoning'].value_counts() == df_final['MSZoning'].value_counts().max()].index
# train_set['MSZoning']=train_set['MSZoning'].fillna('RL')
train_set["LotFrontage"] = train_set["LotFrontage"].fillna(0)
train_set[train_set["GarageArea"].isnull()]
# # Handling test_set null value
# * Functional
# * Exterior1st
# * Exterior2nd
# * KitchenQual
# * SaleType
# * Utilities
# * Functional
# * MSZoning
# Filling null values most freq value
train_set["LotFrontage"] = train_set.groupby("Neighborhood")["LotFrontage"].transform(
lambda val: val.fillna(val.max())
)
# Filling null values most freq value
# train_set['Functional'].value_counts()[train_set['Functional'].value_counts() == train_set['Functional'].value_counts().max()].index
nan_percent = 100 * (train_set.isnull().sum() / len(df_final))
nan_percent = nan_percent[nan_percent > 0].sort_values()
# plot the feature with missing indicating the percent of missing data
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=90)
train_set = train_set.dropna(axis=0, subset=["Electrical", "GarageArea"])
# Filling null values most freq value
# df_final['MSZoning'].value_counts()[df_final['MSZoning'].value_counts() == df_final['MSZoning'].value_counts().max()].index
# df_final['MSZoning']= df_final['MSZoning'].transform(lambda val: val.fillna(val.max()))
df_final.isnull().sum()
# **Finally we check if there is more null value in the dataset**
nan_percent = missing_percent(train_set)
nan_percent = 100 * (train_set.isnull().sum() / len(df_final))
nan_percent = nan_percent[nan_percent > 0].sort_values()
nan_percent
# df = pd.concat([test.assign(ind="test"), train.assign(ind="train")])
train_set.shape
# # Handling Outliers
corr = train_set.corr()
top_corr_features = corr.index[abs(corr["SalePrice"]) > 0.5].sort_values(ascending=True)
top_corr_features
# train_set[(train_set['OverallQual']>8) &(train_set['SalePrice']<200000)][['SalePrice', 'OverallQual']]
sns.scatterplot(x="GrLivArea", y="SalePrice", data=train_set)
plt.axhline(y=200000, color="r")
plt.axvline(x=4000, color="r")
train_set[(train_set["GrLivArea"] > 4000) & (train_set["SalePrice"] < 400000)][
["SalePrice", "GrLivArea"]
]
# Remove the outliers:
index_drop = train_set[
(train_set["GrLivArea"] > 4000) & (train_set["SalePrice"] < 400000)
].index
train_set = train_set.drop(index_drop, axis=0)
sns.scatterplot(x="GrLivArea", y="SalePrice", data=train_set)
plt.axhline(y=200000, color="r")
plt.axvline(x=4000, color="r")
# GrLivArea without outliner
# Remove the outliers:
index_drop = train_set[
(train_set["GrLivArea"] > 4000) & (train_set["SalePrice"] > 400000)
].index
train_set = train_set.drop(index_drop, axis=0)
sns.scatterplot(x="GrLivArea", y="SalePrice", data=train_set)
plt.axhline(y=200000, color="r")
plt.axvline(x=4000, color="r")
sns.scatterplot(x="OverallQual", y="SalePrice", data=train_set)
# no need to remove any data
sns.boxplot(x="GarageCars", y="SalePrice", data=train_set)
plt.axhline(y=680000, color="r")
sns.scatterplot(data=train_set, x="TotRmsAbvGrd", y="SalePrice")
plt.axhline(y=250000, color="r")
plt.axvline(x=12.8, color="r")
train_set[(train_set["TotRmsAbvGrd"] > 12.7) & (train_set["SalePrice"] < 250000)][
["SalePrice", "TotRmsAbvGrd"]
]
# Remove the outliers:
index_drop = train_set[
(train_set["TotRmsAbvGrd"] > 12.7) & (train_set["SalePrice"] < 250000)
].index
train_set = train_set.drop(index_drop, axis=0)
sns.scatterplot(data=train_set, x="TotRmsAbvGrd", y="SalePrice")
sns.scatterplot(data=train_set, x="TotRmsAbvGrd", y="SalePrice")
# # Features that have high correlation (higher than 0.5)
# get correlations of each features in dataset
# Plotting Heat Map to visualise correlation data better.
# Drwan for only features having high correlation
# (>0.5) with Target Variable
corr = train_set.corr()
top_corr_features = corr.index[abs(corr["SalePrice"]) > 0.5]
plt.figure(figsize=(10, 10))
# plot heat map
g = sns.heatmap(train_set[top_corr_features].corr(), annot=True, cmap="YlGnBu")
train_set.groupby("OverallQual")["SalePrice"].median().plot()
plt.xlabel("Year Sold")
plt.ylabel("Median House Price")
plt.title("House Price vs YearSold")
top_corr_features
# # Dealing with Categorical Data
# Convert to String:
train_set["MSSubClass"] = train_set["MSSubClass"].apply(str)
# **Creating Dummy Variables**
train_set.select_dtypes(include="object")
train_set_num = train_set.select_dtypes(exclude="object")
train_set_obj = train_set.select_dtypes(include="object")
# Converting:
train_set_obj = pd.get_dummies(train_set_obj, drop_first=True)
Final_df = pd.concat([train_set_num, train_set_obj], axis=1)
# # **Linear Reggression**
# * we start our path with simple Linear Regression and then we try to improve our model
# Separate features and target from train_df
X = Final_df.drop("SalePrice", axis=1)
y = Final_df["SalePrice"]
# X = X.apply(pd.to_numeric, errors='coerce')
# y = y.apply(pd.to_numeric, errors='coerce')
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
# Split the Dataset to Train & Test
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=100)
# train the model
from sklearn.linear_model import LinearRegression
model = LinearRegression()
model.fit(X_train, y_train)
# predicting test data
y_pred = model.predict(X_test)
# evaluating the model
from sklearn import metrics
MAE = metrics.mean_absolute_error(y_test, y_pred)
MSE = metrics.mean_squared_error(y_test, y_pred)
RMSE = np.sqrt(MSE)
# coeficient matrix
pd.DataFrame(model.coef_, X.columns, columns=["coeficient"])
pd.DataFrame(
data=[MAE, MSE, RMSE], index=["MAE", "MSE", "RMSE"], columns=["LinearRegression"]
)
# # Polynomial Regression improves our model
# * Polynomial Regression adding more relevant features
# *
from sklearn.preprocessing import PolynomialFeatures
polynomial_converter = PolynomialFeatures(degree=2, include_bias=False)
poly_features = polynomial_converter.fit(X)
poly_features = polynomial_converter.transform(X)
# # Poly_Features: X1, X2, X3, X1^2, X2^2, X3^2, X1X2, X1X3, X2X3
# * Split the Data to Train & Test
# * Train the Model
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
poly_features, y, test_size=0.3, random_state=101
)
from sklearn.linear_model import LinearRegression
polymodel = LinearRegression()
polymodel.fit(X_train, y_train)
y_pred = polymodel.predict(X_test)
pd.DataFrame({"Y_Test": y_test, "Y_Pred": y_pred, "Residuals": (y_test - y_pred)}).head(
5
)
# # Polymodel Regression vs Linear Regression
from sklearn import metrics
MAE_Poly = metrics.mean_absolute_error(y_test, y_pred)
MSE_Poly = metrics.mean_squared_error(y_test, y_pred)
RMSE_Poly = np.sqrt(MSE_Poly)
pd.DataFrame(
[MAE_Poly, MSE_Poly, RMSE_Poly], index=["MAE", "MSE", "RMSE"], columns=["metrics"]
)
# **RMSE decresed significeantly**
XS_train, XS_test, ys_train, ys_test = train_test_split(
X, y, test_size=0.3, random_state=101
)
simplemodel = LinearRegression()
simplemodel.fit(XS_train, ys_train)
ys_pred = simplemodel.predict(XS_test)
MAE_simple = metrics.mean_absolute_error(ys_test, ys_pred)
MSE_simple = metrics.mean_squared_error(ys_test, ys_pred)
RMSE_simple = np.sqrt(MSE_simple)
pd.DataFrame(
{
"Poly Metrics": [MAE_Poly, MSE_Poly, RMSE_Poly],
"Simple Metrics": [MAE_simple, MSE_simple, RMSE_simple],
},
index=["MAE", "MSE", "RMSE"],
)
| false | 0 | 4,920 | 0 | 4,920 | 4,920 |
||
69595682
|
import pydicom as dicom
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import cv2
from scipy import ndimage
import os
TRAIN_DIR = "../input/rsna-miccai-brain-tumor-radiogenomic-classification/train"
TEST_DIR = "../input/rsna-miccai-brain-tumor-radiogenomic-classification/test"
TRAIN_LABELS = (
"../input/rsna-miccai-brain-tumor-radiogenomic-classification/train_labels.csv"
)
SUBMISSION = (
"../input/rsna-miccai-brain-tumor-radiogenomic-classification/sample_submission.csv"
)
# for root, dirs, files in os.walk(TRAIN_DIR):
# print(root)
# print("Number of patients: ", len(dirs))
# for patient in dirs:
# patient_dir = os.path.join(root, patient)
# FLAIR = os.path.join(patient_dir, 'FLAIR')
# FLAIR_COUNT = len(os.listdir(FLAIR))
# T1 = os.path.join(patient_dir, 'T1w')
# T1_COUNT = len(os.listdir(T1))
# T1CE = os.path.join(patient_dir, 'T1wCE')
# T1CE_COUNT = len(os.listdir(T1CE))
# T2 = os.path.join(patient_dir, 'T2w')
# T2_COUNT = len(os.listdir(T2))
# print(f'Patient: {patient} || FLAIR: {FLAIR_COUNT} | T1w: {T1_COUNT} | T1wCE: {T1CE_COUNT} | T2w: {T2_COUNT}')
# break
train_labels = pd.read_csv(TRAIN_LABELS)
print(train_labels["MGMT_value"].unique())
train_labels.head()
sub = pd.read_csv(SUBMISSION)
print(sub["MGMT_value"].unique())
sub.head()
save_dir = os.path.join(os.getcwd(), "rsna-slices-npz")
if not os.path.exists(save_dir):
os.makedirs(save_dir)
save_train_dir = os.path.join(save_dir, "train")
save_test_dir = os.path.join(save_dir, "test")
if not os.path.exists(save_train_dir):
os.makedirs(save_train_dir)
if not os.path.exists(save_test_dir):
os.makedirs(save_test_dir)
def combine_scans(input_dir, output_dir):
patients = os.listdir(input_dir)
train_data = {}
j = 0
for patient in patients:
img_types = os.listdir(os.path.join(input_dir, patient))
img_types.sort() # FFLAIR -> T1w -> T1wCE -> T2w
patient_dir = os.path.join(output_dir, patient)
if not os.path.exists(patient_dir):
os.makedirs(patient_dir) # output
for i, img_type in enumerate(img_types):
path = os.path.join(input_dir, patient, img_type) # input
slices = [dicom.read_file(path + "/" + x) for x in os.listdir(path)]
slices.sort(key=lambda x: int(x.ImagePositionPatient[2]))
slices = [x.pixel_array for x in slices if np.sum(x.pixel_array) > 0.0]
if len(slices) > 0:
stacked = np.stack(slices, axis=-1)
img_dir = os.path.join(patient_dir, img_type)
if not os.path.exists(img_dir):
os.makedirs(img_dir)
file_path = os.path.join(img_dir, f"{patient}_{img_type}_Image.npy")
# np.save(file_path, stacked)
else:
print(f"No Images for patient {patient} - {img_type}")
print(j, patient)
j += 1
combine_scans(TRAIN_DIR, save_train_dir)
print("Done with Train Dataset")
print("Start Test Dataset")
combine_scans(TEST_DIR, save_test_dir)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/595/69595682.ipynb
| null | null |
[{"Id": 69595682, "ScriptId": 18909176, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2205999, "CreationDate": "08/02/2021 02:25:24", "VersionNumber": 5.0, "Title": "RSNA_MICCAI_Brain_Tumor", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 88.0, "LinesInsertedFromPrevious": 12.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 76.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import pydicom as dicom
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import cv2
from scipy import ndimage
import os
TRAIN_DIR = "../input/rsna-miccai-brain-tumor-radiogenomic-classification/train"
TEST_DIR = "../input/rsna-miccai-brain-tumor-radiogenomic-classification/test"
TRAIN_LABELS = (
"../input/rsna-miccai-brain-tumor-radiogenomic-classification/train_labels.csv"
)
SUBMISSION = (
"../input/rsna-miccai-brain-tumor-radiogenomic-classification/sample_submission.csv"
)
# for root, dirs, files in os.walk(TRAIN_DIR):
# print(root)
# print("Number of patients: ", len(dirs))
# for patient in dirs:
# patient_dir = os.path.join(root, patient)
# FLAIR = os.path.join(patient_dir, 'FLAIR')
# FLAIR_COUNT = len(os.listdir(FLAIR))
# T1 = os.path.join(patient_dir, 'T1w')
# T1_COUNT = len(os.listdir(T1))
# T1CE = os.path.join(patient_dir, 'T1wCE')
# T1CE_COUNT = len(os.listdir(T1CE))
# T2 = os.path.join(patient_dir, 'T2w')
# T2_COUNT = len(os.listdir(T2))
# print(f'Patient: {patient} || FLAIR: {FLAIR_COUNT} | T1w: {T1_COUNT} | T1wCE: {T1CE_COUNT} | T2w: {T2_COUNT}')
# break
train_labels = pd.read_csv(TRAIN_LABELS)
print(train_labels["MGMT_value"].unique())
train_labels.head()
sub = pd.read_csv(SUBMISSION)
print(sub["MGMT_value"].unique())
sub.head()
save_dir = os.path.join(os.getcwd(), "rsna-slices-npz")
if not os.path.exists(save_dir):
os.makedirs(save_dir)
save_train_dir = os.path.join(save_dir, "train")
save_test_dir = os.path.join(save_dir, "test")
if not os.path.exists(save_train_dir):
os.makedirs(save_train_dir)
if not os.path.exists(save_test_dir):
os.makedirs(save_test_dir)
def combine_scans(input_dir, output_dir):
patients = os.listdir(input_dir)
train_data = {}
j = 0
for patient in patients:
img_types = os.listdir(os.path.join(input_dir, patient))
img_types.sort() # FFLAIR -> T1w -> T1wCE -> T2w
patient_dir = os.path.join(output_dir, patient)
if not os.path.exists(patient_dir):
os.makedirs(patient_dir) # output
for i, img_type in enumerate(img_types):
path = os.path.join(input_dir, patient, img_type) # input
slices = [dicom.read_file(path + "/" + x) for x in os.listdir(path)]
slices.sort(key=lambda x: int(x.ImagePositionPatient[2]))
slices = [x.pixel_array for x in slices if np.sum(x.pixel_array) > 0.0]
if len(slices) > 0:
stacked = np.stack(slices, axis=-1)
img_dir = os.path.join(patient_dir, img_type)
if not os.path.exists(img_dir):
os.makedirs(img_dir)
file_path = os.path.join(img_dir, f"{patient}_{img_type}_Image.npy")
# np.save(file_path, stacked)
else:
print(f"No Images for patient {patient} - {img_type}")
print(j, patient)
j += 1
combine_scans(TRAIN_DIR, save_train_dir)
print("Done with Train Dataset")
print("Start Test Dataset")
combine_scans(TEST_DIR, save_test_dir)
| false | 0 | 1,083 | 0 | 1,083 | 1,083 |
||
69595458
|
# ## 1. Data Preprocessing
# Data Processing
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import time
pd.set_option("display.max_columns", None)
pd.set_option("display.max_rows", None)
pd.set_option("display.width", None)
pd.set_option("display.float_format", lambda x: "%.5f" % x)
# Ignore all future warnings
from warnings import simplefilter
simplefilter(action="ignore", category=FutureWarning)
simplefilter(action="ignore", category=UserWarning)
pd.options.mode.chained_assignment = None # default='warn'
# Visualization
import seaborn as sns
import matplotlib.pyplot as plt
# Satistical Testing
from scipy import stats
# Converting Categorical to Numerical Finite Values
from sklearn.preprocessing import LabelEncoder
# Model Designing and Evaluation
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import jaccard_score
from sklearn.metrics import f1_score
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
# print ('The Pacific Time',time.strftime("%H:%M:%S"))
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Reading the data
df = pd.read_csv(
"/kaggle/input/awccb-2000/Assembled_Workers__Compensation_Claims___Beginning_2000.csv",
low_memory=False,
) # index_col = 0
# Shape of the data
print("Shape of the Data,", df.shape)
df.head(1)
# Columns or Features or Variables
df.columns
# ### 1.1 Data Cleaning
# ### After reading the data, the first step is to find the missing data and remove it. Eliminating any feature having more than 50% missing data or any row with missing values.
# Finding Missing Values
df.isnull().sum()
# Removing Columns with 50% missing data
df.drop(
[
"ANCR Date",
"Controverted Date",
"Section 32 Date",
"PPD Scheduled Loss Date",
"PPD Non-Scheduled Loss Date",
"PTD Date",
"First Appeal Date",
"OIICS Part Of Body Code",
"OIICS Part Of Body Description",
"OIICS Nature of Injury Code",
"OIICS Nature of Injury Description",
"OIICS Injury Source Code",
"OIICS Injury Source Description",
"OIICS Event Exposure Code",
"OIICS Event Exposure Description",
"OIICS Secondary Source Code",
"OIICS Secondary Source Description",
"C-2 Date",
"C-3 Date",
"First Hearing Date",
"IME-4 Count",
"Interval Assembled to ANCR",
],
axis=1,
inplace=True,
)
# Removing Missing Values
df.dropna(inplace=True)
df.reset_index(inplace=True)
df.drop(["index"], axis=1, inplace=True)
# Shape of The Data After Removing Missing Values
print("Shape of the Data after removing missing values and columns", df.shape)
df.head(1)
df["Current Claim Status"].unique()
# # 2. Exploratory Data Analysis
# Finding Type of each feature
df.dtypes
# Label Encoding of Categorical Features
lbl = LabelEncoder()
for item in df.columns:
if type(item) == str:
lbl.fit(list(df[str(item)].values) + list(df[str(item)].values))
df[str(item)] = lbl.transform(list(df[str(item)].values))
df.dtypes
# ## 2.1 Statistical Analysis
# ### In this analysis we first find the correlation among each variable and then after selecting the target variable, we will find the p-value of those variables whose has some relation with the target variable.
# ### Now we have to find the correlation between the variables to see any relation among them.
df.corr()
# ### After finding the correlation values, we have seen that correlation exists between a group of variables. Like, variables related to Part of the body, nature of the injury, injury description, and injury code have somewhat relation among them only. Similarly, 'Medical Fee Region', 'Gender', 'Age at Injury', 'Zip Code', and 'District Name' have correlation among them only.
# ### There are three groups of variables.
# Group 1
group_1 = df[
[
"WCIO Part Of Body Code",
"WCIO Part Of Body Description",
"WCIO Nature of Injury Code",
"WCIO Nature of Injury Description",
"WCIO Cause of Injury Code",
"WCIO Cause of Injury Description",
]
]
group_1.corr()
# Group 2
group_2 = df[
[
"Average Weekly Wage",
"Claim Injury Type",
"Highest Process",
"Hearing Count",
"Closed Count",
"Attorney/Representative",
"Current Claim Status",
"Age at Injury",
"Birth Year",
]
]
group_2.corr()
# Group 3
group_3 = df[
[
"Gender",
"Medical Fee Region",
"Birth Year",
"Age at Injury",
"Zip Code",
"Medical Fee Region",
"District Name",
"County of Injury",
]
]
group_3.corr()
# ### It is clear that in group 2, the correlation values are high.In second group, the feature Attorney/Representative has somewhat realtion with all the other members of the group. So now we will find p-value aganist each independent variable for the target variable.
# ### In statistics, "the p-value is the probability of obtaining results at least as extreme as the observed results of a statistical hypothesis test, assuming that the null hypothesis is correct. A smaller p-value means that there is stronger evidence in favor of the alternative hypothesis".
# ### P-Value is the threshold to check the significance. If P-value < 0.01 or 0.05 or 0.1 (99% or 95% or 90% Significance Levels ) of any independent variable, it is become statistically significant to affect the outcome. In our case, any statistically significant variable will affect the Housing Price.
# ### There are mathematical (Stats library SciPy) and visualization (Heat Map) ways to determine the p-value and the correlation like strong, modest, or weak relation either a positive or negative relationship with the predicted variables.
### View of Correlation Matrix Through head map
corr_matrix = group_3._get_numeric_data().corr()
fig, ax = plt.subplots(figsize=(10, 5)) # Sample figsize in inches
sns.heatmap(
corr_matrix,
annot=False,
linewidths=5,
ax=ax,
xticklabels=corr_matrix.columns.values,
yticklabels=corr_matrix.columns.values,
)
# sns.heatmap(corr, annot=True, fmt=".1f",linewidth=0.5 xticklabels=corr.columns.values,yticklabels=corr.columns.values)
### finding correlation and p_value
pearson_coef, p_value = stats.pearsonr(
df["Average Weekly Wage"], df["Attorney/Representative"]
)
print(
" The Pearson Correlation Coefficient 'Average Weekly Wage' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
pearson_coef, p_value = stats.pearsonr(
df["Claim Injury Type"], df["Attorney/Representative"]
)
print(
" The Pearson Correlation Coefficient 'Claim Injury Type' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
pearson_coef, p_value = stats.pearsonr(
df["Highest Process"], df["Attorney/Representative"]
)
print(
" The Pearson Correlation Coefficient 'Highest Process' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
pearson_coef, p_value = stats.pearsonr(
df["Hearing Count"], df["Attorney/Representative"]
)
print(
" The Pearson Correlation Coefficient 'Hearing Count' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
pearson_coef, p_value = stats.pearsonr(
df["Closed Count"], df["Attorney/Representative"]
)
print(
" The Pearson Correlation Coefficient 'Closed Count' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
pearson_coef, p_value = stats.pearsonr(
df["Current Claim Status"], df["Attorney/Representative"]
)
print(
" The Pearson Correlation Coefficient 'Current Claim Status' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
pearson_coef, p_value = stats.pearsonr(
df["Age at Injury"], df["Attorney/Representative"]
)
print(
" The Pearson Correlation Coefficient 'Age at Injury' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
pearson_coef, p_value = stats.pearsonr(df["Birth Year"], df["Attorney/Representative"])
print(
" The Pearson Correlation Coefficient 'Birth Year' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
# ### Through visualization and statistical findings; most features have a very small p-value which indicates the null hypothesis is true. Or in other words, they are highly significant.
# ### We will pick all the variables here
# # 3. Model Designing & Evaluation
# # 3.1 Feature Selection
# Feature Selection
X = df[
[
"Average Weekly Wage",
"Claim Injury Type",
"Highest Process",
"Hearing Count",
"Closed Count",
"Current Claim Status",
"Age at Injury",
"Birth Year",
]
]
#'Employment Rate',
#'UnEmployment Rate', 'Inflation Rate', 'Growth Rate GDP',
#'Consumer Price Index']]
#'Retail_Distance_From_Property', 'R_Distance_within_0.5M',
#'Subway_Distance_From_Property', 'S_Distance_within_0.5M']]
# Standardized the Features
X = np.asarray(X)
X = preprocessing.StandardScaler().fit(X).transform(X)
# Selecting Target Feature
y = np.asarray(df["Attorney/Representative"]) # .reshape(-1, 1)
# Split the data into training and test data to see results on the test data
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
# Further spliting the Traning set into sub Train and Test Data to see results on training data
# train_X,test_X,train_y,test_y = train_test_split(X_train,y_train,random_state=42)
# ## 3.2 Algorithm Selection
# ### We will choose Random Forest Classifier due to many reasons.
# ### A random forest is a meta estimator that fits a number of classifying decision trees on various sub-samples of the dataset and uses averaging to improve the predictive accuracy and control over-fitting. The sub-sample size is controlled with the max_samples parameter if bootstrap=True (default), otherwise the whole dataset is used to build each tree.
# ### The advantages of random forest are
# 1. Decorrelate trees
# 2. Error reducer
# 3. Good performer on imbalanced dataset
# 4. Good in Handling the huge amount of data
# 5. Little impact of outliers
# 6. Robust to overfitting
# ### Similarly the disadvantages are
# 1. Features need to have some predictive power else they won’t work.
# 2. Predictions of the trees need to be uncorrelated.
# 3. Appears as Black Box: It is tough to know what is happening. You can at best try different parameters and random seeds to change the outcomes and performance.
rf = RandomForestClassifier(
random_state=42,
n_estimators=2000,
max_depth=None,
n_jobs=-1,
min_samples_split=10,
) # 110,325
# Fit Model
rf.fit(X_train, y_train.ravel())
# Make validation predictions
y_pred = rf.predict(X_test)
# Model Evaluation
print("Jaccard Score {:,.5f}".format(jaccard_score(y_test, y_pred)))
# ### Further Analysis
# ### Case 1: 'WCIO Part Of Body Code' plays a role in predicting 'Highest Process'
# First Check the correlation value
pearson_coef, p_value = stats.pearsonr(
df["WCIO Part Of Body Code"], df["Highest Process"]
)
print(
" The Pearson Correlation Coefficient 'WCIO Part Of Body Code' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
# ### There exist a weak correlation and p-value is less than 0.05, so it showing some significance
# Feature Selection
X = df[["WCIO Part Of Body Code"]]
#'Employment Rate',
#'UnEmployment Rate', 'Inflation Rate', 'Growth Rate GDP',
#'Consumer Price Index']]
#'Retail_Distance_From_Property', 'R_Distance_within_0.5M',
#'Subway_Distance_From_Property', 'S_Distance_within_0.5M']]
# Standardized the Features
X = np.asarray(X)
X = preprocessing.StandardScaler().fit(X).transform(X)
# Selecting Target Feature
y = np.asarray(df["Highest Process"]) # .reshape(-1, 1)
# Split the data into training and test data to see results on the test data
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
# Further spliting the Traning set into sub Train and Test Data to see results on training data
# train_X,test_X,train_y,test_y = train_test_split(X_train,y_train,random_state=42)
rf = RandomForestClassifier(
random_state=42,
n_estimators=2000,
max_depth=None,
n_jobs=-1,
min_samples_split=10,
) # 110,325
# Fit Model
rf.fit(X_train, y_train.ravel())
# Make validation predictions
y_pred = rf.predict(X_test)
# Model Evaluation
# Target Variable is multiclass so we will use F1 Score metric
# print ("Jaccard Score {:,.5f}".format(jaccard_score(y_test, y_pred)))
print("F1 Score {:,.5f}".format(f1_score(y_test, y_pred, average="weighted")))
# ### Case 2: 'Attorney/Representative' plays a role in predicting 'Highest Process'
# First Check the correlation value
pearson_coef, p_value = stats.pearsonr(
df["Attorney/Representative"], df["Highest Process"]
)
print(
" The Pearson Correlation Coefficient 'Attorney/Representative' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
# ### There exist a moderate to strong correlation and p-value is less than 0.05, so it showing some significance
# Feature Selection
X = df[["Attorney/Representative"]]
#'Employment Rate',
#'UnEmployment Rate', 'Inflation Rate', 'Growth Rate GDP',
#'Consumer Price Index']]
#'Retail_Distance_From_Property', 'R_Distance_within_0.5M',
#'Subway_Distance_From_Property', 'S_Distance_within_0.5M']]
# Standardized the Features
X = np.asarray(X)
X = preprocessing.StandardScaler().fit(X).transform(X)
# Selecting Target Feature
y = np.asarray(df["Highest Process"]) # .reshape(-1, 1)
# Split the data into training and test data to see results on the test data
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
# Further spliting the Traning set into sub Train and Test Data to see results on training data
# train_X,test_X,train_y,test_y = train_test_split(X_train,y_train,random_state=42)
rf = RandomForestClassifier(
random_state=42,
n_estimators=2000,
max_depth=None,
n_jobs=-1,
min_samples_split=10,
) # 110,325
# Fit Model
rf.fit(X_train, y_train.ravel())
# Make validation predictions
y_pred = rf.predict(X_test)
# Model Evaluation
# Target Variable is multiclass so we will use F1 Score metric
print("F1 Score {:,.5f}".format(f1_score(y_test, y_pred, average="weighted")))
# print ("Jaccard Score {:,.5f}".format(jaccard_score(y_test, y_pred)))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/595/69595458.ipynb
| null | null |
[{"Id": 69595458, "ScriptId": 18936926, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2853990, "CreationDate": "08/02/2021 02:20:28", "VersionNumber": 2.0, "Title": "Assembled Workers Compensation Claims", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 326.0, "LinesInsertedFromPrevious": 102.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 224.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# ## 1. Data Preprocessing
# Data Processing
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import time
pd.set_option("display.max_columns", None)
pd.set_option("display.max_rows", None)
pd.set_option("display.width", None)
pd.set_option("display.float_format", lambda x: "%.5f" % x)
# Ignore all future warnings
from warnings import simplefilter
simplefilter(action="ignore", category=FutureWarning)
simplefilter(action="ignore", category=UserWarning)
pd.options.mode.chained_assignment = None # default='warn'
# Visualization
import seaborn as sns
import matplotlib.pyplot as plt
# Satistical Testing
from scipy import stats
# Converting Categorical to Numerical Finite Values
from sklearn.preprocessing import LabelEncoder
# Model Designing and Evaluation
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import jaccard_score
from sklearn.metrics import f1_score
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
# print ('The Pacific Time',time.strftime("%H:%M:%S"))
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Reading the data
df = pd.read_csv(
"/kaggle/input/awccb-2000/Assembled_Workers__Compensation_Claims___Beginning_2000.csv",
low_memory=False,
) # index_col = 0
# Shape of the data
print("Shape of the Data,", df.shape)
df.head(1)
# Columns or Features or Variables
df.columns
# ### 1.1 Data Cleaning
# ### After reading the data, the first step is to find the missing data and remove it. Eliminating any feature having more than 50% missing data or any row with missing values.
# Finding Missing Values
df.isnull().sum()
# Removing Columns with 50% missing data
df.drop(
[
"ANCR Date",
"Controverted Date",
"Section 32 Date",
"PPD Scheduled Loss Date",
"PPD Non-Scheduled Loss Date",
"PTD Date",
"First Appeal Date",
"OIICS Part Of Body Code",
"OIICS Part Of Body Description",
"OIICS Nature of Injury Code",
"OIICS Nature of Injury Description",
"OIICS Injury Source Code",
"OIICS Injury Source Description",
"OIICS Event Exposure Code",
"OIICS Event Exposure Description",
"OIICS Secondary Source Code",
"OIICS Secondary Source Description",
"C-2 Date",
"C-3 Date",
"First Hearing Date",
"IME-4 Count",
"Interval Assembled to ANCR",
],
axis=1,
inplace=True,
)
# Removing Missing Values
df.dropna(inplace=True)
df.reset_index(inplace=True)
df.drop(["index"], axis=1, inplace=True)
# Shape of The Data After Removing Missing Values
print("Shape of the Data after removing missing values and columns", df.shape)
df.head(1)
df["Current Claim Status"].unique()
# # 2. Exploratory Data Analysis
# Finding Type of each feature
df.dtypes
# Label Encoding of Categorical Features
lbl = LabelEncoder()
for item in df.columns:
if type(item) == str:
lbl.fit(list(df[str(item)].values) + list(df[str(item)].values))
df[str(item)] = lbl.transform(list(df[str(item)].values))
df.dtypes
# ## 2.1 Statistical Analysis
# ### In this analysis we first find the correlation among each variable and then after selecting the target variable, we will find the p-value of those variables whose has some relation with the target variable.
# ### Now we have to find the correlation between the variables to see any relation among them.
df.corr()
# ### After finding the correlation values, we have seen that correlation exists between a group of variables. Like, variables related to Part of the body, nature of the injury, injury description, and injury code have somewhat relation among them only. Similarly, 'Medical Fee Region', 'Gender', 'Age at Injury', 'Zip Code', and 'District Name' have correlation among them only.
# ### There are three groups of variables.
# Group 1
group_1 = df[
[
"WCIO Part Of Body Code",
"WCIO Part Of Body Description",
"WCIO Nature of Injury Code",
"WCIO Nature of Injury Description",
"WCIO Cause of Injury Code",
"WCIO Cause of Injury Description",
]
]
group_1.corr()
# Group 2
group_2 = df[
[
"Average Weekly Wage",
"Claim Injury Type",
"Highest Process",
"Hearing Count",
"Closed Count",
"Attorney/Representative",
"Current Claim Status",
"Age at Injury",
"Birth Year",
]
]
group_2.corr()
# Group 3
group_3 = df[
[
"Gender",
"Medical Fee Region",
"Birth Year",
"Age at Injury",
"Zip Code",
"Medical Fee Region",
"District Name",
"County of Injury",
]
]
group_3.corr()
# ### It is clear that in group 2, the correlation values are high.In second group, the feature Attorney/Representative has somewhat realtion with all the other members of the group. So now we will find p-value aganist each independent variable for the target variable.
# ### In statistics, "the p-value is the probability of obtaining results at least as extreme as the observed results of a statistical hypothesis test, assuming that the null hypothesis is correct. A smaller p-value means that there is stronger evidence in favor of the alternative hypothesis".
# ### P-Value is the threshold to check the significance. If P-value < 0.01 or 0.05 or 0.1 (99% or 95% or 90% Significance Levels ) of any independent variable, it is become statistically significant to affect the outcome. In our case, any statistically significant variable will affect the Housing Price.
# ### There are mathematical (Stats library SciPy) and visualization (Heat Map) ways to determine the p-value and the correlation like strong, modest, or weak relation either a positive or negative relationship with the predicted variables.
### View of Correlation Matrix Through head map
corr_matrix = group_3._get_numeric_data().corr()
fig, ax = plt.subplots(figsize=(10, 5)) # Sample figsize in inches
sns.heatmap(
corr_matrix,
annot=False,
linewidths=5,
ax=ax,
xticklabels=corr_matrix.columns.values,
yticklabels=corr_matrix.columns.values,
)
# sns.heatmap(corr, annot=True, fmt=".1f",linewidth=0.5 xticklabels=corr.columns.values,yticklabels=corr.columns.values)
### finding correlation and p_value
pearson_coef, p_value = stats.pearsonr(
df["Average Weekly Wage"], df["Attorney/Representative"]
)
print(
" The Pearson Correlation Coefficient 'Average Weekly Wage' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
pearson_coef, p_value = stats.pearsonr(
df["Claim Injury Type"], df["Attorney/Representative"]
)
print(
" The Pearson Correlation Coefficient 'Claim Injury Type' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
pearson_coef, p_value = stats.pearsonr(
df["Highest Process"], df["Attorney/Representative"]
)
print(
" The Pearson Correlation Coefficient 'Highest Process' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
pearson_coef, p_value = stats.pearsonr(
df["Hearing Count"], df["Attorney/Representative"]
)
print(
" The Pearson Correlation Coefficient 'Hearing Count' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
pearson_coef, p_value = stats.pearsonr(
df["Closed Count"], df["Attorney/Representative"]
)
print(
" The Pearson Correlation Coefficient 'Closed Count' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
pearson_coef, p_value = stats.pearsonr(
df["Current Claim Status"], df["Attorney/Representative"]
)
print(
" The Pearson Correlation Coefficient 'Current Claim Status' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
pearson_coef, p_value = stats.pearsonr(
df["Age at Injury"], df["Attorney/Representative"]
)
print(
" The Pearson Correlation Coefficient 'Age at Injury' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
pearson_coef, p_value = stats.pearsonr(df["Birth Year"], df["Attorney/Representative"])
print(
" The Pearson Correlation Coefficient 'Birth Year' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
# ### Through visualization and statistical findings; most features have a very small p-value which indicates the null hypothesis is true. Or in other words, they are highly significant.
# ### We will pick all the variables here
# # 3. Model Designing & Evaluation
# # 3.1 Feature Selection
# Feature Selection
X = df[
[
"Average Weekly Wage",
"Claim Injury Type",
"Highest Process",
"Hearing Count",
"Closed Count",
"Current Claim Status",
"Age at Injury",
"Birth Year",
]
]
#'Employment Rate',
#'UnEmployment Rate', 'Inflation Rate', 'Growth Rate GDP',
#'Consumer Price Index']]
#'Retail_Distance_From_Property', 'R_Distance_within_0.5M',
#'Subway_Distance_From_Property', 'S_Distance_within_0.5M']]
# Standardized the Features
X = np.asarray(X)
X = preprocessing.StandardScaler().fit(X).transform(X)
# Selecting Target Feature
y = np.asarray(df["Attorney/Representative"]) # .reshape(-1, 1)
# Split the data into training and test data to see results on the test data
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
# Further spliting the Traning set into sub Train and Test Data to see results on training data
# train_X,test_X,train_y,test_y = train_test_split(X_train,y_train,random_state=42)
# ## 3.2 Algorithm Selection
# ### We will choose Random Forest Classifier due to many reasons.
# ### A random forest is a meta estimator that fits a number of classifying decision trees on various sub-samples of the dataset and uses averaging to improve the predictive accuracy and control over-fitting. The sub-sample size is controlled with the max_samples parameter if bootstrap=True (default), otherwise the whole dataset is used to build each tree.
# ### The advantages of random forest are
# 1. Decorrelate trees
# 2. Error reducer
# 3. Good performer on imbalanced dataset
# 4. Good in Handling the huge amount of data
# 5. Little impact of outliers
# 6. Robust to overfitting
# ### Similarly the disadvantages are
# 1. Features need to have some predictive power else they won’t work.
# 2. Predictions of the trees need to be uncorrelated.
# 3. Appears as Black Box: It is tough to know what is happening. You can at best try different parameters and random seeds to change the outcomes and performance.
rf = RandomForestClassifier(
random_state=42,
n_estimators=2000,
max_depth=None,
n_jobs=-1,
min_samples_split=10,
) # 110,325
# Fit Model
rf.fit(X_train, y_train.ravel())
# Make validation predictions
y_pred = rf.predict(X_test)
# Model Evaluation
print("Jaccard Score {:,.5f}".format(jaccard_score(y_test, y_pred)))
# ### Further Analysis
# ### Case 1: 'WCIO Part Of Body Code' plays a role in predicting 'Highest Process'
# First Check the correlation value
pearson_coef, p_value = stats.pearsonr(
df["WCIO Part Of Body Code"], df["Highest Process"]
)
print(
" The Pearson Correlation Coefficient 'WCIO Part Of Body Code' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
# ### There exist a weak correlation and p-value is less than 0.05, so it showing some significance
# Feature Selection
X = df[["WCIO Part Of Body Code"]]
#'Employment Rate',
#'UnEmployment Rate', 'Inflation Rate', 'Growth Rate GDP',
#'Consumer Price Index']]
#'Retail_Distance_From_Property', 'R_Distance_within_0.5M',
#'Subway_Distance_From_Property', 'S_Distance_within_0.5M']]
# Standardized the Features
X = np.asarray(X)
X = preprocessing.StandardScaler().fit(X).transform(X)
# Selecting Target Feature
y = np.asarray(df["Highest Process"]) # .reshape(-1, 1)
# Split the data into training and test data to see results on the test data
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
# Further spliting the Traning set into sub Train and Test Data to see results on training data
# train_X,test_X,train_y,test_y = train_test_split(X_train,y_train,random_state=42)
rf = RandomForestClassifier(
random_state=42,
n_estimators=2000,
max_depth=None,
n_jobs=-1,
min_samples_split=10,
) # 110,325
# Fit Model
rf.fit(X_train, y_train.ravel())
# Make validation predictions
y_pred = rf.predict(X_test)
# Model Evaluation
# Target Variable is multiclass so we will use F1 Score metric
# print ("Jaccard Score {:,.5f}".format(jaccard_score(y_test, y_pred)))
print("F1 Score {:,.5f}".format(f1_score(y_test, y_pred, average="weighted")))
# ### Case 2: 'Attorney/Representative' plays a role in predicting 'Highest Process'
# First Check the correlation value
pearson_coef, p_value = stats.pearsonr(
df["Attorney/Representative"], df["Highest Process"]
)
print(
" The Pearson Correlation Coefficient 'Attorney/Representative' is",
pearson_coef,
" with a P-value of P =",
p_value,
)
# ### There exist a moderate to strong correlation and p-value is less than 0.05, so it showing some significance
# Feature Selection
X = df[["Attorney/Representative"]]
#'Employment Rate',
#'UnEmployment Rate', 'Inflation Rate', 'Growth Rate GDP',
#'Consumer Price Index']]
#'Retail_Distance_From_Property', 'R_Distance_within_0.5M',
#'Subway_Distance_From_Property', 'S_Distance_within_0.5M']]
# Standardized the Features
X = np.asarray(X)
X = preprocessing.StandardScaler().fit(X).transform(X)
# Selecting Target Feature
y = np.asarray(df["Highest Process"]) # .reshape(-1, 1)
# Split the data into training and test data to see results on the test data
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
# Further spliting the Traning set into sub Train and Test Data to see results on training data
# train_X,test_X,train_y,test_y = train_test_split(X_train,y_train,random_state=42)
rf = RandomForestClassifier(
random_state=42,
n_estimators=2000,
max_depth=None,
n_jobs=-1,
min_samples_split=10,
) # 110,325
# Fit Model
rf.fit(X_train, y_train.ravel())
# Make validation predictions
y_pred = rf.predict(X_test)
# Model Evaluation
# Target Variable is multiclass so we will use F1 Score metric
print("F1 Score {:,.5f}".format(f1_score(y_test, y_pred, average="weighted")))
# print ("Jaccard Score {:,.5f}".format(jaccard_score(y_test, y_pred)))
| false | 0 | 4,442 | 0 | 4,442 | 4,442 |
||
69595125
|
# Import the necessary libraries
import numpy as np
import pandas as pd
import os
import time
import warnings
import os
from six.moves import urllib
import matplotlib
import matplotlib.pyplot as plt
warnings.filterwarnings("ignore")
# Add All the Models Libraries
# Scalers
from sklearn.preprocessing import StandardScaler
from sklearn.utils import shuffle
from sklearn.pipeline import Pipeline
from sklearn.pipeline import FeatureUnion
# Models
from sklearn.linear_model import LogisticRegression # logistic regression
from sklearn.svm import SVC # Support Vector Classifier
from sklearn.ensemble import RandomForestClassifier # Random Forest
from sklearn.neighbors import KNeighborsClassifier # KNN
from sklearn.ensemble import ExtraTreesClassifier
from sklearn.ensemble import VotingClassifier
from sklearn.neural_network import MLPClassifier
from sklearn.tree import DecisionTreeClassifier # Decision Tree
from sklearn.ensemble import BaggingClassifier
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.model_selection import train_test_split # training and testing data split
from sklearn import metrics # accuracy measure
from sklearn.metrics import confusion_matrix # for confusion matrix
from scipy.stats import reciprocal, uniform
from sklearn.ensemble import AdaBoostClassifier
# Cross-validation
from sklearn.model_selection import KFold # for K-fold cross validation
from sklearn.model_selection import cross_val_score # score evaluation
from sklearn.model_selection import cross_val_predict # prediction
from sklearn.model_selection import cross_validate
# GridSearchCV
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import RandomizedSearchCV
# Common data processors
from sklearn.preprocessing import OneHotEncoder, LabelEncoder
from sklearn import feature_selection
from sklearn import model_selection
from sklearn import metrics
from sklearn.base import BaseEstimator, TransformerMixin
import numpy as np
import pandas as pd
from sklearn.metrics import roc_auc_score, log_loss
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import StratifiedKFold, KFold
from sklearn.utils import check_array
from scipy import sparse
# Accuracy Score
from sklearn.metrics import accuracy_score
import lightgbm as lgb
# to make this notebook's output stable across runs
np.random.seed(123)
# To plot pretty figures
plt.rcParams["axes.labelsize"] = 14
plt.rcParams["xtick.labelsize"] = 12
plt.rcParams["ytick.labelsize"] = 12
# merge the data for feature engineering and later split it, just before applying Data Pipeline
TrainFile = pd.read_csv(
"../input/Target-Marketing-Canadians/train.csv"
) # read the data from the csv file.
TestFile = pd.read_csv("../input/Target-Marketing-Canadians/test.csv")
TrainFile.shape
TestFile.shape
TrainFile.info()
TrainFile.head(5)
TestFile.info()
# Interactions
TrainFile["External_Competitive_1"] = (
TrainFile["ExternalAccount1"] * TrainFile["CompetitiveRate1"]
)
TrainFile["External_Competitive_2"] = (
TrainFile["ExternalAccount2"] * TrainFile["CompetitiveRate2"]
)
TrainFile["External_Competitive_3"] = (
TrainFile["ExternalAccount3"] * TrainFile["CompetitiveRate3"]
)
TrainFile["External_Competitive_4"] = (
TrainFile["ExternalAccount4"] * TrainFile["CompetitiveRate4"]
)
TrainFile["External_Competitive_5"] = (
TrainFile["ExternalAccount5"] * TrainFile["CompetitiveRate5"]
)
TrainFile["External_Competitive_6"] = (
TrainFile["ExternalAccount6"] * TrainFile["CompetitiveRate6"]
)
TrainFile["External_Competitive_7"] = (
TrainFile["ExternalAccount7"] * TrainFile["CompetitiveRate7"]
)
TrainFile["Transactions_Mean"] = TrainFile.iloc[:, 9:17].mean(axis=1)
TrainFile["Transactions_std"] = TrainFile.iloc[:, 9:17].std(axis=1)
TrainFile["Transactions_Max"] = TrainFile.iloc[:, 9:17].max(axis=1)
TestFile["External_Competitive_1"] = (
TestFile["ExternalAccount1"] * TestFile["CompetitiveRate1"]
)
TestFile["External_Competitive_2"] = (
TestFile["ExternalAccount2"] * TestFile["CompetitiveRate2"]
)
TestFile["External_Competitive_3"] = (
TestFile["ExternalAccount3"] * TestFile["CompetitiveRate3"]
)
TestFile["External_Competitive_4"] = (
TestFile["ExternalAccount4"] * TestFile["CompetitiveRate4"]
)
TestFile["External_Competitive_5"] = (
TestFile["ExternalAccount5"] * TestFile["CompetitiveRate5"]
)
TestFile["External_Competitive_6"] = (
TestFile["ExternalAccount6"] * TestFile["CompetitiveRate6"]
)
TestFile["External_Competitive_7"] = (
TestFile["ExternalAccount7"] * TestFile["CompetitiveRate7"]
)
TestFile["Transactions_Mean"] = TestFile.iloc[:, 9:17].mean(axis=1)
TestFile["Transactions_std"] = TestFile.iloc[:, 9:17].std(axis=1)
TestFile["Transactions_Max"] = TestFile.iloc[:, 9:17].max(axis=1)
TrainFile.head(2)
# Making Balance bins
TrainFile["Balance_Bin"] = pd.qcut(TrainFile["Balance"], 3)
label = LabelEncoder()
TrainFile["Balance_Bin"] = label.fit_transform(TrainFile["Balance_Bin"])
# Making Balance bins
TestFile["Balance_Bin"] = pd.qcut(TestFile["Balance"], 3)
label = LabelEncoder()
TestFile["Balance_Bin"] = label.fit_transform(TestFile["Balance_Bin"])
TrainFile.tail(2)
# drop Customer_id
TrainFile = TrainFile.drop(["Customer_id", "Balance"], axis=1)
TestFile = TestFile.drop(["Customer_id", "Balance"], axis=1)
# Now define x and y.
# the Y Variable
y_train = TrainFile["Target"].copy()
# the X variables
X_train = TrainFile.drop("Target", axis=1)
X_train.shape
TestFile.shape
y_train.shape
# Pipeline
Features = X_train.columns.tolist()
Features_Test = TestFile.columns.tolist()
# The CategoricalEncoder class will allow us to convert categorical attributes to one-hot vectors.
class CategoricalEncoder(BaseEstimator, TransformerMixin):
def __init__(
self,
encoding="onehot",
categories="auto",
dtype=np.float64,
handle_unknown="error",
):
self.encoding = encoding
self.categories = categories
self.dtype = dtype
self.handle_unknown = handle_unknown
def fit(self, X, y=None):
"""Fit the CategoricalEncoder to X.
Parameters
----------
X : array-like, shape [n_samples, n_feature]
The data to determine the categories of each feature.
Returns
-------
self
"""
if self.encoding not in ["onehot", "onehot-dense", "ordinal"]:
template = (
"encoding should be either 'onehot', 'onehot-dense' "
"or 'ordinal', got %s"
)
raise ValueError(template % self.handle_unknown)
if self.handle_unknown not in ["error", "ignore"]:
template = "handle_unknown should be either 'error' or " "'ignore', got %s"
raise ValueError(template % self.handle_unknown)
if self.encoding == "ordinal" and self.handle_unknown == "ignore":
raise ValueError(
"handle_unknown='ignore' is not supported for" " encoding='ordinal'"
)
X = check_array(X, dtype=np.object, accept_sparse="csc", copy=True)
n_samples, n_features = X.shape
self._label_encoders_ = [LabelEncoder() for _ in range(n_features)]
for i in range(n_features):
le = self._label_encoders_[i]
Xi = X[:, i]
if self.categories == "auto":
le.fit(Xi)
else:
valid_mask = np.in1d(Xi, self.categories[i])
if not np.all(valid_mask):
if self.handle_unknown == "error":
diff = np.unique(Xi[~valid_mask])
msg = (
"Found unknown categories {0} in column {1}"
" during fit".format(diff, i)
)
raise ValueError(msg)
le.classes_ = np.array(np.sort(self.categories[i]))
self.categories_ = [le.classes_ for le in self._label_encoders_]
return self
def transform(self, X):
"""Transform X using one-hot encoding.
Parameters
----------
X : array-like, shape [n_samples, n_features]
The data to encode.
Returns
-------
X_out : sparse matrix or a 2-d array
Transformed input.
"""
X = check_array(X, accept_sparse="csc", dtype=np.object, copy=True)
n_samples, n_features = X.shape
X_int = np.zeros_like(X, dtype=np.int)
X_mask = np.ones_like(X, dtype=np.bool)
for i in range(n_features):
valid_mask = np.in1d(X[:, i], self.categories_[i])
if not np.all(valid_mask):
if self.handle_unknown == "error":
diff = np.unique(X[~valid_mask, i])
msg = (
"Found unknown categories {0} in column {1}"
" during transform".format(diff, i)
)
raise ValueError(msg)
else:
# Set the problematic rows to an acceptable value and
# continue `The rows are marked `X_mask` and will be
# removed later.
X_mask[:, i] = valid_mask
X[:, i][~valid_mask] = self.categories_[i][0]
X_int[:, i] = self._label_encoders_[i].transform(X[:, i])
if self.encoding == "ordinal":
return X_int.astype(self.dtype, copy=False)
mask = X_mask.ravel()
n_values = [cats.shape[0] for cats in self.categories_]
n_values = np.array([0] + n_values)
indices = np.cumsum(n_values)
column_indices = (X_int + indices[:-1]).ravel()[mask]
row_indices = np.repeat(np.arange(n_samples, dtype=np.int32), n_features)[mask]
data = np.ones(n_samples * n_features)[mask]
out = sparse.csc_matrix(
(data, (row_indices, column_indices)),
shape=(n_samples, indices[-1]),
dtype=self.dtype,
).tocsr()
if self.encoding == "onehot-dense":
return out.toarray()
else:
return out
class DataFrameSelector(BaseEstimator, TransformerMixin):
def __init__(self, attribute_names):
self.attribute_names = attribute_names
def fit(self, X, y=None):
return self
def transform(self, X):
return X[self.attribute_names]
Categorical_Features = [
"PreviousCampaignResult",
"Product1",
"Product2",
"Product3",
"Product4",
"Product5",
"Product6",
"ExternalAccount1",
"ExternalAccount2",
"ExternalAccount3",
"ExternalAccount4",
"ExternalAccount5",
"ExternalAccount6",
"ExternalAccount7",
"Balance_Bin",
]
Continuous_Features = [
"Transaction1",
"Transaction2",
"Transaction3",
"Transaction4",
"Transaction5",
"Transaction6",
"Transaction7",
"Transaction8",
"Transaction9",
"ActivityIndicator",
"RegularInteractionIndicator",
"CompetitiveRate1",
"CompetitiveRate2",
"CompetitiveRate3",
"CompetitiveRate4",
"CompetitiveRate5",
"CompetitiveRate6",
"CompetitiveRate7",
"RateBefore",
"ReferenceRate",
"External_Competitive_1",
"External_Competitive_2",
"External_Competitive_3",
"External_Competitive_4",
"External_Competitive_5",
"External_Competitive_6",
"External_Competitive_7",
"Transactions_Mean",
"Transactions_std",
"Transactions_Max",
]
Total_Features = Categorical_Features + Continuous_Features
cat_pipeline = Pipeline([("selector", DataFrameSelector(Categorical_Features))])
num_pipeline = Pipeline(
[
("selector", DataFrameSelector(Continuous_Features)),
("std_scaler", StandardScaler()),
]
)
full_pipeline = FeatureUnion(
transformer_list=[
("cat_pipeline", cat_pipeline),
("num_pipeline", num_pipeline),
]
)
X_train = full_pipeline.fit_transform(X_train)
X_test = full_pipeline.transform(TestFile)
# KNN
# Introduce KNN Classifier
KNeighbours = KNeighborsClassifier()
leaf_size = list(range(1, 25, 5))
n_neighbors = list(range(4, 30, 2))
param_grid_KNeighbours = {
"n_neighbors": n_neighbors,
"algorithm": ["auto"],
"weights": ["uniform", "distance"],
"leaf_size": leaf_size,
}
grid_search_KNeighbours = RandomizedSearchCV(
KNeighbours,
param_grid_KNeighbours,
cv=4,
scoring="roc_auc",
refit=True,
n_jobs=-1,
verbose=2,
)
grid_search_KNeighbours.fit(X_train, y_train)
# Prepare the predictions file
result_test1 = pd.DataFrame()
passenger_id_test = TestFile["Customer_id"].copy()
result_test1["Customer_id"] = passenger_id_test
result_test1["Target"] = y_pred_neighbor_grid
result_test1.to_csv("Knn.csv", index=False)
# Random Forest
forest_class = RandomForestClassifier(random_state=42)
n_estimators = [50, 100, 400, 700, 1000]
max_features = [5, 7, 10]
max_depth = [10, 20]
oob_score = [True, False]
min_samples_split = [2, 4, 10, 12, 16]
min_samples_leaf = [1, 5, 10]
max_leaf_nodes = [2, 10, 20]
param_grid_forest = {
"n_estimators": n_estimators,
"max_features": max_features,
"max_depth": max_depth,
"min_samples_split": min_samples_split,
"oob_score": oob_score,
"min_samples_leaf": min_samples_leaf,
"max_leaf_nodes": max_leaf_nodes,
}
rand_search_forest = RandomizedSearchCV(
forest_class,
param_grid_forest,
cv=4,
scoring="roc_auc",
refit=True,
n_jobs=-1,
verbose=2,
)
rand_search_forest.fit(X_train, y_train)
random_estimator = rand_search_forest.best_estimator_
y_pred_random_estimator = random_estimator.predict(TestFile)
accuracy_score(train_set_y, y_pred_random_estimator)
# Prepare the predictions file
result_test2 = pd.DataFrame()
passenger_id_test = TestFile["Customer_id"].copy()
result_test2["Customer_id"] = passenger_id_test
result_test2["Target"] = y_pred_random_estimator
result_test2.to_csv("Random.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/595/69595125.ipynb
| null | null |
[{"Id": 69595125, "ScriptId": 18977697, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6011915, "CreationDate": "08/02/2021 02:13:24", "VersionNumber": 6.0, "Title": "Baseline model", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 370.0, "LinesInsertedFromPrevious": 68.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 302.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# Import the necessary libraries
import numpy as np
import pandas as pd
import os
import time
import warnings
import os
from six.moves import urllib
import matplotlib
import matplotlib.pyplot as plt
warnings.filterwarnings("ignore")
# Add All the Models Libraries
# Scalers
from sklearn.preprocessing import StandardScaler
from sklearn.utils import shuffle
from sklearn.pipeline import Pipeline
from sklearn.pipeline import FeatureUnion
# Models
from sklearn.linear_model import LogisticRegression # logistic regression
from sklearn.svm import SVC # Support Vector Classifier
from sklearn.ensemble import RandomForestClassifier # Random Forest
from sklearn.neighbors import KNeighborsClassifier # KNN
from sklearn.ensemble import ExtraTreesClassifier
from sklearn.ensemble import VotingClassifier
from sklearn.neural_network import MLPClassifier
from sklearn.tree import DecisionTreeClassifier # Decision Tree
from sklearn.ensemble import BaggingClassifier
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.model_selection import train_test_split # training and testing data split
from sklearn import metrics # accuracy measure
from sklearn.metrics import confusion_matrix # for confusion matrix
from scipy.stats import reciprocal, uniform
from sklearn.ensemble import AdaBoostClassifier
# Cross-validation
from sklearn.model_selection import KFold # for K-fold cross validation
from sklearn.model_selection import cross_val_score # score evaluation
from sklearn.model_selection import cross_val_predict # prediction
from sklearn.model_selection import cross_validate
# GridSearchCV
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import RandomizedSearchCV
# Common data processors
from sklearn.preprocessing import OneHotEncoder, LabelEncoder
from sklearn import feature_selection
from sklearn import model_selection
from sklearn import metrics
from sklearn.base import BaseEstimator, TransformerMixin
import numpy as np
import pandas as pd
from sklearn.metrics import roc_auc_score, log_loss
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import StratifiedKFold, KFold
from sklearn.utils import check_array
from scipy import sparse
# Accuracy Score
from sklearn.metrics import accuracy_score
import lightgbm as lgb
# to make this notebook's output stable across runs
np.random.seed(123)
# To plot pretty figures
plt.rcParams["axes.labelsize"] = 14
plt.rcParams["xtick.labelsize"] = 12
plt.rcParams["ytick.labelsize"] = 12
# merge the data for feature engineering and later split it, just before applying Data Pipeline
TrainFile = pd.read_csv(
"../input/Target-Marketing-Canadians/train.csv"
) # read the data from the csv file.
TestFile = pd.read_csv("../input/Target-Marketing-Canadians/test.csv")
TrainFile.shape
TestFile.shape
TrainFile.info()
TrainFile.head(5)
TestFile.info()
# Interactions
TrainFile["External_Competitive_1"] = (
TrainFile["ExternalAccount1"] * TrainFile["CompetitiveRate1"]
)
TrainFile["External_Competitive_2"] = (
TrainFile["ExternalAccount2"] * TrainFile["CompetitiveRate2"]
)
TrainFile["External_Competitive_3"] = (
TrainFile["ExternalAccount3"] * TrainFile["CompetitiveRate3"]
)
TrainFile["External_Competitive_4"] = (
TrainFile["ExternalAccount4"] * TrainFile["CompetitiveRate4"]
)
TrainFile["External_Competitive_5"] = (
TrainFile["ExternalAccount5"] * TrainFile["CompetitiveRate5"]
)
TrainFile["External_Competitive_6"] = (
TrainFile["ExternalAccount6"] * TrainFile["CompetitiveRate6"]
)
TrainFile["External_Competitive_7"] = (
TrainFile["ExternalAccount7"] * TrainFile["CompetitiveRate7"]
)
TrainFile["Transactions_Mean"] = TrainFile.iloc[:, 9:17].mean(axis=1)
TrainFile["Transactions_std"] = TrainFile.iloc[:, 9:17].std(axis=1)
TrainFile["Transactions_Max"] = TrainFile.iloc[:, 9:17].max(axis=1)
TestFile["External_Competitive_1"] = (
TestFile["ExternalAccount1"] * TestFile["CompetitiveRate1"]
)
TestFile["External_Competitive_2"] = (
TestFile["ExternalAccount2"] * TestFile["CompetitiveRate2"]
)
TestFile["External_Competitive_3"] = (
TestFile["ExternalAccount3"] * TestFile["CompetitiveRate3"]
)
TestFile["External_Competitive_4"] = (
TestFile["ExternalAccount4"] * TestFile["CompetitiveRate4"]
)
TestFile["External_Competitive_5"] = (
TestFile["ExternalAccount5"] * TestFile["CompetitiveRate5"]
)
TestFile["External_Competitive_6"] = (
TestFile["ExternalAccount6"] * TestFile["CompetitiveRate6"]
)
TestFile["External_Competitive_7"] = (
TestFile["ExternalAccount7"] * TestFile["CompetitiveRate7"]
)
TestFile["Transactions_Mean"] = TestFile.iloc[:, 9:17].mean(axis=1)
TestFile["Transactions_std"] = TestFile.iloc[:, 9:17].std(axis=1)
TestFile["Transactions_Max"] = TestFile.iloc[:, 9:17].max(axis=1)
TrainFile.head(2)
# Making Balance bins
TrainFile["Balance_Bin"] = pd.qcut(TrainFile["Balance"], 3)
label = LabelEncoder()
TrainFile["Balance_Bin"] = label.fit_transform(TrainFile["Balance_Bin"])
# Making Balance bins
TestFile["Balance_Bin"] = pd.qcut(TestFile["Balance"], 3)
label = LabelEncoder()
TestFile["Balance_Bin"] = label.fit_transform(TestFile["Balance_Bin"])
TrainFile.tail(2)
# drop Customer_id
TrainFile = TrainFile.drop(["Customer_id", "Balance"], axis=1)
TestFile = TestFile.drop(["Customer_id", "Balance"], axis=1)
# Now define x and y.
# the Y Variable
y_train = TrainFile["Target"].copy()
# the X variables
X_train = TrainFile.drop("Target", axis=1)
X_train.shape
TestFile.shape
y_train.shape
# Pipeline
Features = X_train.columns.tolist()
Features_Test = TestFile.columns.tolist()
# The CategoricalEncoder class will allow us to convert categorical attributes to one-hot vectors.
class CategoricalEncoder(BaseEstimator, TransformerMixin):
def __init__(
self,
encoding="onehot",
categories="auto",
dtype=np.float64,
handle_unknown="error",
):
self.encoding = encoding
self.categories = categories
self.dtype = dtype
self.handle_unknown = handle_unknown
def fit(self, X, y=None):
"""Fit the CategoricalEncoder to X.
Parameters
----------
X : array-like, shape [n_samples, n_feature]
The data to determine the categories of each feature.
Returns
-------
self
"""
if self.encoding not in ["onehot", "onehot-dense", "ordinal"]:
template = (
"encoding should be either 'onehot', 'onehot-dense' "
"or 'ordinal', got %s"
)
raise ValueError(template % self.handle_unknown)
if self.handle_unknown not in ["error", "ignore"]:
template = "handle_unknown should be either 'error' or " "'ignore', got %s"
raise ValueError(template % self.handle_unknown)
if self.encoding == "ordinal" and self.handle_unknown == "ignore":
raise ValueError(
"handle_unknown='ignore' is not supported for" " encoding='ordinal'"
)
X = check_array(X, dtype=np.object, accept_sparse="csc", copy=True)
n_samples, n_features = X.shape
self._label_encoders_ = [LabelEncoder() for _ in range(n_features)]
for i in range(n_features):
le = self._label_encoders_[i]
Xi = X[:, i]
if self.categories == "auto":
le.fit(Xi)
else:
valid_mask = np.in1d(Xi, self.categories[i])
if not np.all(valid_mask):
if self.handle_unknown == "error":
diff = np.unique(Xi[~valid_mask])
msg = (
"Found unknown categories {0} in column {1}"
" during fit".format(diff, i)
)
raise ValueError(msg)
le.classes_ = np.array(np.sort(self.categories[i]))
self.categories_ = [le.classes_ for le in self._label_encoders_]
return self
def transform(self, X):
"""Transform X using one-hot encoding.
Parameters
----------
X : array-like, shape [n_samples, n_features]
The data to encode.
Returns
-------
X_out : sparse matrix or a 2-d array
Transformed input.
"""
X = check_array(X, accept_sparse="csc", dtype=np.object, copy=True)
n_samples, n_features = X.shape
X_int = np.zeros_like(X, dtype=np.int)
X_mask = np.ones_like(X, dtype=np.bool)
for i in range(n_features):
valid_mask = np.in1d(X[:, i], self.categories_[i])
if not np.all(valid_mask):
if self.handle_unknown == "error":
diff = np.unique(X[~valid_mask, i])
msg = (
"Found unknown categories {0} in column {1}"
" during transform".format(diff, i)
)
raise ValueError(msg)
else:
# Set the problematic rows to an acceptable value and
# continue `The rows are marked `X_mask` and will be
# removed later.
X_mask[:, i] = valid_mask
X[:, i][~valid_mask] = self.categories_[i][0]
X_int[:, i] = self._label_encoders_[i].transform(X[:, i])
if self.encoding == "ordinal":
return X_int.astype(self.dtype, copy=False)
mask = X_mask.ravel()
n_values = [cats.shape[0] for cats in self.categories_]
n_values = np.array([0] + n_values)
indices = np.cumsum(n_values)
column_indices = (X_int + indices[:-1]).ravel()[mask]
row_indices = np.repeat(np.arange(n_samples, dtype=np.int32), n_features)[mask]
data = np.ones(n_samples * n_features)[mask]
out = sparse.csc_matrix(
(data, (row_indices, column_indices)),
shape=(n_samples, indices[-1]),
dtype=self.dtype,
).tocsr()
if self.encoding == "onehot-dense":
return out.toarray()
else:
return out
class DataFrameSelector(BaseEstimator, TransformerMixin):
def __init__(self, attribute_names):
self.attribute_names = attribute_names
def fit(self, X, y=None):
return self
def transform(self, X):
return X[self.attribute_names]
Categorical_Features = [
"PreviousCampaignResult",
"Product1",
"Product2",
"Product3",
"Product4",
"Product5",
"Product6",
"ExternalAccount1",
"ExternalAccount2",
"ExternalAccount3",
"ExternalAccount4",
"ExternalAccount5",
"ExternalAccount6",
"ExternalAccount7",
"Balance_Bin",
]
Continuous_Features = [
"Transaction1",
"Transaction2",
"Transaction3",
"Transaction4",
"Transaction5",
"Transaction6",
"Transaction7",
"Transaction8",
"Transaction9",
"ActivityIndicator",
"RegularInteractionIndicator",
"CompetitiveRate1",
"CompetitiveRate2",
"CompetitiveRate3",
"CompetitiveRate4",
"CompetitiveRate5",
"CompetitiveRate6",
"CompetitiveRate7",
"RateBefore",
"ReferenceRate",
"External_Competitive_1",
"External_Competitive_2",
"External_Competitive_3",
"External_Competitive_4",
"External_Competitive_5",
"External_Competitive_6",
"External_Competitive_7",
"Transactions_Mean",
"Transactions_std",
"Transactions_Max",
]
Total_Features = Categorical_Features + Continuous_Features
cat_pipeline = Pipeline([("selector", DataFrameSelector(Categorical_Features))])
num_pipeline = Pipeline(
[
("selector", DataFrameSelector(Continuous_Features)),
("std_scaler", StandardScaler()),
]
)
full_pipeline = FeatureUnion(
transformer_list=[
("cat_pipeline", cat_pipeline),
("num_pipeline", num_pipeline),
]
)
X_train = full_pipeline.fit_transform(X_train)
X_test = full_pipeline.transform(TestFile)
# KNN
# Introduce KNN Classifier
KNeighbours = KNeighborsClassifier()
leaf_size = list(range(1, 25, 5))
n_neighbors = list(range(4, 30, 2))
param_grid_KNeighbours = {
"n_neighbors": n_neighbors,
"algorithm": ["auto"],
"weights": ["uniform", "distance"],
"leaf_size": leaf_size,
}
grid_search_KNeighbours = RandomizedSearchCV(
KNeighbours,
param_grid_KNeighbours,
cv=4,
scoring="roc_auc",
refit=True,
n_jobs=-1,
verbose=2,
)
grid_search_KNeighbours.fit(X_train, y_train)
# Prepare the predictions file
result_test1 = pd.DataFrame()
passenger_id_test = TestFile["Customer_id"].copy()
result_test1["Customer_id"] = passenger_id_test
result_test1["Target"] = y_pred_neighbor_grid
result_test1.to_csv("Knn.csv", index=False)
# Random Forest
forest_class = RandomForestClassifier(random_state=42)
n_estimators = [50, 100, 400, 700, 1000]
max_features = [5, 7, 10]
max_depth = [10, 20]
oob_score = [True, False]
min_samples_split = [2, 4, 10, 12, 16]
min_samples_leaf = [1, 5, 10]
max_leaf_nodes = [2, 10, 20]
param_grid_forest = {
"n_estimators": n_estimators,
"max_features": max_features,
"max_depth": max_depth,
"min_samples_split": min_samples_split,
"oob_score": oob_score,
"min_samples_leaf": min_samples_leaf,
"max_leaf_nodes": max_leaf_nodes,
}
rand_search_forest = RandomizedSearchCV(
forest_class,
param_grid_forest,
cv=4,
scoring="roc_auc",
refit=True,
n_jobs=-1,
verbose=2,
)
rand_search_forest.fit(X_train, y_train)
random_estimator = rand_search_forest.best_estimator_
y_pred_random_estimator = random_estimator.predict(TestFile)
accuracy_score(train_set_y, y_pred_random_estimator)
# Prepare the predictions file
result_test2 = pd.DataFrame()
passenger_id_test = TestFile["Customer_id"].copy()
result_test2["Customer_id"] = passenger_id_test
result_test2["Target"] = y_pred_random_estimator
result_test2.to_csv("Random.csv", index=False)
| false | 0 | 3,944 | 0 | 3,944 | 3,944 |
||
69595703
|
<jupyter_start><jupyter_text>EfficientNet3D
Kaggle dataset identifier: efficientnet3d
<jupyter_script>import torch
import torch.nn as nn
import cv2
import pytorch_lightning as pl
from pytorch_lightning.core.lightning import LightningModule
from torch.utils.data import Dataset, DataLoader
import pydicom
import matplotlib.pyplot as plt
import numpy as np
import os
import sys
import random
import glob
import collections
import time
import pandas as pd
from sklearn.model_selection import train_test_split
import seaborn as sns
if os.path.exists("../input/rsna-miccai-brain-tumor-radiogenomic-classification"):
data_directory = "../input/rsna-miccai-brain-tumor-radiogenomic-classification"
pytorch3dpath = "../input/efficientnet3d/EfficientNet-PyTorch-3D-master"
else:
data_directory = (
"/media/roland/data/kaggle/rsna-miccai-brain-tumor-radiogenomic-classification"
)
pytorch3dpath = "EfficientNet-PyTorch-3D"
mri_types = ["FLAIR", "T1w", "T1wCE", "T2w"]
SIZE = 256
NUM_IMAGES = 64
# sys.path.append(pytorch3dpath)
# from efficientnet_pytorch_3d import EfficientNet3D
df = pd.read_csv(
"../input/rsna-miccai-brain-tumor-radiogenomic-classification/train_labels.csv"
)
df = df.loc[df["BraTS21ID"] != 109]
df = df.loc[df["BraTS21ID"] != 709]
df = df.reset_index(drop=True)
df
sample_df = pd.read_csv(
"../input/rsna-miccai-brain-tumor-radiogenomic-classification/sample_submission.csv"
)
X = df[["BraTS21ID"]]
y = df[["MGMT_value"]]
train_x, test_x, train_y, test_y = train_test_split(
X, y, test_size=0.25, random_state=42, stratify=y["MGMT_value"]
)
test_x, val_x, test_y, val_y = train_test_split(
test_x, test_y, test_size=0.25, random_state=42, stratify=test_y["MGMT_value"]
)
print(len(train_x), "\n", len(test_x), "\n", len(val_x))
def load_dicom_image(path, img_size=SIZE):
dicom = pydicom.read_file(path)
data = dicom.pixel_array
if np.min(data) == np.max(data):
data = np.zeros((img_size, img_size))
return data
data = data - np.min(data)
if np.max(data) != 0:
data = data / np.max(data)
# data = (data * 255).astype(np.uint8)
data = cv2.resize(data, (img_size, img_size))
return data
def load_dicom_images_3d(
scan_id, num_imgs=NUM_IMAGES, img_size=SIZE, mri_type="FLAIR", split="train"
):
files = sorted(glob.glob(f"{data_directory}/{split}/{scan_id}/{mri_type}/*.dcm"))
middle = len(files) // 2
num_imgs2 = num_imgs // 2
p1 = max(0, middle - num_imgs2)
p2 = min(len(files), middle + num_imgs2)
img3d = np.stack([load_dicom_image(f) for f in files[p1:p2]]).T
if img3d.shape[-1] < num_imgs:
n_zero = np.zeros((img_size, img_size, num_imgs - img3d.shape[-1]))
img3d = np.concatenate((img3d, n_zero), axis=-1)
return np.expand_dims(img3d, 0)
load_dicom_images_3d("00002").shape
class RSNA_Dataset(Dataset):
def __init__(
self, paths, targets=None, mri_type="Flair", label_smoothing=0.0, split="train"
):
self.paths = paths
self.targets = targets
self.mri_type = mri_type
self.label_smoothing = label_smoothing
self.split = split
def __len__(self):
return len(self.paths)
def __getitem__(self, index):
scan_id = self.paths[index]
if self.targets is None:
data = load_dicom_images_3d(str(scan_id).zfill(5), split="test")
else:
data = load_dicom_images_3d(str(scan_id).zfill(5))
if self.targets is None:
return torch.tensor(data).float()
else:
y = torch.tensor(
abs(self.targets[index] - self.label_smoothing), dtype=torch.float
)
return torch.tensor(data).float(), y
train_dataset = RSNA_Dataset(train_x["BraTS21ID"].values, train_y["MGMT_value"].values)
test_dataset = RSNA_Dataset(val_x["BraTS21ID"].values, val_y["MGMT_value"].values)
validation_dataset = RSNA_Dataset(
test_x["BraTS21ID"].values, test_y["MGMT_value"].values
)
predict_dataset = RSNA_Dataset(
sample_df["BraTS21ID"].values,
)
class RSNA_DataModule(pl.LightningDataModule):
def __init__(self):
super().__init__()
self.train = train_dataset
self.val = validation_dataset
self.test = test_dataset
self.predict = predict_dataset
def train_dataloader(self):
return DataLoader(self.train, batch_size=28, shuffle=True, num_workers=2)
def val_dataloader(self):
return DataLoader(self.val, batch_size=28, shuffle=False, num_workers=2)
def test_dataloader(self):
return DataLoader(self.test, batch_size=22, shuffle=False, num_workers=2)
def predict_dataloader(self):
return DataLoader(self.predict, batch_size=1, shuffle=False, num_workers=2)
# Sanity Check
image, label = next(iter(DataLoader(train_dataset, batch_size=1, shuffle=True)))
print(image, label)
package_path = "../input/efficientnet3d/EfficientNet-PyTorch-3D-master"
sys.path.append(package_path)
from efficientnet_pytorch_3d import EfficientNet3D
neural_network = EfficientNet3D.from_name(
"efficientnet-b1", override_params={"num_classes": 1}, in_channels=1
)
from sklearn.metrics import roc_curve, auc
probs = nn.Sigmoid()
def get_score(y_pred, y):
probabilities = []
for x in y_pred:
prob = probs(x)
top_p, top_class = prob.topk(1, dim=-1)
probabilities.append(float(top_p))
y = [float(t) for t in y]
logistic_fpr, logistic_tpr, _ = roc_curve(y, probabilities)
aoc_score = auc(logistic_fpr, logistic_tpr)
return aoc_score
class RSNA_Model(pl.LightningModule):
def __init__(self):
super().__init__()
self.neural_net = neural_network
def forward(self, x):
return self.neural_net(x)
def configure_optimizers(self):
optimizer = torch.optim.Adam(self.parameters(), lr=1e-4)
sch = torch.optim.lr_scheduler.StepLR(
optimizer, step_size=5, gamma=0.1, last_epoch=-1, verbose=False
)
return {
"optimizer": optimizer,
"lr_scheduler": {
"scheduler": sch,
# "monitor": "",
},
}
def training_step(self, batch, batch_idx):
x, y = batch
y_pred = self(x)
y = y.unsqueeze(-1)
loss = torch.nn.functional.binary_cross_entropy_with_logits(y_pred, y)
return loss
def validation_step(self, batch, batch_idx):
x, y = batch
y_pred = self(x)
y = y.unsqueeze(-1)
loss = torch.nn.functional.binary_cross_entropy_with_logits(y_pred, y)
print(get_score(y_pred, y))
self.log("auc_score", get_score(y_pred, y))
return loss
def test_step(self, batch, batch_idx):
x, y = batch
y_pred = self(x)
y = y.unsqueeze(-1)
loss = torch.nn.functional.binary_cross_entropy_with_logits(y_pred, y)
print(get_score(y_pred, y))
self.log("test_loss : ", loss)
return loss
from pytorch_lightning.callbacks import ModelCheckpoint
checkpoint_callback = ModelCheckpoint(
monitor="auc_score",
mode="max",
)
from pytorch_lightning import Trainer
model = RSNA_Model()
module = RSNA_DataModule()
model.load_from_checkpoint("../input/efficient3d-checkpoint/FLAIR-Best_Checkpoint.ckpt")
trainer = Trainer(max_epochs=25, callbacks=[checkpoint_callback])
trainer.fit(model, module)
result = trainer.test()
print(result)
predictons = trainer.predict()
probabilities = []
for x in predictons:
prob = probs(x)
top_p, top_class = prob.topk(1, dim=1)
probabilities.append(float(top_p))
trainer.save_checkpoint("FLAIR-Best_Checkpoint.ckpt")
import shutil
shutil.rmtree("./lightning_logs")
data = {"BraTS21ID": list(sample_df["BraTS21ID"]), "MGMT_value": probabilities}
submission = pd.DataFrame(data)
submission.to_csv("submission.csv", index=False)
pd.read_csv("./submission.csv")
sns.displot(submission["MGMT_value"])
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/595/69595703.ipynb
|
efficientnet3d
|
ronaldokun
|
[{"Id": 69595703, "ScriptId": 18952594, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5314291, "CreationDate": "08/02/2021 02:25:50", "VersionNumber": 11.0, "Title": "Flair-Efficientnet-3D", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 250.0, "LinesInsertedFromPrevious": 14.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 236.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92980366, "KernelVersionId": 69595703, "SourceDatasetVersionId": 1523575}]
|
[{"Id": 1523575, "DatasetId": 898203, "DatasourceVersionId": 1557961, "CreatorUserId": 1118320, "LicenseName": "Unknown", "CreationDate": "09/29/2020 00:19:51", "VersionNumber": 1.0, "Title": "EfficientNet3D", "Slug": "efficientnet3d", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 898203, "CreatorUserId": 1118320, "OwnerUserId": 1118320.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1523575.0, "CurrentDatasourceVersionId": 1557961.0, "ForumId": 913831, "Type": 2, "CreationDate": "09/29/2020 00:19:51", "LastActivityDate": "09/29/2020", "TotalViews": 1896, "TotalDownloads": 6, "TotalVotes": 1, "TotalKernels": 1}]
|
[{"Id": 1118320, "UserName": "ronaldokun", "DisplayName": "Ronaldo S.A. Batista", "RegisterDate": "06/09/2017", "PerformanceTier": 2}]
|
import torch
import torch.nn as nn
import cv2
import pytorch_lightning as pl
from pytorch_lightning.core.lightning import LightningModule
from torch.utils.data import Dataset, DataLoader
import pydicom
import matplotlib.pyplot as plt
import numpy as np
import os
import sys
import random
import glob
import collections
import time
import pandas as pd
from sklearn.model_selection import train_test_split
import seaborn as sns
if os.path.exists("../input/rsna-miccai-brain-tumor-radiogenomic-classification"):
data_directory = "../input/rsna-miccai-brain-tumor-radiogenomic-classification"
pytorch3dpath = "../input/efficientnet3d/EfficientNet-PyTorch-3D-master"
else:
data_directory = (
"/media/roland/data/kaggle/rsna-miccai-brain-tumor-radiogenomic-classification"
)
pytorch3dpath = "EfficientNet-PyTorch-3D"
mri_types = ["FLAIR", "T1w", "T1wCE", "T2w"]
SIZE = 256
NUM_IMAGES = 64
# sys.path.append(pytorch3dpath)
# from efficientnet_pytorch_3d import EfficientNet3D
df = pd.read_csv(
"../input/rsna-miccai-brain-tumor-radiogenomic-classification/train_labels.csv"
)
df = df.loc[df["BraTS21ID"] != 109]
df = df.loc[df["BraTS21ID"] != 709]
df = df.reset_index(drop=True)
df
sample_df = pd.read_csv(
"../input/rsna-miccai-brain-tumor-radiogenomic-classification/sample_submission.csv"
)
X = df[["BraTS21ID"]]
y = df[["MGMT_value"]]
train_x, test_x, train_y, test_y = train_test_split(
X, y, test_size=0.25, random_state=42, stratify=y["MGMT_value"]
)
test_x, val_x, test_y, val_y = train_test_split(
test_x, test_y, test_size=0.25, random_state=42, stratify=test_y["MGMT_value"]
)
print(len(train_x), "\n", len(test_x), "\n", len(val_x))
def load_dicom_image(path, img_size=SIZE):
dicom = pydicom.read_file(path)
data = dicom.pixel_array
if np.min(data) == np.max(data):
data = np.zeros((img_size, img_size))
return data
data = data - np.min(data)
if np.max(data) != 0:
data = data / np.max(data)
# data = (data * 255).astype(np.uint8)
data = cv2.resize(data, (img_size, img_size))
return data
def load_dicom_images_3d(
scan_id, num_imgs=NUM_IMAGES, img_size=SIZE, mri_type="FLAIR", split="train"
):
files = sorted(glob.glob(f"{data_directory}/{split}/{scan_id}/{mri_type}/*.dcm"))
middle = len(files) // 2
num_imgs2 = num_imgs // 2
p1 = max(0, middle - num_imgs2)
p2 = min(len(files), middle + num_imgs2)
img3d = np.stack([load_dicom_image(f) for f in files[p1:p2]]).T
if img3d.shape[-1] < num_imgs:
n_zero = np.zeros((img_size, img_size, num_imgs - img3d.shape[-1]))
img3d = np.concatenate((img3d, n_zero), axis=-1)
return np.expand_dims(img3d, 0)
load_dicom_images_3d("00002").shape
class RSNA_Dataset(Dataset):
def __init__(
self, paths, targets=None, mri_type="Flair", label_smoothing=0.0, split="train"
):
self.paths = paths
self.targets = targets
self.mri_type = mri_type
self.label_smoothing = label_smoothing
self.split = split
def __len__(self):
return len(self.paths)
def __getitem__(self, index):
scan_id = self.paths[index]
if self.targets is None:
data = load_dicom_images_3d(str(scan_id).zfill(5), split="test")
else:
data = load_dicom_images_3d(str(scan_id).zfill(5))
if self.targets is None:
return torch.tensor(data).float()
else:
y = torch.tensor(
abs(self.targets[index] - self.label_smoothing), dtype=torch.float
)
return torch.tensor(data).float(), y
train_dataset = RSNA_Dataset(train_x["BraTS21ID"].values, train_y["MGMT_value"].values)
test_dataset = RSNA_Dataset(val_x["BraTS21ID"].values, val_y["MGMT_value"].values)
validation_dataset = RSNA_Dataset(
test_x["BraTS21ID"].values, test_y["MGMT_value"].values
)
predict_dataset = RSNA_Dataset(
sample_df["BraTS21ID"].values,
)
class RSNA_DataModule(pl.LightningDataModule):
def __init__(self):
super().__init__()
self.train = train_dataset
self.val = validation_dataset
self.test = test_dataset
self.predict = predict_dataset
def train_dataloader(self):
return DataLoader(self.train, batch_size=28, shuffle=True, num_workers=2)
def val_dataloader(self):
return DataLoader(self.val, batch_size=28, shuffle=False, num_workers=2)
def test_dataloader(self):
return DataLoader(self.test, batch_size=22, shuffle=False, num_workers=2)
def predict_dataloader(self):
return DataLoader(self.predict, batch_size=1, shuffle=False, num_workers=2)
# Sanity Check
image, label = next(iter(DataLoader(train_dataset, batch_size=1, shuffle=True)))
print(image, label)
package_path = "../input/efficientnet3d/EfficientNet-PyTorch-3D-master"
sys.path.append(package_path)
from efficientnet_pytorch_3d import EfficientNet3D
neural_network = EfficientNet3D.from_name(
"efficientnet-b1", override_params={"num_classes": 1}, in_channels=1
)
from sklearn.metrics import roc_curve, auc
probs = nn.Sigmoid()
def get_score(y_pred, y):
probabilities = []
for x in y_pred:
prob = probs(x)
top_p, top_class = prob.topk(1, dim=-1)
probabilities.append(float(top_p))
y = [float(t) for t in y]
logistic_fpr, logistic_tpr, _ = roc_curve(y, probabilities)
aoc_score = auc(logistic_fpr, logistic_tpr)
return aoc_score
class RSNA_Model(pl.LightningModule):
def __init__(self):
super().__init__()
self.neural_net = neural_network
def forward(self, x):
return self.neural_net(x)
def configure_optimizers(self):
optimizer = torch.optim.Adam(self.parameters(), lr=1e-4)
sch = torch.optim.lr_scheduler.StepLR(
optimizer, step_size=5, gamma=0.1, last_epoch=-1, verbose=False
)
return {
"optimizer": optimizer,
"lr_scheduler": {
"scheduler": sch,
# "monitor": "",
},
}
def training_step(self, batch, batch_idx):
x, y = batch
y_pred = self(x)
y = y.unsqueeze(-1)
loss = torch.nn.functional.binary_cross_entropy_with_logits(y_pred, y)
return loss
def validation_step(self, batch, batch_idx):
x, y = batch
y_pred = self(x)
y = y.unsqueeze(-1)
loss = torch.nn.functional.binary_cross_entropy_with_logits(y_pred, y)
print(get_score(y_pred, y))
self.log("auc_score", get_score(y_pred, y))
return loss
def test_step(self, batch, batch_idx):
x, y = batch
y_pred = self(x)
y = y.unsqueeze(-1)
loss = torch.nn.functional.binary_cross_entropy_with_logits(y_pred, y)
print(get_score(y_pred, y))
self.log("test_loss : ", loss)
return loss
from pytorch_lightning.callbacks import ModelCheckpoint
checkpoint_callback = ModelCheckpoint(
monitor="auc_score",
mode="max",
)
from pytorch_lightning import Trainer
model = RSNA_Model()
module = RSNA_DataModule()
model.load_from_checkpoint("../input/efficient3d-checkpoint/FLAIR-Best_Checkpoint.ckpt")
trainer = Trainer(max_epochs=25, callbacks=[checkpoint_callback])
trainer.fit(model, module)
result = trainer.test()
print(result)
predictons = trainer.predict()
probabilities = []
for x in predictons:
prob = probs(x)
top_p, top_class = prob.topk(1, dim=1)
probabilities.append(float(top_p))
trainer.save_checkpoint("FLAIR-Best_Checkpoint.ckpt")
import shutil
shutil.rmtree("./lightning_logs")
data = {"BraTS21ID": list(sample_df["BraTS21ID"]), "MGMT_value": probabilities}
submission = pd.DataFrame(data)
submission.to_csv("submission.csv", index=False)
pd.read_csv("./submission.csv")
sns.displot(submission["MGMT_value"])
| false | 2 | 2,577 | 0 | 2,600 | 2,577 |
||
69595663
|
<jupyter_start><jupyter_text>yolov5
Kaggle dataset identifier: yolov5
<jupyter_script># # Install Packages
# # Import Libraries
import gc
import os
import sys
import shutil
from copy import deepcopy
from PIL import Image
import pandas as pd
import numpy as np
from glob import glob
from tqdm import tqdm
import pydicom
from pydicom.pixel_data_handlers.util import apply_voi_lut
import efficientnet.tfkeras as efn
import tensorflow as tf
from tensorflow.keras import backend as K
import tensorflow_hub as tfhub
import torch
from numba import cuda
import matplotlib.pyplot as plt
import seaborn as sns
sys.path.append("/kaggle/input/weightedboxesfusion")
from ensemble_boxes import weighted_boxes_fusion, non_maximum_weighted, nms, soft_nms
# # Load Data
def read_prediction_csv(sub_df: pd.DataFrame):
preds_v = []
for image_id, preds in zip(sub_df["id"].values, sub_df["PredictionString"].values):
_cls, bbox, p_det = [], [], []
preds_split = preds.split()
for i in range(0, len(preds_split), 6):
_cls = preds_split[i]
p_det, x_min, y_min, x_max, y_max = [
float(x) for x in preds_split[i + 1 : i + 6]
]
if _cls != "none":
bboxes = np.array([x_min, y_min, x_max, y_max])
_cls.append(1)
bbox.append(bboxes)
p_det.append(p_det)
preds_v.append(
{
"sample_id": image_id,
"cls": np.array(_cls),
"bbox": np.array(bbox),
"p_det": np.array(p_det),
}
)
del _cls, bbox, p_det
gc.collect()
return preds_v
df = pd.read_csv("/kaggle/input/siim-covid19-detection/sample_submission.csv")
if df.shape[0] == 2477:
fast_sub = True
fast_df = pd.DataFrame(
(
[
["00086460a852_study", "negative 1 0 0 1 1"],
["000c9c05fd14_study", "negative 1 0 0 1 1"],
["65761e66de9f_image", "none 1 0 0 1 1"],
["51759b5579bc_image", "none 1 0 0 1 1"],
]
),
columns=["id", "PredictionString"],
)
else:
fast_sub = False
# ## .dcm to .png
def read_xray(path, voi_lut: bool = True, fix_monochrome: bool = True):
dicom = pydicom.read_file(path)
if voi_lut:
data = apply_voi_lut(dicom.pixel_array, dicom)
else:
data = dicom.pixel_array
if fix_monochrome and dicom.PhotometricInterpretation == "MONOCHROME1":
data = np.amax(data) - data
data = data - np.min(data)
data = data / np.max(data)
data = (data * 255).astype(np.uint8)
return data
def resize(array, size, keep_ratio: bool = False, resample=Image.LANCZOS):
im = Image.fromarray(array)
if keep_ratio:
im.thumbnail((size, size), resample)
else:
im = im.resize((size, size), resample)
return im
split = "test"
save_dir = f"/kaggle/tmp/{split}/"
os.makedirs(save_dir, exist_ok=True)
save_dir = f"/kaggle/tmp/{split}/study/"
os.makedirs(save_dir, exist_ok=True)
# ## Load study-level image
STUDY_RES: int = 1024
if fast_sub:
xray = read_xray(
"/kaggle/input/siim-covid19-detection/train/00086460a852/9e8302230c91/65761e66de9f.dcm"
)
im = resize(xray, size=STUDY_RES)
study = "00086460a852" + "_study.png"
im.save(os.path.join(save_dir, study))
xray = read_xray(
"/kaggle/input/siim-covid19-detection/train/000c9c05fd14/e555410bd2cd/51759b5579bc.dcm"
)
im = resize(xray, size=STUDY_RES)
study = "000c9c05fd14" + "_study.png"
im.save(os.path.join(save_dir, study))
else:
for dirname, _, filenames in tqdm(
os.walk(f"../input/siim-covid19-detection/{split}")
):
for file in filenames:
xray = read_xray(os.path.join(dirname, file))
im = resize(xray, size=STUDY_RES)
study = dirname.split("/")[-2] + "_study.png"
im.save(os.path.join(save_dir, study))
# ## Load image-level image
IMAGE_RES: int = 640
image_id = []
dim0 = []
dim1 = []
splits = []
save_dir = f"/kaggle/tmp/{split}/image/"
os.makedirs(save_dir, exist_ok=True)
if fast_sub:
xray = read_xray(
"/kaggle/input/siim-covid19-detection/train/00086460a852/9e8302230c91/65761e66de9f.dcm"
)
im = resize(xray, size=IMAGE_RES)
im.save(os.path.join(save_dir, "65761e66de9f_image.png"))
image_id.append("65761e66de9f.dcm".replace(".dcm", ""))
dim0.append(xray.shape[0])
dim1.append(xray.shape[1])
splits.append(split)
xray = read_xray(
"/kaggle/input/siim-covid19-detection/train/000c9c05fd14/e555410bd2cd/51759b5579bc.dcm"
)
im = resize(xray, size=IMAGE_RES)
im.save(os.path.join(save_dir, "51759b5579bc_image.png"))
image_id.append("51759b5579bc.dcm".replace(".dcm", ""))
dim0.append(xray.shape[0])
dim1.append(xray.shape[1])
splits.append(split)
else:
for dirname, _, filenames in tqdm(
os.walk(f"../input/siim-covid19-detection/{split}")
):
for file in filenames:
xray = read_xray(os.path.join(dirname, file))
im = resize(xray, size=IMAGE_RES)
im.save(os.path.join(save_dir, file.replace(".dcm", "_image.png")))
image_id.append(file.replace(".dcm", ""))
dim0.append(xray.shape[0])
dim1.append(xray.shape[1])
splits.append(split)
meta = pd.DataFrame.from_dict(
{"image_id": image_id, "dim0": dim0, "dim1": dim1, "split": splits}
)
# # Predict study-level image
# ## TF pipeline
def auto_select_accelerator():
try:
tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
tf.config.experimental_connect_to_cluster(tpu)
tf.tpu.experimental.initialize_tpu_system(tpu)
strategy = tf.distribute.experimental.TPUStrategy(tpu)
print("Running on TPU:", tpu.master())
except ValueError:
strategy = tf.distribute.get_strategy()
print(f"Running on {strategy.num_replicas_in_sync} replicas")
return strategy
def build_decoder(with_labels: bool = False, target_size=(640, 640), ext: str = "png"):
def decode(path):
file_bytes = tf.io.read_file(path)
if ext == "png":
img = tf.image.decode_png(file_bytes, channels=3)
elif ext in ["jpg", "jpeg"]:
img = tf.image.decode_jpeg(file_bytes, channels=3)
else:
raise ValueError("Image extension not supported")
img = tf.cast(img, tf.float32) / 255.0
img = tf.image.resize(img, target_size)
return img
return decode
def build_augmenter(img_size: int, with_labels: bool = False):
def augment(img):
# img = tf.image.random_crop(value=img, size=(img_size, img_size, 3))
img = tf.image.random_flip_left_right(img)
# img = tf.image.random_flip_up_down(img)
img = tf.image.random_brightness(img, 0.1)
return img
return augment
def build_dataset(
paths: str,
image_size: int,
bs: int = 16,
decode_fn=None,
augment_fn=None,
augment: bool = False,
repeat: bool = False,
):
if decode_fn is None:
decode_fn = build_decoder(False, (image_size, image_size))
if augment_fn is None:
augment_fn = build_augmenter(image_size, False)
AUTO = tf.data.experimental.AUTOTUNE
dset = tf.data.Dataset.from_tensor_slices(paths)
dset = dset.map(decode_fn, num_parallel_calls=AUTO)
# dset = dset.cache(cache_dir) if cache else dset
dset = dset.map(augment_fn, num_parallel_calls=AUTO) if augment else dset
dset = dset.repeat() if repeat else dset
# dset = dset.shuffle(shuffle) if shuffle else dset
dset = dset.batch(bs).prefetch(AUTO)
return dset
strategy = auto_select_accelerator()
BATCH_SIZE = strategy.num_replicas_in_sync * 16
# ## Models
EFNS = [
efn.EfficientNetB0,
efn.EfficientNetB1,
efn.EfficientNetB2,
efn.EfficientNetB3,
efn.EfficientNetB4,
efn.EfficientNetB5,
efn.EfficientNetB6,
efn.EfficientNetB7,
]
def build_efnet_model(dim: int, ef: int):
inp = tf.keras.layers.Input(shape=(dim, dim, 3))
base = EFNS[ef](input_shape=(dim, dim, 3), weights=None, include_top=False)
x = base(inp)
x = tf.keras.layers.GlobalAveragePooling2D()(x)
head = tf.keras.Sequential([tf.keras.layers.Dropout(0.5), tf.keras.layers.Dense(4)])
x1 = head(x)
x2 = head(x)
x3 = head(x)
x4 = head(x)
x5 = head(x)
x = (x1 + x2 + x3 + x4 + x5) / 5.0
x = tf.keras.layers.Softmax(dtype="float32")(x)
model = tf.keras.Model(inputs=inp, outputs=x)
return model
# ## Make format
if fast_sub:
df = fast_df.copy()
else:
df = pd.read_csv("/kaggle/input/siim-covid19-detection/sample_submission.csv")
df["id_last_str"] = [df.loc[i, "id"][-1] for i in range(df.shape[0])]
study_len = df[df["id_last_str"] == "y"].shape[0]
if fast_sub:
sub_df = fast_df.copy()
else:
sub_df = pd.read_csv("/kaggle/input/siim-covid19-detection/sample_submission.csv")
sub_df = sub_df[:study_len]
test_paths = f"/kaggle/tmp/{split}/study/" + sub_df["id"] + ".png"
sub_df["negative"] = 0
sub_df["typical"] = 0
sub_df["indeterminate"] = 0
sub_df["atypical"] = 0
label_cols = sub_df.columns[2:]
# ## Inference
def infer_efnet_recipe(
test_paths,
model_path: str,
ef: int,
tta: int,
img_size: int,
prefix: str,
do_fastsub: bool,
):
global fast_sub
print(f"[*] recipe ef : {ef} img_size : {img_size} prefix : {prefix}")
dtest = build_dataset(
paths=test_paths,
image_size=img_size,
bs=BATCH_SIZE,
repeat=False if do_fastsub else tta > 1,
augment=False if do_fastsub else tta > 1,
decode_fn=build_decoder(
with_labels=False, target_size=(img_size, img_size), ext="png"
),
)
model_paths = sorted(
glob(os.path.join(model_path, f"effnet*{ef}-{prefix}-res{img_size}-fold*.h5"))
)
model = None
with strategy.scope():
model = build_efnet_model(img_size, ef=ef)
predictions = []
for model_path in model_paths:
print(f" [+] load {model_path}")
with strategy.scope():
model.load_weights(model_path)
if do_fastsub:
pred = model.predict(dtest)
else:
pred = model.predict(dtest, steps=tta * len(test_paths) / BATCH_SIZE)[
: tta * len(test_paths), :
]
pred = np.mean(pred.reshape(tta, len(test_paths), -1), axis=0)
predictions.append(pred)
del model
del dtest
gc.collect()
K.clear_session()
return np.mean(predictions, axis=0)
TTA: int = 1
pred1 = infer_efnet_recipe(
test_paths,
model_path="/kaggle/input/siim-cvoid-19-effnetb7/",
ef=7,
tta=TTA,
img_size=640,
prefix="scce0.05-adam-aug_v3",
do_fastsub=fast_sub,
)
pred2 = infer_efnet_recipe(
test_paths,
model_path="/kaggle/input/siim-cvoid-19-effnetb6/",
ef=6,
tta=TTA,
img_size=800,
prefix="scce0.05-adam",
do_fastsub=fast_sub,
)
sub_df[label_cols] = (pred1 + pred2) / 2.0
del pred1, pred2
sub_df.columns = [
"id",
"PredictionString1",
"negative",
"typical",
"indeterminate",
"atypical",
]
df = pd.merge(df, sub_df, on="id", how="left")
# ## Generate study-string
for i in range(study_len):
negative = df.at[i, "negative"]
typical = df.at[i, "typical"]
indeterminate = df.at[i, "indeterminate"]
atypical = df.at[i, "atypical"]
df.at[
i, "PredictionString"
] = f"negative {negative} 0 0 1 1 typical {typical} 0 0 1 1 indeterminate {indeterminate} 0 0 1 1 atypical {atypical} 0 0 1 1"
df_study = df[["id", "PredictionString"]]
df_study.head()
# ## Predict opacity
if fast_sub:
sub_df = fast_df.copy()
else:
sub_df = pd.read_csv("/kaggle/input/siim-covid19-detection/sample_submission.csv")
sub_df = sub_df[study_len:]
test_paths = f"/kaggle/tmp/{split}/image/" + sub_df["id"] + ".png"
sub_df["none"] = 0
label_cols = sub_df.columns[2]
pred1 = infer_efnet_recipe(
test_paths,
model_path="/kaggle/input/siim-cvoid-19-effnetb7/",
ef=7,
tta=TTA,
img_size=640,
prefix="scce0.05-adam-aug_v3",
do_fastsub=fast_sub,
)
pred2 = infer_efnet_recipe(
test_paths,
model_path="/kaggle/input/siim-cvoid-19-effnetb6/",
ef=6,
tta=TTA,
img_size=800,
prefix="scce0.05-adam",
do_fastsub=fast_sub,
)
preds = (pred1 + pred2) / 2.0
# preds = (pred1 + pred2 + pred3 + pred4) / 4.
sub_df[label_cols] = preds[:, 0]
df_2class = sub_df.reset_index(drop=True)
del pred1, pred2, preds
K.clear_session()
gc.collect()
cuda.select_device(0)
cuda.close()
cuda.select_device(0)
# # Predict image-level image
meta = meta[meta["split"] == "test"]
if fast_sub:
test_df = fast_df.copy()
else:
test_df = pd.read_csv("/kaggle/input/siim-covid19-detection/sample_submission.csv")
test_df = df[study_len:].reset_index(drop=True)
meta["image_id"] = meta["image_id"] + "_image"
meta.columns = ["id", "dim0", "dim1", "split"]
test_df = pd.merge(test_df, meta, on="id", how="left")
test_dir = f"/kaggle/tmp/{split}/image"
shutil.copytree("/kaggle/input/yolov5", "/kaggle/working/yolov5")
os.chdir("/kaggle/working/yolov5")
# ## Utils
# ### yolo2voc
def yolo2voc(image_height, image_width, bboxes):
"""
yolo => [xmid, ymid, w, h] (normalized)
voc => [x1, y1, x2, y1]
"""
bboxes = bboxes.copy().astype(
float
) # otherwise all value will be 0 as voc_pascal dtype is np.int
bboxes[..., [0, 2]] = bboxes[..., [0, 2]] * image_width
bboxes[..., [1, 3]] = bboxes[..., [1, 3]] * image_height
bboxes[..., [0, 1]] = bboxes[..., [0, 1]] - bboxes[..., [2, 3]] / 2.0
bboxes[..., [2, 3]] = bboxes[..., [0, 1]] + bboxes[..., [2, 3]]
return bboxes
# ### bbox
def solve_bbox_problems(bbox_v, scores_v):
bbox_v = np.asarray(bbox_v)
scores_v = np.asarray(scores_v)
to_remove = np.zeros(len(bbox_v), dtype=np.bool)
for i in range(len(bbox_v)):
x1, y1, x2, y2 = bbox_v[i]
if x2 < x1:
x1, x2 = x2, x1
if y2 < y1:
y1, y2 = y2, y1
if x1 < 0:
x1 = 0
if x1 > 1:
x1 = 1
if x2 < 0:
x2 = 0
if x2 > 1:
x2 = 1
if y1 < 0:
y1 = 0
if y1 > 1:
y1 = 1
if y2 < 0:
y2 = 0
if y2 > 1:
y2 = 1
if (x2 - x1) * (y2 - y1) == 0.0:
to_remove[i] = True
bbox_v[i] = x1, y1, x2, y2
if to_remove.sum() > 0:
bbox_v[to_remove] = np.array([0.0, 0.0, 1.0, 1.0])
scores_v[to_remove] = 0.0
return bbox_v, scores_v
def calc_iou(bb0, bb1):
if len(bb0.shape) == 2:
bb0 = bb0.T
if len(bb1.shape) == 2:
bb1 = bb1.T
bb0_x0, bb0_y0, bb0_x1, bb0_y1 = bb0
bb1_x0, bb1_y0, bb1_x1, bb1_y1 = bb1
# determine the coordinates of the intersection rectangle
x_left = np.maximum(bb0_x0, bb1_x0)
y_top = np.maximum(bb0_y0, bb1_y0)
x_right = np.minimum(bb0_x1, bb1_x1)
y_bottom = np.minimum(bb0_y1, bb1_y1)
ret_mask = ~((x_right < x_left) + (y_bottom < y_top))
# The intersection of two axis-aligned bounding boxes is always an
# axis-aligned bounding box
intersection_area = (x_right - x_left) * (y_bottom - y_top)
# compute the area of both AABBs
bb0_area = (bb0_x1 - bb0_x0) * (bb0_y1 - bb0_y0)
bb1_area = (bb1_x1 - bb1_x0) * (bb1_y1 - bb1_y0)
iou = intersection_area / (bb0_area + bb1_area - intersection_area)
return iou * ret_mask
def merge_preds(bbox_v, p_det_v=None, mode: str = "p_det_weight"):
if p_det_v is None:
p_det_v = np.ones(bbox_v.shape[0])
if mode == "p_det_weight" or mode == "p_det_weight_pmean":
typed_p_det_v = p_det_v.astype(bbox_v.dtype)
p_v = (typed_p_det_v / typed_p_det_v.sum())[:, None]
bbox = (bbox_v * p_v).sum(axis=0)
p = p_det_v.mean()
elif mode == "p_det_weight_psum":
typed_p_det_v = p_det_v.astype(bbox_v.dtype)
p_v = (typed_p_det_v / typed_p_det_v.sum())[:, None]
bbox = (bbox_v * p_v).sum(axis=0)
p = p_det_v.sum()
elif mode == "median" or mode == "median_pmean":
bbox = np.median(bbox_v, axis=0)
p = p_det_v.mean()
elif mode == "p_det_max":
i_max = p_det_v.argmax()
bbox = bbox_v[i_max]
p = p_det_v[i_max]
elif mode == "random":
i_max = np.random.randint(0, p_det_v.shape[0])
bbox = bbox_v[i_max]
p = p_det_v[i_max]
else:
raise ValueError(f"Unknown mode {mode}")
return bbox, p
def norm_p_det(pred_v):
p_det_v = [pred_d["p_det"] for pred_d in pred_v if len(pred_d["p_det"]) > 0]
p_det_v = np.concatenate(p_det_v)
p_det_max = p_det_v.max()
print(f"[+] p_det_max = {p_det_max}")
if p_det_max <= 1.0:
print("[*] skipping norm_p_det")
return pred_v
ret_pred_v = deepcopy(pred_v)
for pred_d in ret_pred_v:
if len(pred_d["p_det"]) > 0:
pred_d["p_det"] = pred_d["p_det"] / p_det_max
return ret_pred_v
def fix_boxes(preds_v):
for preds_d in preds_v:
if len(preds_d["cls"]) > 0:
dx_dy = preds_d["bbox"][:, 2:] - preds_d["bbox"][:, :2]
f0 = (dx_dy <= 1).any(axis=-1)
f1 = (preds_d["p_det"] <= 0) + (preds_d["p_det"] > 1.0)
if f0.any() or f1.any():
f = ~(f0 + f1)
for k in ["p_det", "bbox", "cls"]:
preds_d[k] = preds_d[k][f]
# ## Detect Yolov5
import os
import yolov5
from utils.datasets import LoadImages
from utils.general import non_max_suppression, scale_coords, xyxy2xywh
from glob import glob
def yolov5_detect():
device = torch.device("cuda") if torch.cuda.is_available() else torch.device("cpu")
model_paths = [
# YOLOv5x6
"/kaggle/input/siim-covid19-yolov5/yolov5x6-fold0-mAP0.4466.pt",
"/kaggle/input/siim-covid19-yolov5/yolov5x6-fold1-mAP0.49373.pt",
"/kaggle/input/siim-covid19-yolov5/yolov5x6-fold2-mAP0.48003.pt",
"/kaggle/input/siim-covid19-yolov5/yolov5x6-fold3-mAP0.42454.pt",
"/kaggle/input/siim-covid19-yolov5/yolov5x6-fold4-mAP0.46058.pt",
# YOLOv5X6 res640
# '/kaggle/input/siim-covid19-yolov5x6-res640/yolov5x6-res640-fold0-mAP0.4662.pt',
# '/kaggle/input/siim-covid19-yolov5x6-res640/yolov5x6-res640-fold1-mAP0.5044.pt',
# '/kaggle/input/siim-covid19-yolov5x6-res640/yolov5x6-res640-fold2-mAP0.4762.pt',
# '/kaggle/input/siim-covid19-yolov5x6-res640/yolov5x6-res640-fold3-mAP0.4391.pt',
# '/kaggle/input/siim-covid19-yolov5x6-res640/yolov5x6-res640-fold4-mAP0.4676.pt',
# YOLOv5l6
"/kaggle/input/siim-covid19-yolov5l/yolov5l6-res512-fold0-mAP0.42464.pt",
"/kaggle/input/siim-covid19-yolov5l/yolov5l6-res512-fold1-mAP0.39763.pt",
"/kaggle/input/siim-covid19-yolov5l/yolov5l6-res512-fold2-mAP0.42889.pt",
"/kaggle/input/siim-covid19-yolov5l/yolov5l6-res512-fold3-mAP0.39249.pt",
"/kaggle/input/siim-covid19-yolov5l/yolov5l6-res512-fold4-mAP0.4241.pt",
# YOLOv5m6
"/kaggle/input/siim-covid19-yolov5m/yolov5m6-res512-fold0-mAP0.45113.pt",
"/kaggle/input/siim-covid19-yolov5m/yolov5m6-res512-fold1-mAP0.44463.pt",
"/kaggle/input/siim-covid19-yolov5m/yolov5m6-res512-fold2-mAP0.4496.pt",
"/kaggle/input/siim-covid19-yolov5m/yolov5m6-res512-fold3-mAP0.4121.pt",
"/kaggle/input/siim-covid19-yolov5m/yolov5m6-res512-fold4-mAP0.42406.pt",
]
models = [
torch.load(model_path, map_location=device)["model"].to(device).float().eval()
for model_path in model_paths
]
dataset = LoadImages("/kaggle/tmp/test/image", img_size=IMAGE_RES)
all_path = []
all_bboxes = []
all_score = []
for path, img, im0s, _ in dataset:
img = torch.from_numpy(img).to(device).float() / 255.0
if img.ndimension() == 3:
img = img.unsqueeze(0)
bboxes_2, score_2 = [], []
for model in models:
pred = model(img, augment=True)[0]
pred = non_max_suppression(pred, 0.001, 0.5, classes=None, agnostic=False)
bboxes, score = [], []
for i, det in enumerate(pred):
# gain = torch.tensor(im0.shape)[[1, 0, 1, 0]]
if det is not None and len(det):
det[:, :4] = scale_coords(
img.shape[2:], det[:, :4], im0s.shape
).round()
for c in det[:, -1].unique():
n = (det[:, -1] == c).sum()
for *xyxy, conf, _ in det:
bboxes.append(torch.tensor(xyxy).view(-1).numpy())
score.append(conf.cpu().numpy().item())
bboxes_2.append(bboxes)
score_2.append(score)
all_path.append(path)
all_score.append(score_2)
all_bboxes.append(bboxes_2)
del models
del dataset
gc.collect()
torch.cuda.empty_cache()
return all_path, all_score, all_bboxes
# ## Yolov5
def ensemble_pp(boxes, scores, iou_thres: float = 0.25, skip_box_thr: float = 0.01):
labels = [np.ones(len(scores[idx])) for idx in range(len(scores))]
nms_boxes, nms_scores, nms_labels = nms(
boxes, scores, labels, weights=None, iou_thr=iou_thres
)
snms_boxes, snms_scores, snms_labels = soft_nms(
boxes,
scores,
labels,
weights=None,
iou_thr=iou_thres,
sigma=0.1,
thresh=skip_box_thr,
)
nmw_boxes, nmw_scores, nmw_labels = non_maximum_weighted(
boxes,
scores,
labels,
weights=None,
iou_thr=iou_thres,
skip_box_thr=skip_box_thr,
)
wbf_boxes, wbf_scores, wbf_labels = weighted_boxes_fusion(
boxes,
scores,
labels,
weights=None,
iou_thr=iou_thres,
skip_box_thr=skip_box_thr,
)
del labels
boxes, scores, _ = weighted_boxes_fusion(
[nms_boxes, snms_boxes, nmw_boxes, wbf_boxes],
[nms_scores, snms_scores, nmw_scores, wbf_scores],
[nms_labels, snms_labels, nmw_labels, wbf_labels],
weights=[2, 3, 4, 5],
iou_thr=iou_thres,
skip_box_thr=skip_box_thr,
)
del nms_boxes, nms_scores, nms_labels
del snms_boxes, snms_scores, snms_labels
del nmw_boxes, nmw_scores, nmw_labels
del wbf_boxes, wbf_scores, wbf_labels
gc.collect()
return boxes, scores
with torch.no_grad():
yolov5_all_path, yolov5_all_score, yolov5_all_bboxes = yolov5_detect()
yolov5_preds = {}
for row in range(len(yolov5_all_path)):
image_id = yolov5_all_path[row].split("/")[-1].split(".")[0]
boxes = yolov5_all_bboxes[row]
scores = yolov5_all_score[row]
# normalized to [0, 1]
boxes = [[coord / (IMAGE_RES - 1) for coord in box] for box in boxes]
# solve_bbox_problems over the models
# for i in range(len(boxes)):
# boxes, scores = solve_bbox_problems(boxes[i], scores[i])
boxes, scores = ensemble_pp(boxes, scores, iou_thres=0.25, skip_box_thr=0.01)
# unnormalized to [0, IMAGE_RES]
boxes = [
np.asarray([int(coord * (IMAGE_RES - 1)) for coord in box]) for box in boxes
]
yolov5_preds[image_id] = [boxes, scores]
del image_id, boxes, scores
del yolov5_all_path
del yolov5_all_score
del yolov5_all_bboxes
gc.collect()
# ## Convert coordinates
image_ids = []
PredictionStrings = []
for image_id, v in yolov5_preds.items():
w, h = test_df.loc[test_df["id"] == image_id, ["dim1", "dim0"]].values[0]
boxes, scores = v
normalized_boxes = [xyxy2xywh(box[None, :]) / IMAGE_RES for box in boxes]
rescaled_boxes = [np.round(yolo2voc(h, w, x)[0]) for x in normalized_boxes]
string_boxes = [
f"1 {score} {int(box[0])} {int(box[1])} {int(box[2])} {int(box[3])}"
for score, box in zip(scores, rescaled_boxes)
]
image_ids.append(image_id)
PredictionStrings.append(" ".join(string_boxes))
pred_df = pd.DataFrame({"id": image_ids, "PredictionString": PredictionStrings})
test_df = test_df.drop(["PredictionString"], axis=1)
sub_df = pd.merge(test_df, pred_df, on="id", how="left").fillna("none 1 0 0 1 1")
sub_df = sub_df[["id", "PredictionString"]]
for i in range(sub_df.shape[0]):
prediction_string: str = sub_df.at[i, "PredictionString"]
if prediction_string == "none 1 0 0 1 1":
continue
sub_df_split = prediction_string.split()
sub_df_list = []
for j in range(len(sub_df_split) // 6):
sub_df_list.append("opacity")
sub_df_list.append(sub_df_split[6 * j + 1])
sub_df_list.append(sub_df_split[6 * j + 2])
sub_df_list.append(sub_df_split[6 * j + 3])
sub_df_list.append(sub_df_split[6 * j + 4])
sub_df_list.append(sub_df_split[6 * j + 5])
sub_df.at[i, "PredictionString"] = " ".join(sub_df_list)
# ## Post-Processing
def clean_predictions(
preds_v,
iou_th: float = 0.60,
mode: str = "p_det_weight",
consensus_level: int = 1,
n_models2ensemble: int = 1,
):
ret_preds_v = []
for pred_d in preds_v:
cls_v = pred_d["cls"]
bbox_key = "bbox"
bbox_v = pred_d[bbox_key]
if "p_det" in pred_d.keys():
ret_p_det = True
p_det_v = pred_d["p_det"]
else:
ret_p_det = False
p_det_v = np.ones(pred_d["cls"].shape)
model_id_v = np.zeros(pred_d["cls"].shape, dtype=np.int)
new_cls_v = []
new_bbox_v = []
new_p_det_v = []
new_rad_id_v = []
for i_c in np.unique(cls_v):
f_c = cls_v == i_c
n_c = f_c.sum()
if n_c == 1:
if consensus_level > 1 and i_c != -1:
continue
new_cls_v.append(i_c)
new_bbox_v.append(bbox_v[f_c][0])
new_p_det_v.append(p_det_v[f_c][0])
else:
f_cls_v = cls_v[f_c]
f_bbox_v = bbox_v[f_c]
f_p_det_v = p_det_v[f_c]
f_model_id_v = model_id_v[f_c]
to_join_idxs_v = []
for i in range(n_c):
idxs_s = set(
np.argwhere(calc_iou(f_bbox_v[i], f_bbox_v) > iou_th).T[0]
)
for i in range(len(to_join_idxs_v)):
if len(idxs_s.intersection(to_join_idxs_v[i])) > 0:
to_join_idxs_v[i] = to_join_idxs_v[i].union(idxs_s)
break
else:
to_join_idxs_v.append(idxs_s)
for to_join_idxs in to_join_idxs_v:
to_join_idxs = list(to_join_idxs)
if len(to_join_idxs) < consensus_level:
continue
bbox, p_det = merge_preds(
f_bbox_v[to_join_idxs],
f_p_det_v[to_join_idxs],
mode=mode,
)
if n_models2ensemble > 1:
ens_prop = (
len(np.unique(f_model_id_v[to_join_idxs]))
/ n_models2ensemble
)
p_det = p_det * ens_prop
new_cls_v.append(i_c)
new_bbox_v.append(bbox)
new_p_det_v.append(p_det)
if ret_rad_id:
new_rad_id_v.append(
np.concatenate(f_rad_id_v[to_join_idxs], axis=-1)
)
ret_preds_d = {
"cls": np.array(new_cls_v),
bbox_key: np.array(new_bbox_v),
}
if ret_p_det:
ret_preds_d["p_det"] = np.array(new_p_det_v)
for k in pred_d.keys():
if k not in ["cls", bbox_key, "p_det"]:
ret_preds_d[k] = pred_d[k]
ret_preds_v.append(ret_preds_d)
return ret_preds_v
def pred_to_str(pred_d):
bbox_v = pred_d["bbox"]
p_det_v = pred_d["p_det"]
return " ".join(
[
"opacity {:0.05} {} {} {} {}".format(p_det, *bbox)
for p_det, bbox in zip(p_det_v, np.round(bbox_v).astype(np.int))
]
)
# reference : https://www.kaggle.com/morizin/ensemble331-remake/notebook#Final-cleaning
preds_v = read_prediction_csv(sub_df)
fix_boxes(preds_v)
clean_pred_v = clean_predictions(preds_v, iou_th=0.25, mode="p_det_weight_psum")
norm_clean_pred_v = norm_p_det(clean_pred_v)
del preds_v
del clean_pred_v
gc.collect()
pred_summary_d = {"image_id": [], "PredictionString": []}
for pred_d in norm_clean_pred_v:
pred_str = pred_to_str(pred_d)
pred_summary_d["image_id"].append(pred_d["sample_id"])
pred_summary_d["PredictionString"].append(pred_str)
sub_df = pd.DataFrame(pred_summary_d)
low_threshold: float = 0.00
high_threshold: float = 1.00
sub_df["none"] = df_2class["none"]
c0, c1, c2 = 0, 0, 0
for i in range(sub_df.shape[0]):
if sub_df.at[i, "PredictionString"] != "none 1 0 0 1 1":
none_prob: float = sub_df.at[i, "none"]
sub_df.at[i, "PredictionString"] = (
sub_df.at[i, "PredictionString"] + f" none {none_prob} 0 0 1 1"
)
c1 += 1
# if none_prob < low_threshold:
# sub_df.at[i, 'PredictionString'] = sub_df.at[i, 'PredictionString']
# c0 += 1
# elif low_threshold <= none_prob and none_prob < high_threshold:
# sub_df.at[i, 'PredictionString'] = sub_df.at[i, 'PredictionString'] + f' none {none_prob} 0 0 1 1'
# c1 += 1
# else:
# sub_df.at[i, 'PredictionString'] = 'none 1 0 0 1 1'
# c2 += 1
print(f"[+] threshold | low {low_threshold:.3f} high {high_threshold:.3f}")
print(f"[+] total {c0 + c1 + c2} | keep {c0} add {c1} replace {c2}")
sub_df = sub_df[["image_id", "PredictionString"]]
sub_df = sub_df.rename(columns={"image_id": "id"})
# # Submission
df_study = df_study[:study_len]
df_study = df_study.append(sub_df).reset_index(drop=True)
df_study.to_csv("/kaggle/working/submission.csv", index=False)
df_study
shutil.rmtree("/kaggle/working/yolov5")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/595/69595663.ipynb
|
yolov5
|
kozistr
|
[{"Id": 69595663, "ScriptId": 18974254, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1039385, "CreationDate": "08/02/2021 02:24:59", "VersionNumber": 15.0, "Title": "[infer] effnet + yolov5 w/ pp", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 1003.0, "LinesInsertedFromPrevious": 57.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 946.0, "LinesInsertedFromFork": 391.0, "LinesDeletedFromFork": 126.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 612.0, "TotalVotes": 0}]
|
[{"Id": 92980299, "KernelVersionId": 69595663, "SourceDatasetVersionId": 2433256}, {"Id": 92980292, "KernelVersionId": 69595663, "SourceDatasetVersionId": 1666454}, {"Id": 92980298, "KernelVersionId": 69595663, "SourceDatasetVersionId": 2406834}, {"Id": 92980291, "KernelVersionId": 69595663, "SourceDatasetVersionId": 1219292}]
|
[{"Id": 2433256, "DatasetId": 1411992, "DatasourceVersionId": 2475502, "CreatorUserId": 1039385, "LicenseName": "Unknown", "CreationDate": "07/17/2021 05:02:50", "VersionNumber": 2.0, "Title": "yolov5", "Slug": "yolov5", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Automatic Update 2021-07-17", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1411992, "CreatorUserId": 1039385, "OwnerUserId": 1039385.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 4127213.0, "CurrentDatasourceVersionId": 4183575.0, "ForumId": 1431344, "Type": 2, "CreationDate": "06/16/2021 09:10:42", "LastActivityDate": "06/16/2021", "TotalViews": 1155, "TotalDownloads": 3, "TotalVotes": 2, "TotalKernels": 2}]
|
[{"Id": 1039385, "UserName": "kozistr", "DisplayName": "HyeongChan Kim", "RegisterDate": "04/24/2017", "PerformanceTier": 2}]
|
# # Install Packages
# # Import Libraries
import gc
import os
import sys
import shutil
from copy import deepcopy
from PIL import Image
import pandas as pd
import numpy as np
from glob import glob
from tqdm import tqdm
import pydicom
from pydicom.pixel_data_handlers.util import apply_voi_lut
import efficientnet.tfkeras as efn
import tensorflow as tf
from tensorflow.keras import backend as K
import tensorflow_hub as tfhub
import torch
from numba import cuda
import matplotlib.pyplot as plt
import seaborn as sns
sys.path.append("/kaggle/input/weightedboxesfusion")
from ensemble_boxes import weighted_boxes_fusion, non_maximum_weighted, nms, soft_nms
# # Load Data
def read_prediction_csv(sub_df: pd.DataFrame):
preds_v = []
for image_id, preds in zip(sub_df["id"].values, sub_df["PredictionString"].values):
_cls, bbox, p_det = [], [], []
preds_split = preds.split()
for i in range(0, len(preds_split), 6):
_cls = preds_split[i]
p_det, x_min, y_min, x_max, y_max = [
float(x) for x in preds_split[i + 1 : i + 6]
]
if _cls != "none":
bboxes = np.array([x_min, y_min, x_max, y_max])
_cls.append(1)
bbox.append(bboxes)
p_det.append(p_det)
preds_v.append(
{
"sample_id": image_id,
"cls": np.array(_cls),
"bbox": np.array(bbox),
"p_det": np.array(p_det),
}
)
del _cls, bbox, p_det
gc.collect()
return preds_v
df = pd.read_csv("/kaggle/input/siim-covid19-detection/sample_submission.csv")
if df.shape[0] == 2477:
fast_sub = True
fast_df = pd.DataFrame(
(
[
["00086460a852_study", "negative 1 0 0 1 1"],
["000c9c05fd14_study", "negative 1 0 0 1 1"],
["65761e66de9f_image", "none 1 0 0 1 1"],
["51759b5579bc_image", "none 1 0 0 1 1"],
]
),
columns=["id", "PredictionString"],
)
else:
fast_sub = False
# ## .dcm to .png
def read_xray(path, voi_lut: bool = True, fix_monochrome: bool = True):
dicom = pydicom.read_file(path)
if voi_lut:
data = apply_voi_lut(dicom.pixel_array, dicom)
else:
data = dicom.pixel_array
if fix_monochrome and dicom.PhotometricInterpretation == "MONOCHROME1":
data = np.amax(data) - data
data = data - np.min(data)
data = data / np.max(data)
data = (data * 255).astype(np.uint8)
return data
def resize(array, size, keep_ratio: bool = False, resample=Image.LANCZOS):
im = Image.fromarray(array)
if keep_ratio:
im.thumbnail((size, size), resample)
else:
im = im.resize((size, size), resample)
return im
split = "test"
save_dir = f"/kaggle/tmp/{split}/"
os.makedirs(save_dir, exist_ok=True)
save_dir = f"/kaggle/tmp/{split}/study/"
os.makedirs(save_dir, exist_ok=True)
# ## Load study-level image
STUDY_RES: int = 1024
if fast_sub:
xray = read_xray(
"/kaggle/input/siim-covid19-detection/train/00086460a852/9e8302230c91/65761e66de9f.dcm"
)
im = resize(xray, size=STUDY_RES)
study = "00086460a852" + "_study.png"
im.save(os.path.join(save_dir, study))
xray = read_xray(
"/kaggle/input/siim-covid19-detection/train/000c9c05fd14/e555410bd2cd/51759b5579bc.dcm"
)
im = resize(xray, size=STUDY_RES)
study = "000c9c05fd14" + "_study.png"
im.save(os.path.join(save_dir, study))
else:
for dirname, _, filenames in tqdm(
os.walk(f"../input/siim-covid19-detection/{split}")
):
for file in filenames:
xray = read_xray(os.path.join(dirname, file))
im = resize(xray, size=STUDY_RES)
study = dirname.split("/")[-2] + "_study.png"
im.save(os.path.join(save_dir, study))
# ## Load image-level image
IMAGE_RES: int = 640
image_id = []
dim0 = []
dim1 = []
splits = []
save_dir = f"/kaggle/tmp/{split}/image/"
os.makedirs(save_dir, exist_ok=True)
if fast_sub:
xray = read_xray(
"/kaggle/input/siim-covid19-detection/train/00086460a852/9e8302230c91/65761e66de9f.dcm"
)
im = resize(xray, size=IMAGE_RES)
im.save(os.path.join(save_dir, "65761e66de9f_image.png"))
image_id.append("65761e66de9f.dcm".replace(".dcm", ""))
dim0.append(xray.shape[0])
dim1.append(xray.shape[1])
splits.append(split)
xray = read_xray(
"/kaggle/input/siim-covid19-detection/train/000c9c05fd14/e555410bd2cd/51759b5579bc.dcm"
)
im = resize(xray, size=IMAGE_RES)
im.save(os.path.join(save_dir, "51759b5579bc_image.png"))
image_id.append("51759b5579bc.dcm".replace(".dcm", ""))
dim0.append(xray.shape[0])
dim1.append(xray.shape[1])
splits.append(split)
else:
for dirname, _, filenames in tqdm(
os.walk(f"../input/siim-covid19-detection/{split}")
):
for file in filenames:
xray = read_xray(os.path.join(dirname, file))
im = resize(xray, size=IMAGE_RES)
im.save(os.path.join(save_dir, file.replace(".dcm", "_image.png")))
image_id.append(file.replace(".dcm", ""))
dim0.append(xray.shape[0])
dim1.append(xray.shape[1])
splits.append(split)
meta = pd.DataFrame.from_dict(
{"image_id": image_id, "dim0": dim0, "dim1": dim1, "split": splits}
)
# # Predict study-level image
# ## TF pipeline
def auto_select_accelerator():
try:
tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
tf.config.experimental_connect_to_cluster(tpu)
tf.tpu.experimental.initialize_tpu_system(tpu)
strategy = tf.distribute.experimental.TPUStrategy(tpu)
print("Running on TPU:", tpu.master())
except ValueError:
strategy = tf.distribute.get_strategy()
print(f"Running on {strategy.num_replicas_in_sync} replicas")
return strategy
def build_decoder(with_labels: bool = False, target_size=(640, 640), ext: str = "png"):
def decode(path):
file_bytes = tf.io.read_file(path)
if ext == "png":
img = tf.image.decode_png(file_bytes, channels=3)
elif ext in ["jpg", "jpeg"]:
img = tf.image.decode_jpeg(file_bytes, channels=3)
else:
raise ValueError("Image extension not supported")
img = tf.cast(img, tf.float32) / 255.0
img = tf.image.resize(img, target_size)
return img
return decode
def build_augmenter(img_size: int, with_labels: bool = False):
def augment(img):
# img = tf.image.random_crop(value=img, size=(img_size, img_size, 3))
img = tf.image.random_flip_left_right(img)
# img = tf.image.random_flip_up_down(img)
img = tf.image.random_brightness(img, 0.1)
return img
return augment
def build_dataset(
paths: str,
image_size: int,
bs: int = 16,
decode_fn=None,
augment_fn=None,
augment: bool = False,
repeat: bool = False,
):
if decode_fn is None:
decode_fn = build_decoder(False, (image_size, image_size))
if augment_fn is None:
augment_fn = build_augmenter(image_size, False)
AUTO = tf.data.experimental.AUTOTUNE
dset = tf.data.Dataset.from_tensor_slices(paths)
dset = dset.map(decode_fn, num_parallel_calls=AUTO)
# dset = dset.cache(cache_dir) if cache else dset
dset = dset.map(augment_fn, num_parallel_calls=AUTO) if augment else dset
dset = dset.repeat() if repeat else dset
# dset = dset.shuffle(shuffle) if shuffle else dset
dset = dset.batch(bs).prefetch(AUTO)
return dset
strategy = auto_select_accelerator()
BATCH_SIZE = strategy.num_replicas_in_sync * 16
# ## Models
EFNS = [
efn.EfficientNetB0,
efn.EfficientNetB1,
efn.EfficientNetB2,
efn.EfficientNetB3,
efn.EfficientNetB4,
efn.EfficientNetB5,
efn.EfficientNetB6,
efn.EfficientNetB7,
]
def build_efnet_model(dim: int, ef: int):
inp = tf.keras.layers.Input(shape=(dim, dim, 3))
base = EFNS[ef](input_shape=(dim, dim, 3), weights=None, include_top=False)
x = base(inp)
x = tf.keras.layers.GlobalAveragePooling2D()(x)
head = tf.keras.Sequential([tf.keras.layers.Dropout(0.5), tf.keras.layers.Dense(4)])
x1 = head(x)
x2 = head(x)
x3 = head(x)
x4 = head(x)
x5 = head(x)
x = (x1 + x2 + x3 + x4 + x5) / 5.0
x = tf.keras.layers.Softmax(dtype="float32")(x)
model = tf.keras.Model(inputs=inp, outputs=x)
return model
# ## Make format
if fast_sub:
df = fast_df.copy()
else:
df = pd.read_csv("/kaggle/input/siim-covid19-detection/sample_submission.csv")
df["id_last_str"] = [df.loc[i, "id"][-1] for i in range(df.shape[0])]
study_len = df[df["id_last_str"] == "y"].shape[0]
if fast_sub:
sub_df = fast_df.copy()
else:
sub_df = pd.read_csv("/kaggle/input/siim-covid19-detection/sample_submission.csv")
sub_df = sub_df[:study_len]
test_paths = f"/kaggle/tmp/{split}/study/" + sub_df["id"] + ".png"
sub_df["negative"] = 0
sub_df["typical"] = 0
sub_df["indeterminate"] = 0
sub_df["atypical"] = 0
label_cols = sub_df.columns[2:]
# ## Inference
def infer_efnet_recipe(
test_paths,
model_path: str,
ef: int,
tta: int,
img_size: int,
prefix: str,
do_fastsub: bool,
):
global fast_sub
print(f"[*] recipe ef : {ef} img_size : {img_size} prefix : {prefix}")
dtest = build_dataset(
paths=test_paths,
image_size=img_size,
bs=BATCH_SIZE,
repeat=False if do_fastsub else tta > 1,
augment=False if do_fastsub else tta > 1,
decode_fn=build_decoder(
with_labels=False, target_size=(img_size, img_size), ext="png"
),
)
model_paths = sorted(
glob(os.path.join(model_path, f"effnet*{ef}-{prefix}-res{img_size}-fold*.h5"))
)
model = None
with strategy.scope():
model = build_efnet_model(img_size, ef=ef)
predictions = []
for model_path in model_paths:
print(f" [+] load {model_path}")
with strategy.scope():
model.load_weights(model_path)
if do_fastsub:
pred = model.predict(dtest)
else:
pred = model.predict(dtest, steps=tta * len(test_paths) / BATCH_SIZE)[
: tta * len(test_paths), :
]
pred = np.mean(pred.reshape(tta, len(test_paths), -1), axis=0)
predictions.append(pred)
del model
del dtest
gc.collect()
K.clear_session()
return np.mean(predictions, axis=0)
TTA: int = 1
pred1 = infer_efnet_recipe(
test_paths,
model_path="/kaggle/input/siim-cvoid-19-effnetb7/",
ef=7,
tta=TTA,
img_size=640,
prefix="scce0.05-adam-aug_v3",
do_fastsub=fast_sub,
)
pred2 = infer_efnet_recipe(
test_paths,
model_path="/kaggle/input/siim-cvoid-19-effnetb6/",
ef=6,
tta=TTA,
img_size=800,
prefix="scce0.05-adam",
do_fastsub=fast_sub,
)
sub_df[label_cols] = (pred1 + pred2) / 2.0
del pred1, pred2
sub_df.columns = [
"id",
"PredictionString1",
"negative",
"typical",
"indeterminate",
"atypical",
]
df = pd.merge(df, sub_df, on="id", how="left")
# ## Generate study-string
for i in range(study_len):
negative = df.at[i, "negative"]
typical = df.at[i, "typical"]
indeterminate = df.at[i, "indeterminate"]
atypical = df.at[i, "atypical"]
df.at[
i, "PredictionString"
] = f"negative {negative} 0 0 1 1 typical {typical} 0 0 1 1 indeterminate {indeterminate} 0 0 1 1 atypical {atypical} 0 0 1 1"
df_study = df[["id", "PredictionString"]]
df_study.head()
# ## Predict opacity
if fast_sub:
sub_df = fast_df.copy()
else:
sub_df = pd.read_csv("/kaggle/input/siim-covid19-detection/sample_submission.csv")
sub_df = sub_df[study_len:]
test_paths = f"/kaggle/tmp/{split}/image/" + sub_df["id"] + ".png"
sub_df["none"] = 0
label_cols = sub_df.columns[2]
pred1 = infer_efnet_recipe(
test_paths,
model_path="/kaggle/input/siim-cvoid-19-effnetb7/",
ef=7,
tta=TTA,
img_size=640,
prefix="scce0.05-adam-aug_v3",
do_fastsub=fast_sub,
)
pred2 = infer_efnet_recipe(
test_paths,
model_path="/kaggle/input/siim-cvoid-19-effnetb6/",
ef=6,
tta=TTA,
img_size=800,
prefix="scce0.05-adam",
do_fastsub=fast_sub,
)
preds = (pred1 + pred2) / 2.0
# preds = (pred1 + pred2 + pred3 + pred4) / 4.
sub_df[label_cols] = preds[:, 0]
df_2class = sub_df.reset_index(drop=True)
del pred1, pred2, preds
K.clear_session()
gc.collect()
cuda.select_device(0)
cuda.close()
cuda.select_device(0)
# # Predict image-level image
meta = meta[meta["split"] == "test"]
if fast_sub:
test_df = fast_df.copy()
else:
test_df = pd.read_csv("/kaggle/input/siim-covid19-detection/sample_submission.csv")
test_df = df[study_len:].reset_index(drop=True)
meta["image_id"] = meta["image_id"] + "_image"
meta.columns = ["id", "dim0", "dim1", "split"]
test_df = pd.merge(test_df, meta, on="id", how="left")
test_dir = f"/kaggle/tmp/{split}/image"
shutil.copytree("/kaggle/input/yolov5", "/kaggle/working/yolov5")
os.chdir("/kaggle/working/yolov5")
# ## Utils
# ### yolo2voc
def yolo2voc(image_height, image_width, bboxes):
"""
yolo => [xmid, ymid, w, h] (normalized)
voc => [x1, y1, x2, y1]
"""
bboxes = bboxes.copy().astype(
float
) # otherwise all value will be 0 as voc_pascal dtype is np.int
bboxes[..., [0, 2]] = bboxes[..., [0, 2]] * image_width
bboxes[..., [1, 3]] = bboxes[..., [1, 3]] * image_height
bboxes[..., [0, 1]] = bboxes[..., [0, 1]] - bboxes[..., [2, 3]] / 2.0
bboxes[..., [2, 3]] = bboxes[..., [0, 1]] + bboxes[..., [2, 3]]
return bboxes
# ### bbox
def solve_bbox_problems(bbox_v, scores_v):
bbox_v = np.asarray(bbox_v)
scores_v = np.asarray(scores_v)
to_remove = np.zeros(len(bbox_v), dtype=np.bool)
for i in range(len(bbox_v)):
x1, y1, x2, y2 = bbox_v[i]
if x2 < x1:
x1, x2 = x2, x1
if y2 < y1:
y1, y2 = y2, y1
if x1 < 0:
x1 = 0
if x1 > 1:
x1 = 1
if x2 < 0:
x2 = 0
if x2 > 1:
x2 = 1
if y1 < 0:
y1 = 0
if y1 > 1:
y1 = 1
if y2 < 0:
y2 = 0
if y2 > 1:
y2 = 1
if (x2 - x1) * (y2 - y1) == 0.0:
to_remove[i] = True
bbox_v[i] = x1, y1, x2, y2
if to_remove.sum() > 0:
bbox_v[to_remove] = np.array([0.0, 0.0, 1.0, 1.0])
scores_v[to_remove] = 0.0
return bbox_v, scores_v
def calc_iou(bb0, bb1):
if len(bb0.shape) == 2:
bb0 = bb0.T
if len(bb1.shape) == 2:
bb1 = bb1.T
bb0_x0, bb0_y0, bb0_x1, bb0_y1 = bb0
bb1_x0, bb1_y0, bb1_x1, bb1_y1 = bb1
# determine the coordinates of the intersection rectangle
x_left = np.maximum(bb0_x0, bb1_x0)
y_top = np.maximum(bb0_y0, bb1_y0)
x_right = np.minimum(bb0_x1, bb1_x1)
y_bottom = np.minimum(bb0_y1, bb1_y1)
ret_mask = ~((x_right < x_left) + (y_bottom < y_top))
# The intersection of two axis-aligned bounding boxes is always an
# axis-aligned bounding box
intersection_area = (x_right - x_left) * (y_bottom - y_top)
# compute the area of both AABBs
bb0_area = (bb0_x1 - bb0_x0) * (bb0_y1 - bb0_y0)
bb1_area = (bb1_x1 - bb1_x0) * (bb1_y1 - bb1_y0)
iou = intersection_area / (bb0_area + bb1_area - intersection_area)
return iou * ret_mask
def merge_preds(bbox_v, p_det_v=None, mode: str = "p_det_weight"):
if p_det_v is None:
p_det_v = np.ones(bbox_v.shape[0])
if mode == "p_det_weight" or mode == "p_det_weight_pmean":
typed_p_det_v = p_det_v.astype(bbox_v.dtype)
p_v = (typed_p_det_v / typed_p_det_v.sum())[:, None]
bbox = (bbox_v * p_v).sum(axis=0)
p = p_det_v.mean()
elif mode == "p_det_weight_psum":
typed_p_det_v = p_det_v.astype(bbox_v.dtype)
p_v = (typed_p_det_v / typed_p_det_v.sum())[:, None]
bbox = (bbox_v * p_v).sum(axis=0)
p = p_det_v.sum()
elif mode == "median" or mode == "median_pmean":
bbox = np.median(bbox_v, axis=0)
p = p_det_v.mean()
elif mode == "p_det_max":
i_max = p_det_v.argmax()
bbox = bbox_v[i_max]
p = p_det_v[i_max]
elif mode == "random":
i_max = np.random.randint(0, p_det_v.shape[0])
bbox = bbox_v[i_max]
p = p_det_v[i_max]
else:
raise ValueError(f"Unknown mode {mode}")
return bbox, p
def norm_p_det(pred_v):
p_det_v = [pred_d["p_det"] for pred_d in pred_v if len(pred_d["p_det"]) > 0]
p_det_v = np.concatenate(p_det_v)
p_det_max = p_det_v.max()
print(f"[+] p_det_max = {p_det_max}")
if p_det_max <= 1.0:
print("[*] skipping norm_p_det")
return pred_v
ret_pred_v = deepcopy(pred_v)
for pred_d in ret_pred_v:
if len(pred_d["p_det"]) > 0:
pred_d["p_det"] = pred_d["p_det"] / p_det_max
return ret_pred_v
def fix_boxes(preds_v):
for preds_d in preds_v:
if len(preds_d["cls"]) > 0:
dx_dy = preds_d["bbox"][:, 2:] - preds_d["bbox"][:, :2]
f0 = (dx_dy <= 1).any(axis=-1)
f1 = (preds_d["p_det"] <= 0) + (preds_d["p_det"] > 1.0)
if f0.any() or f1.any():
f = ~(f0 + f1)
for k in ["p_det", "bbox", "cls"]:
preds_d[k] = preds_d[k][f]
# ## Detect Yolov5
import os
import yolov5
from utils.datasets import LoadImages
from utils.general import non_max_suppression, scale_coords, xyxy2xywh
from glob import glob
def yolov5_detect():
device = torch.device("cuda") if torch.cuda.is_available() else torch.device("cpu")
model_paths = [
# YOLOv5x6
"/kaggle/input/siim-covid19-yolov5/yolov5x6-fold0-mAP0.4466.pt",
"/kaggle/input/siim-covid19-yolov5/yolov5x6-fold1-mAP0.49373.pt",
"/kaggle/input/siim-covid19-yolov5/yolov5x6-fold2-mAP0.48003.pt",
"/kaggle/input/siim-covid19-yolov5/yolov5x6-fold3-mAP0.42454.pt",
"/kaggle/input/siim-covid19-yolov5/yolov5x6-fold4-mAP0.46058.pt",
# YOLOv5X6 res640
# '/kaggle/input/siim-covid19-yolov5x6-res640/yolov5x6-res640-fold0-mAP0.4662.pt',
# '/kaggle/input/siim-covid19-yolov5x6-res640/yolov5x6-res640-fold1-mAP0.5044.pt',
# '/kaggle/input/siim-covid19-yolov5x6-res640/yolov5x6-res640-fold2-mAP0.4762.pt',
# '/kaggle/input/siim-covid19-yolov5x6-res640/yolov5x6-res640-fold3-mAP0.4391.pt',
# '/kaggle/input/siim-covid19-yolov5x6-res640/yolov5x6-res640-fold4-mAP0.4676.pt',
# YOLOv5l6
"/kaggle/input/siim-covid19-yolov5l/yolov5l6-res512-fold0-mAP0.42464.pt",
"/kaggle/input/siim-covid19-yolov5l/yolov5l6-res512-fold1-mAP0.39763.pt",
"/kaggle/input/siim-covid19-yolov5l/yolov5l6-res512-fold2-mAP0.42889.pt",
"/kaggle/input/siim-covid19-yolov5l/yolov5l6-res512-fold3-mAP0.39249.pt",
"/kaggle/input/siim-covid19-yolov5l/yolov5l6-res512-fold4-mAP0.4241.pt",
# YOLOv5m6
"/kaggle/input/siim-covid19-yolov5m/yolov5m6-res512-fold0-mAP0.45113.pt",
"/kaggle/input/siim-covid19-yolov5m/yolov5m6-res512-fold1-mAP0.44463.pt",
"/kaggle/input/siim-covid19-yolov5m/yolov5m6-res512-fold2-mAP0.4496.pt",
"/kaggle/input/siim-covid19-yolov5m/yolov5m6-res512-fold3-mAP0.4121.pt",
"/kaggle/input/siim-covid19-yolov5m/yolov5m6-res512-fold4-mAP0.42406.pt",
]
models = [
torch.load(model_path, map_location=device)["model"].to(device).float().eval()
for model_path in model_paths
]
dataset = LoadImages("/kaggle/tmp/test/image", img_size=IMAGE_RES)
all_path = []
all_bboxes = []
all_score = []
for path, img, im0s, _ in dataset:
img = torch.from_numpy(img).to(device).float() / 255.0
if img.ndimension() == 3:
img = img.unsqueeze(0)
bboxes_2, score_2 = [], []
for model in models:
pred = model(img, augment=True)[0]
pred = non_max_suppression(pred, 0.001, 0.5, classes=None, agnostic=False)
bboxes, score = [], []
for i, det in enumerate(pred):
# gain = torch.tensor(im0.shape)[[1, 0, 1, 0]]
if det is not None and len(det):
det[:, :4] = scale_coords(
img.shape[2:], det[:, :4], im0s.shape
).round()
for c in det[:, -1].unique():
n = (det[:, -1] == c).sum()
for *xyxy, conf, _ in det:
bboxes.append(torch.tensor(xyxy).view(-1).numpy())
score.append(conf.cpu().numpy().item())
bboxes_2.append(bboxes)
score_2.append(score)
all_path.append(path)
all_score.append(score_2)
all_bboxes.append(bboxes_2)
del models
del dataset
gc.collect()
torch.cuda.empty_cache()
return all_path, all_score, all_bboxes
# ## Yolov5
def ensemble_pp(boxes, scores, iou_thres: float = 0.25, skip_box_thr: float = 0.01):
labels = [np.ones(len(scores[idx])) for idx in range(len(scores))]
nms_boxes, nms_scores, nms_labels = nms(
boxes, scores, labels, weights=None, iou_thr=iou_thres
)
snms_boxes, snms_scores, snms_labels = soft_nms(
boxes,
scores,
labels,
weights=None,
iou_thr=iou_thres,
sigma=0.1,
thresh=skip_box_thr,
)
nmw_boxes, nmw_scores, nmw_labels = non_maximum_weighted(
boxes,
scores,
labels,
weights=None,
iou_thr=iou_thres,
skip_box_thr=skip_box_thr,
)
wbf_boxes, wbf_scores, wbf_labels = weighted_boxes_fusion(
boxes,
scores,
labels,
weights=None,
iou_thr=iou_thres,
skip_box_thr=skip_box_thr,
)
del labels
boxes, scores, _ = weighted_boxes_fusion(
[nms_boxes, snms_boxes, nmw_boxes, wbf_boxes],
[nms_scores, snms_scores, nmw_scores, wbf_scores],
[nms_labels, snms_labels, nmw_labels, wbf_labels],
weights=[2, 3, 4, 5],
iou_thr=iou_thres,
skip_box_thr=skip_box_thr,
)
del nms_boxes, nms_scores, nms_labels
del snms_boxes, snms_scores, snms_labels
del nmw_boxes, nmw_scores, nmw_labels
del wbf_boxes, wbf_scores, wbf_labels
gc.collect()
return boxes, scores
with torch.no_grad():
yolov5_all_path, yolov5_all_score, yolov5_all_bboxes = yolov5_detect()
yolov5_preds = {}
for row in range(len(yolov5_all_path)):
image_id = yolov5_all_path[row].split("/")[-1].split(".")[0]
boxes = yolov5_all_bboxes[row]
scores = yolov5_all_score[row]
# normalized to [0, 1]
boxes = [[coord / (IMAGE_RES - 1) for coord in box] for box in boxes]
# solve_bbox_problems over the models
# for i in range(len(boxes)):
# boxes, scores = solve_bbox_problems(boxes[i], scores[i])
boxes, scores = ensemble_pp(boxes, scores, iou_thres=0.25, skip_box_thr=0.01)
# unnormalized to [0, IMAGE_RES]
boxes = [
np.asarray([int(coord * (IMAGE_RES - 1)) for coord in box]) for box in boxes
]
yolov5_preds[image_id] = [boxes, scores]
del image_id, boxes, scores
del yolov5_all_path
del yolov5_all_score
del yolov5_all_bboxes
gc.collect()
# ## Convert coordinates
image_ids = []
PredictionStrings = []
for image_id, v in yolov5_preds.items():
w, h = test_df.loc[test_df["id"] == image_id, ["dim1", "dim0"]].values[0]
boxes, scores = v
normalized_boxes = [xyxy2xywh(box[None, :]) / IMAGE_RES for box in boxes]
rescaled_boxes = [np.round(yolo2voc(h, w, x)[0]) for x in normalized_boxes]
string_boxes = [
f"1 {score} {int(box[0])} {int(box[1])} {int(box[2])} {int(box[3])}"
for score, box in zip(scores, rescaled_boxes)
]
image_ids.append(image_id)
PredictionStrings.append(" ".join(string_boxes))
pred_df = pd.DataFrame({"id": image_ids, "PredictionString": PredictionStrings})
test_df = test_df.drop(["PredictionString"], axis=1)
sub_df = pd.merge(test_df, pred_df, on="id", how="left").fillna("none 1 0 0 1 1")
sub_df = sub_df[["id", "PredictionString"]]
for i in range(sub_df.shape[0]):
prediction_string: str = sub_df.at[i, "PredictionString"]
if prediction_string == "none 1 0 0 1 1":
continue
sub_df_split = prediction_string.split()
sub_df_list = []
for j in range(len(sub_df_split) // 6):
sub_df_list.append("opacity")
sub_df_list.append(sub_df_split[6 * j + 1])
sub_df_list.append(sub_df_split[6 * j + 2])
sub_df_list.append(sub_df_split[6 * j + 3])
sub_df_list.append(sub_df_split[6 * j + 4])
sub_df_list.append(sub_df_split[6 * j + 5])
sub_df.at[i, "PredictionString"] = " ".join(sub_df_list)
# ## Post-Processing
def clean_predictions(
preds_v,
iou_th: float = 0.60,
mode: str = "p_det_weight",
consensus_level: int = 1,
n_models2ensemble: int = 1,
):
ret_preds_v = []
for pred_d in preds_v:
cls_v = pred_d["cls"]
bbox_key = "bbox"
bbox_v = pred_d[bbox_key]
if "p_det" in pred_d.keys():
ret_p_det = True
p_det_v = pred_d["p_det"]
else:
ret_p_det = False
p_det_v = np.ones(pred_d["cls"].shape)
model_id_v = np.zeros(pred_d["cls"].shape, dtype=np.int)
new_cls_v = []
new_bbox_v = []
new_p_det_v = []
new_rad_id_v = []
for i_c in np.unique(cls_v):
f_c = cls_v == i_c
n_c = f_c.sum()
if n_c == 1:
if consensus_level > 1 and i_c != -1:
continue
new_cls_v.append(i_c)
new_bbox_v.append(bbox_v[f_c][0])
new_p_det_v.append(p_det_v[f_c][0])
else:
f_cls_v = cls_v[f_c]
f_bbox_v = bbox_v[f_c]
f_p_det_v = p_det_v[f_c]
f_model_id_v = model_id_v[f_c]
to_join_idxs_v = []
for i in range(n_c):
idxs_s = set(
np.argwhere(calc_iou(f_bbox_v[i], f_bbox_v) > iou_th).T[0]
)
for i in range(len(to_join_idxs_v)):
if len(idxs_s.intersection(to_join_idxs_v[i])) > 0:
to_join_idxs_v[i] = to_join_idxs_v[i].union(idxs_s)
break
else:
to_join_idxs_v.append(idxs_s)
for to_join_idxs in to_join_idxs_v:
to_join_idxs = list(to_join_idxs)
if len(to_join_idxs) < consensus_level:
continue
bbox, p_det = merge_preds(
f_bbox_v[to_join_idxs],
f_p_det_v[to_join_idxs],
mode=mode,
)
if n_models2ensemble > 1:
ens_prop = (
len(np.unique(f_model_id_v[to_join_idxs]))
/ n_models2ensemble
)
p_det = p_det * ens_prop
new_cls_v.append(i_c)
new_bbox_v.append(bbox)
new_p_det_v.append(p_det)
if ret_rad_id:
new_rad_id_v.append(
np.concatenate(f_rad_id_v[to_join_idxs], axis=-1)
)
ret_preds_d = {
"cls": np.array(new_cls_v),
bbox_key: np.array(new_bbox_v),
}
if ret_p_det:
ret_preds_d["p_det"] = np.array(new_p_det_v)
for k in pred_d.keys():
if k not in ["cls", bbox_key, "p_det"]:
ret_preds_d[k] = pred_d[k]
ret_preds_v.append(ret_preds_d)
return ret_preds_v
def pred_to_str(pred_d):
bbox_v = pred_d["bbox"]
p_det_v = pred_d["p_det"]
return " ".join(
[
"opacity {:0.05} {} {} {} {}".format(p_det, *bbox)
for p_det, bbox in zip(p_det_v, np.round(bbox_v).astype(np.int))
]
)
# reference : https://www.kaggle.com/morizin/ensemble331-remake/notebook#Final-cleaning
preds_v = read_prediction_csv(sub_df)
fix_boxes(preds_v)
clean_pred_v = clean_predictions(preds_v, iou_th=0.25, mode="p_det_weight_psum")
norm_clean_pred_v = norm_p_det(clean_pred_v)
del preds_v
del clean_pred_v
gc.collect()
pred_summary_d = {"image_id": [], "PredictionString": []}
for pred_d in norm_clean_pred_v:
pred_str = pred_to_str(pred_d)
pred_summary_d["image_id"].append(pred_d["sample_id"])
pred_summary_d["PredictionString"].append(pred_str)
sub_df = pd.DataFrame(pred_summary_d)
low_threshold: float = 0.00
high_threshold: float = 1.00
sub_df["none"] = df_2class["none"]
c0, c1, c2 = 0, 0, 0
for i in range(sub_df.shape[0]):
if sub_df.at[i, "PredictionString"] != "none 1 0 0 1 1":
none_prob: float = sub_df.at[i, "none"]
sub_df.at[i, "PredictionString"] = (
sub_df.at[i, "PredictionString"] + f" none {none_prob} 0 0 1 1"
)
c1 += 1
# if none_prob < low_threshold:
# sub_df.at[i, 'PredictionString'] = sub_df.at[i, 'PredictionString']
# c0 += 1
# elif low_threshold <= none_prob and none_prob < high_threshold:
# sub_df.at[i, 'PredictionString'] = sub_df.at[i, 'PredictionString'] + f' none {none_prob} 0 0 1 1'
# c1 += 1
# else:
# sub_df.at[i, 'PredictionString'] = 'none 1 0 0 1 1'
# c2 += 1
print(f"[+] threshold | low {low_threshold:.3f} high {high_threshold:.3f}")
print(f"[+] total {c0 + c1 + c2} | keep {c0} add {c1} replace {c2}")
sub_df = sub_df[["image_id", "PredictionString"]]
sub_df = sub_df.rename(columns={"image_id": "id"})
# # Submission
df_study = df_study[:study_len]
df_study = df_study.append(sub_df).reset_index(drop=True)
df_study.to_csv("/kaggle/working/submission.csv", index=False)
df_study
shutil.rmtree("/kaggle/working/yolov5")
| false | 1 | 11,168 | 0 | 11,187 | 11,168 |
||
69595715
|
<jupyter_start><jupyter_text>Huggingface Roberta Variants
This Dataset contains various variants of Roberta from huggingface
List of Included Datasets:
* **`distilroberta-base`**
* **`roberta-base`**
* **`roberta-large`**
* **`xlm-roberta-large`**
* **`roberta-large-mnli`**
* **`tf-xlm-roberta-base`**
* **`pytorch-xlm-roberta-base`**
Kaggle dataset identifier: huggingface-roberta-variants
<jupyter_script># !pip install tensorflow_addons
# !pip install autokeras
import sys
sys.path.append("../input/autokeras")
import tensorflow as tf
import pandas as pd
import numpy as np
import glob
import tensorflow.keras.layers as layers
import tensorflow_addons as tfa
import random
from transformers import BertTokenizer, TFBertModel
from transformers import RobertaTokenizer, TFRobertaModel
from transformers import AlbertTokenizer, TFAlbertModel
# import keras_tuner as kt
import autokeras as ak
from tensorflow.keras.models import load_model
ds_path = "../input/commonlitreadabilityprize/"
# Turn on tpu
# Detect TPU, return appropriate distribution strategy
strategy = tf.distribute.get_strategy()
try:
tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
print("Running on TPU ", tpu.master())
except ValueError:
tpu = None
if tpu:
tf.config.experimental_connect_to_cluster(tpu)
tf.tpu.experimental.initialize_tpu_system(tpu)
strategy = tf.distribute.experimental.TPUStrategy(tpu)
else:
strategy = tf.distribute.get_strategy()
print("REPLICAS: ", strategy.num_replicas_in_sync)
train_df = pd.read_csv(ds_path + "train.csv")
test_df = pd.read_csv(ds_path + "test.csv")
print(train_df.head())
print(test_df.head())
train_row = 2834
def get_pooling_data(data, tokenizer, model):
inputs = tokenizer(
data, truncation=True, padding="max_length", max_length=350, return_tensors="tf"
)
x = model(inputs)
x = x.last_hidden_state
result = tf.keras.layers.GlobalAveragePooling1D()(x)
return result.numpy().flatten()
def prepare_data(df, training=True):
excerpt = df["excerpt"].to_list()
size = len(excerpt)
model_path_1 = "../input/huggingface-roberta-variants/roberta-large/roberta-large"
tokenizer_1 = RobertaTokenizer.from_pretrained(model_path_1)
model_1 = TFRobertaModel.from_pretrained(model_path_1)
model_path_2 = "../input/tfbert-base-uncased"
tokenizer_2 = BertTokenizer.from_pretrained(model_path_2)
model_2 = TFBertModel.from_pretrained(model_path_2)
model_path_3 = "../input/huggingface-roberta-variants/roberta-base/roberta-base"
tokenizer_3 = RobertaTokenizer.from_pretrained(model_path_3)
model_3 = TFRobertaModel.from_pretrained(model_path_3)
outputs = []
for index, data in enumerate(excerpt):
x_1 = get_pooling_data(data, tokenizer_1, model_1)
x_2 = get_pooling_data(data, tokenizer_2, model_2)
x_3 = get_pooling_data(data, tokenizer_3, model_3)
x = np.concatenate((x_1, x_2, x_3)).flatten() # shape 2834,768
if index % 200 == 0:
print(f"prepared data {index}")
print(x.shape)
outputs.append(x)
outputs = np.asarray(outputs)
if training:
target = np.asarray(df["target"].to_list())
return outputs, target
else:
return outputs
# x_train, y_train = prepare_data(train_df)
x_test = prepare_data(test_df, training=False)
# model_creator = ak.StructuredDataRegressor(max_trials=35, overwrite=True)
# Feed the structured data regressor with training data.
# model_creator.fit(x_train, y_train)
# model = model_creator.export_model()
# model.save("saved_model")
model = load_model(
"../input/commonlitautokeraspretrained/saved_model",
custom_objects=ak.CUSTOM_OBJECTS,
)
predict_data = model.predict(x_test)
print(predict_data)
test_df = test_df.assign(target=predict_data)
selected_column = ["id", "target"]
final_result = test_df[selected_column]
final_result.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/595/69595715.ipynb
|
huggingface-roberta-variants
|
sauravmaheshkar
|
[{"Id": 69595715, "ScriptId": 18917553, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6875167, "CreationDate": "08/02/2021 02:26:02", "VersionNumber": 28.0, "Title": "commonlitreadabilityprize-bert-autokeras", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 116.0, "LinesInsertedFromPrevious": 6.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 110.0, "LinesInsertedFromFork": 64.0, "LinesDeletedFromFork": 159.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 52.0, "TotalVotes": 0}]
|
[{"Id": 92980397, "KernelVersionId": 69595715, "SourceDatasetVersionId": 2236772}, {"Id": 92980398, "KernelVersionId": 69595715, "SourceDatasetVersionId": 2278892}, {"Id": 92980396, "KernelVersionId": 69595715, "SourceDatasetVersionId": 2206810}, {"Id": 92980395, "KernelVersionId": 69595715, "SourceDatasetVersionId": 893616}]
|
[{"Id": 2236772, "DatasetId": 1330744, "DatasourceVersionId": 2278596, "CreatorUserId": 4382914, "LicenseName": "CC0: Public Domain", "CreationDate": "05/16/2021 10:47:13", "VersionNumber": 9.0, "Title": "Huggingface Roberta Variants", "Slug": "huggingface-roberta-variants", "Subtitle": "Various Variants of Roberta from huggingface", "Description": "This Dataset contains various variants of Roberta from huggingface\n\nList of Included Datasets:\n\n* **`distilroberta-base`**\n* **`roberta-base`**\n* **`roberta-large`**\n* **`xlm-roberta-large`**\n* **`roberta-large-mnli`**\n* **`tf-xlm-roberta-base`**\n* **`pytorch-xlm-roberta-base`**", "VersionNotes": "Adds xlm-roberta-large", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1330744, "CreatorUserId": 4382914, "OwnerUserId": 4382914.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 4770448.0, "CurrentDatasourceVersionId": 4833766.0, "ForumId": 1349661, "Type": 2, "CreationDate": "05/10/2021 05:22:05", "LastActivityDate": "05/10/2021", "TotalViews": 5654, "TotalDownloads": 198, "TotalVotes": 26, "TotalKernels": 49}]
|
[{"Id": 4382914, "UserName": "sauravmaheshkar", "DisplayName": "Saurav Maheshkar \u2615\ufe0f", "RegisterDate": "01/25/2020", "PerformanceTier": 2}]
|
# !pip install tensorflow_addons
# !pip install autokeras
import sys
sys.path.append("../input/autokeras")
import tensorflow as tf
import pandas as pd
import numpy as np
import glob
import tensorflow.keras.layers as layers
import tensorflow_addons as tfa
import random
from transformers import BertTokenizer, TFBertModel
from transformers import RobertaTokenizer, TFRobertaModel
from transformers import AlbertTokenizer, TFAlbertModel
# import keras_tuner as kt
import autokeras as ak
from tensorflow.keras.models import load_model
ds_path = "../input/commonlitreadabilityprize/"
# Turn on tpu
# Detect TPU, return appropriate distribution strategy
strategy = tf.distribute.get_strategy()
try:
tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
print("Running on TPU ", tpu.master())
except ValueError:
tpu = None
if tpu:
tf.config.experimental_connect_to_cluster(tpu)
tf.tpu.experimental.initialize_tpu_system(tpu)
strategy = tf.distribute.experimental.TPUStrategy(tpu)
else:
strategy = tf.distribute.get_strategy()
print("REPLICAS: ", strategy.num_replicas_in_sync)
train_df = pd.read_csv(ds_path + "train.csv")
test_df = pd.read_csv(ds_path + "test.csv")
print(train_df.head())
print(test_df.head())
train_row = 2834
def get_pooling_data(data, tokenizer, model):
inputs = tokenizer(
data, truncation=True, padding="max_length", max_length=350, return_tensors="tf"
)
x = model(inputs)
x = x.last_hidden_state
result = tf.keras.layers.GlobalAveragePooling1D()(x)
return result.numpy().flatten()
def prepare_data(df, training=True):
excerpt = df["excerpt"].to_list()
size = len(excerpt)
model_path_1 = "../input/huggingface-roberta-variants/roberta-large/roberta-large"
tokenizer_1 = RobertaTokenizer.from_pretrained(model_path_1)
model_1 = TFRobertaModel.from_pretrained(model_path_1)
model_path_2 = "../input/tfbert-base-uncased"
tokenizer_2 = BertTokenizer.from_pretrained(model_path_2)
model_2 = TFBertModel.from_pretrained(model_path_2)
model_path_3 = "../input/huggingface-roberta-variants/roberta-base/roberta-base"
tokenizer_3 = RobertaTokenizer.from_pretrained(model_path_3)
model_3 = TFRobertaModel.from_pretrained(model_path_3)
outputs = []
for index, data in enumerate(excerpt):
x_1 = get_pooling_data(data, tokenizer_1, model_1)
x_2 = get_pooling_data(data, tokenizer_2, model_2)
x_3 = get_pooling_data(data, tokenizer_3, model_3)
x = np.concatenate((x_1, x_2, x_3)).flatten() # shape 2834,768
if index % 200 == 0:
print(f"prepared data {index}")
print(x.shape)
outputs.append(x)
outputs = np.asarray(outputs)
if training:
target = np.asarray(df["target"].to_list())
return outputs, target
else:
return outputs
# x_train, y_train = prepare_data(train_df)
x_test = prepare_data(test_df, training=False)
# model_creator = ak.StructuredDataRegressor(max_trials=35, overwrite=True)
# Feed the structured data regressor with training data.
# model_creator.fit(x_train, y_train)
# model = model_creator.export_model()
# model.save("saved_model")
model = load_model(
"../input/commonlitautokeraspretrained/saved_model",
custom_objects=ak.CUSTOM_OBJECTS,
)
predict_data = model.predict(x_test)
print(predict_data)
test_df = test_df.assign(target=predict_data)
selected_column = ["id", "target"]
final_result = test_df[selected_column]
final_result.to_csv("submission.csv", index=False)
| false | 0 | 1,110 | 0 | 1,238 | 1,110 |
||
69595032
|
<jupyter_start><jupyter_text>Human Metagenomics
### Microbiome
> The human gut contains trillions of microbial inhabitants, making it one of the most densely populated environments on the planet. The symbiosis between these organisms and the human host is extremely complex, and we are only beginning to understand the impact of the gut microbiota on human biology. Knowledge of the chemical reactions performed and compounds produced by gut microbes will provide new insights into their roles in influencing human health. By studying the gene content of the human gut microbiome and the enzymes encoded by these genes, we hope to better understand the chemical capabilities of this microbial community. However, the activities of the vast majority of enzymes found in microbiomes are unknown.
>Shotgun metagenomic sequencing is a relatively new sequencing approach that allows insight to be gained into community biodiversity and function.
The function of shotgun metagenomic sequencing is to sequence the genomes of untargeted cells in a community in order to elucidate community composition and function.
Research using the method, taps into several fields due to the broad existence of large microbial communities. For example, the study of soil microbiota has led to advances in understanding and treating plant pathogens.
In human gut microbiota, the use of shotgun metagenomics discovered how common antibiotic genes are in our gut bacteria.
By [Sara Ryding](https://www.news-medical.net/life-sciences/Shotgun-Metagenomic-Sequencing.aspx).
### Dataset
This dataset was created by the team of Edoardo Pasolli, Duy Tin Truong, Faizan Malik, Levi Waldron, and Nicola Segata; they published a [research article in July of 2016 ](https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004977), and created [MetAML](https://github.com/segatalab/metaml#metaml---metagenomic-prediction-analysis-based-on-machine-learning) - Metagenomic prediction Analysis based on Machine Learning.
The authors used 8 publicly available metagenomic datasets, and applied MetaPhlAn2 to generate species abundance features. Their goal was to classify diseases using obtained abundance features, and to determine best ML models for this task. Though their experiments they settled on RandomForest as the best classifier for most diseases, with SVM doing better for some diseases.
I transposed abundance data, to 'traditional' view rows-cases/columns-features as opposed to what MethPhlAn2 produced, and saved as csv files for simpler ingestion.
### New approaches
Can we get better predictions? Different models? Ensembling?
Can we determine which sets of species define better predictions, and therefore are related to specific diseases?
Kaggle dataset identifier: human-metagenomics
<jupyter_script># # Introduction
# In this notebook we explore metagenomics data. This dataset was created by the team of Edoardo Pasolli, Duy Tin Truong, Faizan Malik, Levi Waldron, and Nicola Segata; they published [a research article in July of 2016](https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004977). The authors used 8 publicly available metagenomic datasets, and applied [MetaPhlAn2](https://github.com/segatalab/metaml#metaml---metagenomic-prediction-analysis-based-on-machine-learning) to generate species abundance features.
# ## Logistics behind the Input Data
# This notebook was created to further explore the meta-genomics data on kaggle. The link to the data-set is: https://www.kaggle.com/antaresnyc/metagenomics. The datasets include:
# * abundance.txt: a table containing the abundances of each organism type
# * the first 210 features include meta-data about the samples
# * the rest of the features include the abundance data in float-type
# * marker_presence.txt: a table containing the presence of strain-specific markers.
# * the first 210 features include meta-data about the samples (same as abundance.txt)
# * In a previous notebook I converted the marker presence feature data into a sparse matrix for easier downloading. This sparse matrix is found on [kaggle](https://www.kaggle.com/sklasfeld/metagenomics-marker-presence-sparse-matrix).
# * markers2clades_DB.txt: a lookup table to associate each marker identifier to the corresponding species.
# In summary we have 210 samples. We know the abundance of the organisms in the sample. If an organism is in a sample we have strain-specific marker information.
# ## Libraries
# Below I import some librarys that may be useful and then print the input files
import numpy as np # linear algebra
import os # accessing directory structure
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import scipy
import scipy.sparse
import networkx as nx
from sklearn import preprocessing
# plot with matplotlib
import matplotlib.pyplot as plt
# from plotnine import * # used to plot data
# progress bar
from tqdm import tqdm
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
marker_presence_matrix_file = "/kaggle/input/metagenomics-marker-presence-sparse-matrix/marker_presence_matrix.npz"
markers2clades_DB_file = "/kaggle/input/human-metagenomics/markers2clades_DB.csv"
abundance_file = "/kaggle/input/human-metagenomics/abundance.csv"
marker_presence_table_file = "/kaggle/input/human-metagenomics/marker_presence.csv"
# # Cleaning the Data
# The marker matrix is dependent on the abundance table in that strain-specific markers can only appear if a specific strain is abundant. Both tables can be merged together using a join-function on the 210 sample meta-data columns. However these columns are very messy. Therefore let's clean them before we move on to understanding the rest of the data.
# ## Testing the meta data
# The meta data information is given in both the marker_presence and abundance tables. I just wanted to make sure they contain the same information.
samples_df = pd.read_csv(abundance_file, sep=",", dtype=object, usecols=range(0, 210))
if 1 == 0:
samples_df2 = pd.read_csv(
marker_presence_table_file, sep=",", dtype=object, usecols=range(0, 210)
)
if 1 == 0:
samples_df.compare(samples_df2, align_axis=0)
# It looks like they are basically the same so I can move forward using `samples_df`
samples_df.describe()
samples_df.query('dataset_name in ["t2dmeta_long","t2dmeta_short"]')["disease"].unique()
# ## Cleaning meta features
# remove all column with only one value
samples_df = samples_df.loc[:, samples_df.nunique() > 1].copy()
# Next I look at categorical columns (AKA any feature that has 20 possible values or less)
if 1 == 0:
for col in samples_df.loc[:, samples_df.nunique() < 20]:
print("%s:%i" % (col, samples_df[col].nunique()))
print(samples_df[col].unique())
print("")
# It looks like `nd`, `na`, `unknown` and `-` all stands for no data. Therefore let's replace these values all with np.NaN
samples_df = samples_df.replace("nd", np.NaN)
samples_df = samples_df.replace("na", np.NaN)
samples_df = samples_df.replace("-", np.NaN)
samples_df = samples_df.replace(" -", np.NaN)
samples_df = samples_df.replace("unknown", np.NaN)
# We can remove all columns that have only 1 values and NaN. These do not seem to be too informative anyway.
# change the if statement to visualize
if 1 == 0:
for col in samples_df.loc[:, samples_df.nunique() == 1].columns:
samples_df[col].fillna("NaN").value_counts().sort_values().plot(
kind="bar", title=col
)
plt.show()
samples_df = samples_df.loc[:, samples_df.nunique() > 1].copy()
# I want to convert some columns into booleans. For example if the values are either:
# * "yes","no", or null
# * "y","n", or null
# * "positve", "negative", or null
# * "a"(affected), "u" (unaffected), or null
# I want to convert them into `2`, `1`, and `0` respectively.
bool_vals = {"True": 2, "False": 1, "Null": 0}
for col in samples_df.loc[:, samples_df.nunique() < 4]:
if "yes" in samples_df[col].unique() and "no" in samples_df[col].unique():
samples_df[col] = samples_df[col].fillna(bool_vals["Null"])
samples_df = samples_df.replace(
{col: {"yes": bool_vals["True"], "no": bool_vals["False"]}}
)
elif "y" in samples_df[col].unique() and "n" in samples_df[col].unique():
samples_df[col] = samples_df[col].fillna(bool_vals["Null"])
samples_df = samples_df.replace(
{col: {"y": bool_vals["True"], "n": bool_vals["False"]}}
)
elif (
"positive" in samples_df[col].unique()
and "negative" in samples_df[col].unique()
):
samples_df[col] = samples_df[col].fillna(bool_vals["Null"])
samples_df = samples_df.replace(
{col: {"positive": bool_vals["True"], "negative": bool_vals["False"]}}
)
elif "a" in samples_df[col].unique() and "u" in samples_df[col].unique():
samples_df[col] = samples_df[col].fillna(bool_vals["Null"])
samples_df = samples_df.replace(
{col: {"a": bool_vals["True"], "u": bool_vals["False"]}}
)
# Similarly, for columns that contain 2 values (not including null) I will convert the values to numbers. For example, I will change the column named "gender" to "gender:Female|Male". The values will be 1 for Female, 2 for Male, and 0 for null.
for col in samples_df.loc[:, samples_df.nunique() == 2].columns:
if not (True in samples_df[col].unique() and False in samples_df[col].unique()):
val_i = 0
first_val_null = True
first_val = np.NaN
while first_val_null:
first_val = samples_df[col].unique()[val_i]
if first_val == first_val:
first_val_null = False
else:
val_i += 1
val_i += 1
second_val_null = True
second_val = np.NaN
while second_val_null:
second_val = samples_df[col].unique()[val_i]
if second_val == second_val:
second_val_null = False
else:
val_i += 1
new_col_name = "%s:%s|%s" % (col, first_val, second_val)
# change the column name
samples_df = samples_df.rename(columns={col: new_col_name})
# change values in the column
samples_df[new_col_name] = samples_df[new_col_name].fillna(bool_vals["Null"])
samples_df = samples_df.replace(
{
new_col_name: {
first_val: bool_vals["False"],
second_val: bool_vals["True"],
}
}
)
categorical_cols = samples_df.loc[:, samples_df.nunique() < 20].columns
# It was brought to my attention that most samples come from stool. Therefore it makes sense that we remove other types of samples.
samples_df["bodysite"].value_counts().plot(kind="bar")
samples_df["bodysite"] == "stool"
print(np.sum(samples_df.nunique() < 3))
# Unfortonately this didn't help remove any features from the meta data.
stool_samp_df = samples_df.loc[samples_df["bodysite"] == "stool", :].copy()
# ## Cleaning abundance file
# import abundance file without the first 211 columns (since we already dealt with those above)
abundance_df = pd.read_csv(abundance_file, sep=",", dtype=object).iloc[:, 211:]
abundance_df.head()
# I am wondering if we can remove any columns that are redundant. In other words, I would like to remove columns that have identical values.
# In the following for-loop I simply check columns that are consecutive of one another to see if they are identical. I then use the biggest category as the key in `redundant_dict` and all the sub-categories that are equal in the list values.
# seen_list=[]
redundant_dict = {}
remove_cols = []
i = 0
while i < abundance_df.shape[1]:
j = i + 1
next_step = abundance_df.shape[1]
while j < abundance_df.shape[1]:
# print("%i,%i" % (i,j))
col_i = abundance_df.columns[i]
col_j = abundance_df.columns[j]
if col_i in col_j:
# print(abundance_df.iloc[:,i].equals(abundance_df.iloc[:,j]))
if abundance_df.iloc[:, i].equals(abundance_df.iloc[:, j]):
# add redundant column name to data-structure
remove_cols.append(col_j)
if col_i in redundant_dict:
redundant_dict[col_i].append(col_j)
else:
redundant_dict[col_i] = [col_j]
# next look at i vs j+1
else:
# print("next_step: "+ str(next_step))
if next_step > j:
next_step = j
j += 1
if j == abundance_df.shape[1]:
i = j
else:
if next_step < j:
i = next_step
next_step = abundance_df.shape[1]
else:
i = j
j = abundance_df.shape[1]
# drop redundant columns
abundance_df = abundance_df.drop(remove_cols, axis=1)
print(len(remove_cols))
# 1,441 columns were dropped from further analysis since they were redundant with parent columns.
# Note: to get the full abudance file back with cleaning using the following code:
if 1 == 0:
c = samples_df.merge(abundance_df, how="left", left_index=True, right_index=True)
samples_df["dataset_name"]
# ## Cleaning Marker Presence file
# Import this file without the first 211 columns (since we already dealt with those previously). This file could be imported as a sparse numpy matrix. it is very large.
markers_reader = pd.read_csv(
marker_presence_table_file,
sep=",",
dtype=object,
usecols=range(211, 288558),
nrows=10,
)
markers_reader
# ## Construct a graph for genomic part
# In order to capture a tree structure of the genes, we construct a directed graph where an node represents each bacteria and edge represents parent-child relationship. To quantify the presence of each bacteria, I set up a vector as a node property.
def graph_label(samples_df, abundance_df, dataset=None):
if dataset:
dataset = dataset if isinstance(dataset, list) else [dataset]
ids = samples_df["dataset_name"].isin(dataset)
samples_df = samples_df[ids].reset_index(drop=False)
abundance_df = abundance_df[ids].reset_index(drop=False)
le = preprocessing.LabelEncoder()
# target values
y = le.fit_transform(samples_df["disease"])
# get emnedding of all nodes
le_nodes = preprocessing.LabelEncoder()
# encode labels between 0 and n_classes-1 for each bacterial label
le_nodes.fit([col.split("|")[-1] for col in abundance_df.columns])
data_list = []
for i in range(len(abundance_df)):
node_list = [] # list of [$cur_bacteria_name,$abundance_val]
edge_list = [] # list of [$parent_bacteria_name,$cur_bacteria_name]
for key, val in abundance_df.iloc[i].to_dict().items():
if float(val) > 0:
bacteria_list = key.split("|")
node = [le_nodes.transform([bacteria_list[-1]])[0], float(val)]
node_list.append(node)
if len(bacteria_list) >= 2:
edge_list.append(le_nodes.transform(bacteria_list[-2:]))
# convert `y`, `node_list`, and `edge_list` into Tensor formats
edge_array = np.array(edge_list)
edge_index = torch.tensor(
[edge_array[:, 0], edge_array[:, 1]], dtype=torch.long
)
# print(np.array(node_list))
node_features = torch.LongTensor(np.array(node_list))
label = torch.FloatTensor([y[i]])
# set these Tensors into a pytorch Data() object
# which is used to model graphs
data = Data(node_features, edge_index=edge_index, y=label)
data_list.append(data)
return data_list
import torch
print(torch.__version__)
print(torch.version.cuda)
from torch_geometric.utils.convert import from_networkx
from torch_geometric.data import InMemoryDataset
"""
class AbundanceDataset(InMemoryDataset):
def __init__(self, root, transform=None, pre_transform=None,dataset = None):
super(AbundanceDataset, self).__init__(root, transform, pre_transform)
self.dataset = dataset
self.data, self.slices = torch.load(self.processed_paths[0])
@property
def raw_file_names(self):
return []
@property
def processed_file_names(self):
return ['../input/yoochoose_click_binary_1M_sess.dataset']
def download(self):
pass
def process(self):
data_list = []
graph_label_pair = graph_label()
for G,value in zip(graph_label_pair['graph'],graph_label_pair['value']):
data = from_networkx(G)
data.y = torch.float
data_list.append(data)
data, slices = self.collate(data_list)
torch.save((data, slices), self.processed_paths[0])
"""
from torch_geometric.data import DataLoader
from torch_geometric.nn import GINConv, global_add_pool
"""
data_list = []
graph_label_pair = graph_label(samples_df,abundance_df,dataset = ["t2dmeta_long","t2dmeta_short"])
for G,value in zip(graph_label_pair['graph'],graph_label_pair['value']):
print(G)
data = from_networkx(G)
data.y = torch.float(value)
data_list.append(data)
"""
t2dml_samples_df = samples_df.loc[
samples_df["dataset_name"] == "t2dmeta_long", :
].copy()
t2dml_abundance_values_df = abundance_df.iloc[(list(t2dml_samples_df.index))]
# merge meta-data features with abundance features
t2dml_abundance_df = t2dml_samples_df.merge(
abundance_df, how="inner", left_index=True, right_index=True
)
data_list = graph_label(t2dml_samples_df, t2dml_abundance_values_df)
train_datalist = data_list[len(dataset) // 10 :]
test_datalist = data_list[: len(dataset) // 10]
train_loader = DataLoader(train_datalist, batch_size=32, shuffle=True)
test_loader = DataLoader(test_datalist, batch_size=4)
class Net(torch.nn.Module):
def __init__(self, in_channels, dim, out_channels):
super(Net, self).__init__()
self.conv1 = GINConv(
Sequential(
Linear(in_channels, dim),
BatchNorm1d(dim),
ReLU(),
Linear(dim, dim),
ReLU(),
)
)
self.conv2 = GINConv(
Sequential(
Linear(dim, dim), BatchNorm1d(dim), ReLU(), Linear(dim, dim), ReLU()
)
)
self.conv3 = GINConv(
Sequential(
Linear(dim, dim), BatchNorm1d(dim), ReLU(), Linear(dim, dim), ReLU()
)
)
self.conv4 = GINConv(
Sequential(
Linear(dim, dim), BatchNorm1d(dim), ReLU(), Linear(dim, dim), ReLU()
)
)
self.conv5 = GINConv(
Sequential(
Linear(dim, dim), BatchNorm1d(dim), ReLU(), Linear(dim, dim), ReLU()
)
)
self.lin1 = Linear(dim, dim)
self.lin2 = Linear(dim, out_channels)
def forward(self, x, edge_index, batch):
x = self.conv1(x, edge_index)
x = self.conv2(x, edge_index)
x = self.conv3(x, edge_index)
x = self.conv4(x, edge_index)
x = self.conv5(x, edge_index)
x = global_add_pool(x, batch)
x = self.lin1(x).relu()
x = F.dropout(x, p=0.5, training=self.training)
x = self.lin2(x)
return F.log_softmax(x, dim=-1)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model = Net(dataset.num_features, 32, dataset.num_classes).to(device)
optimizer = torch.optim.Adam(model.parameters(), lr=0.01)
def train():
model.train()
total_loss = 0
for data in train_loader:
data = data.to(device)
optimizer.zero_grad()
output = model(data.x, data.edge_index, data.batch)
loss = F.nll_loss(output, data.y)
loss.backward()
optimizer.step()
total_loss += float(loss) * data.num_graphs
return total_loss / len(train_loader.dataset)
@torch.no_grad()
def test(loader):
model.eval()
total_correct = 0
for data in loader:
data = data.to(device)
out = model(data.x, data.edge_index, data.batch)
total_correct += int((out.argmax(-1) == data.y).sum())
return total_correct / len(loader.dataset)
for epoch in range(1, 101):
loss = train()
train_acc = test(train_loader)
test_acc = test(test_loader)
print(
f"Epoch: {epoch:03d}, Loss: {loss:.4f}, Train Acc: {train_acc:.4f} "
f"Test Acc: {test_acc:.4f}"
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/595/69595032.ipynb
|
human-metagenomics
|
antaresnyc
|
[{"Id": 69595032, "ScriptId": 16795565, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1931719, "CreationDate": "08/02/2021 02:10:48", "VersionNumber": 35.0, "Title": "Metagenomics first model - KH", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 447.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 446.0, "LinesInsertedFromFork": 186.0, "LinesDeletedFromFork": 2.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 261.0, "TotalVotes": 0}]
|
[{"Id": 92978859, "KernelVersionId": 69595032, "SourceDatasetVersionId": 1753083}]
|
[{"Id": 1753083, "DatasetId": 1041487, "DatasourceVersionId": 1790253, "CreatorUserId": 2490236, "LicenseName": "CC0: Public Domain", "CreationDate": "12/16/2020 22:21:55", "VersionNumber": 3.0, "Title": "Human Metagenomics", "Slug": "human-metagenomics", "Subtitle": "Data and metadata for 3000+ public available metagenomes", "Description": "### Microbiome\n\n> The human gut contains trillions of microbial inhabitants, making it one of the most densely populated environments on the planet. The symbiosis between these organisms and the human host is extremely complex, and we are only beginning to understand the impact of the gut microbiota on human biology. Knowledge of the chemical reactions performed and compounds produced by gut microbes will provide new insights into their roles in influencing human health. By studying the gene content of the human gut microbiome and the enzymes encoded by these genes, we hope to better understand the chemical capabilities of this microbial community. However, the activities of the vast majority of enzymes found in microbiomes are unknown.\n\n>Shotgun metagenomic sequencing is a relatively new sequencing approach that allows insight to be gained into community biodiversity and function.\nThe function of shotgun metagenomic sequencing is to sequence the genomes of untargeted cells in a community in order to elucidate community composition and function.\nResearch using the method, taps into several fields due to the broad existence of large microbial communities. For example, the study of soil microbiota has led to advances in understanding and treating plant pathogens.\nIn human gut microbiota, the use of shotgun metagenomics discovered how common antibiotic genes are in our gut bacteria. \nBy [Sara Ryding](https://www.news-medical.net/life-sciences/Shotgun-Metagenomic-Sequencing.aspx).\n\n### Dataset\n\nThis dataset was created by the team of Edoardo Pasolli, Duy Tin Truong, Faizan Malik, Levi Waldron, and Nicola Segata; they published a [research article in July of 2016 ](https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004977), and created [MetAML](https://github.com/segatalab/metaml#metaml---metagenomic-prediction-analysis-based-on-machine-learning) - Metagenomic prediction Analysis based on Machine Learning.\n\nThe authors used 8 publicly available metagenomic datasets, and applied MetaPhlAn2 to generate species abundance features. Their goal was to classify diseases using obtained abundance features, and to determine best ML models for this task. Though their experiments they settled on RandomForest as the best classifier for most diseases, with SVM doing better for some diseases.\n\nI transposed abundance data, to 'traditional' view rows-cases/columns-features as opposed to what MethPhlAn2 produced, and saved as csv files for simpler ingestion.\n\n### New approaches\nCan we get better predictions? Different models? Ensembling? \nCan we determine which sets of species define better predictions, and therefore are related to specific diseases?\n\n### Acknowledgements\n\nPasolli E, Truong DT, Malik F, Waldron L, Segata N (2016) Machine Learning Meta-analysis of Large Metagenomic Datasets: Tools and Biological Insights. PLoS Comput Biol 12(7): e1004977.\n[research article ](doi:10.1371/journal.pcbi.1004977)\n\n>Banner image by Sara L\u00f3pez Gilabert/SAPIENS", "VersionNotes": "Uploaded marker presence data", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1041487, "CreatorUserId": 2490236, "OwnerUserId": 2490236.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1753083.0, "CurrentDatasourceVersionId": 1790253.0, "ForumId": 1058441, "Type": 2, "CreationDate": "12/16/2020 17:17:16", "LastActivityDate": "12/16/2020", "TotalViews": 12218, "TotalDownloads": 715, "TotalVotes": 27, "TotalKernels": 7}]
|
[{"Id": 2490236, "UserName": "antaresnyc", "DisplayName": "Alexey Kotlik", "RegisterDate": "11/13/2018", "PerformanceTier": 2}]
|
# # Introduction
# In this notebook we explore metagenomics data. This dataset was created by the team of Edoardo Pasolli, Duy Tin Truong, Faizan Malik, Levi Waldron, and Nicola Segata; they published [a research article in July of 2016](https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004977). The authors used 8 publicly available metagenomic datasets, and applied [MetaPhlAn2](https://github.com/segatalab/metaml#metaml---metagenomic-prediction-analysis-based-on-machine-learning) to generate species abundance features.
# ## Logistics behind the Input Data
# This notebook was created to further explore the meta-genomics data on kaggle. The link to the data-set is: https://www.kaggle.com/antaresnyc/metagenomics. The datasets include:
# * abundance.txt: a table containing the abundances of each organism type
# * the first 210 features include meta-data about the samples
# * the rest of the features include the abundance data in float-type
# * marker_presence.txt: a table containing the presence of strain-specific markers.
# * the first 210 features include meta-data about the samples (same as abundance.txt)
# * In a previous notebook I converted the marker presence feature data into a sparse matrix for easier downloading. This sparse matrix is found on [kaggle](https://www.kaggle.com/sklasfeld/metagenomics-marker-presence-sparse-matrix).
# * markers2clades_DB.txt: a lookup table to associate each marker identifier to the corresponding species.
# In summary we have 210 samples. We know the abundance of the organisms in the sample. If an organism is in a sample we have strain-specific marker information.
# ## Libraries
# Below I import some librarys that may be useful and then print the input files
import numpy as np # linear algebra
import os # accessing directory structure
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import scipy
import scipy.sparse
import networkx as nx
from sklearn import preprocessing
# plot with matplotlib
import matplotlib.pyplot as plt
# from plotnine import * # used to plot data
# progress bar
from tqdm import tqdm
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
marker_presence_matrix_file = "/kaggle/input/metagenomics-marker-presence-sparse-matrix/marker_presence_matrix.npz"
markers2clades_DB_file = "/kaggle/input/human-metagenomics/markers2clades_DB.csv"
abundance_file = "/kaggle/input/human-metagenomics/abundance.csv"
marker_presence_table_file = "/kaggle/input/human-metagenomics/marker_presence.csv"
# # Cleaning the Data
# The marker matrix is dependent on the abundance table in that strain-specific markers can only appear if a specific strain is abundant. Both tables can be merged together using a join-function on the 210 sample meta-data columns. However these columns are very messy. Therefore let's clean them before we move on to understanding the rest of the data.
# ## Testing the meta data
# The meta data information is given in both the marker_presence and abundance tables. I just wanted to make sure they contain the same information.
samples_df = pd.read_csv(abundance_file, sep=",", dtype=object, usecols=range(0, 210))
if 1 == 0:
samples_df2 = pd.read_csv(
marker_presence_table_file, sep=",", dtype=object, usecols=range(0, 210)
)
if 1 == 0:
samples_df.compare(samples_df2, align_axis=0)
# It looks like they are basically the same so I can move forward using `samples_df`
samples_df.describe()
samples_df.query('dataset_name in ["t2dmeta_long","t2dmeta_short"]')["disease"].unique()
# ## Cleaning meta features
# remove all column with only one value
samples_df = samples_df.loc[:, samples_df.nunique() > 1].copy()
# Next I look at categorical columns (AKA any feature that has 20 possible values or less)
if 1 == 0:
for col in samples_df.loc[:, samples_df.nunique() < 20]:
print("%s:%i" % (col, samples_df[col].nunique()))
print(samples_df[col].unique())
print("")
# It looks like `nd`, `na`, `unknown` and `-` all stands for no data. Therefore let's replace these values all with np.NaN
samples_df = samples_df.replace("nd", np.NaN)
samples_df = samples_df.replace("na", np.NaN)
samples_df = samples_df.replace("-", np.NaN)
samples_df = samples_df.replace(" -", np.NaN)
samples_df = samples_df.replace("unknown", np.NaN)
# We can remove all columns that have only 1 values and NaN. These do not seem to be too informative anyway.
# change the if statement to visualize
if 1 == 0:
for col in samples_df.loc[:, samples_df.nunique() == 1].columns:
samples_df[col].fillna("NaN").value_counts().sort_values().plot(
kind="bar", title=col
)
plt.show()
samples_df = samples_df.loc[:, samples_df.nunique() > 1].copy()
# I want to convert some columns into booleans. For example if the values are either:
# * "yes","no", or null
# * "y","n", or null
# * "positve", "negative", or null
# * "a"(affected), "u" (unaffected), or null
# I want to convert them into `2`, `1`, and `0` respectively.
bool_vals = {"True": 2, "False": 1, "Null": 0}
for col in samples_df.loc[:, samples_df.nunique() < 4]:
if "yes" in samples_df[col].unique() and "no" in samples_df[col].unique():
samples_df[col] = samples_df[col].fillna(bool_vals["Null"])
samples_df = samples_df.replace(
{col: {"yes": bool_vals["True"], "no": bool_vals["False"]}}
)
elif "y" in samples_df[col].unique() and "n" in samples_df[col].unique():
samples_df[col] = samples_df[col].fillna(bool_vals["Null"])
samples_df = samples_df.replace(
{col: {"y": bool_vals["True"], "n": bool_vals["False"]}}
)
elif (
"positive" in samples_df[col].unique()
and "negative" in samples_df[col].unique()
):
samples_df[col] = samples_df[col].fillna(bool_vals["Null"])
samples_df = samples_df.replace(
{col: {"positive": bool_vals["True"], "negative": bool_vals["False"]}}
)
elif "a" in samples_df[col].unique() and "u" in samples_df[col].unique():
samples_df[col] = samples_df[col].fillna(bool_vals["Null"])
samples_df = samples_df.replace(
{col: {"a": bool_vals["True"], "u": bool_vals["False"]}}
)
# Similarly, for columns that contain 2 values (not including null) I will convert the values to numbers. For example, I will change the column named "gender" to "gender:Female|Male". The values will be 1 for Female, 2 for Male, and 0 for null.
for col in samples_df.loc[:, samples_df.nunique() == 2].columns:
if not (True in samples_df[col].unique() and False in samples_df[col].unique()):
val_i = 0
first_val_null = True
first_val = np.NaN
while first_val_null:
first_val = samples_df[col].unique()[val_i]
if first_val == first_val:
first_val_null = False
else:
val_i += 1
val_i += 1
second_val_null = True
second_val = np.NaN
while second_val_null:
second_val = samples_df[col].unique()[val_i]
if second_val == second_val:
second_val_null = False
else:
val_i += 1
new_col_name = "%s:%s|%s" % (col, first_val, second_val)
# change the column name
samples_df = samples_df.rename(columns={col: new_col_name})
# change values in the column
samples_df[new_col_name] = samples_df[new_col_name].fillna(bool_vals["Null"])
samples_df = samples_df.replace(
{
new_col_name: {
first_val: bool_vals["False"],
second_val: bool_vals["True"],
}
}
)
categorical_cols = samples_df.loc[:, samples_df.nunique() < 20].columns
# It was brought to my attention that most samples come from stool. Therefore it makes sense that we remove other types of samples.
samples_df["bodysite"].value_counts().plot(kind="bar")
samples_df["bodysite"] == "stool"
print(np.sum(samples_df.nunique() < 3))
# Unfortonately this didn't help remove any features from the meta data.
stool_samp_df = samples_df.loc[samples_df["bodysite"] == "stool", :].copy()
# ## Cleaning abundance file
# import abundance file without the first 211 columns (since we already dealt with those above)
abundance_df = pd.read_csv(abundance_file, sep=",", dtype=object).iloc[:, 211:]
abundance_df.head()
# I am wondering if we can remove any columns that are redundant. In other words, I would like to remove columns that have identical values.
# In the following for-loop I simply check columns that are consecutive of one another to see if they are identical. I then use the biggest category as the key in `redundant_dict` and all the sub-categories that are equal in the list values.
# seen_list=[]
redundant_dict = {}
remove_cols = []
i = 0
while i < abundance_df.shape[1]:
j = i + 1
next_step = abundance_df.shape[1]
while j < abundance_df.shape[1]:
# print("%i,%i" % (i,j))
col_i = abundance_df.columns[i]
col_j = abundance_df.columns[j]
if col_i in col_j:
# print(abundance_df.iloc[:,i].equals(abundance_df.iloc[:,j]))
if abundance_df.iloc[:, i].equals(abundance_df.iloc[:, j]):
# add redundant column name to data-structure
remove_cols.append(col_j)
if col_i in redundant_dict:
redundant_dict[col_i].append(col_j)
else:
redundant_dict[col_i] = [col_j]
# next look at i vs j+1
else:
# print("next_step: "+ str(next_step))
if next_step > j:
next_step = j
j += 1
if j == abundance_df.shape[1]:
i = j
else:
if next_step < j:
i = next_step
next_step = abundance_df.shape[1]
else:
i = j
j = abundance_df.shape[1]
# drop redundant columns
abundance_df = abundance_df.drop(remove_cols, axis=1)
print(len(remove_cols))
# 1,441 columns were dropped from further analysis since they were redundant with parent columns.
# Note: to get the full abudance file back with cleaning using the following code:
if 1 == 0:
c = samples_df.merge(abundance_df, how="left", left_index=True, right_index=True)
samples_df["dataset_name"]
# ## Cleaning Marker Presence file
# Import this file without the first 211 columns (since we already dealt with those previously). This file could be imported as a sparse numpy matrix. it is very large.
markers_reader = pd.read_csv(
marker_presence_table_file,
sep=",",
dtype=object,
usecols=range(211, 288558),
nrows=10,
)
markers_reader
# ## Construct a graph for genomic part
# In order to capture a tree structure of the genes, we construct a directed graph where an node represents each bacteria and edge represents parent-child relationship. To quantify the presence of each bacteria, I set up a vector as a node property.
def graph_label(samples_df, abundance_df, dataset=None):
if dataset:
dataset = dataset if isinstance(dataset, list) else [dataset]
ids = samples_df["dataset_name"].isin(dataset)
samples_df = samples_df[ids].reset_index(drop=False)
abundance_df = abundance_df[ids].reset_index(drop=False)
le = preprocessing.LabelEncoder()
# target values
y = le.fit_transform(samples_df["disease"])
# get emnedding of all nodes
le_nodes = preprocessing.LabelEncoder()
# encode labels between 0 and n_classes-1 for each bacterial label
le_nodes.fit([col.split("|")[-1] for col in abundance_df.columns])
data_list = []
for i in range(len(abundance_df)):
node_list = [] # list of [$cur_bacteria_name,$abundance_val]
edge_list = [] # list of [$parent_bacteria_name,$cur_bacteria_name]
for key, val in abundance_df.iloc[i].to_dict().items():
if float(val) > 0:
bacteria_list = key.split("|")
node = [le_nodes.transform([bacteria_list[-1]])[0], float(val)]
node_list.append(node)
if len(bacteria_list) >= 2:
edge_list.append(le_nodes.transform(bacteria_list[-2:]))
# convert `y`, `node_list`, and `edge_list` into Tensor formats
edge_array = np.array(edge_list)
edge_index = torch.tensor(
[edge_array[:, 0], edge_array[:, 1]], dtype=torch.long
)
# print(np.array(node_list))
node_features = torch.LongTensor(np.array(node_list))
label = torch.FloatTensor([y[i]])
# set these Tensors into a pytorch Data() object
# which is used to model graphs
data = Data(node_features, edge_index=edge_index, y=label)
data_list.append(data)
return data_list
import torch
print(torch.__version__)
print(torch.version.cuda)
from torch_geometric.utils.convert import from_networkx
from torch_geometric.data import InMemoryDataset
"""
class AbundanceDataset(InMemoryDataset):
def __init__(self, root, transform=None, pre_transform=None,dataset = None):
super(AbundanceDataset, self).__init__(root, transform, pre_transform)
self.dataset = dataset
self.data, self.slices = torch.load(self.processed_paths[0])
@property
def raw_file_names(self):
return []
@property
def processed_file_names(self):
return ['../input/yoochoose_click_binary_1M_sess.dataset']
def download(self):
pass
def process(self):
data_list = []
graph_label_pair = graph_label()
for G,value in zip(graph_label_pair['graph'],graph_label_pair['value']):
data = from_networkx(G)
data.y = torch.float
data_list.append(data)
data, slices = self.collate(data_list)
torch.save((data, slices), self.processed_paths[0])
"""
from torch_geometric.data import DataLoader
from torch_geometric.nn import GINConv, global_add_pool
"""
data_list = []
graph_label_pair = graph_label(samples_df,abundance_df,dataset = ["t2dmeta_long","t2dmeta_short"])
for G,value in zip(graph_label_pair['graph'],graph_label_pair['value']):
print(G)
data = from_networkx(G)
data.y = torch.float(value)
data_list.append(data)
"""
t2dml_samples_df = samples_df.loc[
samples_df["dataset_name"] == "t2dmeta_long", :
].copy()
t2dml_abundance_values_df = abundance_df.iloc[(list(t2dml_samples_df.index))]
# merge meta-data features with abundance features
t2dml_abundance_df = t2dml_samples_df.merge(
abundance_df, how="inner", left_index=True, right_index=True
)
data_list = graph_label(t2dml_samples_df, t2dml_abundance_values_df)
train_datalist = data_list[len(dataset) // 10 :]
test_datalist = data_list[: len(dataset) // 10]
train_loader = DataLoader(train_datalist, batch_size=32, shuffle=True)
test_loader = DataLoader(test_datalist, batch_size=4)
class Net(torch.nn.Module):
def __init__(self, in_channels, dim, out_channels):
super(Net, self).__init__()
self.conv1 = GINConv(
Sequential(
Linear(in_channels, dim),
BatchNorm1d(dim),
ReLU(),
Linear(dim, dim),
ReLU(),
)
)
self.conv2 = GINConv(
Sequential(
Linear(dim, dim), BatchNorm1d(dim), ReLU(), Linear(dim, dim), ReLU()
)
)
self.conv3 = GINConv(
Sequential(
Linear(dim, dim), BatchNorm1d(dim), ReLU(), Linear(dim, dim), ReLU()
)
)
self.conv4 = GINConv(
Sequential(
Linear(dim, dim), BatchNorm1d(dim), ReLU(), Linear(dim, dim), ReLU()
)
)
self.conv5 = GINConv(
Sequential(
Linear(dim, dim), BatchNorm1d(dim), ReLU(), Linear(dim, dim), ReLU()
)
)
self.lin1 = Linear(dim, dim)
self.lin2 = Linear(dim, out_channels)
def forward(self, x, edge_index, batch):
x = self.conv1(x, edge_index)
x = self.conv2(x, edge_index)
x = self.conv3(x, edge_index)
x = self.conv4(x, edge_index)
x = self.conv5(x, edge_index)
x = global_add_pool(x, batch)
x = self.lin1(x).relu()
x = F.dropout(x, p=0.5, training=self.training)
x = self.lin2(x)
return F.log_softmax(x, dim=-1)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model = Net(dataset.num_features, 32, dataset.num_classes).to(device)
optimizer = torch.optim.Adam(model.parameters(), lr=0.01)
def train():
model.train()
total_loss = 0
for data in train_loader:
data = data.to(device)
optimizer.zero_grad()
output = model(data.x, data.edge_index, data.batch)
loss = F.nll_loss(output, data.y)
loss.backward()
optimizer.step()
total_loss += float(loss) * data.num_graphs
return total_loss / len(train_loader.dataset)
@torch.no_grad()
def test(loader):
model.eval()
total_correct = 0
for data in loader:
data = data.to(device)
out = model(data.x, data.edge_index, data.batch)
total_correct += int((out.argmax(-1) == data.y).sum())
return total_correct / len(loader.dataset)
for epoch in range(1, 101):
loss = train()
train_acc = test(train_loader)
test_acc = test(test_loader)
print(
f"Epoch: {epoch:03d}, Loss: {loss:.4f}, Train Acc: {train_acc:.4f} "
f"Test Acc: {test_acc:.4f}"
)
| false | 0 | 5,411 | 0 | 6,119 | 5,411 |
||
69921292
|
<jupyter_start><jupyter_text>Breast Histopathology Images
### Context
Invasive Ductal Carcinoma (IDC) is the most common subtype of all breast cancers. To assign an aggressiveness grade to a whole mount sample, pathologists typically focus on the regions which contain the IDC. As a result, one of the common pre-processing steps for automatic aggressiveness grading is to delineate the exact regions of IDC inside of a whole mount slide.
### Content
The original dataset consisted of 162 whole mount slide images of Breast Cancer (BCa) specimens scanned at 40x. From that, 277,524 patches of size 50 x 50 were extracted (198,738 IDC negative and 78,786 IDC positive). Each patch’s file name is of the format: u_xX_yY_classC.png — > example 10253_idx5_x1351_y1101_class0.png . Where u is the patient ID (10253_idx5), X is the x-coordinate of where this patch was cropped from, Y is the y-coordinate of where this patch was cropped from, and C indicates the class where 0 is non-IDC and 1 is IDC.
Kaggle dataset identifier: breast-histopathology-images
<jupyter_script>import numpy as np
import glob
import random
import warnings
warnings.filterwarnings(action="ignore")
import matplotlib.pyplot as plt
from PIL import Image
random.seed(98)
np.random.seed(98)
breast_img = glob.glob(
"/kaggle/input/breast-histopathology-images/IDC_regular_ps50_idx5/**/*.png",
recursive=True,
)
for imgname in breast_img[:3]:
print(imgname)
non_img = []
can_img = []
for img in breast_img:
if img[-5] == "0":
non_img.append(img)
elif img[-5] == "1":
can_img.append(img)
non_num = len(non_img)
can_num = len(can_img)
total_img_num = non_num + can_num
print("Number of Images in IDC (-): {}".format(non_num))
print("Number of Images in IDC (+) : {}".format(can_num))
print("Total Number of Images : {}".format(total_img_num))
from keras.preprocessing import image
plt.figure(figsize=(15, 15))
some_non = np.random.randint(0, len(non_img), 18)
some_can = np.random.randint(0, len(can_img), 18)
s = 0
for num in some_non:
img = image.load_img((non_img[num]), target_size=(100, 100))
img = image.img_to_array(img)
plt.subplot(6, 6, 2 * s + 1)
plt.axis("off")
plt.title("IDC (-)")
plt.imshow(img.astype("uint8"))
s += 1
s = 1
for num in some_can:
img = image.load_img((can_img[num]), target_size=(100, 100))
img = image.img_to_array(img)
plt.subplot(6, 6, 2 * s)
plt.axis("off")
plt.title("IDC (+)")
plt.imshow(img.astype("uint8"))
s += 1
from matplotlib.image import imread
import cv2
some_non_img = random.sample(non_img, len(can_img))
some_can_img = random.sample(can_img, len(can_img))
non_img_arr = []
can_img_arr = []
for img in some_non_img:
n_img = cv2.imread(img, cv2.IMREAD_COLOR)
n_img_size = cv2.resize(n_img, (50, 50), interpolation=cv2.INTER_LINEAR)
non_img_arr.append([n_img_size, 0])
for img in some_can_img:
c_img = cv2.imread(img, cv2.IMREAD_COLOR)
c_img_size = cv2.resize(c_img, (50, 50), interpolation=cv2.INTER_LINEAR)
can_img_arr.append([c_img_size, 1])
X = []
y = []
breast_img_arr = np.concatenate((non_img_arr, can_img_arr))
random.shuffle(breast_img_arr)
for feature, label in breast_img_arr:
X.append(feature)
y.append(label)
X = np.array(X)
y = np.array(y)
print("X shape : {}".format(X.shape))
from sklearn.model_selection import train_test_split
from keras.utils.np_utils import to_categorical
X_train, X_predict, y_train, y_true = train_test_split(
X, y, test_size=0.3, random_state=7
)
rate = 0.5
num = int(X.shape[0] * rate)
X_test = X_train[num:]
X_train = X_train[:num]
y_test = y_train[num:]
y_train = y_train[:num]
y_train = to_categorical(y_train, 2)
y_test = to_categorical(y_test, 2)
y_true = to_categorical(y_true, 2)
print("X_train shape : {}".format(X_train.shape))
print("X_test shape : {}".format(X_test.shape))
print("X_predict shape : {}".format(X_predict.shape))
print("y_train shape : {}".format(y_train.shape))
print("y_test shape : {}".format(y_test.shape))
print("y_true shape : {}".format(y_true.shape))
from keras.models import Sequential
from keras.layers import Conv2D, MaxPooling2D, Dropout, Flatten, Dense
model = Sequential()
model.add(
Conv2D(32, (3, 3), padding="same", activation="relu", input_shape=(50, 50, 3))
)
model.add(MaxPooling2D(2, 2))
model.add(Dropout(0.25))
model.add(
Conv2D(64, (3, 3), padding="same", activation="relu", input_shape=(50, 50, 3))
)
model.add(MaxPooling2D(2, 2))
model.add(Dropout(0.25))
model.add(
Conv2D(128, (3, 3), padding="same", activation="relu", input_shape=(50, 50, 3))
)
model.add(MaxPooling2D(2, 2))
model.add(Dropout(0.25))
model.add(
Conv2D(128, (3, 3), padding="same", activation="relu", input_shape=(50, 50, 3))
)
model.add(MaxPooling2D(2, 2))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(128, activation="relu"))
model.add(Dropout(0.5))
model.add(Dense(2, activation="sigmoid"))
model.summary()
from keras.optimizers import Adam
Adam = Adam(learning_rate=0.0001)
model.compile(loss="binary_crossentropy", optimizer=Adam, metrics=["accuracy"])
history = model.fit(
X_train, y_train, validation_data=(X_test, y_test), epochs=30, batch_size=50
)
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("Model Accuracy")
plt.xlabel("epoch")
plt.ylabel("accuracy")
plt.legend(["train", "test"], loc="upper left")
plt.show()
result = model.evaluate(X_test, y_test, batch_size=50)
print("Test Loss, Test Accuracy :", result)
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Model Loss")
plt.xlabel("epoch")
plt.ylabel("loss")
plt.legend(["train", "test"], loc="upper left")
plt.show()
P = model.predict(X_predict)
true = 0
for i in range(X_predict.shape[0]):
if np.argmax(P[i]) == np.argmax(y_true[i]):
true = true + 1
pre_accuracy = 100 * float(true / X_predict.shape[0])
print("Predict Accuracy: {}".format(pre_accuracy))
import seaborn as sns
false = len(X_predict) - true
predict = [true, false]
label = ["true_predict", "false_predict"]
plt.bar(label[0], predict[0])
plt.bar(label[1], predict[1])
plt.title("Predict Accuracy")
plt.xlabel("predict")
plt.ylabel("count")
plt.legend(["true_predict", "false_predict"], loc="upper left")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/921/69921292.ipynb
|
breast-histopathology-images
|
paultimothymooney
|
[{"Id": 69921292, "ScriptId": 18962282, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7390920, "CreationDate": "08/03/2021 21:09:42", "VersionNumber": 14.0, "Title": "breast_cancer_using_cnn", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 206.0, "LinesInsertedFromPrevious": 7.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 199.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 5}]
|
[{"Id": 93337727, "KernelVersionId": 69921292, "SourceDatasetVersionId": 10564}]
|
[{"Id": 10564, "DatasetId": 7415, "DatasourceVersionId": 10564, "CreatorUserId": 1314380, "LicenseName": "CC0: Public Domain", "CreationDate": "12/19/2017 05:46:40", "VersionNumber": 1.0, "Title": "Breast Histopathology Images", "Slug": "breast-histopathology-images", "Subtitle": "198,738 IDC(-) image patches; 78,786 IDC(+) image patches", "Description": "### Context\n\nInvasive Ductal Carcinoma (IDC) is the most common subtype of all breast cancers. To assign an aggressiveness grade to a whole mount sample, pathologists typically focus on the regions which contain the IDC. As a result, one of the common pre-processing steps for automatic aggressiveness grading is to delineate the exact regions of IDC inside of a whole mount slide.\n\n### Content\n\nThe original dataset consisted of 162 whole mount slide images of Breast Cancer (BCa) specimens scanned at 40x. From that, 277,524 patches of size 50 x 50 were extracted (198,738 IDC negative and 78,786 IDC positive). Each patch\u2019s file name is of the format: u_xX_yY_classC.png \u2014 > example 10253_idx5_x1351_y1101_class0.png . Where u is the patient ID (10253_idx5), X is the x-coordinate of where this patch was cropped from, Y is the y-coordinate of where this patch was cropped from, and C indicates the class where 0 is non-IDC and 1 is IDC.\n\n\n\n### Acknowledgements\n\nThe original files are located here: http://gleason.case.edu/webdata/jpi-dl-tutorial/IDC_regular_ps50_idx5.zip\nCitation: https://www.ncbi.nlm.nih.gov/pubmed/27563488 and http://spie.org/Publications/Proceedings/Paper/10.1117/12.2043872\n\n### Inspiration\n\nBreast cancer is the most common form of cancer in women, and invasive ductal carcinoma (IDC) is the most common form of breast cancer. Accurately identifying and categorizing breast cancer subtypes is an important clinical task, and automated methods can be used to save time and reduce error.", "VersionNotes": "Initial release", "TotalCompressedBytes": 1644892042.0, "TotalUncompressedBytes": 1644892042.0}]
|
[{"Id": 7415, "CreatorUserId": 1314380, "OwnerUserId": 1314380.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 10564.0, "CurrentDatasourceVersionId": 10564.0, "ForumId": 14193, "Type": 2, "CreationDate": "12/19/2017 05:46:40", "LastActivityDate": "02/06/2018", "TotalViews": 333071, "TotalDownloads": 53221, "TotalVotes": 921, "TotalKernels": 157}]
|
[{"Id": 1314380, "UserName": "paultimothymooney", "DisplayName": "Paul Mooney", "RegisterDate": "10/05/2017", "PerformanceTier": 5}]
|
import numpy as np
import glob
import random
import warnings
warnings.filterwarnings(action="ignore")
import matplotlib.pyplot as plt
from PIL import Image
random.seed(98)
np.random.seed(98)
breast_img = glob.glob(
"/kaggle/input/breast-histopathology-images/IDC_regular_ps50_idx5/**/*.png",
recursive=True,
)
for imgname in breast_img[:3]:
print(imgname)
non_img = []
can_img = []
for img in breast_img:
if img[-5] == "0":
non_img.append(img)
elif img[-5] == "1":
can_img.append(img)
non_num = len(non_img)
can_num = len(can_img)
total_img_num = non_num + can_num
print("Number of Images in IDC (-): {}".format(non_num))
print("Number of Images in IDC (+) : {}".format(can_num))
print("Total Number of Images : {}".format(total_img_num))
from keras.preprocessing import image
plt.figure(figsize=(15, 15))
some_non = np.random.randint(0, len(non_img), 18)
some_can = np.random.randint(0, len(can_img), 18)
s = 0
for num in some_non:
img = image.load_img((non_img[num]), target_size=(100, 100))
img = image.img_to_array(img)
plt.subplot(6, 6, 2 * s + 1)
plt.axis("off")
plt.title("IDC (-)")
plt.imshow(img.astype("uint8"))
s += 1
s = 1
for num in some_can:
img = image.load_img((can_img[num]), target_size=(100, 100))
img = image.img_to_array(img)
plt.subplot(6, 6, 2 * s)
plt.axis("off")
plt.title("IDC (+)")
plt.imshow(img.astype("uint8"))
s += 1
from matplotlib.image import imread
import cv2
some_non_img = random.sample(non_img, len(can_img))
some_can_img = random.sample(can_img, len(can_img))
non_img_arr = []
can_img_arr = []
for img in some_non_img:
n_img = cv2.imread(img, cv2.IMREAD_COLOR)
n_img_size = cv2.resize(n_img, (50, 50), interpolation=cv2.INTER_LINEAR)
non_img_arr.append([n_img_size, 0])
for img in some_can_img:
c_img = cv2.imread(img, cv2.IMREAD_COLOR)
c_img_size = cv2.resize(c_img, (50, 50), interpolation=cv2.INTER_LINEAR)
can_img_arr.append([c_img_size, 1])
X = []
y = []
breast_img_arr = np.concatenate((non_img_arr, can_img_arr))
random.shuffle(breast_img_arr)
for feature, label in breast_img_arr:
X.append(feature)
y.append(label)
X = np.array(X)
y = np.array(y)
print("X shape : {}".format(X.shape))
from sklearn.model_selection import train_test_split
from keras.utils.np_utils import to_categorical
X_train, X_predict, y_train, y_true = train_test_split(
X, y, test_size=0.3, random_state=7
)
rate = 0.5
num = int(X.shape[0] * rate)
X_test = X_train[num:]
X_train = X_train[:num]
y_test = y_train[num:]
y_train = y_train[:num]
y_train = to_categorical(y_train, 2)
y_test = to_categorical(y_test, 2)
y_true = to_categorical(y_true, 2)
print("X_train shape : {}".format(X_train.shape))
print("X_test shape : {}".format(X_test.shape))
print("X_predict shape : {}".format(X_predict.shape))
print("y_train shape : {}".format(y_train.shape))
print("y_test shape : {}".format(y_test.shape))
print("y_true shape : {}".format(y_true.shape))
from keras.models import Sequential
from keras.layers import Conv2D, MaxPooling2D, Dropout, Flatten, Dense
model = Sequential()
model.add(
Conv2D(32, (3, 3), padding="same", activation="relu", input_shape=(50, 50, 3))
)
model.add(MaxPooling2D(2, 2))
model.add(Dropout(0.25))
model.add(
Conv2D(64, (3, 3), padding="same", activation="relu", input_shape=(50, 50, 3))
)
model.add(MaxPooling2D(2, 2))
model.add(Dropout(0.25))
model.add(
Conv2D(128, (3, 3), padding="same", activation="relu", input_shape=(50, 50, 3))
)
model.add(MaxPooling2D(2, 2))
model.add(Dropout(0.25))
model.add(
Conv2D(128, (3, 3), padding="same", activation="relu", input_shape=(50, 50, 3))
)
model.add(MaxPooling2D(2, 2))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(128, activation="relu"))
model.add(Dropout(0.5))
model.add(Dense(2, activation="sigmoid"))
model.summary()
from keras.optimizers import Adam
Adam = Adam(learning_rate=0.0001)
model.compile(loss="binary_crossentropy", optimizer=Adam, metrics=["accuracy"])
history = model.fit(
X_train, y_train, validation_data=(X_test, y_test), epochs=30, batch_size=50
)
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("Model Accuracy")
plt.xlabel("epoch")
plt.ylabel("accuracy")
plt.legend(["train", "test"], loc="upper left")
plt.show()
result = model.evaluate(X_test, y_test, batch_size=50)
print("Test Loss, Test Accuracy :", result)
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Model Loss")
plt.xlabel("epoch")
plt.ylabel("loss")
plt.legend(["train", "test"], loc="upper left")
plt.show()
P = model.predict(X_predict)
true = 0
for i in range(X_predict.shape[0]):
if np.argmax(P[i]) == np.argmax(y_true[i]):
true = true + 1
pre_accuracy = 100 * float(true / X_predict.shape[0])
print("Predict Accuracy: {}".format(pre_accuracy))
import seaborn as sns
false = len(X_predict) - true
predict = [true, false]
label = ["true_predict", "false_predict"]
plt.bar(label[0], predict[0])
plt.bar(label[1], predict[1])
plt.title("Predict Accuracy")
plt.xlabel("predict")
plt.ylabel("count")
plt.legend(["true_predict", "false_predict"], loc="upper left")
plt.show()
| false | 0 | 1,984 | 5 | 2,312 | 1,984 |
||
69921431
|
# # Exercises
# Welcome to your first set of Python coding problems!
# If this is your first time using Kaggle Kernels, welcome! For a very quick introduction to the coding environment, [check out this video](https://youtu.be/4C2qMnaIKL4).
# Kernels (also known as notebooks) are composed of blocks (called "cells") of text and code. Each of these is editable, though you'll mainly be editing the code cells to answer some questions.
# To get started, try running the code cell below (by pressing the ► button, or clicking on the cell and pressing ctrl+enter on your keyboard).
print("You've successfully run some Python code")
print("Congratulations!")
print("hello" * 2)
# Try adding another line of code in the cell above and re-running it.
# Now let's get a little fancier: Add a new code cell by clicking on an existing code cell, hitting the escape key, and then hitting the `a` or `b` key. The `a` key will add a cell above the current cell, and `b` adds a cell below.
# Great! Now you know how to use Kernels.
# Each hands-on exercise starts by setting up our feedback and code checking mechanism. Run the code cell below to do that. Then you'll be ready to move on to question 0.
from learntools.core import binder
binder.bind(globals())
from learntools.python.ex1 import *
print("Setup complete! You're ready to start question 0.")
# ## 0.
# *This is a silly question intended as an introduction to the format we use for hands-on exercises throughout all Kaggle courses.*
# **What is your favorite color? **
# To complete this question, create a variable called `color` in the cell below with an appropriate value. The function call `q0.check()` (which we've already provided in the cell below) will check your answer.
# create a variable called color with an appropriate value on the line below
# (Remember, strings in Python must be enclosed in 'single' or "double" quotes)
color = "blue"
q0.check()
# Didn't get the right answer? How do you not even know your own favorite color?!
# Delete the `#` in the line below to make one of the lines run. You can choose between getting a hint or the full answer by choosing which line to remove the `#` from.
# Removing the `#` is called uncommenting, because it changes that line from a "comment" which Python doesn't run to code, which Python does run.
q0.hint()
q0.solution()
# The upcoming questions work the same way. The only thing that will change are the questions numbers. For the next question, you'll call `q1.check()`, `q1.hint()`, `q1.solution()`, for question 5, you'll call `q5.check()`, and so on.
# ## 1.
# Complete the code below. In case it's helpful, here is the table of available arithmatic operations:
# | Operator | Name | Description |
# |--------------|----------------|--------------------------------------------------------|
# | ``a + b`` | Addition | Sum of ``a`` and ``b`` |
# | ``a - b`` | Subtraction | Difference of ``a`` and ``b`` |
# | ``a * b`` | Multiplication | Product of ``a`` and ``b`` |
# | ``a / b`` | True division | Quotient of ``a`` and ``b`` |
# | ``a // b`` | Floor division | Quotient of ``a`` and ``b``, removing fractional parts |
# | ``a % b`` | Modulus | Integer remainder after division of ``a`` by ``b`` |
# | ``a ** b`` | Exponentiation | ``a`` raised to the power of ``b`` |
# | ``-a`` | Negation | The negative of ``a`` |
#
pi = 3.14159 # approximate
diameter = 3
# Create a variable called 'radius' equal to half the diameter
radius = 3 / 2
# Create a variable called 'area', using the formula for the area of a circle: pi times the radius squared
area = pi * radius**2
q1.check()
# Uncomment and run the lines below if you need help.
q1.hint()
q1.solution()
#
# ## 2.
# Add code to the following cell to swap variables `a` and `b` (so that `a` refers to the object previously referred to by `b` and vice versa).
########### Setup code - don't touch this part ######################
# If you're curious, these are examples of lists. We'll talk about
# them in depth a few lessons from now. For now, just know that they're
# yet another type of Python object, like int or float.
a = [1, 2, 3]
b = [3, 2, 1]
q2.store_original_ids()
######################################################################
# Your code goes here. Swap the values to which a and b refer.
# If you get stuck, you can always uncomment one or both of the lines in
# the next cell for a hint, or to peek at the solution.
old_a = a
a = b
b = old_a
######################################################################
q2.check()
# q2.hint()
# q2.solution()
#
# ## 3.
# *Note: some questions, such as this one, don't have a .check() function. But it should be easy to tell if you've succeeded.*
# a) Add parentheses to the following expression so that it evaluates to 1.
(5 - 3) // 2
# q3.a.hint()
# q3.a.solution()
# Questions, like this one, marked a spicy pepper are a bit harder. Don't feel bad if you can't get these.
# b) 🌶️ Add parentheses to the following expression so that it evaluates to 0
#
8 - 3 * 2 - (1 + 1)
# q3.b.hint()
# q3.b.solution()
#
# ## 4.
# Alice, Bob and Carol have agreed to pool their Halloween candy and split it evenly among themselves.
# For the sake of their friendship, any candies left over will be smashed. For example, if they collectively
# bring home 91 candies, they'll take 30 each and smash 1.
# Write an arithmetic expression below to calculate how many candies they must smash for a given haul.
# Variables representing the number of candies collected by alice, bob, and carol
alice_candies = 121
bob_candies = 77
carol_candies = 109
# Your code goes here! Replace the right-hand side of this assignment with an expression
# involving alice_candies, bob_candies, and carol_candies
to_smash = (alice_candies + bob_candies + carol_candies) % 3
q4.check()
# q4.hint()
# q4.solution()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/921/69921431.ipynb
| null | null |
[{"Id": 69921431, "ScriptId": 19119923, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7885390, "CreationDate": "08/03/2021 21:10:51", "VersionNumber": 1.0, "Title": "Exercise: Syntax, Variables, and Numbers Daily", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 162.0, "LinesInsertedFromPrevious": 14.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 148.0, "LinesInsertedFromFork": 14.0, "LinesDeletedFromFork": 11.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 148.0, "TotalVotes": 0}]
| null | null | null | null |
# # Exercises
# Welcome to your first set of Python coding problems!
# If this is your first time using Kaggle Kernels, welcome! For a very quick introduction to the coding environment, [check out this video](https://youtu.be/4C2qMnaIKL4).
# Kernels (also known as notebooks) are composed of blocks (called "cells") of text and code. Each of these is editable, though you'll mainly be editing the code cells to answer some questions.
# To get started, try running the code cell below (by pressing the ► button, or clicking on the cell and pressing ctrl+enter on your keyboard).
print("You've successfully run some Python code")
print("Congratulations!")
print("hello" * 2)
# Try adding another line of code in the cell above and re-running it.
# Now let's get a little fancier: Add a new code cell by clicking on an existing code cell, hitting the escape key, and then hitting the `a` or `b` key. The `a` key will add a cell above the current cell, and `b` adds a cell below.
# Great! Now you know how to use Kernels.
# Each hands-on exercise starts by setting up our feedback and code checking mechanism. Run the code cell below to do that. Then you'll be ready to move on to question 0.
from learntools.core import binder
binder.bind(globals())
from learntools.python.ex1 import *
print("Setup complete! You're ready to start question 0.")
# ## 0.
# *This is a silly question intended as an introduction to the format we use for hands-on exercises throughout all Kaggle courses.*
# **What is your favorite color? **
# To complete this question, create a variable called `color` in the cell below with an appropriate value. The function call `q0.check()` (which we've already provided in the cell below) will check your answer.
# create a variable called color with an appropriate value on the line below
# (Remember, strings in Python must be enclosed in 'single' or "double" quotes)
color = "blue"
q0.check()
# Didn't get the right answer? How do you not even know your own favorite color?!
# Delete the `#` in the line below to make one of the lines run. You can choose between getting a hint or the full answer by choosing which line to remove the `#` from.
# Removing the `#` is called uncommenting, because it changes that line from a "comment" which Python doesn't run to code, which Python does run.
q0.hint()
q0.solution()
# The upcoming questions work the same way. The only thing that will change are the questions numbers. For the next question, you'll call `q1.check()`, `q1.hint()`, `q1.solution()`, for question 5, you'll call `q5.check()`, and so on.
# ## 1.
# Complete the code below. In case it's helpful, here is the table of available arithmatic operations:
# | Operator | Name | Description |
# |--------------|----------------|--------------------------------------------------------|
# | ``a + b`` | Addition | Sum of ``a`` and ``b`` |
# | ``a - b`` | Subtraction | Difference of ``a`` and ``b`` |
# | ``a * b`` | Multiplication | Product of ``a`` and ``b`` |
# | ``a / b`` | True division | Quotient of ``a`` and ``b`` |
# | ``a // b`` | Floor division | Quotient of ``a`` and ``b``, removing fractional parts |
# | ``a % b`` | Modulus | Integer remainder after division of ``a`` by ``b`` |
# | ``a ** b`` | Exponentiation | ``a`` raised to the power of ``b`` |
# | ``-a`` | Negation | The negative of ``a`` |
#
pi = 3.14159 # approximate
diameter = 3
# Create a variable called 'radius' equal to half the diameter
radius = 3 / 2
# Create a variable called 'area', using the formula for the area of a circle: pi times the radius squared
area = pi * radius**2
q1.check()
# Uncomment and run the lines below if you need help.
q1.hint()
q1.solution()
#
# ## 2.
# Add code to the following cell to swap variables `a` and `b` (so that `a` refers to the object previously referred to by `b` and vice versa).
########### Setup code - don't touch this part ######################
# If you're curious, these are examples of lists. We'll talk about
# them in depth a few lessons from now. For now, just know that they're
# yet another type of Python object, like int or float.
a = [1, 2, 3]
b = [3, 2, 1]
q2.store_original_ids()
######################################################################
# Your code goes here. Swap the values to which a and b refer.
# If you get stuck, you can always uncomment one or both of the lines in
# the next cell for a hint, or to peek at the solution.
old_a = a
a = b
b = old_a
######################################################################
q2.check()
# q2.hint()
# q2.solution()
#
# ## 3.
# *Note: some questions, such as this one, don't have a .check() function. But it should be easy to tell if you've succeeded.*
# a) Add parentheses to the following expression so that it evaluates to 1.
(5 - 3) // 2
# q3.a.hint()
# q3.a.solution()
# Questions, like this one, marked a spicy pepper are a bit harder. Don't feel bad if you can't get these.
# b) 🌶️ Add parentheses to the following expression so that it evaluates to 0
#
8 - 3 * 2 - (1 + 1)
# q3.b.hint()
# q3.b.solution()
#
# ## 4.
# Alice, Bob and Carol have agreed to pool their Halloween candy and split it evenly among themselves.
# For the sake of their friendship, any candies left over will be smashed. For example, if they collectively
# bring home 91 candies, they'll take 30 each and smash 1.
# Write an arithmetic expression below to calculate how many candies they must smash for a given haul.
# Variables representing the number of candies collected by alice, bob, and carol
alice_candies = 121
bob_candies = 77
carol_candies = 109
# Your code goes here! Replace the right-hand side of this assignment with an expression
# involving alice_candies, bob_candies, and carol_candies
to_smash = (alice_candies + bob_candies + carol_candies) % 3
q4.check()
# q4.hint()
# q4.solution()
| false | 0 | 1,695 | 0 | 1,695 | 1,695 |
||
69921983
|
import numpy as np
import pandas as pd
import os
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers, callbacks
from sklearn.model_selection import train_test_split
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
train = pd.read_csv("/kaggle/input/tabular-playground-series-aug-2021/train.csv")
train.head()
train.shape
train_y = train["loss"]
train_x = train
train_x.drop(columns=["loss", "id"], inplace=True)
train_x.head()
train_x_train, train_x_valid, train_y_train, train_y_valid = train_test_split(
train_x, train_y, train_size=0.8
)
early_stopping = callbacks.EarlyStopping(min_delta=0.001, restore_best_weights=True)
input_shape = [100]
model = keras.Sequential(
[
layers.Dense(units=6250, activation="relu", input_shape=input_shape),
layers.BatchNormalization(),
layers.Dense(1),
layers.BatchNormalization(),
]
)
model.compile(
optimizer="adam", loss="mse", metrics=[tf.keras.metrics.RootMeanSquaredError()]
)
model.fit(
train_x_train,
train_y_train,
validation_data=(train_x_valid, train_y_valid),
epochs=50,
verbose=1,
callbacks=[early_stopping],
)
test = pd.read_csv("/kaggle/input/tabular-playground-series-aug-2021/test.csv")
test.drop(columns=["id"], inplace=True)
test.head()
predictions = model.predict(test)
sample_submission = pd.read_csv(
"/kaggle/input/tabular-playground-series-aug-2021/sample_submission.csv"
)
sample_submission.head()
sample_submission["loss"] = predictions
sample_submission.reset_index()
sample_submission.head()
sample_submission.to_csv("submission.csv", index=False)
test = pd.read_csv("submission.csv")
test.head()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/921/69921983.ipynb
| null | null |
[{"Id": 69921983, "ScriptId": 19116023, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4686504, "CreationDate": "08/03/2021 21:14:53", "VersionNumber": 6.0, "Title": "Tabular Playground August 2021", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 67.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 66.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
import pandas as pd
import os
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers, callbacks
from sklearn.model_selection import train_test_split
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
train = pd.read_csv("/kaggle/input/tabular-playground-series-aug-2021/train.csv")
train.head()
train.shape
train_y = train["loss"]
train_x = train
train_x.drop(columns=["loss", "id"], inplace=True)
train_x.head()
train_x_train, train_x_valid, train_y_train, train_y_valid = train_test_split(
train_x, train_y, train_size=0.8
)
early_stopping = callbacks.EarlyStopping(min_delta=0.001, restore_best_weights=True)
input_shape = [100]
model = keras.Sequential(
[
layers.Dense(units=6250, activation="relu", input_shape=input_shape),
layers.BatchNormalization(),
layers.Dense(1),
layers.BatchNormalization(),
]
)
model.compile(
optimizer="adam", loss="mse", metrics=[tf.keras.metrics.RootMeanSquaredError()]
)
model.fit(
train_x_train,
train_y_train,
validation_data=(train_x_valid, train_y_valid),
epochs=50,
verbose=1,
callbacks=[early_stopping],
)
test = pd.read_csv("/kaggle/input/tabular-playground-series-aug-2021/test.csv")
test.drop(columns=["id"], inplace=True)
test.head()
predictions = model.predict(test)
sample_submission = pd.read_csv(
"/kaggle/input/tabular-playground-series-aug-2021/sample_submission.csv"
)
sample_submission.head()
sample_submission["loss"] = predictions
sample_submission.reset_index()
sample_submission.head()
sample_submission.to_csv("submission.csv", index=False)
test = pd.read_csv("submission.csv")
test.head()
| false | 0 | 553 | 0 | 553 | 553 |
||
69921677
|
# # Predicting Bike Rental count Depending on weather conditions
# # Introduction :
# #### We need to predict the number of Bikes to be rented based on differrent weather conditions and other effective factors
# # Dataset:
# #### The dataset is about bike rental in seoul korea ,The Data contains weather information ,which are the features (Temperature, Humidity, Wind speed, Visibility, Dew point, Solar radiation, Snowfall, Rainfall),the target is the number of bikes rented per hour and date information.
#
# # Description Of Data Set Columns:
# #### - ID - an ID for this instance
# #### - Date - year-month-day
# #### - Hour - Hour of he day
# #### - Temperature - Temperature in Celsius
# #### - Humidity - %
# #### - Windspeed - m/s
# #### - Visibility - 10m
# #### - Dew point temperature - Celsius
# #### - Solar radiation - MJ/m2
# #### - Rainfall - mm
# #### - Snowfall - cm
# #### - Seasons - Winter, Spring, Summer, Autumn
# #### - Holiday - Holiday/No holiday
# #### - Functional Day - NoFunc(Non Functional Hours), Fun(Functional hours)
# #### - y - Rented Bike count (Target), Count of bikes rented at each hour
# **Hint: You can submit up to 2 submissions per day. You can select only one of the submission you make to be considered in the final ranking.**
# # Data Wrangling Process:
# #### The Data Wrangling Process Consists of 3 phases Gathering ,Assessing and Cleaning the data
# # Data Gathering :
# ## Import necessary libraries
# ##### ● We'll use `pandas` to load and manipulate the data. Other libraries will be imported in the relevant sections.
import pandas as pd
import os
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import OrdinalEncoder
from sklearn import preprocessing # This import for data standarization
from sklearn.metrics import mean_squared_log_error
# # Loading Data:
# ## Read train dataset from train.csv file
dataset_path = "/kaggle/input/seoul-bike-rental-ai-pro-iti/"
df = pd.read_csv(os.path.join(dataset_path, "train.csv"))
print("The shape of the dataset is {}.\n\n".format(df.shape))
df.head(5017)
# We've got 5760 examples in the dataset with 15 featues.
# By looking at the features and a sample from the data, the features look of numerical and catogerical types.
# ### We will change columns' names to be called easily
df.rename(
columns={
"Temperature(�C)": "Temperature",
"Humidity(%)": "Humidity",
"Wind speed (m/s)": "WindSpeed",
"Visibility (10m)": "Visibility",
"Dew point temperature(�C)": "DewPointTemperature",
"Solar Radiation (MJ/m2)": "SolarRadiation",
"Rainfall(mm)": "Rainfall",
"Snowfall (cm)": "Snowfall",
"Functioning Day": "FunctioningDay",
},
inplace=True,
)
df.head()
# # Data Cleaning
# ### We need to convert categorical features into numerical data
# Create Function convert categorical data as:(seasons , Holiday , FunctioningDay) into numerical
def convert_categorical_columns_into_numerical(df, columns):
ord_enc = OrdinalEncoder()
for column in columns:
df[column] = ord_enc.fit_transform(df[[column]])
return df
df = convert_categorical_columns_into_numerical(
df, ["Seasons", "Holiday", "FunctioningDay"]
)
# -----------------------------------------------------------------#
# Another way to convert categorical data into numerical
# Convert columns ('Functioning Day') into (0 , 1)
# df['Functioning Day'] = df['Functioning Day'].replace('Yes', 1)
# df['Functioning Day'] = df['Functioning Day'].replace('No', 0)
# Convert columns ('Holiday') into (0 , 1)
# df['Holiday'] = df['Holiday'].replace('Holiday' , 1)
# df['Holiday'] = df['Holiday'].replace('No Holiday' , 0)
# ------------------------------------------------------------------#
# Convert Date from String to datetime
df["Date"] = pd.to_datetime(df["Date"], format="%d/%m/%Y")
# Create New feature to get DayOfWeek from DateTime
df["DayOfWeek"] = df["Date"].dt.dayofweek
# Create new feature which informs us if it is night or day from Hour
df["LabelDayNight"] = df["Hour"].apply(
lambda x: "Night" if (x > 20 or x < 5) else ("Day")
)
# Convert LabelDayNight Column from Categorical (Day , Night) into Numerical (0,1)
df = convert_categorical_columns_into_numerical(df, ["LabelDayNight"])
# Get Day's Feature from Date
df["Day"] = df["Date"].dt.day
# Get Month's Feature from Date
df["Month"] = df["Date"].dt.month
df.head(600)
# ### Trials to convert categorical features into numerical
# print(type (df['Functioning Day']))
# df['Functioning Day'] = df['Functioning Day'].map({'Yes': 1 , 'No':0})
# df['Functioning Day'] = df['Functioning Day']*1
# df['Functioning Day'] = pd.to_numeric(df['Functioning Day'], errors='coerce')
# (df['COL2'] == 'TRUE').astype(int)
# if df['Holiday']== 'No Holiday':
# df['Holiday'] = 1
# else:
# df['Holiday'] = 0
# ## Describe Data
df.drop(columns="ID").describe()
# ## Correlation between features' data and label's data
df.drop(columns="ID").corr().style.background_gradient()
# ### More visualization of data
sns.set(style="ticks", color_codes=True)
g = sns.pairplot(df.drop(columns="ID"))
plt.show()
# Plot every hour repeated until what!
print(df["y"].value_counts().head())
df.y.value_counts().sort_values(ascending=False).head(15).plot(kind="bar")
plt.xlabel("Number of bikes rented")
plt.ylabel("Counting: How many this number repeated")
plt.show()
# #Drawing same data using Pie Chart
# df['y'].value_counts().sort_values(ascending=False).head(5).plot(kind='pie')
# plt.show()
plt.scatter(df["Hour"], df["y"])
plt.xlabel("Hour Number")
plt.ylabel("Number of Bikes")
plt.show()
# sns.barplot(x=df['Hour'].value_counts() , y=df['y'])
# plt.show()
# ### Check Duplicated data
print(df.duplicated())
# Then, there is no redundancy in data
# ### Check null values in Training Data
df.isnull().sum()
# ### Dropping missing values if existed
print(df.count())
df = df.dropna()
df.count()
# Then, there is no missing values existed in data
# ### Detecting outliers, If existed:
# * Delete these outlier data
# Box plot for number of bikes rented
plt.figure(figsize=(10, 6))
sns.boxplot(x=df["y"])
plt.show()
# Box plot for number of hours
plt.figure(figsize=(10, 6))
sns.boxplot(x=df["Hour"])
plt.show()
# So, it is optimal in distribution
Q1 = df.quantile(0.25)
Q3 = df.quantile(0.75)
IQR = Q3 - Q1
print(IQR)
# # Data Splitting
# Now it's time to split the dataset for the training step. Typically the dataset is split into 3 subsets, namely, the training, validation and test sets. In our case, the test set is already predefined. So we'll split the "training" set into training and validation sets with 0.8:0.2 ratio.
# Note: a good way to generate reproducible results is to set the seed to the algorithms that depends on randomization. This is done with the argument random_state in the following command
from sklearn.model_selection import train_test_split
train_df, val_df = train_test_split(
df, test_size=0.2, random_state=42
) # Try adding `stratify` here
X_train = train_df.drop(columns=["ID", "y"])
y_train = train_df["y"]
X_val = val_df.drop(columns=["ID", "y"])
y_val = val_df["y"]
# This cell is used to select the numerical features only. IT SHOULD BE REMOVED AS YOU DO YOUR WORK.
features = [
"Temperature",
"Hour",
"DewPointTemperature",
"Seasons",
"SolarRadiation",
"Humidity",
"Visibility",
"FunctioningDay",
"Month",
"Rainfall",
"LabelDayNight",
]
X_train = X_train[features]
X_val = X_val[features]
# # Model Training
# * #### Let's train a model with the data! We'll train a Random Forest Classifier to demonstrate the process of making submissions.
from sklearn.ensemble import RandomForestRegressor
from sklearn.linear_model import Ridge
from sklearn.tree import DecisionTreeRegressor
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import GridSearchCV
# # Create an instance of the classifier
# Regressor = RandomForestRegressor(max_depth=2, random_state=0)
# # model = Ridge(alpha=1.0)
# # Train the classifier
# Regressor = Regressor.fit(X_train, y_train)
# # model = model.fit(X_train, y_train)
# print("The accuracy of the classifier (Random Forset Regression) on the training set is ",
# Regressor.score(X_train, y_train))
# * #### Now let's test our classifier on the validation dataset and see the accuracy.
# print("The accuracy of the classifier (Random Forest Regression) on the validation set is ", (Regressor.score(X_val, y_val)))
# * #### We'll train a Linear Regression Classifier to demonstrate the process of making submissions.
# reg = LinearRegression().fit(X_train, y_train)
# print("The accuracy of the classifier (Linear Regression) on the training set is ", (reg.score(X_train, y_train)))
# * #### Now let's test our Linear Regression classifier on the validation dataset and see the accuracy.
# print("The accuracy of the classifier (Linear Regression) on the validation set is ", (reg.score(X_val, y_val)))
# * #### We'll train a Decision Tree Regression Classifier to demonstrate the process of making submissions.
decisionTree = DecisionTreeRegressor()
param = {"max_depth": [1, 4, 5, 6, 7, 10, 15, 20, 8]}
gridSearch_decisionTree = GridSearchCV(decisionTree, param, scoring="r2", cv=6)
gridSearch_decisionTree.fit(X_train, y_train)
best_DecisionTree = gridSearch_decisionTree.best_estimator_
print(
"The accuracy of the classifier(Decision Tree Regression) on the Training set is ",
best_DecisionTree.score(X_train, y_train),
)
# * #### Now let's test our Decision Tree classifier on the validation dataset and see the accuracy.
print(
"The accuracy of the classifier (Decision Tree Regression) on the validation set is ",
best_DecisionTree.score(X_val, y_val),
)
# # Submission File Generation
# We have built a model and we'd like to submit our predictions on the test set! In order to do that, we'll load the test set, predict the class and save the submission file.
# First, we'll load the data.
test_df = pd.read_csv(os.path.join(dataset_path, "test.csv"))
print(test_df.shape)
test_df.head()
# Note that the test set has the same features and doesn't have the `y` column.
# At this stage one must **NOT** forget to apply the same processing done on the training set on the features of the test set.
# **Note** y is `Rented Bike count (Target), Count of bikes rented at each hour` .
# Now we'll add `y` column to the test `DataFrame` and add the values of the predicted class to it.
# **I'll select the numerical features here as I did in the training set. DO NOT forget to change this step as you change the preprocessing of the training data.**
# ### We will change columns' names to be called easily
test_df.rename(
columns={
"Temperature(�C)": "Temperature",
"Humidity(%)": "Humidity",
"Wind speed (m/s)": "WindSpeed",
"Visibility (10m)": "Visibility",
"Dew point temperature(�C)": "DewPointTemperature",
"Solar Radiation (MJ/m2)": "SolarRadiation",
"Rainfall(mm)": "Rainfall",
"Snowfall (cm)": "Snowfall",
"Functioning Day": "FunctioningDay",
},
inplace=True,
)
test_df.head()
test_df = convert_categorical_columns_into_numerical(
test_df, ["Seasons", "Holiday", "FunctioningDay"]
)
# -----------------------------------------------------------------#
# Another way to convert categorical data into numerical
# Convert columns ('Functioning Day') into (0 , 1)
# df['Functioning Day'] = df['Functioning Day'].replace('Yes', 1)
# df['Functioning Day'] = df['Functioning Day'].replace('No', 0)
# Convert columns ('Holiday') into (0 , 1)
# df['Holiday'] = df['Holiday'].replace('Holiday' , 1)
# df['Holiday'] = df['Holiday'].replace('No Holiday' , 0)
# ------------------------------------------------------------------#
# Convert Date from String to datetime
test_df["Date"] = pd.to_datetime(test_df["Date"], format="%d/%m/%Y")
# Create New feature to get DayOfWeek from DateTime
test_df["DayOfWeek"] = test_df["Date"].dt.dayofweek
# Create new feature which informs us if it is night or day from Hour
test_df["LabelDayNight"] = test_df["Hour"].apply(
lambda x: "Night" if (x > 20 or x < 5) else ("Day")
)
# Convert LabelDayNight Column from Categorical (Day , Night) into Numerical (0,1)
test_df = convert_categorical_columns_into_numerical(test_df, ["LabelDayNight"])
# Get Day's Feature from Date
test_df["Day"] = test_df["Date"].dt.day
# Get Month's Feature from Date
test_df["Month"] = test_df["Date"].dt.month
df.head(600)
X_test = test_df.drop(columns=["ID"])
# You should update/remove the next line once you change the features used for training
X_test = X_test[features]
y_test_predicted = best_DecisionTree.predict(X_test)
test_df["y"] = y_test_predicted.astype(int)
test_df.head()
# # Trail for calculating Root Mean Squared Log Error
# def rmsle(y_pred,y_true):
# log1=np.log(y_pred + 1)
# log2=np.log(y_true + 1)
# se = (log1 - log2) ** 2
# mse=np.mean(se)
# return np.sqrt(mse)
# rmsle(predregrtest,test_df['y'])
# * ##### Now we're ready to generate the submission file. The submission file needs the columns ID and y (number of rented bikes) only.
test_df[["ID", "y"]].to_csv("/kaggle/working/submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/921/69921677.ipynb
| null | null |
[{"Id": 69921677, "ScriptId": 18999146, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4166247, "CreationDate": "08/03/2021 21:12:30", "VersionNumber": 13.0, "Title": "Seoul Bike Rental Prediction notebook to start", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 364.0, "LinesInsertedFromPrevious": 16.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 348.0, "LinesInsertedFromFork": 292.0, "LinesDeletedFromFork": 76.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 72.0, "TotalVotes": 0}]
| null | null | null | null |
# # Predicting Bike Rental count Depending on weather conditions
# # Introduction :
# #### We need to predict the number of Bikes to be rented based on differrent weather conditions and other effective factors
# # Dataset:
# #### The dataset is about bike rental in seoul korea ,The Data contains weather information ,which are the features (Temperature, Humidity, Wind speed, Visibility, Dew point, Solar radiation, Snowfall, Rainfall),the target is the number of bikes rented per hour and date information.
#
# # Description Of Data Set Columns:
# #### - ID - an ID for this instance
# #### - Date - year-month-day
# #### - Hour - Hour of he day
# #### - Temperature - Temperature in Celsius
# #### - Humidity - %
# #### - Windspeed - m/s
# #### - Visibility - 10m
# #### - Dew point temperature - Celsius
# #### - Solar radiation - MJ/m2
# #### - Rainfall - mm
# #### - Snowfall - cm
# #### - Seasons - Winter, Spring, Summer, Autumn
# #### - Holiday - Holiday/No holiday
# #### - Functional Day - NoFunc(Non Functional Hours), Fun(Functional hours)
# #### - y - Rented Bike count (Target), Count of bikes rented at each hour
# **Hint: You can submit up to 2 submissions per day. You can select only one of the submission you make to be considered in the final ranking.**
# # Data Wrangling Process:
# #### The Data Wrangling Process Consists of 3 phases Gathering ,Assessing and Cleaning the data
# # Data Gathering :
# ## Import necessary libraries
# ##### ● We'll use `pandas` to load and manipulate the data. Other libraries will be imported in the relevant sections.
import pandas as pd
import os
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import OrdinalEncoder
from sklearn import preprocessing # This import for data standarization
from sklearn.metrics import mean_squared_log_error
# # Loading Data:
# ## Read train dataset from train.csv file
dataset_path = "/kaggle/input/seoul-bike-rental-ai-pro-iti/"
df = pd.read_csv(os.path.join(dataset_path, "train.csv"))
print("The shape of the dataset is {}.\n\n".format(df.shape))
df.head(5017)
# We've got 5760 examples in the dataset with 15 featues.
# By looking at the features and a sample from the data, the features look of numerical and catogerical types.
# ### We will change columns' names to be called easily
df.rename(
columns={
"Temperature(�C)": "Temperature",
"Humidity(%)": "Humidity",
"Wind speed (m/s)": "WindSpeed",
"Visibility (10m)": "Visibility",
"Dew point temperature(�C)": "DewPointTemperature",
"Solar Radiation (MJ/m2)": "SolarRadiation",
"Rainfall(mm)": "Rainfall",
"Snowfall (cm)": "Snowfall",
"Functioning Day": "FunctioningDay",
},
inplace=True,
)
df.head()
# # Data Cleaning
# ### We need to convert categorical features into numerical data
# Create Function convert categorical data as:(seasons , Holiday , FunctioningDay) into numerical
def convert_categorical_columns_into_numerical(df, columns):
ord_enc = OrdinalEncoder()
for column in columns:
df[column] = ord_enc.fit_transform(df[[column]])
return df
df = convert_categorical_columns_into_numerical(
df, ["Seasons", "Holiday", "FunctioningDay"]
)
# -----------------------------------------------------------------#
# Another way to convert categorical data into numerical
# Convert columns ('Functioning Day') into (0 , 1)
# df['Functioning Day'] = df['Functioning Day'].replace('Yes', 1)
# df['Functioning Day'] = df['Functioning Day'].replace('No', 0)
# Convert columns ('Holiday') into (0 , 1)
# df['Holiday'] = df['Holiday'].replace('Holiday' , 1)
# df['Holiday'] = df['Holiday'].replace('No Holiday' , 0)
# ------------------------------------------------------------------#
# Convert Date from String to datetime
df["Date"] = pd.to_datetime(df["Date"], format="%d/%m/%Y")
# Create New feature to get DayOfWeek from DateTime
df["DayOfWeek"] = df["Date"].dt.dayofweek
# Create new feature which informs us if it is night or day from Hour
df["LabelDayNight"] = df["Hour"].apply(
lambda x: "Night" if (x > 20 or x < 5) else ("Day")
)
# Convert LabelDayNight Column from Categorical (Day , Night) into Numerical (0,1)
df = convert_categorical_columns_into_numerical(df, ["LabelDayNight"])
# Get Day's Feature from Date
df["Day"] = df["Date"].dt.day
# Get Month's Feature from Date
df["Month"] = df["Date"].dt.month
df.head(600)
# ### Trials to convert categorical features into numerical
# print(type (df['Functioning Day']))
# df['Functioning Day'] = df['Functioning Day'].map({'Yes': 1 , 'No':0})
# df['Functioning Day'] = df['Functioning Day']*1
# df['Functioning Day'] = pd.to_numeric(df['Functioning Day'], errors='coerce')
# (df['COL2'] == 'TRUE').astype(int)
# if df['Holiday']== 'No Holiday':
# df['Holiday'] = 1
# else:
# df['Holiday'] = 0
# ## Describe Data
df.drop(columns="ID").describe()
# ## Correlation between features' data and label's data
df.drop(columns="ID").corr().style.background_gradient()
# ### More visualization of data
sns.set(style="ticks", color_codes=True)
g = sns.pairplot(df.drop(columns="ID"))
plt.show()
# Plot every hour repeated until what!
print(df["y"].value_counts().head())
df.y.value_counts().sort_values(ascending=False).head(15).plot(kind="bar")
plt.xlabel("Number of bikes rented")
plt.ylabel("Counting: How many this number repeated")
plt.show()
# #Drawing same data using Pie Chart
# df['y'].value_counts().sort_values(ascending=False).head(5).plot(kind='pie')
# plt.show()
plt.scatter(df["Hour"], df["y"])
plt.xlabel("Hour Number")
plt.ylabel("Number of Bikes")
plt.show()
# sns.barplot(x=df['Hour'].value_counts() , y=df['y'])
# plt.show()
# ### Check Duplicated data
print(df.duplicated())
# Then, there is no redundancy in data
# ### Check null values in Training Data
df.isnull().sum()
# ### Dropping missing values if existed
print(df.count())
df = df.dropna()
df.count()
# Then, there is no missing values existed in data
# ### Detecting outliers, If existed:
# * Delete these outlier data
# Box plot for number of bikes rented
plt.figure(figsize=(10, 6))
sns.boxplot(x=df["y"])
plt.show()
# Box plot for number of hours
plt.figure(figsize=(10, 6))
sns.boxplot(x=df["Hour"])
plt.show()
# So, it is optimal in distribution
Q1 = df.quantile(0.25)
Q3 = df.quantile(0.75)
IQR = Q3 - Q1
print(IQR)
# # Data Splitting
# Now it's time to split the dataset for the training step. Typically the dataset is split into 3 subsets, namely, the training, validation and test sets. In our case, the test set is already predefined. So we'll split the "training" set into training and validation sets with 0.8:0.2 ratio.
# Note: a good way to generate reproducible results is to set the seed to the algorithms that depends on randomization. This is done with the argument random_state in the following command
from sklearn.model_selection import train_test_split
train_df, val_df = train_test_split(
df, test_size=0.2, random_state=42
) # Try adding `stratify` here
X_train = train_df.drop(columns=["ID", "y"])
y_train = train_df["y"]
X_val = val_df.drop(columns=["ID", "y"])
y_val = val_df["y"]
# This cell is used to select the numerical features only. IT SHOULD BE REMOVED AS YOU DO YOUR WORK.
features = [
"Temperature",
"Hour",
"DewPointTemperature",
"Seasons",
"SolarRadiation",
"Humidity",
"Visibility",
"FunctioningDay",
"Month",
"Rainfall",
"LabelDayNight",
]
X_train = X_train[features]
X_val = X_val[features]
# # Model Training
# * #### Let's train a model with the data! We'll train a Random Forest Classifier to demonstrate the process of making submissions.
from sklearn.ensemble import RandomForestRegressor
from sklearn.linear_model import Ridge
from sklearn.tree import DecisionTreeRegressor
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import GridSearchCV
# # Create an instance of the classifier
# Regressor = RandomForestRegressor(max_depth=2, random_state=0)
# # model = Ridge(alpha=1.0)
# # Train the classifier
# Regressor = Regressor.fit(X_train, y_train)
# # model = model.fit(X_train, y_train)
# print("The accuracy of the classifier (Random Forset Regression) on the training set is ",
# Regressor.score(X_train, y_train))
# * #### Now let's test our classifier on the validation dataset and see the accuracy.
# print("The accuracy of the classifier (Random Forest Regression) on the validation set is ", (Regressor.score(X_val, y_val)))
# * #### We'll train a Linear Regression Classifier to demonstrate the process of making submissions.
# reg = LinearRegression().fit(X_train, y_train)
# print("The accuracy of the classifier (Linear Regression) on the training set is ", (reg.score(X_train, y_train)))
# * #### Now let's test our Linear Regression classifier on the validation dataset and see the accuracy.
# print("The accuracy of the classifier (Linear Regression) on the validation set is ", (reg.score(X_val, y_val)))
# * #### We'll train a Decision Tree Regression Classifier to demonstrate the process of making submissions.
decisionTree = DecisionTreeRegressor()
param = {"max_depth": [1, 4, 5, 6, 7, 10, 15, 20, 8]}
gridSearch_decisionTree = GridSearchCV(decisionTree, param, scoring="r2", cv=6)
gridSearch_decisionTree.fit(X_train, y_train)
best_DecisionTree = gridSearch_decisionTree.best_estimator_
print(
"The accuracy of the classifier(Decision Tree Regression) on the Training set is ",
best_DecisionTree.score(X_train, y_train),
)
# * #### Now let's test our Decision Tree classifier on the validation dataset and see the accuracy.
print(
"The accuracy of the classifier (Decision Tree Regression) on the validation set is ",
best_DecisionTree.score(X_val, y_val),
)
# # Submission File Generation
# We have built a model and we'd like to submit our predictions on the test set! In order to do that, we'll load the test set, predict the class and save the submission file.
# First, we'll load the data.
test_df = pd.read_csv(os.path.join(dataset_path, "test.csv"))
print(test_df.shape)
test_df.head()
# Note that the test set has the same features and doesn't have the `y` column.
# At this stage one must **NOT** forget to apply the same processing done on the training set on the features of the test set.
# **Note** y is `Rented Bike count (Target), Count of bikes rented at each hour` .
# Now we'll add `y` column to the test `DataFrame` and add the values of the predicted class to it.
# **I'll select the numerical features here as I did in the training set. DO NOT forget to change this step as you change the preprocessing of the training data.**
# ### We will change columns' names to be called easily
test_df.rename(
columns={
"Temperature(�C)": "Temperature",
"Humidity(%)": "Humidity",
"Wind speed (m/s)": "WindSpeed",
"Visibility (10m)": "Visibility",
"Dew point temperature(�C)": "DewPointTemperature",
"Solar Radiation (MJ/m2)": "SolarRadiation",
"Rainfall(mm)": "Rainfall",
"Snowfall (cm)": "Snowfall",
"Functioning Day": "FunctioningDay",
},
inplace=True,
)
test_df.head()
test_df = convert_categorical_columns_into_numerical(
test_df, ["Seasons", "Holiday", "FunctioningDay"]
)
# -----------------------------------------------------------------#
# Another way to convert categorical data into numerical
# Convert columns ('Functioning Day') into (0 , 1)
# df['Functioning Day'] = df['Functioning Day'].replace('Yes', 1)
# df['Functioning Day'] = df['Functioning Day'].replace('No', 0)
# Convert columns ('Holiday') into (0 , 1)
# df['Holiday'] = df['Holiday'].replace('Holiday' , 1)
# df['Holiday'] = df['Holiday'].replace('No Holiday' , 0)
# ------------------------------------------------------------------#
# Convert Date from String to datetime
test_df["Date"] = pd.to_datetime(test_df["Date"], format="%d/%m/%Y")
# Create New feature to get DayOfWeek from DateTime
test_df["DayOfWeek"] = test_df["Date"].dt.dayofweek
# Create new feature which informs us if it is night or day from Hour
test_df["LabelDayNight"] = test_df["Hour"].apply(
lambda x: "Night" if (x > 20 or x < 5) else ("Day")
)
# Convert LabelDayNight Column from Categorical (Day , Night) into Numerical (0,1)
test_df = convert_categorical_columns_into_numerical(test_df, ["LabelDayNight"])
# Get Day's Feature from Date
test_df["Day"] = test_df["Date"].dt.day
# Get Month's Feature from Date
test_df["Month"] = test_df["Date"].dt.month
df.head(600)
X_test = test_df.drop(columns=["ID"])
# You should update/remove the next line once you change the features used for training
X_test = X_test[features]
y_test_predicted = best_DecisionTree.predict(X_test)
test_df["y"] = y_test_predicted.astype(int)
test_df.head()
# # Trail for calculating Root Mean Squared Log Error
# def rmsle(y_pred,y_true):
# log1=np.log(y_pred + 1)
# log2=np.log(y_true + 1)
# se = (log1 - log2) ** 2
# mse=np.mean(se)
# return np.sqrt(mse)
# rmsle(predregrtest,test_df['y'])
# * ##### Now we're ready to generate the submission file. The submission file needs the columns ID and y (number of rented bikes) only.
test_df[["ID", "y"]].to_csv("/kaggle/working/submission.csv", index=False)
| false | 0 | 3,962 | 0 | 3,962 | 3,962 |
||
69921958
|
# 总感觉验证的时候算mAP有问题。为了看看问题出在哪,我拿了两张图出来,把预测结果直接替换成已知的bbox (debug_pred.json),理论上mAP得为1。但两张图分开算但时候是1,放一起评估却出了问题,网上搜了一圈也没意识到问题在哪。
import os, json
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from pycocotools.coco import COCO
from pycocotools.cocoeval import COCOeval
input_dir = "../input/debug-coco"
coco_gt = COCO(os.path.join(input_dir, "debug.json"))
coco_dt = coco_gt.loadRes(os.path.join(input_dir, "debug_pred.json"))
imgIds = coco_gt.getImgIds()
imgIds
cocoEval = COCOeval(coco_gt, coco_dt, "bbox")
cocoEval.params.imgIds = imgIds[0]
cocoEval.evaluate()
cocoEval.accumulate()
cocoEval.summarize()
cocoEval = COCOeval(coco_gt, coco_dt, "bbox")
cocoEval.params.imgIds = imgIds[1]
cocoEval.evaluate()
cocoEval.accumulate()
cocoEval.summarize()
cocoEval = COCOeval(coco_gt, coco_dt, "bbox")
cocoEval.params.imgIds = imgIds
cocoEval.evaluate()
cocoEval.accumulate()
cocoEval.summarize()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/921/69921958.ipynb
| null | null |
[{"Id": 69921958, "ScriptId": 19120771, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7212725, "CreationDate": "08/03/2021 21:14:42", "VersionNumber": 1.0, "Title": "How does the coco eval work?", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 36.0, "LinesInsertedFromPrevious": 36.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# 总感觉验证的时候算mAP有问题。为了看看问题出在哪,我拿了两张图出来,把预测结果直接替换成已知的bbox (debug_pred.json),理论上mAP得为1。但两张图分开算但时候是1,放一起评估却出了问题,网上搜了一圈也没意识到问题在哪。
import os, json
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from pycocotools.coco import COCO
from pycocotools.cocoeval import COCOeval
input_dir = "../input/debug-coco"
coco_gt = COCO(os.path.join(input_dir, "debug.json"))
coco_dt = coco_gt.loadRes(os.path.join(input_dir, "debug_pred.json"))
imgIds = coco_gt.getImgIds()
imgIds
cocoEval = COCOeval(coco_gt, coco_dt, "bbox")
cocoEval.params.imgIds = imgIds[0]
cocoEval.evaluate()
cocoEval.accumulate()
cocoEval.summarize()
cocoEval = COCOeval(coco_gt, coco_dt, "bbox")
cocoEval.params.imgIds = imgIds[1]
cocoEval.evaluate()
cocoEval.accumulate()
cocoEval.summarize()
cocoEval = COCOeval(coco_gt, coco_dt, "bbox")
cocoEval.params.imgIds = imgIds
cocoEval.evaluate()
cocoEval.accumulate()
cocoEval.summarize()
| false | 0 | 377 | 0 | 377 | 377 |
||
69921184
|
<jupyter_start><jupyter_text>House Prices and Images - SoCal
### Context
I created this dataset to predict the house price from its image(s). It has the price and corresponding image. Each house has only one image.
### Content
The data contains 7 columns and over 15000 rows.
Image_id refers to the image in the image folder.
n_citi is the label encode of the citi column.
To clean unwanted images, please go to this link.
github.com/ted2020/House-Price-Prediction-via-Computer-Vision
### Inspiration
I hope to predict the price of a house from its images. For now, the dataset only includes the exterior images of a house.
Kaggle dataset identifier: house-prices-and-images-socal
<jupyter_script># ### **CNN + MLP Model in PyTorch**
# ### **Introduction**
# The following model is intended for predicting house prices based on input images and corresponding numeric data such as beds, baths, etc. There are a few options for achieving this, and here we have chosen to use a hybrid CNN + MLP model. The CNN routinely handles the images, and the numeric data is handled by the MLP. The outputs of the MLP and the fully connected (FC) layers of the CNN are concatenated into another set of FC layers, which have a final output of a price prediction in the continuous range of $[0,1]$. The data used (2000 for training and 1000 for validation) is a cleaned subset of the original which is about 12000. This was done mainly for convenience to speed up computing time. The model is also purposely lightweight in order to run efficiently with the goal of squeezing as much performance as possible out of the system, without large computational demands. Due to the small model size, a 2X performance improvement is obtained with GPU, but not much more. Note that 20 epochs takes about 4 minutes on GPU.
# 
# -*- coding: utf-8 -*-
# Created on Wed Jun 9 2021
# @author: mbadal1996
# ====================================================
# CNN + MLP Model for Image + Numeric Data
# ====================================================
# Comments:
# The following Python code is a hybrid CNN + MLP
# model for combined image data + numeric features
# (meta-data) which further describe the images.The
# output of the model is a continuous float value in the
# range [0,1] which is due to normalization of the
# training label. In that sense it is a regression as
# opposed to a classification. The original purpose of
# the code was to make predictions on housing prices
# (see So-Cal Housing in Kaggle) but this kind of hybrid
# model is useful for various other problems where
# both images and numeric features are combined. In the
# event that a binary or multi-class output is desired
# (instead of a float value regression), then the final
# output layer of the CNN+MLP should be modified for the
# number of classes and then passed through a softmax
# function.
# As an example, the house features (numeric data) CSV
# file is also included in the repository so that the
# user can see the format. House images are not included
# since they are too many and can be easily downloaded
# from Kaggle at:
# https://www.kaggle.com/ted8080/house-prices-and-images-socal
# Useful content at PyTorch forum is acknowledged for
# combining images and numeric data features.
# -----------------------------------------------------
# IMPORTANT NOTE:
# When organizing data in folders to be input to
# dataloader, it is important to keep in mind the
# following for correct loading:
# 1) The train and validation data were separated into
# their own folders by hand by class (one class: house) c
# alled 'socal_pics/train' and 'socal_pics/val'. That
# means the sub-folder 'train' contains one folder: house.
# The same is true for the val data held in the folder
# 'socal_pics/val'. So the organization looks like:
# socal_pics > train > house
# socal_pics > val > house
# Place the metadat CSV file in same folder as Python
# script
# 2) The test data is organized differently since there
# are no labels for those images. Instead, the test data
# are held in the folder 'socal_pics/test' where the
# sub-folder here 'test' just contains one folder called
# 'test'. This is instead of the 'house' folder. So the
# organization looks like:
# socal_pics > test > test
# ======================================================
# ### **Imports and Parameters**
# Below we perform standard imports, choose computing device, define parameters, and set a seed for reproducible results. Note that due to this, the shuffle feature in the data loader is chosen as False to prevent overriding seed. This also ensures that the numeric data loader (defined below) will be in step with the image loader. Approximate timer is also initialized.
# ====================================================
# Python
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import time
# Pytorch
import torch
from torchvision import datasets
import torchvision.transforms as transforms
import torch.nn.functional as F
# Choose Device (GPU or CPU)
dev = "cuda"
# dev = 'cpu'
# =====================================================
# Parameters
# Image Parameters
CH = 3 # number of channels
ratio = 1.5625 # width/height ratio to resize images
imagewidth = 157 # square dimension (size x size)
imageheight = int(np.floor(imagewidth / ratio))
cropsize = imageheight
# cropsize = imagewidth
# Neural Net Parameters
learn_rate = 1e-3
num_epochs = 20 # At least 20 epochs for 100x100 images
batch_size = 100
# Seed for reproduceable random numbers (eg weights
# and biases). NOTE: Seed will be overidden by using
# image transforms like random flip or setting
# shuffle = True in data loader.
torch.manual_seed(1234)
# Start Timing Code (only approximate)
tic = time.perf_counter()
# ======================================================
# ### **Image Transforms**
# Below we set up transforms for the housing images which may or may not be of equal size (which they must be for the CNN). They also need to be reduced in resolution and cropped. Simply for convenience, a size of 100x100 pixels is chosen according to the parameters above. Note that 157/1.5625 = 100.
# The images are not color normalized but this is a potentially useful addition that can be employed to improve model learning.
# ======================================================
# Image Transforms:
# Create transforms for training data augmentation.
# In each epoch, random transforms will be applied
# according to the Compose function. They are random
# since we are explicitly choosing "Random" versions
# of the transforms. To "increase the dataset" one
# should run more epochs, since each epoch has new
# random data.
# NOTE: Augmentation should only be for train data.
# NOTE: For augmentation transforms, best to use
# larger batches
# Transform for training data
transform_train = transforms.Compose(
[
transforms.Resize([imageheight, imagewidth]),
transforms.CenterCrop(cropsize),
# transforms.RandomHorizontalFlip(p=0.5),
# transforms.RandomRotation(degrees = (-20,20)),
# transforms.RandomVerticalFlip(p=0.5),
transforms.ToTensor(),
]
)
# Transform for validation data
transform_val = transforms.Compose(
[
transforms.Resize([imageheight, imagewidth]),
transforms.CenterCrop(cropsize),
transforms.ToTensor(),
]
)
# Transform for test data
# transform_test = transforms.Compose([
# transforms.Resize([imageheight, imagewidth]),
# transforms.CenterCrop(cropsize),
# transforms.ToTensor()])
# ### **Import Numeric Data**
# Below we import the CSV numeric data into a dataframe and then extract the needed values into appropriate tensors. Following this, the data is normalized on a scale of $[0,1]$. One can also standardize the data instead using e.g. the z-norm. Writing a function to handle the repetitive operations below would be a useful modification to the code.
# ====================================================
# DATA IMPORT
# Import train,val, and test data and set up data
# loader. Note that ImageFolder will organize data
# according to class labels of the folders "house,
# etc" as found in the train and val data folder.
# NOTE: When calling a specific image (such as 135)
# from train data, the first XXX images are class 0,
# then the next YYY are class 1, and etc. if more
# than one class existed (which is not the case here).
# Import CSV of Housing Data
# Read Data from File; Create Tensors for train,test,val
rawdata = pd.read_csv("../input/socal-data-cleaned/socal2_cleaned_mod.csv")
# Import all columns in CSV
Xraw = np.column_stack(
(
rawdata["image_id"].values,
rawdata["n_citi"].values,
rawdata["bed"].values,
rawdata["bath"].values,
rawdata["sqft"].values,
rawdata["price"].values,
)
)
# ====================================================
# Prepare Training Data
# ====================================================
# NOTE: Normalization was done after splitting data.
# ====================================================
Xraw_train = Xraw[0:2000, :] # Get required train data
# city_data_train = Xraw_train[:,1] # import city data
bdrm_data_train = Xraw_train[:, 2] # import bdrm data
bath_data_train = Xraw_train[:, 3] # import bath data
sqft_data_train = Xraw_train[:, 4] # import sqft data
yraw_true_train = Xraw_train[:, 5] # import price
# NORMALIZE DATA (COULD STANDARDIZE INSTEAD)
# Normalize data based to scale [0,1]. Could also
# standardize as z = (x - mean)/stddev
# city_train_norm = city_data_train/np.max(city_data_train)
bdrm_train_norm = bdrm_data_train / np.max(bdrm_data_train)
bath_train_norm = bath_data_train / np.max(bath_data_train)
sqft_train_norm = sqft_data_train / np.max(sqft_data_train)
y_true_train_norm = yraw_true_train / np.max(yraw_true_train)
# Convert to torch tensor
# city_train = torch.from_numpy(city_train_norm).float()
bdrm_train = torch.from_numpy(bdrm_train_norm).float()
bath_train = torch.from_numpy(bath_train_norm).float()
sqft_train = torch.from_numpy(sqft_train_norm).float()
y_train = torch.from_numpy(y_true_train_norm).float()
# Combine sqft, bdrm, etc into one meta_data
meta_train = torch.stack((bdrm_train, bath_train, sqft_train), dim=1)
# ===================================================
# Prepare Validation Data
# ===================================================
# NOTE: Normalization was done after splitting data.
# ===================================================
Xraw_val = Xraw[2000:3000, :] # Get required val data
# city_data_val = Xraw_val[:,1] # import city data
bdrm_data_val = Xraw_val[:, 2] # import bdrm data
bath_data_val = Xraw_val[:, 3] # import bath data
sqft_data_val = Xraw_val[:, 4] # import sqft data
yraw_true_val = Xraw_val[:, 5] # import price data
# NORMALIZE DATA (COULD STANDARDIZE INSTEAD)
# Normalize data based to scale [0,1]. Could also
# standardize as z = (x - mean)/stddev
# city_val_norm = city_data_val/np.max(city_data_val)
bdrm_val_norm = bdrm_data_val / np.max(bdrm_data_val)
bath_val_norm = bath_data_val / np.max(bath_data_val)
sqft_val_norm = sqft_data_val / np.max(sqft_data_val)
y_true_val_norm = yraw_true_val / np.max(yraw_true_val)
# Convert to torch tensor
# city_val = torch.from_numpy(city_val_norm).float()
bdrm_val = torch.from_numpy(bdrm_val_norm).float()
bath_val = torch.from_numpy(bath_val_norm).float()
sqft_val = torch.from_numpy(sqft_val_norm).float()
y_val = torch.from_numpy(y_true_val_norm).float()
# Combine sqft, bdrm, etc into one meta_data
meta_val = torch.stack((bdrm_val, bath_val, sqft_val), dim=1)
# ================================================
# ### **Numeric Data Loaders**
# Below we have created data loader functions for pulling out and storing batches of numeric data for train and validation. These batches will be of the same size as the image batches and correspond to the appropriate houses. It is fairly easy to combine into one function but here, for readability, there are two.
# ================================================
# Generate batches of meta_data (house features)
# Metadata (House Feaures) Training Batches
def get_batch_train(batch_size, which_batch, array_len=len(y_train)):
num_batches = int(np.floor(array_len / batch_size))
# Initialize lists
batch_y = []
batch_meta = []
for i in range(num_batches + 1):
batch_y_train = y_train[i * batch_size : (i + 1) * batch_size]
batch_meta_train = meta_train[i * batch_size : (i + 1) * batch_size, :]
# NOTE: batch_y_train and batch_meta_train should
# be enough to pull out batches directly. No need
# to append to lists as below, but is convenient.
batch_y.append(batch_y_train)
batch_meta.append(batch_meta_train)
# Call each batch from ydata_train and metadata_train
ydata_train = torch.FloatTensor(batch_y[which_batch])
metadata_train = torch.FloatTensor(batch_meta[which_batch])
return ydata_train, metadata_train
# Metadata Validation Batches
def get_batch_val(batch_size, which_batch, array_len=len(y_val)):
num_batches = int(np.floor(array_len / batch_size))
# Re-initialize lists
batch_y = []
batch_meta = []
for i in range(num_batches + 1):
batch_y_val = y_val[i * batch_size : (i + 1) * batch_size]
batch_meta_val = meta_val[i * batch_size : (i + 1) * batch_size, :]
# NOTE: batch_y_val and batch_meta_val should
# be enough to pull out batches directly. No need
# to append to lists as below, but is convenient.
batch_y.append(batch_y_val)
batch_meta.append(batch_meta_val)
# Call each batch from ydata_val and metadata_val
ydata_val = torch.FloatTensor(batch_y[which_batch])
metadata_val = torch.FloatTensor(batch_meta[which_batch])
return ydata_val, metadata_val
# ====================================================
# ### **Image Data Loaders**
# Below we create data loaders for the images with shuffle = False, to maintain order with the numeric data loader above.
# ====================================================
# Create Image Data Loader for Train,Validation,Test
# Training Data
images_train = datasets.ImageFolder(
"../input/socal-data-cleaned/socal_pics/train", transform=transform_train
)
loader_train = torch.utils.data.DataLoader(
images_train, shuffle=False, batch_size=batch_size
)
# Validation Data
images_val = datasets.ImageFolder(
"../input/socal-data-cleaned/socal_pics/val", transform=transform_val
)
loader_val = torch.utils.data.DataLoader(
images_val, shuffle=False, batch_size=batch_size
)
# Testing Data
# Can add testing data loader as well if desired
# ### **CNN + MLP Hybrid Model**
# Below we define the CNN+MLP model architecture. This particular model is set up for images of size 100x100. It was found that larger images were not necessarily helpful in improving performance.
# ==================================================
# CNN + MLP Model Architecture
# ==================================================
# Here we have used a combined CNN + MLP. The CNN
# processes image data and the MLP is employed for
# input/learning of numeric data/features. The
# outputs of each are concatenated to form one
# stream of data.
# NOTE NOTE NOTE: The CNN used in this problem
# takes images of 100x100 pixels if linear input
# layer is X * 22 * 22 or 200x200 pixels
# with X * 47 * 47.
# Two convolution network
class Net(torch.nn.Module):
def __init__(self):
super(Net, self).__init__()
# Image CNN
self.pool = torch.nn.MaxPool2d(2, 2)
self.conv1 = torch.nn.Conv2d(3, 10, 5)
self.conv2 = torch.nn.Conv2d(10, 10, 5)
self.fc1 = torch.nn.Linear(10 * 22 * 22, 120)
self.fc2 = torch.nn.Linear(120, 60)
# Data MLP
# 3 inputs (eg bdrm,bath,sqft) to MLP
self.fc3 = torch.nn.Linear(3, 120)
self.fc4 = torch.nn.Linear(120, 60)
# Cat outputs from CNN + MLP
self.fc5 = torch.nn.Linear(60 + 60, 120)
# 1 output (price) from CNN+MLP
self.fc6 = torch.nn.Linear(120, 1)
# NOTE: output is trained as a regression value
# (continuous), in the range [0,1].
def forward(self, x1, x2):
# Image CNN
x1 = self.pool(F.relu(self.conv1(x1)))
x1 = self.pool(F.relu(self.conv2(x1)))
x1 = x1.view(-1, 10 * 22 * 22)
x1 = F.relu(self.fc1(x1))
x1 = F.relu(self.fc2(x1))
# Data MLP
x2 = x2.view(-1, 3)
x2 = F.relu(self.fc3(x2))
x2 = F.relu(self.fc4(x2))
# Cat outputs from CNN + MLP
x3 = torch.cat((x1, x2), dim=1)
x3 = F.relu(self.fc5(x3))
x3 = self.fc6(x3)
return x3
# ### **Loss Function and Optimizer**
# Below we define an instance of the model class 'Net', choose the loss function, and the optimizer. The particular choice of MSE loss is due to the requirement of a regression (continuous) output from the model, which is the price prediction.
# ==============================================
# ==============================================
# Call instance of CNN+MLP NN class
model = Net().to(dev)
# MSE loss func since NN output is contin. in [0,1]
loss_fn = torch.nn.MSELoss(reduction="mean")
# Optimizer used to train parameters
optimizer = torch.optim.Adam(model.parameters(), lr=learn_rate)
# ===============================================
# ### **Training Loop**
# Below is the model training loop as well as the validation loop in each epoch. The average loss values and percent error are computed per epoch and stored in tensors for later plotting after training.
# ===============================================
# Initialize tensor to store loss values
result_vals = torch.zeros(num_epochs, 4)
count_train = 0 # Initialize Counter
count_val = 0
# X_test,y_true_test = loader_test
print(" ")
print("epoch | loss_train | loss_val | err_train | err_val")
print("-------------------------------------------------------")
error_train = torch.zeros(batch_size)
error_val = torch.zeros(batch_size)
for epoch in range(num_epochs):
# New epoch begins
running_loss_train = 0
running_loss_val = 0
running_error_train = 0
running_error_val = 0
num_batches_train = 0
num_batches_val = 0
count_train = 0
count_val = 0
j = 0 # Initialize batch counter
k = 0 # Initialize batch counter
model.train() # Set torch to train
for X_train, _ in loader_train:
X_train = X_train.to(dev)
# (X,y) is a mini-batch:
# X size Nx3xHxW (N: batch_size, 3: three ch )
# y size N
# Get metadata in batches from function
ydata_train, metadata_train = get_batch_train(batch_size, j)
ydata_train, metadata_train = ydata_train.to(dev), metadata_train.to(dev)
# reset gradients to zero
optimizer.zero_grad()
# run model and compute loss
N, C, nX, nY = X_train.size()
y_pred_train = model(X_train.view(N, C, nX, nY), metadata_train)
loss_train = loss_fn(y_pred_train.squeeze(), ydata_train)
# Back propagation
loss_train.backward()
# Update the parameters
optimizer.step()
# Compute and update loss for entire training set
running_loss_train += loss_train.cpu().detach().numpy()
num_batches_train += 1
for i in range(len(ydata_train)):
error_train[i] = (
abs(ydata_train[i].item() - y_pred_train.squeeze()[i].item())
/ ydata_train[i].item()
)
error_train_sum = sum(error_train)
running_error_train = running_error_train + error_train_sum
j = j + 1 # Step batch counter
k = 0 # Re-initialize batch counter for validation
model.eval() # Set torch for evaluation
for X_val, _ in loader_val:
X_val = X_val.to(dev)
# (X,y) is a mini-batch:
# X size Nx3xHxW (N: batch_size, 3: three ch )
# y size N
# Get metadata in batches from function
ydata_val, metadata_val = get_batch_val(batch_size, k)
ydata_val, metadata_val = ydata_val.to(dev), metadata_val.to(dev)
# run model and compute loss
N, C, nX, nY = X_val.size()
y_pred_val = model(X_val.view(N, C, nX, nY), metadata_val)
loss_val = loss_fn(y_pred_val.squeeze(), ydata_val)
# Compute and update loss for entire val set
running_loss_val += loss_val.cpu().detach().numpy()
num_batches_val += 1
for i in range(len(ydata_val)):
error_val[i] = (
abs(ydata_val[i].item() - y_pred_val.squeeze()[i].item())
/ ydata_val[i].item()
)
error_val_sum = sum(error_val)
running_error_val = running_error_val + error_val_sum
k = k + 1 # Step batch counter
ave_loss_train = running_loss_train / num_batches_train
ave_loss_val = running_loss_val / num_batches_val
ave_error_train = (running_error_train.item() / len(y_train)) * 100
ave_error_val = (running_error_val.item() / len(y_val)) * 100
# ============================================
# Store loss to tensor for plotting
result_vals[epoch, 0] = ave_loss_train
result_vals[epoch, 1] = ave_loss_val
result_vals[epoch, 2] = ave_error_train
result_vals[epoch, 3] = ave_error_val
# Print loss every N epochs
# if epoch % 2 == 1:
print(
epoch,
" ",
round(ave_loss_train.item(), 5),
" ",
round(ave_loss_val.item(), 5),
" ",
round(ave_error_train, 5),
" ",
round(ave_error_val, 5),
)
# =================================================
# ### **Conclusion and Plotting Results**
# Below, the computing time is obtained, followed by plotting the loss and percent error in house prediction. Note that with the given (relatively small) data set and limited features (beds, baths, and rooms), this model reaches about 49 percent error (train) and 40 percent error (validation). Obviously this result can be improved, but it is a reasonable starting point. To interpret, if a house costs 200K dollars, the error is about +/- 80K dollars. A better estimate (and better agreement between train and val) can be achieved with k-fold cross validation due to the small size of data set. More images/data can improve the overall accuracy as well if overfitting are kept under control. The CNN+MLP model may also benefit from employing batch-norm and drop-out. Finally, one important data feature which was not included in the CSV file is the house addresses. If these are converted to numeric GPS coordinates and used for training, it should have a significant impact on prediction accuracy since house price is well-known to be influenced by location. This update to the data is forthcoming.
# =================================================
# End Timing Code
toc = time.perf_counter()
# Measure Time
runtime = toc - tic
print(" ")
print("Computing Time")
print(runtime)
# ==============================================
# Plot Loss and Accuracy for train and val sets
# ==============================================
xvals = torch.linspace(0, num_epochs, num_epochs + 1)
plt.plot(xvals[0:num_epochs].cpu().numpy(), result_vals[:, 0].cpu().detach().numpy())
plt.plot(xvals[0:num_epochs].cpu().numpy(), result_vals[:, 1].cpu().detach().numpy())
plt.legend(["loss_train", "loss_val"], loc="upper right")
# plt.xticks(xvals[0:num_epochs])
plt.title("Loss (CNN + MLP Model)")
plt.xlabel("epochs")
plt.ylabel("loss")
plt.tick_params(right=True, labelright=True)
# plt.savefig('loss.pdf', bbox_inches='tight', dpi=2400)
plt.show()
#
#
# For plotting percent error (which needs to be added above)
plt.plot(xvals[0:num_epochs].cpu().numpy(), result_vals[:, 2].cpu().detach().numpy())
plt.plot(xvals[0:num_epochs].cpu().numpy(), result_vals[:, 3].cpu().detach().numpy())
plt.legend(["error_train", "error_val"], loc="upper right")
# plt.xticks(xvals[0:num_epochs])
plt.title("Percent Error (CNN + MLP Model)")
plt.xlabel("epochs")
plt.ylabel("ave_error")
plt.tick_params(right=True, labelright=True)
# plt.ylim(-0.15, 1.0)
plt.show()
## ==============================================
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/921/69921184.ipynb
|
house-prices-and-images-socal
|
ted8080
|
[{"Id": 69921184, "ScriptId": 19116593, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7645509, "CreationDate": "08/03/2021 21:08:57", "VersionNumber": 3.0, "Title": "cnn_mlp_hybrid", "EvaluationDate": NaN, "IsChange": true, "TotalLines": 635.0, "LinesInsertedFromPrevious": 2.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 633.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 93337665, "KernelVersionId": 69921184, "SourceDatasetVersionId": 824623}]
|
[{"Id": 824623, "DatasetId": 434032, "DatasourceVersionId": 847214, "CreatorUserId": 2886790, "LicenseName": "Unknown", "CreationDate": "12/04/2019 00:56:09", "VersionNumber": 1.0, "Title": "House Prices and Images - SoCal", "Slug": "house-prices-and-images-socal", "Subtitle": "Predict House Price from Images", "Description": "### Context\n\nI created this dataset to predict the house price from its image(s). It has the price and corresponding image. Each house has only one image.\n\n\n### Content\n\nThe data contains 7 columns and over 15000 rows.\nImage_id refers to the image in the image folder.\nn_citi is the label encode of the citi column.\n\nTo clean unwanted images, please go to this link. \ngithub.com/ted2020/House-Price-Prediction-via-Computer-Vision\n\n### Inspiration\n\nI hope to predict the price of a house from its images. For now, the dataset only includes the exterior images of a house.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 434032, "CreatorUserId": 2886790, "OwnerUserId": 2886790.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 824623.0, "CurrentDatasourceVersionId": 847214.0, "ForumId": 446704, "Type": 2, "CreationDate": "12/04/2019 00:56:09", "LastActivityDate": "12/04/2019", "TotalViews": 19202, "TotalDownloads": 1953, "TotalVotes": 29, "TotalKernels": 7}]
|
[{"Id": 2886790, "UserName": "ted8080", "DisplayName": "ted8080", "RegisterDate": "03/03/2019", "PerformanceTier": 1}]
|
# ### **CNN + MLP Model in PyTorch**
# ### **Introduction**
# The following model is intended for predicting house prices based on input images and corresponding numeric data such as beds, baths, etc. There are a few options for achieving this, and here we have chosen to use a hybrid CNN + MLP model. The CNN routinely handles the images, and the numeric data is handled by the MLP. The outputs of the MLP and the fully connected (FC) layers of the CNN are concatenated into another set of FC layers, which have a final output of a price prediction in the continuous range of $[0,1]$. The data used (2000 for training and 1000 for validation) is a cleaned subset of the original which is about 12000. This was done mainly for convenience to speed up computing time. The model is also purposely lightweight in order to run efficiently with the goal of squeezing as much performance as possible out of the system, without large computational demands. Due to the small model size, a 2X performance improvement is obtained with GPU, but not much more. Note that 20 epochs takes about 4 minutes on GPU.
# 
# -*- coding: utf-8 -*-
# Created on Wed Jun 9 2021
# @author: mbadal1996
# ====================================================
# CNN + MLP Model for Image + Numeric Data
# ====================================================
# Comments:
# The following Python code is a hybrid CNN + MLP
# model for combined image data + numeric features
# (meta-data) which further describe the images.The
# output of the model is a continuous float value in the
# range [0,1] which is due to normalization of the
# training label. In that sense it is a regression as
# opposed to a classification. The original purpose of
# the code was to make predictions on housing prices
# (see So-Cal Housing in Kaggle) but this kind of hybrid
# model is useful for various other problems where
# both images and numeric features are combined. In the
# event that a binary or multi-class output is desired
# (instead of a float value regression), then the final
# output layer of the CNN+MLP should be modified for the
# number of classes and then passed through a softmax
# function.
# As an example, the house features (numeric data) CSV
# file is also included in the repository so that the
# user can see the format. House images are not included
# since they are too many and can be easily downloaded
# from Kaggle at:
# https://www.kaggle.com/ted8080/house-prices-and-images-socal
# Useful content at PyTorch forum is acknowledged for
# combining images and numeric data features.
# -----------------------------------------------------
# IMPORTANT NOTE:
# When organizing data in folders to be input to
# dataloader, it is important to keep in mind the
# following for correct loading:
# 1) The train and validation data were separated into
# their own folders by hand by class (one class: house) c
# alled 'socal_pics/train' and 'socal_pics/val'. That
# means the sub-folder 'train' contains one folder: house.
# The same is true for the val data held in the folder
# 'socal_pics/val'. So the organization looks like:
# socal_pics > train > house
# socal_pics > val > house
# Place the metadat CSV file in same folder as Python
# script
# 2) The test data is organized differently since there
# are no labels for those images. Instead, the test data
# are held in the folder 'socal_pics/test' where the
# sub-folder here 'test' just contains one folder called
# 'test'. This is instead of the 'house' folder. So the
# organization looks like:
# socal_pics > test > test
# ======================================================
# ### **Imports and Parameters**
# Below we perform standard imports, choose computing device, define parameters, and set a seed for reproducible results. Note that due to this, the shuffle feature in the data loader is chosen as False to prevent overriding seed. This also ensures that the numeric data loader (defined below) will be in step with the image loader. Approximate timer is also initialized.
# ====================================================
# Python
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import time
# Pytorch
import torch
from torchvision import datasets
import torchvision.transforms as transforms
import torch.nn.functional as F
# Choose Device (GPU or CPU)
dev = "cuda"
# dev = 'cpu'
# =====================================================
# Parameters
# Image Parameters
CH = 3 # number of channels
ratio = 1.5625 # width/height ratio to resize images
imagewidth = 157 # square dimension (size x size)
imageheight = int(np.floor(imagewidth / ratio))
cropsize = imageheight
# cropsize = imagewidth
# Neural Net Parameters
learn_rate = 1e-3
num_epochs = 20 # At least 20 epochs for 100x100 images
batch_size = 100
# Seed for reproduceable random numbers (eg weights
# and biases). NOTE: Seed will be overidden by using
# image transforms like random flip or setting
# shuffle = True in data loader.
torch.manual_seed(1234)
# Start Timing Code (only approximate)
tic = time.perf_counter()
# ======================================================
# ### **Image Transforms**
# Below we set up transforms for the housing images which may or may not be of equal size (which they must be for the CNN). They also need to be reduced in resolution and cropped. Simply for convenience, a size of 100x100 pixels is chosen according to the parameters above. Note that 157/1.5625 = 100.
# The images are not color normalized but this is a potentially useful addition that can be employed to improve model learning.
# ======================================================
# Image Transforms:
# Create transforms for training data augmentation.
# In each epoch, random transforms will be applied
# according to the Compose function. They are random
# since we are explicitly choosing "Random" versions
# of the transforms. To "increase the dataset" one
# should run more epochs, since each epoch has new
# random data.
# NOTE: Augmentation should only be for train data.
# NOTE: For augmentation transforms, best to use
# larger batches
# Transform for training data
transform_train = transforms.Compose(
[
transforms.Resize([imageheight, imagewidth]),
transforms.CenterCrop(cropsize),
# transforms.RandomHorizontalFlip(p=0.5),
# transforms.RandomRotation(degrees = (-20,20)),
# transforms.RandomVerticalFlip(p=0.5),
transforms.ToTensor(),
]
)
# Transform for validation data
transform_val = transforms.Compose(
[
transforms.Resize([imageheight, imagewidth]),
transforms.CenterCrop(cropsize),
transforms.ToTensor(),
]
)
# Transform for test data
# transform_test = transforms.Compose([
# transforms.Resize([imageheight, imagewidth]),
# transforms.CenterCrop(cropsize),
# transforms.ToTensor()])
# ### **Import Numeric Data**
# Below we import the CSV numeric data into a dataframe and then extract the needed values into appropriate tensors. Following this, the data is normalized on a scale of $[0,1]$. One can also standardize the data instead using e.g. the z-norm. Writing a function to handle the repetitive operations below would be a useful modification to the code.
# ====================================================
# DATA IMPORT
# Import train,val, and test data and set up data
# loader. Note that ImageFolder will organize data
# according to class labels of the folders "house,
# etc" as found in the train and val data folder.
# NOTE: When calling a specific image (such as 135)
# from train data, the first XXX images are class 0,
# then the next YYY are class 1, and etc. if more
# than one class existed (which is not the case here).
# Import CSV of Housing Data
# Read Data from File; Create Tensors for train,test,val
rawdata = pd.read_csv("../input/socal-data-cleaned/socal2_cleaned_mod.csv")
# Import all columns in CSV
Xraw = np.column_stack(
(
rawdata["image_id"].values,
rawdata["n_citi"].values,
rawdata["bed"].values,
rawdata["bath"].values,
rawdata["sqft"].values,
rawdata["price"].values,
)
)
# ====================================================
# Prepare Training Data
# ====================================================
# NOTE: Normalization was done after splitting data.
# ====================================================
Xraw_train = Xraw[0:2000, :] # Get required train data
# city_data_train = Xraw_train[:,1] # import city data
bdrm_data_train = Xraw_train[:, 2] # import bdrm data
bath_data_train = Xraw_train[:, 3] # import bath data
sqft_data_train = Xraw_train[:, 4] # import sqft data
yraw_true_train = Xraw_train[:, 5] # import price
# NORMALIZE DATA (COULD STANDARDIZE INSTEAD)
# Normalize data based to scale [0,1]. Could also
# standardize as z = (x - mean)/stddev
# city_train_norm = city_data_train/np.max(city_data_train)
bdrm_train_norm = bdrm_data_train / np.max(bdrm_data_train)
bath_train_norm = bath_data_train / np.max(bath_data_train)
sqft_train_norm = sqft_data_train / np.max(sqft_data_train)
y_true_train_norm = yraw_true_train / np.max(yraw_true_train)
# Convert to torch tensor
# city_train = torch.from_numpy(city_train_norm).float()
bdrm_train = torch.from_numpy(bdrm_train_norm).float()
bath_train = torch.from_numpy(bath_train_norm).float()
sqft_train = torch.from_numpy(sqft_train_norm).float()
y_train = torch.from_numpy(y_true_train_norm).float()
# Combine sqft, bdrm, etc into one meta_data
meta_train = torch.stack((bdrm_train, bath_train, sqft_train), dim=1)
# ===================================================
# Prepare Validation Data
# ===================================================
# NOTE: Normalization was done after splitting data.
# ===================================================
Xraw_val = Xraw[2000:3000, :] # Get required val data
# city_data_val = Xraw_val[:,1] # import city data
bdrm_data_val = Xraw_val[:, 2] # import bdrm data
bath_data_val = Xraw_val[:, 3] # import bath data
sqft_data_val = Xraw_val[:, 4] # import sqft data
yraw_true_val = Xraw_val[:, 5] # import price data
# NORMALIZE DATA (COULD STANDARDIZE INSTEAD)
# Normalize data based to scale [0,1]. Could also
# standardize as z = (x - mean)/stddev
# city_val_norm = city_data_val/np.max(city_data_val)
bdrm_val_norm = bdrm_data_val / np.max(bdrm_data_val)
bath_val_norm = bath_data_val / np.max(bath_data_val)
sqft_val_norm = sqft_data_val / np.max(sqft_data_val)
y_true_val_norm = yraw_true_val / np.max(yraw_true_val)
# Convert to torch tensor
# city_val = torch.from_numpy(city_val_norm).float()
bdrm_val = torch.from_numpy(bdrm_val_norm).float()
bath_val = torch.from_numpy(bath_val_norm).float()
sqft_val = torch.from_numpy(sqft_val_norm).float()
y_val = torch.from_numpy(y_true_val_norm).float()
# Combine sqft, bdrm, etc into one meta_data
meta_val = torch.stack((bdrm_val, bath_val, sqft_val), dim=1)
# ================================================
# ### **Numeric Data Loaders**
# Below we have created data loader functions for pulling out and storing batches of numeric data for train and validation. These batches will be of the same size as the image batches and correspond to the appropriate houses. It is fairly easy to combine into one function but here, for readability, there are two.
# ================================================
# Generate batches of meta_data (house features)
# Metadata (House Feaures) Training Batches
def get_batch_train(batch_size, which_batch, array_len=len(y_train)):
num_batches = int(np.floor(array_len / batch_size))
# Initialize lists
batch_y = []
batch_meta = []
for i in range(num_batches + 1):
batch_y_train = y_train[i * batch_size : (i + 1) * batch_size]
batch_meta_train = meta_train[i * batch_size : (i + 1) * batch_size, :]
# NOTE: batch_y_train and batch_meta_train should
# be enough to pull out batches directly. No need
# to append to lists as below, but is convenient.
batch_y.append(batch_y_train)
batch_meta.append(batch_meta_train)
# Call each batch from ydata_train and metadata_train
ydata_train = torch.FloatTensor(batch_y[which_batch])
metadata_train = torch.FloatTensor(batch_meta[which_batch])
return ydata_train, metadata_train
# Metadata Validation Batches
def get_batch_val(batch_size, which_batch, array_len=len(y_val)):
num_batches = int(np.floor(array_len / batch_size))
# Re-initialize lists
batch_y = []
batch_meta = []
for i in range(num_batches + 1):
batch_y_val = y_val[i * batch_size : (i + 1) * batch_size]
batch_meta_val = meta_val[i * batch_size : (i + 1) * batch_size, :]
# NOTE: batch_y_val and batch_meta_val should
# be enough to pull out batches directly. No need
# to append to lists as below, but is convenient.
batch_y.append(batch_y_val)
batch_meta.append(batch_meta_val)
# Call each batch from ydata_val and metadata_val
ydata_val = torch.FloatTensor(batch_y[which_batch])
metadata_val = torch.FloatTensor(batch_meta[which_batch])
return ydata_val, metadata_val
# ====================================================
# ### **Image Data Loaders**
# Below we create data loaders for the images with shuffle = False, to maintain order with the numeric data loader above.
# ====================================================
# Create Image Data Loader for Train,Validation,Test
# Training Data
images_train = datasets.ImageFolder(
"../input/socal-data-cleaned/socal_pics/train", transform=transform_train
)
loader_train = torch.utils.data.DataLoader(
images_train, shuffle=False, batch_size=batch_size
)
# Validation Data
images_val = datasets.ImageFolder(
"../input/socal-data-cleaned/socal_pics/val", transform=transform_val
)
loader_val = torch.utils.data.DataLoader(
images_val, shuffle=False, batch_size=batch_size
)
# Testing Data
# Can add testing data loader as well if desired
# ### **CNN + MLP Hybrid Model**
# Below we define the CNN+MLP model architecture. This particular model is set up for images of size 100x100. It was found that larger images were not necessarily helpful in improving performance.
# ==================================================
# CNN + MLP Model Architecture
# ==================================================
# Here we have used a combined CNN + MLP. The CNN
# processes image data and the MLP is employed for
# input/learning of numeric data/features. The
# outputs of each are concatenated to form one
# stream of data.
# NOTE NOTE NOTE: The CNN used in this problem
# takes images of 100x100 pixels if linear input
# layer is X * 22 * 22 or 200x200 pixels
# with X * 47 * 47.
# Two convolution network
class Net(torch.nn.Module):
def __init__(self):
super(Net, self).__init__()
# Image CNN
self.pool = torch.nn.MaxPool2d(2, 2)
self.conv1 = torch.nn.Conv2d(3, 10, 5)
self.conv2 = torch.nn.Conv2d(10, 10, 5)
self.fc1 = torch.nn.Linear(10 * 22 * 22, 120)
self.fc2 = torch.nn.Linear(120, 60)
# Data MLP
# 3 inputs (eg bdrm,bath,sqft) to MLP
self.fc3 = torch.nn.Linear(3, 120)
self.fc4 = torch.nn.Linear(120, 60)
# Cat outputs from CNN + MLP
self.fc5 = torch.nn.Linear(60 + 60, 120)
# 1 output (price) from CNN+MLP
self.fc6 = torch.nn.Linear(120, 1)
# NOTE: output is trained as a regression value
# (continuous), in the range [0,1].
def forward(self, x1, x2):
# Image CNN
x1 = self.pool(F.relu(self.conv1(x1)))
x1 = self.pool(F.relu(self.conv2(x1)))
x1 = x1.view(-1, 10 * 22 * 22)
x1 = F.relu(self.fc1(x1))
x1 = F.relu(self.fc2(x1))
# Data MLP
x2 = x2.view(-1, 3)
x2 = F.relu(self.fc3(x2))
x2 = F.relu(self.fc4(x2))
# Cat outputs from CNN + MLP
x3 = torch.cat((x1, x2), dim=1)
x3 = F.relu(self.fc5(x3))
x3 = self.fc6(x3)
return x3
# ### **Loss Function and Optimizer**
# Below we define an instance of the model class 'Net', choose the loss function, and the optimizer. The particular choice of MSE loss is due to the requirement of a regression (continuous) output from the model, which is the price prediction.
# ==============================================
# ==============================================
# Call instance of CNN+MLP NN class
model = Net().to(dev)
# MSE loss func since NN output is contin. in [0,1]
loss_fn = torch.nn.MSELoss(reduction="mean")
# Optimizer used to train parameters
optimizer = torch.optim.Adam(model.parameters(), lr=learn_rate)
# ===============================================
# ### **Training Loop**
# Below is the model training loop as well as the validation loop in each epoch. The average loss values and percent error are computed per epoch and stored in tensors for later plotting after training.
# ===============================================
# Initialize tensor to store loss values
result_vals = torch.zeros(num_epochs, 4)
count_train = 0 # Initialize Counter
count_val = 0
# X_test,y_true_test = loader_test
print(" ")
print("epoch | loss_train | loss_val | err_train | err_val")
print("-------------------------------------------------------")
error_train = torch.zeros(batch_size)
error_val = torch.zeros(batch_size)
for epoch in range(num_epochs):
# New epoch begins
running_loss_train = 0
running_loss_val = 0
running_error_train = 0
running_error_val = 0
num_batches_train = 0
num_batches_val = 0
count_train = 0
count_val = 0
j = 0 # Initialize batch counter
k = 0 # Initialize batch counter
model.train() # Set torch to train
for X_train, _ in loader_train:
X_train = X_train.to(dev)
# (X,y) is a mini-batch:
# X size Nx3xHxW (N: batch_size, 3: three ch )
# y size N
# Get metadata in batches from function
ydata_train, metadata_train = get_batch_train(batch_size, j)
ydata_train, metadata_train = ydata_train.to(dev), metadata_train.to(dev)
# reset gradients to zero
optimizer.zero_grad()
# run model and compute loss
N, C, nX, nY = X_train.size()
y_pred_train = model(X_train.view(N, C, nX, nY), metadata_train)
loss_train = loss_fn(y_pred_train.squeeze(), ydata_train)
# Back propagation
loss_train.backward()
# Update the parameters
optimizer.step()
# Compute and update loss for entire training set
running_loss_train += loss_train.cpu().detach().numpy()
num_batches_train += 1
for i in range(len(ydata_train)):
error_train[i] = (
abs(ydata_train[i].item() - y_pred_train.squeeze()[i].item())
/ ydata_train[i].item()
)
error_train_sum = sum(error_train)
running_error_train = running_error_train + error_train_sum
j = j + 1 # Step batch counter
k = 0 # Re-initialize batch counter for validation
model.eval() # Set torch for evaluation
for X_val, _ in loader_val:
X_val = X_val.to(dev)
# (X,y) is a mini-batch:
# X size Nx3xHxW (N: batch_size, 3: three ch )
# y size N
# Get metadata in batches from function
ydata_val, metadata_val = get_batch_val(batch_size, k)
ydata_val, metadata_val = ydata_val.to(dev), metadata_val.to(dev)
# run model and compute loss
N, C, nX, nY = X_val.size()
y_pred_val = model(X_val.view(N, C, nX, nY), metadata_val)
loss_val = loss_fn(y_pred_val.squeeze(), ydata_val)
# Compute and update loss for entire val set
running_loss_val += loss_val.cpu().detach().numpy()
num_batches_val += 1
for i in range(len(ydata_val)):
error_val[i] = (
abs(ydata_val[i].item() - y_pred_val.squeeze()[i].item())
/ ydata_val[i].item()
)
error_val_sum = sum(error_val)
running_error_val = running_error_val + error_val_sum
k = k + 1 # Step batch counter
ave_loss_train = running_loss_train / num_batches_train
ave_loss_val = running_loss_val / num_batches_val
ave_error_train = (running_error_train.item() / len(y_train)) * 100
ave_error_val = (running_error_val.item() / len(y_val)) * 100
# ============================================
# Store loss to tensor for plotting
result_vals[epoch, 0] = ave_loss_train
result_vals[epoch, 1] = ave_loss_val
result_vals[epoch, 2] = ave_error_train
result_vals[epoch, 3] = ave_error_val
# Print loss every N epochs
# if epoch % 2 == 1:
print(
epoch,
" ",
round(ave_loss_train.item(), 5),
" ",
round(ave_loss_val.item(), 5),
" ",
round(ave_error_train, 5),
" ",
round(ave_error_val, 5),
)
# =================================================
# ### **Conclusion and Plotting Results**
# Below, the computing time is obtained, followed by plotting the loss and percent error in house prediction. Note that with the given (relatively small) data set and limited features (beds, baths, and rooms), this model reaches about 49 percent error (train) and 40 percent error (validation). Obviously this result can be improved, but it is a reasonable starting point. To interpret, if a house costs 200K dollars, the error is about +/- 80K dollars. A better estimate (and better agreement between train and val) can be achieved with k-fold cross validation due to the small size of data set. More images/data can improve the overall accuracy as well if overfitting are kept under control. The CNN+MLP model may also benefit from employing batch-norm and drop-out. Finally, one important data feature which was not included in the CSV file is the house addresses. If these are converted to numeric GPS coordinates and used for training, it should have a significant impact on prediction accuracy since house price is well-known to be influenced by location. This update to the data is forthcoming.
# =================================================
# End Timing Code
toc = time.perf_counter()
# Measure Time
runtime = toc - tic
print(" ")
print("Computing Time")
print(runtime)
# ==============================================
# Plot Loss and Accuracy for train and val sets
# ==============================================
xvals = torch.linspace(0, num_epochs, num_epochs + 1)
plt.plot(xvals[0:num_epochs].cpu().numpy(), result_vals[:, 0].cpu().detach().numpy())
plt.plot(xvals[0:num_epochs].cpu().numpy(), result_vals[:, 1].cpu().detach().numpy())
plt.legend(["loss_train", "loss_val"], loc="upper right")
# plt.xticks(xvals[0:num_epochs])
plt.title("Loss (CNN + MLP Model)")
plt.xlabel("epochs")
plt.ylabel("loss")
plt.tick_params(right=True, labelright=True)
# plt.savefig('loss.pdf', bbox_inches='tight', dpi=2400)
plt.show()
#
#
# For plotting percent error (which needs to be added above)
plt.plot(xvals[0:num_epochs].cpu().numpy(), result_vals[:, 2].cpu().detach().numpy())
plt.plot(xvals[0:num_epochs].cpu().numpy(), result_vals[:, 3].cpu().detach().numpy())
plt.legend(["error_train", "error_val"], loc="upper right")
# plt.xticks(xvals[0:num_epochs])
plt.title("Percent Error (CNN + MLP Model)")
plt.xlabel("epochs")
plt.ylabel("ave_error")
plt.tick_params(right=True, labelright=True)
# plt.ylim(-0.15, 1.0)
plt.show()
## ==============================================
| false | 1 | 6,791 | 0 | 6,980 | 6,791 |
||
69921176
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
import re
from wordcloud import WordCloud, STOPWORDS, ImageColorGenerator
from geopy.geocoders import Nominatim
from geopy.extra.rate_limiter import RateLimiter
import folium
from folium import plugins
df_train = pd.read_csv("/kaggle/input/nlp-getting-started/train.csv")
df_test = pd.read_csv("/kaggle/input/nlp-getting-started/test.csv")
df_train.head()
df_train.info()
fig = plt.figure(figsize=(12, 9))
ax1 = fig.add_subplot(121)
sns.barplot(
df_train["keyword"].isnull().value_counts().index,
df_train["keyword"].isnull().value_counts().values,
palette="mako",
ax=ax1,
)
ax1.set_title("Missing Values in Keyword")
ax2 = fig.add_subplot(122)
sns.barplot(
df_train["location"].isnull().value_counts().index,
df_train["location"].isnull().value_counts().values,
palette="mako",
ax=ax2,
)
ax2.set_title("Missing Values in Location")
fig.suptitle("Missing Values")
plt.show()
plt.figure(figsize=(12, 9))
sns.barplot(
df_train["target"].value_counts().index, df_train["target"].value_counts().values
)
plt.title("Target Values")
plt.xlabel("0:not disaster|1:disaster")
plt.show()
df_tgroup = df_train.groupby("target").size()
df_tgroup.plot(
kind="pie",
subplots=True,
figsize=(10, 8),
autopct="%.2f%%",
colors=["blue", "green"],
)
plt.title("Pie chart of Target", fontsize=16)
plt.legend()
plt.show()
data = df_train.location.value_counts()[:20]
data = pd.DataFrame(data)
data = data.reset_index()
data.columns = ["location", "counts"]
geolocator = Nominatim(user_agent="Location Map")
geocode = RateLimiter(geolocator.geocode, min_delay_seconds=1)
dict_lat = {}
dict_long = {}
for i in data.location.values:
print(i)
location = geocode(i)
dict_lat[i] = location.latitude
dict_long[i] = location.longitude
data["latitude"] = data.location.map(dict_lat)
data["longitude"] = data.location.map(dict_long)
location_map = folium.Map(location=[7.0, 7.0], zoom_start=2)
markers = 2
for i, row in data.iterrows():
loss = row["counts"]
if row["counts"] > 0:
count = row["counts"] * 0.4
folium.CircleMarker(
[float(row["latitude"]), float(row["longitude"])],
radius=float(count),
color="red",
fill=True,
).add_to(location_map)
location_map
df_test.head()
import nltk
from nltk.stem.wordnet import WordNetLemmatizer
from nltk.tokenize import TweetTokenizer
from nltk.stem.wordnet import WordNetLemmatizer
nltk.download("wordnet")
nltk.download("stopwords")
nltk.download("punkt")
stop_words = nltk.corpus.stopwords.words(["english"])
lem = WordNetLemmatizer()
print(stop_words)
def cleaning(data):
# remove urls
tweet_without_url = re.sub(r"http\S+", " ", data)
# remove hashtags
tweet_without_hashtag = re.sub(r"#\w+", " ", tweet_without_url)
# 3. Remove mentions and characters that not in the English alphabets
tweet_without_mentions = re.sub(r"@\w+", " ", tweet_without_hashtag)
precleaned_tweet = re.sub("[^A-Za-z]+", " ", tweet_without_mentions)
# 2. Tokenize
tweet_tokens = TweetTokenizer().tokenize(precleaned_tweet)
# 3. Remove Puncs
tokens_without_punc = [w for w in tweet_tokens if w.isalpha()]
# 4. Removing Stopwords
tokens_without_sw = [t for t in tokens_without_punc if t not in stop_words]
# 5. lemma
text_cleaned = [lem.lemmatize(t) for t in tokens_without_sw]
# 6. Joining
return " ".join(text_cleaned)
df_train["cleaned_text"] = df_train["text"].apply(cleaning)
df_test["cleaned_text"] = df_test["text"].apply(cleaning)
df_train.head()
df_test
df_train.dropna(how="any", inplace=True, axis=1)
df_test.dropna(how="any", inplace=True, axis=1)
X_train = df_train["cleaned_text"]
y_train = df_train["target"]
X_test = df_test["cleaned_text"]
# from sklearn.model_selection import train_test_split
# X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25, random_state=1)
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
MAX_VOCAB_SIZE = 30000
tokenizer = Tokenizer(num_words=MAX_VOCAB_SIZE)
tokenizer.fit_on_texts(X_train)
# sequences_train = tokenizer.texts_to_sequences(X_train)
# sequences_test = tokenizer.texts_to_sequences(X_test)
X_train = tokenizer.texts_to_sequences(X_train)
X_test = tokenizer.texts_to_sequences(X_test)
vocab_size = len(tokenizer.word_index) + 1
maxlen = 100
X_train = pad_sequences(X_train, padding="post", maxlen=maxlen)
X_test = pad_sequences(X_test, padding="post", maxlen=maxlen)
from keras.models import Sequential
from keras import layers
embedding_dim = 100
model = Sequential()
model.add(layers.Embedding(vocab_size, embedding_dim, input_length=maxlen))
model.add(layers.Conv1D(128, 5, activation="relu"))
model.add(layers.GlobalMaxPooling1D())
model.add(layers.Dropout(0.5))
model.add(layers.Dense(10, activation="relu"))
model.add(layers.Dropout(0.5))
model.add(layers.Dense(1, activation="sigmoid"))
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
history = model.fit(
X_train,
y_train,
epochs=10,
# validation_data=(X_test, y_test),
batch_size=10,
)
model_eval = model.predict_classes(X_test)
target_val = []
for i in model_eval:
target_val.append(i[0])
df_sample = pd.read_csv("/kaggle/input/nlp-getting-started/sample_submission.csv")
df_sample.head()
test_copy = df_test.copy()
submission = pd.DataFrame({"id": test_copy["id"], "target": target_val})
submission.to_csv("submission.csv", index=False)
submission
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/921/69921176.ipynb
| null | null |
[{"Id": 69921176, "ScriptId": 18956013, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4272003, "CreationDate": "08/03/2021 21:08:53", "VersionNumber": 3.0, "Title": "NLP disaster tweets classification", "EvaluationDate": "08/03/2021", "IsChange": false, "TotalLines": 212.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 212.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
import re
from wordcloud import WordCloud, STOPWORDS, ImageColorGenerator
from geopy.geocoders import Nominatim
from geopy.extra.rate_limiter import RateLimiter
import folium
from folium import plugins
df_train = pd.read_csv("/kaggle/input/nlp-getting-started/train.csv")
df_test = pd.read_csv("/kaggle/input/nlp-getting-started/test.csv")
df_train.head()
df_train.info()
fig = plt.figure(figsize=(12, 9))
ax1 = fig.add_subplot(121)
sns.barplot(
df_train["keyword"].isnull().value_counts().index,
df_train["keyword"].isnull().value_counts().values,
palette="mako",
ax=ax1,
)
ax1.set_title("Missing Values in Keyword")
ax2 = fig.add_subplot(122)
sns.barplot(
df_train["location"].isnull().value_counts().index,
df_train["location"].isnull().value_counts().values,
palette="mako",
ax=ax2,
)
ax2.set_title("Missing Values in Location")
fig.suptitle("Missing Values")
plt.show()
plt.figure(figsize=(12, 9))
sns.barplot(
df_train["target"].value_counts().index, df_train["target"].value_counts().values
)
plt.title("Target Values")
plt.xlabel("0:not disaster|1:disaster")
plt.show()
df_tgroup = df_train.groupby("target").size()
df_tgroup.plot(
kind="pie",
subplots=True,
figsize=(10, 8),
autopct="%.2f%%",
colors=["blue", "green"],
)
plt.title("Pie chart of Target", fontsize=16)
plt.legend()
plt.show()
data = df_train.location.value_counts()[:20]
data = pd.DataFrame(data)
data = data.reset_index()
data.columns = ["location", "counts"]
geolocator = Nominatim(user_agent="Location Map")
geocode = RateLimiter(geolocator.geocode, min_delay_seconds=1)
dict_lat = {}
dict_long = {}
for i in data.location.values:
print(i)
location = geocode(i)
dict_lat[i] = location.latitude
dict_long[i] = location.longitude
data["latitude"] = data.location.map(dict_lat)
data["longitude"] = data.location.map(dict_long)
location_map = folium.Map(location=[7.0, 7.0], zoom_start=2)
markers = 2
for i, row in data.iterrows():
loss = row["counts"]
if row["counts"] > 0:
count = row["counts"] * 0.4
folium.CircleMarker(
[float(row["latitude"]), float(row["longitude"])],
radius=float(count),
color="red",
fill=True,
).add_to(location_map)
location_map
df_test.head()
import nltk
from nltk.stem.wordnet import WordNetLemmatizer
from nltk.tokenize import TweetTokenizer
from nltk.stem.wordnet import WordNetLemmatizer
nltk.download("wordnet")
nltk.download("stopwords")
nltk.download("punkt")
stop_words = nltk.corpus.stopwords.words(["english"])
lem = WordNetLemmatizer()
print(stop_words)
def cleaning(data):
# remove urls
tweet_without_url = re.sub(r"http\S+", " ", data)
# remove hashtags
tweet_without_hashtag = re.sub(r"#\w+", " ", tweet_without_url)
# 3. Remove mentions and characters that not in the English alphabets
tweet_without_mentions = re.sub(r"@\w+", " ", tweet_without_hashtag)
precleaned_tweet = re.sub("[^A-Za-z]+", " ", tweet_without_mentions)
# 2. Tokenize
tweet_tokens = TweetTokenizer().tokenize(precleaned_tweet)
# 3. Remove Puncs
tokens_without_punc = [w for w in tweet_tokens if w.isalpha()]
# 4. Removing Stopwords
tokens_without_sw = [t for t in tokens_without_punc if t not in stop_words]
# 5. lemma
text_cleaned = [lem.lemmatize(t) for t in tokens_without_sw]
# 6. Joining
return " ".join(text_cleaned)
df_train["cleaned_text"] = df_train["text"].apply(cleaning)
df_test["cleaned_text"] = df_test["text"].apply(cleaning)
df_train.head()
df_test
df_train.dropna(how="any", inplace=True, axis=1)
df_test.dropna(how="any", inplace=True, axis=1)
X_train = df_train["cleaned_text"]
y_train = df_train["target"]
X_test = df_test["cleaned_text"]
# from sklearn.model_selection import train_test_split
# X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25, random_state=1)
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
MAX_VOCAB_SIZE = 30000
tokenizer = Tokenizer(num_words=MAX_VOCAB_SIZE)
tokenizer.fit_on_texts(X_train)
# sequences_train = tokenizer.texts_to_sequences(X_train)
# sequences_test = tokenizer.texts_to_sequences(X_test)
X_train = tokenizer.texts_to_sequences(X_train)
X_test = tokenizer.texts_to_sequences(X_test)
vocab_size = len(tokenizer.word_index) + 1
maxlen = 100
X_train = pad_sequences(X_train, padding="post", maxlen=maxlen)
X_test = pad_sequences(X_test, padding="post", maxlen=maxlen)
from keras.models import Sequential
from keras import layers
embedding_dim = 100
model = Sequential()
model.add(layers.Embedding(vocab_size, embedding_dim, input_length=maxlen))
model.add(layers.Conv1D(128, 5, activation="relu"))
model.add(layers.GlobalMaxPooling1D())
model.add(layers.Dropout(0.5))
model.add(layers.Dense(10, activation="relu"))
model.add(layers.Dropout(0.5))
model.add(layers.Dense(1, activation="sigmoid"))
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
history = model.fit(
X_train,
y_train,
epochs=10,
# validation_data=(X_test, y_test),
batch_size=10,
)
model_eval = model.predict_classes(X_test)
target_val = []
for i in model_eval:
target_val.append(i[0])
df_sample = pd.read_csv("/kaggle/input/nlp-getting-started/sample_submission.csv")
df_sample.head()
test_copy = df_test.copy()
submission = pd.DataFrame({"id": test_copy["id"], "target": target_val})
submission.to_csv("submission.csv", index=False)
submission
| false | 0 | 2,061 | 1 | 2,061 | 2,061 |
||
69956580
|
<jupyter_start><jupyter_text>Inadimplência de clientes de cartão de crédito
# Variáveis
Esta pesquisa empregou uma variável binária, pagamento padrão (Sim = 1, Não = 0), como variável de resposta. Este estudo revisou a literatura e usou as seguintes 23 variáveis como variáveis explicativas:
X1: Valor do crédito concedido (dólar NT): inclui tanto o crédito ao consumidor pessoa física quanto o crédito familiar (suplementar).
X2: Gênero (1 = masculino; 2 = feminino).
X3: Educação (1 = pós-graduação; 2 = universidade; 3 = ensino médio; 4 = outros).
X4: Estado civil (1 = casado; 2 = solteiro; 3 = outros).
X5: Idade (ano).
X6 - X11: Histórico de pagamentos anteriores. Rastreamos os registros de pagamentos mensais anteriores (de abril a setembro de 2005) da seguinte maneira: X6 = o status de reembolso em setembro de 2005; X7 = situação de amortização em agosto de 2005; . . .; X11 = estado de reembolso em abril de 2005. A escala de medição para o estado de reembolso é: -1 = pagamento em dia; 1 = atraso no pagamento por um mês; 2 = atraso no pagamento por dois meses; . . .; 8 = atraso no pagamento por oito meses; 9 = atraso no pagamento de nove meses ou mais.
X12-X17: Valor da fatura (dólar NT). X12 = valor da fatura em setembro de 2005; X13 = valor da fatura em agosto de 2005; . . .; X17 = valor da fatura em abril de 2005.
X18-X23: Valor do pagamento anterior (dólar NT). X18 = valor pago em setembro de 2005; X19 = valor pago em agosto de 2005; . . .; X23 = valor pago em abril de 2005.
Fonte: https://archive.ics.uci.edu/ml/datasets/default+of+credit+card+clients
Kaggle dataset identifier: inadimplncia-de-clientes-de-carto-de-crdito
<jupyter_code>import pandas as pd
df = pd.read_csv('inadimplncia-de-clientes-de-carto-de-crdito/default of credit card clients - Data (1).csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 30000 entries, 0 to 29999
Data columns (total 25 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 ID 30000 non-null int64
1 LIMIT_BAL 30000 non-null int64
2 SEX 30000 non-null int64
3 EDUCATION 30000 non-null int64
4 MARRIAGE 30000 non-null int64
5 AGE 30000 non-null int64
6 PAY_0 30000 non-null int64
7 PAY_2 30000 non-null int64
8 PAY_3 30000 non-null int64
9 PAY_4 30000 non-null int64
10 PAY_5 30000 non-null int64
11 PAY_6 30000 non-null int64
12 BILL_AMT1 30000 non-null int64
13 BILL_AMT2 30000 non-null int64
14 BILL_AMT3 30000 non-null int64
15 BILL_AMT4 30000 non-null int64
16 BILL_AMT5 30000 non-null int64
17 BILL_AMT6 30000 non-null int64
18 PAY_AMT1 30000 non-null int64
19 PAY_AMT2 30000 non-null int64
20 PAY_AMT3 30000 non-null int64
21 PAY_AMT4 30000 non-null int64
22 PAY_AMT5 30000 non-null int64
23 PAY_AMT6 30000 non-null int64
24 default payment next month 30000 non-null int64
dtypes: int64(25)
memory usage: 5.7 MB
<jupyter_text>Examples:
{
"ID": 1,
"LIMIT_BAL": 20000,
"SEX": 2,
"EDUCATION": 2,
"MARRIAGE": 1,
"AGE": 24,
"PAY_0": 2,
"PAY_2": 2,
"PAY_3": -1,
"PAY_4": -1,
"PAY_5": -2,
"PAY_6": -2,
"BILL_AMT1": 3913,
"BILL_AMT2": 3102,
"BILL_AMT3": 689,
"BILL_AMT4": 0,
"BILL_AMT5": 0,
"BILL_AMT6": 0,
"PAY_AMT1": 0,
"PAY_AMT2": 689,
"...": "and 5 more columns"
}
{
"ID": 2,
"LIMIT_BAL": 120000,
"SEX": 2,
"EDUCATION": 2,
"MARRIAGE": 2,
"AGE": 26,
"PAY_0": -1,
"PAY_2": 2,
"PAY_3": 0,
"PAY_4": 0,
"PAY_5": 0,
"PAY_6": 2,
"BILL_AMT1": 2682,
"BILL_AMT2": 1725,
"BILL_AMT3": 2682,
"BILL_AMT4": 3272,
"BILL_AMT5": 3455,
"BILL_AMT6": 3261,
"PAY_AMT1": 0,
"PAY_AMT2": 1000,
"...": "and 5 more columns"
}
{
"ID": 3,
"LIMIT_BAL": 90000,
"SEX": 2,
"EDUCATION": 2,
"MARRIAGE": 2,
"AGE": 34,
"PAY_0": 0,
"PAY_2": 0,
"PAY_3": 0,
"PAY_4": 0,
"PAY_5": 0,
"PAY_6": 0,
"BILL_AMT1": 29239,
"BILL_AMT2": 14027,
"BILL_AMT3": 13559,
"BILL_AMT4": 14331,
"BILL_AMT5": 14948,
"BILL_AMT6": 15549,
"PAY_AMT1": 1518,
"PAY_AMT2": 1500,
"...": "and 5 more columns"
}
{
"ID": 4,
"LIMIT_BAL": 50000,
"SEX": 2,
"EDUCATION": 2,
"MARRIAGE": 1,
"AGE": 37,
"PAY_0": 0,
"PAY_2": 0,
"PAY_3": 0,
"PAY_4": 0,
"PAY_5": 0,
"PAY_6": 0,
"BILL_AMT1": 46990,
"BILL_AMT2": 48233,
"BILL_AMT3": 49291,
"BILL_AMT4": 28314,
"BILL_AMT5": 28959,
"BILL_AMT6": 29547,
"PAY_AMT1": 2000,
"PAY_AMT2": 2019,
"...": "and 5 more columns"
}
<jupyter_script>## Trabalho Data Mining - Parte 1: Análise Preliminar da Dase de Dados:
###Importação das bibliotecas e random seed
# Importação dos pacotes
import pandas as pd
import numpy as np
import random
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib.patches as mpatches
# Seed para reprodução de resultados
seed = 1
random.seed(seed)
np.random.seed(seed)
### Importação da base de dados
data = pd.read_csv(
r"../input/inadimplncia-de-clientes-de-carto-de-crdito/default of credit card clients - Data (1).csv"
)
data.head()
# Alterando nome de default payment next month para defaulted para poder fazer as:
data = data.rename(columns={"default payment next month": "defaulted"})
len(data)
### Distribuição
#### Sexo
# Distribuição por Sexo
sexo = data.SEX
sexo = sexo.replace(to_replace=1, value="Homem")
sexo = sexo.replace(to_replace=2, value="Mulher")
sexo = sexo.to_frame()
graph = sns.countplot(
data=sexo, x="SEX", palette="muted", order=sexo["SEX"].value_counts().index
)
plt.title("Distribuição por Sexo")
for idx, bar in enumerate(graph.patches):
height = bar.get_height()
graph.text(
x=bar.get_x() + bar.get_width() / 2.0,
y=height + 500,
s="{:.2%}".format(sexo["SEX"].value_counts(normalize=True)[idx]),
ha="center",
)
graph.set(xlabel=None)
plt.xlabel("Sexo")
plt.ylabel("Quantidade")
plt.ylim(0, 30000)
plt.show()
#### Educação
# Distribuição por Educação
edu = data.EDUCATION
edu = edu.replace(to_replace=0, value="Não Especificado")
edu = edu.replace(to_replace=1, value="Pós-Graduação")
edu = edu.replace(to_replace=2, value="Universidade")
edu = edu.replace(to_replace=3, value="Ensino Médio")
edu = edu.replace(to_replace=4, value="Outros")
edu = edu.replace(to_replace=5, value="Não Especificado")
edu = edu.replace(to_replace=6, value="Não Especificado")
edu = edu.to_frame()
plt.subplots(1, figsize=(10, 5))
graph = sns.countplot(
data=edu,
x="EDUCATION",
palette="muted",
order=edu["EDUCATION"].value_counts().index,
)
for idx, bar in enumerate(graph.patches):
height = bar.get_height()
graph.text(
x=bar.get_x() + bar.get_width() / 2.0,
y=height + 500,
s="{:.2%}".format(edu["EDUCATION"].value_counts(normalize=True)[idx]),
ha="center",
)
plt.title("Distribuição por Nivel Formação")
plt.xlabel("Nivel Escolaridade")
plt.ylabel("Quantidade")
plt.xticks(rotation=45)
graph.set(xlabel=None)
plt.ylim(0, 30000)
plt.show()
#### Estado Civil
# Distribuição por Estado Civil
marriage = data.MARRIAGE
marriage = marriage.replace(to_replace=1, value="Casado")
marriage = marriage.replace(to_replace=2, value="Solteiro")
marriage = marriage.replace(to_replace=3, value="Outros")
marriage = marriage.replace(to_replace=0, value="Não Especificado")
marriage = marriage.to_frame()
plt.subplots(1, figsize=(10, 5))
graph = sns.countplot(
data=marriage,
x="MARRIAGE",
palette="muted",
order=marriage["MARRIAGE"].value_counts().index,
)
for idx, bar in enumerate(graph.patches):
height = bar.get_height()
graph.text(
x=bar.get_x() + bar.get_width() / 2.0,
y=height + 500,
s="{:.2%}".format(marriage["MARRIAGE"].value_counts(normalize=True)[idx]),
ha="center",
)
plt.title("Distribuição por Estado Civil")
plt.xlabel("Nivel Escolaridade")
plt.ylabel("Quantidade")
graph.set(xlabel=None)
plt.ylim(0, 30000)
plt.show()
#### Faixa etária
age = data[["AGE", "defaulted"]]
plt.hist(age["AGE"], bins=20, rwidth=0.9)
plt.title("Distribuição por Faixa Etária")
plt.plot()
# create a list of our conditions
conditions = [
(age["AGE"] < 30),
(age["AGE"] >= 30) & (age["AGE"] < 40),
(age["AGE"] >= 40) & (age["AGE"] < 50),
(age["AGE"] >= 50) & (age["AGE"] < 60),
(age["AGE"] >= 60) & (age["AGE"] < 70),
(age["AGE"] >= 70) & (age["AGE"] < 80),
(age["AGE"] >= 80),
]
# create a list of the values we want to assign for each condition
values = ["20s", "30s", "40s", "50s", "60s", "70s", "maior ou igual a 80"]
# create a new column and use np.select to assign values to it using our lists as arguments
age["new_Age"] = np.select(conditions, values)
# display updated DataFrame
age.head()
age["new_Age"].value_counts()
# Distribuição por Faixa Etária
plt.subplots(1, figsize=(10, 5))
graph = sns.countplot(
data=age, x="new_Age", palette="muted", order=age["new_Age"].value_counts().index
)
for idx, bar in enumerate(graph.patches):
height = bar.get_height()
graph.text(
x=bar.get_x() + bar.get_width() / 2.0,
y=height + 500,
s="{:.2%}".format(age["new_Age"].value_counts(normalize=True)[idx]),
ha="center",
)
plt.title("Distribuição por faixa etária")
plt.xlabel("Faixa etária")
plt.ylabel("Quantidade")
graph.set(xlabel=None)
plt.ylim(0, 30000)
plt.show()
### Distribuição Vs Variável resposta
#### Sexo
# set the figure size
plt.figure(figsize=(8, 6))
sexo = data[["SEX", "defaulted"]]
sexo = sexo.replace({"SEX": {1: "Homem", 2: "Mulher"}})
# from raw value to percentage
total = sexo.groupby("SEX")["defaulted"].count().reset_index()
pay = sexo[sexo.defaulted == 1].groupby("SEX").count().reset_index()
pay["defaulted"] = [i / j * 100 for i, j in zip(pay["defaulted"], total["defaulted"])]
total["defaulted"] = [
i / j * 100 for i, j in zip(total["defaulted"], total["defaulted"])
]
# bar chart 1 -> top bars (group of 'defaulted = 0')
bar1 = sns.barplot(x="SEX", y="defaulted", data=total, color="lightgray")
# bar chart 2 -> bottom bars (group of 'defaulted = 1')
bar2 = sns.barplot(x="SEX", y="defaulted", data=pay, color="green")
# add legend
top_bar = mpatches.Patch(color="lightgray", label="Não Pagou")
bottom_bar = mpatches.Patch(color="green", label="Pagou")
plt.legend(handles=[top_bar, bottom_bar])
plt.xlabel("Sexo")
plt.ylabel("100% da Análise")
# show the graph
plt.show()
#### Educação
# set the figure size
plt.figure(figsize=(18, 6))
edu = data[["EDUCATION", "defaulted"]]
edu = edu.replace(
{
"EDUCATION": {
0: "5-NE",
1: "3-Pós Graduação",
2: "2-Universidade",
3: "1-Ensino Médio",
4: "4-Outros",
5: "5-NE",
6: "5-NE",
}
}
)
# from raw value to percentage
total = edu.groupby("EDUCATION")["defaulted"].count().reset_index()
pay = edu[edu.defaulted == 1].groupby("EDUCATION").count().reset_index()
pay["defaulted"] = [i / j * 100 for i, j in zip(pay["defaulted"], total["defaulted"])]
total["defaulted"] = [
i / j * 100 for i, j in zip(total["defaulted"], total["defaulted"])
]
# bar chart 1 -> top bars (group of 'defaulted = 0')
bar1 = sns.barplot(x="EDUCATION", y="defaulted", data=total, color="lightgray")
# bar chart 2 -> bottom bars (group of 'defaulted = 1')
bar2 = sns.barplot(x="EDUCATION", y="defaulted", data=pay, color="green")
# add legend
top_bar = mpatches.Patch(color="lightgray", label="Não Pagou")
bottom_bar = mpatches.Patch(color="green", label="Pagou")
plt.legend(handles=[top_bar, bottom_bar])
# show the graph
plt.title("Pagantes e não pagantes por Nível de Formação")
plt.xlabel("Nivel de Formação")
plt.ylabel("100% da Análise")
plt.show()
#### Estado civil
# set the figure size
plt.figure(figsize=(18, 6))
marriage = data[["MARRIAGE", "defaulted"]]
marriage = marriage.replace(
{"MARRIAGE": {0: "NE", 1: "2-Casado", 2: "1-Solteiro", 3: "3-Outros"}}
)
# from raw value to percentage
total = marriage.groupby("MARRIAGE")["defaulted"].count().reset_index()
pay = marriage[marriage.defaulted == 1].groupby("MARRIAGE").count().reset_index()
pay["defaulted"] = [i / j * 100 for i, j in zip(pay["defaulted"], total["defaulted"])]
total["defaulted"] = [
i / j * 100 for i, j in zip(total["defaulted"], total["defaulted"])
]
# bar chart 1 -> top bars (group of 'defaulted = 0')
bar1 = sns.barplot(x="MARRIAGE", y="defaulted", data=total, color="lightgray")
# bar chart 2 -> bottom bars (group of 'defaulted = 1')
bar2 = sns.barplot(x="MARRIAGE", y="defaulted", data=pay, color="green")
# add legend
top_bar = mpatches.Patch(color="lightgray", label="Não Pagou")
bottom_bar = mpatches.Patch(color="green", label="Pagou")
plt.legend(handles=[top_bar, bottom_bar])
# show the graph
plt.title("Pagantes e não pagantes por estado civil")
plt.ylabel("100% da Análise")
plt.show()
#### Faixa etária
# set the figure size
plt.figure(figsize=(18, 6))
# from raw value to percentage
total = age.groupby("new_Age")["defaulted"].count().reset_index()
pay = age[marriage.defaulted == 1].groupby("new_Age").count().reset_index()
pay["defaulted"] = [i / j * 100 for i, j in zip(pay["defaulted"], total["defaulted"])]
total["defaulted"] = [
i / j * 100 for i, j in zip(total["defaulted"], total["defaulted"])
]
# bar chart 1 -> top bars (group of 'defaulted = 0')
bar1 = sns.barplot(x="new_Age", y="defaulted", data=total, color="lightgray")
# bar chart 2 -> bottom bars (group of 'defaulted = 1')
bar2 = sns.barplot(x="new_Age", y="defaulted", data=pay, color="green")
# add legend
top_bar = mpatches.Patch(color="lightgray", label="Não Pagou")
bottom_bar = mpatches.Patch(color="green", label="Pagou")
plt.legend(handles=[top_bar, bottom_bar])
# show the graph
plt.title("Pagantes e não pagantes por faixa etária")
plt.ylabel("100% da Análise")
plt.xlabel("Idade")
plt.show()
# PARTE 2 - APLICANDO MODELOS DE MACHINE LEARNING
## Preparando base de dados
#### Download da base de dados
data = pd.read_csv(
r"../input/inadimplncia-de-clientes-de-carto-de-crdito/default of credit card clients - Data (1).csv"
)
data.head()
#### Alterando o nome das Colunas e retirando valores que não estão na descrição da base de dados
data.rename(
{
"SEX": "Sexo",
"EDUCATION": "Escolaridade",
"MARRIAGE": "Estado_Civil",
"AGE": "Idade",
"PAY_6": "Status_Pg_Apr",
"PAY_5": "Status_Pg_May",
"PAY_4": "Status_Pg_Jun",
"PAY_3": "Status_Pg_Jul",
"PAY_2": "Status_Pg_Aug",
"PAY_0": "Status_Pg_Sep",
"BILL_AMT6": "Tot_Apr",
"BILL_AMT5": "Tot_May",
"BILL_AMT4": "Tot_Jun",
"BILL_AMT3": "Tot_Jul",
"BILL_AMT2": "Tot_Aug",
"BILL_AMT1": "Tot_Sep",
"PAY_AMT6": "Pg_Apr",
"PAY_AMT5": "Pg_May",
"PAY_AMT4": "Pg_Jun",
"PAY_AMT3": "Pg_Jul",
"PAY_AMT2": "Pg_Aug",
"PAY_AMT1": "Pg_Sep",
"default payment next month": "defaulted",
},
axis=1,
inplace=True,
)
# Retirando coluna de ID e Agrupando valores para Formação e Estado Civil:
data = data.drop(["ID"], axis=1)
data["Escolaridade"] = data["Escolaridade"].replace([0, 5, 6], [4, 4, 4])
data["Estado_Civil"] = data["Estado_Civil"].replace([0], [3])
len(data)
## Entendendo a base
#### Balanceamento de variável resposta
default = data.defaulted
default = default.replace(to_replace=0, value="Não Pagou")
default = default.replace(to_replace=1, value="Pagou")
default = default.to_frame()
plt.subplots(figsize=(5, 5))
graph = sns.countplot(data=default, x="defaulted")
plt.title("Pagamentos do próximo mês")
plt.xlabel("Status Pagamento ")
plt.ylabel("Quantidade")
for idx, bar in enumerate(graph.patches):
height = bar.get_height()
graph.text(
x=bar.get_x() + bar.get_width() / 2.0,
y=height + 500,
s="{:.2%}".format(
default["defaulted"].value_counts(normalize=True).sort_values()[idx]
),
ha="center",
)
graph.set(xlabel=None)
plt.ylim(0, 30000)
plt.show()
#### Verificando valores nulos
import missingno
missingno.matrix(data)
plt.show()
#### Matriz de Correlação da Base para verificar as variáveis mais correlacionadas com defaulted
mask = np.triu(np.ones_like(data.corr(), dtype=bool))
plt.subplots(figsize=(18, 10))
sns.heatmap(data.corr(), linewidths=0.5, annot=True, cmap="Blues", mask=mask)
plt.title("Correlação entre variáveis do dataset")
plt.show()
#### Identificando Outliers:
sns.set(style="whitegrid", color_codes=True, rc={"figure.figsize": (15, 10)})
sns.boxplot(
data=data.drop(
[
"Sexo",
"Escolaridade",
"Estado_Civil",
"Idade",
"Status_Pg_Sep",
"Status_Pg_Aug",
"Status_Pg_Jul",
"Status_Pg_Jun",
"Status_Pg_May",
"Status_Pg_Apr",
"defaulted",
],
axis=1,
)
)
plt.show()
## Separação em Base de Treino e Base de Teste:
from sklearn.model_selection import train_test_split
X = data.loc[:, data.columns != "defaulted"] # Entrada
y = data["defaulted"] # Saída
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=1)
print(X_train.shape)
print(X_test.shape)
print(y_train.shape)
print(y_test.shape)
## Pré-Processamento dos Dados
### Retirando Outliers
X_train_df = pd.DataFrame(X_train)
from scipy import stats
import numpy as np
z_scores = stats.zscore(X_train_df)
abs_z_scores = np.abs(z_scores)
filtered_entries = (abs_z_scores < 3).all(axis=1)
X_train_out = X_train_df[filtered_entries]
X_train_out
y_train_out = y_train[y_train.index.isin(X_train_out.index)]
y_train_out = pd.DataFrame(y_train_out)
y_train_out
# Boxplot após retirada de outliers
sns.set(style="whitegrid", color_codes=True, rc={"figure.figsize": (15, 10)})
sns.boxplot(
data=X_train_out.drop(
[
"Sexo",
"Escolaridade",
"Estado_Civil",
"Idade",
"Status_Pg_Sep",
"Status_Pg_Aug",
"Status_Pg_Jul",
"Status_Pg_Jun",
"Status_Pg_May",
"Status_Pg_Apr",
],
axis=1,
)
)
plt.show()
### Normalização
# Normalizando os valores das variáveis independentes
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler().fit(X_train_out)
X_train_normalized = scaler.transform(X_train_out)
X_test_normalized = scaler.transform(X_test)
y_train_normalized = y_train_out
X_train_normalized
### Balanceamento na base de Treino:
default = y_train_normalized.defaulted
default = default.replace(to_replace=0, value="Não Pagou")
default = default.replace(to_replace=1, value="Pagou")
default = default.to_frame()
plt.subplots(figsize=(5, 5))
graph = sns.countplot(data=default, x="defaulted")
plt.title("Pagamentos do próximo mês")
plt.xlabel("Status Pagamento ")
plt.ylabel("Quantidade")
for idx, bar in enumerate(graph.patches):
height = bar.get_height()
graph.text(
x=bar.get_x() + bar.get_width() / 2.0,
y=height + 500,
s="{:.2%}".format(
default["defaulted"].value_counts(normalize=True).sort_values()[idx]
),
ha="center",
)
graph.set(xlabel=None)
plt.ylim(0, 30000)
plt.show()
from imblearn.under_sampling import RandomUnderSampler
sampler = RandomUnderSampler(random_state=seed)
X_train_balance, y_train_balance = sampler.fit_resample(
X_train_normalized, y_train_normalized
)
X_train_balance = pd.DataFrame(X_train_balance)
y_train_balance = pd.DataFrame(y_train_balance)
y_train_balance.columns = ["defaulted"]
default = y_train_balance.defaulted
default = default.replace(to_replace=0, value="Não Pagou")
default = default.replace(to_replace=1, value="Pagou")
default = default.to_frame()
plt.subplots(figsize=(5, 5))
graph = sns.countplot(data=default, x="defaulted")
plt.title("Pagamentos do próximo mês")
plt.xlabel("Status Pagamento ")
plt.ylabel("Quantidade")
for idx, bar in enumerate(graph.patches):
height = bar.get_height()
graph.text(
x=bar.get_x() + bar.get_width() / 2.0,
y=height + 500,
s="{:.2%}".format(
default["defaulted"].value_counts(normalize=True).sort_values()[idx]
),
ha="center",
)
graph.set(xlabel=None)
plt.ylim(0, 20000)
plt.show()
## Modelos
#### base de treino com pré-processamento
X_train_clean = X_train_balance
y_train_clean = y_train_balance
X_test_clean = pd.DataFrame(X_test_normalized)
y_test_clean = pd.DataFrame(y_test)
#### base de dados SEM pré-processamento
X_train_zero = X_train
y_train_zero = pd.DataFrame(y_train)
X_test_zero = X_test
y_test_zero = pd.DataFrame(y_test)
#### base de dados com normalização
# Normalizando os valores das variáveis independentes
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler().fit(X_train_zero)
X_train_normalized_zero = scaler.transform(X_train_zero)
X_test_normalized_zero = scaler.transform(X_test_zero)
y_train_normalized_zero = y_train
X_train_normalized_zero
#### fórmula para previsão e avaliação dos modelos
def predict_and_evaluate(X_test, y_test, model):
y_pred = model.predict(X_test)
# ROC
from sklearn.metrics import roc_auc_score
roc = roc_auc_score(y_test, y_pred)
print("ROC: ", roc)
# Acurácia
from sklearn.metrics import accuracy_score
accuracy = accuracy_score(y_test, y_pred)
print("Acurácia: ", accuracy)
# Kappa
from sklearn.metrics import cohen_kappa_score
kappa = cohen_kappa_score(y_test, y_pred)
print("Kappa: ", kappa)
# Recall
from sklearn.metrics import recall_score
recall = recall_score(y_test, y_pred, zero_division=0)
print("Recall: ", recall)
# Precision
from sklearn.metrics import precision_score
precision = precision_score(y_test, y_pred, zero_division=0)
print("Precision: ", precision)
# F1
from sklearn.metrics import f1_score
f1 = f1_score(y_test, y_pred)
print("F1: ", f1)
# Matriz de confusão
from sklearn.metrics import confusion_matrix
confMatrix = confusion_matrix(y_pred, y_test)
ax = plt.subplot()
sns.heatmap(confMatrix, annot=True, fmt=".0f")
plt.xlabel("Real")
plt.ylabel("Previsto")
plt.title("Matriz de Confusão")
# Colocar os nomes
ax.xaxis.set_ticklabels(["Não Pagou (0)", "Pagou (1)"])
ax.yaxis.set_ticklabels(["Não Pagou (0)", "Pagou (1)"])
plt.show()
### Modelo: Machine Learning - Support Vector Machine:
#### fórmula
from sklearn.svm import SVC
def train_SVM(X_train, y_train, seed):
model_SVM = SVC(kernel="rbf", random_state=1)
model_SVM.fit(X_train, y_train)
return model_SVM
#### base com pré-processamento
model_SVM = train_SVM(X_train_clean, y_train_clean, seed)
plt.subplots(figsize=(5, 5))
predict_and_evaluate(X_test_clean, y_test_clean, model_SVM)
#### base com normalização apenas
model_SVM_normalized_zero = train_SVM(
X_train_normalized_zero, y_train_normalized_zero, seed
)
plt.subplots(figsize=(5, 5))
predict_and_evaluate(X_test_normalized_zero, y_test_zero, model_SVM_normalized_zero)
# GridSearch para SVM
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import classification_report
# Set the parameters by cross-validation
tuned_parameters = [
{"kernel": ["rbf"], "gamma": [1e-3, 1e-4, 1e-1], "C": [1, 10, 100, 1000]}
]
model = GridSearchCV(SVC(), tuned_parameters, scoring="f1")
model_SVM_CV = model.fit(X_train_clean, y_train_clean.values.ravel())
# Rodando o modelo com os novos parâmetros, pós GridSearch
predict_and_evaluate(X_test_clean, y_test_clean, model_SVM_CV)
### Modelo: Machine Learning - Regressão Logística:
#### fórmula
from sklearn.linear_model import LogisticRegression
def train_LR(X_train, y_train, seed):
model_LR = LogisticRegression(random_state=1, max_iter=500)
model_LR.fit(X_train, y_train)
return model_LR
#### base com pré-processamento
model_LR = train_LR(X_train_clean, y_train_clean, seed)
plt.subplots(figsize=(5, 5))
predict_and_evaluate(X_test_clean, y_test_clean, model_LR)
#### base com normalização apenas
model_LR_normalized_zero = train_LR(
X_train_normalized_zero, y_train_normalized_zero, seed
)
plt.subplots(figsize=(5, 5))
predict_and_evaluate(X_test_normalized_zero, y_test_zero, model_LR_normalized_zero)
# GridSerach para Logistic Regression
from sklearn.model_selection import GridSearchCV
clf = LogisticRegression()
grid_values = {"penalty": ["l2"], "C": [0.001, 0.01, 0.1, 1, 10, 100, 1000]}
grid_clf_acc = GridSearchCV(clf, param_grid=grid_values, scoring="precision")
model_LR_CV = grid_clf_acc.fit(X_train_clean, y_train_clean)
# Rodando o modelo com os novos parâmetros, pós GridSearch
predict_and_evaluate(X_test_clean, y_test_clean, model_LR_CV)
### Modelo: Machine Learning - Random Forest
#### fórmula
from sklearn.ensemble import RandomForestClassifier
def train_RF(X_train, y_train, seed):
model_RF = RandomForestClassifier(
min_samples_leaf=5, random_state=seed
) # tente mudar parâmetro para evitar overfitting
model_RF.fit(X_train, y_train)
return model_RF
#### base com pré-processamento
model_RF = train_RF(X_train_clean, y_train_clean, seed)
plt.subplots(figsize=(5, 5))
predict_and_evaluate(X_test_clean, y_test_clean, model_RF)
#### base SEM pré-processamento
model_RF = train_RF(X_train_zero, y_train_zero, seed)
plt.subplots(figsize=(5, 5))
predict_and_evaluate(X_test_zero, y_test_zero, model_RF)
####GridSearch para Random Forest
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import classification_report
from sklearn.ensemble import RandomForestClassifier
tuned_parameters = {
"n_estimators": [20, 50, 100, 150, 200, 300, 400, 500],
"min_samples_split": [2, 4, 6, 10],
"min_samples_leaf": [1, 2, 4, 6, 8],
"max_features": [3, 4, 8, 9, 10, 11],
}
print("# Tuning hyper-parameters for F1 score")
print()
model = GridSearchCV(
RandomForestClassifier(n_jobs=-1, verbose=1), tuned_parameters, scoring="f1"
)
model.fit(X_test_clean, y_test_clean.values.ravel())
y_pred = model.predict(X_test_clean)
print(classification_report(y_test_clean, y_pred))
print()
plt.subplots(figsize=(5, 5))
predict_and_evaluate(X_test_clean, y_test_clean, model)
print("Melhor número de árvores: {}".format(model.best_params_["n_estimators"]))
print(
"Melhor número número mínimo de amostras necessárias para dividir um nó interno: {}".format(
model.best_params_["min_samples_split"]
)
)
print(
"Melhor número mínimo de amostras necessárias para estar em um nó da folha: {}".format(
model.best_params_["min_samples_leaf"]
)
)
print(
"Melhor número de variáveis a serem considerados ao procurar a melhor divisão: {}".format(
model.best_params_["max_features"]
)
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/956/69956580.ipynb
|
inadimplncia-de-clientes-de-carto-de-crdito
|
gabrieloliveirasan
|
[{"Id": 69956580, "ScriptId": 19131667, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8060237, "CreationDate": "08/04/2021 02:38:28", "VersionNumber": 3.0, "Title": "Base Credito - ML", "EvaluationDate": "08/04/2021", "IsChange": true, "TotalLines": 705.0, "LinesInsertedFromPrevious": 35.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 670.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 93363363, "KernelVersionId": 69956580, "SourceDatasetVersionId": 1879092}]
|
[{"Id": 1879092, "DatasetId": 1118895, "DatasourceVersionId": 1917190, "CreatorUserId": 3219650, "LicenseName": "Unknown", "CreationDate": "01/25/2021 22:49:31", "VersionNumber": 1.0, "Title": "Inadimpl\u00eancia de clientes de cart\u00e3o de cr\u00e9dito", "Slug": "inadimplncia-de-clientes-de-carto-de-crdito", "Subtitle": NaN, "Description": "# Vari\u00e1veis\n\nEsta pesquisa empregou uma vari\u00e1vel bin\u00e1ria, pagamento padr\u00e3o (Sim = 1, N\u00e3o = 0), como vari\u00e1vel de resposta. Este estudo revisou a literatura e usou as seguintes 23 vari\u00e1veis \u200b\u200bcomo vari\u00e1veis \u200b\u200bexplicativas:\nX1: Valor do cr\u00e9dito concedido (d\u00f3lar NT): inclui tanto o cr\u00e9dito ao consumidor pessoa f\u00edsica quanto o cr\u00e9dito familiar (suplementar).\nX2: G\u00eanero (1 = masculino; 2 = feminino).\nX3: Educa\u00e7\u00e3o (1 = p\u00f3s-gradua\u00e7\u00e3o; 2 = universidade; 3 = ensino m\u00e9dio; 4 = outros).\nX4: Estado civil (1 = casado; 2 = solteiro; 3 = outros).\nX5: Idade (ano).\nX6 - X11: Hist\u00f3rico de pagamentos anteriores. Rastreamos os registros de pagamentos mensais anteriores (de abril a setembro de 2005) da seguinte maneira: X6 = o status de reembolso em setembro de 2005; X7 = situa\u00e7\u00e3o de amortiza\u00e7\u00e3o em agosto de 2005; . . .; X11 = estado de reembolso em abril de 2005. A escala de medi\u00e7\u00e3o para o estado de reembolso \u00e9: -1 = pagamento em dia; 1 = atraso no pagamento por um m\u00eas; 2 = atraso no pagamento por dois meses; . . .; 8 = atraso no pagamento por oito meses; 9 = atraso no pagamento de nove meses ou mais.\nX12-X17: Valor da fatura (d\u00f3lar NT). X12 = valor da fatura em setembro de 2005; X13 = valor da fatura em agosto de 2005; . . .; X17 = valor da fatura em abril de 2005.\nX18-X23: Valor do pagamento anterior (d\u00f3lar NT). X18 = valor pago em setembro de 2005; X19 = valor pago em agosto de 2005; . . .; X23 = valor pago em abril de 2005.\n\nFonte: https://archive.ics.uci.edu/ml/datasets/default+of+credit+card+clients", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1118895, "CreatorUserId": 3219650, "OwnerUserId": 3219650.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1879092.0, "CurrentDatasourceVersionId": 1917190.0, "ForumId": 1136248, "Type": 2, "CreationDate": "01/25/2021 22:49:31", "LastActivityDate": "01/25/2021", "TotalViews": 2804, "TotalDownloads": 189, "TotalVotes": 5, "TotalKernels": 3}]
|
[{"Id": 3219650, "UserName": "gabrieloliveirasan", "DisplayName": "Gabriel Idalino", "RegisterDate": "05/14/2019", "PerformanceTier": 0}]
|
## Trabalho Data Mining - Parte 1: Análise Preliminar da Dase de Dados:
###Importação das bibliotecas e random seed
# Importação dos pacotes
import pandas as pd
import numpy as np
import random
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib.patches as mpatches
# Seed para reprodução de resultados
seed = 1
random.seed(seed)
np.random.seed(seed)
### Importação da base de dados
data = pd.read_csv(
r"../input/inadimplncia-de-clientes-de-carto-de-crdito/default of credit card clients - Data (1).csv"
)
data.head()
# Alterando nome de default payment next month para defaulted para poder fazer as:
data = data.rename(columns={"default payment next month": "defaulted"})
len(data)
### Distribuição
#### Sexo
# Distribuição por Sexo
sexo = data.SEX
sexo = sexo.replace(to_replace=1, value="Homem")
sexo = sexo.replace(to_replace=2, value="Mulher")
sexo = sexo.to_frame()
graph = sns.countplot(
data=sexo, x="SEX", palette="muted", order=sexo["SEX"].value_counts().index
)
plt.title("Distribuição por Sexo")
for idx, bar in enumerate(graph.patches):
height = bar.get_height()
graph.text(
x=bar.get_x() + bar.get_width() / 2.0,
y=height + 500,
s="{:.2%}".format(sexo["SEX"].value_counts(normalize=True)[idx]),
ha="center",
)
graph.set(xlabel=None)
plt.xlabel("Sexo")
plt.ylabel("Quantidade")
plt.ylim(0, 30000)
plt.show()
#### Educação
# Distribuição por Educação
edu = data.EDUCATION
edu = edu.replace(to_replace=0, value="Não Especificado")
edu = edu.replace(to_replace=1, value="Pós-Graduação")
edu = edu.replace(to_replace=2, value="Universidade")
edu = edu.replace(to_replace=3, value="Ensino Médio")
edu = edu.replace(to_replace=4, value="Outros")
edu = edu.replace(to_replace=5, value="Não Especificado")
edu = edu.replace(to_replace=6, value="Não Especificado")
edu = edu.to_frame()
plt.subplots(1, figsize=(10, 5))
graph = sns.countplot(
data=edu,
x="EDUCATION",
palette="muted",
order=edu["EDUCATION"].value_counts().index,
)
for idx, bar in enumerate(graph.patches):
height = bar.get_height()
graph.text(
x=bar.get_x() + bar.get_width() / 2.0,
y=height + 500,
s="{:.2%}".format(edu["EDUCATION"].value_counts(normalize=True)[idx]),
ha="center",
)
plt.title("Distribuição por Nivel Formação")
plt.xlabel("Nivel Escolaridade")
plt.ylabel("Quantidade")
plt.xticks(rotation=45)
graph.set(xlabel=None)
plt.ylim(0, 30000)
plt.show()
#### Estado Civil
# Distribuição por Estado Civil
marriage = data.MARRIAGE
marriage = marriage.replace(to_replace=1, value="Casado")
marriage = marriage.replace(to_replace=2, value="Solteiro")
marriage = marriage.replace(to_replace=3, value="Outros")
marriage = marriage.replace(to_replace=0, value="Não Especificado")
marriage = marriage.to_frame()
plt.subplots(1, figsize=(10, 5))
graph = sns.countplot(
data=marriage,
x="MARRIAGE",
palette="muted",
order=marriage["MARRIAGE"].value_counts().index,
)
for idx, bar in enumerate(graph.patches):
height = bar.get_height()
graph.text(
x=bar.get_x() + bar.get_width() / 2.0,
y=height + 500,
s="{:.2%}".format(marriage["MARRIAGE"].value_counts(normalize=True)[idx]),
ha="center",
)
plt.title("Distribuição por Estado Civil")
plt.xlabel("Nivel Escolaridade")
plt.ylabel("Quantidade")
graph.set(xlabel=None)
plt.ylim(0, 30000)
plt.show()
#### Faixa etária
age = data[["AGE", "defaulted"]]
plt.hist(age["AGE"], bins=20, rwidth=0.9)
plt.title("Distribuição por Faixa Etária")
plt.plot()
# create a list of our conditions
conditions = [
(age["AGE"] < 30),
(age["AGE"] >= 30) & (age["AGE"] < 40),
(age["AGE"] >= 40) & (age["AGE"] < 50),
(age["AGE"] >= 50) & (age["AGE"] < 60),
(age["AGE"] >= 60) & (age["AGE"] < 70),
(age["AGE"] >= 70) & (age["AGE"] < 80),
(age["AGE"] >= 80),
]
# create a list of the values we want to assign for each condition
values = ["20s", "30s", "40s", "50s", "60s", "70s", "maior ou igual a 80"]
# create a new column and use np.select to assign values to it using our lists as arguments
age["new_Age"] = np.select(conditions, values)
# display updated DataFrame
age.head()
age["new_Age"].value_counts()
# Distribuição por Faixa Etária
plt.subplots(1, figsize=(10, 5))
graph = sns.countplot(
data=age, x="new_Age", palette="muted", order=age["new_Age"].value_counts().index
)
for idx, bar in enumerate(graph.patches):
height = bar.get_height()
graph.text(
x=bar.get_x() + bar.get_width() / 2.0,
y=height + 500,
s="{:.2%}".format(age["new_Age"].value_counts(normalize=True)[idx]),
ha="center",
)
plt.title("Distribuição por faixa etária")
plt.xlabel("Faixa etária")
plt.ylabel("Quantidade")
graph.set(xlabel=None)
plt.ylim(0, 30000)
plt.show()
### Distribuição Vs Variável resposta
#### Sexo
# set the figure size
plt.figure(figsize=(8, 6))
sexo = data[["SEX", "defaulted"]]
sexo = sexo.replace({"SEX": {1: "Homem", 2: "Mulher"}})
# from raw value to percentage
total = sexo.groupby("SEX")["defaulted"].count().reset_index()
pay = sexo[sexo.defaulted == 1].groupby("SEX").count().reset_index()
pay["defaulted"] = [i / j * 100 for i, j in zip(pay["defaulted"], total["defaulted"])]
total["defaulted"] = [
i / j * 100 for i, j in zip(total["defaulted"], total["defaulted"])
]
# bar chart 1 -> top bars (group of 'defaulted = 0')
bar1 = sns.barplot(x="SEX", y="defaulted", data=total, color="lightgray")
# bar chart 2 -> bottom bars (group of 'defaulted = 1')
bar2 = sns.barplot(x="SEX", y="defaulted", data=pay, color="green")
# add legend
top_bar = mpatches.Patch(color="lightgray", label="Não Pagou")
bottom_bar = mpatches.Patch(color="green", label="Pagou")
plt.legend(handles=[top_bar, bottom_bar])
plt.xlabel("Sexo")
plt.ylabel("100% da Análise")
# show the graph
plt.show()
#### Educação
# set the figure size
plt.figure(figsize=(18, 6))
edu = data[["EDUCATION", "defaulted"]]
edu = edu.replace(
{
"EDUCATION": {
0: "5-NE",
1: "3-Pós Graduação",
2: "2-Universidade",
3: "1-Ensino Médio",
4: "4-Outros",
5: "5-NE",
6: "5-NE",
}
}
)
# from raw value to percentage
total = edu.groupby("EDUCATION")["defaulted"].count().reset_index()
pay = edu[edu.defaulted == 1].groupby("EDUCATION").count().reset_index()
pay["defaulted"] = [i / j * 100 for i, j in zip(pay["defaulted"], total["defaulted"])]
total["defaulted"] = [
i / j * 100 for i, j in zip(total["defaulted"], total["defaulted"])
]
# bar chart 1 -> top bars (group of 'defaulted = 0')
bar1 = sns.barplot(x="EDUCATION", y="defaulted", data=total, color="lightgray")
# bar chart 2 -> bottom bars (group of 'defaulted = 1')
bar2 = sns.barplot(x="EDUCATION", y="defaulted", data=pay, color="green")
# add legend
top_bar = mpatches.Patch(color="lightgray", label="Não Pagou")
bottom_bar = mpatches.Patch(color="green", label="Pagou")
plt.legend(handles=[top_bar, bottom_bar])
# show the graph
plt.title("Pagantes e não pagantes por Nível de Formação")
plt.xlabel("Nivel de Formação")
plt.ylabel("100% da Análise")
plt.show()
#### Estado civil
# set the figure size
plt.figure(figsize=(18, 6))
marriage = data[["MARRIAGE", "defaulted"]]
marriage = marriage.replace(
{"MARRIAGE": {0: "NE", 1: "2-Casado", 2: "1-Solteiro", 3: "3-Outros"}}
)
# from raw value to percentage
total = marriage.groupby("MARRIAGE")["defaulted"].count().reset_index()
pay = marriage[marriage.defaulted == 1].groupby("MARRIAGE").count().reset_index()
pay["defaulted"] = [i / j * 100 for i, j in zip(pay["defaulted"], total["defaulted"])]
total["defaulted"] = [
i / j * 100 for i, j in zip(total["defaulted"], total["defaulted"])
]
# bar chart 1 -> top bars (group of 'defaulted = 0')
bar1 = sns.barplot(x="MARRIAGE", y="defaulted", data=total, color="lightgray")
# bar chart 2 -> bottom bars (group of 'defaulted = 1')
bar2 = sns.barplot(x="MARRIAGE", y="defaulted", data=pay, color="green")
# add legend
top_bar = mpatches.Patch(color="lightgray", label="Não Pagou")
bottom_bar = mpatches.Patch(color="green", label="Pagou")
plt.legend(handles=[top_bar, bottom_bar])
# show the graph
plt.title("Pagantes e não pagantes por estado civil")
plt.ylabel("100% da Análise")
plt.show()
#### Faixa etária
# set the figure size
plt.figure(figsize=(18, 6))
# from raw value to percentage
total = age.groupby("new_Age")["defaulted"].count().reset_index()
pay = age[marriage.defaulted == 1].groupby("new_Age").count().reset_index()
pay["defaulted"] = [i / j * 100 for i, j in zip(pay["defaulted"], total["defaulted"])]
total["defaulted"] = [
i / j * 100 for i, j in zip(total["defaulted"], total["defaulted"])
]
# bar chart 1 -> top bars (group of 'defaulted = 0')
bar1 = sns.barplot(x="new_Age", y="defaulted", data=total, color="lightgray")
# bar chart 2 -> bottom bars (group of 'defaulted = 1')
bar2 = sns.barplot(x="new_Age", y="defaulted", data=pay, color="green")
# add legend
top_bar = mpatches.Patch(color="lightgray", label="Não Pagou")
bottom_bar = mpatches.Patch(color="green", label="Pagou")
plt.legend(handles=[top_bar, bottom_bar])
# show the graph
plt.title("Pagantes e não pagantes por faixa etária")
plt.ylabel("100% da Análise")
plt.xlabel("Idade")
plt.show()
# PARTE 2 - APLICANDO MODELOS DE MACHINE LEARNING
## Preparando base de dados
#### Download da base de dados
data = pd.read_csv(
r"../input/inadimplncia-de-clientes-de-carto-de-crdito/default of credit card clients - Data (1).csv"
)
data.head()
#### Alterando o nome das Colunas e retirando valores que não estão na descrição da base de dados
data.rename(
{
"SEX": "Sexo",
"EDUCATION": "Escolaridade",
"MARRIAGE": "Estado_Civil",
"AGE": "Idade",
"PAY_6": "Status_Pg_Apr",
"PAY_5": "Status_Pg_May",
"PAY_4": "Status_Pg_Jun",
"PAY_3": "Status_Pg_Jul",
"PAY_2": "Status_Pg_Aug",
"PAY_0": "Status_Pg_Sep",
"BILL_AMT6": "Tot_Apr",
"BILL_AMT5": "Tot_May",
"BILL_AMT4": "Tot_Jun",
"BILL_AMT3": "Tot_Jul",
"BILL_AMT2": "Tot_Aug",
"BILL_AMT1": "Tot_Sep",
"PAY_AMT6": "Pg_Apr",
"PAY_AMT5": "Pg_May",
"PAY_AMT4": "Pg_Jun",
"PAY_AMT3": "Pg_Jul",
"PAY_AMT2": "Pg_Aug",
"PAY_AMT1": "Pg_Sep",
"default payment next month": "defaulted",
},
axis=1,
inplace=True,
)
# Retirando coluna de ID e Agrupando valores para Formação e Estado Civil:
data = data.drop(["ID"], axis=1)
data["Escolaridade"] = data["Escolaridade"].replace([0, 5, 6], [4, 4, 4])
data["Estado_Civil"] = data["Estado_Civil"].replace([0], [3])
len(data)
## Entendendo a base
#### Balanceamento de variável resposta
default = data.defaulted
default = default.replace(to_replace=0, value="Não Pagou")
default = default.replace(to_replace=1, value="Pagou")
default = default.to_frame()
plt.subplots(figsize=(5, 5))
graph = sns.countplot(data=default, x="defaulted")
plt.title("Pagamentos do próximo mês")
plt.xlabel("Status Pagamento ")
plt.ylabel("Quantidade")
for idx, bar in enumerate(graph.patches):
height = bar.get_height()
graph.text(
x=bar.get_x() + bar.get_width() / 2.0,
y=height + 500,
s="{:.2%}".format(
default["defaulted"].value_counts(normalize=True).sort_values()[idx]
),
ha="center",
)
graph.set(xlabel=None)
plt.ylim(0, 30000)
plt.show()
#### Verificando valores nulos
import missingno
missingno.matrix(data)
plt.show()
#### Matriz de Correlação da Base para verificar as variáveis mais correlacionadas com defaulted
mask = np.triu(np.ones_like(data.corr(), dtype=bool))
plt.subplots(figsize=(18, 10))
sns.heatmap(data.corr(), linewidths=0.5, annot=True, cmap="Blues", mask=mask)
plt.title("Correlação entre variáveis do dataset")
plt.show()
#### Identificando Outliers:
sns.set(style="whitegrid", color_codes=True, rc={"figure.figsize": (15, 10)})
sns.boxplot(
data=data.drop(
[
"Sexo",
"Escolaridade",
"Estado_Civil",
"Idade",
"Status_Pg_Sep",
"Status_Pg_Aug",
"Status_Pg_Jul",
"Status_Pg_Jun",
"Status_Pg_May",
"Status_Pg_Apr",
"defaulted",
],
axis=1,
)
)
plt.show()
## Separação em Base de Treino e Base de Teste:
from sklearn.model_selection import train_test_split
X = data.loc[:, data.columns != "defaulted"] # Entrada
y = data["defaulted"] # Saída
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=1)
print(X_train.shape)
print(X_test.shape)
print(y_train.shape)
print(y_test.shape)
## Pré-Processamento dos Dados
### Retirando Outliers
X_train_df = pd.DataFrame(X_train)
from scipy import stats
import numpy as np
z_scores = stats.zscore(X_train_df)
abs_z_scores = np.abs(z_scores)
filtered_entries = (abs_z_scores < 3).all(axis=1)
X_train_out = X_train_df[filtered_entries]
X_train_out
y_train_out = y_train[y_train.index.isin(X_train_out.index)]
y_train_out = pd.DataFrame(y_train_out)
y_train_out
# Boxplot após retirada de outliers
sns.set(style="whitegrid", color_codes=True, rc={"figure.figsize": (15, 10)})
sns.boxplot(
data=X_train_out.drop(
[
"Sexo",
"Escolaridade",
"Estado_Civil",
"Idade",
"Status_Pg_Sep",
"Status_Pg_Aug",
"Status_Pg_Jul",
"Status_Pg_Jun",
"Status_Pg_May",
"Status_Pg_Apr",
],
axis=1,
)
)
plt.show()
### Normalização
# Normalizando os valores das variáveis independentes
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler().fit(X_train_out)
X_train_normalized = scaler.transform(X_train_out)
X_test_normalized = scaler.transform(X_test)
y_train_normalized = y_train_out
X_train_normalized
### Balanceamento na base de Treino:
default = y_train_normalized.defaulted
default = default.replace(to_replace=0, value="Não Pagou")
default = default.replace(to_replace=1, value="Pagou")
default = default.to_frame()
plt.subplots(figsize=(5, 5))
graph = sns.countplot(data=default, x="defaulted")
plt.title("Pagamentos do próximo mês")
plt.xlabel("Status Pagamento ")
plt.ylabel("Quantidade")
for idx, bar in enumerate(graph.patches):
height = bar.get_height()
graph.text(
x=bar.get_x() + bar.get_width() / 2.0,
y=height + 500,
s="{:.2%}".format(
default["defaulted"].value_counts(normalize=True).sort_values()[idx]
),
ha="center",
)
graph.set(xlabel=None)
plt.ylim(0, 30000)
plt.show()
from imblearn.under_sampling import RandomUnderSampler
sampler = RandomUnderSampler(random_state=seed)
X_train_balance, y_train_balance = sampler.fit_resample(
X_train_normalized, y_train_normalized
)
X_train_balance = pd.DataFrame(X_train_balance)
y_train_balance = pd.DataFrame(y_train_balance)
y_train_balance.columns = ["defaulted"]
default = y_train_balance.defaulted
default = default.replace(to_replace=0, value="Não Pagou")
default = default.replace(to_replace=1, value="Pagou")
default = default.to_frame()
plt.subplots(figsize=(5, 5))
graph = sns.countplot(data=default, x="defaulted")
plt.title("Pagamentos do próximo mês")
plt.xlabel("Status Pagamento ")
plt.ylabel("Quantidade")
for idx, bar in enumerate(graph.patches):
height = bar.get_height()
graph.text(
x=bar.get_x() + bar.get_width() / 2.0,
y=height + 500,
s="{:.2%}".format(
default["defaulted"].value_counts(normalize=True).sort_values()[idx]
),
ha="center",
)
graph.set(xlabel=None)
plt.ylim(0, 20000)
plt.show()
## Modelos
#### base de treino com pré-processamento
X_train_clean = X_train_balance
y_train_clean = y_train_balance
X_test_clean = pd.DataFrame(X_test_normalized)
y_test_clean = pd.DataFrame(y_test)
#### base de dados SEM pré-processamento
X_train_zero = X_train
y_train_zero = pd.DataFrame(y_train)
X_test_zero = X_test
y_test_zero = pd.DataFrame(y_test)
#### base de dados com normalização
# Normalizando os valores das variáveis independentes
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler().fit(X_train_zero)
X_train_normalized_zero = scaler.transform(X_train_zero)
X_test_normalized_zero = scaler.transform(X_test_zero)
y_train_normalized_zero = y_train
X_train_normalized_zero
#### fórmula para previsão e avaliação dos modelos
def predict_and_evaluate(X_test, y_test, model):
y_pred = model.predict(X_test)
# ROC
from sklearn.metrics import roc_auc_score
roc = roc_auc_score(y_test, y_pred)
print("ROC: ", roc)
# Acurácia
from sklearn.metrics import accuracy_score
accuracy = accuracy_score(y_test, y_pred)
print("Acurácia: ", accuracy)
# Kappa
from sklearn.metrics import cohen_kappa_score
kappa = cohen_kappa_score(y_test, y_pred)
print("Kappa: ", kappa)
# Recall
from sklearn.metrics import recall_score
recall = recall_score(y_test, y_pred, zero_division=0)
print("Recall: ", recall)
# Precision
from sklearn.metrics import precision_score
precision = precision_score(y_test, y_pred, zero_division=0)
print("Precision: ", precision)
# F1
from sklearn.metrics import f1_score
f1 = f1_score(y_test, y_pred)
print("F1: ", f1)
# Matriz de confusão
from sklearn.metrics import confusion_matrix
confMatrix = confusion_matrix(y_pred, y_test)
ax = plt.subplot()
sns.heatmap(confMatrix, annot=True, fmt=".0f")
plt.xlabel("Real")
plt.ylabel("Previsto")
plt.title("Matriz de Confusão")
# Colocar os nomes
ax.xaxis.set_ticklabels(["Não Pagou (0)", "Pagou (1)"])
ax.yaxis.set_ticklabels(["Não Pagou (0)", "Pagou (1)"])
plt.show()
### Modelo: Machine Learning - Support Vector Machine:
#### fórmula
from sklearn.svm import SVC
def train_SVM(X_train, y_train, seed):
model_SVM = SVC(kernel="rbf", random_state=1)
model_SVM.fit(X_train, y_train)
return model_SVM
#### base com pré-processamento
model_SVM = train_SVM(X_train_clean, y_train_clean, seed)
plt.subplots(figsize=(5, 5))
predict_and_evaluate(X_test_clean, y_test_clean, model_SVM)
#### base com normalização apenas
model_SVM_normalized_zero = train_SVM(
X_train_normalized_zero, y_train_normalized_zero, seed
)
plt.subplots(figsize=(5, 5))
predict_and_evaluate(X_test_normalized_zero, y_test_zero, model_SVM_normalized_zero)
# GridSearch para SVM
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import classification_report
# Set the parameters by cross-validation
tuned_parameters = [
{"kernel": ["rbf"], "gamma": [1e-3, 1e-4, 1e-1], "C": [1, 10, 100, 1000]}
]
model = GridSearchCV(SVC(), tuned_parameters, scoring="f1")
model_SVM_CV = model.fit(X_train_clean, y_train_clean.values.ravel())
# Rodando o modelo com os novos parâmetros, pós GridSearch
predict_and_evaluate(X_test_clean, y_test_clean, model_SVM_CV)
### Modelo: Machine Learning - Regressão Logística:
#### fórmula
from sklearn.linear_model import LogisticRegression
def train_LR(X_train, y_train, seed):
model_LR = LogisticRegression(random_state=1, max_iter=500)
model_LR.fit(X_train, y_train)
return model_LR
#### base com pré-processamento
model_LR = train_LR(X_train_clean, y_train_clean, seed)
plt.subplots(figsize=(5, 5))
predict_and_evaluate(X_test_clean, y_test_clean, model_LR)
#### base com normalização apenas
model_LR_normalized_zero = train_LR(
X_train_normalized_zero, y_train_normalized_zero, seed
)
plt.subplots(figsize=(5, 5))
predict_and_evaluate(X_test_normalized_zero, y_test_zero, model_LR_normalized_zero)
# GridSerach para Logistic Regression
from sklearn.model_selection import GridSearchCV
clf = LogisticRegression()
grid_values = {"penalty": ["l2"], "C": [0.001, 0.01, 0.1, 1, 10, 100, 1000]}
grid_clf_acc = GridSearchCV(clf, param_grid=grid_values, scoring="precision")
model_LR_CV = grid_clf_acc.fit(X_train_clean, y_train_clean)
# Rodando o modelo com os novos parâmetros, pós GridSearch
predict_and_evaluate(X_test_clean, y_test_clean, model_LR_CV)
### Modelo: Machine Learning - Random Forest
#### fórmula
from sklearn.ensemble import RandomForestClassifier
def train_RF(X_train, y_train, seed):
model_RF = RandomForestClassifier(
min_samples_leaf=5, random_state=seed
) # tente mudar parâmetro para evitar overfitting
model_RF.fit(X_train, y_train)
return model_RF
#### base com pré-processamento
model_RF = train_RF(X_train_clean, y_train_clean, seed)
plt.subplots(figsize=(5, 5))
predict_and_evaluate(X_test_clean, y_test_clean, model_RF)
#### base SEM pré-processamento
model_RF = train_RF(X_train_zero, y_train_zero, seed)
plt.subplots(figsize=(5, 5))
predict_and_evaluate(X_test_zero, y_test_zero, model_RF)
####GridSearch para Random Forest
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import classification_report
from sklearn.ensemble import RandomForestClassifier
tuned_parameters = {
"n_estimators": [20, 50, 100, 150, 200, 300, 400, 500],
"min_samples_split": [2, 4, 6, 10],
"min_samples_leaf": [1, 2, 4, 6, 8],
"max_features": [3, 4, 8, 9, 10, 11],
}
print("# Tuning hyper-parameters for F1 score")
print()
model = GridSearchCV(
RandomForestClassifier(n_jobs=-1, verbose=1), tuned_parameters, scoring="f1"
)
model.fit(X_test_clean, y_test_clean.values.ravel())
y_pred = model.predict(X_test_clean)
print(classification_report(y_test_clean, y_pred))
print()
plt.subplots(figsize=(5, 5))
predict_and_evaluate(X_test_clean, y_test_clean, model)
print("Melhor número de árvores: {}".format(model.best_params_["n_estimators"]))
print(
"Melhor número número mínimo de amostras necessárias para dividir um nó interno: {}".format(
model.best_params_["min_samples_split"]
)
)
print(
"Melhor número mínimo de amostras necessárias para estar em um nó da folha: {}".format(
model.best_params_["min_samples_leaf"]
)
)
print(
"Melhor número de variáveis a serem considerados ao procurar a melhor divisão: {}".format(
model.best_params_["max_features"]
)
)
|
[{"inadimplncia-de-clientes-de-carto-de-crdito/default of credit card clients - Data (1).csv": {"column_names": "[\"ID\", \"LIMIT_BAL\", \"SEX\", \"EDUCATION\", \"MARRIAGE\", \"AGE\", \"PAY_0\", \"PAY_2\", \"PAY_3\", \"PAY_4\", \"PAY_5\", \"PAY_6\", \"BILL_AMT1\", \"BILL_AMT2\", \"BILL_AMT3\", \"BILL_AMT4\", \"BILL_AMT5\", \"BILL_AMT6\", \"PAY_AMT1\", \"PAY_AMT2\", \"PAY_AMT3\", \"PAY_AMT4\", \"PAY_AMT5\", \"PAY_AMT6\", \"default payment next month\"]", "column_data_types": "{\"ID\": \"int64\", \"LIMIT_BAL\": \"int64\", \"SEX\": \"int64\", \"EDUCATION\": \"int64\", \"MARRIAGE\": \"int64\", \"AGE\": \"int64\", \"PAY_0\": \"int64\", \"PAY_2\": \"int64\", \"PAY_3\": \"int64\", \"PAY_4\": \"int64\", \"PAY_5\": \"int64\", \"PAY_6\": \"int64\", \"BILL_AMT1\": \"int64\", \"BILL_AMT2\": \"int64\", \"BILL_AMT3\": \"int64\", \"BILL_AMT4\": \"int64\", \"BILL_AMT5\": \"int64\", \"BILL_AMT6\": \"int64\", \"PAY_AMT1\": \"int64\", \"PAY_AMT2\": \"int64\", \"PAY_AMT3\": \"int64\", \"PAY_AMT4\": \"int64\", \"PAY_AMT5\": \"int64\", \"PAY_AMT6\": \"int64\", \"default payment next month\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 30000 entries, 0 to 29999\nData columns (total 25 columns):\n # Column Non-Null Count Dtype\n--- ------ -------------- -----\n 0 ID 30000 non-null int64\n 1 LIMIT_BAL 30000 non-null int64\n 2 SEX 30000 non-null int64\n 3 EDUCATION 30000 non-null int64\n 4 MARRIAGE 30000 non-null int64\n 5 AGE 30000 non-null int64\n 6 PAY_0 30000 non-null int64\n 7 PAY_2 30000 non-null int64\n 8 PAY_3 30000 non-null int64\n 9 PAY_4 30000 non-null int64\n 10 PAY_5 30000 non-null int64\n 11 PAY_6 30000 non-null int64\n 12 BILL_AMT1 30000 non-null int64\n 13 BILL_AMT2 30000 non-null int64\n 14 BILL_AMT3 30000 non-null int64\n 15 BILL_AMT4 30000 non-null int64\n 16 BILL_AMT5 30000 non-null int64\n 17 BILL_AMT6 30000 non-null int64\n 18 PAY_AMT1 30000 non-null int64\n 19 PAY_AMT2 30000 non-null int64\n 20 PAY_AMT3 30000 non-null int64\n 21 PAY_AMT4 30000 non-null int64\n 22 PAY_AMT5 30000 non-null int64\n 23 PAY_AMT6 30000 non-null int64\n 24 default payment next month 30000 non-null int64\ndtypes: int64(25)\nmemory usage: 5.7 MB\n", "summary": "{\"ID\": {\"count\": 30000.0, \"mean\": 15000.5, \"std\": 8660.398374208891, \"min\": 1.0, \"25%\": 7500.75, \"50%\": 15000.5, \"75%\": 22500.25, \"max\": 30000.0}, \"LIMIT_BAL\": {\"count\": 30000.0, \"mean\": 167484.32266666667, \"std\": 129747.66156720239, \"min\": 10000.0, \"25%\": 50000.0, \"50%\": 140000.0, \"75%\": 240000.0, \"max\": 1000000.0}, \"SEX\": {\"count\": 30000.0, \"mean\": 1.6037333333333332, \"std\": 0.48912919609026045, \"min\": 1.0, \"25%\": 1.0, \"50%\": 2.0, \"75%\": 2.0, \"max\": 2.0}, \"EDUCATION\": {\"count\": 30000.0, \"mean\": 1.8531333333333333, \"std\": 0.7903486597207291, \"min\": 0.0, \"25%\": 1.0, \"50%\": 2.0, \"75%\": 2.0, \"max\": 6.0}, \"MARRIAGE\": {\"count\": 30000.0, \"mean\": 1.5518666666666667, \"std\": 0.5219696006132486, \"min\": 0.0, \"25%\": 1.0, \"50%\": 2.0, \"75%\": 2.0, \"max\": 3.0}, \"AGE\": {\"count\": 30000.0, \"mean\": 35.4855, \"std\": 9.217904068090188, \"min\": 21.0, \"25%\": 28.0, \"50%\": 34.0, \"75%\": 41.0, \"max\": 79.0}, \"PAY_0\": {\"count\": 30000.0, \"mean\": -0.0167, \"std\": 1.1238015279973348, \"min\": -2.0, \"25%\": -1.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 8.0}, \"PAY_2\": {\"count\": 30000.0, \"mean\": -0.13376666666666667, \"std\": 1.1971859730345533, \"min\": -2.0, \"25%\": -1.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 8.0}, \"PAY_3\": {\"count\": 30000.0, \"mean\": -0.1662, \"std\": 1.1968675684465735, \"min\": -2.0, \"25%\": -1.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 8.0}, \"PAY_4\": {\"count\": 30000.0, \"mean\": -0.22066666666666668, \"std\": 1.1691386224023375, \"min\": -2.0, \"25%\": -1.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 8.0}, \"PAY_5\": {\"count\": 30000.0, \"mean\": -0.2662, \"std\": 1.1331874060027483, \"min\": -2.0, \"25%\": -1.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 8.0}, \"PAY_6\": {\"count\": 30000.0, \"mean\": -0.2911, \"std\": 1.1499876256079027, \"min\": -2.0, \"25%\": -1.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 8.0}, \"BILL_AMT1\": {\"count\": 30000.0, \"mean\": 51223.3309, \"std\": 73635.86057552956, \"min\": -165580.0, \"25%\": 3558.75, \"50%\": 22381.5, \"75%\": 67091.0, \"max\": 964511.0}, \"BILL_AMT2\": {\"count\": 30000.0, \"mean\": 49179.07516666667, \"std\": 71173.76878252835, \"min\": -69777.0, \"25%\": 2984.75, \"50%\": 21200.0, \"75%\": 64006.25, \"max\": 983931.0}, \"BILL_AMT3\": {\"count\": 30000.0, \"mean\": 47013.1548, \"std\": 69349.38742703684, \"min\": -157264.0, \"25%\": 2666.25, \"50%\": 20088.5, \"75%\": 60164.75, \"max\": 1664089.0}, \"BILL_AMT4\": {\"count\": 30000.0, \"mean\": 43262.94896666666, \"std\": 64332.85613391631, \"min\": -170000.0, \"25%\": 2326.75, \"50%\": 19052.0, \"75%\": 54506.0, \"max\": 891586.0}, \"BILL_AMT5\": {\"count\": 30000.0, \"mean\": 40311.40096666667, \"std\": 60797.15577026487, \"min\": -81334.0, \"25%\": 1763.0, \"50%\": 18104.5, \"75%\": 50190.5, \"max\": 927171.0}, \"BILL_AMT6\": {\"count\": 30000.0, \"mean\": 38871.7604, \"std\": 59554.10753674573, \"min\": -339603.0, \"25%\": 1256.0, \"50%\": 17071.0, \"75%\": 49198.25, \"max\": 961664.0}, \"PAY_AMT1\": {\"count\": 30000.0, \"mean\": 5663.5805, \"std\": 16563.280354025766, \"min\": 0.0, \"25%\": 1000.0, \"50%\": 2100.0, \"75%\": 5006.0, \"max\": 873552.0}, \"PAY_AMT2\": {\"count\": 30000.0, \"mean\": 5921.1635, \"std\": 23040.87040205724, \"min\": 0.0, \"25%\": 833.0, \"50%\": 2009.0, \"75%\": 5000.0, \"max\": 1684259.0}, \"PAY_AMT3\": {\"count\": 30000.0, \"mean\": 5225.6815, \"std\": 17606.961469803104, \"min\": 0.0, \"25%\": 390.0, \"50%\": 1800.0, \"75%\": 4505.0, \"max\": 896040.0}, \"PAY_AMT4\": {\"count\": 30000.0, \"mean\": 4826.076866666666, \"std\": 15666.159744032007, \"min\": 0.0, \"25%\": 296.0, \"50%\": 1500.0, \"75%\": 4013.25, \"max\": 621000.0}, \"PAY_AMT5\": {\"count\": 30000.0, \"mean\": 4799.387633333334, \"std\": 15278.305679144789, \"min\": 0.0, \"25%\": 252.5, \"50%\": 1500.0, \"75%\": 4031.5, \"max\": 426529.0}, \"PAY_AMT6\": {\"count\": 30000.0, \"mean\": 5215.502566666667, \"std\": 17777.4657754353, \"min\": 0.0, \"25%\": 117.75, \"50%\": 1500.0, \"75%\": 4000.0, \"max\": 528666.0}, \"default payment next month\": {\"count\": 30000.0, \"mean\": 0.2212, \"std\": 0.41506180569093337, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}}", "examples": "{\"ID\":{\"0\":1,\"1\":2,\"2\":3,\"3\":4},\"LIMIT_BAL\":{\"0\":20000,\"1\":120000,\"2\":90000,\"3\":50000},\"SEX\":{\"0\":2,\"1\":2,\"2\":2,\"3\":2},\"EDUCATION\":{\"0\":2,\"1\":2,\"2\":2,\"3\":2},\"MARRIAGE\":{\"0\":1,\"1\":2,\"2\":2,\"3\":1},\"AGE\":{\"0\":24,\"1\":26,\"2\":34,\"3\":37},\"PAY_0\":{\"0\":2,\"1\":-1,\"2\":0,\"3\":0},\"PAY_2\":{\"0\":2,\"1\":2,\"2\":0,\"3\":0},\"PAY_3\":{\"0\":-1,\"1\":0,\"2\":0,\"3\":0},\"PAY_4\":{\"0\":-1,\"1\":0,\"2\":0,\"3\":0},\"PAY_5\":{\"0\":-2,\"1\":0,\"2\":0,\"3\":0},\"PAY_6\":{\"0\":-2,\"1\":2,\"2\":0,\"3\":0},\"BILL_AMT1\":{\"0\":3913,\"1\":2682,\"2\":29239,\"3\":46990},\"BILL_AMT2\":{\"0\":3102,\"1\":1725,\"2\":14027,\"3\":48233},\"BILL_AMT3\":{\"0\":689,\"1\":2682,\"2\":13559,\"3\":49291},\"BILL_AMT4\":{\"0\":0,\"1\":3272,\"2\":14331,\"3\":28314},\"BILL_AMT5\":{\"0\":0,\"1\":3455,\"2\":14948,\"3\":28959},\"BILL_AMT6\":{\"0\":0,\"1\":3261,\"2\":15549,\"3\":29547},\"PAY_AMT1\":{\"0\":0,\"1\":0,\"2\":1518,\"3\":2000},\"PAY_AMT2\":{\"0\":689,\"1\":1000,\"2\":1500,\"3\":2019},\"PAY_AMT3\":{\"0\":0,\"1\":1000,\"2\":1000,\"3\":1200},\"PAY_AMT4\":{\"0\":0,\"1\":1000,\"2\":1000,\"3\":1100},\"PAY_AMT5\":{\"0\":0,\"1\":0,\"2\":1000,\"3\":1069},\"PAY_AMT6\":{\"0\":0,\"1\":2000,\"2\":5000,\"3\":1000},\"default payment next month\":{\"0\":1,\"1\":1,\"2\":0,\"3\":0}}"}}]
| true | 1 |
<start_data_description><data_path>inadimplncia-de-clientes-de-carto-de-crdito/default of credit card clients - Data (1).csv:
<column_names>
['ID', 'LIMIT_BAL', 'SEX', 'EDUCATION', 'MARRIAGE', 'AGE', 'PAY_0', 'PAY_2', 'PAY_3', 'PAY_4', 'PAY_5', 'PAY_6', 'BILL_AMT1', 'BILL_AMT2', 'BILL_AMT3', 'BILL_AMT4', 'BILL_AMT5', 'BILL_AMT6', 'PAY_AMT1', 'PAY_AMT2', 'PAY_AMT3', 'PAY_AMT4', 'PAY_AMT5', 'PAY_AMT6', 'default payment next month']
<column_types>
{'ID': 'int64', 'LIMIT_BAL': 'int64', 'SEX': 'int64', 'EDUCATION': 'int64', 'MARRIAGE': 'int64', 'AGE': 'int64', 'PAY_0': 'int64', 'PAY_2': 'int64', 'PAY_3': 'int64', 'PAY_4': 'int64', 'PAY_5': 'int64', 'PAY_6': 'int64', 'BILL_AMT1': 'int64', 'BILL_AMT2': 'int64', 'BILL_AMT3': 'int64', 'BILL_AMT4': 'int64', 'BILL_AMT5': 'int64', 'BILL_AMT6': 'int64', 'PAY_AMT1': 'int64', 'PAY_AMT2': 'int64', 'PAY_AMT3': 'int64', 'PAY_AMT4': 'int64', 'PAY_AMT5': 'int64', 'PAY_AMT6': 'int64', 'default payment next month': 'int64'}
<dataframe_Summary>
{'ID': {'count': 30000.0, 'mean': 15000.5, 'std': 8660.398374208891, 'min': 1.0, '25%': 7500.75, '50%': 15000.5, '75%': 22500.25, 'max': 30000.0}, 'LIMIT_BAL': {'count': 30000.0, 'mean': 167484.32266666667, 'std': 129747.66156720239, 'min': 10000.0, '25%': 50000.0, '50%': 140000.0, '75%': 240000.0, 'max': 1000000.0}, 'SEX': {'count': 30000.0, 'mean': 1.6037333333333332, 'std': 0.48912919609026045, 'min': 1.0, '25%': 1.0, '50%': 2.0, '75%': 2.0, 'max': 2.0}, 'EDUCATION': {'count': 30000.0, 'mean': 1.8531333333333333, 'std': 0.7903486597207291, 'min': 0.0, '25%': 1.0, '50%': 2.0, '75%': 2.0, 'max': 6.0}, 'MARRIAGE': {'count': 30000.0, 'mean': 1.5518666666666667, 'std': 0.5219696006132486, 'min': 0.0, '25%': 1.0, '50%': 2.0, '75%': 2.0, 'max': 3.0}, 'AGE': {'count': 30000.0, 'mean': 35.4855, 'std': 9.217904068090188, 'min': 21.0, '25%': 28.0, '50%': 34.0, '75%': 41.0, 'max': 79.0}, 'PAY_0': {'count': 30000.0, 'mean': -0.0167, 'std': 1.1238015279973348, 'min': -2.0, '25%': -1.0, '50%': 0.0, '75%': 0.0, 'max': 8.0}, 'PAY_2': {'count': 30000.0, 'mean': -0.13376666666666667, 'std': 1.1971859730345533, 'min': -2.0, '25%': -1.0, '50%': 0.0, '75%': 0.0, 'max': 8.0}, 'PAY_3': {'count': 30000.0, 'mean': -0.1662, 'std': 1.1968675684465735, 'min': -2.0, '25%': -1.0, '50%': 0.0, '75%': 0.0, 'max': 8.0}, 'PAY_4': {'count': 30000.0, 'mean': -0.22066666666666668, 'std': 1.1691386224023375, 'min': -2.0, '25%': -1.0, '50%': 0.0, '75%': 0.0, 'max': 8.0}, 'PAY_5': {'count': 30000.0, 'mean': -0.2662, 'std': 1.1331874060027483, 'min': -2.0, '25%': -1.0, '50%': 0.0, '75%': 0.0, 'max': 8.0}, 'PAY_6': {'count': 30000.0, 'mean': -0.2911, 'std': 1.1499876256079027, 'min': -2.0, '25%': -1.0, '50%': 0.0, '75%': 0.0, 'max': 8.0}, 'BILL_AMT1': {'count': 30000.0, 'mean': 51223.3309, 'std': 73635.86057552956, 'min': -165580.0, '25%': 3558.75, '50%': 22381.5, '75%': 67091.0, 'max': 964511.0}, 'BILL_AMT2': {'count': 30000.0, 'mean': 49179.07516666667, 'std': 71173.76878252835, 'min': -69777.0, '25%': 2984.75, '50%': 21200.0, '75%': 64006.25, 'max': 983931.0}, 'BILL_AMT3': {'count': 30000.0, 'mean': 47013.1548, 'std': 69349.38742703684, 'min': -157264.0, '25%': 2666.25, '50%': 20088.5, '75%': 60164.75, 'max': 1664089.0}, 'BILL_AMT4': {'count': 30000.0, 'mean': 43262.94896666666, 'std': 64332.85613391631, 'min': -170000.0, '25%': 2326.75, '50%': 19052.0, '75%': 54506.0, 'max': 891586.0}, 'BILL_AMT5': {'count': 30000.0, 'mean': 40311.40096666667, 'std': 60797.15577026487, 'min': -81334.0, '25%': 1763.0, '50%': 18104.5, '75%': 50190.5, 'max': 927171.0}, 'BILL_AMT6': {'count': 30000.0, 'mean': 38871.7604, 'std': 59554.10753674573, 'min': -339603.0, '25%': 1256.0, '50%': 17071.0, '75%': 49198.25, 'max': 961664.0}, 'PAY_AMT1': {'count': 30000.0, 'mean': 5663.5805, 'std': 16563.280354025766, 'min': 0.0, '25%': 1000.0, '50%': 2100.0, '75%': 5006.0, 'max': 873552.0}, 'PAY_AMT2': {'count': 30000.0, 'mean': 5921.1635, 'std': 23040.87040205724, 'min': 0.0, '25%': 833.0, '50%': 2009.0, '75%': 5000.0, 'max': 1684259.0}, 'PAY_AMT3': {'count': 30000.0, 'mean': 5225.6815, 'std': 17606.961469803104, 'min': 0.0, '25%': 390.0, '50%': 1800.0, '75%': 4505.0, 'max': 896040.0}, 'PAY_AMT4': {'count': 30000.0, 'mean': 4826.076866666666, 'std': 15666.159744032007, 'min': 0.0, '25%': 296.0, '50%': 1500.0, '75%': 4013.25, 'max': 621000.0}, 'PAY_AMT5': {'count': 30000.0, 'mean': 4799.387633333334, 'std': 15278.305679144789, 'min': 0.0, '25%': 252.5, '50%': 1500.0, '75%': 4031.5, 'max': 426529.0}, 'PAY_AMT6': {'count': 30000.0, 'mean': 5215.502566666667, 'std': 17777.4657754353, 'min': 0.0, '25%': 117.75, '50%': 1500.0, '75%': 4000.0, 'max': 528666.0}, 'default payment next month': {'count': 30000.0, 'mean': 0.2212, 'std': 0.41506180569093337, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}}
<dataframe_info>
RangeIndex: 30000 entries, 0 to 29999
Data columns (total 25 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 ID 30000 non-null int64
1 LIMIT_BAL 30000 non-null int64
2 SEX 30000 non-null int64
3 EDUCATION 30000 non-null int64
4 MARRIAGE 30000 non-null int64
5 AGE 30000 non-null int64
6 PAY_0 30000 non-null int64
7 PAY_2 30000 non-null int64
8 PAY_3 30000 non-null int64
9 PAY_4 30000 non-null int64
10 PAY_5 30000 non-null int64
11 PAY_6 30000 non-null int64
12 BILL_AMT1 30000 non-null int64
13 BILL_AMT2 30000 non-null int64
14 BILL_AMT3 30000 non-null int64
15 BILL_AMT4 30000 non-null int64
16 BILL_AMT5 30000 non-null int64
17 BILL_AMT6 30000 non-null int64
18 PAY_AMT1 30000 non-null int64
19 PAY_AMT2 30000 non-null int64
20 PAY_AMT3 30000 non-null int64
21 PAY_AMT4 30000 non-null int64
22 PAY_AMT5 30000 non-null int64
23 PAY_AMT6 30000 non-null int64
24 default payment next month 30000 non-null int64
dtypes: int64(25)
memory usage: 5.7 MB
<some_examples>
{'ID': {'0': 1, '1': 2, '2': 3, '3': 4}, 'LIMIT_BAL': {'0': 20000, '1': 120000, '2': 90000, '3': 50000}, 'SEX': {'0': 2, '1': 2, '2': 2, '3': 2}, 'EDUCATION': {'0': 2, '1': 2, '2': 2, '3': 2}, 'MARRIAGE': {'0': 1, '1': 2, '2': 2, '3': 1}, 'AGE': {'0': 24, '1': 26, '2': 34, '3': 37}, 'PAY_0': {'0': 2, '1': -1, '2': 0, '3': 0}, 'PAY_2': {'0': 2, '1': 2, '2': 0, '3': 0}, 'PAY_3': {'0': -1, '1': 0, '2': 0, '3': 0}, 'PAY_4': {'0': -1, '1': 0, '2': 0, '3': 0}, 'PAY_5': {'0': -2, '1': 0, '2': 0, '3': 0}, 'PAY_6': {'0': -2, '1': 2, '2': 0, '3': 0}, 'BILL_AMT1': {'0': 3913, '1': 2682, '2': 29239, '3': 46990}, 'BILL_AMT2': {'0': 3102, '1': 1725, '2': 14027, '3': 48233}, 'BILL_AMT3': {'0': 689, '1': 2682, '2': 13559, '3': 49291}, 'BILL_AMT4': {'0': 0, '1': 3272, '2': 14331, '3': 28314}, 'BILL_AMT5': {'0': 0, '1': 3455, '2': 14948, '3': 28959}, 'BILL_AMT6': {'0': 0, '1': 3261, '2': 15549, '3': 29547}, 'PAY_AMT1': {'0': 0, '1': 0, '2': 1518, '3': 2000}, 'PAY_AMT2': {'0': 689, '1': 1000, '2': 1500, '3': 2019}, 'PAY_AMT3': {'0': 0, '1': 1000, '2': 1000, '3': 1200}, 'PAY_AMT4': {'0': 0, '1': 1000, '2': 1000, '3': 1100}, 'PAY_AMT5': {'0': 0, '1': 0, '2': 1000, '3': 1069}, 'PAY_AMT6': {'0': 0, '1': 2000, '2': 5000, '3': 1000}, 'default payment next month': {'0': 1, '1': 1, '2': 0, '3': 0}}
<end_description>
| 7,789 | 0 | 10,085 | 7,789 |
69956218
|
<jupyter_start><jupyter_text>Boston Housing
### Context
The dataset for this project originates from the UCI Machine Learning Repository. The Boston housing data was collected in 1978 and each of the 506 entries represent aggregated data about 14 features for homes from various suburbs in Boston, Massachusetts.
Kaggle dataset identifier: bostonhoustingmlnd
<jupyter_code>import pandas as pd
df = pd.read_csv('bostonhoustingmlnd/housing.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 489 entries, 0 to 488
Data columns (total 4 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 RM 489 non-null float64
1 LSTAT 489 non-null float64
2 PTRATIO 489 non-null float64
3 MEDV 489 non-null float64
dtypes: float64(4)
memory usage: 15.4 KB
<jupyter_text>Examples:
{
"RM": 6.575,
"LSTAT": 4.98,
"PTRATIO": 15.3,
"MEDV": 504000.0
}
{
"RM": 6.421,
"LSTAT": 9.14,
"PTRATIO": 17.8,
"MEDV": 453600.0
}
{
"RM": 7.185,
"LSTAT": 4.03,
"PTRATIO": 17.8,
"MEDV": 728700.0
}
{
"RM": 6.998,
"LSTAT": 2.94,
"PTRATIO": 18.7,
"MEDV": 701400.0
}
<jupyter_script># <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#D63B52;
# font-size:220%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 20px;
# color:white;">
# Guide on how to handle Skewed Distribution
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#4c69b9;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# The distribution of a data set is the shape of the graph when all possible values are plotted on a frequency graph (showing how often they occur). Today we will be learning how to handle skewed distribution of numerical data. By skewed data, I mean to say distributions which are not perfect bell shaped. The bell shaped ones are normal distributions. The ones which are not is skewed. Some are jagged as well. We will learn how to handle that today.
#
# 
# importing necessary libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import scipy.stats as stats
import statsmodels.api as sm
sns.set_theme()
sns.set_palette(palette="rainbow")
df = pd.read_csv("../input/bostonhoustingmlnd/housing.csv")
df.head()
# ***
# #### There are 3 ways to know if a data is distributed.
# * QQ Plot
# * Seaborn Histogram and Kdeplot
# * Pandas Skew Function
# ***
# Skew function of Pandas
old_skew = df.skew().sort_values(ascending=False)
old_skew
# ***
# * *Positive value means the distribution is skewed to the right.*
# * *Negative value means the distribution is skewed to the left.*
# * *0 means perfect normal distribution. The bell shaped curve.*
# ***
plt.figure(figsize=(17, 13))
for i in list(enumerate(df.columns)):
plt.subplot(2, 4, i[0] + 1)
sns.histplot(data=df[i[1]], kde=True) # Histogram with KDE line
for i in list(enumerate(df.columns)):
plt.subplot(2, 4, i[0] + 5)
stats.probplot(df[i[1]], dist="norm", plot=plt) # QQ Plot
plt.title("")
plt.tight_layout()
plt.show()
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#4c69b9;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# From the above visualization we can understand that the column RM is more or less normally distributed, with a skew value of 0.08, which is very close to 0. LSTAT and MEDV is skewed to the right, PTRATIO is skewed to the left. We have to find appropriate ways to handle these skewed data and try to turn them to as normally distributed as possible. But we will only do this if your algorithm performs better in a normally distributed data. Some algorithms do not need a normal distribution and in those cases, we can ignore. We are not getting into the details of the data today and our main goal will be to handle the skewed data and turn it into normal distribution.
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#D63B52;
# font-size:220%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 20px;
# color:white;">
# Mathematical Transformers
#
# ***
# *We can handle skewed data using mathematical transformers. I will discuss some of the best transformers that sklearn library
# provides us in order to handle skewed data. There are various types of Mathematical Transformers:*
# * **Function Transformers**
# - Log Transform
# - Reciprocal Transform
# - Square Transform
# - Sq Root Transform
# - Custom Transform
# * **Power Transformers**
# - Box-Cox
# - Yeo-Johnson
# * **Quantile Transformer**
# ***
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#AF65C1;
# font-size:150%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# 1 . Log Transform
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#59729F;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# In this transformation technique, we will use log of each and every value in the feature. So when we use logs of a certain distribution, we will use their exponents instead of the actual value. This makes the X scale exponentially grow in value and that is the reason why only Right Skewed data transforms to a more or less normal distribution. Mind you, only use it in cases where your algorithm will benefit from a normal distribution, for example, in cases of linear regression models. Also, only use it when your data is skewed to the right.
#
#
from sklearn.preprocessing import FunctionTransformer
from sklearn.compose import ColumnTransformer
def logTrans(
feature,
): # function to apply transformer and check the distribution with histogram and kdeplot
logTr = ColumnTransformer(
transformers=[("lg", FunctionTransformer(np.log1p), [feature])]
)
plt.figure(figsize=(15, 6))
plt.subplot(1, 2, 1)
plt.title("Distribution before Transformation", fontsize=15)
sns.histplot(df[feature], kde=True, color="red")
plt.subplot(1, 2, 2)
df_log = pd.DataFrame(logTr.fit_transform(df))
plt.title("Distribution after Transformation", fontsize=15)
sns.histplot(df_log, bins=20, kde=True, legend=False)
plt.xlabel(feature)
plt.show()
print(
f"Skewness was {round(old_skew[feature],2)} before & is {round(df_log.skew()[0],2)} after Log transformation."
)
logTrans(feature="LSTAT")
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#4c69b9;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# The skewness decreased from an extreme right skew to a moderately normal distribution and a negligible left skew.
# Closer the skew value to 0, more normal the distribution is.
#
# Now we will apply Log Transformation to a left skewed data and you will see how the distribution gets more skewed.
#
#
logTrans(feature="PTRATIO")
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#AC0000;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# Hence proved, never use Log transformation for left skewed data. It will mess your data up!
#
# ***
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#AF65C1;
# font-size:150%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# 2. Square Transform
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#59729F;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# In this transformation technique, we will use square of each and every value in the feature. This will only work on Left Skewed data. So, use Square Transform for left skewed data and Log Transform for right skewed data. Simple.
#
#
def squareTrans(
feature,
): # function to apply transformer and check the distribution with histogram and kdeplot
logTr = ColumnTransformer(
transformers=[("lg", FunctionTransformer(np.square), [feature])]
)
plt.figure(figsize=(15, 6))
plt.subplot(1, 2, 1)
plt.title("Distribution before Transformation", fontsize=15)
sns.histplot(df[feature], kde=True, color="red")
plt.subplot(1, 2, 2)
df_square = pd.DataFrame(logTr.fit_transform(df))
plt.title("Distribution after Transformation", fontsize=15)
sns.histplot(df_square, bins=20, kde=True, legend=False)
plt.xlabel(feature)
plt.show()
print(
f"Skewness was {round(old_skew[feature],2)} before & is {round(df_square.skew()[0],2)} after Square transformation."
)
squareTrans(feature="RM")
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#4c69b9;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# RM column was a little bit left skewed, however, the square transformation did not work and the skewness increased by a lot.
#
# Now we will apply Square Transformation to a right skewed data and you will see how the distribution gets more skewed.
#
#
squareTrans(feature="LSTAT")
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#AC0000;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# Hence proved, never use Square transformation for right skewed data. It will mess your data up!
#
# ***
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#AF65C1;
# font-size:150%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# 3. Reciprocal Transform, Squre Root and other Transformers
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#59729F;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# You should already have a clear picture of how we are using column transformer to apply a Function Transformer class of sklearn to the column of the dataframe and we can use any transformation we want. For example, we have tried log and square as of now. We can try, cube, square root, reciprocal or 1/x of the value as well. This is more like a trial and error and comes under fine tuning your feature engineering so that you can optimize your model. Try out as many things as you want! Comment your results.
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#4c69b9;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# For a demonstration I will be using cube transformer in RM feature to show you how it works compared to square transformer.
#
#
def cubeTrans(
feature,
): # function to apply transformer and check the distribution with histogram and kdeplot
logTr = ColumnTransformer(
transformers=[("lg", FunctionTransformer(lambda x: x**3), [feature])]
)
plt.figure(figsize=(15, 6))
plt.subplot(1, 2, 1)
plt.title("Distribution before Transformation", fontsize=15)
sns.histplot(df[feature], kde=True, color="red")
plt.subplot(1, 2, 2)
df_cube = pd.DataFrame(logTr.fit_transform(df))
plt.title("Distribution after Transformation", fontsize=15)
sns.histplot(df_cube, bins=20, kde=True, legend=False)
plt.xlabel(feature)
plt.show()
print(
f"Skewness was {round(old_skew[feature],2)} before & is {round(df_cube.skew()[0],2)} after cube transformation."
)
cubeTrans(feature="RM")
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#AF65C1;
# font-size:150%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# 4. Box-Cox
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#59729F;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# A Box Cox transformation is a transformation of a non-normal dependent variables into a normal shape. So the fun part is, box cox can be used in general for all type of distributions and it will by itself find the lambda value (check the formula below to know what lambda here is). Box-Cox cannot be used for negative values and 0.
#
# #### Formula of Box-Cox Transformation:
# 
from sklearn.preprocessing import PowerTransformer
# #### By default the PowerTransformer will standard scale the column so you need to specifically mention it if you do not want to scale.
def boxcoxtrans(
feature,
): # function to apply transformer and check the distribution with histogram and kdeplot
boxcoxTr = PowerTransformer(method="box-cox", standardize=True)
plt.figure(figsize=(15, 6))
plt.subplot(1, 2, 1)
plt.title("Distribution before Transformation", fontsize=15)
sns.histplot(df[feature], kde=True, color="red")
plt.subplot(1, 2, 2)
df_boxcox = pd.DataFrame(boxcoxTr.fit_transform(df[feature].values.reshape(-1, 1)))
plt.title("Distribution after Transformation", fontsize=15)
sns.histplot(df_boxcox, bins=20, kde=True, legend=False)
plt.xlabel(feature)
plt.show()
print(
f"Skewness was {round(old_skew[feature],2)} before & is {round(df_boxcox.skew()[0],2)} after Box-cox transformation."
)
boxcoxtrans(feature="RM")
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#4c69b9;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# Did you see how it automatically found out the value of the exponent that will be best for this data? Amazing isn't it?
# The next method will blow your mind!
#
# ***
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#AF65C1;
# font-size:150%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# 5. Yeo-Johnson
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#59729F;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# This is Box-cox but better.The limitation on box-cox is it does not work with negative values and 0. Yeo-Johnson fixes that and can be used for any distribution, both positive and negative. So, if you do not understand anything about distribution and know that your algorithm will be benifiting from a normal distribution, then copy pase this code and you are fine. LoL! Never copy paste codes, understand it. Atleast a basic intuition is better than mugging it up.
# In sklearn.preprocessing.PowerTransformer class, this method is the default, so you know it's good! Have fun!
#
# #### Formula of Yeo-Johnson Transformation:
# 
def yeojohntrans(
feature,
): # function to apply transformer and check the distribution with histogram and kdeplot
yeojohnTr = PowerTransformer(
standardize=True
) # not using method attribute as yeo-johnson is the default
plt.figure(figsize=(15, 6))
plt.subplot(1, 2, 1)
plt.title("Distribution before Transformation", fontsize=15)
sns.histplot(df[feature], kde=True, color="red")
plt.subplot(1, 2, 2)
df_yeojohn = pd.DataFrame(
yeojohnTr.fit_transform(df[feature].values.reshape(-1, 1))
)
plt.title("Distribution after Transformation", fontsize=15)
sns.histplot(df_yeojohn, bins=20, kde=True, legend=False)
plt.xlabel(feature)
plt.show()
print(
f"Skewness was {round(old_skew[feature],2)} before & is {round(df_yeojohn.skew()[0],2)} after Yeo-johnson transformation."
)
yeojohntrans(feature="RM")
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#4c69b9;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# By far the best result for the feature RM! Let's check it out on other features as well.
# Let's check the same Yeo Johnson transformation in other features.
#
#
for i in df.columns[1:]:
yeojohntrans(i)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/956/69956218.ipynb
|
bostonhoustingmlnd
|
schirmerchad
|
[{"Id": 69956218, "ScriptId": 18931043, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4347812, "CreationDate": "08/04/2021 02:35:20", "VersionNumber": 6.0, "Title": "How to handle Skewed Distribution \ud83d\udc7d", "EvaluationDate": "08/04/2021", "IsChange": false, "TotalLines": 496.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 496.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 65}]
|
[{"Id": 93363086, "KernelVersionId": 69956218, "SourceDatasetVersionId": 2485}]
|
[{"Id": 2485, "DatasetId": 1379, "DatasourceVersionId": 2485, "CreatorUserId": 602943, "LicenseName": "CC BY-NC-SA 4.0", "CreationDate": "06/11/2017 15:07:11", "VersionNumber": 1.0, "Title": "Boston Housing", "Slug": "bostonhoustingmlnd", "Subtitle": "Concerns housing values in suburbs of Boston", "Description": "### Context\n\nThe dataset for this project originates from the UCI Machine Learning Repository. The Boston housing data was collected in 1978 and each of the 506 entries represent aggregated data about 14 features for homes from various suburbs in Boston, Massachusetts.\n\n### Acknowledgements\n\nhttps://github.com/udacity/machine-learning\n\nhttps://archive.ics.uci.edu/ml/datasets/Housing", "VersionNotes": "Initial release", "TotalCompressedBytes": 12435.0, "TotalUncompressedBytes": 12435.0}]
|
[{"Id": 1379, "CreatorUserId": 602943, "OwnerUserId": 602943.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2485.0, "CurrentDatasourceVersionId": 2485.0, "ForumId": 4057, "Type": 2, "CreationDate": "06/11/2017 15:07:11", "LastActivityDate": "02/04/2018", "TotalViews": 139673, "TotalDownloads": 24289, "TotalVotes": 223, "TotalKernels": 164}]
|
[{"Id": 602943, "UserName": "schirmerchad", "DisplayName": "Chad Schirmer", "RegisterDate": "05/03/2016", "PerformanceTier": 1}]
|
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#D63B52;
# font-size:220%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 20px;
# color:white;">
# Guide on how to handle Skewed Distribution
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#4c69b9;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# The distribution of a data set is the shape of the graph when all possible values are plotted on a frequency graph (showing how often they occur). Today we will be learning how to handle skewed distribution of numerical data. By skewed data, I mean to say distributions which are not perfect bell shaped. The bell shaped ones are normal distributions. The ones which are not is skewed. Some are jagged as well. We will learn how to handle that today.
#
# 
# importing necessary libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import scipy.stats as stats
import statsmodels.api as sm
sns.set_theme()
sns.set_palette(palette="rainbow")
df = pd.read_csv("../input/bostonhoustingmlnd/housing.csv")
df.head()
# ***
# #### There are 3 ways to know if a data is distributed.
# * QQ Plot
# * Seaborn Histogram and Kdeplot
# * Pandas Skew Function
# ***
# Skew function of Pandas
old_skew = df.skew().sort_values(ascending=False)
old_skew
# ***
# * *Positive value means the distribution is skewed to the right.*
# * *Negative value means the distribution is skewed to the left.*
# * *0 means perfect normal distribution. The bell shaped curve.*
# ***
plt.figure(figsize=(17, 13))
for i in list(enumerate(df.columns)):
plt.subplot(2, 4, i[0] + 1)
sns.histplot(data=df[i[1]], kde=True) # Histogram with KDE line
for i in list(enumerate(df.columns)):
plt.subplot(2, 4, i[0] + 5)
stats.probplot(df[i[1]], dist="norm", plot=plt) # QQ Plot
plt.title("")
plt.tight_layout()
plt.show()
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#4c69b9;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# From the above visualization we can understand that the column RM is more or less normally distributed, with a skew value of 0.08, which is very close to 0. LSTAT and MEDV is skewed to the right, PTRATIO is skewed to the left. We have to find appropriate ways to handle these skewed data and try to turn them to as normally distributed as possible. But we will only do this if your algorithm performs better in a normally distributed data. Some algorithms do not need a normal distribution and in those cases, we can ignore. We are not getting into the details of the data today and our main goal will be to handle the skewed data and turn it into normal distribution.
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#D63B52;
# font-size:220%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 20px;
# color:white;">
# Mathematical Transformers
#
# ***
# *We can handle skewed data using mathematical transformers. I will discuss some of the best transformers that sklearn library
# provides us in order to handle skewed data. There are various types of Mathematical Transformers:*
# * **Function Transformers**
# - Log Transform
# - Reciprocal Transform
# - Square Transform
# - Sq Root Transform
# - Custom Transform
# * **Power Transformers**
# - Box-Cox
# - Yeo-Johnson
# * **Quantile Transformer**
# ***
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#AF65C1;
# font-size:150%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# 1 . Log Transform
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#59729F;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# In this transformation technique, we will use log of each and every value in the feature. So when we use logs of a certain distribution, we will use their exponents instead of the actual value. This makes the X scale exponentially grow in value and that is the reason why only Right Skewed data transforms to a more or less normal distribution. Mind you, only use it in cases where your algorithm will benefit from a normal distribution, for example, in cases of linear regression models. Also, only use it when your data is skewed to the right.
#
#
from sklearn.preprocessing import FunctionTransformer
from sklearn.compose import ColumnTransformer
def logTrans(
feature,
): # function to apply transformer and check the distribution with histogram and kdeplot
logTr = ColumnTransformer(
transformers=[("lg", FunctionTransformer(np.log1p), [feature])]
)
plt.figure(figsize=(15, 6))
plt.subplot(1, 2, 1)
plt.title("Distribution before Transformation", fontsize=15)
sns.histplot(df[feature], kde=True, color="red")
plt.subplot(1, 2, 2)
df_log = pd.DataFrame(logTr.fit_transform(df))
plt.title("Distribution after Transformation", fontsize=15)
sns.histplot(df_log, bins=20, kde=True, legend=False)
plt.xlabel(feature)
plt.show()
print(
f"Skewness was {round(old_skew[feature],2)} before & is {round(df_log.skew()[0],2)} after Log transformation."
)
logTrans(feature="LSTAT")
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#4c69b9;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# The skewness decreased from an extreme right skew to a moderately normal distribution and a negligible left skew.
# Closer the skew value to 0, more normal the distribution is.
#
# Now we will apply Log Transformation to a left skewed data and you will see how the distribution gets more skewed.
#
#
logTrans(feature="PTRATIO")
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#AC0000;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# Hence proved, never use Log transformation for left skewed data. It will mess your data up!
#
# ***
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#AF65C1;
# font-size:150%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# 2. Square Transform
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#59729F;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# In this transformation technique, we will use square of each and every value in the feature. This will only work on Left Skewed data. So, use Square Transform for left skewed data and Log Transform for right skewed data. Simple.
#
#
def squareTrans(
feature,
): # function to apply transformer and check the distribution with histogram and kdeplot
logTr = ColumnTransformer(
transformers=[("lg", FunctionTransformer(np.square), [feature])]
)
plt.figure(figsize=(15, 6))
plt.subplot(1, 2, 1)
plt.title("Distribution before Transformation", fontsize=15)
sns.histplot(df[feature], kde=True, color="red")
plt.subplot(1, 2, 2)
df_square = pd.DataFrame(logTr.fit_transform(df))
plt.title("Distribution after Transformation", fontsize=15)
sns.histplot(df_square, bins=20, kde=True, legend=False)
plt.xlabel(feature)
plt.show()
print(
f"Skewness was {round(old_skew[feature],2)} before & is {round(df_square.skew()[0],2)} after Square transformation."
)
squareTrans(feature="RM")
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#4c69b9;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# RM column was a little bit left skewed, however, the square transformation did not work and the skewness increased by a lot.
#
# Now we will apply Square Transformation to a right skewed data and you will see how the distribution gets more skewed.
#
#
squareTrans(feature="LSTAT")
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#AC0000;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# Hence proved, never use Square transformation for right skewed data. It will mess your data up!
#
# ***
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#AF65C1;
# font-size:150%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# 3. Reciprocal Transform, Squre Root and other Transformers
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#59729F;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# You should already have a clear picture of how we are using column transformer to apply a Function Transformer class of sklearn to the column of the dataframe and we can use any transformation we want. For example, we have tried log and square as of now. We can try, cube, square root, reciprocal or 1/x of the value as well. This is more like a trial and error and comes under fine tuning your feature engineering so that you can optimize your model. Try out as many things as you want! Comment your results.
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#4c69b9;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# For a demonstration I will be using cube transformer in RM feature to show you how it works compared to square transformer.
#
#
def cubeTrans(
feature,
): # function to apply transformer and check the distribution with histogram and kdeplot
logTr = ColumnTransformer(
transformers=[("lg", FunctionTransformer(lambda x: x**3), [feature])]
)
plt.figure(figsize=(15, 6))
plt.subplot(1, 2, 1)
plt.title("Distribution before Transformation", fontsize=15)
sns.histplot(df[feature], kde=True, color="red")
plt.subplot(1, 2, 2)
df_cube = pd.DataFrame(logTr.fit_transform(df))
plt.title("Distribution after Transformation", fontsize=15)
sns.histplot(df_cube, bins=20, kde=True, legend=False)
plt.xlabel(feature)
plt.show()
print(
f"Skewness was {round(old_skew[feature],2)} before & is {round(df_cube.skew()[0],2)} after cube transformation."
)
cubeTrans(feature="RM")
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#AF65C1;
# font-size:150%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# 4. Box-Cox
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#59729F;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# A Box Cox transformation is a transformation of a non-normal dependent variables into a normal shape. So the fun part is, box cox can be used in general for all type of distributions and it will by itself find the lambda value (check the formula below to know what lambda here is). Box-Cox cannot be used for negative values and 0.
#
# #### Formula of Box-Cox Transformation:
# 
from sklearn.preprocessing import PowerTransformer
# #### By default the PowerTransformer will standard scale the column so you need to specifically mention it if you do not want to scale.
def boxcoxtrans(
feature,
): # function to apply transformer and check the distribution with histogram and kdeplot
boxcoxTr = PowerTransformer(method="box-cox", standardize=True)
plt.figure(figsize=(15, 6))
plt.subplot(1, 2, 1)
plt.title("Distribution before Transformation", fontsize=15)
sns.histplot(df[feature], kde=True, color="red")
plt.subplot(1, 2, 2)
df_boxcox = pd.DataFrame(boxcoxTr.fit_transform(df[feature].values.reshape(-1, 1)))
plt.title("Distribution after Transformation", fontsize=15)
sns.histplot(df_boxcox, bins=20, kde=True, legend=False)
plt.xlabel(feature)
plt.show()
print(
f"Skewness was {round(old_skew[feature],2)} before & is {round(df_boxcox.skew()[0],2)} after Box-cox transformation."
)
boxcoxtrans(feature="RM")
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#4c69b9;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# Did you see how it automatically found out the value of the exponent that will be best for this data? Amazing isn't it?
# The next method will blow your mind!
#
# ***
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#AF65C1;
# font-size:150%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# 5. Yeo-Johnson
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#59729F;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# This is Box-cox but better.The limitation on box-cox is it does not work with negative values and 0. Yeo-Johnson fixes that and can be used for any distribution, both positive and negative. So, if you do not understand anything about distribution and know that your algorithm will be benifiting from a normal distribution, then copy pase this code and you are fine. LoL! Never copy paste codes, understand it. Atleast a basic intuition is better than mugging it up.
# In sklearn.preprocessing.PowerTransformer class, this method is the default, so you know it's good! Have fun!
#
# #### Formula of Yeo-Johnson Transformation:
# 
def yeojohntrans(
feature,
): # function to apply transformer and check the distribution with histogram and kdeplot
yeojohnTr = PowerTransformer(
standardize=True
) # not using method attribute as yeo-johnson is the default
plt.figure(figsize=(15, 6))
plt.subplot(1, 2, 1)
plt.title("Distribution before Transformation", fontsize=15)
sns.histplot(df[feature], kde=True, color="red")
plt.subplot(1, 2, 2)
df_yeojohn = pd.DataFrame(
yeojohnTr.fit_transform(df[feature].values.reshape(-1, 1))
)
plt.title("Distribution after Transformation", fontsize=15)
sns.histplot(df_yeojohn, bins=20, kde=True, legend=False)
plt.xlabel(feature)
plt.show()
print(
f"Skewness was {round(old_skew[feature],2)} before & is {round(df_yeojohn.skew()[0],2)} after Yeo-johnson transformation."
)
yeojohntrans(feature="RM")
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#4c69b9;
# font-size:110%;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 10px;
# color:white;">
# By far the best result for the feature RM! Let's check it out on other features as well.
# Let's check the same Yeo Johnson transformation in other features.
#
#
for i in df.columns[1:]:
yeojohntrans(i)
|
[{"bostonhoustingmlnd/housing.csv": {"column_names": "[\"RM\", \"LSTAT\", \"PTRATIO\", \"MEDV\"]", "column_data_types": "{\"RM\": \"float64\", \"LSTAT\": \"float64\", \"PTRATIO\": \"float64\", \"MEDV\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 489 entries, 0 to 488\nData columns (total 4 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 RM 489 non-null float64\n 1 LSTAT 489 non-null float64\n 2 PTRATIO 489 non-null float64\n 3 MEDV 489 non-null float64\ndtypes: float64(4)\nmemory usage: 15.4 KB\n", "summary": "{\"RM\": {\"count\": 489.0, \"mean\": 6.240288343558283, \"std\": 0.6436497627572433, \"min\": 3.561, \"25%\": 5.88, \"50%\": 6.185, \"75%\": 6.575, \"max\": 8.398}, \"LSTAT\": {\"count\": 489.0, \"mean\": 12.93963190184049, \"std\": 7.081989789065133, \"min\": 1.98, \"25%\": 7.37, \"50%\": 11.69, \"75%\": 17.12, \"max\": 37.97}, \"PTRATIO\": {\"count\": 489.0, \"mean\": 18.51656441717791, \"std\": 2.111267502630989, \"min\": 12.6, \"25%\": 17.4, \"50%\": 19.1, \"75%\": 20.2, \"max\": 22.0}, \"MEDV\": {\"count\": 489.0, \"mean\": 454342.9447852761, \"std\": 165340.27765266784, \"min\": 105000.0, \"25%\": 350700.0, \"50%\": 438900.0, \"75%\": 518700.0, \"max\": 1024800.0}}", "examples": "{\"RM\":{\"0\":6.575,\"1\":6.421,\"2\":7.185,\"3\":6.998},\"LSTAT\":{\"0\":4.98,\"1\":9.14,\"2\":4.03,\"3\":2.94},\"PTRATIO\":{\"0\":15.3,\"1\":17.8,\"2\":17.8,\"3\":18.7},\"MEDV\":{\"0\":504000.0,\"1\":453600.0,\"2\":728700.0,\"3\":701400.0}}"}}]
| true | 1 |
<start_data_description><data_path>bostonhoustingmlnd/housing.csv:
<column_names>
['RM', 'LSTAT', 'PTRATIO', 'MEDV']
<column_types>
{'RM': 'float64', 'LSTAT': 'float64', 'PTRATIO': 'float64', 'MEDV': 'float64'}
<dataframe_Summary>
{'RM': {'count': 489.0, 'mean': 6.240288343558283, 'std': 0.6436497627572433, 'min': 3.561, '25%': 5.88, '50%': 6.185, '75%': 6.575, 'max': 8.398}, 'LSTAT': {'count': 489.0, 'mean': 12.93963190184049, 'std': 7.081989789065133, 'min': 1.98, '25%': 7.37, '50%': 11.69, '75%': 17.12, 'max': 37.97}, 'PTRATIO': {'count': 489.0, 'mean': 18.51656441717791, 'std': 2.111267502630989, 'min': 12.6, '25%': 17.4, '50%': 19.1, '75%': 20.2, 'max': 22.0}, 'MEDV': {'count': 489.0, 'mean': 454342.9447852761, 'std': 165340.27765266784, 'min': 105000.0, '25%': 350700.0, '50%': 438900.0, '75%': 518700.0, 'max': 1024800.0}}
<dataframe_info>
RangeIndex: 489 entries, 0 to 488
Data columns (total 4 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 RM 489 non-null float64
1 LSTAT 489 non-null float64
2 PTRATIO 489 non-null float64
3 MEDV 489 non-null float64
dtypes: float64(4)
memory usage: 15.4 KB
<some_examples>
{'RM': {'0': 6.575, '1': 6.421, '2': 7.185, '3': 6.998}, 'LSTAT': {'0': 4.98, '1': 9.14, '2': 4.03, '3': 2.94}, 'PTRATIO': {'0': 15.3, '1': 17.8, '2': 17.8, '3': 18.7}, 'MEDV': {'0': 504000.0, '1': 453600.0, '2': 728700.0, '3': 701400.0}}
<end_description>
| 5,028 | 65 | 5,506 | 5,028 |
69956174
|
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#5642C5;
# font-size:33px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 15px;
# color:white;">
# Feature Scaling
#
# Feature Engineering is a big part of Data Science and Machine Learning. Feature Scaling is one of the last steps in the whole life cycle of Feature Engineering. It is a technique to standardize the independent features in a data in a fixed range or scale. Thus the name Feature Scaling.
# In simple words, once we are done with all the other steps of feature engineering, like ecoding variables, handling missing values etc, then we scale all the variable to a very small range of say -1 to +1. So all the data gets squeezed to decimal points between -1 and +1. What it does is keep the distribution of the data, the correlation and covariance absolutely the same however scales every independent or the feature matrix columns to a smaller scale. We do this as most of the ML algorithms problems perform significantly better after scaling.
# 
# ***
# ### Types of Feature Scaling:
# * Standardization:
# - Standard Scaler
# * Normalization:
# - Min Max Scaling
# - Mean Normalization
# - Max Absolute Scaling
# - Robust Scaling *etc.*
# ***
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#5F1666;
# font-size:33px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 15px;
# color:white;">
# Standardization
#
# Normalization is a technique often applied as part of data preparation for machine learning. The goal of normalization is to change the values of numeric columns in the dataset to use a common scale, without distorting differences in the ranges of values or losing information.
# #### Formula of Standardization:
# 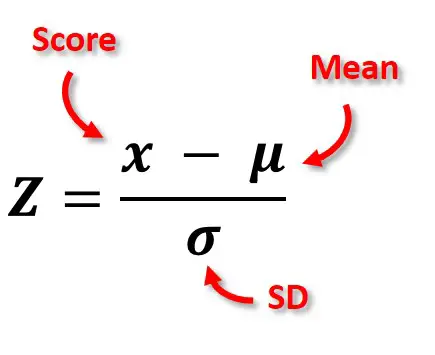
# <div style="font-size:16px;
# display:fill;
# border-radius:5px;
# font-family:Nexa;
# line-height: 1.7em;
# background-color:#FFD0D2">
# <p style="padding: 8px;
# color:black;">
# We will now implement this and see the results for ourselves
#
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_theme(palette="rainbow", style="darkgrid")
df = pd.read_csv("../input/titanic/train.csv", usecols=["Age", "Fare"])
df["Fare"].fillna(value=df["Fare"].mean(), inplace=True)
# importing sklearn StandardScaler class which is for Standardization
from sklearn.preprocessing import StandardScaler
sc = StandardScaler() # creating an instance of the class object
df_new = pd.DataFrame(
sc.fit_transform(df), columns=df.columns
) # fit and transforming StandardScaler the dataframe
# plotting the scatterplot of before and after Standardization
plt.figure(figsize=(14, 7))
plt.subplot(1, 2, 1)
plt.title("Scatterplot Before Standardization", fontsize=18)
sns.scatterplot(data=df, color="blue")
plt.subplot(1, 2, 2)
plt.title("Scatterplot After Standardization", fontsize=18)
sns.scatterplot(data=df_new, color="red")
plt.tight_layout()
plt.show()
# plotting the scatterplot of before and after Standardization
plt.figure(figsize=(14, 7))
plt.subplot(1, 2, 1)
plt.title("PDF Before Standardization", fontsize=18)
sns.kdeplot(data=df, color="blue")
plt.subplot(1, 2, 2)
plt.title("PDF After Standardization", fontsize=18)
sns.kdeplot(data=df_new, color="red")
plt.tight_layout()
plt.show()
#
# Notice how the mean of the distribution is very close to 0 and stadard deviation is exactly 1. This is what Standardization does. We had some outliers in Fare and that's why mean didn't reduce down to 0.
# Also notice that how in the Scatterplot, the scale changed and the distribution came to the centre or 0.
# In the probability density function, the kde plot is exactly the same, so this shows how the distribution is not effected by standardization.
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#5F1666;
# font-size:33px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 15px;
# color:white;">
# Normalization
#
# Normalization is a technique often applied as part of data preparation for machine learning. The goal of normalization is to change the values of numeric columns in the dataset to use a common scale, without distorting differences in the ranges of values or losing information.
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#26785F;
# font-size:18px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 8px;
# color:white;">
# 1 . Min Max Scaling
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#A9C9A9;
# font-size:16px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 8px;
# color:black;">
# Min-max normalization is one of the most common ways to normalize data. For every feature, the minimum value of that feature gets transformed into a 0, the maximum value gets transformed into a 1, and every other value gets transformed into a decimal between 0 and 1.
#
# #### Formula of Min Max Scaling:
#
# importing sklearn Min Max Scaler class which is for Standardization
from sklearn.preprocessing import MinMaxScaler
mm = MinMaxScaler() # creating an instance of the class object
df_new_mm = pd.DataFrame(
mm.fit_transform(df), columns=df.columns
) # fit and transforming MinMaxScaler the dataframe
# plotting the scatterplot of before and after Min Max Scaling
plt.figure(figsize=(14, 7))
plt.subplot(1, 2, 1)
plt.title("Scatterplot Before Min Max Scaling", fontsize=18)
sns.scatterplot(data=df, color="blue")
plt.subplot(1, 2, 2)
plt.title("Scatterplot After Min Max Scaling", fontsize=18)
sns.scatterplot(data=df_new_mm, color="red")
plt.tight_layout()
plt.show()
# plotting the scatterplot of before and after Min Max Scaling
plt.figure(figsize=(14, 7))
plt.subplot(1, 2, 1)
plt.title("PDF Before Min Max Scaling", fontsize=18)
sns.kdeplot(data=df, color="blue")
plt.subplot(1, 2, 2)
plt.title("PDF After Min Max Scaling", fontsize=18)
sns.kdeplot(data=df_new_mm, color="red")
plt.tight_layout()
plt.show()
#
# Min Max Normalization will perform best when the maximum and minimum value is very distinct and known.
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#2A3162;
# font-size:18px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 8px;
# color:white;">
# 2 . Max Absolute Scaling
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#D8E4FF;
# font-size:16px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 8px;
# color:black;">
# Scale each feature by its maximum absolute value.
# This estimator scales and translates each feature individually such that the maximal absolute value of each feature in the training set will be 1.0. It does not shift/center the data, and thus does not destroy any sparsity.
# This scaler can also be applied to sparse CSR or CSC matrices.
#
# #### Formula of Max Absolute Scaling:
#
# importing sklearn Min Max Scaler class which is for Max Absolute Scaling
from sklearn.preprocessing import MaxAbsScaler
ma = MaxAbsScaler() # creating an instance of the class object
df_new_ma = pd.DataFrame(
ma.fit_transform(df), columns=df.columns
) # fit and transforming Max Absolute Scaling the dataframe
# plotting the scatterplot of before and after Max Absolute Scaling
plt.figure(figsize=(14, 7))
plt.subplot(1, 2, 1)
plt.title("Scatterplot Before Max Absolute Scaling", fontsize=18)
sns.scatterplot(data=df, color="blue")
plt.subplot(1, 2, 2)
plt.title("Scatterplot After Max Absolute Scaling", fontsize=18)
sns.scatterplot(data=df_new_ma, color="red")
plt.tight_layout()
plt.show()
# plotting the scatterplot of before and after max Absolute Scaling
plt.figure(figsize=(14, 7))
plt.subplot(1, 2, 1)
plt.title("PDF Before Max Absolute Scaling", fontsize=18)
sns.kdeplot(data=df, color="blue")
plt.subplot(1, 2, 2)
plt.title("PDF After Max Absolute Scaling", fontsize=18)
sns.kdeplot(data=df_new_ma, color="red")
plt.tight_layout()
plt.show()
#
# Max Absolute scaling will perform a lot better in sparse data or when most of the values are 0.
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#4E3626;
# font-size:18px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 8px;
# color:white;">
# 1 . Robust Scaling
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#EEC3B3;
# font-size:16px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 8px;
# color:black;">
# This Scaler removes the median and scales the data according to the quantile range (defaults to IQR: Interquartile Range). The IQR is the range between the 1st quartile (25th quantile) and the 3rd quartile (75th quantile).
#
# #### Formula of Robust Scaling:
#
# importing sklearn Min Max Scaler class which is for Robust scaling
from sklearn.preprocessing import RobustScaler
rs = RobustScaler() # creating an instance of the class object
df_new_rs = pd.DataFrame(
rs.fit_transform(df), columns=df.columns
) # fit and transforming Robust Scaling the dataframe
# plotting the scatterplot of before and after Robust Scaling
plt.figure(figsize=(14, 7))
plt.subplot(1, 2, 1)
plt.title("Scatterplot Before Robust Scaling", fontsize=18)
sns.scatterplot(data=df, color="blue")
plt.subplot(1, 2, 2)
plt.title("Scatterplot After Robust Scaling", fontsize=18)
sns.scatterplot(data=df_new_rs, color="red")
plt.tight_layout()
plt.show()
# plotting the scatterplot of before and after Robust Scaling
plt.figure(figsize=(14, 7))
plt.subplot(1, 2, 1)
plt.title("PDF Before Robust Scaling", fontsize=18)
sns.kdeplot(data=df, color="blue")
plt.subplot(1, 2, 2)
plt.title("PDF After Robust Scaling", fontsize=18)
sns.kdeplot(data=df_new_rs, color="red")
plt.tight_layout()
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/956/69956174.ipynb
| null | null |
[{"Id": 69956174, "ScriptId": 19084181, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4347812, "CreationDate": "08/04/2021 02:34:54", "VersionNumber": 3.0, "Title": "Complete guide to Feature Scaling \ud83e\udd73", "EvaluationDate": "08/04/2021", "IsChange": false, "TotalLines": 369.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 369.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 11}]
| null | null | null | null |
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#5642C5;
# font-size:33px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 15px;
# color:white;">
# Feature Scaling
#
# Feature Engineering is a big part of Data Science and Machine Learning. Feature Scaling is one of the last steps in the whole life cycle of Feature Engineering. It is a technique to standardize the independent features in a data in a fixed range or scale. Thus the name Feature Scaling.
# In simple words, once we are done with all the other steps of feature engineering, like ecoding variables, handling missing values etc, then we scale all the variable to a very small range of say -1 to +1. So all the data gets squeezed to decimal points between -1 and +1. What it does is keep the distribution of the data, the correlation and covariance absolutely the same however scales every independent or the feature matrix columns to a smaller scale. We do this as most of the ML algorithms problems perform significantly better after scaling.
# 
# ***
# ### Types of Feature Scaling:
# * Standardization:
# - Standard Scaler
# * Normalization:
# - Min Max Scaling
# - Mean Normalization
# - Max Absolute Scaling
# - Robust Scaling *etc.*
# ***
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#5F1666;
# font-size:33px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 15px;
# color:white;">
# Standardization
#
# Normalization is a technique often applied as part of data preparation for machine learning. The goal of normalization is to change the values of numeric columns in the dataset to use a common scale, without distorting differences in the ranges of values or losing information.
# #### Formula of Standardization:
# 
# <div style="font-size:16px;
# display:fill;
# border-radius:5px;
# font-family:Nexa;
# line-height: 1.7em;
# background-color:#FFD0D2">
# <p style="padding: 8px;
# color:black;">
# We will now implement this and see the results for ourselves
#
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_theme(palette="rainbow", style="darkgrid")
df = pd.read_csv("../input/titanic/train.csv", usecols=["Age", "Fare"])
df["Fare"].fillna(value=df["Fare"].mean(), inplace=True)
# importing sklearn StandardScaler class which is for Standardization
from sklearn.preprocessing import StandardScaler
sc = StandardScaler() # creating an instance of the class object
df_new = pd.DataFrame(
sc.fit_transform(df), columns=df.columns
) # fit and transforming StandardScaler the dataframe
# plotting the scatterplot of before and after Standardization
plt.figure(figsize=(14, 7))
plt.subplot(1, 2, 1)
plt.title("Scatterplot Before Standardization", fontsize=18)
sns.scatterplot(data=df, color="blue")
plt.subplot(1, 2, 2)
plt.title("Scatterplot After Standardization", fontsize=18)
sns.scatterplot(data=df_new, color="red")
plt.tight_layout()
plt.show()
# plotting the scatterplot of before and after Standardization
plt.figure(figsize=(14, 7))
plt.subplot(1, 2, 1)
plt.title("PDF Before Standardization", fontsize=18)
sns.kdeplot(data=df, color="blue")
plt.subplot(1, 2, 2)
plt.title("PDF After Standardization", fontsize=18)
sns.kdeplot(data=df_new, color="red")
plt.tight_layout()
plt.show()
#
# Notice how the mean of the distribution is very close to 0 and stadard deviation is exactly 1. This is what Standardization does. We had some outliers in Fare and that's why mean didn't reduce down to 0.
# Also notice that how in the Scatterplot, the scale changed and the distribution came to the centre or 0.
# In the probability density function, the kde plot is exactly the same, so this shows how the distribution is not effected by standardization.
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#5F1666;
# font-size:33px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 15px;
# color:white;">
# Normalization
#
# Normalization is a technique often applied as part of data preparation for machine learning. The goal of normalization is to change the values of numeric columns in the dataset to use a common scale, without distorting differences in the ranges of values or losing information.
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#26785F;
# font-size:18px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 8px;
# color:white;">
# 1 . Min Max Scaling
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#A9C9A9;
# font-size:16px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 8px;
# color:black;">
# Min-max normalization is one of the most common ways to normalize data. For every feature, the minimum value of that feature gets transformed into a 0, the maximum value gets transformed into a 1, and every other value gets transformed into a decimal between 0 and 1.
#
# #### Formula of Min Max Scaling:
#
# importing sklearn Min Max Scaler class which is for Standardization
from sklearn.preprocessing import MinMaxScaler
mm = MinMaxScaler() # creating an instance of the class object
df_new_mm = pd.DataFrame(
mm.fit_transform(df), columns=df.columns
) # fit and transforming MinMaxScaler the dataframe
# plotting the scatterplot of before and after Min Max Scaling
plt.figure(figsize=(14, 7))
plt.subplot(1, 2, 1)
plt.title("Scatterplot Before Min Max Scaling", fontsize=18)
sns.scatterplot(data=df, color="blue")
plt.subplot(1, 2, 2)
plt.title("Scatterplot After Min Max Scaling", fontsize=18)
sns.scatterplot(data=df_new_mm, color="red")
plt.tight_layout()
plt.show()
# plotting the scatterplot of before and after Min Max Scaling
plt.figure(figsize=(14, 7))
plt.subplot(1, 2, 1)
plt.title("PDF Before Min Max Scaling", fontsize=18)
sns.kdeplot(data=df, color="blue")
plt.subplot(1, 2, 2)
plt.title("PDF After Min Max Scaling", fontsize=18)
sns.kdeplot(data=df_new_mm, color="red")
plt.tight_layout()
plt.show()
#
# Min Max Normalization will perform best when the maximum and minimum value is very distinct and known.
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#2A3162;
# font-size:18px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 8px;
# color:white;">
# 2 . Max Absolute Scaling
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#D8E4FF;
# font-size:16px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 8px;
# color:black;">
# Scale each feature by its maximum absolute value.
# This estimator scales and translates each feature individually such that the maximal absolute value of each feature in the training set will be 1.0. It does not shift/center the data, and thus does not destroy any sparsity.
# This scaler can also be applied to sparse CSR or CSC matrices.
#
# #### Formula of Max Absolute Scaling:
#
# importing sklearn Min Max Scaler class which is for Max Absolute Scaling
from sklearn.preprocessing import MaxAbsScaler
ma = MaxAbsScaler() # creating an instance of the class object
df_new_ma = pd.DataFrame(
ma.fit_transform(df), columns=df.columns
) # fit and transforming Max Absolute Scaling the dataframe
# plotting the scatterplot of before and after Max Absolute Scaling
plt.figure(figsize=(14, 7))
plt.subplot(1, 2, 1)
plt.title("Scatterplot Before Max Absolute Scaling", fontsize=18)
sns.scatterplot(data=df, color="blue")
plt.subplot(1, 2, 2)
plt.title("Scatterplot After Max Absolute Scaling", fontsize=18)
sns.scatterplot(data=df_new_ma, color="red")
plt.tight_layout()
plt.show()
# plotting the scatterplot of before and after max Absolute Scaling
plt.figure(figsize=(14, 7))
plt.subplot(1, 2, 1)
plt.title("PDF Before Max Absolute Scaling", fontsize=18)
sns.kdeplot(data=df, color="blue")
plt.subplot(1, 2, 2)
plt.title("PDF After Max Absolute Scaling", fontsize=18)
sns.kdeplot(data=df_new_ma, color="red")
plt.tight_layout()
plt.show()
#
# Max Absolute scaling will perform a lot better in sparse data or when most of the values are 0.
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#4E3626;
# font-size:18px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 8px;
# color:white;">
# 1 . Robust Scaling
#
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#EEC3B3;
# font-size:16px;
# font-family:Nexa;
# letter-spacing:0.5px">
# <p style="padding: 8px;
# color:black;">
# This Scaler removes the median and scales the data according to the quantile range (defaults to IQR: Interquartile Range). The IQR is the range between the 1st quartile (25th quantile) and the 3rd quartile (75th quantile).
#
# #### Formula of Robust Scaling:
#
# importing sklearn Min Max Scaler class which is for Robust scaling
from sklearn.preprocessing import RobustScaler
rs = RobustScaler() # creating an instance of the class object
df_new_rs = pd.DataFrame(
rs.fit_transform(df), columns=df.columns
) # fit and transforming Robust Scaling the dataframe
# plotting the scatterplot of before and after Robust Scaling
plt.figure(figsize=(14, 7))
plt.subplot(1, 2, 1)
plt.title("Scatterplot Before Robust Scaling", fontsize=18)
sns.scatterplot(data=df, color="blue")
plt.subplot(1, 2, 2)
plt.title("Scatterplot After Robust Scaling", fontsize=18)
sns.scatterplot(data=df_new_rs, color="red")
plt.tight_layout()
plt.show()
# plotting the scatterplot of before and after Robust Scaling
plt.figure(figsize=(14, 7))
plt.subplot(1, 2, 1)
plt.title("PDF Before Robust Scaling", fontsize=18)
sns.kdeplot(data=df, color="blue")
plt.subplot(1, 2, 2)
plt.title("PDF After Robust Scaling", fontsize=18)
sns.kdeplot(data=df_new_rs, color="red")
plt.tight_layout()
plt.show()
| false | 0 | 3,180 | 11 | 3,180 | 3,180 |
||
69956576
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train_data = pd.read_csv("/kaggle/input/titanic/train.csv")
train_data.head()
test_data = pd.read_csv("/kaggle/input/titanic/test.csv")
test_data.head()
women = train_data.loc[train_data.Sex == "female"]["Survived"]
rate_women = sum(women) / len(women)
print("% of women who survived:", rate_women)
men = train_data.loc[train_data.Sex == "male"]["Survived"]
rate_men = sum(men) / len(men)
print("% of men who survived:", rate_men)
from sklearn.ensemble import RandomForestClassifier
y = train_data["Survived"]
features = ["Pclass", "Sex", "SibSp", "Parch"]
X = pd.get_dummies(train_data[features])
X_test = pd.get_dummies(test_data[features])
model = RandomForestClassifier(n_estimators=100, max_depth=5, random_state=1)
model.fit(X, y)
predictions = model.predict(X_test)
output = pd.DataFrame({"PassengerId": test_data.PassengerId, "Survived": predictions})
output.to_csv("my_submission.csv", index=False)
print("Your submission was successfully saved!")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/956/69956576.ipynb
| null | null |
[{"Id": 69956576, "ScriptId": 19131881, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8006241, "CreationDate": "08/04/2021 02:38:25", "VersionNumber": 2.0, "Title": "Getting Started with Titanic", "EvaluationDate": "08/04/2021", "IsChange": false, "TotalLines": 49.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 49.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 8}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train_data = pd.read_csv("/kaggle/input/titanic/train.csv")
train_data.head()
test_data = pd.read_csv("/kaggle/input/titanic/test.csv")
test_data.head()
women = train_data.loc[train_data.Sex == "female"]["Survived"]
rate_women = sum(women) / len(women)
print("% of women who survived:", rate_women)
men = train_data.loc[train_data.Sex == "male"]["Survived"]
rate_men = sum(men) / len(men)
print("% of men who survived:", rate_men)
from sklearn.ensemble import RandomForestClassifier
y = train_data["Survived"]
features = ["Pclass", "Sex", "SibSp", "Parch"]
X = pd.get_dummies(train_data[features])
X_test = pd.get_dummies(test_data[features])
model = RandomForestClassifier(n_estimators=100, max_depth=5, random_state=1)
model.fit(X, y)
predictions = model.predict(X_test)
output = pd.DataFrame({"PassengerId": test_data.PassengerId, "Survived": predictions})
output.to_csv("my_submission.csv", index=False)
print("Your submission was successfully saved!")
| false | 0 | 520 | 8 | 520 | 520 |
||
69498500
|
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
import pylab
import scipy.stats as stats
from sklearn.preprocessing import MinMaxScaler
data_path = "../input/titanic/train.csv"
df = pd.read_csv(data_path)
test_data_path = "../input/titanic/test.csv"
df_test = pd.read_csv(test_data_path)
df_test.head()
test_passenger_id = df_test["PassengerId"].values
df.head()
print(f"Data frame shape: {df.shape}")
df.isnull().sum()
df_test.isnull().sum()
# There are missing values in the dataset which needs to handled
print(f"Percentage of data missing for Age: {177/891*100}")
# It's not recommended to impute more than 5% of missing data. Imputing more than 5% of data might result in bias
print("Oldest Passenger was of:", df.Age.max(), "Years")
print("Youngest Passenger was of:", df.Age.min(), "Years")
print(
"Average Age of the Passengers in ship:" + "{:.2f}".format(df.Age.mean()), "Years"
)
sns.distplot(df["Age"])
# Age is not normally distributed imputing the mean is not a right approximation. Replacing NaN values with 0 and adding a categorical variable to indicate missing data
df["AgeMissing"] = 0
df.loc[df["Age"].isnull(), "AgeMissing"] = 1
df["Age"].fillna(0, inplace=True)
df_test["AgeMissing"] = 0
df_test.loc[df_test["Age"].isnull(), "AgeMissing"] = 1
df_test["Age"].fillna(0, inplace=True)
# Adding a variable to indicate whether the survivor is child or not.
df["isChild"] = 0
df.loc[df["Age"] <= 10, "isChild"] = 1
df_test["isChild"] = 0
df_test.loc[df_test["Age"] <= 10, "isChild"] = 1
# Embarked has 2 missing values filling it by its mode
df["Embarked"].fillna(df["Embarked"].mode()[0], inplace=True)
# Fill NaN value for fare in test data
df_test["Fare"].fillna(df_test["Fare"].mean(), inplace=True)
df["Initial"] = 0
for i in df:
df["Initial"] = df.Name.str.extract("([A-Za-z]+)\.")
df["Initial"].replace(
[
"Mlle",
"Mme",
"Ms",
"Dr",
"Major",
"Lady",
"Countess",
"Jonkheer",
"Col",
"Rev",
"Capt",
"Sir",
"Don",
"Dona",
],
[
"Miss",
"Miss",
"Miss",
"Mr",
"Mr",
"Mrs",
"Mrs",
"Mrs",
"Mr",
"Mr",
"Mr",
"Mr",
"Mr",
"Mr",
],
inplace=True,
)
df_test["Initial"] = 0
for i in df_test:
df_test["Initial"] = df_test.Name.str.extract("([A-Za-z]+)\.")
df_test["Initial"].replace(
[
"Mlle",
"Mme",
"Ms",
"Dr",
"Major",
"Lady",
"Countess",
"Jonkheer",
"Col",
"Rev",
"Capt",
"Sir",
"Don",
"Dona",
],
[
"Miss",
"Miss",
"Miss",
"Mr",
"Mr",
"Mrs",
"Mrs",
"Mrs",
"Mr",
"Mr",
"Mr",
"Mr",
"Mr",
"Mr",
],
inplace=True,
)
df.groupby("Initial")["Age"].mean().reset_index(name="Mean")
df["Family_Size"] = 0
df["Family_Size"] = df.Parch + df.SibSp
df["Alone"] = 0
df.loc[df["Family_Size"] == 0, "Alone"] = 1
df_test["Family_Size"] = 0
df_test["Family_Size"] = df_test.Parch + df_test.SibSp
df_test["Alone"] = 0
df_test.loc[df_test["Family_Size"] == 0, "Alone"] = 1
# Dropping Name, PassengerId, Ticket, Cabin
df.drop(["Name", "Ticket", "Cabin", "PassengerId"], axis=1, inplace=True)
df_test.drop(["Name", "Ticket", "Cabin", "PassengerId"], axis=1, inplace=True)
# Encoding the string values to integer
one_hot_encoded_df = pd.get_dummies(
df, columns=["Sex", "Embarked", "Pclass", "Initial"]
)
one_hot_encoded_df_test = pd.get_dummies(
df_test, columns=["Sex", "Embarked", "Pclass", "Initial"]
)
one_hot_encoded_df.head()
one_hot_encoded_df_test.head()
scaler = MinMaxScaler()
one_hot_encoded_df.iloc[:, 1:] = scaler.fit_transform(
one_hot_encoded_df.iloc[:, 1:].to_numpy()
)
one_hot_encoded_df_test.iloc[:, 0:] = scaler.fit_transform(
one_hot_encoded_df_test.iloc[:, 0:].to_numpy()
)
one_hot_encoded_df.head()
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import confusion_matrix
from sklearn.svm import SVC
train, val = train_test_split(
one_hot_encoded_df, test_size=0.3, random_state=0, stratify=df["Survived"]
)
train_X = train[train.columns[1:]]
train_Y = train[train.columns[:1]]
val_X = val[val.columns[1:]]
val_Y = val[val.columns[:1]]
model_KNN = KNeighborsClassifier(n_neighbors=5)
model_KNN.fit(train_X, train_Y)
train_pred_KNN = model_KNN.predict(train_X)
val_pred_KNN = model_KNN.predict(val_X)
print(f"KNN Train accuracy:{metrics.accuracy_score(train_pred_KNN, train_Y):.3f}")
print(f"KNN Validation accuracy:{metrics.accuracy_score(val_pred_KNN, val_Y):.3f}")
svclassifier = SVC(kernel="rbf")
svclassifier.fit(train_X, train_Y)
train_pred_SVM = svclassifier.predict(train_X)
pred_SVM = svclassifier.predict(val_X)
print(f"SVM Train accuracy:{metrics.accuracy_score(train_pred_SVM, train_Y):.3f}")
print(f"SVM Validation accuracy:{metrics.accuracy_score(pred_SVM, val_Y):.3f}")
# Performaning variable selection
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import chi2
# feature selection
def select_features(X_train, y_train, X_test):
fs = SelectKBest(score_func=chi2, k="all")
fs.fit(X_train, y_train)
X_train_fs = fs.transform(X_train)
X_test_fs = fs.transform(X_test)
return X_train_fs, X_test_fs, fs
train_fs, X_test_fs, fs = select_features(train_X, train_Y, val_X)
col_names = one_hot_encoded_df.columns.values
col_names = one_hot_encoded_df.columns
for i, col in zip(range(len(fs.scores_)), col_names[1:]):
print(f"{col} : {fs.scores_[i]}")
sns.barplot([i for i in range(len(fs.scores_))], fs.scores_)
plt.show()
# Age, SibSp, Embarked_Q are not so important variable
one_hot_encoded_df.drop(
["Age", "isChild", "AgeMissing", "Embarked_Q"], axis=1, inplace=True
)
one_hot_encoded_df_test.drop(
["Age", "isChild", "AgeMissing", "Embarked_Q"], axis=1, inplace=True
)
one_hot_encoded_df.head()
train, val = train_test_split(
one_hot_encoded_df, test_size=0.3, random_state=0, stratify=df["Survived"]
)
train_X = train[train.columns[1:]]
train_Y = train[train.columns[:1]]
val_X = val[val.columns[1:]]
val_Y = val[val.columns[:1]]
model_KNN = KNeighborsClassifier(n_neighbors=5)
model_KNN.fit(train_X, train_Y)
train_pred_KNN = model_KNN.predict(train_X)
val_pred_KNN = model_KNN.predict(val_X)
print(f"KNN Train accuracy:{metrics.accuracy_score(train_pred_KNN, train_Y):.3f}")
print(f"KNN Validation accuracy:{metrics.accuracy_score(val_pred_KNN, val_Y):.3f}")
svclassifier = SVC(kernel="rbf")
svclassifier.fit(train_X, train_Y)
train_pred_SVM = svclassifier.predict(train_X)
pred_SVM = svclassifier.predict(val_X)
print(f"SVM Train accuracy:{metrics.accuracy_score(train_pred_SVM, train_Y):.3f}")
print(f"SVM Validation accuracy:{metrics.accuracy_score(pred_SVM, val_Y):.3f}")
test_pred = svclassifier.predict(
one_hot_encoded_df_test[one_hot_encoded_df_test.columns[:]]
)
column_names = ["PassengerId", "Survived"]
result = pd.DataFrame(columns=column_names)
result["PassengerId"] = test_passenger_id
result["Survived"] = test_pred
result.to_csv("submission.csv", index=False)
pd.read_csv("./submission.csv")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/498/69498500.ipynb
| null | null |
[{"Id": 69498500, "ScriptId": 18955703, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1626886, "CreationDate": "07/31/2021 18:33:29", "VersionNumber": 3.0, "Title": "Classification using SVM, KNN - Variable Selection", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 214.0, "LinesInsertedFromPrevious": 10.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 204.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 8}]
| null | null | null | null |
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
import pylab
import scipy.stats as stats
from sklearn.preprocessing import MinMaxScaler
data_path = "../input/titanic/train.csv"
df = pd.read_csv(data_path)
test_data_path = "../input/titanic/test.csv"
df_test = pd.read_csv(test_data_path)
df_test.head()
test_passenger_id = df_test["PassengerId"].values
df.head()
print(f"Data frame shape: {df.shape}")
df.isnull().sum()
df_test.isnull().sum()
# There are missing values in the dataset which needs to handled
print(f"Percentage of data missing for Age: {177/891*100}")
# It's not recommended to impute more than 5% of missing data. Imputing more than 5% of data might result in bias
print("Oldest Passenger was of:", df.Age.max(), "Years")
print("Youngest Passenger was of:", df.Age.min(), "Years")
print(
"Average Age of the Passengers in ship:" + "{:.2f}".format(df.Age.mean()), "Years"
)
sns.distplot(df["Age"])
# Age is not normally distributed imputing the mean is not a right approximation. Replacing NaN values with 0 and adding a categorical variable to indicate missing data
df["AgeMissing"] = 0
df.loc[df["Age"].isnull(), "AgeMissing"] = 1
df["Age"].fillna(0, inplace=True)
df_test["AgeMissing"] = 0
df_test.loc[df_test["Age"].isnull(), "AgeMissing"] = 1
df_test["Age"].fillna(0, inplace=True)
# Adding a variable to indicate whether the survivor is child or not.
df["isChild"] = 0
df.loc[df["Age"] <= 10, "isChild"] = 1
df_test["isChild"] = 0
df_test.loc[df_test["Age"] <= 10, "isChild"] = 1
# Embarked has 2 missing values filling it by its mode
df["Embarked"].fillna(df["Embarked"].mode()[0], inplace=True)
# Fill NaN value for fare in test data
df_test["Fare"].fillna(df_test["Fare"].mean(), inplace=True)
df["Initial"] = 0
for i in df:
df["Initial"] = df.Name.str.extract("([A-Za-z]+)\.")
df["Initial"].replace(
[
"Mlle",
"Mme",
"Ms",
"Dr",
"Major",
"Lady",
"Countess",
"Jonkheer",
"Col",
"Rev",
"Capt",
"Sir",
"Don",
"Dona",
],
[
"Miss",
"Miss",
"Miss",
"Mr",
"Mr",
"Mrs",
"Mrs",
"Mrs",
"Mr",
"Mr",
"Mr",
"Mr",
"Mr",
"Mr",
],
inplace=True,
)
df_test["Initial"] = 0
for i in df_test:
df_test["Initial"] = df_test.Name.str.extract("([A-Za-z]+)\.")
df_test["Initial"].replace(
[
"Mlle",
"Mme",
"Ms",
"Dr",
"Major",
"Lady",
"Countess",
"Jonkheer",
"Col",
"Rev",
"Capt",
"Sir",
"Don",
"Dona",
],
[
"Miss",
"Miss",
"Miss",
"Mr",
"Mr",
"Mrs",
"Mrs",
"Mrs",
"Mr",
"Mr",
"Mr",
"Mr",
"Mr",
"Mr",
],
inplace=True,
)
df.groupby("Initial")["Age"].mean().reset_index(name="Mean")
df["Family_Size"] = 0
df["Family_Size"] = df.Parch + df.SibSp
df["Alone"] = 0
df.loc[df["Family_Size"] == 0, "Alone"] = 1
df_test["Family_Size"] = 0
df_test["Family_Size"] = df_test.Parch + df_test.SibSp
df_test["Alone"] = 0
df_test.loc[df_test["Family_Size"] == 0, "Alone"] = 1
# Dropping Name, PassengerId, Ticket, Cabin
df.drop(["Name", "Ticket", "Cabin", "PassengerId"], axis=1, inplace=True)
df_test.drop(["Name", "Ticket", "Cabin", "PassengerId"], axis=1, inplace=True)
# Encoding the string values to integer
one_hot_encoded_df = pd.get_dummies(
df, columns=["Sex", "Embarked", "Pclass", "Initial"]
)
one_hot_encoded_df_test = pd.get_dummies(
df_test, columns=["Sex", "Embarked", "Pclass", "Initial"]
)
one_hot_encoded_df.head()
one_hot_encoded_df_test.head()
scaler = MinMaxScaler()
one_hot_encoded_df.iloc[:, 1:] = scaler.fit_transform(
one_hot_encoded_df.iloc[:, 1:].to_numpy()
)
one_hot_encoded_df_test.iloc[:, 0:] = scaler.fit_transform(
one_hot_encoded_df_test.iloc[:, 0:].to_numpy()
)
one_hot_encoded_df.head()
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import confusion_matrix
from sklearn.svm import SVC
train, val = train_test_split(
one_hot_encoded_df, test_size=0.3, random_state=0, stratify=df["Survived"]
)
train_X = train[train.columns[1:]]
train_Y = train[train.columns[:1]]
val_X = val[val.columns[1:]]
val_Y = val[val.columns[:1]]
model_KNN = KNeighborsClassifier(n_neighbors=5)
model_KNN.fit(train_X, train_Y)
train_pred_KNN = model_KNN.predict(train_X)
val_pred_KNN = model_KNN.predict(val_X)
print(f"KNN Train accuracy:{metrics.accuracy_score(train_pred_KNN, train_Y):.3f}")
print(f"KNN Validation accuracy:{metrics.accuracy_score(val_pred_KNN, val_Y):.3f}")
svclassifier = SVC(kernel="rbf")
svclassifier.fit(train_X, train_Y)
train_pred_SVM = svclassifier.predict(train_X)
pred_SVM = svclassifier.predict(val_X)
print(f"SVM Train accuracy:{metrics.accuracy_score(train_pred_SVM, train_Y):.3f}")
print(f"SVM Validation accuracy:{metrics.accuracy_score(pred_SVM, val_Y):.3f}")
# Performaning variable selection
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import chi2
# feature selection
def select_features(X_train, y_train, X_test):
fs = SelectKBest(score_func=chi2, k="all")
fs.fit(X_train, y_train)
X_train_fs = fs.transform(X_train)
X_test_fs = fs.transform(X_test)
return X_train_fs, X_test_fs, fs
train_fs, X_test_fs, fs = select_features(train_X, train_Y, val_X)
col_names = one_hot_encoded_df.columns.values
col_names = one_hot_encoded_df.columns
for i, col in zip(range(len(fs.scores_)), col_names[1:]):
print(f"{col} : {fs.scores_[i]}")
sns.barplot([i for i in range(len(fs.scores_))], fs.scores_)
plt.show()
# Age, SibSp, Embarked_Q are not so important variable
one_hot_encoded_df.drop(
["Age", "isChild", "AgeMissing", "Embarked_Q"], axis=1, inplace=True
)
one_hot_encoded_df_test.drop(
["Age", "isChild", "AgeMissing", "Embarked_Q"], axis=1, inplace=True
)
one_hot_encoded_df.head()
train, val = train_test_split(
one_hot_encoded_df, test_size=0.3, random_state=0, stratify=df["Survived"]
)
train_X = train[train.columns[1:]]
train_Y = train[train.columns[:1]]
val_X = val[val.columns[1:]]
val_Y = val[val.columns[:1]]
model_KNN = KNeighborsClassifier(n_neighbors=5)
model_KNN.fit(train_X, train_Y)
train_pred_KNN = model_KNN.predict(train_X)
val_pred_KNN = model_KNN.predict(val_X)
print(f"KNN Train accuracy:{metrics.accuracy_score(train_pred_KNN, train_Y):.3f}")
print(f"KNN Validation accuracy:{metrics.accuracy_score(val_pred_KNN, val_Y):.3f}")
svclassifier = SVC(kernel="rbf")
svclassifier.fit(train_X, train_Y)
train_pred_SVM = svclassifier.predict(train_X)
pred_SVM = svclassifier.predict(val_X)
print(f"SVM Train accuracy:{metrics.accuracy_score(train_pred_SVM, train_Y):.3f}")
print(f"SVM Validation accuracy:{metrics.accuracy_score(pred_SVM, val_Y):.3f}")
test_pred = svclassifier.predict(
one_hot_encoded_df_test[one_hot_encoded_df_test.columns[:]]
)
column_names = ["PassengerId", "Survived"]
result = pd.DataFrame(columns=column_names)
result["PassengerId"] = test_passenger_id
result["Survived"] = test_pred
result.to_csv("submission.csv", index=False)
pd.read_csv("./submission.csv")
| false | 0 | 2,689 | 8 | 2,689 | 2,689 |
||
69498296
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory
import os
print(os.listdir("../input"))
# Any results you write to the current directory are saved as output.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import geopandas as gpd
from plotly.offline import init_notebook_mode, iplot
import plotly.graph_objs as go
import plotly.plotly as py
from plotly import tools
import plotly.figure_factory as ff
import folium
from folium import plugins
from io import StringIO
init_notebook_mode(connected=True)
speed_cam_data = pd.read_csv("../input/speed-camera-locations.csv")
speed_cam_data.head()
speed_cam_v_data = pd.read_csv("../input/speed-camera-violations.csv")
speed_cam_v_data.head()
pd.isnull(speed_cam_v_data["LATITUDE"]).count()
# Code need to verfied and modified to taker consider the data with null values for lattitude and longitude.
# These columns can be populated based on address field which is common to both csv files
# **Extract date parameters and as columns to dataframe**
import datetime
speed_cam_v_data["Violation date"] = pd.to_datetime(
speed_cam_v_data["VIOLATION DATE"]
) # timestamp
speed_cam_v_data["day_name"] = speed_cam_v_data[
"Violation date"
].dt.day_name() # get day of the week
speed_cam_v_data["month_name"] = speed_cam_v_data["Violation date"].dt.month_name()
speed_cam_v_data["month"] = speed_cam_v_data["Violation date"].dt.month
speed_cam_v_data["month"] = speed_cam_v_data.month.map("{:02}".format)
speed_cam_v_data["year"] = speed_cam_v_data["Violation date"].dt.year
speed_cam_v_data["yearmonth"] = (
speed_cam_v_data["year"].astype(str) + "-" + speed_cam_v_data["month"]
)
# speed_cam_v_data['day']=speed_cam_v_data['Violation date'].dt.day
speed_cam_v_data.head()
# **Analyze last 7 days of violations data from speed camera**
# To analyze last 7 days data
startdate = max(speed_cam_v_data["Violation date"]) + datetime.timedelta(
days=-6
) # To analyze last 7 days data
lastweek_v = speed_cam_v_data[(speed_cam_v_data["Violation date"] >= startdate)]
# get total violations grouped by ....
lastweek_v_g = (
lastweek_v.groupby(["ADDRESS", "LATITUDE", "LONGITUDE"])["VIOLATIONS"]
.sum()
.sort_values(ascending=False)
.reset_index()
)
max(lastweek_v_g["VIOLATIONS"])
# Note locations with more than half of max violations identified by speed camera.
# Such locations can be closely watched to see for more accidents etc
v_limit = int(max(lastweek_v_g["VIOLATIONS"]) / 2)
lastweek_v_max = lastweek_v_g
v_map = folium.Map(
location=[41.878, -87.62], height=700, tiles="OpenStreetMap", zoom_start=12
)
for i in range(0, len(lastweek_v_max)):
folium.Marker(
[lastweek_v_max.iloc[i]["LATITUDE"], lastweek_v_max.iloc[i]["LONGITUDE"]],
popup=lastweek_v_max.iloc[i]["ADDRESS"],
icon=folium.Icon(
color="red" if lastweek_v_max.iloc[i]["VIOLATIONS"] > v_limit else "green",
icon="circle",
),
).add_to(v_map)
v_map
# **Analyze effectiveness of speed camera checking, are violations coming down fin last 12 months?**
# prepare the data for the last one year and draw a scatter plot
# get the todate for analysis. Assume todate as the last day of the previous month
maxdate = max(speed_cam_v_data["Violation date"])
maxday = maxdate.day
todate = maxdate + datetime.timedelta(days=-maxday) # last day of the previous month
#
from dateutil.relativedelta import relativedelta
# years_ago = datetime.datetime.now() - relativedelta(years=5)
fromdate = todate - relativedelta(years=1)
oneyear_v = speed_cam_v_data[
(speed_cam_v_data["Violation date"] <= todate)
& (speed_cam_v_data["Violation date"] > fromdate)
]
oneyear_v.count()
# get total violations grOuped by
oneyear_v_g = (
oneyear_v.groupby(["CAMERA ID", "ADDRESS", "LATITUDE", "LONGITUDE"])["VIOLATIONS"]
.sum()
.sort_values(ascending=False)
.reset_index()
)
threshold = int(max(oneyear_v_g["VIOLATIONS"]) / 2)
cols = [
"CAMERA ID",
"ADDRESS",
"LATITUDE",
"LONGITUDE",
"year",
"month",
"month_name",
"yearmonth",
"VIOLATIONS",
]
oneyear_vg = oneyear_v[cols]
oneyear_ag = oneyear_vg.groupby(
["CAMERA ID", "yearmonth", "ADDRESS", "LATITUDE", "LONGITUDE"]
).agg({"VIOLATIONS": sum})
oneyear_v_treshold = oneyear_ag.groupby(
["CAMERA ID", "ADDRESS", "LATITUDE", "LONGITUDE"]
).filter(lambda x: x["VIOLATIONS"].sum() > threshold)
oneyear_v_treshold_noind_go = oneyear_v_treshold.reset_index()
# oneyear_v_treshold_noind_go
data = []
for name1, group in oneyear_v_treshold_noind_go[
["CAMERA ID", "yearmonth", "VIOLATIONS"]
].groupby("CAMERA ID"):
data.append(go.Scatter(x=group.yearmonth, y=group.VIOLATIONS, name=name1))
layout = dict(
title="Top violations by month for last 12 months",
xaxis=dict(title="Date", ticklen=5, zeroline=False),
yaxis=dict(title="Violations", ticklen=5, zeroline=False),
)
# create and show our figure
fig = dict(data=data, layout=layout)
iplot(fig)
# For concerned authorities to follow-up the effectiveness of speed camera system
# address of camera will be easier, instead of camera-id
data = []
for name1, group in oneyear_v_treshold_noind_go[
["ADDRESS", "yearmonth", "VIOLATIONS"]
].groupby("ADDRESS"):
data.append(go.Scatter(x=group.yearmonth, y=group.VIOLATIONS, name=name1))
layout = dict(
title="Top violations by month for last 12 months",
xaxis=dict(title="Date", ticklen=5, zeroline=False),
yaxis=dict(title="Violations", ticklen=5, zeroline=False),
)
# create and show our figure
fig = dict(data=data, layout=layout)
iplot(fig)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/498/69498296.ipynb
| null | null |
[{"Id": 69498296, "ScriptId": 2411663, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1241268, "CreationDate": "07/31/2021 18:30:08", "VersionNumber": 888.0, "Title": "Analyze Chicago Red Light and Speed Camera Data", "EvaluationDate": "07/31/2021", "IsChange": false, "TotalLines": 130.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 130.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory
import os
print(os.listdir("../input"))
# Any results you write to the current directory are saved as output.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import geopandas as gpd
from plotly.offline import init_notebook_mode, iplot
import plotly.graph_objs as go
import plotly.plotly as py
from plotly import tools
import plotly.figure_factory as ff
import folium
from folium import plugins
from io import StringIO
init_notebook_mode(connected=True)
speed_cam_data = pd.read_csv("../input/speed-camera-locations.csv")
speed_cam_data.head()
speed_cam_v_data = pd.read_csv("../input/speed-camera-violations.csv")
speed_cam_v_data.head()
pd.isnull(speed_cam_v_data["LATITUDE"]).count()
# Code need to verfied and modified to taker consider the data with null values for lattitude and longitude.
# These columns can be populated based on address field which is common to both csv files
# **Extract date parameters and as columns to dataframe**
import datetime
speed_cam_v_data["Violation date"] = pd.to_datetime(
speed_cam_v_data["VIOLATION DATE"]
) # timestamp
speed_cam_v_data["day_name"] = speed_cam_v_data[
"Violation date"
].dt.day_name() # get day of the week
speed_cam_v_data["month_name"] = speed_cam_v_data["Violation date"].dt.month_name()
speed_cam_v_data["month"] = speed_cam_v_data["Violation date"].dt.month
speed_cam_v_data["month"] = speed_cam_v_data.month.map("{:02}".format)
speed_cam_v_data["year"] = speed_cam_v_data["Violation date"].dt.year
speed_cam_v_data["yearmonth"] = (
speed_cam_v_data["year"].astype(str) + "-" + speed_cam_v_data["month"]
)
# speed_cam_v_data['day']=speed_cam_v_data['Violation date'].dt.day
speed_cam_v_data.head()
# **Analyze last 7 days of violations data from speed camera**
# To analyze last 7 days data
startdate = max(speed_cam_v_data["Violation date"]) + datetime.timedelta(
days=-6
) # To analyze last 7 days data
lastweek_v = speed_cam_v_data[(speed_cam_v_data["Violation date"] >= startdate)]
# get total violations grouped by ....
lastweek_v_g = (
lastweek_v.groupby(["ADDRESS", "LATITUDE", "LONGITUDE"])["VIOLATIONS"]
.sum()
.sort_values(ascending=False)
.reset_index()
)
max(lastweek_v_g["VIOLATIONS"])
# Note locations with more than half of max violations identified by speed camera.
# Such locations can be closely watched to see for more accidents etc
v_limit = int(max(lastweek_v_g["VIOLATIONS"]) / 2)
lastweek_v_max = lastweek_v_g
v_map = folium.Map(
location=[41.878, -87.62], height=700, tiles="OpenStreetMap", zoom_start=12
)
for i in range(0, len(lastweek_v_max)):
folium.Marker(
[lastweek_v_max.iloc[i]["LATITUDE"], lastweek_v_max.iloc[i]["LONGITUDE"]],
popup=lastweek_v_max.iloc[i]["ADDRESS"],
icon=folium.Icon(
color="red" if lastweek_v_max.iloc[i]["VIOLATIONS"] > v_limit else "green",
icon="circle",
),
).add_to(v_map)
v_map
# **Analyze effectiveness of speed camera checking, are violations coming down fin last 12 months?**
# prepare the data for the last one year and draw a scatter plot
# get the todate for analysis. Assume todate as the last day of the previous month
maxdate = max(speed_cam_v_data["Violation date"])
maxday = maxdate.day
todate = maxdate + datetime.timedelta(days=-maxday) # last day of the previous month
#
from dateutil.relativedelta import relativedelta
# years_ago = datetime.datetime.now() - relativedelta(years=5)
fromdate = todate - relativedelta(years=1)
oneyear_v = speed_cam_v_data[
(speed_cam_v_data["Violation date"] <= todate)
& (speed_cam_v_data["Violation date"] > fromdate)
]
oneyear_v.count()
# get total violations grOuped by
oneyear_v_g = (
oneyear_v.groupby(["CAMERA ID", "ADDRESS", "LATITUDE", "LONGITUDE"])["VIOLATIONS"]
.sum()
.sort_values(ascending=False)
.reset_index()
)
threshold = int(max(oneyear_v_g["VIOLATIONS"]) / 2)
cols = [
"CAMERA ID",
"ADDRESS",
"LATITUDE",
"LONGITUDE",
"year",
"month",
"month_name",
"yearmonth",
"VIOLATIONS",
]
oneyear_vg = oneyear_v[cols]
oneyear_ag = oneyear_vg.groupby(
["CAMERA ID", "yearmonth", "ADDRESS", "LATITUDE", "LONGITUDE"]
).agg({"VIOLATIONS": sum})
oneyear_v_treshold = oneyear_ag.groupby(
["CAMERA ID", "ADDRESS", "LATITUDE", "LONGITUDE"]
).filter(lambda x: x["VIOLATIONS"].sum() > threshold)
oneyear_v_treshold_noind_go = oneyear_v_treshold.reset_index()
# oneyear_v_treshold_noind_go
data = []
for name1, group in oneyear_v_treshold_noind_go[
["CAMERA ID", "yearmonth", "VIOLATIONS"]
].groupby("CAMERA ID"):
data.append(go.Scatter(x=group.yearmonth, y=group.VIOLATIONS, name=name1))
layout = dict(
title="Top violations by month for last 12 months",
xaxis=dict(title="Date", ticklen=5, zeroline=False),
yaxis=dict(title="Violations", ticklen=5, zeroline=False),
)
# create and show our figure
fig = dict(data=data, layout=layout)
iplot(fig)
# For concerned authorities to follow-up the effectiveness of speed camera system
# address of camera will be easier, instead of camera-id
data = []
for name1, group in oneyear_v_treshold_noind_go[
["ADDRESS", "yearmonth", "VIOLATIONS"]
].groupby("ADDRESS"):
data.append(go.Scatter(x=group.yearmonth, y=group.VIOLATIONS, name=name1))
layout = dict(
title="Top violations by month for last 12 months",
xaxis=dict(title="Date", ticklen=5, zeroline=False),
yaxis=dict(title="Violations", ticklen=5, zeroline=False),
)
# create and show our figure
fig = dict(data=data, layout=layout)
iplot(fig)
| false | 0 | 1,936 | 0 | 1,936 | 1,936 |
||
69498608
|
<jupyter_start><jupyter_text>Water Quality
# Context
`Access to safe drinking-water is essential to health, a basic human right and a component of effective policy for health protection. This is important as a health and development issue at a national, regional and local level. In some regions, it has been shown that investments in water supply and sanitation can yield a net economic benefit, since the reductions in adverse health effects and health care costs outweigh the costs of undertaking the interventions.`
# Content
The water_potability.csv file contains water quality metrics for 3276 different water bodies.
### 1. pH value:
```PH is an important parameter in evaluating the acid–base balance of water. It is also the indicator of acidic or alkaline condition of water status. WHO has recommended maximum permissible limit of pH from 6.5 to 8.5. The current investigation ranges were 6.52–6.83 which are in the range of WHO standards. ```
### 2. Hardness:
```Hardness is mainly caused by calcium and magnesium salts. These salts are dissolved from geologic deposits through which water travels. The length of time water is in contact with hardness producing material helps determine how much hardness there is in raw water.
Hardness was originally defined as the capacity of water to precipitate soap caused by Calcium and Magnesium.```
### 3. Solids (Total dissolved solids - TDS):
```Water has the ability to dissolve a wide range of inorganic and some organic minerals or salts such as potassium, calcium, sodium, bicarbonates, chlorides, magnesium, sulfates etc. These minerals produced un-wanted taste and diluted color in appearance of water. This is the important parameter for the use of water. The water with high TDS value indicates that water is highly mineralized. Desirable limit for TDS is 500 mg/l and maximum limit is 1000 mg/l which prescribed for drinking purpose. ```
### 4. Chloramines:
```Chlorine and chloramine are the major disinfectants used in public water systems. Chloramines are most commonly formed when ammonia is added to chlorine to treat drinking water. Chlorine levels up to 4 milligrams per liter (mg/L or 4 parts per million (ppm)) are considered safe in drinking water.```
### 5. Sulfate:
```Sulfates are naturally occurring substances that are found in minerals, soil, and rocks. They are present in ambient air, groundwater, plants, and food. The principal commercial use of sulfate is in the chemical industry. Sulfate concentration in seawater is about 2,700 milligrams per liter (mg/L). It ranges from 3 to 30 mg/L in most freshwater supplies, although much higher concentrations (1000 mg/L) are found in some geographic locations. ```
### 6. Conductivity:
```Pure water is not a good conductor of electric current rather’s a good insulator. Increase in ions concentration enhances the electrical conductivity of water. Generally, the amount of dissolved solids in water determines the electrical conductivity. Electrical conductivity (EC) actually measures the ionic process of a solution that enables it to transmit current. According to WHO standards, EC value should not exceeded 400 μS/cm. ```
### 7. Organic_carbon:
```Total Organic Carbon (TOC) in source waters comes from decaying natural organic matter (NOM) as well as synthetic sources. TOC is a measure of the total amount of carbon in organic compounds in pure water. According to US EPA < 2 mg/L as TOC in treated / drinking water, and < 4 mg/Lit in source water which is use for treatment.```
### 8. Trihalomethanes:
```THMs are chemicals which may be found in water treated with chlorine. The concentration of THMs in drinking water varies according to the level of organic material in the water, the amount of chlorine required to treat the water, and the temperature of the water that is being treated. THM levels up to 80 ppm is considered safe in drinking water.```
### 9. Turbidity:
```The turbidity of water depends on the quantity of solid matter present in the suspended state. It is a measure of light emitting properties of water and the test is used to indicate the quality of waste discharge with respect to colloidal matter. The mean turbidity value obtained for Wondo Genet Campus (0.98 NTU) is lower than the WHO recommended value of 5.00 NTU.```
### 10. Potability:
```Indicates if water is safe for human consumption where 1 means Potable and 0 means Not potable.```
Kaggle dataset identifier: water-potability
<jupyter_code>import pandas as pd
df = pd.read_csv('water-potability/water_potability.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 3276 entries, 0 to 3275
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 ph 2785 non-null float64
1 Hardness 3276 non-null float64
2 Solids 3276 non-null float64
3 Chloramines 3276 non-null float64
4 Sulfate 2495 non-null float64
5 Conductivity 3276 non-null float64
6 Organic_carbon 3276 non-null float64
7 Trihalomethanes 3114 non-null float64
8 Turbidity 3276 non-null float64
9 Potability 3276 non-null int64
dtypes: float64(9), int64(1)
memory usage: 256.1 KB
<jupyter_text>Examples:
{
"ph": NaN,
"Hardness": 204.8904554713,
"Solids": 20791.318980747,
"Chloramines": 7.3002118732,
"Sulfate": 368.5164413498,
"Conductivity": 564.3086541722,
"Organic_carbon": 10.379783078100001,
"Trihalomethanes": 86.9909704615,
"Turbidity": 2.9631353806,
"Potability": 0.0
}
{
"ph": 3.7160800754,
"Hardness": 129.4229205149,
"Solids": 18630.0578579703,
"Chloramines": 6.6352458839,
"Sulfate": NaN,
"Conductivity": 592.8853591349,
"Organic_carbon": 15.1800131164,
"Trihalomethanes": 56.3290762845,
"Turbidity": 4.5006562749,
"Potability": 0.0
}
{
"ph": 8.0991241893,
"Hardness": 224.2362593936,
"Solids": 19909.5417322924,
"Chloramines": 9.2758836027,
"Sulfate": NaN,
"Conductivity": 418.6062130645,
"Organic_carbon": 16.8686369296,
"Trihalomethanes": 66.4200925118,
"Turbidity": 3.0559337497,
"Potability": 0.0
}
{
"ph": 8.3167658842,
"Hardness": 214.3733940856,
"Solids": 22018.4174407753,
"Chloramines": 8.0593323774,
"Sulfate": 356.8861356431,
"Conductivity": 363.2665161642,
"Organic_carbon": 18.4365244955,
"Trihalomethanes": 100.3416743651,
"Turbidity": 4.6287705368,
"Potability": 0.0
}
<jupyter_script># basic libraries
import os
import numpy as np
import pandas as pd
import random
# visualization libraries
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
import missingno as msno
## modeling
from sklearn.model_selection import (
train_test_split,
KFold,
StratifiedKFold,
GridSearchCV,
)
from sklearn.preprocessing import (
LabelEncoder,
OneHotEncoder,
StandardScaler,
MinMaxScaler,
)
from sklearn.ensemble import (
RandomForestClassifier,
AdaBoostClassifier,
ExtraTreesClassifier,
GradientBoostingClassifier,
StackingClassifier,
)
from xgboost import XGBClassifier
from lightgbm import LGBMClassifier
# warnings
import warnings
warnings.filterwarnings("ignore")
# import wand
import joypy
#
# # About Dataset:
# **Context**
# Access to safe drinking-water is essential to health, a basic human right and a component of effective policy for health protection. This is important as a health and development issue at a national, regional and local level. In some regions, it has been shown that investments in water supply and sanitation can yield a net economic benefit, since the reductions in adverse health effects and health care costs outweigh the costs of undertaking the interventions.
# **Content**
# The water_potability.csv file contains water quality metrics for 3276 different water bodies.
# **1. pH value:**
# PH is an important parameter in evaluating the acid–base balance of water. It is also the indicator of acidic or alkaline condition of water status. WHO has recommended maximum permissible limit of pH from 6.5 to 8.5. The current investigation ranges were 6.52–6.83 which are in the range of WHO standards.
# **2. Hardness:**
# Hardness is mainly caused by calcium and magnesium salts. These salts are dissolved from geologic deposits through which water travels. The length of time water is in contact with hardness producing material helps determine how much hardness there is in raw water. Hardness was originally defined as the capacity of water to precipitate soap caused by Calcium and Magnesium.
# **3. Solids (Total dissolved solids - TDS):**
# Water has the ability to dissolve a wide range of inorganic and some organic minerals or salts such as potassium, calcium, sodium, bicarbonates, chlorides, magnesium, sulfates etc. These minerals produced un-wanted taste and diluted color in appearance of water. This is the important parameter for the use of water. The water with high TDS value indicates that water is highly mineralized. Desirable limit for TDS is 500 mg/l and maximum limit is 1000 mg/l which prescribed for drinking purpose.
# **4. Chloramines:**
# Chlorine and chloramine are the major disinfectants used in public water systems. Chloramines are most commonly formed when ammonia is added to chlorine to treat drinking water. Chlorine levels up to 4 milligrams per liter (mg/L or 4 parts per million (ppm)) are considered safe in drinking water.
# **5. Sulfate:**
# Sulfates are naturally occurring substances that are found in minerals, soil, and rocks. They are present in ambient air, groundwater, plants, and food. The principal commercial use of sulfate is in the chemical industry. Sulfate concentration in seawater is about 2,700 milligrams per liter (mg/L). It ranges from 3 to 30 mg/L in most freshwater supplies, although much higher concentrations (1000 mg/L) are found in some geographic locations.
# **6. Conductivity:**
# Pure water is not a good conductor of electric current rather’s a good insulator. Increase in ions concentration enhances the electrical conductivity of water. Generally, the amount of dissolved solids in water determines the electrical conductivity. Electrical conductivity (EC) actually measures the ionic process of a solution that enables it to transmit current. According to WHO standards, EC value should not exceeded 400 μS/cm.
# **7. Organic_carbon:**
# Total Organic Carbon (TOC) in source waters comes from decaying natural organic matter (NOM) as well as synthetic sources. TOC is a measure of the total amount of carbon in organic compounds in pure water. According to US EPA < 2 mg/L as TOC in treated / drinking water, and < 4 mg/Lit in source water which is use for treatment.
# **8. Trihalomethanes:**
# THMs are chemicals which may be found in water treated with chlorine. The concentration of THMs in drinking water varies according to the level of organic material in the water, the amount of chlorine required to treat the water, and the temperature of the water that is being treated. THM levels up to 80 ppm is considered safe in drinking water.
# **9. Turbidity:**
# The turbidity of water depends on the quantity of solid matter present in the suspended state. It is a measure of light emitting properties of water and the test is used to indicate the quality of waste discharge with respect to colloidal matter. The mean turbidity value obtained for Wondo Genet Campus (0.98 NTU) is lower than the WHO recommended value of 5.00 NTU.
# **10. Potability:**
# Indicates if water is safe for human consumption where 1 means Potable and 0 means Not potable.
df = pd.read_csv(
"../input/water-potability/water_potability.csv", delimiter=",", encoding="utf-8"
)
df.head()
colors = ["#2C2E43", "#595260", "#B2B1B9", "#FFD523"]
colors = ["#212121", "#323232", "#41AEA9", "#32E0C4"]
# colors = ['#EEEEEE','#A6F6F1','#41AEA9','#213E3B']
sns.palplot(colors, size=3)
plt.text(
-0.75,
-0.75,
"Water Quality: Visualization and Classification",
{"font": "serif", "size": 24, "weight": "bold"},
)
plt.text(
-0.75,
-0.65,
"Lets try to stick to these colors throughout presentation.",
{"font": "serif", "size": 16},
alpha=0.9,
)
for idx, values in enumerate(colors):
plt.text(
idx - 0.25,
0,
colors[idx],
{"font": "serif", "size": 15, "weight": "bold", "color": "white"},
alpha=1,
)
plt.gcf().set_facecolor("#f5f6f6")
plt.gcf().set_dpi(100)
plt.box(None)
plt.axis("off")
plt.text(
2.75,
0.65,
"© Made by bhuvanchennoju/Kaggle",
{"font": "serif", "size": 10, "color": "black"},
alpha=0.7,
)
plt.show()
df.info()
fig, ax = plt.subplots(dpi=100, figsize=(12, 4))
ax.set_facecolor("#f5f6f6")
fig.patch.set_facecolor("#f5f6f6")
# column names
null_col_list = (((df.isnull().sum()).to_frame()).sort_values(by=0)).T.columns.tolist()
color = [colors[2] if (df[col].isnull().sum()) > 0 else "grey" for col in null_col_list]
## plotting
ax.axvspan(xmin=6.5, xmax=9.5, color="grey", alpha=0.3)
msno.bar(df, fontsize=16, color=color, sort="descending", ax=ax, figsize=(12, 6))
## plotsetting
ax.set_xticklabels(
null_col_list,
{"font": "serif", "color": "black", "weight": "bold", "size": 12},
alpha=1,
rotation=90,
)
ax.text(
-1.5,
1.5,
"Water Quality: Visualization of the Nullity",
{"font": "serif", "color": "black", "weight": "bold", "size": 24},
alpha=1,
)
ax.text(
-1.5,
1.35,
"Clearly seen that most of the missing values \nare in Trihalomethanes, Ph, and sulphates.",
{"font": "serif", "color": "black", "weight": "normal", "size": 13},
alpha=1,
)
ax.set_yticklabels("")
ax.spines["bottom"].set_visible(True)
plt.text(
8,
-0.75,
"© Made by bhuvanchennoju/Kaggle",
{"font": "serif", "size": 10, "color": "black"},
alpha=0.7,
)
fig.show()
# # Handling of Null Values
#
for i in (col for col in df.columns if df[col].isnull().sum()):
df[i].fillna(value=df[i].mean(), inplace=True)
print("*" * 30 + " Sanity Check " + "*" * 30 + "\n")
print(
"Is there any null values in the dataset: {}".format(df.isna().sum().any()) + "\n"
)
print(df.isnull().sum())
##### potable values
potable_values = (df["Potability"].value_counts(normalize=True)).round(2)
##### plotting
fig, ax = plt.subplots(figsize=(13, 3.5), dpi=100)
fig.set_facecolor("#f5f6f6")
ax.set_facecolor("#f5f6f6")
# left side values
ax.barh(
potable_values.index[0],
width=potable_values.values[0],
height=0.4,
color=colors[-1],
)
# right side values
ax.barh(
potable_values.index[0],
width=potable_values.values[1],
height=0.4,
left=potable_values.values[0],
color=colors[0],
)
for idx, pa in enumerate(ax.patches):
# annotations
if pa.get_width() < 0.5:
x = -pa.get_width() + 1.16
color = colors[-1]
potable = "Drinkable"
else:
x = pa.get_width() / 2
color = colors[0]
potable = "Non-Drinkable"
ax.text(
x - 0.15,
pa.get_y() + 0.225,
("{}").format(potable),
{"font": "serif", "size": 36, "weight": "normal", "color": color},
alpha=1,
)
ax.text(
x,
pa.get_y() + 0.165,
("{}%").format(str(int(pa.get_width() * 100))),
{"font": "serif", "size": 24, "weight": "normal", "color": color},
alpha=1,
)
## titles
fig.text(
0.05,
1.20,
"Water Quality: How much water is drinkable?",
{"font": "serif", "color": "black", "weight": "bold", "size": 24},
alpha=1,
)
fig.text(
0.05,
1.07,
"""This is class distribution of the potability of water.
So it is clear that, 39% water samples out of 3276 are drinkable.""",
{"font": "serif", "color": "black", "weight": "normal", "size": 13},
alpha=0.9,
)
plt.text(
0.8,
-0.3,
"© Made by bhuvanchennoju/Kaggle",
{"font": "serif", "size": 12, "color": colors[0]},
alpha=0.8,
)
fig.show()
ax.axis("off")
fig.show()
# # univariate analysis
df.head()
from scipy.stats import kurtosis, skew
for col in df.columns:
print("*" * 10 + col + "*" * 10)
print(skew(df[col]), kurtosis(df[col]))
fig, ax = plt.subplots(3, 3, figsize=(12, 10), dpi=100)
axes = ax.ravel()
fig.patch.set_facecolor("#f5f6f6")
for ax in axes:
ax.axes.get_yaxis().set_visible(False)
ax.set_facecolor("#f5f6f6")
for loc in ["left", "right", "top", "bottom"]:
ax.spines[loc].set_visible(False)
temp_nopotable = df[df["Potability"] == 0]
temp_potable = df[df["Potability"] == 1]
for col, ax in zip(df.columns, axes):
sns.kdeplot(
temp_nopotable[col], ax=ax, color=colors[-1], fill=True, alpha=0.5, zorder=3
)
sns.kdeplot(temp_potable[col], ax=ax, color=colors[0], fill=True, alpha=0.5)
temp_df = df.copy()
# df['Solids_cat'] = pd.cut(df['Solids'],bins = [0,5000,10000,100000],labels = ['LowMineralized','DesiredMineralized','HighMineralized'])
temp_df["Nature_of_water"] = pd.cut(
temp_df["ph"], bins=[0, 6.95, 7.05, 14], labels=["Acidic", "Neutral", "Basic"]
)
# https://www.water-research.net/index.php/hardness
temp_df["Hardness_cat"] = pd.cut(
temp_df["Hardness"],
bins=[0, 17.1, 60, 120, 180, 370],
labels=["Soft", "SlightlyHard", "ModeratelyHard", "Hard", "VeryHard"],
)
temp_df["Chloramines_cat"] = pd.cut(
temp_df["Chloramines"], bins=[0, 4, 1000], labels=["Desired", "NotDesired"]
)
temp_df["Sulfate_cat"] = pd.cut(
temp_df["Sulfate"],
bins=[0, 30, 1000, 5000],
labels=["CommonFreshwaters", "RareFreshwaters", "Seawaters"],
)
temp_df["Organic_carbon_cat"] = pd.cut(
temp_df["Organic_carbon"], bins=[0, 2, 100], labels=["Desired", "NotDesired"]
)
temp_df["Trihalomethanes_cat"] = pd.cut(
temp_df["Trihalomethanes"], bins=[0, 80, 1000], labels=["Desired", "NotDesired"]
)
temp_df["Turbidity_cat"] = pd.cut(
temp_df["Turbidity"], bins=[0, 5, 100], labels=["Desired", "NotDesired"]
)
temp_df.dropna(inplace=True)
def plot_dumpbell(column, ax=None, colors=[colors[-1], colors[0], colors[1]]):
value_counts_1 = (
temp_df[temp_df["Potability"] == 0][column].value_counts(normalize=True) * 100
)
value_counts_2 = (
temp_df[temp_df["Potability"] == 1][column].value_counts(normalize=True) * 100
)
if ax == None:
### plot setting
fig, ax = plt.subplots(figsize=(8, 4), dpi=90)
fig.patch.set_facecolor("#f5f6f6")
ax.set_facecolor("#f5f6f6")
else:
ax = ax
ax.set_facecolor("#f5f6f6")
### plotting
# right side plots - potable
ax.barh(
y=value_counts_2.index,
width=value_counts_2.values,
height=0.04,
color=colors[1],
alpha=1,
)
ax.scatter(
y=value_counts_2.index,
x=value_counts_2.values,
s=value_counts_1.values * 10,
c=colors[1],
)
# left side plots
ax.barh(
y=value_counts_2.index,
width=-value_counts_1.values,
height=0.04,
color=colors[0],
alpha=1,
)
ax.scatter(
y=value_counts_2.index,
x=-value_counts_1.values,
s=value_counts_2.values * 10,
c=colors[0],
alpha=1,
)
#### numeric value annotations
for pa in ax.patches:
if pa.get_width() < 0:
value = str(int(np.round(-pa.get_width()))) + "%"
align = "right"
x = pa.get_width() - 10
else:
value = str(int(np.round(pa.get_width()))) + "%"
align = "left"
x = pa.get_width() + 10
ax.text(
x,
pa.get_y(),
value,
horizontalalignment=align,
**{
"font": "serif",
"size": 12,
"weight": "bold",
"verticalalignment": "center",
},
alpha=0.8
)
ax.axvline(
x=0,
ymin=-0.5,
ymax=1.5,
**{"color": "black", "linewidth": 0.6, "linestyle": "--"},
alpha=0.9
)
### setting limits
ax.set_xlim(xmin=-110, xmax=110)
ax.set_ylim(ymin=-1.125, ymax=4.125)
ax.set_yticklabels(
labels=value_counts_1.index.tolist(),
fontdict={
"font": "Serif",
"fontsize": 12,
"fontweight": "bold",
"color": "black",
},
alpha=0.8,
)
# # Ph of the Water
##### plot layout setting #####
fig = plt.figure(figsize=(15, 15), dpi=100)
fig.patch.set_facecolor("#f6f5f5")
gs = fig.add_gridspec(10, 24)
gs.update(wspace=1, hspace=2.5)
ax1 = fig.add_subplot(gs[1:5, 1:-1]) ## distribution plot
ax2 = fig.add_subplot(gs[6:8, 0:11]) ## joyplot1
ax3 = fig.add_subplot(gs[7:9, 0:11]) ## joyplot2
ax4 = fig.add_subplot(gs[6:9, 15:]) ## dumbell plot
###### plot axes control and setting
axes = [ax1, ax2, ax3, ax4]
for ax in axes:
ax.set_facecolor("#f6f5f5")
# ax.axes.get_xaxis().set_visible(False)
ax.axes.get_yaxis().set_visible(False)
for loc in ["left", "right", "top", "bottom"]:
ax.spines[loc].set_visible(False)
## joyplot setting
ax3.patch.set_alpha(0)
ax2.axes.get_xaxis().set_visible(False)
## dumbelplot setting
ax4.axes.get_xaxis().set_visible(False)
######### plotting all the plots together
## disibution plot
sns.kdeplot(df["ph"], ax=ax1, fill=True, color=colors[3], alpha=0.5, zorder=3)
# ranges
bin_labels = ["Acidic", "Neutral", "Basic"]
size_bins = [[0, 6.95], [6.95, 7.05], [7.05, 14]]
col_here = ["black", "black", "black"]
for idx, label in enumerate(bin_labels):
ax1.annotate(
label,
xy=(sum(size_bins[idx]) / 2, 0.01),
xytext=(sum(size_bins[idx]) / 2, 0.01),
va="center",
ha="center",
rotation=0,
**{"font": "serif", "size": 12, "weight": "bold", "color": "white"},
bbox=dict(boxstyle="round4", pad=0.2, color=col_here[idx], alpha=0.6)
)
## adding span over region
ax1.axvline(
x=size_bins[idx][0], ymin=0, ymax=1, color="black", ls="--", alpha=0.4, zorder=0
)
## joyplots
sns.kdeplot(
df[df["Potability"] == 0]["ph"],
ax=ax2,
fill=True,
color=colors[0],
alpha=1,
)
sns.kdeplot(
df[df["Potability"] == 1]["ph"], ax=ax3, fill=True, color=colors[2], alpha=1
)
## dumbell plots for categorization
plot_dumpbell("Nature_of_water", ax=ax4, colors=[colors[0], colors[2]])
###### plot titles and annotations
## titles
fig.text(
0.1,
0.865,
"Water Quality: How Hardness effecting Potability?",
{"font": "serif", "color": "black", "weight": "bold", "size": 24},
alpha=1,
)
fig.text(
0.1,
0.835,
"""Hardness is a mainly caused by salts, they are broadly classified
into five categories, soft, slightly hard,moderately hard,hard, and very hard.""",
{"font": "serif", "color": "black", "weight": "normal", "size": 13},
alpha=0.9,
)
## side title and annotations
## axis1 total distribution plot
ax1.text(
8,
0.015,
"Total",
{
"font": "Serif",
"weight": "bold",
"Size": "20",
"weight": "bold",
"style": "normal",
"color": colors[3],
},
)
ax1.text(
9,
0.015,
"=",
{
"font": "Serif",
"weight": "bold",
"Size": "20",
"weight": "bold",
"style": "normal",
"color": "black",
},
)
ax1.text(
10,
0.015,
"Non-Potable",
{
"font": "Serif",
"weight": "bold",
"Size": "20",
"weight": "bold",
"style": "normal",
"color": colors[0],
},
)
ax1.text(11, 0.015, "+", {"color": "black", "size": "20", "weight": "bold"})
ax1.text(
12,
0.015,
"Potable",
{
"font": "Serif",
"weight": "bold",
"Size": "20",
"style": "normal",
"weight": "bold",
"color": colors[2],
},
)
ax1.set_xlabel(
"Hardness",
fontdict={"font": "Serif", "color": "black", "weight": "bold", "size": 14},
)
## axis2 class distribution
ax2.text(
0,
0.0265,
"Class distribution: Nature of Distribution",
{"font": "serif", "color": "black", "weight": "bold", "size": 16},
alpha=0.8,
)
ax2.text(
10,
0.018,
"Non-Potable",
{
"font": "Serif",
"weight": "bold",
"Size": 14,
"weight": "bold",
"style": "normal",
"color": colors[0],
},
)
ax2.text(11, 0.018, "|", {"color": "black", "size": 14, "weight": "bold"})
ax2.text(
12,
0.018,
"Potable",
{
"font": "Serif",
"weight": "bold",
"Size": 14,
"style": "normal",
"weight": "bold",
"color": colors[2],
},
)
ax3.set_xlabel(
"Hardness",
fontdict={"font": "Serif", "color": "black", "weight": "bold", "size": 14},
)
ax2.text(
45,
0.022,
"""Both potable and non potable samples have
the similar kind of normal distributions.""",
{"font": "serif", "color": "black", "weight": "normal", "size": 13},
alpha=0.9,
)
## axis3 Hardness classification percentages
## axis2 class distribution
ax4.text(
0,
6.45,
"Hardness Classification: Percentage Values",
{"font": "serif", "color": "black", "weight": "bold", "size": 16},
alpha=0.8,
)
ax4.text(
10,
4.75,
"Non-Potable",
{
"font": "Serif",
"weight": "bold",
"Size": 14,
"weight": "bold",
"style": "normal",
"color": colors[0],
},
)
ax4.text(11, 4.75, "|", {"color": "black", "size": 14, "weight": "bold"})
ax4.text(
12,
4.75,
"Potable",
{
"font": "Serif",
"weight": "bold",
"Size": 14,
"style": "normal",
"weight": "bold",
"color": colors[2],
},
)
ax4.text(
0,
5.55,
"""This dumbell plot gives the percentages of each category
of the hardwater in the given samples.""",
{"font": "serif", "color": "black", "weight": "normal", "size": 13},
alpha=0.9,
)
ax4.set_ylabel("")
ax4.axes.get_yaxis().set_visible(True)
fig.text(
0.75,
0.125,
"© Made by bhuvanchennoju/Kaggle",
{"font": "serif", "size": 12, "color": colors[0]},
alpha=0.8,
)
fig.show()
# # Hardness of the Water
##### plot layout setting #####
fig = plt.figure(figsize=(15, 15), dpi=100)
fig.patch.set_facecolor("#f6f5f5")
gs = fig.add_gridspec(10, 24)
gs.update(wspace=1, hspace=2.5)
ax1 = fig.add_subplot(gs[1:5, 1:-1]) ## distribution plot
ax2 = fig.add_subplot(gs[6:8, 0:11]) ## joyplot1
ax3 = fig.add_subplot(gs[7:9, 0:11]) ## joyplot2
ax4 = fig.add_subplot(gs[6:9, 15:]) ## dumbell plot
###### plot axes control and setting
axes = [ax1, ax2, ax3, ax4]
for ax in axes:
ax.set_facecolor("#f6f5f5")
# ax.axes.get_xaxis().set_visible(False)
ax.axes.get_yaxis().set_visible(False)
for loc in ["left", "right", "top", "bottom"]:
ax.spines[loc].set_visible(False)
## joyplot setting
ax3.patch.set_alpha(0)
ax2.axes.get_xaxis().set_visible(False)
## dumbelplot setting
ax4.axes.get_xaxis().set_visible(False)
######### plotting all the plots together
## disibution plot
sns.kdeplot(df["Hardness"], ax=ax1, fill=True, color=colors[3], alpha=0.5, zorder=3)
# ranges
bin_labels = ["Soft", "Slightly\nHard", "Moderately\nHard", "Hard", "Very Hard"]
size_bins = [[0, 17.1], [17.1, 60], [60, 120], [120, 180], [180, 370]]
col_here = ["black", "black", "black", "black", "black", "black"]
for idx, label in enumerate(bin_labels):
ax1.annotate(
label,
xy=(sum(size_bins[idx]) / 2, 0.01),
xytext=(sum(size_bins[idx]) / 2, 0.01),
va="center",
ha="center",
rotation=0,
**{"font": "serif", "size": 12, "weight": "bold", "color": "white"},
bbox=dict(boxstyle="round4", pad=0.2, color=col_here[idx], alpha=0.6)
)
## adding span over region
ax1.axvline(
x=size_bins[idx][0], ymin=0, ymax=1, color="black", ls="--", alpha=0.4, zorder=0
)
## joyplots
sns.kdeplot(
df[df["Potability"] == 0]["Hardness"],
ax=ax2,
fill=True,
color=colors[0],
alpha=1,
)
sns.kdeplot(
df[df["Potability"] == 1]["Hardness"], ax=ax3, fill=True, color=colors[2], alpha=1
)
## dumbell plots for categorization
plot_dumpbell("Hardness_cat", ax=ax4, colors=[colors[0], colors[2]])
###### plot titles and annotations
## titles
fig.text(
0.1,
0.865,
"Water Quality: How Hardness effecting Potability?",
{"font": "serif", "color": "black", "weight": "bold", "size": 24},
alpha=1,
)
fig.text(
0.1,
0.835,
"""Hardness is a mainly caused by salts, they are broadly classified
into five categories, soft, slightly hard,moderately hard,hard, and very hard.""",
{"font": "serif", "color": "black", "weight": "normal", "size": 13},
alpha=0.9,
)
## side title and annotations
## axis1 total distribution plot
ax1.text(
220,
0.015,
"Total",
{
"font": "Serif",
"weight": "bold",
"Size": "20",
"weight": "bold",
"style": "normal",
"color": colors[3],
},
)
ax1.text(
253,
0.015,
"=",
{
"font": "Serif",
"weight": "bold",
"Size": "20",
"weight": "bold",
"style": "normal",
"color": "black",
},
)
ax1.text(
268,
0.015,
"Non-Potable",
{
"font": "Serif",
"weight": "bold",
"Size": "20",
"weight": "bold",
"style": "normal",
"color": colors[0],
},
)
ax1.text(339, 0.015, "+", {"color": "black", "size": "20", "weight": "bold"})
ax1.text(
347,
0.015,
"Potable",
{
"font": "Serif",
"weight": "bold",
"Size": "20",
"style": "normal",
"weight": "bold",
"color": colors[2],
},
)
ax1.set_xlabel(
"Hardness",
fontdict={"font": "Serif", "color": "black", "weight": "bold", "size": 14},
)
## axis2 class distribution
ax2.text(
45,
0.0265,
"Class distribution: Nature of Distribution",
{"font": "serif", "color": "black", "weight": "bold", "size": 16},
alpha=0.8,
)
ax2.text(
205,
0.018,
"Non-Potable",
{
"font": "Serif",
"weight": "bold",
"Size": 14,
"weight": "bold",
"style": "normal",
"color": colors[0],
},
)
ax2.text(280, 0.018, "|", {"color": "black", "size": 14, "weight": "bold"})
ax2.text(
290,
0.018,
"Potable",
{
"font": "Serif",
"weight": "bold",
"Size": 14,
"style": "normal",
"weight": "bold",
"color": colors[2],
},
)
ax3.set_xlabel(
"Hardness",
fontdict={"font": "Serif", "color": "black", "weight": "bold", "size": 14},
)
ax2.text(
45,
0.022,
"""Both potable and non potable samples have
the similar kind of normal distributions.""",
{"font": "serif", "color": "black", "weight": "normal", "size": 13},
alpha=0.9,
)
## axis3 Hardness classification percentages
## axis2 class distribution
ax4.text(
-205,
6.45,
"Hardness Classification: Percentage Values",
{"font": "serif", "color": "black", "weight": "bold", "size": 16},
alpha=0.8,
)
ax4.text(
8,
4.75,
"Non-Potable",
{
"font": "Serif",
"weight": "bold",
"Size": 14,
"weight": "bold",
"style": "normal",
"color": colors[0],
},
)
ax4.text(80, 4.75, "|", {"color": "black", "size": 14, "weight": "bold"})
ax4.text(
85,
4.75,
"Potable",
{
"font": "Serif",
"weight": "bold",
"Size": 14,
"style": "normal",
"weight": "bold",
"color": colors[2],
},
)
ax4.text(
-205,
5.55,
"""This dumbell plot gives the percentages of each category
of the hardwater in the given samples.""",
{"font": "serif", "color": "black", "weight": "normal", "size": 13},
alpha=0.9,
)
ax4.set_ylabel("")
ax4.axes.get_yaxis().set_visible(True)
fig.text(
0.75,
0.125,
"© Made by bhuvanchennoju/Kaggle",
{"font": "serif", "size": 12, "color": colors[0]},
alpha=0.8,
)
fig.show()
##### plot layout setting #####
fig = plt.figure(figsize=(20, 15), dpi=70)
fig.patch.set_facecolor("#f6f5f5")
gs = fig.add_gridspec(10, 24)
gs.update(wspace=1, hspace=0.05)
ax2 = fig.add_subplot(gs[1:4, 0:10]) # distribution plot
ax3 = fig.add_subplot(gs[6:9, 0:10]) # hue distribution plot with cat or
ax1 = fig.add_subplot(gs[1:10, 13:])
# axes list
axes = [ax1, ax2, ax3]
sns.kdeplot(temp_df["Hardness"], ax=ax2)
ax3.barh(
y=temp_df[temp_df["Potability"] == 0]["Hardness_cat"]
.value_counts(normalize=True)
.index,
width=temp_df[temp_df["Potability"] == 0]["Hardness_cat"]
.value_counts(normalize=True)
.values,
height=0.5,
color=colors[0],
)
# sns.kdeplot(temp_df[temp_df['Potability']== 1]['Hardness'].value_counts(normalize = True),ax = ax3,color = colors[-1],fill = True)
sns.kdeplot(
y=temp_df["Hardness"],
ax=ax1,
)
# joypy.joyplot(df, column='Hardness',by = 'Potability',color = [colors[-1],colors[0]],ax=ax1)
fig.show()
temp_df["Hardness_cat"].value_counts()
##### plot layout setting #####
fig = plt.figure(figsize=(24, 10), dpi=100)
fig.patch.set_facecolor("#f6f5f5")
gs = fig.add_gridspec(10, 24)
gs.update(wspace=1, hspace=0.05)
ax2 = fig.add_subplot(gs[1:4, 0:8]) # distribution plot
ax3 = fig.add_subplot(gs[6:9, 0:8]) # hue distribution plot with cat or
ax1 = fig.add_subplot(gs[1:10, 13:])
# axes list
axes = [ax1, ax2, ax3]
# setting of axes; visibility of axes and spines turn off
for ax in axes:
ax.axes.get_yaxis().set_visible(False)
ax.set_facecolor("#f6f5f5")
for loc in ["left", "right", "top", "bottom"]:
ax.spines[loc].set_visible(False)
ax1.axes.get_xaxis().set_visible(False)
ax1.axes.get_yaxis().set_visible(True)
# dumbbell plot of stoke and healthy people
stroke_age = df[df["stroke"] == 1].age_cat.value_counts()
healthy_age = df[df["stroke"] == 0].age_cat.value_counts()
ax1.hlines(
y=["Children", "Teens", "Adults", "Mid Adults", "Elderly"],
xmin=[644, 270, 1691, 1129, 1127],
xmax=[1, 1, 11, 59, 177],
color="grey",
**{"linewidth": 0.5}
)
sns.scatterplot(
y=stroke_age.index,
x=stroke_age.values,
s=stroke_age.values * 2,
color="#fe346e",
ax=ax1,
alpha=1,
)
sns.scatterplot(
y=healthy_age.index,
x=healthy_age.values,
s=healthy_age.values * 2,
color="#512b58",
ax=ax1,
alpha=1,
)
ax1.axes.get_xaxis().set_visible(False)
ax1.set_xlim(xmin=-500, xmax=2250)
ax1.set_ylim(ymin=-1, ymax=5)
ax1.set_yticklabels(
labels=["Children", "Teens", "Adults", "Mid Adults", "Elderly"],
fontdict={"font": "Serif", "fontsize": 16, "fontweight": "bold", "color": "black"},
)
ax1.text(
-950,
5.8,
"How Age Impact on Having Strokes?",
{"font": "Serif", "Size": "25", "weight": "bold", "color": "black"},
alpha=0.9,
)
ax1.text(
1000,
4.8,
"Stroke ",
{
"font": "Serif",
"weight": "bold",
"Size": "16",
"weight": "bold",
"style": "normal",
"color": "#fe346e",
},
)
ax1.text(1300, 4.8, "|", {"color": "black", "size": "16", "weight": "bold"})
ax1.text(
1350,
4.8,
"Healthy",
{
"font": "Serif",
"weight": "bold",
"Size": "16",
"style": "normal",
"weight": "bold",
"color": "#512b58",
},
)
ax1.text(
-950,
5.0,
"Age have significant impact on strokes, and clearly seen that strokes are \nhighest for elderly people and mid age adults, \nwhere as negligible for younger people.",
{"font": "Serif", "size": "16", "color": "black"},
)
ax1.text(
stroke_age.values[0] + 30,
4.05,
stroke_age.values[0],
{"font": "Serif", "Size": 14, "weight": "bold", "color": "#fe346e"},
)
ax1.text(
healthy_age.values[2] - 300,
4.05,
healthy_age.values[2],
{"font": "Serif", "Size": 14, "weight": "bold", "color": "#512b58"},
)
ax1.text(
stroke_age.values[1] + 30,
3.05,
stroke_age.values[1],
{"font": "Serif", "Size": 14, "weight": "bold", "color": "#fe346e"},
)
ax1.text(
healthy_age.values[1] - 300,
3.05,
healthy_age.values[1],
{"font": "Serif", "Size": 14, "weight": "bold", "color": "#512b58"},
)
# distribution plots ---- only single variable
sns.kdeplot(
data=df,
x="age",
ax=ax2,
shade=True,
color="#2c003e",
alpha=1,
)
ax2.set_xlabel(
"Age of a person",
fontdict={"font": "Serif", "color": "black", "size": 16, "weight": "bold"},
)
ax2.text(
-17,
0.025,
"Overall Age Distribution - How skewed is it?",
{"font": "Serif", "color": "black", "weight": "bold", "size": 24},
alpha=0.9,
)
ax2.text(
-17,
0.021,
"Based on Age we have data from infants to elderly people.\nAdult population is the median group.",
{"font": "Serif", "size": "16", "color": "black"},
)
ax2.text(
80,
0.019,
"Total",
{"font": "Serif", "size": "14", "color": "#2c003e", "weight": "bold"},
)
ax2.text(
92, 0.019, "=", {"font": "Serif", "size": "14", "color": "black", "weight": "bold"}
)
ax2.text(
97,
0.019,
"Stroke",
{"font": "Serif", "size": "14", "color": "#fe346e", "weight": "bold"},
)
ax2.text(
113, 0.019, "+", {"font": "Serif", "size": "14", "color": "black", "weight": "bold"}
)
ax2.text(
117,
0.019,
"Healthy",
{"font": "Serif", "size": "14", "color": "#512b58", "weight": "bold"},
)
# distribution plots with hue of strokes
sns.kdeplot(
data=df[df["stroke"] == 0], x="age", ax=ax3, shade=True, alpha=1, color="#512b58"
)
sns.kdeplot(
data=df[df["stroke"] == 1], x="age", ax=ax3, shade=True, alpha=0.8, color="#fe346e"
)
ax3.set_xlabel(
"Age of a person",
fontdict={"font": "Serif", "color": "black", "weight": "bold", "size": 16},
)
ax3.text(
-17,
0.0525,
"Age-Stroke Distribution - How serious is it?",
{"font": "Serif", "weight": "bold", "color": "black", "size": 24},
alpha=0.9,
)
ax3.text(
-17,
0.043,
"From stoke Distribution it is clear that aged people are \nhaving significant number of strokes.",
{"font": "Serif", "color": "black", "size": 14},
)
ax3.text(
100,
0.043,
"Stroke ",
{
"font": "Serif",
"weight": "bold",
"Size": "16",
"weight": "bold",
"style": "normal",
"color": "#fe346e",
},
)
ax3.text(117, 0.043, "|", {"color": "black", "size": "16", "weight": "bold"})
ax3.text(
120,
0.043,
"Healthy",
{
"font": "Serif",
"weight": "bold",
"Size": "16",
"style": "normal",
"weight": "bold",
"color": "#512b58",
},
)
fig.text(
0.25,
1,
"Story of an Aged Heart - Heart Strokes and Age",
{"font": "Serif", "weight": "bold", "color": "black", "size": 35},
)
fig.show()
fig, ax = plt.subplots(figsize=(4, 4), dpi=100)
sns.boxplot(df.ph, ax=ax)
# # understanding data without feature engineering
temp_df = df.copy()
# df['Solids_cat'] = pd.cut(df['Solids'],bins = [0,5000,10000,100000],labels = ['LowMineralized','DesiredMineralized','HighMineralized'])
temp_df["Nature_of_water"] = pd.cut(
temp_df["ph"], bins=[0, 6.5, 7, 14], labels=["Acidic", "Neutral", "Basic"]
)
# https://www.water-research.net/index.php/hardness
temp_df["Hardness_cat"] = pd.cut(
temp_df["Hardness"],
bins=[0, 17.1, 60, 120, 180, 500],
labels=["Soft", "SlightlyHard", "ModeratelyHard", "Hard", "VeryHard"],
)
temp_df["Chloramines_cat"] = pd.cut(
temp_df["Chloramines"], bins=[0, 4, 1000], labels=["Desired", "NotDesired"]
)
temp_df["Sulfate_cat"] = pd.cut(
temp_df["Sulfate"],
bins=[0, 30, 1000, 5000],
labels=["CommonFreshwaters", "RareFreshwaters", "Seawaters"],
)
temp_df["Organic_carbon_cat"] = pd.cut(
temp_df["Organic_carbon"], bins=[0, 2, 100], labels=["Desired", "NotDesired"]
)
temp_df["Trihalomethanes_cat"] = pd.cut(
temp_df["Trihalomethanes"], bins=[0, 80, 1000], labels=["Desired", "NotDesired"]
)
temp_df["Turbidity_cat"] = pd.cut(
temp_df["Turbidity"], bins=[0, 5, 100], labels=["Desired", "NotDesired"]
)
temp_df.dropna(inplace=True)
drink_1 = temp_df[temp_df["Potability"] == 1]["Nature_of_water"].value_counts()
drink_0 = temp_df[temp_df["Potability"] == 0]["Nature_of_water"].value_counts()
fig, ax = plt.subplots(figsize=(12, 6), dpi=100)
ax.barh(y=drink_1.index, width=drink_1.values, height=0.5, color=colors[-1])
ax.barh(y=drink_1.index, width=-drink_0.values, height=0.5, color=colors[0])
fig, ax = plt.subplots(figsize=(12, 12))
sns.heatmap(df.drop(columns="Potability").corr(), annot=True, ax=ax)
plt.show()
df.head()
df.select_dtypes(exclude="int").columns()
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import OneHotEncoder, LabelEncoder, StandardScaler
# preprocessing - label enconding and numerical value scaling
ohe = OneHotEncoder()
ss = StandardScaler()
## feature and target
X = df.drop(columns=["Potability"])
y = df["Potability"]
## column selection
nominal = df.select_dtypes(exclude=["int", "float64"]).columns().tolist()
numerical = df.select_dtypes(exclude=["object"]).columns().tolist()
## normalizing with standard scaler of numerical features
X[numerical] = ss.fit_transform(X[numerical])
## norminal data one hot encoding for categorical features
temp = X.drop(columns=nominal)
dummies = pd.get_dummies(X[nominal])
X = pd.concat([temp, dummies], axis=1)
xtrain, xtest, ytrain, ytest = train_test_split(X, y, stratify=y)
df.isnull().sum()
from sklearn.preprocessing import LabelEncoder, OneHotEncoder
for col in x_train.select_dtypes(exclude=["int", "float64"]):
# le = LabelEncoder()
ohe = OneHotEncoder
x_train[col] = ohe.fit_transform(x_train[col])
x_test[col] = ohe.transform(x_test[col])
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/498/69498608.ipynb
|
water-potability
|
adityakadiwal
|
[{"Id": 69498608, "ScriptId": 18921911, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2845720, "CreationDate": "07/31/2021 18:35:18", "VersionNumber": 24.0, "Title": "notebook46a9816d24", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 742.0, "LinesInsertedFromPrevious": 133.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 609.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92743116, "KernelVersionId": 69498608, "SourceDatasetVersionId": 2157486}]
|
[{"Id": 2157486, "DatasetId": 1292407, "DatasourceVersionId": 2198621, "CreatorUserId": 5454565, "LicenseName": "CC0: Public Domain", "CreationDate": "04/25/2021 10:27:44", "VersionNumber": 3.0, "Title": "Water Quality", "Slug": "water-potability", "Subtitle": "Drinking water potability", "Description": "# Context\n\n`Access to safe drinking-water is essential to health, a basic human right and a component of effective policy for health protection. This is important as a health and development issue at a national, regional and local level. In some regions, it has been shown that investments in water supply and sanitation can yield a net economic benefit, since the reductions in adverse health effects and health care costs outweigh the costs of undertaking the interventions.`\n\n\n# Content\n\n\nThe water_potability.csv file contains water quality metrics for 3276 different water bodies. \n### 1. pH value:\n```PH is an important parameter in evaluating the acid\u2013base balance of water. It is also the indicator of acidic or alkaline condition of water status. WHO has recommended maximum permissible limit of pH from 6.5 to 8.5. The current investigation ranges were 6.52\u20136.83 which are in the range of WHO standards. ```\n\n### 2. Hardness:\n```Hardness is mainly caused by calcium and magnesium salts. These salts are dissolved from geologic deposits through which water travels. The length of time water is in contact with hardness producing material helps determine how much hardness there is in raw water.\nHardness was originally defined as the capacity of water to precipitate soap caused by Calcium and Magnesium.```\n\n### 3. Solids (Total dissolved solids - TDS): \n```Water has the ability to dissolve a wide range of inorganic and some organic minerals or salts such as potassium, calcium, sodium, bicarbonates, chlorides, magnesium, sulfates etc. These minerals produced un-wanted taste and diluted color in appearance of water. This is the important parameter for the use of water. The water with high TDS value indicates that water is highly mineralized. Desirable limit for TDS is 500 mg/l and maximum limit is 1000 mg/l which prescribed for drinking purpose. ```\n\n### 4. Chloramines: \n```Chlorine and chloramine are the major disinfectants used in public water systems. Chloramines are most commonly formed when ammonia is added to chlorine to treat drinking water. Chlorine levels up to 4 milligrams per liter (mg/L or 4 parts per million (ppm)) are considered safe in drinking water.```\n\n### 5. Sulfate: \n```Sulfates are naturally occurring substances that are found in minerals, soil, and rocks. They are present in ambient air, groundwater, plants, and food. The principal commercial use of sulfate is in the chemical industry. Sulfate concentration in seawater is about 2,700 milligrams per liter (mg/L). It ranges from 3 to 30 mg/L in most freshwater supplies, although much higher concentrations (1000 mg/L) are found in some geographic locations. ```\n\n### 6. Conductivity: \n```Pure water is not a good conductor of electric current rather\u2019s a good insulator. Increase in ions concentration enhances the electrical conductivity of water. Generally, the amount of dissolved solids in water determines the electrical conductivity. Electrical conductivity (EC) actually measures the ionic process of a solution that enables it to transmit current. According to WHO standards, EC value should not exceeded 400 \u03bcS/cm. ```\n\n### 7. Organic_carbon: \n ```Total Organic Carbon (TOC) in source waters comes from decaying natural organic matter (NOM) as well as synthetic sources. TOC is a measure of the total amount of carbon in organic compounds in pure water. According to US EPA < 2 mg/L as TOC in treated / drinking water, and < 4 mg/Lit in source water which is use for treatment.```\n\n### 8. Trihalomethanes: \n```THMs are chemicals which may be found in water treated with chlorine. The concentration of THMs in drinking water varies according to the level of organic material in the water, the amount of chlorine required to treat the water, and the temperature of the water that is being treated. THM levels up to 80 ppm is considered safe in drinking water.```\n\n### 9. Turbidity: \n```The turbidity of water depends on the quantity of solid matter present in the suspended state. It is a measure of light emitting properties of water and the test is used to indicate the quality of waste discharge with respect to colloidal matter. The mean turbidity value obtained for Wondo Genet Campus (0.98 NTU) is lower than the WHO recommended value of 5.00 NTU.```\n\n### 10. Potability: \n```Indicates if water is safe for human consumption where 1 means Potable and 0 means Not potable.```", "VersionNotes": "Removed garbage column", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1292407, "CreatorUserId": 5454565, "OwnerUserId": 5454565.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2157486.0, "CurrentDatasourceVersionId": 2198621.0, "ForumId": 1311077, "Type": 2, "CreationDate": "04/24/2021 07:18:57", "LastActivityDate": "04/24/2021", "TotalViews": 422520, "TotalDownloads": 61531, "TotalVotes": 1262, "TotalKernels": 437}]
|
[{"Id": 5454565, "UserName": "adityakadiwal", "DisplayName": "Aditya Kadiwal", "RegisterDate": "07/12/2020", "PerformanceTier": 2}]
|
# basic libraries
import os
import numpy as np
import pandas as pd
import random
# visualization libraries
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
import missingno as msno
## modeling
from sklearn.model_selection import (
train_test_split,
KFold,
StratifiedKFold,
GridSearchCV,
)
from sklearn.preprocessing import (
LabelEncoder,
OneHotEncoder,
StandardScaler,
MinMaxScaler,
)
from sklearn.ensemble import (
RandomForestClassifier,
AdaBoostClassifier,
ExtraTreesClassifier,
GradientBoostingClassifier,
StackingClassifier,
)
from xgboost import XGBClassifier
from lightgbm import LGBMClassifier
# warnings
import warnings
warnings.filterwarnings("ignore")
# import wand
import joypy
#
# # About Dataset:
# **Context**
# Access to safe drinking-water is essential to health, a basic human right and a component of effective policy for health protection. This is important as a health and development issue at a national, regional and local level. In some regions, it has been shown that investments in water supply and sanitation can yield a net economic benefit, since the reductions in adverse health effects and health care costs outweigh the costs of undertaking the interventions.
# **Content**
# The water_potability.csv file contains water quality metrics for 3276 different water bodies.
# **1. pH value:**
# PH is an important parameter in evaluating the acid–base balance of water. It is also the indicator of acidic or alkaline condition of water status. WHO has recommended maximum permissible limit of pH from 6.5 to 8.5. The current investigation ranges were 6.52–6.83 which are in the range of WHO standards.
# **2. Hardness:**
# Hardness is mainly caused by calcium and magnesium salts. These salts are dissolved from geologic deposits through which water travels. The length of time water is in contact with hardness producing material helps determine how much hardness there is in raw water. Hardness was originally defined as the capacity of water to precipitate soap caused by Calcium and Magnesium.
# **3. Solids (Total dissolved solids - TDS):**
# Water has the ability to dissolve a wide range of inorganic and some organic minerals or salts such as potassium, calcium, sodium, bicarbonates, chlorides, magnesium, sulfates etc. These minerals produced un-wanted taste and diluted color in appearance of water. This is the important parameter for the use of water. The water with high TDS value indicates that water is highly mineralized. Desirable limit for TDS is 500 mg/l and maximum limit is 1000 mg/l which prescribed for drinking purpose.
# **4. Chloramines:**
# Chlorine and chloramine are the major disinfectants used in public water systems. Chloramines are most commonly formed when ammonia is added to chlorine to treat drinking water. Chlorine levels up to 4 milligrams per liter (mg/L or 4 parts per million (ppm)) are considered safe in drinking water.
# **5. Sulfate:**
# Sulfates are naturally occurring substances that are found in minerals, soil, and rocks. They are present in ambient air, groundwater, plants, and food. The principal commercial use of sulfate is in the chemical industry. Sulfate concentration in seawater is about 2,700 milligrams per liter (mg/L). It ranges from 3 to 30 mg/L in most freshwater supplies, although much higher concentrations (1000 mg/L) are found in some geographic locations.
# **6. Conductivity:**
# Pure water is not a good conductor of electric current rather’s a good insulator. Increase in ions concentration enhances the electrical conductivity of water. Generally, the amount of dissolved solids in water determines the electrical conductivity. Electrical conductivity (EC) actually measures the ionic process of a solution that enables it to transmit current. According to WHO standards, EC value should not exceeded 400 μS/cm.
# **7. Organic_carbon:**
# Total Organic Carbon (TOC) in source waters comes from decaying natural organic matter (NOM) as well as synthetic sources. TOC is a measure of the total amount of carbon in organic compounds in pure water. According to US EPA < 2 mg/L as TOC in treated / drinking water, and < 4 mg/Lit in source water which is use for treatment.
# **8. Trihalomethanes:**
# THMs are chemicals which may be found in water treated with chlorine. The concentration of THMs in drinking water varies according to the level of organic material in the water, the amount of chlorine required to treat the water, and the temperature of the water that is being treated. THM levels up to 80 ppm is considered safe in drinking water.
# **9. Turbidity:**
# The turbidity of water depends on the quantity of solid matter present in the suspended state. It is a measure of light emitting properties of water and the test is used to indicate the quality of waste discharge with respect to colloidal matter. The mean turbidity value obtained for Wondo Genet Campus (0.98 NTU) is lower than the WHO recommended value of 5.00 NTU.
# **10. Potability:**
# Indicates if water is safe for human consumption where 1 means Potable and 0 means Not potable.
df = pd.read_csv(
"../input/water-potability/water_potability.csv", delimiter=",", encoding="utf-8"
)
df.head()
colors = ["#2C2E43", "#595260", "#B2B1B9", "#FFD523"]
colors = ["#212121", "#323232", "#41AEA9", "#32E0C4"]
# colors = ['#EEEEEE','#A6F6F1','#41AEA9','#213E3B']
sns.palplot(colors, size=3)
plt.text(
-0.75,
-0.75,
"Water Quality: Visualization and Classification",
{"font": "serif", "size": 24, "weight": "bold"},
)
plt.text(
-0.75,
-0.65,
"Lets try to stick to these colors throughout presentation.",
{"font": "serif", "size": 16},
alpha=0.9,
)
for idx, values in enumerate(colors):
plt.text(
idx - 0.25,
0,
colors[idx],
{"font": "serif", "size": 15, "weight": "bold", "color": "white"},
alpha=1,
)
plt.gcf().set_facecolor("#f5f6f6")
plt.gcf().set_dpi(100)
plt.box(None)
plt.axis("off")
plt.text(
2.75,
0.65,
"© Made by bhuvanchennoju/Kaggle",
{"font": "serif", "size": 10, "color": "black"},
alpha=0.7,
)
plt.show()
df.info()
fig, ax = plt.subplots(dpi=100, figsize=(12, 4))
ax.set_facecolor("#f5f6f6")
fig.patch.set_facecolor("#f5f6f6")
# column names
null_col_list = (((df.isnull().sum()).to_frame()).sort_values(by=0)).T.columns.tolist()
color = [colors[2] if (df[col].isnull().sum()) > 0 else "grey" for col in null_col_list]
## plotting
ax.axvspan(xmin=6.5, xmax=9.5, color="grey", alpha=0.3)
msno.bar(df, fontsize=16, color=color, sort="descending", ax=ax, figsize=(12, 6))
## plotsetting
ax.set_xticklabels(
null_col_list,
{"font": "serif", "color": "black", "weight": "bold", "size": 12},
alpha=1,
rotation=90,
)
ax.text(
-1.5,
1.5,
"Water Quality: Visualization of the Nullity",
{"font": "serif", "color": "black", "weight": "bold", "size": 24},
alpha=1,
)
ax.text(
-1.5,
1.35,
"Clearly seen that most of the missing values \nare in Trihalomethanes, Ph, and sulphates.",
{"font": "serif", "color": "black", "weight": "normal", "size": 13},
alpha=1,
)
ax.set_yticklabels("")
ax.spines["bottom"].set_visible(True)
plt.text(
8,
-0.75,
"© Made by bhuvanchennoju/Kaggle",
{"font": "serif", "size": 10, "color": "black"},
alpha=0.7,
)
fig.show()
# # Handling of Null Values
#
for i in (col for col in df.columns if df[col].isnull().sum()):
df[i].fillna(value=df[i].mean(), inplace=True)
print("*" * 30 + " Sanity Check " + "*" * 30 + "\n")
print(
"Is there any null values in the dataset: {}".format(df.isna().sum().any()) + "\n"
)
print(df.isnull().sum())
##### potable values
potable_values = (df["Potability"].value_counts(normalize=True)).round(2)
##### plotting
fig, ax = plt.subplots(figsize=(13, 3.5), dpi=100)
fig.set_facecolor("#f5f6f6")
ax.set_facecolor("#f5f6f6")
# left side values
ax.barh(
potable_values.index[0],
width=potable_values.values[0],
height=0.4,
color=colors[-1],
)
# right side values
ax.barh(
potable_values.index[0],
width=potable_values.values[1],
height=0.4,
left=potable_values.values[0],
color=colors[0],
)
for idx, pa in enumerate(ax.patches):
# annotations
if pa.get_width() < 0.5:
x = -pa.get_width() + 1.16
color = colors[-1]
potable = "Drinkable"
else:
x = pa.get_width() / 2
color = colors[0]
potable = "Non-Drinkable"
ax.text(
x - 0.15,
pa.get_y() + 0.225,
("{}").format(potable),
{"font": "serif", "size": 36, "weight": "normal", "color": color},
alpha=1,
)
ax.text(
x,
pa.get_y() + 0.165,
("{}%").format(str(int(pa.get_width() * 100))),
{"font": "serif", "size": 24, "weight": "normal", "color": color},
alpha=1,
)
## titles
fig.text(
0.05,
1.20,
"Water Quality: How much water is drinkable?",
{"font": "serif", "color": "black", "weight": "bold", "size": 24},
alpha=1,
)
fig.text(
0.05,
1.07,
"""This is class distribution of the potability of water.
So it is clear that, 39% water samples out of 3276 are drinkable.""",
{"font": "serif", "color": "black", "weight": "normal", "size": 13},
alpha=0.9,
)
plt.text(
0.8,
-0.3,
"© Made by bhuvanchennoju/Kaggle",
{"font": "serif", "size": 12, "color": colors[0]},
alpha=0.8,
)
fig.show()
ax.axis("off")
fig.show()
# # univariate analysis
df.head()
from scipy.stats import kurtosis, skew
for col in df.columns:
print("*" * 10 + col + "*" * 10)
print(skew(df[col]), kurtosis(df[col]))
fig, ax = plt.subplots(3, 3, figsize=(12, 10), dpi=100)
axes = ax.ravel()
fig.patch.set_facecolor("#f5f6f6")
for ax in axes:
ax.axes.get_yaxis().set_visible(False)
ax.set_facecolor("#f5f6f6")
for loc in ["left", "right", "top", "bottom"]:
ax.spines[loc].set_visible(False)
temp_nopotable = df[df["Potability"] == 0]
temp_potable = df[df["Potability"] == 1]
for col, ax in zip(df.columns, axes):
sns.kdeplot(
temp_nopotable[col], ax=ax, color=colors[-1], fill=True, alpha=0.5, zorder=3
)
sns.kdeplot(temp_potable[col], ax=ax, color=colors[0], fill=True, alpha=0.5)
temp_df = df.copy()
# df['Solids_cat'] = pd.cut(df['Solids'],bins = [0,5000,10000,100000],labels = ['LowMineralized','DesiredMineralized','HighMineralized'])
temp_df["Nature_of_water"] = pd.cut(
temp_df["ph"], bins=[0, 6.95, 7.05, 14], labels=["Acidic", "Neutral", "Basic"]
)
# https://www.water-research.net/index.php/hardness
temp_df["Hardness_cat"] = pd.cut(
temp_df["Hardness"],
bins=[0, 17.1, 60, 120, 180, 370],
labels=["Soft", "SlightlyHard", "ModeratelyHard", "Hard", "VeryHard"],
)
temp_df["Chloramines_cat"] = pd.cut(
temp_df["Chloramines"], bins=[0, 4, 1000], labels=["Desired", "NotDesired"]
)
temp_df["Sulfate_cat"] = pd.cut(
temp_df["Sulfate"],
bins=[0, 30, 1000, 5000],
labels=["CommonFreshwaters", "RareFreshwaters", "Seawaters"],
)
temp_df["Organic_carbon_cat"] = pd.cut(
temp_df["Organic_carbon"], bins=[0, 2, 100], labels=["Desired", "NotDesired"]
)
temp_df["Trihalomethanes_cat"] = pd.cut(
temp_df["Trihalomethanes"], bins=[0, 80, 1000], labels=["Desired", "NotDesired"]
)
temp_df["Turbidity_cat"] = pd.cut(
temp_df["Turbidity"], bins=[0, 5, 100], labels=["Desired", "NotDesired"]
)
temp_df.dropna(inplace=True)
def plot_dumpbell(column, ax=None, colors=[colors[-1], colors[0], colors[1]]):
value_counts_1 = (
temp_df[temp_df["Potability"] == 0][column].value_counts(normalize=True) * 100
)
value_counts_2 = (
temp_df[temp_df["Potability"] == 1][column].value_counts(normalize=True) * 100
)
if ax == None:
### plot setting
fig, ax = plt.subplots(figsize=(8, 4), dpi=90)
fig.patch.set_facecolor("#f5f6f6")
ax.set_facecolor("#f5f6f6")
else:
ax = ax
ax.set_facecolor("#f5f6f6")
### plotting
# right side plots - potable
ax.barh(
y=value_counts_2.index,
width=value_counts_2.values,
height=0.04,
color=colors[1],
alpha=1,
)
ax.scatter(
y=value_counts_2.index,
x=value_counts_2.values,
s=value_counts_1.values * 10,
c=colors[1],
)
# left side plots
ax.barh(
y=value_counts_2.index,
width=-value_counts_1.values,
height=0.04,
color=colors[0],
alpha=1,
)
ax.scatter(
y=value_counts_2.index,
x=-value_counts_1.values,
s=value_counts_2.values * 10,
c=colors[0],
alpha=1,
)
#### numeric value annotations
for pa in ax.patches:
if pa.get_width() < 0:
value = str(int(np.round(-pa.get_width()))) + "%"
align = "right"
x = pa.get_width() - 10
else:
value = str(int(np.round(pa.get_width()))) + "%"
align = "left"
x = pa.get_width() + 10
ax.text(
x,
pa.get_y(),
value,
horizontalalignment=align,
**{
"font": "serif",
"size": 12,
"weight": "bold",
"verticalalignment": "center",
},
alpha=0.8
)
ax.axvline(
x=0,
ymin=-0.5,
ymax=1.5,
**{"color": "black", "linewidth": 0.6, "linestyle": "--"},
alpha=0.9
)
### setting limits
ax.set_xlim(xmin=-110, xmax=110)
ax.set_ylim(ymin=-1.125, ymax=4.125)
ax.set_yticklabels(
labels=value_counts_1.index.tolist(),
fontdict={
"font": "Serif",
"fontsize": 12,
"fontweight": "bold",
"color": "black",
},
alpha=0.8,
)
# # Ph of the Water
##### plot layout setting #####
fig = plt.figure(figsize=(15, 15), dpi=100)
fig.patch.set_facecolor("#f6f5f5")
gs = fig.add_gridspec(10, 24)
gs.update(wspace=1, hspace=2.5)
ax1 = fig.add_subplot(gs[1:5, 1:-1]) ## distribution plot
ax2 = fig.add_subplot(gs[6:8, 0:11]) ## joyplot1
ax3 = fig.add_subplot(gs[7:9, 0:11]) ## joyplot2
ax4 = fig.add_subplot(gs[6:9, 15:]) ## dumbell plot
###### plot axes control and setting
axes = [ax1, ax2, ax3, ax4]
for ax in axes:
ax.set_facecolor("#f6f5f5")
# ax.axes.get_xaxis().set_visible(False)
ax.axes.get_yaxis().set_visible(False)
for loc in ["left", "right", "top", "bottom"]:
ax.spines[loc].set_visible(False)
## joyplot setting
ax3.patch.set_alpha(0)
ax2.axes.get_xaxis().set_visible(False)
## dumbelplot setting
ax4.axes.get_xaxis().set_visible(False)
######### plotting all the plots together
## disibution plot
sns.kdeplot(df["ph"], ax=ax1, fill=True, color=colors[3], alpha=0.5, zorder=3)
# ranges
bin_labels = ["Acidic", "Neutral", "Basic"]
size_bins = [[0, 6.95], [6.95, 7.05], [7.05, 14]]
col_here = ["black", "black", "black"]
for idx, label in enumerate(bin_labels):
ax1.annotate(
label,
xy=(sum(size_bins[idx]) / 2, 0.01),
xytext=(sum(size_bins[idx]) / 2, 0.01),
va="center",
ha="center",
rotation=0,
**{"font": "serif", "size": 12, "weight": "bold", "color": "white"},
bbox=dict(boxstyle="round4", pad=0.2, color=col_here[idx], alpha=0.6)
)
## adding span over region
ax1.axvline(
x=size_bins[idx][0], ymin=0, ymax=1, color="black", ls="--", alpha=0.4, zorder=0
)
## joyplots
sns.kdeplot(
df[df["Potability"] == 0]["ph"],
ax=ax2,
fill=True,
color=colors[0],
alpha=1,
)
sns.kdeplot(
df[df["Potability"] == 1]["ph"], ax=ax3, fill=True, color=colors[2], alpha=1
)
## dumbell plots for categorization
plot_dumpbell("Nature_of_water", ax=ax4, colors=[colors[0], colors[2]])
###### plot titles and annotations
## titles
fig.text(
0.1,
0.865,
"Water Quality: How Hardness effecting Potability?",
{"font": "serif", "color": "black", "weight": "bold", "size": 24},
alpha=1,
)
fig.text(
0.1,
0.835,
"""Hardness is a mainly caused by salts, they are broadly classified
into five categories, soft, slightly hard,moderately hard,hard, and very hard.""",
{"font": "serif", "color": "black", "weight": "normal", "size": 13},
alpha=0.9,
)
## side title and annotations
## axis1 total distribution plot
ax1.text(
8,
0.015,
"Total",
{
"font": "Serif",
"weight": "bold",
"Size": "20",
"weight": "bold",
"style": "normal",
"color": colors[3],
},
)
ax1.text(
9,
0.015,
"=",
{
"font": "Serif",
"weight": "bold",
"Size": "20",
"weight": "bold",
"style": "normal",
"color": "black",
},
)
ax1.text(
10,
0.015,
"Non-Potable",
{
"font": "Serif",
"weight": "bold",
"Size": "20",
"weight": "bold",
"style": "normal",
"color": colors[0],
},
)
ax1.text(11, 0.015, "+", {"color": "black", "size": "20", "weight": "bold"})
ax1.text(
12,
0.015,
"Potable",
{
"font": "Serif",
"weight": "bold",
"Size": "20",
"style": "normal",
"weight": "bold",
"color": colors[2],
},
)
ax1.set_xlabel(
"Hardness",
fontdict={"font": "Serif", "color": "black", "weight": "bold", "size": 14},
)
## axis2 class distribution
ax2.text(
0,
0.0265,
"Class distribution: Nature of Distribution",
{"font": "serif", "color": "black", "weight": "bold", "size": 16},
alpha=0.8,
)
ax2.text(
10,
0.018,
"Non-Potable",
{
"font": "Serif",
"weight": "bold",
"Size": 14,
"weight": "bold",
"style": "normal",
"color": colors[0],
},
)
ax2.text(11, 0.018, "|", {"color": "black", "size": 14, "weight": "bold"})
ax2.text(
12,
0.018,
"Potable",
{
"font": "Serif",
"weight": "bold",
"Size": 14,
"style": "normal",
"weight": "bold",
"color": colors[2],
},
)
ax3.set_xlabel(
"Hardness",
fontdict={"font": "Serif", "color": "black", "weight": "bold", "size": 14},
)
ax2.text(
45,
0.022,
"""Both potable and non potable samples have
the similar kind of normal distributions.""",
{"font": "serif", "color": "black", "weight": "normal", "size": 13},
alpha=0.9,
)
## axis3 Hardness classification percentages
## axis2 class distribution
ax4.text(
0,
6.45,
"Hardness Classification: Percentage Values",
{"font": "serif", "color": "black", "weight": "bold", "size": 16},
alpha=0.8,
)
ax4.text(
10,
4.75,
"Non-Potable",
{
"font": "Serif",
"weight": "bold",
"Size": 14,
"weight": "bold",
"style": "normal",
"color": colors[0],
},
)
ax4.text(11, 4.75, "|", {"color": "black", "size": 14, "weight": "bold"})
ax4.text(
12,
4.75,
"Potable",
{
"font": "Serif",
"weight": "bold",
"Size": 14,
"style": "normal",
"weight": "bold",
"color": colors[2],
},
)
ax4.text(
0,
5.55,
"""This dumbell plot gives the percentages of each category
of the hardwater in the given samples.""",
{"font": "serif", "color": "black", "weight": "normal", "size": 13},
alpha=0.9,
)
ax4.set_ylabel("")
ax4.axes.get_yaxis().set_visible(True)
fig.text(
0.75,
0.125,
"© Made by bhuvanchennoju/Kaggle",
{"font": "serif", "size": 12, "color": colors[0]},
alpha=0.8,
)
fig.show()
# # Hardness of the Water
##### plot layout setting #####
fig = plt.figure(figsize=(15, 15), dpi=100)
fig.patch.set_facecolor("#f6f5f5")
gs = fig.add_gridspec(10, 24)
gs.update(wspace=1, hspace=2.5)
ax1 = fig.add_subplot(gs[1:5, 1:-1]) ## distribution plot
ax2 = fig.add_subplot(gs[6:8, 0:11]) ## joyplot1
ax3 = fig.add_subplot(gs[7:9, 0:11]) ## joyplot2
ax4 = fig.add_subplot(gs[6:9, 15:]) ## dumbell plot
###### plot axes control and setting
axes = [ax1, ax2, ax3, ax4]
for ax in axes:
ax.set_facecolor("#f6f5f5")
# ax.axes.get_xaxis().set_visible(False)
ax.axes.get_yaxis().set_visible(False)
for loc in ["left", "right", "top", "bottom"]:
ax.spines[loc].set_visible(False)
## joyplot setting
ax3.patch.set_alpha(0)
ax2.axes.get_xaxis().set_visible(False)
## dumbelplot setting
ax4.axes.get_xaxis().set_visible(False)
######### plotting all the plots together
## disibution plot
sns.kdeplot(df["Hardness"], ax=ax1, fill=True, color=colors[3], alpha=0.5, zorder=3)
# ranges
bin_labels = ["Soft", "Slightly\nHard", "Moderately\nHard", "Hard", "Very Hard"]
size_bins = [[0, 17.1], [17.1, 60], [60, 120], [120, 180], [180, 370]]
col_here = ["black", "black", "black", "black", "black", "black"]
for idx, label in enumerate(bin_labels):
ax1.annotate(
label,
xy=(sum(size_bins[idx]) / 2, 0.01),
xytext=(sum(size_bins[idx]) / 2, 0.01),
va="center",
ha="center",
rotation=0,
**{"font": "serif", "size": 12, "weight": "bold", "color": "white"},
bbox=dict(boxstyle="round4", pad=0.2, color=col_here[idx], alpha=0.6)
)
## adding span over region
ax1.axvline(
x=size_bins[idx][0], ymin=0, ymax=1, color="black", ls="--", alpha=0.4, zorder=0
)
## joyplots
sns.kdeplot(
df[df["Potability"] == 0]["Hardness"],
ax=ax2,
fill=True,
color=colors[0],
alpha=1,
)
sns.kdeplot(
df[df["Potability"] == 1]["Hardness"], ax=ax3, fill=True, color=colors[2], alpha=1
)
## dumbell plots for categorization
plot_dumpbell("Hardness_cat", ax=ax4, colors=[colors[0], colors[2]])
###### plot titles and annotations
## titles
fig.text(
0.1,
0.865,
"Water Quality: How Hardness effecting Potability?",
{"font": "serif", "color": "black", "weight": "bold", "size": 24},
alpha=1,
)
fig.text(
0.1,
0.835,
"""Hardness is a mainly caused by salts, they are broadly classified
into five categories, soft, slightly hard,moderately hard,hard, and very hard.""",
{"font": "serif", "color": "black", "weight": "normal", "size": 13},
alpha=0.9,
)
## side title and annotations
## axis1 total distribution plot
ax1.text(
220,
0.015,
"Total",
{
"font": "Serif",
"weight": "bold",
"Size": "20",
"weight": "bold",
"style": "normal",
"color": colors[3],
},
)
ax1.text(
253,
0.015,
"=",
{
"font": "Serif",
"weight": "bold",
"Size": "20",
"weight": "bold",
"style": "normal",
"color": "black",
},
)
ax1.text(
268,
0.015,
"Non-Potable",
{
"font": "Serif",
"weight": "bold",
"Size": "20",
"weight": "bold",
"style": "normal",
"color": colors[0],
},
)
ax1.text(339, 0.015, "+", {"color": "black", "size": "20", "weight": "bold"})
ax1.text(
347,
0.015,
"Potable",
{
"font": "Serif",
"weight": "bold",
"Size": "20",
"style": "normal",
"weight": "bold",
"color": colors[2],
},
)
ax1.set_xlabel(
"Hardness",
fontdict={"font": "Serif", "color": "black", "weight": "bold", "size": 14},
)
## axis2 class distribution
ax2.text(
45,
0.0265,
"Class distribution: Nature of Distribution",
{"font": "serif", "color": "black", "weight": "bold", "size": 16},
alpha=0.8,
)
ax2.text(
205,
0.018,
"Non-Potable",
{
"font": "Serif",
"weight": "bold",
"Size": 14,
"weight": "bold",
"style": "normal",
"color": colors[0],
},
)
ax2.text(280, 0.018, "|", {"color": "black", "size": 14, "weight": "bold"})
ax2.text(
290,
0.018,
"Potable",
{
"font": "Serif",
"weight": "bold",
"Size": 14,
"style": "normal",
"weight": "bold",
"color": colors[2],
},
)
ax3.set_xlabel(
"Hardness",
fontdict={"font": "Serif", "color": "black", "weight": "bold", "size": 14},
)
ax2.text(
45,
0.022,
"""Both potable and non potable samples have
the similar kind of normal distributions.""",
{"font": "serif", "color": "black", "weight": "normal", "size": 13},
alpha=0.9,
)
## axis3 Hardness classification percentages
## axis2 class distribution
ax4.text(
-205,
6.45,
"Hardness Classification: Percentage Values",
{"font": "serif", "color": "black", "weight": "bold", "size": 16},
alpha=0.8,
)
ax4.text(
8,
4.75,
"Non-Potable",
{
"font": "Serif",
"weight": "bold",
"Size": 14,
"weight": "bold",
"style": "normal",
"color": colors[0],
},
)
ax4.text(80, 4.75, "|", {"color": "black", "size": 14, "weight": "bold"})
ax4.text(
85,
4.75,
"Potable",
{
"font": "Serif",
"weight": "bold",
"Size": 14,
"style": "normal",
"weight": "bold",
"color": colors[2],
},
)
ax4.text(
-205,
5.55,
"""This dumbell plot gives the percentages of each category
of the hardwater in the given samples.""",
{"font": "serif", "color": "black", "weight": "normal", "size": 13},
alpha=0.9,
)
ax4.set_ylabel("")
ax4.axes.get_yaxis().set_visible(True)
fig.text(
0.75,
0.125,
"© Made by bhuvanchennoju/Kaggle",
{"font": "serif", "size": 12, "color": colors[0]},
alpha=0.8,
)
fig.show()
##### plot layout setting #####
fig = plt.figure(figsize=(20, 15), dpi=70)
fig.patch.set_facecolor("#f6f5f5")
gs = fig.add_gridspec(10, 24)
gs.update(wspace=1, hspace=0.05)
ax2 = fig.add_subplot(gs[1:4, 0:10]) # distribution plot
ax3 = fig.add_subplot(gs[6:9, 0:10]) # hue distribution plot with cat or
ax1 = fig.add_subplot(gs[1:10, 13:])
# axes list
axes = [ax1, ax2, ax3]
sns.kdeplot(temp_df["Hardness"], ax=ax2)
ax3.barh(
y=temp_df[temp_df["Potability"] == 0]["Hardness_cat"]
.value_counts(normalize=True)
.index,
width=temp_df[temp_df["Potability"] == 0]["Hardness_cat"]
.value_counts(normalize=True)
.values,
height=0.5,
color=colors[0],
)
# sns.kdeplot(temp_df[temp_df['Potability']== 1]['Hardness'].value_counts(normalize = True),ax = ax3,color = colors[-1],fill = True)
sns.kdeplot(
y=temp_df["Hardness"],
ax=ax1,
)
# joypy.joyplot(df, column='Hardness',by = 'Potability',color = [colors[-1],colors[0]],ax=ax1)
fig.show()
temp_df["Hardness_cat"].value_counts()
##### plot layout setting #####
fig = plt.figure(figsize=(24, 10), dpi=100)
fig.patch.set_facecolor("#f6f5f5")
gs = fig.add_gridspec(10, 24)
gs.update(wspace=1, hspace=0.05)
ax2 = fig.add_subplot(gs[1:4, 0:8]) # distribution plot
ax3 = fig.add_subplot(gs[6:9, 0:8]) # hue distribution plot with cat or
ax1 = fig.add_subplot(gs[1:10, 13:])
# axes list
axes = [ax1, ax2, ax3]
# setting of axes; visibility of axes and spines turn off
for ax in axes:
ax.axes.get_yaxis().set_visible(False)
ax.set_facecolor("#f6f5f5")
for loc in ["left", "right", "top", "bottom"]:
ax.spines[loc].set_visible(False)
ax1.axes.get_xaxis().set_visible(False)
ax1.axes.get_yaxis().set_visible(True)
# dumbbell plot of stoke and healthy people
stroke_age = df[df["stroke"] == 1].age_cat.value_counts()
healthy_age = df[df["stroke"] == 0].age_cat.value_counts()
ax1.hlines(
y=["Children", "Teens", "Adults", "Mid Adults", "Elderly"],
xmin=[644, 270, 1691, 1129, 1127],
xmax=[1, 1, 11, 59, 177],
color="grey",
**{"linewidth": 0.5}
)
sns.scatterplot(
y=stroke_age.index,
x=stroke_age.values,
s=stroke_age.values * 2,
color="#fe346e",
ax=ax1,
alpha=1,
)
sns.scatterplot(
y=healthy_age.index,
x=healthy_age.values,
s=healthy_age.values * 2,
color="#512b58",
ax=ax1,
alpha=1,
)
ax1.axes.get_xaxis().set_visible(False)
ax1.set_xlim(xmin=-500, xmax=2250)
ax1.set_ylim(ymin=-1, ymax=5)
ax1.set_yticklabels(
labels=["Children", "Teens", "Adults", "Mid Adults", "Elderly"],
fontdict={"font": "Serif", "fontsize": 16, "fontweight": "bold", "color": "black"},
)
ax1.text(
-950,
5.8,
"How Age Impact on Having Strokes?",
{"font": "Serif", "Size": "25", "weight": "bold", "color": "black"},
alpha=0.9,
)
ax1.text(
1000,
4.8,
"Stroke ",
{
"font": "Serif",
"weight": "bold",
"Size": "16",
"weight": "bold",
"style": "normal",
"color": "#fe346e",
},
)
ax1.text(1300, 4.8, "|", {"color": "black", "size": "16", "weight": "bold"})
ax1.text(
1350,
4.8,
"Healthy",
{
"font": "Serif",
"weight": "bold",
"Size": "16",
"style": "normal",
"weight": "bold",
"color": "#512b58",
},
)
ax1.text(
-950,
5.0,
"Age have significant impact on strokes, and clearly seen that strokes are \nhighest for elderly people and mid age adults, \nwhere as negligible for younger people.",
{"font": "Serif", "size": "16", "color": "black"},
)
ax1.text(
stroke_age.values[0] + 30,
4.05,
stroke_age.values[0],
{"font": "Serif", "Size": 14, "weight": "bold", "color": "#fe346e"},
)
ax1.text(
healthy_age.values[2] - 300,
4.05,
healthy_age.values[2],
{"font": "Serif", "Size": 14, "weight": "bold", "color": "#512b58"},
)
ax1.text(
stroke_age.values[1] + 30,
3.05,
stroke_age.values[1],
{"font": "Serif", "Size": 14, "weight": "bold", "color": "#fe346e"},
)
ax1.text(
healthy_age.values[1] - 300,
3.05,
healthy_age.values[1],
{"font": "Serif", "Size": 14, "weight": "bold", "color": "#512b58"},
)
# distribution plots ---- only single variable
sns.kdeplot(
data=df,
x="age",
ax=ax2,
shade=True,
color="#2c003e",
alpha=1,
)
ax2.set_xlabel(
"Age of a person",
fontdict={"font": "Serif", "color": "black", "size": 16, "weight": "bold"},
)
ax2.text(
-17,
0.025,
"Overall Age Distribution - How skewed is it?",
{"font": "Serif", "color": "black", "weight": "bold", "size": 24},
alpha=0.9,
)
ax2.text(
-17,
0.021,
"Based on Age we have data from infants to elderly people.\nAdult population is the median group.",
{"font": "Serif", "size": "16", "color": "black"},
)
ax2.text(
80,
0.019,
"Total",
{"font": "Serif", "size": "14", "color": "#2c003e", "weight": "bold"},
)
ax2.text(
92, 0.019, "=", {"font": "Serif", "size": "14", "color": "black", "weight": "bold"}
)
ax2.text(
97,
0.019,
"Stroke",
{"font": "Serif", "size": "14", "color": "#fe346e", "weight": "bold"},
)
ax2.text(
113, 0.019, "+", {"font": "Serif", "size": "14", "color": "black", "weight": "bold"}
)
ax2.text(
117,
0.019,
"Healthy",
{"font": "Serif", "size": "14", "color": "#512b58", "weight": "bold"},
)
# distribution plots with hue of strokes
sns.kdeplot(
data=df[df["stroke"] == 0], x="age", ax=ax3, shade=True, alpha=1, color="#512b58"
)
sns.kdeplot(
data=df[df["stroke"] == 1], x="age", ax=ax3, shade=True, alpha=0.8, color="#fe346e"
)
ax3.set_xlabel(
"Age of a person",
fontdict={"font": "Serif", "color": "black", "weight": "bold", "size": 16},
)
ax3.text(
-17,
0.0525,
"Age-Stroke Distribution - How serious is it?",
{"font": "Serif", "weight": "bold", "color": "black", "size": 24},
alpha=0.9,
)
ax3.text(
-17,
0.043,
"From stoke Distribution it is clear that aged people are \nhaving significant number of strokes.",
{"font": "Serif", "color": "black", "size": 14},
)
ax3.text(
100,
0.043,
"Stroke ",
{
"font": "Serif",
"weight": "bold",
"Size": "16",
"weight": "bold",
"style": "normal",
"color": "#fe346e",
},
)
ax3.text(117, 0.043, "|", {"color": "black", "size": "16", "weight": "bold"})
ax3.text(
120,
0.043,
"Healthy",
{
"font": "Serif",
"weight": "bold",
"Size": "16",
"style": "normal",
"weight": "bold",
"color": "#512b58",
},
)
fig.text(
0.25,
1,
"Story of an Aged Heart - Heart Strokes and Age",
{"font": "Serif", "weight": "bold", "color": "black", "size": 35},
)
fig.show()
fig, ax = plt.subplots(figsize=(4, 4), dpi=100)
sns.boxplot(df.ph, ax=ax)
# # understanding data without feature engineering
temp_df = df.copy()
# df['Solids_cat'] = pd.cut(df['Solids'],bins = [0,5000,10000,100000],labels = ['LowMineralized','DesiredMineralized','HighMineralized'])
temp_df["Nature_of_water"] = pd.cut(
temp_df["ph"], bins=[0, 6.5, 7, 14], labels=["Acidic", "Neutral", "Basic"]
)
# https://www.water-research.net/index.php/hardness
temp_df["Hardness_cat"] = pd.cut(
temp_df["Hardness"],
bins=[0, 17.1, 60, 120, 180, 500],
labels=["Soft", "SlightlyHard", "ModeratelyHard", "Hard", "VeryHard"],
)
temp_df["Chloramines_cat"] = pd.cut(
temp_df["Chloramines"], bins=[0, 4, 1000], labels=["Desired", "NotDesired"]
)
temp_df["Sulfate_cat"] = pd.cut(
temp_df["Sulfate"],
bins=[0, 30, 1000, 5000],
labels=["CommonFreshwaters", "RareFreshwaters", "Seawaters"],
)
temp_df["Organic_carbon_cat"] = pd.cut(
temp_df["Organic_carbon"], bins=[0, 2, 100], labels=["Desired", "NotDesired"]
)
temp_df["Trihalomethanes_cat"] = pd.cut(
temp_df["Trihalomethanes"], bins=[0, 80, 1000], labels=["Desired", "NotDesired"]
)
temp_df["Turbidity_cat"] = pd.cut(
temp_df["Turbidity"], bins=[0, 5, 100], labels=["Desired", "NotDesired"]
)
temp_df.dropna(inplace=True)
drink_1 = temp_df[temp_df["Potability"] == 1]["Nature_of_water"].value_counts()
drink_0 = temp_df[temp_df["Potability"] == 0]["Nature_of_water"].value_counts()
fig, ax = plt.subplots(figsize=(12, 6), dpi=100)
ax.barh(y=drink_1.index, width=drink_1.values, height=0.5, color=colors[-1])
ax.barh(y=drink_1.index, width=-drink_0.values, height=0.5, color=colors[0])
fig, ax = plt.subplots(figsize=(12, 12))
sns.heatmap(df.drop(columns="Potability").corr(), annot=True, ax=ax)
plt.show()
df.head()
df.select_dtypes(exclude="int").columns()
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import OneHotEncoder, LabelEncoder, StandardScaler
# preprocessing - label enconding and numerical value scaling
ohe = OneHotEncoder()
ss = StandardScaler()
## feature and target
X = df.drop(columns=["Potability"])
y = df["Potability"]
## column selection
nominal = df.select_dtypes(exclude=["int", "float64"]).columns().tolist()
numerical = df.select_dtypes(exclude=["object"]).columns().tolist()
## normalizing with standard scaler of numerical features
X[numerical] = ss.fit_transform(X[numerical])
## norminal data one hot encoding for categorical features
temp = X.drop(columns=nominal)
dummies = pd.get_dummies(X[nominal])
X = pd.concat([temp, dummies], axis=1)
xtrain, xtest, ytrain, ytest = train_test_split(X, y, stratify=y)
df.isnull().sum()
from sklearn.preprocessing import LabelEncoder, OneHotEncoder
for col in x_train.select_dtypes(exclude=["int", "float64"]):
# le = LabelEncoder()
ohe = OneHotEncoder
x_train[col] = ohe.fit_transform(x_train[col])
x_test[col] = ohe.transform(x_test[col])
|
[{"water-potability/water_potability.csv": {"column_names": "[\"ph\", \"Hardness\", \"Solids\", \"Chloramines\", \"Sulfate\", \"Conductivity\", \"Organic_carbon\", \"Trihalomethanes\", \"Turbidity\", \"Potability\"]", "column_data_types": "{\"ph\": \"float64\", \"Hardness\": \"float64\", \"Solids\": \"float64\", \"Chloramines\": \"float64\", \"Sulfate\": \"float64\", \"Conductivity\": \"float64\", \"Organic_carbon\": \"float64\", \"Trihalomethanes\": \"float64\", \"Turbidity\": \"float64\", \"Potability\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 3276 entries, 0 to 3275\nData columns (total 10 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 ph 2785 non-null float64\n 1 Hardness 3276 non-null float64\n 2 Solids 3276 non-null float64\n 3 Chloramines 3276 non-null float64\n 4 Sulfate 2495 non-null float64\n 5 Conductivity 3276 non-null float64\n 6 Organic_carbon 3276 non-null float64\n 7 Trihalomethanes 3114 non-null float64\n 8 Turbidity 3276 non-null float64\n 9 Potability 3276 non-null int64 \ndtypes: float64(9), int64(1)\nmemory usage: 256.1 KB\n", "summary": "{\"ph\": {\"count\": 2785.0, \"mean\": 7.080794504276835, \"std\": 1.5943195187088104, \"min\": 0.0, \"25%\": 6.09309191422186, \"50%\": 7.036752103833548, \"75%\": 8.06206612314847, \"max\": 13.999999999999998}, \"Hardness\": {\"count\": 3276.0, \"mean\": 196.36949601730151, \"std\": 32.879761476294156, \"min\": 47.432, \"25%\": 176.85053787752437, \"50%\": 196.96762686363076, \"75%\": 216.66745621487073, \"max\": 323.124}, \"Solids\": {\"count\": 3276.0, \"mean\": 22014.092526077104, \"std\": 8768.570827785927, \"min\": 320.942611274359, \"25%\": 15666.69029696465, \"50%\": 20927.833606520187, \"75%\": 27332.762127438615, \"max\": 61227.19600771213}, \"Chloramines\": {\"count\": 3276.0, \"mean\": 7.122276793425786, \"std\": 1.5830848890397096, \"min\": 0.3520000000000003, \"25%\": 6.1274207554913, \"50%\": 7.130298973883081, \"75%\": 8.114887032109028, \"max\": 13.127000000000002}, \"Sulfate\": {\"count\": 2495.0, \"mean\": 333.7757766108135, \"std\": 41.416840461672706, \"min\": 129.00000000000003, \"25%\": 307.69949783471964, \"50%\": 333.073545745888, \"75%\": 359.9501703847443, \"max\": 481.0306423059972}, \"Conductivity\": {\"count\": 3276.0, \"mean\": 426.20511068255325, \"std\": 80.8240640511118, \"min\": 181.483753985146, \"25%\": 365.7344141184627, \"50%\": 421.8849682800544, \"75%\": 481.7923044877282, \"max\": 753.3426195583046}, \"Organic_carbon\": {\"count\": 3276.0, \"mean\": 14.284970247677318, \"std\": 3.308161999126874, \"min\": 2.1999999999999886, \"25%\": 12.065801333613067, \"50%\": 14.218337937208588, \"75%\": 16.557651543843434, \"max\": 28.30000000000001}, \"Trihalomethanes\": {\"count\": 3114.0, \"mean\": 66.39629294676803, \"std\": 16.175008422218657, \"min\": 0.7379999999999995, \"25%\": 55.844535620979954, \"50%\": 66.62248509808484, \"75%\": 77.33747290873062, \"max\": 124.0}, \"Turbidity\": {\"count\": 3276.0, \"mean\": 3.966786169791058, \"std\": 0.7803824084854124, \"min\": 1.45, \"25%\": 3.439710869612912, \"50%\": 3.955027562993039, \"75%\": 4.50031978728511, \"max\": 6.739}, \"Potability\": {\"count\": 3276.0, \"mean\": 0.3901098901098901, \"std\": 0.48784916967025516, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}}", "examples": "{\"ph\":{\"0\":null,\"1\":3.7160800754,\"2\":8.0991241893,\"3\":8.3167658842},\"Hardness\":{\"0\":204.8904554713,\"1\":129.4229205149,\"2\":224.2362593936,\"3\":214.3733940856},\"Solids\":{\"0\":20791.318980747,\"1\":18630.0578579703,\"2\":19909.5417322924,\"3\":22018.4174407753},\"Chloramines\":{\"0\":7.3002118732,\"1\":6.6352458839,\"2\":9.2758836027,\"3\":8.0593323774},\"Sulfate\":{\"0\":368.5164413498,\"1\":null,\"2\":null,\"3\":356.8861356431},\"Conductivity\":{\"0\":564.3086541722,\"1\":592.8853591349,\"2\":418.6062130645,\"3\":363.2665161642},\"Organic_carbon\":{\"0\":10.3797830781,\"1\":15.1800131164,\"2\":16.8686369296,\"3\":18.4365244955},\"Trihalomethanes\":{\"0\":86.9909704615,\"1\":56.3290762845,\"2\":66.4200925118,\"3\":100.3416743651},\"Turbidity\":{\"0\":2.9631353806,\"1\":4.5006562749,\"2\":3.0559337497,\"3\":4.6287705368},\"Potability\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0}}"}}]
| true | 1 |
<start_data_description><data_path>water-potability/water_potability.csv:
<column_names>
['ph', 'Hardness', 'Solids', 'Chloramines', 'Sulfate', 'Conductivity', 'Organic_carbon', 'Trihalomethanes', 'Turbidity', 'Potability']
<column_types>
{'ph': 'float64', 'Hardness': 'float64', 'Solids': 'float64', 'Chloramines': 'float64', 'Sulfate': 'float64', 'Conductivity': 'float64', 'Organic_carbon': 'float64', 'Trihalomethanes': 'float64', 'Turbidity': 'float64', 'Potability': 'int64'}
<dataframe_Summary>
{'ph': {'count': 2785.0, 'mean': 7.080794504276835, 'std': 1.5943195187088104, 'min': 0.0, '25%': 6.09309191422186, '50%': 7.036752103833548, '75%': 8.06206612314847, 'max': 13.999999999999998}, 'Hardness': {'count': 3276.0, 'mean': 196.36949601730151, 'std': 32.879761476294156, 'min': 47.432, '25%': 176.85053787752437, '50%': 196.96762686363076, '75%': 216.66745621487073, 'max': 323.124}, 'Solids': {'count': 3276.0, 'mean': 22014.092526077104, 'std': 8768.570827785927, 'min': 320.942611274359, '25%': 15666.69029696465, '50%': 20927.833606520187, '75%': 27332.762127438615, 'max': 61227.19600771213}, 'Chloramines': {'count': 3276.0, 'mean': 7.122276793425786, 'std': 1.5830848890397096, 'min': 0.3520000000000003, '25%': 6.1274207554913, '50%': 7.130298973883081, '75%': 8.114887032109028, 'max': 13.127000000000002}, 'Sulfate': {'count': 2495.0, 'mean': 333.7757766108135, 'std': 41.416840461672706, 'min': 129.00000000000003, '25%': 307.69949783471964, '50%': 333.073545745888, '75%': 359.9501703847443, 'max': 481.0306423059972}, 'Conductivity': {'count': 3276.0, 'mean': 426.20511068255325, 'std': 80.8240640511118, 'min': 181.483753985146, '25%': 365.7344141184627, '50%': 421.8849682800544, '75%': 481.7923044877282, 'max': 753.3426195583046}, 'Organic_carbon': {'count': 3276.0, 'mean': 14.284970247677318, 'std': 3.308161999126874, 'min': 2.1999999999999886, '25%': 12.065801333613067, '50%': 14.218337937208588, '75%': 16.557651543843434, 'max': 28.30000000000001}, 'Trihalomethanes': {'count': 3114.0, 'mean': 66.39629294676803, 'std': 16.175008422218657, 'min': 0.7379999999999995, '25%': 55.844535620979954, '50%': 66.62248509808484, '75%': 77.33747290873062, 'max': 124.0}, 'Turbidity': {'count': 3276.0, 'mean': 3.966786169791058, 'std': 0.7803824084854124, 'min': 1.45, '25%': 3.439710869612912, '50%': 3.955027562993039, '75%': 4.50031978728511, 'max': 6.739}, 'Potability': {'count': 3276.0, 'mean': 0.3901098901098901, 'std': 0.48784916967025516, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}}
<dataframe_info>
RangeIndex: 3276 entries, 0 to 3275
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 ph 2785 non-null float64
1 Hardness 3276 non-null float64
2 Solids 3276 non-null float64
3 Chloramines 3276 non-null float64
4 Sulfate 2495 non-null float64
5 Conductivity 3276 non-null float64
6 Organic_carbon 3276 non-null float64
7 Trihalomethanes 3114 non-null float64
8 Turbidity 3276 non-null float64
9 Potability 3276 non-null int64
dtypes: float64(9), int64(1)
memory usage: 256.1 KB
<some_examples>
{'ph': {'0': None, '1': 3.7160800754, '2': 8.0991241893, '3': 8.3167658842}, 'Hardness': {'0': 204.8904554713, '1': 129.4229205149, '2': 224.2362593936, '3': 214.3733940856}, 'Solids': {'0': 20791.318980747, '1': 18630.0578579703, '2': 19909.5417322924, '3': 22018.4174407753}, 'Chloramines': {'0': 7.3002118732, '1': 6.6352458839, '2': 9.2758836027, '3': 8.0593323774}, 'Sulfate': {'0': 368.5164413498, '1': None, '2': None, '3': 356.8861356431}, 'Conductivity': {'0': 564.3086541722, '1': 592.8853591349, '2': 418.6062130645, '3': 363.2665161642}, 'Organic_carbon': {'0': 10.3797830781, '1': 15.1800131164, '2': 16.8686369296, '3': 18.4365244955}, 'Trihalomethanes': {'0': 86.9909704615, '1': 56.3290762845, '2': 66.4200925118, '3': 100.3416743651}, 'Turbidity': {'0': 2.9631353806, '1': 4.5006562749, '2': 3.0559337497, '3': 4.6287705368}, 'Potability': {'0': 0, '1': 0, '2': 0, '3': 0}}
<end_description>
| 12,802 | 0 | 15,078 | 12,802 |
69498927
|
# # Test B
# **Test B**
# description
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from tensorflow import keras
from tensorflow.keras import regularizers
from tensorflow.keras import layers
from tensorflow.python.keras.callbacks import EarlyStopping
LABEL = "SalePrice"
train = pd.read_csv("../input/house-prices-advanced-regression-techniques/train.csv")
train_y = train[LABEL]
train_y = train_y.astype("float32")
train.drop(LABEL, axis=1, inplace=True)
train.drop("Id", axis=1, inplace=True)
test = pd.read_csv("../input/house-prices-advanced-regression-techniques/test.csv")
test_id = test.Id
test.drop("Id", axis=1, inplace=True)
ntrain = train.shape[0]
ntest = test.shape[0]
all_data = pd.concat((train, test)).reset_index(drop=True)
for column in all_data:
if all_data[column].dtype != np.object:
all_data[column] = (all_data[column] - all_data[column].mean()) / all_data[
column
].std()
all_data["PoolQC"] = all_data["PoolQC"].fillna("None")
all_data["MiscFeature"] = all_data["MiscFeature"].fillna("None")
all_data["Alley"] = all_data["Alley"].fillna("None")
all_data["Fence"] = all_data["Fence"].fillna("None")
all_data["FireplaceQu"] = all_data["FireplaceQu"].fillna("None")
all_data["LotFrontage"] = all_data.groupby("Neighborhood")["LotFrontage"].transform(
lambda x: x.fillna(x.median())
)
for col in ("GarageType", "GarageFinish", "GarageQual", "GarageCond"):
all_data[col] = all_data[col].fillna("None")
for col in ("GarageYrBlt", "GarageArea", "GarageCars"):
all_data[col] = all_data[col].fillna(0)
for col in (
"BsmtFinSF1",
"BsmtFinSF2",
"BsmtUnfSF",
"TotalBsmtSF",
"BsmtFullBath",
"BsmtHalfBath",
):
all_data[col] = all_data[col].fillna(0)
for col in ("BsmtQual", "BsmtCond", "BsmtExposure", "BsmtFinType1", "BsmtFinType2"):
all_data[col] = all_data[col].fillna("None")
all_data["MasVnrType"] = all_data["MasVnrType"].fillna("None")
all_data["MasVnrArea"] = all_data["MasVnrArea"].fillna(0)
all_data["MSZoning"] = all_data["MSZoning"].fillna(all_data["MSZoning"].mode()[0])
all_data = all_data.drop(["Utilities"], axis=1)
all_data["Functional"] = all_data["Functional"].fillna("Typ")
all_data["Electrical"] = all_data["Electrical"].fillna(all_data["Electrical"].mode()[0])
all_data["KitchenQual"] = all_data["KitchenQual"].fillna(
all_data["KitchenQual"].mode()[0]
)
all_data["Exterior1st"] = all_data["Exterior1st"].fillna(
all_data["Exterior1st"].mode()[0]
)
all_data["Exterior2nd"] = all_data["Exterior2nd"].fillna(
all_data["Exterior2nd"].mode()[0]
)
all_data["SaleType"] = all_data["SaleType"].fillna(all_data["SaleType"].mode()[0])
all_data["MSSubClass"] = all_data["MSSubClass"].fillna("None")
all_data.fillna(0, inplace=True)
all_data = pd.get_dummies(all_data)
features = list(all_data.columns)
print(features)
train = all_data[:ntrain]
test = all_data[ntrain:]
def root_mean_squared_log_error(y_true, y_pred):
return keras.backend.sqrt(
keras.backend.mean(
keras.backend.square(
keras.backend.log(1 + y_pred) - keras.backend.log(1 + y_true)
)
)
)
def build_model(param1):
print("----------New iteration----------")
my_initializer = keras.initializers.RandomNormal(mean=0.0, stddev=0.05, seed=1)
l2 = 1e-7
model = keras.models.Sequential(
[
layers.Dense(
200,
input_shape=[len(features)],
kernel_initializer=my_initializer,
bias_initializer=my_initializer,
activation="relu",
kernel_regularizer=regularizers.l2(l2),
bias_regularizer=regularizers.l2(l2),
),
layers.Dense(
100,
kernel_initializer=my_initializer,
bias_initializer=my_initializer,
activation="relu",
kernel_regularizer=regularizers.l2(l2),
bias_regularizer=regularizers.l2(l2),
),
layers.Dense(
50,
kernel_initializer=my_initializer,
bias_initializer=my_initializer,
activation="relu",
kernel_regularizer=regularizers.l2(l2),
bias_regularizer=regularizers.l2(l2),
),
layers.Dense(
25,
kernel_initializer=my_initializer,
bias_initializer=my_initializer,
activation="relu",
kernel_regularizer=regularizers.l2(l2),
bias_regularizer=regularizers.l2(l2),
),
layers.Dense(
1,
kernel_initializer=my_initializer,
bias_initializer=my_initializer,
kernel_regularizer=regularizers.l2(l2),
bias_regularizer=regularizers.l2(l2),
),
]
)
# for layer in my_model.layers: print(layer.get_config(), layer.get_weights())
model.compile(loss=root_mean_squared_log_error, optimizer="adam")
# validation_split=0.2,
history = model.fit(train, train_y, epochs=124, batch_size=32)
pd.DataFrame(history.history).plot()
plt.grid(True)
plt.gca().set_ylim([0, 0.2])
plt.show()
return history.history, model
param_values = [0]
for val in param_values:
train_history, trained_model = build_model(val)
pd.DataFrame(train_history).plot()
plt.grid(True)
plt.gca().set_ylim([0, 0.3])
plt.show()
# Use the model to make predictions
predicted_prices = trained_model.predict(test)
# We will look at the predicted prices to ensure we have something sensible.
print(predicted_prices)
my_submission = pd.DataFrame({"Id": test_id, LABEL: predicted_prices.ravel()})
# you could use any filename. We choose submission here
my_submission.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/498/69498927.ipynb
| null | null |
[{"Id": 69498927, "ScriptId": 18780948, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7949355, "CreationDate": "07/31/2021 18:42:00", "VersionNumber": 37.0, "Title": "Test B", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 134.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 133.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Test B
# **Test B**
# description
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from tensorflow import keras
from tensorflow.keras import regularizers
from tensorflow.keras import layers
from tensorflow.python.keras.callbacks import EarlyStopping
LABEL = "SalePrice"
train = pd.read_csv("../input/house-prices-advanced-regression-techniques/train.csv")
train_y = train[LABEL]
train_y = train_y.astype("float32")
train.drop(LABEL, axis=1, inplace=True)
train.drop("Id", axis=1, inplace=True)
test = pd.read_csv("../input/house-prices-advanced-regression-techniques/test.csv")
test_id = test.Id
test.drop("Id", axis=1, inplace=True)
ntrain = train.shape[0]
ntest = test.shape[0]
all_data = pd.concat((train, test)).reset_index(drop=True)
for column in all_data:
if all_data[column].dtype != np.object:
all_data[column] = (all_data[column] - all_data[column].mean()) / all_data[
column
].std()
all_data["PoolQC"] = all_data["PoolQC"].fillna("None")
all_data["MiscFeature"] = all_data["MiscFeature"].fillna("None")
all_data["Alley"] = all_data["Alley"].fillna("None")
all_data["Fence"] = all_data["Fence"].fillna("None")
all_data["FireplaceQu"] = all_data["FireplaceQu"].fillna("None")
all_data["LotFrontage"] = all_data.groupby("Neighborhood")["LotFrontage"].transform(
lambda x: x.fillna(x.median())
)
for col in ("GarageType", "GarageFinish", "GarageQual", "GarageCond"):
all_data[col] = all_data[col].fillna("None")
for col in ("GarageYrBlt", "GarageArea", "GarageCars"):
all_data[col] = all_data[col].fillna(0)
for col in (
"BsmtFinSF1",
"BsmtFinSF2",
"BsmtUnfSF",
"TotalBsmtSF",
"BsmtFullBath",
"BsmtHalfBath",
):
all_data[col] = all_data[col].fillna(0)
for col in ("BsmtQual", "BsmtCond", "BsmtExposure", "BsmtFinType1", "BsmtFinType2"):
all_data[col] = all_data[col].fillna("None")
all_data["MasVnrType"] = all_data["MasVnrType"].fillna("None")
all_data["MasVnrArea"] = all_data["MasVnrArea"].fillna(0)
all_data["MSZoning"] = all_data["MSZoning"].fillna(all_data["MSZoning"].mode()[0])
all_data = all_data.drop(["Utilities"], axis=1)
all_data["Functional"] = all_data["Functional"].fillna("Typ")
all_data["Electrical"] = all_data["Electrical"].fillna(all_data["Electrical"].mode()[0])
all_data["KitchenQual"] = all_data["KitchenQual"].fillna(
all_data["KitchenQual"].mode()[0]
)
all_data["Exterior1st"] = all_data["Exterior1st"].fillna(
all_data["Exterior1st"].mode()[0]
)
all_data["Exterior2nd"] = all_data["Exterior2nd"].fillna(
all_data["Exterior2nd"].mode()[0]
)
all_data["SaleType"] = all_data["SaleType"].fillna(all_data["SaleType"].mode()[0])
all_data["MSSubClass"] = all_data["MSSubClass"].fillna("None")
all_data.fillna(0, inplace=True)
all_data = pd.get_dummies(all_data)
features = list(all_data.columns)
print(features)
train = all_data[:ntrain]
test = all_data[ntrain:]
def root_mean_squared_log_error(y_true, y_pred):
return keras.backend.sqrt(
keras.backend.mean(
keras.backend.square(
keras.backend.log(1 + y_pred) - keras.backend.log(1 + y_true)
)
)
)
def build_model(param1):
print("----------New iteration----------")
my_initializer = keras.initializers.RandomNormal(mean=0.0, stddev=0.05, seed=1)
l2 = 1e-7
model = keras.models.Sequential(
[
layers.Dense(
200,
input_shape=[len(features)],
kernel_initializer=my_initializer,
bias_initializer=my_initializer,
activation="relu",
kernel_regularizer=regularizers.l2(l2),
bias_regularizer=regularizers.l2(l2),
),
layers.Dense(
100,
kernel_initializer=my_initializer,
bias_initializer=my_initializer,
activation="relu",
kernel_regularizer=regularizers.l2(l2),
bias_regularizer=regularizers.l2(l2),
),
layers.Dense(
50,
kernel_initializer=my_initializer,
bias_initializer=my_initializer,
activation="relu",
kernel_regularizer=regularizers.l2(l2),
bias_regularizer=regularizers.l2(l2),
),
layers.Dense(
25,
kernel_initializer=my_initializer,
bias_initializer=my_initializer,
activation="relu",
kernel_regularizer=regularizers.l2(l2),
bias_regularizer=regularizers.l2(l2),
),
layers.Dense(
1,
kernel_initializer=my_initializer,
bias_initializer=my_initializer,
kernel_regularizer=regularizers.l2(l2),
bias_regularizer=regularizers.l2(l2),
),
]
)
# for layer in my_model.layers: print(layer.get_config(), layer.get_weights())
model.compile(loss=root_mean_squared_log_error, optimizer="adam")
# validation_split=0.2,
history = model.fit(train, train_y, epochs=124, batch_size=32)
pd.DataFrame(history.history).plot()
plt.grid(True)
plt.gca().set_ylim([0, 0.2])
plt.show()
return history.history, model
param_values = [0]
for val in param_values:
train_history, trained_model = build_model(val)
pd.DataFrame(train_history).plot()
plt.grid(True)
plt.gca().set_ylim([0, 0.3])
plt.show()
# Use the model to make predictions
predicted_prices = trained_model.predict(test)
# We will look at the predicted prices to ensure we have something sensible.
print(predicted_prices)
my_submission = pd.DataFrame({"Id": test_id, LABEL: predicted_prices.ravel()})
# you could use any filename. We choose submission here
my_submission.to_csv("submission.csv", index=False)
| false | 0 | 1,783 | 0 | 1,783 | 1,783 |
||
69498580
|
<jupyter_start><jupyter_text>Source based Fake News Classification
### Context
Social media is a vast pool of content, and among all the content available for users to access, news is an element that is accessed most frequently. These news can be posted by politicians, news channels, newspaper websites, or even common civilians. These posts have to be checked for their authenticity, since spreading misinformation has been a real concern in today’s times, and many firms are taking steps to make the common people aware of the consequences of spread misinformation. The measure of authenticity of the news posted online cannot be definitively measured, since the manual classification of news is tedious and time-consuming, and is also subject to bias.
Published paper: http://www.ijirset.com/upload/2020/june/115_4_Source.PDF
### Content
Data preprocessing has been done on the dataset [Getting Real about Fake News](https://www.kaggle.com/mrisdal/fake-news) and skew has been eliminated.
### Inspiration
In an era where fake WhatsApp forwards and Tweets are capable of influencing naive minds, tools and knowledge have to be put to practical use in not only mitigating the spread of misinformation but also to inform people about the type of news they consume.
Development of practical applications for users to gain insight from the articles they consume, fact-checking websites, built-in plugins and article parsers can
further be refined, made easier to access, and more importantly, should create more awareness.
Kaggle dataset identifier: source-based-news-classification
<jupyter_code>import pandas as pd
df = pd.read_csv('source-based-news-classification/news_articles.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 2096 entries, 0 to 2095
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 author 2096 non-null object
1 published 2096 non-null object
2 title 2096 non-null object
3 text 2050 non-null object
4 language 2095 non-null object
5 site_url 2095 non-null object
6 main_img_url 2095 non-null object
7 type 2095 non-null object
8 label 2095 non-null object
9 title_without_stopwords 2094 non-null object
10 text_without_stopwords 2046 non-null object
11 hasImage 2095 non-null float64
dtypes: float64(1), object(11)
memory usage: 196.6+ KB
<jupyter_text>Examples:
{
"author": "Barracuda Brigade",
"published": "2016-10-26T21:41:00.000+03:00",
"title": "muslims busted they stole millions in govt benefits",
"text": "print they should pay all the back all the money plus interest the entire family and everyone who came in with them need to be deported asap why did it take two years to bust them \nhere we go again another group stealing from the government and taxpayers a group of somalis stole over f...(truncated)",
"language": "english",
"site_url": "100percentfedup.com",
"main_img_url": "http://bb4sp.com/wp-content/uploads/2016/10/Fullscreen-capture-10262016-83501-AM.bmp.jpg",
"type": "bias",
"label": "Real",
"title_without_stopwords": "muslims busted stole millions govt benefits",
"text_without_stopwords": "print pay back money plus interest entire family everyone came need deported asap take two years bust go another group stealing government taxpayers group somalis stole four million government benefits months weve reported numerous cases like one muslim refugeesimmigra...(truncated)",
"hasImage": 1
}
{
"author": "reasoning with facts",
"published": "2016-10-29T08:47:11.259+03:00",
"title": "re why did attorney general loretta lynch plead the fifth",
"text": "why did attorney general loretta lynch plead the fifth barracuda brigade print the administration is blocking congressional probe into cash payments to iran of course she needs to plead the th she either cant recall refuses to answer or just plain deflects the question straight up corr...(truncated)",
"language": "english",
"site_url": "100percentfedup.com",
"main_img_url": "http://bb4sp.com/wp-content/uploads/2016/10/Fullscreen-capture-10282016-102616-PM.bmp.jpg",
"type": "bias",
"label": "Real",
"title_without_stopwords": "attorney general loretta lynch plead fifth",
"text_without_stopwords": "attorney general loretta lynch plead fifth barracuda brigade print administration blocking congressional probe cash payments iran course needs plead th either cant recall refuses answer plain deflects question straight corruption finest percentfedupcom talk covering as...(truncated)",
"hasImage": 1
}
{
"author": "Barracuda Brigade",
"published": "2016-10-31T01:41:49.479+02:00",
"title": "breaking weiner cooperating with fbi on hillary email investigation",
"text": "red state \nfox news sunday reported this morning that anthony weiner is cooperating with the fbi which has reopened yes lefties reopened the investigation into hillary clintons classified emails watch as chris wallace reports the breaking news during the panel segment near the end of ...(truncated)",
"language": "english",
"site_url": "100percentfedup.com",
"main_img_url": "http://bb4sp.com/wp-content/uploads/2016/10/Fullscreen-capture-10302016-60437-PM.bmp.jpg",
"type": "bias",
"label": "Real",
"title_without_stopwords": "breaking weiner cooperating fbi hillary email investigation",
"text_without_stopwords": "red state fox news sunday reported morning anthony weiner cooperating fbi reopened yes lefties reopened investigation hillary clintons classified emails watch chris wallace reports breaking news panel segment near end show news breaking air colleague bret baier sent us...(truncated)",
"hasImage": 1
}
{
"author": "Fed Up",
"published": "2016-11-01T05:22:00.000+02:00",
"title": "pin drop speech by father of daughter kidnapped and killed by isis i have voted for donald j trump percentfedupcom",
"text": "email kayla mueller was a prisoner and tortured by isis while no chance of releasea horrific story her father gave a pin drop speech that was so heartfelt you want to give him a hug carl mueller believes donald trump will be a great presidentepic speech k shares",
"language": "english",
"site_url": "100percentfedup.com",
"main_img_url": "http://100percentfedup.com/wp-content/uploads/2016/10/kayla.jpg",
"type": "bias",
"label": "Real",
"title_without_stopwords": "pin drop speech father daughter kidnapped killed isis voted donald j trump percentfedupcom",
"text_without_stopwords": "email kayla mueller prisoner tortured isis chance releasea horrific story father gave pin drop speech heartfelt want give hug carl mueller believes donald trump great presidentepic speech k shares",
"hasImage": 1
}
<jupyter_script># Exploring the news dataset
# 1. INTRODUCTION
# Social media is a vast pool of content, and among all the content available for users to access, news is an element that is accessed most frequently. These news can be posted by politicians, news channels, newspaper websites, or even common civilians. These posts have to be checked for their authenticity, since spreading misinformation has been a real concern in today’s times, and many firms are taking steps to make the common people aware of the consequences of spread misinformation. The measure of authenticity of the news posted online cannot be definitively measured, since the manual classification of news is tedious and time-consuming, and is also subject to bias.
# 
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import nltk
nltk.download("punkt")
import matplotlib.pyplot as plt
import seaborn as sns
from wordcloud import WordCloud, STOPWORDS
import nltk
import re
from nltk.corpus import stopwords
import seaborn as sns
import gensim
from gensim.utils import simple_preprocess
from gensim.parsing.preprocessing import STOPWORDS
import warnings
warnings.filterwarnings("ignore")
import plotly.express as px
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import roc_auc_score
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier
from sklearn.tree import DecisionTreeClassifier
# Reading the dataset
df = pd.read_csv("../input/source-based-news-classification/news_articles.csv")
print(df.shape)
df.head()
# Checking for null values
df.isnull().sum()
# dropping null values
df.dropna(inplace=True)
# ---
# Exploratory data analysis
# ### Let's visualize the proportion of real and fake news!
# real vs fake
fig = px.pie(df, names="label", title="Proportion of Real vs. Fake News")
fig.show()
# ### Languages of News Articles
sub_tf_df = (
df.groupby("language")
.apply(lambda x: x["language"].count())
.reset_index(name="Counts")
)
fig = px.bar(
sub_tf_df, x="language", y="Counts", color="Counts", barmode="group", height=400
)
fig.show()
# As we can see, English is the most common language in which most news articles are written
# ### Visualizing count of news articles by type
sub_check = (
df.groupby("type").apply(lambda x: x["type"].count()).reset_index(name="Counts")
)
fig = px.bar(
sub_check,
x="type",
y="Counts",
color="Counts",
title="Count of News Articles by type",
)
fig.show()
# here we can se that most news are of type bs (i.e. bullshit)
from wordcloud import WordCloud
wc = WordCloud(
background_color="black",
max_words=100,
max_font_size=256,
random_state=42,
width=1000,
height=1000,
)
wc.generate(" ".join(df["text_without_stopwords"]))
plt.imshow(wc)
plt.axis("off")
plt.show()
# ### Visualizing top 10 unigrams and bigrams
def get_top_n_words(corpus, n=None):
vec = CountVectorizer().fit(corpus)
bag_of_words = vec.transform(corpus)
sum_words = bag_of_words.sum(axis=0)
words_freq = [(word, sum_words[0, idx]) for word, idx in vec.vocabulary_.items()]
words_freq = sorted(words_freq, key=lambda x: x[1], reverse=True)
return words_freq[:n]
def get_top_n_bigram(corpus, n=None):
vec = CountVectorizer(ngram_range=(2, 2)).fit(corpus)
bag_of_words = vec.transform(corpus)
sum_words = bag_of_words.sum(axis=0)
words_freq = [(word, sum_words[0, idx]) for word, idx in vec.vocabulary_.items()]
words_freq = sorted(words_freq, key=lambda x: x[1], reverse=True)
return words_freq[:n]
common_words = get_top_n_words(df["text_without_stopwords"], 10)
df2 = pd.DataFrame(common_words, columns=["word", "count"])
df2.groupby("word").sum()["count"].sort_values(ascending=False)
fig = px.bar(df2, x="word", y="count", color="count", title="Top 10 unigrams")
fig.show()
common_words = get_top_n_bigram(df["text_without_stopwords"], 10)
df2 = pd.DataFrame(common_words, columns=["word", "count"])
df2.groupby("word").sum()["count"].sort_values(ascending=False)
fig = px.bar(df2, x="word", y="count", color="count", title="Top 10 bigrams")
fig.show()
# ### Visualizing top 5 authors
d = df["author"].value_counts().sort_values(ascending=False).head(5)
d = pd.DataFrame(d)
d = d.reset_index() # dataframe with top 5 authors
# Plotting
sns.set()
plt.figure(figsize=(15, 4))
sns.barplot(x="index", y="author", data=d)
plt.xlabel("\n Authors")
plt.ylabel("Number of Articles written")
plt.title("Top 5 authors\n")
plt.show()
# ### Visualizing top 10 fake news site
d = (
df[df["label"] == "Fake"]["site_url"]
.value_counts()
.sort_values(ascending=False)
.head(10)
)
d = pd.DataFrame(d)
d = d.reset_index() # dataframe with top 10 fake news site
# Plotting
sns.set()
plt.figure(figsize=(25, 7))
sns.barplot(x="index", y="site_url", data=d)
plt.xlabel("\n site_url")
plt.ylabel("Number of Articles written")
plt.title("Top 10 Fake news sites\n")
plt.show()
# ### Let's have a look at the top 10 trustworthy news sites
d = (
df[df["label"] == "Real"]["site_url"]
.value_counts()
.sort_values(ascending=False)
.head(10)
)
d = pd.DataFrame(d)
d = d.reset_index() # dataframe with top 10 Trustworthy news site
# Plotting
sns.set()
plt.figure(figsize=(25, 7))
sns.barplot(x="index", y="site_url", data=d)
plt.xlabel("\n site_url")
plt.ylabel("Number of Articles written")
plt.title("Top 10 Trustworthy news sites\n")
plt.show()
# Let's reshuffle the dataset
df = df.sample(frac=1)
# taking the features
features = df[["site_url", "text_without_stopwords"]]
features.head(5)
features["url_text"] = (
features["site_url"].astype(str) + " " + features["text_without_stopwords"]
)
features.drop(["site_url", "text_without_stopwords"], axis=1, inplace=True)
features.head()
X = features
y = df["type"]
y = y.tolist()
# Splitting the dataset and using TF-IDF
from sklearn.feature_extraction.text import TfidfVectorizer
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, random_state=0
)
tfidf_vectorizer = TfidfVectorizer(use_idf=True, stop_words="english")
X_train_tfidf = tfidf_vectorizer.fit_transform(X_train["url_text"])
X_test_tfidf = tfidf_vectorizer.transform(X_test["url_text"])
tfidf_train = pd.DataFrame(
X_train_tfidf.A, columns=tfidf_vectorizer.get_feature_names()
)
tfidf_train.head()
# Above is the representation of tf-idf matrix
# ---
# Modelling
Adab = AdaBoostClassifier(
DecisionTreeClassifier(max_depth=10), n_estimators=5, random_state=1
)
Adab.fit(tfidf_train, y_train)
y_pred3 = Adab.predict(X_test_tfidf)
ABscore = metrics.accuracy_score(y_test, y_pred3)
print("accuracy: %0.3f" % ABscore)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/498/69498580.ipynb
|
source-based-news-classification
|
ruchi798
|
[{"Id": 69498580, "ScriptId": 18883039, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6649075, "CreationDate": "07/31/2021 18:34:51", "VersionNumber": 3.0, "Title": "Fake News Detection", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 229.0, "LinesInsertedFromPrevious": 43.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 186.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 11}]
|
[{"Id": 92743036, "KernelVersionId": 69498580, "SourceDatasetVersionId": 1449505}]
|
[{"Id": 1449505, "DatasetId": 747224, "DatasourceVersionId": 1483057, "CreatorUserId": 3309826, "LicenseName": "CC0: Public Domain", "CreationDate": "08/29/2020 10:20:07", "VersionNumber": 7.0, "Title": "Source based Fake News Classification", "Slug": "source-based-news-classification", "Subtitle": "Classification of news by type and labels", "Description": "### Context\n\nSocial media is a vast pool of content, and among all the content available for users to access, news is an element that is accessed most frequently. These news can be posted by politicians, news channels, newspaper websites, or even common civilians. These posts have to be checked for their authenticity, since spreading misinformation has been a real concern in today\u2019s times, and many firms are taking steps to make the common people aware of the consequences of spread misinformation. The measure of authenticity of the news posted online cannot be definitively measured, since the manual classification of news is tedious and time-consuming, and is also subject to bias.\nPublished paper: http://www.ijirset.com/upload/2020/june/115_4_Source.PDF\n\n\n### Content\n\nData preprocessing has been done on the dataset [Getting Real about Fake News](https://www.kaggle.com/mrisdal/fake-news) and skew has been eliminated.\n\n### Inspiration\n\nIn an era where fake WhatsApp forwards and Tweets are capable of influencing naive minds, tools and knowledge have to be put to practical use in not only mitigating the spread of misinformation but also to inform people about the type of news they consume. \nDevelopment of practical applications for users to gain insight from the articles they consume, fact-checking websites, built-in plugins and article parsers can\nfurther be refined, made easier to access, and more importantly, should create more awareness.\n\n### Acknowledgements\n\n[Getting Real about Fake News](https://www.kaggle.com/mrisdal/fake-news) seemed the most promising for preprocessing, feature extraction, and model classification. \nThe reason is due to the fact that all the other datasets lacked the sources from where the article/statement text was produced and published from. Citing the sources for article text is crucial to check the trustworthiness of the news and further helps in labelling the data as fake or untrustworthy.\n\nThanks to the dataset\u2019s comprehensiveness in terms of citing the source information of the text along with author names, date of publication and labels.", "VersionNotes": "Version 7", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 747224, "CreatorUserId": 3309826, "OwnerUserId": 3309826.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1449505.0, "CurrentDatasourceVersionId": 1483057.0, "ForumId": 762124, "Type": 2, "CreationDate": "06/30/2020 12:30:43", "LastActivityDate": "06/30/2020", "TotalViews": 43003, "TotalDownloads": 4857, "TotalVotes": 159, "TotalKernels": 23}]
|
[{"Id": 3309826, "UserName": "ruchi798", "DisplayName": "Ruchi Bhatia", "RegisterDate": "06/04/2019", "PerformanceTier": 4}]
|
# Exploring the news dataset
# 1. INTRODUCTION
# Social media is a vast pool of content, and among all the content available for users to access, news is an element that is accessed most frequently. These news can be posted by politicians, news channels, newspaper websites, or even common civilians. These posts have to be checked for their authenticity, since spreading misinformation has been a real concern in today’s times, and many firms are taking steps to make the common people aware of the consequences of spread misinformation. The measure of authenticity of the news posted online cannot be definitively measured, since the manual classification of news is tedious and time-consuming, and is also subject to bias.
# 
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import nltk
nltk.download("punkt")
import matplotlib.pyplot as plt
import seaborn as sns
from wordcloud import WordCloud, STOPWORDS
import nltk
import re
from nltk.corpus import stopwords
import seaborn as sns
import gensim
from gensim.utils import simple_preprocess
from gensim.parsing.preprocessing import STOPWORDS
import warnings
warnings.filterwarnings("ignore")
import plotly.express as px
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import roc_auc_score
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier
from sklearn.tree import DecisionTreeClassifier
# Reading the dataset
df = pd.read_csv("../input/source-based-news-classification/news_articles.csv")
print(df.shape)
df.head()
# Checking for null values
df.isnull().sum()
# dropping null values
df.dropna(inplace=True)
# ---
# Exploratory data analysis
# ### Let's visualize the proportion of real and fake news!
# real vs fake
fig = px.pie(df, names="label", title="Proportion of Real vs. Fake News")
fig.show()
# ### Languages of News Articles
sub_tf_df = (
df.groupby("language")
.apply(lambda x: x["language"].count())
.reset_index(name="Counts")
)
fig = px.bar(
sub_tf_df, x="language", y="Counts", color="Counts", barmode="group", height=400
)
fig.show()
# As we can see, English is the most common language in which most news articles are written
# ### Visualizing count of news articles by type
sub_check = (
df.groupby("type").apply(lambda x: x["type"].count()).reset_index(name="Counts")
)
fig = px.bar(
sub_check,
x="type",
y="Counts",
color="Counts",
title="Count of News Articles by type",
)
fig.show()
# here we can se that most news are of type bs (i.e. bullshit)
from wordcloud import WordCloud
wc = WordCloud(
background_color="black",
max_words=100,
max_font_size=256,
random_state=42,
width=1000,
height=1000,
)
wc.generate(" ".join(df["text_without_stopwords"]))
plt.imshow(wc)
plt.axis("off")
plt.show()
# ### Visualizing top 10 unigrams and bigrams
def get_top_n_words(corpus, n=None):
vec = CountVectorizer().fit(corpus)
bag_of_words = vec.transform(corpus)
sum_words = bag_of_words.sum(axis=0)
words_freq = [(word, sum_words[0, idx]) for word, idx in vec.vocabulary_.items()]
words_freq = sorted(words_freq, key=lambda x: x[1], reverse=True)
return words_freq[:n]
def get_top_n_bigram(corpus, n=None):
vec = CountVectorizer(ngram_range=(2, 2)).fit(corpus)
bag_of_words = vec.transform(corpus)
sum_words = bag_of_words.sum(axis=0)
words_freq = [(word, sum_words[0, idx]) for word, idx in vec.vocabulary_.items()]
words_freq = sorted(words_freq, key=lambda x: x[1], reverse=True)
return words_freq[:n]
common_words = get_top_n_words(df["text_without_stopwords"], 10)
df2 = pd.DataFrame(common_words, columns=["word", "count"])
df2.groupby("word").sum()["count"].sort_values(ascending=False)
fig = px.bar(df2, x="word", y="count", color="count", title="Top 10 unigrams")
fig.show()
common_words = get_top_n_bigram(df["text_without_stopwords"], 10)
df2 = pd.DataFrame(common_words, columns=["word", "count"])
df2.groupby("word").sum()["count"].sort_values(ascending=False)
fig = px.bar(df2, x="word", y="count", color="count", title="Top 10 bigrams")
fig.show()
# ### Visualizing top 5 authors
d = df["author"].value_counts().sort_values(ascending=False).head(5)
d = pd.DataFrame(d)
d = d.reset_index() # dataframe with top 5 authors
# Plotting
sns.set()
plt.figure(figsize=(15, 4))
sns.barplot(x="index", y="author", data=d)
plt.xlabel("\n Authors")
plt.ylabel("Number of Articles written")
plt.title("Top 5 authors\n")
plt.show()
# ### Visualizing top 10 fake news site
d = (
df[df["label"] == "Fake"]["site_url"]
.value_counts()
.sort_values(ascending=False)
.head(10)
)
d = pd.DataFrame(d)
d = d.reset_index() # dataframe with top 10 fake news site
# Plotting
sns.set()
plt.figure(figsize=(25, 7))
sns.barplot(x="index", y="site_url", data=d)
plt.xlabel("\n site_url")
plt.ylabel("Number of Articles written")
plt.title("Top 10 Fake news sites\n")
plt.show()
# ### Let's have a look at the top 10 trustworthy news sites
d = (
df[df["label"] == "Real"]["site_url"]
.value_counts()
.sort_values(ascending=False)
.head(10)
)
d = pd.DataFrame(d)
d = d.reset_index() # dataframe with top 10 Trustworthy news site
# Plotting
sns.set()
plt.figure(figsize=(25, 7))
sns.barplot(x="index", y="site_url", data=d)
plt.xlabel("\n site_url")
plt.ylabel("Number of Articles written")
plt.title("Top 10 Trustworthy news sites\n")
plt.show()
# Let's reshuffle the dataset
df = df.sample(frac=1)
# taking the features
features = df[["site_url", "text_without_stopwords"]]
features.head(5)
features["url_text"] = (
features["site_url"].astype(str) + " " + features["text_without_stopwords"]
)
features.drop(["site_url", "text_without_stopwords"], axis=1, inplace=True)
features.head()
X = features
y = df["type"]
y = y.tolist()
# Splitting the dataset and using TF-IDF
from sklearn.feature_extraction.text import TfidfVectorizer
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, random_state=0
)
tfidf_vectorizer = TfidfVectorizer(use_idf=True, stop_words="english")
X_train_tfidf = tfidf_vectorizer.fit_transform(X_train["url_text"])
X_test_tfidf = tfidf_vectorizer.transform(X_test["url_text"])
tfidf_train = pd.DataFrame(
X_train_tfidf.A, columns=tfidf_vectorizer.get_feature_names()
)
tfidf_train.head()
# Above is the representation of tf-idf matrix
# ---
# Modelling
Adab = AdaBoostClassifier(
DecisionTreeClassifier(max_depth=10), n_estimators=5, random_state=1
)
Adab.fit(tfidf_train, y_train)
y_pred3 = Adab.predict(X_test_tfidf)
ABscore = metrics.accuracy_score(y_test, y_pred3)
print("accuracy: %0.3f" % ABscore)
|
[{"source-based-news-classification/news_articles.csv": {"column_names": "[\"author\", \"published\", \"title\", \"text\", \"language\", \"site_url\", \"main_img_url\", \"type\", \"label\", \"title_without_stopwords\", \"text_without_stopwords\", \"hasImage\"]", "column_data_types": "{\"author\": \"object\", \"published\": \"object\", \"title\": \"object\", \"text\": \"object\", \"language\": \"object\", \"site_url\": \"object\", \"main_img_url\": \"object\", \"type\": \"object\", \"label\": \"object\", \"title_without_stopwords\": \"object\", \"text_without_stopwords\": \"object\", \"hasImage\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 2096 entries, 0 to 2095\nData columns (total 12 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 author 2096 non-null object \n 1 published 2096 non-null object \n 2 title 2096 non-null object \n 3 text 2050 non-null object \n 4 language 2095 non-null object \n 5 site_url 2095 non-null object \n 6 main_img_url 2095 non-null object \n 7 type 2095 non-null object \n 8 label 2095 non-null object \n 9 title_without_stopwords 2094 non-null object \n 10 text_without_stopwords 2046 non-null object \n 11 hasImage 2095 non-null float64\ndtypes: float64(1), object(11)\nmemory usage: 196.6+ KB\n", "summary": "{\"hasImage\": {\"count\": 2095.0, \"mean\": 0.7770883054892601, \"std\": 0.41629892386736406, \"min\": 0.0, \"25%\": 1.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}}", "examples": "{\"author\":{\"0\":\"Barracuda Brigade\",\"1\":\"reasoning with facts\",\"2\":\"Barracuda Brigade\",\"3\":\"Fed Up\"},\"published\":{\"0\":\"2016-10-26T21:41:00.000+03:00\",\"1\":\"2016-10-29T08:47:11.259+03:00\",\"2\":\"2016-10-31T01:41:49.479+02:00\",\"3\":\"2016-11-01T05:22:00.000+02:00\"},\"title\":{\"0\":\"muslims busted they stole millions in govt benefits\",\"1\":\"re why did attorney general loretta lynch plead the fifth\",\"2\":\"breaking weiner cooperating with fbi on hillary email investigation\",\"3\":\"pin drop speech by father of daughter kidnapped and killed by isis i have voted for donald j trump percentfedupcom\"},\"text\":{\"0\":\"print they should pay all the back all the money plus interest the entire family and everyone who came in with them need to be deported asap why did it take two years to bust them \\nhere we go again another group stealing from the government and taxpayers a group of somalis stole over four million in government benefits over just months \\nweve reported on numerous cases like this one where the muslim refugeesimmigrants commit fraud by scamming our systemits way out of control more related\",\"1\":\"why did attorney general loretta lynch plead the fifth barracuda brigade print the administration is blocking congressional probe into cash payments to iran of course she needs to plead the th she either cant recall refuses to answer or just plain deflects the question straight up corruption at its finest \\npercentfedupcom talk about covering your ass loretta lynch did just that when she plead the fifth to avoid incriminating herself over payments to irancorrupt to the core attorney general loretta lynch is declining to comply with an investigation by leading members of congress about the obama administrations secret efforts to send iran billion in cash earlier this year prompting accusations that lynch has pleaded the fifth amendment to avoid incriminating herself over these payments according to lawmakers and communications exclusively obtained by the washington free beacon \\nsen marco rubio r fla and rep mike pompeo r kan initially presented lynch in october with a series of questions about how the cash payment to iran was approved and delivered \\nin an oct response assistant attorney general peter kadzik responded on lynchs behalf refusing to answer the questions and informing the lawmakers that they are barred from publicly disclosing any details about the cash payment which was bound up in a ransom deal aimed at freeing several american hostages from iran \\nthe response from the attorney generals office is unacceptable and provides evidence that lynch has chosen to essentially plead the fifth and refuse to respond to inquiries regarding herrole in providing cash to the worlds foremost state sponsor of terrorism rubio and pompeo wrote on friday in a followup letter to lynch more related\",\"2\":\"red state \\nfox news sunday reported this morning that anthony weiner is cooperating with the fbi which has reopened yes lefties reopened the investigation into hillary clintons classified emails watch as chris wallace reports the breaking news during the panel segment near the end of the show \\nand the news is breaking while were on the air our colleague bret baier has just sent us an email saying he has two sources who say that anthony weiner who also had coownership of that laptop with his estranged wife huma abedin is cooperating with the fbi investigation had given them the laptop so therefore they didnt need a warrant to get in to see the contents of said laptop pretty interesting development \\ntargets of federal investigations will often cooperate hoping that they will get consideration from a judge at sentencing given weiners wellknown penchant for lying its hard to believe that a prosecutor would give weiner a deal based on an agreement to testify unless his testimony were very strongly corroborated by hard evidence but cooperation can take many forms and as wallace indicated on this mornings show one of those forms could be signing a consent form to allow the contents of devices that they could probably get a warrant for anyway well see if weiners cooperation extends beyond that more related\",\"3\":\"email kayla mueller was a prisoner and tortured by isis while no chance of releasea horrific story her father gave a pin drop speech that was so heartfelt you want to give him a hug carl mueller believes donald trump will be a great presidentepic speech k shares\"},\"language\":{\"0\":\"english\",\"1\":\"english\",\"2\":\"english\",\"3\":\"english\"},\"site_url\":{\"0\":\"100percentfedup.com\",\"1\":\"100percentfedup.com\",\"2\":\"100percentfedup.com\",\"3\":\"100percentfedup.com\"},\"main_img_url\":{\"0\":\"http:\\/\\/bb4sp.com\\/wp-content\\/uploads\\/2016\\/10\\/Fullscreen-capture-10262016-83501-AM.bmp.jpg\",\"1\":\"http:\\/\\/bb4sp.com\\/wp-content\\/uploads\\/2016\\/10\\/Fullscreen-capture-10282016-102616-PM.bmp.jpg\",\"2\":\"http:\\/\\/bb4sp.com\\/wp-content\\/uploads\\/2016\\/10\\/Fullscreen-capture-10302016-60437-PM.bmp.jpg\",\"3\":\"http:\\/\\/100percentfedup.com\\/wp-content\\/uploads\\/2016\\/10\\/kayla.jpg\"},\"type\":{\"0\":\"bias\",\"1\":\"bias\",\"2\":\"bias\",\"3\":\"bias\"},\"label\":{\"0\":\"Real\",\"1\":\"Real\",\"2\":\"Real\",\"3\":\"Real\"},\"title_without_stopwords\":{\"0\":\"muslims busted stole millions govt benefits\",\"1\":\"attorney general loretta lynch plead fifth\",\"2\":\"breaking weiner cooperating fbi hillary email investigation\",\"3\":\"pin drop speech father daughter kidnapped killed isis voted donald j trump percentfedupcom\"},\"text_without_stopwords\":{\"0\":\"print pay back money plus interest entire family everyone came need deported asap take two years bust go another group stealing government taxpayers group somalis stole four million government benefits months weve reported numerous cases like one muslim refugeesimmigrants commit fraud scamming systemits way control related\",\"1\":\"attorney general loretta lynch plead fifth barracuda brigade print administration blocking congressional probe cash payments iran course needs plead th either cant recall refuses answer plain deflects question straight corruption finest percentfedupcom talk covering ass loretta lynch plead fifth avoid incriminating payments irancorrupt core attorney general loretta lynch declining comply investigation leading members congress obama administrations secret efforts send iran billion cash earlier year prompting accusations lynch pleaded fifth amendment avoid incriminating payments according lawmakers communications exclusively obtained washington free beacon sen marco rubio r fla rep mike pompeo r kan initially presented lynch october series questions cash payment iran approved delivered oct response assistant attorney general peter kadzik responded lynchs behalf refusing answer questions informing lawmakers barred publicly disclosing details cash payment bound ransom deal aimed freeing several american hostages iran response attorney generals office unacceptable provides evidence lynch chosen essentially plead fifth refuse respond inquiries regarding herrole providing cash worlds foremost state sponsor terrorism rubio pompeo wrote friday followup letter lynch related\",\"2\":\"red state fox news sunday reported morning anthony weiner cooperating fbi reopened yes lefties reopened investigation hillary clintons classified emails watch chris wallace reports breaking news panel segment near end show news breaking air colleague bret baier sent us email saying two sources say anthony weiner also coownership laptop estranged wife huma abedin cooperating fbi investigation given laptop therefore didnt need warrant get see contents said laptop pretty interesting development targets federal investigations often cooperate hoping get consideration judge sentencing given weiners wellknown penchant lying hard believe prosecutor would give weiner deal based agreement testify unless testimony strongly corroborated hard evidence cooperation take many forms wallace indicated mornings show one forms could signing consent form allow contents devices could probably get warrant anyway well see weiners cooperation extends beyond related\",\"3\":\"email kayla mueller prisoner tortured isis chance releasea horrific story father gave pin drop speech heartfelt want give hug carl mueller believes donald trump great presidentepic speech k shares\"},\"hasImage\":{\"0\":1.0,\"1\":1.0,\"2\":1.0,\"3\":1.0}}"}}]
| true | 1 |
<start_data_description><data_path>source-based-news-classification/news_articles.csv:
<column_names>
['author', 'published', 'title', 'text', 'language', 'site_url', 'main_img_url', 'type', 'label', 'title_without_stopwords', 'text_without_stopwords', 'hasImage']
<column_types>
{'author': 'object', 'published': 'object', 'title': 'object', 'text': 'object', 'language': 'object', 'site_url': 'object', 'main_img_url': 'object', 'type': 'object', 'label': 'object', 'title_without_stopwords': 'object', 'text_without_stopwords': 'object', 'hasImage': 'float64'}
<dataframe_Summary>
{'hasImage': {'count': 2095.0, 'mean': 0.7770883054892601, 'std': 0.41629892386736406, 'min': 0.0, '25%': 1.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}}
<dataframe_info>
RangeIndex: 2096 entries, 0 to 2095
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 author 2096 non-null object
1 published 2096 non-null object
2 title 2096 non-null object
3 text 2050 non-null object
4 language 2095 non-null object
5 site_url 2095 non-null object
6 main_img_url 2095 non-null object
7 type 2095 non-null object
8 label 2095 non-null object
9 title_without_stopwords 2094 non-null object
10 text_without_stopwords 2046 non-null object
11 hasImage 2095 non-null float64
dtypes: float64(1), object(11)
memory usage: 196.6+ KB
<some_examples>
{'author': {'0': 'Barracuda Brigade', '1': 'reasoning with facts', '2': 'Barracuda Brigade', '3': 'Fed Up'}, 'published': {'0': '2016-10-26T21:41:00.000+03:00', '1': '2016-10-29T08:47:11.259+03:00', '2': '2016-10-31T01:41:49.479+02:00', '3': '2016-11-01T05:22:00.000+02:00'}, 'title': {'0': 'muslims busted they stole millions in govt benefits', '1': 're why did attorney general loretta lynch plead the fifth', '2': 'breaking weiner cooperating with fbi on hillary email investigation', '3': 'pin drop speech by father of daughter kidnapped and killed by isis i have voted for donald j trump percentfedupcom'}, 'text': {'0': 'print they should pay all the back all the money plus interest the entire family and everyone who came in with them need to be deported asap why did it take two years to bust them \nhere we go again another group stealing from the government and taxpayers a group of somalis stole over four million in government benefits over just months \nweve reported on numerous cases like this one where the muslim refugeesimmigrants commit fraud by scamming our systemits way out of control more related', '1': 'why did attorney general loretta lynch plead the fifth barracuda brigade print the administration is blocking congressional probe into cash payments to iran of course she needs to plead the th she either cant recall refuses to answer or just plain deflects the question straight up corruption at its finest \npercentfedupcom talk about covering your ass loretta lynch did just that when she plead the fifth to avoid incriminating herself over payments to irancorrupt to the core attorney general loretta lynch is declining to comply with an investigation by leading members of congress about the obama administrations secret efforts to send iran billion in cash earlier this year prompting accusations that lynch has pleaded the fifth amendment to avoid incriminating herself over these payments according to lawmakers and communications exclusively obtained by the washington free beacon \nsen marco rubio r fla and rep mike pompeo r kan initially presented lynch in october with a series of questions about how the cash payment to iran was approved and delivered \nin an oct response assistant attorney general peter kadzik responded on lynchs behalf refusing to answer the questions and informing the lawmakers that they are barred from publicly disclosing any details about the cash payment which was bound up in a ransom deal aimed at freeing several american hostages from iran \nthe response from the attorney generals office is unacceptable and provides evidence that lynch has chosen to essentially plead the fifth and refuse to respond to inquiries regarding herrole in providing cash to the worlds foremost state sponsor of terrorism rubio and pompeo wrote on friday in a followup letter to lynch more related', '2': 'red state \nfox news sunday reported this morning that anthony weiner is cooperating with the fbi which has reopened yes lefties reopened the investigation into hillary clintons classified emails watch as chris wallace reports the breaking news during the panel segment near the end of the show \nand the news is breaking while were on the air our colleague bret baier has just sent us an email saying he has two sources who say that anthony weiner who also had coownership of that laptop with his estranged wife huma abedin is cooperating with the fbi investigation had given them the laptop so therefore they didnt need a warrant to get in to see the contents of said laptop pretty interesting development \ntargets of federal investigations will often cooperate hoping that they will get consideration from a judge at sentencing given weiners wellknown penchant for lying its hard to believe that a prosecutor would give weiner a deal based on an agreement to testify unless his testimony were very strongly corroborated by hard evidence but cooperation can take many forms and as wallace indicated on this mornings show one of those forms could be signing a consent form to allow the contents of devices that they could probably get a warrant for anyway well see if weiners cooperation extends beyond that more related', '3': 'email kayla mueller was a prisoner and tortured by isis while no chance of releasea horrific story her father gave a pin drop speech that was so heartfelt you want to give him a hug carl mueller believes donald trump will be a great presidentepic speech k shares'}, 'language': {'0': 'english', '1': 'english', '2': 'english', '3': 'english'}, 'site_url': {'0': '100percentfedup.com', '1': '100percentfedup.com', '2': '100percentfedup.com', '3': '100percentfedup.com'}, 'main_img_url': {'0': 'http://bb4sp.com/wp-content/uploads/2016/10/Fullscreen-capture-10262016-83501-AM.bmp.jpg', '1': 'http://bb4sp.com/wp-content/uploads/2016/10/Fullscreen-capture-10282016-102616-PM.bmp.jpg', '2': 'http://bb4sp.com/wp-content/uploads/2016/10/Fullscreen-capture-10302016-60437-PM.bmp.jpg', '3': 'http://100percentfedup.com/wp-content/uploads/2016/10/kayla.jpg'}, 'type': {'0': 'bias', '1': 'bias', '2': 'bias', '3': 'bias'}, 'label': {'0': 'Real', '1': 'Real', '2': 'Real', '3': 'Real'}, 'title_without_stopwords': {'0': 'muslims busted stole millions govt benefits', '1': 'attorney general loretta lynch plead fifth', '2': 'breaking weiner cooperating fbi hillary email investigation', '3': 'pin drop speech father daughter kidnapped killed isis voted donald j trump percentfedupcom'}, 'text_without_stopwords': {'0': 'print pay back money plus interest entire family everyone came need deported asap take two years bust go another group stealing government taxpayers group somalis stole four million government benefits months weve reported numerous cases like one muslim refugeesimmigrants commit fraud scamming systemits way control related', '1': 'attorney general loretta lynch plead fifth barracuda brigade print administration blocking congressional probe cash payments iran course needs plead th either cant recall refuses answer plain deflects question straight corruption finest percentfedupcom talk covering ass loretta lynch plead fifth avoid incriminating payments irancorrupt core attorney general loretta lynch declining comply investigation leading members congress obama administrations secret efforts send iran billion cash earlier year prompting accusations lynch pleaded fifth amendment avoid incriminating payments according lawmakers communications exclusively obtained washington free beacon sen marco rubio r fla rep mike pompeo r kan initially presented lynch october series questions cash payment iran approved delivered oct response assistant attorney general peter kadzik responded lynchs behalf refusing answer questions informing lawmakers barred publicly disclosing details cash payment bound ransom deal aimed freeing several american hostages iran response attorney generals office unacceptable provides evidence lynch chosen essentially plead fifth refuse respond inquiries regarding herrole providing cash worlds foremost state sponsor terrorism rubio pompeo wrote friday followup letter lynch related', '2': 'red state fox news sunday reported morning anthony weiner cooperating fbi reopened yes lefties reopened investigation hillary clintons classified emails watch chris wallace reports breaking news panel segment near end show news breaking air colleague bret baier sent us email saying two sources say anthony weiner also coownership laptop estranged wife huma abedin cooperating fbi investigation given laptop therefore didnt need warrant get see contents said laptop pretty interesting development targets federal investigations often cooperate hoping get consideration judge sentencing given weiners wellknown penchant lying hard believe prosecutor would give weiner deal based agreement testify unless testimony strongly corroborated hard evidence cooperation take many forms wallace indicated mornings show one forms could signing consent form allow contents devices could probably get warrant anyway well see weiners cooperation extends beyond related', '3': 'email kayla mueller prisoner tortured isis chance releasea horrific story father gave pin drop speech heartfelt want give hug carl mueller believes donald trump great presidentepic speech k shares'}, 'hasImage': {'0': 1.0, '1': 1.0, '2': 1.0, '3': 1.0}}
<end_description>
| 2,420 | 11 | 4,482 | 2,420 |
69498243
|
<jupyter_start><jupyter_text>Heart Disease UCI
Kaggle dataset identifier: heart-disease-uci
<jupyter_script>#
#
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("../input/heart-disease-uci/heart.csv")
df.head(5)
df.describe()
df.shape
# Checking for any null values
df.isnull().sum()
# No null values, we are saved! 🙏. Doesn't require converting any null values or dropping rows.
# Making Data Readable
# At present, the dataframe contains nothing but numeric data, however we can see that some features are categorical and it doesn't make sense what the numbers mean 🤔. Let's convert them.
# Reference :: https://pubmed.ncbi.nlm.nih.gov/20494662/
# age: age (in years)
# sex: gender (1 = male; 0 = female)
# cp: chest pain type
# trestbps: resting blood pressure (in mmHg, upon admission to the hospital
# chol: serum cholesterol in mg/dL
# fbs: fasting blood sugar > 120 mg/dL (likely to be diabetic) 1 = true; 0 = false
# restecg: resting electrocardiogram results
# thalach: maximum heart rate achieved
# exang: exercise induced angina (1 = yes; 0 = no)
# oldpeak: ST depression induced by exercise relative to rest (in mm, achieved by subtracting the lowest ST segment points during exercise and rest)
# slope: the slope of the peak exercise ST segment, ST-T abnormalities are considered to be a crucial indicator for identifying presence of ischaemia
# ca: number of major vessels (0-3) colored by fluoroscopy
# thal: thallium test
# target: does that person have disease or not? 0 = no disease, 1 = disease
# The confusing ones 😲..
# Chest Pain Type (cp)
# | No. | Chest Pain Type | Criteria |
# | --------------- | --------------- | --------------- |
# | 0 | Typical Angina | All criteria present |
# | 1 | Atypical Angina | 2 of 3 criteria present |
# | 2 | Non Anginal Pain | Less than one criteria present |
# | 3 | Asymptomatic | None of criteria are satisfied |
# Angina: Discomfort that is noted when the heart does not get enough blood or oxygen
# Non Anginal Pain: Pain in the chest that is not caused by heart disease or a heart attack, usually related to digestive tract
# Asymptomatic: Silent killer which shows no symptoms
# Resting ElectroCardiogram Results (restecg)
# | No. | Results |
# | --------------- | ------------------- |
# | 0 | Normal |
# | 1 | Having ST-T wave abnormality |
# | 2 | Showing probable or definite left ventricular hypertrophy by Estes' criteria |
# Left Ventricular Hypertrophy: A heart's left pumping chamber that has thickened and may not be pumping efficiently
# ST-T wave abnormality: ST segment abnormality (elevation or depression) indicates myocardial ischaemia or infarction i.e. a heart attack
# Slope of peak exercise ST segment (slope)
# | No. | Slope |
# | --------------- | ------------------- |
# | 1 | Upsloping |
# | 2 | Flat |
# | 3 | Downsloping |
# Horizontal or downsloping ST depression ≥ 0.5 mm at the J-point in ≥ 2 contiguous leads indicates myocardial ischaemia or blockage or arteries which eventually leads to heart disease
# Thallium Testing (thal)
# | No. | Results | Meaning |
# | --------------- | ------------------- | ------------------- |
# | 0 | Normal | Passed Thallium Test and condition is normal |
# | 1 | Fixed Defect | Heart tissue can't absorb thallium both under stress and in rest |
# | 2 | Reversible Defect | Heart tissue is unable to absorb thallium only under the exercise portion of the test |
# A thallium stress test is a nuclear medicine study that shows your physician how well blood flows through your heart muscle while you're exercising or at rest and you're basically screwed if the result is a fixed defect or reversible defect. Fixed defect being worse
#
df.loc[df["sex"] == 0, "sex"] = "Female"
df.loc[df["sex"] == 1, "sex"] = "Male"
df.loc[df["cp"] == 0, "cp"] = "Typical Angina"
df.loc[df["cp"] == 1, "cp"] = "Atypical Angina"
df.loc[df["cp"] == 2, "cp"] = "Non Anginal Pain"
df.loc[df["cp"] == 3, "cp"] = "Asymptomatic"
df.loc[df["restecg"] == 0, "restecg"] = "Normal"
df.loc[df["restecg"] == 1, "restecg"] = "ST-T Wave Abnormality"
df.loc[df["restecg"] == 2, "restecg"] = "Left Ventricular Hypertrophy"
df.loc[df["slope"] == 0, "slope"] = "Unsloping"
df.loc[df["slope"] == 1, "slope"] = "Flat"
df.loc[df["slope"] == 2, "slope"] = "Downsloping"
df.loc[df["thal"] == 1, "thal"] = "Normal"
df.loc[df["thal"] == 2, "thal"] = "Fixed Defect"
df.loc[df["thal"] == 3, "thal"] = "Reversible Defect"
df.loc[df["fbs"] == 0, "fbs"] = "> 120mg/dL"
df.loc[df["fbs"] == 1, "fbs"] = "< 120mg/dL"
df.loc[df["exang"] == 0, "exang"] = "No"
df.loc[df["exang"] == 1, "exang"] = "Yes"
df.loc[df["target"] == 0, "target"] = "No heart disease found"
df.loc[df["target"] == 1, "target"] = "Has heart disease"
df.head(5)
# Inspection Time! 🤓
# Get your lab coats ready!
# Relation between Attributes
plt.figure(figsize=(10, 8))
corr = df.corr()
tick_labels = [
"Age",
"Resting BP",
"Cholestrol",
"Max Heart Rate",
"Old Peak",
"Vessels colored",
]
# Getting the Upper Triangle of the co-relation matrix
matrix = np.triu(corr)
# using the upper triangle matrix as mask
corr_heatmap = sns.heatmap(
corr,
annot=True,
mask=matrix,
cmap="viridis",
xticklabels=tick_labels,
yticklabels=tick_labels,
)
plt.yticks(rotation=0)
plt.show()
#
# Amazing!, we can observe many things from this heatmap.
#
# sex: gender (1 = male; 0 = female)
# cp: chest pain type
#
### ST-T slopes for various Rest ECG results
colors = ["#FFF338", "#0CECDD"]
title_style = {"fontname": "monospace", "fontsize": 25}
plt.figure(figsize=(30, 8))
slopes_st_t = df.loc[df["restecg"] == "ST-T Wave Abnormality"]
slopes_ventricular = df.loc[df["restecg"] == "Left Ventricular Hypertrophy"]
slopes_normal = df.loc[df["restecg"] == "Normal"]
plt.subplot(1, 3, 1)
plt.title("ST-T Wave Abnormality", fontdict=title_style)
sns.countplot(x="slope", hue="sex", data=slopes_st_t, palette=colors)
plt.subplot(1, 3, 2)
plt.title("Ventricular Hypertrophy", fontdict=title_style)
sns.countplot(x="slope", hue="sex", data=slopes_ventricular, palette=colors)
plt.subplot(1, 3, 3)
plt.title("Normal", fontdict=title_style)
sns.countplot(x="slope", hue="sex", data=slopes_normal, palette=colors)
plt.show()
# Interestingly, there are no downsloping results when restecg results showed left ventricular hypertrophy. Quite peculiar that all of the patients having flat slope in ventricular hypertrophy are males and unsloping are females 🤔. Count of males in all the three is more perhaps because of majority of patients being males. Let's find that out
plt.title("Count of males vs females")
sns.countplot(x="sex", data=df, palette="magma")
plt.show()
# As expected there are nearly 50 percent more males than females as patients. But how many of them are probably diabetic?. We can tell that by looking at the fbs or fasting blood sugar test which is nothing but measuring sugar levels in blood without eating anything (fun fact : having food causes sugar levels to spike and gives inaccurate results and thus fasting before the test)
plt.title("FBS Count of Patients", fontdict={"fontname": "monospace", "fontsize": 20})
sns.histplot(x="age", hue="fbs", data=df, element="poly")
plt.show()
# We can see that most of the patients admitted have fasting blood sugar levels more than 120mg/L which means they are most probably diabetic and also most of the patients lie in the age group of 50-65 ish. Let's see what results of the Thallium test for the patients who have exercise induced angina and also the maximum heart rate achieved by these patients
exercise_induced_angina = df.loc[(df["exang"] == "Yes") & (df["thal"] != 0)]
plt.figure(figsize=(15, 5))
plt.subplot(1, 2, 1)
plt.title("Thallium Results", fontdict={"fontname": "monospace", "fontsize": 15})
sns.countplot(data=exercise_induced_angina, x="thal", palette="magma")
plt.subplot(1, 2, 2)
plt.title(
"Maximum Heart Rate achieved", fontdict={"fontname": "monospace", "fontsize": 15}
)
plt.ylabel("Maximum heart rate")
sns.lineplot(x="age", y="thalach", data=exercise_induced_angina)
plt.show()
# Most of the people who had exercise induced angina showed reversible defect in thallium test which is true as reversible defect is when heart tissue is unable to absorb thallium only under the exercise portion of the test, followed by fixed defect which means under stress and rest and very small percentage of patients got the result of normal
# Cholestrol and Heart Disease
#
# Cholestrol is in mg/dl according to this data and also dataset hence we can check the range and determine heart disease or not
kwargs = dict(s=30)
fig, ax = plt.subplots(nrows=1, figsize=(10, 8))
sns.scatterplot(x="age", y="chol", hue="target", data=df, palette="plasma", **kwargs)
ax.axhspan(0, 200, alpha=0.2, color="green")
ax.axhspan(200, 239, alpha=0.2, color="yellow")
ax.axhspan(239, 600, alpha=0.2, color="red")
plt.show()
# Quite an unusual result!, the only thing which makes sense here is that most of the patients have cholestrol above the ideal range. However, the patients having heart disease and vice versa is quite spread out.
# Few patients having high cholestrol have been diagnosed as not having a heart disease
# The youngest patient admitted for a check on cardiovascular disease is around 28-29 ish and the oldest one is around 77-78 ish
# Age might contribute slightly to the heart diseases (angina), slightly more concentrated to the right. Still it is distributed across implying age doesn't really matter for a cardiovascular diseases to occur, another reason to enjoy the present :p 💃
# Pain
# ## Checking for outliers
df.plot(kind="box", subplots=True, layout=(2, 3))
plt.figure(figsize=(15, 10))
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/498/69498243.ipynb
|
heart-disease-uci
|
ronitf
|
[{"Id": 69498243, "ScriptId": 18918741, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5540578, "CreationDate": "07/31/2021 18:29:19", "VersionNumber": 2.0, "Title": "Heart Disease UCI EDA", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 295.0, "LinesInsertedFromPrevious": 155.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 140.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92741997, "KernelVersionId": 69498243, "SourceDatasetVersionId": 43520}]
|
[{"Id": 43520, "DatasetId": 33180, "DatasourceVersionId": 45794, "CreatorUserId": 1223413, "LicenseName": "Other (specified in description)", "CreationDate": "06/25/2018 11:33:56", "VersionNumber": NaN, "Title": "Heart Disease UCI", "Slug": "heart-disease-uci", "Subtitle": "https://archive.ics.uci.edu/ml/datasets/Heart+Disease", "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": NaN, "TotalUncompressedBytes": NaN}]
|
[{"Id": 33180, "CreatorUserId": 1223413, "OwnerUserId": 1223413.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 43520.0, "CurrentDatasourceVersionId": 45794.0, "ForumId": 41554, "Type": 2, "CreationDate": "06/25/2018 11:33:56", "LastActivityDate": "06/25/2018", "TotalViews": 1811531, "TotalDownloads": 284703, "TotalVotes": 5708, "TotalKernels": 1999}]
|
[{"Id": 1223413, "UserName": "ronitf", "DisplayName": "ronit", "RegisterDate": "08/20/2017", "PerformanceTier": 0}]
|
#
#
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("../input/heart-disease-uci/heart.csv")
df.head(5)
df.describe()
df.shape
# Checking for any null values
df.isnull().sum()
# No null values, we are saved! 🙏. Doesn't require converting any null values or dropping rows.
# Making Data Readable
# At present, the dataframe contains nothing but numeric data, however we can see that some features are categorical and it doesn't make sense what the numbers mean 🤔. Let's convert them.
# Reference :: https://pubmed.ncbi.nlm.nih.gov/20494662/
# age: age (in years)
# sex: gender (1 = male; 0 = female)
# cp: chest pain type
# trestbps: resting blood pressure (in mmHg, upon admission to the hospital
# chol: serum cholesterol in mg/dL
# fbs: fasting blood sugar > 120 mg/dL (likely to be diabetic) 1 = true; 0 = false
# restecg: resting electrocardiogram results
# thalach: maximum heart rate achieved
# exang: exercise induced angina (1 = yes; 0 = no)
# oldpeak: ST depression induced by exercise relative to rest (in mm, achieved by subtracting the lowest ST segment points during exercise and rest)
# slope: the slope of the peak exercise ST segment, ST-T abnormalities are considered to be a crucial indicator for identifying presence of ischaemia
# ca: number of major vessels (0-3) colored by fluoroscopy
# thal: thallium test
# target: does that person have disease or not? 0 = no disease, 1 = disease
# The confusing ones 😲..
# Chest Pain Type (cp)
# | No. | Chest Pain Type | Criteria |
# | --------------- | --------------- | --------------- |
# | 0 | Typical Angina | All criteria present |
# | 1 | Atypical Angina | 2 of 3 criteria present |
# | 2 | Non Anginal Pain | Less than one criteria present |
# | 3 | Asymptomatic | None of criteria are satisfied |
# Angina: Discomfort that is noted when the heart does not get enough blood or oxygen
# Non Anginal Pain: Pain in the chest that is not caused by heart disease or a heart attack, usually related to digestive tract
# Asymptomatic: Silent killer which shows no symptoms
# Resting ElectroCardiogram Results (restecg)
# | No. | Results |
# | --------------- | ------------------- |
# | 0 | Normal |
# | 1 | Having ST-T wave abnormality |
# | 2 | Showing probable or definite left ventricular hypertrophy by Estes' criteria |
# Left Ventricular Hypertrophy: A heart's left pumping chamber that has thickened and may not be pumping efficiently
# ST-T wave abnormality: ST segment abnormality (elevation or depression) indicates myocardial ischaemia or infarction i.e. a heart attack
# Slope of peak exercise ST segment (slope)
# | No. | Slope |
# | --------------- | ------------------- |
# | 1 | Upsloping |
# | 2 | Flat |
# | 3 | Downsloping |
# Horizontal or downsloping ST depression ≥ 0.5 mm at the J-point in ≥ 2 contiguous leads indicates myocardial ischaemia or blockage or arteries which eventually leads to heart disease
# Thallium Testing (thal)
# | No. | Results | Meaning |
# | --------------- | ------------------- | ------------------- |
# | 0 | Normal | Passed Thallium Test and condition is normal |
# | 1 | Fixed Defect | Heart tissue can't absorb thallium both under stress and in rest |
# | 2 | Reversible Defect | Heart tissue is unable to absorb thallium only under the exercise portion of the test |
# A thallium stress test is a nuclear medicine study that shows your physician how well blood flows through your heart muscle while you're exercising or at rest and you're basically screwed if the result is a fixed defect or reversible defect. Fixed defect being worse
#
df.loc[df["sex"] == 0, "sex"] = "Female"
df.loc[df["sex"] == 1, "sex"] = "Male"
df.loc[df["cp"] == 0, "cp"] = "Typical Angina"
df.loc[df["cp"] == 1, "cp"] = "Atypical Angina"
df.loc[df["cp"] == 2, "cp"] = "Non Anginal Pain"
df.loc[df["cp"] == 3, "cp"] = "Asymptomatic"
df.loc[df["restecg"] == 0, "restecg"] = "Normal"
df.loc[df["restecg"] == 1, "restecg"] = "ST-T Wave Abnormality"
df.loc[df["restecg"] == 2, "restecg"] = "Left Ventricular Hypertrophy"
df.loc[df["slope"] == 0, "slope"] = "Unsloping"
df.loc[df["slope"] == 1, "slope"] = "Flat"
df.loc[df["slope"] == 2, "slope"] = "Downsloping"
df.loc[df["thal"] == 1, "thal"] = "Normal"
df.loc[df["thal"] == 2, "thal"] = "Fixed Defect"
df.loc[df["thal"] == 3, "thal"] = "Reversible Defect"
df.loc[df["fbs"] == 0, "fbs"] = "> 120mg/dL"
df.loc[df["fbs"] == 1, "fbs"] = "< 120mg/dL"
df.loc[df["exang"] == 0, "exang"] = "No"
df.loc[df["exang"] == 1, "exang"] = "Yes"
df.loc[df["target"] == 0, "target"] = "No heart disease found"
df.loc[df["target"] == 1, "target"] = "Has heart disease"
df.head(5)
# Inspection Time! 🤓
# Get your lab coats ready!
# Relation between Attributes
plt.figure(figsize=(10, 8))
corr = df.corr()
tick_labels = [
"Age",
"Resting BP",
"Cholestrol",
"Max Heart Rate",
"Old Peak",
"Vessels colored",
]
# Getting the Upper Triangle of the co-relation matrix
matrix = np.triu(corr)
# using the upper triangle matrix as mask
corr_heatmap = sns.heatmap(
corr,
annot=True,
mask=matrix,
cmap="viridis",
xticklabels=tick_labels,
yticklabels=tick_labels,
)
plt.yticks(rotation=0)
plt.show()
#
# Amazing!, we can observe many things from this heatmap.
#
# sex: gender (1 = male; 0 = female)
# cp: chest pain type
#
### ST-T slopes for various Rest ECG results
colors = ["#FFF338", "#0CECDD"]
title_style = {"fontname": "monospace", "fontsize": 25}
plt.figure(figsize=(30, 8))
slopes_st_t = df.loc[df["restecg"] == "ST-T Wave Abnormality"]
slopes_ventricular = df.loc[df["restecg"] == "Left Ventricular Hypertrophy"]
slopes_normal = df.loc[df["restecg"] == "Normal"]
plt.subplot(1, 3, 1)
plt.title("ST-T Wave Abnormality", fontdict=title_style)
sns.countplot(x="slope", hue="sex", data=slopes_st_t, palette=colors)
plt.subplot(1, 3, 2)
plt.title("Ventricular Hypertrophy", fontdict=title_style)
sns.countplot(x="slope", hue="sex", data=slopes_ventricular, palette=colors)
plt.subplot(1, 3, 3)
plt.title("Normal", fontdict=title_style)
sns.countplot(x="slope", hue="sex", data=slopes_normal, palette=colors)
plt.show()
# Interestingly, there are no downsloping results when restecg results showed left ventricular hypertrophy. Quite peculiar that all of the patients having flat slope in ventricular hypertrophy are males and unsloping are females 🤔. Count of males in all the three is more perhaps because of majority of patients being males. Let's find that out
plt.title("Count of males vs females")
sns.countplot(x="sex", data=df, palette="magma")
plt.show()
# As expected there are nearly 50 percent more males than females as patients. But how many of them are probably diabetic?. We can tell that by looking at the fbs or fasting blood sugar test which is nothing but measuring sugar levels in blood without eating anything (fun fact : having food causes sugar levels to spike and gives inaccurate results and thus fasting before the test)
plt.title("FBS Count of Patients", fontdict={"fontname": "monospace", "fontsize": 20})
sns.histplot(x="age", hue="fbs", data=df, element="poly")
plt.show()
# We can see that most of the patients admitted have fasting blood sugar levels more than 120mg/L which means they are most probably diabetic and also most of the patients lie in the age group of 50-65 ish. Let's see what results of the Thallium test for the patients who have exercise induced angina and also the maximum heart rate achieved by these patients
exercise_induced_angina = df.loc[(df["exang"] == "Yes") & (df["thal"] != 0)]
plt.figure(figsize=(15, 5))
plt.subplot(1, 2, 1)
plt.title("Thallium Results", fontdict={"fontname": "monospace", "fontsize": 15})
sns.countplot(data=exercise_induced_angina, x="thal", palette="magma")
plt.subplot(1, 2, 2)
plt.title(
"Maximum Heart Rate achieved", fontdict={"fontname": "monospace", "fontsize": 15}
)
plt.ylabel("Maximum heart rate")
sns.lineplot(x="age", y="thalach", data=exercise_induced_angina)
plt.show()
# Most of the people who had exercise induced angina showed reversible defect in thallium test which is true as reversible defect is when heart tissue is unable to absorb thallium only under the exercise portion of the test, followed by fixed defect which means under stress and rest and very small percentage of patients got the result of normal
# Cholestrol and Heart Disease
#
# Cholestrol is in mg/dl according to this data and also dataset hence we can check the range and determine heart disease or not
kwargs = dict(s=30)
fig, ax = plt.subplots(nrows=1, figsize=(10, 8))
sns.scatterplot(x="age", y="chol", hue="target", data=df, palette="plasma", **kwargs)
ax.axhspan(0, 200, alpha=0.2, color="green")
ax.axhspan(200, 239, alpha=0.2, color="yellow")
ax.axhspan(239, 600, alpha=0.2, color="red")
plt.show()
# Quite an unusual result!, the only thing which makes sense here is that most of the patients have cholestrol above the ideal range. However, the patients having heart disease and vice versa is quite spread out.
# Few patients having high cholestrol have been diagnosed as not having a heart disease
# The youngest patient admitted for a check on cardiovascular disease is around 28-29 ish and the oldest one is around 77-78 ish
# Age might contribute slightly to the heart diseases (angina), slightly more concentrated to the right. Still it is distributed across implying age doesn't really matter for a cardiovascular diseases to occur, another reason to enjoy the present :p 💃
# Pain
# ## Checking for outliers
df.plot(kind="box", subplots=True, layout=(2, 3))
plt.figure(figsize=(15, 10))
plt.show()
| false | 1 | 3,356 | 0 | 3,380 | 3,356 |
||
69498007
|
from learntools.core import binder
binder.bind(globals())
from learntools.python.ex3 import *
print("Setup complete.")
# # 1.
# Many programming languages have [`sign`](https://en.wikipedia.org/wiki/Sign_function) available as a built-in function. Python doesn't, but we can define our own!
# In the cell below, define a function called `sign` which takes a numerical argument and returns -1 if it's negative, 1 if it's positive, and 0 if it's 0.
# Your code goes here. Define a function called 'sign'
def sign(x):
if x > 0:
return 1
elif x < 0:
return -1
elif x == 0:
return 0
# Check your answer
q1.check()
# q1.solution()
# # 2.
# We've decided to add "logging" to our `to_smash` function from the previous exercise.
def to_smash(total_candies):
"""Return the number of leftover candies that must be smashed after distributing
the given number of candies evenly between 3 friends.
>>> to_smash(91)
1
"""
print("Splitting", total_candies, "candies")
return total_candies % 3
to_smash(91)
# What happens if we call it with `total_candies = 1`?
to_smash(1)
# That isn't great grammar!
# Modify the definition in the cell below to correct the grammar of our print statement. (If there's only one candy, we should use the singular "candy" instead of the plural "candies")
def to_smash(total_candies):
"""Return the number of leftover candies that must be smashed after distributing
the given number of candies evenly between 3 friends.
>>> to_smash(91)
1
"""
if total_candies != 1:
print("Splitting", total_candies, "candies")
if total_candies == 1:
print("Splitting", total_candies, "candie")
return total_candies % 3
to_smash(91)
to_smash(1)
# To get credit for completing this problem, and to see the official answer, run the code cell below.
# Check your answer (Run this code cell to receive credit!)
q2.solution()
# # 3. 🌶️
# In the tutorial, we talked about deciding whether we're prepared for the weather. I said that I'm safe from today's weather if...
# - I have an umbrella...
# - or if the rain isn't too heavy and I have a hood...
# - otherwise, I'm still fine unless it's raining *and* it's a workday
# The function below uses our first attempt at turning this logic into a Python expression. I claimed that there was a bug in that code. Can you find it?
# To prove that `prepared_for_weather` is buggy, come up with a set of inputs where either:
# - the function returns `False` (but should have returned `True`), or
# - the function returned `True` (but should have returned `False`).
# To get credit for completing this question, your code should return a Correct result.
def prepared_for_weather(have_umbrella, rain_level, have_hood, is_workday):
# Don't change this code. Our goal is just to find the bug, not fix it!
return (
(have_umbrella)
or (rain_level < 5 and have_hood)
or not (rain_level > 0 and is_workday)
)
# Change the values of these inputs so they represent a case where prepared_for_weather
# returns the wrong answer.
have_umbrella = False
rain_level = 0.0
have_hood = False
is_workday = False
# Check what the function returns given the current values of the variables above
actual = prepared_for_weather(have_umbrella, rain_level, have_hood, is_workday)
print(actual)
# Check your answer
q3.check()
# q3.hint()
q3.solution()
# # 4.
# The function `is_negative` below is implemented correctly - it returns True if the given number is negative and False otherwise.
# However, it's more verbose than it needs to be. We can actually reduce the number of lines of code in this function by *75%* while keeping the same behaviour.
# See if you can come up with an equivalent body that uses just **one line** of code, and put it in the function `concise_is_negative`. (HINT: you don't even need Python's ternary syntax)
def is_negative(number):
if number < 0:
return True
else:
return False
def concise_is_negative(number):
return number < 0
# Check your answer
q4.check()
q4.hint()
# q4.solution()
# # 5a.
# The boolean variables `ketchup`, `mustard` and `onion` represent whether a customer wants a particular topping on their hot dog. We want to implement a number of boolean functions that correspond to some yes-or-no questions about the customer's order. For example:
def onionless(ketchup, mustard, onion):
"""Return whether the customer doesn't want onions."""
return not onion
def wants_all_toppings(ketchup, mustard, onion):
"""Return whether the customer wants "the works" (all 3 toppings)"""
return ketchup and mustard and onion
# Check your answer
q5.a.check()
q5.a.hint()
# q5.a.solution()
# # 5b.
# For the next function, fill in the body to match the English description in the docstring.
def wants_plain_hotdog(ketchup, mustard, onion):
"""Return whether the customer wants a plain hot dog with no toppings."""
return not ketchup and not mustard and not onion
# Check your answer
q5.b.check()
# q5.b.hint()
# q5.b.solution()
# # 5c.
# You know what to do: for the next function, fill in the body to match the English description in the docstring.
def exactly_one_sauce(ketchup, mustard, onion):
"""Return whether the customer wants either ketchup or mustard, but not both.
(You may be familiar with this operation under the name "exclusive or")
"""
return not (ketchup and mustard) and (ketchup or mustard)
# Check your answer
q5.c.check()
q5.c.hint()
# q5.c.solution()
# # 6. 🌶️
# We’ve seen that calling `bool()` on an integer returns `False` if it’s equal to 0 and `True` otherwise. What happens if we call `int()` on a bool? Try it out in the notebook cell below.
# Can you take advantage of this to write a succinct function that corresponds to the English sentence "does the customer want exactly one topping?"?
def exactly_one_topping(ketchup, mustard, onion):
"""Return whether the customer wants exactly one of the three available toppings
on their hot dog.
"""
return int(ketchup) + int(mustard) + int(onion) == 1
# Check your answer
q6.check()
q6.hint()
# q6.solution()
# # 7. 🌶️ (Optional)
# In this problem we'll be working with a simplified version of [blackjack](https://en.wikipedia.org/wiki/Blackjack) (aka twenty-one). In this version there is one player (who you'll control) and a dealer. Play proceeds as follows:
# - The player is dealt two face-up cards. The dealer is dealt one face-up card.
# - The player may ask to be dealt another card ('hit') as many times as they wish. If the sum of their cards exceeds 21, they lose the round immediately.
# - The dealer then deals additional cards to himself until either:
# - the sum of the dealer's cards exceeds 21, in which case the player wins the round
# - the sum of the dealer's cards is greater than or equal to 17. If the player's total is greater than the dealer's, the player wins. Otherwise, the dealer wins (even in case of a tie).
#
# When calculating the sum of cards, Jack, Queen, and King count for 10. Aces can count as 1 or 11 (when referring to a player's "total" above, we mean the largest total that can be made without exceeding 21. So e.g. A+8 = 19, A+8+8 = 17)
# For this problem, you'll write a function representing the player's decision-making strategy in this game. We've provided a very unintelligent implementation below:
def should_hit(dealer_total, player_total, player_low_aces, player_high_aces):
"""Return True if the player should hit (request another card) given the current game
state, or False if the player should stay.
When calculating a hand's total value, we count aces as "high" (with value 11) if doing so
doesn't bring the total above 21, otherwise we count them as low (with value 1).
For example, if the player's hand is {A, A, A, 7}, we will count it as 11 + 1 + 1 + 7,
and therefore set player_total=20, player_low_aces=2, player_high_aces=1.
"""
return False
# This very conservative agent *always* sticks with the hand of two cards that they're dealt.
# We'll be simulating games between your player agent and our own dealer agent by calling your function.
# Try running the function below to see an example of a simulated game:
q7.simulate_one_game()
# The real test of your agent's mettle is their average win rate over many games. Try calling the function below to simulate 50000 games of blackjack (it may take a couple seconds):
q7.simulate(n_games=50000)
# Our dumb agent that completely ignores the game state still manages to win shockingly often!
# Try adding some more smarts to the `should_hit` function and see how it affects the results.
def should_hit(dealer_total, player_total, player_low_aces, player_high_aces):
"""Return True if the player should hit (request another card) given the current game
state, or False if the player should stay.
When calculating a hand's total value, we count aces as "high" (with value 11) if doing so
doesn't bring the total above 21, otherwise we count them as low (with value 1).
For example, if the player's hand is {A, A, A, 7}, we will count it as 11 + 1 + 1 + 7,
and therefore set player_total=20, player_low_aces=2, player_high_aces=1.
"""
return False
q7.simulate(n_games=50000)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/498/69498007.ipynb
| null | null |
[{"Id": 69498007, "ScriptId": 18976726, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8009223, "CreationDate": "07/31/2021 18:25:38", "VersionNumber": 1.0, "Title": "Exercise: Booleans and Conditionals", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 266.0, "LinesInsertedFromPrevious": 25.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 241.0, "LinesInsertedFromFork": 25.0, "LinesDeletedFromFork": 17.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 241.0, "TotalVotes": 0}]
| null | null | null | null |
from learntools.core import binder
binder.bind(globals())
from learntools.python.ex3 import *
print("Setup complete.")
# # 1.
# Many programming languages have [`sign`](https://en.wikipedia.org/wiki/Sign_function) available as a built-in function. Python doesn't, but we can define our own!
# In the cell below, define a function called `sign` which takes a numerical argument and returns -1 if it's negative, 1 if it's positive, and 0 if it's 0.
# Your code goes here. Define a function called 'sign'
def sign(x):
if x > 0:
return 1
elif x < 0:
return -1
elif x == 0:
return 0
# Check your answer
q1.check()
# q1.solution()
# # 2.
# We've decided to add "logging" to our `to_smash` function from the previous exercise.
def to_smash(total_candies):
"""Return the number of leftover candies that must be smashed after distributing
the given number of candies evenly between 3 friends.
>>> to_smash(91)
1
"""
print("Splitting", total_candies, "candies")
return total_candies % 3
to_smash(91)
# What happens if we call it with `total_candies = 1`?
to_smash(1)
# That isn't great grammar!
# Modify the definition in the cell below to correct the grammar of our print statement. (If there's only one candy, we should use the singular "candy" instead of the plural "candies")
def to_smash(total_candies):
"""Return the number of leftover candies that must be smashed after distributing
the given number of candies evenly between 3 friends.
>>> to_smash(91)
1
"""
if total_candies != 1:
print("Splitting", total_candies, "candies")
if total_candies == 1:
print("Splitting", total_candies, "candie")
return total_candies % 3
to_smash(91)
to_smash(1)
# To get credit for completing this problem, and to see the official answer, run the code cell below.
# Check your answer (Run this code cell to receive credit!)
q2.solution()
# # 3. 🌶️
# In the tutorial, we talked about deciding whether we're prepared for the weather. I said that I'm safe from today's weather if...
# - I have an umbrella...
# - or if the rain isn't too heavy and I have a hood...
# - otherwise, I'm still fine unless it's raining *and* it's a workday
# The function below uses our first attempt at turning this logic into a Python expression. I claimed that there was a bug in that code. Can you find it?
# To prove that `prepared_for_weather` is buggy, come up with a set of inputs where either:
# - the function returns `False` (but should have returned `True`), or
# - the function returned `True` (but should have returned `False`).
# To get credit for completing this question, your code should return a Correct result.
def prepared_for_weather(have_umbrella, rain_level, have_hood, is_workday):
# Don't change this code. Our goal is just to find the bug, not fix it!
return (
(have_umbrella)
or (rain_level < 5 and have_hood)
or not (rain_level > 0 and is_workday)
)
# Change the values of these inputs so they represent a case where prepared_for_weather
# returns the wrong answer.
have_umbrella = False
rain_level = 0.0
have_hood = False
is_workday = False
# Check what the function returns given the current values of the variables above
actual = prepared_for_weather(have_umbrella, rain_level, have_hood, is_workday)
print(actual)
# Check your answer
q3.check()
# q3.hint()
q3.solution()
# # 4.
# The function `is_negative` below is implemented correctly - it returns True if the given number is negative and False otherwise.
# However, it's more verbose than it needs to be. We can actually reduce the number of lines of code in this function by *75%* while keeping the same behaviour.
# See if you can come up with an equivalent body that uses just **one line** of code, and put it in the function `concise_is_negative`. (HINT: you don't even need Python's ternary syntax)
def is_negative(number):
if number < 0:
return True
else:
return False
def concise_is_negative(number):
return number < 0
# Check your answer
q4.check()
q4.hint()
# q4.solution()
# # 5a.
# The boolean variables `ketchup`, `mustard` and `onion` represent whether a customer wants a particular topping on their hot dog. We want to implement a number of boolean functions that correspond to some yes-or-no questions about the customer's order. For example:
def onionless(ketchup, mustard, onion):
"""Return whether the customer doesn't want onions."""
return not onion
def wants_all_toppings(ketchup, mustard, onion):
"""Return whether the customer wants "the works" (all 3 toppings)"""
return ketchup and mustard and onion
# Check your answer
q5.a.check()
q5.a.hint()
# q5.a.solution()
# # 5b.
# For the next function, fill in the body to match the English description in the docstring.
def wants_plain_hotdog(ketchup, mustard, onion):
"""Return whether the customer wants a plain hot dog with no toppings."""
return not ketchup and not mustard and not onion
# Check your answer
q5.b.check()
# q5.b.hint()
# q5.b.solution()
# # 5c.
# You know what to do: for the next function, fill in the body to match the English description in the docstring.
def exactly_one_sauce(ketchup, mustard, onion):
"""Return whether the customer wants either ketchup or mustard, but not both.
(You may be familiar with this operation under the name "exclusive or")
"""
return not (ketchup and mustard) and (ketchup or mustard)
# Check your answer
q5.c.check()
q5.c.hint()
# q5.c.solution()
# # 6. 🌶️
# We’ve seen that calling `bool()` on an integer returns `False` if it’s equal to 0 and `True` otherwise. What happens if we call `int()` on a bool? Try it out in the notebook cell below.
# Can you take advantage of this to write a succinct function that corresponds to the English sentence "does the customer want exactly one topping?"?
def exactly_one_topping(ketchup, mustard, onion):
"""Return whether the customer wants exactly one of the three available toppings
on their hot dog.
"""
return int(ketchup) + int(mustard) + int(onion) == 1
# Check your answer
q6.check()
q6.hint()
# q6.solution()
# # 7. 🌶️ (Optional)
# In this problem we'll be working with a simplified version of [blackjack](https://en.wikipedia.org/wiki/Blackjack) (aka twenty-one). In this version there is one player (who you'll control) and a dealer. Play proceeds as follows:
# - The player is dealt two face-up cards. The dealer is dealt one face-up card.
# - The player may ask to be dealt another card ('hit') as many times as they wish. If the sum of their cards exceeds 21, they lose the round immediately.
# - The dealer then deals additional cards to himself until either:
# - the sum of the dealer's cards exceeds 21, in which case the player wins the round
# - the sum of the dealer's cards is greater than or equal to 17. If the player's total is greater than the dealer's, the player wins. Otherwise, the dealer wins (even in case of a tie).
#
# When calculating the sum of cards, Jack, Queen, and King count for 10. Aces can count as 1 or 11 (when referring to a player's "total" above, we mean the largest total that can be made without exceeding 21. So e.g. A+8 = 19, A+8+8 = 17)
# For this problem, you'll write a function representing the player's decision-making strategy in this game. We've provided a very unintelligent implementation below:
def should_hit(dealer_total, player_total, player_low_aces, player_high_aces):
"""Return True if the player should hit (request another card) given the current game
state, or False if the player should stay.
When calculating a hand's total value, we count aces as "high" (with value 11) if doing so
doesn't bring the total above 21, otherwise we count them as low (with value 1).
For example, if the player's hand is {A, A, A, 7}, we will count it as 11 + 1 + 1 + 7,
and therefore set player_total=20, player_low_aces=2, player_high_aces=1.
"""
return False
# This very conservative agent *always* sticks with the hand of two cards that they're dealt.
# We'll be simulating games between your player agent and our own dealer agent by calling your function.
# Try running the function below to see an example of a simulated game:
q7.simulate_one_game()
# The real test of your agent's mettle is their average win rate over many games. Try calling the function below to simulate 50000 games of blackjack (it may take a couple seconds):
q7.simulate(n_games=50000)
# Our dumb agent that completely ignores the game state still manages to win shockingly often!
# Try adding some more smarts to the `should_hit` function and see how it affects the results.
def should_hit(dealer_total, player_total, player_low_aces, player_high_aces):
"""Return True if the player should hit (request another card) given the current game
state, or False if the player should stay.
When calculating a hand's total value, we count aces as "high" (with value 11) if doing so
doesn't bring the total above 21, otherwise we count them as low (with value 1).
For example, if the player's hand is {A, A, A, 7}, we will count it as 11 + 1 + 1 + 7,
and therefore set player_total=20, player_low_aces=2, player_high_aces=1.
"""
return False
q7.simulate(n_games=50000)
| false | 0 | 2,728 | 0 | 2,728 | 2,728 |
||
69498319
|
# ## House Price Predictor using Different Models
# ## Import Packages
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import sklearn
from sklearn import metrics
import tensorflow as tf
from sklearn.linear_model import LinearRegression
# ## Import Datasets
train = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
)
test = pd.read_csv("/kaggle/input/house-prices-advanced-regression-techniques/test.csv")
# ## Common Functions
# **Evaluation Function**
def evaluate(model, x_val, y_val):
y_pred = model.predict(x_val)
r2 = metrics.r2_score(y_val, y_pred)
mse = metrics.mean_squared_error(y_val, y_pred)
mae = metrics.mean_absolute_error(y_val, y_pred)
msle = metrics.mean_squared_log_error(y_val, y_pred)
mape = np.mean(
tf.keras.metrics.mean_absolute_percentage_error(y_val, y_pred).numpy()
)
rmse = np.sqrt(mse)
print("R2 Score:", r2)
print("MSE:", mse)
print("MAE:", mae)
print("MSLE:", msle)
print("MAPE", mape)
print("MAPE", mape)
print("RMSE:", rmse)
return {"r2": r2, "mse": mse, "mae": mae, "msle": msle, "mape": mape, "rmse": rmse}
# ## Exploratory Data Analysis
# **First 5 rows**
train.head()
# **Its shape**
train.shape
# **Statistic infos**
train.info()
train.describe()
# **Correlation scores**
train.corr()
# **Factors that impact house price most**
train.corr()["SalePrice"].sort_values(ascending=False)
# ## Data Cleaning
# **Features that contains missing values**
null_counts = train.isnull().sum()
null_counts[null_counts > 0]
null_columns = list(pd.DataFrame(null_counts[null_counts > 0]).index)
# **Type of features that has missing values**
train[null_columns].dtypes
# **Solve Missing values**
for column in null_columns:
if train[column].dtype == object:
train[column] = train[[column]].replace(np.NAN, train[column].mode()[0])
else:
train[column] = train[column].replace(np.NAN, train[column].mean())
# ### Do the same for Test data set
null_counts = test.isnull().sum()
null_counts[null_counts > 0]
null_columns = list(pd.DataFrame(null_counts[null_counts > 0]).index)
for column in null_columns:
if test[column].dtype == object:
test[column] = test[[column]].replace(np.NAN, test[column].mode()[0])
else:
test[column] = test[column].replace(np.NAN, test[column].mean())
# ## Now the data are all none nulls
train.info()
train_test_dummied = pd.get_dummies(pd.concat([train, test]))
train_test_dummied.head()
train_dummied = train_test_dummied.iloc[0 : len(train)]
test_dummied = train_test_dummied.iloc[len(train) :]
test_dummied.pop("SalePrice")
train_dummied.head()
test_dummied.head()
# ## Train Validation Split
from sklearn.model_selection import train_test_split
train_dummied, val_dummied = train_test_split(
train_dummied, test_size=0.1, random_state=888
)
train_dummied.corr()
correlated_scores = train_dummied.corr()["SalePrice"]
correlated_scores = correlated_scores[correlated_scores.abs() > 0.05]
correlated_indices = list(correlated_scores.index)
correlated_indices.remove("SalePrice")
y_train_dummied = train_dummied.pop("SalePrice")
x_train_dummied = train_dummied
y_val_dummied = val_dummied.pop("SalePrice")
x_val_dummied = val_dummied
# ## Model Development and Evaluation
# ### Train Linear Regression Model with all features
linear_regression_1 = LinearRegression()
linear_regression_1.fit(x_train_dummied, y_train_dummied)
linear_regression_1_results = evaluate(
linear_regression_1, x_val_dummied, y_val_dummied
)
# ### Train Linear Regression Model with features that's correlated to house price
price_factors = train.corr()["SalePrice"]
linear_regression_2 = LinearRegression()
linear_regression_2.fit(x_train_dummied[correlated_indices], y_train_dummied)
linear_regression_2_results = evaluate(
linear_regression_2, x_val_dummied[correlated_indices], y_val_dummied
)
# ### Train Ridge Model with all features
from sklearn.linear_model import Ridge
ridge1 = Ridge()
ridge1.fit(x_train_dummied, y_train_dummied)
ridge1_results = evaluate(ridge1, x_val_dummied, y_val_dummied)
# Train Ridge Model with correlated features
from sklearn.linear_model import Ridge
ridge2 = Ridge()
ridge2.fit(x_train_dummied[correlated_indices], y_train_dummied)
ridge2_results = evaluate(ridge2, x_val_dummied[correlated_indices], y_val_dummied)
# ## Train Deep Neural Network Model with all features¶
neural_network_model_1 = tf.keras.Sequential(
[
tf.keras.Input(shape=(x_train_dummied.shape[1])),
tf.keras.layers.Dense(
32, activation="relu", kernel_regularizer=tf.keras.regularizers.l1_l2()
),
tf.keras.layers.Dropout(0.3),
tf.keras.layers.Dense(
32, activation="relu", kernel_regularizer=tf.keras.regularizers.l1_l2()
),
tf.keras.layers.Dense(1),
]
)
early_stop = tf.keras.callbacks.EarlyStopping(patience=20)
checkpoint_path = "neural_network_model_1.h5"
checkpoint = tf.keras.callbacks.ModelCheckpoint(checkpoint_path, save_best_only=True)
neural_network_model_1.compile(
loss="mse",
optimizer="adam",
metrics=["mse", "mean_squared_logarithmic_error", "mae", "mape"],
)
history = neural_network_model_1.fit(
x_train_dummied,
y_train_dummied,
epochs=100,
validation_data=(x_val_dummied, y_val_dummied),
callbacks=[early_stop, checkpoint],
)
pd.DataFrame(history.history, columns=["loss", "val_loss"]).plot()
pd.DataFrame(history.history, columns=["mae", "val_mae"]).plot()
pd.DataFrame(history.history, columns=["mape", "val_mape"]).plot()
# ### Train Deep Neural Network Model with with features that's correlated to house price
x_train_subset = x_train_dummied[correlated_indices]
x_val_subset = x_val_dummied[correlated_indices]
neural_network_model_2 = tf.keras.Sequential(
[
tf.keras.Input(shape=(x_train_subset.shape[1])),
tf.keras.layers.Dense(
32, activation="relu", kernel_regularizer=tf.keras.regularizers.l1_l2()
),
tf.keras.layers.Dense(
32, activation="relu", kernel_regularizer=tf.keras.regularizers.l1_l2()
),
tf.keras.layers.Dense(1),
]
)
early_stop = tf.keras.callbacks.EarlyStopping(patience=20)
neural_network_model_2_checkpoint = "neural_network_model_2.h5"
checkpoint = tf.keras.callbacks.ModelCheckpoint(
neural_network_model_2_checkpoint, save_best_only=True
)
neural_network_model_2.compile(
loss="mse", optimizer="adam", metrics=["mse", "mae", "mape"]
)
history = neural_network_model_2.fit(
x_train_subset,
y_train_dummied,
epochs=100,
validation_data=(x_val_subset, y_val_dummied),
callbacks=[early_stop, checkpoint],
)
neural_network_model_2.load_weights
# ## Predict with best Model
# Currently Ridge Model seems to be best Model. So use it to calculate results:
SalePrice = ridge.predict(test_dummied)
submission = pd.DataFrame({"Id": test_dummied["Id"], "SalePrice": SalePrice})
submission.head()
submission.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/498/69498319.ipynb
| null | null |
[{"Id": 69498319, "ScriptId": 18977369, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4562457, "CreationDate": "07/31/2021 18:30:35", "VersionNumber": 1.0, "Title": "notebookab041316c5", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 219.0, "LinesInsertedFromPrevious": 219.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# ## House Price Predictor using Different Models
# ## Import Packages
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import sklearn
from sklearn import metrics
import tensorflow as tf
from sklearn.linear_model import LinearRegression
# ## Import Datasets
train = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
)
test = pd.read_csv("/kaggle/input/house-prices-advanced-regression-techniques/test.csv")
# ## Common Functions
# **Evaluation Function**
def evaluate(model, x_val, y_val):
y_pred = model.predict(x_val)
r2 = metrics.r2_score(y_val, y_pred)
mse = metrics.mean_squared_error(y_val, y_pred)
mae = metrics.mean_absolute_error(y_val, y_pred)
msle = metrics.mean_squared_log_error(y_val, y_pred)
mape = np.mean(
tf.keras.metrics.mean_absolute_percentage_error(y_val, y_pred).numpy()
)
rmse = np.sqrt(mse)
print("R2 Score:", r2)
print("MSE:", mse)
print("MAE:", mae)
print("MSLE:", msle)
print("MAPE", mape)
print("MAPE", mape)
print("RMSE:", rmse)
return {"r2": r2, "mse": mse, "mae": mae, "msle": msle, "mape": mape, "rmse": rmse}
# ## Exploratory Data Analysis
# **First 5 rows**
train.head()
# **Its shape**
train.shape
# **Statistic infos**
train.info()
train.describe()
# **Correlation scores**
train.corr()
# **Factors that impact house price most**
train.corr()["SalePrice"].sort_values(ascending=False)
# ## Data Cleaning
# **Features that contains missing values**
null_counts = train.isnull().sum()
null_counts[null_counts > 0]
null_columns = list(pd.DataFrame(null_counts[null_counts > 0]).index)
# **Type of features that has missing values**
train[null_columns].dtypes
# **Solve Missing values**
for column in null_columns:
if train[column].dtype == object:
train[column] = train[[column]].replace(np.NAN, train[column].mode()[0])
else:
train[column] = train[column].replace(np.NAN, train[column].mean())
# ### Do the same for Test data set
null_counts = test.isnull().sum()
null_counts[null_counts > 0]
null_columns = list(pd.DataFrame(null_counts[null_counts > 0]).index)
for column in null_columns:
if test[column].dtype == object:
test[column] = test[[column]].replace(np.NAN, test[column].mode()[0])
else:
test[column] = test[column].replace(np.NAN, test[column].mean())
# ## Now the data are all none nulls
train.info()
train_test_dummied = pd.get_dummies(pd.concat([train, test]))
train_test_dummied.head()
train_dummied = train_test_dummied.iloc[0 : len(train)]
test_dummied = train_test_dummied.iloc[len(train) :]
test_dummied.pop("SalePrice")
train_dummied.head()
test_dummied.head()
# ## Train Validation Split
from sklearn.model_selection import train_test_split
train_dummied, val_dummied = train_test_split(
train_dummied, test_size=0.1, random_state=888
)
train_dummied.corr()
correlated_scores = train_dummied.corr()["SalePrice"]
correlated_scores = correlated_scores[correlated_scores.abs() > 0.05]
correlated_indices = list(correlated_scores.index)
correlated_indices.remove("SalePrice")
y_train_dummied = train_dummied.pop("SalePrice")
x_train_dummied = train_dummied
y_val_dummied = val_dummied.pop("SalePrice")
x_val_dummied = val_dummied
# ## Model Development and Evaluation
# ### Train Linear Regression Model with all features
linear_regression_1 = LinearRegression()
linear_regression_1.fit(x_train_dummied, y_train_dummied)
linear_regression_1_results = evaluate(
linear_regression_1, x_val_dummied, y_val_dummied
)
# ### Train Linear Regression Model with features that's correlated to house price
price_factors = train.corr()["SalePrice"]
linear_regression_2 = LinearRegression()
linear_regression_2.fit(x_train_dummied[correlated_indices], y_train_dummied)
linear_regression_2_results = evaluate(
linear_regression_2, x_val_dummied[correlated_indices], y_val_dummied
)
# ### Train Ridge Model with all features
from sklearn.linear_model import Ridge
ridge1 = Ridge()
ridge1.fit(x_train_dummied, y_train_dummied)
ridge1_results = evaluate(ridge1, x_val_dummied, y_val_dummied)
# Train Ridge Model with correlated features
from sklearn.linear_model import Ridge
ridge2 = Ridge()
ridge2.fit(x_train_dummied[correlated_indices], y_train_dummied)
ridge2_results = evaluate(ridge2, x_val_dummied[correlated_indices], y_val_dummied)
# ## Train Deep Neural Network Model with all features¶
neural_network_model_1 = tf.keras.Sequential(
[
tf.keras.Input(shape=(x_train_dummied.shape[1])),
tf.keras.layers.Dense(
32, activation="relu", kernel_regularizer=tf.keras.regularizers.l1_l2()
),
tf.keras.layers.Dropout(0.3),
tf.keras.layers.Dense(
32, activation="relu", kernel_regularizer=tf.keras.regularizers.l1_l2()
),
tf.keras.layers.Dense(1),
]
)
early_stop = tf.keras.callbacks.EarlyStopping(patience=20)
checkpoint_path = "neural_network_model_1.h5"
checkpoint = tf.keras.callbacks.ModelCheckpoint(checkpoint_path, save_best_only=True)
neural_network_model_1.compile(
loss="mse",
optimizer="adam",
metrics=["mse", "mean_squared_logarithmic_error", "mae", "mape"],
)
history = neural_network_model_1.fit(
x_train_dummied,
y_train_dummied,
epochs=100,
validation_data=(x_val_dummied, y_val_dummied),
callbacks=[early_stop, checkpoint],
)
pd.DataFrame(history.history, columns=["loss", "val_loss"]).plot()
pd.DataFrame(history.history, columns=["mae", "val_mae"]).plot()
pd.DataFrame(history.history, columns=["mape", "val_mape"]).plot()
# ### Train Deep Neural Network Model with with features that's correlated to house price
x_train_subset = x_train_dummied[correlated_indices]
x_val_subset = x_val_dummied[correlated_indices]
neural_network_model_2 = tf.keras.Sequential(
[
tf.keras.Input(shape=(x_train_subset.shape[1])),
tf.keras.layers.Dense(
32, activation="relu", kernel_regularizer=tf.keras.regularizers.l1_l2()
),
tf.keras.layers.Dense(
32, activation="relu", kernel_regularizer=tf.keras.regularizers.l1_l2()
),
tf.keras.layers.Dense(1),
]
)
early_stop = tf.keras.callbacks.EarlyStopping(patience=20)
neural_network_model_2_checkpoint = "neural_network_model_2.h5"
checkpoint = tf.keras.callbacks.ModelCheckpoint(
neural_network_model_2_checkpoint, save_best_only=True
)
neural_network_model_2.compile(
loss="mse", optimizer="adam", metrics=["mse", "mae", "mape"]
)
history = neural_network_model_2.fit(
x_train_subset,
y_train_dummied,
epochs=100,
validation_data=(x_val_subset, y_val_dummied),
callbacks=[early_stop, checkpoint],
)
neural_network_model_2.load_weights
# ## Predict with best Model
# Currently Ridge Model seems to be best Model. So use it to calculate results:
SalePrice = ridge.predict(test_dummied)
submission = pd.DataFrame({"Id": test_dummied["Id"], "SalePrice": SalePrice})
submission.head()
submission.to_csv("submission.csv", index=False)
| false | 0 | 2,341 | 0 | 2,341 | 2,341 |
||
69498684
|
<jupyter_start><jupyter_text>Phishing
Kaggle dataset identifier: phishing
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np
from sklearn import *
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import accuracy_score
# load the data from 'https://archive.ics.uci.edu/ml/datasets/phishing+websites'
training_data = np.genfromtxt(
"../input/phishing/phishing.csv", delimiter=",", dtype=np.int32
)
training_data
# Identify the inputs (all of the attributes, except for the last one) and the outputs (the last attribute):
inputs = training_data[:, :-1]
outputs = training_data[:, -1]
print(inputs.shape)
print(outputs.shape)
inputs
outputs
# Added by Luiz
# To improve the estimators' accuracy scores, we are going to use the
# sklearn.feature_selection module. This module is used in feature selection or
# dimensionality reduction in the dataset.
# To compute the features' importance, in our case, we are going to use tree-based feature
# selection. Load the sklearn.feature_selection module:
import sklearn
from sklearn.feature_selection import SelectFromModel
from sklearn.ensemble import ExtraTreesClassifier
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from sklearn.model_selection import cross_validate
import matplotlib.pyplot as plt
from collections import Counter
# Added by Luiz
# Obtaining the relevant feature set using ExtraTreesClassifier
featselect = sklearn.ensemble.ExtraTreesClassifier().fit(inputs, outputs)
model = SelectFromModel(featselect, prefit=True)
inputs_new = model.transform(inputs)
print(inputs.shape)
print(inputs_new.shape)
# Added by Luiz
# Dataset normalization
min_max_scaler = preprocessing.MinMaxScaler()
inputs_new_scaled = min_max_scaler.fit_transform(inputs_new)
data_new_features = pd.DataFrame(inputs_new_scaled)
# After normalization
data_new_features.describe()
# Added by Luiz
# Outliers
num_cols = data_new_features.columns
plt.figure(figsize=(18, 9))
data_new_features[num_cols].boxplot()
plt.title("Numerical variables in dataset", fontsize=20)
plt.show()
# Outliers exist only for the first column. Although it seems reasonable to remove those samples, if we remove those that column would be useless for classification, since it would have only a single value, which is zero. So it will remain as-is.
# Added by Luiz
print(
f"Percentage of outlier samples (value=1) for first column: {data_new_features[data_new_features[0]==1].shape[0]/data_new_features.shape[0]*100:.2f}%"
)
print(
f"Percentage of standard samples (value=0) for first column: {data_new_features[data_new_features[0]==0].shape[0]/data_new_features.shape[0]*100:.2f}%"
)
# Added by Luiz
new_inputs = data_new_features.values
new_inputs
# Added by Luiz
print(f"Current size of the inputs: {new_inputs.shape}")
print(f"Current size of the outputs: {outputs.shape}")
# Added by Luiz
# We have roughly the same amount of phishing and non-phishing samples
freq = Counter(outputs)
fishing_perc = freq[1] / (freq[-1] + freq[1])
non_fishing_perc = freq[-1] / (freq[-1] + freq[1])
print(f"Percentage of phishing is: {fishing_perc*100:.2f}%")
print(f"Percentage of non-phishing is: {non_fishing_perc*100:.2f}%")
# Added by Luiz
from imblearn.under_sampling import RandomUnderSampler
from collections import Counter
# Using undersampling to balance the dataset
under_sampler = RandomUnderSampler()
X_res, y_res = under_sampler.fit_resample(new_inputs, outputs)
# Added by Luiz
# We have roughly the same amount of phishing and non-phishing samples
freq = Counter(y_res)
fishing_perc = freq[1] / (freq[-1] + freq[1])
non_fishing_perc = freq[-1] / (freq[-1] + freq[1])
print(f"Percentage of phishing is: {fishing_perc*100:.2f}%")
print(f"Percentage of non-phishing is: {non_fishing_perc*100:.2f}%")
# dividing the dataset into training and testing:
x_train, x_test, y_train, y_test = sklearn.model_selection.train_test_split(
X_res, y_res, test_size=0.2
)
print(x_train.shape)
print(x_test.shape)
print(y_train.shape)
print(y_test.shape)
# Create the scikit-learn logistic regression classifier with standard parameters
classifier1 = LogisticRegression()
# Create the scikit-learn Decision Tree classifier with standard parameters.
classifier2 = DecisionTreeClassifier()
# Train the classifiers:
classifier1.fit(x_train, y_train)
classifier2.fit(x_train, y_train)
# Make predictions:
predictions1 = classifier1.predict(x_test)
predictions2 = classifier2.predict(x_test)
# print out the accuracy of our phishing detector models:
accuracy1 = 100.0 * accuracy_score(y_test, predictions1)
accuracy2 = 100.0 * accuracy_score(y_test, predictions2)
print("The accuracy of your Logistic Regression on testing data is: " + str(accuracy1))
print("The accuracy of your Decision Tree on testing data is: " + str(accuracy2))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/498/69498684.ipynb
|
phishing
|
dscclass
|
[{"Id": 69498684, "ScriptId": 18732022, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7499525, "CreationDate": "07/31/2021 18:36:46", "VersionNumber": 2.0, "Title": "Task #2 - Phishing-Exploratory", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 163.0, "LinesInsertedFromPrevious": 107.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 56.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92743249, "KernelVersionId": 69498684, "SourceDatasetVersionId": 2422455}]
|
[{"Id": 2422455, "DatasetId": 1465878, "DatasourceVersionId": 2464643, "CreatorUserId": 7458594, "LicenseName": "Unknown", "CreationDate": "07/13/2021 18:54:06", "VersionNumber": 1.0, "Title": "Phishing", "Slug": "phishing", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1465878, "CreatorUserId": 7458594, "OwnerUserId": 7458594.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2422455.0, "CurrentDatasourceVersionId": 2464643.0, "ForumId": 1485494, "Type": 2, "CreationDate": "07/13/2021 18:54:06", "LastActivityDate": "07/13/2021", "TotalViews": 1293, "TotalDownloads": 42, "TotalVotes": 3, "TotalKernels": 8}]
|
[{"Id": 7458594, "UserName": "dscclass", "DisplayName": "DSC-class", "RegisterDate": "05/19/2021", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np
from sklearn import *
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import accuracy_score
# load the data from 'https://archive.ics.uci.edu/ml/datasets/phishing+websites'
training_data = np.genfromtxt(
"../input/phishing/phishing.csv", delimiter=",", dtype=np.int32
)
training_data
# Identify the inputs (all of the attributes, except for the last one) and the outputs (the last attribute):
inputs = training_data[:, :-1]
outputs = training_data[:, -1]
print(inputs.shape)
print(outputs.shape)
inputs
outputs
# Added by Luiz
# To improve the estimators' accuracy scores, we are going to use the
# sklearn.feature_selection module. This module is used in feature selection or
# dimensionality reduction in the dataset.
# To compute the features' importance, in our case, we are going to use tree-based feature
# selection. Load the sklearn.feature_selection module:
import sklearn
from sklearn.feature_selection import SelectFromModel
from sklearn.ensemble import ExtraTreesClassifier
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from sklearn.model_selection import cross_validate
import matplotlib.pyplot as plt
from collections import Counter
# Added by Luiz
# Obtaining the relevant feature set using ExtraTreesClassifier
featselect = sklearn.ensemble.ExtraTreesClassifier().fit(inputs, outputs)
model = SelectFromModel(featselect, prefit=True)
inputs_new = model.transform(inputs)
print(inputs.shape)
print(inputs_new.shape)
# Added by Luiz
# Dataset normalization
min_max_scaler = preprocessing.MinMaxScaler()
inputs_new_scaled = min_max_scaler.fit_transform(inputs_new)
data_new_features = pd.DataFrame(inputs_new_scaled)
# After normalization
data_new_features.describe()
# Added by Luiz
# Outliers
num_cols = data_new_features.columns
plt.figure(figsize=(18, 9))
data_new_features[num_cols].boxplot()
plt.title("Numerical variables in dataset", fontsize=20)
plt.show()
# Outliers exist only for the first column. Although it seems reasonable to remove those samples, if we remove those that column would be useless for classification, since it would have only a single value, which is zero. So it will remain as-is.
# Added by Luiz
print(
f"Percentage of outlier samples (value=1) for first column: {data_new_features[data_new_features[0]==1].shape[0]/data_new_features.shape[0]*100:.2f}%"
)
print(
f"Percentage of standard samples (value=0) for first column: {data_new_features[data_new_features[0]==0].shape[0]/data_new_features.shape[0]*100:.2f}%"
)
# Added by Luiz
new_inputs = data_new_features.values
new_inputs
# Added by Luiz
print(f"Current size of the inputs: {new_inputs.shape}")
print(f"Current size of the outputs: {outputs.shape}")
# Added by Luiz
# We have roughly the same amount of phishing and non-phishing samples
freq = Counter(outputs)
fishing_perc = freq[1] / (freq[-1] + freq[1])
non_fishing_perc = freq[-1] / (freq[-1] + freq[1])
print(f"Percentage of phishing is: {fishing_perc*100:.2f}%")
print(f"Percentage of non-phishing is: {non_fishing_perc*100:.2f}%")
# Added by Luiz
from imblearn.under_sampling import RandomUnderSampler
from collections import Counter
# Using undersampling to balance the dataset
under_sampler = RandomUnderSampler()
X_res, y_res = under_sampler.fit_resample(new_inputs, outputs)
# Added by Luiz
# We have roughly the same amount of phishing and non-phishing samples
freq = Counter(y_res)
fishing_perc = freq[1] / (freq[-1] + freq[1])
non_fishing_perc = freq[-1] / (freq[-1] + freq[1])
print(f"Percentage of phishing is: {fishing_perc*100:.2f}%")
print(f"Percentage of non-phishing is: {non_fishing_perc*100:.2f}%")
# dividing the dataset into training and testing:
x_train, x_test, y_train, y_test = sklearn.model_selection.train_test_split(
X_res, y_res, test_size=0.2
)
print(x_train.shape)
print(x_test.shape)
print(y_train.shape)
print(y_test.shape)
# Create the scikit-learn logistic regression classifier with standard parameters
classifier1 = LogisticRegression()
# Create the scikit-learn Decision Tree classifier with standard parameters.
classifier2 = DecisionTreeClassifier()
# Train the classifiers:
classifier1.fit(x_train, y_train)
classifier2.fit(x_train, y_train)
# Make predictions:
predictions1 = classifier1.predict(x_test)
predictions2 = classifier2.predict(x_test)
# print out the accuracy of our phishing detector models:
accuracy1 = 100.0 * accuracy_score(y_test, predictions1)
accuracy2 = 100.0 * accuracy_score(y_test, predictions2)
print("The accuracy of your Logistic Regression on testing data is: " + str(accuracy1))
print("The accuracy of your Decision Tree on testing data is: " + str(accuracy2))
| false | 0 | 1,584 | 0 | 1,602 | 1,584 |
||
69851470
|
# ## Importing necessary Libraries
import numpy as np
import pandas as pd
import os
import tensorflow as tf
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import (
Conv2D,
MaxPooling2D,
Flatten,
Dense,
Dropout,
BatchNormalization,
)
import pandas as pd
import seaborn as sns
# ## Loading the data
train_pd = pd.read_csv("/kaggle/input/digit-recognizer/train.csv")
test_pd = pd.read_csv("/kaggle/input/digit-recognizer/test.csv")
train = np.array(train_pd)
test = np.array(test_pd)
train_images = train[:, 1:].reshape((train.shape[0], 28, 28, 1))
train_labels = train[:, 0].astype(np.uint8)
test_images = test.reshape((test.shape[0], 28, 28, 1))
# ## Exploratory Data Analysis
# **Label Distribution**
train_pd["label"].plot(kind="hist")
# Pixel Distribution
# **Mean image for different labels**
# Calcuate mean image for different labels, they look exactly the label they belong to.
for label in range(10):
plt.imshow(np.mean(train_images[train_labels == label], axis=0))
plt.show()
# ## Model Developmnet
def get_model():
model = Sequential(
[
Conv2D(32, (3, 3), activation="relu", input_shape=(28, 28, 1)),
MaxPooling2D(2, 2),
BatchNormalization(),
Conv2D(32, (3, 3), activation="relu", padding="same"),
MaxPooling2D(2, 2),
BatchNormalization(),
Conv2D(64, (3, 3), activation="relu", padding="same"),
MaxPooling2D(2, 2),
BatchNormalization(),
Flatten(),
Dense(32, activation="relu", kernel_regularizer=tf.keras.regularizers.l2()),
Dropout(0.3),
Dense(10, activation="softmax"),
]
)
model.compile(
optimizer="adam", loss="sparse_categorical_crossentropy", metrics=["accuracy"]
)
return model
model = get_model()
# ## Displaying structure of the model
# We can use summary method to show structure of the model.
model.summary()
# ## Creating Model Checkpoint
# Create a Model Checkpoint to save weights that has the best validation accuracy. So we can use it to generate the best result.
checkpoint_filepath = "best_checkpoint"
model_checkpoint = tf.keras.callbacks.ModelCheckpoint(
filepath=checkpoint_filepath,
save_weights_only=True,
monitor="val_accuracy",
mode="max",
save_best_only=True,
)
# ## Creating Early Stopping Callback
# In order to save time, we want to stop the training when the model stops improving. In this senario. We can use Earaly Stoppping
early_stopping = tf.keras.callbacks.EarlyStopping(monitor="val_accuracy", patience=4)
# ## Train Validation Split
from sklearn.model_selection import train_test_split
train_images, val_images, train_labels, val_labels = train_test_split(
train_images, train_labels
)
# ## Training the Model
# Now we train the model with the training dataset for 50 epochs. We set the validation_split parameter to 0.15. We pass the callbacks we created before.
history = model.fit(
train_images,
train_labels,
epochs=100,
validation_data=(val_images, val_labels),
callbacks=[model_checkpoint, early_stopping],
)
# ## Plot the learning curve
import matplotlib.pyplot as plt
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("Accuracy vs. epochs")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.show()
import matplotlib.pyplot as plt
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Loss vs. epochs")
plt.ylabel("Loss")
plt.xlabel("Epoch")
plt.legend()
plt.show()
# ## Creating the Model again with best weight
model = get_model()
model.load_weights(checkpoint_filepath)
# ## Model Evaluation
import sklearn
val_labels_pred = np.argmax(model.predict(val_images), axis=-1)
(val_labels_pred == val_labels).mean()
# **Confusion Matrix**
cm = sklearn.metrics.confusion_matrix(val_labels, val_labels_pred)
cm
sns.heatmap(cm)
# **Classification report**
print(sklearn.metrics.classification_report(val_labels, val_labels_pred))
# **Show images not correctly classfied**
# As we can see these images are actually hard to reconized by human.
val_images[val_labels != val_labels_pred].shape
miss_classified_images = val_images[val_labels != val_labels_pred]
miss_classified_labels = val_labels[val_labels != val_labels_pred]
for i in range(miss_classified_images.shape[0]):
plt.imshow(miss_classified_images[i])
plt.title(str(miss_classified_labels[i]))
plt.show()
# ## Predicting the data
test_labels = np.argmax(model.predict(test_images), axis=-1)
print(test_labels.shape)
pd.DataFrame(test_labels).hist()
# Now we print first 100 item of test_labels.
print(test_labels[:100])
# ## Sumbit the data
image_ids = np.arange(1, test_labels.shape[0] + 1)
result = np.concatenate(
(
image_ids.reshape(image_ids.shape[0], 1),
test_labels.reshape(test_labels.shape[0], 1),
),
axis=1,
)
df = pd.DataFrame(result, columns=["ImageId", "Label"], dtype="int")
df.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/851/69851470.ipynb
| null | null |
[{"Id": 69851470, "ScriptId": 15596638, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4562457, "CreationDate": "08/03/2021 15:33:36", "VersionNumber": 12.0, "Title": "MNIST EDA and Model Development", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 168.0, "LinesInsertedFromPrevious": 54.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 114.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# ## Importing necessary Libraries
import numpy as np
import pandas as pd
import os
import tensorflow as tf
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import (
Conv2D,
MaxPooling2D,
Flatten,
Dense,
Dropout,
BatchNormalization,
)
import pandas as pd
import seaborn as sns
# ## Loading the data
train_pd = pd.read_csv("/kaggle/input/digit-recognizer/train.csv")
test_pd = pd.read_csv("/kaggle/input/digit-recognizer/test.csv")
train = np.array(train_pd)
test = np.array(test_pd)
train_images = train[:, 1:].reshape((train.shape[0], 28, 28, 1))
train_labels = train[:, 0].astype(np.uint8)
test_images = test.reshape((test.shape[0], 28, 28, 1))
# ## Exploratory Data Analysis
# **Label Distribution**
train_pd["label"].plot(kind="hist")
# Pixel Distribution
# **Mean image for different labels**
# Calcuate mean image for different labels, they look exactly the label they belong to.
for label in range(10):
plt.imshow(np.mean(train_images[train_labels == label], axis=0))
plt.show()
# ## Model Developmnet
def get_model():
model = Sequential(
[
Conv2D(32, (3, 3), activation="relu", input_shape=(28, 28, 1)),
MaxPooling2D(2, 2),
BatchNormalization(),
Conv2D(32, (3, 3), activation="relu", padding="same"),
MaxPooling2D(2, 2),
BatchNormalization(),
Conv2D(64, (3, 3), activation="relu", padding="same"),
MaxPooling2D(2, 2),
BatchNormalization(),
Flatten(),
Dense(32, activation="relu", kernel_regularizer=tf.keras.regularizers.l2()),
Dropout(0.3),
Dense(10, activation="softmax"),
]
)
model.compile(
optimizer="adam", loss="sparse_categorical_crossentropy", metrics=["accuracy"]
)
return model
model = get_model()
# ## Displaying structure of the model
# We can use summary method to show structure of the model.
model.summary()
# ## Creating Model Checkpoint
# Create a Model Checkpoint to save weights that has the best validation accuracy. So we can use it to generate the best result.
checkpoint_filepath = "best_checkpoint"
model_checkpoint = tf.keras.callbacks.ModelCheckpoint(
filepath=checkpoint_filepath,
save_weights_only=True,
monitor="val_accuracy",
mode="max",
save_best_only=True,
)
# ## Creating Early Stopping Callback
# In order to save time, we want to stop the training when the model stops improving. In this senario. We can use Earaly Stoppping
early_stopping = tf.keras.callbacks.EarlyStopping(monitor="val_accuracy", patience=4)
# ## Train Validation Split
from sklearn.model_selection import train_test_split
train_images, val_images, train_labels, val_labels = train_test_split(
train_images, train_labels
)
# ## Training the Model
# Now we train the model with the training dataset for 50 epochs. We set the validation_split parameter to 0.15. We pass the callbacks we created before.
history = model.fit(
train_images,
train_labels,
epochs=100,
validation_data=(val_images, val_labels),
callbacks=[model_checkpoint, early_stopping],
)
# ## Plot the learning curve
import matplotlib.pyplot as plt
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("Accuracy vs. epochs")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.show()
import matplotlib.pyplot as plt
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Loss vs. epochs")
plt.ylabel("Loss")
plt.xlabel("Epoch")
plt.legend()
plt.show()
# ## Creating the Model again with best weight
model = get_model()
model.load_weights(checkpoint_filepath)
# ## Model Evaluation
import sklearn
val_labels_pred = np.argmax(model.predict(val_images), axis=-1)
(val_labels_pred == val_labels).mean()
# **Confusion Matrix**
cm = sklearn.metrics.confusion_matrix(val_labels, val_labels_pred)
cm
sns.heatmap(cm)
# **Classification report**
print(sklearn.metrics.classification_report(val_labels, val_labels_pred))
# **Show images not correctly classfied**
# As we can see these images are actually hard to reconized by human.
val_images[val_labels != val_labels_pred].shape
miss_classified_images = val_images[val_labels != val_labels_pred]
miss_classified_labels = val_labels[val_labels != val_labels_pred]
for i in range(miss_classified_images.shape[0]):
plt.imshow(miss_classified_images[i])
plt.title(str(miss_classified_labels[i]))
plt.show()
# ## Predicting the data
test_labels = np.argmax(model.predict(test_images), axis=-1)
print(test_labels.shape)
pd.DataFrame(test_labels).hist()
# Now we print first 100 item of test_labels.
print(test_labels[:100])
# ## Sumbit the data
image_ids = np.arange(1, test_labels.shape[0] + 1)
result = np.concatenate(
(
image_ids.reshape(image_ids.shape[0], 1),
test_labels.reshape(test_labels.shape[0], 1),
),
axis=1,
)
df = pd.DataFrame(result, columns=["ImageId", "Label"], dtype="int")
df.to_csv("submission.csv", index=False)
| false | 0 | 1,523 | 0 | 1,523 | 1,523 |
||
69851334
|
# # Recurrent Neural Network
# ## Part 1 - Data Preprocessing
# ### Importing the libraries
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
# ### Importing the training set
# We're only grabbing the 'open' price column
# we're using index 1:2 to only get the 'open' column
# the 1 is inclusive and the 2 is exlusive
# the ".values" code is what creates a numpy array
dataset_train = pd.read_csv("../input/rnn-data/Google_Stock_Price_Train.csv")
training_set = dataset_train.iloc[:, 1:2].values
print(training_set)
# ### Feature Scaling
# When you use a Recurrent Neural Network with sigmoid functions, Normalization is better is Standardization
# The MinMaxScalar class can be used for normalization
# Because we're using 0 and 1 as our max and min, all scaled (normalized) values will be between 0 and 1
# The fit_transform method fo the sc object (MinMaxScalar class) gets the min and max from the "fit" and then
# returns a new, transformed dataset
from sklearn.preprocessing import MinMaxScaler
sc = MinMaxScaler(feature_range=(0, 1), copy=True)
training_set_scaled = sc.fit_transform(training_set)
print(training_set_scaled)
print(training_set_scaled.size)
# ### Creating a data structure with 60 timesteps and 1 output
# 60 timesteps means that, at each time "t", our RNN is going to look at the 60 observations before time "t"
# Based on the trends it sees in those 60 timesteps, the RNN will try to predict what is comes next ("t + 1")
# Using too few timestamps will lead to overfitting
# Using too many timestamps will lead to bad answers
# in this example, a timestep is 1 day. there are about 20 business days in a month, so 60 timestamps is about 3 months worth of observations
# This means that each day, we're going to try to use the last 3 months of data to predict the price for the next day.
# X_train is the input for the RNN
# y_train is the output for the RNN
# for each time "t", X_train will contain the 60 observations prior to time "t"
# and y_train will contain the observation for time "t+1"
X_train = []
y_train = []
# we can't start adding observations to our variables until the 61st day of 2012
# for every value in the training_set_scaled, starting with the 61st value (index 60)
# add the previous 60 observations to X_train, and the next observation to y_train
for i in range(60, training_set_scaled.size):
X_train.append(training_set_scaled[i - 60 : i, 0])
y_train.append(training_set_scaled[i, 0])
# reformat X_train and y_train as numpy arrays
X_train, y_train = np.array(X_train), np.array(y_train)
print(X_train)
print(y_train)
# ### Reshaping
# we use the reshape function to add an extra dimension to our numpy array
# the RNN we're using expects a particular shape.
# using the keras documentation, we know that we need the following shape:
# 3D tensor with shape (batchsize, timesteps, input_dim)
# for us batchsize = number of observations (1258)
# timesteps = 60
# input_dim is the number of input parameters (predictive features)
# at the time of inintial publishing, I'm only using 1 predictive feature,
# but if you wanted to add more, this is where you would define that aspect of the shape.
# X_train.shape[0] is the number of observations
# X_train.shape[1] is the number of timestamps
X_train = np.reshape(X_train, (X_train.shape[0], X_train.shape[1], 1))
print(X_train)
# ## Part 2 - Building and Training the RNN
# ### Importing the Keras libraries and packages
# The sequential model is a feed-forward model for the RNN
# The dense layers is for fully connected layers
# The LSTM layers is for the Vanishing Gradient Problem
# The Dropout layers class is to add some dropout regularization
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import LSTM
from keras.layers import Dropout
# ### Initialising the RNN
# we use a regressor because we are predicting a continuous value
# classifier would be for predicting a distict value or category
regressor = Sequential()
# ### Adding the first LSTM layer and some Dropout regularisation
# We add an LSTM layer
# units is the number of LSTM Cells (or memory units)
# we're using 50 because predicting stock prices is very complex
# return_sequences = True because we are creating a "stacked LSTM" which means our RNN will have multiple LSTMs.
# because we have multiple LSTM layers, we need the value of return_sequences = True on LSTM layers that will have more LSTM layers to follow
# you do NOT need to set return_sequences=True on the final LSTM Layer;
# there, it needs to be set equal to False, but this is the default value, so you don't have to do anything.
# input_shape = (timestamps, input dimensions);
# note, you do not have to explicitly declare the number of observations.
# X_train.shape[1] = the number of timestamps (defined above) = 60
# currently, we only have 1 input dimension (predictive feature)
regressor.add(LSTM(units=50, return_sequences=True, input_shape=(X_train.shape[1], 1)))
# adding dropout regularization to our RNN
# Dropout(dropoutRate)
# we use a standard dropoutRate of 0.2 (20%)
# This means that 20% of the neurons will be ignored (dropped out) during each iteration.
regressor.add(Dropout(0.2))
# ### Adding a second LSTM layer and some Dropout regularisation
# exactly the same as above, except we do not need to declare the input shape because this isn't the first layer.
regressor.add(LSTM(units=50, return_sequences=True))
regressor.add(Dropout(0.2))
# ### Adding a third LSTM layer and some Dropout regularisation
# exactly the same as the second layer
regressor.add(LSTM(units=50, return_sequences=True))
regressor.add(Dropout(0.2))
# ### Adding a fourth LSTM layer and some Dropout regularisation
# The same as the third layer, except return_sequences=False becasue there are no more LSTM layers to add.
# We don't need to explicitly declare return_sequences=False because False is the default value
regressor.add(LSTM(units=50))
regressor.add(Dropout(0.2))
# ### Adding the output layer
# we just use a Dense layer that will be fully connected to the previous layer
# units=1 because we're predicting a single value
regressor.add(Dense(units=1))
# ### Compiling the RNN
# Optimizer options: RMSprop and Adam
# we're using the adam optimizer
# because this is regression and not classificaiton, we won't be using crossentropy
# for regression models, we use Mean Squared Error
regressor.compile(optimizer="adam", loss="mean_squared_error")
# ### Fitting the RNN to the Training set
# the fit method attaches the dataset to the model and starts training it
# regressor.fit(input, output, epochs, batch_size)
# epochs of 100. you might need to play with this to find the point where you start to see convergence.
# we're using batch_size = 32. this means the model will be trained on 32 observations before making adjustments.
regressor.fit(X_train, y_train, epochs=100, batch_size=32)
# ## Part 3 - Making the predictions and visualising the results
# ### Getting the real stock price of 2017
# import the data file for the stock price in 2017
dataset_test = pd.read_csv("../input/rnn-data/Google_Stock_Price_Test.csv")
real_stock_price = dataset_test.iloc[:, 1:2].values
print(real_stock_price)
# ### Getting the predicted stock price of 2017
# In order to make predictions, we will need observations from the previous 60 days
# In order to get those observations, we will need a complete dataset with the training set and the test set
# dataset_total = concatenation of dataset_train and dataset_test
# we only need the "Open" column from both datasets
# pd.concat((dataset1, dataset2), axis)
# axis = 0 for vertical concatenation
# axis = 1 for horizontal concatenation
# we're using vertical concatenation because we just want to add on dataset right beneath the first
dataset_total = pd.concat((dataset_train["Open"], dataset_test["Open"]), axis=0)
# for each day, get the observations for the previous 60 days
# first, we need the index of the start date
index_start = len(dataset_total) - len(dataset_test)
# now we can grab all observations we'll need to make our predictions
inputs = dataset_total[index_start - 60 :].values
print(inputs)
# we need to reshape our inputs array to become a 2D array with one column
# we use the reshape method with arguments (-1,1)
inputs = inputs.reshape(-1, 1)
print(inputs)
# we need to scale our inputs
# we don't want to use fit_transform because we don't want to change our scalar, we just want to apply it.
inputs = sc.transform(inputs)
print(inputs)
# we need to create a 3D structure, which is what our model expects
# we don't need a "y_test" because we're making "real" predictions. We don't know the answer.
X_test = []
# we're still using 60 timesteps
for i in range(60, inputs.size):
X_test.append(inputs[i - 60 : i, 0])
# reformat X_test as a numpy arrays
X_test = np.array(X_test)
# we use the reshape function to add an extra dimension to our numpy array
# the RNN we're using expects a particular shape.
# using the keras documentation, we know that we need the following shape:
# 3D tensor with shape (batchsize, timesteps, input_dim)
# for us batchsize = number of observations (80)
# timesteps = 60
# input_dim is the number of input parameters (predictive features)
# at the time of inintial publishing, I'm only using 1 predictive feature,
# but if you wanted to add more, this is where you would define that aspect of the shape.
# X_test.shape[0] is the number of observations (80)
# X_test.shape[1] is the number of timestamps (60)
X_test = np.reshape(X_test, (X_test.shape[0], X_test.shape[1], 1))
print(X_test)
# time to finally make the prediction
predicted_stock_price = regressor.predict(X_test)
print(predicted_stock_price)
# we have to inverse the scaling of the predicted stock price to get a real price
predicted_stock_price = sc.inverse_transform(predicted_stock_price)
print(predicted_stock_price)
# ### Visualising the results
#
plt.plot(real_stock_price, color="red", label="Real Google Stock Price")
plt.plot(predicted_stock_price, color="blue", label="Predicted Googel Stock Price")
plt.title("Google Stock Price Prediction (Jan 2017)")
plt.xlabel("Time (Days in Jan 2017)")
plt.ylabel("Google Stock Price")
plt.legend()
plt.show()
# ## Evaluating the RNN
# we evaluate the performance of the RNN using a metric called Root Mean Squared Error
import math
from sklearn.metrics import mean_squared_error
rmse = math.sqrt(mean_squared_error(real_stock_price, predicted_stock_price))
print(rmse)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/851/69851334.ipynb
| null | null |
[{"Id": 69851334, "ScriptId": 19048891, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7118846, "CreationDate": "08/03/2021 15:32:47", "VersionNumber": 1.0, "Title": "Recurrent Neural Network - Python", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 350.0, "LinesInsertedFromPrevious": 350.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Recurrent Neural Network
# ## Part 1 - Data Preprocessing
# ### Importing the libraries
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
# ### Importing the training set
# We're only grabbing the 'open' price column
# we're using index 1:2 to only get the 'open' column
# the 1 is inclusive and the 2 is exlusive
# the ".values" code is what creates a numpy array
dataset_train = pd.read_csv("../input/rnn-data/Google_Stock_Price_Train.csv")
training_set = dataset_train.iloc[:, 1:2].values
print(training_set)
# ### Feature Scaling
# When you use a Recurrent Neural Network with sigmoid functions, Normalization is better is Standardization
# The MinMaxScalar class can be used for normalization
# Because we're using 0 and 1 as our max and min, all scaled (normalized) values will be between 0 and 1
# The fit_transform method fo the sc object (MinMaxScalar class) gets the min and max from the "fit" and then
# returns a new, transformed dataset
from sklearn.preprocessing import MinMaxScaler
sc = MinMaxScaler(feature_range=(0, 1), copy=True)
training_set_scaled = sc.fit_transform(training_set)
print(training_set_scaled)
print(training_set_scaled.size)
# ### Creating a data structure with 60 timesteps and 1 output
# 60 timesteps means that, at each time "t", our RNN is going to look at the 60 observations before time "t"
# Based on the trends it sees in those 60 timesteps, the RNN will try to predict what is comes next ("t + 1")
# Using too few timestamps will lead to overfitting
# Using too many timestamps will lead to bad answers
# in this example, a timestep is 1 day. there are about 20 business days in a month, so 60 timestamps is about 3 months worth of observations
# This means that each day, we're going to try to use the last 3 months of data to predict the price for the next day.
# X_train is the input for the RNN
# y_train is the output for the RNN
# for each time "t", X_train will contain the 60 observations prior to time "t"
# and y_train will contain the observation for time "t+1"
X_train = []
y_train = []
# we can't start adding observations to our variables until the 61st day of 2012
# for every value in the training_set_scaled, starting with the 61st value (index 60)
# add the previous 60 observations to X_train, and the next observation to y_train
for i in range(60, training_set_scaled.size):
X_train.append(training_set_scaled[i - 60 : i, 0])
y_train.append(training_set_scaled[i, 0])
# reformat X_train and y_train as numpy arrays
X_train, y_train = np.array(X_train), np.array(y_train)
print(X_train)
print(y_train)
# ### Reshaping
# we use the reshape function to add an extra dimension to our numpy array
# the RNN we're using expects a particular shape.
# using the keras documentation, we know that we need the following shape:
# 3D tensor with shape (batchsize, timesteps, input_dim)
# for us batchsize = number of observations (1258)
# timesteps = 60
# input_dim is the number of input parameters (predictive features)
# at the time of inintial publishing, I'm only using 1 predictive feature,
# but if you wanted to add more, this is where you would define that aspect of the shape.
# X_train.shape[0] is the number of observations
# X_train.shape[1] is the number of timestamps
X_train = np.reshape(X_train, (X_train.shape[0], X_train.shape[1], 1))
print(X_train)
# ## Part 2 - Building and Training the RNN
# ### Importing the Keras libraries and packages
# The sequential model is a feed-forward model for the RNN
# The dense layers is for fully connected layers
# The LSTM layers is for the Vanishing Gradient Problem
# The Dropout layers class is to add some dropout regularization
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import LSTM
from keras.layers import Dropout
# ### Initialising the RNN
# we use a regressor because we are predicting a continuous value
# classifier would be for predicting a distict value or category
regressor = Sequential()
# ### Adding the first LSTM layer and some Dropout regularisation
# We add an LSTM layer
# units is the number of LSTM Cells (or memory units)
# we're using 50 because predicting stock prices is very complex
# return_sequences = True because we are creating a "stacked LSTM" which means our RNN will have multiple LSTMs.
# because we have multiple LSTM layers, we need the value of return_sequences = True on LSTM layers that will have more LSTM layers to follow
# you do NOT need to set return_sequences=True on the final LSTM Layer;
# there, it needs to be set equal to False, but this is the default value, so you don't have to do anything.
# input_shape = (timestamps, input dimensions);
# note, you do not have to explicitly declare the number of observations.
# X_train.shape[1] = the number of timestamps (defined above) = 60
# currently, we only have 1 input dimension (predictive feature)
regressor.add(LSTM(units=50, return_sequences=True, input_shape=(X_train.shape[1], 1)))
# adding dropout regularization to our RNN
# Dropout(dropoutRate)
# we use a standard dropoutRate of 0.2 (20%)
# This means that 20% of the neurons will be ignored (dropped out) during each iteration.
regressor.add(Dropout(0.2))
# ### Adding a second LSTM layer and some Dropout regularisation
# exactly the same as above, except we do not need to declare the input shape because this isn't the first layer.
regressor.add(LSTM(units=50, return_sequences=True))
regressor.add(Dropout(0.2))
# ### Adding a third LSTM layer and some Dropout regularisation
# exactly the same as the second layer
regressor.add(LSTM(units=50, return_sequences=True))
regressor.add(Dropout(0.2))
# ### Adding a fourth LSTM layer and some Dropout regularisation
# The same as the third layer, except return_sequences=False becasue there are no more LSTM layers to add.
# We don't need to explicitly declare return_sequences=False because False is the default value
regressor.add(LSTM(units=50))
regressor.add(Dropout(0.2))
# ### Adding the output layer
# we just use a Dense layer that will be fully connected to the previous layer
# units=1 because we're predicting a single value
regressor.add(Dense(units=1))
# ### Compiling the RNN
# Optimizer options: RMSprop and Adam
# we're using the adam optimizer
# because this is regression and not classificaiton, we won't be using crossentropy
# for regression models, we use Mean Squared Error
regressor.compile(optimizer="adam", loss="mean_squared_error")
# ### Fitting the RNN to the Training set
# the fit method attaches the dataset to the model and starts training it
# regressor.fit(input, output, epochs, batch_size)
# epochs of 100. you might need to play with this to find the point where you start to see convergence.
# we're using batch_size = 32. this means the model will be trained on 32 observations before making adjustments.
regressor.fit(X_train, y_train, epochs=100, batch_size=32)
# ## Part 3 - Making the predictions and visualising the results
# ### Getting the real stock price of 2017
# import the data file for the stock price in 2017
dataset_test = pd.read_csv("../input/rnn-data/Google_Stock_Price_Test.csv")
real_stock_price = dataset_test.iloc[:, 1:2].values
print(real_stock_price)
# ### Getting the predicted stock price of 2017
# In order to make predictions, we will need observations from the previous 60 days
# In order to get those observations, we will need a complete dataset with the training set and the test set
# dataset_total = concatenation of dataset_train and dataset_test
# we only need the "Open" column from both datasets
# pd.concat((dataset1, dataset2), axis)
# axis = 0 for vertical concatenation
# axis = 1 for horizontal concatenation
# we're using vertical concatenation because we just want to add on dataset right beneath the first
dataset_total = pd.concat((dataset_train["Open"], dataset_test["Open"]), axis=0)
# for each day, get the observations for the previous 60 days
# first, we need the index of the start date
index_start = len(dataset_total) - len(dataset_test)
# now we can grab all observations we'll need to make our predictions
inputs = dataset_total[index_start - 60 :].values
print(inputs)
# we need to reshape our inputs array to become a 2D array with one column
# we use the reshape method with arguments (-1,1)
inputs = inputs.reshape(-1, 1)
print(inputs)
# we need to scale our inputs
# we don't want to use fit_transform because we don't want to change our scalar, we just want to apply it.
inputs = sc.transform(inputs)
print(inputs)
# we need to create a 3D structure, which is what our model expects
# we don't need a "y_test" because we're making "real" predictions. We don't know the answer.
X_test = []
# we're still using 60 timesteps
for i in range(60, inputs.size):
X_test.append(inputs[i - 60 : i, 0])
# reformat X_test as a numpy arrays
X_test = np.array(X_test)
# we use the reshape function to add an extra dimension to our numpy array
# the RNN we're using expects a particular shape.
# using the keras documentation, we know that we need the following shape:
# 3D tensor with shape (batchsize, timesteps, input_dim)
# for us batchsize = number of observations (80)
# timesteps = 60
# input_dim is the number of input parameters (predictive features)
# at the time of inintial publishing, I'm only using 1 predictive feature,
# but if you wanted to add more, this is where you would define that aspect of the shape.
# X_test.shape[0] is the number of observations (80)
# X_test.shape[1] is the number of timestamps (60)
X_test = np.reshape(X_test, (X_test.shape[0], X_test.shape[1], 1))
print(X_test)
# time to finally make the prediction
predicted_stock_price = regressor.predict(X_test)
print(predicted_stock_price)
# we have to inverse the scaling of the predicted stock price to get a real price
predicted_stock_price = sc.inverse_transform(predicted_stock_price)
print(predicted_stock_price)
# ### Visualising the results
#
plt.plot(real_stock_price, color="red", label="Real Google Stock Price")
plt.plot(predicted_stock_price, color="blue", label="Predicted Googel Stock Price")
plt.title("Google Stock Price Prediction (Jan 2017)")
plt.xlabel("Time (Days in Jan 2017)")
plt.ylabel("Google Stock Price")
plt.legend()
plt.show()
# ## Evaluating the RNN
# we evaluate the performance of the RNN using a metric called Root Mean Squared Error
import math
from sklearn.metrics import mean_squared_error
rmse = math.sqrt(mean_squared_error(real_stock_price, predicted_stock_price))
print(rmse)
| false | 0 | 3,018 | 0 | 3,018 | 3,018 |
||
69851269
|
<jupyter_start><jupyter_text>Appliances energy prediction Data Set
### Context
Experimental data used to create regression models of appliances energy use in a low energy building.
### Content
Data Set Characteristics:
Multivariate, Time-Series, Regression
Number of Instances(Rows):
19735
Number of Attributes(Columns):
29
Associated Tasks:
Regression
Source:
Luis Candanedo, luismiguel.candanedoibarra '@' umons.ac.be, University of Mons (UMONS).
Data Set Information:
Given in Metadata tab about the sources and collection methodology.
## Attribute Information:
date time year-month-day hour:minute:second
Appliances, energy use in Wh (**target variable for prediction**)
lights, energy use of light fixtures in the house in Wh
T1, Temperature in kitchen area, in Celsius
RH_1, Humidity in kitchen area, in %
T2, Temperature in living room area, in Celsius
RH_2, Humidity in living room area, in %
T3, Temperature in laundry room area
RH_3, Humidity in laundry room area, in %
T4, Temperature in office room, in Celsius
RH_4, Humidity in office room, in %
T5, Temperature in bathroom, in Celsius
RH_5, Humidity in bathroom, in %
T6, Temperature outside the building (north side), in Celsius
RH_6, Humidity outside the building (north side), in %
T7, Temperature in ironing room , in Celsius
RH_7, Humidity in ironing room, in %
T8, Temperature in teenager room 2, in Celsius
RH_8, Humidity in teenager room 2, in %
T9, Temperature in parents room, in Celsius
RH_9, Humidity in parents room, in %
To, Temperature outside (from Chievres weather station), in Celsius
Pressure (from Chievres weather station), in mm Hg
RH_out, Humidity outside (from Chievres weather station), in %
Wind speed (from Chievres weather station), in m/s
Visibility (from Chievres weather station), in km
Tdewpoint (from Chievres weather station), °C
rv1, Random variable 1, nondimensional
rv2, Random variable 2, nondimensional
Where indicated, hourly data (then interpolated) from the nearest airport weather station (Chievres Airport, Belgium) was downloaded from a public data set from Reliable Prognosis, rp5.ru. Permission was obtained from Reliable Prognosis for the distribution of the 4.5 months of weather data.
Kaggle dataset identifier: appliances-energy-prediction-data-set
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
# ## Dataset Loading
path = "../input/appliances-energy-prediction-data-set/energydata_complete.csv"
dataLoad = pd.read_csv(path, index_col="date")
# ## Basic Data Exploration
dataLoad.head()
dataLoad.info()
dataLoad.describe()
dataLoad.isnull().sum()
# ## Exploratory Data Analysis
# ### Univariate
dataVisual = dataLoad.drop(["rv1", "rv2"], axis=1)
# Distribution plot for each features
ls = []
colName = [j for j in dataVisual.columns]
a = 0
while a < 7:
for i in range(5):
ls.append((a, i))
a += 1
fig, ax = plt.subplots(6, 5, figsize=(28, 30))
for k in range(26):
sns.histplot(ax=ax[ls[k][0], ls[k][1]], x=colName[k], data=dataVisual)
a = 0
while a < 7:
for i in range(5):
ls.append((a, i))
a += 1
fig, ax = plt.subplots(6, 5, figsize=(28, 30))
for k in range(26):
sns.boxplot(ax=ax[ls[k][0], ls[k][1]], y=colName[k], data=dataVisual)
# ## Multivariate
b = 0
while b < 7:
for i in range(5):
ls.append((b, i))
b += 1
fig, ax = plt.subplots(5, 5, figsize=(28, 30))
for k in range(25):
sns.scatterplot(
ax=ax[ls[k][0], ls[k][1]], x=colName[k + 1], y=colName[0], data=dataVisual
)
corr = dataVisual.corr()
plt.figure(figsize=(30, 30))
sns.heatmap(corr, annot=True)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/851/69851269.ipynb
|
appliances-energy-prediction-data-set
|
sohommajumder21
|
[{"Id": 69851269, "ScriptId": 18982839, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7924426, "CreationDate": "08/03/2021 15:32:27", "VersionNumber": 4.0, "Title": "HouseAppliancesEnergy", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 83.0, "LinesInsertedFromPrevious": 23.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 60.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 93300289, "KernelVersionId": 69851269, "SourceDatasetVersionId": 2325641}]
|
[{"Id": 2325641, "DatasetId": 1403770, "DatasourceVersionId": 2367158, "CreatorUserId": 4429509, "LicenseName": "Database: Open Database, Contents: \u00a9 Original Authors", "CreationDate": "06/12/2021 02:45:24", "VersionNumber": 1.0, "Title": "Appliances energy prediction Data Set", "Slug": "appliances-energy-prediction-data-set", "Subtitle": "https://archive.ics.uci.edu/ml/datasets/Appliances+energy+prediction", "Description": "### Context\n\nExperimental data used to create regression models of appliances energy use in a low energy building.\n\n\n### Content\n\nData Set Characteristics: \n\nMultivariate, Time-Series, Regression\n\nNumber of Instances(Rows):\n\n19735\n\nNumber of Attributes(Columns):\n\n29\n\n\nAssociated Tasks:\n\nRegression\n\n\nSource:\n\nLuis Candanedo, luismiguel.candanedoibarra '@' umons.ac.be, University of Mons (UMONS).\n\n\nData Set Information:\nGiven in Metadata tab about the sources and collection methodology.\n\n\n## Attribute Information:\n\ndate time year-month-day hour:minute:second\n\nAppliances, energy use in Wh (**target variable for prediction**)\n\nlights, energy use of light fixtures in the house in Wh\n\nT1, Temperature in kitchen area, in Celsius\n\nRH_1, Humidity in kitchen area, in %\n\nT2, Temperature in living room area, in Celsius\n\nRH_2, Humidity in living room area, in %\n\nT3, Temperature in laundry room area\n\nRH_3, Humidity in laundry room area, in %\n\nT4, Temperature in office room, in Celsius\n\nRH_4, Humidity in office room, in %\n\nT5, Temperature in bathroom, in Celsius\n\nRH_5, Humidity in bathroom, in %\n\nT6, Temperature outside the building (north side), in Celsius\n\nRH_6, Humidity outside the building (north side), in %\n\nT7, Temperature in ironing room , in Celsius\n\nRH_7, Humidity in ironing room, in %\n\nT8, Temperature in teenager room 2, in Celsius\n\nRH_8, Humidity in teenager room 2, in %\n\nT9, Temperature in parents room, in Celsius\n\nRH_9, Humidity in parents room, in %\n\n\nTo, Temperature outside (from Chievres weather station), in Celsius\n\nPressure (from Chievres weather station), in mm Hg\n\nRH_out, Humidity outside (from Chievres weather station), in %\n\nWind speed (from Chievres weather station), in m/s\n\nVisibility (from Chievres weather station), in km\n\nTdewpoint (from Chievres weather station), \u00c2\u00b0C\n\nrv1, Random variable 1, nondimensional\n\nrv2, Random variable 2, nondimensional\n\nWhere indicated, hourly data (then interpolated) from the nearest airport weather station (Chievres Airport, Belgium) was downloaded from a public data set from Reliable Prognosis, rp5.ru. Permission was obtained from Reliable Prognosis for the distribution of the 4.5 months of weather data.\n\n\n\n\n### Acknowledgements\n\nLuis M. Candanedo, Veronique Feldheim, Dominique Deramaix, Data driven prediction models of energy use of appliances in a low-energy house, Energy and Buildings, Volume 140, 1 April 2017, Pages 81-97, ISSN 0378-7788, [Web Link](https://www.sciencedirect.com/science/article/abs/pii/S0378778816308970?via%3Dihub).\n\n### Citation\n\nDua, D. and Graff, C. (2019). UCI Machine Learning Repository [http://archive.ics.uci.edu/ml]. Irvine, CA: University of California, School of Information and Computer Science.\n\n\n### Inspiration\n\n1) This is a regression task, You should predict the \"appliances\" column. Column descriptions are given above. Please read them before proceeding.\n2) Appropriate time series analysis with regression is preferred more.\n3) Exploratory data analysis with charts and plots.\n\nHave fun!", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1403770, "CreatorUserId": 4429509, "OwnerUserId": 4429509.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2325641.0, "CurrentDatasourceVersionId": 2367158.0, "ForumId": 1423076, "Type": 2, "CreationDate": "06/12/2021 02:45:24", "LastActivityDate": "06/12/2021", "TotalViews": 13594, "TotalDownloads": 1164, "TotalVotes": 26, "TotalKernels": 5}]
|
[{"Id": 4429509, "UserName": "sohommajumder21", "DisplayName": "Sohom Majumder", "RegisterDate": "02/03/2020", "PerformanceTier": 3}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
# ## Dataset Loading
path = "../input/appliances-energy-prediction-data-set/energydata_complete.csv"
dataLoad = pd.read_csv(path, index_col="date")
# ## Basic Data Exploration
dataLoad.head()
dataLoad.info()
dataLoad.describe()
dataLoad.isnull().sum()
# ## Exploratory Data Analysis
# ### Univariate
dataVisual = dataLoad.drop(["rv1", "rv2"], axis=1)
# Distribution plot for each features
ls = []
colName = [j for j in dataVisual.columns]
a = 0
while a < 7:
for i in range(5):
ls.append((a, i))
a += 1
fig, ax = plt.subplots(6, 5, figsize=(28, 30))
for k in range(26):
sns.histplot(ax=ax[ls[k][0], ls[k][1]], x=colName[k], data=dataVisual)
a = 0
while a < 7:
for i in range(5):
ls.append((a, i))
a += 1
fig, ax = plt.subplots(6, 5, figsize=(28, 30))
for k in range(26):
sns.boxplot(ax=ax[ls[k][0], ls[k][1]], y=colName[k], data=dataVisual)
# ## Multivariate
b = 0
while b < 7:
for i in range(5):
ls.append((b, i))
b += 1
fig, ax = plt.subplots(5, 5, figsize=(28, 30))
for k in range(25):
sns.scatterplot(
ax=ax[ls[k][0], ls[k][1]], x=colName[k + 1], y=colName[0], data=dataVisual
)
corr = dataVisual.corr()
plt.figure(figsize=(30, 30))
sns.heatmap(corr, annot=True)
| false | 0 | 685 | 0 | 1,381 | 685 |
||
69851605
|
<jupyter_start><jupyter_text>Indian Earthquakes Dataset(2018 onwards)
### Context
The [National Center for Seismology](https://seismo.gov.in/) is the nodal agency of the Government of India for monitoring earthquake activity in the country. NCS maintains the National Seismological Network of 115 stations each having state of art equipment and spreading all across the country.NCS monitors earthquake activity all across the country through its 24x7 round-the-clock monitoring center. NCS also monitors earthquake swarm and aftershock through deploying a temporary observatory close to the affected region.
### Content
This dataset includes a record of the date, time, location, depth, magnitude, and source of every Indian earthquake since 2018.
Kaggle dataset identifier: indian-earthquakes-dataset2018-onwards
<jupyter_code>import pandas as pd
df = pd.read_csv('indian-earthquakes-dataset2018-onwards/Indian_earthquake_data.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 2719 entries, 0 to 2718
Data columns (total 6 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Origin Time 2719 non-null object
1 Latitude 2719 non-null float64
2 Longitude 2719 non-null float64
3 Depth 2719 non-null float64
4 Magnitude 2719 non-null float64
5 Location 2719 non-null object
dtypes: float64(4), object(2)
memory usage: 127.6+ KB
<jupyter_text>Examples:
{
"Origin Time": "2021-07-31 09:43:23 IST",
"Latitude": 29.06,
"Longitude": 77.42,
"Depth": 5,
"Magnitude": 2.5,
"Location": "53km NNE of New Delhi, India"
}
{
"Origin Time": "2021-07-30 23:04:57 IST",
"Latitude": 19.93,
"Longitude": 72.92,
"Depth": 5,
"Magnitude": 2.4,
"Location": "91km W of Nashik, Maharashtra, India"
}
{
"Origin Time": "2021-07-30 21:31:10 IST",
"Latitude": 31.5,
"Longitude": 74.37,
"Depth": 33,
"Magnitude": 3.4,
"Location": "49km WSW of Amritsar, Punjab, India"
}
{
"Origin Time": "2021-07-30 13:56:31 IST",
"Latitude": 28.34,
"Longitude": 76.23,
"Depth": 5,
"Magnitude": 3.1,
"Location": "50km SW of Jhajjar, Haryana"
}
<jupyter_script># Geospatial data refers to the time-based data that is related to a specific location on the Earth’s surface. It is useful since it can reveal vital patterns and trends in the landscape. There are many libraries available to analyze such data, but often there is a lot of manual effort involved. Memory management of geospatial data is another issue. This means the magnitude of such data makes it a little difficult to process and analyze it easily.
# [Kepler.gl](https://kepler.gl/) is a powerful open source geospatial analysis tool for large-scale data sets. Developed by Uber, Kepler.gl is a data-agnostic and high-performance web-based application for large-scale geolocation data sets. You very easily find patterns in a dataset, combine different datasets, perform aggregations and manipulations in real-time. If a dataset has a timestamp, geopoints, and some meta information, Kepler.gl can automatically extract the information and visualize it.
# 🗒️ Incase you want to know more, here is an article that goes deeper into the theory behind the library along with other useful resources: [Visualizing India’s Seismic activity](https://towardsdatascience.com/visualizing-indias-seismic-activity-4ed390de298c?sk=12befb1ffd589de4bb5251892b161a25)
##Installation
# Importing necessary libaries
import pandas as pd
from keplergl import KeplerGl
# importing the dataset
df = pd.read_csv(
"../input/indian-earthquakes-dataset2018-onwards/Indian_earthquake_data.csv"
)
df.head()
# preprocessing dataset
df["Origin Time"] = pd.to_datetime(df["Origin Time"])
df["Place"] = df.apply(lambda x: x["Location"].split(",")[1], axis=1)
df.head()
# Visualizing with Kepler.gl
map_1 = KeplerGl(height=600)
map_1.add_data(data=df, name="Indian Earthquake Visualization")
map_1
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/851/69851605.ipynb
|
indian-earthquakes-dataset2018-onwards
|
parulpandey
|
[{"Id": 69851605, "ScriptId": 19092210, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 391404, "CreationDate": "08/03/2021 15:34:26", "VersionNumber": 2.0, "Title": "Visualizing India\u2019s Seismic activity", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 33.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 33.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 3}]
|
[{"Id": 93300555, "KernelVersionId": 69851605, "SourceDatasetVersionId": 2495302}]
|
[{"Id": 2495302, "DatasetId": 1510756, "DatasourceVersionId": 2537917, "CreatorUserId": 391404, "LicenseName": "Other (specified in description)", "CreationDate": "08/03/2021 14:02:06", "VersionNumber": 1.0, "Title": "Indian Earthquakes Dataset(2018 onwards)", "Slug": "indian-earthquakes-dataset2018-onwards", "Subtitle": "Date, time, and location of all Indian earthquakes after 2018", "Description": "### Context\n\nThe [National Center for Seismology](https://seismo.gov.in/) is the nodal agency of the Government of India for monitoring earthquake activity in the country. NCS maintains the National Seismological Network of 115 stations each having state of art equipment and spreading all across the country.NCS monitors earthquake activity all across the country through its 24x7 round-the-clock monitoring center. NCS also monitors earthquake swarm and aftershock through deploying a temporary observatory close to the affected region.\n\n### Content\n\nThis dataset includes a record of the date, time, location, depth, magnitude, and source of every Indian earthquake since 2018.\n\n### Acknowledgements\n\nOriginal Image by [Tumisu](https://pixabay.com/users/tumisu-148124/?utm_source=link-attribution&utm_medium=referral&utm_campaign=image&utm_content=3167693) from [Pixabay](https://pixabay.com/?utm_source=link-attribution&utm_medium=referral&utm_campaign=image&utm_content=3167693)\n\n## [Starter Notebookl](https://www.kaggle.com/parulpandey/visualizing-india-s-seismic-activity)", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1510756, "CreatorUserId": 391404, "OwnerUserId": 391404.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2495302.0, "CurrentDatasourceVersionId": 2537917.0, "ForumId": 1530515, "Type": 2, "CreationDate": "08/03/2021 14:02:06", "LastActivityDate": "08/03/2021", "TotalViews": 6820, "TotalDownloads": 722, "TotalVotes": 14, "TotalKernels": 3}]
|
[{"Id": 391404, "UserName": "parulpandey", "DisplayName": "Parul Pandey", "RegisterDate": "07/26/2015", "PerformanceTier": 4}]
|
# Geospatial data refers to the time-based data that is related to a specific location on the Earth’s surface. It is useful since it can reveal vital patterns and trends in the landscape. There are many libraries available to analyze such data, but often there is a lot of manual effort involved. Memory management of geospatial data is another issue. This means the magnitude of such data makes it a little difficult to process and analyze it easily.
# [Kepler.gl](https://kepler.gl/) is a powerful open source geospatial analysis tool for large-scale data sets. Developed by Uber, Kepler.gl is a data-agnostic and high-performance web-based application for large-scale geolocation data sets. You very easily find patterns in a dataset, combine different datasets, perform aggregations and manipulations in real-time. If a dataset has a timestamp, geopoints, and some meta information, Kepler.gl can automatically extract the information and visualize it.
# 🗒️ Incase you want to know more, here is an article that goes deeper into the theory behind the library along with other useful resources: [Visualizing India’s Seismic activity](https://towardsdatascience.com/visualizing-indias-seismic-activity-4ed390de298c?sk=12befb1ffd589de4bb5251892b161a25)
##Installation
# Importing necessary libaries
import pandas as pd
from keplergl import KeplerGl
# importing the dataset
df = pd.read_csv(
"../input/indian-earthquakes-dataset2018-onwards/Indian_earthquake_data.csv"
)
df.head()
# preprocessing dataset
df["Origin Time"] = pd.to_datetime(df["Origin Time"])
df["Place"] = df.apply(lambda x: x["Location"].split(",")[1], axis=1)
df.head()
# Visualizing with Kepler.gl
map_1 = KeplerGl(height=600)
map_1.add_data(data=df, name="Indian Earthquake Visualization")
map_1
|
[{"indian-earthquakes-dataset2018-onwards/Indian_earthquake_data.csv": {"column_names": "[\"Origin Time\", \"Latitude\", \"Longitude\", \"Depth\", \"Magnitude\", \"Location\"]", "column_data_types": "{\"Origin Time\": \"object\", \"Latitude\": \"float64\", \"Longitude\": \"float64\", \"Depth\": \"float64\", \"Magnitude\": \"float64\", \"Location\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 2719 entries, 0 to 2718\nData columns (total 6 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Origin Time 2719 non-null object \n 1 Latitude 2719 non-null float64\n 2 Longitude 2719 non-null float64\n 3 Depth 2719 non-null float64\n 4 Magnitude 2719 non-null float64\n 5 Location 2719 non-null object \ndtypes: float64(4), object(2)\nmemory usage: 127.6+ KB\n", "summary": "{\"Latitude\": {\"count\": 2719.0, \"mean\": 29.93943324751747, \"std\": 7.361563580475768, \"min\": 0.12, \"25%\": 25.7, \"50%\": 31.21, \"75%\": 36.39, \"max\": 40.0}, \"Longitude\": {\"count\": 2719.0, \"mean\": 80.9056377344612, \"std\": 10.139075349818116, \"min\": 60.3, \"25%\": 71.81, \"50%\": 76.61, \"75%\": 92.515, \"max\": 99.96}, \"Depth\": {\"count\": 2719.0, \"mean\": 53.40047811695477, \"std\": 68.23973746619275, \"min\": 0.8, \"25%\": 10.0, \"50%\": 15.0, \"75%\": 82.0, \"max\": 471.0}, \"Magnitude\": {\"count\": 2719.0, \"mean\": 3.7721956601691793, \"std\": 0.768075773849231, \"min\": 1.5, \"25%\": 3.2, \"50%\": 3.9, \"75%\": 4.3, \"max\": 7.0}}", "examples": "{\"Origin Time\":{\"0\":\"2021-07-31 09:43:23 IST\",\"1\":\"2021-07-30 23:04:57 IST\",\"2\":\"2021-07-30 21:31:10 IST\",\"3\":\"2021-07-30 13:56:31 IST\"},\"Latitude\":{\"0\":29.06,\"1\":19.93,\"2\":31.5,\"3\":28.34},\"Longitude\":{\"0\":77.42,\"1\":72.92,\"2\":74.37,\"3\":76.23},\"Depth\":{\"0\":5.0,\"1\":5.0,\"2\":33.0,\"3\":5.0},\"Magnitude\":{\"0\":2.5,\"1\":2.4,\"2\":3.4,\"3\":3.1},\"Location\":{\"0\":\"53km NNE of New Delhi, India\",\"1\":\"91km W of Nashik, Maharashtra, India\",\"2\":\"49km WSW of Amritsar, Punjab, India\",\"3\":\"50km SW of Jhajjar, Haryana\"}}"}}]
| true | 1 |
<start_data_description><data_path>indian-earthquakes-dataset2018-onwards/Indian_earthquake_data.csv:
<column_names>
['Origin Time', 'Latitude', 'Longitude', 'Depth', 'Magnitude', 'Location']
<column_types>
{'Origin Time': 'object', 'Latitude': 'float64', 'Longitude': 'float64', 'Depth': 'float64', 'Magnitude': 'float64', 'Location': 'object'}
<dataframe_Summary>
{'Latitude': {'count': 2719.0, 'mean': 29.93943324751747, 'std': 7.361563580475768, 'min': 0.12, '25%': 25.7, '50%': 31.21, '75%': 36.39, 'max': 40.0}, 'Longitude': {'count': 2719.0, 'mean': 80.9056377344612, 'std': 10.139075349818116, 'min': 60.3, '25%': 71.81, '50%': 76.61, '75%': 92.515, 'max': 99.96}, 'Depth': {'count': 2719.0, 'mean': 53.40047811695477, 'std': 68.23973746619275, 'min': 0.8, '25%': 10.0, '50%': 15.0, '75%': 82.0, 'max': 471.0}, 'Magnitude': {'count': 2719.0, 'mean': 3.7721956601691793, 'std': 0.768075773849231, 'min': 1.5, '25%': 3.2, '50%': 3.9, '75%': 4.3, 'max': 7.0}}
<dataframe_info>
RangeIndex: 2719 entries, 0 to 2718
Data columns (total 6 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Origin Time 2719 non-null object
1 Latitude 2719 non-null float64
2 Longitude 2719 non-null float64
3 Depth 2719 non-null float64
4 Magnitude 2719 non-null float64
5 Location 2719 non-null object
dtypes: float64(4), object(2)
memory usage: 127.6+ KB
<some_examples>
{'Origin Time': {'0': '2021-07-31 09:43:23 IST', '1': '2021-07-30 23:04:57 IST', '2': '2021-07-30 21:31:10 IST', '3': '2021-07-30 13:56:31 IST'}, 'Latitude': {'0': 29.06, '1': 19.93, '2': 31.5, '3': 28.34}, 'Longitude': {'0': 77.42, '1': 72.92, '2': 74.37, '3': 76.23}, 'Depth': {'0': 5.0, '1': 5.0, '2': 33.0, '3': 5.0}, 'Magnitude': {'0': 2.5, '1': 2.4, '2': 3.4, '3': 3.1}, 'Location': {'0': '53km NNE of New Delhi, India', '1': '91km W of Nashik, Maharashtra, India', '2': '49km WSW of Amritsar, Punjab, India', '3': '50km SW of Jhajjar, Haryana'}}
<end_description>
| 498 | 3 | 1,299 | 498 |
69826100
|
<jupyter_start><jupyter_text>Formula 1 (F1) trending tweets 🏎 🏁

- Formula One (also known as Formula 1 or F1) is the highest class of international auto racing for single-seater formula racing cars sanctioned by the Fédération Internationale de l'Automobile (FIA). The World Drivers' Championship, which became the FIA Formula One World Championship in 1981, has been one of the premier forms of racing around the world since its inaugural season in 1950. The word formula in the name refers to the set of rules to which all participants' cars must conform. A Formula One season consists of a series of races, known as Grands Prix, which take place worldwide on both purpose-built circuits and closed public roads.
- The craze for F1 among the fans is astonishing, which has been creating quite a buzz in major social media platforms like Twitter. The dataset brings you such tweets posted with the #f1 hashtag.
### Information regarding the data
- The data totally consists of 50k+ records with 13 columns. The collection started on 25/7/2020 and will be updated regularly. The description of the features is given below.
### Inspiration
"I am an artist, the track is my canvas and the car is my brush." – Graham Hill
Kaggle dataset identifier: formula-1-trending-tweets
<jupyter_code>import pandas as pd
df = pd.read_csv('formula-1-trending-tweets/F1_tweets.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 632388 entries, 0 to 632387
Data columns (total 13 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 user_name 632382 non-null object
1 user_location 443577 non-null object
2 user_description 592917 non-null object
3 user_created 632388 non-null object
4 user_followers 632388 non-null float64
5 user_friends 632388 non-null object
6 user_favourites 632388 non-null object
7 user_verified 632388 non-null object
8 date 632388 non-null object
9 text 632388 non-null object
10 hashtags 632360 non-null object
11 source 632384 non-null object
12 is_retweet 632384 non-null object
dtypes: float64(1), object(12)
memory usage: 62.7+ MB
<jupyter_text>Examples:
{
"user_name": "Zack Shephard",
"user_location": null,
"user_description": "My opinions are mine ALONE. Venting is the only way that I can get everthing off my chest. #BeKind #FoxNewsIsGoingToHell #BlackLivesMatter",
"user_created": "2011-12-03 20:36:55",
"user_followers": 89,
"user_friends": 2598,
"user_favourites": 212886,
"user_verified": false,
"date": "2021-08-01 23:59:18",
"text": "The next great #F1 duo!!! @ESPNF1 https://t.co/WMhaoAdSxF",
"hashtags": "['F1']",
"source": "Twitter for iPhone",
"is_retweet": false
}
{
"user_name": "Matteo Vannucci",
"user_location": null,
"user_description": "Free speech, free ideas, random thoughts",
"user_created": "2010-05-28 01:02:32",
"user_followers": 10,
"user_friends": 40,
"user_favourites": 8,
"user_verified": false,
"date": "2021-08-01 23:59:10",
"text": "@F1 What if\u2026Hamilton would have gone to the pit\u2026empty grid for a start?!\ud83e\udd14 something to think about #HungarianGP #F1",
"hashtags": "['HungarianGP', 'F1']",
"source": "Twitter for iPhone",
"is_retweet": false
}
{
"user_name": "Michelle",
"user_location": "The Burbs",
"user_description": "likes- movies, candy, being lazy, formula 1, 65\u00b0F, watching various sports & reality shows\n\ndislikes- phone calls, bugs, hot weather, most foods, & group texts",
"user_created": "2009-03-04 16:53:13",
"user_followers": 231,
"user_friends": 110,
"user_favourites": 428,
"user_verified": false,
"date": "2021-08-01 23:58:48",
"text": "This has been a tiring #f1 day... https://t.co/34ggWgAlT6",
"hashtags": "['f1']",
"source": "Twitter for Android",
"is_retweet": false
}
{
"user_name": "JoshWFC \ud83c\udde7\ud83c\uddf7",
"user_location": null,
"user_description": "| JP10\u2019s biggest fan | pronouns : Jo\u00e3o/Pedro | @watfordfc | @RamsNFL | @GeorgeRussell63",
"user_created": "2021-04-05 16:14:53",
"user_followers": 156,
"user_friends": 443,
"user_favourites": 2994,
"user_verified": false,
"date": "2021-08-01 23:58:11",
"text": "Valteri Bottass tracking down Max Verstappen on the way home\n\n#F1 https://t.co/epTHlDyjU6",
"hashtags": "['F1']",
"source": "Twitter for iPhone",
"is_retweet": false
}
<jupyter_script># 
#
# - Formula One (also known as Formula 1 or F1) is the highest class of international auto racing for single-seater formula racing cars sanctioned by the Fédération Internationale de l'Automobile (FIA). The World Drivers' Championship, which became the FIA Formula One World Championship in 1981, has been one of the premier forms of racing around the world since its inaugural season in 1950. The word formula in the name refers to the set of rules to which all participants' cars must conform. A Formula One season consists of a series of races, known as Grands Prix, which take place worldwide on both purpose-built circuits and closed public roads.
# - The craze for F1 among the fans is astonishing, which has been creating quite a buzz in major social media platforms like Twitter. The dataset brings you such tweets posted with the #f1 hashtag.
# This notebook analyses the tweets with the trending #f1 hashtag. So grab your gloves and fasten your seatbelts and let's analyze the impact of f1 in social media platforms like Twitter
#
# 
# Quick navigation
# * [1. Required Libraries](#1)
# * [2. Dataset Quick Overview](#2)
# * [3. Tweets EDA](#3)
# * [4. Tweets text analysis](#4)
# Kindly, Upvote the notebook!
# Required Libraries
import numpy as np
import pandas as pd
import os
import itertools
# plots
import matplotlib.pyplot as plt
import plotly.express as px
import plotly.graph_objects as go
import plotly.figure_factory as ff
from plotly.colors import n_colors
from plotly.subplots import make_subplots
from wordcloud import WordCloud, STOPWORDS, ImageColorGenerator
from sklearn.feature_extraction.text import CountVectorizer
from PIL import Image
from nltk.corpus import stopwords
stop = set(stopwords.words("english"))
from nltk.util import ngrams
import re
from collections import Counter
import nltk
from nltk.corpus import stopwords
import requests
import json
import seaborn as sns
sns.set(rc={"figure.figsize": (11.7, 8.27)})
import warnings
warnings.filterwarnings("ignore")
#
# Dataset Quick Overview
# ## Let's get some basic information about the data!
f1 = pd.read_csv("../input/formula-1-trending-tweets/F1_tweets.csv")
f1.info()
f1.shape
# ## Let's visualize some missing values!
import missingno as mno
mno.matrix(f1)
missed = pd.DataFrame()
missed["column"] = f1.columns
missed["percent"] = [
round(100 * f1[col].isnull().sum() / len(f1), 2) for col in f1.columns
]
missed = missed.sort_values("percent", ascending=False)
missed = missed[missed["percent"] > 0]
fig = sns.barplot(
x=missed["percent"], y=missed["column"], orientation="horizontal"
).set_title("Missed values percent for every column")
#
# Tweets EDA
# ## Lets Visualize the top 20 users by number of tweets
#
ds = f1["user_name"].value_counts().reset_index()
ds.columns = ["user_name", "tweets_count"]
ds = ds.sort_values(["tweets_count"], ascending=False)
f1 = pd.merge(f1, ds, on="user_name")
fig = sns.barplot(
x=ds.head(20)["tweets_count"], y=ds.head(20)["user_name"], orientation="horizontal"
).set_title("Top 20 users by number of tweets")
# ## Users created - yearwise
#
f1["user_created"] = pd.to_datetime(
f1["user_created"], infer_datetime_format=True, errors="coerce"
)
f1["year_created"] = f1["user_created"].dt.year
data = f1.drop_duplicates(subset="user_name", keep="first")
data = data[data["year_created"] > 1970]
data = data["year_created"].value_counts().reset_index()
data.columns = ["year", "number"]
fig = sns.barplot(
x=data["year"],
y=data["number"],
orientation="vertical"
# title='',
).set_title("User created year by year")
# ## Top 20 Users location based on the number of tweets
ds = f1["user_location"].value_counts().reset_index()
ds.columns = ["user_location", "count"]
ds = ds[ds["user_location"] != "NA"]
ds = ds.sort_values(["count"], ascending=False)
fig = sns.barplot(
x=ds.head(20)["count"], y=ds.head(20)["user_location"], orientation="horizontal"
).set_title("Top 20 user locations by number of tweets")
# ## Visualizing the number of tweets per location!!
from plotly.offline import init_notebook_mode, iplot
def pie_count(data, field, percent_limit, title):
data[field] = data[field].fillna("NA")
data = data[field].value_counts().to_frame()
total = data[field].sum()
data["percentage"] = 100 * data[field] / total
percent_limit = percent_limit
otherdata = data[data["percentage"] < percent_limit]
others = otherdata["percentage"].sum()
maindata = data[data["percentage"] >= percent_limit]
data = maindata
other_label = "Others(<" + str(percent_limit) + "% each)"
data.loc[other_label] = pd.Series({field: otherdata[field].sum()})
labels = data.index.tolist()
datavals = data[field].tolist()
trace = go.Pie(labels=labels, values=datavals)
layout = go.Layout(title=title, height=600, width=600)
fig = go.Figure(data=[trace], layout=layout)
iplot(fig)
pie_count(f1, "user_location", 0.5, "Number of tweets per location")
# ## Top 10 user sources by number of tweets
ds = f1["source"].value_counts().reset_index()
ds.columns = ["source", "count"]
ds = ds.sort_values(["count"], ascending=False)
fig = sns.barplot(
x=ds.head(10)["count"],
y=ds.head(10)["source"],
orientation="horizontal",
# title='Top 40 user sources by number of tweets',
# width=800,
# height=800
).set_title("Top 10 user sources by number of tweets")
# ## Total number of tweets for users and number of hashtags in every tweet
f1["hashtags"] = f1["hashtags"].fillna("[]")
f1["hashtags_count"] = f1["hashtags"].apply(lambda x: len(x.split(",")))
f1.loc[f1["hashtags"] == "[]", "hashtags_count"] = 0
fig = sns.scatterplot(x=f1["hashtags_count"], y=f1["tweets_count"]).set_title(
"Total number of tweets for users and number of hashtags in every tweet"
)
#
# * users who post 100 tweets use a range of 1 to a maximum of 33 hastags!
# ## Number of hashtags used in each tweet
ds = f1["hashtags_count"].value_counts().reset_index()
ds.columns = ["hashtags_count", "count"]
ds = ds.sort_values(["count"], ascending=False)
ds["hashtags_count"] = ds["hashtags_count"].astype(str) + " tags"
fig = sns.barplot(
x=ds["count"], y=ds["hashtags_count"], orientation="horizontal"
).set_title("Distribution of number of hashtags in tweets")
# * Most users use 2 hastag followed by 1 hashtag
# * Very less amount of people use more than 5 hashtags in their post
# ## Number of unqiue users each day!
f1["date"] = pd.to_datetime(f1["date"], infer_datetime_format=True, errors="coerce")
df = f1.sort_values(["date"])
df["day"] = df["date"].astype(str).str.split(" ", expand=True)[0]
df["time"] = df["date"].astype(str).str.split(" ", expand=True)[1]
df.head()
ds = df.groupby(["day", "user_name"])["hashtags_count"].count().reset_index()
ds = ds.groupby(["day"])["user_name"].count().reset_index()
ds.columns = ["day", "number_of_users"]
ds["day"] = ds["day"].astype(str)
fig = sns.barplot(
x=ds["day"],
y=ds["number_of_users"],
orientation="vertical",
# title='Number of unique users per day',
# width=800,
# height=800
).set_title("Number of unique users per day")
# fig.show()
plt.xticks(rotation=90)
# ## Tweets distribution over days present in dataset
ds = df["day"].value_counts().reset_index()
ds.columns = ["day", "count"]
ds = ds.sort_values("count", ascending=False)
ds["day"] = ds["day"].astype(str)
fig = sns.barplot(
x=ds["count"],
y=ds["day"],
orientation="horizontal",
).set_title("Tweets distribution over days present in dataset")
# ## Tweets per day
f1["tweet_date"] = f1["date"].dt.date
tweet_date = (
f1["tweet_date"]
.value_counts()
.to_frame()
.reset_index()
.rename(columns={"index": "date", "tweet_date": "count"})
)
tweet_date["date"] = pd.to_datetime(
tweet_date["date"], infer_datetime_format=True, errors="coerce"
)
tweet_date = tweet_date.sort_values("date", ascending=False)
fig = go.Figure(
go.Scatter(
x=tweet_date["date"],
y=tweet_date["count"],
mode="markers+lines",
name="Submissions",
marker_color="dodgerblue",
)
)
f1_dummy = f1.dropna(subset=["tweet_date"])
fig.update_layout(
title_text="Tweets per Day : ({} - {})".format(
f1_dummy["tweet_date"].sort_values()[0].strftime("%d/%m/%Y"),
f1_dummy["tweet_date"].sort_values().iloc[-1].strftime("%d/%m/%Y"),
),
template="plotly_dark",
title_x=0.5,
)
fig.show()
# ## Tweet distribution - hourly
f1["hour"] = f1["date"].dt.hour
ds = f1["hour"].value_counts().reset_index()
ds.columns = ["hour", "count"]
ds["hour"] = "Hour " + ds["hour"].astype(str)
fig = sns.barplot(
x=ds["hour"],
y=ds["count"],
orientation="vertical",
).set_title("Tweets distribution over hours")
plt.xticks(rotation="vertical")
# ### Top 10 hastags used in the tweet!
def split_hashtags(x):
return str(x).replace("[", "").replace("]", "").split(",")
tweets_df = f1.copy()
tweets_df["hashtag"] = tweets_df["hashtags"].apply(lambda row: split_hashtags(row))
tweets_df = tweets_df.explode("hashtag")
tweets_df["hashtag"] = (
tweets_df["hashtag"]
.astype(str)
.str.lower()
.str.replace("'", "")
.str.replace(" ", "")
)
tweets_df.loc[tweets_df["hashtag"] == "", "hashtag"] = "NO HASHTAG"
# tweets_df
ds = tweets_df["hashtag"].value_counts().reset_index()
ds.columns = ["hashtag", "count"]
ds = ds.sort_values(["count"], ascending=False)
fig = sns.barplot(
x=ds.head(10)["count"],
y=ds.head(10)["hashtag"],
orientation="horizontal",
# title='Top 20 hashtags',
# width=800,
# height=700
).set_title("Top 10 hashtags")
# fig.show()
#
# Tweets text analysis
# ## Prevalent words in tweets
def build_wordcloud(df, title):
wordcloud = WordCloud(
background_color="black",
colormap="Oranges",
stopwords=set(STOPWORDS),
max_words=50,
max_font_size=40,
random_state=666,
).generate(str(df))
fig = plt.figure(1, figsize=(14, 14))
plt.axis("off")
fig.suptitle(title, fontsize=16)
fig.subplots_adjust(top=2.3)
plt.imshow(wordcloud)
plt.show()
build_wordcloud(f1["text"], "Prevalent words in tweets for all dataset")
# ## Prevalent words in tweets from India
india_df = f1.loc[
(f1.user_location == "United Kingdom") | (f1.user_location == "London, England")
]
build_wordcloud(india_df["text"], title="Prevalent words in tweets from UK")
india_df = f1.loc[f1.user_location == "Paris"]
build_wordcloud(india_df["text"], title="Prevalent words in tweets from Paris")
india_df = f1.loc[f1.user_location == "India"]
build_wordcloud(india_df["text"], title="Prevalent words in tweets from India")
# ## Refining the text (Important step)
def remove_tag(string):
text = re.sub("<.*?>", "", string)
return text
def remove_mention(text):
line = re.sub(r"@\w+", "", text)
return line
def remove_hash(text):
line = re.sub(r"#\w+", "", text)
return line
def remove_newline(string):
text = re.sub("\n", "", string)
return text
def remove_url(string):
text = re.sub(
"http[s]?://(?:[a-zA-Z]|[0-9]|[$-_@.&+]|[!*\(\),]|(?:%[0-9a-fA-F][0-9a-fA-F]))+",
"",
string,
)
return text
def remove_number(text):
line = re.sub(r"[0-9]+", "", text)
return line
def remove_punct(text):
line = re.sub(r'[!"\$%&\'()*+,\-.\/:;=#@?\[\\\]^_`{|}~]*', "", text)
return line
def text_strip(string):
line = re.sub("\s{2,}", " ", string.strip())
return line
def remove_thi_amp_ha_words(string):
line = re.sub(r"\bamp\b|\bthi\b|\bha\b", " ", string)
return line
f1["refine_text"] = f1["text"].str.lower()
f1["refine_text"] = f1["refine_text"].apply(lambda x: remove_tag(str(x)))
f1["refine_text"] = f1["refine_text"].apply(lambda x: remove_mention(str(x)))
f1["refine_text"] = f1["refine_text"].apply(lambda x: remove_hash(str(x)))
f1["refine_text"] = f1["refine_text"].apply(lambda x: remove_newline(x))
f1["refine_text"] = f1["refine_text"].apply(lambda x: remove_url(x))
f1["refine_text"] = f1["refine_text"].apply(lambda x: remove_number(x))
f1["refine_text"] = f1["refine_text"].apply(lambda x: remove_punct(x))
f1["refine_text"] = f1["refine_text"].apply(lambda x: remove_thi_amp_ha_words(x))
f1["refine_text"] = f1["refine_text"].apply(lambda x: text_strip(x))
f1["text_length"] = f1["refine_text"].str.split().map(lambda x: len(x))
# ## The average length for a f1 Tweet using violin plot
fig = go.Figure(
data=go.Violin(
y=f1["text_length"],
box_visible=True,
line_color="black",
meanline_visible=True,
fillcolor="royalblue",
opacity=0.6,
x0="Tweet Text Length",
)
)
fig.update_layout(
yaxis_zeroline=False, title="Distribution of Text length", template="ggplot2"
)
fig.show()
# * Average length of the f12020 tweet: 14.36
# * Median length of the f1 2020 tweet:11
# * Interquartile lie between : 6 and 19
# * Min: 1
# * Max: 58
# ## N-GRAM
# ## Listing below the top N-gram sequential words used in f1 tweets
def ngram_df(corpus, nrange, n=None):
vec = CountVectorizer(stop_words="english", ngram_range=nrange).fit(corpus)
bag_of_words = vec.transform(corpus)
sum_words = bag_of_words.sum(axis=0)
words_freq = [(word, sum_words[0, idx]) for word, idx in vec.vocabulary_.items()]
words_freq = sorted(words_freq, key=lambda x: x[1], reverse=True)
total_list = words_freq[:n]
df = pd.DataFrame(total_list, columns=["text", "count"])
return df
unigram_df = ngram_df(f1["refine_text"], (1, 1), 20)
bigram_df = ngram_df(f1["refine_text"], (2, 2), 20)
trigram_df = ngram_df(f1["refine_text"], (3, 3), 20)
fig = make_subplots(
rows=3,
cols=1,
subplot_titles=("Unigram", "Bigram", "Trigram"),
specs=[[{"type": "scatter"}], [{"type": "scatter"}], [{"type": "scatter"}]],
)
fig.add_trace(
go.Bar(
y=unigram_df["text"][::-1],
x=unigram_df["count"][::-1],
marker={"color": "blue"},
text=unigram_df["count"],
textposition="outside",
orientation="h",
name="Months",
),
row=1,
col=1,
)
fig.add_trace(
go.Bar(
y=bigram_df["text"][::-1],
x=bigram_df["count"][::-1],
marker={"color": "blue"},
text=bigram_df["count"],
name="Days",
textposition="outside",
orientation="h",
),
row=2,
col=1,
)
fig.add_trace(
go.Bar(
y=trigram_df["text"][::-1],
x=trigram_df["count"][::-1],
marker={"color": "blue"},
text=trigram_df["count"],
name="Days",
orientation="h",
textposition="outside",
),
row=3,
col=1,
)
fig.update_xaxes(showline=True, linewidth=2, linecolor="black", mirror=True)
fig.update_yaxes(showline=True, linewidth=2, linecolor="black", mirror=True)
fig.update_layout(
title_text="Top N Grams",
xaxis_title=" ",
yaxis_title=" ",
showlegend=False,
title_x=0.5,
height=1200,
template="plotly_dark",
)
fig.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/826/69826100.ipynb
|
formula-1-trending-tweets
|
kaushiksuresh147
|
[{"Id": 69826100, "ScriptId": 19072419, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1430847, "CreationDate": "08/03/2021 13:10:07", "VersionNumber": 2.0, "Title": "Formula 1 trending tweets EDA \ud83c\udfce\ud83c\udfc1\ud83c\udfc6", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 488.0, "LinesInsertedFromPrevious": 3.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 485.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 93278913, "KernelVersionId": 69826100, "SourceDatasetVersionId": 2491379}]
|
[{"Id": 2491379, "DatasetId": 1508196, "DatasourceVersionId": 2533955, "CreatorUserId": 1430847, "LicenseName": "CC0: Public Domain", "CreationDate": "08/02/2021 12:08:50", "VersionNumber": 1.0, "Title": "Formula 1 (F1) trending tweets \ud83c\udfce \ud83c\udfc1", "Slug": "formula-1-trending-tweets", "Subtitle": "Tweets posted with the trending #f1 hashtag", "Description": "\n\n\n- Formula One (also known as Formula 1 or F1) is the highest class of international auto racing for single-seater formula racing cars sanctioned by the F\u00e9d\u00e9ration Internationale de l'Automobile (FIA). The World Drivers' Championship, which became the FIA Formula One World Championship in 1981, has been one of the premier forms of racing around the world since its inaugural season in 1950. The word formula in the name refers to the set of rules to which all participants' cars must conform. A Formula One season consists of a series of races, known as Grands Prix, which take place worldwide on both purpose-built circuits and closed public roads.\n\n- The craze for F1 among the fans is astonishing, which has been creating quite a buzz in major social media platforms like Twitter. The dataset brings you such tweets posted with the #f1 hashtag. \n\n\n\n### Information regarding the data \n\n- The data totally consists of 50k+ records with 13 columns. The collection started on 25/7/2020 and will be updated regularly. The description of the features is given below.\n\n### Inspiration\n \"I am an artist, the track is my canvas and the car is my brush.\" \u2013 Graham Hill", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1508196, "CreatorUserId": 1430847, "OwnerUserId": 1430847.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 4108887.0, "CurrentDatasourceVersionId": 4165152.0, "ForumId": 1527943, "Type": 2, "CreationDate": "08/02/2021 12:08:50", "LastActivityDate": "08/02/2021", "TotalViews": 6537, "TotalDownloads": 232, "TotalVotes": 11, "TotalKernels": 1}]
|
[{"Id": 1430847, "UserName": "kaushiksuresh147", "DisplayName": "Kash", "RegisterDate": "11/19/2017", "PerformanceTier": 3}]
|
# 
#
# - Formula One (also known as Formula 1 or F1) is the highest class of international auto racing for single-seater formula racing cars sanctioned by the Fédération Internationale de l'Automobile (FIA). The World Drivers' Championship, which became the FIA Formula One World Championship in 1981, has been one of the premier forms of racing around the world since its inaugural season in 1950. The word formula in the name refers to the set of rules to which all participants' cars must conform. A Formula One season consists of a series of races, known as Grands Prix, which take place worldwide on both purpose-built circuits and closed public roads.
# - The craze for F1 among the fans is astonishing, which has been creating quite a buzz in major social media platforms like Twitter. The dataset brings you such tweets posted with the #f1 hashtag.
# This notebook analyses the tweets with the trending #f1 hashtag. So grab your gloves and fasten your seatbelts and let's analyze the impact of f1 in social media platforms like Twitter
#
# 
# Quick navigation
# * [1. Required Libraries](#1)
# * [2. Dataset Quick Overview](#2)
# * [3. Tweets EDA](#3)
# * [4. Tweets text analysis](#4)
# Kindly, Upvote the notebook!
# Required Libraries
import numpy as np
import pandas as pd
import os
import itertools
# plots
import matplotlib.pyplot as plt
import plotly.express as px
import plotly.graph_objects as go
import plotly.figure_factory as ff
from plotly.colors import n_colors
from plotly.subplots import make_subplots
from wordcloud import WordCloud, STOPWORDS, ImageColorGenerator
from sklearn.feature_extraction.text import CountVectorizer
from PIL import Image
from nltk.corpus import stopwords
stop = set(stopwords.words("english"))
from nltk.util import ngrams
import re
from collections import Counter
import nltk
from nltk.corpus import stopwords
import requests
import json
import seaborn as sns
sns.set(rc={"figure.figsize": (11.7, 8.27)})
import warnings
warnings.filterwarnings("ignore")
#
# Dataset Quick Overview
# ## Let's get some basic information about the data!
f1 = pd.read_csv("../input/formula-1-trending-tweets/F1_tweets.csv")
f1.info()
f1.shape
# ## Let's visualize some missing values!
import missingno as mno
mno.matrix(f1)
missed = pd.DataFrame()
missed["column"] = f1.columns
missed["percent"] = [
round(100 * f1[col].isnull().sum() / len(f1), 2) for col in f1.columns
]
missed = missed.sort_values("percent", ascending=False)
missed = missed[missed["percent"] > 0]
fig = sns.barplot(
x=missed["percent"], y=missed["column"], orientation="horizontal"
).set_title("Missed values percent for every column")
#
# Tweets EDA
# ## Lets Visualize the top 20 users by number of tweets
#
ds = f1["user_name"].value_counts().reset_index()
ds.columns = ["user_name", "tweets_count"]
ds = ds.sort_values(["tweets_count"], ascending=False)
f1 = pd.merge(f1, ds, on="user_name")
fig = sns.barplot(
x=ds.head(20)["tweets_count"], y=ds.head(20)["user_name"], orientation="horizontal"
).set_title("Top 20 users by number of tweets")
# ## Users created - yearwise
#
f1["user_created"] = pd.to_datetime(
f1["user_created"], infer_datetime_format=True, errors="coerce"
)
f1["year_created"] = f1["user_created"].dt.year
data = f1.drop_duplicates(subset="user_name", keep="first")
data = data[data["year_created"] > 1970]
data = data["year_created"].value_counts().reset_index()
data.columns = ["year", "number"]
fig = sns.barplot(
x=data["year"],
y=data["number"],
orientation="vertical"
# title='',
).set_title("User created year by year")
# ## Top 20 Users location based on the number of tweets
ds = f1["user_location"].value_counts().reset_index()
ds.columns = ["user_location", "count"]
ds = ds[ds["user_location"] != "NA"]
ds = ds.sort_values(["count"], ascending=False)
fig = sns.barplot(
x=ds.head(20)["count"], y=ds.head(20)["user_location"], orientation="horizontal"
).set_title("Top 20 user locations by number of tweets")
# ## Visualizing the number of tweets per location!!
from plotly.offline import init_notebook_mode, iplot
def pie_count(data, field, percent_limit, title):
data[field] = data[field].fillna("NA")
data = data[field].value_counts().to_frame()
total = data[field].sum()
data["percentage"] = 100 * data[field] / total
percent_limit = percent_limit
otherdata = data[data["percentage"] < percent_limit]
others = otherdata["percentage"].sum()
maindata = data[data["percentage"] >= percent_limit]
data = maindata
other_label = "Others(<" + str(percent_limit) + "% each)"
data.loc[other_label] = pd.Series({field: otherdata[field].sum()})
labels = data.index.tolist()
datavals = data[field].tolist()
trace = go.Pie(labels=labels, values=datavals)
layout = go.Layout(title=title, height=600, width=600)
fig = go.Figure(data=[trace], layout=layout)
iplot(fig)
pie_count(f1, "user_location", 0.5, "Number of tweets per location")
# ## Top 10 user sources by number of tweets
ds = f1["source"].value_counts().reset_index()
ds.columns = ["source", "count"]
ds = ds.sort_values(["count"], ascending=False)
fig = sns.barplot(
x=ds.head(10)["count"],
y=ds.head(10)["source"],
orientation="horizontal",
# title='Top 40 user sources by number of tweets',
# width=800,
# height=800
).set_title("Top 10 user sources by number of tweets")
# ## Total number of tweets for users and number of hashtags in every tweet
f1["hashtags"] = f1["hashtags"].fillna("[]")
f1["hashtags_count"] = f1["hashtags"].apply(lambda x: len(x.split(",")))
f1.loc[f1["hashtags"] == "[]", "hashtags_count"] = 0
fig = sns.scatterplot(x=f1["hashtags_count"], y=f1["tweets_count"]).set_title(
"Total number of tweets for users and number of hashtags in every tweet"
)
#
# * users who post 100 tweets use a range of 1 to a maximum of 33 hastags!
# ## Number of hashtags used in each tweet
ds = f1["hashtags_count"].value_counts().reset_index()
ds.columns = ["hashtags_count", "count"]
ds = ds.sort_values(["count"], ascending=False)
ds["hashtags_count"] = ds["hashtags_count"].astype(str) + " tags"
fig = sns.barplot(
x=ds["count"], y=ds["hashtags_count"], orientation="horizontal"
).set_title("Distribution of number of hashtags in tweets")
# * Most users use 2 hastag followed by 1 hashtag
# * Very less amount of people use more than 5 hashtags in their post
# ## Number of unqiue users each day!
f1["date"] = pd.to_datetime(f1["date"], infer_datetime_format=True, errors="coerce")
df = f1.sort_values(["date"])
df["day"] = df["date"].astype(str).str.split(" ", expand=True)[0]
df["time"] = df["date"].astype(str).str.split(" ", expand=True)[1]
df.head()
ds = df.groupby(["day", "user_name"])["hashtags_count"].count().reset_index()
ds = ds.groupby(["day"])["user_name"].count().reset_index()
ds.columns = ["day", "number_of_users"]
ds["day"] = ds["day"].astype(str)
fig = sns.barplot(
x=ds["day"],
y=ds["number_of_users"],
orientation="vertical",
# title='Number of unique users per day',
# width=800,
# height=800
).set_title("Number of unique users per day")
# fig.show()
plt.xticks(rotation=90)
# ## Tweets distribution over days present in dataset
ds = df["day"].value_counts().reset_index()
ds.columns = ["day", "count"]
ds = ds.sort_values("count", ascending=False)
ds["day"] = ds["day"].astype(str)
fig = sns.barplot(
x=ds["count"],
y=ds["day"],
orientation="horizontal",
).set_title("Tweets distribution over days present in dataset")
# ## Tweets per day
f1["tweet_date"] = f1["date"].dt.date
tweet_date = (
f1["tweet_date"]
.value_counts()
.to_frame()
.reset_index()
.rename(columns={"index": "date", "tweet_date": "count"})
)
tweet_date["date"] = pd.to_datetime(
tweet_date["date"], infer_datetime_format=True, errors="coerce"
)
tweet_date = tweet_date.sort_values("date", ascending=False)
fig = go.Figure(
go.Scatter(
x=tweet_date["date"],
y=tweet_date["count"],
mode="markers+lines",
name="Submissions",
marker_color="dodgerblue",
)
)
f1_dummy = f1.dropna(subset=["tweet_date"])
fig.update_layout(
title_text="Tweets per Day : ({} - {})".format(
f1_dummy["tweet_date"].sort_values()[0].strftime("%d/%m/%Y"),
f1_dummy["tweet_date"].sort_values().iloc[-1].strftime("%d/%m/%Y"),
),
template="plotly_dark",
title_x=0.5,
)
fig.show()
# ## Tweet distribution - hourly
f1["hour"] = f1["date"].dt.hour
ds = f1["hour"].value_counts().reset_index()
ds.columns = ["hour", "count"]
ds["hour"] = "Hour " + ds["hour"].astype(str)
fig = sns.barplot(
x=ds["hour"],
y=ds["count"],
orientation="vertical",
).set_title("Tweets distribution over hours")
plt.xticks(rotation="vertical")
# ### Top 10 hastags used in the tweet!
def split_hashtags(x):
return str(x).replace("[", "").replace("]", "").split(",")
tweets_df = f1.copy()
tweets_df["hashtag"] = tweets_df["hashtags"].apply(lambda row: split_hashtags(row))
tweets_df = tweets_df.explode("hashtag")
tweets_df["hashtag"] = (
tweets_df["hashtag"]
.astype(str)
.str.lower()
.str.replace("'", "")
.str.replace(" ", "")
)
tweets_df.loc[tweets_df["hashtag"] == "", "hashtag"] = "NO HASHTAG"
# tweets_df
ds = tweets_df["hashtag"].value_counts().reset_index()
ds.columns = ["hashtag", "count"]
ds = ds.sort_values(["count"], ascending=False)
fig = sns.barplot(
x=ds.head(10)["count"],
y=ds.head(10)["hashtag"],
orientation="horizontal",
# title='Top 20 hashtags',
# width=800,
# height=700
).set_title("Top 10 hashtags")
# fig.show()
#
# Tweets text analysis
# ## Prevalent words in tweets
def build_wordcloud(df, title):
wordcloud = WordCloud(
background_color="black",
colormap="Oranges",
stopwords=set(STOPWORDS),
max_words=50,
max_font_size=40,
random_state=666,
).generate(str(df))
fig = plt.figure(1, figsize=(14, 14))
plt.axis("off")
fig.suptitle(title, fontsize=16)
fig.subplots_adjust(top=2.3)
plt.imshow(wordcloud)
plt.show()
build_wordcloud(f1["text"], "Prevalent words in tweets for all dataset")
# ## Prevalent words in tweets from India
india_df = f1.loc[
(f1.user_location == "United Kingdom") | (f1.user_location == "London, England")
]
build_wordcloud(india_df["text"], title="Prevalent words in tweets from UK")
india_df = f1.loc[f1.user_location == "Paris"]
build_wordcloud(india_df["text"], title="Prevalent words in tweets from Paris")
india_df = f1.loc[f1.user_location == "India"]
build_wordcloud(india_df["text"], title="Prevalent words in tweets from India")
# ## Refining the text (Important step)
def remove_tag(string):
text = re.sub("<.*?>", "", string)
return text
def remove_mention(text):
line = re.sub(r"@\w+", "", text)
return line
def remove_hash(text):
line = re.sub(r"#\w+", "", text)
return line
def remove_newline(string):
text = re.sub("\n", "", string)
return text
def remove_url(string):
text = re.sub(
"http[s]?://(?:[a-zA-Z]|[0-9]|[$-_@.&+]|[!*\(\),]|(?:%[0-9a-fA-F][0-9a-fA-F]))+",
"",
string,
)
return text
def remove_number(text):
line = re.sub(r"[0-9]+", "", text)
return line
def remove_punct(text):
line = re.sub(r'[!"\$%&\'()*+,\-.\/:;=#@?\[\\\]^_`{|}~]*', "", text)
return line
def text_strip(string):
line = re.sub("\s{2,}", " ", string.strip())
return line
def remove_thi_amp_ha_words(string):
line = re.sub(r"\bamp\b|\bthi\b|\bha\b", " ", string)
return line
f1["refine_text"] = f1["text"].str.lower()
f1["refine_text"] = f1["refine_text"].apply(lambda x: remove_tag(str(x)))
f1["refine_text"] = f1["refine_text"].apply(lambda x: remove_mention(str(x)))
f1["refine_text"] = f1["refine_text"].apply(lambda x: remove_hash(str(x)))
f1["refine_text"] = f1["refine_text"].apply(lambda x: remove_newline(x))
f1["refine_text"] = f1["refine_text"].apply(lambda x: remove_url(x))
f1["refine_text"] = f1["refine_text"].apply(lambda x: remove_number(x))
f1["refine_text"] = f1["refine_text"].apply(lambda x: remove_punct(x))
f1["refine_text"] = f1["refine_text"].apply(lambda x: remove_thi_amp_ha_words(x))
f1["refine_text"] = f1["refine_text"].apply(lambda x: text_strip(x))
f1["text_length"] = f1["refine_text"].str.split().map(lambda x: len(x))
# ## The average length for a f1 Tweet using violin plot
fig = go.Figure(
data=go.Violin(
y=f1["text_length"],
box_visible=True,
line_color="black",
meanline_visible=True,
fillcolor="royalblue",
opacity=0.6,
x0="Tweet Text Length",
)
)
fig.update_layout(
yaxis_zeroline=False, title="Distribution of Text length", template="ggplot2"
)
fig.show()
# * Average length of the f12020 tweet: 14.36
# * Median length of the f1 2020 tweet:11
# * Interquartile lie between : 6 and 19
# * Min: 1
# * Max: 58
# ## N-GRAM
# ## Listing below the top N-gram sequential words used in f1 tweets
def ngram_df(corpus, nrange, n=None):
vec = CountVectorizer(stop_words="english", ngram_range=nrange).fit(corpus)
bag_of_words = vec.transform(corpus)
sum_words = bag_of_words.sum(axis=0)
words_freq = [(word, sum_words[0, idx]) for word, idx in vec.vocabulary_.items()]
words_freq = sorted(words_freq, key=lambda x: x[1], reverse=True)
total_list = words_freq[:n]
df = pd.DataFrame(total_list, columns=["text", "count"])
return df
unigram_df = ngram_df(f1["refine_text"], (1, 1), 20)
bigram_df = ngram_df(f1["refine_text"], (2, 2), 20)
trigram_df = ngram_df(f1["refine_text"], (3, 3), 20)
fig = make_subplots(
rows=3,
cols=1,
subplot_titles=("Unigram", "Bigram", "Trigram"),
specs=[[{"type": "scatter"}], [{"type": "scatter"}], [{"type": "scatter"}]],
)
fig.add_trace(
go.Bar(
y=unigram_df["text"][::-1],
x=unigram_df["count"][::-1],
marker={"color": "blue"},
text=unigram_df["count"],
textposition="outside",
orientation="h",
name="Months",
),
row=1,
col=1,
)
fig.add_trace(
go.Bar(
y=bigram_df["text"][::-1],
x=bigram_df["count"][::-1],
marker={"color": "blue"},
text=bigram_df["count"],
name="Days",
textposition="outside",
orientation="h",
),
row=2,
col=1,
)
fig.add_trace(
go.Bar(
y=trigram_df["text"][::-1],
x=trigram_df["count"][::-1],
marker={"color": "blue"},
text=trigram_df["count"],
name="Days",
orientation="h",
textposition="outside",
),
row=3,
col=1,
)
fig.update_xaxes(showline=True, linewidth=2, linecolor="black", mirror=True)
fig.update_yaxes(showline=True, linewidth=2, linecolor="black", mirror=True)
fig.update_layout(
title_text="Top N Grams",
xaxis_title=" ",
yaxis_title=" ",
showlegend=False,
title_x=0.5,
height=1200,
template="plotly_dark",
)
fig.show()
|
[{"formula-1-trending-tweets/F1_tweets.csv": {"column_names": "[\"user_name\", \"user_location\", \"user_description\", \"user_created\", \"user_followers\", \"user_friends\", \"user_favourites\", \"user_verified\", \"date\", \"text\", \"hashtags\", \"source\", \"is_retweet\"]", "column_data_types": "{\"user_name\": \"object\", \"user_location\": \"object\", \"user_description\": \"object\", \"user_created\": \"object\", \"user_followers\": \"float64\", \"user_friends\": \"object\", \"user_favourites\": \"object\", \"user_verified\": \"object\", \"date\": \"object\", \"text\": \"object\", \"hashtags\": \"object\", \"source\": \"object\", \"is_retweet\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 632388 entries, 0 to 632387\nData columns (total 13 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 user_name 632382 non-null object \n 1 user_location 443577 non-null object \n 2 user_description 592917 non-null object \n 3 user_created 632388 non-null object \n 4 user_followers 632388 non-null float64\n 5 user_friends 632388 non-null object \n 6 user_favourites 632388 non-null object \n 7 user_verified 632388 non-null object \n 8 date 632388 non-null object \n 9 text 632388 non-null object \n 10 hashtags 632360 non-null object \n 11 source 632384 non-null object \n 12 is_retweet 632384 non-null object \ndtypes: float64(1), object(12)\nmemory usage: 62.7+ MB\n", "summary": "{\"user_followers\": {\"count\": 632388.0, \"mean\": 45581.93898998716, \"std\": 490349.57707797847, \"min\": 0.0, \"25%\": 107.0, \"50%\": 438.0, \"75%\": 2039.0, \"max\": 24096107.0}}", "examples": "{\"user_name\":{\"0\":\"Zack Shephard\",\"1\":\"Matteo Vannucci\",\"2\":\"Michelle\",\"3\":\"JoshWFC \\ud83c\\udde7\\ud83c\\uddf7\"},\"user_location\":{\"0\":null,\"1\":null,\"2\":\"The Burbs\",\"3\":null},\"user_description\":{\"0\":\"My opinions are mine ALONE. Venting is the only way that I can get everthing off my chest. #BeKind #FoxNewsIsGoingToHell #BlackLivesMatter\",\"1\":\"Free speech, free ideas, random thoughts\",\"2\":\"likes- movies, candy, being lazy, formula 1, 65\\u00b0F, watching various sports & reality shows\\n\\ndislikes- phone calls, bugs, hot weather, most foods, & group texts\",\"3\":\"| JP10\\u2019s biggest fan | pronouns : Jo\\u00e3o\\/Pedro | @watfordfc | @RamsNFL | @GeorgeRussell63\"},\"user_created\":{\"0\":\"2011-12-03 20:36:55\",\"1\":\"2010-05-28 01:02:32\",\"2\":\"2009-03-04 16:53:13\",\"3\":\"2021-04-05 16:14:53\"},\"user_followers\":{\"0\":89.0,\"1\":10.0,\"2\":231.0,\"3\":156.0},\"user_friends\":{\"0\":2598,\"1\":40,\"2\":110,\"3\":443},\"user_favourites\":{\"0\":212886,\"1\":8,\"2\":428,\"3\":2994},\"user_verified\":{\"0\":false,\"1\":false,\"2\":false,\"3\":false},\"date\":{\"0\":\"2021-08-01 23:59:18\",\"1\":\"2021-08-01 23:59:10\",\"2\":\"2021-08-01 23:58:48\",\"3\":\"2021-08-01 23:58:11\"},\"text\":{\"0\":\"The next great #F1 duo!!! @ESPNF1 https:\\/\\/t.co\\/WMhaoAdSxF\",\"1\":\"@F1 What if\\u2026Hamilton would have gone to the pit\\u2026empty grid for a start?!\\ud83e\\udd14 something to think about #HungarianGP #F1\",\"2\":\"This has been a tiring #f1 day... https:\\/\\/t.co\\/34ggWgAlT6\",\"3\":\"Valteri Bottass tracking down Max Verstappen on the way home\\n\\n#F1 https:\\/\\/t.co\\/epTHlDyjU6\"},\"hashtags\":{\"0\":\"['F1']\",\"1\":\"['HungarianGP', 'F1']\",\"2\":\"['f1']\",\"3\":\"['F1']\"},\"source\":{\"0\":\"Twitter for iPhone\",\"1\":\"Twitter for iPhone\",\"2\":\"Twitter for Android\",\"3\":\"Twitter for iPhone\"},\"is_retweet\":{\"0\":false,\"1\":false,\"2\":false,\"3\":false}}"}}]
| true | 1 |
<start_data_description><data_path>formula-1-trending-tweets/F1_tweets.csv:
<column_names>
['user_name', 'user_location', 'user_description', 'user_created', 'user_followers', 'user_friends', 'user_favourites', 'user_verified', 'date', 'text', 'hashtags', 'source', 'is_retweet']
<column_types>
{'user_name': 'object', 'user_location': 'object', 'user_description': 'object', 'user_created': 'object', 'user_followers': 'float64', 'user_friends': 'object', 'user_favourites': 'object', 'user_verified': 'object', 'date': 'object', 'text': 'object', 'hashtags': 'object', 'source': 'object', 'is_retweet': 'object'}
<dataframe_Summary>
{'user_followers': {'count': 632388.0, 'mean': 45581.93898998716, 'std': 490349.57707797847, 'min': 0.0, '25%': 107.0, '50%': 438.0, '75%': 2039.0, 'max': 24096107.0}}
<dataframe_info>
RangeIndex: 632388 entries, 0 to 632387
Data columns (total 13 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 user_name 632382 non-null object
1 user_location 443577 non-null object
2 user_description 592917 non-null object
3 user_created 632388 non-null object
4 user_followers 632388 non-null float64
5 user_friends 632388 non-null object
6 user_favourites 632388 non-null object
7 user_verified 632388 non-null object
8 date 632388 non-null object
9 text 632388 non-null object
10 hashtags 632360 non-null object
11 source 632384 non-null object
12 is_retweet 632384 non-null object
dtypes: float64(1), object(12)
memory usage: 62.7+ MB
<some_examples>
{'user_name': {'0': 'Zack Shephard', '1': 'Matteo Vannucci', '2': 'Michelle', '3': 'JoshWFC 🇧🇷'}, 'user_location': {'0': None, '1': None, '2': 'The Burbs', '3': None}, 'user_description': {'0': 'My opinions are mine ALONE. Venting is the only way that I can get everthing off my chest. #BeKind #FoxNewsIsGoingToHell #BlackLivesMatter', '1': 'Free speech, free ideas, random thoughts', '2': 'likes- movies, candy, being lazy, formula 1, 65°F, watching various sports & reality shows\n\ndislikes- phone calls, bugs, hot weather, most foods, & group texts', '3': '| JP10’s biggest fan | pronouns : João/Pedro | @watfordfc | @RamsNFL | @GeorgeRussell63'}, 'user_created': {'0': '2011-12-03 20:36:55', '1': '2010-05-28 01:02:32', '2': '2009-03-04 16:53:13', '3': '2021-04-05 16:14:53'}, 'user_followers': {'0': 89.0, '1': 10.0, '2': 231.0, '3': 156.0}, 'user_friends': {'0': 2598, '1': 40, '2': 110, '3': 443}, 'user_favourites': {'0': 212886, '1': 8, '2': 428, '3': 2994}, 'user_verified': {'0': False, '1': False, '2': False, '3': False}, 'date': {'0': '2021-08-01 23:59:18', '1': '2021-08-01 23:59:10', '2': '2021-08-01 23:58:48', '3': '2021-08-01 23:58:11'}, 'text': {'0': 'The next great #F1 duo!!! @ESPNF1 https://t.co/WMhaoAdSxF', '1': '@F1 What if…Hamilton would have gone to the pit…empty grid for a start?!🤔 something to think about #HungarianGP #F1', '2': 'This has been a tiring #f1 day... https://t.co/34ggWgAlT6', '3': 'Valteri Bottass tracking down Max Verstappen on the way home\n\n#F1 https://t.co/epTHlDyjU6'}, 'hashtags': {'0': "['F1']", '1': "['HungarianGP', 'F1']", '2': "['f1']", '3': "['F1']"}, 'source': {'0': 'Twitter for iPhone', '1': 'Twitter for iPhone', '2': 'Twitter for Android', '3': 'Twitter for iPhone'}, 'is_retweet': {'0': False, '1': False, '2': False, '3': False}}
<end_description>
| 5,089 | 0 | 6,818 | 5,089 |
69826889
|
# ------------------------------------------------------------------------------------------------------------------------------
import pandas as pd # Importing for panel data analysis
from pandas_profiling import (
ProfileReport,
) # Import Pandas Profiling (To generate Univariate Analysis)
pd.set_option(
"display.max_columns", None
) # Unfolding hidden features if the cardinality is high
pd.set_option(
"display.max_rows", None
) # Unfolding hidden data points if the cardinality is high
pd.set_option(
"mode.chained_assignment", None
) # Removing restriction over chained assignments operations
# -------------------------------------------------------------------------------------------------------------------------------
import numpy as np # Importing package numpys (For Numerical Python)
# -------------------------------------------------------------------------------------------------------------------------------
import matplotlib.pyplot as plt # Importing pyplot interface using matplotlib
import seaborn as sns # Importin seaborm library for interactive visualization
# -------------------------------------------------------------------------------------------------------------------------------
from sklearn.preprocessing import (
StandardScaler,
) # To scaled data with mean 0 and variance 1
from sklearn.model_selection import (
train_test_split,
) # To split the data in training and testing part
from sklearn.tree import DecisionTreeClassifier # To implement decision tree classifier
from sklearn.tree import export_graphviz
from sklearn.metrics import classification_report # To generate classification report
from sklearn.metrics import plot_confusion_matrix # To plot confusion matrix
from IPython.display import Image # To generate image using pydot file
from sklearn.model_selection import (
GridSearchCV,
) # To find best hyperparamter setting for the algorithm
# -------------------------------------------------------------------------------------------------------------------------------
import warnings # Importing warning to disable runtime warnings
warnings.filterwarnings("ignore") # Warnings will appear only once
# Load the data
data = pd.read_csv(filepath_or_buffer="../input/titanic/train.csv")
# Get the dimesions of data
print("Shape of the dataset:", data.shape)
# Output first 5 data rows
data.head()
data.describe()
# **Observations:**
# **Survived:**
# More than 50% did not survive the accident.
# **Pclass:**
# There are a lot more 3rd class passengers than 1st and 2nd class.
# We can also see that there are more 2nd class passengers than 1st class passengers.
# **SibSp:**
# More than 50% of passengers are not travelling with their siblings or a spouse.
# There are some passengers who are travelling with as maximum as 8 siblings and spouse.
# **Parch:**
# More than 75% passengers are not travelling with a parent or children
# But there are some passengers who have a maximum number of 6 children and/or parents with them on the ship.
# We observe that a vast majority of passengers are not travelling with their family members.
# **Age:**
# The average age of passengers is around 29 years while the minimum and maximum ages are 0.4 years and 80 years respectively.
# There is some missing data in the Age feature.
# **Fare:**
# The average price of ticket seems to be £32.2. Minimum price of the ticket is recorded as £0 and maximum price recorded as high as £512.32.
# More than 50% of the passengers have paid atleast £14
# More than 75% passengers have paid atleast £7 for their ticket whereas less than 25% have paid for more than £31.
# We have to replace the minimum value in the Fare feature with a reasonable value.
profile = ProfileReport(df=data)
profile.to_file(output_file="Pre Profiling Report.html")
print("Accomplished!")
# Observations:
# The report shows that there are a total of 12 features out of which 7 are numerical and 5 are categorical.
# Only 342 passengers out of 891 survived the accident.
# Name, Ticket, and Cabin features have high cardinality and are uniformly distributed.
# PassengerId is having a uniformly distribution in the dataset.
# Fare feature is highly skewed towards right while Age feature is faily symmetrical.
# There are no duplicate rows in the dataset while a lot of zeros are present in Fare, Sibsp and Parch features.
# We can observe that 8.1% of data in cells is missing:
# Age (17 missing values) → Fill with median.
# Cabin (687 missing values) → Fill with median.
# Embarked (2 missing values) → Fill with mode.
# Embarked feature has just 2 missing values.
# For detailed information, check the Pre Profiling Report.html file.
# Filling the missing values of Embarked feature with the mode of the feature.
data["Embarked"] = data["Embarked"].fillna(value=data["Embarked"].mode()[0])
# Filling the missing values of Age feature with the median age.
data["Age"].fillna(value=data["Age"].median(), inplace=True)
# Dropping the Cabin feature
data.drop(labels="Cabin", axis=1, inplace=True)
post_profile = ProfileReport(df=data)
post_profile.to_file(output_file="Post Profiling Report.html")
print("Accomplished!")
# Observations:
# You can compare the two reports, i.e Pre Profiling Report.html and Post Profiling Report.html.
# Observations in Post Profiling Report.html:
# In the Dataset info, Total Missing = 0.0%
# Number of features = 11
# You can see the difference in the Age feature in both the reports.
# A lot of zeros are present in Sibsp and Parch features. They won't be removed as they are necessary.
# We can observe that Pclass and Fare are highly correlated to each other inversely.
# A lot of inverse correlations are observed among the features.
# For detailed information, check the Post Profiling Report.html file.
# We can now begin the Exploratory Data Analysis.
# Creating a new feature FamilySize from Sibsp and Parch
data["FamilySize"] = data["SibSp"] + data["Parch"] + 1
data.head()
# Performing one hot encoding over sex, embarked, title
data = pd.get_dummies(data=data, columns=["Sex", "Embarked"])
data.head(2)
# Instatiatig input and output data by dropping unnecessary data features
X = data.drop(
labels=["PassengerId", "Name", "Ticket", "Age", "Fare", "Survived"], axis=1
)
y = data["Survived"]
# Instatiate a scaler object and performing transformation on age and fare
scaler = StandardScaler()
scaled_data = scaler.fit_transform(data[["Age", "Fare"]])
data2 = pd.DataFrame(data=scaled_data, columns=["Age", "Fare"])
data2.head(2)
finalX = pd.concat(objs=[X, data2], axis=1)
finalX.drop(["SibSp", "Parch"], axis=1, inplace=True)
finalX.head()
# Splitting data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(
finalX, y, test_size=0.20, random_state=42, stratify=y
)
# Display the shape of training and testing data
print("X_train shape: ", X_train.shape)
print("y_train shape: ", y_train.shape)
print("X_test shape: ", X_test.shape)
print("y_test shape: ", y_test.shape)
# Instantiate a decision tree classifier
dtc = DecisionTreeClassifier(random_state=42, class_weight="balanced")
dtc.fit(X_train, y_train)
# Predicting training and testing labels
y_train_pred_count = dtc.predict(X_train)
y_test_pred_count = dtc.predict(X_test)
# Plotting confusion maxtrix of train and test data
fig, (ax1, ax2) = plt.subplots(nrows=1, ncols=2, sharex=False, figsize=(15, 7))
plot_confusion_matrix(
estimator=dtc, X=X_train, y_true=y_train, values_format=".5g", cmap="YlGnBu", ax=ax1
)
plot_confusion_matrix(
estimator=dtc, X=X_test, y_true=y_test, values_format=".5g", cmap="YlGnBu", ax=ax2
)
ax1.set_title(label="Train Data", size=14)
ax2.set_title(label="Test Data", size=14)
ax1.grid(b=False)
ax2.grid(b=False)
plt.suptitle(t="Confusion Matrix", size=16)
plt.show()
# Observation:
# Train Data:
# Model predicted 430 instances correctly for negative class while 269 instances were predicted correctly for positive class.
# Model identified 4 instances negative but in actual they were positive.
# Model identified 9 instances positive but in actual they were negative.
# Test Data:
# Model predicted 90 instances correctly for negative class while 52 instances were predicted correctly for positive class.
# Model identified 17 instance negative but in actual it was positive.
# Model identified 20 instances positive but in actual they were negative.
train_report = classification_report(y_train, y_train_pred_count)
test_report = classification_report(y_test, y_test_pred_count)
print(" Training Report ")
print(train_report)
print(" Testing Report ")
print(test_report)
# Load the actual test data
testdata = pd.read_csv(filepath_or_buffer="../input/titanic/test.csv")
# Get the dimesions of data
print("Shape of the dataset:", testdata.shape)
# Output first 5 data rows
testdata.head()
testdata.describe()
testdata.isnull().sum()
# Filling the missing values of Age feature with the median age.
testdata["Age"].fillna(value=testdata["Age"].median(), inplace=True)
# Dropping the Cabin feature
testdata.drop(labels="Cabin", axis=1, inplace=True)
# Filling the missing value of Fare feature with the median Fare.
testdata["Fare"].fillna(value=testdata["Fare"].median(), inplace=True)
testdata.isnull().sum()
# Creating a new feature FamilySize from Sibsp and Parch
testdata["FamilySize"] = testdata["SibSp"] + testdata["Parch"] + 1
testdata = pd.get_dummies(data=testdata, columns=["Sex", "Embarked"])
testdata.head(2)
# dropping unnecessary data features
test = testdata.drop(
labels=["PassengerId", "SibSp", "Parch", "Name", "Ticket", "Age", "Fare"], axis=1
)
# performing transformation on age and fare
scaled_testdata = scaler.transform(testdata[["Age", "Fare"]])
data3 = pd.DataFrame(data=scaled_testdata, columns=["Age", "Fare"])
data3.head(2)
finaltest = pd.concat(objs=[test, data3], axis=1)
finaltest.head()
finaltest_pred = dtc.predict(finaltest)
finaltest_pred
finaltest.join(
pd.DataFrame(finaltest_pred)
) # = pd.concat(objs=[finaltest, finaltest_pred], axis=1)
print("Thank you")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/826/69826889.ipynb
| null | null |
[{"Id": 69826889, "ScriptId": 19081455, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7939984, "CreationDate": "08/03/2021 13:14:50", "VersionNumber": 1.0, "Title": "30 Days of Kaggle [Day 1]", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 249.0, "LinesInsertedFromPrevious": 249.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": 249.0, "LinesDeletedFromFork": 51.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 0.0, "TotalVotes": 2}]
| null | null | null | null |
# ------------------------------------------------------------------------------------------------------------------------------
import pandas as pd # Importing for panel data analysis
from pandas_profiling import (
ProfileReport,
) # Import Pandas Profiling (To generate Univariate Analysis)
pd.set_option(
"display.max_columns", None
) # Unfolding hidden features if the cardinality is high
pd.set_option(
"display.max_rows", None
) # Unfolding hidden data points if the cardinality is high
pd.set_option(
"mode.chained_assignment", None
) # Removing restriction over chained assignments operations
# -------------------------------------------------------------------------------------------------------------------------------
import numpy as np # Importing package numpys (For Numerical Python)
# -------------------------------------------------------------------------------------------------------------------------------
import matplotlib.pyplot as plt # Importing pyplot interface using matplotlib
import seaborn as sns # Importin seaborm library for interactive visualization
# -------------------------------------------------------------------------------------------------------------------------------
from sklearn.preprocessing import (
StandardScaler,
) # To scaled data with mean 0 and variance 1
from sklearn.model_selection import (
train_test_split,
) # To split the data in training and testing part
from sklearn.tree import DecisionTreeClassifier # To implement decision tree classifier
from sklearn.tree import export_graphviz
from sklearn.metrics import classification_report # To generate classification report
from sklearn.metrics import plot_confusion_matrix # To plot confusion matrix
from IPython.display import Image # To generate image using pydot file
from sklearn.model_selection import (
GridSearchCV,
) # To find best hyperparamter setting for the algorithm
# -------------------------------------------------------------------------------------------------------------------------------
import warnings # Importing warning to disable runtime warnings
warnings.filterwarnings("ignore") # Warnings will appear only once
# Load the data
data = pd.read_csv(filepath_or_buffer="../input/titanic/train.csv")
# Get the dimesions of data
print("Shape of the dataset:", data.shape)
# Output first 5 data rows
data.head()
data.describe()
# **Observations:**
# **Survived:**
# More than 50% did not survive the accident.
# **Pclass:**
# There are a lot more 3rd class passengers than 1st and 2nd class.
# We can also see that there are more 2nd class passengers than 1st class passengers.
# **SibSp:**
# More than 50% of passengers are not travelling with their siblings or a spouse.
# There are some passengers who are travelling with as maximum as 8 siblings and spouse.
# **Parch:**
# More than 75% passengers are not travelling with a parent or children
# But there are some passengers who have a maximum number of 6 children and/or parents with them on the ship.
# We observe that a vast majority of passengers are not travelling with their family members.
# **Age:**
# The average age of passengers is around 29 years while the minimum and maximum ages are 0.4 years and 80 years respectively.
# There is some missing data in the Age feature.
# **Fare:**
# The average price of ticket seems to be £32.2. Minimum price of the ticket is recorded as £0 and maximum price recorded as high as £512.32.
# More than 50% of the passengers have paid atleast £14
# More than 75% passengers have paid atleast £7 for their ticket whereas less than 25% have paid for more than £31.
# We have to replace the minimum value in the Fare feature with a reasonable value.
profile = ProfileReport(df=data)
profile.to_file(output_file="Pre Profiling Report.html")
print("Accomplished!")
# Observations:
# The report shows that there are a total of 12 features out of which 7 are numerical and 5 are categorical.
# Only 342 passengers out of 891 survived the accident.
# Name, Ticket, and Cabin features have high cardinality and are uniformly distributed.
# PassengerId is having a uniformly distribution in the dataset.
# Fare feature is highly skewed towards right while Age feature is faily symmetrical.
# There are no duplicate rows in the dataset while a lot of zeros are present in Fare, Sibsp and Parch features.
# We can observe that 8.1% of data in cells is missing:
# Age (17 missing values) → Fill with median.
# Cabin (687 missing values) → Fill with median.
# Embarked (2 missing values) → Fill with mode.
# Embarked feature has just 2 missing values.
# For detailed information, check the Pre Profiling Report.html file.
# Filling the missing values of Embarked feature with the mode of the feature.
data["Embarked"] = data["Embarked"].fillna(value=data["Embarked"].mode()[0])
# Filling the missing values of Age feature with the median age.
data["Age"].fillna(value=data["Age"].median(), inplace=True)
# Dropping the Cabin feature
data.drop(labels="Cabin", axis=1, inplace=True)
post_profile = ProfileReport(df=data)
post_profile.to_file(output_file="Post Profiling Report.html")
print("Accomplished!")
# Observations:
# You can compare the two reports, i.e Pre Profiling Report.html and Post Profiling Report.html.
# Observations in Post Profiling Report.html:
# In the Dataset info, Total Missing = 0.0%
# Number of features = 11
# You can see the difference in the Age feature in both the reports.
# A lot of zeros are present in Sibsp and Parch features. They won't be removed as they are necessary.
# We can observe that Pclass and Fare are highly correlated to each other inversely.
# A lot of inverse correlations are observed among the features.
# For detailed information, check the Post Profiling Report.html file.
# We can now begin the Exploratory Data Analysis.
# Creating a new feature FamilySize from Sibsp and Parch
data["FamilySize"] = data["SibSp"] + data["Parch"] + 1
data.head()
# Performing one hot encoding over sex, embarked, title
data = pd.get_dummies(data=data, columns=["Sex", "Embarked"])
data.head(2)
# Instatiatig input and output data by dropping unnecessary data features
X = data.drop(
labels=["PassengerId", "Name", "Ticket", "Age", "Fare", "Survived"], axis=1
)
y = data["Survived"]
# Instatiate a scaler object and performing transformation on age and fare
scaler = StandardScaler()
scaled_data = scaler.fit_transform(data[["Age", "Fare"]])
data2 = pd.DataFrame(data=scaled_data, columns=["Age", "Fare"])
data2.head(2)
finalX = pd.concat(objs=[X, data2], axis=1)
finalX.drop(["SibSp", "Parch"], axis=1, inplace=True)
finalX.head()
# Splitting data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(
finalX, y, test_size=0.20, random_state=42, stratify=y
)
# Display the shape of training and testing data
print("X_train shape: ", X_train.shape)
print("y_train shape: ", y_train.shape)
print("X_test shape: ", X_test.shape)
print("y_test shape: ", y_test.shape)
# Instantiate a decision tree classifier
dtc = DecisionTreeClassifier(random_state=42, class_weight="balanced")
dtc.fit(X_train, y_train)
# Predicting training and testing labels
y_train_pred_count = dtc.predict(X_train)
y_test_pred_count = dtc.predict(X_test)
# Plotting confusion maxtrix of train and test data
fig, (ax1, ax2) = plt.subplots(nrows=1, ncols=2, sharex=False, figsize=(15, 7))
plot_confusion_matrix(
estimator=dtc, X=X_train, y_true=y_train, values_format=".5g", cmap="YlGnBu", ax=ax1
)
plot_confusion_matrix(
estimator=dtc, X=X_test, y_true=y_test, values_format=".5g", cmap="YlGnBu", ax=ax2
)
ax1.set_title(label="Train Data", size=14)
ax2.set_title(label="Test Data", size=14)
ax1.grid(b=False)
ax2.grid(b=False)
plt.suptitle(t="Confusion Matrix", size=16)
plt.show()
# Observation:
# Train Data:
# Model predicted 430 instances correctly for negative class while 269 instances were predicted correctly for positive class.
# Model identified 4 instances negative but in actual they were positive.
# Model identified 9 instances positive but in actual they were negative.
# Test Data:
# Model predicted 90 instances correctly for negative class while 52 instances were predicted correctly for positive class.
# Model identified 17 instance negative but in actual it was positive.
# Model identified 20 instances positive but in actual they were negative.
train_report = classification_report(y_train, y_train_pred_count)
test_report = classification_report(y_test, y_test_pred_count)
print(" Training Report ")
print(train_report)
print(" Testing Report ")
print(test_report)
# Load the actual test data
testdata = pd.read_csv(filepath_or_buffer="../input/titanic/test.csv")
# Get the dimesions of data
print("Shape of the dataset:", testdata.shape)
# Output first 5 data rows
testdata.head()
testdata.describe()
testdata.isnull().sum()
# Filling the missing values of Age feature with the median age.
testdata["Age"].fillna(value=testdata["Age"].median(), inplace=True)
# Dropping the Cabin feature
testdata.drop(labels="Cabin", axis=1, inplace=True)
# Filling the missing value of Fare feature with the median Fare.
testdata["Fare"].fillna(value=testdata["Fare"].median(), inplace=True)
testdata.isnull().sum()
# Creating a new feature FamilySize from Sibsp and Parch
testdata["FamilySize"] = testdata["SibSp"] + testdata["Parch"] + 1
testdata = pd.get_dummies(data=testdata, columns=["Sex", "Embarked"])
testdata.head(2)
# dropping unnecessary data features
test = testdata.drop(
labels=["PassengerId", "SibSp", "Parch", "Name", "Ticket", "Age", "Fare"], axis=1
)
# performing transformation on age and fare
scaled_testdata = scaler.transform(testdata[["Age", "Fare"]])
data3 = pd.DataFrame(data=scaled_testdata, columns=["Age", "Fare"])
data3.head(2)
finaltest = pd.concat(objs=[test, data3], axis=1)
finaltest.head()
finaltest_pred = dtc.predict(finaltest)
finaltest_pred
finaltest.join(
pd.DataFrame(finaltest_pred)
) # = pd.concat(objs=[finaltest, finaltest_pred], axis=1)
print("Thank you")
| false | 0 | 2,804 | 2 | 2,804 | 2,804 |
||
69826733
|
<jupyter_start><jupyter_text>Medical Cost Personal Datasets
## Context
Machine Learning with R by Brett Lantz is a book that provides an introduction to machine learning using R. As far as I can tell, Packt Publishing does not make its datasets available online unless you buy the book and create a user account which can be a problem if you are checking the book out from the library or borrowing the book from a friend. All of these datasets are in the public domain but simply needed some cleaning up and recoding to match the format in the book.
## Content
**Columns**
- age: age of primary beneficiary
- sex: insurance contractor gender, female, male
- bmi: Body mass index, providing an understanding of body, weights that are relatively high or low relative to height,
objective index of body weight (kg / m ^ 2) using the ratio of height to weight, ideally 18.5 to 24.9
- children: Number of children covered by health insurance / Number of dependents
- smoker: Smoking
- region: the beneficiary's residential area in the US, northeast, southeast, southwest, northwest.
- charges: Individual medical costs billed by health insurance
## Acknowledgements
The dataset is available on GitHub [here](https://github.com/stedy/Machine-Learning-with-R-datasets).
## Inspiration
Can you accurately predict insurance costs?
Kaggle dataset identifier: insurance
<jupyter_code>import pandas as pd
df = pd.read_csv('insurance/insurance.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 1338 entries, 0 to 1337
Data columns (total 7 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 age 1338 non-null int64
1 sex 1338 non-null object
2 bmi 1338 non-null float64
3 children 1338 non-null int64
4 smoker 1338 non-null object
5 region 1338 non-null object
6 charges 1338 non-null float64
dtypes: float64(2), int64(2), object(3)
memory usage: 73.3+ KB
<jupyter_text>Examples:
{
"age": 19,
"sex": "female",
"bmi": 27.9,
"children": 0,
"smoker": "yes",
"region": "southwest",
"charges": 16884.924
}
{
"age": 18,
"sex": "male",
"bmi": 33.77,
"children": 1,
"smoker": "no",
"region": "southeast",
"charges": 1725.5523
}
{
"age": 28,
"sex": "male",
"bmi": 33.0,
"children": 3,
"smoker": "no",
"region": "southeast",
"charges": 4449.462
}
{
"age": 33,
"sex": "male",
"bmi": 22.705,
"children": 0,
"smoker": "no",
"region": "northwest",
"charges": 21984.47061
}
<jupyter_script>import numpy as np
import pandas as pd
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
data = pd.read_csv("/kaggle/input/insurance/insurance.csv")
data.head()
# The dataset above is about the cost of treatment of patients.
# The cost of treatment according to dataset may depend upon :
# -> Age
# -> Sex
# -> BMI
# -> Children
# -> Smoking Habit
# -> Region
data.isnull().sum()
# Good to see there are no "NaN" in the data.
# ### Encoding Categorical Features
from sklearn.preprocessing import LabelEncoder
# sex
le = LabelEncoder()
data.sex = le.fit_transform(data.sex)
# smoker
data.smoker = le.fit_transform(data.smoker)
# region
data_region = pd.get_dummies(data.region)
data = pd.concat([data, data_region], axis=1)
data = data.drop(["region"], axis=1)
data.head()
data.corr()["charges"].sort_values()
import seaborn as sns
import matplotlib.pyplot as plt
f, ax = plt.subplots(figsize=(12, 10))
corr = data.corr()
sns.heatmap(
corr,
mask=np.zeros_like(corr, dtype=np.bool),
cmap=sns.diverging_palette(240, 10, as_cmap=True),
square=True,
ax=ax,
)
# A strong correlation is observed only with the fact of smoking the patient. To be honest, I expected a higher correlation with bmi. Well. We investigate smoking in more detail.
#
fig, ax = plt.subplots(figsize=(10, 7))
ax.hist(data.charges)
plt.style.use("ggplot")
# Show plot
plt.show()
f = plt.figure(figsize=(12, 5))
ax = f.add_subplot(121)
sns.distplot(data[(data.smoker == 1)]["charges"], color="c", ax=ax)
ax.set_title("Distribution of charges for smokers")
ax = f.add_subplot(122)
sns.distplot(data[(data.smoker == 0)]["charges"], color="b", ax=ax)
ax.set_title("Distribution of charges for non-smokers")
plt.tight_layout()
# Smoking patients spend more on treatment. But there is a feeling that the number of non-smoking patients is greater. Going to check it.
sns.catplot(x="smoker", kind="count", hue="sex", palette="pink", data=data)
# Please note that women are coded with the symbol " 1 "and men - "0". Thus non-smoking people and the truth more.
sns.catplot(
x="sex", y="charges", hue="smoker", kind="violin", data=data, palette="magma"
)
plt.figure(figsize=(12, 5))
plt.title("Box plot for charges of women")
sns.boxplot(
y="smoker", x="charges", data=data[(data.sex == 1)], orient="h", palette="magma"
)
plt.figure(figsize=(12, 5))
plt.title("Box plot for charges of men")
sns.boxplot(
y="smoker", x="charges", data=data[(data.sex == 0)], orient="h", palette="rainbow"
)
# Now let's pay attention to the age of the patients. First, let's look at how age affects the cost of treatment, and also look at patients of what age more in our data set.
plt.figure(figsize=(12, 5))
plt.title("Distribution of age")
ax = sns.distplot(data["age"], color="g")
sns.lmplot(x="age", y="charges", hue="smoker", data=data, palette="inferno_r", size=7)
ax.set_title("Smokers and non-smokers")
plt.figure(figsize=(12, 5))
plt.title("Distribution of bmi")
ax = sns.distplot(data["bmi"], color="m")
plt.figure(figsize=(10, 6))
ax = sns.scatterplot(x="bmi", y="charges", data=data, palette="magma", hue="smoker")
ax.set_title("Scatter plot of charges and bmi")
sns.lmplot(x="bmi", y="charges", hue="smoker", data=data, palette="magma", size=8)
sns.catplot(x="children", kind="count", palette="ch:.25", data=data, size=6)
sns.catplot(
x="smoker",
kind="count",
palette="rainbow",
hue="sex",
data=data[(data.children > 0)],
size=6,
)
ax.set_title("Smokers and non-smokers who have childrens")
# Now we are going to predict the cost of treatment. Let's start with the usual linear regression.
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import PolynomialFeatures
from sklearn.metrics import r2_score, mean_squared_error
from sklearn.ensemble import RandomForestRegressor
x = data.drop(["charges"], axis=1)
y = data.charges
x_train, x_test, y_train, y_test = train_test_split(x, y, random_state=0)
lr = LinearRegression().fit(x_train, y_train)
y_train_pred = lr.predict(x_train)
y_test_pred = lr.predict(x_test)
print(lr.score(x_test, y_test))
X = data.drop(["charges"], axis=1)
Y = data.charges
quad = PolynomialFeatures(degree=2)
x_quad = quad.fit_transform(X)
X_train, X_test, Y_train, Y_test = train_test_split(x_quad, Y, random_state=0)
plr = LinearRegression().fit(X_train, Y_train)
Y_train_pred = plr.predict(X_train)
Y_test_pred = plr.predict(X_test)
print(plr.score(X_test, Y_test))
forest = RandomForestRegressor(
n_estimators=100, criterion="mse", random_state=1, n_jobs=-1
)
forest.fit(x_train, y_train)
forest_train_pred = forest.predict(x_train)
forest_test_pred = forest.predict(x_test)
print(
"MSE train data: %.3f, MSE test data: %.3f"
% (
mean_squared_error(y_train, forest_train_pred),
mean_squared_error(y_test, forest_test_pred),
)
)
print(
"R2 train data: %.3f, R2 test data: %.3f"
% (r2_score(y_train, forest_train_pred), r2_score(y_test, forest_test_pred))
)
plt.figure(figsize=(10, 6))
plt.scatter(
forest_train_pred,
forest_train_pred - y_train,
c="black",
marker="o",
s=35,
alpha=0.5,
label="Train data",
)
plt.scatter(
forest_test_pred,
forest_test_pred - y_test,
c="c",
marker="o",
s=35,
alpha=0.7,
label="Test data",
)
plt.xlabel("Predicted values")
plt.ylabel("Tailings")
plt.legend(loc="upper left")
plt.hlines(y=0, xmin=0, xmax=60000, lw=2, color="red")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/826/69826733.ipynb
|
insurance
|
mirichoi0218
|
[{"Id": 69826733, "ScriptId": 16311420, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5236306, "CreationDate": "08/03/2021 13:13:53", "VersionNumber": 29.0, "Title": "\ud83e\udd12Patient Charges || Regression", "EvaluationDate": "08/03/2021", "IsChange": false, "TotalLines": 191.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 191.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 93279436, "KernelVersionId": 69826733, "SourceDatasetVersionId": 18513}]
|
[{"Id": 18513, "DatasetId": 13720, "DatasourceVersionId": 18513, "CreatorUserId": 1616098, "LicenseName": "Database: Open Database, Contents: Database Contents", "CreationDate": "02/21/2018 00:15:14", "VersionNumber": 1.0, "Title": "Medical Cost Personal Datasets", "Slug": "insurance", "Subtitle": "Insurance Forecast by using Linear Regression", "Description": "## Context\nMachine Learning with R by Brett Lantz is a book that provides an introduction to machine learning using R. As far as I can tell, Packt Publishing does not make its datasets available online unless you buy the book and create a user account which can be a problem if you are checking the book out from the library or borrowing the book from a friend. All of these datasets are in the public domain but simply needed some cleaning up and recoding to match the format in the book.\n\n## Content\n**Columns**\n - age: age of primary beneficiary \n\n - sex: insurance contractor gender, female, male \n\n - bmi: Body mass index, providing an understanding of body, weights that are relatively high or low relative to height,\n objective index of body weight (kg / m ^ 2) using the ratio of height to weight, ideally 18.5 to 24.9 \n\n - children: Number of children covered by health insurance / Number of dependents\n\n - smoker: Smoking\n\n - region: the beneficiary's residential area in the US, northeast, southeast, southwest, northwest.\n\n - charges: Individual medical costs billed by health insurance\n\n## Acknowledgements\n\nThe dataset is available on GitHub [here](https://github.com/stedy/Machine-Learning-with-R-datasets).\n\n## Inspiration\nCan you accurately predict insurance costs?", "VersionNotes": "Initial release", "TotalCompressedBytes": 55628.0, "TotalUncompressedBytes": 55628.0}]
|
[{"Id": 13720, "CreatorUserId": 1616098, "OwnerUserId": 1616098.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 18513.0, "CurrentDatasourceVersionId": 18513.0, "ForumId": 21253, "Type": 2, "CreationDate": "02/21/2018 00:15:14", "LastActivityDate": "02/21/2018", "TotalViews": 1223332, "TotalDownloads": 195148, "TotalVotes": 2334, "TotalKernels": 1185}]
|
[{"Id": 1616098, "UserName": "mirichoi0218", "DisplayName": "Miri Choi", "RegisterDate": "02/07/2018", "PerformanceTier": 2}]
|
import numpy as np
import pandas as pd
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
data = pd.read_csv("/kaggle/input/insurance/insurance.csv")
data.head()
# The dataset above is about the cost of treatment of patients.
# The cost of treatment according to dataset may depend upon :
# -> Age
# -> Sex
# -> BMI
# -> Children
# -> Smoking Habit
# -> Region
data.isnull().sum()
# Good to see there are no "NaN" in the data.
# ### Encoding Categorical Features
from sklearn.preprocessing import LabelEncoder
# sex
le = LabelEncoder()
data.sex = le.fit_transform(data.sex)
# smoker
data.smoker = le.fit_transform(data.smoker)
# region
data_region = pd.get_dummies(data.region)
data = pd.concat([data, data_region], axis=1)
data = data.drop(["region"], axis=1)
data.head()
data.corr()["charges"].sort_values()
import seaborn as sns
import matplotlib.pyplot as plt
f, ax = plt.subplots(figsize=(12, 10))
corr = data.corr()
sns.heatmap(
corr,
mask=np.zeros_like(corr, dtype=np.bool),
cmap=sns.diverging_palette(240, 10, as_cmap=True),
square=True,
ax=ax,
)
# A strong correlation is observed only with the fact of smoking the patient. To be honest, I expected a higher correlation with bmi. Well. We investigate smoking in more detail.
#
fig, ax = plt.subplots(figsize=(10, 7))
ax.hist(data.charges)
plt.style.use("ggplot")
# Show plot
plt.show()
f = plt.figure(figsize=(12, 5))
ax = f.add_subplot(121)
sns.distplot(data[(data.smoker == 1)]["charges"], color="c", ax=ax)
ax.set_title("Distribution of charges for smokers")
ax = f.add_subplot(122)
sns.distplot(data[(data.smoker == 0)]["charges"], color="b", ax=ax)
ax.set_title("Distribution of charges for non-smokers")
plt.tight_layout()
# Smoking patients spend more on treatment. But there is a feeling that the number of non-smoking patients is greater. Going to check it.
sns.catplot(x="smoker", kind="count", hue="sex", palette="pink", data=data)
# Please note that women are coded with the symbol " 1 "and men - "0". Thus non-smoking people and the truth more.
sns.catplot(
x="sex", y="charges", hue="smoker", kind="violin", data=data, palette="magma"
)
plt.figure(figsize=(12, 5))
plt.title("Box plot for charges of women")
sns.boxplot(
y="smoker", x="charges", data=data[(data.sex == 1)], orient="h", palette="magma"
)
plt.figure(figsize=(12, 5))
plt.title("Box plot for charges of men")
sns.boxplot(
y="smoker", x="charges", data=data[(data.sex == 0)], orient="h", palette="rainbow"
)
# Now let's pay attention to the age of the patients. First, let's look at how age affects the cost of treatment, and also look at patients of what age more in our data set.
plt.figure(figsize=(12, 5))
plt.title("Distribution of age")
ax = sns.distplot(data["age"], color="g")
sns.lmplot(x="age", y="charges", hue="smoker", data=data, palette="inferno_r", size=7)
ax.set_title("Smokers and non-smokers")
plt.figure(figsize=(12, 5))
plt.title("Distribution of bmi")
ax = sns.distplot(data["bmi"], color="m")
plt.figure(figsize=(10, 6))
ax = sns.scatterplot(x="bmi", y="charges", data=data, palette="magma", hue="smoker")
ax.set_title("Scatter plot of charges and bmi")
sns.lmplot(x="bmi", y="charges", hue="smoker", data=data, palette="magma", size=8)
sns.catplot(x="children", kind="count", palette="ch:.25", data=data, size=6)
sns.catplot(
x="smoker",
kind="count",
palette="rainbow",
hue="sex",
data=data[(data.children > 0)],
size=6,
)
ax.set_title("Smokers and non-smokers who have childrens")
# Now we are going to predict the cost of treatment. Let's start with the usual linear regression.
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import PolynomialFeatures
from sklearn.metrics import r2_score, mean_squared_error
from sklearn.ensemble import RandomForestRegressor
x = data.drop(["charges"], axis=1)
y = data.charges
x_train, x_test, y_train, y_test = train_test_split(x, y, random_state=0)
lr = LinearRegression().fit(x_train, y_train)
y_train_pred = lr.predict(x_train)
y_test_pred = lr.predict(x_test)
print(lr.score(x_test, y_test))
X = data.drop(["charges"], axis=1)
Y = data.charges
quad = PolynomialFeatures(degree=2)
x_quad = quad.fit_transform(X)
X_train, X_test, Y_train, Y_test = train_test_split(x_quad, Y, random_state=0)
plr = LinearRegression().fit(X_train, Y_train)
Y_train_pred = plr.predict(X_train)
Y_test_pred = plr.predict(X_test)
print(plr.score(X_test, Y_test))
forest = RandomForestRegressor(
n_estimators=100, criterion="mse", random_state=1, n_jobs=-1
)
forest.fit(x_train, y_train)
forest_train_pred = forest.predict(x_train)
forest_test_pred = forest.predict(x_test)
print(
"MSE train data: %.3f, MSE test data: %.3f"
% (
mean_squared_error(y_train, forest_train_pred),
mean_squared_error(y_test, forest_test_pred),
)
)
print(
"R2 train data: %.3f, R2 test data: %.3f"
% (r2_score(y_train, forest_train_pred), r2_score(y_test, forest_test_pred))
)
plt.figure(figsize=(10, 6))
plt.scatter(
forest_train_pred,
forest_train_pred - y_train,
c="black",
marker="o",
s=35,
alpha=0.5,
label="Train data",
)
plt.scatter(
forest_test_pred,
forest_test_pred - y_test,
c="c",
marker="o",
s=35,
alpha=0.7,
label="Test data",
)
plt.xlabel("Predicted values")
plt.ylabel("Tailings")
plt.legend(loc="upper left")
plt.hlines(y=0, xmin=0, xmax=60000, lw=2, color="red")
plt.show()
|
[{"insurance/insurance.csv": {"column_names": "[\"age\", \"sex\", \"bmi\", \"children\", \"smoker\", \"region\", \"charges\"]", "column_data_types": "{\"age\": \"int64\", \"sex\": \"object\", \"bmi\": \"float64\", \"children\": \"int64\", \"smoker\": \"object\", \"region\": \"object\", \"charges\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1338 entries, 0 to 1337\nData columns (total 7 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 age 1338 non-null int64 \n 1 sex 1338 non-null object \n 2 bmi 1338 non-null float64\n 3 children 1338 non-null int64 \n 4 smoker 1338 non-null object \n 5 region 1338 non-null object \n 6 charges 1338 non-null float64\ndtypes: float64(2), int64(2), object(3)\nmemory usage: 73.3+ KB\n", "summary": "{\"age\": {\"count\": 1338.0, \"mean\": 39.20702541106129, \"std\": 14.049960379216154, \"min\": 18.0, \"25%\": 27.0, \"50%\": 39.0, \"75%\": 51.0, \"max\": 64.0}, \"bmi\": {\"count\": 1338.0, \"mean\": 30.66339686098655, \"std\": 6.098186911679014, \"min\": 15.96, \"25%\": 26.29625, \"50%\": 30.4, \"75%\": 34.69375, \"max\": 53.13}, \"children\": {\"count\": 1338.0, \"mean\": 1.0949177877429, \"std\": 1.205492739781914, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 2.0, \"max\": 5.0}, \"charges\": {\"count\": 1338.0, \"mean\": 13270.422265141257, \"std\": 12110.011236694001, \"min\": 1121.8739, \"25%\": 4740.28715, \"50%\": 9382.033, \"75%\": 16639.912515, \"max\": 63770.42801}}", "examples": "{\"age\":{\"0\":19,\"1\":18,\"2\":28,\"3\":33},\"sex\":{\"0\":\"female\",\"1\":\"male\",\"2\":\"male\",\"3\":\"male\"},\"bmi\":{\"0\":27.9,\"1\":33.77,\"2\":33.0,\"3\":22.705},\"children\":{\"0\":0,\"1\":1,\"2\":3,\"3\":0},\"smoker\":{\"0\":\"yes\",\"1\":\"no\",\"2\":\"no\",\"3\":\"no\"},\"region\":{\"0\":\"southwest\",\"1\":\"southeast\",\"2\":\"southeast\",\"3\":\"northwest\"},\"charges\":{\"0\":16884.924,\"1\":1725.5523,\"2\":4449.462,\"3\":21984.47061}}"}}]
| true | 1 |
<start_data_description><data_path>insurance/insurance.csv:
<column_names>
['age', 'sex', 'bmi', 'children', 'smoker', 'region', 'charges']
<column_types>
{'age': 'int64', 'sex': 'object', 'bmi': 'float64', 'children': 'int64', 'smoker': 'object', 'region': 'object', 'charges': 'float64'}
<dataframe_Summary>
{'age': {'count': 1338.0, 'mean': 39.20702541106129, 'std': 14.049960379216154, 'min': 18.0, '25%': 27.0, '50%': 39.0, '75%': 51.0, 'max': 64.0}, 'bmi': {'count': 1338.0, 'mean': 30.66339686098655, 'std': 6.098186911679014, 'min': 15.96, '25%': 26.29625, '50%': 30.4, '75%': 34.69375, 'max': 53.13}, 'children': {'count': 1338.0, 'mean': 1.0949177877429, 'std': 1.205492739781914, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 2.0, 'max': 5.0}, 'charges': {'count': 1338.0, 'mean': 13270.422265141257, 'std': 12110.011236694001, 'min': 1121.8739, '25%': 4740.28715, '50%': 9382.033, '75%': 16639.912515, 'max': 63770.42801}}
<dataframe_info>
RangeIndex: 1338 entries, 0 to 1337
Data columns (total 7 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 age 1338 non-null int64
1 sex 1338 non-null object
2 bmi 1338 non-null float64
3 children 1338 non-null int64
4 smoker 1338 non-null object
5 region 1338 non-null object
6 charges 1338 non-null float64
dtypes: float64(2), int64(2), object(3)
memory usage: 73.3+ KB
<some_examples>
{'age': {'0': 19, '1': 18, '2': 28, '3': 33}, 'sex': {'0': 'female', '1': 'male', '2': 'male', '3': 'male'}, 'bmi': {'0': 27.9, '1': 33.77, '2': 33.0, '3': 22.705}, 'children': {'0': 0, '1': 1, '2': 3, '3': 0}, 'smoker': {'0': 'yes', '1': 'no', '2': 'no', '3': 'no'}, 'region': {'0': 'southwest', '1': 'southeast', '2': 'southeast', '3': 'northwest'}, 'charges': {'0': 16884.924, '1': 1725.5523, '2': 4449.462, '3': 21984.47061}}
<end_description>
| 1,944 | 0 | 2,790 | 1,944 |
69826220
|
<jupyter_start><jupyter_text>Cirrhosis Prediction Dataset
### Similar Datasets
- Hepatitis C Dataset: [LINK](https://www.kaggle.com/fedesoriano/hepatitis-c-dataset)
- Body Fat Prediction Dataset: [LINK](https://www.kaggle.com/fedesoriano/body-fat-prediction-dataset)
- Stroke Prediction Dataset: [LINK](https://www.kaggle.com/fedesoriano/stroke-prediction-dataset)
- Wind Speed Prediction Dataset: [LINK](https://www.kaggle.com/datasets/fedesoriano/wind-speed-prediction-dataset)
- Spanish Wine Quality Dataset: [LINK](https://www.kaggle.com/datasets/fedesoriano/spanish-wine-quality-dataset)
### Context
Cirrhosis is a late stage of scarring (fibrosis) of the liver caused by many forms of liver diseases and conditions, such as hepatitis and chronic alcoholism. The following data contains the information collected from the Mayo Clinic trial in primary biliary cirrhosis (PBC) of the liver conducted between 1974 and 1984. A description of the clinical background for the trial and the covariates recorded here is in Chapter 0, especially Section 0.2 of Fleming and Harrington, Counting
Processes and Survival Analysis, Wiley, 1991. A more extended discussion can be found in Dickson, et al., Hepatology 10:1-7 (1989) and in Markus, et al., N Eng J of Med 320:1709-13 (1989).
A total of 424 PBC patients, referred to Mayo Clinic during that ten-year interval, met eligibility criteria for the randomized placebo-controlled trial of the drug D-penicillamine. The first 312 cases in the dataset participated in the randomized trial and contain largely complete data. The additional 112 cases did not participate in the clinical trial but consented to have basic measurements recorded and to be followed for survival. Six of those cases were lost to follow-up shortly after diagnosis, so the data here are on an additional 106 cases as well as the 312 randomized participants.
### Attribute Information
1) ID: unique identifier
2) N\_Days: number of days between registration and the earlier of death, transplantation, or study analysis time in July 1986
3) Status: status of the patient C (censored), CL (censored due to liver tx), or D (death)
4) Drug: type of drug D-penicillamine or placebo
5) Age: age in [days]
6) Sex: M (male) or F (female)
7) Ascites: presence of ascites N (No) or Y (Yes)
8) Hepatomegaly: presence of hepatomegaly N (No) or Y (Yes)
9) Spiders: presence of spiders N (No) or Y (Yes)
10) Edema: presence of edema N (no edema and no diuretic therapy for edema), S (edema present without diuretics, or edema resolved by diuretics), or Y (edema despite diuretic therapy)
11) Bilirubin: serum bilirubin in [mg/dl]
12) Cholesterol: serum cholesterol in [mg/dl]
13) Albumin: albumin in [gm/dl]
14) Copper: urine copper in [ug/day]
15) Alk\_Phos: alkaline phosphatase in [U/liter]
16) SGOT: SGOT in [U/ml]
17) Triglycerides: triglicerides in [mg/dl]
18) Platelets: platelets per cubic [ml/1000]
19) Prothrombin: prothrombin time in seconds [s]
20) Stage: histologic stage of disease (1, 2, 3, or 4)
Kaggle dataset identifier: cirrhosis-prediction-dataset
<jupyter_code>import pandas as pd
df = pd.read_csv('cirrhosis-prediction-dataset/cirrhosis.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 418 entries, 0 to 417
Data columns (total 20 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 ID 418 non-null int64
1 N_Days 418 non-null int64
2 Status 418 non-null object
3 Drug 312 non-null object
4 Age 418 non-null int64
5 Sex 418 non-null object
6 Ascites 312 non-null object
7 Hepatomegaly 312 non-null object
8 Spiders 312 non-null object
9 Edema 418 non-null object
10 Bilirubin 418 non-null float64
11 Cholesterol 284 non-null float64
12 Albumin 418 non-null float64
13 Copper 310 non-null float64
14 Alk_Phos 312 non-null float64
15 SGOT 312 non-null float64
16 Tryglicerides 282 non-null float64
17 Platelets 407 non-null float64
18 Prothrombin 416 non-null float64
19 Stage 412 non-null float64
dtypes: float64(10), int64(3), object(7)
memory usage: 65.4+ KB
<jupyter_text>Examples:
{
"ID": 1,
"N_Days": 400,
"Status": "D",
"Drug": "D-penicillamine",
"Age": 21464,
"Sex": "F",
"Ascites": "Y",
"Hepatomegaly": "Y",
"Spiders": "Y",
"Edema": "Y",
"Bilirubin": 14.5,
"Cholesterol": 261,
"Albumin": 2.6,
"Copper": 156,
"Alk_Phos": 1718.0,
"SGOT": 137.95,
"Tryglicerides": 172,
"Platelets": 190,
"Prothrombin": 12.2,
"Stage": 4
}
{
"ID": 2,
"N_Days": 4500,
"Status": "C",
"Drug": "D-penicillamine",
"Age": 20617,
"Sex": "F",
"Ascites": "N",
"Hepatomegaly": "Y",
"Spiders": "Y",
"Edema": "N",
"Bilirubin": 1.1,
"Cholesterol": 302,
"Albumin": 4.14,
"Copper": 54,
"Alk_Phos": 7394.8,
"SGOT": 113.52,
"Tryglicerides": 88,
"Platelets": 221,
"Prothrombin": 10.6,
"Stage": 3
}
{
"ID": 3,
"N_Days": 1012,
"Status": "D",
"Drug": "D-penicillamine",
"Age": 25594,
"Sex": "M",
"Ascites": "N",
"Hepatomegaly": "N",
"Spiders": "N",
"Edema": "S",
"Bilirubin": 1.4,
"Cholesterol": 176,
"Albumin": 3.48,
"Copper": 210,
"Alk_Phos": 516.0,
"SGOT": 96.1,
"Tryglicerides": 55,
"Platelets": 151,
"Prothrombin": 12.0,
"Stage": 4
}
{
"ID": 4,
"N_Days": 1925,
"Status": "D",
"Drug": "D-penicillamine",
"Age": 19994,
"Sex": "F",
"Ascites": "N",
"Hepatomegaly": "Y",
"Spiders": "Y",
"Edema": "S",
"Bilirubin": 1.8,
"Cholesterol": 244,
"Albumin": 2.54,
"Copper": 64,
"Alk_Phos": 6121.8,
"SGOT": 60.63,
"Tryglicerides": 92,
"Platelets": 183,
"Prothrombin": 10.3,
"Stage": 4
}
<jupyter_script># # Bilgiler
# Hepatomegaly = karaciğer büyümesi enfeksiyon kapma, doğrudan zehirlenme, karaciğer tümörü ve metabolik bozukluklar gibi birçok nedenden kaynaklanabilen tıbbi bir belirtidir. Genellikle karındaki bir kitle olarak hissedilir, nedene bağlı olarak bazen de kendini sarılıkla birlikte gösterir.
# spiders = Örümcek telangiektazi (Spider telangiectasia), derialtı arterlerinin ya da arteriollerinin genişlemesinin sonucudur. Ortadaki bir odaktan çevreye doğru ışınsal çizgilenmeler yapan geniş damarlar örümceksi bir görünüm oluştururlar. Yüz, boyun ve göğüs bölgesinde görece sıktır. Gebelerde ve siroz hastalarında sık görülmeleri nedeniyle östrojen düzeyindeki yükselmenin etkili olduğu düşünülmektedir
# Edema = ödem ,aşırı sıvı birikmesi
# Bilirubin = Bilirubin, kan elemanlarından olan alyuvarların yıkımı sonucu ortaya çıkan bir maddedir. Bilirubin yüksek olursa ne olur?
# Bilirubin yüksekliği, sarılık olarak tanımlanan, ciltte ve göz akında sararma şeklinde fark edilen duruma yol açar. Bilirubin temel olarak karaciğerde alyuvarlar yıkımı sonrası açığa çıkan artık madde olduğu için, bilirubin yüksekliği vücutta anormal hızlı alyuvar yıkımı olduğu anlamına gelebilir.
# Kolesterol, hayvanların vücut dokularındaki hücre zarlarında bulunan ve kan plazmasında taşınan bir sterol, yani bir steroid ve alkol birleşimidir. Daha düşük miktarlarda bitkilerde de bulunur. İlk defa 1754'te safra taşlarında kolesterol bulunduğu için bu maddenin ismi Yunanca chole- (safra) ve steros (katı) sözcükleri ile kimyadaki -ol ekinden türetilmiştir.
# Cholesterol = Kolesterol, özellikle hayvansal gıdalarda bulunur ama vücuttaki kolesterolun ancak ufak bir kısmı gıda kaynaklıdır; çoğu vücut tarafından sentezlenir. Vücudun her hücresinde bulunmakla beraber, onun sentezlendiği veya hücre zarlarının daha çok olduğu organ ve dokularda, örneğin karaciğer, omurilik ve beyinde, ayrıca ateromlarda, kolesterolun yoğunluğu daha yüksektir. Kolesterol kanda normalden fazla bulunması halinde damarlarda birikerek damar sertleşmesine (ateroskleroz) yol açar. Bazen de safra pigmentleri ile birleşerek safra taşlarının oluşumunda rol oynar.
# Albumin = Kısaca albümin diye de bilinen serum albümini, insan ve diğer memeli hayvanların kan plazmasında bulunan en yaygın proteindir. Kanda bulunan proteinlerin %60'ını oluşturur.
# copper = bakır
# Bakır vazgeçilmez bir iz elementtir. Hücresel enerji metabolizması, antioksidan savunma, demir taşımacılığı ve fibrogenezde yer alan enzimler için bir kofaktör görevi görür. Bu süreçler karaciğer rahatsızlıklarının patogenezinde merkezi olmasına rağmen, çok az çalışma bunları bakır eksikliğine bağlamaktadır.
# (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6671688/)
# Alk_Phos = Alkalen fosfataz veya ALP Bu enzim kemik, karaciğer, bağırsak ve plasenta tarafından oluşturulan enzim, safra yoluyla itrah edilir. Serumda alkalin fosfataz aktivitesi; kemik, karaciğer ve safra yolları hastalıklarında anlamlı olarak değiştiğinden klinikte bu enzimin değeri önem taşır. Kemik tümörleri, karaciğer hasarı, safra yollarının tıkanıklığı gibi patolojilerde bu enzimin seviyeleri artar.
# SGOT = Aspartat transaminaz ,amino asit metabolizmasında önemli bir enzimdir. AST karaciğer, kalp, iskelet kası, böbrekler, beyin ve kırmızı kan hücrelerinde bulunur. Serum AST düzeyi, serum ALT(alanin transaminaz)düzeyi ve bunların oranı(AST/ALT oranı)klinik olarak karaciğer sağlığı için biyobelirteç olarak ölçülür. Testler kan panellerinin bir parçasıdır..
# AST, her iki enzimin de karaciğer parenkimal hücreleriyle ilişkili olması nedeniyle alanin transaminaz (ALT) ile benzer. Fark, ALT'ın ağırlıklı olarak karaciğerde, böbreklerde, kalpte ve iskelet kasında klinik olarak ihmal edilebilir miktarlarda bulunurken, AST'nin karaciğerde, kalpte(kalp kası),iskelet kasında, böbreklerde, beyinde ve kırmızı kan hücrelerinde bulunmasıdır. Sonuç olarak, ALT, AST'den daha spesifik bir karaciğer iltihabı göstergesidir, çünkü AST miyokard enfarktüsü, akut pankreatit, akut hemolitik anemi, şiddetli yanıklar, akut böbrek hastalığı , kas-iskelet sistemi hastalıkları ve travma gibi diğer organları etkileyen hastalıklarda da yükselebilir.
# Tryglicerides = trigliseritler, Kandaki trigliserid düzeyinin 150 mg/dL'nin üzerinde bulunması, trigliserid yüksekliği (hipertrigliseridemi) olarak adlandırılır.
# 50 mg/dl'den düşük olan trigliserid seviyesine, trigliserid düşüklüğü denmektedir. 35 mg/dl'den daha düşük trigliserid seviyesine de aşırı trigliserid düşüklüğü denir.
# (https://www.milliyet.com.tr/pembenar/trigliserid-nedir-trigliserid-kac-olmali-yuksekligi-ve-dusuklugu-nedenleri-6169751#:~:text=50%20mg%2Fdl'den%20d%C3%BC%C5%9F%C3%BCk,ya%C4%9Fl%C4%B1%20g%C4%B1dalarla%20beslenme%20durumlar%C4%B1nda%20olabilir.)
# (https://www.medicalpark.com.tr/trigliserid/hg-2181)
# Platelets = trombositler, Trombosit veya kan pulcukları kan pıhtılarının oluşumunda görev alan hücre parçalarına verilen isimdir. Platelet olarak da adlandırılır. Düşük trombosit seviyeleri veya fonksiyon anormallikleri (disfonksiyon) kanamayı yatkınlaştırırken, yüksek trombosit seviyeleri -çoğunlukla asemptomatik- tromboz (damarda kanın pıhtılaşması) riskini yükseltir
# Sağlıklı bir kişide kan pulcukları değerinin 150, 000 ila 450, 000 hücre/ml aralığında olması beklenir. Trombosit değerinin taban değeri altına inmesiyle trombositopeni adı verilen trombosit düşüklüğü durumu söz konusu olur. Tavan değeri üstüne çıktığı takdirde de trombositoz adı verilen trombosit yüksekliği durumu gündemde olur.
# Prothrombin = Protrombin zamanı (PT), pıhtılaşmanın ortak ve ekstrinsik yolunu değerlendirmede kullanılan bir testtir. Bu testte plazmaya kalsiyum ve tromboplastin (doku faktörü) eklenerek, ekstrinsik yoldan fibrin pıhtısı oluşana dek geçen süre ölçülür. PT, FII, FVII, FX, protrombin ve fibrinojenin eksikliklerinin saptanması ile K vitamini antagonisti (coumadin) tedavisinin takibinde kullanılabilir
# Stage: hastalığın aşaması ,derecesi(1, 2, 3, or 4)
# bilgilerin çoğunluğu vikipediden elde edilmiştir
# # Kodlar
veri.info()
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sbn
veri = pd.read_csv("../input/cirrhosis-prediction-dataset/cirrhosis.csv")
veri
plt.subplots(figsize=(20, 10))
sbn.heatmap(veri.isnull(), cbar=False)
veri.isnull().sum()
veri.describe()
plt.subplots(figsize=(20, 10))
sbn.heatmap(veri.corr(), annot=True, cbar=False, linewidths=0.9)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/826/69826220.ipynb
|
cirrhosis-prediction-dataset
|
fedesoriano
|
[{"Id": 69826220, "ScriptId": 19043619, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7957274, "CreationDate": "08/03/2021 13:10:52", "VersionNumber": 1.0, "Title": "siroz tahmini", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 59.0, "LinesInsertedFromPrevious": 59.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 93279008, "KernelVersionId": 69826220, "SourceDatasetVersionId": 2492225}]
|
[{"Id": 2492225, "DatasetId": 1508604, "DatasourceVersionId": 2534803, "CreatorUserId": 6402661, "LicenseName": "Data files \u00a9 Original Authors", "CreationDate": "08/02/2021 15:36:59", "VersionNumber": 2.0, "Title": "Cirrhosis Prediction Dataset", "Slug": "cirrhosis-prediction-dataset", "Subtitle": "18 clinical features for predicting liver cirrhosis stage", "Description": "### Similar Datasets\n\n- Hepatitis C Dataset: [LINK](https://www.kaggle.com/fedesoriano/hepatitis-c-dataset)\n- Body Fat Prediction Dataset: [LINK](https://www.kaggle.com/fedesoriano/body-fat-prediction-dataset)\n- Stroke Prediction Dataset: [LINK](https://www.kaggle.com/fedesoriano/stroke-prediction-dataset)\n- Wind Speed Prediction Dataset: [LINK](https://www.kaggle.com/datasets/fedesoriano/wind-speed-prediction-dataset)\n- Spanish Wine Quality Dataset: [LINK](https://www.kaggle.com/datasets/fedesoriano/spanish-wine-quality-dataset)\n\n\n### Context\n\nCirrhosis is a late stage of scarring (fibrosis) of the liver caused by many forms of liver diseases and conditions, such as hepatitis and chronic alcoholism. The following data contains the information collected from the Mayo Clinic trial in primary biliary cirrhosis (PBC) of the liver conducted between 1974 and 1984. A description of the clinical background for the trial and the covariates recorded here is in Chapter 0, especially Section 0.2 of Fleming and Harrington, Counting\nProcesses and Survival Analysis, Wiley, 1991. A more extended discussion can be found in Dickson, et al., Hepatology 10:1-7 (1989) and in Markus, et al., N Eng J of Med 320:1709-13 (1989). \n\nA total of 424 PBC patients, referred to Mayo Clinic during that ten-year interval, met eligibility criteria for the randomized placebo-controlled trial of the drug D-penicillamine. The first 312 cases in the dataset participated in the randomized trial and contain largely complete data. The additional 112 cases did not participate in the clinical trial but consented to have basic measurements recorded and to be followed for survival. Six of those cases were lost to follow-up shortly after diagnosis, so the data here are on an additional 106 cases as well as the 312 randomized participants.\n\n\n### Attribute Information\n\n1) ID: unique identifier\n2) N\\_Days: number of days between registration and the earlier of death, transplantation, or study analysis time in July 1986\n3) Status: status of the patient C (censored), CL (censored due to liver tx), or D (death)\n4) Drug: type of drug D-penicillamine or placebo\n5) Age: age in [days]\n6) Sex: M (male) or F (female)\n7) Ascites: presence of ascites N (No) or Y (Yes)\n8) Hepatomegaly: presence of hepatomegaly N (No) or Y (Yes)\n9) Spiders: presence of spiders N (No) or Y (Yes)\n10) Edema: presence of edema N (no edema and no diuretic therapy for edema), S (edema present without diuretics, or edema resolved by diuretics), or Y (edema despite diuretic therapy)\n11) Bilirubin: serum bilirubin in [mg/dl]\n12) Cholesterol: serum cholesterol in [mg/dl]\n13) Albumin: albumin in [gm/dl]\n14) Copper: urine copper in [ug/day]\n15) Alk\\_Phos: alkaline phosphatase in [U/liter]\n16) SGOT: SGOT in [U/ml]\n17) Triglycerides: triglicerides in [mg/dl]\n18) Platelets: platelets per cubic [ml/1000]\n19) Prothrombin: prothrombin time in seconds [s]\n20) Stage: histologic stage of disease (1, 2, 3, or 4)\n\n### Acknowledgements\n\nThe dataset can be found in appendix D of:\n> Fleming, T.R. and Harrington, D.P. (1991) Counting Processes and Survival Analysis. Wiley Series in Probability and Mathematical Statistics: Applied Probability and Statistics, John Wiley and Sons Inc., New York.\n\nIf you want to cite this data:\n> fedesoriano. (August 2021). Cirrhosis Prediction Dataset. Retrieved [Date Retrieved] from https://www.kaggle.com/fedesoriano/cirrhosis-prediction-dataset.", "VersionNotes": "v2", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1508604, "CreatorUserId": 6402661, "OwnerUserId": 6402661.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2492225.0, "CurrentDatasourceVersionId": 2534803.0, "ForumId": 1528351, "Type": 2, "CreationDate": "08/02/2021 14:47:06", "LastActivityDate": "08/02/2021", "TotalViews": 96634, "TotalDownloads": 8794, "TotalVotes": 127, "TotalKernels": 24}]
|
[{"Id": 6402661, "UserName": "fedesoriano", "DisplayName": "fedesoriano", "RegisterDate": "12/18/2020", "PerformanceTier": 4}]
|
# # Bilgiler
# Hepatomegaly = karaciğer büyümesi enfeksiyon kapma, doğrudan zehirlenme, karaciğer tümörü ve metabolik bozukluklar gibi birçok nedenden kaynaklanabilen tıbbi bir belirtidir. Genellikle karındaki bir kitle olarak hissedilir, nedene bağlı olarak bazen de kendini sarılıkla birlikte gösterir.
# spiders = Örümcek telangiektazi (Spider telangiectasia), derialtı arterlerinin ya da arteriollerinin genişlemesinin sonucudur. Ortadaki bir odaktan çevreye doğru ışınsal çizgilenmeler yapan geniş damarlar örümceksi bir görünüm oluştururlar. Yüz, boyun ve göğüs bölgesinde görece sıktır. Gebelerde ve siroz hastalarında sık görülmeleri nedeniyle östrojen düzeyindeki yükselmenin etkili olduğu düşünülmektedir
# Edema = ödem ,aşırı sıvı birikmesi
# Bilirubin = Bilirubin, kan elemanlarından olan alyuvarların yıkımı sonucu ortaya çıkan bir maddedir. Bilirubin yüksek olursa ne olur?
# Bilirubin yüksekliği, sarılık olarak tanımlanan, ciltte ve göz akında sararma şeklinde fark edilen duruma yol açar. Bilirubin temel olarak karaciğerde alyuvarlar yıkımı sonrası açığa çıkan artık madde olduğu için, bilirubin yüksekliği vücutta anormal hızlı alyuvar yıkımı olduğu anlamına gelebilir.
# Kolesterol, hayvanların vücut dokularındaki hücre zarlarında bulunan ve kan plazmasında taşınan bir sterol, yani bir steroid ve alkol birleşimidir. Daha düşük miktarlarda bitkilerde de bulunur. İlk defa 1754'te safra taşlarında kolesterol bulunduğu için bu maddenin ismi Yunanca chole- (safra) ve steros (katı) sözcükleri ile kimyadaki -ol ekinden türetilmiştir.
# Cholesterol = Kolesterol, özellikle hayvansal gıdalarda bulunur ama vücuttaki kolesterolun ancak ufak bir kısmı gıda kaynaklıdır; çoğu vücut tarafından sentezlenir. Vücudun her hücresinde bulunmakla beraber, onun sentezlendiği veya hücre zarlarının daha çok olduğu organ ve dokularda, örneğin karaciğer, omurilik ve beyinde, ayrıca ateromlarda, kolesterolun yoğunluğu daha yüksektir. Kolesterol kanda normalden fazla bulunması halinde damarlarda birikerek damar sertleşmesine (ateroskleroz) yol açar. Bazen de safra pigmentleri ile birleşerek safra taşlarının oluşumunda rol oynar.
# Albumin = Kısaca albümin diye de bilinen serum albümini, insan ve diğer memeli hayvanların kan plazmasında bulunan en yaygın proteindir. Kanda bulunan proteinlerin %60'ını oluşturur.
# copper = bakır
# Bakır vazgeçilmez bir iz elementtir. Hücresel enerji metabolizması, antioksidan savunma, demir taşımacılığı ve fibrogenezde yer alan enzimler için bir kofaktör görevi görür. Bu süreçler karaciğer rahatsızlıklarının patogenezinde merkezi olmasına rağmen, çok az çalışma bunları bakır eksikliğine bağlamaktadır.
# (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6671688/)
# Alk_Phos = Alkalen fosfataz veya ALP Bu enzim kemik, karaciğer, bağırsak ve plasenta tarafından oluşturulan enzim, safra yoluyla itrah edilir. Serumda alkalin fosfataz aktivitesi; kemik, karaciğer ve safra yolları hastalıklarında anlamlı olarak değiştiğinden klinikte bu enzimin değeri önem taşır. Kemik tümörleri, karaciğer hasarı, safra yollarının tıkanıklığı gibi patolojilerde bu enzimin seviyeleri artar.
# SGOT = Aspartat transaminaz ,amino asit metabolizmasında önemli bir enzimdir. AST karaciğer, kalp, iskelet kası, böbrekler, beyin ve kırmızı kan hücrelerinde bulunur. Serum AST düzeyi, serum ALT(alanin transaminaz)düzeyi ve bunların oranı(AST/ALT oranı)klinik olarak karaciğer sağlığı için biyobelirteç olarak ölçülür. Testler kan panellerinin bir parçasıdır..
# AST, her iki enzimin de karaciğer parenkimal hücreleriyle ilişkili olması nedeniyle alanin transaminaz (ALT) ile benzer. Fark, ALT'ın ağırlıklı olarak karaciğerde, böbreklerde, kalpte ve iskelet kasında klinik olarak ihmal edilebilir miktarlarda bulunurken, AST'nin karaciğerde, kalpte(kalp kası),iskelet kasında, böbreklerde, beyinde ve kırmızı kan hücrelerinde bulunmasıdır. Sonuç olarak, ALT, AST'den daha spesifik bir karaciğer iltihabı göstergesidir, çünkü AST miyokard enfarktüsü, akut pankreatit, akut hemolitik anemi, şiddetli yanıklar, akut böbrek hastalığı , kas-iskelet sistemi hastalıkları ve travma gibi diğer organları etkileyen hastalıklarda da yükselebilir.
# Tryglicerides = trigliseritler, Kandaki trigliserid düzeyinin 150 mg/dL'nin üzerinde bulunması, trigliserid yüksekliği (hipertrigliseridemi) olarak adlandırılır.
# 50 mg/dl'den düşük olan trigliserid seviyesine, trigliserid düşüklüğü denmektedir. 35 mg/dl'den daha düşük trigliserid seviyesine de aşırı trigliserid düşüklüğü denir.
# (https://www.milliyet.com.tr/pembenar/trigliserid-nedir-trigliserid-kac-olmali-yuksekligi-ve-dusuklugu-nedenleri-6169751#:~:text=50%20mg%2Fdl'den%20d%C3%BC%C5%9F%C3%BCk,ya%C4%9Fl%C4%B1%20g%C4%B1dalarla%20beslenme%20durumlar%C4%B1nda%20olabilir.)
# (https://www.medicalpark.com.tr/trigliserid/hg-2181)
# Platelets = trombositler, Trombosit veya kan pulcukları kan pıhtılarının oluşumunda görev alan hücre parçalarına verilen isimdir. Platelet olarak da adlandırılır. Düşük trombosit seviyeleri veya fonksiyon anormallikleri (disfonksiyon) kanamayı yatkınlaştırırken, yüksek trombosit seviyeleri -çoğunlukla asemptomatik- tromboz (damarda kanın pıhtılaşması) riskini yükseltir
# Sağlıklı bir kişide kan pulcukları değerinin 150, 000 ila 450, 000 hücre/ml aralığında olması beklenir. Trombosit değerinin taban değeri altına inmesiyle trombositopeni adı verilen trombosit düşüklüğü durumu söz konusu olur. Tavan değeri üstüne çıktığı takdirde de trombositoz adı verilen trombosit yüksekliği durumu gündemde olur.
# Prothrombin = Protrombin zamanı (PT), pıhtılaşmanın ortak ve ekstrinsik yolunu değerlendirmede kullanılan bir testtir. Bu testte plazmaya kalsiyum ve tromboplastin (doku faktörü) eklenerek, ekstrinsik yoldan fibrin pıhtısı oluşana dek geçen süre ölçülür. PT, FII, FVII, FX, protrombin ve fibrinojenin eksikliklerinin saptanması ile K vitamini antagonisti (coumadin) tedavisinin takibinde kullanılabilir
# Stage: hastalığın aşaması ,derecesi(1, 2, 3, or 4)
# bilgilerin çoğunluğu vikipediden elde edilmiştir
# # Kodlar
veri.info()
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sbn
veri = pd.read_csv("../input/cirrhosis-prediction-dataset/cirrhosis.csv")
veri
plt.subplots(figsize=(20, 10))
sbn.heatmap(veri.isnull(), cbar=False)
veri.isnull().sum()
veri.describe()
plt.subplots(figsize=(20, 10))
sbn.heatmap(veri.corr(), annot=True, cbar=False, linewidths=0.9)
|
[{"cirrhosis-prediction-dataset/cirrhosis.csv": {"column_names": "[\"ID\", \"N_Days\", \"Status\", \"Drug\", \"Age\", \"Sex\", \"Ascites\", \"Hepatomegaly\", \"Spiders\", \"Edema\", \"Bilirubin\", \"Cholesterol\", \"Albumin\", \"Copper\", \"Alk_Phos\", \"SGOT\", \"Tryglicerides\", \"Platelets\", \"Prothrombin\", \"Stage\"]", "column_data_types": "{\"ID\": \"int64\", \"N_Days\": \"int64\", \"Status\": \"object\", \"Drug\": \"object\", \"Age\": \"int64\", \"Sex\": \"object\", \"Ascites\": \"object\", \"Hepatomegaly\": \"object\", \"Spiders\": \"object\", \"Edema\": \"object\", \"Bilirubin\": \"float64\", \"Cholesterol\": \"float64\", \"Albumin\": \"float64\", \"Copper\": \"float64\", \"Alk_Phos\": \"float64\", \"SGOT\": \"float64\", \"Tryglicerides\": \"float64\", \"Platelets\": \"float64\", \"Prothrombin\": \"float64\", \"Stage\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 418 entries, 0 to 417\nData columns (total 20 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 ID 418 non-null int64 \n 1 N_Days 418 non-null int64 \n 2 Status 418 non-null object \n 3 Drug 312 non-null object \n 4 Age 418 non-null int64 \n 5 Sex 418 non-null object \n 6 Ascites 312 non-null object \n 7 Hepatomegaly 312 non-null object \n 8 Spiders 312 non-null object \n 9 Edema 418 non-null object \n 10 Bilirubin 418 non-null float64\n 11 Cholesterol 284 non-null float64\n 12 Albumin 418 non-null float64\n 13 Copper 310 non-null float64\n 14 Alk_Phos 312 non-null float64\n 15 SGOT 312 non-null float64\n 16 Tryglicerides 282 non-null float64\n 17 Platelets 407 non-null float64\n 18 Prothrombin 416 non-null float64\n 19 Stage 412 non-null float64\ndtypes: float64(10), int64(3), object(7)\nmemory usage: 65.4+ KB\n", "summary": "{\"ID\": {\"count\": 418.0, \"mean\": 209.5, \"std\": 120.81045760473994, \"min\": 1.0, \"25%\": 105.25, \"50%\": 209.5, \"75%\": 313.75, \"max\": 418.0}, \"N_Days\": {\"count\": 418.0, \"mean\": 1917.7822966507176, \"std\": 1104.6729923907321, \"min\": 41.0, \"25%\": 1092.75, \"50%\": 1730.0, \"75%\": 2613.5, \"max\": 4795.0}, \"Age\": {\"count\": 418.0, \"mean\": 18533.351674641148, \"std\": 3815.8450545514697, \"min\": 9598.0, \"25%\": 15644.5, \"50%\": 18628.0, \"75%\": 21272.5, \"max\": 28650.0}, \"Bilirubin\": {\"count\": 418.0, \"mean\": 3.2208133971291866, \"std\": 4.407506384141372, \"min\": 0.3, \"25%\": 0.8, \"50%\": 1.4, \"75%\": 3.4, \"max\": 28.0}, \"Cholesterol\": {\"count\": 284.0, \"mean\": 369.51056338028167, \"std\": 231.944545037874, \"min\": 120.0, \"25%\": 249.5, \"50%\": 309.5, \"75%\": 400.0, \"max\": 1775.0}, \"Albumin\": {\"count\": 418.0, \"mean\": 3.4974401913875592, \"std\": 0.4249716057796193, \"min\": 1.96, \"25%\": 3.2425, \"50%\": 3.53, \"75%\": 3.77, \"max\": 4.64}, \"Copper\": {\"count\": 310.0, \"mean\": 97.64838709677419, \"std\": 85.61391990897141, \"min\": 4.0, \"25%\": 41.25, \"50%\": 73.0, \"75%\": 123.0, \"max\": 588.0}, \"Alk_Phos\": {\"count\": 312.0, \"mean\": 1982.6557692307692, \"std\": 2140.388824451761, \"min\": 289.0, \"25%\": 871.5, \"50%\": 1259.0, \"75%\": 1980.0, \"max\": 13862.4}, \"SGOT\": {\"count\": 312.0, \"mean\": 122.55634615384616, \"std\": 56.699524863313016, \"min\": 26.35, \"25%\": 80.6, \"50%\": 114.7, \"75%\": 151.9, \"max\": 457.25}, \"Tryglicerides\": {\"count\": 282.0, \"mean\": 124.70212765957447, \"std\": 65.14863866583947, \"min\": 33.0, \"25%\": 84.25, \"50%\": 108.0, \"75%\": 151.0, \"max\": 598.0}, \"Platelets\": {\"count\": 407.0, \"mean\": 257.02457002457004, \"std\": 98.32558454996843, \"min\": 62.0, \"25%\": 188.5, \"50%\": 251.0, \"75%\": 318.0, \"max\": 721.0}, \"Prothrombin\": {\"count\": 416.0, \"mean\": 10.731730769230769, \"std\": 1.0220003464104215, \"min\": 9.0, \"25%\": 10.0, \"50%\": 10.6, \"75%\": 11.1, \"max\": 18.0}, \"Stage\": {\"count\": 412.0, \"mean\": 3.0242718446601944, \"std\": 0.8820420919404809, \"min\": 1.0, \"25%\": 2.0, \"50%\": 3.0, \"75%\": 4.0, \"max\": 4.0}}", "examples": "{\"ID\":{\"0\":1,\"1\":2,\"2\":3,\"3\":4},\"N_Days\":{\"0\":400,\"1\":4500,\"2\":1012,\"3\":1925},\"Status\":{\"0\":\"D\",\"1\":\"C\",\"2\":\"D\",\"3\":\"D\"},\"Drug\":{\"0\":\"D-penicillamine\",\"1\":\"D-penicillamine\",\"2\":\"D-penicillamine\",\"3\":\"D-penicillamine\"},\"Age\":{\"0\":21464,\"1\":20617,\"2\":25594,\"3\":19994},\"Sex\":{\"0\":\"F\",\"1\":\"F\",\"2\":\"M\",\"3\":\"F\"},\"Ascites\":{\"0\":\"Y\",\"1\":\"N\",\"2\":\"N\",\"3\":\"N\"},\"Hepatomegaly\":{\"0\":\"Y\",\"1\":\"Y\",\"2\":\"N\",\"3\":\"Y\"},\"Spiders\":{\"0\":\"Y\",\"1\":\"Y\",\"2\":\"N\",\"3\":\"Y\"},\"Edema\":{\"0\":\"Y\",\"1\":\"N\",\"2\":\"S\",\"3\":\"S\"},\"Bilirubin\":{\"0\":14.5,\"1\":1.1,\"2\":1.4,\"3\":1.8},\"Cholesterol\":{\"0\":261.0,\"1\":302.0,\"2\":176.0,\"3\":244.0},\"Albumin\":{\"0\":2.6,\"1\":4.14,\"2\":3.48,\"3\":2.54},\"Copper\":{\"0\":156.0,\"1\":54.0,\"2\":210.0,\"3\":64.0},\"Alk_Phos\":{\"0\":1718.0,\"1\":7394.8,\"2\":516.0,\"3\":6121.8},\"SGOT\":{\"0\":137.95,\"1\":113.52,\"2\":96.1,\"3\":60.63},\"Tryglicerides\":{\"0\":172.0,\"1\":88.0,\"2\":55.0,\"3\":92.0},\"Platelets\":{\"0\":190.0,\"1\":221.0,\"2\":151.0,\"3\":183.0},\"Prothrombin\":{\"0\":12.2,\"1\":10.6,\"2\":12.0,\"3\":10.3},\"Stage\":{\"0\":4.0,\"1\":3.0,\"2\":4.0,\"3\":4.0}}"}}]
| true | 1 |
<start_data_description><data_path>cirrhosis-prediction-dataset/cirrhosis.csv:
<column_names>
['ID', 'N_Days', 'Status', 'Drug', 'Age', 'Sex', 'Ascites', 'Hepatomegaly', 'Spiders', 'Edema', 'Bilirubin', 'Cholesterol', 'Albumin', 'Copper', 'Alk_Phos', 'SGOT', 'Tryglicerides', 'Platelets', 'Prothrombin', 'Stage']
<column_types>
{'ID': 'int64', 'N_Days': 'int64', 'Status': 'object', 'Drug': 'object', 'Age': 'int64', 'Sex': 'object', 'Ascites': 'object', 'Hepatomegaly': 'object', 'Spiders': 'object', 'Edema': 'object', 'Bilirubin': 'float64', 'Cholesterol': 'float64', 'Albumin': 'float64', 'Copper': 'float64', 'Alk_Phos': 'float64', 'SGOT': 'float64', 'Tryglicerides': 'float64', 'Platelets': 'float64', 'Prothrombin': 'float64', 'Stage': 'float64'}
<dataframe_Summary>
{'ID': {'count': 418.0, 'mean': 209.5, 'std': 120.81045760473994, 'min': 1.0, '25%': 105.25, '50%': 209.5, '75%': 313.75, 'max': 418.0}, 'N_Days': {'count': 418.0, 'mean': 1917.7822966507176, 'std': 1104.6729923907321, 'min': 41.0, '25%': 1092.75, '50%': 1730.0, '75%': 2613.5, 'max': 4795.0}, 'Age': {'count': 418.0, 'mean': 18533.351674641148, 'std': 3815.8450545514697, 'min': 9598.0, '25%': 15644.5, '50%': 18628.0, '75%': 21272.5, 'max': 28650.0}, 'Bilirubin': {'count': 418.0, 'mean': 3.2208133971291866, 'std': 4.407506384141372, 'min': 0.3, '25%': 0.8, '50%': 1.4, '75%': 3.4, 'max': 28.0}, 'Cholesterol': {'count': 284.0, 'mean': 369.51056338028167, 'std': 231.944545037874, 'min': 120.0, '25%': 249.5, '50%': 309.5, '75%': 400.0, 'max': 1775.0}, 'Albumin': {'count': 418.0, 'mean': 3.4974401913875592, 'std': 0.4249716057796193, 'min': 1.96, '25%': 3.2425, '50%': 3.53, '75%': 3.77, 'max': 4.64}, 'Copper': {'count': 310.0, 'mean': 97.64838709677419, 'std': 85.61391990897141, 'min': 4.0, '25%': 41.25, '50%': 73.0, '75%': 123.0, 'max': 588.0}, 'Alk_Phos': {'count': 312.0, 'mean': 1982.6557692307692, 'std': 2140.388824451761, 'min': 289.0, '25%': 871.5, '50%': 1259.0, '75%': 1980.0, 'max': 13862.4}, 'SGOT': {'count': 312.0, 'mean': 122.55634615384616, 'std': 56.699524863313016, 'min': 26.35, '25%': 80.6, '50%': 114.7, '75%': 151.9, 'max': 457.25}, 'Tryglicerides': {'count': 282.0, 'mean': 124.70212765957447, 'std': 65.14863866583947, 'min': 33.0, '25%': 84.25, '50%': 108.0, '75%': 151.0, 'max': 598.0}, 'Platelets': {'count': 407.0, 'mean': 257.02457002457004, 'std': 98.32558454996843, 'min': 62.0, '25%': 188.5, '50%': 251.0, '75%': 318.0, 'max': 721.0}, 'Prothrombin': {'count': 416.0, 'mean': 10.731730769230769, 'std': 1.0220003464104215, 'min': 9.0, '25%': 10.0, '50%': 10.6, '75%': 11.1, 'max': 18.0}, 'Stage': {'count': 412.0, 'mean': 3.0242718446601944, 'std': 0.8820420919404809, 'min': 1.0, '25%': 2.0, '50%': 3.0, '75%': 4.0, 'max': 4.0}}
<dataframe_info>
RangeIndex: 418 entries, 0 to 417
Data columns (total 20 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 ID 418 non-null int64
1 N_Days 418 non-null int64
2 Status 418 non-null object
3 Drug 312 non-null object
4 Age 418 non-null int64
5 Sex 418 non-null object
6 Ascites 312 non-null object
7 Hepatomegaly 312 non-null object
8 Spiders 312 non-null object
9 Edema 418 non-null object
10 Bilirubin 418 non-null float64
11 Cholesterol 284 non-null float64
12 Albumin 418 non-null float64
13 Copper 310 non-null float64
14 Alk_Phos 312 non-null float64
15 SGOT 312 non-null float64
16 Tryglicerides 282 non-null float64
17 Platelets 407 non-null float64
18 Prothrombin 416 non-null float64
19 Stage 412 non-null float64
dtypes: float64(10), int64(3), object(7)
memory usage: 65.4+ KB
<some_examples>
{'ID': {'0': 1, '1': 2, '2': 3, '3': 4}, 'N_Days': {'0': 400, '1': 4500, '2': 1012, '3': 1925}, 'Status': {'0': 'D', '1': 'C', '2': 'D', '3': 'D'}, 'Drug': {'0': 'D-penicillamine', '1': 'D-penicillamine', '2': 'D-penicillamine', '3': 'D-penicillamine'}, 'Age': {'0': 21464, '1': 20617, '2': 25594, '3': 19994}, 'Sex': {'0': 'F', '1': 'F', '2': 'M', '3': 'F'}, 'Ascites': {'0': 'Y', '1': 'N', '2': 'N', '3': 'N'}, 'Hepatomegaly': {'0': 'Y', '1': 'Y', '2': 'N', '3': 'Y'}, 'Spiders': {'0': 'Y', '1': 'Y', '2': 'N', '3': 'Y'}, 'Edema': {'0': 'Y', '1': 'N', '2': 'S', '3': 'S'}, 'Bilirubin': {'0': 14.5, '1': 1.1, '2': 1.4, '3': 1.8}, 'Cholesterol': {'0': 261.0, '1': 302.0, '2': 176.0, '3': 244.0}, 'Albumin': {'0': 2.6, '1': 4.14, '2': 3.48, '3': 2.54}, 'Copper': {'0': 156.0, '1': 54.0, '2': 210.0, '3': 64.0}, 'Alk_Phos': {'0': 1718.0, '1': 7394.8, '2': 516.0, '3': 6121.8}, 'SGOT': {'0': 137.95, '1': 113.52, '2': 96.1, '3': 60.63}, 'Tryglicerides': {'0': 172.0, '1': 88.0, '2': 55.0, '3': 92.0}, 'Platelets': {'0': 190.0, '1': 221.0, '2': 151.0, '3': 183.0}, 'Prothrombin': {'0': 12.2, '1': 10.6, '2': 12.0, '3': 10.3}, 'Stage': {'0': 4.0, '1': 3.0, '2': 4.0, '3': 4.0}}
<end_description>
| 2,644 | 0 | 4,966 | 2,644 |
69826392
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# To use pandas we need to import pandas library
import pandas as pd
# **Creating DataFrame**
# A DataFrame is a table. It contains an array of individual entries, each of which has a certain value. Each entry corresponds to a row (or record) and a column
# **pd.DataFrame()** = constructor to generate DataFrame objects. The syntax for declaring a new one is similar to a dictionary whose keys are the column names , and whose values are a list of entries.
pd.DataFrame({"CLASS": [1, 2, 3, 4, 5], "No. of Students": [23, 28, 45, 34, 21]})
#
# DataFrame entries are not limited to integers. For example, here's a DataFrame whose values are strings:-
pd.DataFrame(
{
"Customer_name": ["Jyoti", "Alice", "Bob"],
"Feedback": ["I like the content", "content is ok", "Insightful content"],
}
)
# Let's assign our own values for index
pd.DataFrame(
{
"Customer_name": ["Jyoti", "Alice", "Bob"],
"Feedback": ["I like the content", "content is ok", "Insightful content"],
},
index=["Notebook1", "Notebook2", "Notebook3"],
)
# Lets create a DataFrame which is more similar to real world data i.e. contains various datatypes such as integer, string, float etc.
pd.DataFrame(
{
"Items": ["Milk", "Bread", "Butter"],
"Quantity": [1.5, 2, 1],
"Amount": [4, 5, 6],
},
index=["Customer A", "Customer B", "Customer C"],
)
# **Series**
# A Series, by contrast, is a sequence of data values. If a DataFrame is a table, a Series is a list.
pd.Series([1, 2, 3, 4, 5])
# A Series is, in layman terms is a single column of a DataFrame. We can assign column values to the Series the same way as before, using an index parameter. However, a Series does not have a column name, it only has one overall name:
d.Series(
[30, 35, 40], index=["2019 Sales", "2020 Sales", "2021 Sales"], name="Notebook A"
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/826/69826392.ipynb
| null | null |
[{"Id": 69826392, "ScriptId": 19085358, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6848542, "CreationDate": "08/03/2021 13:11:51", "VersionNumber": 3.0, "Title": "Getting-Started-with-Pandas", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 51.0, "LinesInsertedFromPrevious": 4.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 47.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# To use pandas we need to import pandas library
import pandas as pd
# **Creating DataFrame**
# A DataFrame is a table. It contains an array of individual entries, each of which has a certain value. Each entry corresponds to a row (or record) and a column
# **pd.DataFrame()** = constructor to generate DataFrame objects. The syntax for declaring a new one is similar to a dictionary whose keys are the column names , and whose values are a list of entries.
pd.DataFrame({"CLASS": [1, 2, 3, 4, 5], "No. of Students": [23, 28, 45, 34, 21]})
#
# DataFrame entries are not limited to integers. For example, here's a DataFrame whose values are strings:-
pd.DataFrame(
{
"Customer_name": ["Jyoti", "Alice", "Bob"],
"Feedback": ["I like the content", "content is ok", "Insightful content"],
}
)
# Let's assign our own values for index
pd.DataFrame(
{
"Customer_name": ["Jyoti", "Alice", "Bob"],
"Feedback": ["I like the content", "content is ok", "Insightful content"],
},
index=["Notebook1", "Notebook2", "Notebook3"],
)
# Lets create a DataFrame which is more similar to real world data i.e. contains various datatypes such as integer, string, float etc.
pd.DataFrame(
{
"Items": ["Milk", "Bread", "Butter"],
"Quantity": [1.5, 2, 1],
"Amount": [4, 5, 6],
},
index=["Customer A", "Customer B", "Customer C"],
)
# **Series**
# A Series, by contrast, is a sequence of data values. If a DataFrame is a table, a Series is a list.
pd.Series([1, 2, 3, 4, 5])
# A Series is, in layman terms is a single column of a DataFrame. We can assign column values to the Series the same way as before, using an index parameter. However, a Series does not have a column name, it only has one overall name:
d.Series(
[30, 35, 40], index=["2019 Sales", "2020 Sales", "2021 Sales"], name="Notebook A"
)
| false | 0 | 738 | 0 | 738 | 738 |
||
69826582
|
<jupyter_start><jupyter_text>COVID-19 Dataset
[](https://forthebadge.com) [](https://forthebadge.com)
### Context
- A new coronavirus designated 2019-nCoV was first identified in Wuhan, the capital of China's Hubei province
- People developed pneumonia without a clear cause and for which existing vaccines or treatments were not effective.
- The virus has shown evidence of human-to-human transmission
- Transmission rate (rate of infection) appeared to escalate in mid-January 2020
- As of 30 January 2020, approximately 8,243 cases have been confirmed
### Content
> * **full_grouped.csv** - Day to day country wise no. of cases (Has County/State/Province level data)
> * **covid_19_clean_complete.csv** - Day to day country wise no. of cases (Doesn't have County/State/Province level data)
> * **country_wise_latest.csv** - Latest country level no. of cases
> * **day_wise.csv** - Day wise no. of cases (Doesn't have country level data)
> * **usa_county_wise.csv** - Day to day county level no. of cases
> * **worldometer_data.csv** - Latest data from https://www.worldometers.info/
Kaggle dataset identifier: corona-virus-report
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Importing Libraries
# Data Processing
import pandas as pd
import numpy as np
# Data Visulaisation
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
import os
# # Loading Data
files = os.listdir(r"../input/corona-virus-report")
files
# Function to read the data
def read_data(path, file):
return pd.read_csv(path + "/" + file)
path = r"../input/corona-virus-report"
world = read_data(path, "worldometer_data.csv")
world.head()
world.columns
# Countries with highest number of Cases
features = ["TotalCases", "TotalDeaths", "TotalRecovered", "ActiveCases"]
# tab_colors=['#03201A','#042823','#05302D','#073838','#0C4043','#13484F','#1C505B','#275868','#345F74','#426680','#516D8C','#627498','#737AA2','#8680AC','#9A86B5','#AE8BBD','#C390C4','#D995C9','#EE99CD','#000000'
# ]
tab_colors = [
"#1D2533",
"#242838",
"#2B2B3C",
"#322E41",
"#3A3145",
"#423349",
"#4B364C",
"#53384F",
"#5C3B52",
"#653D54",
"#6E3F55",
"#764257",
"#7F4457",
"#874758",
"#904A57",
"#984D57",
"#9F5056",
"#A65455",
"#AD5853",
"#B35D51",
]
for i in features:
fig = px.treemap(
world.iloc[0:20],
values=i,
path=["Country/Region"],
template="plotly_dark",
title="Tree Map depicting Impact of Covid-19 w.r.t {}".format(i),
color_discrete_sequence=tab_colors,
)
fig.show()
# A quick view of the daily trends
daily_records = read_data(path, "day_wise.csv")
daily_records.head()
daily_records.columns
line_col = ["#DDA2D2", "#C53595", "#2D6EAC", "#409BC5"]
px.line(
daily_records,
x="Date",
y=["Confirmed", "Deaths", "Recovered", "Active"],
template="plotly_dark",
title="Daily trends of Covid-19 cases",
labels={"Date": "Month", "value": "Statistics"},
color_discrete_sequence=line_col,
)
# % of Population tested in countries with highest number of cases
pop_test_ratio = ((world["TotalTests"] / world["Population"]) * 100).iloc[0:20]
fig1 = px.bar(
world.iloc[0:20],
x="Country/Region",
y=pop_test_ratio,
template="plotly_dark",
color="Country/Region",
title="% of Population tested",
labels={"y": "Test to Population Ratio"},
text=pop_test_ratio,
color_discrete_sequence=[
"#1D2533",
"#242838",
"#2B2B3C",
"#322E41",
"#3A3145",
"#423349",
"#4B364C",
"#53384F",
"#5C3B52",
"#653D54",
"#6E3F55",
"#764257",
"#7F4457",
"#874758",
"#904A57",
"#984D57",
"#9F5056",
"#A65455",
"#AD5853",
"#B35D51",
],
)
fig1.update_traces(texttemplate="%{text:0.1fs%}", textposition="outside")
fig1.update_xaxes(tickangle=270)
fig1.show()
# Countries that are most severly affected by Covid-19
world.columns
bar_color = ["#2B293A", "#42526C", "#4D819E", "#4BB4CE", "#41EBF6"]
bar_color = bar_color[::-1]
fig2 = px.bar(
world.iloc[0:20],
x="Country/Region",
y=[
"TotalCases",
"TotalRecovered",
"ActiveCases",
"TotalDeaths",
"Serious,Critical",
],
template="plotly_dark",
title="Severly Hit Countries",
color_discrete_sequence=bar_color,
)
fig2.update_xaxes(tickangle=270)
fig2.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/826/69826582.ipynb
|
corona-virus-report
|
imdevskp
|
[{"Id": 69826582, "ScriptId": 19078056, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6341134, "CreationDate": "08/03/2021 13:13:03", "VersionNumber": 3.0, "Title": "notebook48a1f670e2", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 118.0, "LinesInsertedFromPrevious": 16.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 102.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 93279296, "KernelVersionId": 69826582, "SourceDatasetVersionId": 1402868}]
|
[{"Id": 1402868, "DatasetId": 494766, "DatasourceVersionId": 1435700, "CreatorUserId": 1302389, "LicenseName": "Other (specified in description)", "CreationDate": "08/07/2020 03:47:47", "VersionNumber": 166.0, "Title": "COVID-19 Dataset", "Slug": "corona-virus-report", "Subtitle": "Number of Confirmed, Death and Recovered cases every day across the globe", "Description": "[](https://forthebadge.com) [](https://forthebadge.com)\n\n### Context\n\n- A new coronavirus designated 2019-nCoV was first identified in Wuhan, the capital of China's Hubei province\n- People developed pneumonia without a clear cause and for which existing vaccines or treatments were not effective. \n- The virus has shown evidence of human-to-human transmission\n- Transmission rate (rate of infection) appeared to escalate in mid-January 2020\n- As of 30 January 2020, approximately 8,243 cases have been confirmed\n\n\n### Content\n\n> * **full_grouped.csv** - Day to day country wise no. of cases (Has County/State/Province level data) \n> * **covid_19_clean_complete.csv** - Day to day country wise no. of cases (Doesn't have County/State/Province level data) \n> * **country_wise_latest.csv** - Latest country level no. of cases \n> * **day_wise.csv** - Day wise no. of cases (Doesn't have country level data) \n> * **usa_county_wise.csv** - Day to day county level no. of cases \n> * **worldometer_data.csv** - Latest data from https://www.worldometers.info/ \n\n\n### Acknowledgements / Data Source\n\n> https://github.com/CSSEGISandData/COVID-19\n> https://www.worldometers.info/\n\n### Collection methodology\n\n> https://github.com/imdevskp/covid_19_jhu_data_web_scrap_and_cleaning\n\n### Cover Photo\n\n> Photo from National Institutes of Allergy and Infectious Diseases\n> https://www.niaid.nih.gov/news-events/novel-coronavirus-sarscov2-images\n> https://blogs.cdc.gov/publichealthmatters/2019/04/h1n1/\n\n### Similar Datasets\n\n> * COVID-19 - https://www.kaggle.com/imdevskp/corona-virus-report \n> * MERS - https://www.kaggle.com/imdevskp/mers-outbreak-dataset-20122019\n> * Ebola Western Africa 2014 Outbreak - https://www.kaggle.com/imdevskp/ebola-outbreak-20142016-complete-dataset\n> * H1N1 | Swine Flu 2009 Pandemic Dataset - https://www.kaggle.com/imdevskp/h1n1-swine-flu-2009-pandemic-dataset\n> * SARS 2003 Pandemic - https://www.kaggle.com/imdevskp/sars-outbreak-2003-complete-dataset\n> * HIV AIDS - https://www.kaggle.com/imdevskp/hiv-aids-dataset", "VersionNotes": "update", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 494766, "CreatorUserId": 1302389, "OwnerUserId": 1302389.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1402868.0, "CurrentDatasourceVersionId": 1435700.0, "ForumId": 507860, "Type": 2, "CreationDate": "01/30/2020 14:46:58", "LastActivityDate": "01/30/2020", "TotalViews": 1009073, "TotalDownloads": 271389, "TotalVotes": 2056, "TotalKernels": 642}]
|
[{"Id": 1302389, "UserName": "imdevskp", "DisplayName": "Devakumar K. P.", "RegisterDate": "09/30/2017", "PerformanceTier": 3}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Importing Libraries
# Data Processing
import pandas as pd
import numpy as np
# Data Visulaisation
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
import os
# # Loading Data
files = os.listdir(r"../input/corona-virus-report")
files
# Function to read the data
def read_data(path, file):
return pd.read_csv(path + "/" + file)
path = r"../input/corona-virus-report"
world = read_data(path, "worldometer_data.csv")
world.head()
world.columns
# Countries with highest number of Cases
features = ["TotalCases", "TotalDeaths", "TotalRecovered", "ActiveCases"]
# tab_colors=['#03201A','#042823','#05302D','#073838','#0C4043','#13484F','#1C505B','#275868','#345F74','#426680','#516D8C','#627498','#737AA2','#8680AC','#9A86B5','#AE8BBD','#C390C4','#D995C9','#EE99CD','#000000'
# ]
tab_colors = [
"#1D2533",
"#242838",
"#2B2B3C",
"#322E41",
"#3A3145",
"#423349",
"#4B364C",
"#53384F",
"#5C3B52",
"#653D54",
"#6E3F55",
"#764257",
"#7F4457",
"#874758",
"#904A57",
"#984D57",
"#9F5056",
"#A65455",
"#AD5853",
"#B35D51",
]
for i in features:
fig = px.treemap(
world.iloc[0:20],
values=i,
path=["Country/Region"],
template="plotly_dark",
title="Tree Map depicting Impact of Covid-19 w.r.t {}".format(i),
color_discrete_sequence=tab_colors,
)
fig.show()
# A quick view of the daily trends
daily_records = read_data(path, "day_wise.csv")
daily_records.head()
daily_records.columns
line_col = ["#DDA2D2", "#C53595", "#2D6EAC", "#409BC5"]
px.line(
daily_records,
x="Date",
y=["Confirmed", "Deaths", "Recovered", "Active"],
template="plotly_dark",
title="Daily trends of Covid-19 cases",
labels={"Date": "Month", "value": "Statistics"},
color_discrete_sequence=line_col,
)
# % of Population tested in countries with highest number of cases
pop_test_ratio = ((world["TotalTests"] / world["Population"]) * 100).iloc[0:20]
fig1 = px.bar(
world.iloc[0:20],
x="Country/Region",
y=pop_test_ratio,
template="plotly_dark",
color="Country/Region",
title="% of Population tested",
labels={"y": "Test to Population Ratio"},
text=pop_test_ratio,
color_discrete_sequence=[
"#1D2533",
"#242838",
"#2B2B3C",
"#322E41",
"#3A3145",
"#423349",
"#4B364C",
"#53384F",
"#5C3B52",
"#653D54",
"#6E3F55",
"#764257",
"#7F4457",
"#874758",
"#904A57",
"#984D57",
"#9F5056",
"#A65455",
"#AD5853",
"#B35D51",
],
)
fig1.update_traces(texttemplate="%{text:0.1fs%}", textposition="outside")
fig1.update_xaxes(tickangle=270)
fig1.show()
# Countries that are most severly affected by Covid-19
world.columns
bar_color = ["#2B293A", "#42526C", "#4D819E", "#4BB4CE", "#41EBF6"]
bar_color = bar_color[::-1]
fig2 = px.bar(
world.iloc[0:20],
x="Country/Region",
y=[
"TotalCases",
"TotalRecovered",
"ActiveCases",
"TotalDeaths",
"Serious,Critical",
],
template="plotly_dark",
title="Severly Hit Countries",
color_discrete_sequence=bar_color,
)
fig2.update_xaxes(tickangle=270)
fig2.show()
| false | 0 | 1,490 | 0 | 1,896 | 1,490 |
||
69826638
|
<jupyter_start><jupyter_text>COVID-19 in India
### Context
Coronaviruses are a large family of viruses which may cause illness in animals or humans. In humans, several coronaviruses are known to cause respiratory infections ranging from the common cold to more severe diseases such as Middle East Respiratory Syndrome (MERS) and Severe Acute Respiratory Syndrome (SARS). The most recently discovered coronavirus causes coronavirus disease COVID-19 - World Health Organization
The number of new cases are increasing day by day around the world. This dataset has information from the states and union territories of India at daily level.
State level data comes from [Ministry of Health & Family Welfare](https://www.mohfw.gov.in/)
Testing data and vaccination data comes from [covid19india](https://www.covid19india.org/). Huge thanks to them for their efforts!
Update on April 20, 2021: Thanks to the [Team at ISIBang](https://www.isibang.ac.in/~athreya/incovid19/), I was able to get the historical data for the periods that I missed to collect and updated the csv file.
### Content
COVID-19 cases at daily level is present in `covid_19_india.csv` file
Statewise testing details in `StatewiseTestingDetails.csv` file
Travel history dataset by @dheerajmpai - https://www.kaggle.com/dheerajmpai/covidindiatravelhistory
Kaggle dataset identifier: covid19-in-india
<jupyter_code>import pandas as pd
df = pd.read_csv('covid19-in-india/covid_vaccine_statewise.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 7845 entries, 0 to 7844
Data columns (total 24 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Updated On 7845 non-null object
1 State 7845 non-null object
2 Total Doses Administered 7621 non-null float64
3 Sessions 7621 non-null float64
4 Sites 7621 non-null float64
5 First Dose Administered 7621 non-null float64
6 Second Dose Administered 7621 non-null float64
7 Male (Doses Administered) 7461 non-null float64
8 Female (Doses Administered) 7461 non-null float64
9 Transgender (Doses Administered) 7461 non-null float64
10 Covaxin (Doses Administered) 7621 non-null float64
11 CoviShield (Doses Administered) 7621 non-null float64
12 Sputnik V (Doses Administered) 2995 non-null float64
13 AEFI 5438 non-null float64
14 18-44 Years (Doses Administered) 1702 non-null float64
15 45-60 Years (Doses Administered) 1702 non-null float64
16 60+ Years (Doses Administered) 1702 non-null float64
17 18-44 Years(Individuals Vaccinated) 3733 non-null float64
18 45-60 Years(Individuals Vaccinated) 3734 non-null float64
19 60+ Years(Individuals Vaccinated) 3734 non-null float64
20 Male(Individuals Vaccinated) 160 non-null float64
21 Female(Individuals Vaccinated) 160 non-null float64
22 Transgender(Individuals Vaccinated) 160 non-null float64
23 Total Individuals Vaccinated 5919 non-null float64
dtypes: float64(22), object(2)
memory usage: 1.4+ MB
<jupyter_text>Examples:
{
"Updated On": "16/01/2021",
"State": "India",
"Total Doses Administered": 48276,
"Sessions": 3455,
" Sites ": 2957,
"First Dose Administered": 48276,
"Second Dose Administered": 0,
"Male (Doses Administered)": NaN,
"Female (Doses Administered)": NaN,
"Transgender (Doses Administered)": NaN,
" Covaxin (Doses Administered)": 579,
"CoviShield (Doses Administered)": 47697,
"Sputnik V (Doses Administered)": NaN,
"AEFI": NaN,
"18-44 Years (Doses Administered)": NaN,
"45-60 Years (Doses Administered)": NaN,
"60+ Years (Doses Administered)": NaN,
"18-44 Years(Individuals Vaccinated)": NaN,
"45-60 Years(Individuals Vaccinated)": NaN,
"60+ Years(Individuals Vaccinated)": NaN,
"...": "and 4 more columns"
}
{
"Updated On": "17/01/2021",
"State": "India",
"Total Doses Administered": 58604,
"Sessions": 8532,
" Sites ": 4954,
"First Dose Administered": 58604,
"Second Dose Administered": 0,
"Male (Doses Administered)": NaN,
"Female (Doses Administered)": NaN,
"Transgender (Doses Administered)": NaN,
" Covaxin (Doses Administered)": 635,
"CoviShield (Doses Administered)": 57969,
"Sputnik V (Doses Administered)": NaN,
"AEFI": NaN,
"18-44 Years (Doses Administered)": NaN,
"45-60 Years (Doses Administered)": NaN,
"60+ Years (Doses Administered)": NaN,
"18-44 Years(Individuals Vaccinated)": NaN,
"45-60 Years(Individuals Vaccinated)": NaN,
"60+ Years(Individuals Vaccinated)": NaN,
"...": "and 4 more columns"
}
{
"Updated On": "18/01/2021",
"State": "India",
"Total Doses Administered": 99449,
"Sessions": 13611,
" Sites ": 6583,
"First Dose Administered": 99449,
"Second Dose Administered": 0,
"Male (Doses Administered)": NaN,
"Female (Doses Administered)": NaN,
"Transgender (Doses Administered)": NaN,
" Covaxin (Doses Administered)": 1299,
"CoviShield (Doses Administered)": 98150,
"Sputnik V (Doses Administered)": NaN,
"AEFI": NaN,
"18-44 Years (Doses Administered)": NaN,
"45-60 Years (Doses Administered)": NaN,
"60+ Years (Doses Administered)": NaN,
"18-44 Years(Individuals Vaccinated)": NaN,
"45-60 Years(Individuals Vaccinated)": NaN,
"60+ Years(Individuals Vaccinated)": NaN,
"...": "and 4 more columns"
}
{
"Updated On": "19/01/2021",
"State": "India",
"Total Doses Administered": 195525,
"Sessions": 17855,
" Sites ": 7951,
"First Dose Administered": 195525,
"Second Dose Administered": 0,
"Male (Doses Administered)": NaN,
"Female (Doses Administered)": NaN,
"Transgender (Doses Administered)": NaN,
" Covaxin (Doses Administered)": 3017,
"CoviShield (Doses Administered)": 192508,
"Sputnik V (Doses Administered)": NaN,
"AEFI": NaN,
"18-44 Years (Doses Administered)": NaN,
"45-60 Years (Doses Administered)": NaN,
"60+ Years (Doses Administered)": NaN,
"18-44 Years(Individuals Vaccinated)": NaN,
"45-60 Years(Individuals Vaccinated)": NaN,
"60+ Years(Individuals Vaccinated)": NaN,
"...": "and 4 more columns"
}
<jupyter_code>import pandas as pd
df = pd.read_csv('covid19-in-india/StatewiseTestingDetails.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 16336 entries, 0 to 16335
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Date 16336 non-null object
1 State 16336 non-null object
2 TotalSamples 16336 non-null float64
3 Negative 6969 non-null object
4 Positive 5662 non-null float64
dtypes: float64(2), object(3)
memory usage: 638.2+ KB
<jupyter_text>Examples:
{
"Date": "2020-04-17 00:00:00",
"State": "Andaman and Nicobar Islands",
"TotalSamples": 1403,
"Negative": 1210.0,
"Positive": 12
}
{
"Date": "2020-04-24 00:00:00",
"State": "Andaman and Nicobar Islands",
"TotalSamples": 2679,
"Negative": NaN,
"Positive": 27
}
{
"Date": "2020-04-27 00:00:00",
"State": "Andaman and Nicobar Islands",
"TotalSamples": 2848,
"Negative": NaN,
"Positive": 33
}
{
"Date": "2020-05-01 00:00:00",
"State": "Andaman and Nicobar Islands",
"TotalSamples": 3754,
"Negative": NaN,
"Positive": 33
}
<jupyter_code>import pandas as pd
df = pd.read_csv('covid19-in-india/covid_19_india.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 18110 entries, 0 to 18109
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Sno 18110 non-null int64
1 Date 18110 non-null object
2 Time 18110 non-null object
3 State/UnionTerritory 18110 non-null object
4 ConfirmedIndianNational 18110 non-null object
5 ConfirmedForeignNational 18110 non-null object
6 Cured 18110 non-null int64
7 Deaths 18110 non-null int64
8 Confirmed 18110 non-null int64
dtypes: int64(4), object(5)
memory usage: 1.2+ MB
<jupyter_text>Examples:
{
"Sno": 1,
"Date": "2020-01-30 00:00:00",
"Time": "6:00 PM",
"State/UnionTerritory": "Kerala",
"ConfirmedIndianNational": 1,
"ConfirmedForeignNational": 0,
"Cured": 0,
"Deaths": 0,
"Confirmed": 1
}
{
"Sno": 2,
"Date": "2020-01-31 00:00:00",
"Time": "6:00 PM",
"State/UnionTerritory": "Kerala",
"ConfirmedIndianNational": 1,
"ConfirmedForeignNational": 0,
"Cured": 0,
"Deaths": 0,
"Confirmed": 1
}
{
"Sno": 3,
"Date": "2020-02-01 00:00:00",
"Time": "6:00 PM",
"State/UnionTerritory": "Kerala",
"ConfirmedIndianNational": 2,
"ConfirmedForeignNational": 0,
"Cured": 0,
"Deaths": 0,
"Confirmed": 2
}
{
"Sno": 4,
"Date": "2020-02-02 00:00:00",
"Time": "6:00 PM",
"State/UnionTerritory": "Kerala",
"ConfirmedIndianNational": 3,
"ConfirmedForeignNational": 0,
"Cured": 0,
"Deaths": 0,
"Confirmed": 3
}
<jupyter_script>import numpy as np
import pandas as pd
from scipy.stats import ttest_ind
from statsmodels.stats.proportion import proportions_ztest
import geopandas as gpd
import matplotlib.dates as mdates
from matplotlib.dates import DateFormatter
state_df = pd.read_csv("../input/covid19-in-india/StatewiseTestingDetails.csv")
country_df = pd.read_csv("../input/covid19-in-india/covid_19_india.csv")
state_df.head()
state_df.head()
state_df[state_df["State"] == "Tamil Nadu"].head()
country_df[
(
(country_df["Date"] == "2020-03-30")
& (country_df["State/UnionTerritory"] == "Tamil Nadu")
)
]
# Copying the contry_df data to df for easy reference
df = country_df.copy()
# Keeping it simple, we will drop the rows with state name ending with "***" as it is seems to be rows with incomplete values.
def drop_star(df):
for i in df["State/UnionTerritory"].iteritems():
if i[1][-3:] == "***":
df.drop(i[0], inplace=True)
drop_star(df)
df["State/UnionTerritory"].unique()
# lets convert the Date feature to Date&time datatype
df["Date"] = pd.to_datetime(df["Date"], format="%Y-%m-%d")
# Time is not required as it doesnt make much difference
df.drop(["Time"], axis=1, inplace=True)
# Renaming State/UnionTerritory to States for easy reference
df.rename(columns={"State/UnionTerritory": "States"}, inplace=True)
df["Active_cases"] = df["Confirmed"] - (df["Cured"] + df["Deaths"])
df["Discharge_Rate"] = np.round(
(df["Cured"] / df["Confirmed"]) * 100, decimals=4
) # create instances for 'death_rate and discharge_rate'
df["Death_Rate"] = np.round((df["Deaths"] / df["Confirmed"]) * 100, decimals=4)
df.head()
df[df["States"] == "Tamil Nadu"].tail()
df[df["States"] == "Maharashtra"].tail()
df[df["States"] == "West Bengal"].tail()
import matplotlib.pyplot as plt
import matplotlib.dates as mtd
import seaborn as sns
from matplotlib.ticker import ScalarFormatter
colors = ["#0C68C7", "#3A6794", "#00FAF3", "#FA643C", "#C71D12"]
sns.set(palette=colors, style="white")
sns.palplot(colors)
# Current date from the record
current = df[df.Date == "2021-07-06"]
# Sorting data in descending ordrer (Confirmed)
max_confirmed_cases = current.sort_values(by="Confirmed", ascending=False)
max_confirmed_cases.head()
# Selecting top 10 States/U.T
top_cases = max_confirmed_cases[:10]
top_cases.head()
for feature in top_cases[
["Confirmed", "Cured", "Deaths", "Active_cases", "Discharge_Rate", "Death_Rate"]
]:
fig = plt.figure(figsize=(15, 5))
plt.title("Top 10 highly impacted sates as on 7th June", size=10)
ax = sns.barplot(
data=top_cases, y=top_cases[feature], x="States", linewidth=0, edgecolor="black"
)
plt.xlabel("States", size=15)
plt.ylabel(feature, size=15)
for i in ax.patches:
ax.text(x=i.get_x(), y=i.get_height(), s=i.get_height())
plt.show()
states = [
"Kerala",
"Tamil Nadu",
"Maharashtra",
"Tamil Nadu",
"Andhra Pradesh",
"Uttar Pradesh",
]
mh = df[df["States"] == "Maharashtra"]
kl = df[df["States"] == "Kerala"]
ka = df[df["States"] == "Karnataka"]
tn = df[df["States"] == "Tamil Nadu"]
ap = df[df["States"] == "Andhra Pradesh"]
up = df[df["States"] == "Uttar Pradesh"]
fig, ax = plt.subplots(
nrows=3,
ncols=3,
figsize=(23, 10),
squeeze=False,
sharex=True,
sharey=False,
constrained_layout=True,
)
plt.suptitle("Comparison of Active, Cured & Deaths for top States", size=25)
sns.lineplot(data=tn, x="Date", y="Active_cases", ax=ax[0, 0], color=colors[1])
ax[0, 0].set_title("Maharashtra", size=20)
sns.lineplot(data=tn, x="Date", y="Cured", ax=ax[1, 0], color=colors[1])
sns.lineplot(data=tn, x="Date", y="Deaths", ax=ax[2, 0], color=colors[1])
sns.lineplot(data=kl, x="Date", y="Active_cases", ax=ax[0, 1], color=colors[2])
ax[0, 1].set_title("Kerala", size=20)
sns.lineplot(data=kl, x="Date", y="Cured", ax=ax[1, 1], color=colors[2])
sns.lineplot(data=kl, x="Date", y="Deaths", ax=ax[2, 1], color=colors[2])
sns.lineplot(data=mh, x="Date", y="Active_cases", ax=ax[0, 2], color=colors[3])
ax[0, 2].set_title("Karnataka", size=20)
sns.lineplot(data=mh, x="Date", y="Cured", ax=ax[1, 2], color=colors[3])
sns.lineplot(data=mh, x="Date", y="Deaths", ax=ax[2, 2], color=colors[3])
plt.show()
fig, ax = plt.subplots(
nrows=3,
ncols=3,
figsize=(23, 10),
squeeze=False,
sharex=True,
sharey=False,
constrained_layout=True,
)
sns.lineplot(data=tn, x="Date", y="Active_cases", ax=ax[0, 0], color=colors[1])
ax[0, 0].set_title("Tamil Nadu", size=20)
sns.lineplot(data=tn, x="Date", y="Cured", ax=ax[1, 0], color=colors[1])
sns.lineplot(data=tn, x="Date", y="Deaths", ax=ax[2, 0], color=colors[1])
sns.lineplot(data=kl, x="Date", y="Active_cases", ax=ax[0, 1], color=colors[2])
ax[0, 1].set_title("Andhra Pradesh", size=20)
sns.lineplot(data=kl, x="Date", y="Cured", ax=ax[1, 1], color=colors[2])
sns.lineplot(data=kl, x="Date", y="Deaths", ax=ax[2, 1], color=colors[2])
sns.lineplot(data=mh, x="Date", y="Active_cases", ax=ax[0, 2], color=colors[3])
ax[0, 2].set_title("Uttar Pradesh", size=20)
sns.lineplot(data=mh, x="Date", y="Cured", ax=ax[1, 2], color=colors[3])
sns.lineplot(data=mh, x="Date", y="Deaths", ax=ax[2, 2], color=colors[3])
plt.show()
df["Date"] = pd.to_datetime(df["Date"]) # Date is converted to DateTime format.
data_20 = df[df["Date"].dt.year == 2020] # Considering data of only the year 2020.
data_21 = df[df["Date"].dt.year == 2021] # Considering data of only the year 2021.
data_20["Month"] = data_20[
"Date"
].dt.month # Month is accessed from the DateTime object.
data_21["Month"] = data_21["Date"].dt.month
# Year 2020
data_confirm_20 = data_20["Confirmed"].groupby(data_20["Month"]).sum()
data_dis_20 = (
data_20["Cured"].groupby(data_20["Month"]).sum()
) # creating instances for 'confirmed','deaths','discharged' by month column
data_death_20 = data_20["Deaths"].groupby(data_20["Month"]).sum()
# Year 2021
data_confirm_21 = data_21["Confirmed"].groupby(data_21["Month"]).sum()
data_dis_21 = (
data_21["Cured"].groupby(data_21["Month"]).sum()
) # creating instances for 'confirmed','deaths','discharged' by month column
data_death_21 = data_21["Deaths"].groupby(data_21["Month"]).sum()
cols_20 = [data_confirm_20, data_dis_20, data_death_20]
data_20 = pd.concat(cols_20, axis=1)
cols_21 = [data_confirm_21, data_dis_21, data_death_21]
data_21 = pd.concat(cols_21, axis=1)
# Year 2020
data_20["discharge_rate_20"] = np.round(
(data_20["Cured"] / data_20["Confirmed"]) * 100, decimals=4
) # create instances for 'death_rate and discharge_rate'
data_20["death_rate_20"] = np.round(
(data_20["Deaths"] / data_20["Confirmed"]) * 100, decimals=4
)
# Year 2020
data_21["discharge_rate_21"] = np.round(
(data_21["Cured"] / data_21["Confirmed"]) * 100, decimals=4
) # create instances for 'death_rate and discharge_rate'
data_21["death_rate_21"] = np.round(
(data_21["Deaths"] / data_21["Confirmed"]) * 100, decimals=4
)
# Year 2020
data_20.reset_index(inplace=True)
data_20.head()
# Year 2021
data_21.reset_index(inplace=True)
data_21.head()
plt.figure(figsize=(10, 5))
sns.lineplot(
x="Month",
y="discharge_rate_20",
data=data_20,
color="g",
lw=3,
marker="o",
markersize=10,
)
plt.title("DISCHARGE RATE PER MONTH IN 2020")
plt.show()
plt.figure(figsize=(10, 5))
sns.lineplot(
x="Month",
y="discharge_rate_21",
data=data_21,
color="g",
lw=3,
marker="o",
markersize=10,
)
plt.title("DISCHARGE RATE PER MONTH IN 2021")
plt.show()
plt.figure(figsize=(10, 5))
sns.lineplot(
x="Month",
y="death_rate_20",
data=data_20,
color="r",
lw=3,
marker="o",
markersize=10,
)
plt.title("DEATH RATE PER MONTH IN 2020")
plt.show()
plt.figure(figsize=(10, 5))
sns.lineplot(
x="Month",
y="death_rate_21",
data=data_21,
color="r",
lw=3,
marker="o",
markersize=10,
)
plt.title("DEATH RATE PER MONTH IN 2021")
plt.show()
tn = df[df["States"] == "Tamil Nadu"]["Cured"]
mh = df[df["States"] == "Maharashtra"]["Cured"]
kl = df[df["States"] == "Kerala"]["Cured"]
df.head()
print(
"Total number of Active Covid-19 cases across India : {}".format(
max_confirmed_cases["Active_cases"].sum()
)
)
top_cases = (
max_confirmed_cases.groupby("States")["Active_cases"]
.max()
.sort_values(ascending=False)
.to_frame()
)
top_cases.style.background_gradient(cmap="flare")
from fbprophet import Prophet
confirmed = df.groupby("Date").sum()["Confirmed"].reset_index()
recovered = df.groupby("Date").sum()["Cured"].reset_index()
deaths = df.groupby("Date").sum()["Deaths"].reset_index()
confirmed.head()
confirmed.columns = ["ds", "y"]
confirmed["ds"] = pd.to_datetime(confirmed["ds"])
m = Prophet(interval_width=0.95)
m.fit(confirmed)
future = m.make_future_dataframe(periods=7) # Making future prediction for next 7 days
future.tail(10)
forecast = m.predict(future)
forecast[["ds", "yhat", "yhat_lower", "yhat_upper"]].tail()
confirmed_forecast_plot = m.plot(forecast)
confirmed_forecast_plot = m.plot_components(forecast)
print(
"Average recovery rate Covid-19 cases across India : {}".format(
max_confirmed_cases["Discharge_Rate"].mean()
)
)
top_cases = (
max_confirmed_cases.groupby("States")["Discharge_Rate"]
.max()
.sort_values(ascending=False)
.to_frame()
)
top_cases.style.background_gradient(cmap="flare")
discharge_rate = df[["Date", "Discharge_Rate"]]
discharge_rate.head()
discharge_rate.columns = ["ds", "y"]
discharge_rate["ds"] = pd.to_datetime(discharge_rate["ds"])
m = Prophet(interval_width=0.50)
m.fit(discharge_rate)
future = m.make_future_dataframe(periods=7)
forecast = m.predict(future)
forecast[["ds", "yhat", "yhat_lower", "yhat_upper"]].tail()
confirmed_forecast_plot = m.plot(forecast)
confirmed_forecast_plot = m.plot_components(forecast)
print(
"Average recovery rate Covid-19 cases across India : {}".format(
max_confirmed_cases["Discharge_Rate"].mean()
)
)
top_cases = (
max_confirmed_cases.groupby("States")["Death_Rate"]
.max()
.sort_values(ascending=False)
.to_frame()
)
top_cases.style.background_gradient(cmap="flare")
discharge_rate = df[["Date", "Death_Rate"]]
discharge_rate.head()
discharge_rate.columns = ["ds", "y"]
discharge_rate["ds"] = pd.to_datetime(discharge_rate["ds"])
m = Prophet(interval_width=0.50)
m.fit(discharge_rate)
future = m.make_future_dataframe(periods=7) # Making future prediction for next 7 days
future.tail(10)
forecast = m.predict(future)
forecast[["ds", "yhat", "yhat_lower", "yhat_upper"]].tail()
df.head()
state_cases = max_confirmed_cases.drop(
["Sno", "Date", "ConfirmedIndianNational", "ConfirmedForeignNational"], axis=1
)
state_wise_cases = (
state_cases.sort_values("Confirmed", ascending=False)
.fillna(0)
.style.background_gradient(cmap="Blues", subset=["Confirmed"])
.background_gradient(cmap="Reds", subset=["Deaths"])
.background_gradient(cmap="Greens", subset=["Cured"])
.background_gradient(cmap="Oranges", subset=["Active_cases"])
.background_gradient(cmap="RdYlBu", subset=["Death_Rate"])
.background_gradient(cmap="Accent", subset=["Discharge_Rate"])
)
state_wise_cases
# Loading the vaccination data
vaccine_df = pd.read_csv("../input/covid19-in-india/covid_vaccine_statewise.csv")
vacc_df = vaccine_df.copy()
vaccine_df = vaccine_df[
[
"Updated On",
"State",
"Total Doses Administered",
"Total Sessions Conducted",
"First Dose Administered",
"Second Dose Administered",
"Male(Individuals Vaccinated)",
"Female(Individuals Vaccinated)",
"Transgender(Individuals Vaccinated)",
"Total Covaxin Administered",
"Total CoviShield Administered",
"Total Sputnik V Administered",
"18-45 years (Age)",
"45-60 years (Age)",
"60+ years (Age)",
"Total Individuals Vaccinated",
]
]
vaccine_df.columns = [
"Date",
"States",
"Total_Doses_Administered",
"Total_Sessions_Conducted",
"First_Dose_Administered",
"Second_Dose_Administered",
"Male_Vaccinated",
"Female_Vaccinated",
"Transgender_Vaccinated",
"Total_Covaxin_Administered",
"Total_CoviShield_Administered",
"Total_SputnikV_Administered",
"18-45_Years",
"45-60_Years",
"60+_Years",
"Total_Individuals_Vaccinated",
]
vaccine_df.head()
# Checking the null values
vaccine_df.isnull().sum()
vaccine_df = vaccine_df[vaccine_df["Total_Individuals_Vaccinated"].notna()]
vaccine_df["Date"] = pd.to_datetime(vaccine_df["Date"], format="%d/%m/%Y")
latest_date = max(vaccine_df["Date"])
print("Current Date : ", latest_date)
vaccine_df[vaccine_df["States"] == "Dadra and Nagar Haveli and Daman and Diu"]
fig, ax = plt.subplots(ncols=1, nrows=1, dpi=100, figsize=(6, 4))
sns.lineplot(
data=vaccine_df[vaccine_df["States"] == "India"],
x="Date",
y="Total_Individuals_Vaccinated",
ax=ax,
)
ax.xaxis.set_major_locator(mdates.MonthLocator(interval=1))
ax.xaxis.set_major_formatter(DateFormatter("%b%y"))
ax.set_ylabel("Total Indians Vaccinated so far in millions", fontsize=10)
ax.set_yticklabels([0, 0, 20, 40, 60, 80, 100, 120])
Xstart, Xend = ax.get_xlim()
plt.title("India's Vaccination Performance")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/826/69826638.ipynb
|
covid19-in-india
|
sudalairajkumar
|
[{"Id": 69826638, "ScriptId": 19082139, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6267564, "CreationDate": "08/03/2021 13:13:23", "VersionNumber": 1.0, "Title": "Covid 19 Analysis and Prediction", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 302.0, "LinesInsertedFromPrevious": 7.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 295.0, "LinesInsertedFromFork": 7.0, "LinesDeletedFromFork": 5.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 295.0, "TotalVotes": 5}]
|
[{"Id": 93279358, "KernelVersionId": 69826638, "SourceDatasetVersionId": 2492403}]
|
[{"Id": 2492403, "DatasetId": 557629, "DatasourceVersionId": 2534981, "CreatorUserId": 71388, "LicenseName": "CC0: Public Domain", "CreationDate": "08/02/2021 16:29:41", "VersionNumber": 236.0, "Title": "COVID-19 in India", "Slug": "covid19-in-india", "Subtitle": "Dataset on Novel Corona Virus Disease 2019 in India", "Description": "### Context\n\nCoronaviruses are a large family of viruses which may cause illness in animals or humans. In humans, several coronaviruses are known to cause respiratory infections ranging from the common cold to more severe diseases such as Middle East Respiratory Syndrome (MERS) and Severe Acute Respiratory Syndrome (SARS). The most recently discovered coronavirus causes coronavirus disease COVID-19 - World Health Organization\n\nThe number of new cases are increasing day by day around the world. This dataset has information from the states and union territories of India at daily level.\n\nState level data comes from [Ministry of Health & Family Welfare](https://www.mohfw.gov.in/)\n\nTesting data and vaccination data comes from [covid19india](https://www.covid19india.org/). Huge thanks to them for their efforts!\n\nUpdate on April 20, 2021: Thanks to the [Team at ISIBang](https://www.isibang.ac.in/~athreya/incovid19/), I was able to get the historical data for the periods that I missed to collect and updated the csv file.\n\n### Content\n\nCOVID-19 cases at daily level is present in `covid_19_india.csv` file\n\nStatewise testing details in `StatewiseTestingDetails.csv` file \n\nTravel history dataset by @dheerajmpai - https://www.kaggle.com/dheerajmpai/covidindiatravelhistory\n\n\n### Acknowledgements\n\nThanks to Indian [Ministry of Health & Family Welfare](https://www.mohfw.gov.in/) for making the data available to general public. \n\nThanks to [covid19india.org](http://portal.covid19india.org/) for making the individual level details, testing details, vaccination details available to general public.\n\nThanks to [Wikipedia](https://en.wikipedia.org/wiki/List_of_states_and_union_territories_of_India_by_population) for population information.\n\nThanks to the [Team at ISIBang](https://www.isibang.ac.in/~athreya/incovid19/)\n\nPhoto Courtesy - https://hgis.uw.edu/virus/\n\n### Inspiration\n\nLooking for data based suggestions to stop / delay the spread of virus", "VersionNotes": "new data", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 557629, "CreatorUserId": 71388, "OwnerUserId": 71388.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2516524.0, "CurrentDatasourceVersionId": 2559305.0, "ForumId": 571269, "Type": 2, "CreationDate": "03/16/2020 06:24:37", "LastActivityDate": "03/16/2020", "TotalViews": 894555, "TotalDownloads": 204666, "TotalVotes": 1906, "TotalKernels": 548}]
|
[{"Id": 71388, "UserName": "sudalairajkumar", "DisplayName": "SRK", "RegisterDate": "11/28/2012", "PerformanceTier": 4}]
|
import numpy as np
import pandas as pd
from scipy.stats import ttest_ind
from statsmodels.stats.proportion import proportions_ztest
import geopandas as gpd
import matplotlib.dates as mdates
from matplotlib.dates import DateFormatter
state_df = pd.read_csv("../input/covid19-in-india/StatewiseTestingDetails.csv")
country_df = pd.read_csv("../input/covid19-in-india/covid_19_india.csv")
state_df.head()
state_df.head()
state_df[state_df["State"] == "Tamil Nadu"].head()
country_df[
(
(country_df["Date"] == "2020-03-30")
& (country_df["State/UnionTerritory"] == "Tamil Nadu")
)
]
# Copying the contry_df data to df for easy reference
df = country_df.copy()
# Keeping it simple, we will drop the rows with state name ending with "***" as it is seems to be rows with incomplete values.
def drop_star(df):
for i in df["State/UnionTerritory"].iteritems():
if i[1][-3:] == "***":
df.drop(i[0], inplace=True)
drop_star(df)
df["State/UnionTerritory"].unique()
# lets convert the Date feature to Date&time datatype
df["Date"] = pd.to_datetime(df["Date"], format="%Y-%m-%d")
# Time is not required as it doesnt make much difference
df.drop(["Time"], axis=1, inplace=True)
# Renaming State/UnionTerritory to States for easy reference
df.rename(columns={"State/UnionTerritory": "States"}, inplace=True)
df["Active_cases"] = df["Confirmed"] - (df["Cured"] + df["Deaths"])
df["Discharge_Rate"] = np.round(
(df["Cured"] / df["Confirmed"]) * 100, decimals=4
) # create instances for 'death_rate and discharge_rate'
df["Death_Rate"] = np.round((df["Deaths"] / df["Confirmed"]) * 100, decimals=4)
df.head()
df[df["States"] == "Tamil Nadu"].tail()
df[df["States"] == "Maharashtra"].tail()
df[df["States"] == "West Bengal"].tail()
import matplotlib.pyplot as plt
import matplotlib.dates as mtd
import seaborn as sns
from matplotlib.ticker import ScalarFormatter
colors = ["#0C68C7", "#3A6794", "#00FAF3", "#FA643C", "#C71D12"]
sns.set(palette=colors, style="white")
sns.palplot(colors)
# Current date from the record
current = df[df.Date == "2021-07-06"]
# Sorting data in descending ordrer (Confirmed)
max_confirmed_cases = current.sort_values(by="Confirmed", ascending=False)
max_confirmed_cases.head()
# Selecting top 10 States/U.T
top_cases = max_confirmed_cases[:10]
top_cases.head()
for feature in top_cases[
["Confirmed", "Cured", "Deaths", "Active_cases", "Discharge_Rate", "Death_Rate"]
]:
fig = plt.figure(figsize=(15, 5))
plt.title("Top 10 highly impacted sates as on 7th June", size=10)
ax = sns.barplot(
data=top_cases, y=top_cases[feature], x="States", linewidth=0, edgecolor="black"
)
plt.xlabel("States", size=15)
plt.ylabel(feature, size=15)
for i in ax.patches:
ax.text(x=i.get_x(), y=i.get_height(), s=i.get_height())
plt.show()
states = [
"Kerala",
"Tamil Nadu",
"Maharashtra",
"Tamil Nadu",
"Andhra Pradesh",
"Uttar Pradesh",
]
mh = df[df["States"] == "Maharashtra"]
kl = df[df["States"] == "Kerala"]
ka = df[df["States"] == "Karnataka"]
tn = df[df["States"] == "Tamil Nadu"]
ap = df[df["States"] == "Andhra Pradesh"]
up = df[df["States"] == "Uttar Pradesh"]
fig, ax = plt.subplots(
nrows=3,
ncols=3,
figsize=(23, 10),
squeeze=False,
sharex=True,
sharey=False,
constrained_layout=True,
)
plt.suptitle("Comparison of Active, Cured & Deaths for top States", size=25)
sns.lineplot(data=tn, x="Date", y="Active_cases", ax=ax[0, 0], color=colors[1])
ax[0, 0].set_title("Maharashtra", size=20)
sns.lineplot(data=tn, x="Date", y="Cured", ax=ax[1, 0], color=colors[1])
sns.lineplot(data=tn, x="Date", y="Deaths", ax=ax[2, 0], color=colors[1])
sns.lineplot(data=kl, x="Date", y="Active_cases", ax=ax[0, 1], color=colors[2])
ax[0, 1].set_title("Kerala", size=20)
sns.lineplot(data=kl, x="Date", y="Cured", ax=ax[1, 1], color=colors[2])
sns.lineplot(data=kl, x="Date", y="Deaths", ax=ax[2, 1], color=colors[2])
sns.lineplot(data=mh, x="Date", y="Active_cases", ax=ax[0, 2], color=colors[3])
ax[0, 2].set_title("Karnataka", size=20)
sns.lineplot(data=mh, x="Date", y="Cured", ax=ax[1, 2], color=colors[3])
sns.lineplot(data=mh, x="Date", y="Deaths", ax=ax[2, 2], color=colors[3])
plt.show()
fig, ax = plt.subplots(
nrows=3,
ncols=3,
figsize=(23, 10),
squeeze=False,
sharex=True,
sharey=False,
constrained_layout=True,
)
sns.lineplot(data=tn, x="Date", y="Active_cases", ax=ax[0, 0], color=colors[1])
ax[0, 0].set_title("Tamil Nadu", size=20)
sns.lineplot(data=tn, x="Date", y="Cured", ax=ax[1, 0], color=colors[1])
sns.lineplot(data=tn, x="Date", y="Deaths", ax=ax[2, 0], color=colors[1])
sns.lineplot(data=kl, x="Date", y="Active_cases", ax=ax[0, 1], color=colors[2])
ax[0, 1].set_title("Andhra Pradesh", size=20)
sns.lineplot(data=kl, x="Date", y="Cured", ax=ax[1, 1], color=colors[2])
sns.lineplot(data=kl, x="Date", y="Deaths", ax=ax[2, 1], color=colors[2])
sns.lineplot(data=mh, x="Date", y="Active_cases", ax=ax[0, 2], color=colors[3])
ax[0, 2].set_title("Uttar Pradesh", size=20)
sns.lineplot(data=mh, x="Date", y="Cured", ax=ax[1, 2], color=colors[3])
sns.lineplot(data=mh, x="Date", y="Deaths", ax=ax[2, 2], color=colors[3])
plt.show()
df["Date"] = pd.to_datetime(df["Date"]) # Date is converted to DateTime format.
data_20 = df[df["Date"].dt.year == 2020] # Considering data of only the year 2020.
data_21 = df[df["Date"].dt.year == 2021] # Considering data of only the year 2021.
data_20["Month"] = data_20[
"Date"
].dt.month # Month is accessed from the DateTime object.
data_21["Month"] = data_21["Date"].dt.month
# Year 2020
data_confirm_20 = data_20["Confirmed"].groupby(data_20["Month"]).sum()
data_dis_20 = (
data_20["Cured"].groupby(data_20["Month"]).sum()
) # creating instances for 'confirmed','deaths','discharged' by month column
data_death_20 = data_20["Deaths"].groupby(data_20["Month"]).sum()
# Year 2021
data_confirm_21 = data_21["Confirmed"].groupby(data_21["Month"]).sum()
data_dis_21 = (
data_21["Cured"].groupby(data_21["Month"]).sum()
) # creating instances for 'confirmed','deaths','discharged' by month column
data_death_21 = data_21["Deaths"].groupby(data_21["Month"]).sum()
cols_20 = [data_confirm_20, data_dis_20, data_death_20]
data_20 = pd.concat(cols_20, axis=1)
cols_21 = [data_confirm_21, data_dis_21, data_death_21]
data_21 = pd.concat(cols_21, axis=1)
# Year 2020
data_20["discharge_rate_20"] = np.round(
(data_20["Cured"] / data_20["Confirmed"]) * 100, decimals=4
) # create instances for 'death_rate and discharge_rate'
data_20["death_rate_20"] = np.round(
(data_20["Deaths"] / data_20["Confirmed"]) * 100, decimals=4
)
# Year 2020
data_21["discharge_rate_21"] = np.round(
(data_21["Cured"] / data_21["Confirmed"]) * 100, decimals=4
) # create instances for 'death_rate and discharge_rate'
data_21["death_rate_21"] = np.round(
(data_21["Deaths"] / data_21["Confirmed"]) * 100, decimals=4
)
# Year 2020
data_20.reset_index(inplace=True)
data_20.head()
# Year 2021
data_21.reset_index(inplace=True)
data_21.head()
plt.figure(figsize=(10, 5))
sns.lineplot(
x="Month",
y="discharge_rate_20",
data=data_20,
color="g",
lw=3,
marker="o",
markersize=10,
)
plt.title("DISCHARGE RATE PER MONTH IN 2020")
plt.show()
plt.figure(figsize=(10, 5))
sns.lineplot(
x="Month",
y="discharge_rate_21",
data=data_21,
color="g",
lw=3,
marker="o",
markersize=10,
)
plt.title("DISCHARGE RATE PER MONTH IN 2021")
plt.show()
plt.figure(figsize=(10, 5))
sns.lineplot(
x="Month",
y="death_rate_20",
data=data_20,
color="r",
lw=3,
marker="o",
markersize=10,
)
plt.title("DEATH RATE PER MONTH IN 2020")
plt.show()
plt.figure(figsize=(10, 5))
sns.lineplot(
x="Month",
y="death_rate_21",
data=data_21,
color="r",
lw=3,
marker="o",
markersize=10,
)
plt.title("DEATH RATE PER MONTH IN 2021")
plt.show()
tn = df[df["States"] == "Tamil Nadu"]["Cured"]
mh = df[df["States"] == "Maharashtra"]["Cured"]
kl = df[df["States"] == "Kerala"]["Cured"]
df.head()
print(
"Total number of Active Covid-19 cases across India : {}".format(
max_confirmed_cases["Active_cases"].sum()
)
)
top_cases = (
max_confirmed_cases.groupby("States")["Active_cases"]
.max()
.sort_values(ascending=False)
.to_frame()
)
top_cases.style.background_gradient(cmap="flare")
from fbprophet import Prophet
confirmed = df.groupby("Date").sum()["Confirmed"].reset_index()
recovered = df.groupby("Date").sum()["Cured"].reset_index()
deaths = df.groupby("Date").sum()["Deaths"].reset_index()
confirmed.head()
confirmed.columns = ["ds", "y"]
confirmed["ds"] = pd.to_datetime(confirmed["ds"])
m = Prophet(interval_width=0.95)
m.fit(confirmed)
future = m.make_future_dataframe(periods=7) # Making future prediction for next 7 days
future.tail(10)
forecast = m.predict(future)
forecast[["ds", "yhat", "yhat_lower", "yhat_upper"]].tail()
confirmed_forecast_plot = m.plot(forecast)
confirmed_forecast_plot = m.plot_components(forecast)
print(
"Average recovery rate Covid-19 cases across India : {}".format(
max_confirmed_cases["Discharge_Rate"].mean()
)
)
top_cases = (
max_confirmed_cases.groupby("States")["Discharge_Rate"]
.max()
.sort_values(ascending=False)
.to_frame()
)
top_cases.style.background_gradient(cmap="flare")
discharge_rate = df[["Date", "Discharge_Rate"]]
discharge_rate.head()
discharge_rate.columns = ["ds", "y"]
discharge_rate["ds"] = pd.to_datetime(discharge_rate["ds"])
m = Prophet(interval_width=0.50)
m.fit(discharge_rate)
future = m.make_future_dataframe(periods=7)
forecast = m.predict(future)
forecast[["ds", "yhat", "yhat_lower", "yhat_upper"]].tail()
confirmed_forecast_plot = m.plot(forecast)
confirmed_forecast_plot = m.plot_components(forecast)
print(
"Average recovery rate Covid-19 cases across India : {}".format(
max_confirmed_cases["Discharge_Rate"].mean()
)
)
top_cases = (
max_confirmed_cases.groupby("States")["Death_Rate"]
.max()
.sort_values(ascending=False)
.to_frame()
)
top_cases.style.background_gradient(cmap="flare")
discharge_rate = df[["Date", "Death_Rate"]]
discharge_rate.head()
discharge_rate.columns = ["ds", "y"]
discharge_rate["ds"] = pd.to_datetime(discharge_rate["ds"])
m = Prophet(interval_width=0.50)
m.fit(discharge_rate)
future = m.make_future_dataframe(periods=7) # Making future prediction for next 7 days
future.tail(10)
forecast = m.predict(future)
forecast[["ds", "yhat", "yhat_lower", "yhat_upper"]].tail()
df.head()
state_cases = max_confirmed_cases.drop(
["Sno", "Date", "ConfirmedIndianNational", "ConfirmedForeignNational"], axis=1
)
state_wise_cases = (
state_cases.sort_values("Confirmed", ascending=False)
.fillna(0)
.style.background_gradient(cmap="Blues", subset=["Confirmed"])
.background_gradient(cmap="Reds", subset=["Deaths"])
.background_gradient(cmap="Greens", subset=["Cured"])
.background_gradient(cmap="Oranges", subset=["Active_cases"])
.background_gradient(cmap="RdYlBu", subset=["Death_Rate"])
.background_gradient(cmap="Accent", subset=["Discharge_Rate"])
)
state_wise_cases
# Loading the vaccination data
vaccine_df = pd.read_csv("../input/covid19-in-india/covid_vaccine_statewise.csv")
vacc_df = vaccine_df.copy()
vaccine_df = vaccine_df[
[
"Updated On",
"State",
"Total Doses Administered",
"Total Sessions Conducted",
"First Dose Administered",
"Second Dose Administered",
"Male(Individuals Vaccinated)",
"Female(Individuals Vaccinated)",
"Transgender(Individuals Vaccinated)",
"Total Covaxin Administered",
"Total CoviShield Administered",
"Total Sputnik V Administered",
"18-45 years (Age)",
"45-60 years (Age)",
"60+ years (Age)",
"Total Individuals Vaccinated",
]
]
vaccine_df.columns = [
"Date",
"States",
"Total_Doses_Administered",
"Total_Sessions_Conducted",
"First_Dose_Administered",
"Second_Dose_Administered",
"Male_Vaccinated",
"Female_Vaccinated",
"Transgender_Vaccinated",
"Total_Covaxin_Administered",
"Total_CoviShield_Administered",
"Total_SputnikV_Administered",
"18-45_Years",
"45-60_Years",
"60+_Years",
"Total_Individuals_Vaccinated",
]
vaccine_df.head()
# Checking the null values
vaccine_df.isnull().sum()
vaccine_df = vaccine_df[vaccine_df["Total_Individuals_Vaccinated"].notna()]
vaccine_df["Date"] = pd.to_datetime(vaccine_df["Date"], format="%d/%m/%Y")
latest_date = max(vaccine_df["Date"])
print("Current Date : ", latest_date)
vaccine_df[vaccine_df["States"] == "Dadra and Nagar Haveli and Daman and Diu"]
fig, ax = plt.subplots(ncols=1, nrows=1, dpi=100, figsize=(6, 4))
sns.lineplot(
data=vaccine_df[vaccine_df["States"] == "India"],
x="Date",
y="Total_Individuals_Vaccinated",
ax=ax,
)
ax.xaxis.set_major_locator(mdates.MonthLocator(interval=1))
ax.xaxis.set_major_formatter(DateFormatter("%b%y"))
ax.set_ylabel("Total Indians Vaccinated so far in millions", fontsize=10)
ax.set_yticklabels([0, 0, 20, 40, 60, 80, 100, 120])
Xstart, Xend = ax.get_xlim()
plt.title("India's Vaccination Performance")
plt.show()
|
[{"covid19-in-india/covid_vaccine_statewise.csv": {"column_names": "[\"Updated On\", \"State\", \"Total Doses Administered\", \"Sessions\", \" Sites \", \"First Dose Administered\", \"Second Dose Administered\", \"Male (Doses Administered)\", \"Female (Doses Administered)\", \"Transgender (Doses Administered)\", \" Covaxin (Doses Administered)\", \"CoviShield (Doses Administered)\", \"Sputnik V (Doses Administered)\", \"AEFI\", \"18-44 Years (Doses Administered)\", \"45-60 Years (Doses Administered)\", \"60+ Years (Doses Administered)\", \"18-44 Years(Individuals Vaccinated)\", \"45-60 Years(Individuals Vaccinated)\", \"60+ Years(Individuals Vaccinated)\", \"Male(Individuals Vaccinated)\", \"Female(Individuals Vaccinated)\", \"Transgender(Individuals Vaccinated)\", \"Total Individuals Vaccinated\"]", "column_data_types": "{\"Updated On\": \"object\", \"State\": \"object\", \"Total Doses Administered\": \"float64\", \"Sessions\": \"float64\", \" Sites \": \"float64\", \"First Dose Administered\": \"float64\", \"Second Dose Administered\": \"float64\", \"Male (Doses Administered)\": \"float64\", \"Female (Doses Administered)\": \"float64\", \"Transgender (Doses Administered)\": \"float64\", \" Covaxin (Doses Administered)\": \"float64\", \"CoviShield (Doses Administered)\": \"float64\", \"Sputnik V (Doses Administered)\": \"float64\", \"AEFI\": \"float64\", \"18-44 Years (Doses Administered)\": \"float64\", \"45-60 Years (Doses Administered)\": \"float64\", \"60+ Years (Doses Administered)\": \"float64\", \"18-44 Years(Individuals Vaccinated)\": \"float64\", \"45-60 Years(Individuals Vaccinated)\": \"float64\", \"60+ Years(Individuals Vaccinated)\": \"float64\", \"Male(Individuals Vaccinated)\": \"float64\", \"Female(Individuals Vaccinated)\": \"float64\", \"Transgender(Individuals Vaccinated)\": \"float64\", \"Total Individuals Vaccinated\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 7845 entries, 0 to 7844\nData columns (total 24 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Updated On 7845 non-null object \n 1 State 7845 non-null object \n 2 Total Doses Administered 7621 non-null float64\n 3 Sessions 7621 non-null float64\n 4 Sites 7621 non-null float64\n 5 First Dose Administered 7621 non-null float64\n 6 Second Dose Administered 7621 non-null float64\n 7 Male (Doses Administered) 7461 non-null float64\n 8 Female (Doses Administered) 7461 non-null float64\n 9 Transgender (Doses Administered) 7461 non-null float64\n 10 Covaxin (Doses Administered) 7621 non-null float64\n 11 CoviShield (Doses Administered) 7621 non-null float64\n 12 Sputnik V (Doses Administered) 2995 non-null float64\n 13 AEFI 5438 non-null float64\n 14 18-44 Years (Doses Administered) 1702 non-null float64\n 15 45-60 Years (Doses Administered) 1702 non-null float64\n 16 60+ Years (Doses Administered) 1702 non-null float64\n 17 18-44 Years(Individuals Vaccinated) 3733 non-null float64\n 18 45-60 Years(Individuals Vaccinated) 3734 non-null float64\n 19 60+ Years(Individuals Vaccinated) 3734 non-null float64\n 20 Male(Individuals Vaccinated) 160 non-null float64\n 21 Female(Individuals Vaccinated) 160 non-null float64\n 22 Transgender(Individuals Vaccinated) 160 non-null float64\n 23 Total Individuals Vaccinated 5919 non-null float64\ndtypes: float64(22), object(2)\nmemory usage: 1.4+ MB\n", "summary": "{\"Total Doses Administered\": {\"count\": 7621.0, \"mean\": 9188170.544023095, \"std\": 37461801.169766985, \"min\": 7.0, \"25%\": 135657.0, \"50%\": 818202.0, \"75%\": 6625243.0, \"max\": 513228400.0}, \"Sessions\": {\"count\": 7621.0, \"mean\": 479235.7989765123, \"std\": 1911511.192594298, \"min\": 0.0, \"25%\": 6004.0, \"50%\": 45470.0, \"75%\": 342869.0, \"max\": 35010311.0}, \" Sites \": {\"count\": 7621.0, \"mean\": 2282.8720640335914, \"std\": 7275.973729842437, \"min\": 0.0, \"25%\": 69.0, \"50%\": 597.0, \"75%\": 1708.0, \"max\": 73933.0}, \"First Dose Administered\": {\"count\": 7621.0, \"mean\": 7414415.300354284, \"std\": 29952087.78002959, \"min\": 7.0, \"25%\": 116632.0, \"50%\": 661459.0, \"75%\": 5387805.0, \"max\": 400150406.0}, \"Second Dose Administered\": {\"count\": 7621.0, \"mean\": 1773755.2436688098, \"std\": 7570382.4018907, \"min\": 0.0, \"25%\": 12831.0, \"50%\": 138818.0, \"75%\": 1166434.0, \"max\": 113077994.0}, \"Male (Doses Administered)\": {\"count\": 7461.0, \"mean\": 3620156.0107224234, \"std\": 17379382.717492737, \"min\": 0.0, \"25%\": 56555.0, \"50%\": 389785.0, \"75%\": 2735777.0, \"max\": 270163622.0}, \"Female (Doses Administered)\": {\"count\": 7461.0, \"mean\": 3168416.360407452, \"std\": 15153103.672247777, \"min\": 2.0, \"25%\": 52107.0, \"50%\": 334238.0, \"75%\": 2561513.0, \"max\": 239518609.0}, \"Transgender (Doses Administered)\": {\"count\": 7461.0, \"mean\": 1162.9780190323013, \"std\": 5931.353995293084, \"min\": 0.0, \"25%\": 8.0, \"50%\": 113.0, \"75%\": 800.0, \"max\": 98275.0}, \" Covaxin (Doses Administered)\": {\"count\": 7621.0, \"mean\": 1044669.3220049862, \"std\": 4452258.870168184, \"min\": 0.0, \"25%\": 0.0, \"50%\": 11851.0, \"75%\": 757930.0, \"max\": 62367416.0}, \"CoviShield (Doses Administered)\": {\"count\": 7621.0, \"mean\": 8126552.9236320695, \"std\": 32984142.5435157, \"min\": 7.0, \"25%\": 133134.0, \"50%\": 756736.0, \"75%\": 6007817.0, \"max\": 446825051.0}, \"Sputnik V (Doses Administered)\": {\"count\": 2995.0, \"mean\": 9655.57061769616, \"std\": 43882.536177337286, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 2519.0, \"max\": 588039.0}, \"AEFI\": {\"count\": 5438.0, \"mean\": 1139.4025376976829, \"std\": 3454.608045574211, \"min\": 0.0, \"25%\": 109.25, \"50%\": 294.0, \"75%\": 808.0, \"max\": 26542.0}, \"18-44 Years (Doses Administered)\": {\"count\": 1702.0, \"mean\": 8773958.21386604, \"std\": 26608287.5871943, \"min\": 26624.0, \"25%\": 434484.25, \"50%\": 3095970.0, \"75%\": 7366240.75, \"max\": 224330364.0}, \"45-60 Years (Doses Administered)\": {\"count\": 1702.0, \"mean\": 7442161.202115159, \"std\": 22259992.51020605, \"min\": 16815.0, \"25%\": 232627.5, \"50%\": 2695938.0, \"75%\": 6969726.5, \"max\": 166757453.0}, \"60+ Years (Doses Administered)\": {\"count\": 1702.0, \"mean\": 5641605.495299648, \"std\": 16816496.62130506, \"min\": 9994.0, \"25%\": 128560.5, \"50%\": 1805696.5, \"75%\": 5294762.75, \"max\": 118692689.0}, \"18-44 Years(Individuals Vaccinated)\": {\"count\": 3733.0, \"mean\": 1395894.5357621217, \"std\": 5501454.261410582, \"min\": 1059.0, \"25%\": 56554.0, \"50%\": 294727.0, \"75%\": 910516.0, \"max\": 92243148.0}, \"45-60 Years(Individuals Vaccinated)\": {\"count\": 3734.0, \"mean\": 2916514.789769684, \"std\": 9567607.054644102, \"min\": 1136.0, \"25%\": 92482.25, \"50%\": 833039.5, \"75%\": 2499280.5, \"max\": 90968877.0}, \"60+ Years(Individuals Vaccinated)\": {\"count\": 3734.0, \"mean\": 2627444.0565077662, \"std\": 8192225.1807218585, \"min\": 558.0, \"25%\": 56159.75, \"50%\": 788742.5, \"75%\": 2337874.0, \"max\": 67310981.0}, \"Male(Individuals Vaccinated)\": {\"count\": 160.0, \"mean\": 44616867.8625, \"std\": 39507492.96552456, \"min\": 23757.0, \"25%\": 5739350.0, \"50%\": 37165905.0, \"75%\": 74416634.5, \"max\": 134941971.0}, \"Female(Individuals Vaccinated)\": {\"count\": 160.0, \"mean\": 39510179.6, \"std\": 34176840.95163573, \"min\": 24517.0, \"25%\": 5023407.25, \"50%\": 33654024.5, \"75%\": 66853682.25, \"max\": 115668447.0}, \"Transgender(Individuals Vaccinated)\": {\"count\": 160.0, \"mean\": 12370.54375, \"std\": 12485.026752752348, \"min\": 2.0, \"25%\": 1278.75, \"50%\": 8007.5, \"75%\": 19851.0, \"max\": 46462.0}, \"Total Individuals Vaccinated\": {\"count\": 5919.0, \"mean\": 4547841.557357661, \"std\": 18341821.27664394, \"min\": 7.0, \"25%\": 74275.5, \"50%\": 402288.0, \"75%\": 3501562.0, \"max\": 250656880.0}}", "examples": "{\"Updated On\":{\"0\":\"16\\/01\\/2021\",\"1\":\"17\\/01\\/2021\",\"2\":\"18\\/01\\/2021\",\"3\":\"19\\/01\\/2021\"},\"State\":{\"0\":\"India\",\"1\":\"India\",\"2\":\"India\",\"3\":\"India\"},\"Total Doses Administered\":{\"0\":48276.0,\"1\":58604.0,\"2\":99449.0,\"3\":195525.0},\"Sessions\":{\"0\":3455.0,\"1\":8532.0,\"2\":13611.0,\"3\":17855.0},\" Sites \":{\"0\":2957.0,\"1\":4954.0,\"2\":6583.0,\"3\":7951.0},\"First Dose Administered\":{\"0\":48276.0,\"1\":58604.0,\"2\":99449.0,\"3\":195525.0},\"Second Dose Administered\":{\"0\":0.0,\"1\":0.0,\"2\":0.0,\"3\":0.0},\"Male (Doses Administered)\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"Female (Doses Administered)\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"Transgender (Doses Administered)\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\" Covaxin (Doses Administered)\":{\"0\":579.0,\"1\":635.0,\"2\":1299.0,\"3\":3017.0},\"CoviShield (Doses Administered)\":{\"0\":47697.0,\"1\":57969.0,\"2\":98150.0,\"3\":192508.0},\"Sputnik V (Doses Administered)\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"AEFI\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"18-44 Years (Doses Administered)\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"45-60 Years (Doses Administered)\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"60+ Years (Doses Administered)\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"18-44 Years(Individuals Vaccinated)\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"45-60 Years(Individuals Vaccinated)\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"60+ Years(Individuals Vaccinated)\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"Male(Individuals Vaccinated)\":{\"0\":23757.0,\"1\":27348.0,\"2\":41361.0,\"3\":81901.0},\"Female(Individuals Vaccinated)\":{\"0\":24517.0,\"1\":31252.0,\"2\":58083.0,\"3\":113613.0},\"Transgender(Individuals Vaccinated)\":{\"0\":2.0,\"1\":4.0,\"2\":5.0,\"3\":11.0},\"Total Individuals Vaccinated\":{\"0\":48276.0,\"1\":58604.0,\"2\":99449.0,\"3\":195525.0}}"}}, {"covid19-in-india/StatewiseTestingDetails.csv": {"column_names": "[\"Date\", \"State\", \"TotalSamples\", \"Negative\", \"Positive\"]", "column_data_types": "{\"Date\": \"object\", \"State\": \"object\", \"TotalSamples\": \"float64\", \"Negative\": \"object\", \"Positive\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 16336 entries, 0 to 16335\nData columns (total 5 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Date 16336 non-null object \n 1 State 16336 non-null object \n 2 TotalSamples 16336 non-null float64\n 3 Negative 6969 non-null object \n 4 Positive 5662 non-null float64\ndtypes: float64(2), object(3)\nmemory usage: 638.2+ KB\n", "summary": "{\"TotalSamples\": {\"count\": 16336.0, \"mean\": 5376466.053317825, \"std\": 8780337.764526756, \"min\": 58.0, \"25%\": 172909.75, \"50%\": 930758.0, \"75%\": 7284795.25, \"max\": 67897856.0}, \"Positive\": {\"count\": 5662.0, \"mean\": 56526.53585305546, \"std\": 167310.77901611742, \"min\": 0.0, \"25%\": 536.25, \"50%\": 4771.0, \"75%\": 33618.75, \"max\": 1638961.0}}", "examples": "{\"Date\":{\"0\":\"2020-04-17\",\"1\":\"2020-04-24\",\"2\":\"2020-04-27\",\"3\":\"2020-05-01\"},\"State\":{\"0\":\"Andaman and Nicobar Islands\",\"1\":\"Andaman and Nicobar Islands\",\"2\":\"Andaman and Nicobar Islands\",\"3\":\"Andaman and Nicobar Islands\"},\"TotalSamples\":{\"0\":1403.0,\"1\":2679.0,\"2\":2848.0,\"3\":3754.0},\"Negative\":{\"0\":\"1210\",\"1\":null,\"2\":null,\"3\":null},\"Positive\":{\"0\":12.0,\"1\":27.0,\"2\":33.0,\"3\":33.0}}"}}, {"covid19-in-india/covid_19_india.csv": {"column_names": "[\"Sno\", \"Date\", \"Time\", \"State/UnionTerritory\", \"ConfirmedIndianNational\", \"ConfirmedForeignNational\", \"Cured\", \"Deaths\", \"Confirmed\"]", "column_data_types": "{\"Sno\": \"int64\", \"Date\": \"object\", \"Time\": \"object\", \"State/UnionTerritory\": \"object\", \"ConfirmedIndianNational\": \"object\", \"ConfirmedForeignNational\": \"object\", \"Cured\": \"int64\", \"Deaths\": \"int64\", \"Confirmed\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 18110 entries, 0 to 18109\nData columns (total 9 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Sno 18110 non-null int64 \n 1 Date 18110 non-null object\n 2 Time 18110 non-null object\n 3 State/UnionTerritory 18110 non-null object\n 4 ConfirmedIndianNational 18110 non-null object\n 5 ConfirmedForeignNational 18110 non-null object\n 6 Cured 18110 non-null int64 \n 7 Deaths 18110 non-null int64 \n 8 Confirmed 18110 non-null int64 \ndtypes: int64(4), object(5)\nmemory usage: 1.2+ MB\n", "summary": "{\"Sno\": {\"count\": 18110.0, \"mean\": 9055.5, \"std\": 5228.051023086901, \"min\": 1.0, \"25%\": 4528.25, \"50%\": 9055.5, \"75%\": 13582.75, \"max\": 18110.0}, \"Cured\": {\"count\": 18110.0, \"mean\": 278637.5180563225, \"std\": 614890.8944243209, \"min\": 0.0, \"25%\": 3360.25, \"50%\": 33364.0, \"75%\": 278869.75, \"max\": 6159676.0}, \"Deaths\": {\"count\": 18110.0, \"mean\": 4052.402263942573, \"std\": 10919.076411131335, \"min\": 0.0, \"25%\": 32.0, \"50%\": 588.0, \"75%\": 3643.75, \"max\": 134201.0}, \"Confirmed\": {\"count\": 18110.0, \"mean\": 301031.40182219766, \"std\": 656148.8729651569, \"min\": 0.0, \"25%\": 4376.75, \"50%\": 39773.5, \"75%\": 300149.75, \"max\": 6363442.0}}", "examples": "{\"Sno\":{\"0\":1,\"1\":2,\"2\":3,\"3\":4},\"Date\":{\"0\":\"2020-01-30\",\"1\":\"2020-01-31\",\"2\":\"2020-02-01\",\"3\":\"2020-02-02\"},\"Time\":{\"0\":\"6:00 PM\",\"1\":\"6:00 PM\",\"2\":\"6:00 PM\",\"3\":\"6:00 PM\"},\"State\\/UnionTerritory\":{\"0\":\"Kerala\",\"1\":\"Kerala\",\"2\":\"Kerala\",\"3\":\"Kerala\"},\"ConfirmedIndianNational\":{\"0\":\"1\",\"1\":\"1\",\"2\":\"2\",\"3\":\"3\"},\"ConfirmedForeignNational\":{\"0\":\"0\",\"1\":\"0\",\"2\":\"0\",\"3\":\"0\"},\"Cured\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"Deaths\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"Confirmed\":{\"0\":1,\"1\":1,\"2\":2,\"3\":3}}"}}]
| true | 3 |
<start_data_description><data_path>covid19-in-india/covid_vaccine_statewise.csv:
<column_names>
['Updated On', 'State', 'Total Doses Administered', 'Sessions', ' Sites ', 'First Dose Administered', 'Second Dose Administered', 'Male (Doses Administered)', 'Female (Doses Administered)', 'Transgender (Doses Administered)', ' Covaxin (Doses Administered)', 'CoviShield (Doses Administered)', 'Sputnik V (Doses Administered)', 'AEFI', '18-44 Years (Doses Administered)', '45-60 Years (Doses Administered)', '60+ Years (Doses Administered)', '18-44 Years(Individuals Vaccinated)', '45-60 Years(Individuals Vaccinated)', '60+ Years(Individuals Vaccinated)', 'Male(Individuals Vaccinated)', 'Female(Individuals Vaccinated)', 'Transgender(Individuals Vaccinated)', 'Total Individuals Vaccinated']
<column_types>
{'Updated On': 'object', 'State': 'object', 'Total Doses Administered': 'float64', 'Sessions': 'float64', ' Sites ': 'float64', 'First Dose Administered': 'float64', 'Second Dose Administered': 'float64', 'Male (Doses Administered)': 'float64', 'Female (Doses Administered)': 'float64', 'Transgender (Doses Administered)': 'float64', ' Covaxin (Doses Administered)': 'float64', 'CoviShield (Doses Administered)': 'float64', 'Sputnik V (Doses Administered)': 'float64', 'AEFI': 'float64', '18-44 Years (Doses Administered)': 'float64', '45-60 Years (Doses Administered)': 'float64', '60+ Years (Doses Administered)': 'float64', '18-44 Years(Individuals Vaccinated)': 'float64', '45-60 Years(Individuals Vaccinated)': 'float64', '60+ Years(Individuals Vaccinated)': 'float64', 'Male(Individuals Vaccinated)': 'float64', 'Female(Individuals Vaccinated)': 'float64', 'Transgender(Individuals Vaccinated)': 'float64', 'Total Individuals Vaccinated': 'float64'}
<dataframe_Summary>
{'Total Doses Administered': {'count': 7621.0, 'mean': 9188170.544023095, 'std': 37461801.169766985, 'min': 7.0, '25%': 135657.0, '50%': 818202.0, '75%': 6625243.0, 'max': 513228400.0}, 'Sessions': {'count': 7621.0, 'mean': 479235.7989765123, 'std': 1911511.192594298, 'min': 0.0, '25%': 6004.0, '50%': 45470.0, '75%': 342869.0, 'max': 35010311.0}, ' Sites ': {'count': 7621.0, 'mean': 2282.8720640335914, 'std': 7275.973729842437, 'min': 0.0, '25%': 69.0, '50%': 597.0, '75%': 1708.0, 'max': 73933.0}, 'First Dose Administered': {'count': 7621.0, 'mean': 7414415.300354284, 'std': 29952087.78002959, 'min': 7.0, '25%': 116632.0, '50%': 661459.0, '75%': 5387805.0, 'max': 400150406.0}, 'Second Dose Administered': {'count': 7621.0, 'mean': 1773755.2436688098, 'std': 7570382.4018907, 'min': 0.0, '25%': 12831.0, '50%': 138818.0, '75%': 1166434.0, 'max': 113077994.0}, 'Male (Doses Administered)': {'count': 7461.0, 'mean': 3620156.0107224234, 'std': 17379382.717492737, 'min': 0.0, '25%': 56555.0, '50%': 389785.0, '75%': 2735777.0, 'max': 270163622.0}, 'Female (Doses Administered)': {'count': 7461.0, 'mean': 3168416.360407452, 'std': 15153103.672247777, 'min': 2.0, '25%': 52107.0, '50%': 334238.0, '75%': 2561513.0, 'max': 239518609.0}, 'Transgender (Doses Administered)': {'count': 7461.0, 'mean': 1162.9780190323013, 'std': 5931.353995293084, 'min': 0.0, '25%': 8.0, '50%': 113.0, '75%': 800.0, 'max': 98275.0}, ' Covaxin (Doses Administered)': {'count': 7621.0, 'mean': 1044669.3220049862, 'std': 4452258.870168184, 'min': 0.0, '25%': 0.0, '50%': 11851.0, '75%': 757930.0, 'max': 62367416.0}, 'CoviShield (Doses Administered)': {'count': 7621.0, 'mean': 8126552.9236320695, 'std': 32984142.5435157, 'min': 7.0, '25%': 133134.0, '50%': 756736.0, '75%': 6007817.0, 'max': 446825051.0}, 'Sputnik V (Doses Administered)': {'count': 2995.0, 'mean': 9655.57061769616, 'std': 43882.536177337286, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 2519.0, 'max': 588039.0}, 'AEFI': {'count': 5438.0, 'mean': 1139.4025376976829, 'std': 3454.608045574211, 'min': 0.0, '25%': 109.25, '50%': 294.0, '75%': 808.0, 'max': 26542.0}, '18-44 Years (Doses Administered)': {'count': 1702.0, 'mean': 8773958.21386604, 'std': 26608287.5871943, 'min': 26624.0, '25%': 434484.25, '50%': 3095970.0, '75%': 7366240.75, 'max': 224330364.0}, '45-60 Years (Doses Administered)': {'count': 1702.0, 'mean': 7442161.202115159, 'std': 22259992.51020605, 'min': 16815.0, '25%': 232627.5, '50%': 2695938.0, '75%': 6969726.5, 'max': 166757453.0}, '60+ Years (Doses Administered)': {'count': 1702.0, 'mean': 5641605.495299648, 'std': 16816496.62130506, 'min': 9994.0, '25%': 128560.5, '50%': 1805696.5, '75%': 5294762.75, 'max': 118692689.0}, '18-44 Years(Individuals Vaccinated)': {'count': 3733.0, 'mean': 1395894.5357621217, 'std': 5501454.261410582, 'min': 1059.0, '25%': 56554.0, '50%': 294727.0, '75%': 910516.0, 'max': 92243148.0}, '45-60 Years(Individuals Vaccinated)': {'count': 3734.0, 'mean': 2916514.789769684, 'std': 9567607.054644102, 'min': 1136.0, '25%': 92482.25, '50%': 833039.5, '75%': 2499280.5, 'max': 90968877.0}, '60+ Years(Individuals Vaccinated)': {'count': 3734.0, 'mean': 2627444.0565077662, 'std': 8192225.1807218585, 'min': 558.0, '25%': 56159.75, '50%': 788742.5, '75%': 2337874.0, 'max': 67310981.0}, 'Male(Individuals Vaccinated)': {'count': 160.0, 'mean': 44616867.8625, 'std': 39507492.96552456, 'min': 23757.0, '25%': 5739350.0, '50%': 37165905.0, '75%': 74416634.5, 'max': 134941971.0}, 'Female(Individuals Vaccinated)': {'count': 160.0, 'mean': 39510179.6, 'std': 34176840.95163573, 'min': 24517.0, '25%': 5023407.25, '50%': 33654024.5, '75%': 66853682.25, 'max': 115668447.0}, 'Transgender(Individuals Vaccinated)': {'count': 160.0, 'mean': 12370.54375, 'std': 12485.026752752348, 'min': 2.0, '25%': 1278.75, '50%': 8007.5, '75%': 19851.0, 'max': 46462.0}, 'Total Individuals Vaccinated': {'count': 5919.0, 'mean': 4547841.557357661, 'std': 18341821.27664394, 'min': 7.0, '25%': 74275.5, '50%': 402288.0, '75%': 3501562.0, 'max': 250656880.0}}
<dataframe_info>
RangeIndex: 7845 entries, 0 to 7844
Data columns (total 24 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Updated On 7845 non-null object
1 State 7845 non-null object
2 Total Doses Administered 7621 non-null float64
3 Sessions 7621 non-null float64
4 Sites 7621 non-null float64
5 First Dose Administered 7621 non-null float64
6 Second Dose Administered 7621 non-null float64
7 Male (Doses Administered) 7461 non-null float64
8 Female (Doses Administered) 7461 non-null float64
9 Transgender (Doses Administered) 7461 non-null float64
10 Covaxin (Doses Administered) 7621 non-null float64
11 CoviShield (Doses Administered) 7621 non-null float64
12 Sputnik V (Doses Administered) 2995 non-null float64
13 AEFI 5438 non-null float64
14 18-44 Years (Doses Administered) 1702 non-null float64
15 45-60 Years (Doses Administered) 1702 non-null float64
16 60+ Years (Doses Administered) 1702 non-null float64
17 18-44 Years(Individuals Vaccinated) 3733 non-null float64
18 45-60 Years(Individuals Vaccinated) 3734 non-null float64
19 60+ Years(Individuals Vaccinated) 3734 non-null float64
20 Male(Individuals Vaccinated) 160 non-null float64
21 Female(Individuals Vaccinated) 160 non-null float64
22 Transgender(Individuals Vaccinated) 160 non-null float64
23 Total Individuals Vaccinated 5919 non-null float64
dtypes: float64(22), object(2)
memory usage: 1.4+ MB
<some_examples>
{'Updated On': {'0': '16/01/2021', '1': '17/01/2021', '2': '18/01/2021', '3': '19/01/2021'}, 'State': {'0': 'India', '1': 'India', '2': 'India', '3': 'India'}, 'Total Doses Administered': {'0': 48276.0, '1': 58604.0, '2': 99449.0, '3': 195525.0}, 'Sessions': {'0': 3455.0, '1': 8532.0, '2': 13611.0, '3': 17855.0}, ' Sites ': {'0': 2957.0, '1': 4954.0, '2': 6583.0, '3': 7951.0}, 'First Dose Administered': {'0': 48276.0, '1': 58604.0, '2': 99449.0, '3': 195525.0}, 'Second Dose Administered': {'0': 0.0, '1': 0.0, '2': 0.0, '3': 0.0}, 'Male (Doses Administered)': {'0': None, '1': None, '2': None, '3': None}, 'Female (Doses Administered)': {'0': None, '1': None, '2': None, '3': None}, 'Transgender (Doses Administered)': {'0': None, '1': None, '2': None, '3': None}, ' Covaxin (Doses Administered)': {'0': 579.0, '1': 635.0, '2': 1299.0, '3': 3017.0}, 'CoviShield (Doses Administered)': {'0': 47697.0, '1': 57969.0, '2': 98150.0, '3': 192508.0}, 'Sputnik V (Doses Administered)': {'0': None, '1': None, '2': None, '3': None}, 'AEFI': {'0': None, '1': None, '2': None, '3': None}, '18-44 Years (Doses Administered)': {'0': None, '1': None, '2': None, '3': None}, '45-60 Years (Doses Administered)': {'0': None, '1': None, '2': None, '3': None}, '60+ Years (Doses Administered)': {'0': None, '1': None, '2': None, '3': None}, '18-44 Years(Individuals Vaccinated)': {'0': None, '1': None, '2': None, '3': None}, '45-60 Years(Individuals Vaccinated)': {'0': None, '1': None, '2': None, '3': None}, '60+ Years(Individuals Vaccinated)': {'0': None, '1': None, '2': None, '3': None}, 'Male(Individuals Vaccinated)': {'0': 23757.0, '1': 27348.0, '2': 41361.0, '3': 81901.0}, 'Female(Individuals Vaccinated)': {'0': 24517.0, '1': 31252.0, '2': 58083.0, '3': 113613.0}, 'Transgender(Individuals Vaccinated)': {'0': 2.0, '1': 4.0, '2': 5.0, '3': 11.0}, 'Total Individuals Vaccinated': {'0': 48276.0, '1': 58604.0, '2': 99449.0, '3': 195525.0}}
<end_description>
<start_data_description><data_path>covid19-in-india/StatewiseTestingDetails.csv:
<column_names>
['Date', 'State', 'TotalSamples', 'Negative', 'Positive']
<column_types>
{'Date': 'object', 'State': 'object', 'TotalSamples': 'float64', 'Negative': 'object', 'Positive': 'float64'}
<dataframe_Summary>
{'TotalSamples': {'count': 16336.0, 'mean': 5376466.053317825, 'std': 8780337.764526756, 'min': 58.0, '25%': 172909.75, '50%': 930758.0, '75%': 7284795.25, 'max': 67897856.0}, 'Positive': {'count': 5662.0, 'mean': 56526.53585305546, 'std': 167310.77901611742, 'min': 0.0, '25%': 536.25, '50%': 4771.0, '75%': 33618.75, 'max': 1638961.0}}
<dataframe_info>
RangeIndex: 16336 entries, 0 to 16335
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Date 16336 non-null object
1 State 16336 non-null object
2 TotalSamples 16336 non-null float64
3 Negative 6969 non-null object
4 Positive 5662 non-null float64
dtypes: float64(2), object(3)
memory usage: 638.2+ KB
<some_examples>
{'Date': {'0': '2020-04-17', '1': '2020-04-24', '2': '2020-04-27', '3': '2020-05-01'}, 'State': {'0': 'Andaman and Nicobar Islands', '1': 'Andaman and Nicobar Islands', '2': 'Andaman and Nicobar Islands', '3': 'Andaman and Nicobar Islands'}, 'TotalSamples': {'0': 1403.0, '1': 2679.0, '2': 2848.0, '3': 3754.0}, 'Negative': {'0': '1210', '1': None, '2': None, '3': None}, 'Positive': {'0': 12.0, '1': 27.0, '2': 33.0, '3': 33.0}}
<end_description>
<start_data_description><data_path>covid19-in-india/covid_19_india.csv:
<column_names>
['Sno', 'Date', 'Time', 'State/UnionTerritory', 'ConfirmedIndianNational', 'ConfirmedForeignNational', 'Cured', 'Deaths', 'Confirmed']
<column_types>
{'Sno': 'int64', 'Date': 'object', 'Time': 'object', 'State/UnionTerritory': 'object', 'ConfirmedIndianNational': 'object', 'ConfirmedForeignNational': 'object', 'Cured': 'int64', 'Deaths': 'int64', 'Confirmed': 'int64'}
<dataframe_Summary>
{'Sno': {'count': 18110.0, 'mean': 9055.5, 'std': 5228.051023086901, 'min': 1.0, '25%': 4528.25, '50%': 9055.5, '75%': 13582.75, 'max': 18110.0}, 'Cured': {'count': 18110.0, 'mean': 278637.5180563225, 'std': 614890.8944243209, 'min': 0.0, '25%': 3360.25, '50%': 33364.0, '75%': 278869.75, 'max': 6159676.0}, 'Deaths': {'count': 18110.0, 'mean': 4052.402263942573, 'std': 10919.076411131335, 'min': 0.0, '25%': 32.0, '50%': 588.0, '75%': 3643.75, 'max': 134201.0}, 'Confirmed': {'count': 18110.0, 'mean': 301031.40182219766, 'std': 656148.8729651569, 'min': 0.0, '25%': 4376.75, '50%': 39773.5, '75%': 300149.75, 'max': 6363442.0}}
<dataframe_info>
RangeIndex: 18110 entries, 0 to 18109
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Sno 18110 non-null int64
1 Date 18110 non-null object
2 Time 18110 non-null object
3 State/UnionTerritory 18110 non-null object
4 ConfirmedIndianNational 18110 non-null object
5 ConfirmedForeignNational 18110 non-null object
6 Cured 18110 non-null int64
7 Deaths 18110 non-null int64
8 Confirmed 18110 non-null int64
dtypes: int64(4), object(5)
memory usage: 1.2+ MB
<some_examples>
{'Sno': {'0': 1, '1': 2, '2': 3, '3': 4}, 'Date': {'0': '2020-01-30', '1': '2020-01-31', '2': '2020-02-01', '3': '2020-02-02'}, 'Time': {'0': '6:00 PM', '1': '6:00 PM', '2': '6:00 PM', '3': '6:00 PM'}, 'State/UnionTerritory': {'0': 'Kerala', '1': 'Kerala', '2': 'Kerala', '3': 'Kerala'}, 'ConfirmedIndianNational': {'0': '1', '1': '1', '2': '2', '3': '3'}, 'ConfirmedForeignNational': {'0': '0', '1': '0', '2': '0', '3': '0'}, 'Cured': {'0': 0, '1': 0, '2': 0, '3': 0}, 'Deaths': {'0': 0, '1': 0, '2': 0, '3': 0}, 'Confirmed': {'0': 1, '1': 1, '2': 2, '3': 3}}
<end_description>
| 4,963 | 5 | 8,498 | 4,963 |
69492148
|
<jupyter_start><jupyter_text>resized_plant2021
# Plant Pathology 2021 - FGVC8 Dataset resized
In this dataset, I have included multiple resized version of the dataset. There are four different sizes of the dataset: 256, 384, 512, 640.
Each *folder_name* contains the image size.
The original images in this competition are pretty big (~(2672, 4000)). It takes a lot of time to read all these. I saw **massive performance** boost by using resized images. To give you some numbers, with original dimensions it was taking 45 mins to train a ResNet50. But it took only 6 mins using 512 x 512 dataset.
The following code was used to resize the images:
```python
from fastai.vision.all import *
path = Path('../input/plant-pathology-2021-fgvc8')
for sz in [256, 384, 512, 640]:
resize_images(path/'train_images', max_size=sz, dest=f'img_sz_{sz}')
print(f'{sz} - Done!')
```
Kaggle dataset identifier: resized-plant2021
<jupyter_script>import pandas as pd
import os
import cv2
import matplotlib.pyplot as plt
from torch.utils.data import Dataset, DataLoader
from sklearn.model_selection import train_test_split
import numpy as np
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
import torchvision.models as models
import albumentations as A
from albumentations.pytorch import ToTensorV2
from torchvision import transforms as T
import gc
train_df = pd.read_csv("../input/plant-pathology-2021-fgvc8/train.csv")
# folder_path_256 = "../input/resized-plant2021/img_sz_256"
# data_paths_256 = os.listdir(folder_path_256)
train_df_cp = train_df.copy()
train_df_cp["label_list"] = train_df_cp["labels"].str.split(" ")
def lbl_lgc(col, lbl_list):
if col in lbl_list:
res = 1
else:
res = 0
return res
lbls = ["healthy", "complex", "rust", "frog_eye_leaf_spot", "powdery_mildew", "scab"]
for x in lbls:
train_df_cp[x] = 0
for x in lbls:
train_df_cp[x] = np.vectorize(lbl_lgc)(x, train_df_cp["label_list"])
class Plant_Dataset(Dataset):
def __init__(
self,
folder_path,
data_paths,
data_df=train_df_cp,
size=224,
transforms=None,
train=True,
):
self.folder_path = folder_path
self.data_paths = data_paths
self.data_df = data_df
self.transforms = transforms
self.train = train
self.size = size
def __getitem__(self, idx):
img_path = os.path.join(self.folder_path, self.data_paths[idx])
image = cv2.imread(img_path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.resize(image, (self.size, self.size), interpolation=cv2.INTER_AREA)
image = np.asarray(image)
if self.train: # for train or validation data
# label = self.data_df.loc[self.data_df['image']==self.data_paths[idx]].values[0][1]
j = 0
vector = [0] * 6
values = self.data_df.loc[
self.data_df["image"] == self.data_paths[idx]
].values
for i in range(3, 9):
num = values[0][i]
vector[j] = num
j = j + 1
vector = np.asarray(vector)
if self.transforms:
image = self.transforms(image=image)["image"]
if self.train:
return image, vector # train or validation data
else:
return image, self.data_paths[idx] # test data
def __len__(self):
return len(self.data_paths)
def submission(images, predictions):
str_preds = []
img_names = []
j = 0
for vec in predictions:
labels = []
for i in range(len(vec[0])):
if vec[0][i] == 1:
labels.append(lbls[i])
l = " ".join(labels)
str_preds.append(l)
img_names.append(images[j][0])
j += 1
output = pd.DataFrame({"image": img_names, "labels": str_preds})
output.to_csv("submission.csv", index=False)
def model_test(model, test_loader):
model.eval()
images = []
predictions = []
treshold = 0.5
for i, (img, img_name) in enumerate(test_loader):
images.append(img_name)
img = img.float()
img = img.cuda()
with torch.no_grad():
output = torch.sigmoid(model(img)).float()
output = torch.where(output > treshold, 1, 0)
predictions.append(output)
del img
del output
gc.collect()
torch.cuda.empty_cache()
return images, predictions
def load_model(path, model, optimizer):
checkpoint = torch.load(path)
model.load_state_dict(checkpoint["model_state_dict"])
optimizer.load_state_dict(checkpoint["optimizer_state_dict"])
epoch = checkpoint["epoch"]
train_loss = checkpoint["train_loss"]
val_loss = checkpoint["val_loss"]
train_acc = checkpoint["train_acc"]
val_acc = checkpoint["val_acc"]
return train_loss, val_loss, train_acc, val_acc, epoch
test_path = "../input/plant-pathology-2021-fgvc8/test_images"
test_data_paths = os.listdir(test_path)
gc.collect()
torch.cuda.empty_cache()
transforms = A.Compose([A.Normalize(), ToTensorV2()])
test_data = Plant_Dataset(
test_path, test_data_paths, transforms=transforms, train=False
)
test_loader = DataLoader(test_data, batch_size=1, shuffle=False, num_workers=0)
model = models.vgg19_bn(pretrained=True)
model.classifier[6] = nn.Linear(4096, 6)
optimi = optim.AdamW(model.parameters(), lr=1e-4)
# load_model(model1_path,model,optimi)
load_model("../input/model-5/vgg19bn_1", model, optimi)
model.cuda()
images, predictions = model_test(model, test_loader)
submission(images, predictions)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/492/69492148.ipynb
|
resized-plant2021
|
ankursingh12
|
[{"Id": 69492148, "ScriptId": 18850085, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6157237, "CreationDate": "07/31/2021 16:56:04", "VersionNumber": 11.0, "Title": "model_submission", "EvaluationDate": "07/31/2021", "IsChange": false, "TotalLines": 155.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 155.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92727615, "KernelVersionId": 69492148, "SourceDatasetVersionId": 2032065}]
|
[{"Id": 2032065, "DatasetId": 1216613, "DatasourceVersionId": 2071889, "CreatorUserId": 1537159, "LicenseName": "Unknown", "CreationDate": "03/17/2021 08:45:09", "VersionNumber": 1.0, "Title": "resized_plant2021", "Slug": "resized-plant2021", "Subtitle": "Plant-2021 resized images dataset", "Description": "# Plant Pathology 2021 - FGVC8 Dataset resized\n\nIn this dataset, I have included multiple resized version of the dataset. There are four different sizes of the dataset: 256, 384, 512, 640. \n\nEach *folder_name* contains the image size. \n\nThe original images in this competition are pretty big (~(2672, 4000)). It takes a lot of time to read all these. I saw **massive performance** boost by using resized images. To give you some numbers, with original dimensions it was taking 45 mins to train a ResNet50. But it took only 6 mins using 512 x 512 dataset.\n\nThe following code was used to resize the images:\n```python\n\nfrom fastai.vision.all import *\n\npath = Path('../input/plant-pathology-2021-fgvc8')\nfor sz in [256, 384, 512, 640]:\n resize_images(path/'train_images', max_size=sz, dest=f'img_sz_{sz}')\n print(f'{sz} - Done!')\n\n```", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1216613, "CreatorUserId": 1537159, "OwnerUserId": 1537159.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2032065.0, "CurrentDatasourceVersionId": 2071889.0, "ForumId": 1234689, "Type": 2, "CreationDate": "03/17/2021 08:45:09", "LastActivityDate": "03/17/2021", "TotalViews": 5352, "TotalDownloads": 1467, "TotalVotes": 49, "TotalKernels": 156}]
|
[{"Id": 1537159, "UserName": "ankursingh12", "DisplayName": "AnkurSingh", "RegisterDate": "01/08/2018", "PerformanceTier": 2}]
|
import pandas as pd
import os
import cv2
import matplotlib.pyplot as plt
from torch.utils.data import Dataset, DataLoader
from sklearn.model_selection import train_test_split
import numpy as np
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
import torchvision.models as models
import albumentations as A
from albumentations.pytorch import ToTensorV2
from torchvision import transforms as T
import gc
train_df = pd.read_csv("../input/plant-pathology-2021-fgvc8/train.csv")
# folder_path_256 = "../input/resized-plant2021/img_sz_256"
# data_paths_256 = os.listdir(folder_path_256)
train_df_cp = train_df.copy()
train_df_cp["label_list"] = train_df_cp["labels"].str.split(" ")
def lbl_lgc(col, lbl_list):
if col in lbl_list:
res = 1
else:
res = 0
return res
lbls = ["healthy", "complex", "rust", "frog_eye_leaf_spot", "powdery_mildew", "scab"]
for x in lbls:
train_df_cp[x] = 0
for x in lbls:
train_df_cp[x] = np.vectorize(lbl_lgc)(x, train_df_cp["label_list"])
class Plant_Dataset(Dataset):
def __init__(
self,
folder_path,
data_paths,
data_df=train_df_cp,
size=224,
transforms=None,
train=True,
):
self.folder_path = folder_path
self.data_paths = data_paths
self.data_df = data_df
self.transforms = transforms
self.train = train
self.size = size
def __getitem__(self, idx):
img_path = os.path.join(self.folder_path, self.data_paths[idx])
image = cv2.imread(img_path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.resize(image, (self.size, self.size), interpolation=cv2.INTER_AREA)
image = np.asarray(image)
if self.train: # for train or validation data
# label = self.data_df.loc[self.data_df['image']==self.data_paths[idx]].values[0][1]
j = 0
vector = [0] * 6
values = self.data_df.loc[
self.data_df["image"] == self.data_paths[idx]
].values
for i in range(3, 9):
num = values[0][i]
vector[j] = num
j = j + 1
vector = np.asarray(vector)
if self.transforms:
image = self.transforms(image=image)["image"]
if self.train:
return image, vector # train or validation data
else:
return image, self.data_paths[idx] # test data
def __len__(self):
return len(self.data_paths)
def submission(images, predictions):
str_preds = []
img_names = []
j = 0
for vec in predictions:
labels = []
for i in range(len(vec[0])):
if vec[0][i] == 1:
labels.append(lbls[i])
l = " ".join(labels)
str_preds.append(l)
img_names.append(images[j][0])
j += 1
output = pd.DataFrame({"image": img_names, "labels": str_preds})
output.to_csv("submission.csv", index=False)
def model_test(model, test_loader):
model.eval()
images = []
predictions = []
treshold = 0.5
for i, (img, img_name) in enumerate(test_loader):
images.append(img_name)
img = img.float()
img = img.cuda()
with torch.no_grad():
output = torch.sigmoid(model(img)).float()
output = torch.where(output > treshold, 1, 0)
predictions.append(output)
del img
del output
gc.collect()
torch.cuda.empty_cache()
return images, predictions
def load_model(path, model, optimizer):
checkpoint = torch.load(path)
model.load_state_dict(checkpoint["model_state_dict"])
optimizer.load_state_dict(checkpoint["optimizer_state_dict"])
epoch = checkpoint["epoch"]
train_loss = checkpoint["train_loss"]
val_loss = checkpoint["val_loss"]
train_acc = checkpoint["train_acc"]
val_acc = checkpoint["val_acc"]
return train_loss, val_loss, train_acc, val_acc, epoch
test_path = "../input/plant-pathology-2021-fgvc8/test_images"
test_data_paths = os.listdir(test_path)
gc.collect()
torch.cuda.empty_cache()
transforms = A.Compose([A.Normalize(), ToTensorV2()])
test_data = Plant_Dataset(
test_path, test_data_paths, transforms=transforms, train=False
)
test_loader = DataLoader(test_data, batch_size=1, shuffle=False, num_workers=0)
model = models.vgg19_bn(pretrained=True)
model.classifier[6] = nn.Linear(4096, 6)
optimi = optim.AdamW(model.parameters(), lr=1e-4)
# load_model(model1_path,model,optimi)
load_model("../input/model-5/vgg19bn_1", model, optimi)
model.cuda()
images, predictions = model_test(model, test_loader)
submission(images, predictions)
| false | 1 | 1,449 | 0 | 1,757 | 1,449 |
||
69492234
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import datetime as dt
import pandas as pd
import numpy as np
import requests
import os.path
from urllib.parse import urlparse
from newspaper import Article
from bs4 import BeautifulSoup
from urllib.parse import unquote, urlparse
from pathlib import PurePosixPath
# # Key Note!
# Because the assessment system and psychological test results may ONLY be known by psychologists, we need to assume what **Dreams**, **motivations**, and **user test results** are to make it easier to input as data
class data:
dream = "teknik" # asumsi jurusan impian
motivation = "menentukan pilihan jurusan yang tepat" # motivasi user using potenZ
psikotes = "teknik elektro" # asumsi luaran psikotes
# # Key Note!
# In this project, i just used 1 input variable, it's working and just to make it simple. And this section, i just used the result of psychological test.
# scraping raw data
PATH = "https://medium.com/tag/" + "-".join(data.psikotes.split()) + "/" + "archive"
print(PATH)
# makes a request to the web page and gets its HTML
html_article = requests.get(PATH).content
soup = BeautifulSoup(html_article, "html.parser")
titles = [] # link untuk setiap judul dari tagging
title = soup.find_all("div", {"class": "postArticle-readMore"})
for i in title:
titles.append(i.find("a")["href"])
print(titles)
# scraping
def tagging(url):
html_tag = requests.get(url).content
soup2 = BeautifulSoup(html_tag, "html.parser")
tags = []
for k in soup2.find_all("a"):
tags.append(k.get("href"))
alltags = []
for i in tags:
try:
indexing = PurePosixPath(unquote(urlparse(i).path)).parts[
-2
] # nama tagged/tag
if indexing == "tagged":
alltags.append(PurePosixPath(unquote(urlparse(i).path)).parts[-1])
elif indexing == "tag":
alltags.append(PurePosixPath(unquote(urlparse(i).path)).parts[-1])
except:
pass
return alltags
# get the summary and keyword
# ML scraping (uji coba)
for link in titles:
med_articles = Article(link, language="id") # medium artikel
med_articles.download()
med_articles.parse()
med_articles.nlp()
article_sum = med_articles.summary
# To extract summary
print("Article's Summary:")
print(article_sum)
article_keyword = med_articles.keywords
# To extract keywords
print("Article's Keywords:")
print(article_keyword)
# To extract tags
print("Article's tags:")
print(tagging(link))
print("\n")
# summary to input >>> recognize behaviour
# ML scraping
splitting_input = data.psikotes.split() # teknik elektro
articles = [] # final keywords
for links in titles:
med_articles = Article(links, language="id") # medium artikel
med_articles.download()
med_articles.parse()
med_articles.nlp()
splitting_output = med_articles.summary.split() # summary of articles
splitting_output2 = med_articles.keywords # keywords of articles
splitting_output3 = tagging(link) # tagged in articles
total_splitting_output = splitting_output + splitting_output2 + splitting_output3
dummy = list(set([]))
for b in total_splitting_output:
z = b.lower()
dummy.append(z)
articles.append(dummy)
print(articles[0])
# # Key Note!
# Thi section just calculate matches of user keywords with article keywords. The highest three will be a recommendation an articles for user.
# recommend system
# classification the same thing with the higher value
final_comparassion = {}
for idxi, i in enumerate(splitting_input):
for idxj, j in enumerate(articles):
if idxi == 0:
final_comparassion["Article {}".format(idxj + 1)] = j.count(i)
else:
final_comparassion["Article {}".format(idxj + 1)] = final_comparassion[
"Article {}".format(idxj + 1)
] + j.count(i)
print(final_comparassion)
# urutin 3 terbaik
val_dict = list(final_comparassion.values())
val_dict.sort(reverse=True)
print(val_dict)
# make a sequences of data to suggest
key_list = list(final_comparassion.keys())
val_list = list(final_comparassion.values())
final_seq = []
for i in range(0, 3):
position = val_list.index(val_dict[i])
final_seq.append(key_list[position].split()[-1])
# top 3 recommendation
for final in final_seq:
print(titles[int(final) - 1])
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/492/69492234.ipynb
| null | null |
[{"Id": 69492234, "ScriptId": 18955535, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7330269, "CreationDate": "07/31/2021 16:57:09", "VersionNumber": 3.0, "Title": "webscrap2", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 168.0, "LinesInsertedFromPrevious": 6.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 162.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import datetime as dt
import pandas as pd
import numpy as np
import requests
import os.path
from urllib.parse import urlparse
from newspaper import Article
from bs4 import BeautifulSoup
from urllib.parse import unquote, urlparse
from pathlib import PurePosixPath
# # Key Note!
# Because the assessment system and psychological test results may ONLY be known by psychologists, we need to assume what **Dreams**, **motivations**, and **user test results** are to make it easier to input as data
class data:
dream = "teknik" # asumsi jurusan impian
motivation = "menentukan pilihan jurusan yang tepat" # motivasi user using potenZ
psikotes = "teknik elektro" # asumsi luaran psikotes
# # Key Note!
# In this project, i just used 1 input variable, it's working and just to make it simple. And this section, i just used the result of psychological test.
# scraping raw data
PATH = "https://medium.com/tag/" + "-".join(data.psikotes.split()) + "/" + "archive"
print(PATH)
# makes a request to the web page and gets its HTML
html_article = requests.get(PATH).content
soup = BeautifulSoup(html_article, "html.parser")
titles = [] # link untuk setiap judul dari tagging
title = soup.find_all("div", {"class": "postArticle-readMore"})
for i in title:
titles.append(i.find("a")["href"])
print(titles)
# scraping
def tagging(url):
html_tag = requests.get(url).content
soup2 = BeautifulSoup(html_tag, "html.parser")
tags = []
for k in soup2.find_all("a"):
tags.append(k.get("href"))
alltags = []
for i in tags:
try:
indexing = PurePosixPath(unquote(urlparse(i).path)).parts[
-2
] # nama tagged/tag
if indexing == "tagged":
alltags.append(PurePosixPath(unquote(urlparse(i).path)).parts[-1])
elif indexing == "tag":
alltags.append(PurePosixPath(unquote(urlparse(i).path)).parts[-1])
except:
pass
return alltags
# get the summary and keyword
# ML scraping (uji coba)
for link in titles:
med_articles = Article(link, language="id") # medium artikel
med_articles.download()
med_articles.parse()
med_articles.nlp()
article_sum = med_articles.summary
# To extract summary
print("Article's Summary:")
print(article_sum)
article_keyword = med_articles.keywords
# To extract keywords
print("Article's Keywords:")
print(article_keyword)
# To extract tags
print("Article's tags:")
print(tagging(link))
print("\n")
# summary to input >>> recognize behaviour
# ML scraping
splitting_input = data.psikotes.split() # teknik elektro
articles = [] # final keywords
for links in titles:
med_articles = Article(links, language="id") # medium artikel
med_articles.download()
med_articles.parse()
med_articles.nlp()
splitting_output = med_articles.summary.split() # summary of articles
splitting_output2 = med_articles.keywords # keywords of articles
splitting_output3 = tagging(link) # tagged in articles
total_splitting_output = splitting_output + splitting_output2 + splitting_output3
dummy = list(set([]))
for b in total_splitting_output:
z = b.lower()
dummy.append(z)
articles.append(dummy)
print(articles[0])
# # Key Note!
# Thi section just calculate matches of user keywords with article keywords. The highest three will be a recommendation an articles for user.
# recommend system
# classification the same thing with the higher value
final_comparassion = {}
for idxi, i in enumerate(splitting_input):
for idxj, j in enumerate(articles):
if idxi == 0:
final_comparassion["Article {}".format(idxj + 1)] = j.count(i)
else:
final_comparassion["Article {}".format(idxj + 1)] = final_comparassion[
"Article {}".format(idxj + 1)
] + j.count(i)
print(final_comparassion)
# urutin 3 terbaik
val_dict = list(final_comparassion.values())
val_dict.sort(reverse=True)
print(val_dict)
# make a sequences of data to suggest
key_list = list(final_comparassion.keys())
val_list = list(final_comparassion.values())
final_seq = []
for i in range(0, 3):
position = val_list.index(val_dict[i])
final_seq.append(key_list[position].split()[-1])
# top 3 recommendation
for final in final_seq:
print(titles[int(final) - 1])
| false | 0 | 1,439 | 0 | 1,439 | 1,439 |
||
69492505
|
<jupyter_start><jupyter_text>word2vec_js
Kaggle dataset identifier: word2vec-js
<jupyter_script># # Imports and Var decleration
## Imports
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
from nltk import word_tokenize
from gensim.models.doc2vec import Doc2Vec
from gensim.models import Word2Vec
from gensim.models.doc2vec import TaggedDocument
from pickle import dump, load
from random import choices
from multiprocessing import Pool
import multiprocessing
import time
import tensorflow as tf
import matplotlib.pyplot as plt
from sklearn.metrics import (
confusion_matrix,
recall_score,
accuracy_score,
precision_score,
roc_auc_score,
auc,
roc_curve,
)
import seaborn as sns
import random
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import StratifiedKFold
from shutil import copyfile
import multiprocessing
from termcolor import colored
## Var decleration
path_train = "/kaggle/input/javascript-files/data/train"
path_test = "/kaggle/input/javascript-files/data/test"
paths = [path_train, path_test]
# # Preprocess Section - all the preprocess necessary for data
## Helping Functions
def pickle_something(obj, filename):
# Save object as pickle
with open(filename, "wb") as f:
dump(obj, f)
def read_pickle(path):
# Read pickle object from file
with open(path, "rb") as file:
return load(file)
def get_all_data_from_file(tokens, k=10000):
# Given file names, read the tokenize data and labels
data = []
files = [10000, 80000]
all_files = {tokens: [], "labels": []}
for f in files:
curr = read_pickle(f"../input/tags-tokens/test_{tokens}_{f}.pickle")
all_files[tokens] += curr[tokens]
all_files["labels"] += curr["labels"]
return all_files
def read_data_pickle(files):
# given a list of files, go over the files and read each file from disk
# After reading the JS file from disk, tokenize the data and add label
texts, labels = [], []
length = len(files)
for idx, file in enumerate(files):
if idx % 1000 == 0:
print(f"{idx}/{length}")
# Check whether file is in text format or not
if file.endswith(".txt"):
# call read text file function
texts.append(read_text_file(file))
labels.append(0 if "benign" in file else 1)
tokenized_text = [t.replace("\n", "").split(" ") for t in texts]
obj = {"tokens": tokenized_text, "labels": labels}
return obj
def load_tokens():
# After files were tokenized and save to disk, read them
base_path = "../input/train-test-tokens"
test_mal_tokens = read_pickle(f"{base_path}/test_mal_tokens")
train_mal_tokens = read_pickle(f"{base_path}/train_mal_tokens")
train_benign_tokens = read_pickle(f"{base_path}/train_benign_tokens")
test_benign_tokens = read_pickle(f"{base_path}/test_benign_tokens")
return test_mal_tokens, train_mal_tokens, train_benign_tokens, test_benign_tokens
def read_text_file(file_path):
# Read one JS file from disk as text
with open(file_path, "r", encoding="utf-8") as f:
try:
text = f.read()
return text
except:
with open(file_path, "r", encoding="windows-1252") as f:
try:
text2 = f.read()
return text2
except:
return ""
def read_data(path, mal):
texts = []
files = path
length = len(files)
for idx, file in enumerate(files):
if idx % 1000 == 0:
print(f"{idx}/{length}")
# Check whether file is in text format or not
if file.endswith(".txt"):
file_path = f"{path}" + "/" + f"{file}"
# call read text file function
texts.append(read_text_file(file))
if idx % 10000 == 0 and idx != 0:
print("Tokenizing")
texts = [t.replace("\n", "").split(" ") for t in texts]
print("Pickeling")
labels = np.ones(len(texts)) if mal else np.zeros(len(texts))
tag = "mal" if mal else "benign"
pickle_something(
{"tokens": texts, "labels": labels},
f"./{tag}_tokens_labels_{idx//10000}",
)
texts = []
return texts
def get_all_data_tokenized(paths, mal=True):
# A more general function that read all files from path and tokenize it
data = []
for path in paths:
# print(len(path))
data += read_data(path, mal)
return
print("tokenizing")
data = [t.replace("\n", "").split(" ") for t in data]
return data
return tags
def get_files():
# Get all malicious and all benign files
mal_files = [
f"../input/javascript-files/data/test/{file}"
for file in os.listdir(path_test)
if "mal" in file
][:40001]
benign_files = [
f"../input/javascript-files/data/test/{file}"
for file in os.listdir(path_test)
if "ben" in file
][:40001]
return mal_files, benign_files
def Word2Vecing():
# Performing word2vec and save the model to disk
cpus = multiprocessing.cpu_count()
tokens = "tokens"
min_count = 10
vector_size = 100
window = 5
print(vector_size, window)
path = "../input/tags-tokens"
for idx, file in enumerate(os.listdir(path)):
pick = read_pickle(f"{path}/{file}")
if idx == 0:
model = Word2Vec(
sentences=pick["tokens"],
vector_size=vector_size,
window=window,
min_count=min_count,
workers=cpus,
)
print(model.wv.vectors.shape[0])
# return model
else:
print("vocab")
model.build_vocab(pick["tokens"], update=True)
print("train")
model.train(pick["tokens"], total_examples=len(pick["tokens"]), epochs=1)
print(model.wv.vectors.shape[0])
model_file = f"Word2vec_v{vector_size}_w{window}.pickle"
with open(model_file, "wb") as f:
dump(model, f)
return model
def load_word2vec(vector_size, window):
path_to_word2vec = f"../input/word2vec-js/Word2vec_v100_w5.pickle"
return Word2Vec.load(path_to_word2vec)
def avg_doc(token_embeddings):
# Basically perform Doc2Vec by taking the mean of each feature
return np.array(token_embeddings).mean(axis=0)
def embed_docs(model, tokens):
# The entire process of the embed the files
print("Embedding")
vocab = model.wv
docs = []
for token in tokens:
token_embeddings = []
for word in token:
if word not in vocab:
continue
token_embeddings.append(vocab[word])
token_embeddings = [vocab["["]] if token_embeddings == [] else token_embeddings
docs.append(avg_doc(token_embeddings))
docs = np.array(docs)
docs = np.array([np.array(doc) for doc in docs])
return docs
def load_tokens():
# Load the tokens from files
base_path = "../input/train-test-tokens"
test_mal_tokens = read_pickle(f"{base_path}/test_mal_tokens")
train_mal_tokens = read_pickle(f"{base_path}/train_mal_tokens")
train_benign_tokens = read_pickle(f"{base_path}/train_benign_tokens")
test_benign_tokens = read_pickle(f"{base_path}/test_benign_tokens")
return test_mal_tokens, train_mal_tokens, train_benign_tokens, test_benign_tokens
def load_embeddings(vector_size, window):
# load the embedded files
base_path = "../input/javascript-embeddings"
test_mal_embd = read_pickle(
f"{base_path}/test_mal_embd_v{vector_size}_w{window}.pickle"
)
train_mal_embd = read_pickle(
f"{base_path}/train_mal_embd_v{vector_size}_w{window}.pickle"
)
train_benign_embd = read_pickle(
f"{base_path}/train_benign_embd_v{vector_size}_w{window}.pickle"
)
test_benign_embd = read_pickle(
f"{base_path}/test_benign_embd_v{vector_size}_w{window}.pickle"
)
return test_mal_embd, train_mal_embd, train_benign_embd, test_benign_embd
def load_word2vec(vector_size, window):
# Load the pretrained Word2Vec model
path_to_word2vec = f"../input/word2vec-js/Word2vec_v100_w5.pickle"
return Word2Vec.load(path_to_word2vec)
def avg_doc(token_embeddings):
# Basically perform Doc2Vec by taking the mean of each feature
return np.array(token_embeddings).mean(axis=0)
def load_embedding(mal, num):
token = "mal" if mal else "benign"
return read_pickle(f"../input/tags-tokens/{token}_{num}.pickle")
print(colored("Preprocess steps (For Training and for inference):", "red"))
print(colored("1. read JS file from disk", "blue"))
print(colored("2. Tokenize the file", "blue"))
print(
colored("3. Use the pretrained Word2Vec model to embed the tokenized text", "blue")
)
print(
colored(
"* Note: we trained Word2Vec model on all the malicious and benign JS files upfront",
"green",
)
)
# # Training RF classifier
print(colored("Training steps:", "red"))
print(colored("1. Load the pretrained word2vec model", "blue"))
print(colored("2. Load the embedded train files (malicious and benign)", "blue"))
print(colored("3. Train the RF classifier", "blue"))
model = load_word2vec(100, 5)
vector_size, window = 100, 5
x_1, x_2 = load_embedding(True, 1), load_embedding(False, 1)
x_train = np.concatenate([x_1["embds"], x_2["embds"]])
y_train = np.concatenate([x_1["labels"], x_2["labels"]])
clf = RandomForestClassifier(n_estimators=200, max_depth=10)
clf.fit(x_train, y_train)
# # Predicting & Score
print(colored("Predicting steps:", "red"))
print(colored("1. Load the embedded test files (malicious and benign)", "blue"))
print(colored("2. Predict class based on the RF classifier", "blue"))
print(colored("3. Evaluate results based on pre-defined measurements", "blue"))
# Load the Test files with data and labels
embds = np.zeros((1, 100))
labels = np.array([0])
for tf in [True, False]:
for num in [2, 3, 4]:
curr_file = load_embedding(tf, num)
embds = np.concatenate([embds, curr_file["embds"]])
labels = np.concatenate([labels, curr_file["labels"]])
# Get the predictions
preds = clf.predict(embds)
# Calculate confusion_matrix and plot it
conf_matrix = confusion_matrix(labels, preds)
plt.figure(figsize=(4, 4))
LABELS = ["Benign", "Malicious"]
sns.heatmap(conf_matrix, xticklabels=LABELS, yticklabels=LABELS, annot=True, fmt="d")
plt.title("Confusion matrix")
plt.ylabel("True class")
plt.xlabel("Predicted class")
plt.show()
print(" Accuracy: ", accuracy_score(labels, preds))
print(" Recall: ", recall_score(labels, preds))
print(" Precision: ", precision_score(labels, preds))
print(" ROC: ", roc_auc_score(labels, preds))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/492/69492505.ipynb
|
word2vec-js
|
yr12345
|
[{"Id": 69492505, "ScriptId": 18372529, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2395947, "CreationDate": "07/31/2021 17:01:04", "VersionNumber": 53.0, "Title": "javascript_project", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 284.0, "LinesInsertedFromPrevious": 8.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 276.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92728720, "KernelVersionId": 69492505, "SourceDatasetVersionId": 2448815}, {"Id": 92728721, "KernelVersionId": 69492505, "SourceDatasetVersionId": 2449000}]
|
[{"Id": 2448815, "DatasetId": 1482060, "DatasourceVersionId": 2491161, "CreatorUserId": 2395947, "LicenseName": "Unknown", "CreationDate": "07/21/2021 15:05:42", "VersionNumber": 1.0, "Title": "word2vec_js", "Slug": "word2vec-js", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1482060, "CreatorUserId": 2395947, "OwnerUserId": 2395947.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2448815.0, "CurrentDatasourceVersionId": 2491161.0, "ForumId": 1501735, "Type": 2, "CreationDate": "07/21/2021 15:05:42", "LastActivityDate": "07/21/2021", "TotalViews": 889, "TotalDownloads": 3, "TotalVotes": 0, "TotalKernels": 1}]
|
[{"Id": 2395947, "UserName": "yr12345", "DisplayName": "Yarden Rotem", "RegisterDate": "10/23/2018", "PerformanceTier": 0}]
|
# # Imports and Var decleration
## Imports
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
from nltk import word_tokenize
from gensim.models.doc2vec import Doc2Vec
from gensim.models import Word2Vec
from gensim.models.doc2vec import TaggedDocument
from pickle import dump, load
from random import choices
from multiprocessing import Pool
import multiprocessing
import time
import tensorflow as tf
import matplotlib.pyplot as plt
from sklearn.metrics import (
confusion_matrix,
recall_score,
accuracy_score,
precision_score,
roc_auc_score,
auc,
roc_curve,
)
import seaborn as sns
import random
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import StratifiedKFold
from shutil import copyfile
import multiprocessing
from termcolor import colored
## Var decleration
path_train = "/kaggle/input/javascript-files/data/train"
path_test = "/kaggle/input/javascript-files/data/test"
paths = [path_train, path_test]
# # Preprocess Section - all the preprocess necessary for data
## Helping Functions
def pickle_something(obj, filename):
# Save object as pickle
with open(filename, "wb") as f:
dump(obj, f)
def read_pickle(path):
# Read pickle object from file
with open(path, "rb") as file:
return load(file)
def get_all_data_from_file(tokens, k=10000):
# Given file names, read the tokenize data and labels
data = []
files = [10000, 80000]
all_files = {tokens: [], "labels": []}
for f in files:
curr = read_pickle(f"../input/tags-tokens/test_{tokens}_{f}.pickle")
all_files[tokens] += curr[tokens]
all_files["labels"] += curr["labels"]
return all_files
def read_data_pickle(files):
# given a list of files, go over the files and read each file from disk
# After reading the JS file from disk, tokenize the data and add label
texts, labels = [], []
length = len(files)
for idx, file in enumerate(files):
if idx % 1000 == 0:
print(f"{idx}/{length}")
# Check whether file is in text format or not
if file.endswith(".txt"):
# call read text file function
texts.append(read_text_file(file))
labels.append(0 if "benign" in file else 1)
tokenized_text = [t.replace("\n", "").split(" ") for t in texts]
obj = {"tokens": tokenized_text, "labels": labels}
return obj
def load_tokens():
# After files were tokenized and save to disk, read them
base_path = "../input/train-test-tokens"
test_mal_tokens = read_pickle(f"{base_path}/test_mal_tokens")
train_mal_tokens = read_pickle(f"{base_path}/train_mal_tokens")
train_benign_tokens = read_pickle(f"{base_path}/train_benign_tokens")
test_benign_tokens = read_pickle(f"{base_path}/test_benign_tokens")
return test_mal_tokens, train_mal_tokens, train_benign_tokens, test_benign_tokens
def read_text_file(file_path):
# Read one JS file from disk as text
with open(file_path, "r", encoding="utf-8") as f:
try:
text = f.read()
return text
except:
with open(file_path, "r", encoding="windows-1252") as f:
try:
text2 = f.read()
return text2
except:
return ""
def read_data(path, mal):
texts = []
files = path
length = len(files)
for idx, file in enumerate(files):
if idx % 1000 == 0:
print(f"{idx}/{length}")
# Check whether file is in text format or not
if file.endswith(".txt"):
file_path = f"{path}" + "/" + f"{file}"
# call read text file function
texts.append(read_text_file(file))
if idx % 10000 == 0 and idx != 0:
print("Tokenizing")
texts = [t.replace("\n", "").split(" ") for t in texts]
print("Pickeling")
labels = np.ones(len(texts)) if mal else np.zeros(len(texts))
tag = "mal" if mal else "benign"
pickle_something(
{"tokens": texts, "labels": labels},
f"./{tag}_tokens_labels_{idx//10000}",
)
texts = []
return texts
def get_all_data_tokenized(paths, mal=True):
# A more general function that read all files from path and tokenize it
data = []
for path in paths:
# print(len(path))
data += read_data(path, mal)
return
print("tokenizing")
data = [t.replace("\n", "").split(" ") for t in data]
return data
return tags
def get_files():
# Get all malicious and all benign files
mal_files = [
f"../input/javascript-files/data/test/{file}"
for file in os.listdir(path_test)
if "mal" in file
][:40001]
benign_files = [
f"../input/javascript-files/data/test/{file}"
for file in os.listdir(path_test)
if "ben" in file
][:40001]
return mal_files, benign_files
def Word2Vecing():
# Performing word2vec and save the model to disk
cpus = multiprocessing.cpu_count()
tokens = "tokens"
min_count = 10
vector_size = 100
window = 5
print(vector_size, window)
path = "../input/tags-tokens"
for idx, file in enumerate(os.listdir(path)):
pick = read_pickle(f"{path}/{file}")
if idx == 0:
model = Word2Vec(
sentences=pick["tokens"],
vector_size=vector_size,
window=window,
min_count=min_count,
workers=cpus,
)
print(model.wv.vectors.shape[0])
# return model
else:
print("vocab")
model.build_vocab(pick["tokens"], update=True)
print("train")
model.train(pick["tokens"], total_examples=len(pick["tokens"]), epochs=1)
print(model.wv.vectors.shape[0])
model_file = f"Word2vec_v{vector_size}_w{window}.pickle"
with open(model_file, "wb") as f:
dump(model, f)
return model
def load_word2vec(vector_size, window):
path_to_word2vec = f"../input/word2vec-js/Word2vec_v100_w5.pickle"
return Word2Vec.load(path_to_word2vec)
def avg_doc(token_embeddings):
# Basically perform Doc2Vec by taking the mean of each feature
return np.array(token_embeddings).mean(axis=0)
def embed_docs(model, tokens):
# The entire process of the embed the files
print("Embedding")
vocab = model.wv
docs = []
for token in tokens:
token_embeddings = []
for word in token:
if word not in vocab:
continue
token_embeddings.append(vocab[word])
token_embeddings = [vocab["["]] if token_embeddings == [] else token_embeddings
docs.append(avg_doc(token_embeddings))
docs = np.array(docs)
docs = np.array([np.array(doc) for doc in docs])
return docs
def load_tokens():
# Load the tokens from files
base_path = "../input/train-test-tokens"
test_mal_tokens = read_pickle(f"{base_path}/test_mal_tokens")
train_mal_tokens = read_pickle(f"{base_path}/train_mal_tokens")
train_benign_tokens = read_pickle(f"{base_path}/train_benign_tokens")
test_benign_tokens = read_pickle(f"{base_path}/test_benign_tokens")
return test_mal_tokens, train_mal_tokens, train_benign_tokens, test_benign_tokens
def load_embeddings(vector_size, window):
# load the embedded files
base_path = "../input/javascript-embeddings"
test_mal_embd = read_pickle(
f"{base_path}/test_mal_embd_v{vector_size}_w{window}.pickle"
)
train_mal_embd = read_pickle(
f"{base_path}/train_mal_embd_v{vector_size}_w{window}.pickle"
)
train_benign_embd = read_pickle(
f"{base_path}/train_benign_embd_v{vector_size}_w{window}.pickle"
)
test_benign_embd = read_pickle(
f"{base_path}/test_benign_embd_v{vector_size}_w{window}.pickle"
)
return test_mal_embd, train_mal_embd, train_benign_embd, test_benign_embd
def load_word2vec(vector_size, window):
# Load the pretrained Word2Vec model
path_to_word2vec = f"../input/word2vec-js/Word2vec_v100_w5.pickle"
return Word2Vec.load(path_to_word2vec)
def avg_doc(token_embeddings):
# Basically perform Doc2Vec by taking the mean of each feature
return np.array(token_embeddings).mean(axis=0)
def load_embedding(mal, num):
token = "mal" if mal else "benign"
return read_pickle(f"../input/tags-tokens/{token}_{num}.pickle")
print(colored("Preprocess steps (For Training and for inference):", "red"))
print(colored("1. read JS file from disk", "blue"))
print(colored("2. Tokenize the file", "blue"))
print(
colored("3. Use the pretrained Word2Vec model to embed the tokenized text", "blue")
)
print(
colored(
"* Note: we trained Word2Vec model on all the malicious and benign JS files upfront",
"green",
)
)
# # Training RF classifier
print(colored("Training steps:", "red"))
print(colored("1. Load the pretrained word2vec model", "blue"))
print(colored("2. Load the embedded train files (malicious and benign)", "blue"))
print(colored("3. Train the RF classifier", "blue"))
model = load_word2vec(100, 5)
vector_size, window = 100, 5
x_1, x_2 = load_embedding(True, 1), load_embedding(False, 1)
x_train = np.concatenate([x_1["embds"], x_2["embds"]])
y_train = np.concatenate([x_1["labels"], x_2["labels"]])
clf = RandomForestClassifier(n_estimators=200, max_depth=10)
clf.fit(x_train, y_train)
# # Predicting & Score
print(colored("Predicting steps:", "red"))
print(colored("1. Load the embedded test files (malicious and benign)", "blue"))
print(colored("2. Predict class based on the RF classifier", "blue"))
print(colored("3. Evaluate results based on pre-defined measurements", "blue"))
# Load the Test files with data and labels
embds = np.zeros((1, 100))
labels = np.array([0])
for tf in [True, False]:
for num in [2, 3, 4]:
curr_file = load_embedding(tf, num)
embds = np.concatenate([embds, curr_file["embds"]])
labels = np.concatenate([labels, curr_file["labels"]])
# Get the predictions
preds = clf.predict(embds)
# Calculate confusion_matrix and plot it
conf_matrix = confusion_matrix(labels, preds)
plt.figure(figsize=(4, 4))
LABELS = ["Benign", "Malicious"]
sns.heatmap(conf_matrix, xticklabels=LABELS, yticklabels=LABELS, annot=True, fmt="d")
plt.title("Confusion matrix")
plt.ylabel("True class")
plt.xlabel("Predicted class")
plt.show()
print(" Accuracy: ", accuracy_score(labels, preds))
print(" Recall: ", recall_score(labels, preds))
print(" Precision: ", precision_score(labels, preds))
print(" ROC: ", roc_auc_score(labels, preds))
| false | 0 | 3,186 | 0 | 3,209 | 3,186 |
||
69492376
|
<jupyter_start><jupyter_text>AMZ data
Kaggle dataset identifier: amz-data
<jupyter_script>import numpy as np
import pandas as pd
from tqdm import tqdm
tqdm.pandas()
import missingno as msno
import matplotlib.pyplot as plt
import seaborn as sns
from nltk.corpus import stopwords
from nltk.stem import WordNetLemmatizer
from nltk import word_tokenize, RegexpTokenizer, TweetTokenizer, PorterStemmer
from re import sub
import tokenization
from tokenization import FullTokenizer
from sklearn.preprocessing import LabelEncoder
import tensorflow as tf
from sklearn.model_selection import train_test_split
df_train = pd.read_csv(
"../input/amz-data/dataset/train.csv", escapechar="\\", quoting=3
)
df_mini = df_train[:40000]
df_test = pd.read_csv("../input/amz-data/dataset/test.csv", escapechar="\\", quoting=3)
df = pd.concat([df_train, df_test])
df.reset_index(inplace=True)
print(df_train.shape, df_test.shape, df.shape)
df_mini.head(3)
df_train.isna().sum(), df_test.isna().sum()
msno.matrix(df_mini)
df = df.replace(np.nan, "error", regex=True)
df.isna().sum()
df.head(5)
# ## NLP Processing
def remove_URL(text):
url = RegexpTokenizer(r"https?://\S+|www\.\S+", gaps=True)
return " ".join(url.tokenize(text))
def stopWords(tweet):
stop_words, toker = stopwords.words("english"), TweetTokenizer()
words_tokens = toker.tokenize(tweet)
return " ".join([word for word in words_tokens if not word in stop_words])
def remove_pontucations(text):
tokenizer_dots = RegexpTokenizer(r"\w+")
return " ".join(tokenizer_dots.tokenize(text))
def remove_words_min(text, param):
tmp = text
for x in tmp.split():
if len(x) < param:
tmp = tmp.replace(x, "")
return " ".join(tmp.split())
def clean(data, col="TITLE"): # print('make text lowercase')
data[col] = data[col].apply(lambda x: x.lower()) # print('delete excess spaces')
data[col] = data[col].apply(
lambda x: " ".join(x.split())
) # print('delete numbers')
data[col] = data[col].apply(
lambda x: sub(r"\d+", "", x)
) # print('remove punctuation and special character')
data[col] = data[col].progress_apply(
lambda x: remove_pontucations(x)
) # print('remove stopWords')
data[col] = data[col].progress_apply(
lambda x: stopWords(x)
) # print('remove _ attatched words')
data[col] = data[col].apply(lambda x: x.replace("_", " "))
# data[col] = data[col].apply(lambda x: remove_words_min(x, 2))
# BEFORE
print(df.TITLE[0])
print(df.DESCRIPTION[0])
print(df.BULLET_POINTS[0])
print(df.BRAND[0])
clean(df, col="TITLE")
clean(df, col="DESCRIPTION")
clean(df, col="BULLET_POINTS")
clean(df, col="BRAND")
# AFTER
print(df.TITLE[0])
print(df.DESCRIPTION[0])
print(df.BULLET_POINTS[0])
print(df.BRAND[0])
df.to_csv("amz_df.csv", index=False)
df_train.BROWSE_NODE_ID.unique().shape
plt.bar(
df_train.BROWSE_NODE_ID.value_counts().index, df_train.BROWSE_NODE_ID.value_counts()
)
plt.show()
sns.kdeplot(data=df_train, x="BROWSE_NODE_ID")
plt.show()
# ## EMB. MOdel
import tensorflow_hub as hub
import tensorflow as tf
model = "https://tfhub.dev/google/nnlm-en-dim50/2"
hub_layer = hub.KerasLayer(model, input_shape=[], dtype=tf.string, trainable=True)
df.TITLE[0:3].values
hub_layer(df.TITLE[0:3].values)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/492/69492376.ipynb
|
amz-data
|
akhileshdkapse
|
[{"Id": 69492376, "ScriptId": 18962222, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4684168, "CreationDate": "07/31/2021 16:59:10", "VersionNumber": 4.0, "Title": "AMZ Data Processing", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 113.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 112.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92728278, "KernelVersionId": 69492376, "SourceDatasetVersionId": 2482576}]
|
[{"Id": 2482576, "DatasetId": 1502602, "DatasourceVersionId": 2525120, "CreatorUserId": 4684168, "LicenseName": "Unknown", "CreationDate": "07/31/2021 05:06:01", "VersionNumber": 1.0, "Title": "AMZ data", "Slug": "amz-data", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1502602, "CreatorUserId": 4684168, "OwnerUserId": 4684168.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2482576.0, "CurrentDatasourceVersionId": 2525120.0, "ForumId": 1522340, "Type": 2, "CreationDate": "07/31/2021 05:06:01", "LastActivityDate": "07/31/2021", "TotalViews": 850, "TotalDownloads": 2, "TotalVotes": 1, "TotalKernels": 2}]
|
[{"Id": 4684168, "UserName": "akhileshdkapse", "DisplayName": "Akhilesh D. Kapse", "RegisterDate": "03/17/2020", "PerformanceTier": 2}]
|
import numpy as np
import pandas as pd
from tqdm import tqdm
tqdm.pandas()
import missingno as msno
import matplotlib.pyplot as plt
import seaborn as sns
from nltk.corpus import stopwords
from nltk.stem import WordNetLemmatizer
from nltk import word_tokenize, RegexpTokenizer, TweetTokenizer, PorterStemmer
from re import sub
import tokenization
from tokenization import FullTokenizer
from sklearn.preprocessing import LabelEncoder
import tensorflow as tf
from sklearn.model_selection import train_test_split
df_train = pd.read_csv(
"../input/amz-data/dataset/train.csv", escapechar="\\", quoting=3
)
df_mini = df_train[:40000]
df_test = pd.read_csv("../input/amz-data/dataset/test.csv", escapechar="\\", quoting=3)
df = pd.concat([df_train, df_test])
df.reset_index(inplace=True)
print(df_train.shape, df_test.shape, df.shape)
df_mini.head(3)
df_train.isna().sum(), df_test.isna().sum()
msno.matrix(df_mini)
df = df.replace(np.nan, "error", regex=True)
df.isna().sum()
df.head(5)
# ## NLP Processing
def remove_URL(text):
url = RegexpTokenizer(r"https?://\S+|www\.\S+", gaps=True)
return " ".join(url.tokenize(text))
def stopWords(tweet):
stop_words, toker = stopwords.words("english"), TweetTokenizer()
words_tokens = toker.tokenize(tweet)
return " ".join([word for word in words_tokens if not word in stop_words])
def remove_pontucations(text):
tokenizer_dots = RegexpTokenizer(r"\w+")
return " ".join(tokenizer_dots.tokenize(text))
def remove_words_min(text, param):
tmp = text
for x in tmp.split():
if len(x) < param:
tmp = tmp.replace(x, "")
return " ".join(tmp.split())
def clean(data, col="TITLE"): # print('make text lowercase')
data[col] = data[col].apply(lambda x: x.lower()) # print('delete excess spaces')
data[col] = data[col].apply(
lambda x: " ".join(x.split())
) # print('delete numbers')
data[col] = data[col].apply(
lambda x: sub(r"\d+", "", x)
) # print('remove punctuation and special character')
data[col] = data[col].progress_apply(
lambda x: remove_pontucations(x)
) # print('remove stopWords')
data[col] = data[col].progress_apply(
lambda x: stopWords(x)
) # print('remove _ attatched words')
data[col] = data[col].apply(lambda x: x.replace("_", " "))
# data[col] = data[col].apply(lambda x: remove_words_min(x, 2))
# BEFORE
print(df.TITLE[0])
print(df.DESCRIPTION[0])
print(df.BULLET_POINTS[0])
print(df.BRAND[0])
clean(df, col="TITLE")
clean(df, col="DESCRIPTION")
clean(df, col="BULLET_POINTS")
clean(df, col="BRAND")
# AFTER
print(df.TITLE[0])
print(df.DESCRIPTION[0])
print(df.BULLET_POINTS[0])
print(df.BRAND[0])
df.to_csv("amz_df.csv", index=False)
df_train.BROWSE_NODE_ID.unique().shape
plt.bar(
df_train.BROWSE_NODE_ID.value_counts().index, df_train.BROWSE_NODE_ID.value_counts()
)
plt.show()
sns.kdeplot(data=df_train, x="BROWSE_NODE_ID")
plt.show()
# ## EMB. MOdel
import tensorflow_hub as hub
import tensorflow as tf
model = "https://tfhub.dev/google/nnlm-en-dim50/2"
hub_layer = hub.KerasLayer(model, input_shape=[], dtype=tf.string, trainable=True)
df.TITLE[0:3].values
hub_layer(df.TITLE[0:3].values)
| false | 2 | 1,099 | 0 | 1,120 | 1,099 |
||
69492790
|
import math
import random
import numpy as np
class Tic_Tac_Toe:
def __init__(self):
self.dim = 0
self.AI = None
self.human = None
def set_players(self, player, n):
self.dim = n
self.AI = player
if self.AI == "X":
self.human = "O"
elif self.AI == "O":
self.human = "X"
def get_human(self):
return self.human
def checker(self, board1):
dim = int(math.sqrt(len(board1)))
board = np.array([0] * len(board1))
for i in range(len(board1)):
if board1[i] == "X":
board[i] = 1
elif board1[i] == "O":
board[i] = 0
elif board1[i] == " ":
board[i] = -10
board = board.reshape(dim, dim)
row = 0
daigonal2 = 0
daigonal1 = np.trace(board)
for i in range(0, len(board[0])):
daigonal2 += board[i][len(board[0]) - i - 1]
data = list()
data.append(daigonal1)
data.append(daigonal2)
for i in range(len(board[0])):
for j in range(len(board[0])):
row += board[i][j]
data.append(row)
row = 0
board = board.T
row = 0
for i in range(len(board[0])):
for j in range(len(board[0])):
row += board[i][j]
data.append(row)
row = 0
sum1 = dim
sum2 = 0
if sum1 in data:
return "X"
elif sum2 in data:
return "O"
return "-"
def alpha_beta(self, board, player, alpha=-np.inf, beta=np.inf):
winner = self.checker(board)
if winner == self.AI:
return 5
elif winner == self.human:
return -5
elif " " not in board:
return 0
if player == self.AI: # Alpha
best = -np.inf
for i in range(len(board)):
if board[i] == " ":
board[i] = self.AI
val = self.alpha_beta(board, self.human, alpha, beta)
board[i] = " "
best = max(val, best)
alpha = max(best, alpha)
if alpha >= beta:
break
elif player == self.human: # Beta
best = np.inf
for i in range(len(board)):
if board[i] == " ":
board[i] = self.human
val = self.alpha_beta(board, self.AI, alpha, beta)
board[i] = " "
best = min(val, best)
beta = min(best, beta)
if alpha >= beta:
break
return best
def filled_space(self, board):
count = 0
for i in board:
if i != " ":
count += 1
return count
def rand_move(self, board):
empty_slotes = list()
for i in range(len(board)):
for j in range(len(board)):
if board[i][j] == " ":
empty_slotes.append((i, j))
return random.choice(empty_slotes)
def Agent(Currentboard, n, AI):
game = Tic_Tac_Toe()
game.set_players(AI, n)
board = np.array(Currentboard)
free = game.filled_space(board.reshape(-1))
if len(set(board.reshape(-1))) == 1:
board[0][0] = AI
return board
elif free < 4 and n == 4:
move = game.rand_move(board)
board[move] = AI
return board
elif free < 7 and n == 5:
move = game.rand_move(board)
board[move] = AI
return board
elif free < 9 and n == 6:
move = game.rand_move(board)
board[move] = AI
return board
move = None
score = -np.inf
for i in range(n):
for j in range(n):
if board[i][j] == " ":
board[i][j] = AI
bestscore = game.alpha_beta(board.reshape(-1), game.get_human())
board[i][j] = " "
if bestscore > score:
score = bestscore
move = (i, j)
board[move] = AI
return board
def end(board):
for i in range(len(board)):
for j in range(len(board)):
if board[i][j] == " ":
return True
return False
no = int(input("Enter Value of n to make nxn Board : "))
game1 = Tic_Tac_Toe()
board = np.array([[" "] * no] * no)
print(board)
print(" ")
print("Enter Value from 1 to ", no * no)
print(" ")
for i in range(9):
if end(board):
check = game1.checker(board.reshape(-1))
if check == "X":
print("X won")
break
elif check == "O":
print("O won")
break
player1 = input("Enter position : ")
player1 = int(player1)
board = board.reshape(-1)
board[player1 - 1] = "O"
board = board.reshape(no, no)
print("Human : ")
print(board)
print("")
if end(board):
check = game1.checker(board.reshape(-1))
if check == "X":
print("X won")
break
elif check == "O":
print("O won")
break
board = Agent(board, no, "X")
print("Agent: ")
print(board)
print("")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/492/69492790.ipynb
| null | null |
[{"Id": 69492790, "ScriptId": 18975924, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7863466, "CreationDate": "07/31/2021 17:05:42", "VersionNumber": 2.0, "Title": "Tic Tac Toe AI Agent", "EvaluationDate": "07/31/2021", "IsChange": false, "TotalLines": 191.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 191.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
| null | null | null | null |
import math
import random
import numpy as np
class Tic_Tac_Toe:
def __init__(self):
self.dim = 0
self.AI = None
self.human = None
def set_players(self, player, n):
self.dim = n
self.AI = player
if self.AI == "X":
self.human = "O"
elif self.AI == "O":
self.human = "X"
def get_human(self):
return self.human
def checker(self, board1):
dim = int(math.sqrt(len(board1)))
board = np.array([0] * len(board1))
for i in range(len(board1)):
if board1[i] == "X":
board[i] = 1
elif board1[i] == "O":
board[i] = 0
elif board1[i] == " ":
board[i] = -10
board = board.reshape(dim, dim)
row = 0
daigonal2 = 0
daigonal1 = np.trace(board)
for i in range(0, len(board[0])):
daigonal2 += board[i][len(board[0]) - i - 1]
data = list()
data.append(daigonal1)
data.append(daigonal2)
for i in range(len(board[0])):
for j in range(len(board[0])):
row += board[i][j]
data.append(row)
row = 0
board = board.T
row = 0
for i in range(len(board[0])):
for j in range(len(board[0])):
row += board[i][j]
data.append(row)
row = 0
sum1 = dim
sum2 = 0
if sum1 in data:
return "X"
elif sum2 in data:
return "O"
return "-"
def alpha_beta(self, board, player, alpha=-np.inf, beta=np.inf):
winner = self.checker(board)
if winner == self.AI:
return 5
elif winner == self.human:
return -5
elif " " not in board:
return 0
if player == self.AI: # Alpha
best = -np.inf
for i in range(len(board)):
if board[i] == " ":
board[i] = self.AI
val = self.alpha_beta(board, self.human, alpha, beta)
board[i] = " "
best = max(val, best)
alpha = max(best, alpha)
if alpha >= beta:
break
elif player == self.human: # Beta
best = np.inf
for i in range(len(board)):
if board[i] == " ":
board[i] = self.human
val = self.alpha_beta(board, self.AI, alpha, beta)
board[i] = " "
best = min(val, best)
beta = min(best, beta)
if alpha >= beta:
break
return best
def filled_space(self, board):
count = 0
for i in board:
if i != " ":
count += 1
return count
def rand_move(self, board):
empty_slotes = list()
for i in range(len(board)):
for j in range(len(board)):
if board[i][j] == " ":
empty_slotes.append((i, j))
return random.choice(empty_slotes)
def Agent(Currentboard, n, AI):
game = Tic_Tac_Toe()
game.set_players(AI, n)
board = np.array(Currentboard)
free = game.filled_space(board.reshape(-1))
if len(set(board.reshape(-1))) == 1:
board[0][0] = AI
return board
elif free < 4 and n == 4:
move = game.rand_move(board)
board[move] = AI
return board
elif free < 7 and n == 5:
move = game.rand_move(board)
board[move] = AI
return board
elif free < 9 and n == 6:
move = game.rand_move(board)
board[move] = AI
return board
move = None
score = -np.inf
for i in range(n):
for j in range(n):
if board[i][j] == " ":
board[i][j] = AI
bestscore = game.alpha_beta(board.reshape(-1), game.get_human())
board[i][j] = " "
if bestscore > score:
score = bestscore
move = (i, j)
board[move] = AI
return board
def end(board):
for i in range(len(board)):
for j in range(len(board)):
if board[i][j] == " ":
return True
return False
no = int(input("Enter Value of n to make nxn Board : "))
game1 = Tic_Tac_Toe()
board = np.array([[" "] * no] * no)
print(board)
print(" ")
print("Enter Value from 1 to ", no * no)
print(" ")
for i in range(9):
if end(board):
check = game1.checker(board.reshape(-1))
if check == "X":
print("X won")
break
elif check == "O":
print("O won")
break
player1 = input("Enter position : ")
player1 = int(player1)
board = board.reshape(-1)
board[player1 - 1] = "O"
board = board.reshape(no, no)
print("Human : ")
print(board)
print("")
if end(board):
check = game1.checker(board.reshape(-1))
if check == "X":
print("X won")
break
elif check == "O":
print("O won")
break
board = Agent(board, no, "X")
print("Agent: ")
print(board)
print("")
| false | 0 | 1,457 | 1 | 1,457 | 1,457 |
||
69492407
|
# # MLB Player Digital Engagement Forecasting
# - In this notebook, we have used features from playersBoxScores, teamBoxScores, transactions, standings and awards from train dataset.
# - Simple NN model was used for making predictions on MLB player digital engagement scores.
# ## Data Preprocessing I : Train.csv
# - We have separated each features in the train set and used different approaches on each features - for example, filling missing values or changing monthly data into daily view using interpolation
# Data Preprocessing
import pandas as pd
import numpy as np
import json
import os
import time
from datetime import datetime, timedelta
# Modeling
import tensorflow as tf
from tensorflow import keras
import matplotlib.pyplot as plt
import random
from tensorflow.keras import models
from tensorflow.keras import layers
# - To reduce time for the data processing, we will leave the code how we read and processed train_updated.csv, however the entire process will be done by reading csv files which were separated before creating this notebook.
# fileDir = "../input/mlb-player-digital-engagement-forecasting/"
# train_path = os.path.join(fileDir, "train_updated.csv")
# train = pd.read_csv(train_path)
# def dict_to_df(df):
# result = pd.DataFrame()
# for i in range(len(df)):
# temp_dict = df[i]
# # some records have NaN
# if type(temp_dict) == float:
# pass
# else:
# temp_df = pd.DataFrame.from_dict(json.loads(temp_dict))
# result = result.append(temp_df, ignore_index = True)
# return(result)
# start = time.time()
# rosters = dict_to_df(train["rosters"])
# games = dict_to_df(train["games"])
# playerBoxScores = dict_to_df(train["playerBoxScores"])
# teamBoxScores = dict_to_df(train["teamBoxScores"])
# transactions = dict_to_df(train["transactions"])
# standings = dict_to_df(train["standings"])
# awards = dict_to_df(train["awards"])
# events = dict_to_df(train["events"])
# playerTwitterFollowers = dict_to_df(train["playerTwitterFollowers"])
# teamTwitterFollowers = dict_to_df(train["teamTwitterFollowers"])
# target = dict_to_df(train["nextDayPlayerEngagement"])
# print(time.time() - start, "seconds")
# ## Data Preprocessing II : Merge csv files
# - For those datasets not included in train.csv, we have loaded each data separately and took another steps of data preprocessing.
# - For players.csv, we filtered playerIds which are chosen for the test set.
# - For seasons.csv, we labeled each date for different types (Preseason, Regular season, etc.) to indicate different event types.
# - For awards.csv, we mutated data to use the information of number of awards received by each players.
fileDir = "../input/mlb-player-digital-engagement-forecasting/"
dfs = {}
for fileName in ["players.csv", "seasons.csv", "teams.csv"]:
key = fileName.split(".")[0]
idx = pd.read_csv(fileDir + fileName)
dfs[key] = idx
# ### Players
players = dfs["players"]
playersForTest = players[players["playerForTestSetAndFuturePreds"] == True]
print("Players for test set:", len(playersForTest))
# ### Seasons
seasons = dfs["seasons"]
seasons_df = pd.DataFrame()
for i in range(len(seasons)):
seasonId = seasons["seasonId"][i]
yearStart = str(seasonId) + "-01-01"
yearEnd = str(seasonId + 1) + "-01-01"
yearDates = pd.date_range(yearStart, yearEnd).date
df = pd.DataFrame(
{
"date": yearDates,
"seasonIdx": np.full_like(len(yearDates), np.nan, dtype=np.double()),
}
)
for index in df.index:
df.at[index, "date"] = str(df.at[index, "date"])
preSeasonStartDate = seasons["preSeasonStartDate"][i]
preSeasonEndDate = seasons["preSeasonEndDate"][i]
regularSeasonStartDate = seasons["regularSeasonStartDate"][i]
regularSeasonEndDate = seasons["regularSeasonEndDate"][i]
allStarDate = seasons["allStarDate"][i]
postSeasonStartDate = seasons["postSeasonStartDate"][i]
postSeasonEndDate = seasons["postSeasonEndDate"][i]
df[df["date"] == preSeasonStartDate] = df[df["date"] == preSeasonStartDate].fillna(
1
)
df[df["date"] == preSeasonEndDate] = df[df["date"] == preSeasonEndDate].fillna(1)
df[df["date"] == regularSeasonStartDate] = df[
df["date"] == regularSeasonStartDate
].fillna(2)
df[df["date"] == regularSeasonEndDate] = df[
df["date"] == regularSeasonEndDate
].fillna(2)
df[df["date"] == postSeasonStartDate] = df[
df["date"] == postSeasonStartDate
].fillna(4)
df[df["date"] == postSeasonEndDate] = df[df["date"] == postSeasonEndDate].fillna(4)
df = df.fillna(method="ffill")
df[df["date"] == allStarDate] = 3
df = df.fillna(0)
seasons_df = seasons_df.append(df, ignore_index=True)
seasons_df.loc[seasons_df["seasonIdx"] > 0]
# deleting dfs and train to save RAM memories.
# del dfs
# del train
del seasons
# ### Awards
def date_playerId(df, date, playerId="playerId"):
df["date_playerId"] = (
df[date].astype(str).str.replace("-", "")
+ "_"
+ df[playerId].astype(str).str.replace("\.0", "", regex=True)
)
return df
fileDir = "../input/mlbcsv"
awards = pd.read_csv(fileDir + "/awards.csv")
awards = date_playerId(awards, "awardDate")[["date_playerId", "playerId", "awardId"]]
awards["isAwarded"] = np.ones(len(awards))
awards["cumulativeAwardScore"] = awards.groupby("playerId")["isAwarded"].transform(
pd.Series.cumsum
)
awards = awards.drop(columns=["awardId"])
awards.head()
# - For the rest of the features in train.csv, we have dropped unnecessary columns and created primary key date_playerId for further uses
# ### Transactions
fileDir = "../input/mlbcsv"
transactions = pd.read_csv(fileDir + "/transactions.csv")
transactions = transactions.dropna(subset=["playerId"]).reset_index(drop=True)
transactions = date_playerId(transactions, "date")[
["date_playerId", "fromTeamId", "toTeamId", "effectiveDate", "typeCode"]
]
transactions.head()
# ### Standings
fileDir = "../input/mlbcsv"
standings = pd.read_csv(fileDir + "/standings.csv")
standings = standings[
[
"season",
"gameDate",
"teamId",
"streakCode",
"divisionRank",
"leagueRank",
"wins",
"losses",
"pct",
"divisionChamp",
"divisionLeader",
]
]
standings.head()
divisionRank_byDates = {}
for index in standings.index:
gamedate = (
standings.at[index, "gameDate"][:4]
+ standings.at[index, "gameDate"][5:7]
+ standings.at[index, "gameDate"][8:10]
)
teamid = standings.at[index, "teamId"]
streakCode = standings.at[index, "streakCode"]
divisionRank = standings.at[index, "divisionRank"]
leagueRank = standings.at[index, "leagueRank"]
pct = standings.at[index, "pct"]
divisionChamp = standings.at[index, "divisionChamp"]
divisionLeader = standings.at[index, "divisionLeader"]
if teamid not in divisionRank_byDates.keys():
divisionRank_byDates[teamid] = {
gamedate: [
streakCode,
divisionRank,
leagueRank,
pct,
divisionChamp,
divisionLeader,
]
}
else:
divisionRank_byDates[teamid][gamedate] = [
streakCode,
divisionRank,
leagueRank,
pct,
divisionChamp,
divisionLeader,
]
# ### Team Twitter Followers
fileDir = "../input/mlbcsv"
teamTwitterFollowers = pd.read_csv(fileDir + "/teamTwitterFollowers.csv")
teamTwitterFollowers = teamTwitterFollowers[["date", "teamId", "numberOfFollowers"]]
teamTwitterFollowers = teamTwitterFollowers.rename(
columns={"numberOfFollowers": "teamFollowers"}
)
teamTwitterFollowers["date"] = pd.to_datetime(teamTwitterFollowers["date"])
teamTwitter = pd.DataFrame()
minDate = min(teamTwitterFollowers["date"])
maxDate = max(teamTwitterFollowers["date"]) + timedelta(days=30)
for teamId in teamTwitterFollowers["teamId"].unique():
df = teamTwitterFollowers[teamTwitterFollowers["teamId"] == teamId]
df = df.set_index(df["date"]).drop(columns=["date"])
idx = pd.date_range(minDate, maxDate)
df = df.reindex(idx, fill_value=np.nan)
df["teamId"] = df["teamId"].interpolate(method="pad")
df["teamFollowers"] = df["teamFollowers"].interpolate(
method="linear", limit_direction="forward"
)
teamTwitter = teamTwitter.append(df)
del teamTwitterFollowers
teamTwitter = teamTwitter.reset_index()
teamTwitter = teamTwitter.rename(columns={"index": "date"})
# converting timestamp to string (up to YYYY-MM-DD)
teamTwitter["date"] = teamTwitter["date"].astype(str)
teamTwitter.head()
# ### Player Twitter Followers
fileDir = "../input/mlbcsv"
playerTwitterFollowers = pd.read_csv(fileDir + "/playerTwitterFollowers.csv")
playerTwitterFollowers = playerTwitterFollowers[
["date", "playerId", "numberOfFollowers"]
]
playerTwitterFollowers["date"] = pd.to_datetime(playerTwitterFollowers["date"])
playerTwitter = pd.DataFrame()
minDate = min(playerTwitterFollowers["date"])
maxDate = max(playerTwitterFollowers["date"]) + timedelta(days=30)
for playerId in playerTwitterFollowers["playerId"].unique():
df = playerTwitterFollowers[playerTwitterFollowers["playerId"] == playerId]
df = df.set_index(df["date"]).drop(columns=["date"])
idx = pd.date_range(minDate, maxDate)
df = df.reindex(idx, fill_value=np.nan)
df["playerId"] = df["playerId"].interpolate(method="pad")
df["numberOfFollowers"] = df["numberOfFollowers"].interpolate(
method="linear", limit_direction="forward"
)
playerTwitter = playerTwitter.append(df)
del playerTwitterFollowers
playerTwitter.head()
# ### Rosters
fileDir = "../input/mlbcsv"
rosters = pd.read_csv(fileDir + "/rosters.csv").iloc[:, 1:]
rosters = date_playerId(rosters, "gameDate").drop(columns=["status", "gameDate"])
rosters.head()
# ### Box Scores
# - For features in player box scores, we have separated features in to 3 different types: batting, pitching and defense.
# - For each features, we have applied PCA to reduce the dimensions which will be used in later modeling.
fileDir = "../input/mlbcsv"
playerBoxScores = pd.read_csv(fileDir + "/playerBoxScores.csv").iloc[:, 1:]
playerBoxScores.columns
battingStats = [
"flyOuts",
"groundOuts",
"runsScored",
"doubles",
"triples",
"homeRuns",
"strikeOuts",
"baseOnBalls",
"intentionalWalks",
"hits",
"hitByPitch",
"atBats",
"caughtStealing",
"stolenBases",
"groundIntoDoublePlay",
"groundIntoTriplePlay",
"plateAppearances",
"totalBases",
"rbi",
"leftOnBase",
"sacBunts",
"sacFlies",
"catchersInterference",
"pickoffs",
]
pitchingStats = [
"flyOutsPitching",
"airOutsPitching",
"groundOutsPitching",
"runsPitching",
"doublesPitching",
"triplesPitching",
"homeRunsPitching",
"strikeOutsPitching",
"baseOnBallsPitching",
"intentionalWalksPitching",
"hitsPitching",
"hitByPitchPitching",
"atBatsPitching",
"caughtStealingPitching",
"stolenBasesPitching",
"inningsPitched",
"saveOpportunities",
"earnedRuns",
"battersFaced",
"outsPitching",
"pitchesThrown",
"balls",
"strikes",
"hitBatsmen",
"balks",
"wildPitches",
"pickoffsPitching",
"rbiPitching",
"gamesFinishedPitching",
"inheritedRunners",
"inheritedRunnersScored",
"catchersInterferencePitching",
"sacBuntsPitching",
"sacFliesPitching",
"saves",
"holds",
"blownSaves",
]
defStats = ["assists", "putOuts", "errors", "chances"]
def high_corr_cols(df, threshold=0.7):
for col in range(len(df)):
for row in range(col + 1, len(df)):
if 0.7 <= df.iloc[row, col] < 1:
print(df.index[row], df.columns[col], df.iloc[row, col])
battingCorr = playerBoxScores[battingStats].corr().abs()
high_corr_cols(battingCorr)
# drop totalBases, plateAppearances
pitchingCorr = playerBoxScores[pitchingStats].corr().abs()
high_corr_cols(pitchingCorr)
# drop atBatsPitching, airOutsPitching, pitchesThrown, inningsPitched, outsPitching, battersFaced
defCorr = playerBoxScores[defStats].corr().abs()
high_corr_cols(defCorr)
# drop chances
playerBoxScores = playerBoxScores.drop(
columns=[
"gameTimeUTC",
"teamName",
"playerName",
"jerseyNum",
"positionName",
"positionType",
"totalBases",
"plateAppearances",
"atBatsPitching",
"airOutsPitching",
"pitchesThrown",
"inningsPitched",
"outsPitching",
"battersFaced",
"chances",
]
)
playerBoxScores.tail()
playerBoxScores = date_playerId(playerBoxScores, "gameDate")
playerBoxScores.head()
playerBoxScores.columns
battingFeat = [
"flyOuts",
"groundOuts",
"runsScored",
"doubles",
"triples",
"homeRuns",
"strikeOuts",
"baseOnBalls",
"intentionalWalks",
"hits",
"hitByPitch",
"atBats",
"caughtStealing",
"stolenBases",
"groundIntoDoublePlay",
"groundIntoTriplePlay",
"rbi",
"leftOnBase",
"sacBunts",
"sacFlies",
"catchersInterference",
"pickoffs",
]
batPCA = playerBoxScores[battingFeat].fillna(0)
from sklearn.decomposition import PCA
import matplotlib.pyplot as plt
n_comp, var_ratio = [], []
for i in range(1, 21):
pca = PCA(n_components=i, random_state=1)
pca.fit(batPCA)
n_comp.append(i)
var_ratio.append(pca.explained_variance_ratio_.sum())
plt.plot(n_comp, var_ratio)
plt.show()
# n = 7
bat_pca = PCA(n_components=7, random_state=1)
bat_pca.fit(playerBoxScores[battingFeat].fillna(0))
batPCAdf = pd.concat(
[
playerBoxScores[playerBoxScores["gamesPlayedBatting"] == 1]["date_playerId"],
playerBoxScores[playerBoxScores["gamesPlayedBatting"] == 1][battingFeat],
],
axis=1,
)
batPCAdf = batPCAdf.fillna(0).reset_index(drop=True)
bat_df = pd.concat(
[batPCAdf.iloc[:, 0], pd.DataFrame(bat_pca.fit_transform(batPCAdf.iloc[:, 1:]))],
axis=1,
)
bat_df = bat_df.rename(
columns={
0: "bat0",
1: "bat1",
2: "bat2",
3: "bat3",
4: "bat4",
5: "bat5",
6: "bat6",
}
)
bat_df
pitchingFeat = [
"gamesStartedPitching",
"completeGamesPitching",
"shutoutsPitching",
"winsPitching",
"lossesPitching",
"flyOutsPitching",
"groundOutsPitching",
"runsPitching",
"doublesPitching",
"triplesPitching",
"homeRunsPitching",
"strikeOutsPitching",
"baseOnBallsPitching",
"intentionalWalksPitching",
"hitsPitching",
"hitByPitchPitching",
"caughtStealingPitching",
"stolenBasesPitching",
"saveOpportunities",
"earnedRuns",
"balls",
"strikes",
"hitBatsmen",
"balks",
"wildPitches",
"pickoffsPitching",
"rbiPitching",
"gamesFinishedPitching",
"inheritedRunners",
"inheritedRunnersScored",
"catchersInterferencePitching",
"sacBuntsPitching",
"sacFliesPitching",
"saves",
"holds",
"blownSaves",
]
pitchPCA = playerBoxScores[pitchingFeat].fillna(0)
n_comp, var_ratio = [], []
for i in range(1, 21):
pca = PCA(n_components=i, random_state=1)
pca.fit(pitchPCA)
n_comp.append(i)
var_ratio.append(pca.explained_variance_ratio_.sum())
plt.plot(n_comp, var_ratio)
# plt.show()
# n = 4
pitch_pca = PCA(n_components=4, random_state=1)
pitch_pca.fit(playerBoxScores[pitchingFeat].fillna(0))
pitchPCAdf = pd.concat(
[
playerBoxScores[playerBoxScores["gamesPlayedPitching"] == 1]["date_playerId"],
playerBoxScores[playerBoxScores["gamesPlayedPitching"] == 1][pitchingFeat],
],
axis=1,
)
pitchPCAdf = pitchPCAdf.fillna(0).reset_index(drop=True)
pitch_df = pd.concat(
[
pitchPCAdf.iloc[:, 0],
pd.DataFrame(pitch_pca.fit_transform(pitchPCAdf.iloc[:, 1:])),
],
axis=1,
)
pitch_df = pitch_df.rename(columns={0: "pitch0", 1: "pitch1", 2: "pitch2", 3: "pitch4"})
pitch_df
# ### Games
# - For games, we dropped highly correlated columns and leave those necessary to include game results into the model.
fileDir = "../input/mlbcsv"
games = pd.read_csv(fileDir + "/games.csv").iloc[:, 1:]
games.head()
games.columns
games_corr = (
games[[col for col in games.columns if "home" in col or "away" in col]].corr().abs()
)
high_corr_cols(games_corr)
# drop all wins/losses and use pct instead
games_df = games[
[
"gameDate",
"homeId",
"homeWinPct",
"homeWinner",
"homeScore",
"awayId",
"awayWinPct",
"awayWinner",
"awayScore",
]
]
del games
games_df.head()
# ## Standings
fileDir = "../input/mlbcsv"
standings = pd.read_csv(fileDir + "/standings.csv").iloc[:, 1:]
standings = standings[
[
"season",
"gameDate",
"teamId",
"streakCode",
"divisionRank",
"leagueRank",
"wins",
"losses",
"pct",
"divisionChamp",
"divisionLeader",
]
]
standings.head()
divisionRank_byDates = {}
for index in standings.index:
gamedate = (
standings.at[index, "gameDate"][:4]
+ standings.at[index, "gameDate"][5:7]
+ standings.at[index, "gameDate"][8:10]
)
teamid = standings.at[index, "teamId"]
streakCode = standings.at[index, "streakCode"]
divisionRank = standings.at[index, "divisionRank"]
leagueRank = standings.at[index, "leagueRank"]
pct = standings.at[index, "pct"]
divisionChamp = standings.at[index, "divisionChamp"]
divisionLeader = standings.at[index, "divisionLeader"]
if teamid not in divisionRank_byDates.keys():
divisionRank_byDates[teamid] = {
gamedate: [
streakCode,
divisionRank,
leagueRank,
pct,
divisionChamp,
divisionLeader,
]
}
else:
divisionRank_byDates[teamid][gamedate] = [
streakCode,
divisionRank,
leagueRank,
pct,
divisionChamp,
divisionLeader,
]
# ### Target
fileDir = "../input/mlbcsv"
target = pd.read_csv(fileDir + "/target.csv").iloc[:, 1:]
target.head()
target = date_playerId(target, "engagementMetricsDate")
target.head()
target_df = target[target["playerId"].isin(playersForTest["playerId"])]
del target
target_df.head()
targetShift = pd.DataFrame()
for playerId in target_df["playerId"].unique():
df = target_df[target_df["playerId"] == playerId]
df = df.set_index("engagementMetricsDate").shift(1).dropna()
targetShift = targetShift.append(df)
targetShift = targetShift.reset_index()
targetShift.head()
# ### Delete train and target_df
del target_df
# ### Merge All
# Merge Season
seasons_df = seasons_df.drop_duplicates()
targetShift = targetShift.merge(
seasons_df.drop_duplicates(),
how="left",
left_on="engagementMetricsDate",
right_on="date",
).drop(columns="date")
# Merge Awards
targetShift = (
targetShift.merge(awards, how="left", on="date_playerId")
.fillna(0)
.drop(columns=["playerId_y"])
)
# Merge playerTwitter
playerTwitter_df = date_playerId(playerTwitter.reset_index(), "index")
playerTwitter_df = playerTwitter_df.drop(columns=["index", "playerId"])
targetShift = targetShift.merge(playerTwitter_df, how="left", on="date_playerId")
for index in targetShift.index:
if pd.isna(targetShift.at[index, "numberOfFollowers"]):
targetShift.at[index, "numberOfFollowers"] = 0
# Merge boxScore
targetShift = targetShift.merge(pitch_df, how="left", on="date_playerId").fillna(0)
targetShift = targetShift.merge(bat_df, how="left", on="date_playerId").fillna(0)
targetShift = targetShift.merge(
playerBoxScores[["date_playerId", "home", "positionCode"]],
how="left",
on="date_playerId",
)
# Merge rosters
targetShift = targetShift.merge(
rosters, how="left", on="date_playerId"
) # .drop(columns = ["playerId_y"])
# standings
targetShift = targetShift.merge(
standings,
how="left",
left_on=["engagementMetricsDate", "teamId"],
right_on=["gameDate", "teamId"],
)
# Merge games
targetShift = targetShift.merge(
games_df,
how="left",
left_on=["gameDate", "teamId"],
right_on=["gameDate", "homeId"],
)
targetShift.head()
# Merge teamTwitterFollowers
targetShift = targetShift.merge(
teamTwitter, how="left", left_on=["gameDate", "teamId"], right_on=["date", "teamId"]
)
for index in targetShift.index:
if pd.isna(targetShift.at[index, "teamFollowers"]):
targetShift.at[index, "teamFollowers"] = 0.0
targetShift.head()
for index in targetShift.index:
if pd.isna(targetShift.at[index, "teamId"]):
targetShift.at[index, "teamId"] = 0
for index in targetShift.index:
teamId = targetShift.at[index, "teamId"]
divisionRank = targetShift.at[index, "divisionRank"]
leagueRank = targetShift.at[index, "leagueRank"]
pct = targetShift.at[index, "pct"]
date = targetShift.at[index, "date_playerId"][:8]
if pd.isna(divisionRank):
targetShift.at[index, "divisionRank"] = 6
if pd.isna(leagueRank):
targetShift.at[index, "leagueRank"] = 16
if pd.isna(leagueRank):
targetShift.at[index, "pct"] = -1
if teamId in divisionRank_byDates.keys():
if date in divisionRank_byDates[teamId].keys():
targetShift.at[index, "streakCode"] = divisionRank_byDates[teamId][date][0]
targetShift.at[index, "divisionRank"] = int(
divisionRank_byDates[teamId][date][1]
)
targetShift.at[index, "leagueRank"] = int(
divisionRank_byDates[teamId][date][2]
)
targetShift.at[index, "pct"] = float(divisionRank_byDates[teamId][date][3])
targetShift.head()
currentPlayer = targetShift.at[0, "playerId"]
currentdivisionRank = 6.0
currentleagueRank = 16.0
currentpct = -1.0
for index in targetShift.index:
playerid = targetShift.at[index, "playerId"]
# date = targetShift.at[index,"date_playerId"][:8]
divisionRank = float(targetShift.at[index, "divisionRank"])
leagueRank = float(targetShift.at[index, "leagueRank"])
pct = float(targetShift.at[index, "pct"])
# teamId = targetShift.at[index,"teamId"]
if currentPlayer == playerid:
if currentdivisionRank != divisionRank:
# when standing update happens
if divisionRank < 6:
currentdivisionRank = divisionRank
targetShift.at[index, "divisionRank"] = currentdivisionRank
# updating all 6 after / when divisionRank = 6
elif currentdivisionRank < 6:
targetShift.at[index, "divisionRank"] = currentdivisionRank
if currentleagueRank != leagueRank:
# when standing update happens
if leagueRank < 16:
currentleagueRank = leagueRank
targetShift.at[index, "leagueRank"] = currentleagueRank
# updating all 16 after / when divisionRank = 16
elif currentleagueRank < 16:
targetShift.at[index, "leagueRank"] = currentleagueRank
if currentpct != -1:
# when standing update happens
if pct > -1:
currentpct = pct
targetShift.at[index, "pct"] = currentpct
# updating all 16 after / when divisionRank = 16
elif currentpct > -1:
targetShift.at[index, "pct"] = currentpct
else:
currentPlayer = playerid
currentdivisionRank = 6
currentleagueRank = 16
currentpct = -1
for index in targetShift.index:
# if record is empty for streakCode, then replace them to 0
if pd.isna(targetShift.at[index, "streakCode"]):
targetShift.at[index, "streakCode"] = 0.0
else:
if "W" in targetShift.at[index, "streakCode"]:
targetShift.at[index, "streakCode"] = float(
targetShift.at[index, "streakCode"][1]
)
elif "L" in targetShift.at[index, "streakCode"]:
targetShift.at[index, "streakCode"] = -(
float(targetShift.at[index, "streakCode"][1])
)
targetShift = targetShift[
[
"engagementMetricsDate",
"date_playerId",
"teamId",
"seasonIdx",
"isAwarded",
"cumulativeAwardScore",
"numberOfFollowers",
"teamFollowers",
"pitch0",
"pitch1",
"pitch2",
"pitch4",
"bat0",
"bat1",
"bat2",
"bat3",
"bat4",
"bat5",
"bat6",
"streakCode",
"divisionRank",
"leagueRank",
"wins",
"losses",
"pct",
"divisionChamp",
"divisionLeader",
"homeWinPct",
"homeWinner",
"homeScore",
"awayWinPct",
"awayScore",
"target1",
"target2",
"target3",
"target4",
]
] #'home','awayWinner'
targetShift.head()
for index in targetShift.index:
homeWinPct = targetShift.at[index, "homeWinPct"]
homeWinner = targetShift.at[index, "homeWinner"]
homeScore = targetShift.at[index, "homeScore"]
awayWinPct = targetShift.at[index, "awayWinPct"]
awayScore = targetShift.at[index, "awayScore"]
seasonIdx = targetShift.at[index, "seasonIdx"]
streakCode = targetShift.at[index, "streakCode"]
divisionRank = targetShift.at[index, "divisionRank"]
leagueRank = targetShift.at[index, "leagueRank"]
wins = targetShift.at[index, "wins"]
losses = targetShift.at[index, "losses"]
pct = targetShift.at[index, "pct"]
divisionChamp = targetShift.at[index, "divisionChamp"]
divisionLeader = targetShift.at[index, "divisionLeader"]
# off-season
# if seasonIdx == 0:
if pd.isna(homeWinPct):
targetShift.at[index, "homeWinPct"] = -1.0
if pd.isna(homeWinner):
targetShift.at[index, "homeWinner"] = -1.0
elif homeWinner == False:
targetShift.at[index, "homeWinner"] = 0.0
elif homeWinner == True:
targetShift.at[index, "homeWinner"] = 1.0
if pd.isna(homeScore):
targetShift.at[index, "homeScore"] = -1.0
if pd.isna(awayWinPct):
targetShift.at[index, "awayWinPct"] = -1.0
if pd.isna(awayScore):
targetShift.at[index, "awayScore"] = -1.0
if pd.isna(streakCode):
targetShift.at[index, "streakCode"] = 0.0
if pd.isna(divisionRank):
targetShift.at[index, "divisionRank"] = 6.0
if pd.isna(leagueRank):
targetShift.at[index, "leagueRank"] = 16.0
if pd.isna(wins):
targetShift.at[index, "wins"] = -1.0
if pd.isna(losses):
targetShift.at[index, "losses"] = -1.0
if pd.isna(pct):
targetShift.at[index, "pct"] = -1.0
if divisionChamp == True:
targetShift.at[index, "divisionChamp"] = 1.0
else:
targetShift.at[index, "divisionChamp"] = 0.0
if divisionLeader == True:
targetShift.at[index, "divisionLeader"] = 1.0
else:
targetShift.at[index, "divisionLeader"] = 0.0
targetShift
# targetShift.to_csv("ANN_7_July30th.csv", index=False)
# ## Modeling
# - We trained simple NN model on the sampled dataset from the train dataset created above.
maxplayerFollowers = targetShift["numberOfFollowers"].max()
maxteamFollowers = targetShift["teamFollowers"].max()
print(maxplayerFollowers, maxteamFollowers)
targetShift["numberOfFollowers"] = (
targetShift["numberOfFollowers"] / maxplayerFollowers
) * 100
targetShift["teamFollowers"] = (targetShift["teamFollowers"] / maxteamFollowers) * 100
targetShift.rename(
{
"numberOfFollowers": "norm_playerFollowers",
"teamFollowers": "norm_teamFollowers",
},
axis=1,
inplace=True,
)
targetShift.head()
# ### ANN approach
train = targetShift[targetShift["engagementMetricsDate"] < "2021-04-25"]
test = targetShift[targetShift["engagementMetricsDate"] >= "2021-04-25"]
print(len(train), len(test))
del targetShift
n_sample = 500000
train_sample = train.sample(n=n_sample)
X_train = train_sample[
[
"isAwarded",
"cumulativeAwardScore",
"norm_playerFollowers",
"norm_teamFollowers",
"pitch0",
"pitch1",
"pitch2",
"pitch4",
"bat0",
"bat1",
"bat2",
"bat3",
"bat4",
"bat5",
"bat6",
"streakCode",
"divisionRank",
"leagueRank",
"wins",
"losses",
"pct",
"divisionChamp",
"divisionLeader",
"homeWinPct",
"homeWinner",
"homeScore",
"awayWinPct",
"awayScore",
]
].fillna(0)
Y_train = train_sample[["target1", "target2", "target3", "target4"]]
print("Train Data Shape : {}".format(X_train.shape))
print("Train Label Shape : {}".format(Y_train.shape))
feature_size = X_train.shape[1]
def build_network(input_shape=(feature_size,)):
model = models.Sequential()
model.add(layers.Dense(28, activation="relu", input_shape=input_shape))
model.add(layers.Dropout(0.2))
model.add(layers.Dense(28, activation="relu"))
model.add(layers.Dropout(0.2))
model.add(layers.Dense(28, activation="relu"))
model.add(layers.Dropout(0.2))
model.add(layers.Dense(14, activation="relu"))
model.add(layers.Dropout(0.2))
model.add(layers.Dense(1))
model.compile(optimizer="adam", loss="mse", metrics=["mae"])
return model
model1 = build_network((feature_size,))
model2 = build_network((feature_size,))
model3 = build_network((feature_size,))
model4 = build_network((feature_size,))
model1.summary()
model1.fit(X_train.values, Y_train.iloc[:, 0].values, epochs=20)
model2.fit(X_train.values, Y_train.iloc[:, 1].values, epochs=20)
model3.fit(X_train.values, Y_train.iloc[:, 2].values, epochs=20)
model4.fit(X_train.values, Y_train.iloc[:, 3].values, epochs=20)
# ### Predictions
# - From the models and test set created above, we will make a prediction and save the result into dataframe.
test_id = test["date_playerId"]
X_test = test[
[
"isAwarded",
"cumulativeAwardScore",
"norm_playerFollowers",
"norm_teamFollowers",
"pitch0",
"pitch1",
"pitch2",
"pitch4",
"bat0",
"bat1",
"bat2",
"bat3",
"bat4",
"bat5",
"bat6",
"streakCode",
"divisionRank",
"leagueRank",
"wins",
"losses",
"pct",
"divisionChamp",
"divisionLeader",
"homeWinPct",
"homeWinner",
"homeScore",
"awayWinPct",
"awayScore",
]
].fillna(0)
Y_pred1 = model1.predict(X_test.values)
Y_pred2 = model2.predict(X_test.values)
Y_pred3 = model3.predict(X_test.values)
Y_pred4 = model4.predict(X_test.values)
prediction_df = pd.DataFrame()
prediction_df["date_playerId"] = test_id
prediction_df["target1"] = Y_pred1
prediction_df["target2"] = Y_pred2
prediction_df["target3"] = Y_pred3
prediction_df["target4"] = Y_pred4
prediction_df = prediction_df.reset_index(drop=True)
prediction_df.head()
import mlb
env = mlb.make_env() # initialize the environment
iter_test = env.iter_test() # iterator which loops over each date in test set
for test_df, prediction_df in iter_test:
env.predict(prediction_df)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/492/69492407.ipynb
| null | null |
[{"Id": 69492407, "ScriptId": 18970201, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 804214, "CreationDate": "07/31/2021 16:59:39", "VersionNumber": 2.0, "Title": "MLB Player Digital Eng. Forecasting - Simple ANN", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 786.0, "LinesInsertedFromPrevious": 37.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 749.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # MLB Player Digital Engagement Forecasting
# - In this notebook, we have used features from playersBoxScores, teamBoxScores, transactions, standings and awards from train dataset.
# - Simple NN model was used for making predictions on MLB player digital engagement scores.
# ## Data Preprocessing I : Train.csv
# - We have separated each features in the train set and used different approaches on each features - for example, filling missing values or changing monthly data into daily view using interpolation
# Data Preprocessing
import pandas as pd
import numpy as np
import json
import os
import time
from datetime import datetime, timedelta
# Modeling
import tensorflow as tf
from tensorflow import keras
import matplotlib.pyplot as plt
import random
from tensorflow.keras import models
from tensorflow.keras import layers
# - To reduce time for the data processing, we will leave the code how we read and processed train_updated.csv, however the entire process will be done by reading csv files which were separated before creating this notebook.
# fileDir = "../input/mlb-player-digital-engagement-forecasting/"
# train_path = os.path.join(fileDir, "train_updated.csv")
# train = pd.read_csv(train_path)
# def dict_to_df(df):
# result = pd.DataFrame()
# for i in range(len(df)):
# temp_dict = df[i]
# # some records have NaN
# if type(temp_dict) == float:
# pass
# else:
# temp_df = pd.DataFrame.from_dict(json.loads(temp_dict))
# result = result.append(temp_df, ignore_index = True)
# return(result)
# start = time.time()
# rosters = dict_to_df(train["rosters"])
# games = dict_to_df(train["games"])
# playerBoxScores = dict_to_df(train["playerBoxScores"])
# teamBoxScores = dict_to_df(train["teamBoxScores"])
# transactions = dict_to_df(train["transactions"])
# standings = dict_to_df(train["standings"])
# awards = dict_to_df(train["awards"])
# events = dict_to_df(train["events"])
# playerTwitterFollowers = dict_to_df(train["playerTwitterFollowers"])
# teamTwitterFollowers = dict_to_df(train["teamTwitterFollowers"])
# target = dict_to_df(train["nextDayPlayerEngagement"])
# print(time.time() - start, "seconds")
# ## Data Preprocessing II : Merge csv files
# - For those datasets not included in train.csv, we have loaded each data separately and took another steps of data preprocessing.
# - For players.csv, we filtered playerIds which are chosen for the test set.
# - For seasons.csv, we labeled each date for different types (Preseason, Regular season, etc.) to indicate different event types.
# - For awards.csv, we mutated data to use the information of number of awards received by each players.
fileDir = "../input/mlb-player-digital-engagement-forecasting/"
dfs = {}
for fileName in ["players.csv", "seasons.csv", "teams.csv"]:
key = fileName.split(".")[0]
idx = pd.read_csv(fileDir + fileName)
dfs[key] = idx
# ### Players
players = dfs["players"]
playersForTest = players[players["playerForTestSetAndFuturePreds"] == True]
print("Players for test set:", len(playersForTest))
# ### Seasons
seasons = dfs["seasons"]
seasons_df = pd.DataFrame()
for i in range(len(seasons)):
seasonId = seasons["seasonId"][i]
yearStart = str(seasonId) + "-01-01"
yearEnd = str(seasonId + 1) + "-01-01"
yearDates = pd.date_range(yearStart, yearEnd).date
df = pd.DataFrame(
{
"date": yearDates,
"seasonIdx": np.full_like(len(yearDates), np.nan, dtype=np.double()),
}
)
for index in df.index:
df.at[index, "date"] = str(df.at[index, "date"])
preSeasonStartDate = seasons["preSeasonStartDate"][i]
preSeasonEndDate = seasons["preSeasonEndDate"][i]
regularSeasonStartDate = seasons["regularSeasonStartDate"][i]
regularSeasonEndDate = seasons["regularSeasonEndDate"][i]
allStarDate = seasons["allStarDate"][i]
postSeasonStartDate = seasons["postSeasonStartDate"][i]
postSeasonEndDate = seasons["postSeasonEndDate"][i]
df[df["date"] == preSeasonStartDate] = df[df["date"] == preSeasonStartDate].fillna(
1
)
df[df["date"] == preSeasonEndDate] = df[df["date"] == preSeasonEndDate].fillna(1)
df[df["date"] == regularSeasonStartDate] = df[
df["date"] == regularSeasonStartDate
].fillna(2)
df[df["date"] == regularSeasonEndDate] = df[
df["date"] == regularSeasonEndDate
].fillna(2)
df[df["date"] == postSeasonStartDate] = df[
df["date"] == postSeasonStartDate
].fillna(4)
df[df["date"] == postSeasonEndDate] = df[df["date"] == postSeasonEndDate].fillna(4)
df = df.fillna(method="ffill")
df[df["date"] == allStarDate] = 3
df = df.fillna(0)
seasons_df = seasons_df.append(df, ignore_index=True)
seasons_df.loc[seasons_df["seasonIdx"] > 0]
# deleting dfs and train to save RAM memories.
# del dfs
# del train
del seasons
# ### Awards
def date_playerId(df, date, playerId="playerId"):
df["date_playerId"] = (
df[date].astype(str).str.replace("-", "")
+ "_"
+ df[playerId].astype(str).str.replace("\.0", "", regex=True)
)
return df
fileDir = "../input/mlbcsv"
awards = pd.read_csv(fileDir + "/awards.csv")
awards = date_playerId(awards, "awardDate")[["date_playerId", "playerId", "awardId"]]
awards["isAwarded"] = np.ones(len(awards))
awards["cumulativeAwardScore"] = awards.groupby("playerId")["isAwarded"].transform(
pd.Series.cumsum
)
awards = awards.drop(columns=["awardId"])
awards.head()
# - For the rest of the features in train.csv, we have dropped unnecessary columns and created primary key date_playerId for further uses
# ### Transactions
fileDir = "../input/mlbcsv"
transactions = pd.read_csv(fileDir + "/transactions.csv")
transactions = transactions.dropna(subset=["playerId"]).reset_index(drop=True)
transactions = date_playerId(transactions, "date")[
["date_playerId", "fromTeamId", "toTeamId", "effectiveDate", "typeCode"]
]
transactions.head()
# ### Standings
fileDir = "../input/mlbcsv"
standings = pd.read_csv(fileDir + "/standings.csv")
standings = standings[
[
"season",
"gameDate",
"teamId",
"streakCode",
"divisionRank",
"leagueRank",
"wins",
"losses",
"pct",
"divisionChamp",
"divisionLeader",
]
]
standings.head()
divisionRank_byDates = {}
for index in standings.index:
gamedate = (
standings.at[index, "gameDate"][:4]
+ standings.at[index, "gameDate"][5:7]
+ standings.at[index, "gameDate"][8:10]
)
teamid = standings.at[index, "teamId"]
streakCode = standings.at[index, "streakCode"]
divisionRank = standings.at[index, "divisionRank"]
leagueRank = standings.at[index, "leagueRank"]
pct = standings.at[index, "pct"]
divisionChamp = standings.at[index, "divisionChamp"]
divisionLeader = standings.at[index, "divisionLeader"]
if teamid not in divisionRank_byDates.keys():
divisionRank_byDates[teamid] = {
gamedate: [
streakCode,
divisionRank,
leagueRank,
pct,
divisionChamp,
divisionLeader,
]
}
else:
divisionRank_byDates[teamid][gamedate] = [
streakCode,
divisionRank,
leagueRank,
pct,
divisionChamp,
divisionLeader,
]
# ### Team Twitter Followers
fileDir = "../input/mlbcsv"
teamTwitterFollowers = pd.read_csv(fileDir + "/teamTwitterFollowers.csv")
teamTwitterFollowers = teamTwitterFollowers[["date", "teamId", "numberOfFollowers"]]
teamTwitterFollowers = teamTwitterFollowers.rename(
columns={"numberOfFollowers": "teamFollowers"}
)
teamTwitterFollowers["date"] = pd.to_datetime(teamTwitterFollowers["date"])
teamTwitter = pd.DataFrame()
minDate = min(teamTwitterFollowers["date"])
maxDate = max(teamTwitterFollowers["date"]) + timedelta(days=30)
for teamId in teamTwitterFollowers["teamId"].unique():
df = teamTwitterFollowers[teamTwitterFollowers["teamId"] == teamId]
df = df.set_index(df["date"]).drop(columns=["date"])
idx = pd.date_range(minDate, maxDate)
df = df.reindex(idx, fill_value=np.nan)
df["teamId"] = df["teamId"].interpolate(method="pad")
df["teamFollowers"] = df["teamFollowers"].interpolate(
method="linear", limit_direction="forward"
)
teamTwitter = teamTwitter.append(df)
del teamTwitterFollowers
teamTwitter = teamTwitter.reset_index()
teamTwitter = teamTwitter.rename(columns={"index": "date"})
# converting timestamp to string (up to YYYY-MM-DD)
teamTwitter["date"] = teamTwitter["date"].astype(str)
teamTwitter.head()
# ### Player Twitter Followers
fileDir = "../input/mlbcsv"
playerTwitterFollowers = pd.read_csv(fileDir + "/playerTwitterFollowers.csv")
playerTwitterFollowers = playerTwitterFollowers[
["date", "playerId", "numberOfFollowers"]
]
playerTwitterFollowers["date"] = pd.to_datetime(playerTwitterFollowers["date"])
playerTwitter = pd.DataFrame()
minDate = min(playerTwitterFollowers["date"])
maxDate = max(playerTwitterFollowers["date"]) + timedelta(days=30)
for playerId in playerTwitterFollowers["playerId"].unique():
df = playerTwitterFollowers[playerTwitterFollowers["playerId"] == playerId]
df = df.set_index(df["date"]).drop(columns=["date"])
idx = pd.date_range(minDate, maxDate)
df = df.reindex(idx, fill_value=np.nan)
df["playerId"] = df["playerId"].interpolate(method="pad")
df["numberOfFollowers"] = df["numberOfFollowers"].interpolate(
method="linear", limit_direction="forward"
)
playerTwitter = playerTwitter.append(df)
del playerTwitterFollowers
playerTwitter.head()
# ### Rosters
fileDir = "../input/mlbcsv"
rosters = pd.read_csv(fileDir + "/rosters.csv").iloc[:, 1:]
rosters = date_playerId(rosters, "gameDate").drop(columns=["status", "gameDate"])
rosters.head()
# ### Box Scores
# - For features in player box scores, we have separated features in to 3 different types: batting, pitching and defense.
# - For each features, we have applied PCA to reduce the dimensions which will be used in later modeling.
fileDir = "../input/mlbcsv"
playerBoxScores = pd.read_csv(fileDir + "/playerBoxScores.csv").iloc[:, 1:]
playerBoxScores.columns
battingStats = [
"flyOuts",
"groundOuts",
"runsScored",
"doubles",
"triples",
"homeRuns",
"strikeOuts",
"baseOnBalls",
"intentionalWalks",
"hits",
"hitByPitch",
"atBats",
"caughtStealing",
"stolenBases",
"groundIntoDoublePlay",
"groundIntoTriplePlay",
"plateAppearances",
"totalBases",
"rbi",
"leftOnBase",
"sacBunts",
"sacFlies",
"catchersInterference",
"pickoffs",
]
pitchingStats = [
"flyOutsPitching",
"airOutsPitching",
"groundOutsPitching",
"runsPitching",
"doublesPitching",
"triplesPitching",
"homeRunsPitching",
"strikeOutsPitching",
"baseOnBallsPitching",
"intentionalWalksPitching",
"hitsPitching",
"hitByPitchPitching",
"atBatsPitching",
"caughtStealingPitching",
"stolenBasesPitching",
"inningsPitched",
"saveOpportunities",
"earnedRuns",
"battersFaced",
"outsPitching",
"pitchesThrown",
"balls",
"strikes",
"hitBatsmen",
"balks",
"wildPitches",
"pickoffsPitching",
"rbiPitching",
"gamesFinishedPitching",
"inheritedRunners",
"inheritedRunnersScored",
"catchersInterferencePitching",
"sacBuntsPitching",
"sacFliesPitching",
"saves",
"holds",
"blownSaves",
]
defStats = ["assists", "putOuts", "errors", "chances"]
def high_corr_cols(df, threshold=0.7):
for col in range(len(df)):
for row in range(col + 1, len(df)):
if 0.7 <= df.iloc[row, col] < 1:
print(df.index[row], df.columns[col], df.iloc[row, col])
battingCorr = playerBoxScores[battingStats].corr().abs()
high_corr_cols(battingCorr)
# drop totalBases, plateAppearances
pitchingCorr = playerBoxScores[pitchingStats].corr().abs()
high_corr_cols(pitchingCorr)
# drop atBatsPitching, airOutsPitching, pitchesThrown, inningsPitched, outsPitching, battersFaced
defCorr = playerBoxScores[defStats].corr().abs()
high_corr_cols(defCorr)
# drop chances
playerBoxScores = playerBoxScores.drop(
columns=[
"gameTimeUTC",
"teamName",
"playerName",
"jerseyNum",
"positionName",
"positionType",
"totalBases",
"plateAppearances",
"atBatsPitching",
"airOutsPitching",
"pitchesThrown",
"inningsPitched",
"outsPitching",
"battersFaced",
"chances",
]
)
playerBoxScores.tail()
playerBoxScores = date_playerId(playerBoxScores, "gameDate")
playerBoxScores.head()
playerBoxScores.columns
battingFeat = [
"flyOuts",
"groundOuts",
"runsScored",
"doubles",
"triples",
"homeRuns",
"strikeOuts",
"baseOnBalls",
"intentionalWalks",
"hits",
"hitByPitch",
"atBats",
"caughtStealing",
"stolenBases",
"groundIntoDoublePlay",
"groundIntoTriplePlay",
"rbi",
"leftOnBase",
"sacBunts",
"sacFlies",
"catchersInterference",
"pickoffs",
]
batPCA = playerBoxScores[battingFeat].fillna(0)
from sklearn.decomposition import PCA
import matplotlib.pyplot as plt
n_comp, var_ratio = [], []
for i in range(1, 21):
pca = PCA(n_components=i, random_state=1)
pca.fit(batPCA)
n_comp.append(i)
var_ratio.append(pca.explained_variance_ratio_.sum())
plt.plot(n_comp, var_ratio)
plt.show()
# n = 7
bat_pca = PCA(n_components=7, random_state=1)
bat_pca.fit(playerBoxScores[battingFeat].fillna(0))
batPCAdf = pd.concat(
[
playerBoxScores[playerBoxScores["gamesPlayedBatting"] == 1]["date_playerId"],
playerBoxScores[playerBoxScores["gamesPlayedBatting"] == 1][battingFeat],
],
axis=1,
)
batPCAdf = batPCAdf.fillna(0).reset_index(drop=True)
bat_df = pd.concat(
[batPCAdf.iloc[:, 0], pd.DataFrame(bat_pca.fit_transform(batPCAdf.iloc[:, 1:]))],
axis=1,
)
bat_df = bat_df.rename(
columns={
0: "bat0",
1: "bat1",
2: "bat2",
3: "bat3",
4: "bat4",
5: "bat5",
6: "bat6",
}
)
bat_df
pitchingFeat = [
"gamesStartedPitching",
"completeGamesPitching",
"shutoutsPitching",
"winsPitching",
"lossesPitching",
"flyOutsPitching",
"groundOutsPitching",
"runsPitching",
"doublesPitching",
"triplesPitching",
"homeRunsPitching",
"strikeOutsPitching",
"baseOnBallsPitching",
"intentionalWalksPitching",
"hitsPitching",
"hitByPitchPitching",
"caughtStealingPitching",
"stolenBasesPitching",
"saveOpportunities",
"earnedRuns",
"balls",
"strikes",
"hitBatsmen",
"balks",
"wildPitches",
"pickoffsPitching",
"rbiPitching",
"gamesFinishedPitching",
"inheritedRunners",
"inheritedRunnersScored",
"catchersInterferencePitching",
"sacBuntsPitching",
"sacFliesPitching",
"saves",
"holds",
"blownSaves",
]
pitchPCA = playerBoxScores[pitchingFeat].fillna(0)
n_comp, var_ratio = [], []
for i in range(1, 21):
pca = PCA(n_components=i, random_state=1)
pca.fit(pitchPCA)
n_comp.append(i)
var_ratio.append(pca.explained_variance_ratio_.sum())
plt.plot(n_comp, var_ratio)
# plt.show()
# n = 4
pitch_pca = PCA(n_components=4, random_state=1)
pitch_pca.fit(playerBoxScores[pitchingFeat].fillna(0))
pitchPCAdf = pd.concat(
[
playerBoxScores[playerBoxScores["gamesPlayedPitching"] == 1]["date_playerId"],
playerBoxScores[playerBoxScores["gamesPlayedPitching"] == 1][pitchingFeat],
],
axis=1,
)
pitchPCAdf = pitchPCAdf.fillna(0).reset_index(drop=True)
pitch_df = pd.concat(
[
pitchPCAdf.iloc[:, 0],
pd.DataFrame(pitch_pca.fit_transform(pitchPCAdf.iloc[:, 1:])),
],
axis=1,
)
pitch_df = pitch_df.rename(columns={0: "pitch0", 1: "pitch1", 2: "pitch2", 3: "pitch4"})
pitch_df
# ### Games
# - For games, we dropped highly correlated columns and leave those necessary to include game results into the model.
fileDir = "../input/mlbcsv"
games = pd.read_csv(fileDir + "/games.csv").iloc[:, 1:]
games.head()
games.columns
games_corr = (
games[[col for col in games.columns if "home" in col or "away" in col]].corr().abs()
)
high_corr_cols(games_corr)
# drop all wins/losses and use pct instead
games_df = games[
[
"gameDate",
"homeId",
"homeWinPct",
"homeWinner",
"homeScore",
"awayId",
"awayWinPct",
"awayWinner",
"awayScore",
]
]
del games
games_df.head()
# ## Standings
fileDir = "../input/mlbcsv"
standings = pd.read_csv(fileDir + "/standings.csv").iloc[:, 1:]
standings = standings[
[
"season",
"gameDate",
"teamId",
"streakCode",
"divisionRank",
"leagueRank",
"wins",
"losses",
"pct",
"divisionChamp",
"divisionLeader",
]
]
standings.head()
divisionRank_byDates = {}
for index in standings.index:
gamedate = (
standings.at[index, "gameDate"][:4]
+ standings.at[index, "gameDate"][5:7]
+ standings.at[index, "gameDate"][8:10]
)
teamid = standings.at[index, "teamId"]
streakCode = standings.at[index, "streakCode"]
divisionRank = standings.at[index, "divisionRank"]
leagueRank = standings.at[index, "leagueRank"]
pct = standings.at[index, "pct"]
divisionChamp = standings.at[index, "divisionChamp"]
divisionLeader = standings.at[index, "divisionLeader"]
if teamid not in divisionRank_byDates.keys():
divisionRank_byDates[teamid] = {
gamedate: [
streakCode,
divisionRank,
leagueRank,
pct,
divisionChamp,
divisionLeader,
]
}
else:
divisionRank_byDates[teamid][gamedate] = [
streakCode,
divisionRank,
leagueRank,
pct,
divisionChamp,
divisionLeader,
]
# ### Target
fileDir = "../input/mlbcsv"
target = pd.read_csv(fileDir + "/target.csv").iloc[:, 1:]
target.head()
target = date_playerId(target, "engagementMetricsDate")
target.head()
target_df = target[target["playerId"].isin(playersForTest["playerId"])]
del target
target_df.head()
targetShift = pd.DataFrame()
for playerId in target_df["playerId"].unique():
df = target_df[target_df["playerId"] == playerId]
df = df.set_index("engagementMetricsDate").shift(1).dropna()
targetShift = targetShift.append(df)
targetShift = targetShift.reset_index()
targetShift.head()
# ### Delete train and target_df
del target_df
# ### Merge All
# Merge Season
seasons_df = seasons_df.drop_duplicates()
targetShift = targetShift.merge(
seasons_df.drop_duplicates(),
how="left",
left_on="engagementMetricsDate",
right_on="date",
).drop(columns="date")
# Merge Awards
targetShift = (
targetShift.merge(awards, how="left", on="date_playerId")
.fillna(0)
.drop(columns=["playerId_y"])
)
# Merge playerTwitter
playerTwitter_df = date_playerId(playerTwitter.reset_index(), "index")
playerTwitter_df = playerTwitter_df.drop(columns=["index", "playerId"])
targetShift = targetShift.merge(playerTwitter_df, how="left", on="date_playerId")
for index in targetShift.index:
if pd.isna(targetShift.at[index, "numberOfFollowers"]):
targetShift.at[index, "numberOfFollowers"] = 0
# Merge boxScore
targetShift = targetShift.merge(pitch_df, how="left", on="date_playerId").fillna(0)
targetShift = targetShift.merge(bat_df, how="left", on="date_playerId").fillna(0)
targetShift = targetShift.merge(
playerBoxScores[["date_playerId", "home", "positionCode"]],
how="left",
on="date_playerId",
)
# Merge rosters
targetShift = targetShift.merge(
rosters, how="left", on="date_playerId"
) # .drop(columns = ["playerId_y"])
# standings
targetShift = targetShift.merge(
standings,
how="left",
left_on=["engagementMetricsDate", "teamId"],
right_on=["gameDate", "teamId"],
)
# Merge games
targetShift = targetShift.merge(
games_df,
how="left",
left_on=["gameDate", "teamId"],
right_on=["gameDate", "homeId"],
)
targetShift.head()
# Merge teamTwitterFollowers
targetShift = targetShift.merge(
teamTwitter, how="left", left_on=["gameDate", "teamId"], right_on=["date", "teamId"]
)
for index in targetShift.index:
if pd.isna(targetShift.at[index, "teamFollowers"]):
targetShift.at[index, "teamFollowers"] = 0.0
targetShift.head()
for index in targetShift.index:
if pd.isna(targetShift.at[index, "teamId"]):
targetShift.at[index, "teamId"] = 0
for index in targetShift.index:
teamId = targetShift.at[index, "teamId"]
divisionRank = targetShift.at[index, "divisionRank"]
leagueRank = targetShift.at[index, "leagueRank"]
pct = targetShift.at[index, "pct"]
date = targetShift.at[index, "date_playerId"][:8]
if pd.isna(divisionRank):
targetShift.at[index, "divisionRank"] = 6
if pd.isna(leagueRank):
targetShift.at[index, "leagueRank"] = 16
if pd.isna(leagueRank):
targetShift.at[index, "pct"] = -1
if teamId in divisionRank_byDates.keys():
if date in divisionRank_byDates[teamId].keys():
targetShift.at[index, "streakCode"] = divisionRank_byDates[teamId][date][0]
targetShift.at[index, "divisionRank"] = int(
divisionRank_byDates[teamId][date][1]
)
targetShift.at[index, "leagueRank"] = int(
divisionRank_byDates[teamId][date][2]
)
targetShift.at[index, "pct"] = float(divisionRank_byDates[teamId][date][3])
targetShift.head()
currentPlayer = targetShift.at[0, "playerId"]
currentdivisionRank = 6.0
currentleagueRank = 16.0
currentpct = -1.0
for index in targetShift.index:
playerid = targetShift.at[index, "playerId"]
# date = targetShift.at[index,"date_playerId"][:8]
divisionRank = float(targetShift.at[index, "divisionRank"])
leagueRank = float(targetShift.at[index, "leagueRank"])
pct = float(targetShift.at[index, "pct"])
# teamId = targetShift.at[index,"teamId"]
if currentPlayer == playerid:
if currentdivisionRank != divisionRank:
# when standing update happens
if divisionRank < 6:
currentdivisionRank = divisionRank
targetShift.at[index, "divisionRank"] = currentdivisionRank
# updating all 6 after / when divisionRank = 6
elif currentdivisionRank < 6:
targetShift.at[index, "divisionRank"] = currentdivisionRank
if currentleagueRank != leagueRank:
# when standing update happens
if leagueRank < 16:
currentleagueRank = leagueRank
targetShift.at[index, "leagueRank"] = currentleagueRank
# updating all 16 after / when divisionRank = 16
elif currentleagueRank < 16:
targetShift.at[index, "leagueRank"] = currentleagueRank
if currentpct != -1:
# when standing update happens
if pct > -1:
currentpct = pct
targetShift.at[index, "pct"] = currentpct
# updating all 16 after / when divisionRank = 16
elif currentpct > -1:
targetShift.at[index, "pct"] = currentpct
else:
currentPlayer = playerid
currentdivisionRank = 6
currentleagueRank = 16
currentpct = -1
for index in targetShift.index:
# if record is empty for streakCode, then replace them to 0
if pd.isna(targetShift.at[index, "streakCode"]):
targetShift.at[index, "streakCode"] = 0.0
else:
if "W" in targetShift.at[index, "streakCode"]:
targetShift.at[index, "streakCode"] = float(
targetShift.at[index, "streakCode"][1]
)
elif "L" in targetShift.at[index, "streakCode"]:
targetShift.at[index, "streakCode"] = -(
float(targetShift.at[index, "streakCode"][1])
)
targetShift = targetShift[
[
"engagementMetricsDate",
"date_playerId",
"teamId",
"seasonIdx",
"isAwarded",
"cumulativeAwardScore",
"numberOfFollowers",
"teamFollowers",
"pitch0",
"pitch1",
"pitch2",
"pitch4",
"bat0",
"bat1",
"bat2",
"bat3",
"bat4",
"bat5",
"bat6",
"streakCode",
"divisionRank",
"leagueRank",
"wins",
"losses",
"pct",
"divisionChamp",
"divisionLeader",
"homeWinPct",
"homeWinner",
"homeScore",
"awayWinPct",
"awayScore",
"target1",
"target2",
"target3",
"target4",
]
] #'home','awayWinner'
targetShift.head()
for index in targetShift.index:
homeWinPct = targetShift.at[index, "homeWinPct"]
homeWinner = targetShift.at[index, "homeWinner"]
homeScore = targetShift.at[index, "homeScore"]
awayWinPct = targetShift.at[index, "awayWinPct"]
awayScore = targetShift.at[index, "awayScore"]
seasonIdx = targetShift.at[index, "seasonIdx"]
streakCode = targetShift.at[index, "streakCode"]
divisionRank = targetShift.at[index, "divisionRank"]
leagueRank = targetShift.at[index, "leagueRank"]
wins = targetShift.at[index, "wins"]
losses = targetShift.at[index, "losses"]
pct = targetShift.at[index, "pct"]
divisionChamp = targetShift.at[index, "divisionChamp"]
divisionLeader = targetShift.at[index, "divisionLeader"]
# off-season
# if seasonIdx == 0:
if pd.isna(homeWinPct):
targetShift.at[index, "homeWinPct"] = -1.0
if pd.isna(homeWinner):
targetShift.at[index, "homeWinner"] = -1.0
elif homeWinner == False:
targetShift.at[index, "homeWinner"] = 0.0
elif homeWinner == True:
targetShift.at[index, "homeWinner"] = 1.0
if pd.isna(homeScore):
targetShift.at[index, "homeScore"] = -1.0
if pd.isna(awayWinPct):
targetShift.at[index, "awayWinPct"] = -1.0
if pd.isna(awayScore):
targetShift.at[index, "awayScore"] = -1.0
if pd.isna(streakCode):
targetShift.at[index, "streakCode"] = 0.0
if pd.isna(divisionRank):
targetShift.at[index, "divisionRank"] = 6.0
if pd.isna(leagueRank):
targetShift.at[index, "leagueRank"] = 16.0
if pd.isna(wins):
targetShift.at[index, "wins"] = -1.0
if pd.isna(losses):
targetShift.at[index, "losses"] = -1.0
if pd.isna(pct):
targetShift.at[index, "pct"] = -1.0
if divisionChamp == True:
targetShift.at[index, "divisionChamp"] = 1.0
else:
targetShift.at[index, "divisionChamp"] = 0.0
if divisionLeader == True:
targetShift.at[index, "divisionLeader"] = 1.0
else:
targetShift.at[index, "divisionLeader"] = 0.0
targetShift
# targetShift.to_csv("ANN_7_July30th.csv", index=False)
# ## Modeling
# - We trained simple NN model on the sampled dataset from the train dataset created above.
maxplayerFollowers = targetShift["numberOfFollowers"].max()
maxteamFollowers = targetShift["teamFollowers"].max()
print(maxplayerFollowers, maxteamFollowers)
targetShift["numberOfFollowers"] = (
targetShift["numberOfFollowers"] / maxplayerFollowers
) * 100
targetShift["teamFollowers"] = (targetShift["teamFollowers"] / maxteamFollowers) * 100
targetShift.rename(
{
"numberOfFollowers": "norm_playerFollowers",
"teamFollowers": "norm_teamFollowers",
},
axis=1,
inplace=True,
)
targetShift.head()
# ### ANN approach
train = targetShift[targetShift["engagementMetricsDate"] < "2021-04-25"]
test = targetShift[targetShift["engagementMetricsDate"] >= "2021-04-25"]
print(len(train), len(test))
del targetShift
n_sample = 500000
train_sample = train.sample(n=n_sample)
X_train = train_sample[
[
"isAwarded",
"cumulativeAwardScore",
"norm_playerFollowers",
"norm_teamFollowers",
"pitch0",
"pitch1",
"pitch2",
"pitch4",
"bat0",
"bat1",
"bat2",
"bat3",
"bat4",
"bat5",
"bat6",
"streakCode",
"divisionRank",
"leagueRank",
"wins",
"losses",
"pct",
"divisionChamp",
"divisionLeader",
"homeWinPct",
"homeWinner",
"homeScore",
"awayWinPct",
"awayScore",
]
].fillna(0)
Y_train = train_sample[["target1", "target2", "target3", "target4"]]
print("Train Data Shape : {}".format(X_train.shape))
print("Train Label Shape : {}".format(Y_train.shape))
feature_size = X_train.shape[1]
def build_network(input_shape=(feature_size,)):
model = models.Sequential()
model.add(layers.Dense(28, activation="relu", input_shape=input_shape))
model.add(layers.Dropout(0.2))
model.add(layers.Dense(28, activation="relu"))
model.add(layers.Dropout(0.2))
model.add(layers.Dense(28, activation="relu"))
model.add(layers.Dropout(0.2))
model.add(layers.Dense(14, activation="relu"))
model.add(layers.Dropout(0.2))
model.add(layers.Dense(1))
model.compile(optimizer="adam", loss="mse", metrics=["mae"])
return model
model1 = build_network((feature_size,))
model2 = build_network((feature_size,))
model3 = build_network((feature_size,))
model4 = build_network((feature_size,))
model1.summary()
model1.fit(X_train.values, Y_train.iloc[:, 0].values, epochs=20)
model2.fit(X_train.values, Y_train.iloc[:, 1].values, epochs=20)
model3.fit(X_train.values, Y_train.iloc[:, 2].values, epochs=20)
model4.fit(X_train.values, Y_train.iloc[:, 3].values, epochs=20)
# ### Predictions
# - From the models and test set created above, we will make a prediction and save the result into dataframe.
test_id = test["date_playerId"]
X_test = test[
[
"isAwarded",
"cumulativeAwardScore",
"norm_playerFollowers",
"norm_teamFollowers",
"pitch0",
"pitch1",
"pitch2",
"pitch4",
"bat0",
"bat1",
"bat2",
"bat3",
"bat4",
"bat5",
"bat6",
"streakCode",
"divisionRank",
"leagueRank",
"wins",
"losses",
"pct",
"divisionChamp",
"divisionLeader",
"homeWinPct",
"homeWinner",
"homeScore",
"awayWinPct",
"awayScore",
]
].fillna(0)
Y_pred1 = model1.predict(X_test.values)
Y_pred2 = model2.predict(X_test.values)
Y_pred3 = model3.predict(X_test.values)
Y_pred4 = model4.predict(X_test.values)
prediction_df = pd.DataFrame()
prediction_df["date_playerId"] = test_id
prediction_df["target1"] = Y_pred1
prediction_df["target2"] = Y_pred2
prediction_df["target3"] = Y_pred3
prediction_df["target4"] = Y_pred4
prediction_df = prediction_df.reset_index(drop=True)
prediction_df.head()
import mlb
env = mlb.make_env() # initialize the environment
iter_test = env.iter_test() # iterator which loops over each date in test set
for test_df, prediction_df in iter_test:
env.predict(prediction_df)
| false | 0 | 9,516 | 0 | 9,516 | 9,516 |
||
69492411
|
<jupyter_start><jupyter_text>London bike sharing dataset
### License
These licence terms and conditions apply to TfL's free transport data service and are based on version 2.0 of the Open Government Licence with specific amendments for Transport for London (the "Licence"). TfL may at any time revise this Licence without notice. It is up to you ("You") to regularly review the Licence, which will be available on this website, in case there are any changes. Your continued use of the transport data feeds You have opted to receive ("Information") after a change has been made to the Licence will be treated as Your acceptance of that change.
Using Information under this Licence
TfL grants You a worldwide, royalty-free, perpetual, non-exclusive Licence to use the Information subject to the conditions below (as varied from time to time).
This Licence does not affect Your freedom under fair dealing or fair use or any other copyright or database right exceptions and limitations.
This Licence shall apply from the date of registration and shall continue for the period the Information is provided to You or You breach the Licence.
Rights
You are free to:
Copy, publish, distribute and transmit the Information
Adapt the Information and
Exploit the Information commercially and non-commercially for example, by combining it with other Information, or by including it in Your own product or application
Requirements
You must, where You do any of the above:
Acknowledge TfL as the source of the Information by including the following attribution statement 'Powered by TfL Open Data'
Acknowledge that this Information contains Ordnance Survey derived data by including the following attribution statement: 'Contains OS data © Crown copyright and database rights 2016' and Geomni UK Map data © and database rights [2019]
Ensure our intellectual property rights, including all logos, design rights, patents and trademarks, are protected by following our design and branding guidelines
Limit traffic requests up to a maximum of 300 calls per minute per data feed. TfL reserves the right to throttle or limit access to feeds when it is believed the overall service is being degraded by excessive use and
Ensure the information You provide on registration is accurate
These are important conditions of this Licence and if You fail to comply with them the rights granted to You under this Licence, or any similar licence granted by TfL, will end automatically.
Exemptions
This Licence does not:
Transfer any intellectual property rights in the Information to You or any third party
Include personal data in the Information
Provide any rights to use the Information after this Licence has ended
Provide any rights to use any other intellectual property rights, including patents, trade marks, and design rights or permit You to:
Use data from the Oyster, Congestion Charging and Santander Cycles websites to populate or update any other software or database or
Use any automated system, software or process to extract content and/or data, including trawling, data mining and screen scraping
in relation to the Oyster, Congestion Charging and Santander Cycles websites, except where expressly permitted under a written licence agreement with TfL.
These are important conditions of this Licence and, if You fail to comply with them, the rights granted to You under this Licence, or any similar licence granted by TfL, will end automatically.
Non-endorsement
This Licence does not grant You any right to use the Information in a way that suggests any official status or that TfL endorses You or Your use of the Information.
### Context
The purpose is to try predict the future bike shares.
### Content
The data is acquired from 3 sources:
- Https://cycling.data.tfl.gov.uk/ 'Contains OS data © Crown copyright and database rights 2016' and Geomni UK Map data © and database rights [2019] 'Powered by TfL Open Data'
- freemeteo.com - weather data
- https://www.gov.uk/bank-holidays
From 1/1/2015 to 31/12/2016
The data from cycling dataset is grouped by "Start time", this represent the count of new bike shares grouped by hour. The long duration shares are not taken in the count.
### Metadata:
"timestamp" - *timestamp field for grouping the data*
"cnt" - *the count of a new bike shares*
"t1" - *real temperature in C*
"t2" - *temperature in C "feels like"*
"hum" - *humidity in percentage*
"wind_speed" - *wind speed in km/h*
"weather_code" - *category of the weather*
"is_holiday" - *boolean field - 1 holiday / 0 non holiday*
"is_weekend" - *boolean field - 1 if the day is weekend*
"season" - *category field meteorological seasons: 0-spring ; 1-summer; 2-fall; 3-winter.*
"weathe_code" category description:
*1 = Clear ; mostly clear but have some values with haze/fog/patches of fog/ fog in vicinity
2 = scattered clouds / few clouds
3 = Broken clouds
4 = Cloudy
7 = Rain/ light Rain shower/ Light rain
10 = rain with thunderstorm
26 = snowfall
94 = Freezing Fog*
Kaggle dataset identifier: london-bike-sharing-dataset
<jupyter_code>import pandas as pd
df = pd.read_csv('london-bike-sharing-dataset/london_merged.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 17414 entries, 0 to 17413
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 timestamp 17414 non-null object
1 cnt 17414 non-null int64
2 t1 17414 non-null float64
3 t2 17414 non-null float64
4 hum 17414 non-null float64
5 wind_speed 17414 non-null float64
6 weather_code 17414 non-null float64
7 is_holiday 17414 non-null float64
8 is_weekend 17414 non-null float64
9 season 17414 non-null float64
dtypes: float64(8), int64(1), object(1)
memory usage: 1.3+ MB
<jupyter_text>Examples:
{
"timestamp": "2015-01-04 00:00:00",
"cnt": 182,
"t1": 3.0,
"t2": 2.0,
"hum": 93.0,
"wind_speed": 6,
"weather_code": 3,
"is_holiday": 0,
"is_weekend": 1,
"season": 3
}
{
"timestamp": "2015-01-04 01:00:00",
"cnt": 138,
"t1": 3.0,
"t2": 2.5,
"hum": 93.0,
"wind_speed": 5,
"weather_code": 1,
"is_holiday": 0,
"is_weekend": 1,
"season": 3
}
{
"timestamp": "2015-01-04 02:00:00",
"cnt": 134,
"t1": 2.5,
"t2": 2.5,
"hum": 96.5,
"wind_speed": 0,
"weather_code": 1,
"is_holiday": 0,
"is_weekend": 1,
"season": 3
}
{
"timestamp": "2015-01-04 03:00:00",
"cnt": 72,
"t1": 2.0,
"t2": 2.0,
"hum": 100.0,
"wind_speed": 0,
"weather_code": 1,
"is_holiday": 0,
"is_weekend": 1,
"season": 3
}
<jupyter_script># # Let's start with importing essential libraries.
import numpy as np
import pandas as pd
import plotly
import plotly.express as px
import plotly.graph_objects as go
from plotly.subplots import make_subplots
import warnings
warnings.filterwarnings("ignore")
# # Now, let's dive into our data.
df = pd.read_csv("../input/london-bike-sharing-dataset/london_merged.csv")
df.head()
# ## See if there are any dublicated or NaN values
df.duplicated().value_counts()
df.isnull().sum()
# - Not a single missing value! PERFECT!
# ## Now, Let's plot the distribution of various discrete features such as season, holiday, weekend and weathercode.
fig = px.bar(
x=df["season"].value_counts().index,
y=df["season"].value_counts().values,
title="Seasons",
labels={"y": "Count", "x": "Seasons"},
)
fig.update_layout(xaxis={"categoryorder": "total descending"})
fig.show()
# ### Seems like season column distributed normally. Let's check value counts of this column for more clear info.
df.season.value_counts()
# - Values almost equal as expected.
# ### Now let's check *`is_holiday`* column.
weekend = (
df.groupby("is_weekend")["cnt"]
.mean()
.reset_index()
.rename(
columns={"is_weekend": "Weekend", "cnt": "Number of Bike Shared"},
)
)
weekend["Weekend"] = weekend["Weekend"].replace({0: "Weekday", 1: "Weekend"})
fig = px.bar(
weekend,
x="Weekend",
y="Number of Bike Shared",
color="Weekend",
)
fig.update_layout(xaxis={"categoryorder": "total descending"})
fig.show()
# - As expected highly 'not a holiday' distribution. Most likely *`is_weekend`* column is also in the same situation. Let's check.
# ### Now, look at *`weather_code`* column.
fig = px.pie(
df,
values=df["weather_code"].value_counts().values,
names=[
"Clear",
"Scattered Clouds",
"Broken Clouds",
"Cloudy" "Rain",
"Rain With Thunderstorm",
"Snowfall",
"Freezing Fog",
],
)
fig.show()
# ### Let's transform `timestamp` column to `datetime` in type, and set it as index.
df["timestamp"] = pd.to_datetime(df["timestamp"])
df = df.set_index("timestamp")
df.head()
# ### Now it is time to make feature engineering. Let's extract new columns (day of the week, day of the month, hour, month, season, year etc.) by using new index.
# We can use strftime() function to get year, month, day, weekday and hour of the index.
df["year_month"] = df.index.strftime("%Y-%m")
df["year"] = df.index.year
df["month"] = df.index.month
df["day_of_month"] = df.index.day
df["day_of_week"] = df.index.weekday
df["hour"] = df.index.hour
df.head()
# ### Everything seems perfect. Now, let's visualize the correlation with a heatmap.
fig = px.imshow(df.corr(), title="Correlation Heat Map")
fig.show()
# ### For better understanding, let's see the correlation between our target variable which is *`cnt`* and the others.
fig = px.imshow(df.corr()[["cnt"]], title="Correlation Heat Map")
fig.show()
# #### We understand that the count of a new bike shares(*`cnt`*) column has a positive correlation with *`t1`*, *`t2`* and *`hour`* columns. Also *`hum`* column, which gives information about humidity in percentage, has a fairly high negative correlation with *`cnt`*.
# ### For more clear understanding, let's visualize the correlation of the target variable and the other features with barplot
fig = px.bar(
y=df.corr()["cnt"].index,
x=df.corr()["cnt"].values,
title="Correlation (CNT)",
labels={"y": "Variables", "x": "Values"},
)
fig.update_layout(yaxis={"categoryorder": "total descending"})
fig.show()
# ### Now it is time to plot bike shares over time with lineplot.
fig = go.Figure(data=go.Scatter(x=df.index, y=df["cnt"]))
fig.update_layout(
title="Bike Shares Over Time",
xaxis_title="Date",
yaxis_title="Count of Bike Shares",
)
fig.show()
# - There are days with unusually high count of a new bike shares. Let's find out which days are they.
df[df["cnt"] > 7000]
# In 2015-07-09 and 2015-08-06 count of a new bike shares increases. There must be something about those days.
# This is a great example of getting information great insights by visualization.
# ### It is time to plot bike shares by months and year_of_month to understand the correlation between bike shares and months.
year_month = df.groupby("year_month").sum().reset_index()
fig = go.Figure(data=go.Scatter(x=year_month["year_month"], y=year_month["cnt"]))
fig.update_layout(
title="Bike Shares by Month", xaxis_title="Date", yaxis_title="Count of Bike Shares"
)
fig.show()
# As expected, in summer bike shares is increasing. Let's see this relation better by different plot.
px.line(
df.groupby("month").mean(),
x=df.groupby("month").mean().index,
y=df.groupby("month").mean()["cnt"],
)
px.bar(
df.groupby("month").mean(),
x=df.groupby("month").mean().index,
y=df.groupby("month").mean()["cnt"],
)
# In those two plots, we can clearly see the bike share difference by months. Bike share leans to increase in summer.
# ### What about correlation between bike shares and hours? It would be great to see the difference when it is a holiday too right! Let's plot bike shares by hours.
fig = go.Figure()
fig.add_trace(
go.Scatter(
x=df[df["is_holiday"] == 0].groupby(["hour"]).mean()[["cnt"]].index,
y=df[df["is_holiday"] == 0].groupby(["hour"]).mean()[["cnt"]]["cnt"],
name="Not Holiday",
)
)
fig.add_trace(
go.Scatter(
x=df[df["is_holiday"] == 1].groupby(["hour"]).mean()[["cnt"]].index,
y=df[df["is_holiday"] == 1].groupby(["hour"]).mean()[["cnt"]]["cnt"],
name="Holiday",
)
)
fig.update_layout(
title="Bike Shares in Holidays By Hour",
xaxis_title="Hour",
yaxis_title="Count of Bike Shares",
)
fig.show()
# We can clearly see that when it is not holiday, bike shares tends to increase 8AM and 7PM.
# This means people use bikes when going to work.
# Also difference by seasons plot confirms our conclusion. People tends to use bikes more when it is spring.
# ### Let's plot bike shares by day of week to understand better.
day_of_week = df.groupby("day_of_week").sum()[["cnt"]]
day_of_week["days"] = [
"Monday",
"Tuesday",
"Wednesday",
"Thursday",
"Friday",
"Saturday",
"Sunday",
]
day_of_week = day_of_week.set_index("days")
fig = px.bar(
x=day_of_week.index,
y=day_of_week["cnt"],
color=day_of_week.index,
title="Bike Shares by Day",
labels={"x": "Days", "y": "Count of Bike Shares"},
)
fig.show()
# People use bike in weekdays more than weekends.
# ### Let's see the difference by seasons.
fig = go.Figure()
for i in range(0, 4):
fig.add_trace(
go.Scatter(
x=df[df["season"] == i].groupby(["day_of_week"]).mean()[["cnt"]].index,
y=df[df["season"] == i].groupby(["day_of_week"]).mean()[["cnt"]]["cnt"],
)
)
fig.update_layout(
title="Bike Shares in Seasons By Hour",
xaxis_title="Day of Week",
yaxis_title="Count of Bike Shares",
)
fig.show()
# ### Plot bike shares by day of month
day_of_month = df.groupby("day_of_month").mean()[["cnt"]].astype("int")
fig = px.line(
x=day_of_month.index,
y=day_of_month.cnt,
title="Bike Shares by Day of Month",
labels={"x": "Day of Month", "y": "Count of Bike Shares"},
)
fig.show()
# ### It is time to plot bike shares by year and by seasons.
df.groupby("year").mean()[["cnt"]]
fig = px.bar(
x=df.groupby("year").mean()[["cnt"]].index,
y=df.groupby("year").mean()[["cnt"]]["cnt"],
title="Bike Share by Year",
labels={"y": "Count of Bike Shares", "x": "Year"},
)
fig.show()
# It does seem like in 2017 bike share dropped heavily. But that is not true. Because our data does not contain
# all information about 2017. This plot may dislead us.
fig = px.histogram(df, x="season", y="cnt", color="season")
fig.show()
# We can clearly see from this plot that people use bike most in summer.
# ### Now, let's visualize the distribution of bike shares by weekday/weekend with barplot
holiday = (
df.groupby("is_holiday")["cnt"]
.mean()
.reset_index()
.rename(
columns={"is_holiday": "Holiday", "cnt": "Number of Bike Shared"},
)
)
holiday["Holiday"] = holiday["Holiday"].replace({0: "Normal Day", 1: "Holiday"})
fig = px.bar(
holiday,
x="Holiday",
y="Number of Bike Shared",
color="Holiday",
)
fig.update_layout(xaxis={"categoryorder": "total descending"})
fig.show()
# People use bikes in weekdays more than in weekends. Before we saw that in 7AM and also in 5PM bike usage increase.
# This addresses that people use bike when going to work and also when coming back to their home.
# ### Visualize the continuous variables with scatterplot
fig = px.scatter(x=df["t1"], y=df["hum"], color=df["season"])
fig.show()
fig = px.scatter(x=df["t1"], y=df["wind_speed"], color=df["season"])
fig.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/492/69492411.ipynb
|
london-bike-sharing-dataset
|
hmavrodiev
|
[{"Id": 69492411, "ScriptId": 18975839, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7390227, "CreationDate": "07/31/2021 16:59:41", "VersionNumber": 1.0, "Title": "PLOTLY - Beginner Friendly Data Visualization", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 221.0, "LinesInsertedFromPrevious": 221.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 23}]
|
[{"Id": 92728374, "KernelVersionId": 69492411, "SourceDatasetVersionId": 731448}]
|
[{"Id": 731448, "DatasetId": 376751, "DatasourceVersionId": 751930, "CreatorUserId": 1963674, "LicenseName": "Other (specified in description)", "CreationDate": "10/10/2019 12:49:37", "VersionNumber": 1.0, "Title": "London bike sharing dataset", "Slug": "london-bike-sharing-dataset", "Subtitle": "Historical data for bike sharing in London 'Powered by TfL Open Data'", "Description": "### License\nThese licence terms and conditions apply to TfL's free transport data service and are based on version 2.0 of the Open Government Licence with specific amendments for Transport for London (the \"Licence\"). TfL may at any time revise this Licence without notice. It is up to you (\"You\") to regularly review the Licence, which will be available on this website, in case there are any changes. Your continued use of the transport data feeds You have opted to receive (\"Information\") after a change has been made to the Licence will be treated as Your acceptance of that change.\n\nUsing Information under this Licence\nTfL grants You a worldwide, royalty-free, perpetual, non-exclusive Licence to use the Information subject to the conditions below (as varied from time to time).\n\nThis Licence does not affect Your freedom under fair dealing or fair use or any other copyright or database right exceptions and limitations.\n\nThis Licence shall apply from the date of registration and shall continue for the period the Information is provided to You or You breach the Licence. \n\nRights\nYou are free to:\n\nCopy, publish, distribute and transmit the Information\nAdapt the Information and\nExploit the Information commercially and non-commercially for example, by combining it with other Information, or by including it in Your own product or application\nRequirements\nYou must, where You do any of the above:\n\nAcknowledge TfL as the source of the Information by including the following attribution statement 'Powered by TfL Open Data'\nAcknowledge that this Information contains Ordnance Survey derived data by including the following attribution statement: 'Contains OS data \u00a9 Crown copyright and database rights 2016' and Geomni UK Map data \u00a9 and database rights [2019]\nEnsure our intellectual property rights, including all logos, design rights, patents and trademarks, are protected by following our design and branding guidelines\nLimit traffic requests up to a maximum of 300 calls per minute per data feed. TfL reserves the right to throttle or limit access to feeds when it is believed the overall service is being degraded by excessive use and\nEnsure the information You provide on registration is accurate \nThese are important conditions of this Licence and if You fail to comply with them the rights granted to You under this Licence, or any similar licence granted by TfL, will end automatically.\n\nExemptions\nThis Licence does not:\n\nTransfer any intellectual property rights in the Information to You or any third party\nInclude personal data in the Information\nProvide any rights to use the Information after this Licence has ended \nProvide any rights to use any other intellectual property rights, including patents, trade marks, and design rights or permit You to:\nUse data from the Oyster, Congestion Charging and Santander Cycles websites to populate or update any other software or database or\nUse any automated system, software or process to extract content and/or data, including trawling, data mining and screen scraping\nin relation to the Oyster, Congestion Charging and Santander Cycles websites, except where expressly permitted under a written licence agreement with TfL.\nThese are important conditions of this Licence and, if You fail to comply with them, the rights granted to You under this Licence, or any similar licence granted by TfL, will end automatically.\n\nNon-endorsement\nThis Licence does not grant You any right to use the Information in a way that suggests any official status or that TfL endorses You or Your use of the Information.\n\n\n### Context\n\nThe purpose is to try predict the future bike shares.\n\n### Content\n\n\nThe data is acquired from 3 sources: \n- Https://cycling.data.tfl.gov.uk/ 'Contains OS data \u00a9 Crown copyright and database rights 2016' and Geomni UK Map data \u00a9 and database rights [2019] 'Powered by TfL Open Data' \n - freemeteo.com - weather data \n - https://www.gov.uk/bank-holidays \nFrom 1/1/2015 to 31/12/2016 \n\nThe data from cycling dataset is grouped by \"Start time\", this represent the count of new bike shares grouped by hour. The long duration shares are not taken in the count.\n\n### Metadata:\n\"timestamp\" - *timestamp field for grouping the data* \n\"cnt\" - *the count of a new bike shares* \n\"t1\" - *real temperature in C* \n\"t2\" - *temperature in C \"feels like\"* \n\"hum\" - *humidity in percentage* \n\"wind_speed\" - *wind speed in km/h* \n\"weather_code\" - *category of the weather* \n\"is_holiday\" - *boolean field - 1 holiday / 0 non holiday* \n\"is_weekend\" - *boolean field - 1 if the day is weekend* \n\"season\" - *category field meteorological seasons: 0-spring ; 1-summer; 2-fall; 3-winter.* \n\n \n \n \n\n\n\"weathe_code\" category description: \n*1 = Clear ; mostly clear but have some values with haze/fog/patches of fog/ fog in vicinity \n2 = scattered clouds / few clouds \n3 = Broken clouds \n4 = Cloudy \n7 = Rain/ light Rain shower/ Light rain \n10 = rain with thunderstorm \n26 = snowfall \n94 = Freezing Fog*", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 376751, "CreatorUserId": 1963674, "OwnerUserId": 1963674.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 731448.0, "CurrentDatasourceVersionId": 751930.0, "ForumId": 388652, "Type": 2, "CreationDate": "10/10/2019 12:49:37", "LastActivityDate": "10/10/2019", "TotalViews": 194568, "TotalDownloads": 23002, "TotalVotes": 398, "TotalKernels": 118}]
|
[{"Id": 1963674, "UserName": "hmavrodiev", "DisplayName": "Hristo Mavrodiev", "RegisterDate": "06/03/2018", "PerformanceTier": 1}]
|
# # Let's start with importing essential libraries.
import numpy as np
import pandas as pd
import plotly
import plotly.express as px
import plotly.graph_objects as go
from plotly.subplots import make_subplots
import warnings
warnings.filterwarnings("ignore")
# # Now, let's dive into our data.
df = pd.read_csv("../input/london-bike-sharing-dataset/london_merged.csv")
df.head()
# ## See if there are any dublicated or NaN values
df.duplicated().value_counts()
df.isnull().sum()
# - Not a single missing value! PERFECT!
# ## Now, Let's plot the distribution of various discrete features such as season, holiday, weekend and weathercode.
fig = px.bar(
x=df["season"].value_counts().index,
y=df["season"].value_counts().values,
title="Seasons",
labels={"y": "Count", "x": "Seasons"},
)
fig.update_layout(xaxis={"categoryorder": "total descending"})
fig.show()
# ### Seems like season column distributed normally. Let's check value counts of this column for more clear info.
df.season.value_counts()
# - Values almost equal as expected.
# ### Now let's check *`is_holiday`* column.
weekend = (
df.groupby("is_weekend")["cnt"]
.mean()
.reset_index()
.rename(
columns={"is_weekend": "Weekend", "cnt": "Number of Bike Shared"},
)
)
weekend["Weekend"] = weekend["Weekend"].replace({0: "Weekday", 1: "Weekend"})
fig = px.bar(
weekend,
x="Weekend",
y="Number of Bike Shared",
color="Weekend",
)
fig.update_layout(xaxis={"categoryorder": "total descending"})
fig.show()
# - As expected highly 'not a holiday' distribution. Most likely *`is_weekend`* column is also in the same situation. Let's check.
# ### Now, look at *`weather_code`* column.
fig = px.pie(
df,
values=df["weather_code"].value_counts().values,
names=[
"Clear",
"Scattered Clouds",
"Broken Clouds",
"Cloudy" "Rain",
"Rain With Thunderstorm",
"Snowfall",
"Freezing Fog",
],
)
fig.show()
# ### Let's transform `timestamp` column to `datetime` in type, and set it as index.
df["timestamp"] = pd.to_datetime(df["timestamp"])
df = df.set_index("timestamp")
df.head()
# ### Now it is time to make feature engineering. Let's extract new columns (day of the week, day of the month, hour, month, season, year etc.) by using new index.
# We can use strftime() function to get year, month, day, weekday and hour of the index.
df["year_month"] = df.index.strftime("%Y-%m")
df["year"] = df.index.year
df["month"] = df.index.month
df["day_of_month"] = df.index.day
df["day_of_week"] = df.index.weekday
df["hour"] = df.index.hour
df.head()
# ### Everything seems perfect. Now, let's visualize the correlation with a heatmap.
fig = px.imshow(df.corr(), title="Correlation Heat Map")
fig.show()
# ### For better understanding, let's see the correlation between our target variable which is *`cnt`* and the others.
fig = px.imshow(df.corr()[["cnt"]], title="Correlation Heat Map")
fig.show()
# #### We understand that the count of a new bike shares(*`cnt`*) column has a positive correlation with *`t1`*, *`t2`* and *`hour`* columns. Also *`hum`* column, which gives information about humidity in percentage, has a fairly high negative correlation with *`cnt`*.
# ### For more clear understanding, let's visualize the correlation of the target variable and the other features with barplot
fig = px.bar(
y=df.corr()["cnt"].index,
x=df.corr()["cnt"].values,
title="Correlation (CNT)",
labels={"y": "Variables", "x": "Values"},
)
fig.update_layout(yaxis={"categoryorder": "total descending"})
fig.show()
# ### Now it is time to plot bike shares over time with lineplot.
fig = go.Figure(data=go.Scatter(x=df.index, y=df["cnt"]))
fig.update_layout(
title="Bike Shares Over Time",
xaxis_title="Date",
yaxis_title="Count of Bike Shares",
)
fig.show()
# - There are days with unusually high count of a new bike shares. Let's find out which days are they.
df[df["cnt"] > 7000]
# In 2015-07-09 and 2015-08-06 count of a new bike shares increases. There must be something about those days.
# This is a great example of getting information great insights by visualization.
# ### It is time to plot bike shares by months and year_of_month to understand the correlation between bike shares and months.
year_month = df.groupby("year_month").sum().reset_index()
fig = go.Figure(data=go.Scatter(x=year_month["year_month"], y=year_month["cnt"]))
fig.update_layout(
title="Bike Shares by Month", xaxis_title="Date", yaxis_title="Count of Bike Shares"
)
fig.show()
# As expected, in summer bike shares is increasing. Let's see this relation better by different plot.
px.line(
df.groupby("month").mean(),
x=df.groupby("month").mean().index,
y=df.groupby("month").mean()["cnt"],
)
px.bar(
df.groupby("month").mean(),
x=df.groupby("month").mean().index,
y=df.groupby("month").mean()["cnt"],
)
# In those two plots, we can clearly see the bike share difference by months. Bike share leans to increase in summer.
# ### What about correlation between bike shares and hours? It would be great to see the difference when it is a holiday too right! Let's plot bike shares by hours.
fig = go.Figure()
fig.add_trace(
go.Scatter(
x=df[df["is_holiday"] == 0].groupby(["hour"]).mean()[["cnt"]].index,
y=df[df["is_holiday"] == 0].groupby(["hour"]).mean()[["cnt"]]["cnt"],
name="Not Holiday",
)
)
fig.add_trace(
go.Scatter(
x=df[df["is_holiday"] == 1].groupby(["hour"]).mean()[["cnt"]].index,
y=df[df["is_holiday"] == 1].groupby(["hour"]).mean()[["cnt"]]["cnt"],
name="Holiday",
)
)
fig.update_layout(
title="Bike Shares in Holidays By Hour",
xaxis_title="Hour",
yaxis_title="Count of Bike Shares",
)
fig.show()
# We can clearly see that when it is not holiday, bike shares tends to increase 8AM and 7PM.
# This means people use bikes when going to work.
# Also difference by seasons plot confirms our conclusion. People tends to use bikes more when it is spring.
# ### Let's plot bike shares by day of week to understand better.
day_of_week = df.groupby("day_of_week").sum()[["cnt"]]
day_of_week["days"] = [
"Monday",
"Tuesday",
"Wednesday",
"Thursday",
"Friday",
"Saturday",
"Sunday",
]
day_of_week = day_of_week.set_index("days")
fig = px.bar(
x=day_of_week.index,
y=day_of_week["cnt"],
color=day_of_week.index,
title="Bike Shares by Day",
labels={"x": "Days", "y": "Count of Bike Shares"},
)
fig.show()
# People use bike in weekdays more than weekends.
# ### Let's see the difference by seasons.
fig = go.Figure()
for i in range(0, 4):
fig.add_trace(
go.Scatter(
x=df[df["season"] == i].groupby(["day_of_week"]).mean()[["cnt"]].index,
y=df[df["season"] == i].groupby(["day_of_week"]).mean()[["cnt"]]["cnt"],
)
)
fig.update_layout(
title="Bike Shares in Seasons By Hour",
xaxis_title="Day of Week",
yaxis_title="Count of Bike Shares",
)
fig.show()
# ### Plot bike shares by day of month
day_of_month = df.groupby("day_of_month").mean()[["cnt"]].astype("int")
fig = px.line(
x=day_of_month.index,
y=day_of_month.cnt,
title="Bike Shares by Day of Month",
labels={"x": "Day of Month", "y": "Count of Bike Shares"},
)
fig.show()
# ### It is time to plot bike shares by year and by seasons.
df.groupby("year").mean()[["cnt"]]
fig = px.bar(
x=df.groupby("year").mean()[["cnt"]].index,
y=df.groupby("year").mean()[["cnt"]]["cnt"],
title="Bike Share by Year",
labels={"y": "Count of Bike Shares", "x": "Year"},
)
fig.show()
# It does seem like in 2017 bike share dropped heavily. But that is not true. Because our data does not contain
# all information about 2017. This plot may dislead us.
fig = px.histogram(df, x="season", y="cnt", color="season")
fig.show()
# We can clearly see from this plot that people use bike most in summer.
# ### Now, let's visualize the distribution of bike shares by weekday/weekend with barplot
holiday = (
df.groupby("is_holiday")["cnt"]
.mean()
.reset_index()
.rename(
columns={"is_holiday": "Holiday", "cnt": "Number of Bike Shared"},
)
)
holiday["Holiday"] = holiday["Holiday"].replace({0: "Normal Day", 1: "Holiday"})
fig = px.bar(
holiday,
x="Holiday",
y="Number of Bike Shared",
color="Holiday",
)
fig.update_layout(xaxis={"categoryorder": "total descending"})
fig.show()
# People use bikes in weekdays more than in weekends. Before we saw that in 7AM and also in 5PM bike usage increase.
# This addresses that people use bike when going to work and also when coming back to their home.
# ### Visualize the continuous variables with scatterplot
fig = px.scatter(x=df["t1"], y=df["hum"], color=df["season"])
fig.show()
fig = px.scatter(x=df["t1"], y=df["wind_speed"], color=df["season"])
fig.show()
|
[{"london-bike-sharing-dataset/london_merged.csv": {"column_names": "[\"timestamp\", \"cnt\", \"t1\", \"t2\", \"hum\", \"wind_speed\", \"weather_code\", \"is_holiday\", \"is_weekend\", \"season\"]", "column_data_types": "{\"timestamp\": \"object\", \"cnt\": \"int64\", \"t1\": \"float64\", \"t2\": \"float64\", \"hum\": \"float64\", \"wind_speed\": \"float64\", \"weather_code\": \"float64\", \"is_holiday\": \"float64\", \"is_weekend\": \"float64\", \"season\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 17414 entries, 0 to 17413\nData columns (total 10 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 timestamp 17414 non-null object \n 1 cnt 17414 non-null int64 \n 2 t1 17414 non-null float64\n 3 t2 17414 non-null float64\n 4 hum 17414 non-null float64\n 5 wind_speed 17414 non-null float64\n 6 weather_code 17414 non-null float64\n 7 is_holiday 17414 non-null float64\n 8 is_weekend 17414 non-null float64\n 9 season 17414 non-null float64\ndtypes: float64(8), int64(1), object(1)\nmemory usage: 1.3+ MB\n", "summary": "{\"cnt\": {\"count\": 17414.0, \"mean\": 1143.1016423567244, \"std\": 1085.1080679362178, \"min\": 0.0, \"25%\": 257.0, \"50%\": 844.0, \"75%\": 1671.75, \"max\": 7860.0}, \"t1\": {\"count\": 17414.0, \"mean\": 12.468090808162016, \"std\": 5.571817562920526, \"min\": -1.5, \"25%\": 8.0, \"50%\": 12.5, \"75%\": 16.0, \"max\": 34.0}, \"t2\": {\"count\": 17414.0, \"mean\": 11.52083572604418, \"std\": 6.615144673127893, \"min\": -6.0, \"25%\": 6.0, \"50%\": 12.5, \"75%\": 16.0, \"max\": 34.0}, \"hum\": {\"count\": 17414.0, \"mean\": 72.32495405995176, \"std\": 14.313186095310977, \"min\": 20.5, \"25%\": 63.0, \"50%\": 74.5, \"75%\": 83.0, \"max\": 100.0}, \"wind_speed\": {\"count\": 17414.0, \"mean\": 15.913063244133072, \"std\": 7.894570329361161, \"min\": 0.0, \"25%\": 10.0, \"50%\": 15.0, \"75%\": 20.5, \"max\": 56.5}, \"weather_code\": {\"count\": 17414.0, \"mean\": 2.722751808889399, \"std\": 2.341163284645666, \"min\": 1.0, \"25%\": 1.0, \"50%\": 2.0, \"75%\": 3.0, \"max\": 26.0}, \"is_holiday\": {\"count\": 17414.0, \"mean\": 0.02205122315378431, \"std\": 0.1468543671527596, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"is_weekend\": {\"count\": 17414.0, \"mean\": 0.28540254967267714, \"std\": 0.45161891757102296, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}, \"season\": {\"count\": 17414.0, \"mean\": 1.4920753416791088, \"std\": 1.1189108748277623, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 2.0, \"max\": 3.0}}", "examples": "{\"timestamp\":{\"0\":\"2015-01-04 00:00:00\",\"1\":\"2015-01-04 01:00:00\",\"2\":\"2015-01-04 02:00:00\",\"3\":\"2015-01-04 03:00:00\"},\"cnt\":{\"0\":182,\"1\":138,\"2\":134,\"3\":72},\"t1\":{\"0\":3.0,\"1\":3.0,\"2\":2.5,\"3\":2.0},\"t2\":{\"0\":2.0,\"1\":2.5,\"2\":2.5,\"3\":2.0},\"hum\":{\"0\":93.0,\"1\":93.0,\"2\":96.5,\"3\":100.0},\"wind_speed\":{\"0\":6.0,\"1\":5.0,\"2\":0.0,\"3\":0.0},\"weather_code\":{\"0\":3.0,\"1\":1.0,\"2\":1.0,\"3\":1.0},\"is_holiday\":{\"0\":0.0,\"1\":0.0,\"2\":0.0,\"3\":0.0},\"is_weekend\":{\"0\":1.0,\"1\":1.0,\"2\":1.0,\"3\":1.0},\"season\":{\"0\":3.0,\"1\":3.0,\"2\":3.0,\"3\":3.0}}"}}]
| true | 1 |
<start_data_description><data_path>london-bike-sharing-dataset/london_merged.csv:
<column_names>
['timestamp', 'cnt', 't1', 't2', 'hum', 'wind_speed', 'weather_code', 'is_holiday', 'is_weekend', 'season']
<column_types>
{'timestamp': 'object', 'cnt': 'int64', 't1': 'float64', 't2': 'float64', 'hum': 'float64', 'wind_speed': 'float64', 'weather_code': 'float64', 'is_holiday': 'float64', 'is_weekend': 'float64', 'season': 'float64'}
<dataframe_Summary>
{'cnt': {'count': 17414.0, 'mean': 1143.1016423567244, 'std': 1085.1080679362178, 'min': 0.0, '25%': 257.0, '50%': 844.0, '75%': 1671.75, 'max': 7860.0}, 't1': {'count': 17414.0, 'mean': 12.468090808162016, 'std': 5.571817562920526, 'min': -1.5, '25%': 8.0, '50%': 12.5, '75%': 16.0, 'max': 34.0}, 't2': {'count': 17414.0, 'mean': 11.52083572604418, 'std': 6.615144673127893, 'min': -6.0, '25%': 6.0, '50%': 12.5, '75%': 16.0, 'max': 34.0}, 'hum': {'count': 17414.0, 'mean': 72.32495405995176, 'std': 14.313186095310977, 'min': 20.5, '25%': 63.0, '50%': 74.5, '75%': 83.0, 'max': 100.0}, 'wind_speed': {'count': 17414.0, 'mean': 15.913063244133072, 'std': 7.894570329361161, 'min': 0.0, '25%': 10.0, '50%': 15.0, '75%': 20.5, 'max': 56.5}, 'weather_code': {'count': 17414.0, 'mean': 2.722751808889399, 'std': 2.341163284645666, 'min': 1.0, '25%': 1.0, '50%': 2.0, '75%': 3.0, 'max': 26.0}, 'is_holiday': {'count': 17414.0, 'mean': 0.02205122315378431, 'std': 0.1468543671527596, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'is_weekend': {'count': 17414.0, 'mean': 0.28540254967267714, 'std': 0.45161891757102296, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}, 'season': {'count': 17414.0, 'mean': 1.4920753416791088, 'std': 1.1189108748277623, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 2.0, 'max': 3.0}}
<dataframe_info>
RangeIndex: 17414 entries, 0 to 17413
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 timestamp 17414 non-null object
1 cnt 17414 non-null int64
2 t1 17414 non-null float64
3 t2 17414 non-null float64
4 hum 17414 non-null float64
5 wind_speed 17414 non-null float64
6 weather_code 17414 non-null float64
7 is_holiday 17414 non-null float64
8 is_weekend 17414 non-null float64
9 season 17414 non-null float64
dtypes: float64(8), int64(1), object(1)
memory usage: 1.3+ MB
<some_examples>
{'timestamp': {'0': '2015-01-04 00:00:00', '1': '2015-01-04 01:00:00', '2': '2015-01-04 02:00:00', '3': '2015-01-04 03:00:00'}, 'cnt': {'0': 182, '1': 138, '2': 134, '3': 72}, 't1': {'0': 3.0, '1': 3.0, '2': 2.5, '3': 2.0}, 't2': {'0': 2.0, '1': 2.5, '2': 2.5, '3': 2.0}, 'hum': {'0': 93.0, '1': 93.0, '2': 96.5, '3': 100.0}, 'wind_speed': {'0': 6.0, '1': 5.0, '2': 0.0, '3': 0.0}, 'weather_code': {'0': 3.0, '1': 1.0, '2': 1.0, '3': 1.0}, 'is_holiday': {'0': 0.0, '1': 0.0, '2': 0.0, '3': 0.0}, 'is_weekend': {'0': 1.0, '1': 1.0, '2': 1.0, '3': 1.0}, 'season': {'0': 3.0, '1': 3.0, '2': 3.0, '3': 3.0}}
<end_description>
| 2,788 | 23 | 4,831 | 2,788 |
69492739
|
<jupyter_start><jupyter_text>dart-model
Kaggle dataset identifier: dart-model
<jupyter_script>import os
import itertools
import pandas as pd
import datatable as dt
import numpy as np
import mlb
import pickle as pkl
from tqdm import tqdm
from itertools import product
import lightgbm as lgb
from fuzzywuzzy import fuzz
import re
import numba as nb
from numba import njit
import xgboost as xgb
@njit
def nb_cumsum(arr):
return arr.cumsum()
@njit
def nb_sum(arr):
return arr.sum()
def reduce_mem_usage(df, verbose=True):
numerics = ["int16", "int32", "int64", "float16", "float32", "float64"]
start_mem = df.memory_usage().sum() / 1024**2
for col in df.columns:
col_type = df[col].dtypes
if col_type in numerics:
c_min = df[col].min()
c_max = df[col].max()
if str(col_type)[:3] == "int":
if c_min > np.iinfo(np.int8).min and c_max < np.iinfo(np.int8).max:
df[col] = df[col].astype(np.int16)
elif c_min > np.iinfo(np.int16).min and c_max < np.iinfo(np.int16).max:
df[col] = df[col].astype(np.int32)
elif c_min > np.iinfo(np.int32).min and c_max < np.iinfo(np.int32).max:
df[col] = df[col].astype(np.int64)
elif c_min > np.iinfo(np.int64).min and c_max < np.iinfo(np.int64).max:
df[col] = df[col].astype(np.int64)
else:
if (
c_min > np.finfo(np.float16).min
and c_max < np.finfo(np.float16).max
):
df[col] = df[col].astype(np.float32)
elif (
c_min > np.finfo(np.float32).min
and c_max < np.finfo(np.float32).max
):
df[col] = df[col].astype(np.float64)
else:
df[col] = df[col].astype(np.float64)
end_mem = df.memory_usage().sum() / 1024**2
if verbose:
print(
"Mem. usage decreased to {:5.2f} Mb ({:.1f}% reduction)".format(
end_mem, 100 * (start_mem - end_mem) / start_mem
)
)
return df
def unnest(data, name):
try:
date_nested_table = data[["date", name]]
date_nested_table = date_nested_table[
~pd.isna(date_nested_table[name])
].reset_index(drop=True)
daily_dfs_collection = []
for date_index, date_row in date_nested_table.iterrows():
daily_df = pd.read_json(date_row[name])
daily_df["dailyDataDate"] = date_row["date"]
daily_dfs_collection = daily_dfs_collection + [daily_df]
if daily_dfs_collection:
# Concatenate all daily dfs into single df for each row
unnested_table = (
pd.concat(daily_dfs_collection, ignore_index=True)
.
# Set and reset index to move 'dailyDataDate' to front of df
set_index("dailyDataDate")
.reset_index()
)
return reduce_mem_usage(unnested_table, False)
else:
return pd.DataFrame()
except Exception as e:
print(e)
print(f"unnest failed for {name}. returning empty dataframe")
return pd.DataFrame()
def get_unnested_data_dict(data, daily_data_nested_df_names):
df_dict = {}
for df_name in daily_data_nested_df_names:
df_dict[df_name] = unnest(data, df_name)
return df_dict
def get_unnested_data(data, colnames):
return (unnest(data, df_name) for df_name in colnames)
## Find win expectancy and volatility given inning, out, base, run situation.
## no. of runs that score with HR in diff. base situations
baseHr = {1: 1, 2: 2, 3: 2, 4: 3, 5: 2, 6: 3, 7: 3, 8: 4}
def getRunsInn(rpinn):
runsinn = {
0: 1 / ((rpinn * 0.761) + 1),
1: (rpinn * (0.761**2)) / (((rpinn * 0.761) + 1) ** 2),
}
for i in range(2, 11):
v = (rpinn * (0.761**2) * (((rpinn * 0.761) - 0.761 + 1) ** (i - 1))) / (
((rpinn * 0.761) + 1) ** (i + 1)
)
runsinn[i] = v
return runsinn
def getRunExp(rpinn, runsinn):
runExp = {"10": runsinn}
for i in range(0, 3):
for j in range(1, 9):
k = str(j) + str(i)
if k == "10":
continue
runExp[k] = {0: ((tangoRunExp[k]["m"] * rpinn) + tangoRunExp[k]["b"])}
for r in range(1, 11):
runExp[k][r] = (1 - runExp[k][0]) * tangoRunExp[k][r]
return runExp
def getInnWinexp(runExp):
## Chance of home team winning with zero
## outs at the beg. of each inning
innWinexp = {"101": {0: 0.5}}
for i in range(-25, 0):
innWinexp["101"][i] = 0
for i in range(1, 26):
innWinexp["101"][i] = 1
for i in range(9, 0, -1):
for j in range(2, 0, -1):
if j == 2:
next = str(i + 1) + "1"
else:
next = str(i) + "2"
this = str(i) + str(j)
innWinexp[this] = {}
if j == 2:
for k in range(-25, 26):
p = 0
if i == 9 and k > 0:
innWinexp[this][k] = 1
continue
else:
pass
for m in range(0, 11):
if k + m > 25:
iw = 1
else:
iw = innWinexp[next][k + m]
p += runExp["10"][m] * iw
innWinexp[this][k] = p
else:
for k in range(-25, 26):
p = 0
for m in range(0, 11):
if k - m < -25:
iw = 0
else:
iw = innWinexp[next][k - m]
p += runExp["10"][m] * iw
innWinexp[this][k] = p
return innWinexp
def getWinexp(innWinexp, runExp, inn, half, base, outs, rdiff):
if inn > 9:
inn = 9
innkey = str(inn) + str(half)
if outs > 2:
outs = 2
sitkey = str(base) + str(outs)
if half == 2:
next = str(inn + 1) + "1"
else:
next = str(inn) + "2"
if sitkey == "10": ## beginning of half inning
if rdiff > 25:
rdiff = 25
elif rdiff < -25:
rdiff = -25
else:
pass
Winexp = innWinexp[innkey][rdiff]
elif half == 1:
Winexp = 0
for i in range(10, -1, -1):
if rdiff - i < -25:
iw = 0
elif rdiff - i > 25:
iw = 1
else:
iw = innWinexp[next][rdiff - i]
Winexp += runExp[sitkey][i] * iw
else:
Winexp = 0
for i in range(0, 11):
if rdiff - i < -25:
iw = 0
elif rdiff + i > 25:
iw = 1
else:
iw = innWinexp[next][rdiff + i]
Winexp += runExp[sitkey][i] * iw
return Winexp
def getVol(innWinexp, runExp, inn, half, base, outs, rdiff):
## changes if strikeout:
if outs == 2:
outsK = 0
baseK = 1
if half == 1:
halfK = 2
innK = inn
else:
halfK = 1
innK = inn + 1
else:
outsK = outs + 1
baseK, halfK, innK = base, half, inn
WinexpK = getWinexp(innWinexp, runExp, innK, halfK, baseK, outsK, rdiff)
## changes if homerun
if half == 1:
rdiff -= baseHr[base]
else:
rdiff += baseHr[base]
base = 1
WinexpHr = getWinexp(innWinexp, runExp, inn, half, base, outs, rdiff)
return (abs(WinexpHr - WinexpK)) / 0.133
def rpgToInnWinexp(rpg):
rpinn = float(rpg) / 9 ## r/inn
runsinn = getRunsInn(rpinn)
runExp = getRunExp(rpinn, runsinn)
innWinexp = getInnWinexp(runExp)
return innWinexp, runExp
def winnexp_feature(x):
return getWinexp(
innWinexp,
runExp,
x["inning"],
x["halfInning_index"],
x["base_state"],
x["outs_beg"],
x["run_diff"],
)
##################################################################################################
## Functions for extracting and matching ejected player names and getting their playerId
##################################################################################################
# Need to map names to the players.csv or playerBoxScores playerIds
def find_closest_playerName(playerName, players):
players["fuzz_score"] = [fuzz.WRatio(playerName, x) for x in players["playerName"]]
best_match = players.loc[
players["fuzz_score"] == players["fuzz_score"].max(), "playerName"
].iloc[0]
return best_match
def find_playerId(x, players, rosters_players):
# rosters_players is a merge of the rosters df and the players df on the playerId
tmp = players[players["playerName"] == x["playerName"]]
if tmp.shape[0] == 1:
return tmp["playerId"].iloc[0]
else:
# If there are two players with the same name in players, then use the daily roster data to find the player on the matching team
return rosters_players.loc[
(rosters_players["dailyDataDate"] == x["dailyDataDate"])
& (rosters_players["teamId"] == x["teamId"])
& (rosters_players["playerName"] == x["playerName"]),
"playerId",
].iloc[0]
##################################################################################################
# Set up win expectancy variables
rpg = 4.5
innWinexp, runExp = rpgToInnWinexp(rpg)
def game_score_james(x):
"""
# • Start with 50 points
# • Add 1 point for each out recorded (or 3 points per inning)
# • Add 2 points for each inning completed after the fourth
# • Add 1 additional point for every strikeout
# • Remove 2 points for each hit allowed
# • Remove 4 points for each earned run allowed
# • Remove 2 points for each unearned run allowed
# • Remove 1 point for each walk allowed
"""
score = 50
score += x["outsPitching"]
score += 2 * (x["inningsPitched"] - 4)
score += x["strikeOutsPitching"]
score -= 2 * x["hitsPitching"]
score -= 4 * x["earnedRuns"]
score -= 2 * (x["runsPitching"] - x["earnedRuns"])
score -= x["baseOnBallsPitching"] + x["hitByPitchPitching"]
# score = 50 + x['outsPitching'] + 2*(x['inningsPitched'] - 4) + x['strikeOutsPitching'] - 2*x['hitsPitching'] - 4*x['earnedRuns'] - 2*(x['runsPitching'] - x['earnedRuns']) - (x['baseOnBallsPitching']+x['hitByPitchPitching'])
return score
def game_score_tango(x):
"""
Game Score formula (updated by Tom Tango)
# • Start with 40 points
# • Add 2 points for each out recorded (or 6 points per inning)
# • Add 1 additional point for every strikeout
# • Remove 2 points for each walk allowed
# • Remove 2 points for each hit allowed
# • Remove 3 points for each run allowed (earned or unearned)
# • Remove 6 additional points for each home run allowed
"""
score = 40
score += 2 * x["outsPitching"]
score += x["strikeOutsPitching"]
score -= 2 * (x["baseOnBallsPitching"] + x["hitByPitchPitching"])
score -= 2 * x["hitsPitching"]
score -= 3 * x["runsPitching"]
score -= 6 * x["homeRunsPitching"]
return score
pitching_features = [
"gamesPlayedPitching",
"gamesStartedPitching",
"completeGamesPitching",
"shutoutsPitching",
"winsPitching",
"lossesPitching",
"flyOutsPitching",
"airOutsPitching",
"groundOutsPitching",
"runsPitching",
"doublesPitching",
"triplesPitching",
"homeRunsPitching",
"strikeOutsPitching",
"baseOnBallsPitching",
"intentionalWalksPitching",
"hitsPitching",
"hitByPitchPitching",
"atBatsPitching",
"caughtStealingPitching",
"stolenBasesPitching",
"inningsPitched",
"saveOpportunities",
"earnedRuns",
"battersFaced",
"outsPitching",
"pitchesThrown",
"balls",
"strikes",
"hitBatsmen",
"balks",
"wildPitches",
"pickoffsPitching",
"rbiPitching",
"gamesFinishedPitching",
"inheritedRunners",
"inheritedRunnersScored",
"catchersInterferencePitching",
"sacBuntsPitching",
"sacFliesPitching",
"saves",
"holds",
"blownSaves",
"assists",
"putOuts",
"errors",
"chances",
]
train = dt.fread(
"../input/mlb-player-digital-engagement-forecasting/train_updated.csv"
).to_pandas()
test = True
if test:
last_date = train["date"].max()
else:
last_date = 20210430
eng = unnest(train, "nextDayPlayerEngagement")
p_box_scores_og = unnest(train, "playerBoxScores")
teams = pd.read_csv("../input/mlb-player-digital-engagement-forecasting/teams.csv")
players = pd.read_csv("../input/mlb-player-digital-engagement-forecasting/players.csv")
awards_history = pd.read_csv(
"../input/mlb-player-digital-engagement-forecasting/awards.csv"
)
##################################################################################################
## Regex's for extracting and matching ejected player names and getting their playerId
##################################################################################################
team_names = list(teams["teamName"].unique()) + ["Diamondbacks"]
team_regex = re.compile("|".join(map(re.escape, team_names)))
team_full_names = list(teams["name"].unique()) + list(teams["teamName"].unique())
team_full_regex = re.compile("|".join(map(re.escape, team_full_names)))
coaching_names = [
"Assistant Hitting Coach",
"Manager",
"Bench Coach",
"Interim Manager",
"Hitting Coach",
"First Base Coach",
"Pitching Coach",
"bench caoch",
"assistant hitting coach",
"Third Base Coach",
"catching coach",
"field coordinator",
"first base coach",
"hitting coach",
"major league coach",
"manager",
"pitching coach",
"third base coach",
"bench coach",
]
coaching_regex = re.compile("|".join(map(re.escape, coaching_names)))
positions = [
"pitcher",
"catcher",
"first baseman",
"second baseman",
"third baseman",
"shortstop",
"left fielder",
"center fielder",
"right fielder",
"designated hitter",
]
pos_regex = re.compile("|".join(map(re.escape, positions)))
##################################################################################################
colnames = [
"games",
"rosters",
"playerBoxScores",
"teamBoxScores",
"transactions",
"standings",
"awards",
"events",
"playerTwitterFollowers",
"teamTwitterFollowers",
]
hitter_history_feats = [
"hits",
"doubles",
"triples",
"homeRuns",
"rbi",
"totalBases",
"plateAppearances",
"strikeOuts",
"baseOnBalls",
"hitByPitch",
"atBats",
"sacFlies",
]
pitcher_history_feats = [
"gamesPlayedPitching",
"gamesStartedPitching",
"inningsPitched",
"pitchesThrown",
"winsPitching",
"runsPitching",
"homeRunsPitching",
"strikeOutsPitching",
"earnedRuns",
"blownSaves",
"holds",
]
fielder_history_feats = ["errors"]
keep_awards = [
"NLPOW",
"ALPOW",
"NLROM",
"ALROM",
"NLPOM",
"ALPOM",
"NLRRELMON",
"ALRRELMON",
"ALPITOM",
"NLPITOM",
"MLBPLAYOW",
]
keep_annual_awards = [
"ALMVP",
"NLMVP",
"ALCY",
"NLCY",
"ALROY",
"NLROY",
"ALPG",
"NLPG",
"ALSS",
"NLSS",
"ALGG",
"NLGG",
]
##################################################################################################
## Mappings
##################################################################################################
team_mapping = teams.set_index("teamName")["id"].to_dict()
team_mapping["Diamondbacks"] = 109
player_mapping = p_box_scores_og[["playerId", "playerName"]].drop_duplicates()
##################################################################################################
pitchers = players[players["primaryPositionName"] == "Pitcher"]
players["value"] = 1
player_country_dummies = pd.pivot_table(
players,
values="value",
index=["playerId"],
columns=["birthCountry"],
aggfunc="sum",
fill_value=0,
).reset_index()
player_country_dummies.columns = player_country_dummies.columns.str.replace(" ", "_")
quantile_20 = lambda x: x.quantile(0.20)
quantile_20.__name__ = "quantile_20"
quantile_80 = lambda x: x.quantile(0.80)
quantile_80.__name__ = "quantile_80"
# agg_list = ['median','var', quantile_20, quantile_80]
# player_aggs = eng.groupby('playerId')[['target1','target2','target3','target4']].agg({'target1': agg_list,
# 'target2': agg_list,
# 'target3': agg_list,
# 'target4': agg_list}).round(6)
# player_aggs.columns = ["_".join(x) for x in player_aggs.columns.ravel()]
# player_aggs = player_aggs.reset_index()
# player_medians = eng.groupby('playerId')[['target1','target2','target3','target4']].median().round(6).reset_index()
# player_medians = player_medians.rename({'target1': 'target1_p_median',
# 'target2': 'target2_p_median',
# 'target3': 'target3_p_median',
# 'target4': 'target4_p_median'}, axis=1)
# player_variances = eng.groupby('playerId')[['target1','target2','target3','target4']].var().round(6).reset_index()
# player_variances = player_variances.rename({'target1': 'target1_p_var',
# 'target2': 'target2_p_var',
# 'target3': 'target3_p_var',
# 'target4': 'target4_p_var'}, axis=1)
# game_day_player_means = eng.merge(p_box_scores_og[['dailyDataDate','playerId', 'gamePk']], how='left')
# game_day_player_means['game_played'] = game_day_player_means['gamePk'].notnull().astype(int)
# off_day_player_means = game_day_player_means[game_day_player_means['game_played']==0].groupby(['playerId'])[['target1','target2','target3','target4']].mean().round(6).reset_index()
# off_day_player_means = off_day_player_means.rename({'target1': 'target1_p_mean_off_day',
# 'target2': 'target2_p_mean_off_day',
# 'target3': 'target3_p_mean_off_day',
# 'target4': 'target4_p_mean_off_day'}, axis=1)
# game_day_player_means = game_day_player_means[game_day_player_means['game_played']==1].groupby(['playerId'])[['target1','target2','target3','target4']].mean().round(6).reset_index()
# game_day_player_means = game_day_player_means.rename({'target1': 'target1_p_mean_game_day',
# 'target2': 'target2_p_mean_game_day',
# 'target3': 'target3_p_mean_game_day',
# 'target4': 'target4_p_mean_game_day'}, axis=1)
# game_day_player_vars = eng.merge(p_box_scores_og[['dailyDataDate','playerId']])
# game_day_player_vars = game_day_player_vars.groupby('playerId')[['target1','target2','target3','target4']].var().round(6).reset_index()
# game_day_player_vars = game_day_player_vars.rename({'target1': 'target1_p_var_game_day',
# 'target2': 'target2_p_var_game_day',
# 'target3': 'target3_p_var_game_day',
# 'target4': 'target4_p_var_game_day'}, axis=1)
eng["ddd_month"] = np.floor(eng.dailyDataDate / 100).astype(int)
months = eng.ddd_month.unique()
## add one extra month to grab entire data set's TEs
months = np.append(months, months[months.size - 1] + 1)
MAX_MONTH = months[months.size - 1]
ttl_player_medians = []
ttl_player_variances = []
ttl_player_means = []
ttl_roll12_player_medians = []
ttl_roll12_player_variances = []
ttl_roll12_player_means = []
for i in range(months.size - 1):
month = months[i + 1]
roll_month = months[0]
if i > 11:
roll_month = months[i - 12]
# print(str(month) + ' roll:' + str(roll_month))
player_medians = (
eng[eng.ddd_month < month]
.groupby("playerId")[["target1", "target2", "target3", "target4"]]
.median()
.round(6)
.reset_index()
)
player_medians = player_medians.rename(
{
"target1": "target1_p_median",
"target2": "target2_p_median",
"target3": "target3_p_median",
"target4": "target4_p_median",
},
axis=1,
)
player_medians["ddd_month"] = month
roll12_player_medians = (
eng[(eng.ddd_month < month) & (eng.ddd_month >= roll_month)]
.groupby("playerId")[["target1", "target2", "target3", "target4"]]
.median()
.round(6)
.reset_index()
)
roll12_player_medians = roll12_player_medians.rename(
{
"target1": "roll12_target1_p_median",
"target2": "roll12_target2_p_median",
"target3": "roll12_target3_p_median",
"target4": "roll12_target4_p_median",
},
axis=1,
)
roll12_player_medians["ddd_month"] = month
player_variances = (
eng[eng.ddd_month < month]
.groupby("playerId")[["target1", "target2", "target3", "target4"]]
.var()
.round(6)
.reset_index()
)
player_variances = player_variances.rename(
{
"target1": "target1_p_var",
"target2": "target2_p_var",
"target3": "target3_p_var",
"target4": "target4_p_var",
},
axis=1,
)
player_variances["ddd_month"] = month
roll12_player_variances = (
eng[(eng.ddd_month < month) & (eng.ddd_month >= roll_month)]
.groupby("playerId")[["target1", "target2", "target3", "target4"]]
.var()
.round(6)
.reset_index()
)
roll12_player_variances = roll12_player_variances.rename(
{
"target1": "roll12_target1_p_var",
"target2": "roll12_target2_p_var",
"target3": "roll12_target3_p_var",
"target4": "roll12_target4_p_var",
},
axis=1,
)
roll12_player_variances["ddd_month"] = month
player_means = (
eng[eng.ddd_month < month]
.groupby("playerId")[["target1", "target2", "target3", "target4"]]
.mean()
.round(6)
.reset_index()
)
player_means = player_means.rename(
{
"target1": "target1_p_mean",
"target2": "target2_p_mean",
"target3": "target3_p_mean",
"target4": "target4_p_mean",
},
axis=1,
)
player_means["ddd_month"] = month
roll12_player_means = (
eng[(eng.ddd_month < month) & (eng.ddd_month >= roll_month)]
.groupby("playerId")[["target1", "target2", "target3", "target4"]]
.mean()
.round(6)
.reset_index()
)
roll12_player_means = roll12_player_means.rename(
{
"target1": "roll12_target1_p_mean",
"target2": "roll12_target2_p_mean",
"target3": "roll12_target3_p_mean",
"target4": "roll12_target4_p_mean",
},
axis=1,
)
roll12_player_means["ddd_month"] = month
ttl_player_medians.append(player_medians)
ttl_player_variances.append(player_variances)
ttl_player_means.append(player_means)
ttl_roll12_player_medians.append(roll12_player_medians)
ttl_roll12_player_variances.append(roll12_player_variances)
ttl_roll12_player_means.append(roll12_player_means)
dt_player_medians = pd.concat(ttl_player_medians)
dt_player_variances = pd.concat(ttl_player_variances)
dt_player_means = pd.concat(ttl_player_means)
dt_roll12_player_medians = pd.concat(ttl_roll12_player_medians)
dt_roll12_player_variances = pd.concat(ttl_roll12_player_variances)
dt_roll12_player_means = pd.concat(ttl_roll12_player_means)
dt_player_aggregations = dt_player_medians.merge(
dt_player_variances, how="left", on=["playerId", "ddd_month"]
)
dt_player_aggregations = dt_player_aggregations.merge(
dt_player_means, how="left", on=["playerId", "ddd_month"]
)
dt_player_aggregations = dt_player_aggregations.merge(
dt_roll12_player_medians, how="left", on=["playerId", "ddd_month"]
)
dt_player_aggregations = dt_player_aggregations.merge(
dt_roll12_player_variances, how="left", on=["playerId", "ddd_month"]
)
dt_player_aggregations = dt_player_aggregations.merge(
dt_roll12_player_means, how="left", on=["playerId", "ddd_month"]
)
played_game = (
p_box_scores_og.groupby(["playerId", "dailyDataDate"])["gamePk"]
.count()
.reset_index()
)
played_game = played_game.rename({"gamePk": "played_game"}, axis=1)
eng_box = eng.merge(played_game, how="left", on=["dailyDataDate", "playerId"])
eng_box["played_game"] = eng_box["played_game"].fillna(0).clip(upper=1)
ttl_player_gameday_medians = []
ttl_player_gameday_variances = []
ttl_player_gameday_means = []
ttl_roll12_player_gameday_medians = []
ttl_roll12_player_gameday_variances = []
ttl_roll12_player_gameday_means = []
for i in range(months.size - 1):
month = months[i + 1]
roll_month = months[0]
if i > 11:
roll_month = months[i - 12]
# print(str(month) + ' roll:' + str(roll_month))
player_gameday_medians = (
eng_box[eng_box.ddd_month < month]
.groupby(["playerId", "played_game"])[
["target1", "target2", "target3", "target4"]
]
.median()
.round(6)
.reset_index()
)
player_gameday_medians = player_gameday_medians.rename(
{
"target1": "target1_p_gameday_median",
"target2": "target2_p_gameday_median",
"target3": "target3_p_gameday_median",
"target4": "target4_p_gameday_median",
},
axis=1,
)
player_gameday_medians["ddd_month"] = month
roll12_player_gameday_medians = (
eng_box[(eng_box.ddd_month < month) & (eng_box.ddd_month >= roll_month)]
.groupby(["playerId", "played_game"])[
["target1", "target2", "target3", "target4"]
]
.median()
.round(6)
.reset_index()
)
roll12_player_gameday_medians = roll12_player_gameday_medians.rename(
{
"target1": "roll12_target1_p_gameday_median",
"target2": "roll12_target2_p_gameday_median",
"target3": "roll12_target3_p_gameday_median",
"target4": "roll12_target4_p_gameday_median",
},
axis=1,
)
roll12_player_gameday_medians["ddd_month"] = month
player_gameday_variances = (
eng_box[eng_box.ddd_month < month]
.groupby(["playerId", "played_game"])[
["target1", "target2", "target3", "target4"]
]
.var()
.round(6)
.reset_index()
)
player_gameday_variances = player_gameday_variances.rename(
{
"target1": "target1_p_gameday_var",
"target2": "target2_p_gameday_var",
"target3": "target3_p_gameday_var",
"target4": "target4_p_gameday_var",
},
axis=1,
)
player_gameday_variances["ddd_month"] = month
roll12_player_gameday_variances = (
eng_box[(eng_box.ddd_month < month) & (eng_box.ddd_month >= roll_month)]
.groupby(["playerId", "played_game"])[
["target1", "target2", "target3", "target4"]
]
.var()
.round(6)
.reset_index()
)
roll12_player_gameday_variances = roll12_player_gameday_variances.rename(
{
"target1": "roll12_target1_p_gameday_var",
"target2": "roll12_target2_p_gameday_var",
"target3": "roll12_target3_p_gameday_var",
"target4": "roll12_target4_p_gameday_var",
},
axis=1,
)
roll12_player_gameday_variances["ddd_month"] = month
player_gameday_means = (
eng_box[eng_box.ddd_month < month]
.groupby(["playerId", "played_game"])[
["target1", "target2", "target3", "target4"]
]
.mean()
.round(6)
.reset_index()
)
player_gameday_means = player_gameday_means.rename(
{
"target1": "target1_p_gameday_mean",
"target2": "target2_p_gameday_mean",
"target3": "target3_p_gameday_mean",
"target4": "target4_p_gameday_mean",
},
axis=1,
)
player_gameday_means["ddd_month"] = month
roll12_player_gameday_means = (
eng_box[(eng_box.ddd_month < month) & (eng_box.ddd_month >= roll_month)]
.groupby(["playerId", "played_game"])[
["target1", "target2", "target3", "target4"]
]
.mean()
.round(6)
.reset_index()
)
roll12_player_gameday_means = roll12_player_gameday_means.rename(
{
"target1": "roll12_target1_p_gameday_mean",
"target2": "roll12_target2_p_gameday_mean",
"target3": "roll12_target3_p_gameday_mean",
"target4": "roll12_target4_p_gameday_mean",
},
axis=1,
)
roll12_player_gameday_means["ddd_month"] = month
ttl_player_gameday_medians.append(player_gameday_medians)
ttl_player_gameday_variances.append(player_gameday_variances)
ttl_player_gameday_means.append(player_gameday_means)
ttl_roll12_player_gameday_medians.append(roll12_player_gameday_medians)
ttl_roll12_player_gameday_variances.append(roll12_player_gameday_variances)
ttl_roll12_player_gameday_means.append(roll12_player_gameday_means)
dt_player_gameday_medians = pd.concat(ttl_player_gameday_medians)
dt_player_gameday_variances = pd.concat(ttl_player_gameday_variances)
dt_player_gameday_means = pd.concat(ttl_player_gameday_means)
dt_roll12_player_gameday_medians = pd.concat(ttl_roll12_player_gameday_medians)
dt_roll12_player_gameday_variances = pd.concat(ttl_roll12_player_gameday_variances)
dt_roll12_player_gameday_means = pd.concat(ttl_roll12_player_gameday_means)
dt_player_game_aggregations = dt_player_gameday_medians.merge(
dt_player_gameday_variances, how="left", on=["playerId", "ddd_month", "played_game"]
)
dt_player_game_aggregations = dt_player_game_aggregations.merge(
dt_player_gameday_means, how="left", on=["playerId", "ddd_month", "played_game"]
)
dt_player_game_aggregations = dt_player_game_aggregations.merge(
dt_roll12_player_gameday_medians,
how="left",
on=["playerId", "ddd_month", "played_game"],
)
dt_player_game_aggregations = dt_player_game_aggregations.merge(
dt_roll12_player_gameday_variances,
how="left",
on=["playerId", "ddd_month", "played_game"],
)
dt_player_game_aggregations = dt_player_game_aggregations.merge(
dt_roll12_player_gameday_means,
how="left",
on=["playerId", "ddd_month", "played_game"],
)
position_freq = (
p_box_scores_og["positionType"].fillna(-999).value_counts(normalize=True).to_dict()
)
position_target_agg = eng.merge(
p_box_scores_og[
["dailyDataDate", "playerId", "gamePk", "gameTimeUTC", "positionType"]
],
how="left",
)
dh_games = (
position_target_agg[
position_target_agg[["dailyDataDate", "playerId"]].duplicated(keep=False)
]
.sort_values("gameTimeUTC")[["dailyDataDate", "playerId", "gamePk"]]
.reset_index(drop=True)
)
dh_last_game = dh_games[
dh_games[["dailyDataDate", "playerId"]].duplicated(keep="first")
] # games to remove
position_target_agg = position_target_agg[
~(
position_target_agg["playerId"].isin(dh_last_game["playerId"])
& position_target_agg["gamePk"].isin(dh_last_game["gamePk"])
)
]
position_freq = (
position_target_agg["positionType"]
.fillna(-999)
.value_counts(normalize=True)
.to_dict()
)
position_target_agg = (
position_target_agg.groupby("positionType")[
["target1", "target2", "target3", "target4"]
]
.agg(
{
"target1": ["median", "var"],
"target2": ["median", "var"],
"target3": ["median", "var"],
"target4": ["median", "var"],
}
)
.round(6)
)
position_target_agg.columns = [
"_".join(x + ("position",)) for x in position_target_agg.columns.ravel()
]
position_target_agg = position_target_agg.reset_index()
last_day = train[train["date"] == last_date]
(
eng,
games,
rosters,
p_box_scores,
t_box_scores,
transactions,
standings,
awards,
events,
p_twitter,
t_twitter,
) = get_unnested_data(last_day, ["nextDayPlayerEngagement"] + colnames)
# eng_lag = eng.copy()
# eng_lag = eng[['playerId','target1','target2','target3','target4']].copy()
# eng_lag = eng_lag.rename({'target1': 'target1_lag',
# 'target2': 'target2_lag',
# 'target3': 'target3_lag',
# 'target4': 'target4_lag'}, axis=1)
try:
if not p_box_scores.empty:
t_tmp = eng.merge(
p_box_scores[
["dailyDataDate", "playerId", "positionCode", "pitchesThrown"]
],
how="left",
on=["dailyDataDate", "playerId"],
)
t_tmp["position_player_pitching"] = (
(t_tmp["positionCode"] > 1) & (t_tmp["pitchesThrown"] > 0)
).astype(int)
prior_day_pos_player_pitching = t_tmp.loc[
t_tmp["position_player_pitching"] == 1,
["playerId", "position_player_pitching"],
].fillna(0)
else:
eng["position_player_pitching"] = 0
prior_data_pos_player_pitching = eng[["playerId", "position_player_pitching"]]
except Exception as e:
print(e)
eng["position_player_pitching"] = 0
prior_data_pos_player_pitching = eng[["playerId", "position_player_pitching"]]
# Instead of using specific date, find latest date with twitter data available
last_twitter_date = train.loc[train["playerTwitterFollowers"].notnull(), "date"].max()
second_last_twitter_date = train.loc[
train["playerTwitterFollowers"].notnull(), "date"
].iloc[-2]
last_twitter_update = train[train["date"] == last_twitter_date]
second_last_twitter_update = train[train["date"] == second_last_twitter_date]
p_twitter, _ = get_unnested_data(
last_twitter_update, ["playerTwitterFollowers", "teamTwitterFollowers"]
)
p_twitter_recent = p_twitter.copy()
p_twitter_second_last, _ = get_unnested_data(
second_last_twitter_update, ["playerTwitterFollowers", "teamTwitterFollowers"]
)
p_twitter_recent = p_twitter_recent.set_index("playerId")
p_twitter_second_last = p_twitter_second_last.set_index("playerId")
p_twitter_delta = (
(p_twitter_recent["numberOfFollowers"] - p_twitter_second_last["numberOfFollowers"])
.reset_index()
.rename(columns={"numberOfFollowers": "numberOfFollowers_delta"})
)
p_twitter_recent = p_twitter_recent.reset_index()
# games = unnest(train, 'games')
# schedule_21 = pd.read_csv("../input/mlbdata/schedule_2021.csv")
# schedule_21['gameDate'] = pd.to_datetime(schedule_21['gameDate'])
# games['gameDate'] = pd.to_datetime(games['gameDate'])
# games = games.sort_values('gameDate')
# schedule = pd.concat([games[['dailyDataDate', 'homeId', 'gameDate']].rename({'homeId': 'teamId'}, axis=1),
# games[['dailyDataDate', 'awayId', 'gameDate']].rename({'awayId': 'teamId'}, axis=1)])
# schedule = schedule[schedule['dailyDataDate']<20210401]
# schedule = pd.concat([schedule, schedule_21[['dailyDataDate','teamId','gameDate']]])
# schedule['gameDate'] = pd.to_datetime(schedule['gameDate'])
# all_dates = pd.DataFrame(list(itertools.product(pd.date_range(start="2018-01-01", end="2021-12-31"), schedule['teamId'].unique())), columns=['gameDate', 'teamId'])
# all_dates = all_dates.merge(schedule, how='outer', on=['gameDate','teamId'])
# all_dates = all_dates.sort_values(['teamId','gameDate']).drop_duplicates()
# all_dates['dailyDataDate_lead'] = all_dates.groupby('teamId')['dailyDataDate'].shift(1)
# all_dates = all_dates[all_dates['dailyDataDate_lead'].notnull()].reset_index(drop=True)
# all_dates['nextDayGame'] = 1
awards_dict = (
awards_history[awards_history["awardId"].isin(keep_awards + keep_annual_awards)]
.groupby(["playerId", "awardId"])
.size()
.to_dict()
)
awards_dict_tmp = {
n: grp.to_dict("list")
for n, grp in awards_history.loc[
awards_history["awardId"].isin(keep_awards + keep_annual_awards),
["awardId", "playerId"],
].groupby("playerId")
}
awards_dict = {}
for k, v in awards_dict_tmp.items():
if not k in awards_dict:
awards_dict[k] = {}
counts = np.unique(v["awardId"], return_counts=True)
for feat, value in zip(counts[0], counts[1]):
awards_dict[k][feat] = value
hitter_history_dict = {}
fielder_history_dict = {}
pitcher_history_dict = {}
for i, data in tqdm(train[train["date"] <= last_date].iterrows()):
try:
data = data.to_frame().T
daily_data_date = data["date"].iloc[0]
season = int(str(daily_data_date)[:4])
p_box_scores, games, rosters, awards = get_unnested_data(
data, ["playerBoxScores", "games", "rosters", "awards"]
)
if rosters.empty:
rosters = prior_day_rosters
prior_day_rosters = rosters.copy()
if not games.empty:
games_filtered = games.loc[
games["gameType"].isin(["R", "F", "D", "L", "W", "C", "P"])
& ~games["detailedGameState"].isin(["Postponed"])
]
if not games_filtered.empty:
schedule_day = pd.concat(
[
games_filtered[
[
"dailyDataDate",
"gamePk",
"homeId",
"gameDate",
"gameTimeUTC",
"homeWinner",
]
].rename({"homeId": "teamId", "homeWinner": "winner"}, axis=1),
games_filtered[
[
"dailyDataDate",
"gamePk",
"awayId",
"gameDate",
"gameTimeUTC",
"awayWinner",
]
].rename({"awayId": "teamId", "awayWinner": "winner"}, axis=1),
]
)
schedule_day = schedule_day.sort_values("gameTimeUTC")
if not schedule_day.empty and not p_box_scores.empty:
game_rosters = schedule_day.merge(
rosters, how="left", on=["gameDate", "teamId"]
)
game_rosters = game_rosters[
game_rosters["playerId"].notnull()
] # missing roster for Nationals 20200910
game_rosters["playerId"] = game_rosters["playerId"].astype(int)
p_box_scores = p_box_scores.sort_values("gameTimeUTC")
p_box_scores["gameDate"] = pd.to_datetime(p_box_scores["gameDate"])
p_box_scores["season"] = p_box_scores["gameDate"].dt.year
player_history_daily = game_rosters.merge(
p_box_scores, how="left", on=["gamePk", "playerId"]
)
player_history_daily["gameTimeUTC_y"] = player_history_daily[
"gameTimeUTC_y"
].fillna(player_history_daily["gameTimeUTC_x"])
# NOTE: dailyDataDate==2020918 gamePk==631122 Start time of 2020-09-18T03:33:00Z is not accurate; that would imply the game started the day before at ~11:30PM local time
player_history_daily = player_history_daily.sort_values(
["playerId", "gameTimeUTC_y"]
) # SORT BY gameTimeUTC from p_box_scores. `gameTimeUTC` is not accurate from the `games` data
player_history_daily[hitter_history_feats] = player_history_daily[
hitter_history_feats
].fillna(0)
hitter_history_tmp = {
n: grp.to_dict("list")
for n, grp in player_history_daily[
hitter_history_feats + ["season", "playerId"]
].groupby("playerId")
}
for k, v in hitter_history_tmp.items():
if not k in hitter_history_dict:
hitter_history_dict[k] = v
else:
for feat in hitter_history_feats + ["season"]:
hitter_history_dict[k][feat].extend(v[feat])
# For hitters, only use games they played in. Pitchers need off days filled in because it's important to account for rest/off days
# Fill in days with 0 if hitter isn't in daily box scores
# for k,v in hitter_history_dict.items():
# if not k in hitter_history_tmp:
# for feat in hitter_history_feats + ['season']:
# hitter_history_dict[k][feat].append(season if feat=='season' else 0.0)
fielder_history_tmp = {
n: grp.to_dict("list")
for n, grp in player_history_daily[
fielder_history_feats + ["season", "playerId"]
].groupby("playerId")
}
for k, v in fielder_history_tmp.items():
if not k in fielder_history_dict:
fielder_history_dict[k] = v
else:
for feat in fielder_history_feats + ["season"]:
fielder_history_dict[k][feat].extend(v[feat])
pitcher_history_tmp = {
n: grp.to_dict("list")
for n, grp in p_box_scores.loc[
p_box_scores["positionName"] == "Pitcher",
pitcher_history_feats + ["season", "playerId"],
].groupby("playerId")
}
for k, v in pitcher_history_tmp.items():
if not k in pitcher_history_dict:
pitcher_history_dict[k] = v
else:
for feat in pitcher_history_feats + ["season"]:
pitcher_history_dict[k][feat].extend(v[feat])
# Fill in days with 0 if pitcher isn't in daily box scores
for k, v in pitcher_history_dict.items():
if not k in pitcher_history_tmp:
for feat in pitcher_history_feats + ["season"]:
pitcher_history_dict[k][feat].append(
season if feat == "season" else 0.0
)
except Exception as e:
# If fails, just move on to the next day
print(f"history dicts loop failed: {e}")
pass
try:
if not awards.empty:
awards_filtered = awards[
awards["awardId"].isin(keep_awards + keep_annual_awards)
].reset_index(drop=True)
# Update awards counts
awards_dict_tmp = {
n: grp.to_dict("list")
for n, grp in awards_filtered[["awardId", "playerId"]].groupby(
"playerId"
)
}
for k, v in awards_dict_tmp.items():
try:
if not k in awards_dict:
awards_dict[k] = {}
counts = np.unique(v["awardId"], return_counts=True)
for feat, value in zip(counts[0], counts[1]):
if feat in awards_dict[k]:
awards_dict[k][feat] += value
else:
awards_dict[k][feat] = value
except:
# If fails, move on to the next one
pass
except Exception as e:
# If fails, don't worry about updating dict
print(e)
pass
games_og = unnest(train, "games")
schedule_og = pd.concat(
[
games_og.loc[
games_og["gameType"].isin(["R", "F", "D", "L", "W", "C", "P"])
& ~games_og["detailedGameState"].isin(["Postponed"]),
[
"dailyDataDate",
"gamePk",
"homeId",
"gameDate",
"gameTimeUTC",
"homeWinner",
],
].rename({"homeId": "teamId", "homeWinner": "winner"}, axis=1),
games_og.loc[
games_og["gameType"].isin(["R", "F", "D", "L", "W", "C", "P"])
& ~games_og["detailedGameState"].isin(["Postponed"]),
[
"dailyDataDate",
"gamePk",
"awayId",
"gameDate",
"gameTimeUTC",
"awayWinner",
],
].rename({"awayId": "teamId", "awayWinner": "winner"}, axis=1),
]
)
schedule_og = schedule_og.sort_values("gameTimeUTC")
schedule_og = schedule_og[schedule_og["dailyDataDate"] <= last_date]
schedule_og["gameDate"] = pd.to_datetime(schedule_og["gameDate"])
team_win_history = {}
team_win_dict = schedule_og.groupby("teamId")["winner"].apply(list).to_dict()
for k, v in team_win_dict.items():
if not k in team_win_history:
team_win_history[k] = v
else:
team_win_history[k].extend(v)
win_streaks = {
k: v[::-1].index(0) if 0 in v else len(v) for k, v in team_win_history.items()
}
# Load models
lgb_target1 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target1_v30_full.txt"
)
lgb_target2 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target2_v30_full.txt"
)
lgb_target3 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target3_v30_full.txt"
)
lgb_target4 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target4_v30_full.txt"
)
lgb_dblsqrt_target1 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target1_dblsqrt_full.txt"
)
lgb_dblsqrt_target2 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target2_dblsqrt_full.txt"
)
lgb_dblsqrt_target3 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target3_dblsqrt_full.txt"
)
lgb_dblsqrt_target4 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target4_dblsqrt_full.txt"
)
lgb_bfa_target1 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target1_v30_bfa_full.txt"
)
lgb_bfa_target2 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target2_v30_bfa_full.txt"
)
lgb_bfa_target3 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target3_v30_bfa_full.txt"
)
lgb_bfa_target4 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target4_v30_bfa_full.txt"
)
xgb_target1 = xgb.Booster()
xgb_target2 = xgb.Booster()
xgb_target3 = xgb.Booster()
xgb_target4 = xgb.Booster()
xgb_target1.load_model("../input/d/brandenkmurray/mlbmodels/xgb_target1_v30_full.txt")
xgb_target2.load_model("../input/d/brandenkmurray/mlbmodels/xgb_target2_v30_full.txt")
xgb_target3.load_model("../input/d/brandenkmurray/mlbmodels/xgb_target3_v30_full.txt")
xgb_target4.load_model("../input/d/brandenkmurray/mlbmodels/xgb_target4_v30.txt")
lgb_john_target1 = lgb.Booster(
model_file="../input/mlb-models-and-files/lgb_target1_dubs_tripsX_all.txt"
)
lgb_john_target2 = lgb.Booster(
model_file="../input/mlb-models-and-files/lgb_target2_dubs_tripsX_all.txt"
)
lgb_john_target3 = lgb.Booster(
model_file="../input/mlb-models-and-files/lgb_target3_dubs_tripsX_all.txt"
)
lgb_john_target4 = lgb.Booster(
model_file="../input/mlb-models-and-files/lgb_target4_dubs_tripsX_all.txt"
)
lgb_dart_target1 = lgb.Booster(
model_file="../input/dart-model/lgb_target1_dubs_trips_dart_full_data.txt"
)
lgb_dart_target2 = lgb.Booster(
model_file="../input/dart-model/lgb_target2_dubs_trips_dart_full_data.txt"
)
lgb_dart_target3 = lgb.Booster(
model_file="../input/dart-model/lgb_target3_dubs_trips_dart_full_data.txt"
)
lgb_dart_target4 = lgb.Booster(
model_file="../input/dart-model/lgb_target4_dubs_trips_dart_full_data.txt"
)
yesterday = pd.DataFrame()
t = []
sub_list = []
env = mlb.make_env() # initialize the environment
iter_test = env.iter_test() # iterator which loops over each date in test set
for i, (data, sub) in enumerate(iter_test):
# for i, (i2, data) in enumerate(train[(train['date']>=20210501) & (train['date']<=20210731)].iloc[1:].iterrows()):
### REMOVE below
# data = data.to_frame().T
# sub = unnest(data, 'nextDayPlayerEngagement')
# sub = sub.rename(columns={'target1': 'target1_act', 'target2': 'target2_act', 'target3': 'target3_act', 'target4': 'target4_act'})
# sub['date_playerId'] = pd.to_datetime(sub['engagementMetricsDate']).dt.strftime("%Y%m%d") + "_" + sub['playerId'].astype(str)
### REMOVE above
### UNCOMMENT BELOW
sub = sub.reset_index()
sub = sub.rename({"date": "dailyDataDate"}, axis=1)
sub["playerId"] = sub["date_playerId"].apply(lambda x: int(x.split("_")[1]))
data = data.reset_index()
data = data.rename({"index": "date"}, axis=1)
### UNCOMMENT ABOVE
try:
season = int(str(data["date"].iloc[0])[:4])
except:
season = 2021.0
try:
(
games,
rosters,
p_box_scores,
t_box_scores,
transactions,
standings,
awards,
events,
p_twitter,
t_twitter,
) = get_unnested_data(data, colnames)
eng_shape = sub.shape
t_tmp = sub.copy()
if rosters.empty:
rosters = prior_day_rosters
prior_day_rosters = rosters.copy()
if not p_twitter.empty:
# Get twitter follower delta if not the first month
if not p_twitter_recent.empty:
p_twitter = p_twitter.set_index("playerId")
p_twitter_recent = p_twitter_recent.set_index("playerId")
p_twitter_delta = (
(
p_twitter["numberOfFollowers"]
- p_twitter_recent["numberOfFollowers"]
)
.reset_index()
.rename(columns={"numberOfFollowers": "numberOfFollowers_delta"})
)
p_twitter = p_twitter.reset_index()
p_twitter_recent = p_twitter
if not games.empty:
schedule_daily = pd.concat(
[
games.loc[
games["gameType"].isin(["R", "F", "D", "L", "W", "C", "P"])
& ~games["detailedGameState"].isin(["Postponed"]),
[
"dailyDataDate",
"gamePk",
"homeId",
"gameDate",
"gameTimeUTC",
"homeWinner",
],
].rename({"homeId": "teamId", "homeWinner": "winner"}, axis=1),
games.loc[
games["gameType"].isin(["R", "F", "D", "L", "W", "C", "P"])
& ~games["detailedGameState"].isin(["Postponed"]),
[
"dailyDataDate",
"gamePk",
"awayId",
"gameDate",
"gameTimeUTC",
"awayWinner",
],
].rename({"awayId": "teamId", "awayWinner": "winner"}, axis=1),
]
)
schedule_daily = schedule_daily.sort_values("gameTimeUTC")
team_win_dict = (
schedule_daily.groupby("teamId")["winner"].apply(list).to_dict()
)
for k, v in team_win_dict.items():
if not k in team_win_history:
team_win_history[k] = v
else:
team_win_history[k].extend(v)
win_streaks = {
k: v[::-1].index(0) if 0 in v else len(v)
for k, v in team_win_history.items()
}
if not schedule_daily.empty and not p_box_scores.empty:
game_rosters = schedule_daily.merge(
rosters, how="left", on=["gameDate", "teamId"]
)
game_rosters = game_rosters[
game_rosters["playerId"].notnull()
] # missing roster for Nationals 20200910
game_rosters["playerId"] = game_rosters["playerId"].astype(int)
p_box_scores = p_box_scores.sort_values("gameTimeUTC")
p_box_scores["gameDate"] = pd.to_datetime(p_box_scores["gameDate"])
p_box_scores["season"] = p_box_scores["gameDate"].dt.year
player_history_daily = game_rosters.merge(
p_box_scores, how="left", on=["gamePk", "playerId"]
)
player_history_daily["gameTimeUTC_y"] = player_history_daily[
"gameTimeUTC_y"
].fillna(player_history_daily["gameTimeUTC_x"])
# NOTE: dailyDataDate==2020918 gamePk==631122 Start time of 2020-09-18T03:33:00Z is not accurate; that would imply the game started the day before at ~11:30PM local time
player_history_daily = player_history_daily.sort_values(
["playerId", "gameTimeUTC_y"]
) # SORT BY gameTimeUTC from p_box_scores. `gameTimeUTC` is not accurate from the `games` data
player_history_daily[hitter_history_feats] = player_history_daily[
hitter_history_feats
].fillna(0)
hitter_history_tmp = {
n: grp.to_dict("list")
for n, grp in player_history_daily[
hitter_history_feats + ["season", "playerId"]
].groupby("playerId")
}
for k, v in hitter_history_tmp.items():
if not k in hitter_history_dict:
hitter_history_dict[k] = v
else:
for feat in hitter_history_feats + ["season"]:
hitter_history_dict[k][feat].extend(v[feat])
# For hitters, only use games they played in. Pitchers need off days filled in because it's important to account for rest/off days
# Fill in days with 0 if hitter isn't in daily box scores
# for k,v in hitter_history_dict.items():
# if not k in hitter_history_tmp:
# for feat in hitter_history_feats + ['season']:
# hitter_history_dict[k][feat].append(season if feat=='season' else 0.0)
fielder_history_tmp = {
n: grp.to_dict("list")
for n, grp in player_history_daily[
fielder_history_feats + ["season", "playerId"]
].groupby("playerId")
}
for k, v in fielder_history_tmp.items():
if not k in fielder_history_dict:
fielder_history_dict[k] = v
else:
for feat in fielder_history_feats + ["season"]:
fielder_history_dict[k][feat].extend(v[feat])
pitcher_history_tmp = {
n: grp.to_dict("list")
for n, grp in p_box_scores.loc[
p_box_scores["positionName"] == "Pitcher",
pitcher_history_feats + ["season", "playerId"],
].groupby("playerId")
}
for k, v in pitcher_history_tmp.items():
if not k in pitcher_history_dict:
pitcher_history_dict[k] = v
else:
for feat in pitcher_history_feats + ["season"]:
pitcher_history_dict[k][feat].extend(v[feat])
# Fill in days with 0 if pitcher isn't in daily box scores
for k, v in pitcher_history_dict.items():
if not k in pitcher_history_tmp:
for feat in pitcher_history_feats + ["season"]:
pitcher_history_dict[k][feat].append(
season if feat == "season" else 0.0
)
days_of_history = list(
range(2, 21)
) # [2,3,4,5,7,10,20] #also could be games_of_history depending how its used
max_days_of_history = np.max(days_of_history)
hitting_history_features = {}
pitching_history_features = {}
fielding_history_features = {}
for k, v in hitter_history_dict.items():
# only need to include players in the current eng
hitting_history_features[k] = {}
hitting_history_features[k]["hit_streak"] = (
v["hits"][::-1].index(0) if 0 in v["hits"] else len(v["hits"])
)
for feat in hitter_history_feats:
d = hitter_history_dict[k][feat]
hitting_history_features[k][f"{feat}_season"] = sum(
[
f
for seas, f in zip(hitter_history_dict[k]["season"], d)
if seas == season
]
)
if feat not in ["sacFlies", "atBats"]:
d_padded = np.zeros(max_days_of_history)
d_padded[: np.minimum(max_days_of_history, len(d))] = d[
-np.minimum(max_days_of_history, len(d)) :
][::-1]
d_cumsum = nb_cumsum(d_padded)
for day in days_of_history:
hitting_history_features[k][f"{feat}_last{day}"] = d_cumsum[
day - 1
]
# hitting_history_features[k][f'{feat}_{day-1}_games_ago'] = d_padded[day-1]
hitting_history_df = (
pd.DataFrame.from_dict(hitting_history_features, orient="index")
.reset_index()
.rename({"index": "playerId"}, axis=1)
)
if "homeRuns_season" in hitting_history_df.columns:
hitting_history_df["homeRuns_rank"] = hitting_history_df[
"homeRuns_season"
].rank(method="min", ascending=False)
hitting_history_df["BA"] = (
hitting_history_df["hits_season"] / hitting_history_df["atBats_season"]
)
hitting_history_df["OBP"] = hitting_history_df[
["hits_season", "baseOnBalls_season", "hitByPitch_season"]
].sum(axis=1) / hitting_history_df[
[
"atBats_season",
"baseOnBalls_season",
"hitByPitch_season",
"sacFlies_season",
]
].sum(
axis=1
)
hitting_history_df["SLG"] = (
(
hitting_history_df["hits_season"]
- hitting_history_df[
["doubles_season", "triples_season", "homeRuns_season"]
].sum(axis=1)
)
+ 2 * hitting_history_df["doubles_season"]
+ 3 * hitting_history_df["triples_season"]
+ 4 * hitting_history_df["homeRuns_season"]
) / hitting_history_df["atBats_season"]
for k, v in fielder_history_dict.items():
# only need to include players in the current eng
fielding_history_features[k] = {}
for feat in fielder_history_feats:
d = fielder_history_dict[k][feat]
d_padded = np.zeros(max_days_of_history)
d_padded[: np.minimum(max_days_of_history, len(d))] = d[
-np.minimum(max_days_of_history, len(d)) :
][::-1]
# d_padded = np.pad(d[-days_of_history:], (np.maximum(0, days_of_history-len(d)+1), 0))[::-1]
d_cumsum = nb_cumsum(d_padded)
for day in days_of_history:
# fielding_history_features[k][f'{feat}_last{day}'] = d_cumsum[day-1]
fielding_history_features[k][
f"{feat}_{day-1}_games_ago"
] = d_padded[day - 1]
fielding_history_df = (
pd.DataFrame.from_dict(fielding_history_features, orient="index")
.reset_index()
.rename({"index": "playerId"}, axis=1)
)
for k, v in pitcher_history_dict.items():
# only need to include players in the current eng
pitching_history_features[k] = {}
season_starts = [
starts
for seas, starts in zip(
pitcher_history_dict[k]["season"],
pitcher_history_dict[k]["gamesStartedPitching"],
)
if seas == season
]
season_played = [
played
for seas, played in zip(
pitcher_history_dict[k]["season"],
pitcher_history_dict[k]["gamesPlayedPitching"],
)
if seas == season
]
pitching_history_features[k]["season_starts_to_date"] = sum(season_starts)
pitching_history_features[k]["days_since_last_start"] = (
season_starts[::-1].index(1.0)
if 1 in season_starts
else len(season_starts)
)
pitching_history_features[k]["days_since_last_played"] = (
season_played[::-1].index(1.0)
if 1 in season_played
else len(season_played)
)
for feat in [
"gamesPlayedPitching",
"gamesStartedPitching",
"inningsPitched",
"pitchesThrown",
"winsPitching",
"runsPitching",
"homeRunsPitching",
"strikeOutsPitching",
"earnedRuns",
"blownSaves",
"holds",
]:
d = pitcher_history_dict[k][feat]
pitching_history_features[k][f"{feat}_season"] = sum(
[
f
for seas, f in zip(pitcher_history_dict[k]["season"], d)
if seas == season
]
)
d_padded = np.pad(d, (np.maximum(0, max_days_of_history - len(d)), 0))[
::-1
]
d_cumsum = nb_cumsum(d_padded)
for day in days_of_history:
pitching_history_features[k][f"{feat}_last{day}"] = d_cumsum[
day - 1
]
pitching_history_features[k][
f"{feat}_{day-1}_games_ago"
] = d_padded[day - 1]
pitching_history_df = (
pd.DataFrame.from_dict(pitching_history_features, orient="index")
.reset_index()
.rename({"index": "playerId"}, axis=1)
)
if not p_twitter.empty:
p_twitter_recent = p_twitter
# How to handle doubleheaders? Taking stats from first game for now
if not p_box_scores.empty and not t_box_scores.empty:
t_tmp = t_tmp.merge(
p_box_scores, how="left", on=["dailyDataDate", "playerId"]
)
dh_games = (
t_tmp[t_tmp[["dailyDataDate", "playerId"]].duplicated(keep=False)]
.sort_values("gameTimeUTC")[["dailyDataDate", "playerId", "gamePk"]]
.reset_index(drop=True)
)
dh_last_game = dh_games[
dh_games[["dailyDataDate", "playerId"]].duplicated(keep="first")
] # games to remove
t_tmp = t_tmp[
~(
t_tmp["playerId"].isin(dh_last_game["playerId"])
& t_tmp["gamePk"].isin(dh_last_game["gamePk"])
)
]
t_tmp["game_score_james"] = game_score_james(t_tmp)
t_tmp["game_score_tango"] = game_score_tango(t_tmp)
t_tmp["position_player_pitching"] = (
(t_tmp["positionCode"] > 1) & (t_tmp["pitchesThrown"] > 0)
).astype(int)
t_tmp["pitcher_hit_home_run"] = (
(t_tmp["positionCode"] == 1) & (t_tmp["homeRuns"] > 0)
).astype(int)
# t_tmp['pos_player_pitched_prior_day'] = 0
# if not prior_day_pos_player_pitching.empty:
# t_tmp['pos_player_pitched_prior_day'] = t_tmp['playerId'].map(dict(zip(prior_day_pos_player_pitching.playerId, prior_day_pos_player_pitching.position_player_pitching)))
t_tmp["no_hitter"] = (
(t_tmp["inningsPitched"] >= 9) & (t_tmp["hitsPitching"] == 0)
).astype(int)
t_tmp["no_hitter_league"] = t_tmp["no_hitter"].max()
t_tmp["position_player_pitching_league"] = t_tmp[
"position_player_pitching"
].max()
t_tmp["game_hour"] = (
pd.to_datetime(t_tmp["gameTimeUTC"]) + pd.Timedelta(hours=-5)
).dt.hour
t_tmp = t_tmp.merge(
t_box_scores,
how="left",
on=["gamePk", "teamId"],
suffixes=["", "_team_box_score"],
)
t_tmp["positionType_freq"] = (
t_tmp["positionType"].fillna(-999).map(position_freq)
)
if "positionType" in t_tmp.columns:
t_tmp = t_tmp.merge(position_target_agg, how="left", on="positionType")
if t_tmp.shape[0] != eng_shape[0]:
print(
"t_tmp length does not match engagement frame length, check for duplicated data"
)
t_tmp = t_tmp[~t_tmp[["playerId"]].duplicated()]
else:
if "teamId" not in t_tmp.columns and not rosters.empty:
t_tmp = t_tmp.merge(
rosters[["playerId", "teamId"]], how="left", on="playerId"
)
else:
t_tmp["teamId"] = np.nan
if t_tmp.shape[0] != eng_shape[0]:
print(
"teamId: t_tmp length does not match engagement frame length, check for duplicated data"
)
t_tmp = t_tmp[~t_tmp[["playerId"]].duplicated()]
# Did player have a walk-off hit/home run?
if not events.empty:
events = events.sort_values(
["inning", "halfInning", "atBatIndex", "eventId"],
ascending=[True, False, True, True],
)
last_play = events.groupby("gamePk").tail(1)
# filter out top of inning because one game was ended after the top of the inning
walk_offs = last_play[
(last_play["halfInning"] == "bottom") & (last_play["rbi"] > 0)
][["dailyDataDate", "hitterId", "pitcherId", "rbi", "event"]]
walk_offs.columns = [
"dailyDataDate",
"hitterId",
"pitcherId",
"walk_off_rbi",
"walk_off_hr",
]
walk_offs["walk_off_hr"] = (
walk_offs["walk_off_hr"].isin(["Home Run"])
).astype(int)
t_tmp = t_tmp.merge(
walk_offs[
["dailyDataDate", "hitterId", "walk_off_hr", "walk_off_rbi"]
].rename({"hitterId": "playerId"}, axis=1),
how="left",
on=["dailyDataDate", "playerId"],
)
t_tmp = t_tmp.merge(
walk_offs[
["dailyDataDate", "pitcherId", "walk_off_hr", "walk_off_rbi"]
].rename({"pitcherId": "playerId"}, axis=1),
how="left",
on=["dailyDataDate", "playerId"],
suffixes=["", "_pitcher"],
)
t_tmp[
[
"walk_off_rbi",
"walk_off_hr",
"walk_off_hr_pitcher",
"walk_off_rbi_pitcher",
]
] = t_tmp[
[
"walk_off_rbi",
"walk_off_hr",
"walk_off_hr_pitcher",
"walk_off_rbi_pitcher",
]
].fillna(
0
)
t_tmp["walk_off_league"] = t_tmp["walk_off_rbi"].max()
hr_dist = (
events[events["event"] == "Home Run"]
.groupby("hitterId")["totalDistance"]
.max()
.reset_index()
)
hr_launchSpeed = (
events[events["event"] == "Home Run"]
.groupby("hitterId")["launchSpeed"]
.max()
.reset_index()
)
t_tmp = t_tmp.merge(
hr_dist.rename({"hitterId": "playerId"}, axis=1),
how="left",
on="playerId",
)
t_tmp = t_tmp.merge(
hr_launchSpeed.rename({"hitterId": "playerId"}, axis=1),
how="left",
on="playerId",
)
# How long did a starting pitcher go without a hit? (Did they start picking up potential no-hitter hype?)
starters = events[events["isStarter"] == 1].reset_index(drop=True)
starters["hit"] = (
starters["event"]
.isin(["Single", "Double", "Triple", "Home Run"])
.astype(int)
)
starters["hits_cumsum"] = starters.groupby("pitcherId")["hit"].cumsum()
starters_first_hit_inning = (
starters[starters["hits_cumsum"] == 1]
.groupby("pitcherId")
.first()[["inning", "outs"]]
.reset_index()
)
starters_first_hit_inning["inning"] = (
starters_first_hit_inning["inning"]
+ starters_first_hit_inning["outs"] / 10
)
starters_first_hit_inning = starters_first_hit_inning.rename(
{"inning": "pitcher_first_hit_inning"}, axis=1
)
t_tmp = t_tmp.merge(
starters_first_hit_inning[["pitcherId", "pitcher_first_hit_inning"]],
how="left",
left_on="playerId",
right_on="pitcherId",
)
starters_first_mob_inning = starters[
~starters["menOnBase"].isin([None, "Empty"])
]
starters_first_mob_inning = (
starters_first_mob_inning.groupby("pitcherId")
.first()[["inning", "outs"]]
.reset_index()
)
starters_first_mob_inning["inning"] = (
starters_first_mob_inning["inning"]
+ starters_first_mob_inning["outs"] / 10
)
starters_first_mob_inning = starters_first_mob_inning.rename(
{"inning": "pitcher_first_mob_inning"}, axis=1
)
t_tmp = t_tmp.merge(
starters_first_mob_inning[["pitcherId", "pitcher_first_mob_inning"]],
how="left",
left_on="playerId",
right_on="pitcherId",
)
# Pitch features
nastyFactor_features = (
events[events["type"] == "pitch"]
.groupby("pitcherId")["nastyFactor"]
.agg(["mean", "median", "min", "max"])
.reset_index()
.rename(
columns={
f: f"nastyFactor_{f}" for f in ["mean", "median", "max", "min"]
}
)
.rename(columns={"pitcherId": "playerId"})
)
t_tmp = t_tmp.merge(nastyFactor_features, how="left", on="playerId")
# Calculate player Win Probability Added
# need to get assign 100% WPA to winning team to assign WPA scores to correct player/team
player_wpa = pd.Series(dtype=float)
for gamePk, game in events.groupby("gamePk"):
game = game.reset_index(drop=True)
game["run_diff"] = game["homeScore"] - game["awayScore"]
game["halfInning_index"] = game["halfInning"].map(
{"top": 1, "bottom": 2}
)
game["base_state"] = game["menOnBase"].map(
{None: np.nan, "Empty": 1, "Men_On": 2, "RISP": 3, "Loaded": 8}
)
game["base_state"] = game["base_state"].ffill().fillna(1).astype(int)
game["outs_beg"] = np.maximum(game["outs"] - 1, 0)
game["win_exp"] = game.apply(winnexp_feature, axis=1)
game["win_exp_lag"] = game["win_exp"].shift(-1)
game.loc[game.shape[0] - 1, "win_exp_lag"] = (
1
if game.loc[game.shape[0] - 1, "homeScore"]
> game.loc[game.shape[0] - 1, "awayScore"]
else 0
)
game["win_exp_delta"] = game["win_exp_lag"] - game["win_exp"]
# Increases in the top of the inning are assigned to the pitcher
# Increases in the bottom of the inning are assigned to the hitter
pitcher_wpa_top = (
game.loc[
(game["halfInning"] == "top") & (game["win_exp_delta"] > 0),
["pitcherId", "win_exp_delta"],
]
.groupby("pitcherId")["win_exp_delta"]
.sum()
)
hitter_wpa_top = (
game.loc[
(game["halfInning"] == "top") & (game["win_exp_delta"] > 0),
["hitterId", "win_exp_delta"],
]
.groupby("hitterId")["win_exp_delta"]
.sum()
)
hitter_wpa_top = -hitter_wpa_top
pitcher_wpa_bot = (
game.loc[
(game["halfInning"] == "bottom") & (game["win_exp_delta"] > 0),
["pitcherId", "win_exp_delta"],
]
.groupby("pitcherId")["win_exp_delta"]
.sum()
)
hitter_wpa_bot = (
game.loc[
(game["halfInning"] == "bottom") & (game["win_exp_delta"] > 0),
["hitterId", "win_exp_delta"],
]
.groupby("hitterId")["win_exp_delta"]
.sum()
)
pitcher_wpa_bot = -pitcher_wpa_bot
player_wpa = player_wpa.add(pitcher_wpa_top, fill_value=0)
player_wpa = player_wpa.add(hitter_wpa_top, fill_value=0)
player_wpa = player_wpa.add(pitcher_wpa_bot, fill_value=0)
player_wpa = player_wpa.add(hitter_wpa_bot, fill_value=0)
player_wpa = player_wpa.reset_index()
player_wpa = player_wpa.rename({"index": "playerId", 0: "wpa"}, axis=1)
t_tmp = t_tmp.merge(player_wpa, how="left", on="playerId")
t_tmp["wpa_daily_max"] = t_tmp["wpa"].max()
t_tmp["wpa_rank"] = t_tmp["wpa"].rank(method="min", ascending=False)
# get ejections
ejections = events.loc[
events["event"] == "Ejection", ["dailyDataDate", "description"]
].reset_index(drop=True)
if not ejections.empty:
ejections["description"] = [
x.split(" ejected by")[0] for x in ejections["description"]
]
# Get team; needed for coach_ejected feature
ejections["teamName"] = [
team_regex.findall(x)[0] if team_regex.findall(x) else None
for x in ejections["description"]
] # else None to account for names not spelled in a way that matches the regex
ejections["teamId"] = ejections["teamName"].map(team_mapping)
ejections["coach_ejected"] = [
1 if coaching_regex.search(x) else 0
for x in ejections["description"]
]
ejections["player_ejected"] = 1 - ejections["coach_ejected"]
# Get player name
ejections["playerName"] = [
team_full_regex.sub("", " ".join(x.split()))
for x in ejections["description"]
]
ejections["playerName"] = [
coaching_regex.sub("", " ".join(x.split()))
for x in ejections["playerName"]
]
ejections["playerName"] = [
pos_regex.sub("", " ".join(x.split())).strip()
for x in ejections["playerName"]
]
# If there is no match for a player use fuzzywuzzy to find the closest match
ejections.loc[
(ejections["player_ejected"] == 1), "playerName"
] = ejections.loc[
(ejections["player_ejected"] == 1), "playerName"
].apply(
lambda x: find_closest_playerName(x, players)
)
ejections.loc[
(ejections["player_ejected"] == 1), "playerId"
] = ejections.loc[(ejections["player_ejected"] == 1)].apply(
lambda x: find_playerId(x, players, rosters), axis=1
)
t_tmp = t_tmp.merge(
ejections.groupby("teamId")["coach_ejected"].sum().reset_index(),
how="left",
on="teamId",
)
t_tmp["coach_ejected"] = t_tmp["coach_ejected"].fillna(0)
t_tmp = t_tmp.merge(
ejections.loc[
ejections["player_ejected"] == 1, ["playerId", "player_ejected"]
],
how="left",
on="playerId",
)
t_tmp["player_ejected"] = t_tmp["player_ejected"].fillna(0)
else:
t_tmp["coach_ejected"] = 0
t_tmp["player_ejected"] = 0
if not rosters.empty:
# if 'teamId' not in t_tmp.columns:
# t_tmp = t_tmp.merge(rosters[['playerId','teamId']], how='left', on='playerId')
# t_tmp = t_tmp.merge(all_dates[['dailyDataDate_lead','teamId','nextDayGame']], how='left', left_on=['dailyDataDate', 'teamId'], right_on=['dailyDataDate_lead','teamId'])
# t_tmp['nextDayGame'] = t_tmp['nextDayGame'].fillna(0)
roster_dummies = pd.concat(
[
rosters[["dailyDataDate", "playerId"]],
pd.get_dummies(rosters["statusCode"]),
],
axis=1,
)
roster_dummies = (
roster_dummies.groupby(["dailyDataDate", "playerId"])
.sum()
.reset_index()
)
for col in [
"A",
"BRV",
"D10",
"D60",
"D7",
"DEC",
"FME",
"PL",
"RES",
"RM",
"SU",
]:
if col not in roster_dummies.columns:
roster_dummies[col] = 0
t_tmp = t_tmp.merge(
roster_dummies, how="left", on=["dailyDataDate", "playerId"]
)
else:
t_tmp[
["A", "BRV", "D10", "D60", "D7", "DEC", "FME", "PL", "RES", "RM", "SU"]
] = 0
t_tmp[
"nextDayGame"
] = 0 # There should be a better way to handle this. Don't want to miss this just because rosters is missing
if not transactions.empty:
transactions_dummies = pd.concat(
[
transactions[["dailyDataDate", "playerId"]],
pd.get_dummies(transactions["typeCode"]),
],
axis=1,
)
transactions_dummies = (
transactions_dummies.groupby(["dailyDataDate", "playerId"])
.sum()
.reset_index()
)
for col in [
"ASG",
"CLW",
"CU",
"DES",
"DFA",
"NUM",
"OPT",
"OUT",
"REL",
"RET",
"RTN",
"SC",
"SE",
"SFA",
"SGN",
"TR",
]:
if col not in transactions_dummies.columns:
transactions_dummies[col] = 0
t_tmp = t_tmp.merge(
transactions_dummies, how="left", on=["dailyDataDate", "playerId"]
)
else:
t_tmp[
[
"ASG",
"CLW",
"CU",
"DES",
"DFA",
"NUM",
"OPT",
"OUT",
"REL",
"RET",
"RTN",
"SC",
"SE",
"SFA",
"SGN",
"TR",
]
] = 0
if not awards.empty:
awards_filtered = awards[
awards["awardId"].isin(keep_awards + keep_annual_awards)
].reset_index(drop=True)
# Update awards counts
awards_dict_tmp = {
n: grp.to_dict("list")
for n, grp in awards_filtered[["awardId", "playerId"]].groupby(
"playerId"
)
}
for k, v in awards_dict_tmp.items():
if not k in awards_dict:
awards_dict[k] = {}
counts = np.unique(v["awardId"], return_counts=True)
for feat, value in zip(counts[0], counts[1]):
if feat in awards_dict[k]:
awards_dict[k][feat] += value
else:
awards_dict[k][feat] = value
awards_filtered = awards[awards["awardId"].isin(keep_awards)].reset_index(
drop=True
)
if not awards_filtered.empty:
awards_dummies = pd.concat(
[
awards_filtered[["dailyDataDate", "playerId"]],
pd.get_dummies(awards_filtered["awardId"]),
],
axis=1,
)
awards_dummies = (
awards_dummies.groupby(["dailyDataDate", "playerId"])
.sum()
.reset_index()
)
for col in keep_awards:
if col not in awards_dummies.columns:
awards_dummies[col] = 0
t_tmp = t_tmp.merge(
awards_dummies, how="left", on=["dailyDataDate", "playerId"]
)
else:
t_tmp[keep_awards] = 0
else:
t_tmp[keep_awards] = 0
if t_tmp.shape[0] != eng_shape[0]:
print(
"awards: t_tmp length does not match engagement frame length, check for duplicated data"
)
t_tmp = t_tmp[~t_tmp[["playerId"]].duplicated()]
awards_df = pd.DataFrame.from_dict(awards_dict, orient="index").fillna(0)
awards_df.columns = [f"{x}_career" for x in awards_df.columns]
t_tmp = t_tmp.merge(
awards_df.reset_index().rename(columns={"index": "playerId"}),
how="left",
on="playerId",
)
t_tmp[awards_df.columns] = t_tmp[awards_df.columns].fillna(0)
if not standings.empty:
standings = standings.replace("-", 0.0)
object_cols = standings.select_dtypes(exclude=["float"]).columns
standings[object_cols] = standings[object_cols].apply(
pd.to_numeric, downcast="float", errors="coerce"
)
bool_cols = standings.select_dtypes(include=["boolean"]).columns
standings[bool_cols] = standings[bool_cols].astype(int)
t_tmp = t_tmp.merge(
standings, how="left", on=["teamId"], suffixes=["", "_team_standings"]
)
t_tmp["team_games_played"] = t_tmp["wins"] + t_tmp["losses"]
if t_tmp.shape[0] != eng_shape[0]:
print(
"standings: t_tmp length does not match engagement frame length, check for duplicated data"
)
t_tmp = t_tmp[~t_tmp[["playerId"]].duplicated()]
if len(win_streaks) > 0:
t_tmp["team_win_streak"] = t_tmp["teamId"].map(win_streaks)
if not hitting_history_df.empty:
t_tmp = t_tmp.merge(hitting_history_df, how="left", on="playerId")
t_tmp["hr_rank"] = t_tmp["homeRuns_season"].rank(ascending=False)
if t_tmp.shape[0] != eng_shape[0]:
print(
"hitting_history_df: t_tmp length does not match engagement frame length, check for duplicated data"
)
t_tmp = t_tmp[~t_tmp[["playerId"]].duplicated()]
if not pitching_history_df.empty:
t_tmp = t_tmp.merge(pitching_history_df, how="left", on="playerId")
# Calculate ERA
# there are no more standings after season end so team_games_played is no longer known
if "team_games_played" in t_tmp.columns:
t_tmp["era"] = 9 * (
t_tmp["earnedRuns_season"] / t_tmp["inningsPitched_season"]
)
t_tmp["era_rank"] = t_tmp.loc[
t_tmp["inningsPitched_season"] >= t_tmp["team_games_played"], "era"
].rank(method="min")
if t_tmp.shape[0] != eng_shape[0]:
print(
"pitching_history_df: t_tmp length does not match engagement frame length, check for duplicated data"
)
t_tmp = t_tmp[~t_tmp[["playerId"]].duplicated()]
if not fielding_history_df.empty:
t_tmp = t_tmp.merge(fielding_history_df, how="left", on="playerId")
if t_tmp.shape[0] != eng_shape[0]:
print(
"fielding_history_df: t_tmp length does not match engagement frame length, check for duplicated data"
)
t_tmp = t_tmp[~t_tmp[["playerId"]].duplicated()]
player_countries = [
"Aruba",
"Australia",
"Bahamas",
"Brazil",
"Canada",
"China",
"Colombia",
"Cuba",
"Curacao",
"Dominican Republic",
"Germany",
"Honduras",
"Japan",
"Lithuania",
"Mexico",
"Netherlands",
"Nicaragua",
"Northern Ireland",
"Panama",
"Peru",
"Puerto Rico",
"Saudi Arabia",
"South Africa",
"South Korea",
"Taiwan",
"U.S. Virgin Islands",
"USA",
"Venezuela",
]
t_tmp = t_tmp.merge(player_country_dummies, how="left", on="playerId")
# Add games features
# if not games.empty and not p_box_scores.empty:
# games['dayNight'] = games['dayNight'].map({'day': 0, 'night': 1})
# games['homeWinner'] = games['homeWinner'].fillna(-1).astype(float)
# t_tmp = t_tmp.merge(games[['gamePk', 'dayNight','homeWinPct','awayWinPct','homeScore','awayScore','homeWinner']], how='left', on='gamePk')
# Add Twitter features
if not p_twitter_recent.empty:
t_tmp = t_tmp.merge(
p_twitter_recent[["playerId", "numberOfFollowers"]],
how="left",
on=["playerId"],
)
if not p_twitter_delta.empty:
t_tmp = t_tmp.merge(p_twitter_delta, how="left", on=["playerId"])
### TRAILING AGGREGATION MERGES
t_tmp["ddd_month"] = (
np.floor(t_tmp.dailyDataDate / 100).clip(upper=MAX_MONTH).astype(int)
)
t_tmp = t_tmp.merge(
dt_player_aggregations, how="left", on=["playerId", "ddd_month"]
)
if "gamePk" in t_tmp.columns:
t_tmp["played_game"] = t_tmp["gamePk"].notnull().astype(float)
else:
t_tmp["played_game"] = 0.0
t_tmp = t_tmp.merge(
dt_player_game_aggregations,
how="left",
on=["playerId", "ddd_month", "played_game"],
)
# t_tmp = t_tmp.merge(recent_player_means, how='left', on='playerId')
t_tmp["monthday"] = t_tmp["dailyDataDate"].astype(str).str[4:].astype(int)
t_tmp["dayofweek"] = pd.to_datetime(
t_tmp["date_playerId"].str.split("_", expand=True)[0]
).dt.dayofweek
t_tmp["data_dayofmonth"] = (
t_tmp["dailyDataDate"].astype(str).str[6:].astype(int)
)
t_tmp["eng_dayofmonth"] = pd.to_datetime(
t_tmp["date_playerId"].str.split("_", expand=True)[0]
).dt.day
# Fill season values with 2021
t_tmp["season"] = season
# t.append(t_tmp)
use_cols = lgb_target1.feature_name()
# Add any missing columns so that it does not crash
missing_cols = [col for col in use_cols if col not in t_tmp.columns]
for col in missing_cols:
print(f"lgb_target1 missing: {col}")
t_tmp[col] = np.nan
sub["target1_v30"] = np.clip(lgb_target1.predict(t_tmp[use_cols]), 0, 100)
sub["target2_v30"] = np.clip(lgb_target2.predict(t_tmp[use_cols]), 0, 100)
sub["target3_v30"] = np.clip(lgb_target3.predict(t_tmp[use_cols]), 0, 100)
sub["target4_v30"] = np.clip(lgb_target4.predict(t_tmp[use_cols]), 0, 100)
use_cols = lgb_bfa_target1.feature_name()
# Add any missing columns so that it does not crash
missing_cols = [col for col in use_cols if col not in t_tmp.columns]
for col in missing_cols:
print(f"lgb_bfa_target1 missing: {col}")
t_tmp[col] = np.nan
sub["target1_v30_bfa"] = np.clip(
lgb_bfa_target1.predict(t_tmp[use_cols]), 0, 100
)
sub["target2_v30_bfa"] = np.clip(
lgb_bfa_target2.predict(t_tmp[use_cols]), 0, 100
)
sub["target3_v30_bfa"] = np.clip(
lgb_bfa_target3.predict(t_tmp[use_cols]), 0, 100
)
sub["target4_v30_bfa"] = np.clip(
lgb_bfa_target4.predict(t_tmp[use_cols]), 0, 100
)
use_cols = lgb_dblsqrt_target1.feature_name()
# Add any missing columns so that it does not crash
missing_cols = [col for col in use_cols if col not in t_tmp.columns]
for col in missing_cols:
print(f"lgb_dblsqrt_target1 missing: {col}")
t_tmp[col] = np.nan
sub["target1_v30_dblsqrt"] = np.clip(
lgb_dblsqrt_target1.predict(t_tmp[use_cols]) ** 2**2, 0, 100
)
sub["target2_v30_dblsqrt"] = np.clip(
lgb_dblsqrt_target2.predict(t_tmp[use_cols]) ** 2**2, 0, 100
)
sub["target3_v30_dblsqrt"] = np.clip(
lgb_dblsqrt_target3.predict(t_tmp[use_cols]) ** 2**2, 0, 100
)
sub["target4_v30_dblsqrt"] = np.clip(
lgb_dblsqrt_target4.predict(t_tmp[use_cols]) ** 2**2, 0, 100
)
dart_use_cols = lgb_dart_target1.feature_name()
# Add any missing columns so that it does not crash
missing_cols = [col for col in dart_use_cols if col not in t_tmp.columns]
for col in missing_cols:
print(f"lgb_dart_target1 missing: {col}")
t_tmp[col] = np.nan
sub["target1_dart"] = np.clip(
lgb_dart_target1.predict(t_tmp[dart_use_cols]), 0, 100
)
sub["target2_dart"] = np.clip(
lgb_dart_target2.predict(t_tmp[dart_use_cols]), 0, 100
)
sub["target3_dart"] = np.clip(
lgb_dart_target3.predict(t_tmp[dart_use_cols]), 0, 100
)
sub["target4_dart"] = np.clip(
lgb_dart_target4.predict(t_tmp[dart_use_cols]), 0, 100
)
with open("../input/d/brandenkmurray/mlbmodels/xgb_v30_use_cols.txt") as f:
xgb_use_cols = [x.rstrip() for x in f.readlines()]
# xgb_use_cols = xgb_target1.feature_names
missing_cols = [col for col in xgb_use_cols if col not in t_tmp.columns]
missing_cols_filled = [x.replace(" ", "_") for x in missing_cols]
t_tmp = t_tmp.rename(
columns={k: v for k, v in zip(missing_cols_filled, missing_cols)}
)
for col in missing_cols:
print(f"{col} is missing for XGB model. Adding and filling with NaN")
t_tmp[col] = np.nan
sub["target1_xgb"] = np.clip(
xgb_target1.predict(
xgb.DMatrix(t_tmp[xgb_use_cols].fillna(-99999).replace(np.inf, -99999))
)
** 2
** 2,
0,
100,
)
sub["target2_xgb"] = np.clip(
xgb_target2.predict(
xgb.DMatrix(t_tmp[xgb_use_cols].fillna(-99999).replace(np.inf, -99999))
)
** 2
** 2,
0,
100,
)
sub["target3_xgb"] = np.clip(
xgb_target3.predict(
xgb.DMatrix(t_tmp[xgb_use_cols].fillna(-99999).replace(np.inf, -99999))
)
** 2
** 2,
0,
100,
)
sub["target4_xgb"] = np.clip(
xgb_target4.predict(
xgb.DMatrix(t_tmp[xgb_use_cols].fillna(-99999).replace(np.inf, -99999))
)
** 2
** 2,
0,
100,
)
correlates = [
"hitBatsmen",
"no_hitter",
"home_team_box_score",
"hitBatsmen_team_box_score",
"season_team_standings",
"sportGamesBack",
"nlWins",
"nlLosses",
"errors_1_games_ago",
]
t_tmp = t_tmp.drop(columns=correlates)
use_cols = lgb_john_target1.feature_name()
missing_cols = [col for col in use_cols if col not in t_tmp.columns]
for col in missing_cols:
print(col + " missing for John's model")
t_tmp[col] = np.nan
sub["target1_john"] = np.clip(lgb_john_target1.predict(t_tmp[use_cols]), 0, 100)
sub["target2_john"] = np.clip(lgb_john_target2.predict(t_tmp[use_cols]), 0, 100)
sub["target3_john"] = np.clip(lgb_john_target3.predict(t_tmp[use_cols]), 0, 100)
sub["target4_john"] = np.clip(lgb_john_target4.predict(t_tmp[use_cols]), 0, 100)
sub["target1"] = (
(sub["target1_john"] * 0.4)
+ (
(
sub["target1_v30_bfa"] * 0.2
+ sub["target1_v30"] * 0.1
+ sub["target1_v30_dblsqrt"] * 0.7
)
* 0.1
)
+ (sub["target1_xgb"] * 0.1)
+ (sub["target1_dart"] * 0.4)
)
sub["target2"] = (
(sub["target2_john"] * 0.4)
+ (
(
sub["target2_v30_bfa"] * 0.2
+ sub["target2_v30"] * 0.1
+ sub["target2_v30_dblsqrt"] * 0.7
)
* 0.1
)
+ (sub["target2_xgb"] * 0.1)
+ (sub["target2_dart"] * 0.4)
)
sub["target3"] = (
(sub["target3_john"] * 0.4)
+ (
(
sub["target3_v30_bfa"] * 0.2
+ sub["target3_v30"] * 0.1
+ sub["target3_v30_dblsqrt"] * 0.7
)
* 0.1
)
+ (sub["target3_xgb"] * 0.1)
+ (sub["target3_dart"] * 0.4)
)
sub["target4"] = (
(sub["target4_john"] * 0.4)
+ (
(
sub["target4_v30_bfa"] * 0.2
+ sub["target4_v30"] * 0.1
+ sub["target4_v30_dblsqrt"] * 0.7
)
* 0.1
)
+ (sub["target4_xgb"] * 0.1)
+ (sub["target4_dart"] * 0.4)
)
except Exception as e:
# If all else fails try to use player means
print(f"Main loop failed: {e}")
try:
print("Using player rolling12 means")
sub["ddd_month"] = (
np.floor(sub.dailyDataDate / 100).clip(upper=MAX_MONTH).astype(int)
)
sub = sub.drop(["target1", "target2", "target3", "target4"], axis=1)
sub = sub.merge(
dt_player_aggregations[
[
"playerId",
"ddd_month",
"roll12_target1_p_gameday_median",
"roll12_target2_p_gameday_median",
"roll12_target3_p_gameday_median",
"roll12_target4_p_gameday_median",
]
],
how="left",
on=["playerId", "ddd_month"],
)
sub = sub.rename(
{
k: v
for k, v in zip(
[
"roll12_target1_p_gameday_median",
"roll12_target2_p_gameday_median",
"roll12_target3_p_gameday_median",
"roll12_target4_p_gameday_median",
],
["target1", "target2", "target3", "target4"],
)
},
axis=1,
)
except Exception as e:
print(e)
# If player means fail, use overall means
print("Player means failed. Using overall means")
sub["target1"] = 0.001046
sub["target2"] = 0.521472
sub["target3"] = 0.001735
sub["target4"] = 0.226034
# Do a final check to ensure there are no duplicate players that will cause a scoring error
sub = sub[~sub[["playerId"]].duplicated()]
# sub_list.append(sub)
env.predict(sub[["date_playerId", "target1", "target2", "target3", "target4"]])
# eng_lag = sub[['playerId','target1','target2','target3','target4']].copy()
# eng_lag = eng_lag.rename({'target1': 'target1_lag',
# 'target2': 'target2_lag',
# 'target3': 'target3_lag',
# 'target4': 'target4_lag'}, axis=1)
# sub_all = pd.concat(sub_list)
# include_players = players[players['playerForTestSetAndFuturePreds']==1]['playerId'].tolist()
# sub_all = sub_all[sub_all['playerId'].isin(include_players)]
# print("v30")
# may_mae_list = []
# june_mae_list = []
# for target in ['target1', 'target2', 'target3', 'target4']:
# may_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_v30'] - sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_act'])))
# june_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_v30'] - sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_act'])))
# # print(may_mae_list)
# print(f"May MAE: {np.mean(may_mae_list)}")
# print(f"June MAE: {np.mean(june_mae_list)}")
# print("v30_bfa")
# may_mae_list = []
# june_mae_list = []
# for target in ['target1', 'target2', 'target3', 'target4']:
# may_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_v30_bfa'] - sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_act'])))
# june_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_v30_bfa'] - sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_act'])))
# # print(may_mae_list)
# print(f"May MAE: {np.mean(may_mae_list)}")
# print(f"June MAE: {np.mean(june_mae_list)}")
# print("v30_dblsqrt")
# may_mae_list = []
# june_mae_list = []
# for target in ['target1', 'target2', 'target3', 'target4']:
# may_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_v30_dblsqrt'] - sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_act'])))
# june_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_v30_dblsqrt'] - sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_act'])))
# # print(may_mae_list)
# print(f"May MAE: {np.mean(may_mae_list)}")
# print(f"June MAE: {np.mean(june_mae_list)}")
# print("john")
# may_mae_list = []
# june_mae_list = []
# for target in ['target1', 'target2', 'target3', 'target4']:
# may_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_john'] - sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_act'])))
# june_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_john'] - sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_act'])))
# # print(may_mae_list)
# print(f"May MAE: {np.mean(may_mae_list)}")
# print(f"June MAE: {np.mean(june_mae_list)}")
# print("xgb")
# may_mae_list = []
# june_mae_list = []
# for target in ['target1', 'target2', 'target3', 'target4']:
# may_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_xgb'] - sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_act'])))
# june_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_xgb'] - sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_act'])))
# # print(may_mae_list)
# print(f"May MAE: {np.mean(may_mae_list)}")
# print(f"June MAE: {np.mean(june_mae_list)}")
# print("dart")
# may_mae_list = []
# june_mae_list = []
# for target in ['target1', 'target2', 'target3', 'target4']:
# may_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_dart'] - sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_act'])))
# june_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_dart'] - sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_act'])))
# # print(may_mae_list)
# print(f"May MAE: {np.mean(may_mae_list)}")
# print(f"June MAE: {np.mean(june_mae_list)}")
# print("blend")
# may_mae_list = []
# june_mae_list = []
# for target in ['target1', 'target2', 'target3', 'target4']:
# may_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target] - sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_act'])))
# june_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target] - sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_act'])))
# # print(may_mae_list)
# print(f"May MAE: {np.mean(may_mae_list)}")
# print(f"June MAE: {np.mean(june_mae_list)}")
# t_df = pd.concat(t)
# t_df.to_csv("./train_features.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/492/69492739.ipynb
|
dart-model
|
kazanova
|
[{"Id": 69492739, "ScriptId": 18957801, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 150865, "CreationDate": "07/31/2021 17:04:54", "VersionNumber": 12.0, "Title": "mlb-predict-final", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 1494.0, "LinesInsertedFromPrevious": 6.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 1488.0, "LinesInsertedFromFork": 345.0, "LinesDeletedFromFork": 301.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 1149.0, "TotalVotes": 0}]
|
[{"Id": 92729261, "KernelVersionId": 69492739, "SourceDatasetVersionId": 2484106}, {"Id": 92729260, "KernelVersionId": 69492739, "SourceDatasetVersionId": 2482609}]
|
[{"Id": 2484106, "DatasetId": 1499698, "DatasourceVersionId": 2526660, "CreatorUserId": 111640, "LicenseName": "CC0: Public Domain", "CreationDate": "07/31/2021 13:22:07", "VersionNumber": 3.0, "Title": "dart-model", "Slug": "dart-model", "Subtitle": NaN, "Description": NaN, "VersionNotes": "added models with full data", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1499698, "CreatorUserId": 111640, "OwnerUserId": 111640.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2484106.0, "CurrentDatasourceVersionId": 2526660.0, "ForumId": 1519428, "Type": 2, "CreationDate": "07/29/2021 20:59:38", "LastActivityDate": "07/29/2021", "TotalViews": 1331, "TotalDownloads": 4, "TotalVotes": 0, "TotalKernels": 1}]
|
[{"Id": 111640, "UserName": "kazanova", "DisplayName": "\u039c\u03b1\u03c1\u03b9\u03bf\u03c2 \u039c\u03b9\u03c7\u03b1\u03b7\u03bb\u03b9\u03b4\u03b7\u03c2 KazAnova", "RegisterDate": "06/24/2013", "PerformanceTier": 4}]
|
import os
import itertools
import pandas as pd
import datatable as dt
import numpy as np
import mlb
import pickle as pkl
from tqdm import tqdm
from itertools import product
import lightgbm as lgb
from fuzzywuzzy import fuzz
import re
import numba as nb
from numba import njit
import xgboost as xgb
@njit
def nb_cumsum(arr):
return arr.cumsum()
@njit
def nb_sum(arr):
return arr.sum()
def reduce_mem_usage(df, verbose=True):
numerics = ["int16", "int32", "int64", "float16", "float32", "float64"]
start_mem = df.memory_usage().sum() / 1024**2
for col in df.columns:
col_type = df[col].dtypes
if col_type in numerics:
c_min = df[col].min()
c_max = df[col].max()
if str(col_type)[:3] == "int":
if c_min > np.iinfo(np.int8).min and c_max < np.iinfo(np.int8).max:
df[col] = df[col].astype(np.int16)
elif c_min > np.iinfo(np.int16).min and c_max < np.iinfo(np.int16).max:
df[col] = df[col].astype(np.int32)
elif c_min > np.iinfo(np.int32).min and c_max < np.iinfo(np.int32).max:
df[col] = df[col].astype(np.int64)
elif c_min > np.iinfo(np.int64).min and c_max < np.iinfo(np.int64).max:
df[col] = df[col].astype(np.int64)
else:
if (
c_min > np.finfo(np.float16).min
and c_max < np.finfo(np.float16).max
):
df[col] = df[col].astype(np.float32)
elif (
c_min > np.finfo(np.float32).min
and c_max < np.finfo(np.float32).max
):
df[col] = df[col].astype(np.float64)
else:
df[col] = df[col].astype(np.float64)
end_mem = df.memory_usage().sum() / 1024**2
if verbose:
print(
"Mem. usage decreased to {:5.2f} Mb ({:.1f}% reduction)".format(
end_mem, 100 * (start_mem - end_mem) / start_mem
)
)
return df
def unnest(data, name):
try:
date_nested_table = data[["date", name]]
date_nested_table = date_nested_table[
~pd.isna(date_nested_table[name])
].reset_index(drop=True)
daily_dfs_collection = []
for date_index, date_row in date_nested_table.iterrows():
daily_df = pd.read_json(date_row[name])
daily_df["dailyDataDate"] = date_row["date"]
daily_dfs_collection = daily_dfs_collection + [daily_df]
if daily_dfs_collection:
# Concatenate all daily dfs into single df for each row
unnested_table = (
pd.concat(daily_dfs_collection, ignore_index=True)
.
# Set and reset index to move 'dailyDataDate' to front of df
set_index("dailyDataDate")
.reset_index()
)
return reduce_mem_usage(unnested_table, False)
else:
return pd.DataFrame()
except Exception as e:
print(e)
print(f"unnest failed for {name}. returning empty dataframe")
return pd.DataFrame()
def get_unnested_data_dict(data, daily_data_nested_df_names):
df_dict = {}
for df_name in daily_data_nested_df_names:
df_dict[df_name] = unnest(data, df_name)
return df_dict
def get_unnested_data(data, colnames):
return (unnest(data, df_name) for df_name in colnames)
## Find win expectancy and volatility given inning, out, base, run situation.
## no. of runs that score with HR in diff. base situations
baseHr = {1: 1, 2: 2, 3: 2, 4: 3, 5: 2, 6: 3, 7: 3, 8: 4}
def getRunsInn(rpinn):
runsinn = {
0: 1 / ((rpinn * 0.761) + 1),
1: (rpinn * (0.761**2)) / (((rpinn * 0.761) + 1) ** 2),
}
for i in range(2, 11):
v = (rpinn * (0.761**2) * (((rpinn * 0.761) - 0.761 + 1) ** (i - 1))) / (
((rpinn * 0.761) + 1) ** (i + 1)
)
runsinn[i] = v
return runsinn
def getRunExp(rpinn, runsinn):
runExp = {"10": runsinn}
for i in range(0, 3):
for j in range(1, 9):
k = str(j) + str(i)
if k == "10":
continue
runExp[k] = {0: ((tangoRunExp[k]["m"] * rpinn) + tangoRunExp[k]["b"])}
for r in range(1, 11):
runExp[k][r] = (1 - runExp[k][0]) * tangoRunExp[k][r]
return runExp
def getInnWinexp(runExp):
## Chance of home team winning with zero
## outs at the beg. of each inning
innWinexp = {"101": {0: 0.5}}
for i in range(-25, 0):
innWinexp["101"][i] = 0
for i in range(1, 26):
innWinexp["101"][i] = 1
for i in range(9, 0, -1):
for j in range(2, 0, -1):
if j == 2:
next = str(i + 1) + "1"
else:
next = str(i) + "2"
this = str(i) + str(j)
innWinexp[this] = {}
if j == 2:
for k in range(-25, 26):
p = 0
if i == 9 and k > 0:
innWinexp[this][k] = 1
continue
else:
pass
for m in range(0, 11):
if k + m > 25:
iw = 1
else:
iw = innWinexp[next][k + m]
p += runExp["10"][m] * iw
innWinexp[this][k] = p
else:
for k in range(-25, 26):
p = 0
for m in range(0, 11):
if k - m < -25:
iw = 0
else:
iw = innWinexp[next][k - m]
p += runExp["10"][m] * iw
innWinexp[this][k] = p
return innWinexp
def getWinexp(innWinexp, runExp, inn, half, base, outs, rdiff):
if inn > 9:
inn = 9
innkey = str(inn) + str(half)
if outs > 2:
outs = 2
sitkey = str(base) + str(outs)
if half == 2:
next = str(inn + 1) + "1"
else:
next = str(inn) + "2"
if sitkey == "10": ## beginning of half inning
if rdiff > 25:
rdiff = 25
elif rdiff < -25:
rdiff = -25
else:
pass
Winexp = innWinexp[innkey][rdiff]
elif half == 1:
Winexp = 0
for i in range(10, -1, -1):
if rdiff - i < -25:
iw = 0
elif rdiff - i > 25:
iw = 1
else:
iw = innWinexp[next][rdiff - i]
Winexp += runExp[sitkey][i] * iw
else:
Winexp = 0
for i in range(0, 11):
if rdiff - i < -25:
iw = 0
elif rdiff + i > 25:
iw = 1
else:
iw = innWinexp[next][rdiff + i]
Winexp += runExp[sitkey][i] * iw
return Winexp
def getVol(innWinexp, runExp, inn, half, base, outs, rdiff):
## changes if strikeout:
if outs == 2:
outsK = 0
baseK = 1
if half == 1:
halfK = 2
innK = inn
else:
halfK = 1
innK = inn + 1
else:
outsK = outs + 1
baseK, halfK, innK = base, half, inn
WinexpK = getWinexp(innWinexp, runExp, innK, halfK, baseK, outsK, rdiff)
## changes if homerun
if half == 1:
rdiff -= baseHr[base]
else:
rdiff += baseHr[base]
base = 1
WinexpHr = getWinexp(innWinexp, runExp, inn, half, base, outs, rdiff)
return (abs(WinexpHr - WinexpK)) / 0.133
def rpgToInnWinexp(rpg):
rpinn = float(rpg) / 9 ## r/inn
runsinn = getRunsInn(rpinn)
runExp = getRunExp(rpinn, runsinn)
innWinexp = getInnWinexp(runExp)
return innWinexp, runExp
def winnexp_feature(x):
return getWinexp(
innWinexp,
runExp,
x["inning"],
x["halfInning_index"],
x["base_state"],
x["outs_beg"],
x["run_diff"],
)
##################################################################################################
## Functions for extracting and matching ejected player names and getting their playerId
##################################################################################################
# Need to map names to the players.csv or playerBoxScores playerIds
def find_closest_playerName(playerName, players):
players["fuzz_score"] = [fuzz.WRatio(playerName, x) for x in players["playerName"]]
best_match = players.loc[
players["fuzz_score"] == players["fuzz_score"].max(), "playerName"
].iloc[0]
return best_match
def find_playerId(x, players, rosters_players):
# rosters_players is a merge of the rosters df and the players df on the playerId
tmp = players[players["playerName"] == x["playerName"]]
if tmp.shape[0] == 1:
return tmp["playerId"].iloc[0]
else:
# If there are two players with the same name in players, then use the daily roster data to find the player on the matching team
return rosters_players.loc[
(rosters_players["dailyDataDate"] == x["dailyDataDate"])
& (rosters_players["teamId"] == x["teamId"])
& (rosters_players["playerName"] == x["playerName"]),
"playerId",
].iloc[0]
##################################################################################################
# Set up win expectancy variables
rpg = 4.5
innWinexp, runExp = rpgToInnWinexp(rpg)
def game_score_james(x):
"""
# • Start with 50 points
# • Add 1 point for each out recorded (or 3 points per inning)
# • Add 2 points for each inning completed after the fourth
# • Add 1 additional point for every strikeout
# • Remove 2 points for each hit allowed
# • Remove 4 points for each earned run allowed
# • Remove 2 points for each unearned run allowed
# • Remove 1 point for each walk allowed
"""
score = 50
score += x["outsPitching"]
score += 2 * (x["inningsPitched"] - 4)
score += x["strikeOutsPitching"]
score -= 2 * x["hitsPitching"]
score -= 4 * x["earnedRuns"]
score -= 2 * (x["runsPitching"] - x["earnedRuns"])
score -= x["baseOnBallsPitching"] + x["hitByPitchPitching"]
# score = 50 + x['outsPitching'] + 2*(x['inningsPitched'] - 4) + x['strikeOutsPitching'] - 2*x['hitsPitching'] - 4*x['earnedRuns'] - 2*(x['runsPitching'] - x['earnedRuns']) - (x['baseOnBallsPitching']+x['hitByPitchPitching'])
return score
def game_score_tango(x):
"""
Game Score formula (updated by Tom Tango)
# • Start with 40 points
# • Add 2 points for each out recorded (or 6 points per inning)
# • Add 1 additional point for every strikeout
# • Remove 2 points for each walk allowed
# • Remove 2 points for each hit allowed
# • Remove 3 points for each run allowed (earned or unearned)
# • Remove 6 additional points for each home run allowed
"""
score = 40
score += 2 * x["outsPitching"]
score += x["strikeOutsPitching"]
score -= 2 * (x["baseOnBallsPitching"] + x["hitByPitchPitching"])
score -= 2 * x["hitsPitching"]
score -= 3 * x["runsPitching"]
score -= 6 * x["homeRunsPitching"]
return score
pitching_features = [
"gamesPlayedPitching",
"gamesStartedPitching",
"completeGamesPitching",
"shutoutsPitching",
"winsPitching",
"lossesPitching",
"flyOutsPitching",
"airOutsPitching",
"groundOutsPitching",
"runsPitching",
"doublesPitching",
"triplesPitching",
"homeRunsPitching",
"strikeOutsPitching",
"baseOnBallsPitching",
"intentionalWalksPitching",
"hitsPitching",
"hitByPitchPitching",
"atBatsPitching",
"caughtStealingPitching",
"stolenBasesPitching",
"inningsPitched",
"saveOpportunities",
"earnedRuns",
"battersFaced",
"outsPitching",
"pitchesThrown",
"balls",
"strikes",
"hitBatsmen",
"balks",
"wildPitches",
"pickoffsPitching",
"rbiPitching",
"gamesFinishedPitching",
"inheritedRunners",
"inheritedRunnersScored",
"catchersInterferencePitching",
"sacBuntsPitching",
"sacFliesPitching",
"saves",
"holds",
"blownSaves",
"assists",
"putOuts",
"errors",
"chances",
]
train = dt.fread(
"../input/mlb-player-digital-engagement-forecasting/train_updated.csv"
).to_pandas()
test = True
if test:
last_date = train["date"].max()
else:
last_date = 20210430
eng = unnest(train, "nextDayPlayerEngagement")
p_box_scores_og = unnest(train, "playerBoxScores")
teams = pd.read_csv("../input/mlb-player-digital-engagement-forecasting/teams.csv")
players = pd.read_csv("../input/mlb-player-digital-engagement-forecasting/players.csv")
awards_history = pd.read_csv(
"../input/mlb-player-digital-engagement-forecasting/awards.csv"
)
##################################################################################################
## Regex's for extracting and matching ejected player names and getting their playerId
##################################################################################################
team_names = list(teams["teamName"].unique()) + ["Diamondbacks"]
team_regex = re.compile("|".join(map(re.escape, team_names)))
team_full_names = list(teams["name"].unique()) + list(teams["teamName"].unique())
team_full_regex = re.compile("|".join(map(re.escape, team_full_names)))
coaching_names = [
"Assistant Hitting Coach",
"Manager",
"Bench Coach",
"Interim Manager",
"Hitting Coach",
"First Base Coach",
"Pitching Coach",
"bench caoch",
"assistant hitting coach",
"Third Base Coach",
"catching coach",
"field coordinator",
"first base coach",
"hitting coach",
"major league coach",
"manager",
"pitching coach",
"third base coach",
"bench coach",
]
coaching_regex = re.compile("|".join(map(re.escape, coaching_names)))
positions = [
"pitcher",
"catcher",
"first baseman",
"second baseman",
"third baseman",
"shortstop",
"left fielder",
"center fielder",
"right fielder",
"designated hitter",
]
pos_regex = re.compile("|".join(map(re.escape, positions)))
##################################################################################################
colnames = [
"games",
"rosters",
"playerBoxScores",
"teamBoxScores",
"transactions",
"standings",
"awards",
"events",
"playerTwitterFollowers",
"teamTwitterFollowers",
]
hitter_history_feats = [
"hits",
"doubles",
"triples",
"homeRuns",
"rbi",
"totalBases",
"plateAppearances",
"strikeOuts",
"baseOnBalls",
"hitByPitch",
"atBats",
"sacFlies",
]
pitcher_history_feats = [
"gamesPlayedPitching",
"gamesStartedPitching",
"inningsPitched",
"pitchesThrown",
"winsPitching",
"runsPitching",
"homeRunsPitching",
"strikeOutsPitching",
"earnedRuns",
"blownSaves",
"holds",
]
fielder_history_feats = ["errors"]
keep_awards = [
"NLPOW",
"ALPOW",
"NLROM",
"ALROM",
"NLPOM",
"ALPOM",
"NLRRELMON",
"ALRRELMON",
"ALPITOM",
"NLPITOM",
"MLBPLAYOW",
]
keep_annual_awards = [
"ALMVP",
"NLMVP",
"ALCY",
"NLCY",
"ALROY",
"NLROY",
"ALPG",
"NLPG",
"ALSS",
"NLSS",
"ALGG",
"NLGG",
]
##################################################################################################
## Mappings
##################################################################################################
team_mapping = teams.set_index("teamName")["id"].to_dict()
team_mapping["Diamondbacks"] = 109
player_mapping = p_box_scores_og[["playerId", "playerName"]].drop_duplicates()
##################################################################################################
pitchers = players[players["primaryPositionName"] == "Pitcher"]
players["value"] = 1
player_country_dummies = pd.pivot_table(
players,
values="value",
index=["playerId"],
columns=["birthCountry"],
aggfunc="sum",
fill_value=0,
).reset_index()
player_country_dummies.columns = player_country_dummies.columns.str.replace(" ", "_")
quantile_20 = lambda x: x.quantile(0.20)
quantile_20.__name__ = "quantile_20"
quantile_80 = lambda x: x.quantile(0.80)
quantile_80.__name__ = "quantile_80"
# agg_list = ['median','var', quantile_20, quantile_80]
# player_aggs = eng.groupby('playerId')[['target1','target2','target3','target4']].agg({'target1': agg_list,
# 'target2': agg_list,
# 'target3': agg_list,
# 'target4': agg_list}).round(6)
# player_aggs.columns = ["_".join(x) for x in player_aggs.columns.ravel()]
# player_aggs = player_aggs.reset_index()
# player_medians = eng.groupby('playerId')[['target1','target2','target3','target4']].median().round(6).reset_index()
# player_medians = player_medians.rename({'target1': 'target1_p_median',
# 'target2': 'target2_p_median',
# 'target3': 'target3_p_median',
# 'target4': 'target4_p_median'}, axis=1)
# player_variances = eng.groupby('playerId')[['target1','target2','target3','target4']].var().round(6).reset_index()
# player_variances = player_variances.rename({'target1': 'target1_p_var',
# 'target2': 'target2_p_var',
# 'target3': 'target3_p_var',
# 'target4': 'target4_p_var'}, axis=1)
# game_day_player_means = eng.merge(p_box_scores_og[['dailyDataDate','playerId', 'gamePk']], how='left')
# game_day_player_means['game_played'] = game_day_player_means['gamePk'].notnull().astype(int)
# off_day_player_means = game_day_player_means[game_day_player_means['game_played']==0].groupby(['playerId'])[['target1','target2','target3','target4']].mean().round(6).reset_index()
# off_day_player_means = off_day_player_means.rename({'target1': 'target1_p_mean_off_day',
# 'target2': 'target2_p_mean_off_day',
# 'target3': 'target3_p_mean_off_day',
# 'target4': 'target4_p_mean_off_day'}, axis=1)
# game_day_player_means = game_day_player_means[game_day_player_means['game_played']==1].groupby(['playerId'])[['target1','target2','target3','target4']].mean().round(6).reset_index()
# game_day_player_means = game_day_player_means.rename({'target1': 'target1_p_mean_game_day',
# 'target2': 'target2_p_mean_game_day',
# 'target3': 'target3_p_mean_game_day',
# 'target4': 'target4_p_mean_game_day'}, axis=1)
# game_day_player_vars = eng.merge(p_box_scores_og[['dailyDataDate','playerId']])
# game_day_player_vars = game_day_player_vars.groupby('playerId')[['target1','target2','target3','target4']].var().round(6).reset_index()
# game_day_player_vars = game_day_player_vars.rename({'target1': 'target1_p_var_game_day',
# 'target2': 'target2_p_var_game_day',
# 'target3': 'target3_p_var_game_day',
# 'target4': 'target4_p_var_game_day'}, axis=1)
eng["ddd_month"] = np.floor(eng.dailyDataDate / 100).astype(int)
months = eng.ddd_month.unique()
## add one extra month to grab entire data set's TEs
months = np.append(months, months[months.size - 1] + 1)
MAX_MONTH = months[months.size - 1]
ttl_player_medians = []
ttl_player_variances = []
ttl_player_means = []
ttl_roll12_player_medians = []
ttl_roll12_player_variances = []
ttl_roll12_player_means = []
for i in range(months.size - 1):
month = months[i + 1]
roll_month = months[0]
if i > 11:
roll_month = months[i - 12]
# print(str(month) + ' roll:' + str(roll_month))
player_medians = (
eng[eng.ddd_month < month]
.groupby("playerId")[["target1", "target2", "target3", "target4"]]
.median()
.round(6)
.reset_index()
)
player_medians = player_medians.rename(
{
"target1": "target1_p_median",
"target2": "target2_p_median",
"target3": "target3_p_median",
"target4": "target4_p_median",
},
axis=1,
)
player_medians["ddd_month"] = month
roll12_player_medians = (
eng[(eng.ddd_month < month) & (eng.ddd_month >= roll_month)]
.groupby("playerId")[["target1", "target2", "target3", "target4"]]
.median()
.round(6)
.reset_index()
)
roll12_player_medians = roll12_player_medians.rename(
{
"target1": "roll12_target1_p_median",
"target2": "roll12_target2_p_median",
"target3": "roll12_target3_p_median",
"target4": "roll12_target4_p_median",
},
axis=1,
)
roll12_player_medians["ddd_month"] = month
player_variances = (
eng[eng.ddd_month < month]
.groupby("playerId")[["target1", "target2", "target3", "target4"]]
.var()
.round(6)
.reset_index()
)
player_variances = player_variances.rename(
{
"target1": "target1_p_var",
"target2": "target2_p_var",
"target3": "target3_p_var",
"target4": "target4_p_var",
},
axis=1,
)
player_variances["ddd_month"] = month
roll12_player_variances = (
eng[(eng.ddd_month < month) & (eng.ddd_month >= roll_month)]
.groupby("playerId")[["target1", "target2", "target3", "target4"]]
.var()
.round(6)
.reset_index()
)
roll12_player_variances = roll12_player_variances.rename(
{
"target1": "roll12_target1_p_var",
"target2": "roll12_target2_p_var",
"target3": "roll12_target3_p_var",
"target4": "roll12_target4_p_var",
},
axis=1,
)
roll12_player_variances["ddd_month"] = month
player_means = (
eng[eng.ddd_month < month]
.groupby("playerId")[["target1", "target2", "target3", "target4"]]
.mean()
.round(6)
.reset_index()
)
player_means = player_means.rename(
{
"target1": "target1_p_mean",
"target2": "target2_p_mean",
"target3": "target3_p_mean",
"target4": "target4_p_mean",
},
axis=1,
)
player_means["ddd_month"] = month
roll12_player_means = (
eng[(eng.ddd_month < month) & (eng.ddd_month >= roll_month)]
.groupby("playerId")[["target1", "target2", "target3", "target4"]]
.mean()
.round(6)
.reset_index()
)
roll12_player_means = roll12_player_means.rename(
{
"target1": "roll12_target1_p_mean",
"target2": "roll12_target2_p_mean",
"target3": "roll12_target3_p_mean",
"target4": "roll12_target4_p_mean",
},
axis=1,
)
roll12_player_means["ddd_month"] = month
ttl_player_medians.append(player_medians)
ttl_player_variances.append(player_variances)
ttl_player_means.append(player_means)
ttl_roll12_player_medians.append(roll12_player_medians)
ttl_roll12_player_variances.append(roll12_player_variances)
ttl_roll12_player_means.append(roll12_player_means)
dt_player_medians = pd.concat(ttl_player_medians)
dt_player_variances = pd.concat(ttl_player_variances)
dt_player_means = pd.concat(ttl_player_means)
dt_roll12_player_medians = pd.concat(ttl_roll12_player_medians)
dt_roll12_player_variances = pd.concat(ttl_roll12_player_variances)
dt_roll12_player_means = pd.concat(ttl_roll12_player_means)
dt_player_aggregations = dt_player_medians.merge(
dt_player_variances, how="left", on=["playerId", "ddd_month"]
)
dt_player_aggregations = dt_player_aggregations.merge(
dt_player_means, how="left", on=["playerId", "ddd_month"]
)
dt_player_aggregations = dt_player_aggregations.merge(
dt_roll12_player_medians, how="left", on=["playerId", "ddd_month"]
)
dt_player_aggregations = dt_player_aggregations.merge(
dt_roll12_player_variances, how="left", on=["playerId", "ddd_month"]
)
dt_player_aggregations = dt_player_aggregations.merge(
dt_roll12_player_means, how="left", on=["playerId", "ddd_month"]
)
played_game = (
p_box_scores_og.groupby(["playerId", "dailyDataDate"])["gamePk"]
.count()
.reset_index()
)
played_game = played_game.rename({"gamePk": "played_game"}, axis=1)
eng_box = eng.merge(played_game, how="left", on=["dailyDataDate", "playerId"])
eng_box["played_game"] = eng_box["played_game"].fillna(0).clip(upper=1)
ttl_player_gameday_medians = []
ttl_player_gameday_variances = []
ttl_player_gameday_means = []
ttl_roll12_player_gameday_medians = []
ttl_roll12_player_gameday_variances = []
ttl_roll12_player_gameday_means = []
for i in range(months.size - 1):
month = months[i + 1]
roll_month = months[0]
if i > 11:
roll_month = months[i - 12]
# print(str(month) + ' roll:' + str(roll_month))
player_gameday_medians = (
eng_box[eng_box.ddd_month < month]
.groupby(["playerId", "played_game"])[
["target1", "target2", "target3", "target4"]
]
.median()
.round(6)
.reset_index()
)
player_gameday_medians = player_gameday_medians.rename(
{
"target1": "target1_p_gameday_median",
"target2": "target2_p_gameday_median",
"target3": "target3_p_gameday_median",
"target4": "target4_p_gameday_median",
},
axis=1,
)
player_gameday_medians["ddd_month"] = month
roll12_player_gameday_medians = (
eng_box[(eng_box.ddd_month < month) & (eng_box.ddd_month >= roll_month)]
.groupby(["playerId", "played_game"])[
["target1", "target2", "target3", "target4"]
]
.median()
.round(6)
.reset_index()
)
roll12_player_gameday_medians = roll12_player_gameday_medians.rename(
{
"target1": "roll12_target1_p_gameday_median",
"target2": "roll12_target2_p_gameday_median",
"target3": "roll12_target3_p_gameday_median",
"target4": "roll12_target4_p_gameday_median",
},
axis=1,
)
roll12_player_gameday_medians["ddd_month"] = month
player_gameday_variances = (
eng_box[eng_box.ddd_month < month]
.groupby(["playerId", "played_game"])[
["target1", "target2", "target3", "target4"]
]
.var()
.round(6)
.reset_index()
)
player_gameday_variances = player_gameday_variances.rename(
{
"target1": "target1_p_gameday_var",
"target2": "target2_p_gameday_var",
"target3": "target3_p_gameday_var",
"target4": "target4_p_gameday_var",
},
axis=1,
)
player_gameday_variances["ddd_month"] = month
roll12_player_gameday_variances = (
eng_box[(eng_box.ddd_month < month) & (eng_box.ddd_month >= roll_month)]
.groupby(["playerId", "played_game"])[
["target1", "target2", "target3", "target4"]
]
.var()
.round(6)
.reset_index()
)
roll12_player_gameday_variances = roll12_player_gameday_variances.rename(
{
"target1": "roll12_target1_p_gameday_var",
"target2": "roll12_target2_p_gameday_var",
"target3": "roll12_target3_p_gameday_var",
"target4": "roll12_target4_p_gameday_var",
},
axis=1,
)
roll12_player_gameday_variances["ddd_month"] = month
player_gameday_means = (
eng_box[eng_box.ddd_month < month]
.groupby(["playerId", "played_game"])[
["target1", "target2", "target3", "target4"]
]
.mean()
.round(6)
.reset_index()
)
player_gameday_means = player_gameday_means.rename(
{
"target1": "target1_p_gameday_mean",
"target2": "target2_p_gameday_mean",
"target3": "target3_p_gameday_mean",
"target4": "target4_p_gameday_mean",
},
axis=1,
)
player_gameday_means["ddd_month"] = month
roll12_player_gameday_means = (
eng_box[(eng_box.ddd_month < month) & (eng_box.ddd_month >= roll_month)]
.groupby(["playerId", "played_game"])[
["target1", "target2", "target3", "target4"]
]
.mean()
.round(6)
.reset_index()
)
roll12_player_gameday_means = roll12_player_gameday_means.rename(
{
"target1": "roll12_target1_p_gameday_mean",
"target2": "roll12_target2_p_gameday_mean",
"target3": "roll12_target3_p_gameday_mean",
"target4": "roll12_target4_p_gameday_mean",
},
axis=1,
)
roll12_player_gameday_means["ddd_month"] = month
ttl_player_gameday_medians.append(player_gameday_medians)
ttl_player_gameday_variances.append(player_gameday_variances)
ttl_player_gameday_means.append(player_gameday_means)
ttl_roll12_player_gameday_medians.append(roll12_player_gameday_medians)
ttl_roll12_player_gameday_variances.append(roll12_player_gameday_variances)
ttl_roll12_player_gameday_means.append(roll12_player_gameday_means)
dt_player_gameday_medians = pd.concat(ttl_player_gameday_medians)
dt_player_gameday_variances = pd.concat(ttl_player_gameday_variances)
dt_player_gameday_means = pd.concat(ttl_player_gameday_means)
dt_roll12_player_gameday_medians = pd.concat(ttl_roll12_player_gameday_medians)
dt_roll12_player_gameday_variances = pd.concat(ttl_roll12_player_gameday_variances)
dt_roll12_player_gameday_means = pd.concat(ttl_roll12_player_gameday_means)
dt_player_game_aggregations = dt_player_gameday_medians.merge(
dt_player_gameday_variances, how="left", on=["playerId", "ddd_month", "played_game"]
)
dt_player_game_aggregations = dt_player_game_aggregations.merge(
dt_player_gameday_means, how="left", on=["playerId", "ddd_month", "played_game"]
)
dt_player_game_aggregations = dt_player_game_aggregations.merge(
dt_roll12_player_gameday_medians,
how="left",
on=["playerId", "ddd_month", "played_game"],
)
dt_player_game_aggregations = dt_player_game_aggregations.merge(
dt_roll12_player_gameday_variances,
how="left",
on=["playerId", "ddd_month", "played_game"],
)
dt_player_game_aggregations = dt_player_game_aggregations.merge(
dt_roll12_player_gameday_means,
how="left",
on=["playerId", "ddd_month", "played_game"],
)
position_freq = (
p_box_scores_og["positionType"].fillna(-999).value_counts(normalize=True).to_dict()
)
position_target_agg = eng.merge(
p_box_scores_og[
["dailyDataDate", "playerId", "gamePk", "gameTimeUTC", "positionType"]
],
how="left",
)
dh_games = (
position_target_agg[
position_target_agg[["dailyDataDate", "playerId"]].duplicated(keep=False)
]
.sort_values("gameTimeUTC")[["dailyDataDate", "playerId", "gamePk"]]
.reset_index(drop=True)
)
dh_last_game = dh_games[
dh_games[["dailyDataDate", "playerId"]].duplicated(keep="first")
] # games to remove
position_target_agg = position_target_agg[
~(
position_target_agg["playerId"].isin(dh_last_game["playerId"])
& position_target_agg["gamePk"].isin(dh_last_game["gamePk"])
)
]
position_freq = (
position_target_agg["positionType"]
.fillna(-999)
.value_counts(normalize=True)
.to_dict()
)
position_target_agg = (
position_target_agg.groupby("positionType")[
["target1", "target2", "target3", "target4"]
]
.agg(
{
"target1": ["median", "var"],
"target2": ["median", "var"],
"target3": ["median", "var"],
"target4": ["median", "var"],
}
)
.round(6)
)
position_target_agg.columns = [
"_".join(x + ("position",)) for x in position_target_agg.columns.ravel()
]
position_target_agg = position_target_agg.reset_index()
last_day = train[train["date"] == last_date]
(
eng,
games,
rosters,
p_box_scores,
t_box_scores,
transactions,
standings,
awards,
events,
p_twitter,
t_twitter,
) = get_unnested_data(last_day, ["nextDayPlayerEngagement"] + colnames)
# eng_lag = eng.copy()
# eng_lag = eng[['playerId','target1','target2','target3','target4']].copy()
# eng_lag = eng_lag.rename({'target1': 'target1_lag',
# 'target2': 'target2_lag',
# 'target3': 'target3_lag',
# 'target4': 'target4_lag'}, axis=1)
try:
if not p_box_scores.empty:
t_tmp = eng.merge(
p_box_scores[
["dailyDataDate", "playerId", "positionCode", "pitchesThrown"]
],
how="left",
on=["dailyDataDate", "playerId"],
)
t_tmp["position_player_pitching"] = (
(t_tmp["positionCode"] > 1) & (t_tmp["pitchesThrown"] > 0)
).astype(int)
prior_day_pos_player_pitching = t_tmp.loc[
t_tmp["position_player_pitching"] == 1,
["playerId", "position_player_pitching"],
].fillna(0)
else:
eng["position_player_pitching"] = 0
prior_data_pos_player_pitching = eng[["playerId", "position_player_pitching"]]
except Exception as e:
print(e)
eng["position_player_pitching"] = 0
prior_data_pos_player_pitching = eng[["playerId", "position_player_pitching"]]
# Instead of using specific date, find latest date with twitter data available
last_twitter_date = train.loc[train["playerTwitterFollowers"].notnull(), "date"].max()
second_last_twitter_date = train.loc[
train["playerTwitterFollowers"].notnull(), "date"
].iloc[-2]
last_twitter_update = train[train["date"] == last_twitter_date]
second_last_twitter_update = train[train["date"] == second_last_twitter_date]
p_twitter, _ = get_unnested_data(
last_twitter_update, ["playerTwitterFollowers", "teamTwitterFollowers"]
)
p_twitter_recent = p_twitter.copy()
p_twitter_second_last, _ = get_unnested_data(
second_last_twitter_update, ["playerTwitterFollowers", "teamTwitterFollowers"]
)
p_twitter_recent = p_twitter_recent.set_index("playerId")
p_twitter_second_last = p_twitter_second_last.set_index("playerId")
p_twitter_delta = (
(p_twitter_recent["numberOfFollowers"] - p_twitter_second_last["numberOfFollowers"])
.reset_index()
.rename(columns={"numberOfFollowers": "numberOfFollowers_delta"})
)
p_twitter_recent = p_twitter_recent.reset_index()
# games = unnest(train, 'games')
# schedule_21 = pd.read_csv("../input/mlbdata/schedule_2021.csv")
# schedule_21['gameDate'] = pd.to_datetime(schedule_21['gameDate'])
# games['gameDate'] = pd.to_datetime(games['gameDate'])
# games = games.sort_values('gameDate')
# schedule = pd.concat([games[['dailyDataDate', 'homeId', 'gameDate']].rename({'homeId': 'teamId'}, axis=1),
# games[['dailyDataDate', 'awayId', 'gameDate']].rename({'awayId': 'teamId'}, axis=1)])
# schedule = schedule[schedule['dailyDataDate']<20210401]
# schedule = pd.concat([schedule, schedule_21[['dailyDataDate','teamId','gameDate']]])
# schedule['gameDate'] = pd.to_datetime(schedule['gameDate'])
# all_dates = pd.DataFrame(list(itertools.product(pd.date_range(start="2018-01-01", end="2021-12-31"), schedule['teamId'].unique())), columns=['gameDate', 'teamId'])
# all_dates = all_dates.merge(schedule, how='outer', on=['gameDate','teamId'])
# all_dates = all_dates.sort_values(['teamId','gameDate']).drop_duplicates()
# all_dates['dailyDataDate_lead'] = all_dates.groupby('teamId')['dailyDataDate'].shift(1)
# all_dates = all_dates[all_dates['dailyDataDate_lead'].notnull()].reset_index(drop=True)
# all_dates['nextDayGame'] = 1
awards_dict = (
awards_history[awards_history["awardId"].isin(keep_awards + keep_annual_awards)]
.groupby(["playerId", "awardId"])
.size()
.to_dict()
)
awards_dict_tmp = {
n: grp.to_dict("list")
for n, grp in awards_history.loc[
awards_history["awardId"].isin(keep_awards + keep_annual_awards),
["awardId", "playerId"],
].groupby("playerId")
}
awards_dict = {}
for k, v in awards_dict_tmp.items():
if not k in awards_dict:
awards_dict[k] = {}
counts = np.unique(v["awardId"], return_counts=True)
for feat, value in zip(counts[0], counts[1]):
awards_dict[k][feat] = value
hitter_history_dict = {}
fielder_history_dict = {}
pitcher_history_dict = {}
for i, data in tqdm(train[train["date"] <= last_date].iterrows()):
try:
data = data.to_frame().T
daily_data_date = data["date"].iloc[0]
season = int(str(daily_data_date)[:4])
p_box_scores, games, rosters, awards = get_unnested_data(
data, ["playerBoxScores", "games", "rosters", "awards"]
)
if rosters.empty:
rosters = prior_day_rosters
prior_day_rosters = rosters.copy()
if not games.empty:
games_filtered = games.loc[
games["gameType"].isin(["R", "F", "D", "L", "W", "C", "P"])
& ~games["detailedGameState"].isin(["Postponed"])
]
if not games_filtered.empty:
schedule_day = pd.concat(
[
games_filtered[
[
"dailyDataDate",
"gamePk",
"homeId",
"gameDate",
"gameTimeUTC",
"homeWinner",
]
].rename({"homeId": "teamId", "homeWinner": "winner"}, axis=1),
games_filtered[
[
"dailyDataDate",
"gamePk",
"awayId",
"gameDate",
"gameTimeUTC",
"awayWinner",
]
].rename({"awayId": "teamId", "awayWinner": "winner"}, axis=1),
]
)
schedule_day = schedule_day.sort_values("gameTimeUTC")
if not schedule_day.empty and not p_box_scores.empty:
game_rosters = schedule_day.merge(
rosters, how="left", on=["gameDate", "teamId"]
)
game_rosters = game_rosters[
game_rosters["playerId"].notnull()
] # missing roster for Nationals 20200910
game_rosters["playerId"] = game_rosters["playerId"].astype(int)
p_box_scores = p_box_scores.sort_values("gameTimeUTC")
p_box_scores["gameDate"] = pd.to_datetime(p_box_scores["gameDate"])
p_box_scores["season"] = p_box_scores["gameDate"].dt.year
player_history_daily = game_rosters.merge(
p_box_scores, how="left", on=["gamePk", "playerId"]
)
player_history_daily["gameTimeUTC_y"] = player_history_daily[
"gameTimeUTC_y"
].fillna(player_history_daily["gameTimeUTC_x"])
# NOTE: dailyDataDate==2020918 gamePk==631122 Start time of 2020-09-18T03:33:00Z is not accurate; that would imply the game started the day before at ~11:30PM local time
player_history_daily = player_history_daily.sort_values(
["playerId", "gameTimeUTC_y"]
) # SORT BY gameTimeUTC from p_box_scores. `gameTimeUTC` is not accurate from the `games` data
player_history_daily[hitter_history_feats] = player_history_daily[
hitter_history_feats
].fillna(0)
hitter_history_tmp = {
n: grp.to_dict("list")
for n, grp in player_history_daily[
hitter_history_feats + ["season", "playerId"]
].groupby("playerId")
}
for k, v in hitter_history_tmp.items():
if not k in hitter_history_dict:
hitter_history_dict[k] = v
else:
for feat in hitter_history_feats + ["season"]:
hitter_history_dict[k][feat].extend(v[feat])
# For hitters, only use games they played in. Pitchers need off days filled in because it's important to account for rest/off days
# Fill in days with 0 if hitter isn't in daily box scores
# for k,v in hitter_history_dict.items():
# if not k in hitter_history_tmp:
# for feat in hitter_history_feats + ['season']:
# hitter_history_dict[k][feat].append(season if feat=='season' else 0.0)
fielder_history_tmp = {
n: grp.to_dict("list")
for n, grp in player_history_daily[
fielder_history_feats + ["season", "playerId"]
].groupby("playerId")
}
for k, v in fielder_history_tmp.items():
if not k in fielder_history_dict:
fielder_history_dict[k] = v
else:
for feat in fielder_history_feats + ["season"]:
fielder_history_dict[k][feat].extend(v[feat])
pitcher_history_tmp = {
n: grp.to_dict("list")
for n, grp in p_box_scores.loc[
p_box_scores["positionName"] == "Pitcher",
pitcher_history_feats + ["season", "playerId"],
].groupby("playerId")
}
for k, v in pitcher_history_tmp.items():
if not k in pitcher_history_dict:
pitcher_history_dict[k] = v
else:
for feat in pitcher_history_feats + ["season"]:
pitcher_history_dict[k][feat].extend(v[feat])
# Fill in days with 0 if pitcher isn't in daily box scores
for k, v in pitcher_history_dict.items():
if not k in pitcher_history_tmp:
for feat in pitcher_history_feats + ["season"]:
pitcher_history_dict[k][feat].append(
season if feat == "season" else 0.0
)
except Exception as e:
# If fails, just move on to the next day
print(f"history dicts loop failed: {e}")
pass
try:
if not awards.empty:
awards_filtered = awards[
awards["awardId"].isin(keep_awards + keep_annual_awards)
].reset_index(drop=True)
# Update awards counts
awards_dict_tmp = {
n: grp.to_dict("list")
for n, grp in awards_filtered[["awardId", "playerId"]].groupby(
"playerId"
)
}
for k, v in awards_dict_tmp.items():
try:
if not k in awards_dict:
awards_dict[k] = {}
counts = np.unique(v["awardId"], return_counts=True)
for feat, value in zip(counts[0], counts[1]):
if feat in awards_dict[k]:
awards_dict[k][feat] += value
else:
awards_dict[k][feat] = value
except:
# If fails, move on to the next one
pass
except Exception as e:
# If fails, don't worry about updating dict
print(e)
pass
games_og = unnest(train, "games")
schedule_og = pd.concat(
[
games_og.loc[
games_og["gameType"].isin(["R", "F", "D", "L", "W", "C", "P"])
& ~games_og["detailedGameState"].isin(["Postponed"]),
[
"dailyDataDate",
"gamePk",
"homeId",
"gameDate",
"gameTimeUTC",
"homeWinner",
],
].rename({"homeId": "teamId", "homeWinner": "winner"}, axis=1),
games_og.loc[
games_og["gameType"].isin(["R", "F", "D", "L", "W", "C", "P"])
& ~games_og["detailedGameState"].isin(["Postponed"]),
[
"dailyDataDate",
"gamePk",
"awayId",
"gameDate",
"gameTimeUTC",
"awayWinner",
],
].rename({"awayId": "teamId", "awayWinner": "winner"}, axis=1),
]
)
schedule_og = schedule_og.sort_values("gameTimeUTC")
schedule_og = schedule_og[schedule_og["dailyDataDate"] <= last_date]
schedule_og["gameDate"] = pd.to_datetime(schedule_og["gameDate"])
team_win_history = {}
team_win_dict = schedule_og.groupby("teamId")["winner"].apply(list).to_dict()
for k, v in team_win_dict.items():
if not k in team_win_history:
team_win_history[k] = v
else:
team_win_history[k].extend(v)
win_streaks = {
k: v[::-1].index(0) if 0 in v else len(v) for k, v in team_win_history.items()
}
# Load models
lgb_target1 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target1_v30_full.txt"
)
lgb_target2 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target2_v30_full.txt"
)
lgb_target3 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target3_v30_full.txt"
)
lgb_target4 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target4_v30_full.txt"
)
lgb_dblsqrt_target1 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target1_dblsqrt_full.txt"
)
lgb_dblsqrt_target2 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target2_dblsqrt_full.txt"
)
lgb_dblsqrt_target3 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target3_dblsqrt_full.txt"
)
lgb_dblsqrt_target4 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target4_dblsqrt_full.txt"
)
lgb_bfa_target1 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target1_v30_bfa_full.txt"
)
lgb_bfa_target2 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target2_v30_bfa_full.txt"
)
lgb_bfa_target3 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target3_v30_bfa_full.txt"
)
lgb_bfa_target4 = lgb.Booster(
model_file="../input/d/brandenkmurray/mlbmodels/lgb_target4_v30_bfa_full.txt"
)
xgb_target1 = xgb.Booster()
xgb_target2 = xgb.Booster()
xgb_target3 = xgb.Booster()
xgb_target4 = xgb.Booster()
xgb_target1.load_model("../input/d/brandenkmurray/mlbmodels/xgb_target1_v30_full.txt")
xgb_target2.load_model("../input/d/brandenkmurray/mlbmodels/xgb_target2_v30_full.txt")
xgb_target3.load_model("../input/d/brandenkmurray/mlbmodels/xgb_target3_v30_full.txt")
xgb_target4.load_model("../input/d/brandenkmurray/mlbmodels/xgb_target4_v30.txt")
lgb_john_target1 = lgb.Booster(
model_file="../input/mlb-models-and-files/lgb_target1_dubs_tripsX_all.txt"
)
lgb_john_target2 = lgb.Booster(
model_file="../input/mlb-models-and-files/lgb_target2_dubs_tripsX_all.txt"
)
lgb_john_target3 = lgb.Booster(
model_file="../input/mlb-models-and-files/lgb_target3_dubs_tripsX_all.txt"
)
lgb_john_target4 = lgb.Booster(
model_file="../input/mlb-models-and-files/lgb_target4_dubs_tripsX_all.txt"
)
lgb_dart_target1 = lgb.Booster(
model_file="../input/dart-model/lgb_target1_dubs_trips_dart_full_data.txt"
)
lgb_dart_target2 = lgb.Booster(
model_file="../input/dart-model/lgb_target2_dubs_trips_dart_full_data.txt"
)
lgb_dart_target3 = lgb.Booster(
model_file="../input/dart-model/lgb_target3_dubs_trips_dart_full_data.txt"
)
lgb_dart_target4 = lgb.Booster(
model_file="../input/dart-model/lgb_target4_dubs_trips_dart_full_data.txt"
)
yesterday = pd.DataFrame()
t = []
sub_list = []
env = mlb.make_env() # initialize the environment
iter_test = env.iter_test() # iterator which loops over each date in test set
for i, (data, sub) in enumerate(iter_test):
# for i, (i2, data) in enumerate(train[(train['date']>=20210501) & (train['date']<=20210731)].iloc[1:].iterrows()):
### REMOVE below
# data = data.to_frame().T
# sub = unnest(data, 'nextDayPlayerEngagement')
# sub = sub.rename(columns={'target1': 'target1_act', 'target2': 'target2_act', 'target3': 'target3_act', 'target4': 'target4_act'})
# sub['date_playerId'] = pd.to_datetime(sub['engagementMetricsDate']).dt.strftime("%Y%m%d") + "_" + sub['playerId'].astype(str)
### REMOVE above
### UNCOMMENT BELOW
sub = sub.reset_index()
sub = sub.rename({"date": "dailyDataDate"}, axis=1)
sub["playerId"] = sub["date_playerId"].apply(lambda x: int(x.split("_")[1]))
data = data.reset_index()
data = data.rename({"index": "date"}, axis=1)
### UNCOMMENT ABOVE
try:
season = int(str(data["date"].iloc[0])[:4])
except:
season = 2021.0
try:
(
games,
rosters,
p_box_scores,
t_box_scores,
transactions,
standings,
awards,
events,
p_twitter,
t_twitter,
) = get_unnested_data(data, colnames)
eng_shape = sub.shape
t_tmp = sub.copy()
if rosters.empty:
rosters = prior_day_rosters
prior_day_rosters = rosters.copy()
if not p_twitter.empty:
# Get twitter follower delta if not the first month
if not p_twitter_recent.empty:
p_twitter = p_twitter.set_index("playerId")
p_twitter_recent = p_twitter_recent.set_index("playerId")
p_twitter_delta = (
(
p_twitter["numberOfFollowers"]
- p_twitter_recent["numberOfFollowers"]
)
.reset_index()
.rename(columns={"numberOfFollowers": "numberOfFollowers_delta"})
)
p_twitter = p_twitter.reset_index()
p_twitter_recent = p_twitter
if not games.empty:
schedule_daily = pd.concat(
[
games.loc[
games["gameType"].isin(["R", "F", "D", "L", "W", "C", "P"])
& ~games["detailedGameState"].isin(["Postponed"]),
[
"dailyDataDate",
"gamePk",
"homeId",
"gameDate",
"gameTimeUTC",
"homeWinner",
],
].rename({"homeId": "teamId", "homeWinner": "winner"}, axis=1),
games.loc[
games["gameType"].isin(["R", "F", "D", "L", "W", "C", "P"])
& ~games["detailedGameState"].isin(["Postponed"]),
[
"dailyDataDate",
"gamePk",
"awayId",
"gameDate",
"gameTimeUTC",
"awayWinner",
],
].rename({"awayId": "teamId", "awayWinner": "winner"}, axis=1),
]
)
schedule_daily = schedule_daily.sort_values("gameTimeUTC")
team_win_dict = (
schedule_daily.groupby("teamId")["winner"].apply(list).to_dict()
)
for k, v in team_win_dict.items():
if not k in team_win_history:
team_win_history[k] = v
else:
team_win_history[k].extend(v)
win_streaks = {
k: v[::-1].index(0) if 0 in v else len(v)
for k, v in team_win_history.items()
}
if not schedule_daily.empty and not p_box_scores.empty:
game_rosters = schedule_daily.merge(
rosters, how="left", on=["gameDate", "teamId"]
)
game_rosters = game_rosters[
game_rosters["playerId"].notnull()
] # missing roster for Nationals 20200910
game_rosters["playerId"] = game_rosters["playerId"].astype(int)
p_box_scores = p_box_scores.sort_values("gameTimeUTC")
p_box_scores["gameDate"] = pd.to_datetime(p_box_scores["gameDate"])
p_box_scores["season"] = p_box_scores["gameDate"].dt.year
player_history_daily = game_rosters.merge(
p_box_scores, how="left", on=["gamePk", "playerId"]
)
player_history_daily["gameTimeUTC_y"] = player_history_daily[
"gameTimeUTC_y"
].fillna(player_history_daily["gameTimeUTC_x"])
# NOTE: dailyDataDate==2020918 gamePk==631122 Start time of 2020-09-18T03:33:00Z is not accurate; that would imply the game started the day before at ~11:30PM local time
player_history_daily = player_history_daily.sort_values(
["playerId", "gameTimeUTC_y"]
) # SORT BY gameTimeUTC from p_box_scores. `gameTimeUTC` is not accurate from the `games` data
player_history_daily[hitter_history_feats] = player_history_daily[
hitter_history_feats
].fillna(0)
hitter_history_tmp = {
n: grp.to_dict("list")
for n, grp in player_history_daily[
hitter_history_feats + ["season", "playerId"]
].groupby("playerId")
}
for k, v in hitter_history_tmp.items():
if not k in hitter_history_dict:
hitter_history_dict[k] = v
else:
for feat in hitter_history_feats + ["season"]:
hitter_history_dict[k][feat].extend(v[feat])
# For hitters, only use games they played in. Pitchers need off days filled in because it's important to account for rest/off days
# Fill in days with 0 if hitter isn't in daily box scores
# for k,v in hitter_history_dict.items():
# if not k in hitter_history_tmp:
# for feat in hitter_history_feats + ['season']:
# hitter_history_dict[k][feat].append(season if feat=='season' else 0.0)
fielder_history_tmp = {
n: grp.to_dict("list")
for n, grp in player_history_daily[
fielder_history_feats + ["season", "playerId"]
].groupby("playerId")
}
for k, v in fielder_history_tmp.items():
if not k in fielder_history_dict:
fielder_history_dict[k] = v
else:
for feat in fielder_history_feats + ["season"]:
fielder_history_dict[k][feat].extend(v[feat])
pitcher_history_tmp = {
n: grp.to_dict("list")
for n, grp in p_box_scores.loc[
p_box_scores["positionName"] == "Pitcher",
pitcher_history_feats + ["season", "playerId"],
].groupby("playerId")
}
for k, v in pitcher_history_tmp.items():
if not k in pitcher_history_dict:
pitcher_history_dict[k] = v
else:
for feat in pitcher_history_feats + ["season"]:
pitcher_history_dict[k][feat].extend(v[feat])
# Fill in days with 0 if pitcher isn't in daily box scores
for k, v in pitcher_history_dict.items():
if not k in pitcher_history_tmp:
for feat in pitcher_history_feats + ["season"]:
pitcher_history_dict[k][feat].append(
season if feat == "season" else 0.0
)
days_of_history = list(
range(2, 21)
) # [2,3,4,5,7,10,20] #also could be games_of_history depending how its used
max_days_of_history = np.max(days_of_history)
hitting_history_features = {}
pitching_history_features = {}
fielding_history_features = {}
for k, v in hitter_history_dict.items():
# only need to include players in the current eng
hitting_history_features[k] = {}
hitting_history_features[k]["hit_streak"] = (
v["hits"][::-1].index(0) if 0 in v["hits"] else len(v["hits"])
)
for feat in hitter_history_feats:
d = hitter_history_dict[k][feat]
hitting_history_features[k][f"{feat}_season"] = sum(
[
f
for seas, f in zip(hitter_history_dict[k]["season"], d)
if seas == season
]
)
if feat not in ["sacFlies", "atBats"]:
d_padded = np.zeros(max_days_of_history)
d_padded[: np.minimum(max_days_of_history, len(d))] = d[
-np.minimum(max_days_of_history, len(d)) :
][::-1]
d_cumsum = nb_cumsum(d_padded)
for day in days_of_history:
hitting_history_features[k][f"{feat}_last{day}"] = d_cumsum[
day - 1
]
# hitting_history_features[k][f'{feat}_{day-1}_games_ago'] = d_padded[day-1]
hitting_history_df = (
pd.DataFrame.from_dict(hitting_history_features, orient="index")
.reset_index()
.rename({"index": "playerId"}, axis=1)
)
if "homeRuns_season" in hitting_history_df.columns:
hitting_history_df["homeRuns_rank"] = hitting_history_df[
"homeRuns_season"
].rank(method="min", ascending=False)
hitting_history_df["BA"] = (
hitting_history_df["hits_season"] / hitting_history_df["atBats_season"]
)
hitting_history_df["OBP"] = hitting_history_df[
["hits_season", "baseOnBalls_season", "hitByPitch_season"]
].sum(axis=1) / hitting_history_df[
[
"atBats_season",
"baseOnBalls_season",
"hitByPitch_season",
"sacFlies_season",
]
].sum(
axis=1
)
hitting_history_df["SLG"] = (
(
hitting_history_df["hits_season"]
- hitting_history_df[
["doubles_season", "triples_season", "homeRuns_season"]
].sum(axis=1)
)
+ 2 * hitting_history_df["doubles_season"]
+ 3 * hitting_history_df["triples_season"]
+ 4 * hitting_history_df["homeRuns_season"]
) / hitting_history_df["atBats_season"]
for k, v in fielder_history_dict.items():
# only need to include players in the current eng
fielding_history_features[k] = {}
for feat in fielder_history_feats:
d = fielder_history_dict[k][feat]
d_padded = np.zeros(max_days_of_history)
d_padded[: np.minimum(max_days_of_history, len(d))] = d[
-np.minimum(max_days_of_history, len(d)) :
][::-1]
# d_padded = np.pad(d[-days_of_history:], (np.maximum(0, days_of_history-len(d)+1), 0))[::-1]
d_cumsum = nb_cumsum(d_padded)
for day in days_of_history:
# fielding_history_features[k][f'{feat}_last{day}'] = d_cumsum[day-1]
fielding_history_features[k][
f"{feat}_{day-1}_games_ago"
] = d_padded[day - 1]
fielding_history_df = (
pd.DataFrame.from_dict(fielding_history_features, orient="index")
.reset_index()
.rename({"index": "playerId"}, axis=1)
)
for k, v in pitcher_history_dict.items():
# only need to include players in the current eng
pitching_history_features[k] = {}
season_starts = [
starts
for seas, starts in zip(
pitcher_history_dict[k]["season"],
pitcher_history_dict[k]["gamesStartedPitching"],
)
if seas == season
]
season_played = [
played
for seas, played in zip(
pitcher_history_dict[k]["season"],
pitcher_history_dict[k]["gamesPlayedPitching"],
)
if seas == season
]
pitching_history_features[k]["season_starts_to_date"] = sum(season_starts)
pitching_history_features[k]["days_since_last_start"] = (
season_starts[::-1].index(1.0)
if 1 in season_starts
else len(season_starts)
)
pitching_history_features[k]["days_since_last_played"] = (
season_played[::-1].index(1.0)
if 1 in season_played
else len(season_played)
)
for feat in [
"gamesPlayedPitching",
"gamesStartedPitching",
"inningsPitched",
"pitchesThrown",
"winsPitching",
"runsPitching",
"homeRunsPitching",
"strikeOutsPitching",
"earnedRuns",
"blownSaves",
"holds",
]:
d = pitcher_history_dict[k][feat]
pitching_history_features[k][f"{feat}_season"] = sum(
[
f
for seas, f in zip(pitcher_history_dict[k]["season"], d)
if seas == season
]
)
d_padded = np.pad(d, (np.maximum(0, max_days_of_history - len(d)), 0))[
::-1
]
d_cumsum = nb_cumsum(d_padded)
for day in days_of_history:
pitching_history_features[k][f"{feat}_last{day}"] = d_cumsum[
day - 1
]
pitching_history_features[k][
f"{feat}_{day-1}_games_ago"
] = d_padded[day - 1]
pitching_history_df = (
pd.DataFrame.from_dict(pitching_history_features, orient="index")
.reset_index()
.rename({"index": "playerId"}, axis=1)
)
if not p_twitter.empty:
p_twitter_recent = p_twitter
# How to handle doubleheaders? Taking stats from first game for now
if not p_box_scores.empty and not t_box_scores.empty:
t_tmp = t_tmp.merge(
p_box_scores, how="left", on=["dailyDataDate", "playerId"]
)
dh_games = (
t_tmp[t_tmp[["dailyDataDate", "playerId"]].duplicated(keep=False)]
.sort_values("gameTimeUTC")[["dailyDataDate", "playerId", "gamePk"]]
.reset_index(drop=True)
)
dh_last_game = dh_games[
dh_games[["dailyDataDate", "playerId"]].duplicated(keep="first")
] # games to remove
t_tmp = t_tmp[
~(
t_tmp["playerId"].isin(dh_last_game["playerId"])
& t_tmp["gamePk"].isin(dh_last_game["gamePk"])
)
]
t_tmp["game_score_james"] = game_score_james(t_tmp)
t_tmp["game_score_tango"] = game_score_tango(t_tmp)
t_tmp["position_player_pitching"] = (
(t_tmp["positionCode"] > 1) & (t_tmp["pitchesThrown"] > 0)
).astype(int)
t_tmp["pitcher_hit_home_run"] = (
(t_tmp["positionCode"] == 1) & (t_tmp["homeRuns"] > 0)
).astype(int)
# t_tmp['pos_player_pitched_prior_day'] = 0
# if not prior_day_pos_player_pitching.empty:
# t_tmp['pos_player_pitched_prior_day'] = t_tmp['playerId'].map(dict(zip(prior_day_pos_player_pitching.playerId, prior_day_pos_player_pitching.position_player_pitching)))
t_tmp["no_hitter"] = (
(t_tmp["inningsPitched"] >= 9) & (t_tmp["hitsPitching"] == 0)
).astype(int)
t_tmp["no_hitter_league"] = t_tmp["no_hitter"].max()
t_tmp["position_player_pitching_league"] = t_tmp[
"position_player_pitching"
].max()
t_tmp["game_hour"] = (
pd.to_datetime(t_tmp["gameTimeUTC"]) + pd.Timedelta(hours=-5)
).dt.hour
t_tmp = t_tmp.merge(
t_box_scores,
how="left",
on=["gamePk", "teamId"],
suffixes=["", "_team_box_score"],
)
t_tmp["positionType_freq"] = (
t_tmp["positionType"].fillna(-999).map(position_freq)
)
if "positionType" in t_tmp.columns:
t_tmp = t_tmp.merge(position_target_agg, how="left", on="positionType")
if t_tmp.shape[0] != eng_shape[0]:
print(
"t_tmp length does not match engagement frame length, check for duplicated data"
)
t_tmp = t_tmp[~t_tmp[["playerId"]].duplicated()]
else:
if "teamId" not in t_tmp.columns and not rosters.empty:
t_tmp = t_tmp.merge(
rosters[["playerId", "teamId"]], how="left", on="playerId"
)
else:
t_tmp["teamId"] = np.nan
if t_tmp.shape[0] != eng_shape[0]:
print(
"teamId: t_tmp length does not match engagement frame length, check for duplicated data"
)
t_tmp = t_tmp[~t_tmp[["playerId"]].duplicated()]
# Did player have a walk-off hit/home run?
if not events.empty:
events = events.sort_values(
["inning", "halfInning", "atBatIndex", "eventId"],
ascending=[True, False, True, True],
)
last_play = events.groupby("gamePk").tail(1)
# filter out top of inning because one game was ended after the top of the inning
walk_offs = last_play[
(last_play["halfInning"] == "bottom") & (last_play["rbi"] > 0)
][["dailyDataDate", "hitterId", "pitcherId", "rbi", "event"]]
walk_offs.columns = [
"dailyDataDate",
"hitterId",
"pitcherId",
"walk_off_rbi",
"walk_off_hr",
]
walk_offs["walk_off_hr"] = (
walk_offs["walk_off_hr"].isin(["Home Run"])
).astype(int)
t_tmp = t_tmp.merge(
walk_offs[
["dailyDataDate", "hitterId", "walk_off_hr", "walk_off_rbi"]
].rename({"hitterId": "playerId"}, axis=1),
how="left",
on=["dailyDataDate", "playerId"],
)
t_tmp = t_tmp.merge(
walk_offs[
["dailyDataDate", "pitcherId", "walk_off_hr", "walk_off_rbi"]
].rename({"pitcherId": "playerId"}, axis=1),
how="left",
on=["dailyDataDate", "playerId"],
suffixes=["", "_pitcher"],
)
t_tmp[
[
"walk_off_rbi",
"walk_off_hr",
"walk_off_hr_pitcher",
"walk_off_rbi_pitcher",
]
] = t_tmp[
[
"walk_off_rbi",
"walk_off_hr",
"walk_off_hr_pitcher",
"walk_off_rbi_pitcher",
]
].fillna(
0
)
t_tmp["walk_off_league"] = t_tmp["walk_off_rbi"].max()
hr_dist = (
events[events["event"] == "Home Run"]
.groupby("hitterId")["totalDistance"]
.max()
.reset_index()
)
hr_launchSpeed = (
events[events["event"] == "Home Run"]
.groupby("hitterId")["launchSpeed"]
.max()
.reset_index()
)
t_tmp = t_tmp.merge(
hr_dist.rename({"hitterId": "playerId"}, axis=1),
how="left",
on="playerId",
)
t_tmp = t_tmp.merge(
hr_launchSpeed.rename({"hitterId": "playerId"}, axis=1),
how="left",
on="playerId",
)
# How long did a starting pitcher go without a hit? (Did they start picking up potential no-hitter hype?)
starters = events[events["isStarter"] == 1].reset_index(drop=True)
starters["hit"] = (
starters["event"]
.isin(["Single", "Double", "Triple", "Home Run"])
.astype(int)
)
starters["hits_cumsum"] = starters.groupby("pitcherId")["hit"].cumsum()
starters_first_hit_inning = (
starters[starters["hits_cumsum"] == 1]
.groupby("pitcherId")
.first()[["inning", "outs"]]
.reset_index()
)
starters_first_hit_inning["inning"] = (
starters_first_hit_inning["inning"]
+ starters_first_hit_inning["outs"] / 10
)
starters_first_hit_inning = starters_first_hit_inning.rename(
{"inning": "pitcher_first_hit_inning"}, axis=1
)
t_tmp = t_tmp.merge(
starters_first_hit_inning[["pitcherId", "pitcher_first_hit_inning"]],
how="left",
left_on="playerId",
right_on="pitcherId",
)
starters_first_mob_inning = starters[
~starters["menOnBase"].isin([None, "Empty"])
]
starters_first_mob_inning = (
starters_first_mob_inning.groupby("pitcherId")
.first()[["inning", "outs"]]
.reset_index()
)
starters_first_mob_inning["inning"] = (
starters_first_mob_inning["inning"]
+ starters_first_mob_inning["outs"] / 10
)
starters_first_mob_inning = starters_first_mob_inning.rename(
{"inning": "pitcher_first_mob_inning"}, axis=1
)
t_tmp = t_tmp.merge(
starters_first_mob_inning[["pitcherId", "pitcher_first_mob_inning"]],
how="left",
left_on="playerId",
right_on="pitcherId",
)
# Pitch features
nastyFactor_features = (
events[events["type"] == "pitch"]
.groupby("pitcherId")["nastyFactor"]
.agg(["mean", "median", "min", "max"])
.reset_index()
.rename(
columns={
f: f"nastyFactor_{f}" for f in ["mean", "median", "max", "min"]
}
)
.rename(columns={"pitcherId": "playerId"})
)
t_tmp = t_tmp.merge(nastyFactor_features, how="left", on="playerId")
# Calculate player Win Probability Added
# need to get assign 100% WPA to winning team to assign WPA scores to correct player/team
player_wpa = pd.Series(dtype=float)
for gamePk, game in events.groupby("gamePk"):
game = game.reset_index(drop=True)
game["run_diff"] = game["homeScore"] - game["awayScore"]
game["halfInning_index"] = game["halfInning"].map(
{"top": 1, "bottom": 2}
)
game["base_state"] = game["menOnBase"].map(
{None: np.nan, "Empty": 1, "Men_On": 2, "RISP": 3, "Loaded": 8}
)
game["base_state"] = game["base_state"].ffill().fillna(1).astype(int)
game["outs_beg"] = np.maximum(game["outs"] - 1, 0)
game["win_exp"] = game.apply(winnexp_feature, axis=1)
game["win_exp_lag"] = game["win_exp"].shift(-1)
game.loc[game.shape[0] - 1, "win_exp_lag"] = (
1
if game.loc[game.shape[0] - 1, "homeScore"]
> game.loc[game.shape[0] - 1, "awayScore"]
else 0
)
game["win_exp_delta"] = game["win_exp_lag"] - game["win_exp"]
# Increases in the top of the inning are assigned to the pitcher
# Increases in the bottom of the inning are assigned to the hitter
pitcher_wpa_top = (
game.loc[
(game["halfInning"] == "top") & (game["win_exp_delta"] > 0),
["pitcherId", "win_exp_delta"],
]
.groupby("pitcherId")["win_exp_delta"]
.sum()
)
hitter_wpa_top = (
game.loc[
(game["halfInning"] == "top") & (game["win_exp_delta"] > 0),
["hitterId", "win_exp_delta"],
]
.groupby("hitterId")["win_exp_delta"]
.sum()
)
hitter_wpa_top = -hitter_wpa_top
pitcher_wpa_bot = (
game.loc[
(game["halfInning"] == "bottom") & (game["win_exp_delta"] > 0),
["pitcherId", "win_exp_delta"],
]
.groupby("pitcherId")["win_exp_delta"]
.sum()
)
hitter_wpa_bot = (
game.loc[
(game["halfInning"] == "bottom") & (game["win_exp_delta"] > 0),
["hitterId", "win_exp_delta"],
]
.groupby("hitterId")["win_exp_delta"]
.sum()
)
pitcher_wpa_bot = -pitcher_wpa_bot
player_wpa = player_wpa.add(pitcher_wpa_top, fill_value=0)
player_wpa = player_wpa.add(hitter_wpa_top, fill_value=0)
player_wpa = player_wpa.add(pitcher_wpa_bot, fill_value=0)
player_wpa = player_wpa.add(hitter_wpa_bot, fill_value=0)
player_wpa = player_wpa.reset_index()
player_wpa = player_wpa.rename({"index": "playerId", 0: "wpa"}, axis=1)
t_tmp = t_tmp.merge(player_wpa, how="left", on="playerId")
t_tmp["wpa_daily_max"] = t_tmp["wpa"].max()
t_tmp["wpa_rank"] = t_tmp["wpa"].rank(method="min", ascending=False)
# get ejections
ejections = events.loc[
events["event"] == "Ejection", ["dailyDataDate", "description"]
].reset_index(drop=True)
if not ejections.empty:
ejections["description"] = [
x.split(" ejected by")[0] for x in ejections["description"]
]
# Get team; needed for coach_ejected feature
ejections["teamName"] = [
team_regex.findall(x)[0] if team_regex.findall(x) else None
for x in ejections["description"]
] # else None to account for names not spelled in a way that matches the regex
ejections["teamId"] = ejections["teamName"].map(team_mapping)
ejections["coach_ejected"] = [
1 if coaching_regex.search(x) else 0
for x in ejections["description"]
]
ejections["player_ejected"] = 1 - ejections["coach_ejected"]
# Get player name
ejections["playerName"] = [
team_full_regex.sub("", " ".join(x.split()))
for x in ejections["description"]
]
ejections["playerName"] = [
coaching_regex.sub("", " ".join(x.split()))
for x in ejections["playerName"]
]
ejections["playerName"] = [
pos_regex.sub("", " ".join(x.split())).strip()
for x in ejections["playerName"]
]
# If there is no match for a player use fuzzywuzzy to find the closest match
ejections.loc[
(ejections["player_ejected"] == 1), "playerName"
] = ejections.loc[
(ejections["player_ejected"] == 1), "playerName"
].apply(
lambda x: find_closest_playerName(x, players)
)
ejections.loc[
(ejections["player_ejected"] == 1), "playerId"
] = ejections.loc[(ejections["player_ejected"] == 1)].apply(
lambda x: find_playerId(x, players, rosters), axis=1
)
t_tmp = t_tmp.merge(
ejections.groupby("teamId")["coach_ejected"].sum().reset_index(),
how="left",
on="teamId",
)
t_tmp["coach_ejected"] = t_tmp["coach_ejected"].fillna(0)
t_tmp = t_tmp.merge(
ejections.loc[
ejections["player_ejected"] == 1, ["playerId", "player_ejected"]
],
how="left",
on="playerId",
)
t_tmp["player_ejected"] = t_tmp["player_ejected"].fillna(0)
else:
t_tmp["coach_ejected"] = 0
t_tmp["player_ejected"] = 0
if not rosters.empty:
# if 'teamId' not in t_tmp.columns:
# t_tmp = t_tmp.merge(rosters[['playerId','teamId']], how='left', on='playerId')
# t_tmp = t_tmp.merge(all_dates[['dailyDataDate_lead','teamId','nextDayGame']], how='left', left_on=['dailyDataDate', 'teamId'], right_on=['dailyDataDate_lead','teamId'])
# t_tmp['nextDayGame'] = t_tmp['nextDayGame'].fillna(0)
roster_dummies = pd.concat(
[
rosters[["dailyDataDate", "playerId"]],
pd.get_dummies(rosters["statusCode"]),
],
axis=1,
)
roster_dummies = (
roster_dummies.groupby(["dailyDataDate", "playerId"])
.sum()
.reset_index()
)
for col in [
"A",
"BRV",
"D10",
"D60",
"D7",
"DEC",
"FME",
"PL",
"RES",
"RM",
"SU",
]:
if col not in roster_dummies.columns:
roster_dummies[col] = 0
t_tmp = t_tmp.merge(
roster_dummies, how="left", on=["dailyDataDate", "playerId"]
)
else:
t_tmp[
["A", "BRV", "D10", "D60", "D7", "DEC", "FME", "PL", "RES", "RM", "SU"]
] = 0
t_tmp[
"nextDayGame"
] = 0 # There should be a better way to handle this. Don't want to miss this just because rosters is missing
if not transactions.empty:
transactions_dummies = pd.concat(
[
transactions[["dailyDataDate", "playerId"]],
pd.get_dummies(transactions["typeCode"]),
],
axis=1,
)
transactions_dummies = (
transactions_dummies.groupby(["dailyDataDate", "playerId"])
.sum()
.reset_index()
)
for col in [
"ASG",
"CLW",
"CU",
"DES",
"DFA",
"NUM",
"OPT",
"OUT",
"REL",
"RET",
"RTN",
"SC",
"SE",
"SFA",
"SGN",
"TR",
]:
if col not in transactions_dummies.columns:
transactions_dummies[col] = 0
t_tmp = t_tmp.merge(
transactions_dummies, how="left", on=["dailyDataDate", "playerId"]
)
else:
t_tmp[
[
"ASG",
"CLW",
"CU",
"DES",
"DFA",
"NUM",
"OPT",
"OUT",
"REL",
"RET",
"RTN",
"SC",
"SE",
"SFA",
"SGN",
"TR",
]
] = 0
if not awards.empty:
awards_filtered = awards[
awards["awardId"].isin(keep_awards + keep_annual_awards)
].reset_index(drop=True)
# Update awards counts
awards_dict_tmp = {
n: grp.to_dict("list")
for n, grp in awards_filtered[["awardId", "playerId"]].groupby(
"playerId"
)
}
for k, v in awards_dict_tmp.items():
if not k in awards_dict:
awards_dict[k] = {}
counts = np.unique(v["awardId"], return_counts=True)
for feat, value in zip(counts[0], counts[1]):
if feat in awards_dict[k]:
awards_dict[k][feat] += value
else:
awards_dict[k][feat] = value
awards_filtered = awards[awards["awardId"].isin(keep_awards)].reset_index(
drop=True
)
if not awards_filtered.empty:
awards_dummies = pd.concat(
[
awards_filtered[["dailyDataDate", "playerId"]],
pd.get_dummies(awards_filtered["awardId"]),
],
axis=1,
)
awards_dummies = (
awards_dummies.groupby(["dailyDataDate", "playerId"])
.sum()
.reset_index()
)
for col in keep_awards:
if col not in awards_dummies.columns:
awards_dummies[col] = 0
t_tmp = t_tmp.merge(
awards_dummies, how="left", on=["dailyDataDate", "playerId"]
)
else:
t_tmp[keep_awards] = 0
else:
t_tmp[keep_awards] = 0
if t_tmp.shape[0] != eng_shape[0]:
print(
"awards: t_tmp length does not match engagement frame length, check for duplicated data"
)
t_tmp = t_tmp[~t_tmp[["playerId"]].duplicated()]
awards_df = pd.DataFrame.from_dict(awards_dict, orient="index").fillna(0)
awards_df.columns = [f"{x}_career" for x in awards_df.columns]
t_tmp = t_tmp.merge(
awards_df.reset_index().rename(columns={"index": "playerId"}),
how="left",
on="playerId",
)
t_tmp[awards_df.columns] = t_tmp[awards_df.columns].fillna(0)
if not standings.empty:
standings = standings.replace("-", 0.0)
object_cols = standings.select_dtypes(exclude=["float"]).columns
standings[object_cols] = standings[object_cols].apply(
pd.to_numeric, downcast="float", errors="coerce"
)
bool_cols = standings.select_dtypes(include=["boolean"]).columns
standings[bool_cols] = standings[bool_cols].astype(int)
t_tmp = t_tmp.merge(
standings, how="left", on=["teamId"], suffixes=["", "_team_standings"]
)
t_tmp["team_games_played"] = t_tmp["wins"] + t_tmp["losses"]
if t_tmp.shape[0] != eng_shape[0]:
print(
"standings: t_tmp length does not match engagement frame length, check for duplicated data"
)
t_tmp = t_tmp[~t_tmp[["playerId"]].duplicated()]
if len(win_streaks) > 0:
t_tmp["team_win_streak"] = t_tmp["teamId"].map(win_streaks)
if not hitting_history_df.empty:
t_tmp = t_tmp.merge(hitting_history_df, how="left", on="playerId")
t_tmp["hr_rank"] = t_tmp["homeRuns_season"].rank(ascending=False)
if t_tmp.shape[0] != eng_shape[0]:
print(
"hitting_history_df: t_tmp length does not match engagement frame length, check for duplicated data"
)
t_tmp = t_tmp[~t_tmp[["playerId"]].duplicated()]
if not pitching_history_df.empty:
t_tmp = t_tmp.merge(pitching_history_df, how="left", on="playerId")
# Calculate ERA
# there are no more standings after season end so team_games_played is no longer known
if "team_games_played" in t_tmp.columns:
t_tmp["era"] = 9 * (
t_tmp["earnedRuns_season"] / t_tmp["inningsPitched_season"]
)
t_tmp["era_rank"] = t_tmp.loc[
t_tmp["inningsPitched_season"] >= t_tmp["team_games_played"], "era"
].rank(method="min")
if t_tmp.shape[0] != eng_shape[0]:
print(
"pitching_history_df: t_tmp length does not match engagement frame length, check for duplicated data"
)
t_tmp = t_tmp[~t_tmp[["playerId"]].duplicated()]
if not fielding_history_df.empty:
t_tmp = t_tmp.merge(fielding_history_df, how="left", on="playerId")
if t_tmp.shape[0] != eng_shape[0]:
print(
"fielding_history_df: t_tmp length does not match engagement frame length, check for duplicated data"
)
t_tmp = t_tmp[~t_tmp[["playerId"]].duplicated()]
player_countries = [
"Aruba",
"Australia",
"Bahamas",
"Brazil",
"Canada",
"China",
"Colombia",
"Cuba",
"Curacao",
"Dominican Republic",
"Germany",
"Honduras",
"Japan",
"Lithuania",
"Mexico",
"Netherlands",
"Nicaragua",
"Northern Ireland",
"Panama",
"Peru",
"Puerto Rico",
"Saudi Arabia",
"South Africa",
"South Korea",
"Taiwan",
"U.S. Virgin Islands",
"USA",
"Venezuela",
]
t_tmp = t_tmp.merge(player_country_dummies, how="left", on="playerId")
# Add games features
# if not games.empty and not p_box_scores.empty:
# games['dayNight'] = games['dayNight'].map({'day': 0, 'night': 1})
# games['homeWinner'] = games['homeWinner'].fillna(-1).astype(float)
# t_tmp = t_tmp.merge(games[['gamePk', 'dayNight','homeWinPct','awayWinPct','homeScore','awayScore','homeWinner']], how='left', on='gamePk')
# Add Twitter features
if not p_twitter_recent.empty:
t_tmp = t_tmp.merge(
p_twitter_recent[["playerId", "numberOfFollowers"]],
how="left",
on=["playerId"],
)
if not p_twitter_delta.empty:
t_tmp = t_tmp.merge(p_twitter_delta, how="left", on=["playerId"])
### TRAILING AGGREGATION MERGES
t_tmp["ddd_month"] = (
np.floor(t_tmp.dailyDataDate / 100).clip(upper=MAX_MONTH).astype(int)
)
t_tmp = t_tmp.merge(
dt_player_aggregations, how="left", on=["playerId", "ddd_month"]
)
if "gamePk" in t_tmp.columns:
t_tmp["played_game"] = t_tmp["gamePk"].notnull().astype(float)
else:
t_tmp["played_game"] = 0.0
t_tmp = t_tmp.merge(
dt_player_game_aggregations,
how="left",
on=["playerId", "ddd_month", "played_game"],
)
# t_tmp = t_tmp.merge(recent_player_means, how='left', on='playerId')
t_tmp["monthday"] = t_tmp["dailyDataDate"].astype(str).str[4:].astype(int)
t_tmp["dayofweek"] = pd.to_datetime(
t_tmp["date_playerId"].str.split("_", expand=True)[0]
).dt.dayofweek
t_tmp["data_dayofmonth"] = (
t_tmp["dailyDataDate"].astype(str).str[6:].astype(int)
)
t_tmp["eng_dayofmonth"] = pd.to_datetime(
t_tmp["date_playerId"].str.split("_", expand=True)[0]
).dt.day
# Fill season values with 2021
t_tmp["season"] = season
# t.append(t_tmp)
use_cols = lgb_target1.feature_name()
# Add any missing columns so that it does not crash
missing_cols = [col for col in use_cols if col not in t_tmp.columns]
for col in missing_cols:
print(f"lgb_target1 missing: {col}")
t_tmp[col] = np.nan
sub["target1_v30"] = np.clip(lgb_target1.predict(t_tmp[use_cols]), 0, 100)
sub["target2_v30"] = np.clip(lgb_target2.predict(t_tmp[use_cols]), 0, 100)
sub["target3_v30"] = np.clip(lgb_target3.predict(t_tmp[use_cols]), 0, 100)
sub["target4_v30"] = np.clip(lgb_target4.predict(t_tmp[use_cols]), 0, 100)
use_cols = lgb_bfa_target1.feature_name()
# Add any missing columns so that it does not crash
missing_cols = [col for col in use_cols if col not in t_tmp.columns]
for col in missing_cols:
print(f"lgb_bfa_target1 missing: {col}")
t_tmp[col] = np.nan
sub["target1_v30_bfa"] = np.clip(
lgb_bfa_target1.predict(t_tmp[use_cols]), 0, 100
)
sub["target2_v30_bfa"] = np.clip(
lgb_bfa_target2.predict(t_tmp[use_cols]), 0, 100
)
sub["target3_v30_bfa"] = np.clip(
lgb_bfa_target3.predict(t_tmp[use_cols]), 0, 100
)
sub["target4_v30_bfa"] = np.clip(
lgb_bfa_target4.predict(t_tmp[use_cols]), 0, 100
)
use_cols = lgb_dblsqrt_target1.feature_name()
# Add any missing columns so that it does not crash
missing_cols = [col for col in use_cols if col not in t_tmp.columns]
for col in missing_cols:
print(f"lgb_dblsqrt_target1 missing: {col}")
t_tmp[col] = np.nan
sub["target1_v30_dblsqrt"] = np.clip(
lgb_dblsqrt_target1.predict(t_tmp[use_cols]) ** 2**2, 0, 100
)
sub["target2_v30_dblsqrt"] = np.clip(
lgb_dblsqrt_target2.predict(t_tmp[use_cols]) ** 2**2, 0, 100
)
sub["target3_v30_dblsqrt"] = np.clip(
lgb_dblsqrt_target3.predict(t_tmp[use_cols]) ** 2**2, 0, 100
)
sub["target4_v30_dblsqrt"] = np.clip(
lgb_dblsqrt_target4.predict(t_tmp[use_cols]) ** 2**2, 0, 100
)
dart_use_cols = lgb_dart_target1.feature_name()
# Add any missing columns so that it does not crash
missing_cols = [col for col in dart_use_cols if col not in t_tmp.columns]
for col in missing_cols:
print(f"lgb_dart_target1 missing: {col}")
t_tmp[col] = np.nan
sub["target1_dart"] = np.clip(
lgb_dart_target1.predict(t_tmp[dart_use_cols]), 0, 100
)
sub["target2_dart"] = np.clip(
lgb_dart_target2.predict(t_tmp[dart_use_cols]), 0, 100
)
sub["target3_dart"] = np.clip(
lgb_dart_target3.predict(t_tmp[dart_use_cols]), 0, 100
)
sub["target4_dart"] = np.clip(
lgb_dart_target4.predict(t_tmp[dart_use_cols]), 0, 100
)
with open("../input/d/brandenkmurray/mlbmodels/xgb_v30_use_cols.txt") as f:
xgb_use_cols = [x.rstrip() for x in f.readlines()]
# xgb_use_cols = xgb_target1.feature_names
missing_cols = [col for col in xgb_use_cols if col not in t_tmp.columns]
missing_cols_filled = [x.replace(" ", "_") for x in missing_cols]
t_tmp = t_tmp.rename(
columns={k: v for k, v in zip(missing_cols_filled, missing_cols)}
)
for col in missing_cols:
print(f"{col} is missing for XGB model. Adding and filling with NaN")
t_tmp[col] = np.nan
sub["target1_xgb"] = np.clip(
xgb_target1.predict(
xgb.DMatrix(t_tmp[xgb_use_cols].fillna(-99999).replace(np.inf, -99999))
)
** 2
** 2,
0,
100,
)
sub["target2_xgb"] = np.clip(
xgb_target2.predict(
xgb.DMatrix(t_tmp[xgb_use_cols].fillna(-99999).replace(np.inf, -99999))
)
** 2
** 2,
0,
100,
)
sub["target3_xgb"] = np.clip(
xgb_target3.predict(
xgb.DMatrix(t_tmp[xgb_use_cols].fillna(-99999).replace(np.inf, -99999))
)
** 2
** 2,
0,
100,
)
sub["target4_xgb"] = np.clip(
xgb_target4.predict(
xgb.DMatrix(t_tmp[xgb_use_cols].fillna(-99999).replace(np.inf, -99999))
)
** 2
** 2,
0,
100,
)
correlates = [
"hitBatsmen",
"no_hitter",
"home_team_box_score",
"hitBatsmen_team_box_score",
"season_team_standings",
"sportGamesBack",
"nlWins",
"nlLosses",
"errors_1_games_ago",
]
t_tmp = t_tmp.drop(columns=correlates)
use_cols = lgb_john_target1.feature_name()
missing_cols = [col for col in use_cols if col not in t_tmp.columns]
for col in missing_cols:
print(col + " missing for John's model")
t_tmp[col] = np.nan
sub["target1_john"] = np.clip(lgb_john_target1.predict(t_tmp[use_cols]), 0, 100)
sub["target2_john"] = np.clip(lgb_john_target2.predict(t_tmp[use_cols]), 0, 100)
sub["target3_john"] = np.clip(lgb_john_target3.predict(t_tmp[use_cols]), 0, 100)
sub["target4_john"] = np.clip(lgb_john_target4.predict(t_tmp[use_cols]), 0, 100)
sub["target1"] = (
(sub["target1_john"] * 0.4)
+ (
(
sub["target1_v30_bfa"] * 0.2
+ sub["target1_v30"] * 0.1
+ sub["target1_v30_dblsqrt"] * 0.7
)
* 0.1
)
+ (sub["target1_xgb"] * 0.1)
+ (sub["target1_dart"] * 0.4)
)
sub["target2"] = (
(sub["target2_john"] * 0.4)
+ (
(
sub["target2_v30_bfa"] * 0.2
+ sub["target2_v30"] * 0.1
+ sub["target2_v30_dblsqrt"] * 0.7
)
* 0.1
)
+ (sub["target2_xgb"] * 0.1)
+ (sub["target2_dart"] * 0.4)
)
sub["target3"] = (
(sub["target3_john"] * 0.4)
+ (
(
sub["target3_v30_bfa"] * 0.2
+ sub["target3_v30"] * 0.1
+ sub["target3_v30_dblsqrt"] * 0.7
)
* 0.1
)
+ (sub["target3_xgb"] * 0.1)
+ (sub["target3_dart"] * 0.4)
)
sub["target4"] = (
(sub["target4_john"] * 0.4)
+ (
(
sub["target4_v30_bfa"] * 0.2
+ sub["target4_v30"] * 0.1
+ sub["target4_v30_dblsqrt"] * 0.7
)
* 0.1
)
+ (sub["target4_xgb"] * 0.1)
+ (sub["target4_dart"] * 0.4)
)
except Exception as e:
# If all else fails try to use player means
print(f"Main loop failed: {e}")
try:
print("Using player rolling12 means")
sub["ddd_month"] = (
np.floor(sub.dailyDataDate / 100).clip(upper=MAX_MONTH).astype(int)
)
sub = sub.drop(["target1", "target2", "target3", "target4"], axis=1)
sub = sub.merge(
dt_player_aggregations[
[
"playerId",
"ddd_month",
"roll12_target1_p_gameday_median",
"roll12_target2_p_gameday_median",
"roll12_target3_p_gameday_median",
"roll12_target4_p_gameday_median",
]
],
how="left",
on=["playerId", "ddd_month"],
)
sub = sub.rename(
{
k: v
for k, v in zip(
[
"roll12_target1_p_gameday_median",
"roll12_target2_p_gameday_median",
"roll12_target3_p_gameday_median",
"roll12_target4_p_gameday_median",
],
["target1", "target2", "target3", "target4"],
)
},
axis=1,
)
except Exception as e:
print(e)
# If player means fail, use overall means
print("Player means failed. Using overall means")
sub["target1"] = 0.001046
sub["target2"] = 0.521472
sub["target3"] = 0.001735
sub["target4"] = 0.226034
# Do a final check to ensure there are no duplicate players that will cause a scoring error
sub = sub[~sub[["playerId"]].duplicated()]
# sub_list.append(sub)
env.predict(sub[["date_playerId", "target1", "target2", "target3", "target4"]])
# eng_lag = sub[['playerId','target1','target2','target3','target4']].copy()
# eng_lag = eng_lag.rename({'target1': 'target1_lag',
# 'target2': 'target2_lag',
# 'target3': 'target3_lag',
# 'target4': 'target4_lag'}, axis=1)
# sub_all = pd.concat(sub_list)
# include_players = players[players['playerForTestSetAndFuturePreds']==1]['playerId'].tolist()
# sub_all = sub_all[sub_all['playerId'].isin(include_players)]
# print("v30")
# may_mae_list = []
# june_mae_list = []
# for target in ['target1', 'target2', 'target3', 'target4']:
# may_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_v30'] - sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_act'])))
# june_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_v30'] - sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_act'])))
# # print(may_mae_list)
# print(f"May MAE: {np.mean(may_mae_list)}")
# print(f"June MAE: {np.mean(june_mae_list)}")
# print("v30_bfa")
# may_mae_list = []
# june_mae_list = []
# for target in ['target1', 'target2', 'target3', 'target4']:
# may_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_v30_bfa'] - sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_act'])))
# june_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_v30_bfa'] - sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_act'])))
# # print(may_mae_list)
# print(f"May MAE: {np.mean(may_mae_list)}")
# print(f"June MAE: {np.mean(june_mae_list)}")
# print("v30_dblsqrt")
# may_mae_list = []
# june_mae_list = []
# for target in ['target1', 'target2', 'target3', 'target4']:
# may_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_v30_dblsqrt'] - sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_act'])))
# june_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_v30_dblsqrt'] - sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_act'])))
# # print(may_mae_list)
# print(f"May MAE: {np.mean(may_mae_list)}")
# print(f"June MAE: {np.mean(june_mae_list)}")
# print("john")
# may_mae_list = []
# june_mae_list = []
# for target in ['target1', 'target2', 'target3', 'target4']:
# may_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_john'] - sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_act'])))
# june_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_john'] - sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_act'])))
# # print(may_mae_list)
# print(f"May MAE: {np.mean(may_mae_list)}")
# print(f"June MAE: {np.mean(june_mae_list)}")
# print("xgb")
# may_mae_list = []
# june_mae_list = []
# for target in ['target1', 'target2', 'target3', 'target4']:
# may_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_xgb'] - sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_act'])))
# june_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_xgb'] - sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_act'])))
# # print(may_mae_list)
# print(f"May MAE: {np.mean(may_mae_list)}")
# print(f"June MAE: {np.mean(june_mae_list)}")
# print("dart")
# may_mae_list = []
# june_mae_list = []
# for target in ['target1', 'target2', 'target3', 'target4']:
# may_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_dart'] - sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_act'])))
# june_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_dart'] - sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_act'])))
# # print(may_mae_list)
# print(f"May MAE: {np.mean(may_mae_list)}")
# print(f"June MAE: {np.mean(june_mae_list)}")
# print("blend")
# may_mae_list = []
# june_mae_list = []
# for target in ['target1', 'target2', 'target3', 'target4']:
# may_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target] - sub_all.loc[(sub_all['dailyDataDate']>=20210501) & (sub_all['dailyDataDate']<=20210531), target + '_act'])))
# june_mae_list.append(np.mean(np.abs(sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target] - sub_all.loc[(sub_all['dailyDataDate']>=20210601) & (sub_all['dailyDataDate']<=20210630), target + '_act'])))
# # print(may_mae_list)
# print(f"May MAE: {np.mean(may_mae_list)}")
# print(f"June MAE: {np.mean(june_mae_list)}")
# t_df = pd.concat(t)
# t_df.to_csv("./train_features.csv", index=False)
| false | 3 | 31,607 | 0 | 31,627 | 31,607 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.