hexsha
stringlengths 40
40
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 11
148
| max_stars_repo_name
stringlengths 11
79
| max_stars_repo_licenses
stringclasses 11
values | content
stringlengths 3.39k
756k
| avg_line_length
float64 26
3.16k
| max_line_length
int64 1k
734k
|
|---|---|---|---|---|---|---|---|---|
dfa9d4a1fe919b41b4366f18bc260c7ee6aaa726
|
py
|
python
|
benchmarks/matbench_v0.1_Ax_CrabNet_v1.2.1/notebook.ipynb
|
sparks-baird/matbench
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# # Perform Bayesian optimization of CrabNet hyperparameters using Ax
# ###### Created January 8, 2022
# # Description
# We use [(my fork of) CrabNet](https://github.com/sgbaird/CrabNet) to adjust various hyperparameters for the experimental band gap matbench task (`matbench_expt_gap`). We chose this task because `CrabNet` is currently (2021-01-08) listed at the top of this leaderboard (with `MODNet` just marginally worse) which is likely related to it being a composition-only dataset (`CrabNet` is a composition-only model).
#
# The question we're asking in this additional `matbench` submission is:
# **When a model whose defaults already produce state-of-the-art property prediction performance, to what extent can it benefit from hyperparameter optimization (i.e. tuning parameters such as Neural Network dimensions, learning rates, etc.)?**
#
# Eventually, I plan to incorporate this into (my fork of) `CrabNet`, but for now this can serve as an illustrative example of hyperparameter optimization using Bayesian adaptive design and could certainly be adapted to other models, especially expensive-to-train models (e.g. neural networks) that have not undergone much by way of parameter tuning.
#
# [facebook/Ax](https://github.com/facebook/Ax) is used as the backend for performing Bayesian adaptive design.
#
# For additional files related to this `matbench` submission, see the [crabnet-hyperparameter](https://github.com/sparks-baird/crabnet-hyperparameter) repository.
# # Benchmark name
# Matbench v0.1
# # Package versions
# - ax_platform==0.2.3
# - crabnet==1.2.1
# - scikit_learn==1.0.2
# - matbench==0.5
# - kaleido==0.2.1
# # Algorithm description
# Use Ax Bayesian adaptive design to simultaneously optimize 23 hyperparameters of CrabNet. `100` sequential design iterations were used, and parameters were chosen based on a combination of intuition and algorithm/data constraints (e.g. elemental featurizers which were missing elements contained in the dataset were removed). The first `46` iterations (`23*2` parameters) were based on SOBOL sampling to create a rough initial model, while the remaining `56` iterations were Bayesian adaptive design iterations. For the inner loops (where hyperparameter optimization is performed), the average MAE across each of the *five inner folds* was used as Ax's objective to minimize. The best parameter set was then trained on all the inner fold data and used to predict on the test set (unknown during hyperparameter optimization). This is nested cross-validation (CV), and is computationally expensive. See [automatminer: running a benchmark](https://hackingmaterials.lbl.gov/automatminer/advanced.html#running-a-benchmark) for more information on nested CV.
# ## Imports
# +
import pprint
from os.path import join
from pathlib import Path
import numpy as np
import pandas as pd
import plotly.graph_objects as go
import gc
import torch
from ax.storage.json_store.save import save_experiment
from ax.plot.trace import optimization_trace_single_method
from ax.service.managed_loop import optimize
import crabnet
from crabnet.train_crabnet import get_model
from sklearn.metrics import mean_absolute_error
from sklearn.model_selection import KFold
from matbench.bench import MatbenchBenchmark
# -
# ## Setup
# `dummy` lets you swap between a fast run and a more comprehensive run. The more comprehensive run was used for this matbench submission.
dummy = False
if dummy:
n_splits = 2
total_trials = 2
else:
n_splits = 5
total_trials = 100
# Specify directories where you want to save things and make sure they exist.
# create dir https://stackoverflow.com/a/273227/13697228
experiment_dir = "experiments"
figure_dir = "figures"
Path(experiment_dir).mkdir(parents=True, exist_ok=True)
Path(figure_dir).mkdir(parents=True, exist_ok=True)
# ## Helper Functions
# ### matplotlibify
# The following code makes a Plotly figure look more like a matplotlib figure to make it easier to include in a manuscript.
def matplotlibify(fig, size=24, width_inches=3.5, height_inches=3.5, dpi=142):
# make it look more like matplotlib
# modified from: https://medium.com/swlh/formatting-a-plotly-figure-with-matplotlib-style-fa56ddd97539)
font_dict = dict(family="Arial", size=size, color="black")
fig.update_layout(
font=font_dict,
plot_bgcolor="white",
width=width_inches * dpi,
height=height_inches * dpi,
margin=dict(r=40, t=20, b=10),
)
fig.update_yaxes(
showline=True, # add line at x=0
linecolor="black", # line color
linewidth=2.4, # line size
ticks="inside", # ticks outside axis
tickfont=font_dict, # tick label font
mirror="allticks", # add ticks to top/right axes
tickwidth=2.4, # tick width
tickcolor="black", # tick color
)
fig.update_xaxes(
showline=True,
showticklabels=True,
linecolor="black",
linewidth=2.4,
ticks="inside",
tickfont=font_dict,
mirror="allticks",
tickwidth=2.4,
tickcolor="black",
)
fig.update(layout_coloraxis_showscale=False)
width_default_px = fig.layout.width
targ_dpi = 300
scale = width_inches / (width_default_px / dpi) * (targ_dpi / dpi)
return fig, scale
# ### correct_parameterization
# The following function is very important for interfacing Ax with CrabNet. The information in `parameterization` (a Python `dict`) as an input and `parameterization` as an output (please excuse the faux pas) is essentially the same, but Ax needs a representation for the parameter space and CrabNet has a certain API for the parameters. `correct_parameterization` just converts from the Ax representation of CrabNet parameters to the CrabNet API for parameters.
def correct_parameterization(parameterization):
pprint.pprint(parameterization)
parameterization["out_hidden"] = [
parameterization.get("out_hidden4") * 8,
parameterization.get("out_hidden4") * 4,
parameterization.get("out_hidden4") * 2,
parameterization.get("out_hidden4"),
]
parameterization.pop("out_hidden4")
parameterization["betas"] = (
parameterization.get("betas1"),
parameterization.get("betas2"),
)
parameterization.pop("betas1")
parameterization.pop("betas2")
d_model = parameterization["d_model"]
# make heads even (unless it's 1) (because d_model must be even)
heads = parameterization["heads"]
if np.mod(heads, 2) != 0:
heads = heads + 1
parameterization["heads"] = heads
# NOTE: d_model must be divisible by heads
d_model = parameterization["heads"] * round(d_model / parameterization["heads"])
parameterization["d_model"] = d_model
parameterization["pos_scaler_log"] = (
1 - parameterization["emb_scaler"] - parameterization["pos_scaler"]
)
parameterization["epochs"] = parameterization["epochs_step"] * 4
return parameterization
# ## Hyperparameter Optimization (and matbench recording)
# Note that `crabnet_mae` is defined inside of the loop so that `train_val_df` object is updated for each outer fold. This is due to a limitation of Ax where `crabnet_mae` (the objective function) can *only* take parameterization as an input (no additional parameters, kwargs, etc.). This was my workaround. Note that for the inner loop where Ax optimizes the CrabNet parameters for each outer fold, five inner folds are used, and the objective that Ax optimizes is the average MAE across the five inner folds. Additionally, hyperparameter optimization plots are produced for each of the five outer folds (Ax objective vs. iteration) using `100` sequential iterations. In other words, for a single `matbench` task (this notebook), `CrabNet` undergoes model instantiation and fitting `5*5*100 --> 2500` times. This took a few days to run on a single machine.
# +
mb = MatbenchBenchmark(autoload=False, subset=["matbench_expt_gap"])
kf = KFold(n_splits=n_splits, shuffle=True, random_state=18012019)
task = list(mb.tasks)[0]
task.load()
for i, fold in enumerate(task.folds):
train_inputs, train_outputs = task.get_train_and_val_data(fold)
# TODO: treat train_val_df as Ax fixed_parameter
train_val_df = pd.DataFrame(
{"formula": train_inputs.values, "target": train_outputs.values}
)
if dummy:
train_val_df = train_val_df[:100]
def crabnet_mae(parameterization):
"""Compute the mean absolute error of a CrabNet model.
Assumes that `train_df` and `val_df` are predefined.
Parameters
----------
parameterization : dict
Dictionary of the parameters passed to `get_model()` after some slight
modification.
Returns
-------
results: dict
Dictionary of `{"rmse": rmse}` where `rmse` is the root-mean-square error of the
CrabNet model.
"""
parameterization = correct_parameterization(parameterization)
mae = 0.0
for train_index, val_index in kf.split(train_val_df):
train_df, val_df = (
train_val_df.loc[train_index],
train_val_df.loc[val_index],
)
crabnet_model = get_model(
mat_prop="expt_gap",
train_df=train_df,
learningcurve=False,
force_cpu=False,
**parameterization
)
val_true, val_pred, val_formulas, val_sigma = crabnet_model.predict(val_df)
# rmse = mean_squared_error(val_true, val_pred, squared=False)
mae = mae + mean_absolute_error(val_true, val_pred)
# deallocate CUDA memory https://discuss.pytorch.org/t/how-can-we-release-gpu-memory-cache/14530/28
del crabnet_model
gc.collect()
torch.cuda.empty_cache()
mae = mae / n_splits
results = {"mae": mae}
return results
best_parameters, values, experiment, model = optimize(
parameters=[
{"name": "batch_size", "type": "range", "bounds": [32, 256]},
{"name": "fudge", "type": "range", "bounds": [0.0, 0.1]},
{"name": "d_model", "type": "range", "bounds": [100, 1024]},
{"name": "N", "type": "range", "bounds": [1, 10]},
{"name": "heads", "type": "range", "bounds": [1, 10]},
{"name": "out_hidden4", "type": "range", "bounds": [32, 512]},
{"name": "emb_scaler", "type": "range", "bounds": [0.0, 1.0]},
{"name": "pos_scaler", "type": "range", "bounds": [0.0, 1.0]},
{"name": "bias", "type": "choice", "values": [False, True]},
{"name": "dim_feedforward", "type": "range", "bounds": [1024, 4096],},
{"name": "dropout", "type": "range", "bounds": [0.0, 1.0]},
# jarvis and oliynyk don't have enough elements
# ptable contains str, which isn't a handled case
{
"name": "elem_prop",
"type": "choice",
"values": [
"mat2vec",
"magpie",
"onehot",
], # "jarvis", "oliynyk", "ptable"
},
{"name": "epochs_step", "type": "range", "bounds": [5, 20]},
{"name": "pe_resolution", "type": "range", "bounds": [2500, 10000]},
{"name": "ple_resolution", "type": "range", "bounds": [2500, 10000],},
{
"name": "criterion",
"type": "choice",
"values": ["RobustL1", "RobustL2"],
},
{"name": "lr", "type": "range", "bounds": [0.0001, 0.006]},
{"name": "betas1", "type": "range", "bounds": [0.5, 0.9999]},
{"name": "betas2", "type": "range", "bounds": [0.5, 0.9999]},
{"name": "eps", "type": "range", "bounds": [0.0000001, 0.0001]},
{"name": "weight_decay", "type": "range", "bounds": [0.0, 1.0]},
# {"name": "adam", "type": "choice", "values": [False, True]}, # issues with onehot
# {"name": "min_trust", "type": "range", "bounds": [0.0, 1.0]}, #issues with onehot
{"name": "alpha", "type": "range", "bounds": [0.0, 1.0]},
{"name": "k", "type": "range", "bounds": [2, 10]},
],
experiment_name="crabnet-hyperparameter",
evaluation_function=crabnet_mae,
objective_name="mae",
minimize=True,
parameter_constraints=["betas1 <= betas2", "emb_scaler + pos_scaler <= 1"],
total_trials=total_trials,
)
print(best_parameters)
print(values)
experiment_fpath = join(experiment_dir, "experiment" + str(i) + ".json")
save_experiment(experiment, experiment_fpath)
# TODO: save plot, save experiment
test_inputs, test_outputs = task.get_test_data(fold, include_target=True)
test_df = pd.DataFrame({"formula": test_inputs, "target": test_outputs})
default_model = get_model(
mat_prop="expt_gap",
train_df=train_val_df,
learningcurve=False,
force_cpu=False,
)
default_true, default_pred, default_formulas, default_sigma = default_model.predict(
test_df
)
# rmse = mean_squared_error(val_true, val_pred, squared=False)
default_mae = mean_absolute_error(default_true, default_pred)
# deallocate CUDA memory https://discuss.pytorch.org/t/how-can-we-release-gpu-memory-cache/14530/28
del default_model
gc.collect()
torch.cuda.empty_cache()
best_parameterization = correct_parameterization(best_parameters)
test_model = get_model(
mat_prop="expt_gap",
train_df=train_val_df,
learningcurve=False,
force_cpu=False,
**best_parameterization
)
# TODO: update CrabNet predict function to allow for no target specified
test_true, test_pred, test_formulas, test_sigma = test_model.predict(test_df)
# rmse = mean_squared_error(val_true, val_pred, squared=False)
test_mae = mean_absolute_error(test_true, test_pred)
# deallocate CUDA memory https://discuss.pytorch.org/t/how-can-we-release-gpu-memory-cache/14530/28
del test_model
gc.collect()
torch.cuda.empty_cache()
trials = experiment.trials.values()
best_objectives = np.array([[trial.objective_mean for trial in trials]])
parameter_strs = [
pprint.pformat(trial.arm.parameters).replace("\n", "<br>") for trial in trials
]
best_objective_plot = optimization_trace_single_method(
y=best_objectives,
optimization_direction="minimize",
ylabel="MAE (eV)",
hover_labels=parameter_strs,
plot_trial_points=True,
)
figure_fpath = join(figure_dir, "best_objective_plot_" + str(i))
data = best_objective_plot[0]["data"]
data.append(
go.Scatter(
x=(1, total_trials),
y=(default_mae, default_mae),
mode="lines",
line={"dash": "dash"},
name="default MAE",
yaxis="y1",
)
)
data.append(
go.Scatter(
x=(1, total_trials),
y=(test_mae, test_mae),
mode="lines",
line={"dash": "dash"},
name="best model test MAE",
yaxis="y1",
)
)
layout = best_objective_plot[0]["layout"]
fig = go.Figure({"data": data, "layout": layout})
fig.show()
fig.write_html(figure_fpath + ".html")
fig.to_json(figure_fpath + ".json")
fig.update_layout(
legend=dict(
font=dict(size=16),
yanchor="top",
y=0.99,
xanchor="right",
x=0.99,
bgcolor="rgba(0,0,0,0)",
)
)
fig, scale = matplotlibify(fig)
fig.write_image(figure_fpath + ".png")
task.record(fold, test_pred, params=best_parameterization)
# -
# ## Export matbench file
my_metadata = {"algorithm_version": crabnet.__version__}
mb.add_metadata(my_metadata)
mb.to_file("expt_gap_benchmark.json.gz")
# ## Static Plots
# For convenience (and so you don't have to run this notebook over the course of a few days), objective vs. iteration static plots are given for each of the five outer folds. If you'd like a more interactive experience, open the corresponding HTML files in your browser of choice from [`crabnet-hyperparameter/figures`](https://github.com/sparks-baird/crabnet-hyperparameter/tree/main/figures). Note that the first 46 (`2*23 parameters`) iterations are [SOBOL iterations](https://en.wikipedia.org/wiki/Sobol_sequence) (quasi-random sequence), and the remaining 54 are actual Bayesian optimization iterations.
#
# In the last four out of five folds, the hyperparameter optimization results in a slightly better test MAE (i.e. MAE for data which was unknown during the hyperparameter optimization).
# ### Objective vs. iteration for outer fold 0
# The best model test MAE is somewhat worse that the default test MAE, and both are close to the best validation MAE.
# 
# ### Objective vs. iteration for outer fold 1
# The best model test MAE is slightly better than the default test MAE.
# 
# ### Objective vs. iteration for outer fold 2
# The best model test MAE is somewhat better than the default test MAE.
# 
# ### Objective vs. iteration for outer fold 3
# The best model test MAE is somewhat better than the default test MAE.
# 
# ### Objective vs. iteration for outer fold 4
# 
| 39.389755
| 1,054
|
c7cd02d4a619bcaa7b94f54552b64e64fed21ad1
|
py
|
python
|
Assignments/Assgn_Dataframes1.ipynb
|
ColdCoffee21/Foundations-of-Data-Science
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/ColdCoffee21/Foundations-of-Data-Science/blob/master/Assgn_Dataframes1.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + id="4vmAw0iAYEp7"
import pandas as pd
# + id="2_EqCLBWZCHc"
# Colab library to upload files to notebook
from google.colab import files
# + id="kY2BnhPuZJUi" colab={"resources": {"http://localhost:8080/nbextensions/google.colab/files.js": {"data": "Ly8gQ29weXJpZ2h0IDIwMTcgR29vZ2xlIExMQwovLwovLyBMaWNlbnNlZCB1bmRlciB0aGUgQXBhY2hlIExpY2Vuc2UsIFZlcnNpb24gMi4wICh0aGUgIkxpY2Vuc2UiKTsKLy8geW91IG1heSBub3QgdXNlIHRoaXMgZmlsZSBleGNlcHQgaW4gY29tcGxpYW5jZSB3aXRoIHRoZSBMaWNlbnNlLgovLyBZb3UgbWF5IG9idGFpbiBhIGNvcHkgb2YgdGhlIExpY2Vuc2UgYXQKLy8KLy8gICAgICBodHRwOi8vd3d3LmFwYWNoZS5vcmcvbGljZW5zZXMvTElDRU5TRS0yLjAKLy8KLy8gVW5sZXNzIHJlcXVpcmVkIGJ5IGFwcGxpY2FibGUgbGF3IG9yIGFncmVlZCB0byBpbiB3cml0aW5nLCBzb2Z0d2FyZQovLyBkaXN0cmlidXRlZCB1bmRlciB0aGUgTGljZW5zZSBpcyBkaXN0cmlidXRlZCBvbiBhbiAiQVMgSVMiIEJBU0lTLAovLyBXSVRIT1VUIFdBUlJBTlRJRVMgT1IgQ09ORElUSU9OUyBPRiBBTlkgS0lORCwgZWl0aGVyIGV4cHJlc3Mgb3IgaW1wbGllZC4KLy8gU2VlIHRoZSBMaWNlbnNlIGZvciB0aGUgc3BlY2lmaWMgbGFuZ3VhZ2UgZ292ZXJuaW5nIHBlcm1pc3Npb25zIGFuZAovLyBsaW1pdGF0aW9ucyB1bmRlciB0aGUgTGljZW5zZS4KCi8qKgogKiBAZmlsZW92ZXJ2aWV3IEhlbHBlcnMgZm9yIGdvb2dsZS5jb2xhYiBQeXRob24gbW9kdWxlLgogKi8KKGZ1bmN0aW9uKHNjb3BlKSB7CmZ1bmN0aW9uIHNwYW4odGV4dCwgc3R5bGVBdHRyaWJ1dGVzID0ge30pIHsKICBjb25zdCBlbGVtZW50ID0gZG9jdW1lbnQuY3JlYXRlRWxlbWVudCgnc3BhbicpOwogIGVsZW1lbnQudGV4dENvbnRlbnQgPSB0ZXh0OwogIGZvciAoY29uc3Qga2V5IG9mIE9iamVjdC5rZXlzKHN0eWxlQXR0cmlidXRlcykpIHsKICAgIGVsZW1lbnQuc3R5bGVba2V5XSA9IHN0eWxlQXR0cmlidXRlc1trZXldOwogIH0KICByZXR1cm4gZWxlbWVudDsKfQoKLy8gTWF4IG51bWJlciBvZiBieXRlcyB3aGljaCB3aWxsIGJlIHVwbG9hZGVkIGF0IGEgdGltZS4KY29uc3QgTUFYX1BBWUxPQURfU0laRSA9IDEwMCAqIDEwMjQ7CgpmdW5jdGlvbiBfdXBsb2FkRmlsZXMoaW5wdXRJZCwgb3V0cHV0SWQpIHsKICBjb25zdCBzdGVwcyA9IHVwbG9hZEZpbGVzU3RlcChpbnB1dElkLCBvdXRwdXRJZCk7CiAgY29uc3Qgb3V0cHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKG91dHB1dElkKTsKICAvLyBDYWNoZSBzdGVwcyBvbiB0aGUgb3V0cHV0RWxlbWVudCB0byBtYWtlIGl0IGF2YWlsYWJsZSBmb3IgdGhlIG5leHQgY2FsbAogIC8vIHRvIHVwbG9hZEZpbGVzQ29udGludWUgZnJvbSBQeXRob24uCiAgb3V0cHV0RWxlbWVudC5zdGVwcyA9IHN0ZXBzOwoKICByZXR1cm4gX3VwbG9hZEZpbGVzQ29udGludWUob3V0cHV0SWQpOwp9CgovLyBUaGlzIGlzIHJvdWdobHkgYW4gYXN5bmMgZ2VuZXJhdG9yIChub3Qgc3VwcG9ydGVkIGluIHRoZSBicm93c2VyIHlldCksCi8vIHdoZXJlIHRoZXJlIGFyZSBtdWx0aXBsZSBhc3luY2hyb25vdXMgc3RlcHMgYW5kIHRoZSBQeXRob24gc2lkZSBpcyBnb2luZwovLyB0byBwb2xsIGZvciBjb21wbGV0aW9uIG9mIGVhY2ggc3RlcC4KLy8gVGhpcyB1c2VzIGEgUHJvbWlzZSB0byBibG9jayB0aGUgcHl0aG9uIHNpZGUgb24gY29tcGxldGlvbiBvZiBlYWNoIHN0ZXAsCi8vIHRoZW4gcGFzc2VzIHRoZSByZXN1bHQgb2YgdGhlIHByZXZpb3VzIHN0ZXAgYXMgdGhlIGlucHV0IHRvIHRoZSBuZXh0IHN0ZXAuCmZ1bmN0aW9uIF91cGxvYWRGaWxlc0NvbnRpbnVlKG91dHB1dElkKSB7CiAgY29uc3Qgb3V0cHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKG91dHB1dElkKTsKICBjb25zdCBzdGVwcyA9IG91dHB1dEVsZW1lbnQuc3RlcHM7CgogIGNvbnN0IG5leHQgPSBzdGVwcy5uZXh0KG91dHB1dEVsZW1lbnQubGFzdFByb21pc2VWYWx1ZSk7CiAgcmV0dXJuIFByb21pc2UucmVzb2x2ZShuZXh0LnZhbHVlLnByb21pc2UpLnRoZW4oKHZhbHVlKSA9PiB7CiAgICAvLyBDYWNoZSB0aGUgbGFzdCBwcm9taXNlIHZhbHVlIHRvIG1ha2UgaXQgYXZhaWxhYmxlIHRvIHRoZSBuZXh0CiAgICAvLyBzdGVwIG9mIHRoZSBnZW5lcmF0b3IuCiAgICBvdXRwdXRFbGVtZW50Lmxhc3RQcm9taXNlVmFsdWUgPSB2YWx1ZTsKICAgIHJldHVybiBuZXh0LnZhbHVlLnJlc3BvbnNlOwogIH0pOwp9CgovKioKICogR2VuZXJhdG9yIGZ1bmN0aW9uIHdoaWNoIGlzIGNhbGxlZCBiZXR3ZWVuIGVhY2ggYXN5bmMgc3RlcCBvZiB0aGUgdXBsb2FkCiAqIHByb2Nlc3MuCiAqIEBwYXJhbSB7c3RyaW5nfSBpbnB1dElkIEVsZW1lbnQgSUQgb2YgdGhlIGlucHV0IGZpbGUgcGlja2VyIGVsZW1lbnQuCiAqIEBwYXJhbSB7c3RyaW5nfSBvdXRwdXRJZCBFbGVtZW50IElEIG9mIHRoZSBvdXRwdXQgZGlzcGxheS4KICogQHJldHVybiB7IUl0ZXJhYmxlPCFPYmplY3Q+fSBJdGVyYWJsZSBvZiBuZXh0IHN0ZXBzLgogKi8KZnVuY3Rpb24qIHVwbG9hZEZpbGVzU3RlcChpbnB1dElkLCBvdXRwdXRJZCkgewogIGNvbnN0IGlucHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKGlucHV0SWQpOwogIGlucHV0RWxlbWVudC5kaXNhYmxlZCA9IGZhbHNlOwoKICBjb25zdCBvdXRwdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQob3V0cHV0SWQpOwogIG91dHB1dEVsZW1lbnQuaW5uZXJIVE1MID0gJyc7CgogIGNvbnN0IHBpY2tlZFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgaW5wdXRFbGVtZW50LmFkZEV2ZW50TGlzdGVuZXIoJ2NoYW5nZScsIChlKSA9PiB7CiAgICAgIHJlc29sdmUoZS50YXJnZXQuZmlsZXMpOwogICAgfSk7CiAgfSk7CgogIGNvbnN0IGNhbmNlbCA9IGRvY3VtZW50LmNyZWF0ZUVsZW1lbnQoJ2J1dHRvbicpOwogIGlucHV0RWxlbWVudC5wYXJlbnRFbGVtZW50LmFwcGVuZENoaWxkKGNhbmNlbCk7CiAgY2FuY2VsLnRleHRDb250ZW50ID0gJ0NhbmNlbCB1cGxvYWQnOwogIGNvbnN0IGNhbmNlbFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgY2FuY2VsLm9uY2xpY2sgPSAoKSA9PiB7CiAgICAgIHJlc29sdmUobnVsbCk7CiAgICB9OwogIH0pOwoKICAvLyBXYWl0IGZvciB0aGUgdXNlciB0byBwaWNrIHRoZSBmaWxlcy4KICBjb25zdCBmaWxlcyA9IHlpZWxkIHsKICAgIHByb21pc2U6IFByb21pc2UucmFjZShbcGlja2VkUHJvbWlzZSwgY2FuY2VsUHJvbWlzZV0pLAogICAgcmVzcG9uc2U6IHsKICAgICAgYWN0aW9uOiAnc3RhcnRpbmcnLAogICAgfQogIH07CgogIGNhbmNlbC5yZW1vdmUoKTsKCiAgLy8gRGlzYWJsZSB0aGUgaW5wdXQgZWxlbWVudCBzaW5jZSBmdXJ0aGVyIHBpY2tzIGFyZSBub3QgYWxsb3dlZC4KICBpbnB1dEVsZW1lbnQuZGlzYWJsZWQgPSB0cnVlOwoKICBpZiAoIWZpbGVzKSB7CiAgICByZXR1cm4gewogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbXBsZXRlJywKICAgICAgfQogICAgfTsKICB9CgogIGZvciAoY29uc3QgZmlsZSBvZiBmaWxlcykgewogICAgY29uc3QgbGkgPSBkb2N1bWVudC5jcmVhdGVFbGVtZW50KCdsaScpOwogICAgbGkuYXBwZW5kKHNwYW4oZmlsZS5uYW1lLCB7Zm9udFdlaWdodDogJ2JvbGQnfSkpOwogICAgbGkuYXBwZW5kKHNwYW4oCiAgICAgICAgYCgke2ZpbGUudHlwZSB8fCAnbi9hJ30pIC0gJHtmaWxlLnNpemV9IGJ5dGVzLCBgICsKICAgICAgICBgbGFzdCBtb2RpZmllZDogJHsKICAgICAgICAgICAgZmlsZS5sYXN0TW9kaWZpZWREYXRlID8gZmlsZS5sYXN0TW9kaWZpZWREYXRlLnRvTG9jYWxlRGF0ZVN0cmluZygpIDoKICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgJ24vYSd9IC0gYCkpOwogICAgY29uc3QgcGVyY2VudCA9IHNwYW4oJzAlIGRvbmUnKTsKICAgIGxpLmFwcGVuZENoaWxkKHBlcmNlbnQpOwoKICAgIG91dHB1dEVsZW1lbnQuYXBwZW5kQ2hpbGQobGkpOwoKICAgIGNvbnN0IGZpbGVEYXRhUHJvbWlzZSA9IG5ldyBQcm9taXNlKChyZXNvbHZlKSA9PiB7CiAgICAgIGNvbnN0IHJlYWRlciA9IG5ldyBGaWxlUmVhZGVyKCk7CiAgICAgIHJlYWRlci5vbmxvYWQgPSAoZSkgPT4gewogICAgICAgIHJlc29sdmUoZS50YXJnZXQucmVzdWx0KTsKICAgICAgfTsKICAgICAgcmVhZGVyLnJlYWRBc0FycmF5QnVmZmVyKGZpbGUpOwogICAgfSk7CiAgICAvLyBXYWl0IGZvciB0aGUgZGF0YSB0byBiZSByZWFkeS4KICAgIGxldCBmaWxlRGF0YSA9IHlpZWxkIHsKICAgICAgcHJvbWlzZTogZmlsZURhdGFQcm9taXNlLAogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbnRpbnVlJywKICAgICAgfQogICAgfTsKCiAgICAvLyBVc2UgYSBjaHVua2VkIHNlbmRpbmcgdG8gYXZvaWQgbWVzc2FnZSBzaXplIGxpbWl0cy4gU2VlIGIvNjIxMTU2NjAuCiAgICBsZXQgcG9zaXRpb24gPSAwOwogICAgd2hpbGUgKHBvc2l0aW9uIDwgZmlsZURhdGEuYnl0ZUxlbmd0aCkgewogICAgICBjb25zdCBsZW5ndGggPSBNYXRoLm1pbihmaWxlRGF0YS5ieXRlTGVuZ3RoIC0gcG9zaXRpb24sIE1BWF9QQVlMT0FEX1NJWkUpOwogICAgICBjb25zdCBjaHVuayA9IG5ldyBVaW50OEFycmF5KGZpbGVEYXRhLCBwb3NpdGlvbiwgbGVuZ3RoKTsKICAgICAgcG9zaXRpb24gKz0gbGVuZ3RoOwoKICAgICAgY29uc3QgYmFzZTY0ID0gYnRvYShTdHJpbmcuZnJvbUNoYXJDb2RlLmFwcGx5KG51bGwsIGNodW5rKSk7CiAgICAgIHlpZWxkIHsKICAgICAgICByZXNwb25zZTogewogICAgICAgICAgYWN0aW9uOiAnYXBwZW5kJywKICAgICAgICAgIGZpbGU6IGZpbGUubmFtZSwKICAgICAgICAgIGRhdGE6IGJhc2U2NCwKICAgICAgICB9LAogICAgICB9OwogICAgICBwZXJjZW50LnRleHRDb250ZW50ID0KICAgICAgICAgIGAke01hdGgucm91bmQoKHBvc2l0aW9uIC8gZmlsZURhdGEuYnl0ZUxlbmd0aCkgKiAxMDApfSUgZG9uZWA7CiAgICB9CiAgfQoKICAvLyBBbGwgZG9uZS4KICB5aWVsZCB7CiAgICByZXNwb25zZTogewogICAgICBhY3Rpb246ICdjb21wbGV0ZScsCiAgICB9CiAgfTsKfQoKc2NvcGUuZ29vZ2xlID0gc2NvcGUuZ29vZ2xlIHx8IHt9OwpzY29wZS5nb29nbGUuY29sYWIgPSBzY29wZS5nb29nbGUuY29sYWIgfHwge307CnNjb3BlLmdvb2dsZS5jb2xhYi5fZmlsZXMgPSB7CiAgX3VwbG9hZEZpbGVzLAogIF91cGxvYWRGaWxlc0NvbnRpbnVlLAp9Owp9KShzZWxmKTsK", "ok": true, "headers": [["content-type", "application/javascript"]], "status": 200, "status_text": ""}}, "base_uri": "https://localhost:8080/", "height": 71} outputId="35b2fe43-ecdb-4942-c201-fa91078a8104"
# Upload the dataset to collab
uploaded = files.upload()
# + id="bwjYfMkUbZMZ"
#Q1
df = pd.read_csv('datasets_1189_2137_degrees-that-pay-back.csv')
# + id="f3WhXaaSbiZw" colab={"base_uri": "https://localhost:8080/", "height": 150} outputId="16cf2dce-0d59-446c-f1c8-e11cb1b3806f"
#Q2
df.columns
# + id="Zqxk_K1Fbvct" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="d09a145c-b0e3-4611-97b3-338686798bd2"
#Q3
df.size
# + id="oWAH-y8cb5eF" colab={"base_uri": "https://localhost:8080/", "height": 140} outputId="27bfb0d1-b820-4020-bbad-3489fa03d325"
#Q4
df.rename(columns={
'Undergraduate Major': 'major', 'Starting Median Salary': 'Start Med Sal',
'Mid-Career Median Salary':'Mid Career Med Sal',
'Percent change from Starting to Mid-Career Salary': 'Start-Mid Career Sal Change',
'Mid-Career 10th Percentile Salary':'10%tile Sal',
'Mid-Career 25th Percentile Salary':'25%tile Sal',
'Mid-Career 75th Percentile Salary':'75%tile Sal',
'Mid-Career 90th Percentile Salary':'90%tileSal'
}).head(3)
# + id="ssWVRp-ib_mN" colab={"base_uri": "https://localhost:8080/", "height": 166} outputId="44f4434e-4092-4fe4-9551-c6cb1a7a79bd"
#Q5
df.dtypes
# + id="TodhA7oggAKB" colab={"base_uri": "https://localhost:8080/", "height": 166} outputId="c4747ec8-527a-4640-a618-07c03688c346"
#Q6
df[df.columns[1:]] = df[df.columns[1:]].replace('[\$,]', '', regex=True).astype(float)
df.dtypes
# + id="qD3KAdA9giH6" colab={"base_uri": "https://localhost:8080/", "height": 364} outputId="23368f04-42d5-4f01-e383-01b725128571"
#Q7
df.loc[df['Percent change from Starting to Mid-Career Salary']>70, 'Undergraduate Major']
# + id="ASon23vhBuWZ" colab={"base_uri": "https://localhost:8080/", "height": 1000} outputId="6d8e80ac-2a57-4098-96e2-6a3398aecd5f"
#Q8
df.loc[df['Mid-Career 90th Percentile Salary']-df['Mid-Career 10th Percentile Salary']>40000, 'Starting Median Salary':'Mid-Career Median Salary']
# + id="l8Hl570zC3O1" colab={"resources": {"http://localhost:8080/nbextensions/google.colab/files.js": {"data": "Ly8gQ29weXJpZ2h0IDIwMTcgR29vZ2xlIExMQwovLwovLyBMaWNlbnNlZCB1bmRlciB0aGUgQXBhY2hlIExpY2Vuc2UsIFZlcnNpb24gMi4wICh0aGUgIkxpY2Vuc2UiKTsKLy8geW91IG1heSBub3QgdXNlIHRoaXMgZmlsZSBleGNlcHQgaW4gY29tcGxpYW5jZSB3aXRoIHRoZSBMaWNlbnNlLgovLyBZb3UgbWF5IG9idGFpbiBhIGNvcHkgb2YgdGhlIExpY2Vuc2UgYXQKLy8KLy8gICAgICBodHRwOi8vd3d3LmFwYWNoZS5vcmcvbGljZW5zZXMvTElDRU5TRS0yLjAKLy8KLy8gVW5sZXNzIHJlcXVpcmVkIGJ5IGFwcGxpY2FibGUgbGF3IG9yIGFncmVlZCB0byBpbiB3cml0aW5nLCBzb2Z0d2FyZQovLyBkaXN0cmlidXRlZCB1bmRlciB0aGUgTGljZW5zZSBpcyBkaXN0cmlidXRlZCBvbiBhbiAiQVMgSVMiIEJBU0lTLAovLyBXSVRIT1VUIFdBUlJBTlRJRVMgT1IgQ09ORElUSU9OUyBPRiBBTlkgS0lORCwgZWl0aGVyIGV4cHJlc3Mgb3IgaW1wbGllZC4KLy8gU2VlIHRoZSBMaWNlbnNlIGZvciB0aGUgc3BlY2lmaWMgbGFuZ3VhZ2UgZ292ZXJuaW5nIHBlcm1pc3Npb25zIGFuZAovLyBsaW1pdGF0aW9ucyB1bmRlciB0aGUgTGljZW5zZS4KCi8qKgogKiBAZmlsZW92ZXJ2aWV3IEhlbHBlcnMgZm9yIGdvb2dsZS5jb2xhYiBQeXRob24gbW9kdWxlLgogKi8KKGZ1bmN0aW9uKHNjb3BlKSB7CmZ1bmN0aW9uIHNwYW4odGV4dCwgc3R5bGVBdHRyaWJ1dGVzID0ge30pIHsKICBjb25zdCBlbGVtZW50ID0gZG9jdW1lbnQuY3JlYXRlRWxlbWVudCgnc3BhbicpOwogIGVsZW1lbnQudGV4dENvbnRlbnQgPSB0ZXh0OwogIGZvciAoY29uc3Qga2V5IG9mIE9iamVjdC5rZXlzKHN0eWxlQXR0cmlidXRlcykpIHsKICAgIGVsZW1lbnQuc3R5bGVba2V5XSA9IHN0eWxlQXR0cmlidXRlc1trZXldOwogIH0KICByZXR1cm4gZWxlbWVudDsKfQoKLy8gTWF4IG51bWJlciBvZiBieXRlcyB3aGljaCB3aWxsIGJlIHVwbG9hZGVkIGF0IGEgdGltZS4KY29uc3QgTUFYX1BBWUxPQURfU0laRSA9IDEwMCAqIDEwMjQ7CgpmdW5jdGlvbiBfdXBsb2FkRmlsZXMoaW5wdXRJZCwgb3V0cHV0SWQpIHsKICBjb25zdCBzdGVwcyA9IHVwbG9hZEZpbGVzU3RlcChpbnB1dElkLCBvdXRwdXRJZCk7CiAgY29uc3Qgb3V0cHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKG91dHB1dElkKTsKICAvLyBDYWNoZSBzdGVwcyBvbiB0aGUgb3V0cHV0RWxlbWVudCB0byBtYWtlIGl0IGF2YWlsYWJsZSBmb3IgdGhlIG5leHQgY2FsbAogIC8vIHRvIHVwbG9hZEZpbGVzQ29udGludWUgZnJvbSBQeXRob24uCiAgb3V0cHV0RWxlbWVudC5zdGVwcyA9IHN0ZXBzOwoKICByZXR1cm4gX3VwbG9hZEZpbGVzQ29udGludWUob3V0cHV0SWQpOwp9CgovLyBUaGlzIGlzIHJvdWdobHkgYW4gYXN5bmMgZ2VuZXJhdG9yIChub3Qgc3VwcG9ydGVkIGluIHRoZSBicm93c2VyIHlldCksCi8vIHdoZXJlIHRoZXJlIGFyZSBtdWx0aXBsZSBhc3luY2hyb25vdXMgc3RlcHMgYW5kIHRoZSBQeXRob24gc2lkZSBpcyBnb2luZwovLyB0byBwb2xsIGZvciBjb21wbGV0aW9uIG9mIGVhY2ggc3RlcC4KLy8gVGhpcyB1c2VzIGEgUHJvbWlzZSB0byBibG9jayB0aGUgcHl0aG9uIHNpZGUgb24gY29tcGxldGlvbiBvZiBlYWNoIHN0ZXAsCi8vIHRoZW4gcGFzc2VzIHRoZSByZXN1bHQgb2YgdGhlIHByZXZpb3VzIHN0ZXAgYXMgdGhlIGlucHV0IHRvIHRoZSBuZXh0IHN0ZXAuCmZ1bmN0aW9uIF91cGxvYWRGaWxlc0NvbnRpbnVlKG91dHB1dElkKSB7CiAgY29uc3Qgb3V0cHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKG91dHB1dElkKTsKICBjb25zdCBzdGVwcyA9IG91dHB1dEVsZW1lbnQuc3RlcHM7CgogIGNvbnN0IG5leHQgPSBzdGVwcy5uZXh0KG91dHB1dEVsZW1lbnQubGFzdFByb21pc2VWYWx1ZSk7CiAgcmV0dXJuIFByb21pc2UucmVzb2x2ZShuZXh0LnZhbHVlLnByb21pc2UpLnRoZW4oKHZhbHVlKSA9PiB7CiAgICAvLyBDYWNoZSB0aGUgbGFzdCBwcm9taXNlIHZhbHVlIHRvIG1ha2UgaXQgYXZhaWxhYmxlIHRvIHRoZSBuZXh0CiAgICAvLyBzdGVwIG9mIHRoZSBnZW5lcmF0b3IuCiAgICBvdXRwdXRFbGVtZW50Lmxhc3RQcm9taXNlVmFsdWUgPSB2YWx1ZTsKICAgIHJldHVybiBuZXh0LnZhbHVlLnJlc3BvbnNlOwogIH0pOwp9CgovKioKICogR2VuZXJhdG9yIGZ1bmN0aW9uIHdoaWNoIGlzIGNhbGxlZCBiZXR3ZWVuIGVhY2ggYXN5bmMgc3RlcCBvZiB0aGUgdXBsb2FkCiAqIHByb2Nlc3MuCiAqIEBwYXJhbSB7c3RyaW5nfSBpbnB1dElkIEVsZW1lbnQgSUQgb2YgdGhlIGlucHV0IGZpbGUgcGlja2VyIGVsZW1lbnQuCiAqIEBwYXJhbSB7c3RyaW5nfSBvdXRwdXRJZCBFbGVtZW50IElEIG9mIHRoZSBvdXRwdXQgZGlzcGxheS4KICogQHJldHVybiB7IUl0ZXJhYmxlPCFPYmplY3Q+fSBJdGVyYWJsZSBvZiBuZXh0IHN0ZXBzLgogKi8KZnVuY3Rpb24qIHVwbG9hZEZpbGVzU3RlcChpbnB1dElkLCBvdXRwdXRJZCkgewogIGNvbnN0IGlucHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKGlucHV0SWQpOwogIGlucHV0RWxlbWVudC5kaXNhYmxlZCA9IGZhbHNlOwoKICBjb25zdCBvdXRwdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQob3V0cHV0SWQpOwogIG91dHB1dEVsZW1lbnQuaW5uZXJIVE1MID0gJyc7CgogIGNvbnN0IHBpY2tlZFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgaW5wdXRFbGVtZW50LmFkZEV2ZW50TGlzdGVuZXIoJ2NoYW5nZScsIChlKSA9PiB7CiAgICAgIHJlc29sdmUoZS50YXJnZXQuZmlsZXMpOwogICAgfSk7CiAgfSk7CgogIGNvbnN0IGNhbmNlbCA9IGRvY3VtZW50LmNyZWF0ZUVsZW1lbnQoJ2J1dHRvbicpOwogIGlucHV0RWxlbWVudC5wYXJlbnRFbGVtZW50LmFwcGVuZENoaWxkKGNhbmNlbCk7CiAgY2FuY2VsLnRleHRDb250ZW50ID0gJ0NhbmNlbCB1cGxvYWQnOwogIGNvbnN0IGNhbmNlbFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgY2FuY2VsLm9uY2xpY2sgPSAoKSA9PiB7CiAgICAgIHJlc29sdmUobnVsbCk7CiAgICB9OwogIH0pOwoKICAvLyBXYWl0IGZvciB0aGUgdXNlciB0byBwaWNrIHRoZSBmaWxlcy4KICBjb25zdCBmaWxlcyA9IHlpZWxkIHsKICAgIHByb21pc2U6IFByb21pc2UucmFjZShbcGlja2VkUHJvbWlzZSwgY2FuY2VsUHJvbWlzZV0pLAogICAgcmVzcG9uc2U6IHsKICAgICAgYWN0aW9uOiAnc3RhcnRpbmcnLAogICAgfQogIH07CgogIGNhbmNlbC5yZW1vdmUoKTsKCiAgLy8gRGlzYWJsZSB0aGUgaW5wdXQgZWxlbWVudCBzaW5jZSBmdXJ0aGVyIHBpY2tzIGFyZSBub3QgYWxsb3dlZC4KICBpbnB1dEVsZW1lbnQuZGlzYWJsZWQgPSB0cnVlOwoKICBpZiAoIWZpbGVzKSB7CiAgICByZXR1cm4gewogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbXBsZXRlJywKICAgICAgfQogICAgfTsKICB9CgogIGZvciAoY29uc3QgZmlsZSBvZiBmaWxlcykgewogICAgY29uc3QgbGkgPSBkb2N1bWVudC5jcmVhdGVFbGVtZW50KCdsaScpOwogICAgbGkuYXBwZW5kKHNwYW4oZmlsZS5uYW1lLCB7Zm9udFdlaWdodDogJ2JvbGQnfSkpOwogICAgbGkuYXBwZW5kKHNwYW4oCiAgICAgICAgYCgke2ZpbGUudHlwZSB8fCAnbi9hJ30pIC0gJHtmaWxlLnNpemV9IGJ5dGVzLCBgICsKICAgICAgICBgbGFzdCBtb2RpZmllZDogJHsKICAgICAgICAgICAgZmlsZS5sYXN0TW9kaWZpZWREYXRlID8gZmlsZS5sYXN0TW9kaWZpZWREYXRlLnRvTG9jYWxlRGF0ZVN0cmluZygpIDoKICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgJ24vYSd9IC0gYCkpOwogICAgY29uc3QgcGVyY2VudCA9IHNwYW4oJzAlIGRvbmUnKTsKICAgIGxpLmFwcGVuZENoaWxkKHBlcmNlbnQpOwoKICAgIG91dHB1dEVsZW1lbnQuYXBwZW5kQ2hpbGQobGkpOwoKICAgIGNvbnN0IGZpbGVEYXRhUHJvbWlzZSA9IG5ldyBQcm9taXNlKChyZXNvbHZlKSA9PiB7CiAgICAgIGNvbnN0IHJlYWRlciA9IG5ldyBGaWxlUmVhZGVyKCk7CiAgICAgIHJlYWRlci5vbmxvYWQgPSAoZSkgPT4gewogICAgICAgIHJlc29sdmUoZS50YXJnZXQucmVzdWx0KTsKICAgICAgfTsKICAgICAgcmVhZGVyLnJlYWRBc0FycmF5QnVmZmVyKGZpbGUpOwogICAgfSk7CiAgICAvLyBXYWl0IGZvciB0aGUgZGF0YSB0byBiZSByZWFkeS4KICAgIGxldCBmaWxlRGF0YSA9IHlpZWxkIHsKICAgICAgcHJvbWlzZTogZmlsZURhdGFQcm9taXNlLAogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbnRpbnVlJywKICAgICAgfQogICAgfTsKCiAgICAvLyBVc2UgYSBjaHVua2VkIHNlbmRpbmcgdG8gYXZvaWQgbWVzc2FnZSBzaXplIGxpbWl0cy4gU2VlIGIvNjIxMTU2NjAuCiAgICBsZXQgcG9zaXRpb24gPSAwOwogICAgd2hpbGUgKHBvc2l0aW9uIDwgZmlsZURhdGEuYnl0ZUxlbmd0aCkgewogICAgICBjb25zdCBsZW5ndGggPSBNYXRoLm1pbihmaWxlRGF0YS5ieXRlTGVuZ3RoIC0gcG9zaXRpb24sIE1BWF9QQVlMT0FEX1NJWkUpOwogICAgICBjb25zdCBjaHVuayA9IG5ldyBVaW50OEFycmF5KGZpbGVEYXRhLCBwb3NpdGlvbiwgbGVuZ3RoKTsKICAgICAgcG9zaXRpb24gKz0gbGVuZ3RoOwoKICAgICAgY29uc3QgYmFzZTY0ID0gYnRvYShTdHJpbmcuZnJvbUNoYXJDb2RlLmFwcGx5KG51bGwsIGNodW5rKSk7CiAgICAgIHlpZWxkIHsKICAgICAgICByZXNwb25zZTogewogICAgICAgICAgYWN0aW9uOiAnYXBwZW5kJywKICAgICAgICAgIGZpbGU6IGZpbGUubmFtZSwKICAgICAgICAgIGRhdGE6IGJhc2U2NCwKICAgICAgICB9LAogICAgICB9OwogICAgICBwZXJjZW50LnRleHRDb250ZW50ID0KICAgICAgICAgIGAke01hdGgucm91bmQoKHBvc2l0aW9uIC8gZmlsZURhdGEuYnl0ZUxlbmd0aCkgKiAxMDApfSUgZG9uZWA7CiAgICB9CiAgfQoKICAvLyBBbGwgZG9uZS4KICB5aWVsZCB7CiAgICByZXNwb25zZTogewogICAgICBhY3Rpb246ICdjb21wbGV0ZScsCiAgICB9CiAgfTsKfQoKc2NvcGUuZ29vZ2xlID0gc2NvcGUuZ29vZ2xlIHx8IHt9OwpzY29wZS5nb29nbGUuY29sYWIgPSBzY29wZS5nb29nbGUuY29sYWIgfHwge307CnNjb3BlLmdvb2dsZS5jb2xhYi5fZmlsZXMgPSB7CiAgX3VwbG9hZEZpbGVzLAogIF91cGxvYWRGaWxlc0NvbnRpbnVlLAp9Owp9KShzZWxmKTsK", "ok": true, "headers": [["content-type", "application/javascript"]], "status": 200, "status_text": ""}}, "base_uri": "https://localhost:8080/", "height": 71} outputId="36d9eb74-0887-48f4-d8d5-a66e93059703"
##PART 2
#Q1
uploaded = files.upload()
# + id="qnhvIfwYDHNT" colab={"base_uri": "https://localhost:8080/", "height": 190} outputId="808342e1-a5dd-4b60-8e23-cc0bd2ca0fcc"
df = pd.read_csv('salaries-by-college-type.csv')
df.head(3)
# + id="U-nr34vZDSt2" colab={"base_uri": "https://localhost:8080/", "height": 166} outputId="5361fc80-bc86-4d2b-dc69-55914ce5284f"
#Q2
df[df.columns[2:]] = df[df.columns[2:]].replace('[\$,]', '', regex=True).astype(float)
df.dtypes
# + id="gPaj3vtuF8Yh"
import numpy as np
# + id="Iu1khnruGBp7" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="c1203e28-691d-44d4-e1a8-48c0bc35eb25"
#Q4
g = df.groupby('School Type')
g.mean().loc['Liberal Arts','Starting Median Salary']
# + id="yiKl0XfyIXvn" colab={"base_uri": "https://localhost:8080/", "height": 50} outputId="087db02d-3cc7-4ed5-8182-f509ccfd4b5b"
#Q5
df['School Name'].loc[((df['Mid-Career 25th Percentile Salary']==df['Mid-Career 25th Percentile Salary'].max() )& (df['School Type']=='Engineering'))]
# + id="Dm07B43DrEzv" colab={"base_uri": "https://localhost:8080/", "height": 67} outputId="f771f75f-977d-4200-aa25-fdb20b728e1a"
#Q6
df['Starting Median Salary'].loc[(df['School Type']=='Party')].nlargest(2) #highest 2
# + id="FH-LA9oRrGMX" colab={"base_uri": "https://localhost:8080/", "height": 67} outputId="717af330-95c0-423b-f079-c04c7e1b2f84"
df['Starting Median Salary'].loc[(df['School Type']=='Ivy League')].nlargest(2)
# + id="uHGPx0gtsa5_" colab={"base_uri": "https://localhost:8080/", "height": 67} outputId="a02a04ae-1669-4099-9d52-f3436dbb1491"
df['Starting Median Salary'].loc[(df['School Type']=='Party')].nsmallest(2)
# + id="pxn9omegssTr" colab={"base_uri": "https://localhost:8080/", "height": 67} outputId="1e947db6-6271-4145-b046-01161a5b1c3b"
df['Starting Median Salary'].loc[(df['School Type']=='Ivy League')].nsmallest(2)
# + id="9ByFscmNsvJm" colab={"base_uri": "https://localhost:8080/", "height": 50} outputId="e9a7499b-cd9c-419d-8f52-d4ab25c1ec16"
#Q7
df['School Type'].loc[(df['Starting Median Salary']==df['Starting Median Salary'].max() )]
# + id="fKKajur_s58c" colab={"base_uri": "https://localhost:8080/", "height": 83} outputId="e1a823d1-3d61-4951-ff2c-8dca0955f915"
#Q8
df['School Name'].loc[(df['School Type']=='Engineering')&(df['Starting Median Salary']>65000)]
# + id="F-G715g0uIDx" colab={"base_uri": "https://localhost:8080/", "height": 477} outputId="3b745733-39ef-414c-86b1-984b73daf1ba"
#Q3 Fill na values with school type wise average
for col in df.columns[2:]:
#print(df[df[col].isnull()]['School Type'])
df[col] = df.groupby('School Type').transform(lambda x: x.fillna(x.mean()))
df
# + id="Dfm28itHvNgX"
| 158.491935
| 7,233
|
5cd88b305e459378b445c74e76bdc668375a1194
|
py
|
python
|
notebooks/introduction.ipynb
|
mscarey/AuthoritySpoke
|
['FTL', 'CNRI-Python', 'Apache-1.1']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: authorityspoke
# language: python
# name: authorityspoke
# ---
# # Legal Rules as Data: An Introduction to AuthoritySpoke
#
# > "Details, unnumbered, shifting, sharp, disordered, unchartable, jagged."
# >
# > Karl N. Llewellyn, The Bramble Bush: On Our Law and Its Study (1930).
#
# AuthoritySpoke is a Python package that will help you make legal authority readable by computers.
#
# This notebook will provide an overview of AuthoritySpoke's most important features. AuthoritySpoke is still in an alpha state, so many features have yet to be implemented, and some others still have limited functionality.
#
# AuthoritySpoke is open source software (as well as [Ethical Source](https://ethicalsource.dev/definition/) software). That mean you have the opportunity to reuse AuthoritySpoke in your own projects. You can also [participate in its development](https://github.com/mscarey/AuthoritySpoke) by submitting issues, bug reports, and pull requests.
# ## 0. Getting Example Data
#
# AuthoritySpoke helps you work with three kinds of data: court opinions, legislative enactments, and structured annotations of legal procedural rules.
#
# To help you obtain court opinions, AuthoritySpoke provides an interface to the [Caselaw Access Project](https://case.law/) API, a project of the Harvard Law School Library Innovation Lab. You'll need to [register for an API key](https://case.law/user/register/).
#
# To provide you with legislation text, AuthoritySpoke imports the [Legislice](https://pypi.org/project/legislice/) Python package, which provides an interface to the Legislice API at [authorityspoke.com](https://authorityspoke.com/). This API currently provides access to recent versions of the United States Code, plus the United States Constitution. You'll need to [sign up](https://authorityspoke.com/account/signup/) for an account and then obtain a Legislice API key from your account page. The Legislice API key is not the same as the Caselaw Access Project API key.
#
# In the current version, you mostly have to create your own procedural rule annotations, but the `example_data` folder of the [GitHub repository for AuthoritySpoke](https://github.com/mscarey/AuthoritySpoke) contains example annotations for several cases. The rest of this tutorial depends on having access to the `example_data` folder, so if you're running the tutorial code interactively, you'll need to either clone the AuthoritySpoke repository to your computer and run the tutorial from there, or else run the tutorial from a cloud environment like [Binder](https://mybinder.org/v2/gh/mscarey/AuthoritySpoke/master). If you've only installed AuthoritySpoke from `pip`, you won't have access to the example data files.
# ## 1. Importing the Package
#
# If you want to use AuthoritySpoke in your own Python environment, be sure you have installed AuthoritySpoke using a command like `pip install AuthoritySpoke` on the command line. Visit [the Python Package Index](https://pypi.org/project/AuthoritySpoke/) for more details.
#
# With a Python environment activated, let's import AuthoritySpoke by running the cell below. If you're running this code on your own machine but you don't want to obtain API keys or make real API calls over the Internet, you can change the two `True` variables to `False` to usefake versions of the APIs. The notebook will try to find the data for the fake APIs in the `example_data` folder alongside a `notebooks` folder where this notebook is running.
# +
import authorityspoke
USE_REAL_CASE_API = False
USE_REAL_LEGISLICE_API = False
# -
# If you executed that cell with no error messages, then it worked!
#
# If you got a message like `No module named 'authorityspoke'`, then AuthoritySpoke is probably not installed in your current Python environment. In that case, check the [Python documentation](https://docs.python.org/3/installing/index.html) for help on installing modules.
# ### 1.1 Optional: Skip the Downloads and Load Decisions from a File
# To use the cell below to access `Decision` objects from a file rather than an API, be sure the `USE_REAL_CASE_API` variable is set to `False`. This should work if you're running the tutorial in a notebook in a cloud environment like Binder, or if you've cloned AuthoritySpoke's GitHub repository to your hard drive and you're using `jupyter` to run the tutorial in from the `notebooks` folder of the repository. The notebook will try to find the data for the fake APIs in the ``example_data`` folder alongside a ``notebooks`` folder where this notebook is running.
# +
from authorityspoke.io.loaders import load_decision
from authorityspoke import Decision
if not USE_REAL_CASE_API:
oracle_case = Decision(**load_decision("oracle_h.json"))
lotus_case = Decision(**load_decision("lotus_h.json"))
# -
# ## 2. Downloading and Importing Decisions
#
# If you didn't load court opinions from the GitHub repository as described in section 1.1, then you'll be using the Caselaw Access Project (CAP) API to get court opinions to load into AuthoritySpoke. To download full cases from CAP, you'll need to [register for a CAP API key](https://case.law/user/register/).
# One good way to use an API key in a Jupyter Notebook or other Python working file is to save the API key in a file called `.env`. The `.env` file should contain a line that looks like `CAP_API_KEY=your-api-key-here`. Then you can use the `dotenv` Python package to load the API key as an environment variable without ever writing the API key in the notebook. That makes it easier to keep your API key secret, even if you publish your copy of the notebook and make it visible on the internet.
#
# However, if you're viewing this tutorial in a cloud environment like Binder, you probably won't be able to create an environment variable. Instead, you could replace `os.getenv('CAP_API_KEY')` with a string containing your own API key.
# +
import os
from dotenv import load_dotenv
load_dotenv()
CAP_API_KEY = os.getenv('CAP_API_KEY')
# -
# Next we need to download some cases for analysis.
#
# The CAP API limits users to downloading 500 full cases per day. If you accidentally make a query that returns hundreds of full cases, you could hit your limit before you know it. You should first try out your API queries without the `"full_case": "true"` parameter, and then only request full cases once you're confident you'll receive what you expect.
#
# Let's download Oracle America v. Google, 750 F.3d 1339 (2014), a landmark opinion in which the Federal Circuit Court of Appeals held that the interface of the Java language was copyrightable. And since we'll want to compare the Oracle case to a related case, let's also download Lotus Development Corporation v. Borland International, 49 F.3d 807 (1995). In that case, the First Circuit Court of Appeals held that the menu structure of a spreadsheet program called Lotus 1-2-3 was uncopyrightable because it was a "method of operation" under the Copyright Act. As we'll see, the Oracle case discusses and disagrees with the Lotus case.
#
# If you already loaded `Opinion`s from a file, running the cells below with `USE_REAL_CASE_API` set to True will attempt to overwrite them with data from the API. You should be able to run the rest of the tutorial code either way.
# +
from authorityspoke import CAPClient
if USE_REAL_CASE_API:
case_client = CAPClient(api_token=CAP_API_KEY)
oracle_case = case_client.read_cite(cite="750 F.3d 1339")
# -
# Now we have a record representing the _Oracle_ case, which can also be found in the "example_data/opinions" folder under the filename "oracle_h.json". Let's look at a field from the API response.
oracle_case.name
# Yes, this is the correct case name. But if we had provided the API key and used the `full_case` flag, we could have received more information, like whether there are any non-majority opinions in the case, and the names of the opinion authors. So let's request the _Oracle_ case with `full_case=True`.
if USE_REAL_CASE_API:
oracle_case = case_client.read_cite(
cite="750 F.3d 1339", full_case=True)
# And then do the same for the _Lotus_ case.
if USE_REAL_CASE_API:
lotus_case = case_client.read_cite(
cite="49 F.3d 807", full_case=True)
# Now let's take a look at the objects we made.
print(oracle_case)
print(lotus_case)
# One judicial `Decision` can include multiple `Opinion`s. The Lotus `Decision` has a concurring opinion as well as a majority opinion. Access the `majority` attribute of the `Decision` object to get the majority opinion.
print(oracle_case.majority)
print(lotus_case.majority)
# ## 3. Downloading Enactments
# AuthoritySpoke also includes an interface for downloading provisions of the United States Constitution and US Code from the API at authorityspoke.com. To use this, first create a LegisClient object that holds your API key. Then you can use the `Client.fetch` method to fetch JSON representing the provision at a specified citation on a specified date (or the most recent version, if you don't specify a date). Or you can use `Client.read`, which also fetches the JSON but then loads it into an instance of the `Enactment` class.
# +
from authorityspoke import LegisClient
from authorityspoke.io.fake_enactments import FakeClient
if USE_REAL_LEGISLICE_API:
LEGISLICE_API_TOKEN = os.getenv("LEGISLICE_API_TOKEN")
legis_client = LegisClient(api_token=LEGISLICE_API_TOKEN)
else:
legis_client = FakeClient.from_file("usc.json")
# -
# ## 4. Importing and Exporting Legal Holdings
# Now we can link some legal analysis to each majority `Opinion` by using `Decision.posit` or `Opinion.posit`. The parameter we pass to this function is a `Holding` or list of `Holding`s posited by the `Opinion`. You can think of a `Holding` as a statement about whether a `Rule` is or is not valid law. A `Holding` may exist in the abstract, or it may be **posited** by one or more `Opinion`s, which means that the `Opinion` adopts the `Holding` as its own. An `Opinion` may posit more than one `Holding`.
#
# Sadly, the labor of creating data about `Holding`s falls mainly to the user rather than the computer, at least in this early version of AuthoritySpoke. AuthoritySpoke loads `Holding`s from structured descriptions that need to be created outside of AuthoritySpoke as JSON files. For more information on creating these JSON files, see the [guide to creating Holding data](https://authorityspoke.readthedocs.io/en/latest/guides/create_holding_data.html). The guide includes a [JSON specification](https://authorityspoke.readthedocs.io/en/latest/guides/create_holding_data.html#json-api-specification) describing the required data format.
#
# For now, this introduction will rely on example YAML files that have already been created. AuthoritySpoke should find them and convert them to AuthoritySpoke objects when we call the `read_holdings_from_file` function. If you pass in a `client` parameter, AuthoritySpoke will make calls to the API at [authorityspoke.com](https://authorityspoke.com/) to find and link the statutes or other `Enactment`s cited in the `Holding`.
#
# +
from authorityspoke.io.loaders import read_holdings_from_file
oracle_holdings = read_holdings_from_file("holding_oracle.yaml", client=legis_client)
print(oracle_holdings[0])
# -
# You can also convert Holdings back to JSON, or to a Python dictionary, using the ``dump`` module.
# +
from authorityspoke.io.dump import to_json, to_dict
to_dict(oracle_holdings[0])["rule"]["procedure"]
# -
# ## 5. Linking Holdings to Opinions
# If you want annotation anchors to link each Holding to a passage in the Opinion, you can use the ``read_anchored_holdings_from_file`` method. The result is a type of NamedTuple called `AnchoredHoldings`. You can pass this NamedTuple as the only argument to the `Opinion.posit()` method to assign the `Holding`s to the majority `Opinion`. This will also link the correct text passages from the Opinion to each Holding.
# +
from authorityspoke.decisions import DecisionReading
from authorityspoke.io.loaders import read_anchored_holdings_from_file
oracle_holdings = read_anchored_holdings_from_file("holding_oracle.yaml", client=legis_client)
lotus_holdings = read_anchored_holdings_from_file("holding_lotus.yaml", client=legis_client)
oracle = DecisionReading(decision=oracle_case)
oracle.posit(oracle_holdings)
lotus = DecisionReading(decision=lotus_case)
lotus.posit(lotus_holdings)
# -
# You can pass either one Holding or a list of Holdings to `Opinion.posit()`. The `Opinion.posit()` method also has a `text_links` parameter that takes a dict indicating what text spans in the Opinion should be linked to which Holding.
# ## 6. Viewing an Opinion's Holdings
# If you take a look in [holding_oracle.json](https://github.com/mscarey/AuthoritySpoke/blob/master/example_data/holdings/holding_oracle.json) in AuthoritySpoke's git repository, you’ll see that it would be loaded in Python as a list of 20 dicts, each representing a holding. (In case you aren't familiar with how Python handles JSON, the outersquare brackets represent the beginning and end of the list. The start and end of each dict in the list is shown by a matched pair of curly brackets.)
#
#
# Let's make sure that the .posit() method linked all of those holdings to our `oracle` Opinion object.
len(oracle.holdings)
# Now let's see the string representation of the AuthoritySpoke Holding object we created from the structured JSON we saw above.
print(oracle.holdings[0])
# Instead of the terms "inputs" and "outputs" we saw in the JSON file, we now have "GIVEN" and "RESULT". And the "RESULT" comes first, because it's hard to understand anything else about a legal rule until you understand what it does. Also, notice the separate heading "GIVEN the ENACTMENT". This indicates that the existence of statutory text (or another kind of enactment such as a constitution) can also be a precondition for a `Rule` to apply. So the two preconditions that must be present to apply this `Rule` are "the Fact it is false that the Java API was an original work" and the statutory text creating copyright protection.
#
# It's also important to notice that a `Rule` can be purely hypothetical from the point of view of the Opinion that posits it. In this case, the court finds that there would be a certain legal significance if it was "GIVEN" that `it is false that <the Java API> was an original work`, but the court isn't going to find that precondition applies, so it's also not going to accept the "RESULT" that `it is false that <the Java API> was copyrightable`.
# We can also access just the inputs of a `Holding`, just the `Enactment`s, etc.
print(oracle.holdings[0].inputs[0])
print(oracle.holdings[0].enactments[0])
# ## 7. Generic Factors
#
# The two instances of the phrase "the Java API" are in angle brackets to indicate that the Java API is a generic `Entity` mentioned in the `Fact`.
oracle.holdings[0].generic_terms()
# A generic `Entity` is "generic" in the sense that in the context of the `Factor` where the `Entity` appears, it could be replaced with some other generic `Entity` without changing the meaning of the `Factor` or the `Rule` where it appears.
#
# Let's illustrate this idea with the first `Holding` from the _Lotus_ case.
print(lotus.holdings[0])
# What if we wanted to generalize this `Holding` about copyright and apply it in a different context, such as a case about books or television shows instead of computer programs? First we could look at the "generic" `Factor`s of the `Holding`, which were marked off in angle brackets in the string representation of the `Holding`.
lotus.holdings[0].generic_terms()
# The same `Rule`s and `Holding`s may be relevant to more than one `Opinion`. Let's try applying the idea from `lotus.holdings[0]` to a different copyright case that's also about a derivative work. In [_Castle Rock Entertainment, Inc. v. Carol Publishing Group Inc._](https://en.wikipedia.org/wiki/Castle_Rock_Entertainment,_Inc._v._Carol_Publishing_Group_Inc.) (1998), a United States Court of Appeals found that a publisher infringed the copyright in the sitcom _Seinfeld_ by publishing a trivia book called _SAT: The Seinfeld Aptitude Test_.
#
# Maybe we'd like to see how the `Holding` from the _Lotus_ case could have applied in the context of the _Castle Rock Entertainment_ case, under 17 USC 102. We can check that by using the `Holding.new_context()` method to replace the generic factors from the _Lotus_ `Holding`. One way to do this is by passing a tuple containing a list of factors that need to be replaced, followed by a list of their replacements.
# +
from authorityspoke import Entity
seinfeld_holding = lotus.holdings[0].new_context(
terms_to_replace=[
Entity("Borland International"),
Entity("the Lotus menu command hierarchy"),
],
changes=[Entity("Carol Publishing Group"), Entity("Seinfeld")]
)
# -
# The `new_context` method returns a new `Holding` object, which we've assigned to the name `seinfeld_holding`, but the `Holding` that we used as a basis for the new object also still exists, and it's unchanged.
print(seinfeld_holding)
# Even though these `Holding`s have different generic factors and don't evaluate equal to one another, the `Holding.means()` method shows that they have the same meaning. In other words, they both endorse exactly the same legal Rule. If Holding A `means` Holding B, then Holding A also necessarily `implies` Holding B.
lotus.holdings[0] == seinfeld_holding
lotus.holdings[0].means(seinfeld_holding)
# ## 8. Enactment Objects and Implication
# Sometimes it's useful to know whether one `Rule` or `Holding` implies another. Basically, one legal `Holding` implies a second `Holding` if its meaning entirely includes the meaning of the second `Holding`. To illustrate this idea, let's look at the `Enactment` that needs to be present to trigger the `Holding` at `oracle.holdings[0]`.
copyright_provision = oracle.holdings[0].enactments[0]
print(copyright_provision)
# The `Enactment` object refers to part of the text of subsection 102(a) from [Title 17 of the United States Code](https://www.copyright.gov/title17/).
# Next, let's create a new `Enactment` object representing a shorter passage of text from the same provision. We select some text from the provision by calling the `select` method with the string `works_of_authorship_passage`, which exactly matches some text that can be found in subsection 102(a).
# +
from authorityspoke import Enactment
works_of_authorship_passage = (
"Copyright protection subsists, in accordance with this title, "
+ "in original works of authorship"
)
works_of_authorship_clause = legis_client.read("/us/usc/t17/s102/a")
works_of_authorship_clause.select(works_of_authorship_passage)
# -
# Now we can create a new `Holding` object that cites to our new `Enactment` object rather than the old one. This time, instead of using the `new_context` method to create a new `Holding` object, we'll use Python's built-in `deepcopy` function. This method gives us an identical copy of the `Holding` object that we can change without changing the original. Then we can use the `set_enactments` method to change what `Enactment` is cited by the new `Holding`.
# +
from copy import deepcopy
holding_with_shorter_enactment = deepcopy(oracle.holdings[0])
holding_with_shorter_enactment.set_enactments(works_of_authorship_clause)
# -
print(holding_with_shorter_enactment)
# Now let's try comparing this new `Holding` with the real `Holding` from the _Oracle_ case, to see whether one implies the other. When you're comparing AuthoritySpoke objects, the greater than sign `>` means "implies, but is not equal to".
holding_with_shorter_enactment > oracle.holdings[0]
# You can also use the greater than or equal sign `>=` to mean "implies or is equal to". You can also use lesser than signs to test whether an object on the right side of the expression implies the object on the left. Thus, `<=` would mean "is implied by or is equal to".
holding_with_shorter_enactment <= oracle.holdings[0]
# By comparing the string representations of the original `Holding` from the _Oracle_ case and `holding_with_shorter_enactment`, can you tell why the latter implies the former, and not the other way around?
#
# If you guessed that it was because `holding_with_shorter_enactment` has a shorter `Enactment`, you're right. `Rule`s that require fewer, or less specific, inputs are _broader_ than `Rule`s that have more inputs, because there's a larger set of situations where those `Rule`s can be triggered.
#
# If this relationship isn't clear to you, imagine some "Enactment A" containing only a subset of the text of "Enactment B", and then imagine what would happen if a legislature amended some of the statutory text that was part of Enactment B but not of Enactment A. A requirement to cite Enactment B would no longer be possible to satisfy, because some of that text would no longer be available. Thus a requirement to cite Enactment A could be satisfied in every situation where a requirement to cite Enactment B could be satisfied, and then some.
# ## 9. Checking for Contradictions
# Let's turn back to the _Lotus_ case.
#
# It says that under a statute providing that "In no case does copyright protection for an original work of authorship extend to any...method of operation", the fact that a Lotus menu command hierarchy was a "method of operation" meant that it was also uncopyrightable, despite a couple of `Fact`s that might tempt some courts to rule the other way.
print(lotus.holdings[6])
# _Lotus_ was a case relied upon by Google in the _Oracle v. Google_ case, but Oracle was the winner in that decision. So we might wonder whether the _Oracle_ Opinion contradicts the _Lotus_ Opinion. Let's check.
oracle.contradicts(lotus)
# That's good to know, but we don't want to take it on faith that a contradiction exists. Let's use the `explain_contradiction` method to find the contradictory `Holding`s posited by the _Oracle_ and _Lotus_ cases, and to generate a rudimentary explanation of why they contradict.
explanation = lotus.explain_contradiction(oracle)
print(explanation)
# That's a really complicated holding! Good thing we have AuthoritySpoke to help us grapple with it.
#
# We can use the `explanations_contradiction` method directly on `Holding`s to generate all available "explanations" of why a contradiction is possible between these lotus.holdings[6] and oracle.holdings[10]. Each `Explanation` includes a mapping that shows how the context factors of the `Holding` on the left can be mapped onto the `Holding` on the right. The explanation we've already been given is that these two `Holding`s contradict each other if you consider 'the Lotus menu command hierarchy' to be analagous to 'the Java API'. The other possible explanation AuthoritySpoke could have given would have been that 'the Lotus menu command hierarchy' is analagous to 'the Java language'. Let's see if the other possible `Explanation` also appears in `explanations`.
explanations = list(lotus.holdings[6].explanations_contradiction(oracle.holdings[10]))
len(explanations)
# No, there’s only one `Explanation` given for how these rules can contradict each other. (The `Holding.explain_contradiction` method
# returns only one one `Explanation`, but `explanations_contradiction`
# is a generator that yields every `Explanation` it can find.) If you read the *Oracle* case, is makes sense that ‘the Lotus menu command hierarchy’ is not considered analagous to ‘the Java language’. The *Oracle* case is only about infringing the copyright in the Java API, not the copyright in the whole Java language. A statement about infringement of ‘the Java
# language’ would be irrelevant, not contradictory.
#
# But what exactly is the contradiction between the two `Holding`s?
# The first obvious contrast between `lotus.holdings[6]` and `oracle.holdings[10]` is that the `Holding` from the _Lotus_ case is relatively succinct and categorical. The _Lotus_ court interprets Section 102(b) of the Copyright Act to mean that if a work is a "method of operation", it's simply impossible for that work to be copyrighted, so it's not necessary to consider a lot of case-specific facts to reach a conclusion.
#
# The Federal Circuit's _Oracle_ decision complicates that view significantly. The Federal Circuit believes that the fact that an API is, or hypothetically might be, a "method of operation" is only one of many factors that a court can consider in deciding copyrightability. The following quotation, repeated in the _Oracle_ case, illustrates the Federal Circuit's view.
#
# > “Section 102(b) does not extinguish the protection accorded a particular expression of an idea merely because that expression is embodied in a method of operation.” Mitel, Inc. v. Iqtel, Inc., 124 F.3d 1366, 1372 (10th Cir.1997)
#
# And that's why AuthoritySpoke finds a contradiction between these two `Rule`s. The _Oracle_ opinion says that courts can sometimes accept the result `the Fact that <the Java API> was copyrightable` despite the `Fact` `<the Java API> was a method of operation`. The _Lotus_ Opinion would consider that impossible.
#
# By the way, AuthoritySpoke does not draw on any Natural Language Understanding technologies to determine the meaning of each `Fact`. AuthoritySpoke mostly won't recognize that `Fact`s have the same meaning unless their `content` values are exactly the same string. As discussed above, they can also differ in their references to generic factors, which are the phrases that appear in brackets when you use the `str()` command on them. Also, AuthoritySpoke has a limited ability to compare numerical statements in `Fact`s using [pint](https://pint.readthedocs.io/en/stable/), an amazing Python library that performs dimensional analysis.
# ## 10. Adding Holdings to One Another
# To try out the addition operation, let's load another case from the `example_data` folder.
# +
from authorityspoke.io.loaders import load_decision_as_reading
if USE_REAL_CASE_API:
feist_case = case_client.read_cite('499 U.S. 340')
feist = DecisionReading(decision=feist_case)
else:
feist = load_decision_as_reading("feist_h.json")
feist.posit(read_anchored_holdings_from_file("holding_feist.json", client=legis_client))
# -
# [_Feist Publications, Inc. v. Rural Telephone Service Co._](https://en.wikipedia.org/wiki/Feist_Publications,_Inc.,_v._Rural_Telephone_Service_Co.) was a case that held that the listings in a telephone directory did not qualify as "an original work" and that only original works are eligible for protection under the Copyright Act. This is a two-step analysis.
#
# The first step results in the `Fact` it is false that a generic `Entity` was "an original work":
print(feist.holdings[10])
# And the second step relies on the result of the first step to reach the further result of "absence of the Fact that" a generic `Entity` was "copyrightable".
print(feist.holdings[3])
# In this situation, anytime the first Holding (feist.holdings[10]) is applied, the second Holding (feist.holdings[3]) can be applied as well. That means the two Holdings can be added together to make a single Holding that captures the whole process.
listings_not_copyrightable = feist.holdings[10] + feist.holdings[3]
print(listings_not_copyrightable)
# The difference between `feist.holdings[10]` and the newly-created Holding `listings_not_copyrightable` is that `listings_not_copyrightable` has two Factors under its "RESULT", not just one. Notice that it doesn't matter that the two original Holdings reference different generic Entities ("Rural's telephone directory" versus "Rural's telephone listings"). Because they're generic, they're interchangeable for this purpose.
# You might recall that oracle.holdings[0] also was also about the relationship between originality and copyrightability. Let's see what happens when we add oracle.holdings[0] to feist.holdings[10].
print(feist.holdings[10] + oracle.holdings[0])
# Can you guess why it's not possible to add these two Holdings together? Here's a hint:
feist.holdings[10].exclusive
oracle.holdings[0].exclusive
feist.holdings[3].exclusive
# `feist.holdings[10]` and `oracle.holdings[0]` are both Holdings that purport to apply in only "SOME" cases where the specified inputs are present, while `feist.holdings[3]` purports to be the "EXCLUSIVE" way to reach its output, which indicates a statement about "ALL" cases.
#
# You can't infer that there's any situation where `feist.holdings[10]` and `oracle.holdings[0]` can actually be applied together, because there might not be any overlap between the "SOME" cases where one applies and the "SOME" cases where the other applies. But if `feist.holdings[10]` and `feist.holdings[3]` are both valid law, then we know they can both apply together in any of the "SOME" cases where `feist.holdings[10]` applies.
# ## 11. Set Operations with Holdings
# In AuthoritySpoke, the union operation is different from the addition operation, and it usually gives different results.
# +
result_of_adding = feist.holdings[10] + feist.holdings[3]
result_of_union = feist.holdings[10] | feist.holdings[3]
result_of_adding == result_of_union
# -
# Although the existence of the union operation might suggest that there should also be an intersection operation, an intersection operation is not yet implemented in AuthoritySpoke 0.4.
# Apply the union operator to two `Holding`s to get a new `Holding` with all of the inputs and all of the outputs of both of the two original `Holding`s. However, you only get such a new `Holding` if it can be inferred by accepting the truth of the two original `Holding`s. If the two original `Holding`s contradict one another, the operation returns `None`. Likewise, if the two original `Holding`s both have the value `False` for the parameter `universal`, the operation will return `None` if it's possible that the "SOME" cases where one of the original `Holding`s applies don't overlap with the "SOME" cases where the other applies.
#
# In this example, we'll look at a `Holding` from _Oracle_, then a `Holding` from _Feist_, and then the union of both of them.
print(oracle.holdings[1])
print(feist.holdings[2])
print(oracle.holdings[1] | feist.holdings[2])
# It's not obvious that a litigant could really establish all the "GIVEN" Factors listed above in a single case in a court where `oracle.holdings[1]` and `feist.holdings[2]` were both valid law, but if they could, then it seems correct for AuthoritySpoke to conclude that the court would have to find both `the Fact that <the Java API> was an original work` and `the Fact it is false that <the Java API> was copyrightable`.
#
# The union operator is useful for searching for contradictions in a collection of `Holding`s. When two `Holding`s are combined together with the union operator, their union might contradict other `Holding`s that neither of the two original `Holding`s would have contradicted on their own.
# ## 12. Nuances of Meaning in Holdings
# Let's look at one more sentence from the _Oracle_ `Opinion`, so I can point out a few more design decisions AuthoritySpoke makes in representing procedural `Holding`s.
#
# > In the Ninth Circuit, while questions regarding originality are considered questions of copyrightability, concepts of merger and scenes a faire are affirmative defenses to claims of infringement.
#
# (The "merger" doctrine says that a work is deemed to be "merged" with an uncopyrightable idea if it's essentially the only way to express the idea. "Scenes a faire" is a concept applied mostly to works of fiction, and it means that conventional genre tropes are not copyrightable.)
#
# The quoted sentence is fairly ordinary, as court opinions go, but I found several interesting challenges in creating structered data about its procedural meaning.
#
# 1. The sentence describes what the law is "In the Ninth Circuit". You might remember that the court that issued the _Oracle_ opinion was the Federal Circuit, not the Ninth Circuit. So the Federal Circuit is deciding what it thinks that the Ninth Circuit thinks that Congress meant by enacting the statute. The middle layer of this interpretation, in which the Federal Circuit attributes a belief to the Ninth Circuit, is simply absent from the AuthoritySpoke model of the `Holding`. However, future updates to AuthoritySpoke might make it possible to capture this information.
#
# 2. The sentence uses the concept of an "affirmative defense", which generally means a defense that the defendant has the burden of proof to establish. I chose to model this concept by writing that if one of the facts that would establish the affirmative defense is present, then it could be established that the copyright was not infringed, but if they are both absent, then the copyright could have been infringed. I'm sure some legal experts would find this too simplistic, and would argue that it's not possible to formalize the concept of an affirmative defense without explicitly mentioning procedural concepts like a burden of proof.
#
# 3. The sentence seems to have something to say about what happens if either of two Factors are present, or if both of them are absent. That makes three different Rules. It's not ideal for one sentence to explode into multiple different Python objects when it's formalized, and it's worth wondering whether there would have been a way to pack all the information into a single object.
#
# 4. I noticed that the concept of a copyrighted work being "merged" or being a "scene a faire" are both characteristics intrinsic in the copyrighted work, and don't depend on the characteristics of the allegedly infringing work. So if a work that's "merged" or is a "scene a faire" can't be infringed, but those concepts aren't relevant to copyrightability, then that means there are some works that are "copyrightable" but that can never be infringed by any other work. I suspect that the court's interpretation of these legal categories could confuse future courts and parties, with the result that the "merger" or "scene a faire" concepts could fall through the cracks and be ignored. Would there be a useful way to have AuthoritySpoke flag such anomalies?
#
# The three Holding objects used to represent the sentence from the _Oracle_ opinion can be found in the Appendix below. They're `oracle.holdings[11]` through `oracle.holdings[13]`.
# ## Appendix: Holding Objects in *Oracle v. Google* and *Lotus v. Borland*
# This Appendix will list all of the Holding objects in `oracle.holdings` and `lotus.holdings`. Each `Holding` will be preceded by a passage from the `Opinion` that indicates the `Opinion` has endorsed the `Holding`. In future versions, AuthoritySpoke will give users the ability to explore the text passages in `Opinion`s that provide support for each `Holding`, but that's currently not fully implemented.
#
# To find the full text of the opinions, look in the example_data/opinions/ folder. The text delivered by the CAP API was collected from print sources, so it will contain some Optical Character Recognition errors.
# ### *Lotus v. Borland* 49 F.3d 807 (1995)
# > To establish copyright infringement, a plaintiff must prove "(1) ownership of a valid copyright, and (2) copying of constituent elements of the work that are original."
print(lotus.holdings[0])
# > To, show ownership of a valid copyright and therefore satisfy Feist’s first prong, a plaintiff must prove that the work as a whole is original and that the plaintiff complied with applicable statutory formalities.
print(lotus.holdings[1])
# > In judicial proceedings, a certificate of copyright registration constitutes prima facie evidence of copyrightability and shifts the burden to the defendant to demonstrate why the copyright is not valid.
print(lotus.holdings[2])
# > To show actionable copying and therefore satisfy Feist’s second prong, a plaintiff must first prove that the alleged infringer copied plaintiffs copyrighted work as a factual matter; to do this, he or she may either present direct evidence of factual copying or...
print(lotus.holdings[3])
# > To show actionable copying and therefore satisfy Feist’s second prong, a plaintiff must first prove that the alleged infringer copied plaintiffs copyrighted work as a factual matter; to do this, he or she may either present direct evidence of factual copying or, if that is unavailable, evidence that the alleged infringer had access to the copyrighted work and that the offending and copyrighted works are so similar that the court may infer that there was factual copying (i.e., probative similarity).
print(lotus.holdings[4])
# > To show actionable copying and therefore satisfy Feist’s second prong, a plaintiff must first prove that the alleged infringer copied plaintiffs copyrighted work as a factual matter...The plaintiff must then prove that the copying of copyrighted material was so extensive that it rendered the offending and copyrighted works substantially similar.
print(lotus.holdings[5])
# > Section 102(b) states: “In no case does copyright protection for an original work of authorship extend to any idea, procedure, process, system, method of operation, concept, principle, or discovery, regardless of the form in which it is described, explained, illustrated, or embodied in such work.” Because we conclude that the Lotus menu command hierarchy is a method of operation, we do not consider whether it could also be a system, process, or procedure...while original expression is necessary for copyright protection, we do not think that it is alone sufficient. Courts must still inquire whether original expression falls within one of the categories foreclosed from copyright protection by § 102(b), such as being a “method of operation.”
print(lotus.holdings[6])
# > We hold that the Lotus menu command hierarchy is an uneopyrightable “method of operation.” The Lotus menu command hierarchy provides the means by which users control and operate Lotus 1-2-3. If users wish to copy material, for example, they use the “Copy” command. If users wish to print material, they use the “Print” command. Users must use the command terms to tell the computer what to do. Without the menu command hierarchy, users would not be able to access and control, or indeed make use of, Lotus 1-2-3’s functional capabilities.
print(lotus.holdings[7])
# > We do not think that “methods of operation” are limited to abstractions; rather, they are the means by which a user operates something.
print(lotus.holdings[8])
# > In other words, to offer the same capabilities as Lotus 1-2-3, Borland did not have to copy Lotus’s underlying code (and indeed it did not); to 'allow users to operate its programs in substantially the same way, however, Bor-land had to copy the Lotus menu command hierarchy. Thus the Lotus 1-2-3 code is not a uncopyrightable “method of operation.”
print(lotus.holdings[9])
# ### *Oracle v. Google* 750 F.3d 1339 (2014)
# > By statute, a work must be “original” to qualify for copyright protection. 17 U.S.C. § 102(a).
print(oracle.holdings[0])
# > Original, as the term is used in copyright, means only that the work was independently created by the author (as opposed to copied from other works), and that it possesses at least some minimal degree of creativity.
print(oracle.holdings[1])
# > Copyright protection extends only to the expression of an idea — not to the underlying idea itself...In the Ninth Circuit, while questions regarding originality are considered questions of copyrightability, concepts of merger and scenes a faire are affirmative defenses to claims of infringement.
print(oracle.holdings[2])
# > The literal elements of a computer program are the source code and object code.
print(oracle.holdings[3])
print("\n")
print(oracle.holdings[4])
# > It is well established that copyright protection can extend to both literal and non-literal elements of a computer program. See Altai 982 F.2d at 702.
print(oracle.holdings[5])
# > The non-literal components of a computer program include, among other things, the program’s sequence, structure, and organization, as well as the program’s user interface.
print(oracle.holdings[6])
print("\n")
print(oracle.holdings[7])
# > It is well established that copyright protection can extend to both literal and non-literal elements of a computer program...As discussed below, whether the non-literal elements of a program “are protected depends on whether, on the particular facts of each case, the component in question qualifies as an expression of an idea, or an idea itself.”
print(oracle.holdings[8])
print("\n")
print(oracle.holdings[9])
# > On appeal, Oracle argues that the district court’s reliance on Lotus is misplaced because it is distinguishable on its facts and is inconsistent with Ninth Circuit law. We agree. First, while the defendant in Lotus did not copy any of the underlying code, Google concedes that it copied portions of Oracle’s declaring source code verbatim. Second, the Lotus court found that the commands at issue there (copy, print, etc.) were not creative, but it is undisputed here that the declaring code and the structure and organization of the API packages are both creative and original. Finally, while the court in Lotus found the commands at issue were “essential to operating” the system, it is undisputed that— other than perhaps as to the three core packages — Google did not need to copy the structure, sequence, and organization of the Java API packages to write programs in the Java language.\nMore importantly, however, the Ninth Circuit has not adopted the court’s “method of operation” reasoning in Lotus, and we conclude that it is inconsistent with binding precedent.
print(oracle.holdings[10])
# > In the Ninth Circuit, while questions regarding originality are considered questions of copyrightability, concepts of merger and scenes a faire are affirmative defenses to claims of infringement.
print(oracle.holdings[11])
# > In the Ninth Circuit, while questions regarding originality are considered questions of copyrightability, concepts of merger and scenes a faire are affirmative defenses to claims of infringement...Under the merger doctrine, a court will not protect a copyrighted work from infringement if the idea contained therein can be expressed in only one way.
print(oracle.holdings[12])
print("\n")
print(oracle.holdings[13])
# #### A Missing Holding
#
# The following text represents a rule posited by the Oracle court, but it's not currently possible to create a corresponding Holding object, because AuthoritySpoke doesn't yet include "Argument" objects.
#
# > Google responds that Oracle waived its right to assert copyrightability based on the 7,000 lines of declaring code by failing “to object to instructions and a verdict form that effectively eliminated that theory from the case.” Appellee Br. 67...We find that Oracle did not waive arguments based on Google’s literal copying of the declaring code.
# > Regardless of when the analysis occurs, we conclude that merger does not apply on the record before us...We have recognized, however, applying Ninth Circuit law, that the “unique arrangement of computer program expression ... does not merge with the process so long as alternate expressions are available.”...The evidence showed that Oracle had “unlimited options as to the selection and arrangement of the 7000 lines Google copied.”...This was not a situation where Oracle was selecting among preordained names and phrases to create its packages.
print(oracle.holdings[14])
# > the relevant question for copyright-ability purposes is not whether the work at issue contains short phrases — as literary works often do — but, rather, whether those phrases are creative.
print(oracle.holdings[15])
# > In the computer context, “the scene a faire doctrine denies protection to program elements that are dictated by external factors such as ‘the mechanical specifications of the computer on which a particular program is intended to run’ or ‘widely accepted programming practices within the computer industry. Like merger, the focus of the scenes a faire doctrine is on the circumstances presented to the creator, not the copier.
print(oracle.holdings[16])
# > Specifically, we find that Lotus is inconsistent with Ninth Circuit case law recognizing that the structure, sequence, and organization of a computer program is eligible for copyright protection where it qualifies as an expression of an idea, rather than the idea itself.
print(oracle.holdings[17])
# > an original work — even one that serves a function — is entitled to copyright protection as long as the author had multiple ways to express the underlying idea. Section 102(b) does not, as Google seems to suggest, automatically deny copyright protection to elements of a computer program that are functional.
print(oracle.holdings[18])
# > Until either the Supreme Court or Congress tells us otherwise, we are bound to respect the Ninth Circuit’s decision to afford software programs protection under the copyright laws. We thus decline any invitation to declare that protection of software programs should be the domain of patent law, and only patent law.
print(oracle.holdings[19])
| 79.567138
| 1,075
|
15ef7f2d0ac41434d4f919fc0f63a2ccd7537374
|
py
|
python
|
Simple_regression_task.ipynb
|
SoumyadeepB/DeepLearning
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] colab_type="text" id="QjlEFdXpMgvw"
# # Deep learning programming I-A: Regression
#
# ## Introduction
# This programming exercise is the first of a series of exercises, which are intended as a supplement to the theoretical part of the Deep Learning course. The goal is to introduce you to basic tasks and applications of methods you have encountered in the lecture. After completing the exercise you should be familiar with the basic ideas and one, possibly simple, way of solving the respective task. It is worth mentioning that most of the tasks can be solved in many different, not necessarily deep learning based, ways and the solution presented here is just one of them.
#
# ## Regression
#
# In this exercise we consider the problem of regression, where we are interested in modeling a functional dependence between different variables with, possibly noisy, observations of input-output pairs. Mathematically such a dependence can be formulated as
#
# $\mathbf{y}=f(\mathbf{x})+\boldsymbol{\epsilon}$,
#
# where $\mathbf{y}\in\mathbb{R}^{M}$ and $\mathbf{x}\in\mathbb{R}^{N}$ are the input and output observations, $f:\mathbb{R}^{N}\rightarrow\mathbb{R}^{M}$ is the function mapping inputs to outputs and $\boldsymbol{\epsilon}\in\mathbb{R}^{M}$ is a random vector, which models noise in our observations. Note that this assumes additive noise that only acts on the output and not on the input variable, which might not be true in all practical applications but is a reasonable approximation. For regression we are now interested in estimating the functional relationship $f$ between the inputs and outputs. This can be done in many different ways, not just with neural networks, but for this exercise we focus on approximating this relationship with a neural network $g_{\boldsymbol{\theta}}:\mathbb{R}^{N}\rightarrow\mathbb{R}^{M}$ with parameter vector $\boldsymbol{\theta}$. The task is now to choose the parameters of the neural network in a way that results in a "good" approximation of $f$ with $g_{\boldsymbol{\theta}}$.
#
# In order to quantify how "good" our neural network can approximate $f$, we adopt a probabilistic view. For this we make the assumption that the noise $\boldsymbol{\epsilon}$ is a random vector drawn from a known dustribution, which enables us to derive a suitable cost function for training our neural network and also for quantifying a "good" approximation.
#
# ### Mathematical formulation
# If we assume that the noise $\boldsymbol{\epsilon}$ is drawn from a gaussian distribution, e.g. $\boldsymbol{\epsilon}\sim\mathcal{N}(\mathbf{0},\sigma^{2}\mathbf{I})$, we can use
#
# $\mathbf{y}=g_{\boldsymbol{\theta}}(\mathbf{x})+\boldsymbol{\epsilon}\Rightarrow \mathbf{y}-g_{\boldsymbol{\theta}}(\mathbf{x})=\boldsymbol{\epsilon}$
#
# to derive a log likelihood. Since the probability density function (pdf) of a multivariate normal distribution is given by
#
# $p(\mathbf{x})=\dfrac{1}{\sqrt{(2\pi)^{D}\vert\mathbf{C}\vert}}\mathrm{e}^{-\dfrac{1}{2}(\mathbf{x}-\boldsymbol{\mu})\mathbf{C}^{-1}(\mathbf{x}-\boldsymbol{\mu})^{T}}$,
#
# we get
#
# $\ln{p(\boldsymbol{\epsilon})}=\ln{\dfrac{1}{\sqrt{(2\pi)^{M}\sigma^{2}}}\mathrm{e}^{-\dfrac{1}{2\sigma^{2}}\Vert\boldsymbol{\epsilon}\Vert_{2}^{2}}}=-\dfrac{1}{2\sigma^{2}}\Vert\boldsymbol{\epsilon}\Vert_{2}^{2}-\dfrac{1}{2}\ln{(2\pi)^{M}\sigma^{2}}$.
#
# Replacing $\boldsymbol{\epsilon}$ by $\mathbf{y}-g_{\boldsymbol{\theta}}(\mathbf{x})$ yields the log likelihood for one particular input-output pair:
#
# $\mathcal{L}(\mathbf{x},\mathbf{y},\boldsymbol{\theta})=\ln {p(\mathbf{y}\vert\mathbf{x},\boldsymbol{\theta})}=-\dfrac{1}{2\sigma^{2}}\Vert\boldsymbol{\mathbf{y}-g_{\boldsymbol{\theta}}(\mathbf{x})}\Vert_{2}^{2}-\dfrac{1}{2}\ln{(2\pi)^{M}\sigma^{2}}$
#
# This log likelihood measures how likely the input-output pair is and we can use it to train our neural network. For this we maximize the expected log likelihood over all input-output pairs under the assumption that the noise is idependent and identically distributed (i.i.d.) over all input-output pairs. This corresponds to finding the parameters $\boldsymbol{\theta}^{\star}$ of our neural network, which maximize the the expected probability for observing the corresponding input-output pairs. Mathematically the optimal parameters for our neural network are given by
#
# $\boldsymbol{\theta}^{\star}=\arg\max_{\boldsymbol{\theta}}\mathbb{E}\left[\mathcal{L}(\mathbf{x},\mathbf{y},\boldsymbol{\theta})\right]=\arg\max_{\boldsymbol{\theta}}\mathbb{E}\left[-\Vert\boldsymbol{\mathbf{y}-g_{\boldsymbol{\theta}}(\mathbf{x})}\Vert_{2}^{2}\right]\approx\arg\min_{\boldsymbol{\theta}}\dfrac{1}{N_{D}}\sum_{i=1}^{N_{D}}\Vert\boldsymbol{\mathbf{y}_{i}-g_{\boldsymbol{\theta}}(\mathbf{x}_{i})}\Vert_{2}^{2}$,
#
# where all terms, which are independent of $\boldsymbol{\theta}$, are ignored and the expectation operator is approximated by the mean over all $N_{D}$ input-output pairs. In other words we are maximizing the log likelihood by minimizing the mean squared error loss over all input-output pairs in our dataset, hence this approach is called Maximum Likelihood (ML) estimation. For solving this optimization problem and obtaining the optimal network parameters, stochastic gradient descent (SGD) or one of it's many variants is typically used.
#
# It is worth noting, that choosing different distributions for the noise $\boldsymbol{\epsilon}$ leads to different log likelihoods and therefore different cost functions for training the neural network. Another commonly used distribution for modelling the noise in regression tasks is the laplace distribution. Deriving the log likelihood and the corresponding costfunction leads to the mean absolute error, which is given by the $l_{1}$-norm of the difference between observations predictions of the neural network. This cost function is considered more robust against outliers since these have less influence on the averall loss compared to the mean squared error.
#
# ### Implementation
#
# In the following we consider a simple regression task, implement a neural network and train it based on the mathematical fomrulation above. For this we first need to create a set of input-output pairs, which then needs to be partitioned into a training, validation and test set. We also define some constants to be used for partitioning the data and the hyperparameters for our neural network.
#
# But before we can start, we need to import the necessary packages tensorflow, numpy and matplotlib.
# + colab_type="code" id="CPuVp2lyNK2J" colab={}
import tensorflow as tf
import matplotlib.pyplot as plt
import numpy as np
# + [markdown] colab_type="text" id="J84v9uucMgv5"
# Next we define our constants and set the random seeds of tensorflow and numpy in order to get reproducable results.
# + colab_type="code" id="xwC-1OnHMgv7" colab={}
N_train_samples = 600
N_validation_samples = 100
N_test_samples = 100
N_samples = N_train_samples + N_validation_samples + N_test_samples
noise_sig = 0.1
N_epochs = 150
batch_size = 8
learning_rate = 0.01
tf.random.set_seed(0)
np.random.seed(0)
# + [markdown] colab_type="text" id="ngFFSyG-MgwA"
# We create $600$ training samples, $100$ validation samples to optimize our hyperparameters and $100$ test samples, which are used to check if our model can generalize to unseen data. Furthermore we set the level of noise added to the observations. For training the model we plan to train it for $150$ epochs with a batch size of $8$ and a learning rate of $0.01$. Next we create the actual input-output pairs $\mathbf{x},\mathbf{y}$ for which we want to learn the regression model and plot them. In this simple example we choose scalar inputs as well as output but in general $\mathbf{x}$ and $\mathbf{y}$ can be vectors.
# + colab_type="code" id="4s8AtsdQMgwB" outputId="8c2a68b3-e1a4-48b2-e460-934e59bf04d5" colab={"base_uri": "https://localhost:8080/", "height": 265}
x = np.linspace(0.0, 3.0, N_samples, dtype=np.float32)
y = np.expand_dims(np.sin(1.0+x*x) + noise_sig*np.random.randn(N_samples).astype(np.float32), axis=-1)
y_true = np.sin(1.0+x*x)
plt.plot(x, y)
plt.plot(x, y_true)
plt.legend(["Observation", "Ground truth"])
plt.show()
# + [markdown] colab_type="text" id="Y3wx8vJlMgwI"
# With the input-output pairs created, your first task is now to partition the data in the training, validation and test sets. Keep in mind that we have created the data in a structured way, i.e. the input-output pairs are ordered. This means you need to shuffle the data before partitioning it.
# + colab_type="code" id="kDIMUZs0MgwK" colab={}
""" Shuffle and partition the data set accordingly. you can use the predefined constants "N_train_samples", "N_validation_samples" and "N_test_samples". Use the variable names that are already in the below code
to store the final shuffled and partitioned data. Hint: Shuffle the data and the labels in such a way that the pairing between an image and it's label is preserved."""
# Shuffle the data
np.random.seed(0)
np.random.shuffle(x)
np.random.seed(0)
np.random.shuffle(y)
np.random.seed(0)
np.random.shuffle(y_true)
# Partition the data
x_train = x[:N_train_samples].reshape([-1,1])
y_train = y[:N_train_samples].reshape([-1,1])
x_test = x[N_train_samples:N_train_samples+N_test_samples].reshape([-1,1])
y_test = y[N_train_samples:N_train_samples+N_test_samples].reshape([-1,1])
x_validation = x[N_train_samples+N_test_samples:N_samples].reshape([-1,1])
y_validation = y[N_train_samples+N_test_samples:N_samples].reshape([-1,1])
# + [markdown] colab_type="text" id="-ucvKRhOMgwN"
# In order to feed the data to our model, we will use the Dataset class provided by Tensorflow. This class is simple to use and provides all the functionality we need for shuffling, batching and feeding the data to our model. It is also tightly integrated into the Tensorflow framework, which makes it very performant. Performance is not an aspect we need to worry about in this exercise, but it is important in more demanding applications.
#
# In this exercise we instantiate a separate Dataset object for the training, validation and test data sets, where we shuffle and repeat just the training data set. Shuffling the validation and test data sets is not necessary, since we only evaluate the loss on those data sets and do not perform SGD on it. Please fill in the missing part of the code.
# + colab_type="code" id="HifQ63iPMgwP" colab={}
""" Create three tensorflow Dataset objects that can be used to feed the training test and validation data to a neural network. Hint: For the training data set use shuffling, batching with the size according to
the predefined constant "batch_size" and repeat the data set indefinetly. For the validation and test data sets no shuffling or batching is needed."""
train_ds = tf.data.Dataset.from_tensor_slices((x_train,y_train)).shuffle(N_train_samples).batch(batch_size).repeat() #Repeat indefinitely
validation_ds = tf.data.Dataset.from_tensor_slices((x_validation,y_validation))
test_ds = tf.data.Dataset.from_tensor_slices((x_test,y_test))
# + id="FupUrcS6PGcT" colab_type="code" outputId="bdb45d21-cb06-4447-f793-130f1bbfe9b3" colab={"base_uri": "https://localhost:8080/", "height": 34}
train_ds
# + [markdown] colab_type="text" id="5Crr6fIkMgwT"
# In this exercise we will create a a simple neural network with two hidden layers containing $10$ neurons. For creating a model and keeping track of its weights a class called MyModel is used. When initializing an instance of this class the necessary variables are created and stored in a list called "trainable_variables". This makes it easy to get all trainable variables of the model. We also override the \__call__ method of this class in order to implement the forward pass of the neural network. This method should accept the inputs to the neural network and should return the result of the forward pass as an output. Please fill in the missing part of the code and select suitable activation functions for the different layers.
# + colab_type="code" id="Nq8ri416MgwX" colab={}
""" Implement a neural network with two hidden dense layers containing 10 neurons each. As an activation function use the tangens hyperbolicus (tf.nn.tanh()). Since we are not using Keras, we need to create and
manage all the variables that we need ourselves. The varaibles are created in the constructor of our model class. Since we want to be able to just call the class with some inputs in order to make a prediction,
we implement a __call__ method which computes the forward pass and returns the output of the network."""
class MyModel(object):
def __init__(self):
# Create model variables
input_dim = 1
hidden_dim = 10
output_dim = 1
self.W0 = tf.Variable(tf.random.normal([input_dim,hidden_dim]))
self.b0 = tf.Variable(tf.random.normal([hidden_dim]))
self.W1 = tf.Variable(tf.random.normal([hidden_dim,hidden_dim]))
self.b1 = tf.Variable(tf.random.normal([hidden_dim]))
self.W2 = tf.Variable(tf.random.normal([hidden_dim,output_dim]))
self.b2 = tf.Variable(tf.random.normal([output_dim]))
self.trainable_variables = [self.W0, self.b0, self.W1, self.b1, self.W2, self.b2]
def __call__(self, inputs):
# Compute forward pass
output = tf.reshape(inputs, [-1, 1])
output = tf.nn.tanh(tf.linalg.matmul(output,self.W0) + self.b0)
output = tf.nn.tanh(tf.linalg.matmul(output,self.W1) + self.b1)
output = tf.linalg.matmul(output,self.W2) + self.b2
return output
# + [markdown] colab_type="text" id="uf6m8zXoMgwb"
# Now after the model class is defined we can instantiate a MyModel object by running
# + colab_type="code" id="nSfI8wjLMgwb" colab={}
mdl = MyModel()
# + [markdown] colab_type="text" id="KraeH6MsMgwh"
# We can now use the model to make predictions by calling it. In the following we predict on the inputs an plot the result.
# + colab_type="code" id="K1at5RObMgwi" outputId="cdbb2c19-48d4-4b9f-8f06-65c01be5d27e" colab={"base_uri": "https://localhost:8080/", "height": 265}
""" We want to plot a prediction on the complete data set with a model before training. For this make a prediction on the variable "x". """
y_pred = mdl.__call__(x)
plt.plot(x, y,'b.')
plt.plot(x, y_true,'r.')
plt.plot(x, y_pred.numpy(),'g.')
plt.legend(["Observation", "Target", "Prediction"])
plt.show()
# + [markdown] colab_type="text" id="utFkv4F3NULQ"
# Since we have initialized the variables of the neural network randomly, it's prediction is also random. In order to fit the model we need to minimize the expected mean squared error over all input-ouput pairs in our training data set. For this we need to create a function, that performs a training step when provided with the model, an optimizer and a batch of input-ouput pairs.
# + colab_type="code" id="o4zFN0-kOdu7" colab={}
""" For training we need to implement a function that executes one training step. Fill in the missing code pieces for this function."""
def train_step(model, optimizer, x, y):
with tf.GradientTape() as tape:
tape.watch(model.trainable_variables)
y_pred = model.__call__(x) # Compute a prediction with "model" on the input "x"
loss_val = tf.reduce_mean((y - y_pred)**2) # Compute the Mean Squared Error (MSE) for the prediction "y_pred" and the targets "y"
grads = tape.gradient(loss_val, model.trainable_variables)
optimizer.apply_gradients(zip(grads, model.trainable_variables))
return loss_val
# + [markdown] colab_type="text" id="Ax-Kfd-tOm7I"
# This function uses the GradientTape to record the operations for which gradients have to be calculated. In our case this is the forward pass through our model and the computation of the loss function. After these operations are recoded we can get their gradients and apply these through the use of an optimizer. Finally we return the loss value in order to print it.
#
# With the training step function defined we now need to choose a suitable optimizer. Tensorflow offers a wide variety of optimizers but in this exercise we will use the RMSprop optimizer.
# + colab_type="code" id="PahsqMscPcKD" colab={}
opt = tf.optimizers.RMSprop(learning_rate)
# + [markdown] colab_type="text" id="8_WjHrAwQB9e"
# We now have everything we need to start training the model. For this we repeatedly sample a batch of input-output pairs from our training data set and use the train_step function to minimize the loss function over this batch. We repeat this until we have iterated over the complete training data set once. After this we compute the loss on the validation data set, print it and repeat with another epoch until we have reached $N\_epochs$ epochs.
# + colab_type="code" id="x3LqS3d_QrX6" outputId="7d7081b3-699b-4753-da95-206925c7b169" colab={"base_uri": "https://localhost:8080/", "height": 1000}
""" We can now use the train_step function to perform the training. Fill in the missing code parts."""
epoch = 0
train_iters = 0
train_loss = 0.0
for x_t, y_t in train_ds:
train_loss += train_step(mdl, opt, x_t, y_t) # Perform a training step with the model "mdl" and the optimizer "opt" on the inputs "x_t" and the corresponding targets "y_t"
train_iters += 1
if train_iters==(N_train_samples/batch_size): # An epoch is completed
validation_loss = 0.0
for x_v, y_v in validation_ds:
y_pred = mdl.__call__(x_v)# Compute a prediction with "mdl" on the input "x_v"
validation_loss += ((y_v-y_pred)**2)# Compute the Mean Squared Error (MSE) for the prediction "y_pred" and the targets "y_v"
#print("Epoch: {} Train loss: {:.5} Validation loss: {:.5}".format(epoch, train_loss/train_iters, validation_loss/N_validation_samples))
print("------ Epoch: ",epoch)
print("Train loss: ",(train_loss/train_iters).numpy())
print("Validation loss: ",(validation_loss/N_validation_samples).numpy())
train_iters = 0
train_loss = 0.0
epoch += 1
if (epoch == N_epochs):
break
# + [markdown] colab_type="text" id="DduaR_pCQ0k-"
# After completion of the training process we use the test data set to test the models generalization to unseen data.
# + colab_type="code" id="3ofClNnHRAUy" outputId="edb0356c-554b-4e04-d494-3e21d2cfe34b" colab={"base_uri": "https://localhost:8080/", "height": 34}
test_loss = 0
for x_t, y_t in test_ds:
y_pred = mdl.__call__(x_t) # Compute a prediction with "mdl" on the input "x_t"
test_loss += (y_t - y_pred)**2 # Compute the Mean Squared Error (MSE) for the prediction "y_pred" and the targets "y_t"
test_loss = test_loss.numpy()
print("Test loss: {:.5}".format(test_loss[0][0]/N_test_samples))
# + [markdown] colab_type="text" id="NzJ7wOZmRbez"
# After we have verified that our model achieves a similar loss on the test as on the validation and training data set, we can conclude that our model is not overfitting or underfitting and generalizes to unseen data. We can now predict on the inputs again and plot the results.
# + colab_type="code" id="tyvFN03bR8ez" outputId="e2578289-892e-417e-dc02-678331189192" colab={"base_uri": "https://localhost:8080/", "height": 265}
""" Now we want to plot the prediction after training. Predict on the variable "x" again. """
y_pred = mdl.__call__(x)# Compute a prediction on the variable "x"
plt.plot(x, y,'b.')
plt.plot(x, y_true,'r.')
plt.plot(x, y_pred.numpy(),'g.')
plt.legend(["Observation", "Target", "Prediction"])
plt.show()
# + [markdown] colab_type="text" id="WKmuAAmLSmiR"
# Now our model has learned to approximate the function mapping from the input to the output. The capability of neural networks to learn from input-ouput pairs alone and approximate an arbitrary function, see universal approximation theorem, can be very useful if the mapping between the input and output is too complex to be captured with model based approaches. But learning from input-ouput pairs alone implies that the model will only be able to make accurate predictions over input ranges it has seen during training. In order to demonstrate this we will predict on an interval that exeeds the $\left[0,3\right]$ interval the model was trained on, i.e. we will predict on the interval $\left[-2,5\right]$.
# + colab_type="code" id="ZRF8hAmmVFcH" outputId="9faa7db8-9155-4701-9bcc-d2086463f597" colab={"base_uri": "https://localhost:8080/", "height": 265}
x_generalize = np.linspace(-2.0, 5.0, N_samples, dtype=np.float32)
y_generalize = np.sin(1.0+x_generalize*x_generalize) + noise_sig*np.random.randn(N_samples).astype(np.float32)
y_truey_generalize = np.sin(1.0+x_generalize*x_generalize)
y_pred = mdl(x_generalize)
plt.plot(x_generalize, y_generalize)
plt.plot(x_generalize, y_truey_generalize)
plt.plot(x_generalize, y_pred.numpy())
plt.legend(["Observation", "Target", "Prediction"])
plt.show()
# + [markdown] colab_type="text" id="J2yr-0UCVIli"
# As expected, the model is able to make farely accurate predictions on the interval it was trained on but makes unreliable predictions outside this interval.
#
# ### Regularization
# In this section we will explore the concept of regularization. As there is no theorem that can be used to determine the required size and structure of a neural network given a certain task, one has to find a suitable neural architecture by trial and error. This can result in choosing a architecture with a capacity that is higher than required for solving the given task and hence overfitting might occur. A common way to prevent large neural networks networks from overfitting is to employ some sort of regularization, e.g. weight norm penalty, dropout, early stopping and data augmentation. In this section we will focus on weight norm penalty as a regularization and derive a probabilistic interpretation for some of those.
#
# We start by restating the conditional probability of the output $\mathbf{y}$ given the input $\mathbf{x}$ and the networks parameters $\boldsymbol{\theta}$
#
# $p(\mathbf{y}\vert\mathbf{x},\boldsymbol{\theta})=\dfrac{1}{\sqrt{(2\pi)^{M}\sigma^{2}}}\mathrm{e}^{-\dfrac{1}{2\sigma^{2}}\Vert\boldsymbol{\mathbf{y}-g_{\boldsymbol{\theta}}(\mathbf{x})}\Vert_{2}^{2}}$,
#
# which we used to derive the log likelihood. If we have some prior knowledge about the parameters of the neural network, which is given by a pdf $p(\boldsymbol{\theta})$ over the weights, we can use Bayes theorem to derive a posterior distribution
#
# $p(\boldsymbol{\theta}\vert\mathbf{x},\mathbf{y})=\dfrac{p(\boldsymbol{\theta},\mathbf{y}\vert\mathbf{x})}{p(\mathbf{y})}=\dfrac{p(\mathbf{y}\vert\mathbf{x},\boldsymbol{\theta})p(\boldsymbol{\theta})}{p(\mathbf{y})}$
#
# where $p(\boldsymbol{\theta})$ is the prior over the networks parameters. Similar to the derivation at the beginning of the exercise, we can use this posterior distribution to derive a cost function for training the neural network. In this case, however, we are not maximizing the log likelihood but the posterior distribution over the weights, hence this approach is called Maximum A Posteori (MAP) estimation of the parameters. Mathematically we can formulate this as
#
# $\boldsymbol{\theta}^{\star}=\arg\max_{\boldsymbol{\theta}}\mathbb{E}\left[p(\boldsymbol{\theta}\vert\mathbf{x},\mathbf{y})\right]=\arg\max_{\boldsymbol{\theta}}\mathbb{E}\left[\ln{p(\boldsymbol{\theta}\vert\mathbf{x},\mathbf{y})}\right]=\arg\max_{\boldsymbol{\theta}}\ln{p(\boldsymbol{\theta})}+\mathbb{E}\left[\ln{p(\mathbf{y}\vert\mathbf{x},\boldsymbol{\theta})}\right]$,
#
# where we used the fact that applying a strictly increasing function, e.g. $\ln{}$, does not change the position of the maximum of a cost function, ignored $p(\mathbf{y})$, since it is independent of the network parameters and dropped the expectation operator for $\ln{p(\boldsymbol{\theta})}$, since it is not depending on the random variable $\mathbf{x}$. Comparing the MAP estimate of the parameters with the ML estimate we derived above, shows that the only difference is the addition of $\ln{p(\boldsymbol{\theta})}$. This term is the regularization, i.e. the weight norm penalty. Depending on the distribution over $\boldsymbol{\theta}$ it can have different forms. If we choose a standard normal distribution, i.e. $\boldsymbol{\theta\sim\mathcal{N}(\mathbf{0},\mathbf{I})}$, we get
#
# $\boldsymbol{\theta}^{\star}=\arg\min_{\boldsymbol{\theta}}\lambda\Vert\boldsymbol{\theta}\Vert_{2}^{2}+\dfrac{1}{N_{D}}\sum_{i=1}^{N_{D}}\Vert\boldsymbol{\mathbf{y}_{i}-g_{\boldsymbol{\theta}}(\mathbf{x}_{i})}\Vert_{2}^{2}$,
#
# where we have made the same simplifications as for the ML estimation and also introduced the parameter $\lambda=\sigma^{2}$, which is used to control the strength of the regularization. This form of regularization is commonly known as $l_{2}$-norm or weight decay regularization. Choosing a prior where the weights follow an i.i.d laplacian distribution, i.e. $p(\boldsymbol{\theta})=\prod_{j}\dfrac{1}{2}\mathrm{e}^{\vert\theta_{j}\vert}$, leads to
#
# $\boldsymbol{\theta}^{\star}=\arg\min_{\boldsymbol{\theta}}\lambda\Vert\boldsymbol{\theta}\Vert_{1}+\dfrac{1}{N_{D}}\sum_{i=1}^{N_{D}}\Vert\boldsymbol{\mathbf{y}_{i}-g_{\boldsymbol{\theta}}(\mathbf{x}_{i})}\Vert_{2}^{2}$,
#
# where the strength of the regularization is again controlled by $\lambda=\sigma^{2}$. This type of regularization is known as $l_{1}$-norm and it has the property to induce sparsity in the parameters of the network.
#
# With this theoretical background on regularization we can now implement it and observe it's effects on the regression problem covered in this exercise. For this we will define a model with a high capacity and train it for a extended time to provoke overfitting. For this, we will increase the number of hidden neurons in both hidden layers to $100$ and $50$.
# + colab_type="code" id="T2Fv2J-VK_vw" colab={}
""" Implement a bigger model with again two hidden layers contatining 100 and 50 neurons. As an activation use the tangens hyperbolicus function where it is appropiate. """
class MyBigModel(object):
def __init__(self):
input_dim = 1
hidden_dim1 = 100
hidden_dim2 = 50
output_dim = 1
# Create model variables
self.W0 = tf.Variable(tf.random.normal([input_dim,hidden_dim1]))
self.b0 = tf.Variable(tf.random.normal([hidden_dim1]))
self.W1 = tf.Variable(tf.random.normal([hidden_dim1,hidden_dim2]))
self.b1 = tf.Variable(tf.random.normal([hidden_dim2]))
self.W2 = tf.Variable(tf.random.normal([hidden_dim2,output_dim]))
self.b2 = tf.Variable(tf.random.normal([output_dim]))
self.trainable_variables = [self.W0, self.b0, self.W1, self.b1, self.W2, self.b2]
def __call__(self, inputs):
# Compute forward pass
output = tf.reshape(inputs, [-1, 1])
output = tf.nn.tanh(tf.linalg.matmul(output,self.W0) + self.b0)
output = tf.nn.tanh(tf.linalg.matmul(output,self.W1) + self.b1)
output = tf.linalg.matmul(output,self.W2) +self.b2
return output
# + [markdown] colab_type="text" id="Y1XpaZTNLV-p"
# After creating one instance of this class we can again train it on our data set. We will also create a new optimizer for training this bigger model, since some optimizers adapt the learning rates for individual parameters during a training process and we do not want to train our bigger model with learning rates adopted from an earlier training run.
# + colab_type="code" id="b7Yq-a1FLi0X" colab={}
big_mdl = MyBigModel()
big_opt = tf.optimizers.SGD(learning_rate)
# + [markdown] colab_type="text" id="dBrf0I68MnMy"
# Now we are ready to train this bigger model using the same training step and training loop. In order to provoke overfitting we also reduce the number of samples in the training data set a lot, increase the batch size and train for a more epochs.
# + colab_type="code" id="Nz_lXM8LMwXo" outputId="1ffcc30c-4f74-40b0-cd95-b94ecc94b397" colab={"base_uri": "https://localhost:8080/", "height": 1000}
""" Implement the training for the bigger model similar to the training of the small model before. """
N_train_samples_overfit = 30
N_epochs = 1000
batch_size = 30
sel_idx = np.arange(0, N_train_samples)
sel_idx = np.random.choice(sel_idx, N_train_samples_overfit)
x_train_overfit = x_train[sel_idx]
y_train_overfit = y_train[sel_idx]
train_overfit_ds = tf.data.Dataset.from_tensor_slices((x_train_overfit, y_train_overfit)).shuffle(N_train_samples_overfit).batch(batch_size).repeat()
epoch = 0
train_iters = 0
train_loss = 0.0
for x_t, y_t in train_overfit_ds:
train_loss += train_step(big_mdl,big_opt,x_t,y_t) # Perform a training step with the model "big_mdl" and the optimizer "big_opt" on the inputs "x_t" and the corresponding targets "y_t"
train_iters += 1
if train_iters == N_train_samples_overfit/batch_size: # An epoch is completed
for x_v, y_v in validation_ds:
y_pred = big_mdl.__call__(x_v) # Compute a prediction with "big_mdl" on the input "x_v"
validation_loss += (y_v - y_pred)**2 # Compute the Mean Squared Error (MSE) for the prediction "y_pred" and the targets "y_v"
print("------ Epoch: ",epoch)
print("Train loss: ",(train_loss/train_iters).numpy())
print("Validation loss: ",(validation_loss/N_validation_samples).numpy())
train_iters = 0
train_loss = 0.0
validation_loss = 0
epoch += 1
if (epoch == N_epochs):
break
# + [markdown] colab_type="text" id="gkaUDmA8Ps6w"
# Predicting with this model shows overfitting. For recognizing overfitting a comparison of the validation and training loss is very useful. If the training loss decreases during training while the validation loss consistently increases, the model you are training is probably overfitting. Plotting the models prediction and the target also shows that there is a significant discrepancy between the target and the prediction of the model.
# + colab_type="code" id="3Sq-9xhWPxI4" outputId="f6e78562-420d-4ab4-df11-c16990ef17b2" colab={"base_uri": "https://localhost:8080/", "height": 265}
y_pred =big_mdl.__call__(x) # Predict on x with "big_mdl"
plt.scatter(x_train_overfit, y_train_overfit)
plt.plot(x, y_true,'r.')
plt.plot(x, y_pred.numpy(),'g.')
plt.legend(["Target", "Prediction", "Training samples"])
plt.show()
# + [markdown] colab_type="text" id="lZ36J2EN85b9"
# In order to implement a regularization we need to modify the loss function. Since the loss function in this exercise is computed during the training step, we define a new training step with a regularization.
# + colab_type="code" id="sLbcWwlt9Jwl" colab={}
""" In order to avoid overfitting we implement a training step that also includes a regularization on the weights of our big model. For this we use the Frobenius/squared l2-norm of each weight matrix/vector.
Hint: Use the tf.reduce_sum() function on a list of individual regularization terms for each matrix/vector of the network."""
def regularized_train_step(model, optimizer, x, y, lmbd):
with tf.GradientTape() as tape:
tape.watch(model.trainable_variables)
y_pred = model.__call__(x) # Compute a prediction with "model" on the input "x"
loss_val = tf.reduce_mean((y-y_pred)**2) # Compute the Mean Squared Error (MSE) for the prediction "y_pred" and the targets "y"
regul_val = tf.reduce_sum([tf.norm(param) for param in model.trainable_variables])# Compute the regularization based on the list "model.trainable_variables"
total_loss = loss_val + lmbd * regul_val # Add the loss with a the regularization term weighted by "lmbd"
grads = tape.gradient(total_loss, model.trainable_variables)
optimizer.apply_gradients(zip(grads, model.trainable_variables))
return loss_val
# + [markdown] colab_type="text" id="ExK4FIw89s0M"
# We can now set the strength of the regularization and retrain the big model with a regularization. We create another instance of the big model in order to compare the big model with and without regularization.
# + colab_type="code" id="_PUUBGi496Vt" outputId="6354b156-4bba-4d27-fb04-8cfead1fda04" colab={"base_uri": "https://localhost:8080/", "height": 1000}
""" Implement the training for the bigger model with the regularized_train_step function. Note: We are plotting the MSE loss without the regularization in order to compare it with the unregularized model. """
lmbd = 0.005
big_reg_mdl = MyBigModel()
big_opt = tf.optimizers.SGD(learning_rate)
epoch = 0
train_iters = 0
train_loss = 0.0
for x_t, y_t in train_overfit_ds:
train_loss += regularized_train_step(big_reg_mdl, big_opt, x_t, y_t, lmbd) # Perform a regularized training step with the model "big_mdl" and the optimizer "big_opt" on the inputs "x_t" and the corresponding targets "y_t" with the regularization parameter being "lmbd"
train_iters += 1
if train_iters == N_train_samples_overfit/batch_size: # An epoch is completed
for x_v, y_v in validation_ds:
y_pred = big_reg_mdl.__call__(x_v)# Compute a prediction with "big_mdl" on the input "x_v"
validation_loss += (y_v- y_pred)**2 # Compute the Mean Squared Error (MSE) for the prediction "y_pred" and the targets "y_v"
print("------ Epoch: ",epoch)
print("Train loss: ",(train_loss/train_iters).numpy())
print("Validation loss: ",(validation_loss/N_validation_samples).numpy())
train_iters = 0
train_loss = 0.0
train_reg = 0.0
validation_loss = 0
epoch += 1
if (epoch == N_epochs):
break
# + [markdown] colab_type="text" id="ylr1C2ovWXbO"
# During the training of the regularized model we can already notice, that, although there is still a difference between training and validation loss, the validation loss decreases as the training loss dreases. The effect of the regularization becomes even more evident if we plot the predictions of the regularized model and the overfitting model.
# + colab_type="code" id="UdMRV0vAWLmm" outputId="e100c956-4d43-42a2-a391-b627da9f37af" colab={"base_uri": "https://localhost:8080/", "height": 265}
""" We now want to plot the prediction of the regularized and unregularized big model. """
y_pred = big_reg_mdl.__call__(x) # Predict with "big_reg_mdl" on "x"
y_pred_overfit = big_mdl.__call__(x)# Predict with "reg_mdl" on "x"
plt.scatter(x_train_overfit, y_train_overfit)
plt.plot(x, y_true,'r.')
plt.plot(x, y_pred.numpy(),'g.')
plt.plot(x, y_pred_overfit.numpy(),'b.')
plt.legend(["Target", "Regularization", "No regularization", "Training samples"])
plt.show()
# + [markdown] colab_type="text" id="sePQBaGqZzT6"
# The model with regularization seems to follow the overall trend of the data, while the model without any regularization very precisely fits the training samples. This is espacilly evident in the interval $\left[0,0.5\right]$, where the prediction of the unregularized model shows an oscillating behavior. Such oscillations are however not present in the ground truth and therefore undesirable. The regularized model on the other hand is not as flexible as the unregularized model and therefore does not fit the target function well in the interval $\left[2.25, 3.0\right]$.
#
# ## Conclusion
# In this exercise we revisited the mathematical background for a simple regression task and covered it's practical implementation in Tensorflow 2. We also explored the phenomenon of overfitting and derived different regularizations from a probabilistic perspective. This exercise covers a very simple task with a very basic neural architecture and is intended as a primer for the second part of the regression exercise, which is dealing with a bigger and more realistic problem. In this second part we will consider the problem of estimating the age of a person from a potrait picture.
| 77.277056
| 1,024
|
1ac4f24bfe4a1fa1b471743067b896a1bd090e8c
|
py
|
python
|
Code/ICCICT_Demo.ipynb
|
Infernolia/ICCICT
|
['Apache-2.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + id="8GlaCwmIUMOK"
import pandas as pd
import numpy as np
# + id="djelMo2qUMOQ"
word =pd.read_csv("finalwords.csv")
# + id="4jDfyoNBUMOQ"
word.drop(columns = ["Unnamed: 0"],inplace=True)
# + id="37rlbTjdUMOR"
word.columns = ["words"]
# + colab={"base_uri": "https://localhost:8080/", "height": 402} id="KOjwi0HDUMOR" outputId="bd833e0a-7b34-447c-ac10-fc85a654bab6"
word
# + id="kzErrUSqUMOS"
words = word["words"].unique()
# + id="IT4DS4YdUMOT" colab={"base_uri": "https://localhost:8080/"} outputId="dbe80607-167a-4d3a-8c90-45044ea26a40"
words
# + id="pP3N0e6vUMOT"
data= pd.read_csv("clean_df.csv")
# + id="LbmxnbahUMOT" colab={"base_uri": "https://localhost:8080/", "height": 303} outputId="dea61154-8939-43a3-c956-ca032f45553f"
data.drop(columns = ["Unnamed: 0","lang","created_at","time"],inplace=True)
# + colab={"base_uri": "https://localhost:8080/", "height": 402} id="YLv7Pke9UMOU" outputId="e5709c39-8e30-4607-a397-4bb4fb964031"
data
# + id="HLTviJDTUMOU"
data["topics"]=" "
# + colab={"base_uri": "https://localhost:8080/"} id="SMoA--r8UMOU" outputId="0c0dfc69-1e5d-432f-cfa7-8a6c559f5885"
import nltk.data
from nltk.tokenize import word_tokenize
nltk.download('punkt')
# + id="ekRPPDTdUMOV"
for i in data.index:
stri = data.at[i,"text"]
l = word_tokenize(stri)
for a in l:
if a in words:
data.at[i,"topics"] = data.at[i,"topics"] +"," + a
# + colab={"base_uri": "https://localhost:8080/", "height": 402} id="8UlMSh2FUMOV" outputId="b570d57a-a0d0-4e06-c180-7af8fb420b45"
data
# + id="s7iMiG24UMOV"
for i in data.index:
data.at[i,"topics"] = data.at[i,"topics"][2:]
# + colab={"base_uri": "https://localhost:8080/", "height": 402} id="nB-uot-jUMOV" outputId="ddfc19d0-5c12-45ce-89c6-87d407a63fce"
data
# + id="9iQ-CEiqUMOW"
alltopics = ""
for i in data.index:
alltopics = alltopics + str(data.at[i,"topics"])
# + id="9KAs41x1UMOW"
all_topics = alltopics.split(",")
# + id="s3JDgTz4UMOW"
all_topics
# + colab={"base_uri": "https://localhost:8080/"} id="MkrzsusHUMOW" outputId="70a6423d-b414-4c51-c4b6-d59a6f5e4d56"
import collections
counts_no_urls = collections.Counter(all_topics)
counts_no_urls.most_common(15)
len(counts_no_urls)
# + id="0rwyJ4F4UMOX"
clean_tweets_no_urls = pd.DataFrame(counts_no_urls.most_common(10),
columns=['words', 'count'])
# + colab={"base_uri": "https://localhost:8080/", "height": 195} id="mvkWRDKaUMOX" outputId="fe64def1-b636-455e-98f5-2385deb08f0b"
clean_tweets_no_urls.head()
# + id="Egu5IP5jbBxP"
clean_tweets_no_urls.to_csv("a.csv")
# + id="V9LYpTkCUMOX"
term_freq_df = clean_tweets_no_urls
# + id="kYeH0vYfUMOX"
term_freq_df.columns = ["words","total"]
# + colab={"base_uri": "https://localhost:8080/", "height": 34} id="AnYdEBLHUMOY" outputId="a5128118-94d4-44a0-ff47-e1460bc013cc"
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
y_pos = np.arange(500)
plt.figure(figsize=(10,8))
s = 1
term_freq_df.drop(columns=["words"],inplace=True)
plt.show()
# + id="zEqLLsfdUMOZ"
#india = data[data["country_code"]=="IN"]
# + id="Nt0zcrzlUMOZ"
india = data
# + colab={"base_uri": "https://localhost:8080/"} id="tN8O_gyWUMOa" outputId="2bca806e-c11f-4ad4-953f-cea51d7ae759"
india.info()
# + colab={"base_uri": "https://localhost:8080/", "height": 402} id="F1ucFPLaCOJ-" outputId="baa76df6-531e-4786-e8a6-a83d63adacee"
india
# + id="4qZ5AVx3CQJK"
india["date"] = ""
# + id="OVHL930NUMOa"
from datetime import datetime
for i in india.index:
india.at[i,"date"] = str(india.at[i,"created_at"]).split(' ')[0]
# + id="QQ7ADVl0DAsM"
india = india.drop(columns=["created_at"])
# + colab={"base_uri": "https://localhost:8080/", "height": 402} id="TMzqpkVbDGRC" outputId="481d3500-2246-42a0-eb97-a518b0acc7c3"
india
# + id="kHL52olqDTxW"
india["date"] = india["date"].astype('datetime64[ns]')
# + id="nolr0tyuDTrc"
# + id="cKbHM1W0UMOa"
la1 = '2020-03-25'
la2 = '2020-04-14'
lb1 = '2020-04-15'
lb2 = '2020-05-03'
lc1 = '2020-05-04'
lc2 = '2020-05-17'
ld1 = '2020-05-18'
ld2 = '2020-05-31'
a1 = datetime.strptime(la1, '%Y-%m-%d')
a2 = datetime.strptime(la2, '%Y-%m-%d')
b1 = datetime.strptime(lb1, '%Y-%m-%d')
b2 = datetime.strptime(lb2, '%Y-%m-%d')
c1 = datetime.strptime(lc1, '%Y-%m-%d')
c2 = datetime.strptime(lc2, '%Y-%m-%d')
d1 = datetime.strptime(ld1, '%Y-%m-%d')
d2 = datetime.strptime(ld2, '%Y-%m-%d')
# + id="wwgZdLgmUMOa"
indialockdown1 = india[(india.date>=a1) & (india.date<=a2)]
indialockdown2 = india[(india.date>=b1) & (india.date<=b2)]
indialockdown3 = india[(india.date>=c1) & (india.date<=c2)]
indialockdown4 = india[(india.date>=d1) & (india.date<=d2)]
# + colab={"base_uri": "https://localhost:8080/"} id="GPRTaQHGdUTh" outputId="9292fbf2-7da2-48cc-ca68-2e44b94ce98d"
from textblob import TextBlob
def get_sentiment(text):
blob = TextBlob(text)
return [blob.sentiment.polarity, blob.sentiment.subjectivity]
indialockdown1.reset_index(inplace = True)
polarity,subjectivity = [], []
for i in range(len(indialockdown1)):
pol , sub = get_sentiment(indialockdown1['text'][i])
polarity.append(pol)
subjectivity.append(sub)
indialockdown1['polarity'] = polarity
indialockdown1['subjectivity'] = subjectivity
indialockdown2.reset_index(inplace = True)
polarity,subjectivity = [], []
for i in range(len(indialockdown2)):
pol , sub = get_sentiment(indialockdown2['text'][i])
polarity.append(pol)
subjectivity.append(sub)
indialockdown2['polarity'] = polarity
indialockdown2['subjectivity'] = subjectivity
indialockdown3.reset_index(inplace = True)
polarity,subjectivity = [], []
for i in range(len(indialockdown3)):
pol , sub = get_sentiment(indialockdown3['text'][i])
polarity.append(pol)
subjectivity.append(sub)
indialockdown3['polarity'] = polarity
indialockdown3['subjectivity'] = subjectivity
indialockdown4.reset_index(inplace = True)
polarity,subjectivity = [], []
for i in range(len(indialockdown4)):
pol , sub = get_sentiment(indialockdown4['text'][i])
polarity.append(pol)
subjectivity.append(sub)
indialockdown4['polarity'] = polarity
indialockdown4['subjectivity'] = subjectivity
# + colab={"base_uri": "https://localhost:8080/", "height": 195} id="C-nxGzRvdUPw" outputId="dc24b108-20a3-4f96-cf07-b727b3b7c447"
indialockdown1.head()
# + id="9yb5KnYIdUMN"
poscount1 = 0
negcount1 = 0
neutcount1 = 0
poscount2 = 0
negcount2 = 0
neutcount2 = 0
poscount3 = 0
negcount3 = 0
neutcount3 = 0
poscount4 = 0
negcount4 = 0
neutcount4 = 0
for i in indialockdown1.index:
if (indialockdown1.at[i,"polarity"]>0):
poscount1 = poscount1 + 1
elif (indialockdown1.at[i,"polarity"]==0):
neutcount1 = neutcount1 + 1
else:
negcount1 = negcount1 + 1
for i in indialockdown2.index:
if (indialockdown2.at[i,"polarity"]>0):
poscount2 = poscount2 + 1
elif (indialockdown2.at[i,"polarity"]==0):
neutcount2 = neutcount2 + 1
else:
negcount2 = negcount2 + 1
for i in indialockdown3.index:
if (indialockdown3.at[i,"polarity"]>0):
poscount3 = poscount3 + 1
elif (indialockdown3.at[i,"polarity"]==0):
neutcount3 = neutcount3 + 1
else:
negcount3 = negcount3 + 1
for i in indialockdown4.index:
if(indialockdown4.at[i,"polarity"]>0):
poscount4 = poscount4 + 1
elif (indialockdown4.at[i,"polarity"]==0):
neutcount4 = neutcount4 + 1
else:
negcount4 = negcount4 + 1
# + id="ghfYptVhdUH0"
draw = pd.DataFrame(columns=["Lockdown","Positive","Negative","Neutral"])
draw = draw.append({'Lockdown': "Lockdown 1", 'Positive': poscount1, 'Negative': negcount1, 'Neutral': neutcount1}, ignore_index=True)
draw = draw.append({'Lockdown': "Lockdown 2",'Positive': poscount2, 'Negative': negcount2, 'Neutral': neutcount2}, ignore_index=True)
draw = draw.append({'Lockdown': "Lockdown 3",'Positive': poscount3, 'Negative': negcount3, 'Neutral': neutcount3}, ignore_index=True)
draw = draw.append({'Lockdown': "Lockdown 4",'Positive': poscount4, 'Negative': negcount4, 'Neutral': neutcount4}, ignore_index=True)
# + id="ab3rivyunMBO"
# + colab={"base_uri": "https://localhost:8080/", "height": 166} id="KFaE-ZQRfMV-" outputId="50397b92-7ebb-4106-ba5f-2138bae02485"
draw
# + colab={"base_uri": "https://localhost:8080/"} id="fO9sYF7rmBw-" outputId="5c689835-c079-46f7-b2b4-b0967bc8f220"
draw.sum(axis=0)
# + colab={"base_uri": "https://localhost:8080/"} id="EAG0YYM3jIrR" outputId="559412e2-6491-4d44-d9d1-ad0da42fa2e0"
indialockdown1['sentiment'] = ""
for i in indialockdown1.index:
if (indialockdown1.at[i,"polarity"]>0):
indialockdown1.at[i,'sentiment']= 'Positive'
elif (indialockdown1.at[i,"polarity"]==0):
indialockdown1.at[i,'sentiment']= 'Neutral'
else:
indialockdown1.at[i,'sentiment']= 'Negative'
# + colab={"base_uri": "https://localhost:8080/"} id="OYLqSNaqocl8" outputId="9af60596-41fc-42e9-9550-40f93948cb49"
indialockdown2['sentiment'] = ""
for i in indialockdown2.index:
if (indialockdown2.at[i,"polarity"]>0):
indialockdown2.at[i,'sentiment']= 'Positive'
elif (indialockdown2.at[i,"polarity"]==0):
indialockdown2.at[i,'sentiment']= 'Neutral'
else:
indialockdown2.at[i,'sentiment']= 'Negative'
# + colab={"base_uri": "https://localhost:8080/"} id="e9MHu6t-EDGc" outputId="f7583413-9b73-4624-af3b-99cecbdc7b72"
indialockdown3['sentiment'] = ""
for i in indialockdown3.index:
if (indialockdown3.at[i,"polarity"]>0):
indialockdown3.at[i,'sentiment']= 'Positive'
elif (indialockdown3.at[i,"polarity"]==0):
indialockdown3.at[i,'sentiment']= 'Neutral'
else:
indialockdown3.at[i,'sentiment']= 'Negative'
# + colab={"base_uri": "https://localhost:8080/"} id="Tdz1ZnkeEDCm" outputId="1e388b37-dcc9-42f5-812e-346ed67a7f4b"
indialockdown4['sentiment'] = ""
for i in indialockdown4.index:
if (indialockdown4.at[i,"polarity"]>0):
indialockdown4.at[i,'sentiment']= 'Positive'
elif (indialockdown4.at[i,"polarity"]==0):
indialockdown4.at[i,'sentiment']= 'Neutral'
else:
indialockdown4.at[i,'sentiment']= 'Negative'
# + id="KX8BHVYYjoDJ"
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.feature_extraction.text import CountVectorizer
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
from keras.models import Sequential
from keras.layers import Dense, Embedding, LSTM, SpatialDropout1D
from sklearn.model_selection import train_test_split
from keras.utils.np_utils import to_categorical
import re
# + id="cU9RDuCsEdGF"
# + id="r7pDfCBEsVf5"
df1 = indialockdown1[['text','sentiment']]
df1.columns = ['statement','sentiment']
df2 = indialockdown2[['text','sentiment']]
df2.columns = ['statement','sentiment']
df3 = indialockdown3[['text','sentiment']]
df3.columns = ['statement','sentiment']
df4 = indialockdown4[['text','sentiment']]
df4.columns = ['statement','sentiment']
# + colab={"base_uri": "https://localhost:8080/", "height": 402} id="P0GBUKiLEW8e" outputId="d2e551fb-7a39-4f42-b95a-15dad5db9615"
df = df1.append(df2)
df = df.append(df3)
df = df.append(df4)
df
# + id="yf0QnmkJsVcx"
train,eva = train_test_split(df,test_size = 0.2)
# + id="QYvrO51RsVZM"
# !pip install simpletransformers
# + id="qLglACtJt7YC"
# !pip install simpletransformers[classification]
# + colab={"base_uri": "https://localhost:8080/"} id="MFZcBXpGsVVh" outputId="70395762-53ed-40b0-a0db-0d49dfc5bc70"
from simpletransformers.classification import ClassificationModel
# Create a TransformerModel
model = ClassificationModel('bert', 'bert-base-cased', num_labels=3, args={'reprocess_input_data': True, 'overwrite_output_dir': True},use_cuda=False)
# + colab={"base_uri": "https://localhost:8080/"} id="0E6X4GLz1HXX" outputId="793c5612-3328-49c4-f896-ba477de03738"
# Create a TransformerModel
model = ClassificationModel('roberta', 'roberta-base', num_labels=3, args={'reprocess_input_data': True, 'overwrite_output_dir': True},use_cuda=False)
# + colab={"base_uri": "https://localhost:8080/"} id="TTABGAlnsVR_" outputId="db2375c7-c29a-464f-bc83-b2c38e3051a3"
def making_label(st):
if(st=='Positive'):
return 0
elif(st=='Neutral'):
return 2
else:
return 1
train['label'] = train['sentiment'].apply(making_label)
eva['label'] = eva['sentiment'].apply(making_label)
print(train.shape)
# + id="Gi7WQ4MhuFBk"
train_df = pd.DataFrame({
'text': train['statement'][:1500].replace(r'\n', ' ', regex=True),
'label': train['label'][:1500]
})
eval_df = pd.DataFrame({
'text': eva['statement'][-400:].replace(r'\n', ' ', regex=True),
'label': eva['label'][-400:]
})
# + colab={"base_uri": "https://localhost:8080/", "height": 183, "referenced_widgets": ["0cc18213a0894f709f7e4c762b50056a", "d6fcea00de0a484e8f85fd09366a522f", "19cf8527e6d44da89a195b7c98098254", "0d9f23ea6c6d4d2887e9adf20a632cd2", "8c9a44c4b36f44f282cdabd1e50a4a2d", "3bc4cdea46d34e38b5df82fd23e9cd7c", "172e3c465c654de0ba9e0cb323f7e252", "94455e0359f04fa9a9e75d780d57ca1e", "32cad1016109465dae79f9b8cae4511c", "6b971b818458486983e32fa8a5464fb1", "b8d9e12c31ab46a694694de93acf740f", "1cef315fb07f49ab9c9e7d3c6f31f901", "d3c5885febc5448e8bc423c79a78ceb3", "578b0994e762482bbfbdae6a04c5440d", "619e799898b44e5890b4c472f09fc700", "fefa4cd46f2243f1b30516875d75428f", "70bc569b6fa446a4ac7ab4b4e0fbf8a2", "56b8787bf54e497e8d286e750244c59f", "cc10dcb43b8d408a9ac8d22e88abe2fe", "36bfc1ad9ab94082b3775ee479add205", "614e7a454d154faf8ca4b7e5a3099e81", "9592a8ad4c3d42bebe4e130738dfb191", "2a67d2e780bc4633b56a87416d505896", "59bf76d4b31845d5af8745c42ac8f6c3", "9ea69c26bb1649a59416672180e127eb", "630d0ff925e74419bf65674ef0e4563a", "cd89ea5d97954a8a87cc706651b3f53a", "2bc5b2ccdab74d0ca471fb05fca95948", "7dddaf0288dc4d988b0655e53e3d3c17", "f0d07f9b5fd145a0befd3f6a0b7b9ec4", "252c81acb63a46b29205b7e6d68ff6d7", "698b1610640448c7b49659dc848da79b", "7eebf9c2250e4b24b9a5357a404a4d36"]} id="MQ6dJFZluE9W" outputId="238c606e-4b0c-41ba-acff-6ddcea0e56d9"
model.train_model(train_df)
# + colab={"base_uri": "https://localhost:8080/", "height": 134, "referenced_widgets": ["4d5ca4d56d7a4e4097763ef7db427cdf", "daaa3afa5a634a90bc15854267b92bbd", "5dd591ded25e43f9b7e527c795cdfa32", "b56f930d46c647edb82271b788fd95bd", "016e5bad514f47f282f1eaf91a90df6a", "a86ec260faa84f78bb53d5d24ba09be4", "3881bd069bf8435e81fe1ad606e5062f", "99c5fc821de444faa41f769fea3a175f", "ea562dfee7f54518b04fcdbc2c8ca86f", "423fd6c42e7e419d9c74d66c8f351953", "e7eb2828b9794467bb04191f2bf9179d", "3e4e08dd29664c2b85b3296e4e7d86ee", "2b43edba03de4947a9e30c613610f587", "de18dbe1d6e44418993703ee2f55fb00", "f0af658c51f749a5af523758d6a96ced", "54cf6cc3cb7f425389a6e55a282cab70", "0e92dbbec7804c0b91906343b5721e06", "b2b81b0b2b094aaeb2f4398ae6c27999", "09bcb77d875e4c2b8bdc571e0ae4c314", "4bd4120325a04eb8b4d3308fd096cbb6", "24e280f717db4a1d8d60e0e194a3a39b", "c71b2ae59e9146748a6be9c8d5951b2b"]} id="NW4MAP-EuE52" outputId="3bdfc0f9-d8f2-4263-b6c2-251f608a26b6"
result, model_outputs, wrong_predictions = model.eval_model(eval_df)
# + colab={"base_uri": "https://localhost:8080/"} id="vrEjeA4fomOU" outputId="e253f4d3-76cf-4fbd-a81e-df4c7e097e2d"
result
# + colab={"base_uri": "https://localhost:8080/"} id="XpXVxS88vVhe" outputId="6ef8f1d7-a512-41d8-f4c0-80d6461687d4"
model_outputs
# + id="lV6gVYearRkk"
# + id="MZfi53ocvVeV"
lst = []
for arr in model_outputs:
lst.append(np.argmax(arr))
# + id="TK1GM-ScvVbS"
true = eval_df['label'].tolist()
predicted = lst
# + colab={"base_uri": "https://localhost:8080/"} id="Lg076SygvVZM" outputId="9458e407-07aa-4a85-cdef-58f24674b3dd"
import sklearn
mat = sklearn.metrics.confusion_matrix(true , predicted)
mat
# + colab={"base_uri": "https://localhost:8080/", "height": 265} id="N0Q2KO8ZvVW2" outputId="6c87573f-5de4-407f-b2bd-41115ae8fb7e"
df_cm = pd.DataFrame(mat, range(3), range(3))
sns.heatmap(df_cm, annot=True)
plt.show()
# + colab={"base_uri": "https://localhost:8080/"} id="LjZrQlfQvVS-" outputId="0f878f9e-8603-4009-dc96-6995810ada03"
print(sklearn.metrics.classification_report(true,predicted,target_names=['positive','neutral','negative']))
# + colab={"base_uri": "https://localhost:8080/"} id="hTJhW1EEvVP7" outputId="c878192a-4492-4c16-e038-b5d1350dd049"
sklearn.metrics.accuracy_score(true,predicted)
# + id="3p0i0dUGw01a"
# + id="b8077NjvDkKj" colab={"base_uri": "https://localhost:8080/", "height": 195} outputId="c1aac8f6-12e9-435b-a503-99a683796432"
indialockdown1.head()
# + id="Bgrh-XUPBNPh"
indialockdown1.to_csv("a1.csv")
indialockdown2.to_csv("a2.csv")
# + id="ynTN4mtq27x2" colab={"base_uri": "https://localhost:8080/", "height": 555} outputId="f81c4992-67db-44c7-8818-19690c6cd8f0"
poscount1 = 0
negcount1 = 0
neutcount1 = 0
poscount2 = 0
negcount2 = 0
neutcount2 = 0
poscount3 = 0
negcount3 = 0
neutcount3 = 0
poscount4 = 0
negcount4 = 0
neutcount4 = 0
for i in indialockdown1.index:
if (indialockdown1.at[i,"pred"]=="positive"):
poscount1 = poscount1 + 1
elif (indialockdown1.at[i,"pred"]=="neutral"):
neutcount1 = neutcount1 + 1
else:
negcount1 = negcount1 + 1
for i in indialockdown2.index:
if (indialockdown2.at[i,"pred"]=="positive"):
poscount2 = poscount2 + 1
elif (indialockdown2.at[i,"pred"]=="neutral"):
neutcount2 = neutcount2 + 1
else:
negcount2 = negcount2 + 1
for i in indialockdown3.index:
if (indialockdown3.at[i,"pred"]=="positive"):
poscount3 = poscount3 + 1
elif (indialockdown3.at[i,"pred"]=="neutral"):
neutcount3 = neutcount3 + 1
else:
negcount3 = negcount3 + 1
for i in indialockdown4.index:
if (indialockdown4.at[i,"pred"]=="positive"):
poscount4 = poscount4 + 1
elif (indialockdown4.at[i,"pred"]=="neutral"):
neutcount4 = neutcount4 + 1
else:
negcount4 = negcount4 + 1
# + id="ttQIfxqd3Tx3"
x = len(indialockdown1)
y = len(indialockdown2)
poscount1 = (poscount1/x) *100
negcount1 = (negcount1/x) *100
neutcount1 = (neutcount1/x) *100
poscount2= (poscount2/y) *100
negcount2= (negcount2/y) *100
neutcount2= (neutcount2/y) *100
# + id="nB7nhV2rv5_e"
indialockdown1.head()
# + id="HL0eoAMEv9Dy"
indialockdown2.head()
# + id="Y7QIOwuwwLai"
draw = pd.DataFrame(columns=["Lockdown","Positive","Negative","Neutral"])
draw = draw.append({'Lockdown': "Lockdown 1", 'Positive': poscount1, 'Negative': negcount1, 'Neutral': neutcount1}, ignore_index=True)
draw = draw.append({'Lockdown': "Lockdown 2",'Positive': poscount2, 'Negative': negcount2, 'Neutral': neutcount2}, ignore_index=True)
draw.head()
# + colab={"base_uri": "https://localhost:8080/", "height": 309} id="be44oPp5fMMd" outputId="93b88496-17b7-4cc8-fea9-4e619297e662"
draw.plot(kind="bar")
plt.title("Sentiment Analysis")
plt.xlabel("Lockdown")
plt.ylabel("Number of Tweets")
# + colab={"base_uri": "https://localhost:8080/", "height": 499} id="JXADmEmbUMOa" outputId="c52fce34-f4fa-4c6d-f3bd-f77664331d5e"
lockdown1 = ""
for i in indialockdown1.index:
lockdown1 = lockdown1 + str(indialockdown1.at[i,"topics"])
all_topicslockdown1 = lockdown1.split(",")
counts_no_urls = collections.Counter(all_topicslockdown1)
counts_no_urls.most_common(15)
len(counts_no_urls)
clean_tweets_no_urls = pd.DataFrame(counts_no_urls.most_common(10),
columns=['words', 'count'])
fig, ax = plt.subplots(figsize=(8, 8))
# Plot horizontal bar graph
clean_tweets_no_urls.sort_values(by='count').plot.barh(x='words',
y='count',
ax=ax,
color="red")
ax.set_title("Common Topics Found in Tweets (Including All Words) for India Lockdown 1")
plt.show()
# + id="AM23K9muUMOb" colab={"base_uri": "https://localhost:8080/", "height": 499} outputId="bfcfc18f-13dd-49bf-b373-ce74cd75eae5"
lockdown2 = ""
for i in indialockdown2.index:
lockdown2 = lockdown2 + str(indialockdown2.at[i,"topics"])
all_topicslockdown2 = lockdown2.split(",")
counts_no_urls = collections.Counter(all_topicslockdown2)
counts_no_urls.most_common(15)
len(counts_no_urls)
clean_tweets_no_urls = pd.DataFrame(counts_no_urls.most_common(10),
columns=['words', 'count'])
fig, ax = plt.subplots(figsize=(8, 8))
# Plot horizontal bar graph
clean_tweets_no_urls.sort_values(by='count').plot.barh(x='words',
y='count',
ax=ax,
color="green")
ax.set_title("Common Topics Found in Tweets (Including All Words) for India Lockdown 2")
plt.show()
# + id="lEK4uOdxUMOb" colab={"base_uri": "https://localhost:8080/", "height": 499} outputId="71ca86b0-6e2b-40d8-e46c-3887547ce656"
lockdown3 = ""
for i in indialockdown3.index:
lockdown3 = lockdown3 + str(indialockdown3.at[i,"topics"])
all_topicslockdown3 = lockdown3.split(",")
counts_no_urls = collections.Counter(all_topicslockdown3)
clean_tweets_no_urls = pd.DataFrame(counts_no_urls.most_common(1),
columns=['words', 'count'])
fig, ax = plt.subplots(figsize=(8, 8))
# Plot horizontal bar graph
clean_tweets_no_urls.sort_values(by='count').plot.barh(x='words',
y='count',
ax=ax,
color="blue")
ax.set_title("Common Topics Found in Tweets (Including All Words) for India Lockdown 3")
plt.show()
# + colab={"base_uri": "https://localhost:8080/", "height": 499} id="8OXrOMbRP_4U" outputId="528a5ff6-3363-43e1-c0e0-6ecde4098b3f"
lockdown4 = ""
for i in indialockdown4.index:
lockdown4 = lockdown4 + str(indialockdown4.at[i,"topics"])
all_topicslockdown4 = lockdown4.split(",")
counts_no_urls = collections.Counter(all_topicslockdown4)
clean_tweets_no_urls = pd.DataFrame(counts_no_urls.most_common(1),
columns=['words', 'count'])
fig, ax = plt.subplots(figsize=(8, 8))
# Plot horizontal bar graph
clean_tweets_no_urls.sort_values(by='count').plot.barh(x='words',
y='count',
ax=ax,
color="blue")
ax.set_title("Common Topics Found in Tweets (Including All Words) for India Lockdown 4")
plt.show()
# + colab={"base_uri": "https://localhost:8080/", "height": 402} id="CKVInpAMQJoJ" outputId="d62a969a-0229-4a43-e5d3-fc49a8579700"
indialockdown4
# + id="rSBvGA9fUMOb" colab={"base_uri": "https://localhost:8080/", "height": 402} outputId="d00ca120-833f-4913-d711-eaf5673b3ca7"
indialockdown3
# + id="bOf1xHpzUMOc" colab={"base_uri": "https://localhost:8080/", "height": 402} outputId="b309a31c-4558-42ff-e740-fd5d0f4ea66b"
indialockdown4
# + id="59p6pj-sUMOc" outputId="f1638b6b-addb-44fa-c41d-bacdf566f73a"
usa = data[data["country_code"]=="US"]
usa
# + id="cjmks7toUMOc"
india = usa
# + id="qaPtLUsXUMOc"
from datetime import datetime
for i in india.index:
india.at[i,"date"] = datetime.strptime(india.at[i,"date"], '%Y-%m-%d')
indialockdown1 = india[(india.date>=a1) & (india.date<=a2)]
indialockdown2 = india[(india.date>=b1) & (india.date<=b2)]
indialockdown3 = india[(india.date>=c1) & (india.date<=c2)]
indialockdown4 = india[(india.date>=d1) & (india.date<=d2)]
# + id="NS9B1tCVUMOc" outputId="3750f757-84a6-4b75-85c2-f39fa5cb4cd5"
lockdown1 = ""
for i in indialockdown1.index:
lockdown1 = lockdown1 + str(indialockdown1.at[i,"topics"])
all_topicslockdown1 = lockdown1.split(",")
counts_no_urls = collections.Counter(all_topicslockdown1)
counts_no_urls.most_common(15)
len(counts_no_urls)
clean_tweets_no_urls = pd.DataFrame(counts_no_urls.most_common(10),
columns=['words', 'count'])
fig, ax = plt.subplots(figsize=(8, 8))
# Plot horizontal bar graph
clean_tweets_no_urls.sort_values(by='count').plot.barh(x='words',
y='count',
ax=ax,
color="blue")
ax.set_title("Common Topis Found in Tweets (Including All Words)")
plt.show()
# + id="n3S-hdHhUMOd" outputId="10c18bb1-ced9-4e02-de46-430289e72389"
lockdown2 = ""
for i in indialockdown2.index:
lockdown2 = lockdown2 + str(indialockdown2.at[i,"topics"])
all_topicslockdown2 = lockdown2.split(",")
counts_no_urls = collections.Counter(all_topicslockdown2)
counts_no_urls.most_common(15)
len(counts_no_urls)
clean_tweets_no_urls = pd.DataFrame(counts_no_urls.most_common(10),
columns=['words', 'count'])
fig, ax = plt.subplots(figsize=(8, 8))
# Plot horizontal bar graph
clean_tweets_no_urls.sort_values(by='count').plot.barh(x='words',
y='count',
ax=ax,
color="black")
ax.set_title("Common Topis Found in Tweets (Including All Words)")
plt.show()
# + id="ty3t27x-UMOe" colab={"base_uri": "https://localhost:8080/", "height": 499} outputId="8c209a82-cc32-4d1c-85f3-e41b1da52b9b"
lockdown3 = ""
for i in indialockdown3.index:
lockdown3 = lockdown3 + str(indialockdown3.at[i,"topics"])
all_topicslockdown3 = lockdown3.split(",")
counts_no_urls = collections.Counter(all_topicslockdown3)
clean_tweets_no_urls = pd.DataFrame(counts_no_urls.most_common(1),
columns=['words', 'count'])
fig, ax = plt.subplots(figsize=(8, 8))
# Plot horizontal bar graph
clean_tweets_no_urls.sort_values(by='count').plot.barh(x='words',
y='count',
ax=ax,
color="pink")
ax.set_title("Common Topis Found in Tweets (Including All Words)")
plt.show()
# + id="97aGZQy7UMOf" outputId="1840df28-f467-48d0-a8ed-9b6c1dcd6a44"
data["country_code"].unique()
# + id="2qC7l0LYUMOf"
import seaborn as sns
# + id="0QZJcBgSUMOg" outputId="921de784-d1b2-4f2a-c973-02f6dee88d3e"
sns.countplot(data['country_code'])
# + id="R24xPN9BUMOg"
df = data
freq_vals = df['country_code'].value_counts()[:].index.tolist()
freq_df = df[df['country_code'].isin(freq_vals)]
# + id="VlgysW1wUMOg"
count = df['country_code'].value_counts()
# + id="ccMn_TzUUMOh" outputId="c028b25d-8859-453e-9c02-068a7024a337"
count
# + id="F2F0tGMdUMOh"
count.to_csv("a.csv")
# + id="bQJsExf1UMOh" outputId="331fc317-07e6-4832-af02-e6647e122fbc"
# + id="kN2r_zLDUMOi"
| 31.808219
| 1,342
|
0075cf2da726ae3d68699a439b02aac6c54ef898
|
py
|
python
|
Customer Segments/customer_segments.ipynb
|
simmy88/UdacityMLND
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# # Machine Learning Engineer Nanodegree
# ## Unsupervised Learning
# ## Project: Creating Customer Segments
# Welcome to the third project of the Machine Learning Engineer Nanodegree! In this notebook, some template code has already been provided for you, and it will be your job to implement the additional functionality necessary to successfully complete this project. Sections that begin with **'Implementation'** in the header indicate that the following block of code will require additional functionality which you must provide. Instructions will be provided for each section and the specifics of the implementation are marked in the code block with a `'TODO'` statement. Please be sure to read the instructions carefully!
#
# In addition to implementing code, there will be questions that you must answer which relate to the project and your implementation. Each section where you will answer a question is preceded by a **'Question X'** header. Carefully read each question and provide thorough answers in the following text boxes that begin with **'Answer:'**. Your project submission will be evaluated based on your answers to each of the questions and the implementation you provide.
#
# >**Note:** Code and Markdown cells can be executed using the **Shift + Enter** keyboard shortcut. In addition, Markdown cells can be edited by typically double-clicking the cell to enter edit mode.
# ## Getting Started
#
# In this project, you will analyze a dataset containing data on various customers' annual spending amounts (reported in *monetary units*) of diverse product categories for internal structure. One goal of this project is to best describe the variation in the different types of customers that a wholesale distributor interacts with. Doing so would equip the distributor with insight into how to best structure their delivery service to meet the needs of each customer.
#
# The dataset for this project can be found on the [UCI Machine Learning Repository](https://archive.ics.uci.edu/ml/datasets/Wholesale+customers). For the purposes of this project, the features `'Channel'` and `'Region'` will be excluded in the analysis — with focus instead on the six product categories recorded for customers.
#
# Run the code block below to load the wholesale customers dataset, along with a few of the necessary Python libraries required for this project. You will know the dataset loaded successfully if the size of the dataset is reported.
# +
# Import libraries necessary for this project
import numpy as np
import pandas as pd
from IPython.display import display # Allows the use of display() for DataFrames
# Import supplementary visualizations code visuals.py
import visuals as vs
# Pretty display for notebooks
# %matplotlib inline
# Load the wholesale customers dataset
try:
data = pd.read_csv("customers.csv")
data.drop(['Region', 'Channel'], axis = 1, inplace = True)
print "Wholesale customers dataset has {} samples with {} features each.".format(*data.shape)
except:
print "Dataset could not be loaded. Is the dataset missing?"
# -
# ## Data Exploration
# In this section, you will begin exploring the data through visualizations and code to understand how each feature is related to the others. You will observe a statistical description of the dataset, consider the relevance of each feature, and select a few sample data points from the dataset which you will track through the course of this project.
#
# Run the code block below to observe a statistical description of the dataset. Note that the dataset is composed of six important product categories: **'Fresh'**, **'Milk'**, **'Grocery'**, **'Frozen'**, **'Detergents_Paper'**, and **'Delicatessen'**. Consider what each category represents in terms of products you could purchase.
# Display a description of the dataset
display(data.describe())
# ### Implementation: Selecting Samples
# To get a better understanding of the customers and how their data will transform through the analysis, it would be best to select a few sample data points and explore them in more detail. In the code block below, add **three** indices of your choice to the `indices` list which will represent the customers to track. It is suggested to try different sets of samples until you obtain customers that vary significantly from one another.
# +
# TODO: Select three indices of your choice you wish to sample from the dataset
indices = [100,200,300]
# Create a DataFrame of the chosen samples
samples = pd.DataFrame(data.loc[indices], columns = data.keys()).reset_index(drop = True)
print "Chosen samples of wholesale customers dataset:"
display(samples)
# -
# ### Question 1
# Consider the total purchase cost of each product category and the statistical description of the dataset above for your sample customers.
# *What kind of establishment (customer) could each of the three samples you've chosen represent?*
# **Hint:** Examples of establishments include places like markets, cafes, and retailers, among many others. Avoid using names for establishments, such as saying *"McDonalds"* when describing a sample customer as a restaurant.
# **Answer:** Sample 0 has a values which are significantly higher than the mean in the categories of Grocery and Detergents_Paper but values which are much closer to the mean in the other categories. This sample could represent supermarket which sells freshly prepared food. Sample 1 has a much higher than mean value for Milk and Grocery and significantly below mean value for Fresh and Delicatessen. This sample could represent a supermarket - selling mostly Groceries, Milk. Sample 2 has very high values, compared to the mean, for Fresh and very low values for Frozen. This sample could represent a Cafe or Restaurant which sells a lot of fresh food but little frozen food.
# ### Implementation: Feature Relevance
# One interesting thought to consider is if one (or more) of the six product categories is actually relevant for understanding customer purchasing. That is to say, is it possible to determine whether customers purchasing some amount of one category of products will necessarily purchase some proportional amount of another category of products? We can make this determination quite easily by training a supervised regression learner on a subset of the data with one feature removed, and then score how well that model can predict the removed feature.
#
# In the code block below, you will need to implement the following:
# - Assign `new_data` a copy of the data by removing a feature of your choice using the `DataFrame.drop` function.
# - Use `sklearn.cross_validation.train_test_split` to split the dataset into training and testing sets.
# - Use the removed feature as your target label. Set a `test_size` of `0.25` and set a `random_state`.
# - Import a decision tree regressor, set a `random_state`, and fit the learner to the training data.
# - Report the prediction score of the testing set using the regressor's `score` function.
# +
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeRegressor
from sklearn.metrics import r2_score
# TODO: Make a copy of the DataFrame, using the 'drop' function to drop the given feature
new_data = data.drop(['Milk'], axis = 1)
# TODO: Split the data into training and testing sets using the given feature as the target
X_train, X_test, y_train, y_test = train_test_split(new_data, data['Milk'], test_size=0.25, random_state=23)
# TODO: Create a decision tree regressor and fit it to the training set
regressor = DecisionTreeRegressor()
regressor = regressor.fit(X_train, y_train)
# TODO: Report the score of the prediction using the testing set
y_pred = regressor.predict(X_test)
score = r2_score(y_test, y_pred)
print "R^2 score for prediction using testing set:"
print score
# -
# ### Question 2
# *Which feature did you attempt to predict? What was the reported prediction score? Is this feature necessary for identifying customers' spending habits?*
# **Hint:** The coefficient of determination, `R^2`, is scored between 0 and 1, with 1 being a perfect fit. A negative `R^2` implies the model fails to fit the data.
# **Answer:** I tried to predict the Fresh feature from the data and the score was -1.56. This implies that the remaining data was unable to model the Fresh feature. This implies that the feature is necessary for identifying customer spending habits. By contrast the Milk feature was predicted with a score of 0.47. This implies that the feature can be modelled using the other features to a reasonable extent and might not be necessary to predict customer spending habits.
# ### Visualize Feature Distributions
# To get a better understanding of the dataset, we can construct a scatter matrix of each of the six product features present in the data. If you found that the feature you attempted to predict above is relevant for identifying a specific customer, then the scatter matrix below may not show any correlation between that feature and the others. Conversely, if you believe that feature is not relevant for identifying a specific customer, the scatter matrix might show a correlation between that feature and another feature in the data. Run the code block below to produce a scatter matrix.
# Produce a scatter matrix for each pair of features in the data
pd.plotting.scatter_matrix(data, alpha = 0.3, figsize = (14,8), diagonal = 'kde');
# ### Question 3
# *Are there any pairs of features which exhibit some degree of correlation? Does this confirm or deny your suspicions about the relevance of the feature you attempted to predict? How is the data for those features distributed?*
# **Hint:** Is the data normally distributed? Where do most of the data points lie?
# **Answer:** There seems to be a correlation between Grocery and Milk as well as between Detergents_Paper and Grocery. The correlation between Milk and Grocery seems to be weaker than between Detergents_Paper and Grocery. This confirms why the Milk feature was hard to predict. The data is not normally distributed, with a significant number of points lying at the lower end of the scale.
# ## Data Preprocessing
# In this section, you will preprocess the data to create a better representation of customers by performing a scaling on the data and detecting (and optionally removing) outliers. Preprocessing data is often times a critical step in assuring that results you obtain from your analysis are significant and meaningful.
# ### Implementation: Feature Scaling
# If data is not normally distributed, especially if the mean and median vary significantly (indicating a large skew), it is most [often appropriate](http://econbrowser.com/archives/2014/02/use-of-logarithms-in-economics) to apply a non-linear scaling — particularly for financial data. One way to achieve this scaling is by using a [Box-Cox test](http://scipy.github.io/devdocs/generated/scipy.stats.boxcox.html), which calculates the best power transformation of the data that reduces skewness. A simpler approach which can work in most cases would be applying the natural logarithm.
#
# In the code block below, you will need to implement the following:
# - Assign a copy of the data to `log_data` after applying logarithmic scaling. Use the `np.log` function for this.
# - Assign a copy of the sample data to `log_samples` after applying logarithmic scaling. Again, use `np.log`.
# +
# TODO: Scale the data using the natural logarithm
log_data = np.log(data)
# TODO: Scale the sample data using the natural logarithm
log_samples = np.log(samples)
# Produce a scatter matrix for each pair of newly-transformed features
pd.plotting.scatter_matrix(log_data, alpha = 0.3, figsize = (14,8), diagonal = 'kde');
# -
# ### Observation
# After applying a natural logarithm scaling to the data, the distribution of each feature should appear much more normal. For any pairs of features you may have identified earlier as being correlated, observe here whether that correlation is still present (and whether it is now stronger or weaker than before).
#
# Run the code below to see how the sample data has changed after having the natural logarithm applied to it.
# Display the log-transformed sample data
display(log_samples)
# ### Implementation: Outlier Detection
# Detecting outliers in the data is extremely important in the data preprocessing step of any analysis. The presence of outliers can often skew results which take into consideration these data points. There are many "rules of thumb" for what constitutes an outlier in a dataset. Here, we will use [Tukey's Method for identfying outliers](http://datapigtechnologies.com/blog/index.php/highlighting-outliers-in-your-data-with-the-tukey-method/): An *outlier step* is calculated as 1.5 times the interquartile range (IQR). A data point with a feature that is beyond an outlier step outside of the IQR for that feature is considered abnormal.
#
# In the code block below, you will need to implement the following:
# - Assign the value of the 25th percentile for the given feature to `Q1`. Use `np.percentile` for this.
# - Assign the value of the 75th percentile for the given feature to `Q3`. Again, use `np.percentile`.
# - Assign the calculation of an outlier step for the given feature to `step`.
# - Optionally remove data points from the dataset by adding indices to the `outliers` list.
#
# **NOTE:** If you choose to remove any outliers, ensure that the sample data does not contain any of these points!
# Once you have performed this implementation, the dataset will be stored in the variable `good_data`.
# +
# For each feature find the data points with extreme high or low values
for feature in log_data.keys():
# TODO: Calculate Q1 (25th percentile of the data) for the given feature
Q1 = np.percentile(log_data[feature], 25)
# TODO: Calculate Q3 (75th percentile of the data) for the given feature
Q3 = np.percentile(log_data[feature],75)
# TODO: Use the interquartile range to calculate an outlier step (1.5 times the interquartile range)
step = (Q3-Q1)*1.5
# Display the outliers
print "Data points considered outliers for the feature '{}':".format(feature)
display(log_data[~((log_data[feature] >= Q1 - step) & (log_data[feature] <= Q3 + step))])
# OPTIONAL: Select the indices for data points you wish to remove
outliers = [65, 66, 75, 128, 154]
# Remove the outliers, if any were specified
good_data = log_data.drop(log_data.index[outliers]).reset_index(drop = True)
# -
# ### Question 4
# *Are there any data points considered outliers for more than one feature based on the definition above? Should these data points be removed from the dataset? If any data points were added to the `outliers` list to be removed, explain why.*
# **Answer:** There were three points which were considered outliers for more than one feature. These were points 65, 66, 75, 128, 154. These points were removed as they might skew the correlations and affect the location of the cluster boundaries between these features. I decided not to remove any of the other outliers as they would affect the location of the cluster centers but shouldn't affect the boundaries between the clusters.
# ## Feature Transformation
# In this section you will use principal component analysis (PCA) to draw conclusions about the underlying structure of the wholesale customer data. Since using PCA on a dataset calculates the dimensions which best maximize variance, we will find which compound combinations of features best describe customers.
# ### Implementation: PCA
#
# Now that the data has been scaled to a more normal distribution and has had any necessary outliers removed, we can now apply PCA to the `good_data` to discover which dimensions about the data best maximize the variance of features involved. In addition to finding these dimensions, PCA will also report the *explained variance ratio* of each dimension — how much variance within the data is explained by that dimension alone. Note that a component (dimension) from PCA can be considered a new "feature" of the space, however it is a composition of the original features present in the data.
#
# In the code block below, you will need to implement the following:
# - Import `sklearn.decomposition.PCA` and assign the results of fitting PCA in six dimensions with `good_data` to `pca`.
# - Apply a PCA transformation of `log_samples` using `pca.transform`, and assign the results to `pca_samples`.
# +
from sklearn.decomposition import PCA
# TODO: Apply PCA by fitting the good data with the same number of dimensions as features
pca = PCA(n_components=6).fit(good_data)
# TODO: Transform log_samples using the PCA fit above
pca_samples = pca.transform(log_samples)
# Generate PCA results plot
pca_results = vs.pca_results(good_data, pca)
# -
# ### Question 5
# *How much variance in the data is explained* ***in total*** *by the first and second principal component? What about the first four principal components? Using the visualization provided above, discuss what the first four dimensions best represent in terms of customer spending.*
# **Hint:** A positive increase in a specific dimension corresponds with an *increase* of the *positive-weighted* features and a *decrease* of the *negative-weighted* features. The rate of increase or decrease is based on the individual feature weights.
# **Answer:** The first two components account for approximately 71.6% of the variance. When this is extended to the first four components they account for approximately 93.4% of the variance. The first four dimensions probably represent different types of customer spending, i.e. shop types. The first dimension has large weights between Detergents_Paper, Milk, and Grocery, which are negatively correlated to Fresh and Frozen. This could indicate purchasing of household items. Dimension 2 has strong weights in Fresh, Frozen, and Delicatessen. This could indicate purchasing of food items which are generally bought together. Dimension 3 indicates there is a negative relationship between Fresh and Delicatessen, which could be a measure which shows people who buy a lot of Delicatessen items are unlikely to spend a lot on Fresh food. Dimension 4 shows strong negative correlations between Frozen and Delicatessen. Indicating that people who spend a large amount of Frozen spend less on Delicatessen, and vice versa.
# ### Observation
# Run the code below to see how the log-transformed sample data has changed after having a PCA transformation applied to it in six dimensions. Observe the numerical value for the first four dimensions of the sample points. Consider if this is consistent with your initial interpretation of the sample points.
# Display sample log-data after having a PCA transformation applied
display(pd.DataFrame(np.round(pca_samples, 4), columns = pca_results.index.values))
# ### Implementation: Dimensionality Reduction
# When using principal component analysis, one of the main goals is to reduce the dimensionality of the data — in effect, reducing the complexity of the problem. Dimensionality reduction comes at a cost: Fewer dimensions used implies less of the total variance in the data is being explained. Because of this, the *cumulative explained variance ratio* is extremely important for knowing how many dimensions are necessary for the problem. Additionally, if a signifiant amount of variance is explained by only two or three dimensions, the reduced data can be visualized afterwards.
#
# In the code block below, you will need to implement the following:
# - Assign the results of fitting PCA in two dimensions with `good_data` to `pca`.
# - Apply a PCA transformation of `good_data` using `pca.transform`, and assign the results to `reduced_data`.
# - Apply a PCA transformation of `log_samples` using `pca.transform`, and assign the results to `pca_samples`.
# +
# TODO: Apply PCA by fitting the good data with only two dimensions
pca = PCA(n_components=2).fit(good_data)
# TODO: Transform the good data using the PCA fit above
reduced_data = pca.transform(good_data)
# TODO: Transform log_samples using the PCA fit above
pca_samples = pca.transform(log_samples)
# Create a DataFrame for the reduced data
reduced_data = pd.DataFrame(reduced_data, columns = ['Dimension 1', 'Dimension 2'])
# -
# ### Observation
# Run the code below to see how the log-transformed sample data has changed after having a PCA transformation applied to it using only two dimensions. Observe how the values for the first two dimensions remains unchanged when compared to a PCA transformation in six dimensions.
# Display sample log-data after applying PCA transformation in two dimensions
display(pd.DataFrame(np.round(pca_samples, 4), columns = ['Dimension 1', 'Dimension 2']))
# ## Visualizing a Biplot
# A biplot is a scatterplot where each data point is represented by its scores along the principal components. The axes are the principal components (in this case `Dimension 1` and `Dimension 2`). In addition, the biplot shows the projection of the original features along the components. A biplot can help us interpret the reduced dimensions of the data, and discover relationships between the principal components and original features.
#
# Run the code cell below to produce a biplot of the reduced-dimension data.
# Create a biplot
vs.biplot(good_data, reduced_data, pca)
# ### Observation
#
# Once we have the original feature projections (in red), it is easier to interpret the relative position of each data point in the scatterplot. For instance, a point the lower right corner of the figure will likely correspond to a customer that spends a lot on `'Milk'`, `'Grocery'` and `'Detergents_Paper'`, but not so much on the other product categories.
#
# From the biplot, which of the original features are most strongly correlated with the first component? What about those that are associated with the second component? Do these observations agree with the pca_results plot you obtained earlier?
# ## Clustering
#
# In this section, you will choose to use either a K-Means clustering algorithm or a Gaussian Mixture Model clustering algorithm to identify the various customer segments hidden in the data. You will then recover specific data points from the clusters to understand their significance by transforming them back into their original dimension and scale.
# ### Question 6
# *What are the advantages to using a K-Means clustering algorithm? What are the advantages to using a Gaussian Mixture Model clustering algorithm? Given your observations about the wholesale customer data so far, which of the two algorithms will you use and why?*
# **Answer:** K-Means clustering uses a hard assignment of points where as the Gaussian Mixture Model uses soft assignment. Given that the customer data seems to not have clear and distinct boundaries I will use the Gaussian Mixture Model as it assigns points to clusters probablistically rather than using hard assignments.
# ### Implementation: Creating Clusters
# Depending on the problem, the number of clusters that you expect to be in the data may already be known. When the number of clusters is not known *a priori*, there is no guarantee that a given number of clusters best segments the data, since it is unclear what structure exists in the data — if any. However, we can quantify the "goodness" of a clustering by calculating each data point's *silhouette coefficient*. The [silhouette coefficient](http://scikit-learn.org/stable/modules/generated/sklearn.metrics.silhouette_score.html) for a data point measures how similar it is to its assigned cluster from -1 (dissimilar) to 1 (similar). Calculating the *mean* silhouette coefficient provides for a simple scoring method of a given clustering.
#
# In the code block below, you will need to implement the following:
# - Fit a clustering algorithm to the `reduced_data` and assign it to `clusterer`.
# - Predict the cluster for each data point in `reduced_data` using `clusterer.predict` and assign them to `preds`.
# - Find the cluster centers using the algorithm's respective attribute and assign them to `centers`.
# - Predict the cluster for each sample data point in `pca_samples` and assign them `sample_preds`.
# - Import `sklearn.metrics.silhouette_score` and calculate the silhouette score of `reduced_data` against `preds`.
# - Assign the silhouette score to `score` and print the result.
# +
from sklearn.mixture import GaussianMixture
from sklearn.metrics import silhouette_score
# TODO: Apply your clustering algorithm of choice to the reduced data
GM = GaussianMixture(n_components = 2)
clusterer = GM.fit(reduced_data)
# TODO: Predict the cluster for each data point
preds = clusterer.predict(reduced_data)
# TODO: Find the cluster centers
centers = clusterer.means_
# TODO: Predict the cluster for each transformed sample data point
sample_preds = clusterer.predict(pca_samples)
# TODO: Calculate the mean silhouette coefficient for the number of clusters chosen
score = silhouette_score(reduced_data, preds)
print score
# -
# ### Question 7
# *Report the silhouette score for several cluster numbers you tried. Of these, which number of clusters has the best silhouette score?*
# **Answer:** 2 clusters has the best silhouette score of 0.422 which is slightly better than 3 clusters, which had a score of 0.403. 4 clusters had a significantly smaller score of 0.333
# ### Cluster Visualization
# Once you've chosen the optimal number of clusters for your clustering algorithm using the scoring metric above, you can now visualize the results by executing the code block below. Note that, for experimentation purposes, you are welcome to adjust the number of clusters for your clustering algorithm to see various visualizations. The final visualization provided should, however, correspond with the optimal number of clusters.
# Display the results of the clustering from implementation
vs.cluster_results(reduced_data, preds, centers, pca_samples)
# ### Implementation: Data Recovery
# Each cluster present in the visualization above has a central point. These centers (or means) are not specifically data points from the data, but rather the *averages* of all the data points predicted in the respective clusters. For the problem of creating customer segments, a cluster's center point corresponds to *the average customer of that segment*. Since the data is currently reduced in dimension and scaled by a logarithm, we can recover the representative customer spending from these data points by applying the inverse transformations.
#
# In the code block below, you will need to implement the following:
# - Apply the inverse transform to `centers` using `pca.inverse_transform` and assign the new centers to `log_centers`.
# - Apply the inverse function of `np.log` to `log_centers` using `np.exp` and assign the true centers to `true_centers`.
#
# +
# TODO: Inverse transform the centers
log_centers = pca.inverse_transform(centers)
# TODO: Exponentiate the centers
true_centers = np.exp(log_centers)
# Display the true centers
segments = ['Segment {}'.format(i) for i in range(0,len(centers))]
true_centers = pd.DataFrame(np.round(true_centers), columns = data.keys())
true_centers.index = segments
display(true_centers)
# -
# ### Question 8
# Consider the total purchase cost of each product category for the representative data points above, and reference the statistical description of the dataset at the beginning of this project. *What set of establishments could each of the customer segments represent?*
# **Hint:** A customer who is assigned to `'Cluster X'` should best identify with the establishments represented by the feature set of `'Segment X'`.
# **Answer:** Segment 1 has high values for Fresh and Frozen, but lower values for Milk, Grocery, and Detergents_Paper, when compared to Segment 0. This indicates that Segment 1 could represent Food Markets where shoppers spend mostly on Fresh and Frozen items. Segment 0, with higher values of Milk, Grocery, and Detergents_Paper, could indicate Supermarkets where people are buying household staples.
# ### Question 9
# *For each sample point, which customer segment from* ***Question 8*** *best represents it? Are the predictions for each sample point consistent with this?*
#
# Run the code block below to find which cluster each sample point is predicted to be.
# Display the predictions
for i, pred in enumerate(sample_preds):
print "Sample point", i, "predicted to be in Cluster", pred
# **Answer:** The points are all predicted to be in Cluster 0. Analysing the scatter plot, one of them is quite close to the boundary with Cluster 1 and this is likely to be Sample point 2, which has a smaller Grocery value and significant higher Fresh value than the other two points. When compared to the cluster centers, the two points which are firmly in Cluster 0 have values which generally correlate well with Cluster 0's center, particularly values of Milk and Grocery for point 0 and point 1 follows the correlation between the two features. The point which is closer to the boundary has high values of Fresh, middling values of Milk and Grocery, but a low value of Frozen. This combination of features results in it not clearly representing either Cluster particularly well.
# ## Conclusion
# In this final section, you will investigate ways that you can make use of the clustered data. First, you will consider how the different groups of customers, the ***customer segments***, may be affected differently by a specific delivery scheme. Next, you will consider how giving a label to each customer (which *segment* that customer belongs to) can provide for additional features about the customer data. Finally, you will compare the ***customer segments*** to a hidden variable present in the data, to see whether the clustering identified certain relationships.
# ### Question 10
# Companies will often run [A/B tests](https://en.wikipedia.org/wiki/A/B_testing) when making small changes to their products or services to determine whether making that change will affect its customers positively or negatively. The wholesale distributor is considering changing its delivery service from currently 5 days a week to 3 days a week. However, the distributor will only make this change in delivery service for customers that react positively. *How can the wholesale distributor use the customer segments to determine which customers, if any, would react positively to the change in delivery service?*
# **Hint:** Can we assume the change affects all customers equally? How can we determine which group of customers it affects the most?
# **Answer:** The wholesale distributor can test the change in delivery service on both groups and see which, if any, reacts positively to the change in delivery service.
# ### Question 11
# Additional structure is derived from originally unlabeled data when using clustering techniques. Since each customer has a ***customer segment*** it best identifies with (depending on the clustering algorithm applied), we can consider *'customer segment'* as an **engineered feature** for the data. Assume the wholesale distributor recently acquired ten new customers and each provided estimates for anticipated annual spending of each product category. Knowing these estimates, the wholesale distributor wants to classify each new customer to a ***customer segment*** to determine the most appropriate delivery service.
# *How can the wholesale distributor label the new customers using only their estimated product spending and the* ***customer segment*** *data?*
# **Hint:** A supervised learner could be used to train on the original customers. What would be the target variable?
# **Answer:** Using a supervised learner to associate the points with the customer segment label that had been arributed using the unsupervised learner. The trained supervised learner can then be used to label the new points.
# ### Visualizing Underlying Distributions
#
# At the beginning of this project, it was discussed that the `'Channel'` and `'Region'` features would be excluded from the dataset so that the customer product categories were emphasized in the analysis. By reintroducing the `'Channel'` feature to the dataset, an interesting structure emerges when considering the same PCA dimensionality reduction applied earlier to the original dataset.
#
# Run the code block below to see how each data point is labeled either `'HoReCa'` (Hotel/Restaurant/Cafe) or `'Retail'` the reduced space. In addition, you will find the sample points are circled in the plot, which will identify their labeling.
# Display the clustering results based on 'Channel' data
vs.channel_results(reduced_data, outliers, pca_samples)
# ### Question 12
# *How well does the clustering algorithm and number of clusters you've chosen compare to this underlying distribution of Hotel/Restaurant/Cafe customers to Retailer customers? Are there customer segments that would be classified as purely 'Retailers' or 'Hotels/Restaurants/Cafes' by this distribution? Would you consider these classifications as consistent with your previous definition of the customer segments?*
# **Answer:** The clustering algorithm I chose, with 2 clusters, matched the underlying distribution in the data reasonably well despite having one additional cluster. I would consider the classification in the data to be consistent with my pervious definitions.
# > **Note**: Once you have completed all of the code implementations and successfully answered each question above, you may finalize your work by exporting the iPython Notebook as an HTML document. You can do this by using the menu above and navigating to
# **File -> Download as -> HTML (.html)**. Include the finished document along with this notebook as your submission.
| 84.116915
| 1,020
|
7b4d37736aea44e194da2dca14f03c964b628bca
|
py
|
python
|
src/ganPred.ipynb
|
avani17101/goal-oriented-next-best-activity-recomendation
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# +
import numpy as np
import pandas as pd
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.nn.utils.rnn import pad_sequence
from dfg import check_dfg_compliance
def model_eval_test(dataset_name, csvwriter=None):
'''
This module is for validation and testing the Generator
@param modelG: Generator neural network
@param mode: 'validation', 'test', 'test-validation'
@param obj: A data object created from "Input" class that contains the required information
@return: The accuracy of the Generator
'''
# set the evaluation mode (this mode is necessary if you train with batch, since in test the size of batch is different)
batch = 1
#events = list(np.arange(0, len(obj.unique_event) + 1))
predicted = []
accuracy = []
mae = []
dfg_compliance_pred = []
dfg_compliance_gt = []
y_truth_list = []
y_pred_last_event_list = []
accuracy_last1 = []
accuracy_last2 = []
accuracy_last3 = []
lastk = {}
df = pd.read_pickle("dataset/preprocessed/"+dataset_name+"_design_mat.pkl")
unique_event = [0] + sorted(df['class'].unique())
events = list(np.arange(0, len(unique_event)))
print("events",events)
max_activities = len(unique_event)
dur = 0
dur_gt = 0
num_overall_goal_satisfied = 0
num_overall_goal_not_satisfied = 0
num_overall_goal_satisfied_gt = 0
num_overall_goal_not_satisfied_gt = 0
row = []
selected_columns = np.arange(0,max_activities+1)
y_pred_last_event_list_prev = None
group = df.groupby('CaseID')
cur_prefix_len = 1
prev_event = None
max_prefix_len = 13
pred_events = []
thresh = 13.89
if dataset_name == "helpdesk":
thresh = 13.89
max_prefix_len = 13
if dataset_name == "bpi_12_w":
thresh = 18.28
max_prefix_len = 73
if dataset_name == "traffic_ss":
thresh = 607.04
max_prefix_len = 16
for name,gr in group:
gr = gr.reset_index(drop=True)
new_row = [0] * gr.shape[1]
gr.loc[gr.shape[0]] = new_row
gr.iloc[gr.shape[0] - 1, gr.columns.get_loc('0')] = 1 # End of line is denoted by class 0
temp = torch.tensor(gr.values, dtype=torch.float, requires_grad=False)
temp_shifted = torch.tensor(gr[['duration_time','class']].values, dtype=torch.float, requires_grad=False)
x = pad_sequence([temp], batch_first=True)
y_truth = pad_sequence([temp_shifted], batch_first=True)
cur_trace_len = int(x.shape[1])
if cur_trace_len > max_prefix_len:
continue
activites = torch.argmax(x[:, :, events])
# When we create mini batches, the length of the last one probably is less than the batch size, and it makes problem for the LSTM, therefore we skip it.
if (x.size()[0] < batch):
continue
for cur_prefix_len in range(1,cur_trace_len):
# Separating event and timestamp
y_truth_timestamp = y_truth[:, cur_prefix_len, 0].view(batch, 1, -1)
y_truth_event = y_truth[:, cur_prefix_len, 1].view(batch, 1, -1)
# Executing LSTM
if cur_prefix_len == 1:
x_inn = x[:, :cur_prefix_len, selected_columns]
prev_event = x[:, :cur_prefix_len, len(selected_columns)+1].ravel().detach().numpy().astype("int")
else:
x_inn = x[:, :cur_prefix_len, selected_columns]
rnnG = torch.load("checkpoints/"+dataset_name+"/event_timestamp_prediction/prefix_"+str(cur_prefix_len)+"/rnnG.m")
rnnG.eval()
y_pred = rnnG(x_inn[:, :cur_prefix_len, selected_columns])
# Just taking the last predicted element from each the batch
y_pred_last = y_pred[0: batch, cur_prefix_len - 1, :]
y_pred_last = y_pred_last.view((batch, 1, -1))
y_pred_last_event = torch.argmax(F.softmax(y_pred_last[:, :, events], dim=2), dim=2)
#Storing list of predictions and corresponding ground truths (to be used for f1score)
y_truth_list += list(y_truth_event.flatten().data.cpu().numpy().astype(int))
y_pred_last_event_list += list(y_pred_last_event.flatten().data.cpu().numpy().astype(int))
# checking dfg compliance for predicted event
if not(int(prev_event[0]) == 0 and int(y_pred_last_event.detach()[0][0])==0):
dfg_compliance_bool = check_dfg_compliance(prev_event , y_pred_last_event.detach(), dset=dataset_name)
dfg_compliance_pred.append(int(dfg_compliance_bool))
dfg_compliance_gt_bool = check_dfg_compliance(prev_event , y_truth_event.detach().reshape(y_pred_last_event.shape), dset=dataset_name)
dfg_compliance_gt.append(int(dfg_compliance_gt_bool))
if y_pred_last_event.flatten().data.cpu().numpy().astype(int)==y_truth_event.flatten().data.cpu().numpy().astype(int):
accuracy.append(1)
else:
accuracy.append(0)
pred_events.append(int(y_pred_last_event.detach()[0][0]))
# Computing MAE for the timestamp
y_pred_timestamp = y_pred_last[:, :, len(events)].view((batch, 1, -1))
mae.append(torch.abs(y_truth_timestamp - y_pred_timestamp).mean().detach())
dur += max(y_pred_timestamp.detach().numpy()[0][0],0) #adding to total proccess duration, making sure y_pred is non-negative
dur_gt += max(y_truth_timestamp.detach().numpy()[0][0],0)
prev_event = y_pred_last_event.ravel().detach().numpy().astype("int")
if prev_event ==0: #model predicted 0
break
# GS cases
try:
if dur < thresh:
num_overall_goal_satisfied += 1
else:
num_overall_goal_not_satisfied += 1
if dur_gt < thresh:
num_overall_goal_satisfied_gt += 1
else:
num_overall_goal_not_satisfied_gt += 1
except Exception as e: print(e)
# print(pred_events)
lenn = min(5, len(pred_events))
if lenn<=2:
lenn = 3
if len(pred_events)>=2:
for i in range(2,lenn):
if pred_events[-i] in lastk:
lastk[pred_events[-i]] += 1
else:
lastk[pred_events[-i]] = 1
if i == 1+1:
if(pred_events[-i] == y_truth_list[-i]): #action chosen by RL agent = gt
accuracy_last1.append(1)
accuracy_last2.append(1)
accuracy_last3.append(1)
else:
accuracy_last1.append(0)
accuracy_last2.append(0)
accuracy_last3.append(0)
if i == 2+1:
if(pred_events[-i] == y_truth_list[-i]): #action chosen by RL agent = gt
accuracy_last2.append(1)
accuracy_last3.append(1)
else:
accuracy_last2.append(0)
accuracy_last3.append(0)
if i == 3+1:
if(pred_events[-i] == y_truth_list[-i]): #action chosen by RL agent = gt
accuracy_last3.append(1)
else:
accuracy_last3.append(0)
dur = 0
dur_gt = 0
pred_events = []
tot = num_overall_goal_satisfied+ num_overall_goal_not_satisfied
gs = num_overall_goal_satisfied/tot
gv = num_overall_goal_not_satisfied/tot
tot_gt = num_overall_goal_satisfied_gt+ num_overall_goal_not_satisfied_gt
gs_gt = num_overall_goal_satisfied_gt/tot_gt
gv_gt = num_overall_goal_not_satisfied_gt/tot_gt
print("lastk",lastk)
print("compliance pred", np.mean(np.array(dfg_compliance_pred)))
print("compliance gt", np.mean(np.array(dfg_compliance_gt)))
print("gs ", gs)
print("gv ", gv)
print("gs gt ", gs_gt)
print("gv gt ", gv_gt)
print("acc ",np.mean(np.array(accuracy)))
print("acc last 3",np.mean(np.array(accuracy_last3)))
print("acc last 2",np.mean(np.array(accuracy_last2)))
print("acc last 1",np.mean(np.array(accuracy_last1)))
print("mae ", np.mean(np.array(mae)))
# -
datasets = ["helpdesk", "bpi_12_w","traffic_ss"]
for dataset in datasets:
print("dataset: ",dataset)
model_eval_test(dataset_name=dataset, csvwriter=None)
# +
# import numpy as np
# import pandas as pd
# import torch
# import torch.nn as nn
# import torch.nn.functional as F
# from torch.nn.utils.rnn import pad_sequence
# from dfg import check_dfg_compliance
# def model_eval_test(dataset_name, csvwriter=None):
# '''
# This module is for validation and testing the Generator
# @param modelG: Generator neural network
# @param mode: 'validation', 'test', 'test-validation'
# @param obj: A data object created from "Input" class that contains the required information
# @return: The accuracy of the Generator
# '''
# # set the evaluation mode (this mode is necessary if you train with batch, since in test the size of batch is different)
# batch = 1
# #events = list(np.arange(0, len(obj.unique_event) + 1))
# predicted = []
# accuracy = []
# mae = []
# dfg_compliance_pred = []
# dfg_compliance_gt = []
# y_truth_list = []
# y_pred_last_event_list = []
# df = pd.read_pickle("dataset/preprocessed/"+dataset_name+"_design_mat.pkl")
# unique_event = [0] + sorted(df['class'].unique())
# events = list(np.arange(0, len(unique_event)))
# print("events",events)
# max_activities = len(unique_event)
# dur = 0
# dur_gt = 0
# num_overall_goal_satisfied = 0
# num_overall_goal_not_satisfied = 0
# num_overall_goal_satisfied_gt = 0
# num_overall_goal_not_satisfied_gt = 0
# row = []
# selected_columns = np.arange(0,max_activities+1)
# y_pred_last_event_list_prev = None
# group = df.groupby('CaseID')
# cur_prefix_len = 1
# prev_event = None
# max_prefix_len = 13
# thresh = 13.89
# if dataset_name == "helpdesk":
# thresh = 13.89
# max_prefix_len = 13
# if dataset_name == "bpi_12_w":
# thresh = 18.28
# max_prefix_len = 73
# if dataset_name == "traffic_ss":
# thresh = 607.04
# max_prefix_len = 16
# for name,gr in group:
# gr = gr.reset_index(drop=True)
# new_row = [0] * gr.shape[1]
# gr.loc[gr.shape[0]] = new_row
# gr.iloc[gr.shape[0] - 1, gr.columns.get_loc('0')] = 1 # End of line is denoted by class 0
# temp = torch.tensor(gr.values, dtype=torch.float, requires_grad=False)
# temp_shifted = torch.tensor(gr[['duration_time','class']].values, dtype=torch.float, requires_grad=False)
# x = pad_sequence([temp], batch_first=True)
# y_truth = pad_sequence([temp_shifted], batch_first=True)
# cur_trace_len = int(x.shape[1])
# if cur_trace_len > max_prefix_len:
# continue
# activites = torch.argmax(x[:, :, events])
# # When we create mini batches, the length of the last one probably is less than the batch size, and it makes problem for the LSTM, therefore we skip it.
# if (x.size()[0] < batch):
# continue
# for cur_prefix_len in range(1,cur_trace_len):
# # Separating event and timestamp
# y_truth_timestamp = y_truth[:, cur_prefix_len, 0].view(batch, 1, -1)
# y_truth_event = y_truth[:, cur_prefix_len, 1].view(batch, 1, -1)
# # Executing LSTM
# if cur_prefix_len == 1:
# x_inn = x[:, :cur_prefix_len, selected_columns]
# prev_event = x[:, :cur_prefix_len, len(selected_columns)+1].ravel().detach().numpy().astype("int")
# else:
# x_inn = x[:, :cur_prefix_len, selected_columns]
# rnnG = torch.load("checkpoints/"+dataset_name+"/event_timestamp_prediction/prefix_"+str(cur_prefix_len)+"/rnnG.m")
# rnnG.eval()
# y_pred = rnnG(x_inn[:, :cur_prefix_len, selected_columns])
# # Just taking the last predicted element from each the batch
# y_pred_last = y_pred[0: batch, cur_prefix_len - 1, :]
# y_pred_last = y_pred_last.view((batch, 1, -1))
# y_pred_last_event = torch.argmax(F.softmax(y_pred_last[:, :, events], dim=2), dim=2)
# #Storing list of predictions and corresponding ground truths (to be used for f1score)
# y_truth_list += list(y_truth_event.flatten().data.cpu().numpy().astype(int))
# y_pred_last_event_list += list(y_pred_last_event.flatten().data.cpu().numpy().astype(int))
# # checking dfg compliance for predicted event
# if not(int(prev_event[0]) == 0 and int(y_pred_last_event.detach()[0][0])==0):
# dfg_compliance_bool = check_dfg_compliance(prev_event , y_pred_last_event.detach(), dset=dataset_name)
# dfg_compliance_pred.append(int(dfg_compliance_bool))
# dfg_compliance_gt_bool = check_dfg_compliance(prev_event , y_truth_event.detach().reshape(y_pred_last_event.shape), dset=dataset_name)
# dfg_compliance_gt.append(int(dfg_compliance_gt_bool))
# if y_pred_last_event.flatten().data.cpu().numpy().astype(int)==y_truth_event.flatten().data.cpu().numpy().astype(int):
# accuracy.append(1)
# else:
# accuracy.append(0)
# # Computing MAE for the timestamp
# y_pred_timestamp = y_pred_last[:, :, len(events)].view((batch, 1, -1))
# mae.append(torch.abs(y_truth_timestamp - y_pred_timestamp).mean().detach())
# dur += max(y_pred_timestamp.detach().numpy()[0][0],0) #adding to total proccess duration, making sure y_pred is non-negative
# dur_gt += max(y_truth_timestamp.detach().numpy()[0][0],0)
# prev_event = y_pred_last_event.ravel().detach().numpy().astype("int")
# # GS cases
# try:
# if dur < thresh:
# num_overall_goal_satisfied += 1
# else:
# num_overall_goal_not_satisfied += 1
# if dur_gt < thresh:
# num_overall_goal_satisfied_gt += 1
# else:
# num_overall_goal_not_satisfied_gt += 1
# except Exception as e: print(e)
# dur = 0
# dur_gt = 0
# tot = num_overall_goal_satisfied+ num_overall_goal_not_satisfied
# gs = num_overall_goal_satisfied/tot
# gv = num_overall_goal_not_satisfied/tot
# tot_gt = num_overall_goal_satisfied_gt+ num_overall_goal_not_satisfied_gt
# gs_gt = num_overall_goal_satisfied_gt/tot_gt
# gv_gt = num_overall_goal_not_satisfied_gt/tot_gt
# print("compliance pred", np.mean(np.array(dfg_compliance_pred)))
# print("compliance gt", np.mean(np.array(dfg_compliance_gt)))
# print("gs ", gs)
# print("gv ", gv)
# print("gs gt", gs_gt)
# print("gv gt", gv_gt)
# print("acc ",np.mean(np.array(accuracy)))
# print("mae ", np.mean(np.array(mae)))
# datasets = ["traffic_ss"]
# for dataset in datasets:
# print("dataset: ",dataset)
# model_eval_test(dataset_name=dataset, csvwriter=None)
| 53
| 2,046
|
58bc10c9c9941a9f3f6a482f5cc8cd366a45d021
|
py
|
python
|
Code/model_part3.ipynb
|
yogeshwaran-shanmuganathan/Success-Prediction-Analysis-for-Startups
|
['Apache-2.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="F3pxxsK-ynO_"
# <p align="center">
# <img src="https://www.dbs.ie/images/default-source/logos/dbs-logo-2019-small.png" />
# </p>
# This code is submitted by Yogeshwaran Shanmuganathan as a part of my final dissertation or thesis for the completion of "Master of Science in Data Analytics" at Dublin Business School, Dublin, Ireland.
# + colab={"base_uri": "https://localhost:8080/"} id="cK10uzzbM-OF" outputId="09cf158e-88dc-4088-f724-e1e2ccf4b7d0"
# !pip install pandas
# !pip install numpy
# !pip install matplotlib
# !pip install pyspark
# + id="j3xTpH-jMyb9"
import pandas as pd
import numpy as np
# Load functionality to manipulate dataframes
from pyspark.sql import functions as fn
import matplotlib.pyplot as plt
from pyspark.sql.functions import stddev, mean, col
from pyspark.sql import SQLContext
from pyspark.sql import SparkSession
# Functionality for computing features
from pyspark.ml import feature, regression, classification, Pipeline
from pyspark.ml.classification import LogisticRegression
from pyspark.ml import feature, regression, classification, Pipeline
from pyspark.ml.feature import Tokenizer, VectorAssembler, HashingTF, Word2Vec, StringIndexer, OneHotEncoder
from pyspark.ml import clustering
from itertools import chain
from pyspark.ml.linalg import Vectors, VectorUDT
from pyspark.ml import classification
from pyspark.ml.classification import LogisticRegression, RandomForestClassifier, DecisionTreeClassifier, GBTClassifier
from pyspark.ml import evaluation
from pyspark.ml.evaluation import BinaryClassificationEvaluator,MulticlassClassificationEvaluator
from pyspark import keyword_only
from pyspark.ml import Transformer
from pyspark.ml.param.shared import HasInputCol, HasOutputCol, Param
#Classification Report
from sklearn.metrics import classification_report, confusion_matrix
# + colab={"resources": {"http://localhost:8080/nbextensions/google.colab/files.js": {"data": "Ly8gQ29weXJpZ2h0IDIwMTcgR29vZ2xlIExMQwovLwovLyBMaWNlbnNlZCB1bmRlciB0aGUgQXBhY2hlIExpY2Vuc2UsIFZlcnNpb24gMi4wICh0aGUgIkxpY2Vuc2UiKTsKLy8geW91IG1heSBub3QgdXNlIHRoaXMgZmlsZSBleGNlcHQgaW4gY29tcGxpYW5jZSB3aXRoIHRoZSBMaWNlbnNlLgovLyBZb3UgbWF5IG9idGFpbiBhIGNvcHkgb2YgdGhlIExpY2Vuc2UgYXQKLy8KLy8gICAgICBodHRwOi8vd3d3LmFwYWNoZS5vcmcvbGljZW5zZXMvTElDRU5TRS0yLjAKLy8KLy8gVW5sZXNzIHJlcXVpcmVkIGJ5IGFwcGxpY2FibGUgbGF3IG9yIGFncmVlZCB0byBpbiB3cml0aW5nLCBzb2Z0d2FyZQovLyBkaXN0cmlidXRlZCB1bmRlciB0aGUgTGljZW5zZSBpcyBkaXN0cmlidXRlZCBvbiBhbiAiQVMgSVMiIEJBU0lTLAovLyBXSVRIT1VUIFdBUlJBTlRJRVMgT1IgQ09ORElUSU9OUyBPRiBBTlkgS0lORCwgZWl0aGVyIGV4cHJlc3Mgb3IgaW1wbGllZC4KLy8gU2VlIHRoZSBMaWNlbnNlIGZvciB0aGUgc3BlY2lmaWMgbGFuZ3VhZ2UgZ292ZXJuaW5nIHBlcm1pc3Npb25zIGFuZAovLyBsaW1pdGF0aW9ucyB1bmRlciB0aGUgTGljZW5zZS4KCi8qKgogKiBAZmlsZW92ZXJ2aWV3IEhlbHBlcnMgZm9yIGdvb2dsZS5jb2xhYiBQeXRob24gbW9kdWxlLgogKi8KKGZ1bmN0aW9uKHNjb3BlKSB7CmZ1bmN0aW9uIHNwYW4odGV4dCwgc3R5bGVBdHRyaWJ1dGVzID0ge30pIHsKICBjb25zdCBlbGVtZW50ID0gZG9jdW1lbnQuY3JlYXRlRWxlbWVudCgnc3BhbicpOwogIGVsZW1lbnQudGV4dENvbnRlbnQgPSB0ZXh0OwogIGZvciAoY29uc3Qga2V5IG9mIE9iamVjdC5rZXlzKHN0eWxlQXR0cmlidXRlcykpIHsKICAgIGVsZW1lbnQuc3R5bGVba2V5XSA9IHN0eWxlQXR0cmlidXRlc1trZXldOwogIH0KICByZXR1cm4gZWxlbWVudDsKfQoKLy8gTWF4IG51bWJlciBvZiBieXRlcyB3aGljaCB3aWxsIGJlIHVwbG9hZGVkIGF0IGEgdGltZS4KY29uc3QgTUFYX1BBWUxPQURfU0laRSA9IDEwMCAqIDEwMjQ7CgpmdW5jdGlvbiBfdXBsb2FkRmlsZXMoaW5wdXRJZCwgb3V0cHV0SWQpIHsKICBjb25zdCBzdGVwcyA9IHVwbG9hZEZpbGVzU3RlcChpbnB1dElkLCBvdXRwdXRJZCk7CiAgY29uc3Qgb3V0cHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKG91dHB1dElkKTsKICAvLyBDYWNoZSBzdGVwcyBvbiB0aGUgb3V0cHV0RWxlbWVudCB0byBtYWtlIGl0IGF2YWlsYWJsZSBmb3IgdGhlIG5leHQgY2FsbAogIC8vIHRvIHVwbG9hZEZpbGVzQ29udGludWUgZnJvbSBQeXRob24uCiAgb3V0cHV0RWxlbWVudC5zdGVwcyA9IHN0ZXBzOwoKICByZXR1cm4gX3VwbG9hZEZpbGVzQ29udGludWUob3V0cHV0SWQpOwp9CgovLyBUaGlzIGlzIHJvdWdobHkgYW4gYXN5bmMgZ2VuZXJhdG9yIChub3Qgc3VwcG9ydGVkIGluIHRoZSBicm93c2VyIHlldCksCi8vIHdoZXJlIHRoZXJlIGFyZSBtdWx0aXBsZSBhc3luY2hyb25vdXMgc3RlcHMgYW5kIHRoZSBQeXRob24gc2lkZSBpcyBnb2luZwovLyB0byBwb2xsIGZvciBjb21wbGV0aW9uIG9mIGVhY2ggc3RlcC4KLy8gVGhpcyB1c2VzIGEgUHJvbWlzZSB0byBibG9jayB0aGUgcHl0aG9uIHNpZGUgb24gY29tcGxldGlvbiBvZiBlYWNoIHN0ZXAsCi8vIHRoZW4gcGFzc2VzIHRoZSByZXN1bHQgb2YgdGhlIHByZXZpb3VzIHN0ZXAgYXMgdGhlIGlucHV0IHRvIHRoZSBuZXh0IHN0ZXAuCmZ1bmN0aW9uIF91cGxvYWRGaWxlc0NvbnRpbnVlKG91dHB1dElkKSB7CiAgY29uc3Qgb3V0cHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKG91dHB1dElkKTsKICBjb25zdCBzdGVwcyA9IG91dHB1dEVsZW1lbnQuc3RlcHM7CgogIGNvbnN0IG5leHQgPSBzdGVwcy5uZXh0KG91dHB1dEVsZW1lbnQubGFzdFByb21pc2VWYWx1ZSk7CiAgcmV0dXJuIFByb21pc2UucmVzb2x2ZShuZXh0LnZhbHVlLnByb21pc2UpLnRoZW4oKHZhbHVlKSA9PiB7CiAgICAvLyBDYWNoZSB0aGUgbGFzdCBwcm9taXNlIHZhbHVlIHRvIG1ha2UgaXQgYXZhaWxhYmxlIHRvIHRoZSBuZXh0CiAgICAvLyBzdGVwIG9mIHRoZSBnZW5lcmF0b3IuCiAgICBvdXRwdXRFbGVtZW50Lmxhc3RQcm9taXNlVmFsdWUgPSB2YWx1ZTsKICAgIHJldHVybiBuZXh0LnZhbHVlLnJlc3BvbnNlOwogIH0pOwp9CgovKioKICogR2VuZXJhdG9yIGZ1bmN0aW9uIHdoaWNoIGlzIGNhbGxlZCBiZXR3ZWVuIGVhY2ggYXN5bmMgc3RlcCBvZiB0aGUgdXBsb2FkCiAqIHByb2Nlc3MuCiAqIEBwYXJhbSB7c3RyaW5nfSBpbnB1dElkIEVsZW1lbnQgSUQgb2YgdGhlIGlucHV0IGZpbGUgcGlja2VyIGVsZW1lbnQuCiAqIEBwYXJhbSB7c3RyaW5nfSBvdXRwdXRJZCBFbGVtZW50IElEIG9mIHRoZSBvdXRwdXQgZGlzcGxheS4KICogQHJldHVybiB7IUl0ZXJhYmxlPCFPYmplY3Q+fSBJdGVyYWJsZSBvZiBuZXh0IHN0ZXBzLgogKi8KZnVuY3Rpb24qIHVwbG9hZEZpbGVzU3RlcChpbnB1dElkLCBvdXRwdXRJZCkgewogIGNvbnN0IGlucHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKGlucHV0SWQpOwogIGlucHV0RWxlbWVudC5kaXNhYmxlZCA9IGZhbHNlOwoKICBjb25zdCBvdXRwdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQob3V0cHV0SWQpOwogIG91dHB1dEVsZW1lbnQuaW5uZXJIVE1MID0gJyc7CgogIGNvbnN0IHBpY2tlZFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgaW5wdXRFbGVtZW50LmFkZEV2ZW50TGlzdGVuZXIoJ2NoYW5nZScsIChlKSA9PiB7CiAgICAgIHJlc29sdmUoZS50YXJnZXQuZmlsZXMpOwogICAgfSk7CiAgfSk7CgogIGNvbnN0IGNhbmNlbCA9IGRvY3VtZW50LmNyZWF0ZUVsZW1lbnQoJ2J1dHRvbicpOwogIGlucHV0RWxlbWVudC5wYXJlbnRFbGVtZW50LmFwcGVuZENoaWxkKGNhbmNlbCk7CiAgY2FuY2VsLnRleHRDb250ZW50ID0gJ0NhbmNlbCB1cGxvYWQnOwogIGNvbnN0IGNhbmNlbFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgY2FuY2VsLm9uY2xpY2sgPSAoKSA9PiB7CiAgICAgIHJlc29sdmUobnVsbCk7CiAgICB9OwogIH0pOwoKICAvLyBXYWl0IGZvciB0aGUgdXNlciB0byBwaWNrIHRoZSBmaWxlcy4KICBjb25zdCBmaWxlcyA9IHlpZWxkIHsKICAgIHByb21pc2U6IFByb21pc2UucmFjZShbcGlja2VkUHJvbWlzZSwgY2FuY2VsUHJvbWlzZV0pLAogICAgcmVzcG9uc2U6IHsKICAgICAgYWN0aW9uOiAnc3RhcnRpbmcnLAogICAgfQogIH07CgogIGNhbmNlbC5yZW1vdmUoKTsKCiAgLy8gRGlzYWJsZSB0aGUgaW5wdXQgZWxlbWVudCBzaW5jZSBmdXJ0aGVyIHBpY2tzIGFyZSBub3QgYWxsb3dlZC4KICBpbnB1dEVsZW1lbnQuZGlzYWJsZWQgPSB0cnVlOwoKICBpZiAoIWZpbGVzKSB7CiAgICByZXR1cm4gewogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbXBsZXRlJywKICAgICAgfQogICAgfTsKICB9CgogIGZvciAoY29uc3QgZmlsZSBvZiBmaWxlcykgewogICAgY29uc3QgbGkgPSBkb2N1bWVudC5jcmVhdGVFbGVtZW50KCdsaScpOwogICAgbGkuYXBwZW5kKHNwYW4oZmlsZS5uYW1lLCB7Zm9udFdlaWdodDogJ2JvbGQnfSkpOwogICAgbGkuYXBwZW5kKHNwYW4oCiAgICAgICAgYCgke2ZpbGUudHlwZSB8fCAnbi9hJ30pIC0gJHtmaWxlLnNpemV9IGJ5dGVzLCBgICsKICAgICAgICBgbGFzdCBtb2RpZmllZDogJHsKICAgICAgICAgICAgZmlsZS5sYXN0TW9kaWZpZWREYXRlID8gZmlsZS5sYXN0TW9kaWZpZWREYXRlLnRvTG9jYWxlRGF0ZVN0cmluZygpIDoKICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgJ24vYSd9IC0gYCkpOwogICAgY29uc3QgcGVyY2VudCA9IHNwYW4oJzAlIGRvbmUnKTsKICAgIGxpLmFwcGVuZENoaWxkKHBlcmNlbnQpOwoKICAgIG91dHB1dEVsZW1lbnQuYXBwZW5kQ2hpbGQobGkpOwoKICAgIGNvbnN0IGZpbGVEYXRhUHJvbWlzZSA9IG5ldyBQcm9taXNlKChyZXNvbHZlKSA9PiB7CiAgICAgIGNvbnN0IHJlYWRlciA9IG5ldyBGaWxlUmVhZGVyKCk7CiAgICAgIHJlYWRlci5vbmxvYWQgPSAoZSkgPT4gewogICAgICAgIHJlc29sdmUoZS50YXJnZXQucmVzdWx0KTsKICAgICAgfTsKICAgICAgcmVhZGVyLnJlYWRBc0FycmF5QnVmZmVyKGZpbGUpOwogICAgfSk7CiAgICAvLyBXYWl0IGZvciB0aGUgZGF0YSB0byBiZSByZWFkeS4KICAgIGxldCBmaWxlRGF0YSA9IHlpZWxkIHsKICAgICAgcHJvbWlzZTogZmlsZURhdGFQcm9taXNlLAogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbnRpbnVlJywKICAgICAgfQogICAgfTsKCiAgICAvLyBVc2UgYSBjaHVua2VkIHNlbmRpbmcgdG8gYXZvaWQgbWVzc2FnZSBzaXplIGxpbWl0cy4gU2VlIGIvNjIxMTU2NjAuCiAgICBsZXQgcG9zaXRpb24gPSAwOwogICAgd2hpbGUgKHBvc2l0aW9uIDwgZmlsZURhdGEuYnl0ZUxlbmd0aCkgewogICAgICBjb25zdCBsZW5ndGggPSBNYXRoLm1pbihmaWxlRGF0YS5ieXRlTGVuZ3RoIC0gcG9zaXRpb24sIE1BWF9QQVlMT0FEX1NJWkUpOwogICAgICBjb25zdCBjaHVuayA9IG5ldyBVaW50OEFycmF5KGZpbGVEYXRhLCBwb3NpdGlvbiwgbGVuZ3RoKTsKICAgICAgcG9zaXRpb24gKz0gbGVuZ3RoOwoKICAgICAgY29uc3QgYmFzZTY0ID0gYnRvYShTdHJpbmcuZnJvbUNoYXJDb2RlLmFwcGx5KG51bGwsIGNodW5rKSk7CiAgICAgIHlpZWxkIHsKICAgICAgICByZXNwb25zZTogewogICAgICAgICAgYWN0aW9uOiAnYXBwZW5kJywKICAgICAgICAgIGZpbGU6IGZpbGUubmFtZSwKICAgICAgICAgIGRhdGE6IGJhc2U2NCwKICAgICAgICB9LAogICAgICB9OwogICAgICBwZXJjZW50LnRleHRDb250ZW50ID0KICAgICAgICAgIGAke01hdGgucm91bmQoKHBvc2l0aW9uIC8gZmlsZURhdGEuYnl0ZUxlbmd0aCkgKiAxMDApfSUgZG9uZWA7CiAgICB9CiAgfQoKICAvLyBBbGwgZG9uZS4KICB5aWVsZCB7CiAgICByZXNwb25zZTogewogICAgICBhY3Rpb246ICdjb21wbGV0ZScsCiAgICB9CiAgfTsKfQoKc2NvcGUuZ29vZ2xlID0gc2NvcGUuZ29vZ2xlIHx8IHt9OwpzY29wZS5nb29nbGUuY29sYWIgPSBzY29wZS5nb29nbGUuY29sYWIgfHwge307CnNjb3BlLmdvb2dsZS5jb2xhYi5fZmlsZXMgPSB7CiAgX3VwbG9hZEZpbGVzLAogIF91cGxvYWRGaWxlc0NvbnRpbnVlLAp9Owp9KShzZWxmKTsK", "headers": [["content-type", "application/javascript"]], "ok": true, "status": 200, "status_text": ""}}, "base_uri": "https://localhost:8080/", "height": 72} id="2Fl2cEb9O9YI" outputId="bfd1e6fe-bcd4-41d5-e4f6-7e1d2221afdb"
from google.colab import files
uploaded = files.upload()
# + id="TGAOtNcnPK50"
MAX_MEMORY = "45g"
spark = SparkSession \
.builder \
.appName("how to read csv file") \
.config("spark.executor.memory", MAX_MEMORY) \
.config("spark.driver.memory", MAX_MEMORY) \
.getOrCreate()
# + id="NBpeEo-0PXZP"
# load master dataset
dfmaster = spark.read.format("csv").load("master.csv", delimiter = ",", header = True)
# + [markdown] id="2LB5xRi2QFU2"
#
# Preparations and Understanding before Modeling
# + id="mIkm1LgHQCCk"
# create a 0/1 column for acquistions
dfmaster = dfmaster.\
withColumn("labelacq", fn.when(col("status") == "acquired","1").otherwise("0"))
# + colab={"base_uri": "https://localhost:8080/"} id="ZknlsZTQQIhG" outputId="6477bf55-482b-4307-b7a0-87987c27f255"
# number of rows in master table
print(dfmaster.count())
# + colab={"base_uri": "https://localhost:8080/"} id="b4V3vRukgnqD" outputId="fd232e7d-d43a-43ed-9e5f-9150c6b21042"
dfmaster
# + [markdown] id="gQ-F_CvoQKqZ"
# NAs and market column (with too many levels) handeling
# + colab={"base_uri": "https://localhost:8080/"} id="lQjuF2P-QNvT" outputId="61256d46-a9dc-4fb2-fa14-9a29b057f919"
# check for missing values
dfmaster.toPandas().isnull().sum()
# + id="AiaaUGk0QcQB"
# drop market columns because of too many level and better breakdown with the category_final column
dfmaster1 = dfmaster.drop("market")
# + colab={"base_uri": "https://localhost:8080/", "height": 560} id="kjOYJTFidt0c" outputId="95bc42c5-974f-4170-8284-e203b1d86180"
dfmaster1 = dfmaster1.toPandas()
dfmaster1
# + id="ASU37NVbh1LN"
# Replace NaN with mode for categorical variables
dfmaster1['total_raised_usd'] = dfmaster1['total_raised_usd'].fillna(dfmaster1['total_raised_usd'].mode()[0])
dfmaster1['time_to_first_funding'] = dfmaster1['time_to_first_funding'].fillna(dfmaster1['time_to_first_funding'].mode()[0])
dfmaster1['founded_year'] = dfmaster1['founded_year'].fillna(dfmaster1['founded_year'].mode()[0])
dfmaster1['age'] = dfmaster1['age'].fillna(dfmaster1['age'].mode()[0])
dfmaster1['status'] = dfmaster1['status'].fillna(dfmaster1['status'].mode()[0])
dfmaster1['country_code'] = dfmaster1['country_code'].fillna(dfmaster1['country_code'].mode()[0])
dfmaster1['city'] = dfmaster1['city'].fillna(dfmaster1['city'].mode()[0])
dfmaster1['quarter_new'] = dfmaster1['quarter_new'].fillna(dfmaster1['quarter_new'].mode()[0])
dfmaster1['investor_country_codes'] = dfmaster1['investor_country_codes'].fillna(dfmaster1['investor_country_codes'].mode()[0])
dfmaster1['funding_round_types'] = dfmaster1['funding_round_types'].fillna(dfmaster1['funding_round_types'].mode()[0])
dfmaster1['permaround'] = dfmaster1['permaround'].fillna(dfmaster1['permaround'].mode()[0])
dfmaster1['investor_country_code'] = dfmaster1['investor_country_code'].fillna(dfmaster1['investor_country_code'].mode()[0])
dfmaster1['funding_round_type'] = dfmaster1['funding_round_type'].fillna(dfmaster1['funding_round_type'].mode()[0])
dfmaster1['category_final'] = dfmaster1['category_final'].fillna(dfmaster1['category_final'].mode()[0])
dfmaster1['perma'] = dfmaster1['perma'].fillna(dfmaster1['perma'].mode()[0])
# + colab={"base_uri": "https://localhost:8080/"} id="wZyY7LbojbJ7" outputId="f8f06ddd-2d5d-49c2-d6b5-7b855d6ea20d"
# check for missing values
dfmaster1.isnull().sum()
# + id="9zAO9vMnp1JM"
# drop rows with missing values
dfmaster1drop = dfmaster1.dropna()
# + colab={"base_uri": "https://localhost:8080/"} id="yyf7mW3xqOWu" outputId="d72eecee-46d1-445e-c2a1-542dc1288a38"
print(dfmaster1drop.count())
# + id="4hQFr6Ymqsya"
sql = SQLContext(spark)
# + id="1aCy3ZSIrCsT"
dfmaster2 = sql.createDataFrame(dfmaster1drop)
# + colab={"base_uri": "https://localhost:8080/", "height": 53} id="bfy-lv0nrSAk" outputId="38bbee81-828c-4942-ab8d-da7b85321fd9"
display(dfmaster2)
# + [markdown] id="bBzVi9rNQmhv"
# String indexer, one hot encoder and casting to numerics
# + id="z3w-8St8QkEA"
# create index for categorical variables
# use pipline to apply indexer
list1 = ["country_code","city","quarter_new","investor_country_code","funding_round_type","category_final"]
indexers = [StringIndexer(inputCol=column, outputCol=column+"_index").fit(dfmaster2) for column in list1]
pipelineindex = Pipeline(stages=indexers).fit(dfmaster2)
dfmasternew = pipelineindex.transform(dfmaster2)
# + id="ofJqkkyJQqyL"
# convert string to double for numerical variables
dfmasternew = dfmasternew.\
withColumn("numeric_funding_rounds", dfmasternew["funding_rounds"].cast("int")).\
withColumn("numeric_age", dfmasternew["age"].cast("int")).\
withColumn("numeric_count_investor", dfmasternew["count_investor"].cast("int")).\
withColumn("numeric_time_to_first_funding", dfmasternew["time_to_first_funding"].cast("int")).\
withColumn("numeric_total_raised_usd", dfmasternew["total_raised_usd"].cast("int")).\
withColumn("label", dfmasternew["labelacq"].cast("int"))
# + id="v_vGMKkl2vor"
dfmasternew = dfmasternew.\
withColumn("funding_round_type", dfmasternew["funding_round_type"].cast("double")).\
withColumn("country_code_index", dfmasternew["country_code_index"].cast("double")).\
withColumn("city_index", dfmasternew["city_index"].cast("double")).\
withColumn("quarter_new_index", dfmasternew["quarter_new_index"].cast("double")).\
withColumn("labelacq", dfmasternew["labelacq"].cast("double"))
# + id="M9frd6bGQ0h7"
# save
dfone = dfmasternew
# + colab={"base_uri": "https://localhost:8080/", "height": 53} id="waO0Uf_aQ5Fr" outputId="c10f3599-1aa9-4c13-a4e6-25403b56a9d4"
display(dfone)
# + colab={"base_uri": "https://localhost:8080/"} id="b22orTWg_Ep8" outputId="e291d5d5-597c-4143-cd9b-0bfd2dfed9bf"
print(dfone.count())
# + colab={"base_uri": "https://localhost:8080/"} id="z-ocjzD4Q6Yb" outputId="98733877-cdad-43a8-96d3-cf4456342ebd"
# list of index columns of categorical variables for the onehotencoder
list2 = dfone.columns[24:30]
list2
# + id="fbl2eaDKRFwz"
# create sparse matrix of indexed categorical columns
# use pipline to apply the encoder
onehotencoder_stages = [OneHotEncoder(inputCol=c, outputCol='onehotencoded_' + c) for c in list2]
pipelineonehot = Pipeline(stages=onehotencoder_stages)
pipeline_mode = pipelineonehot.fit(dfone)
df_coded = pipeline_mode.transform(dfone)
# + colab={"base_uri": "https://localhost:8080/"} id="18vlEgxt3och" outputId="01f30982-5912-4f2a-da80-036fd69a073f"
df_coded.show()
# + [markdown] id="FUEDzSq13rsG"
#
# Data split, defining vector assemblers & standard scaler and creating labellist
# + id="ZcsTHOZr3vTD"
# split dataset into training, validaiton and testing dataset
training_df, validation_df, testing_df = df_coded.randomSplit([0.6, 0.3, 0.1])
# + colab={"base_uri": "https://localhost:8080/"} id="o9LmQ1xN30F9" outputId="28d8af0a-54ca-472b-e9df-e00acc75f7b3"
training_df.columns[30:35]
# + colab={"base_uri": "https://localhost:8080/"} id="ezy5qwM84oTe" outputId="471f799e-e2d2-4e72-c942-aab1db4a2166"
training_df.columns[36:42]
# + id="oUeZlYsO5Haw"
# define vector assembler with the features for the modelling
vanum = VectorAssembler(). \
setInputCols(training_df.columns[30:35]). \
setOutputCol('features_nonstd')
# + id="oTA1QXoP5LpN"
# define vector assembler with the features for the modelling
vacate = VectorAssembler(). \
setInputCols(training_df.columns[36:42]). \
setOutputCol('featurescate')
# + id="U38mX6ex5OIV"
va = VectorAssembler(). \
setInputCols(['featuresnum','featurescate']). \
setOutputCol('features')
# + id="WXfCTaHu5QkA"
std = feature.StandardScaler(withMean=True, withStd=True).setInputCol('features_nonstd').setOutputCol('featuresnum')
# + id="SSogkbno5UBy"
# suffix for investor country code because intersection with county_code of the companies
invcc = ['{}_{}'.format(a, "investor") for a in indexers[3].labels]
# + id="Xh_gzS3f5XDD"
# define labellist by using the indexer stages for displaying the weights & loadings
labellist = training_df.columns[30:35] + indexers[0].labels + indexers[1].labels + indexers[2].labels + invcc + indexers[4].labels + indexers[5].labels
# + colab={"base_uri": "https://localhost:8080/"} id="wi6qlUeU5hq-" outputId="38c3d097-5b91-4abc-b6c7-fa790f4b75c6"
# null dummy for onehotencoded_country_code_index
print("null dummy for onehotencoded_country_code_index")
print(len(indexers[0].labels))
print(indexers[0].labels)
# null dummy for onehotencoded_city_index
print("null dummy for onehotencoded_city_index")
print(indexers[1].labels)
print(len(indexers[1].labels))
# null dummy for onehotencoded_quarter_new_index
print("null dummy for onehotencoded_quarter_new_index")
print(len(indexers[2].labels))
print(indexers[2].labels)
# null dummy for onehotencoded_investor_country_code_index
print("null dummy for onehotencoded_investor_country_code_index")
print(len(invcc))
print(invcc)
# null dummy for onehotencoded_funding_round_type_index
print("null dummy for onehotencoded_funding_round_type_index")
print(len(indexers[4].labels))
print(indexers[4].labels)
# null dummy for onehotencoded_category_final_index
print("null dummy for onehotencoded_category_final_index")
print(len(indexers[5].labels))
print(indexers[5].labels)
# + [markdown] id="jP7-JY6C6OVS"
# # MODELLING
# + [markdown] id="m1ikKpVyiVmN"
# ## Gradient Boosted Trees
# + id="0cmI4oeBiS3X"
gbt = GBTClassifier(labelCol="label", featuresCol="features", maxIter=250)
# + id="KO-7-tGYjKNT"
gbt_pipeline = Pipeline(stages=[vanum, std, vacate, va, gbt]).fit(training_df)
# + id="vbRAxNZcjPFW"
dfgbt = gbt_pipeline.transform(validation_df)
# + id="a9mmJQ-twWUb"
# define multiclass classification evaluator
mce = MulticlassClassificationEvaluator()
# + colab={"base_uri": "https://localhost:8080/"} id="XcWwPOTRjVs0" outputId="1212ac2a-7ddf-4225-a240-c9c78b9ee078"
# print the areas under the curve for the gradient boosted trees model pipeline
print("Gradient Boosted Trees: AUC = {}".format(mce.evaluate(dfgbt)))
# + colab={"base_uri": "https://localhost:8080/"} id="NFdJDJmRjcWV" outputId="8f812222-4517-47ea-c430-2a5d7277c56a"
# print the accuracies for the gradient boosted trees model pipeline
print(dfgbt.select(fn.expr('float(label = prediction)').alias('correct')).select(fn.avg('correct').alias("Accuracy for Gradient Boosted Tree")).show())
# + [markdown] id="5-IW7cEq1u01"
# # Neural Networks
# + [markdown] id="orCgUTvB11fo"
# ### Standard Model
# + id="19k4FqWt10TG"
# define neural networks (MultilayerPerceptron) classifier
mlp = classification.MultilayerPerceptronClassifier(seed=0).\
setStepSize(0.2).\
setMaxIter(200).\
setLayers([4399, 2]).\
setFeaturesCol('features')
# + id="vFKTs6YG1-Mg"
# define and fit neural network pipeline
mlp_simple_model = Pipeline(stages=[vanum, std, vacate, va, mlp]).fit(training_df)
# + id="LhuLx08c2BAa"
# define evaluators for accuracy and area under the curve
evaluator = MulticlassClassificationEvaluator().setMetricName("accuracy")
evaluatorauc = MulticlassClassificationEvaluator()
# + id="M7P_g5cx2Db7"
# apply pipeline to validation dataset
dfnn = mlp_simple_model.transform(validation_df)
# + colab={"base_uri": "https://localhost:8080/"} id="6h36no5A4pqP" outputId="3c846327-9910-4ed6-9645-6ed18ec2cc85"
# print accuracy and area under the curve for NN
print(evaluator.evaluate(dfnn))
print(evaluatorauc.evaluate(dfnn))
# + [markdown] id="2yduyVQ42NBu"
# ### Models with Hidden Layers
# + id="q13r9KOk2KTw"
# define neural networks (MultilayerPerceptron) classifier with hidden layers
mlp2 = classification.MultilayerPerceptronClassifier(seed=0).\
setStepSize(0.2).\
setMaxIter(200).\
setFeaturesCol('features').\
setLayers([4399,30,30, 2])
mlp3 = classification.MultilayerPerceptronClassifier(seed=0).\
setStepSize(0.2).\
setMaxIter(200).\
setFeaturesCol('features').\
setLayers([4399,10,10, 2])
mlp4 = classification.MultilayerPerceptronClassifier(seed=0).\
setStepSize(0.2).\
setMaxIter(200).\
setFeaturesCol('features').\
setLayers([4399,20,20, 2])
mlp5 = classification.MultilayerPerceptronClassifier(seed=0).\
setStepSize(0.2).\
setMaxIter(200).\
setFeaturesCol('features').\
setLayers([4399,30, 2])
mlp6 = classification.MultilayerPerceptronClassifier(seed=0).\
setStepSize(0.2).\
setMaxIter(200).\
setFeaturesCol('features').\
setLayers([4399,30,30,30, 2])
# + colab={"background_save": true} id="RNAzkxe92VF4"
# define and fit pipeline
mlp2_model = Pipeline(stages=[vanum, std, vacate, va, mlp2]).fit(training_df)
mlp3_model = Pipeline(stages=[vanum, std, vacate, va, mlp3]).fit(training_df)
mlp4_model = Pipeline(stages=[vanum, std, vacate, va, mlp4]).fit(training_df)
mlp5_model = Pipeline(stages=[vanum, std, vacate, va, mlp5]).fit(training_df)
mlp6_model = Pipeline(stages=[vanum, std, vacate, va, mlp6]).fit(training_df)
# + colab={"background_save": true} id="Gtm8Kufq2X5l"
# apply and fit model to validation dataset
dfnn2 = mlp2_model.transform(validation_df)
dfnn3 = mlp3_model.transform(validation_df)
dfnn4 = mlp4_model.transform(validation_df)
dfnn5 = mlp5_model.transform(validation_df)
dfnn6 = mlp6_model.transform(validation_df)
# + [markdown] id="dAIZweU72cDZ"
# ## Performance
# + colab={"background_save": true} id="JPIXR8hd2bmw" outputId="18f89ab5-a5bf-4a7a-b3e8-cb2adefa69c2"
# print accuracy and area under the curve
print("NN Model with 2 hidden layers and 30 neurons each: Accuracy = {}".format(evaluator.evaluate(dfnn2)))
print("NN Model with 2 hidden layers and 30 neurons each: AUC = {}".format(evaluatorauc.evaluate(dfnn2)))
print("____________________________________________________________________________")
print("NN Model with 2 hidden layers and 10 neurons each: Accuracy = {}".format(evaluator.evaluate(dfnn3)))
print("NN Model with 2 hidden layers and 10 neurons each: AUC = {}".format(evaluatorauc.evaluate(dfnn3)))
print("____________________________________________________________________________")
print("NN Model with 2 hidden layers and 20 neurons each: Accuracy = {}".format(evaluator.evaluate(dfnn4)))
print("NN Model with 2 hidden layers and 20 neurons each: AUC = {}".format(evaluatorauc.evaluate(dfnn4)))
print("____________________________________________________________________________")
print("NN Model with 1 hidden layer and 30 neurons: Accuracy = {}".format(evaluator.evaluate(dfnn5)))
print("NN Model with 1 hidden layer and 30 neurons: AUC = {}".format(evaluatorauc.evaluate(dfnn5)))
print("____________________________________________________________________________")
print("NN Model with 3 hidden layers and 30 neurons each: Accuracy = {}".format(evaluator.evaluate(dfnn6)))
print("NN Model with 3 hidden layers and 30 neurons each: AUC = {}".format(evaluatorauc.evaluate(dfnn6)))
# + [markdown] id="HHKK4XQn6YLb"
# # TESTING PERFORMANCE
# + [markdown] id="CHlbqkc56b5i"
# We tested with the best performing (AUC and Accuracy) Gradient Bossted Trees and Neural Network Model.
# + colab={"background_save": true} id="YVp1Ug4h6Vk5" outputId="dbba1e9c-0123-43a1-dc3c-c4de96be318f"
# Gradient Boosted Trees Classifier
dfgbt_test = gbt_pipeline.transform(testing_df)
print("Gradient Boosted Trees: AUC = {}".format(mce.evaluate(dfgbt_test)))
print(dfgbt_test.select(fn.expr('float(label = prediction)').alias('correct')).select(fn.avg('correct').alias("Accuracy for Gradient Boosted Trees")).show())
# + colab={"background_save": true} id="ArlJEAHiyptt" outputId="b2642a10-bb9c-47c8-a87b-f43cdd1b2fd8"
gbt_true = dfgbt_test.select(['label']).collect()
gbt_pred = dfgbt_test.select(['prediction']).collect()
print(classification_report(gbt_true, gbt_pred))
# + colab={"background_save": true} id="rlCDzogTyvxl" outputId="5f73fd30-09b9-4eb8-b725-d01257da384f"
# Best performing neural network model
dfnntest = mlp6_model.transform(testing_df)
print("Neural Network Model with 3 hidden layers and 30 neurons each: Accuracy = {}".format(evaluator.evaluate(dfnntest)))
print("Neural Network Model with 3 hidden layers and 30 neurons each: AUC = {}".format(evaluatorauc.evaluate(dfnntest)))
# + colab={"background_save": true} id="MysDwf_syy8K" outputId="565e3d3e-0056-45eb-8830-8f13dc094e66"
nn_true = dfnntest.select(['label']).collect()
nn_pred = dfnntest.select(['prediction']).collect()
print(classification_report(nn_true, nn_pred))
| 60.739759
| 7,233
|
4ace7617a92e8920c0c0083b682c523a051e88f5
|
py
|
python
|
Big-Data-Clusters/CU9/public/content/cert-management/cer043-install-master-certs.ipynb
|
WilliamAntonRohm/tigertoolbox
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] tags=[]
# # CER043 - Install signed Master certificates
#
# This notebook installs into the Big Data Cluster the certificates signed
# using:
#
# - [CER033 - Sign Master certificates with generated
# CA](../cert-management/cer033-sign-master-generated-certs.ipynb)
#
# ## Steps
#
# ### Parameters
# + tags=["parameters"]
app_name = "master"
scaledset_name = "master"
container_name = "mssql-server"
common_name = "master-svc"
user = "mssql"
group = "mssql"
mode = "550"
prefix_keyfile_name = "sql"
certificate_names = {"master-0" : "master-0-certificate.pem", "master-1" : "master-1-certificate.pem", "master-2" : "master-2-certificate.pem"}
key_names = {"master-0" : "master-0-privatekey.pem", "master-1" : "master-1-privatekey.pem", "master-2" : "master-2-privatekey.pem"}
test_cert_store_root = "/var/opt/secrets/test-certificates"
timeout = 600 # amount of time to wait before cluster is healthy: default to 10 minutes
check_interval = 10 # amount of time between health checks - default 10 seconds
min_pod_count = 10 # minimum number of healthy pods required to assert health
# + [markdown] tags=[]
# ### Common functions
#
# Define helper functions used in this notebook.
# + tags=["hide_input"]
# Define `run` function for transient fault handling, suggestions on error, and scrolling updates on Windows
import sys
import os
import re
import platform
import shlex
import shutil
import datetime
from subprocess import Popen, PIPE
from IPython.display import Markdown
retry_hints = {} # Output in stderr known to be transient, therefore automatically retry
error_hints = {} # Output in stderr where a known SOP/TSG exists which will be HINTed for further help
install_hint = {} # The SOP to help install the executable if it cannot be found
def run(cmd, return_output=False, no_output=False, retry_count=0, base64_decode=False, return_as_json=False):
"""Run shell command, stream stdout, print stderr and optionally return output
NOTES:
1. Commands that need this kind of ' quoting on Windows e.g.:
kubectl get nodes -o jsonpath={.items[?(@.metadata.annotations.pv-candidate=='data-pool')].metadata.name}
Need to actually pass in as '"':
kubectl get nodes -o jsonpath={.items[?(@.metadata.annotations.pv-candidate=='"'data-pool'"')].metadata.name}
The ' quote approach, although correct when pasting into Windows cmd, will hang at the line:
`iter(p.stdout.readline, b'')`
The shlex.split call does the right thing for each platform, just use the '"' pattern for a '
"""
MAX_RETRIES = 5
output = ""
retry = False
# When running `azdata sql query` on Windows, replace any \n in """ strings, with " ", otherwise we see:
#
# ('HY090', '[HY090] [Microsoft][ODBC Driver Manager] Invalid string or buffer length (0) (SQLExecDirectW)')
#
if platform.system() == "Windows" and cmd.startswith("azdata sql query"):
cmd = cmd.replace("\n", " ")
# shlex.split is required on bash and for Windows paths with spaces
#
cmd_actual = shlex.split(cmd)
# Store this (i.e. kubectl, python etc.) to support binary context aware error_hints and retries
#
user_provided_exe_name = cmd_actual[0].lower()
# When running python, use the python in the ADS sandbox ({sys.executable})
#
if cmd.startswith("python "):
cmd_actual[0] = cmd_actual[0].replace("python", sys.executable)
# On Mac, when ADS is not launched from terminal, LC_ALL may not be set, which causes pip installs to fail
# with:
#
# UnicodeDecodeError: 'ascii' codec can't decode byte 0xc5 in position 4969: ordinal not in range(128)
#
# Setting it to a default value of "en_US.UTF-8" enables pip install to complete
#
if platform.system() == "Darwin" and "LC_ALL" not in os.environ:
os.environ["LC_ALL"] = "en_US.UTF-8"
# When running `kubectl`, if AZDATA_OPENSHIFT is set, use `oc`
#
if cmd.startswith("kubectl ") and "AZDATA_OPENSHIFT" in os.environ:
cmd_actual[0] = cmd_actual[0].replace("kubectl", "oc")
# To aid supportability, determine which binary file will actually be executed on the machine
#
which_binary = None
# Special case for CURL on Windows. The version of CURL in Windows System32 does not work to
# get JWT tokens, it returns "(56) Failure when receiving data from the peer". If another instance
# of CURL exists on the machine use that one. (Unfortunately the curl.exe in System32 is almost
# always the first curl.exe in the path, and it can't be uninstalled from System32, so here we
# look for the 2nd installation of CURL in the path)
if platform.system() == "Windows" and cmd.startswith("curl "):
path = os.getenv('PATH')
for p in path.split(os.path.pathsep):
p = os.path.join(p, "curl.exe")
if os.path.exists(p) and os.access(p, os.X_OK):
if p.lower().find("system32") == -1:
cmd_actual[0] = p
which_binary = p
break
# Find the path based location (shutil.which) of the executable that will be run (and display it to aid supportability), this
# seems to be required for .msi installs of azdata.cmd/az.cmd. (otherwise Popen returns FileNotFound)
#
# NOTE: Bash needs cmd to be the list of the space separated values hence shlex.split.
#
if which_binary == None:
which_binary = shutil.which(cmd_actual[0])
# Display an install HINT, so the user can click on a SOP to install the missing binary
#
if which_binary == None:
print(f"The path used to search for '{cmd_actual[0]}' was:")
print(sys.path)
if user_provided_exe_name in install_hint and install_hint[user_provided_exe_name] is not None:
display(Markdown(f'HINT: Use [{install_hint[user_provided_exe_name][0]}]({install_hint[user_provided_exe_name][1]}) to resolve this issue.'))
raise FileNotFoundError(f"Executable '{cmd_actual[0]}' not found in path (where/which)")
else:
cmd_actual[0] = which_binary
start_time = datetime.datetime.now().replace(microsecond=0)
print(f"START: {cmd} @ {start_time} ({datetime.datetime.utcnow().replace(microsecond=0)} UTC)")
print(f" using: {which_binary} ({platform.system()} {platform.release()} on {platform.machine()})")
print(f" cwd: {os.getcwd()}")
# Command-line tools such as CURL and AZDATA HDFS commands output
# scrolling progress bars, which causes Jupyter to hang forever, to
# workaround this, use no_output=True
#
# Work around a infinite hang when a notebook generates a non-zero return code, break out, and do not wait
#
wait = True
try:
if no_output:
p = Popen(cmd_actual)
else:
p = Popen(cmd_actual, stdout=PIPE, stderr=PIPE, bufsize=1)
with p.stdout:
for line in iter(p.stdout.readline, b''):
line = line.decode()
if return_output:
output = output + line
else:
if cmd.startswith("azdata notebook run"): # Hyperlink the .ipynb file
regex = re.compile(' "(.*)"\: "(.*)"')
match = regex.match(line)
if match:
if match.group(1).find("HTML") != -1:
display(Markdown(f' - "{match.group(1)}": "{match.group(2)}"'))
else:
display(Markdown(f' - "{match.group(1)}": "[{match.group(2)}]({match.group(2)})"'))
wait = False
break # otherwise infinite hang, have not worked out why yet.
else:
print(line, end='')
if wait:
p.wait()
except FileNotFoundError as e:
if install_hint is not None:
display(Markdown(f'HINT: Use {install_hint} to resolve this issue.'))
raise FileNotFoundError(f"Executable '{cmd_actual[0]}' not found in path (where/which)") from e
exit_code_workaround = 0 # WORKAROUND: azdata hangs on exception from notebook on p.wait()
if not no_output:
for line in iter(p.stderr.readline, b''):
try:
line_decoded = line.decode()
except UnicodeDecodeError:
# NOTE: Sometimes we get characters back that cannot be decoded(), e.g.
#
# \xa0
#
# For example see this in the response from `az group create`:
#
# ERROR: Get Token request returned http error: 400 and server
# response: {"error":"invalid_grant",# "error_description":"AADSTS700082:
# The refresh token has expired due to inactivity.\xa0The token was
# issued on 2018-10-25T23:35:11.9832872Z
#
# which generates the exception:
#
# UnicodeDecodeError: 'utf-8' codec can't decode byte 0xa0 in position 179: invalid start byte
#
print("WARNING: Unable to decode stderr line, printing raw bytes:")
print(line)
line_decoded = ""
pass
else:
# azdata emits a single empty line to stderr when doing an hdfs cp, don't
# print this empty "ERR:" as it confuses.
#
if line_decoded == "":
continue
print(f"STDERR: {line_decoded}", end='')
if line_decoded.startswith("An exception has occurred") or line_decoded.startswith("ERROR: An error occurred while executing the following cell"):
exit_code_workaround = 1
# inject HINTs to next TSG/SOP based on output in stderr
#
if user_provided_exe_name in error_hints:
for error_hint in error_hints[user_provided_exe_name]:
if line_decoded.find(error_hint[0]) != -1:
display(Markdown(f'HINT: Use [{error_hint[1]}]({error_hint[2]}) to resolve this issue.'))
# Verify if a transient error, if so automatically retry (recursive)
#
if user_provided_exe_name in retry_hints:
for retry_hint in retry_hints[user_provided_exe_name]:
if line_decoded.find(retry_hint) != -1:
if retry_count < MAX_RETRIES:
print(f"RETRY: {retry_count} (due to: {retry_hint})")
retry_count = retry_count + 1
output = run(cmd, return_output=return_output, retry_count=retry_count)
if return_output:
if base64_decode:
import base64
return base64.b64decode(output).decode('utf-8')
else:
return output
elapsed = datetime.datetime.now().replace(microsecond=0) - start_time
# WORKAROUND: We avoid infinite hang above in the `azdata notebook run` failure case, by inferring success (from stdout output), so
# don't wait here, if success known above
#
if wait:
if p.returncode != 0:
raise SystemExit(f'Shell command:\n\n\t{cmd} ({elapsed}s elapsed)\n\nreturned non-zero exit code: {str(p.returncode)}.\n')
else:
if exit_code_workaround !=0 :
raise SystemExit(f'Shell command:\n\n\t{cmd} ({elapsed}s elapsed)\n\nreturned non-zero exit code: {str(exit_code_workaround)}.\n')
print(f'\nSUCCESS: {elapsed}s elapsed.\n')
if return_output:
if base64_decode:
import base64
return base64.b64decode(output).decode('utf-8')
else:
return output
# Hints for tool retry (on transient fault), known errors and install guide
#
retry_hints = {'azdata': ['Endpoint sql-server-master does not exist', 'Endpoint livy does not exist', 'Failed to get state for cluster', 'Endpoint webhdfs does not exist', 'Adaptive Server is unavailable or does not exist', 'Error: Address already in use', 'Login timeout expired (0) (SQLDriverConnect)', 'SSPI Provider: No Kerberos credentials available', ], 'kubectl': ['A connection attempt failed because the connected party did not properly respond after a period of time, or established connection failed because connected host has failed to respond', ], 'python': [ ], }
error_hints = {'azdata': [['Please run \'azdata login\' to first authenticate', 'SOP028 - azdata login', '../common/sop028-azdata-login.ipynb'], ['The token is expired', 'SOP028 - azdata login', '../common/sop028-azdata-login.ipynb'], ['Reason: Unauthorized', 'SOP028 - azdata login', '../common/sop028-azdata-login.ipynb'], ['Max retries exceeded with url: /api/v1/bdc/endpoints', 'SOP028 - azdata login', '../common/sop028-azdata-login.ipynb'], ['Look at the controller logs for more details', 'TSG027 - Observe cluster deployment', '../diagnose/tsg027-observe-bdc-create.ipynb'], ['provided port is already allocated', 'TSG062 - Get tail of all previous container logs for pods in BDC namespace', '../log-files/tsg062-tail-bdc-previous-container-logs.ipynb'], ['Create cluster failed since the existing namespace', 'SOP061 - Delete a big data cluster', '../install/sop061-delete-bdc.ipynb'], ['Failed to complete kube config setup', 'TSG067 - Failed to complete kube config setup', '../repair/tsg067-failed-to-complete-kube-config-setup.ipynb'], ['Data source name not found and no default driver specified', 'SOP069 - Install ODBC for SQL Server', '../install/sop069-install-odbc-driver-for-sql-server.ipynb'], ['Can\'t open lib \'ODBC Driver 17 for SQL Server', 'SOP069 - Install ODBC for SQL Server', '../install/sop069-install-odbc-driver-for-sql-server.ipynb'], ['Control plane upgrade failed. Failed to upgrade controller.', 'TSG108 - View the controller upgrade config map', '../diagnose/tsg108-controller-failed-to-upgrade.ipynb'], ['NameError: name \'azdata_login_secret_name\' is not defined', 'SOP013 - Create secret for azdata login (inside cluster)', '../common/sop013-create-secret-for-azdata-login.ipynb'], ['ERROR: No credentials were supplied, or the credentials were unavailable or inaccessible.', 'TSG124 - \'No credentials were supplied\' error from azdata login', '../repair/tsg124-no-credentials-were-supplied.ipynb'], ['Please accept the license terms to use this product through', 'TSG126 - azdata fails with \'accept the license terms to use this product\'', '../repair/tsg126-accept-license-terms.ipynb'], ], 'kubectl': [['no such host', 'TSG010 - Get configuration contexts', '../monitor-k8s/tsg010-get-kubernetes-contexts.ipynb'], ['No connection could be made because the target machine actively refused it', 'TSG056 - Kubectl fails with No connection could be made because the target machine actively refused it', '../repair/tsg056-kubectl-no-connection-could-be-made.ipynb'], ], 'python': [['Library not loaded: /usr/local/opt/unixodbc', 'SOP012 - Install unixodbc for Mac', '../install/sop012-brew-install-odbc-for-sql-server.ipynb'], ['WARNING: You are using pip version', 'SOP040 - Upgrade pip in ADS Python sandbox', '../install/sop040-upgrade-pip.ipynb'], ], }
install_hint = {'azdata': [ 'SOP063 - Install azdata CLI (using package manager)', '../install/sop063-packman-install-azdata.ipynb' ], 'kubectl': [ 'SOP036 - Install kubectl command line interface', '../install/sop036-install-kubectl.ipynb' ], }
print('Common functions defined successfully.')
# + [markdown] tags=[]
# ### Get the Kubernetes namespace for the big data cluster
#
# Get the namespace of the Big Data Cluster use the kubectl command line
# interface .
#
# **NOTE:**
#
# If there is more than one Big Data Cluster in the target Kubernetes
# cluster, then either:
#
# - set \[0\] to the correct value for the big data cluster.
# - set the environment variable AZDATA_NAMESPACE, before starting Azure
# Data Studio.
# + tags=["hide_input"]
# Place Kubernetes namespace name for BDC into 'namespace' variable
if "AZDATA_NAMESPACE" in os.environ:
namespace = os.environ["AZDATA_NAMESPACE"]
else:
try:
namespace = run(f'kubectl get namespace --selector=MSSQL_CLUSTER -o jsonpath={{.items[0].metadata.name}}', return_output=True)
except:
from IPython.display import Markdown
print(f"ERROR: Unable to find a Kubernetes namespace with label 'MSSQL_CLUSTER'. SQL Server Big Data Cluster Kubernetes namespaces contain the label 'MSSQL_CLUSTER'.")
display(Markdown(f'HINT: Use [TSG081 - Get namespaces (Kubernetes)](../monitor-k8s/tsg081-get-kubernetes-namespaces.ipynb) to resolve this issue.'))
display(Markdown(f'HINT: Use [TSG010 - Get configuration contexts](../monitor-k8s/tsg010-get-kubernetes-contexts.ipynb) to resolve this issue.'))
display(Markdown(f'HINT: Use [SOP011 - Set kubernetes configuration context](../common/sop011-set-kubernetes-context.ipynb) to resolve this issue.'))
raise
print(f'The SQL Server Big Data Cluster Kubernetes namespace is: {namespace}')
# + [markdown] tags=[]
# ### Create a temporary directory to stage files
# + tags=["hide_input"]
# Create a temporary directory to hold configuration files
import tempfile
temp_dir = tempfile.mkdtemp()
print(f"Temporary directory created: {temp_dir}")
# + [markdown] tags=[]
# ### Helper function to save configuration files to disk
# + tags=["hide_input"]
# Define helper function 'save_file' to save configuration files to the temporary directory created above
import os
import io
def save_file(filename, contents):
with io.open(os.path.join(temp_dir, filename), "w", encoding='utf8', newline='\n') as text_file:
text_file.write(contents)
print("File saved: " + os.path.join(temp_dir, filename))
print("Function `save_file` defined successfully.")
# + [markdown] tags=[]
# ### Instantiate Kubernetes client
# + tags=["hide_input"]
# Instantiate the Python Kubernetes client into 'api' variable
import os
from IPython.display import Markdown
try:
from kubernetes import client, config
from kubernetes.stream import stream
except ImportError:
# Install the Kubernetes module
import sys
# !{sys.executable} -m pip install kubernetes
try:
from kubernetes import client, config
from kubernetes.stream import stream
except ImportError:
display(Markdown(f'HINT: Use [SOP059 - Install Kubernetes Python module](../install/sop059-install-kubernetes-module.ipynb) to resolve this issue.'))
raise
if "KUBERNETES_SERVICE_PORT" in os.environ and "KUBERNETES_SERVICE_HOST" in os.environ:
config.load_incluster_config()
else:
try:
config.load_kube_config()
except:
display(Markdown(f'HINT: Use [TSG118 - Configure Kubernetes config](../repair/tsg118-configure-kube-config.ipynb) to resolve this issue.'))
raise
api = client.CoreV1Api()
print('Kubernetes client instantiated')
# + [markdown] tags=[]
# ### Helper functions for waiting for the cluster to become healthy
# + tags=["hide_input"]
import threading
import time
import sys
import os
from IPython.display import Markdown
isRunning = True
def all_containers_ready(pod):
"""helper method returns true if all the containers within the given pod are ready
Arguments:
pod {v1Pod} -- Metadata retrieved from the api call to.
"""
return all(map(lambda c: c.ready is True, pod.status.container_statuses))
def pod_is_ready(pod):
"""tests that the pod, and all containers are ready
Arguments:
pod {v1Pod} -- Metadata retrieved from api call.
"""
return "job-name" in pod.metadata.labels or (pod.status.phase == "Running" and all_containers_ready(pod))
def waitReady():
"""Waits for all pods, and containers to become ready.
"""
while isRunning:
try:
time.sleep(check_interval)
pods = get_pods()
allReady = len(pods.items) >= min_pod_count and all(map(pod_is_ready, pods.items))
if allReady:
return True
else:
display(Markdown(get_pod_failures(pods)))
display(Markdown(f"cluster not healthy, rechecking in {check_interval} seconds."))
except Exception as ex:
last_error_message = str(ex)
display(Markdown(last_error_message))
time.sleep(check_interval)
def get_pod_failures(pods=None):
"""Returns a status message for any pods that are not ready.
"""
results = ""
if not pods:
pods = get_pods()
for pod in pods.items:
if "job-name" not in pod.metadata.labels:
if pod.status and pod.status.container_statuses:
for container in filter(lambda c: c.ready is False, pod.status.container_statuses):
results = results + "Container {0} in Pod {1} is not ready. Reported status: {2} <br/>".format(container.name, pod.metadata.name, container.state)
else:
results = results + "Pod {0} is not ready. <br/>".format(pod.metadata.name)
return results
def get_pods():
"""Returns a list of pods by namespace, or all namespaces if no namespace is specified
"""
pods = None
if namespace is not None:
display(Markdown(f'Checking namespace {namespace}'))
pods = api.list_namespaced_pod(namespace, _request_timeout=30)
else:
display(Markdown('Checking all namespaces'))
pods = api.list_pod_for_all_namespaces(_request_timeout=30)
return pods
def wait_for_cluster_healthy():
isRunning = True
mt = threading.Thread(target=waitReady)
mt.start()
mt.join(timeout=timeout)
if mt.is_alive():
raise SystemExit("Timeout waiting for all cluster to be healthy.")
isRunning = False
# + [markdown] tags=[]
# ### Get name of the ‘Running’ `controller` `pod`
# + tags=["hide_input"]
# Place the name of the 'Running' controller pod in variable `controller`
controller = run(f'kubectl get pod --selector=app=controller -n {namespace} -o jsonpath={{.items[0].metadata.name}} --field-selector=status.phase=Running', return_output=True)
print(f"Controller pod name: {controller}")
# + [markdown] tags=[]
# ### Get the name of the `master` `pods`
# + tags=["hide_input"]
# Place the name of the master pods in variable `pods`
podNames = run(f'kubectl get pod --selector=app=master -n {namespace} -o jsonpath={{.items[*].metadata.name}}', return_output=True)
pods = podNames.split(" ")
print(f"Master pod names: {pods}")
# + [markdown] tags=[]
# ### Validate certificate common name and alt names
# + tags=[]
import json
from urllib.parse import urlparse
kubernetes_default_record_name = 'kubernetes.default'
kubernetes_default_svc_prefix = 'kubernetes.default.svc'
default_dns_suffix = 'svc.cluster.local'
dns_suffix = ''
nslookup_output=run(f'kubectl exec {controller} -c controller -n {namespace} -- bash -c "nslookup {kubernetes_default_record_name} > /tmp/nslookup.out; cat /tmp/nslookup.out; rm /tmp/nslookup.out" ', return_output=True)
name = re.findall('Name:\s+(.[^,|^\s|^\n]+)', nslookup_output)
if not name or kubernetes_default_svc_prefix not in name[0]:
dns_suffix = default_dns_suffix
else:
dns_suffix = 'svc' + name[0].replace(kubernetes_default_svc_prefix, '')
pods.sort()
for pod_name in pods:
alt_names = ""
bdc_fqdn = ""
alt_names += f"DNS.1 = {common_name}\n"
alt_names += f"DNS.2 = {common_name}.{namespace}.{dns_suffix} \n"
hdfs_vault_svc = "hdfsvault-svc"
bdc_config = run("azdata bdc config show", return_output=True)
bdc_config = json.loads(bdc_config)
dns_counter = 3 # DNS.1 and DNS.2 are already in the certificate template
# Stateful set related DNS names
#
if app_name == "gateway" or app_name == "master":
alt_names += f'DNS.{str(dns_counter)} = {pod_name}.{common_name}\n'
dns_counter = dns_counter + 1
alt_names += f'DNS.{str(dns_counter)} = {pod_name}.{common_name}.{namespace}.{dns_suffix}\n'
dns_counter = dns_counter + 1
# AD related DNS names
#
if "security" in bdc_config["spec"] and "activeDirectory" in bdc_config["spec"]["security"]:
domain_dns_name = bdc_config["spec"]["security"]["activeDirectory"]["domainDnsName"]
subdomain_name = bdc_config["spec"]["security"]["activeDirectory"]["subdomain"]
if subdomain_name:
bdc_fqdn = f"{subdomain_name}.{domain_dns_name}"
else:
bdc_fqdn = f"{namespace}.{domain_dns_name}"
alt_names += f"DNS.{str(dns_counter)} = {common_name}.{bdc_fqdn}\n"
dns_counter = dns_counter + 1
if app_name == "gateway" or app_name == "master":
alt_names += f'DNS.{str(dns_counter)} = {pod_name}.{bdc_fqdn}\n'
dns_counter = dns_counter + 1
# Endpoint DNS names for bdc certificates
#
if app_name in bdc_config["spec"]["resources"]:
app_name_endpoints = bdc_config["spec"]["resources"][app_name]["spec"]["endpoints"]
for endpoint in app_name_endpoints:
if "dnsName" in endpoint:
alt_names += f'DNS.{str(dns_counter)} = {endpoint["dnsName"]}\n'
dns_counter = dns_counter + 1
# Endpoint DNS names for control plane certificates
#
if app_name == "controller" or app_name == "mgmtproxy":
bdc_endpoint_list = run("azdata bdc endpoint list", return_output=True)
bdc_endpoint_list = json.loads(bdc_endpoint_list)
# Parse the DNS host name from:
#
# "endpoint": "https://monitor.aris.local:30777"
#
for endpoint in bdc_endpoint_list:
if endpoint["name"] == app_name:
url = urlparse(endpoint["endpoint"])
alt_names += f"DNS.{str(dns_counter)} = {url.hostname}\n"
dns_counter = dns_counter + 1
# Special case for the controller certificate
#
if app_name == "controller":
alt_names += f"DNS.{str(dns_counter)} = localhost\n"
dns_counter = dns_counter + 1
# Add hdfsvault-svc host for key management calls.
#
alt_names += f"DNS.{str(dns_counter)} = {hdfs_vault_svc}\n"
dns_counter = dns_counter + 1
# Add hdfsvault-svc FQDN for key management calls.
#
if bdc_fqdn:
alt_names += f"DNS.{str(dns_counter)} = {hdfs_vault_svc}.{bdc_fqdn}\n"
dns_counter = dns_counter + 1
required_dns_names = re.findall('DNS\.[0-9] = ([^,|^\s|^\n]+)', alt_names)
# Get certificate common name and DNS names
# use nameopt compat, to generate CN= format on all versions of openssl
#
cert = run(f'kubectl exec {controller} -c controller -n {namespace} -- openssl x509 -nameopt compat -in {test_cert_store_root}/{app_name}/{certificate_names[pod_name]} -text -noout', return_output=True)
subject = re.findall('Subject:(.+)', cert)[0]
certficate_common_name = re.findall('CN=(.[^,|^\s|^\n]+)', subject)[0]
certficate_dns_names = re.findall('DNS:(.[^,|^\s|^\n]+)', cert)
# Validate the common name
#
if (common_name != certficate_common_name):
run(f'kubectl exec {controller} -c controller -n {namespace} -- bash -c "rm -rf {test_cert_store_root}/{app_name}"')
raise SystemExit(f'Certficate common name does not match the expected one: {common_name}')
# Validate the DNS names
#
if not all(dns_name in certficate_dns_names for dns_name in required_dns_names):
run(f'kubectl exec {controller} -c controller -n {namespace} -- bash -c "rm -rf {test_cert_store_root}/{app_name}"')
raise SystemExit(f'Certficate does not have all required DNS names: {required_dns_names}')
# + [markdown] tags=[]
# ### Copy certifcate files from `controller` to local machine
# + tags=[]
import os
cwd = os.getcwd()
os.chdir(temp_dir) # Use chdir to workaround kubectl bug on Windows, which incorrectly processes 'c:\' on kubectl cp cmd line
for pod_name in pods:
run(f'kubectl cp {controller}:{test_cert_store_root}/{app_name}/{certificate_names[pod_name]} {certificate_names[pod_name]} -c controller -n {namespace}')
run(f'kubectl cp {controller}:{test_cert_store_root}/{app_name}/{key_names[pod_name]} {key_names[pod_name]} -c controller -n {namespace}')
os.chdir(cwd)
# + [markdown] tags=[]
# ### Copy certifcate files from local machine to `controldb`
# + tags=[]
import os
cwd = os.getcwd()
os.chdir(temp_dir) # Workaround kubectl bug on Windows, can't put c:\ on kubectl cp cmd line
for pod_name in pods:
run(f'kubectl cp {certificate_names[pod_name]} controldb-0:/var/opt/mssql/{certificate_names[pod_name]} -c mssql-server -n {namespace}')
run(f'kubectl cp {key_names[pod_name]} controldb-0:/var/opt/mssql/{key_names[pod_name]} -c mssql-server -n {namespace}')
os.chdir(cwd)
# + [markdown] tags=[]
# ### Get the `controller-db-rw-secret` secret
#
# Get the controller SQL symmetric key password for decryption.
# + tags=[]
import base64
controller_db_rw_secret = run(f'kubectl get secret/controller-db-rw-secret -n {namespace} -o jsonpath={{.data.encryptionPassword}}', return_output=True)
controller_db_rw_secret = base64.b64decode(controller_db_rw_secret).decode('utf-8')
print("controller_db_rw_secret retrieved")
# + [markdown] tags=[]
# ### Update the files table with the certificates through opened SQL connection
# + tags=[]
import os
sql = f"""
OPEN SYMMETRIC KEY ControllerDbSymmetricKey DECRYPTION BY PASSWORD = '{controller_db_rw_secret}'
DECLARE @FileData VARBINARY(MAX), @Key uniqueidentifier;
SELECT @Key = KEY_GUID('ControllerDbSymmetricKey');
"""
for pod_name in pods:
insert = f"""
SELECT TOP 1 @FileData = doc.BulkColumn FROM OPENROWSET(BULK N'/var/opt/mssql/{certificate_names[pod_name]}', SINGLE_BLOB) AS doc;
EXEC [dbo].[sp_set_file_data_encrypted] @FilePath = '/config/scaledsets/{scaledset_name}/pods/{pod_name}/containers/{container_name}/files/{prefix_keyfile_name}-certificate.pem',
@Data = @FileData,
@KeyGuid = @Key,
@Version = '0',
@User = '{user}',
@Group = '{group}',
@Mode = '{mode}';
SELECT TOP 1 @FileData = doc.BulkColumn FROM OPENROWSET(BULK N'/var/opt/mssql/{key_names[pod_name]}', SINGLE_BLOB) AS doc;
EXEC [dbo].[sp_set_file_data_encrypted] @FilePath = '/config/scaledsets/{scaledset_name}/pods/{pod_name}/containers/{container_name}/files/{prefix_keyfile_name}-privatekey.pem',
@Data = @FileData,
@KeyGuid = @Key,
@Version = '0',
@User = '{user}',
@Group = '{group}',
@Mode = '{mode}';
"""
sql += insert
save_file("insert_certificates.sql", sql)
cwd = os.getcwd()
os.chdir(temp_dir) # Workaround kubectl bug on Windows, can't put c:\ on kubectl cp cmd line
run(f'kubectl cp insert_certificates.sql controldb-0:/var/opt/mssql/insert_certificates.sql -c mssql-server -n {namespace}')
run(f"""kubectl exec controldb-0 -c mssql-server -n {namespace} -- bash -c "SQLCMDPASSWORD=`cat /var/run/secrets/credentials/mssql-sa-password/password` /opt/mssql-tools/bin/sqlcmd -b -U sa -d controller -i /var/opt/mssql/insert_certificates.sql" """)
# Clean up
run(f"""kubectl exec controldb-0 -c mssql-server -n {namespace} -- bash -c "rm /var/opt/mssql/insert_certificates.sql" """)
for pod_name in pods:
run(f"""kubectl exec controldb-0 -c mssql-server -n {namespace} -- bash -c "rm /var/opt/mssql/{certificate_names[pod_name]}" """)
run(f"""kubectl exec controldb-0 -c mssql-server -n {namespace} -- bash -c "rm /var/opt/mssql/{key_names[pod_name]}" """)
os.chdir(cwd)
# + [markdown] tags=[]
# ### Clear out the controller_db_rw_secret variable
# + tags=[]
controller_db_rw_secret= ""
# + [markdown] tags=[]
# ### Get the name of the `master` `pods`
# + tags=["hide_input"]
# Place the name of the master pods in variable `pods`
podNames = run(f'kubectl get pod --selector=app=master -n {namespace} -o jsonpath={{.items[*].metadata.name}}', return_output=True)
pods = podNames.split(" ")
print(f"Master pod names: {pods}")
# + [markdown] tags=[]
# ### Restart Pods
# + tags=[]
import threading
import time
if len(pods) == 1:
# One master pod indicates non-HA environment, just delete it
run(f'kubectl delete pod {pods[0]} -n {namespace}')
wait_for_cluster_healthy()
else:
# HA setup, delete secondaries before primary
timeout_s = 300
check_interval_s = 20
master_primary_svc_ip = run(f'kubectl get service master-p-svc -n {namespace} -o jsonpath={{.spec.clusterIP}}', return_output=True)
master_password = run(f'kubectl exec master-0 -c mssql-server -n {namespace} -- cat /var/run/secrets/credentials/pool/mssql-system-password', return_output=True)
def get_number_of_unsynchronized_replicas(result):
cmd = 'select count(*) from sys.dm_hadr_database_replica_states where synchronization_state <> 2'
res = run(f"kubectl exec controldb-0 -c mssql-server -n {namespace} -- /opt/mssql-tools/bin/sqlcmd -S {master_primary_svc_ip} -U system -P {master_password} -h -1 -q \"SET NOCOUNT ON; {cmd}\" ", return_output=True)
rows = res.strip().split("\n")
result[0] = int(rows[0])
return True
def get_primary_replica():
cmd = 'select distinct replica_server_name from sys.dm_hadr_database_replica_states s join sys.availability_replicas r on s.replica_id = r.replica_id where is_primary_replica = 1'
res = run(f"kubectl exec controldb-0 -c mssql-server -n {namespace} -- /opt/mssql-tools/bin/sqlcmd -S {master_primary_svc_ip} -U system -P {master_password} -h -1 -q \"SET NOCOUNT ON; {cmd}\" ", return_output=True)
rows = res.strip().split("\n")
return rows[0]
def get_secondary_replicas():
cmd = 'select distinct replica_server_name from sys.dm_hadr_database_replica_states s join sys.availability_replicas r on s.replica_id = r.replica_id where is_primary_replica = 0'
res = run(f"kubectl exec controldb-0 -c mssql-server -n {namespace} -- /opt/mssql-tools/bin/sqlcmd -S {master_primary_svc_ip} -U system -P {master_password} -h -1 -q \"SET NOCOUNT ON; {cmd}\" ", return_output=True)
rows = res.strip().split("\n")
res = []
for row in rows:
if (row != "" and "Sqlcmd: Warning" not in row):
res.append(row.strip())
return res
def all_replicas_syncrhonized():
while True:
unsynchronized_replicas_cnt = len(pods)
rows = [None]
time.sleep(check_interval_s)
getNumberOfReplicasThread = threading.Thread(target=get_number_of_unsynchronized_replicas, args=(rows,) )
getNumberOfReplicasThread.start()
getNumberOfReplicasThread.join(timeout=timeout_s)
if getNumberOfReplicasThread.is_alive():
raise SystemExit("Timeout getting the number of unsynchronized replicas.")
unsynchronized_replicas_cnt = rows[0]
if (unsynchronized_replicas_cnt == 0):
return True
def wait_for_replicas_to_synchronize():
waitForReplicasToSynchronizeThread = threading.Thread(target=all_replicas_syncrhonized)
waitForReplicasToSynchronizeThread.start()
waitForReplicasToSynchronizeThread.join(timeout=timeout_s)
if waitForReplicasToSynchronizeThread.is_alive():
raise SystemExit("Timeout waiting for all replicas to be synchronized.")
secondary_replicas = get_secondary_replicas()
for replica in secondary_replicas:
wait_for_replicas_to_synchronize()
run(f'kubectl delete pod {replica} -n {namespace}')
primary_replica = get_primary_replica()
wait_for_replicas_to_synchronize()
key = "/var/run/secrets/certificates/sqlha/mssql-ha-operator-controller-client/mssql-ha-operator-controller-client-privatekey.pem"
cert = "/var/run/secrets/certificates/sqlha/mssql-ha-operator-controller-client/mssql-ha-operator-controller-client-certificate.pem"
content_type_header = "Content-Type: application/json"
authorization_header = "Authorization: Certificate"
data = f'{{"TargetReplicaName":"{secondary_replicas[0]}","ForceFailover":"false"}}'
request_url = f'https://controller-svc:443/internal/api/v1/bdc/services/sql/resources/master/availabilitygroups/containedag/failover'
manual_failover_api_command = f"curl -sS --key {key} --cert {cert} -X POST --header '{content_type_header}' --header '{authorization_header}' --data '{data}' {request_url}"
operator_pod = run(f'kubectl get pod --selector=app=mssql-operator -n {namespace} -o jsonpath={{.items[0].metadata.name}}', return_output=True)
run(f'kubectl exec {operator_pod} -c mssql-ha-operator -n {namespace} -- {manual_failover_api_command}')
wait_for_replicas_to_synchronize()
run(f'kubectl delete pod {primary_replica} -n {namespace}')
wait_for_replicas_to_synchronize()
# + [markdown] tags=[]
# ### Clean up certificate staging area
#
# Remove the certificate files generated on disk (they have now been
# placed in the controller database).
# + tags=[]
cmd = f"rm -r {test_cert_store_root}/{app_name}"
run(f'kubectl exec {controller} -c controller -n {namespace} -- bash -c "{cmd}"')
# + [markdown] tags=[]
# ### Clean up temporary directory for staging configuration files
# + tags=["hide_input"]
# Delete the temporary directory used to hold configuration files
import shutil
shutil.rmtree(temp_dir)
print(f'Temporary directory deleted: {temp_dir}')
# + tags=[]
print("Notebook execution is complete.")
# + [markdown] tags=[]
# Related
# -------
#
# - [CER023 - Create Master certificates](../cert-management/cer023-create-master-certs.ipynb)
# - [CER033 - Sign Master certificates with generated CA](../cert-management/cer033-sign-master-generated-certs.ipynb)
# - [CER044 - Install signed Controller certificate](../cert-management/cer044-install-controller-cert.ipynb)
#
| 42.11804
| 2,802
|
929144a1da7d3be60b986e1a973a63ae8db80cfa
|
py
|
python
|
scripts/rock_paper_scissors_cnn.ipynb
|
arryaaas/Machine-Learning-Image-Classifier-CNN
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# # Rock, Paper, Scissors Classification
#
# Mochammad Arya Salsabila / Learn Machine Learning for Beginners
# ### Download dataset
# + colab={"base_uri": "https://localhost:8080/"} id="UwMxBe-iHKSA" outputId="d050419d-e83c-4e69-f90f-4f51e4378278"
# !wget --no-check-certificate \
# https://github.com/dicodingacademy/assets/releases/download/release/rockpaperscissors.zip \
# -O /tmp/rockpaperscissors.zip
# -
# ### Extract dataset
# + id="4nsAvLJYIrGj"
import os
import zipfile
local_zip = "/tmp/rockpaperscissors.zip"
zip_ref = zipfile.ZipFile(local_zip, "r")
zip_ref.extractall("/tmp")
zip_ref.close()
# -
# ### See the contents of the dataset
# + colab={"base_uri": "https://localhost:8080/"} id="LqE4BIeWJEBP" outputId="f35193b9-d132-4089-a683-f6ed5788f551"
os.listdir("/tmp/rockpaperscissors")
# -
# ### See the number of datasets for each class
# + colab={"base_uri": "https://localhost:8080/"} id="t272N41hJJ8d" outputId="b420ddd7-0632-4267-e7d0-bd7a5962da40"
print("Number of Rock images: {}".format(len(os.listdir("/tmp/rockpaperscissors/rock"))))
print("Number of Paper images: {}".format(len(os.listdir("/tmp/rockpaperscissors/paper"))))
print("Number of Scissors images: {}".format(len(os.listdir("/tmp/rockpaperscissors/scissors"))))
# -
# ### Create directory
# + id="jVsFKWIbJciD"
base_dir = "/tmp/rockpaperscissors"
train_dir = os.path.join(base_dir, "train")
validation_dir = os.path.join(base_dir, "validation")
train_rock_dir = os.path.join(train_dir, "rock")
train_paper_dir = os.path.join(train_dir, "paper")
train_scissors_dir = os.path.join(train_dir, "scissors")
validation_rock_dir = os.path.join(validation_dir, "rock")
validation_paper_dir = os.path.join(validation_dir, "paper")
validation_scissors_dir = os.path.join(validation_dir, "scissors")
try:
os.mkdir(train_dir)
os.mkdir(train_rock_dir)
os.mkdir(train_paper_dir)
os.mkdir(train_scissors_dir)
os.mkdir(validation_dir)
os.mkdir(validation_rock_dir)
os.mkdir(validation_paper_dir)
os.mkdir(validation_scissors_dir)
except OSError as error:
print(error)
# -
# ### Split dataset
# + id="PEdNsKY_LYGV"
import shutil
from sklearn.model_selection import train_test_split
def copy_files(train_or_validation_size, source_dir, destination_dir):
for i in train_or_validation_size:
shutil.copy(os.path.join(source_dir, i), os.path.join(destination_dir, i))
rock_dir = os.path.join(base_dir, "rock")
paper_dir = os.path.join(base_dir, "paper")
scissors_dir = os.path.join(base_dir, "scissors")
train_rock_size, validation_rock_size = train_test_split(os.listdir(rock_dir), test_size = 0.4)
train_paper_size, validation_paper_size = train_test_split(os.listdir(paper_dir), test_size = 0.4)
train_scissors_size, validation_scissors_size = train_test_split(os.listdir(scissors_dir), test_size = 0.4)
copy_files(train_rock_size, rock_dir, train_rock_dir)
copy_files(train_paper_size, paper_dir, train_paper_dir)
copy_files(train_scissors_size, scissors_dir, train_scissors_dir)
copy_files(validation_rock_size, rock_dir, validation_rock_dir)
copy_files(validation_paper_size, paper_dir, validation_paper_dir)
copy_files(validation_scissors_size, scissors_dir, validation_scissors_dir)
# -
# ### Image augmentation
# + id="ewGDGfJgZkzj"
import tensorflow as tf
train_datagen = tf.keras.preprocessing.image.ImageDataGenerator(
rescale = 1./255,
rotation_range = 30,
width_shift_range = 0.1,
height_shift_range = 0.2,
shear_range = 0.2,
zoom_range = 0.2,
horizontal_flip = True,
fill_mode = "nearest"
)
validation_datagen = tf.keras.preprocessing.image.ImageDataGenerator(
rescale = 1./255
)
# -
# ### Load training and validation data into memory
# + colab={"base_uri": "https://localhost:8080/"} id="dOPQb0HJbRwM" outputId="60330f9d-5675-4236-ec66-aa3b8c092b3e"
train_generator = train_datagen.flow_from_directory(
train_dir,
target_size = (150, 150),
batch_size = 64,
class_mode = "categorical"
)
validation_generator = validation_datagen.flow_from_directory(
validation_dir,
target_size = (150, 150),
batch_size = 64,
class_mode = "categorical"
)
# -
# ### Build a Convolutional Neural Network (CNN) model
# + colab={"base_uri": "https://localhost:8080/"} id="REzPegv1bePv" outputId="7b787193-10af-45b1-d6d9-53208dd4b5a1"
model = tf.keras.models.Sequential([
tf.keras.layers.Conv2D(32, (3,3), padding="same", activation="relu", input_shape=(150, 150, 3)),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(64,(3,3), padding="same", activation="relu"),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(128,(3,3), padding="same", activation="relu"),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(256,(3,3), padding="same", activation="relu"),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Flatten(),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Dense(512, activation="relu"),
tf.keras.layers.Dense(3, activation="softmax")
])
model.summary()
# -
# ### Compile model
# + id="O_hIQKCQbsDj"
model.compile(
loss = 'categorical_crossentropy',
optimizer = tf.optimizers.Adam(),
metrics = ['accuracy']
)
# -
# ### Create callbacks
# + id="1lKVvthHeQ8Y"
reduce_learning_rate = tf.keras.callbacks.ReduceLROnPlateau(
monitor = "val_loss",
factor = 0.2,
patience = 5,
verbose = 0,
mode = "auto",
min_lr = 1.5e-5,
)
early_stopping = tf.keras.callbacks.EarlyStopping(
monitor = "val_loss",
patience = 10,
verbose = 0,
mode = "auto",
baseline = None,
restore_best_weights=True
)
# -
# ### Train model
# + colab={"base_uri": "https://localhost:8080/"} id="yXlXqD48cb7q" outputId="345dfee8-c956-46a9-8d7a-4cc4d2077b5b"
STEP_PER_EPOCH = train_generator.n // train_generator.batch_size
VALIDATION_STEPS = validation_generator.n // validation_generator.batch_size
history = model.fit(
train_generator,
steps_per_epoch = STEP_PER_EPOCH,
epochs = 100,
validation_data = validation_generator,
validation_steps = VALIDATION_STEPS,
verbose = 1,
callbacks = [reduce_learning_rate, early_stopping]
)
# -
# ### Visualize the history of the model
# + colab={"base_uri": "https://localhost:8080/", "height": 352} id="vTqSSmARgsMT" outputId="4770773b-eaea-42d9-ef5f-a63c15c8956a"
import matplotlib.pyplot as plt
acc = history.history["accuracy"]
val_acc = history.history["val_accuracy"]
loss = history.history["loss"]
val_loss = history.history["val_loss"]
plt.style.use("seaborn")
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(15, 5))
ax[0].plot(acc, label="Training Accuracy")
ax[0].plot(val_acc, label="Validation Accuracy")
ax[0].legend(loc="lower right")
ax[0].set_title("Training and Validation Accuracy", fontsize=16)
ax[0].set_xlabel("Epoch")
ax[0].set_ylabel("Accuracy")
ax[1].plot(loss, label="Training Loss")
ax[1].plot(val_loss, label="Validation Loss")
ax[1].legend(loc="upper right")
ax[1].set_title("Training and Validation Loss", fontsize=16)
ax[1].set_xlabel("Epoch")
ax[1].set_ylabel("Loss")
plt.show()
# -
# ### Save model
# + id="Y1UHcQgwjSZ_"
model.save_weights("model_weights.h5")
model.save("model.h5")
# -
# ### Predict image
# + colab={"base_uri": "https://localhost:8080/", "height": 441, "resources": {"http://localhost:8080/nbextensions/google.colab/files.js": {"data": "Ly8gQ29weXJpZ2h0IDIwMTcgR29vZ2xlIExMQwovLwovLyBMaWNlbnNlZCB1bmRlciB0aGUgQXBhY2hlIExpY2Vuc2UsIFZlcnNpb24gMi4wICh0aGUgIkxpY2Vuc2UiKTsKLy8geW91IG1heSBub3QgdXNlIHRoaXMgZmlsZSBleGNlcHQgaW4gY29tcGxpYW5jZSB3aXRoIHRoZSBMaWNlbnNlLgovLyBZb3UgbWF5IG9idGFpbiBhIGNvcHkgb2YgdGhlIExpY2Vuc2UgYXQKLy8KLy8gICAgICBodHRwOi8vd3d3LmFwYWNoZS5vcmcvbGljZW5zZXMvTElDRU5TRS0yLjAKLy8KLy8gVW5sZXNzIHJlcXVpcmVkIGJ5IGFwcGxpY2FibGUgbGF3IG9yIGFncmVlZCB0byBpbiB3cml0aW5nLCBzb2Z0d2FyZQovLyBkaXN0cmlidXRlZCB1bmRlciB0aGUgTGljZW5zZSBpcyBkaXN0cmlidXRlZCBvbiBhbiAiQVMgSVMiIEJBU0lTLAovLyBXSVRIT1VUIFdBUlJBTlRJRVMgT1IgQ09ORElUSU9OUyBPRiBBTlkgS0lORCwgZWl0aGVyIGV4cHJlc3Mgb3IgaW1wbGllZC4KLy8gU2VlIHRoZSBMaWNlbnNlIGZvciB0aGUgc3BlY2lmaWMgbGFuZ3VhZ2UgZ292ZXJuaW5nIHBlcm1pc3Npb25zIGFuZAovLyBsaW1pdGF0aW9ucyB1bmRlciB0aGUgTGljZW5zZS4KCi8qKgogKiBAZmlsZW92ZXJ2aWV3IEhlbHBlcnMgZm9yIGdvb2dsZS5jb2xhYiBQeXRob24gbW9kdWxlLgogKi8KKGZ1bmN0aW9uKHNjb3BlKSB7CmZ1bmN0aW9uIHNwYW4odGV4dCwgc3R5bGVBdHRyaWJ1dGVzID0ge30pIHsKICBjb25zdCBlbGVtZW50ID0gZG9jdW1lbnQuY3JlYXRlRWxlbWVudCgnc3BhbicpOwogIGVsZW1lbnQudGV4dENvbnRlbnQgPSB0ZXh0OwogIGZvciAoY29uc3Qga2V5IG9mIE9iamVjdC5rZXlzKHN0eWxlQXR0cmlidXRlcykpIHsKICAgIGVsZW1lbnQuc3R5bGVba2V5XSA9IHN0eWxlQXR0cmlidXRlc1trZXldOwogIH0KICByZXR1cm4gZWxlbWVudDsKfQoKLy8gTWF4IG51bWJlciBvZiBieXRlcyB3aGljaCB3aWxsIGJlIHVwbG9hZGVkIGF0IGEgdGltZS4KY29uc3QgTUFYX1BBWUxPQURfU0laRSA9IDEwMCAqIDEwMjQ7CgpmdW5jdGlvbiBfdXBsb2FkRmlsZXMoaW5wdXRJZCwgb3V0cHV0SWQpIHsKICBjb25zdCBzdGVwcyA9IHVwbG9hZEZpbGVzU3RlcChpbnB1dElkLCBvdXRwdXRJZCk7CiAgY29uc3Qgb3V0cHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKG91dHB1dElkKTsKICAvLyBDYWNoZSBzdGVwcyBvbiB0aGUgb3V0cHV0RWxlbWVudCB0byBtYWtlIGl0IGF2YWlsYWJsZSBmb3IgdGhlIG5leHQgY2FsbAogIC8vIHRvIHVwbG9hZEZpbGVzQ29udGludWUgZnJvbSBQeXRob24uCiAgb3V0cHV0RWxlbWVudC5zdGVwcyA9IHN0ZXBzOwoKICByZXR1cm4gX3VwbG9hZEZpbGVzQ29udGludWUob3V0cHV0SWQpOwp9CgovLyBUaGlzIGlzIHJvdWdobHkgYW4gYXN5bmMgZ2VuZXJhdG9yIChub3Qgc3VwcG9ydGVkIGluIHRoZSBicm93c2VyIHlldCksCi8vIHdoZXJlIHRoZXJlIGFyZSBtdWx0aXBsZSBhc3luY2hyb25vdXMgc3RlcHMgYW5kIHRoZSBQeXRob24gc2lkZSBpcyBnb2luZwovLyB0byBwb2xsIGZvciBjb21wbGV0aW9uIG9mIGVhY2ggc3RlcC4KLy8gVGhpcyB1c2VzIGEgUHJvbWlzZSB0byBibG9jayB0aGUgcHl0aG9uIHNpZGUgb24gY29tcGxldGlvbiBvZiBlYWNoIHN0ZXAsCi8vIHRoZW4gcGFzc2VzIHRoZSByZXN1bHQgb2YgdGhlIHByZXZpb3VzIHN0ZXAgYXMgdGhlIGlucHV0IHRvIHRoZSBuZXh0IHN0ZXAuCmZ1bmN0aW9uIF91cGxvYWRGaWxlc0NvbnRpbnVlKG91dHB1dElkKSB7CiAgY29uc3Qgb3V0cHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKG91dHB1dElkKTsKICBjb25zdCBzdGVwcyA9IG91dHB1dEVsZW1lbnQuc3RlcHM7CgogIGNvbnN0IG5leHQgPSBzdGVwcy5uZXh0KG91dHB1dEVsZW1lbnQubGFzdFByb21pc2VWYWx1ZSk7CiAgcmV0dXJuIFByb21pc2UucmVzb2x2ZShuZXh0LnZhbHVlLnByb21pc2UpLnRoZW4oKHZhbHVlKSA9PiB7CiAgICAvLyBDYWNoZSB0aGUgbGFzdCBwcm9taXNlIHZhbHVlIHRvIG1ha2UgaXQgYXZhaWxhYmxlIHRvIHRoZSBuZXh0CiAgICAvLyBzdGVwIG9mIHRoZSBnZW5lcmF0b3IuCiAgICBvdXRwdXRFbGVtZW50Lmxhc3RQcm9taXNlVmFsdWUgPSB2YWx1ZTsKICAgIHJldHVybiBuZXh0LnZhbHVlLnJlc3BvbnNlOwogIH0pOwp9CgovKioKICogR2VuZXJhdG9yIGZ1bmN0aW9uIHdoaWNoIGlzIGNhbGxlZCBiZXR3ZWVuIGVhY2ggYXN5bmMgc3RlcCBvZiB0aGUgdXBsb2FkCiAqIHByb2Nlc3MuCiAqIEBwYXJhbSB7c3RyaW5nfSBpbnB1dElkIEVsZW1lbnQgSUQgb2YgdGhlIGlucHV0IGZpbGUgcGlja2VyIGVsZW1lbnQuCiAqIEBwYXJhbSB7c3RyaW5nfSBvdXRwdXRJZCBFbGVtZW50IElEIG9mIHRoZSBvdXRwdXQgZGlzcGxheS4KICogQHJldHVybiB7IUl0ZXJhYmxlPCFPYmplY3Q+fSBJdGVyYWJsZSBvZiBuZXh0IHN0ZXBzLgogKi8KZnVuY3Rpb24qIHVwbG9hZEZpbGVzU3RlcChpbnB1dElkLCBvdXRwdXRJZCkgewogIGNvbnN0IGlucHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKGlucHV0SWQpOwogIGlucHV0RWxlbWVudC5kaXNhYmxlZCA9IGZhbHNlOwoKICBjb25zdCBvdXRwdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQob3V0cHV0SWQpOwogIG91dHB1dEVsZW1lbnQuaW5uZXJIVE1MID0gJyc7CgogIGNvbnN0IHBpY2tlZFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgaW5wdXRFbGVtZW50LmFkZEV2ZW50TGlzdGVuZXIoJ2NoYW5nZScsIChlKSA9PiB7CiAgICAgIHJlc29sdmUoZS50YXJnZXQuZmlsZXMpOwogICAgfSk7CiAgfSk7CgogIGNvbnN0IGNhbmNlbCA9IGRvY3VtZW50LmNyZWF0ZUVsZW1lbnQoJ2J1dHRvbicpOwogIGlucHV0RWxlbWVudC5wYXJlbnRFbGVtZW50LmFwcGVuZENoaWxkKGNhbmNlbCk7CiAgY2FuY2VsLnRleHRDb250ZW50ID0gJ0NhbmNlbCB1cGxvYWQnOwogIGNvbnN0IGNhbmNlbFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgY2FuY2VsLm9uY2xpY2sgPSAoKSA9PiB7CiAgICAgIHJlc29sdmUobnVsbCk7CiAgICB9OwogIH0pOwoKICAvLyBXYWl0IGZvciB0aGUgdXNlciB0byBwaWNrIHRoZSBmaWxlcy4KICBjb25zdCBmaWxlcyA9IHlpZWxkIHsKICAgIHByb21pc2U6IFByb21pc2UucmFjZShbcGlja2VkUHJvbWlzZSwgY2FuY2VsUHJvbWlzZV0pLAogICAgcmVzcG9uc2U6IHsKICAgICAgYWN0aW9uOiAnc3RhcnRpbmcnLAogICAgfQogIH07CgogIGNhbmNlbC5yZW1vdmUoKTsKCiAgLy8gRGlzYWJsZSB0aGUgaW5wdXQgZWxlbWVudCBzaW5jZSBmdXJ0aGVyIHBpY2tzIGFyZSBub3QgYWxsb3dlZC4KICBpbnB1dEVsZW1lbnQuZGlzYWJsZWQgPSB0cnVlOwoKICBpZiAoIWZpbGVzKSB7CiAgICByZXR1cm4gewogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbXBsZXRlJywKICAgICAgfQogICAgfTsKICB9CgogIGZvciAoY29uc3QgZmlsZSBvZiBmaWxlcykgewogICAgY29uc3QgbGkgPSBkb2N1bWVudC5jcmVhdGVFbGVtZW50KCdsaScpOwogICAgbGkuYXBwZW5kKHNwYW4oZmlsZS5uYW1lLCB7Zm9udFdlaWdodDogJ2JvbGQnfSkpOwogICAgbGkuYXBwZW5kKHNwYW4oCiAgICAgICAgYCgke2ZpbGUudHlwZSB8fCAnbi9hJ30pIC0gJHtmaWxlLnNpemV9IGJ5dGVzLCBgICsKICAgICAgICBgbGFzdCBtb2RpZmllZDogJHsKICAgICAgICAgICAgZmlsZS5sYXN0TW9kaWZpZWREYXRlID8gZmlsZS5sYXN0TW9kaWZpZWREYXRlLnRvTG9jYWxlRGF0ZVN0cmluZygpIDoKICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgJ24vYSd9IC0gYCkpOwogICAgY29uc3QgcGVyY2VudCA9IHNwYW4oJzAlIGRvbmUnKTsKICAgIGxpLmFwcGVuZENoaWxkKHBlcmNlbnQpOwoKICAgIG91dHB1dEVsZW1lbnQuYXBwZW5kQ2hpbGQobGkpOwoKICAgIGNvbnN0IGZpbGVEYXRhUHJvbWlzZSA9IG5ldyBQcm9taXNlKChyZXNvbHZlKSA9PiB7CiAgICAgIGNvbnN0IHJlYWRlciA9IG5ldyBGaWxlUmVhZGVyKCk7CiAgICAgIHJlYWRlci5vbmxvYWQgPSAoZSkgPT4gewogICAgICAgIHJlc29sdmUoZS50YXJnZXQucmVzdWx0KTsKICAgICAgfTsKICAgICAgcmVhZGVyLnJlYWRBc0FycmF5QnVmZmVyKGZpbGUpOwogICAgfSk7CiAgICAvLyBXYWl0IGZvciB0aGUgZGF0YSB0byBiZSByZWFkeS4KICAgIGxldCBmaWxlRGF0YSA9IHlpZWxkIHsKICAgICAgcHJvbWlzZTogZmlsZURhdGFQcm9taXNlLAogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbnRpbnVlJywKICAgICAgfQogICAgfTsKCiAgICAvLyBVc2UgYSBjaHVua2VkIHNlbmRpbmcgdG8gYXZvaWQgbWVzc2FnZSBzaXplIGxpbWl0cy4gU2VlIGIvNjIxMTU2NjAuCiAgICBsZXQgcG9zaXRpb24gPSAwOwogICAgZG8gewogICAgICBjb25zdCBsZW5ndGggPSBNYXRoLm1pbihmaWxlRGF0YS5ieXRlTGVuZ3RoIC0gcG9zaXRpb24sIE1BWF9QQVlMT0FEX1NJWkUpOwogICAgICBjb25zdCBjaHVuayA9IG5ldyBVaW50OEFycmF5KGZpbGVEYXRhLCBwb3NpdGlvbiwgbGVuZ3RoKTsKICAgICAgcG9zaXRpb24gKz0gbGVuZ3RoOwoKICAgICAgY29uc3QgYmFzZTY0ID0gYnRvYShTdHJpbmcuZnJvbUNoYXJDb2RlLmFwcGx5KG51bGwsIGNodW5rKSk7CiAgICAgIHlpZWxkIHsKICAgICAgICByZXNwb25zZTogewogICAgICAgICAgYWN0aW9uOiAnYXBwZW5kJywKICAgICAgICAgIGZpbGU6IGZpbGUubmFtZSwKICAgICAgICAgIGRhdGE6IGJhc2U2NCwKICAgICAgICB9LAogICAgICB9OwoKICAgICAgbGV0IHBlcmNlbnREb25lID0gZmlsZURhdGEuYnl0ZUxlbmd0aCA9PT0gMCA/CiAgICAgICAgICAxMDAgOgogICAgICAgICAgTWF0aC5yb3VuZCgocG9zaXRpb24gLyBmaWxlRGF0YS5ieXRlTGVuZ3RoKSAqIDEwMCk7CiAgICAgIHBlcmNlbnQudGV4dENvbnRlbnQgPSBgJHtwZXJjZW50RG9uZX0lIGRvbmVgOwoKICAgIH0gd2hpbGUgKHBvc2l0aW9uIDwgZmlsZURhdGEuYnl0ZUxlbmd0aCk7CiAgfQoKICAvLyBBbGwgZG9uZS4KICB5aWVsZCB7CiAgICByZXNwb25zZTogewogICAgICBhY3Rpb246ICdjb21wbGV0ZScsCiAgICB9CiAgfTsKfQoKc2NvcGUuZ29vZ2xlID0gc2NvcGUuZ29vZ2xlIHx8IHt9OwpzY29wZS5nb29nbGUuY29sYWIgPSBzY29wZS5nb29nbGUuY29sYWIgfHwge307CnNjb3BlLmdvb2dsZS5jb2xhYi5fZmlsZXMgPSB7CiAgX3VwbG9hZEZpbGVzLAogIF91cGxvYWRGaWxlc0NvbnRpbnVlLAp9Owp9KShzZWxmKTsK", "headers": [["content-type", "application/javascript"]], "ok": true, "status": 200, "status_text": ""}}} id="rF0zksIxjV4M" outputId="d155ea31-1b26-4b9d-a385-37d01ee622a6"
import numpy as np
from google.colab import files
uploaded = files.upload()
for fn in uploaded.keys():
path = fn
img = tf.keras.utils.load_img(path, target_size=(150,150))
img_plot = plt.imshow(img)
img_array = tf.keras.utils.img_to_array(img)
x = tf.keras.utils.img_to_array(img)
x = np.expand_dims(x, axis=0)
images = np.vstack([x])
classes = model.predict(images, batch_size=10)
print(fn)
if classes[0, 0] != 0:
print("Paper")
elif classes[0, 1] != 0:
print("Rock")
else:
print("Scissors")
| 53.469751
| 7,350
|
418690a216286e6fb9c2756e661c1e90ac12716f
|
py
|
python
|
Report/crypto_arbitrage.ipynb
|
ipopester/Bitcoin-Arbitrage-Analysis
|
['Unlicense']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ## Crypto Arbitrage
#
# In this Challenge, you'll take on the role of an analyst at a high-tech investment firm. The vice president (VP) of your department is considering arbitrage opportunities in Bitcoin and other cryptocurrencies. As Bitcoin trades on markets across the globe, can you capitalize on simultaneous price dislocations in those markets by using the powers of Pandas?
#
# For this assignment, you’ll sort through historical trade data for Bitcoin on two exchanges: Bitstamp and Coinbase. Your task is to apply the three phases of financial analysis to determine if any arbitrage opportunities exist for Bitcoin.
#
# This aspect of the Challenge will consist of 3 phases.
#
# 1. Collect the data.
#
# 2. Prepare the data.
#
# 3. Analyze the data.
#
#
# ### Import the required libraries and dependencies.
import pandas as pd
from pathlib import Path
# %matplotlib inline
import matplotlib.pyplot as plt
# ## Collect the Data
#
# To collect the data that you’ll need, complete the following steps:
#
# Instructions.
#
# 1. Using the Pandas `read_csv` function and the `Path` module, import the data from `bitstamp.csv` file, and create a DataFrame called `bitstamp`. Set the DatetimeIndex as the Timestamp column, and be sure to parse and format the dates.
#
# 2. Use the `head` (and/or the `tail`) function to confirm that Pandas properly imported the data.
#
# 3. Repeat Steps 1 and 2 for `coinbase.csv` file.
# ### Step 1: Using the Pandas `read_csv` function and the `Path` module, import the data from `bitstamp.csv` file, and create a DataFrame called `bitstamp`. Set the DatetimeIndex as the Timestamp column, and be sure to parse and format the dates.
# Read in the CSV file called "bitstamp.csv" using the Path module.
# The CSV file is located in the Resources folder.
# Set the index to the column "Date"
# Set the parse_dates and infer_datetime_format parameters
bitstamp = pd.read_csv(
Path('./Resources/bitstamp.csv'),
index_col = 'Timestamp',
parse_dates = True,
infer_datetime_format = True
)
# ### Step 2: Use the `head` (and/or the `tail`) function to confirm that Pandas properly imported the data.
# Use the head (and/or tail) function to confirm that the data was imported properly.
bitstamp.head()
# ### Step 3: Repeat Steps 1 and 2 for `coinbase.csv` file.
# Read in the CSV file called "coinbase.csv" using the Path module.
# The CSV file is located in the Resources folder.
# Set the index to the column "Timestamp"
# Set the parse_dates and infer_datetime_format parameters
coinbase = pd.read_csv(
Path('./Resources/coinbase.csv'),
index_col = 'Timestamp',
parse_dates = True,
infer_datetime_format = True
)
# Use the head (and/or tail) function to confirm that the data was imported properly.
coinbase.head()
# ## Prepare the Data
#
# To prepare and clean your data for analysis, complete the following steps:
#
# 1. For the bitstamp DataFrame, replace or drop all `NaN`, or missing, values in the DataFrame.
#
# 2. Use the `str.replace` function to remove the dollar signs ($) from the values in the Close column.
#
# 3. Convert the data type of the Close column to a `float`.
#
# 4. Review the data for duplicated values, and drop them if necessary.
#
# 5. Repeat Steps 1–4 for the coinbase DataFrame.
# ### Step 1: For the bitstamp DataFrame, replace or drop all `NaN`, or missing, values in the DataFrame.
# For the bitstamp DataFrame, replace or drop all NaNs or missing values in the DataFrame
bitstamp.isnull().sum()
bitstamp = bitstamp.dropna().copy()
bitstamp.isnull().sum()
# ### Step 2: Use the `str.replace` function to remove the dollar signs ($) from the values in the Close column.
# Use the str.replace function to remove the dollar sign, $
bitstamp['Close'] = bitstamp['Close'].str.replace("$", "", regex=False)
bitstamp['Close']
# ### Step 3: Convert the data type of the Close column to a `float`.
# Convert the Close data type to a float
bitstamp['Close'] = bitstamp['Close'].astype('float')
# ### Step 4: Review the data for duplicated values, and drop them if necessary.
# Review the data for duplicate values, and drop them if necessary
bitstamp.info()
# ### Step 5: Repeat Steps 1–4 for the coinbase DataFrame.
# Repeat Steps 1–4 for the coinbase DataFrame
coinbase.isnull().sum()
coinbase = coinbase.dropna().copy()
coinbase.isnull().sum()
coinbase['Close'] = coinbase['Close'].str.replace("$","", regex=False)
coinbase['Close'] = coinbase['Close'].astype('float')
coinbase.info()
coinbase['Open'] = coinbase['Open'].astype('float')
coinbase['High'] = coinbase['High'].astype('float')
coinbase['Low'] = coinbase['Low'].astype('float')
coinbase['BTC Volume'] = coinbase['BTC Volume'].astype('float')
coinbase['USD Volume'] = coinbase['USD Volume'].astype('float')
coinbase['Weighted Price'] = coinbase['Weighted Price'].astype('float')
coinbase.info()
coinbase.head()
# ## Analyze the Data
#
# Your analysis consists of the following tasks:
#
# 1. Choose the columns of data on which to focus your analysis.
#
# 2. Get the summary statistics and plot the data.
#
# 3. Focus your analysis on specific dates.
#
# 4. Calculate the arbitrage profits.
# ### Step 1: Choose columns of data on which to focus your analysis.
#
# Select the data you want to analyze. Use `loc` or `iloc` to select the following columns of data for both the bitstamp and coinbase DataFrames:
#
# * Timestamp (index)
#
# * Close
#
# +
# Use loc or iloc to select `Timestamp (the index)` and `Close` from bitstamp DataFrame
bitstamp_sliced = bitstamp.loc[:, 'Close']
# Review the first five rows of the DataFrame
bitstamp_sliced.head()
# +
# Use loc or iloc to select `Timestamp (the index)` and `Close` from coinbase DataFrame
coinbase_sliced = coinbase.loc[:, 'Close']
# Review the first five rows of the DataFrame
coinbase_sliced.head()
# -
# ### Step 2: Get summary statistics and plot the data.
#
# Sort through the time series data associated with the bitstamp and coinbase DataFrames to identify potential arbitrage opportunities. To do so, complete the following steps:
#
# 1. Generate the summary statistics for each DataFrame by using the `describe` function.
#
# 2. For each DataFrame, create a line plot for the full period of time in the dataset. Be sure to tailor the figure size, title, and color to each visualization.
#
# 3. In one plot, overlay the visualizations that you created in Step 2 for bitstamp and coinbase. Be sure to adjust the legend and title for this new visualization.
#
# 4. Using the `loc` and `plot` functions, plot the price action of the assets on each exchange for different dates and times. Your goal is to evaluate how the spread between the two exchanges changed across the time period that the datasets define. Did the degree of spread change as time progressed?
# Generate the summary statistics for the bitstamp DataFrame
bitstamp_sliced.describe()
# Generate the summary statistics for the coinbase DataFrame
coinbase_sliced.describe()
# Create a line plot for the bitstamp DataFrame for the full length of time in the dataset
# Be sure that the figure size, title, and color are tailored to each visualization
bitstamp_sliced.plot(
figsize=(15, 7), color='orange')
plt.title("Bitcoin Price: Bitstamp ")
plt.ylim(5000,18000)
plt.xlabel("DateTime")
plt.ylabel("USD")
plt.show()
# + tags=[]
# Create a line plot for the coinbase DataFrame for the full length of time in the dataset
# Be sure that the figure size, title, and color are tailored to each visualization
coinbase_sliced.plot(
figsize =(15, 7), color ='blue')
plt.title("Bitcoin Price: Coinbase")
plt.ylim(5000,18000)
plt.xlabel("DateTime")
plt.ylabel("USD")
plt.show()
# -
# Overlay the visualizations for the bitstamp and coinbase DataFrames in one plot
# The plot should visualize the prices over the full lenth of the dataset
# Be sure to include the parameters: legend, figure size, title, and color and label
plt.figure(figsize=(15,7))
bitstamp_sliced.plot(
legend=True, label='Bitstamp', color='orange')
coinbase_sliced.plot(
legend=True, label='Coinbase', color='blue')
plt.title("Bitcoin Price: Coinbase and Bitstamp")
plt.ylim(5000,18000)
plt.xlabel("DateTime")
plt.ylabel("USD")
plt.show()
# Using the loc and plot functions, create an overlay plot that visualizes
# the price action of both DataFrames for a one month period early in the dataset
# Be sure to include the parameters: legend, figure size, title, and color and label
plt.figure(figsize=(15,7))
bitstamp_sliced.loc['2018-01-01':'2018-01-31'].plot(
legend=True, label='Bitstamp', color='orange')
coinbase_sliced.loc['2018-01-01':'2018-01-31'].plot(
legend=True, label='Coinbase', color='blue')
plt.title("Bitcoin Price: Coinbase and Bitstamp - January 2018")
plt.xlabel("DateTime")
plt.ylabel("USD")
plt.show()
# Using the loc and plot functions, create an overlay plot that visualizes
# the price action of both DataFrames for a one month period later in the dataset
# Be sure to include the parameters: legend, figure size, title, and color and label
plt.figure(figsize=(15,7))
bitstamp_sliced.loc['2018-03-01':'2018-03-30'].plot(
legend=True, label='Bitstamp', color='orange')
coinbase_sliced.loc['2018-03-01':'2018-03-30'].plot(
legend=True, label='Coinbase', color='blue')
plt.title("Bitcoin Price: Coinbase and Bitstamp - March 2018")
plt.ylim(6000,12000)
plt.xlabel("DateTime")
plt.ylabel("USD")
plt.show()
# **Question** Based on the visualizations of the different time periods, has the degree of spread change as time progressed?
#
# **Answer** The degree of spread does change as time progressed over the time period studied: January through March 2018. The spread was much higher during the January compared to February and March.
# ### Step 3: Focus Your Analysis on Specific Dates
#
# Focus your analysis on specific dates by completing the following steps:
#
# 1. Select three dates to evaluate for arbitrage profitability. Choose one date that’s early in the dataset, one from the middle of the dataset, and one from the later part of the time period.
#
# 2. For each of the three dates, generate the summary statistics and then create a box plot. This big-picture view is meant to help you gain a better understanding of the data before you perform your arbitrage calculations. As you compare the data, what conclusions can you draw?
# +
# Create an overlay plot that visualizes the two dataframes over a period of one day early in the dataset.
# Be sure that the plots include the parameters `legend`, `figsize`, `title`, `color` and `label`
bitstamp_day_early = bitstamp_sliced.loc['2018-01-28']
coinbase_day_early = coinbase_sliced.loc['2018-01-28']
plt.figure(figsize=(15,7))
bitstamp_day_early.plot(
legend=True, label='Bitstamp', color='orange')
coinbase_day_early.plot(
legend=True, label='Coinbase', color='blue')
plt.title("Bitcoin Price: Coinbase and Bitstamp - January 28, 2018")
plt.xlabel("DateTime")
plt.ylabel("Price")
plt.show()
# + tags=[]
# Using the early date that you have selected, calculate the arbitrage spread
# by subtracting the bitstamp lower closing prices from the coinbase higher closing prices
arbitrage_spread_early = bitstamp_day_early - coinbase_day_early
# Generate summary statistics for the early DataFrame
arbitrage_spread_early.describe()
# -
# Visualize the arbitrage spread from early in the dataset in a box plot
arbitrage_spread_early.plot(
kind='box', figsize=(10, 7), label='---', color='green')
plt.title("Bitcoin Arbitrage Spread: January 28, 2018")
plt.ylim(0, 500)
plt.xlabel(None)
plt.ylabel("Price")
plt.show()
# +
# Create an overlay plot that visualizes the two dataframes over a period of one day from the middle of the dataset.
# Be sure that the plots include the parameters `legend`, `figsize`, `title`, `color` and `label`
bitstamp_day_middle = bitstamp_sliced.loc['2018-02-20']
coinbase_day_middle = coinbase_sliced.loc['2018-02-20']
plt.figure(figsize=(15,7))
bitstamp_day_middle.plot(
legend=True, label='Bitstamp', color='orange')
coinbase_day_middle.plot(
legend=True, label='Coinbase', color='blue')
plt.title("Bitcoin Price: Coinbase and Bitstamp - February 20, 2018")
plt.xlabel("DateTIme")
plt.ylabel("USD")
plt.show()
# +
# Using the date in the middle that you have selected, calculate the arbitrage spread
# by subtracting the bitstamp lower closing prices from the coinbase higher closing prices
arbitrage_spread_middle = bitstamp_day_middle - coinbase_day_middle
# Generate summary statistics
arbitrage_spread_middle.describe()
# -
# Visualize the arbitrage spread from the middle of the dataset in a box plot
arbitrage_spread_middle.plot(
kind='box', figsize=(10, 7), label='---', color='green')
plt.title("Bitcoin Arbitrage Spread: February 20, 2018")
plt.ylim(-100,100)
plt.xlabel(None)
plt.ylabel("USD")
plt.show()
# +
# Create an overlay plot that visualizes the two dataframes over a period of one day from late in the dataset.
# Be sure that the plots include the parameters `legend`, `figsize`, `title`, `color` and `label`
bitstamp_day_late = bitstamp_sliced.loc['2018-03-20']
coinbase_day_late = coinbase_sliced.loc['2018-03-20']
plt.figure(figsize=(15,7))
bitstamp_day_late.plot(
legend=True, label='Bitstamp', color='orange')
coinbase_day_late.plot(
legend=True, label='Coinbase', color='blue')
plt.title("Bitcoin Price: Coinbase and Bitstamp - March 20, 2018")
plt.xlabel("DateTIme")
plt.ylabel("USD")
plt.show()
# +
# Using the date from the late that you have selected, calculate the arbitrage spread
# by subtracting the bitstamp lower closing prices from the coinbase higher closing prices
arbitrage_spread_late = bitstamp_day_late - coinbase_day_late
# Generate summary statistics for the late DataFrame
arbitrage_spread_late.describe()
# -
# Visualize the arbitrage spread from late in the dataset in a box plot
arbitrage_spread_late.plot(
kind='box', legend=True, figsize=(10, 7), label='---', color='green')
plt.title("Bitcoin Arbitrage Spread: March 20, 2018")
plt.ylim(-50,50)
plt.xlabel(None)
plt.ylabel("USD")
plt.show()
# ### Step 4: Calculate the Arbitrage Profits
#
# Calculate the potential profits for each date that you selected in the previous section. Your goal is to determine whether arbitrage opportunities still exist in the Bitcoin market. Complete the following steps:
#
# 1. For each of the three dates, measure the arbitrage spread between the two exchanges by subtracting the lower-priced exchange from the higher-priced one. Then use a conditional statement to generate the summary statistics for each arbitrage_spread DataFrame, where the spread is greater than zero.
#
# 2. For each of the three dates, calculate the spread returns. To do so, divide the instances that have a positive arbitrage spread (that is, a spread greater than zero) by the price of Bitcoin from the exchange you’re buying on (that is, the lower-priced exchange). Review the resulting DataFrame.
#
# 3. For each of the three dates, narrow down your trading opportunities even further. To do so, determine the number of times your trades with positive returns exceed the 1% minimum threshold that you need to cover your costs.
#
# 4. Generate the summary statistics of your spread returns that are greater than 1%. How do the average returns compare among the three dates?
#
# 5. For each of the three dates, calculate the potential profit, in dollars, per trade. To do so, multiply the spread returns that were greater than 1% by the cost of what was purchased. Make sure to drop any missing values from the resulting DataFrame.
#
# 6. Generate the summary statistics, and plot the results for each of the three DataFrames.
#
# 7. Calculate the potential arbitrage profits that you can make on each day. To do so, sum the elements in the profit_per_trade DataFrame.
#
# 8. Using the `cumsum` function, plot the cumulative sum of each of the three DataFrames. Can you identify any patterns or trends in the profits across the three time periods?
#
# (NOTE: The starter code displays only one date. You'll want to do this analysis for two additional dates).
# #### 1. For each of the three dates, measure the arbitrage spread between the two exchanges by subtracting the lower-priced exchange from the higher-priced one. Then use a conditional statement to generate the summary statistics for each arbitrage_spread DataFrame, where the spread is greater than zero.
#
# *NOTE*: For illustration, only one of the three dates is shown in the starter code below.
# +
# For the date early in the dataset, measure the arbitrage spread between the two exchanges
# by subtracting the lower-priced exchange from the higher-priced one
#arbitrage_spead_early = bitstamp_day_early - coinbase_day_early
# Use a conditional statement to generate the summary statistics for each arbitrage_spread DataFrame
arbitrage_spread_early.describe()
# -
arbitrage_spread_middle.describe()
arbitrage_spread_late.describe()
# #### 2. For each of the three dates, calculate the spread returns. To do so, divide the instances that have a positive arbitrage spread (that is, a spread greater than zero) by the price of Bitcoin from the exchange you’re buying on (that is, the lower-priced exchange). Review the resulting DataFrame.
# For the date early in the dataset, calculate the spread returns by dividing the instances when the arbitrage spread is positive (> 0)
# by the price of Bitcoin from the exchange you are buying on (the lower-priced exchange).
#spread_return_early = arbitrage_spread_early[arbitrage_spread_early > 0] / coinbase_sliced.loc['2018-01-15 00:00:00':'2018-01-15 23:59:59']# YOUR CODE HERE
spread_return_early = arbitrage_spread_early[arbitrage_spread_early > 0] / coinbase_day_early
# Review the spread return DataFrame
spread_return_early.describe(include='all').round(5)
spread_return_middle = arbitrage_spread_middle[arbitrage_spread_middle > 0] / coinbase_day_middle
spread_return_middle.describe(include='all').round(5)
spread_return_late = arbitrage_spread_late[arbitrage_spread_late > 0] / bitstamp_day_late
spread_return_late.describe(include='all').round(5)
# #### 3. For each of the three dates, narrow down your trading opportunities even further. To do so, determine the number of times your trades with positive returns exceed the 1% minimum threshold that you need to cover your costs.
# +
# For the date early in the dataset, determine the number of times your trades with positive returns
# exceed the 1% minimum threshold (.01) that you need to cover your costs
profitable_trades_early = spread_return_early[spread_return_early > 0.01]
# Review the first five profitable trades
profitable_trades_early.head()
# -
profitable_trades_middle = spread_return_middle[spread_return_middle > 0.01]
profitable_trades_middle.head()
profitable_trades_late = spread_return_late[spread_return_late > 0.01]
profitable_trades_late.head()
# #### 4. Generate the summary statistics of your spread returns that are greater than 1%. How do the average returns compare among the three dates?
# For the date early in the dataset, generate the summary statistics for the profitable trades
# or you trades where the spread returns are are greater than 1%
profitable_trades_early[profitable_trades_early > 0.01].describe(include='all')
profitable_trades_middle[profitable_trades_middle > 0.01].describe(include='all')
profitable_trades_late[profitable_trades_late > 0.01].describe(include='all')
# #### 5. For each of the three dates, calculate the potential profit, in dollars, per trade. To do so, multiply the spread returns that were greater than 1% by the cost of what was purchased. Make sure to drop any missing values from the resulting DataFrame.
# +
# For the date early in the dataset, calculate the potential profit per trade in dollars
# Multiply the profitable trades by the cost of the Bitcoin that was purchased
profit_early = profitable_trades_early * coinbase_day_early
# Drop any missing values from the profit DataFrame
profit_per_trade_early = profit_early.dropna()
# View the early profit DataFrame
profit_per_trade_early
# -
profit_middle = profitable_trades_middle * coinbase_day_middle
profit_per_trade_middle = profit_middle.dropna()
profit_per_trade_middle
profit_late = profitable_trades_late * coinbase_day_late
profit_per_trade_late = profit_late.dropna()
profit_per_trade_late
# #### 6. Generate the summary statistics, and plot the results for each of the three DataFrames.
# Generate the summary statistics for the early profit per trade DataFrame
profit_per_trade_early.describe(include='all')
profit_per_trade_middle.describe(include='all')
# + tags=[]
profit_per_trade_late.describe(include='all')
# -
# Plot the results for the early profit per trade DataFrame
profit_per_trade_early.plot(
kind='line', rot=45, figsize =(15, 7))
plt.title("Opportunities for Profit: January 28, 2018")
plt.xlabel('DateTime')
plt.ylabel('USD')
plt.show()
# #### Middle profit per trade: profit per trade was negligible
# #### Late profit per trade: profit per trade was negligible
#
# #### 7. Calculate the potential arbitrage profits that you can make on each day. To do so, sum the elements in the profit_per_trade DataFrame.
# Calculate the sum of the potential profits for the early profit per trade DataFrame
total_profit_early = profit_per_trade_early.sum().round(2)
total_profit_early
total_profit_middle = profit_per_trade_middle.sum()
total_profit_middle
total_profit_late = profit_per_trade_late.sum()
total_profit_late
# #### 8. Using the `cumsum` function, plot the cumulative sum of each of the three DataFrames. Can you identify any patterns or trends in the profits across the three time periods?
# Use the cumsum function to calculate the cumulative profits over time for the early profit per trade DataFrame
cumulative_profit_early = profit_per_trade_early.cumsum()
cumulative_profit_middle = profit_per_trade_middle.cumsum()
cumulative_profit_late = profit_per_trade_late.cumsum()
# Plot the cumulative sum of profits for the early profit per trade DataFrame
cumulative_profit_early.plot(kind='line', rot=45, figsize =(10, 5))
plt.title("Cumulative Profit: January 28, 2018")
plt.xlabel('DateTime')
plt.ylabel('USD')
plt.show()
# #### Middle profit per trade: profit per trade was negligible to display
# #### Late profit per trade: profit per trade was negligible to display
# **Question:** After reviewing the profit information across each date from the different time periods, can you identify any patterns or trends?
#
# **Answer:** There is a clear pattern and trend with regard to the disparity of the price, or spread, of Bitcoin on Coinbase and Bitstamp from January through March 2018. In general, the spread decreased over time. The opportunities to exploit the spread were highly limited during most of the time period evaluated, occuring once every few weeks, particularly during February and March. The spread was highest in late January and peaked around January 28. Based on a threshold of greater than 1% profit (to cover trading fees), the spread on January 28 presented a unique opportunity for arbitrage that was highly profitable. If optimally traded, cumulative profits on January 28 would be around $350,000. With the exception of this short window of time, few opportunities for arbitrage were presented. The decrease in spread over time could be attributed to an increase in algorithmic bot trading designed for these types of arbitrage opportunities, which brought the price of Bitcoin closer to parody. To further test this hypothesis, data spanning a longer time frame should be analyzed.
| 38.993344
| 1,092
|
7bd81f4c52360f5271ed19536470b7da0c804cff
|
py
|
python
|
prediction/multitask/fine-tuning/function documentation generation/java/small_model.ipynb
|
victory-hash/CodeTrans
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/agemagician/CodeTrans/blob/main/prediction/multitask/fine-tuning/function%20documentation%20generation/java/small_model.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="c9eStCoLX0pZ"
# **<h3>Predict the documentation for java code using codeTrans mulititask finetuning model</h3>**
# <h4>You can make free prediction online through this
# <a href="https://huggingface.co/SEBIS/code_trans_t5_small_code_documentation_generation_java_multitask_finetune">Link</a></h4> (When using the prediction online, you need to parse and tokenize the code first.)
# + [markdown] id="6YPrvwDIHdBe"
# **1. Load necessry libraries including huggingface transformers**
# + colab={"base_uri": "https://localhost:8080/"} id="6FAVWAN1UOJ4" outputId="1d3075d0-b269-4bb7-92b1-d2bcf233c44b"
# !pip install -q transformers sentencepiece
# + id="53TAO7mmUOyI"
from transformers import AutoTokenizer, AutoModelWithLMHead, SummarizationPipeline
# + [markdown] id="xq9v-guFWXHy"
# **2. Load the token classification pipeline and load it into the GPU if avilabile**
# + colab={"base_uri": "https://localhost:8080/", "height": 316, "referenced_widgets": ["b5884b26654c42b2975049d6f521985d", "af14fee2d4444025869a205544b531c5", "60b355fcf7d94ba4806bde223e11393e", "c286cf6c5e1c437abacb2a7dc2a0d576", "ab29305d5d7b46c180a3e2c3d8cd766d", "794545870ea04ae9bd8726a732469470", "e79044f52b7443409787e58244aebab1", "723265d5815d4b03ae23f1efde39fd3a", "23e00fc0cf614f3c8774ce2b89e93111", "1dfa23815d394e8eaad1df58fca4d7c0", "48e1a8ae90de47d282e160ab9bae8ecf", "5db39bd6c30545fa86e26e1568a89f9e", "da9810f726dd48548f80d625bb373004", "82d8ac4697f14d0e97806a27adcba482", "7566c034764c48bbae85697793fdb381", "9e4523a50414481ca0fe282a8d104721", "71ee0637ddb048f08acb00836da7374e", "18b03909022841f9bb9780164963851d", "f9d4326476ed447c99a1af69ae071101", "c99c8867d560480e8b11cc46285655f9", "412639d4f12f43bd8ff9b6518bd1eada", "09dc045158a14531b8412cb6f74206fa", "a363c8600b434114b5fd7fdf3552d41d", "3917a8aeae5e4e8a8b0b0d17882f60e3", "2a907945ee9f44c5b35b4f60c1cd612a", "32cc372920dd4f67bfdd11086b2fbb37", "1bbaddd2ed0547188312234835062220", "fe983d9796614a129ea98a0371c8e93e", "e042d480056347eaacc70dbe6ee285c6", "45e26260db384575a4f01abe62cd7c6b", "634299a71399429ebf1ffcbbc910c647", "c49f58daff564b2d8e534c890bb21ac6", "da5f491fc59e49079051085bd8745a49", "b213d7c185d84149b24093a579f0a683", "62b5219e3bd349a0bf4637b1fbd0d105", "2ed34c8edf0b44bba70e30157bcd353b", "72e39d963a1b49af95772d0cc87d2496", "5106ea5468534c8ebd1d71e491f2ef48", "a8b20751164a418c80c9ef57e0b49073", "c005ccbff00d46d683c3598686ce80b8"]} id="5ybX8hZ3UcK2" outputId="548eec3a-bb63-44d2-a396-29090847d592"
pipeline = SummarizationPipeline(
model=AutoModelWithLMHead.from_pretrained("SEBIS/code_trans_t5_small_code_documentation_generation_java_multitask_finetune"),
tokenizer=AutoTokenizer.from_pretrained("SEBIS/code_trans_t5_small_code_documentation_generation_java_multitask_finetune", skip_special_tokens=True),
device=0
)
# + [markdown] id="hkynwKIcEvHh"
# **3 Give the code for summarization, parse and tokenize it**
# + id="nld-UUmII-2e"
code = """public static <T, U> Function<T, U> castFunction(Class<U> target) {\n return new CastToClass<T, U>(target);\n }""" #@param {type:"raw"}
# + id="cJLeTZ0JtsB5" colab={"base_uri": "https://localhost:8080/"} outputId="c15b0a16-7e6d-4cb3-98b0-6e195abc747b"
# !pip install tree_sitter
# !git clone https://github.com/tree-sitter/tree-sitter-java
# + id="hqACvTcjtwYK"
from tree_sitter import Language, Parser
Language.build_library(
'build/my-languages.so',
['tree-sitter-java']
)
JAVA_LANGUAGE = Language('build/my-languages.so', 'java')
parser = Parser()
parser.set_language(JAVA_LANGUAGE)
# + id="LLCv2Yb8t_PP"
def my_traverse(node, code_list):
lines = code.split('\n')
if node.child_count == 0:
line_start = node.start_point[0]
line_end = node.end_point[0]
char_start = node.start_point[1]
char_end = node.end_point[1]
if line_start != line_end:
code_list.append(' '.join([lines[line_start][char_start:]] + lines[line_start+1:line_end] + [lines[line_end][:char_end]]))
else:
code_list.append(lines[line_start][char_start:char_end])
else:
for n in node.children:
my_traverse(n, code_list)
return ' '.join(code_list)
# + id="BhF9MWu1uCIS" colab={"base_uri": "https://localhost:8080/"} outputId="538f4867-557c-40f9-870b-a65eab694df4"
tree = parser.parse(bytes(code, "utf8"))
code_list=[]
tokenized_code = my_traverse(tree.root_node, code_list)
print("Output after tokenization: " + tokenized_code)
# + [markdown] id="sVBz9jHNW1PI"
# **4. Make Prediction**
# + colab={"base_uri": "https://localhost:8080/"} id="KAItQ9U9UwqW" outputId="3b6b3eaa-a058-4339-bcc1-b7f70a97bb28"
pipeline([tokenized_code])
| 55.652174
| 1,594
|
745ebcb9ff5d28380733d5b0b7f333fa2a08b7ac
|
py
|
python
|
chatbot.ipynb
|
bowenwen/transit_chatbot
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Transit Chatbot
#
# This chatbot is designed to help customers find transit-related information quickly and efficiently. At this stage, it provides information on the next bus's arrival time at a given stop and step-by-step directions from point A to point B.
#
# This project was done as part of a team effort at a one-day hackaton at [TransLink](www.translink.ca) and was awarded the "face-lift" prize along with two other projects.
#
# The chatbot is built using [Rasa](https://rasa.com/), an open-source library for conversational AI. The [Rasa Stack](https://rasa.com/products/rasa-stack/) consists of two main components: Rasa NLU and Rasa Core. The figure below decribes Rasa's ecosystem:
#
# 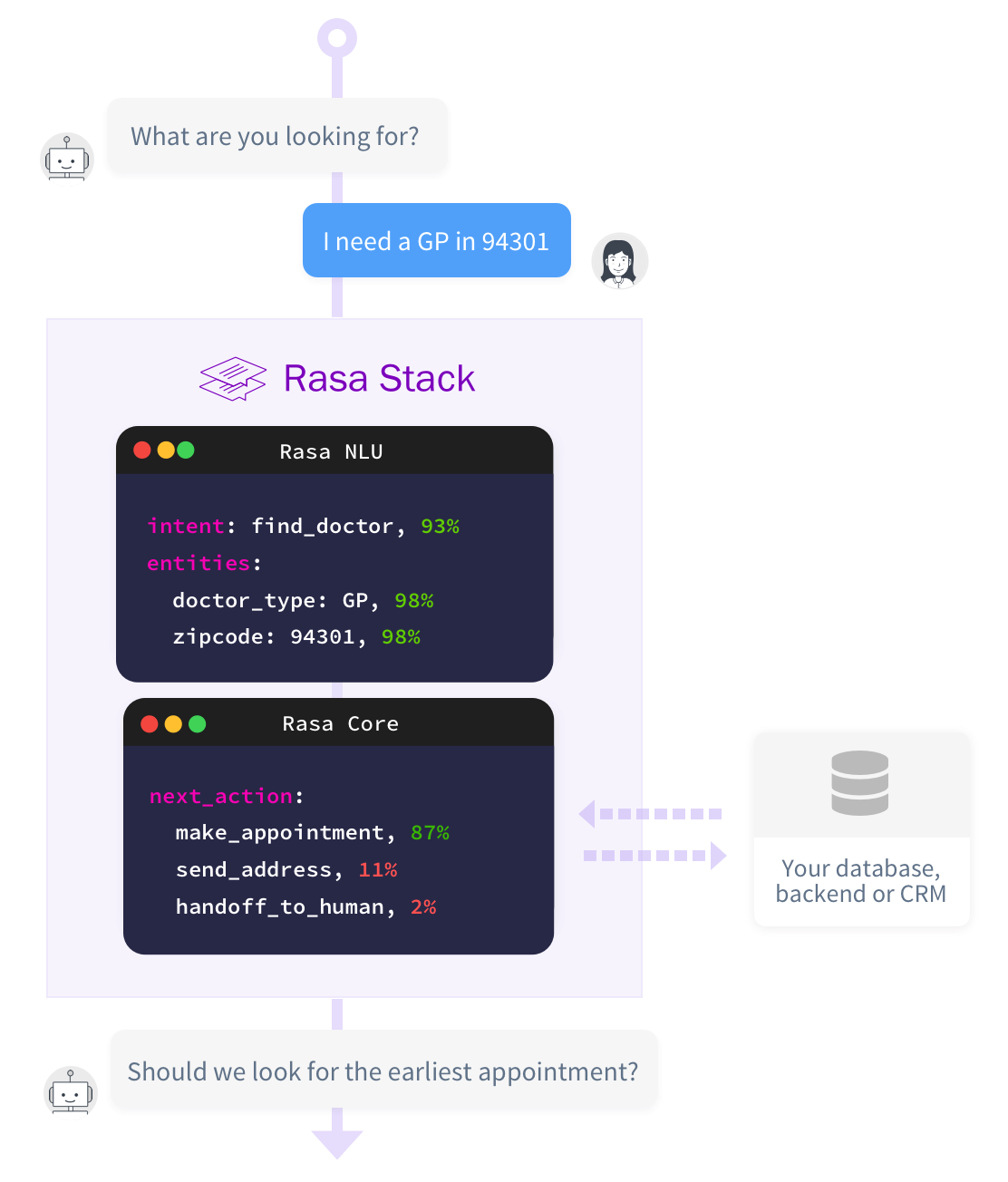
#
#
# ### Rasa NLU
#
# Rasa NLU is a natural language understanding tool that allows machines to interpert natual language inputs in the form of text or speech. This is acheived by performing two tasks: intent classification and entity extraction. For example, a user input: "When is the next bus #8 coming at stop number 50234?" can be classified with an intent of checking the next bus's arrival time while the bus number and the stop number are the extracted entities. Rasa NLU provides a flexibile framework that allows the users to choose from pre-defined pipelines or defined their own custom pipelines. The [official documenation](https://rasa.com/docs/nlu/0.13.8/choosing_pipeline/) describes this in detail. In short, choosing an intent classification model or an entity extraction model is fairly straightforward. In this project, since the training data is relatively small (<1000 training examples) and we are developing the bot to operate in English (which is available as a [SpaCy](https://spacy.io/) langaue model), we chose the `spacy_sklearn` pipeline. SpaCy is an open-source library for advanced natural language processing (NLP) - it provides the underlying text processing capabilities for our chatbot. As described by its developers: "_SpaCy comes with pre-trained statistical models and word vectors, and currently supports tokenization for 30+ languages. It features the fastest syntactic parser in the world, convolutional neural network models for tagging, parsing and named entity recognition and easy deep learning integration. It's commercial open-source software, released under the MIT license._" An introduction to SpaCy can be found [here](https://spacy.io/usage/spacy-101#section-features). We specifically use the following Spacy's NLP tools: the language model represented by word vectors - multi-dimensional meaning representations of words that allows us to determine how similar they are to each other, a tokenizer - which splits the sentence into tokens, and a featurizer - which transforms the sentence into a vector representation. We could use the named entity recognition (ner) pre-trained models, however, since we are dealing with custom entities, we trained a conditional random field model. We also trained a sklearn's support vector machine (SVM) model to classify users' intents. Rasa NLU provides a nice wrapper for these models as defined in the processing pipeline which defines how structured data is extracted from unstructured user inputs.
#
# ### Rasa Core
#
# Rasa Core is the dialogue engine that generates responses to users based on state of the coversation. Rasa Core achieves this by utilizing machine learning rather than state machines or rigid rules. This is how Rasa Core works:
#
# 
#
# _The steps are (according to the [documentation](https://rasa.com/docs/core/architecture/)):_
# 1. The message is received and passed to an Interpreter, which converts it into a dictionary including the original text, the intent, and any entities that were found.
# 2. The Tracker is the object which keeps track of conversation state. It receives the info that a new message has come in.
# 3. The policy receives the current state of the tracker.
# 4. The policy chooses which action to take next.
# 5. The chosen action is logged by the tracker.
# 6. A response is sent to the user.
#
# .... to be continued
#
# ### Installation
# You will need to install [Rasa NLU](https://rasa.com/docs/nlu/installation/), [Rasa Core](https://rasa.com/docs/core/installation/) and a [SpaCy language model](https://spacy.io/usage/models#section-install).
#
# These resourses were very useful while working on this project:
# 1. https://rasa.com/docs/ ##Rasa documentation
# 2. https://spacy.io/ ##SpaCy documentation
# 3. https://github.com/RasaHQ/rasa-workshop-pydata-berlin ##Workshop at Pydata by @JustinaPetr
# 4. https://github.com/RasaHQ/starter-pack-rasa-stack ##Rasa Stack starter-pack
# 5. https://developer.translink.ca/ServicesRtti/ApiReference ##TansLink next bus API documentation
# 6. https://developers.google.com/maps/documentation/directions/intro ##Google Maps directions API documentation
# +
import pandas as pd
import numpy as np
import matplotlib
import rasa_nlu
import rasa_core
import spacy
import nbmultitask
import sklearn_crfsuite
print("pandas_" + pd.__version__)
print("numpy_" + np.__version__)
print("matplotlib_" + matplotlib.__version__)
print("rasa_nlu_" + rasa_nlu.__version__)
print("rasa_core_" + rasa_core.__version__)
print("spacy_" + spacy.__version__)
print("sklearn-crfsuite_" + "0.3.6")
print("nbmultitask_" + "0.1.0")
# -
# ## Jupyter notebook configuration
import matplotlib
# +
# %matplotlib inline
import logging, io, json, warnings
logging.basicConfig(level="INFO")
warnings.filterwarnings('ignore')
# helper function to pretty print json
def pprint(o):
print(json.dumps(o, indent=2))
# -
# ## Import Rasa NLU, Rasa Core, and Spacy
# +
import rasa_nlu
import rasa_core
import spacy
print("rasa_nlu: {} rasa_core: {}".format(rasa_nlu.__version__, rasa_core.__version__))
print("Loading spaCy language model...")
print(spacy.load("en")("Hello world!"))
# -
# ## Rasa NLU Model
# We will build an NLU model as described above to teach our chatbot how to understand user inputs.
# ### Configuring the NLU model
# We define the processing pipeline by specifiying the tools/models we will use to turnunstructured user inputs to structured data. As we discussed above, we will use the `spacy_sklearn` pipeline.
# +
config = """
language: "en"
pipeline: "spacy_sklearn"
"""
# %store config > config.yml
# -
# ### Creating training data for the NLU model
#
#
# ... details to be added
# ### Training the NLU model
# We use the training data and the model configuration to develp the NLU model.
# +
from rasa_nlu.training_data import load_data
from rasa_nlu.config import RasaNLUModelConfig
from rasa_nlu.model import Trainer
from rasa_nlu import config
# loading the nlu training samples
training_data = load_data("data/data.json")
# trainer to educate our pipeline
trainer = Trainer(config.load("config.yml"))
# train the model!
interpreter = trainer.train(training_data)
# store it for future use
model_directory = trainer.persist("./models/nlu", fixed_model_name="current")
# -
# ### Evaluating the model
# +
#on the training data
from rasa_nlu.evaluate import run_evaluation
run_evaluation("data/data.json", model_directory)
# +
#using cross validation
import sys
python = sys.executable
# !{python} -m rasa_nlu.evaluate \
# --data data/data.json \
# --config config.yml \
# --mode crossvalidation
# -
# ### Testing the model
pprint(interpreter.parse("how can I go to 41 and 22 from 161 and 4th west?"))
#pprint(interpreter.parse("How can I go from Metrotown to Rogers Arena?"))
# ## Rasa Core Model
# ### Configure the domain file
# +
#https://rasa.com/docs/core/domains/
domain_yml = """
intents:
- greet
- affirm
- thanks
- goodbye
- search_bus_time
- search_trip
- garbage
slots:
line_number:
type: text
stop_number:
type: text
location_start:
type: text
location_end:
type: text
entities:
- line_number
- stop_number
- location_start
- location_end
actions:
- utter_greet
- utter_goodbye
- utter_thanks
- utter_garbage
- utter_affirm
- utter_ask_line_number
- utter_ask_stop_number
- utter_ask_location_start
- utter_ask_location_end
- action_next_bus
- action_trip
templates:
utter_greet:
- text: "Hey! How can help you?"
- text: "Hello! How can help you?"
- text: "Hi, traveller! How can help you?"
- text: "Greetings, welcome to the future! How can help you?"
utter_affirm:
- text: "Great!"
- text: "Awesome!"
- text: "Perfect!"
utter_thanks:
- text: "You are very welcome!"
- text: "Anytime!"
- text: "My pleasure!"
utter_goodbye:
- text: "Bye"
- text: "See you soon!"
- text: "Goodbye!"
- text: "Safe trip!"
- text: "Happy travelling!"
utter_garbage:
- text: "I am not sure what you are aiming for. Please give me more specific information such as the stop number and the bus line or the origin and destination for your trip."
- text: "Huh, come again?! Please give me more specific information such as the stop number and the bus line or the origin and destination for your trip."
- text: "Hmm ... not sure what you're asking? Please provide more specific information such as the stop number and the bus line or the origin and destination for your trip."
- text: "Hmm ... I am still learning! Can you rephrase that please? It's easier for me if you give me specific information such as the stop number and the bus line or the origin and destination for your trip."
- text: "What do you mean? It'd be easier for me if you included specific information such as the stop number and the bus line or the origin and destination for your trip."
- text: "Well .. it depends! Can you be more specific? Please provide me with the stop number and the bus line or the origin and destination for your trip."
utter_ask_line_number:
- text: "Sure, what is the bus number? For example, you can say: 'Bus number 8'"
- text: "I need the bus number. For example, you can say: 'Bus number 8'"
- text: "Which bus line do you plan on taking? For example, you can say: 'Bus number 8'"
utter_ask_stop_number:
- text: "Sure, what is the stop number? For example, you can say: 'Stop number 50234'"
- text: "I can help you with that! I need the stop number. For example, you can say: 'Stop number 50234'"
- text: "Which stop number? For example, you can say: 'Stop number 50234'"
utter_ask_location_start:
- text: "Sure, what is the start address?"
- text: "I can help you with that! I just need to know where you're trip is starting from."
- text: "I can help you with that! I need the starting address."
utter_ask_location_end:
- text: "Sure, what is the address of your destination?"
- text: "I can help you with that! I just need the location or address of where you're going."
- text: "To which destination?"
"""
# %store domain_yml > domain.yml
# -
# ### Specify action endpoint
# +
actions_py = """
from rasa_core_sdk import Action
from rasa_core_sdk.events import SlotSet
#from rasa_core.actions import Action
#from rasa_core.events import SlotSet
# nextbus api action
import requests
import xml.etree.ElementTree as ET
import pandas as pd
import datetime
# google maps api action
import json
import urllib.request, urllib.parse, urllib.error
import time
import re
from IPython.core.display import display, HTML
class NextbusApiAction(Action):
def name(self):
return "action_next_bus"
def run(self, dispatcher, tracker, domain):
with open('keys/nextbus_api_key.txt', 'r') as myfile:
nextbus_api_key = myfile.read()
line_number = tracker.get_slot('line_number')
stop_number = tracker.get_slot('stop_number')
req = 'http://api.translink.ca/rttiapi/v1/stops/'+stop_number+'/estimates?apikey='+nextbus_api_key+'&routeNo='+line_number
resp = requests.get(req)
root = ET.fromstring(resp.text)
xmlstr = str(ET.tostring(root, encoding='utf8', method='text'))
if '200' in str(resp): #valid response code
nbs = []
for nb in root:
r = nb.find('RouteNo').text
for s in nb.find('Schedules'):
t = s.find('ExpectedLeaveTime').text
nbs.append((r,t))
nbs=pd.DataFrame(nbs,columns=('route','lvtime'))
nbs.sort_values(['route','lvtime'])
lvtime = nbs.iloc[0].lvtime
dispatcher.utter_message("Your next bus will arrive at: {}".format(lvtime))
else:
dispatcher.utter_message(\"\"\"Hmm ... something went wrong :(\nInvalid bus number and/or stop number combination specified. Make sure that this bus number runs at the stop number you specified.\nPlease use a valid bus number is 3 digits, for example 003, 590. For community shuttle or night bus routes, please use \"C\" or \"N\" followed by up to 2 digits, i.e. N9, C22\"\"\")
class TripApiAction(Action):
#define regex to strip postal code from address
postalcode_re = re.compile(r'[A-Z]\d[A-Z] *\d[A-Z]\d')
def remove_postalcode(text):
return postalcode_re.sub('', text)
#define regex to strip html tags from instructions
htmltags_re = re.compile(r'<[^>]+>')
def remove_htmltags(text):
return htmltags_re.sub(' ', text)
#base url for api call
url_base = 'https://maps.googleapis.com/maps/api/directions/json?'
#mode of travel
mode = 'transit'
#set departure time as now -- can be changed later to provide directions in the future
departure_time = int(time.time())
def name(self):
return "action_trip"
def run(self, dispatcher, tracker, domain):
with open('keys/googlemaps_api_key.txt', 'r') as myfile:
googlemaps_api_key = myfile.read()
location_start = tracker.get_slot('location_start')
location_end = tracker.get_slot('location_end')
try:
location_start = location_start + ' bc, Canada'
except:
pass
try:
location_end = location_end + ' bc, Canada'
except:
pass
def direction(location_start, location_end, departure_time):
'''
Function to retreive Google Maps directions based on start and end locations, mode, and departure time.
Mode is defined as transit and departure time is set to the current time of the request.
'''
#define query arguments
query_args = {
'origin':location_start,
'destination':location_end,
'mode': mode,
'departure_time': departure_time
}
#encode the arguments
encoded_args = urllib.parse.urlencode(query_args)
#create the url with the encoded arguments and api key
url = url_base + encoded_args
encoded_url= url+'&key='+googlemaps_api_key
#make the request and save the json response
resp=urllib.request.urlopen(encoded_url).read()
data = json.loads(resp)
return(data)
if location_start == '' or location_end == '' or location_start is None or location_end is None or location_start == location_end:
print("Hmm ... Something went wrong! I can't find the location :(")
else:
data = direction(location_start, location_end, departure_time)
#fallback if the resquest failed
if data['status'] != 'OK':
instructions = 'No route found! Please give me more specific information about the locations of your trip origin and destination.'
dispatcher.utter_message(instructions)
else:
#grab some information form the json response
start_address = data['routes'][0]['legs'][0]['start_address']
end_address = data['routes'][0]['legs'][0]['end_address']
start_address = remove_postalcode(start_address).replace(', BC', '').replace(', Canada', '')
end_address = remove_postalcode(end_address).replace(', BC', '').replace(', Canada', '')
#create a map url
map_url = 'https://www.google.com/maps?'+urllib.parse.urlencode({'saddr': start_address, 'daddr': end_address, 'dirflg': 'r'})
travel_time = data['routes'][0]['legs'][0]['duration']['text']
if travel_time == 0:
arrival_time = departure_time
else:
arrival_time = data['routes'][0]['legs'][0]['arrival_time']['text']
#grab instructions (step-by-step directions)
#html_instructions can be found under routes/legs/steps or routes/legs/steps/steps
#loop through all the steps and save instructions
instructions = []
num_iters = max(len(data['routes'][0]['legs'][0]['steps']), len(data['routes'][0]['legs'][0]['steps'][0]['steps']))
for i in range (0, num_iters):
try:
instructions.append(remove_postalcode(data['routes'][0]['legs'][0]['steps'][i]['html_instructions'].replace('Subway towards', 'Take SkyTrain')).replace(', BC', '').replace(', Canada', '').replace('U-turn', 'turn'))
except:
pass
for j in range (0, num_iters):
try:
instructions.append(remove_postalcode(remove_htmltags(data['routes'][0]['legs'][0]['steps'][i]['steps'][j]['html_instructions'])).replace(', BC', '').replace(', Canada', '').replace('U-turn', 'turn'))
except:
pass
dispatcher.utter_message(\"\"\"Here are the directions from {} to {}:\n{}\n\nIf you leave now, you should arrive at {} by {}. Your trip should take around {}.\n\"\"\".format(start_address, end_address, (\"\"\"\n\"\"\".join(instructions)), end_address, arrival_time, travel_time))
dispatcher.utter_message(\"\"\"Click on the following link to view the direction on Google Maps:\n{}\"\"\".format(map_url))
#display(HTML("<a href="+map_url+">Google Maps Directions</a>"))
"""
# # %store actions_py > actions.py
# +
endpoints_yml = """
action_endpoint:
url: "http://localhost:5055/webhook"
"""
# %store endpoints_yml > endpoints.yml
# +
# run this in a separate notebook session
# # !python -m rasa_core_sdk.endpoint --actions actions
# # alternatively, run the custome action endpoint on a separate thread
# # # !pip install --user nbmultitask
# from nbmultitask import ThreadWithLogAndControls
# from time import sleep
#
# # the target function will be passed a function called `thread_print`
# def fn(thread_print):
# # !python -m rasa_core_sdk.endpoint --actions actions
# task = ThreadWithLogAndControls(target=fn, name="run action end point")
# task.control_panel()
# -
# ### Training The Dialogue Model
# +
# https://rasa.com/docs/core/policies/#policy-file
policy_config_yml = """
policies:
- name: KerasPolicy
epochs: 200
max_history: 2
validation_split: 0.2
batch_size: 32
- name: FallbackPolicy
fallback_action_name: 'utter_garbage'
core_threshold: 0.1
nlu_threshold: 0.1
- name: MemoizationPolicy
max_history: 5
- name: FormPolicy
"""
# %store policy_config_yml > policy_config.yml
# default changed #
#policies:
# - name: KerasPolicy
# epochs: 100
# max_history: 5
# - name: FallbackPolicy
# fallback_action_name: 'action_default_fallback'
# - name: MemoizationPolicy
# max_history: 5
# - name: FormPolicy
# -
#from rasa_core.policies import FallbackPolicy, KerasPolicy, MemoizationPolicy
#from rasa_core.agent import Agent
#
## this will catch predictions the model isn't very certain about
#fallback = FallbackPolicy(fallback_action_name="utter_garbage",
# core_threshold=0.1,
# nlu_threshold=0.1)
#
#agent = Agent(domain='domain.yml',
# policies=[MemoizationPolicy(max_history=2),
# KerasPolicy(epochs = 200, batch_size = 32, validation_split = 0.2),
# fallback])
#
## loading our neatly defined training dialogues
#training_data = agent.load_data('data/stories.md')
#
#agent.train(training_data)
#
#agent.persist('models/dialogue')
# alternatively, run directly via python
# !python -m rasa_core.train -d domain.yml -s data/stories.md -o models/dialogue -c policy_config.yml
# ### Evaluating the model
#### Evaluation of the dialogue model on the *traning data*
#from rasa_core.evaluate import run_story_evaluation
#
#run_story_evaluation("data/stories.md", "models/dialogue")#,
#nlu_model_path=None,
#max_stories=None,
#out_file_plot="models/dialogue/story_eval.pdf")
# +
# #!pip install --user service_identity==18.1.0
# -
# !python -m rasa_core.evaluate --core models/dialogue --stories data/stories.md --nlu models/nlu/default/current --output results
# ## Loading the model
from rasa_core.agent import Agent
agent = Agent.load('models/dialogue', interpreter=model_directory)
# ## Testing the model
# +
print("TransLink's Trip Planner bot is ready to serve you! Type your messages here or send 'stop' to end the session.")
while True:
a = input()
if a == 'stop':
break
responses = agent.handle_text(a)
for response in responses:
print(response["text"])
## Use the following line to debug when needed
#python -m rasa_core.run -d models/dialogue -u models/nlu/default/current --endpoints endpoints.yml --debug
# -
# # Interactive Training Session
# +
# from __future__ import absolute_import
# from __future__ import division
# from __future__ import print_function
# from __future__ import unicode_literals
# import logging
# from rasa_core import utils
# from rasa_core.agent import Agent
# from rasa_core.channels.console import ConsoleInputChannel
# from rasa_core.interpreter import RasaNLUInterpreter
# from rasa_core.policies import FallbackPolicy, KerasPolicy, MemoizationPolicy
# logger = logging.getLogger(__name__)
# def run_online(input_channel, interpreter,
# domain_file="domain.yml",
# training_data_file='stories.md'):
# fallback = FallbackPolicy(fallback_action_name="utter_garbage",
# core_threshold=0.1,
# nlu_threshold=0.1)
# agent = Agent('domain.yml', policies=[MemoizationPolicy(), KerasPolicy(), fallback], interpreter = interpreter)
# training_data = agent.load_data(training_data_file)
# agent.train_online(training_data,
# input_channel=input_channel,
# epochs=200)
# return agent
# utils.configure_colored_logging(loglevel="INFO")
# nlu_interpreter = RasaNLUInterpreter('models/nlu/default/current')
# run_online(ConsoleInputChannel(), nlu_interpreter)
# -
# ## Slack setup
#
# Useful resources:
#
# https://rasa.com/docs/core/connectors/#slack-setup
#
# https://github.com/JustinaPetr/Rasa-Slack-Connector
#
# https://www.fullstackpython.com/blog/build-first-slack-bot-python.html
#
# https://api.slack.com/slack-apps
#
# setup ngrok ... to be added.
# +
from rasa_core.channels import HttpInputChannel
from rasa_core.agent import Agent
from rasa_core.interpreter import RasaNLUInterpreter
from rasa_slack_connector import SlackInput
import pandas as pd
slack_tokens = pd.read_csv("keys/slack_tokens.txt", header=0, sep='\t')
app_verification_token = slack_tokens[slack_tokens['token'] == 'app_verification_token']['key'].values[0]
bot_verification_token = slack_tokens[slack_tokens['token'] == 'bot_verification_token']['key'].values[0]
slack_verification_token = slack_tokens[slack_tokens['token'] == 'slack_verification_token']['key'].values[0]
nlu_interpreter = RasaNLUInterpreter('./models/nlu/default/current')
agent = Agent.load('./models/dialogue', interpreter = nlu_interpreter)
input_channel = SlackInput(app_verification_token, bot_verification_token, slack_verification_token, True)
agent.handle_channel(HttpInputChannel(5004, '/', input_channel))
| 36.915902
| 2,476
|
1a1ffec2f4b9daba937988c5f45aa4050ac3ff36
|
py
|
python
|
tutorials/Tutorial6_Better_Retrieval_via_DPR.ipynb
|
yingzwang/haystack
|
['Apache-2.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] colab_type="text" id="bEH-CRbeA6NU"
# # Better Retrieval via "Dense Passage Retrieval"
#
# [](https://colab.research.google.com/github/deepset-ai/haystack/blob/master/tutorials/Tutorial6_Better_Retrieval_via_DPR.ipynb)
#
# ### Importance of Retrievers
#
# The Retriever has a huge impact on the performance of our overall search pipeline.
#
#
# ### Different types of Retrievers
# #### Sparse
# Family of algorithms based on counting the occurrences of words (bag-of-words) resulting in very sparse vectors with length = vocab size.
#
# **Examples**: BM25, TF-IDF
#
# **Pros**: Simple, fast, well explainable
#
# **Cons**: Relies on exact keyword matches between query and text
#
#
# #### Dense
# These retrievers use neural network models to create "dense" embedding vectors. Within this family there are two different approaches:
#
# a) Single encoder: Use a **single model** to embed both query and passage.
# b) Dual-encoder: Use **two models**, one to embed the query and one to embed the passage
#
# Recent work suggests that dual encoders work better, likely because they can deal better with the different nature of query and passage (length, style, syntax ...).
#
# **Examples**: REALM, DPR, Sentence-Transformers
#
# **Pros**: Captures semantinc similarity instead of "word matches" (e.g. synonyms, related topics ...)
#
# **Cons**: Computationally more heavy, initial training of model
#
#
# ### "Dense Passage Retrieval"
#
# In this Tutorial, we want to highlight one "Dense Dual-Encoder" called Dense Passage Retriever.
# It was introdoced by Karpukhin et al. (2020, https://arxiv.org/abs/2004.04906.
#
# Original Abstract:
#
# _"Open-domain question answering relies on efficient passage retrieval to select candidate contexts, where traditional sparse vector space models, such as TF-IDF or BM25, are the de facto method. In this work, we show that retrieval can be practically implemented using dense representations alone, where embeddings are learned from a small number of questions and passages by a simple dual-encoder framework. When evaluated on a wide range of open-domain QA datasets, our dense retriever outperforms a strong Lucene-BM25 system largely by 9%-19% absolute in terms of top-20 passage retrieval accuracy, and helps our end-to-end QA system establish new state-of-the-art on multiple open-domain QA benchmarks."_
#
# Paper: https://arxiv.org/abs/2004.04906
# Original Code: https://fburl.com/qa-dpr
#
#
# *Use this* [link](https://colab.research.google.com/github/deepset-ai/haystack/blob/master/tutorials/Tutorial6_Better_Retrieval_via_DPR.ipynb) *to open the notebook in Google Colab.*
#
# + [markdown] colab_type="text" id="3K27Y5FbA6NV"
# ### Prepare environment
#
# #### Colab: Enable the GPU runtime
# Make sure you enable the GPU runtime to experience decent speed in this tutorial.
# **Runtime -> Change Runtime type -> Hardware accelerator -> GPU**
#
# <img src="https://raw.githubusercontent.com/deepset-ai/haystack/master/docs/_src/img/colab_gpu_runtime.jpg">
# + colab={"base_uri": "https://localhost:8080/", "height": 357} colab_type="code" id="JlZgP8q1A6NW" outputId="c893ac99-b7a0-4d49-a8eb-1a9951d364d9"
# Make sure you have a GPU running
# !nvidia-smi
# + colab={"base_uri": "https://localhost:8080/", "height": 1000} colab_type="code" id="NM36kbRFA6Nc" outputId="af1a9d85-9557-4d68-ea87-a01f00c584f9"
# Install the latest release of Haystack in your own environment
# #! pip install farm-haystack
# Install the latest master of Haystack
# !pip install grpcio-tools==1.34.1
# !pip install git+https://github.com/deepset-ai/haystack.git
# If you run this notebook on Google Colab, you might need to
# restart the runtime after installing haystack.
# + colab={} colab_type="code" id="xmRuhTQ7A6Nh"
from haystack.utils import clean_wiki_text, convert_files_to_dicts, fetch_archive_from_http, print_answers
from haystack.nodes import FARMReader, TransformersReader
# + [markdown] colab_type="text" id="q3dSo7ZtA6Nl"
# ### Document Store
#
# #### Option 1: FAISS
#
# FAISS is a library for efficient similarity search on a cluster of dense vectors.
# The `FAISSDocumentStore` uses a SQL(SQLite in-memory be default) database under-the-hood
# to store the document text and other meta data. The vector embeddings of the text are
# indexed on a FAISS Index that later is queried for searching answers.
# The default flavour of FAISSDocumentStore is "Flat" but can also be set to "HNSW" for
# faster search at the expense of some accuracy. Just set the faiss_index_factor_str argument in the constructor.
# For more info on which suits your use case: https://github.com/facebookresearch/faiss/wiki/Guidelines-to-choose-an-index
# + colab={"base_uri": "https://localhost:8080/", "height": 51} colab_type="code" id="1cYgDJmrA6Nv" outputId="a8aa6da1-9acf-43b1-fa3c-200123e9bdce" pycharm={"name": "#%%\n"}
from haystack.document_stores import FAISSDocumentStore
document_store = FAISSDocumentStore(faiss_index_factory_str="Flat")
# + [markdown] pycharm={"name": "#%% md\n"}
# #### Option 2: Milvus
#
# Milvus is an open source database library that is also optimized for vector similarity searches like FAISS.
# Like FAISS it has both a "Flat" and "HNSW" mode but it outperforms FAISS when it comes to dynamic data management.
# It does require a little more setup, however, as it is run through Docker and requires the setup of some config files.
# See [their docs](https://milvus.io/docs/v1.0.0/milvus_docker-cpu.md) for more details.
# + pycharm={"name": "#%%\n"}
from haystack.utils import launch_milvus
from haystack.document_stores import MilvusDocumentStore
launch_milvus()
document_store = MilvusDocumentStore()
# + [markdown] colab_type="text" id="06LatTJBA6N0" pycharm={"name": "#%% md\n"}
# ### Cleaning & indexing documents
#
# Similarly to the previous tutorials, we download, convert and index some Game of Thrones articles to our DocumentStore
# + colab={"base_uri": "https://localhost:8080/", "height": 156} colab_type="code" id="iqKnu6wxA6N1" outputId="bb5dcc7b-b65f-49ed-db0b-842981af213b" pycharm={"name": "#%%\n"}
# Let's first get some files that we want to use
doc_dir = "data/article_txt_got"
s3_url = "https://s3.eu-central-1.amazonaws.com/deepset.ai-farm-qa/datasets/documents/wiki_gameofthrones_txt.zip"
fetch_archive_from_http(url=s3_url, output_dir=doc_dir)
# Convert files to dicts
dicts = convert_files_to_dicts(dir_path=doc_dir, clean_func=clean_wiki_text, split_paragraphs=True)
# Now, let's write the dicts containing documents to our DB.
document_store.write_documents(dicts)
# + [markdown] colab_type="text" id="wgjedxx_A6N6"
# ### Initalize Retriever, Reader & Pipeline
#
# #### Retriever
#
# **Here:** We use a `DensePassageRetriever`
#
# **Alternatives:**
#
# - The `ElasticsearchRetriever`with custom queries (e.g. boosting) and filters
# - Use `EmbeddingRetriever` to find candidate documents based on the similarity of embeddings (e.g. created via Sentence-BERT)
# - Use `TfidfRetriever` in combination with a SQL or InMemory Document store for simple prototyping and debugging
# + colab={"base_uri": "https://localhost:8080/", "height": 1000, "referenced_widgets": ["20affb86c4574e3a9829136fdfe40470", "7f8c2c86bbb74a18ac8bd24046d99d34", "84311c037c6e44b5b621237f59f027a0", "05d793fc179746e9b74cbcbc1a3389eb", "ad2ce6a8b4f844ac93b425f1261c131f", "bb45d5e4c9944fcd87b408e2fbfea440", "248d02e01dea4a63a3296e28e4537eaf", "74a9c43eb61a43aa973194b0b70e18f5", "58fc3339f13644aea1d4c6d8e1d43a65", "460bef2bfa7d4aa480639095555577ac", "8553a48fb3144739b99fa04adf8b407c", "babe35bb292f4010b64104b2b5bc92af", "887412c45ce744efbcc875b563770c29", "b4b950d899df4e3fbed9255b281e988a", "89535c589aa64648b82a9794a2888e78", "f35430501bb14fba8dbd5fb797c2e509", "eb5d93a8416a437e9cb039650756ac74", "5b8d5975d2674e7e9ada64e77c463c0a", "4afa2be1c2c5483f932a42ea4a7897af", "0e7186eeb5fa47d89c8c111ebe43c5af", "fa946133dfcc4a6ebc6fef2ef9dd92f7", "518b6a993e42490297289f2328d0270a", "cea074a636d34a75b311569fc3f0b3ab", "2630fd2fa91d498796af6d7d8d73aba4"]} colab_type="code" id="kFwiPP60A6N7" outputId="07249856-3222-4898-9246-68e9ecbf5a1b" pycharm={"is_executing": true}
from haystack.nodes import DensePassageRetriever
retriever = DensePassageRetriever(document_store=document_store,
query_embedding_model="facebook/dpr-question_encoder-single-nq-base",
passage_embedding_model="facebook/dpr-ctx_encoder-single-nq-base",
max_seq_len_query=64,
max_seq_len_passage=256,
batch_size=16,
use_gpu=True,
embed_title=True,
use_fast_tokenizers=True)
# Important:
# Now that after we have the DPR initialized, we need to call update_embeddings() to iterate over all
# previously indexed documents and update their embedding representation.
# While this can be a time consuming operation (depending on corpus size), it only needs to be done once.
# At query time, we only need to embed the query and compare it the existing doc embeddings which is very fast.
document_store.update_embeddings(retriever)
# + [markdown] colab_type="text" id="rnVR28OXA6OA"
# #### Reader
#
# Similar to previous Tutorials we now initalize our reader.
#
# Here we use a FARMReader with the *deepset/roberta-base-squad2* model (see: https://huggingface.co/deepset/roberta-base-squad2)
#
#
#
# ##### FARMReader
# + colab={"base_uri": "https://localhost:8080/", "height": 739, "referenced_widgets": ["3d273d2d3b25435ba4eb4ffd8e812b6f", "5104b7cddf6d4d0f92d3dd142b9f4c42", "e0510255a31d448497af3ca0f4915cb4", "670270fd06274932adad4d42c8a1912e", "6ca292cd3f46417ea296684e48863af9", "75578e0466cd4b84ba7dfee1028ae4cd", "cbe09b984b804402b1fe82739cbc375c", "4fd0caca56bd415b8c31860ba542145a", "9960be4cc1c64905917b5fd7ea6bb294", "2f3d901b3acb4841a4b03b2c5cd4393b", "04644b74bb2a45a7a6fcf86151b5bf8c", "5efa895c53284b72adec629a6fc59fa9", "182e5db14fac427b90380b5213f57825", "243600e420f449089c1b5ed0d2715339", "466222c8b2e1403ca69c8130423f0a8b", "a458be4cc49240e4b9bc1c95c05551e8", "d9ee08fa621d4b558bd1a415e3ee6f62", "1b905c5551b940ed9bc5320e1e5a9213", "64fc7775a84e425c8082a545f7c2a0c1", "66cd72dae82d434a87b638236784fd4b", "36b1b48aea02494a8bc94020a15d7417", "5934bc4db2a94c20b5c55f1c017024ab", "f9289caeac404087ad4973a646e3a117", "7e121f0fdb1746c094bff218a4f623ab", "98781635b86244aca5d22be4280c32de", "e148b28d946549a9b5eb09294ebe124e", "4b8b29c1b1a243808de4cc1cae3f6bd6", "bbef597f804e4ca580aee665399a3bc1", "345f49b2b42c40278478d30e8a691768", "e3724385769d443cb4ea39b92e0b2abd", "d05fbb94014840cab4584c4781a590c1", "b8d52b604dad43c18ba00c935b961422", "e625a32fc81b42fb9e0fff7ce766fcdc", "885390f24e08495db6a1febd661531e0", "c2a614f48e974fb8b13a3c5d7cafaed6", "ada8fa1c88954ef8b839f29090de9e79", "427b07b356e44c68b47178b277aaa16f", "1b4166bda5ae48aa8539e0fa5521007a", "fd30d43909874239b2183c5fb61241fe", "09a647660cf94131a1c140d06eb293ab", "3e482e9ef4d34d93b4ba4f7f07b0e44f", "66450cab654d40ae8ed1c32fa733397a", "aa4becf2e33d4f1e9fdac70236d48f6e", "78d087ed952e429b97eb3d8fcdc7c8ec", "5020846874ae473bbfa7038fe98de474", "08c736f4ad424330a82df1b5dc047b2c", "9169ca606bf64d41aa08fb42876bd2ab", "c8f1f7e8462d4d14a507816f67953eae"]} colab_type="code" id="fyIuWVwhA6OB" outputId="33113253-8b95-4604-f9e5-1aa28ee66a91"
# Load a local model or any of the QA models on
# Hugging Face's model hub (https://huggingface.co/models)
reader = FARMReader(model_name_or_path="deepset/roberta-base-squad2", use_gpu=True)
# + [markdown] colab_type="text" id="unhLD18yA6OF"
# ### Pipeline
#
# With a Haystack `Pipeline` you can stick together your building blocks to a search pipeline.
# Under the hood, `Pipelines` are Directed Acyclic Graphs (DAGs) that you can easily customize for your own use cases.
# To speed things up, Haystack also comes with a few predefined Pipelines. One of them is the `ExtractiveQAPipeline` that combines a retriever and a reader to answer our questions.
# You can learn more about `Pipelines` in the [docs](https://haystack.deepset.ai/docs/latest/pipelinesmd).
# + colab={} colab_type="code" id="TssPQyzWA6OG"
from haystack.pipelines import ExtractiveQAPipeline
pipe = ExtractiveQAPipeline(reader, retriever)
# + [markdown] colab_type="text" id="bXlBBxKXA6OL"
# ## Voilà! Ask a question!
# + colab={"base_uri": "https://localhost:8080/", "height": 275} colab_type="code" id="Zi97Hif2A6OM" outputId="5eb9363d-ba92-45d5-c4d0-63ada3073f02"
# You can configure how many candidates the reader and retriever shall return
# The higher top_k for retriever, the better (but also the slower) your answers.
prediction = pipe.run(
query="Who created the Dothraki vocabulary?", params={"Retriever": {"top_k": 10}, "Reader": {"top_k": 5}}
)
# -
print_answers(prediction, details="minimal")
# ## About us
#
# This [Haystack](https://github.com/deepset-ai/haystack/) notebook was made with love by [deepset](https://deepset.ai/) in Berlin, Germany
#
# We bring NLP to the industry via open source!
# Our focus: Industry specific language models & large scale QA systems.
#
# Some of our other work:
# - [German BERT](https://deepset.ai/german-bert)
# - [GermanQuAD and GermanDPR](https://deepset.ai/germanquad)
# - [FARM](https://github.com/deepset-ai/FARM)
#
# Get in touch:
# [Twitter](https://twitter.com/deepset_ai) | [LinkedIn](https://www.linkedin.com/company/deepset-ai/) | [Slack](https://haystack.deepset.ai/community/join) | [GitHub Discussions](https://github.com/deepset-ai/haystack/discussions) | [Website](https://deepset.ai)
#
# By the way: [we're hiring!](https://www.deepset.ai/jobs)
| 59.299145
| 1,900
|
58113bb02d0d13424204a547592142366f48e90a
|
py
|
python
|
ResourceWatch/Api_definition/layer_definition.ipynb
|
resource-watch/notebooks
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] toc="true"
# # Table of Contents
# <p><div class="lev1 toc-item"><a href="#Layer-object-definition" data-toc-modified-id="Layer-object-definition-1"><span class="toc-item-num">1 </span>Layer object definition</a></div><div class="lev2 toc-item"><a href="#Index" data-toc-modified-id="Index-11"><span class="toc-item-num">1.1 </span>Index</a></div><div class="lev2 toc-item"><a href="#Basic-object-definition-" data-toc-modified-id="Basic-object-definition--12"><span class="toc-item-num">1.2 </span>Basic object definition </a></div><div class="lev2 toc-item"><a href="#Layer-Provider-&-Layer-Type-definition-" data-toc-modified-id="Layer-Provider-&-Layer-Type-definition--13"><span class="toc-item-num">1.3 </span>Layer Provider & Layer Type definition </a></div><div class="lev2 toc-item"><a href="#Definition:-layerConfig-object-" data-toc-modified-id="Definition:-layerConfig-object--14"><span class="toc-item-num">1.4 </span>Definition: layerConfig object </a></div><div class="lev2 toc-item"><a href="#interactionConfig-definition--" data-toc-modified-id="interactionConfig-definition---15"><span class="toc-item-num">1.5 </span>interactionConfig definition </a></div><div class="lev2 toc-item"><a href="#Definition:-legendConfig-object-" data-toc-modified-id="Definition:-legendConfig-object--16"><span class="toc-item-num">1.6 </span>Definition: legendConfig object </a></div>
# -
# # Layer object definition
# ## Index
#
# * [Basic object definition](#Basic object definition)
# * [Layer Provider](#Layer Provider)
# * Definition: [layerConfig](#layerConfig)
# * Definition: [pulseConfig](#pulseConfig)
# * Definition: [interactionConfig](#interactionConfig)
# * Definition: [legendConfig](#legendConfig)
# The aim of this document is to define a common layer object across all projects powered by the RW-API (for our own sanity)
#
# Right now we have identify 6 types of layers:
#
# * Raster tile
# * WMS
# * Vector tile
# * Geojson
# * Image overlay
# * Canvas layer
#
# We are going to try to cover all of them on our proposal.
# For more information here there is a compendium of documantation:
# [RW postman collection](https://www.getpostman.com/collections/5f3e83c82ad5a6066657)
# [RW documentation](https://resource-watch.github.io/doc-api/)
# [Leaflet](http://leafletjs.com/reference-1.0.3.html)
# [CARTOcss](https://carto.com/docs/carto-engine/cartocss/properties/)
# [Wms sld styling](http://server.arcgis.com/es/server/latest/publish-services/linux/customizing-a-wms-getfeatureinfo-response.htm)
# [Feature and image service styling](http://resources.arcgis.com/en/help/arcgis-rest-api/#/The_ArcGIS_REST_API/02r300000054000000/)
# ## Basic object definition <a name="Basic object definition"></a>
# Our proposal for the new basic layer definition is:
#
# ```json
# { "id": "<layer-id>",
# "type": "layer",
# "attributes": {
# "slug": "<slug>",
# "userId": "<user-id>",
# "dataset": "<dataset-id>",
# "application":["apps"],
# "name":"Example layer",
# "default": true,
# "published": true,
# "layerProvider": "carto",
# "layerType": "raster-tile",
# "layerConfig":{
# "body":{...},
# "pulseConfig": {...},
# ...
# },
# "legendConfig":{...},
# "interactionConfig":{...}
# }
# }
# ```
#
# Where each key parameter:
#
# | Field | Description | Type | Accepted values | Required |
# |:-------|:-------------|:------|:--------:|:----------|
# |**application**|Application to which the layer belongs|Array|gfw, forest-atlas, rw, prep, aqueduct, data4sdg|Yes|
# |**name**|Administrative name of the layer|Text|Any Text|Yes|
# |**default**|Especifies if the layer is the main layer visualization of the dataset. There can only be one by default per dataset and per application|Boolean|true, false|Yes|
# |**published**|If it is published within the app |Boolean|true, false|Yes|
# |**layerProvider**|Service used to retrive the visualization|Text|Any Text|Yes|
# |**layerType**|Type of layer|Text|Any Text|Yes|
# |**layerConfig**|Layer definition|Object|Valid object|Yes|
# |**legendConfig**|Legend configuration|Object|Valid object|Yes|
# |**interactionConfig**|Interaction configuration for the layer|Object|Valid object|Yes|
#
#
# This will work like:
# 1.- Each ```layerProvider``` will have its own ```layerType```
# 2.- Each ```layerType``` will define ```layerConfig``` possibilities
# 3.- ```layerType``` will also define the ```inteactionConfig```
# 4.- Depend on the data we will have a different set of ```legendConfig``` definition
#
#
#
# ## Layer Provider & Layer Type definition <a name="Layer Provider"></a>
# |layerProvider|layerType accepted values|
# |:------------|:------------------------|
# |carto|tile, canvas, geojson|
# |esriFeatureService|geojson|
# |esriMapService|tile|
# |esriImageService|tile, canvas, overlay|
# |esriVectorService|vector|
# |tileService|tile, canvas|
# |wmsService|wms, wfs|
# |mapbox|vector|
# |gee (not yet especified)|tile, canvas, geojson|
# |nasaGibs|tile|
# ## Definition: layerConfig object <a name="layerConfig"></a>
# ```json
# {
# "body":{...},
# "pulseConfig": {...},
# ...
# }
# ```
#
# Depending on the ```layerProvider``` and ```layerType``` there are variations over whats inside body and pulseConfig and also other parameters can be added.
# |provider|type (accepted values)|
# |:------------|:------------------------|
# |cartodb|Carto tile layers,tileLayer, canvas, geojson, wms|
# |featureservice|featureLayer, dynamicMapLayer, imageMapLayer, Vector.Layer, tileLayer, wms|
# |esriMapService|tile|
# |esriImageService|tile, canvas, overlay|
# |esriVectorService|vector|
# |tileService|tile, canvas|
# |wmsService|wms, wfs|
# |mapbox|vector|
# |gee (not yet especified)|tile, canvas, geojson|
# |nasaGibs|tile|
#
#
#
# ### Diferent layers definition
# #### [Carto tile layer](https://carto.com/docs/carto-engine/maps-api/mapconfig)
#
# This layers relays on the carto maps api definition. Inside body, we should use it. outside, we should have the account, and depend on the app there can be other params associated like in aqueduct or global forest watch.
# This layer configuration accepts gridjson as interactivity.
#
# ```json
# {
# "account": "insight",
# "body":{
# "maxzoom": 18,
# "minzoom": 3,
# "layers": [{
# "type": "cartodb",
# "options": {
# "sql": "",
# "cartocss": "",
# "cartocss_version": "2.3.0",
# "interactivity": ["cartodb_id", "iso3"]
# ...
# }
# }]
# },
# "pulseConfig": {...}
# }
# ```
#
#
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```account```|Carto account hosting data|text|Any valid account name|yes|
# |```body```|Visualization configuration|object|[Valid Layergroup configurations](https://carto.com/docs/carto-engine/maps-api/mapconfig)|yes|
# |```pulseConfig```|if exists how to visualize this layer in planet pulse|object|valid planet pulse object|no|
#
# #### Carto canvas.
# #### [esriFeatureService Layer](http://esri.github.io/esri-leaflet/api-reference/layers/feature-layer.html)
#
# ```json
# {
# "type": "featureLayer",
# "body": {
# "url": "http://services5.arcgis.com/jPWpe1SD1RQyeOMy/ArcGIS/rest/services/Areas_de_Risco/FeatureServer/0",
# "style": "function (feature) { var c,o = 0.8; switch (feature.properties.GRAU_RISCO) { case 'Alto': c = '#ffdb5d'; break; case 'Muito Alto': c = '#c15467'; break;default: c = '#ffdb5d'; return {color: c, opacity: o, weight: 5};}}",
# "useCors": false
# },
# "pulseConfig": {...}
# }
# ```
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```type```|Contructor name|Text|"featureLayer"|yes|
# |```body```|Visualization configuration|object|[Valid configuration](http://esri.github.io/esri-leaflet/api-reference/layers/feature-layer.html)|yes|
# |```pulseConfig```|if exists how to visualize this layer in planet pulse|object|valid planet pulse object|no|
#
# #### [esriMapService tiles](http://esri.github.io/esri-leaflet/api-reference/layers/dynamic-map-layer.html)
#
# ```json
# {
# "type": "dynamicMapLayer",
# "body": {
# "url": "https://services.arcgisonline.com/ArcGIS/rest/services/USA_Topo_Maps/MapServer",
# "layers": [8],
# "useCors": false
# }
# "pulseConfig": {...}
# }
# ```
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```type```|Contructor name|Text|"dynamicMapLayer"|yes|
# |```body```|Visualization configuration|object|[Valid configuration](http://esri.github.io/esri-leaflet/api-reference/layers/dynamic-map-layer.html)|yes|
# |```pulseConfig```|if exists how to visualize this layer in planet pulse|object|valid planet pulse object|no|
#
# #### [esriImageService](http://esri.github.io/esri-leaflet/api-reference/layers/image-map-layer.html)
#
# ```json
# {
# "type": "imageMapLayer",
# "body": {
# "url": "http://sampleserver3.arcgisonline.com/ArcGIS/rest/services/World/Temperature/ImageServer",
# "mosaicRule": {
# "mosaicMethod": "esriMosaicLockRaster",
# "lockRasterIds": [8]
# },
# "useCors": false
# },
# "pulseConfig": {...}
# }
# ```
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```type```|Contructor name|Text|"imageMapLayer"|yes|
# |```body```|Visualization configuration|object|[Valid configuration](http://esri.github.io/esri-leaflet/api-reference/layers/image-map-layer.html)|yes|
# |```pulseConfig```|if exists how to visualize this layer in planet pulse|object|valid planet pulse object|no|
# #### [esriVectorService](http://esri.github.io/esri-leaflet/api-reference/layers/vector-layer.html)
#
# ```json
# {
# "type": "Vector.Layer",
# "body": {
# "id":"bd505ce3efff479bb4e87b182f180159"
# },
# "pulseConfig": {...}
# }
# ```
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```type```|Contructor name|Text|"Vector.Layer"|yes|
# |```body```|Visualization configuration|object|[Valid configuration](http://esri.github.io/esri-leaflet/api-reference/layers/vector-layer.html)|yes|
# |```pulseConfig```|if exists how to visualize this layer in planet pulse|object|valid planet pulse object|no|
# #### [tileService](http://leafletjs.com/reference-1.0.3.html#tilelayer)
#
# ```json
# {
# "type": "tileLayer",
# "url": "https://storage.googleapis.com/global-surface-water/maptiles/transitions/{z}/{x}/{y}.png",
# "body": {
# "format": "image/png",
# "maxZoom": 13,
# "errorTileUrl" : "https://storage.googleapis.com/global-surface-water/downloads_ancillary/blank.png",
# "attribution": "2016 EC JRC/Google",
# "transparent": true
# },
# "pulseConfig": {...}
# }
# ```
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```type```|Contructor name|Text|"tileLayer"|yes|
# |```url```|Tileset url|Text|valid url|yes|
# |```body```|Visualization configuration options|object|[Valid configuration](http://leafletjs.com/reference-1.0.3.html#tilelayer)|yes|
# |```pulseConfig```|if exists how to visualize this layer in planet pulse|object|valid planet pulse object|no|
# #### [wmsService](http://leafletjs.com/reference-1.0.3.html#tilelayer-wms)
# ```json
# {
# "type": "wms",
# "url": "http://raster.nationalmap.gov/arcgis/services/LandCover/USGS_EROS_LandCover_NLCD/MapServer/WMSServer",
# "body": {
# "layers": "5",
# "format": "image/png",
# "transparent": true
# },
# "pulseConfig": {...}
# }
# ```
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```type```|Contructor name|Text|"wms"|yes|
# |```url```|Tileset url|Text|valid url|yes|
# |```body```|Visualization configuration options|object|[Valid configuration](http://leafletjs.com/reference-1.0.3.html#tilelayer-wms)|yes|
# |```pulseConfig```|if exists how to visualize this layer in planet pulse|object|valid planet pulse object|no|
# #### [Vector tiles](http://leaflet.github.io/Leaflet.VectorGrid/vectorgrid-api-docs.html#vectorgrid-protobuf)
# ```json
# {
# "type": "vector",
# "url": "https://www.tilehosting.com/data/v3/{z}/{x}/{y}.pbf",
# "body": {
# "vectorTileLayerStyles": { ... },
# "maxNativeZoom": 14
# },
# "pulseConfig": {...}
# }
# ```
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```type```|Contructor name|Text|"vector"|yes|
# |```url```|Tileset url|Text|valid url|yes|
# |```body```|Visualization configuration options|object|[Valid configuration](http://leaflet.github.io/Leaflet.VectorGrid/vectorgrid-api-docs.html#vectorgrid-protobuf)|yes|
# |```pulseConfig```|if exists how to visualize this layer in planet pulse|object|valid planet pulse object|no|
# #### [NasaGIBS](https://github.com/aparshin/leaflet-GIBS)
# ```json
# {
# "type": "nasaGibs",
# "gibsLayerId": "MODIS_Aqua_SurfaceReflectance_Bands721",
# "body": {
# "date": "2015/04/01",
# "transparent": 14
# },
# "pulseConfig": {...}
# }
# ```
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```type```|Contructor name|Text|"wms"|yes|
# |```gibsLayerId```|datasetId|Text|[valid name](https://wiki.earthdata.nasa.gov/display/GIBS/GIBS+Available+Imagery+Products)|yes|
# |```body```|Visualization configuration options|object|[Valid configuration](https://github.com/aparshin/leaflet-GIBS)|yes|
# |```pulseConfig```|if exists how to visualize this layer in planet pulse|object|valid planet pulse object|no|
# #### [EE livetiles](https://earthengine.google.com)
# ```json
# {
# "type": "gee",
# "asset_id": "landsat/blabla",
# "body": {
# "style_type": "standard",
# "sld_value": "sld definition"
# "standard_value": {}
# },
# "pulseConfig": {...}
# }
# ```
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```type```|Contructor name|Text|"gee"|yes|
# |```asset_id```|datasetId|Text|[valid name](https://wiki.earthdata.nasa.gov/display/GIBS/GIBS+Available+Imagery+Products)|yes|
# |```body```|Visualization configuration options|object|[Valid configuration]()|yes|
# |```pulseConfig```|if exists how to visualize this layer in planet pulse|object|valid planet pulse object|no|
#
# ****To be able to access it we will load a tilelayer: `api.resourcewatch.org/v1/layer/<layer-id>/tile/gee/{z}/{x}/{y}`
# ##### Valid ee layer configuration
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```style_type```|style type name|Text|"standard"/"sld"|yes|
# |```sld_value```|datasetId|Text|[valid name](https://wiki.earthdata.nasa.gov/display/GIBS/GIBS+Available+Imagery+Products)|yes|
# #### [NEXGDDP layers]()
# ```json
# {
# "type": "gee",
# "asset_id": "landsat/blabla",
# "body": {
# "style_type": "standard",
# "sld_value": "sld definition"
# "standard_value": {}
# },
# "pulseConfig": {...}
# }
# ```
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```type```|Contructor name|Text|"gee"|yes|
# |```asset_id```|datasetId|Text|[valid name](https://wiki.earthdata.nasa.gov/display/GIBS/GIBS+Available+Imagery+Products)|yes|
# |```body```|Visualization configuration options|object|[Valid configuration]()|yes|
# |```pulseConfig```|if exists how to visualize this layer in planet pulse|object|valid planet pulse object|no|
#
# ***To be able to access it we will load a tilelayer: `api.resourcewatch.org/v1/layer/<layer-id>/tile/gee/{z}/{x}/{y}`
# ##### Valid ee layer configuration
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```style_type```|style type name|Text|"standard"/"sld"|yes|
# |```sld_value```|datasetId|Text|[valid name](https://wiki.earthdata.nasa.gov/display/GIBS/GIBS+Available+Imagery+Products)|yes|
# ### Definition: pulseConfig object <a name="pulseConfig"></a>
#
# #### Image overlay
# * **layerProvider**: carto
# ```json
# {
# "type":"imageOverlay",
# "values": {
# "format": "png",
# "bbox": [-110, -65, 110, 65],
# "width": 2048,
# "height": 1024
# },
# "sql":"",
# "urlTemplate": "https://{{account}}.carto.com/api/v1/map/static/bbox/{{token_groupid}}/{{bbox}}/{{width}}/{{height}}.{{format}}"
# }
# ```
#
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```type```|Pulse object type|Text|"imageOverlay"|yes|
# |```values```|params to be substitute|object||yes|
# |-> ```format```|image format|Text|"png"|yes|
# |-> ```bbox```|bbox|Text|[-110, -65, 110, 65]|yes|
# |-> ```width```|output image width|Text|2048|yes|
# |-> ```height```|output image height|Text|1024|yes|
# |```urlTemplate```|url template|text|https://{{account}}.carto.com/api/v1/map/static/bbox/{{token_groupid}}/{{bbox}}/{{width}}/{{height}}.{{format}}|yes|
# |```sql```|reproyect sql|text|valid planet pulse sql|yes|
#
# * **layerProvider**: [esriImageService](http://resources.arcgis.com/en/help/arcgis-rest-api/#/Export_Image/02r3000000wm000000/)
# ```json
# {
# "type":"imageOverlay",
# "url":"http://gis-gfw.wri.org/arcgis/rest/services/image_services/glad_alerts/ImageServer/exportImage",
# "params":{
# "f":"image"
# "format":"png8",
# "pixelType":"U1"
# "bbox":[-180,-90,180,90],
# "bboxSR":{"wkid":4326},
# "size":"2048, 1024",
# "imageSR":{"wkid":4326},
# "noData":0,
# "interpolation":"RSP_Majority",
# "renderingRule":{"rasterFunction" : "Colormap", "rasterFunctionArguments" : { "Colormap" : [[0, 0, 0, 0, 0], [1, 219,101,152,1], [1000, 219,101,152,1]]}, "variableName" : "Raster"}
# }
# }
# ```
#
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```type```|Pulse object type|Text|"imageOverlay"|yes|
# |```params```|params to be substitute|object|[valid request parameters](http://resources.arcgis.com/en/help/arcgis-rest-api/#/Export_Image/02r3000000wm000000/)|yes|
# |```url```|url|text|valid Arcgis export image|yes|
#
#
# * **layerProvider**: s3
# ```json
# {
# "type":"imageOverlay",
# "url":"https://s3.amazonaws.com/mybucket/image.png",
# }
# ```
#
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```type```|Pulse object type|Text|"imageOverlay"|yes|
# |```url```|s3 url to image|text|valid s3 url|yes|
#
# #### 3d data overlay
# * **layerProvider**: carto
# ```json
# {
# "type":"3d",
# "url": "",
# "markerType": "<type>"
# }
# ```
#
#
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```type```|Pulse object type|Text|"3d"|yes|
# |```url```|url call from where to fetch the data|text|valid url|yes|
# |```markerType```|Valid marker type visualization for 3d rendering|text|`"hemisphere","bar","volcano"`|yes|
# ## interactionConfig definition <a name="interactionConfig"></a>
# This object will define the interaction and will control the information appearance.
#
#
# ```json
# {
# "type": "gridjson",
# "config": {...},
# "output": [...]
# }
# ```
#
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```type```|type of interaction for the layer|text|null, gridjson, intersection, geojson|yes|
# |```config```|how to access the interaction|object|valid object|yes|
# |```output```|visual format representation for the infowindow|array of objects||yes|
# ### config object definition
# + [markdown] heading_collapsed=true
# #### gridjson
#
# ```json
# {
# // if the grid layer doesnt belong to carto provider we will add this property
# "url":"<>/x/y/z.json"
# }
# ```
#
#
# #### intersection
#
# ```json
# {
# "url": "queryURL"
# }
# ```
#
#
# #### geojson
#
# ```json
# {
#
# }
# ```
#
#
# -
# ### Output object definition
# ```json
# // We will need to move this as an object instead of an array
# [{
# "column": "name",
# "property":"name of whatever:"
# "prefix":"",
# "suffix: "",
# "type": "string",
# "format": null
# }, {
# "column": "value",
# "property":"name of whatever:"
# "type": "number",
# "format": "$ %s M"
# },{
# "column": "date",
# "property":"name of whatever:"
# "type": "date",
# "format": "YYYY"
# },
# ...
# ]
# ```
# |Field|Description|Type|Accepted values|Required|
# |:----|:----------|:---|:--------------|:-------|
# |```column```|property or column name to get the data from|text|valid column name|yes|
# |```property```|displayable text for the property|text|valid text|yes|
# |```type```|data type|text|valid datatype|yes|
# |```format```|data representation format to be displayed in the infowindow|text|valid format syntax|yes|
#
# ## Definition: legendConfig object <a name="legendConfig"></a>
# * **Basic legend**: This type of legend can be used for single and for categorical legends
# ```json
# {
# "type": "basic",
# "items": [{
# "name": "name",
# "color": "#ff0000",
# "icon": "URL/SVG/string"},
# ...
# ],
# "unit": "m."
# }
# ```
# * **Gradient**: This type of legend can be used for gradient blending legends
# ```json
# {
# "type": "gradient",
# "items": [{
# "name": "name",
# "value": "1",
# "color": "#ff0000"},
# ...
# ],
# "unit": "m."
# }
# ```
# * **Choropleth**:
# ```json
# {
# "type": "choropleth",
# "items": [{
# "name": "name",
# "value": "1",
# "color": "#ff0000"},
# ...
# ],
# "unit": "m."
# }
# ```
#
#
| 35.464286
| 1,442
|
74bd375e0d99cc658a77f3e23e521feadef9446e
|
py
|
python
|
content/ch-machine-learning/machine-learning-qiskit-pytorch.ipynb
|
duartefrazao/qiskit-textbook
|
['Apache-2.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] tags=["remove_cell"]
# # Hybrid quantum-classical Neural Networks with PyTorch and Qiskit
# -
# Machine learning (ML) has established itself as a successful interdisciplinary field which seeks to mathematically extract generalizable information from data. Throwing in quantum computing gives rise to interesting areas of research which seek to leverage the principles of quantum mechanics to augment machine learning or vice-versa. Whether you're aiming to enhance classical ML algorithms by outsourcing difficult calculations to a quantum computer or optimise quantum algorithms using classical ML architectures - both fall under the diverse umbrella of quantum machine learning (QML).
#
# In this chapter, we explore how a classical neural network can be partially quantized to create a hybrid quantum-classical neural network. We will code up a simple example that integrates **Qiskit** with a state-of-the-art open-source software package - **[PyTorch](https://pytorch.org/)**. The purpose of this example is to demonstrate the ease of integrating Qiskit with existing ML tools and to encourage ML practitioners to explore what is possible with quantum computing.
#
# ## Contents
#
# 1. [How Does it Work?](#how)
# 1.1 [Preliminaries](#prelims)
# 2. [So How Does Quantum Enter the Picture?](#quantumlayer)
# 3. [Let's code!](#code)
# 3.1 [Imports](#imports)
# 3.2 [Create a "Quantum Class" with Qiskit](#q-class)
# 3.3 [Create a "Quantum-Classical Class" with PyTorch](#qc-class)
# 3.4 [Data Loading and Preprocessing](#data-loading-preprocessing)
# 3.5 [Creating the Hybrid Neural Network](#hybrid-nn)
# 3.6 [Training the Network](#training)
# 3.7 [Testing the Network](#testing)
# 4. [What Now?](#what-now)
# ## 1. How does it work? <a id='how'></a>
# <img src="hybridnetwork.png" />
#
# **Fig.1** Illustrates the framework we will construct in this chapter. Ultimately, we will create a hybrid quantum-classical neural network that seeks to classify hand drawn digits. Note that the edges shown in this image are all directed downward; however, the directionality is not visually indicated.
# ### 1.1 Preliminaries <a id='prelims'></a>
# The background presented here on classical neural networks is included to establish relevant ideas and shared terminology; however, it is still extremely high-level. __If you'd like to dive one step deeper into classical neural networks, see the well made video series by youtuber__ [3Blue1Brown](https://youtu.be/aircAruvnKk). Alternatively, if you are already familiar with classical networks, you can [skip to the next section](#quantumlayer).
#
# ###### Neurons and Weights
# A neural network is ultimately just an elaborate function that is built by composing smaller building blocks called neurons. A ***neuron*** is typically a simple, easy-to-compute, and nonlinear function that maps one or more inputs to a single real number. The single output of a neuron is typically copied and fed as input into other neurons. Graphically, we represent neurons as nodes in a graph and we draw directed edges between nodes to indicate how the output of one neuron will be used as input to other neurons. It's also important to note that each edge in our graph is often associated with a scalar-value called a [***weight***](https://en.wikipedia.org/wiki/Artificial_neural_network#Connections_and_weights). The idea here is that each of the inputs to a neuron will be multiplied by a different scalar before being collected and processed into a single value. The objective when training a neural network consists primarily of choosing our weights such that the network behaves in a particular way.
#
# ###### Feed Forward Neural Networks
# It is also worth noting that the particular type of neural network we will concern ourselves with is called a **[feed-forward neural network (FFNN)](https://en.wikipedia.org/wiki/Feedforward_neural_network)**. This means that as data flows through our neural network, it will never return to a neuron it has already visited. Equivalently, you could say that the graph which describes our neural network is a **[directed acyclic graph (DAG)](https://en.wikipedia.org/wiki/Directed_acyclic_graph)**. Furthermore, we will stipulate that neurons within the same layer of our neural network will not have edges between them.
#
# ###### IO Structure of Layers
# The input to a neural network is a classical (real-valued) vector. Each component of the input vector is multiplied by a different weight and fed into a layer of neurons according to the graph structure of the network. After each neuron in the layer has been evaluated, the results are collected into a new vector where the i'th component records the output of the i'th neuron. This new vector can then be treated as an input for a new layer, and so on. We will use the standard term ***hidden layer*** to describe all but the first and last layers of our network.
#
# ## 2. So How Does Quantum Enter the Picture? <a id='quantumlayer'> </a>
#
# To create a quantum-classical neural network, one can implement a hidden layer for our neural network using a parameterized quantum circuit. By "parameterized quantum circuit", we mean a quantum circuit where the rotation angles for each gate are specified by the components of a classical input vector. The outputs from our neural network's previous layer will be collected and used as the inputs for our parameterized circuit. The measurement statistics of our quantum circuit can then be collected and used as inputs for the following layer. A simple example is depicted below:
#
# <img src="neuralnetworkQC.png" />
#
# Here, $\sigma$ is a [nonlinear function](https://en.wikipedia.org/wiki/Activation_function) and $h_i$ is the value of neuron $i$ at each hidden layer. $R(h_i)$ represents any rotation gate about an angle equal to $h_i$ and $y$ is the final prediction value generated from the hybrid network.
#
# ### What about backpropagation?
# If you're familiar with classical ML, you may immediately be wondering *how do we calculate gradients when quantum circuits are involved?* This would be necessary to enlist powerful optimisation techniques such as **[gradient descent](https://en.wikipedia.org/wiki/Gradient_descent)**. It gets a bit technical, but in short, we can view a quantum circuit as a black box and the gradient of this black box with respect to its parameters can be calculated as follows:
#
# <img src="quantumgradient.png" />
#
# where $\theta$ represents the parameters of the quantum circuit and $s$ is a macroscopic shift. The gradient is then simply the difference between our quantum circuit evaluated at $\theta+s$ and $\theta - s$. Thus, we can systematically differentiate our quantum circuit as part of a larger backpropogation routine. This closed form rule for calculating the gradient of quantum circuit parameters is known as **[the parameter shift rule](https://arxiv.org/pdf/1905.13311.pdf)**.
# ## 3. Let's code! <a id='code'></a>
#
#
# ### 3.1 Imports <a id='imports'></a>
# First, we import some handy packages that we will need, including Qiskit and PyTorch.
# +
import numpy as np
import matplotlib.pyplot as plt
import torch
from torch.autograd import Function
from torchvision import datasets, transforms
import torch.optim as optim
import torch.nn as nn
import torch.nn.functional as F
import qiskit
from qiskit import transpile, assemble
from qiskit.visualization import *
# -
# ### 3.2 Create a "Quantum Class" with Qiskit <a id='q-class'></a>
# We can conveniently put our Qiskit quantum functions into a class. First, we specify how many trainable quantum parameters and how many shots we wish to use in our quantum circuit. In this example, we will keep it simple and use a 1-qubit circuit with one trainable quantum parameter $\theta$. We hard code the circuit for simplicity and use a $RY-$rotation by the angle $\theta$ to train the output of our circuit. The circuit looks like this:
#
# <img src="1qubitcirc.png" width="400"/>
#
# In order to measure the output in the $z-$basis, we calculate the $\sigma_\mathbf{z}$ expectation.
# $$\sigma_\mathbf{z} = \sum_i z_i p(z_i)$$
# We will see later how this all ties into the hybrid neural network.
class QuantumCircuit:
"""
This class provides a simple interface for interaction
with the quantum circuit
"""
def __init__(self, n_qubits, backend, shots):
# --- Circuit definition ---
self._circuit = qiskit.QuantumCircuit(n_qubits)
all_qubits = [i for i in range(n_qubits)]
self.theta = qiskit.circuit.Parameter('theta')
self._circuit.h(all_qubits)
self._circuit.barrier()
self._circuit.ry(self.theta, all_qubits)
self._circuit.measure_all()
# ---------------------------
self.backend = backend
self.shots = shots
def run(self, thetas):
t_qc = transpile(self._circuit,
self.backend)
qobj = assemble(t_qc,
shots=self.shots,
parameter_binds = [{self.theta: theta} for theta in thetas])
job = self.backend.run(qobj)
result = job.result().get_counts(self._circuit)
counts = np.array(list(result.values()))
states = np.array(list(result.keys())).astype(float)
# Compute probabilities for each state
probabilities = counts / self.shots
# Get state expectation
expectation = np.sum(states * probabilities)
return np.array([expectation])
# Let's test the implementation
# +
simulator = qiskit.Aer.get_backend('qasm_simulator')
circuit = QuantumCircuit(1, simulator, 100)
print('Expected value for rotation pi {}'.format(circuit.run([np.pi])[0]))
circuit._circuit.draw()
# -
# ### 3.3 Create a "Quantum-Classical Class" with PyTorch <a id='qc-class'></a>
# Now that our quantum circuit is defined, we can create the functions needed for backpropagation using PyTorch. [The forward and backward passes](http://www.ai.mit.edu/courses/6.034b/backprops.pdf) contain elements from our Qiskit class. The backward pass directly computes the analytical gradients using the finite difference formula we introduced above.
# +
class HybridFunction(Function):
""" Hybrid quantum - classical function definition """
@staticmethod
def forward(ctx, input, quantum_circuit, shift):
""" Forward pass computation """
ctx.shift = shift
ctx.quantum_circuit = quantum_circuit
expectation_z = ctx.quantum_circuit.run(input[0].tolist())
result = torch.tensor([expectation_z])
ctx.save_for_backward(input, result)
return result
@staticmethod
def backward(ctx, grad_output):
""" Backward pass computation """
input, expectation_z = ctx.saved_tensors
input_list = np.array(input.tolist())
shift_right = input_list + np.ones(input_list.shape) * ctx.shift
shift_left = input_list - np.ones(input_list.shape) * ctx.shift
gradients = []
for i in range(len(input_list)):
expectation_right = ctx.quantum_circuit.run(shift_right[i])
expectation_left = ctx.quantum_circuit.run(shift_left[i])
gradient = torch.tensor([expectation_right]) - torch.tensor([expectation_left])
gradients.append(gradient)
gradients = np.array([gradients]).T
return torch.tensor([gradients]).float() * grad_output.float(), None, None
class Hybrid(nn.Module):
""" Hybrid quantum - classical layer definition """
def __init__(self, backend, shots, shift):
super(Hybrid, self).__init__()
self.quantum_circuit = QuantumCircuit(1, backend, shots)
self.shift = shift
def forward(self, input):
return HybridFunction.apply(input, self.quantum_circuit, self.shift)
# -
# ### 3.4 Data Loading and Preprocessing <a id='data-loading-preprocessing'></a>
# ##### Putting this all together:
# We will create a simple hybrid neural network to classify images of two types of digits (0 or 1) from the [MNIST dataset](http://yann.lecun.com/exdb/mnist/). We first load MNIST and filter for pictures containing 0's and 1's. These will serve as inputs for our neural network to classify.
# #### Training data
# +
# Concentrating on the first 100 samples
n_samples = 100
X_train = datasets.MNIST(root='./data', train=True, download=True,
transform=transforms.Compose([transforms.ToTensor()]))
# Leaving only labels 0 and 1
idx = np.append(np.where(X_train.targets == 0)[0][:n_samples],
np.where(X_train.targets == 1)[0][:n_samples])
X_train.data = X_train.data[idx]
X_train.targets = X_train.targets[idx]
train_loader = torch.utils.data.DataLoader(X_train, batch_size=1, shuffle=True)
# +
n_samples_show = 6
data_iter = iter(train_loader)
fig, axes = plt.subplots(nrows=1, ncols=n_samples_show, figsize=(10, 3))
while n_samples_show > 0:
images, targets = data_iter.__next__()
axes[n_samples_show - 1].imshow(images[0].numpy().squeeze(), cmap='gray')
axes[n_samples_show - 1].set_xticks([])
axes[n_samples_show - 1].set_yticks([])
axes[n_samples_show - 1].set_title("Labeled: {}".format(targets.item()))
n_samples_show -= 1
# -
# #### Testing data
# +
n_samples = 50
X_test = datasets.MNIST(root='./data', train=False, download=True,
transform=transforms.Compose([transforms.ToTensor()]))
idx = np.append(np.where(X_test.targets == 0)[0][:n_samples],
np.where(X_test.targets == 1)[0][:n_samples])
X_test.data = X_test.data[idx]
X_test.targets = X_test.targets[idx]
test_loader = torch.utils.data.DataLoader(X_test, batch_size=1, shuffle=True)
# -
# So far, we have loaded the data and coded a class that creates our quantum circuit which contains 1 trainable parameter. This quantum parameter will be inserted into a classical neural network along with the other classical parameters to form the hybrid neural network. We also created backward and forward pass functions that allow us to do backpropagation and optimise our neural network. Lastly, we need to specify our neural network architecture such that we can begin to train our parameters using optimisation techniques provided by PyTorch.
#
#
# ### 3.5 Creating the Hybrid Neural Network <a id='hybrid-nn'></a>
# We can use a neat PyTorch pipeline to create a neural network architecture. The network will need to be compatible in terms of its dimensionality when we insert the quantum layer (i.e. our quantum circuit). Since our quantum in this example contains 1 parameter, we must ensure the network condenses neurons down to size 1. We create a typical Convolutional Neural Network with two fully-connected layers at the end. The value of the last neuron of the fully-connected layer is fed as the parameter $\theta$ into our quantum circuit. The circuit measurement then serves as the final prediction for 0 or 1 as provided by a $\sigma_z$ measurement.
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.conv1 = nn.Conv2d(1, 6, kernel_size=5)
self.conv2 = nn.Conv2d(6, 16, kernel_size=5)
self.dropout = nn.Dropout2d()
self.fc1 = nn.Linear(256, 64)
self.fc2 = nn.Linear(64, 1)
self.hybrid = Hybrid(qiskit.Aer.get_backend('qasm_simulator'), 100, np.pi / 2)
def forward(self, x):
x = F.relu(self.conv1(x))
x = F.max_pool2d(x, 2)
x = F.relu(self.conv2(x))
x = F.max_pool2d(x, 2)
x = self.dropout(x)
x = x.view(1, -1)
x = F.relu(self.fc1(x))
x = self.fc2(x)
x = self.hybrid(x)
return torch.cat((x, 1 - x), -1)
# ### 3.6 Training the Network <a id='training'></a>
# We now have all the ingredients to train our hybrid network! We can specify any [PyTorch optimiser](https://pytorch.org/docs/stable/optim.html), [learning rate](https://en.wikipedia.org/wiki/Learning_rate) and [cost/loss function](https://en.wikipedia.org/wiki/Loss_function) in order to train over multiple epochs. In this instance, we use the [Adam optimiser](https://arxiv.org/abs/1412.6980), a learning rate of 0.001 and the [negative log-likelihood loss function](https://pytorch.org/docs/stable/_modules/torch/nn/modules/loss.html).
# +
model = Net()
optimizer = optim.Adam(model.parameters(), lr=0.001)
loss_func = nn.NLLLoss()
epochs = 20
loss_list = []
model.train()
for epoch in range(epochs):
total_loss = []
for batch_idx, (data, target) in enumerate(train_loader):
optimizer.zero_grad()
# Forward pass
output = model(data)
# Calculating loss
loss = loss_func(output, target)
# Backward pass
loss.backward()
# Optimize the weights
optimizer.step()
total_loss.append(loss.item())
loss_list.append(sum(total_loss)/len(total_loss))
print('Training [{:.0f}%]\tLoss: {:.4f}'.format(
100. * (epoch + 1) / epochs, loss_list[-1]))
# -
# Plot the training graph
plt.plot(loss_list)
plt.title('Hybrid NN Training Convergence')
plt.xlabel('Training Iterations')
plt.ylabel('Neg Log Likelihood Loss')
# ### 3.7 Testing the Network <a id='testing'></a>
model.eval()
with torch.no_grad():
correct = 0
for batch_idx, (data, target) in enumerate(test_loader):
output = model(data)
pred = output.argmax(dim=1, keepdim=True)
correct += pred.eq(target.view_as(pred)).sum().item()
loss = loss_func(output, target)
total_loss.append(loss.item())
print('Performance on test data:\n\tLoss: {:.4f}\n\tAccuracy: {:.1f}%'.format(
sum(total_loss) / len(total_loss),
correct / len(test_loader) * 100)
)
# +
n_samples_show = 6
count = 0
fig, axes = plt.subplots(nrows=1, ncols=n_samples_show, figsize=(10, 3))
model.eval()
with torch.no_grad():
for batch_idx, (data, target) in enumerate(test_loader):
if count == n_samples_show:
break
output = model(data)
pred = output.argmax(dim=1, keepdim=True)
axes[count].imshow(data[0].numpy().squeeze(), cmap='gray')
axes[count].set_xticks([])
axes[count].set_yticks([])
axes[count].set_title('Predicted {}'.format(pred.item()))
count += 1
# -
# ## 4. What Now? <a id='what-now'></a>
#
# #### While it is totally possible to create hybrid neural networks, does this actually have any benefit?
#
# In fact, the classical layers of this network train perfectly fine (in fact, better) without the quantum layer. Furthermore, you may have noticed that the quantum layer we trained here **generates no entanglement**, and will, therefore, continue to be classically simulatable as we scale up this particular architecture. This means that if you hope to achieve a quantum advantage using hybrid neural networks, you'll need to start by extending this code to include a more sophisticated quantum layer.
#
#
# The point of this exercise was to get you thinking about integrating techniques from ML and quantum computing in order to investigate if there is indeed some element of interest - and thanks to PyTorch and Qiskit, this becomes a little bit easier.
import qiskit
qiskit.__qiskit_version__
| 49.787179
| 1,015
|
1a37e0f0c0a5b38f0c568a62c1f4be6a5174d4d3
|
py
|
python
|
prob-stats-data-analysis/brain-teasers-paradoxes-carefulness/use-of-aggregated-metrics.ipynb
|
walkenho/tales-science-data
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# %run ../../common/import_all.py
from common.setup_notebook import set_css_style, setup_matplotlib, config_ipython
config_ipython()
setup_matplotlib()
set_css_style()
# -
# # Using aggregated metrics
#
# Sometimes the wrong metric is used, sometimes the language is too sloppy. For the description of the things described in this notebook, head over to ["Distributions and probability measures"](../distributions-measures/).
# ## When people talk average
#
# Technically speaking, an average is a mean.
#
# But.
#
# The word *average* in common (street) English is usually used to mean (oh, the irony!) an aggregated number which is supposed to represent a collection of data points as it's "central" value. We're talking about common talk, not the one among professionals of data, and this often happens in politics and (sloppy) journalism, with consequences that can go from funny to disastrous. This aggregated number, the *average* is not necessarily calculated as the (arithmetic) mean, and this can lead to awkward situations where people don't know what they're exactly talking about and whether that number is really representative of their data. Sometimes they mean the mean, sometimes they mean the median, for one. But they can mean other different things as well: an example is when to derive the "average" value of a set of fractions, you compute the [mediant](https://en.wikipedia.org/wiki/Mediant_(mathematics) instead of averaging said fractions. Some other times, the mode is intended when people talk about an average.
#
# I find this one of the interesting peculiarities of the English language, and it wouldn't really matter if it weren't for the fact that in many occasions it is quite necessary to go look at a properly defined metric in order to understand what the data says. See the lovely lil' book **How to lie with Statistics** [[1]](#1), which is very old but a nice read. Also, this discussion on Quora [[2]](#2) is interesting.
# ## Is the mean always the best descriptor of a distribution?
#
# If you have data distributed in a gaussian way, a good descriptor of the typical situation of your data is the mean. If the distribution is skewed though, and in particular in the case of [power laws](../distribution-measures/power-laws.ipynb), the median is a much better indicator to represent the distribution. Have a look at this video on the matter.
#
# The mean is very sensible to outliers in this cases as when you add a data point , the median is not so it's a much more robust statistic to represent the distribution.
# ## References
#
# 1. <a name="lie-stats"></a> [**How to lie with Statistics**](https://en.wikipedia.org/wiki/How_to_Lie_with_Statistics)
# 2. <a name="quora"></a> [**Quora** on mean and average](https://www.quora.com/What-is-difference-between-the-mean-and-the-average)
# 3. <a name="cbs"></a> [**Calling Bullshit** on mean vs. median](https://www.youtube.com/watch?v=mc-6-v2c4WM)
# 4. <a name="meanmedian"></a> [A nice example of when using a mean is a misuse of statistics: income distributions](http://www.conceptstew.co.uk/pages/mean_or_median.html)
#
| 64.25
| 1,022
|
00fdf30b8cbd33d62e698b212be49505207cff9f
|
py
|
python
|
master/tutorial_img_ds.ipynb
|
lrittner/ia979
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] toc="true"
# # Table of Contents
# <p><div class="lev1 toc-item"><a href="#ATENÇÃO:-este-notebook-ainda-não-está-pronto" data-toc-modified-id="ATENÇÃO:-este-notebook-ainda-não-está-pronto-1"><span class="toc-item-num">1 </span>ATENÇÃO: este notebook ainda não está pronto</a></div><div class="lev1 toc-item"><a href="#Representação,-Leitura-e-Visualização-de-Imagens" data-toc-modified-id="Representação,-Leitura-e-Visualização-de-Imagens-2"><span class="toc-item-num">2 </span>Representação, Leitura e Visualização de Imagens</a></div><div class="lev2 toc-item"><a href="#Imagem-como-matriz" data-toc-modified-id="Imagem-como-matriz-21"><span class="toc-item-num">2.1 </span>Imagem como matriz</a></div><div class="lev2 toc-item"><a href="#Leitura-de-uma-imagem" data-toc-modified-id="Leitura-de-uma-imagem-22"><span class="toc-item-num">2.2 </span>Leitura de uma imagem</a></div><div class="lev2 toc-item"><a href="#Visualização-de-uma-imagem" data-toc-modified-id="Visualização-de-uma-imagem-23"><span class="toc-item-num">2.3 </span>Visualização de uma imagem</a></div><div class="lev2 toc-item"><a href="#Visualizando-numericamente-uma-pequena-região-de-interesse-da-imagem" data-toc-modified-id="Visualizando-numericamente-uma-pequena-região-de-interesse-da-imagem-24"><span class="toc-item-num">2.4 </span>Visualizando numericamente uma pequena região de interesse da imagem</a></div><div class="lev2 toc-item"><a href="#Criando-legenda-da-imagem-com-impressão-de-variáveis" data-toc-modified-id="Criando-legenda-da-imagem-com-impressão-de-variáveis-25"><span class="toc-item-num">2.5 </span>Criando legenda da imagem com impressão de variáveis</a></div>
# -
# # ATENÇÃO: este notebook ainda não está pronto
#
#
#
# # Representação, Leitura e Visualização de Imagens
#
# Uma imagem digital pode ser representada por uma matriz bidimensional, onde os seus elementos são chamados de pixels
# (abreviatura de *picture elements*). Existem vários pacotes de processamento de imagens onde a imagem é representada
# por uma estrutura de dados específica. No nosso caso, no Adessowiki iremos utilizar a matriz disponível no *ndarray NumPy*.
# A vantagem é que todas as operações disponíveis para processamento matricial podem ser utilizados como processamento de
# imagens. Este é um dos principais objetivos deste curso: como utilizar linguagens de processamento matricial para fazermos
# processamento de imagens.
# ## Imagem como matriz
#
# Neste curso, uma imagem é definida pelo seu cabeçalho (tamanho da matriz e tipo de pixel) e pelos pixels em si. Estas
# informações são inerentes ao tipo ``ndarray`` do NumPy.
#
# O tamanho da matriz é caracterizado pelas suas dimensões: vertical e horizontal.
# A dimensão vertical é definida pelo número de linhas (*rows*) ou altura H (*height*) e a dimensão
# horizontal é definida pelo número de colunas (*cols*) ou largura W (*width*). No NumPy, as dimensões são armazenadas
# no ``shape`` da matriz como uma tupla (H,W).
#
# Uma imagem pode ter valores de pixels que podem ser armazenados em vários tipos de dados:
# a imagem binária tem apenas dois valores possíveis,
# muitas vezes atribuídos a preto e branco; uma imagem em nível de cinza tem valores inteiros positivos, muitas vezes, de 0 a um
# valor máximo. É possível ter pixels com valores negativos, com números reais, e até mesmo pixels com valores complexos.
# Um exemplo de uma imagem com valores de pixel negativos são imagens térmicas com temperaturas negativas.
# As imagens com pixels que são números reais podem ser encontradas nas imagens que representam uma onda senóide com valores que
# variam de -1 a +1. As imagens com os valores de pixel complexos podem ser encontrados em algumas transformações da imagem como
# a Transformada Discreta de Fourier.
#
# Como as imagens usualmente possuem centenas de milhares ou milhões de pixels, é importante escolher a menor representação do
# pixel para economizar o uso da memória do computador e usar a representação que seja mais eficiente para processamento.
# No Numpy, o tipo do pixel é armazenado no ``dtype`` que pode assumir vários tipos. Os quatro tipos que mais usaremos neste curso
# são indicados na tabela:
#
# ====== ===============================
# dtype valores
# ====== ===============================
# bool True, False
# uint8 8 bits sem sinal, de 0 a 255
# uint16 16 bits sem sinal, de 0 a 65535
# int 64 bits com sinal
# float ponto flutuante
# ====== ===============================
#
# ## Leitura de uma imagem
#
# Neste curso iremos trabalhar com imagens criadas sinteticamente e com imagens guardadas em arquivos. A leitura de uma
# imagem no Adessowiki é feita pelas funções ``adread`` e ``adreadgray`` que utilizam o pacote
# `http://effbot.org/imagingbook/ PIL` de processamento de imagens. Neste curso não utilizaremos as funções de processamento
# de imagens do PIL, mas sim utilizaremos as operações matriciais do NumPy. Existem diversas formas de salvar uma imagem em
# arquivo e utilizaremos as mais comuns: png, jpg, tif. As imagens disponíveis podem ser visualizadas na toolbox ia636 do Adessowiki:
# `ia636:iaimages`.
#
# Veja a seguir um exemplo de leitura de imagem e a impressão de seu cabeçalho e de seus pixels:
#
# +
# %matplotlib inline
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import numpy as np
# !ls ../data
# +
f = mpimg.imread('../data/cameraman.tif')
print('Tamanho de f: ', f.shape)
print('Tipo do pixel:', f.dtype)
print('Número total de pixels:', f.size)
print('Pixels:\n', f)
# -
# Note que a imagem possui 174 linhas e 314 colunas, totalizando mais de 54 mil pixels. A representação do pixel é pelo tipo
# ``uint8``, isto é, valores de 8 bits sem sinal, de 0 a 255. Note também que a impressão de todos os pixels é feita de
# forma especial. Se todos os 54 mil pixels tivessem que ser impressos, o resultado da impressão seria proibitivo. Neste caso, quando
# a imagem (matriz) for muito grande, o NumPy imprime apenas os pixels dos quatro cantos da imagem.
# ## Visualização de uma imagem
#
# No Adessowiki, a visualização de uma imagem é feita unicamente pela função ``adshow``, que internamente utiliza o pacote PIL já
# mencionado. O processo de exibição de uma imagem cria uma representação gráfica desta matriz
# em que os valores do pixel é atribuído a um nível de cinza (imagem monocromática) ou a uma cor particular. Quando o pixel da imagem
# é ``uint8``, o valor zero é atribuído ao preto e o valor 255 ao branco e gerando um tom de cinza proporcional ao valor do pixel.
#
# Veja abaixo a visualização da imagem ``cookies.tif`` já lida no trecho de programa anterior. Note que a função ``adshow`` possui
# dois parâmetros, a imagem e um string para ser exibido na legenda da visualização da imagem.
plt.imshow(f,cmap='gray')
# O segundo tipo de imagem que o ``adshow`` visualiza é a imagem com pixels do tipo booleano. Como ilustração, faremos uma
# operação comparando cada pixel da imagem cookies com o valor 128 gerando assim uma nova imagem ``f_bin`` onde cada pixel será
# ``True`` ou ``False`` dependendo do resultado da comparação. O ``adshow`` mapeia os pixels verdadeiros como branco e os pixels
# falsos como preto:
f_bin = f > 128
print('Tipo do pixel:', f_bin.dtype)
plt.imshow(f_bin,cmap='gray')
plt.colorbar()
print(f_bin.min(), f_bin.max())
f_f = f_bin.astype(np.float)
f_i = f_bin.astype(np.int)
print(f_f.min(),f_f.max())
print(f_i.min(),f_i.max())
#
# Por fim, além destes dois modos de exibição, o ``adshow`` pode também exibir imagens coloridas no formato RGB e tipo de pixel ``uint8``.
# No NumPy a imagem RGB é representada como três images armazenadas na dimensão profundidade. Neste caso o ``array`` tem 3
# dimensões e seu ``shape`` tem o formato (3,H,W).
f_cor = mpimg.imread('../data/boat.tif')
print('Dimensões: ', f_cor.shape)
print('Tipo do pixel:', f_cor.dtype)
plt.imshow(f_cor)
f_roi = f_cor[:2,:3,:]
print(f_roi)
# Neste curso, por motivos didáticos, o ``adshow`` somente visualiza estes 3 tipos de imagens. Qualquer outro tipo de imagem,
# seja de valores maiores que 255, negativos ou complexos, precisam ser explicitamente convertidos para os valores entre 0 e 255
# ou ``True`` e ``False``.
#
# Maiores informações no uso do ``adshow`` podem ser vistas em `ia636:adshow`.
#
# .. note:: Uma das principais causas de erro em processamento de imagens é não prestar atenção no tipo do pixel ou nas dimensões da
# imagem. Recomenda-se verificar esta informações. Uma função que é bastante útil é a `ia636:iaimginfo` que foi criada para
# verificar rapidamente o tipo de pixel, dimensões e os valores mínimo e máximo da imagem. Veja a seguir um exemplo do seu uso
# nas três imagens processadas anteriormente:
#
#
# import ia636
# print 'f: ', ia636.iaimginfo(f)
# print 'f_bin:', ia636.iaimginfo(f_bin)
# print 'f_cor:', ia636.iaimginfo(f_cor)
# ## Visualizando numericamente uma pequena região de interesse da imagem
#
# Para verificar que a imagem lida é composta de valores entre 0 e 255, vamos imprimir numericamente
# apenas uma pequena região de 7 linhas e 10 colunas do canto superior esquerdo da imagem. Fazemos isto
# com fatiamento:
f= mpimg.imread('../data/gull.pgm')
plt.imshow(f,cmap='gray')
g = f[:7,:10]
print('g=')
print(g)
# ## Criando legenda da imagem com impressão de variáveis
#
# É oportuno colocar informações na legenda da imagem na hora de mostrá-la pelo ``adshow``.
# Constrói-se um string contendo a informação desejada. O Python possui diversas facilidades
# para criar este string. Abaixo é utilizado a formatação via %, onde o string é formatado
# utilizando campos especiais: %s (string), %d (inteiro), %f (ponto flutuante), entre outros.
# A variáveis a serem preenchidas nestes campos são colocadas na forma de tupla do Python.
# Veja o exemplo abaixo:
#
# .. code:: python
#
# f = adreadgray('astablet.tif')
# H,W = f.shape
# legenda = 'astablet dimensões: (%d,%d)' % (H,W)
# adshow(f, legenda)
| 54.844086
| 1,706
|
585212fb4b005d55dafc26d2a9acbb7a9be40ec5
|
py
|
python
|
text classification pytorch lightning.ipynb
|
kstathou/covid19-classifier
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + colab={"base_uri": "https://localhost:8080/"} id="TK5TZoUoErli" executionInfo={"status": "ok", "timestamp": 1613930847200, "user_tz": 0, "elapsed": 8683, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="25b245ea-89d4-4f93-bff6-c6b60cb4012e"
# !pip install pytorch-lightning==1.1.8
# !pip install transformers
# !pip install wandb
# + id="mKME0SgcrokG" executionInfo={"status": "ok", "timestamp": 1613930857323, "user_tz": 0, "elapsed": 1865, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
import torch
from torch import nn
import pytorch_lightning as pl
from pytorch_lightning.loggers import WandbLogger
from torch.utils.data import Dataset, DataLoader
from transformers import BertModel, BertTokenizer, AdamW, get_linear_schedule_with_warmup
from sklearn.model_selection import train_test_split
import numpy as np
import pandas as pd
import wandb
# + [markdown] id="d6vmr27etdnX"
# We will leverage [Orion, a knowledge discovery tool for scientific publications](https://github.com/orion-search/orion), to get a subset of publications from [biorXiv](https://www.biorxiv.org/). Then, we will keep those related to COVID-19 as well as a random sample of non-COVID-19 publications using [MAG's Fields of Study](https://arxiv.org/abs/1805.12216).
# + id="J4of5X-Hroh9" executionInfo={"status": "ok", "timestamp": 1613930862863, "user_tz": 0, "elapsed": 373, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
# Read abstracts from Microsoft Academic Graph
df = pd.read_csv("drive/MyDrive/Colab Notebooks/mag_papers.csv")
# + colab={"base_uri": "https://localhost:8080/", "height": 206} id="R8rBpauNrofq" executionInfo={"status": "ok", "timestamp": 1613930863449, "user_tz": 0, "elapsed": 457, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="b140844c-03cb-4292-ec9d-7d96bbb2566e"
df.head()
# + colab={"base_uri": "https://localhost:8080/"} id="Xw7nQH_ptYnT" executionInfo={"status": "ok", "timestamp": 1613930863808, "user_tz": 0, "elapsed": 733, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="d23523d2-39cc-472a-b077-b53eb846bb53"
print(f"Number of Covid-19 publications: {df[df.is_Covid19==1].shape[0]}")
print(f"Number of non-Covid-19 publications: {df[df.is_Covid19!=1].shape[0]}")
# + colab={"base_uri": "https://localhost:8080/"} id="5tZRahj1rodS" executionInfo={"status": "ok", "timestamp": 1613930863808, "user_tz": 0, "elapsed": 655, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="abe9a479-49b1-4ea6-ea04-8afc268a0a18"
# Set a seed with Pytorch Lightning
pl.seed_everything(42)
# Split the dataframe to training and evaluation sets
df_train, df_val = train_test_split(df, test_size=.05)
# Split the dataframe to training and test sets
df_train, df_test = train_test_split(df_train, test_size=.05)
# + id="x6NoLW6lvs_S" executionInfo={"status": "ok", "timestamp": 1613932252052, "user_tz": 0, "elapsed": 1155, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
class CovidDataset(torch.utils.data.Dataset):
def __init__(self, data, tokenizer, max_token_len=128):
self.data = data
self.tokenizer = tokenizer
self.max_token_len = max_token_len
def __len__(self):
return len(self.data)
def __getitem__(self, index):
# Grab a single row from the dataframe
data_row = self.data.iloc[index]
# Grab text and label
abstract_text = data_row.abstract
labels = torch.tensor(data_row.is_Covid19)
encoding = tokenizer.encode_plus(
abstract_text,
add_special_tokens=True,
max_length=self.max_token_len,
return_token_type_ids=False,
padding="max_length",
truncation=True,
return_attention_mask=True,
return_tensors="pt"
)
return dict(
abstract_text=abstract_text,
input_ids=encoding["input_ids"].flatten(),
attention_mask=encoding["attention_mask"].flatten(),
labels=labels
)
# + id="Ae2mnmNevs8Z" executionInfo={"status": "ok", "timestamp": 1613932252053, "user_tz": 0, "elapsed": 881, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
class CovidDataModule(pl.LightningDataModule):
def __init__(self, df_train, df_val, df_test, tokenizer, batch_size=8, max_token_len=512):
super().__init__()
self.df_train = df_train
self.df_val = df_val
self.df_test = df_test
self.tokenizer = tokenizer
self.batch_size = batch_size
self.max_token_len = max_token_len
def setup(self):
self.train_dataset = CovidDataset(self.df_train, self.tokenizer, self.max_token_len)
self.val_dataset = CovidDataset(self.df_val, self.tokenizer, self.max_token_len)
self.test_dataset = CovidDataset(self.df_val, self.tokenizer, self.max_token_len)
def train_dataloader(self):
return DataLoader(self.train_dataset, batch_size=self.batch_size, shuffle=True, num_workers=4)
def val_dataloader(self):
return DataLoader(self.val_dataset, batch_size=self.batch_size)
def test_dataloader(self):
return DataLoader(self.test_dataset, batch_size=self.batch_size)
# + id="k7689wJ9vs5p" executionInfo={"status": "ok", "timestamp": 1613932252845, "user_tz": 0, "elapsed": 1418, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
# Hyperparameters
N_EPOCHS = 4
BATCH_SIZE = 16
# Pick a transformer from Huggingface
TRANSFORMER_MODEL_NAME = "bert-base-uncased"
# Instantiate the tokenizer
tokenizer = BertTokenizer.from_pretrained(TRANSFORMER_MODEL_NAME)
# Create a Pytorch Lightning DataModule
data_module = CovidDataModule(df_train, df_val, df_test, tokenizer, batch_size=BATCH_SIZE, max_token_len=128)
data_module.setup()
# + id="H5M_Q49ivs3R" executionInfo={"status": "ok", "timestamp": 1613932790061, "user_tz": 0, "elapsed": 420, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
class CovidClassifier(pl.LightningModule):
def __init__(self, n_classes, lr=2e-5, steps_per_epoch=None, n_epochs=None):
super().__init__()
self.bert_model = BertModel.from_pretrained(TRANSFORMER_MODEL_NAME, return_dict=True)
self.n_classes = n_classes
self.classifier = nn.Linear(self.bert_model.config.hidden_size, self.n_classes)
self.steps_per_epoch = steps_per_epoch
self.n_epochs = n_epochs
self.lr = lr
self.criterion = nn.CrossEntropyLoss()
# self.criterion = nn.BCELoss()
self.save_hyperparameters()
def forward(self, input_ids, attention_mask, labels=None):
output = self.bert_model(input_ids, attention_mask=attention_mask)
output = self.classifier(output.pooler_output)
# output = torch.sigmoid(output)
loss = 0
if labels is not None:
loss = self.criterion(output.view(-1, self.n_classes), labels.view(-1))
# loss = self.criterion(output, labels)
return loss, output
def training_step(self, batch, batch_idx):
input_ids = batch["input_ids"]
attention_mask = batch["attention_mask"]
labels = batch["labels"]
loss, outputs = self(input_ids, attention_mask, labels)
self.log("train_loss", loss, prog_bar=True)
return {"loss":loss, "predictions":outputs, "labels":labels}
def validation_step(self, batch, batch_idx):
input_ids = batch["input_ids"]
attention_mask = batch["attention_mask"]
labels = batch["labels"]
loss, outputs = self(input_ids, attention_mask, labels)
self.log("val_loss", loss, prog_bar=True)
return loss
def test_step(self, batch, batch_idx):
input_ids = batch["input_ids"]
attention_mask = batch["attention_mask"]
labels = batch["labels"]
loss, outputs = self(input_ids, attention_mask, labels)
self.log("test_loss", loss, prog_bar=True)
return loss
def configure_optimizers(self):
optimizer = AdamW(self.parameters(), lr=self.lr)
warmup_steps = self.steps_per_epoch // 3
total_steps = self.steps_per_epoch * self.n_epochs - warmup_steps
scheduler = get_linear_schedule_with_warmup(
optimizer, warmup_steps, total_steps
)
return [optimizer], [scheduler]
# + id="RnJL6A6StY0Z" executionInfo={"status": "ok", "timestamp": 1613932252848, "user_tz": 0, "elapsed": 919, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
# wandb.login()
# + colab={"base_uri": "https://localhost:8080/", "height": 121} id="gPM6D7i8vs0q" executionInfo={"status": "ok", "timestamp": 1613932284488, "user_tz": 0, "elapsed": 10932, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="c1dcb434-c955-4a7f-d69d-23ab29e41f4a"
model = CovidClassifier(n_classes=2, steps_per_epoch=len(df_train) // BATCH_SIZE, n_epochs=N_EPOCHS)
wandb_logger = WandbLogger(
project="testing",
save_code=False,
tags=["covid-classifier"],
reinit=True,
)
wandb_logger.watch(model, log='all')
# + colab={"base_uri": "https://localhost:8080/", "height": 36} id="VYg6aROO3IB2" executionInfo={"status": "ok", "timestamp": 1613932430483, "user_tz": 0, "elapsed": 472, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="01c5077f-9281-4bab-aae3-9a8d0e09f655"
wandb.run.name
# + colab={"base_uri": "https://localhost:8080/"} id="5-djz0bWvsx_" executionInfo={"status": "ok", "timestamp": 1613932443484, "user_tz": 0, "elapsed": 470, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="f8726f73-0783-46ef-f825-87942e994c88"
trainer = pl.Trainer(max_epochs=N_EPOCHS, gpus=1, progress_bar_refresh_rate=30, logger=wandb_logger)
# + colab={"base_uri": "https://localhost:8080/", "height": 324, "referenced_widgets": ["06813a876abc4d93b7c178268cb15155", "91607f77e1f74323a2bccfa40eaf0faa", "36b8a8e5036b4e69896d5ce9caf452a6", "31c14b935bed44ac958bf945eea0b08c", "61c38c8f11174fdead21dc14833f5b52", "5b7533cefa4245bcbc26c87291f8b5f0", "15c6973ac4084ace8cc7ab93b95df829", "7956b9a4629c47d197b2caf122df35b4", "70a6a26ba9254ff3b45af7c98f32c2c3", "7c180e98ba974051acd6a077fd2effc5", "26e822e9b20c44e09b15cc7918771bfb", "c833c45b7d324ef48616ae726fef068b", "76ecfc5376f840d78e9ac2ece89501b5", "ffb70ec164974b8fbab5ab1a8c8a9809", "2e598e38acd849d580939a0db90a8ae7", "aba44fd379354ac5944951c21ede7136", "41b3585711014675b4401999d84ffd4d", "8aed8dc8810b446a861c6e22fc240ff0", "06a15e67612b49a085a62e1b3239147b", "bc8854f53bf245a2bf93d297753cfb68", "726e33c723f0445fba4f2b25bbe690ee", "cbec3a5b42db472a8ddceb7002ac2d7b", "bb111e76f9ce4f798e278f88d19277e6", "b0e515778e454d1098cd31a8566a1f0d", "feee28944e7b4c748362ffc6788a861c", "05c4c4234bcb4f609e76fad4d6b25784", "742dd1a55c10440f9caa7f3b348705ef", "9e1b9876e3f940789e7d42820b00b554", "abe3c1d4dfc941c88abe3d029d3585a4", "f9c35f85717643a99cfb5cb2f2734df0", "6346bd039224491d95b8237cce591404", "375387be454c4cbcaf49dcbbc6741a4f", "4534a68824f84a0db07f40074e228840", "02e97356c74d48a3942f8bf46e6a502f", "b84f6a3cf0284963808114b954dc7ab6", "6fb31ebfcdc34f1f8d01d7c9555aca84", "674feb33a3a7442fa5a08275e43c4ead", "49493dc360774a778bce5c938d9c1975", "4899d6f3a22f4e81b41fe5f955a80acb", "870d328535ac4e879582b4f08729b76a", "0e6fb769e5894ae689788b4d7d510ba4", "bd50eb0cfd1d43ff956192e31f554e27", "7712309aba294faab82709bfda30d6f6", "3c01f6961e044b69a39adf264392e0ec", "3877346cd1c3438db20d3b6a35c61b1e", "4eac280a96e847349e41a1f002034c5d", "d5d38d25765f482f90aea8e1e3147e4a", "223947902b694f6cba26903506a911e8"]} id="ObxpIp1hroLE" executionInfo={"status": "ok", "timestamp": 1613932653236, "user_tz": 0, "elapsed": 206497, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="29203cde-a5c5-4287-dbe9-a0e71240b1a8"
trainer.fit(model, data_module)
# + colab={"base_uri": "https://localhost:8080/", "height": 153, "referenced_widgets": ["19c69f6f7ffb4e6590b73762c0de7b28", "58501b9a8040491bb3ac7cbce6f96877", "2d4c5e70546a48b58d8c00228c32a9ab", "e7c688c53d1c4409a7bbde795c9ac3a0", "4bf32f142c3e40df862b2e5307baa213", "75c56295cfa248ee89a0b884294b2c2a", "66dafb22505f4fe6a08e9ad055ceb341", "72d588c73d294e7481943a9313c2e18b"]} id="wCOSrxATxAQg" executionInfo={"status": "ok", "timestamp": 1613932731870, "user_tz": 0, "elapsed": 2763, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="6df53022-6e3c-4956-a7a3-c8bb1cdd64fe"
trainer.test()
# + colab={"base_uri": "https://localhost:8080/", "height": 422, "referenced_widgets": ["407a7c2c60ba4c01a16dbe94884b1574", "343d825a99e84f79852d26ff86d613df", "153f42aa2d1047b8a34a48451cf4605b", "7db8cc07931e4be1aa6749c70d2e1d3d", "ea414bb11bcb4e9690500e4dff5e9aa6", "129352413d13457fb58521586c8fb193", "20ac5039e21744be8d6c9ed8d2731370", "3c9aa717ed984171a51117437a648e31"]} id="pQHczbY5zqSe" executionInfo={"status": "ok", "timestamp": 1613932755030, "user_tz": 0, "elapsed": 20797, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="bff482be-9a7c-4bdf-ca44-903450eb2f13"
wandb.finish()
# + colab={"base_uri": "https://localhost:8080/"} id="gUFNXf8ayyy0" executionInfo={"status": "ok", "timestamp": 1613914438355, "user_tz": 0, "elapsed": 12708, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="603cf3ba-2a48-4c6a-9f39-d0408bb5b774"
trainer.save_checkpoint("covid_classifier.ckpt")
new_model = CovidClassifier.load_from_checkpoint(n_classes=2, checkpoint_path="covid_classifier.ckpt")
new_model.freeze()
# + colab={"base_uri": "https://localhost:8080/", "height": 136, "referenced_widgets": ["e1a6d3e1b41d46cfa713a418b046720a", "2b814eaa1b814846ac567137e2ef24ce", "140f6dca50ea4c85b3e40ce6106f73e3", "ca472c353ddf4c3d82f0fb9c688138f5", "702b32a7605e46e0a5cd0c077ff19228", "1b0a768841784d9fa7ae33a67a2f42e9", "784507b49c0f4ed193deb48a205a9642", "d105a624d384463594b68112adc7a4da"]} id="nio7ZvPwyywI" executionInfo={"status": "ok", "timestamp": 1613914546829, "user_tz": 0, "elapsed": 30864, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="10e514a1-63cd-41f5-80fa-8753923fb558"
x = trainer.test()
# + colab={"base_uri": "https://localhost:8080/"} id="W4QENGl3zTvx" executionInfo={"status": "ok", "timestamp": 1613914552341, "user_tz": 0, "elapsed": 774, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="88e1965a-b9b4-43e9-aa31-0bc0e0eafd76"
x
# + colab={"base_uri": "https://localhost:8080/"} id="ymxRMhwszTsa" executionInfo={"status": "ok", "timestamp": 1613914810313, "user_tz": 0, "elapsed": 458, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="a8c2cfd6-c701-42ca-fe85-7bb72e28581c"
(len(df_train) // BATCH_SIZE) * 1 * BATCH_SIZE
# + colab={"base_uri": "https://localhost:8080/"} id="0zYp8J_HzTpj" executionInfo={"status": "ok", "timestamp": 1613914801911, "user_tz": 0, "elapsed": 549, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="1fa00fe6-1f69-4029-b837-ece141dec36e"
df_train.shape
# + id="wzNZIjAtzTm7"
# + id="ULdX6g18zTkU"
# + id="lU8ucwbSzTeW"
# + id="EZHmNLGrzTbi"
# + id="6n9rNykxzTY2"
# + id="aBgwJpueyytY"
# + [markdown] id="2TtHYB1ArrZf"
# # code scraps
# + id="I80Dwkq_9Qed" executionInfo={"status": "ok", "timestamp": 1613904244260, "user_tz": 0, "elapsed": 21637, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
import torch
from torch import nn
import pytorch_lightning as pl
from torch.utils.data import Dataset, DataLoader
from transformers import BertModel, BertTokenizer, AdamW, get_linear_schedule_with_warmup
from sklearn.model_selection import train_test_split
import numpy as np
import pandas as pd
# + id="Qo2el-yjDEy9" executionInfo={"status": "ok", "timestamp": 1613904246082, "user_tz": 0, "elapsed": 22725, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
# Read abstracts from Microsoft Academic Graph
df = pd.read_csv("drive/MyDrive/Colab Notebooks/mag_papers.csv")
# + colab={"base_uri": "https://localhost:8080/", "height": 206} id="QbLfnNK_DGg_" executionInfo={"status": "ok", "timestamp": 1613904246089, "user_tz": 0, "elapsed": 22294, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="aea5e6c2-659c-4eb8-bf76-4810063866c3"
df.head()
# + colab={"base_uri": "https://localhost:8080/"} id="yp333YBoEHE5" executionInfo={"status": "ok", "timestamp": 1613904246089, "user_tz": 0, "elapsed": 21161, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="d860c3d7-1c09-4667-ffa3-95586520f229"
# Split the dataframe to training and evaluation sets
pl.seed_everything(42)
df_train, df_val = train_test_split(df, test_size=.05)
# + colab={"base_uri": "https://localhost:8080/"} id="3Hhp7yu0EHCL" executionInfo={"status": "ok", "timestamp": 1613904246090, "user_tz": 0, "elapsed": 19036, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="1c1d4ca9-0166-4a63-8f4c-f085b8c7e812"
print(f"% of Covid-19 publications: {df[df.is_Covid19==1].shape[0]}")
print(f"% of non-Covid-19 publications: {df[df.is_Covid19!=1].shape[0]}")
# + colab={"base_uri": "https://localhost:8080/"} id="XHCCTHcAEHAF" executionInfo={"status": "ok", "timestamp": 1613904246091, "user_tz": 0, "elapsed": 18813, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="5fafea34-22d3-4824-dd8f-ac3fa119e7b0"
df_train.shape
# + id="xAaoTVDoEG97" executionInfo={"status": "ok", "timestamp": 1613904246092, "user_tz": 0, "elapsed": 17959, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
# Shuffle the training data
df_train = df_train.sample(frac=1.)
# + colab={"base_uri": "https://localhost:8080/"} id="0yzGzXq0EG5p" executionInfo={"status": "ok", "timestamp": 1613329356230, "user_tz": 0, "elapsed": 14789, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="e28b237c-936b-4b26-f027-52ccbc6a1475"
sample_row = df.iloc[15]
sample_abstract = sample_row.abstract
sample_label = sample_row.is_Covid19
print(sample_abstract)
print()
print(sample_label)
# + colab={"base_uri": "https://localhost:8080/", "height": 66, "referenced_widgets": ["01bd4f186d93483991fe5f8bf3f7f4fe", "a0bf47204db24c17a22cc3d70d36e0c4", "d3a3fc6cd18047a69e2eb9fc72d4a79c", "aebfb9c49f2e44a9b7d76bf26401f005", "926f193a43fb484ba388f234f7740582", "00b9d850f306420cadf045ac2777798b", "8520fc8902484084b8c7cc76804b2f01", "720055d4d0fd405e8b8dd8ee703e7b17"]} id="DGu-eI5FEG1R" executionInfo={"status": "ok", "timestamp": 1613329491791, "user_tz": 0, "elapsed": 1278, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="f3301d9f-5f71-43f9-c24b-dc33c1c9291a"
# Instantiate the tokenizer
TRANSFORMER_MODEL_NAME = "bert-base-uncased"
tokenizer = BertTokenizer.from_pretrained(TRANSFORMER_MODEL_NAME)
# + id="J0E7HEQGEGxS"
encoding = tokenizer.encode_plus(
sample_abstract,
add_special_tokens=True,
max_length=512,
return_token_type_ids=True,
padding="max_length",
return_attention_mask=True,
return_tensors="pt"
)
# + colab={"base_uri": "https://localhost:8080/"} id="ltAGYRgBFLRz" executionInfo={"status": "ok", "timestamp": 1613326688878, "user_tz": 0, "elapsed": 377, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="c2a6d524-73d3-4c65-9f8c-29b56ba19aef"
encoding.keys()
# + colab={"base_uri": "https://localhost:8080/"} id="K7XaPcTFFLPn" executionInfo={"status": "ok", "timestamp": 1613326689147, "user_tz": 0, "elapsed": 413, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="e7ed5b48-c973-41f3-978f-a3404db58d89"
len(encoding["token_type_ids"].squeeze(0))
# + colab={"base_uri": "https://localhost:8080/"} id="psPjqUzIFLNs" executionInfo={"status": "ok", "timestamp": 1613326689558, "user_tz": 0, "elapsed": 463, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="bae702df-3577-4906-f797-4be2c8c90295"
encoding["input_ids"].shape
# + colab={"base_uri": "https://localhost:8080/"} id="HrPysdUEFLMN" executionInfo={"status": "ok", "timestamp": 1613326689799, "user_tz": 0, "elapsed": 487, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="88e05e79-3762-4e80-f57c-a09afdfaedc8"
encoding["attention_mask"]
# + colab={"base_uri": "https://localhost:8080/"} id="J6Z5JTqcMsQf" executionInfo={"status": "ok", "timestamp": 1613326690075, "user_tz": 0, "elapsed": 572, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="8997dbcb-de72-4d86-f885-c9ca8a523ef6"
type(encoding["input_ids"])
# + id="IK_uskAaFLHT" colab={"base_uri": "https://localhost:8080/", "height": 66, "referenced_widgets": ["9c9becde39dd4a92a91f61faf13f41b7", "24aa3932a394476ca19e00c3598968cb", "9b31c40bc52e4bffb1abb2f06277dc9a", "f35eb7c8a91b4e509aa976e83044b63d", "2c25e4e60a8149069b258a6b7c7c656b", "ace2992cf35744719345061dca7f1737", "bcb41474df5547c09f536b5f2160dcbc", "239de735cf29402fa4d88497ab360e54"]} executionInfo={"status": "ok", "timestamp": 1613904246545, "user_tz": 0, "elapsed": 10786, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="b4137fe6-38df-4a28-a8a4-b131e35cbeba"
#Instantiate the tokenizer
TRANSFORMER_MODEL_NAME = "bert-base-uncased"
tokenizer = BertTokenizer.from_pretrained(TRANSFORMER_MODEL_NAME)
class CovidDataset(torch.utils.data.Dataset):
def __init__(self, data, tokenizer, max_token_len=128):
self.data = data
self.tokenizer = tokenizer
self.max_token_len = max_token_len
def __len__(self):
return len(self.data)
def __getitem__(self, index):
# Grab a single row from the dataframe
data_row = self.data.iloc[index]
# Grab text and label
abstract_text = data_row.abstract
labels = torch.tensor(data_row.is_Covid19)
# print(torch.FloatTensor(labels))
encoding = tokenizer.encode_plus(
abstract_text,
add_special_tokens=True,
max_length=self.max_token_len,
return_token_type_ids=False,
padding="max_length",
truncation=True,
return_attention_mask=True,
return_tensors="pt"
)
return dict(
abstract_text=abstract_text,
input_ids=encoding["input_ids"].flatten(),
attention_mask=encoding["attention_mask"].flatten(),
labels=labels
)
# + id="O8xt_t7lM6n0" executionInfo={"status": "ok", "timestamp": 1613901749742, "user_tz": 0, "elapsed": 450, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
train_dataset = CovidDataset(df_train, tokenizer, max_token_len=512)
# + id="n7Kqvh-wM6l5" executionInfo={"status": "ok", "timestamp": 1613901749917, "user_tz": 0, "elapsed": 438, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
sample_items = train_dataset[0]
# + colab={"base_uri": "https://localhost:8080/"} id="J6fYgFymM6jl" executionInfo={"status": "ok", "timestamp": 1613901750143, "user_tz": 0, "elapsed": 515, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="8e25d034-8e99-4023-e350-1ade60579ac8"
sample_items.keys()
# + colab={"base_uri": "https://localhost:8080/"} id="Eam1iipGM6hQ" executionInfo={"status": "ok", "timestamp": 1613901751551, "user_tz": 0, "elapsed": 434, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="552c296a-95c1-4971-d2e2-9132554d3eaa"
sample_items['labels']
# + colab={"base_uri": "https://localhost:8080/", "height": 106} id="YNCCSaG621du" executionInfo={"status": "ok", "timestamp": 1613901753111, "user_tz": 0, "elapsed": 428, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="d7a8f418-5cf9-4f57-ba73-651e7e3c0426"
sample_items['abstract_text']
# + id="xFfGowTR21Ye"
# + id="qKMhFtDnFLFC"
# load model
bert_model = BertModel.from_pretrained(TRANSFORMER_MODEL_NAME, return_dict=True)
# + colab={"base_uri": "https://localhost:8080/"} id="XwWmaHbKQxCE" executionInfo={"status": "ok", "timestamp": 1613326911917, "user_tz": 0, "elapsed": 627, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="1e27ce5e-f771-403c-c546-115126b4de8b"
sample_items["input_ids"].unsqueeze(dim=0).shape
# + id="1dymwLOeFLCg"
prediction = bert_model(sample_items["input_ids"].unsqueeze(dim=0), sample_items["attention_mask"].unsqueeze(dim=0))
# + colab={"base_uri": "https://localhost:8080/"} id="h4BUWYEGQUE-" executionInfo={"status": "ok", "timestamp": 1613326917240, "user_tz": 0, "elapsed": 2153, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="d41c9580-ece6-4b09-e399-312872618bc4"
prediction.last_hidden_state.shape
# + id="E973ijszQUC4" executionInfo={"status": "ok", "timestamp": 1613905855019, "user_tz": 0, "elapsed": 465, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
class CovidDataModule(pl.LightningDataModule):
def __init__(self, df_train, df_val, tokenizer, batch_size=8, max_token_len=512):
super().__init__()
self.df_train = df_train
self.df_val = df_val
self.tokenizer = tokenizer
self.batch_size = batch_size
self.max_token_len = max_token_len
def setup(self):
self.train_dataset = CovidDataset(self.df_train, self.tokenizer, self.max_token_len)
self.val_dataset = CovidDataset(self.df_val, self.tokenizer, self.max_token_len)
self.test_dataset = CovidDataset(self.df_val, self.tokenizer, self.max_token_len)
def train_dataloader(self):
return DataLoader(self.train_dataset, batch_size=self.batch_size, shuffle=True, num_workers=4)
def val_dataloader(self):
return DataLoader(self.val_dataset, batch_size=self.batch_size)
def test_dataloader(self):
return DataLoader(self.val_dataset, batch_size=self.batch_size)
# + id="LYrjGAIeQUBC" executionInfo={"status": "ok", "timestamp": 1613905866698, "user_tz": 0, "elapsed": 474, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
N_EPOCHS = 4
BATCH_SIZE = 12
data_module = CovidDataModule(df_train, df_val, tokenizer, batch_size=BATCH_SIZE, max_token_len=128)
data_module.setup()
# + colab={"base_uri": "https://localhost:8080/"} id="iQn48ua9HUQo" executionInfo={"status": "ok", "timestamp": 1613903004449, "user_tz": 0, "elapsed": 442, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="464728f0-7cbb-4ff1-93d4-62b8d78b55e8"
# + [markdown] id="Fmyzv13bg6SS"
# ## Modelling
# + colab={"base_uri": "https://localhost:8080/"} id="vbHTV-PwSbkP" executionInfo={"status": "ok", "timestamp": 1613905920901, "user_tz": 0, "elapsed": 432, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="5bd97a51-a805-4ce7-f612-92e8b2418c4a"
target = torch.ones([10, 64], dtype=torch.float32) # 64 classes, batch size = 10
output = torch.full([10, 64], 1.5) # A prediction (logit)
pos_weight = torch.ones([64]) # All weights are equal to 1
criterion = torch.nn.BCEWithLogitsLoss(pos_weight=pos_weight)
criterion(output, target) # -log(sigmoid(1.5))
# + colab={"base_uri": "https://localhost:8080/"} id="lV23KOhCSZBn" executionInfo={"status": "ok", "timestamp": 1613905899547, "user_tz": 0, "elapsed": 573, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="85599bb2-ff76-44b7-bc1c-3e22d8ae5f42"
nn.BCEWithLogitsLoss
# + id="pTniHyBDfFP7" executionInfo={"status": "ok", "timestamp": 1613906002881, "user_tz": 0, "elapsed": 460, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
class CovidClassifier(pl.LightningModule):
def __init__(self, n_classes, steps_per_epoch=None, n_epochs=None):
super().__init__()
self.bert_model = BertModel.from_pretrained(TRANSFORMER_MODEL_NAME, return_dict=True)
self.n_classes = n_classes
self.classifier = nn.Linear(self.bert_model.config.hidden_size, self.n_classes)
self.steps_per_epoch = steps_per_epoch
self.n_epochs = n_epochs
self.criterion = nn.CrossEntropyLoss()
# self.criterion = nn.BCELoss()
def forward(self, input_ids, attention_mask, labels=None):
output = self.bert_model(input_ids, attention_mask=attention_mask)
output = self.classifier(output.pooler_output)
# output = torch.sigmoid(output)
loss = 0
if labels is not None:
loss = self.criterion(output.view(-1, self.n_classes), labels.view(-1))
# loss = self.criterion(output, labels)
return loss, output
def training_step(self, batch, batch_idx):
input_ids = batch["input_ids"]
attention_mask = batch["attention_mask"]
labels = batch["labels"]
loss, outputs = self(input_ids, attention_mask, labels)
self.log("train_loss", loss, prog_bar=True, logger=False)
return {"loss":loss, "predictions":outputs, "labels":labels}
def validation_step(self, batch, batch_idx):
input_ids = batch["input_ids"]
attention_mask = batch["attention_mask"]
labels = batch["labels"]
loss, outputs = self(input_ids, attention_mask, labels)
self.log("val_loss", loss, prog_bar=True, logger=False)
return loss
def test_step(self, batch, batch_idx):
input_ids = batch["input_ids"]
attention_mask = batch["attention_mask"]
labels = batch["labels"]
loss, outputs = self(input_ids, attention_mask, labels)
self.log("test_loss", loss, prog_bar=True, logger=False)
return loss
def configure_optimizers(self):
optimizer = AdamW(self.parameters(), lr=2e-5)
warmup_steps = self.steps_per_epoch // 3
total_steps = self.steps_per_epoch * self.n_epochs - warmup_steps
scheduler = get_linear_schedule_with_warmup(
optimizer, warmup_steps, total_steps
)
return [optimizer], [scheduler]
# + id="bqdUxpx0iW6h" executionInfo={"status": "ok", "timestamp": 1613906007011, "user_tz": 0, "elapsed": 2943, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
model = CovidClassifier(n_classes=2, steps_per_epoch=len(df_train) // BATCH_SIZE, n_epochs=N_EPOCHS)
# + id="zq1EfWWmiW14" executionInfo={"status": "ok", "timestamp": 1613906007013, "user_tz": 0, "elapsed": 2273, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
# # predictions with the untrained model
# _, predictions = model(sample_items["input_ids"].unsqueeze(dim=0), sample_items["attention_mask"].unsqueeze(dim=0))
# predictions
# + colab={"base_uri": "https://localhost:8080/"} id="86C7NM3wiWzy" executionInfo={"status": "ok", "timestamp": 1613906007013, "user_tz": 0, "elapsed": 1941, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="60d857b0-e122-41ab-e0ff-afdc07a63d90"
trainer = pl.Trainer(max_epochs=N_EPOCHS, gpus=1, progress_bar_refresh_rate=30)
# + colab={"base_uri": "https://localhost:8080/", "height": 342, "referenced_widgets": ["3f807452ce184d8f96e00194d7e90660", "2ee4cb2621c74785863b5a8159d2d52b", "70a5e8a7b5444ffeb1874dbe6084ab57", "62b2b63b70884fc99d44b85506bddf0f", "268f3dcb554d4a0f97ba54977bb8da10", "a8b35aa629f04ae785b6e5d2ef48c9b3", "36a7e0ef63324335880b724fa97f25b6", "9dc5cccdfe234ff98f431f52ee9adec4", "0d14f097fdc84b8b8692bbb863ea5996", "627bcba2ce8343c994eaa3fc42976ac8", "2f7a841a99424e82af80ee44adc1ec5e", "c0e40e3481964af7956a498fb6b326a7", "a16945e71b354273935525b93fe9747b", "9ca4ceb0a7bd43df85e4ed928916ae7d", "eba61e6491ab41a089e1dcaa2ca5b0ee", "93cbca2db5e1454caf64468fa891314e", "aa775070f7894f98975acd43109c15dc", "35c13842731947089fd98f25646c8b53", "f71396cbd381406aacd82ddf43f40cc7", "1f012f41691541f98d328a2c1f0acef1", "13b2a78bbf8b418a8a97b62c9565c01c", "55ca5d2760ff4883a46dcd8bdfe101eb", "c2f20217f84d4ee49d48a58afe53f0e0", "fdddae4200de4826aadfbd2662cfa66d", "dfad88e2af0c43ce885c425e2979f1b3", "cbaf7269618842feb33e091352385fd7", "c46385a245934e22911992c05e5dd1fd", "89f545b844354bf482ae9aae8c44e70d", "31222f8e20734d95bdd8b60e981fbdbe", "7dd419438cd946cfbf202904be0c76a7", "c59199f9efae4ce38d24426577e6795e", "557b953af7c4429e95bd24f64e36ebf9", "6ae7ed4800784f798faf405be8101cbe", "6070f51faea8449483cbaf2dff58b3bc", "6b6ed30c011d4ec78698a4ba5aea2eff", "67772a913aa94a039215f04c947193f4", "a882434308194a97ad6df1c7fd2bb250", "4e26ecd81140428f9eb7e8e3b0e0eaa9", "ebf18c6dbf1e4c078d32e2359cbf13bf", "5fe1e917ae6b43f5983d6406dcad80b1", "3b8fca1060b64a95909febcf59879fff", "a048bf8d6bff4585b69a4cb8dcb4fafd", "7f3282444e5f4529bf490d5f5751c279", "e0525366268e433eaa3c2a9ed8956a68", "4dafbd4b11dd44178b68f7eda17b953c", "f9510d76cc4245959f374dbed0ec6584", "7804f84db032442e9ed974f1a864bafa", "61aaf8f6d9004268ac58fdd6fcd4e7e6"]} id="Vtg1g79tiWxi" executionInfo={"status": "ok", "timestamp": 1613906333551, "user_tz": 0, "elapsed": 327996, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="9a3d74b7-465d-4d00-c1dc-854a2c18e5a9"
trainer.fit(model, data_module)
# + colab={"base_uri": "https://localhost:8080/", "height": 153, "referenced_widgets": ["af047fb9b3154853b61401cc761f7914", "5ac46b6e304f405692c731197892b0a1", "563802aa82604f4f9003639cc4a93e25", "28e165a9185e4034b4c0574ff232f994", "847c9586320741fc97ce693db8ecf30d", "05af886c4dfe4cf5bc4afb41a99fbbd0", "c285e4d6b4ab48f3b65f2992712a784c", "32c1724bfab5495abca4459d64877356"]} id="K7ksDbBhGUik" executionInfo={"status": "ok", "timestamp": 1613906401952, "user_tz": 0, "elapsed": 3179, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="f481ccae-42f0-4fde-abaf-838582b3988d"
trainer.test()
# + colab={"base_uri": "https://localhost:8080/"} id="D1KzEC6ufFNL" executionInfo={"status": "ok", "timestamp": 1613330642788, "user_tz": 0, "elapsed": 3180, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="45cfd265-65a9-477d-c99b-4ee2c6fb2c20"
# # predictions with the untrained model
# _, predictions = model(sample_items["input_ids"].unsqueeze(dim=0), sample_items["attention_mask"].unsqueeze(dim=0))
# print(predictions)
# softmax = nn.Softmax(-1)
# print(softmax(predictions))
# + colab={"base_uri": "https://localhost:8080/"} id="z0TL_5-4_xz7" executionInfo={"status": "ok", "timestamp": 1613906617827, "user_tz": 0, "elapsed": 13687, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="65bfaf8e-4e8f-4298-92e4-ba7fbbdbb125"
trainer.save_checkpoint("covid_classifier.ckpt")
new_model = CovidClassifier.load_from_checkpoint(n_classes=2, checkpoint_path="covid_classifier.ckpt")
new_model.freeze()
# + id="45nB567kGE7h"
# + id="-KzTyQ6GGE4_"
# + id="48sdGGC3GE2v"
# + id="-BmQZjhQGEvl"
# + id="EPsz3ZqKAoss" executionInfo={"status": "ok", "timestamp": 1613906622873, "user_tz": 0, "elapsed": 548, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
sample_abstract = "COVID-19 is a deadly coronavirus that broke out last year."
# + id="-EckVhhjFoeX" executionInfo={"status": "ok", "timestamp": 1613906622874, "user_tz": 0, "elapsed": 346, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}}
encoding = tokenizer.encode_plus(
sample_abstract,
add_special_tokens=True,
max_length=128,
return_token_type_ids=True,
padding="max_length",
return_attention_mask=True,
return_tensors="pt"
)
# + colab={"base_uri": "https://localhost:8080/"} id="wqpYSq53_xvI" executionInfo={"status": "ok", "timestamp": 1613906624476, "user_tz": 0, "elapsed": 942, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="687bbb92-eca2-4b5a-ab82-26de43749243"
_, predictions = new_model(encoding["input_ids"], encoding["attention_mask"])
print(predictions)
softmax = nn.Softmax(-1)
print(softmax(predictions))
# + id="AE-wJ68k_xqV" colab={"base_uri": "https://localhost:8080/"} executionInfo={"status": "ok", "timestamp": 1613902517235, "user_tz": 0, "elapsed": 442, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="21d9111f-d9bb-409d-d18b-b2576ad1d64a"
sample_items["labels"]
# + colab={"base_uri": "https://localhost:8080/", "height": 123} id="OrrlB2ueQT39" executionInfo={"status": "ok", "timestamp": 1613902651436, "user_tz": 0, "elapsed": 423, "user": {"displayName": "Kostas Stathoulopoulos", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj6adaPSsxjG4KL-vKtFxpnbR8udIWR-Qr1QPQr=s64", "userId": "04447701163448939568"}} outputId="bae9717a-6175-4262-d886-20167f5f2df3"
df_train.iloc[8][0]
# + id="T5SvT5DnHdk3"
# + id="7Mq32HFMHdie"
# + id="s0FCKyimHdgU"
# + id="kUSPKEHoHddw"
# + id="3FfG1GyFHdbb"
# + id="6mwnXV7GHdZB"
# + id="slYjYXDtHdWX"
| 70.138889
| 2,160
|
9200630c427703c8178025ede5ed870d0827d82f
|
py
|
python
|
week4/Regex Practice Part II.ipynb
|
Emmayyyyy/dso-560-nlp-and-text-analytics
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] toc=true
# <h1>Table of Contents<span class="tocSkip"></span></h1>
# <div class="toc"><ul class="toc-item"><li><span><a href="#Import-Dependencies-and-Load-in-Dataset" data-toc-modified-id="Import-Dependencies-and-Load-in-Dataset-1"><span class="toc-item-num">1 </span>Import Dependencies and Load in Dataset</a></span></li><li><span><a href="#Pandas-Exploratory-Data-Analysis" data-toc-modified-id="Pandas-Exploratory-Data-Analysis-2"><span class="toc-item-num">2 </span>Pandas Exploratory Data Analysis</a></span><ul class="toc-item"><li><ul class="toc-item"><li><span><a href="#Show-the-First/Last-Rows-of-Dataset" data-toc-modified-id="Show-the-First/Last-Rows-of-Dataset-2.0.1"><span class="toc-item-num">2.0.1 </span>Show the First/Last Rows of Dataset</a></span></li><li><span><a href="#Drop-Unnecessary-Pandas-Column" data-toc-modified-id="Drop-Unnecessary-Pandas-Column-2.0.2"><span class="toc-item-num">2.0.2 </span>Drop Unnecessary Pandas Column</a></span></li><li><span><a href="#Compute-a-New-Pandas-Series-and-Attach-to-Dataframe-as-Column" data-toc-modified-id="Compute-a-New-Pandas-Series-and-Attach-to-Dataframe-as-Column-2.0.3"><span class="toc-item-num">2.0.3 </span>Compute a New Pandas Series and Attach to Dataframe as Column</a></span></li><li><span><a href="#Count-Number-of-Rows-in-Dataframe-Meet-Some-Criteria" data-toc-modified-id="Count-Number-of-Rows-in-Dataframe-Meet-Some-Criteria-2.0.4"><span class="toc-item-num">2.0.4 </span>Count Number of Rows in Dataframe Meet Some Criteria</a></span></li><li><span><a href="#Filter-DataFrame-for-a-Subset-of-Rows" data-toc-modified-id="Filter-DataFrame-for-a-Subset-of-Rows-2.0.5"><span class="toc-item-num">2.0.5 </span>Filter DataFrame for a Subset of Rows</a></span></li></ul></li><li><span><a href="#Difference-Between-extractall-and-findall" data-toc-modified-id="Difference-Between-extractall-and-findall-2.1"><span class="toc-item-num">2.1 </span>Difference Between <code>extractall</code> and <code>findall</code></a></span></li></ul></li><li><span><a href="#Regex-Character-Classes" data-toc-modified-id="Regex-Character-Classes-3"><span class="toc-item-num">3 </span>Regex Character Classes</a></span><ul class="toc-item"><li><ul class="toc-item"><li><span><a href="#Find-all-Tweets-That-Start-with-a-Number" data-toc-modified-id="Find-all-Tweets-That-Start-with-a-Number-3.0.1"><span class="toc-item-num">3.0.1 </span>Find all Tweets That Start with a Number</a></span></li><li><span><a href="#Find-all-@-Mentions" data-toc-modified-id="[email protected]"><span class="toc-item-num">3.0.2 </span>Find all @ Mentions</a></span><ul class="toc-item"><li><span><a href="#Alternative-Method-to-Work-With-Lists-in-Pandas" data-toc-modified-id="Alternative-Method-to-Work-With-Lists-in-Pandas-3.0.2.1"><span class="toc-item-num">3.0.2.1 </span>Alternative Method to Work With Lists in Pandas</a></span></li></ul></li></ul></li></ul></li><li><span><a href="#Quantifiers" data-toc-modified-id="Quantifiers-4"><span class="toc-item-num">4 </span>Quantifiers</a></span><ul class="toc-item"><li><ul class="toc-item"><li><span><a href="#Match-All-Phone-Numbers-(Link)" data-toc-modified-id="Match-All-Phone-Numbers-(Link)-4.0.1"><span class="toc-item-num">4.0.1 </span>Match All Phone Numbers (<a href="https://regexr.com/50v17" target="_blank">Link</a>)</a></span></li><li><span><a href="#Parse-Out-Zip-Codes-(Link)" data-toc-modified-id="Parse-Out-Zip-Codes-(Link)-4.0.2"><span class="toc-item-num">4.0.2 </span>Parse Out Zip Codes (<a href="https://regexr.com/50v1g" target="_blank">Link</a>)</a></span></li></ul></li></ul></li><li><span><a href="#Capture-Groups" data-toc-modified-id="Capture-Groups-5"><span class="toc-item-num">5 </span>Capture Groups</a></span><ul class="toc-item"><li><ul class="toc-item"><li><span><a href="#Example:-Parsing-out-Weekday-from-Timestamp-String-(Link)" data-toc-modified-id="Example:-Parsing-out-Weekday-from-Timestamp-String-(Link)-5.0.1"><span class="toc-item-num">5.0.1 </span>Example: Parsing out Weekday from Timestamp String (<a href="https://regexr.com/50v0l" target="_blank">Link</a>)</a></span></li><li><span><a href="#Example:-Parsing-Out-Domain-Names" data-toc-modified-id="Example:-Parsing-Out-Domain-Names-5.0.2"><span class="toc-item-num">5.0.2 </span>Example: Parsing Out Domain Names</a></span></li></ul></li><li><span><a href="#Non-Capture-Groups" data-toc-modified-id="Non-Capture-Groups-5.1"><span class="toc-item-num">5.1 </span>Non-Capture Groups</a></span><ul class="toc-item"><li><span><a href="#Example-1-(Link)" data-toc-modified-id="Example-1-(Link)-5.1.1"><span class="toc-item-num">5.1.1 </span>Example 1 <a href="https://regexr.com/50t7c" target="_blank">(Link)</a></a></span></li><li><span><a href="#Example-2-(Link)" data-toc-modified-id="Example-2-(Link)-5.1.2"><span class="toc-item-num">5.1.2 </span>Example 2 <a href="https://regexr.com/50t7c" target="_blank">(Link)</a></a></span></li><li><span><a href="#Example-3-(Link)" data-toc-modified-id="Example-3-(Link)-5.1.3"><span class="toc-item-num">5.1.3 </span>Example 3 (<a href="https://regexr.com/50ush" target="_blank">Link</a>)</a></span></li></ul></li></ul></li><li><span><a href="#Named-Capture-Groups" data-toc-modified-id="Named-Capture-Groups-6"><span class="toc-item-num">6 </span>Named Capture Groups</a></span><ul class="toc-item"><li><ul class="toc-item"><li><span><a href="#Using-Named-Capture-Groups-in-Pandas-for-Feature-Engineering" data-toc-modified-id="Using-Named-Capture-Groups-in-Pandas-for-Feature-Engineering-6.0.1"><span class="toc-item-num">6.0.1 </span>Using Named Capture Groups in Pandas for Feature Engineering</a></span></li></ul></li></ul></li><li><span><a href="#Finding-Repeat-Words-in-Reviews-Using-Backreferences" data-toc-modified-id="Finding-Repeat-Words-in-Reviews-Using-Backreferences-7"><span class="toc-item-num">7 </span>Finding Repeat Words in Reviews Using Backreferences</a></span><ul class="toc-item"><li><ul class="toc-item"><li><span><a href="#Using-Named-Backreferences" data-toc-modified-id="Using-Named-Backreferences-7.0.1"><span class="toc-item-num">7.0.1 </span>Using Named Backreferences</a></span></li><li><span><a href="#Using-Negative-Lookaheads" data-toc-modified-id="Using-Negative-Lookaheads-7.0.2"><span class="toc-item-num">7.0.2 </span>Using Negative Lookaheads</a></span></li><li><span><a href="#Negative-Lookbehind" data-toc-modified-id="Negative-Lookbehind-7.0.3"><span class="toc-item-num">7.0.3 </span>Negative Lookbehind</a></span><ul class="toc-item"><li><span><a href="#Find-all-tweets-that-do-not-begin-with-a-hashtag-or-a-mention." data-toc-modified-id="Find-all-tweets-that-do-not-begin-with-a-hashtag-or-a-mention.-7.0.3.1"><span class="toc-item-num">7.0.3.1 </span>Find all tweets that do not begin with a hashtag or a mention.</a></span></li></ul></li></ul></li></ul></li><li><span><a href="#Exercises" data-toc-modified-id="Exercises-8"><span class="toc-item-num">8 </span>Exercises</a></span><ul class="toc-item"><li><ul class="toc-item"><li><ul class="toc-item"><li><span><a href="#Use-named-capture-groups-to-find-the-subject-headings-for-these-emails." data-toc-modified-id="Use-named-capture-groups-to-find-the-subject-headings-for-these-emails.-8.0.0.1"><span class="toc-item-num">8.0.0.1 </span>Use named capture groups to find the subject headings for these emails.</a></span></li><li><span><a href="#Find-all-hashtags-or-references-mentioned-in-the-tweets_df-dataset.-Store-them-as-two-separate-columns,-one-for-whether-it-is-either-a-@-or-#,-and-the-other-for-the-actual-content-(ie.-the-hello-part-of-#hello)." data-toc-modified-id="Find-all-hashtags-or-references-mentioned-in-the-tweets_df-dataset.-Store-them-as-two-separate-columns,-one-for-whether-it-is-either-a-@-or-#,-and-the-other-for-the-actual-content-(ie.-the-hello-part-of-#hello).-8.0.0.2"><span class="toc-item-num">8.0.0.2 </span>Find all hashtags or references mentioned in the <code>tweets_df</code> dataset. Store them as two separate columns, one for whether it is either a <code>@</code> or <code>#</code>, and the other for the actual content (ie. the <code>hello</code> part of <code>#hello</code>).</a></span></li><li><span><a href="#Use-named-capture-groups-to-provide-a-list-of-email-addresses-for-your-security-administrator-to-blacklist-from-your-company's-email-servers.-The-output-should-be-a-list-of-dictionaries:" data-toc-modified-id="Use-named-capture-groups-to-provide-a-list-of-email-addresses-for-your-security-administrator-to-blacklist-from-your-company's-email-servers.-The-output-should-be-a-list-of-dictionaries:-8.0.0.3"><span class="toc-item-num">8.0.0.3 </span>Use named capture groups to provide a <strong>list of email addresses</strong> for your security administrator to blacklist from your company's email servers. The output should be a list of dictionaries:</a></span></li><li><span><a href="#Use-a-negative-lookahead-to-capture-all-emails-except-for-the-ones-from-yahoo.com." data-toc-modified-id="Use-a-negative-lookahead-to-capture-all-emails-except-for-the-ones-from-yahoo.com.-8.0.0.4"><span class="toc-item-num">8.0.0.4 </span>Use a negative lookahead to capture all emails except for the ones from <code>yahoo.com</code>.</a></span></li><li><span><a href="#Identify-any-IP-addresses-that-should-be-blacklisted" data-toc-modified-id="Identify-any-IP-addresses-that-should-be-blacklisted-8.0.0.5"><span class="toc-item-num">8.0.0.5 </span>Identify any IP addresses that should be blacklisted</a></span></li><li><span><a href="#The-word-"AT&T"-is-not-spelled-correctly-in-the-tweets_df-dataset.-Correct-the-misspelling." data-toc-modified-id="The-word-"AT&T"-is-not-spelled-correctly-in-the-tweets_df-dataset.-Correct-the-misspelling.-8.0.0.6"><span class="toc-item-num">8.0.0.6 </span>The word "AT&T" is not spelled correctly in the <code>tweets_df</code> dataset. Correct the misspelling.</a></span></li></ul></li></ul></li></ul></li></ul></div>
# -
# # Import Dependencies and Load in Dataset
import pandas as pd
tweets_df: pd.DataFrame = pd.read_csv("tweets_pandas.csv")
# # Pandas Exploratory Data Analysis
# + [markdown] solution2="shown" solution2_first=true
# ### Show the First/Last Rows of Dataset
# + solution2="shown"
# + solution2="shown"
# + [markdown] solution2="shown" solution2_first=true
# ### Drop Unnecessary Pandas Column
# + solution2="shown"
# get rid of some columns we don't care about
# preview the dataset
# + [markdown] solution2="shown" solution2_first=true
# ### Compute a New Pandas Series and Attach to Dataframe as Column
# + solution2="shown"
# get length of tweets in characters
# + [markdown] solution2="shown" solution2_first=true
# ### Count Number of Rows in Dataframe Meet Some Criteria
# + solution2="shown"
# count number of times Obama appears in tweets
# + [markdown] solution2="shown" solution2_first=true
# ### Filter DataFrame for a Subset of Rows
# + solution2="shown"
# filter dataframe for only tweets that contain
# -
# ## Difference Between `extractall` and `findall`
#
# The `extractall` method will return a fanned-out multi-dimensional index. For example, in the example below, the primary index is `0`, but there are sub-indices for `stellargirl` (`0`), `loooooooovvvvvveee` (`1`), etc.
# The `findall` method will return a list of results (or an empty list if there is no match).
# # Regex Character Classes
#
# There are often shortcut keywords you can use instead of typing out every possible character you want to match against. For instance, instead of `[a-zA-Z0-9]`, for all practical purposes, you can type out `/w`.
#
# 
# + [markdown] solution2="shown" solution2_first=true
# ### Find all Tweets That Start with a Number
# + [markdown] solution2="shown"
# * [Non-Match Example](https://regexr.com/50ur7)
# * [Match Example](https://regexr.com/50urj)
# + solution2="shown"
# + [markdown] solution2="shown" solution2_first=true
# ### Find all @ Mentions
# We'll use the `\w` character class to match for mentions (ie. `@ychennay`).
# + solution2="shown"
# + [markdown] solution2="shown"
# #### Alternative Method to Work With Lists in Pandas
# + solution2="shown"
# Convert the mentions column to boolean. Because an empty list is False, and a non-empty list is True, this filters
# out all non-empty lists.
# -
# # Quantifiers
#
# Quantifiers let you specify how many times a character or group should be matched.
# 
# + [markdown] solution2="shown" solution2_first=true
# ### Match All Phone Numbers ([Link](https://regexr.com/50v17))
# In the unwanted callers dataset (`unwanted_calls.csv`), parse out all phone numbers.
#
# We'll only consider for the time being phone numbers that follow the format `123-456-7890`.
# + solution2="shown"
import pandas as pd
# + [markdown] solution2="shown" solution2_first=true
# ### Parse Out Zip Codes ([Link](https://regexr.com/50v1g))
# In the `location_center_point_of_the_zip_code` field, we store both zip codes as well as geolocation data (latitudes and longitudes). We'll only consider zip codes with 5 digits (not the +4 digit delivery route).
# + solution2="shown"
# -
# # Capture Groups
#
# [Oracle documentation on Capture Groups:](https://docs.oracle.com/javase/tutorial/essential/regex/groups.html)
# > Capturing groups are a way to treat **multiple characters as a single unit**. They are created by placing the characters to be grouped inside a set of parentheses. For example, the regular expression `(dog)` creates a single group containing the letters `d`, `o`, and `g`.
# + [markdown] solution2="shown" solution2_first=true
# ### Example: Parsing out Weekday from Timestamp String ([Link](https://regexr.com/50v0l))
#
# In the `date` field of `tweets_df`, we have timestamp strings that look like this: `Mon May 11 03:22:30 UTC 2009`. We want to parse out the weekday from this string.
# + solution2="shown"
# + [markdown] solution2="shown" solution2_first=true
# ### Example: Parsing Out Domain Names
# We want to capture the domain names of different websites. Here, we need to escape the `.` part of `www.google.com`. In regex, `.` means "anything". To actually indicate we want to match for the literal `.` period character, we need to escape it, using `\.`.
# + solution2="shown"
# + solution2="shown"
# -
# ## Non-Capture Groups
# Sometimes you want to group multiple characters into a single unit to apply regex operations on them, but you don't
# want to actually capture or return their result.
# + [markdown] solution2="shown" solution2_first=true
# ### Example 1 [(Link)](https://regexr.com/50t7c)
# Using non-capture groups to match for optional text:
#
# You want to capture both `child` and `children` in your text:
# + solution2="shown"
import re
text = "The word child is singular, but the word children is plural."
# + [markdown] solution2="shown" solution2_first=true
# ### Example 2 [(Link)](https://regexr.com/50t7c)
# Find all the mentions in the tweets. Mentions start with `@`.
# + solution2="shown"
# + [markdown] solution2="shown" solution2_first=true
# ### Example 3 ([Link](https://regexr.com/50ush))
# Here we want to match for the dollar amount, but we don't want to include the currency notation (`$` or `USD`).
# + solution2="shown"
text = "Average fast food wage is $9.08, but inflation has increased USD9.23"
# -
# # Named Capture Groups
#
# Often, we might have a hard time keeping track of all the capture groups in our regex. We can use **named capture groups** instead.
# +
import re
file = open("mcdonalds-yelp-negative-reviews.csv", encoding="latin1")
reviews = file.read()
# what's wrong with this expression?
# +
# this looks much better
# +
# -
# ### Using Named Capture Groups in Pandas for Feature Engineering
import pandas as pd
reviews_df = pd.read_csv("mcdonalds-yelp-negative-reviews.csv", encoding="latin1")
# # Finding Repeat Words in Reviews Using Backreferences
# You can refer to capture groups earlier in your expression using **backreferences**. For instance, `\1` means the first capture group in your expression.
regex_pattern = r'(\b\w*burgers?\b).+\b\1\b'
file.seek(0)
reviews = file.readlines()
# ### Using Named Backreferences
#
# If you named a group `BURGER`, you can reference it later with `(?P=BURGER)`.
# +
# -
# ### Using Negative Lookaheads
#
# Whenever you are thinking *I need to match this expression, but only when the piece of text doesn't say X*, you can use **negative lookaheads**.
#
#
# For instance, to match only the timestamps in McDonalds reviews that happen before **12pm**:
# +
# -
# If we still want to capture the time of day indicator:
# +
# -
# ### Negative Lookbehind
# #### Find all tweets that do not begin with a hashtag or a mention.
# +
# -
# # Exercises
#
# You'll be using the the `tweets_df` Pandas dataframe and `fraudulent_emails.txt` text files for these examples.
email_text = open("fraudulent_emails.txt").read()
# + [markdown] solution2="shown" solution2_first=true
# #### Use named capture groups to find the subject headings for these emails.
# **Hint**: Look for the subject line within the email.
# + solution2="shown"
# Find the subject headings for these emails.
# + [markdown] solution2="hidden" solution2_first=true
# #### Find all hashtags or references mentioned in the `tweets_df` dataset. Store them as two separate columns, one for whether it is either a `@` or `#`, and the other for the actual content (ie. the `hello` part of `#hello`).
# + solution2="hidden"
# +
# + [markdown] solution2="shown" solution2_first=true
# #### Use named capture groups to provide a **list of email addresses** for your security administrator to blacklist from your company's email servers. The output should be a list of dictionaries:
#
# ```python
# [
# {
# "username": "yuchen",
# "domain": "gmail.com
# },
# # ...
# ]
# ```
#
# * Not all emails are malicious! Provide only the list of email addresses from where the email originates from. **Hint**: identify the pattern in the emails that tells you the source of the email.
#
# + solution2="shown"
# -
# #### Use a negative lookahead to capture all emails except for the ones from `yahoo.com`.
# + [markdown] solution2="hidden" solution2_first=true
# #### The word "AT&T" is not spelled correctly in the `tweets_df` dataset. Correct the misspelling.
# -
# + solution2="shown"
# -
| 51.704918
| 10,308
|
743a9cda0d69db2d21f8b4abdc8723bd8b795f61
|
py
|
python
|
Copy_of_DL.ipynb
|
nnayar7/FEELVOS
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/nnayar7/FEELVOS/blob/master/Copy_of_DL.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + colab={"resources": {"http://localhost:8080/nbextensions/google.colab/files.js": {"data": "Ly8gQ29weXJpZ2h0IDIwMTcgR29vZ2xlIExMQwovLwovLyBMaWNlbnNlZCB1bmRlciB0aGUgQXBhY2hlIExpY2Vuc2UsIFZlcnNpb24gMi4wICh0aGUgIkxpY2Vuc2UiKTsKLy8geW91IG1heSBub3QgdXNlIHRoaXMgZmlsZSBleGNlcHQgaW4gY29tcGxpYW5jZSB3aXRoIHRoZSBMaWNlbnNlLgovLyBZb3UgbWF5IG9idGFpbiBhIGNvcHkgb2YgdGhlIExpY2Vuc2UgYXQKLy8KLy8gICAgICBodHRwOi8vd3d3LmFwYWNoZS5vcmcvbGljZW5zZXMvTElDRU5TRS0yLjAKLy8KLy8gVW5sZXNzIHJlcXVpcmVkIGJ5IGFwcGxpY2FibGUgbGF3IG9yIGFncmVlZCB0byBpbiB3cml0aW5nLCBzb2Z0d2FyZQovLyBkaXN0cmlidXRlZCB1bmRlciB0aGUgTGljZW5zZSBpcyBkaXN0cmlidXRlZCBvbiBhbiAiQVMgSVMiIEJBU0lTLAovLyBXSVRIT1VUIFdBUlJBTlRJRVMgT1IgQ09ORElUSU9OUyBPRiBBTlkgS0lORCwgZWl0aGVyIGV4cHJlc3Mgb3IgaW1wbGllZC4KLy8gU2VlIHRoZSBMaWNlbnNlIGZvciB0aGUgc3BlY2lmaWMgbGFuZ3VhZ2UgZ292ZXJuaW5nIHBlcm1pc3Npb25zIGFuZAovLyBsaW1pdGF0aW9ucyB1bmRlciB0aGUgTGljZW5zZS4KCi8qKgogKiBAZmlsZW92ZXJ2aWV3IEhlbHBlcnMgZm9yIGdvb2dsZS5jb2xhYiBQeXRob24gbW9kdWxlLgogKi8KKGZ1bmN0aW9uKHNjb3BlKSB7CmZ1bmN0aW9uIHNwYW4odGV4dCwgc3R5bGVBdHRyaWJ1dGVzID0ge30pIHsKICBjb25zdCBlbGVtZW50ID0gZG9jdW1lbnQuY3JlYXRlRWxlbWVudCgnc3BhbicpOwogIGVsZW1lbnQudGV4dENvbnRlbnQgPSB0ZXh0OwogIGZvciAoY29uc3Qga2V5IG9mIE9iamVjdC5rZXlzKHN0eWxlQXR0cmlidXRlcykpIHsKICAgIGVsZW1lbnQuc3R5bGVba2V5XSA9IHN0eWxlQXR0cmlidXRlc1trZXldOwogIH0KICByZXR1cm4gZWxlbWVudDsKfQoKLy8gTWF4IG51bWJlciBvZiBieXRlcyB3aGljaCB3aWxsIGJlIHVwbG9hZGVkIGF0IGEgdGltZS4KY29uc3QgTUFYX1BBWUxPQURfU0laRSA9IDEwMCAqIDEwMjQ7CgpmdW5jdGlvbiBfdXBsb2FkRmlsZXMoaW5wdXRJZCwgb3V0cHV0SWQpIHsKICBjb25zdCBzdGVwcyA9IHVwbG9hZEZpbGVzU3RlcChpbnB1dElkLCBvdXRwdXRJZCk7CiAgY29uc3Qgb3V0cHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKG91dHB1dElkKTsKICAvLyBDYWNoZSBzdGVwcyBvbiB0aGUgb3V0cHV0RWxlbWVudCB0byBtYWtlIGl0IGF2YWlsYWJsZSBmb3IgdGhlIG5leHQgY2FsbAogIC8vIHRvIHVwbG9hZEZpbGVzQ29udGludWUgZnJvbSBQeXRob24uCiAgb3V0cHV0RWxlbWVudC5zdGVwcyA9IHN0ZXBzOwoKICByZXR1cm4gX3VwbG9hZEZpbGVzQ29udGludWUob3V0cHV0SWQpOwp9CgovLyBUaGlzIGlzIHJvdWdobHkgYW4gYXN5bmMgZ2VuZXJhdG9yIChub3Qgc3VwcG9ydGVkIGluIHRoZSBicm93c2VyIHlldCksCi8vIHdoZXJlIHRoZXJlIGFyZSBtdWx0aXBsZSBhc3luY2hyb25vdXMgc3RlcHMgYW5kIHRoZSBQeXRob24gc2lkZSBpcyBnb2luZwovLyB0byBwb2xsIGZvciBjb21wbGV0aW9uIG9mIGVhY2ggc3RlcC4KLy8gVGhpcyB1c2VzIGEgUHJvbWlzZSB0byBibG9jayB0aGUgcHl0aG9uIHNpZGUgb24gY29tcGxldGlvbiBvZiBlYWNoIHN0ZXAsCi8vIHRoZW4gcGFzc2VzIHRoZSByZXN1bHQgb2YgdGhlIHByZXZpb3VzIHN0ZXAgYXMgdGhlIGlucHV0IHRvIHRoZSBuZXh0IHN0ZXAuCmZ1bmN0aW9uIF91cGxvYWRGaWxlc0NvbnRpbnVlKG91dHB1dElkKSB7CiAgY29uc3Qgb3V0cHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKG91dHB1dElkKTsKICBjb25zdCBzdGVwcyA9IG91dHB1dEVsZW1lbnQuc3RlcHM7CgogIGNvbnN0IG5leHQgPSBzdGVwcy5uZXh0KG91dHB1dEVsZW1lbnQubGFzdFByb21pc2VWYWx1ZSk7CiAgcmV0dXJuIFByb21pc2UucmVzb2x2ZShuZXh0LnZhbHVlLnByb21pc2UpLnRoZW4oKHZhbHVlKSA9PiB7CiAgICAvLyBDYWNoZSB0aGUgbGFzdCBwcm9taXNlIHZhbHVlIHRvIG1ha2UgaXQgYXZhaWxhYmxlIHRvIHRoZSBuZXh0CiAgICAvLyBzdGVwIG9mIHRoZSBnZW5lcmF0b3IuCiAgICBvdXRwdXRFbGVtZW50Lmxhc3RQcm9taXNlVmFsdWUgPSB2YWx1ZTsKICAgIHJldHVybiBuZXh0LnZhbHVlLnJlc3BvbnNlOwogIH0pOwp9CgovKioKICogR2VuZXJhdG9yIGZ1bmN0aW9uIHdoaWNoIGlzIGNhbGxlZCBiZXR3ZWVuIGVhY2ggYXN5bmMgc3RlcCBvZiB0aGUgdXBsb2FkCiAqIHByb2Nlc3MuCiAqIEBwYXJhbSB7c3RyaW5nfSBpbnB1dElkIEVsZW1lbnQgSUQgb2YgdGhlIGlucHV0IGZpbGUgcGlja2VyIGVsZW1lbnQuCiAqIEBwYXJhbSB7c3RyaW5nfSBvdXRwdXRJZCBFbGVtZW50IElEIG9mIHRoZSBvdXRwdXQgZGlzcGxheS4KICogQHJldHVybiB7IUl0ZXJhYmxlPCFPYmplY3Q+fSBJdGVyYWJsZSBvZiBuZXh0IHN0ZXBzLgogKi8KZnVuY3Rpb24qIHVwbG9hZEZpbGVzU3RlcChpbnB1dElkLCBvdXRwdXRJZCkgewogIGNvbnN0IGlucHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKGlucHV0SWQpOwogIGlucHV0RWxlbWVudC5kaXNhYmxlZCA9IGZhbHNlOwoKICBjb25zdCBvdXRwdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQob3V0cHV0SWQpOwogIG91dHB1dEVsZW1lbnQuaW5uZXJIVE1MID0gJyc7CgogIGNvbnN0IHBpY2tlZFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgaW5wdXRFbGVtZW50LmFkZEV2ZW50TGlzdGVuZXIoJ2NoYW5nZScsIChlKSA9PiB7CiAgICAgIHJlc29sdmUoZS50YXJnZXQuZmlsZXMpOwogICAgfSk7CiAgfSk7CgogIGNvbnN0IGNhbmNlbCA9IGRvY3VtZW50LmNyZWF0ZUVsZW1lbnQoJ2J1dHRvbicpOwogIGlucHV0RWxlbWVudC5wYXJlbnRFbGVtZW50LmFwcGVuZENoaWxkKGNhbmNlbCk7CiAgY2FuY2VsLnRleHRDb250ZW50ID0gJ0NhbmNlbCB1cGxvYWQnOwogIGNvbnN0IGNhbmNlbFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgY2FuY2VsLm9uY2xpY2sgPSAoKSA9PiB7CiAgICAgIHJlc29sdmUobnVsbCk7CiAgICB9OwogIH0pOwoKICAvLyBXYWl0IGZvciB0aGUgdXNlciB0byBwaWNrIHRoZSBmaWxlcy4KICBjb25zdCBmaWxlcyA9IHlpZWxkIHsKICAgIHByb21pc2U6IFByb21pc2UucmFjZShbcGlja2VkUHJvbWlzZSwgY2FuY2VsUHJvbWlzZV0pLAogICAgcmVzcG9uc2U6IHsKICAgICAgYWN0aW9uOiAnc3RhcnRpbmcnLAogICAgfQogIH07CgogIGNhbmNlbC5yZW1vdmUoKTsKCiAgLy8gRGlzYWJsZSB0aGUgaW5wdXQgZWxlbWVudCBzaW5jZSBmdXJ0aGVyIHBpY2tzIGFyZSBub3QgYWxsb3dlZC4KICBpbnB1dEVsZW1lbnQuZGlzYWJsZWQgPSB0cnVlOwoKICBpZiAoIWZpbGVzKSB7CiAgICByZXR1cm4gewogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbXBsZXRlJywKICAgICAgfQogICAgfTsKICB9CgogIGZvciAoY29uc3QgZmlsZSBvZiBmaWxlcykgewogICAgY29uc3QgbGkgPSBkb2N1bWVudC5jcmVhdGVFbGVtZW50KCdsaScpOwogICAgbGkuYXBwZW5kKHNwYW4oZmlsZS5uYW1lLCB7Zm9udFdlaWdodDogJ2JvbGQnfSkpOwogICAgbGkuYXBwZW5kKHNwYW4oCiAgICAgICAgYCgke2ZpbGUudHlwZSB8fCAnbi9hJ30pIC0gJHtmaWxlLnNpemV9IGJ5dGVzLCBgICsKICAgICAgICBgbGFzdCBtb2RpZmllZDogJHsKICAgICAgICAgICAgZmlsZS5sYXN0TW9kaWZpZWREYXRlID8gZmlsZS5sYXN0TW9kaWZpZWREYXRlLnRvTG9jYWxlRGF0ZVN0cmluZygpIDoKICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgJ24vYSd9IC0gYCkpOwogICAgY29uc3QgcGVyY2VudCA9IHNwYW4oJzAlIGRvbmUnKTsKICAgIGxpLmFwcGVuZENoaWxkKHBlcmNlbnQpOwoKICAgIG91dHB1dEVsZW1lbnQuYXBwZW5kQ2hpbGQobGkpOwoKICAgIGNvbnN0IGZpbGVEYXRhUHJvbWlzZSA9IG5ldyBQcm9taXNlKChyZXNvbHZlKSA9PiB7CiAgICAgIGNvbnN0IHJlYWRlciA9IG5ldyBGaWxlUmVhZGVyKCk7CiAgICAgIHJlYWRlci5vbmxvYWQgPSAoZSkgPT4gewogICAgICAgIHJlc29sdmUoZS50YXJnZXQucmVzdWx0KTsKICAgICAgfTsKICAgICAgcmVhZGVyLnJlYWRBc0FycmF5QnVmZmVyKGZpbGUpOwogICAgfSk7CiAgICAvLyBXYWl0IGZvciB0aGUgZGF0YSB0byBiZSByZWFkeS4KICAgIGxldCBmaWxlRGF0YSA9IHlpZWxkIHsKICAgICAgcHJvbWlzZTogZmlsZURhdGFQcm9taXNlLAogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbnRpbnVlJywKICAgICAgfQogICAgfTsKCiAgICAvLyBVc2UgYSBjaHVua2VkIHNlbmRpbmcgdG8gYXZvaWQgbWVzc2FnZSBzaXplIGxpbWl0cy4gU2VlIGIvNjIxMTU2NjAuCiAgICBsZXQgcG9zaXRpb24gPSAwOwogICAgd2hpbGUgKHBvc2l0aW9uIDwgZmlsZURhdGEuYnl0ZUxlbmd0aCkgewogICAgICBjb25zdCBsZW5ndGggPSBNYXRoLm1pbihmaWxlRGF0YS5ieXRlTGVuZ3RoIC0gcG9zaXRpb24sIE1BWF9QQVlMT0FEX1NJWkUpOwogICAgICBjb25zdCBjaHVuayA9IG5ldyBVaW50OEFycmF5KGZpbGVEYXRhLCBwb3NpdGlvbiwgbGVuZ3RoKTsKICAgICAgcG9zaXRpb24gKz0gbGVuZ3RoOwoKICAgICAgY29uc3QgYmFzZTY0ID0gYnRvYShTdHJpbmcuZnJvbUNoYXJDb2RlLmFwcGx5KG51bGwsIGNodW5rKSk7CiAgICAgIHlpZWxkIHsKICAgICAgICByZXNwb25zZTogewogICAgICAgICAgYWN0aW9uOiAnYXBwZW5kJywKICAgICAgICAgIGZpbGU6IGZpbGUubmFtZSwKICAgICAgICAgIGRhdGE6IGJhc2U2NCwKICAgICAgICB9LAogICAgICB9OwogICAgICBwZXJjZW50LnRleHRDb250ZW50ID0KICAgICAgICAgIGAke01hdGgucm91bmQoKHBvc2l0aW9uIC8gZmlsZURhdGEuYnl0ZUxlbmd0aCkgKiAxMDApfSUgZG9uZWA7CiAgICB9CiAgfQoKICAvLyBBbGwgZG9uZS4KICB5aWVsZCB7CiAgICByZXNwb25zZTogewogICAgICBhY3Rpb246ICdjb21wbGV0ZScsCiAgICB9CiAgfTsKfQoKc2NvcGUuZ29vZ2xlID0gc2NvcGUuZ29vZ2xlIHx8IHt9OwpzY29wZS5nb29nbGUuY29sYWIgPSBzY29wZS5nb29nbGUuY29sYWIgfHwge307CnNjb3BlLmdvb2dsZS5jb2xhYi5fZmlsZXMgPSB7CiAgX3VwbG9hZEZpbGVzLAogIF91cGxvYWRGaWxlc0NvbnRpbnVlLAp9Owp9KShzZWxmKTsK", "ok": true, "headers": [["content-type", "application/javascript"]], "status": 200, "status_text": "OK"}}, "base_uri": "https://localhost:8080/", "height": 144} id="1peq2qBX__6z" outputId="3c0f5c17-9c59-431a-9554-4c1a809d83c0"
from google.colab import files
denseupl = files.upload()
dataupl = files.upload()
# + colab={"base_uri": "https://localhost:8080/"} id="5NU4BHJtAdIk" outputId="9ba42e14-3bbe-47d8-dbea-e2eb9aa6a2ce"
# !unzip DenseTorch.zip
# !unzip datasets.zip
# + colab={"base_uri": "https://localhost:8080/"} id="5My3zjetAf4S" outputId="8ce0e1f4-1684-4947-c53e-bd990da75386"
# !pip install -r requirements.txt
# + colab={"base_uri": "https://localhost:8080/"} id="yEoiuHryAfmi" outputId="34fecf8c-156a-4c05-b4a9-2fb5292b9e9e"
# !pip install -e .
# + colab={"base_uri": "https://localhost:8080/"} id="uPqihXCdAlEO" outputId="56fe074d-16fc-4534-c105-7d1689fd671e"
# !python setup.py build_ext --inplace
# + colab={"base_uri": "https://localhost:8080/"} id="GFMEcxywAmv0" outputId="c5b28bf7-bb6a-4c4c-fb43-37a1094b2222"
# cd datasets/
# + colab={"base_uri": "https://localhost:8080/"} id="gmaVXUqjAmj_" outputId="262c7d36-ba27-4de1-ffba-594d084e1cca"
# !unzip nyud.zip
#Now move the folders into nyudv2 created in dataset
# + colab={"base_uri": "https://localhost:8080/"} id="bpxtloUDAwYx" outputId="c087c79a-0ae8-40ac-b97a-0c71e5738997"
# cd ..
# + colab={"base_uri": "https://localhost:8080/"} id="1RwTm0TtAY2h" outputId="f2628d03-4008-4438-8304-d22140451d11"
# !python train.py
# + colab={"base_uri": "https://localhost:8080/"} id="Oun5h71fWNVD" outputId="31c5974a-a6de-4245-c68d-890b6f1595a9"
# cd tests/
# + id="jgbP_H4NWQsG"
# !python test_networks.py
| 172.471698
| 7,236
|
927622d16fee2538a0af08d153ac7c3cb4eae3e1
|
py
|
python
|
Clustering-New York Job Posting Data.ipynb
|
Data-Citadel/IDS_GROUP006
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: dsenv
# language: python
# name: dsenv
# ---
# # IDS Assignment 1
#
# This is a Jupyter Notebook created for the analysis on the given dataset for the IDS assignment #1. The assignment is done collaborately by the group of 4 members. The details are as follows,
#
# Group ID : `IDS_GROUP006`
#
# Group Members:
# 1. Venkataramanan K
# 2. Bala Kavin Pon
# 3. Ponvani
# 4. Poornima J
#
#
# ## Problem statement
#
#
# **Business Context**
#
# Using the given data set for New York City Current Job Posting data.
#
# **Business Problem Understanding**
#
# Focus on applying the learnt data analytics concepts and try to share your findings on following aspects:
# 1. What are the highest paid Skills in the US market?
# 2. What are the job categories, which involve above mentioned niche skills?
# 3. Applying clustering concepts, please depict visually what are the different salary ranges based on job category and years of experience
#
# The analysis on the data is done in the following sequence of steps.
# ## Step 0: Import libraries
# +
import pandas as pd
import numpy as np
# Graphs and Plotting related dependencies
import matplotlib.pyplot as plt
import seaborn as sns
# %matplotlib inline
# Plot stylesheet
plt.style.use('ggplot')
plt.rcParams['font.family'] = 'sans-serif'
plt.rcParams['font.sans-serif'] = 'Helvetica'
# Text preprocessing related libraries
import re
import nltk
# -
# ## Step 1: Data Preparation
#
# ### Loading a data from file
#
# +
# Reading data from the CSV file
job_data = pd.read_csv("input/jobs.csv", delimiter=",")
# Gist of data
job_data.head()
# -
# <a id="datainfo"></a>
# ### Information about the data
#
# The scheme of the data can be explored by examining the information about the dataset such as number of entries, column count, data type of the columns and the null constraints.
#
job_data.info()
print("---"*40)
job_data.describe()
# Checking whether there are any missing values in the given dataset using `isnull()` method.
# +
def plot_null_data(dframe):
# Figure size
sns.set(rc={'figure.figsize':(9,8)})
ax = sns.heatmap(dframe.isnull(), cbar=False)
ax.set_title("Dataset columns with null values")
job_data.isnull().sum()
# -
# Figure size
plot_null_data(job_data)
# ### Step 2: Identification of Variables
#
# The variables need to be identified for further processing and analysis. The variables can be identified two ways,
# 1. Variables with minimum percentage of null values. Here **30%** of total dataset volume will be allowed as null or NaN values. The variables that are more than 30% will not be considered for analysis.
# 2. The date attributes will also not contribute for analysis so they will be removed.
#
# Casting the date fields from string to datetime
job_data['Posting Date'] = pd.to_datetime(job_data['Posting Date'])
job_data['Process Date'] = pd.to_datetime(job_data['Process Date'])
job_data['Post Until'] = pd.to_datetime(job_data['Post Until'])
job_data['Posting Updated'] = pd.to_datetime(job_data['Posting Updated'])
# +
# Identifying suitable variables based on the percentage of null values and the data type
null_limit = job_data.shape[0] * 0.3 # Minimum # number of records without null or NaN values
identified_cols = list()
data_with_null = job_data.isnull().sum() # Columns with null value count
try:
for items in data_with_null.iteritems(): # Iterate through the pd.Series object
if items[1] < null_limit:
if job_data[items[0]].dtype in ('int64','object','float64'):
identified_cols.append(items[0])
except ValueError as ve:
pass
print("Identified variables:\n{0}".format(identified_cols))
# -
# ## Step 3: Imputing the missing values
#
# The variables like Job Category, Full-Time/Part-Time indicator,Minimum Qual Requirements, Preferred Skills,Residency Requirement have missing values(NaN).
#
# These have to be handled by filling meaningful values. These variables are categorical in nature, hence they can be imputed either by using Mode strategy or constant value strategy.
# +
from sklearn.impute import SimpleImputer
imputer = SimpleImputer(strategy="most_frequent")
nan_cols = ['Job Category', 'Full-Time/Part-Time indicator','Minimum Qual Requirements', 'Preferred Skills','Residency Requirement', 'To Apply']
cols = [col for col in identified_cols if col not in nan_cols]
# print(cols)
# Apply the imputation on the dataset
imputed_data = pd.DataFrame(imputer.fit_transform(job_data[nan_cols]), columns=nan_cols)
# # imputed_data.head()
posting_data = pd.concat([job_data[cols], imputed_data], axis=1)
# Empty values plot
# combined_data.isnull().sum()
plot_null_data(posting_data)
# -
# The above plot shows a clean dark background without any white lines across the column names, which implies that there are not NaN values in the dataset.
# ## Step 4: Feature Engineering
#
# There are few variables in the dataset are of type text(refer the columns <a href="#datainfo">info</a>). We need perform certain feature engineering operations to clean, transform and reduce those features.
#
# The first and foremost text preprocessing step is to remove unwanted characters such as special characters, unwanted whitespaces and punctuation. The following functions will be used to remove the special charaters from the text data.
#
pattern = r'(zero|one|two|three|four|five|six|seven|eight|nine|ten)'
doc = "I have seven years of experience"
num_dict = {'zero':0, 'one':1, 'two':2, 'three':3, 'four':4,'five':5,'six':6,'seven':7,'eight':8,'nine':9,'ten':10}
match = re.search(pattern, doc, re.I|re.A)
if match:
statement = re.sub(pattern, str(num_dict[match.group(0)]), doc, re.I|re.A)
print(statement)
# +
def extract_years_exp(doc):
phrase = ""
num_year = 1
try:
str_num_pattern = r'(zero|one|two|three|four|five|six|seven|eight|nine|ten)'
num_dict = {'zero':0, 'one':1, 'two':2, 'three':3, 'four':4,'five':5,'six':6,'seven':7,'eight':8,'nine':9,'ten':10}
# Search for text numeric value
match = re.search(str_num_pattern, doc, re.IGNORECASE)
if match:
statement = re.sub(str_num_pattern, str(num_dict[match.group(0).lower()]), doc.lower(), re.IGNORECASE)
statement = statement.strip()
exp_pattern = r'.?[(0-9)|(0-9\+)]\s+years'
exp_phrase = re.search(exp_pattern, statement, re.IGNORECASE)
if exp_phrase:
opr_pattern = r'(\+|\)|\(|\-|years)'
phrase = exp_phrase.group(0).strip()
year_val = re.sub(opr_pattern, "", phrase).strip()
if year_val:
num_year = int(year_val)
if int(year_val) > 10:
num_year = int(year_val[0])
else:
num_year
# print(num_year)
else:
phrase = np.nan
except IndexError as ie:
pass
return num_year
def extract_skills(doc):
statement = ""
num_year = 1
try:
patterns = (r'=?.(with|proficiency)\s+\w{3}+', r'\w{2}+\s+(skills|acquired)')
# Search for text numeric value
for ptrn in patterns:
match = re.search(ptrn, doc, re.IGNORECASE)
if match:
statement = match.group(0).lower().strip()
break
except IndexError as ie:
pass
return statement
'''
Cleanse the special characters, white space and punctuations.
'''
def cleanse_text_impurities(text_val):
try:
# remove the special characters
text_val = re.sub(r'[^(a-zA-Z)|(0-9)\s]', ' ', text_val, re.I|re.A)
text_val = text_val.strip()
return text_val
except ValueError as ve:
print("Error while cleasing the data: {0}".format(ve))
'''
Removes the english language stop words
'''
def remove_stopwords(text_val):
try:
word_tokenizer = nltk.WordPunctTokenizer()
stopwords = nltk.corpus.stopwords.words("english")
# tokenize the sentences
tokens = word_tokenizer.tokenize(text_val)
# Filter stop words
filterd_tokens = [tok for tok in tokens if tok not in stopwords]
return ' '.join(filtered_tokens)
except ValueError as ve:
print("Error while cleasing the data: {0}".format(ve))
# -
# #### Cleansing of text value
#
# The cleansing activity of text value usually involves with removal of whitespaces, special characters and other ASCII characters. Here, the cleansing function is applied for all the given text fields.
# +
str_cols = ['Job Description', 'Full-Time/Part-Time indicator', 'Minimum Qual Requirements','Preferred Skills','Residency Requirement']
# Iterate through the dataframe and apply the cleansing function
for col in str_cols:
posting_data[col] = posting_data[col].apply(cleanse_text_impurities)
posting_data[str_cols]
# -
# #### Extracting year of experience
#
# The year of experience is a required information to understand more on the relationship between the salary and years of experience. However, the data is hidden in the text data of `Preferred Skills` field. We need to perform a search to identify the pattern of years and transform the extracted information into a numerical values.
# posting_data['Experience Phrase'] = posting_data.head(50)['Preferred Skills'].apply(extract_years_exp)
posting_data['Years of Experience'] = posting_data['Preferred Skills'].apply(extract_years_exp)
posting_data['Years of Experience']
# +
# documents = ['Ability to code .NET and Java script from scratch 2+ years of experience with Dynamics CRM 2010/2013 Experience Developing Plugins 3 -5 years .NET and C# Coding 2+ years of SQL 2008/2012 experience with store procedure / SSIS/SSRS/ database design/maintenance',
# 'Five years of managerial and supervisory experience. 2. Excellent verbal and written communication skills. 3. Ability to work collaboratively with others. 4. Ability to perform detailed work under time-sensitive deadlines.',
# 'Minimum 5 years of experience planning, designing, configuring, installing, troubleshooting and maintaining data and voice networks. Experience in performance and capacity monitoring required. Cisco Certified Network Professional certification preferred. Familiarity with Cisco network technology (routers, switches, IOS, NX-OS, Firewall ASA, Intrusion Prevention System, Secure Access Control System and Fiber Channel Over IP) and with Avaya telephone systems strongly preferred. Experience with Voice Over IP (VoIP), disaster recovery, automated failover, resilient networks, performance and capacity monitoring, load balancing, network security, and wireless networks (Wi-Fi) are desirable.',
# 'The preferred candidate should possess the following: A Bachelor’s degree in a related IT field; 3+ years experience in a specialized role that includes implementation, support, and maintenance of large scale n-tier web applications; 3+ years hands-on experience with large scale data warehouses and analytics products; 3+ years of Relational and dimensional database experience; 3+ years PL/SQL experience; extremely proficient; 3+ years experience in business intelligence; performance tuning experience; data modeling experience; knowledge of HTML, XML and CSS and scripting; experience with MS SQL reporting; experience utilizing SAP business objects products for reports and administrating business object environment; knowledge of the implications of developing for high-availability clustered environments; experience MS SQL Reporting Services; experience with UNIX shell scripting; strong knowledge of server and application architectures; ability to work in cross functional teams to provide the best solution; strong customer and quality-focus; sound problem resolution, judgment, and decision-making skills; ability to work directly with customers to elicit and document reporting requirements; ability to develop clear and actionable reporting specifications based on these requirements; demonstrated experience working with technical and non-technical staff; outstanding collaboration and team building skills; strong written and verbal communication skills; excellent analytic, organization, presentation and facilitation skills; experience with WebLogic cluster environment; Database experience with MS SQL; experience with MS IIS Web Server and other J2EE application server such as Tomcat, JBOSS, WebSphere; and the ability to handle multiple tasks under tight deadlines.',
# 'Experience Developing Plugins 3 -5 years .NET and C# Coding 2',
# 'information systems management, computer science, or equivalent experience;6+ years of DevOps or similar production / platform engineering experience, 1-2 of which in'
# ]
# print(documents[5])
# years_phrases = [extract_years_exp(doc) for doc in documents]
# preferred_skills = [extract_skills(doc) for doc in documents]
# print("Phrase matched:{0} ".format(years_phrases))
# print("Skills:{0} ".format(preferred_skills))
# -
# ## Step 5: Analysis
#
# The analysis are done on the following questions,
#
# * a. What are the highest paid skills in the US market?
# * b. What are the highest paid Job Category in the US market?
#
# +
# highest_paid_skills = posting_data.groupby(['Job Category','Preferred Skills'])['Salary Range From','Salary Range To'].mean()
highest_paid_skills = posting_data.groupby(['Job Category','Preferred Skills'])['Salary Range From'].mean()
highest_paid_skills = highest_paid_skills.reset_index()
top_paid_list = highest_paid_skills.nlargest(10, 'Salary Range From')
# top_paid_list.sort_values(by='Salary Range From', ascending=False)
# -
# The following table shows the top 10 preferred skills and job category in the US job market.
# +
# Lambda expression to split the comma delimited category name and extract only the first token
short_job_name = lambda x: x.split(',')[0]
top_paid_list.index = top_paid_list['Job Category'].apply(short_job_name)
top_paid_list.drop(['Job Category'], axis=1)
# Print the top paid skillset list
# top_paid_list
# +
fig, barax = plt.subplots(figsize=(7,4))
top_paid_list.plot(ax=barax, kind='barh', legend = False)
barax.set_title("Top 10 Highest Paid Skills", fontsize=18)
barax.set_ylabel("Job Category", fontsize=14)
barax.set_xlabel("Salary Range From", fontsize=14)
# barax.legend(bbox_to_anchor=(1.0, 1.00))
barax.invert_yaxis()
plt.show()
# -
# `Note: The preferred skills is a text blob field, which has more words which can not be ploted in the graph. Hence the data are plotted against the job category.`
print("The highest paid skills in the US job market is :\n {0}".format(top_paid_list['Preferred Skills'][0]))
print("==="*40)
print("The Job Category is- {0}".format(top_paid_list['Job Category'][0]))
# ### 5.c Clustering Technique
jobs_by_category
jobs_by_category.index.values
fig, job_ax = plt.subplots(figsize=(8,7))
plt.scatter(jobs_by_category.index.values, jobs_by_category['Job ID'])
plt.show()
| 41.569061
| 1,813
|
2d17421b8559186511393473fa020c3cd12962e8
|
py
|
python
|
WAR!!.ipynb
|
53nicholsonsam/AutomatedWarPlayer
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + id="grfZpyDixQvg" colab_type="code" outputId="5ea16193-6c64-4ee4-9a72-4a3f779ef0a9" colab={"base_uri": "https://localhost:8080/", "height": 1000} tags=["outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend", "outputPrepend"]
### Sam Nicholson 2020
### Script to play war with itself
# %reset -f
from collections import defaultdict
import random
import statistics
import pandas as pd
import matplotlib.pyplot as plt
itteration=0
handcount=[]
for loop in range(0,100001):
Cardvalues=[]
IndexNumbers =list(range(1,53))
Dummyarray =list(range(1,14))
i=0
while i<52:
for ii in range(0,len(Dummyarray)):
Cardvalues.append(Dummyarray[ii])
#print("still working")
i+=1
#print(i)
##Merge the lists into 1 list to creat the dictionary with
listtodict=[]
for ii in range(0,len(Cardvalues)):
temparray=[]
temparray.append(IndexNumbers[ii])
temparray.append(Cardvalues[ii])
listtodict.append(temparray)
#print(IndexNumbers)
#print(Cardvalues)
deck=dict(listtodict)
#print(listtodict)
##Dealing
Indextodealfrom=list(range(1,53))
random.shuffle(Indextodealfrom)
middlindex=int(len(Indextodealfrom)/2)
Player1index=Indextodealfrom[middlindex:]
Player2index=Indextodealfrom[:middlindex]
#print(Player1index)
#print(Player2index)
#print(Indextodealfrom)
#print(deck)
iii=0
Player1deck=[]
Player2deck=[]
while iii<26:
Player1deck.append(deck[Player1index[iii]])
Player2deck.append(deck[Player2index[iii]])
iii+=1
#print("Player 1 Deck:" + str(Player1deck))
#print("Player 2 Deck:" + str(Player2deck))
##Playing war
count=0
while len(Player1deck)>0 and len(Player2deck)>0: #While someone still has cards
#print("Player 1's card: " + str(Player1deck[0]) + " v "+ str(Player2deck[0])+ ": Player 2's card")
if Player1deck[0]>Player2deck[0]: #Player 1 take the card
Player1deck.append(Player1deck[0]) #Move the card card played to the bottem
Player1deck.append(Player2deck[0]) #Move the taken card to the bottem
Player1deck.pop(0) #Move the card played to the bottem
Player2deck.pop(0) #Move the card played to the other player
elif Player1deck[0]<Player2deck[0]: #Player 2 takes the card
Player2deck.append(Player2deck[0])
Player2deck.append(Player1deck[0])
Player1deck.pop(0)
Player2deck.pop(0)
elif Player1deck[0]==Player2deck[0]: #There is a war
Player1War=[]
Player2War=[]
test=0
while test==0:
if (len(Player1deck)<5 or len(Player2deck)<5): #Where there is a war and one player has less than 4 cards left
for vi in range(0,max(1,min(len(Player1deck)-1,3))): #Saves 1 card to use putting in at least 1
#print(vi)
#print ("Player1deck len:"+str(len(Player1deck)))
if len(Player1deck)!=1:
Player1War.append(Player1deck[vi])
for viii in range(0,max(1,min(len(Player1deck)-1,3))):
#print(viii)
#print ("Player1deck len:"+str(len(Player1deck)))
if len(Player1deck)!=1:
Player1deck.pop(0)
for vii in range(0,max(1,min(len(Player2deck)-1,3))):
#print(vii)
#print ("Player2deck len:"+str(len(Player2deck)))
if len(Player2deck)!=1:
Player2War.append(Player2deck[vii])
for ix in range(0,max(1,min(len(Player2deck)-1,3))):
#print(ix)
#print ("Player2deck len:"+str(len(Player2deck)))
if len(Player2deck)!=1:
Player2deck.pop(0)
#print("There's a war!")
#print("Player 1's card: " + str(Player1deck[0]) + " v "+ str(Player2deck[0])+ ": Player 2's card")
if Player1deck[0]>Player2deck[0]: #Player 1 wins and gets the cards
Player1deck.append(Player1deck[0])
Player1deck.append(Player2deck[0])
Player1deck.pop(0)
Player2deck.pop(0)
for iv in range(0,len(Player1War)):
Player1deck.append(Player1War[iv])
for v in range(0,len(Player2War)):
Player1deck.append(Player2War[v])
test=1
elif Player1deck[0]<Player2deck[0]: #Player 2 wins and gets the cards
Player2deck.append(Player2deck[0])
Player2deck.append(Player1deck[0])
Player1deck.pop(0)
Player2deck.pop(0)
for iv in range(0,len(Player1War)):
Player2deck.append(Player1War[iv])
for v in range(0,len(Player2War)):
Player2deck.append(Player2War[v])
test=1
else:
Player1War.append(Player1deck[0]) #Each are putting their turned up card and 2 more cards into the war pile
Player1War.append(Player1deck[1])
Player1War.append(Player1deck[2])
Player1deck.pop(0)
Player1deck.pop(1)
Player1deck.pop(2)
Player2War.append(Player2deck[0])
Player2War.append(Player2deck[1])
Player2War.append(Player2deck[2])
Player2deck.pop(0)
Player2deck.pop(1)
Player2deck.pop(2)
#Checking the next card to see who wins the war
#print("There's a war!")
#print("Player 1's card: " + str(Player1deck[0]) + " v "+ str(Player2deck[0])+ ": Player 2's card")
if Player1deck[0]>Player2deck[0]: #Player 1 wins and gets the cards
Player1deck.append(Player1deck[0])
Player1deck.append(Player2deck[0])
Player1deck.pop(0)
Player2deck.pop(0)
for iv in range(0,len(Player1War)):
Player1deck.append(Player1War[iv])
for v in range(0,len(Player2War)):
Player1deck.append(Player2War[v])
test=1
elif Player1deck[0]<Player2deck[0]: #Player 2 wins and gets the cards
Player2deck.append(Player2deck[0])
Player2deck.append(Player1deck[0])
Player1deck.pop(0)
Player2deck.pop(0)
for iv in range(0,len(Player1War)):
Player2deck.append(Player1War[iv])
for v in range(0,len(Player2War)):
Player2deck.append(Player2War[v])
test=1
#They keep playing
count+=1
itteration+=1
print("Trial: "+ str(itteration))
### For printing results of 1 game###
#if len(Player1deck)>0:
# print("Player 1 Wins!")
#else:
# print("Player 2 Wins!")
#print("It took " +str(count) + " hands to finish the game")
### For gathering data ###
handcount.append(count)
#print(handcount)
plt.hist(handcount, bins= 75)
plt.title("Length of game in hands for 100000 War Games")
plt.xlabel("Counts")
plt.ylabel("Number of Hands")
plt.grid(axis="y",alpha=0.75)
print("Mean of Sample Game Length is % s" % (round(statistics.mean(handcount),3)))
print("Standard Deviation of Sample Game Length is % s" % (round(statistics.stdev(handcount),3)))
print("Max number of hands in Sample Game Length is % s" % (max(handcount)))
print("Minimum number of hands in Sample Game Length is % s" % (min(handcount)))
# -
| 54.321053
| 3,385
|
7b1bb3b8c854bdfeddf4d124c9c20b9405c8e008
|
py
|
python
|
JavaScripts/FeatureCollection/Buffer.ipynb
|
c11/earthengine-py-notebooks
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <table class="ee-notebook-buttons" align="left">
# <td><a target="_blank" href="https://github.com/giswqs/earthengine-py-notebooks/tree/master/JavaScripts/FeatureCollection/Buffer.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" /> View source on GitHub</a></td>
# <td><a target="_blank" href="https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/JavaScripts/FeatureCollection/Buffer.ipynb"><img width=26px src="https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png" />Notebook Viewer</a></td>
# <td><a target="_blank" href="https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/JavaScripts/FeatureCollection/Buffer.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" /> Run in Google Colab</a></td>
# </table>
# ## Install Earth Engine API and geemap
# Install the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geemap](https://github.com/giswqs/geemap). The **geemap** Python package is built upon the [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) and [folium](https://github.com/python-visualization/folium) packages and implements several methods for interacting with Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, and `Map.centerObject()`.
# The following script checks if the geemap package has been installed. If not, it will install geemap, which automatically installs its [dependencies](https://github.com/giswqs/geemap#dependencies), including earthengine-api, folium, and ipyleaflet.
#
# **Important note**: A key difference between folium and ipyleaflet is that ipyleaflet is built upon ipywidgets and allows bidirectional communication between the front-end and the backend enabling the use of the map to capture user input, while folium is meant for displaying static data only ([source](https://blog.jupyter.org/interactive-gis-in-jupyter-with-ipyleaflet-52f9657fa7a)). Note that [Google Colab](https://colab.research.google.com/) currently does not support ipyleaflet ([source](https://github.com/googlecolab/colabtools/issues/60#issuecomment-596225619)). Therefore, if you are using geemap with Google Colab, you should use [`import geemap.eefolium`](https://github.com/giswqs/geemap/blob/master/geemap/eefolium.py). If you are using geemap with [binder](https://mybinder.org/) or a local Jupyter notebook server, you can use [`import geemap`](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py), which provides more functionalities for capturing user input (e.g., mouse-clicking and moving).
# +
# Installs geemap package
import subprocess
try:
import geemap
except ImportError:
print('geemap package not installed. Installing ...')
subprocess.check_call(["python", '-m', 'pip', 'install', 'geemap'])
# Checks whether this notebook is running on Google Colab
try:
import google.colab
import geemap.eefolium as emap
except:
import geemap as emap
# Authenticates and initializes Earth Engine
import ee
try:
ee.Initialize()
except Exception as e:
ee.Authenticate()
ee.Initialize()
# -
# ## Create an interactive map
# The default basemap is `Google Satellite`. [Additional basemaps](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py#L13) can be added using the `Map.add_basemap()` function.
Map = emap.Map(center=[40,-100], zoom=4)
Map.add_basemap('ROADMAP') # Add Google Map
Map
# ## Add Earth Engine Python script
# +
# Add Earth Engine dataset
# Feature buffer example.
# Display the area within 2 kilometers of San Francisco BART stations.
# Instantiate a FeatureCollection of BART locations in Downtown San Francisco
# (points).
stations = [
ee.Feature(
ee.Geometry.Point(-122.42, 37.77), {'name': '16th St. Mission (16TH)'}),
ee.Feature(
ee.Geometry.Point(-122.42, 37.75), {'name': '24th St. Mission (24TH)'}),
ee.Feature(
ee.Geometry.Point(-122.41, 37.78),
{'name': 'Civic Center/UN Plaza (CIVC)'})
]
bartStations = ee.FeatureCollection(stations)
# Map a function over the collection to buffer each feature.
def func_dky(f):
return f.buffer(2000, 100); # Note that the errorMargin is set to 100.
buffered = bartStations.map(func_dky)
Map.addLayer(buffered, {'color': '800080'})
Map.setCenter(-122.4, 37.7, 11)
# -
# ## Display Earth Engine data layers
Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.
Map
| 46.97
| 1,023
|
8f0cd1f43d7c879b3427b2d19e6ed6745a4bf7af
|
py
|
python
|
JavaScripts/Image/HoughTransform.ipynb
|
mllzl/earthengine-py-notebooks
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <table class="ee-notebook-buttons" align="left">
# <td><a target="_blank" href="https://github.com/giswqs/earthengine-py-notebooks/tree/master/JavaScripts/Image/HoughTransform.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" /> View source on GitHub</a></td>
# <td><a target="_blank" href="https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/JavaScripts/Image/HoughTransform.ipynb"><img width=26px src="https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png" />Notebook Viewer</a></td>
# <td><a target="_blank" href="https://mybinder.org/v2/gh/giswqs/earthengine-py-notebooks/master?filepath=JavaScripts/Image/HoughTransform.ipynb"><img width=58px src="https://mybinder.org/static/images/logo_social.png" />Run in binder</a></td>
# <td><a target="_blank" href="https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/JavaScripts/Image/HoughTransform.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" /> Run in Google Colab</a></td>
# </table>
# ## Install Earth Engine API and geemap
# Install the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geemap](https://github.com/giswqs/geemap). The **geemap** Python package is built upon the [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) and [folium](https://github.com/python-visualization/folium) packages and implements several methods for interacting with Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, and `Map.centerObject()`.
# The following script checks if the geemap package has been installed. If not, it will install geemap, which automatically installs its [dependencies](https://github.com/giswqs/geemap#dependencies), including earthengine-api, folium, and ipyleaflet.
#
# **Important note**: A key difference between folium and ipyleaflet is that ipyleaflet is built upon ipywidgets and allows bidirectional communication between the front-end and the backend enabling the use of the map to capture user input, while folium is meant for displaying static data only ([source](https://blog.jupyter.org/interactive-gis-in-jupyter-with-ipyleaflet-52f9657fa7a)). Note that [Google Colab](https://colab.research.google.com/) currently does not support ipyleaflet ([source](https://github.com/googlecolab/colabtools/issues/60#issuecomment-596225619)). Therefore, if you are using geemap with Google Colab, you should use [`import geemap.eefolium`](https://github.com/giswqs/geemap/blob/master/geemap/eefolium.py). If you are using geemap with [binder](https://mybinder.org/) or a local Jupyter notebook server, you can use [`import geemap`](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py), which provides more functionalities for capturing user input (e.g., mouse-clicking and moving).
# +
# Installs geemap package
import subprocess
try:
import geemap
except ImportError:
print('geemap package not installed. Installing ...')
subprocess.check_call(["python", '-m', 'pip', 'install', 'geemap'])
# Checks whether this notebook is running on Google Colab
try:
import google.colab
import geemap.eefolium as emap
except:
import geemap as emap
# Authenticates and initializes Earth Engine
import ee
try:
ee.Initialize()
except Exception as e:
ee.Authenticate()
ee.Initialize()
# -
# ## Create an interactive map
# The default basemap is `Google Satellite`. [Additional basemaps](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py#L13) can be added using the `Map.add_basemap()` function.
Map = emap.Map(center=[40,-100], zoom=4)
Map.add_basemap('ROADMAP') # Add Google Map
Map
# ## Add Earth Engine Python script
# +
# Add Earth Engine dataset
# An example finding linear features using the HoughTransform.
# Load an image and compute NDVI.
image = ee.Image('LANDSAT/LC08/C01/T1_TOA/LC08_033032_20170719')
ndvi = image.normalizedDifference(['B5', 'B4'])
# Apply a Canny edge detector.
canny = ee.Algorithms.CannyEdgeDetector({
'image': ndvi,
'threshold': 0.4
}).multiply(255)
# Apply the Hough transform.
h = ee.Algorithms.HoughTransform({
'image': canny,
'gridSize': 256,
'inputThreshold': 50,
'lineThreshold': 100
})
# Display.
Map.setCenter(-103.80140, 40.21729, 13)
Map.addLayer(image, {'bands': ['B4', 'B3', 'B2'], 'max': 0.3}, 'source_image')
Map.addLayer(canny.updateMask(canny), {'min': 0, 'max': 1, 'palette': 'blue'}, 'canny')
Map.addLayer(h.updateMask(h), {'min': 0, 'max': 1, 'palette': 'red'}, 'hough')
# -
# ## Display Earth Engine data layers
Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.
Map
| 50.680412
| 1,023
|
2232c962033502bc50eba73aa0e02e1e5df7bf0f
|
py
|
python
|
py-pkgs/02-setup.ipynb
|
UBC-MDS/py-pkgs
|
['CC-BY-4.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# (02:System-setup)=
# # System setup
# <hr style="height:1px;border:none;color:#666;background-color:#666;" />
# If you intend to follow along with the code presented in this book, we recommend you follow these setup instructions so that you will run into fewer technical issues.
# ## The command-line interface
# A command-line interface\index{command-line interface} (CLI) is a text-based interface used to interact with your computer. We'll be using a CLI for various tasks throughout this book. We'll assume Mac and Linux users are using the "Terminal"\index{Terminal} and Windows users are using the "Anaconda Prompt"\index{Anaconda Prompt} (which we'll install in the next section) as a CLI.
# ## Installing software
# (02:Installing-Python)=
# ### Installing Python
# We recommend installing the latest version of Python via the Miniconda\index{Miniconda} distribution by following the instructions in the Miniconda [documentation](https://docs.conda.io/en/latest/miniconda.html). Miniconda is a lightweight version of the popular Anaconda\index{Anaconda} distribution. If you have previously installed the Anaconda or Miniconda distribution feel free to skip to **{numref}`02:Install-packaging-software`**.
#
# If you are unfamiliar with Miniconda and Anaconda, they are distributions of Python that also include the `conda`\index{conda} package and environment manager, and a number of other useful packages. The difference between Anaconda and Miniconda is that Anaconda automatically installs over 250 additional packages (many of which you might never use), while Miniconda is a much smaller distribution that comes bundled with just a few key packages; you can then install additional packages as you need them using the command `conda install`.
#
# `conda` is a piece of software that supports the process of installing and updating software (like Python packages). It is also an environment manager which is the key function we'll be using it for in this book. An environment manager helps you create "virtual environments\index{virtual environment}" on your machine where you can safely install different packages and their dependencies in an isolated location. Installing all the packages you need in the same place (i.e., the system default location) can be problematic because different packages often depend on different versions of the same dependencies; as you install more packages, you'll inevitably get conflicts between dependencies and your code will start to break. Virtual environments help you compartmentalize and isolate the packages you are using for different projects to avoid this issue. You can read more about virtual environments in the `conda` [documentation](https://conda.io/projects/conda/en/latest/user-guide/concepts/environments.html). While alternative package and environment managers exist, we choose to use `conda` in this book because of its popularity, ease-of-use, and ability to handle any software stack (not just Python).
# (02:Install-packaging-software)=
# ### Install packaging software
# Once you've installed the Miniconda\index{Miniconda} distribution, ensure that Python and `conda`\index{conda} are up to date by running the following command at the command-line:
#
# ```{prompt} bash \$ auto
# $ conda update --all
# ```
#
# Then, use `conda` to install the two main pieces of software we'll be using to help us create Python packages in this book:
#
# 1. [`poetry`\index{poetry}](https://python-poetry.org/): software that will help us build our own Python packages; and,
# 2. [`cookiecutter`\index{cookiecutter}](https://github.com/cookiecutter/cookiecutter): software that will help us create packages from pre-made templates:
#
# ```{prompt} bash \$ auto
# $ conda install -c conda-forge poetry cookiecutter
# ```
#
# ```{note}
# You may also choose to install `poetry` and/or `cookiecutter` using alternative tools that you're familiar with, such as `pip`, but be sure to properly manage your virtual environments.
# ```
# (02:Register-for-a-PyPI-account)=
# ## Register for a PyPI account
# The Python Package Index (PyPI)\index{PyPI} is the official online software repository for Python. A software repository\index{software repository} is a storage location for downloadable software, like Python packages. In this book we'll be publishing a package to PyPI. Before publishing packages to PyPI, it is typical to "test drive" their publication on TestPyPI\index{TestPyPI} which is a test version of PyPI. To follow along with this book, you should register for a TestPyPI account on the [TestPyPI website](https://test.pypi.org/account/register/) and a PyPI account on the [PyPI website](https://pypi.org/account/register/).
# (02:Set-up-Git-and-GitHub)=
# ## Set up Git and GitHub
# If you're not using a version control\index{version control} system, we highly recommend you get into the habit! A version control system tracks changes to the file(s) of your project in a clear and organized way (no more "document_1.doc", "document_1_new.doc", "document_final.doc", etc.). As a result, a version control system contains a full history of all the revisions made to your project which you can view and retrieve at any time. You don't *need* to use or be familiar with version control to read this book, but if you're serious about creating Python packages, version control will become an invaluable part of your workflow, so now is a good time to learn!
#
# There are many version control systems available, but the most common is Git\index{Git} and we'll be using it throughout this book. You can download Git by following the instructions in the [Git documentation](https://git-scm.com/book/en/v2/Getting-Started-Installing-Git). Git helps track changes to a project on a local computer, but what if we want to collaborate with others? Or, what happens if your computer crashes and you lose all your work? That's where GitHub\index{GitHub} comes in. GitHub is one of many online services for hosting Git-managed projects. GitHub helps you create an online copy of your local Git repository which acts as a back-up of your local work and allows others to easily and transparently collaborate on your project. You can sign up for a free GitHub account on the [GitHub website](https://www.github.com).
#
# ```{attention}
# This book assume that those who choose to follow the optional version control sections of this book have basic familiarity with Git and GitHub (or equivalent). Two excellent learning resources are [Happy Git and GitHub for the useR](https://happygitwithr.com){cite:p}`bryan2021` and [Research Software Engineering with Python: Using Git at the Command Line](https://merely-useful.tech/py-rse/git-cmdline.html){cite:p}`rsep2021`.
# ```
# ## Python integrated development environments
# A Python integrated development environment\index{integrated development environment} (IDE) will make the process of creating Python packages significantly easier. An IDE is a piece of software that provides advanced functionality for code development such as directory and file creation and navigation, autocomplete, debugging, and syntax highlighting, to name a few. An IDE will save you time and help you write better code. Commonly used free Python IDEs include [Visual Studio Code\index{Visual Studio Code}](https://code.visualstudio.com/), [Atom](https://atom.io/), [Sublime Text](https://www.sublimetext.com/), [Spyder](https://www.spyder-ide.org/), and [PyCharm Community Edition](https://www.jetbrains.com/pycharm/). For those more familiar with the Jupyter\index{Jupyter} ecosystem, [JupyterLab](https://jupyter.org/) is a suitable browser-based IDE. Finally, for the R\index{R} community, the [RStudio\index{RStudio} IDE](https://rstudio.com/products/rstudio/download/) also supports Python.
#
# You'll be able to follow along with the examples presented in this book regardless of what IDE you choose to develop your Python code in. If you don't know which IDE to use, we recommend starting with Visual Studio Code. Below we briefly describe how to set up Visual Studio Code, JupyterLab, and RStudio as Python IDEs (these are the IDEs we personally use in our day-to-day work).
# ### Visual Studio Code
# You can download Visual Studio Code\index{Visual Studio Code} (VS Code) from the Visual Studio Code [website](https://code.visualstudio.com/). Once you've installed VS Code, you should install the "Python" extension from the VS Code Marketplace. To do this, follow the steps listed below and illustrated in {numref}`02-vscode-1-fig`:
#
# 1. Open the Marketplace by clicking the *Extensions* tab on the VS Code activity bar;
# 2. Search for "Python" in the search bar;
# 3. Select the extension named "Python" and then click *Install*.
#
# ```{figure} images/02-vscode-1.png
# ---
# width: 100%
# name: 02-vscode-1-fig
# alt: Installing the Python extension in Visual Studio Code.
# ---
# Installing the Python extension in Visual Studio Code.
# ```
#
# Once this is done, you have everything you need to start creating packages! For example, you can create files and directories from the *File Explorer* tab on the VS Code activity bar and you can open up an integrated CLI by selecting *Terminal* from the *View* menu. {numref}`02-vscode-2-fig` shows an example of executing a Python *.py* file from the command line in VS Code.
#
# ```{figure} images/02-vscode-2.png
# ---
# width: 100%
# name: 02-vscode-2-fig
# alt: Executing a simple Python file called hello-world.py from the integrated terminal in Visual Studio Code.
# ---
# Executing a simple Python file called *hello-world.py* from the integrated terminal in Visual Studio Code.
# ```
#
# We recommend you take a look at the VS Code [Getting Started Guide](https://code.visualstudio.com/docs) to learn more about using VS Code. While you don't need to install any additional extensions to start creating packages in VS Code, there are many extensions available that can support and streamline your programming workflows in VS Code. Below are a few we recommend installing to support the workflows we use in this book (you can search for and install these from the "Marketplace" as we did earlier):
#
# - [Python Docstring Generator](https://marketplace.visualstudio.com/items?itemName=njpwerner.autodocstring): an extension to quickly generate documentation strings (docstrings\index{docstring}) for Python functions.
# - [Markdown All in One](https://marketplace.visualstudio.com/items?itemName=yzhang.markdown-all-in-one): an extension that provides keyboard shortcuts, automatic table of contents, and preview functionality for Markdown\index{Markdown} files. [Markdown](https://www.markdownguide.org) is a plain-text markup language that we'll use and learn about in this book.
# - [markdownlint](https://marketplace.visualstudio.com/items?itemName=DavidAnson.vscode-markdownlint): an extension that enables automatic style checking of Markdown files.
# ### JupyterLab
# For those comfortable in the Jupyter\index{Jupyter} ecosystem feel free to stay there to create your Python packages! JupyterLab is a browser-based IDE that supports all of the core functionality we need to create packages. As per the JupyterLab [installation instructions](https://jupyterlab.readthedocs.io/en/stable/getting_started/installation.html), you can install JupyterLab with:
#
# ```{prompt} bash \$ auto
# $ conda install -c conda-forge jupyterlab
# ```
#
# Once installed, you can launch JupyterLab from your current directory by typing the following command in your terminal:
#
# ```{prompt} bash \$ auto
# $ jupyter lab
# ```
#
# In JupyterLab, you can create files and directories from the *File Browser* and can open up an integrated terminal from the *File* menu. {numref}`02-jupyterlab-fig` shows an example of executing a Python *.py* file from the command line in JupyterLab.
#
# ```{figure} images/02-jupyterlab.png
# ---
# width: 100%
# name: 02-jupyterlab-fig
# alt: Executing a simple Python file called hello-world.py from a terminal in JupyterLab.
# ---
# Executing a simple Python file called *hello-world.py* from a terminal in JupyterLab.
# ```
#
# We recommend you take a look at the JupyterLab [documentation](https://jupyterlab.readthedocs.io/en/stable/index.html) to learn more about how to use Jupyterlab. In particular, we'll note that, like VS Code, JupyterLab supports an ecosystem of extensions that can add additional functionality to the IDE. We won't install any here, but you can browse them in the JupyterLab *Extension Manager* if you're interested.
# ### RStudio
# Users with an R\index{R} background may prefer to stay in the RStudio\index{RStudio} IDE. We recommend installing the most recent version of the IDE from the RStudio [website](https://rstudio.com/products/rstudio/download/preview/) (we recommend installing at least version ^1.4) and then installing the most recent version of R from [CRAN](https://cran.r-project.org/). To use Python in RStudio, you will need to install the [reticulate\index{reticulate}](https://rstudio.github.io/reticulate/) R package by typing the following in the R console inside RStudio:
#
# ```r
# install.packages("reticulate")
# ```
#
# When installing reticulate, you may be prompted to install the Anaconda distribution. We already installed the Miniconda distribution of Python in **{numref}`02:Installing-Python`**, so answer "no" to this prompt. Before being able to use Python in RStudio, you will need to configure `reticulate`. We will briefly describe how to do this for different operating systems below, but encourage you to look at the `reticulate` [documentation](https://rstudio.github.io/reticulate/) for more help.
# **Mac and Linux**
# 1. Find the path to the Python interpreter installed with Miniconda by typing `which python` at the command line;
# 2. Open (or create) an `.Rprofile` file in your HOME directory and add the line `Sys.setenv(RETICULATE_PYTHON = "path_to_python")`, where `"path_to_python"` is the path identified in step 1;
# 3. Open (or create) a `.bash_profile` file in your HOME directory and add the line `export PATH="/opt/miniconda3/bin:$PATH"`, replacing `/opt/miniconda3/bin` with the path you identified in step 1 but without the `python` at the end.
# 4. Restart R.
# 5. Try using Python in RStudio by running the following in the R console:
#
# ```r
# library(reticulate)
# repl_python()
# ```
# **Windows**
# 1. Find the path to the Python interpreter installed with Miniconda by opening an Anaconda Prompt from the Start Menu and typing `where python` in a terminal;
# 2. Open (or create) an `.Rprofile` file in your HOME directory and add the line `Sys.setenv(RETICULATE_PYTHON = "path_to_python")`, where `"path_to_python"` is the path identified in step 1. Note that in Windows, you need `\\` instead of `\` to separate the directories; for example your path might look like: `C:\\Users\\miniconda3\\python.exe`;
# 3. Open (or create) a `.bash_profile` file in your HOME directory and add the line `export PATH="/opt/miniconda3/bin:$PATH"`, replacing `/opt/miniconda3/bin` with the path you identified in step 1 but without the `python` at the end.
# 4. Restart R.
# 5. Try using Python in RStudio by running the following in the R console:
#
# ```r
# library(reticulate)
# repl_python()
# ```
#
# {numref}`02-rstudio-fig` shows an example of executing Python code interactively in RStudio.
#
# ```{figure} images/02-rstudio.png
# ---
# width: 100%
# name: 02-rstudio-fig
# alt: Executing Python code in the RStudio.
# ---
# Executing Python code in RStudio.
# ```
| 84.112903
| 1,216
|
585640c58a1bba8a399e65b17da1dbce441e1518
|
py
|
python
|
quests/dei/census/income_xgboost.ipynb
|
lkuligin/training-data-analyst
|
['Apache-2.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# Only execute if you haven't already. Make sure to restart the kernel if these libraries have not been previously installed.
# !pip install xgboost==0.82 --user
# !pip install scikit-learn==0.20.4 --user
# # Import Python packages
#
# Execute the command below (__Shift + Enter__) to load all the python libraries we'll need for the lab.
# +
import datetime
import pickle
import os
import pandas as pd
import xgboost as xgb
import numpy as np
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import FeatureUnion, make_pipeline
from sklearn.utils import shuffle
from sklearn.base import clone
from sklearn.model_selection import train_test_split
from witwidget.notebook.visualization import WitWidget, WitConfigBuilder
import custom_transforms
import warnings
warnings.filterwarnings(action='ignore', category=DeprecationWarning)
# -
# Before we continue, note that we'll be using your Qwiklabs project id a lot in this notebook. For convenience, set it as an environment variable using the command below:
os.environ['QWIKLABS_PROJECT_ID'] = ''
# # Download and process data
#
# The models you'll build will predict the income level, whether it's less than or equal to $50,000 per year, of individuals given 14 data points about each individual. You'll train your models on this UCI [Census Income Dataset](https://archive.ics.uci.edu/ml/datasets/Adult).
#
# We'll read the data into a Pandas DataFrame to see what we'll be working with. It's important to shuffle our data in case the original dataset is ordered in a specific way. We use an sklearn utility called shuffle to do this, which we imported in the first cell:
# +
train_csv_path = 'https://archive.ics.uci.edu/ml/machine-learning-databases/adult/adult.data'
COLUMNS = (
'age',
'workclass',
'fnlwgt',
'education',
'education-num',
'marital-status',
'occ|upation',
'relationship',
'race',
'sex',
'capital-gain',
'capital-loss',
'hours-per-week',
'native-country',
'income-level'
)
raw_train_data = pd.read_csv(train_csv_path, names=COLUMNS, skipinitialspace=True)
raw_train_data = shuffle(raw_train_data, random_state=4)
# -
# `data.head()` lets us preview the first five rows of our dataset in Pandas.
raw_train_data.head()
# The `income-level` column is the thing our model will predict. This is the binary outcome of whether the individual makes more than $50,000 per year. To see the distribution of income levels in the dataset, run the following:
print(raw_train_data['income-level'].value_counts(normalize=True))
# As explained in [this paper](http://cseweb.ucsd.edu/classes/sp15/cse190-c/reports/sp15/048.pdf), each entry in the dataset contains the following information
# about an individual:
#
# * __age__: the age of an individual
# * __workclass__: a general term to represent the employment status of an individual
# * __fnlwgt__: final weight. In other words, this is the number of people the census believes
# the entry represents...
# * __education__: the highest level of education achieved by an individual.
# * __education-num__: the highest level of education achieved in numerical form.
# * __marital-status__: marital status of an individual.
# * __occupation__: the general type of occupation of an individual
# * __relationship__: represents what this individual is relative to others. For example an
# individual could be a Husband. Each entry only has one relationship attribute and is
# somewhat redundant with marital status.
# * __race__: Descriptions of an individual’s race
# * __sex__: the biological sex of the individual
# * __capital-gain__: capital gains for an individual
# * __capital-loss__: capital loss for an individual
# * __hours-per-week__: the hours an individual has reported to work per week
# * __native-country__: country of origin for an individual
# * __income-level__: whether or not an individual makes more than $50,000 annually
# An important concept in machine learning is train / test split. We'll take the majority of our data and use it to train our model, and we'll set aside the rest for testing our model on data it's never seen before. There are many ways to create training and test datasets. Fortunately, for our census data we can simply download a pre-defined test set.
test_csv_path = 'https://archive.ics.uci.edu/ml/machine-learning-databases/adult/adult.test'
raw_test_data = pd.read_csv(test_csv_path, names=COLUMNS, skipinitialspace=True, skiprows=1)
raw_test_data.head()
# Since we don't want to train a model on our labels, we're going to separate them from the features in both the training and test datasets. Also, notice that `income-level` is a string datatype. For machine learning, it's better to convert this to an binary integer datatype. We do this in the next cell.
# +
raw_train_features = raw_train_data.drop('income-level', axis=1).values
raw_test_features = raw_test_data.drop('income-level', axis=1).values
# Create training labels list
train_labels = (raw_train_data['income-level'] == '>50K').values.astype(int)
test_labels = (raw_test_data['income-level'] == '>50K.').values.astype(int)
# -
# Now you're ready to build and train your first model!
# # Build a First Model
#
# The model we build closely follows a template for the [census dataset found on AI Hub](https://aihub.cloud.google.com/p/products%2F526771c4-9b36-4022-b9c9-63629e9e3289). For our model we use an XGBoost classifier. However, before we train our model we have to pre-process the data a little bit. We build a processing pipeline using [Scikit-Learn's Pipeline constructor](https://scikit-learn.org/stable/modules/generated/sklearn.pipeline.Pipeline.html). We appl some custom transformations that are defined in `custom_transforms.py`. Open the file `custom_transforms.py` and inspect the code. Out features are either numerical or categorical. The numerical features are `age-num`, and `hours-per-week`. These features will be processed by applying [Scikit-Learn's StandardScaler function](https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.StandardScaler.html). The categorical features are `workclass`, `education`, `marital-status`, and `relationship`. These features are [one-hot encoded](https://machinelearningmastery.com/why-one-hot-encode-data-in-machine-learning/).
# +
numerical_indices = [0, 12]
categorical_indices = [1, 3, 5, 7]
p1 = make_pipeline(
custom_transforms.PositionalSelector(categorical_indices),
custom_transforms.StripString(),
custom_transforms.SimpleOneHotEncoder()
)
p2 = make_pipeline(
custom_transforms.PositionalSelector(numerical_indices),
StandardScaler()
)
p3 = FeatureUnion([
('numericals', p1),
('categoricals', p2),
])
# -
# To finalize the pipeline we attach an XGBoost classifier at the end. The complete pipeline object takes the raw data we loaded from csv files, processes the categorical features, processes the numerical features, concatenates the two, and then passes the result through the XGBoost classifier.
pipeline = make_pipeline(
p3,
xgb.sklearn.XGBClassifier(max_depth=4)
)
# We train our model with one function call using the fit() method. We pass the fit() method our training data.
pipeline.fit(raw_train_features, train_labels)
# Let's go ahead and save our model as a pickle file. Executing the command below will save the trained model in the file `model.pkl` in the same directory as this notebook.
with open('model.pkl', 'wb') as model_file:
pickle.dump(pipeline, model_file)
# # Save Trained Model to AI Platform
#
# We've got our model working locally, but it would be nice if we could make predictions on it from anywhere (not just this notebook!). In this step we'll deploy it to the cloud. For detailed instructions on how to do this visit [the official documenation](https://cloud.google.com/ai-platform/prediction/docs/exporting-for-prediction). Note that since we have custom components in our data pipeline we need to go through a few extra steps.
# ## Create a Cloud Storage bucket for the model
#
# We first need to create a storage bucket to store our pickled model file. We'll point Cloud AI Platform at this file when we deploy. Run this gsutil command to create a bucket. This will ensure the name of the cloud storage bucket you create will be globally unique.
# !gsutil mb gs://$QWIKLABS_PROJECT_ID
# ## Package custom transform code
#
# Since we're using custom transformation code we need to package it up and direct AI Platform to it when we ask it make predictions. To package our custom code we create a source distribution. The following code creates this distribution and then ports the distribution and the model file to the bucket we created. Ignore the warnings about missing meta data.
# + language="bash"
#
# python setup.py sdist --formats=gztar
#
# gsutil cp model.pkl gs://$QWIKLABS_PROJECT_ID/original/
# gsutil cp dist/custom_transforms-0.1.tar.gz gs://$QWIKLABS_PROJECT_ID/
# -
# ## Create and Deploy Model
#
# The following ai-platform gcloud command will create a new model in your project. We'll call this one `census_income_classifier`.
# !gcloud ai-platform models create census_income_classifier --regions us-central1
# Now it's time to deploy the model. We can do that with this gcloud command:
# + language="bash"
#
# MODEL_NAME="census_income_classifier"
# VERSION_NAME="original"
# MODEL_DIR="gs://$QWIKLABS_PROJECT_ID/original/"
# CUSTOM_CODE_PATH="gs://$QWIKLABS_PROJECT_ID/custom_transforms-0.1.tar.gz"
#
# gcloud beta ai-platform versions create $VERSION_NAME \
# --model $MODEL_NAME \
# --runtime-version 1.15 \
# --python-version 3.7 \
# --origin $MODEL_DIR \
# --package-uris $CUSTOM_CODE_PATH \
# --prediction-class predictor.MyPredictor
# -
# While this is running, check the [models section](https://console.cloud.google.com/ai-platform/models) of your AI Platform console. You should see your new version deploying there. When the deploy completes successfully you'll see a green check mark where the loading spinner is. The deploy should take 2-3 minutes. In the command above, notice we specify `prediction-class`. The reason we must specify a prediction class is that by default, AI Platform prediction will call a Scikit-Learn model's `predict` method, which in this case returns either 0 or 1. However, the What-If Tool requires output from a model in line with a Scikit-Learn model's `predict_proba` method. Consequently, we must write a [custom prediction routine](https://cloud.google.com/ai-platform/prediction/docs/custom-prediction-routines) that basically renames `predict_proba` as `predict`. The custom prediction method can be found in the file `predictor.py`. This file was packaged in the section __Package custom transform code__. By specifying `prediction-class` we're telling AI Platform to call our custom prediction method--basically, `predict_proba`-- instead of the default `predict` method.
# ## Test the deployed model
#
# To make sure your deployed model is working, test it out using gcloud to make a prediction. First, save a JSON file with one test instance for prediction:
# %%writefile predictions.json
[25, "Private", 226802, "11th", 7, "Never-married", "Machine-op-inspct", "Own-child", "Black", "Male", 0, 0, 40, "United-States"]
# Test your model by running this code:
# !gcloud ai-platform predict --model=census_income_classifier --json-instances=predictions.json --version=original
# You should see your model's prediction in the output. The first entry in the output is the model's probability that the individual makes under \\$50K while the second entry is the model's confidence that the individual makes over \\$50k. The two entries sum to 1.
# # What-If Tool
# To connect the What-if Tool to your AI Platform models, you need to pass it a subset of your test examples along with the ground truth values for those examples. Let's create a Numpy array of 2000 of our test examples.
# +
num_datapoints = 2000
test_examples = np.hstack(
(raw_test_features[:num_datapoints],
test_labels[:num_datapoints].reshape(-1,1)
)
)
# -
# Instantiating the What-if Tool is as simple as creating a WitConfigBuilder object and passing it the AI Platform model we built. Note that it'll take a minute to load the visualization.
# +
config_builder = (
WitConfigBuilder(test_examples.tolist(), COLUMNS)
.set_ai_platform_model(os.environ['QWIKLABS_PROJECT_ID'], 'census_income_classifier', 'original')
.set_target_feature('income-level')
.set_model_type('classification')
.set_label_vocab(['Under 50K', 'Over 50K'])
)
WitWidget(config_builder, height=800)
# -
# The default view on the What-if Tool is the __Datapoint editor__ tab. Here, you can click on any individual data point to see its features and even change feature values. Navigate to the __Performance & Fairness__ tab in the What-if Tool. By slicing on a feature you can view the model error for individual feature values. Finally, navigate to the __Features__ tab in the What-if Tool. This shows you the distribution of values for each feature in your dataset. You can use this tab to make sure your dataset is balanced. For example, if we only had Asians in a population, the model's predictions wouldn't necessarily reflect real world data. This tab gives us a good opportunity to see where our dataset might fall short, so that we can go back and collect more data to make it balanced.
#
# In the __Features__ tab, we can look to see the distribution of values for each feature in the dataset. We can see that of the 2000 test datapoints, 1346 are from men and 1702 are from caucasions. Women and minorities seem under-represented in this dataset. That may lead to the model not learning an accurate representation of the world in which it is trying to make predictions (of course, even if it does learn an accurate representation, is that what we want the model to perpetuate? This is a much deeper question still falling under the ML fairness umbrella and worthy of discussion outside of WIT). Predictions on those under-represented groups are more likely to be inaccurate than predictions on the over-represented groups.
#
# The features in this visualization can be sorted by a number of different metrics, including non-uniformity. With this sorting, the features that have the most non-uniform distributions are shown first. For numeric features, capital gain is very non-uniform, with most datapoints having it set to 0, but a small number having non-zero capital gains, all the way up to a maximum of 100k. For categorical features, country is the most non-uniform with most datapoints being from the USA, but there is a long tail of 40 other countries which are not well represented.
#
# Back in the __Performance & Fairness__ tab, we can set an input feature (or set of features) with which to slice the data. For example, setting this to `sex` allows us to see the breakdown of model performance on male datapoints versus female datapoints. We can see that the model is more accurate (has less false positives and false negatives) on females than males. We can also see that the model predicts high income for females much less than it does for males (8.0% of the time for females vs 27.1% of the time for males). __Note, your numbers will be slightly different due to the random elements of model training__.
#
# Imagine a scenario where this simple income classifier was used to approve or reject loan applications (not a realistic example but it illustrates the point). In this case, 28% of men from the test dataset have their loans approved but only 10% of women have theirs approved. If we wished to ensure than men and women get their loans approved the same percentage of the time, that is a fairness concept called "demographic parity". One way to achieve demographic parity would be to have different classification thresholds for males and females in our model. You'll notice there is a button on the tool labeled "demographic parity". When we press this button, the tool will take the cost ratio into account, and come up with ideal separate thresholds for men and women that will achieve demographic parity over the test dataset.
#
# In this case, demographic parity can be found with both groups getting loans 16% of the time by having the male threshold at 0.67 and the female threshold at 0.31. Because of the vast difference in the properties of the male and female training data in this 1994 census dataset, we need quite different thresholds to achieve demographic parity. Notice how with the high male threshold there are many more false negatives than before, and with the low female threshold there are many more false positives than before. This is necessary to get the percentage of positive predictions to be equal between the two groups. WIT has buttons to optimize for other fairness constraints as well, such as "equal opportunity" and "equal accuracy".
#
# The use of these features can help shed light on subsets of your data on which your classifier is performing very differently. Understanding biases in your datasets and data slices on which your model has disparate performance are very important parts of analyzing a model for fairness. There are many approaches to improving fairness, including augmenting training data, building fairness-related loss functions into your model training procedure, and post-training inference adjustments like those seen in WIT. We think that WIT provides a great interface for furthering ML fairness learning, but of course there is no silver bullet to improving ML fairness.
# # Training on a more balanced dataset
#
# Using the What-If Tool we saw that the model we trained on the census dataset wouldn't be very considerate in a production environment. What if we retrained the model on a dataset that was more balanced? Fortunately, we have such a dataset. Let's train a new model on this balanced dataset and compare it to our original dataset using the What-If Tool.
# First, let's load the balanced dataset into a Pandas dataframe.
bal_data_path = 'https://storage.googleapis.com/cloud-training/dei/balanced_census_data.csv'
bal_data = pd.read_csv(bal_data_path, names=COLUMNS, skiprows=1)
bal_data.head()
# Execute the command below to see the distribution of gender in the data.
bal_data['sex'].value_counts(normalize=True)
# Unlike the original dataset, this dataset has an equal number of rows for both males and females. Execute the command below to see the distriubtion of rows in the dataset of both `sex` and `income-level`.
bal_data.groupby(['sex', 'income-level'])['sex'].count()
# We see that not only is the dataset balanced across gender, it's also balanced across income. Let's train a model on this data. We'll use exactly the same model pipeline as in the previous section. Scikit-Learn has a convenient utility function for copying model pipelines, `clone`. The `clone` function copies a pipeline architecture without saving learned parameter values.
# +
bal_data['income-level'] = bal_data['income-level'].isin(['>50K', '>50K.']).values.astype(int)
raw_bal_features = bal_data.drop('income-level', axis=1).values
bal_labels = bal_data['income-level'].values
# -
pipeline_bal = clone(pipeline)
pipeline_bal.fit(raw_bal_features, bal_labels)
# As before, we save our trained model to a pickle file. Note, when we version this model in AI Platform the model in this case must be named `model.pkl`. It's ok to overwrite the existing `model.pkl` file since we'll be uploading it to Cloud Storage anyway.
with open('model.pkl', 'wb') as model_file:
pickle.dump(pipeline_bal, model_file)
# Deploy the model to AI Platform using the following bash script:
# + language="bash"
#
# gsutil cp model.pkl gs://$QWIKLABS_PROJECT_ID/balanced/
#
# MODEL_NAME="census_income_classifier"
# VERSION_NAME="balanced"
# MODEL_DIR="gs://$QWIKLABS_PROJECT_ID/balanced/"
# CUSTOM_CODE_PATH="gs://$QWIKLABS_PROJECT_ID/custom_transforms-0.1.tar.gz"
#
# gcloud beta ai-platform versions create $VERSION_NAME \
# --model $MODEL_NAME \
# --runtime-version 1.15 \
# --python-version 3.7 \
# --origin $MODEL_DIR \
# --package-uris $CUSTOM_CODE_PATH \
# --prediction-class predictor.MyPredictor
# -
# Now let's instantiate the What-if Tool by configuring a WitConfigBuilder. Here, we want to compare the original model we built with the one trained on the balanced census dataset. To achieve this we utilize the `set_compare_ai_platform_model` method.
# +
config_builder = (
WitConfigBuilder(test_examples.tolist(), COLUMNS)
.set_ai_platform_model(os.environ['QWIKLABS_PROJECT_ID'], 'census_income_classifier', 'balanced')
.set_compare_ai_platform_model(os.environ['QWIKLABS_PROJECT_ID'], 'census_income_classifier', 'original')
.set_target_feature('income-level')
.set_model_type('classification')
.set_label_vocab(['Under 50K', 'Over 50K'])
)
WitWidget(config_builder, height=800)
# -
# Once the WIT widget loads, click on the __Performance & Fairness__ tab. In the __Slice by__ field select `sex` and wait a minute for the graphics to load. For females, the model trained on the balanced dataset is 3 times more likely to predict an income of over 50k than the model trained on the original dataset. How else does the model trained on balanced data perform differently when compared to the original model?
| 61.965217
| 1,176
|
c514561b305834b0fc43e23596f5ed76d5387feb
|
py
|
python
|
Chatbot/Chatbot.ipynb
|
Elyrie/Hacktoberfest-2021
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + id="nMcYmPQF3krn" colab_type="code" colab={"base_uri": "https://localhost:8080/"} outputId="91c9a3cd-14f1-4348-f3af-88716870378b"
# Libraries needed for NLP
import nltk
nltk.download('punkt')
from nltk.stem.lancaster import LancasterStemmer
stemmer = LancasterStemmer()
# Libraries needed for Tensorflow processing
import tensorflow as tf
import numpy as np
import tflearn
import random
import json
# + id="zitZRhjq9Vqu" colab_type="code" colab={"resources": {"http://localhost:8080/nbextensions/google.colab/files.js": {"data": "Ly8gQ29weXJpZ2h0IDIwMTcgR29vZ2xlIExMQwovLwovLyBMaWNlbnNlZCB1bmRlciB0aGUgQXBhY2hlIExpY2Vuc2UsIFZlcnNpb24gMi4wICh0aGUgIkxpY2Vuc2UiKTsKLy8geW91IG1heSBub3QgdXNlIHRoaXMgZmlsZSBleGNlcHQgaW4gY29tcGxpYW5jZSB3aXRoIHRoZSBMaWNlbnNlLgovLyBZb3UgbWF5IG9idGFpbiBhIGNvcHkgb2YgdGhlIExpY2Vuc2UgYXQKLy8KLy8gICAgICBodHRwOi8vd3d3LmFwYWNoZS5vcmcvbGljZW5zZXMvTElDRU5TRS0yLjAKLy8KLy8gVW5sZXNzIHJlcXVpcmVkIGJ5IGFwcGxpY2FibGUgbGF3IG9yIGFncmVlZCB0byBpbiB3cml0aW5nLCBzb2Z0d2FyZQovLyBkaXN0cmlidXRlZCB1bmRlciB0aGUgTGljZW5zZSBpcyBkaXN0cmlidXRlZCBvbiBhbiAiQVMgSVMiIEJBU0lTLAovLyBXSVRIT1VUIFdBUlJBTlRJRVMgT1IgQ09ORElUSU9OUyBPRiBBTlkgS0lORCwgZWl0aGVyIGV4cHJlc3Mgb3IgaW1wbGllZC4KLy8gU2VlIHRoZSBMaWNlbnNlIGZvciB0aGUgc3BlY2lmaWMgbGFuZ3VhZ2UgZ292ZXJuaW5nIHBlcm1pc3Npb25zIGFuZAovLyBsaW1pdGF0aW9ucyB1bmRlciB0aGUgTGljZW5zZS4KCi8qKgogKiBAZmlsZW92ZXJ2aWV3IEhlbHBlcnMgZm9yIGdvb2dsZS5jb2xhYiBQeXRob24gbW9kdWxlLgogKi8KKGZ1bmN0aW9uKHNjb3BlKSB7CmZ1bmN0aW9uIHNwYW4odGV4dCwgc3R5bGVBdHRyaWJ1dGVzID0ge30pIHsKICBjb25zdCBlbGVtZW50ID0gZG9jdW1lbnQuY3JlYXRlRWxlbWVudCgnc3BhbicpOwogIGVsZW1lbnQudGV4dENvbnRlbnQgPSB0ZXh0OwogIGZvciAoY29uc3Qga2V5IG9mIE9iamVjdC5rZXlzKHN0eWxlQXR0cmlidXRlcykpIHsKICAgIGVsZW1lbnQuc3R5bGVba2V5XSA9IHN0eWxlQXR0cmlidXRlc1trZXldOwogIH0KICByZXR1cm4gZWxlbWVudDsKfQoKLy8gTWF4IG51bWJlciBvZiBieXRlcyB3aGljaCB3aWxsIGJlIHVwbG9hZGVkIGF0IGEgdGltZS4KY29uc3QgTUFYX1BBWUxPQURfU0laRSA9IDEwMCAqIDEwMjQ7Ci8vIE1heCBhbW91bnQgb2YgdGltZSB0byBibG9jayB3YWl0aW5nIGZvciB0aGUgdXNlci4KY29uc3QgRklMRV9DSEFOR0VfVElNRU9VVF9NUyA9IDMwICogMTAwMDsKCmZ1bmN0aW9uIF91cGxvYWRGaWxlcyhpbnB1dElkLCBvdXRwdXRJZCkgewogIGNvbnN0IHN0ZXBzID0gdXBsb2FkRmlsZXNTdGVwKGlucHV0SWQsIG91dHB1dElkKTsKICBjb25zdCBvdXRwdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQob3V0cHV0SWQpOwogIC8vIENhY2hlIHN0ZXBzIG9uIHRoZSBvdXRwdXRFbGVtZW50IHRvIG1ha2UgaXQgYXZhaWxhYmxlIGZvciB0aGUgbmV4dCBjYWxsCiAgLy8gdG8gdXBsb2FkRmlsZXNDb250aW51ZSBmcm9tIFB5dGhvbi4KICBvdXRwdXRFbGVtZW50LnN0ZXBzID0gc3RlcHM7CgogIHJldHVybiBfdXBsb2FkRmlsZXNDb250aW51ZShvdXRwdXRJZCk7Cn0KCi8vIFRoaXMgaXMgcm91Z2hseSBhbiBhc3luYyBnZW5lcmF0b3IgKG5vdCBzdXBwb3J0ZWQgaW4gdGhlIGJyb3dzZXIgeWV0KSwKLy8gd2hlcmUgdGhlcmUgYXJlIG11bHRpcGxlIGFzeW5jaHJvbm91cyBzdGVwcyBhbmQgdGhlIFB5dGhvbiBzaWRlIGlzIGdvaW5nCi8vIHRvIHBvbGwgZm9yIGNvbXBsZXRpb24gb2YgZWFjaCBzdGVwLgovLyBUaGlzIHVzZXMgYSBQcm9taXNlIHRvIGJsb2NrIHRoZSBweXRob24gc2lkZSBvbiBjb21wbGV0aW9uIG9mIGVhY2ggc3RlcCwKLy8gdGhlbiBwYXNzZXMgdGhlIHJlc3VsdCBvZiB0aGUgcHJldmlvdXMgc3RlcCBhcyB0aGUgaW5wdXQgdG8gdGhlIG5leHQgc3RlcC4KZnVuY3Rpb24gX3VwbG9hZEZpbGVzQ29udGludWUob3V0cHV0SWQpIHsKICBjb25zdCBvdXRwdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQob3V0cHV0SWQpOwogIGNvbnN0IHN0ZXBzID0gb3V0cHV0RWxlbWVudC5zdGVwczsKCiAgY29uc3QgbmV4dCA9IHN0ZXBzLm5leHQob3V0cHV0RWxlbWVudC5sYXN0UHJvbWlzZVZhbHVlKTsKICByZXR1cm4gUHJvbWlzZS5yZXNvbHZlKG5leHQudmFsdWUucHJvbWlzZSkudGhlbigodmFsdWUpID0+IHsKICAgIC8vIENhY2hlIHRoZSBsYXN0IHByb21pc2UgdmFsdWUgdG8gbWFrZSBpdCBhdmFpbGFibGUgdG8gdGhlIG5leHQKICAgIC8vIHN0ZXAgb2YgdGhlIGdlbmVyYXRvci4KICAgIG91dHB1dEVsZW1lbnQubGFzdFByb21pc2VWYWx1ZSA9IHZhbHVlOwogICAgcmV0dXJuIG5leHQudmFsdWUucmVzcG9uc2U7CiAgfSk7Cn0KCi8qKgogKiBHZW5lcmF0b3IgZnVuY3Rpb24gd2hpY2ggaXMgY2FsbGVkIGJldHdlZW4gZWFjaCBhc3luYyBzdGVwIG9mIHRoZSB1cGxvYWQKICogcHJvY2Vzcy4KICogQHBhcmFtIHtzdHJpbmd9IGlucHV0SWQgRWxlbWVudCBJRCBvZiB0aGUgaW5wdXQgZmlsZSBwaWNrZXIgZWxlbWVudC4KICogQHBhcmFtIHtzdHJpbmd9IG91dHB1dElkIEVsZW1lbnQgSUQgb2YgdGhlIG91dHB1dCBkaXNwbGF5LgogKiBAcmV0dXJuIHshSXRlcmFibGU8IU9iamVjdD59IEl0ZXJhYmxlIG9mIG5leHQgc3RlcHMuCiAqLwpmdW5jdGlvbiogdXBsb2FkRmlsZXNTdGVwKGlucHV0SWQsIG91dHB1dElkKSB7CiAgY29uc3QgaW5wdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQoaW5wdXRJZCk7CiAgaW5wdXRFbGVtZW50LmRpc2FibGVkID0gZmFsc2U7CgogIGNvbnN0IG91dHB1dEVsZW1lbnQgPSBkb2N1bWVudC5nZXRFbGVtZW50QnlJZChvdXRwdXRJZCk7CiAgb3V0cHV0RWxlbWVudC5pbm5lckhUTUwgPSAnJzsKCiAgY29uc3QgcGlja2VkUHJvbWlzZSA9IG5ldyBQcm9taXNlKChyZXNvbHZlKSA9PiB7CiAgICBpbnB1dEVsZW1lbnQuYWRkRXZlbnRMaXN0ZW5lcignY2hhbmdlJywgKGUpID0+IHsKICAgICAgcmVzb2x2ZShlLnRhcmdldC5maWxlcyk7CiAgICB9KTsKICB9KTsKCiAgY29uc3QgY2FuY2VsID0gZG9jdW1lbnQuY3JlYXRlRWxlbWVudCgnYnV0dG9uJyk7CiAgaW5wdXRFbGVtZW50LnBhcmVudEVsZW1lbnQuYXBwZW5kQ2hpbGQoY2FuY2VsKTsKICBjYW5jZWwudGV4dENvbnRlbnQgPSAnQ2FuY2VsIHVwbG9hZCc7CiAgY29uc3QgY2FuY2VsUHJvbWlzZSA9IG5ldyBQcm9taXNlKChyZXNvbHZlKSA9PiB7CiAgICBjYW5jZWwub25jbGljayA9ICgpID0+IHsKICAgICAgcmVzb2x2ZShudWxsKTsKICAgIH07CiAgfSk7CgogIC8vIENhbmNlbCB1cGxvYWQgaWYgdXNlciBoYXNuJ3QgcGlja2VkIGFueXRoaW5nIGluIHRpbWVvdXQuCiAgY29uc3QgdGltZW91dFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgc2V0VGltZW91dCgoKSA9PiB7CiAgICAgIHJlc29sdmUobnVsbCk7CiAgICB9LCBGSUxFX0NIQU5HRV9USU1FT1VUX01TKTsKICB9KTsKCiAgLy8gV2FpdCBmb3IgdGhlIHVzZXIgdG8gcGljayB0aGUgZmlsZXMuCiAgY29uc3QgZmlsZXMgPSB5aWVsZCB7CiAgICBwcm9taXNlOiBQcm9taXNlLnJhY2UoW3BpY2tlZFByb21pc2UsIHRpbWVvdXRQcm9taXNlLCBjYW5jZWxQcm9taXNlXSksCiAgICByZXNwb25zZTogewogICAgICBhY3Rpb246ICdzdGFydGluZycsCiAgICB9CiAgfTsKCiAgaWYgKCFmaWxlcykgewogICAgcmV0dXJuIHsKICAgICAgcmVzcG9uc2U6IHsKICAgICAgICBhY3Rpb246ICdjb21wbGV0ZScsCiAgICAgIH0KICAgIH07CiAgfQoKICBjYW5jZWwucmVtb3ZlKCk7CgogIC8vIERpc2FibGUgdGhlIGlucHV0IGVsZW1lbnQgc2luY2UgZnVydGhlciBwaWNrcyBhcmUgbm90IGFsbG93ZWQuCiAgaW5wdXRFbGVtZW50LmRpc2FibGVkID0gdHJ1ZTsKCiAgZm9yIChjb25zdCBmaWxlIG9mIGZpbGVzKSB7CiAgICBjb25zdCBsaSA9IGRvY3VtZW50LmNyZWF0ZUVsZW1lbnQoJ2xpJyk7CiAgICBsaS5hcHBlbmQoc3BhbihmaWxlLm5hbWUsIHtmb250V2VpZ2h0OiAnYm9sZCd9KSk7CiAgICBsaS5hcHBlbmQoc3BhbigKICAgICAgICBgKCR7ZmlsZS50eXBlIHx8ICduL2EnfSkgLSAke2ZpbGUuc2l6ZX0gYnl0ZXMsIGAgKwogICAgICAgIGBsYXN0IG1vZGlmaWVkOiAkewogICAgICAgICAgICBmaWxlLmxhc3RNb2RpZmllZERhdGUgPyBmaWxlLmxhc3RNb2RpZmllZERhdGUudG9Mb2NhbGVEYXRlU3RyaW5nKCkgOgogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAnbi9hJ30gLSBgKSk7CiAgICBjb25zdCBwZXJjZW50ID0gc3BhbignMCUgZG9uZScpOwogICAgbGkuYXBwZW5kQ2hpbGQocGVyY2VudCk7CgogICAgb3V0cHV0RWxlbWVudC5hcHBlbmRDaGlsZChsaSk7CgogICAgY29uc3QgZmlsZURhdGFQcm9taXNlID0gbmV3IFByb21pc2UoKHJlc29sdmUpID0+IHsKICAgICAgY29uc3QgcmVhZGVyID0gbmV3IEZpbGVSZWFkZXIoKTsKICAgICAgcmVhZGVyLm9ubG9hZCA9IChlKSA9PiB7CiAgICAgICAgcmVzb2x2ZShlLnRhcmdldC5yZXN1bHQpOwogICAgICB9OwogICAgICByZWFkZXIucmVhZEFzQXJyYXlCdWZmZXIoZmlsZSk7CiAgICB9KTsKICAgIC8vIFdhaXQgZm9yIHRoZSBkYXRhIHRvIGJlIHJlYWR5LgogICAgbGV0IGZpbGVEYXRhID0geWllbGQgewogICAgICBwcm9taXNlOiBmaWxlRGF0YVByb21pc2UsCiAgICAgIHJlc3BvbnNlOiB7CiAgICAgICAgYWN0aW9uOiAnY29udGludWUnLAogICAgICB9CiAgICB9OwoKICAgIC8vIFVzZSBhIGNodW5rZWQgc2VuZGluZyB0byBhdm9pZCBtZXNzYWdlIHNpemUgbGltaXRzLiBTZWUgYi82MjExNTY2MC4KICAgIGxldCBwb3NpdGlvbiA9IDA7CiAgICB3aGlsZSAocG9zaXRpb24gPCBmaWxlRGF0YS5ieXRlTGVuZ3RoKSB7CiAgICAgIGNvbnN0IGxlbmd0aCA9IE1hdGgubWluKGZpbGVEYXRhLmJ5dGVMZW5ndGggLSBwb3NpdGlvbiwgTUFYX1BBWUxPQURfU0laRSk7CiAgICAgIGNvbnN0IGNodW5rID0gbmV3IFVpbnQ4QXJyYXkoZmlsZURhdGEsIHBvc2l0aW9uLCBsZW5ndGgpOwogICAgICBwb3NpdGlvbiArPSBsZW5ndGg7CgogICAgICBjb25zdCBiYXNlNjQgPSBidG9hKFN0cmluZy5mcm9tQ2hhckNvZGUuYXBwbHkobnVsbCwgY2h1bmspKTsKICAgICAgeWllbGQgewogICAgICAgIHJlc3BvbnNlOiB7CiAgICAgICAgICBhY3Rpb246ICdhcHBlbmQnLAogICAgICAgICAgZmlsZTogZmlsZS5uYW1lLAogICAgICAgICAgZGF0YTogYmFzZTY0LAogICAgICAgIH0sCiAgICAgIH07CiAgICAgIHBlcmNlbnQudGV4dENvbnRlbnQgPQogICAgICAgICAgYCR7TWF0aC5yb3VuZCgocG9zaXRpb24gLyBmaWxlRGF0YS5ieXRlTGVuZ3RoKSAqIDEwMCl9JSBkb25lYDsKICAgIH0KICB9CgogIC8vIEFsbCBkb25lLgogIHlpZWxkIHsKICAgIHJlc3BvbnNlOiB7CiAgICAgIGFjdGlvbjogJ2NvbXBsZXRlJywKICAgIH0KICB9Owp9CgpzY29wZS5nb29nbGUgPSBzY29wZS5nb29nbGUgfHwge307CnNjb3BlLmdvb2dsZS5jb2xhYiA9IHNjb3BlLmdvb2dsZS5jb2xhYiB8fCB7fTsKc2NvcGUuZ29vZ2xlLmNvbGFiLl9maWxlcyA9IHsKICBfdXBsb2FkRmlsZXMsCiAgX3VwbG9hZEZpbGVzQ29udGludWUsCn07Cn0pKHNlbGYpOwo=", "ok": true, "headers": [["content-type", "application/javascript"]], "status": 200, "status_text": ""}}, "base_uri": "https://localhost:8080/", "height": 112} outputId="5c43e603-42f3-4c1f-81d7-45269708f0bb"
from google.colab import files
files.upload()
# + id="sBEuh9TEGBo7" colab_type="code" colab={}
# import our chat-bot intents file
with open('intents.json') as json_data:
intents = json.load(json_data)
# + id="mpFsVWPKGKbI" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 1012} outputId="d7adb195-dcff-496b-8acf-f3c44af24724"
intents
# + id="jUNlDhkSGVL1" colab_type="code" colab={}
words = []
classes = []
documents = []
ignore = ['?']
# loop through each sentence in the intent's patterns
for intent in intents['intents']:
for pattern in intent['patterns']:
# tokenize each and every word in the sentence
w = nltk.word_tokenize(pattern)
# add word to the words list
words.extend(w)
# add word(s) to documents
documents.append((w, intent['tag']))
# add tags to our classes list
if intent['tag'] not in classes:
classes.append(intent['tag'])
# + id="GO7b9xTwHJMJ" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 90} outputId="72a2271e-9ba6-4b01-d8ab-bee99e1ccf71"
# Perform stemming and lower each word as well as remove duplicates
words = [stemmer.stem(w.lower()) for w in words if w not in ignore]
words = sorted(list(set(words)))
# remove duplicate classes
classes = sorted(list(set(classes)))
print (len(documents), "documents")
print (len(classes), "classes", classes)
print (len(words), "unique stemmed words", words)
# + id="vziuGlP1Iq-P" colab_type="code" colab={}
# create training data
training = []
output = []
# create an empty array for output
output_empty = [0] * len(classes)
# create training set, bag of words for each sentence
for doc in documents:
# initialize bag of words
bag = []
# list of tokenized words for the pattern
pattern_words = doc[0]
# stemming each word
pattern_words = [stemmer.stem(word.lower()) for word in pattern_words]
# create bag of words array
for w in words:
bag.append(1) if w in pattern_words else bag.append(0)
# output is '1' for current tag and '0' for rest of other tags
output_row = list(output_empty)
output_row[classes.index(doc[1])] = 1
training.append([bag, output_row])
# shuffling features and turning it into np.array
random.shuffle(training)
training = np.array(training)
# creating training lists
train_x = list(training[:,0])
train_y = list(training[:,1])
# + id="ZX8rC1x9PVTb" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 105} outputId="bf3f14ab-0dae-4518-b4f6-914761545e40"
# resetting underlying graph data
tf.reset_default_graph()
# Building neural network
net = tflearn.input_data(shape=[None, len(train_x[0])])
net = tflearn.fully_connected(net, 10)
net = tflearn.fully_connected(net, 10)
net = tflearn.fully_connected(net, len(train_y[0]), activation='softmax')
net = tflearn.regression(net)
# Defining model and setting up tensorboard
model = tflearn.DNN(net, tensorboard_dir='tflearn_logs')
# Start training
model.fit(train_x, train_y, n_epoch=1000, batch_size=8, show_metric=True)
model.save('model.tflearn')
# + id="ha2RGmK1Zz5l" colab_type="code" colab={}
import pickle
pickle.dump( {'words':words, 'classes':classes, 'train_x':train_x, 'train_y':train_y}, open( "training_data", "wb" ) )
# + id="ux9WbDZvbAzf" colab_type="code" colab={}
# restoring all the data structures
data = pickle.load( open( "training_data", "rb" ) )
words = data['words']
classes = data['classes']
train_x = data['train_x']
train_y = data['train_y']
# + id="Ms17AFyEbEjt" colab_type="code" colab={}
with open('intents.json') as json_data:
intents = json.load(json_data)
# + id="WKYk9_FXbLfd" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 90} outputId="268b990e-6bd9-421d-f2f4-9716d204e2bb"
# load the saved model
model.load('./model.tflearn')
# + id="VB27k_vQbhu4" colab_type="code" colab={}
def clean_up_sentence(sentence):
# tokenizing the pattern
sentence_words = nltk.word_tokenize(sentence)
# stemming each word
sentence_words = [stemmer.stem(word.lower()) for word in sentence_words]
return sentence_words
# returning bag of words array: 0 or 1 for each word in the bag that exists in the sentence
def bow(sentence, words, show_details=False):
# tokenizing the pattern
sentence_words = clean_up_sentence(sentence)
# generating bag of words
bag = [0]*len(words)
for s in sentence_words:
for i,w in enumerate(words):
if w == s:
bag[i] = 1
if show_details:
print ("found in bag: %s" % w)
return(np.array(bag))
# + id="3lToEtkTb5Pr" colab_type="code" colab={}
ERROR_THRESHOLD = 0.30
def classify(sentence):
# generate probabilities from the model
results = model.predict([bow(sentence, words)])[0]
# filter out predictions below a threshold
results = [[i,r] for i,r in enumerate(results) if r>ERROR_THRESHOLD]
# sort by strength of probability
results.sort(key=lambda x: x[1], reverse=True)
return_list = []
for r in results:
return_list.append((classes[r[0]], r[1]))
# return tuple of intent and probability
return return_list
def response(sentence, userID='123', show_details=False):
results = classify(sentence)
# if we have a classification then find the matching intent tag
if results:
# loop as long as there are matches to process
while results:
for i in intents['intents']:
# find a tag matching the first result
if i['tag'] == results[0][0]:
# a random response from the intent
return print(random.choice(i['responses']))
results.pop(0)
# + id="eal_MyIEcC9X" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="c0111154-0224-4074-cfa2-5e767034d429"
classify('What are you hours of operation?')
# + id="zvFA4ef4cW96" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="db7a0790-0cbf-47c6-b20e-2c2750c4f280"
response('What are you hours of operation?')
# + id="9ox0fJIlOIJ-" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="af4b9344-f19d-45ed-d6b4-8e1a1f839c06"
response('What is menu for today?')
# + id="4_QtpNdkc5wD" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="0f6beb67-c11b-4859-e32a-ba6059bf012d"
#Some of other context free responses.
response('Do you accept Credit Card?')
# + id="xp7OJvpHKXuw" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="39a166d8-80e7-4210-d7e3-d96ae8a7fc43"
response('Where can we locate you?')
# + id="3PNLGpl0gKCF" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="3e086203-35bc-49e5-af83-2c6848d884ba"
response('That is helpful')
# + id="wzeIGchZK2fJ" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="9ea5fc7c-522d-4ae7-dc8e-40107893e046"
response('Bye')
# + id="p_NgjLGwggdI" colab_type="code" colab={}
#Adding some context to the conversation i.e. Contexualization for altering question and intents etc.
# create a data structure to hold user context
context = {}
ERROR_THRESHOLD = 0.25
def classify(sentence):
# generate probabilities from the model
results = model.predict([bow(sentence, words)])[0]
# filter out predictions below a threshold
results = [[i,r] for i,r in enumerate(results) if r>ERROR_THRESHOLD]
# sort by strength of probability
results.sort(key=lambda x: x[1], reverse=True)
return_list = []
for r in results:
return_list.append((classes[r[0]], r[1]))
# return tuple of intent and probability
return return_list
def response(sentence, userID='123', show_details=False):
results = classify(sentence)
# if we have a classification then find the matching intent tag
if results:
# loop as long as there are matches to process
while results:
for i in intents['intents']:
# find a tag matching the first result
if i['tag'] == results[0][0]:
# set context for this intent if necessary
if 'context_set' in i:
if show_details: print ('context:', i['context_set'])
context[userID] = i['context_set']
# check if this intent is contextual and applies to this user's conversation
if not 'context_filter' in i or \
(userID in context and 'context_filter' in i and i['context_filter'] == context[userID]):
if show_details: print ('tag:', i['tag'])
# a random response from the intent
return print(random.choice(i['responses']))
results.pop(0)
# + id="Gqyn3Q-LGpfe" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="24be5520-1f5b-40d6-dd13-d2b151ba8fc4"
response('Can you please let me know the delivery options?')
# + id="NtqhOcjlG6O4" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="08d40e82-2e3b-47c8-8a37-219ffb0b4fb9"
response('What is menu for today?')
# + id="kX-CBzN7KAII" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="6c30fe4e-cc37-4c00-b65c-0d58d9176450"
context
# + id="TBZyzI1SLeMu" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 70} outputId="b98a9f1b-aece-48e8-ad80-fe40585069da"
response("Hi there!", show_details=True)
# + id="JB_FgfW4aYua" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="25144d80-d5be-48bf-b7f5-ee4c8bfefbe5"
response('What is menu for today?')
# + id="aBejUcxra0UV" colab_type="code" colab={}
# + id="xYwHSa_Bb6rM" colab_type="code" colab={}
| 64.342857
| 7,664
|
58220b057e4128b119a13632e631671c11d415c2
|
py
|
python
|
doc/ipython-notebooks/metric/LMNN.ipynb
|
Khalifa1997/shogun
|
['BSD-3-Clause']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Metric Learning with the Shogun Machine Learning Toolbox
# #### *By Fernando J. Iglesias Garcia (GitHub ID: [iglesias](https://github.com/iglesias)) as project report for GSoC 2013 ([project details](http://www.google-melange.com/gsoc/project/google/gsoc2013/iglesias/62013)).*
# This notebook illustrates <a href="http://en.wikipedia.org/wiki/Statistical_classification">classification</a> and <a href="http://en.wikipedia.org/wiki/Feature_selection">feature selection</a> using <a href="http://en.wikipedia.org/wiki/Similarity_learning#Metric_learning">metric learning</a> in Shogun. To overcome the limitations of <a href="http://en.wikipedia.org/wiki/K-nearest_neighbors_algorithm">knn</a> with Euclidean distance as the distance measure, <a href="http://en.wikipedia.org/wiki/Large_margin_nearest_neighbor">Large Margin Nearest Neighbour</a>(LMNN) is discussed. This is consolidated by applying LMNN over the metagenomics data set.
# ## Building up the intuition to understand LMNN
# First of all, let us introduce LMNN through a simple example. For this purpose, we will be using the following two-dimensional toy data set:
# +
import numpy as np
import os
import shogun as sg
import matplotlib.pyplot as plt
# %matplotlib inline
SHOGUN_DATA_DIR=os.getenv('SHOGUN_DATA_DIR', '../../../data')
x = np.array([[0,0],[-1,0.1],[0.3,-0.05],[0.7,0.3],[-0.2,-0.6],[-0.15,-0.63],[-0.25,0.55],[-0.28,0.67]])
y = np.array([0,0,0,0,1,1,2,2])
# -
# That is, there are eight feature vectors where each of them belongs to one out of three different classes (identified by either 0, 1, or 2). Let us have a look at this data:
# +
def plot_data(feats,labels,axis,alpha=1.0):
# separate features according to their class
X0,X1,X2 = feats[labels==0], feats[labels==1], feats[labels==2]
# class 0 data
axis.plot(X0[:,0], X0[:,1], 'o', color='green', markersize=12, alpha=alpha)
# class 1 data
axis.plot(X1[:,0], X1[:,1], 'o', color='red', markersize=12, alpha=alpha)
# class 2 data
axis.plot(X2[:,0], X2[:,1], 'o', color='blue', markersize=12, alpha=alpha)
# set axes limits
axis.set_xlim(-1.5,1.5)
axis.set_ylim(-1.5,1.5)
axis.set_aspect('equal')
axis.set_xlabel('x')
axis.set_ylabel('y')
figure,axis = plt.subplots(1,1)
plot_data(x,y,axis)
axis.set_title('Toy data set')
plt.show()
# -
# In the figure above, we can see that two of the classes are represented by two points that are, for each of these classes, very close to each other. The third class, however, has four points that are close to each other with respect to the y-axis, but spread along the x-axis.
# If we were to apply kNN (*k-nearest neighbors*) in a data set like this, we would expect quite some errors using the standard Euclidean distance. This is due to the fact that the spread of the data is not similar amongst the feature dimensions. The following piece of code plots an ellipse on top of the data set. The ellipse in this case is in fact a circunference that helps to visualize how the Euclidean distance weights equally both feature dimensions.
# +
def make_covariance_ellipse(covariance):
import matplotlib.patches as patches
import scipy.linalg as linalg
# the ellipse is centered at (0,0)
mean = np.array([0,0])
# eigenvalue decomposition of the covariance matrix (w are eigenvalues and v eigenvectors),
# keeping only the real part
w,v = linalg.eigh(covariance)
# normalize the eigenvector corresponding to the largest eigenvalue
u = v[0]/linalg.norm(v[0])
# angle in degrees
angle = 180.0/np.pi*np.arctan(u[1]/u[0])
# fill Gaussian ellipse at 2 standard deviation
ellipse = patches.Ellipse(mean, 2*w[0]**0.5, 2*w[1]**0.5, 180+angle, color='orange', alpha=0.3)
return ellipse
# represent the Euclidean distance
figure,axis = plt.subplots(1,1)
plot_data(x,y,axis)
ellipse = make_covariance_ellipse(np.eye(2))
axis.add_artist(ellipse)
axis.set_title('Euclidean distance')
plt.show()
# -
# A possible workaround to improve the performance of kNN in a data set like this would be to input to the kNN routine a distance measure. For instance, in the example above a good distance measure would give more weight to the y-direction than to the x-direction to account for the large spread along the x-axis. Nonetheless, it would be nicer (and, in fact, much more useful in practice) if this distance could be learnt automatically from the data at hand. Actually, LMNN is based upon this principle: given a number of neighbours *k*, find the Mahalanobis distance measure which maximizes kNN accuracy (using the given value for *k*) in a training data set. As we usually do in machine learning, under the assumption that the training data is an accurate enough representation of the underlying process, the distance learnt will not only perform well in the training data, but also have good generalization properties.
# Now, let us use the [LMNN class](http://www.shogun-toolbox.org/doc/en/latest/classshogun_1_1LMNN.html) implemented in Shogun to find the distance and plot its associated ellipse. If everything goes well, we will see that the new ellipse only overlaps with the data points of the green class.
# First, we need to wrap the data into Shogun's feature and label objects:
feats = sg.features(x.T)
labels = sg.labels(y.astype(np.float64))
# Secondly, perform LMNN training:
# +
# number of target neighbours per example
k = 1
lmnn = sg.LMNN(feats,labels,k)
# set an initial transform as a start point of the optimization
init_transform = np.eye(2)
lmnn.put('maxiter', 2000)
lmnn.train(init_transform)
# -
# LMNN is an iterative algorithm. The argument given to [`train`](http://www.shogun-toolbox.org/doc/en/latest/classshogun_1_1LMNN.html#ab1b8bbdb8390415ac3ae7dc655cb512d) represents the initial state of the solution. By default, if no argument is given, then LMNN uses [PCA](http://en.wikipedia.org/wiki/Principal_component_analysis) to obtain this initial value.
# Finally, we retrieve the distance measure learnt by LMNN during training and visualize it together with the data:
# +
# get the linear transform from LMNN
L = lmnn.get('linear_transform')
# square the linear transform to obtain the Mahalanobis distance matrix
M = np.matrix(np.dot(L.T,L))
# represent the distance given by LMNN
figure,axis = plt.subplots(1,1)
plot_data(x,y,axis)
ellipse = make_covariance_ellipse(M.I)
axis.add_artist(ellipse)
axis.set_title('LMNN distance')
plt.show()
# -
# ## Beyond the main idea
# LMNN is one of the so-called linear metric learning methods. What this means is that we can understand LMNN's output in two different ways: on the one hand, as a distance measure, this was explained above; on the other hand, as a linear transformation of the input data. Like any other linear transformation, LMNN's output can be written as a matrix, that we will call $L$. In other words, if the input data is represented by the matrix $X$, then LMNN can be understood as the data transformation expressed by $X'=L X$. We use the convention that each column is a feature vector; thus, the number of rows of $X$ is equal to the input dimension of the data, and the number of columns is equal to the number of vectors.
#
# So far, so good. But, if the output of the same method can be interpreted in two different ways, then there must be a relation between them! And that is precisely the case! As mentioned above, the ellipses that were plotted in the previous section represent a distance measure. This distance measure can be thought of as a matrix $M$, being the distance between two vectors $\vec{x_i}$ and $\vec{x_j}$ equal to $d(\vec{x_i},\vec{x_j})=(\vec{x_i}-\vec{x_j})^T M (\vec{x_i}-\vec{x_j})$. In general, this type of matrices are known as *Mahalanobis* matrices. In LMNN, the matrix $M$ is precisely the 'square' of the linear transformation $L$, i.e. $M=L^T L$. Note that a direct consequence of this is that $M$ is guaranteed to be positive semi-definite (PSD), and therefore define a valid metric.
#
# This distance measure/linear transform duality in LMNN has its own advantages. An important one is that the optimization problem can go back and forth between the $L$ and the $M$ representations, giving raise to a very efficient solution.
# Let us now visualize LMNN using the linear transform interpretation. In the following figure we have taken our original toy data, transform it using $L$ and plot both the before and after versions of the data together.
# +
# project original data using L
lx = np.dot(L,x.T)
# represent the data in the projected space
figure,axis = plt.subplots(1,1)
plot_data(lx.T,y,axis)
plot_data(x,y,axis,0.3)
ellipse = make_covariance_ellipse(np.eye(2))
axis.add_artist(ellipse)
axis.set_title('LMNN\'s linear transform')
plt.show()
# -
# In the figure above, the transparent points represent the original data and are shown to ease the visualization of the LMNN transformation. Note also that the ellipse plotted is the one corresponding to the common Euclidean distance. This is actually an important consideration: if we think of LMNN as a linear transformation, the distance considered in the projected space is the Euclidean distance, and no any Mahalanobis distance given by M. To sum up, we can think of LMNN as a linear transform of the input space, or as method to obtain a distance measure to be used in the input space. It is an error to apply **both** the projection **and** the learnt Mahalanobis distance.
# ### Neighbourhood graphs
# An alternative way to visualize the effect of using the distance found by LMNN together with kNN consists of using neighbourhood graphs. Despite the fancy name, these are actually pretty simple. The idea is just to construct a graph in the Euclidean space, where the points in the data set are the nodes of the graph, and a directed edge from one point to another denotes that the destination node is the 1-nearest neighbour of the origin node. Of course, it is also possible to work with neighbourhood graphs where $k \gt 1$. Here we have taken the simplification of $k = 1$ so that the forthcoming plots are not too cluttered.
# Let us define a data set for which the Euclidean distance performs considerably bad. In this data set there are several levels or layers in the y-direction. Each layer is populated by points that belong to the same class spread along the x-direction. The layers are close to each other in pairs, whereas the spread along x is larger. Let us define a function to generate such a data set and have a look at it.
# +
def sandwich_data():
# number of distinct classes
num_classes = 6
# number of points per class
num_points = 9
# distance between layers, the points of each class are in a layer
dist = 0.7
# memory pre-allocation
x = np.zeros((num_classes*num_points, 2))
y = np.zeros(num_classes*num_points)
for i,j in zip(range(num_classes), range(-num_classes//2, num_classes//2 + 1)):
for k,l in zip(range(num_points), range(-num_points//2, num_points//2 + 1)):
x[i*num_points + k, :] = np.array([np.random.normal(l, 0.1), np.random.normal(dist*j, 0.1)])
y[i*num_points:i*num_points + num_points] = i
return x,y
def plot_sandwich_data(x, y, axis=plt, cols=['r', 'b', 'g', 'm', 'k', 'y']):
for idx,val in enumerate(np.unique(y)):
xi = x[y==val]
axis.scatter(xi[:,0], xi[:,1], s=50, facecolors='none', edgecolors=cols[idx])
x, y = sandwich_data()
figure, axis = plt.subplots(1, 1, figsize=(5,5))
plot_sandwich_data(x, y, axis)
axis.set_aspect('equal')
axis.set_title('"Sandwich" toy data set')
axis.set_xlabel('x')
axis.set_ylabel('y')
plt.show()
# -
# Let the fun begin now! In the following block of code, we create an instance of a kNN classifier, compute the nearest neighbours using the Euclidean distance and, afterwards, using the distance computed by LMNN. The data set in the space result of the linear transformation given by LMNN is also shown.
# +
def plot_neighborhood_graph(x, nn, axis=plt, cols=['r', 'b', 'g', 'm', 'k', 'y']):
for i in range(x.shape[0]):
xs = [x[i,0], x[nn[1,i], 0]]
ys = [x[i,1], x[nn[1,i], 1]]
axis.plot(xs, ys, cols[int(y[i])])
feats = sg.features(x.T)
labels = sg.MulticlassLabels(y)
fig, axes = plt.subplots(1, 3, figsize=(15, 10))
# use k = 2 instead of 1 because otherwise the method nearest_neighbors just returns the same
# points as their own 1-nearest neighbours
k = 2
distance = sg.distance('EuclideanDistance')
distance.init(feats, feats)
knn = sg.machine("KNN",k=k, distance=distance, labels=labels)
knn.train(feats)
plot_sandwich_data(x, y, axes[0])
plot_neighborhood_graph(x, knn.get("nearest_neighbors"), axes[0])
axes[0].set_title('Euclidean neighbourhood in the input space')
lmnn = sg.LMNN(feats, labels, k)
# set a large number of iterations. The data set is small so it does not cost a lot, and this way
# we ensure a robust solution
lmnn.put('maxiter', 3000)
lmnn.train()
knn.put('distance', lmnn.get_distance())
plot_sandwich_data(x, y, axes[1])
plot_neighborhood_graph(x, knn.get("nearest_neighbors"), axes[1])
axes[1].set_title('LMNN neighbourhood in the input space')
# plot features in the transformed space, with the neighbourhood graph computed using the Euclidean distance
L = lmnn.get('linear_transform')
xl = np.dot(x, L.T)
feats = sg.features(xl.T)
dist = sg.distance('EuclideanDistance')
dist.init(feats, feats)
knn.put('distance', dist)
plot_sandwich_data(xl, y, axes[2])
plot_neighborhood_graph(xl, knn.get("nearest_neighbors"), axes[2])
axes[2].set_ylim(-3, 2.5)
axes[2].set_title('Euclidean neighbourhood in the transformed space')
[axes[i].set_xlabel('x') for i in range(len(axes))]
[axes[i].set_ylabel('y') for i in range(len(axes))]
[axes[i].set_aspect('equal') for i in range(len(axes))]
plt.show()
# -
# Notice how all the lines that go across the different layers in the left hand side figure have disappeared in the figure in the middle. Indeed, LMNN did a pretty good job here. The figure in the right hand side shows the disposition of the points in the transformed space; from which the neighbourhoods in the middle figure should be clear. In any case, this toy example is just an illustration to give an idea of the power of LMNN. In the next section we will see how after applying a couple methods for feature normalization (e.g. scaling, whitening) the Euclidean distance is not so sensitive against different feature scales.
# ## Real data sets
# ### Feature selection in metagenomics
# Metagenomics is a modern field in charge of the study of the DNA of microorganisms. The data set we have chosen for this section contains information about three different types of apes; in particular, gorillas, chimpanzees, and bonobos. Taking an approach based on metagenomics, the main idea is to study the DNA of the microorganisms (e.g. bacteria) which live inside the body of the apes. Owing to the many chemical reactions produced by these microorganisms, it is not only the DNA of the host itself important when studying, for instance, sickness or health, but also the DNA of the microorganisms inhabitants.
# First of all, let us load the ape data set. This data set contains features taken from the bacteria inhabitant in the gut of the apes.
ape_features = sg.features(sg.CSVFile(os.path.join(SHOGUN_DATA_DIR, 'multiclass/fm_ape_gut.dat')))
ape_labels = sg.labels(sg.CSVFile(os.path.join(SHOGUN_DATA_DIR, 'multiclass/label_ape_gut.dat')))
# It is of course important to have a good insight of the data we are dealing with. For instance, how many examples and different features do we have?
print('Number of examples = %d, number of features = %d.' % (ape_features.get("num_vectors"), ape_features.get("num_features")))
# So, 1472 features! Those are quite many features indeed. In other words, the feature vectors at hand lie on a 1472-dimensional space. We cannot visualize in the input feature space how the feature vectors look like. However, in order to gain a little bit more of understanding of the data, we can apply dimension reduction, embed the feature vectors in a two-dimensional space, and plot the vectors in the embedded space. To this end, we are going to use one of the [many](http://www.shogun-toolbox.org/doc/en/latest/classshogun_1_1EmbeddingConverter.html) methods for dimension reduction included in Shogun. In this case, we are using t-distributed stochastic neighbour embedding (or [t-dsne](http://jmlr.org/papers/v9/vandermaaten08a.html)). This method is particularly suited to produce low-dimensional embeddings (two or three dimensions) that are straightforward to visualize.
# +
def visualize_tdsne(features, labels):
converter = sg.transformer("TDistributedStochasticNeighborEmbedding",
target_dim=2, perplexity=25)
embedding = converter.transform(features)
x = embedding.get('feature_matrix')
y = labels.get('labels')
plt.scatter(x[0, y==0], x[1, y==0], color='green')
plt.scatter(x[0, y==1], x[1, y==1], color='red')
plt.scatter(x[0, y==2], x[1, y==2], color='blue')
plt.show()
visualize_tdsne(ape_features, ape_labels)
# -
# In the figure above, the green points represent chimpanzees, the red ones bonobos, and the blue points gorillas. Providing the results in the figure, we can rapidly draw the conclusion that the three classes of apes are somewhat easy to discriminate in the data set since the classes are more or less well separated in two dimensions. Note that t-dsne use randomness in the embedding process. Thus, the figure result of the experiment in the previous block of code will be different after different executions. Feel free to play around and observe the results after different runs! After this, it should be clear that the bonobos form most of the times a very compact cluster, whereas the chimpanzee and gorillas clusters are more spread. Also, there tends to be a chimpanzee (a green point) closer to the gorillas' cluster. This is probably a outlier in the data set.
# Even before applying LMNN to the ape gut data set, let us apply kNN classification and study how it performs using the typical Euclidean distance. Furthermore, since this data set is rather small in terms of number of examples, the kNN error above may vary considerably (I have observed variation of almost 20% a few times) across different runs. To get a robust estimate of how kNN performs in the data set, we will perform cross-validation using [Shogun's framework](http://www.shogun-toolbox.org/doc/en/latest/classshogun_1_1CrossValidation.html) for evaluation. This will give us a reliable result regarding how well kNN performs in this data set.
# +
# set up the classifier
knn = sg.machine("KNN", k=3, distance=sg.distance('EuclideanDistance'))
# set up 5-fold cross-validation
splitting = sg.splitting_strategy("StratifiedCrossValidationSplitting",
labels=ape_labels, num_subsets=5)
# evaluation method
evaluator = sg.evaluation("MulticlassAccuracy")
cross_validation = sg.machine_evaluation("CrossValidation",
machine=knn,
features=ape_features,
labels=ape_labels,
splitting_strategy=splitting,
evaluation_criterion=evaluator)
# number of experiments, the more we do, the less variance in the result
num_runs = 200
cross_validation.put('num_runs', num_runs)
# perform cross-validation and print the result!
result = cross_validation.evaluate()
print('kNN mean accuracy in a total of %d runs is %.4f.' % (num_runs, result.get('mean')))
# -
# Finally, we can say that KNN performs actually pretty well in this data set. The average test classification error is less than between 2%. This error rate is already low and we should not really expect a significant improvement applying LMNN. This ought not be a surprise. Recall that the points in this data set have more than one thousand features and, as we saw before in the dimension reduction experiment, only two dimensions in an embedded space were enough to discern arguably well the chimpanzees, gorillas and bonobos.
# Note that we have used stratified splitting for cross-validation. Stratified splitting divides the folds used during cross-validation so that the proportion of the classes in the initial data set is approximately maintained for each of the folds. This is particular useful in *skewed* data sets, where the number of examples among classes varies significantly.
# Nonetheless, LMNN may still turn out to be very useful in a data set like this one. Making a small modification of the vanilla LMNN algorithm, we can enforce that the linear transform found by LMNN is diagonal. This means that LMNN can be used to weight each of the features and, once the training is performed, read from these weights which features are relevant to apply kNN and which ones are not. This is indeed a form of *feature selection*. Using Shogun, it is extremely easy to switch to this so-called *diagonal* mode for LMNN: just call the method [`set_diagonal(use_diagonal)`](http://www.shogun-toolbox.org/doc/en/latest/classshogun_1_1LMNN.html#ad2f03dbad3ad08ab76aecbb656a486e6) with `use_diagonal` set to `True`.
# The following experiment takes about five minutes until it is completed (using Shogun Release, i.e. compiled with optimizations enabled). This is mostly due to the high dimension of the data (1492 features) and the fact that, during training, LMNN has to compute many outer products of feature vectors, which is a computation whose time complexity is proportional to the square of the number of features. For the illustration purposes of this notebook, in the following cell we are just going to use a small subset of all the features so that the training finishes faster.
# +
# to make training faster, use a portion of the features
fm = ape_features.get('feature_matrix')
ape_features_subset = sg.features(fm[:150, :])
# number of targer neighbours in LMNN, here we just use the same value that was used for KNN before
k = 3
lmnn = sg.LMNN(ape_features_subset, ape_labels, k)
lmnn.put('m_diagonal', True)
lmnn.put('maxiter', 1000)
init_transform = np.eye(ape_features_subset.get("num_features"))
lmnn.train(init_transform)
diagonal = np.diag(lmnn.get('linear_transform'))
print('%d out of %d elements are non-zero.' % (np.sum(diagonal != 0), diagonal.size))
# -
# So only 64 out of the 150 first features are important according to the result transform! The rest of them have been given a weight exactly equal to zero, even if all of the features were weighted equally with a value of one at the beginnning of the training. In fact, if all the 1472 features were used, only about 158 would have received a non-zero weight. Please, feel free to experiment using all the features!
# It is a fair question to ask how did we know that the maximum number of iterations in this experiment should be around 1200 iterations. Well, the truth is that we know this only because we have run this experiment with this same data beforehand, and we know that after this number of iterations the algorithm has converged. This is not something nice, and the ideal case would be if one could completely forget about this parameter, so that LMNN uses as many iterations as it needs until it converges. Nevertheless, this is not practical at least because of two reasons:
#
# - If you are dealing with many examples or with very high dimensional feature vectors, you might not want to wait until the algorithm converges and have a look at what LMNN has found before it has completely converged.
# - As with any other algorithm based on gradient descent, the termination criteria can be tricky. Let us illustrate this further:
statistics = lmnn.get_statistics()
plt.plot(statistics.obj.get())
plt.grid(True)
plt.xlabel('Number of iterations')
plt.ylabel('LMNN objective')
plt.show()
# Along approximately the first three hundred iterations, there is not much variation in the objective. In other words, the objective curve is pretty much flat. If we are not careful and use termination criteria that are not demanding enough, training could be stopped at this point. This would be wrong, and might have terrible results as the training had not clearly converged yet at that moment.
# In order to avoid disastrous situations, in Shogun we have implemented LMNN with really demanding criteria for automatic termination of the training process. Albeit, it is possible to tune the termination criteria using the methods [`set_stepsize_threshold`](http://www.shogun-toolbox.org/doc/en/latest/classshogun_1_1LMNN.html#a76b6914cf9d1a53b0c9ecd828c7edbcb) and [`set_obj_threshold`](http://www.shogun-toolbox.org/doc/en/latest/classshogun_1_1LMNN.html#af78c7dd9ed2307c0d53e7383cdc01a24). These methods can be used to modify the lower bound required in the step size and the increment in the objective (relative to its absolute value), respectively, to stop training. Also, it is possible to set a hard upper bound on the number of iterations using [`set_maxiter`](http://www.shogun-toolbox.org/doc/en/latest/classshogun_1_1LMNN.html#afcf319806eb710a0d9535fbeddf93795) as we have done above. In case the internal termination criteria did not fire before the maximum number of iterations was reached, you will receive a warning message, similar to the one shown above. This is not a synonym that the training went wrong; but it is strongly recommended at this event to have a look at the objective plot as we have done in the previous block of code.
# ### Multiclass classification
# In addition to feature selection, LMNN can be of course used for multiclass classification. I like to think about LMNN in multiclass classification as a way to empower kNN. That is, the idea is basically to apply kNN using the distance found by LMNN $-$ in contrast with using one of the other most common distances, such as the Euclidean one. To this end we will use the wine data set from the [UCI Machine Learning repository](http://archive.ics.uci.edu/ml/datasets/Wine "Wine data set").
# +
wine_features = sg.features(sg.CSVFile(os.path.join(SHOGUN_DATA_DIR, 'uci/wine/fm_wine.dat')))
wine_labels = sg.labels(sg.CSVFile(os.path.join(SHOGUN_DATA_DIR, 'uci/wine/label_wine.dat')))
assert(wine_features.get("num_vectors") == wine_labels.get("num_labels"))
print('%d feature vectors with %d features from %d different classes.' % (wine_features.get("num_vectors"), \
wine_features.get("num_features"), wine_labels.get("num_classes")))
# -
# First, let us evaluate the performance of kNN in this data set using the same cross-validation setting used in the previous section:
# +
# kNN classifier
k = 5
knn = sg.machine("KNN", k=k, distance=sg.distance("EuclideanDistance"))
splitting = sg.splitting_strategy("StratifiedCrossValidationSplitting",
labels=wine_labels, num_subsets=5)
evaluator = sg.evaluation("MulticlassAccuracy")
cross_validation = sg.machine_evaluation("CrossValidation",
machine=knn,
features=wine_features,
labels=wine_labels,
splitting_strategy=splitting,
evaluation_criterion=evaluator,
num_runs=200)
result = cross_validation.evaluate()
euclidean_means = np.zeros(3)
euclidean_means[0] = result.get('mean')
print('kNN accuracy with the Euclidean distance %.4f.' % result.get('mean'))
# -
# Seconly, we will use LMNN to find a distance measure and use it with kNN:
# +
# train LMNN
lmnn = sg.LMNN(wine_features, wine_labels, k)
lmnn.put('maxiter', 1500)
lmnn.train()
# evaluate kNN using the distance learnt by LMNN
knn.put("distance", lmnn.get_distance())
result = cross_validation.evaluate()
lmnn_means = np.zeros(3)
lmnn_means[0] = result.get('mean')
print('kNN accuracy with the distance obtained by LMNN %.4f.' % result.get('mean'))
# -
# The warning is fine in this case, we have made sure that the objective variation was really small after 1500 iterations. In any case, do not hesitate to check it yourself studying the objective plot as it was shown in the previous section.
# As the results point out, LMNN really helps here to achieve better classification performance. However, this comparison is not entirely fair since the Euclidean distance is very sensitive to the scaling that different feature dimensions may have, whereas LMNN can adjust to this during training. Let us have a closer look to this fact. Next, we are going to retrieve the feature matrix and see what are the maxima and minima for every dimension.
print('minima = ' + str(np.min(wine_features.get("feature_matrix"), axis=1)))
print('maxima = ' + str(np.max(wine_features.get("feature_matrix"), axis=1)))
# Examine the second and the last dimensions, for instance. The second dimension has values ranging from 0.74 to 5.8, while the values of the last dimension range from 278 to 1680. This will cause that the Euclidean distance works specially wrong in this data set. You can realize of this considering that the total distance between two points will almost certainly just take into account the contributions of the dimensions with largest range.
# In order to produce a more fair comparison, we will rescale the data so that all the feature dimensions are within the interval [0,1]. Luckily, there is a [preprocessor](http://www.shogun-toolbox.org/doc/en/latest/classshogun_1_1Preprocessor.html) class in Shogun that makes this straightforward.
# +
# preprocess features so that all of them vary within [0,1]
preprocessor = sg.transformer("RescaleFeatures")
preprocessor.fit(wine_features)
wine_features = preprocessor.transform(wine_features)
# sanity check
assert(np.min(wine_features.get("feature_matrix")) >= 0.0 and np.max(wine_features.get("feature_matrix")) <= 1.0)
# perform kNN classification after the feature rescaling
knn.put('distance', sg.distance("EuclideanDistance"))
result = cross_validation.evaluate()
euclidean_means[1] = result.get('mean')
print('kNN accuracy with the Euclidean distance after feature rescaling %.4f.' % result.get('mean'))
# train kNN in the new features and classify with kNN
lmnn.train()
knn.put('distance', lmnn.get_distance())
result = cross_validation.evaluate()
lmnn_means[1] = result.get('mean')
print('kNN accuracy with the distance obtained by LMNN after feature rescaling %.4f.' % result.get('mean'))
# -
# Another different preprocessing that can be applied to the data is called *whitening*. Whitening, which is explained in an [article in wikipedia](http://en.wikipedia.org/wiki/Whitening_transformation "Whitening transform"), transforms the covariance matrix of the data into the identity matrix.
# +
import scipy.linalg as linalg
# shorthand for the feature matrix -- this makes a copy of the feature matrix
data = wine_features.get('feature_matrix')
# remove mean
data = data.T
data-= np.mean(data, axis=0)
# compute the square of the covariance matrix and its inverse
M = linalg.sqrtm(np.cov(data.T))
# keep only the real part, although the imaginary that pops up in the sqrtm operation should be equal to zero
N = linalg.inv(M).real
# apply whitening transform
white_data = np.dot(N, data.T)
wine_white_features = sg.features(white_data)
# -
# The covariance matrices before and after the transformation can be compared to see that the covariance really becomes the identity matrix.
fig, axarr = plt.subplots(1,2)
axarr[0].matshow(np.cov(data))
axarr[1].matshow(np.cov(white_data))
plt.show()
# Finally, we evaluate again the performance obtained with kNN using the Euclidean distance and the distance found by LMNN using the whitened features.
# +
wine_features = wine_white_features
# perform kNN classification after whitening
knn.put("distance", sg.distance("EuclideanDistance"))
result = cross_validation.evaluate()
euclidean_means[2] = result.get('mean')
print('kNN accuracy with the Euclidean distance after whitening %.4f.' % result.get('mean'))
# train kNN in the new features and classify with kNN
lmnn.train()
knn.put('distance', lmnn.get_distance())
result = cross_validation.evaluate()
lmnn_means[2] = result.get('mean')
print('kNN accuracy with the distance obtained by LMNN after whitening %.4f.' % result.get('mean'))
# -
# As it can be seen, it did not really help to whiten the features in this data set with respect to only applying feature rescaling; the accuracy was already rather large after rescaling. In any case, it is good to know that this transformation exists, as it can become useful with other data sets, or before applying other machine learning algorithms.
# Let us summarize the results obtained in this section with a bar chart grouping the accuracy results by distance (Euclidean or the one found by LMNN), and feature preprocessing:
# +
assert(euclidean_means.shape[0] == lmnn_means.shape[0])
N = euclidean_means.shape[0]
# the x locations for the groups
ind = 0.5*np.arange(N)
# bar width
width = 0.15
figure, axes = plt.subplots()
figure.set_size_inches(6, 5)
euclidean_rects = axes.bar(ind, euclidean_means, width, color='y')
lmnn_rects = axes.bar(ind+width, lmnn_means, width, color='r')
# attach information to chart
axes.set_ylabel('Accuracies')
axes.set_ylim(top=1.4)
axes.set_title('kNN accuracy by distance and feature preprocessing')
axes.set_xticks(ind+width)
axes.set_xticklabels(('Raw', 'Rescaling', 'Whitening'))
axes.legend(( euclidean_rects[0], lmnn_rects[0]), ('Euclidean', 'LMNN'), loc='upper right')
def autolabel(rects):
# attach text labels to bars
for rect in rects:
height = rect.get_height()
axes.text(rect.get_x()+rect.get_width()/2., 1.05*height, '%.3f' % height,
ha='center', va='bottom')
autolabel(euclidean_rects)
autolabel(lmnn_rects)
plt.show()
# -
# ## References
# - Weinberger, K. Q., Saul, L. K. Distance Metric Learning for Large Margin Nearest Neighbor Classification. [(Link to paper in JMLR)](http://jmlr.org/papers/v10/weinberger09a.html).
| 60.272085
| 1,255
|
a615e166bc90f06819f8af24ce9760cc33cb3776
|
py
|
python
|
Cal2-PythonPandas.ipynb
|
dissina/introduction_to_stats
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# <center>
# <a href="http://www.insa-toulouse.fr/" ><img src="http://www.math.univ-toulouse.fr/~besse/Wikistat/Images/logo-insa.jpg" style="float:left; max-width: 120px; display: inline" alt="INSA"/></a>
#
# <a href="http://wikistat.fr/" ><img src="http://www.math.univ-toulouse.fr/~besse/Wikistat/Images/wikistat.jpg" style="max-width: 250px; display: inline" alt="Wikistat"/></a>
#
# <a href="http://www.math.univ-toulouse.fr/" ><img src="http://www.math.univ-toulouse.fr/~besse/Wikistat/Images/logo_imt.jpg" style="float:right; max-width: 250px; display: inline" alt="IMT"/> </a>
# </center>
# # <a href="https://www.python.org/"><img src="https://upload.wikimedia.org/wikipedia/commons/thumb/f/f8/Python_logo_and_wordmark.svg/390px-Python_logo_and_wordmark.svg.png" style="max-width: 200px; display: inline" alt="Python"/></a> [pour Statistique et Science des Données](https://github.com/wikistat/Intro-Python)
# # Trafic de données avec <a href="https://www.python.org/"><img src="https://upload.wikimedia.org/wikipedia/commons/thumb/f/f8/Python_logo_and_wordmark.svg/390px-Python_logo_and_wordmark.svg.png" style="max-width: 150px; display: inline" alt="Python"/></a> & <a href="http://pandas.pydata.org/"><img src="http://pandas.pydata.org/_static/pandas_logo.png" style="max-width: 250px; display: inline" alt="Pandas"/></a>
#
# **Résumé**: Utilisation de Python pour la préparation (*data munging* ou *wrangling* ou trafic) de données pas trop massives: qui tiennent en mémoire une fois réorganisées. Cette étape est abordée par l'initiation aux fonctionnalités de la librairie `pandas` et à la classe `DataFrame`; lire et écrire des fichiers, gérer une table de données et les types des variables, échantillonner, discrétiser, regrouper des modalités, description élémentaires uni et bi-variées; concaténation et jointure de tables.
# ## 1 Introduction
# ### 1.1 Objectifs
# Le *data munging* ou *wrangling* (traduit ici par *trafic*) de données est l'ensemble des opérations permettant de passer de données brutes à une table (*data frame*) correcte et adaptée aux objectifs à atteindre par des méthodes statistiques d'analyse, exploration, modélisation ou apprentissage.
#
# En présence de données complexes, peu ou mal organisées, présentant des trous, trop massives pour tenir en mémoire... la qualité de cette étape est fondamentale (*garbage in garbage out*) pour la bonne réalisation d'une étude. Compte tenu de la diversité des situations envisageables, il serait vain de vouloir exposer tous les outils et techniques qui peuvent s'avérer nécessaires. Tâchons néanmoins de résumer les problèmes qui peuvent être rencontrés.
# ### 1.2 Croissance du volume
# Le volume des données et sa croissance occasionnent schématiquement trois situations.
# 1. Le fichier initial des données brutes peut être chargé intégralement en mémoire moyennant éventuellement de sauter quelques colonnes ou lignes du fichier (cf. section 3.1). C'est la situation courante, tout logiciel statistique comme R peut réaliser les traitements. *C'est l'objet des sections 2 à 6*.
# 2. Le fichier initial est très volumineux mais la table (*DataFrame*), qui résulte de quelques trafics (*munging*) appropriés, tient en mémoire. Cette situations nécessite: lecture, analyse, transformation, ré-écriture, séquentielles du fichier ligne à ligne ou par bloc. Il existe des astuces avec R mais il est préférable d'utiliser des outils plus adaptés. Tout langage de programmation (java, c, perl, ruby...) peut être utilisé pour écrire le ou les programmes réalisant ce travail. Néanmoins Python, et plus précisément la librairie [`pandas`](http://pandas.pydata.org/), offrent un ensemble d'outils efficaces pour accomplir ces tâches sans avoir à ré-inventer la roue et ré-écrire tout un ensemble de fonctionnalités relativement basiques. Remarque : les procédures `univariate` et `freq` et l'étape `data` de SAS sont adaptées car elles ne chargent pas les données en mémoire pour réaliser des traitements rudimentaires. Néanmoins pour tout un tas de raisons trop longues à exposer, notamment de coût annuel de location, SAS perd régulièrement des parts de marché sur ce créneau. *Cette approche est introduite ci-dessous en section 7 et consiste à enchâsser dans une même structure itérative et séquentielle les étapes précédentes des sections 2 à 6*.
# 3. Lorsque les données, très massives, sont archivées sur un système de données distribuées (*Hadoop Distributed File System* ou HDFS), trafic et prétraitement des données doivent tenir compte de cet environnement. L'environnement *Spark* et l'API `PySpark` permettant de gérer en python des données distribuées est à favoriser. *Cf. le [calepin](http://wikistat.fr/Notebooks/Cal6-PythonSpark.html)* concerné.
#
# ### 1.3 Quelques problèmes
# Liste non exhaustive des problèmes pouvant être rencontrés et dont la résolution nécessite simultanément des compétences en Informatique, Statistique, Mathématiques et aussi "métier" du domaine de l'étude.
# - Identifier les "individus" $\times$ "variables" (*instances*$\times$*features* en langue informatique) de la table à mettre en forme à partir de bases de données variées; *i.e.* logs d'un site web, listes d'incidents, localisations...
# - Donnés atypiques (*outliers*): correction, suppression, transformation des variables ou méthode statistique robuste?
# - Variable qualitative avec beaucoup de modalités dont certaines très peu fréquentes: suppression, modalité `autres`, recodage aléatoire, regroupement "métier" ou méthode tolérante?
# - Distributions a-normales (log-normale, Poisson, multimodales...) et problèmes d'hétéroscédasticité: transformation, discrétisation ou méthodes tolérantes?
# - Données manquantes: suppressions (ligne ou colonne), imputation ou méthodes tolérantes ?
# - Représentations (splines, Fourier, ondelettes) et recalage (*time warping*) de données fonctionnelles.
# - Représentation de trajectoires, de chemins sur un graphe ?
# - Choix d'une distance (quadratique, absolue, géodésique...) entre les objets étudiés.
# - ...
# Bien entendu les "bons" choix dépendent directement de l'objectif poursuivi et des méthodes mises en oeuvre par la suite. D'où l'importance d'intégrer de façon précoce, dès la planification du recueil des données, les compétences statistiques nécessaires au sein d'une équipe.
# ### 1.4 Fonctionnalités de `pandas`
# La richesse des fonctionnalités de la librairie `pandas` est une des raisons, si ce n'est la principale, d'utiliser Python pour extraire, préparer, éventuellement analyser, des données. En voici un bref aperçu.
# - *Objets*: les classes `Series` et `DataFrame` ou *table de données*.
# - *Lire, écrire* création et exportation de tables de données à partir de fichiers textes (séparateurs, `.csv`, format fixe, compressés), binaires (HDF5 avec `Pytable`), HTML, XML, JSON, MongoDB, SQL...
# - *Gestion* d'une table: sélection des lignes, colonnes, transformations, réorganisation par niveau d'un facteur, discrétisation de variables quantitatives, exclusion ou imputation élémentaire de données manquantes, permutation et échantillonnage aléatoire, variables indicatrices, chaînes de caractères...
# - *Statistiques* élémentaires uni et bivariées, tri à plat (nombre de modalités, de valeurs nulles, de valeurs manquantes...), graphiques associés, statistiques par groupe, détection élémentaire de valeurs atypiques...
# - *Manipulation* de tables: concaténations, fusions, jointures, tri, gestion des types et formats...
# ### 1.5 Références
# Ce tutoriel élémentaire s'inspire largement du livre de référence (Mc Kinney, 2013) et de la [documentation en ligne](http://pandas.pydata.org/pandas-docs/stable/) à consulter sans modération. Cette documentation inclut également des [tutoriels](http://pandas.pydata.org/pandas-docs/stable/tutorials.html) à exécuter pour compléter et approfondir la première ébauche d'un sujet relativement technique et qui peut prendre des tournures très diverses en fonction de la qualité et des types de données traitées.
# ### 1.6 Exemple
# Les données choisies pour illustrer ce tutoriel sont issues d'une compétition du site [Kaggle](https://www.kaggle.com/): [Titanic: Machine learnic from Disaster](https://www.kaggle.com/c/titanic-gettingStarted). Le concours est terminé mais les [données](https://www.kaggle.com/c/titanic-gettingStarted/data) sont toujours disponibles sur le site avec des tutoriels utilisant Excel, Python ou R.
#
# Une des raisons du drame, qui provoqua la mort de 1502 personnes sur les 2224 passagers et membres d'équipage, fut le manque de canots de sauvetage. Il apparaît que les chances de survie dépendaient de différents facteurs (sexe, âge, classe...). Le but du concours est de construire un modèle de prévision (classification supervisée) de survie en fonction de ces facteurs. Les données sont composées d'un échantillon d'apprentissage (891) et d'un échantillon test (418) chacun décrit par 11 variables dont la première indiquant la survie ou non lors du naufrage.
# Liste des variables
#
# Label | Intitulé
# ----------|-------------
# survival | Survival (0 = No; 1 = Yes)
# pclass | Passenger Class (1 = 1st; 2 = 2nd; 3 = 3rd)
# name | Name
# sex | Sex
# age | Age
# sibsp | Number of Siblings/Spouses Aboard
# parch | Number of Parents/Children Aboard
# ticket | Ticket Number
# fare | Passenger Fare
# cabin | Cabin
# embarked | Port of Embarkation (C = Cherbourg; Q = Queenstown; S = Southampton)
# ## 2 Les classes `Series` et `DataFrame`
# De même que la librairie `Numpy` introduit le type `array` indispensable à la manipulation de matrices en calcul scientifique, celle `pandas` introduit les classes `Series` (séries chronologiques) et `DataFrame` ou table de données indispensables en statistique.
#
# ### 2.1 *Series*
# La classe `Series` est l'association de deux `arrays` unidimensionnels. Le premier est un ensemble de valeurs indexées par le 2ème qui est souvent une série temporelle. Ce type est introduit principalement pour des applications en Econométrie et Finance où Python est largement utilisé.
# ### 2.2 *DataFrame*
# Cette classe est proche de celle du même nom dans le langage R, il s'agit d'associer avec le même index de lignes des colonnes ou variables de types différents (entier, réel, booléen, caractère). C'est un tableau bi-dimensionnel avec des index de lignes et de colonnes mais il peut également être vu comme une liste de `Series` partageant le même index. L'index de colonne (noms des variables) est un objet de type `dict` (dictionnaire). C'est la classe qui sera principalement utilisée dans ce tutoriel.
# Exemple de data frame
import pandas as pd
data = {"state": ["Ohio", "Ohio", "Ohio",
"Nevada", "Nevada"],
"year": [2000, 2001, 2002, 2001, 2002],
"pop": [1.5, 1.7, 3.6, 2.4, 2.9]}
frame = pd.DataFrame(data)
# ordre des colonnes
pd.DataFrame(data, columns=["year", "state", "pop"])
# index des lignes et valeurs manquantes (NaN)
frame2=pd.DataFrame(data, columns=["year", "state", "pop", "debt"],
index=["one", "two", "three", "four", "five"])
# liste des colonnes
frame.columns
# valeurs d'une colonnes
frame["state"]
frame.year
# "imputation"
frame2["debt"] = 16.5
frame2
# créer une variable
frame2["eastern"] = frame2.state == "Ohio"
frame2
frame2.columns
# supprimer une variable
del frame2[u"eastern"]
frame2.columns
# ### 2.3 Index
# Les index peuvent être définis par emboîtement et beaucoup d'autres fonctionnalités sur la gestion des index sont décrites par Mac Kinney (2013) (chapitre 5):
# - `append` nouvel index par concaténation,
# - `diff` différence ensembliste,
# - `intersection` intersection ensembliste,
# - `union` union ensembliste
# - `isin` vrai si la valeur est dans la liste,
# - `delete` suppression de l'index $i$,
# - `drop` suppression d'une valeur d'index,
# - `is_monotonic` vrai si les valeurs sont croissantes,
# - `is_unique` vrai si toutes les valeurs sont différentes,
# - `nique` tableau des valeurs uniques de l'index.
# ## 3 Lire écrire des tables de données
# `Pandas` offre des outils efficaces pour lire écrire des fichiers selon différents formats (csv, texte, fixe, compressé, xml, html, hdf5) ou interagir avec des bases de données SQL, MongoDB, des APIs web. Ce document se contente de décrire les fonctions les plus utiles `read_csv` et `read_table` pour lire des fichiers textes et générer un objet de classe ` DataFrame`.
#
# En principe ces fonctions font appel à un code écrit en C dont très rapide à l'exécution sauf pour l'emploi de certaines options (`skip\_footer, sep`} autre qu'un seul caractère), à éviter, qui provoquent une exécution en Python (`engine=Python`).
#
# La réciproque pour l'écriture est obtenue par les commandes `data.to_csv` ou `_table` avec des options similaires.
#
# ### 3.1 Syntaxe
# L'exemple de base est donné pour lire un fichier au format `.csv` dont les valeurs sont séparées par des "," et dont la première ligne contient le nom des variables.
# ``
# import pandas as pd
# data=pd.read_csv("fichier.csv")
# data=pd.read_table("fichier.csv", sep=",")
# ``
#
# Il est important de connaître la liste des possibilités et options offertes par cette simple commande. Voici les principales ci-dessous et un lien à la [liste complète](http://pandas.pydata.org/pandas-docs/stable/io.html#io-read-csv-table).
# - `path` chemin ou non du fichier ou URL.
# - `sep` délimiteur comme \verb+ , ; | \t + ou \verb# \s+ # pour un nombre variable d'espaces.
# - `header` défaut 0, la première ligne contient le nom des variables; si {\tt None} les noms sont générés ou définis par ailleurs.
# - `index_col` noms ou numéros de colonnes définissant les index de lignes, index pouvant être hiérarchisés comme les facteurs d'un plan d'expérience.
# - `names` si {\tt header=None}, liste des noms des variables.
# - `nrows` utile pour tester et limiter le nombre de ligne à lire.
# - `skiprow` liste de lignes à sauter en lecture.
# - `skip_footer` nombre de lignes à sauter en fin de fichier.
# - `na_values` définition du ou des codes signalant des valeurs manquantes. Ils peuvent être définis dans un dictionnaire pour associer variables et codes de valeurs manquantes spécifiques.
# - `usecols` sélectionne une liste des variable à lire pour éviter de lire des champs ou variables volumineuses et inutiles.
# - `skip_blan_lines` à `True` pour sauter les lignes blanches.
# - `converters` appliquer une fonction à une colonne ou variable.
# - `day_first` par défaut `False`, pour des dates françaises au format `7/06/2013`.
# - `chunksize` taille des morceaux à lire itérativement.
# - `verbose` imprime des informations comme le nombre de valeurs manquantes des variables non numériques.
# - `encoding` type d'encodage comme "utf-8" ou "latin-1"
# - `thousand` séparateur des miliers: "." ou ",".
#
# Remarques:
# - De nombreuses options de gestion des dates et séries ne sont pas citées.
# - `chunksize` provoque la lecture d'un gros fichiers par morceaux de même taille (nombre de lignes). Des fonctions (comptage, dénombrement...) peuvent ensuite s'appliquer itérativement sur les morceaux.
# \end{itemize}
#
# ### 3.2 Exemple
# Les données du naufrage du Titanic illustrent l'utilisation de `pandas`. Elles sont lues directement à partir de leur URL ou sinon les charger [ici](http://www.math.univ-toulouse.fr/~besse/Wikistat/data) vers le répertoire de travail de Python.
# Importations
import pandas as pd
import numpy as np
# tester la lecture
# path=""
path='http://www.math.univ-toulouse.fr/~besse/Wikistat/data/'
df = pd.read_csv(path+'titanic-train.csv',nrows=5)
df
df.tail()
# tout lire
df = pd.read_csv(path+"titanic-train.csv")
df.head()
# Des variables sont inexploitables
# Choisir les colonnes utiles
df=pd.read_csv(path+"titanic-train.csv",
usecols=[1,2,4,5,6,7,9,11],nrows=5)
df.head()
# À partir de la version 0.15, `pandas`, inclut un type `category` assez proche de celui ` factor` de R. Il devrait normalement être déclaré dans un dictionnaire au moment par exemple de la lecture (`dtype={"Surv":pd.Categorical...}`) mais ce n'est pas le cas, c'est donc le type objet qui est déclaré puis modifié. Il est vivement recommandé de bien affecter les bons types à chaque variable ne serait-ce que pour éviter de faire des opérations douteuses, par exemple arithmétiques sur des codes de modalités.
df=pd.read_csv(path+"titanic-train.csv",skiprows=1,header=None,usecols=[1,2,4,5,9,11],
names=["Surv","Classe","Genre","Age","Prix","Port"],dtype={"Surv":object,
"Classe":object,"Genre":object,"Port":object})
df.head()
df.dtypes
# Redéfinition des bons types.
df["Surv"]=pd.Categorical(df["Surv"],ordered=False)
df["Classe"]=pd.Categorical(df["Classe"],ordered=False)
df["Genre"]=pd.Categorical(df["Genre"],ordered=False)
df["Port"]=pd.Categorical(df["Port"],ordered=False)
df.dtypes
# Remarque: il est également possible de tout lire avant de laisser "tomber" les variable inexploitables. C'est le rôle de la commande:
#
# `df = df.drop(["Name", "Ticket", "Cabin"], axis=1)`
# ### 3.3 Echantillonnage simple
# Comme dans R, le type `DataFrame` de Python est chargé en mémoire. Si, malgré les options précédentes permettant de sélectionner, les colonnes, les types des variables... le fichier est encore trop gros, il reste possible, avant de chercher une configuration matérielle lourde et en première approximation, de tirer un échantillon aléatoire simple selon une distribution uniforme. Un tirage stratifié demanderait plus de travail. Cela suppose de connaître le nombre de ligne du fichier ou une valeur inférieure proche.
# pour les données titanic:
N=891 # taille du fichier
n=200 # taille de l'échantillon
lin2skipe=[0] # ne pas lire la première ligne
# ne pas lire N-n lignes tirées aléatoirement
lin2skipe.extend(np.random.choice(np.arange(1,N+1),
(N-n),replace=False))
df_small=pd.read_csv(path+"titanic-train.csv",
skiprows=lin2skipe,header=None,
usecols=[1,2,4,5,9,11],
names=["Surv","Classe","Genre","Age",
"Prix","Port"])
df_small
# ## 3 Gérer une table de données
# ### 3.1 Discrétisation d'une variable quantitative
# Pour la discrétisation d'une variable quantitative. Il est d'un bon usage de définir les bornes des classes à des quantiles, plutôt qu'également espacées, afin de construire des classes d'effectifs sensiblement égaux. Ceci est obtenu par la fonction `qcut`. La fonction `cut` propose par défaut des bornes équi-réparties à moins de fournir une liste de ces bornes.
df["AgeQ"]=pd.qcut(df.Age,3,labels=["Ag1","Ag2",
"Ag3"])
df["PrixQ"]=pd.qcut(df.Prix,3,labels=["Pr1","Pr2",
"Pr3"])
df["PrixQ"].describe()
# ### 3.2 Modifier / regrouper des modalités
# Le recodage des variables qualitatives ou renommage en clair des modalités est obtenu simplement.
df["Surv"]=df["Surv"].cat.rename_categories(
["Vnon","Voui"])
df["Classe"]=df["Classe"].cat.rename_categories(
["Cl1","Cl2","Cl3"])
df["Genre"]=df["Genre"].cat.rename_categories(
["Gfem","Gmas"])
df["Port"]=df["Port"].cat.rename_categories(
["Pc","Pq","Ps"])
df.head()
# Il est possible d'associer recodage et regroupement des modalités en définissant un dictionnaire de transformation.
data = pd.DataFrame({"food":["bacon","pulled pork",
"bacon", "Pastrami",
"corned beef", "Bacon", "pastrami", "honey ham",
"nova lox"],
"ounces": [4, 3, 12, 6, 7.5, 8, 3, 5, 6]})
data
meat_to_animal = {
"bacon": "pig",
"pulled pork": "pig",
"pastrami": "cow",
"corned beef": "cow",
"honey ham": "pig",
"nova lox": "salmon"
}
# Eviter les mélanges de majuscules minuscules
# en mettant tout en minuscule
data["animal"] = data["food"].map(
str.lower).map(meat_to_animal)
data
data["food"].map(lambda x: meat_to_animal[x.lower()])
dfs = pd.DataFrame({"key": ["b", "b", "a", "c",
"a", "b"],"data1": range(6)})
pd.get_dummies(dfs["key"])
# ### 3.3 Variables indicatrices
# Générer des indicatrices des modalités ou *dummy variables*.
dummies = pd.get_dummies(dfs['key'], prefix='key')
df_with_dummy = dfs[['data1']].join(dummies)
df_with_dummy
# ### 3.4 Permutation et tirage aléatoires
# Permutation aléatoire:
dfs = pd.DataFrame(np.arange(5 * 4).reshape(5, 4))
sampler = np.random.permutation(5)
sampler
dfs
dfs.take(sampler)
# Tirage aléatoire avec remplacement ou \emph{bootstrap} ; celui sans remplacement est traité section 3.3.
bag = np.array([5, 7, -1, 6, 4])
sampler = np.random.randint(0, len(bag), size=10)
draws = bag.take(sampler)
draws
# ### 3.5 Transformations, opérations
# Les opérations arithmétiques entre `Series` et `DataFrame` sont possibles au même titre qu'entre `array`. Si les index ne correspondent pas, des valeurs manquantes (NAN) sont créées à moins d'utiliser des méthodes d'arithmétique `flexible` (`add, sub, div, mul`) autorisant la complétion par une valeur par défaut, généralement 0.
#
# Une fonction quelconque (`lambda`) peut être appliquée avec une même commande qu'`apply` de R.
# la table de données
frame = pd.DataFrame(np.random.randn(4,3),
columns=list("bde"),
index=["Utah", "Ohio", "Texas", "Oregon"])
# une fonction
f = lambda x: x.max() - x.min()
frame.apply(f, axis=1)
# ### 3.6 Tri et rangs
# Trier une table selon les valeurs d'une variable ou d'un index.
frame = pd.DataFrame(np.arange(8).reshape((2,4)),
index=["three", "one"],
columns=["d", "a", "b", "c"])
frame.sort_index()
frame.sort_index(axis=1)
frame.sort_index(axis=1, ascending=False)
frame.sort_values(by="b")
# La commande `rank` remplace les valeurs par leur rang dans l'ordre des lignes ou des colonnes.
frame = pd.DataFrame({"b": [4.3, 7, -3, 2],
"a": [0, 1, 0, 1],"c": [-2, 5, 8, -2.5]})
frame.rank(axis=1)
frame.rank(axis=0)
# ## 4 Statistiques descriptives élémentaires
# Continuer l'étude des données sur le naufrage du Titanic. Les commandes ci-dessous permettent des premiers diagnostics sur la qualité des données.
# ### 4.1 Description univariée
df.dtypes
df.describe()
df.head()
import matplotlib.pyplot as plt
# %matplotlib inline
df["Age"].hist()
plt.show()
df["Age"].plot(kind="box")
plt.show()
df["Prix"].plot(kind="hist")
plt.show()
# qualitatif
df["Surv"].value_counts()
df["Classe"].value_counts()
df["Genre"].value_counts()
df["Port"].value_counts()
# ### 4.2 Description bivariée
df.plot(kind="scatter",x="Age",y="Prix")
plt.show()
# afficher une sélection
df[df["Age"]>60][["Genre","Classe","Age","Surv"]]
df.boxplot(column="Age",by="Classe")
plt.show()
df.boxplot(column="Prix",by="Surv")
plt.show()
# table de contingence
table=pd.crosstab(df["Surv"],df["Classe"])
print(table)
# Mosaic plot
from statsmodels.graphics.mosaicplot import mosaic
mosaic(df,["Classe","Genre"])
plt.show()
from statsmodels.graphics.mosaicplot import mosaic
mosaic(df,["Surv","Classe"])
plt.show()
# ### 4.3 Imputation de données manquantes
# La gestion des données manquantes est souvent un point délicat. De nombreuses stratégies ont été élaborées, les principales sont décrites dans une [vignette](http://wikistat.fr/pdf/st-m-app-idm.pdf). Nous ne décrivons ici que les plus élémentaires à [mettre en oeuvre](http://pandas.pydata.org/pandas-docs/version/0.15.2/missing_data.html) avec `pandas`.
#
# Il est ainsi facile de supprimer toutes les observations présentant des données manquantes lorsque celles-ci sont peu nombreuses et majoritairement regroupées sur certaines lignes ou colonnes.
#
# ``
# df = df.dropna(axis=0)
# df = df.dropna(axis=1)
# ``
#
# `Pandas` permet également de faire le choix pour une variable qualitative de considérer ` `np.nan` comme une modalité spécifique ou d'ignorer l'observation correspondante.
#
# Autres stratégies:
# * Cas quantitatif: une valeur manquante est imputée par la moyenne ou la médiane.
# * Cas d'une série chronologique: imputation par la valeur précédente ou suivante ou par interpolation linéaire, polynomiale ou encore lissage spline.
# * Cas qualitatif: modalité la plus fréquente ou répartition aléatoire selon les fréquences observées des modalités.
#
# La variable âge contient de nombreuses données manquantes. La fonction `fillna` présente plusieurs options d'imputation.
# Remplacement par la médiane d'une variable quantitative
df=df.fillna(df.median())
df.describe()
# par la modalité "médiane" de AgeQ
df.info()
df.AgeQ=df["AgeQ"].fillna("Ag2")
# par le port le plus fréquent
df["Port"].value_counts()
df.Port=df["Port"].fillna("Ps")
df.info()
# Ces imputations sont pour le moins très rudimentaires et d'autres sont à privilégier pour des modélisations plus soignées mais ces méthodes font généralement appel à R.
#
# D'autres fonctions (Mac Kinney, 2013) sont proposées pour supprimer les duplicatas (` drop\_duplicates`), modifier les dimensions, traquer des atypiques unidimensionnels selon un modèle gaussien ou par rapport à des quantiles.
# ## 5 Manipuler des tables de données
# ### 5.1 Jointure
# Il s'agit de "jointer" deux tables partageant la même clef ou encore de concaténer horizontalement les lignes en faisant correspondre les valeurs d'une variable clef qui peuvent ne pas être uniques.
# tables
df1 = pd.DataFrame({"key": ["b", "b", "a", "c",
"a","a", "b"],"data1": range(7)})
df2 = pd.DataFrame({"key": ["a", "b", "d"],
"data2": range(3)})
pd.merge(df1,df2,on="key")
# La gestion des clefs manquantes est en option: entre autres, ne pas introduire de ligne (ci-dessus), insérer des valeurs manquantes ci-dessous.
# valeurs manquantes
pd.merge(df1,df2,on="key", how="outer")
# ### 5.2 Concaténation selon un axe
# Concaténation verticale (axis=0) ou horizontales (axis=1) de tables. La concaténation horizontale est similaire à la jointure (option `outer`).
# tables
df1 = pd.DataFrame({"key": ["b", "b", "a", "c",
"a", "a", "b"],"var": range(7)})
df2 = pd.DataFrame({"key": ["a", "b", "d"],
"var": range(3)})
# concaténation verticales
pd.concat([df1,df2],axis=0)
# concaténation horizontale
pd.concat([df1,df2],axis=1)
# ## 6 Trafic séquentiel de gros fichiers
# Étape suivante associée de la croissance du volume: les fichiers des données brutes ne tiennent pas en mémoire. Il "suffit" d'intégrer ou enchâsser les étapes des sections précédentes dans la lecture séquentielle d'un gros fichier. En apparence, simple d'un point de vue méthodologique, cette étape peut consommer beaucoup de temps par tests et remises en cause incessantes des choix de sélection, transformation, recodage... des variables. Il est crucial de se doter d'outils efficaces.
#
# Il s'agit donc de lire les données par morceau (nombre fixé de lignes) ou ligne à ligne, traiter chaque morceau, le ré-écrire dans un fichier de format binaire plutôt que texte; le choix du format HDF5 semble le plus efficace du point de vue technique et pour servir d'interface à d'autres environnements: C, java, Matlab... et R car une librairie ({\tt rhdf5} de Bioconductor) gère ce format.
#
# La procédure est comparable à une étape `Data` de SAS, qui lit/écrit les tables ligne à ligne.
#
# Deux librairies: `h5py` et `PyTables` gèrent le format HDF5 en Python. Pour simplifier la tâche, `pandas` intègre une classe `HDFStore` utilisant `PyTables` qui doit donc être installée.
#
# **Attention**: ce format n'est pas adapté à une gestion *parallélisée*, notamment en écriture.
#
#
# ### 6.1 Lecture séquentielle
# L'exemple est ici donné pour lire un fichier texte mais beaucoup d'autres formats (excel, hdf, sql, json, msgpack, html, gbq, stata, clipboard, pickle) sont connus de `pandas`.
# importations
import pandas as pd
import numpy as np
# lire tout le fichier par morceaux
# avec l'option chunksize
Partition=pd.read_csv(path+"titanic-train.csv",skiprows=1,
header=None,usecols=[1,2,4,5,9,11],
names=["Surv","Classe","Genre","Age",
"Prix","Port"],dtype={"Surv":object,
"Classe":object,"Genre":object,"Port":object},
chunksize=100)
# ouverture du fichier HDF5
stock=pd.HDFStore("titan.h5")
# boucle de lecture
for Part in Partition:
# "nettoyage" préliminaire des données
#Part=Part.drop(["Name","Ticket","Cabin"],axis=1)
# ... autres opérations
# création de la table "df" dans "stock" puis
# extension de celle-ci par chaque "Part"
stock.append("df",Part)
# dernier morceau lu et ajouté
Part.head()
# Il est généralement utile de fermer le fichier
stock.close()
# **Attention** aux types implicites des variables. Si, par exemple, une donnée manquante n'apparaît pas dans une colonne du 1er morceau mais dans le 2ème, cela peut engendrer un conflit de type. Expliciter systématiquement les types et noms des variables dans un dictionnaire en paramètre.
#
# ### 6.2 Utilisation d'une table HDF5
# Ouverture du fichier
Archiv=pd.HDFStore("titan.h5")
# sélection de la table et affichage de l'entête
Archiv.select("df").head()
# Cette partie est à développer pour illustrer les fonctionnalités de `pandas` permettant d'interroger / requêter (*querying* notamment SQL) une table archivée dans un fichier HDF5. Consulter la [documentation en ligne](http://pandas.pydata.org/pandas-docs/dev/io.html#hdf5-pytables) à ce sujet.
#
# ### 6.3 Echantillon aléatoire simple
# Le fichier créé au format HDF5 peut être encore très volumineux. Par souci d'efficacité, son raffinement, son exploitation, voire même son analyse pour modélisation, peuvent ou même doivent être opérés sur un simple échantillon aléatoire.
# extraction du nombre de lignes / individus
nrows = Archiv.get_storer("df").nrows
# génération des index aléatoires
r = np.random.randint(0,nrows,size=10)
print(r)
# extraction des lignes d'index fixés
df_ech=Archiv.select("df",where=pd.Index(r))
df_ech
# Il "suffit" alors d'appliquer les outils des sections 4 à 6 précédentes.
# ### 6.4 Echanges entre R et Pyhton
# Les données ayant été préparées, nettoyées, le transfert de la table dans R permet de déployer toute la richesse des librairies développées dans cet environnement plus familier au statisticien. Il est possible d'appeler des commandes R à partir de Python avec la librairie `rpy2` et réciproquement d'appeler des commandes Python de R avec la librairie `rpython`.
#
# La librairie `rpy2` définit la commande `load_data` qui charge un `data.frame` de R dans un `DataFrame` tandis que celle `convert_to_r_dataframe` génère un objet R.
#
# Le plus efficace mais sans doute pas le plus simple, consisterait à lire directement, à partir de R, le fichier intermédiaire précédent au format binaire HDF5 en utilisant la librairie ` rhdf5` de Bioconductor. Cette démarche pose des problèmes pour la gestions des variables qualitatives et plus généralement celle de la classe `DataFrame`. Une alternative simple consiste à construire un fichier intermédiaire au format classique `.csv`.
#
#
# ## À suivre...
# Ces traitements font appel à de très nombreuses opérations de lectures / écritures sur un seul ordinateur, un seul disque au regard du volume des calculs; ils ne sont pas adaptés à une parallélisation sur un ordinateur multiprocesseur. La gestion et l'analyse de plus gros volumes de données nécessite une distribution de celles-ci sur plusieurs serveurs / disques. D'autres technologies doivent être utilisées; c'est actuellement le couple *Spark/Hadoop* le plus en vogue.
#
# **Intérêt**: *Spark* est utilisable avec java, Scala et aussi Python. L'investissement dans ce langage est donc rentable.
# ## Références
#
# **Mac Kinney W.** (2013). *Python for Data Analysis*, O’Reilly. [pdf](http://it-ebooks.info/book/104)
| 56.069892
| 1,264
|
e8b5fd15891bdf8ecaaaaadbf9d632278a7f9aaa
|
py
|
python
|
BERT_Paper_Replication/BERT_biLSTM.ipynb
|
hafezgh/Hate-Speech-Detection-in-Social-Media
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] colab_type="text" id="view-in-github"
# <a href="https://colab.research.google.com/github/hafezgh/Hate-Speech-Detection-in-Social-Media/blob/main/BERT_Paper_Replication/BERT_biLSTM.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="s4ga5LLSSPQN"
# # Imports
# + colab={"base_uri": "https://localhost:8080/"} id="lqOFKm6QLoua" outputId="aac8984b-11c8-48fd-cbc7-068040708a10"
# !pip install transformers==3.0.0
# !pip install emoji
import gc
import os
import emoji as emoji
import re
import string
import numpy as np
import pandas as pd
import torch
import torch.nn as nn
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report, accuracy_score, confusion_matrix, ConfusionMatrixDisplay
from transformers import AutoModel
from transformers import BertModel, BertTokenizer
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings('ignore')
# + colab={"base_uri": "https://localhost:8080/"} id="f3lx2XKTSOn6" outputId="ac08fc25-b434-4cbe-b68b-dcf2ceaf1dce"
# !git clone https://github.com/hafezgh/Hate-Speech-Detection-in-Social-Media
# + [markdown] id="7tylYXo4SRoi"
# # Model
# + id="CUcU4tEVLvun"
class BERT_Arch(nn.Module):
def __init__(self, bert, mode='cnn'):
super(BERT_Arch, self).__init__()
self.bert = BertModel.from_pretrained('bert-base-uncased')
self.mode = mode
if mode == 'cnn':
# CNN
self.conv = nn.Conv2d(in_channels=13, out_channels=13, kernel_size=(3, 768), padding='valid')
self.relu = nn.ReLU()
# change the kernel size either to (3,1), e.g. 1D max pooling
# or remove it altogether
self.pool = nn.MaxPool2d(kernel_size=(3, 1), stride=1)
self.dropout = nn.Dropout(0.1)
# be careful here, this needs to be changed according to your max pooling
# without pooling: 443, with 3x1 pooling: 416
# FC
self.fc = nn.Linear(416, 3)
self.flat = nn.Flatten()
elif mode == 'rnn':
### RNN
self.lstm = nn.LSTM(768, 256, batch_first=True, bidirectional=True)
## FC
self.fc = nn.Linear(256*2, 3)
elif mode == 'shallow_fc':
self.fc = nn.Linear(768, 3)
elif mode == 'deep_fc':
self.leaky_relu = nn.LeakyReLU(0.01)
self.fc1 = nn.Linear(768, 768)
self.dropout1 = nn.Dropout(0.1)
self.fc2 = nn.Linear(768, 768)
self.dropout2 = nn.Dropout(0.1)
self.fc3 = nn.Linear(768, 3)
else:
raise NotImplementedError("Unsupported extension!")
self.softmax = nn.LogSoftmax(dim=1)
def forward(self, sent_id, mask):
sequence_output, _, all_layers = self.bert(sent_id, attention_mask=mask, output_hidden_states=True)
if self.mode == 'cnn':
x = torch.transpose(torch.cat(tuple([t.unsqueeze(0) for t in all_layers]), 0), 0, 1)
x = self.pool(self.dropout(self.relu(self.conv(self.dropout(x)))))
x = self.fc(self.dropout(self.flat(self.dropout(x))))
elif self.mode == 'rnn':
lstm_output, (h,c) = self.lstm(sequence_output)
hidden = torch.cat((lstm_output[:,-1, :256],lstm_output[:,0, 256:]),dim=-1)
x = self.fc(hidden.view(-1,256*2))
elif self.mode == 'shallow_fc':
x = self.fc(sequence_output[:,0,:])
elif self.mode == 'deep_fc':
x = self.fc1(sequence_output[:,0,:])
x = self.leaky_relu(x)
x = self.dropout1(x)
x = self.fc2(x)
x = self.leaky_relu(x)
x = self.dropout2(x)
x = self.fc3(x)
else:
raise NotImplementedError("Unsupported extension!")
gc.collect()
torch.cuda.empty_cache()
del all_layers
c = self.softmax(x)
return c
def read_dataset():
data = pd.read_csv("/content/Hate-Speech-Detection-in-Social-Media/Irirs_kedro/get-started/data/01_raw/labeled_data.csv")
data = data.drop(['count', 'hate_speech', 'offensive_language', 'neither'], axis=1)
print(len(data))
return data['tweet'].tolist(), data['class']
def pre_process_dataset(values):
new_values = list()
# Emoticons
emoticons = [':-)', ':)', '(:', '(-:', ':))', '((:', ':-D', ':D', 'X-D', 'XD', 'xD', 'xD', '<3', '</3', ':\*',
';-)',
';)', ';-D', ';D', '(;', '(-;', ':-(', ':(', '(:', '(-:', ':,(', ':\'(', ':"(', ':((', ':D', '=D',
'=)',
'(=', '=(', ')=', '=-O', 'O-=', ':o', 'o:', 'O:', 'O:', ':-o', 'o-:', ':P', ':p', ':S', ':s', ':@',
':>',
':<', '^_^', '^.^', '>.>', 'T_T', 'T-T', '-.-', '*.*', '~.~', ':*', ':-*', 'xP', 'XP', 'XP', 'Xp',
':-|',
':->', ':-<', '$_$', '8-)', ':-P', ':-p', '=P', '=p', ':*)', '*-*', 'B-)', 'O.o', 'X-(', ')-X']
for value in values:
# Remove dots
text = value.replace(".", "").lower()
text = re.sub(r"[^a-zA-Z?.!,¿]+", " ", text)
users = re.findall("[@]\w+", text)
for user in users:
text = text.replace(user, "<user>")
urls = re.findall(r'(https?://[^\s]+)', text)
if len(urls) != 0:
for url in urls:
text = text.replace(url, "<url >")
for emo in text:
if emo in emoji.UNICODE_EMOJI:
text = text.replace(emo, "<emoticon >")
for emo in emoticons:
text = text.replace(emo, "<emoticon >")
numbers = re.findall('[0-9]+', text)
for number in numbers:
text = text.replace(number, "<number >")
text = text.replace('#', "<hashtag >")
text = re.sub(r"([?.!,¿])", r" ", text)
text = "".join(l for l in text if l not in string.punctuation)
text = re.sub(r'[" "]+', " ", text)
new_values.append(text)
return new_values
def data_process(data, labels):
input_ids = []
attention_masks = []
bert_tokenizer = BertTokenizer.from_pretrained("bert-base-uncased")
for sentence in data:
bert_inp = bert_tokenizer.__call__(sentence, max_length=64,
padding='max_length', pad_to_max_length=True,
truncation=True, return_token_type_ids=False)
input_ids.append(bert_inp['input_ids'])
attention_masks.append(bert_inp['attention_mask'])
input_ids = np.asarray(input_ids)
attention_masks = np.array(attention_masks)
labels = np.array(labels)
return input_ids, attention_masks, labels
def load_and_process():
data, labels = read_dataset()
num_of_labels = len(labels.unique())
input_ids, attention_masks, labels = data_process(pre_process_dataset(data), labels)
return input_ids, attention_masks, labels
# function to train the model
def train():
model.train()
total_loss, total_accuracy = 0, 0
# empty list to save model predictions
total_preds = []
# iterate over batches
total = len(train_dataloader)
for i, batch in enumerate(train_dataloader):
step = i+1
percent = "{0:.2f}".format(100 * (step / float(total)))
lossp = "{0:.2f}".format(total_loss/(total*batch_size))
filledLength = int(100 * step // total)
bar = '█' * filledLength + '>' *(filledLength < 100) + '.' * (99 - filledLength)
print(f'\rBatch {step}/{total} |{bar}| {percent}% complete, loss={lossp}, accuracy={total_accuracy}', end='')
# push the batch to gpu
batch = [r.to(device) for r in batch]
sent_id, mask, labels = batch
del batch
gc.collect()
torch.cuda.empty_cache()
# clear previously calculated gradients
model.zero_grad()
# get model predictions for the current batch
preds = model(sent_id, mask)
# compute the loss between actual and predicted values
loss = cross_entropy(preds, labels)
# add on to the total loss
total_loss += float(loss.item())
# backward pass to calculate the gradients
loss.backward()
# clip the the gradients to 1.0. It helps in preventing the exploding gradient problem
torch.nn.utils.clip_grad_norm_(model.parameters(), 1.0)
# update parameters
optimizer.step()
# append the model predictions
total_preds.append(preds.detach().cpu().numpy())
gc.collect()
torch.cuda.empty_cache()
# compute the training loss of the epoch
avg_loss = total_loss / (len(train_dataloader)*batch_size)
# predictions are in the form of (no. of batches, size of batch, no. of classes).
# reshape the predictions in form of (number of samples, no. of classes)
total_preds = np.concatenate(total_preds, axis=0)
# returns the loss and predictions
return avg_loss, total_preds
# function for evaluating the model
def evaluate():
print("\n\nEvaluating...")
# deactivate dropout layers
model.eval()
total_loss, total_accuracy = 0, 0
# empty list to save the model predictions
total_preds = []
# iterate over batches
total = len(val_dataloader)
for i, batch in enumerate(val_dataloader):
step = i+1
percent = "{0:.2f}".format(100 * (step / float(total)))
lossp = "{0:.2f}".format(total_loss/(total*batch_size))
filledLength = int(100 * step // total)
bar = '█' * filledLength + '>' * (filledLength < 100) + '.' * (99 - filledLength)
print(f'\rBatch {step}/{total} |{bar}| {percent}% complete, loss={lossp}, accuracy={total_accuracy}', end='')
# push the batch to gpu
batch = [t.to(device) for t in batch]
sent_id, mask, labels = batch
del batch
gc.collect()
torch.cuda.empty_cache()
# deactivate autograd
with torch.no_grad():
# model predictions
preds = model(sent_id, mask)
# compute the validation loss between actual and predicted values
loss = cross_entropy(preds, labels)
total_loss += float(loss.item())
#preds = preds.detach().cpu().numpy()
#total_preds.append(preds)
total_preds.append(preds.detach().cpu().numpy())
gc.collect()
torch.cuda.empty_cache()
# compute the validation loss of the epoch
avg_loss = total_loss / (len(val_dataloader)*batch_size)
# reshape the predictions in form of (number of samples, no. of classes)
total_preds = np.concatenate(total_preds, axis=0)
return avg_loss, total_preds
# + [markdown] id="5v8P9TN4So5Y"
# # Train
# + id="0Y3L2lH1mROW"
### Extension mode
MODE = 'rnn'
# + colab={"base_uri": "https://localhost:8080/", "height": 148, "referenced_widgets": ["1b152337cc8b4646878469e02062925c", "f4712961d1474deb87b342f245b94618", "ddda6da2e15e40159db5fd8e4e57db6f", "fef3232d038242cf95460ef1dd9ab0bf", "7440c9676ea94c7aaf3a429358a745a1", "33a99c5551c3417b834c75f86aa6d916", "74b799a818624278944912fbf82596e7", "a57199db692140d8ba77a47a43e92a56", "371c3e33f51748ca97db215834d74bed", "9f60fcc001944b049831cf167dee612d", "faff5bf69f2d4abcab8631328674d709", "f7f8fcf35a3e4857b9ba43d19a477e3e", "c150356cd1ae4c8b80b44fdd5e9a863b", "5b3bffad5cd44bd49de101f3f6b324d1", "793396721bea43149262d623af3f8a4a", "bd8e32a4620e47338b0fc69c9bcd6a2d", "664bf3568546466b92eaed8c61331deb", "a05644dd84cd480ba63f101b05f5d81c", "b19a351d121c42c089823b7b81373db4", "51316b9d5b124ccfad6cf8adf68a7170", "679adf788bdd4c4f8b939080cb3ba2c9", "42f47aacede34d2e8a12b8b46eadca1c", "aed12a63cfa54a4196c6d0d2df25a996", "5c4bb8e4ff9442b6844867dcc086e129", "39eec80dd6fd4bca98403380ceb6e610", "69febcbe218a4fec9da52979fb9cf70c", "1441f8bbcf1849e7a161bd1af23c884e", "9ac132ebacd645619ec5266aba0db2f7", "702f2d1686144988a4cee9e7d1238405", "59288451a42646a9a4dc357e75204423", "106328416a684261b374fee5eff098cd", "7da52cd672a8478da39b2bd2c6b0b39d", "53700556e7f34dfeaa549d674255116a"]} id="EH3HDzr9WDgY" outputId="a1fcce98-55a7-4ccd-94c1-781e393a102c"
# Specify the GPU
# Setting up the device for GPU usage
device = 'cuda' if torch.cuda.is_available() else 'cpu'
print(device)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ Load Data-set ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~#
input_ids, attention_masks, labels = load_and_process()
df = pd.DataFrame(list(zip(input_ids, attention_masks)), columns=['input_ids', 'attention_masks'])
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~#
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ class distribution ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~#
# class = class label for majority of CF users. 0 - hate speech 1 - offensive language 2 - neither
# ~~~~~~~~~~ Split train data-set into train, validation and test sets ~~~~~~~~~~#
train_text, temp_text, train_labels, temp_labels = train_test_split(df, labels,
random_state=2018, test_size=0.2, stratify=labels)
val_text, test_text, val_labels, test_labels = train_test_split(temp_text, temp_labels,
random_state=2018, test_size=0.5, stratify=temp_labels)
del temp_text
gc.collect()
torch.cuda.empty_cache()
train_count = len(train_labels)
test_count = len(test_labels)
val_count = len(val_labels)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~#
# ~~~~~~~~~~~~~~~~~~~~~ Import BERT Model~~~~~~~~~~~~~~~~~~~~~#
# import BERT-base pretrained model
bert = AutoModel.from_pretrained('bert-base-uncased')
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~#
# + colab={"base_uri": "https://localhost:8080/"} id="bUtglOzvL6bU" outputId="9d13eff9-6d32-4dd1-ca94-5636b0696d3d"
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ Tokenization ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~#
# for train set
train_seq = torch.tensor(train_text['input_ids'].tolist())
train_mask = torch.tensor(train_text['attention_masks'].tolist())
train_y = torch.tensor(train_labels.tolist())
# for validation set
val_seq = torch.tensor(val_text['input_ids'].tolist())
val_mask = torch.tensor(val_text['attention_masks'].tolist())
val_y = torch.tensor(val_labels.tolist())
# for test set
test_seq = torch.tensor(test_text['input_ids'].tolist())
test_mask = torch.tensor(test_text['attention_masks'].tolist())
test_y = torch.tensor(test_labels.tolist())
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~#
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ Create DataLoaders ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~#
from torch.utils.data import TensorDataset, DataLoader, RandomSampler, SequentialSampler
# define a batch size
batch_size = 32
# wrap tensors
train_data = TensorDataset(train_seq, train_mask, train_y)
# sampler for sampling the data during training
train_sampler = RandomSampler(train_data)
# dataLoader for train set
train_dataloader = DataLoader(train_data, sampler=train_sampler, batch_size=batch_size)
# wrap tensors
val_data = TensorDataset(val_seq, val_mask, val_y)
# sampler for sampling the data during training
val_sampler = SequentialSampler(val_data)
# dataLoader for validation set
val_dataloader = DataLoader(val_data, sampler=val_sampler, batch_size=batch_size)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~#
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~ Freeze BERT Parameters ~~~~~~~~~~~~~~~~~~~~~~~~~~~~#
# freeze all the parameters
for param in bert.parameters():
param.requires_grad = False
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~#
# pass the pre-trained BERT to our define architecture
model = BERT_Arch(bert, mode=MODE)
# push the model to GPU
model = model.to(device)
# optimizer from hugging face transformers
from transformers import AdamW
# define the optimizer
optimizer = AdamW(model.parameters(), lr=2e-5)
# loss function
cross_entropy = nn.NLLLoss()
# set initial loss to infinite
best_valid_loss = float('inf')
# number of training epochs
epochs = 3
current = 1
# for each epoch
while current <= epochs:
print(f'\nEpoch {current} / {epochs}:')
# train model
train_loss, _ = train()
# evaluate model
valid_loss, _ = evaluate()
# save the best model
if valid_loss < best_valid_loss:
best_valid_loss = valid_loss
#torch.save(model.state_dict(), 'saved_weights.pth')
# append training and validation loss
print(f'\n\nTraining Loss: {train_loss:.3f}')
print(f'Validation Loss: {valid_loss:.3f}')
current = current + 1
# get predictions for test data
gc.collect()
torch.cuda.empty_cache()
with torch.no_grad():
preds = model(test_seq.to(device), test_mask.to(device))
preds = preds.detach().cpu().numpy()
print("Performance:")
# model's performance
preds = np.argmax(preds, axis=1)
print('Classification Report')
print(classification_report(test_y, preds))
print("Accuracy: " + str(accuracy_score(test_y, preds)))
# + colab={"base_uri": "https://localhost:8080/", "height": 296} id="6zUbAqAGIVgt" outputId="801e4511-d3d8-4d6e-af9c-cbdb8439670d"
cm = confusion_matrix(test_y, preds, labels=[0,1,2])
disp_cm = ConfusionMatrixDisplay(cm, display_labels = ['hate','offensive','neither'])
disp_cm.plot(cmap='Blues')
| 36.707265
| 1,342
|
5b21fdbf173c6a56feb7caff14426c9340cd398e
|
py
|
python
|
examples/Logica_example_Avengers_and_PostgreSQL.ipynb
|
sefgit/logica
|
['Apache-2.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/EvgSkv/logica/blob/main/examples/Logica_example_Avengers_and_PostgreSQL.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="S26GH8bkUrmK"
# # Logica example: Avengers and PostgreSQL
#
# At the moment only BigQuery is fully supported in Logica, but in the future we plan to support other engines.
# There is already an **experimental** support for PostgreSQL.
#
# Engine is controlled with `@Engine` annotation.
#
# In this CoLab we give an example of how mortality of Avengers can be computed
# with Logica on PostgreSQL engine.
#
# The Avengers dataset published by [FiveThirtyEight](https://fivethirtyeight.com/) is used.
# + [markdown] id="ltqvl1qmVkfR"
# ## Install Logica and PostgreSQL.
# + id="W7NuEqRJPMzt" outputId="c3585f04-587b-457d-8732-a4182270b8fe" colab={"base_uri": "https://localhost:8080/", "height": 119}
# Install Logica.
# !pip install logica
# Install postgresql server.
# !sudo apt-get -y -qq update
# !sudo apt-get -y -qq install postgresql
# !sudo service postgresql start
# Prepare database for Logica.
# !sudo -u postgres psql -c "CREATE USER logica WITH SUPERUSER"
# !sudo -u postgres psql -c "ALTER USER logica PASSWORD 'logica';"
# !sudo -u postgres psql -U postgres -c 'CREATE DATABASE logica;'
# Connect to the database.
from logica import colab_logica
from sqlalchemy import create_engine
import pandas
engine = create_engine('postgresql+psycopg2://logica:[email protected]', pool_recycle=3600);
connection = engine.connect();
colab_logica.SetDbConnection(connection)
# + [markdown] id="Yt6mtsC8Vnww"
# ## Load dataset
# + id="J4zesAMzNQXv"
import urllib.request
import pandas
import io
url = urllib.request.urlopen('https://github.com/fivethirtyeight/data/blob/master/avengers/avengers.csv?raw=True')
text = url.read().decode('utf-8', errors='ignore').lower()
f = io.StringIO(text)
avengers_data = pandas.read_csv(f)
avengers_data.to_sql('avengers', engine, if_exists='replace')
# + [markdown] id="VcA_QMUAVpjJ"
# ## Calculate mortality
# + id="NkehJ8LhPdFI" outputId="d1c5d09a-0f8c-45c0-e5f1-ea9d7ae0259b" colab={"resources": {"http://localhost:8080/nbextensions/google.colab/tabbar.css": {"data": "Lmdvb2ctdGFie3Bvc2l0aW9uOnJlbGF0aXZlO3BhZGRpbmc6NHB4IDhweDtjb2xvcjojMDBjO3RleHQtZGVjb3JhdGlvbjp1bmRlcmxpbmU7Y3Vyc29yOmRlZmF1bHR9Lmdvb2ctdGFiLWJhci10b3AgLmdvb2ctdGFie21hcmdpbjoxcHggNHB4IDAgMDtib3JkZXItYm90dG9tOjA7ZmxvYXQ6bGVmdH0uZ29vZy10YWItYmFyLXRvcDphZnRlciwuZ29vZy10YWItYmFyLWJvdHRvbTphZnRlcntjb250ZW50OiIgIjtkaXNwbGF5OmJsb2NrO2hlaWdodDowO2NsZWFyOmJvdGg7dmlzaWJpbGl0eTpoaWRkZW59Lmdvb2ctdGFiLWJhci1ib3R0b20gLmdvb2ctdGFie21hcmdpbjowIDRweCAxcHggMDtib3JkZXItdG9wOjA7ZmxvYXQ6bGVmdH0uZ29vZy10YWItYmFyLXN0YXJ0IC5nb29nLXRhYnttYXJnaW46MCAwIDRweCAxcHg7Ym9yZGVyLXJpZ2h0OjB9Lmdvb2ctdGFiLWJhci1lbmQgLmdvb2ctdGFie21hcmdpbjowIDFweCA0cHggMDtib3JkZXItbGVmdDowfS5nb29nLXRhYi1ob3ZlcntiYWNrZ3JvdW5kOiNlZWV9Lmdvb2ctdGFiLWRpc2FibGVke2NvbG9yOiM2NjZ9Lmdvb2ctdGFiLXNlbGVjdGVke2NvbG9yOiMwMDA7YmFja2dyb3VuZDojZmZmO3RleHQtZGVjb3JhdGlvbjpub25lO2ZvbnQtd2VpZ2h0OmJvbGQ7Ym9yZGVyOjFweCBzb2xpZCAjNmI5MGRhfS5nb29nLXRhYi1iYXItdG9we3BhZGRpbmctdG9wOjVweCFpbXBvcnRhbnQ7cGFkZGluZy1sZWZ0OjVweCFpbXBvcnRhbnQ7Ym9yZGVyLWJvdHRvbToxcHggc29saWQgIzZiOTBkYSFpbXBvcnRhbnR9Lmdvb2ctdGFiLWJhci10b3AgLmdvb2ctdGFiLXNlbGVjdGVke3RvcDoxcHg7bWFyZ2luLXRvcDowO3BhZGRpbmctYm90dG9tOjVweH0uZ29vZy10YWItYmFyLWJvdHRvbSAuZ29vZy10YWItc2VsZWN0ZWR7dG9wOi0xcHg7bWFyZ2luLWJvdHRvbTowO3BhZGRpbmctdG9wOjVweH0uZ29vZy10YWItYmFyLXN0YXJ0IC5nb29nLXRhYi1zZWxlY3RlZHtsZWZ0OjFweDttYXJnaW4tbGVmdDowO3BhZGRpbmctcmlnaHQ6OXB4fS5nb29nLXRhYi1iYXItZW5kIC5nb29nLXRhYi1zZWxlY3RlZHtsZWZ0Oi0xcHg7bWFyZ2luLXJpZ2h0OjA7cGFkZGluZy1sZWZ0OjlweH0uZ29vZy10YWItYmFye21hcmdpbjowO2JvcmRlcjowO3BhZGRpbmc6MDtsaXN0LXN0eWxlOm5vbmU7Y3Vyc29yOmRlZmF1bHQ7b3V0bGluZTpub25lO2JhY2tncm91bmQ6I2ViZWZmOX0uZ29vZy10YWItYmFyLWNsZWFye2NsZWFyOmJvdGg7aGVpZ2h0OjA7b3ZlcmZsb3c6aGlkZGVufS5nb29nLXRhYi1iYXItc3RhcnR7ZmxvYXQ6bGVmdH0uZ29vZy10YWItYmFyLWVuZHtmbG9hdDpyaWdodH0qIGh0bWwgLmdvb2ctdGFiLWJhci1zdGFydHttYXJnaW4tcmlnaHQ6LTNweH0qIGh0bWwgLmdvb2ctdGFiLWJhci1lbmR7bWFyZ2luLWxlZnQ6LTNweH0=", "ok": true, "headers": [["content-type", "text/css"]], "status": 200, "status_text": ""}, "http://localhost:8080/nbextensions/google.colab/tabbar_main.min.js": {"data": "Ly8gY2xhbmctZm9ybWF0IG9mZgooZnVuY3Rpb24oKXt2YXIgaCxhYT1mdW5jdGlvbihhKXt2YXIgYj0wO3JldHVybiBmdW5jdGlvbigpe3JldHVybiBiPGEubGVuZ3RoP3tkb25lOiExLHZhbHVlOmFbYisrXX06e2RvbmU6ITB9fX0sYmE9ZnVuY3Rpb24oYSl7dmFyIGI9InVuZGVmaW5lZCIhPXR5cGVvZiBTeW1ib2wmJlN5bWJvbC5pdGVyYXRvciYmYVtTeW1ib2wuaXRlcmF0b3JdO3JldHVybiBiP2IuY2FsbChhKTp7bmV4dDphYShhKX19LGw9dGhpc3x8c2VsZixjYT1mdW5jdGlvbigpe30sZGE9ZnVuY3Rpb24oYSl7YS5CYT12b2lkIDA7YS5SPWZ1bmN0aW9uKCl7cmV0dXJuIGEuQmE/YS5CYTphLkJhPW5ldyBhfX0sZWE9ZnVuY3Rpb24oYSl7dmFyIGI9dHlwZW9mIGE7aWYoIm9iamVjdCI9PWIpaWYoYSl7aWYoYSBpbnN0YW5jZW9mIEFycmF5KXJldHVybiJhcnJheSI7aWYoYSBpbnN0YW5jZW9mIE9iamVjdClyZXR1cm4gYjt2YXIgYz1PYmplY3QucHJvdG90eXBlLnRvU3RyaW5nLmNhbGwoYSk7aWYoIltvYmplY3QgV2luZG93XSI9PWMpcmV0dXJuIm9iamVjdCI7CmlmKCJbb2JqZWN0IEFycmF5XSI9PWN8fCJudW1iZXIiPT10eXBlb2YgYS5sZW5ndGgmJiJ1bmRlZmluZWQiIT10eXBlb2YgYS5zcGxpY2UmJiJ1bmRlZmluZWQiIT10eXBlb2YgYS5wcm9wZXJ0eUlzRW51bWVyYWJsZSYmIWEucHJvcGVydHlJc0VudW1lcmFibGUoInNwbGljZSIpKXJldHVybiJhcnJheSI7aWYoIltvYmplY3QgRnVuY3Rpb25dIj09Y3x8InVuZGVmaW5lZCIhPXR5cGVvZiBhLmNhbGwmJiJ1bmRlZmluZWQiIT10eXBlb2YgYS5wcm9wZXJ0eUlzRW51bWVyYWJsZSYmIWEucHJvcGVydHlJc0VudW1lcmFibGUoImNhbGwiKSlyZXR1cm4iZnVuY3Rpb24ifWVsc2UgcmV0dXJuIm51bGwiO2Vsc2UgaWYoImZ1bmN0aW9uIj09YiYmInVuZGVmaW5lZCI9PXR5cGVvZiBhLmNhbGwpcmV0dXJuIm9iamVjdCI7cmV0dXJuIGJ9LGZhPWZ1bmN0aW9uKGEpe3JldHVybiJhcnJheSI9PWVhKGEpfSxoYT1mdW5jdGlvbihhKXt2YXIgYj1lYShhKTtyZXR1cm4iYXJyYXkiPT1ifHwib2JqZWN0Ij09CmImJiJudW1iZXIiPT10eXBlb2YgYS5sZW5ndGh9LGlhPWZ1bmN0aW9uKGEpe3JldHVybiJmdW5jdGlvbiI9PWVhKGEpfSxtPWZ1bmN0aW9uKGEpe3ZhciBiPXR5cGVvZiBhO3JldHVybiJvYmplY3QiPT1iJiZudWxsIT1hfHwiZnVuY3Rpb24iPT1ifSxqYT0iY2xvc3VyZV91aWRfIisoMUU5Kk1hdGgucmFuZG9tKCk+Pj4wKSxrYT0wLGxhPWZ1bmN0aW9uKGEsYil7dmFyIGM9QXJyYXkucHJvdG90eXBlLnNsaWNlLmNhbGwoYXJndW1lbnRzLDEpO3JldHVybiBmdW5jdGlvbigpe3ZhciBkPWMuc2xpY2UoKTtkLnB1c2guYXBwbHkoZCxhcmd1bWVudHMpO3JldHVybiBhLmFwcGx5KHRoaXMsZCl9fSxuPWZ1bmN0aW9uKGEsYil7ZnVuY3Rpb24gYygpe31jLnByb3RvdHlwZT1iLnByb3RvdHlwZTthLmk9Yi5wcm90b3R5cGU7YS5wcm90b3R5cGU9bmV3IGM7YS5wcm90b3R5cGUuY29uc3RydWN0b3I9YX07dmFyIG1hLG5hPXtlYjoiYWN0aXZlZGVzY2VuZGFudCIsamI6ImF0b21pYyIsa2I6ImF1dG9jb21wbGV0ZSIsbWI6ImJ1c3kiLHBiOiJjaGVja2VkIixxYjoiY29saW5kZXgiLHZiOiJjb250cm9scyIseGI6ImRlc2NyaWJlZGJ5IixBYjoiZGlzYWJsZWQiLENiOiJkcm9wZWZmZWN0IixEYjoiZXhwYW5kZWQiLEViOiJmbG93dG8iLEdiOiJncmFiYmVkIixLYjoiaGFzcG9wdXAiLE1iOiJoaWRkZW4iLE9iOiJpbnZhbGlkIixQYjoibGFiZWwiLFFiOiJsYWJlbGxlZGJ5IixSYjoibGV2ZWwiLFdiOiJsaXZlIixrYzoibXVsdGlsaW5lIixsYzoibXVsdGlzZWxlY3RhYmxlIixwYzoib3JpZW50YXRpb24iLHFjOiJvd25zIixyYzoicG9zaW5zZXQiLHRjOiJwcmVzc2VkIix4YzoicmVhZG9ubHkiLHpjOiJyZWxldmFudCIsQWM6InJlcXVpcmVkIixFYzoicm93aW5kZXgiLEhjOiJzZWxlY3RlZCIsSmM6InNldHNpemUiLExjOiJzb3J0IixaYzoidmFsdWVtYXgiLCRjOiJ2YWx1ZW1pbiIsYWQ6InZhbHVlbm93IiwKYmQ6InZhbHVldGV4dCJ9O3ZhciBvYT1mdW5jdGlvbihhLGIsYyl7Zm9yKHZhciBkIGluIGEpYi5jYWxsKGMsYVtkXSxkLGEpfSxwYT1mdW5jdGlvbihhLGIpe2Zvcih2YXIgYyBpbiBhKWlmKGFbY109PWIpcmV0dXJuITA7cmV0dXJuITF9LHFhPWZ1bmN0aW9uKGEsYixjKXtpZihudWxsIT09YSYmYiBpbiBhKXRocm93IEVycm9yKCdUaGUgb2JqZWN0IGFscmVhZHkgY29udGFpbnMgdGhlIGtleSAiJytiKyciJyk7YVtiXT1jfSxzYT1mdW5jdGlvbihhKXt2YXIgYj17fSxjO2ZvcihjIGluIGEpYlthW2NdXT1jO3JldHVybiBifSx0YT0iY29uc3RydWN0b3IgaGFzT3duUHJvcGVydHkgaXNQcm90b3R5cGVPZiBwcm9wZXJ0eUlzRW51bWVyYWJsZSB0b0xvY2FsZVN0cmluZyB0b1N0cmluZyB2YWx1ZU9mIi5zcGxpdCgiICIpLHVhPWZ1bmN0aW9uKGEsYil7Zm9yKHZhciBjLGQsZT0xO2U8YXJndW1lbnRzLmxlbmd0aDtlKyspe2Q9YXJndW1lbnRzW2VdO2ZvcihjIGluIGQpYVtjXT1kW2NdO2Zvcih2YXIgZj0wO2Y8dGEubGVuZ3RoO2YrKyljPQp0YVtmXSxPYmplY3QucHJvdG90eXBlLmhhc093blByb3BlcnR5LmNhbGwoZCxjKSYmKGFbY109ZFtjXSl9fTt2YXIgdmE9e2ZiOiJhbGVydCIsZ2I6ImFsZXJ0ZGlhbG9nIixoYjoiYXBwbGljYXRpb24iLGliOiJhcnRpY2xlIixsYjoiYmFubmVyIixuYjoiYnV0dG9uIixvYjoiY2hlY2tib3giLHJiOiJjb2x1bW5oZWFkZXIiLHNiOiJjb21ib2JveCIsdGI6ImNvbXBsZW1lbnRhcnkiLHViOiJjb250ZW50aW5mbyIsd2I6ImRlZmluaXRpb24iLHliOiJkaWFsb2ciLHpiOiJkaXJlY3RvcnkiLEJiOiJkb2N1bWVudCIsRmI6ImZvcm0iLEhiOiJncmlkIixJYjoiZ3JpZGNlbGwiLEpiOiJncm91cCIsTGI6ImhlYWRpbmciLE5iOiJpbWciLFNiOiJsaW5rIixUYjoibGlzdCIsVWI6Imxpc3Rib3giLFZiOiJsaXN0aXRlbSIsWGI6ImxvZyIsWWI6Im1haW4iLFpiOiJtYXJxdWVlIiwkYjoibWF0aCIsYWM6Im1lbnUiLGJjOiJtZW51YmFyIixjYzoibWVudWl0ZW0iLGRjOiJtZW51aXRlbWNoZWNrYm94IixlYzoibWVudWl0ZW1yYWRpbyIsbWM6Im5hdmlnYXRpb24iLG5jOiJub3RlIixvYzoib3B0aW9uIiwKc2M6InByZXNlbnRhdGlvbiIsdWM6InByb2dyZXNzYmFyIix2YzoicmFkaW8iLHdjOiJyYWRpb2dyb3VwIix5YzoicmVnaW9uIixCYzoicm93IixDYzoicm93Z3JvdXAiLERjOiJyb3doZWFkZXIiLEZjOiJzY3JvbGxiYXIiLEdjOiJzZWFyY2giLEljOiJzZXBhcmF0b3IiLEtjOiJzbGlkZXIiLE1jOiJzcGluYnV0dG9uIixOYzoic3RhdHVzIixPYzoidGFiIixQYzoidGFibGlzdCIsUWM6InRhYnBhbmVsIixSYzoidGV4dGJveCIsU2M6InRleHRpbmZvIixUYzoidGltZXIiLFVjOiJ0b29sYmFyIixWYzoidG9vbHRpcCIsV2M6InRyZWUiLFhjOiJ0cmVlZ3JpZCIsWWM6InRyZWVpdGVtIn07dmFyIHdhPWZ1bmN0aW9uKGEpe2lmKEVycm9yLmNhcHR1cmVTdGFja1RyYWNlKUVycm9yLmNhcHR1cmVTdGFja1RyYWNlKHRoaXMsd2EpO2Vsc2V7dmFyIGI9RXJyb3IoKS5zdGFjaztiJiYodGhpcy5zdGFjaz1iKX1hJiYodGhpcy5tZXNzYWdlPVN0cmluZyhhKSl9O24od2EsRXJyb3IpO3dhLnByb3RvdHlwZS5uYW1lPSJDdXN0b21FcnJvciI7dmFyIHhhO3ZhciB5YT1mdW5jdGlvbihhLGIpe2E9YS5zcGxpdCgiJXMiKTtmb3IodmFyIGM9IiIsZD1hLmxlbmd0aC0xLGU9MDtlPGQ7ZSsrKWMrPWFbZV0rKGU8Yi5sZW5ndGg/YltlXToiJXMiKTt3YS5jYWxsKHRoaXMsYythW2RdKX07bih5YSx3YSk7eWEucHJvdG90eXBlLm5hbWU9IkFzc2VydGlvbkVycm9yIjsKdmFyIHphPWZ1bmN0aW9uKGEsYixjLGQpe3ZhciBlPSJBc3NlcnRpb24gZmFpbGVkIjtpZihjKXtlKz0iOiAiK2M7dmFyIGY9ZH1lbHNlIGEmJihlKz0iOiAiK2EsZj1iKTt0aHJvdyBuZXcgeWEoIiIrZSxmfHxbXSk7fSxxPWZ1bmN0aW9uKGEsYixjKXthfHx6YSgiIixudWxsLGIsQXJyYXkucHJvdG90eXBlLnNsaWNlLmNhbGwoYXJndW1lbnRzLDIpKTtyZXR1cm4gYX0sQWE9ZnVuY3Rpb24oYSxiLGMpe20oYSkmJjE9PWEubm9kZVR5cGV8fHphKCJFeHBlY3RlZCBFbGVtZW50IGJ1dCBnb3QgJXM6ICVzLiIsW2VhKGEpLGFdLGIsQXJyYXkucHJvdG90eXBlLnNsaWNlLmNhbGwoYXJndW1lbnRzLDIpKX0sQ2E9ZnVuY3Rpb24oYSxiLGMsZCl7YSBpbnN0YW5jZW9mIGJ8fHphKCJFeHBlY3RlZCBpbnN0YW5jZW9mICVzIGJ1dCBnb3QgJXMuIixbQmEoYiksQmEoYSldLGMsQXJyYXkucHJvdG90eXBlLnNsaWNlLmNhbGwoYXJndW1lbnRzLDMpKX0sQmE9ZnVuY3Rpb24oYSl7cmV0dXJuIGEgaW5zdGFuY2VvZgpGdW5jdGlvbj9hLmRpc3BsYXlOYW1lfHxhLm5hbWV8fCJ1bmtub3duIHR5cGUgbmFtZSI6YSBpbnN0YW5jZW9mIE9iamVjdD9hLmNvbnN0cnVjdG9yLmRpc3BsYXlOYW1lfHxhLmNvbnN0cnVjdG9yLm5hbWV8fE9iamVjdC5wcm90b3R5cGUudG9TdHJpbmcuY2FsbChhKTpudWxsPT09YT8ibnVsbCI6dHlwZW9mIGF9O3ZhciBEYT1BcnJheS5wcm90b3R5cGUuaW5kZXhPZj9mdW5jdGlvbihhLGIpe3EobnVsbCE9YS5sZW5ndGgpO3JldHVybiBBcnJheS5wcm90b3R5cGUuaW5kZXhPZi5jYWxsKGEsYix2b2lkIDApfTpmdW5jdGlvbihhLGIpe2lmKCJzdHJpbmciPT09dHlwZW9mIGEpcmV0dXJuInN0cmluZyIhPT10eXBlb2YgYnx8MSE9Yi5sZW5ndGg/LTE6YS5pbmRleE9mKGIsMCk7Zm9yKHZhciBjPTA7YzxhLmxlbmd0aDtjKyspaWYoYyBpbiBhJiZhW2NdPT09YilyZXR1cm4gYztyZXR1cm4tMX0scj1BcnJheS5wcm90b3R5cGUuZm9yRWFjaD9mdW5jdGlvbihhLGIsYyl7cShudWxsIT1hLmxlbmd0aCk7QXJyYXkucHJvdG90eXBlLmZvckVhY2guY2FsbChhLGIsYyl9OmZ1bmN0aW9uKGEsYixjKXtmb3IodmFyIGQ9YS5sZW5ndGgsZT0ic3RyaW5nIj09PXR5cGVvZiBhP2Euc3BsaXQoIiIpOmEsZj0wO2Y8ZDtmKyspZiBpbiBlJiZiLmNhbGwoYyxlW2ZdLGYsYSl9LEVhPUFycmF5LnByb3RvdHlwZS5maWx0ZXI/CmZ1bmN0aW9uKGEsYil7cShudWxsIT1hLmxlbmd0aCk7cmV0dXJuIEFycmF5LnByb3RvdHlwZS5maWx0ZXIuY2FsbChhLGIsdm9pZCAwKX06ZnVuY3Rpb24oYSxiKXtmb3IodmFyIGM9YS5sZW5ndGgsZD1bXSxlPTAsZj0ic3RyaW5nIj09PXR5cGVvZiBhP2Euc3BsaXQoIiIpOmEsZz0wO2c8YztnKyspaWYoZyBpbiBmKXt2YXIgaz1mW2ddO2IuY2FsbCh2b2lkIDAsayxnLGEpJiYoZFtlKytdPWspfXJldHVybiBkfSxGYT1BcnJheS5wcm90b3R5cGUuZXZlcnk/ZnVuY3Rpb24oYSxiKXtxKG51bGwhPWEubGVuZ3RoKTtyZXR1cm4gQXJyYXkucHJvdG90eXBlLmV2ZXJ5LmNhbGwoYSxiLHZvaWQgMCl9OmZ1bmN0aW9uKGEsYil7Zm9yKHZhciBjPWEubGVuZ3RoLGQ9InN0cmluZyI9PT10eXBlb2YgYT9hLnNwbGl0KCIiKTphLGU9MDtlPGM7ZSsrKWlmKGUgaW4gZCYmIWIuY2FsbCh2b2lkIDAsZFtlXSxlLGEpKXJldHVybiExO3JldHVybiEwfSxHYT1mdW5jdGlvbihhLGIpe3JldHVybiAwPD0KRGEoYSxiKX0sSGE9ZnVuY3Rpb24oYSxiKXtiPURhKGEsYik7dmFyIGM7aWYoYz0wPD1iKXEobnVsbCE9YS5sZW5ndGgpLEFycmF5LnByb3RvdHlwZS5zcGxpY2UuY2FsbChhLGIsMSk7cmV0dXJuIGN9LElhPWZ1bmN0aW9uKGEpe3JldHVybiBBcnJheS5wcm90b3R5cGUuY29uY2F0LmFwcGx5KFtdLGFyZ3VtZW50cyl9LEphPWZ1bmN0aW9uKGEpe3ZhciBiPWEubGVuZ3RoO2lmKDA8Yil7Zm9yKHZhciBjPUFycmF5KGIpLGQ9MDtkPGI7ZCsrKWNbZF09YVtkXTtyZXR1cm4gY31yZXR1cm5bXX0sTGE9ZnVuY3Rpb24oYSxiLGMsZCl7cShudWxsIT1hLmxlbmd0aCk7QXJyYXkucHJvdG90eXBlLnNwbGljZS5hcHBseShhLEthKGFyZ3VtZW50cywxKSl9LEthPWZ1bmN0aW9uKGEsYixjKXtxKG51bGwhPWEubGVuZ3RoKTtyZXR1cm4gMj49YXJndW1lbnRzLmxlbmd0aD9BcnJheS5wcm90b3R5cGUuc2xpY2UuY2FsbChhLGIpOkFycmF5LnByb3RvdHlwZS5zbGljZS5jYWxsKGEsYixjKX07dmFyIE1hPVN0cmluZy5wcm90b3R5cGUudHJpbT9mdW5jdGlvbihhKXtyZXR1cm4gYS50cmltKCl9OmZ1bmN0aW9uKGEpe3JldHVybi9eW1xzXHhhMF0qKFtcc1xTXSo/KVtcc1x4YTBdKiQvLmV4ZWMoYSlbMV19LE5hPS8mL2csT2E9LzwvZyxQYT0vPi9nLFFhPS8iL2csUmE9LycvZyxTYT0vXHgwMC9nLFRhPS9bXHgwMCY8PiInXS8sdD1mdW5jdGlvbihhLGIpe3JldHVybi0xIT1hLmluZGV4T2YoYil9LFVhPWZ1bmN0aW9uKGEsYil7cmV0dXJuIGE8Yj8tMTphPmI/MTowfTt2YXIgdTthOnt2YXIgVmE9bC5uYXZpZ2F0b3I7aWYoVmEpe3ZhciBXYT1WYS51c2VyQWdlbnQ7aWYoV2Epe3U9V2E7YnJlYWsgYX19dT0iIn07LyoKCiBDb3B5cmlnaHQgVGhlIENsb3N1cmUgTGlicmFyeSBBdXRob3JzLgogU1BEWC1MaWNlbnNlLUlkZW50aWZpZXI6IEFwYWNoZS0yLjAKKi8KdmFyIFhhPWZ1bmN0aW9uKGEpe1RhLnRlc3QoYSkmJigtMSE9YS5pbmRleE9mKCImIikmJihhPWEucmVwbGFjZShOYSwiJmFtcDsiKSksLTEhPWEuaW5kZXhPZigiPCIpJiYoYT1hLnJlcGxhY2UoT2EsIiZsdDsiKSksLTEhPWEuaW5kZXhPZigiPiIpJiYoYT1hLnJlcGxhY2UoUGEsIiZndDsiKSksLTEhPWEuaW5kZXhPZignIicpJiYoYT1hLnJlcGxhY2UoUWEsIiZxdW90OyIpKSwtMSE9YS5pbmRleE9mKCInIikmJihhPWEucmVwbGFjZShSYSwiJiMzOTsiKSksLTEhPWEuaW5kZXhPZigiXHgwMCIpJiYoYT1hLnJlcGxhY2UoU2EsIiYjMDsiKSkpO3JldHVybiBhfTt2YXIgWWE9ZnVuY3Rpb24oYSl7WWFbIiAiXShhKTtyZXR1cm4gYX07WWFbIiAiXT1jYTt2YXIgJGE9ZnVuY3Rpb24oYSxiKXt2YXIgYz1aYTtyZXR1cm4gT2JqZWN0LnByb3RvdHlwZS5oYXNPd25Qcm9wZXJ0eS5jYWxsKGMsYSk/Y1thXTpjW2FdPWIoYSl9O3ZhciBhYj10KHUsIk9wZXJhIiksdj10KHUsIlRyaWRlbnQiKXx8dCh1LCJNU0lFIiksYmI9dCh1LCJFZGdlIikseD10KHUsIkdlY2tvIikmJiEodCh1LnRvTG93ZXJDYXNlKCksIndlYmtpdCIpJiYhdCh1LCJFZGdlIikpJiYhKHQodSwiVHJpZGVudCIpfHx0KHUsIk1TSUUiKSkmJiF0KHUsIkVkZ2UiKSx5PXQodS50b0xvd2VyQ2FzZSgpLCJ3ZWJraXQiKSYmIXQodSwiRWRnZSIpLHo9dCh1LCJNYWNpbnRvc2giKSxjYj1mdW5jdGlvbigpe3ZhciBhPWwuZG9jdW1lbnQ7cmV0dXJuIGE/YS5kb2N1bWVudE1vZGU6dm9pZCAwfSxkYjsKYTp7dmFyIGViPSIiLGZiPWZ1bmN0aW9uKCl7dmFyIGE9dTtpZih4KXJldHVybi9ydjooW15cKTtdKykoXCl8OykvLmV4ZWMoYSk7aWYoYmIpcmV0dXJuL0VkZ2VcLyhbXGRcLl0rKS8uZXhlYyhhKTtpZih2KXJldHVybi9cYig/Ok1TSUV8cnYpWzogXShbXlwpO10rKShcKXw7KS8uZXhlYyhhKTtpZih5KXJldHVybi9XZWJLaXRcLyhcUyspLy5leGVjKGEpO2lmKGFiKXJldHVybi8oPzpWZXJzaW9uKVsgXC9dPyhcUyspLy5leGVjKGEpfSgpO2ZiJiYoZWI9ZmI/ZmJbMV06IiIpO2lmKHYpe3ZhciBnYj1jYigpO2lmKG51bGwhPWdiJiZnYj5wYXJzZUZsb2F0KGViKSl7ZGI9U3RyaW5nKGdiKTticmVhayBhfX1kYj1lYn0KdmFyIGhiPWRiLFphPXt9LEE9ZnVuY3Rpb24oYSl7cmV0dXJuICRhKGEsZnVuY3Rpb24oKXtmb3IodmFyIGI9MCxjPU1hKFN0cmluZyhoYikpLnNwbGl0KCIuIiksZD1NYShTdHJpbmcoYSkpLnNwbGl0KCIuIiksZT1NYXRoLm1heChjLmxlbmd0aCxkLmxlbmd0aCksZj0wOzA9PWImJmY8ZTtmKyspe3ZhciBnPWNbZl18fCIiLGs9ZFtmXXx8IiI7ZG97Zz0vKFxkKikoXEQqKSguKikvLmV4ZWMoZyl8fFsiIiwiIiwiIiwiIl07az0vKFxkKikoXEQqKSguKikvLmV4ZWMoayl8fFsiIiwiIiwiIiwiIl07aWYoMD09Z1swXS5sZW5ndGgmJjA9PWtbMF0ubGVuZ3RoKWJyZWFrO2I9VWEoMD09Z1sxXS5sZW5ndGg/MDpwYXJzZUludChnWzFdLDEwKSwwPT1rWzFdLmxlbmd0aD8wOnBhcnNlSW50KGtbMV0sMTApKXx8VWEoMD09Z1syXS5sZW5ndGgsMD09a1syXS5sZW5ndGgpfHxVYShnWzJdLGtbMl0pO2c9Z1szXTtrPWtbM119d2hpbGUoMD09Yil9cmV0dXJuIDA8PWJ9KX0saWI7CmliPWwuZG9jdW1lbnQmJnY/Y2IoKTp2b2lkIDA7dmFyIGpiPSF2fHw5PD1OdW1iZXIoaWIpO3ZhciBsYj1mdW5jdGlvbihhLGIpe29hKGIsZnVuY3Rpb24oYyxkKXtjJiYib2JqZWN0Ij09dHlwZW9mIGMmJmMuZGQmJihjPWMuY2QoKSk7InN0eWxlIj09ZD9hLnN0eWxlLmNzc1RleHQ9YzoiY2xhc3MiPT1kP2EuY2xhc3NOYW1lPWM6ImZvciI9PWQ/YS5odG1sRm9yPWM6a2IuaGFzT3duUHJvcGVydHkoZCk/YS5zZXRBdHRyaWJ1dGUoa2JbZF0sYyk6MD09ZC5sYXN0SW5kZXhPZigiYXJpYS0iLDApfHwwPT1kLmxhc3RJbmRleE9mKCJkYXRhLSIsMCk/YS5zZXRBdHRyaWJ1dGUoZCxjKTphW2RdPWN9KX0sa2I9e2NlbGxwYWRkaW5nOiJjZWxsUGFkZGluZyIsY2VsbHNwYWNpbmc6ImNlbGxTcGFjaW5nIixjb2xzcGFuOiJjb2xTcGFuIixmcmFtZWJvcmRlcjoiZnJhbWVCb3JkZXIiLGhlaWdodDoiaGVpZ2h0IixtYXhsZW5ndGg6Im1heExlbmd0aCIsbm9uY2U6Im5vbmNlIixyb2xlOiJyb2xlIixyb3dzcGFuOiJyb3dTcGFuIix0eXBlOiJ0eXBlIix1c2VtYXA6InVzZU1hcCIsdmFsaWduOiJ2QWxpZ24iLAp3aWR0aDoid2lkdGgifSxtYj1mdW5jdGlvbihhLGIsYyl7ZnVuY3Rpb24gZChrKXtrJiZiLmFwcGVuZENoaWxkKCJzdHJpbmciPT09dHlwZW9mIGs/YS5jcmVhdGVUZXh0Tm9kZShrKTprKX1mb3IodmFyIGU9MjtlPGMubGVuZ3RoO2UrKyl7dmFyIGY9Y1tlXTtpZighaGEoZil8fG0oZikmJjA8Zi5ub2RlVHlwZSlkKGYpO2Vsc2V7YTp7aWYoZiYmIm51bWJlciI9PXR5cGVvZiBmLmxlbmd0aCl7aWYobShmKSl7dmFyIGc9ImZ1bmN0aW9uIj09dHlwZW9mIGYuaXRlbXx8InN0cmluZyI9PXR5cGVvZiBmLml0ZW07YnJlYWsgYX1pZihpYShmKSl7Zz0iZnVuY3Rpb24iPT10eXBlb2YgZi5pdGVtO2JyZWFrIGF9fWc9ITF9cihnP0phKGYpOmYsZCl9fX0sbmI9ZnVuY3Rpb24oYSxiKXtiPVN0cmluZyhiKTsiYXBwbGljYXRpb24veGh0bWwreG1sIj09PWEuY29udGVudFR5cGUmJihiPWIudG9Mb3dlckNhc2UoKSk7cmV0dXJuIGEuY3JlYXRlRWxlbWVudChiKX0sb2I9ZnVuY3Rpb24oYSl7YSYmCmEucGFyZW50Tm9kZSYmYS5wYXJlbnROb2RlLnJlbW92ZUNoaWxkKGEpfSxwYj1mdW5jdGlvbihhLGIpe2lmKCFhfHwhYilyZXR1cm4hMTtpZihhLmNvbnRhaW5zJiYxPT1iLm5vZGVUeXBlKXJldHVybiBhPT1ifHxhLmNvbnRhaW5zKGIpO2lmKCJ1bmRlZmluZWQiIT10eXBlb2YgYS5jb21wYXJlRG9jdW1lbnRQb3NpdGlvbilyZXR1cm4gYT09Ynx8ISEoYS5jb21wYXJlRG9jdW1lbnRQb3NpdGlvbihiKSYxNik7Zm9yKDtiJiZhIT1iOyliPWIucGFyZW50Tm9kZTtyZXR1cm4gYj09YX0scWI9ZnVuY3Rpb24oYSl7cShhLCJOb2RlIGNhbm5vdCBiZSBudWxsIG9yIHVuZGVmaW5lZC4iKTtyZXR1cm4gOT09YS5ub2RlVHlwZT9hOmEub3duZXJEb2N1bWVudHx8YS5kb2N1bWVudH0scmI9ZnVuY3Rpb24oYSxiKXtiP2EudGFiSW5kZXg9MDooYS50YWJJbmRleD0tMSxhLnJlbW92ZUF0dHJpYnV0ZSgidGFiSW5kZXgiKSl9LHNiPWZ1bmN0aW9uKGEpe3JldHVybiB2JiYhQSgiOSIpPyhhPWEuZ2V0QXR0cmlidXRlTm9kZSgidGFiaW5kZXgiKSwKbnVsbCE9YSYmYS5zcGVjaWZpZWQpOmEuaGFzQXR0cmlidXRlKCJ0YWJpbmRleCIpfSx0Yj1mdW5jdGlvbihhKXthPWEudGFiSW5kZXg7cmV0dXJuIm51bWJlciI9PT10eXBlb2YgYSYmMDw9YSYmMzI3Njg+YX0sdWI9ZnVuY3Rpb24oYSl7dGhpcy5hPWF8fGwuZG9jdW1lbnR8fGRvY3VtZW50fTt1Yi5wcm90b3R5cGUuZj1mdW5jdGlvbihhKXtyZXR1cm4ic3RyaW5nIj09PXR5cGVvZiBhP3RoaXMuYS5nZXRFbGVtZW50QnlJZChhKTphfTsKdWIucHJvdG90eXBlLmI9ZnVuY3Rpb24oYSxiLGMpe3ZhciBkPXRoaXMuYSxlPWFyZ3VtZW50cyxmPVN0cmluZyhlWzBdKSxnPWVbMV07aWYoIWpiJiZnJiYoZy5uYW1lfHxnLnR5cGUpKXtmPVsiPCIsZl07Zy5uYW1lJiZmLnB1c2goJyBuYW1lPSInLFhhKGcubmFtZSksJyInKTtpZihnLnR5cGUpe2YucHVzaCgnIHR5cGU9IicsWGEoZy50eXBlKSwnIicpO3ZhciBrPXt9O3VhKGssZyk7ZGVsZXRlIGsudHlwZTtnPWt9Zi5wdXNoKCI+Iik7Zj1mLmpvaW4oIiIpfWY9bmIoZCxmKTtnJiYoInN0cmluZyI9PT10eXBlb2YgZz9mLmNsYXNzTmFtZT1nOmZhKGcpP2YuY2xhc3NOYW1lPWcuam9pbigiICIpOmxiKGYsZykpOzI8ZS5sZW5ndGgmJm1iKGQsZixlKTtyZXR1cm4gZn07dmFyIHZiPWZ1bmN0aW9uKGEsYil7Yj8ocShwYSh2YSxiKSwiTm8gc3VjaCBBUklBIHJvbGUgIitiKSxhLnNldEF0dHJpYnV0ZSgicm9sZSIsYikpOmEucmVtb3ZlQXR0cmlidXRlKCJyb2xlIil9LHhiPWZ1bmN0aW9uKGEsYixjKXtmYShjKSYmKGM9Yy5qb2luKCIgIikpO3ZhciBkPXdiKGIpOyIiPT09Y3x8dm9pZCAwPT1jPyhtYXx8KG1hPXthdG9taWM6ITEsYXV0b2NvbXBsZXRlOiJub25lIixkcm9wZWZmZWN0OiJub25lIixoYXNwb3B1cDohMSxsaXZlOiJvZmYiLG11bHRpbGluZTohMSxtdWx0aXNlbGVjdGFibGU6ITEsb3JpZW50YXRpb246InZlcnRpY2FsIixyZWFkb25seTohMSxyZWxldmFudDoiYWRkaXRpb25zIHRleHQiLHJlcXVpcmVkOiExLHNvcnQ6Im5vbmUiLGJ1c3k6ITEsZGlzYWJsZWQ6ITEsaGlkZGVuOiExLGludmFsaWQ6ImZhbHNlIn0pLGM9bWEsYiBpbiBjP2Euc2V0QXR0cmlidXRlKGQsY1tiXSk6YS5yZW1vdmVBdHRyaWJ1dGUoZCkpOmEuc2V0QXR0cmlidXRlKGQsCmMpfSx3Yj1mdW5jdGlvbihhKXtxKGEsIkFSSUEgYXR0cmlidXRlIGNhbm5vdCBiZSBlbXB0eS4iKTtxKHBhKG5hLGEpLCJObyBzdWNoIEFSSUEgYXR0cmlidXRlICIrYSk7cmV0dXJuImFyaWEtIithfTt2YXIgeWI9T2JqZWN0LmZyZWV6ZXx8ZnVuY3Rpb24oYSl7cmV0dXJuIGF9O3ZhciBDPWZ1bmN0aW9uKCl7dGhpcy5VPXRoaXMuVTt0aGlzLko9dGhpcy5KfTtDLnByb3RvdHlwZS5VPSExO0MucHJvdG90eXBlLk49ZnVuY3Rpb24oKXt0aGlzLlV8fCh0aGlzLlU9ITAsdGhpcy51KCkpfTt2YXIgemI9ZnVuY3Rpb24oYSxiKXthLlU/YigpOihhLkp8fChhLko9W10pLGEuSi5wdXNoKGIpKX07Qy5wcm90b3R5cGUudT1mdW5jdGlvbigpe2lmKHRoaXMuSilmb3IoO3RoaXMuSi5sZW5ndGg7KXRoaXMuSi5zaGlmdCgpKCl9O3ZhciBBYj1mdW5jdGlvbihhKXthJiYiZnVuY3Rpb24iPT10eXBlb2YgYS5OJiZhLk4oKX07dmFyIEJiPWZ1bmN0aW9uKGEpe3JldHVybiJzdHJpbmciPT10eXBlb2YgYS5jbGFzc05hbWU/YS5jbGFzc05hbWU6YS5nZXRBdHRyaWJ1dGUmJmEuZ2V0QXR0cmlidXRlKCJjbGFzcyIpfHwiIn0sRD1mdW5jdGlvbihhKXtyZXR1cm4gYS5jbGFzc0xpc3Q/YS5jbGFzc0xpc3Q6QmIoYSkubWF0Y2goL1xTKy9nKXx8W119LENiPWZ1bmN0aW9uKGEsYil7InN0cmluZyI9PXR5cGVvZiBhLmNsYXNzTmFtZT9hLmNsYXNzTmFtZT1iOmEuc2V0QXR0cmlidXRlJiZhLnNldEF0dHJpYnV0ZSgiY2xhc3MiLGIpfSxEYj1mdW5jdGlvbihhLGIpe3JldHVybiBhLmNsYXNzTGlzdD9hLmNsYXNzTGlzdC5jb250YWlucyhiKTpHYShEKGEpLGIpfSxFYj1mdW5jdGlvbihhLGIpe2lmKGEuY2xhc3NMaXN0KWEuY2xhc3NMaXN0LmFkZChiKTtlbHNlIGlmKCFEYihhLGIpKXt2YXIgYz1CYihhKTtDYihhLGMrKDA8Yy5sZW5ndGg/IiAiK2I6YikpfX0sRmI9ZnVuY3Rpb24oYSxiKXtpZihhLmNsYXNzTGlzdClyKGIsCmZ1bmN0aW9uKGUpe0ViKGEsZSl9KTtlbHNle3ZhciBjPXt9O3IoRChhKSxmdW5jdGlvbihlKXtjW2VdPSEwfSk7cihiLGZ1bmN0aW9uKGUpe2NbZV09ITB9KTtiPSIiO2Zvcih2YXIgZCBpbiBjKWIrPTA8Yi5sZW5ndGg/IiAiK2Q6ZDtDYihhLGIpfX0sR2I9ZnVuY3Rpb24oYSxiKXthLmNsYXNzTGlzdD9hLmNsYXNzTGlzdC5yZW1vdmUoYik6RGIoYSxiKSYmQ2IoYSxFYShEKGEpLGZ1bmN0aW9uKGMpe3JldHVybiBjIT1ifSkuam9pbigiICIpKX0sSGI9ZnVuY3Rpb24oYSxiKXthLmNsYXNzTGlzdD9yKGIsZnVuY3Rpb24oYyl7R2IoYSxjKX0pOkNiKGEsRWEoRChhKSxmdW5jdGlvbihjKXtyZXR1cm4hR2EoYixjKX0pLmpvaW4oIiAiKSl9O3ZhciBJYj0hdnx8OTw9TnVtYmVyKGliKSxKYj0hdnx8OTw9TnVtYmVyKGliKSxLYj12JiYhQSgiOSIpLExiPWZ1bmN0aW9uKCl7aWYoIWwuYWRkRXZlbnRMaXN0ZW5lcnx8IU9iamVjdC5kZWZpbmVQcm9wZXJ0eSlyZXR1cm4hMTt2YXIgYT0hMSxiPU9iamVjdC5kZWZpbmVQcm9wZXJ0eSh7fSwicGFzc2l2ZSIse2dldDpmdW5jdGlvbigpe2E9ITB9fSk7dHJ5e2wuYWRkRXZlbnRMaXN0ZW5lcigidGVzdCIsY2EsYiksbC5yZW1vdmVFdmVudExpc3RlbmVyKCJ0ZXN0IixjYSxiKX1jYXRjaChjKXt9cmV0dXJuIGF9KCk7dmFyIEU9ZnVuY3Rpb24oYSxiKXt0aGlzLnR5cGU9YTt0aGlzLmE9dGhpcy50YXJnZXQ9Yjt0aGlzLmg9ITE7dGhpcy5LYT0hMH07RS5wcm90b3R5cGUuaj1mdW5jdGlvbigpe3RoaXMuaD0hMH07RS5wcm90b3R5cGUuZz1mdW5jdGlvbigpe3RoaXMuS2E9ITF9O3ZhciBGPXtMOiJtb3VzZWRvd24iLE06Im1vdXNldXAiLGFhOiJtb3VzZWNhbmNlbCIsaGM6Im1vdXNlbW92ZSIsamM6Im1vdXNlb3ZlciIsaWM6Im1vdXNlb3V0IixmYzoibW91c2VlbnRlciIsZ2M6Im1vdXNlbGVhdmUifTt2YXIgRz1mdW5jdGlvbihhLGIpe0UuY2FsbCh0aGlzLGE/YS50eXBlOiIiKTt0aGlzLnJlbGF0ZWRUYXJnZXQ9dGhpcy5hPXRoaXMudGFyZ2V0PW51bGw7dGhpcy5idXR0b249dGhpcy5zY3JlZW5ZPXRoaXMuc2NyZWVuWD10aGlzLmNsaWVudFk9dGhpcy5jbGllbnRYPTA7dGhpcy5rZXk9IiI7dGhpcy5jPTA7dGhpcy5BPXRoaXMubWV0YUtleT10aGlzLnNoaWZ0S2V5PXRoaXMuYWx0S2V5PXRoaXMuY3RybEtleT0hMTt0aGlzLnBvaW50ZXJJZD0wO3RoaXMucG9pbnRlclR5cGU9IiI7dGhpcy5iPW51bGw7aWYoYSl7dmFyIGM9dGhpcy50eXBlPWEudHlwZSxkPWEuY2hhbmdlZFRvdWNoZXMmJmEuY2hhbmdlZFRvdWNoZXMubGVuZ3RoP2EuY2hhbmdlZFRvdWNoZXNbMF06bnVsbDt0aGlzLnRhcmdldD1hLnRhcmdldHx8YS5zcmNFbGVtZW50O3RoaXMuYT1iO2lmKGI9YS5yZWxhdGVkVGFyZ2V0KXtpZih4KXthOnt0cnl7WWEoYi5ub2RlTmFtZSk7dmFyIGU9ITA7YnJlYWsgYX1jYXRjaChmKXt9ZT0KITF9ZXx8KGI9bnVsbCl9fWVsc2UibW91c2VvdmVyIj09Yz9iPWEuZnJvbUVsZW1lbnQ6Im1vdXNlb3V0Ij09YyYmKGI9YS50b0VsZW1lbnQpO3RoaXMucmVsYXRlZFRhcmdldD1iO2Q/KHRoaXMuY2xpZW50WD12b2lkIDAhPT1kLmNsaWVudFg/ZC5jbGllbnRYOmQucGFnZVgsdGhpcy5jbGllbnRZPXZvaWQgMCE9PWQuY2xpZW50WT9kLmNsaWVudFk6ZC5wYWdlWSx0aGlzLnNjcmVlblg9ZC5zY3JlZW5YfHwwLHRoaXMuc2NyZWVuWT1kLnNjcmVlbll8fDApOih0aGlzLmNsaWVudFg9dm9pZCAwIT09YS5jbGllbnRYP2EuY2xpZW50WDphLnBhZ2VYLHRoaXMuY2xpZW50WT12b2lkIDAhPT1hLmNsaWVudFk/YS5jbGllbnRZOmEucGFnZVksdGhpcy5zY3JlZW5YPWEuc2NyZWVuWHx8MCx0aGlzLnNjcmVlblk9YS5zY3JlZW5ZfHwwKTt0aGlzLmJ1dHRvbj1hLmJ1dHRvbjt0aGlzLmM9YS5rZXlDb2RlfHwwO3RoaXMua2V5PWEua2V5fHwiIjt0aGlzLmN0cmxLZXk9YS5jdHJsS2V5O3RoaXMuYWx0S2V5PQphLmFsdEtleTt0aGlzLnNoaWZ0S2V5PWEuc2hpZnRLZXk7dGhpcy5tZXRhS2V5PWEubWV0YUtleTt0aGlzLkE9ej9hLm1ldGFLZXk6YS5jdHJsS2V5O3RoaXMucG9pbnRlcklkPWEucG9pbnRlcklkfHwwO3RoaXMucG9pbnRlclR5cGU9InN0cmluZyI9PT10eXBlb2YgYS5wb2ludGVyVHlwZT9hLnBvaW50ZXJUeXBlOk1iW2EucG9pbnRlclR5cGVdfHwiIjt0aGlzLmI9YTthLmRlZmF1bHRQcmV2ZW50ZWQmJnRoaXMuZygpfX07bihHLEUpO3ZhciBOYj15YihbMSw0LDJdKSxNYj15Yih7MjoidG91Y2giLDM6InBlbiIsNDoibW91c2UifSksT2I9ZnVuY3Rpb24oYSl7cmV0dXJuIEliPzA9PWEuYi5idXR0b246ImNsaWNrIj09YS50eXBlPyEwOiEhKGEuYi5idXR0b24mTmJbMF0pfTtHLnByb3RvdHlwZS5qPWZ1bmN0aW9uKCl7Ry5pLmouY2FsbCh0aGlzKTt0aGlzLmIuc3RvcFByb3BhZ2F0aW9uP3RoaXMuYi5zdG9wUHJvcGFnYXRpb24oKTp0aGlzLmIuY2FuY2VsQnViYmxlPSEwfTsKRy5wcm90b3R5cGUuZz1mdW5jdGlvbigpe0cuaS5nLmNhbGwodGhpcyk7dmFyIGE9dGhpcy5iO2lmKGEucHJldmVudERlZmF1bHQpYS5wcmV2ZW50RGVmYXVsdCgpO2Vsc2UgaWYoYS5yZXR1cm5WYWx1ZT0hMSxLYil0cnl7aWYoYS5jdHJsS2V5fHwxMTI8PWEua2V5Q29kZSYmMTIzPj1hLmtleUNvZGUpYS5rZXlDb2RlPS0xfWNhdGNoKGIpe319O3ZhciBQYj0iY2xvc3VyZV9saXN0ZW5hYmxlXyIrKDFFNipNYXRoLnJhbmRvbSgpfDApLFFiPWZ1bmN0aW9uKGEpe3JldHVybiEoIWF8fCFhW1BiXSl9LFJiPTA7dmFyIFNiPWZ1bmN0aW9uKGEsYixjLGQsZSl7dGhpcy5saXN0ZW5lcj1hO3RoaXMuYT1udWxsO3RoaXMuc3JjPWI7dGhpcy50eXBlPWM7dGhpcy5jYXB0dXJlPSEhZDt0aGlzLm1hPWU7dGhpcy5rZXk9KytSYjt0aGlzLlg9dGhpcy5oYT0hMX0sVGI9ZnVuY3Rpb24oYSl7YS5YPSEwO2EubGlzdGVuZXI9bnVsbDthLmE9bnVsbDthLnNyYz1udWxsO2EubWE9bnVsbH07dmFyIFViPWZ1bmN0aW9uKGEpe3RoaXMuc3JjPWE7dGhpcy5hPXt9O3RoaXMuYj0wfTtVYi5wcm90b3R5cGUuYWRkPWZ1bmN0aW9uKGEsYixjLGQsZSl7dmFyIGY9YS50b1N0cmluZygpO2E9dGhpcy5hW2ZdO2F8fChhPXRoaXMuYVtmXT1bXSx0aGlzLmIrKyk7dmFyIGc9VmIoYSxiLGQsZSk7LTE8Zz8oYj1hW2ddLGN8fChiLmhhPSExKSk6KGI9bmV3IFNiKGIsdGhpcy5zcmMsZiwhIWQsZSksYi5oYT1jLGEucHVzaChiKSk7cmV0dXJuIGJ9Owp2YXIgV2I9ZnVuY3Rpb24oYSxiKXt2YXIgYz1iLnR5cGU7YyBpbiBhLmEmJkhhKGEuYVtjXSxiKSYmKFRiKGIpLDA9PWEuYVtjXS5sZW5ndGgmJihkZWxldGUgYS5hW2NdLGEuYi0tKSl9LFhiPWZ1bmN0aW9uKGEsYixjLGQsZSl7YT1hLmFbYi50b1N0cmluZygpXTtiPS0xO2EmJihiPVZiKGEsYyxkLGUpKTtyZXR1cm4tMTxiP2FbYl06bnVsbH0sVmI9ZnVuY3Rpb24oYSxiLGMsZCl7Zm9yKHZhciBlPTA7ZTxhLmxlbmd0aDsrK2Upe3ZhciBmPWFbZV07aWYoIWYuWCYmZi5saXN0ZW5lcj09YiYmZi5jYXB0dXJlPT0hIWMmJmYubWE9PWQpcmV0dXJuIGV9cmV0dXJuLTF9O3ZhciBZYj0iY2xvc3VyZV9sbV8iKygxRTYqTWF0aC5yYW5kb20oKXwwKSxaYj17fSwkYj0wLGJjPWZ1bmN0aW9uKGEsYixjLGQsZSl7aWYoZCYmZC5vbmNlKXJldHVybiBhYyhhLGIsYyxkLGUpO2lmKGZhKGIpKXtmb3IodmFyIGY9MDtmPGIubGVuZ3RoO2YrKyliYyhhLGJbZl0sYyxkLGUpO3JldHVybiBudWxsfWM9Y2MoYyk7cmV0dXJuIFFiKGEpP2RjKGEsYixjLG0oZCk/ISFkLmNhcHR1cmU6ISFkLGUpOmVjKGEsYixjLCExLGQsZSl9LGVjPWZ1bmN0aW9uKGEsYixjLGQsZSxmKXtpZighYil0aHJvdyBFcnJvcigiSW52YWxpZCBldmVudCB0eXBlIik7dmFyIGc9bShlKT8hIWUuY2FwdHVyZTohIWUsaz1mYyhhKTtrfHwoYVtZYl09az1uZXcgVWIoYSkpO2M9ay5hZGQoYixjLGQsZyxmKTtpZihjLmEpcmV0dXJuIGM7ZD1nYygpO2MuYT1kO2Quc3JjPWE7ZC5saXN0ZW5lcj1jO2lmKGEuYWRkRXZlbnRMaXN0ZW5lcilMYnx8KGU9Zyksdm9pZCAwPT09ZSYmKGU9ITEpLGEuYWRkRXZlbnRMaXN0ZW5lcihiLnRvU3RyaW5nKCksCmQsZSk7ZWxzZSBpZihhLmF0dGFjaEV2ZW50KWEuYXR0YWNoRXZlbnQoaGMoYi50b1N0cmluZygpKSxkKTtlbHNlIGlmKGEuYWRkTGlzdGVuZXImJmEucmVtb3ZlTGlzdGVuZXIpcSgiY2hhbmdlIj09PWIsIk1lZGlhUXVlcnlMaXN0IG9ubHkgaGFzIGEgY2hhbmdlIGV2ZW50IiksYS5hZGRMaXN0ZW5lcihkKTtlbHNlIHRocm93IEVycm9yKCJhZGRFdmVudExpc3RlbmVyIGFuZCBhdHRhY2hFdmVudCBhcmUgdW5hdmFpbGFibGUuIik7JGIrKztyZXR1cm4gY30sZ2M9ZnVuY3Rpb24oKXt2YXIgYT1pYyxiPUpiP2Z1bmN0aW9uKGMpe3JldHVybiBhLmNhbGwoYi5zcmMsYi5saXN0ZW5lcixjKX06ZnVuY3Rpb24oYyl7Yz1hLmNhbGwoYi5zcmMsYi5saXN0ZW5lcixjKTtpZighYylyZXR1cm4gY307cmV0dXJuIGJ9LGFjPWZ1bmN0aW9uKGEsYixjLGQsZSl7aWYoZmEoYikpe2Zvcih2YXIgZj0wO2Y8Yi5sZW5ndGg7ZisrKWFjKGEsYltmXSxjLGQsZSk7cmV0dXJuIG51bGx9Yz1jYyhjKTtyZXR1cm4gUWIoYSk/CmEuaC5hZGQoU3RyaW5nKGIpLGMsITAsbShkKT8hIWQuY2FwdHVyZTohIWQsZSk6ZWMoYSxiLGMsITAsZCxlKX0samM9ZnVuY3Rpb24oYSxiLGMsZCxlKXtpZihmYShiKSlmb3IodmFyIGY9MDtmPGIubGVuZ3RoO2YrKylqYyhhLGJbZl0sYyxkLGUpO2Vsc2UgZD1tKGQpPyEhZC5jYXB0dXJlOiEhZCxjPWNjKGMpLFFiKGEpPyhhPWEuaCxiPVN0cmluZyhiKS50b1N0cmluZygpLGIgaW4gYS5hJiYoZj1hLmFbYl0sYz1WYihmLGMsZCxlKSwtMTxjJiYoVGIoZltjXSkscShudWxsIT1mLmxlbmd0aCksQXJyYXkucHJvdG90eXBlLnNwbGljZS5jYWxsKGYsYywxKSwwPT1mLmxlbmd0aCYmKGRlbGV0ZSBhLmFbYl0sYS5iLS0pKSkpOmEmJihhPWZjKGEpKSYmKGM9WGIoYSxiLGMsZCxlKSkmJmtjKGMpfSxrYz1mdW5jdGlvbihhKXtpZigibnVtYmVyIiE9PXR5cGVvZiBhJiZhJiYhYS5YKXt2YXIgYj1hLnNyYztpZihRYihiKSlXYihiLmgsYSk7ZWxzZXt2YXIgYz1hLnR5cGUsZD1hLmE7Yi5yZW1vdmVFdmVudExpc3RlbmVyPwpiLnJlbW92ZUV2ZW50TGlzdGVuZXIoYyxkLGEuY2FwdHVyZSk6Yi5kZXRhY2hFdmVudD9iLmRldGFjaEV2ZW50KGhjKGMpLGQpOmIuYWRkTGlzdGVuZXImJmIucmVtb3ZlTGlzdGVuZXImJmIucmVtb3ZlTGlzdGVuZXIoZCk7JGItLTsoYz1mYyhiKSk/KFdiKGMsYSksMD09Yy5iJiYoYy5zcmM9bnVsbCxiW1liXT1udWxsKSk6VGIoYSl9fX0saGM9ZnVuY3Rpb24oYSl7cmV0dXJuIGEgaW4gWmI/WmJbYV06WmJbYV09Im9uIithfSxtYz1mdW5jdGlvbihhLGIsYyxkKXt2YXIgZT0hMDtpZihhPWZjKGEpKWlmKGI9YS5hW2IudG9TdHJpbmcoKV0pZm9yKGI9Yi5jb25jYXQoKSxhPTA7YTxiLmxlbmd0aDthKyspe3ZhciBmPWJbYV07ZiYmZi5jYXB0dXJlPT1jJiYhZi5YJiYoZj1sYyhmLGQpLGU9ZSYmITEhPT1mKX1yZXR1cm4gZX0sbGM9ZnVuY3Rpb24oYSxiKXt2YXIgYz1hLmxpc3RlbmVyLGQ9YS5tYXx8YS5zcmM7YS5oYSYma2MoYSk7cmV0dXJuIGMuY2FsbChkLGIpfSxpYz1mdW5jdGlvbihhLApiKXtpZihhLlgpcmV0dXJuITA7aWYoIUpiKXtpZighYilhOntiPVsid2luZG93IiwiZXZlbnQiXTtmb3IodmFyIGM9bCxkPTA7ZDxiLmxlbmd0aDtkKyspaWYoYz1jW2JbZF1dLG51bGw9PWMpe2I9bnVsbDticmVhayBhfWI9Y31kPWI7Yj1uZXcgRyhkLHRoaXMpO2M9ITA7aWYoISgwPmQua2V5Q29kZXx8dm9pZCAwIT1kLnJldHVyblZhbHVlKSl7YTp7dmFyIGU9ITE7aWYoMD09ZC5rZXlDb2RlKXRyeXtkLmtleUNvZGU9LTE7YnJlYWsgYX1jYXRjaChnKXtlPSEwfWlmKGV8fHZvaWQgMD09ZC5yZXR1cm5WYWx1ZSlkLnJldHVyblZhbHVlPSEwfWQ9W107Zm9yKGU9Yi5hO2U7ZT1lLnBhcmVudE5vZGUpZC5wdXNoKGUpO2E9YS50eXBlO2ZvcihlPWQubGVuZ3RoLTE7IWIuaCYmMDw9ZTtlLS0pe2IuYT1kW2VdO3ZhciBmPW1jKGRbZV0sYSwhMCxiKTtjPWMmJmZ9Zm9yKGU9MDshYi5oJiZlPGQubGVuZ3RoO2UrKyliLmE9ZFtlXSxmPW1jKGRbZV0sYSwhMSxiKSxjPWMmJmZ9cmV0dXJuIGN9cmV0dXJuIGxjKGEsCm5ldyBHKGIsdGhpcykpfSxmYz1mdW5jdGlvbihhKXthPWFbWWJdO3JldHVybiBhIGluc3RhbmNlb2YgVWI/YTpudWxsfSxuYz0iX19jbG9zdXJlX2V2ZW50c19mbl8iKygxRTkqTWF0aC5yYW5kb20oKT4+PjApLGNjPWZ1bmN0aW9uKGEpe3EoYSwiTGlzdGVuZXIgY2FuIG5vdCBiZSBudWxsLiIpO2lmKGlhKGEpKXJldHVybiBhO3EoYS5oYW5kbGVFdmVudCwiQW4gb2JqZWN0IGxpc3RlbmVyIG11c3QgaGF2ZSBoYW5kbGVFdmVudCBtZXRob2QuIik7YVtuY118fChhW25jXT1mdW5jdGlvbihiKXtyZXR1cm4gYS5oYW5kbGVFdmVudChiKX0pO3JldHVybiBhW25jXX07dmFyIG9jPWZ1bmN0aW9uKGEpe0MuY2FsbCh0aGlzKTt0aGlzLmI9YTt0aGlzLmE9e319O24ob2MsQyk7CnZhciBwYz1bXSxIPWZ1bmN0aW9uKGEsYixjLGQpe2ZhKGMpfHwoYyYmKHBjWzBdPWMudG9TdHJpbmcoKSksYz1wYyk7Zm9yKHZhciBlPTA7ZTxjLmxlbmd0aDtlKyspe3ZhciBmPWJjKGIsY1tlXSxkfHxhLmhhbmRsZUV2ZW50LCExLGEuYnx8YSk7aWYoIWYpYnJlYWs7YS5hW2Yua2V5XT1mfXJldHVybiBhfSxxYz1mdW5jdGlvbihhLGIsYyxkLGUsZil7aWYoZmEoYykpZm9yKHZhciBnPTA7ZzxjLmxlbmd0aDtnKyspcWMoYSxiLGNbZ10sZCxlLGYpO2Vsc2UgZD1kfHxhLmhhbmRsZUV2ZW50LGU9bShlKT8hIWUuY2FwdHVyZTohIWUsZj1mfHxhLmJ8fGEsZD1jYyhkKSxlPSEhZSxjPVFiKGIpP1hiKGIuaCxTdHJpbmcoYyksZCxlLGYpOmI/KGI9ZmMoYikpP1hiKGIsYyxkLGUsZik6bnVsbDpudWxsLGMmJihrYyhjKSxkZWxldGUgYS5hW2Mua2V5XSk7cmV0dXJuIGF9LHJjPWZ1bmN0aW9uKGEpe29hKGEuYSxmdW5jdGlvbihiLGMpe3RoaXMuYS5oYXNPd25Qcm9wZXJ0eShjKSYma2MoYil9LAphKTthLmE9e319O29jLnByb3RvdHlwZS51PWZ1bmN0aW9uKCl7b2MuaS51LmNhbGwodGhpcyk7cmModGhpcyl9O29jLnByb3RvdHlwZS5oYW5kbGVFdmVudD1mdW5jdGlvbigpe3Rocm93IEVycm9yKCJFdmVudEhhbmRsZXIuaGFuZGxlRXZlbnQgbm90IGltcGxlbWVudGVkIik7fTt2YXIgST1mdW5jdGlvbigpe0MuY2FsbCh0aGlzKTt0aGlzLmg9bmV3IFViKHRoaXMpO3RoaXMuTWE9dGhpczt0aGlzLnBhPW51bGx9O24oSSxDKTtJLnByb3RvdHlwZVtQYl09ITA7SS5wcm90b3R5cGUucmE9ZnVuY3Rpb24oYSl7dGhpcy5wYT1hfTtJLnByb3RvdHlwZS5yZW1vdmVFdmVudExpc3RlbmVyPWZ1bmN0aW9uKGEsYixjLGQpe2pjKHRoaXMsYSxiLGMsZCl9Owp2YXIgdWM9ZnVuY3Rpb24oYSxiKXtzYyhhKTt2YXIgYz1hLnBhO2lmKGMpe3ZhciBkPVtdO2Zvcih2YXIgZT0xO2M7Yz1jLnBhKWQucHVzaChjKSxxKDFFMz4rK2UsImluZmluaXRlIGxvb3AiKX1hPWEuTWE7Yz1iLnR5cGV8fGI7InN0cmluZyI9PT10eXBlb2YgYj9iPW5ldyBFKGIsYSk6YiBpbnN0YW5jZW9mIEU/Yi50YXJnZXQ9Yi50YXJnZXR8fGE6KGU9YixiPW5ldyBFKGMsYSksdWEoYixlKSk7ZT0hMDtpZihkKWZvcih2YXIgZj1kLmxlbmd0aC0xOyFiLmgmJjA8PWY7Zi0tKXt2YXIgZz1iLmE9ZFtmXTtlPXRjKGcsYywhMCxiKSYmZX1iLmh8fChnPWIuYT1hLGU9dGMoZyxjLCEwLGIpJiZlLGIuaHx8KGU9dGMoZyxjLCExLGIpJiZlKSk7aWYoZClmb3IoZj0wOyFiLmgmJmY8ZC5sZW5ndGg7ZisrKWc9Yi5hPWRbZl0sZT10YyhnLGMsITEsYikmJmU7cmV0dXJuIGV9OwpJLnByb3RvdHlwZS51PWZ1bmN0aW9uKCl7SS5pLnUuY2FsbCh0aGlzKTtpZih0aGlzLmgpe3ZhciBhPXRoaXMuaCxiPTAsYztmb3IoYyBpbiBhLmEpe2Zvcih2YXIgZD1hLmFbY10sZT0wO2U8ZC5sZW5ndGg7ZSsrKSsrYixUYihkW2VdKTtkZWxldGUgYS5hW2NdO2EuYi0tfX10aGlzLnBhPW51bGx9O3ZhciBkYz1mdW5jdGlvbihhLGIsYyxkLGUpe3NjKGEpO3JldHVybiBhLmguYWRkKFN0cmluZyhiKSxjLCExLGQsZSl9LHRjPWZ1bmN0aW9uKGEsYixjLGQpe2I9YS5oLmFbU3RyaW5nKGIpXTtpZighYilyZXR1cm4hMDtiPWIuY29uY2F0KCk7Zm9yKHZhciBlPSEwLGY9MDtmPGIubGVuZ3RoOysrZil7dmFyIGc9YltmXTtpZihnJiYhZy5YJiZnLmNhcHR1cmU9PWMpe3ZhciBrPWcubGlzdGVuZXIscD1nLm1hfHxnLnNyYztnLmhhJiZXYihhLmgsZyk7ZT0hMSE9PWsuY2FsbChwLGQpJiZlfX1yZXR1cm4gZSYmMCE9ZC5LYX0sc2M9ZnVuY3Rpb24oYSl7cShhLmgsIkV2ZW50IHRhcmdldCBpcyBub3QgaW5pdGlhbGl6ZWQuIERpZCB5b3UgY2FsbCB0aGUgc3VwZXJjbGFzcyAoZ29vZy5ldmVudHMuRXZlbnRUYXJnZXQpIGNvbnN0cnVjdG9yPyIpfTt2YXIgeGM9ZnVuY3Rpb24oYSxiLGMsZCxlLGYpe2lmKHkmJiFBKCI1MjUiKSlyZXR1cm4hMDtpZih6JiZlKXJldHVybiB2YyhhKTtpZihlJiYhZClyZXR1cm4hMTtpZigheCl7Im51bWJlciI9PT10eXBlb2YgYiYmKGI9d2MoYikpO3ZhciBnPTE3PT1ifHwxOD09Ynx8eiYmOTE9PWI7aWYoKCFjfHx6KSYmZ3x8eiYmMTY9PWImJihkfHxmKSlyZXR1cm4hMX1pZigoeXx8YmIpJiZkJiZjKXN3aXRjaChhKXtjYXNlIDIyMDpjYXNlIDIxOTpjYXNlIDIyMTpjYXNlIDE5MjpjYXNlIDE4NjpjYXNlIDE4OTpjYXNlIDE4NzpjYXNlIDE4ODpjYXNlIDE5MDpjYXNlIDE5MTpjYXNlIDE5MjpjYXNlIDIyMjpyZXR1cm4hMX1pZih2JiZkJiZiPT1hKXJldHVybiExO3N3aXRjaChhKXtjYXNlIDEzOnJldHVybiB4P2Z8fGU/ITE6IShjJiZkKTohMDtjYXNlIDI3OnJldHVybiEoeXx8YmJ8fHgpfXJldHVybiB4JiYoZHx8ZXx8Zik/ITE6dmMoYSl9LHZjPWZ1bmN0aW9uKGEpe2lmKDQ4PD1hJiY1Nz49YXx8Cjk2PD1hJiYxMDY+PWF8fDY1PD1hJiY5MD49YXx8KHl8fGJiKSYmMD09YSlyZXR1cm4hMDtzd2l0Y2goYSl7Y2FzZSAzMjpjYXNlIDQzOmNhc2UgNjM6Y2FzZSA2NDpjYXNlIDEwNzpjYXNlIDEwOTpjYXNlIDExMDpjYXNlIDExMTpjYXNlIDE4NjpjYXNlIDU5OmNhc2UgMTg5OmNhc2UgMTg3OmNhc2UgNjE6Y2FzZSAxODg6Y2FzZSAxOTA6Y2FzZSAxOTE6Y2FzZSAxOTI6Y2FzZSAyMjI6Y2FzZSAyMTk6Y2FzZSAyMjA6Y2FzZSAyMjE6Y2FzZSAxNjM6Y2FzZSA1ODpyZXR1cm4hMDtjYXNlIDE3MzpyZXR1cm4geDtkZWZhdWx0OnJldHVybiExfX0sd2M9ZnVuY3Rpb24oYSl7aWYoeClhPXljKGEpO2Vsc2UgaWYoeiYmeSlzd2l0Y2goYSl7Y2FzZSA5MzphPTkxfXJldHVybiBhfSx5Yz1mdW5jdGlvbihhKXtzd2l0Y2goYSl7Y2FzZSA2MTpyZXR1cm4gMTg3O2Nhc2UgNTk6cmV0dXJuIDE4NjtjYXNlIDE3MzpyZXR1cm4gMTg5O2Nhc2UgMjI0OnJldHVybiA5MTtjYXNlIDA6cmV0dXJuIDIyNDsKZGVmYXVsdDpyZXR1cm4gYX19O3ZhciBKPWZ1bmN0aW9uKGEsYil7SS5jYWxsKHRoaXMpO2EmJnpjKHRoaXMsYSxiKX07bihKLEkpO2g9Si5wcm90b3R5cGU7aC5TPW51bGw7aC5uYT1udWxsO2guQ2E9bnVsbDtoLm9hPW51bGw7aC53PS0xO2guRj0tMTtoLnVhPSExOwp2YXIgQWM9ezM6MTMsMTI6MTQ0LDYzMjMyOjM4LDYzMjMzOjQwLDYzMjM0OjM3LDYzMjM1OjM5LDYzMjM2OjExMiw2MzIzNzoxMTMsNjMyMzg6MTE0LDYzMjM5OjExNSw2MzI0MDoxMTYsNjMyNDE6MTE3LDYzMjQyOjExOCw2MzI0MzoxMTksNjMyNDQ6MTIwLDYzMjQ1OjEyMSw2MzI0NjoxMjIsNjMyNDc6MTIzLDYzMjQ4OjQ0LDYzMjcyOjQ2LDYzMjczOjM2LDYzMjc1OjM1LDYzMjc2OjMzLDYzMjc3OjM0LDYzMjg5OjE0NCw2MzMwMjo0NX0sQmM9e1VwOjM4LERvd246NDAsTGVmdDozNyxSaWdodDozOSxFbnRlcjoxMyxGMToxMTIsRjI6MTEzLEYzOjExNCxGNDoxMTUsRjU6MTE2LEY2OjExNyxGNzoxMTgsRjg6MTE5LEY5OjEyMCxGMTA6MTIxLEYxMToxMjIsRjEyOjEyMywiVSswMDdGIjo0NixIb21lOjM2LEVuZDozNSxQYWdlVXA6MzMsUGFnZURvd246MzQsSW5zZXJ0OjQ1fSxDYz0heXx8QSgiNTI1IiksRGM9eiYmeDsKSi5wcm90b3R5cGUuYT1mdW5jdGlvbihhKXtpZih5fHxiYilpZigxNz09dGhpcy53JiYhYS5jdHJsS2V5fHwxOD09dGhpcy53JiYhYS5hbHRLZXl8fHomJjkxPT10aGlzLncmJiFhLm1ldGFLZXkpdGhpcy5GPXRoaXMudz0tMTstMT09dGhpcy53JiYoYS5jdHJsS2V5JiYxNyE9YS5jP3RoaXMudz0xNzphLmFsdEtleSYmMTghPWEuYz90aGlzLnc9MTg6YS5tZXRhS2V5JiY5MSE9YS5jJiYodGhpcy53PTkxKSk7Q2MmJiF4YyhhLmMsdGhpcy53LGEuc2hpZnRLZXksYS5jdHJsS2V5LGEuYWx0S2V5LGEubWV0YUtleSk/dGhpcy5oYW5kbGVFdmVudChhKToodGhpcy5GPXdjKGEuYyksRGMmJih0aGlzLnVhPWEuYWx0S2V5KSl9O0oucHJvdG90eXBlLmI9ZnVuY3Rpb24oYSl7dGhpcy5GPXRoaXMudz0tMTt0aGlzLnVhPWEuYWx0S2V5fTsKSi5wcm90b3R5cGUuaGFuZGxlRXZlbnQ9ZnVuY3Rpb24oYSl7dmFyIGI9YS5iLGM9Yi5hbHRLZXk7aWYodiYmImtleXByZXNzIj09YS50eXBlKXt2YXIgZD10aGlzLkY7dmFyIGU9MTMhPWQmJjI3IT1kP2Iua2V5Q29kZTowfWVsc2UoeXx8YmIpJiYia2V5cHJlc3MiPT1hLnR5cGU/KGQ9dGhpcy5GLGU9MDw9Yi5jaGFyQ29kZSYmNjMyMzI+Yi5jaGFyQ29kZSYmdmMoZCk/Yi5jaGFyQ29kZTowKTphYiYmIXk/KGQ9dGhpcy5GLGU9dmMoZCk/Yi5rZXlDb2RlOjApOigia2V5cHJlc3MiPT1hLnR5cGU/KERjJiYoYz10aGlzLnVhKSxiLmtleUNvZGU9PWIuY2hhckNvZGU/MzI+Yi5rZXlDb2RlPyhkPWIua2V5Q29kZSxlPTApOihkPXRoaXMuRixlPWIuY2hhckNvZGUpOihkPWIua2V5Q29kZXx8dGhpcy5GLGU9Yi5jaGFyQ29kZXx8MCkpOihkPWIua2V5Q29kZXx8dGhpcy5GLGU9Yi5jaGFyQ29kZXx8MCkseiYmNjM9PWUmJjIyND09ZCYmKGQ9MTkxKSk7dmFyIGY9ZD13YyhkKTtkPzYzMjMyPD0KZCYmZCBpbiBBYz9mPUFjW2RdOjI1PT1kJiZhLnNoaWZ0S2V5JiYoZj05KTpiLmtleUlkZW50aWZpZXImJmIua2V5SWRlbnRpZmllciBpbiBCYyYmKGY9QmNbYi5rZXlJZGVudGlmaWVyXSk7eCYmQ2MmJiJrZXlwcmVzcyI9PWEudHlwZSYmIXhjKGYsdGhpcy53LGEuc2hpZnRLZXksYS5jdHJsS2V5LGMsYS5tZXRhS2V5KXx8KGE9Zj09dGhpcy53LHRoaXMudz1mLGI9bmV3IEVjKGYsZSxhLGIpLGIuYWx0S2V5PWMsdWModGhpcyxiKSl9O0oucHJvdG90eXBlLmY9ZnVuY3Rpb24oKXtyZXR1cm4gdGhpcy5TfTsKdmFyIHpjPWZ1bmN0aW9uKGEsYixjKXthLm9hJiZGYyhhKTthLlM9YjthLm5hPWJjKGEuUywia2V5cHJlc3MiLGEsYyk7YS5DYT1iYyhhLlMsImtleWRvd24iLGEuYSxjLGEpO2Eub2E9YmMoYS5TLCJrZXl1cCIsYS5iLGMsYSl9LEZjPWZ1bmN0aW9uKGEpe2EubmEmJihrYyhhLm5hKSxrYyhhLkNhKSxrYyhhLm9hKSxhLm5hPW51bGwsYS5DYT1udWxsLGEub2E9bnVsbCk7YS5TPW51bGw7YS53PS0xO2EuRj0tMX07Si5wcm90b3R5cGUudT1mdW5jdGlvbigpe0ouaS51LmNhbGwodGhpcyk7RmModGhpcyl9O3ZhciBFYz1mdW5jdGlvbihhLGIsYyxkKXtHLmNhbGwodGhpcyxkKTt0aGlzLnR5cGU9ImtleSI7dGhpcy5jPWE7dGhpcy5yZXBlYXQ9Y307bihFYyxHKTt2YXIgR2M9eD8iTW96VXNlclNlbGVjdCI6eXx8YmI/IldlYmtpdFVzZXJTZWxlY3QiOm51bGwsSGM9ZnVuY3Rpb24oYSxiKXtiPWI/bnVsbDphLmdldEVsZW1lbnRzQnlUYWdOYW1lKCIqIik7aWYoR2Mpe3ZhciBjPSJub25lIjthLnN0eWxlJiYoYS5zdHlsZVtHY109Yyk7aWYoYil7YT0wO2Zvcih2YXIgZDtkPWJbYV07YSsrKWQuc3R5bGUmJihkLnN0eWxlW0djXT1jKX19ZWxzZSBpZih2fHxhYilpZihjPSJvbiIsYS5zZXRBdHRyaWJ1dGUoInVuc2VsZWN0YWJsZSIsYyksYilmb3IoYT0wO2Q9YlthXTthKyspZC5zZXRBdHRyaWJ1dGUoInVuc2VsZWN0YWJsZSIsYyl9O3ZhciBJYz1mdW5jdGlvbigpe307ZGEoSWMpO0ljLnByb3RvdHlwZS5hPTA7dmFyIEs9ZnVuY3Rpb24oYSl7SS5jYWxsKHRoaXMpO3RoaXMuaj1hfHx4YXx8KHhhPW5ldyB1Yik7dGhpcy5xYT1KYzt0aGlzLlk9bnVsbDt0aGlzLm09ITE7dGhpcy5hPW51bGw7dGhpcy5BPXZvaWQgMDt0aGlzLmc9dGhpcy5jPXRoaXMuYj1udWxsO3RoaXMuR2E9ITF9O24oSyxJKTtLLnByb3RvdHlwZS5PYT1JYy5SKCk7CnZhciBKYz1udWxsLEtjPWZ1bmN0aW9uKGEsYil7c3dpdGNoKGEpe2Nhc2UgMTpyZXR1cm4gYj8iZGlzYWJsZSI6ImVuYWJsZSI7Y2FzZSAyOnJldHVybiBiPyJoaWdobGlnaHQiOiJ1bmhpZ2hsaWdodCI7Y2FzZSA0OnJldHVybiBiPyJhY3RpdmF0ZSI6ImRlYWN0aXZhdGUiO2Nhc2UgODpyZXR1cm4gYj8ic2VsZWN0IjoidW5zZWxlY3QiO2Nhc2UgMTY6cmV0dXJuIGI/ImNoZWNrIjoidW5jaGVjayI7Y2FzZSAzMjpyZXR1cm4gYj8iZm9jdXMiOiJibHVyIjtjYXNlIDY0OnJldHVybiBiPyJvcGVuIjoiY2xvc2UifXRocm93IEVycm9yKCJJbnZhbGlkIGNvbXBvbmVudCBzdGF0ZSIpO30sTGM9ZnVuY3Rpb24oYSl7cmV0dXJuIGEuWXx8KGEuWT0iOiIrKGEuT2EuYSsrKS50b1N0cmluZygzNikpfSxNYz1mdW5jdGlvbihhLGIpe2lmKGEuYiYmYS5iLmcpe3ZhciBjPWEuYi5nLGQ9YS5ZO2QgaW4gYyYmZGVsZXRlIGNbZF07cWEoYS5iLmcsYixhKX1hLlk9Yn07Sy5wcm90b3R5cGUuZj1mdW5jdGlvbigpe3JldHVybiB0aGlzLmF9Owp2YXIgTmM9ZnVuY3Rpb24oYSl7YT1hLmE7cShhLCJDYW4gbm90IGNhbGwgZ2V0RWxlbWVudFN0cmljdCBiZWZvcmUgcmVuZGVyaW5nL2RlY29yYXRpbmcuIik7cmV0dXJuIGF9LE9jPWZ1bmN0aW9uKGEpe2EuQXx8KGEuQT1uZXcgb2MoYSkpO3JldHVybiBxKGEuQSl9O0sucHJvdG90eXBlLnJhPWZ1bmN0aW9uKGEpe2lmKHRoaXMuYiYmdGhpcy5iIT1hKXRocm93IEVycm9yKCJNZXRob2Qgbm90IHN1cHBvcnRlZCIpO0suaS5yYS5jYWxsKHRoaXMsYSl9O0sucHJvdG90eXBlLmlhPWZ1bmN0aW9uKCl7dGhpcy5hPW5iKHRoaXMuai5hLCJESVYiKX07CnZhciBQYz1mdW5jdGlvbihhLGIpe2lmKGEubSl0aHJvdyBFcnJvcigiQ29tcG9uZW50IGFscmVhZHkgcmVuZGVyZWQiKTtpZihiJiZhLnhhKGIpKXthLkdhPSEwO3ZhciBjPXFiKGIpO2EuaiYmYS5qLmE9PWN8fChhLmo9Yj9uZXcgdWIocWIoYikpOnhhfHwoeGE9bmV3IHViKSk7YS52YShiKTthLkQoKX1lbHNlIHRocm93IEVycm9yKCJJbnZhbGlkIGVsZW1lbnQgdG8gZGVjb3JhdGUiKTt9O2g9Sy5wcm90b3R5cGU7aC54YT1mdW5jdGlvbigpe3JldHVybiEwfTtoLnZhPWZ1bmN0aW9uKGEpe3RoaXMuYT1hfTtoLkQ9ZnVuY3Rpb24oKXt0aGlzLm09ITA7UWModGhpcyxmdW5jdGlvbihhKXshYS5tJiZhLmYoKSYmYS5EKCl9KX07aC5QPWZ1bmN0aW9uKCl7UWModGhpcyxmdW5jdGlvbihhKXthLm0mJmEuUCgpfSk7dGhpcy5BJiZyYyh0aGlzLkEpO3RoaXMubT0hMX07CmgudT1mdW5jdGlvbigpe3RoaXMubSYmdGhpcy5QKCk7dGhpcy5BJiYodGhpcy5BLk4oKSxkZWxldGUgdGhpcy5BKTtRYyh0aGlzLGZ1bmN0aW9uKGEpe2EuTigpfSk7IXRoaXMuR2EmJnRoaXMuYSYmb2IodGhpcy5hKTt0aGlzLmI9dGhpcy5hPXRoaXMuZz10aGlzLmM9bnVsbDtLLmkudS5jYWxsKHRoaXMpfTtoLnNhPWZ1bmN0aW9uKGEsYil7dGhpcy50YShhLFJjKHRoaXMpLGIpfTsKaC50YT1mdW5jdGlvbihhLGIsYyl7cSghIWEsIlByb3ZpZGVkIGVsZW1lbnQgbXVzdCBub3QgYmUgbnVsbC4iKTtpZihhLm0mJihjfHwhdGhpcy5tKSl0aHJvdyBFcnJvcigiQ29tcG9uZW50IGFscmVhZHkgcmVuZGVyZWQiKTtpZigwPmJ8fGI+UmModGhpcykpdGhyb3cgRXJyb3IoIkNoaWxkIGNvbXBvbmVudCBpbmRleCBvdXQgb2YgYm91bmRzIik7dGhpcy5nJiZ0aGlzLmN8fCh0aGlzLmc9e30sdGhpcy5jPVtdKTtpZihhLmI9PXRoaXMpe3ZhciBkPUxjKGEpO3RoaXMuZ1tkXT1hO0hhKHRoaXMuYyxhKX1lbHNlIHFhKHRoaXMuZyxMYyhhKSxhKTtpZihhPT10aGlzKXRocm93IEVycm9yKCJVbmFibGUgdG8gc2V0IHBhcmVudCBjb21wb25lbnQiKTtpZihkPXRoaXMmJmEuYiYmYS5ZKXt2YXIgZT1hLmI7ZD1hLlk7ZS5nJiZkPyhlPWUuZyxkPShudWxsIT09ZSYmZCBpbiBlP2VbZF06dm9pZCAwKXx8bnVsbCk6ZD1udWxsfWlmKGQmJmEuYiE9dGhpcyl0aHJvdyBFcnJvcigiVW5hYmxlIHRvIHNldCBwYXJlbnQgY29tcG9uZW50Iik7CmEuYj10aGlzO0suaS5yYS5jYWxsKGEsdGhpcyk7TGEodGhpcy5jLGIsMCxhKTtpZihhLm0mJnRoaXMubSYmYS5iPT10aGlzKWM9dGhpcy5qYSgpLGI9Yy5jaGlsZE5vZGVzW2JdfHxudWxsLGIhPWEuZigpJiZjLmluc2VydEJlZm9yZShhLmYoKSxiKTtlbHNlIGlmKGMpe3RoaXMuYXx8dGhpcy5pYSgpO2M9TCh0aGlzLGIrMSk7Yj10aGlzLmphKCk7Yz1jP2MuYTpudWxsO2lmKGEubSl0aHJvdyBFcnJvcigiQ29tcG9uZW50IGFscmVhZHkgcmVuZGVyZWQiKTthLmF8fGEuaWEoKTtiP2IuaW5zZXJ0QmVmb3JlKGEuYSxjfHxudWxsKTphLmouYS5ib2R5LmFwcGVuZENoaWxkKGEuYSk7YS5iJiYhYS5iLm18fGEuRCgpfWVsc2UgdGhpcy5tJiYhYS5tJiZhLmEmJmEuYS5wYXJlbnROb2RlJiYxPT1hLmEucGFyZW50Tm9kZS5ub2RlVHlwZSYmYS5EKCl9O2guamE9ZnVuY3Rpb24oKXtyZXR1cm4gdGhpcy5hfTsKdmFyIFNjPWZ1bmN0aW9uKGEpe2lmKG51bGw9PWEucWEpe3ZhciBiPWEubT9hLmE6YS5qLmEuYm9keTthOnt2YXIgYz1xYihiKTtpZihjLmRlZmF1bHRWaWV3JiZjLmRlZmF1bHRWaWV3LmdldENvbXB1dGVkU3R5bGUmJihjPWMuZGVmYXVsdFZpZXcuZ2V0Q29tcHV0ZWRTdHlsZShiLG51bGwpKSl7Yz1jLmRpcmVjdGlvbnx8Yy5nZXRQcm9wZXJ0eVZhbHVlKCJkaXJlY3Rpb24iKXx8IiI7YnJlYWsgYX1jPSIifWEucWE9InJ0bCI9PShjfHwoYi5jdXJyZW50U3R5bGU/Yi5jdXJyZW50U3R5bGUuZGlyZWN0aW9uOm51bGwpfHxiLnN0eWxlJiZiLnN0eWxlLmRpcmVjdGlvbil9cmV0dXJuIGEucWF9LFJjPWZ1bmN0aW9uKGEpe3JldHVybiBhLmM/YS5jLmxlbmd0aDowfSxMPWZ1bmN0aW9uKGEsYil7cmV0dXJuIGEuYz9hLmNbYl18fG51bGw6bnVsbH0sUWM9ZnVuY3Rpb24oYSxiLGMpe2EuYyYmcihhLmMsYixjKX0sVGM9ZnVuY3Rpb24oYSxiKXtyZXR1cm4gYS5jJiZiP0RhKGEuYyxiKTotMX07dmFyIFZjPWZ1bmN0aW9uKGEsYil7aWYoIWEpdGhyb3cgRXJyb3IoIkludmFsaWQgY2xhc3MgbmFtZSAiK2EpO2lmKCFpYShiKSl0aHJvdyBFcnJvcigiSW52YWxpZCBkZWNvcmF0b3IgZnVuY3Rpb24gIitiKTtVY1thXT1ifSxXYz17fSxVYz17fTt2YXIgWGM9ZnVuY3Rpb24oYSl7dGhpcy5oPWF9O2RhKFhjKTt2YXIgWWM9ZnVuY3Rpb24oYSxiKXthJiYoYS50YWJJbmRleD1iPzA6LTEpfSwkYz1mdW5jdGlvbihhLGIsYyl7Yy5pZCYmTWMoYixjLmlkKTt2YXIgZD1hLmIoKSxlPSExLGY9RChjKTtmJiZyKGYsZnVuY3Rpb24oZyl7Zz09ZD9lPSEwOmcmJnRoaXMuZyhiLGcsZCl9LGEpO2V8fEViKGMsZCk7WmMoYixjKTtyZXR1cm4gY307WGMucHJvdG90eXBlLmc9ZnVuY3Rpb24oYSxiLGMpe2I9PWMrIi1kaXNhYmxlZCI/YS5aKCExKTpiPT1jKyItaG9yaXpvbnRhbCI/YWQoYSwiaG9yaXpvbnRhbCIpOmI9PWMrIi12ZXJ0aWNhbCImJmFkKGEsInZlcnRpY2FsIil9Owp2YXIgWmM9ZnVuY3Rpb24oYSxiKXtpZihiKWZvcih2YXIgYz1iLmZpcnN0Q2hpbGQsZDtjJiZjLnBhcmVudE5vZGU9PWI7KXtkPWMubmV4dFNpYmxpbmc7aWYoMT09Yy5ub2RlVHlwZSl7YTp7dmFyIGU9YztxKGUpO2U9RChlKTtmb3IodmFyIGY9MCxnPWUubGVuZ3RoO2Y8ZztmKyspe3ZhciBrPWVbZl07aWYoaz1rIGluIFVjP1VjW2tdKCk6bnVsbCl7ZT1rO2JyZWFrIGF9fWU9bnVsbH1lJiYoZS5hPWMsYS5pc0VuYWJsZWQoKXx8ZS5aKCExKSxhLnNhKGUpLFBjKGUsYykpfWVsc2UgYy5ub2RlVmFsdWUmJiIiIT1NYShjLm5vZGVWYWx1ZSl8fGIucmVtb3ZlQ2hpbGQoYyk7Yz1kfX0sYmQ9ZnVuY3Rpb24oYSxiKXtiPWIuZigpO3EoYiwiVGhlIGNvbnRhaW5lciBET00gZWxlbWVudCBjYW5ub3QgYmUgbnVsbC4iKTtIYyhiLHgpO3YmJihiLmhpZGVGb2N1cz0hMCk7KGE9YS5oKSYmdmIoYixhKX07WGMucHJvdG90eXBlLmI9ZnVuY3Rpb24oKXtyZXR1cm4iZ29vZy1jb250YWluZXIifTsKWGMucHJvdG90eXBlLmM9ZnVuY3Rpb24oYSl7dmFyIGI9dGhpcy5iKCksYz1bYiwiaG9yaXpvbnRhbCI9PWEuSD9iKyItaG9yaXpvbnRhbCI6YisiLXZlcnRpY2FsIl07YS5pc0VuYWJsZWQoKXx8Yy5wdXNoKGIrIi1kaXNhYmxlZCIpO3JldHVybiBjfTt2YXIgTT1mdW5jdGlvbigpe30sY2Q7ZGEoTSk7dmFyIGRkPXtidXR0b246InByZXNzZWQiLGNoZWNrYm94OiJjaGVja2VkIixtZW51aXRlbToic2VsZWN0ZWQiLG1lbnVpdGVtY2hlY2tib3g6ImNoZWNrZWQiLG1lbnVpdGVtcmFkaW86ImNoZWNrZWQiLHJhZGlvOiJjaGVja2VkIix0YWI6InNlbGVjdGVkIix0cmVlaXRlbToic2VsZWN0ZWQifTtNLnByb3RvdHlwZS5oPWZ1bmN0aW9uKCl7fTtNLnByb3RvdHlwZS5jPWZ1bmN0aW9uKGEpe3JldHVybiBhLmouYigiRElWIixlZCh0aGlzLGEpLmpvaW4oIiAiKSxhLmJhKX07dmFyIGdkPWZ1bmN0aW9uKGEsYixjKXtpZihhPWEuZj9hLmYoKTphKXt2YXIgZD1bYl07diYmIUEoIjciKSYmKGQ9ZmQoRChhKSxiKSxkLnB1c2goYikpOyhjP0ZiOkhiKShhLGQpfX07Ck0ucHJvdG90eXBlLmc9ZnVuY3Rpb24oYSxiKXtiLmlkJiZNYyhhLGIuaWQpO2ImJmIuZmlyc3RDaGlsZD9oZChhLGIuZmlyc3RDaGlsZC5uZXh0U2libGluZz9KYShiLmNoaWxkTm9kZXMpOmIuZmlyc3RDaGlsZCk6YS5iYT1udWxsO3ZhciBjPTAsZD10aGlzLmEoKSxlPXRoaXMuYSgpLGY9ITEsZz0hMSxrPSExLHA9SmEoRChiKSk7cihwLGZ1bmN0aW9uKEIpe2Z8fEIhPWQ/Z3x8QiE9ZT9jfD1pZCh0aGlzLEIpOmc9ITA6KGY9ITAsZT09ZCYmKGc9ITApKTsxPT1pZCh0aGlzLEIpJiYoQWEoYiksc2IoYikmJnRiKGIpJiZyYihiLCExKSl9LHRoaXMpO2Eubz1jO2Z8fChwLnB1c2goZCksZT09ZCYmKGc9ITApKTtnfHxwLnB1c2goZSk7KGE9YS53YSkmJnAucHVzaC5hcHBseShwLGEpO2lmKHYmJiFBKCI3Iikpe3ZhciB3PWZkKHApOzA8dy5sZW5ndGgmJihwLnB1c2guYXBwbHkocCx3KSxrPSEwKX1mJiZnJiYhYSYmIWt8fENiKGIscC5qb2luKCIgIikpO3JldHVybiBifTsKdmFyIGpkPWZ1bmN0aW9uKGEsYil7aWYoYT1hLmgoKSl7cShiLCJUaGUgZWxlbWVudCBwYXNzZWQgYXMgYSBmaXJzdCBwYXJhbWV0ZXIgY2Fubm90IGJlIG51bGwuIik7dmFyIGM9Yi5nZXRBdHRyaWJ1dGUoInJvbGUiKXx8bnVsbDthIT1jJiZ2YihiLGEpfX0sa2Q9ZnVuY3Rpb24oYSxiKXt2YXIgYztpZihhLnYmMzImJihjPWEuZigpKSl7aWYoIWImJmEubyYzMil7dHJ5e2MuYmx1cigpfWNhdGNoKGQpe31hLm8mMzImJmEuRmEobnVsbCl9KHNiKGMpJiZ0YihjKSkhPWImJnJiKGMsYil9fSxsZD1mdW5jdGlvbihhLGIsYyl7Y2R8fChjZD17MToiZGlzYWJsZWQiLDg6InNlbGVjdGVkIiwxNjoiY2hlY2tlZCIsNjQ6ImV4cGFuZGVkIn0pO3EoYSwiVGhlIGVsZW1lbnQgcGFzc2VkIGFzIGEgZmlyc3QgcGFyYW1ldGVyIGNhbm5vdCBiZSBudWxsLiIpO2I9Y2RbYl07dmFyIGQ9YS5nZXRBdHRyaWJ1dGUoInJvbGUiKXx8bnVsbDtkJiYoZD1kZFtkXXx8YixiPSJjaGVja2VkIj09Ynx8InNlbGVjdGVkIj09CmI/ZDpiKTtiJiZ4YihhLGIsYyl9O00ucHJvdG90eXBlLmE9ZnVuY3Rpb24oKXtyZXR1cm4iZ29vZy1jb250cm9sIn07CnZhciBlZD1mdW5jdGlvbihhLGIpe3ZhciBjPWEuYSgpLGQ9W2NdLGU9YS5hKCk7ZSE9YyYmZC5wdXNoKGUpO2M9Yi5vO2ZvcihlPVtdO2M7KXt2YXIgZj1jJi1jO2UucHVzaChtZChhLGYpKTtjJj1+Zn1kLnB1c2guYXBwbHkoZCxlKTsoYT1iLndhKSYmZC5wdXNoLmFwcGx5KGQsYSk7diYmIUEoIjciKSYmZC5wdXNoLmFwcGx5KGQsZmQoZCkpO3JldHVybiBkfSxmZD1mdW5jdGlvbihhLGIpe3ZhciBjPVtdO2ImJihhPUlhKGEsW2JdKSk7cihbXSxmdW5jdGlvbihkKXshRmEoZCxsYShHYSxhKSl8fGImJiFHYShkLGIpfHxjLnB1c2goZC5qb2luKCJfIikpfSk7cmV0dXJuIGN9LG1kPWZ1bmN0aW9uKGEsYil7YS5ifHxuZChhKTtyZXR1cm4gYS5iW2JdfSxpZD1mdW5jdGlvbihhLGIpe2Euanx8KGEuYnx8bmQoYSksYS5qPXNhKGEuYikpO2E9cGFyc2VJbnQoYS5qW2JdLDEwKTtyZXR1cm4gaXNOYU4oYSk/MDphfSxuZD1mdW5jdGlvbihhKXt2YXIgYj1hLmEoKSxjPSF0KGIucmVwbGFjZSgvXHhhMHxccy9nLAoiICIpLCIgIik7cShjLCJDb250cm9sUmVuZGVyZXIgaGFzIGFuIGludmFsaWQgY3NzIGNsYXNzOiAnIitiKyInIik7YS5iPXsxOmIrIi1kaXNhYmxlZCIsMjpiKyItaG92ZXIiLDQ6YisiLWFjdGl2ZSIsODpiKyItc2VsZWN0ZWQiLDE2OmIrIi1jaGVja2VkIiwzMjpiKyItZm9jdXNlZCIsNjQ6YisiLW9wZW4ifX07dmFyIE49ZnVuY3Rpb24oYSxiLGMpe0suY2FsbCh0aGlzLGMpO2lmKCFiKXtiPXRoaXMuY29uc3RydWN0b3I7Zm9yKHZhciBkO2I7KXtkPWJbamFdfHwoYltqYV09KytrYSk7aWYoZD1XY1tkXSlicmVhaztiPWIuaT9iLmkuY29uc3RydWN0b3I6bnVsbH1iPWQ/aWEoZC5SKT9kLlIoKTpuZXcgZDpudWxsfXRoaXMuQj1iO3RoaXMuYmE9dm9pZCAwIT09YT9hOm51bGx9O24oTixLKTtoPU4ucHJvdG90eXBlO2guYmE9bnVsbDtoLm89MDtoLnY9Mzk7aC5LPTA7aC5JYT0hMDtoLndhPW51bGw7aC55YT0hMDtoLmlhPWZ1bmN0aW9uKCl7dmFyIGE9dGhpcy5CLmModGhpcyk7dGhpcy5hPWE7amQodGhpcy5CLGEpO0hjKGEsIXYmJiFhYik7dGhpcy5pc1Zpc2libGUoKXx8KGEuc3R5bGUuZGlzcGxheT0ibm9uZSIsYSYmeGIoYSwiaGlkZGVuIiwhMCkpfTtoLmphPWZ1bmN0aW9uKCl7cmV0dXJuIHRoaXMuZigpfTtoLnhhPWZ1bmN0aW9uKCl7cmV0dXJuITB9OwpoLnZhPWZ1bmN0aW9uKGEpe3RoaXMuYT1hPXRoaXMuQi5nKHRoaXMsYSk7amQodGhpcy5CLGEpO0hjKGEsIXYmJiFhYik7dGhpcy5JYT0ibm9uZSIhPWEuc3R5bGUuZGlzcGxheX07CmguRD1mdW5jdGlvbigpe04uaS5ELmNhbGwodGhpcyk7dmFyIGE9TmModGhpcyk7cSh0aGlzKTtxKGEpO3RoaXMuaXNWaXNpYmxlKCl8fHhiKGEsImhpZGRlbiIsIXRoaXMuaXNWaXNpYmxlKCkpO3RoaXMuaXNFbmFibGVkKCl8fGxkKGEsMSwhdGhpcy5pc0VuYWJsZWQoKSk7dGhpcy52JjgmJmxkKGEsOCwhISh0aGlzLm8mOCkpO3RoaXMudiYxNiYmbGQoYSwxNiwhISh0aGlzLm8mMTYpKTt0aGlzLnYmNjQmJmxkKGEsNjQsISEodGhpcy5vJjY0KSk7YT10aGlzLkI7U2ModGhpcykmJmdkKHRoaXMuZigpLGEuYSgpKyItcnRsIiwhMCk7dGhpcy5pc0VuYWJsZWQoKSYma2QodGhpcyx0aGlzLmlzVmlzaWJsZSgpKTtpZih0aGlzLnYmLTImJih0aGlzLnlhJiZvZCh0aGlzLCEwKSx0aGlzLnYmMzImJihhPXRoaXMuZigpKSkpe3ZhciBiPXRoaXMuV3x8KHRoaXMuVz1uZXcgSik7emMoYixhKTtIKEgoSChPYyh0aGlzKSxiLCJrZXkiLHRoaXMuZmEpLGEsImZvY3VzIix0aGlzLk5hKSxhLCJibHVyIiwKdGhpcy5GYSl9fTt2YXIgb2Q9ZnVuY3Rpb24oYSxiKXt2YXIgYz1PYyhhKSxkPWEuZigpO2I/KEgoSChIKEgoYyxkLEYuTCxhLmthKSxkLFtGLk0sRi5hYV0sYS5sYSksZCwibW91c2VvdmVyIixhLkFhKSxkLCJtb3VzZW91dCIsYS56YSksYS5lYSE9Y2EmJkgoYyxkLCJjb250ZXh0bWVudSIsYS5lYSksdiYmKEEoOSl8fEgoYyxkLCJkYmxjbGljayIsYS5KYSksYS5nYXx8KGEuZ2E9bmV3IHBkKGEpLHpiKGEsbGEoQWIsYS5nYSkpKSkpOihxYyhxYyhxYyhxYyhjLGQsRi5MLGEua2EpLGQsW0YuTSxGLmFhXSxhLmxhKSxkLCJtb3VzZW92ZXIiLGEuQWEpLGQsIm1vdXNlb3V0IixhLnphKSxhLmVhIT1jYSYmcWMoYyxkLCJjb250ZXh0bWVudSIsYS5lYSksdiYmKEEoOSl8fHFjKGMsZCwiZGJsY2xpY2siLGEuSmEpLEFiKGEuZ2EpLGEuZ2E9bnVsbCkpfTsKTi5wcm90b3R5cGUuUD1mdW5jdGlvbigpe04uaS5QLmNhbGwodGhpcyk7dGhpcy5XJiZGYyh0aGlzLlcpO3RoaXMuaXNWaXNpYmxlKCkmJnRoaXMuaXNFbmFibGVkKCkmJmtkKHRoaXMsITEpfTtOLnByb3RvdHlwZS51PWZ1bmN0aW9uKCl7Ti5pLnUuY2FsbCh0aGlzKTt0aGlzLlcmJih0aGlzLlcuTigpLGRlbGV0ZSB0aGlzLlcpO2RlbGV0ZSB0aGlzLkI7dGhpcy5nYT10aGlzLndhPXRoaXMuYmE9bnVsbH07dmFyIGhkPWZ1bmN0aW9uKGEsYil7YS5iYT1ifTtOLnByb3RvdHlwZS5pc1Zpc2libGU9ZnVuY3Rpb24oKXtyZXR1cm4gdGhpcy5JYX07Ti5wcm90b3R5cGUuaXNFbmFibGVkPWZ1bmN0aW9uKCl7cmV0dXJuISh0aGlzLm8mMSl9OwpOLnByb3RvdHlwZS5aPWZ1bmN0aW9uKGEpe3ZhciBiPXRoaXMuYjtiJiYiZnVuY3Rpb24iPT10eXBlb2YgYi5pc0VuYWJsZWQmJiFiLmlzRW5hYmxlZCgpfHwhTyh0aGlzLDEsIWEpfHwoYXx8KHFkKHRoaXMsITEpLFAodGhpcywhMSkpLHRoaXMuaXNWaXNpYmxlKCkmJmtkKHRoaXMsYSksUSh0aGlzLDEsIWEsITApKX07CnZhciBQPWZ1bmN0aW9uKGEsYil7TyhhLDIsYikmJlEoYSwyLGIpfSxxZD1mdW5jdGlvbihhLGIpe08oYSw0LGIpJiZRKGEsNCxiKX0scmQ9ZnVuY3Rpb24oYSxiKXtPKGEsOCxiKSYmUShhLDgsYil9LHNkPWZ1bmN0aW9uKGEsYil7TyhhLDY0LGIpJiZRKGEsNjQsYil9LFE9ZnVuY3Rpb24oYSxiLGMsZCl7aWYoIWQmJjE9PWIpYS5aKCFjKTtlbHNlIGlmKGEudiZiJiZjIT0hIShhLm8mYikpe3ZhciBlPWEuQjtpZihkPWEuZigpKShlPW1kKGUsYikpJiZnZChhLGUsYyksbGQoZCxiLGMpO2Eubz1jP2Eub3xiOmEubyZ+Yn19LHRkPWZ1bmN0aW9uKGEsYixjKXtpZihhLm0mJmEubyZiJiYhYyl0aHJvdyBFcnJvcigiQ29tcG9uZW50IGFscmVhZHkgcmVuZGVyZWQiKTshYyYmYS5vJmImJlEoYSxiLCExKTthLnY9Yz9hLnZ8YjphLnYmfmJ9LFI9ZnVuY3Rpb24oYSxiKXtyZXR1cm4hISgyNTUmYikmJiEhKGEudiZiKX0sTz1mdW5jdGlvbihhLGIsYyl7cmV0dXJuISEoYS52JmIpJiYhIShhLm8mCmIpIT1jJiYoIShhLksmYil8fHVjKGEsS2MoYixjKSkpJiYhYS5VfTtoPU4ucHJvdG90eXBlO2guQWE9ZnVuY3Rpb24oYSl7KCFhLnJlbGF0ZWRUYXJnZXR8fCFwYih0aGlzLmYoKSxhLnJlbGF0ZWRUYXJnZXQpKSYmdWModGhpcywiZW50ZXIiKSYmdGhpcy5pc0VuYWJsZWQoKSYmUih0aGlzLDIpJiZQKHRoaXMsITApfTtoLnphPWZ1bmN0aW9uKGEpe2EucmVsYXRlZFRhcmdldCYmcGIodGhpcy5mKCksYS5yZWxhdGVkVGFyZ2V0KXx8IXVjKHRoaXMsImxlYXZlIil8fChSKHRoaXMsNCkmJnFkKHRoaXMsITEpLFIodGhpcywyKSYmUCh0aGlzLCExKSl9O2guZWE9Y2E7Cmgua2E9ZnVuY3Rpb24oYSl7aWYodGhpcy5pc0VuYWJsZWQoKSYmKFIodGhpcywyKSYmUCh0aGlzLCEwKSxPYihhKSYmISh5JiZ6JiZhLmN0cmxLZXkpKSl7Uih0aGlzLDQpJiZxZCh0aGlzLCEwKTt2YXIgYjtpZihiPXRoaXMuQil7dmFyIGM7Yj10aGlzLnYmMzImJihjPXRoaXMuZigpKT9zYihjKSYmdGIoYyk6ITF9YiYmdGhpcy5mKCkuZm9jdXMoKX0hT2IoYSl8fHkmJnomJmEuY3RybEtleXx8YS5nKCl9O2gubGE9ZnVuY3Rpb24oYSl7dGhpcy5pc0VuYWJsZWQoKSYmKFIodGhpcywyKSYmUCh0aGlzLCEwKSx0aGlzLm8mNCYmdWQodGhpcyxhKSYmUih0aGlzLDQpJiZxZCh0aGlzLCExKSl9O2guSmE9ZnVuY3Rpb24oYSl7dGhpcy5pc0VuYWJsZWQoKSYmdWQodGhpcyxhKX07CnZhciB1ZD1mdW5jdGlvbihhLGIpe2lmKFIoYSwxNikpe3ZhciBjPSEoYS5vJjE2KTtPKGEsMTYsYykmJlEoYSwxNixjKX1SKGEsOCkmJnJkKGEsITApO1IoYSw2NCkmJnNkKGEsIShhLm8mNjQpKTtjPW5ldyBFKCJhY3Rpb24iLGEpO2ImJihjLmFsdEtleT1iLmFsdEtleSxjLmN0cmxLZXk9Yi5jdHJsS2V5LGMubWV0YUtleT1iLm1ldGFLZXksYy5zaGlmdEtleT1iLnNoaWZ0S2V5LGMuQT1iLkEpO3JldHVybiB1YyhhLGMpfTtOLnByb3RvdHlwZS5OYT1mdW5jdGlvbigpe1IodGhpcywzMikmJk8odGhpcywzMiwhMCkmJlEodGhpcywzMiwhMCl9O04ucHJvdG90eXBlLkZhPWZ1bmN0aW9uKCl7Uih0aGlzLDQpJiZxZCh0aGlzLCExKTtSKHRoaXMsMzIpJiZPKHRoaXMsMzIsITEpJiZRKHRoaXMsMzIsITEpfTsKTi5wcm90b3R5cGUuZmE9ZnVuY3Rpb24oYSl7cmV0dXJuIHRoaXMuaXNWaXNpYmxlKCkmJnRoaXMuaXNFbmFibGVkKCkmJjEzPT1hLmMmJnVkKHRoaXMsYSk/KGEuZygpLGEuaigpLCEwKTohMX07aWYoIWlhKE4pKXRocm93IEVycm9yKCJJbnZhbGlkIGNvbXBvbmVudCBjbGFzcyAiK04pO2lmKCFpYShNKSl0aHJvdyBFcnJvcigiSW52YWxpZCByZW5kZXJlciBjbGFzcyAiK00pO3ZhciB2ZD1OW2phXXx8KE5bamFdPSsra2EpO1djW3ZkXT1NO1ZjKCJnb29nLWNvbnRyb2wiLGZ1bmN0aW9uKCl7cmV0dXJuIG5ldyBOKG51bGwpfSk7dmFyIHBkPWZ1bmN0aW9uKGEpe0MuY2FsbCh0aGlzKTt0aGlzLmI9YTt0aGlzLmE9ITE7dGhpcy5jPW5ldyBvYyh0aGlzKTt6Yih0aGlzLGxhKEFiLHRoaXMuYykpO2E9TmModGhpcy5iKTtIKEgoSCh0aGlzLmMsYSxGLkwsdGhpcy5oKSxhLEYuTSx0aGlzLmopLGEsImNsaWNrIix0aGlzLmcpfTtuKHBkLEMpO3ZhciB3ZD0hdnx8OTw9TnVtYmVyKGliKTsKcGQucHJvdG90eXBlLmg9ZnVuY3Rpb24oKXt0aGlzLmE9ITF9O3BkLnByb3RvdHlwZS5qPWZ1bmN0aW9uKCl7dGhpcy5hPSEwfTt2YXIgeGQ9ZnVuY3Rpb24oYSxiKXtpZighd2QpcmV0dXJuIGEuYnV0dG9uPTAsYS50eXBlPWIsYTt2YXIgYz1kb2N1bWVudC5jcmVhdGVFdmVudCgiTW91c2VFdmVudHMiKTtjLmluaXRNb3VzZUV2ZW50KGIsYS5idWJibGVzLGEuY2FuY2VsYWJsZSxhLnZpZXd8fG51bGwsYS5kZXRhaWwsYS5zY3JlZW5YLGEuc2NyZWVuWSxhLmNsaWVudFgsYS5jbGllbnRZLGEuY3RybEtleSxhLmFsdEtleSxhLnNoaWZ0S2V5LGEubWV0YUtleSwwLGEucmVsYXRlZFRhcmdldHx8bnVsbCk7cmV0dXJuIGN9OwpwZC5wcm90b3R5cGUuZz1mdW5jdGlvbihhKXtpZih0aGlzLmEpdGhpcy5hPSExO2Vsc2V7dmFyIGI9YS5iLGM9Yi5idXR0b24sZD1iLnR5cGUsZT14ZChiLCJtb3VzZWRvd24iKTt0aGlzLmIua2EobmV3IEcoZSxhLmEpKTtlPXhkKGIsIm1vdXNldXAiKTt0aGlzLmIubGEobmV3IEcoZSxhLmEpKTt3ZHx8KGIuYnV0dG9uPWMsYi50eXBlPWQpfX07cGQucHJvdG90eXBlLnU9ZnVuY3Rpb24oKXt0aGlzLmI9bnVsbDtwZC5pLnUuY2FsbCh0aGlzKX07dmFyIFM9ZnVuY3Rpb24oYSxiLGMpe0suY2FsbCh0aGlzLGMpO3RoaXMuZGE9Ynx8WGMuUigpO3RoaXMuSD1hfHwidmVydGljYWwifTtuKFMsSyk7aD1TLnByb3RvdHlwZTtoLkRhPW51bGw7aC5jYT1udWxsO2guZGE9bnVsbDtoLkg9bnVsbDtoLlQ9ITA7aC5PPSEwO2gubD0tMTtoLnM9bnVsbDtoLlY9ITE7aC5JPW51bGw7dmFyIHlkPWZ1bmN0aW9uKGEpe3JldHVybiBhLkRhfHxhLmYoKX07aD1TLnByb3RvdHlwZTtoLmlhPWZ1bmN0aW9uKCl7dGhpcy5hPXRoaXMuai5iKCJESVYiLHRoaXMuZGEuYyh0aGlzKS5qb2luKCIgIikpfTtoLmphPWZ1bmN0aW9uKCl7cmV0dXJuIHRoaXMuZigpfTtoLnhhPWZ1bmN0aW9uKGEpe3JldHVybiJESVYiPT1hLnRhZ05hbWV9O2gudmE9ZnVuY3Rpb24oYSl7dGhpcy5hPSRjKHRoaXMuZGEsdGhpcyxhKTsibm9uZSI9PWEuc3R5bGUuZGlzcGxheSYmKHRoaXMuVD0hMSl9OwpoLkQ9ZnVuY3Rpb24oKXtTLmkuRC5jYWxsKHRoaXMpO1FjKHRoaXMsZnVuY3Rpb24oYil7Yi5tJiZ6ZCh0aGlzLGIpfSx0aGlzKTt2YXIgYT10aGlzLmYoKTtiZCh0aGlzLmRhLHRoaXMpO0FkKHRoaXMsdGhpcy5UKTtIKEgoSChIKEgoSChIKEgoT2ModGhpcyksdGhpcywiZW50ZXIiLHRoaXMuV2EpLHRoaXMsImhpZ2hsaWdodCIsdGhpcy5YYSksdGhpcywidW5oaWdobGlnaHQiLHRoaXMuY2IpLHRoaXMsIm9wZW4iLHRoaXMuWWEpLHRoaXMsImNsb3NlIix0aGlzLlVhKSxhLEYuTCx0aGlzLlNhKSxxYihhKSxbRi5NLEYuYWFdLHRoaXMuVmEpLGEsW0YuTCxGLk0sRi5hYSwibW91c2VvdmVyIiwibW91c2VvdXQiLCJjb250ZXh0bWVudSJdLHRoaXMuVGEpO0JkKHRoaXMpfTt2YXIgQmQ9ZnVuY3Rpb24oYSl7dmFyIGI9T2MoYSksYz15ZChhKTtIKEgoSChiLGMsImZvY3VzIixhLkhhKSxjLCJibHVyIixhLlFhKSxhLmNhfHwoYS5jYT1uZXcgSih5ZChhKSkpLCJrZXkiLGEuUmEpfTtoPVMucHJvdG90eXBlOwpoLlA9ZnVuY3Rpb24oKXtDZCh0aGlzLC0xKTt0aGlzLnMmJnNkKHRoaXMucywhMSk7dGhpcy5WPSExO1MuaS5QLmNhbGwodGhpcyl9O2gudT1mdW5jdGlvbigpe1MuaS51LmNhbGwodGhpcyk7dGhpcy5jYSYmKHRoaXMuY2EuTigpLHRoaXMuY2E9bnVsbCk7dGhpcy5kYT10aGlzLnM9dGhpcy5JPXRoaXMuRGE9bnVsbH07aC5XYT1mdW5jdGlvbigpe3JldHVybiEwfTsKaC5YYT1mdW5jdGlvbihhKXt2YXIgYj1UYyh0aGlzLGEudGFyZ2V0KTtpZigtMTxiJiZiIT10aGlzLmwpe3ZhciBjPUwodGhpcyx0aGlzLmwpO2MmJlAoYywhMSk7dGhpcy5sPWI7Yz1MKHRoaXMsdGhpcy5sKTt0aGlzLlYmJnFkKGMsITApO3RoaXMucyYmYyE9dGhpcy5zJiYoYy52JjY0P3NkKGMsITApOnNkKHRoaXMucywhMSkpfWI9dGhpcy5mKCk7cShiLCJUaGUgRE9NIGVsZW1lbnQgZm9yIHRoZSBjb250YWluZXIgY2Fubm90IGJlIG51bGwuIik7bnVsbCE9YS50YXJnZXQuZigpJiZ4YihiLCJhY3RpdmVkZXNjZW5kYW50IixhLnRhcmdldC5mKCkuaWQpfTtoLmNiPWZ1bmN0aW9uKGEpe2EudGFyZ2V0PT1MKHRoaXMsdGhpcy5sKSYmKHRoaXMubD0tMSk7YT10aGlzLmYoKTtxKGEsIlRoZSBET00gZWxlbWVudCBmb3IgdGhlIGNvbnRhaW5lciBjYW5ub3QgYmUgbnVsbC4iKTthLnJlbW92ZUF0dHJpYnV0ZSh3YigiYWN0aXZlZGVzY2VuZGFudCIpKX07CmguWWE9ZnVuY3Rpb24oYSl7KGE9YS50YXJnZXQpJiZhIT10aGlzLnMmJmEuYj09dGhpcyYmKHRoaXMucyYmc2QodGhpcy5zLCExKSx0aGlzLnM9YSl9O2guVWE9ZnVuY3Rpb24oYSl7YS50YXJnZXQ9PXRoaXMucyYmKHRoaXMucz1udWxsKTt2YXIgYj10aGlzLmYoKSxjPWEudGFyZ2V0LmYoKTtiJiZhLnRhcmdldC5vJjImJmMmJihhPSIiLGMmJihhPWMuaWQscShhLCJUaGUgYWN0aXZlIGVsZW1lbnQgc2hvdWxkIGhhdmUgYW4gaWQuIikpLHhiKGIsImFjdGl2ZWRlc2NlbmRhbnQiLGEpKX07aC5TYT1mdW5jdGlvbihhKXt0aGlzLk8mJih0aGlzLlY9ITApO3ZhciBiPXlkKHRoaXMpO2ImJnNiKGIpJiZ0YihiKT9iLmZvY3VzKCk6YS5nKCl9O2guVmE9ZnVuY3Rpb24oKXt0aGlzLlY9ITF9OwpoLlRhPWZ1bmN0aW9uKGEpe2E6e3ZhciBiPWEudGFyZ2V0O2lmKHRoaXMuSSlmb3IodmFyIGM9dGhpcy5mKCk7YiYmYiE9PWM7KXt2YXIgZD1iLmlkO2lmKGQgaW4gdGhpcy5JKXtiPXRoaXMuSVtkXTticmVhayBhfWI9Yi5wYXJlbnROb2RlfWI9bnVsbH1pZihiKXN3aXRjaChhLnR5cGUpe2Nhc2UgRi5MOmIua2EoYSk7YnJlYWs7Y2FzZSBGLk06Y2FzZSBGLmFhOmIubGEoYSk7YnJlYWs7Y2FzZSAibW91c2VvdmVyIjpiLkFhKGEpO2JyZWFrO2Nhc2UgIm1vdXNlb3V0IjpiLnphKGEpO2JyZWFrO2Nhc2UgImNvbnRleHRtZW51IjpiLmVhKGEpfX07aC5IYT1mdW5jdGlvbigpe307aC5RYT1mdW5jdGlvbigpe0NkKHRoaXMsLTEpO3RoaXMuVj0hMTt0aGlzLnMmJnNkKHRoaXMucywhMSl9OwpoLlJhPWZ1bmN0aW9uKGEpe3JldHVybiB0aGlzLmlzRW5hYmxlZCgpJiZ0aGlzLmlzVmlzaWJsZSgpJiYoMCE9UmModGhpcyl8fHRoaXMuRGEpJiZEZCh0aGlzLGEpPyhhLmcoKSxhLmooKSwhMCk6ITF9Owp2YXIgRGQ9ZnVuY3Rpb24oYSxiKXt2YXIgYz1MKGEsYS5sKTtpZihjJiYiZnVuY3Rpb24iPT10eXBlb2YgYy5mYSYmYy5mYShiKXx8YS5zJiZhLnMhPWMmJiJmdW5jdGlvbiI9PXR5cGVvZiBhLnMuZmEmJmEucy5mYShiKSlyZXR1cm4hMDtpZihiLnNoaWZ0S2V5fHxiLmN0cmxLZXl8fGIubWV0YUtleXx8Yi5hbHRLZXkpcmV0dXJuITE7c3dpdGNoKGIuYyl7Y2FzZSAyNzp5ZChhKS5ibHVyKCk7YnJlYWs7Y2FzZSAzNjpFZChhKTticmVhaztjYXNlIDM1OkZkKGEpO2JyZWFrO2Nhc2UgMzg6aWYoInZlcnRpY2FsIj09YS5IKUdkKGEpO2Vsc2UgcmV0dXJuITE7YnJlYWs7Y2FzZSAzNzppZigiaG9yaXpvbnRhbCI9PWEuSClTYyhhKT9IZChhKTpHZChhKTtlbHNlIHJldHVybiExO2JyZWFrO2Nhc2UgNDA6aWYoInZlcnRpY2FsIj09YS5IKUhkKGEpO2Vsc2UgcmV0dXJuITE7YnJlYWs7Y2FzZSAzOTppZigiaG9yaXpvbnRhbCI9PWEuSClTYyhhKT9HZChhKTpIZChhKTtlbHNlIHJldHVybiExOwpicmVhaztkZWZhdWx0OnJldHVybiExfXJldHVybiEwfSx6ZD1mdW5jdGlvbihhLGIpe3ZhciBjPWIuZigpO2M9Yy5pZHx8KGMuaWQ9TGMoYikpO2EuSXx8KGEuST17fSk7YS5JW2NdPWJ9O1MucHJvdG90eXBlLnNhPWZ1bmN0aW9uKGEsYil7Q2EoYSxOLCJUaGUgY2hpbGQgb2YgYSBjb250YWluZXIgbXVzdCBiZSBhIGNvbnRyb2wiKTtTLmkuc2EuY2FsbCh0aGlzLGEsYil9O1MucHJvdG90eXBlLnRhPWZ1bmN0aW9uKGEsYixjKXtDYShhLE4pO2EuS3w9MjthLkt8PTY0O3RkKGEsMzIsITEpO2EubSYmMCE9YS55YSYmb2QoYSwhMSk7YS55YT0hMTt2YXIgZD1hLmI9PXRoaXM/VGModGhpcyxhKTotMTtTLmkudGEuY2FsbCh0aGlzLGEsYixjKTthLm0mJnRoaXMubSYmemQodGhpcyxhKTthPWQ7LTE9PWEmJihhPVJjKHRoaXMpKTthPT10aGlzLmw/dGhpcy5sPU1hdGgubWluKFJjKHRoaXMpLTEsYik6YT50aGlzLmwmJmI8PXRoaXMubD90aGlzLmwrKzphPHRoaXMubCYmYj50aGlzLmwmJnRoaXMubC0tfTsKdmFyIGFkPWZ1bmN0aW9uKGEsYil7aWYoYS5mKCkpdGhyb3cgRXJyb3IoIkNvbXBvbmVudCBhbHJlYWR5IHJlbmRlcmVkIik7YS5IPWJ9O1MucHJvdG90eXBlLmlzVmlzaWJsZT1mdW5jdGlvbigpe3JldHVybiB0aGlzLlR9O3ZhciBBZD1mdW5jdGlvbihhLGIpe2EuVD1iO3ZhciBjPWEuZigpO2MmJihjLnN0eWxlLmRpc3BsYXk9Yj8iIjoibm9uZSIsWWMoeWQoYSksYS5PJiZhLlQpKX07Uy5wcm90b3R5cGUuaXNFbmFibGVkPWZ1bmN0aW9uKCl7cmV0dXJuIHRoaXMuT307Uy5wcm90b3R5cGUuWj1mdW5jdGlvbihhKXt0aGlzLk8hPWEmJnVjKHRoaXMsYT8iZW5hYmxlIjoiZGlzYWJsZSIpJiYoYT8odGhpcy5PPSEwLFFjKHRoaXMsZnVuY3Rpb24oYil7Yi5MYT9kZWxldGUgYi5MYTpiLlooITApfSkpOihRYyh0aGlzLGZ1bmN0aW9uKGIpe2IuaXNFbmFibGVkKCk/Yi5aKCExKTpiLkxhPSEwfSksdGhpcy5WPXRoaXMuTz0hMSksWWMoeWQodGhpcyksYSYmdGhpcy5UKSl9Owp2YXIgQ2Q9ZnVuY3Rpb24oYSxiKXsoYj1MKGEsYikpP1AoYiwhMCk6LTE8YS5sJiZQKEwoYSxhLmwpLCExKX0sRWQ9ZnVuY3Rpb24oYSl7SWQoYSxmdW5jdGlvbihiLGMpe3JldHVybihiKzEpJWN9LFJjKGEpLTEpfSxGZD1mdW5jdGlvbihhKXtJZChhLGZ1bmN0aW9uKGIsYyl7Yi0tO3JldHVybiAwPmI/Yy0xOmJ9LDApfSxIZD1mdW5jdGlvbihhKXtJZChhLGZ1bmN0aW9uKGIsYyl7cmV0dXJuKGIrMSklY30sYS5sKX0sR2Q9ZnVuY3Rpb24oYSl7SWQoYSxmdW5jdGlvbihiLGMpe2ItLTtyZXR1cm4gMD5iP2MtMTpifSxhLmwpfSxJZD1mdW5jdGlvbihhLGIsYyl7Yz0wPmM/VGMoYSxhLnMpOmM7dmFyIGQ9UmMoYSk7Yz1iLmNhbGwoYSxjLGQpO2Zvcih2YXIgZT0wO2U8PWQ7KXt2YXIgZj1MKGEsYyksZztpZihnPWYpZz1mLmlzVmlzaWJsZSgpJiZmLmlzRW5hYmxlZCgpJiYhIShmLnYmMik7aWYoZyl7YS5FYShjKTticmVha31lKys7Yz1iLmNhbGwoYSxjLGQpfX07ClMucHJvdG90eXBlLkVhPWZ1bmN0aW9uKGEpe0NkKHRoaXMsYSl9O3ZhciBUPWZ1bmN0aW9uKCl7fTtuKFQsTSk7ZGEoVCk7VC5wcm90b3R5cGUuYT1mdW5jdGlvbigpe3JldHVybiJnb29nLXRhYiJ9O1QucHJvdG90eXBlLmg9ZnVuY3Rpb24oKXtyZXR1cm4idGFiIn07VC5wcm90b3R5cGUuYz1mdW5jdGlvbihhKXt2YXIgYj1ULmkuYy5jYWxsKHRoaXMsYSk7KGE9YS5QYSkmJmImJihiLnRpdGxlPWF8fCIiKTtyZXR1cm4gYn07VC5wcm90b3R5cGUuZz1mdW5jdGlvbihhLGIpe2I9VC5pLmcuY2FsbCh0aGlzLGEsYik7dmFyIGM9Yi50aXRsZXx8IiI7YyYmKGEuUGE9Yyk7YS5vJjgmJihjPWEuYikmJmlhKGMuRykmJihRKGEsOCwhMSksYy5HKGEpKTtyZXR1cm4gYn07dmFyIEpkPWZ1bmN0aW9uKGEsYixjKXtOLmNhbGwodGhpcyxhLGJ8fFQuUigpLGMpO3RkKHRoaXMsOCwhMCk7dGhpcy5LfD05fTtuKEpkLE4pO1ZjKCJnb29nLXRhYiIsZnVuY3Rpb24oKXtyZXR1cm4gbmV3IEpkKG51bGwpfSk7dmFyIFU9ZnVuY3Rpb24oKXt0aGlzLmg9InRhYmxpc3QifTtuKFUsWGMpO2RhKFUpO1UucHJvdG90eXBlLmI9ZnVuY3Rpb24oKXtyZXR1cm4iZ29vZy10YWItYmFyIn07VS5wcm90b3R5cGUuZz1mdW5jdGlvbihhLGIsYyl7dGhpcy5qfHwodGhpcy5hfHxLZCh0aGlzKSx0aGlzLmo9c2EodGhpcy5hKSk7dmFyIGQ9dGhpcy5qW2JdO2Q/KGFkKGEsTGQoZCkpLGEuQj1kKTpVLmkuZy5jYWxsKHRoaXMsYSxiLGMpfTtVLnByb3RvdHlwZS5jPWZ1bmN0aW9uKGEpe3ZhciBiPVUuaS5jLmNhbGwodGhpcyxhKTt0aGlzLmF8fEtkKHRoaXMpO2IucHVzaCh0aGlzLmFbYS5CXSk7cmV0dXJuIGJ9O3ZhciBLZD1mdW5jdGlvbihhKXt2YXIgYj1hLmIoKTthLmE9e3RvcDpiKyItdG9wIixib3R0b206YisiLWJvdHRvbSIsc3RhcnQ6YisiLXN0YXJ0IixlbmQ6YisiLWVuZCJ9fTt2YXIgVj1mdW5jdGlvbihhLGIsYyl7YT1hfHwidG9wIjthZCh0aGlzLExkKGEpKTt0aGlzLkI9YTtTLmNhbGwodGhpcyx0aGlzLkgsYnx8VS5SKCksYyk7TWQodGhpcyl9O24oVixTKTtoPVYucHJvdG90eXBlO2guQz1udWxsO2guRD1mdW5jdGlvbigpe1YuaS5ELmNhbGwodGhpcyk7TWQodGhpcyl9O2gudT1mdW5jdGlvbigpe1YuaS51LmNhbGwodGhpcyk7dGhpcy5DPW51bGx9O2guRWE9ZnVuY3Rpb24oYSl7Vi5pLkVhLmNhbGwodGhpcyxhKTt0aGlzLkcoTCh0aGlzLGEpKX07aC5HPWZ1bmN0aW9uKGEpe2E/cmQoYSwhMCk6dGhpcy5DJiZyZCh0aGlzLkMsITEpfTsKdmFyIE5kPWZ1bmN0aW9uKGEsYil7aWYoYiYmYj09YS5DKXtmb3IodmFyIGM9VGMoYSxiKSxkPWMtMTtiPUwoYSxkKTtkLS0pe3ZhciBlPWI7aWYoZS5pc1Zpc2libGUoKSYmZS5pc0VuYWJsZWQoKSl7YS5HKGIpO3JldHVybn19Zm9yKGMrPTE7Yj1MKGEsYyk7YysrKWlmKGQ9YixkLmlzVmlzaWJsZSgpJiZkLmlzRW5hYmxlZCgpKXthLkcoYik7cmV0dXJufWEuRyhudWxsKX19O2g9Vi5wcm90b3R5cGU7aC5hYj1mdW5jdGlvbihhKXt0aGlzLkMmJnRoaXMuQyE9YS50YXJnZXQmJnJkKHRoaXMuQywhMSk7dGhpcy5DPWEudGFyZ2V0fTtoLmJiPWZ1bmN0aW9uKGEpe2EudGFyZ2V0PT10aGlzLkMmJih0aGlzLkM9bnVsbCl9O2guWmE9ZnVuY3Rpb24oYSl7TmQodGhpcyxhLnRhcmdldCl9O2guJGE9ZnVuY3Rpb24oYSl7TmQodGhpcyxhLnRhcmdldCl9O2guSGE9ZnVuY3Rpb24oKXtMKHRoaXMsdGhpcy5sKXx8Q2QodGhpcyxUYyh0aGlzLHRoaXMuQ3x8TCh0aGlzLDApKSl9Owp2YXIgTWQ9ZnVuY3Rpb24oYSl7SChIKEgoSChPYyhhKSxhLCJzZWxlY3QiLGEuYWIpLGEsInVuc2VsZWN0IixhLmJiKSxhLCJkaXNhYmxlIixhLlphKSxhLCJoaWRlIixhLiRhKX0sTGQ9ZnVuY3Rpb24oYSl7cmV0dXJuInN0YXJ0Ij09YXx8ImVuZCI9PWE/InZlcnRpY2FsIjoiaG9yaXpvbnRhbCJ9O1ZjKCJnb29nLXRhYi1iYXIiLGZ1bmN0aW9uKCl7cmV0dXJuIG5ldyBWfSk7ZnVuY3Rpb24gT2QoYSl7dmFyIGI9e3RvcDoiYm90dG9tIixib3R0b206InRvcCIsc3RhcnQ6InJpZ2h0IixlbmQ6ImxlZnQifVthLmxvY2F0aW9uXSxjPWEuZWxlbWVudElkLGQ9ZG9jdW1lbnQuY3JlYXRlRWxlbWVudCgic3R5bGUiKTtkLnRleHRDb250ZW50PSJcbiAgICBmaWVsZHNldCB7XG4gICAgICBwYWRkaW5nOiAxMHB4O1xuICAgICAgYm9yZGVyOiAxcHggc29saWQgIzM2OTtcbiAgICB9XG5cbiAgICAjIitjKyIgLmdvb2ctdGFiLWNvbnRlbnQge1xuICAgICAgbWluLWhlaWdodDogM2VtO1xuICAgICAgbWFyZ2luOiAwO1xuICAgICAgYm9yZGVyOiAiK2EuYm9yZGVyKyIgc29saWQgIithLmJvcmRlckNvbG9yKyI7XG4gICAgICBib3JkZXItdG9wOiAwO1xuICAgICAgaGVpZ2h0OiAiK2EuY29udGVudEhlaWdodCsiO1xuICAgICAgcGFkZGluZzogNHB4IDhweDtcbiAgICAgIG1hcmdpbi1yaWdodDogNHB4O1xuICAgICAgYmFja2dyb3VuZDogdmFyKC0tY29sYWItcHJpbWFyeS1zdXJmYWNlLWNvbG9yKTtcbiAgICAgIG92ZXJmbG93OiBhdXRvO1xuICAgIH1cblxuICAgICMiKwpjKyIgLmdvb2ctdGFiLWJhci0iK2EubG9jYXRpb24rIiAuZ29vZy10YWItc2VsZWN0ZWQge1xuICAgICAgYmFja2dyb3VuZC1jb2xvcjogdmFyKC0tY29sYWItcHJpbWFyeS1zdXJmYWNlLWNvbG9yKTtcbiAgICAgIGJvcmRlcjogMXB4IHNvbGlkICIrYS5ib3JkZXJDb2xvcisiO1xuICAgICAgYm9yZGVyLSIrYisiOiAwcHg7XG4gICAgfVxuXG4gICAgIyIrYysiIC5nb29nLXRhYi1iYXItIithLmxvY2F0aW9uKyIge1xuICAgICAgcGFkZGluZy0iK2EubG9jYXRpb24rIjogNXB4ICFpbXBvcnRhbnQ7XG4gICAgICBib3JkZXItIitiKyI6IDFweCBzb2xpZCAiK2EuYm9yZGVyQ29sb3IrIiAhaW1wb3J0YW50O1xuICAgICAgYmFja2dyb3VuZDogdmFyKC0tY29sYWItcHJpbWFyeS1zdXJmYWNlLWNvbG9yKTtcbiAgICB9XG5cbiAgICAjIitjKyIgLmdvb2ctdGFiLWJhciB7XG4gICAgICAgbWFyZ2luOiAwO1xuICAgICAgIGJvcmRlcjogMDtcbiAgICAgICBwYWRkaW5nOiAwO1xuICAgICAgIGxpc3Qtc3R5bGU6IG5vbmU7XG4gICAgICAgY3Vyc29yOiBkZWZhdWx0O1xuICAgICAgIG91dGxpbmU6IG5vbmU7XG4gICAgICAgYmFja2dyb3VuZDogdmFyKC0tY29sYWItcHJpbWFyeS1zdXJmYWNlLWNvbG9yKTtcbiAgICAgICBtYXJnaW4tcmlnaHQ6IDRweDtcbiAgICAgIH1cblxuICAgICAjIisKYysiIC5nb29nLXRhYiB7XG4gICAgICAgcG9zaXRpb246IHJlbGF0aXZlO1xuICAgICAgIHBhZGRpbmc6IDRweCA4cHg7XG4gICAgICAgY29sb3I6IHZhcigtLWNvbGFiLXByaW1hcnktdGV4dC1jb2xvcik7XG4gICAgICAgdGV4dC1kZWNvcmF0aW9uOiBpbml0aWFsO1xuICAgICAgIGN1cnNvcjogZGVmYXVsdDtcbiAgICAgIH1cblxuICAgICAgIyIrYysiIC5nb29nLXRhYi1ob3ZlciB7XG4gICAgICAgIGJhY2tncm91bmQtY29sb3I6IHZhcigtLWNvbGFiLWhpZ2hsaWdodGVkLXN1cmZhY2UtY29sb3IpO1xuICAgICAgfVxuICAgICAgIjtyZXR1cm4gZH0KdmFyIFBkPWZ1bmN0aW9uKGEpe3ZhciBiPWEuZWxlbWVudElkLGM9YS50YWJOYW1lcyxkPWEuc2VsZWN0ZWRJbmRleDsiY29udGVudEJvcmRlciJpbiBhfHwoYS5jb250ZW50Qm9yZGVyPSIwcHgiKTsiY29udGVudEhlaWdodCJpbiBhfHwoYS5jb250ZW50SGVpZ2h0PSJpbml0aWFsIik7ImJvcmRlckNvbG9yImluIGF8fChhLmJvcmRlckNvbG9yPSJ2YXIoLS1jb2xhYi1ib3JkZXItY29sb3IpIik7YS5sb2NhdGlvbnx8KGEubG9jYXRpb249InRvcCIpO3ZhciBlPWRvY3VtZW50LnF1ZXJ5U2VsZWN0b3IoIiMiK2IpLGY9ZG9jdW1lbnQuY3JlYXRlRWxlbWVudCgiZGl2Iik7Zi5jbGFzc0xpc3QuYWRkKCJnb29nLXRhYi1iYXIiKTt2YXIgZz1hLmxvY2F0aW9uO2YuY2xhc3NMaXN0LmFkZCgiZ29vZy10YWItYmFyLSIrZyk7Zm9yKHZhciBrPVtdLHA9YmEoYyksdz1wLm5leHQoKTshdy5kb25lO3c9cC5uZXh0KCkpe3c9dy52YWx1ZTt2YXIgQj1kb2N1bWVudC5jcmVhdGVFbGVtZW50KCJkaXYiKTsKQi5jbGFzc0xpc3QuYWRkKCJnb29nLXRhYiIpO0IudGV4dENvbnRlbnQ9dztmLmFwcGVuZENoaWxkKEIpO2sucHVzaChCKX0iYm90dG9tIiE9ZyYmZS5hcHBlbmRDaGlsZChmKTtwPW51bGw7aWYoInRvcCI9PWd8fCJib3R0b20iPT1nKXA9ZG9jdW1lbnQuY3JlYXRlRWxlbWVudCgiZGl2IikscC5jbGFzc0xpc3QuYWRkKCJnb29nLXRhYi1iYXItY2xlYXIiKTsidG9wIj09ZyYmcCYmZS5hcHBlbmRDaGlsZChwKTtCPWRvY3VtZW50LmNyZWF0ZUVsZW1lbnQoImRpdiIpO0IuY2xhc3NMaXN0LmFkZCgiZ29vZy10YWItY29udGVudCIpO3ZhciByYT1bXTtjPWJhKGMpO2Zvcih3PWMubmV4dCgpOyF3LmRvbmU7dz1jLm5leHQoKSl3PWRvY3VtZW50LmNyZWF0ZUVsZW1lbnQoImRpdiIpLHcuaWQ9ZS5pZCsiX2NvbnRlbnRfIityYS5sZW5ndGgsdy5zdHlsZS5kaXNwbGF5PSJub25lIixCLmFwcGVuZENoaWxkKHcpLHJhLnB1c2godyk7ZS5hcHBlbmRDaGlsZChCKTsiYm90dG9tIj09ZyYmKHAmJgplLmFwcGVuZENoaWxkKHApLGUuYXBwZW5kQ2hpbGQoZikpO3ZhciBYPW5ldyBWKGcpO1BjKFgsZik7dmFyIFk9LTE7ZGMoWCwic2VsZWN0IixmdW5jdGlvbihaKXtaPWsuaW5kZXhPZihaLnRhcmdldC5mKCkpO1ohPVkmJigwPD1ZJiZZPHJhLmxlbmd0aCYmKHJhW1ldLnN0eWxlLmRpc3BsYXk9Im5vbmUiLFk9LTEpLDA8PVomJlo8cmEubGVuZ3RoJiYoWT1aLHJhW1ldLnN0eWxlLmRpc3BsYXk9ImlubGluZSIsd2luZG93LmRpc3BhdGNoRXZlbnQobmV3IEV2ZW50KCJyZXNpemUiKSksZ29vZ2xlLmNvbGFiLm91dHB1dC5yZXNpemVJZnJhbWVUb0NvbnRlbnQoKSksWC5HKEwoWCxZKSkpfSk7WC5HKEwoWCxkKSk7d2luZG93W2JdPXtzZXRTZWxlY3RlZFRhYkluZGV4OmZ1bmN0aW9uKFope1guRyhMKFgsWikpfX07ZG9jdW1lbnQuaGVhZC5hcHBlbmRDaGlsZChPZChhKSl9LFFkPVsiY29sYWJfbGliIiwiY3JlYXRlVGFiQmFyIl0sVz1sOwpRZFswXWluIFd8fCJ1bmRlZmluZWQiPT10eXBlb2YgVy5leGVjU2NyaXB0fHxXLmV4ZWNTY3JpcHQoInZhciAiK1FkWzBdKTtmb3IodmFyIFJkO1FkLmxlbmd0aCYmKFJkPVFkLnNoaWZ0KCkpOylRZC5sZW5ndGh8fHZvaWQgMD09PVBkP1dbUmRdJiZXW1JkXSE9PU9iamVjdC5wcm90b3R5cGVbUmRdP1c9V1tSZF06Vz1XW1JkXT17fTpXW1JkXT1QZDt9KS5jYWxsKHRoaXMpOwovLyBjbGFuZy1mb3JtYXQgb24K", "ok": true, "headers": [["content-type", "application/javascript"]], "status": 200, "status_text": ""}}, "base_uri": "https://localhost:8080/", "height": 239}
# %%logica AvengerMortality
@Engine("psql");
Probability(x) = Sum(if x == "yes" then 1.0 else 0.0) / Sum(1.0);
AvengerMortality(gender:, death_probability? Probability= death) distinct :-
avengers(gender: avenger_gender, death1: death),
(gender == avenger_gender | gender == "all");
| 798.333333
| 62,041
|
3de60d31886188f4fee3fca79cdb0f59247dffa6
|
py
|
python
|
content/06-inequality/.ipynb_checkpoints/historical-inequality-checkpoint.ipynb
|
d8a-88/econ-models-textbook
|
['BSD-3-Clause']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + tags=["remove_cell"]
import pandas as pd
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
from datascience import *
# %matplotlib inline
plt.style.use('seaborn-muted')
mpl.rcParams['figure.figsize'] = (10.0, 10.0)
# -
# # Income Inequality Historically
#
# <!-- Written by Amal Bhatnagar -->
#
# In the last chart on the previous page, you may have noticed that income inequality was rising in the United States in the last few decades. We will examine this in more detail, and also observe global trends in inequality.
# ## The United States
#
# Let's look at historical trends of income inequality in the US over the last 100 years. The data has been collected from [The World Inequality Database](https://wid.world/), which is co-directed by Berkeley Economics professors Emanuel Saez and Gabriel Zucman. Specifically, we will observe income distributions for the bottom 50 percent, top 10 percent, and top 1 percent.
us_hist = Table.read_table("US_inequality.csv")
us_hist.show(5)
us_hist.take(np.arange(100,105))
# Let's begin with some data cleaning: it seems like our 3 brackets are 'vertically stacked' on top of each other. Instead, we would like a table with 5 columns: `Year`, `bottom 50% income share`, `top 10% income share`, and `top 1% income share`.
bottom_50_us = us_hist.where("Percentile", "p0p50").drop("Percentile").relabeled("Income Share", "Bottom 50% Share")
top_10_us = us_hist.where("Percentile", "p90p100").drop("Percentile").relabeled("Income Share", "Top 10% Share")
top_1_us = us_hist.where("Percentile", "p99p100").drop("Percentile").relabeled("Income Share", "Top 1% Share")
us_hist_joined = bottom_50_us.join("Year", top_10_us).join("Year", top_1_us)
us_hist_joined
# Oh no, there are some `nan` values! NaN (not a number) values are very common in real world datasets: often, we may not have some observations simply because no data was collected, or perhaps the data collected was faulty. Sometimes, we can try to impute or replace NaN values in order to avoid having gaps in our data, but for now let's ignore NaNs and when plotting to see what's going on:
# + tags=["remove_input"]
# mpl.rcParams['figure.dpi'] = 120
us_hist_joined.plot("Year", width=11, height=7)
plt.title("Income Share over Time", fontsize = 16)
plt.ylabel("Proportion", fontsize = 14)
plt.xlabel("Year", fontsize = 14)
plt.show()
# -
# # Income Inequality for the Rest of the World
# Now let's examine the trends of income inequality in other parts of the world.
world_hist = Table.read_table("World_Inequality.csv")
bottom_50_world = world_hist.where("Percentile", "p0p50").drop("Percentile")
top_10_world = world_hist.where("Percentile", "p90p100").drop("Percentile")
top_1_world = world_hist.where("Percentile", "p99p100").drop("Percentile")
top_10_world
# + tags=["remove_input"]
top_10_world.plot("Year", width=11, height=7)
plt.ylabel("Gini Coefficient", fontsize=14)
plt.xlabel("Year", fontsize=14)
plt.title("Income Inequality over Time", fontsize=18);
# -
# Just like the US, it seems global inequality has been rising around the world, especially in China, India, Russia, and across Europe. However, in absolute terms, the level of income inequality in Europe is much lower than that in the United States.
#
# Also look at Russia: income inequality spiked up around 1991. This was likely caused by the fall of the USSR: the failing Soviet state left the ownership of state assets uncontested, which allowed former USSR officials to acquire state property through informal deals. This led to the rise of many Russian oligarchs - those who rapidly accumulated wealth during the era of Russian privatization directly follwing the dissolution of the Soviet Union.
top_10_world.select("Year", "USA", "Europe").plot("Year", width=11, height=7)
plt.ylabel("Gini Coefficient", fontsize=14)
plt.xlabel("Year", fontsize=14)
plt.title("Income Inequality over Time", fontsize=18);
# ## The Elephant Graph
# ```{figure} elephant_curve.jpg
# ---
# width: 500px
# name: elephant-curve
# ---
# The elephant curve {cite}`10inequality-elephantCurve`
# ```
# The elephant curve is a graph that shows the real income growth per adult across each income group’s percentile around the world.
#
# There are 3 key features of the elephant curve: a hump for the world’s poorest, valley for the middle class, and trunk for the upper class. The thump is made of the world’s poorest countries, most likely those from developing countries. The valley comprises the working class from the developed world and upper class from developing countries. The trunk is made of people from the upper class from developed countries. The hump and valley indicate growth among emerging countries, and the top global 1%’s growth is higher than any other income group, thus explaining the positively sloped shape of the trunk.
#
#
# A study done by the Brookings Institution] found that “poorer countries, and the lower income groups within those countries, have grown most rapidly in the past 20 years” {cite}`10inequality-brookings`. This supports the World Bank’s claim that inequality between countries and within countries is decreasing. The Brookings Institution used only household surveys, however, which usually excludes the top and bottom percentile of the population, due to non-response bias. Still, the study is useful in corroborating the trends and growth in global income inequality.
# # Factors that Affect Income Inequality
#
# Economists have isolated multiple factors that influence a country's income inequality
# - top marginal tax rates
# - unemployment rates
# - population growth
#
# We will look at each of these scenarios independently and see its overall trends
# ## Top marginal tax rates
#
# Let's also take a look at the top marginal tax rates in the United States throughout this time. Overall, the United States (and most of the rest of the world) has a progressive tax system, which means that the more income you earn, the higher percentage you will be taxed at. A good way to reduce income inequality is through progressive taxation; having the richer paying a higher portion of their income will help increase equality. Currently, the top marginal tax rate is 37%, as we can see in the table below.
# ```{figure} MTR.png
# ---
# width: 500px
# name: irs
# ---
# Marginal tax rates. Image from the IRS
# ```
# The top marginal tax rate only applies to the portion of your income above a certain income level. For example, if you earned 19501 dollars in 2019, then you will pay 1940 dollars plus 12% of $19501-19401$, i.e. 12 dollars. For another example, if you earned 80000 dollars, then you will pay $9086 + 0.22(80000-78950) = 9317$ dollars in tax, effectively a $\frac{9317}{80000} = 11.6\%$ tax rate.
#
# In general, the idea is you will pay a lower tax rate for your first $x$ dollars, but a higher rate for dollars earned over $x$.
#
# Now let's look at the historical trends in marginal top tax rates, which is the % taxed at the highest tax bracket.
toptax = Table.read_table("toptaxrate.csv")
toptax
# + tags=["remove_input"]
# mpl.rcParams['figure.dpi'] = 120
toptax.plot(0,1, width=11, height=7)
plt.title("Top Marginal Tax Rate over Time", fontsize = 18)
plt.show()
# + tags=["remove_input"]
# mpl.rcParams['figure.dpi'] = 120
us_hist_joined.plot("Year", width=11, height=7)
plt.title("Income Share over Time", fontsize = 18)
plt.ylabel("Proportion", fontsize = 14)
plt.xlabel("Year", fontsize = 14)
plt.show()
# -
# This graph depicts income inequality decreasing between 1910 and 1970 and increasing from 1970 to present.
#
# In 1913, Congress implemented the current income tax to promote equality. Originally meant to help compensate for revenue lost from reducing high tariffs, the new policy essentially made the top 1% start contributing to taxes. Additionally, the top marginal tax rate increased from 7% in 1913 to 73% in 1918, thus helping reduce income inequality. Right before the Great Depression, income inequality peaked, where the richest 1% possessed 19.6% of all income. During the Great Depression, top marginal tax rates increased, peaking at 94% in 1944. The top marginal tax rate decreased but remained high over subsequent decades, where it was 70% in 1965 and 50% in 1982. These high top marginal tax rates are correlated with low income inequality. During the Great Depression, the richest 1% had about 15% of total income. For the 10 years after the Great Depression, the top 1% had below 10% of total income and 8% for the 30 years afterwards. This period was known as the Great Compression, as income differentials between the top 1% and the rest of the country decreased.
#
# In the 1970s, the economy took a turn for the worse with high unemployment and inflation (stagflation) and low growth. In order to stimulate economic growth, the government reduced top marginal tax rates (70% to 38.5% in 1980s), deregulated corporate institutions, and attacked labor union memberships (membership decreased by half within 30 years). Although these policies improved economic growth, it resulted in higher income inequality.
#
# The graph below better shows that the share of income earned by the bottom 50% percentile steadily decreased, while the share earned by the top 1% increased steadily. This means that the top 1% has more wealth than the entire bottom 50% of the population. Suppose a class of 100 people has \$100 in aggregate. In a world with perfect equality, each person would have \$1. With this current level of income inequality, one person would have more wealth than 50 people combined.
#
# The media continues to report on the nation's significant income disparity. [The Washington Post wrote a story](https://www.washingtonpost.com/business/2019/09/26/income-inequality-america-highest-its-been-since-census-started-tracking-it-data-show/) and found that “The number of families earning \$15,000 or less has fallen since 2007, according to the latest census data, while the number of households bringing in \$250,000 a year or more has grown more than 15 percent.”
#
# Can we conclude that high marginal tax rates lead to low income inequality but slow economic growth?
# ## Unemployment rates
#
# Economists believe that unemployment is one of the leading factors that leads to income inequality. When looking at what influences the Gini coefficient, a paper from [Princeton](https://rpds.princeton.edu/sites/rpds/files/media/menendez_unemployment_ar.pdf) found that the unemployment rate had the largest effect on the income inequality rates
#
# Below, we look at the unemployment rates for the past 20 years across many different countries. These are the same countries and regions that we will further study below.
unemployment = Table.read_table("Unemployment.csv")
unemployment
# As we can see from the graph, the unemployment rates for China, India and the rest of the world have stayed somewhat steady. On the other hand, Brazil, the US, Russia and Europe are encountering drastically different unemployment situations than before.
# + tags=["remove_input"]
# mpl.rcParams['figure.dpi'] = 120
unemployment.plot("Year", width=11, height=7)
plt.ylabel("Unemployment Rate (%)", fontsize = 14)
plt.xlabel("Year", fontsize =14)
plt.title("Unemployment Rate over Time", fontsize = 18)
plt.show()
# + tags=["remove_input"]
top_10_world.plot("Year", width=11, height=7)
plt.ylabel("Gini Coefficient", size=14)
plt.xlabel("Year", fontsize=14)
plt.title("International Income Inequality over Time", fontsize=18)
plt.show()
# -
# The graphs above show a positive correlation between unemployment and income inequality. As unemployment increases, income inequality also increases. In 2011, Hanan Morsy, an Egyptian economist who serves as the Director of Macroeconomic Policy, Forecasting and Research at the African Development Bank, actually researched this topic {cite}`10inequality-morsy`. Her group examined the member nations of the Organization for Economic Cooperation and Development (OECD) between 1980 and 2005. She found that specific groups that were vulnerable to the economic shocks that led to an increase in income inequality:
# - young workers
# - low-skilled workers
# - workers who had been out of work for a long time
#
# Her solution was to increase job creation opportunities for temporary, recently fired, and recently hired workers, provide job assistance and training to prevent long-term unemployment, and improve incentives for working by aligning incentives with productivity of labor. Morsy's research found that the most vulnerable groups to economic shocks were young, low-skilled and temporary workers. Creating opportunities for these different demographics would help them be more protected from potential shocks and thus decrease income inequality.
# ## Population growth
#
# As the number of people in a country's population increase, it becomes more difficult for a country to distribute its public goods to everyone. This leads to many social consequences in which resources are not fairly distributed to all members of the population, cause inaccessibility for different parts of the population
#
# The table below shows how the population growth has changed for the same countries we saw above. We are only looking at data for the past 10 years.
pop_growth = Table.read_table("Population Growth.csv")
pop_growth
# + tags=["remove_input"]
pop_growth.plot("Year", width=11, height=7)
plt.title("Population Growth over Time", fontsize = 18)
plt.ylabel("Population Growth (%)", fontsize = 14)
plt.xlabel("Year", fontsize = 14)
plt.show()
# + tags=["remove_input"]
top_10_world.plot("Year", width=11, height=7)
plt.ylabel("Gini Coefficient", fontsize=14)
plt.xlabel("Year", fontsize=14)
plt.title("International Income Inequality over Time", fontsize=18)
plt.show()
# -
# The graphs above show that most countries high population growth between 1-2% during the 1980's. The effects of this can be seen in the rising income inequality during the 90's.
#
# Recent research by University of Toronto's Marijn Bolhuis and Univeristy of Oxford's Alexandra de Pleijt shows that there is a strong correlation between a country's population growth (measured by birth rates) and its income inequality {cite}`10inequality-popGrowth`. Their most recent study in 2016 analyzed income inequality and birth rates data between 1870 and 2000 across 67 countries. They concluded that if a country had 50% higher income inequality, then that country's birth rate would be about twice as high as another country with the same level of economic development. Bolhuis says that these higher birth rates mean that economic growth has to be equal to or greater than the birth rate to offset the implications of higher birth rates.
#
# This is part of a larger debate about the relationship between birth rates and income inequality. Economist Thomas Piketty finds that low birth rates, rather than high birth rates, are causing today's income inequality. With lower birth rates, fewer children per couple are being borne, so these children get more of their parents' inheritance.
#
| 67.78125
| 1,074
|
d046fc7ae264d23b73881ae5f1236328fbad494c
|
py
|
python
|
notebooks/thesis_experiments/20200924_eMVFTS_Wind_Energy_Raw.ipynb
|
cseveriano/spatio-temporal-forecasting
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/cseveriano/spatio-temporal-forecasting/blob/master/notebooks/thesis_experiments/20200924_eMVFTS_Wind_Energy_Raw.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="WiQufqAuISUg"
# ## Forecasting experiments for GEFCOM 2012 Wind Dataset
# + [markdown] id="B98rDdRxx1F3"
#
# ## Install Libs
#
# + id="O_UcD8t_x-XW" colab={"base_uri": "https://localhost:8080/", "height": 1000} outputId="09528ca2-7809-4423-a7fc-4fc0d0111c5b"
# !pip3 install -U git+https://github.com/PYFTS/pyFTS
# !pip3 install -U git+https://github.com/cseveriano/spatio-temporal-forecasting
# !pip3 install -U git+https://github.com/cseveriano/evolving_clustering
# !pip3 install -U git+https://github.com/cseveriano/fts2image
# !pip3 install -U hyperopt
# !pip3 install -U pyts
# + id="wHu_JMgegEAY"
import pandas as pd
import numpy as np
from hyperopt import hp
from spatiotemporal.util import parameter_tuning, sampling
from spatiotemporal.util import experiments as ex
from sklearn.metrics import mean_squared_error
from google.colab import files
import matplotlib.pyplot as plt
import pickle
import math
from pyFTS.benchmarks import Measures
from pyts.decomposition import SingularSpectrumAnalysis
from google.colab import files
import warnings
warnings.filterwarnings("ignore", category=DeprecationWarning)
import datetime
# + [markdown] id="g9BRMrQ_P51z"
# ## Aux Functions
# + id="lBr4-k-8iISF"
def normalize(df):
mindf = df.min()
maxdf = df.max()
return (df-mindf)/(maxdf-mindf)
def denormalize(norm, _min, _max):
return [(n * (_max-_min)) + _min for n in norm]
def getRollingWindow(index):
pivot = index
train_start = pivot.strftime('%Y-%m-%d')
pivot = pivot + datetime.timedelta(days=20)
train_end = pivot.strftime('%Y-%m-%d')
pivot = pivot + datetime.timedelta(days=1)
test_start = pivot.strftime('%Y-%m-%d')
pivot = pivot + datetime.timedelta(days=6)
test_end = pivot.strftime('%Y-%m-%d')
return train_start, train_end, test_start, test_end
def calculate_rolling_error(cv_name, df, forecasts, order_list):
cv_results = pd.DataFrame(columns=['Split', 'RMSE', 'SMAPE'])
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
for i in np.arange(len(forecasts)):
train_start, train_end, test_start, test_end = getRollingWindow(index)
test = df[test_start : test_end]
yhat = forecasts[i]
order = order_list[i]
rmse = Measures.rmse(test.iloc[order:], yhat[:-1])
smape = Measures.smape(test.iloc[order:], yhat[:-1])
res = {'Split' : index.strftime('%Y-%m-%d') ,'RMSE' : rmse, 'SMAPE' : smape}
cv_results = cv_results.append(res, ignore_index=True)
cv_results.to_csv(cv_name+".csv")
index = index + datetime.timedelta(days=7)
return cv_results
def get_final_forecast(norm_forecasts):
forecasts_final = []
for i in np.arange(len(norm_forecasts)):
f_raw = denormalize(norm_forecasts[i], min_raw, max_raw)
forecasts_final.append(f_raw)
return forecasts_final
# + id="ZlkR5-ya4JCM"
from spatiotemporal.test import methods_space_oahu as ms
from spatiotemporal.util import parameter_tuning, sampling
from spatiotemporal.util import experiments as ex
from sklearn.metrics import mean_squared_error
import numpy as np
from hyperopt import fmin, tpe, hp, STATUS_OK, Trials
from hyperopt import space_eval
import traceback
from . import sampling
import pickle
def calculate_error(loss_function, test_df, forecast, offset):
error = loss_function(test_df.iloc[(offset):], forecast)
print("Error : "+str(error))
return error
def method_optimize(experiment, forecast_method, train_df, test_df, space, loss_function, max_evals):
def objective(params):
print(params)
try:
_output = list(params['output'])
forecast = forecast_method(train_df, test_df, params)
_step = params.get('step', 1)
offset = params['order'] + _step - 1
error = calculate_error(loss_function, test_df[_output], forecast, offset)
except Exception:
traceback.print_exc()
error = 1000
return {'loss': error, 'status': STATUS_OK}
print("Running experiment: " + experiment)
trials = Trials()
best = fmin(objective, space, algo=tpe.suggest, max_evals=max_evals, trials=trials)
print('best parameters: ')
print(space_eval(space, best))
pickle.dump(best, open("best_" + experiment + ".pkl", "wb"))
pickle.dump(trials, open("trials_" + experiment + ".pkl", "wb"))
def run_search(methods, data, train, loss_function, max_evals=100, resample=None):
if resample:
data = sampling.resample_data(data, resample)
train_df, test_df = sampling.train_test_split(data, train)
for experiment, method, space in methods:
method_optimize(experiment, method, train_df, test_df, space, loss_function, max_evals)
# + [markdown] id="hZ4KkWX7Jb5g"
# ## Load Dataset
# + id="HKOTmAwrQM2G"
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import math
from sklearn.metrics import mean_squared_error
# + id="aHWBkv_BJfWZ"
#columns names
wind_farms = ['wp1', 'wp2', 'wp3', 'wp4', 'wp5', 'wp6', 'wp7']
# read raw dataset
import pandas as pd
df = pd.read_csv('https://query.data.world/s/3zx2jusk4z6zvlg2dafqgshqp3oao6', parse_dates=['date'], index_col=0)
df.index = pd.to_datetime(df.index, format="%Y%m%d%H")
interval = ((df.index >= '2009-07') & (df.index <= '2010-08'))
df = df.loc[interval]
#Normalize Data
# Save Min-Max for Denorm
min_raw = df.min()
max_raw = df.max()
# Perform Normalization
norm_df = normalize(df)
# Tuning split
tuning_df = norm_df["2009-07-01":"2009-07-31"]
norm_df = norm_df["2009-08-01":"2010-08-30"]
df = df["2009-08-01":"2010-08-30"]
# + [markdown] id="VS9lU7sIXrJl"
# ## Forecasting Methods
# + [markdown] id="jEustxeTa7Ub"
# ### Persistence
# + id="uLxMBBLWa6du"
def persistence_forecast(train, test, step):
predictions = []
for t in np.arange(0,len(test), step):
yhat = [test.iloc[t]] * step
predictions.extend(yhat)
return predictions
def rolling_cv_persistence(df, step):
forecasts = []
lags_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
test = df[test_start : test_end]
yhat = persistence_forecast(train, test, step)
lags_list.append(1)
forecasts.append(yhat)
return forecasts, lags_list
# + id="31nxRmc4bCYn" colab={"base_uri": "https://localhost:8080/", "height": 1000} outputId="500ee19d-b9ef-46c5-85bb-060ae9856fa4"
forecasts_raw, order_list = rolling_cv_persistence(norm_df, 1)
forecasts_final = get_final_forecast(forecasts_raw)
calculate_rolling_error("rolling_cv_wind_raw_persistence", norm_df, forecasts_final, order_list)
# + id="H9z4GDsu6kDU" colab={"base_uri": "https://localhost:8080/", "height": 17} outputId="2883ea15-8cc1-4d99-8f6a-bce3c79512b7"
files.download('rolling_cv_wind_raw_persistence.csv')
# + [markdown] id="DdMi7e1scYu4"
# ### VAR
# + id="1MxApA3Acf7m"
from statsmodels.tsa.api import VAR, DynamicVAR
# + id="7MpVUQPbKgYb"
def evaluate_VAR_models(test_name, train, validation,target, maxlags_list):
var_results = pd.DataFrame(columns=['Order','RMSE'])
best_score, best_cfg, best_model = float("inf"), None, None
for lgs in maxlags_list:
model = VAR(train)
results = model.fit(maxlags=lgs, ic='aic')
order = results.k_ar
forecast = []
for i in range(len(validation)-order) :
forecast.extend(results.forecast(validation.values[i:i+order],1))
forecast_df = pd.DataFrame(columns=validation.columns, data=forecast)
rmse = Measures.rmse(validation[target].iloc[order:], forecast_df[target].values)
if rmse < best_score:
best_score, best_cfg, best_model = rmse, order, results
res = {'Order' : str(order) ,'RMSE' : rmse}
print('VAR (%s) RMSE=%.3f' % (str(order),rmse))
var_results = var_results.append(res, ignore_index=True)
var_results.to_csv(test_name+".csv")
print('Best VAR(%s) RMSE=%.3f' % (best_cfg, best_score))
return best_model
# + id="SK-a1a0ZcjBi"
def var_forecast(train, test, params):
order = params['order']
step = params['step']
model = VAR(train.values)
results = model.fit(maxlags=order)
lag_order = results.k_ar
print("Lag order:" + str(lag_order))
forecast = []
for i in np.arange(0,len(test)-lag_order+1,step) :
forecast.extend(results.forecast(test.values[i:i+lag_order],step))
forecast_df = pd.DataFrame(columns=test.columns, data=forecast)
return forecast_df.values, lag_order
# + id="YnAuyYY2Ku0M"
def rolling_cv_var(df, params):
forecasts = []
order_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
test = df[test_start : test_end]
# Concat train & validation for test
yhat, lag_order = var_forecast(train, test, params)
forecasts.append(yhat)
order_list.append(lag_order)
return forecasts, order_list
# + id="2ALT505EcomF" colab={"base_uri": "https://localhost:8080/", "height": 1000} outputId="20e13148-d19a-401e-fa4b-8370d7fcdaaf"
params_raw = {'order': 4, 'step': 1}
forecasts_raw, order_list = rolling_cv_var(norm_df, params_raw)
forecasts_final = get_final_forecast(forecasts_raw)
calculate_rolling_error("rolling_cv_wind_raw_var", df, forecasts_final, order_list)
# + id="QzigtkZ-Repi" colab={"base_uri": "https://localhost:8080/", "height": 17} outputId="8f44c614-c57f-4cb5-d3c4-c474751944cd"
files.download('rolling_cv_wind_raw_var.csv')
# + [markdown] id="WCvsy-tHdTuN"
# ### e-MVFTS
# + id="lCx9NiYEjSZr"
from spatiotemporal.models.clusteredmvfts.fts import evolvingclusterfts
# + id="iq9njQOmdtHU"
def evolvingfts_forecast(train_df, test_df, params, train_model=True):
_variance_limit = params['variance_limit']
_defuzzy = params['defuzzy']
_t_norm = params['t_norm']
_membership_threshold = params['membership_threshold']
_order = params['order']
_step = params['step']
model = evolvingclusterfts.EvolvingClusterFTS(variance_limit=_variance_limit, defuzzy=_defuzzy, t_norm=_t_norm,
membership_threshold=_membership_threshold)
model.fit(train_df.values, order=_order, verbose=False)
forecast = model.predict(test_df.values, steps_ahead=_step)
forecast_df = pd.DataFrame(data=forecast, columns=test_df.columns)
return forecast_df.values
# + id="NNFgSmALkRra"
def rolling_cv_evolving(df, params):
forecasts = []
order_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
first_time = True
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
test = df[test_start : test_end]
# Concat train & validation for test
yhat = list(evolvingfts_forecast(train, test, params, train_model=first_time))
#yhat.append(yhat[-1]) #para manter o formato do vetor de metricas
forecasts.append(yhat)
order_list.append(params['order'])
first_time = False
return forecasts, order_list
# + id="u9HSOzyzXNJq" colab={"base_uri": "https://localhost:8080/", "height": 1000} outputId="ac86e391-b4af-4585-a79a-534622375179"
params_raw = {'variance_limit': 0.001, 'order': 2, 'defuzzy': 'weighted', 't_norm': 'threshold', 'membership_threshold': 0.6, 'step':1}
forecasts_raw, order_list = rolling_cv_evolving(norm_df, params_raw)
forecasts_final = get_final_forecast(forecasts_raw)
calculate_rolling_error("rolling_cv_wind_raw_emvfts", df, forecasts_final, order_list)
# + id="M-_n_uWMjPw7" colab={"base_uri": "https://localhost:8080/", "height": 17} outputId="0782e819-9728-43c0-8e16-b0ea8b8725b1"
files.download('rolling_cv_wind_raw_emvfts.csv')
# + [markdown] id="8lbu8aLan_Yt"
# ### MLP
# + id="LwBHQp9er2BG"
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import LSTM
from keras.layers import Dropout
from keras.constraints import maxnorm
from keras.models import Sequential
from keras.layers.core import Dense, Dropout, Activation
from keras.layers.normalization import BatchNormalization
# + id="OhmFnGmHoB5X"
# convert series to supervised learning
def series_to_supervised(data, n_in=1, n_out=1, dropnan=True):
n_vars = 1 if type(data) is list else data.shape[1]
df = pd.DataFrame(data)
cols, names = list(), list()
# input sequence (t-n, ... t-1)
for i in range(n_in, 0, -1):
cols.append(df.shift(i))
names += [('var%d(t-%d)' % (j+1, i)) for j in range(n_vars)]
# forecast sequence (t, t+1, ... t+n)
for i in range(0, n_out):
cols.append(df.shift(-i))
if i == 0:
names += [('var%d(t)' % (j+1)) for j in range(n_vars)]
else:
names += [('var%d(t+%d)' % (j+1, i)) for j in range(n_vars)]
# put it all together
agg = pd.concat(cols, axis=1)
agg.columns = names
# drop rows with NaN values
if dropnan:
agg.dropna(inplace=True)
return agg
# + [markdown] id="M67t6lilUlYC"
# #### MLP Parameter Tuning
# + id="nJBFQgGhU_0z"
from spatiotemporal.util import parameter_tuning, sampling
from spatiotemporal.util import experiments as ex
from sklearn.metrics import mean_squared_error
from hyperopt import hp
import numpy as np
# + id="YNRVPkeDUzu3"
mlp_space = {'choice':
hp.choice('num_layers',
[
{'layers': 'two',
},
{'layers': 'three',
'units3': hp.choice('units3', [8, 16, 64, 128, 256, 512]),
'dropout3': hp.choice('dropout3', [0, 0.25, 0.5, 0.75])
}
]),
'units1': hp.choice('units1', [8, 16, 64, 128, 256, 512]),
'units2': hp.choice('units2', [8, 16, 64, 128, 256, 512]),
'dropout1': hp.choice('dropout1', [0, 0.25, 0.5, 0.75]),
'dropout2': hp.choice('dropout2', [0, 0.25, 0.5, 0.75]),
'batch_size': hp.choice('batch_size', [28, 64, 128, 256, 512]),
'order': hp.choice('order', [1, 2, 3]),
'input': hp.choice('input', [wind_farms]),
'output': hp.choice('output', [wind_farms]),
'epochs': hp.choice('epochs', [100, 200, 300])}
# + id="aeB6ImDOVGsY"
def mlp_tuning(train_df, test_df, params):
_input = list(params['input'])
_nlags = params['order']
_epochs = params['epochs']
_batch_size = params['batch_size']
nfeat = len(train_df.columns)
nsteps = params.get('step',1)
nobs = _nlags * nfeat
output_index = -nfeat*nsteps
train_reshaped_df = series_to_supervised(train_df[_input], n_in=_nlags, n_out=nsteps)
train_X, train_Y = train_reshaped_df.iloc[:, :nobs].values, train_reshaped_df.iloc[:, output_index:].values
test_reshaped_df = series_to_supervised(test_df[_input], n_in=_nlags, n_out=nsteps)
test_X, test_Y = test_reshaped_df.iloc[:, :nobs].values, test_reshaped_df.iloc[:, output_index:].values
# design network
model = Sequential()
model.add(Dense(params['units1'], input_dim=train_X.shape[1], activation='relu'))
model.add(Dropout(params['dropout1']))
model.add(BatchNormalization())
model.add(Dense(params['units2'], activation='relu'))
model.add(Dropout(params['dropout2']))
model.add(BatchNormalization())
if params['choice']['layers'] == 'three':
model.add(Dense(params['choice']['units3'], activation='relu'))
model.add(Dropout(params['choice']['dropout3']))
model.add(BatchNormalization())
model.add(Dense(train_Y.shape[1], activation='sigmoid'))
model.compile(loss='mse', optimizer='adam')
# includes the call back object
model.fit(train_X, train_Y, epochs=_epochs, batch_size=_batch_size, verbose=False, shuffle=False)
# predict the test set
forecast = model.predict(test_X, verbose=False)
return forecast
# + id="xfCg9twkiDPh" colab={"base_uri": "https://localhost:8080/", "height": 1000} outputId="ec265dbe-b131-4cae-b31f-338a4b6ddcd3"
methods = []
methods.append(("EXP_OAHU_MLP", mlp_tuning, mlp_space))
train_split = 0.6
run_search(methods, tuning_df, train_split, Measures.rmse, max_evals=30, resample=None)
# + [markdown] id="DZ9toU2mVac2"
# #### MLP Forecasting
# + id="5oahrWS9r-zE"
def mlp_multi_forecast(train_df, test_df, params):
nfeat = len(train_df.columns)
nlags = params['order']
nsteps = params.get('step',1)
nobs = nlags * nfeat
output_index = -nfeat*nsteps
train_reshaped_df = series_to_supervised(train_df, n_in=nlags, n_out=nsteps)
train_X, train_Y = train_reshaped_df.iloc[:, :nobs].values, train_reshaped_df.iloc[:, output_index:].values
test_reshaped_df = series_to_supervised(test_df, n_in=nlags, n_out=nsteps)
test_X, test_Y = test_reshaped_df.iloc[:, :nobs].values, test_reshaped_df.iloc[:, output_index:].values
# design network
model = designMLPNetwork(train_X.shape[1], train_Y.shape[1], params)
# fit network
model.fit(train_X, train_Y, epochs=500, batch_size=1000, verbose=False, shuffle=False)
forecast = model.predict(test_X)
# fcst = [f[0] for f in forecast]
fcst = forecast
return fcst
# + id="c4ZEEfNLsJp6"
def designMLPNetwork(input_shape, output_shape, params):
model = Sequential()
model.add(Dense(params['units1'], input_dim=input_shape, activation='relu'))
model.add(Dropout(params['dropout1']))
model.add(BatchNormalization())
model.add(Dense(params['units2'], activation='relu'))
model.add(Dropout(params['dropout2']))
model.add(BatchNormalization())
if params['choice']['layers'] == 'three':
model.add(Dense(params['choice']['units3'], activation='relu'))
model.add(Dropout(params['choice']['dropout3']))
model.add(BatchNormalization())
model.add(Dense(output_shape, activation='sigmoid'))
model.compile(loss='mse', optimizer='adam')
return model
# + id="t8btj2aDsVyk"
def rolling_cv_mlp(df, params):
forecasts = []
order_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
test = df[test_start : test_end]
# Perform forecast
yhat = list(mlp_multi_forecast(train, test, params))
yhat.append(yhat[-1]) #para manter o formato do vetor de metricas
forecasts.append(yhat)
order_list.append(params['order'])
return forecasts, order_list
# + id="pVEGpB98sq23" colab={"base_uri": "https://localhost:8080/", "height": 1000} outputId="1afff3bd-94ed-467a-d004-6c31cd6ecb89"
# Enter best params
params_raw = {'batch_size': 64, 'choice': {'layers': 'two'}, 'dropout1': 0.25, 'dropout2': 0.5, 'epochs': 200, 'input': ('wp1', 'wp2', 'wp3', 'wp4', 'wp5', 'wp6', 'wp7'), 'order': 2, 'output': ('wp1', 'wp2', 'wp3', 'wp4', 'wp5', 'wp6', 'wp7'), 'units1': 128, 'units2': 128}
forecasts_raw, order_list = rolling_cv_mlp(norm_df, params_raw)
forecasts_final = get_final_forecast(forecasts_raw)
calculate_rolling_error("rolling_cv_wind_raw_mlp_multi", df, forecasts_final, order_list)
# + id="7nKc-SoUG7Ob" colab={"base_uri": "https://localhost:8080/", "height": 17} outputId="6d1f8d75-89f0-4e67-9f79-f7f6fda9bbf2"
files.download('rolling_cv_wind_raw_mlp_multi.csv')
# + [markdown] id="8TrCf5o3wRtf"
# ### Granular FTS
# + id="8XLpD3ADwUKl"
from pyFTS.models.multivariate import granular
from pyFTS.partitioners import Grid, Entropy
from pyFTS.models.multivariate import variable
from pyFTS.common import Membership
from pyFTS.partitioners import Grid, Entropy
# + [markdown] id="Sy91cSv-bPe-"
# #### Granular Parameter Tuning
# + id="VSznTw3qdZm5"
granular_space = {
'npartitions': hp.choice('npartitions', [100, 150, 200]),
'order': hp.choice('order', [1, 2]),
'knn': hp.choice('knn', [1, 2, 3, 4, 5]),
'alpha_cut': hp.choice('alpha_cut', [0, 0.1, 0.2, 0.3]),
'input': hp.choice('input', [['wp1', 'wp2', 'wp3']]),
'output': hp.choice('output', [['wp1', 'wp2', 'wp3']])}
# + id="pDf07Ua7bOkd"
def granular_tuning(train_df, test_df, params):
_input = list(params['input'])
_output = list(params['output'])
_npartitions = params['npartitions']
_order = params['order']
_knn = params['knn']
_alpha_cut = params['alpha_cut']
_step = params.get('step',1)
## create explanatory variables
exp_variables = []
for vc in _input:
exp_variables.append(variable.Variable(vc, data_label=vc, alias=vc,
npart=_npartitions, func=Membership.trimf,
data=train_df, alpha_cut=_alpha_cut))
model = granular.GranularWMVFTS(explanatory_variables=exp_variables, target_variable=exp_variables[0], order=_order,
knn=_knn)
model.fit(train_df[_input], num_batches=1)
if _step > 1:
forecast = pd.DataFrame(columns=test_df.columns)
length = len(test_df.index)
for k in range(0,(length -(_order + _step - 1))):
fcst = model.predict(test_df[_input], type='multivariate', start_at=k, steps_ahead=_step)
forecast = forecast.append(fcst.tail(1))
else:
forecast = model.predict(test_df[_input], type='multivariate')
return forecast[_output].values
# + id="LJ9SITzDdM4K"
methods = []
methods.append(("EXP_WIND_GRANULAR", granular_tuning, granular_space))
# + id="OnCvUcmOdlQw" colab={"base_uri": "https://localhost:8080/", "height": 445} outputId="39b3df5d-0090-48fd-a48d-3f1424be6b54"
train_split = 0.6
run_search(methods, tuning_df, train_split, Measures.rmse, max_evals=10, resample=None)
# + [markdown] id="5FZfm7NjbVc0"
# #### Granular Forecasting
# + id="c2bZmI1obYUh"
def granular_forecast(train_df, test_df, params):
_input = list(params['input'])
_output = list(params['output'])
_npartitions = params['npartitions']
_knn = params['knn']
_alpha_cut = params['alpha_cut']
_order = params['order']
_step = params.get('step',1)
## create explanatory variables
exp_variables = []
for vc in _input:
exp_variables.append(variable.Variable(vc, data_label=vc, alias=vc,
npart=_npartitions, func=Membership.trimf,
data=train_df, alpha_cut=_alpha_cut))
model = granular.GranularWMVFTS(explanatory_variables=exp_variables, target_variable=exp_variables[0], order=_order,
knn=_knn)
model.fit(train_df[_input], num_batches=1)
if _step > 1:
forecast = pd.DataFrame(columns=test_df.columns)
length = len(test_df.index)
for k in range(0,(length -(_order + _step - 1))):
fcst = model.predict(test_df[_input], type='multivariate', start_at=k, steps_ahead=_step)
forecast = forecast.append(fcst.tail(1))
else:
forecast = model.predict(test_df[_input], type='multivariate')
return forecast[_output].values
# + id="zRRkdE-Jg4X1"
def rolling_cv_granular(df, params):
forecasts = []
order_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
test = df[test_start : test_end]
# Perform forecast
yhat = list(granular_forecast(train, test, params))
yhat.append(yhat[-1]) #para manter o formato do vetor de metricas
forecasts.append(yhat)
order_list.append(params['order'])
return forecasts, order_list
# + id="2iS4VCWa0QcL"
def granular_get_final_forecast(forecasts_raw, input):
forecasts_final = []
l_min = df[input].min()
l_max = df[input].max()
for i in np.arange(len(forecasts_raw)):
f_raw = denormalize(forecasts_raw[i], l_min, l_max)
forecasts_final.append(f_raw)
return forecasts_final
# + id="TxmOkURxcJ79" colab={"base_uri": "https://localhost:8080/", "height": 1000} outputId="1f4597cc-852b-47db-d176-daf2ac30c338"
# Enter best params
params_raw = {'alpha_cut': 0.3, 'input': ('wp1', 'wp2', 'wp3'), 'knn': 5, 'npartitions': 200, 'order': 2, 'output': ('wp1', 'wp2', 'wp3')}
forecasts_raw, order_list = rolling_cv_granular(norm_df, params_raw)
forecasts_final = granular_get_final_forecast(forecasts_raw, list(params_raw['input']))
calculate_rolling_error("rolling_cv_wind_raw_granular", df[list(params_raw['input'])], forecasts_final, order_list)
# + id="yhEjDeFNfS_w" colab={"base_uri": "https://localhost:8080/", "height": 17} outputId="7e20a078-d01f-401d-fdf2-f3c7523c92d7"
files.download('rolling_cv_wind_raw_granular.csv')
# + [markdown] id="8lFQJ2-BU9aK"
# ## Result Analysis
# + id="9T-7825NVCcG"
import pandas as pd
from google.colab import files
# + id="cJlDztPk8FxF" colab={"resources": {"http://localhost:8080/nbextensions/google.colab/files.js": {"data": "Ly8gQ29weXJpZ2h0IDIwMTcgR29vZ2xlIExMQwovLwovLyBMaWNlbnNlZCB1bmRlciB0aGUgQXBhY2hlIExpY2Vuc2UsIFZlcnNpb24gMi4wICh0aGUgIkxpY2Vuc2UiKTsKLy8geW91IG1heSBub3QgdXNlIHRoaXMgZmlsZSBleGNlcHQgaW4gY29tcGxpYW5jZSB3aXRoIHRoZSBMaWNlbnNlLgovLyBZb3UgbWF5IG9idGFpbiBhIGNvcHkgb2YgdGhlIExpY2Vuc2UgYXQKLy8KLy8gICAgICBodHRwOi8vd3d3LmFwYWNoZS5vcmcvbGljZW5zZXMvTElDRU5TRS0yLjAKLy8KLy8gVW5sZXNzIHJlcXVpcmVkIGJ5IGFwcGxpY2FibGUgbGF3IG9yIGFncmVlZCB0byBpbiB3cml0aW5nLCBzb2Z0d2FyZQovLyBkaXN0cmlidXRlZCB1bmRlciB0aGUgTGljZW5zZSBpcyBkaXN0cmlidXRlZCBvbiBhbiAiQVMgSVMiIEJBU0lTLAovLyBXSVRIT1VUIFdBUlJBTlRJRVMgT1IgQ09ORElUSU9OUyBPRiBBTlkgS0lORCwgZWl0aGVyIGV4cHJlc3Mgb3IgaW1wbGllZC4KLy8gU2VlIHRoZSBMaWNlbnNlIGZvciB0aGUgc3BlY2lmaWMgbGFuZ3VhZ2UgZ292ZXJuaW5nIHBlcm1pc3Npb25zIGFuZAovLyBsaW1pdGF0aW9ucyB1bmRlciB0aGUgTGljZW5zZS4KCi8qKgogKiBAZmlsZW92ZXJ2aWV3IEhlbHBlcnMgZm9yIGdvb2dsZS5jb2xhYiBQeXRob24gbW9kdWxlLgogKi8KKGZ1bmN0aW9uKHNjb3BlKSB7CmZ1bmN0aW9uIHNwYW4odGV4dCwgc3R5bGVBdHRyaWJ1dGVzID0ge30pIHsKICBjb25zdCBlbGVtZW50ID0gZG9jdW1lbnQuY3JlYXRlRWxlbWVudCgnc3BhbicpOwogIGVsZW1lbnQudGV4dENvbnRlbnQgPSB0ZXh0OwogIGZvciAoY29uc3Qga2V5IG9mIE9iamVjdC5rZXlzKHN0eWxlQXR0cmlidXRlcykpIHsKICAgIGVsZW1lbnQuc3R5bGVba2V5XSA9IHN0eWxlQXR0cmlidXRlc1trZXldOwogIH0KICByZXR1cm4gZWxlbWVudDsKfQoKLy8gTWF4IG51bWJlciBvZiBieXRlcyB3aGljaCB3aWxsIGJlIHVwbG9hZGVkIGF0IGEgdGltZS4KY29uc3QgTUFYX1BBWUxPQURfU0laRSA9IDEwMCAqIDEwMjQ7CgpmdW5jdGlvbiBfdXBsb2FkRmlsZXMoaW5wdXRJZCwgb3V0cHV0SWQpIHsKICBjb25zdCBzdGVwcyA9IHVwbG9hZEZpbGVzU3RlcChpbnB1dElkLCBvdXRwdXRJZCk7CiAgY29uc3Qgb3V0cHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKG91dHB1dElkKTsKICAvLyBDYWNoZSBzdGVwcyBvbiB0aGUgb3V0cHV0RWxlbWVudCB0byBtYWtlIGl0IGF2YWlsYWJsZSBmb3IgdGhlIG5leHQgY2FsbAogIC8vIHRvIHVwbG9hZEZpbGVzQ29udGludWUgZnJvbSBQeXRob24uCiAgb3V0cHV0RWxlbWVudC5zdGVwcyA9IHN0ZXBzOwoKICByZXR1cm4gX3VwbG9hZEZpbGVzQ29udGludWUob3V0cHV0SWQpOwp9CgovLyBUaGlzIGlzIHJvdWdobHkgYW4gYXN5bmMgZ2VuZXJhdG9yIChub3Qgc3VwcG9ydGVkIGluIHRoZSBicm93c2VyIHlldCksCi8vIHdoZXJlIHRoZXJlIGFyZSBtdWx0aXBsZSBhc3luY2hyb25vdXMgc3RlcHMgYW5kIHRoZSBQeXRob24gc2lkZSBpcyBnb2luZwovLyB0byBwb2xsIGZvciBjb21wbGV0aW9uIG9mIGVhY2ggc3RlcC4KLy8gVGhpcyB1c2VzIGEgUHJvbWlzZSB0byBibG9jayB0aGUgcHl0aG9uIHNpZGUgb24gY29tcGxldGlvbiBvZiBlYWNoIHN0ZXAsCi8vIHRoZW4gcGFzc2VzIHRoZSByZXN1bHQgb2YgdGhlIHByZXZpb3VzIHN0ZXAgYXMgdGhlIGlucHV0IHRvIHRoZSBuZXh0IHN0ZXAuCmZ1bmN0aW9uIF91cGxvYWRGaWxlc0NvbnRpbnVlKG91dHB1dElkKSB7CiAgY29uc3Qgb3V0cHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKG91dHB1dElkKTsKICBjb25zdCBzdGVwcyA9IG91dHB1dEVsZW1lbnQuc3RlcHM7CgogIGNvbnN0IG5leHQgPSBzdGVwcy5uZXh0KG91dHB1dEVsZW1lbnQubGFzdFByb21pc2VWYWx1ZSk7CiAgcmV0dXJuIFByb21pc2UucmVzb2x2ZShuZXh0LnZhbHVlLnByb21pc2UpLnRoZW4oKHZhbHVlKSA9PiB7CiAgICAvLyBDYWNoZSB0aGUgbGFzdCBwcm9taXNlIHZhbHVlIHRvIG1ha2UgaXQgYXZhaWxhYmxlIHRvIHRoZSBuZXh0CiAgICAvLyBzdGVwIG9mIHRoZSBnZW5lcmF0b3IuCiAgICBvdXRwdXRFbGVtZW50Lmxhc3RQcm9taXNlVmFsdWUgPSB2YWx1ZTsKICAgIHJldHVybiBuZXh0LnZhbHVlLnJlc3BvbnNlOwogIH0pOwp9CgovKioKICogR2VuZXJhdG9yIGZ1bmN0aW9uIHdoaWNoIGlzIGNhbGxlZCBiZXR3ZWVuIGVhY2ggYXN5bmMgc3RlcCBvZiB0aGUgdXBsb2FkCiAqIHByb2Nlc3MuCiAqIEBwYXJhbSB7c3RyaW5nfSBpbnB1dElkIEVsZW1lbnQgSUQgb2YgdGhlIGlucHV0IGZpbGUgcGlja2VyIGVsZW1lbnQuCiAqIEBwYXJhbSB7c3RyaW5nfSBvdXRwdXRJZCBFbGVtZW50IElEIG9mIHRoZSBvdXRwdXQgZGlzcGxheS4KICogQHJldHVybiB7IUl0ZXJhYmxlPCFPYmplY3Q+fSBJdGVyYWJsZSBvZiBuZXh0IHN0ZXBzLgogKi8KZnVuY3Rpb24qIHVwbG9hZEZpbGVzU3RlcChpbnB1dElkLCBvdXRwdXRJZCkgewogIGNvbnN0IGlucHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKGlucHV0SWQpOwogIGlucHV0RWxlbWVudC5kaXNhYmxlZCA9IGZhbHNlOwoKICBjb25zdCBvdXRwdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQob3V0cHV0SWQpOwogIG91dHB1dEVsZW1lbnQuaW5uZXJIVE1MID0gJyc7CgogIGNvbnN0IHBpY2tlZFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgaW5wdXRFbGVtZW50LmFkZEV2ZW50TGlzdGVuZXIoJ2NoYW5nZScsIChlKSA9PiB7CiAgICAgIHJlc29sdmUoZS50YXJnZXQuZmlsZXMpOwogICAgfSk7CiAgfSk7CgogIGNvbnN0IGNhbmNlbCA9IGRvY3VtZW50LmNyZWF0ZUVsZW1lbnQoJ2J1dHRvbicpOwogIGlucHV0RWxlbWVudC5wYXJlbnRFbGVtZW50LmFwcGVuZENoaWxkKGNhbmNlbCk7CiAgY2FuY2VsLnRleHRDb250ZW50ID0gJ0NhbmNlbCB1cGxvYWQnOwogIGNvbnN0IGNhbmNlbFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgY2FuY2VsLm9uY2xpY2sgPSAoKSA9PiB7CiAgICAgIHJlc29sdmUobnVsbCk7CiAgICB9OwogIH0pOwoKICAvLyBXYWl0IGZvciB0aGUgdXNlciB0byBwaWNrIHRoZSBmaWxlcy4KICBjb25zdCBmaWxlcyA9IHlpZWxkIHsKICAgIHByb21pc2U6IFByb21pc2UucmFjZShbcGlja2VkUHJvbWlzZSwgY2FuY2VsUHJvbWlzZV0pLAogICAgcmVzcG9uc2U6IHsKICAgICAgYWN0aW9uOiAnc3RhcnRpbmcnLAogICAgfQogIH07CgogIGNhbmNlbC5yZW1vdmUoKTsKCiAgLy8gRGlzYWJsZSB0aGUgaW5wdXQgZWxlbWVudCBzaW5jZSBmdXJ0aGVyIHBpY2tzIGFyZSBub3QgYWxsb3dlZC4KICBpbnB1dEVsZW1lbnQuZGlzYWJsZWQgPSB0cnVlOwoKICBpZiAoIWZpbGVzKSB7CiAgICByZXR1cm4gewogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbXBsZXRlJywKICAgICAgfQogICAgfTsKICB9CgogIGZvciAoY29uc3QgZmlsZSBvZiBmaWxlcykgewogICAgY29uc3QgbGkgPSBkb2N1bWVudC5jcmVhdGVFbGVtZW50KCdsaScpOwogICAgbGkuYXBwZW5kKHNwYW4oZmlsZS5uYW1lLCB7Zm9udFdlaWdodDogJ2JvbGQnfSkpOwogICAgbGkuYXBwZW5kKHNwYW4oCiAgICAgICAgYCgke2ZpbGUudHlwZSB8fCAnbi9hJ30pIC0gJHtmaWxlLnNpemV9IGJ5dGVzLCBgICsKICAgICAgICBgbGFzdCBtb2RpZmllZDogJHsKICAgICAgICAgICAgZmlsZS5sYXN0TW9kaWZpZWREYXRlID8gZmlsZS5sYXN0TW9kaWZpZWREYXRlLnRvTG9jYWxlRGF0ZVN0cmluZygpIDoKICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgJ24vYSd9IC0gYCkpOwogICAgY29uc3QgcGVyY2VudCA9IHNwYW4oJzAlIGRvbmUnKTsKICAgIGxpLmFwcGVuZENoaWxkKHBlcmNlbnQpOwoKICAgIG91dHB1dEVsZW1lbnQuYXBwZW5kQ2hpbGQobGkpOwoKICAgIGNvbnN0IGZpbGVEYXRhUHJvbWlzZSA9IG5ldyBQcm9taXNlKChyZXNvbHZlKSA9PiB7CiAgICAgIGNvbnN0IHJlYWRlciA9IG5ldyBGaWxlUmVhZGVyKCk7CiAgICAgIHJlYWRlci5vbmxvYWQgPSAoZSkgPT4gewogICAgICAgIHJlc29sdmUoZS50YXJnZXQucmVzdWx0KTsKICAgICAgfTsKICAgICAgcmVhZGVyLnJlYWRBc0FycmF5QnVmZmVyKGZpbGUpOwogICAgfSk7CiAgICAvLyBXYWl0IGZvciB0aGUgZGF0YSB0byBiZSByZWFkeS4KICAgIGxldCBmaWxlRGF0YSA9IHlpZWxkIHsKICAgICAgcHJvbWlzZTogZmlsZURhdGFQcm9taXNlLAogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbnRpbnVlJywKICAgICAgfQogICAgfTsKCiAgICAvLyBVc2UgYSBjaHVua2VkIHNlbmRpbmcgdG8gYXZvaWQgbWVzc2FnZSBzaXplIGxpbWl0cy4gU2VlIGIvNjIxMTU2NjAuCiAgICBsZXQgcG9zaXRpb24gPSAwOwogICAgd2hpbGUgKHBvc2l0aW9uIDwgZmlsZURhdGEuYnl0ZUxlbmd0aCkgewogICAgICBjb25zdCBsZW5ndGggPSBNYXRoLm1pbihmaWxlRGF0YS5ieXRlTGVuZ3RoIC0gcG9zaXRpb24sIE1BWF9QQVlMT0FEX1NJWkUpOwogICAgICBjb25zdCBjaHVuayA9IG5ldyBVaW50OEFycmF5KGZpbGVEYXRhLCBwb3NpdGlvbiwgbGVuZ3RoKTsKICAgICAgcG9zaXRpb24gKz0gbGVuZ3RoOwoKICAgICAgY29uc3QgYmFzZTY0ID0gYnRvYShTdHJpbmcuZnJvbUNoYXJDb2RlLmFwcGx5KG51bGwsIGNodW5rKSk7CiAgICAgIHlpZWxkIHsKICAgICAgICByZXNwb25zZTogewogICAgICAgICAgYWN0aW9uOiAnYXBwZW5kJywKICAgICAgICAgIGZpbGU6IGZpbGUubmFtZSwKICAgICAgICAgIGRhdGE6IGJhc2U2NCwKICAgICAgICB9LAogICAgICB9OwogICAgICBwZXJjZW50LnRleHRDb250ZW50ID0KICAgICAgICAgIGAke01hdGgucm91bmQoKHBvc2l0aW9uIC8gZmlsZURhdGEuYnl0ZUxlbmd0aCkgKiAxMDApfSUgZG9uZWA7CiAgICB9CiAgfQoKICAvLyBBbGwgZG9uZS4KICB5aWVsZCB7CiAgICByZXNwb25zZTogewogICAgICBhY3Rpb246ICdjb21wbGV0ZScsCiAgICB9CiAgfTsKfQoKc2NvcGUuZ29vZ2xlID0gc2NvcGUuZ29vZ2xlIHx8IHt9OwpzY29wZS5nb29nbGUuY29sYWIgPSBzY29wZS5nb29nbGUuY29sYWIgfHwge307CnNjb3BlLmdvb2dsZS5jb2xhYi5fZmlsZXMgPSB7CiAgX3VwbG9hZEZpbGVzLAogIF91cGxvYWRGaWxlc0NvbnRpbnVlLAp9Owp9KShzZWxmKTsK", "ok": true, "headers": [["content-type", "application/javascript"]], "status": 200, "status_text": ""}}, "base_uri": "https://localhost:8080/", "height": 262} outputId="def592a8-6b87-497a-c77d-5929dfc38bba"
files.upload()
# + id="Z4JWb2PkjwOQ"
def createBoxplot(filename, data, xticklabels, ylabel):
# Create a figure instance
fig = plt.figure(1, figsize=(9, 6))
# Create an axes instance
ax = fig.add_subplot(111)
# Create the boxplot
bp = ax.boxplot(data, patch_artist=True)
## change outline color, fill color and linewidth of the boxes
for box in bp['boxes']:
# change outline color
box.set( color='#7570b3', linewidth=2)
# change fill color
box.set( facecolor = '#AACCFF' )
## change color and linewidth of the whiskers
for whisker in bp['whiskers']:
whisker.set(color='#7570b3', linewidth=2)
## change color and linewidth of the caps
for cap in bp['caps']:
cap.set(color='#7570b3', linewidth=2)
## change color and linewidth of the medians
for median in bp['medians']:
median.set(color='#FFE680', linewidth=2)
## change the style of fliers and their fill
for flier in bp['fliers']:
flier.set(marker='o', color='#e7298a', alpha=0.5)
## Custom x-axis labels
ax.set_xticklabels(xticklabels)
ax.set_ylabel(ylabel)
plt.show()
fig.savefig(filename, bbox_inches='tight')
# + id="FcyiPw-8VHW2"
var_results = pd.read_csv("rolling_cv_wind_raw_var.csv")
evolving_results = pd.read_csv("rolling_cv_wind_raw_emvfts.csv")
mlp_results = pd.read_csv("rolling_cv_wind_raw_mlp_multi.csv")
granular_results = pd.read_csv("rolling_cv_wind_raw_granular.csv")
# + id="3OB8fPYW81R6" colab={"base_uri": "https://localhost:8080/", "height": 374} outputId="d91795ac-38af-4e5e-da91-b6d9a041864e"
metric = 'RMSE'
results_data = [evolving_results[metric],var_results[metric], mlp_results[metric], granular_results[metric]]
xticks = ['e-MVFTS','VAR','MLP','FIG-FTS']
ylab = 'RMSE'
createBoxplot("e-mvfts_boxplot_rmse_solar", results_data, xticks, ylab)
# + id="hUCDYHyMFwl3"
pd.options.display.float_format = '{:.2f}'.format
# + id="qACWp2l5_iJ4"
metric = 'RMSE'
rmse_df = pd.DataFrame(columns=['e-MVFTS','VAR','MLP','FIG-FTS'])
rmse_df["e-MVFTS"] = evolving_results[metric]
rmse_df["VAR"] = var_results[metric]
rmse_df["MLP"] = mlp_results[metric]
rmse_df["FIG-FTS"] = granular_results[metric]
# + id="ioz9IYOdAN2Z" colab={"base_uri": "https://localhost:8080/", "height": 102} outputId="0a34dd80-b6f1-4759-b387-beaa9b81ee3d"
rmse_df.std()
# + id="fqZ9gjtr-Eq2" colab={"base_uri": "https://localhost:8080/", "height": 376} outputId="d741e254-afd3-4b07-d4c5-114c5996c2a0"
metric = 'SMAPE'
results_data = [evolving_results[metric],var_results[metric], mlp_results[metric], granular_results[metric]]
xticks = ['e-MVFTS','VAR','MLP','FIG-FTS']
ylab = 'SMAPE'
createBoxplot("e-mvfts_boxplot_smape_solar", results_data, xticks, ylab)
# + id="0yajGeb7HPUG"
metric = 'SMAPE'
smape_df = pd.DataFrame(columns=['e-MVFTS','VAR','MLP','FIG-FTS'])
smape_df["e-MVFTS"] = evolving_results[metric]
smape_df["VAR"] = var_results[metric]
smape_df["MLP"] = mlp_results[metric]
smape_df["FIG-FTS"] = granular_results[metric]
# + id="466gaNDyHYSJ" colab={"base_uri": "https://localhost:8080/", "height": 102} outputId="b5868803-de90-4e96-9aac-36d3e7ac920d"
smape_df.std()
# + id="hkXC-k92VKoc" colab={"base_uri": "https://localhost:8080/", "height": 720} outputId="61c1e4a0-bb3d-47c3-c08a-8cff1effd918"
metric = "RMSE"
data = pd.DataFrame(columns=["VAR", "Evolving", "MLP", "Granular"])
data["VAR"] = var_results[metric]
data["Evolving"] = evolving_results[metric]
data["MLP"] = mlp_results[metric]
data["Granular"] = granular_results[metric]
ax = data.plot(figsize=(18,6))
ax.set(xlabel='Window', ylabel=metric)
fig = ax.get_figure()
#fig.savefig(path_images + exp_id + "_prequential.png")
x = np.arange(len(data.columns.values))
names = data.columns.values
values = data.mean().values
plt.figure(figsize=(5,6))
plt.bar(x, values, align='center', alpha=0.5, width=0.9)
plt.xticks(x, names)
#plt.yticks(np.arange(0, 1.1, 0.1))
plt.ylabel(metric)
#plt.savefig(path_images + exp_id + "_bars.png")
# + id="ghhKlmDwVOhr"
metric = "SMAPE"
data = pd.DataFrame(columns=["VAR", "Evolving", "MLP", "Granular"])
data["VAR"] = var_results[metric]
data["Evolving"] = evolving_results[metric]
data["MLP"] = mlp_results[metric]
data["Granular"] = granular_results[metric]
ax = data.plot(figsize=(18,6))
ax.set(xlabel='Window', ylabel=metric)
fig = ax.get_figure()
#fig.savefig(path_images + exp_id + "_prequential.png")
x = np.arange(len(data.columns.values))
names = data.columns.values
values = data.mean().values
plt.figure(figsize=(5,6))
plt.bar(x, values, align='center', alpha=0.5, width=0.9)
plt.xticks(x, names)
#plt.yticks(np.arange(0, 1.1, 0.1))
plt.ylabel(metric)
#plt.savefig(path_images + exp_id + "_bars.png")
| 39.042355
| 7,234
|
c7027a260a51639ee3e2bc904fd16a2705393175
|
py
|
python
|
WGAN_GP_FMNIST.ipynb
|
JungWoo-Chae/Generative_Models
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/JungWoo-Chae/Generative_Models/blob/main/WGAN_GP_FMNIST.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="1fEvAcwbfJvR"
# # **WGAN GP with FMNIST**
#
# + [markdown] id="181Mi0DSwREM"
# ## **Imports**
# + id="Nx2pe892jknQ"
import numpy as np
import matplotlib.pyplot as plt
import torch
import torch.nn as nn
import torch.nn.functional as F
import torchvision
from torch.autograd import Variable
from torch import autograd
# + id="pX4Yv853s9Sx"
device = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')
# + [markdown] id="ErZZIQ_zUjXv"
# ## **Model**
# + id="WJmq2i6Iq2PS"
class Generator(torch.nn.Module):
def __init__(self, channels, nf):
super().__init__()
self.main_module = nn.Sequential(
nn.ConvTranspose2d(in_channels=100, out_channels=nf*4, kernel_size=4, stride=1, padding=0),
nn.BatchNorm2d(num_features=nf*4),
nn.ReLU(True),
nn.ConvTranspose2d(in_channels=nf*4, out_channels=nf*2, kernel_size=4, stride=1, padding=0),
nn.BatchNorm2d(num_features=nf*2),
nn.ReLU(True),
nn.ConvTranspose2d(in_channels=nf*2, out_channels=nf, kernel_size=4, stride=2, padding=1),
nn.BatchNorm2d(num_features=nf),
nn.ReLU(True),
nn.ConvTranspose2d(in_channels=nf , out_channels=channels, kernel_size=4, stride=2, padding=1))
self.output = nn.Tanh()
def forward(self, x):
x = self.main_module(x)
return self.output(x)
# + id="Ng9MOjRgAt5-"
class Discriminator(torch.nn.Module):
def __init__(self, channels, nf):
super().__init__()
self.main_module = nn.Sequential(
nn.Conv2d(in_channels=channels, out_channels=nf, kernel_size=4, stride=2, padding=1),
nn.InstanceNorm2d(nf, affine=True),
nn.LeakyReLU(0.2, inplace=True),
nn.Conv2d(in_channels=nf, out_channels=nf*2, kernel_size=4, stride=2, padding=1),
nn.InstanceNorm2d(nf*2, affine=True),
nn.LeakyReLU(0.2, inplace=True),
nn.Conv2d(in_channels=nf*2, out_channels=nf*4, kernel_size=4, stride=2, padding=1),
nn.InstanceNorm2d(nf*4, affine=True),
nn.LeakyReLU(0.2, inplace=True))
self.output = nn.Sequential(
nn.Conv2d(in_channels=nf*4, out_channels=1, kernel_size=3, stride=1, padding=0)
)
def forward(self, x):
x = self.main_module(x)
return self.output(x)
# + id="0Fw8FrBfR6Vr"
z_dim = 100
channels = 1
nf = 64
# + id="rPscO_2k2eEj"
D = Discriminator(channels, nf)
D = D.to(device)
G = Generator(channels, nf)
G = G.to(device)
# + id="8ZuJmBW_JuFD"
transform = torchvision.transforms.Compose([
torchvision.transforms.ToTensor(),
torchvision.transforms.Normalize((0.5), (0.5)),
])
# + id="dSK9-8o_v1S-" outputId="ef0a9db7-c34d-4f25-be0e-dd8068e87c00" colab={"base_uri": "https://localhost:8080/", "height": 417, "referenced_widgets": ["69acc9c624ef497e99cf639bcc41f1f5", "711b641ee736497bb318a19b62852839", "fde573e2b429493ea6c89271f946dfe3", "f68e9c825e224f0aa966e6806f7819e5", "e8b478e9c1c24a9fb85b1800aab56094", "a26357de97ca44f1ac00c455c2255b3c", "5b2bfe8f258b4d81acc2a2090f81c6d3", "6842359e9b8740a8a1060636acf8c63e", "c5f937d0d8ce4da6bb3246a6fd06ce54", "6c28b9cc1f834262bf8a2ae17470c1bf", "c9498726372e4728b244bb5763a6732d", "abaff5fb2e284c65b902000ce139c465", "adb8a75544e94a968ac66108309eafbd", "9d7effd75f454b45900c071b7ac07be3", "bd7c627e4fa841869c3d87eda1a95f95", "93f27d6905bf4b059f7ea2d4864bfe05", "1c56cbe4e6a04df68fcfe25e2a3b9264", "ba1b9e8b62524370b580a758a532d4cf", "7336d4874d5b4b57a10ac46798ebe00a", "e8d415831c95437bb823b1366269041c", "594a8b4c7b754ecf8c0d8db01c59fda2", "fd9553347f0c46edab4268d21ef35284", "0e4ee69b27544aff97a87e784fe618cf", "2bbabc45c813468eab3e57e89ed22e34", "45514254d29646718f2404720d7bd97f", "7420ee9e301844eda82776a99ebdd6bb", "89a81d473eac4c0e85d731f82d828212", "eb021ccf1139410499ee3ffde94eb4e1", "0079a54451f4450dbcb82e94333f4a01", "2e79286f97314597ac8cd40c32bc3fdf", "e0b73166b7de4bb7af96f336dd772f58", "e336e4a121c74f348a93fa8081a5594e"]}
dataset = torchvision.datasets.FashionMNIST(root='./data', train=True,
download=True, transform=transform)
# + [markdown] id="efo2XO6vziLs"
# # Training
# + id="SlDUNudeausp"
losses = []
checkpoints = []
# + id="gmjrprUfeK52"
def compute_gradient_penalty(D, real_samples, fake_samples):
alpha = torch.randn(real_samples.size(0), 1, 1, 1, device=device)
interpolates = (alpha * real_samples + ((1 - alpha) * fake_samples)).requires_grad_(True)
d_interpolates = D(interpolates)
fake = torch.ones(d_interpolates.size()).to(device).requires_grad_(False)
gradients = autograd.grad(
outputs=d_interpolates,
inputs=interpolates,
grad_outputs=fake,
create_graph=True,
retain_graph=True,
only_inputs=True,
)[0]
gradients = gradients.view(gradients.size(0), -1)
gradient_penalty = ((gradients.norm(2, dim=1) - 1) ** 2).mean()
return gradient_penalty
# + id="7des2_Ll7aFq"
losses = []
checkpoints = []
def train(epochs, batch_size, sample_interval):
D.train()
G.train()
train_loader = torch.utils.data.DataLoader(dataset, batch_size=batch_size,
shuffle=True, num_workers=4, pin_memory=True)
d_optimizer = torch.optim.Adam(D.parameters(), lr=0.0002, betas=(0.5, 0.999))
g_optimizer = torch.optim.Adam(G.parameters(), lr=0.0002, betas=(0.5, 0.999))
for epoch in range(epochs):
iteration = 0
for imgs, _ in train_loader:
imgs = imgs.to(device)
if len(imgs) != batch_size:
break
for p in D.parameters():
p.requires_grad = True
D.zero_grad()
z = torch.randn(batch_size, z_dim,1,1, device=device)
gen_imgs = G(z)
gradient_penalty = compute_gradient_penalty(D, imgs.data, gen_imgs.data)
d_loss = -1 * torch.mean(D(imgs)) + torch.mean(D(gen_imgs)) + 10 * gradient_penalty
d_loss.backward()
d_optimizer.step()
if iteration%5==0:
for p in D.parameters():
p.requires_grad = False
G.zero_grad()
z = torch.randn(batch_size, z_dim,1,1, device=device)
gen_imgs = G(z)
g_loss = - torch.mean(D(gen_imgs))
g_loss.backward()
g_optimizer.step()
iteration +=1
print("%d/%d [D loss: %f] [G loss: %f]" % (epoch + 1, epochs, d_loss.item(), g_loss.item()))
if (epoch+1) % sample_interval == 0:
losses.append((d_loss.cpu().detach().numpy(), g_loss.cpu().detach().numpy()))
checkpoints.append(epoch + 1)
sample_images(G)
# + id="k8lTQYfI2sZO"
def sample_images(generator, t=3, image_grid_rows=2, image_grid_columns=5):
z = torch.randn(image_grid_rows * image_grid_columns, z_dim, 1, 1, device=device)
labels = np.arange(0, 10)
labels = torch.tensor(labels).to(device)
gen_imgs = generator(z).cpu().detach()
gen_imgs = 0.5 * gen_imgs + 0.5
fig, axs = plt.subplots(image_grid_rows,
image_grid_columns,
figsize=(10, 4),
sharey=True,
sharex=True)
cnt = 0
for i in range(image_grid_rows):
for j in range(image_grid_columns):
axs[i,j].imshow(gen_imgs[cnt].view(28,28), cmap='gray')
axs[i, j].axis('off')
cnt += 1
# + id="9whoTeMn6Ol_" outputId="c334a910-c175-4ccf-9782-e69e21b8cf76" colab={"base_uri": "https://localhost:8080/", "height": 1000}
epochs = 30
batch_size = 1024
sample_interval = 5
train(epochs, batch_size, sample_interval)
# + id="mcQYpxZLcAmg" outputId="76eca66b-b802-4fb7-c5fb-c231deb4d9c9" colab={"base_uri": "https://localhost:8080/", "height": 368}
losses_ = np.array(losses)
plt.figure(figsize=(15, 5))
plt.plot(checkpoints, losses_.T[0], label="Discriminator loss")
plt.plot(checkpoints, losses_.T[1], label="Generator loss")
plt.title("Training Loss")
plt.xlabel("epoch")
plt.ylabel("Loss")
plt.legend()
| 34.75
| 1,306
|
a608981bfb49a6886a30ff79124e4698773b1e44
|
py
|
python
|
3_nlp/classification/text_classification sent_transformer.ipynb
|
DukeAIPI/AIPI540-Deep-Learning-Applications
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="8glhZbJYm44L"
# # Text Classification using Sentence Transformers
# In this notebook we will be doing text classification using document embeddings obtained using a pre-trained [Sentence Transformer](https://www.sbert.net) model. SentenceTransformers is a framework for sentence / text embeddings which works particularly well for shorter text. It was developed in 2019 and uses Siamese-BERT to develop semantically meaningful sentence embeddings which can be compared using cosine similarity. You can use a [pretrained embedding model](https://www.sbert.net/docs/pretrained_models.html) or can train your own on a corpus.
#
# Our goal will be to classify the articles in the AgNews dataset into their correct category: "World", "Sports", "Business", or "Sci/Tec".
#
# **Notes:**
# - This must be run using GPU acceleration
#
# **References:**
# - Read the [Sentence-BERT paper](https://arxiv.org/abs/1908.10084) by Reimers & Gurevych
# + colab={"base_uri": "https://localhost:8080/"} id="gM37lju3m44O" outputId="aaebdb59-e2d1-48b5-9527-e55216268f83"
import os
import numpy as np
import pandas as pd
import string
import time
import urllib.request
import zipfile
import torch
from sklearn.linear_model import LogisticRegression
# #!pip install sentence_transformers
from sentence_transformers import SentenceTransformer
import warnings
warnings.filterwarnings('ignore')
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# + [markdown] id="KmcwQwFIm44P"
# ## Download and prepare data
# + colab={"base_uri": "https://localhost:8080/", "height": 204} id="rPF1ISIMm44Q" outputId="6d0fb779-6fca-42f7-d6da-641f84f17c78"
# Download the data
if not os.path.exists('../data'):
os.mkdir('../data')
if not os.path.exists('../data/agnews'):
url = 'https://storage.googleapis.com/aipi540-datasets/agnews.zip'
urllib.request.urlretrieve(url,filename='../data/agnews.zip')
zip_ref = zipfile.ZipFile('../data/agnews.zip', 'r')
zip_ref.extractall('../data/agnews')
zip_ref.close()
train_df = pd.read_csv('../data/agnews/train.csv')
test_df = pd.read_csv('../data/agnews/test.csv')
# Combine title and description of article to use as input documents for model
train_df['full_text'] = train_df.apply(lambda x: ' '.join([x['Title'],x['Description']]),axis=1)
test_df['full_text'] = test_df.apply(lambda x: ' '.join([x['Title'],x['Description']]),axis=1)
# Create dictionary to store mapping of labels
ag_news_label = {1: "World",
2: "Sports",
3: "Business",
4: "Sci/Tec"}
train_df.head()
# + colab={"base_uri": "https://localhost:8080/"} id="xRb3dY56m44S" outputId="58b921fd-1545-4798-e88b-7a217540c081"
# View a couple of the documents
for i in range(5):
print(train_df.iloc[i]['full_text'])
print()
# + [markdown] id="r7mmGw3Gm44T"
# ## Create document embeddings
# We will load a pre-trained model [('all-MiniLM-L6-v2')](https://huggingface.co/sentence-transformers/all-MiniLM-L6-v2) which we will then use to create embeddings for our training and test set text. The MiniLM-L6-v2 model was trained on 1.1 billion sentence pairs to produce high-quality sentence / short document embeddings in 384 dimensions which can be used for example to calculate similarity between documents.
# + colab={"base_uri": "https://localhost:8080/", "height": 465, "referenced_widgets": ["d2799a32f0bb4979b5a8cc14579f301a", "273f2d709b6f4d64b1b9f6e5c61f927e", "3bfc61af763447df88b0b2998d0d004a", "10dbb52297ae4c41b2847a699410280b", "606ec11713cd48349c100ca996c8f31e", "37d1279f10b44952832f23cabd0fdee3", "5c0b276823de45ffab0ac52fad2989dd", "2454552c15854fa995adbc9adc9c2214", "f66c021a18b849cc945f4ab6ed17c52e", "7208483505a54f749baf755d6a0a2735", "7c6599ec06064fe482fce478bf633348", "ee0b782f0bbf437087788461959422a0", "80ea8bf9a976453c92f3aac6738a8fea", "19f01b2754ea46f0aac95dc567375df6", "41a029351eae4b23a9f9fa3b0573aef4", "9925cc02ae384e76906b20e973586d1a", "14ca492fd95141759380d796fa69af55", "1cd9944c37124c60a2054ad8a36aba33", "84ec01e8575d40dbae59bc5d5fcb7311", "dd30bf2447d348a5a4eac9069a01d679", "2074588f772944398b12a99bf21a2762", "f145bafad2ac43ac9bbc48b063328d81", "de7cb00fe9024e1ca773fd25bcb2e878", "3be500ed846a4ee6b955fe4f6a656507", "09acef5b55964cf6af4047dacbc84871", "ed43507285ac46409fa2f6b374423830", "87028292430c4d5ea4ad1c55ad8b0e19", "0f6654f3af9d436fb11dbfa12d2e1749", "7cd034a0f99d4cb6a8527bfca7d617ef", "1ae6fcc8bae845e78ac79065b99ba102", "917c490805ed4e6793e42eb1b7abf4f3", "d2f2a25e7f344259b019ccbf13c8ea8e", "e4fd43ea52aa494187842f7604d3c912", "e86178184aa94c8081152290b5fc299f", "ea1deb2f53084124a2d240cf415387fc", "f060288b6d764cfb8a5a19388abb13fb", "f559457b6d074b06ac193e613f3755af", "75caed7e3fe845029a38c701cc6240d5", "e29d4ed403d84f8fa7a3ff4f4abf3bab", "882204d755d84f9a97c56af869c4cdd0", "590599e8c0e54fa9a2d5eb26f59ebceb", "f091172ca80d4e7d9ce01192d7c6a018", "c96c31ebd1f94c42810e7cfcc98b9987", "8d15981dc3304399a28027f886fb5647", "d5b25e261acc4fca9933b2b85c779aed", "a1c41b8e6c434aa6976099fdf64a2e25", "5e6493d494ee436999f85edb594abcd2", "6d25561ea197467a9928ce42eee66f3a", "d60c2e62fe644141b7be1fbf09264676", "9291c4476d844acca06e562200d38c09", "d0316f1e6017473ba981ee0757f6f2a3", "fa804523d6c04b429f1c2d781c2af706", "153bb46d92794605a066376b0ce04cea", "b774d13697cc4ad3bdf75548fb775df2", "292c72f1a0bb460da9a63f07e74abb0c", "6b668475a3cf4161b9569f28eb93f4c3", "d190bae155964a02ad130c43b0f20bb5", "f1f1bb3db87d4f36a0862410379b4862", "025b5c352e9f444a9fe5b780c9913478", "9beb500b36634e91b1df040308a08b39", "412fceec18714affad7862c754b0e3db", "19e228afef4a41c2adf813b2080b99ce", "69a1731124524a30a0dbbc9698551310", "c8fa28a55c794c1fb6c6da425cf36453", "00552fa4e3f84fc9910ebe65c1cb27bf", "67d34001e73949028d265ee98615b918", "0593a83d01fa40808a78635393e88515", "aabe03787de340caaa605912e26cd4b6", "bd1d220f9c5548f8b5255cb1df81764b", "598f08bfc0a14448ba102a5944a186a2", "566f9c1d331046199baddc36ee4a3976", "49e6bfb30ce84233a78cba6fd768b861", "6bf2f00f9a8a4719810aff657ebee0cb", "854b6beaf0a04a4a8c7aee1f4b13549e", "073cf81da1384b389f0d629a00eb7eee", "bad37717f8b64f2d8e923c2c4146f79b", "b0c57bbfa3534aa38db21ccf002a1de7", "d7d9f64cfb3d4b91a55eaa97cf012f66", "e607595e440b45259674673de85fcc93", "4566c75932c845009a1d4fd43e7f27f5", "4e6d6b8a80e747529b8eceb75deb283e", "6729a2d1cc4f4d64aa192efc7532ba87", "d91af57bbf5441b482ceae72a312a8e3", "91604b4153554ff581e8e43512e281eb", "db1282f2c8e44f81a3d9c78e82295634", "75f3757256ea4ff9aa689cc29a4a5c6a", "4603687b148246a28b4676ad91ec1109", "a359ddb4f89f498fb76844d70217917a", "fa723f384ad5458d99a07b51fe773c93", "f955d945911c4160b5f8e1e6b49f6f17", "291fac885524499eaf7669d0c7ec0488", "1d93cca370014675b2e9fcee7281e105", "7d7937310c364f9a9c2155e1aecad0c9", "cf72e61a1bc144b89dcafd47faa0afd3", "b61963c619f94ba8b838bf4f1a17fd74", "ee3608c6783d4fb8a2d14c14f24b98a2", "7a93b72553e643ae9dd4a3a30297ea40", "a729db8c481d4595aa44f217470c15fe", "ce396640fa4b4cf6b529c20295aebfca", "d7b73c4d327d4cb19dbb5d8691c338d6", "aec13e9eaaf542eb825e4050bd6864ae", "846b0d2c90044046bfdb22999aaf0ed4", "de7aaa23198d4b50ab54a5bc204668fc", "02b14a24405e4e929d19d30ec6946729", "86e7bbb2afb74b0ba7ed8a3be90d86d1", "738a1db2ceec47d9ad07ba86bd0f9d25", "fa609ad583d9481ea88b98bb7dc1005c", "2ea09ee1cec84ad4a01a9b120ecc668a", "924d0612193f411d929d5b41f9be6571", "8a0f9225134a48bcaa9ca73679d2dec8", "668fa4c7dc734d4fa894cd970ced5e02", "77e92ad2d70d499b87288b8155a24a3f", "b173052022c84deebafadb43e8853c22", "7ff4540900354c90ad107485c82cb845", "ceac76a1d03f44d19f238fa530dbb21b", "be2859c0d4ee4e3583552e58d7bc2077", "f9af745470424f2db6c82dfeec9fac51", "f23cb54c359d4f7bab1c3b9a637a271c", "80591907881e45849a5b7f4e2492d0b1", "ce853398e73f495ba4290a07038bdae5", "3499459c936f4b64a32c4f2204bdc564", "1d5781ad73bd4812afb7db7b4e0b3d28", "bbbe866b2de44786bb75ce0e1e61449e", "39b220429cb3403386787e77e313aaa9", "e399b2ebd4d842a1b104dcffab291298", "d5c5fe469ca14bd59d68cb04c7702315", "a5fdd82900664fec890688ed827de75d", "a0e14b978c164ed697abdc14ddaec197", "74b49502c02c408f986aa812d9675b1d", "158d7b03dbf244a9918094e9de01229f", "0bc6fb823c1c42b8bceef2b5343ea802", "c29514395a5d4ce8953b7825d3eda0e6", "1e6d912ebe0f4c58b5ae91648ff9a00a", "b3fa353a926c44479cf794745e21cac5", "e9b8b1c619124e13b3e12054b6ec32d4", "04afd41023b4446c976d45d2f385c1c1", "ecb652ff469b47d0b370671eb78b4b69", "32c7b55f6ec84bf3a08df05575e4c476", "df9cf26cf9324ceeb8870915193e843d", "55210bba7dd64cbdbddadb1e26e82a2b", "c66f93860294474692b22bb4aa1ff21d", "a26285fdd17e4fcaaf206f12278fefb0", "4a760fa8103b4ac097416857a141cfac", "f731b13928224706830407d546434497", "cc3239e865f34432b4cd525cc85e06a5", "5be10a258b8948679acced67ef3e4d45", "82d78aae04b1494c8a19888c64060e5f", "126608cb106f4a0f97cb14d68735903d", "868ce03db2624c2585f30d49c984ac5c", "5f43605e598f4ce29c07bb203481259e", "6ceaf8e7189843a0b6c5a90c1c083a2e", "1cf106f2f81c4e8abf765e82b6ba77e6", "7d64420a637e4366bcea4c9bdb8f27f8", "f5647afd297242cbb27ead8182414c39"]} id="KXjJ9WuNm44U" outputId="0bf9a5bc-8978-40d4-cb38-e43e25ad437b"
# Load pre-trained model
senttrans_model = SentenceTransformer('all-MiniLM-L6-v2',device=device)
# Create embeddings for training set text
X_train = train_df['full_text'].values.tolist()
X_train = [senttrans_model.encode(doc) for doc in X_train]
# Create embeddings for test set text
X_test = test_df['full_text'].values.tolist()
X_test = [senttrans_model.encode(doc) for doc in X_test]
# + [markdown] id="mXvjOrrWm44V"
# ## Train classification model
# Finally, we will used our embeddings as features to train a softmax regression model to classify the documents.
# + colab={"base_uri": "https://localhost:8080/"} id="z4h9Pkzxm44W" outputId="d8289431-5db5-441e-addc-70b3322f31e2"
# Train a classification model using logistic regression classifier
y_train = train_df['Class Index']
logreg_model = LogisticRegression(solver='saga')
logreg_model.fit(X_train,y_train)
preds = logreg_model.predict(X_train)
acc = sum(preds==y_train)/len(y_train)
print('Accuracy on the training set is {:.3f}'.format(acc))
# + [markdown] id="i2dlajjum44W"
# ## Evaluate model performance
# + colab={"base_uri": "https://localhost:8080/"} id="DLA5ybaHm44X" outputId="a370b8c1-8064-489c-eb93-74e9ac043893"
# Evaluate performance on the test set
y_test = test_df['Class Index']
preds = logreg_model.predict(X_test)
acc = sum(preds==y_test)/len(y_test)
print('Accuracy on the test set is {:.3f}'.format(acc))
| 89.389831
| 5,698
|
2d6031d6856a34c7395c34ac236d2e7c126b89cf
|
py
|
python
|
examples/workshops/FOSS4G_2021.ipynb
|
Piphi5/leafmap
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# [](https://gishub.org/foss4g-colab)
# [](https://gishub.org/foss4g-binder)
# [](https://gishub.org/foss4g-binder-nb)
# 
# **Using Leafmap for Geospatial Analysis and Data Visualization**
#
#
# This notebook was developed for the [leafmap workshop](https://callforpapers.2021.foss4g.org/foss4g-2021-workshop/talk/VAHX9A/) taking place on September 27, 2021 at the [FOSS4G 2021 Conference](https://2021.foss4g.org/).
#
# Author: [Qiusheng Wu](https://github.com/giswqs)
#
# Launch this notebook to execute code interactively using:
# - Google Colab: https://gishub.org/foss4g-colab
# - Pangeo Binder JupyterLab: https://gishub.org/foss4g-binder
# - Pangeo Binder Jupyter Notebook: https://gishub.org/foss4g-binder-nb
#
#
# ## Introduction
#
# ### Workshop description
#
# [Leafmap](https://leafmap.org) is a Python package for interactive mapping and geospatial analysis with minimal coding in a Jupyter environment. It is built upon a number of open-source packages, such as [folium](https://github.com/python-visualization/folium) and [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) (for creating interactive maps), [WhiteboxTools](https://github.com/jblindsay/whitebox-tools) and [whiteboxgui](https://github.com/giswqs/whiteboxgui) (for analyzing geospatial data), and [ipywidgets](https://github.com/jupyter-widgets/ipywidgets) (for designing interactive graphical user interface). The WhiteboxTools library currently contains 480+ tools for advanced geospatial analysis. Leafmap provides many convenient functions for loading and visualizing geospatial data with only one line of code. Users can also use the interactive user interface to load geospatial data without coding. Anyone with a web browser and Internet connection can use leafmap to perform geospatial analysis and data visualization in the cloud with minimal coding. The topics that will be covered in this workshop include:
#
# 1. Creating interactive maps
# 2. Changing basemaps
# 3. Loading and visualizing vector/raster data
# 4. Using Cloud Optimized GeoTIFF (COG) and SpatialTemporal Asset Catalog (STAC)
# 5. Downloading OpenStreetMap data
# 6. Loading data from a PostGIS database
# 7. Creating custom legends and colorbars
# 8. Creating split-panel maps and linked maps
# 9. Visualizing Planet global monthly/quarterly mosaic
# 10. Designing and publishing interactive web apps
# 11. Performing geospatial analysis (e.g., hydrological analysis and LiDAR data analysis) using whiteboxgui.
#
# This workshop is intended for scientific programmers, data scientists, geospatial analysts, and concerned citizens of Earth. The attendees are expected to have a basic understanding of Python and the Jupyter ecosystem. Familiarity with Earth science and geospatial datasets is useful but not required. More information about leafmap can be found at https://leafmap.org
#
#
# ### Jupyter keyboard shortcuts
#
# - Shift+Enter: run cell, select below
# - Ctrl+Enter: : run selected cells
# - Alt+Enter: run cell and insert below
# - Tab: code completion or indent
# - Shift+Tab: tooltip
# - Ctrl+/: comment out code
# ## Set up environment
#
# ### Required Python packages:
# * [leafmap](https://github.com/giswqs/leafmap) - A Python package for interactive mapping and geospatial analysis with minimal coding in a Jupyter environment
# * [geopandas](https://geopandas.org) - An open source project to make working with geospatial data in python easier.
# * [keplergl](https://docs.kepler.gl/docs/keplergl-jupyter) - A high-performance web-based application for visual exploration of large-scale geolocation data sets
# * [datapane](https://datapane.com) - A Python library for building interactive reports in seconds
# * [xarray-leaflet](https://github.com/davidbrochart/xarray_leaflet) - An xarray extension for tiled map plotting
#
# ### Required API keys
# - [HERE Map](https://developer.here.com) API key
# - [datapane](https://datapane.com) API key
# - [Planet](https://www.planet.com/nicfi) API key
#
# ### Use Google Colab
#
# Click the button below to open this notebook in Google Colab and execute code interactively.
#
# [](https://githubtocolab.com/giswqs/leafmap/blob/master/examples/workshops/foss4g_2021.ipynb)
import os
import subprocess
import sys
# +
import warnings
warnings.filterwarnings("ignore")
# -
# A function for installing Python packages.
def install(package):
subprocess.check_call([sys.executable, "-m", "pip", "install", package])
# Install required Python packages in Google Colab.
pkgs = [
'leafmap',
'geopandas',
'keplergl',
'datapane',
'xarray_leaflet',
'osmnx',
'pygeos',
'imageio',
'tifffile',
]
if "google.colab" in sys.modules:
for pkg in pkgs:
install(pkg)
# ### Use Pangeo Binder
#
# Click the buttons below to open this notebook in JupyterLab (first button) or Jupyter Notebook (second button) and execute code interactively.
#
# [](https://gishub.org/foss4g-binder)
# [](https://gishub.org/foss4g-binder-nb)
#
# - JupyterLab: https://gishub.org/foss4g-binder
# - Jupyter Notebook: https://gishub.org/foss4g-binder-nb
# ### Use Miniconda/Anaconda
#
# If you have
# [Anaconda](https://www.anaconda.com/distribution/#download-section) or [Miniconda](https://docs.conda.io/en/latest/miniconda.html) installed on your computer, you can install leafmap using the following commands. Leafmap has an optional dependency - [geopandas](https://geopandas.org), which can be challenging to install on some computers, especially Windows. It is highly recommended that you create a fresh conda environment to install geopandas and leafmap. Follow the commands below to set up a conda env and install geopandas, leafmap, datapane, keplergl, and xarray_leaflet.
#
# ```
# conda create -n geo python=3.8
# conda activate geo
# conda install geopandas
# conda install mamba -c conda-forge
# mamba install leafmap datapane keplergl xarray_leaflet -c conda-forge
# mamba install osmnx pygeos imageio tifffile -c conda-forge
# pip install -U folium
# jupyter lab
# ```
try:
import leafmap
except ImportError:
install('leafmap')
# ## Create an interactive map
#
# `leafmap` has four plotting backends: [folium](https://github.com/python-visualization/folium), [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet), [here-map](https://github.com/heremaps/here-map-widget-for-jupyter), and [kepler.gl](https://docs.kepler.gl/docs/keplergl-jupyter). Note that the backends do not offer equal functionality. Some interactive functionality in `ipyleaflet` might not be available in other plotting backends. To use a specific plotting backend, use one of the following:
#
# - `import leafmap.leafmap as leafmap`
# - `import leafmap.foliumap as leafmap`
# - `import leafmap.heremap as leafmap`
# - `import leafmap.kepler as leafmap`
#
#
# ### Use ipyleaflet
# +
import leafmap
m = leafmap.Map()
m
# -
# ### Use folium
# +
import leafmap.foliumap as leafmap
m = leafmap.Map()
m
# -
# ### Use kepler.gl
# +
import leafmap.kepler as leafmap
m = leafmap.Map()
m
# -
# If you encounter an error saying `Error displaying widget: model not found` when trying to display the map, you can use `m.static_map()` as a workaround until this [kepler.gl bug](https://github.com/keplergl/kepler.gl/issues/1165) has been resolved.
#
m.static_map(width=1280, height=600)
# ### Use here-map
#
# Before you run the below cells make sure you have:
# - A HERE developer account, free and available under [HERE Developer Portal](https://developer.here.com)
# - An [API key](https://developer.here.com/documentation/identity-access-management/dev_guide/topics/dev-apikey.html) from the [HERE Developer Portal](https://developer.here.com)
#
import os
import leafmap.heremap as leafmap
# +
if os.environ.get("HEREMAPS_API_KEY") is None:
os.environ["HEREMAPS_API_KEY"] = 'your-api-key'
m = leafmap.Map()
m
# -
# ## Customize the default map
#
# ### Specify map center and zoom level
import leafmap
m = leafmap.Map(center=(40, -100), zoom=4) # center=[lat, lon]
m
m = leafmap.Map(center=(51.5, -0.15), zoom=17)
m
# ### Change map size
m = leafmap.Map(height="400px", width="800px")
m
# ### Set control visibility
#
# When creating a map, set the following controls to either `True` or `False` as appropriate.
#
# * attribution_control
# * draw_control
# * fullscreen_control
# * layers_control
# * measure_control
# * scale_control
# * toolbar_control
m = leafmap.Map(
draw_control=False,
measure_control=False,
fullscreen_control=False,
attribution_control=False,
)
m
# Remove all controls from the map.
m = leafmap.Map()
m.clear_controls()
m
# ## Change basemaps
#
# Specify a Google basemap to use, can be one of ["ROADMAP", "TERRAIN", "SATELLITE", "HYBRID"].
import leafmap
m = leafmap.Map(google_map="TERRAIN") # HYBIRD, ROADMAP, SATELLITE, TERRAIN
m
# Add a basemap using the `add_basemap()` function.
m = leafmap.Map()
m.add_basemap("Esri.NatGeoWorldMap")
m
# Print out the list of available basemaps.
for basemap in leafmap.leafmap_basemaps:
print(basemap)
# ## Add tile layers
#
# ### Add XYZ tile layer
import leafmap
m = leafmap.Map()
m.add_tile_layer(
url="https://mt1.google.com/vt/lyrs=y&x={x}&y={y}&z={z}",
name="Google Satellite",
attribution="Google",
)
m
# ### Add WMS tile layer
#
# More WMS basemaps can be found at the following websites:
#
# - USGS National Map: https://viewer.nationalmap.gov/services
# - MRLC NLCD Land Cover data: https://www.mrlc.gov/data-services-page
# - FWS NWI Wetlands data: https://www.fws.gov/wetlands/Data/Web-Map-Services.html
m = leafmap.Map()
naip_url = 'https://services.nationalmap.gov/arcgis/services/USGSNAIPImagery/ImageServer/WMSServer?'
m.add_wms_layer(
url=naip_url, layers='0', name='NAIP Imagery', format='image/png', shown=True
)
m
# ### Add xyzservices provider
#
# Add a layer from [xyzservices](https://github.com/geopandas/xyzservices) provider object.
import os
import xyzservices.providers as xyz
basemap = xyz.HEREv3.basicMap
basemap
basemap['apiKey'] = os.environ["HEREMAPS_API_KEY"]
m = leafmap.Map()
m.add_basemap(basemap)
m
# ## Add vector tile layer
import leafmap
m = leafmap.Map()
# The URL to the vector tile.
url = 'https://tile.nextzen.org/tilezen/vector/v1/512/all/{z}/{x}/{y}.mvt?api_key=gCZXZglvRQa6sB2z7JzL1w'
# Attribution of the vector tile.
attribution = "Nextzen"
# One can customize the vector tile layer style if needed. More info can be found at https://ipyleaflet.readthedocs.io/en/latest/api_reference/vector_tile.html
vector_tile_layer_styles = {}
# Add the vector tile layer to the map.
m.add_vector_tile_layer(url, attribution, vector_tile_layer_styles)
m
# ## Add COG/STAC layers
#
# A Cloud Optimized GeoTIFF (COG) is a regular GeoTIFF file, aimed at being hosted on a HTTP file server, with an internal organization that enables more efficient workflows on the cloud. It does this by leveraging the ability of clients issuing HTTP GET range requests to ask for just the parts of a file they need.
#
# More information about COG can be found at <https://www.cogeo.org/in-depth.html>
#
# Some publicly available Cloud Optimized GeoTIFFs:
#
# * https://stacindex.org/
# * https://cloud.google.com/storage/docs/public-datasets/landsat
# * https://www.digitalglobe.com/ecosystem/open-data
# * https://earthexplorer.usgs.gov/
#
# For this demo, we will use data from https://www.maxar.com/open-data/california-colorado-fires for mapping California and Colorado fires. A list of COGs can be found [here](https://github.com/giswqs/leafmap/blob/master/examples/data/cog_files.txt).
#
# ### Add COG layer
import leafmap
# +
m = leafmap.Map()
url = 'https://opendata.digitalglobe.com/events/california-fire-2020/pre-event/2018-02-16/pine-gulch-fire20/1030010076004E00.tif'
url2 = 'https://opendata.digitalglobe.com/events/california-fire-2020/post-event/2020-08-14/pine-gulch-fire20/10300100AAC8DD00.tif'
m.add_cog_layer(url, name="Fire (pre-event)")
m.add_cog_layer(url2, name="Fire (post-event)")
m
# -
# Retrieve the bounding box coordinates of the COG file.
leafmap.cog_bounds(url)
# Retrieve the centroid coordinates of the COG file.
leafmap.cog_center(url)
# Retrieves the tile layer URL of the COG file.
leafmap.cog_tile(url)
# ### Add STAC layer
#
# The SpatioTemporal Asset Catalog (STAC) specification provides a common language to describe a range of geospatial information, so it can more easily be indexed and discovered. A 'spatiotemporal asset' is any file that represents information about the earth captured in a certain space and time. The initial focus is primarily remotely-sensed imagery (from satellites, but also planes, drones, balloons, etc), but the core is designed to be extensible to SAR, full motion video, point clouds, hyperspectral, LiDAR and derived data like NDVI, Digital Elevation Models, mosaics, etc. More information about STAC can be found at https://stacspec.org/
#
# Some publicly available SpatioTemporal Asset Catalog (STAC):
#
# * https://stacindex.org
#
# For this demo, we will use STAC assets from https://stacindex.org/catalogs/spot-orthoimages-canada-2005#/?t=catalogs
m = leafmap.Map()
url = 'https://canada-spot-ortho.s3.amazonaws.com/canada_spot_orthoimages/canada_spot5_orthoimages/S5_2007/S5_11055_6057_20070622/S5_11055_6057_20070622.json'
m.add_stac_layer(url, bands=['B3', 'B2', 'B1'], name='False color')
m
# Retrieve the bounding box coordinates of the STAC file.
leafmap.stac_bounds(url)
# Retrieve the centroid coordinates of the STAC file.
leafmap.stac_center(url)
# Retrieve the band names of the STAC file.
leafmap.stac_bands(url)
# Retrieve the tile layer URL of the STAC file.
leafmap.stac_tile(url, bands=['B3', 'B2', 'B1'])
# ## Add local raster datasets
#
# The `add_raster` function relies on the `xarray_leaflet` package and is only available for the ipyleaflet plotting backend. Therefore, Google Colab is not supported. Note that `xarray_leaflet` does not work properly on Windows ([source](https://github.com/davidbrochart/xarray_leaflet/issues/30)).
import os
import leafmap
# Download samples raster datasets
#
# More datasets can be downloaded from https://viewer.nationalmap.gov/basic/
# +
out_dir = os.getcwd()
landsat = os.path.join(out_dir, 'landsat.tif')
dem = os.path.join(out_dir, 'dem.tif')
# -
# Download a small Landsat imagery.
if not os.path.exists(landsat):
landsat_url = 'https://drive.google.com/file/d/1EV38RjNxdwEozjc9m0FcO3LFgAoAX1Uw/view?usp=sharing'
leafmap.download_from_gdrive(landsat_url, 'landsat.tif', out_dir, unzip=False)
# Download a small DEM dataset.
if not os.path.exists(dem):
dem_url = 'https://drive.google.com/file/d/1vRkAWQYsLWCi6vcTMk8vLxoXMFbdMFn8/view?usp=sharing'
leafmap.download_from_gdrive(dem_url, 'dem.tif', out_dir, unzip=False)
m = leafmap.Map()
# Add local raster datasets to the map
#
# More colormap can be found at https://matplotlib.org/3.1.0/tutorials/colors/colormaps.html
m.add_raster(dem, colormap='terrain', layer_name='DEM')
m.add_raster(landsat, bands=[5, 4, 3], layer_name='Landsat')
m
# ## Add legend
#
# ### Add built-in legend
import leafmap
# List all available built-in legends.
legends = leafmap.builtin_legends
for legend in legends:
print(legend)
# Add a WMS layer and built-in legend to the map.
m = leafmap.Map()
url = "https://www.mrlc.gov/geoserver/mrlc_display/NLCD_2019_Land_Cover_L48/wms?"
m.add_wms_layer(
url,
layers="NLCD_2019_Land_Cover_L48",
name="NLCD 2019 CONUS Land Cover",
format="image/png",
transparent=True,
)
m.add_legend(builtin_legend='NLCD')
m
# Add U.S. National Wetlands Inventory (NWI). More info at https://www.fws.gov/wetlands.
# +
m = leafmap.Map(google_map="HYBRID")
url1 = "https://www.fws.gov/wetlands/arcgis/services/Wetlands/MapServer/WMSServer?"
m.add_wms_layer(
url1, layers="1", format='image/png', transparent=True, name="NWI Wetlands Vector"
)
url2 = "https://www.fws.gov/wetlands/arcgis/services/Wetlands_Raster/ImageServer/WMSServer?"
m.add_wms_layer(
url2, layers="0", format='image/png', transparent=True, name="NWI Wetlands Raster"
)
m.add_legend(builtin_legend="NWI")
m
# -
# ### Add custom legend
#
# There are two ways you can add custom legends:
#
# 1. Define legend labels and colors
# 2. Define legend dictionary
#
# Define legend keys and colors
# +
m = leafmap.Map()
labels = ['One', 'Two', 'Three', 'Four', 'ect']
# color can be defined using either hex code or RGB (0-255, 0-255, 0-255)
colors = ['#8DD3C7', '#FFFFB3', '#BEBADA', '#FB8072', '#80B1D3']
# colors = [(255, 0, 0), (127, 255, 0), (127, 18, 25), (36, 70, 180), (96, 68, 123)]
m.add_legend(title='Legend', labels=labels, colors=colors)
m
# -
# Define a legend dictionary.
# +
m = leafmap.Map()
url = "https://www.mrlc.gov/geoserver/mrlc_display/NLCD_2019_Land_Cover_L48/wms?"
m.add_wms_layer(
url,
layers="NLCD_2019_Land_Cover_L48",
name="NLCD 2019 CONUS Land Cover",
format="image/png",
transparent=True,
)
legend_dict = {
'11 Open Water': '466b9f',
'12 Perennial Ice/Snow': 'd1def8',
'21 Developed, Open Space': 'dec5c5',
'22 Developed, Low Intensity': 'd99282',
'23 Developed, Medium Intensity': 'eb0000',
'24 Developed High Intensity': 'ab0000',
'31 Barren Land (Rock/Sand/Clay)': 'b3ac9f',
'41 Deciduous Forest': '68ab5f',
'42 Evergreen Forest': '1c5f2c',
'43 Mixed Forest': 'b5c58f',
'51 Dwarf Scrub': 'af963c',
'52 Shrub/Scrub': 'ccb879',
'71 Grassland/Herbaceous': 'dfdfc2',
'72 Sedge/Herbaceous': 'd1d182',
'73 Lichens': 'a3cc51',
'74 Moss': '82ba9e',
'81 Pasture/Hay': 'dcd939',
'82 Cultivated Crops': 'ab6c28',
'90 Woody Wetlands': 'b8d9eb',
'95 Emergent Herbaceous Wetlands': '6c9fb8',
}
m.add_legend(legend_title="NLCD Land Cover Classification", legend_dict=legend_dict)
m
# -
# ## Add colorbar
#
# ### Continuous color
#
# Add a continuous colorbar with a custom palette to the map.
import leafmap
m = leafmap.Map()
m.add_basemap('USGS 3DEP Elevation')
colors = ['006633', 'E5FFCC', '662A00', 'D8D8D8', 'F5F5F5']
m.add_colorbar(colors=colors, vmin=0, vmax=4000)
m
# ### Categorical colorbar
# +
m = leafmap.Map()
url = "https://elevation.nationalmap.gov/arcgis/services/3DEPElevation/ImageServer/WMSServer?"
m.add_wms_layer(
url,
layers="3DEPElevation:Hillshade Elevation Tinted",
name="USGS 3DEP Elevation",
format="image/png",
transparent=True,
)
colors = ['006633', 'E5FFCC', '662A00', 'D8D8D8', 'F5F5F5']
m.add_colorbar(colors=colors, vmin=0, vmax=4000, categorical=True, step=4)
m
# -
# ## Add colormap
#
# The colormap functionality requires the ipyleaflet plotting backend. Folium is not supported.
import leafmap
import leafmap.colormaps as cm
# ### Common colormaps
#
# Color palette for DEM data.
cm.palettes.dem
# Show the DEM palette.
cm.plot_colormap(colors=cm.palettes.dem, axis_off=True)
# Color palette for NDVI data.
cm.palettes.ndvi
# Show the NDVI palette.
cm.plot_colormap(colors=cm.palettes.ndvi)
# ### Custom colormaps
#
# Specify the number of classes for a palette.
cm.get_palette('terrain', n_class=8)
# Show the terrain palette with 8 classes.
cm.plot_colormap(colors=cm.get_palette('terrain', n_class=8))
# Create a palette with custom colors, label, and font size.
cm.plot_colormap(colors=["red", "green", "blue"], label="Temperature", font_size=12)
# Create a discrete color palette.
cm.plot_colormap(
colors=["red", "green", "blue"], discrete=True, label="Temperature", font_size=12
)
# Specify the width and height for the palette.
cm.plot_colormap(
'terrain',
label="Elevation",
width=8.0,
height=0.4,
orientation='horizontal',
vmin=0,
vmax=1000,
)
# Change the orentation of the colormap to be vertical.
cm.plot_colormap(
'terrain',
label="Elevation",
width=0.4,
height=4,
orientation='vertical',
vmin=0,
vmax=1000,
)
# ### Horizontal colormap
#
# Add a horizontal colorbar to an interactive map.
m = leafmap.Map()
m.add_basemap("OpenTopoMap")
m.add_colormap(
'terrain',
label="Elevation",
width=8.0,
height=0.4,
orientation='horizontal',
vmin=0,
vmax=4000,
)
m
# ### Vertical colormap
#
# Add a vertical colorbar to an interactive map.
m = leafmap.Map()
m.add_basemap("OpenTopoMap")
m.add_colormap(
'terrain',
label="Elevation",
width=0.4,
height=4,
orientation='vertical',
vmin=0,
vmax=4000,
)
m
# ### List of available colormaps
cm.plot_colormaps(width=12, height=0.4)
# ## Add vector datasets
#
# ### Add CSV
#
# Read a CSV as a Pandas DataFrame.
import os
import leafmap
in_csv = 'https://raw.githubusercontent.com/giswqs/data/main/world/world_cities.csv'
df = leafmap.csv_to_pandas(in_csv)
df
# Create a point layer from a CSV file containing lat/long information.
m = leafmap.Map()
m.add_xy_data(in_csv, x="longitude", y="latitude", layer_name="World Cities")
m
# Set the output directory.
out_dir = os.getcwd()
out_shp = os.path.join(out_dir, 'world_cities.shp')
# Convert a CSV file containing lat/long information to a shapefile.
leafmap.csv_to_shp(in_csv, out_shp)
# Convert a CSV file containing lat/long information to a GeoJSON.
out_geojson = os.path.join(out_dir, 'world_cities.geojson')
leafmap.csv_to_geojson(in_csv, out_geojson)
# Convert a CSV file containing lat/long information to a GeoPandas GeoDataFrame.
gdf = leafmap.csv_to_gdf(in_csv)
gdf
# ### Add GeoJSON
#
# Add a GeoJSON to the map.
m = leafmap.Map(center=[0, 0], zoom=2)
in_geojson = 'https://raw.githubusercontent.com/giswqs/leafmap/master/examples/data/cable-geo.geojson'
m.add_geojson(in_geojson, layer_name="Cable lines", info_mode='on_hover')
m
# Add a GeoJSON with random filled color to the map.
m = leafmap.Map(center=[0, 0], zoom=2)
url = "https://raw.githubusercontent.com/giswqs/leafmap/master/examples/data/countries.geojson"
m.add_geojson(
url, layer_name="Countries", fill_colors=['red', 'yellow', 'green', 'orange']
)
m
# Use the `style_callback` function for assigning a random color to each polygon.
# +
import random
m = leafmap.Map(center=[0, 0], zoom=2)
url = "https://raw.githubusercontent.com/giswqs/leafmap/master/examples/data/countries.geojson"
def random_color(feature):
return {
'color': 'black',
'fillColor': random.choice(['red', 'yellow', 'green', 'orange']),
}
m.add_geojson(url, layer_name="Countries", style_callback=random_color)
m
# -
# Use custom `style` and `hover_style` functions.
m = leafmap.Map(center=[0, 0], zoom=2)
url = "https://raw.githubusercontent.com/giswqs/leafmap/master/examples/data/countries.geojson"
style = {
"stroke": True,
"color": "#0000ff",
"weight": 2,
"opacity": 1,
"fill": True,
"fillColor": "#0000ff",
"fillOpacity": 0.1,
}
hover_style = {"fillOpacity": 0.7}
m.add_geojson(url, layer_name="Countries", style=style, hover_style=hover_style)
m
# ### Add shapefile
m = leafmap.Map(center=[0, 0], zoom=2)
in_shp = 'https://github.com/giswqs/leafmap/raw/master/examples/data/countries.zip'
m.add_shp(in_shp, layer_name="Countries")
m
# ### Add KML
import leafmap
m = leafmap.Map()
in_kml = 'https://raw.githubusercontent.com/giswqs/leafmap/master/examples/data/us-states.kml'
m.add_kml(in_kml, layer_name="US States KML")
m
# ### Add GeoDataFrame
import geopandas as gpd
m = leafmap.Map()
gdf = gpd.read_file(
"https://github.com/giswqs/leafmap/raw/master/examples/data/cable-geo.geojson"
)
m.add_gdf(gdf, layer_name="Cable lines")
m
# Read the GeoPandas sample dataset as a GeoDataFrame.
path_to_data = gpd.datasets.get_path("nybb")
gdf = gpd.read_file(path_to_data)
gdf
m = leafmap.Map()
m.add_gdf(gdf, layer_name="New York boroughs", fill_colors=["red", "green", "blue"])
m
# + [markdown] tags=[]
# ### Add point layer
#
# Add a point layer using the interactive GUI.
#
# 
# -
m = leafmap.Map()
m
# Add a point layer programmatically.
m = leafmap.Map()
url = "https://raw.githubusercontent.com/giswqs/leafmap/master/examples/data/us_cities.geojson"
m.add_point_layer(url, popup=["name", "pop_max"], layer_name="US Cities")
m
# ### Add vector
#
# The `add_vector` function supports any vector data format supported by GeoPandas.
m = leafmap.Map(center=[0, 0], zoom=2)
url = "https://raw.githubusercontent.com/giswqs/leafmap/master/examples/data/countries.geojson"
m.add_vector(
url, layer_name="Countries", fill_colors=['red', 'yellow', 'green', 'orange']
)
m
# ## Download OSM data
#
# ### OSM from geocode
#
# Add OSM data of place(s) by name or ID to the map. Note that the leafmap custom layer control does not support GeoJSON, we need to use the ipyleaflet built-in layer control.
import leafmap
m = leafmap.Map(toolbar_control=False, layers_control=True)
m.add_osm_from_geocode("New York City", layer_name='NYC')
m
m = leafmap.Map(toolbar_control=False, layers_control=True)
m.add_osm_from_geocode("Chicago, Illinois", layer_name='Chicago, IL')
m
# ### OSM from place
#
# Add OSM entities within boundaries of geocodable place(s) to the map.
m = leafmap.Map(toolbar_control=False, layers_control=True)
place = "Bunker Hill, Los Angeles, California"
tags = {"building": True}
m.add_osm_from_place(place, tags, layer_name="Los Angeles, CA")
m
# Show OSM feature tags.
# https://wiki.openstreetmap.org/wiki/Map_features
# +
# leafmap.osm_tags_list()
# -
# ### OSM from address
m = leafmap.Map(toolbar_control=False, layers_control=True)
m.add_osm_from_address(
address="New York City", tags={"amenity": "bar"}, dist=1500, layer_name="NYC bars"
)
m
m = leafmap.Map(toolbar_control=False, layers_control=True)
m.add_osm_from_address(
address="New York City",
tags={"landuse": ["retail", "commercial"], "building": True},
dist=1000,
layer_name="NYC buildings",
)
m
# ### OSM from bbox
m = leafmap.Map(toolbar_control=False, layers_control=True)
north, south, east, west = 40.7551, 40.7454, -73.9738, -73.9965
m.add_osm_from_bbox(
north, south, east, west, tags={"amenity": "bar"}, layer_name="NYC bars"
)
m
# ### OSM from point
#
# Add OSM entities within some distance N, S, E, W of a point to the map.
m = leafmap.Map(
center=[46.7808, -96.0156], zoom=12, toolbar_control=False, layers_control=True
)
m.add_osm_from_point(
center_point=(46.7808, -96.0156),
tags={"natural": "water"},
dist=10000,
layer_name="Lakes",
)
m
m = leafmap.Map(
center=[39.9170, 116.3908], zoom=15, toolbar_control=False, layers_control=True
)
m.add_osm_from_point(
center_point=(39.9170, 116.3908),
tags={"building": True, "natural": "water"},
dist=1000,
layer_name="Beijing",
)
m
# ### OSM from view
#
# Add OSM entities within the current map view to the map.
m = leafmap.Map(toolbar_control=False, layers_control=True)
m.set_center(-73.9854, 40.7500, 16)
m
m.add_osm_from_view(tags={"amenity": "bar", "building": True}, layer_name="New York")
# Create a GeoPandas GeoDataFrame from place.
gdf = leafmap.osm_gdf_from_place("New York City", tags={"amenity": "bar"})
gdf
# ## Use WhiteboxTools
#
# Use the built-in toolbox to perform geospatial analysis. For example, you can perform depression filling using the sample DEM dataset downloaded in the above step.
#
# 
import os
import leafmap
import urllib.request
# Download a sample DEM dataset.
url = 'https://github.com/giswqs/whitebox-python/raw/master/whitebox/testdata/DEM.tif'
urllib.request.urlretrieve(url, "dem.tif")
m = leafmap.Map()
m
# Display the toolbox using the default mode.
leafmap.whiteboxgui()
# Display the toolbox using the collapsible tree mode. Note that the tree mode does not support Google Colab.
leafmap.whiteboxgui(tree=True)
# Perform geospatial analysis using the [whitebox](https://github.com/giswqs/whitebox-python) package.
import os
import whitebox
wbt = whitebox.WhiteboxTools()
wbt.verbose = False
wbt.version()
data_dir = os.getcwd()
wbt.set_working_dir(data_dir)
wbt.feature_preserving_smoothing("dem.tif", "smoothed.tif", filter=9)
wbt.breach_depressions("smoothed.tif", "breached.tif")
wbt.d_inf_flow_accumulation("breached.tif", "flow_accum.tif")
# +
import matplotlib.pyplot as plt
import imageio
# %matplotlib inline
# -
original = imageio.imread(os.path.join(data_dir, 'dem.tif'))
smoothed = imageio.imread(os.path.join(data_dir, 'smoothed.tif'))
breached = imageio.imread(os.path.join(data_dir, 'breached.tif'))
flow_accum = imageio.imread(os.path.join(data_dir, 'flow_accum.tif'))
# +
fig = plt.figure(figsize=(16, 11))
ax1 = fig.add_subplot(2, 2, 1)
ax1.set_title('Original DEM')
plt.imshow(original)
ax2 = fig.add_subplot(2, 2, 2)
ax2.set_title('Smoothed DEM')
plt.imshow(smoothed)
ax3 = fig.add_subplot(2, 2, 3)
ax3.set_title('Breached DEM')
plt.imshow(breached)
ax4 = fig.add_subplot(2, 2, 4)
ax4.set_title('Flow Accumulation')
plt.imshow(flow_accum)
plt.show()
# -
# ## Create basemap gallery
import leafmap
for basemap in leafmap.leafmap_basemaps:
print(basemap)
layers = list(leafmap.leafmap_basemaps.keys())[17:117]
leafmap.linked_maps(rows=20, cols=5, height="200px", layers=layers, labels=layers)
# ## Create linked map
import leafmap
leafmap.leafmap_basemaps.keys()
layers = ['ROADMAP', 'HYBRID']
leafmap.linked_maps(rows=1, cols=2, height='400px', layers=layers)
layers = ['Stamen.Terrain', 'OpenTopoMap']
leafmap.linked_maps(rows=1, cols=2, height='400px', layers=layers)
# Create a 2 * 2 linked map to visualize land cover change. Specify the `center` and `zoom` parameters to change the default map center and zoom level.
layers = [str(f"NLCD {year} CONUS Land Cover") for year in [2001, 2006, 2011, 2016]]
labels = [str(f"NLCD {year}") for year in [2001, 2006, 2011, 2016]]
leafmap.linked_maps(
rows=2,
cols=2,
height='300px',
layers=layers,
labels=labels,
center=[36.1, -115.2],
zoom=9,
)
# ## Create split-panel map
#
# Create a split-panel map by specifying the `left_layer` and `right_layer`, which can be chosen from the basemap names, or any custom XYZ tile layer.
import leafmap
leafmap.split_map(left_layer="ROADMAP", right_layer="HYBRID")
# Hide the zoom control from the map.
leafmap.split_map(
left_layer="Esri.WorldTopoMap", right_layer="OpenTopoMap", zoom_control=False
)
# Add labels to the map and change the default map center and zoom level.
leafmap.split_map(
left_layer="NLCD 2001 CONUS Land Cover",
right_layer="NLCD 2019 CONUS Land Cover",
left_label="2001",
right_label="2019",
label_position="bottom",
center=[36.1, -114.9],
zoom=10,
)
# ## Create heat map
#
# Specify the file path to the CSV. It can either be a file locally or on the Internet.
import leafmap
m = leafmap.Map(layers_control=True)
in_csv = "https://raw.githubusercontent.com/giswqs/leafmap/master/examples/data/world_cities.csv"
m.add_heatmap(
in_csv,
latitude="latitude",
longitude='longitude',
value="pop_max",
name="Heat map",
radius=20,
)
m
# Use the folium plotting backend.
from leafmap import foliumap
m = foliumap.Map()
in_csv = "https://raw.githubusercontent.com/giswqs/leafmap/master/examples/data/world_cities.csv"
m.add_heatmap(
in_csv,
latitude="latitude",
longitude='longitude',
value="pop_max",
name="Heat map",
radius=20,
)
colors = ['blue', 'lime', 'red']
m.add_colorbar(colors=colors, vmin=0, vmax=10000)
m.add_title("World Population Heat Map", font_size="20px", align="center")
m
# ## Save map to HTML
import leafmap
m = leafmap.Map()
m.add_basemap("Esri.NatGeoWorldMap")
m
# Specify the output HTML file name to save the map as a web page.
m.to_html("mymap.html")
# If the output HTML file name is not provided, the function will return a string containing contain the source code of the HTML file.
html = m.to_html()
# +
# print(html)
# -
# ## Use kepler plotting backend
import leafmap.kepler as leafmap
# ### Create an interactive map
#
# Create an interactive map. You can specify various parameters to initialize the map, such as `center`, `zoom`, `height`, and `widescreen`.
m = leafmap.Map(center=[40, -100], zoom=2, height=600, widescreen=False)
m
# If you encounter an error saying `Error displaying widget: model not found` when trying to display the map, you can use `m.static_map()` as a workaround until this [kepler.gl bug](https://github.com/keplergl/kepler.gl/issues/1165) has been resolved.
m.static_map(width=1280, height=600)
# ### Add CSV
#
# Add a CSV to the map. If you have a map config file, you can directly apply config to the map.
m = leafmap.Map(center=[37.7621, -122.4143], zoom=12)
in_csv = (
'https://raw.githubusercontent.com/giswqs/leafmap/master/examples/data/hex_data.csv'
)
config = 'https://raw.githubusercontent.com/giswqs/leafmap/master/examples/data/hex_config.json'
m.add_csv(in_csv, layer_name="hex_data", config=config)
m
m.static_map(width=1280, height=600)
# ### Save map config
#
# Save the map configuration as a JSON file.
m.save_config("cache/config.json")
# ### Save map as html
#
# Save the map to an interactive html.
m.to_html(outfile="cache/kepler_hex.html")
# ### Add GeoJONS
#
# Add a GeoJSON with US state boundaries to the map.
m = leafmap.Map(center=[50, -110], zoom=2)
polygons = 'https://raw.githubusercontent.com/giswqs/leafmap/master/examples/data/us-states.json'
m.add_geojson(polygons, layer_name="Countries")
m
m.static_map(width=1280, height=600)
# ### Add shapefile
#
# Add a shapefile to the map.
m = leafmap.Map(center=[20, 0], zoom=1)
in_shp = "https://github.com/giswqs/leafmap/raw/master/examples/data/countries.zip"
m.add_shp(in_shp, "Countries")
m
m.static_map(width=1280, height=600)
# ### Add GeoDataFrame
#
# Add a GeoPandas GeoDataFrame to the map.
import geopandas as gpd
gdf = gpd.read_file(
"https://raw.githubusercontent.com/giswqs/leafmap/master/examples/data/world_cities.geojson"
)
gdf
m = leafmap.Map(center=[20, 0], zoom=1)
m.add_gdf(gdf, "World cities")
m
m.static_map(width=1280, height=600)
# ## Publish maps
#
# To follow this tutorial, you will need to [sign up](https://datapane.com/accounts/signup/) for an account with <https://datapane.com>, then install and authenticate the `datapane` Python package. More information can be found [here](https://docs.datapane.com/tutorials/tut-getting-started).
#
# - `pip install datapane`
# - `datapane login`
# - `datapane ping`
#
# 
#
# If you encounter folium version errors, please uncomment the following line to update folium and restart the kernel.
# +
# # !pip install -U folium
# -
import os
import datapane as dp
import leafmap.foliumap as leafmap
if os.environ.get("DP_TOKEN") is None:
os.environ["DP_TOKEN"] = 'your-api-key'
dp.login(token=os.environ['DP_TOKEN'])
# ### Elevation map
m = leafmap.Map()
m.add_basemap('USGS 3DEP Elevation')
colors = ['006633', 'E5FFCC', '662A00', 'D8D8D8', 'F5F5F5']
vmin = 0
vmax = 4000
m.add_colorbar(colors=colors, vmin=vmin, vmax=vmax)
m
m.publish(name="Elevation Map of North America")
# ### Land cover map
m = leafmap.Map()
m.add_basemap("NLCD 2019 CONUS Land Cover")
m.add_legend(builtin_legend='NLCD')
m
m.publish(name="National Land Cover Database (NLCD) 2019")
# ### Population heat map
m = leafmap.Map()
in_csv = "https://raw.githubusercontent.com/giswqs/leafmap/master/examples/data/world_cities.csv"
m.add_heatmap(
in_csv,
latitude="latitude",
longitude='longitude',
value="pop_max",
name="Heat map",
radius=20,
)
colors = ['blue', 'lime', 'red']
vmin = 0
vmax = 10000
m.add_colorbar(colors=colors, vmin=vmin, vmax=vmax)
m
m.publish(name="World Population Heat Map")
# ## Use planet imagery
#
# First, you need to sign up a Planet account and get an API key. See https://www.planet.com/nicfi & https://developers.planet.com/quickstart/apis.
# Uncomment the following line to pass in your API key.
import os
import leafmap
if os.environ.get("PLANET_API_KEY") is None:
os.environ["PLANET_API_KEY"] = 'your-api-key'
biannual_tiles = leafmap.planet_biannual_tiles_tropical()
for tile in biannual_tiles:
print(tile)
monthly_tiles = leafmap.planet_monthly_tiles_tropical()
for tile in monthly_tiles:
print(tile)
# Add a Planet monthly mosaic by specifying year and month.
m = leafmap.Map()
layer = monthly_tiles['Planet_2021-08']
m.add_layer(layer)
m
m = leafmap.Map()
layer = biannual_tiles['Planet_2020-06_2020-08']
m.add_layer(layer)
m
# ## Use timeseries inspector
import os
import leafmap
if os.environ.get("PLANET_API_KEY") is None:
os.environ["PLANET_API_KEY"] = 'your-api-key'
biannual_tiles = leafmap.planet_biannual_tiles_tropical()
leafmap.ts_inspector(biannual_tiles, center=[8.5, -80], zoom=5)
monthly_tiles = leafmap.planet_monthly_tiles_tropical()
leafmap.ts_inspector(monthly_tiles, center=[8.5, -80], zoom=5)
tiles = leafmap.planet_tiles_tropical()
leafmap.ts_inspector(tiles, center=[8.5, -80], zoom=5)
# ## Use time slider
# Use the time slider to visualize Planet quarterly mosaic.
import os
import leafmap
if os.environ.get("PLANET_API_KEY") is None:
os.environ["PLANET_API_KEY"] = 'your-api-key'
m = leafmap.Map(center=[8.5, -80], zoom=5)
layers_dict = leafmap.planet_monthly_tiles_tropical()
m.add_time_slider(layers_dict, time_interval=1)
m
m = leafmap.Map(center=[8.5, -80], zoom=5)
layers_dict = leafmap.planet_biannual_tiles_tropical()
m.add_time_slider(layers_dict, time_interval=1)
m
# Use the time slider to visualize basemaps.
m = leafmap.Map()
m.clear_layers()
layers_dict = leafmap.basemap_xyz_tiles()
m.add_time_slider(layers_dict, time_interval=1)
m
# ## Use PostGIS
#
# Setting up the conda env:
#
# ```
# conda create -n geo python=3.8
# conda activate geo
# conda install geopandas
# conda install mamba -c conda-forge
# mamba install leafmap sqlalchemy psycopg2 -c conda-forge
# ```
#
# Sample dataset:
# - [nyc_data.zip](https://github.com/giswqs/postgis/raw/master/data/nyc_data.zip) (Watch this [video](https://youtu.be/fROzLrjNDrs) to load data into PostGIS)
#
# ### Connect to the database
#
# You can directly pass in the user name and password to access the database. Alternative, you can define environment variables. The default environment variables for user and password are `SQL_USER` and `SQL_PASSWORD`, respectively.
#
# The `try...except...` statements are only used for building the documentation website (https://leafmap.org) because the PostGIS database is not available on GitHub. If you are running the notebook with Jupyter installed locally and PostGIS set up properly, you don't need these `try...except...` statements.
try:
con = leafmap.connect_postgis(
database="nyc", host="localhost", user=None, password=None, use_env_var=True
)
except:
pass
# ### Perform SQL queries
#
# Create a GeoDataFrame from a sql query.
sql = 'SELECT * FROM nyc_neighborhoods'
try:
gdf = leafmap.read_postgis(sql, con)
display(gdf)
except:
pass
# ### Display data on the map
try:
m = leafmap.Map()
m.add_gdf_from_postgis(
sql, con, layer_name="NYC Neighborhoods", fill_colors=["red", "green", "blue"]
)
display(m)
except:
pass
# 
# ## Add widget to the map
import leafmap
import numpy as np
import matplotlib.pyplot as plt
from ipywidgets import Output
from ipyleaflet import WidgetControl
# +
m = leafmap.Map()
# Data for plotting
t = np.arange(0.0, 2.0, 0.01)
s = 1 + np.sin(2 * np.pi * t)
fig, ax = plt.subplots()
ax.plot(t, s)
ax.set(
xlabel='time (s)', ylabel='voltage (mV)', title='About as simple as it gets, folks'
)
ax.grid()
# Create an output widget to host the plot
output_widget = Output()
# Show the plot on the widget
with output_widget:
output_widget.clear_output()
plt.show()
# Add the widget as a control to the map
output_control = WidgetControl(widget=output_widget, position="bottomright")
m.add_control(output_control)
# -
m
# ## Develop custom widgets
import leafmap
import ipywidgets as widgets
from ipyleaflet import WidgetControl
# ### Create a toolbar button
# +
widget_width = "250px"
padding = "0px 0px 0px 5px" # upper, right, bottom, left
toolbar_button = widgets.ToggleButton(
value=False,
tooltip="Toolbar",
icon="gears",
layout=widgets.Layout(width="28px", height="28px", padding=padding),
)
close_button = widgets.ToggleButton(
value=False,
tooltip="Close the tool",
icon="times",
button_style="primary",
layout=widgets.Layout(height="28px", width="28px", padding=padding),
)
# -
toolbar = widgets.HBox([toolbar_button])
toolbar
# ### Add toolbar event
# +
def toolbar_click(change):
if change["new"]:
toolbar.children = [toolbar_button, close_button]
else:
toolbar.children = [toolbar_button]
toolbar_button.observe(toolbar_click, "value")
# +
def close_click(change):
if change["new"]:
toolbar_button.close()
close_button.close()
toolbar.close()
close_button.observe(close_click, "value")
toolbar
# -
# ### Add a toolbar grid
rows = 2
cols = 2
grid = widgets.GridspecLayout(
rows, cols, grid_gap="0px", layout=widgets.Layout(width="65px")
)
# icons: https://fontawesome.com/v4.7.0/icons/
# +
icons = ["folder-open", "map", "info", "question"]
for i in range(rows):
for j in range(cols):
grid[i, j] = widgets.Button(
description="",
button_style="primary",
icon=icons[i * rows + j],
layout=widgets.Layout(width="28px", padding="0px"),
)
grid
# -
toolbar = widgets.VBox([toolbar_button])
# +
def toolbar_click(change):
if change["new"]:
toolbar.children = [widgets.HBox([close_button, toolbar_button]), grid]
else:
toolbar.children = [toolbar_button]
toolbar_button.observe(toolbar_click, "value")
toolbar
# -
# ### Add toolbar to leafmap
toolbar_ctrl = WidgetControl(widget=toolbar, position="topright")
m = leafmap.Map()
m.add_control(toolbar_ctrl)
m
output = widgets.Output()
output_ctrl = WidgetControl(widget=output, position="bottomright")
m.add_control(output_ctrl)
def tool_click(b):
with output:
output.clear_output()
print(f"You clicked the {b.icon} button")
for i in range(rows):
for j in range(cols):
tool = grid[i, j]
tool.on_click(tool_click)
# 
| 25.971857
| 1,136
|
5ccfbc644b5ac458333ded29a3bff276bb69524a
|
py
|
python
|
chapter08_computer-vision/object-detection.ipynb
|
vishaalkapoor/mxnet-the-straight-dope
|
['Apache-2.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Environment (conda_mxnet_p27)
# language: python
# name: conda_mxnet_p27
# ---
# # Object Detection Using Convolutional Neural Networks
#
# So far, when we've talked about making predictions based on images,
# we were concerned only with classification.
# We asked questions like is this digit a "0", "1", ..., or "9?"
# or, does this picture depict a "cat" or a "dog"?
# Object detection is a more challenging task.
# Here our goal is not only to say *what* is in the image
# but also to recognize *where* it is in the image.
# As an example, consider the following image, which depicts two dogs and a cat together with their locations.
#
# 
#
# So object defers from image classification in a few ways.
# First, while a classifier outputs a single category per image,
# an object detector must be able to recognize multiple objects in a single image.
# Technically, this task is called *multiple object detection*,
# but most research in the area addresses the multiple object setting,
# so we'll abuse terminology just a little.
# Second, while classifiers need only to output probabilities over classes,
# object detectors must output both probabilities of class membership
# and also the coordinates that identify the location of the objects.
#
#
# On this chapter we'll demonstrate the single shot multiple box object detector (SSD),
# a popular model for object detection that was first described in [this paper](https://arxiv.org/abs/1512.02325),
# and is straightforward to implement in MXNet Gluon.
#
# ## SSD: Single Shot MultiBox Detector
#
# The SSD model predicts anchor boxes at multiple scales. The model architecture is illustrated in the following figure.
#
# 
#
# We first use a `body` network to extract the image features,
# which are used as the input to the first scale (scale 0). The class labels and the corresponding anchor boxes
# are predicted by `class_predictor` and `box_predictor`, respectively.
# We then downsample the representations to the next scale (scale 1).
# Again, at this new resolution, we predict both classes and anchor boxes.
# This downsampling and predicting routine
# can be repeated in multiple times to obtain results on multiple resolution scales.
# Let's walk through the components one by one in a bit more detail.
#
# ### Default anchor boxes
#
# Since an anchor box can have arbituary shape,
# we sample a set of anchor boxes as the candidate.
# In particular, for each pixel, we sample multiple boxes
# centered at this pixel but have various sizes and ratios.
# Assume the input size is $w \times h$,
# - for size $s\in (0,1]$, the generated box shape will be $ws \times hs$
# - for ratio $r > 0$, the generated box shape will be $w\sqrt{r} \times \frac{h}{\sqrt{r}}$
#
# We can sample the boxes using the operator `MultiBoxPrior`.
# It accepts *n* sizes and *m* ratios to generate *n+m-1* boxes for each pixel.
# The first *i* boxes are generated from `sizes[i], ratios[0]`
# if $i \le n$ otherwise `sizes[0], ratios[i-n]`.
# +
# %matplotlib inline
import mxnet as mx
from mxnet import nd
from mxnet.contrib.ndarray import MultiBoxPrior
n = 40
# shape: batch x channel x height x weight
x = nd.random_uniform(shape=(1, 3, n, n))
y = MultiBoxPrior(x, sizes=[.5, .25, .1], ratios=[1, 2, .5])
# the first anchor box generated for pixel at (20,20)
# its format is (x_min, y_min, x_max, y_max)
boxes = y.reshape((n, n, -1, 4))
print('The first anchor box at row 21, column 21:', boxes[20, 20, 0, :])
# -
# We can visualize all anchor boxes generated for one pixel on a certain size feature map.
import matplotlib.pyplot as plt
def box_to_rect(box, color, linewidth=3):
"""convert an anchor box to a matplotlib rectangle"""
box = box.asnumpy()
return plt.Rectangle(
(box[0], box[1]), (box[2]-box[0]), (box[3]-box[1]),
fill=False, edgecolor=color, linewidth=linewidth)
colors = ['blue', 'green', 'red', 'black', 'magenta']
plt.imshow(nd.ones((n, n, 3)).asnumpy())
anchors = boxes[20, 20, :, :]
for i in range(anchors.shape[0]):
plt.gca().add_patch(box_to_rect(anchors[i,:]*n, colors[i]))
plt.show()
# ### Predict classes
#
# For each anchor box, we want to predict the associated class label.
# We make this prediction by using a convolution layer.
# We choose a kernel of size $3\times 3$ with padding size $(1, 1)$
# so that the output will have the same width and height as the input.
# The confidence scores for the anchor box class labels are stored in channels.
# In particular, for the *i*-th anchor box:
#
# - channel `i*(num_class+1)` store the scores for this box contains only background
# - channel `i*(num_class+1)+1+j` store the scores for this box contains an object from the *j*-th class
# +
from mxnet.gluon import nn
def class_predictor(num_anchors, num_classes):
"""return a layer to predict classes"""
return nn.Conv2D(num_anchors * (num_classes + 1), 3, padding=1)
cls_pred = class_predictor(5, 10)
cls_pred.initialize()
x = nd.zeros((2, 3, 20, 20))
print('Class prediction', cls_pred(x).shape)
# -
# ### Predict anchor boxes
#
# The goal is predict how to transfer the current anchor box to the correct box. That is, assume $b$ is one of the sampled default box, while $Y$ is the ground truth, then we want to predict the delta positions $\Delta(Y, b)$, which is a 4-length vector.
#
# More specifically, the we define the delta vector as:
# [$t_x$, $t_y$, $t_{width}$, $t_{height}$], where
#
# - $t_x = (Y_x - b_x) / b_{width}$
# - $t_y = (Y_y - b_y) / b_{height}$
# - $t_{width} = (Y_{width} - b_{width}) / b_{width}$
# - $t_{height} = (Y_{height} - b_{height}) / b_{height}$
#
# Normalizing the deltas with box width/height tends to result in better convergence behavior.
#
# Similar to classes, we use a convolution layer here. The only difference is that the output channel size is now `num_anchors * 4`, with the predicted delta positions for the *i*-th box stored from channel `i*4` to `i*4+3`.
# +
def box_predictor(num_anchors):
"""return a layer to predict delta locations"""
return nn.Conv2D(num_anchors * 4, 3, padding=1)
box_pred = box_predictor(10)
box_pred.initialize()
x = nd.zeros((2, 3, 20, 20))
print('Box prediction', box_pred(x).shape)
# -
# ### Down-sample features
#
# Each time, we downsample the features by half.
# This can be achieved by a simple pooling layer with pooling size 2.
# We may also stack two convolution, batch normalization and ReLU blocks
# before the pooling layer to make the network deeper.
# +
def down_sample(num_filters):
"""stack two Conv-BatchNorm-Relu blocks and then a pooling layer
to halve the feature size"""
out = nn.HybridSequential()
for _ in range(2):
out.add(nn.Conv2D(num_filters, 3, strides=1, padding=1))
out.add(nn.BatchNorm(in_channels=num_filters))
out.add(nn.Activation('relu'))
out.add(nn.MaxPool2D(2))
return out
blk = down_sample(10)
blk.initialize()
x = nd.zeros((2, 3, 20, 20))
print('Before', x.shape, 'after', blk(x).shape)
# -
# ### Manage preditions from multiple layers
#
# A key property of SSD is that predictions are made
# at multiple layers with shrinking spatial size.
# Thus, we have to handle predictions from multiple feature layers.
# One idea is to concatenate them along convolutional channels,
# with each one predicting a corresponding value (class or box) for each default anchor.
# We give class predictor as an example, and box predictor follows the same rule.
# a certain feature map with 20x20 spatial shape
feat1 = nd.zeros((2, 8, 20, 20))
print('Feature map 1', feat1.shape)
cls_pred1 = class_predictor(5, 10)
cls_pred1.initialize()
y1 = cls_pred1(feat1)
print('Class prediction for feature map 1', y1.shape)
# down-sample
ds = down_sample(16)
ds.initialize()
feat2 = ds(feat1)
print('Feature map 2', feat2.shape)
cls_pred2 = class_predictor(3, 10)
cls_pred2.initialize()
y2 = cls_pred2(feat2)
print('Class prediction for feature map 2', y2.shape)
# +
def flatten_prediction(pred):
return nd.flatten(nd.transpose(pred, axes=(0, 2, 3, 1)))
def concat_predictions(preds):
return nd.concat(*preds, dim=1)
flat_y1 = flatten_prediction(y1)
print('Flatten class prediction 1', flat_y1.shape)
flat_y2 = flatten_prediction(y2)
print('Flatten class prediction 2', flat_y2.shape)
print('Concat class predictions', concat_predictions([flat_y1, flat_y2]).shape)
# -
# ### Body network
#
# The body network is used to extract features from the raw pixel inputs. Common choices follow the architectures of the state-of-the-art convolution neural networks for image classification. For demonstration purpose, we just stack several down sampling blocks to form the body network.
# +
from mxnet import gluon
def body():
"""return the body network"""
out = nn.HybridSequential()
for nfilters in [16, 32, 64]:
out.add(down_sample(nfilters))
return out
bnet = body()
bnet.initialize()
x = nd.zeros((2, 3, 256, 256))
print('Body network', [y.shape for y in bnet(x)])
# -
# ### Create a toy SSD model
#
# Now, let's create a toy SSD model that takes images of resolution $256 \times 256$ as input.
# +
def toy_ssd_model(num_anchors, num_classes):
"""return SSD modules"""
downsamples = nn.Sequential()
class_preds = nn.Sequential()
box_preds = nn.Sequential()
downsamples.add(down_sample(128))
downsamples.add(down_sample(128))
downsamples.add(down_sample(128))
for scale in range(5):
class_preds.add(class_predictor(num_anchors, num_classes))
box_preds.add(box_predictor(num_anchors))
return body(), downsamples, class_preds, box_preds
print(toy_ssd_model(5, 2))
# -
# ### Forward
#
# Given an input and the model, we can run the forward pass.
def toy_ssd_forward(x, body, downsamples, class_preds, box_preds, sizes, ratios):
# extract feature with the body network
x = body(x)
# for each scale, add anchors, box and class predictions,
# then compute the input to next scale
default_anchors = []
predicted_boxes = []
predicted_classes = []
for i in range(5):
default_anchors.append(MultiBoxPrior(x, sizes=sizes[i], ratios=ratios[i]))
predicted_boxes.append(flatten_prediction(box_preds[i](x)))
predicted_classes.append(flatten_prediction(class_preds[i](x)))
if i < 3:
x = downsamples[i](x)
elif i == 3:
# simply use the pooling layer
x = nd.Pooling(x, global_pool=True, pool_type='max', kernel=(4, 4))
return default_anchors, predicted_classes, predicted_boxes
# ### Put all things together
from mxnet import gluon
class ToySSD(gluon.Block):
def __init__(self, num_classes, **kwargs):
super(ToySSD, self).__init__(**kwargs)
# anchor box sizes for 4 feature scales
self.anchor_sizes = [[.2, .272], [.37, .447], [.54, .619], [.71, .79], [.88, .961]]
# anchor box ratios for 4 feature scales
self.anchor_ratios = [[1, 2, .5]] * 5
self.num_classes = num_classes
with self.name_scope():
self.body, self.downsamples, self.class_preds, self.box_preds = toy_ssd_model(4, num_classes)
def forward(self, x):
default_anchors, predicted_classes, predicted_boxes = toy_ssd_forward(x, self.body, self.downsamples,
self.class_preds, self.box_preds, self.anchor_sizes, self.anchor_ratios)
# we want to concatenate anchors, class predictions, box predictions from different layers
anchors = concat_predictions(default_anchors)
box_preds = concat_predictions(predicted_boxes)
class_preds = concat_predictions(predicted_classes)
# it is better to have class predictions reshaped for softmax computation
class_preds = nd.reshape(class_preds, shape=(0, -1, self.num_classes + 1))
return anchors, class_preds, box_preds
# ### Outputs of ToySSD
# instantiate a ToySSD network with 10 classes
net = ToySSD(2)
net.initialize()
x = nd.zeros((1, 3, 256, 256))
default_anchors, class_predictions, box_predictions = net(x)
print('Outputs:', 'anchors', default_anchors.shape, 'class prediction', class_predictions.shape, 'box prediction', box_predictions.shape)
# ## Dataset
#
# For demonstration purposes, we'll train our model to detect Pikachu in the wild.
# We generated a synthetic toy dataset by rendering images from open-sourced 3D Pikachu models.
# The dataset consists of 1000 pikachus with random pose/scale/position in random background images.
# The exact locations are recorded as ground-truth for training and validation.
#
# 
#
# ### Download dataset
# +
from mxnet.test_utils import download
import os.path as osp
def verified(file_path, sha1hash):
import hashlib
sha1 = hashlib.sha1()
with open(file_path, 'rb') as f:
while True:
data = f.read(1048576)
if not data:
break
sha1.update(data)
matched = sha1.hexdigest() == sha1hash
if not matched:
print('Found hash mismatch in file {}, possibly due to incomplete download.'.format(file_path))
return matched
url_format = 'https://apache-mxnet.s3-accelerate.amazonaws.com/gluon/dataset/pikachu/{}'
hashes = {'train.rec': 'e6bcb6ffba1ac04ff8a9b1115e650af56ee969c8',
'train.idx': 'dcf7318b2602c06428b9988470c731621716c393',
'val.rec': 'd6c33f799b4d058e82f2cb5bd9a976f69d72d520'}
for k, v in hashes.items():
fname = 'pikachu_' + k
target = osp.join('data', fname)
url = url_format.format(k)
if not osp.exists(target) or not verified(target, v):
print('Downloading', target, url)
download(url, fname=fname, dirname='data', overwrite=True)
# -
# ### Load dataset
# +
import mxnet.image as image
data_shape = 256
batch_size = 32
def get_iterators(data_shape, batch_size):
class_names = ['pikachu']
num_class = len(class_names)
train_iter = image.ImageDetIter(
batch_size=batch_size,
data_shape=(3, data_shape, data_shape),
path_imgrec='./data/pikachu_train.rec',
path_imgidx='./data/pikachu_train.idx',
shuffle=True,
mean=True,
rand_crop=1,
min_object_covered=0.95,
max_attempts=200)
val_iter = image.ImageDetIter(
batch_size=batch_size,
data_shape=(3, data_shape, data_shape),
path_imgrec='./data/pikachu_val.rec',
shuffle=False,
mean=True)
return train_iter, val_iter, class_names, num_class
train_data, test_data, class_names, num_class = get_iterators(data_shape, batch_size)
batch = train_data.next()
print(batch)
# -
# ### Illustration
#
# Let's display one image loaded by ImageDetIter.
# +
import numpy as np
img = batch.data[0][0].asnumpy() # grab the first image, convert to numpy array
img = img.transpose((1, 2, 0)) # we want channel to be the last dimension
img += np.array([123, 117, 104])
img = img.astype(np.uint8) # use uint8 (0-255)
# draw bounding boxes on image
for label in batch.label[0][0].asnumpy():
if label[0] < 0:
break
print(label)
xmin, ymin, xmax, ymax = [int(x * data_shape) for x in label[1:5]]
rect = plt.Rectangle((xmin, ymin), xmax - xmin, ymax - ymin, fill=False, edgecolor=(1, 0, 0), linewidth=3)
plt.gca().add_patch(rect)
plt.imshow(img)
plt.show()
# -
# ## Train
# ### Losses
#
# Network predictions will be penalized for incorrect class predictions and wrong box deltas.
from mxnet.contrib.ndarray import MultiBoxTarget
def training_targets(default_anchors, class_predicts, labels):
class_predicts = nd.transpose(class_predicts, axes=(0, 2, 1))
z = MultiBoxTarget(*[default_anchors, labels, class_predicts])
box_target = z[0] # box offset target for (x, y, width, height)
box_mask = z[1] # mask is used to ignore box offsets we don't want to penalize, e.g. negative samples
cls_target = z[2] # cls_target is an array of labels for all anchors boxes
return box_target, box_mask, cls_target
# Pre-defined losses are provided in `gluon.loss` package, however, we can define losses manually.
#
# First, we need a Focal Loss for class predictions.
# +
class FocalLoss(gluon.loss.Loss):
def __init__(self, axis=-1, alpha=0.25, gamma=2, batch_axis=0, **kwargs):
super(FocalLoss, self).__init__(None, batch_axis, **kwargs)
self._axis = axis
self._alpha = alpha
self._gamma = gamma
def hybrid_forward(self, F, output, label):
output = F.softmax(output)
pt = F.pick(output, label, axis=self._axis, keepdims=True)
loss = -self._alpha * ((1 - pt) ** self._gamma) * F.log(pt)
return F.mean(loss, axis=self._batch_axis, exclude=True)
# cls_loss = gluon.loss.SoftmaxCrossEntropyLoss()
cls_loss = FocalLoss()
print(cls_loss)
# -
# Next, we need a SmoothL1Loss for box predictions.
# +
class SmoothL1Loss(gluon.loss.Loss):
def __init__(self, batch_axis=0, **kwargs):
super(SmoothL1Loss, self).__init__(None, batch_axis, **kwargs)
def hybrid_forward(self, F, output, label, mask):
loss = F.smooth_l1((output - label) * mask, scalar=1.0)
return F.mean(loss, self._batch_axis, exclude=True)
box_loss = SmoothL1Loss()
print(box_loss)
# -
# ### Evaluation metrics
#
# Here, we define two metrics that we'll use to evaluate our performance whien training.
# You're already familiar with accuracy unless you've been naughty and skipped straight to object detection.
# We use the accuracy metric to assess the quality of the class predictions.
# Mean absolute error (MAE) is just the L1 distance, introduced in our [linear algebra chapter](../chapter01_crashcourse/linear-algebra.ipynb).
# We use this to determine how close the coordinates of the predicted bounding boxes are to the ground-truth coordinates.
# Because we are jointly solving both a classification problem and a regression problem, we need an appropriate metric for each task.
cls_metric = mx.metric.Accuracy()
box_metric = mx.metric.MAE() # measure absolute difference between prediction and target
### Set context for training
ctx = mx.gpu() # it may takes too long to train using CPU
try:
_ = nd.zeros(1, ctx=ctx)
# pad label for cuda implementation
train_data.reshape(label_shape=(3, 5))
train_data = test_data.sync_label_shape(train_data)
except mx.base.MXNetError as err:
print('No GPU enabled, fall back to CPU, sit back and be patient...')
ctx = mx.cpu()
# ### Initialize parameters
net = ToySSD(num_class)
net.initialize(mx.init.Xavier(magnitude=2), ctx=ctx)
# ### Set up trainer
net.collect_params().reset_ctx(ctx)
trainer = gluon.Trainer(net.collect_params(), 'sgd', {'learning_rate': 0.1, 'wd': 5e-4})
# ### Start training
#
# Optionally we load pretrained model for demonstration purpose. One can set `from_scratch = True` to training from scratch, which may take more than 30 mins to finish using a single capable GPU.
epochs = 150 # set larger to get better performance
log_interval = 20
from_scratch = False # set to True to train from scratch
if from_scratch:
start_epoch = 0
else:
start_epoch = 148
pretrained = 'ssd_pretrained.params'
sha1 = 'fbb7d872d76355fff1790d864c2238decdb452bc'
url = 'https://apache-mxnet.s3-accelerate.amazonaws.com/gluon/models/ssd_pikachu-fbb7d872.params'
if not osp.exists(pretrained) or not verified(pretrained, sha1):
print('Downloading', pretrained, url)
download(url, fname=pretrained, overwrite=True)
net.load_params(pretrained, ctx)
# +
import time
from mxnet import autograd as ag
for epoch in range(start_epoch, epochs):
# reset iterator and tick
train_data.reset()
cls_metric.reset()
box_metric.reset()
tic = time.time()
# iterate through all batch
for i, batch in enumerate(train_data):
btic = time.time()
# record gradients
with ag.record():
x = batch.data[0].as_in_context(ctx)
y = batch.label[0].as_in_context(ctx)
default_anchors, class_predictions, box_predictions = net(x)
box_target, box_mask, cls_target = training_targets(default_anchors, class_predictions, y)
# losses
loss1 = cls_loss(class_predictions, cls_target)
loss2 = box_loss(box_predictions, box_target, box_mask)
# sum all losses
loss = loss1 + loss2
# backpropagate
loss.backward()
# apply
trainer.step(batch_size)
# update metrics
cls_metric.update([cls_target], [nd.transpose(class_predictions, (0, 2, 1))])
box_metric.update([box_target], [box_predictions * box_mask])
if (i + 1) % log_interval == 0:
name1, val1 = cls_metric.get()
name2, val2 = box_metric.get()
print('[Epoch %d Batch %d] speed: %f samples/s, training: %s=%f, %s=%f'
%(epoch ,i, batch_size/(time.time()-btic), name1, val1, name2, val2))
# end of epoch logging
name1, val1 = cls_metric.get()
name2, val2 = box_metric.get()
print('[Epoch %d] training: %s=%f, %s=%f'%(epoch, name1, val1, name2, val2))
print('[Epoch %d] time cost: %f'%(epoch, time.time()-tic))
# we can save the trained parameters to disk
net.save_params('ssd_%d.params' % epochs)
# -
# ## Test
# Testing is similar to training, except that we don't need to compute gradients and training targets. Instead, we take the predictions from network output, and combine them to get the real detection output.
# ### Prepare the test data
# +
import numpy as np
import cv2
def preprocess(image):
"""Takes an image and apply preprocess"""
# resize to data_shape
image = cv2.resize(image, (data_shape, data_shape))
# swap BGR to RGB
image = image[:, :, (2, 1, 0)]
# convert to float before subtracting mean
image = image.astype(np.float32)
# subtract mean
image -= np.array([123, 117, 104])
# organize as [batch-channel-height-width]
image = np.transpose(image, (2, 0, 1))
image = image[np.newaxis, :]
# convert to ndarray
image = nd.array(image)
return image
image = cv2.imread('img/pikachu.jpg')
x = preprocess(image)
print('x', x.shape)
# -
# ### Network inference
#
# In a single line of code!
# if pre-trained model is provided, we can load it
# net.load_params('ssd_%d.params' % epochs, ctx)
anchors, cls_preds, box_preds = net(x.as_in_context(ctx))
print('anchors', anchors)
print('class predictions', cls_preds)
print('box delta predictions', box_preds)
# ### Convert predictions to real object detection results
from mxnet.contrib.ndarray import MultiBoxDetection
# convert predictions to probabilities using softmax
cls_probs = nd.SoftmaxActivation(nd.transpose(cls_preds, (0, 2, 1)), mode='channel')
# apply shifts to anchors boxes, non-maximum-suppression, etc...
output = MultiBoxDetection(*[cls_probs, box_preds, anchors], force_suppress=True, clip=False)
print(output)
# Each row in the output corresponds to a detection box, as in format [class_id, confidence, xmin, ymin, xmax, ymax].
#
# Most of the detection results are -1, indicating that they either have very small confidence scores, or been suppressed through non-maximum-suppression.
# ### Display results
# +
def display(img, out, thresh=0.5):
import random
import matplotlib as mpl
mpl.rcParams['figure.figsize'] = (10,10)
pens = dict()
plt.clf()
plt.imshow(img)
for det in out:
cid = int(det[0])
if cid < 0:
continue
score = det[1]
if score < thresh:
continue
if cid not in pens:
pens[cid] = (random.random(), random.random(), random.random())
scales = [img.shape[1], img.shape[0]] * 2
xmin, ymin, xmax, ymax = [int(p * s) for p, s in zip(det[2:6].tolist(), scales)]
rect = plt.Rectangle((xmin, ymin), xmax - xmin, ymax - ymin, fill=False,
edgecolor=pens[cid], linewidth=3)
plt.gca().add_patch(rect)
text = class_names[cid]
plt.gca().text(xmin, ymin-2, '{:s} {:.3f}'.format(text, score),
bbox=dict(facecolor=pens[cid], alpha=0.5),
fontsize=12, color='white')
plt.show()
display(image[:, :, (2, 1, 0)], output[0].asnumpy(), thresh=0.45)
# -
# ## Conclusion
#
# Detection is harder than classification, since we want not only class probabilities, but also localizations of different objects including potential small objects. Using sliding window together with a good classifier might be an option, however, we have shown that with a properly designed convolutional neural network, we can do single shot detection which is blazing fast and accurate!
#
# For whinges or inquiries, [open an issue on GitHub.](https://github.com/zackchase/mxnet-the-straight-dope)
| 52.308029
| 11,197
|
1ad1893994b16ab614d59347785fd35158b308f2
|
py
|
python
|
_ipynb/Statistics as Toolbox and Statistics as Detective Work.ipynb
|
simkovic/simkovic.github.io
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# I will write lot about how statistical inference is done in psychological research. I think however that it will be helpfull to first point out few issues which I think are paramount to all my other opinions. In this post I write about the general attitude towards statistics that is imprinted on psychology students in their introductory stats classes.
# ## Toolbox
# This is how the statistics works for psychologists.
#
# 1. Formulate hypothesis
# 2. Design a study to test it
# 3. Collect data
# 4. Evaluate data
# 5. Draw a conclusion and publish
#
# When we zoom in at point 4 we get following instructions. First, see whether the data are at categorical, ordinal or nominal scale. Second, what design do we have? How many conditions/groups do we have? How many levels does each variable have? Do we have repeated measurements or independent samples? Once we have determined the answers we look at our toolbox and choose the appropriate method. This is what our toolbox looks like.
#
#
# <img src="/assets/nominaltable.png" width="600">
#
# Then students are told how to perform the analyses in the cells with a statistical software, which values to copy from the software output and how one does report them. In almost all cases p-values are available in the output and are reported along with the test statistic. Since late 90s most software also offers effect size estimates and students are told where to look for them.
#
# Let's go back to the toolbox table. As an example if we measured the performance of two groups (treatment group and control) at three consecutive time points then we have one nominal DV and two IVs: two independent groups and three repeated measurements. Looking at the table, we see that we have to perform Mixed-design Anova.
#
# In the case where the scale of DV is not nominal the following alternative table is provided. Ordinal scale is assumed.
#
# <img src="/assets/ordinaltable.png" width="600">
#
# Finally, psychologists are also taught $\chi^2$ test which is applied when the DV is dichotomic categorical (i.e. count data).
#
# That's the toolbox approach in a nutshell. It has problems.
# ## Problems with toolbox
# First, there is a discrepancy between how it is taught and how it is applied. The procedure sketched above is slavishly obeyed even when it doesn't make any sense. For instance, Anova is used as default model even in context where it is inappropriate (i.e. the assumptions of linearity, normality or heteroskedasticity are not satisfied).
#
# Second, the above mentioned approach is intended as a one-way street. You can go only in one direction from step 1 to 5. This is extremely inflexible. The toolbox approach does not allow for the fitted model to be discarded. The model is fitted and the obtained estimates are reported. The 1972 APA manual captures the toolbox spirit: "Caution: Do not infer trends from data that fail by a small margin to meet the usual levels of significance. Such results are best interpreted as caused by chance and are best reported as such. Treat the result section like an income tax return. Take what's coming to you, but no more"
# One may protest that too much flexibility is a bad thing. Obviously, too much rigidity - reporting models that are (in hindsight but nevertheless) incorrect is not the solution.
#
# Third, the toolbox implicitly claims to be exhaustive - it aplies as a tool to all research problems. Of course this doesn't happen and as a consequence two cases arise. First, inappropriate models are being fit and reported. We discussed this already in the previous paragraph. Second, the problem is defined in more general terms, such that not all available information (provided by data or prior knowledge) is used. That is, we throw away information so that some tool becomes available, because we would have no tool available if we included the information in the analysis. Good example are non-normal measurements (e.g. skewed response times) which are handled by rank tests listed in the second table. This is done even where it would be perfectly appropriate to fit a parametric model at the nominal scale. For instance we could fit response times with Gamma regression. Unfortunately, Gamma regression didn't make it into the toolbox. At other times structural information is discarded. In behavioral experiments we mostly obtain data with hierarchical structure. We have several subjects and many consecutive trials for each subject and condition. The across-trials variability (due to learning or order effects) can be difficult to analyze with the tools in the toolbox (i.e. time series methods are not in the toolbox). Common strategy is to build single score for each subject (e.g. average performance across trials) and then to analyze the obtained scores across subjects and conditions.
#
# There is one notable strategy to ensure that you have the appropriate tool in the toolbox. If you can't fit a model to the data, then ensure that your data fit some tool in the toolbox. Psychologists devise experiments with manipulations that map the hypothesis onto few measured variables. Ideally the critical comparison is mapped onto single dimension and can be evaluated with a simple t-test. For example, we test two conditions which are identical except for a single manipulation. In this case we discard all the additional information and structure of the data since this is the same across the two conditions (and we do not expect that it will interact with the manipulation).
#
# Unfortunately, there are other more important considerations which should influence the choice of design than the limits of our toolbox. Ecological validity is more important than convenient analysis. In fact, I think that this is the biggest trouble with the toolbox approach. It not only cripples the analysis, it also cripples the experiment design and in turn the choice of the research question.
# ## Detective Work
# Let's now have a look at the detective approach. The most vocal recent advocate has been Andrew Gelman (Shalizi & Gelman, 2013) but the idea goes back to George Box and John Tukey (1969). This approach has been most prevalent in fields that heavily rely on observational data - econometry, sociology and political science. Here, researchers were not able to off-load their problems to experimental design. Instead they had to tackle the problems head on by developing flexible data analysis methods.
# While the toolbox approach is a one-way street, the detective approach contains a loop that iterates between model estimation and model checking. The purpose of the model checking part is to see whether the model describes the data appropriately. This can be done for instance by looking at the residuals and whether their distribution does not deviate from the distribution postulated by the model. Another option (the so-called predictive checking) is to generate data from the model and to look whether these are reasonably similar to the actual data. In any case, model checking is informal and doesn't even need to be quantitative. Whether a model is appropriate depends on the purpose of the analysis and which aspects of the data are crucial. Still, model checking is part of the results. It should be transparent and replicable. Even if it is informal there are instances which are rather formal up to the degree that can be written down as an algorithm (e.g. the Box-Jenkins method for analyzing time series). Once an appropriate model has been identified this model is used to estimate the quantities of interest. Often however the (structure of the) model itself is of theoretical interest.
# ## An Application of Detective Approach
# I already presented an example of detective approach when I discussed modeling of data with skewed distributions. Here, let's take a look at a non-psychological example which illustrates the logic of the detective approach more clearly. The example is taken from Montgomery, Jennings and Kulahci (2008). The data are available from the [companion website](ftp://ftp.wiley.com/public/sci_tech_med/time_series/).
# %pylab inline
d= np.loadtxt('b5.dat')
t=np.arange(0,d.size)/12.+1992
plt.plot(t,d)
plt.gca().set_xticks(np.arange(0,d.size/12)+1992)
plt.xlabel('year')
plt.ylabel('product shipments');
# The data are U.S. Beverage Manufacturer Product Shipments from 1992 to 2006 as provided by www.census.gov. The shipments show rising trend and yearly seasonal fluctuations. We first get rid of the trend to obtain a better look at the seasonal changes.
# +
res=np.linalg.lstsq(np.concatenate(
[np.atleast_2d(np.ones(d.size)),np.atleast_2d(t)]).T,d)
y=res[0][0]+res[0][1]*t
plt.plot(t, y)
plt.plot(t,d)
plt.gca().set_xticks(np.arange(0,d.size/12)+1992);
# -
# The fit of the linear curve is acceptable and we continue with model building. We now subtract the trend from the data to obtain the residuals. These show the remaining paterns in the data that require modeling.
# +
plt.plot(t,d-y)
plt.gca().set_xticks(np.arange(0,d.size/12)+1992);
plt.ylim([-1000,1000]);
plt.ylabel('residuals')
plt.xlabel('year')
plt.figure()
plt.plot(np.mod(range(d.size),12)+1,d-y,'o')
plt.xlim([0.5,12.5])
plt.grid(False,axis='x')
plt.xticks(range(1,13))
plt.ylim([-1000,1000]);
plt.ylabel('residuals')
plt.xlabel('month');
# -
# We add the yearly fluctuations to the model. The rough idea is to use sinusoid $\alpha sin(2\pi (t+\phi))$ where $\alpha$ is the amplitude and $\phi$ is the shift. Here is a sketch what we are looking for. (The parameter values were found through trial and error).
plt.plot(np.mod(range(d.size),12)+1,d-y,'o')
plt.xlim([0.5,12.5])
plt.grid(False,axis='x')
plt.xticks(range(1,13))
plt.ylim([-1000,1000])
plt.ylabel('residuals')
plt.xlabel('month')
tt=np.arange(1,13,0.1)
plt.plot(tt,600*np.sin(2*np.pi*tt/12.+4.3));
# We fit the model. We simplify the fitting process by writing
# $$\alpha sin(2\pi (t+\phi))= \alpha cos(2\pi \phi) sin(2\pi t)+\alpha cos(2\pi t) sin(2\pi \phi)= \beta_1 sin(2\pi t)+\beta_2 cos(2\pi t) $$
x=np.concatenate([np.atleast_2d(np.cos(2*np.pi*t)),
np.atleast_2d(np.sin(2*np.pi*t))]).T
res=np.linalg.lstsq(x,d-y)
plt.plot(t,y+x.dot(res[0]))
plt.plot(t,d,'-')
plt.gca().set_xticks(np.arange(0,d.size/12)+1992);
# The results look already good. We zoom in at the residuals, to see how it can be further improved.
ynew=y+x.dot(res[0])
from scipy import stats
plt.figure()
plt.hist(d-ynew,15,normed=True);
plt.xlabel('residuals')
print np.std(d-y)
r=range(-600,600)
plt.plot(r,stats.norm.pdf(r,0,np.std(d-ynew)))
plt.figure()
stats.probplot(d-ynew,dist='norm',plot=plt);
# The residuals are well described by normal distribution with $\sigma=233$. We can summarize our model as $d=\beta_0+\beta_1 t + \beta_2sin 2\pi t + \beta_3sin 2\pi t + \epsilon$ with error $\epsilon \sim \mathcal{N}(0,\sigma)$
#
# The plots below suggest some additional improvements.
# +
plt.plot(t,d-ynew)
plt.ylabel('residuals')
plt.xlabel('year')
plt.gca().set_xticks(np.arange(0,d.size/12)+1992);
plt.figure()
plt.acorr(d-ynew,maxlags=90);
plt.xlabel('month lag')
plt.ylabel('autocorrelation');
plt.figure()
plt.plot(d,d-ynew,'o');
plt.ylabel('residuals')
plt.xlabel('predicted shipments');
# -
# The first two plots suggests a 9-year cycle. The third plot suggests that variance is increasing with time. Both suggestions are however difficult to evaluate because we don't have enough data.
#
# Once we are satisfied with the obtained model we can look at the estimated parameter values if they conform to our predictions. The model can be also used to predict future shipments. For our purpose it was important to see the model building iteration and especially the model checking part. In this case model checking was done by inspecting the residuals. The presented case was rather straight-forward. If we tried to extend the model with a 9 year cycle or evolving variance this would require more thought and more detective work, which is the usual case with psychological data.
# ## Better Results but More Work
# Let's now evaluate detective approach by reviewing the problems caused by toolbox.
#
# First, it seems difficult to make turn detective approach into ritual. Almost by definition it excludes this possibility. (Although I don't want to underestimate psychologist's ability to abuse any statistical method.)
#
# Second, the approach is iterative. We are not required to stick to a model that we had in mind at the outset of our analysis.
#
# Third, modeling is open-ended. The only limit is the computational tractability of the models, we would like to fit and the information afforded by the evidence.
#
# Second and third point mean that detective approach is much more flexible and this flexibility can be abused. As stated above model building is an integral part of the reported results and all decisions should be made transparent, replicable accountable.
#
# Finally detective approach requires more work. In particular, it requires the kind of work that researchers don't have time for - thinking. With toolbox you can get your students compute the p values. But he probably won't be able to accomplish the model building. This extra work is only a corollary of a general maxim that better results require more work. In future posts I will provide further illustrations that the better results afforded by detective approach are sorely needed.
| 75.949438
| 1,505
|
a69fc3be94c0a65b90f3cb42e94580c4457a1bd8
|
py
|
python
|
The Bare Minimum about Floating Point Arithmetic.ipynb
|
rcorless/ComputationalDiscoveryOnJupyterBetas
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <H1> The Bare Minimum about Floating-Point </H1>
#
# <H2>Warning: Reading this requires care.</H2>
#
# To a first approximation, no-one likes thinking carefully about floating-point numbers. Most people think that floats are finicky, tedious, and boring. Isn't that why we have computers, to do that sort of thing? Most everyone just wants floating-point arithmetic to work, fast and without giving problems. But there comes a time when you <i>have</i> to think about them. This notebook tries to give the absolute minimum that you need to know, so that (most of the time) you can forget about them (but still be ready when their peculiarities strike). <b>In fact, we're going to assume that you have already seen something weird about decimal arithmetic on a computer, and are looking here for answers.</b> You'll get them, but it's not that simple.
#
# To make it more complicated, there is a <A HREF="https://en.wikipedia.org/wiki/IEEE_754">standard for floating-point arithmetic laid out by the IEEE that is now over 35 years old </A>: it's very well-known, and very very good; but unfortunately, </i>not</i> everyone knows about it or follows it, and so not everyone supports it (we do---it makes life so much easier). Indeed a lot of machines and computer languages do, and so we use it here, because you need to know something about it. But when it is <b>not</b> supported, it's like barking your shin on a piece of furniture in the dark: not fun. Sadly, we will see an example below, with Python native floating-point arithmetic (which is, quite surprisingly, not the same as NumPy floating-point arithmetic, which is different from SymPy floating-point arithmetic. Yay.).
#
# Reading about this topic requires close attention. You have to read every word, and not just the words, but also the actual code and its output. You have to look at the graphs and think about why they look the way they do (instead of how they would look if we used theoretical real arithmetic, or arbitrary precision arithmetic). You have to look at the fine details of the numerical output (and sometimes you can only see what's going on if you use <i>hexadecimal</i> notation). Understanding floating-point arithmetic even requires <i>a painful reversal of understanding</i> and <i>unlearning</i> some things. Sometimes, this will not seem like fun. (...although, there's a certain satisfaction in understanding what's really going on.)
#
# Most writers don't like it, either. Most start off thinking that they can convey the essentials of floating point in a really short time and space with a minimum of fuss, concentrating on just exactly what you can use to avoid learning all the horrible details. <b>Um, no. We can't do it either</b>. This section wound up being much longer than we wanted to write, and it's almost certainly longer than you want to read. And, it's hard to read, too, being full of ugly details. Well, we'll try to point you at some useful videos, too. You can start with one by <A HREF="https://youtu.be/PZRI1IfStY0"> Computerphile </A>, which is very good. That may help you to read what follows. But, be warned: you'll need to take a deep breath each time you start to read again.
#
# So if (when!) you want a break from learning about floating-point arithmetic, go for it. Take a break, then come back and re-read. For whimsical relaxation that's still (sort of) on topic, we suggest listening to the music of <A HREF="https://youtu.be/yYqu5NdFMf8"> Floating Points </A>. We think it's seriously good: Sam Shepard, the artist, has a PhD in Neuroscience from University College London; whether he chose the name with any reference to floating-point arithmetic we don't know, but maybe. Listening might help you to come back in the mood for more details. Or, well, there's always <A HREF="https://youtu.be/Zqd_R5Rq6KM"> Rachmaninov, played by the great Leslie Howard.</A>
#
#
# <H3> What you will want to get from this notebook </H3>
#
# <OL>
# <li> Why we use floating-point arithmetic, even though it causes headaches (In short: it forestalls or alleviates much worse trouble) </li>
# <LI> A sense in which floats are better models of reality than the real numbers are </li>
# <LI> How to recognize when floating-point effects ("rounding errors") are adding up and making a difference </LI>
# <li> What's a rounding error, anyway?</LI>
# <LI> What happens when your significant figures cancel each other, and other ways to lose accuracy </LI>
# <LI> The difference between accuracy and precision </LI>
# <LI> How to predict when problems <i>might</i> arise, and when instead to be confident (actually, most of the time!) </LI>
# <LI> How to model floating-point arithmetic with the epsilon-delta tools from real analysis (yes, that's right: the tools of real analysis make numerical analysis <i>easier</i>---don't be dismayed, this is actually kind of cool). </LI>
# <LI> The basic constants: unit roundoff $\mu$, machine epsilon $\varepsilon$, the largest representable real number (often called realmax), the smallest representable positive real number (often called realmin)</LI>
# <LI> What happens when the numbers <i>overflow</i> (get bigger than realmax) or <i>underflow</i> (get smaller than realmin)
# </LI>
# <LI> Why machine epsilon $\varepsilon$ is usually more important than the (much smaller) realmin. </LI>
# <LI> Where to look for more information (when you wind up really needing it).
# <OL> <li>Yes, <A HREF="https://en.wikipedia.org/wiki/Floating-point_arithmetic"> Wikipedia </A> is a really good place to start (although, good grief, there is a lot of information densely packed there). </li>
# <li>Then there is the <A HREF="https://docs.python.org/3/tutorial/floatingpoint.html">Python floating-point tutorial</A> </li>
# <li>and the old classic <A HREF="https://dl.acm.org/doi/10.1145/103162.103163">What every computer scientist should know about Floating-Point Arithmetic</A>. </li>
# <li>An excellent slim textbook (maybe the best reference) is <A HREF="https://cs.nyu.edu/overton/book/">Michael Overton, Numerical Computing with IEEE Floating-Point Arithmetic</A>.</li>
# <li> Many references at <A HREF="https://people.eecs.berkeley.edu/~wkahan/ieee754status/"> William (Velvel) Kahan's page </A> </li>
# <li> The wonderful book <A HREF="https://epubs.siam.org/doi/book/10.1137/1.9780898718027">Accuracy and Stability of Numerical Algorithms</A> by <A HREF="https://nhigham.com/">Nicholas J. Higham</A> </li>
# <li> The book <A HREF="https://www.springer.com/gp/book/9781461484523"> A Graduate Introduction to Numerical Methods </A> by Corless and Fillion.</li>
# </ol> </LI>
# </OL>
# <H4> Floats versus the Real World </H4>
#
# Let's begin by thinking of something really, really small: the nanofibers making up the fine strands of the finest spider silk, that of the (venomous, of course) brown recluse spider. According to <A href="https://pubs.acs.org/doi/10.1021/acsmacrolett.8b00678"> this article from ACS Macro Letters </A> which has some really cool pictures (of fine silk fibres, not so many of spiders, thankfully enough if you don't like spiders much), the nanofibers average about $20$nm in width---in scientific notation, that is about $20\cdot 10^{-9}$ metres wide. Very hard to see, even with the best equipment.
#
# As an aside, if you <b>do</b> like spiders, we recommend some <A HREF="https://youtu.be/d_yYC5r8xMI">really cool videos with dancing Peacock Spiders (genus Maratus) by Jürgen Otto</A>. The spiders are typically smaller than your little fingernail (length 4--5mm), absolutely harmless, and really cool to watch. Anyway, back to work. (Sorry if we sent you down a rabbit hole (spider hole)).
#
# Machine epsilon (we'll learn what that means) for <i>IEEE Standard 754 double precision</i> floating point arithmetic is $2^{-52} \approx 2.2\cdot 10^{-16}$, or about $20 \cdot 10^{-17}$. Machine epsilon, compared to the unit $1$, is therefore $10^8$ (one hundred million) times finer than those nanofibers of spider silk are when compared to a metre. You can compare double precision machine epsilon to the size of a proton, even, which has a width about $10^{-15}$m. In other words, double precision can resolve things about ten times smaller than a proton, compared to everyday scales.
#
# <b> A basic fact: </b> Floating-point numbers are represented by a fixed number of bits. IEEE double-precision numbers use just $64$ bits, which means that there are at most $2^{64} \approx 1.85\cdot 10^{19}$ distinct such numbers. (Actually there are fewer than that, because there are some special codes sprinkled in there). That sounds like a lot, but it means that there is a <i>largest representable number</i>, a <i>smallest positive number</i>, and other strange things that are not true of the real number system, including weirdnesses such as nonzero numbers $x$ that are so small that adding them to $1$ doesn't change it: $1 + x = 1$ with $x$ <i>not</i> being zero. This causes brain-hurt, so if you want to take a break for a bit, go ahead.
#
# (In fact, the machine epsilon $\varepsilon$ is often (sort of) defined as the smallest machine number for which $1+\varepsilon$ winds up being bigger than $1$; it's actually a bit trickier than that, as you will see below.)
# +
import numpy as np
import sys
unity = 1.0
small = 1.0e-22
ouch = unity + small
print( 'one plus 10**(-22) equals ', ouch, ' in floats; in hexadecimal, we can see it really is:', ouch.hex() )
eps = sys.float_info.epsilon
print('machine epsilon in decimal is ', eps, 'and in hex it is 2**(-52): ', eps.hex() )
ah = unity + eps
print( 'one plus machine epsilon equals ', ah, 'in floats: in hex we can see it exactly:', ah.hex())
eh = unity + 2*eps/3
print( 'adding 2/3 of epsilon to one gets', eh.hex())
uh = unity + eps/2
print( 'adding half of epsilon to one gets ', uh.hex() )
# -
# Most floating-point arithmetic nowadays uses double precision; sometimes double-double (or "quad") precision is used, which is absurdly more precise; less frequently but sometimes just <i>single</i> precision is used, which has machine epsilon $2^{-23} \approx 1.2\cdot 10^{-7}$ (so about six times the width of an average spider nanofibre); and nowadays especially in applications where there is a <i>lot</i> of data, <i>half</i>-precision can be used, which has machine epsilon $2^{-10} \approx 10^{-3}$; note that the average width of a human hair is about $1.8\cdot 10^{-4}$m, so half precision has a fineness about five human hairs' width. Compare that to spider silk, and we see that the comparison human hair::half precision is more or less the same as spider fibre::single precision. Double precision takes that down to nuclear structure fineness.
#
# So how could that degree of fineness ever cause problems in ordinary arithmetic? Well, most of the time---really, very much most of the time---it does not. The IEEE 754 standard was designed to do as good a job as possible, given the constraint that we want memory access to be predictable (and therefore memory operations on computers will be fast).
# But sometimes it goes off the rails, and we need to understand how, and why.
#
# <H4> Rounding Errors Can Grow in Relative Importance (sometimes quickly) </H4>
#
# The fundamental reason it goes off the rails is that it <i>has to</i>. We give you no less an Authority than Aristotle himself: in "On the Heavens" written 350BCE we find
#
# <A HREF="http://classics.mit.edu/Aristotle/heavens.1.i.html"> Admit, for instance, the existence of a minimum magnitude, and you will find that the minimum which you have introduced, small as it is, causes the greatest truths of mathematics to totter. </A>
#
# What could this mean for modern computers? Consider the following simple little bits of arithmetic.
( -7.35e22 + 7.35e22) + 100.0
-7.35e22 + (7.35e22 + 100.0 )
# Those answers are different, although to your eyes those instructions should give the same answer.
#
# All we did there was do the operations in a different order. Real arithmetic is associative: (a+b)+c = a+(b+c), end of story. But in floating-point, this is <i>not so</i>. Both those answers are correct (even IEEE standard!), and actually what we <i>want</i> when we do floating-point arithmetic (that may be hard to believe, when the second answer is so clearly "wrong" in real arithmetic).
#
# But ok, you say. Big deal, you say. Those numbers are different on a nuclear scale. Or, rather, on an astronomical scale: if the $100$ represents the mass of a person in kilograms, then $7.35e22$ is about the mass of the Earth's Moon. If we threw an anti-moon at the moon and it all vanished in a blaze of energy, it wouldn't matter much if there was a person standing on the moon or not when that happened; but if you put the human there after the anti-moon destroyed the moon, you'd still have the person. (Well, if the spacesuit was good, anyway.)
#
# Incidentally, the main benefit of floating-point arithmetic (as opposed to fixed-point arithmetic) is the <i>dynamic range</i>. It is equally useful talking about things on astronomical scales, normal scales, or nuclear scales, or indeed even more scales: there is an <i>exponent</i>, as in scientific notation (which floating-point translates to, readily), and a number of significant "digits" (actually, bits) called the <i>significand</i> or <i>mantissa</i>. More soon.
# The really annoying thing is that floating-point differences (called "rounding errors") can sometimes add up rapidly and show up unexpectedly, even with very few innocuous-looking operations. Let's look at another example, namely plotting $(x-1/3)^2$, and let's zoom in on the region around $x=1/3$. We all remember from high school (right, we all remember) what the graph <i> should</i> look like, and indeed on a "human" scale it does:
from matplotlib import pyplot as plt
poly = np.poly1d([1.0, -2.0/3.0, 1.0/9.0])
x = np.linspace(-1/3,1)
y = poly(x)
plt.plot(x,y)
plt.show()
# Now let's zoom in: instead of a nice smooth curve (even on a nuclear scale) we see flat spots---on the spider silk scale in the x-direction!
poly = np.poly1d([1.0, -2.0/3.0, 1.0/9.0])
x = np.linspace(1/3-1.0e-8,1/3+1.0e-8)
y = poly(x)
plt.plot(x,y,".")
plt.show()
# That's actually not bad, and kind of pretty. You can see that the floating-point numbers, which are discrete, seem to be <i>trying</i> to represent a nice smooth function. There <i>are no double-precision floating point numbers</i> on the vertical scale between the flat layers, but in spite of that the sense of the curve is conveyed fairly well.
#
# Let's try a more complicated polynomial (after all, for that last example we hardly needed a computer). This next one one is $(3x-1)^5(x-2)$, expanded. On a human scale, all looks more or less well.
poly = np.poly1d([243., -891., 1080., -630., 195., -31., 2.])
x = np.linspace(0,1)
y = poly(x)
plt.plot(x,y)
plt.show()
# Now zoom in on the zero at $x=1/10$.
poly = np.poly1d([243., -891., 1080., -630., 195., -31., 2.])
c = 0.3333
delt = 4.0e-4
x = np.linspace(c-delt,c+delt,2021)
y = poly(x)
plt.plot(x,y,"k,")
plt.xticks([c-delt, c, c+delt])
plt.show()
# Now, on the scale of about <b>the width of four or five human hairs</b> (even though we are using nuclear-fine arithmetic!), we see all <i>kinds</i> of weirdness. This is what rounding error looks like, when you zoom in. (Don't connect the dots, it just looks like a mess.)
#
# Even though the values are all fairly small on the $y$-axis, this is kind of a disaster. Evaluating that polynomial takes only six operations at each point; we have evaluated it at each of $2021$ points, near to the zero at $x=1/3$. But deciding from the plot where the zero is is now pretty difficult. If we didn't know ahead of time where it was, we'd be out of luck. Even next-door points might go seemingly unpredictably to one flat layer or another. The x-scale is $\pm 4\cdot 10^{-4}$, about the width of a few human hairs, as we said.
#
# And we haven't gone beyond a degree six polynomial, and the coefficients are all modestly sized. How much more difficulty are we going to find in real-life-sized problems?
#
# Actually, there are some real difficulties with floating-point in real-life problems. <A HREF="https://en.wikipedia.org/wiki/Cluster_(spacecraft)">Maybe the most famous is the loss of the Ariane 5</A> although that was a conversion error that caused a cascading failure, not a rounding error. But floating-point doesn't cause all that many problems, really, and <i>most</i> of those are going to show up in obvious ways. But there are a nasty few that can cause the worst kinds of problems and give you wrong but <i>plausible</i> answers. These are the ones you need to know about.
# <b> An example with no subtractions </b>
#
# Here's a nasty one. Take a simple number, such as 6.0. Take its square root, then take the square root of that, and so on 52 times. Then take the result of that, and square it. Then square that, then square the result, and so on 52 times, again. Square root and square are inverse operations, so we <i>ought</i> to get back to where we started from. We don't, and this time, even though we are using nuclear-scale precision, we get answers that are wrong on a human scale (never mind being off by a hair, we are off by almost fifty percent). This result, being done with such simple operations, ought to shock you.
#
# The appearance of a number so close to $e$, the base of the natural logarithms, is not an accident; but we won't stop to explain that here.
# +
x = 6.0
for i in range(52):
x = np.sqrt(x)
print( 'After ', i+1, 'square roots, x is ', x, 'or in hex ', x.hex() )
for i in range(52):
x = x*x
print( 'x started as 6.0, and is now ', x)
# -
# It's easier to see what's happening in hex; after a while, not enough decimals are printed, base 10, but in hex we see everything. We'll see in the next cell just what those hex symbols mean.
#
# We did not print the results when squaring---they're kind of the same but backwards, and different in detail. We have all we need, now, to explain what is going on.
#
# The problem is <i>loss of precision</i> causing loss of accuracy. The information about where we started from ($x=6.0$) is carried in hex digits further and further to the right, as we progressively take square roots, as you can see above as the zeros creep out from the "decimal" point. A similar thing happens in decimal: That information drops off the right end of the numbers each time the result is stored in the variable $x$. When we start squaring again, we are starting from a number that has lost almost all information about where it started. Indeed, one more square root and it would have rounded down to exactly $1$, and then when we started squaring we wouldn't have got anywhere.
#
# This loop is a simple iteration, executing a very simple dynamical system such as are studied frequently in real-life applications. There are other examples of similar disasters.
# <H3> What are the rules? </H3>
#
# Computers work in binary. Hexadecimal is basically the same thing except four times shorter: one hex digit takes four bits, so 64 bits in binary is only 16 hexadecimal places. An IEEE 754 double precision float occupies 64 bits, or 16 hex digits. Let's try to follow the addition $\sqrt{2} + \pi$ in double precision.
import math
root2float = np.sqrt(2.0)
pifloat = math.pi
modest = 100.35
print( root2float.hex() )
print( pifloat.hex() )
print( modest.hex() )
# What you are looking at there is a human-readable version of the internal representation of the two floating-point approximations to $\sqrt2$ and to $\pi$, together with a modestly bigger number chosen more or less at random. Neither of the first two numbers can be represented exactly as a finite decimal, binary, or hexadecimal number, of course (although both are "computable" in a technical sense: we can get as many digits of either as we could possibly want). The third has a terminating decimal representation but a <i>nonterminating</i> hex representation (this was a lucky accident). Asking Matlab to use "format hex" actually shows the internal representations: $\sqrt2$ is represented as "3ff6a09e667f3bcd" using exactly 16 hexadecimal digits, and $\pi$ is represented as "400921fb54442d18" which at first glance look baffling, and bafflingly different to those representations above. But the first three hex "digits" of each of those are used for the <i>sign bit</i> and the 11-bit <i>exponent</i> in a <i>biased format</i> which we'll ignore for a moment. After those three "digits" we see the sequence "6a09e667f3bcd" from Matlab and that's exactly the string after the "decimal" point for $\sqrt2$; similarly, the $13$ hexadecimal digits after the "decimal" point for $\pi$ show up in the Matlab string as the final $13$ hexadecimal digits in Python.
#
# Hexadecimal digits are in a one-to-one correspondence with the following numbers. We put them in decimal--hex--binary.
# Here they are typed by hand:
#
# 00--1--0000
#
# 01--1--0001
#
# 02--2--0010
#
# 03--3--0011
#
# 04--4--0100
#
# 05--5--0101
#
# 06--6--0110
#
# 07--7--0111
#
# 08--8--1000
#
# 09--9--1001
#
# 10--a--1010
#
# 11--b--1011
#
# 12--c--1100
#
# 13--d--1101
#
# 14--e--1110
#
# 15--f--1111
#
# and here they are tabulated automatically (with decorations 0x meaning hex and 0b meaning binary). The leading binary 0 bits are trimmed below, which makes the correspondence with hex somewhat less intelligible: four bits to a hex digit is the point.
#
# +
from tabulate import tabulate
results = [(n, hex(n), bin(n)) for n in range(16)]
print(tabulate(results, headers=["decimal", "hex", "binary"]))
# -
# You can also use capital letters, A (hex) for 10 (decimal) and 1010 (binary), and so on, to F for 15. 16 in decimal is thus 10 in hex or 10000 in binary. Translating groups of four bits to hex is easy, and translating a hex "digit" to binary is just as easy. Binary to decimal and decimal to binary (with fractional parts), on the other hand, is annoyingly full of special cases that cause headaches.
#
# So what does the Python format 0x1.6a09e667f3bcdp+0 mean? This seems weird at first, and then gets a bit weirder, because it actually uses binary in the exponent, p+0 means times 2 to the 0 and p+1 means $\cdot 2^1$ and p+6 means $\cdot 2^6$ so that number is bigger than $64$ (which we knew, of course). As we said before, the 0x means hex. The 1. means there is a 1 before the "decimal" point. (For "normal" numbers this is always true so we don't even have to store it---this gives us what is called a "hidden bit"). Then after the "decimal" point comes a string of hex digits, meaning $6/16 + a/16^2 + 0/16^3 + 9/16^4 + e/16^5 + 6/16^6 + 6/16^7 + 7/16^8 + f/16^9 + 3/16^{10} + b/16^{11} + c/16^{12} + d/16^{13}$. Using exact rational arithmetic, that adds up to $6369051672525773/4503599627370496$ (including the leading 1).
#
# Ick. There's the other reason we like scientific notation. It's hard even to see if that exact rational is bigger than $1$ or not. We'll convert it to decimal in a minute, and we'll see that it makes more sense to humans that way. If we square it exactly as a rational number (mercifully keeping the computation to ourselves, but go ahead and do it yourself if you like), we get a rational number <i>just</i> a little bigger than $2$; it is $2$ plus $5545866846675497/20282409603651670423947251286016$, which is about $2.73\cdot 10^{-16}$. It's no coincidence that this is about the same size as the machine epsilon for double precision. If we convert the rational number $6369051672525773/4503599627370496$ to a decimal number, we get $1.4142135623730951\ldots$ which, if we were going to report only to five significant figures, we would round to $1.4142$. That accuracy after rounding is enough to get us the distance corner-to-corner of a square one kilometer on a side accurate to better than $10$cm. If we hadn't rounded, and instead used all figures, it would be precise to a tenth of a billionth of a millimeter. We don't say <i>accurate</i> there because how could we measure the angles of a square to that much accuracy? That's about 100 protons wide! Precision can be spurious.
#
# But keeping all precision <i>during</i> the computation guards against growth of rounding errors. So, as a general rule, try to convert to decimals and round your answers only at the very end.
#
#
# Now that we have talked about binary, hexadecimal, and the <b>fact that we have to approximate</b> we can talk about simple arithmetic. Here's the IEEE rule:
#
# <H4> Do the arithmetic (+, -, *, divide) on two operands <i>exactly correctly</i>, then round the results to the <i>nearest</i> machine representable number. In the case of a tie, round to the <i>nearest machine representable number whose last bit is 0</i>. </H4>
# This tie-breaking rule (which only rarely happens) is called "round-to-even" and is done to eliminate bias in long computations. (There is only one place in this notebook where we know that it happened. Good exercise to find it.)
#
# This is a pretty good principle, and really, how could it be better? It's saying that the only error that's allowed is the one at the end, which you can't escape, once you have decided to store your numbers in just 64 bits (or 32 if you're using single precision, or 16 if you're using half precision).
#
root2float = np.sqrt(2.0)
pifloat = math.pi
added = root2float + pifloat
print( root2float.hex() )
print( pifloat.hex() )
print( 'sqrt(2) + pi = ', added, 'which is a few decimals translated from ', added.hex(), 'exactly in the internal hex' )
# You should notice that Python only prints 12 significant figures there in decimal (seems to be a default printing width. In Maple, working to 20 decimal digits, we get $4.5558062159628882873$). In order to <i>reliably</i> convert binary (or hex) to and from IEEE double precision and back again, to get exactly the same number, you have to keep about $18$ decimal places (not just sixteen as suggested by the machine epsilon; we're not sure where "twelve" comes from in that example). There are worse problems with conversion to decimal, such as the one called <A HREF="http://perso.ens-lyon.fr/jean-michel.muller/Intro-to-TMD.htm">"The Table-Maker's Dilemma"</A> which has to do with how many bits you have to carry when evaluating an elementary function just to be sure you are going to round correctly: it's more than you think, and (perhaps very surprisingly) for some functions the full answer is not even known! But we are <i>not</i> going to go into that. That's a hard problem for mathematical software designers. We're just going to walk by, whistling.
#
# Back to the sum $\sqrt2 + \pi$ above. That hex answer is the <i>exact</i> sum of the two rational numbers represented in hex by 0x1.6a09e667f3bcdp+0 and 0x1.921fb54442d18p+1, correctly rounded back into the IEEE double precision format. You can see already that there have been some changes from real arithmetic. First, $\sqrt2$ had to be rounded, to $13$ hex digits plus a hidden bit; then $\pi$ had to suffer the same kind of rounding. Then even though the arithmetic is carried out correctly, we aren't adding $\sqrt2$ and $\pi$ any more, we're adding the rational approximations thereof. Then one final rounding happens when we put the result of that addition into a double precision float.
#
# The details of how computer manufacturers guarantee that the arithmetic is done correctly, and the correct rounding is done, are beyond the scope of almost anyone's interest. We are willing to trust the manufacturers to do it right. But <A HREF="https://en.wikipedia.org/wiki/Pentium_FDIV_bug"> remembering the Intel Pentium bug uncovered by Professor Thomas R. Nicely </A> we remember that sometimes that trust is misplaced. Intel paid over a third of a billion US dollars at the time on the recall. Ow.
#
#
# <H4>Translating the IEEE guarantee into the language of real analysis </H4>
#
# We have the following rules: if $\varepsilon$ is the <i>machine epsilon</i>, which is the distance to the next larger representable machine number (after $1$, although we can talk about this for other numbers as well), and $\mu$ is the <i>unit roundoff</i>, which is half of that, namely $\mu = \varepsilon/2$, then we have the mathematical statements: if the results do not <i>overflow</i> or <i>underflow</i> then
#
# <OL>
# <li> fl$(a+b) = (a+b)(1+\delta)$ where $\delta$ is some real number at most as big as $\mu$: $|\delta| \le \mu$ </li>
# <li> fl$(a-b) = (a-b)(1+\delta)$ where $\delta$ is some real number at most as big as $\mu$: $|\delta| \le \mu$ </li>
# <li> fl$(a*b) = (a\cdot b)(1+\delta)$ where $\delta$ is some real number at most as big as $\mu$: $|\delta| \le \mu$ </li>
# <li> fl$(a/b) = (a/b)(1+\delta)$ where $\delta$ is some real number at most as big as $\mu$: $|\delta| \le \mu$ </li>
# </OL>
# That last rule applies only if $b \ne 0$. The notation fl$(\cdot)$ means "the floating-point result of".
#
# A result <i>overflows</i> if it is larger than realmax. A result underflows if it is positive but smaller than realmin. On reflection, the same thing happens for negative numbers. (Complex numbers are, well, slightly more complex.)
#
#
# Using these rules, we can <i>prove</i> some facts about floating-point arithmetic. For instance, if $a$, $b$ and $c$ are three positive real numbers, then if operations are performed left-to-right fl$(a+b+c) = $fl(fl$(a+b)+c) = ((a+b)(1+\delta_1)+c)(1+\delta_2)$. From there you can expand to get $(a+b+c) + $ rounding error; the rounding error can be written as a formula, which is $(a+b+c)\delta_2 + (a+b)\delta_1(1+\delta_2) $. By the triangle inequality, the absolute value of the error is less than $|(a+b+c)\delta_2| + |(a+b)\delta_1(1+\delta_2)|$. Since $a$, $b$, and $c$ are positive, this is less than $(a+b+c)(|\delta_2| + |\delta_1(1+\delta_2)|$, which by hypothesis is less than $(a+b+c)(\mu + \mu(1+\mu))$ or $(a+b+c)(2\mu + \mu^2)$. This means that the relative error in adding <i>three</i> positive numbers is, ignoring the $\mu^2$ term, $2\mu$. This is a nice result and says that for adding positive numbers, the errors grow essentially only linearly.
#
# That kind of analysis is <i>not</i> to everyone's taste. You should try to see what happens when one of the $a$, $b$, or $c$ is zero, though. In particular, if $a=-b$, say. You should be able to convince yourself that the resulting relative error might be infinite! In fact, this is <i>what happened</i> in our first example, above.
# +
import sys
print( 'The largest representable double precision number is ', sys.float_info.max, 'which in hex is', sys.float_info.max.hex() )
print( 'The smallest normal positive double precision number is ', sys.float_info.min, 'which in hex is ', sys.float_info.min.hex() )
# -
# <H4> NaN, or what you get when the limits are exceeded </H4>
#
# A NaN, or "Not a Number," is a special code which results when you perform something like $0/0$ or $\infty-\infty$ or $0\cdot \infty$. Maybe surprisingly, IEEE arithmetic can do some useful things with infinity (which can result from <i>overflow</i>). And even NaNs can be useful: if you compute a whole array of results, with a few NaNs in them, and then plot the results, you still get a plot (maybe missing some spots---the NaNs won't be plotted and will leave holes).
#
# What happens when you try to make a number smaller than realmin? Say, by squaring realmin? You get <i>underflow</i> (default behaviour is to replace it by zero). This is frequently, but not always, what you want.
#
# Sadly, Python actually gives an error message if you try to do 0/0 in its native arithmetic, which we think is nasty. Doubtless there will be a way to alter that behaviour, but we won't fuss now (numpy helps). <b>The following statement is a mistake of this author (RMC) caused by his forgetting that ^ in Python does not mean power:</b> We think that Python's $0^0$ is worse---Donald Knuth argues <i>very</i> persuasively that it should be $1$! See <A HREF="https://en.wikipedia.org/wiki/Zero_to_the_power_of_zero"> the Wikipedia article </A> which even says "There do not seem to be any authors assigning a specific value of $0^0$ other than $1$". <b> End of mistake </b> When we use the power operator ** correctly, Python is fine. In my defence (this is RMC speaking, not EYSC) in markdown the ^ is power, as it is in Maple and Matlab and every other language I use. Just Python now insists on $**$ (I think even Fortran allows ^ now, although it's been decades since my last Fortan program). Grump, grump.
# +
big = float.fromhex('0x1.fffffffffffffp+1023')
small = float.fromhex('0x1.0000000000000p-1022')
print('Yes, it\'s big: ', big, 'in hex ', big.hex())
# Cut and paste the hex string, then edit the last f to be an e (this is the next smaller number)
notquiteasbig = float.fromhex('0x1.ffffffffffffep+1023')
stepback = big - notquiteasbig
print( 'distance to next smaller number is ', stepback, 'in hex ', stepback.hex() )
print( 'exponent difference ', 1023 - 971 )
jumpingoffthecliff = big + stepback
print('jump over the cliff, whee! ', jumpingoffthecliff, '---in hex: ', jumpingoffthecliff.hex() )
infminusinf = jumpingoffthecliff - jumpingoffthecliff
print('Surely it must be zero: but (this is good) 0*inf is: ', infminusinf )
print('small*big should actually be ok: ', small*big )
print('Here is an underflow: ', small*small )
bad = 0
worse = bad*jumpingoffthecliff
print('What\'s 0*inf = ', worse )
print('what\'s 0^0 (We don\'t like this one)', bad^bad )
print('Well if we use the proper syntax, 0**0 is ', bad**bad )
badfloat = 0.0
print('what\'s 0.0**0.0? ', badfloat**badfloat )
print('But 0/0 generates an error (commented out, so the notebook runs to completion)')
# bad/bad
# -
# We also wanted to show the internal representation of "inf" when we used .hex(), but instead of "7ff0000000000000" it just gave inf again. Anyway inf is the normal result when you overflow: you get a "machine infinity".
#
# We see that realmax is about 4 times bigger than realmin is small (their product is 4, but not exactly 4). That seems weird, until you realize that realmax has a whole string of F's in its expansion and is <i>not quite</i> a pure power of $2$.
#
# You will at first hate NaNs, especially when your output is nothing but NaNs, but eventually come to accept them. They actually are quite useful.
# <H4> Revealing earlier rounding errors by subtractive cancellation </H4>
#
# Consider the familiar quadratic formula for the roots of a quadratic equation. Let's make it simpler by considering a quadratic of the form $x^2 - 2b x + 1 = 0$. Factoring this gets $(x-r_1)(x-r_2)$ where (say) $r_1 = b + \sqrt{b^2-1}$ and $r_2 = b - \sqrt{b^2-1}$. Feel free to check the algebra, maybe by adding the roots to get $2b$ (as we should) or multiplying them to get $(b+\sqrt{b^2-1})(b-\sqrt{b^2-1}) = b^2 - (b^2-1) = 1$, again as we should.
# +
n = 100
barr = np.logspace( 1.0, 2.0, num=n )
unity = np.array(range(n),dtype=float)
for i in range(n):
b = barr[i]
delt = np.sqrt(b*b-1)
r1 = b + delt
r2 = b - delt
unity[i] = r1*r2
plt.plot( barr, unity, '.')
plt.show()
# +
n = 250
barr = np.logspace( 6.0, 8.0, num=n )
unity = np.array(range(n),dtype=float)
for i in range(n):
b = barr[i]
delt = np.sqrt(b*b-1)
r1 = b + delt
r2 = b - delt
unity[i] = r1*r2
plt.plot( barr, unity, '.')
plt.show()
# -
# Inspection of those two plots ought to surprise you. The first one is ok---the results are all $1$ to plus or minus $10^{-12}$, which is maybe a <i>bit</i> bigger rounding error than the $10^{-15}$ or so that we might be expecting, but it's not all that bad. The product of the two computed roots is $1 \pm O(10^{-12})$. Fine.
#
# The second plot is, however, a shocker. The vertical scale runs from $0$ to $3$. Some of the products of the computed roots are as big as $3$, almost; and many of them are actually zero (which means that one of the computed roots is zero). Some of those blasted straight lines overlap, too. Those results do not look to the unskilled eye like they reveal any rounding errors. But that's exactly what's happening.
#
# This, from the venerable quadratic formula, known since Babylonian times.
#
# In detail: for large $b$, $b^2$ will be even larger. Then, subtracting $1$ from $b^2$ will leave it close to the same; taking the square root again gets it nearly back to $b$, but not quite (even in exact arithmetic). Then, subtracting this from $b$ will leave a small number. Let's take a specific example, and track this in hex.
b = 2.0e5
print( 'b is ', b, 'which is ', b.hex(), ' in hex ')
delt = np.sqrt(b*b-1)
print( 'delt is ', delt, 'which is ', delt.hex(), ' in hex ')
r1 = b + delt
print( 'r1 is ', r1, 'which is ', r1.hex(), 'in hex')
r2 = b - delt
print( 'r2 is ', r2, 'which is ', r2.hex(), 'in hex')
# We see that even for $b = 2\cdot 10^5$ that $\Delta = \sqrt{b^2-1}$ is not so different to $b$ itself, being about $2\cdot 10^5 (1- 10^{-11})$ (if we have counted the decimal 9s correctly). In hex we see the digits 1.86a000000000000 and 1.869ffffffeb075. The f's are all in binary 1111, so we have a string of binary 1s that is 24 bits long; adding a 1 to the end of that causes a carry to chain all along those 24 bits, making the 9 turn into an a. Subtracting those two things (which we apparently have to do to get $r_2$) will cancel out the 1.869ffffff with the 1.86a000000 which will cost us nine hexadecimal digits of significance and bring the lower 3 hexadecimal digits up to the front of the new number, the result. That is, we subtracted two nearly equal numbers, each about $2^{17}$, leaving a result about the size of $2^{-19}$ (to see where we got the $17$ and the $19$ just look at the values in the +p parts). This means we have lost about 36 bits of precision (leaving just approximately 16 bits left). We have made rounding errors earlier in computing $\Delta$: one when we multiplied the b's together, another when we subtracted the 1, and more when we took the square root. All that information that would have wound up sitting below the 13 hexadecimal digits was lost when we formed $\Delta$. Now when we subtract, the first nine hexadecimal digits are the same and cancel, leaving only four good hex digits and those rounding errors to move upward in significance.
#
# This is called <i>catastrophic cancellation</i> when subtraction reveals earlier rounding errors.
#
# Trying it for larger $b$, say $b=10^8$, gets zero for $r_2$ because <i>all</i> the good digits cancel out; then there are no significant digits left.
#
# It might seem like a cheat, but the solution to this for this particular problem is not to do the subtraction at all. Compute $r_1$ which adds instead of subtracts---there's still a rounding error in $\Delta$ but it's not revealed. Then compute $r_2$ by $r_2 = 1/r_1$. This gets us a perfectly accurate small root.
#
# This trick is important enough to be part of the canon. For more general problems, we have to use other tricks.
#
# Welcome to the seamy underbelly of numerical analysis. (Don't worry too much---most of the time we can treat floats as if they were just real numbers; it's only occasionally that weird stuff happens.)
b = 1.0e16
print( 'b in decimal is ', b, 'and in hex is ', b.hex() )
bigger = float.fromhex('0x1.1c37937e08001p+53')
print( 'The next machine representable number is ', bigger )
b = 1.0
print( 'b in decimal is ', b, 'and in hex is ', b.hex() )
bigger = float.fromhex('0x1.0000000000001p+0')
print( 'The next machine representable number is ', bigger )
print('The machine epsilon at 1 is ', bigger - b )
# This will work in Python 3.9; until installed, commented out
#from math import nextafter, inf
#b = 1.0e16
#print( 'b in decimal is ', b, 'and in hex is ', b.hex() )
#bigger = nextafter(b, inf )
#print( 'next floating point number after b is ', bigger, 'and in hex is ', bigger.hex() )
# Now let's try an example from a 1981 paper by W. Kahan, "Why do we need a floating-point arithmetic standard?" namely the evaluation of the rational function
#
# $$
# R(z) = 1 - \frac{3}{z-2 - \frac{1}{z-7 + \frac{10}{z-2 - \frac{2}{z-3}}}}
# $$
#
# which we may wish to write as $1 - 1/(z-2 - 1/(z-7 + 10/(z-2 - 2/(z-3))))$ which ought to translate nicely into Python. Kahan calls this a one-liner, that ought to work just fine if the conventions described above are followed.
#
# Unfortunately, we walk straight into a bog. Python apparently <i>does not</i> follow the IEEE 754 standard, but instead breaks it on purpose. On one discussion board from 2008, we find quotes that make RMC groan in pain: "If you're doing such serious number-crunching that you really want to handle NANs, you're probably not writing in Python anyway."
#
# This isn't serious number crunching, it's just wanting to plot something that has removable discontinuities :(
#
# and "For the vast majority of programming, division by zero is a mistake and not merely a degenerate case, so Python decided to treat it like one."
#
# This attitude just makes life harder for Python programmers :(
#
# The Matlab one-line program
#
# R = @(z) 1 - 1/(z-2 - 1/(z-7 + 10/(z-2 - 2/(z-3))))
#
# and the Maple one-line program
#
# R := z -> 1 - 1/(z - 2 - 1/(z - 7 + 10/(z - 2 - 2/(z - 3))))
#
# both work perfectly. The Maple one is maybe the best, because if you call it with floats, e.g. R(1.0), you get the right answer by the IEEE arithmetic (to the <i>last bit</i>) whereas if you call it with integers, e.g. R(1), you get the error message "division by zero"
#
# However, the <b>plot worked</b> even though it called R(1), R(2), R(3), R(4), and R(5). This is because numpy (as opposed to plain Python) actually does use NaNs intelligently. Whew! And, looking at the plot, numpy got the right answers. The difference is in the use of arrays, apparently (Thanks to Jack Betteridge for pointing this out).
# +
R = lambda z: 1.0-1.0/(z-2.0-1.0/(z-7.0+10.0/(z-2.0-2.0/(z-3.0))))
n = 13
x = np.linspace(0,6,num=n)
y = np.array(range(n),dtype=float)
for i in range(n):
y[i] = R(x[i])
#print( x[i], y[i])
plt.plot( x, y, '.' )
plt.show()
r1 = R( x[2] )
r2 = R( x[4] )
r3 = R( x[6] )
r4 = R( x[8] )
print( 'R(1) = ', r1, 'R(2) = ', r2, 'R(3) = ', r3, 'R(4) = ', r4 )
#np.seterr(divide='ignore') # This command seems to be ignored its own self :(
worked = R( np.float64(1.0) )
print( 'It worked with NumPy', worked )
print( 'But would fail with native Python floats, which are different for some reason. Commented out')
#didntwork = R( 1.0 )
# Should have been 2
#R(2)
# Should have been 1
#R(3)
# Should have been 1/5
#R(4)
# Should have been 1/2
# +
#
# Here's an example where the 0/0 occurs naturally in a plot
# and we DON'T want to fuss with special cases. It's nice
# that NaNs allow this to work (in NumPy)
n = 201
x = np.linspace(-4*math.pi,4*math.pi,num=n)
y = np.array(range(n),dtype=float)
#np.seterr(divide='ignore')
for i in range(n):
y[i] = math.sin(x[i])/x[i]
#print( x[i], y[i])
plt.plot( x, y )
plt.show()
| 86.045817
| 1,493
|
d82e07dd7dd6ae033373649ba087ecb0da059710
|
py
|
python
|
Taller4.ipynb
|
AngieCat26/MujeresDigitales
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/AngieCat26/MujeresDigitales/blob/main/Taller4.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="SUIcZJyyd6Vw"
# # Investigando los accidentes de tránsito en Nueva York
# + id="0qKbKxgBd6Vz" colab={"resources": {"http://localhost:8080/nbextensions/google.colab/files.js": {"data": "Ly8gQ29weXJpZ2h0IDIwMTcgR29vZ2xlIExMQwovLwovLyBMaWNlbnNlZCB1bmRlciB0aGUgQXBhY2hlIExpY2Vuc2UsIFZlcnNpb24gMi4wICh0aGUgIkxpY2Vuc2UiKTsKLy8geW91IG1heSBub3QgdXNlIHRoaXMgZmlsZSBleGNlcHQgaW4gY29tcGxpYW5jZSB3aXRoIHRoZSBMaWNlbnNlLgovLyBZb3UgbWF5IG9idGFpbiBhIGNvcHkgb2YgdGhlIExpY2Vuc2UgYXQKLy8KLy8gICAgICBodHRwOi8vd3d3LmFwYWNoZS5vcmcvbGljZW5zZXMvTElDRU5TRS0yLjAKLy8KLy8gVW5sZXNzIHJlcXVpcmVkIGJ5IGFwcGxpY2FibGUgbGF3IG9yIGFncmVlZCB0byBpbiB3cml0aW5nLCBzb2Z0d2FyZQovLyBkaXN0cmlidXRlZCB1bmRlciB0aGUgTGljZW5zZSBpcyBkaXN0cmlidXRlZCBvbiBhbiAiQVMgSVMiIEJBU0lTLAovLyBXSVRIT1VUIFdBUlJBTlRJRVMgT1IgQ09ORElUSU9OUyBPRiBBTlkgS0lORCwgZWl0aGVyIGV4cHJlc3Mgb3IgaW1wbGllZC4KLy8gU2VlIHRoZSBMaWNlbnNlIGZvciB0aGUgc3BlY2lmaWMgbGFuZ3VhZ2UgZ292ZXJuaW5nIHBlcm1pc3Npb25zIGFuZAovLyBsaW1pdGF0aW9ucyB1bmRlciB0aGUgTGljZW5zZS4KCi8qKgogKiBAZmlsZW92ZXJ2aWV3IEhlbHBlcnMgZm9yIGdvb2dsZS5jb2xhYiBQeXRob24gbW9kdWxlLgogKi8KKGZ1bmN0aW9uKHNjb3BlKSB7CmZ1bmN0aW9uIHNwYW4odGV4dCwgc3R5bGVBdHRyaWJ1dGVzID0ge30pIHsKICBjb25zdCBlbGVtZW50ID0gZG9jdW1lbnQuY3JlYXRlRWxlbWVudCgnc3BhbicpOwogIGVsZW1lbnQudGV4dENvbnRlbnQgPSB0ZXh0OwogIGZvciAoY29uc3Qga2V5IG9mIE9iamVjdC5rZXlzKHN0eWxlQXR0cmlidXRlcykpIHsKICAgIGVsZW1lbnQuc3R5bGVba2V5XSA9IHN0eWxlQXR0cmlidXRlc1trZXldOwogIH0KICByZXR1cm4gZWxlbWVudDsKfQoKLy8gTWF4IG51bWJlciBvZiBieXRlcyB3aGljaCB3aWxsIGJlIHVwbG9hZGVkIGF0IGEgdGltZS4KY29uc3QgTUFYX1BBWUxPQURfU0laRSA9IDEwMCAqIDEwMjQ7CgpmdW5jdGlvbiBfdXBsb2FkRmlsZXMoaW5wdXRJZCwgb3V0cHV0SWQpIHsKICBjb25zdCBzdGVwcyA9IHVwbG9hZEZpbGVzU3RlcChpbnB1dElkLCBvdXRwdXRJZCk7CiAgY29uc3Qgb3V0cHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKG91dHB1dElkKTsKICAvLyBDYWNoZSBzdGVwcyBvbiB0aGUgb3V0cHV0RWxlbWVudCB0byBtYWtlIGl0IGF2YWlsYWJsZSBmb3IgdGhlIG5leHQgY2FsbAogIC8vIHRvIHVwbG9hZEZpbGVzQ29udGludWUgZnJvbSBQeXRob24uCiAgb3V0cHV0RWxlbWVudC5zdGVwcyA9IHN0ZXBzOwoKICByZXR1cm4gX3VwbG9hZEZpbGVzQ29udGludWUob3V0cHV0SWQpOwp9CgovLyBUaGlzIGlzIHJvdWdobHkgYW4gYXN5bmMgZ2VuZXJhdG9yIChub3Qgc3VwcG9ydGVkIGluIHRoZSBicm93c2VyIHlldCksCi8vIHdoZXJlIHRoZXJlIGFyZSBtdWx0aXBsZSBhc3luY2hyb25vdXMgc3RlcHMgYW5kIHRoZSBQeXRob24gc2lkZSBpcyBnb2luZwovLyB0byBwb2xsIGZvciBjb21wbGV0aW9uIG9mIGVhY2ggc3RlcC4KLy8gVGhpcyB1c2VzIGEgUHJvbWlzZSB0byBibG9jayB0aGUgcHl0aG9uIHNpZGUgb24gY29tcGxldGlvbiBvZiBlYWNoIHN0ZXAsCi8vIHRoZW4gcGFzc2VzIHRoZSByZXN1bHQgb2YgdGhlIHByZXZpb3VzIHN0ZXAgYXMgdGhlIGlucHV0IHRvIHRoZSBuZXh0IHN0ZXAuCmZ1bmN0aW9uIF91cGxvYWRGaWxlc0NvbnRpbnVlKG91dHB1dElkKSB7CiAgY29uc3Qgb3V0cHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKG91dHB1dElkKTsKICBjb25zdCBzdGVwcyA9IG91dHB1dEVsZW1lbnQuc3RlcHM7CgogIGNvbnN0IG5leHQgPSBzdGVwcy5uZXh0KG91dHB1dEVsZW1lbnQubGFzdFByb21pc2VWYWx1ZSk7CiAgcmV0dXJuIFByb21pc2UucmVzb2x2ZShuZXh0LnZhbHVlLnByb21pc2UpLnRoZW4oKHZhbHVlKSA9PiB7CiAgICAvLyBDYWNoZSB0aGUgbGFzdCBwcm9taXNlIHZhbHVlIHRvIG1ha2UgaXQgYXZhaWxhYmxlIHRvIHRoZSBuZXh0CiAgICAvLyBzdGVwIG9mIHRoZSBnZW5lcmF0b3IuCiAgICBvdXRwdXRFbGVtZW50Lmxhc3RQcm9taXNlVmFsdWUgPSB2YWx1ZTsKICAgIHJldHVybiBuZXh0LnZhbHVlLnJlc3BvbnNlOwogIH0pOwp9CgovKioKICogR2VuZXJhdG9yIGZ1bmN0aW9uIHdoaWNoIGlzIGNhbGxlZCBiZXR3ZWVuIGVhY2ggYXN5bmMgc3RlcCBvZiB0aGUgdXBsb2FkCiAqIHByb2Nlc3MuCiAqIEBwYXJhbSB7c3RyaW5nfSBpbnB1dElkIEVsZW1lbnQgSUQgb2YgdGhlIGlucHV0IGZpbGUgcGlja2VyIGVsZW1lbnQuCiAqIEBwYXJhbSB7c3RyaW5nfSBvdXRwdXRJZCBFbGVtZW50IElEIG9mIHRoZSBvdXRwdXQgZGlzcGxheS4KICogQHJldHVybiB7IUl0ZXJhYmxlPCFPYmplY3Q+fSBJdGVyYWJsZSBvZiBuZXh0IHN0ZXBzLgogKi8KZnVuY3Rpb24qIHVwbG9hZEZpbGVzU3RlcChpbnB1dElkLCBvdXRwdXRJZCkgewogIGNvbnN0IGlucHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKGlucHV0SWQpOwogIGlucHV0RWxlbWVudC5kaXNhYmxlZCA9IGZhbHNlOwoKICBjb25zdCBvdXRwdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQob3V0cHV0SWQpOwogIG91dHB1dEVsZW1lbnQuaW5uZXJIVE1MID0gJyc7CgogIGNvbnN0IHBpY2tlZFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgaW5wdXRFbGVtZW50LmFkZEV2ZW50TGlzdGVuZXIoJ2NoYW5nZScsIChlKSA9PiB7CiAgICAgIHJlc29sdmUoZS50YXJnZXQuZmlsZXMpOwogICAgfSk7CiAgfSk7CgogIGNvbnN0IGNhbmNlbCA9IGRvY3VtZW50LmNyZWF0ZUVsZW1lbnQoJ2J1dHRvbicpOwogIGlucHV0RWxlbWVudC5wYXJlbnRFbGVtZW50LmFwcGVuZENoaWxkKGNhbmNlbCk7CiAgY2FuY2VsLnRleHRDb250ZW50ID0gJ0NhbmNlbCB1cGxvYWQnOwogIGNvbnN0IGNhbmNlbFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgY2FuY2VsLm9uY2xpY2sgPSAoKSA9PiB7CiAgICAgIHJlc29sdmUobnVsbCk7CiAgICB9OwogIH0pOwoKICAvLyBXYWl0IGZvciB0aGUgdXNlciB0byBwaWNrIHRoZSBmaWxlcy4KICBjb25zdCBmaWxlcyA9IHlpZWxkIHsKICAgIHByb21pc2U6IFByb21pc2UucmFjZShbcGlja2VkUHJvbWlzZSwgY2FuY2VsUHJvbWlzZV0pLAogICAgcmVzcG9uc2U6IHsKICAgICAgYWN0aW9uOiAnc3RhcnRpbmcnLAogICAgfQogIH07CgogIGNhbmNlbC5yZW1vdmUoKTsKCiAgLy8gRGlzYWJsZSB0aGUgaW5wdXQgZWxlbWVudCBzaW5jZSBmdXJ0aGVyIHBpY2tzIGFyZSBub3QgYWxsb3dlZC4KICBpbnB1dEVsZW1lbnQuZGlzYWJsZWQgPSB0cnVlOwoKICBpZiAoIWZpbGVzKSB7CiAgICByZXR1cm4gewogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbXBsZXRlJywKICAgICAgfQogICAgfTsKICB9CgogIGZvciAoY29uc3QgZmlsZSBvZiBmaWxlcykgewogICAgY29uc3QgbGkgPSBkb2N1bWVudC5jcmVhdGVFbGVtZW50KCdsaScpOwogICAgbGkuYXBwZW5kKHNwYW4oZmlsZS5uYW1lLCB7Zm9udFdlaWdodDogJ2JvbGQnfSkpOwogICAgbGkuYXBwZW5kKHNwYW4oCiAgICAgICAgYCgke2ZpbGUudHlwZSB8fCAnbi9hJ30pIC0gJHtmaWxlLnNpemV9IGJ5dGVzLCBgICsKICAgICAgICBgbGFzdCBtb2RpZmllZDogJHsKICAgICAgICAgICAgZmlsZS5sYXN0TW9kaWZpZWREYXRlID8gZmlsZS5sYXN0TW9kaWZpZWREYXRlLnRvTG9jYWxlRGF0ZVN0cmluZygpIDoKICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgJ24vYSd9IC0gYCkpOwogICAgY29uc3QgcGVyY2VudCA9IHNwYW4oJzAlIGRvbmUnKTsKICAgIGxpLmFwcGVuZENoaWxkKHBlcmNlbnQpOwoKICAgIG91dHB1dEVsZW1lbnQuYXBwZW5kQ2hpbGQobGkpOwoKICAgIGNvbnN0IGZpbGVEYXRhUHJvbWlzZSA9IG5ldyBQcm9taXNlKChyZXNvbHZlKSA9PiB7CiAgICAgIGNvbnN0IHJlYWRlciA9IG5ldyBGaWxlUmVhZGVyKCk7CiAgICAgIHJlYWRlci5vbmxvYWQgPSAoZSkgPT4gewogICAgICAgIHJlc29sdmUoZS50YXJnZXQucmVzdWx0KTsKICAgICAgfTsKICAgICAgcmVhZGVyLnJlYWRBc0FycmF5QnVmZmVyKGZpbGUpOwogICAgfSk7CiAgICAvLyBXYWl0IGZvciB0aGUgZGF0YSB0byBiZSByZWFkeS4KICAgIGxldCBmaWxlRGF0YSA9IHlpZWxkIHsKICAgICAgcHJvbWlzZTogZmlsZURhdGFQcm9taXNlLAogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbnRpbnVlJywKICAgICAgfQogICAgfTsKCiAgICAvLyBVc2UgYSBjaHVua2VkIHNlbmRpbmcgdG8gYXZvaWQgbWVzc2FnZSBzaXplIGxpbWl0cy4gU2VlIGIvNjIxMTU2NjAuCiAgICBsZXQgcG9zaXRpb24gPSAwOwogICAgZG8gewogICAgICBjb25zdCBsZW5ndGggPSBNYXRoLm1pbihmaWxlRGF0YS5ieXRlTGVuZ3RoIC0gcG9zaXRpb24sIE1BWF9QQVlMT0FEX1NJWkUpOwogICAgICBjb25zdCBjaHVuayA9IG5ldyBVaW50OEFycmF5KGZpbGVEYXRhLCBwb3NpdGlvbiwgbGVuZ3RoKTsKICAgICAgcG9zaXRpb24gKz0gbGVuZ3RoOwoKICAgICAgY29uc3QgYmFzZTY0ID0gYnRvYShTdHJpbmcuZnJvbUNoYXJDb2RlLmFwcGx5KG51bGwsIGNodW5rKSk7CiAgICAgIHlpZWxkIHsKICAgICAgICByZXNwb25zZTogewogICAgICAgICAgYWN0aW9uOiAnYXBwZW5kJywKICAgICAgICAgIGZpbGU6IGZpbGUubmFtZSwKICAgICAgICAgIGRhdGE6IGJhc2U2NCwKICAgICAgICB9LAogICAgICB9OwoKICAgICAgbGV0IHBlcmNlbnREb25lID0gZmlsZURhdGEuYnl0ZUxlbmd0aCA9PT0gMCA/CiAgICAgICAgICAxMDAgOgogICAgICAgICAgTWF0aC5yb3VuZCgocG9zaXRpb24gLyBmaWxlRGF0YS5ieXRlTGVuZ3RoKSAqIDEwMCk7CiAgICAgIHBlcmNlbnQudGV4dENvbnRlbnQgPSBgJHtwZXJjZW50RG9uZX0lIGRvbmVgOwoKICAgIH0gd2hpbGUgKHBvc2l0aW9uIDwgZmlsZURhdGEuYnl0ZUxlbmd0aCk7CiAgfQoKICAvLyBBbGwgZG9uZS4KICB5aWVsZCB7CiAgICByZXNwb25zZTogewogICAgICBhY3Rpb246ICdjb21wbGV0ZScsCiAgICB9CiAgfTsKfQoKc2NvcGUuZ29vZ2xlID0gc2NvcGUuZ29vZ2xlIHx8IHt9OwpzY29wZS5nb29nbGUuY29sYWIgPSBzY29wZS5nb29nbGUuY29sYWIgfHwge307CnNjb3BlLmdvb2dsZS5jb2xhYi5fZmlsZXMgPSB7CiAgX3VwbG9hZEZpbGVzLAogIF91cGxvYWRGaWxlc0NvbnRpbnVlLAp9Owp9KShzZWxmKTsK", "ok": true, "headers": [["content-type", "application/javascript"]], "status": 200, "status_text": ""}}, "base_uri": "https://localhost:8080/", "height": 39} outputId="bba948db-a082-4b29-9c56-29103286c3ba"
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import datetime
# %matplotlib inline
from google.colab import files
uploaded = files.upload()
# + colab={"base_uri": "https://localhost:8080/", "height": 189} id="F98X4eqr-FjI" outputId="055d295d-a50a-432d-92a8-fa31fd1e69a7"
df = pd.read_csv('accidents_sample.csv', sep= ';')
df.head(1)
# + [markdown] id="CciJWSAqd6V0"
# ## Introducción
#
# **Contexto empresarial.** La ciudad de Nueva York ha experimentado un aumento en el número de accidentes en las vías de la ciudad. Quieren saber si el número de accidentes ha aumentado en las últimas semanas. Para todos los accidentes reportados, han recopilado detalles para cada accidente y han estado manteniendo registros durante el último año y medio (desde enero de 2018 hasta agosto de 2019).
#
# La ciudad te ha contratado para que construyas visualizaciones que les ayuden a identificar patrones en accidentes, lo que les ayudaría a tomar acciones preventivas para reducir la cantidad de accidentes en el futuro. Tienen ciertos parámetros como comuna, hora del día, motivo del accidente, etc. De los que se preocupan y de los que les gustaría obtener información específica.
#
# **Problema empresarial.** Su tarea es formatear los datos proporcionados y proporcionar visualizaciones que respondan a las preguntas específicas que tiene el cliente, que se mencionan a continuación.
#
# **Contexto analítico.** Se le proporciona un archivo CSV que contiene detalles sobre cada accidente como fecha, hora, ubicación del accidente, motivo del accidente, tipos de vehículos involucrados, recuento de lesiones y muertes, etc. El delimitador en el archivo CSV dado es; en lugar del predeterminado,. Realizará las siguientes tareas con los datos:
#
# * Leer, transformar y preparar datos para su visualización
# * Realizar análisis y construir visualizaciones de los datos para identificar patrones en el conjunto de datos.
#
# El cliente tiene un conjunto específico de preguntas a las que le gustaría obtener respuestas. Deberá proporcionar visualizaciones para acompañar estos:
#
# 1. ¿Cómo ha fluctuado el número de accidentes durante el último año y medio? ¿Han aumentado con el tiempo?
# 2. Para un día en particular, ¿durante qué horas es más probable que ocurran accidentes?
# 3. ¿Hay más accidentes entre semana que los fines de semana?
# 4. ¿Cuál es la proporción de accidentes por área por comuna? ¿Qué distritos tienen un número desproporcionadamente grande de accidentes para su tamaño?
# 5. Para cada comuna, ¿durante qué horas es más probable que ocurran accidentes?
# 6. ¿Cuáles son las 5 principales causas de accidentes en la ciudad?
# + [markdown] id="QP-Y4A-3d6V2"
# Tenemos las siguientes columnas
#
# Borough: el comuna en el que ocurrió el accidente.
# COLLISION_ID: un identificador único para esta colisión
# CONTRIBUTING FACTOR VEHICLE (1, 2, 3, 4, 5): Motivos del accidente
# CROSS STREET : Calle transversal más cercana al lugar de los accidentes
# DATE: Fecha del accidente
# TIME: Hora del accidente
# DATETIME: la columna que creamos anteriormente con la combinación de fecha y hora
# LATITUDE: Latitud del accidente
# LONGITUDE: Longitud del accidente
# NUMBER OF (CYCLIST, MOTORIST, PEDESTRIANS) INJURED: Lesión por categoría
# NUMBER OF (CYCLIST, MOTORIST, PEDESTRIANS) KILLED: Categoría muerte sabia
# ON STREET NAME: Calle donde ocurrió el accidente
# TOTAL INJURED: Total de heridos por el accidente
# TOTAL KILLED: Total de bajas en el accidente
# VEHICLE TYPE CODE (1, 2, 3, 4, 5): Tipos de vehículos involucrados en el accidente
# ZIP CODE: código postal del lugar del accidente
# + [markdown] id="KsHqg54gd6V3"
# Primero cargue los datos del csv a un dataframe.
#
# **Nota:** El archivo se encuentra separado por punto y coma ( ; ) en lugar de coma ( , ), así que se debe ingresar el parámetro sep para leer el archivo. Aquí puede encontrar información:
#
# https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.read_csv.html#pandas.read_csv
# + id="a3aS_HjiFe_i" colab={"base_uri": "https://localhost:8080/", "height": 225} outputId="df66eba5-0e71-49a0-d9c6-fc1d97a5ffc7"
##Intento 2
df['date_format'] = pd.to_datetime(df['DATE'])
df['ano-mes'] = df['date_format'].dt.to_period('M')
# + [markdown] id="fP2uBUny362i"
#
# + [markdown] id="icNi1Weed6V4"
# ## Parte 1: Accidentes a lo largo del tiempo
#
# Agrupe los datos disponibles mensualmente y genere una gráfica lineal de accidentes a lo largo del tiempo. ¿Ha aumentado el número de accidentes durante el último año y medio?
# + [markdown] id="3UtkblnF36mh"
#
# + [markdown] id="5rYaj1Bt35-f"
#
# + id="ZTRWb-ml0tEj"
# + id="aK3EN6uPGW-4" colab={"base_uri": "https://localhost:8080/", "height": 244} outputId="841c1643-4b6c-4ada-8a22-d20caf29bf56"
df['DATE'] =pd.to_datetime(df['DATE'])
dx=df.grouphy(df['DATE'].dt.to_period('H')).size()
dx.plot.line(color='green',title='ACCIDENTES POR MES. NY', xlabel='fecha',ylabel='No HAY ACCIDENTES')
# + id="61pElde8pVwg" colab={"base_uri": "https://localhost:8080/", "height": 434} outputId="c262c721-4f5a-45d6-fb29-116bdf234a85"
df["DATETIME"].groupby(df["DATETIME"].dt.to_period("m")).agg("count").plot(kind="line",figsize=(8,5))
plt.title("Accidentes entre 2018 y 2019",fontsize=18,color="red",)
plt.xlabel("Meses por año",fontsize=15,color="blue")
plt.ylabel("Numero de accidentes", fontsize=15,color="yellow",)
plt.xticks(fontsize=14)
# + [markdown] id="NqOw_sajd6V7"
# ## Parte 2: Puntos calientes de accidentes en un día
#
# ¿Cómo varía el número de accidentes a lo largo de un solo día? Cree una nueva columna HORA basada en los datos de la columna DATETIME, luego trace un gráfico de barras de la distribución por hora a lo largo del día.
# + id="IC6Do3s3d6V8" colab={"base_uri": "https://localhost:8080/", "height": 371} outputId="33b74919-d118-4fcc-f4ff-4af84e4b0c11"
df['TIME']=pd.to_datatime(df['TIME'])
DF['HORA'] = df['TIME'].dt.hour
dx=df.groupby('HORA').size()
plt.bar(dx.index,dx.values, color='gray')
plt.title('ACCIDENTES POR HORA - NY')
plt.ylabel('NO HAY ACCIDENTES')
plt-xlabel('HORA')
# + id="wzrHdilkKhgT"
# Escriba su código aquí
areas = {'A': 40, 'B':20}
df2['areas'] = df2['barrio'.map(areas)]
df2
# + [markdown] id="DbVnzWWGd6V8"
# ## Parte 3: Accidentes por día laborable
#
# ¿Cómo varía el número de accidentes en una sola semana? Trace un gráfico de barras basado en el recuento de accidentes por día de la semana.
# + id="wjmKw6mGd6V8" colab={"base_uri": "https://localhost:8080/", "height": 345} outputId="4be12722-10c0-4c7b-fb40-dcbf464083d0"
df['DATE']=pd.to_datetime(df['DATE'])
dx=df.groupby(df['DATE'].dt.day)_name()).size()
plt.bar(dx.index,dx,value, color = 'blue')
plt.title('ACCIDENTES POR DIA EN LA SEMANA')
plt.ylabel('NO HAY ACCIDENTES ')
plt.xlabel('DIA DE LA SEMANA ')
# + [markdown] id="7fNI3lqed6V9"
# ## Parte 4: Análisis de comuna
#
# Trace un gráfico de barras del número total de accidentes en cada comuna, así como uno de los accidentes por kilómetro cuadrado por comuna. ¿Qué puedes concluir?
# + id="pf5H533Td6V1" colab={"base_uri": "https://localhost:8080/", "height": 130} outputId="7e840722-fe77-423c-fe60-a81a138ff440"
# Use la siguiente informacipón de cada comuna ("borough") para realizar los análisis
borough_data = {'the bronx': {'name': 'the bronx', 'population': 1471160.0, 'area': 42.1},
'brooklyn': {'name': 'brooklyn', 'population': 2648771.0, 'area': 70.82},
'manhattan': {'name': 'manhattan', 'population': 1664727.0, 'area': 22.83},
'queens': {'name': 'queens', 'population': 2358582.0, 'area': 108.53},
'staten island': {'name': 'staten island', 'population': 479458.0, 'area': 58.37}}
dfc = pd-DatFrame(borough_data).transpose()
dfc
# + colab={"base_uri": "https://localhost:8080/", "height": 577} id="r0ZSpsZ_o1kq" outputId="e471df7e-a29a-4d3c-ef4d-8d9be1b6889b"
fig.ca = plt.subplots(1,2, figsize=(14,6))
fig.suptitle("Accidentes VS km2 por comuna", fontsize=18, color="blue")
df["BOROUGH"].value_counts().plot.bar(ax=ca[0],fontsize=10,color="purple")
ca[0].set_title("Accidentes por comuna",fontsize=15,color="pink")
ca[0].set_xlabel(" comuna",fontsize=15,color="pink")
ca[0].set_ylabel("Numero de accidentes",fontsize=15,color="pink")
dfc["area"].sort_values(ascending=False).plot.bar(ax=ca[1],fontsize=10,color="purple")
ca[1].set_title("Area por comuna",fontsize=15,color="pink")
ca[1].set_xlabel("comuna",fontsize=15,color="pink")
ca[1].set_tylabel("Km 2",fontsize=15,color="pink")
# + id="ZDt6VtoTd6V9"
# Escriba su código aquí
# + [markdown] id="T7GhKEKRd6V9"
# ## Parte 5: Análisis por hora del comuna
#
# ¿Qué horas tienen más accidentes en cada comuna? Trace un gráfico de barras para cada comuna que muestre el número de accidentes por cada hora del día.
# + id="26MV8Ak0d6V9"
# Escriba su código aquí
#ejemplo
areas = {'A': 40, 'B':20}
df2['areas'] = df2['barrio'.map(areas)]
df2
# + id="W4nxjB-TtWuC"
b=df.loc[(df["BOROUGHT"] == "MANHATAN") & (df["HOUR"]).count()]
b.head(3)
# + [markdown] id="6PPahsuVd6V-"
# ## Parte 6: Causa de accidentes
#
# ¿Qué factores provocan la mayoría de los accidentes?
# + id="C2TiU-2gd6V-" colab={"base_uri": "https://localhost:8080/", "height": 605} outputId="60c9bcf5-8dd4-4810-e56d-ea8de6342471"
fa=df['CONTRIBUTTING FACTOR VEHICULO 1'].value_conunts().plot(kind="bar",figsize=(23,6),color="violet")
plt.title("Factores que provocan la mayoria de accidentes",fontsize=20,color="dark")
plt.xlabel("Causantes de accidentes",fontsize=15,color="blue")
plt.ylabel("Numero de accidentes",fontsize=15,color="orange")
Text(0, 0.5, 'Numero de accidentes')
| 76.933921
| 7,349
|
5bb5c19ae66d5c08cdfbf6a338bc8219e2467c2a
|
py
|
python
|
Sentence_classification_task.ipynb
|
fathyshalaby/Somemoredeeplearning
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="qhI4EoTlcg46"
# #Sentence classification task
# + id="Qto-YKerTm4M"
# RUN ONCE load models + data
# #!python -m spacy download en_core_web_md
#import nltk
#nltk.download('stopwords')
# get data via wget if it doesn't exist
# #!wget -nc http://öä.eu/sst5.data.txt
# #!wget -nc http://öä.eu/sst5.labels.txt
# other option: load data via gdrive mounting
# drive.mount('/gdrive')
# # %cd /gdrive/My Drive/sst5/<
# + id="wr1hE0V4uTWw"
# %matplotlib inline
import numpy as np
import pandas as pd
import seaborn as sns
import numpy as np
import spacy
import sklearn
import nltk
from nltk.corpus import stopwords
from nltk.stem.porter import *
from collections import Counter
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
from sklearn.metrics import accuracy_score, confusion_matrix
from sklearn.base import TransformerMixin
from sklearn.pipeline import Pipeline
from sklearn.svm import LinearSVC
from sklearn.model_selection import train_test_split
from sklearn.decomposition import TruncatedSVD, PCA
from spacy.lang.en.stop_words import STOP_WORDS
from spacy.lang.en import English
from spacy.lemmatizer import Lemmatizer
from spacy.tokens import Doc, Span, Token
from matplotlib import pyplot as plt
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.linear_model import SGDClassifier
from sklearn.model_selection import GridSearchCV
flatten = lambda l: [item for sublist in l for item in sublist]
mapl = lambda f, i: list(map(f, i))
filterl = lambda f, i: list(filter(f, i))
SEED = 42
# + [markdown] id="2T6-zHimCDaG"
#
# **Data Exploration**
# + [markdown] id="eg_r_IXB6uAm"
# ***sst-5 is a sentiment analysis collection, created based on the movie reviews in the IMDB
# website [1]. The provided dataset in this assignment consists of a set of sentences, and their
# corresponding labels. The labels are in 5 levels, annotated from very negative (-2) to very
# positive (+2). The dataset is available for downloading in http://drive.jku.at/ssf/s/
# readFile/share/14324/4544703232741430166/publicLink/sst5.zip***
# + id="pBR6YUNajnJy" outputId="141f6f6a-b3fa-48e7-e3c2-a4ab6b8ad641" colab={"base_uri": "https://localhost:8080/", "height": 407}
df = pd.read_csv('sst5.data.txt')
X = np.array(df['text'])
Y = np.array(df['label'])
df
# + [markdown] id="-1AuNyyGndul"
# The data also has the issue, that the classes -1 and 1 have significantly more members: Over 6000 in these 2 vs 500 in the other 3 classes.
# + id="TrO3IUZNndul" outputId="4dc9181a-dba5-422f-b3c3-aa6748265d7b"
df['label'].hist()
# + id="w7S469CIkB2Y" outputId="5b61fe91-cf52-4fd9-8965-39a3f660ea36" colab={"base_uri": "https://localhost:8080/", "height": 108}
labels = pd.read_csv('sst5.labels.txt',header=None)
label_id = {lab[0]: lab[1] for lab in labels.to_numpy()}; label_id
# + [markdown] id="qjJYTAF5fOFq"
# **Manual data inspection for adjectives**
#
# + id="IJzo6PlY3hR_" outputId="6dbfa851-1e1f-464b-a7b2-a81f2a16c763" colab={"base_uri": "https://localhost:8080/", "height": 72}
# loading a prebuilt the english language processing pipeline - that starts out with 3 components. this can take some time
nlp = spacy.load("en_core_web_md")
print(*nlp.pipeline, sep = "\n")
# + id="CUeWXo3ryBvh" outputId="5a450521-17b0-4d39-fb51-19401b82818e" colab={"base_uri": "https://localhost:8080/", "height": 35}
# Loop to get just the adjectives from each review
docl = list( nlp.pipe(X)) # returns just an iterator -> make list
adj_filter = lambda tok: tok.pos_ == 'ADJ'
words_adj = [filterl(adj_filter, line) for line in docl]
df['words_adj'] = pd.Series(words_adj)
df.head() # we can see that for sentence 1 that adj were found which could be because of the default dictionary not having all words
max_sent_len=max(len(docl[i]) for i in range(0,len(docl)))
print('maximum sentence length = ', max_sent_len)
# + [markdown] id="mYj45Ooj3ppj"
# Analyze the most common adjectives and plot histograms
# + id="AxDkOiUT1HcV" outputId="530e2f4e-5d0a-45d5-9aff-ec0c13b92d0c" colab={"base_uri": "https://localhost:8080/", "height": 1000}
# Print the most common adjectives
adj_count = dict()
for id in label_id:
adj = df[df['label']==id]['words_adj']
print(adj)
adj = flatten(adj)
adj = mapl(lambda tok: tok.lemma_, adj)
adj_count[id] = Counter(adj).most_common(10)
print(label_id[id],adj_count[id])
#Potting of all review types
def plot_reviews(adj_count,reviewtype):
labels, values = zip(*adj_count)
indexes = np.arange(len(labels))
#sns.set(style="whitegrid")
ax = sns.barplot(indexes, values, palette="BuGn_r")
plt.xticks(indexes + 0.1, labels, rotation=30);
plt.tick_params(axis='x', labelsize=13)
plt.title("Most Common "+str(reviewtype)+" Adjectives in Reviews")
plt.show()
for id in label_id:
plot_reviews(adj_count[id],label_id[id])
# + [markdown] id="-bSdME23cuyb"
# ## Task1: Dataset Preparation
# + [markdown] id="iOUif-4Jndu4"
# If you want class-balancing for task 4.3, rerun this notebook with this cell enabled
# + [markdown] id="5ZU56mY7ndu4"
# minl = sum(df["label"]==-2)
# subsamples = [df[df["label"]==i][:minl] for i in range(-2, 3)]
# df = pd.concat(subsamples)
# df['label'].hist()
#
# X = np.array(df['text'])
# Y = np.array(df['label'])
# docl = list( nlp.pipe(X)) # create new smaller docl
# + [markdown] id="HDGeVzJ2c9uy"
# ### Preprocessing.
#
# Most of this is done by the en_core_web_sm piepline, that contains an english tokenizer, a tagger that cleans up unneccesary spaces, a parser and a named entity recognizer and has built in stopword detection.
#
# But we can improve it by adding NLTK stemming and building a custom dict.
# + [markdown] id="1NY91v4vqPK0"
# Stopwords - can be checked with tok.is_stop()
# + id="L0arQqSEqJ9j" outputId="30098c7d-4f76-4ad6-e4ee-4d566cafc4c3" colab={"base_uri": "https://localhost:8080/", "height": 74}
# use spacy and nltk stopwords and put them into the default nlp stopword detector
print("Adding nltk stopwords", set(nltk.corpus.stopwords.words('english')).difference(nlp.Defaults.stop_words))
nlp.Defaults.stop_words = nlp.Defaults.stop_words.union(nltk.corpus.stopwords.words('english'));
print("Total list of stopwords", ", ".join(sorted(nlp.Defaults.stop_words)))
# + [markdown] id="fuGM-2zC4lFC"
# To add a custom stemmer, you can extend tokens with getters
# + id="Ha0ScbQ2t9-k"
stem_function = PorterStemmer().stem
Token.set_extension("stem", getter=lambda tok: stem_function(tok.lemma_), force=True) # force=True to be able to rerun the line
# + [markdown] id="CLIS9STY4wQy"
# Print 3 sentences with to show lemmatization, stemming, stopwords and more:
# + id="tCpN9OAwgq-c" outputId="d1300dec-e7d1-4365-bb05-f277bd6bcb58" colab={"base_uri": "https://localhost:8080/", "height": 890}
print("Word\tLemma\tStem\tstop")
for d in docl[:3]:
for word in d:
print(word, word.lemma_, word._.stem, word.is_stop or word.is_punct, sep="\t")
print() # newline between sentences
# + [markdown] id="sbJ3B4vsdFC5"
# ### Creating a dictionary.
# To reduce the dict size we use lemmatization and stopword/punctuation removal. This leads to 250k -> 15k -> 12k vocabulary size.
# + id="rNmLjvOxdq31" outputId="8d9b9f42-e2e7-4780-a6c2-f662aa69c530" colab={"base_uri": "https://localhost:8080/", "height": 146}
vocab0 = set(flatten(docl))
print("Size without any filtering/stemming", len(vocab0))
print("Examples", list(vocab0)[:50])
vocab1 = set(flatten([map(lambda tok: tok.lemma_, line) for line in docl]))
print("Size with lemmatization alone", len(vocab1))
print("Examples", list(vocab1)[:50])
stemf = lambda tok: tok._.stem
tokfilt = lambda tok: not tok.is_stop and not tok.is_punct
stem_doc = [ mapl(stemf, # only use stem
filter(tokfilt, line)) # filter out punctuation
for line in docl]
vocab2 = set(flatten(stem_doc)) # make unique set out of list of list
print("Size with filtering/stemming", len(vocab2))
print("Examples", list(vocab2)[:50])
# + [markdown] id="ht1jsb02dFR2"
# ### Splitting the dataset.
# + id="ufW42nLFdwRf" outputId="5af5a50d-23aa-4b32-ecf1-4d97f294da9c" colab={"base_uri": "https://localhost:8080/", "height": 219}
text_train, text_test, label_train, label_test = train_test_split(docl,Y,test_size = 0.2,random_state = SEED)
print(*zip(label_train[:10], text_train[:10]), sep="\n")
# + [markdown] id="3AIVztgFdsdZ"
# ## Task2: Feature Extraction
# + [markdown] id="ZmyQDsyyd6Rd"
# ### Creating sentence vectors.
# We are using [TF-IDF (Term-Frequency, Inverse-Document-Frequency)](https://en.wikipedia.org/wiki/Tf%E2%80%93idf) appraoch which includes the term and document frequencies for term weighting.
#
# Our 2nd approach is the spacy builtin [word2vec](https://en.wikipedia.org/wiki/Word2vec) which only has implicit pretrained weighting.
#
# + [markdown] id="CUCio4_OcPgo"
# **word-ids**
# + id="nE72vONDcTRP"
sec2vec = {word:i for i,word in enumerate(vocab0)}
# + [markdown] id="88OujLDWcKNF"
# ### TF-idf with scikit learn
# + id="TpuAJ33UeGOk" outputId="a5ae6c97-bff6-4aa9-fcc0-55af44076961" colab={"base_uri": "https://localhost:8080/", "height": 290}
# as our data is already tokenized, we can skip tokenization by setting the tokenizer to the identity function and skipping lowercase
tfvectorizer = TfidfVectorizer(vocabulary=vocab2, tokenizer=lambda x: x, lowercase=False);
tfdoc = tfvectorizer.fit_transform(stem_doc)
print(tfvectorizer); tfdoc
# + id="c8ajLBj7aHNv" outputId="c7e7f0a3-0db6-40c3-afcd-c3c01af14752" colab={"base_uri": "https://localhost:8080/", "height": 35}
print("The sparcity rate (rate of emtpy elements) is", 100*(1 - len(tfdoc.nonzero()[0])/np.prod(tfdoc.shape)), "%")
# + [markdown] id="_Q8mXs4pb7id"
# As this array is 11844x12081, you can't use PCA which is why we can only use TruncatedSVD
# + [markdown] id="L8XvHILXXw5o"
# ### Word2Vec with spacy
# Spacy also offers a built in word2vec in all models bigger than small, and word2vec is the state of the art for text vectorization. The vec size is 96
#
# + id="YdIk3rwFXv1w" outputId="e653288d-a582-48cc-8e94-7cd8b7484637" colab={"base_uri": "https://localhost:8080/", "height": 35}
w2v = np.array(mapl(lambda tok: tok.vector, docl)); w2v.shape
# + [markdown] id="jyaEcfe8d-lJ"
# ### Dimensionality reduction.
#
#
# + id="B5LsYcGWcpMV"
tf_20 = TruncatedSVD(n_components=20).fit_transform(tfdoc)
w2v_20 = PCA(n_components=20).fit_transform(w2v)
# + [markdown] id="asFqSrb8eP9t"
# ## Task3: Classification
# + [markdown] id="B53Lx24IeR-s"
# ### Training classifiers.
# + id="krQkMCQuHGLa"
def train(clf,x_train,y_train,hyperparameter):
"""
clf: The specified model
x_train:The sentence vector from the training dataset
y_train:The corresponding labels from the training dataset
hyperparameter:Dictionary with parameters names (string) as keys and lists of parameter settings to try as values.
"""
classifier = GridSearchCV(clf, hyperparameter,n_jobs=8,cv=10)
grid_result = classifier.fit(x_train,y_train)
print('Score: ',grid_result.best_score_)
return grid_result.best_estimator_
# + id="_WutqH2Guec6" outputId="2ff6ea4b-74b7-4875-ccb3-e1aea3fcd2c7" colab={"base_uri": "https://localhost:8080/", "height": 35}
print('Baseline = ',max(list(Counter(Y).values()))/sum(list(Counter(Y).values())))
# + [markdown] id="fdbO5AZ_wREq"
# **Classifers**
# + id="ty_ZOXNkB6KB"
names = ['Gradient boosting','Random Forest','Linear Support vector','SGD Classifier']
model_list = [GradientBoostingClassifier(),RandomForestClassifier(),LinearSVC(),SGDClassifier()]
hyperparameters = [{'learning_rate':[0.1,0.01,0.001]},
{'n_estimators':[100,1000],'random_state':[42],'class_weight':("balanced",None)},
{'loss':('hinge','squared_hinge'),'C':[1.0,8.0,10.0],'random_state':[42]},
{'loss':('hinge','squared_epsilon_insensitive'),'epsilon':[0.1,0.25,0.01]}]
# + [markdown] id="FwH_LWdQe2Rj"
# ### Results and comparison.
# Report the evaluation results of the models on both validation and test sets in a table. Compare different models, and conclude the best performing approach on the dataset.
# + id="GThjff0y_eG4" outputId="e5db70f5-1d8a-413b-ee81-197216762e89" colab={"base_uri": "https://localhost:8080/", "height": 459, "referenced_widgets": ["cf20713d6d5b475c94cd08ccc8c334f0", "eaf50e9f64714bde9669d355d789122f", "1717f15018f541078076a4bff97a0cba", "d5ecef9880bb4cc69af25cdc91f19fcf", "697c652d864f4b0494b97f604c96bd89", "8af81d353c45443290fa6c3a36356bac", "7792ec1821b74ce39419e8f90bee6d4f", "1299c68936af46f3b78ac1520ceb2397", "9190d69ab6c9478797f5144c34242ebd", "8ed1389830924befabdd65210ee301b4", "0f00aa6cb3474d58a34218ab26d0316d", "36f2a940988245669c9e5d570fb42377", "939436f9c67e4ba39718cf42c077a5d6", "b4a8964dc81a45fd81ce6db9d93f25b2", "cd10f63e4ff54570af1ab9f640be1d36", "9782f5302572480f9901ca791b841f05"]}
def scores(clf, X_val, y_val,X_test,y_test):
"""
Input:
clf: The model
X_val:The sentence vector from the validation dataset
y_val: The corresponding labels from the validation dataset
X_test:The sentence vector from the test dataset
y_test:The corresponding labels from the test dataset
Output:
acc_val: Accuracy for the validation set
acc_test: Accuracy for the test set
conf_mat: Confusion matrix for the validation set
conf_mat_test: confusion matrix for the test set
"""
confidence = clf.score(X_val,y_val)
y_pred = clf.predict(X_val)
acc_val = accuracy_score(y_val, y_pred)
conf_mat = confusion_matrix(y_val, y_pred)
test_labels_pred = clf.predict(X_test)
acc_test = accuracy_score(y_test, test_labels_pred)
conf_mat_test = confusion_matrix(y_test, test_labels_pred)
return acc_val,acc_test,conf_mat,conf_mat_test
def evaluate(model_list,vectors,hyperparameters,names):
"""
Input:
model_list: A list which contains all initilized models
vectors: A dictionary which contains the name of the sentence vector as keys and sentence vector variable as a value
hyperparameters: A list of dictionaries that contain the parameters for each model
names: A list containg the names of the models as strings
Output:
table: pandas dataframe which displays the classifers the corrresp[onding sentence vectors and how they performed
conf_list: A list which contains the corresponding confusion matrix for each classifier
"""
test_acc_list = []
val_acc_list=[]
sentence_vector_list = []
conf_list = []
name_list = []
for key in vectors.keys():
i=0
print('Sentence Vector',key)
x_train,x_test,y_train,y_test = train_test_split(vectors[key],Y,test_size=.2,random_state=42)
x_train,x_val,y_train,y_val = train_test_split(x_train,y_train,test_size=.2,random_state=42)
for model in model_list:
print('classifier = ',names[i])
clf = train(model,x_train,y_train,hyperparameters[i])
#print('finished training')
val_acc,test_acc,conf_val,conf_test= scores(clf,x_val,y_val,x_test,y_test)
test_acc_list.append(test_acc)
val_acc_list.append(val_acc)
sentence_vector_list.append(key)
name_list.append(names[i])
conf_list.append((conf_val,conf_test))
i+=1
table = pd.DataFrame({'Model':model_list+model_list,
'sentence vector':sentence_vector_list,
'Validation Accuracy ':val_acc_list,
'Test Accuracy':test_acc_list})
return table, conf_list
vectors = {'tf-id':tf_20,'word2vec':w2v_20}
result, conf_list= evaluate(model_list,vectors,hyperparameters,names)
result
# + id="WHHKyQ_0-Iyf" outputId="ad4aaec0-da1a-45f3-e8fa-9bc5801fed7b"
def plot_confusion_matrix(num_classifer,conf_list):
"""
Input:
num_classifer: The classifer which we want to see the confusion matrix of
conf_list: A list of confusion matrices corresponding to each classifier in the table above
Output:
Two confusion matrix plots of both the validation dataset and the test dataset
"""
conf_mat, conf_mat_test = conf_list[num_classifer]
for name, data in [("Validation Data", conf_mat), ("Test data", conf_mat_test)]:
# Plot Confusion Matrix Data as a Matrix
plt.matshow(data)
plt.title('Confusion Matrix for '+name)
plt.colorbar()
plt.ylabel('True label')
plt.xlabel('Predicted label')
plt.show()
print(len(conf_list))
plot_confusion_matrix(6,conf_list) # Plots the confusion matrix of the best model based on the test accuracy score shown above
# + [markdown] id="lZwngz53ndvc"
# ### Summary Findings: Explanation of behaviour
# Looking at the matrix above and the results table in the Results and comparison section, we can see that the accuracy scores that we got when evaluating the model are due to our model focusing on two classes mainly out of the 5 classes in this dataset. We can see that our model predicts the best on class 1(negative) and class 3 (positive) best. This performance can be due to many factors that include the data distribution and the vocabulary in the specific classes. As shown during our Data exploration section of our notebook there are many overlapping words between the classes which further contributed to the accuracy score that we got for this task. Furthermore we can also see that the model somewhat performs well on class 4 also (very positive) which again could be due to the overlap in features between the classes. The model doesnt perform so well in predicting coirrectly for the class 0 and 2 which are very negative and neutral respectively, which could be due to the vocab in these classes doesn't occur enough in the samples or they include characteristucs like sarcasm which is not easy to detect.
# + [markdown] id="Bq6p8gJfsWFU"
# ## **Task4: Visualization and Analysis**
# + [markdown] id="WWJo5ljosdw8"
#
# + [markdown] id="AIE6JFlJndvd"
# ### Questions
# We want to find out two things about our preprocessing:
# 1. Does the stemming + lemmatization preprocessing (used with TF-IDF) actually help?
# 2. Does applying PCA to to our word2vec help?
# 3. Would balacing the classes help?
# + id="mbuk6vAyndve" outputId="ed5bb40f-e812-400b-e673-1a582b0aaa19"
#tF_idf + random forest, trying some other preprocessing like the none-preprocessing
tfvectorizer = TfidfVectorizer(vocabulary=vocab0, tokenizer=lambda x: x, lowercase=False)
tfdoc = tfvectorizer.fit_transform(stem_doc)
tf_20 = TruncatedSVD(n_components=20).fit_transform(tfdoc)
# word2vec, if it performs differently if we don't PCA it in between
names = ['Gradient boosting','Random Forest','Linear Support vector','SGD Classifier']
model_list = [GradientBoostingClassifier(),RandomForestClassifier(),LinearSVC(),SGDClassifier()]
hyperparameters = [{'learning_rate':[0.1,0.01,0.001]},
{'n_estimators':[100,1000],'random_state':[42],'class_weight':("balanced",None)},
{'loss':('hinge','squared_hinge'),'C':[1.0,8.0,10.0],'random_state':[42]},
{'loss':('hinge','squared_epsilon_insensitive'),'epsilon':[0.1,0.25,0.01]}]
vectors = {'tf-id':tf_20,'word2vec':w2v}
result,conf_list= evaluate(model_list,vectors,hyperparameters,names); result
# + id="1XQbhmrMndvf" outputId="eeec59af-d331-43b1-cec0-218458942e65"
plot_confusion_matrix(1,conf_list) #confusion matrix for Random Forest + TF-ID
# + id="Y371v0_endvh" outputId="3ad136c2-efd6-4917-c099-3bbc2f80f129"
plot_confusion_matrix(5,conf_list) #confusion matrix for best model
# + [markdown] id="zP-hztRPndvi"
# ### Results for balanced classes
#
# When rerunning the notebook with the 1st cell in preprocessing enabled for ballanced classes, we get the following results (included as a screenshot image):
#
# <img src="data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAAA7YAAAD1CAYAAABk3mnHAAAVTHpUWHRSYXcgcHJvZmlsZSB0eXBlIGV4aWYAAHjarZpZkhw5kkT/cYo+AmDYj4NVZG4wx++niCCrWEtPtcgwhJnJSA93uJmaLnC687//c92/+JO9NZdybaWX4vmTeuo2+KH5z5/+vgaf3tf3ZxYfv+/+8r67+fsh4y0d8jnM1/H5Hgbv598+8OMaYf76vmvf31j7nij8PPH7E3Vl/bx/v0jet8/7IX1P1M/nh9Jb/eUWvida3wPfUr5/089lfb7p3+6XNypV2pSO69mJIfr3tX1WED9/B38jXy12jgvv5xzN8S3E/j0ZBfnl9n589/73BfqlyD9+cn+sfrK/Lr6N7xHxD7Us3xrxw1/+IuQ/vB9/Xt9+f+H4c0X26y/KDvan2/n+vXe3e8/n7kYqVLR8EfWKHX6chgMnNxffxwqvyt/Mz/W9Oq/mh1+0fPvlJ68VejC6cl1IYYcRbjjv+wqLJSY7Vvlutiy+91qs1m1F9SnpFa7V2OOOjf4tO472pWg/1xLedfu73gqNK+/AoRY4WeAjf/ty/+mX/83L3btUovAQa69WrMtUcJahzukrR9GQcL99y6/AP17f9vvfAQuo0sH8yty4weHn5xQzh9+wFV+fI8dlvn9GKLi6vydgQVw7s5gQ6YAvIeZQgq9mNQTq2GjQYOUWk006EHK2zSItxVjMVWuma/OZGt6xlq2Y3oabaESOJVZ60+OgWSll8FNTA0Mjx5xyziXX3FzueZRYUsmllFpEcqPGmmqupdbaaq+jxZZabqXV1lpvo1uPcGDupdfeeu9jmBtcaHCuwfGDd6bNONPMs8w62+xzLOCz0sqrrLra6mts23FDE7vsutvue5zgDkxx0smnnHra6WdcsHbjTTffcuttt9/xs2vfrv7p9V90LXy7Zq9TOq7+7Brvulp/nCKITrJ6RscsBTpe1QEAbeqZbyElU+fUM9+NocjGIrN643ZQx2hhOsHyDT9791vn/lHfXG7/qG/2f3XOqXX/H51ztO7PffuLrm3p3Hod+0yhauoj08cxYXm3cshc3DKwim0tH9os/LrBV+n2Sp0rUzHrhXys91tqvvy+nx71fV9OxHcn4dh+2OzMzR7Fwop5TO6sxRv9lEQDrVVO3HPfmalyvr2HxAW7txK567ObW6nTkOVPbafCoWHulKHZM7enuLuzrHCX2c3tpFrPvSMDltF87HdaWse31byzs3stN25/16pxcWg8nRNumCCzqtNhylJ2STZqX22q3Fw2tHV1+KiF1S43OyQw4QpYuIZ66EKrV6fEIZQDVczFi/uYrWWDQDqEbsj7XK0tXyjrie2428c6+VLgMMcu+dQJf7OG2GjW2PHENOHzV5YBlPLyefYSgNGhp003cPZ2CZeR+eS8XHUKYHlHILFzTiHOoRXdxFV9v5fu3TxamLnOHmjEaZt783UehzDdDZgNhLdwY+VjMN0qdsKi7yrDTaPHsw4TDATo9rxHLWex3oPgltZyDPKoedupcVN0xjGeSTsvB5R6y8jr7FzuWWDipO5zPflV+9y2ArcPskvf7u8AFpgUrUboCH5VSz1Q2G5Fbdy7B0Fmo4qeFl1zh+pSjtK4ZJ6o87m+7cY8M3k3czuDc549+t622qAVHdWtILiYHwdWomdzOZU+nZ1m2POIThBgNJqCwmz7jdLhgDujaPImSv7qQ73aljOsTFgvzZ1YuX8GPPvFogDfOJnlrojfOYArqWS43QMUVp0sooeKMSxM8Ex21+w3r+6AwA0UaJUipMFpOEBmoS8mlvOxMqguH96Kuwy4RXOy1r4FU5NVINBTsceQC34G3NK3e2AkXyCcOiNmKAAoiLVnlmZlX0vTQ6yRAQAFtqa/OZyeITw3dm0UKMCo+2xfIiOg1fYWGSHmSXpPSXetCO/GYG0Pc6U11EAMzV6xM3fuhn3hmUsbGXExdW/ghQJR6l37PQYeMfB9p1H8WKn0WcsC21KNuHS1fpcrY+WRN4OWqxXfRzBwllrPlXECDo3Vc3Q8w0MgheWGMwAE1JX8QQIWJiyYs2l4LxgHylglMkpiY5btmSxuYI367N7iZBR93UzxzgL5/kOJY6IQoTokjVm3rea/3FD97GeKzOaNdIn19JhwkNQ7FmZpM7LtDBZ8H3F1SKJTbExd6lCrrnk5DFakq6nsozYmX0bqNfu5xSx7z7TLgsIwjz1VqAoyQB2OS1cvaHFBQyjBOOdYKqMxkje0VuzNuZXZ287zmL9Adtdsffg2aTwdP3s5D/hh7waW8K2ZWdt0N4S9TqI8G2qwdGxoJC3vfQAk9w7wg60VwylNYSU4uOdAVyhUGDnPOUeZMfXOpMeJj6v0qdM5esfKgVVAXO5eeAt+7imDBZ3c9Qe5hZhGGQ9xdL+NFkeSJeuCf2/scHgBxpphLN+u9G50pqaAvVPUDLe9cAELUhgWkUSNg8aMiAPxGb09CSDQE2DLlOQ4T1dqhX/hHqRiMPOnOB/pEzeGRwefusHbmdMFjogCqCO6sCAIec63dswIVOKnpAtl44P7KS0lTZ+bCjen0Qr0XlHmhEEZCBfMOuk1orBwK9mPq0mDcdEi4s09oArHNBygqx0dRaoSiobQ7uipC3YWFO8MyKzA33jg4LcIN9GtdCVHq4/TRui4n+5G4sjduN8Bs9R9/DncItfp0BaLT/iejluCeXNneRVDMOrE0w1IEFZegxLr1hqiLU3CyMFfmgEAUWbqg6nEW0D4dhJySE4CBDiAG6DN3WCn1qgjNDKis0Cxe2YWUlhVnIAw1oCKA4RSIEhM2smGYfOgh7JlUTXsOOIp+AVkaKJ+7oQ8UecnQFgUZmNwt3hDQjPgjtRfd1V1G9AsY8QAzHMPOCDSMZus2ld8NvBDP0EzM0SAg0c1Jv4w140PVUYOo4Fty1e7E/Apjg+1A0KDSlXFd/DjCHxRhiyb3INhtwAkbgsDl5muASThHK598ZPS1Ma91JQQHju5J0wGBv9Uh0pjKJBnzB8kOBCkcdM8R66c2oyeIUdsBf6yNGC5kJKJfaRxez1TRe3aBUf0nfq0GUOuT3U3ZYSBKu54wrELH32nf8U1/QLDwVwB+QlvXcB/+i5OFN9vfXLOQbp37hlW5QsrmaAH39PoJOvFNlPNQwjYiyaDdgyXOl2qQ7nQci43Sw7TUB1sPdxn9akRNMcUCBMyvvlGG3i1LYHwDPElWjCyuE+XInYbVMFYFQM4mXT1FcZCdQA78tp8yjDfAh7hloa/fn5FY50l2UAgm8PkkSu24sTlhsjoZU+1nBbj47i2R1R73UhQRg6P3MVmKcuToClKQPwM5y99RF2xZCQXOJLerXFK7kwrdeML3gkWVKQqQiDmRT6VNTLoJmlhFP1wiBttBGv1u0j8PwbOMNuWQbP/EN+mM98FhgI01jLaFidgSIkmHkk26ovebZpNjBl3QpU7MYMbc+rx4o17xQxqa4MZgM4gJtzqwI4k+cCxSeGuYus89W3+LMWcnIhKCZGIllGZMUeHVwF/fBtCJ6rfUx6cJbauQI9t7QE+wtd02EFOHm9ZTHuJi4sQHdFj+QkEkG5ovdIy8NENQdV59trUWuLA9BM2BsES7iTaRIOxqqSoymbmkhp3Qj1j2MwxXmldJCFoh60OwEGMurj47WbEgiZiycTa6xzyIkGjDDszgxAIBEFWPoeERPaEzExhF0ewWBu2r6IMw+FwYcU0yb1EHX5iCvHt6ANmXK2+5zlgqNiGkuTEM8XVkTdSK/DzzEXtxHXmGiH1CowZvoE0OqrGtU5C0EhJ8lY4l7mr3TGYgtnwXDAITLd3k3VaNl3iRNQdM6au4D9ICiZKKtgGjQpHXlwX8CoDru74QtaGCapkZEQM5DEUx0FeXQ640GuoaYxYO4gjHGihuC7yfSEU17BA6+zJJJsl4j0L05pxiECwFBfBv+fTMpY0in5mufMEjTPiBcVgap8T57yP28GZ7rQ0u01KOolTwtFVsld2nilg42URUlISzDXaRLsu1wzEiaI4sWvAHEvDA0yBwsGZxMsdnZgCx51ALDkF/sF71AzE7Ej941ZwOHTKi1oyNgLRp0q4O8X405qi5DFHNGHiO3IJ3sE3VI0I8SkAkrkHBoTEEIZUCErGDnlCyyTJsTya4qFJLCm2Bk2Z2PAulcKnXx2ohYSkGRj7CS+xoGPn0JYtL4aUKrPqZhK5F11MDjaaRAvtwcgOQq7GTGdATQoqivxiuRJJM0AJIiBQYBeUBA6MiavDmeLAXccBwZdjMKIoDr5yctOMOAZG2XgpnryA5LE3oIZQBSPCAvEkCCJZQ0NJ2UdiRTIcdUUTYPH0KBj5mYFdxCMBH/dkEFCGKkArbggdBL7aCidvYRbRfoQT8sOOVkjZVqWZqUCPYibCCkqWPAvAEXl8yzM3UGgjXnO36WqjHTbC1pBW+GV5Ro7JV3in7m/bB1JPNLUTyJbsMJ3QxgvJERuhXiyo74yucIsbgVZYfxn0Le5QL6JE0tqk7WuaFwgIp02ZGJhaCbQKZUnKTFbUJr+dNq0oQR6EFcerRyu9RNEKUKbWDEIWwyJB1IcC0jwSQRJSE3XU5vY0XOau/jg8N01nNcggOTEA/64gy2kJ3lE5JHM+j0cn4lExTurxLvj8fbAvA19gpV+njRtFemgGeNy5IIq3AQGyT8TTQ2rmt/bRPR9AyIApVMIvKQpgOBKenR34ZJAz9oGUU7VvCKjePtYgwx9xqR7GXCxfVWVJtdj5jbPsIj7NVRUmoJEY6I9/+wVwb2QEtA93GO80Z2Uotbs2iO7IlUIuv9M2AsXCYsIlsn90Dd/OoqMIBlTchm+bYKBIJvHU3DVqrhWTKcjRCHJHo1jmYJ1mDZuKr0COwERFEOBWEB5JjmgalplilAQ7emz8wEtPD3+eS07m9AMpuJJimSoL2DIjZmV4FEu7seSyLWB5wuOHydIjg8pQNNWHrEgaUjokrgxyAv7A5MpI/UjidXYRYPzfwqeRhKBz7qttMnsl6d4CPUGl9Xll6oSqEuKxwPLfqCcrVJby2Jr1IG7CJFUMJOUhtijwlW5a8EFopLCiMnAVgvmGPCOM2ACVn/5nx/XhmXGYCuI2viRHWW0PENfQaVARue5ZE6E6i68xUCGjBxtU4orecHpXSKzqUpLf4i0Qh0pIHZs26AAnowGP87728ZJEjlvHnZaNObWMhQpv+pW2CSI13/Kc2EL8NkmyIjsYBiRkyM7jiOYO2reTJOWLuCqOiBSnyu2y9mrDCwxUR9uEKATktuUTq+fz5lkddcQt4G6UzhoMZ8h+4q4RirHKTm5e2FLGQNfZiM/VExtW9MSQ5ISgwQXTNF6kBm3Q0cRCJM4LG8XAyLhi/V5OtQGVjdQw85A7OFjWF2Zwv7zqJRdMLFeDGCpEgEUgEFD4PIgdMOxxVLlAY5h1bgoKjBRLe0TXync3sLxVgoRB6+DHC/wuHJUHgwKscGfU1WVsJRKP1G5CGgwMcF+UQskIA4eYC6yqoqDn8guDfZH/SXTR9nN52UfWj+ZfFCuT0mBbFtESmda8BizoTGp53oeYKQFZ+HBCUbxR2gRIsBjo6c2uVIVxnO5cW0pu9xwCKAYtYj35ALdAovSUFwOcm2ycj/hn6FzxTWxEdBgOtuDeRmFMg6xjpQumzSUQZdaxWdo2Oli6UJhuxiNo7xtLBW5klOXZWZybkxQ/C4kPlfPSdJyZx9tp4x6lwtlEZA4/0KFYNDTiRcmoDDGDA/4GPA2WiVkBkALfwrKhbFwi8IUKiDJggMyHWQtYqIjta1LRbpjrKUui4qIkep5RHYaaZS7cHQZGbuLt452HUJE1Q4sCXNiTvs6UgQU1oPZYKLk6yA8ui8lxCviOvl/IsZetR6K13Co3cyIygZXHYnc/bLdn8jYU3VBlssnb3+55QA4OV5enB/6yvQiWkgsJk2zOC7mEgMeLee2w5GF6dBiVMgbnQmw9WRMbQ4KEAJOe78A9txsWBl7AACJLlG5C6/gpSmV+6qlSJR7aMji7WGGYseDYbCbGXfWIeIFR2AXPqYfk8CiKEaimdpqxNMrraEHAxgOG7ImpLSr5kuWXLAhKy6iNMiENakepKU1KeohEncDmk64ledH94nABDW6bVRE5cqMLc8oM9u0O6pQVuhMOkfKoZVO26u2yW4GwARV+KGMDo/YaDqZBO5mABVuG40H/ybRZj6Y6wl8M8Ytv67QsD2tkq7B2iSsKR3QBFwIzfqSceP10tGYMPvEpOGqDK2MJUf+3YvfeMNIFVZXTH+Ng8xjEGjvdgmtIphAK0nRE828TAnoY4zqoiOyEE12QNjlGLgHbjrPDBxw5vWRkhWv5FYTMtjyz06WCYPJtE5MamuMda9q22IgH1cSPfm+wGpVFebCK2ErRDMfh95E/gQtcwpTBt7luW9F5PSQbegSg/dJWNUPinDEQYuj59LO0Du2tsAQZV1/1kIAJM7pC4bqemrrp59IjGIVemKaBLbgWhTsEHHl8v01WlXGBBxNzOp6jPtpOB2Kw3/HQmJNgaSsDpMwcz+GIEgkqzxtr90Ex4OrRCMdoX0LPmS5z19E0hlCPMBQLHIEGjmsY0yGPR6jSgzbqh1hHojCBNuJl0UkCOLeBBY1qG8blnFO1dasdW9xIxC7XSlAy/TcJ+V0UUMm6M4p6VLGHFKWjDXpcUwpxhYFu2sEzPcHVfwxi1o7SB9MMwIYSGZ43ah+Tu8WcYYmOHkximunR0M58wDLAINqX0zyIGZBt7WjJEVVWJY2EOdAAGh+vNlpwLRd+UZTDKslU6DkABbdyYTbcGBYX0zTSdto4QxYDLv4WzA+2g8sIM+SamGXORQeCsMmd4EG0T68ARs7So1P6AyM60ifChmjvMoFNsWC7r25V28KFlWG2Mcdo+dFzCRwRLp53i/4rE5dMxKEIjB3p6ortyCFmeOiAUkdCNy6KH4KeRGnzCTpkoODUl/RQUpoIizA6hYjMXBBqhNXo9bzlbZayEo0zrSNKk+nqR/nbaOgIXnXSS1BWAHKepu/aQo7a9TuZTN0hiPb2HJKeY22xIQNSoQ1wRjpnzDbxFAeOEufCmOPoUYayM0mgOogrbancQKHxi006fAuhkTUXPamdV+rPGBZS0IxqXQlyllQ2bG0O4vabg/ZwzqmwOtbGouQvEKgGLjwtD2NqD6NjjzzOZ+LsQFrLWPg4EcwbtkSiu8vQI387I6O2VR8sDTkmozjh/eD/0Xf3Tw/8T9+TXZBNo8loDGK5yLUe+pM2uhcMiEQDZiemyKVi0xhE92+eZXpnoO8lEwAAAYRpQ0NQSUNDIHByb2ZpbGUAAHicfZE9SMNAHMVfW6WilQ52EFHIUJ0sSBVx1CoUoUKoFVp1MLn0C5o0JCkujoJrwcGPxaqDi7OuDq6CIPgB4uTopOgiJf4vKbSI8eC4H+/uPe7eAf5Ghalm1wSgapaRTiaEbG5VCL4igBH0IY6wxEx9ThRT8Bxf9/Dx9S7Gs7zP/Tn6lbzJAJ9APMt0wyLeIJ7etHTO+8QRVpIU4nPicYMuSPzIddnlN85Fh/08M2Jk0vPEEWKh2MFyB7OSoRJPEUcVVaN8f9ZlhfMWZ7VSY6178heG8trKMtdpDiOJRSxBhAAZNZRRgYUYrRopJtK0n/DwDzl+kVwyucpg5FhAFSokxw/+B7+7NQuTcTcplAC6X2z7YxQI7gLNum1/H9t28wQIPANXWttfbQAzn6TX21r0CAhvAxfXbU3eAy53gMEnXTIkRwrQ9BcKwPsZfVMOGLgFetfc3lr7OH0AMtRV6gY4OATGipS97vHuns7e/j3T6u8HeDFyqdsP0c0AAAAGYktHRAD/AP8A/6C9p5MAAAAJcEhZcwAACxMAAAsTAQCanBgAAAAHdElNRQfkAx0LNBV+cP5bAAAgAElEQVR42uydd3jN1//AX/dm78gQI0FiBkmM2COJWhHUjFpVNUttRdGqPWrvfpUqitoqNg1BbIKITYggJLL3vff8/rjcJEjEaPHreT3PfZ7kfM7nfM55n/k+7zMUQgiBRCKRSCQSiUQikUgknyjK1LsJUgoSiUQikUgkEolEIvl0FdtHG65IKUgkEolEIpFIJBKJ5NNVbI0czKUUJBKJRCKRSCQSiUTyyaKQe2wlEolEIpFIJBKJRPIpo5QikEgkEolEIpFIJBLJJ63YalJVUgoSiUQikUgkEolEIvl0Fdu7C89KKUgkEolEIpFIJBKJ5NNVbKUIJBKJRCKRSCQSiUTyKSMPj5JIJBKJRCKRSCQSySeNtNhKJBKJRCKRSCQSiUQqthKJRCKRSCQSiUQikUjFViKRSCQSiUQikUgkEqnYSiQSiUQikUgk/wypqam0aNECKysr2rdv/9bh/PHHHzRu3PiTl4evry+///77W707duxY7OzsKFSo0AdNQ3h4OAqFApVK9do0vej3TZkyZQo9e/aUFUkqthKJRCKRSCQSyetZu3Ytnp6emJubU7hwYXx9fTl69Og7h7tp0yaioqKIiYlh48aNbx1O586d2bdv33tP96FDh1AoFLRp0yaH+4ULF1AoFHh7e+crnJ9++okuXbq81t/u3bvp1q3bG8czIiKCWbNmERYWxqNHj9453eXKlWPFihUvuc+bNw9PT883Cutt0/SqvHB0dMzhNnr0aH799df3nu8rV65ET08Pc3NzzM3NcXZ2pnv37ly/fj3fYXz11VeMHTtWKrYSiUSSG3u+/BwfHx98fHyIzFBrHUUmzRt+ho+PD83aznzjMDMST+Lj48NXc8Ly9JcWsw0fHx/6rrv90cgjKiqKJzHp/+/yOSX6CY8fx/wr3zo3rhM+Pj5sjUnLKhMJR/Dx8cF/6LE8323XuCEtOm/Ulc0GnzV9pb/Hp7/Dx8eHn+8n5hme0CQTFRVFbLq2bN/bMQAfHx/WPE55/wkXGXRo0hAfHx/m30v8T7UjCXd/1LUjL/5adT/6j5XZu9v64+Pjg5//EtmYf0LMnj2bwYMHM3r0aKKiorh37x79+vVj+/bt7xz23bt3KVOmDPr6+h9t+u3t7QkODiYmJqt8//7775QpU+b9NUdCoNFo3kmOtra2FCxY8I3ffZW1tFu3bqxateol99WrV78XJfVToFatWiQlJREfH8+BAwcwMTGhatWqhIaG/ncqv5BIJJJ/kBVlbAQgADH8ZqwQQojkR7/p3MwL9XzjMFOf7hSAcO0bnKe/pAcLBSA8p134aOQBCLvyW//f5fPkElbCwLT8v/KtJ+f7C0DUnH9Z53Z7Y0MBiK5HHuT5bmFDPWHtMlsIIcTuri2FT4Mmr/R3b08jAYivrz/NM7z48DECEF7rbwohhLj717fC29tbrI5Kfu/pjgkboas3pb7Y/59qR+LDfxDe3t7C29tblDc1EICoVNdLeHt7i8+/OvKPldmRJawEIBRKA3EmMUM26J8AcXFxwszMTGzYsCFXP2lpaWLQoEGicOHConDhwmLQoEEiLS1NCCFEYGCgKFq0qJg5c6awt7cXhQoVEitWrBBCCPHjjz8KAwMDoa+vL8zMzMSvv/4qxo0bJzp37qwL+86dOwIQmZmZQgghfvvtN+Hs7CzMzc1FiRIlxJo1a3TuderU0b137Ngx4enpKSwtLYWnp6c4duyY7pmXl5cYO3asqF27tjA3NxeNGjUST548eWXanse/T58+YuHChUIIIVQqlShatKgYP3688PLy0vkdOHCgcHR0FBYWFqJKlSoiKChI2zbu3p0jne7u7rp4jB49WtSuXVsYGxuLGzduCC8vL7Fs2TIhhBB9+/YVbdu21YU/YsQI0aBBA6HRaHLEcf/+/cLY2FgoFAphZmYmunXrJoQQYvv27aJ8+fLCyspKeHl5ibCwMN07xYsXF9OmTRNubm7C0NBQJ9/nRERECD09PREeHq5zCwsLEwYGBuLJkyciICBAVKpUSVhYWAhHR0cxbty4XPMse5pUKpUYNmyYsLW1Fc7OzmLhwoU5/K5YsUKUK1dOmJubC2dnZ7F06VLt+CMpKUcazczMRGRk5Evl5XVp/vnnn4Wbm5uwtLQU/v7+IjU19ZX5/mJ5eo6fn1+OPGnXrp1wcHAQlpaWol69eiI0NFQIIcQvv/wi9PX1hYGBgWjsbSqquhvl69ekSZOPqv7rI5FIJP8CrqYGBK6/C2OseXzyD/SNXdDLuJPDz8MzO1m/9zh3nqrwqNuK7q1r6paVqNPusHzRKq480adNr/I5A9eksf33ZRy/eBebMrXp0asVtvrvviBFnXqfP35ZzYU70dgWL0+Hnl9S0tIAgMzk6/y2bBNX7idRvn4rerasjgKIOrKcX4Me0fSbLzg6/3/czTCjUdeB+Lpac3L7ZgDSE44SEFSL5vUdcg0neNEsDicZM6BDGWYs2onG2plug/pR2tzgmTwiWLtyI5euPaJQudr07vk55noKAOKv7mX5xkNEppnSsH1vfCs55LQaH1vBr4ceUqv/cBpYG6FKvcr02ZuxKtWZbzuUyPP9W4c3senvUzzFBh//3jStYMOT4zu5lJyJRh3Ppm1HaNeqHprMaLauXs+ZSzfRK1iK9j164lHQGIDJkydjYteazh63WHnYkZEjK79Rvti6TcPF5BeuzP4TBowHYP/kC+gZFmZ2DQeEJpndK5cRFHoHhYUDXq170rTSy1aBIrW9aPg0U/d/4u2D/PL7bmKNy9LJI6clIjPhJit/Wc+ViKdYFSlNu57dKV8gky07tKsGngTv5Eij3pR39KRhw0KUMNZ2r3nJ4XV5/CJBw9ajUJrQzBr27RhMmiYUY2WWNffg+t85fD4MfbtSdOjTi7JWhnmWlb3zfyYkswgjh3UGIOyX2WyLyWT06JGkxx9m5sKjFGv5Dc43NnPesRUDqtsTum8VmwIvkCDMKF+nOT1bVM/1+2XM05g6fSGGFrUYPsBHa9HespjVV2LpOnwUxYz08p3nlsUnEBio/Xu9qx0dr8YwO2AvPlZGedZHgFtH/uSP3adIwJJKDdrTpWH5V5bZF0mP28/PdxMoWNedx0cvMmzjHQ51z7J4JYUf5feNf3PziQq3+m35urlHnvVEaJKZMnUuFsU7MrCLCwCLpk8lzaIBw/rV0JWHIV/XZPmyffQf/f0ry10FO+Nc81Uvdiezf7lAwZpf0+uzwgBsmzuDa2onRg7r+J/oa44fP05aWhqtW7fO1c/kyZM5ceIEISEhKBQKPv/8cyZNmsTEiRMBePToEfHx8URGRrJ//37atWtHq1atGD9+PAqFgps3b7JmzRpAu2Q3N5KTkxk4cCCnT5+mbNmyPHz4kKdPn77k7+nTp/j5+TF//nw6duzIxo0b8fPz4+bNm9ja2gLapdW7d+/GyckJX19fZs6cybRp03L99pdffsmQIUPo378/e/fupUKFChQpUiSHn2rVqvHjjz9iZWXFvHnzaN++PeHh4TRt2pTRo0fnSGd2C+ju3bspW7Ys2rnaLGbNmkWlSpVYuXIlJUuWZPny5ToZZ6dhw4bs3r2bLl26cP/+fQCuX79Ox44d2bZtG97e3syZM4cWLVoQFhaGoaG2LVu3bh07d+7Ezs7uJYu5o6MjPj4+rF69WrecdtWqVTRr1gw7OzvMzMxYtWoVFSpUIDQ0lEaNGlGpUiVatWqVZ3latmwZAQEBnD9/HjMzM9q2bZvjecGCBQkICMDFxYWgoCB8fX2pVq0aVapUeSmNL5KfNG/YsIE9e/ZgbGxMnTp1WLlyJX379s13fWjTpg3ff/+97n9fX19WrFiBoaEhI0eOpHPnzoSEhNC7d2+Cg4NxdHRk946ZnNzrmK/wazaPlhZbiUTy37PYTvMuImzKLhBCCLGrsZOwKbtAmCgVOott+NZBwkCpEEYFSgiPktp3qvbWzrir0yOFn7OlUCgMRIlSRYWRVbEsi60mUwzzKiqU+gWEV8P6wlpfKeyq9BCp6ne02GrSRUdnS6FnYC/qetUVhY31hZF1dXEvTSUyEk+LanYmwtimvPisrpsARO1BO4UQQpz/qYoARFlnK1G0RDGhr1AIfeMS4lpKppjQQmtVNDB1FW2Hnc4znMklrIRS31JUsLIVJRwLCEDYuA7XyiMjSvgVsxBKPQvhXtVdGCgVolC9H4VGCBEVPFkU0FcK24p1RO0KNkKhNBaj997Paf26M0UAotrPF4UQQtw/2EoAomNgZJ7vX/+tm1AoFMKqREVRvqiZUCiNxdyQaBHyUwdRwlhfKPVMxWfNvhMaVZzoUqGAUCiUorSHuyigrxQGpqXFzscpOqu1qX0HYamvFLbl1r1VudrerLhQKI3FjdRMoVHFCQdDPVHUWxvW6g4lhUKhJ8pXrSFK2xoLpZ6FWPUo+SWL7YoyNkKhNBFCCJFwe5UoZqQv9AxtRKkiFsLS2VxnsdWoEoVfYTOhZ2AnqtWoJmwN9ISxjZd4mnxH1KrmJABhVc5TfHc5RlxZWlsAYtK9hNfKIa88fhF1ZoxwMtIXNq4zxMUZ1QQghobG6J7/0r6sAISLW1VRxFhfGFlVE1dSMvMsK98WMRfG1p/pwlhXzlYnj+eW6KLNqwpA1Ft5Xdxa/6UARMFylUWVcnYCEO1WXM/z+wOdLISegZ14kqEW4tk3TWyavVObsq6crQDE33FaC1te9ejezoECEA4Va4qaHo4CEE1nXHipzL6KC9M8BSCm3LwrihrpCctiw3TPEu+tFYUN9YShhZOoWrGIAETDqSfzrCfqjIdamXrv0YXjYqyvK4+TS1gJPcPComlBU6Fv7JJruYtXaXLN18zUm8JSXymsXcZrrU1pd4SJUiFKtAz4z/Q5a9asEQ4ODnn6cXFxETt37tT9v2fPHlG8eHGdxdPY2DiHRdDe3l4cP35cCCFesrjlZbFNSkoSVlZWYtOmTSIlJSVXC9uqVatEtWrVcjyvWbOm+O2333QWxIkTJ+qeLVq0KFdL2XOLrRBClCpVSly9elV06NBBrFmzRixbtiyHxfZFrK2tRUhIyCvT9TweP/zww0tuz62bQghx8uRJUaBAAVGsWDGxdu3aXL+VPZ5CCDFhwgTRvn37rDZPrRZFihQRgYGBOuvl8uXL88zX1atXizJlyujed3JyElu2bHml30GDBonBgwe/1mLr4+MjlixZontv7969Ofy+yOeffy7mzp37yjS+KNf8pHn16tW65999953o06fPG1lsd+/eLfT19V/5TmxsrABEXFycEEKIbt26iTFjxogq7oYi7YFzvn5Vq1Z9bZ3s3r27sLe3FxUqVNC5nT9/XtSoUUN4eHiIqlWripMnT+qeTZkyRZQsWVKUKVNG7Nmz543qv9xjK5FI/hXKD6pIwt3ZqAT8dj4G5245LSR9e/yCwrQyoZE3CLnxiPFV7Dm3rBNHEzK4/WdXdt5J4PPfQ7hz4z57hmRZ3+JujmbW4Ug+W32aQ/sPc269L9HnltP7dNQ7xTct9gDr7iRQqP4YNgQEcunEdLq2cyc4IYMLk3pwOjqV1VdOc+DIRdY0duL4gjacT86y/pWfcpz7d+4SPL4yqrRwlj1K5oe/9gNgVWIKm2Z6vjYcoU5m+qVw7kTEMMHFmthr80jWCCL2fM3Oe4m03nKRC2cu8PegaqRdms+GJ6nM7TyDFNNa3L1whKMX7+FtCXO7jshp/SoxEi9rI64t+AOAkxNPo2dYkDl1CuX5fvfB6zG2aUnEzYtcuH4SJxtTFg36G49x6+lVyAw9oxIc2DmDyMAerLkcS72Zx7gecoEbp+eQmXKD/j0P6+KQkXiS346GcuVwq7fKnzrTWyM0aYw+9Zi4mxOIylDTfOZnAJw2qknP4fu4fOYEh7e1QaNO5PczT/IMb3OnEURk6vHntXBuRDymn6lhVlyTTmPh1YIxey5w6sQpVrdzJu3pYY6kFmLPxi8BqPTTemaUt8kRZn7kkFsev0jU8cFEpKuoMaU9pXtorRGbhgc9K6u76LvpOs6t13Hr4hlCjgzCRHGD4QEReZaV/JB8rTJnr0ewpb0zD84a0fnLwdwPO8eJ0wcAOLL4ep7f7z+hKurMaMaExpCZfJGlD5Mp03vCe21b8qpHlyZuAGD28j8IDrnHlG/7Ujr61ktl9lVMmRWGkWUdRpYsxuzahUiMmMP+OO3++P3dRvAoU8nO29c5cymSAa6OXJj/bZ71JF+rRDIeUnbmbu6GB+da7g7Fpeear5sTizCrqj3x4VO5nqoiOuQHUjWCL6bX/s/0Nba2tkRHR+d5au2DBw8oXry47v/ixYvz4MGDHGFktwiampqSlJT0xnExMzPjzz//ZOnSpRQuXBg/Pz+uXr362vg8j1NkZKTu/+wnB+c3Pl27dmXhwoUEBga+0oI9a9YsXF1dsbKywtramvj4eKKj87bAOTk55fm8evXquLi4IITA398/37J6UQZKpRInJ6ccMnjdt9u0acPDhw85ceIEhw4dIiUlBT8/P20/d1J7Noe9vT1WVlYsXbr0tWl9Hq/s330xn3bv3k3NmjWxsbHB2tqaXbt25Svc/Kb5bfI9Rz8UGYmNjbZvUqvVjBo1ipIlS2JpaUmJEiUAXoqvADSIfP3yw1dffcWePXtyuI0YMYJx48YREhLChAkTGDFCO84ICwtj/fr1XL58mT179tCvXz/UanW+0ysVW4lE8q9gV6U3qrQ7/BERypboVBr4l9A902REsudpGrbuEyhlog8KA778wQ0hVPz2KJlbK65pB5pttcsAq/XJWobz6G/tYUEXfmqJm5sbzcecBeDSjsh3iq+RtRf+FWyIPDgYR2trGg0OomiVlrS3N+Hi1vsoFArGf1YDNzc3xl+MQWjSWZ/twCA/72IAFK2vVcKfql4+ZON14egZFcPPyRxQUL2YGUKTSaJaELHpBgB96muXGtadfZLY2Fg62JuwMjIJVdotanq44+5Rk6spmaQ++ZN0kbPpn9atFIn3Z3M5OZ3xp5/gUHMODgbKXN9PSb3Nsfh0bCr0xUJPgb5pBe4+ieH6oZevu7i38TIAI7+qpB0kVhpIZXNDos/szFKui42hTQ1X7J8ty31TbMpPpqypAcE/BhM6LQB9Y2dmVLIDYNzw9pg+WYhnBRdcvP/UdtSqvDvg1VfjMLVrR9sSFqA0pvt3Wcvdjax8GNG2Jnfmd6NcCQf8/rwFgOo1nXp+5JBbHr/I9iF7tcvI9K5x7LwZNS2NeHBoEHEqQUrUaoQQlPlWO1lk7zmT2NhYAto751lW8oPr4KFUKe2Inak+NX4YQnn7RBrVcKOQfTWdXPP6vnP7ORgpFewbd5rHJ35CJQQjhpZ/r21LXvWowsh2mCgVdK5REiunioSkW/H5N41er9A//B9/PknBunwLAg8ehBbOCKFh7HKtPAOuxGJoVZuGz5YFzw+L4PGDU6jT8l9PXoWeYUHmdq1PEQeHPMtdXvnaYk4z7aTP8SjOTjiCoXkVxpct8J/pa2rVqoWxsTHbtm3L1U+RIkW4e/duVl29d++lZbpvorympGS1/S+e8NukSRP279/Pw4cPKVeuHL169XptfJ7HqWjRou8ki65du7J48WKaNWuGqalpjmdHjhxh+vTpbNiwgdjYWOLi4rCystItL35x+fBzcnN/zqJFi0hPT6dIkSLMmDEj33F9UQZCCCIiInLI4HXfNjU1pV27dqxatYrVq1fzxRdf6Jb0durUiZYtWxIREUF8fDx9+/Z9aSn1qyhcuDARERE58uU56enptG3bluHDhxMVFUVcXBzNmjV7rQzfJM3vytatW6lXT9s2r127lu3bt3PgwAHi4+MJDw/XfTd7fAWCTKHO1y8/1K9fX6dcZ8/LhIQEAOLj43X1b/v27XzxxRcYGRnh7OxMqVKlOHXqVL7TK/fYSiSSfwVjmxa4mRmwYMMYNApDBjtZsPB5A6dnjoFSQWZ8llUt9aHWomSpr8TQWtsxRWaocTXVR53+MGsgaKLdp+c3dnKOwbp5sZLA8beOr0Jpxp+XHvH94Z1s27aNrRs3Mr7fdq6WvU9LQyUojZk5M+eJzk4OZmRktdqvl0ke4WiHZK8OQ99c23RHZ2qV5fSYi5wIjaVEzbqY6CkwsW7FzJmt8pzFdP9+KGJeD4ZuncTF5Ax6zWwIkOv7enqmKBUKMhOzTuM9d/QIqYYlqFM95yz68/y6rzsFO4OYTA1Kfcts8jV/p/KkUJoyq1FRWu8bzbRL9ynSYB2WegqE6ik1a7TncZEOrPr1T8rZraOs25zXK8r6StQZWRabtIdZJy4n3Z9DlfZDKddjOsu2LICNX1B/2oXXhpkfOeSWx9lRpV7nuxDtjPrAltlPcb7H8HOP+bmA+bNyoLUmalTRHDl2GQtnzzzLinYAk2XVSn2FpVjfLGuYMM/bi3Fh5iz6YxkNarjj6uSgq7+5fb9KsUpMKW/D94FjCIp9jJlDVzrlU6nOd9uSRz0q3moBMZH92L5tO9u3bWXz8hns2HKZlOgdeYZ5YeI8AKJOjKJhwyz3yz/PhGErMddTosnM2isZc+44l5MyqVurTK71pFZl7d5pkW3i4kWZZ68XeZW7vPK1WLXZlDBezbGxfxMR+pASbX7DUPHf6WusrKyYMGEC/fv3R19fn8aNG2NgYMCBAwcIDAxkxowZdOzYkUmTJlGtWjUUCgUTJkzI19U2r6JSpUpMnz6de/fuYWVlxdSpU3XPoqKiWLJkCWvXrkWj0VCiRAn09F7eW96sWTMGDBjA2rVr8ff3Z/PmzYSFhdG8efN3koWzszOHDx/GxUW7p3vlypWcOnWKihUrMmPGDPT19bG3t0elUjFt2jSdogHg4ODA/v370Wg0KJX5s4Ndv36dsWPHcujQIUxNTalevTq+vr5UqlTpJb9PnjwhOjoaV1dXlEolrVq1YufOnRw8eJD69eszb948jIyMqF37zVYbdOvWjTZt2pCZmcnBgwd17omJidjY2GBsbMypU6dYu3btS/cI165dm/T0dG7evImlpSU9e/bE39+f+fPn07x5c8zMzHLsa87IyCA9PR17e3v09fXZvXs3+/bto2LFijoZxsTEEB8fj5WV1Utx9ff3Z9q0aXmmWaPRULlyZYoWLZrva4vUajX37t1j9uzZHDp0iOPHj+tkYGRkhK2tLSkpKYwePTrHew4ODty+rb1FIr/W2Ldl7ty5NGnShOHDh6PRaAgODtaO8yIjqVmzps6fo6NjDgv265AWW4lE8u+gUDC0lDXnxwRg5tCNIoZZzY9Cz4pRZQrw9OpAFu47z51LfzNw7Hn0jRwZWtScCt+3AKDfgCVcDDvL5G6Ls2Y8G2sHI2fOJFO1tif6d9czfPhwjivfrVGOudQfp2IuTLxuxcDxc1mxULuM60lMOlW+rYBQp3IgyZ7anmW5/sc0RoyahIXe60ePegoFqvQ7aNTircMp+XUDAMaPXsOdyDtMbtyQBp/58TRTw/CKtqTF7UNTzIMqpWxZNW40P80OweCFIE0dvqaDvSl/95mGvnEJfq5iD5Dr+0aGhehV1JyYsCFsOn2TsMCf8azvRfeFWiuSUgGazCckqdSU6qE9XGNSj5+5cjecrVNbcy9dRalubd9rkao1tT2ZqTfY9TSVdjO0M9KZKVe5kZqJWTFPqpS2Y+/0HfkKa0CHEqTFBfL1kl2EntpF/2mXshSMe9rBkWPN2rhYJrFo9a2XutGUe4m8aJN/X3K4FzCIJLUGr4XbOHr0KEePHiVwn/b6mV1D92FRdAhW+krOfTeei3fus3taS7y9vVn0ICnPslLKRJ+MxFMsP3iaE/tW8tO9hDzjcSg8CT3jYtStVo57u4ejejbLn9f3Afxn+ZCRFEKP4IeUH/7de29a8qpHAyqXoaLPCFz9erD497V0sDNFnXbvpTKbA6Fi2B+3MLKso5P30aNHmVW7EMlRv7P+SSr+zR3JTL7EN6uCuH/rJD71vWjWYRlKg9zriVLfDmt9JbFhiwg6d4EdS/ryMCN3i0de5S6vfFXoWzO/sSNRJ/twKjGDnhM9/3PdzdChQ5k9ezaTJk3C3t4eJycnFi5cqDsoaOzYsXh6euLu7o6bmxtVqlR56/s7GzVqRIcOHXB3d6dq1ao5lFGVSsX06dN59OgR0dHRnDhxgmHDhr0Uhq2tLQEBAcyaNQtbW1tmzJhBQEAAdnZ27yyLunXr6qxhderUwd3dHdBakn19fSlTpgzFixfH2Ng4x5Lb9u3b6+JWpUqV10/AqVR06dKFkSNH4uHhQenSpZkyZQpdu3YlPf3lK+709PSwsrLiypUrnDhxgo0bNzJt2jQGDBiAnZ0dO3bsYMeOHTqLa36pX78+VlZWFC1alGrVquncFy9ezI8//oiFhQUTJkx45TLp/fv3c+HCBTw9PQkNDeXEiRP06tWLJk2a4OHhQZUqVXLcD2xhYcH8+fPx9/enQIECrF27lpYtW+qelytXjo4dO+Li4oK1tXWO5e4AZcuWZc2aNXmmeevWrbi6uuYr7cePH8fc3BxLS0u8vb1JSEjg9OnTuLm5AdoDxYoXL07RokUpX758DgUSoEePHoSFhSEANSJfv+joaDw9PXW///3vf/mK65IlS5gzZw4RERHMmTOHHj16aJvfV1jRFYo3mJmTR9tIJJJ/4/Coc0kZImxRLQGIkh0OCSFEjsOjkiIDRJ1i5rrrTPSNi4oft9x+fqSC+Kmlq+5Z8YYtclz3s2VEM6GnUDy7lsNQNPl2uVCLdzs8Sp0ZLb6u7aT7JiAKV/UX4WkqocmMFf29S2SLaxExfO3lHIdH/frssKLIQ01zXBvTw9lSAKJY0/15hjO5hJXQN3bJOtzEu6gAxMMMtRBCI+Z0raF7T6lvKXotPCeEECIlao+oVcQs6zolpxpi8434V6YxZIr2YKBivlnXD39mh0cAACAASURBVOX1fmzYb8LV2kj3zM7jc3EtRXuARlDvigIQJjZ+2gN+hjUTBkqFzm/JBv3F42cHCAFvfWhUzkxKFRXMDISBWQWRotY5iiE1HLRlQaEUDbq3E4Ao81VAnodHZSRdEE1KWure+6xdMV2+ZSSdE+7P0q1nWEh07egiANHrwH2RkXhOOBjqCUD4nXiU4/Co18kh7zzOYqyLtVDomYgrKTkPK+nmoD1YKDJdLUKW9BYWekrdd2p0nCm0oeReVu4FDBImetq4GdvUEL0Lm790eFS9ldd13wsa3UAXTonPvhFFjfSEWcHOQiVEHt8XQp3+QBQ01BMKhULsfZr2ztn+4uFRedWj2xtHCHsDPd0zPQNb0XfZ5VeWWd2BKte0aS/XMyiHe9SprwUgPEafEZkp10Q7DztduEYFyov/hT59bT3Z2jsrLwrVHZCjPL5YHvIqd3nlqzYNY7X5at1AqGU39MEIDg4WjRs3znEozpQpUz5onO7cuZPjEJ+PhZYtW4p9+/Z9FHFJTk4WlStXFidOnPig8YiIiBANGjQQBw8eFH5+fv/adz3cDcTjyCL5+uXn8KhXlTtLS0vdNVAajUZYWFi8so40btxYBAcH5zvuUrGVSCQfD+pUcevSKXH45EURk6p66fGjG+dF4LEQkal5+dWEh7fE0cAj4mpE4vvtWMLOiAP79omjZ6+KnJ/ViPvXL4jAIyfF/eTM/Ccx/aE4+vdBce5W4juFI4QQTyOuiaDAY+J2VM577TTqJHH1/DFx5PQlkazWvHGa83pfnREnLp85Io6fvyrSsz/SZIrzxwJF0KlbWYODx3fFiUOHxeU7j/7lcpQizgUfEqHhMW+Y8HRx7WyQOHEp8qVHmckR4tihY+JebPpLz1IeXBYH9/8tbqe8Ov/+LTmkx0aK00GHxIUbj/JdVhLCL4tDfx8WEUn5K3vhF06I4Iu3hOoNv9+7sLmwdBryD6Y+93qUmfRAnAj6W+w7eFjczp5/ryizb/bJTBEedlYEHjsrotJU+asnQojbF0+Kv4+cyVfdzKvc5ZWvqrRwAYiKgz7swPy/zsaNG0WPHj10/69atUr0799fKraviJOTk5OIj4//oPFQqVTCw8NDmJmZiREjRnxwubRt21acOXNGBAYG/quKrbu7gXh4v3C+fm+r2JYrV053+vOBAwdElSpVhBBChIaGCnd3d5GWliZu374tnJ2dhUqlynfcFUIIgUQikUgkEsl7Ju1pAD17TeePLUdpteEWW9u7SKH8w9zfM4H+E5ay42Q0Wx4n0MrWWArlA7Fx40b27t3Lr7/+CmjvgD116hQLFiz4YHEKDw+nefPmhIaGfhQySkpKwsvLizFjxuRY5vshiYuLo3Xr1ixYsEC3X/bfJiAggF27drF48WIOHTrEzJkzCQgI+Fe+7eFhyO5d+VsG3/LzIpw5cyZPPx07duTQoUNER0fj4ODA+PHjKVu2LIMGDUKlUmFsbMzixYupWrUqoL1nesWKFejr6zN37lx8fX3zHXd5eJREIpFIJJJ/BIXSBBObCkxYMIxR7aRS+2+gZ2RIQY/2LB3TTyq1HxhHR8ccJ+rev3//rU9f/v9IZmYmbdu2pXPnzh+NUgtgbW2Nt7c3e/bs+WCK7bFjx/jrr7/YtWsXaWlpJCQk0KVLF9asWfOPf1s82z/7vli3bt0r3c+ePftK9zFjxjBmzJi363OkxVYikUgkEolEInm/qFQqypQpw8GDB3WHGa1du5YKFSp8sDh9LBZbIQTdunXDxsaGuXPnfvC8evLkCQYGBlhbW5Oamkrjxo0ZOXLkO59M/T74ty22bu4GbM+nxda/VdHXWmz/TeSpyBKJRCKRSCQSyXtGX1+fhQsX0qRJE1xdXfH39/+gSm3Hjh2pVasW165dw9HRkeXLl3+wuBw7dozVq1fz999/U6lSJSpVqsSuXbs+WHwePnyIj48P7u7uVKtWjUaNGn0USu2HQYE6n7+PLubSYiuRSCQSiUQikUgkkoruhmzemT+LbefWRT4qi63cYyuRSCQSiUQikUgkEoCP0horFVuJRCKRSCQSiUQikeQLIRVbiUQikUgkEolEIpF86miEVGwlEolEIpFIJBKJRPKJ8p+12CoUCpn7EolEIpFIJJ/qIFaeISqRSHIotgoyhd5/T7FNTk6WuS+RSCQSiUQikeTC//73P3r37i3jI+PzScTlU7bYyntsJRKJRCKRSCSSf1A5kfGR8fl04qJALZT5+n1syD22EolEIpFIJBKJRCJBAJpP1PYpFVuJRCKRSCQSieQVGFmbYFLI4p3CyIhLxbpcwfcSH/Ujw3cOIzMjGQtrx/cSH2Wa6p3DyFCnYmVc6N0VsvSM95KmTNKxVNi8czgK5bsrhxkiDSs9u3cOJ1WTRIZIy385k9f9SCQSiUQikUgk/38wKWSB96/tPpr4xPzs/FHJx+xq9EcTF/Wtux+VbJQmxh9NXE6kBOR/gkAoPsplxlKxlUgkEolEIpFIJBJJvtFIi61EIpFIJBKJRCKRSD5VBAoyxKepIv5jdubgDUsZ/k13Rk6cy7HIj+haIJHBgd8n09q3AZXcKtHAtzU/rzmI5j0EnXR/Bv69DgOwv6c/cyOT3iGeGjTPrpZLipxL06ZNdb9mLVszdOpqYlX/zN1zarVa93fnFn7v/J3Yizvo16kZld0rUKN+YwZMW038szDT4vbSquPP7z0N2eN9bMEwmjXxJVkjWN2tLeufpLxVmAfG9uVBhvqDF+HIA9/x9bTQ1/ob0LoFt97D3pfs3Diwiu8H9mbUxDlcjEt/Z39vSo56pdG8l3oreUuyyV+VeoMhnVvRbdLFl7xF7BpG7wVXXxnEvE6t2RuXJmX5D9bF4D8X0r93d/oPG832849zPAvbtYaxg3szdMzPXE3I2puWeCuQqWMG0++7cWw9+/Iyw8ykc/QbfT7Xb64e0ottMam6/5+c28wPw75hwKiJbD0T9UnKW50WztIZP9K733CWbjyVq7+06PP8PHoIX3/9DRPm/Knrh0IW/cyECRNy/CZOnKp77+SGhQz9phejp8zjfExWnUh7GsrMH0fQo/dAZq48QOYruuPE8PX0Hrr7jfNJIpFIXq3Yag+Pys/vP6HYXpzbikbdh7H/WjR7F0/Ar7IXYSmqjyLBy76uSddZp/li4Hg279rK1KHtODaxEw1GHXznsDNTr3D8jHbgYF+5Gi4mbz/bETrHi9rjzj8bNF7jzGULxo0bx7hx4xg1sBuJW7/Hu9fef0RGDtZWpD/rPD1r1sLgHVYjJN5Zh4dPH8y9e7J66y6Wz/oe48M/4dN/i3ZsnPGI4ONX33sasuKtoe3YXxm7eBmmSgVOVWtQ1OjNL51OiviNEY9aUcTww19YnRp1gTNX417r71zwUZLU72/yI/rcJGp2XoBznRY4ZgbS0LMryRrx1v7ehuz1aoG7I2PCE2QP9IHILv8HgYMJiG3Bz9+We8mfqUNFKpU0f2UY108E8yhDTk/8U3XxxoovaP3jAao09MenohnDG1ZkyY14AG6t64XPkC2Urvc55fSO413rGwSQmXyZOjXbE+3gSYOyRvRvXIXd2ZTU1Kd3+GXI1+wKfPTKuN3e2Ie+/1vL9VTtJGD0mWlU9J1BQc9mNHQzY7xfVda+y6TvB+L7evVYfd+C5p+58dewZvTe/vJePo0qlpaVGnJe35n2HXxJPT4Bz2baiVuhVqNSqXS/5CeBzF9+Vjv5MK4R7WafpVbz9pTSu0yTKq2JUWnQqJ7SvpI3l43L0v5zL24s70Hj8Sdf+OZTejcaRMC+mzn7idfkk0QikeSFWijy9fvY+EfszP2mHsa8cF9C/p5Fwo2ZFKk0jmHbw9ndsdQHTWzcjSmM2GVGyL3NFH+m3JR0cmZtkAHu9YbxdNI5bPSVHA/8mxp13di1fhOeHfpgp35AwJY93ItV41SxLq28K+hWnidHnGXH/lPoFa5IPZdsymHl6hgaPxevhuP7thNyIwaXmk1oUtUJgLToc4TElaZ08nE2BN3A3tWbdg0rkJl8kRM3EklKPsnxqy646oOeQUFq1aqlC79CoU24ttwONNUq1Uk32LUjiMeaAtT29aOCjZHOb/y1YHYeCUFpV4omzRtRQF+hU5C27gwmUVmABq3bUNrKkMtHAlEDBw8coGHDhnjWqoWhUoEm8wlBJxOoVSGN9RsCEdaladeuMeZ62rDSn4ayffsxjF3q0LhCBiFxpalZyoKlHUZQafYhZnQro41MyeJM3riO30u2J3JBKwq8qEDePcPWvSeJxxwPrxbUK2vzTAF+QsCWXdyL01DR63O8XfN2fx7vkEP7SNIoSblzFbWLI05VqqM0zDtfUqPOcjG5LCXiT7AnoizdWjqx85upfLPsnC6eGfFX2RlwmARDJ7z9GlPcVD9X96fnj3K/TA30zu3n6JWnVKzfgjrlrPIxZZbOsZ1bOH87hgJFXGnRugGWejkbkZDDgZSqVYXTe3ZyI9EU75Z+lLEwyDboiWPb71uJVJnRqF17ylgZ5innvNjefxk1FgTS298FaEHI6oL8cDWW2eVt3srfi8SGHWPn0YuYFnejWaO6GCvhyfEgEirVJubQnzyq0ppqz+pVXOgxrqeqeXoskLCCfpQ3lbsq/tW2NJv8L1mU4vyRx+iZpHL9bjIFrXOe2GlcyI3KZs9OFBUqju7ZxqXITDw/ayEF+Zbkt44tnhJIlx1X6OFqA/jiFLqVXlMu8c1vdeg/dDM/nLxHt2Lm0NaP2MwR3EhVwYZBJFVcwKyBHQEoePxPhky6hO+c6iRFzqVRuz/JTHwIrzgkNu1pIK1GR9HAOuvAlIB+i6my+DAD2moPvHHN2EmrwcfptLHRJyPv5EfLWHbPkcenvsNAATWLHcOt00L+93nO1Ubxd0ZzwaQHURMGAtDYx5MltmW5njqUygNHUTmrZWZ07dJM2LUF0PD1wlPMvvYX7e1MwK8xmq3FGRj8iF9KbiBYVY2Y0T0AqOV6itINfoGfauhC+mtIY+LaOcGObP3oa/JJIpFI8hx+okD9Hm2fX3/9NQEBARQsWJDQ0KwVhwsWLGDhwoXo6+vj5+fHjBkzAJg6dSrLly9HT0+P+fPn06RJk3x/671bbFUpYZxPysCu2ucoAAvn7gCEb7r/wTMqdMp6XDrN0im1zzF18Ofm9RBs9LXi+Na/DbM6NOW3fadJyYijdcUq/BZ8m8yUx6zo3wD/3248G4j/QuXKfuy7/JDT22ZQ4+tAXZgH+nVmwYMkQM3EdpX5Zsk+4uLvMfOLqnSdHwJA1ImxdB0+nC7zDqIWiczrWo+eO++RmRLGuTuJJEWc58yNly1SmoxYts29SHE/f60iFr0Xr3L12HTqLg8u7KJR+Rqsv6udEY/YOZYKXv0JuZ/Aia0TqVijIw8zNKTG7MDdvT1nIhKIurKbmmUb8CBDzdXjx9AIOBoURIZG8FXb1sSqNKQnBtOu0xi+6jKL6EwNR5b0oWY3bU+anhCEt7sfQTejObyyP769OzB41U00GVFMvRLHLH+XHPE3NK/O46i7FH3B+pkStYEKnh04G5lA0sNL9K3ryu+PUxDqJDq7V2TlsRukxtxgYP2yLA1PzNUd0MU79HgwCA1BQUGoBKzt+QUbo1PzzJeHR8bQ/cfR+PSbR9jdZIQ6gdHn7elV2PxZPAOoX6Ex+0KfcPPoCqqWa8K9dHWu7hfG96TTgKb0WbaXqDsn+LJuBaadfPLa8rqqY1W6LwkkOTOF4LWD8fx8+Ut+JnZuz7ct6rIm6Bo3g5ZTu/xnnEnKWlI4tUMXQp5k8PjUr9Tx6JCnnF8bn9sJ+Hs56P7vWt2eU7si39pfdsK3fo9b09FceRLP/kV98Gg5E4AT33Zh6I/NGbg0gIhUta5exV05w61UFfdPHed6aqbshf5txTab/K/GXuf09QRSHoZwJuzllQSRe0bSf8lVQDDdvyqdJm3i0cNL/NDMk2OJ6VKYb0F+61ibeb8yxCVrEi36USrmJS1ITzjK8XQ7+trFc3TfXwSdvsLIqTMoY6LPzVW3KdnDU/eO64ByPNx/HADzooM5fvw42+ZWf+VE3OgmPej310oKGmS17REJGdi4ZFnsrVxtiDm/55OS99OQ7ZgXGaxbvVTAtR8pUavIeMFIbmzdlZW/DMmyeKTfQ+hZYm+Qc6h1Z2N3dlVazDflCyA0qTxIV1Mu2wovl8KmXNkcgXGBJjhkXmDd2fuoM5LY+79D2Nf01fl7dGQCw6+1Zc3zieNn5JlPEolEkg80QpmvX3746quv2LMnZ7sfGBjI9u3buXjxIpcvX2b48OEAhIWFsX79ei5fvsyePXvo169fji2Sr+O9mzlU6fe0DXwh7YytUt8WY6WC5LuxHzyT7ofEUnB81j1isWHzGDUvTPd/o8lzaWdnohVsz21sbuZEZtIZKnSczLRJvQDo4h1CzWG3oHtpfu86kVLTDrGip3b5Xc0O5Rl0Pec3Y6+NZWGIFw9vzkcJDOlbl4Il25L6rXbZUPxFJbtuT0cBNLE8QOu1dzH1+4J+jRdxLq4vA1o4EXcTkh+vwspqjbawqdXomZRh3Qnt4OP4t0NRd9/O6onaWdw2RXxo1WMXXxzw57tvltJj3zXGu9sCo3CsWYzef91lockS0ouPYfa4bgBUKj6fqAwNbUeMpc/EaYybMAGjF1YYpMcFMnrDfdzMDEhuZ0qJGpuAlpwcMgD9IQEsHOYGjGFsxcIcqAgZSWdQ6dtTOp9LslMfPKHn5N380FvbSdc4H8DMc9H4Vw9hd0xhHsybiKlSQYcKLgTHZ5JmefSV7tnpMmo0facsYsKECW+UL4+DHhMRvgMTJSTdn0maXQ/du/u+HorpiF0sGVgRgNIRn7HkbiI1hrzavTGQeNWP0OCh2pmrBml4dpvGqKuz8pTHQ+dm7B03FWdjPTL6eOFQcgTQ8yV/EX5rOTjQDYD6X7nR58cQzs7WDmgK/bSKn2o6AP3ZYGvNjVQVNrnIuVvTYlw6dZIkdc6loUo9c2pUd+NGmorSplnWYPOSZiTdfHlJYX79ZadXv18YFXyXb50tEOpv8a3XkNtp2gHirYd9CN3eGoDVz/yXaD8I3/HTeTBkNK1sTWQP9C+TXf7tS1hSuc0Sjp3qxeCOuV9DkXR/NtOCzLl5fy22BkrS+jegcPFWUphvQX7rWD0/rVVcaFLYNm84fYMc2PtLeTITfgGlES2qt8GhbhVizvdniMtQzq4bwv2YdMxLZpn5DK2Ko0698do4HZ/agnO+vzHb1ZrT2dxb93LHt+80buycQMH0m/zY5xjqjE9rhUXSrSRMi9pnDZxMS6NRJ/EoQ02xbBPlJva18W3wbPLn2kH6d/oS75GbKKCfNfjTqGLoMfQkq6/+CoBCacaYKnb0m7yZv35oTVxYAAOCHiA+S0PftAbLR1WhkVd5hpnpkax2ZMfV5oB2yXj7LgFsvHwMRWQ3WSkkEsl7Q8AbWGxfr3TWr1+f8PDwHG5Llixh1KhRGBlpV5gWLKjVz7Zv384XX3yBkZERzs7OlCpVilOnTuVYtfqvKrZ6BtrGX63bU6vRHnbwEazDtrTQJ/l21kFWRjaVaNBAeyH0vpEDOPv9zzrF9qs6WgEbmHsytH0s82dM4Oat25w7chRhpVUu1t9P5rvWxXXh1R5cDvrl/OaDXcfAwIZv+vTJGiiooziTlEExoEDF9rplzYZ2hohc9kSaFfySqNuLng1S0ri482e8avlwLfIU+89F4zPVVefXuX0LEmZvR6gaEfA0g18rZC1Pa9bJmd82RVB42RjKJrTB3XsLTXy88G3VhcrmBnnKz8iyNm5mWj9KQ3sQWgVoR9Aj6v2QJYcWnxXmAKA0tEOjiidFIzBV5sz/yKthZBYvQ/brym0r98Pv4Tqm/fQbt27fICj4ESV7CkwKNKF3jTG4uNejeSMfGjVtRQcPGxTi1e75Ia98KQoUKN8Tk2d1Ou3pWUwLZym2m0NjafG/Err/v9yi3Z/dNRf3QKBkdz+de6G6I0mKrI5KzEI/j2oxaPiXrPt1Dktu3OJqyBHg1cuX27XPUiZqfVeVB/5BgFax7Vjh+WJvBTb6SkQecgbY+edaIl44cErf2Ika1d0wUSpIzqb0qpJVKI1ebvjy6+85mcmXOJFizD5n7WBaoWfOnuATAFwGSvWpKXuZT4j9Pf2ZdV+70uSHjX/xXB2IPnsAq1LfYfvMemVs0xAvKyMpsLfgTerYo+Nr6fHNCDIqd+fQ5T24mhuQlKBBnXaH3vuiaF3EDKGeQRPH4vx0uwcllQpU2c7EEOpEUJjmGZ/E8N/osKogZ05XJTk5GZUQpKcmk5ZpRoUhW/khug9f1HdH2Jbhq++92Tra+pOSt56x3gsy0U4iGClebsCFKpalY/oyaeN9Bk/7i+/8q+Z4fnNVF576/qLrSwGGBGznSa8R1KnwI4UqNmBwm+IsUhoQe3k6LWdlEHjjEdUKm3Bq7be0qNOTyKt/sKR9KyrODqCMIoPYFBVCZJCcnIyZmZmsIBKJ5B0VWwWZIn9nykRHR+HpmbXKp3fv3vTu3fu1712/fp0jR44wZswYjI2NmTlzJtWqVSMyMpKaNbPGfY6OjkRGRuY77u9dsdU3KYuhUkHC9SfPBq2hqIXA2s3ug2dU2W9cuTF5HQyeBIBpIS86dACEit+H9MuhZFk/m2GNuz4btyabGTN7JN1bdGHkoHTqPNNxLJUKklTZBhevWBKpNFBSwLUt335bSef27bffUsLMkKeA/lvsDVQojfFo8QNOmpnsepqKiVLBk2yDHKFOAaUhKAzQQ5CqEbq9sOpUNUojBYYWddgfFs6F4MMcCQpkaKPytN57lXGV7fL47qs7TD0FpGU7uESTpgErMDSvQXUzwcwb8fxY1jrHQKl+rZp8fSGSIdnurr4wtSmtdxVjxqjOtOo2mC+VjZgKoDBiRsAlBoQGc/jIUdaNasHiQ1s5PLVGru6vI698eQIYZtsnqDQ0QpPtgBt9BWQ/7ybtcThPjIvk6v584KlLvyYFhUIfZR5KrTotnHrlP8Nr1HRafenL0HEDcHXt9Uq/ydkmyzTp6SgUWXE3fcVHcpUz0KH3Nzny8nl5A6hjacSp+EwaP9s/9zA0Hrueti+Fn19/uvAV+iDUqAU6Rf/x7ZsYFSupnVyyNpC9zCdE7YlzKJupLZT2ZgZEPHM3sDBAk572QtkVUmBvQX7r2KOgKVTpvIeZG07SqVZRnbuBmQdKw8K0LmL2bDLJgi8KmrDtXiKfVbDm6amn8JkjAAnX72Fs2zXP+MTfPIh+RjA1Pdy17zxNRa9RTc6M2sP2vmXpPXUNvZ81Mne2NmVOkU/LUl+gcmFSZl8EGgOQkXgKfZOSOBi+MJmgSWNYw8pcqvwdF659g53By5MNk8afYeBJzxxuhpYezPxzLzOf/b/Bx4VC/Qpxa9UmCjeeQ7XC2omF6p0WYtDPkuDEdPZHKLn8nS97vwOhiicpPgh3j1+5dfOKrCASieTdFFsB6nwuM7azs+PMmTNv/A2VSkVsbCwnTpzg9OnT+Pv7c/v2bYQQrxgn5t84+t732Cr0zPmhtDUxl8aw+cgZ1k8ZBEC9/mU+eEaVaPMLZaMX8+WCA1mGc5HOxsmtCc5lr1fUoQBs605kYKeWVHZ14dbWrJMQe3rYsnB20LNwVKz54eWrLoo29yX6zA6sy5THzc2N4sbn6PDlLCz0Xp9JIreTZIWKsL0ziMCJxgWMadqsKAd/2Kl7HDh9Ew51OqHQM6dnUVOG7bjzTKFMYsGvN/D8uiRh89rQdMQ5POo25dvR05lW356/j2ZdBaER+R9wft6kCH/PCtZ9Y9GerJmVRT/UY9HnfTkVnTWgPbmsB7HGtRnqmPNUi3N/Xqb6rEl08POmXHFL9ofEANqlwFXqDsOxYm26fDOCJfPrcHPX+Vzd88Ob5ItRAS9So7I2u3eq48DG+cd181qTPqvN5PDEXN0Bri1eqDuh+NKqUViU+DbPypcau4s7hvWZOeQr6latgPLm9lz9/jE9MKsMfneS4u3q55n23OQMsHzGNCZPnpzjN22Gdm9vb9+ibJmtTZ86PYLJl2Lp21g7WH589gTnbia+kT/dRIGpK75WasYcffRsgugmdarV4HGmOl+N74vhajIecuTIEd21GKeOHdUeiiP5Rzq/FzErXJRixYpRrFgxTLJNrNh4fE3C7bGceXY1zdNLCwlOkHts34b81rFx3efSY8taPne3Jjk5meTkZFLTNRhZedHUPJZ5z+q+KvUmix8m07Z8AcoO9ubm8l90+0c3TgqlyvC892o6NlzD7du3db/PbU0ZdvQS2/uW5erShlTssEHXP8wbE4LPT3U+KXlblx2JeDiHs4na8wuurZ1DQc/RL7U3j88MZ93jjmyb9BUmGak6mT+vJimP1xCQUZmvC+WcJF5a3ZnOf2nHFulxJxl9KZnRTYpiX6c4j09uJvXZhGni7XUkKAvhZmrAjkvXdPI+u78JFk5jpFIrkUjelzaHJp+/t8XR0ZE2bdqgUCioXr06SqWS6OhoHB0diYiI0Pm7f/8+RYoUyXe4/8hGl14bZ7OuXl++bOoFQLVuM5jnWfCDZ5OekRM7jq2iU9uvKDLPlsquRXh86yqVv5rD9jbhvOo4C6dWA9H88BUdezZDRF1Br0wx4m6NY2FIdXr+sYzfanWgekgdbJMuYVa/AS8GYlliJHM6tMTTow4NPItx4e8LdF0T+Nq4mjpZc2fJT0wsP5cB1XPusVWgh4OzO1M37KOooR4Ok9ZSo3FjKjXcRCn9SI5Hl2PHIa3sf9g6C98m9Wj6hw+q8MMk1x7M4fpFULn24KFHKxrcaEghvccEXSvGtpXaU6tLmRrQuU9fVi5dmi+5VpvxJ+XrNcCrZV3MYsOpWNuO+w5aS4Jr/y1MutUG37KlKVfFA5OklMNsdwAAIABJREFUW4Q8smb+/l2YKCH7kUXeYz5nVLtW9PKrwMMrt3HxsObixJFc27sAj4yqlK1/lZrFTDj192WGbV6EWSFDPDLcX3LPD2+SL2YFu1I4zoMMMRRDBXj98jsu9VpQu1ldCmWEcs2xC+fcbFHk4h4M2NSMoVLNxlQumEHg+XQWBw3JM35m9p1pWGAi9f27U9rgKREGHphnXGbglIsMLJ7Tb5knE6neZCU2SRe4ovTh8Ci3PMPOTc7nvFczafnKXN+rMfN3ClZpSrPHvmhu7KXE4HW0eba/dW+Pdszx2sy5eTXy7S87izZPptHntbjiVZvEC3/j8e0mSpvok9dFUIUKGrNw2GDKL12IIlu4qU//omnTodyOS8TBQEnH5s1ofPoeS0pZy77qPZJd/rXz4d/EthXrhvxJk/I18KrrxK0rhvQtbC4F+Rbkp46dnVWWjTEZqBtWYH62d53b7CLktzosXD+aWo2rcsCrOo/PBFGu/2q6FTQF+7n0q+BB5cb+VDIK54xtV063cX7ruJb+6n+UWlyHWs03YfrwBCmewznyWdFPSt4GZm5s+d4LvxpNaFbVir+PG/LHKa3VOXt7c3/rWVIiwyhSZHGO908/iqGMiT7/x955RlV1dA34oXdFRFREEAVRuoLYBbHFhth7V2zYS0xs0Wgs0dhLjC2aWGKiYu9dUUQEFaWKSrMA0vu99/txkSJF9NP31bzzrHXX4syZM2fYU/fsPXMivXagZz25WPr998xnXasWdPujMc9ve9Nm4XFaVVBD1nkXo/a2xczBlZbm2ty9EsykzSeL7NkVCASCT42M8ltsPxZ3d3cuXryIi4sLISEhZGdno6+vj5ubGwMGDGDatGnExMQQGhqKk1P5D8JTkMlkH+0Llp5e+kmq0uwEfH0eomNsRX3jyl9coSVEBvMoJhtre0t01ZSQ5WaSo6SOagmLDzkpT/H1j6GahT2mBpo8vHkVZcvG1NNVQ5abwmP/AJSq2WBhVLGM94Xw6GkiJvYNqKlTDtdKWQ53b9xEsaYDDUzKMfmTZREeeJ/XUl3srM2KWEokWQk8CghEobIZ1nWqFwp/SYBfMFlqlbG3t8x/Jj36AT7hqbRo2bTMPaBvSXxwg1C9umhEhaFbz4EHQ2zZOu0ih50LVljSX4biFxyFgnoV7B2t0CrFDzc5IpCA2GzqN7SlsnIK168/wKZFC3SVcnkU4EdcuhJm9vYYvnXhluWUHF7uelC+crk6oQFnPC+ypL58z6pMmk7ofX+SlA1wsDbLt76WFH7JvR4/jzrDX4653I9IwaKBDZXV3793QSZJxd/nHsoG5tjUqUbCo9sESyxoalOgoPU00mfI/Ugc44J4nqlDQxuzYod+fZCc31fg0gxC/O8hq2qFRY2KHxxPJs2g1dIQrs2xK/ZIbtpr7vsHo25ohaVppff+D5LMGG56h1KzaQtqqSsh+M/ysfJPig7lYXQWtg2t0FFWEIL8WMrbFssgNz2OB/4haNeywdywqAdNbOg9InMq42hp/P927ZJJUrnvew/NmvbF3vM1kfYqnICnmdg6WOZv7/lU5KRE4hcQQy17B6pqFx3HYsMfEvYiCzNbW6rrfPqtGZqamqI9lYFuPQNctvX6YvIT/7PpFyUfraC4L2dcCn/2RclGUUP9i8nLrfTjJEnKV1Ym1jp8+49jueLuGJjyXlfk/v37c/nyZeLi4qhatSoLFy5k8ODBjBgxAn9/f1RVVVm5ciWurvLT95YsWcKOHTtQVlZmzZo1dOzY8b+v2Ar+d4g660mTOVK2rxpI7gsfpk7aycHwu9hp/Xv2RmYlX8O572NunfL44GffKrYnuxQ1tV7fsrZUV8yanUcXOvipdN4qtt30vo6TgR9uX8qrHlNxraQuGo5AIBB8AfyvKbanT59m8uTJSCQSRo0axezZs4ViKxRbodgWwti6ArP+Lp9iu2tQ8kftsf1cfF1n7gu+SIzab+BY9jYOHjuIVMOIHd43/1VKLYBahZbsGXKHmGwJhqofZh00bNeLb4yLWynse/Sldq605PdVqlCutNsMGISJ6tfTjK1HficajEAgEAj+O4qPRMKECRM4d+4cRkZGNGrUCDc3NywtLYVwBILCbYWvc8uDUGwFn4QGXUbRoMu/+3807z/to56zmLAYixLCtQ2q8f/dYTh+xRpR+QQCgUAgKAc+Pj6YmZlRu3ZtAPr164eXl5dQbAWCQnzI536+NMQJBAKBQCAQCASCfz3R0dHUrFkz//pDv5EpEPxvKLYglSmW6/elISy2AoFAIBAIBIJ//4S9nN/I3Lp1K1u3bgUgOzFDCE7wP4eEr/NwR6HYCgQCgUAgEAj+9ZT3G5keHh54eMgPi9StZyAEJ/ifQiZT+CKtseVBuCILBAKBQCAQCP71NGrUiNDQUCIiIsjOzmb//v24ubkJwQgE7yCRKZbr96UhLLYCgUAgEAgEgn89ysrKbNiwgQ4dOiCRSBgxYgRWVlZCMAJBIWSAVLgiCwQCgUAgEAgEXy6dOnWiU6dOQhACQakofJHW2M+u2P6vfdRbIBAIBAKBQCAQCP6tyOCr/dyPsNgKBAKBQCAQCAQCgQAZCkhlwhVZIBAIBAKBQCAQCARfMVL+B12RBQKBQCAQCAQCgUDw70AmA4mw2AoEAoFAIBAIBAKB4GtGuCILBAKBQCAQCAQCgeCrRb7HVrgiCwQCgUAgEAgEAoHgK0YivmNbnOwUH35ccYw5C39E/RMr/rcn9GL2o/j8ayVVbWrVa8LEBTOw01P7JO+Q5rygfZdpnD+z95OkF3NpKgMX+RcLt572O+u7Gn++gpBJkaKIYqE6evfASn7ccoDAZ0lUNq5N99Hz+HZgcwAuDu3G5UV7WGRS4ZNlITP+CD3GPufkwUlkJ99i7OB5yL5Zzwb3S/Sboc+xP3t/cJpZiZeZ8KcB2yZY8n23b+j2x1Ea66h+mgxLpUgVFcveOl+CXD8XYb97MHpXaIn3mmw6xNL6lf6VHWvmm5N0H+nPqUPfi1HmS6NQG8nNCGHy4MnEWy1l/0L7ItGeH5vI/LBx7JpqWSyJlT07YbntEJ0qqQt5fiAhZ3aw9fBVpAaWDJk6GftKJY971/9cw+/n7oCOIR2HzaCHQ1UAXt38lfWnovLjKShqsGjh9/it/YnDcRlF0lBQUGHRovkAJD46zaY9JwhJVKH72Dl0s6ucH+/hsV3sOXWZNO26jJ8zA8uKRfvj5Ig/mLRKl10bunx9k7zMCDav2opvRBqObQfg2a9JyX3W67v8smIHj2IzMbV1Zeq0AegpyweJl3cP8/POE8Tl6uDSx5NhrnXeW5631/zE8fiC8tCo3JXvpzjJ5Rl2njVbD/E8S5dvBk2lV6MqxfITvHs6OytOZlk3Y9FoBAJB+dQGFMiVfp2f+/k8dmZZDrHBN5nZzZ3FixeTJZN98lck+N3kRb3hLF68mMWLFzN/tidVnm2jZYMxn1AfzOTK1dufLL2MV/fwC2+an+e3Pw+nKp+1kO+vaEzD7+/mXz/6rT8txu2ny5QVnLp0ilWzB3JiUlvGHX4mn/D43CAwPefTVjTV6jRzkg/iwb9O4prOSFYMqIOSuilNHWt8VJq/dh/FiMFm8v/x+lXic6WfLL+rzSszKyLpg+T6OanuOiG/vrglBRJTe2j+9ZCa2v9e3Sk7lms3HotR5gukcBuJPj8erzfurJ1SXHnVrGZLQzOdEtMIvnmN2GypEOYH8tp3AXa9VlO7pTvGORdoYd2HNGnxcTZ4a3c6fncGxw79aWurxaQWdVgfkghA0MaV/BmSQG5ubv4PQCaRFAlLfXWeVb/eASAt9gD1nEYirdWczo66jGzegOvJ2QCE7RlK0wkHqevSA0ulGzRtMJLCOZLmxjOs5Ti8ToV+lTKf3qgROyN16NbejsMTXRl26Gnx/io3gfb1WnBXuQ79BnQh48Y8rF1/kssuZi/1Wk2hiuM3dHc2Y727NUvuxr23PDcu+4ng1KxCZSJvLzlpD3Cwc+N1NSfa1VdjdCtLjr+zIJEWc4QWo9ZyIShRNBqBQPBh8y8UyvUrDyNGjMDAwABra+ti91auXImCggJxcXH5YUuXLsXMzAwLCwvOnDnzQfn+LBbb/ZbV6R8U/9mFrl3LnubNbfOvG9msYFmVPsTl7kBfWZHUpz4cPOlNEtrYt3bHpb58Zfm19yUS7FuSfGEfN56kYdOuL23yLF6SrOcc/+cML2UGtO5YdJKWkxLMsSOXeSnVo0UXN2wqqxWkZ9eM5167eRAHHYYNo07WfX7fexWVqnYM7uOKSl7ZK2uY07x58xL/n8TH1zl6xQ/FKuZ06vZN/irvjfPnaOpsx7E/DuA0cALVVeHGqUP4hcRRp1knOjUqWImN9j3Fce9gVKvUpWfvjmhkBnAzJJmUVG9uPKpDU/Msuk4+xLaQeAYayRUiM5M6HDh8GruRm9ncfdk72n0W147+xd2weCoZWeLeqx0VleT5ynhxj4NHr5OiWIm2vfpgoataariSqiFNnSqQneTHFd94FLUTCX2RjpOxKU0cC6w16dF38Tp9k1TVmnTt05VqakolykA3xYufkqfyokKBRUCWm8ihHYeIVdDHpbs7Vnn5kUnTOH/Ui8ex2dg27YyLfZUyy/TN/WsEZeQSf/UCD6u6Ya2p/H65mmVw+VYmTeulcPDIU4Z4uJVa//wunqduC0dunThKSLIWrt3dqFdBpcy6rlXTjuY15X+nVlRF29ie5s3llrFXNy4R1rAlcRf+JNaxN46ht0h1aEV9TXnzjrx2sdC1tNS6UxrZiY846nWJJDVj2rh1pFZeujJJMme9vAhJUKKJa1ca1dYhO8mP289MsVYK5MilB+hbt6Kri1U5VpFKr2dvib97lch6TVHyPc3VwHhsXdxpaakrRp//AoXbSIBOXe5eeYmSRgbBT1OpWkmvSFyN6nY4aOm8baRcPfEPAVHZOLV3F4L8SA6N3kyzrd6M718HcMdvpzbfPkpgg3XlIvHWLTzPsLNPGWNVGeiC8f2/GfLDfSbubUWwbwIdji9mqXlRbw+HafNwKDS1mdnQiGUXTgBwduhMTBefZe4YeZs2jcvEJzadFhVUGDXxAD8GvGakiQ70cSMhewrBGbnU05D3F4cnOJPY1wSOfH3yTovdzOanNUm9/z0qCtDM5BrmPVezq8faomN4+Ez8NcaQvHQaAB3bOrFOw5igjG/JPbAWrUa/8u2wbwCocW8r/Vc+Ys6+VmWUpx5n32TxYMUyqqgUtUOE7x1Hqu2vrJ82GACDG3/iuSCALhub5PXPqUxw8aSHkz5+oskIBIIP4FOfijxs2DA8PT0ZMmRIkfDIyEjOnTuHsXHBXPTRo0fs37+fwMBAYmJiaNu2LSEhISgplc+C/Fkstm4X/Hn69Ck7Gxj8Rwsi8fFVNPTaoa+sSPqLfdS2cudOVBIpMQGMcDRl+8t0AG6P78fo6W6su/qU3IQA3O1NOZmQSW5GEO3rWrH4yF0i752kvc3g/LQzXp+kcS0n9t96SvS9Y7SqbcsfT1Py0+vbrx0nwpJ5dXsDTW3c+abbSpKlueyf6kaHTe+3OD0/+i11mozGLzKJmwfnY2bXg5g8S4ZHt84sc2/Nb6dukyHNZX7Xeoxcf4o3ic9Y2t2SPqvkw1bEQQ/quf9MTHIGd/+aTr3WK8hJe4jvkxRSn9/FJySZpIjFvKzQP1+pfUv1Vn/yKnRZsXzt6GHJgPUXSM1J4/ru8Vh/86tcHnFHMDd3w+d5Ei8Cj2Nv0ozobEmp4ZkJR3EfsIns5AfcfZ5KyhNf7oSlkPbiN3oOOwRA3N01WFn342ZYIo8ubaRu7XYEZ+SWIAMZIVuWYDWnU5G8/ty5KV4B0YRc+Q2HWo04FZeBTJLMyGZ1mPb7ZRJjHjDV1YyxO4LKLNPER7cJy8gl8tZ1gtNzyiXXzKRLuPX5ge7NR3Du3pMy69/83m6MbufI75eCCLn8Kw1rN8cnJfuj6/1Njz54zm7L2A1ePM/I5fiIXmyMTc2/X3AtKbXulEb6Cy+c6jhz6v4rQq9sxdLEmWdZEmS5CQxtbMbCw3dIfOpNL1sTNoYm8SZoPu6DRuE0YjVhr57w88DGdPj+6nv/h9LqWWHuzR1MzzEuDN90khfhN+nrWIcfvV+JEei/QOE28jghiNtBSaTH+HH7YXHLUOSJqYxe/xiQsbibJT0X7Cc25j6zXa25lpwlhPkR7AhPon/ragWThiYG3DoWVSxen817mGVWsPjzOjYdHXP5IsOZhEyq39zNzEmTWbJ6By9yilvOn+wfwLGG25hoLV+s2OT7msG9q3Lv2mmOnr9J7elLGG+hS1bSFW5kVsGzShJXTx3m0u1A5q5ak6/Uxl6Zx6THffhrpMVXKe94v0PoGM3IX6DWs5pE2osdZL9jJNeoNJy9O2flX0synyFTqoiBiiLVXNrwJnAT91+mkZX4hJ3HImnUo2aZ5ZmT6k+Ccl2ubV3GpEmzWL/7Im9LKXRHOGZjGuc/YzXVkpjT1/Ovz33bmujRRxhYU0c0GIFA8MFIZYrl+pWHVq1aoaenVyx86tSprFixAgWFAiXay8uLfv36oaamhqmpKWZmZvj4+JQ735/FYqtpaIQJUFXt8/pnB28dRevTWgDkZiRx/8Eb/rgfIJ+QR79i7M8XWTS+HgBN73qxzPc1IzubyAf4nOlcXdEGgCp/HeS3qFRqnhnKI6PFxP41GYBRLftg1u81ANc9PJGMPs1fy5oC0LtGUzoOOsag6wMAUG21kTUzbJBm92O5mhHf38ukfSU1+pqfpenSZzChPgBvwsagoTE+/39Q123Nm9iTTB65njFXnvOTvT4wj5r2VRh2+Cln+9YGIHDsKY53NSbh8XRW+7mSFL0FRWCWZyu0DbuQMTWah8uPYrfwCgtHWiDNGY7J2r/RNBjEpG/W4pvoyVR3Y56deIxaxQHllnFM7a5cWbKK2upKZE9wRafGFGAsL2+uJ8v0BzYsHglAQ9NfeJEtJaeU8LdTK+2aQ/F0Wc1D3UlM7mxEUkTBu5b1XIDbiWDWNpMP8I06mzBkSxC3p1oXkQHAml1P6Hq5qAu3bMgxfveUx3Xtb8ZEz1tcHfcrh+IGknhrFQCTh9bAoMEANo/wK7VMY67PoMucJUTPWkBPfQ2OlUOu6a8h/fUBZj5KpI2eOnF315ZZ/553+4cb0+wAaN3fjOHf+fFgjTnet4OKlYGKliVO9mXvoQ2N8ST8dC8ANpcSJ+HxrFLrjkYpG4VPDfJEc84Ftk+Te0bUfd6c9RHJTAwZyqncGbzeI5/E9a31milbQ+nVC1LCHxH4JpBqqopkTetClWrdCZn3kroayh9cz94l5ZEb4X7fyhc72mVi3e9H5j1bL0ag/zCm/QraSD/Tijj0Xse1W+OYObh2qc+kRC7nx0s6RCf8g76KIpmT21LRoKMQ5kcQkpGLhWaBl4eOuTapoanF4jm7ya3iMmk6/6ycxPDL1biyyxpkuVxIzOTZhReMb29P0PH1WKw7RmTYISrkeUpIc+MY5OnNgWd78tPzT81BsY8Tx4ycqZjqz+CxBtx9eJJqqf6gqEZ7m85UdXYk/q4XnnVmEXhoFjlpD3Dr7cXRJ3dRiOz3Vco7JSwFTaOChXplTQukklRisyWYFJrnaBi0oEs7+d9vHp9ldK9+tJl7DD1lRbBfyIz6xjSoYYC2YhZKdYbztEetMssz881VcrMiOPFandYNanFkVW9+PbuWh38MIjIuK3+RAkBVtxa5GSHy+Y3vzww535zIe454DxDtRSAQfBjyU5E/7wEyR48epUaNGtjZ2RUJj46OpkmTgjMMjIyMiI6O/u8qtv8parT3ZPEg+b5NaVYKx1aMZva8a/TY2xl9h0m4xezhxznbCAsL5tK1WMzHFiyv1h5R4B5ZWU0JiUzGk71PqT/XLT+8eutJwFAAzvjG0WZVgWty7f7uJK04BMhHjWrt5MqYooo+AO3zDn5Q01dDJil4byWzzbx6NCz/WkFBAVluAl7x2ey2KXAj6zqkNr/tfwZ5iu3IlvIDP6KPXUNBpTIjhw8vGNAkL/BJycZx5QSedW5Ey0OdcXVtS69ho4rJTK2yBpLsEiqILIeA+48wt7EtEjzjuxHs2bKCdcFhPPa7DDK5imro8gP1kjph3uQgndq2pkvPYThoq5BdSnhayvub0ZaoVNps/pbhv8lXgJLeZBJzPAbyFNu3MgDwTspirHZR991BvUzz/27+fSPiul4jsupDTHrOyA+vaDYFzcyZ+KfmvLdM31IeuQKoVWhOGz25W/X76l+/fnWK5DW62yUky1TZtWtXsXR1ak58r2Jb17PZe9tLWXXHuWLJB88cuJ+A+64CuY44cQOAS3MCqT10acH7R+3nJPDyNugYzaCaqrwM1XSd6aGbxZ5X6fxYxmFkpdWzdzEbXah9Os8lJcqGXNl6lBXEQPSlcHpIN5ZHJgOwyOsMb5ef4u6cRbfu9+jnuVSqV+6Aq66aENhHoKGoQKqkwMKam5qLolrJK+exN3YzeMRUshxGcyv8MlbaKoCEgPCnGBvn7W8YOJigKhWY+jiB7XnuzCE7+hDfZSd2WgX9rBQZymNOc2JIXQB2tK1JnyUBXB0rRZL5hPFXU+hVQwuZZDXOlQ2YEz4GPY+O2G44Rz2FbBLSc5HJskhLS0NLS+urkbeSuhK5abkFo5VEPqCpKRTveGS5CWyYOYIF+yOZ8csZvu/fCACfec5sUxpLTPo8DJTS2DLSEadhxwja41ZqeWpVH82z557UNJQrsH162qFRqTUR2/qjoahQPE8Kmkizo+nZaR2/+vqTnZ5GRq4UaXYG6Rm5aGqID2F8DJnJagSdM/9i8mOYkvlFyUdS+cs550M5Rf+Lko00OeWrrffl3T8bFxeHo6Nj/rWHhwceHh5lPpOens6SJUs4e/Zs8T60hHOZFBTKP8n7qnu5d/fY2tUZwUbHnUBn7i1yodOxWqyeN4SeI2cwQrEliwoPVJrFrclqWsrkpBYcmiSVpBaZSLwqNPDIJOmg+DGTMsVifuIymSpKyMiQytDJWy2XpEuKTFQq5U0GFVUVqWTVhylTGubfmzJlCqbaqlRwmUfE61FcPX+RS2eP0NhkKbdfF7UA6tYdTVqMB5FZ31Oz0EpzSuQqGjouJykzoaCzyoygUe3mtJ67ml4jumC+eCq1askVfdUKLbkW8Yp71y9y5dJ5JrY0pdeVZyx2KDn8O8P3yUUBFQUYOXFyoRXwKahomhaTAYCmogKZ7xyWklpoAUGalYWiih7KmsrkpuQWnnWQLQM1RYVyl2n1csgVQFG5wM3iffXv3bwqKKiirGHOlClTijdS9ffvg1XRLX2P7ttDtcqqO6V2EApQ+GyfzJcRvFKvgaKyItJCN6TZL3gSrYnOO+0GIE0iQ6WMTqmsevYuuamFJnLSNBQUlP8jp1ILyk/LZRupnyMBwEBbhWdv66iOCtKsjFLbrOADZFxRjVtJ2XTMO0065n4i+mOLT+hiLy3EsvcJ1noFMKS5UaG+LhUNzUL7cRWU6aynzqHXBRPmBXNvMz2gcZH07LVU6dKu4LC/Ju5GzDvwCpXp9iiqGtKrhlxZVVCqwCADTf55moLsmSIPJrtycrL8HISUxMuY191CTPTTr0beeg6GpC/3B+QeBtnJt1DWMMtfwCvoCDOZ2KI+AQ7fE/x8YpF9sQd/f0jzE0epqqoI6DB48Sim2awD3EotT0m6AloVNQoWIis0p7KSlCeZuZja6BJ/Kx7ayxcnkoOeoaE/jOzUAEKVchnbWL4gnJ0YR8qZLjQ8sZigW2NE4xEIBO9FBuW22Orr6+Pr6/tB6YeHhxMREZFvrY2KiqJhw4b4+PhgZGREZGRkftyoqCgMDQ3Lnbbiv6kglFQNyc2ST6N8/3xIk/XLGejWBstaFTl97/2HWdWdZMuDRWvzJ1v+v/2Yf69zVyPOfns0//rC4gNUazn4k+RbQUmbsUZaTDzyJG/SkcLqLcE09jArFteoWxde+xyhUj1r7OzsqKXhi3u/ZVRQUmBNC1tmPFbG1W0gP244gKt6DKcTMvKUAPn/pK7nxtKmSrgOXU5CrizvXgabx26iRtt1aBc6sCcj4Rjhqq1ZN2sUrRrZoBh6KP/ew5WdcZniS4NWnZmyYDWrWhtw7urLUsPLwzQzXfY8VsXOzg47O1suzRzIqpi0EuO2qqrJreSi+1K3Lb+cr7xun3gT81GtMOnnTMSBZcTlKXeRZych0+2Kpabye8v0rd5cHrm+y/vq3+7FF/LzumuyN7X6tSYn/RE//PBDsd/PW0I+qD5pKCsQ+Vwut5y0QLbk7bctq+5kxPpz/VZYsbSGtKzG/lU38ru6Bc0b8kNEMmYeToRs2UJG3v9/e1FHOi+8L59gPV/EjXj5BPlN4Fa8knUYWa1060xZ9exdgtatISWvfQbsmE4F06koAi/v3MQ3NDlPyY7hypUr5OQVza1rVwnJyBUj1edY0S2h+msZGmFiYoKJiUkRF/fKDTxICvsWnzfyfbXxAWu4niT22H4ME7rU5OByebuUZD3nh4AEJnaUK66F28L3A39mzIl/6GlfibS0NNLS0sjIkpL55iw1TBrmn2GQmx7Mmph0RtvIF+fSX+7CK9sBj+pF2+101+rs3uGf33f9tSMci6GmqOm60lkngZV+8lMtczNCWReTSh9rPc6GPSc2NpbY2FgCr3aigvEPX5VSC1Cp/lxkMT9zJ2/MebxnBVUbLyjW37z0mcQfLwdzesUoNLMz8mUuAxrXrcij7QWTvyfHz6JZvU2Z5Rl5agB1my5EkvdM3L2fSdZwwrmiGvVntiHk1435+3z3LbiP43dNUdfrlC/v2NhYDrvXwm7OBaHUCgSCD9Bs5Z/7Kc/vY7CxseHVq1c8ffqUp0+fYmRkhJ+fH9WqVcPNzY39+/eTlZVeoeJ3AAAgAElEQVRFREQEoaGhODk5lTvtf5VfimqFZuSmTmD38xTaLOzB9K4dGepmQ0xgGHXsdfGfPxXfNn+V+rxpzwOM2GmPsW1zWhhKCdbqiL7KHwA4rvibpq2csWhxgLrKUdyIq89Zb9dPlvdFJ9fh6tKI1r+3ISfiEmktZnK7dfHP4FQ0ncvGAR2wqutAOycT7p2/x/C/vAFwn9IKu5YNCOvYgpwob546TuafGjpEmVQifP0c5lttYtGQOkw5dYmgbzpSo9pWnBrWJTHoFvHVu3L2Uv+iE1SDIXSoNB+nbgOxUInnmUoDdLIfMHahP7+MHUOMxTc0C+lAdaWXXH5swqm9damdXnI4qVfeK4Mpp36lfdMWNPmjNXrpAQSrdcO/RbUS4zafWp/fTkazcFyBe2uPuO+wb70F3SQ/QrTcuDvVGn3V1Szu2gIzq+a0sdDiytVoVp65+t4yrV5VndUTJ2C9Y2u55Dr7na2C76t/Fq/mY+u8jcqp9whUbMvteXaoqitx8ODB/3ddajmnK+PcXBjUuynRAfGMsK1MwnvqzrOjE+i4qCEp0UX3q7ru3E8dp/Y0bNOK6ln3eWw8lMd2+qixi2mNGmHcwAVnEwUu3FPn+EMnCAJN/Y5MtHfCsEFV7p2/w4QtN6mhWvoaWtn1rGhcvWZx1LNvhUPVbM7fzWKbz0wATg7qys+ux3m0uSkZ8YdxcfEkNltCNRVFerR1pePD12w3ryQGq09I4TbSsjwLLvo9OTTrT5xr2+HaypjQQFU8DbWFID+Cpuv2YVC/NW1edkESchLTmYfora9RpC0Erq/HvrhsJC1qs6rQs3X6XCToz97sHbcTR1NH2jSrSdj1mzSZd4qeeWk8O7SVyjYzir237c69LLVsh8NFFyq+8SWm+kC8h8ndkrceXkCDVlacdW3CS5/L1J/yFyOrav4r5K2iZceJ+a1pa+dM10a6nLuhxt/35WcaFO5vIv++Q1rUQ3R1i56W/CA5gy5/7WF7yx5YtWpMHbUEroao8NtlzzLLU9pzH302O2DU8AZNDRW56f2C5cfOo6wAVZ02MdmmLvVadaOhegQ++sN50Lu2aBwCgeD/r9dSflfk8tC/f38uX75MXFwcRkZGLFy4kJEjR5YY18rKij59+mBpaYmysjIbN24s94nIAAoymexf6wuW9OQB/jHZWDrao6+czNUr97F1bkWlMjfkSXj2yI9YaRUaWdeiiChlWYQ98OeVtBINbM1LPXDnY5FkxfPw3kMU9M2xNSvb7B7/PIjAiERqNXDAuNCnYjJiQ/ELjkLDoA4NLfNcWGU53Ll2DUVjJxxqFUwkox/7EhydjE61OjSyNim5cktS8Lt1F+WqFtiZVSf+oTdBkno0t6uEJOsF93wfk6WmT8OG1vnyKC28PEhzEnl07z4Z6jVoZFun1Hg5af7UtttHZNjyIk0x5M4N4tSq08imTv4JlgAvIwIJisnCsoEdVQq7oZdSppLMaK5dD8akhTOm6kofJNf31b/BVbUYERKP0+tHPMusgKOdOWqf2J32TVgA96JSsW7aBIN3DnErre6scl7H9CuTitcBaTrB/n4kKVelka15ETeP6BB/whLAxtEOPWUFXt7ugu3YsURfb8S9u2FUqd+QWlU03t+JllHP3nK+owlLx17ByykX/yfJ1HOwQ19dCcF/h3fbSHlJjArmQVQWdo42VBCboz8eaQZBfneRVbOmvtHHffYq/fUz/B9FY1i/IbUM1Ms34ZFmEOJ/lzRNYxrWK7pNIjf9NQF+weiY2lK3RoV/ncjTXoZxLyID+0bWRbybyl9mmQTf9+d1rg52DSzztx69rzxfRTzk8YtcrBrYFOvzYkLu8jxbHydrk3+XC94XhLphTWqNnvbF5Mfw2pe1x1Yp88vxiFJ++vKLks2XtMf2VsYJkiRx5YpbqZ4BLtt7lytu5OTbH+yK/Dn5Vyu2gn83x0dbEbfgNsOMvi6rT5fKmowIiadHZY0vJk8JAX+y66UL09rX+H+l81axfXmvS5HwF5e2sf1myQOOhn53po2xfG/abxXbC91qicovEAgEAqHYCsVWKLafQbHVrWeA87Y+5YobPeXWF6XYiiPyBF8tHTecYtXBFzDI7KvKd/vBw6il9mU1PT27gXyKYVvDoD0Duxc/7KqygxvD6uaU+Iyiil650q7xTT86mYhvMgoEAoFAIBB8VqVc9nV6VAmLrUAgEAgEAoFAUALCYls2wmJbhnL4lVpsK9arSoutfcsV9+W0m8JiKxAIBAKBQCAQCASCLwwZ5Mq+zl37QrEVCAQCgUAgEAgEAsEHfcdWKLYCgUAgEAgEAoFAIPgiEYqtQCAQCAQCgUAgEAi+WmQoCMVWIBAIBAKBQCAQCARfuXIrFFuBQCAQCAQCgUAgEHzNSPkfVGzT09NFyQsEAoFAIBB8pWhqagohCASCfGSyr3ePraIoPoFAIBAIBALBv50RI0ZgYGCAtbW1EIZAUCoKSKSK5foJxVYgEAgEAoFAIPgPM2zYME6fPi0EIRC8B5lMoVw/odgKBAKBQCAQCAT/YVq1aoWenp4QhEBQllKL3BW5PL8vDXF4lEAgEAgEAoFAkMfWrVvZunUrAJL0NCEQwf+cZiuTfZ1ZFxZbgUAgEAgEAoEgDw8PD3x9ffH19UVJU0sIRPA/hxSFcv2+NITFViAQCAQCgUAgEAgEyCj/d2y/NNX28yi2smxOb1vDiduPUNCrScchU+hoXfmjkto0qDvxP+1mnrFOkfDIk9P5MXw0WyfW+48I6urOxaw6cJ6nkW+oWKMu7qPnMK23PWF7JjD3lhv7N3YoEv/B8hGsypjArh8cyE4JZOWc5Ry77k+6UkXqO7Rj9tJvsa+klh8/5tJ8jpvMwKN2BQDu/b2WZdv+5nFkMno1a+E2fDbT+jb9oDwv8vyB+Rt+KPV+SvglNuzwIjK7Iu36TaS7g/5HySb6/Ezm+Q5lx+wv/5TBzIRj9J8UyeE/xpcZb2L3rkzZd5g66sWbiEQiQUlJ6b3vWtK3G2G2s9g5p3lB08h9Q8cu/f+rh1dIc17StedsThzd+XEyTDxDv3H3ObJvZpnxvvnmmxLD1So0xuuvhWLk+NqRSpEqKqII5GaEMnPUTBLqL+L3ubbl7qvXDuhOvU376KCrLuT5gYSe382Oo9eRVanPgInjsdVVKzHezQMb+PPCXdCpTvtBU+jWwIDs1Dss++VUsbiGbcbi6P87R+Mzik5cFFSYN+87slNusmz1+SL32k2dTVMd1SJhe6aORmfuOtwrawDw6t5RVu85TXyuDq16jmGQc+2vTt6SzKf8tm4Hfk/Tadi6D2N7O5XcP8bdY/0vu3n8IpNaNi5MnNiHSsoK5ZJDTqofk39SYNNPDYql6/ujJ7Kpv9BIW7VQ/FB2bt3HnaA4bDsOY2L3hgCsXrKYFIm0yPMaldozc2IT0XAEAkG51NXy7p/90lx/P0t+jnk0pueUhdx4Fs+dv7fQu6k5ax+9+ai0wm/d5HFGTrFwzarW2NfR/o8IKersWHr+4I3Hj1u5dOMcG+b25shEV+bceEn1do6c3DueuJyig8iC9Uep1N0ESXY0ve1bc0XRiV92HeKfXWtorXebDk1HkiqR5Ska8QyfGMMIU7lSG7RzGG0nH6Sj5xIOnzrM0un9OD29C5OPPi/fAJyVyNnt41l98FypcXLSAmnepDdxVR1xtVBjQvuGnHpnMlNeMl4G4BuU+FU0VUXVajRxfP+kyu/m9fzyeZequhXJKsfeg8feN/lneTf2PU8tUGxl2Vy7du2/uxInzeT6jTsfr89kv+Cmd9B74y1YsIAFCxYwZ4Yb165dy7/+ftYAMWb8C1hva8Scp8kAxFyawvE3XfnZs94H9dUht27yIlsqhPmBxPktpsnA9Zg274pRziXaOg4mTVq8Uwrd0Y/u88/TsG0fWltrMaOtNZtDkwAZubm5RX6XN6/BJz0bmURSJDzt9SXWbb8LQMrzrWzcHVrk/rul9+TgGMZu3UtIhgSAtNi/sG8/iyoN2+HWojab+zRixb34r07m37VsyZ4oHbq0seHo9E54eD0r3jfmvsHNvi33lE3p3bcjGd6LcOz0c7nkkJEQwa9TR3Dy0ot3OuxsHl/bS//Vu4nJlhR5Vz/75lxO1ad7JydOT2nPt1dj5XMASdGyjbq0g92+CaLhAP3796dp06YEBwdjZGTE9u3bhVAEgpLmelKFcv3KQ0mf2Zo5cyb16tXD1taW7t27k5hYoEcsXboUMzMzLCwsOHPmzAfl+zNYbGVM+jscXbNF+J2bTlbSZfQMO7Nxph+TT7T5ZG9Rr2ZDAy25FTfu9lXe2DYj+fJBvCPSsG7TCxcL3fy40XcucdY3CC0TB7p3dEIlrxxSn/ly+MxtktDGzrkrLS3kJ+W99r5Ksn0z4i8f4EXD7mj+cgHz0X/TuVFdAPRa9WH78lPM2PcUrQ3D6VlxFt/eecX2ZtXkA9jL37mQaUi0dWUer3Eh0HwxYWs88vNj9tNxrh80Yml4Ekvq6vLs6Bikk5egrADSnFf0munFpoBI+taQTwbrGNdm9/6zNB7/G2vdfixb+pJUXFp3Iic3GahYaryIvyaTar2eVZP6A2DgfYCpix/QcbVTmelnJwVx4vgVklVr4tK5PSaaxatQxK3TnPUNQalCNZzd3DHXVc1TiF5z/NBJnidKsXbuhkt9vTLDS2xouQlcuhpIG9eWAMTfvUZgjgWtmhgAcPvyRQybOlNTTanEcldSqU5jhwr56WUlPMTL6wbqtZvT3iob/0Rzmpjp5L0rkSO/HyY6V4t2vXpTt6IqgdcuIQEunD9P27ZtUX1Pmx4ypxnTOs2i54NNJcaVSdO4dOIEQS+ysWn8DS1t9f9fdbr0xY5ITnmd55WsCq3aFygfqc/v8DDLgibmFYpfy7K4ceIQ957EU8mwPl27u1JBqfxOJ02byj0MspJzilxLs19w5fpTnCxSOXzsGX0HWnPNJxuXluZ5eX1W5Do95h7Hz90iTdWITj06UVVNSYw4/2USH94gJENCwo1LPNAx4961VyhpZBDyLA0DXdVS+2pkuVw/fYQH0Tk4tukqBPmReE34jcbrL+HRpzbQFf89BswLesMvlkX7zk0/XWLQsceMrK8HdKTmw8OM/ukB43a2YNEip0KK8s/s9Z7JhXZGKLSbTYG9UMr3zcxZdPKQPN6N59R0X86iRY1KzFdmwiXcv3+JayEL/LO/N6LlsJ5pg9oBYBiwg2FrHzNrV4uvRt5pL37jt+dGvPKZiYoCNDG+gc2ADWzt9nOReEkR3xOgMZKXiyYB0L61I5srWxCSMY3cMuSQGr2Gdr0OkJMSC0Wd09jcszO7Y1NJyi26cBF9zoM7evN4Pl/ufdTYOImN3onQqjoz5v+QHy8n7SEt99zg8Mn2ouEA+/btE0IQCN6nycn4pJ/yGTZsGJ6engwZMiQ/rF27dixduhRlZWW+/fZbli5dyvLly3n06BH79+8nMDCQmJgY2rZtS0hISLk8JeFzWGxlOUxduJgli/sAoKRqmDc+ftrjtaJPf8uEzXKr0Z0pQ5jwXW82XX+G5M0D+japz5k3mXJFZ9VQnEat5lnSG86sG4p9r+XyyfLLv7By7Mvd6GRSYx8wtkV9fn+VDsAtz0FMm9+FSVuOE5khwbCTCcFbZrDd6xrxmfIVU/OhO/Ha0Fi+6jDNkouzC1xLQzZuxqjtz2grKfDnr0E4L+/xTu4V2BMazZK6ckXl0A83Gd/DGIDkp8t5pdM7X6l9S7UWO3l2/8f3ykVBSZtrN29y+eyqMuOF7X5CnZGO+df1J9Yj9px3mc+kvzxOK6v2nH34mrDrO3Co14HnWZIiccL3DqfxwDW8Ssvh2d1DNLVsT6pEhkySykBba3bdCCUjPpRJrSzY8jSl1PBSK6ySFlP7dOFKUhYAP/fvRc++cpfY3PQgOrgPRFVBodRyz3xzgr7Dt+YpXFdxse3M1bA4ruyaQEePvkzZHVawYtR3EP6vs3nls43mdn0BCPK+gVQG169eJbscdbruoD8YpnmMnpselrAIkcI4V2u+/fMqSbGBzOpozaTdIR9dp0sjNyOErnaOLD92jyj/M7g1GpV/L/LYTCZuCynxend/B4ZvvkRaTjo3907BsdunWdnOTL5K78FL6NdmLBcCIsiI96LvyF359wtfx9/bgKPjUG6HJxF05VdsrboQmpErRp3/tmL72JfwjFyifLwJehPCnZBk0mP98X2UWEZfLWN5HwcGLP6bF7EPmNfJkRspWUKYH8HuJ8n0ca6afz3YqQo+J6OLxeuxdhtTaxcscMa9yEC7jk6xxdAxvbay55/pxfZKRRwczkn7TYyzrATAy4svUat+nx/nzGDe0jV4xxQ6LVaWxfcdRjL+6C4MVAomIFVbuZD4aCsPX6WRlRTBnpNROLgZfVXyTvD3QttwSv4CYqX640l/uZvsd4YAdd3B7Pp1aqEFxefIlCpQRUWxTDlo15iCt7c3R9YUX1ged+gc3t7e2GqqFAkP3hCA+bgOPA+8zdETF0k0HcX8CfWLPf/3qD403rkHMw1xpIpAICg/n/JzPyV9Zqt9+/YoK8v7pSZNmhAVFQWAl5cX/fr1Q01NDVNTU8zMzPDx8Sl3vj99T6egyqRJ8tVKmTSNjeN6o6CkwcTVjp+1AF7nTObsEhcA9P85xM7oNNpqvqLnTxc59fwJtloqyGZNpWtNI9ZET2Dwq9eMWnKKeR5yK2zje8dZ6RfH0G/kCmZ47BgeenWX/x+ex1iTPpcdi0YyZXASNs2c6dSjPzNGdUddEeoMW8yb+X14njUYYzUlVu0Kp/cFudJ7Mymb/gZl7B+T5bLmhQbP8vYiJYcHoVqxb9mTyvu+PE4r7p5t0agxesrlW6uIis8qMsFRrWiCJCO0zGfOjpiG5qyTbJ4kdyUwj2zD5mcpjCwUJzW6JkvPrWNkbXnaj/7R50pSFq5c5FR8dWLW/oimogJ9rWpzMymHzArXSwwvvX6pMc9enzW3XtGytQq70x2on3OcgLQcDINWoF17DvqyKBxKKffRhRZ8bk+diPLU42yYbgPMYa51dc4X2iZc7Yfd/NCkKjCBvyrrEpqRS89Zcxnz4zIWLFqEiiQBb+/gYllU0aqPo22edVVBkXnHNmFu1ZVLAx7jXGi94sVNT7zi+xF7eSkAEwZWx6TZMNYOuflRdXpKjZLdPYN/HU1Qjfk82TNBvnLWfDA2Q+PeW0diTTtxZsFSTNWVyB7jTNU6s4BRn6Qupsf9w1S/GFwqqZMas67UeCsHLKbzoQB+biKfxDv0qMeobSFcmWgpRp3/IrV6T6bjwuXETP2e3rUq0KDHZm74jGZKf9NSn0mN+oVlV7UJi9pLZRVFMie4Ut3EXQjzIwjNzMW8kKKjXUeL1LDUYvFadu6aNxanc2TtDMZercqZX4u2nZCdA4jstYOmFYpa2qW58Yycdps9Qdvywx4EvuH5s+sYjO9AVsQF3G0b8Ntjf9yqaOK9tCt+HXfyS31dCm90qGw7l8n1LGhqVgstxSyUTAcT1M3kq5J3angqmjWqFEycNM2RSlJ5kS3BuJAHiUaVZnR0zesbgy8wYcAQXL79m0rKivCJ5fA8Ko3nxz1w/8eQxlUkjB8xiQVXbzDaomKhNvcHMx60I8rFUDQagUDwQfwnP/ezY8cO+vaV6z7R0dE0aVJwHoCRkRHR0dH/RcU2j7SY63j2HcShQFXm/3mbifV0P6tQTIcUrFTqqSkhkcnIeL2fFHTYOM0z/16mVMblpylMaT6ezrH7WPbDTsKfhHL15gvqjCooRbMxBUJVUNRg8OxVDJ69itTYEC6cPcmmn8by900p/rt6olahJTNqKjDzcgy7HLw5kWXBKzP5/6uvokRMCXvI0qNCicytTu0qT0lXqZdvOlfT00CSHVNCDcvhwcMg6lhZE3Xib/6IKm7VHG3jiJ52+RRbDUUFctMLLF8ySQooaJb5zD8P39B1a6386yGHLgAQVmgWYzt1Ks/27WH+1lCeBN/nSlImQ5ChUakDHo3nUNu2JV3atabdN+70tdNDQVZyeFk0m+PId0t9Sa7tjbbtBGZIQ/nlYQIjN97BetacMst9dJ2CdI5dfUHLeQUTi65tqlP4WJT+VpXe1gD0lBV5t41LMp/zxx9/FMufjtG4AsUW0KzalSMz1tLbfSXh50YXLC4cDcS42+T86wp1PNHInMP91JyPq9OlKLYRB55h8W3nAoXdeRzg8d46MnnGEPZtW83m0HCC/K9Rkmv7x9ZFNZ0muFR634FBMrZHp+Hy21zG7JSnlZyYSeypWBCK7RfLuVF9WBUl33s77+BR3qoDcXfPU9FsJpVV5GWprtcW54pqQmAfgYaiAmmFDgfKTctFUa3k9vbCey8jx80iu8FwLgeepr62SqF+P5Whc++wLbz4oUJhuweR0PFXbLQK4g847cOQajXQUVIA+lEnyIo5CwJoPSuIvrsN8L3jQFpaGrkyGVkZaWTmaPHwpw7sUhxFeNxsqiils21cC1p6nMR/e+evRt5K6krvjJfyRQQ1heLWClnuG7bMGcvig1FMWXaUmX0cAPBd9GnlIAUUsofgd3wEADMO9KBl77WMvj8/P86+wfPptt37C/wgh0Ag+PIV2/L1HHFxcTg6FhgvPTw88PDwKPd7lixZgrKyMgMHDsx7b3GNWkGh/L3YZ1Fsk0P30qTJOBIM23EsYBetan7+Q56UNEvwvVZQRUXTCk/PAiUAT0+0TfUJWPoN3U8as2L2QNyHTmGIYjuWFnpURbdgMJ/ZtQOuu73oWEkd7ep16Ta0Lq6tpRg7rgB6AjBocRNafn+I0F5/YeK+LN9laZB1JVb8Hsai+Q2LZO23Li4cHHCay5M0kckK3PEqmg8nPdaTqKyZGBVaCU6NWkezFquITYimVt+ReGYVd8es+QGuRrWsdEnwSYA2cleo5JDnqFceXHZlUYDCOnrmq6e8Vi+6Ery4TQPO24xldt8BjJy2GDWnPAuOghorjj9g4sObXLl2nX2zu7Lp8mGuLG1canhpGDjNIzHAg7AduVhOnkhDRRu+W/sYyY0XzFpvBKmllzuFjBpKCnKlMH+ikCktortpKpbdkJQ1zIq+4224Ws1iYY4zj9BiWx1GH25b6HllclNzC8+IyJGBWt57P7ROl6pEaimTW8iqKpWU/rH57DfZeUr7U1patsF59nLch3Rk2oKJ1K8/ung9+si6qKhc+uKFLCcuf0FBWQGGjptQyCriiYpGLTHifME0+3E1FjnyLQpVtFSIfNun6qggzcosugAqkQmBfQTNK6jhk5RD+7y9rLEPk9AfVfzLAy+u/kTDgadZ+ddtBjStUfz+zam8MF+ErZZKsXuLF/oy6bZjEVUqR0WVSoX22ddpZUDakTSSwi6gnH2TJnbyE7GTEzJQatcE39mnsfozkKaHDmKgqgho03/BMGY32gR8PYptpQbVSf/lPiDfp5qd4oOyRh2qqr6zmCDNZHrbBjxoMJOA4HHoqxTcP/SJ5WBiqEXNHgX7lKu2dCXz9fGCvjz1DnMf6RHuaCAajEAg+DClFoVyK7b6+vr4+vp+1Ht+//13jh8/zoULF/KVVyMjIyIjI/PjREVFYWhYfq+Tz3Iq8vRvpvAsM5cWXay5vmcdP/30E+t+D/+PF4ymwUD0Mq7yRKsWNjY2WNbVZWb/3ryUyfA7EIjTqsX07exCPZMKnPMv/ZTGhrII5sw7TOHdpMHHLqFp2KVgIt9hJZIni5mzIYjhcwuO3mi7ZRZha/uy40ZsflhCwB6WRGQwY6QZyuq1qSENIj1PwVKv1JlFjZXo5PELb/IOi5BJM/ht0lYMW69CW0mB0G2rWbJkSbFfSadHFybjxX1u+sjLwWKKC2Hbf83fI3Rw8UMaznAqFq8wA5pX5eA67/xqv7hNM5a8sx92+/0Efl42jY4tG2Go+YzbKVnIZJAatZKGLaZjZN2MQeNmsXldc8JO3is1vCxUtKwZXiGcyTtC8XCqQpXGU4i5MIezCp1xrahWZrkXplsHQy6ukrv9yiSpbDxdflcHqUxGTsbjEsth9bbiLt0KStpsOLGQI+OH5ocZ927Js79XEZ8rXy2IujADWcVO1NNU/qg6Lc2O5dq1a+S8oyuYjbPm4dJN+ac839+5rMiCUGr4q3zFev9fT+V14M1JIlRbsXLqMFo4WKEY5lVifj62LhaRjaIGkvRH+aerBmwrOAFvUu2K7AtWxcbGBhsba658P5y1sXLF3OfG9fz9tq/u3sIvTF4X35VD4XiCT72iWzxMq3oNjI2NMTY2RqPQ4pCe3QiSn8zFN1G+kJfwYAM3k8Ue24/Bo2MNDv0i74slWZEsefCGse1rFGsLC4avYeShvXSz1SUtLY20tDQysgpWJy98d44G8zsWSz/91R8cz27AiGpaRcJ72FiwJiBvrJRlc3BXOLaj62DU9g+ePHmS/+tWWZPp1x/gNdaCRmYVefy7X34aT0+dR6Na669K3roW3yKLXc3dFPnCX/De1Rg4fl+sv3nlO4N9r/pzZPEwNLIz8mUug08uB8tZzQn9bX/+GP5o/2EqWvTJv//86FwqNFyItpKw1woEgo9Rbsv3+1hOnz7N8uXLOXr0KJqaBR6jbm5u7N+/n6ysLCIiIggNDcXJyanc6X5yi21uRgh7X8gnnqfW/czbL+VVc3Jk0tA6H5XmMSfTIk6QVRv9zfH+5dDaVapwYtMY2jd1YrurA0n+F6nSeztNdFSpOqcbs3u5M7qzFbGPn1DbTpf7P36Ln8ueYun03PcXxzv1xNhmPY5mVUh5EUpgUg22nyk4JEJJvTbL7HWYGmLJ0UIWap1ao7m09gndu9mw1tyemhUyuH0nisGrLuR/42+WmTK7XqYxvrr8uQmHTxHs3h0z0x042puRGOJDQrVOHDvVG4AGPz4+Eo4AACAASURBVG3ij4+QY+SJqXRfas/LsFUYOK5hvJUdDdr3wV7tKb6VB3Onh2mxeIVx/vV3arfsSrNOLaiW/ZBgo0H42VQm8kFBnJ/c69G7Qz861tciJEJGo8rqLJ+5nG5bp2CXbYtFqyCaGGvgczGQ6f9sRKuaaonh72PQ0DpsW69Ep0rqQCucVYKI6rnmveWeVshY2WjFASxbuuLs1gKtN0+xbqZPVNX3f0/TTFOFgWPGsmvLlhJdkUufHI1ld7+t9N0lv65ss5z5ndpi7diG1uaaXLsew09Hz310nU57cZRvvpnGk8QUqhayFNRy38PQPU2wcGpDs+pSQrXaU1l5v3xlrPNYcmYOw32YOyovHmA2yATiQavKQNpW+pFWfYZjrpJApIod2tmBTPrpPssKbbP92LpYRFnX741zhbk07DKIhtpxJNbuAsgte55H1tO1dRtc9jlTKeMBoapduNVMvt+2f5dOtL/znM1mupwZ2YvVzv/gt7YxGQlF5VA4nuDTUc1AnQ3Tp2C5ZQPNyhFfo7I7+6YeoINlY5xb1CT8sSpjq2sLQX4EjVf+jkHDb+j0qiPS0DPUmrKPHnnjydu2cHeVBQfjs5G0taLwLnbTHifx39kcpJn89PgNP5fw/fJIrx3oWU8utg6+d/sUXNs34rJLY9IibpBR34Oz3WuVmdeOf2zn97b9cGjfiNqqb7gepsLG02O/KnmraNlw6DtnOjfuQCeHilz0VuVPH/n+8ML9TdThu6RHP8LQcFOR5++8iP/kcqjhsoUheg2o2/QGjSpncP2ZDn9cLJgYXVr9mHo/2IjGIhAIPkqrlUk/3aJY//79uXz5MnFxcRgZGbFw4UKWLl1KVlYW7drJT4pv0qQJW7ZswcrKij59+mBpaYmysjIbN24s94nIAAoy2cdvD05PT/8qyic7MZqAB0/QrmlH/VoFn3pJjggkIDab+g1tqaycwvXrD7Bp0QJd5ZILMzroHuGxiahXNqKhjTnKH1DmkozX+Pk9Jl2qSt0GDlQvtM/p1e3JdDkyBJ+lDkWeiQn2IzQmBe2qpjhYGn8W2cSG3iMypzKOlsZFzPfrOmxi0pnxxeu6NJ3Q+/4kKRvgYG1Wosk/7P5tXsv0aGBjjkJSELcCc3BuYQOyHB4F+BGXroSZvT2Gby2TpYV/pnJ/S+KDG4Tq1UUjKgzdeg48GGLL1mkXOexctstDevQDfMJTadGy6QfVgdJ49fQx/8feWUdVlXUB/PdIKQVEEAUUGxBRCQu7Cxu7u7vHLuweHduxdcYYuxUDE8HAABVJCQGl4cX3x2MeMgqig9/gzPmt9dZ679zz9j13n9z7xH0enoqNgz0muup/69l85tSl/Iyr6HySMTKCnvnwVm6Co20JPr5L2vvX3PUJoXAFJyqY6WTmtSwBnzsP0DAti33posT43ea5rDw17PPeQFTIPuB92weFYVmcbM2zXJOnv+eZ7yOSCxTDsWLmO4g/BE5juWQqc0oYiE7oH0CWEsZNL38sa7hSskDuO533of48Dk2lUlU7DDTEbNI3I0/mhc8DFGZ2lC9e6P+Y77E88XmChllZbK3NcpnWFPwfPyRKakAlhwo/7CxiYuRLfANTqORo+23PkOd6UPA24BEvP2ji8DfkfTxbIviUAsUsKTlwXL5JT7FrKflKP+op+WdFlEZgRP5qpj/E55u03Eo+yXtZdO7KfOniWHoMzVXcQouPfvNS5O/Bf8Kwzf+ekTQGO7dh0a2TuT7V+HsS++gAuyJrM6rhP3WSoowlS7J/XVHz4WOzHGjyrYScG0H16XK2Lu+O9O0dxo7azqGX93HIA9n/BOmJD1mzOZrxYxr8J6rNhkVL6TlpglhqJxAIBH8DYdgKw1YYtsKw/atha7Eod4at4ZL8ZdiKF5vlC/eCFssO9eV0WBLuVv/80jwj+86M+mebSXr06JF9Jcqj9/FZNFnH8bQtHDp+CLmOBdu8bv6wRi2Apl4lxo/571SboVMnirZDIBAI/kOMHDkyxxNS16xZI5QkEPxNFOT+VOT8hjBs8wkG1u64CzWo+JoT0P4OVVoNoEoroW+BQCAQCPI7H79WRCAQfE/LVhi2AoFAIBAIBALBd6F3795ZficmJqKnpycUIxDktW37g76NT01knUAgEAgEAoHgR8HLywtbW1tsbGwA8PX1ZdiwYUIxAkGeWbZ83/f9fCfEjK1AIBAIBAKB4IdhzJgxnD17Fjc3NwAcHBzw9PT8voP8fEJCca18lRcGIflHOXITo3ylmyTnEvlHN1cvfkVsSZ6+7kcYtgKBQCAQCAQCQTZYWlpm+f0177oUCAQ5oBCHRwkEAoFAIBAIBP8Xo/bmzZtIJBLS0tJYs2aNalmyQCDIG+P2R0TssRUIBAKBQCAQ/DBs3LiR9evXExoaSvHixfHx8WH9+vVCMQJBniHJ5Sd/8bdmbMVLvQUCgUAgEAgE/09MTEzYs2ePUIRA8L0QM7YCgUAgEAgEAsH35dWrV7Ru3ZoiRYpgampKmzZtePXqlVCMQJCXhu0PeCqyMGwFAoFAIBAIBD8M3bp1w93dnfDwcMLCwujUqRNdu3YVihEI8sioVcglufoIw1YgEAgEAoFAIPjWcbdCQc+ePdHQ0EBDQ4MePXogkUiEYgSCPDRuxXtsBQKBQCAQCASC70BMTAwA9evXx8PDgy5duiCRSDhw4AAtW7YUChII8sywFa/7EQgEAoFAIBAIvguOjo5IJBIUCuVU0S+//KK6JpFImDFjhlCSQJAHSH7Qw6OEYSsQCAQCgUAgyPe8fv1aKEEg+N7k02XG/5hhq5DFs2fNYs7f9UfHtDQdB0+kkY3Rd3mA8Os7mbBoJw/934CBOfXaDGLxTz3RVcuYQlekcXbbQlbvOcfLoBgKW5WiVa8xTOnTRLXBuFvTRoSnyQBQUy9AcRsnhkyeQk0Lvcxnkiexa/EUtv52mZB4BSXLOTF05hI6upiqZHj8cRYrbfU8e7ag4yOZGTCUHWNtiby5gX4zDtB01wmanhzDCv1pbOxa6qtlhl6YyjHrKQwrXSjbOCFnRzPlTn92z6iUbZy0D9dw63WRM0dn52HBSef4ppWcvPkcc4f6jBndnULqkq+KJ0t5zYblm7j3OhGnRt0Y0aW66m8fAi6watNhglINadZjLB2di2QRG3VvPhsUg5n5l/B/M32bNyYwRZolTF2zMBfO/fa3ykdekRJ7inb9fTh9eJroaPIbcjlyNTXUAGnyC0b3HM07u0Xsn1M523bsryzr0ALbLYdpYVRA6PMreXF2G5uOeCI3taXX2NFUNtL+bLzre1ax8/xdMChG8z4TaO9oBkDkzV9YezpEFU+ipsPcOcp6lp7wnM3rd3P7aRSVWw1gbEcnVbzbB1fx69n7YGhN15GTcC2pn9H2hrBx8Vru+kdStHwNRkzsj0UBZX+4YNZMUuSZoyTLFiMZVMP0h9J3Tn1LljYr6j4rlmzDLzwF60oNGDuuG8YaWfuxD693M2q5ITvWtQJgyexZxMvkWeLoGDdn2tiaJAReZfG6g7x+J6Ni7Q5M6NuYv4j7Knn/Jh4/foyfnx8pKSmqsF69eonGQSD420jydClyv379OHHiBKampjx+/BhQbivo3LkzgYGBlCxZkoMHD2JkpLQVFy1axNatW1FXV2fNmjU0bdo01/f6LodHbWlrT6/xSwh4l8DV7ctp5lCOM7GpeX6f1LgLVGw8Foc+cznvdZdjvy5EfmoiNcZcUsXZ0KMynT1u033cQk5eOsXySZ25NrMDNcedy+yoPa9SfdxPzJ8/nxkTBmKrfp/65atw9l1mY7muvT2TjycyZeUOLp06wNj2JRhQrwonMuLc9rxKkjxv3Ru6RStRtYwBAHt7TcNw7Gp6memhX7IKlT8yunM9Dk2PptvgUAaVKpRjvOS3D7jtF5dzJ58exbUbT/L0ea9PcaXvz8+o064p709PxrnHwa+ON97Zme3BBrRp4sCRkQ3oczhQOVBLfISjgxtRRV1obKPNwDq2nIhOVv0vwv8Go9yXcjUs8T/VdN3xvEqVMdOZP3++6jN3zuS/XT7yzHZKC+fajaeij8mHrCxbmEmv32c4zIZxLLYtq8fY5tiO/ZXnN68RniYXyvxKou7NwqHjSkrVbotV+kVcK7qT+Jn+5/mmdjSfehanpl1pVEmPUa6lWftCWXefrV/GnhcxSKVS1QdALo2hXXlHLiYUoWPr6pwcVoexl8OUxvS2TjSedIXqrbpQzTKephVdeJSk/N80l8ocijGmXY8O6L9cg1P9BQBIk58za/G2LPeRyn+8qYDs+pYs7ZU0hiYVXLmvUZou3VqRfGMGFRss/Eucd/SpPZRjp/0z+1OZNIt+gi9sYtvtd0iTn+Nk34YUCyd6tK+P71J3Gi3y+WZ5/ybmzJnDyJEjGTlyJJcvX2bSpEn88ccfonEQCPKKPDw8qk+fPpw5cyZLmIeHBw0bNsTf35+GDRvi4eEBgJ+fH/v37+fJkyecOXOGYcOGIZPJcp3svJ+xVaQx+lQQFg1/58bZdkTc6ULRagdYcTuSZs0s8/RW73yXIys6jkmdXJUBheuw5OhcGnY+BDQk9vkcxhzX43nUCUpmzKSWsSrF73e0KOs8kndLnlJYQ2nbl3GpQS0zXQDqNWuPXVRZBvU8yZtTHXj/agkTLxcl5N1mTDLilygzhwNnDzJ98WNaLXHKki5pcjDHDp4kMFZGCfs6dGhoz59+j9B7pznh9RytIuXo0Kk5BTNmGj8XrmPugKOeAe/ue3I3Pg2NuFeEpdpRvGQVKmtkGrYhty9w+s5T9Es607FVdTQlSoPgyq0UalSI59DRQHoNciPwSF/kE5aovL05pRNAIXvPxWtB1KmiyR+HL5JsWJb2bRqjp5YZK/WdD7v2XUZhWJYuXVtikPE8CYF3OHTKi/foU7l+W+rZFP5CuUml71pv1gZepqupLu5NbFhfqAaBOzqq8u5L8YrEbGJDoCUJD6ehKYGaJa5RtsNKdrRfzcu9Q0mo9Atrx/UEwPTGHkbM8qXV+uo8WdmZHr++4ENEMiVzXc5TufbHQe4HvMPIwpa2HRtTUJLEufPXqd+kKVoZKop54Mkz3crULF+QpND7HDtzkwQtS1q7t6ZoxnPduHCeGnUdOL77AC7dh2Oumf6J7D9npFPfPeTw4WsUKF2b5vZpeMeVo2bZggDZyv8SpV1qUMv8U0fJl8qH0vCM5NjB47yJk2Nfvz0N7QrnWC5zVKnsA+eOHeNFjDrVG7TGudSnxtBXlyvBdyH24TWeJUt553kRX4Ny3L8agbpOMs8DEzAzMs4S9892TJnJUjxP/o5vSBouTdoKRX4jhwduoOYmL4Z1LQ20xXu7PpP9YlhXMWt9WDPnAn3OBTLYrjDQCquHv9Fr9kNG7q3D83sxND0xn0Vls66mCjnTh9uF5xE1bxQANUq8Z/WNWKhfjPOLLlJz+yN61i8OtOT5ykLMeRXHPutQVvgbkOw7GQ0JtKpXgZm65YiRzkAr9iy6RdxZtGjRD6vvxPAN2fYtHxP3ciI+OoP5sGgcAM0bubBGx4pnyZOpoKMcbh0ZXpe4ziXgaOb/ps5boPqenvgQ5+2enL7UnBi/LkRbrmTpmL4AOJU8TqkWe2Ba5qqIr5H3b+K3337D19eXKlWqsH37diIiIhgwYIBoHASCvCIPfc516tQhMDCrM/DYsWNcuXIFgN69e1OvXj0WL17MsWPH6NKlC9ra2lhbW1OmTBnu3LlDjRo1cnWvvJ+xlahz9+EjLu9uBsjxORuARE2bjvZ5vxS5YOkmxIcsZcq6A/hHKmff9IoO5tbVjQA8mrubMj3XZjWMAN2iXQkNfq4yaj9HvcWDiLixDIDna3Zh1XqRyqj9k6YHnnLvL0atQhpHi9I2bL7+kvTECDYNrEWbzc8BeH1oEBXaLiXsQzL3D46nQv0lOYYHnxzLwLVPiX3oxZsUGYFe1/FPluK/bRgjDypfRO7l0YVKPZcSGBfDqRVdKN96PgAp7y/j5j6bdrX6cf6BMu7BadcZ3ankF9Op6hCT/Gjp1o8WTn25EfCWM2sHUK7eWFVZT0/yw63DEqKlCq6u60flrkpvadLbfZSya8vdkPfEh/nSz8marRFJOeZlSuxZAhWWdDVVOhc0dO1pZajg14jEXMd7530YA4sJKgPK2G4UiW+3kaYA/20vKTO4mkqO3Vhbws5cz/h+gAcPHrCukkmuy9629rZ0W3uRhPRErv86jIrNfkGipseG7u2Y9zJzNnNc8xYcSU8n+v4q7Cp24WZAHH6X11OuVGOeJytnOga1aYlH2/psPn2bZLnis7IBUt9fpnq5Rlz2j+LS1oHU79OWYduUXvqc5H+TfyoX5UMhi6dj2TJsufaCpOgXDHGxYt3rDzmWy+zvF0PvamWYc+QucYFedKxUgvX+77PE+ZZyJfg+xPndJiBZSvCt6zyNecbtZ+9JCvPm9uNPZ/L/bMdAwfw2tnSYtZ/wsIdMaVCRax9ShTK/gW0v39O1ftFMb3h1U24dD/kknvuGXUwqY6j6HRWehEFZpZPhbEwK5jd/ZeKo0SxYuY236cqW/emqB5Qb2Zw3j7w48sd54koNYd5oOwCqtrbm2aqDRCfLiHp+joMxGnQz10NDpwwvX9xSOU0jfI6iVag2RhoSksKuoGlgxtq5Uxk9eTb7PIN+OH3n1LdkceIY9WXv9kmq37KUNyjUC2GqqRw7hF+dwain7hzsXz7be+3v2YYaew5SVkcD4wrr8b3cVTXKfHgyAKOKLqq4Xyvv34SOjg5qampoaGjw4cMHTE1NefXq1Rf/FxwcTP369bGxscHOzo7Vq1eLBkUg+GRQhnIpcm4+30hERATm5uYAmJubExkZCUBoaCiWlpkToRYWFoSGhuZa7ndo6dSxs1N2go3N9LgQmUSJNmsYWFw/z++kbzGWOzvTmf3zAiqP6Y6pXU2aterA6KnDqKCvSZB3LGYLzVTxYx4vY/zyzOWzzZb8TOciOp+VrWXgRFriLOVg4EYUJv1zt+9SmvyCij2XsmLxUOWAo6E3lUcGwMDyPF78Bw5zrjKnf3nk6X0psVq5jzG78D8p03cyDWYvRHvKbNwKF+D+n51m6htazbnApehwKutpopg+iSYmxiwNGc1wbUiKOsBEvzgaGhcAhZRl4TpEmuh8MZ0fkxZ/D7cHcYwqXQgUM+hkZsRY35/wsABZahBLTvrgoKdJYhc9zCofAdqQFBrJkKWXmDusAgA17h/D414UfZvq43X72Sc609SzxdbkBRo65bKEl9fR4MVfjDNpUvbx4gPi0bXI3LeloVseuSyB8DQZwdGpqgEdgJZhSaTJL7657IWVas3VBcspVUCdtOENMCg+BhjCrJEVcJ/pzby9DUiO2sf+lKok2BVmivUs3E4+Z3VN5WDUuWUJem18xu2xFQF4MuQ0J1pbAbA3G9k3RwxGY+J5Nk1xAGYzubQhZzO2uXp0+Lx8z94S7j391NgwtHHCzli5J29sKRMmfHStyblX/FY5+IvlIyXuAieiixG3wQNdNQnd7UtzLS49x3I50eLzS1KDTvXmtHQCUbuUg8LOJaMYs8mfvh8lLLty1b9lCdEJ/Z+x7jKBVtMXEDppFl2sC+HYaQ3Xbg1lYs/s9/3HBy9m3mUDQmN+x0RTjZTRjShk2lwo8xt4kSylvK6m6rdBWX0S/BM+iVfXTTkrrpAn8fuyUfS9UpSrOyqCQsrFuBTeXHzLsCaVeXZiLeXXHCc44DBvghJ5c6wPzQ8Wp4apjAHdhzD/zn2G2hjiNG8nRYs5YW4yD0lyHHbDf6d9YWWfYmWphzTpMSVK1SE8MoHxxwKQABFXQkgI8SfeaCyVDENY0Ko8lzb7sbmz9Q+j75z6lhIfOc51TF1p1Vj5PfbpOQZ27ELDn45jrKFGeuIj3Dod449X95EEd/nsfRKCdzDatxnvGhZX3kfPjOJ68M6vL7a19hGbVpI/3ij30X6LvH8TTk5OxMXFMXDgQBwdHdHX18fFxeXLg14NDZYvX07VqlWJj4/H0dGRxo0bY2trKxoWgeAjcnsqcnR0NE5OmZN8gwYNYtCgQd9uUys+vfHXvKP6u7rw1py6yLXfZzB40SjctjbleP9yeX4Px26TON5tEtLECG5cPMOutbNxtrlExJujFDLQICEgs7MvULgqjRopvQNnxg3m7sxV2Rq2spQ3aGhZAKBtok1yWPKncVKD8AuQYW+X2UFrGrgwuUsMKxbM4EVAAPeueKIwHKJsiJcN501LZ2ofbkmDBo3o2GdAjuFfIilyNx8wYPWIzAKULFdw6XU8wyuAdsFaSqMW5exqopataoo+p3RmcVNomSmNWgCJFqOaFGfMsRAYDtqFauOgpxxcqWkWAYVyDbyJ4yjcwnYxb/oWAgKec/laOGWHKJClvGHHjh2f3MPAciTzBhRAIc86MIuXydH+S2GWqGUfT72AOtLETENYIYtX5p9Ego6a5NNrEt1vLncTpvZj18YlrHkewFPvK6BQzorYjJhJkPUkpIp7PFnmQYXh29CQKNgYkkDDDZPpu1mZA+9jUwg7EQYZhm3/2mZflH308lvqzS2pite2STHOZrjWspOfUP8RO3Z86kyoMLyCyrBd7h/BsI+WIkvU1VHD/IvlQ8e4BcNqTsS8rDNtmjWiacsOdK9SmITgjdmWy+wM21c7n1Cqd+ZSxXID9nMKSIrwVYVlV64E+YszvdqwOFg5cz/32Fn+dAlG3z2HYblpmGTMXhUo3JQGhtpCYd+AjpqEhI8OB5ImSFHT/vwKpPAbv9Kz31hSHQdy6+UV7PQ1ARm+LwOxssrwinfvybMiBRn7NAZnQJLWD7/zyvo7dU9LnN2WM9R/HtNr1afg9MOkTGqBLNaPvlWqM6ilE5sythlp6FYkJCyMp5d2ULO1Cx0ig6jS9yhvBhehqIGyr2hq8xib3ovZ3HnjD6PvnPqWTwZl0hjWTezHrP3BTFhxlmldnZXjIbfmVFp3ngqSNGKSpCgUqSQmJqKnl9n27nKfSvvdDz7Z8lHYdjthYSs5vaE37k4D+BC452/J+zfw888/AzBkyBCaNWvGhw8fqFTpywcampubq2aJDAwMsLGxITQ0VBi2AsEnjVnuopmYmHDv3r2vFm9mZkZ4eDjm5uaEh4djaqp0HlpYWBAcHKyKFxISQrFixf45w1aW8pL5i3djWn0AQ5tWp0Ll3xjiYcSddc8gjw3b2yPbsKPFOjY0t0RDz4y6br2p06ojJ3QKcSImhaoj7XgxezdMXAyArnkDuncHFFK2jRiAWQ6yX+7eRKEywwAoM9iOwAmHYW7VLHH81nSkzi/tiQ2YogqLfbaYMvUOMmftTwxs24efxqfi2DOjQa03g9dRA/C8cInL545SrcQibkc9wz6bcM0veVMkWmjq2TNmzJjMwDFjMChVBFJATcP4I2NFF4UiJVfpzFKu5SlIFWTuy02UopYxMJWoff4Aqwdz69HieElWzuhFh/4T6KdWm7mAhk7ZrGn9sxAWsEKrkIL0xBnEyxSqfbq349MYVijrwFerUO1s4xk7FiNpsQ+gnAVK+3ALDZ0yFNVSw9rekHe33kET5QDsw7M36Jj0+cYy/hrnUrWo/9NKOvZrRdn5YylZsnfGYL0t/Q16suh1HDe2BLA4oCIgQVMC/UeO/si7PwZN3UyHiFGGTnOSrS5RGoh/Ik/5c2CbvfyCRcszZsynMzn6VgVV39XU1VFXz7pcP1flQ6LNqvMBjHt4nUtXr7J7fBPWXDzFxbE5lMtsUNNQQ/7RIULytLe8CtWlmO6Xy5Ugf1HbYz026Uonl6m+Jm9UzjRN5KlZHYQJMuGY+CYdF9Lm1vs0mmecJh32MA6TIZ9upQi/PAfbTidZfcyXXrUsPjLMEtDR/Wg/rkSDlsYFOByVQicLPaw61VFdKlqvMclRR1FI41jqG43fzSaoA+pGtkwbY0vTxX4kO95k5W5Lpo2tiUStALaNhtDZaCK/hCey2lgDA73MoYahbU2kycd/KH3n1LdkQZ7CSFcbfB2n8TxoJEU0M6+ffaPGo9ENODVaudUjPu4KZcttJCw0UCkz/jaTHxcmzCVzZPLm0CpOF+3KkNpmqOsY0nz4WuLHlyA0bdc3yfs34O3tneO1qlWr5lpWYGAgDx48oFq1aqJREQj+z7i5ubFz506mTJnCzp07adOmjSq8W7dujBs3jrCwMPz9/XO1GuO7GbZqmiZsXTSfmMKPqHBsBnHXFqFQKDBvlvdLYUxqydk77CdmPNtGsYwBfeyTfcRKzHAtqI2Z+3YqjChH55UN2Du2KeoAilT2z27NtQ+pVPms0RLL/XPbcJ/hzbjryj2jVi22Uq63Nb1X1mXnWOU6o/R4P8Z7PKbu+n1Z/v/20jFM6ngwrpcy3uXZgaprq1wrEbj6IqvcutPArSsP9ulzJiaZi01qfTa89ReeX9esN4WT5vJS35r2pQsiSw2ioW1tFj7wp+gnxmNpLGRPSZIr0FWT5JjOLP209D2jzofwcxMLZCmvmHwxlKaLrYCAbNN1b89jqu88QPfqRUGezNQH76CLctZ49uxln8QvVHIKW5Y60quIlKme4ayrX4y4Z+vwpTI9MvbS3rrmibFTTcrpOWQbT6H/E4qw+tz9MB7nglo83bUEs2rK5eQ2Exvyov160mZsRksC+2Y9xGnqlzeiq+770f6k5JjjvNSqz6NJypn1iFtZzatRMyvTcEh/kotMpmnGKzjGlTFk11MtDvWuCChY1cSeR9PPs7VswSz/zUl2hxbF6e9xHTa3QiGLZ9XJYOhPjvI94nYye8mjT57LftLPOb7WKDflIz54EdXahfHk3lr6VHKlSaXbVBx4H12P7MtlybQwrnn5U7NO3SyHSZUZ5MKLARtJnrgeHTUJt+c2p0/IWh4s/nK5Aoi4e5Ngw4o4lS2IhO7CJAAAIABJREFU/C/3+FweCvKGzx1uq1fMgs+5vApXGcT7gFHcie2Ei5E273xXcf19Kn2EGr+a4a0sGbL4BnM2NEeWGsRs3xhmNrf4pC5M676UwSef0qGiEYmJiRlOJB2IP0fxEjN4FP2Y8joaSJOesyosiXn2xlScVpsXk/aQNmAeWhJ4vPsQhhV6I9EoRK2C2my5E8nyesUAOecOB1GsuTkKRRSzZwyi5zBfLLXVkSY95WxsKouNCnCynR2rmv3O7enK9tZ3xwFMqo78ofRtZJN93/JxexNzZxS7I3oSumQApCWTmJbRV+vpcS4gc2/xu6cdKNPKlbCXYzON2COTKOi8EP2PXnGXnnacmWNSGHx/ChIg5tE2tPSrYK6l9k3y/g2MHz8+B2e/hEuXLuVKTkJCAh06dGDVqlUULFjwk+ubNm1i06ZNynFhUqJodAT/OSR56Hfu2rUrV65cITo6GgsLC+bMmcOUKVNwd3dn69atWFlZcejQIQDs7Oxwd3fH1tYWDQ0N1q9f/8nky//VsJWoF+L8lrHU67+CBs6/A1Cidi9+n1U5z5Ve2v0gU082pnTRMtRwsYHESO75RjL+16sU01IDrDh3/wAdW3XDaFlhqtpZEOnvh+OA9Zzp9IpTH8kaZFGIIYBCoY55eSdG7PRmclWlB1xNy5yzXjto3rw9puusqVzKGL+btyjdZRGnOpfOkqYSHcYjn9yN9r1ao3j7BPXyJYj1n8Yq7+q0HVMHh9pVCGjuSnqIF4FOo/m9uAHh2YR/6TXkapqmXNwygjqVK/FLY2fivC9g2nUXNQtqkRT1Sc4wvZwGW94mMqqYfo7pbJnFIC5B3NS6NFhjxzvvi+h3Wcn8coYk5/DmgIZz2jO+dXN6u9kT9iSA0pUN8Zk5locND6oK7udYcnQOVRq5Eu7mxLNTnsw77K2aKW7fqAHNH0extaxR9vH0HDg5sz6NHOrS2tmQ8ze0+e1hRwDMXH5mtH05KtRpQ9UCr7lj0pdHnb78HuCP76sauJv2oqnRTFzadKe85jveaFbBIO0RQ+b4sHFWZUp1W0rU0Jq0PZ15KMWY07/QpIYr1XfXxzjJl+fabfBxLfqpUZCD7HWrjmDnXItqTeugH/Mae9ciBBctkKN8A/WZ5KDybMlN+dA3H0LltHJYuTylZgkdbl14xOQTm3Msl4nhR6hXbwThaTKKfjSbUbzBDsY5O2NVpR51S0i4+KAAJx67QMrzL5arew0P8qhHa5Y2OIHfhhokv8t6j8/loeDvY25WgJUjh1Nx2yZq5yK+jkkHDk/aQ91SDjSoY4X/Ey1GFNMXivwGaqzZh6lNfRpGtEL24hTWEw/TKeP8hFMZdeHJ2grsi05D5lqK5Vn6zUs829OJvUO342TtRMOalgRcv0n1GafpYKIDDbfTr3AFLKt4Us0kGc/XBTl4sycg4eBvc6jfzp4HNeugeHuHNwWbc3WcHbraldjUbSM2pZxpVN2Cl9eu4TTmAJ2L6JD02zbWODbF+Wp9TNOCuB1Tkj88O/xQ+tbMoW/5uL0J/u0uiSGPMTTMeiDRow/JqlORs+PCUj9sFzhkCSvlvofGKyph5XIeZzM1blx7zbS9l3J16ufn5P0buHz58t+WkZ6eTocOHejevTvt27f/bJyP9wkWKGYpGh3Bf488fI/tvn37Pht+8eLFz4ZPnz6d6dOnf6NB/rldunlA2vtQfB/6o1mkJA4VSn7XPR4JYc948DwEubYxFZ0rU1jz02b/XdBTnoSmYV+1Ikba6iikKaSrF1C9liVXyFPwu3+fsA9Sipergo1lwc83mh9ec8c7FHObqpQy0+Xh9cto2NXE1kib5HB/vJ+HoGNamqq2Vqr/ZBeeK13HhvDg4Uv0rSpjZ539O2ojvIbS+Pd+PFzm/MV0AqTFe2FQbAQp76/xzPs+aYZlcShTNFdpev/qET5hadg6VcZE4wOeVx9SqW4djDRyVrgsKQrv+y8p7uBIsYKa3xQvMSKAB6+Tqexc8RNvddiL+wSlmeBSsUSuBgfvX09ksWQGC0tmzWuFLB7vW/fRMCuPQxlz3j324pmsArUcsjee5Olx+D14SHKB4jhXKp19W5KNbFse86JweXSC/TGycca3S1l+nnSTUw2Kf5X8XHf+XygfysSm8/jBPaKS1ClXtSrFdTW+WC69f6qGzdzb6HwmA0Jf+BAQA/ZODhh/pqx8a7kS5D2ylFCuXX9OCde6WBfIvTc1LuQ5j0JScXCyp6DIt29Hnswz7/soilbExsLwm0QkRb3Bxy+UYjZVKWla4ONWiHB/XwLea1K5iq1q2weALDkS3wd+YFSSKjZZ+/Zo/wc8DknGsrwDpYvpZek7Xz5+QJTChCqVyqL9g2Z7Tn3Ld6xpPL93m/C0AthUroSZrlh58rfG6goFvXv3xtjYmFWrVuXqPwWKWVJywLh88wwFX+evd38bhOSf0+013iXnK90kWRfMN2nxubqa+LiQXMXVtrSk+LixuYprvGfvN+2x/V58N8NWkB9b9DT62jdjmc+FHF91pDJMMgzb1Pj7/1mVrZ27kL7Tp+aL5VzBpwdReZKcXWt6Iw33YvjQzfwR5kcVPc0fRp/piT4s/zmKKRMbi/ooEAgEgv8r169fp3bt2tjb26OmphwHLVy4kBYtWgjDVhi2wrD9yLC1GJs7w9Zob/4ybIXr77+ERIvVfwzgRGgi3UoYfDG6uqYp/fq0/U+rbOTMafkmLZbNN3EubSP7j+5DpmvJngfeP5RRC6CpV5kpE0VVFAgEAsH/H1dXV8R8jkCQC37QaiIM2/8YBUt1o1su46oXKM2GtTOE0vIRjm2G4NhG6EEgEAgE/+Ext0LBnj17ePXqFTNnziQoKIi3b99+1empAoHg32fYqomcEwgEAoFAIBD8KAwbNgwvLy/VoTQGBgYMHz5cKEYgyAMkitx/8htixlYgEAgEAoFA8MNw+/ZtvL29qVJF+eJGIyMj0tLShGIEgrxC8WOe8icMW4FAIBAIBALBD4OmpiYymQyJRDn4joqKUh0GJRAI8sKw/TGTLVoBgUAgEAgEAsEPw6hRo2jXrh2RkZFMnz4dV1dXpk2bJhQjEOQREnnuPvkNMWMrEAgEAoFAIPhh6N69O46Ojly8eBGFQsHRo0exsbERihEI8oJ8un9WGLYCgUAgEAgEgn8VQUFB6Orq0rp16yxhVlZWQjkCQR4Zt/85wzYpKUlkvEAgEAgEAsEPiq6u7g+X5pYtWyKRSFAoFKSkpPD69WvKly/PkydPRIYKBMKwFQgEAoFAIBAI8j+PHj3K8tvb25tffvlFKEYgyCMk4vAogUAgEAgEAoHg/0vVqlW5e/euUIRA8B9HzNgKBAKBQCAQCH4YVqxYofoul8vx9vamSJEiQjECQV4hliILBAKBQCAQCATfl/j4+MyBrIYGLVu2pEOHDkIxAkEeGbX58VU+wrAVCAQCgUAgEPxrkMlkJCQksHTp0v/L/bSj07De8Sb/2ByF9PNXhoS+zTdJSapRLl+pJsJZPd+kJf2e5KuNW2HYCgQCgUAgEAgE3wGpVIqGhgbe3t5CGQLBd0KCeI/tZ0mJucSSddfR0LFm2sSe+eOJFWlc+HUp6/df5HVIDMYW1jTvPoLxPRr+7ZO0EkKW0G9ONQ5ursv5Ae48mbONMcW/0bOmkCNHDTUJJISuomP/M6pLalo6VKjRnhkTe2CkIclzFclkMtTVlV6m7q1bsu7Iib91n9iHx5nusQGvx2/QMiyOS4uezJ/Qg0IaElLiztJl6EOO7puYp8/wcbpvrB3PghN+HDp9isN9O6K5bBddinz96w0u/DQE25nrKab1z3rgQi9MZMa93mybUjHHeCPbtWbMviOULpB31dz/wq9s++M6iiI2dBs5jEqG2n8r3teSpV7J5cjV1MQJeN+J1d3aUeHnfTQ1LEBK9APWrviVp29TKGlfj5Ej3b9L2yPI47qokHJu60pO3H2KhnEpOg0dQw0rfdIS7uKx4vQn0Ys1HMKAWqakJ/izfdM+7j6LplLzPoxsVzVLvPQEb0YvlPDzwiqZ/UZKKFtWbOB+QBRm5aoxZExvihdQtpVR3r+zZs8F4jSL0aDjINo5mf1w+palBLJ5zTa8A5OoWt+dIZ1cPj8OeHONFRsPExgjw65WW0b3bMCfVSU7vabF38Rj5YUschqPnUINA60c5UU++IOVu87wTmpAnQ6D6VG31Bfl/ei4uLjg7e1N5cqVcXNzo1OnTujp6amut2/fXjQOAkGe2Et5J2rlypVs2bIFiUSCvb0927dvJykpic6dOxMYGEjJkiU5ePAgRkZGf/te33VMuK5jfxYvXszylfvzTT5t7lednsvv0mXUHH4/dYRF4zpyY143Gky5+Ldlpyc/xeteJABFqjhTSufbDYrHK+tSc9YDAKTJz7n3xIBZs2Yxa9YspozqTfyRqdQbePa76MjMsBCpGQXaqXoNNP/G+DX+9T4c6g9Gv94Adh05xdblUylwdTb1hx8GQJ72lptez/L8GTLTLafDT1v46efN6KpJsHSsRnHtrzdME4K3M+lt23/cqAVIjvDl3rO4L8bzvnmdBFnetUzR3vOp3n0t1rVaY5F+mUZOPUmUK7453rfwcb1aW8mC6YEfROfznXhx6yZv0+TIpbG4VW7EAw1rOnVuTrLXXJxaLBUK+gfJbR27u6AhQw6E4Nq6C1VN39LCuTGBKTJAgVQqzfK5smEVd5LSkEtj6VK5FlcSTGjXwoUzY5ow2TM8s/2Jec0vY/tx6nLW5Yez6lTnSKwxbl3bov/qZ1ybL1Gm9Z4HFZsvwdSpBY3s9ZjT0pG9oQk/nM6n1q7NrhADWjW054/xLRh07NOlqdJkf1xd3EkpXpWuberwaGUPWi17qOzrctBrfNAm1v/qnyU/5F+Qlxh+kMpNJlGkamPcXEuxwd2ZJQ/e5Sjv30RMTAyFCxfm0qVLnDhxguPHj3PixAnROAgEeWTUSnL5+RKhoaGsWbOGe/fu8fjxY2QyGfv378fDw4OGDRvi7+9Pw4YN8fDwyJOkf7cZ27fXprLgiRWtC8dzXpo/8inOfyGTTunhE/Q7JTKMm9KW1uz11KRS7fHEzPfGWEMNr8uXqOZqz6n9v+HUeTAmsjBOHD5DUKwMy4qutK1nx5+2XmLwfY6fv4O6eUVql/rIOKzigpZqpkyO17lj+Pi/o1T1pjR1tAQgJdobn7iylE304qCnP0Vs6tGxkR3piQ+55R9PQuJtvJ6VwkYD1DVNqVGjhkq+XdHfsHE7BjRTGtUJ/pw67kmk3IiazVtiZ5zpvX///CYnr/mgZlKGpq0aq2ZakiN8OXLyJvFqRjRo156yhbR4cu0yMuDihQs0atQIpxo10FKTIE+PwvP2B2rYpbD/4GUUhmXp2LEJ+upKWakxjzl27AYFStWiiV0aPnFlqV7GgI2dJ1F5xRWW9M7Y91C6BAsO7WNn6U6Erm3LX30zCW/uceTsbd6jj0Pd1tQub5xhAEdx4vApguLkVKzbhno2OYf/mW6fK+dIkKuR9PoZslIWWFZ1QU0r53xJjrjPw8TylHx/izPB5entZsnJoYsYujlz6VPa+2ecPHGVD1qW1GvZhBK6GtmGxzy4Tki5aqh7n+f60xgq1mlNrQqFctGwpHLj5GEevHqHUTEbWrdrQEH1rF4Gn6uXKVOjKnfPnMQ/Xpd6bi0pZ6Cpui6XxnF05xFCpXo07tiJcoW0ctRzThwbvplqay8zyL0U0BqfXabMeBbLClvjb4r3V2L9bnDy+kN0S9jTorErBdQgysuTD5Vr8u7KAd5WbYdzRr2Ke3yDF8kyYm5cxs+0Jba6/71dFfevXsK0el0stdVJjXvAjQcpNKivbCOibnkSaOGIs4VetvX/r+1cUU05188c5VFoOk4NW2e2H6+n4avTn4i5owBoUt+JDYXL4xfdhMinutSrnbmn6aHnZQo416acjgahdy9z7t4z9Eo40q65i8pBppDFc/HECfxj1XGp2wJHa30EX0du69iazX5MvX0K96J6QBOurzVldWgCK0u7MHeuy0eG8lL2ek3kYmMLQk534q7xDIJmDgOgmtV71nvFQR1zEkJX0bjjAdLjw8HgI6du4iPWvjTg3e1xaEigee1yzDNxIFY6hRPDfqbqz1cZ2cEaAJu0k7Qd40W3Q41/GH0nvt3M5iALIu9MRFMC1a1uYN9tHZvaZHXwxD6bzbviS1g4QrlCrWqJU9i22w8TKxF6flC2eo2+EYRl28XMneucRV7Ug+zlvfltPXqOaxnXQ6nHYr7b6LP6KZN2uGYr799AZGQkK1asoGLFikgkEhSKzJG1RCJWkQgEeWnc5hVSqZTk5GQ0NTVJSkqiWLFiLFq0iCtXrgDQu3dv6tWrx+LFi//2vb7LjK1CGsuArpvovWsPxbXyz8bpxwv3U6rbcpVR+ye6Zu4EvPDBWEOpjhHu7VneuRnbz90lKS2OdhWrsv3mK9KTItk2vAHu2/0zBuK/UKVKS849Cefu0SVU63dZJfPCsO6sDUsAZMzrWIWhG84R9z6IZV0c6bnGB4CIWz/Rc8IEeqy+iEwRz+qetRlwMoj0JD+8X8eTEPyAe/6fzkjJ02I5uuohJVq6Kw2x6LPUrVCb3+68Icz3FI1tq7H/jdIjHnzyJ+zqDscn5AO3jsyjYrWuhKfJSX53nEqVOnEv+AMRT09TvXwDwtJkPPO6gVwB1z09SZMr6NOhHbFSOanxN+nYbTp9eiwnOl3OtQ2Dqd77uNKo/eBJvUot8QyI5uqO4TQf1JkxvwYgT4tg0dM4lruXypJ+LX0XIiPefFI2kiIOYufUmfuhH0gIf8QQVxt2RiahkCXQvVJFdtzwJ/mdP6PqlGdjYHy24YAq3Y+9boJCjqenJ1IF7B3QhUPRyTnmS/i16fSdOY36w1bj9yYRhewD0x4UYaC5fkY6T1DHrgnnHkcRcH0bjhWaEpQqyzbcd84Auo1sxuDNZ4l4fYternZ43I76Ynn9tasjfTdcJjE9iZt7x+DUZusnceZ178SI1q7s9nxOgOdWato25F5Cmur6os498IlKI/LOFmo5dM5Rz19Mz6sPuNfNXELY06UId06FfnO8jwk8MhX7ZtN4GvWe8+sH4+C2DIBbI3owbmYrRm08QXCyTFWv4p7e42WylJA7XrxITv9P9jm+U/oy9rpy1uzh/IG4tW7Co0SlLqZ27cjR5PRs6/9f27lkuZzF7o50m/8bb8MfMaOFEzfiUwEoYNiTHb+MVd1XlhqEQr0gJurJtHdrTFS6PMO4eUKjtr3Qkki4vbw3LgNW8uZ9LGfX9KZyx8WqvmFg3Yos/OM+79/cpnu18vwSIGbdv5bc1rEl173paaqX4Xx8wv1EaGScdcmyQpbA4I6b2PX7eCTA83W+lB3alKAnt/nj5CXirAcwc7gNAPrFx+Dl5cXRVVmX4WrolOKx75XMJbIPT6BVsBaGGhKCP6RhXCrTeVHIxph3D878UPqO8TmGfrExKueMkc0wkiJ+Je0vAz+jciu5faaTynH6+MwrjGyd4At6jbgUgbb5Q+ZNn8CMRavwCkv8ojyzOvWI89vE48hEUt+/ZtepEBzdLHKU92/gz4OjEhISiI+PV33/8yMQCPIGiTx3ny9RvHhxJkyYgJWVFebm5hQqVIgmTZoQERGBubk5AObm5kRGRuZJur/LNMe9xe14UnYiJ5pYMD4fZVKITyymc0wzvat+q5my2k/1u/GCVXQ00QHAb8BRfm9hSXrCPey6LsBj/kAAetTzofr4l9C3LDt7zqOMxxW2DagAQPXOtox+kfWesc9/Yp1PXcID1qAGjB3iimnpDiSPCADg/UM1Tr1ajARoWvAC7fa+QbdlF4Y1WY933BBGtrYkLgASI3+lUKHdyu5NJkNdpxz7bik7OK8R45D1PcauedUAaF+sPm37n6LLBXcmDt1I/3PPmVOpMDAFi+pWDPrjDet0NpBaYjorZvUGoHKJNUSkyekw6ScGz/Ng1ty5aP/F+Zkad5lpB0Ow19MksaMuJav9Brhxe+xINMaeYN14e2A6P1U050JFSEu4h1SjCGVzuSQ7OSyKAQtOM2OQcgao2oMTLPOOxt3Fh9PvzAlbPQ9dNQmd7Upx8306KQWvfzb8Y3pMmcaQheuZO3fuV+VLpGckwYHH0VGDhJBlpJj0V/33XL9x6E46xYZRyj2uZYMbsuFNPNXGfj68CRD/rCWPb44DoF+DFJx6ezDl2fIc9RFu3YKzsxZhXUCdtMF1MSs9CRjwSbzglnu5OMoegDp97Bk804f7K5QDz6Kzf2V2dTNgOAcLG+KfLMU4Gz33bmbFozu3SZBlbanU1PWp5mKPf4qUsrqZs8H6pfVICPh0IJHbeB8zcNgvTLn5hhHWBihkI2heuxGvUpTG1MvwwTw+1g6AXRnxS3YaTfM5iwkbO422hXX+k51Og0n2LFrmDQ2Lc+SPYPrbFGLZk1i2Vozm9/eGBJcqxICGn6//xztaZ2nnEkKW4+GpT0DIXgprqpEyvAHmJdoCoFOkJs0bKO8Z9/wiw7v1ot7k3zA1qsFEKylT70expboZQScnYVhzBZaSUFwXXuJ00Csq6WmimDSW1pYWrAodTnufgZyTjiFoqzJvO5SIZtK2AAYvrCpGEV9BbuuYuaVyFcoIZ1v2PgvCrv9uWhoVyBLnxfZuBHfcRo2CytUcQSGJBJ0YRNvfi1GtiIxh/UYxy/MGA8tnv8pEoqaHpYUe0iQ/Ktg15m1UIqMOPkICtBtYieZDPPA/ORfT1ABmDr6BLO3HWmGR8DIB3eKZ70jV0C2LXJbA2zQZVh85yjX0TCmmBzHPBlO1wSHi0q049Kz5F/X66EksQW+uYzqsKamvL9K2UhU2P/XBrUj28gpX+onRFcpTo0xJ9NRSUbfuybM2JQBykKf7w5d9c3NzZs6cKRoBgeB7k8sZ2+joaJycnFS/Bw0axKBBgzLH27GxHDt2jNevX2NoaEinTp3YvXv3d0t2nvcuaQl3abf0Pt03TeDChQsEp8qQy2K4cv0l9VxL/6N5VNBAg8RXmZ5LbePKNGhQVGmsTB7J/alLVYZtn1pKA1hT34lxnWJZs2QuAS9f4X3tOopCSuNif0giE9uVUMmrOaYCDMt6z7BTN0DTmKGDB6vCtGQR3EtIwwowqthJtaxZy0QLRTZ7IvVMexHxar2yrMlTeHhyKXVr1Od56B3Oe0dTf5GNKq51p9Z8WHEMhbQxJ2LS2GKXuTytRTdrtv8WjPnm6ZT/0J5K9Q7TtH5dmrftQRV9zRz1p12wJvZ6yjhqWkVAoTSAjnu+pfaMTD20bmjOBUBNywS59D1JcgW6almt5NBnfqSXKIfpR2GFqwyjZfg+PGZv5+UrfzxvvqX0AAU6Rk0ZVG06pSrVplXj+jRu1pbODsZIFJ8Pzw055UtxwMh2ADoZ6xlSYu6ja55p2P7+OJbWm0qqfvc6rNyf3TOb8MtA6b4tVeFFXSeTEOqCVLGcnM7fGT2hF/u2rGSD/0ue+VwDPj+w7NjJWvW9xkRHwtw9AaVh29Xuz8XeEow11FDkoGeAkwf2EpySde+ARgFLqrnYo6MmIfEjo1eaKEVN+9NFH7mN9yfpiY+4lVSAc9bKtY0SdX3O3LwFwBOgzODqooP53ACv4WSih/yELM2FHYmVubVahxar/YgZsxND+1noKeKyrf9kGLZ/tnPR9y9QqMxECmsq86mAcSPqFsqc2VNIY9k4fQjzD4UwxuMPJro7KsvXHGcazrgC5zuzc7Y33Y/uITlqE/EYsH7cCNX/U+QKrgTGU3XPU6y7ZzqZyvbZyRGRlV/N19axdXcfM/PlLQbWd2Ncx+qscFX2ewpZAr1/usuWl5l1TA5I0nrhfaIfABMOtKd2p9UMfPhlY0JD1xb/ly95fnU39TvWoe2b5ziPPcKM6MF0qVMJReFy9JlajyPTDH8ofasXUEealNkuKmRKJ4J2NktfjSv8wsuXSzi3eSA9XIcR8XR7jnrtduYOvYoWx0BdAnSh9DM7ps/yxe3nGtnKuze3KTvUBvAyegpF1JPYMtSV2oNO4bO15Rfl/dBjbYVCNAACwf/DqM1lVTMxMeHevXvZXr9w4QLW1tYUKaJ0DrZv356bN29iZmZGeHg45ubmhIeHY2pqmidJz/OlyGkfvIhNl7Oub2fatGnDyZhk0hIe0mXA3n88n8oPtcF/yz7Vb92idencuTOd3TsQnibLEtcwY1ly3IsV2DedDRaV6TtmJgd+a5BpKKtJSJB+NLj4zJJINU01jGw6MGLECNXn8g0vKutpZQwEvt63IFErgEPrGVjK/TkVk/zJIEchSwI1LZBooo6C5I8OFZEly1DTlqBlUIvzfoHsmj+UEpoRjGtsy5wH0V+4r97nO32JcuCqGhilKNOipV8NFz0Fy/zfZ60vsnjq1KjOnqjkLOG+i5rRbtFlrJ2bMn7Oera0sMq4sTZLTjzi7v4l1CtnwL4prak39Xb24bkp+F/IFy3DzNMj1bS0kadl6ldDAh/9JCUykOAPadmG/znwVD2/PAmJRAO1HIxaWUogtW0b8iTdBLdeI9h05GC2cRM/Krry1FQkksy0637mJtnqGeg8aGgWnYwYMYKhg5RLmGsV1ObORzPi4Y/fY1K98CfycxtPVa4kGqCQ8bFPJ/JVAO+lygBNQ03RyXzW0VSbtjp+7PRegJ79CMxqjCTi2mb8Vt3FaXrdHOv/X9s5TQNN5KkpfylXij8rNOMbVeFwWh18n19TGbUAFk2XkvRgOuHvrrDhvSM/lTMEiRaaunZZytDKc1dZ6WCCREMNeXpmJZGnRfAyY/uAIPfkpo7Jpe+YN98jY2yihmnpmkxpacHlA4GqOG9vjuVt2blU0susYyWK6WHZ3lX126x2A1KibuaYnuTo31i69paqf6pQfwAdDZPYGp4PqCvWAAAgAElEQVSERE2PQYt2c9/vJd7XTtNaOwa9YvV+KH0bVTEnKeJh5jgn/g4aOqUx08o6hAo6vI4tN5TL6dR1CtFk8HISgg4SlibPQa9y0jW1MoxQJaXrmJL4KjFHeYf3PKHGssGYaqkhUden66w+BJ/+OUd5/wYuXrwoGgCB4P9AXh0eZWVlxa1bt0hKSkKhUHDx4kVsbGxwc3Nj586dAOzcuZM2bdrkT8NWz6wf/v7+qk8PUz20C9bAx3PsP55JJdv/Qvnon+m19gIqW0CRyqEF7biZsZ/sr0RcOUFh13mM6uZGFZtSvDySeRLiAIfCrFvhmSFHyu4ZDz/5f/FWzYm+dxzDcrbY29tTooA3nXstz9LpZOswye4kWYUUv7NLCMaSJkYFaNaiOBdnnFRdvrz4N8xqdUOirs+A4rqMP/46w6BMYO0Wf5z6lcZvdXuaTfLGwbUZI6YtxqNOES5dz1zfLv8Kr2ibpsW4tPym6h7rz2Tu9Vo/ozbr2wzhTnTmoPn25v7EFqjJOAuDLHK8DzzBZfl8OresR4USBTnvozzhMSFkGVVdx2NRsSY9hk5iw5paBJx6kG14bviafNE2qktyxGPV7261zDi0xkvl1prfsCYLAuOzDQd4/vM61QnFj36dgkHJETlWvuTYU7zWqsOysX1wdbRDLeBYtnH3LL6cWQYn3qZExzo5Pnt2egbYusSDBQsWZPl4LFHu7R3UvDiHVyifT5YazIJHsQxpUlxpiN6/hXdA/FfFUzkKdG1oXkjG9Iz9otLkAGo5VyMyXfblOpJRTD+WK08L59q1a6RnXLtz4zr+ydJ/ZccztJ0VM8f8js0oB7QNG+IiO8v0C2+ZUcssx/r/V4wd+vHh1U/ci1O2gzGP1nHzg/J75L0J7IvsytH5fdBJSyYxMZHExEQUgIZOWeaWl9J96kjKjViIOqBr2h3jZE9e6ZXE3t4e23KGTOzaiQiFgtL9nPDfskVlbN9d1JYOCx+JEcRXkps6pqZhxLE1Hmz807GokHLSKwrL2ple8YtTz1NlZvMssm0n1cJ/837V/lG//UcoVN79C/VQyoJ5IwhJVdZZadJzLsSl0thIm2cbG1Gx80FVGVw93Yf6s2v9UPo2LD8ZRfhK7scrHZXP967E1GnaJ+1Nevop5k3apZroiH3yK1r6DhTVUstRr+3ty7PKN6MdVqRxaMdLKg0snaM85zKFeLoz80DDwNMX0ClaP0d5/waMjY1FAyAQ/D9Q5PLzBapVq0bHjh2pWrUq9vb2yOVyBg0axJQpUzh//jxly5bl/PnzTJkyJU+SnedLkSXq+hQrlnlQhL66BIlEm6Km//zJl+ralhy/8SvdOvSh2OrCVLEpRuTLZ1Tps5Jj7QP53HEWlm1HIZ/Rh64DWqCIeIp6OSviXs5inY8LA/ZsZnuNzrj41KJwwiP06jTgr0IKlpzMys5uODnUooGTFb6XfOm5+/IX06pracjrDbOZZ7uKkS5Z99hKUMfMuhKLDp6juJY6ZvP3Uq1JEyo3+o0yGqF4RVfg+JW6AMw4spzmTWvTbE99pIFXSaw5hqt1iiG16U+4Q1sa+DeiqHokns+tOLqjDABldDXpPngIOzZuzJVenZcc+B975x0V5fE14GfpVQERRBEVxII0BVHEgiAq9t57j73HaNTYImqMGo0tdpMYY2JvsWGvSLOiWFCq9A4L7H5/LFIUEPPDL5LMc86ewzs7OzPcuVPuzJ15sWrhRqsuzdGOf4l1M0NCjRXnuOpPOMjSZz3wrGtJvUZ2aKY8wz9Sjx/OnkRTCQpeWeQ6rytzenVjdMcGRDx6jrmdHoFLviTor/XYSR2o2/IxTc00uX3hATP+/BHtKmrYSW3fCy8NH1Mv2kaDMUmwQyqfjpoEWm3ZjXmLzjTr0Jwq0vsEmQ7C16YSkmLCrwMGTWOxb9qWhkZSvP0y2fiBhR7tygNpo7+Eln2GY6kax2tVO3SkD5j8bSCTaxSOWyd6CU7tdmGQEsAjpdZcmmNTYtrFydnXdS9Lt+8qvnP6bjdGjdrT4Y0nsqd/UXPqPnrknm/9a2Qv1rT6E991TUodryA//rkMj67OPGrVjOSAC9hN/ANLTRVKehFUFSMNNsyYitXmDUgKpJsed5T27afzPCEZY1Ul+nfqQNs7r9hUW+9fN+7Um9yF+K0rGdVUYaxMcTJk8P2ueTtwxbX/d9Gs1I190/bTzqoJrZpX59kjNcblXpb2+tBd0sIeUrXqxkK/uRMZSx1NFTp5tWaa5yFOhddVrJaqVubExrG0dXZiu5sDif4XqNx7O0111aDVFiY7tKCuc3tamEnwDtDgTx9HMYH4SErbxn7bMIaWzRrzl5sj6SHXiak1gHM9co8uyDL49lE8qxwMCy/6uW5miEFD6jhfo3GldK6G6PLzhf4lj1eV+7GhzzYaNWhBa6dqPL92jUYT9tDLUJOcYVupvdEF505/oBVxkzTHmVxxr1au5K2qbcPBr1rRsUk7OjhU5MINNX65rTiDXrC/qdVjJ24/OFG35XkcjJS4cS2EWTtPolSiXJX4dftU3No25qJrE1JfXCO9/hjOdK+JRk7x6Xn+vJ3dbfrh0LYx5mrxXA1W5cfT40pMTyAQCEptz5Wh1/+iRYtYtGhRoTB1dfVP4oEhkf8PBxbS0tLKbYXFvQ7iYbgUa3sr9NSVkWdnkKWsgVoRG6lZyS/x8Q+nSl17ahlpcf/6ZVSsmlBPTx15djKP/ANQrmJDXdOKJeT3hIcvE6hh35DquqVwrZRncffadZSqO9CwRikWBeSZPHsQSLRMDzvr2mgWcEHNyYzjYcADJJVqY21hUiA8igDfIDLVK2Fvb5X3m7Swe9x+lkLzFs4lngF9S8K9azw1qINmaDB69Ry4N8SWrdMvcKhV/gQ6LeopvkGhSDQqY+/YAO1i/HCTXjwgIEJK/Ua2VFJJ5urVe9g0b46ecjYPA3yJSVOmtr09Vd+6cMuzig4vtR6Url4uT2jIXxMvsKy+4syqXJbG00B/ElWMcLCunbf7WlS4d7d6rBr1F787ZhP4Ipm6DW2opPHh28LlOSn43/ZDxcgSG4sqxD28RVBOXZxt8g20nqaGDAl8jWPMY15l6NLIpvZ7l359lJw/VOGydJ74+yE3bkDdahU/Op5clk7L5U+4Ms/uvZ9kp0YT6B+ERtUGWNX68Eu6czLCuX7jKdWdm1NTQxlBMXIqpv0XRWLYU+6HZWLbqAG6Kv/bqzOkCWEE3HuOTnU76tesUOi78KeBPIsH60Y2ea8fEnwkpWyLmXHP8bkfSsWqFljXLq1BKScy+B7PklSxs6uX91q3DxEbHMDD8HSqWdpgbqJdqC8L9PFDq7o9llV1y63IU988I+BlBrYOViXIJIenvneIlGpQ19Yao0JjUvFyzcmI54H/A1SMLbGqZVy69GQZPL0fSHS2LralTq+IhQktLdGeSqCimjHNqvT/bMojr/iZvSItLPKzKUqac53PSjSRzp/PRXmvNq0hI+x1qeJqVqlO7YHTSxVX/dIvJZ6x/f83yP+jhq2g7Ag9M5Gm82RsXz2Q7MjbTJu8kwPP7mKn/e85G5mZdIVWfR9x89SYj/7tW8P2ZKfCW61XN6/Lc/d8l+odRxe4+Kl43hq2XQ3Kx83A97cv502Pabi9czOrQCAQCP4ZhGErDFth2ArDtlCfYFx6w1bt8udl2JavO/cFnyWmbTdwTLqNA8cOINM0ZceN6/8qoxYUF/XsHXKHcGkOVT/y3cxVPXrR3uz9XQr7Hn0xzy76JWDq+hVKlbb7gEHUUCs/zdh65FeiwQgEAoFAIBB8zpTTC8iFYSsoExp2GkXDTv/u/9Gy//S/9bu6E5ZSt4hwHaMq/K/rruNXrhXKJxAIBAKBQCAoMyTl1LBVElUnEAgEAoFAIPi3k5GRgZOTE3Z2djRo0ICFCxcKoQgERVFGtyL/fyN2bAUCgUAgEAgE/3rU1dW5cOECOjo6ZGVl0bx5czw9PWnatKkQjkDwrmFbDhGGrUAgEAgEAoHgX49EIkFHR3EIKCsri6ysLCQScTO7QPCuUStckQUCgUAgEAgEgs+YnJwc7O3tMTIywsPDgyZNmgihCARFGLfl0RVZGLYCgUAgEAgEgv8EysrK+Pv7Exoayu3bt7l///57cbZu3YqjoyOOjo5IZelCaIL/HBJZ6T7CsBUIBAKBQCAQCP5B9PT0cHV15fTp0+99N2bMGHx8fPDx8UFNSVMIS/DfM2zlpft8bvxPZ2zFS70FAoFAIBAIBOWB6OhoVFVV0dPTIz09nXPnzvHll18KwQgEBflM3Yw/uWErEAgEAoFAIBCUByIiIhg6dCg5OTnIZDL69OlDp06dhGAEgqKMW2HYCgQCgUAgEAgEnx+2trb4+fkJQQgEJSCh/N6KLAxbgUAgEAgEAoFAIBAoEIatQCAQCAQCgUAgEAjKMxJ5+bRshWErEAgEAoFAIBAIBAKQl/5VPp+b+Ste9yMQCAQCgUAgEAgEgnyLtTSfUpCQkECvXr2oV68e9evX58aNG8TFxeHh4YGlpSUeHh7Ex8eXSbE/yY6tNPEyS747WyjMcuRshtTULbM8bk3oxZyHsXnPymo61KzXlEkLZ2JnoF4meciyImnbaTrn/vq1TNIL957GwMX+74VbT9/N+s5mn1A5ZchQQkmSH3R3/3cs2byfByGJVDIzp/vo+Xw50AWAC0O7cnHxXhbXqFBmRciIPUyPca84eWAy0qSbjBs8H3n79Wzo5k2/mYYc+6X3R6eZmXCRCb8YsW2CFXO7tqfrz0dpoqtWNgWWyZApKZW88lOEXD8VwbvHMHrX0yK/a7rxIMvr6/8r+9WM+JN0H+nPqYNzxSDz/8x3PTtgte0gHfQ1yIi+y/crd/AwIoNatm5Mmz4AAxWJENI/yJO/drD10GVkRlYMmTYFe/0ixj15Nqe2rOTIrYeoVLKg/6RZuNTQAeDN9S2sPxWaF1WipMniRYXbWdKLn5m8Wo9dGxS3xkqTrrJk1V+F4rSfNR+XCmrIcxLZ+/1qLt2LwNyhJV9MGJSnI1F3D7Fq5wlisnVx7TORYW4W5U7eORkv2LR6Kz4vUnFsM4CJ/ZoWGS/l5SVWbPidF7E5WLfoyczhHqhIwHfdtxyKSS8UVyJRZfHiBQBc/WUtu8/eAd2qeA6bSQ8H41LnO3/MPJZsXVZ4IvnwNBv3nuBJgirdx82jq10l0WgEAkGpKe3lUaWJNmXKFNq3b88ff/yBVColLS2Nb7/9Fnd3d+bMmYOXlxdeXl6sWLHify73J9mxjQ9ay7Jly/IK6uXlxe+vkss0jzjf60TWG87SpUtZunQpC+ZMpHLINlo0HFuG9mAGly7fKrP00t/44fvMOa/Mbz9jnCp/UuUMXNmERnPv5j0//Kk/zb/4jU5TV3LK+xSr5wzkxOQ2fHEoRDHhuX2NB2lZZVoGJTUTmjkpJjNBWyZzRXckKwdYoKxRC2fHan8rzS3dRzFicG3F/3j1MrHZsjIr7xrLSsx+kfhRcv2UmLhNyNOXLokPCDcfmvc8pLrOv7ZjlUkjuHLtkRhh/gGCrl8hQipDlh1H23rNuatiQb8BnUi/Nh9rt2+FgP5Bon0WYtdrDeYtumGWdZ7m1n1Ilb0/vbj1jQsjfnlNy24DcTSKwM26BS8ycgB4/ON3/PIkjuzs7LxPobaXHcuwFl9w5FT+glpSyI+s3RFU6Dey3KnNjKaW/PBAla79O5F5cwUNu20BIDX8V+q1nEplx/Z0b1Wb9d2sWXY3ptzJfEbjxux8rUvXtnYcmuTGsIMv34uTnR6Eo01XMkwdGdSjNQGr+tBmuWIxW56TU0huKW/OsXrLHUVb29odz6/+wrFdf9rYajO5uQXrnyR8MN+czHhObRnFyn2nC5UjNWI/9ZxGIqvpQkdHPUa6NORqklQ0HIFA8BFGEGWyY5uUlMTly5cZOXIkAGpqaujp6XHkyBGGDh0KwNChQzl8+HCZFPuT7NiGHX+BrulsEl95fVKZ69S0x8XFNu+5sc1KvCr3ISZ7B4YqSqS8vM2BkzdIRAf71t1wra9YsYy+4U2cfQuSzu/j2vNUbDz64p6745WT+Yrjf/5FlNyI1p5WhfLLSg7i2OGLRMkMaN6pCzaV1PPTs2vGqyN7uBcD7YYNwyIzkN2/XkbV2I7BfdxQzd3cUNG0xMXFpcj/J+HRVY5e8kWpsiUdurbPW+2+du4szq3sOPbzfpwGTsBEDa6dOojvkxgsmnWgQ+P83d4wn1McvxGEWuU69OztiWZGANefJJGccoNrDy1wtsyk85SDbHsSy0BThUFUu4YF+w+dxm7kJjZ1f6fO5JlcOfo7d4Nj0Te1olsvDyoqK8qVHunHgaNXSVbSp02vPtTVUys2XFmtKs5OFZAm+nLJJxYlnQSeRqbhZFaLpo4aedmlhd3lyOnrpKhVp3OfzlRRVy5SBnrJR/g2aRqRFfJ3aOXZCRzccZAIiSGu3bvRILc8clkq544e4VGEFFvnjrjaVy6xTuMDr/A4PZvYy+e5b9wFay2VD8u1djoXb2bgXC+ZA4dfMmRMl2L1z/fCOeo0d+TmiaM8SdLGrXsX6lVQLVHXtavb4VJd8XdKRTV0zOxxcbFXLERc8ya4UQtizv9ChGNvHJ/eJMWhJfW1FM379ZULBZ5lxepOcUgTHnL0iDeJ6ma4d/GkZm668pwkzhw5wpM4ZZq6daaxuS7SRF9uhdTCWvkBh73vYWjdks6uDUrRgRavZ2+JvXuZ1/WcUfY5zeUHsdi6dqOFld5/dsy5c+Esxi5umKkrkxl/l8t3M/Boo+hb3lz35kV1J5pU1y59v6Iq4/KJPwkIleLUtlt+v/RsFv6aY0laPh0AzzZO/KBpxv1oT6IeauHeql5eXH/vc2g0daWepgqht85x6vYjdGo2plenpnl9YFF6I/g4Do7eRLOtNxjf3wLohu9OHb58GMcG68K7cqs33WdBwAUGmGgDnlxeo8N3ocn8WFuPIJ842h1fynLLor09Dk1oRULfGlBgrhF9JYQaPdewfHmTdwypTfwYVIOk2/NRl0Dntm4cqGDItaQRVNy/Du3GW/hyWHsAqvltpf93D5m3r2W5kXdqxCY2vaxOSuBcVCXQrMYVLHuuYVePdYXixT2cR0z1NayaOhwAx5rHMO/wC8y1x2H6fBzylw2Y1cgUr/MnAPhh0TmGnXnJ2AaVgE6YBf7BkG8CGbH6QbH5ynNSaOrsTlZ2IlC4HzwzdBa1lp7h67GKvrdWTAa3I9JoXkFNNB6BQFAqo7asXvfz/PlzKleuzPDhwwkICMDBwYF169YRFRWFiYkJACYmJrx586ZM8vskO7bPT0agpCllXP9udOk3ki3nXvy/1EPCo8toGnhgqKJEWuQ+zBt0405oIsnhAYxwrMX2qDTFKvb4foye0YUfLr8kOy6Abva1OBmXQXb6Y9rWacDSw3d57XeStjaD89JOjz5Jk5pO/HbzJWF+x2hpbsvPL5Pz0uvbz4MTwUm8ubUBZ5tutO/6HUmybH6b1oV2Gz+84/Tq6JdYNB2N7+tErh9YQG27HoRLFWvhY7p2xKtba346dYt0WTYLOtdj5PpTxCeEsLy7FX1W+wLw4sAY6nVbRXhSOnd/n0G91ivJSr2Pz/NkUl7d5faTJBJfLCWqQv88o/YtJi1/4c3T9xcidvSwYsD686RkpXJ1z3is2ytW4dNjDmNp2YXbrxKJfHAc+xrNCJPmFBueEXeUbgM2Ik26x91XKSQ/9+FOcDKpkT/Rc9hBAGLurqWBdT+uByfw0PtH6ph7EJSeXYQM5DzZvIwG8zoUKuuqjs4cCQjjyaWfcKjZmFMx6chzkhjZzILpuy+SEH6PaW61GbfjcYl1mvDwFsHp2by+eZWgtKxSyTUj0Zsufb6hu8sIzvo9L1H/FvTuwmgPR3Z7P+bJxS00MnfhdvLfX02/PqYPE+e0YdyGI7xKz+b4iF78GJGS933+c06xulMcaZFHcLJoxanANzy9tBWrGq0IycxBnh3H0Ca1WXToDgkvb9DLtgY/Pk0k/vECug0ahdOINQS/ec6qgU1oN/fyB/+H4vSsIH5fD6bnWFeGbzxJ5LPr9HW0YMmNN//ZccdvxkAmXIpQGJQLh9KubUsCUhWeFjN7dObPdOlH9Csylna1oufC34gID2SOmzVXkjIB0NQfzq87Z+flm5MRgly5IobKaXRs25I3WYr0slLv0dyzH+oSCTe8+mE7eBUvE+I4+X0/6nZemrv4VLTeCD6OHc8S6d+6St7zsKZG3DwW+l68tT6PGG6sDUBm7D3upEA7A8VC4l9xGZhc38OsyVNYtmYHkVn5Hi8Rl+Yz+VEffh9Zt1B6kWcjUa/qz4LZk5mzeBXXwlIByM54hopGLdRzFy8kyrpYaCizIyqVKq7uxD/YSGBUKpkJz9l57DWNe1QvV/KO9T2IrunMvMUZgwaTSY3cgfSdiZ9BvR8J8O6fZ7wGnghG39rp/TnSbwM41mgbk6wNAOizaS+za+cbp9ERaeha6paYr0RZhzu+vty8tP699Df6RDO4tzF+V05z9Nx1zGcsY3xdPdFwBALBRxm3pfnExMTg6OiY99m6dWuhZLKzs/H19eWLL77Az88PbW1tvLw+3cbnJ9mxPR6aQlLsTqKbdiPh0gG++H03sQFvmGtjUKb5BG0dRevTikE7Oz2RwHvx/BwYoJiQh71h3KoLLB6v2E1wvnsEL59oRnasoRg4smZweaU7AJV/P8BPoSlU/2soD02XEvH7FABGtehD7X7RAFwdM5Gc0af53csZgN7VnPEcdIxBVwcAoNbyR9bOtEEm7ccKdVPm+mXQVl+dvpZncF4eAhPqAxAfPBZNzfF5/4OGXmviI04yZeR6xl56xbf2hsB8qttXZtihl5zpaw7Ag3GnON7ZjLhHM1jj60Zi2GaUgNkTW6JTtRPp08K4v+IodosusWhkXWRZw6mx7g+0jAYxuf06fBImMq2bGSEnHqFecUCpZRxu3plLy1ZjrqGMdIIbutWmAuOIur6ezFrfsGGpwrWgUa3viZTKyCom/O2QqlN9KBNd13BfbzJTOpqSWGDNw6vnQrqcCGJdM8WErXHHGgzZ/Jhb06wLyQBg7a7ndL5Y2IVbPuQYuycq4rr1r82kiTe5/MUWDsYMJOHmagCmDK2GUcMBbBrhW2ydhl+dSad5ywibvZCehpocK4Vc06IhLXo/sx4m4G6gQczddSXq36uuf3Jtuh0ArfvXZvhXvtxba8mNW4/fqwNVbSuc7Es+Q/s0fCLPTvcCYFMxceIezS5WdzSLOSh8atBEtOadZ/t0hWdEnVcurH+RxKQnQzmVPZPovQqDp2/NaKZufUqvXpD87CEP4h9QRU2JzOmdqFylO0/mR1FHU+Wj9exdkh924ZnvlwrDzCMD635LmB+y/j855njMs2Oxlw+0NeWPQ68Y20CP5ffi+Nk2mv0J+sRa6DHYpXT9SvJrL5Z46xIW9yeGqkpkTGlDRSNPhWFr1JxOHoo84x+dYXSvfrh/fYwqBi7MrZHNzDtv2NOsCiFHpqHfYgNmklAcF53jQkwE9tqqyOfNpq2hAatCp9DHt2i9mbDKUUwiPoIn6dnU1cr38tC11CHlacp78aqaKfrLMTa12PMwBJuxB+hioAHybM4nZBByPpLxbe15fHw9dX84xuvgg2hm3KdL7yMcfX4Xyet+hdILvBdPyMtLGE/uQObzs7S3rMeel0F0qToFi7TazP/Tl7mdLPHZN48Tcel0SsnC0H4RM+ub0bCaETpKmShbDOdlj5rlSt7JwclomRrlT5y06iLLSSFCmkONXK8iABVtY6ppQ+zD4Vi57CNeWpOjIZ0KpSXLjmHQxBvsD9mbF9aqi8JDQi5L48/vJjP8YhUu7bImeXvp8n0X/5QslPo4ccy0FRVT/Bk8zoi7909SW0NZNB6BQPBBJJR+x9bQ0BAfH59ivzc1NcXU1JQmTRSePr169cLLywtjY2MiIiIwMTEhIiICIyOjz9ewnXL4LOM1LHGyNyQzYTAa+u5smnaHuefalWk+1dpOZOkgxblNWWYyx1aOZs78K/T4tSOGDpPpEr6XJfO2ERwchPeVCCzH5deS+Yh898hK6srkyOU8//Ul9b/ukhdu0noyoPD//ssnBvfV+a7J5v27kbjyIKAwEqt4KIwxJVVDANrmXuShbqiOPCc/X/3am3jzcFi+8kgkyLPjOBIrZY9NvhtZ5yHm/PRbCOROQEe2UFwkEXbsChLVSowcPjwvrlpOJLeTpTh+N4GQjo1pcbAjbm5t6DVs1HsyU6+kSY40rIiVmSwCAh9iaWNbKHjmVyPYu3klPwQF88j3IsgVJmpV12+ol9gBy6YH6NCmNZ16DsNBRxVpMeGpHzxiLWdzaArum75k+E8KR4LE+AzCj4dDrmH7VgYANxIzGadT2H13UK9aeX+7zG1MTOcrvDa+T42eM/PCK9aeilbGLPxTsj5Yp28pjVwB1Cu44J67G/Ih/evXz6JQWcO6epPjpcauXbveS1e3+qQPGrZ1Jjb7YHspSXdaVSz6wrX9gXF025Uv1xEnrgHgPe8B5kOX5+c/6jdOAlG3QNd0JlXUFHWorteKHnqZ7H2TxpISLiMrTs/epfboAu2z1dckh9qQLV/Pf/Eeo6oeXxM9YjY50qZsS22I/yYt3L67T+ys7ejbLUVbHl/qfiXmzhn06szFUFVRbxqV2uGml68T8uw4NswawcLfXjPz+7+Y278xAIOXN8Hly/NwZSDb5/kw9NQB0t5sJAld1k0ck/f7dJmcCy+ScdxdtN4IPg5NJQkpOfk7rNkp2SipF++AtfXeM5YEX2eoc1sm9mvGhpZGBDx7iZlZ7s7pwME8rlyBaY/isJriie2Gs9STSIlLy0YuzyQ1NRVtbW0GXwxkuIkpFZQlwCBqPzRn9mVFPqMAACAASURBVFw/um9z4cKFHQydNgKLqek4dviCMSY6JGuqcHt+K7YpjyM8bT5GyqlsHumI07BjPN7bpdzIW1lDmezU/DPI8hzFgKYuKbrjqWS1k/DwNZzaNJQ+jqNIevlL/qLEjj7EdtqJnXbh8Svi2h4Gj5hGpsNobj67SAMdVR5/ZL55xjNyVMae5sSQOgDsaFOdPssC8F3SSDQegUBQOuNWVja+yFWqVKF69eoEBQVRt25dzp8/j5WVFVZWVuzevZs5c+awe/duunbt+nkatrLsOKQSJbQraykmzhWaoCKRkPkmucyF/u4ZWzuLEfzouBPoiN9iVzocq8ma+UPoOXImI5RasLjgQKX1/sqlurYKWSn5lybJclIKTSTeFJhIyHPSQOnv3L6shLJy4bzlcjWUkZMuk6Obe64wJy2n0ERFP3fCqaSmhH6DPkydmj9ATZ06lVo6alRwnc+L6FFcPncB7zOHaVJjObeiC+8A6tUZTWr4GF5nzqV6gRXf5NeraeS4gsSMuLywnIwXNDZ3ofXXa+g1ohOWS6dRs+bQ3HptwZUXb/C7eoFL3ueY1KIWvS6FsNSh6PCvqn54fUhVAiMnTSmwEj0VVa1a78kAQEtJQsY7jS6lwAKCLDMTJVUDVLRUyE4ucCmKPBupHNSVJKWuU5NSyBVASSXfI+FD+vduWSUSNVQ0LZk6der7jVTjw+dgVfWKP6P79lKtknSn2A5CAtICd3JlRL3gjUY1lFSUkBX4QiaN5HmYFrrvtBuA1Bw5qiVMxErSs3fJTikwyZOlIpGo/L/cSv05ol6xFb0077Pd5xu0badRxUWLyOGbuI8fTt+sAEnp+xVVXVVkmelFtydZBpOa1yfAYS5BryZRuUA7rN5hHWmDWxMeU4UfEhuTVE+f9FA1VLVtCuvy1KnomlfmdTF6U7tWBQSlp0VFdW4mSvHUVyykhQcmYDjO8J2xOIZvlmxi0aL5SFDCuHZz5ncxY9QvL5C7aKGpVeA8rkSFjgYaHIzO4HWIEvemuHFyiuLeguSEi1jW2Ux42HOyVNUwKHD2vXZrY1L+ULR3Q6eBnLg2MM+0avjrl8w00ODA7vu4nDiKsZoSoMvgpaOYbvMDUH4MWwOHqqSt8AcUXgzSpJuoaNbOW8B7S8iBtZyq0p9xLYxR1tTDc8J6kmfUIEy6l2q5cRd+fYsZAYXPKEd4L8Kq9wnWHQlgiIvpR+f7LvbaanTyyL+UsWk3U+bvfyMajkAgKB0f8Sqf0rB+/XoGDhyIVCrF3NycnTt3IpPJ6NOnD9u3b8fMzIwDBw6USV5lfsZWnp1IG5dmtPSYyU3/APYt6UG2XI7dFNtPXg/KalXJzlTc7Ovzy32arl/BwC7uWNWsyGm/2A/+vs5kW+4tXpc3ofP/aUnedx07m3Lmy6N5z+eX7qdKi8FlsyqirMM4U20mHX6ea2Als2ZzEE3G1H4vrmnXTkTfPox+PWvs7OyoqelDt35eVFCWsLa5LTMfqeDWZSBLNuzHTSOc03HpuUaA4n/SMOjCcmdl3IauIC5bnvtdOpvGbaRamx/QKTBpSY87xjO11vwwexQtG9ug9PRg3nf3v+uI61QfGrbsyNSFa1jd2oizl6OKDS8N02vrsfeRGnZ2dtjZ2eI9ayCrw1OLjNvSWIub79zyuG3FxTzjdfuk61iOakmNfq14sd+LmFzj7vWZycj1OmOlpfLBOn1rN5dGru/yIf3bs/R8Xll3TblBzX6tyUp7yDfffPPeZ9XmJx+lT5oqEl6/UsgtK/UBm3PP25akO+kR/ly9GfxeWkNaVOG31dfyerqFLo345kUStcc48WTzZtJz//9biz3puCgQgKRXi7kWmwFA/IOtHEnSZWQV7WLLW5KevcvjH9aSnNs+A3bMoEKtaSgBUXeu4/M0KddYCufSpUtk5VbNzSuXeZKe/a8ceyb1qslX43+nwYyGqOu3xTnnJLPPRLC4ZZWP6lcqNRxDYvCX3I5XnKuNDVjL1UTF31G3J/Nz1GBOrxyFljSd1NRUUlNTkQMqmnVYXj+bXjPHUm/qKpQBLeOhVErz5plOLezs7LCup8+UHl2IlMtL1BtB6ZnQqToHVijaZU7mK74JiGOSp8IgetsWlFQMOLh6CRuCEvL6mqNX32DmakxG/Bmq1WiUd4dBdloQa8PTGG1jwJngV0RERBAREcGDyx2oYPYN4WEvFeOgpRmr/HJvNJZL+W1bMPbjLZHL0jDX1eZUtKJfDDs7myCN9gww0qJJnYo83J7vpvb8+Bm0TNzLlbz163+NPHwVd3LHnEd7V2LcZOF7/U2W9BgLpu7Mmw/G3duBmk5DTHIN0bSoXRyROjDGpHB/OHfgKsae+JOe9vp57Ss9U1ZiviUxw82EPTv88+r99x3PqDu0lmg4AoGg9LaJrHSf0mBvb4+Pjw+BgYEcPnwYfX19KlWqxPnz53n69Cnnz5/HwKBsjquW+Y6tskYtzq4ajufsLTg3VJz2a9J/AYeGWX7ySlCr0IzslAnseZWM+6IezOjsydAuNoQ/CMbCXg//BdPwcf+92N/X6rmfETvtMbN1oXlVGUHanhiq/gyA48o/cG7ZirrN91NHJZRrMfU5c8OtzMq++OQPuLk2pvVud7JeeJPafBa3Wr//GpyKtb7mxwHtaFDHAQ+nGvid82P47zcA6Da1JXYtGhLs2Zys0Bu8dJzCn9V0Ca2hz7P181jQYCOLh1gw9ZQ3j9t7Uq3KVpwa1SHh8U1iTTpzJu/SCwXaRkNop78Ap64DqasaS4hqQ3Sl9xi3yJ/vx40lvG57mj1ph4lyFBcf1eDUr3UwTys6nJRLH5TB1FNbaOvcnKY/t8YgLYAg9a74N69SZFyXafX56WQYi77I3+npEfMV9q03o5foyxPtLtydZo2h2hqWdm5O7QYuuNfV5tLlML776/IH69TEWIM1kyZgvWNrqeQ6x7Nw+T6kf3XfLMC21TYqpfjxQKkNt+bboaahXCYrVi3mdeaLLq4M6u1MWEAsI2wrEfcB3Qk5OgHPxY1IDit8XtVt529YOLWlkXtLTDIDeWQ2lEd2hqizi+mNG2PW0JVWNSSc99Pg+H0neAxahp5MsneiakNj/M7dYcLm63m7FUVRsp4VjmvQLIZ69i1xMJZy7m4m227PAuDkoM6scjvOw03OpMcewtV1IhHSHKqoKtGjjRue96PZbvnve9dv/Rndidu4jHHNFC7FM5pWpm9gD+xz3RxL269oGvbk4OxfaGVuh1tLM54+UGNiVcUFc6/+uENq6H309ArfAHsvKZ16mip0/d6dCa3/4EK84h4BJVUjzm+bSEt7W7Z4NCbB9xxG/ffSrIIauBWjN4KPwvmHfRjVb417VCdynpyk1qyD9DbUfK8tHNo6HqdGNpz0cCLt5VViLAZzpbc5mioW/PrFThxrOeLerDrBV6/TdP4peuamUdw6+J97Z9GspQ0X3J1JeX6FdKvxXO5VC4mShIPLu9OyvhOtnQ25dTWC789fQwJ0+n0v21v0oEHLJliox3H5iSo/XZxYruStqm3HiQWtaWPXis6N9Th7TZ0/AhV3GhTsb8z7/ILH97aYOZ2lsbES1668YO6vF/J2EEIObqWSzcx3NgPi2RcjJae5OasLhFv0ucDjX1oUm29JtNn5K8utPHC44ErFeB/CTQZyY1gd0XD+JvKsLLJDwz6b8ijFaX1eBpCmxufTVtM+r0Xsx6N++mzK4nQg+iMVv5wa5HK5/JMUPSP6Jf4PX1GxphX1axj+I/9c4vN7+IdLsXK0x1AlicuXArFt1RL9Eg/k5RDy0JcIWWUaW9ekkNOwPJPge/68kenT0Nay2At3/i45mbHc97uPxNAS29ol++7GvnrMgxcJ1GzogFmBV8WkRzzFNygUTSMLGlmZve2VuXPlCkpmTjjUzL8NOeyRD0FhSehWsaCxdY2i9TonGd+bd1ExrotdbRNi79/gcU49XOz0ycmMxM/nEZnqhjRqZJ0nj+LCS4MsK4GHfoGka1Sjsa1FsfGyUv0xt9vH6+AVhVrhkzvXiFE3obGNRd5NkgBRLx7wODwTq4Z2VC7ohl5MneZkhHHlahA1mreilobyR8n1Q/o32FibEU9icYp+SEhGBRztLPNuEy0r4oMD8AtNwdq5KUbvXDJSnO6sbvUDMy5Nfl8HZGkE+fuSqGJMY1vLQm4eYU/8CY4DG0c7DFQkRN3qhO24cYRdbYzf3WAq129EzcqaH+4/S9Czt5zzrMHycZc44pSN//Mk6jnYYSguQynTfiUhNIh7oZnYOdpQ4X88uCyND8Uv8Bk6ZvY0qFWx0Hfv6o3gbyBL57HvXeRVrKlvWvyNt5mxz7gd+IqKppbYWpoW+i4tOgT/h2FUrd+Imkalm5zmZMRxz/c+KlXqYG1eeOEx8cV9AqLkODhZo12w35dlEBToT3S2LnYNrfJc48sbqVHB+L1Ix76xdSHvpnfnEEE+t4iQalDf3hZjLZX/p3zf7bfTeeJ/l1QtMxrVMxPt5X+ggsSAJpLPx8tASUsYtsXODYuZy/5TnN238/MxbNu9xicgo1RxdQyqY+c+tVRxM1/sK/HyqH+NYSsQfGqOj25AzMJbDDPVKVfl7lRJixFPYulRSfOzKVNcwC/sinJlettq/1M6bw3bKL/CN4FGem9j+/WiXdI1DbszfazVB9N+a9ie71pTKL9AIBAIhGErDFth2H4Kw1a/OnbuU0pn2L787bMybFUQCMopnhtOsfpAJAyqXa7K3XbwMGqqf15Nz8BuINPLIB1No7YM7P7+7kAlhy4Mq5NV9CCtWrpzFdXa96NDDV2h+AKBQCAQCASfkNKen/3syi12bAUCgUAgEAgEgvcRO7YfMCTEjm2xlNcdW1396ti7lm7HNv212LEVCAQCgUAgEAgEAsHnhlyu+JRDhGErEAgEAoFAIBAIBAIAJOXUn1cYtgKBQCAQCAQCgUAgUCAMW4FAIBAIBAKBQCAQlGfEjq1AIBAIBAKBQCAQCMovckAmztgKBAKBQCAQCAQCgaAcU15f9/M/GbZpaWmi5gUCgUAgEAjKKVqf2etjBALBZ4C4FVkgEAgEAoFAIBAIBOUZccZWIBAIBAKBQCAQCATlFzniVmSBQCAQCAQCgUAgEJRfJICknLoiK4nqEwgEAoFAIBD8V8jJyaFhw4Z06tRJCEMgKApZKT/CsBUIBAKBQCAQCP4Z1q1bR/369YUgBIJikMjlpfoIw1YgEAgEAoFAIPgHCA0N5cSJE4waNUoIQyAoCrlc8R7b0nxKybteEnFxcXh4eGBpaYmHhwfx8fFlUvRPdsb21eXfWLfvDEnqJrToOIwhHpZ/K52Ng7oT++0e5pvpFgp/fXIGS56NZuukev8vdXx551JW7z/Hy9fxVKxWh26j5zG9tz3Beyfw9c0u/PZju0Lx760Ywer0Cez6xgFp8gO+m7eCY1f9SVOuSH0HD+Ys/xJ7ffW8+OHeCzheYyZjzCsA4PfHOry2/cGj10kYVK9Jl+FzmN7X+aPKvHjiNyzY8E2x3yc/82bDjiO8llbEo98kujsY/i3ZhJ2bxXyfoeyYY/3Zt9WMuGP0n/yaQz+PLzHepO6dmbrvEBYaKkU2TmVl5Q/mtaxvV4JtZ7Nznkt+X5Edj2en/pw+ffofk4EsK4rOPedw4ujOvyfDhL/o90Ugh/fNKjFe+/btiwxXr9CEI78vEgPHZ866Ad2pt3Ef7fQ0yIjxY/33e3gUmUFNG1cmTeqDvopECOkf5Om5Pew4ehV55foMmDQeWz31IiYn2ZzZvobjdx6hYmBO7y+m4mymgzTlDl7fn3ovelX3cYxyMSLa909++OUcCapVces1hu6Oxu/n/+tX7KkwniWdqgMQfXM7m86E5X0vUdJk/teKPuL5xd1sOXSdBPRo2XMcA1vWKnfyzsl4yU8/7MD3ZRqNWvdhXG+nIuOlhFzh+80HeRmXQwOXbkwZ7IaKBPx/XMXR2PRCcSUSVebP/6pEuZaUb3FjuDT5Ol5rzhVK12PaHJx11UTDAaZOncrKlStJTk4WwhAIiqGsb0V+6yWRlJQEgJeXF+7u7syZMwcvLy+8vLxYsWLF/5zPJ9mxjbq2FJsOozjoE8rLq7sZ392BZYFxfyutZzev8yg9671wLWNr7C10/l8qN/TMOHp+c4MxS7bife0sG77uzeFJbsy7FoWJhyMnfx1PTFZhR/OF64+i370GOdIwetu35pKSE9/vOsifu9bS2uAW7ZxHkpIjzzU0Yhk+KZwRtRRG7eOdw2gz5QCeE5dx6NQhls/ox+kZnZhy9FXpBuDMBM5sH8+aA2eLjZOV+gCXpr2JMXbEra46E9o24tQ7g25pSY8KwOdxQrloqEpqVWjqaP7BeL7Xr+bVz7sY61UksxQN/tGN6/y5oiv7XqUUWASTcuXKlX92IU6WwdVrd/6+YSyN5PqNxx+Mt3DhQhYuXMi8mV24cuVK3vPc2QPEiFEOeHLzOpFSGbLseLrYt8FPpRa9+3qSfmMxjh1WCQH9g8T4LqXpwPXUcumMaZY3bRwHk1rEyvmdZe6M2x9K8879aGQUSYfGHrzMyAHkZGdnF/pc3LSW22lSYny8sPZciZFjB9rYaLOoowO/hqUUSjc14hju43/k4pPEvLCgLevYHxxfKE2AuHuraDxgNxbOnejiUp1VPR3Z8DSx3Mn8qxYt2BuqSyd3G47O6MCYIyHvxclOf0pzpz5kVGtE/64tubdmEJ2+C1T0uzk5hWSTGu3ND9vvflCuxeVb0hie/GorP+55Wig/mWg2ABw/fhwjIyMcHBxKjLd161YcHR1xdHQki0whOMF/D7m8dJ/S2FFFeEkcOXKEoUOHAjB06FAOHz5cJsX+JDu234/cgFqlHjy4tRuN7Ah+2nkMg9ScMs1Do4oNDbUVu7gxty4Tb9uMpIsHuPEiFWv3XrjW1cuLG3bHmzM+j9Gu4UB3TydUczcaUkJ8OPTXLRLRwa5VZ1rUNQAg+sZlkuybEXtxP5GNuqP1/XksR/9Bx8Z1ADBo2YftK04xc99LtDcMp2fF2Xx55w3bm1VRDE5RuzmfUZUw60o8WuvKA8ulBK8dk1ee2t8e5+oBU5Y/S2RZHT1Cjo5FNmUZKhKQZb2h16wjbAx4Td9qCsPdwsycPb+docn4n1jXZUnJepiTgmvrDmRlJwEVi4334vcppFivZ/Xk/gAY3djPtKX38FzjVGL60sTHnDh+iSS16rh2bEsNrfdV6MXN05zxeYJyhSq06tINSz21XIMomuMHT/IqQYZ1q6641jcoMbxIoyo7Du/LD3B3awFA7N0rPMiqS8umRgDcuniBqs6tqK6uXGS9K6ua0MShQl56mXH3OXLkGhrmLrRtIMU/wZKmtXVz80rg8O5DhGVr49GrN3UqqvHgijc5wPlz52jTpg1qH9i0GjKvGdM7zKbnvY1FxpXLUvE+cYLHkVJsmrSnha3h/6TTxS92vObUkXO8kVemZdt8L4eUV3e4n1mXppYV3n+WZ3LtxEH8nseiX7U+nbu7UUG59Lt0zs4KD4PMpKxCzzJpJJeuvsSpbgqHjoXQd6A1V25LcW1hmVvWkELPaeF+HD97k1Q1Uzr06ICxurIYcIC7ly5g1FSh65kJflzzy8CttULG0Tcv89LUgcam2iQGXefEFX+UDGvTrpNH3k7rDe8LNGluw8nf/sCx71iqqMq4evow98KycHTvnJdP4ou5BGiOJGrxZADatnZkU6W6PIxpy5tHWri2qJMXN/CyNxqNW1BHU6VYHZXnJHP++HGexivj1KoDDrV0RGV+JEcm/EST9d6M6WMOdMZ/rxHzH8fzvVXhvvOHnx7y1a2T9KmiDbTl6noj1oWlsMbCicWLnQoYyqv49cYsznuYsttpI402XmJST8Wuan3pCbpNvcGAAx55Y8y09tPp6lgJ/wJ5PfWLo80fC1lcW69QGe4v30XDZYcZ00/RnvXPbmPKtqdMXOFYbuSdGvkTP70y5c3tWahKoKnZNWwGbGBr18ILPPGPvyG22kq+nTgYgEY1TmLV/TeYZUvDyXNomD+SMbeZJYtPHiw0dr8r15LyLWkMj7n2iurdVrB4cWPRWN7h2rVrHD16lJMnT5KRkUFSUhKDBg3i559/LhRvzJgxjBmjmLdVkBgIwQn+Y0YtSMpwNawoL4moqChMTEwAMDEx4c2bN2WSV5nv2MpzUtgaloKBXV+Ob/Bi9sLtVGrYld7Olcs0n7DTXzJhk2LX6M7UIUz4qjcbr4aQE3+Pvk3r81d8hsLQWT0Up1FrCEmM568fhmLfS7HNnRb1Ow0c+3I3LImUiHuMa16f3W/SALg5cRDTF3Ri8ubjvE7PoWqHGgRtnsn2I1eIzVAY6JZDd3JkQxMAZk234sKcfNfSJz9uwrTNKnSUJfyy5TGtVvR4p/QS9j4NY1kdxQTg4DfXGd/DDICklyt4o9s7z6h9S5XmOwkJXPJBuUiUdbhy/ToXz6wuMV7wnudYjMyfWNSfVI+IszdK/E1a1HFaNmjLmfvRBF/dgUO9drzKLLxg8ezX4TQZuJY3qVmE3D2Is1VbUnLkyHNSGGhrza5rT0mPfcrklnXZ/DK52PBiFVZZm2l9OnEpUbGCuqp/L3r2Vbi7Zac9pl23gahJJMXWe0b8CfoO35prcF3G1bYjl4NjuLRrAp5j+jJ1T3BeXsv7DsI/Wsqb29twsesLwOMb15DJ4erly0hLcbagzqCfGaZ1jJ4b7xfRVpL5ws2aL3+5TGLEA2Z7WjN5z5O/rdPFkZ3+hM52jqw45keo/190aZy/Yvb62CwmbXtS5POe/g4M3+RNalYa13+dimPX7WXSdjOSLtN78DL6uY/jfMAL0mOP0HfkrrzvCz7H+m3A0XEot54l8vjSFmwbdOJperYYdICAOcOZdjVSYVAuHU2Xzm25l6pYRPiqfy8Op2fx+sTXNGg1Af/QJG4eWoJ1k/5ESBWj1cQ+PVjdtz07z9whXSZjRR8HBiz9g8iIe8zv4Mi1ZEUb09AbzK4t0woskrxCrlwBQ+V0enTxIDrXWyUr9QFtug0psf3Js+MZ3cqab4/eJTHkFgOb1GVLcJKozI9kz/Mk+rTKdw8e7FSZ2yfD3ou38qovg420cxfxHnA3FdoYqL83Zo/ttZW9f85AArxOkmJgnj/+VKxvQKxf/vh24WtPwofvp69p4eNB5+IyqXLrF+bOnMXK9XuIytULx9Vn2DfAPK/P874fh4VL5XIl7zj/I+hUnZq3OKNffzxpUXuQvjME6NdZw63TvfOM1/unn6Nv9b4B/+LAcE7ab+QLK/0S5VpSviWN4VEXolA3CWTJvJnMX76WG+GpotG8HdeXLyc0NJSXL1/y22+/4ebm9p5RKxAIKPWObUxMTJ53g6OjI1u3bi2UTGm9JMqKMt+xzc54hlQmJ+LKAL6KbIRuZCCbN6wh/PZzJtfX+2T/SHTWFM4scwXA8M+D7AxLpY3WG3p+e4FTr55jq62KfPY0Olc3ZW3YBAa/iWbUslPMH6PYbWjid5zvfGMY2l5hYD6LGMv9I90VdTvxGGvTvmbH4pFMHZyITbNWdOjRn5mjuqOhBBbDlhK/oA+vMgdjpq7M6l3P6H1eYfReT5TS30ijBMXJZm2kJiGVNBWG7bPHqFXsW+L/mhDow6PU992z6zZugoFK6dYqQmMz0bHIH0DVKtYgJ/1pib85M2I6WrNPsmmy4iyt5Wt3NoUkM7JAnJSw6iw/+wMjzRVpP/zTkEuJmbhxgVOxJoSvW4KWkoS+Dcy5nphFRoWrRYYXb7mrM9/ekLU339CitSp70hyon3WcgNQsqj5eiY75PAzloTgUU++jC2z23Zo2CZVpx9kwwwaYx9fWJpwrcEy4yjd7+KapMTCB3yvp8TQ9m56zv2bsEi8WLl6Mak4cN24EvVdEVe36ONrm6rpEifnHNmLZoDPeAx7RqsB6ReT1iRyJ7UfExeUATBhoQo1mw1g35Prf0ump1Yre+QraMprH1RbwfO8EAIa5DMZmaMwHdSSiVgf+WricWhrKSMe2wthiNjCqTHQxLeZPpvmG46qvQUr4D8XG+27AUjoeDGBVU8Uk3qFHPUZte8KlSVb/+THHbbYNy7/zBfdqHDr6mpH1K/Ldg3i2W8fwZ6Ier80rMsp9MyPPBLHIthIwB9OmZow5GsKxXorduIejDvNnh+qkhK7G67IOwaG/UklViYwJbpjU6AaAZuVmeLrl1nfQeSYMGILrl39gpO/MLLNsvrobzbamxrw6MRu9Zt9TXRJG82J0tIf/aM5kT+XVdoWh3LNGDLN3BDP220ZiEvERPM3IxlJLNe9Zx0KblOCU9+KZVFec05zY2IpfH7+iwcif6ahfeDx6snMAr3vtwLmCwrOm+2hbPMd58fTEYowyg1kw9ho5UsVUIcZ3LaO8nXlyoxG3hhcex7wTM3jlHcWYNrYEndqEnd1Jnt7bh65JNbSAm+Nb0/23uyhb9ONxR7NyJe+UZyloVcs3xlW0LJHlpBApzcGsgAeJirYRVbUh7vFYGrkdICHLjAOPPQulJcuOZeT0W+x9vC0vrDi5lpRvSWP4vQfxvAq5itH4dmS+OE8324b89MifLpW1ROMRCASlNGxLF83Q0BAfH59ivy/OS8LY2JiIiAhMTEyIiIjAyMjo8zRslZQVk2vdGjN4ensBOSkBmFZtztrxl5js3fWTyb/WkPxr2w3UlcmRy0mP/o1kdPlx+sS87zJkci6+TGaqy3g6RuzD65udPHv+lMvXI7EYlV+Ltcc2zbellDQZPGc1g+esJiXiCefPnGTjt+P447oM/109Ua/QgpnVJcy6GM4uhxucyKzLm1x3LENVZcKl7+/np4U+5XW2CeaVX5KmWi9v61zdQJMcaXgRCpbFvfuPsWhgTeiJP/g59P1dzdE2jhjolM6w1VSSkJ2Wv/MlnCJG9gAAIABJREFUz0kGScmD3p/34+m8tWbe85CD5wEILnBc03baNEL27WXB1qc8DwrkUmIGQ5Cjqd+OMU3mYW7bgk4erfFo342+dgZI5EWHl0SzeY58tdyHJPMb6NhOYKbsKd/fj2Pkj3ewnj2vxHofbZGfzrHLkbSYXyPvubO7CQWv2+jf4O1qugQDFaX32nhOxqsiV3p1Tb/IN2wBLePOHJ65jt7dvuPZ2dH5iwtHH2DWdUrecwWLiWhmzCMwJevv6XQxhu2L/SHU/bJjvsHe6gtgzAd1ZMrMIezbtoZNT5/x2P8KRbm2/11dVNdtiqu+xgd71e1hqbj+9DVjdyrSSkrIIOJUBAjDFhP3L4kZ9zU5Uid2pdpzc50mHdY9JG7qbvRsFqItT+B4nJRtDfLbU4cBtdj5x2vINWyHuSgGkpi756hYexaVVBVy1jBoQ6uK6gXslng2zxvH0gOhTPU6yqw+ipXX/osa4z7/Ipzty+5vfBl4+BfSo7cWq6ONfnlErYGL88Ith+3mkJg+fDSaShJSc/LHlezUbJTUi29vG+7cZ8Gzm4xu3YXpvZryffMquf1+CkO/vsO2Z/njXYNph5gfM5Z+LW2RV6rDsK9cOTRXD5k0nAHdN7L+6k2kaalkZMuQSdNJS89GS1PCrfuPqF7dVJFI3/48MavC7KB4NuW6Rzfd6E3w0pf8OKodrSZ6c3eTW7mRt7KG8jvjpWIRQV1S9NEMg3pbePZsJWd+Gs2g5uOJepR/UV/wnkHEeW7BRluxMFGSXEvKt6QxfMDp2wypUg1dZQnQD4vHDZi3MIAuG51F4ymAq6srrq6uQhACQRGU1at8li9fzvLlig2cixcv8t133/Hzzz8za9Ysdu/ezZw5c9i9ezddu5aNjVjmhq2yRi20lZXQs3JFCVDSsaOaujJv4j+tu5myVhHn7iRqqGo1YOLE/AkWEyeiU8uQgOXt6X7SjJVzBtJt6FSGKHmwvMBPVfXyV8NndW6H254jeOproGNSh65D6+DWWoaZ40qgJwCDljalxdyDPO31OzW6eeW5Dg2y1mfl7mAWLyi8I/FTJ1cODDjNxclayOX5FxNUtBxOWsREQjNnYVpgJTgl9AeaNV9NRFwYNfuOZGLm++6Y1TVLX501G+gRdzsO3BUTkaQnr9CoNLhkZZFAQRs9481LojWqFoqz1L0h52zGMafvAEZOX4q6U+7tlxJ1Vh6/x6T717l05Sr75nRm48VDXFrepNjw4jBymk9CwBiCd2RjNWUSjZRs+GrdI3KuRTJ7vSmkFF/vFNjUUJYoJtxvkWXICtluWkolnydV0axdOI+34erV3wtznHWY5tssGH2oTYHfq5CdUqAe5dlkyUE9N9+P1elijUhtFbIL7KrKcop3S5PGS3ON9pe0sHKn1ZwVdBviyfSFk6hff/T7evQ3dVFJpfjFC3lWTN6CgooEhn4xocCuyERUNWuKEQdQr9CCbpoP2e27DG2biRg7axE19iceEoDjvCUgUUUZOekyOTq5Z6Nz0nNQUs/Xa73cXXVVXVVkmRmF0k99e3maLIMZbRpyr+EsAoK+wFA134AybbeKtFGeRMQasynRgcg6eqSHFa+joSpKyApctCeTRvEiXAuLmrqiQj8Clwrq3E7Moq2eYnEo4n4ihqMqFYojy45lmddPfP31HCQoYWTRjDkdTRm//yXkGraR16cRabkYW23VAgu52oxZ/jNjcgfEF4fas6ZqN6Sp9whWzmZyK4X7qzQxlpSzPWl2eiF+53ujqVWgTUtUaG+gzpGYDA5/70WlUdNoUUEdbYOajPZqweoOx4DyY9jqNzQh7ftAoK3if0++jYqmBcZqhRcTXh3cwBnjPoxyMUJZsyJtx64m5at6hEu3UzU37tJFPky+le9CXJJcz35ffL7Fj+EyslTV0C9wH4JFSyNSDwt3ZIFAUErkQM6nfUftnDlz6NOnD9u3b8fMzIwDBw6USbqf4FZkJWbW1SP8/BwOX/PjxOYxBKVlYTns/9/VTMtoIAbpl3muXROb/2vvvMOiOr4+/l1670oRERBE+lJFAQUUsIu99xorxhKNJfau2DW2mPCz99h7w46AgAgCglKlt6XvnvePxYUNRTT6RpP5PM99nt25s3NnZ86UM3PmXGtrWLRSw5zB/fGeCCHHXsJ54woM7OaB1i1UcD0su9507CkBCxadQc3TpDHnb0NBr3v1RN53A/hvVmDB9miMXljtIqLT7rmI2zIQBx6kicJyXgRiZUIJZo81gZScMZoJolFcpWDJqXfDsjaS6DphE3IrhWEkKMHe6Xug57kRSpIcxO4LwMqVK2tddXmPrklJejgePo0HAJj5eyBu/6+iM0InVkTCfrZzrXg1GeKqjRNbH4mkfkXHdlj5l/Ow+8NzsH7Nj+ji7gQ9hbd4UlgGIqAoeQPs3WZB36odhv0wF7u2uiLuUmi94Q0hrWiF0SrxmHEgFhOcm6BJG3+k3lyAa5xu8FKVbbDea9LLVw+3NgrNfolfhB1XUhotWwIiVJS8qrMeAvbVNunmSCph+8WlODt5pCjMoL873p7ciOxK4UQ/+eZskGpXtFaQ+iyZFpSn4f79+6j4S19k8oMVIlfvFHl5Dv9tjdiCUFF8hkixPno8USgDuZeQINMeG2aOgpuDJSTiztWZn8+VRbGykZAHvzhK5LXzxb6ronvTjVVxJEYG1tbWsLa2wt2fR2NLmnCC9vRBkOi8bcbzxwiJK6xSlsTLoWa8fxs/9DbAYv9TMJ9uC1m1jnDmX8WCG+lY5KoNjqQSxjVTwKzzCSIZ37YvFo5jWtZKR8N2DAreLERwnnCRLSdiOx4WCD9nBM/GkYzBOLtiFOTLS8Dj8cDj8UAApORNscysEkPnT0Orqasg+REZbTnGEbH79qGkqr97ttoPfVdFsEnEJzKhSzOc3iTsi/llSVgZkYtJPs3E2oKElDrObV2D3R88EFMlLj7KRHP3anOvm/Ovw26xuKls9O5OsBp4XCQzWxaEwXOJK+TUffHmzRvRdbRHC1j/dAlhd8aiNO8GTFq3E7WzyuJYbE8rwWhLdRT/uQe//F7tRf3VsZdQMnT5rspbzewnUFoAnhcKF/5iDgegqePPtfqbiopLWD43UGTdk/vyD8go2UKnSqktzvgfLpTbYYyOoijthsq1oec2NIb3sTbD5hdVcxoqx4mD8bAZ35I1HAaD0bh5GQgcatz1KXh4eODChQsAAE1NTdy8eROxsbG4efMmNDS+jJO2r+IVecrZ3bjoPgpDfdwAAIaeE3B8ivlnp3fe2UjMCFLb6SQuDG6Eii3dBBd3ToRPW2fs93JAftgtNOm/Hy7KMtBe0Avz+vlhfDdLpL16A2NbNYQv/wkhHoG10ul75DgudO0LA+ttcDRpgsL0WLzMb4b9V6sdqkjKGWMNVxkzX1vgz+bVJqHKhuNxe8sb9O5ljS2mXDRXKcGTZ8kYvvEm/KrO1c41kcLB9zxM1hX+bsqZy4jx6w0TowNw5Jog7/VT5Oh0xfnLQqcUdqt24nNcHSRdnIneq7l4H7cRTR03Y7KlLex8BoArm4hgzeF41seoVryadPj1dxi790C7rm7QKY9EjP4whFhrIqnGvHSVX2v09x2ELuaKeJ1AcNKUw9o5a9Frjz9sy21g1j4aLgbyeHrrJWad2gFFHZk6wz/GsJEtsW+bJLqqywFojw7S0Ujuu/mj9c6rsWjttO4YLNy90KGnGxRzE2HVTgvJ2nIffbaJgjSGTpyEg7t3f5LTCTWzSfhj0B4MPCj8rmm9Fou7doKVY0d4mirgflAqVv15/bNlmpf+Jzp3/hFv8gqhXWNXzdAvECMDXWDm3BHtdAWIVfSBptRRAIB+t0momDMKfqP8IJ0eAZNhLYBsQLHJUHRSX472A0bDVDoHSdK2UCp/iemrwrGmxjHbz5VFMWVdqz86qCyEffdhsFfKQp5xdwDC3cOpZ7ehh2dHeBzpAPWSCMTKdMfjdsLztoO7d4XPs3fYZaKGq2P7IaDDKYRsaYOSHPFyqBnv30br6T2Ru2cdxlV5BZ/hrIXhkb1EO3CLzmxEF193dD7kicrEu+C188fd9nq10pHX9MORmcfga9EGHdyaI/6VDCZV9UdJZ56jOCUKeno7xX7zLD0breSl0H2NJ2Z2OYPLqWYflVF0+BXTHdxh1rYz3A04uP1CDqeCHcH4NNps+B1N7Tuja0YXCGKvwtD/CPpUjSc128LR7RPQvp0Trno5ouTtQ2QZDcGNqn4eglKsepWL9X95f7npqD0w2emKtt1PQiHtMYodZ+N+x2YN5kdesw9+GxcINws3eLjoI/7hYzjPOws/TXkUBW7EFhdveF7rAI3KFDxKVcfv13t8V+UtrWiN0/M7oFsbX3R1UMWtRzI49FR4Br1mf2PU5zd4bXWGWfubcGgqgUcP3mLOb5dEOwhJ5w5Aw2rGF3lu/WM4B4f3+8PLxwl3PNqAl/AAJeYTcK23IWs4DAaj8RB9l9nmEH1+zouLixsokEq8iQxGvowebM0Mvs4LcxtJeV4KXkS8gVJzW5gbVr/qpSDhJV6klcPc3gaaUoUICoqAtZsb1KTqNkFNiQ5FfFoe5DT1YW9tCqnGv/kE/JJMhIS8QrFABq3sHKCrVG36lfFkBrqfHYGnq8U9hqXGhCA2tRBK2kZwsPg6zjbSYkORVKEJRwvxOtrquxPTr06uXa2CYsSGhyFfqikcrEzqrNe48CfIJA3YWZuCkx+Nxy8r0MHNGqAKRL0IQVaxJEy4XOh92JmsL/wr1fsH8iIeIFajFeST46DW2gERI2yw58dbONNBr2G5T4nA0/giuLm3/SQZqI+MxFeISSuDua01tBQk/9Z/C1vaAWaL7kK+VsXw8S46DOkCLThYtEDNp5TnJ+BZWDI0WzuitbZ8dV3zixD2NBRSTU1h3VIHOVFPEMM3Q1vrL68gEr8AIU/CQGqmcLTQFbsnqMhH9IsIlMjpwcGq+h3EBYk/YyNnPpa2YGasDfY9ZTmIevESHE0TWLXUbTBufkosIlPKYGNvCeW/KdwNtb/U2HDE5wJW9tai1w8xPhFBCV6HhYK0LWHWrP5Xu5XlvEFwZDJU9VrCyqRZI9tjEcKDQ6HQnAtTvca3r+Ksdwh/lQrd1ly0aFK9SCiozEPI03AIlHRgZWX60WMe3yq8jHi8SCyFjYOFyLy/jhaH2JBnSC+Xg5mNFZp+gfGsoefWN4bzS3PxMuwlpLRNYWGk3fDiogJzKtUQKhwNtOF0/GbyI/GN1RdHXu6byUuFVYtvqmyuH/ntm8mLs28Sgl+UNiquqqIeXFqPb1TcbM7FBp1H/XsUW8YnzOrLMdGpF1Y/vthor8Zfk9yIYwjMcMf0jnr/1FQc69bV/7qiLlNmihxv/B2Sr02FywIB9m8cisr0p5g5/TeciH8O2y+Q9j/SofPCsXVvFmb5e/0nms2u1esxfO7sBiaYDAaDwfgYTLFlii1TbJliK6bYKujBxayRiq3kt6XYSoHxDfQKMthwYjQupxZjgIHSP54ddeuBmP6P5kASw4YNq/eumvyXEVt9n+04X74PJ86fgEBeHwcePfxulVoAkFa0wSz//06z+WH+HNZ3MBgMBoPBYHxp1eQ7NUVmiu03grLRAAxgxSBCT+//Z7fYrvs42HVn5c1gMBgMBoPBYAD4bs/YMsWWwWAwGAwGg8FgMBhCpVYgYIotg8FgMBgMBoPBYDC+Y75PvZYptgwGg8FgMBgMBoPBEMLO2DIYDAaDwWAwGAwG4/uGKbYMBoPBYDAYDAaDwfh+lVoAAqbYMhgMBoPBYDAY/xpktSSR2yLhb6WRlZUFLS2tb+Y//WvzU/j8m8qPc3fVbyYvb1PzPk2z/S/u2LKXejMYDAaDwWAw/q1kZmb+7TQcHR0RHBz8zfwnlp/vJz//WF6YKTKDwWAwGAwGg8FgML5bCACfve6HwWAwGAwGg8FgMBjfs2ZLTLFlMBgMBoPBYDAYNZgwYQLLD8vP95WX79QUWYJ1NwwGg8FgMBgMxn9Dcfvhhx/A5XJhZWWF/v37o7i4+LPTGjVqFE6ePAkAGDduHKKiouqNe+fOHTx8+PCTy8fQ0BBZWVmNDq+JkpLSJ/2fJUuWoKCg4L8tOx+8IjfmYootg8FgMBgMBoPB+CeQl5dHWFgYIiMjISMjg927d4vd5/P5n5Xuvn37YGFh8cmKLeMbhKhx10dISkqCp6cnzM3NYWlpiS1btgAAcnJy4O3tDVNTU3h7eyM3N5cptgwGg8FgMBgMBuPzcHd3R1xcHO7cuQNPT08MGTIE1tbW4PP5mDNnDpycnGBjY4Nff/21St8hTJ06FRYWFujWrRsyMjJEaXl4eIg8+F65cgX29vawtbVFx44dkZiYiN27dyMgIABcLhf3799HZmYm+vbtCycnJzg5OeHBgwcAgOzsbPj4+MDOzg4TJ04ENUKB8vPzg4ODAywtLbFnzx6xe7NmzYK9vT06duwo8nIdHx+Pzp07w8HBAe7u7oiOjmbC8BUUWykpKWzcuBGvXr3C48ePsWPHDkRFRWHNmjXo2LEjYmNj0bFjR6xZs+aLZPuLn7GtLI7C0tVHaoVLK1pi8bxBX7zc04J+x+zVvyM89i2grAuPXhOwduFwKEhwqiqmHFcPrMKWQ9cQ/y4HmgbG6D7CH/NG+Yi0+iG+nZBWLlydkpCUQzNzR0z6aR7a6StW16+gGIFr52H/ydtILiQYtnLED4vXoZ9zU1Eaa/68CgNZyS/2396dn4bFcT/g4EwLZDzchTGLjsE38AJ8L/pjk9LP2D3Y+JPTTLkxH+eM5mFyy/rfrZV8dQbmPR2L/y2yqTdOecF99BxxE1fOLvmCjagC5/cE4OLDGOjaesJ/xlCoSnI+KR6/NAG7Nu5BcAIPjp2GYOogF9HPCuJuYPOe03hXpobOw2ain1MTsWQzg1dgF03E4r+E/5sZ3cUbiaWVYmGS0pq4ce3k35KPL0Vp7iX0HhuGy6d/ZoPM/zMb+naFxb7T6Kouh9LM59i07gCi0kphZOOFmT8OgYYUhxXSP8jrqwew58w9CJpaYMTMGeCqy9bRV1bi8q/rcO5JFKQ0W2LwtDlwbaFUdSsXRwI24caLJDRp5YJpc8dDX044fr1/fgbrf7uIrEpleAyYilFeLav6/SAsX39V7BGd5yyCq4oMACDy/EEEXr4DnlIrTF4wGxaqMgjZsgpnskrEfsPhSGPZssXfVXk3NLbUpCjxLtZuP46EbD6s3Pti9mhvfGgqK39ZjNIapnvNu07DhLbCOcSjw5tx6GYo5A2sMWjKVDhoyYk/vzwFk0bNw97DgbWeuWjCAizfs1IsLC/qCnYGXsTrPGn0nrQAvWw1WaNh1J6zV1bi8uXL6Ny5MwDg6dOniIyMhJGREfbs2QNVVVU8e/YMZWVlcHV1hY+PD0JDQxETE4OIiAi8f/8eFhYWGDNmjPh8KjMT48ePx71792BkZIScnBxoaGhg0qRJUFJSwuzZs4Vz5yFDMHPmTLi5ueHdu3fw9fXFq1evsHTpUri5uWHx4sW4ePFiLUW1Lg4cOAANDQ2UlJTAyckJffv2haamJng8Huzt7bFx40YsW7YMS5cuxfbt2zFhwgTs3r0bpqamePLkCSZPnoxbt24xofig1H7mrv1f0dXVha6uLgBAWVkZ5ubmSElJwblz53Dnzh0AwMiRI+Hh4YG1a9f+7ed98R3bipIYrFmzRuxasWIF1gac/+LlXpZ3A1beM2E7ahmuP3qGc3+sguDSHLT1rxbMXcO4GLjmCYb+uAoXb13CxrkDcX9xX7T78ZoozpN7d+Hy40KsWLECi2aPh4Xkc3ia2eFqdqkozvY+1vjpPA/zAg7i1qVjmNmnBcZ52OFCVZwn9+6i+Avbmivo2MDeRBkAcHjEz1CbuQUjtBWhZGgHbg2lu7EIKrIwZGIKJhg3/MLokvRQPIlq+EXO/IpM3H/w8ov+36B5bhi9Mxrte/si//JPcBp2/JPjzXJywm9JyujlY4sz07ww6nSiUC55EXCw7YlMHWd4m8tifHsLXKgx2Xof+wDTB6zH3VTef6rvenrvLuz8F2DFihWia9nSn/62fHwpBOVpuP/gFRtk/gFiHt5HWrkAgsoc+LR2w3Oplhg0pDtKHiyCldcqVkD/IJnBv8C2XwCM3f1gUHETblYDwKtj/HmyxBVjDiWhvd9QODZNg5eVOxJKhZOVvX3t8MszAXoN6QuNtN9g13YOAICXehit2/ujiWNn9O5ggm1+Vlj5XHiGreDtDmw+EIPKykrR9cFvZlzgSLSdcgKtPPrAQvIB2tqNBQEgPl8sflHGDWz89dl3V+b1jS1iSkJJDByte6FU3xHD+njixfoB6LQ6THTvl7UHxMqisqrOgn52R491T+HWaxBMJSPRwaIrsirFPZL+b7Qn9h87JT4Ol+Xi8q/jsO7IFbFwXtoxtHYeC4GhK7o5qmGsqx2CCspZw2FUj+MlJeByuXB0dISBgQHGjh0LAHB2doaRkREA4Nq1a/jjjz/A5XLRpk0bZGdnIzY2Fvfu3cPgwYMhKSkJPT09eHl51Ur/8ePHaN++vSgtDQ2NOvNx48YNTJ06FVwuFz179kRBQQEKCwtx7949DBs2DADQrVs3qKurf/Q/bd26Fba2tnBxcUFSUhJiY2OFio6EBAYOHAgAGDZsGIKCglBUVISHDx+if//+4HK5mDhxItLS0phg/FW5/QI7tjVJTExEaGgo2rRpg/fv34sUXl1dXbGd/7+Z76/Lu4uzSIIjQT9fTf7iaafc6UyqhsvFworSdlOb9hOJiCgnegnJKDtSQmmlWBxe2mHS029FWRV8IiIylpOifek8sTh/DjEhgy4niYgoL34tyaq0o8yq+B+4MqA1Ocx5JkrjVXEFERFVFL+jkwd30YaA7XTiRjgJavwm+dkl2r01gA4cuUj5lYIGw4uSnlDQy1zKCr5LQ5oq0IjA0xTFq6D81w/o0ZtC0W+THl+nPVu30uE/H1F5VZL8slS6efcNFb9/Qb//eo6IiOKPdSe3nVGi39WXz9cH3clk0F0SVObR9dvhVJb3ik4c2E5/nL5KRXxhrOKsU6Sg1Y9Ks0Jp77ZNtCfwPBXU+D+FCU/owI7NFLBjH92Oyvp4ZQpKyUReig6/F9ZDBS+cZKQUa9VdQ/GKUneSjBJXVAbpj0eScrPpRET0ao8r6bj8IUrm1jBTspj8iIiIIjcNIC6XS8YK0uR1NqFxwicopXtn/6CADQF08OhVyqsUkIBfRFeuXKGyGhWeHXKXHkTnC+UuOZgO79tKe/44Q2k1/lfQ9WvEL39PZw9spdQyfp1pf6A06wUd3rOdTt98QSUZz+jB6/xqua4n/YawUJCm7alFdd77mHwI5ew9nQ7cRwHb9tCNSPF6rksuGyzSyny6cuoP2rr3ED2NLxD+p/R9pNh02OfL1b+Ypzev0duqei7NCaZr14NE994/uEWP3wnrNTfqPv2+awsFnrxE2RWCBuSugu6eP0pbd/1Bj+MLaJyOEu1L51F29BhSbjZD9LvKsmSSlpCgiIzndOPOK7E8hd66LuoH66v/uuqZ8Wns5jYhr8Nxou/DtRVpSkTt9tC/iQLtrNG+R+so0uTYXCIiUpWSoBu5pVV1UkCSHA5F8SooYpMzNetwWfSbZ3NsRO09akdbspz2uK7WSx1UZSkgsbo+l/84QyQL1fBptp0ubY3I/q7Ku6GxpSbvg/uSpvkB0ffM8KGkrD9b2HelbCFl/Zl1pM4nAzkpOpJRLArZZduE+t6qnjO9uzCD9DpNI46EfI12VEiOdnZka21MMkr2Yime9m5OLgGR1XW4Zj7tiM5lDYchQlFRsVbY7du3qVu3bqLvffr0oStXrtSKN2PGDDpwoFrOe/fuTSdOnCAiog4dOtCzZ8/o3LlzNHTo0Fq//eWXX2j9+vWi75qamlRcXFwrnq2tLb1580b0XV1dnTIzM2vFa9GiBWVmZtLt27fJ1dWVeDyeKB+3b98mIiIJCQmqqBD2RfHx8cTlcik/P590dHTqLJu/5vG/iIpUE+rcdFKjrhYtWpCDg4Po+vXXX+tMs7CwkOzt7enUqVPCMUhVVey+mpraF8n7Vz1jW8GLgHf/rTAZeRQrfZp98fRVWvqgMHk95m0/htgM4e6bos5EPL4rPAQfsex/MBm+DYZ/MQ9W0BmMlKQYaErV//c91k7A+wcbhDsXWwNh0GM1tP4S3/fYKwSvcxRfKKjMQ9eW5tgbFI8K3nvsGe+KXntjAAAJJyagtd96pBaU4PnxWWjtua7B8KSLMzF+2yvkhj/C21I+Eh8FIbakErEHJmPa8TdC86U1g2AzfD0S83JwadMgmPVYAQAozb+NngOWoLfrGFwPFcY9/nMQZvQ3/Gg+RfVXHIVuPcegq+NoPIhLx5Vt49DKY6Zohb6iOAo9+65DViXh7vYx4A7+EwBQnH4ExpZ+eJacj8LUFxjjaIT97xv2uFeaexWJ1ByDmyoAAKQUrNFdjfDHe16j42WHnIay/mxIV5l9aVhOBy/9AMoJiD0QD5OJbUTpWM60QOqVoKrPxxAaGortNlqNlr0DfSwwZNtNFFXwEPTHZFh1/hUcCUXsGtoby+OrdzN/7NIVZyoqkPV8MyytBuFhXB6ibu9AK2NvxJQITYAn9OqGNX6e2Hv5CUoEVGfaAFCWfxsurTrhdmwmbu0fD89Rfph8QLgi2VD6n7Xg1Qj5IH4h+pmaYN/91yjOeo1JzgbYnlDQoFzW/7wcjGxjgqVnniEv8RH62bTAjth8sTifI1f/ZkJnDcWUu8IV5rBfRsLXpz1e8CoAALP79MCpknK8+/MntHQZj5CkfDw8sRgmtn2QWi6oQ+4EWNHLAn1/OYq01HDM87LC/YIyAIC8+mgc/m1u9S5R6VuQpCq0JIvRzac9MioEov7ercsmBMu9AAAXNUlEQVQgyHI49dZ/Y+qZ0Yj+Jz4fgz11RN9HuTTF4/PJteJtDn6F0dpC656y7Ag8KwJ8NYQmrsNNVLHr6GOUCSoRcX4FZJt0hLG8FHQ8OiL35U6Ev+ehLO8NfjufBKc+zQEA6dfTIasXhsVzp2PesvV4kMKr6pvu4kFpE0xtko97l8/g9pOXWLhxM1rLi592enN0CM7b78M0K43vqrwbGltqotF6B17cHvzB3gThF+OgbuUs7L9S70BaWRvbls3HjJ+W4Mi9d8I2IShBShkf5grVZdVSTwFRx4X3y/MfocvYZ7h+dLzYsziSSngWEoLHd7fVyu/O4EwM76+N0PtX8OeNhzCetRKTzdRYw2F8Er6+vti1axcqKoTjyuvXr8Hj8dC+fXscPXoUfD4faWlpuH37dq3ftm3bFnfv3kVCQgIAoaMgQGiKWlhYKIrn4+OD7du3i76HhQktHNq3b49Dhw4BAC5fvvxRx0L5+flQV1eHgoICoqOj8fjx42rLL4FA5LX58OHDcHNzg4qKCoyMjHDixIkPm3x48eIFq/TqWVmjvSJraWkhODhYdNXlxbmiogJ9+/bF0KFD0adPHwCAtra2aJc8LS0NTZs2/fZ3bM8OMSEpOUOKLan4as8IPrSWurtak4KkJBnauNOknzfTq8JyIiIKbK1JnqerV3yyI9bTqFGjRNfRqhXSunZsS/NuEUdSuDp6wVGb2u6KajAfH3Zsywue0My5O0XhaY+6kbbjBeEusIM2ue6LFq7RlqfRuvXbGgx/tbsdmU96SEREiwxUaMU74Wp48DxbclzzgipLE0lDTpNCi4T/V8Avpk7qcrQuqYB4GYeIIyFLN7JLqpZ3K0hTQZc+7Dk3lM8PO3JlBQ8JAG2Jy6tKo4z6NVGg6WGZVJx1iiQklSis6tlFabtJUXukcJU6eDMt2lG9k3O9swF1vJBI/IosCgoKqnU9Cc2hwqT1JKfuK1am85ur0NBo8ZX9huJFbm5DzTtdq17R5hcRAEosraQdJurUJShVdC83fgYp6Y4XS+eSi26jd2yX/ziD4kuEu2VlBQ9JRtmZiIhCltiRyeCbwl3tjMMkq+pOFQKiWS1UaPqDNNHvD3U1IOdNEaJd0yF/vv1o2reGmZLj6jBRvLnGqmQ7L5iI6k+/NDuyzjKPzC4VPVtaTo7kalw976U2Sj6Ks06TtIIZ8ap28RNP7qLAkKwG5bI+Es91Jy3btaLvMXsHUpfZz8R2bOuTq/8qb050omaeZ4iIaLa+Mk2y0qCBj9KpghdBMrL6VFgpID8teZofWr3KvcpWi7yPxteSu4J3q0lGyZ4yy4U9REnWFZKR4NTqF3OirlJfC3XyXSrcHV5qqk7Dq+Qu9lBH0u90pMH6r6+eGZ+GmpQE3csvq969n21NZqOD6o0/3sqQZCU45PjDyerdxOebSVaCQwpqSsThSNBPl99VdZzltNhdhyQkFUhFWpLUW48TWRFtbqlGmtzBtP1AIG1cOIKU5PXp9HseFSYHkJR8S/I0tqFBo8eQt40mWfReK74vWZFJbTUNRGPG90RDY0tdZL0cRU3VZElawYwuZwrnGeHrnUhG2YpWbt1P+7YsJUtlORp3VDg/We7UlBxnB1J2cSnFPz1GRnJSZNjzJhFV0jxnbVr6IJ0qil+J7dh+oCTnYq0dWy1pSfJpZ0SdB4yigV25pNLSh2JLKlnDYXzSji2fz6f58+eTlZUVWVpakoeHB+Xl5ZFAIKApU6aQubk59erVi3r16lVrx5aI6NKlS8TlcsnGxoY6deok7PNjYsja2ppsbW3p3r17lJmZSQMGDCBra2syNzeniROFFpdZWVnk7e1NdnZ25O/vTwYGBg3u2JaWllLnzp3J2tqa+vXrJ7Zjq6ioSAsXLiR7e3vy9PSkjIwM4Rj65g35+vqSjY0NmZub09KlS9mO7YcdW0kt8tUc36jLwcGhwbQEAgENHz6cZsyYIRY+e/ZsWr16NRERrV69mubMmfNF8i71tXT9Cl4ERhx/A7Mf7sFE7qs9Bg5D5uL8kLmo5L3Hg5tXELhtCZzMb+H927NQVZZCUVyRKK6cpj06dRLac1/5cSKeLd6MgU3k60yXX/oWUjL6AABZLVmUpJbUjlP2DlFxfFhbGonCpJWd8dOgHGxauQiv4+IQfOceSG0SAMBxwxS87eYE99Pd4OXVCf1GjWsw/GMUZ/wPBVDGlqnVqyMlAsKthEJMaQ3IqriiY9XqfEVxFHgyFqJD1Q3lsyaSMtqY/sHRFEcG032awf9cMjAFkFV1h62itPAMg3QTgIRnt7QcpqNnaiCWL9iHuLgY3L6fBtNJBH7pWxw8eLDWM5SbT8PycXIgQZFYeCFfAFmOuIMajkT98STlJFHJqxTbUQQAWQ4H8hKc2vc4Cp8td7Pnj0Hg7nXYGhOHVyF3ABKuhptPXYx3RnNRScF4uWENWk85ACkOYXdyETru+gmj9wprID+3FKkXUoGZVgCAse7aH0377O10eCwzFMXz89HD1aqVtfrSL/KMwMGDtT39tZ7SGpYaQmczG2PfY7KuYo2dAElIQPej8iGv0RWT282BrqkTenXuBN9ufTHUThNFSbvrlcs5+sp1lueb31/CeORq0fdW447iEoDi99UrqPXJ1X8VPe+FyBwzF/xyF+zj2SFslwK8NkQie85+qNuugCLl4lx2Of6wrnYa02OEMfYefQsMNBaTu6xn16DW6mdoSUtU9ZW+8FKTFdtR3z5nDH45moTZm67i58FOwl2/1W3g+tNN4P5Q7F8QjJGXT6A4Y2e99e9YTz0zPg15CQ6K+NVnMCuLKiEhW78F0p6IeCyPe4iRbX0wdVA7bHYqhGv7BVh/MwHTPFogM+wkuG5O6Jj8FqobPLFPchJSixehqSQPu8c6wnnUeUQH9sTwO+EYrasPFUkOgGEwiTLG3J9D4b1UAH7pG0y+V4h+zRRB/AB00GyKBfETsbJq/Hh9YACyu/8mGjO+JxoaW+pC0+I3pKYG4PKukRjgOA4FiYfQavRZvJ3YBDrKwv/vax4J85FrsXfgbsy9fgUZI/3hYDwfujbemDPAEJslZBC1pQdOtdqAUFsl8HjJAAg8Hg8yCoqi3eO6EIAgNfEKLo5oJdzh79QcA1a+QMhye9Z4GACAoqKi2paKHh7w8PAQfZeQkMCqVauwalVtnwo1d1lr8sEZEAB06dIFXbp0EbvfqlUrhIeHi4UdO3asdhvS1MS1a9W+cAICAup8XmJioujz5cuXG/yvy5cvFws3MjLClStXasVfsmQJExDgi72j9sGDBwgMDIS1tTW4XC4AYNWqVZg3bx4GDBiA/fv3w8DAQLR7/nf5ahpn8uVZKKgUYMp87lcr8yfTeuFg1+3Y1aU5pBS10aHnSLTv3g8X5FVxIacU9tMs8XrJ/4A5Qi9bCrpeGDoUAFXiwNRx0G4g7fj/7YGqyWQAgMlESyTOPg0sEx8Uorb2Q/tf+yA3bp4oLDd6LUw8jmPptoUY7zcKC2eVwWG48J6uxyIkZI7DvRu3cPvaWbRpsRpPMqNhXU/4x4Z/DkcG0orW8Pf3rw7094eycROgFJCQ0qihrCiAqLRR+RTb0ReUopIg8upYyauERNXklyNRtwOr0GUe6HreEAGLRqDv2NkYI+GOZQCk5E3F8/pBCOUMIKNKqOAtQiGfoFzl4fhJYTkmq4p7+pRRda83noaDHorXhgEQdqTlBY8hJW8CHRkJGFmrIftxNuAjNKkriH4Lea1RnyV3/NIEOBm7wnNhAPqN6Q7TFTNhaDiySiHww1jl4VidkIcH++KwNs4KAAfSHGDstBloITKL94e0QvWCiHpVmTaUtiRHqCCI+pzSDxPb+tNX0TGDv3/tAUzJQKV68JKUhKSkuLl+o+SDI4vN1+PwY3gQbt29i//N8sHWm5dwc2YDclkPElISEJRXT9QF5el4k6IAPYWPy9V/FVnVDugnH4n9wUugaDMTOq4KSB+9C5EIhfOStQBHBpIglAiq2wq/mC+mAH2QO2llaQjKxBfvivj0QdAwzc0cLxx+Rsy7aWgiXf375l23oHi4J1KzdLA13wkFrdVRklx//SfVU88mRipsEvEJuKvK4nF+ObqoCxcuU8PzoDVJ/CiFoDILS5bvwtKli8CBBLRN3LCopwHGHUpAkd4hpCj6YZpHCwBAE24/LNCdgE0vsmH1eyRcL/4JbRkJAMoYvmIcfrTeCqA7KqRloFHDU72JpzaKThZBWpELCRk99GumWDXeqGBYUwWcSiwEqhTbXxY+wawXbb7L8m5obKnJ2xObcVlnMCa5a0NSXg1dpmxD4awWSCkPhDJJQVmxesqlZtEOlSXnq8Y1O2w9exdbq+4daasLvRm6SNmfhMLIOTAxmQNQJUBlMDExwU/BsfBvplRvfrmKMujuXX38y8VPH4uOZbCGw2AwGg99GcXWzc2t3tc13bx582vk++twyUufJKRUqELw9bbK4450JxXDEZRSwxwoO3wvycjqUUoZnypL31IbFVkasOkKiWIISunIYm+SluDQrDd5tUyRK0ty6Mm5DdRCXpZWPReaPfDLUslZRZZGbKo2RSoveEneGvLU60icmCly1I62ZNi9Ot6tX+yoKfc8EREFuFrTjOAMkcOIbhrytC6poN7wj5ki88vfk768Ep2KEzoQqix9Sx2MDehBfhnxMg6RglY/MeceLZU0RGajDeXzr6bIP1xNqiqbeHJSlqEFMbki51EfKM46JTIZ3dNKg3o8qjKL5RfTPDN18jqbQGUFj6lfv361rrGzhea0o3SVaMqtFKGp8KttJKPsLJKfR/fuUkyVI5L64pUXhZGKrDo9rTLRCwloS808AqucfYwnJb1xIsdOq+2aUJcjcR81Ra75XJE5dMoWktfsIfqe/mipmCnYq91upOfdh9TNlorClrXWoH4HI0R1EeBtSWPupIpMQkOqzPMaSjtoQmsyG3de5OylXxMFkSlyfelnPF1aZ5kvfZohenZdzqMaIx8F71aRucNUkVOplDs9SN10W4NyyS9LoTt37tRyJpV8YzCpGP5AxVXy+XABl1qNvC9milyfXBERpT99QM+qHGn99Rl11eG/hUeTLUjDVp18/hSaZHdUlyd7ZXmRGfDU5so04HisSGZGNlOiITeTa8ldceZJkpbVoyc5QhP1rLAAAkD70nmU/mg8qRrOoqKiIrHrQxXutG9KbUe2JPuloVXHKeqv//rqmfFp3B1tRuaTLonK10pRlo5XmbxWtwU+WSpK09YPToMEFTS3lTr5HI6jkuxzJC2rQ09zhX1lZVkyuajI0srEfDrhpU+W06udR73Y1Yk0zFcREZ8clGVoXUim6GjKIlst6nZMaNreU0ue1leNmRXFr8lSUVo0rvLSfyNZVffvtrwbGltq9jex//OiJvarRW0j49kvJKNkR3wiOuymS84rHorSDFrhQvpeh4iIaJu1NvU9LezLSnMekp68Et3KKxXLw6eYIl/sY0QuK4JE9f6LXRPy3BvNGg6DwWicKbKEJvkqj2rU9TFT5P9vvppiO1ZHkeS1en/d3POLafUwV5JTMyRPny7k6epAykrNaf6xWFGU/Niz5G2mQcp6ptTB25PMDbVp2IqTdHNgSzHFVkJKiqSkpEhSUpb0LVxpzfGXYo/KfXmEXAyUqImxNXl36kDNFGSp/ZhNIsXrg2LLSz9JBkpa1Hv4aPLzdqa+U/1IWtGaAp5nUsKJKaQi34y69RlIPs4G1MpnHpUKqN7wjym2REQxh+aRtlIL8undj5xbqFH3eULlo7ZiS3TAviltSSmsmmjUn8+aiq2UXAsabG9Mnt16kI2uArUbv0OkyNan2MYfGUfKWlwaMWY4dWrTliYONCINGz96Vtjw2aqMJxupmbIR9RnanyzUtWnt9RTRPV0ZSRrzOuej8e6v7EMqhi40tH9naqrnQfeyqs8YL/A1IiO3ntS3kzU1954r5m24PsW25nNFSwQVueRnokpOPYfQsL6+5D5oHmnKSNPEJcKJfVnBI5LmcKj/lSTRbwoSTpCLjiq16eRHXdoZkbGnv8iLdE0Fo6G0K4pfUR9LDXL28SMvR1ua0bsFtdkc+dH0G6I+xbYx8iGoyKHB1lqk79SRBvTrTgZqLWhNUHqDclmUup0AUFo5/y+HMMpoSX8b0rJpT317dCA1fRcKyisTU2wbkqsDrTREbeWvz6irDv8t5MUvIAB0ukrOL3UxIOVm/qL7ORG/EVdTjTy69SVXCw3iDlwqWtypKXdEROcX9SY5NTPq2tObTFt2o6l6Qq/IT2dxSaqqf6x5ffB4m3JnMHE40nS7xkS8vvqvr54Zn6hoFYZQB31V8uo9lDpYqlOPJRer+/kabeH1oRmkpqBPnXv1ofa2Tcmiy3SRZ+xLSwaTmroZde/dk7i6ytRlmtDLaUnWbepsrk4W7p2pRydnUjVwpeNvhGNP4tkFpKekQ5179SY3ay1yGLhYtFiafn8N6So2Je8ePclGW4X6LThTvVC2sy3puZ36rsu8vrGlZn/DL0+jIfZNSN/Ji3p370RNVVvSkqpz7Lz0y+TSTJkcvXtS1w5c0rT2owdVXqlzo/aQobIm+fbsTq2bqNOozbU9T3+KYluW/4jcmimRvVd38rTTIbOu/pTzNXcZGAzGv06x9VEc0ajrW1NsOUT03R9SK0qNRmhMMgSyGrBy4kJTuvZZo+x3r/AypRzW9lZQl5UEVZaiQlIOMpxPsTcvRdTz50gtqESzVnYwb163+VxFQQKehqRA19wextoKCA+6DSnLdrBQl0VJWixCYpIh37Ql7C0MRL+pL7wxlOcmIzQ8HkoGXFga1f+O2vePfoD3qTEI3+D00XwCQHnhIyjrTUVp/n1EhzxHuZopbE10GpWn/DcRCEsth4UjF1pSBbh3Nxw2HdpDXarhAucXZyLkeTya2TpAT0X6s+Lx3schNKEEXCcrKEmKPy/19XO8K9eCs1WLRr3EOT9hDtZyFmGVoXhdE78QIY+fQ0rbDLYmusiOfIRofmu42tb/rjVBRR6iQsNRItcMTjYt67eiqCdtC0TitaYZ5JNioW7uhBeDTLFz7kNc8mr2Sek3lo/JhzCzFYgMDUZmsSRa2dujWQ3PnvXJZcjCNjBf9gTydVRAyuswxOUA1o620KhDVj5Xrv7L8MuyERkaCY6WKWxM9BqMm5ccg4jkMtg6WkPlb5ZpQ/3Sx+qZ0ZjxqATRIc9BOlYw16/f421Zdjyehr+Dqr4pbEz1xe6VpL9BaHQqtFpao1VzVbGxLiY8DJmVyrC1sxCZsguPS+QgIiQSUjqtYGUsPh5UFmfiRUgMlI1s0KrZv8+8vKGxpUaLQ0zwE6SVy8GcawPtGn0iBKWIjwxFJmnBzsYUspya/e07BIelwMjeCTpKf/+UGAlK8DrsOXgKBrBvbcDaC4PBaDSqkppwkevWqLjZ5lEIDg7+ZvL+r1BsGY0d6cox2rozNoTdaPBVR6KJaZViW1b4/D9bZNuWrcLoBfMbmMT8/5F0eQK4cwUI3DoSlWmPMOWHvfgzNQp235EzlgpeGDbuzMS8Od6sPTIYDAaDwWB8a4qthCZcZLs2TrG1fMUUW8Y/R8Gbw7gg2QNDWih/NC6/NB5T5xzGrm2LWMF9Izw/txtHb4WDr9AcfuOnor2xMisUBoPBYDAYDMaXU2xlOjdOsbWKYYotg8FgMBgMBoPBYDC+LVQkNOEi5duouDk2r78pxVaKVR+DwWAwGAwGg8FgMEAEkOC7zDpTbBkMBoPBYDAYDAaDIdRt+Xym2DIYDAaDwWAwGAwG4/ukna8zsrISGhVXS0vrm8o7O2PLYDAYDAaDwWAwGIzvGglWBAwGg8FgMBgMBoPBYIotg8FgMBgMBoPBYDAYTLFlMBgMBoPBYDAYDAaDKbYMBoPBYDAYDAaDwfiP8X8Qs52MLijOjQAAAABJRU5ErkJggg==" />
#
# The confusion-matrices for all the classifiers would look roughly the same and just like in the non-normalized case, the validation-testset looks roughly the same and performs ~2-3% worse.
#
# However, this is to a 20% baseline, so on this limited subsample of data, it performs slightly better on the sub-sampled data, but now strongly over-predicts the -2 and +2 classes, as they are easier to separate.
#
# If we use the classifier on the unbalanced data (rerun cell for realoading the original data + rerun validation), it would still over-predict the rather small -2 and +2 classes. This would lead to a sub-26%-baseline result.
# + [markdown] id="X7kCjyi3ndvi"
# ### Summary Findings
# 1. the TF-IDF accuracies are significantly worse: Roughly 26% without preprocessing vs 27-35% with preprocessing.
# 2. using raw word2vec without PCA yields 42% instead of 40% (with vectors of dimensionality 300 vs 20 PCA) - while increasing computation times 5-fold (as there is more data)
# 3. rerunning the notebook with enabling the balancing-cell would not lead to slightly better performing classes - the over-prediction of the bigger classes is a natural consequence of the data.
#
# But the principle problems of classifiers over-predicting into the classes -1 and 1 gets even worse here, our tf-idf only predicted the majority-class -1.
| 296.512035
| 114,382
|
5791e36982fdc83a3a8ae0caade1b8b44e8ee94f
|
py
|
python
|
lecture-07/lab.ipynb
|
fralvarezz/modern-ai-course
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="zaAJl_PZBcmB"
# # Convolutional Neural Networks
# A CNN is made up of basic building blocks defined as tensor, neurons, layers and kernel weights and biases. In this lab, we use PyTorch to build a image classifier using CNN. The objective is to learn CNN using PyTorch framework.
# Please refer to the link below for know more about CNN
# https://poloclub.github.io/cnn-explainer/
#
#
# ## Import necessary libraries
# + id="V9zNOw6E-mVI"
import torch
import torchvision
import numpy as np
import matplotlib.pyplot as plt
import torch.nn as nn
import torch.nn.functional as F
from torchvision.datasets import MNIST
from torchvision.transforms import ToTensor
from torchvision import datasets, transforms
from torchvision.utils import make_grid
from torch.utils.data.dataloader import DataLoader
from torch.optim.lr_scheduler import StepLR
from torch.utils.data import random_split
from torch.utils.data.sampler import SubsetRandomSampler
# %matplotlib inline
# + [markdown] id="DwZhpV-DBoUu"
# ### Download the MNIST dataset.
# + colab={"base_uri": "https://localhost:8080/", "height": 467, "referenced_widgets": ["4d539869566942e2b311340b077500da", "8569cfe202be4a68850a5d2715f6b3fe", "1eb92d6ae26542bb987032527a61a341", "cb65b2f30d37451fa9c865140bca7e5e", "aed4fb085a9448508f091fd915eccb67", "158d7389801f4f17acd8738cc150e531", "be89f810f0f34391aec551f30f5ff364", "936bc613bcda4f8ea6b5463bc29de3cd", "65c427dd44db40078bd996d852d822b3", "81f7755f2d7d49f18119b9f5131f2064", "f0470fb9d1b74f219b05590d4b39e805", "623d23ec70034257a7cf75ebb7149b8c", "4af10895ee134857ace052045e244048", "f3ec755299c44c35bb264759673b6611", "88ed1c56f660444ca1bf334be8d1620c", "d1d277a9880d4023882d97591256be66", "289939067756489b891cb32bc6788387", "75631933f0954a258f87365c55cd4df1", "e53cc84bbb214640a5bace4b7a00a4b4", "c5908dcd7aa9494cb742ac0b3008ac2f", "abcb01d7d8264aea8ff33e09a4d92c59", "c16a408413b34db58858abb8ef2e74e3", "59cc2bf3d23f4c5e8d46b4521b056b89", "fd2cab6bea2d4a4a940778dece28e9a7", "84b3bceeb2bd4fa284d903a1b8daa72f", "393143f52a70418a8c4a89f9cc2d2205", "41e7de1ecc784c788143eb872869066a", "c083ea4cdbe84932bcae03bd87c0789c", "28ab9376d5584afda7059d84f639b393", "2ae9c1e7a3e844ee8382e50a71bcc701", "210e707a89384a13829476b1e93f8c14", "37f3c5c6112842efbe69a32eb34b6e0f", "685bb4e5ceed4b779c1b8aa77f3fc21f", "979b2ffaf3364c158ac6b43ffaff2d10", "c5dc3c830d26426786c5aaab319da648", "89c2164b7e0b4bbc9d1e04046d57f177", "f13c8ad55b57400cac8f0e57966eb879", "257f98c5521849c199909caf9eac165a", "421357b4e2be45c088c6d3861c2275d5", "850c9587acc74cc1abe9645f204a6dd5", "91e1331c08f04228b70ceed587ed07e1", "5daefca82efa40f885f3d57088f3cfdf", "0875bc90380945d5b4f17fcf5e3386b3", "9ea010d0c2244d648446cdaf42d45852"]} id="bGflK2dO7UoS" outputId="809442e4-d03b-45b9-e536-725fed0d9130"
# choose the training and test datasets
train_data = datasets.MNIST(root='data', train=True,download=True, transform=ToTensor())
test_data = datasets.MNIST(root='data', train=False,download=True, transform=ToTensor())
# + [markdown] id="9z2X_j6h9P7H"
# ##Chopping training datasets in train set and validation set. This is done to avoid overfitting on the test set.
#
# Use simple algorithm to create validation set:
# First, create a list of indices of the training data. Then randomly shuffle those indices. Lastly,split the indices in 80-20.
#
#
# + id="4CKFaoARkF5n"
indices = np.arange(len(train_data))
np.random.shuffle(indices)
train_indices = indices[:int(len(indices)*0.8)]
test_indices = indices[len(train_indices):]
# + [markdown] id="nPbg9anSy_wW"
# ## Print data size
# + colab={"base_uri": "https://localhost:8080/"} id="6un0Yv1wyVku" outputId="803dfafd-8c7f-4089-9350-99beef3c2b7d"
print(train_data)
print(test_data)
# + [markdown] id="DnFpmxo70TgG"
# ## Data Visualization
# + colab={"base_uri": "https://localhost:8080/", "height": 482} id="sMEdkcEle0mn" outputId="876f818f-834e-49fc-8b0e-fd2e19ea51ef"
figure = plt.figure(figsize=(10, 8))
cols, rows = 5, 5
for i in range(1, cols * rows + 1):
sample_idx = torch.randint(len(train_data), size=(1,)).item()
img, label = train_data[sample_idx]
figure.add_subplot(rows, cols, i)
plt.title(label)
plt.axis("off")
plt.imshow(img.squeeze(), cmap="gray")
plt.show()
# + [markdown] id="-fgoof86ByQ_"
# ## Data preparation for training with PyTorch DataLoaders
# + colab={"base_uri": "https://localhost:8080/"} id="NM-r0rCtB3QG" outputId="8b07728d-4e68-45c3-f89f-440755586134"
# Obtaining training and validation batches
train_batch = SubsetRandomSampler(train_indices)
val_batch = SubsetRandomSampler(test_indices)
# Samples per batch to load
batch_size = 256
# Training Set
train_loader = torch.utils.data.DataLoader(dataset=train_data, batch_size=batch_size,sampler=train_batch,num_workers=4,pin_memory=True)
# Validation Set
val_loader = torch.utils.data.DataLoader(dataset=train_data,batch_size=batch_size, sampler=val_batch, num_workers=4,pin_memory=True)
# Test Set
test_loader = torch.utils.data.DataLoader(dataset=test_data,batch_size=batch_size,num_workers=4,pin_memory=True)
# + [markdown] id="Le1Q_knpOodf"
# ## Data normalization step: Calculate Mean and Std
# + colab={"base_uri": "https://localhost:8080/"} id="C1dN24PjOnb6" outputId="57a03c21-abda-4f04-a23e-db8ad732c382"
train_mean = 0.
train_std = 0.
for images, _ in train_loader:
batch_samples = images.size(0) # batch size (the last batch can have smaller size!)
images = images.view(batch_samples, images.size(1), -1)
train_mean += images.mean(2).sum(0)
train_std += images.std(2).sum(0)
train_mean /= len(train_loader.dataset)
train_std /= len(train_loader.dataset)
print('Mean: ', train_mean)
print('Std: ', train_std)
# + [markdown] id="oHNipERCMlJa"
# ## Data Augmentation:
# It is usually done to increase the performance of the CNN based classifiers. Consider this is preprocess of the data. PyTorch inculdes lots of pre-built data augumentation and data transformation features such as Below are the list of transformations that come pre-built with PyTorch: ToTensor, Normalize, Scale, RandomCrop, LinearTransformation, RandomGrayscale, etc. Try to use atleaset one the data.
# + id="po1v_9XqN__V"
# Your code
# Check the data and see weither suggested augumenation is done. Also check for normalization transformation.
# + [markdown] id="2T8jj4KCD-VJ"
# ## Evaluation Metrics
# prediction acuracy
# + id="hu3hdE5xDIuf"
def accuracy(outputs, labels):
_, preds = torch.max(outputs, dim=1)
return torch.tensor(torch.sum(preds == labels).item() / len(preds))
## Use different form of evaluation meterics
# + [markdown] id="gEOvCUn5DPrA"
# ## Loss Function: Cross Entropy
# For each output row, pick the predicted probability for the correct label. E.g. if the predicted probabilities for an image are [0.1, 0.3, 0.2, ...] and the correct label is 1, we pick the corresponding element 0.3 and ignore the rest.
#
# Then, take the logarithm of the picked probability. If the probability is high i.e. close to 1, then its logarithm is a very small negative value, close to 0. And if the probability is low (close to 0), then the logarithm is a very large negative value. We also multiply the result by -1, which results is a large postive value of the loss for poor predictions.
#
# Finally, take the average of the cross entropy across all the output rows to get the overall loss for a batch of data.
#
# + id="DvZ205h7CWs7"
class MnistModelBase(nn.Module):
def training_step(self, batch):
pass
# your code
def validation_step(self, batch):
pass
# your code
def validation_epoch_end(self, outputs):
pass
#your code
def epoch_end(self, epoch, result,LR):
pass
#your code
# + [markdown] id="MHcZeg50CjRe"
# ## Convolutional Neural Network model
# we will use a convolutional neural network, using the nn.Conv2d class from PyTorch. The activation function we'll use here is called a Rectified Linear Unit or ReLU, and it has a really simple formula: relu(x) = max(0,x) i.e. if an element is negative, we replace it by 0, otherwise we leave it unchanged. To define the model, we extend the nn.Module class
# + id="LksbwKNXD9oQ"
class MnistModel(MnistModelBase):
"""Feedfoward neural network with 2 hidden layer"""
def __init__(self):
super().__init__()
self.conv1 = nn.Sequential(
nn.Conv2d(in_channels = 1, out_channels = 16, kernel_size=3), #RF - 3x3 # 26x26
nn.ReLU(),
nn.BatchNorm2d(16),
nn.Dropout2d(0.1),
nn.Conv2d(16, 16, 3), #RF - 5x5 # 24x24
nn.ReLU(),
nn.BatchNorm2d(16),
nn.Dropout2d(0.1),
nn.Conv2d(16, 32, 3), #RF - 7x7 # 22x22
nn.ReLU(),
nn.BatchNorm2d(32),
nn.Dropout2d(0.1),
)
# translation layer
# input - 22x22x64; output - 11x11x32
self.trans1 = nn.Sequential(
# RF - 7x7
nn.Conv2d(32, 20, 1), # 22x22
nn.ReLU(),
nn.BatchNorm2d(20),
# RF - 14x14
nn.MaxPool2d(2, 2), # 11x11
)
self.conv2 = nn.Sequential(
nn.Conv2d(20,20,3,padding=1), #RF - 16x16 #output- 9x9
nn.ReLU(),
nn.BatchNorm2d(20),
nn.Dropout2d(0.1),
nn.Conv2d(20,16,3), #RF - 16x16 #output- 9x9
nn.ReLU(),
nn.BatchNorm2d(16),
nn.Dropout2d(0.1),
nn.Conv2d(16, 16, 3), #RF - 18x18 #output- 7x7
nn.ReLU(),
nn.BatchNorm2d(16),
nn.Dropout2d(0.1),
)
self.conv3 = nn.Sequential(
nn.Conv2d(16,16,3), #RF - 20x20 #output- 5x5
nn.ReLU(),
nn.BatchNorm2d(16),
nn.Dropout2d(0.1),
#nn.Conv2d(16,10,1), #RF - 20x20 #output- 7x7
)
# GAP Layer
self.avg_pool = nn.Sequential(
# # RF - 22x22
nn.AvgPool2d(5)
) ## output_size=1
self.conv4 = nn.Sequential(
nn.Conv2d(16,10,1), #RF - 20x20 #output- 7x7
)
def forward(self, xb):
x = self.conv1(xb)
x = self.trans1(x)
x = self.conv2(x)
x = self.conv3(x)
x = self.avg_pool(x)
x = self.conv4(x)
x = x.view(-1, 10)
return x
# + [markdown] id="fY3SuML1FI8b"
# ### Using a GPU
#
# + id="tVvVT7oMFE2i"
#function to ensure that our code uses the GPU if available, and defaults to using the CPU if it isn't.
def get_default_device():
"""Pick GPU if available, else CPU"""
if torch.cuda.is_available():
return torch.device('cuda')
else:
return torch.device('cpu')
# a function that can move data and model to a chosen device.
def to_device(data, device):
"""Move tensor(s) to chosen device"""
if isinstance(data, (list,tuple)):
return [to_device(x, device) for x in data]
return data.to(device, non_blocking=True)
#Finally, we define a DeviceDataLoader class to wrap our existing data loaders and move data to the selected device,
#as a batches are accessed. Interestingly, we don't need to extend an existing class to create a PyTorch dataloader.
#All we need is an __iter__ method to retrieve batches of data, and an __len__ method to get the number of batches.
class DeviceDataLoader():
"""Wrap a dataloader to move data to a device"""
def __init__(self, dl, device):
self.dl = dl
self.device = device
def __iter__(self):
"""Yield a batch of data after moving it to device"""
for b in self.dl:
yield to_device(b, self.device)
def __len__(self):
"""Number of batches"""
return len(self.dl)
# + [markdown] id="8_I-NkpjHKoq"
# We can now wrap our data loaders using DeviceDataLoader.
# + id="LRCONOQTG7gD"
device = get_default_device()
train_loader = DeviceDataLoader(train_loader, device)
val_loader = DeviceDataLoader(val_loader, device)
test_loader = DeviceDataLoader(test_loader, device)
# + [markdown] id="NsUaPke8HV96"
# ### Model Training
#
#
#
# + id="2c0xgvskHM2s"
from torch.optim.lr_scheduler import OneCycleLR
@torch.no_grad()
def evaluate(model, val_loader):
model.eval()
outputs = [model.validation_step(batch) for batch in val_loader]
return model.validation_epoch_end(outputs)
def fit(epochs, lr, model, train_loader, val_loader, opt_func=torch.optim.SGD):
torch.cuda.empty_cache()
history = []
optimizer = opt_func(model.parameters(), lr)
scheduler = OneCycleLR(optimizer, lr, epochs=epochs,steps_per_epoch=len(train_loader))
for epoch in range(epochs):
# Training Phase
model.train()
train_losses = []
for batch in train_loader:
loss = model.training_step(batch)
train_losses.append(loss)
loss.backward()
optimizer.step()
optimizer.zero_grad()
scheduler.step()
# Validation phase
result = evaluate(model, val_loader)
result['train_loss'] = torch.stack(train_losses).mean().item()
model.epoch_end(epoch, result,scheduler.get_lr())
history.append(result)
return history
# + [markdown] id="M6S-2843Hmhy"
# Before we train the model, we need to ensure that the data and the model's parameters (weights and biases) are on the same device (CPU or GPU). We can reuse the to_device function to move the model's parameters to the right device.
# + colab={"base_uri": "https://localhost:8080/"} id="DQLS_8CBHqml" outputId="30c07f9c-e4be-4a2d-ad2e-cc23ebac5a67"
# Model (on GPU)
model = MnistModel()
to_device(model, device)
# + [markdown] id="XQypas1CIOcU"
# Print Summary of the model
# + colab={"base_uri": "https://localhost:8080/"} id="S2Kh76K3HsCQ" outputId="2f4c5a9c-57bb-4506-8929-9d913aa938e2"
from torchsummary import summary
# print the summary of the model
summary(model, input_size=(1, 28, 28), batch_size=-1)
# + [markdown] id="ImcS-ZRTIhMR"
# Let's see how the model performs on the validation set with the initial set of weights and biases.
# + colab={"base_uri": "https://localhost:8080/"} id="3HPI7rYUIXBG" outputId="8a8713b3-8e94-487a-a3d0-523ade1a1406"
history = [evaluate(model, val_loader)]
history
# + [markdown] id="NurAbj8GIq1d"
# The initial accuracy is around 10%, which is what one might expect from a randomly intialized model (since it has a 1 in 10 chance of getting a label right by guessing randomly).
#
# We are now ready to train the model. Let's train for 5 epochs and look at the results. We can use a relatively higher learning of 0.01.
# + colab={"base_uri": "https://localhost:8080/"} id="09oYD6v6Ii-F" outputId="8386d532-312a-4285-f6d6-5359bd3b2462"
history += fit(10, 0.01, model, train_loader, val_loader)
# + [markdown] id="HqsXzHmNJMPj"
# ## Plot Metrics
# + id="T6ca6rzNI-KW"
def plot_scores(history):
# scores = [x['val_score'] for x in history]
acc = [x['val_acc'] for x in history]
plt.plot(acc, '-x')
plt.xlabel('epoch')
plt.ylabel('acc')
plt.title('acc vs. No. of epochs');
# + id="Yo8WpVtuJd0B"
def plot_losses(history):
train_losses = [x.get('train_loss') for x in history]
val_losses = [x['val_loss'] for x in history]
plt.plot(train_losses, '-bx')
plt.plot(val_losses, '-rx')
plt.xlabel('epoch')
plt.ylabel('loss')
plt.legend(['Training', 'Validation'])
plt.title('Loss vs. No. of epochs');
# + colab={"base_uri": "https://localhost:8080/", "height": 295} id="4uZ48YR2JepG" outputId="4bbeae3c-dcbd-406c-a298-df022be164ed"
plot_losses(history)
# + colab={"base_uri": "https://localhost:8080/", "height": 295} id="Ay2lZl22Jgfw" outputId="0f2a33be-0e6e-47f0-9d49-32f7bed04bab"
plot_scores(history)
# + id="xSSscNJqT2hA"
def get_misclassified(model, test_loader):
misclassified = []
misclassified_pred = []
misclassified_target = []
# put the model to evaluation mode
model.eval()
# turn off gradients
with torch.no_grad():
for data, target in test_loader:
# do inferencing
output = model(data)
# get the predicted output
pred = output.argmax(dim=1, keepdim=True)
# get the current misclassified in this batch
list_misclassified = (pred.eq(target.view_as(pred)) == False)
batch_misclassified = data[list_misclassified]
batch_mis_pred = pred[list_misclassified]
batch_mis_target = target.view_as(pred)[list_misclassified]
# batch_misclassified =
misclassified.append(batch_misclassified)
misclassified_pred.append(batch_mis_pred)
misclassified_target.append(batch_mis_target)
# group all the batches together
misclassified = torch.cat(misclassified)
misclassified_pred = torch.cat(misclassified_pred)
misclassified_target = torch.cat(misclassified_target)
return list(map(lambda x, y, z: (x, y, z), misclassified, misclassified_pred, misclassified_target))
# + colab={"base_uri": "https://localhost:8080/"} id="iqY-D72KWCNK" outputId="3d19aace-39a1-4e90-8943-d42cefb3bc8b"
misclassified = get_misclassified(model, test_loader)
# + colab={"base_uri": "https://localhost:8080/", "height": 873} id="smqb4GwBWN5P" outputId="3edc17c9-5c3b-4939-be61-c7c4b485eb02"
import random
num_images = 25
fig = plt.figure(figsize=(12, 12))
for idx, (image, pred, target) in enumerate(random.choices(misclassified, k=num_images)):
image, pred, target = image.cpu().numpy(), pred.cpu(), target.cpu()
ax = fig.add_subplot(5, 5, idx+1)
ax.axis('off')
ax.set_title('target {}\npred {}'.format(target.item(), pred.item()), fontsize=12)
ax.imshow(image.squeeze())
plt.tight_layout()
plt.show()
| 37.529661
| 1,738
|
5cce858d8a30025c7adcd90c8fa001dfc6c07adb
|
py
|
python
|
intro-to-pytorch/Part 3 - Training Neural Networks (Exercises).ipynb
|
faber6911/deep-learning-v2-pytorch
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + id="4ETIAZvCOrDm" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 122} outputId="78370618-d82b-4de9-8da6-614df8237a65" executionInfo={"status": "ok", "timestamp": 1586121924311, "user_tz": -120, "elapsed": 31757, "user": {"displayName": "Fabrizio D'Intinosante", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhrjCQVLGrXDlJRniViKxzD_05EWfYGhPN_t37-=s64", "userId": "05737257329450160630"}}
from google.colab import drive
ROOT = "/content/drive"
drive.mount(ROOT)
# + id="BGgRaEGrOzdA" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="2c9a9362-0e88-47fe-bda7-86875af4693a" executionInfo={"status": "ok", "timestamp": 1586121950443, "user_tz": -120, "elapsed": 2452, "user": {"displayName": "Fabrizio D'Intinosante", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhrjCQVLGrXDlJRniViKxzD_05EWfYGhPN_t37-=s64", "userId": "05737257329450160630"}}
# %cd "/content/drive/My Drive/Learning/deep-learning-v2-pytorch/intro-to-pytorch"
# + [markdown] id="T90fBnVvOo4u" colab_type="text"
# # Training Neural Networks
#
# The network we built in the previous part isn't so smart, it doesn't know anything about our handwritten digits. Neural networks with non-linear activations work like universal function approximators. There is some function that maps your input to the output. For example, images of handwritten digits to class probabilities. The power of neural networks is that we can train them to approximate this function, and basically any function given enough data and compute time.
#
# <img src="/content/drive/My Drive/Learning/deep-learning-v2-pytorch/intro-to-pytorch/assets/function_approx.png">
#
# At first the network is naive, it doesn't know the function mapping the inputs to the outputs. We train the network by showing it examples of real data, then adjusting the network parameters such that it approximates this function.
#
# To find these parameters, we need to know how poorly the network is predicting the real outputs. For this we calculate a **loss function** (also called the cost), a measure of our prediction error. For example, the mean squared loss is often used in regression and binary classification problems
#
# $$
# \large \ell = \frac{1}{2n}\sum_i^n{\left(y_i - \hat{y}_i\right)^2}
# $$
#
# where $n$ is the number of training examples, $y_i$ are the true labels, and $\hat{y}_i$ are the predicted labels.
#
# By minimizing this loss with respect to the network parameters, we can find configurations where the loss is at a minimum and the network is able to predict the correct labels with high accuracy. We find this minimum using a process called **gradient descent**. The gradient is the slope of the loss function and points in the direction of fastest change. To get to the minimum in the least amount of time, we then want to follow the gradient (downwards). You can think of this like descending a mountain by following the steepest slope to the base.
#
# <img src='assets/gradient_descent.png' width=350px>
# + [markdown] id="EopBUo7IOo4z" colab_type="text"
# ## Backpropagation
#
# For single layer networks, gradient descent is straightforward to implement. However, it's more complicated for deeper, multilayer neural networks like the one we've built. Complicated enough that it took about 30 years before researchers figured out how to train multilayer networks.
#
# Training multilayer networks is done through **backpropagation** which is really just an application of the chain rule from calculus. It's easiest to understand if we convert a two layer network into a graph representation.
#
# <img src='assets/backprop_diagram.png' width=550px>
#
# In the forward pass through the network, our data and operations go from bottom to top here. We pass the input $x$ through a linear transformation $L_1$ with weights $W_1$ and biases $b_1$. The output then goes through the sigmoid operation $S$ and another linear transformation $L_2$. Finally we calculate the loss $\ell$. We use the loss as a measure of how bad the network's predictions are. The goal then is to adjust the weights and biases to minimize the loss.
#
# To train the weights with gradient descent, we propagate the gradient of the loss backwards through the network. Each operation has some gradient between the inputs and outputs. As we send the gradients backwards, we multiply the incoming gradient with the gradient for the operation. Mathematically, this is really just calculating the gradient of the loss with respect to the weights using the chain rule.
#
# $$
# \large \frac{\partial \ell}{\partial W_1} = \frac{\partial L_1}{\partial W_1} \frac{\partial S}{\partial L_1} \frac{\partial L_2}{\partial S} \frac{\partial \ell}{\partial L_2}
# $$
#
# **Note:** I'm glossing over a few details here that require some knowledge of vector calculus, but they aren't necessary to understand what's going on.
#
# We update our weights using this gradient with some learning rate $\alpha$.
#
# $$
# \large W^\prime_1 = W_1 - \alpha \frac{\partial \ell}{\partial W_1}
# $$
#
# The learning rate $\alpha$ is set such that the weight update steps are small enough that the iterative method settles in a minimum.
# + [markdown] id="g96Uc2n4Oo41" colab_type="text"
# ## Losses in PyTorch
#
# Let's start by seeing how we calculate the loss with PyTorch. Through the `nn` module, PyTorch provides losses such as the cross-entropy loss (`nn.CrossEntropyLoss`). You'll usually see the loss assigned to `criterion`. As noted in the last part, with a classification problem such as MNIST, we're using the softmax function to predict class probabilities. With a softmax output, you want to use cross-entropy as the loss. To actually calculate the loss, you first define the criterion then pass in the output of your network and the correct labels.
#
# Something really important to note here. Looking at [the documentation for `nn.CrossEntropyLoss`](https://pytorch.org/docs/stable/nn.html#torch.nn.CrossEntropyLoss),
#
# > This criterion combines `nn.LogSoftmax()` and `nn.NLLLoss()` in one single class.
# >
# > The input is expected to contain scores for each class.
#
# This means we need to pass in the raw output of our network into the loss, not the output of the softmax function. This raw output is usually called the *logits* or *scores*. We use the logits because softmax gives you probabilities which will often be very close to zero or one but floating-point numbers can't accurately represent values near zero or one ([read more here](https://docs.python.org/3/tutorial/floatingpoint.html)). It's usually best to avoid doing calculations with probabilities, typically we use log-probabilities.
# + id="KWpxHvVSOo42" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 315, "referenced_widgets": ["7cb7e532e4c540099afeab27ab40463b", "ff420d47f31c4af48a270aa602eb1df5", "9b918a73a3e94f8d96664422f1c94d50", "e2410ca888ad483bbe642aea953cd57f", "2cddc5f5fe614d2d8b3016c1aa1e87d9", "59ceaf4f44a0475f88b0ce515dd703dc", "aec9f32b4e7c461086ff8219b915a2d3", "e80c7c349aa8473c8c980960d5a9eea3", "28448276506042a68600ca7c78b433a5", "bf524fa48ba843409af77519d43be2cb", "c96aa36e1d174b97b3e422c6a7bea2a5", "196519f153cc42c7a9a7abae1e557d0f", "1bb8a0406ca14153930e055cac021343", "1836e3467f304e4c9104a13911d82500", "51be64ab0d4748dca41b195ebdfa977c", "fbdb62a0354e4e93b561b5de5fe351b0", "5388137ff87348adb0174f5fa0e0f028", "d86273ed922b4434a2da315f95dedba9", "2dc192fd9f8249fd82a4f2546ca1b74e", "e1a4fb887b574792875ed82c8190a69d", "f02abb75bb3542c8ac933e3703f20eed", "c7a28534abcc4409b79b427b1d2d7c49", "5e0a2f1602dd44d095515c18371e0237", "ca579be6ec7245ecb21897e7c65418fd", "e7fdc0cb77a748f1a5fe2f6000e0e832", "b4fc520a884a474c924f1b04ee211c95", "c39b3b7c91994d1abaf0e57eb297e634", "5c53196d242243cabeebff76e5302343", "a547b0c0ea7d44a1848a15e8ec992fa7", "75d42132fc5c48ca8b8a70adfddbbc86", "8c195a3d23d74772b2513723b41ea31c", "a80a1c3db5db454989535942ee3c4d9c"]} outputId="d46a6511-1d40-4ca5-8c84-37fdfaca9840" executionInfo={"status": "ok", "timestamp": 1586122847939, "user_tz": -120, "elapsed": 10531, "user": {"displayName": "Fabrizio D'Intinosante", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhrjCQVLGrXDlJRniViKxzD_05EWfYGhPN_t37-=s64", "userId": "05737257329450160630"}}
import torch
from torch import nn
import torch.nn.functional as F
from torchvision import datasets, transforms
# Define a transform to normalize the data
transform = transforms.Compose([transforms.ToTensor(),
transforms.Normalize((0.5,), (0.5,)),
])
# Download and load the training data
trainset = datasets.MNIST('~/.pytorch/MNIST_data/', download=True, train=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)
# + [markdown] id="s67taNnwOo4_" colab_type="text"
# ### Note
# If you haven't seen `nn.Sequential` yet, please finish the end of the Part 2 notebook.
# + id="WFx75DVXOo5A" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="cbd02c83-4d11-4e8d-c535-e75fb8ec9fac" executionInfo={"status": "ok", "timestamp": 1586122967944, "user_tz": -120, "elapsed": 1163, "user": {"displayName": "Fabrizio D'Intinosante", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhrjCQVLGrXDlJRniViKxzD_05EWfYGhPN_t37-=s64", "userId": "05737257329450160630"}}
# Build a feed-forward network
model = nn.Sequential(nn.Linear(784, 128),
nn.ReLU(),
nn.Linear(128, 64),
nn.ReLU(),
nn.Linear(64, 10))
# Define the loss
criterion = nn.CrossEntropyLoss()
# Get our data
images, labels = next(iter(trainloader))
# Flatten images
images = images.view(images.shape[0], -1)
# Forward pass, get our logits
logits = model(images)
# Calculate the loss with the logits and the labels
loss = criterion(logits, labels)
print(loss)
# + [markdown] id="RQYUluZuOo5I" colab_type="text"
# In my experience it's more convenient to build the model with a log-softmax output using `nn.LogSoftmax` or `F.log_softmax` ([documentation](https://pytorch.org/docs/stable/nn.html#torch.nn.LogSoftmax)). Then you can get the actual probabilities by taking the exponential `torch.exp(output)`. With a log-softmax output, you want to use the negative log likelihood loss, `nn.NLLLoss` ([documentation](https://pytorch.org/docs/stable/nn.html#torch.nn.NLLLoss)).
#
# >**Exercise:** Build a model that returns the log-softmax as the output and calculate the loss using the negative log likelihood loss. Note that for `nn.LogSoftmax` and `F.log_softmax` you'll need to set the `dim` keyword argument appropriately. `dim=0` calculates softmax across the rows, so each column sums to 1, while `dim=1` calculates across the columns so each row sums to 1. Think about what you want the output to be and choose `dim` appropriately.
# + id="B8Dn99ciTuUd" colab_type="code" colab={}
from collections import OrderedDict
# + id="cSFpeohIOo5J" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="ab95ef4d-f45c-480a-d74c-678626e422eb" executionInfo={"status": "ok", "timestamp": 1586124070266, "user_tz": -120, "elapsed": 1243, "user": {"displayName": "Fabrizio D'Intinosante", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhrjCQVLGrXDlJRniViKxzD_05EWfYGhPN_t37-=s64", "userId": "05737257329450160630"}}
# TODO: Build a feed-forward network
model = nn.Sequential(OrderedDict([
('fc1', nn.Linear(784, 128)),
('act1', nn.ReLU()),
('fc2', nn.Linear(128, 64)),
('act2', nn.ReLU()),
('fc3', nn.Linear(64, 10)),
('output', nn.LogSoftmax(dim = 1))
]))
# TODO: Define the loss
criterion = nn.NLLLoss()
### Run this to check your work
# Get our data
images, labels = next(iter(trainloader))
# Flatten images
images = images.view(images.shape[0], -1)
# Forward pass, get our logits
logits = model(images)
# Calculate the loss with the logits and the labels
loss = criterion(logits, labels)
print(loss)
# + [markdown] id="FLE3yIlNOo5R" colab_type="text"
# ## Autograd
#
# Now that we know how to calculate a loss, how do we use it to perform backpropagation? Torch provides a module, `autograd`, for automatically calculating the gradients of tensors. We can use it to calculate the gradients of all our parameters with respect to the loss. Autograd works by keeping track of operations performed on tensors, then going backwards through those operations, calculating gradients along the way. To make sure PyTorch keeps track of operations on a tensor and calculates the gradients, you need to set `requires_grad = True` on a tensor. You can do this at creation with the `requires_grad` keyword, or at any time with `x.requires_grad_(True)`.
#
# You can turn off gradients for a block of code with the `torch.no_grad()` content:
# ```python
# x = torch.zeros(1, requires_grad=True)
# >>> with torch.no_grad():
# ... y = x * 2
# >>> y.requires_grad
# False
# ```
#
# Also, you can turn on or off gradients altogether with `torch.set_grad_enabled(True|False)`.
#
# The gradients are computed with respect to some variable `z` with `z.backward()`. This does a backward pass through the operations that created `z`.
# + id="RH1xBb5fOo5T" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 51} outputId="af2ebe81-6274-40fa-c2bc-3a56932b7ac7" executionInfo={"status": "ok", "timestamp": 1586125055136, "user_tz": -120, "elapsed": 1373, "user": {"displayName": "Fabrizio D'Intinosante", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhrjCQVLGrXDlJRniViKxzD_05EWfYGhPN_t37-=s64", "userId": "05737257329450160630"}}
x = torch.randn(2,2, requires_grad=True)
print(x)
# + id="8vG7oA45Oo5a" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 51} outputId="8e4bd2a4-cc40-46e8-cf2d-ffe4a2283c00" executionInfo={"status": "ok", "timestamp": 1586125066421, "user_tz": -120, "elapsed": 843, "user": {"displayName": "Fabrizio D'Intinosante", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhrjCQVLGrXDlJRniViKxzD_05EWfYGhPN_t37-=s64", "userId": "05737257329450160630"}}
y = x**2
print(y)
# + [markdown] id="qqDT01srOo5h" colab_type="text"
# Below we can see the operation that created `y`, a power operation `PowBackward0`.
# + id="sNls0FTkOo5i" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="2241e6d0-26fa-4121-db9d-cb50052a90fd" executionInfo={"status": "ok", "timestamp": 1586125552882, "user_tz": -120, "elapsed": 1591, "user": {"displayName": "Fabrizio D'Intinosante", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhrjCQVLGrXDlJRniViKxzD_05EWfYGhPN_t37-=s64", "userId": "05737257329450160630"}}
## grad_fn shows the function that generated this variable
print(y.grad_fn)
# + [markdown] id="LNi2VCazOo5u" colab_type="text"
# The autograd module keeps track of these operations and knows how to calculate the gradient for each one. In this way, it's able to calculate the gradients for a chain of operations, with respect to any one tensor. Let's reduce the tensor `y` to a scalar value, the mean.
# + id="ZpyFnSW3Oo5v" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="027270bc-42e3-4f90-b4d3-9c892f0aec94" executionInfo={"status": "ok", "timestamp": 1586125581003, "user_tz": -120, "elapsed": 985, "user": {"displayName": "Fabrizio D'Intinosante", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhrjCQVLGrXDlJRniViKxzD_05EWfYGhPN_t37-=s64", "userId": "05737257329450160630"}}
z = y.mean()
print(z)
# + [markdown] id="RHoJ2bQCOo55" colab_type="text"
# You can check the gradients for `x` and `y` but they are empty currently.
# + id="vKKIoTtUOo57" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="3b7df32f-e878-4d96-ab62-712cb4d7b114" executionInfo={"status": "ok", "timestamp": 1586125691238, "user_tz": -120, "elapsed": 928, "user": {"displayName": "Fabrizio D'Intinosante", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhrjCQVLGrXDlJRniViKxzD_05EWfYGhPN_t37-=s64", "userId": "05737257329450160630"}}
print(x.grad)
# + [markdown] id="9KX0ZilfOo6D" colab_type="text"
# To calculate the gradients, you need to run the `.backward` method on a Variable, `z` for example. This will calculate the gradient for `z` with respect to `x`
#
# $$
# \frac{\partial z}{\partial x} = \frac{\partial}{\partial x}\left[\frac{1}{n}\sum_i^n x_i^2\right] = \frac{x}{2}
# $$
# + id="xqAtv3pgOo6F" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 85} outputId="fe009e6c-5478-4ff6-b2f2-d067894300ea" executionInfo={"status": "ok", "timestamp": 1586125707099, "user_tz": -120, "elapsed": 1336, "user": {"displayName": "Fabrizio D'Intinosante", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhrjCQVLGrXDlJRniViKxzD_05EWfYGhPN_t37-=s64", "userId": "05737257329450160630"}}
z.backward()
print(x.grad)
print(x/2)
# + [markdown] id="w9BQ5yj9Oo6U" colab_type="text"
# These gradients calculations are particularly useful for neural networks. For training we need the gradients of the cost with respect to the weights. With PyTorch, we run data forward through the network to calculate the loss, then, go backwards to calculate the gradients with respect to the loss. Once we have the gradients we can make a gradient descent step.
# + [markdown] id="81dwVgPlOo6c" colab_type="text"
# ## Loss and Autograd together
#
# When we create a network with PyTorch, all of the parameters are initialized with `requires_grad = True`. This means that when we calculate the loss and call `loss.backward()`, the gradients for the parameters are calculated. These gradients are used to update the weights with gradient descent. Below you can see an example of calculating the gradients using a backwards pass.
# + id="PX-L8HmtOo6f" colab_type="code" colab={}
# Build a feed-forward network
model = nn.Sequential(nn.Linear(784, 128),
nn.ReLU(),
nn.Linear(128, 64),
nn.ReLU(),
nn.Linear(64, 10),
nn.LogSoftmax(dim=1))
criterion = nn.NLLLoss()
images, labels = next(iter(trainloader))
images = images.view(images.shape[0], -1)
logits = model(images)
loss = criterion(logits, labels)
# + id="pJ0VOJRROo6t" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 187} outputId="1b49c1eb-ce2b-4e5a-f8f9-399e923ddd08" executionInfo={"status": "ok", "timestamp": 1586126528228, "user_tz": -120, "elapsed": 1702, "user": {"displayName": "Fabrizio D'Intinosante", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhrjCQVLGrXDlJRniViKxzD_05EWfYGhPN_t37-=s64", "userId": "05737257329450160630"}}
print('Before backward pass: \n', model[0].weight.grad)
loss.backward()
print('After backward pass: \n', model[0].weight.grad)
# + [markdown] id="aCLlNuOJOo61" colab_type="text"
# ## Training the network!
#
# There's one last piece we need to start training, an optimizer that we'll use to update the weights with the gradients. We get these from PyTorch's [`optim` package](https://pytorch.org/docs/stable/optim.html). For example we can use stochastic gradient descent with `optim.SGD`. You can see how to define an optimizer below.
# + id="ccTocT6AOo63" colab_type="code" colab={}
from torch import optim
# Optimizers require the parameters to optimize and a learning rate
optimizer = optim.SGD(model.parameters(), lr=0.01)
# + [markdown] id="3fPokeIqOo6_" colab_type="text"
# Now we know how to use all the individual parts so it's time to see how they work together. Let's consider just one learning step before looping through all the data. The general process with PyTorch:
#
# * Make a forward pass through the network
# * Use the network output to calculate the loss
# * Perform a backward pass through the network with `loss.backward()` to calculate the gradients
# * Take a step with the optimizer to update the weights
#
# Below I'll go through one training step and print out the weights and gradients so you can see how it changes. Note that I have a line of code `optimizer.zero_grad()`. When you do multiple backwards passes with the same parameters, the gradients are accumulated. This means that you need to zero the gradients on each training pass or you'll retain gradients from previous training batches.
# + id="W3COLsbEOo7Q" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 392} outputId="a32ae133-9489-438d-8063-d74857f5645a" executionInfo={"status": "ok", "timestamp": 1586126841151, "user_tz": -120, "elapsed": 904, "user": {"displayName": "Fabrizio D'Intinosante", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhrjCQVLGrXDlJRniViKxzD_05EWfYGhPN_t37-=s64", "userId": "05737257329450160630"}}
print('Initial weights - ', model[0].weight)
images, labels = next(iter(trainloader))
images.resize_(64, 784)
# Clear the gradients, do this because gradients are accumulated
optimizer.zero_grad()
# Forward pass, then backward pass, then update weights
output = model(images)
loss = criterion(output, labels)
loss.backward()
print('Gradient -', model[0].weight.grad)
# + id="ivB1lpOZOo7c" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 170} outputId="cfd8571b-0e26-44c7-9788-bf9f8c0787ad" executionInfo={"status": "ok", "timestamp": 1586126862125, "user_tz": -120, "elapsed": 857, "user": {"displayName": "Fabrizio D'Intinosante", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhrjCQVLGrXDlJRniViKxzD_05EWfYGhPN_t37-=s64", "userId": "05737257329450160630"}}
# Take an update step and few the new weights
optimizer.step()
print('Updated weights - ', model[0].weight)
# + [markdown] id="V_fz0ZUEOo7m" colab_type="text"
# ### Training for real
#
# Now we'll put this algorithm into a loop so we can go through all the images. Some nomenclature, one pass through the entire dataset is called an *epoch*. So here we're going to loop through `trainloader` to get our training batches. For each batch, we'll doing a training pass where we calculate the loss, do a backwards pass, and update the weights.
#
# >**Exercise:** Implement the training pass for our network. If you implemented it correctly, you should see the training loss drop with each epoch.
# + id="3B9myvv8klXz" colab_type="code" colab={}
import matplotlib.pyplot as plt
# + id="I5ovLmcdOo7q" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 366} outputId="79ceaa7b-3c44-4290-ad87-a89f0d986497" executionInfo={"status": "ok", "timestamp": 1586127887108, "user_tz": -120, "elapsed": 36796, "user": {"displayName": "Fabrizio D'Intinosante", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhrjCQVLGrXDlJRniViKxzD_05EWfYGhPN_t37-=s64", "userId": "05737257329450160630"}}
## Your solution here
model = nn.Sequential(nn.Linear(784, 128),
nn.ReLU(),
nn.Linear(128, 64),
nn.ReLU(),
nn.Linear(64, 10),
nn.LogSoftmax(dim=1))
criterion = nn.NLLLoss()
optimizer = optim.SGD(model.parameters(), lr=0.003)
total_loss = []
epochs = 5
for e in range(epochs):
running_loss = 0
for images, labels in trainloader:
# Flatten MNIST images into a 784 long vector
images = images.view(images.shape[0], -1)
optimizer.zero_grad()
# TODO: Training pass
output = model(images)
loss = criterion(output, labels)
loss.backward()
optimizer.step()
running_loss += loss.item()
else:
print(f"Training loss: {running_loss/len(trainloader)}")
total_loss.append(running_loss/len(trainloader))
plt.plot(total_loss)
# + [markdown] id="LBLjI6sjOo70" colab_type="text"
# With the network trained, we can check out it's predictions.
# + id="7w5nq3tOOo71" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 252} outputId="00652a73-9410-4a67-c605-42d22bab0b7c" executionInfo={"status": "ok", "timestamp": 1586127294563, "user_tz": -120, "elapsed": 1878, "user": {"displayName": "Fabrizio D'Intinosante", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhrjCQVLGrXDlJRniViKxzD_05EWfYGhPN_t37-=s64", "userId": "05737257329450160630"}}
# %matplotlib inline
import helper
images, labels = next(iter(trainloader))
img = images[0].view(1, 784)
# Turn off gradients to speed up this part
with torch.no_grad():
logps = model(img)
# Output of the network are log-probabilities, need to take exponential for probabilities
ps = torch.exp(logps)
helper.view_classify(img.view(1, 28, 28), ps)
# + [markdown] id="KfG5I65tOo8B" colab_type="text"
# Now our network is brilliant. It can accurately predict the digits in our images. Next up you'll write the code for training a neural network on a more complex dataset.
| 70.610169
| 1,604
|
e7d30d30ab318ac728ba7cfb4b475393bf667797
|
py
|
python
|
colab-analysis-training.ipynb
|
NISH1001/earth-science-text-classification
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# + colab={"base_uri": "https://localhost:8080/"} id="-6341Ltu1uye" outputId="9a868d93-b1bf-4974-8d78-f6ad076dd2eb"
from google.colab import drive
drive.mount('/content/drive')
# + id="144XyoTb2afg"
import os
# + id="UOQxwYNk2Fwb"
DRIVE_BASE = "/content/drive/MyDrive/Colab Notebooks/uah-ra/"
# + id="2BCoS8CD2Yrh"
KW_PATH = os.path.join(DRIVE_BASE, "data/keywords.txt")
DATA_PATH = os.path.join(DRIVE_BASE, "data/data.csv")
# + id="VCDwpVdb2mxE"
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# %matplotlib inline
# + id="SY8DZlml2qPE"
def load_keywords(path):
res = []
with open(path) as f:
text = f.read().strip()
tags_str = text.split(",")
res = map(lambda t: [_.strip().lower() for _ in t.split(">")], tags_str)
res = filter(lambda x: len(x) > 0, res)
res = list(res)
return res
# + colab={"base_uri": "https://localhost:8080/"} id="WO41bXr62rpr" outputId="87243a01-2133-4c66-dfd7-646ca5a6e72a"
KEYWORDS = load_keywords(KW_PATH)
len(KEYWORDS), len(set([kw for kws in KEYWORDS for kw in kws]))
# + colab={"base_uri": "https://localhost:8080/"} id="OhYJHdL-HI2V" outputId="8cee126f-ead9-48e9-f0e3-8425870f87ac"
KEYWORDS
# + [markdown] id="WWESiiOA23c2"
# # Tag Analysis
# + colab={"base_uri": "https://localhost:8080/"} id="fva3vYHv2wKT" outputId="17a48e07-07fa-462e-b7f5-ef8d38ae92fe"
# !pip install loguru
# + id="AxzBe9bN266E"
from loguru import logger
# + id="SsrH5dwl28PE"
from collections import Counter, defaultdict
# + id="r_b0vt7T28w8"
def get_counts(keywords, level=0):
kws = map(lambda x: x[level if level<len(x) else len(x)-1], keywords)
kws = list(kws)
# kws = list(map(str.lower, kws))
counter = Counter(kws)
return counter
# + id="XilboUHy2-Ts"
def analyze_kws(keywords, topn=10):
plt.figure(figsize=(15, 8))
for level in [0, 1, 2, 3, -1]:
_ = get_counts(KEYWORDS, level=level)
logger.debug(f"[Level={level}, NKWs={len(_)}] : {_.most_common(10)}")
df = pd.DataFrame(_.most_common(topn), columns=["kw", "frequency"])
ax = sns.barplot(
x="frequency", y="kw",
data=df,
linewidth=2.5,
facecolor=(1, 1, 1, 0),
errcolor=".2",
edgecolor=".2"
)
plt.title(f"Level={level}, topn={topn}")
plt.figure(figsize=(15, 8))
# + colab={"base_uri": "https://localhost:8080/", "height": 105} id="v86lPBpLJwNY" outputId="7c016032-fe43-4b6d-82f6-579b91d9f606"
", ".join(list(get_counts(KEYWORDS, level=1).keys()))
# + colab={"base_uri": "https://localhost:8080/", "height": 1000} id="R-xYzCP42_gU" outputId="6ff6a241-76af-4de2-b9d0-62537b9cc949"
analyze_kws(KEYWORDS, topn=20)
# + [markdown] id="__MOlrWI3M6X"
# # Data Analysis
# + id="BOIb9Zuz3AxN"
def parse_kws(kw_str, level=2):
res = kw_str.split(",")
res = map(lambda kw: [_.strip().lower() for _ in kw.split(">")], res)
res = map(lambda x: x[level if level<len(x) else len(x)-1], res)
return list(set(res))
def load_data(path, level=0):
logger.info(f"Loading data from {path}. [KW Level={level}]")
df = pd.read_csv(path)
df["desc"] = df["desc"].apply(str.strip)
df["labels"] = df["keywords"].apply(lambda x: parse_kws(x, level))
df["textlen"] = df["desc"].apply(len)
return df
# + colab={"base_uri": "https://localhost:8080/"} id="5RjiG10u3QY4" outputId="f40a0f5b-ff6a-4207-90b8-892b018d4047"
DATA = load_data(DATA_PATH, level=1)
# + colab={"base_uri": "https://localhost:8080/"} id="PaxosUAt3TD4" outputId="a5ed4520-91b6-4057-d6f9-5973a1221318"
DATA.shape
# + colab={"base_uri": "https://localhost:8080/", "height": 537} id="5MkKxnFj3XUZ" outputId="9e15107e-4b92-4430-9814-06b789e1db4f"
DATA.head(10)
# + id="64Ljd-338tuZ"
# + id="s5PuxtYENMdx"
def analyze_labels(df):
df = df.copy()
labels = [l for ls in df["labels"] for l in ls]
uniques = set(labels)
logger.info(f"{len(uniques)} unique labels")
# + colab={"base_uri": "https://localhost:8080/"} id="3jIV_bSzNkh5" outputId="78b6d955-f0eb-4a03-e5d8-f26be08fa43c"
analyze_labels(DATA)
# + id="_L9GNTFRJfPr"
# idx = 2
# _data.iloc[2].keywords_processed
# + id="H0MMa2S83YXo"
_data = DATA.copy()
_data = _data[_data["textlen"]>0]
# + colab={"base_uri": "https://localhost:8080/"} id="DUUXNTr_MpTR" outputId="794e86fc-d2c9-4f6b-f4b4-7515b00512f2"
_data.shape
# + colab={"base_uri": "https://localhost:8080/"} id="yBXBekFk3eK9" outputId="4635f376-b6f8-467c-dc23-6683e7dc8dd8"
# BERT can only process 512 tokens at once
len(_data[_data["textlen"] <= 512]) / len(_data), len(_data[_data["textlen"] <= 1024]) / len(_data)
# + colab={"base_uri": "https://localhost:8080/", "height": 719} id="HKIu_vkz3hmz" outputId="9fa7b042-547c-4846-a3a2-f62ec9df747c"
plt.figure(figsize=(20, 15))
sns.histplot(data=_data, x="textlen", bins=100).set(xlim=(0, 3000))
# + [markdown] id="vr9N-4l6B_tZ"
# # Baseline Model
# + [markdown] id="-QSLlF9rPMkw"
# # Encode Labels
# + id="wee39IU_3tw2"
from sklearn.preprocessing import MultiLabelBinarizer
# + id="2WxySRkbPp-r"
DATA_TO_USE = DATA.copy()
DATA_TO_USE = DATA_TO_USE[DATA_TO_USE["textlen"]<=500]
# + colab={"base_uri": "https://localhost:8080/"} id="7_Zgwb2yQOVl" outputId="f4845d29-d39f-44a5-ccca-7b7b50f99295"
DATA_TO_USE.shape
# + colab={"base_uri": "https://localhost:8080/", "height": 293} id="xnwUllgfQn6P" outputId="ecb3881d-960e-405f-b29f-dd40be29d2fd"
DATA_TO_USE.head()
# + colab={"base_uri": "https://localhost:8080/"} id="AH2HldquYGMZ" outputId="315ef217-d87a-45a1-d50f-fcd06e9d47d4"
analyze_labels(DATA_TO_USE)
# + id="usXW0OvzOK2M"
LE = MultiLabelBinarizer()
LABELS_ENCODED = LE.fit_transform(DATA_TO_USE["labels"])
# + colab={"base_uri": "https://localhost:8080/"} id="Vt5zUAwjOXw0" outputId="42932ca6-bd6c-4c78-b840-8a74a82738e8"
LABELS_ENCODED.shape
# + colab={"base_uri": "https://localhost:8080/"} id="9UBL0SGFOZL8" outputId="77000e40-cfc1-488e-e8b6-49582ec896e6"
LE.classes_
# + colab={"base_uri": "https://localhost:8080/"} id="3ly-VQUMOfwG" outputId="f733a533-7a09-4dab-d814-bc775499d9c8"
LE.inverse_transform(LABELS_ENCODED[0].reshape(1,-1))
# + id="7nZ6d7ywO7d7"
DATA_TO_USE["labels_encoded"] = list(LABELS_ENCODED)
# + colab={"base_uri": "https://localhost:8080/", "height": 293} id="yEdAijQ_PEmt" outputId="413d9d98-80a6-4600-9f35-22af33266eb6"
DATA_TO_USE.head()
# + [markdown] id="Q4xoTt6pPP45"
# # Split Dataset
# + id="UYATgxlLOkW8"
from sklearn.model_selection import train_test_split
# + id="NFcCLWGrOxXA"
X_train, X_test, Y_train, Y_test = train_test_split(DATA_TO_USE["desc"].to_numpy(), LABELS_ENCODED, test_size=0.1, random_state=42)
X_train, X_val, Y_train, Y_val = train_test_split(X_train, Y_train, test_size=0.1, random_state=42)
# + colab={"base_uri": "https://localhost:8080/"} id="E8Lz-NaCQ_lZ" outputId="04537c76-dbf7-4d15-9bea-ba358200998c"
X_train.shape, X_val.shape, X_test.shape
# + colab={"base_uri": "https://localhost:8080/"} id="JCcU1mUORDJo" outputId="7aeafefc-aa7d-4078-92e4-942b6075bd95"
Y_train.shape, Y_val.shape, Y_test.shape
# + colab={"base_uri": "https://localhost:8080/"} id="817C1t86XlCz" outputId="60dab30a-1693-4740-b36d-1053601228a5"
X_test
# + [markdown] id="2j8gOjlSRUnf"
# # CreateDataset
# + colab={"base_uri": "https://localhost:8080/"} id="UV0N9YJdTARH" outputId="5313937a-d566-4210-dc71-a212cc3424ea"
# ! pip install pytorch_lightning
# + id="EE-yRTzORJ5e"
import torch
from torch.utils.data import DataLoader, Dataset
# + id="3cEAMBSRSxt2"
import pytorch_lightning as pl
# + id="-5VC2YyxRafR"
class TagDataset (Dataset):
def __init__(self,texts, tags, tokenizer, max_len=512):
self.tokenizer = tokenizer
self.texts = texts
self.labels = tags
self.max_len = max_len
def __len__(self):
return len(self.texts)
def __getitem__(self, item_idx):
text = self.texts[item_idx]
inputs = self.tokenizer.encode_plus(
text,
None,
add_special_tokens=True,
max_length= self.max_len,
padding = 'max_length',
return_token_type_ids= False,
return_attention_mask= True,
truncation=True,
return_tensors = 'pt'
)
input_ids = inputs['input_ids'].flatten()
attn_mask = inputs['attention_mask'].flatten()
return {
'input_ids': input_ids ,
'attention_mask': attn_mask,
'label': torch.tensor(self.labels[item_idx], dtype=torch.float)
}
# + id="FC1hc9THRwB6"
class TagDataModule (pl.LightningDataModule):
def __init__(self, x_train, y_train, x_val, y_val, x_test, y_test,tokenizer, batch_size=16, max_token_len=512):
super().__init__()
self.train_text = x_train
self.train_label = y_train
self.val_text = x_val
self.val_label = y_val
self.test_text = x_test
self.test_label = y_test
self.tokenizer = tokenizer
self.batch_size = batch_size
self.max_token_len = max_token_len
def setup(self):
self.train_dataset = TagDataset(texts=self.train_text, tags=self.train_label, tokenizer=self.tokenizer,max_len = self.max_token_len)
self.val_dataset = TagDataset(texts=self.val_text,tags=self.val_label,tokenizer=self.tokenizer,max_len = self.max_token_len)
self.test_dataset = TagDataset(texts=self.test_text,tags=self.test_label,tokenizer=self.tokenizer,max_len = self.max_token_len)
def train_dataloader(self):
return DataLoader (self.train_dataset, batch_size = self.batch_size,shuffle = True , num_workers=2)
def val_dataloader(self):
return DataLoader (self.val_dataset, batch_size= 16)
def test_dataloader(self):
return DataLoader (self.test_dataset, batch_size= 16)
# + [markdown] id="_q9_AkKpVBO2"
# # Transformers
# + colab={"base_uri": "https://localhost:8080/"} id="D1T2m83vVLA8" outputId="685cd43d-86ec-4d46-ad5b-9ab1ebbc5f82"
# !pip install transformers
# + id="nQQAhtDSVAHM"
from transformers import AutoTokenizer, AutoModel
# + id="7BRQKG6EU9hQ"
TOKENIZER = AutoTokenizer.from_pretrained("bert-base-uncased")
# BASE_MODEL = AutoModel.from_pretrained("bert-base-uncased")
BASE_MODEL = None
# + id="AqAsw6szUYcT"
# Initialize the parameters that will be use for training
EPOCHS = 10
BATCH_SIZE = 4
MAX_LEN = 512
LR = 1e-03
# + id="kqbqmoBfUkFI"
TAG_DATA_MODULE = TagDataModule(
X_train, Y_train,
X_val, Y_val,
X_test, Y_test,
TOKENIZER,
BATCH_SIZE,
MAX_LEN
)
TAG_DATA_MODULE.setup()
# + [markdown] id="JvS2SYIuV5bX"
# # Model
# + id="yBjSwKl9Wwfu"
from pytorch_lightning.callbacks import ModelCheckpoint
# + id="RK66R1jyXEnk"
from transformers import AdamW, get_linear_schedule_with_warmup
# + id="aUeDES7kVrLm"
class TagClassifier(pl.LightningModule):
# Set up the classifier
def __init__(self, base_model=None, n_classes=10, steps_per_epoch=None, n_epochs=5, lr=1e-5 ):
super().__init__()
self.model = base_model or AutoModel.from_pretrained("bert-base-uncased", return_dict=True)
self.classifier = torch.nn.Linear(self.model.config.hidden_size,n_classes)
self.steps_per_epoch = steps_per_epoch
self.n_epochs = n_epochs
self.lr = lr
self.criterion = torch.nn.BCEWithLogitsLoss()
def forward(self,input_ids, attn_mask):
output = self.model(input_ids = input_ids ,attention_mask = attn_mask)
output = self.classifier(output.pooler_output)
return output
def training_step(self,batch,batch_idx):
input_ids = batch['input_ids']
attention_mask = batch['attention_mask']
labels = batch['label']
outputs = self(input_ids,attention_mask)
loss = self.criterion(outputs,labels)
self.log('train_loss',loss , prog_bar=True,logger=True)
return {"loss" :loss, "predictions":outputs, "labels": labels }
def validation_step(self,batch,batch_idx):
input_ids = batch['input_ids']
attention_mask = batch['attention_mask']
labels = batch['label']
outputs = self(input_ids,attention_mask)
loss = self.criterion(outputs,labels)
self.log('val_loss',loss , prog_bar=True,logger=True)
return loss
def test_step(self,batch,batch_idx):
input_ids = batch['input_ids']
attention_mask = batch['attention_mask']
labels = batch['label']
outputs = self(input_ids,attention_mask)
loss = self.criterion(outputs,labels)
self.log('test_loss',loss , prog_bar=True,logger=True)
return loss
def configure_optimizers(self):
optimizer = AdamW(self.parameters() , lr=self.lr)
warmup_steps = self.steps_per_epoch//3
total_steps = self.steps_per_epoch * self.n_epochs - warmup_steps
scheduler = get_linear_schedule_with_warmup(optimizer,warmup_steps,total_steps)
return [optimizer], [scheduler]
# + colab={"base_uri": "https://localhost:8080/"} id="oymr96pwWXfs" outputId="10f936aa-4c89-4640-804f-000612a0d4ca"
steps_per_epoch = len(X_train)//BATCH_SIZE
MODEL = TagClassifier(BASE_MODEL, n_classes=22, steps_per_epoch=steps_per_epoch,n_epochs=EPOCHS,lr=LR)
# + id="V2HTWTqKWaT3"
# # saves a file like: input/QTag-epoch=02-val_loss=0.32.ckpt
# checkpoint_callback = ModelCheckpoint(
# monitor='val_loss',# monitored quantity
# filename='QTag-{epoch:02d}-{val_loss:.2f}',
# save_top_k=3, # save the top 3 models
# mode='min', # mode of the monitored quantity for optimization
# )
# + colab={"base_uri": "https://localhost:8080/"} id="l3ZuSky-WqWk" outputId="a18a947a-4f29-4d8e-9cd0-547ad0978d83"
trainer = pl.Trainer(max_epochs = EPOCHS , gpus = 1, callbacks=[], progress_bar_refresh_rate = 30)
# + colab={"base_uri": "https://localhost:8080/", "height": 431, "referenced_widgets": ["c68fbd329c1a4caf85e2d62a52cab8b2", "c4d073fd5fc045eb869d7adb4f2c392e", "8673d9a795694b2baa04f1cbb1c5c550", "aae83a643f5b4902a894900a9ce89fa5", "f28aa4d2e1bd400f96acb1f5177c21db", "21bb4306ee06496a81338fd675195a4e", "96784c7028af4483ae63b64199741f7d", "f262bf1165414f27bd0830aaadbf6753", "07cb2e0a81d54fc68150efcdd1a44f51", "5993acafaa254c60bf226ead66ff833c", "3fee823208d0448fb1b3176c16f9943d", "cde7348a657045b78cb19398c179fe1a", "1e810d9811934ac4b26832113ea78064", "ab69cec849f54e6ca1355c4c7140f5ed", "df5eb007449d455ca59609a89307c8a9", "495cad4c2e82465187d9c3657bab6b5f", "a91524699c2343bd8f570a5b59f9f44a", "ab9edcc67f9745789804d83d7710c5e4", "8755fdb8e5244b2d80ce514255cc8c39", "118a4aecb97543b29d6a93b8809dc077", "8e736341094b4667b1d742ca34fa1e85", "9d1d776ca3b04cf59e550af52873df4f", "dedb945e912847dba8afcae5db6c6cfb", "eabb18e3c27a4d68a7e002e8074cd04b", "95f6daaddf8d4966acfc68832f9953c0", "050de9e38b824cebacc9b19f0e1ff403", "fd7daead85da44baa168d1471ad49ff0", "853fb6a163554e41bb6cbd87f182413b", "1cb8bbfdcf4e4349adafa47e1338b254", "c6e54a31c99f45938b20af546af7bf0d", "2488634a62c647dbb6256328b6fa42d8", "e5da749956844f5eb2471e3e82b9380b", "de17d16b0cb842bea9c54483a432ef7c", "27a55078f33649c2ad663c86e91d8cd6", "40b1f70cc9a64eb49517e758251d6865", "1863cdc405c2419f86d4f52f7741544c", "01722e8150234ad7a9b5caee75d6f5ac", "4e432a73e63a42ae93faf3e4e0cb6848", "bbe9f0a6641142bfaa43879ea341747f", "c433d16c52ef4c2591d9e495162cca9f", "56d9456c4b18454cbc2ac5ff8eee6522", "8357bfeed71642b99e215888e434b8b4", "73a37e3d58304a9392d2a94a483615a4", "53f5921591f84e0cb20a6051106917ae", "fe2e39c874704eb6a2a905bdbaae5380", "c55c2cd26f444781b5df0a87ad80bfeb", "93b44574bb0b4626afaada52add4e511", "a796c415b3bb47f79b140344d98f1768", "bceb05f6669647b6b1c3ea30a2115f55", "4b86af91997347c195316d2553f520d2", "a577d864481646f78a2a5473ecca51d5", "854d6dd740d9415bb979ba8868c8c856", "306adbff49da4d9185c1e8298777c921", "af34952bc0bc4c50986c246cde4fa612", "d7016a99278b4bcb9f9aca0581116f71", "0c07a72138af4348adfe9061d57fcb55", "306de37fcafc4ed994d78debeb3ac251", "49ab0b3dd7e1428d8818a204a4b372da", "c383be911d614b5791a9e7d545023f87", "5b2c9a8afcf9428091f2e14d39c8ab8f", "00e25c54285b477090c101299569dc2f", "686866a39afb4dfaaf4908b38f17b157", "06300c870fed49bd84ea6f353f3a9256", "20611a8808fa4023ac65fae48ea77802", "8b2063a0607b44d29eac4ab92ddeb265", "bc0dffeccce74cc8b8431f68f2c217d1", "2b740eb56c114ec4bd795cf61ec09d04", "dad85699ad0848eb90458e3085f739ce", "b1e167adca814100ab5ad14832dade63", "6efbce18ff084e59be6e77279c68b442", "f693d59cfd5e4b7488f46d2fdbc4265e", "32dc33784a1c4f0c892a3b1b1f1d3e73", "d57dffcbcdb04cc48bf00468c6415f3e", "510b62f1e2b24ba2953e68151e3e1ff3", "7b674856797044ea984d29235857e917", "cf08f979bf1b4bbeb1408e05046303d9", "0f5c2313b3a145c4b83f61053062c3ef", "4511fbd002e94c379ab8b8f2469d9c1e", "cf65e06590b14966a3329c961971f368", "b80780d591684c14b90b948571651de8", "4a1ac85d686f44a88d7a2c4e5da7639f", "de012e7724344974b2b8cd89204e5210", "23e754ffaf8e47aabb24028e77b56a1f", "fd15d3928233407d8580919713fef925", "a871a4825c7e413db8b0252da6cbda80", "ce9fb305f0f745208a0f07bfe979385e", "ba1e8d1e83cd4eaaae29605ed0b7ee2b", "6577c97dfd3d4f46b2ef6a24bed609ee", "c3009db6a04e4c73b00fc8e6e31a3812", "213d56542b0d4a6a804fcc043f2993ce", "08b562ebe42b43a68fb1335befa137fc", "1b90f21777bc459c82352dd5e5cc1eb3", "ac528010035e486badfef2ceb3ca173c", "3986743bbabe4fcfa8b0158f27de696a", "536ddf1fef824ef0bbb5077f8ef7d5d1", "4d8cd643b6c74027920a4dd0f0ea674b"]} id="UpLb5TuuW3hk" outputId="2b8e0922-d166-4a03-a7be-17465055ce21"
trainer.fit(MODEL, TAG_DATA_MODULE)
# + id="YNUr9rnpW8AL"
# !nvidia-smi
# + id="lz-K0iVvmodp"
trainer.save_checkpoint("model-10.ckpt")
# + id="lzCEikNknJiK"
# !mkdir "$DRIVE_BASE/checkpoints/"
# + id="BvKUNunSmogw"
# ! cp "/content/model-10.ckpt" "$DRIVE_BASE/checkpoints"
# + colab={"base_uri": "https://localhost:8080/"} id="z-K8eT06mokD" outputId="c6c2512d-78c8-4292-becd-600a5fd86c15"
# !ls "$DRIVE_BASE/checkpoints"
# + [markdown] id="GgDoJrnQngS-"
# # Test
# + colab={"base_uri": "https://localhost:8080/", "height": 260, "referenced_widgets": ["db7f49a8ce864cdba113ac776db29157", "a2f78077352c49fd82d0d13efdbd2c0d", "fab1751b40e94735846120a3311d40d2", "7ccdd56bf3b24012bc1dadd265cd3d1b", "6dadf58f1d24499fb965b29e7cf5a657", "7b5e0fec92c5413bad13bb5b884a0cf5", "56115b0b564f4bf3912a042cc6127c47", "ba5987f2e0944251931b5d5ee8d7770f"]} id="vi-NADzene9q" outputId="8aa5231e-7d84-491d-b79c-d38fb67ecc73"
trainer.test(MODEL,datamodule=TAG_DATA_MODULE)
# + [markdown] id="OUHvgUvro8fm"
# # Inference
# + colab={"base_uri": "https://localhost:8080/"} id="_4oHH6yqnfAw" outputId="c405f9df-3353-49d8-f176-c20a39562f45"
MODEL.eval()
# + id="1PbjW3QsnfEr"
import pickle
# + id="oCb3psGFnfHu"
with open("le.pkl", "wb") as f:
pickle.dump(LE, f)
# + id="3K0aqvvIrM1e"
from torch.utils.data import TensorDataset, SequentialSampler
# + id="sB3zFgm1r2Jy"
DEVICE = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
# + colab={"base_uri": "https://localhost:8080/"} id="Y_AMFEdBr4Lr" outputId="1445366f-5152-427a-d281-7abe383ad522"
MODEL.to(DEVICE)
# + id="umEa6uxLsA4y"
# + id="aUCHK1zgqc13"
def inference(model, texts, tokenizer, batch_size=2):
# model.eval()
if isinstance(texts, str):
texts = [texts]
input_ids, attention_masks = [], []
for text in texts:
text_encoded = tokenizer.encode_plus(
text,
None,
add_special_tokens=True,
max_length= MAX_LEN,
padding = 'max_length',
return_token_type_ids= False,
return_attention_mask= True,
truncation=True,
return_tensors = 'pt'
)
input_ids.append(text_encoded["input_ids"])
attention_masks.append(text_encoded["attention_mask"])
input_ids = torch.cat(input_ids, dim=0)
attention_masks = torch.cat(attention_masks, dim=0)
pred_data = TensorDataset(input_ids, attention_masks)
pred_sampler = SequentialSampler(pred_data)
pred_dataloader = DataLoader(pred_data, sampler=pred_sampler, batch_size=batch_size)
pred_outs = []
for batch in pred_dataloader:
# Add batch to GPU
batch = tuple(t.to(DEVICE) for t in batch)
# Unpack the inputs from our dataloader
b_input_ids, b_attn_mask = batch
with torch.no_grad():
# Forward pass, calculate logit predictions
pred_out = model(b_input_ids,b_attn_mask)
pred_out = torch.sigmoid(pred_out)
# Move predicted output and labels to CPU
pred_out = pred_out.detach().cpu().numpy()
pred_outs.append(pred_out)
return pred_outs
# + id="QlZ8cLNlqc5F"
_texts = X_test[:10]
_pred_outs = inference(MODEL, _texts, TOKENIZER)
# + colab={"base_uri": "https://localhost:8080/"} id="ppOd0i-yyTTt" outputId="1ad4e0f3-71c6-48c5-ae5f-d1d46507c8af"
_pred_outs
# + colab={"base_uri": "https://localhost:8080/"} id="wKHVYLcJw2BB" outputId="4a86d513-be94-4b9b-92cd-b4684729abe7"
_texts
# + colab={"base_uri": "https://localhost:8080/"} id="Txr_Bya3qc8a" outputId="7c33bfdf-eb7b-43b8-9193-e384973c0b71"
thresh = 0.3
for _txt, _yt, _p in zip(_texts, Y_test, _pred_outs.copy()):
_p = _p.flatten()
confs = _p[_p>thresh]
_p[_p<thresh] = 0
_p[_p>=thresh] = 1
print(confs)
pred_tag = LE.inverse_transform(np.array([_p]))[0]
gt_tag = LE.inverse_transform(np.array([_yt]))[0]
print(_txt[:50], gt_tag, pred_tag)
# + [markdown] id="C5jKurUmzp3x"
# # Custom Evaluation
# + id="Sb91Fsed0Mz7"
def inference2(model, tokenizer, texts, gts, threshold=0.3):
_pred_outs = inference(model, texts, tokenizer, batch_size=1)
res = []
for txt, gt, pred in zip(texts, gts, _pred_outs):
p = pred.flatten().copy()
confs = p[p>threshold]
p[p<threshold] = 0
p[p>=threshold] = 1
p = np.array([p])
gt = np.array([gt])
pred_tags = LE.inverse_transform(p)[0]
gt_tags = LE.inverse_transform(gt)[0]
res.append({"gts": gt_tags, "preds": pred_tags, "text": txt})
return res
# + id="yw7pPTa_3KYt"
def compute_jaccard(tokens1, tokens2):
if not tokens1 or not tokens2:
return 0
intersection = set(tokens1).intersection(tokens2)
union = set(tokens1).union(tokens2)
return len(intersection)/len(union)
# + colab={"base_uri": "https://localhost:8080/", "height": 168} id="pO4f7ero3nEA" outputId="bd6c5afc-318f-44b1-d1d8-1c5678f86514"
compute_jaccard([1, 2], [1, 2, 3])
# + id="Zm97PTJT3nKj"
import json
# + colab={"base_uri": "https://localhost:8080/", "height": 349} id="xJU3EwYF7-_m" outputId="f994890e-00a2-4763-c649-39e0f0a7757f"
# + id="VPCH4IQ14W1z"
# !mkdir "$DRIVE_BASE/outputs/"
# + id="uwI11bwFvFlC"
def evaluate_jaccard(model, tokenizer, texts, gts, threshold=0.3):
"""
Jaccard Evaluation. SIimlar to IoU
"""
predictions = inference2(model, tokenizer, texts, gts, threshold)
with open("inference.json", "w") as f:
json.dump(predictions, f)
metrics = []
for pmap in predictions:
metrics.append(compute_jaccard(pmap["gts"], pmap["preds"]))
return metrics
# + id="WTvb3Na-0l_d"
_ = evaluate_jaccard(MODEL, TOKENIZER, X_test[:50], Y_test[:50], threshold=0.3)
# + colab={"base_uri": "https://localhost:8080/"} id="mZL6KGjr4okM" outputId="c96b707d-2fb5-4dd6-f9cc-e4baf83157c7"
_
# + id="YsV6XYf-4bw3"
# !cp "inference.json" "$DRIVE_BASE/outputs/"
# + [markdown] id="Gxo3ouitd722"
# # Reference
#
# - https://discuss.pytorch.org/t/using-bcewithlogisloss-for-multi-label-classification/67011/2
# - https://medium.com/analytics-vidhya/finetune-distilbert-for-multi-label-text-classsification-task-994eb448f94c
# + [markdown] id="OhxU7UcQgxfG"
#
# + [markdown] id="NfOai7uxgxal"
#
# + id="xwRFwYG6d5Nz"
| 38.090323
| 3,610
|
c78d491d3203fc4c9dbd8df782d5cd0e0efe123a
|
py
|
python
|
src/case_study_2/preprocess.ipynb
|
aljabadi/SARS-CoV-2
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Preprocessing data from the COVID-19 multiomics project
#
# This notebook is provided as a similar case study for our pipeline. [It is conceptually similar to our previous integrative analysis of proteome and translatome data, but contains a larger quantity of omics data and samples](https://gitlab.com/tyagilab/sars-cov-2/-/blob/master/README.md).
#
# Full details of the original study can be found in the original publication: [https://dx.doi.org/10.1101%2F2020.07.17.20156513](https://dx.doi.org/10.1101%2F2020.07.17.20156513)
#
# [Authors provided a sql database with accession number `MSV000085703`](ftp://massive.ucsd.edu/MSV000085703/other/Covid-19%20Study%20DB.sqlite) from which multiomics data measurements were extracted.
#
# ## Summary of the original study
#
# *This information is taken from the original study by [Overmyer et al 2020](https://dx.doi.org/10.1101%2F2020.07.17.20156513)*
#
# The authors intended to investigate (a) biological insight into the host's response to SARS-CoV-2, and (b) pathways influencing its severity. Authors integrated molecule measurements with clinical attributes and identified features associated with (1) COVID-19 status and (2) `HFD-45` (hospital free days, this is a composite metric).
#
# The authors performed multiple experiments to obtain their results (summarised below). We focus specifically on the experiment of `covid severity`. As it has a relatively straightforward experimental design, we illustrate the usage of our pipeline on this case study.
#
# ### Covid state
#
# To identify differentially abundant molecules, authors performed **ANOVA and log-likelihood ratio tests** to discover:
# - 2,537 leukocyte transcripts,
# - 146 plasma proteins,
# - 168 plasma lipids,
# - 13 plasma metabolites
# associated with COVID-19 status (Table S1).
#
# To discover enriched biological processes associated with the differing biomolecules, the authors used **GO and molecular class enrichment analysis** (Tables S2A and S2B). The authors showed that these included:
# - mitotic cell cycle,
# - phagocytosis recognition,
# - positive recognition of B cell activation,
# - complement activation (classical pathway),
# - innate immune response.
#
# ### Covid severity
#
# To discover biomolecules associated with severity, authors used **univariate regression** of HFD-45 against abundance of each biomolecule. Authors accounted for sex and age and found disease associated molcules (Table S1):
# - 6,202 transcripts,
# - 189 plasma proteins,
# - 218 plasma lipids,
# - 35 plasma small molecules
#
# To further refine these authors performed **multivariate linear regression** on HFD-45 using the elastic net penalty (Zou and Hastie, 2005) as predictive features for HFD-45 (Table S1):
# - 497 transcripts,
# - 382 proteins,
# - 140 lipids,
# - 60 metabolites
#
# ### Combined information
#
# To generate a list of 219 features that were most significantly associated with COVID-19 status and severity (Figure 2C; Table S1), the authors combined:
# 1. significance with COVID-19 status,
# 2. significance with HFD-45,
# 3. elastic net feature selection
#
# ## Summary of our analysis
#
# We integrated lipidomics, metabolomics, proteomics and transcriptomics data for 100 samples. Two classes were included: `less severe` vs `more severe` covid states, represented by the variable `HFD-45`. Classes are balanced but age and sex of patients are not consistent.
#
# This jupyter notebook describes the steps taken to download and parse the input data as well as metadata, resulting in matrices of continuous values suitable for input into our pipeline. The appendix contains suggestions for handling cases where experimental design is less straightforward.
import os
import matplotlib.pyplot as plt
import pandas as pd
from functools import reduce
from scipy import stats
from scipy.stats import ttest_ind
from statsmodels.stats.multitest import multipletests
# %matplotlib inline
# Tables in sqlite database:
#
# ```
# biomolecules
# deidentified_patient_metadata
# lipidomics_measurements proteomics_measurements
# lipidomics_runs proteomics_runs
# metabolomics_measurements pvalues
# metabolomics_runs rawfiles
# metadata transcriptomics_measurements
# omes transcriptomics_runs
# ```
#
# `.csv` files were extracted from the publicly accessible `sqlite` database. The following set of commands were repeated for each of the tables listed above.
#
# ```
# .file 'Covid-19 Study DB.sqlite'
# .header on
# .mode csv
#
# .output ${TABLE}.csv
# select * from ${TABLE};
# ```
#
# `md5` sums are shown below for reproducibility:
# !for i in $(find ../../data/MSV000085703 -name *csv.gz | sort); do md5 $i; done
# !find ../../data/MSV000085703 -name *csv.gz -exec gzip -d {} \;
files = [x for x in os.listdir("../../data/MSV000085703") if x.endswith("csv")]
data = [pd.read_csv("/".join(["../../data/MSV000085703", x]), sep=",") for x in files]
data = dict(zip(files, data))
data.keys()
# Data is spread across individual `sql` tables. We remap the numerical codes to biologically meaningful identifiers. Database schema is available in Supp Fig S3 of the original manuscript.
# +
def biomolecules_to_omes(data):
gene = pd.DataFrame([[5, "Gene"]], columns=["omics_id", "omics_name"])
omes = pd.concat([data["omes.csv"], gene])
biomolecules = data["biomolecules.csv"]
biomolecules = biomolecules.merge(omes, left_on="omics_id", right_on="omics_id", how="outer")
return biomolecules
biomolecules = biomolecules_to_omes(data)
keep = biomolecules[biomolecules["keep"] == "1"]["biomolecule_id"].tolist()
biomolecules
# -
# ## Selection of Outcome Measure
#
# The authors constructed a composite variable `HFD-45` (hospital-free days at day 45). This assigns zero value to patients requiring admission longer than 45 days or who die during the admission, and progressively more free days depending on the hospitalization length. A lower value indicates higher severity.
#
# The variable is intended to:
#
# 1. be able to combine severity of disease with mortality in one single metric;
# 2. be amenable to both ICU and medical floor populations;
# 3. use a timeframe that accounts for the fact that COVID-19 patients with respiratory failure require longer hospitalizations compared with non-COVID-19 individuals (Wang et al., 2020a, 2020b);
# 4. consider that COVID-19 causes a linear disease’s deterioration pattern that transition from mild respiratory compromise to respiratory failure, followed by respiratory distress requiring mechanical ventilatory support and eventually death.
#
# ## Examining the raw data
patient_metadata = data["deidentified_patient_metadata.csv"]
raw_files = data["rawfiles.csv"]
patient_files = raw_files.merge(patient_metadata, left_on="sample_id", right_on="sample_id", how="left")
patient_files.rename(columns={"keep": "keep_patient"}, inplace=True)
patient_files
# The raw data is assigned to individual categories based on their omics category (lipidome, metabolome, proteome, transcriptome).
# +
def biomolecules_to_data(biomolecules, data):
# biomolecules = biomolecules[biomolecules["keep"] == "1"]
return data.merge(biomolecules, left_on="biomolecule_id", right_on="biomolecule_id", how="left")
def measurements_to_runs(data, runs):
return data.merge(runs, left_on="replicate_id", right_on="replicate_id", how="left")
def runs_to_patient(runs, patient):
return runs.merge(patient, left_on="rawfile_id", right_on="rawfile_id", how="left")
omics = {"L": data['lipidomics_measurements.csv'],
"M": data['metabolomics_measurements.csv'],
"P": data['proteomics_measurements.csv'],
"T": data['transcriptomics_measurements.csv'],}
runs = {"L": data['lipidomics_runs.csv'],
"M": data['metabolomics_runs.csv'],
"P": data['proteomics_runs.csv'],
"T": data['transcriptomics_runs.csv'],}
omics = [measurements_to_runs(omics[i], runs[i]) for i in omics.keys()]
omics = dict(zip(runs.keys(), omics))
omics = [runs_to_patient(omics[i], patient_files) for i in omics.keys()]
omics = dict(zip(runs.keys(), omics))
data_omics = [biomolecules_to_data(biomolecules, omics[i]) for i in omics.keys()]
patient_status = "Albany_sampleID" # unique_identifier
# -
# ## Lipidomics
#
# ### Experiment notes
#
# "The LC–MS data were processed using Compound Discoverer 2.1 (Thermo Scientific) and LipiDex (Hutchins et al., 2018) (v. 1.1.0). All peaks between 1 min and 45 min retention time and 100 Da to 5000 Da MS1 precursor mass were grouped into distinct chromatographic profiles (i.e., compound groups) and aligned using a 10-ppm mass and 0.3 min retention time tolerance. Profiles not reaching a minimum peak intensity of 5x10ˆ5, a maximum peak-width of 0.75, a signal-to-noise (S/N) ratio of 3, and a 3-fold intensity increase over blanks were excluded from further processing. MS/MS spectra were searched against an in-silico generated lipid spectral library containing 35,000 unique molecular compositions representing 48 distinct lipid classes (LipiDex library “LipiDex_HCD_Formic”, with a full range of acyl-chains included). Spectral matches with a dot product score greater than 500 and a reverse dot product score greater than 700 were retained for further analysis, with a minimum 75% spectral purity for designating fatty acid composition. Removed from the data set were adducts, class IDs greater than 3.5 median absolute retention time deviation (M.A.D. RT) of each other, and features found in less than 3 files. Data were additionally searched with Compound Discoverer 3.1 with the discovery metabolomics nodes for additional spectral matching to mzCloud and mzVault libraries but retaining the feature group and peak picking settings as detailed for the Compound Discoverer 2.1 analysis." - [Overmyer et al 2020](https://dx.doi.org/10.1101%2F2020.07.17.20156513)
#
# ### Data notes
#
# Data is a matrix of continuous values. Column names correspond to lipid names and row names correspond to unique sample identifiers. Lipids with no known annotation are also recorded and assigned a unique identifier based on their mass and charge.
# +
l = data_omics[0][["unique_identifier", "standardized_name", "normalized_abundance", "keep"]]
l = l[l["keep"] == "1"]
l.drop("keep", axis=1, inplace=True)
l = l.pivot(index="unique_identifier", columns="standardized_name")
# this maps the sample to patient
l_id = data_omics[0][["unique_identifier", patient_status]]
l_id.drop_duplicates(inplace=True)
l_id.set_index("unique_identifier", inplace=True)
l = l.merge(l_id, left_index=True, right_index=True, how="left")
l.reset_index(inplace=True)
l.set_index(["unique_identifier", patient_status], inplace=True)
l.sort_index(level=patient_status, inplace=True)
# remove null and assign sample groups
l_class = l.reset_index()
l_class.dropna(inplace=True)
l_class["covid_state"] = l_class[patient_status]
l_class["covid_state"].replace(regex=r'^C.*', value='Covid', inplace=True)
l_class["covid_state"].replace(regex=r'^NC.*', value='NonCovid', inplace=True)
l_map = l_class[["unique_identifier", patient_status, "covid_state"]]
l_data = l_class.drop([patient_status, "covid_state"], axis=1)
l_data.set_index("unique_identifier", inplace=True)
l_data.columns = [y for x, y in l_data.columns]
l_data
# -
# ## Metabolomics
#
# ### Experiment notes
#
# "GC-MS raw files were processed using a software suite developed in-house that is available at https://github.com/coongroup. Following data acquisition, raw EI-GC/MS spectral data was deconvolved into chromatographic features and then grouped into features based on co-elution. Only features with at least 10 fragment ions and present in 33% of samples were kept. Feature groups from samples and background were compared, and only feature groups greater than 3-fold higher than background were retained. Compound identifications for the metabolites analyzed were assigned by comparing deconvolved high-resolution spectra against unit-resolution reference spectra present in the NIST 12 MS/EI library as well as to authentic standards run in-house. To calculate spectral similarity between experimental and reference spectra, a weighted dot product calculation was used. Metabolites lacking a confident identification were classified as “Unknown metabolites” and appended a unique identifier based on retention time. Peak heights of specified quant m/z were used to represent feature (metabolite) abundance. The data set was also processed through, where we applied a robust linear regression approach, rlm() function (Marazzi et al., 1993), non- log2 transformed intensity values versus run order, to normalize for run order effects on signal. AEX-LC-MS/MS: raw files were processed using Xcalibur Qual Browser (v4.0.27.10, Thermo Scientific) with results exported and further processed using Microsoft Excel 2010. The prepared standard solution was used to locate appropriate peaks for peak area analysis." - [Overmyer et al 2020](https://dx.doi.org/10.1101%2F2020.07.17.20156513)
#
# ### Data notes
#
# Data is a matrix of continuous values. Column names correspond to metabolite names and row names correspond to unique sample identifiers. Metabolites with no known annotation are also recorded and assigned a unique identifier based on their mass and charge. There are two subsets of metabolites - discovery and targeted. These were originally split into two sets for preliminary investigation but are later recombined before entering the pipeline.
# +
m = data_omics[1][["unique_identifier", "standardized_name", "normalized_abundance", "keep", "omics_id"]]
m = m[m["keep"] == "1"]
m.drop("keep", axis=1, inplace=True)
m_d = m[m["omics_id"] == 3]
m_d.drop("omics_id", axis=1, inplace=True)
m_t = m[m["omics_id"] == 4]
m_t.drop("omics_id", axis=1, inplace=True)
m_d = m_d.pivot(index="unique_identifier", columns="standardized_name")
m_d.columns = [x[1] for x in m_d.columns]
# this maps the sample to patient
m_id = data_omics[1][["unique_identifier", patient_status]]
m_id.drop_duplicates(inplace=True)
m_id.set_index("unique_identifier", inplace=True)
m_d = m_d.merge(m_id, left_index=True, right_index=True, how="left")
m_d.reset_index(inplace=True)
m_d.set_index(["unique_identifier", patient_status], inplace=True)
m_d.sort_index(level=patient_status, inplace=True)
# remove null and assign sample groups
m_d_class = m_d.reset_index()
m_d_class.dropna(inplace=True)
m_d_class["covid_state"] = m_d_class[patient_status]
m_d_class["covid_state"].replace(regex=r'^C.*', value='Covid', inplace=True)
m_d_class["covid_state"].replace(regex=r'^NC.*', value='NonCovid', inplace=True)
m_d_data = m_d_class.drop([patient_status, "covid_state"], axis=1)
m_d_data.set_index("unique_identifier", inplace=True)
keep = ~m_d_data.eq(m_d_data.iloc[:, 0], axis=0).all(1)
m_d_data = m_d_data[keep]
m_d_map = m_d_class[["unique_identifier", patient_status, "covid_state"]]
m_d_map.set_index("unique_identifier", inplace=True)
m_d_map = m_d_map[keep].reset_index()
m_d_data
# +
m_t = m_t.pivot(index="unique_identifier", columns="standardized_name")
m_t.columns = [x[1] for x in m_t.columns]
m_t = m_t.merge(m_id, left_index=True, right_index=True, how="left")
m_t.reset_index(inplace=True)
m_t.set_index(["unique_identifier", patient_status], inplace=True)
m_t.sort_index(level=patient_status, inplace=True)
# remove null and assign sample groups
m_t_class = m_t.reset_index()
m_t_class.dropna(inplace=True)
m_t_class["covid_state"] = m_t_class[patient_status]
m_t_class["covid_state"].replace(regex=r'^C.*', value='Covid', inplace=True)
m_t_class["covid_state"].replace(regex=r'^NC.*', value='NonCovid', inplace=True)
m_t_map = m_t_class[["unique_identifier", patient_status, "covid_state"]]
m_t_data = m_t_class.drop([patient_status, "covid_state"], axis=1)
m_t_data.set_index("unique_identifier", inplace=True)
m_t_data
# -
# ## Proteomics
#
# ### Experiment notes
#
# "Shotgun proteomics raw files were searched using MaxQuant quantitative software package (Cox et al., 2014) (version 1.6.10.43) against UniProt Homo Sapiens database (downloaded on 6.18.2019), containing protein isoforms and computationally predicted proteins. If not specified, default MaxQuant settings were used. LFQ quantification was performed using LFQ minimum ratio count of 1 and no MS/MS requirement for LFQ comparisons. iBAQ quantitation and “match between runs” were enabled with default settings. ITMS MS/MS tolerance was set to 0.35 Da. Lists of quantified protein groups were filtered to remove reverse identifications, potential contaminants, and proteins identified only by a modification site. LFQ abundance values were log2 transformed. Missing quantitative values were imputed by randomly drawing values from the left tail of the normal distribution of all measured protein abundance values (Tyanova et al., 2016). Protein groups that contained more than 50% missing values were removed from final analyses. Relative standard deviations (RSDs) for each protein group quantified across all seven technical replicates of healthy plasma controls were calculated, and proteins with RSD greater than 30% were removed from final analyses. PRM: identification and quantification of targeted peptides for PRM analysis were performed using Skyline open access software package (version 20.1). 4-5 most intense and specific transitions were used to quantify peptide abundances, and area-under-the-curve measurements for each peptide were exported for further analysis." - [Overmyer et al 2020](https://dx.doi.org/10.1101%2F2020.07.17.20156513)
#
# ### Data notes
#
# Data is a matrix of continuous values. Column names correspond to protein names and row names correspond to unique sample identifiers.
# +
p = data_omics[2][["unique_identifier", "standardized_name", "normalized_abundance", "keep"]]
p = p[p["keep"] == "1"]
p.drop("keep", axis=1, inplace=True)
p = p.pivot(index="unique_identifier", columns="standardized_name")
p.columns = [x[1] for x in p.columns]
# this maps the sample to patient
p_id = data_omics[2][["unique_identifier", patient_status]]
p_id.drop_duplicates(inplace=True)
p_id.set_index("unique_identifier", inplace=True)
p = p.merge(p_id, left_index=True, right_index=True, how="left")
p.reset_index(inplace=True)
p.set_index(["unique_identifier", patient_status], inplace=True)
p.sort_index(level=patient_status, inplace=True)
# remove null and assign sample groups
p_class = p.reset_index()
p_class.dropna(inplace=True)
p_class["covid_state"] = p_class[patient_status]
p_class["covid_state"].replace(regex=r'^C.*', value='Covid', inplace=True)
p_class["covid_state"].replace(regex=r'^NC.*', value='NonCovid', inplace=True)
p_map = p_class[["unique_identifier", patient_status, "covid_state"]]
p_data = p_class.drop([patient_status, "covid_state"], axis=1)
p_data.set_index("unique_identifier", inplace=True)
p_data
# -
# ## RNA-Seq Data Processing
#
# "All RNA transcripts were downloaded from the NCBI refseq ftp site (wget ftp://ftp.ncbi.nlm.nih.gov/refseq/H_sapiens/mRNA_Prot/∗.rna.fna.gz ). Only mRNA (accessions NM_xxxx and XM_xxxx) and rRNA (excluding 5.8S) was then extracted, and immunoglobulin transcripts were downloaded from ENSEMBL (IG_C, IG_D, IG_J and IG_V ). We created a file mapping accession numbers to gene symbols, and then used rsem-prepare-reference to build a bowtie-2 reference database. Fastq files were trimmed and filtered using a custom algorithm tailored to improve quality scores and maximize retained reads in paired-end data. RNA-Seq expression estimation was performed by RSEM v 1.3.0 (parameters: seed-length=20, no-qualities, bowtie2-k=200, bowtie2-sensitivity-level=sensitive) (Li and Dewey, 2011), with bowtie-2 (v 2.3.4.1) for the alignment step (Langmead and Salzberg, 2012), using the custom hg38 reference described above. After the collation of expression estimates, hemoglobin transcripts were removed from further analysis, and TPM values were rescaled to total 1,000,000 in each sample. Differential Expression analysis was performed using the EBSeq package (v 1.26.0) (Leng et al., 2013) in R (v 3.6.2)." - [Overmyer et al 2020](https://dx.doi.org/10.1101%2F2020.07.17.20156513)
#
# ### Data notes
#
# Data is a matrix of continuous values. Zero values exist. Column names correspond to unique gene names and row names correspond to sample identifiers.
# +
t = data_omics[3][["unique_identifier", "standardized_name", "normalized_abundance", "keep"]]
t = t[t["keep"] == "1"]
t.drop("keep", axis=1, inplace=True)
t = t.pivot(index="unique_identifier", columns="standardized_name")
t.columns = [x[1] for x in t.columns]
# this maps the sample to patient
t_id = data_omics[3][["unique_identifier", patient_status]]
t_id.drop_duplicates(inplace=True)
t_id.set_index("unique_identifier", inplace=True)
t = t.merge(t_id, left_index=True, right_index=True, how="left")
t.reset_index(inplace=True)
t.set_index(["unique_identifier", patient_status], inplace=True)
t.sort_index(level=patient_status, inplace=True)
# remove null and assign sample groups
t_class = t.reset_index()
t_class.dropna(inplace=True)
t_class["covid_state"] = t_class[patient_status]
t_class["covid_state"].replace(regex=r'^C.*', value='Covid', inplace=True)
t_class["covid_state"].replace(regex=r'^NC.*', value='NonCovid', inplace=True)
t_map = t_class[["unique_identifier", patient_status, "covid_state"]]
t_data = t_class.drop([patient_status, "covid_state"], axis=1)
t_data.set_index("unique_identifier", inplace=True)
t_data
# -
# ## Sample filtering
#
# Our pipeline and underlying methods require all samples to be identical across the blocks of omics data. These steps filter out all samples which are not represented in each omics data block.
# our framework only ingests data with matched samples
data_clean = [l_data, m_d_data, m_t_data, p_data, t_data]
maps = [l_map, m_d_map, m_t_map, p_map, t_map]
maps = [x.set_index("Albany_sampleID") for x in maps]
common = [set(x.index.tolist()) for x in maps]
common = set.intersection(*common)
maps = [x.loc[list(common)] for x in maps]
matched = [i.loc[j["unique_identifier"]] for i, j in tuple(zip(data_clean, maps))]
[x.shape for x in matched]
# ## Sample mapping (Covid state)
#
# Unique sample identifiers are matched to their biologically relevant sample categories. Long feature names are also shortened, and mapping files containing the original names are generated for later recovery if necessary.
# +
# remap identifiers to sample types
def map_common_id(metadata, data):
metadata = pd.DataFrame(metadata.set_index("unique_identifier")["Albany_sampleID"])
data = pd.merge(metadata, data, left_index=True, right_index=True)
data = data.reset_index().set_index("Albany_sampleID")
data.index.name = None
return data.drop("unique_identifier", axis=1)
# long feature names break the pipeline
def shorten_colnames(data):
name_long = pd.Series(data.columns, name="long")
data.columns = [x[:24] for x in data.columns]
name_short = pd.Series(data.columns, name="short")
name_map = pd.concat([name_short, name_long], axis=1)
return data, name_map
classes_clean = [l_class, m_d_class, m_t_class, p_class, t_class]
data_final = [map_common_id(x, y) for x, y in tuple(zip(classes_clean, matched))]
data_short = [shorten_colnames(x) for x in data_final]
data_map = [x[1] for x in data_short]
data_final = [x[0] for x in data_short]
# -
# ## Data for pipeline input
#
# Output the preprocessed data into the relevant files.
# +
outcovid = [
"../../data/MSV000085703/covid/data_lipidomics.tsv" ,
"../../data/MSV000085703/covid/data_metabolomicsdiscovery.tsv" ,
"../../data/MSV000085703/covid/data_metabolomicstargeted.tsv" ,
"../../data/MSV000085703/covid/data_proteomics.tsv" ,
"../../data/MSV000085703/covid/data_transcriptomics.tsv" ,
]
outhfd45 = [
"../../data/MSV000085703/hfd45/data_lipidomics.tsv" ,
"../../data/MSV000085703/hfd45/data_metabolomicsdiscovery.tsv" ,
"../../data/MSV000085703/hfd45/data_metabolomicstargeted.tsv" ,
"../../data/MSV000085703/hfd45/data_proteomics.tsv" ,
"../../data/MSV000085703/hfd45/data_transcriptomics.tsv" ,
]
# !mkdir -p ../../data/MSV000085703/covid ../../data/MSV000085703/hfd45
[x.to_csv(y, sep="\t") for x, y in tuple(zip(data_final, outcovid))]
[x[:100].to_csv(y, sep="\t") for x, y in tuple(zip(data_final, outhfd45))]
# +
outmeta = [
"../../data/MSV000085703/lipidomics_class.tsv" ,
"../../data/MSV000085703/metabolomicsdiscovery_class.tsv" ,
"../../data/MSV000085703/metabolomicstargeted_class.tsv" ,
"../../data/MSV000085703/proteomics_class.tsv" ,
"../../data/MSV000085703/transcriptomics_class.tsv" ,
]
def map_sample_type(metadata, data):
data = pd.merge(metadata, data.set_index("Albany_sampleID"), left_index=True, right_index=True)
data = data[~data.index.duplicated(keep='first')]
return pd.DataFrame(data["covid_state"])
classes_final = [map_sample_type(x, y) for x, y in tuple(zip(data_final, classes_clean))]
[x.to_csv(y, sep="\t") for x, y in tuple(zip(classes_final, outmeta))]
# all these files are identical, choose one to copy to avoid confusion
classes_final[0].to_csv("../../data/MSV000085703/covid/classes_diablo.tsv", sep="\t")
classes_final[0][:100].to_csv("../../data/MSV000085703/hfd45/classes_diablo.tsv", sep="\t")
# +
outmap = [
"../../data/MSV000085703/lipidomics_featuremap.tsv" ,
"../../data/MSV000085703/metabolomicsdiscovery_featuremap.tsv" ,
"../../data/MSV000085703/metabolomicstargeted_featuremap.tsv" ,
"../../data/MSV000085703/proteomics_featuremap.tsv" ,
"../../data/MSV000085703/transcriptomics_featuremap.tsv" ,
]
[x.to_csv(y, sep="\t") for x, y in tuple(zip(data_map, outmap))]
# -
# ## Combine metabolomics data
#
# The authors combined both sets of `discovery` and `targeted` metabolomics data. These were originally separated to investigate the possibility that the individual blocks of omics data would be informative during integration.
# +
m_covid = "../../data/MSV000085703/covid/data_metabolomics.tsv"
m_hfd45 = "../../data/MSV000085703/hfd45/data_metabolomics.tsv"
metabolomics = data_final[1].merge(data_final[2], left_index=True, right_index=True)
metabolomics.to_csv(m_covid, sep="\t")
metabolomics[:100].to_csv(m_hfd45, sep="\t")
# -
# ## Sample mapping (Covid severity)
#
# The `HFD-45` metric created by the authors measures the days a patient is in hospital. The authors use this to classify patients by disease severity.
# +
# transcriptomics covid vs noncovid
dge_path_hfd45 = "../../data/MSV000085703/hfd45/data_transcriptomics_dge_hfd45.tsv"
def extract_hfd45(outpath, data):
data = data[:100]
data.to_csv(outpath, sep="\t")
return data
hfd45_paths = [
"../../data/MSV000085703/hfd45/data_lipidomics.tsv",
"../../data/MSV000085703/hfd45/data_metabolomicsdiscovery.tsv",
"../../data/MSV000085703/hfd45/data_metabolomicstargeted.tsv",
"../../data/MSV000085703/hfd45/data_proteomics.tsv",
"../../data/MSV000085703/hfd45/data_transcriptomics.tsv",
]
# transcriptomics hfd45 spectrum
patient_info = patient_metadata.set_index("Albany_sampleID")
patient_info = classes_final[4].merge(patient_info, left_index=True, right_index=True)
patient_info = patient_info[["covid_state", "Hospital_free_days_45"]]
patient_info = patient_info[patient_info["covid_state"] == "Covid"].drop("covid_state", axis=1)
# assign severity score based on median value (26) reported by authors
patient_info[patient_info["Hospital_free_days_45"] <= 26] = 1
patient_info[patient_info["Hospital_free_days_45"] > 26] = 0
patient_info[patient_info["Hospital_free_days_45"] == 1] = "More severe"
patient_info[patient_info["Hospital_free_days_45"] == 0] = "Less severe"
patient_info.to_csv("../../data/MSV000085703/hfd45/classes_diablo.tsv", sep="\t")
# subset data accordingly (take only the 100 covid patients)
covid_only = patient_info.index
hfd45_data = [extract_hfd45(x, y) for x, y in list(zip(hfd45_paths, data_final))]
# -
# ## Data imputation
#
# Transcriptomics data was sparse, and was imputed as part of the pipeline. For reproducibility, we included the imputed data file used, which can be provided as an input file directly.
#
# Note that the `--icomp` flag can be enabled in the main pipeline script, which will perform the imputation step internally given `N` components.
# # Appendix
#
# This appendix is included for completeness only and describes a few ways to work around common problems in data.
#
# ## Handling class imbalance in data
#
# There are class imbalances between `COVID` and `NON-COVID` samples. It is possible to subsampled a set of samples from the `COVID` sample type. In this example, we first investigated if the distribution of the samples are equal. The subset contains `25` samples while the full set contains `100` samples.
# +
# check that the distribution of data in all covid cases is similar to the subset
def check_dist(data, meta):
f = plt.figure(figsize=(10,3))
ax1 = f.add_subplot(121)
ax1.set_title(meta + " subset")
ax2 = f.add_subplot(122)
ax2.set_title(meta + " full set")
pd.DataFrame(data[75:100].values.flatten()).boxplot(ax=ax1)
pd.DataFrame(data[:100].values.flatten()).boxplot(ax=ax2)
plt.show()
meta = [
"lipidomics",
"metabolomics discovery",
"metabolomics targeted",
"proteomics",
"transcriptomics",
]
[check_dist(x, y) for x, y in list(zip(data_final, meta))]
# -
# ## Feature selection
#
# To improve run time and performance, feature selection can be performed. Ideally, features can be iteratively removed and the classification accuracy observed. However, in biological datasets where features exceed samples (for example a sample may have 10000 associated genes), this is not always possible.
# Features were selected by performing a t test on the data after checking assumptions of independence, distributions and variance. Because of the absence of raw patient data, the t test was performed on normalised abundance measures and did not go through a conventional differential expression pipeline with `edgeR` and `limma`. We note that the purpose of this step is to lower the feature count and not to use these results in downstream analysis.
# +
dge_path_covid = "../../data/MSV000085703/data_transcriptomics_dge_covid.tsv"
dge_path_covid_balanced = "../../data/MSV000085703/balanced/data_transcriptomics_dge_covid.tsv"
# we want to reduce features in transcriptomics data only
df = classes_final[4].merge(data_final[4], left_index=True, right_index=True)
df = df.set_index("covid_state")
# perform a welch t test for covid vs noncovid
ttest = stats.ttest_ind(df.loc["Covid"], df.loc["NonCovid"], equal_var=False)
mean_covid = df.loc["Covid"].mean()
mean_noncovid = df.loc["NonCovid"].mean()
# get bh adjusted p values
ttest = pd.DataFrame(multipletests(ttest[1], method="fdr_bh")[1])
ttest.index = df.columns
dgelist = ttest[ttest < 0.05].dropna().index
# independent (no patients are represented in more than 1 sample group)
# variances (similar but not identical)
stats.levene(mean_covid, mean_noncovid, center="mean")
# normal distribution
df.loc["Covid"].mean().hist(alpha=0.5)
df.loc["NonCovid"].mean().hist(alpha=0.5, color="orange")
plt.show()
sampling_difference = mean_covid - mean_noncovid
stats.shapiro(sampling_difference)
stats.probplot(sampling_difference, plot=plt, rvalue= True)
plt.show()
# -
| 50.85342
| 1,683
|
5ccfcd2d46440db11ba35f6b3add0f686cc61f94
|
py
|
python
|
modulo1_tema4_Py_50_gest_dat.ipynb
|
griu/msc_python
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: mufib_env383
# language: python
# name: mufib_env383
# ---
# + [markdown] toc=true
# <h1>Table of Contents<span class="tocSkip"></span></h1>
# <div class="toc"><ul class="toc-item"><li><span><a href="#Preparación-del-entorno" data-toc-modified-id="Preparación-del-entorno-1"><span class="toc-item-num">1 </span>Preparación del entorno</a></span></li><li><span><a href="#8---Data-Management" data-toc-modified-id="8---Data-Management-2"><span class="toc-item-num">2 </span>8 - Data Management</a></span><ul class="toc-item"><li><ul class="toc-item"><li><span><a href="#8.2.-Import-and-Export-Data" data-toc-modified-id="8.2.-Import-and-Export-Data-2.0.1"><span class="toc-item-num">2.0.1 </span>8.2. Import and Export Data</a></span><ul class="toc-item"><li><span><a href="#8.2.1.-Reading-Text-with-Separator" data-toc-modified-id="8.2.1.-Reading-Text-with-Separator-2.0.1.1"><span class="toc-item-num">2.0.1.1 </span>8.2.1. Reading Text with Separator</a></span></li><li><span><a href="#8.2.2.-Writing-Text-with-Separator" data-toc-modified-id="8.2.2.-Writing-Text-with-Separator-2.0.1.2"><span class="toc-item-num">2.0.1.2 </span>8.2.2. Writing Text with Separator</a></span></li><li><span><a href="#8.2.3.-Reading-and-Writing-in-PICKLE-Format" data-toc-modified-id="8.2.3.-Reading-and-Writing-in-PICKLE-Format-2.0.1.3"><span class="toc-item-num">2.0.1.3 </span>8.2.3. Reading and Writing in PICKLE Format</a></span></li></ul></li><li><span><a href="#8.3.-Crossing-between-tables" data-toc-modified-id="8.3.-Crossing-between-tables-2.0.2"><span class="toc-item-num">2.0.2 </span>8.3. Crossing between tables</a></span><ul class="toc-item"><li><span><a href="#8.3.1.-Crosses-by-Ordination-Without-Shared-Index" data-toc-modified-id="8.3.1.-Crosses-by-Ordination-Without-Shared-Index-2.0.2.1"><span class="toc-item-num">2.0.2.1 </span>8.3.1. Crosses by Ordination Without Shared Index</a></span></li><li><span><a href="#8.3.2.-Table-Sorting" data-toc-modified-id="8.3.2.-Table-Sorting-2.0.2.2"><span class="toc-item-num">2.0.2.2 </span>8.3.2. Table Sorting</a></span></li><li><span><a href="#8.3.3.-Union-by-Columns-Without-Shared-Index" data-toc-modified-id="8.3.3.-Union-by-Columns-Without-Shared-Index-2.0.2.3"><span class="toc-item-num">2.0.2.3 </span>8.3.3. Union by Columns Without Shared Index</a></span></li><li><span><a href="#8.3.4.-Union-by-rows" data-toc-modified-id="8.3.4.-Union-by-rows-2.0.2.4"><span class="toc-item-num">2.0.2.4 </span>8.3.4. Union by rows</a></span></li><li><span><a href="#8.3.5.-Key-Field-Crossing" data-toc-modified-id="8.3.5.-Key-Field-Crossing-2.0.2.5"><span class="toc-item-num">2.0.2.5 </span>8.3.5. Key Field Crossing</a></span></li></ul></li><li><span><a href="#8.4.-Aggregate-Summaries" data-toc-modified-id="8.4.-Aggregate-Summaries-2.0.3"><span class="toc-item-num">2.0.3 </span>8.4. Aggregate Summaries</a></span><ul class="toc-item"><li><span><a href="#8.4.1.-describe" data-toc-modified-id="8.4.1.-describe-2.0.3.1"><span class="toc-item-num">2.0.3.1 </span>8.4.1. describe</a></span></li><li><span><a href="#8.4.2.-Basic-Statistics" data-toc-modified-id="8.4.2.-Basic-Statistics-2.0.3.2"><span class="toc-item-num">2.0.3.2 </span>8.4.2. Basic Statistics</a></span></li><li><span><a href="#8.4.3.-Tables-of-Frequencies-or-Counts" data-toc-modified-id="8.4.3.-Tables-of-Frequencies-or-Counts-2.0.3.3"><span class="toc-item-num">2.0.3.3 </span>8.4.3. Tables of Frequencies or Counts</a></span></li></ul></li><li><span><a href="#8.5.-Aggregated-by-Subgroups" data-toc-modified-id="8.5.-Aggregated-by-Subgroups-2.0.4"><span class="toc-item-num">2.0.4 </span>8.5. Aggregated by Subgroups</a></span><ul class="toc-item"><li><span><a href="#8.5.1.-Grouping-or-GROUPBY" data-toc-modified-id="8.5.1.-Grouping-or-GROUPBY-2.0.4.1"><span class="toc-item-num">2.0.4.1 </span>8.5.1. Grouping or <em>GROUPBY</em></a></span></li><li><span><a href="#8.5.2.-Aggregate" data-toc-modified-id="8.5.2.-Aggregate-2.0.4.2"><span class="toc-item-num">2.0.4.2 </span>8.5.2. Aggregate</a></span></li><li><span><a href="#8.5.3.-Filtered-out" data-toc-modified-id="8.5.3.-Filtered-out-2.0.4.3"><span class="toc-item-num">2.0.4.3 </span>8.5.3. Filtered out</a></span></li><li><span><a href="#8.5.4.-Transformations" data-toc-modified-id="8.5.4.-Transformations-2.0.4.4"><span class="toc-item-num">2.0.4.4 </span>8.5.4. Transformations</a></span></li><li><span><a href="#8.5.5.-Application-of-functions" data-toc-modified-id="8.5.5.-Application-of-functions-2.0.4.5"><span class="toc-item-num">2.0.4.5 </span>8.5.5. Application of functions</a></span></li></ul></li><li><span><a href="#8.6.-Pivot-Tables" data-toc-modified-id="8.6.-Pivot-Tables-2.0.5"><span class="toc-item-num">2.0.5 </span>8.6. Pivot Tables</a></span></li><li><span><a href="#8.7.-Management-of-Dates-and-Times-(DATETIME)" data-toc-modified-id="8.7.-Management-of-Dates-and-Times-(DATETIME)-2.0.6"><span class="toc-item-num">2.0.6 </span>8.7. Management of Dates and Times (DATETIME)</a></span><ul class="toc-item"><li><span><a href="#8.7.1.-Create-Dates-Hours" data-toc-modified-id="8.7.1.-Create-Dates-Hours-2.0.6.1"><span class="toc-item-num">2.0.6.1 </span>8.7.1. Create Dates Hours</a></span></li><li><span><a href="#8.7.2.-Get-Components" data-toc-modified-id="8.7.2.-Get-Components-2.0.6.2"><span class="toc-item-num">2.0.6.2 </span>8.7.2. Get Components</a></span></li><li><span><a href="#8.7.3.-Datetime-and-Numpy" data-toc-modified-id="8.7.3.-Datetime-and-Numpy-2.0.6.3"><span class="toc-item-num">2.0.6.3 </span>8.7.3. Datetime and Numpy</a></span></li><li><span><a href="#8.7.4.-Datetime-and-Pandas" data-toc-modified-id="8.7.4.-Datetime-and-Pandas-2.0.6.4"><span class="toc-item-num">2.0.6.4 </span>8.7.4. Datetime and Pandas</a></span></li><li><span><a href="#8.7.5.-Panda-Time-Series" data-toc-modified-id="8.7.5.-Panda-Time-Series-2.0.6.5"><span class="toc-item-num">2.0.6.5 </span>8.7.5. Panda Time Series</a></span></li><li><span><a href="#8.7.6.-Date-Sequences" data-toc-modified-id="8.7.6.-Date-Sequences-2.0.6.6"><span class="toc-item-num">2.0.6.6 </span>8.7.6. Date Sequences</a></span></li><li><span><a href="#8.7.7.-Tables-of-Counts-or-Frequencies" data-toc-modified-id="8.7.7.-Tables-of-Counts-or-Frequencies-2.0.6.7"><span class="toc-item-num">2.0.6.7 </span>8.7.7. Tables of Counts or Frequencies</a></span></li></ul></li></ul></li></ul></li></ul></div>
# -
# <img align="left" style="padding-right:10px;" width="150" src="https://upload.wikimedia.org/wikipedia/commons/thumb/6/6c/Star_Wars_Logo.svg/320px-Star_Wars_Logo.svg.png" />
#
# *made by Ferran Carrascosa Mallafrè.*
# < [Flow control](modulo1_tema4_Py_40_contr_flujo.ipynb) | [Table of contents](modulo1_tema4_Py_00_indice.ipynb) | [Anex](modulo1_tema4_Py_60_anexo.ipynb) >
#
# __[Open in Colab](https://colab.research.google.com/github/griu/msc_python/blob/master/modulo1_tema4_Py_50_gest_dat.ipynb)__ *: <span style="color:rgba(255, 99, 71, 0.8)">Padawan! When you login to Colab, prepare the environment by running the following code.</span>*
# + [markdown] id="rwi61M3czOFJ"
# # Preparación del entorno
#
# ¡Padawan! Cuando inicies sesión en Colab, prepara el entorno ejecutando el siguiente código:
# + colab={"base_uri": "https://localhost:8080/"} executionInfo={"elapsed": 25678, "status": "ok", "timestamp": 1604317087349, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="2SBF5dlSTi-h" outputId="b85ff6ea-725c-464c-abf4-fc481a6882de"
if 'google.colab' in str(get_ipython()):
# !git clone https://github.com/griu/msc_python.git /content/msc_python
# !git -C /content/msc_python pull
# %cd /content/msc_python
# + [markdown] id="J9-tONdozOF2"
# # 8 - Data Management
#
# Next, we present the functions for reading/writing data, crossing and building summary tables.
#
# At the end of the chapter, we will introduce how to handle temporary data.
# + [markdown] id="NZv9wCxdzOF7"
# ##### 8.1. Guided Activity 2.5
#
# It is about analyzing the characters of the series:
#
# > "He would rather be a monster who believes in something, who would sacrifice everything to improve the galaxy, than be someone who stands on the sidelines and looks like it has no impact on them."
# ―Princess Leia Organa
#
#
# This activity consists of crossing data of characters and planets to build descriptive summary of the data of characters.
#
# The first step is to load the data of the characters and planets:
# + colab={"base_uri": "https://localhost:8080/", "height": 458} executionInfo={"elapsed": 25855, "status": "ok", "timestamp": 1604393639069, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="gYgda8kXzOF9" outputId="ca317a7a-8ae1-4db7-bad7-394d26df0ece"
# %matplotlib inline
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns; sns.set() # for graphics style
entidades = ['planets','starships','vehicles','people','species']
entidades_df = {x: pd.read_pickle('www/' + x + '_df.pkl') for x in entidades}
# planetas
planets_df = entidades_df['planets'][["climate","temperate_tropical","population","url"]].dropna()
# Datos principales
people_df = entidades_df['people'][["height","mass","eye_color","birth_year","gender","homeworld"]].dropna()
display(people_df.head(),planets_df.head())
# + [markdown] id="jzpPEaAlzOGn"
# ### 8.2. Import and Export Data
#
# The simplest way to import structured data (in the form of a matrix of rows and columns), is through DataFrames. The reason is simple, these objects allow you to store data of different types in a single object or data table.
#
#
# + [markdown] id="sTnlKrG84dG8"
# #### 8.2.1. Reading Text with Separator
#
# To read the following text file:
# + colab={"base_uri": "https://localhost:8080/"} eval=false executionInfo={"elapsed": 25834, "status": "ok", "timestamp": 1604393639075, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="jIAnGhYVzOGv" outputId="051f8115-535e-4bdc-ff38-930399ba5c31"
# show the first 5 rows
n=5
with open('www/mtcars.csv') as f:
muestra_texto = ""
for i in range(5):
muestra_texto +=f.readline()
print(muestra_texto)
# + [markdown] id="Wf5nTbxBzOHM"
# Since this is a character-separated file, the generic function for such files is `pd.read_table()`:
# + colab={"base_uri": "https://localhost:8080/", "height": 206} executionInfo={"elapsed": 25810, "status": "ok", "timestamp": 1604393639079, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="gwF4VPPwzOHS" outputId="8e05a5c9-1fd9-41af-a8db-3d114554fd9d"
mtcars = pd.read_table("www/mtcars.csv",sep=',', decimal=".")
mtcars.head()
# + [markdown] id="fhGY-VcrzOH-"
# We can customize the load with the following parameters:
#
# - `decimal`: the decimal separator.
# - `sep`: the column separator.
#
# Also, it is common to use `enconding="latin_1"` when the file has been created with Windows.
#
# Given the structure of the file, with `pd.read_csv()` the loading is simpler:
# + id="tmnVMx3zzOID"
mtcars = pd.read_csv("www/mtcars.csv")
# + [markdown] id="8ro7EDvDzOIa"
# Aside from `.head()`, it's good practice to check your payload with `.shape()`, and `describe()`:
# + colab={"base_uri": "https://localhost:8080/"} executionInfo={"elapsed": 25785, "status": "ok", "timestamp": 1604393639087, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="TV3CxR8IzOIc" outputId="afa022bf-1ad6-4ed9-8d32-390eb60047c6"
mtcars.shape
# + colab={"base_uri": "https://localhost:8080/", "height": 300} eval=false executionInfo={"elapsed": 25766, "status": "ok", "timestamp": 1604393639093, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="0Adljra4zOIq" outputId="03510b5d-9dfd-4e77-a642-9810a053d1d3"
mtcars.describe()
# + [markdown] id="8ADL7S9ezOI3"
# #### 8.2.2. Writing Text with Separator
#
# For writing, you can use their equivalents: `.to_csv()`. A few minor variations are worth noting:
# + id="LxTs7yMDzOI6"
mtcars.to_csv("www/mtcars2.csv", decimal=",", sep=";", index=False, encoding="latin_1")
# + [markdown] id="SeERnxNJzOJI"
# - `index`: Logical field True, False. By default, it inserts the row number.
#
# Notice how the new csv is now in the european csv format with windows encoding.
# + colab={"base_uri": "https://localhost:8080/"} eval=false executionInfo={"elapsed": 25744, "status": "ok", "timestamp": 1604393639103, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="HSxqIa6KzOJL" outputId="78097c19-6cc6-4782-98b7-ada6c1a709c1"
# show the first 5 rows
n=5
with open('www/mtcars2.csv') as f:
muestra_texto = ""
for i in range(5):
muestra_texto +=f.readline()
print(muestra_texto)
# + [markdown] id="zTVxszNXzOJa"
# To read fixed-width text, check out the [pd.read_fwf()](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.read_fwf.html) function.
# + [markdown] id="PNXSv5R6zOJc"
# #### 8.2.3. Reading and Writing in PICKLE Format
#
# Python objects can be made persistent on disk in the pickle format.
#
# > **Did you know**: pickle means pepinillo o vinagreta. That is, the pickle format means that we are putting the Python objects into preserves.
#
# To save a DataFrame in pickle format, use the `.to_pickle("name.pkl")` method:
# + colab={"base_uri": "https://localhost:8080/"} eval=false executionInfo={"elapsed": 25732, "status": "ok", "timestamp": 1604393639107, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="ir-dmCbfzOJe" outputId="3fbe85ef-fb78-4e9d-8264-ebc5c86d893c"
mtcars.to_pickle("www/mtcars.pkl")
# !dir www/mtcars*
# + [markdown] id="JiCFYTXjzOJw"
# To load a pickle file (remove it from the preserve), we use `pd.read_pickle()`:
# + colab={"base_uri": "https://localhost:8080/"} executionInfo={"elapsed": 25722, "status": "ok", "timestamp": 1604393639112, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="uMfxbRw2zOJ1" outputId="036fb9ae-a1e9-48fd-d49e-cea89801b629"
mtcars_pkl = pd.read_pickle("www/mtcars.pkl")
mtcars_pkl.shape
# + [markdown] id="FKyo1m5KzOKE"
# ### 8.3. Crossing between tables
#
# To define a crossover in pandas, it is very important to be aware of the presence of the row and column indexes of the source tables. In this sense, the crossing can be carried out by 3 different systems:
#
# - By indices.
# - By ordination.
# - By key fields.
#
# Let's look at the last two, since index matching is the default option for all functions to be displayed.
# + [markdown] id="O_dLyCbJzOKG"
# #### 8.3.1. Crosses by Ordination Without Shared Index
#
# One way to cross two DataFrames is from the shared ordering, either of the rows or the columns.
# + [markdown] id="dfaI0KpjzOKI"
# #### 8.3.2. Table Sorting
#
# To sort an array, we use the `.sort.values()` method:
# + colab={"base_uri": "https://localhost:8080/"} executionInfo={"elapsed": 25709, "status": "ok", "timestamp": 1604393639115, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="qZkc-4ZyzOKL" outputId="66245f68-5e85-4139-a790-bdc16f1f56af"
a = people_df.birth_year.sort_values(ascending=False)
a.head()
# + [markdown] id="srHYOhrSzOKf"
# - `ascending`: Logical field. By default it sorts in ascending order.
#
# In DataFrames, it's not much different, but now you have to indicate the sort field in the ` by ` parameter:
# + colab={"base_uri": "https://localhost:8080/", "height": 238} executionInfo={"elapsed": 25694, "status": "ok", "timestamp": 1604393639118, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="yQMkawI2zOKh" outputId="eca1b061-4a3d-4907-df95-af98c8db58cc"
people_df_Ord = people_df.sort_values(by=["gender","height"], ascending=[True, False])
people_df_Ord.head()
# + [markdown] id="NytSxMPNzOKt"
# Notice how `ascending` allows you to choose a different sort for each sort field.
#
# In this way, we know that *Padmé Amidala*, at 185 cm, was the tallest woman in the series.
# + [markdown] id="QJ--QLDGzOKv"
# #### 8.3.3. Union by Columns Without Shared Index
#
# To join the columns of two DataFrames that share sorting, use `pd.concat()`.
#
# To see an example, first we are going to prepare 2 example tables.
#
# For the first, we select the first four columns and reset the index with `.reset_index()`:
# + colab={"base_uri": "https://localhost:8080/", "height": 206} executionInfo={"elapsed": 25673, "status": "ok", "timestamp": 1604393639121, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="gT7W5mfWzOKx" outputId="0aab56ee-3bca-4766-bb10-b61ec903813a"
people_df1 = people_df.iloc[:,:4].reset_index()
people_df1.head()
# + [markdown] id="WxaD-mPOzOLC"
# > **Important**: when resetting the index with `.reset_index()`, this has become a new field **name** of the table.
#
# Now, the second table with the rest of the fields:
# + colab={"base_uri": "https://localhost:8080/", "height": 238} executionInfo={"elapsed": 25656, "status": "ok", "timestamp": 1604393639125, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="DWUbDXNNzOLE" outputId="51b17863-a304-4d19-d282-1e7460e4ffb4"
people_df2 = people_df.iloc[:,4:]
people_df2.head()
# + [markdown] id="mcHJ1Oq6zOLP"
# The two tables `people_df1` and `people_df2` share a sort, but not the same index. To perform the crossover `ignore_index=True` prevents the function from using the indices:
# + colab={"base_uri": "https://localhost:8080/", "height": 206} executionInfo={"elapsed": 25644, "status": "ok", "timestamp": 1604393639130, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="sNqHKknVzOLW" outputId="6ec5db38-cded-4e7a-e7fd-5be657301e87"
a = pd.concat([people_df1, people_df2], ignore_index=True)
a.head()
# + [markdown] id="nzp2o2xNzOLk"
# #### 8.3.4. Union by rows
#
# To join two DataFrames with the same column ordering, we can use pd.concat, although it is more direct with the `.append()` method:
# + colab={"base_uri": "https://localhost:8080/", "height": 459} executionInfo={"elapsed": 25632, "status": "ok", "timestamp": 1604393639135, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="P6goaQjWzOLn" outputId="09686c15-4170-43b4-c3a6-33aa33f6848d"
a1 = people_df.iloc[0:2,:2]
a2 = people_df.iloc[3:5,:2]
a = a1.append(a2)
display(a1,a2,a)
# + [markdown] eval=false id="8UEtuovxzOLy"
# > **Important**: In case you don't share the same column names, you can still use the following parameter: `ignore_index=True`.
# + [markdown] id="vqtIszTVzOL0"
# #### 8.3.5. Key Field Crossing
#
# Before performing a key field match, it is important to know if it has duplicate values.
#
# ##### KEY VECTOR
#
# To get the unique keys of a String or DataFrame, use `.unique()`:
# + colab={"base_uri": "https://localhost:8080/"} executionInfo={"elapsed": 25619, "status": "ok", "timestamp": 1604393639139, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="Gk2564mVzOL4" outputId="c67134b2-1120-431b-e036-c84f27bcb849"
a = people_df.homeworld.unique()
a
# + [markdown] id="tkTjg7IRzOMF"
# To find out which rows in a DataFrame are duplicates, use `.duplicated()`:
# + colab={"base_uri": "https://localhost:8080/"} executionInfo={"elapsed": 25608, "status": "ok", "timestamp": 1604393639142, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="35r3S7MRzOMI" outputId="de2dcc9d-46f3-4884-ad56-8bbf3cf7bb04"
people_df_dup = people_df.iloc[[1,1,2,3,3,4,5,5,6],:]
people_df_dup.duplicated()
# + [markdown] id="tVtQRyOEzOMZ"
# > **Note**: The `.duplicated()` function marks the first copy as `False` and the second copy as `True`.
#
# To remove duplicates, `.drop_duplicates()` is used.
#
# With `keep=False` all rows with duplicates are removed:
# + colab={"base_uri": "https://localhost:8080/", "height": 175} executionInfo={"elapsed": 25594, "status": "ok", "timestamp": 1604393639144, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="5Zw0rfQEzOMd" outputId="201c2fa2-a859-4a65-979e-19c6d43c304a"
people_df_dup.drop_duplicates(keep=False)
# + [markdown] id="fK-34ZbJzOMr"
# ##### 8.3.5.1. Fusion with Keys
#
# To cross 2 tables with keys, use `pd.merge()`.
# + [markdown] id="Qzlw_lvwzOMt"
# ##### 8.3.5.2. Inner Join
#
# Recall that inner join consists of building a table that has matching keys in the tables.
#
# To see an example, we are going to select, on the one hand, planets with a bad climate, that is, that is not temperate or tropical:
# + colab={"base_uri": "https://localhost:8080/", "height": 457} executionInfo={"elapsed": 25584, "status": "ok", "timestamp": 1604393639147, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="mTQSVBbJzOMx" outputId="e4e93849-5c01-4f5e-cf75-f020bb5ce7a4"
planets_clima_df = planets_df[planets_df.temperate_tropical==0]
planets_clima_df
# + [markdown] id="Hf-A47YOzOM-"
# On the other hand, we select characters with blue eye color:
# + colab={"base_uri": "https://localhost:8080/", "height": 363} executionInfo={"elapsed": 25572, "status": "ok", "timestamp": 1604393639150, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="R2i1X6nezONA" outputId="9f8c2ef7-f7e9-4ced-8e87-7c47d91efc32"
people_eyes_df = people_df[people_df.eye_color=="blue"]
people_eyes_df
# + [markdown] id="emRgfLpLzONM"
# The key match field for the match is `url` which is equivalent to the `homeworld` field of people_df:
# + colab={"base_uri": "https://localhost:8080/", "height": 175} executionInfo={"elapsed": 25560, "status": "ok", "timestamp": 1604393639152, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="xgyJxqGyzONO" outputId="b8e0bd36-29da-4268-ce7b-d1522077264d"
a_inner = pd.merge(people_eyes_df, planets_clima_df, left_on=["homeworld"], right_on=["url"])
a_inner.head()
# + [markdown] eval=false id="ruhPBPl_zONc"
# We see that there are four blue-eyed characters who were born on planets with bad weather. The problem is that, with the crossing, the names of both tables contained in the indexes have been lost. Let's rescue them, before the crossover, with the reset_index() function:
# + colab={"base_uri": "https://localhost:8080/", "height": 175} executionInfo={"elapsed": 25793, "status": "ok", "timestamp": 1604393639401, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="td_FBxnLzONe" outputId="16ac4897-e540-417b-d175-6890b0b2c0ad" tags=["remove_input", "remove_output"] warning=false
people_eyes_df = people_eyes_df.reset_index()
planets_clima_df = planets_clima_df.reset_index()
a_inner = pd.merge(people_eyes_df, planets_clima_df, left_on=["homeworld"], right_on=["url"])
a_inner.head()
# + [markdown] id="AmT95E3HzONm"
# > **Note**: the `name` field was duplicated in both tables without being a key field. `pd.merge()` has included both in the final table by adding the suffix `_x` for left table (people_df) and `_y` right table (planets_df).
#
# The crossing has been done, implicitly, with the `how="inner"` option:
# + [markdown] id="iY86G6LSzONo"
# ##### 8.3.5.3. Outer Join
#
# To keep **all the records from the original tables**, both from the left table and the right table, whether they match or not, use the `how = "outer"` parameter:
# + colab={"base_uri": "https://localhost:8080/", "height": 677} executionInfo={"elapsed": 25781, "status": "ok", "timestamp": 1604393639406, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="7oUmkblRzONq" outputId="6c2beaa1-8d67-400d-ca2a-f7ef96ff1856"
a_outer = pd.merge(people_eyes_df, planets_clima_df, how = "outer", left_on=["homeworld"], right_on=["url"])
a_outer
# + [markdown] id="-CUnH-PyzON0"
# You notice that, now, you have not dropped any records from the source tables. In addition, you have padded with `NaN` the fields that do not match. This behavior in SQL is known as *FULL JOIN* or *OUTTER JOIN*.
#
# + [markdown] id="KKiuW75ABbL-"
# ##### 8.3.5.4. Left Join
#
# To force it to keep **all source values from the left table** and drop those from the right table that are not shared, use `how="left"`:
# + colab={"base_uri": "https://localhost:8080/", "height": 332} executionInfo={"elapsed": 25719, "status": "ok", "timestamp": 1604393639408, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="_3K9xv06zON1" outputId="64a424c4-f1aa-4ed3-da52-2e998d363528"
a_left = pd.merge(people_eyes_df, planets_clima_df, how="left", left_on=["homeworld"], right_on=["url"])
a_left
# + [markdown] id="1xYTlV9czON-"
# Now, he has retained all the characters.
# + [markdown] id="-fg10B6IzOOA"
# ##### 8.3.5.5. Right Join
#
# To preserve **all values from the right table**, use `how="right"`:
# + colab={"base_uri": "https://localhost:8080/", "height": 520} executionInfo={"elapsed": 25708, "status": "ok", "timestamp": 1604393639410, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="VkJAuAHLzOOC" outputId="b1a58aa3-0f03-4677-85ce-04e5035e9dc7"
a_right = pd.merge(people_eyes_df, planets_clima_df, how="right", left_on=["homeworld"], right_on=["url"])
a_right
# + [markdown] id="4QO10oDmzOOQ"
# Now, it has preserved all the planets with bad weather.
# + [markdown] id="7h-6c6p4zOOX"
# ##### 8.3.5.6. Definition of the Keys
#
# When the keys are common fields, it is not necessary to define the `left_on` and `right_on` fields.
#
# On the other hand, if one of the key fields is contained in the DataFrame index, the `left_index=True` or `right_index=True` parameter or both can be used.
# + [markdown] id="YZ8EUDhBzOOZ"
# ### 8.4. Aggregate Summaries
#
# The numpy and pandas libraries implement an extensive collection of digest functions.
# + [markdown] id="G06KsW3WzOOa"
# #### 8.4.1. describe
#
# To get a first impression of the statistics of a DataFrame, use `.describe()`.
#
# Before we add to people_df some new columns of different types and, in addition, some missings:
# + colab={"base_uri": "https://localhost:8080/", "height": 238} executionInfo={"elapsed": 697, "status": "ok", "timestamp": 1604393646855, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="WmvFkwRHzOOf" outputId="657b9c66-4241-4b03-cf68-bce55cc94bba"
people_dfSumm = people_df.copy()
people_dfSumm.loc[people_dfSumm.index[[0,2,4,8,20]],'height'] = np.nan
people_dfSumm.loc[people_dfSumm.index[[0,2,4,8,20]],'eye_color'] = np.nan
people_dfSumm["Alto"] = people_dfSumm.height>188
people_dfSumm["Fecha_hoy"] = np.datetime64('2020-09-06')
people_dfSumm.head()
# + colab={"base_uri": "https://localhost:8080/", "height": 300} executionInfo={"elapsed": 671, "status": "ok", "timestamp": 1604393646864, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="LTfqHwAHzOOt" outputId="f2da7007-9a1a-4b28-dcae-827a3383ae37"
people_df.describe()
# + [markdown] id="SQxmGmU_zOO3"
# By default, it calculates the following basic statistics on numeric columns:
#
# - `count`: number of reported values (other than NaN).
# - `mean`: mean.
# - `str`: standard deviation.
# - `min`: minimum.
# - `25%`: 25% quantile.
# - `50%`: 50% quantile or median.
# - `75%`: 75% quantile.
# - `max`: maximum.
#
# To include them all, use `include='all'`:
# + colab={"base_uri": "https://localhost:8080/", "height": 514} executionInfo={"elapsed": 654, "status": "ok", "timestamp": 1604393646869, "user": {"displayName": "alumnos bigdata", "photoUrl": "", "userId": "10710302360204190833"}, "user_tz": -60} id="bytcavIczOO5" outputId="2f43bae3-729d-4d23-b6ab-08143e235845"
people_dfSumm.describe(include='all')
# + [markdown] id="3cXmknWzzOPE"
# Now, for non-numeric columns, it reports the `count` and, in addition,:
#
# - `unique`: number of unique values.
# - `top`: most frequent value.
# - `freq`: observed frequency of the top value.
# - `first` and `last`: for datetimes, the first and last date are reported.
# + [markdown] id="JWdTMOdUzOPH"
# #### 8.4.2. Basic Statistics
#
# To calculate the statistics displayed in the `.describe()` function with numpy functions:
# + id="ZUecbUjNzOPI" outputId="e180e009-073d-43dc-83fd-957c935526b1"
def resumen_numericas_numpy(x):
return {
"count":np.sum(~np.isnan(x))
,"mean":np.nanmean(x)
,"std":np.nanstd(x)
,"min":np.min(x)
,"quantile 25, 50, 75":np.nanquantile(x, [0.25,.5,.75])
,"max":np.max(x)}
resumen_numericas_numpy(people_dfSumm.height)
# + [markdown] id="yRV43iwfzOPQ"
# > **Important**: The equivalent functions np.mean, np.std or np.quantile return NaN when the array contains any NaN values.
#
# Now with equivalent functions in pandas:
# + id="X7bvIUhOzOPQ" outputId="18dac0b1-1dbb-4187-f292-0f6792c01270"
def resumen_numericas_pandas(x):
return {
"count":x.count()
,"mean":x.mean()
,"std":x.std()
,"min":x.min()
,"quantile 25, 50, 75":x.quantile([0.25,.5,.75])
,"max":x.max()}
resumen_numericas_pandas(people_dfSumm.height)
# + [markdown] id="g9zpCcwPzOPX"
# Note that the default option in pandas is to remove NaNs:
# + id="akvpqcbdzOPZ" outputId="d4a4fcb9-7067-45dd-a2ae-f7878d80334a"
def resumen_no_numericas_numpy(x):
value, counts = np.unique(x[~pd.isnull(x)], return_counts=True)
return {
"count":np.sum(~pd.isnull(x))
,"unique": len(np.unique(x[~pd.isnull(x)]))
,"top": value[np.argmax(counts)]
,"freq":np.max(counts)}
resumen_no_numericas_numpy(people_dfSumm.eye_color)
# + [markdown] id="uZXNal3mzOPi"
# > **Important**: For non-numeric columns, None or NaN values have been filtered out with pd.isnull().
#
# Now, with pandas functions:
# + id="05HGqJkozOPj" outputId="92c32404-6f02-4744-e3b7-cbbd487b1b05"
def resumen_no_numericas_pandas(x):
value, counts = np.unique(x[~pd.isnull(x)], return_counts=True)
return {
"count":x.count()
,"unique": x.nunique()
,"top": list(x.mode())[0]
,"freq":(x==list(x.mode())[0]).sum()}
resumen_no_numericas_pandas(people_dfSumm.eye_color)
# + [markdown] id="hIoRiuQizOPq"
# #### 8.4.3. Tables of Frequencies or Counts
#
# ##### FREQUENCIES OR COUNTS OF A COLUMN
#
# To get the frequencies of a pandas series, use `.value_counts()`:
# + eval=false id="qXfFTHgkzOPs" outputId="34e02782-243d-4394-c0dd-b10b3780d8df"
people_dfSumm.eye_color.value_counts()
# + [markdown] id="sTI-tTz9zOP0"
# By default, it sorts from most to least frequent.
#
# We can later sort it by index with `.sort_index()`:
# + id="a94nPRoyzOP2" outputId="b48d07bd-02b7-4658-af01-3eb8003a2f8a"
people_dfSumm.eye_color.value_counts().sort_index()
# + [markdown] id="jjQk0CpPzOP-"
# ##### 8.4.3.1. Cross Tables
#
# To get the counts or frequencies of two columns, use `pd.crosstab()`:
# + eval=false id="FlbsDAMJzOQC" outputId="af3dd75f-76ed-418d-e328-a07ef9f3c2af"
pd.crosstab(people_dfSumm.eye_color,people_dfSumm.gender)
# + [markdown] id="uORXJsKHzOQK"
# ##### 8.4.3.2. Columns cutting
#
# In pandas series, you can cut a column with the `pd.cut()` function:
# + id="kTn6icSrzOQL" outputId="89e03664-db03-45c6-f641-48343d9f81c9"
people_dfSumm["Altura_Cat"] = pd.cut(people_dfSumm.height,[-np.infty,171,189,np.infty], right=False)
people_dfSumm[["height","Altura_Cat"]].head(8)
# + [markdown] id="zfFO7Zo1zOQT"
# Notice how pd.cut() has plotted, at intervals, the `height` column. With `right=False` we ask that the interval be closed on the left and open on the right.
#
# The frequencies of each category:
# + eval=false id="gauw9L8czOQU" outputId="d9e63203-aba9-42de-b947-12c72d3f52f0"
people_dfSumm["Altura_Cat"].value_counts().sort_index()
# + [markdown] id="G-dewkJCzOQb"
# > **Important**: `pd.cut()` creates a new column of type `pd.Categorical`. This data type contains .categories() and .codes(), similar to the `factors` seen in R:
# + id="vza3Ia6BzOQf" outputId="b08cd4e2-023d-4121-d71d-90cf675194ec"
people_dfSumm["Altura_Cat"].dtype
# + [markdown] id="CA_TMcu-zOQm"
# To retrieve the categories, use `.cat.categories`:
# + id="4RfBgUAkzOQo" outputId="39d49423-a047-46c3-9042-d826cb3f86ce"
people_dfSumm["Altura_Cat"].cat.categories
# + [markdown] id="QXrtZ4ZkzOQw"
# The inner codes of the `.cat.codes` values:
# + id="oKuyX-X1zOQx" outputId="8d67c5d1-189f-494f-da80-ee79ebadc1ea"
people_dfSumm["Altura_Cat"].cat.codes.head()
# + [markdown] id="sBRhAI0PzOQ8"
# To modify the labels, you can directly modify the category indices:
# + id="mYh6o6I4zOQ9" outputId="93440d76-582f-497d-be6b-00701d6decfa"
people_dfSumm["Altura_Cat"].cat.categories = ["Bajo/a","Mediano/a","Alto/a"]
people_dfSumm[["height","Altura_Cat"]].head(8)
# + [markdown] id="RV3EKBcCzORG"
# ##### 8.4.3.3. Guided Activity 2.5
#
# Let's see now, a summary of the height of the characters by the type of climate of their planet.
#
# The steps to perform are:
#
# 1. We cross the tables of characters and planets.
# 1. We build a new hatched column with a height of 171 cm and 189 cm at cutoff points.
# 1. We cross the height with the climate of the planet.
# + id="tfDxHPeEzORH" outputId="665f8e54-6980-4803-c466-b9d18fa6f904"
personajes_df = pd.merge(people_df.reset_index(),
planets_df.reset_index(), left_on=["homeworld"], right_on=["url"])
personajes_df.index= personajes_df.name_x # indexamos por nombre del personaje
personajes_df.head()
# + id="DGh9Gf6EzORQ" outputId="40595954-4bea-4248-d8db-38981f552299"
personajes_df["Altura_Cat"] = pd.cut(personajes_df.height,[-np.infty,171,189,np.infty], right=False)
personajes_df.Altura_Cat.cat.categories = ["Bajo/a","Mediano/a","Alto/a"]
personajes_df.Altura_Cat.value_counts().sort_index()
# + id="l6ZccKtQzORW" outputId="92c4b213-7918-4804-89ef-df14caaefe79"
summ_altura_clima = pd.crosstab(personajes_df.Altura_Cat,personajes_df.temperate_tropical)
summ_altura_clima
# + [markdown] id="jUl22nytzOR2"
# To find out the % of characters of each height that live in a temperate-tropical climate or not, use `normalize="index"`. In this way each row will add 1 (that is, 100%):
# + id="aioc6_pozOR4" outputId="967f7e2b-739a-4856-bb09-529224a3e405"
summ_altura_clima = pd.crosstab(personajes_df.Altura_Cat,personajes_df.temperate_tropical, normalize="index")
summ_altura_clima.sort_index(axis=1, ascending=False)
# + [markdown] id="Ww45ihT-zOR9"
# Gráficamente:
# + id="9YhJ37OCzOR-" outputId="08c9d538-db33-471e-97e7-0fb79cd37ea7"
summ_altura_clima = summ_altura_clima.sort_index(axis=1, ascending=False) # cambiamos orden columnas para el gráfico
summ_altura_clima
# + id="eFMRchBvzOSE" outputId="7e9389c9-9129-4403-b260-28d2e8e465cb"
g = summ_altura_clima.plot.bar(stacked=True,include_bool=True, alpha=0.75, rot=0)
g.legend(bbox_to_anchor=(1, 0.8),title="temperate or tropical")
plt.gcf().subplots_adjust(bottom=0.15,right=0.7)
plt.title("% characters in each climate by Height")
plt.ylabel("%")
plt.xlabel("Height");
# + [markdown] id="0vwVJpNnzOSM"
# Note that **70% of short characters** were born in temperate or tropical climates, compared to **60% of tall characters**.
# + [markdown] id="cfgrSrdCzOSM"
# ### 8.5. Aggregated by Subgroups
#
# In addition to frequencies, it is necessary to calculate more relevant statistics. For example, the median age (birth_year), the minimum weight (mass) of the characters, or the average population of their planets by type of climate and height range (high, medium low) of the characters .
# + [markdown] id="IHCa835QzOSO"
# #### 8.5.1. Grouping or *GROUPBY*
#
# In order to provide an answer to the problem posed, it is first necessary to introduce the concept of Grouping or GroupBy.
#
# The solution proposed in pandas is to define a division of the table using `.grouby()`. Once the request is defined, summary statistics can be calculated:
# + id="-N_xz14-zOSP" outputId="18bb4c67-d927-4f20-9b07-74b207b92a8b"
summ1_altura_clima = personajes_df.groupby(["temperate_tropical"
,"Altura_Cat"])[["mass","birth_year","population"]].median()
summ1_altura_clima
# + [markdown] id="_3P6jxNEzOSV"
# Observe how the mean function has been applied to each of the columns for each combination of the climate of the planets and the height of the people.
#
# Also, a new element appears. It is the ability of DataFrames to have an index composed of more than one column, in this case `temperate_tropical` and `Altura_Cat`.
#
# One way to get around multiple indexes is to add them as columns with `.reset_index()`:
# + id="l0qSj-ryzOSX" outputId="09cbe67b-f05b-40d7-a7b7-fee1666a76ea"
summ1_altura_clima = summ1_altura_clima.reset_index()
summ1_altura_clima
# + [markdown] id="Wek7fZXuzOSd"
# Now the 2 indexes have become columns of the table.
# + [markdown] id="n1T4FStTzOSd"
# #### 8.5.2. Aggregate
#
# To apply different aggregation functions to different columns, use `.aggregate()` or `.agg()` for short in combination with `.groupby()`.
#
# A first pass consists of applying several functions to all the selected columns of the data frame:
# + id="UFjizovvzOSe" outputId="badeb40a-57b7-43dd-c1a2-c968bc9f6976"
summ2_altura_clima = personajes_df.groupby(
["temperate_tropical","Altura_Cat"])[
["mass","birth_year","population"]].agg(["min","median","mean"])
summ2_altura_clima
# + [markdown] id="gFW6X1tvzOSk"
# Notice that there are now multiple column indexes:
# + id="KCNzs4ZOzOSl" outputId="f23dba6a-1cda-4399-e023-9b4da47ee622"
summ2_altura_clima.columns
# + [markdown] id="XbonJ2n9zOSq"
# One way to combine the two levels of the index into a single name of type `Statistic_Variable`, is by using the function vectorization function `.map()` (you can see more information at: [map](https://www.w3schools.com/python/ref_func_map.asp)). The function to combine the 2 levels is: `"_".join`:
# + id="VtjX_fH-zOSs" outputId="dae4e85f-b30c-4b49-9415-d51680a3237d"
summ2_altura_clima.columns = summ2_altura_clima.columns.map("_".join) # combina nombres columna
summ2_altura_clima = summ2_altura_clima.reset_index() # pasamos los indices fila a columnas.
summ2_altura_clima
# + [markdown] id="4KJFRCUazOSz"
# To resolve the question posed, we need to be able to decide which statistic we want to apply to each column.
#
# We can do this using a dictionary:
# + id="fEuZ3AvAzOS0" outputId="f8a756b5-961d-4abe-9b43-d47a272a89ce"
summ3_altura_clima = personajes_df.groupby(
["temperate_tropical","Altura_Cat"]).agg({'name_x': 'count', 'birth_year': 'median'
,'mass':'min','population':'mean'})
summ3_altura_clima = summ3_altura_clima.reset_index()
summ3_altura_clima
# + [markdown] id="AuqIVLPkzOS7"
# Observe, for example, that there are 2 characters of short stature who were born on a planet with a non-temperate or tropical climate, with a median age of 79.5 years BBY, a minimum weight of 75 Kg and who live on planets with an average of 200,000 inhabitants.
#
# In order to identify each row with a label, we are going to combine the tropical_temperature column with the height of the character:
# + id="xG832W5DzOS8" outputId="eaa3d229-2ce4-476c-98de-10ae92b1b7bd"
summ3_altura_clima["clima_altura"] = pd.Series(
np.where(summ3_altura_clima["temperate_tropical"]==0,"Clima Malo","Clima Bueno")
) + "-" + summ3_altura_clima["Altura_Cat"].astype(str)
summ3_altura_clima
# + [markdown] id="4PIaYN9EzOTK"
# To represent this Pandas graphically, we can directly use `plot.scatter()`:
# + id="psBCMEARzOTL" outputId="cb612beb-e476-470f-93a0-2ddbcfb60657"
summ3_altura_clima.plot.scatter(x="birth_year",y="mass",s=20*np.log(summ3_altura_clima.population)
,c="temperate_tropical");
plt.gcf().subplots_adjust(bottom=0.15, left=0.15)
plt.title("Groups by climate and height of the characters");
plt.xlabel("Age");
plt.ylabel("Weight");
plt.text(50,20,"Age")
for index,x in summ3_altura_clima.iterrows():
plt.text(x["birth_year"],x["mass"]+4,x["clima_altura"],horizontalalignment='center',size=8)
plt.show()
# + [markdown] id="7Up-TX3QzOTR"
# Note that the 3 Bad weather groups (white balls) have higher weight characters.
# + [markdown] id="mtZRf11MzOTT"
# #### 8.5.3. Filtered out
#
# Apart from adding `.groupby()`, we can use it, in combination, with the `.filter()` function to select groups from the original table that meet certain conditions.
#
# For example, we select the group of characters with the same climate and altitude range where the median age of this group is greater than 90 years:
# + id="-tl90DF_zOTU" outputId="54349950-b621-4a20-dbb8-4b4f0c758202"
seleccion1 = personajes_df.groupby(
["temperate_tropical","Altura_Cat"]).filter(lambda fila: fila['birth_year'].median() > 90)
seleccion1
# + [markdown] id="9Lhvm4XNzOTe"
# We see how the group formed by "Jar Jar Binks", "Chewbacca", "Ki-Adi-Mundi" are part of the same climate and altitude group and have a median age of over 90 years.
# + [markdown] id="yBnGI3yAzOTh"
# #### 8.5.4. Transformations
#
# Another utility of `.groupby()` combined with `.transform()` is to be able to transform the original data with aggregates, in a simple way.
#
# We compare the age of each character, with respect to the median of their climate group and height range:
# + id="XNer6Th_zOTi" outputId="873169ed-123c-4671-d293-08284ae91c92"
compara1 = personajes_df.groupby(
["temperate_tropical","Altura_Cat"]).birth_year.transform(lambda x: np.abs(x - x.median()))
compara1.sort_values(ascending=False).head()
# + [markdown] id="I7w590pSzOT0"
# Notice how "Jabba Desilijic Tiure" has a difference of 554 years over the average of his group.
# + [markdown] id="eh9ljA8-zOT3"
# #### 8.5.5. Application of functions
#
# Finally, to gain even more flexibility in transforming data, `.groupby()` is combined with `.apply()` to allow you to selectively apply transformations to certain columns in the DataFrame:
# + id="iL2nKl4mzOT8" outputId="128b9404-3aba-4bb5-e5ea-ce718e9efc12"
def centrado(x):
x['Edad_c'] = x['birth_year']-x['birth_year'].median()
return x
personajes_df2 = personajes_df.groupby(["temperate_tropical","Altura_Cat"]).apply(centrado)
personajes_df2.head()
# + [markdown] id="XLpSygszzOUF"
# Notice that the new Age_c column lets you know that Luke is 19 years below the median for his climate group (bad) and altitude range (medium).
# + [markdown] id="r6lP2TbOzOUI"
# ### 8.6. Pivot Tables
#
# `.pivot_table()` pivot tables are an intermediate solution between `pd.crosstabs()`, aimed at obtaining cross tables (rows x columns) and the aggregations performed with `.groupby()`.
#
# That is, apply different aggregation functions, on predefined row and column crossings:
# + id="mNSp812MzOUL" outputId="be67cdd7-dbea-4ade-e2e6-10b5f83b4f1a"
personajes_df.pivot_table("birth_year", index='Altura_Cat', columns='temperate_tropical')
# + [markdown] id="DsATkOSPzOUZ"
# This table informs us of the average age BBY ('birth_year') over the crossing of the altitude and climate categories.
#
# Pivot table, allows to create multiple index in rows and columns:
# + id="PooTwY6wzOUb" outputId="70ed9484-c39d-4afc-de7a-c194160b4377"
personajes_df.pivot_table("birth_year", index=['gender','Altura_Cat'], columns='temperate_tropical')
# + [markdown] id="-eBUXMxGzOUn"
# Also, it allows selecting which aggregation function is required at each defined junction:
#
# + id="6k1MHtTdzOUq" outputId="1990bd70-e819-4b11-d5b2-84834d3d734b"
personalizarDict = {'name_x': 'count', 'birth_year': 'median'
,'mass':'min','population':'mean'}
personajes_df.pivot_table(index='Altura_Cat', columns='temperate_tropical'
, aggfunc=personalizarDict)
# + [markdown] id="xFTChbQqzOVC"
# Notice how the same table as in the exercise with `.agg()` has been obtained, but now, with a different orientation of rows and columns.
# + [markdown] id="a-xhLCgMzOVE"
# ### 8.7. Management of Dates and Times (DATETIME)
#
# The library for managing dates and times in Python is `datetime`. In addition, the `dateutil` extended functionality library is also used:
# + id="9P_nlszTzOVF"
from datetime import datetime
from dateutil import parser
# + [markdown] id="TEQ5OyARzOVO"
# #### 8.7.1. Create Dates Hours
#
# To create dates:
# + id="77v8ce1UzOVT"
fecha = datetime(year=2020, month=9, day=7) # con datetime
# + [markdown] id="IzibXTlozOVg"
# #### 8.7.2. Get Components
#
# To obtain the months and days of the week in Spanish:
# + id="KVisUAhvzOVh" outputId="1a3d8e88-77ed-4ed3-d168-fceb0c00c49c"
import locale
locale.setlocale(locale.LC_TIME, 'es_ES.UTF-8')
fecha.strftime('%A')
# + [markdown] id="09EJOhPwzOVs"
# More information about other components can be found in the [strftime section](https://docs.python.org/3/library/datetime.html#strftime-and-strptime-behavior), or in the [datetime] documentation (https://docs.python.org/3/library/datetime.html) from Python.
# + [markdown] id="-AO1V8QuzOVu"
# #### 8.7.3. Datetime and Numpy
#
# We can define arrays of datetime64 data type:
# + id="QJ0CweW_zOVy" outputId="953cf9c0-4213-4993-b67a-d5132b99082f"
fecha = np.array('2020-09-07', dtype=np.datetime64)
fecha
# + [markdown] id="cF4TS4ztzOWI"
# Now, we can optimally perform vectorized operations in numpy:
# + id="YTYeB6eKzOWL" outputId="07afb21d-6ecf-422d-a98a-9ca18a8a22c9"
fecha + np.arange(12)
# + [markdown] id="2lhSBaQEzOWV"
# Also, we can declare datetime with date and time:
# + id="dHjjMI5WzOWX" outputId="7e04cac1-e25f-4677-c1d0-a5330d35639c"
np.datetime64('2020-09-07 12:00')
# + [markdown] id="m6mWlNWTzOWj"
# #### 8.7.4. Datetime and Pandas
#
# To define dates:
# + id="0gz8KdBXzOWl" outputId="aac3d3ea-e810-499b-b479-908929eac2f7"
fecha = pd.to_datetime("2020-9-7")
fecha
# + [markdown] id="SzmL7ZsrzOWt"
# Turn into:
# + id="NL1y64wDzOWv" outputId="14a7776f-d67f-41d1-e81f-8567ea49c7d9"
fecha.strftime("%A, %d de %m de %Y")
# + [markdown] id="AaCbm0_8zOW4"
# Vectorize operations:
# + id="N8U1GjlozOW5" outputId="2d968f4b-c1de-4ef3-e119-d73dd643de9f"
fecha + pd.to_timedelta(np.arange(12), 'D')
# + [markdown] id="ot2NN58MzOXB"
# ##### 8.7.4.1. Pandas with Datetime Indexing
#
# Combining both features is very useful:
# + id="yqisnWqIzOXC" outputId="f47fbcdb-f458-46fa-f413-165caf122c5c"
index = pd.DatetimeIndex(['2019-08-07', '2019-09-07',
'2020-08-07', '2020-09-07'])
fecha = pd.Series([0, 1, 2, 3], index=index)
fecha
# + [markdown] id="6Z5QlBgZzOXL"
# It can be selected by ranges:
# + id="FkF6br79zOXM" outputId="58a2e3a3-cc12-448a-d63d-50482edbb1f8"
fecha['2019-08-07':'2020-08-07']
# + [markdown] id="TAbBCN-UzOXo"
# Even just by year:
# + id="RU_lyuhdzOXp" outputId="d41ef070-a2fb-41c4-9420-34bb50c47af6"
fecha['2020']
# + [markdown] id="apUPvWG_zOXx"
# #### 8.7.5. Panda Time Series
#
# To define a pandas datetime series:
# + id="iMmRgxPSzOXz" outputId="86beb82f-3ed0-4f02-b392-6480918d0473"
fecha = pd.to_datetime(['2019-08-07', '2019-09-07',
'2020-08-07', '2020-09-07'])
fecha
# + [markdown] id="9QzhLsJIzOX6"
# Convert to daily rate:
# + id="XtJ5R8kDzOX8" outputId="36d02536-0412-4fdb-e3e7-abb84a70f5f4"
fecha.to_period('D')
# + [markdown] id="BUFt2ZGGzOYE"
# subtract dates:
# + id="cwv0U2lBzOYH" outputId="45fb4e21-8882-4978-b5b6-08d204705766"
fecha - fecha[0]
# + [markdown] id="cA0uGSVHzOYQ"
# #### 8.7.6. Date Sequences
#
# To define a sequence:
# + id="KV2dOQgyzOYS" outputId="2e318ab2-42b5-4eea-eb47-be3ec1cfecdb"
pd.date_range('2020-09-03', '2020-09-07')
# + [markdown] id="mt1MPipXzOYb"
# Or, specifying the length of the sequence and the periodicity:
# + id="cBKBnI_azOYc" outputId="30ab4156-b64d-4147-b45f-a257c8c464d7"
pd.period_range('2020-07', periods=8, freq='M')
# + [markdown] id="WW-1HJCWzOYk"
# #### 8.7.7. Tables of Counts or Frequencies
#
# To generate frequencies use `pd.timedelta_range()`:
# + id="X_WUBkODzOYm" outputId="0bea5c86-feef-47af-e3c6-eacbdecf4e29"
pd.timedelta_range(0, periods=9, freq="2H30T")
# + [markdown] id="YcRqpnYnzOYt"
# You can learn more in the ["Time Series/Date"](https://pandas.pydata.org/pandas-docs/stable/user_guide/timeseries.html) section of the Pandas documentation.
# -
# < [Flow control](modulo1_tema4_Py_40_contr_flujo.ipynb) | [Table of contents](modulo1_tema4_Py_00_indice.ipynb) | [Anex](modulo1_tema4_Py_60_anexo.ipynb) >
#
# __[Open in Colab](https://colab.research.google.com/github/griu/msc_python/blob/master/modulo1_tema4_Py_50_gest_dat.ipynb)__
| 51.735232
| 6,566
|
1ad1396c585f6941d5b0534cee1d23b3ac7b9897
|
py
|
python
|
docs/use cases/Workflow Single Model - Gredient Booster Classifier - Diabetes/Workflow-Single-Model-ReadMe.ipynb
|
em3ndez/MLW
|
['Apache-2.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <h1>Workflow Single Model - Diabetes Usecase</h1>
# How can We create logical chain or Model pipeline so that We can perform sequence of tasks along with Model scoring and or training. For example, How can We build, train and or score single Model to perform list of activities such as</font><br/>
# <ol>
# <li>Process an input data set and create runtime features like feature1,feature2 .. feature[n]</li>
# <li>Score and or Train on new data set with Model [X]</li>
# <li>When Model [X] finishes its scoring/training process, <br/>We want to do some process like creating some output data file or aggregate some data set fields.</li>
#
# MLW has the Pipeline Model feature named as "Workflow Single Model". We can associate scripts as pre-processing and post-processing to the Model so that We can create logical Workflow to perform as Single Model or Pipeline.</font>
# ### To jump start, Use UMOYA (Repo).
# UMOYA does versioning resources (like model, data and code) and manages its dependencies for ai projects.
# MLW Solution Team publishes Data, Model and Code for this Usecase on UMOYA (Repo) so We can easily grab them all in one click.<br/>
# <img src="data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAAA0AAAAKtCAYAAAAOzg6qAAAAAXNSR0IArs4c6QAAAARnQU1BAACxjwv8YQUAAAAJcEhZcwAAEnQAABJ0Ad5mH3gAAM62SURBVHhe7f0J3BxF3a+Ns2RjTQQUgUDCDmFJWAwEhERBUETDEwVEwLCvYoBAiGxZQQQ0YROIQEAWWQJJ2AwogiBBcAkgArLFBR/0ec85nnP+7znved/zPKf+/avunqmqrp7p6el7pqfnuj6f7yf3dHdVV/f0ndSVqu5eQwEAAAAAAPQJCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCBAAAAAAAPQNCFDZ+B+vE0IIKSL/8T+iv1gBAADqIEBl4z8vVeo/PUAIIaTdIEAAAOABASobCBAhhBQTBAgAADwgQGUDASKEkGKCAAEAgAcEqGwgQIQQUkwQIAAA8IAAlQ0EiBBCigkCBAAAHhCgsoEAEUJIMUGAAADAAwJUNhAgQggpJggQAAB4QIDKBgJECCHFBAECAAAPCFDZQIAIIaSYIEAAAOABASobCBAhhBQTBAgAADwgQGUDASKEkGKCAAEAgAcEqGwgQIQQUkwQIAAA8IAAlY0eE6CPFo1Xa6yxhlpj8jnqI896ycqZwfpgm/kromWvn6OmmJ+t3KrunRxsP3OeZ908NT9tX1GdUxbdGu3vaLXS3SZDdFnvsUTt8tYbtcvb5hay4ujG7dbrpQ1RG5ttP2CJz4URzznT57Ldc5JIuG/5nv3rM6Z2LtPPX3xtt72vPG3W1/N4de/rnnV5kqgvuGbN76zh72QPBwECAAAPCFDZ6DEBCoVhvJrilYYgUcdrSrBNrfOlO5/pnbu0jnMoNv5Oa7hOljcSqObRnV7PsdREz9PucF37ndXwXKacxyDueWm2/cAklD27M99IDgtOQR318HsOrsvUNkfHGWzTthTkaHN4TQ3U+fQIWddkeoCDAAEAgAcEqGz0lABFHamZR6d2JKWTPn+FdCbr6+uykty+tt4VmKgT6RWgaF3YoQs7rnk7rX4BCuv0C1C0ru2RjmajBFGHvLa+PdHLG//5kbjtG6A0keesCa+xo4M2++uS45yyaF5wjtvfV542D6zcJn9H0r/XHg8CBAAAHhCgstFLAlT7n23pUHmERjp+QQddd65qHfXmHXdf5y+WpnulLmdfuv7aMqctljg13q/ErstcFnRgF3k6sonObXR8tX265yXufNa384pbrd3j1U3flf0bdepjCLe3hKPRsXo64eE59SxrcI585yeOVTaxv/rxhfsN44pqWH+03jjOeLvk/pudb1/ic5ciOHIe5fqTY/Beh3E8ZZ32SLt956xxPY1/R9LPQXJZeH0Y9envJd6vZ715DaXsv6eCAAEAgAcEqGz0kgDVOrnSofR34uKOfr2j7um4O9EdPLPjGXXKdJmUjrW3vqhcvZPdfN9h/WZHMixjdg7r9RkdR3fbqHzY0TXq03U4UwIl1nEl6/GfE6OOqGPrHmutDnd7Wa+ngLnn0mmXm+gcuB16N2nfodlGtyPvfpZzJ+fJ/L71+XTPt7Efvd6RlkSitsTXZv2chdH7Da4Re1/Rd23UHbbXPF/u9SCfj1bzffU0OO74uNx21eJeo3I8+rt0ltU+2/UlvptovT7X8fLaOYq36dGUUIAmTZpE+jAPPfRQdAUAQBlAgMpGDwlQvePm6UhKJ006U25HKkPHyu2g6Y5o3JmzRMHXeTTidhSDJDt/TpwyYf3R/ty2O23xd76dzqwukzz+eruiTn2twxzG7owHsdoZdWBdsfNsY7YjMQIi2zc6N3Gi86BHCSwBqMdtr3Ue4+3M9uk63bqic1FrU3idxcdpXRdxMlxf4X7DfUkd1nnT5aVO+5z6rzO7Pd5t9L7cbfzHWWuH91wYcdZLne4orN5PfP6d7RPXUnTO7GvOaVOvpoQCJNfDtGnTSB9FvvM111xTPfzww9FVAADdBgEqGz0kQGZHSn6udzqlYxh1uIzOpl7nfvbE6khGnTNbIOLyTTppcccu0XFtkEb1W22J/ie/1mkMt012vDN0kmt1yT0pbkdUkjxOfe5jMdBt9hyj1fG127dypiwP92seT7L9jRLWGYqQuX/7mCWJTne8LCpnHU8t8XmJylnnP3lOwqR9D/Xo7yDal/zstlOX9XzXyX2Zx5WyTcNrJo5d1n+NGLG+Vykr28pxm991/HMQ6/rwtNP3O2m1u4dTUgH68Y9/rD744APSJ5HvfO+991Zrr722Wro0+DceALoOAlQ2ekaA7A6odARrnSrpUEWdPLdjqz836twFMTuAbnmzY9a0oyiJtk920lNidAaT9RvH7HYafZ1It0zwud5hTm4TtlHSrDNqd6QT5yiO1Saj4yv16e3DZbpevY8M58ebqP3xcSXaa5+DMOYx2MfjbmNdV/Hx6J+DMilJ77jb+9LfsdluQ4xq333ieOIYdaVto5dHbY628bVXYl0jvu+zlvB86n3JedDtl2XGuTHK2/UZZaP1+lid/WX63eqFIECkBJHv/I477lCHHHKIGjx4sFq2bFl0NQBAt0CAykavCJDT4ZMOU9iBk06h3emud2yjzw07d0F05zbofPk6lbVlvk51g9Q6n006dXo7ab+v/niZTB0zjytI3ObatlGsY0h2PnWizny4L89+9fr4nErsevxSlezEynZSb/27CpdJPeYyf8LvLm0ba19ue2vntL69fZwpdVvnLtpHfO2kne+mcc6v1BOdu/hcxD/X9uVtv8SoK2Ub67yk1mOm8XkOE24jba23WZaFdZvHEW9buz48bdDH6lw/1vH3chAgUoLEAvT73/9eS9DQoUPVo48+Gl0RANANEKCy0SsC5HRydUdPOlHSwap1ptzOvPs5JVHdU6Tj5nbCdAcuqENu2G7QAbY6nnGisvXOoScN64/ar9c5HVlPx1KSpSNtdeyDuJ1Pd71bT2K9TvJcS72hvNWPK1x2TrBt+rmMo9vldJTjmG1w26M/u/U714+v7rBcyjbec+l09n2Jvt/aNSDt0G0NzletzW494bl0rxvruNx6daJ6avX669HH1WQbO2G9+j8BnDbPD75L+51cdn3J7yJcb/9OZjiPvRIEiJQgpgDFErTOOuuoxx9/PLoqAKDTIEBlo0cEyO60BYn+J/2jRUfXO6VOJ9ffSfRElwvq924bdthkXUORiuowt/F2xN1EbfTXH3UMZX2ic5jsNLod+LBNyf0nOv/OefOvN+pJnNeoLVZHOGpPIG91QQ3r1kKXOB5PPOfUXB7v321vov1BwrY410/iXAXlascZHlN938nzrffT5PtNXANy7qQdwf5q589zneq6E+1tfM7D9njaaGyTuEb0vo3PKZF65Htzz4e9LIhTX+K8e47VL0U9GgSIlCDynZsCFEvQuuuuq5588snoygCAToIAlY2eEKBk5zPuSJmdpkRns9ap9cXT+TU7arXEAtS4o6uT2F+GMtFx+LeNjju1gxqv9+8v0fnUafY/8FHH1lif6IxLau2O4giHJPw+PB37DB3ueuoCWo95nG57mx1fvKzePp1gnW5bvI23o+6cb+/1Yid57qLjMduir5vkOQnPVRzfObPPTfIR8GHsejzXSIbrNPm9xefCKauPxVhW+52IyrrrJfpct3JNlDgIEClB5Dt3BSiWoPXXX1+tWLEiujoAoFMgQGWjZx6CQMhAxidOhLQYBIiUIGkCFEvQhhtuqJ5++unoCgGAToAAlQ0EiPRbPCMOWUdCCGkYBIiUII0ESCISNGLECPWzn/0sukoAYKBBgMoGAkT6MNb0Nx3khxQQBIiUIM0ESCIStNFGG6lnnnkmulIAYCBBgMoGAkQIIcUEASIlSBYBkogEbbLJJurZZ5+NrhYAGCgQoLKBABFCSDFBgEgJklWAJCJBH//4x9UvfvGL6IoBgIEAASobCBAhhBQTBIiUIK0IkEQkaNNNN1UvvPBCdNUAQNEgQGUDASKEkGKCAJESpFUBkogEbbbZZurFF1+MrhwAKBIEqGy0I0CJ9964700ZoDjvErHe31Jk5GlhTet13g1TS5ffa5Kp7e0n08MEuvGeF/d9M7oNRhsHvE2+9xcF8b07KEdbku8Xyp+VMzvx3dTPR+rfEfF3VMRx+d431CSF/D2CAJESRL7zVgVIIhK0xRZbqJdeeim6egCgKBCgspFLgFJeghh3YAa4411k5y89YYetqdBFx+xu193HKmdse1uJOrTO95B8YWY5HjGt2zDg14yR6D8H3O+gmGvX/8LTPOnYeYkkb0rQ7rTrMjw3wTYF/P3R+nkOr+e2zykCREqQvAIkEQnacsst1a9+9avoCgKAIkCAykYOAWrYudAdv4H8H+VIvgZ6dCPr/8qnHW/W8gORAd93o+8gua6YTn876dA1YyRd+oqQ0+IEV383nTgv0YjM/GB/XsmQaza4RlYG5619scvxfevfmQLOKQJESpB2BEgiErTVVlupV155JbqKAKBdEKCy0aoANRWcsPNR78TUP+vOVvAXc61jEnU69DJzuZnof9J1dCfa+Z9aT3tq+9HxtbXegTS3jTs/Yec1udyX1I5uioQ0b1v29te2MeQive1Rp7C2zm1zfE7q26V1RNM79771ns6o+7175Kjpd9Dw2jGuEfP6keh9ZWhT4lrMfn5qx+w5Lon+fmv15zg/8TW/wtjOt68M58i7D+ec+Y4zy3VsRm8f7MMvOHIOpA5pk1NXhrZY2+jjML7/eJtm3298TpscR9MgQKQEke+8HQGSiASNHj1a/eY3v4muJABoBwSobLQkQJ7OWtOEnZrE1Jeo01LvpHg6LW6nJPg8ZWYQY5nuKNc6b3HntN4593bWjc6QLT317exOanrijp3d+fSdp2iZsW24T7PTlWUb9zwl95Vse1jGrDfRbn1O5Htq1gn0HVujhPuufffRuU+ImVFf4jtzr4NEHc45iY6ltr0ub14DTpuyXIuZz4/EU96Ife7znp9kHdb+Wjgm83jiutPbk7x2Et9XIvVj0Nu61460Vepzvqew3uT3njxO+7t2/47Ici70vszfh7xBgEgJIt95uwIkEQnaZptt1O9+97voagKAvCBAZaMlAXI6a1ni67RE9bgdIbsj5emwxR0po5NkdvYTHSadlM6Ou53V+Qr3Y+/bl+g4PHHL2scWx95Plm18ndZmbdfnyK3X7dhGncTm322L14DTXvP7iqOP2+h8Jrdxjsk63jBWHc56t367TVmuxSCZz08Qva17HdZjCUTe89Po+Fs6JuNzdE24167VXu/vRrNrIlyvy8g+ze8iqk/KJs9Lsk7fcdptCeurH1e2c+E777mCAJESRL7zIgRIIhK03XbbqVdffTW6ogAgDwhQ2WhFgJzOWn2ZdDjMGNv4OoNpHUSzQ+Z2zqKkd8aijk+iE5PssPk6O3qZ02lq2tlN6aQl25JsQ5x6W7JsI5+jDp3v/Bnr623ydRKT2yU6x2nxXQMNkqVe+zuNy5jH7KR2zfnrtTvJnusicZ01uRaDZD4/QZpta32fKde5Gfv8eI4niLXPVo4pcd6TbXG30+3xXlMpMa8Z+dmoS7dJH4t9XO4+azGPIeXcWecr07lI+x3JEQSIlCDynRclQBKRoO233169/vrr0VUFAK2CAJWNdgXIibcz63Rk4g6UP2GnxK3HKht3/qKOsO7E1zrF/tQ7N77OjtOpzHCcOmmdq9q6qINlttPazthvlm1qy8JjCI/N2b/bdt2OeNtk4v1Z57VRsp6bKO73mPrdO/sOO+P+dTrR+Qq3MTvB7vlKft9mm1Lbo1OvN/P58X5fZmzxbP382OXjmL9n+Y4pare3TBDnd9HcRzNxCL9L83chboPsM7qWrOs//RyadbnnLo55XJnOhbXvNoMAkRJEvvOLLrpILV68uLB86lOf0k+HW7lyZXRlAUArIEBlo9ApcFHH3OlUuR0ku+Plj38bpzNrCkjWjrl3O1+99Y5iUiLCdVbHzk1CgHxtM/abZRtreX2dJUHetqe0sZawHut7TTlm77ZWzPX29+8/X42OLy4T7D/teok6r277avVF6+vttTvXWa7FPOcn7Xjs78NuS6bz471O8hyT+7vpfs6WWDAaldPb1ERFjic6RjkXcTv1eYmPK70tZl3+47TPV6ZzYX0nbQYBIiWI/XdTsdl9992jKwsAWgEBKhstCVCTDkWis+npOAbRHT33f26dsnanKUrU6Yy3sevx78utx9vJtDpfKftOxO50urHraHAeam3JsI3TzjCeDp/Z9gwdZv826dH7yHHc3nKejq+7jXkOvN+fee24x5Lo3Npt0vW537VzLbZ0frzfURz3O271/MTH79TvtDf7MZltybJ/+1qLY3/nbtzvVOoI6zNfwOp+r962OG327le3t7Vz4d0mbxAgfx6eHrTjULVwpbN85UJ1aNC+Q6972V7+wRI1PVg+/WH7c9wJ1zlvibH9B+rl6w4Nlk9XS4xldqI6nHJ2PPuppVHd/ZFp06apiRMnRlcWALQCAlQ2WhSguPPg76TKPxJG50xv63TWasvNzlfYsbLqdDoytf0a9bmdJLdDFLbJ3r+vY+V2gLydr0T8nUFJ6n7NTpZ7fFm28Z17q4Pqa3tSKvQ2phToOhypaJSUayBRr/P9h+fFXS9lnPZb585pf3ROzPNu1esci/vdum2K29D8Wsx2fhLnoBbP9ZL7/Jjt80hjlmNyz4PEuZbieuptjvZlHV/674FOoi1Sh+wjKNfoGNy2pH4vnt8PzzE0Ohf6nJr1thMEKCWu0IQRaTn0sEBcDluoXjaWh8IUCUckSXbZl9XCw4LvzSinBSio61C3rihLzpN1QZkMAuS2U7LkPHt//RgECCA/CFDZaFWAdOKOkJ1EJ6hRx7HWWUkpGyTsEEbbSCfWqi9sg1uu1kHUcfft66wlO1/1/aa0XeK0347TsYxity3nNon92tv42+58X6YQxGWcZc3juQacTmSiQ58oE6zTx2Mfg30Ogrid06jjW099H+6xJDq3vmuyybWY/fx4zkktzj4libY0Oz/hemlf/XsO4uu8Nzmmmgi465xzWxeHOMljTNZtRNfn+37N4/b9XgZx2uLbj3UeEn9HRGl4Ltx9h5+Tx50xCFBKQmGxR3pk2aFq4cMiOPboUCgzoWyYP9fLBtFiVC8Xb7cwEJ3ESJOIjV4XXAM5BcjdXz8GAQLIDwJUNnIJECGEVDMfLTo6+R8PWYMApSYhMlooZJQnlKO6dNiypMtlmH5Wq19GjzzT46Q+PYpThABFbV+o2xbUWTuuaGRKlulUa9ocAgSQHwSobCBAhBAS5VZ178xWR0ONIEDpMae1BdHCEsmILSauhISfY6lI3i8Upi5Ysr0pHtFIUyAu+QUoEptYdLQAuXWFZc32ZZW3XgkCBJAfBKhsIECEEKJjPpghVxCgBhFBqE8hExmpiYYpR44oxQlloi5CElNU6gLk1C2yYizPIkDufnRqozxBIgGyREnabW6jY49m9XoQIID8IEBlAwEihJBiggA1jAhIKAPuKE1djsyRofTEolIXKlOAtIxEdcjyWEDyjwA5MafDRct8ghYHAQIABKhsIECEEFJMEKCGqclNYrSkPlJSlyS7bDKhrMTbWgJUE6z69DfZxhQg/bMhKaH05BcgXV9iBMhIPG0uTqNtSxoECCA/CFDZQIAIIaSYIECNoyUgfHiAKzmhHE0PBMQUC1ty7NjTy2wBCoVk+nXB/pxlAzsCVJ37fXxBgADygwCVjT4TIOuxuVbanPvftdQfjexfL6k/8jgR3yOU206wv6aPje50m7qVLN9PgXEe+SwpbN++R0w3TZZroUJBgJokFAzvS1H1vT+yzpaIUCw8UuLcK+QKUFzOlKeBFKDasZn1R6M+TevrkSBAAPlBgMpGXwlQ9A4TTwc7FKNWO3clSNThbfjeEs+7WMKEElJs5zxjh7+jbeqT+M5pLERdkcoOy18ZggA1TfpUMY9AxHGnj0mcOlwB8knKwAqQJBa8eqoiPxIECCA/CFDZ6CsBatC51h3FHhwFShWJehrJXeJFoW0nPMfNXiTZ2Tb1QxrIRoZrZGCS7VqoVBAgUuEgQAD5QYDKRj8JkO4IpnTILAGKO27RiFHwc61jGdURJ9v/btfrCZPsjGohkKlC5hSmxNQhux45juYjV1GZlGlICdnIcHzhm/zjGMeS+dxkb1PtvNTqjo/VPae+cxB+j7VtPPtLPZZamtXReL37/WT7ns06wzbVylnbmYnOh1cc4+u5vixsV7wPzzrZl3XOwzrs34NgedpxZL4WKhYEiFQ4CBBAfhCgstFHAuR2RlPXRTI0ZbLdIY47jbXOYtT58wpVLcmOqa8dtY54rRPpdDgT9cjno9V8V2AScetJXxe2yzjm6PjMsrqdRkfXPRb9uWFHXZK9TbI/+R7sYwy3MZeF5888p+4+fOfPLpP8XprV0Wx91K5EO4Okfs/O59q1aNfjS9j+rNsZx6llpf69e8951I54m9q+nOMwy2S7FioWBIhUOAgQQH4QoLLRNwIUdU59HbLof6trIuN+lnhkQKI7tI06efH/lJvLnM6kr/MYtzfeX7JzHiRqp9smK75j0XHOR3R87nZ2J9ZuU5iw7XE5t8PvTdY2pXxn/nOeoR3GdxGeT1tw0+TD2sb8PputT5yv5t+zr92xbCTPVzLxtjqJcxTE12arDf5zbh9X1E7v9ehs0+xaqFoQIFLhIEAA+UGAykbfCFDU+fTG7hD6ZMO3rLbc19FsFN1RNPbp65RGQmJ2SusdaXubRh1jq0Psxuicph6Ht+PraYtOSjudZG1TQkiMZWnyFG6btk2cqJOf6Jy77Y+vGVcY4mRbX2tHs+857ft0r5cs0WWS57T59Rq22f0O7XIp37N1rWS7FioXBIhUOAgQQH4QoLLRLwKUQRbiJP/nOuowxx1KN406lGZH1EpdKkIhcOTK7PSmtd3XoXaij6WpoKUJgb9tsQRJ7A5uM/EIk61NQXzHlyoDRsc9dZso0fm0v496fMcUrkvfr3e9046m33Nau/Vyp1zW6LLxd5L+PdfivaaccmnXndXObNdC5YIAkQoHAQLIDwJUNvpEgLydT298Hbec/5sddbTdTqArAEnhctqb0uFsfkyGFHjXx0k/vkayEotQrVxax9hK1jZFx+fuO00GzHPdTBgytdNNLDpp5ZLr3e+n6fec0u5G34FOw+Mx5aX5dew95+53ptuZ3J/VzlznuAJBgEiFgwAB5AcBKht9IkBNO5FxUjpuvs5rWkfQXu92aF0B8AmB73/c/VLWvGOc7X/hvcdnlfe10z6vzYUsSCFtatL59n0vVrnwWNw2NK3DPAfN1gef7fZn/Z79dSbOgxX/8YQxpcfZXxSznVnOefg9+9sZtyHTtVDFIECtpfbyU08avq+HdCMIEEB+EKCy0RcClKUTGcUrLfFyo9MXdeRdIbASbVPvmEYdULNcg05vopwhO7qj2uSY/B3VlCQ69O45i9tunptwm/hY/KMHdrK3yey4J5ebx52s026X7/u3ZCdIoo7ou7POr3mOmq1329/K95w4577zYCe8Htz64+X17yw8TvdzSpvjOL8ToSTJEwiddhrnQtfb5FqoZBCg1qIFaLpaklgXvlD00OtedpaTbgYBAsgPAlQ2+kGAEiKSnoYdN90RDDukksz1WWXsTqbbIdVJyIgk6sSn1OOL28lvGuf4knXXO+TebWrlkx3xONnb5MqBGbcdzvnTsc+X7zyFctCgjlhyanGOq+F6u/35vudg3QqfOKXE+f50TEGLYl+TZpv859z+nTCuO3N/7n4yXAuVDALUWlIF6AP18nWHqjUOW6he9qwj3QkCBJAfBKhs9MtDEAjpwXjFqZtp4T8T+jIIUGvJKED65/MWqoWHhcJdGxlauVAdqkU7SmLa3Mu1MmEOVQtXGuublNf7NdbbI1LhKFV9vf84qhQECCA/CFDZQIAIKUXCqWXmSEo4ItNolK/j0SM7JRKysgUBai1pAhSJSSwcsYhMf9jYJrp/qL4sEpKaxETyY4wihfVE+9PlTSGKto/Lu23TbYq3D7c1hWjJeea+qxkECCA/CFDZQIAIKUnsaXuSUslPEHs6HEkEAWotkcT4YsqFJS5RvMJhSoslLMlIeXtEJ4hRxrfPevrzHiUECCA/CFDZQIAIIaSYIECtxR1lSYmWEet+oOQITBgRk0h6GtbtTo0zE0uTPcXN3Vc8KhWm+TFUIQgQQH4QoLKBABFCSDFBgFpL0QJkjvo0rLu1ERxTdvzSZYuQHp2qLXOm7vVwECCA/CBAZQMBIoSQYoIAtZbcAtTuFDjnfp+M8bWjnlCEqiI7viBAAPlBgMoGAkQIIcUEAWotbQhQWDbDQxBMyUmMENnCEo70hO0xfzbrC0eAPCNIGY+ll4MAAeQHASobCBAhhBQTBKi1tCNAEi004TQzncSIjnuvjzMiFElQPXZb3KlsSZky16eNNlUnCBBAfhCgsoEAEUJIMUGASIWDAAHkBwEqGwgQIYQUEwSIVDgIEEB+EKCygQARQkgxQYBIhVNKAfo//05Ie+kQCFDZQIAIIaSYIECkwimlAP3fv/b/LhKSJf/+X6MLaeBBgMoGAkQIIcUEASIVDgJEKhcEqI9BgAghpJggQKTCQYBI5YIA9TEIECGEFBMEKEzi8dJxko+89r7QNDVFvWx0iVpS4ReWDlQQIFK5IEB9DAJECCHFBAEKk/J+n/Dlou28L6cIASpKovovCBCpXBCgPgYBIoSQYoIAhWnwglM94uN7qWmmIEDdDAJEKhcEqI9BgAghpJggQGEaCNAHKxeqQ41RoMQUOM/0uUOvezlaH8nLdVJHfX1CZpw63PK1dcZ+dTtq69xRKqdc2rHpNGmjbptTv3NOyhoEiFQuCFAf8z/fIqTUef+NJ9RvXrjHu46QUgUBCtNIgJwRGEuAPCJgT5uLRcSo2xGKcHtz32GZhETVpOlltfAwow0Sq85wfb18s/uW4jYax2HV57YnanPuUbHOBQEilQsCBABlZfHixfofXgBonZ4SIF8sKXLlJUy9jlBW3PV2e5w6dP3JttbrTApL4/i3N49T/1wTnqRglTUIEKlcECAAAIDq0ZsCFG4TTx+zR0+MkZUo9REUt5yZFAHSbfVtHyRqVziqFC9PO644Tv1RrFEec0SoR6a/SRAgUrkgQAAAANVDOu3luweovs4SIL0uFI3aiEhiBKiBAEXlEyNAVmxBCeWmmdTEMQUrrUwGATJGfezl5Q4CRCoXBAgAysrq1avV7Nmzo08A0AplEyB3xMf87JUBc7QkRS7qdYTrG08nc+qw6s8afzvMdY2mwEn0sZ63sGemv0kQIFK5IEAAUFZWrVpVvn90AXqEMglQONpiy0ZCgMxytREhW4CsbRyBSd2HZ/QlXB9+tsXLlBiP0DQQvHobjTb4JCtxbOUPAkQqFwQIAACgenRPgKRz7yYpDe7IiP5c217kICkj9iOmkwIRSpBRjzOqVFvvSJFZxhIeY2peGHOfrlBla6NfvModBIhULggQAABA9ZAOeMcFqK8TCZB3epwZV5zKHwSIVC4IEACUGfmHd8GCBdEnAMgKAtTpZBQgPaqUNo2unEGASOWCAAFAmXn22Wf1vUAA0BoIUKfTXIDiaX7NR4nKFQSIVC4IEAAAQPVAgEhRqb4AzVPzg98X+Z1JZOY8z/YF5fVz1BTfPoNMWXSrv0wvRh/neHXv65513QoCBABlR0aBJACQHelEIUCkiFRegFYcHfy++DrooRgNlIx8tGh8sN+j1Up3XSRG81c4y0lxQYAAoOxMnTpV/wMMANlBgEhRqboApYpIkJUz1xiwUSBd9+Rz1EeJdbeqeydXbBSobEGAAAAAqsM///lP/QJhBIgUlWoLUCgbfhFxBEiPFAWiVJu6Fo8aRXXoZRK/TNmJpt155coWIC1o0j69f6N+dwqdty5nel/iODO0vel+Gu0jqt8oUzses95Eu8w6w/NcK2dtlzMIEAD0CtKxA4B05ImJ66+/fq0jggCRIlJtAWo0zc1eF3bAx6spVic86qgby9JHdow0mubmrJP6pgT7tcQjkqF6+agdlpy4x+bKSIa2J9rp1pltH+Zx6n1Y+21Sp26DnHez3jaDAAFArzB69Gi1dOnS6BMAxCxevFh98pOfVIMHD67JDwJEikqlBSghEnGijrwrB84IiW9ZQ7mJEk67S7/vqN7RT7YjKQxR4hGq6LNumysMxjaZ2u7UKbFGYiI5sY7DLJNY7x6fJDzG+Hh87Q7PV+Nz2lIQIADoFeRBCIwCAdQR8dlss80S4hMHASJFpMoCFHesvfF00u1lKSISLW/UWQ/lwx+7Ps8+PFKiY8lGszZkbHskRN796YTb+2UuiNtWnzBF+9BtiX5OtFvXk7KPPEGAAKDXEAlChKCfEfHZfPPNU8VHRoPkTwSIFJEqC5AWEWt0JS0eodCd8uTvX5x0+fDJVEo8wpDaZlMSmglDK22vSZDEJ0KxBEkatzUUTqeOLO3Wy9MkLEcQIADoNeSpcBKAfkSufbOj4sv++++v/0SASBGprgCljYJ44uuA5+2Up41yeGJNN4vimyImseSiWdvytL0mQmnl3NGgpOj52p6l3dlFNWMQIADoNRgBgn5Grv1GEjR06FA1a9Ys/TMCRIpIZQWoTRHxjc5kGt3RnXzPKIcnqcKQkAFH5nz7MNuboe2WmMQxz5n3OMx2hD/Xz69POJ3z5W1XJFaNzmmrQYAAoJc54ogj9HQggH5Drvt1111XDRo0SMtOHPks6xAgUlSqKkBhB9/tbPvjH3VJGeFoMrKSZZswYf2JESpTQoztbClyZcOViAxt14Jj79+Soqgd1nkxpciVmQZykzgWt51OO9oOAgQAvYw8FW7VqlX6Z0aGoN8Qwdlhhx0SEiQPDJE/ESBSRKoqQLrDn2laVYqIGOtqv39N60uKR3pcOTASy0ccb32R9ERJtj9D2yMJqsc/IlRfbwiOLlvf3jui1GAUqVbfCp84tRkECACqgvyv9/Dhw6NPvDcIqo2I/4gRI/R1Lx3Uegck/OdW/kSASBGp7j1ApBfiFad2gwABQJWIR4ME6RzG7w1avXq1fjt+TPxZ/oxBmKCXGDdunJo8eXL0KXxM/HrrraefACcgQKSoiACNGTNGX2Mxzz33XO3vTPlTPsfI36vm38Xys/l3bSEgQJVMcqphOBpU6PQ3CQIEAFVF/sGN/4GWf4DN/8GMP8f/SMuf0mGMt5d/6M1/7AHKhIi9jHa60i6f5QEIQj8K0Pvvv6/efvtt9Yc//EG98cYbVmS5rPeVM/PWW28lyrZSvooRAZIXUZv3W8r1Ff8dKX/K5xi5Bs2/b+Xn+LoU2ik7adKk8O9tBKiisaftSQqXHwkCBAAQEsuQsGDBAv2Pfkzh/3sJ0Cau/LhIx6GfBEgE5ac//am69tpr1VlnnaVOPvlkKzfddJN67bXXvGXNfPe731WnnHKKVfbMM89UV111lVqxYoXej69clVP0FDiRnvj6lT9jGRKajR6JDOnPCBBpJwgQAEBj4tEhJAh6iX4SIBm1uf322/W0wPh/jd0ce+yx6qWXXvKWN3PIIYeotdZaK1FelskDJ2655Ra9P1/ZqoZ7gEjlggABADTH/B/JZv/zDjBQmNdhM6TT3g8C9O6776ply5apnXbaKSEtZtoVIMmaa66ptttuO3XPPfd4y1Y1CBCpXBAgAIDsiPyMGjXKmgsP0Alk6o/vvp80pMPeDwL0+uuvq3PPPTchK26KECDJsGHD1Fe+8hX1zjvveMtXMQgQqVwQIACA1mAqHHSaeBpm/FTDLMj2/SBAL7/8sr4x3pSUoUOH6qfhbbnllrWcffbZ6pVXXvHWYeaYY47R/8kRl9tiiy3UkCFDrPr33ntv9atf/cpbvopBgLqX5FPRmkceG+19d1AiA/SENSP6EdYttr8jQYAAAPIhImQ+WhtgoJDO59SpU6NP2ZCOej8I0MqVKxP3/sg0tRkzZugHIsR5+OGH9dPhfHWYkeltZrl58+ZpITLrHzt2rH7ggq98FYMA9VCcl4+mp9HLXQuMflFqwS8xLSIIEABAPuR/5VvtlALkQaa9tXrvmXTU+0WAREhMQZkwYYJ65plnvNu3Gnly3F577WXVv/vuu6unn37au30VgwD1TvSIy+Rz1EeedWaybtd+QtHKNiLVwSBAAAAA1UM66giQv0wrQYAQoEzRIx31ayQ57ct5v40lH/XRGD3lLS6vR3Pi0RPPNkHqYhGur9XfaBQo06hMs/2lSVTUDuP480zjG/AgQAAA7SFT4XgoAhSNXFftPHFQOisIkL9MK0GAEKCmieSnLgfuvTXuZ1cSZP14NSVYli4YdYGKt9HrLZEJtzHr8EULSdPRnwz78023s6QtDAIEAFBB5CV+pescQM8j97VIxzMv0nFBgPxlWgkChAA1jUcETHnxCoBZJhKouiCFscp5t6mP0tS3aTayk02S8u0vOfojySZcHQ4CBAAAUC6kw9nKI699SEcdAfKXaSUIEALUNJEs+KedZRAOz6hJQjZ8oy2ZtnGSZRtJlv25x5ZSNyNAAAAA0JD4kdcystgOUgcC5C/TShAgBChTahIkMSRAS0HjURnvvTRRfbFc+CXCFhJvPU6ybCPJsj/7s7vOLoMAAQBUFOkgtPKeFoA0injXlHTEECB/mVaCACFALcUdDWo64pIiDpY4pUhEJkmyk200Jtv+JFKfbnvqcYajREkx6nIQIACAYhD54SWpUBako44A+cu0EgQIAWqU8MEATsffFAXfCJBeHy/zT5GzR2rCbVwh8W3TTDSyCVCW/RnLZp6TMvoTJMMIWFeCAAEAAHSfouVZOuoIkL9MK0GAEKCG0R18u/NvS5ErJo5cWDJUjyUq0TbzZx5d3y4hFmG9rki5ySRAmfZnLpffC/8oVzbh6kIQIACAYpAb1hcuXNjWjevQn8j9PtKJKFKCpL5+EKAXX3xR7bDDDlEnLMxAC9Cuu+6qHnvsMe/2VQwC1CQ1CYjjykAkPVGskRJd1t3emRZX2yaamqbrcWXEWNdAOFJHccw2ZNpflGi0yzv6kyJ3pQgCBABQHNJJkJvYAbIiwjx69GjdySwS6bRUXYDee+89tXz5crX22mtHnbQw++2334AK0BZbbKGuvvpq7/ZVDALU3ehRlAwPLsgUr3DZaWV/CXkyUtrRHwkCBAAA0D2kcylTuIoeOZSOetUF6Ne//rX66le/WhOTOJ/+9KfVL37xC2+ZViMCtM8++1j1Dxo0SH3uc59TL7/8snr//fe95aoUBKibcUaD2o6MRjUalWllfw2m3YloFSVtAxEECAAAoHuI+AzEqKF01KsiQCIZ7777rnrnnXd03nzzTT317bTTTlNrrbWWJSfy+fDDD1d/+MMfvHW1mjfeeEN9/etft/YhWX/99dXXvvY1PX1R9hW3TdpZNSlCgLqZbPf2tBIZmUkXnCz7q0/p89aTYZSp60GAAACKQzoKixcvjj4BdA/pnFRFgGQU5u6779a/W3fccYeaNWuWGj9+vFpzzTVrQiKRzzI9be7cud568kSE5s4771QbbLCBtS/J0KFD1Z577qlmzpypFi1apNsm5/yVV17x1tWrQYBI5YIAAQAUh3TQFixYEH0C8COjPgP9sAzpoFdFgOQR88OHD08IjxkZ+fnEJz6hpk6dqqfG+erJGxGwM844Q3384x9PjDi5kfu5RIZ89fRqECBSuSBAAAAAnUU6lJMnT44+DQzSGa+KAC1btkwLkCsbceSenG233VZdcMEFAzb6ItMUL7nkEv3EOfehC2YQoA6BAJF2ggABgIvMaT9w4iRCMuerXz0yunqgGTKaIR1l+T0bSGQf/SJAw4YNU5/97GfVD37wA/Wb3/zGW0c7kXt6ZBTolltuUYcddpie+uZrhwQB6hAIEGknCBAAuMTvJPnM8dMJaZo9DjlKXy9TvvLV6AqCNGTa24gRI/Q9LAONfCdVESB51PWmm26q1llnHS07vhEYmZq25ZZb6lEaGa3x1ZM38iCEefPmqVGjRiX2K5H2SLukfTvuuKO6/fbbvfX0ahAgUrkgQADgEgvQvJ/+jZCmOenaJfp62XybHZGgDAz0yE+MfCdVEaCVK1eqGTNm6I645Pjjj1e77babnvoWS4hE7hGSqXA33nijt548kYcgPPTQQ2rkyJHWviQyEjRmzBj9NLhzzjlHt03k9mc/+5m3rl4NAkQqFwQIAFwQINJKYgE657ZnkaASId9JVQTIzeuvv66fzDZlyhQ1ePDgmpBIhgwZoo466ij11ltvecu2Gnnk9plnnmntQyKPwf785z+vR3t++9vfVvp9QAgQqVwQIABwQYBIK4kFSH5GgvwM9BPffMh3UlUBkrz33nvq6aef1vf+xFIS58ADD9SjRr5yrUZkSzr/Zv0y8rTffvupRx99lBehdgsEiLSTKgnQ7Nmz1cRJE0mfROZkw8CAAJFWYgqQBAmyEfmRG+M7NfUtRr6TKguQRF48Ku/ecafCiZz8/Oc/95ZpNfLwg7322suqXx63femll/aF/EgQIFK5VE2A9tpvV3XKBUeRikf+Adpq1JZI0ACBAJFW4gqQBAmqc8QRR6ixY8d2fBRIvpOqC5Dk+eef1w8/kOONM2HCBPXMM894t281PgHaaaed1IMPPujdvopBgEjlUkUBeunvS0jFI/8A7Rl81zuN2REJGgAQINJKfAIkQYLCF+PK45vlqWSdRr6TfhAgmeomD0SQ440z0AK0++67qxUrVni3r2IQIFK5IECkFyP/AN30yBx10Jf3R4IGAASItJI0AZL0uwTJqI+896cbyHfSLwIkI2xyvHE6IUBy/5Fv+yoGASKVCwJEejHyD5AIkPyMBBUPAkRaSSMBkjAS1B3kO0GA/GVaCQJUYgH67y8Qki8IEOnFyD9AsQBJkKBiQYBIK2kmQBIkqPPId4IA+cu0EgSopAIE0CMgQKSwyD9ApgBJkKDiQIBIK8kiQJJ+kSCZ8jZu3LiuPPraRL4TBMhfppUgQAgQQDsgQKSwyD9ArgBJkKBiQIBIK8kqQJKqS9Dq1avViBEj1KxZs6Il3UO+EwTIX6aVIEAIEEA7IECksMg/QD4BkiBB7YMAkVbSigBJqixBMvJTlo6ifCcIkL9MK0GAECCAdkCASGGRf4DSBEiCBLUHAkRaSasCJKmqBMn0t25PfYuR7wQB8pdpJQgQAgTQDggQKSzyD1AjAZIgQflBgEgrySNAkliC/oUHIwwI8p0gQP4yrQQBQoAA2gEBIoVF/gFqJkASJCgfCBBpJXkFSIIEDRzynfSLAMnUQzneOPL59ttv1y8rjfPSSy+pd955x1uHmV/+8pfqqaeeqpV7+OGH1ZgxY6z6RbgQIADIAgJECov8A5RFgCRIUOsgQKSVxAJ08veW5MqUCxeqTTbfUn32oIOjK7C3OOKII9TixYujT+VBvpN+EKBf/epXar/99tPHG2fIkCFqk002UZtttlktZ5xxhnr55Ze9dZg58sgj1RZbbFErt+mmm6rBgwdb9e+5557qhRde8JavYhAggPwgQKSwyD9AWQVIggS1BgJEWkksQO1myLB19d/jvcSCBQvU8OHD1apVq6Il5UHOaT8IkJz7k08+OXE9uTn22GP1KJCvDjOHHHKIWmuttbx1SESGvvCFL6i33nrLW76KQYAA8oMAkcIi/wi1IkASJCg7CBDpRrbfY7+eEiDpeMvvSRlHfwRpWz8IkExru/fee9VWW22ljzktRQiQLN966621+PrKVjUIEEB+ECBSWOQfolYFSIIEZQMBIt1IrwmQPO1NOsJlRX6H+0GAJK+//rqaN2+e2mmnndSgQYNqwmKmXQGSerfffnt10UUXqd///vfeslUNAgSQHwSIFBb5xyiPAEmQoOYgQKQb6TUBKjvyO9wvAiQRCVq0aJE68cQT1eTJk9Xhhx9u5Tvf+Y4etfOVNfPtb39bfelLX7LKfvnLX1ZTp05VN9xwg34qnK9clYMAAeQHASKFRf5hP3feieoHS+fmyp7776q2Gr0lEpQCAkS6EQSoWPpNgCTvvfeeFiF5ktsvfvELK7/97W/Vu+++6y1n5pVXXkmUlQcevPrqq+r999/3lql6ECCA/CBApLDIP+xFZOLEA6OrB0wQINKN9IIArV69Wk2aNEn/WXbkd7jfBIgMTBAggPwgQKRUOeWCoxCgFBAg0o30ggDJ+2V6pSOIAJGiggAB5AcBIqUKApQOAkS6kbIL0KxZs/Qjr+XhB70AAkSKCgIEkB8EiJQqCFA6CBDpRsouQEuXLtXpFRAgUlQQIID8IECkVEGA0qmmAK1SRx64hlpj6qOJdadNlXvCLlKnOcsvuvgwtcaBN6qLnOXZ8qg6ODiHB1/vWxdG1x9ss8Yah6kjb7xRjZE/7/Nv2w/hIQjFggCRooIAAeQHASKlCgKUTlVHgPxCI6JymBoTyJEtK6Ewjbl4lbGslTQTIGf9fQgQAlQsCBApKggQQH4QIFKqIEDpVHYK3PUXBcfljPTIsqmPhqNA1uhQ8xGcxmlS3hUeBKiUAiQvOi3zy04bgQCRooIAAeQHASKlCgKUTnXvAUpKiYiPHuURETJHhxKyFE2hC8qHMddJvYG8XCxl4nXuvsLPerrb3Hi7KCJeHgEKp+bFqa9LjmR5Rqt8slfylE2A5KWZcu4XL14cLektpO0IECkiCBBAfhAgUqogQOlUV4BcUYjERcTCERBbMiJ5MUaI7PuGovWWlJgClCzfeAQoki2jvvB+IX9bw8+e9lkjWuVPmQRInvQmj7yWt//3KggQKSoIEEB+EKDMuU3N+VLQeZlxZWLdD2cEy9c4Rv3QWf74zfuoNb40TT0eL3viGP2Pn5nTnzDLpO+jlqgOu1x1ggClU10BikQiFgMRh5pkmHLkiJJ3NCUpONYITG29R34kjQTIXaeT3j45pjEX3xjsxxay1Ol3JU3ZBEg6fb3yyGsf8juMAJEiggAB5AcBaiEJodG5Up2+xj5qUiAuPpmZdPNt+rMu60rSqmlqUvCPYbxNrcyXgvpqy8wE+5J1QRkEqP+osgCFchGKQigOdWmpy5EIRF1A9HJrdCdMbfqcVzgi8Yliy1GQRgKUMn3NlDfzZ2nHwdeHUqTbYByjWb7s4SEIxYIAkaKCAAHkBwFqJXr0xZEYWTbjynAUyBq5ETGKRcWWITO2VMXbTQtExxWtIMG+9LpavdULApROpQWoJjeGMMTrYvFwBMIvQOYoTAMBEknR9TojOjkEyJrWVttG9hNuK+2U9phyFNZbFzGfyJUlCFCxIECkqCBAAPlBgFqKKTVhRHy02IgIJaa7xbKUYWqbTl2UpN7kiNI+as6qZBuqFAQonWoLUDxyY04ZixPK0cFTDYGQeIXElJ50AQqXhbJkyUcjAXLX6djT3mr1Tw3aZsrOgRepg63teidlEKATTjhBP/ygCiBApKggQAD5QYBaijuSE05/m7Mq+FlPZ4t+DtY1vv+nvp0do/5oZKm2TurX9SFA/UrVBUiPkMjvhyk5OpGoBOtsgQhlw9xej8bUpKiZAAWJRmJq9TYSII8whW22pShsQ1KKEqNNPZJuC5B08oYPH97T9/2YyLWBAJEiggAB5AcBajFabGIxqUmJrDPlyBWlOOFy3cmrxZxSZ5YT0amvk/3WlyNA/UjVBSiWEVtYwvhEI0xdjsKYI0IZBChILF56WUMBChMLThhPmzxT63SZEk9za5RuClB8zcufVUGOBwEiRQQBAsgPAtRq9EhPKCZ1KQnX1eXIGBmKy/kSjwp5JSqcXheKjiyP60OA+pXKCxApZbopQEuXLlWzZs2KPlUDBIgUFQQIID8IUMuJ5SaUFUtE4vt+DEmyy3qiy8RyYwtQbRqcNdJkClD4s5YonYz7LHEQoHQQINKN8BCEYkGASFFBgADygwDlSPjgA3kamyscoRydPsOYJiexJMeJlqUUAdL1HaPmWCNNpgBVLwhQOggQ6UYQoGKR3+F9992XkEKCAAHkAwHKET3VLfhHzJIcnVBgZJ05Na6+PDlCYz8+2xWguJwpTwhQv4IAkW6k0wIkT3ursnDJsRFSZACgdRCgPNGjNn4JCeXIP9pTEycjPlEyl+ky5tPkEKC+BQEi3UgnBUie9DZ69Gg1derUaAkAAEDxIECkVEGA0kGASDfSSQGS9/2MGjWqMo+8BgCAcoIAkVIFAUqnlwSo9k6fWpzHRXseL91WiqjP8/hqO54Xp+aIfiR24l1HafE9yruz6aQATZ48uTIvPAUAgPKCAJFSBQFKp1cEyPvOGy0XA9iRL1CAxhxovkuoHpG6MQcGYocAAQAA9DQIEClVEKB0ekOA0jvsrXX8W0xhAnSROjIQnWT7ZfQnXIcAAQAA9DYIEClVEKB0ekmAxly8yrPOiCUscSc//DOcMpesw55W58iIR4C0aNS2zyBHkQCdJnX5RrACadFtcNY124+1Pih7pCtAuu3GNpYcVV+AFixYwLQ3AADoKAgQKVUQoHR6ZQqcKyq+6WQ+AbK2jaaj1WTC+zko4xWg6F4dUyTc8r7obaQNUt7eViRGJMQWoOQ9QeGx18uGn+vHVTs3cdsS7XLbXm0Biq9p+RMAAKBTIED6JaVBhyOR5Dt7ko+kbpSiHld9pfphRR957QsClE6vCFAYezRHxxw58QiQPeJjdvxDKXBHhPTIik+A9M9J8dLbW6MrTmoCFIpKfX/Slvpyv3TFMdvqkxdbcKRN7nH5zk0VBUie9DZixAj9NnsAAIBOggBpAUqRnZT3+WRLEQJU7Xf++IIApdNbAmQnMfLRtJNvLvNLQKqMxKNDvmQUIF1fXLcsj8pZ+zS3N6K3ke29gmSKWCRDvnb2gQDJ9cxb7AEAoBsgQCkCJPnhjKAjknnExw0ClCcIUDq9LECSUIIMwRggAbL2401YX102om0toRE5CeuLp79J2SwCVBOcpgIUtiMxAmTFOHZdn9HuuB0DHB6CAAAAVQMBaiBAL62apiYZo0CJKXCe6XOTbr4tKh/Jy81SR319Qmb0Pow6ZlxplU8uj8Ssts4dpXLKpR1bSYMApdMLAtRQPtwRlswC1OIUOL2fpHg0jSM0cixjLr4xaIe9zLvPaH1rU+DMn81tzPjq6GwQIAAAqBoIUCMBckZgLAFy5Ki2vrYsFhGjbr0vo4z7+e+3qTlfCso4ElSXJnd9EKuOcH1dwiJZMrcveRCgdHpjBCjssCdHJxyJaUmAgrhSoz8b+7Hqi8TCakNYX8PRFkeAavswBMUSIM9+QgGst9MVwvCzUWe0D/PY7TLVEiC572fhwoXRJwAAgO6AAOUVIF8sKXLlJYwpJPKzKSs6jerQ65JtrdcZbp+os4eCAKXTS1Pg9OhM0FYzVie+VQEKUpMHnWaPwY7kpLZ9s6lmQVwBSmuDJVbusZptMMrE64Oyicdgx6JVS+M2dDpFCtARRxyhxo4dq0UIAACgWyBAbQtQuE2982LKizm6E6ZeRzSaY5WNkyJAuq2+7YNEUhWOQsXL046rvEGA0un1e4CKjhaLhtPHSBEpSoAWL16sr1/e+QMAAN0GAWokQM6IiyVAel0oGrURl8ToTSMBCuWm8WiNLUCh3GSVmrCsKUL2vUPJ0akyBAFKp38FyHOvjB7x6e7ISL+kKAGaOnWqfukpAABAt0GAGgiQe/+MKUCWDMXRdaWM3kSp1+m5nycRpw6r/qzxt6OsQYDS6e8RoHAqmCnwyE9nwkMQAACgaiBAKQIUjrbYspEQILNcbUTIFiBrG1dg9GdbTux63YcaRNJkiVe4n3Ab8+dofcrxlTUIUDpMgSPdCAIEAABVAwGKJCSZFCky5MOeUiZik5QR+zHYntGbxP7t/YZCFCyv7Td575AlPMbUvDCefZY4CFA6CBDpRtoRILlmuecHAADKBgJEShUEKB0EiHQjeQVInvQ2YsQINWvWrGgJAABAOUCASKmCAKWDAJFuJK8ATZo0ST/yGgAAoGwgQKRUQYDSKacAJd+3Eyfx3p3Eu3oax/fOnTy56PpH266j1GnxvLaaPAIk095GjRqlVq9eHS0BAAAoDwgQKVUQoHTKLEDJl4xGT21r4z09RQhQURJV6pRQgAAAAMoMAkRKFQQond4SoCBtvqsHAcoYBAgAAKAlECBSqiBA6fScAAU5baoxCuR21CNBiqfM6RgjRrG8HCl1xOsTMuNOwbtInWaWry039nv9RcbyZNvtcunHJgmPL2ij2YbEC1uDfV9s7jNsoy7rKeOrU7fBOl/ueSyHAN15553RTwAAAOUFASKlCgKUTi8KkDUCY3XUwyly1uhQJCbxslhE6nVHslOThbAOc99hGUeCDGly1yfq0G0w1jeRi1hi6m1w6oulpdaGurDVjj3aJv4c11lbHwubcRx6G+95LT5ZBWjBggW6ndz3AwAAZQcBcpN4j04y5otL8+U29cMnjHf3kFoQoHSqJUC+2FLkyouOWYeIgbveaY9dR7iuJhZxDOlJClLjWCISx5QoR24kyeOyjztZp70+UUcJBEgeeiCPvBYJAgAAKDsIUJPol53OuNK7Ll/CF5laLy8ltSBA6VRTgOojInEsATKnk+nUZSCUFbtsHL8AhWV929elx94m7bjiaFlx22gep+eY7TZJPAJk1Vl+AZJ3/UyePDn6BAAAUG4QoCZBgDobBCidXr0HqLbO6qgbouFMacsqQN7RFydJUbBFolF02aiNjY4PAQqRF58CAAD0AghQk6QJkF4edY7WWGMfNWeVuT6UnOR6Z/mXpqnHjTrNPH7zPkZ5W5hS1z1xTPD5GPXDaLswvSVcCFA6PSdAbsfc/Ozea1Nb7wiQKzhGHaGgNJ6uZtcRikSzUR033nZE8UpYYgpcfwgQAABAr4AANUlSgCKJMZdp8ahLkFtGC0tNdpoLSSg4hlRF9yXpe49cydHr4m2vVKcH21l1W+vLHwQond4SoLDTbnXkEwJkdtqj7YNYAhR8rtcdTZer1Zm2D0MWHNEK67RlwZSYcL0pVc1HuOw2hm2qfa6wAMnDDuTeHwAAgF4DAWqShABpoXBHWcztPIJkpZkANV4fylFy/9Z6Y2TJ/Vz2IEDplFmApF1uEtLgdNRjwYlz8PW24MSdfOsx2JYYSOriVK/Hv94Vq1osGalLTS3mPh2hCmXF88jqePsKC9CkSZO47wcAAHoSBKhJEgKkR2DqnR0r8XbWNu7oSzMBCkdx0p80F66P60/UY434NB9tKlsQoHTKKUD9naSsVC8+AZKHHgwfPpxHXgMAQE+CADWJdzpbgxEYO9FokJaVrFLSTIDqCdviipBRf8poVZmDAKWDAJUv/ShA8rADkZ+lS5dGSwAAAHoLBKhJ/CNArd5TY0pPMwFqtj6ZtGlvc9y290BaEaB++99nBKh86dcRIJ74BgAAvQwC1CQJAYoExb6vxnz4QLTeLOM8iCBZp53kKFNdihqtq9dRnybX/ktbO5ssAiQiMGHCBDVu3LhoSX+AAJFuJO0eIAAAgF4FAWoSv6xEkhNJhiRNQOJYIlK7RygUGd8+zOltErN+vb2xzidT4Ta9Nf1N0kiAYvGJjxsBImTgEwuQXH8AAABVAAEipYpPgFzxQYD8HVVCBiIiQGeccYYaMWIE9/0AAEAlQIBIqWIKUJr4xEGACBn4bBcI0Cc/+UkeeQ0AAJUBASKligjQ2LG7NxSfOJtuuqk64ogjdL7yla9EV1zIa6+9VlsnefDBB6M1IRdffHEhZadMmRItDXn99detsg888EC0JqSdsp/+9Kf1cfs6qYQMVLbZfR/91DcefAAAAFUBASKligjQPvuMV1OnTtWd/aFDh1rSY2aLLbZQ06ZN0zn//POjKy7kj3/8Y22d5KmnnorWhFx33XW1deedd160NKRZ2euvvz617DvvvFNY2RUrVkRrQkTU5Lh9nVRCBioyBe7cc8/V1yD3AQEAQBVAgEipYk6Bk8dcNxIhpsARMvAxH4Ig19/ChQujKxLyIOeR9F8AoFwgQKRU8T0EIU2EECBCBj7mY7AXL16sp8OdcMIJ+jO0Tvz3F+mvfPGLX4yuAAAoAwgQKVV8AhRjitBaa62FAJFMmf/TD9WVy19Tl973orp82R/0Z992xB9TgIRVq1bpKZqQD/kd/vGPf6w++OAD0ieJJehrX/tadBUAQLdBgEip0kiAYmIR2nvvvaMl/QEClC9zfrJafenIr6uNNvmE+timW6htd9lDnTbtQnXxDx9Xc598DyFqEleAoD0QoP6LfOdnnnmm/vPrX/96dCUAQDdBgEipkkWAYvrtqVQIUL7Mevw99YnNt1RrrrmmPn/y5+DBQ9T6wz+mrnjgl8E2CFCjNBMgGRHiCXHZQYD6L/Kd33HHHeraa6/VPx933HHR1QAA3QIBIqVKKwLUbyBA+TL3oZfVuhsM1+fOzHrBsosf/K23DKmnmQBNnDhRT0dFgrIh1x4C1F+R71wE6Pe//31Ngr7xjW9EVwQAdAMEiJQqCFA6CFC+HHXeFWrosHX0uTMzbs+91WXL3/KWIfU0EyARn7Fjx2oJktEgaIxcewhQf0W+81iATAniYSIA3QMBIqUKApQOApQvBx59lho0eIg+d2ZOOmuamvPk+94ypJ4s9wCJBMl9eQhQc+TaQ4D6K/KdmwJkStCJJ54YXRkA0EkQIFKqIEDpIECt54qf/lUd8dWvqUGDButzF0fuA/rKuVeq2U+u9pYj9fAQhGKR6w8B6q/Id+4KkClBJ598cnR1AECnQIAKyMq/PaieXXmTemjJFep7P5iprgpyRbP88Gp13ZJ71FPv/069+I8XvPX2YxCgdBCgFvP0h2r2E++prbbeVj82Xc6dRORnyJAhasbNy9Tcp/7iL0tqyStAcr1CErkGEaD+inznPgEyJejUU0+NrhAA6AQIUJt58YMfqedeWajuvv5cddnpR6nJnztIHRrkoGY5/Hj11TO/q2546g312DuvqRc/ekqt/PtS7z76KQhQOv0sQHNW/FnNfniVd11a5j79V/WdJ95S66y3vj5vcUSAhq2znprzyGvBdvET4D5U333qPTX38XeQIid5BEgeVS/nWl6cCjZyXhCg/op852kCJIkl6PTTT4+uEgAYaBCgNvPc05epJTP2U8ccPFaN22UnteN226ntg2zXLDvsrHbaY5La+6jZauadz6pn/vb3QIJ+6t1HPwUBSqdfBWjWEx+oM77/oBq92/jgc/ZHVovIzF2wSA0eMlSftzgiQJ/cYks167F36tsGsnTerT9R+086WM24Z6UWLrOufk7eESCRHznfSJCNnBMEqL8i33kjAZLEEiTvCwKAgQcByplXgrz3j6Xq53dfoOZNHK3232FLNXLkyNYyens1creD1cEnzQ8k6Dm15LVX1C8+fNa7v34JApROPwqQ3KMz7ablav0RG6vNt9tVT2vzbeeLSMxuEz6j1nbu/5HpcCeddqaaHYhVvK3I0pnXLVXD1llXjd1rvLpkyWtBeUaCJO3cAyTyI0+IgzpyDSJA/RX5zpsJkCSWoLPPPju6WgBgoECAcuZ3f39Y/fd/W66euXG6Oj6QmbGu3LSS3Q5Sux9xiZp377PqvpVPq2W/vTVffvdDtey1u9VP339Evfj3J4J2PpJod9mDAKXTbwIkUvLN65eqDTf+hJaYlgXoJ39Sm22zs1pr7bX1eYsjny+97XE11xjliQVIHpctgrTr3vtpCZrbwv6qGh6CUCxyDSJA/RX5zrMIkCSWoHPOOSe6YgBgIECAcuZ3Qf57IEHP3HBu+wI0egc1eue91acOPFgdeMhB6jOf3z9fvnyY+sxJF6tZP35K/fxvf1UvfrTC2/YyBwFKp68EKBCPU6++Vw1ZZ109Za1lAZLtlr6qPrnZ5okHIKy3/gbq4h//yrrXxxQg2WattdZWu+w+Tl34oxftevswRQoQL0tFgPox8p1nFSBJLEHTpk2LrhoAKBoEKGdCAXooEKBp7QtQUdl6RzVy7BfUF2fcruY/975asfon3raXOQhQOv0iQLN/8if1zeseVkOGBfITyUurAjRHhObGx9XgIUO00EgdEpGhHXceo+Y+9rZVlytAep9rD1I77bKbuvihV4P1f7Xq76cUKUATJ05UkyZN6msRkmsLAeqvyHfeigBJYgk699xzoysHAIoEAWqQV/7+sHr5bw+p59+7V/38nbvVM+/dp57/cJl68aNlXgEatfW2avtdxqqdd9tF7bL7zp3NztuonbYdqbbacqQa/eXz1YELfqHuf2O597jKHAQonX4QIHngwZnff0BtsNHHAxGpj9y0KkBzV/xJXfmDu73T37Ydu6+a8xP7/T8+AdL7FQkau7f6diBB/fpghCIFSF6UKvcEjRs3rm8lSK4rBKi/It95qwIkiSXo/PPPj64eACgKBKhBPvjHcvXu6/eoH145TV064xR14ZUXq9t++Zp68oPfegVon88fpU6/cYX67v03q0WPXdnR3HzVF9WlR49Uu+0wUm150BlqwuXL1Y9ffdh7XGUOApRO1QVo1hPvq1O+e68aus56+jjNtCpAs4O6/uWrR9VGkOIMGjxYffqwo/QIkbm9jPCc5REgidSx0667qwvuekHfV2SW64cUfQ+QiI9I0NKlS6Ml/YVcUwhQf0W+8zwCJIkl6IILLoiuIAAoAgSoQf71H8vUn19cpOZP+YL66n4T1Oe/coya99i7ask7b3sF6LNfP0fN/+X/UMvf+6W3voHMymXHqbu/NVLtvctINXLiqWqfix5SP17l37bMQYDSqbIAzX7yA3XOjcvV0HWT8iMZNHiI2nLHsZkF6PLH3lEjNpZRJFtmhgwdpq5e/EhiSps8BvvcW55U666/oXXPUBypZ+ddx6qZD67qu5EgHoJQLHI9IUD9FfnO8wqQJJagGTNmRFcRALQLAtQg/xoIzgfPfE9dtO84dWAgOLtP+IyavuQP6t4/+gXo4OPPVVf99j/Uo6tf8tY3kPn1Y8epB88fqT6FAFWWqgqQCIVMe9two0/o43MjIzAbfmxjddLV93vLu5kf5NZn3lbrBDKVEKB11lOXLnnV83S3D9Xsx99Vn/vqVC1JbjmJlqA9xvfdPUGdEKB+mg4n1xIC1F+R77wdAZLEEjRz5szoSgKAdkCAGgQB6nwQoHSqKEAiEiddsTiQjnXUGinSMXyjj6vjZt+W+ZHUIlSnzLpRDRo0yKpLRnZ2H7enmv+T91NHkqTsZ796kpYgs2wckbHtdhqjp8P5ylcxAy1Acl2PGDFC3x/UD8h1hAD1V+Q7b1eAJLEEXXzxxdHVBAB5QYAaBAHqfBCgdKomQPKS03Ouf0TLj/nAAzPyAtTTFi5tadqZ1PuZ489LjOKIEH3jpNPU/KcbveD0Q30v0hePPV0NGRZImVE+jjxIYYedd1EzH/idfniCv57qpBMjQFOnTtUSJC9OrTpyDSFA/RX5zosQIEksQZdcckl0RQFAHhCgBkGAOh8EKJ3qCFAoGedcv1Rt8LFNvNPN1lhjTbXhRh9Xp3zvIf1YbH89/sx+/D31paOOT9Qp9xF98bRvq/nB/n3lann6Q3XZ0j+oLx13hn4ogluPRCRo+133UN9e8lrlH4zQqXuAFixYoEaNGhV9qi5y/XRegF5WCw8Lrt3zliTWLTlPrunpaomz/OXrDlVrHLZQvRwve3i69Tsgmf6wWSZ9H7VEddjlnHj2U0ujunPk5YeX1I9vACNtL0qAJLEEXXbZZdFVBQCtggA1CALU+SBA6VRDgAL5CQTlpCvu1C85rXVsjMhLSIdv/HF13JzbWpYfyWXL3lLb7bhTol55utyZVy32lkkkkCBp58FHnaxHgtLuCdphl93V+Xc+n6udvZJOPgShH+4FkmunGyNACaHRWaKmr3GoOjQQF5/MHHrdy/qzLutK0sqF6tDgWOJtamUOC+qrLTMT7EvWBWWaC1BSyMK2mvtrL/7zMTCR73ynnXZSn/rUpwrLtttuq+s9/fTToysLAFoBAWoQBKjzQYDSqYIAycjPmdf+OJCRFPlZe221/vCN1OnXLUs8qjprvr3kde+jtNfdcIS69JE3vGW8CSRInk532HFnpk7TEwnabsed1Yz7Xmlpml4vpZMC1A/IddOVKXA+sZBl5y0JR4Gs0ZVQNkJRsWXIjC0R8XYLA9HxiEWwL72uVm9KUgWoWGnppABNmzZtQLLPPvvoAEDrIEANggB1PghQOr0uQDJVbNqN4bQ3UyLixE97O33hUm/5THn6r+o7t8t9RfZDDERUNttqazXrsXf85VITPh1u8tSzGo4Ebb/rnlq8qvh0uG4JkHTwTjjhhOhTdZBrpjv3AJlSE0bER4uNSIcpA5aEZJjaplMXJak3OaJ0qFq4MtmGRFoRoGgUqvb76LRRb2+sjyXOXi7tqpfppcjvyMSJE6MrCwBaAQFqEASo80GA0ullAZKHBZww7w41eIj/6WqSERt/Qn0j2Cbru358mfXEB2rcpz+XEBV5AMLJZ5yt5q3IN1Vt7oo/q0OPOS31wQiyv1Hb7qDOv7N6T4frlgDJU+GGDx9eOQmS66U7AuSO5IiMRJ1/LRJ1EUiIhpaS+HpPEwajftnelBGpX9fXhgC5U+6iNtXrCuuu7detp9kx9mAQIID8IEANggB1PghQOr0pQB+qOU9+oE7/7t2B/AxNHUGRR12ftuARNWdFe09Vm/vEe2rUzuMS+xgydKi6b+mT6opmD0BoEHm63OQTzkmfvrfW2mqr0dtE0+Gq83S4bk6BiyVo1qxZ0ZLeR66V7ghQ1OmPBaEmJbLOlCNXlOKEy+3r3hQVs5zISH2d7Le+PIsAufsJY7YpOW0vLhvuNxzl8YhUFAQIoL9BgBoEAep8EKB0ek+AwgcJnHnNvWr4xzZKlZ+NP7GZOuvGx9Wcn7R/D82cJb9RW229TWI/666/gbrgjmf1S1J95bIlOJ5H31bHfPNiNcxzj5FE3jU0avsxauZDr1XmwQjdvgeoag9GkOukWwIUjoLUBcEUirocGSNDcTlfYlHxSlQoKKHoyPK4vjZGgKykSZrZ9nBf8e+muy0CBNDfIEANggB1PghQOr0mQHOf/qs6/7afqfU2HFHrhLiRhx4cNW2eHl3x1dFqzr7pCbX+hsMT+9lxzK5q1iOvecu0lg/V5Y/+Ue15oPzvsr2POCJ1O40brx+l7a+jt8JDEIpFrpGuCVBNEEKBsEQkFg9DkuyynugysXA4UiLrRKiskSZTgGxBqUlPOwLkTHOLE44GhfuJy9gClNKWkgcBAsgPAtQgCFDngwCl04sCdOnSN9UBh35ZDRo82Ohc1CMCtMunDlCXLHmt/QcIPP2hOv3a+9WgIUMT+9lx7wPV7Cfe85drIfKkt3Nvf1ZtvOnmqSNa66y3gTr7u3fq0S8RJl89vZQyCZBMiRs9erT+s1eR66R7AhQ/+ECexuZ28kM5mn5ePBIULbckx4klHK6USH3T1UJrpMkUoJRkEqDmU+Cs5VFM6WEECKC/QYAaBAHqfBCgdHrxHiCRoMuWv60+e8Sxau21BwXtT0qDSNB2u+2tvv3jV/T2vnqyZO6KP6mvfu1Y/cADq/611lIHfvmYtkeZ5EEOp37vQbXRJzZLlR+RrxPn3Kofn10F+ZGUbQRo6tSpasSIET0rQXKtdFOAaqMhrjxEAiPr7JGVeHlSLGwJcQUoLmfKU3ECFG5n1hWN4kTtCY/TrMdpX9b9lDgIEEB+EKAGQYA6HwQond58CIJI0If6/T+fnfKNSILCTpYZkaDd9jlQzbznRW8dWXLZo++qDTf6eKLuoUOHqmt+eJ+a34ZcSU78zj36fiWf/EiGrbueOmH2LVqUfOV7NWWcAicSNGrUqOhTbyHXSjcFKBy18UtIKA3+0Z6aOBnxiZK5LDnKUqAASaJjqbXJkTotaKnrI2EK0rA9JQ4CBJAfBKhBEKDOBwFKp1cFSCISdNmjf9TT4eRpcFanJMqgwUPUmL3209PhfHU0zaNvqnU3SN7/M2TYuurih171l8mSoO3Tbvu5+tjHP6lFza1fIvc5nXnVndHIj6eOHk5Z7wFavXp19FNvIddLVwWIVCYIEEB+EKAGQYA6HwQonV4WIEl8T9BnjpBpasl7guIpZFvvPFZ9+/7ftDwd7oIF93gfUS0PQLh8+dveMs0ij7M+7ftL9MjSWimjVyJYp8y/vTL3/LjhIQjFItcMAkSKCAIEkB8EqEFaFaCDjv2WumLl/6OWv/+it76BzK+WH6fumTZS7Y0AVZZeFyCJPOjgkqV/UAcdeVK6BA0erMbsvb+68K7nM08nk8dbf+4b56rBQ4Yk6pxy4jfV7Cfe95ZrlDkr/qROuOIutcmmm+t3/Lj1Soatt4E6cc4idXlF5UfSCwIk7wlauHBh9KncyHWDAJEiggAB5AcBapCmAvSPR9SzN56nTthypBoXrD/gX05SM5a8pRa/uEw9/tptHc2yO45UC04O2rFzIECfOU3te/EjCFDFqIIASfRI0LI31We/dGQoLJ57akQ4dtpjXzVTRoKySFCwzZSvHafWdh6AIEJ1+JmzW34nz9wVf1Y3Pfi02ujjm1r1mfXKtLezv/djfX9TVeVH0gsCtHjxYv3C1BNOOCFaUl7k+kGASBFBgADygwA1SEMBCuTnv/+3n6mX7rhUnb/9SDV+1Ei1/Zixau/PfllN/MJB6uDDD+xoDjpwF7Xf2JFq66Ado754rjrgmmfU/b9f5j2uMgcBSqcqAiSpPR1u8jGBtAz2PlhAJGj0zuPUzPtebjwd7ukP1ezH31OjttlWP/HNrEOeCHfq/Nv046u9ZX0JZGruoiVquDxQwdMuaeuQoeuoU4J6Z/+kmPcXlTm9MgVOngonEiSjQWVGriEEiBQRBAggPwhQgzQSoF8HAvSnf65Qq37yXXXPCZPUEeN2VDsG24zsZkZtp0aOOUBN+ub16vzH31SPv/eE97jKHAQonSoJkI6IyxPvq88ddVJi5CaOSNCOe+yrLr3neX8dQUSOLnnkDTV0nfWssiIq66y7nvrO0t+q+RlHaGQq3dlX36VHfnxSJpH7jGTa27wW71Hq1fTSPUAiQf/85z+jT+VEriEEiBQRBAggPwhQgzQSoHibP7x5i/rgsRlq5pQD1cQxO6gdu5mgo7jjF85V5y/6mfrZ3/6zevGjn1rH0wtBgNKpnABJAgm6/NF31KFTjlVDhw2zRCOOjBBtu+te6tL7X/HWIVPk7nrsOTVkqF1+zTXXUiM2/ria9dg73nJuvvOzD9UPHviJ2miTj6fe87PBiI3V2dfc0xcjP3F4CEKxyHWEAJEiggAB5AcBapAsAvTrv96nfvf2bWrpT69Rdyydp27uZpZ/V928YpF6+LXl6pcfPadW/p0pcFWikgIURD8ie9lb6rCvnawfhW0KR5g11eBAbnbaa38tTInyK/6s7ycSUTLLySOrTz3jLDUng6zIKNKMO59VH9vkE6nvKpIRptOu+lHl7/lx06sCJCNBck9Q2UaE5FoqrQBFLxf1JvHiVNLtIEAA+UGAGiSLAJFigwClU1UBksQPRvj810/3SpDIzebb7eoVoDlPfqC22H7XxDt6RGTm375MC5Jbxo08ne6s65eqocPW8U59G7bu+upk/ajr/pIfSS8L0NixY9W4ceP01LiyINdTuQXI9xLS8KWh9otPSbeDAAHkBwFqEK8APYQADWQQoHSqLEASkaDLlr+lvvi1E9XQocMsEUkVoODznKWvqS1GbqXWdB6AIFPizl/8XKanyMk2Z16XFCCZRifT3qZd96Ca9YS85LS/5EfSy1PgRIKkgzhixIjSjATJddV7AvSBevm6Q9Uahy1UL3vWke4EAQLIDwLUIK4A7bbPJHXu3b9TP3rj997tSftBgNKpugBJRILkniCRIBkJimUkTYDmrviLOv/2Z9SwdRxxCWRo85Fbqssffk3XaZbxxSdAIj/yktMzr75bzWnxMdpVShXuAZLfnbIg11avC5D++byFauFh4e9bbWRo5UJ1aPQ7qJOYNvdyrUyYQ9XClcb6JuX1fo319ohUOEpVX+8/jioFAQLIDwLUIP/6fy1Xf3r+RjX703uog7YaqXbcfW/1lQuvVzN+8AO18L7LupeHr1ELf/qgWv6Wv929HAQonX4QIB0Z1XnyA/XF487UL0WVY04TIBGTUxc8khj9WWvtQWrULnsFgpRNXHwCJE97O+XKO5OjTn0WHoJQLHJt9ZwARWISC0csItMfNraJ7h+qL4uEpCYxkfwYo0hhPdH+dHlTiKLt4/Ju23Sb4u3DbU0hWnKeue9qBgECyA8C1CD/+t9+qj78zY/UdQfvpQ7fZqQaueWWaqtRo9Wo0aPV6K1HdS+f+oIafeI9av5j/nb3chCgdPpGgCSBdMx6/F31pWNP1aM7aQI0+8n31fSLvm2N/khk9GjiF4/MNP1N4gqQvAPovBuWtPb+oIqmagIkL02VdAu5PnvxIQimXFjiEsUrHKa0WMKSjJS3R3SCGGV8+6ynP+9RQoAA8oMANcjv/69H1Ztv/Ug9v/BkNfNL+6o9Ro5UWwfxvoOnk9njUDXyG/eoeY/6293LQYDS6SsBkgSyc/mjf1STjz9dDVt3Pa8AySOuR2y0SUKARGR+cO+yzKM3pgCts94G6lsLHwjkSu758W/fT6miAMk10i0Jkn334hQ4M1pGrPuBkiMwYURMIulpWLc7Nc5MLE32FDd3X/GoVJjmx1CFIEAA+UGAmuTXf7pHvf3yterOa05Vpxx5sPqXIF/sdk45WX1x/n3qB7/wt7mXgwCl03cCFCR8MMLb6l9OvUCNGrOnJTTyctPvPfaqGrbOusF5cQRonfXUFY+/1YIA/VWdc9Nj6mMf31SdXnvUtX/bfksVp8B1U4Jkv30jQOaoT8O6WxvBMWXHL122COnRqdoyZ+peDwcBAsgPAtQsHz2kXv7bA+qF1feqZ975kfppGfLuveqnHzyknv/Q094eDwKUTj8KkEQkaPYT76mz5t5kr3vqL+rC63+shgyxH5u91lprqU9usaWa9fh7mQVItpv90G/UJYuf7usHHvhS1XuAli5dqlavXh196hxyjVZPgNqdAufc75MxvnbUE4pQVWTHFwQIID8IEClVEKB0+lWA0jL7ydXqi6ddkrz/Z9BgdcIpp6n5Ge//iTM/im9dP4eHIBRLVQUoLJvhIQim5CRGiGxhCUd6wvaYP5v1hSNAnhGkjMfSy0GAAPKDAJFSBQFKBwGyM/uJ99WXTzhHnxMzg4YMVYecNNNbhrSefhGghQsXduRdQXKNVlKAJFpojN/HxIiOe6+PMyIUSVA9dlvcqWxJmTLXp402VScIEEB+ECBSqiBA6SBAdi5b9qbaZfdxRocnjDww4fR5t3rLkNbTDwIkU+HGjh2rxo0bN+ASJNdoaQWI9FQQIID8IECkVEGA0kGA6pGpapcseVXLjik/kvU3HKFu+cnvvOVI6+mXESARn05IkFyjCBApIggQQH4QIFKqIEDpIED1zH/6r+oHD65QQ4YOteRH7gdaf8TG+h1CvnKk9fTTPUAiPrNmzYo+DQwIECkqCBBAfhAgUqogQOkgQEZW/EkddfI5avDgwZYArb32IPXFI76q5vxktb8caTk8BKFYECBSVBAggPwgQKRUQYDSQYDqmRsI0BkXzVWbbj5SDVt3fTV02Lpq8JCh+iWmZ930hH5HkK8caT39LECrVq3SKRIEiBQVBAggPwgQKVUQoHQQIDtyH9B3f/ZXNX/562rmbT9RZ867WU05fYaacd8r3u1JvvSzAEkHc8SIEYW+MBUBIkUFAQLIDwJEShUEKB0EKC0f6heZygtT5z4VJOvLT0mm9PsUuAULFujfu6IkqCsClHi8dJzkI6+9LzRNTVEvG12illT4haUDlVIK0P/7V0Ly59//a3QhDTwIEClVEKB0ECDSjXAPkNLyU9RUOPkd7o4AJWUnfLloO+/LKUKAipKo/kspBej//rVS/+kBQvIFASL9GgQoHQSIdCMIULGUSYAkesTH91LTTEGAuhkEiFQuCBDp1yBA6SBApBtBgJLceeed0U+tUzYB+mDlQnWoMQqUmALnmT536HUvR+sjeblO6qivT8iMU4dbvrbO2K9uR22dO0rllEs7Np0mbdRtc+p3zklZgwCRygUBIv0aBCgdBIh0IwiQjbwraPjw4eqEE06IlrSG/A6XSoCcERhLgDwiYE+bi0XEqNsRinB7c99hmYRE1aTpZbXwMKMNEqvOcH29fLP7luI2Gsdh1ee2J2pz7lGxzgUBIpULAkT6NQhQOggQ6UYQoCRyP5BI0Lhx47QQtUJPCZAvlhS58hKmXkcoK+56uz1OHbr+ZFvrdSaFpXH825vHqX+uCU9SsMoaBIhULggQ6dcgQOkgQKQbQYD8rF69Wk2dOrVPBCjcRtoexhQgY2QlSn0ExS1nJkWAdFt92weJ2hWOKsXL044rjlN/FGuUxxwR6pHpbxIEiFQuCBDp1yBA6SBApBtBgPIjI0WzZs2KPoXI73D57gGqr7MESK8LRaM2IpIYAWogQFH5xAiQFVtQQrlpJjVxTMFKK5NBgIxRH3t5uYMAkcoFASL9GgQoHQSIdCMIUDZkJMh8VLb8vN566+mYlE2A3BEf87NXBszRkhS5qNcRrm88ncypw6o/a/ztMNc1mgIn0cd63sKemf4mQYBI5YIAkX4NApQOAkS6EQQoGzLSM2LECC0+kvXXX1//vg4ePNh6iWqZBCgcbbFlIyFAZrnaiJAtQNY2jsCk7sMz+hKuDz/b4mVKjEdoGghevY1GG3ySlTi28gcBIpULAkT6NQhQOggQ6UYQoOzIPUHycIRRo0apQYMG6d9XyRe+8IVoi24KUNgWO0lpcEdG9Ofa9iIHSRmxHzGdFIhQgox6nFGl2npHiswylvAYU/PCmPt0hSpbG/3iVe4gQKRyQYBIvwYBSgcBIt0IApQdmQa3ySab6FGfeuc8jDw0QZCfOy5AfZ1IgLzT48y44lT+IECkckGASL8GAUoHASLdCAKUDZGfXXbZRQ0bNswSH8nQoUPVggUL9HbyGQHqZDIKkB5VSptGV84gQKRyQYBIvwYBSgcBIt0IApSNww8/XIuOKT5mNttsM72d/IwAdTLNBSie5td8lKhcQYBI5YIAkX4NApQOAkS6EQSoOdIRld9N874fX+ThCPInAkSKCAJEKhcEiPRrEKB0ECDSjSBA2ZDfT+mQ7rDDDvr31J0KJ6ND8pAE+RkBIkUEASKVS+UEaP9do+xm/OxblvazL62sb7ZtmZL3HPi2TVvfrN7uBgHygwCRbgQBah154IE8+vqwww5T6667rv69XXvttfU7gRAgUlSqKkArZ9ojp/UcrVZ6tm83K2eOV/e+7l/XN1lx9ICd35ZSNQEipNVAEgSIdCMIUPssXbpUd1blPiD5HUaASBGppgDNU/OD35Epi25NrNNiNPkc9ZGzvJ18tGh84XWSNlIlAQKAYkCASDeCABULAkSKSiUF6PVz1JTgd2T+Cs+6ARil0FI1c553HelCEKDqI1Mk5IZYgKwgQKQbQYCKBQEiRaWKAqRHZNZImZKWEKBb1b2Tm0yRi4Sqtk1NdsKRptry1FGgcB8yImVOzbMFzbNNvB/d5no538hWeMz1bRLyl3oM9bRXhzvqVj8es95E281j0+evXs7arpUgQNVn1qxZ5fuLC0oNAkS6EQSoWOR3GAEiRaSKAtRompu9LhIYoyMfyochQYnRJKejr9c3u/+nLkpxPUlJk23GqylB598Uj8R2UXtMQQi3MdqspcIoE0lGvYwrKxnqaPk81I+5to23XfXPug2Tg3Ng7SdHECAAcIkFaNtx+xHSsYgAnXHGGdFVCO2CAJGiUj0BijreqSMc9Q63X5TC8rUOuO6k26NCYUc9KudZn4hHWswRktRtEtIRxtp/QjwkZt3+8+EKjz4X1jZO+1o9D/qz03arrc551gn32fR8NgsCBAAuIkDyP/GEdDKf//zn9SOdmbJbDAgQKSqVE6BIGuR3JBmzY+3rgEv8YpLWKbckIC1eSfIJhi0yqXUb9TXdv6fe+vJ6m0IhCo7TI446LZ4HX7vCfYRtCX9O1tVo9C5zEKDqI51ZeUwqAAw80pGfNGlS9AlaRTpZ48aNU//85z+jJZAX6aggQKSIVE6A0jr8blK3C8XINxKTlCjfyEky/m1sAUoKQzQa4qm7Lg/p28TR+6613Y19LGG90TpfnannwW2Hv12m3KSJTpbz2TQIUPUR+ZG/vABg4EGA2kPEZ9SoUfynTQFIBwQBIkWkagKU1rFOxBkBqSXq5CdHhurr6uVsifEnRVKs/fjqSa+7fozN959HKGoilFYucR5caWwwvS2q098uj3zmCQIEAFAcCFD7MPpTDAgQKSrVEqAU2fBFd+KTI0CmQHmnaZniYklMWsJOvdsme8THJwwpkuARJ3cbs5y9nygZ6jCPPdt5MM6lb3TNOVfedulyyfPQchAgAIDiQICgLCBApKhUSoCiTna2EYRkxz/s6Lsdebs+Swbcjr8v0TbzZx6dLghp9SREIilTrpwkjsERD18dWpis/Tjnptl50OudNnjlxpAot13RZ7sdOYMAVR8egw0AvYo8EIERoXwgQKSoVEqAEsLQLFFHX3e8JUYHPU7U+fdvE8lEkFTpqnX8zX05bXTlwIyzf99+QhmJt/HUU5OL9DpCCTLijAg1Og+28PhHlMyRtdpys85g3UqfOOUJAlR9eAgCAPQqY8eOVUcccUT0CVpBOgz9JkDvvfeeevPNN9Vrr72mXn31VSt/+MMf9HpfOTNvvPFGoqzUJ/VmKV/FiADttdde+sXqMV1/WmM7D0EoWbwdf+KJX5xyBQECAICyIp2s4cOHqwULFkRLICv9JkAiKE8++aS68sor1amnnqqmTp1q5frrr9ci4ytrRsqfcMIJVlmpb968eeqxxx7T+/GVq3JEgEaMGKFnlMTI9SX/wSrE746LcWeeyM9mWXnSYyxQ8qc5bdh9cJP8bP4nrkwz1iJWGQEKO/Wpo0N9G889T3o0qIDpbxIECACgOOQfZwkUh3R+RIKgNfpJgERKbrnlFrX77rvr4/bl2GOPVS+99JK3vJlDDjlErbXWWonysmy77bZTN9xwQ99JkG8KnExNjaenyp+xDAkiKOYIkfxsjh7Jf2jEn+VPU47cWSvys1m3tEPXXRkB8j/cgARJTKkrSH4kCFD14R4ggM5x1llnqa985SvRJygK7gNqHekw9IMAvfvuu+qRRx5RO+ywg9NZstOuAEnWXHNNtc0226g777zTW7aq8QlQ16nQFDjShSBA1cf9nxgAGDiOPPJIdeaZZ0afALqHdNj7QYBkWts555yTkBU3RQiQZOjQoWrKlCnqj3/8o7d8FYMAkcoFAQIAKI5DDz1UzZgxI/oEA4E5HQbSkc56PwjQyy+/rDvnpqQMGzZMbbHFFmr06NG1iCS98sor3jrMiCjJKE9cbqutttLSY9a/9957q1/96lfe8lUMAkQqFwSo+jACBNA5pHNOB33gWLp0qe6Aco6bI+epHwRo5cqV+qZ6U1C23357dckll6jrrruulmXLlmW6d+f++++3yl111VVahMz65emEP/3pT73lqxgEqHsxX1iaNfLI6Wz3FIX3Hw3kAxj046+LeGpb0UGAqg/3AAFAlZDOmDyRivuCGiMd9X4RIBESU1AmTJignnnmGe/2rUam2MkjoM365WELTz/9tHf7KgYB6qHoBwekvC/ISoeePpf2AtduBwECAIBeQzq85pOjIIl01BEgf5lWggAhQL0U+4Wj6cm6XfsJRat0T7lDgAAAAKqHdNQRIH+ZVoIAIUCZokc66tdIctpXON2stt6Sj/pojJ7yFpe33nvj2SZIXSzC9bX6G40CZRqVaba/NImK2mEcf55pfAMeBKj6uC8VA4CBQ17ox3uAoAxIZwUB8pdpJQgQAtQ0kfzU5cC9t8b97EqCrB+vpgTL0gWjLlDxNnq9JTLhNmYdvmghaTr6k2F/vul2npeVIkAAABUHAeo87ksWIUQ6LgiQv0wrQYAQoKbxiIApL14BMMtEAlUXpDBWOe829VGa+jbNRnaySVK+/SVHfyTZhKvDQYAAAIoDAeo80jGTp4CBjXTUESB/mVaCACFATRPJgn/aWQbh8IyaJGTDN9qSaRsnWbaRZNmfe2wpdTMCBABQcRCgziNPgxs+fLg64YQToiUgSEcdAfKXaSUIEAKUKTUJkhgSoKWg8aiM916aqL5YLvwSYQuJtx4nWbaRZNmf/dldZ5dBgKDj8BhsAKg6Mg1OJIhHY9eRjhgC5C/TShAgBKiluKNBTUdcUsTBEqcUicgkSXayjcZk259E6tNtTz3OcJQoKUZdDgIEAABQPaSjjgD5y7QSBAgBapTwwQBOx98UBd8IkF4fL/NPkbNHasJtXCHxbdNMNLIJUJb9GctmnpMy+hMkwwhYV4IAAQAAVA/pqPeDAL344otqhx120McbZ6AFaNddd1WPPfaYd/sqBgFqEN3Btzv/thS5YuLIhSVD9ViiEm0zf+bR9e0SYhHW64qUm0wClGl/5nL5vfCPcmUTri4EAao+PAYboHPI/T/cA9R97rzzzuin/kU6JVUXoPfee0898sgjau211446YWH222+/ARWgLbbYQl111VXq/fff95apWhCgJqlJQBxXBiLpiWKNlOiy7vbOtLjaNtHUNF2PKyPGugbCkTqKY7Yh0/6iRKNd3tGfFLkrRRCg6oMAAXQOHoLQfeL7gRYsWBAt6U+k01JlARL5eOmll9S//Mu/RB20ej796U+r559/3luu1YgA7bvvvlb9gwYNUgcddJAefeoHCUKAuhs9ipLhwQWZ4hUuO63sLyFPRko7+iNBgAAAigMBKgfyHz/SURUZ6lfk+KsiQCIZ77zzjvrjH/+o8/vf/14999xz6sQTT1RrrbWWPtY4Mhr05S9/Wf3hD3/w1tVq3njjDXXcccdZ+5Csv/766sgjj1RPPfWUev3119Xbb7+t2ybtrJoUIUDdjDMa1HZkNKrRqEwr+2sw7U5EqyhpG4ggQAAAxYEAlYepU6f29ei3dNKrIkAisrfddpu6+eab1Q9+8AP17W9/W+29995qzTXXtKREPo8cOVJdccUV3nry5N1331U/+tGP1IgRI6x9SYYOHarfQTV9+nR1ww036Pbddddd6le/+pW3rl4NAtTNZLu3p5XIyEy64GTZX31Kn7eeDKNMXQ8CVH2YAgfQOZ599lkdgG4jnZOqCNDSpUv1tEZXeMzIyM8nP/lJdcopp6jf/OY33nryRkZ4vvnNb+r63REnN6NHj1aLFi3y1tOrQYBI5YIAVR/pjIkEAQBA/yCd8aoI0LJly7QAubIRR+7JkSfBzZw5U/3617/21tFuXn31Vf1evTFjxiQeumAGAeoQCBBpJwgQAABUHZlC1W8jc9IZ7xcBGjZsmDrwwAPVwoUL1SuvvOKto53IPT1yDV1//fXqkEMOUUOGDPG2Q4IAdQgEiLQTBKj6rF69uq9vBAYAkA6c3MPxz3/+M1pSfaQzXhUBWr58udp0003VOuusoyMjPq54yNQ0uf/noosuUr/73e+89eSNPHTh8ssvV1tuuWVivxJpT9y2nXbaSd1+++3eeno1CBCpXBCg6iND9qX7iwugosj7R6699troE5SJsWPH6odU9AvSMa+KAMlDBURARG5mzJihTj31VLXHHnskREjuEdp6663Vdddd560nT+QhCPfff7/afPPNrX1JZORp99131w/cuPDCC3X75AEMP//5z7119WoQIFK5IEAAAMWx/fbbc89dSZHR8FGjRuk/+wHpoFdFgNzIiMw999yjjj76aDV48GBLSmR62le/+lX15ptvesu2Gnmc9umnn27tQ7LBBhuoww8/XD/1TUacqvw+IASIVC4IEABAccg0qyVLlkSfALqHdNKrKkASEY6f/exn+p4cV07kfiB5SamvXKuRJ8BJfWb9MvJ0wAEHqCeeeKLS4hMHASKVCwJUfbgHCKBzyBQreTEiQLeRjnqVBUgiLx2VERh3KpxMRS1qGtprr72m9tprL6t+uR9Jppf3g/xIECBSuSBA1Yd7gAAAklT9hbXSUa+6AEmef/55tdVWW1mCMmHCBPXMM894t281PgGSBx089NBD3u2rGASIVC59I0D/5/9T6p8/6cv8f//2qPp/PlrqXdcX+d//OboIAABC5Glw8lhl6dhVFemo94MArVy5Uu22226WoAy0AMmDD1asWOHdvopBgEjl0lcC5DsBpPpBgADAg7wXSDqzVX1ohRxbvwiQPOHPFJROCNDTTz/t3b6KKaUA/T/vEpI/CBCpfBAg6CAyrarfXrjZyyxYsKCyo0DSUUeA/GVaCQJUUgEC6BEQINKdIEDQQaRzhABBGZBrEQHyl2klCBACBNAOCBDpThAg6CDSOUKAoAzItYgA+cu0EgQIAQJoBwSIdCcIEHQQ6RwhQL2JvC6gSvcDybWIAPnLtBIECAECaAcEiHQnCBB0EJEfBKg3kfuB5EW2VXlvmnTUESB/mVaCACFAAO2AAJHuBAECgIxMnTpVjR49Wj8mu9eRjjoC5C/TShAgBAigHRAg0p0gQACQEREf6ehVYRRIOuoIkL9MK0GAECCAdkCASHeCAAFAHyId9X4RoHHjxiUE5eabb1bLly+v5YUXXlB//OMfvXWYkSmsZrn77rtP7bTTTlb9IlwIEABkAQEi3QkCBB1EOkfcAwRlQK7FfhCgX/3qV2r//fe3BGXIkCFq4403Vp/85Cd1Nt10U3X66aerl19+2VuHmSOPPFJtttlmtbIf//jH1eDBg636ZUTol7/8pbd8FYMAAeQHASLdCQIEHUQ6RwhQdTj33HN79n4guRb7QYBeffVVdeqpp1qC4suxxx6rXnrpJW8dZg455BC11lpreeuQiFx98YtfVG+//ba3fBWDAAHkBwEi3QkCBB1EOkgIUHWQqU6TJk2KPvUWci32gwC98847+ji33nprS1TcFCFAsnybbbZRN9xwg7dsVYMAAeSn0gL00aLx3r8s56/wb98889T8yeeoj7zrWszr56gpTrumLLrVv23TZGiXtb+j1UrfNp0MAgQdRDrLCFB1WL16tRo+fLiaNWtWtKR3kL+D+0GAJK+//rq66qqr1K677qoGDRpU+7fOTLsCJPXKvUCXXnqpeuONN7xlqxoECCA/FRagW9W9k4O/IF0xiESgdQkK68svKUZWHB38xT1e3fu6sSwWlJnz7G2bJlu7tAwWJW9FBAECgDZYunSpfjx2ryGd9n4RIMnvf/97dccdd6gzzjhD38czZcoUK9dcc42eLucra+ayyy5TX/nKV6yyX/3qV9Vpp52mbr31Vi1bvnJVDgIEkJ8KC9A8NT/4h8YnBvlkIKwv/+hRnAbC4hOjpsnSrkgGW5arAQwCBAB9SL8JkOS9997TIiRPhpOHFMiT3+L87ne/U++++663nJlf//rXibJSnzwO+/333/eWqXoQIID8VFeAtEykiIEebUkZgYljClJUVxxTXlbONMoEaS5IjWTEIzNuu8xyDdqVto15XHbbXfGqi1ptuyIFCgECgD5E/i7tNwEiAxMECCA/lRWg8P6flHtd3Glw7mePpPhGjbQYJESp+QhO7d6kZkIRyUtdbJKjWplGs3Q95rmIjs8oF7bJbLvsa7yaEmzXXOpyBAGCDvG//tf/UrNnz9b/WwzVRF6Q2iv3AyFApKggQAD5qagANZny5QiPFhlnW1csEtvoOlzZyT5NriZBEq/AhHV522XIjK/tbtxjcesIUx/x0Z+jc+QdVSoiCBB0iH/7t3/Tv2dvvvlmtASqhjzgQr5juS+o7CBApKggQAD5qagAJUdKrHjlxY49uuPIQVoSI0kZYk5RM0UmbTTJGs3J0i5XBtPLWDKVtv+iggBBh5D7D+T36y9/+Uu0BKqIjACNGDFCPyGuzCBApKggQAD5qaYANem8e0d3YgkxU5MGz8hOJDvJcjmlIRIha1QqUXecWICyjDg5MpgqabYoueeo8CBA0CF++9vf6t+bXn1xJmRHngon0+HKjFyLCBApIggQQH4qKUD+KV5x7BEQ/7Y+aTDFxj/C1Hi/QRqOPNkCkmVqW+P6orjCk1rGPKb0UaLCggBBh5DpUZJ///d/j5YAdA8EiBQVBAggPxUUIFsk3HhHf9xtnRGkhNh4JaLxfsM0GrHxiJk7AuPITFPhklhT5iT+Nth1NWpnQUGAAKAPQYBIUUGAAPJTPQGKJME3euETBr/cBCLjCpAlI8kRoNqUtSajNuF2yRGYcHmyHXUJCfdp1p9sVzK+bfS+zGXO9Du/4BUcBAgABpBzzz23lNPhECBSVBAggPxUT4Cizrwv/ild0chNbbtAQlwBqNXpWxZG6vaOJvnia6OvXE3GwiTa72uXk7Q2hcIVxymv67VFsfAgQAAwgEyePFmNGzeudPd+yd+5CBApIggQQH6q+RAEUv4gQNAh4kckQ38h4jN27Fh1xBFHREvKAQJEigoCBJAfBIh0JwgQdAgEqH+RKXDSSSzTKBACRIoKAgSQHwSIdCcIEHQIBAjKBAJEigoCBJAfBIh0JwgQdAgRoEmTJkWfALoLAkSKCgIEkB8EiHQnCBAAdBCZDnfCCSdEn7oHAkSKCgIEkB8EiHQnCBAAdBARoOHDh6tZs2ZFS7oDAkSKCgIEkB8EKBH3sdj++B+p3Wrmqfnm+3g68f6dsgQBAoAOs3TpUv33t0yL7BayfwSIFBEECCA/CFDTJF96WkxC0Sq+3h4JAgQdQjq7s2fPjj5BvyMjQAgQqUIQIID8IEDNEr2MdP4Kz7q2EopV8fX2SBAg6BA8BQ7KhFyL++67LyGFBAECyAcC1CQfLRof/IPlmZa24mj9D1kc30hOWLa+TU12vGWjqXcz59nlZYpcJGF6e3PKnE4oUmFdYTtr5aztShYECDoEAgRlQkYjCSkyANA6CFCTrJyZlI5YbGpC4xklCrc5Wq2MPofSUxeppKQkR4T0vkVsatu50/Gcz9E9RFMckSplECDoEAgQpHHuuefq+4IAAKC/QIAaJjkqE8uOO+LjipL+bEmIfc9PYn0kL/WRpmhkp5U6giTkrKxBgKBDiAB1854PKC9yP9CIESPU6tWroyUAANAPIEAN4464eEZ2zOWGAMUi4gpKGM8DEPQIkVFvQojiZVG56OeE6DgjTaUNAgQAJUDuoRg3blz0CQAA+gEEqFESMhGNCInY+JIyVU7HEqGU6W4JgXJEy2xPmui4IlXWIEAAUAL++c9/6qdpyZ8AANAfIEANkpQQz8hNhiRGgxKjO5FYGZKUPr0tak+K6LgiVdogQAAAAADQBRCgBvHJhE9M7NGYpMxITHlJipU7IpScepeo1zdFznvfUEmDAEGHiF9+CQAAACAgQKlJkQl36pl5X060jZakBiM8WoBMsXJlpoHc1CXJFa3os9OW0gYBgg6xePFitfnmm0efANJZtWqVOuKII5gOBwBQcRCgtERi432ampagUDYkvm1CCTJiilStfCQ5znS25AhRvI1fimp1rfCJU0mDAEGHuO6669Ree+0VfQJIR8Rn1KhR6oQTToiWAABAFUGAKhSvOJU1CBB0iHnz5qlJkyZFnwAaI6NAw4cPVwsWLIiWAABA1UCAejTJe5F89w2VOAgQAJQUmTbJu6MAAKoLAtSzMae/hekZ+ZEgQAAAAADQBRAg0p0gQAAAAADQBRAg0p0gQNAhZs+erQOQh1mzZnE/EABAxUCASHeCAEGHEPnhIQiQF7kfSKYYc08QAEB1QIBId4IAQYdAgKBdpk2bpkaMGMH7gQAAKgICRLoTBAg6BAIERSBT4RAgAIBqgACR7gQBgg4hU5eYvgQAAAAxCBDpThAgAAAAAOgC3Reg/7Kc9GMQIADoQVavXq3GjRun/wQAgN6kuwIEAADQY0ycOFFLEAAA9CYIEABUGh6CAEUjD0MYPny4fjocAAD0HggQAFQaBAgGAh6uAQDQuyBAAFBpECAAAAAwQYAAoNIgQAAAAGCCAAEAALSBvCT1hBNOiD4BAEDZQYAAAADaYNWqVWqNNdZQixcvjpYAAECZQYAAAADaZMGCBWrEiBFahgAAoNwgQABQaeQeIAnAQCMjQPKIbAAAKDcIEABUmrPOOkt95StfiT4BAABAv4MAAUClOfLII9WZZ54ZfQIAAIB+BwECgMog91/EU97i7LTTTuqYY45Rzz33nBWmKsFAIdfW6NGjuR8IAKCkIEAAUBmk47neeuupoUOHqg033FANHz5cDRkyRD+hy8ymm26KAMGAMnXqVP1QBK4zAIDygQABQKWQd7IMGzYsIT1xBg8ezOOKoSOMHTuWl/ACAJQQBAgAKoX8j/u6667rlR+JjP4AdILVq1erZ599NvpkI9fp0qVLo08AANBJECAAqBwyCiTT4Fz5YfQHyoBIkUzVnDZtWrQEAAA6CQIEAJUjbRTok5/8ZLQFQHcQOY+vx0996lPRUgAA6CQIEABUEulomg9AkJ8Z/YFuceWVV+qHcsgoZHxNSgAAoPPwty8AVBJ3FGizzTaL1gB0Fnkcttx75nsiIY/KBgDoPAgQAFQWucdCOp2M/kC3WLBgQUJ64gwaNIjrEgCgCyBAAFBZ5Clc0tHcfPPNoyUAnUFGII844oiE9JgRMZepmgAA0FkQIICKIE+WIskceuih+n/hfev6PTCwyDk+/PDDtey49/7E2XfffaOtAQCgUyBAABVh/wMmejtYhKTl08E1AwOPjEQef/zx+py79wHJ47ABAKCzIEAAFUEE6DPHT1fzfvo3Qppm9O4T1GZb76iOmPLV6AqCgUZEaOrUqVp8hg0bVpMgWQ4AAJ0DAQKoCAgQaSUiQJ85/ny112e/jAR1GFOEJEuXLo3WAABAJ+iIAMk86NmzZ5M+yRtvvBF989BJECDSSmIBkp+RoO4Qi9CkSZO8f5eS/gkAdJaOCZD8L9fe++9OKp5NN99Ebbb5J5GgLoAAkVZiCpAECeoe8u/j2LFjSR9myy231N//3Llzo6sBADpBRwXopb8vIRXPHhPGqD3320XtNGZHJKjDIECklbgCJEGCuoP8+3jFFVeoRx99lPRZjjnmmJoEzZ8/P7oiAGCgQYBIoREBOvmCo9QpFxyNBHUYBIi0Ep8ASZCgzoMA9W9EgHbddVd10UUX6evgO9/5TnRVAMBAggCRQhMLkPyMBHUWBIi0kjQBkiBBnQUB6t/EAiQ/xxL03e9+N7oyAGCgQIBIoTEFSIIEdQ4EiLSSRgIkQYI6BwLUvzEFSBJL0LXXXhtdHQAwECBApNC4AiRBgjoDAkRaSTMBkiBBnQEB6t+4AiSJJej73/9+dIUAQNEgQKTQ+ARIggQNPAgQaSVZBEiCBA08CFD/xidAkliCFi5cGF0lAFAkCBApNGkCJEGCBhYEiLSSrAIkQYIGFgSof5MmQJJYgm644YboSgGAokCASKFpJEASJGjgQIBIK2lFgCRI0MCBAPVvGgmQJJagm266KbpaAKAIECBSaJoJkAQJGhgQINJKWhUgCRI0MCBA/ZtmAiSJJejmm2+OrhgAaBcEiBSaLAIkQYKKBwEirSSPAEmQoOJBgPo3WQRIEkvQokWLoqsGANoBASKFJqsASZCgYkGASCvJK0ASJKhYEKD+TVYBksQSdNttt0VXDgDkBQEihaYVAZIgQcWBAJFW0o4ASZCg4kCA+jetCJAklqA77rgjunoAIA8IECk0rQqQBAkqBgSItBIRoBGbbqm2Hjshdzb42CbqMwcdHF2BkBcEqH/TqgBJYgm66667oisIAFoFASKFJo8ASZCg9kGASCuZcuECPQLUTvY45Cj9dzu0BwLUv8kjQJJYgu6+++7oKgKAVkCASKHJK0ASJKg9ECDS6Zx07RIEqAAQoP5NXgGSxBJ03333RVcSAGQFASKFph0BkiBB+UGASKeDABUDAtS/aUeAJLEE3X///dHVBABZQIBIoREBku+63Ww1asvo6oGsIECk00GAikHOIQLUnxEBMv/tayfS1wKAbCBApND86GfXqpsemdNWzp17or5eoDUQINLpIEDFIOcQAerPyCOt5btvNwgQQGsgQKR0EQmiU9U6CBDpdBCgYihGgO5S0/dfQ61x7NWJdVcfKyMEx6mrneV3XTBBrbH/dHWXszxbrlbHBe0+7hrfujC6/mCbNdaYoKZ/b7qaIH/e4d92YBK2MWxDFOf86HPjOWd5U3R9WSPHhgABZAcBIqULApQPBIh0OghQMcg5bF+A0oRGJGCCmhDIkS0roTBNuOAuY1kraSZAzvo7OixAen/+Y84vfc2DAAH0BggQKV0QoHwgQKTTQYCKoSgBevSa44K6nJEeWRZ0yJMd8+YjOI3TpLwrPB0WoNTRrQFuBwIE0BsgQKR0QYDygQCRTgcBKobCBMgjJdIh16M8IkKmECRkKRodCcqHMddJvYE0XCBl4nXuvsLPerrbJfF2UUQIPOKhZaG2nbHOI3IJsWgiMuH0O0cGPTHrjaXp6trUPYlbR3ycYSZcMN06D/521rcfKDmSuhEggOwgQKR0QYDygQCRTgcBKgY5h8UIkDutLRIXkQRHGOwRkqhTb3TOQzmJO//RemtEJVwWdvyT5ROCYn1OTkULhSVeb9Zd/5zY3mqPm6hMlLSpfgkBsraN2lk7rpTPQRmvAGmRM85BonxxkTYgQADZQYBI6YIA5QMBIp0OAlQMcg6LEaCoEx93sEU6apIQdr7Dzr35c7DOM+LiExxbIuL1kWi4nfpGAuSu07HblBCJY6cH6+MyTvsbJJYaM3Wx8gmQfR4s0fKdJ30sfgGSnxNt9B57+5HjQoAAsoMA5cwPZ9h/oa6xxj5qzipzm9vUnC+524Q5/Qlzu3oev3kfZ9tj1A8921U9CFA+uiZA14cv4jMz5uJV/m3bzX03qjFrHKaOvC/8fNrUYH9TH01u10Yuuv5RdZFneS2e49U58MbG5QYiQVtP8y3vUBCgYpBzWJQAhR3ssJMunXezA16XI5GWeifc6uQbqXfgTRmKE4lPlKYdffOzV7jM9kXbRG2Kj8NuTx6JiNtcL5sQIOc8mMv858k+N/X6otEe4xzVk6ftjSP1IkAA2UGAWs2qaWqS/AU240p7+RPhy8wm3XxbtCwUoPpneztbgq5Up0udX5qmHje2DYXIFavqBwHKR+cFaJU68kD5x/wipxP+qDpYrueBEAJHgIrORRcf1rzdWoDcY47ORSclyNuOzgYBKgY5h4UJUE0Owg64JS2xeBiSJMv9HXtzlMXu5IeJZEI6+7pep1OfQ4BMGalvL+0wysX7i9urt5O/h6IkjsNN2O5Y2AZOgOz9+KLri9vdZNtmkfIIEEB2EKCWEo3quPITR8tNPGqTIkCe5Xo0yZGfLOuqGgQoH50WIC0LqR3wUIIOvt63ro2UVoAaLB+oIECVQc5hcQIUdsLDm/NdyZBO+QR13LFBxzsWDYlXSMyOvd3JT66PRjtMOWgkQO46HVO4jM/HHmdM45N9Hhe0P9iX2f5Ewrb5hcLeTysC5D1PkYAlBSg6Jw3bWVwQIIDWQIBaiR79yToik1WAwtGftGlx/RgEKB+dFaBwxCPrVDctFlNvjEaMjHJaaMJlOu50Nmf9wRc3mQLXsL5YyqIRqihxW0Khi5c3kKxWBKjZ8dVG0eI45XWdxvq4vLO8LpqN6pPjDo7r4rhs+/KEABWDnMMiBag2spDofEed8mCdLQehMJjb6458rbPfTICCRCJQq7eRAMXtMGQjbLMtRcnjqLffbksycdnEdo7EtCRACalJtsesL9yX3YawXa5sth/ZDwIEkB0EqIXoKWmZR2NSBEhGicw6WpKq/ggClI+OClCLIzGxXFgjQroTb9YRdd5rkhCKSl2yYnFJEaCM9Vkdf6dM/hGgcF/J4zOXRft321P7HB1PXHfiHIfb1+pLtCNcb0ppeN7jbaL9FThNDwEqBjmHRQqQOyphJuyAu6MvknpnPozZSc8gQEEs6WgoQGFCyYrjaZPnOFoSiKh8fR9BHMFpTYAkkSxGafoY7EiC6ilefiRSNwIEkB0EqIV4BSi6p8dMOJoTCpC7LozxcAMtQP35sIO0IED56L4ARR1sM1Hn3u6Ih5HOfmIEyajXKyOOsJgC1Kw+nyC4UpFdgJzjjGLWbcmZVTY6DwmBkRjtcY41Ebe8fE60PRSzsF2+428vCFAxyDksVIBIByNC5BPKzkauIQQIIDsIUAtpPgJkTmdLmwLnPPCAEaBEEKB8lG0EyBSApFi4U7XMhPV6BcLZb32b5vW5shMmrwC54hJLXlyXKR7mdrK/sD1p+6qLnHNMjWQqSLx/X0wBso+/vSBAxSDnEAHqgejRHM8oljNq1I3INYQAAWQHAWolTWUliwAFsR6W0OQeIGOEyH1MtrfuCgQBykfZ7gFqLEDNRyNaE6Dm9fkFwF5mtzNcV/+dazRyIzHPScr5aTbClVZO7zNqR3xOnHboc9FQ3pLH70pT4/OXDAJUDHIOEaDeSDzNr5YSyI9E2oIAAWQHAWop0bS2tKfA5RKgxiNLPAUOstJZAYo7zz4RCNNYgKLRDVdwjHgFQXf6G4wANaivdQFKSSYBShE4s6y3Hl8bjTQo3+z7aFp3jiBAxSDnEAEi7QQBAmgNBKjlOFPY4sT3AtWWpwmQb7m/znDEh/cAQTY6LUAS3ck3hCRM2NE2l6fLjG9EIu7Eh/XURyWS9VqSkbE+WwCcZV4pcZK2jbs80Z6o/TUpcj/H5zOqQ5e3z611vIlpiMn6wm3iNviOv70gQMUg5xABIu1EriEECCA7CFDO6JGZ4JjqcUUlGi2ytgmTNnUtWWd/PhwBAcpHNwRIJ+rom3GnUqWOrCTKumIRdeqjNH0MdsP6fALgLqvvL1UUPMcbxiNFkYDUtjHbqhONXKXUEQqcsd43ihYsT0piPe6xIUDlQ84hAkTaiVxDCBBAdhAgUrogQPnomgCRvg0CVAzdFSD33TaeeN5n03I8j8FuFN8jqe24j+2201ZbM6Z5GzsXOWYECCA7CBApXRCgfCBApNNBgIpBzmG3BWjC/hOcl6PGuVodJ+uCNpZRgLxtLkLYMgQBAuhdECBSuiBA+UCASKeDABWDnMOuC5C80NPXmQ9kwn3ZZ650UoCCJF5IOgBBgFL4j/9JSOvpMAgQKV0QoHwgQKTTQYCKoRwCdJeWBltyZJ1Ii7zs01mnhcaYdubKhrP+uECiEgIUjdTEMWWmPQEK19ltipbV9necutos4x6PxDkm3aba+uPUdATIz//6s1L/6QFCsue/LIsuns6BAJHSBQHKBwJEOh0EqBjkHJZBgLSQmJ1+kQLdwXcEKDHFLFxfLxt+rstJtN4QoFAmTAmxy7QlQLp9pmy57XH3H663BM89RrfOaD0C5AEBIq0GASIEAcoLAkQ6HQSoGOQclkKAtAjUpUQkob68LgPe6WVaCMKyXnmxBCLcpyUctW0a1GHFHdEx4xlpStTVQKB0zGP2b6vPAwKUBAEirQYB6lRaeUS1+XLT5mn0UtNW8vgTV/bVy0/NIED5qLwAuY+UDtL4kdYDmLR38EimPpp8PHdFgwAVg5zDcghQ2Kmvd/pjkWguA+E24fZeQbLuAQrri3+P7bQmQFY7olEZV6zC0R7fvtzjCOs014d12QJo1YsAJUGASKtBgDqVtJeURi8knXGlszx7ihCgoiSqV4MA5aPKAhS+INR+KWgsRO47cDoiQE5S33NU8SBAxSDnsCwCpCVC5KU2/U22ySBAhuA0FSD9c1IozJhy4QpMuG9/O+Jtzbqbj9QYQuZM48sqQHofcR2ebQc6sk8EiPRsEKBOJU2AgqyapiYFbc064uMGAWo/CFA+KitA+sWjjvx413VPgPplxMcNAlQMcg5LI0C6sx/e4G8vq3fqvYLTbPqaXm+PACUkyoi3DispIhYtr9/fE0tR/XMiRttryyxJ8++ruVh1LggQ6ekgQJ1KAwEK8sMZwV9qtVEgdwqcb/rcMeqHUdlYXuZIHfF6j8zofdTK76PmrDLKe5bHYlZb54xS2eXSj60XggDlo6oClF0uXAFapY48sP47EeYidZpRRo/cGOvro0kSY1qbjlG2NgXO3UcoY4k2u9P3EusuUkfGbemhkSQEqBjkHJZHgGKBiGVF4oyAJKaahevd0RNXoMw6Qykx92ELRX4BChLJS2L/prSZgmPJmbF9vF6WudtE5wAB8oAAkVaDAHUqjQXIHoGxBciWo/r6eFksIvW6I2GqlXE/B3nimKCMI0GmNDnrE3Xo9XUJC2XJ3L63ggDlo5oCFAqGLSZpsQUoKU6R0MTL9OiRT2rkc3K/Vn3Wtsl9WZ8TI1iRNFl12eV7JQhQMcg5LI8A+eQjFIK68ASJBELaruOOCBkSIfE9BjuUIKMOY59tCVCQeEpaQtKMmMfjtuW4ayIRNI8rlh4dHoOdCgJEWg0C1KnkFyBfTClKyIvEFBL9syErURrVIesSbTXqDKUrWWevBgHKR5UFKNu0tuZT4EwxCUd/7BGhesK6UsWrBQGSnxP1mOUjAerG1L12gwAVg5zD7gkQqUIQINLTQYA6lfYFKJSOoJMTx5QXc3RHx6hDj9YY5cx4BSga7fFtXxvliUahoqQdV68EAcoHI0B+AXKnudVFJRoRiuLuwy7niFJmAYpGe4z91GMKkDlC1DtBgIpBziECRNqJXEMIEOnZIECdSmMBskdcbAHSIzXBZ3PEJTF600CAQnFqPFpjC1BYNqvUhPWHbdRl3HuH3NGpEgYBykd/3gNkjhDZAqTL6eu+Li9pdZmyk5QtU5SiujILUJORJIlHgOptD1PW0SEEqBjkHCJApJ3INYQAkZ4NAtSpNBCgxP0zpgDZMhQmGqHxjt5EMetM3M+TjF2HXX/WeNvRI0GA8lFVAWo4QpL6FDhbhsI49954okUo9SEERp2tjgA12CcjQIAAkXaDAJGeDgLUqaQJUCg4tmwkBcgsVxsRMgXI2sYVmOizJSdOve5DDaJpc6Z4hfsJtzF/DtenHV9vBAHKR2UFKEg4IuJIghYHc3QlKUDmyEttVCWSkeQ9QOZ0u2R566EJmQUoLmfLmLVvBKjvkXOIAJF2ItdQrwvQR4vG6+NYY/I56iPPesnKmeHf4/NXRMteP0dNMT9buVXdOznYfuY8z7p5an7avqI6pyy6KNzGWz5b4vZ60+A4i094LqYsutWzLsqKo6O2Ha1W+tYHib+jhvXkCQLUqUQSEl+ERtKkqCYf7pSyQGwSMhLIjfUY7MToTXL/9n4jEQtS22/i3iF7Gl19al6UxD57JwhQPqosQDqRSJixR3icUZ9IkGrbB0LiSk9NioxtavW55U1JaUWAJIm2G+KFAPU9cg4RINJO5BrqdQHSsjB5vJqSJgZaTGT9eHXv69Ey3Wk3PjvRdXoEpi4myc5+uC5Y3lCusqSRgDVaNwDJcCxabuT8pwpQKI1y/vOfk5QgQIQgQHmpvACR0gUBKgY5hwgQaSdyDfW2AEUjFDOPTu2Ai5jMXyGd8Pr6mqwY25nxClAkA3LOEmVroz+3RnKVXnfzRMKQNlrSdv0tpIkoSsJzdXTQZv92IkhTFs0LvqfG9eQKAkQIApQXBIh0OghQMcg57C0B8rwjx03iZamtxvPuoQFM8/cOlTtyrntagGojFLbg1CId+EBk9ChFTWiaj6LoTr0zohRL0716Ope9r3CKV7Qs2qe9TsqGaToK0mzUxSNAYdvi+EXD3sZXf3RejPXWcXkTy1qK4MixyHmUNnvPZ5yccoQAEYIA5QUBIp0OAlQMcg57UYAm7D8h5UWkgbzIuuC4OiUw7QYBKpA8AlQboZCOuNuJDjv081dEo0S1EZUmIyxBdMff7LAnRnjMfaXXlxCIRNlkwjLp29hyFktLfR8+abHLBEm0w5VC+Xy0mi/lGohiXdbic22vl/3KedH7dwW0SZszBQEiBAHKCwJEOh0EqBh6VoAumB6IjkcarjkuXBccFwLUmfS6ANU7zp4OeDzq4I6oNBthCaLrNYQhHK2IOuiOPDTqvNsdf0nYzkbylZAVI+G+6m0PP7uy5AiZPl7/NnY9zjHo42zcVvNcxLJTW6f3K3Xa7cnU5qxBgAhBgPKCAJFOBwEqBjmHvSlAd6mrj3UlR9ZNUNPv8Exhu2O6HhWS49VpNIXOmQIn+1nj2Onh1LtEef+UvLBMuCz+Wf8ZlzeEJxag6Snryx5pby8LkCkY8nNdakQ0ok62O9rhfvbEEgJXmKzyjTvusbA0HEWxEo+OpMWUlGjbRN3NJcs+ppTtWxRF+dmso/Z9ePaVq82+IECEIEB5QYBIp4MAFYOcw14VIH2vjykeIjlaHBwB0vcEiRhFn1OkpR6PAAWf61PuwvXxZy0waxynrm5Svi5kvvJm/c3aV65I23tXgGz5sEYgRFIMMUodzUmJKUBuebNDb4lSSmoSJGkqQn6hSrRBErWjVreTWh2p20USlyY6enkjUbRlRh9nfHxS1hAjVyaTbQmDAEUgQKSVIED5QIBIp4MAFYOcw54VIC0SdfEQkagvtwWkLhdR9IiQKUVmPALjjshoqYr2HY0u1QTHXBfEV14viwTHOwWuYfvKFbmGelaAnI67dLTDDrR0zOOOuzviEH12ZcKNHuUJOu0+Oagta23aVk2EGkmQb3864b6ssnrbxiNZaUKVlJJkPdY23jh1O9IZH4Mlb5na3EIQoM4m8e4cN9bLSluJ8+4g0lIQoHyUUoA87+4JY76ANEzi3TkN47zzJ3ceVae1XUe509p5bS0IUDHIOexdAQpFIhQPWR4Lgykw0WiK/t1304IAuaMxlqDYIzbu9r7yCQFy63faUObIuexZAXKmsukOu3TApZNdEwVXADJKS1T3FJ8sRZIi77VJFwRXvMI0k4pwvV8QkuvCY3FlqblwOG2LjseuJ4MouuXknOntg3bVyrnnIUObWwkC1MXoF5zuo+as8qwjHQ0ClI/yClBSdsIXkrbz8s8iBKgoiSp3EKDyI+ewlwWoNg2uNv1NtjHlIfw5MQLUMK0KUJDaqE9SXBCgDtKiACU6zdEIxEeLjq53+B1J8nf2PdHlgvq924adeFnXSKR0+yz58EuRmYYiEIuXsU93+zRJSpSR9tfakZSd5DbJJGRO2id1BOeuds4857t5m1sIAtTFIEClCQKUj14SIInumB94o7rIs655EKCsQYDKj5zDnhYgLQrHqem16W/xslge7NGZbPEIUKMpcDpSJhCiC9zlGQWIKXDF0JIAeWTCIwiJTrohNskYnfB4O6+MxALUaHpYmJpIxGkgFJkFydmvvQ9Pm5xjlvOjy1j7qUudJPno8GRckanVYdar952Um6ZtzhoEqItJEyC9/Bg15+Z9wi84nhb3xDHGlx5m0s23ReXsKXB6qt2MK+0pd02n14V11Os/Rv2w6brb1Jwvme2Iottqli93EKB89JoAzbvvRjXGGAVyO+r6s3WdmyNGkbxcLHXUt0nIjDMFb8zFq6zytXW1/a5SRx5oLE+03SmXdmyS+PguNttgj3qFx3yjtU/dRl3WU8ZbZ9gG63y55xEBKjVyDntbgCLBsWTBGT3RsmKPpmjpcESlHo8ABZ9dwXJHleLtGsmOb1nYFs8xOWXKGml7r44AEYIAdTMNBSj4SzAQmEbbPq4FKV7mESDjc7w+ISq1RIJj7FPXoaUpKTmxYMnPYTts2THX90IQoHz0nAA5IzBmR11PkbNGhyIxqS2LRcSoW++rLgvhNDtz32EZV4Lq0uSud+sI22CubygXNYmpt8FtUywttTbEwmYcu94m/hzXWVtfF7ZaHdE2vvNadBCgYpBz2NsC5BtBcQRIEklQPab8uHV6BMh5DLYrPzoe0ZJkEqCg/dZjsHtEfiTSXgSI9GwQoC6miQA1faCBVd4jQM6IT0MpaThik02eXNlq2v4SBQHKR5UEyBdbilx5CVOvIxQDd73dHqcOWWeIRxhTepKC1DCOiISxJcqSG53kcVnH7anTPi+S1s5rO0GAikHOYW8JUOfjExhvEtPi+iMIEOnpIEBdTEMB8izXiUZqamkgQI7sNBIgPYrTYIpcOMoT7zMpSlbdPTb9TYIA5aOSAhSPiMSxBMieTiapy0BYt1W2Fr8AhaMzvu3rwmJvk3ZcUbSsJNtoHmfymO02SZICZNeJAPU+cg4RoMbJKkCynXdkqOKRawgBIj0bBKiLaUWA9DLpABkjMdZ2AytA9ZgCZkiOIT2yn/TRonIGAcpHb94DVF9ndtTrouFMacsqQN7RFzceUbBEolHCsmEbGx0fAgTNkXOIADVOUwHSDywItnEfZNAnkWsIASI9GwSoi2lBgLyCoqWjGAFqfdTG3l/t880px1TyIED56DUBcjvm9c/Je21q6y0BSgpOvY5wfePpanYdoXQ1GdVJxN8OHa+EeabAIUB9DwJE2g0CRHo6CFAX06oAmYKitwk6GUUJUFTeGrmpSVGjddHnILVpcplGksoVBCgfvSRAoWzYHXlXgMxOe7h9sMwRIKtuva96nan7qNXhilZUpykLlsSE6y2pajrCZbcxbFP9MwIEgpxDBIi0E7mGECDSs0GAupgWBEiiBSY4pjCy3hSTVgXI9/jqsI76PnzCFSet3b03/U2CAOWjvAJkXqtxktJgd9RjwYkiHXxLcKJOvvUYbFsMJDVximOJgrG+ttzZbxBTRupSE8fcpyNUsax4Hlkd14cAgSDnEAEi7USuod4VoPC9M01fajogCfbtfUdQAYneZ5T2PqDEu40yZuXM1l82Gu4r/neonkbvB2o3yXcUNQgCREh2AZLratKkSdEnKKUA9XM8slK1IEDFIOcQASLtRK6hnhUgLQqtd+rbT/OXhLaV2otLfZLT/GWpvmiRaVnYUvYVCdrAiGcotZnPLQJESHMBkuvpgAMOiP5iofMVgwCVLAgQZETOIQJE2olcQ70qQHlHQtrPwI486RGQQFbuDf5MikCLghClpVGVWtL2NYAC2KrUIkCEpAuQKT6DBg3Sf/q261cQoJIFAYKMyDlEgEg7kWuoVwUoFoX6qIavw26PYtRGQmqjLI4YpHTAa7JllnP2pdtTW+fWUZeG2nZeITHaK/tyR218oy/Rstq+rXrDc1JbZ9bX4Fh0Ukd6/ALU+PgzbqPbVJfa2hQ89zzEQYAISQpQmvjEgRAEiHQ6CFAxyDlEgEg7kWuoNwXIFhsdr7zYozUJ+Yg6+fXOvL29uSzeJjmdLGpLouNujk5JHePVlGC7xiNH5r7CMubxhPUayyKJqdcZlm92XmKxqJXzyY6uOykpyWOLjt84J4l2ZtrGkdrEsXmCABFSF6Bm4hPnhRdeqOW//bf/Fl11Sv2f//N/rHW///3vozUhf/rTn6z1nSr75z//2VpfVFkEiHQ6CFAxyDlEgEg7kWto3Lhx0RUV9rsWL14cfVL6Z1OQZs+erVavXq1/lj/lc4yvbLyt8Nxzz6l//vOf+mf5Uz7HyHarXv6Jv5PrjS0lkmTHPIjViU92wmvlasvCbRrVqzvphmD4OvKJ9iVEKyWOiMi+zDKWIHjOgY4zipL4nNIWu+7oc7BdIr7z55535zxm2cb6nEV+JAgQIXUBWm+99fSfrUSkIOZ//+//ba076KCDojUhl156qbXeLPvv//7v1rpmZZ9//vloTbLsZz/72WhNyGWXXWatN8v+x3/8h7WuWdlf/OIX0RoEiHQ+CFAxyDlEgEg7kWto6tSp0RUVSsu0adOiT0r/bErNxIkT1apVq/TP8qd8jvGVnTVrVvQpvF5jmYr7dzGy3cQD9vV3cn3xjFi4YiKx5SYUBrdTbW/j1uOW8Xfa3f0mtrNErEHc7Sx5cfblik0cfW7qdbjH55cRd7u043LLu+ejnvp5zLJN8Dn+TmdmlB8JAkRIXYDkf5bkL1MRoWHDhullvojoxJERFBNznYiJiciGub7XyyJApNNBgIpBziECRNqJXEOxlHSdVqbAJYTCNxqSTRi8ghB9dtclhCjqtMf9Cjdxe5L1+KOFIHV/9jEmt43inBtb6KJzYrTRSq0+3/mMY7TJI6JhjHOfZRv5rNvta0uDIECEJO8BaiZCEIIAkU4HASoGOYcIEGkncg31ogDpTr0pMwkhktiyYo9cxHE64RKpS3e+7fI6zuhK4rM34T78MmHG05Ygut0ekbDFxtk+dYQmY1tSpUXiCpDv+A2ByrJN8Dn8TqPtvN+nJwgQIUkBikkTIQhBgHLm6b+quU9+oOb85E/6Z+82xBsEqBjkHCJApJ3INVR2AUqKS1IUfCMsYbl6JzohTRJf51wvO1rdm1qnWYdHkoLofTmjKX6ZMGMLQS3SnqCulc6+fcecqEMfi71vrzg5wpE8TiPWtv5js8tn2cYVs5Rz4QYBIiRdgGJMEaLzVQcBypGnP1Tfe/R3asIBn1FfOfPb6qzv36/Ov/N5demS19ScJ94LtvkwWYbUggAVg5xDBIi0E7mGSj8C5I4G6M+eTr0hA2Hn2lwWdbBnHq3mN+1kh8vdfUh80uHft9He1BEQJx5ZCSNtD58gZ+07sX0kholtnH275zOqxzwP7jHVkmXbDN9PcpukJKW2wQwCREhzAYqJRQhCEKDWM2fFn9WJ825Xa6+9tlpzzTXVWmutpTbYYEM1cuvt1ddP/Zaaz4hQwyBAxSDnEAEi7USuodILUBDdGQ7aGicpCXVpkUxZNM87oiDlanKkt3PlR+IRiThRx90SiCB2+5yRE10mZTTFjCsmRmptdkduIiGp7TsxJa5+XqxjrR1HGPt8RsdvrDfjO2f28fuPoeE2vmNPCJ4nCBAh2QUIbBCg1iPT3r4249roL/J61lp7kNrr80eruQhQwyBAxSDnEAEi7USuoV4QoLbTQC4SydLxJuUIAkQIApQXBKj1zHr8XXXI4Ufo683MoMFD1BdPPC/YhilwjYIAFYOcQwSItBO5hvpBgHxT19KiRyo8DxcgJQwCRAgClBcEqPVcuvxttfEnNtPXm5lh66yr5tzyoLcMqQcBKgY5hwgQaSdyDfWDAGWRmtoUrYyiREoQBIgQBCgvCFBrmf/TD9UNP3ldrbNu8oW766y3gbrmsde95Ug9CFAxyDlEgEg7kWuoHwSIVDQIECEIUF4QoBbz1F/UBd+9RQ0ZMkRfb3HkQQg77bK7mvukPAXOU47UggAVg5xDBIi0E7mGECDSs0GACEGA8tLPAjT3qb+quU+3dr/O7Cc/UOO/cLR++ptcb3EGDR6szr14rpofCFJ9+6BueV8QD0WwggAVg5xDBIi0E7mGECDSs0GACEGA8tKvAiTyc9aCh9Rly9/2rk/LnCfeVeMnfk5fa2YGDxmqjpt1i739U39WF869Vl1y/yv63UHWuj4OAlQMcg77RYB+9KMf6WOdOXOmuuiii2qRz/fcc4+3jJk777xTv/7ALXvllVfqupctW+YtV/XINYQAkZ4NAtTbee43t6rlj1yprrs5+Mv4ugvVnGa5aa6ad9ut6uFXX1S//MdvgjoeTtTZj0GA8tGPAjRnxV/UKd+5U22w0cfVhT/+jXebtHz7oVfVFlttra81M+tvsKE6+/ql1rYyWrTnvp9WO+86Vl34oxeQoCgIUDHIOay6AC1dulTNmDFD7bnnnmrkyJFq0003tbLZZpupBQsWeMuamTdvntptt90S5aXOsWPHqosvvlg99NBD3rJVjlxDCFCbqb1TJ/1dP/F7fPzvHWox+jHdGR/pXfUgQL2ZlX++R72w6nr149suUPO++TV19GGfU4ce9Bn1mWb5/BR10Femq1l3PqMeeO1N9cK/PqtW/v0x7z76KQhQPvpNgGY/uVqde+NSte4Gw9Ww9TZQF9z7ine7tFx436/1ww7Cf/Dq+cRmI9X8pausbUWAdttzvBo8eIjafsxu6uJAnkS+zG36MQhQMcg5rLoAfetb31KjRo3SLx02f9/iyL13V199tbesmcsuu0xtvvnmqXV84hOf0BL08MMPe8tXNXL8CFB7CR+xPT6QkjQBCl9GOiXYpoh3C4UyleHFqv0QBKg38/yvv6+enPMZdfbhu6vddt5B7bDdtmrbbbdR2zTN9mrbHXZVu33+bPWNKx9WK/7839XzH73k3Uc/BQHKRz8J0Kwn3lcnf+dHap31N9T38Axdd73WBOjpv6qr7npCDR46VF9rcaSuzbfZUc1+/B1r+1iApIO1VtCB22n3PdUFP/qlfpGquV2/BQEqBjmHVRagW2+9VY/OyO+P+ftmpggBiiMjRIsWLfKWr2rkuBGg9hI+YvvoQHL8ozIiLFMWzVP3BgJUxKgN7ykyggD1Xt7+xyPqNz+/Rl03eTd1+K5b6WH41rKlGrnzfupTk7+lTlvwE3X7Cy+pZ/+6Uq38+1Lv/vohCFA++kWARH5OuvJONXSd+uOrWxWgOT9ZrSYderhae+1BtTokaw8apE4867yE2JgCJNuJKO04Zjd1/uLn+lqCEKBikHNYZQGaPn26nuIW/575UqQAySjTd77znb66H0iOGwFqJ9HoTprgyHQ1ea+QTJNz3i9Ue+9QFHd0qPbyVmuK3a3BfsypdOH+zfLeESJdR9y+sMz8FWFdYd09OqUOAeq9/OPflqq3nrhGnTd2jNrfKzgZs+On1MhPn6HOuuZ+dcczz6gHXrxFPbjyxnx5ZbF67M2H1IsfPR2I1HJvu8scBCgf/SBAMu3tW9ctUetsMDz6yz5MqwI067F31UaeF6AOGTpMnffDn+oRInN7V4AkIkHb7rSL+vaDq9ScFX+2tu+XIEDFIOewygJ0/PHHq+HD7d/ZHXfcUR1wwAFq4sSJOpMmTVI333yzt7yZ73//++rLX/5yrZxkm222UYMHD7bqnzZtmlqyZIm3jipGjhkBaiP6fpy6TLgSI5IjsuKO2ujPphBZgmKUDaTKGu2p7U8+RwLjEytnhKgmU/I5qsPcny7Ti9PqEKDeyz+CvPX4d9oXoK22ViO33VXttvd+at/gL/T9Ju2j9v/M+Nbz2Qlq/6PPVmcsuF/99MP/ql746Jfedpc5CFA+qi5Ac5/6izr5ijv1tLe4kxOnVQG66sEX1IbDP5aoZ9h666tvP/C7xPY+AZJoCdp5Nz0drtXHcFchCFAxyDmssgAde+yxasMN7d/bCy+8UD3yyCPe7VvNySefrD72Mfv3+Zvf/GZfPQxBjhkBaiOGuGhhMR9yoEVDpCIeJYrW6eXuiEs8KhN/9stNuL/6SFBivbsvnWjbWIqiESVb1tz990gQoN7LP/7+kHrrsSsDAdq5PQEqKltupUaOOVDtN3Wumv7Eh+rBN5/ztrvMQYDyUWUBkhGWE+fdoQYPXUdLh1wfZloVoOMvv0UNGbZOop5dx+6hLlv2VmJ7EaA9xu+XECDJWmutrXbceRc1ffFzfSdBCFAxyDlEgPIHAUKA2o05shLe61MXDxEiLRTWqE1KEtv4RCYaqZl8tJrvlZ8gGeTKGg1K2aZnggCVL6/8/WH1248eVs+/e4/62R/uVCuC/PxPj6gX/jV8WpsrQKNGb6122G1Ptduee6o9PrVbRzNu3I5q7E4j1TajR6qtDjxKbXvZC+p7zz+eOKayBwHKRzUF6EM164kP1DnXP6IlZ801/TdRD99kU3XRg696yidzxc/+pv7l/Kv1+37cesZ/4Wtq1uPvJcrI6NPx0y5VQ9dZN1FGIg9G2HbHMdHT4fpnOhwCVAxyDhGg/EGAEKD2Yo+saLGIR1lERAwxsqaXRbJjXndhDHHxiky0P6OMKyyJfUms6XVhHa5YIUDZQYCa5O1/LFP/+W+PqgdvukRdft6J6vQgCx/9mVr27h/Vix8tTQjQruMnqm9850F1xYP3qjufvqajuWPRVHXT6SPVQXuPVFvuc4Ta6ozl6tqfPuI9rjIHAcpH9QToQ3V5ICMnzb8jvOfHM/Kz5lprqRGB/Jx8zQNqdsaHEcxd8Sf19W+cqB94YNYlN05/6aTp+gEJyXIf6hetHvaNc9IlKGjLtjvtqi6465eZ29LrQYCKQc4hApQ/CBAC1F6cURoRjUiAaqM/0c/1ERf/yI4rLt5Rmlic9D6caW06vmVuXf7925LUQ0GAypf3/7E0+F1+WC361nHq2An7qH2CTF/0lLr7zX8NBGhZQoD2nHiYumDJ2+q+N1/z1jeg+cV09ezskeqIT49UI/f+shp54oPqmqc825U8CFA+qiVAH+qRmKlzFqkhDUZdNhixsZafVkZdLn/sXTV8o00SU+mGDBmqZt3yoJr3VMr7fZ6W0aj31ReO/6aePucbjdL3BO2wkzrvtmf6QoIQoGKQc4gA5Q8ChAC1lUhIaqMmIhFaNALJqAmHIyWNRnYMcdHS5IhMKCmOJFmjPb5RnHCZvf+UqXXu/nohCFD58n6Q//XBPeq6Yw5VX4juszlpwTK16A//qn7pEaC9PnO4mvnYanX/O3/w1jeQeeX56eqleSPVVw5AgPqRKgmQTHs7e+FDati66+trwU0sP+fdKQ8fsJ/Y1jiBxCx/y1vvsHXXU5c+tErND7bxl5V8qO8HOuKExhK0zfY7qW/3wXQ4BKgY5BwiQPmDACFA7SQhICIXIj6BqNQkxJUkzwiMlg+5/moC4p+mlhgVcutO2ZfUXVsWSdT8mY1EqoeCAJUvCFDngwDloxoC9GH4wIP5d6h1nUddxxHpGL7xJ9TJ1z7oKd8kT3+oLrzqJjXU8wCEzbbYSl2+/G1/OScyTe6L3zg7kiDP1Lxg2dY77qIuvHtli4LWW0GAikHOIQKUPwgQAtRO7KltEme0ReKbWqaX1a85ER17BMY3kuOf3ua2IZSZuO5AanyjRnr7uhz1rPxIEKDyBQHqfBCgfFRBgOSlosdf/gP9gII0sZB7fk659oFcYiFPadvnC0ertQfZ7wyRnHjuJXp0x1fOF2nrF6d+K/WeILk/afQ226lzb3umshKEABWDnEMEKH8QIASov+KXqJ4OAlS+IECdDwKUj14XILm/5qzv3a8GpciPREZ+zr/rxfxTy576i9rl019Qa69tPwBB9nfcnNu01HjLefOhmv3EB+qrp01v8GCEtdWWo7dW337otUpOh0OAikHOIQKUPwgQAtRf8Y0s9XgQoPIFAep8EKB89K4AfagfTDB17g/VuhuMqHVgzMhoykaf+KQ69fsPB5KSTyRkFObyx95RI0eN1vXV6g7kZ9iwYWraDx7Tj7v2lU3N00HbH/2jOuKUUIK8o1bBvkZtv7O64O6X1OwnfU+Y690gQMUg57DKAvT1r38dARrgyDEjQH0SfY9QDz7prVEQoPIFAep8EKB89KYAydPe3lXHz/K/mFQiozV62luLT3tzI3Jz6SNv6BEms355fPXHNtpYXfv461pofGUbJ3wwwpdPOj+UIEOu4ogYbTl6G/WtRT+tlAQhQMUg57CqArRs2TL1hS98QQ0dav/eDbQAHXXUUeq+++7zbl/FyDEjQKRngwCVLwhQ54MA5aMXBWj2E++ra2+9u6H8fCyQn/Pvkqe95ZGTeuYG8rTi58+rQYPt+39kmtpGm26u5gYSM99TLmtEzuRlqfI0OZEqcx8SkaCRo7ZWF93/2xan2pU3CFAxyDmsqgB9//vfV2PGjLF+FyQXXXRRYQJ0yimnJARo6623VjfeeKNavny5t0zVIseMAJGeDQJUviBAnQ8ClI9eE6C5T/1VnSjT3tZPe9rbmmqTTTdTZ1631Fu+1cjIy5Qjv6ZfeGruR16IOvnIrxciJfKS1a+ecVGq0GkJ2mZHNeO+V7zley0IUDHIOayiAN18883qc5/7nFp3XfseOfk9uPrqq/XokK9cqznvvPPUZpttZu1jUPB7ffDBB6vbb7+9LyRIjhkBIj0bBKh8QYA6HwQoH70lQOHjrg8//iw1xJkaE0dGUbbbbS81896VnvKt5/LH39f3GLlT1OSJcz966pXW7/9JyalX36tGbPxx3ckz9xNnw40+rs669l41rwJPhkOAikHOYS8LkAjGAw88oO666y519913q0WLFqkLLrhATZgwQa2/fvKdW9tss40WE19deXLDDTeo3XbbLbGfddZZR40fP15Nnz5dt+3OO+/Uf1ZxapwcLwJEejYIUPmCAHU+CFA+em4K3NMfqsse/aP67ORjAgkZUuu01LOmGjR4iNpq+13UJQ+tynl/TpRANq599HdqWNAhcsVknXXXV7OWv6lHpLxlM0bKn33T42rDj22iX9Rq7qO2r/U2UGd89y799DhfHb0WBKgY5Bz2sgCJUMg0tG233VZn9OjRapNNNlFDPL/X8sCRb33rW4U+oECm0sn+N95448T+ZCRI2iLSJdluu+3U5MmTvfX0cuRYESDSs0GAyhcEqPNBgPLRi/cAyZPZLl32lvr8cWcn7s2RiKyIBG290+7qoh89n1tSZLTpxCvuTEx/k1GmnXfZVd//045gSf3fmHu7Gr7Rx1PlZ8iwddXJV96pZj3+XlCmvfuZyhIEqBjkHPayAMmoz5QpU6zr3ZcttthCnX322eree+8tfFqaSNiZZ56pttxyS+89eHHk75R99tnHW0cvR44NASI9GwSofEGAOh8EKB+9KECSWIK+cOwZ/ulwQYdF7tPZaY991UV3/zLXVDW5v+eob9+QqFvqPfDzX1bz25iONjuo+6Tv3KOfVJf2BLgNPraJOv3qeyolPxIEqBjkHFZdgHbaaSd12WWX6alyA3VPjowqzZo1S22//faJ/+yIgwB1AASItBoEqHxBgDofBCgfvSpAEpGgy5a/rQ4+4utq8OAhiWlqEhlZGb3zWDXz/t+0LEHytLlzL5iZqFNGlyZ8+XhvmSyRkZ8zb3hUv6DV1+Y111xLT3uTe37kRa9tTeMrYRCgYpBzWHUBEiGRJ7WdfvrpWoJ89bQTkZ8TTzxRbbrppowAdRsEiLQaBKh8QYA6HwQoH70sQJLwRaXvqs8ddYpae9BgPfITd1riiARtHUjQhXf+Qm/vq8eXy5e/pUZtvW2ivmHrrKvm3fqgt0yzyHS8E+bfEciP/4EHskyeBnfyFXdoUfLV0etBgIpBzmEvC9A999yjjj32WLXRRhvpyEtP5d4b8/chjoiQPJRgyZIl3rry5qSTTko8CjuO/C7KOmmb3Cd00EEHeevo5chxIkCkZ4MAlS8IUOeDAOWj1wVI5+kP9WjN579+evrT4YIO1A677x1I0HP+OpzIu32uXr5Kv5/HrWud9TdUly99w1uuUa565m/q1CsXq+EbbeKVH4l+2tv3fpxryl6vBAEqBjmHvSxAZh577DH18MMP66loe+yxh/dBCPKQhMWLF3vL54k8BW733XdP7EdkS95BNG/ePN0uX9mqRI63VAL0Xx4jpLV0GASoSRCgzgcBykclBEgSSNDlj/5RHfKV41MlSEaIttxujPr2A79RzV6QKi9Anf6dm9Vg5yELIi6bbrGlmvX4u95yabni6b+os65frtYf/jElL1E164yz3oYfU+cseEC/e8hXR1WCABWDnMOqCJAZeey0jLYM9fweX3PNNYW9B+j8889Xm2++uVW/TIP77Gc/q0enfGWqFjnm0ggQQA+AADVJqwK056TD1IUP/1Hd99Zr3voGNL+Yrp6bPVId8WkEqB+pjAAFiR+McPjUc/R9OnGnppZAXtYOhGbktjurCxY/23CUZfaTH6j9J38jcV+ATNH5xkmnaUHylXMjI0nyotNT59+mNvzYRqnyM3Sd9dSpV/0ovOfHqaNqQYCKQc5hFQVI8r3vfU/tvPPO1u+IZMaMGfrx1b4yrUYege1Of5Onwd1yyy3e7asYOWYECCA7CFCTNBWgfzyi3n7yu+rCPcaoA7YcqXaf8Fl16vUr1MIVj6lHXrm5o3nox6epH31rpDp0fNDO8ZPVlqcuVdc8/bD3uMocBCgfVRIgSSxBX/zGN9XQYcNqHZt61lRrrT1I7bbH3urcRU+niszsx99T+35ucqK8vAD1uEtvCMSm+YMJRH6ueGq1+vb3blMbbZL2wIM19TuAzrwmeuBBhZ72lhYEqBjkHFZVgGTq2aGHHpoYgb3wwgsLE6CTTz45IUBHHXVUJV94mhY5ZgQIIDsIUJM0FaD/+pR6/9mb1ZUTdlGfGz1Sjd52e7Xbvp9R+35mkpr4uQkdzYEH7K4+PW6k2mGbkWqrA45U21z8C/W95x/zHleZgwDlo2oCJNEPRnj0j+pLx56qJcgnHjKys/2Y3dVlS//grWPW8rfUbuP2TJRbd/0N1NkLHvCWSSRox/X3PqZGbLSxX36CNqw/fCN17o1L9YhTP8iPBAEqBjmHVRUgiTwgQR6MYP7ODLQAnXPOOYW+bLXskWNGgACygwA1STMBeuc/P6He/t3t6okZ/6JOP3B3NSZYPyrarivZcis1cqcJavzXL1GnP/wnde/vf+49rjIHAcpHFQVI5+kP9YjKF48/S0+H8wnI0HXXUxfc+4q3/MUP/FYNW3f9RJmNP7Gp+t6jq7xl3IjU7LnPp70vOZVHXctLTk//7t2VfuCBLwhQMcg5RIDyxydA3/zmNxEgAEgFAWqSZgIk26x6/0714Qtz1Y3TjlCH7bGL2jPIbt3Knrur3b5wqvrGlferpz/8n+qFj1YmjqnsQYDyUVkB0vlQzQkk5EtTz1FDhyanw6UJ0Hx5QMLix70PU/jYpiPV5Y+9kyjjiwjQbnuOT9xHJImnvbXyWO6qBAEqBjmHCFD+IEAIEECrIEBNkkWAXv7bA2rVB3eqp1Zer+554ip1R5Dbupbvqtt+dot68LcPBfLzC7Xy70yB6xeqLUBBZCTosXf1dDh5f0/c0ZGkCdCcn6xWh//LkYm3wsvnCRMPzvyUtjQBGr7Rx9V5NzwS7OdP3nJVDwJUDHIOEaD8QYAQIIBWQYCaJIsAkWKDAOWj8gIURB55LfcEHXHyefohBnFnJ02AZIRno023SEybk7KX3f6EfpmpW8YXnwANW299dfaCh/Q6X5l+CAJUDHIOEaD8QYAQIIBWQYCaxCtA3w8E6I2/IUADFAQoH/0gQBKZanbZ8rfVlNMurD0YIXUE6OHf6ff1xJ2iOOttMFxd/GC2+38kpgDJ/vRLTr8v7/npX/mRIEDFIOcQAcofBAgBAmgVBKhJfAI09aoH1c2/Xa1++a9LvWVIe0GA8tEvAiTRI0GPvaO+etI31TrrrquGrbeBV4DOuObeYP16tU5RnN3G7hlI1FuJ7dMSC9Dagwap4Rt/Qp13o0x7q/ZLTrMEASoGOYcIUP4gQAgQQKsgQE3y/r8tU//rw0fUrd84TB0xKhSgQ06Yoc669hZ11R2XqKsXf7s7uXu2uvqxu9S9v/W3u5eDAOWjnwRIJ5AgEZMpJ5+n1h+xsbrwvl8723yovnLB97wvUp30paPVnCezv6hU9rP73vvqkaOzv/9A5qlzVQ8CVAxyDhGg/EGAECCAVkGAmuT9//IT9f/+l2fVj08/Qh2/XShApcj249TIKd9Tp9/+kLfdvRwEKB99J0BR5v5ktTr2vDnqkodft5Zf8fRf1GlnfjPxAkbJvpNPULMyPgBBIg85OOakM9W06x/W7wTybdOPQYCKQc4hApQ/CBACBNAqCFCTrPq3ZeqP/1iuXr73QnXdiQer8YF8bO/KSDeCAIFDvwqQjATJ+3fsx1B/qGY9/q7absedEw9AGDp0qLrgmtvUvJbe2fOhuu7ZP/fde36aBQEqBjmHCFD+IEAIEECrIEAZ8qt/fVD94dUb1ON3nq++eezn1deD/Eu3c+JR6l/m/EBd8bi/zb0cBCgffStAvgRSdOkjb6h11tug1iGKM3Sd9dSc5X8ItvswWY60FASoGOQcVlmAjj/+eDV8+HDr93C77bZT++67r5owYYLOfvvtp2666SZveTPf+9731GGHHVYrJxk1alRipHfatGlqyZIl3jqqGDlmBAggOwhQxvzqo4fUyg8fUM//6T71iyDPdT0/Vs/95SH1wt/87e3lIED5QIDqkYckXHLTA2rI0HVqHSKdNddUo7bZTl3+aLYXoJLGQYCKQc5hlQXo3HPPVZtuuqn1uyhPVBw0aFAtQ4YMUddcc423vJnLL79cbbnlllZZ9/1cMuor53PZsmXeOqoYOW4ECCA7CBApXRCgfCBARp7+q9rvS8eqQc7/CkvH6MRTz1Czn8j+AASSHgSoGOQcVlmAfvjDH6pddtnF+l10IxJz9dVXe8ubueyyy9Tmm2/urSPOmDFj1C233OItX9XIcSNAANlBgEjpggDlAwEy8tRf1P6f+5Jae+1BtU6RZM0111InzLlVzV3xJ3850lIQoGKQc1hlAZJMnz49MQpkpigBknuNLrnkksLuL+qVyLEjQADZQYBI6YIA5QMBiiIPRXjyfTV23B6JqTEiRCdddY+as+LP/rKkpSBAxSDnsOoCtHz5cjV79mx974/cryO/m2ZkKluWKXAiQPIgILe8ZMcdd9T7ePjhh71lqxy5hhAggOwgQKR0QYDygQCFkafBzX/sLbX5FmEnSa4lify83gYbqu88+rqWJF9Z0loQoGKQc1h1AYojDya4/vrr1Xe+8x19zGbuv/9+bxkz99xzjxYls9yVV16pbrzxRvXggw96y/RD5BpCgACygwCR0gUBygcCFOdDNXfFn9XMB36rrrz7KXXBxbPUxIMOUZ/YbKTaaNOR+p0+/nKk1SBAxdBPAkQGJggQQGsgQKR0QYDygQDZmS95+q/6fp/Zj7+n5j2ySs28Z6XzviDSThCgYkCASLtBgABaAwEipQsClA8EiHQ6CFAxIECk3SBAAK2BAJHSBQHKBwJEOh0EqBgQINJuECCA1kCASOmCAOUDASKdDgJUDAgQaTcIEEBrIECkdEGA8oEAkU4HASoGBIi0GwQIoDUQIFK6IED5QIBIp4MAFQMCRNoNAgTQGggQKV0QoHwgQKTTQYCKAQEi7QYBAmgNBIiULghQPhAg0ukgQMWAAJF2gwABtAYCREoXBCgfCBDpdBCgYkCASLtBgABaAwEipQsClA8EiHQ6CFAxIECk3SBAAK2BAJHSBQHKBwJEOh0EqBgQINJuECCA1kCASOmCAOUDASKdDgJUDAgQaTcIEEBrIECkdEGA8oEAkU4HASoGBIi0GwQIoDUQIFK6IED5QIBIp4MAFQMCRNoNAgTQGggQKV0QoHwgQKTTQYCKAQEi7QYBAmiNjgrQKRccRUim0KlqHQSIdDoIUDHIOTzmmGMIyR0ECKA1OiZAB048gDjZc6891YgRw73r+j0TJx4YXT2QFQSIdDoIUDEceOCB6oADDiCkrSBAANnhXy6AioAAkU4HAQIAgF6Ef7kAKgICRDodBAgAAHoR/uXqIqtXr1azZ8+OPgG0BwJEOh0ECAAAehH+5eoiq1atUhMnTow+AbQHAkQ6HQQIAAB6Ef7lAqgIsQBJp5SQTgUBAgCAXoN/uQAqwqcPnKg7o4R0OgAAAL0E/3J1Ee4BAgAAAADoLAhQF+EeIAAAAACAzoIAAQAAAABA34AAAQAAAABA34AAdZFnn32WG4gBAAAAADoIve8u8s9//lNLEAAAAAAAdAYECAAAAAAA+gYEqIvICNBzzz0XfQIAAAAAgIEGAeoi3AMEAAAAANBZ6H13Ee4BAgAAAADoLAgQAAAAAAD0DQhQF+EeIAAAAACAzoIAdZFVq1apiRMnRp8AAAAAAGCgQYAAAAAAAKBvQIAAAAAAAKBvQIC6iEyBmzRpUvQJAAAAAAAGGgSoi6xevVrNmjUr+gQAAAAAAAMNAgQAAAAAAH0DAgQAAAAAAH0DAtRFnn32WbXGGnwFAAAAAACdgt53F5EXoYoEAQAAAABAZ0CAAAAAAACgb0CAAAAAAACgb0CAugj3AAEAAAAAdJZy9L7/fy/3Zf754c/Us0/e7F3XF/mfb0cXAAAAAABAZyiHAP3Xp5T6Tw+QfgsCBAAAAAAdBgEi3QsCBAAAAAAdBgEi3QsCBAAAAAAdBgEi3QsCBAAAAAAdBgEi3QsCBAAAAAAdBgEi3QsCBAAAAAAdBgEi3QsCBAAAAAAdBgEi3QsCBAAAAAAdBgEi3QsCBAAAAAAdBgEi3QsCBAAAAAAdBgEi3QsCBAAAAAAdBgEi3QsCBAAAAAAdpvoCtOJotcYaayQyZdGt/u0zZOXM8ere1/3rWsut6t7JTtsmn6M+8m7bPM3bZe9v/grfNh0MAgQAAAAAHabyArRypnT2j1YrreXz1PxAAPJI0EeLxrclKfX42hALitve5snUrtfPUVPWKEreCggCBAAAAAAdpuICFAnFzHnJdXpkqHXR0ELlq6/FpAtLPjnL0i69zxzHPGBBgAAAAACgw1RbgPSIR5pMhKJhTwNzp6SZoyXh9rV1pry40+wyCFIjGUnKjNsus1yDdqVtYx6X03b3XNVErbZdgQKFAAEAAABAh6m2AOlOe9qUL3ekJTlalJAU3xSyxD7celMSyVlzoYjkxRAbLUim6GSa2hbWYwpfeHxGOY8wyr6mTA62K2DUKxEECAAAAAA6TKUFKCEKVhxR0SLjyIgrFoltQmlyZSfLdDSdmgRJ/AKj6/K2y5AZX9vduMfi1hGlNuKjP0dSmHoO2wwCBAAAAAAdpsIClBzRsZMcEUnEGd2x5SAtzfbrSzTKo2OKjCNpzvK47Vna5Y5mpZaxZCpt/wUFAQIAAACADlNdAUoZ4ajFHTXRn2MJMVPfJjmyE8mOp1w+aYhEKN5HapvCxMeWZcTJHg1LlzRLlNxRo6KDAAEAAABAh6muADmjN24saUiRJZ80mGJjr4/STLw89ZixBMSVNG8a1xfGFZ70MuYxZRlZaisIEAAAAAB0mMoKkFdO4nhHf1zRcKZ/JcTGLxHuVDNfLPlyYkmHdwTGkZmmwiWxp8xJvG1w6mrUzkKCAAEAAABAh6moADlTycz4RoZS5EammdkCZJdLSJauW8o1GbWJtkuMwETLE+0wjkPv06zfK0lOfNskzoN7ztJHiQoLAgQAAAAAHaaaAhQJTSgjTlJGNMKRm/p281e4AhAJQhDfMh2RIS0WTQRIxymr4ytXl7HaPqz1vnY5SWtTJFxx7PJhvY1HltoMAgQAAAAAHaa69wCR8gcBAgAAAIAOgwCR7gUBAgAAAIAOgwCR7gUBAgAAAIAOgwCR7gUBAgAAAIAOgwCR7gUBAgAAAIAOgwCR7gUBAgAAAIAOgwD54jwe2p8sj7pOSeZHZdcz4C8l7UYQIAAAAADoMAhQhoTvCGpDeIg/CBAAAAAAdBgEKEP06EviBaSk7SBAAAAAANBhEKCmmafmy5Q3Z/qZHhUSKapNl4tHiG5V906Wz3HGq3tfr5eL65uy6Fbr8/wVkWhF5eRzrYzeh1FPbQpd1DYdd4TKboeuL8fUuwENAgQAAAAAHQYBapbXz1FTXCEJIrIyZXIgQZYYRdJhLEtMn9P1GTIT1W9Kj1umJlvmZ10m3sbdr/tZRGl80N5gWZlGshAgAAAAAOgwCFCzuKMvOpFguDLhG2FxhcfZJpQZp35nG/sBCP59m9u4wqSj6zRHnkoQBAgAAAAAOgwC1CTuaEwYdxpbgzgC5cqJLTfGMmd0xzdlrl4mmgqn60lpW8pIVleDAAEAAABAh0GAGsadShbFHdWJE42yJGPLTL0+n6w427ji4ghVYps00UlrczeDAAEAAABAh0GAGsY/muKdYpYiHno0p7atU59XSpxtmowgJbZJER3/SFaXgwABAAAAQIdBgBqlkdS4o0JaQppMlXPkxCslDYXHPyLlkyy7zVE5V5y6HQQIAAAAADoMAtQgoaC4oymhTLijQklZiqQjMZpTFx6fSLkjPPY26XJjtscWouhzUC4hbd0OAgQAAAAAHQYBahBXJML4JCRMKEyRbOhtbDmx5cYZHdKJpKkmKo7cOKNDOt5Rqrp8he1Ib3NXgwABAAAAQIdBgPohPnEqQxAgAAAAAOgwCFDF4k6hS44qlSgIEAAAAAB0GASocrGnv+mUUX4kCBAAAAAAdBgEiHQvCBAAAAAAdBgEiHQvCBAAAAAAdBgEiHQvCBAAAAAAdBgEiHQvCBAAAAAAdBgEiHQvCBAAAAAAdBgEiHQvCBAAAAAAdBgEiHQvCBAAAAAAdBgEiHQvCBAAAAAAdJhyCNB//E/SrwEAAAAA6CDlECAAAAAAAIAOgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAEDfgAABAAAAAECfoNT/H32Bq1GWrNQsAAAAAElFTkSuQmCC" width="70%" height="70%"/>
# <br/></br> There are two ways to download latest version of Workflow Single Model and its dependents.
# 1. Download resource(s) from an MLW tenant UI.
# <ol>
# <li>Go to "Repo" Tab.</li>
# <li>Select "Workflow-Single-Model-Diabetes.pmml" </li>
# <li>Click on "Download" </li>
# </ol>
# 2. Use UMOYA CLI.
# Go to ZMOD resource directory in terminal, Give command : <br/>umoya add Workflow-Single-Model-Diabetes.pmml
# ### <font color=darkgreen>» About Model and Its dependent resource(s) :
# <ul><li>Once We download the Model "Workflow-Single-Model-Diabetes.pmml", <br/>We have the trained Model "Gredient-Booster-Classifier-Diabetes.pmml" along with training Data-Set "GBC-Diabetes-Train.csv". We have used MLW's Auto ML to train and create this Model using Gredient Booster Classifier algorithm.</li>
# <li>From Model Tab, We have the main Model "Workflow-Single-Model-Diabetes.pmml".<br/>We can click on "Edit" and View the architecture.<br/> This Model workflow contains the Gredient Booster Classifier Model and Pre/Post Processing Script.<br/>Pre-Processing Script creates feature1 and feature2 from the input Data-Set when scoring and Once Gredient Booster Classifier Model performed scoring,Post-Processing Script does prediction counts and stores into file : predictionscounts.csv</li></ul>
# ### » Test Workflow Single Model :
# 1. Load "Workflow-Single-Model-Diabetes.pmml" to ZMK1.
# 2. Select "WSM-Diabetes-Test.csv" in "Data" Tab.
# 3. Click on "Predect Data" and Select "Machine Learning Workbench".
# 4. Selct Model "Workflow-Single-Model-Diabetes.pmml" and submit.
#
# It will create a new file with column feature1 and feature2 and target. Target field has value with 1 or 0 to indecate that respective patient has diabetes or not. As part of Post-Processing Script, It also creates file : predictionscounts.csv
# <br/><br/>
# Further Reference(s) :
# <ul>
# <li><a href="https://mlw.ai">MLW Documentation</a>
# <li><a href="https://github.com/SoftwareAG/MLW">MLW Souce</a>
# <li><a href="https://github.com/Umoya-ai/UMOYA/tree/master/docs/cli">UMOYA documentation</a>
# </ul>
| 1,259.20339
| 70,763
|
929fa7f782a64751d412f1fa166d354f2294b476
|
py
|
python
|
chapter-9/Fourier-A.ipynb
|
subblue/applied-maths-in-chem-book
|
['CC0-1.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] deletable=false editable=false
# Godfrey Beddard 'Applying Maths in the Chemical & Biomolecular Sciences an example-based approach' Chapter 9
# -
# import all python add-ons etc that will be needed later on
# %matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
from sympy import *
init_printing() # allows printing of SymPy results in typeset maths format
plt.rcParams.update({'font.size': 14}) # set font size for plots
# + [markdown] deletable=false editable=false
# # Chapter 9 Fourier Series and Transforms
# + [markdown] deletable=false editable=false
# This chapter describes several related topics: the Fourier series and Fourier transforms, as well as autocorrelations, convolutions, and their numerical calculation.
#
# Fourier series are used in solving partial differential equations, as explained in Chapter 10.5, for example, the diffusion and Schroedinger equation. Fourier transforms are used to produce every NMR spectrum, MRI scan, X-ray structure, and IR spectrum recorded; a good grasp of this topic is therefore essential for the molecular scientist.
#
# In many experiments the measuring instrument distorts the data and, in this case, what is measured is convoluted with the response of the instrument. Here Fourier transforms can unravel the data to produce the true response. At the end of the chapter a different type of transform, the Hadamard transform is also described and some examples of its use in time-resolved measurements, such as time-resolved kinetics and x-ray diffraction, given.
#
# ## 1 Motivation and concept
#
# The Taylor and Maclaurin series reconstruct functions as an infinite series in the powers $x^n$, and the coefficients needed to do this are the derivatives of the function. These series have rather tight restrictions placed upon them; the function must be differentiable $n$ times over and the remainder must approach zero. These are described in Chapter 5. In a Fourier series, the expansion is performed instead, as trigonometric series in sines and cosines, with two sets of coefficients, $a_n$ and $b_n$, to describe the $n^\text{th}$ term, and which are evaluated by integration. The series formally extends to infinity, but in practice, at most only a few tens of terms are needed to replicate most functions to an acceptable level of approximation. The advantage of using a Fourier rather than a Taylor/Maclaurin series is that a wide class of functions can be described by the series, including discontinuous ones. However, by their very nature, Fourier series can only represent _periodic functions_, and this must not be forgotten. Periodic means that the function repeats itself; the repeat interval is normally taken to be $-\pi$ to $+\pi$ but can be extended to the range $-L$ to $L$ and $L$ can even be made infinite.
#
# To illustrate that it is possible to make an arbitrary shaped function by adding a number of sine and cosine waves, four such waves are shown in Fig.1. It is possible to imagine, without any difficulty, that waveforms that are more complex can be formed by using more sine or cosine terms. The lowest and most complicated waveform is simply the sum of the individual waves, and repeats itself with a period of 2$\pi$. This waveform could be the signal that is measured on an oscilloscope or spectrometer and recorded on a computer. It might alternatively, be part of an image that is formed from adjacent columns of different waveforms. Periodic components present in a signal represent information in the waveform or image can be retrieved by performing a Fourier transform. This unravelling process is discussed later on but now the Fourier series is considered.
#
# <img src = 'fourier-fig1.png' alt='Drawing' style='width:300px;'/>
#
# **Fig 1**. A complex and _periodic_ waveform or function is constructed out of the sum of sine and/or cosine waves. The complicated waveform, repeats itself with a period of 2$\pi$. In the Fourier series, the reconstruction of this waveform will require many more sine and cosine terms to reconstruct its form than are used to generate it, because the Fourier series only represents a function _exactly_ when an infinite number of terms are included in the summation.
# + [markdown] deletable=false editable=false
# ## 1.1 The Fourier Series.
#
# A Fourier series aims to reconstruct a function, such as a waveform, using the weighted sums of sine and cosine functions or their exponential representations. The great importance and usefulness of the Fourier series is that they represents the best fit, in a least-squares way, to a function $f(x)$ because $\displaystyle \int_{-L}^{L}[f(x)-g(x)]^2dx$ is minimized when $g(x)$ is the Fourier series expansion of $f(x)$.
#
# Because the Fourier series can extend to infinity, many more sine and cosine terms can be used to exactly reproduce a waveform, such as that shown in Fig. 1, than were used to produce it in the first place. The Fourier series cannot somehow magically pick out just those sine and cosine waves used to make the original function, therefore very many terms each with a unique frequency are needed to faithfully reconstruct a waveform.
#
# If the waveform shown in Fig. 1 were truncated so that it was zero outside $x = \pm \pi$, for example, then its Fourier series would have to match this new waveform which suddenly becomes zero and do so a series must contain a sufficiently large number terms not only to cancel out to zero outside the range $x = \pm \pi$, but also to reproduce the function inside the range.
#
# Suppose that, over the range $\pm\pi$, a Fourier series $g(x)$ approximates a 'victim' or target function $f(x)$, which might be $x^3$ or $\displaystyle (e^{-x} - 1)^2$ or any 'normal' function, then the Fourier series $g(x)$ that approximates the true function $f(x)$ can be written down in general and in quite a straightforward manner and is
#
# $$\displaystyle g(x) = \frac{a_0}{2}+ \sum\limits_{n=1}^\infty a_n\cos(nx) + \sum\limits_{n=1}^\infty b_n\sin(nx) \tag{1}$$
#
# where $g(x)$ approximates $f(x)$. The summations start at index $n$ = 1, and $n$ is a positive integer. The $a_n$ and $b_n$ coefficients are the integrals
#
# $$\displaystyle a_n= \frac{1}{\pi}\int\limits_{-\pi}^{+\pi}f(x)\cos(nx)dx \qquad (n \ge 0) \tag{2} $$
# $$\displaystyle b_n= \frac{1}{\pi}\int\limits_{-\pi}^{+\pi}f(x)\sin(nx)dx \qquad (n \gt 0) \tag{3} $$
#
# which are normalized by 1/$\pi$.
#
# When the number of terms in the series is large, then $g(x) \rightarrow f (x)$, and when infinite, $g(x) = f (x)$. At each value of $x$, the Fourier series consists of a constant term, $a_0$/2 plus the sum of an infinite number of oscillating terms in integer multiples of $x$. Notice, that the target function $f(x)$ appears as part of the expansion coefficients only, and must, therefore, be capable of being integrated. The target function $f(x)$ determines the weighting to be placed on each term in the expansion, and this is how information about the shape of $f(x)$ is included in the expansion.
#
# If $f(x)$ is periodic in time, then the variable $x$ would normally be changed to $\omega t$ where $\omega = 2\pi\nu$ is the angular frequency in units of radian s$^{-1}$, and $\nu$ is the frequency in s$^{-1}$. If the dimension is spatial, then $x$ is often replaced with $2\pi x/L$ of which $2\pi/L$ can be interpreted as a spatial frequency, with units of radians m${-1}$ by analogy with 'normal' frequencies. Often this spatial frequency is called the wavevector and given the symbol $k$.
#
# Before a worked example, it is worthwhile examining limits other than $\pm\pi$, describe the exponential form of the series and also simplifying some series using symmetry. The $a_n$ and $b_n$ constants are also derived.
#
# ### 1.2 Series limits from -$L$ to $L$
#
# Over the range $−L$ to $L$, the equations to use for $f(x)$ are similar to equations (1)–(3) but $x$ is
# changed to $\pi x/L$. The series is
#
# $$\displaystyle g(x) = \frac{a_0}{2}+ \sum\limits_{n=1}^\infty a_n\cos \left(\frac{n\pi x}{L}\right) + \sum\limits_{n=1}^\infty b_n\sin\left(\frac{n\pi x}{L}\right) \tag{4}$$
#
# and the coefficients are changed similarly,
#
# $$\displaystyle a_n= \frac{1}{L}\int\limits_{-L}^{L}f(x)\cos \left(\frac{n\pi x}{L}\right)dx \qquad (n \ge 0) \tag{5} $$
# $$\displaystyle b_n= \frac{1}{L}\int\limits_{-L}^{L}f(x)\sin \left(\frac{n\pi x}{L}\right)dx \qquad (n \gt 0) \tag{6} $$
#
# Notice that the arguments, limits, and normalization are each changed compared to those when the range is $\pm \pi$. The integral now has limits $\pm L$ instead of $\pm \pi$, and normalization 1/$L$ rather than 1/$\pi$.
#
# ### 1.3 Exponential representation
#
# Because the sine and cosine functions can be represented as sums and differences of complex exponential terms, for example, $\displaystyle \cos(x) = (e^{ix} + e^{-ix})/2$, the most general way of describing the Fourier series is to use the complex exponential form;
#
# $$\displaystyle g(x) = \sum\limits_{n=-\infty}^\infty c_ne^{+i\, n\pi x/L} \tag{7}$$
#
# and the coefficients become
#
# $$\displaystyle c_n=\frac{1}{2L}\int\limits_{-L}^L f(x)e^{-i\,n\pi x/L}dx \tag{8} $$
#
# Note the change in sign and limits in the second exponential, and that $i=\sqrt{-1}$. The set of coefficients $c_n$, are sometimes called the _amplitude spectrum_ of the transform. Note also that for practical purposes the expansion $g(x)$, and the true function $f(x)$ are considered equivalent.
# + [markdown] deletable=false editable=false
# ### 1.4 Deriving the integral describing the $a$ and $b$ coefficients
#
# The integrals describing the coefficients $a_n$ are obtained by multiplying each term in the Fourier series by $\cos(mx)$, where $m$ is an integer, then integrating term by term using the orthogonality of sine and cosine integrals as necessary. The coefficients $b_n$ can be obtained similarly by multiplying the series by $\sin(mx)$ and integrating.
#
# First, the form of the cosine integrals is examined. The integrals containing a product of sine and cosine are all zero for any $n$ and $m$ because the integral has 'odd' symmetry because of the sine, and the limits are symmetrical;
#
# $$\displaystyle \int\limits_{-\infty}^{+\infty}\cos(mx)\sin(nx)dx = 0 \tag{9}$$
#
# To convince you that the symmetry makes the integral zero, which is the area under the curve, a curve is plotted below, see figure 2. The curve, while complicated, is inverted about $x$ = 0 so that the area $\gt$ 0 is equal and opposite to that $\lt$ 0. More details on odd and even functions is given in the next section.
#
# <img src='fourier-fig1a.png' alt='Drawing' style='width:350px;'/>
# Figure 2. An odd function such as shown above has zero integral when evaluated over a symmetrical region about zero.
# ____
#
# The product of two cosines makes the integral
#
# $$\displaystyle \int\limits_{-\pi}^\pi \cos(mx)\cos(nx)dx = \pi\delta_{n,m} \tag{10}$$
#
# where $m$ and $n$ are integers and the (Kronecker) delta function $\delta_{n,m}$ is zero only if $n \ne m$, and is $1$ if $n = m$.
#
# To calculate the $a_n$ coefficients, $f(x)$ is represented as the Fourier series $g(x)$
#
# $$\displaystyle f(x) \approx g(x) = \frac{a_0}{2}+ \sum\limits_{n=1}^\infty a_n\cos(nx) + \sum\limits_{n=1}^\infty b_n\sin(nx) \tag{11}$$
#
# and this is now multiplied by $\cos(mx)$ and integrated. The cosine integrals with $m\gt$ 0 are,
#
# $$\int\limits_{-\pi}^\pi f(x) \cos(mx)dx = \frac{a_0}{2}\int\limits_{-\pi}^\pi \cos(mx)dx +\int\limits_{-\pi}^\pi \left [\sum\limits_{n=1} a_n\cos(nx)+ \sum\limits_{n=1} b_n\sin(nx ) \right] \cos(mx)dx \tag{12}$$
#
# These integrals can be easily evaluated: The first is zero by symmetry, and by direct integration, $\displaystyle \int_{-\pi}^\pi \cos(mx)dx=\left. \frac{\sin(mx)}{m}\right|_{-\pi}^\pi =0$. The integral containing $b_n$ is given by eqn. 10 and is zero also when $n \ne m$, therefore eqn. 12 becomes
#
#
# $$\displaystyle \int\limits_{-\pi}^\pi f(x) \cos(mx)dx =\pi\sum\limits_{n=1}a_n\delta_{n,m} = \pi a_n$$
#
# because the delta function picks out just one term from the summation. Rearranging gives
#
# $$\displaystyle a_n = \frac{1}{\pi}\int\limits_{-\pi}^\pi f(x) \cos(mx)dx \quad (n \gt 0) $$
#
# The index has been changed to $n$ from $m$ because both $n$ and $m$ are integers and only one index is needed; equation (11) was written initially with $n$, so this is again chosen. When $m = n =$ 0, equation (12) becomes
#
# $$\displaystyle \int\limits_{-\pi}^\pi f(x)dx =\frac{a_0}{2}\int\limits_{-\pi}^\pi 1dx=\pi a_0 $$
#
# making $\displaystyle a_0= \pi^{-1}\int\limits_{-\pi}^\pi f(x)dx$. Similar arguments lead to the calculation of $b_n$.
#
# ### 1.5 Odd and even functions
#
# When functions are either even or odd, the Fourier series is simplified to cosine or sine series respectively. An even function has the property $f (-x) = f (x)$ and the sine integration (eqn. 6) producing the Fourier coefficient $b_n$ is odd and therefore $b_n$ = 0, but only because the integration limits are symmetrical. The cosine integral (eqn. 5) is not zero, and can be written as
#
# $$\displaystyle a_n=\frac{2}{\pi}\int\limits_0^\pi f(x)\cos(nx)dx \tag{13}$$
#
# When $f(x)$ is odd $f(-x) = -f(x)$, the opposite situation arises; $a_n$ = 0 and only the sine terms remain;
#
# $$\displaystyle b_n=\frac{2}{\pi}\int\limits_0^\pi f(x)\sin(nx)dx \tag{14}$$
#
# In cases when $f(x)$ is neither odd nor even, for example $f(x) = \pi/2 - x$, both coefficients have to be evaluated.
#
# As an example, consider calculating the series of $f(x) = x^2$ over the range $\pm \pi$. Because this is an even function, the sine integral should be zero and $b_n$ = 0. The $a$ coefficients are
#
# $$\displaystyle a_n=\frac{2}{\pi}\int\limits_0^\pi x^2\cos(nx)dx $$
#
# and when $n$ = 0,
#
# $$\displaystyle a_0=\frac{2}{\pi}\int\limits_0^\pi x^2dx =\frac{2}{3}\pi^2 $$
#
# When $n\gt$ 0, $a_n$ can be evaluated using integration by parts.
# -
#using SymPy to do the indefinite integration
x ,n = symbols('x n', positive = True)
func = x**2*cos(n*x)/pi
an = integrate(func,x, conds='none') # coefficients a_n
an
# + [markdown] deletable=false editable=false
# Adding the limits $\pm \pi$ produces zero for the sine terms and the remaining cosine produces $\displaystyle a_n=(-1)^n\frac{4}{n^3}$. All the $b_n$=0 thus using eqn. 4. the Fourier series for $x^2$ is
#
# $$\displaystyle x^2 \approx g(x)= \frac{\pi^2}{3}-4\cos(x)+\cos(2x)-\frac{4}{9}\cos(3x) \cdots \ = \frac{\pi^2}{3}+4\sum\limits_{n=1}^\infty \frac{(-1)^n}{n^2}\cos(nx) \tag{15} $$
#
# which, as shown in the next figure, is a good representation of $x^2$. The following Python code is included as an example of a Fourier series calculation
# +
fig1= plt.figure(figsize=(12.5, 5.0))
plt.rcParams.update({'font.size': 16}) # set font size for plots
ax0 = fig1.add_subplot(1,3,1)
ax1 = fig1.add_subplot(1,3,2)
ax2 = fig1.add_subplot(1,3,3)
gx = lambda x, k : (np.pi**2)/3.0 + 4.0*sum( (-1)**n *np.cos(n*x)/n**2 for n in range(1,k)) # series summation
x = np.linspace(-2*np.pi,2*np.pi,200) # define x values, -pi to pi with 200 data points
for i,ax in enumerate([ax0,ax1]):
j = 20
if i == 0 : j = 3 # number of terms in summation
ax.plot(x/np.pi, gx(x,j),linestyle='dashed',color='black',label='max n = '+str(j)) # plot as x/pi , 4 terms
ax.plot(x/np.pi,x**2,color='red')
ax.set_xlabel(r'$x/\pi$')
ax.set_ylim([-1,15])
ax.axhline(0)
ax.legend()
kk = 10
nvals = [i for i in range(kk)] # make list of values 0, 1, 2,...
fn = lambda n : (np.pi**2)/3.0 if n ==0 else (-1)**n/n**2 if n >0 else 0
an = [fn(i) for i in range(kk)]
ax2.axhline(0,linewidth=1, color='grey')
ax2.bar(nvals,an,label=r'$a_n$ coefficients')
ax2.set_ylim([-2,4])
ax2.set_xlabel('n')
ax2.legend()
plt.tight_layout()
plt.show()
# + [markdown] deletable=false editable=false
# **Figure 3**. Left: Plot of $x^2$ and its Fourier series to $n$ = 3 showing a poor fit to the true function. More terms produce a better fit but only over the range $\pm \pi$ as shown in the centre panel where 20 terms are included in the summation. This plot also shows how the fit is only over the range $\pm \pi$ and then it repeats itself. On the right are shown the coefficients $a_n$ and shows how rapidly their value decreases as $n$ increases.
# -
| 76.162037
| 1,233
|
d82d0b493f382aea6e8ecc08836e604dcdc588b9
|
py
|
python
|
S9/2 - Upgraded Sentiment Analysis.ipynb
|
pankaj90382/TSAI-2
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="vmNDI_KoM9rz"
# # 2 - Updated Sentiment Analysis
#
# In the previous notebook, we got the fundamentals down for sentiment analysis. In this notebook, we'll actually get decent results.
#
# We will use:
# - packed padded sequences
# - pre-trained word embeddings
# - different RNN architecture
# - bidirectional RNN
# - multi-layer RNN
# - regularization
# - a different optimizer
#
# This will allow us to achieve ~84% test accuracy.
# + [markdown] id="fjhtilz3M9r0"
# ## Preparing Data
#
# As before, we'll set the seed, define the `Fields` and get the train/valid/test splits.
#
# We'll be using *packed padded sequences*, which will make our RNN only process the non-padded elements of our sequence, and for any padded element the `output` will be a zero tensor. To use packed padded sequences, we have to tell the RNN how long the actual sequences are. We do this by setting `include_lengths = True` for our `TEXT` field. This will cause `batch.text` to now be a tuple with the first element being our sentence (a numericalized tensor that has been padded) and the second element being the actual lengths of our sentences.
# + id="SYOWQKhSM9r1" executionInfo={"status": "ok", "timestamp": 1602999474509, "user_tz": -330, "elapsed": 8192, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}}
import torch
from torchtext import data
from torchtext import datasets
SEED = 1234
torch.manual_seed(SEED)
torch.backends.cudnn.deterministic = True
TEXT = data.Field(tokenize = 'spacy', include_lengths = True)
LABEL = data.LabelField(dtype = torch.float)
# + [markdown] id="fMvdkH2tM9r6"
# We then load the IMDb dataset.
# + id="N0QSdklaM9r7" executionInfo={"status": "ok", "timestamp": 1602999666563, "user_tz": -330, "elapsed": 102964, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}} outputId="0f48c2da-d896-448a-c2cd-a6264cea7522" colab={"base_uri": "https://localhost:8080/", "height": 50}
from torchtext import datasets
train_data, test_data = datasets.IMDB.splits(TEXT, LABEL)
# + [markdown] id="0YnB7iPsM9r-"
# Then create the validation set from our training set.
# + id="Muwv-UVaM9r_" executionInfo={"status": "ok", "timestamp": 1602999666568, "user_tz": -330, "elapsed": 93252, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}}
import random
train_data, valid_data = train_data.split(random_state = random.seed(SEED))
# + [markdown] id="xI7QHG8FM9sB"
# Next is the use of pre-trained word embeddings. Now, instead of having our word embeddings initialized randomly, they are initialized with these pre-trained vectors.
# We get these vectors simply by specifying which vectors we want and passing it as an argument to `build_vocab`. `TorchText` handles downloading the vectors and associating them with the correct words in our vocabulary.
#
# Here, we'll be using the `"glove.6B.100d" vectors"`. `glove` is the algorithm used to calculate the vectors, go [here](https://nlp.stanford.edu/projects/glove/) for more. `6B` indicates these vectors were trained on 6 billion tokens and `100d` indicates these vectors are 100-dimensional.
#
# You can see the other available vectors [here](https://github.com/pytorch/text/blob/master/torchtext/vocab.py#L113).
#
# The theory is that these pre-trained vectors already have words with similar semantic meaning close together in vector space, e.g. "terrible", "awful", "dreadful" are nearby. This gives our embedding layer a good initialization as it does not have to learn these relations from scratch.
#
# **Note**: these vectors are about 862MB, so watch out if you have a limited internet connection.
#
# By default, TorchText will initialize words in your vocabulary but not in your pre-trained embeddings to zero. We don't want this, and instead initialize them randomly by setting `unk_init` to `torch.Tensor.normal_`. This will now initialize those words via a Gaussian distribution.
# + id="T6XpRpMrM9sC" executionInfo={"status": "ok", "timestamp": 1603000105963, "user_tz": -330, "elapsed": 471800, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}} outputId="b8816c54-ef7c-4fe0-aa5a-c1ae23bdb710" colab={"base_uri": "https://localhost:8080/", "height": 50}
MAX_VOCAB_SIZE = 25_000
TEXT.build_vocab(train_data,
max_size = MAX_VOCAB_SIZE,
vectors = "glove.6B.100d",
unk_init = torch.Tensor.normal_)
LABEL.build_vocab(train_data)
# + [markdown] id="LBahvmxrM9sE"
# As before, we create the iterators, placing the tensors on the GPU if one is available.
#
# Another thing for packed padded sequences all of the tensors within a batch need to be sorted by their lengths. This is handled in the iterator by setting `sort_within_batch = True`.
# + id="XBv6BrS0M9sF" executionInfo={"status": "ok", "timestamp": 1603000107419, "user_tz": -330, "elapsed": 1441, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}}
BATCH_SIZE = 64
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
train_iterator, valid_iterator, test_iterator = data.BucketIterator.splits(
(train_data, valid_data, test_data),
batch_size = BATCH_SIZE,
sort_within_batch = True,
device = device)
# + [markdown] id="WoaQ2x7tM9sH"
# ## Build the Model
#
# The model features the most drastic changes.
#
# ### Different RNN Architecture
#
# We'll be using a different RNN architecture called a Long Short-Term Memory (LSTM). Why is an LSTM better than a standard RNN? Standard RNNs suffer from the [vanishing gradient problem](https://en.wikipedia.org/wiki/Vanishing_gradient_problem). LSTMs overcome this by having an extra recurrent state called a _cell_, $c$ - which can be thought of as the "memory" of the LSTM - and the use use multiple _gates_ which control the flow of information into and out of the memory. For more information, go [here](https://colah.github.io/posts/2015-08-Understanding-LSTMs/). We can simply think of the LSTM as a function of $x_t$, $h_t$ and $c_t$, instead of just $x_t$ and $h_t$.
#
# $$(h_t, c_t) = \text{LSTM}(x_t, h_t, c_t)$$
#
# Thus, the model using an LSTM looks something like (with the embedding layers omitted):
#
# 
#
# The initial cell state, $c_0$, like the initial hidden state is initialized to a tensor of all zeros. The sentiment prediction is still, however, only made using the final hidden state, not the final cell state, i.e. $\hat{y}=f(h_T)$.
#
# ### Bidirectional RNN
#
# The concept behind a bidirectional RNN is simple. As well as having an RNN processing the words in the sentence from the first to the last (a forward RNN), we have a second RNN processing the words in the sentence from the **last to the first** (a backward RNN). At time step $t$, the forward RNN is processing word $x_t$, and the backward RNN is processing word $x_{T-t+1}$.
#
# In PyTorch, the hidden state (and cell state) tensors returned by the forward and backward RNNs are stacked on top of each other in a single tensor.
#
# We make our sentiment prediction using a concatenation of the last hidden state from the forward RNN (obtained from final word of the sentence), $h_T^\rightarrow$, and the last hidden state from the backward RNN (obtained from the first word of the sentence), $h_T^\leftarrow$, i.e. $\hat{y}=f(h_T^\rightarrow, h_T^\leftarrow)$
#
# The image below shows a bi-directional RNN, with the forward RNN in orange, the backward RNN in green and the linear layer in silver.
#
# 
#
# ### Multi-layer RNN
#
# Multi-layer RNNs (also called *deep RNNs*) are another simple concept. The idea is that we add additional RNNs on top of the initial standard RNN, where each RNN added is another *layer*. The hidden state output by the first (bottom) RNN at time-step $t$ will be the input to the RNN above it at time step $t$. The prediction is then made from the final hidden state of the final (highest) layer.
#
# The image below shows a multi-layer unidirectional RNN, where the layer number is given as a superscript. Also note that each layer needs their own initial hidden state, $h_0^L$.
#
# 
#
# ### Regularization
#
# Although we've added improvements to our model, each one adds additional parameters. Without going into overfitting into too much detail, the more parameters you have in in your model, the higher the probability that your model will overfit (memorize the training data, causing a low training error but high validation/testing error, i.e. poor generalization to new, unseen examples). To combat this, we use regularization. More specifically, we use a method of regularization called *dropout*. Dropout works by randomly *dropping out* (setting to 0) neurons in a layer during a forward pass. The probability that each neuron is dropped out is set by a hyperparameter and each neuron with dropout applied is considered indepenently. One theory about why dropout works is that a model with parameters dropped out can be seen as a "weaker" (less parameters) model. The predictions from all these "weaker" models (one for each forward pass) get averaged together withinin the parameters of the model. Thus, your one model can be thought of as an ensemble of weaker models, none of which are over-parameterized and thus should not overfit.
#
# ### Implementation Details
#
# Another addition to this model is that we are not going to learn the embedding for the `<pad>` token. This is because we want to explitictly tell our model that padding tokens are irrelevant to determining the sentiment of a sentence. This means the embedding for the pad token will remain at what it is initialized to (we initialize it to all zeros later). We do this by passing the index of our pad token as the `padding_idx` argument to the `nn.Embedding` layer.
#
# To use an LSTM instead of the standard RNN, we use `nn.LSTM` instead of `nn.RNN`. Also, note that the LSTM returns the `output` and a tuple of the final `hidden` state and the final `cell` state, whereas the standard RNN only returned the `output` and final `hidden` state.
#
# As the final hidden state of our LSTM has both a forward and a backward component, which will be concatenated together, the size of the input to the `nn.Linear` layer is twice that of the hidden dimension size.
#
# Implementing bidirectionality and adding additional layers are done by passing values for the `num_layers` and `bidirectional` arguments for the RNN/LSTM.
#
# Dropout is implemented by initializing an `nn.Dropout` layer (the argument is the probability of dropping out each neuron) and using it within the `forward` method after each layer we want to apply dropout to. **Note**: never use dropout on the input or output layers (`text` or `fc` in this case), you only ever want to use dropout on intermediate layers. The LSTM has a `dropout` argument which adds dropout on the connections between hidden states in one layer to hidden states in the next layer.
#
# As we are passing the lengths of our sentences to be able to use packed padded sequences, we have to add a second argument, `text_lengths`, to `forward`.
#
# Before we pass our embeddings to the RNN, we need to pack them, which we do with `nn.utils.rnn.packed_padded_sequence`. This will cause our RNN to only process the non-padded elements of our sequence. The RNN will then return `packed_output` (a packed sequence) as well as the `hidden` and `cell` states (both of which are tensors). Without packed padded sequences, `hidden` and `cell` are tensors from the last element in the sequence, which will most probably be a pad token, however when using packed padded sequences they are both from the last non-padded element in the sequence.
#
# We then unpack the output sequence, with `nn.utils.rnn.pad_packed_sequence`, to transform it from a packed sequence to a tensor. The elements of `output` from padding tokens will be zero tensors (tensors where every element is zero). Usually, we only have to unpack output if we are going to use it later on in the model. Although we aren't in this case, we still unpack the sequence just to show how it is done.
#
# The final hidden state, `hidden`, has a shape of _**[num layers * num directions, batch size, hid dim]**_. These are ordered: **[forward_layer_0, backward_layer_0, forward_layer_1, backward_layer 1, ..., forward_layer_n, backward_layer n]**. As we want the final (top) layer forward and backward hidden states, we get the top two hidden layers from the first dimension, `hidden[-2,:,:]` and `hidden[-1,:,:]`, and concatenate them together before passing them to the linear layer (after applying dropout).
# + id="MaXgsAXvM9sH" executionInfo={"status": "ok", "timestamp": 1603000929241, "user_tz": -330, "elapsed": 1304, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}}
import torch.nn as nn
class RNN(nn.Module):
def __init__(self, vocab_size, embedding_dim, hidden_dim, output_dim, n_layers,
bidirectional, dropout, pad_idx):
super().__init__()
self.embedding = nn.Embedding(vocab_size, embedding_dim, padding_idx = pad_idx)
self.rnn = nn.LSTM(embedding_dim,
hidden_dim,
num_layers=n_layers,
bidirectional=bidirectional,
dropout=dropout)
self.fc = nn.Linear(hidden_dim * 2, output_dim)
self.dropout = nn.Dropout(dropout)
def forward(self, text, text_lengths):
#text = [sent len, batch size]
embedded = self.dropout(self.embedding(text))
#embedded = [sent len, batch size, emb dim]
#pack sequence
packed_embedded = nn.utils.rnn.pack_padded_sequence(embedded, text_lengths)
packed_output, (hidden, cell) = self.rnn(packed_embedded)
#unpack sequence
output, output_lengths = nn.utils.rnn.pad_packed_sequence(packed_output)
#output = [sent len, batch size, hid dim * num directions]
#output over padding tokens are zero tensors
#hidden = [num layers * num directions, batch size, hid dim]
#cell = [num layers * num directions, batch size, hid dim]
#concat the final forward (hidden[-2,:,:]) and backward (hidden[-1,:,:]) hidden layers
#and apply dropout
hidden = self.dropout(torch.cat((hidden[-2,:,:], hidden[-1,:,:]), dim = 1))
#hidden = [batch size, hid dim * num directions]
return self.fc(hidden)
# + [markdown] id="rQR71AH8M9sN"
# Like before, we'll create an instance of our RNN class, with the new parameters and arguments for the number of layers, bidirectionality and dropout probability.
#
# To ensure the pre-trained vectors can be loaded into the model, the `EMBEDDING_DIM` must be equal to that of the pre-trained GloVe vectors loaded earlier.
#
# We get our pad token index from the vocabulary, getting the actual string representing the pad token from the field's `pad_token` attribute, which is `<pad>` by default.
# + id="wYEHmkgsM9sO" executionInfo={"status": "ok", "timestamp": 1603000962489, "user_tz": -330, "elapsed": 2246, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}}
INPUT_DIM = len(TEXT.vocab)
EMBEDDING_DIM = 100
HIDDEN_DIM = 256
OUTPUT_DIM = 1
N_LAYERS = 2
BIDIRECTIONAL = True
DROPOUT = 0.5
PAD_IDX = TEXT.vocab.stoi[TEXT.pad_token]
model = RNN(INPUT_DIM,
EMBEDDING_DIM,
HIDDEN_DIM,
OUTPUT_DIM,
N_LAYERS,
BIDIRECTIONAL,
DROPOUT,
PAD_IDX)
# + [markdown] id="zPLhjNfZM9sQ"
# We'll print out the number of parameters in our model.
#
# Notice how we have almost twice as many parameters as before!
# + id="0di0vXeBM9sQ" executionInfo={"status": "ok", "timestamp": 1603000966168, "user_tz": -330, "elapsed": 1817, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}} outputId="b5f10038-2a6d-4867-ae5a-afe650eee612" colab={"base_uri": "https://localhost:8080/", "height": 34}
def count_parameters(model):
return sum(p.numel() for p in model.parameters() if p.requires_grad)
print(f'The model has {count_parameters(model):,} trainable parameters')
# + [markdown] id="jBTl1-0LM9sU"
# The final addition is copying the pre-trained word embeddings we loaded earlier into the `embedding` layer of our model.
#
# We retrieve the embeddings from the field's vocab, and check they're the correct size, _**[vocab size, embedding dim]**_
# + id="Ti2371zpM9sU" executionInfo={"status": "ok", "timestamp": 1603000986493, "user_tz": -330, "elapsed": 1265, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}} outputId="0a94c4df-8220-4ecf-ed38-28d5d3a52969" colab={"base_uri": "https://localhost:8080/", "height": 34}
pretrained_embeddings = TEXT.vocab.vectors
print(pretrained_embeddings.shape)
# + [markdown] id="rmG6grimM9sW"
# We then replace the initial weights of the `embedding` layer with the pre-trained embeddings.
#
# **Note**: this should always be done on the `weight.data` and not the `weight`!
# + id="Jobn_BnIM9sX" executionInfo={"status": "ok", "timestamp": 1603000989609, "user_tz": -330, "elapsed": 1312, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}} outputId="76ebe31b-5788-42fe-c179-d838d98369b2" colab={"base_uri": "https://localhost:8080/", "height": 134}
model.embedding.weight.data.copy_(pretrained_embeddings)
# + [markdown] id="ovry0LEUM9sa"
# As our `<unk>` and `<pad>` token aren't in the pre-trained vocabulary they have been initialized using `unk_init` (an $\mathcal{N}(0,1)$ distribution) when building our vocab. It is preferable to initialize them both to all zeros to explicitly tell our model that, initially, they are irrelevant for determining sentiment.
#
# We do this by manually setting their row in the embedding weights matrix to zeros. We get their row by finding the index of the tokens, which we have already done for the padding index.
#
# **Note**: like initializing the embeddings, this should be done on the `weight.data` and not the `weight`!
# + id="YMSkJFJ9M9sb" executionInfo={"status": "ok", "timestamp": 1603001000845, "user_tz": -330, "elapsed": 1265, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}} outputId="4e0702eb-36b7-4260-d669-805b2ace03df" colab={"base_uri": "https://localhost:8080/", "height": 134}
UNK_IDX = TEXT.vocab.stoi[TEXT.unk_token]
model.embedding.weight.data[UNK_IDX] = torch.zeros(EMBEDDING_DIM)
model.embedding.weight.data[PAD_IDX] = torch.zeros(EMBEDDING_DIM)
print(model.embedding.weight.data)
# + [markdown] id="-4uUZiMEM9sf"
# We can now see the first two rows of the embedding weights matrix have been set to zeros. As we passed the index of the pad token to the `padding_idx` of the embedding layer it will remain zeros throughout training, however the `<unk>` token embedding will be learned.
# + [markdown] id="-XykhCNQM9sf"
# ## Train the Model
# + [markdown] id="tr-FDqjQM9sg"
# Now to training the model.
#
# The only change we'll make here is changing the optimizer from `SGD` to `Adam`. SGD updates all parameters with the same learning rate and choosing this learning rate can be tricky. `Adam` adapts the learning rate for each parameter, giving parameters that are updated more frequently lower learning rates and parameters that are updated infrequently higher learning rates. More information about `Adam` (and other optimizers) can be found [here](http://ruder.io/optimizing-gradient-descent/index.html).
#
# To change `SGD` to `Adam`, we simply change `optim.SGD` to `optim.Adam`, also note how we do not have to provide an initial learning rate for Adam as PyTorch specifies a sensibile default initial learning rate.
# + id="DQddWMX9M9sg" executionInfo={"status": "ok", "timestamp": 1603001004704, "user_tz": -330, "elapsed": 1254, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}}
import torch.optim as optim
optimizer = optim.Adam(model.parameters())
# + [markdown] id="o_rLT03YM9si"
# The rest of the steps for training the model are unchanged.
#
# We define the criterion and place the model and criterion on the GPU (if available)...
# + id="R5KZ-Ea_M9sj" executionInfo={"status": "ok", "timestamp": 1603001016924, "user_tz": -330, "elapsed": 10861, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}}
criterion = nn.BCEWithLogitsLoss()
model = model.to(device)
criterion = criterion.to(device)
# + [markdown] id="OO6yOEYHM9sl"
# We implement the function to calculate accuracy...
# + id="GQEwjWwxM9sm" executionInfo={"status": "ok", "timestamp": 1603001033665, "user_tz": -330, "elapsed": 1266, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}}
def binary_accuracy(preds, y):
"""
Returns accuracy per batch, i.e. if you get 8/10 right, this returns 0.8, NOT 8
"""
#round predictions to the closest integer
rounded_preds = torch.round(torch.sigmoid(preds))
correct = (rounded_preds == y).float() #convert into float for division
acc = correct.sum() / len(correct)
return acc
# + [markdown] id="2a5-JqvkM9so"
# We define a function for training our model.
#
# As we have set `include_lengths = True`, our `batch.text` is now a tuple with the first element being the numericalized tensor and the second element being the actual lengths of each sequence. We separate these into their own variables, `text` and `text_lengths`, before passing them to the model.
#
# **Note**: as we are now using dropout, we must remember to use `model.train()` to ensure the dropout is "turned on" while training.
# + id="8voMvXUfM9sp" executionInfo={"status": "ok", "timestamp": 1603001053158, "user_tz": -330, "elapsed": 1403, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}}
def train(model, iterator, optimizer, criterion):
epoch_loss = 0
epoch_acc = 0
model.train()
for batch in iterator:
optimizer.zero_grad()
text, text_lengths = batch.text
predictions = model(text, text_lengths).squeeze(1)
loss = criterion(predictions, batch.label)
acc = binary_accuracy(predictions, batch.label)
loss.backward()
optimizer.step()
epoch_loss += loss.item()
epoch_acc += acc.item()
return epoch_loss / len(iterator), epoch_acc / len(iterator)
# + [markdown] id="Ua1GtvjQM9ss"
# Then we define a function for testing our model, again remembering to separate `batch.text`.
#
# **Note**: as we are now using dropout, we must remember to use `model.eval()` to ensure the dropout is "turned off" while evaluating.
# + id="KgaoY7m9M9st" executionInfo={"status": "ok", "timestamp": 1603001053160, "user_tz": -330, "elapsed": 828, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}}
def evaluate(model, iterator, criterion):
epoch_loss = 0
epoch_acc = 0
model.eval()
with torch.no_grad():
for batch in iterator:
text, text_lengths = batch.text
predictions = model(text, text_lengths).squeeze(1)
loss = criterion(predictions, batch.label)
acc = binary_accuracy(predictions, batch.label)
epoch_loss += loss.item()
epoch_acc += acc.item()
return epoch_loss / len(iterator), epoch_acc / len(iterator)
# + [markdown] id="lC7UyehdM9sv"
# And also create a nice function to tell us how long our epochs are taking.
# + id="oyfYa_LGM9sw" executionInfo={"status": "ok", "timestamp": 1603001056754, "user_tz": -330, "elapsed": 1265, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}}
import time
def epoch_time(start_time, end_time):
elapsed_time = end_time - start_time
elapsed_mins = int(elapsed_time / 60)
elapsed_secs = int(elapsed_time - (elapsed_mins * 60))
return elapsed_mins, elapsed_secs
# + [markdown] id="WMBShtE5M9sy"
# Finally, we train our model...
# + id="bc0BtpPQM9sy" executionInfo={"status": "ok", "timestamp": 1603001268322, "user_tz": -330, "elapsed": 211122, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}} outputId="e2208f96-66ec-4613-8e0b-fb2eec3528ae" colab={"base_uri": "https://localhost:8080/", "height": 269}
N_EPOCHS = 5
best_valid_loss = float('inf')
for epoch in range(N_EPOCHS):
start_time = time.time()
train_loss, train_acc = train(model, train_iterator, optimizer, criterion)
valid_loss, valid_acc = evaluate(model, valid_iterator, criterion)
end_time = time.time()
epoch_mins, epoch_secs = epoch_time(start_time, end_time)
if valid_loss < best_valid_loss:
best_valid_loss = valid_loss
torch.save(model.state_dict(), 'tut2-model.pt')
print(f'Epoch: {epoch+1:02} | Epoch Time: {epoch_mins}m {epoch_secs}s')
print(f'\tTrain Loss: {train_loss:.3f} | Train Acc: {train_acc*100:.2f}%')
print(f'\t Val. Loss: {valid_loss:.3f} | Val. Acc: {valid_acc*100:.2f}%')
# + [markdown] id="gnTTw8z-M9s0"
# ...and get our new and vastly improved test accuracy!
# + id="wGSksHOEM9s1" executionInfo={"status": "ok", "timestamp": 1603001336866, "user_tz": -330, "elapsed": 15780, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}} outputId="9311b081-2cb0-479a-da49-1ad3bee1f907" colab={"base_uri": "https://localhost:8080/", "height": 34}
model.load_state_dict(torch.load('tut2-model.pt'))
test_loss, test_acc = evaluate(model, test_iterator, criterion)
print(f'Test Loss: {test_loss:.3f} | Test Acc: {test_acc*100:.2f}%')
# + [markdown] id="CZtzU0h1M9s3"
# ## User Input
#
# We can now use our model to predict the sentiment of any sentence we give it. As it has been trained on movie reviews, the sentences provided should also be movie reviews.
#
# When using a model for inference it should always be in evaluation mode. If this tutorial is followed step-by-step then it should already be in evaluation mode (from doing `evaluate` on the test set), however we explicitly set it to avoid any risk.
#
# Our `predict_sentiment` function does a few things:
# - sets the model to evaluation mode
# - tokenizes the sentence, i.e. splits it from a raw string into a list of tokens
# - indexes the tokens by converting them into their integer representation from our vocabulary
# - gets the length of our sequence
# - converts the indexes, which are a Python list into a PyTorch tensor
# - add a batch dimension by `unsqueeze`ing
# - converts the length into a tensor
# - squashes the output prediction from a real number between 0 and 1 with the `sigmoid` function
# - converts the tensor holding a single value into an integer with the `item()` method
#
# We are expecting reviews with a negative sentiment to return a value close to 0 and positive reviews to return a value close to 1.
# + id="NgZoWbeBM9s3" executionInfo={"status": "ok", "timestamp": 1603001369841, "user_tz": -330, "elapsed": 2851, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}}
import spacy
nlp = spacy.load('en')
def predict_sentiment(model, sentence):
model.eval()
tokenized = [tok.text for tok in nlp.tokenizer(sentence)]
indexed = [TEXT.vocab.stoi[t] for t in tokenized]
length = [len(indexed)]
tensor = torch.LongTensor(indexed).to(device)
tensor = tensor.unsqueeze(1)
length_tensor = torch.LongTensor(length)
prediction = torch.sigmoid(model(tensor, length_tensor))
return prediction.item()
# + [markdown] id="ULZdRWoqM9s5"
# An example negative review...
# + id="89oS4HFHM9s6" executionInfo={"status": "ok", "timestamp": 1603001382344, "user_tz": -330, "elapsed": 2649, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}} outputId="f6b9e0d4-86c5-4999-89de-0a7cd217e244" colab={"base_uri": "https://localhost:8080/", "height": 34}
predict_sentiment(model, "This film is terrible")
# + [markdown] id="o1kmC1GyM9s-"
# An example positive review...
# + id="eZEuHPGVM9s_" executionInfo={"status": "ok", "timestamp": 1603001412458, "user_tz": -330, "elapsed": 2431, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}} outputId="16c8f2bf-68bc-49e1-cbf8-edf1f17a087d" colab={"base_uri": "https://localhost:8080/", "height": 34}
predict_sentiment(model, "This film is great")
# + [markdown] id="_0-tWEzEVKXm"
# ## Save the Model
# + id="8Q0tJV_jVNqG" executionInfo={"status": "ok", "timestamp": 1603001457908, "user_tz": -330, "elapsed": 1300, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}}
cpu_model = model.to('cpu')
torch.save(cpu_model.state_dict(), 'upgraded_sentiment_analysis.pt')
# + id="VYjPQBqzVSdX" executionInfo={"status": "ok", "timestamp": 1603001472146, "user_tz": -330, "elapsed": 1272, "user": {"displayName": "SACHIN SALMAN", "photoUrl": "", "userId": "18077240758423476746"}}
import pickle
with open('upgraded_sentiment_analysis_metadata.pkl', 'wb') as f:
metadata = {
'input_stoi': TEXT.vocab.stoi,
'label_itos': LABEL.vocab.itos,
}
pickle.dump(metadata, f)
# + [markdown] id="fdtLag4VM9tB"
# ## Next Steps
#
# We've now built a decent sentiment analysis model for movie reviews! In the next notebook we'll implement a model that gets comparable accuracy with far fewer parameters and trains much, much faster.
| 57.272381
| 1,138
|
8fbfdf456b44812c1b37495d67d190da0726e49b
|
py
|
python
|
analysis/.ipynb_checkpoints/task5-checkpoint.ipynb
|
data301-2020-winter2/course-project-group_1047
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# **Exploratory Data Analysis of Netflix Movie/Film Data**
# - **RQ #1: What's the distribution of Netflix Titles across all the countires Netflix streams in?
# - **RQ #2: Is there a specialisation for directors based on the audiences they make movies for?
# - **RQ #3: How are ratings distributed across Netflix titles?
import pandas as pd
import numpy as np
import matplotlib.pylab as plt
import seaborn as sns
import sys
import os
import re
# imported all the necessary libraries needed for data analysis and visualization
sys.path.append("../")
import Scripts
from Scripts import project_functions #relative import
df = project_functions.load_and_process("netflix_titles.csv")
df
df.info()
df.head()
# ---
# We will try to explore the first research question by trying to count how many titles are available in every country
s = df['country'].value_counts()
print (s)
# As you can see from the data we extracted above, the US has the most entries with 2555, India in second with 923, and so on. From this data we found we were able to graph it in a counterplot to show the count on every country
t = df['country'].value_counts()
print(s)
sns.countplot(data = df, y = "country",order=pd.value_counts(df['country']).iloc[:10].index)
# Unsurprisingly the US has the most entries because of Hollywood and Netflix. The Netflix is a US based company, and its main selling demographic is people in America.So Netflix tries to meet the demand in America in order to gain more success. The next most was India. India has one of the largest contributions in this industry. Firstly the population is really high and bollywood is nearly as big as hollywood, it has a really high budget like hollywood reaching the 80 million mark. Our analysis is worldwide, meaning countries with high populations,like India, will be high in the list, however Netflix is not available in China for political reasons. This stops them from being on the list let alone be the highest. In third place is the United Kingdom, one of the major leaders in the industry. With classics such as Sherlock Holmes which has a number of seasons and a Movie. Simultaneously one of the most viewed shows on Netflix according to Kimberly Bond ,a radio times journalist, https://www.radiotimes.com/tv/drama/peaky-blinders-gets-its-highest-audience-figures-ever/ is peaky blinders. This is a collab involving Netflix and the bbc which is the most well known broadcasting Corporations in the uk and the world.
# Because Netflix has aggressively tried to grow in the last decade by collaborating with various production houses, there must be some figures that could show us whether Netflix tried to increase its video library. This could easily be done through the use of a countplot that can show how Netflix added titles based on the years.
sns.countplot(data = df, x = "release_year",order=pd.value_counts(df['release_year']).iloc[0:14].index)
plt.title('Number of Netflix Titles available in a given year')
df = df.sort_values("date_added")
df.head()
# From the data obtained above, it is interesting to see the earliest dates that the movies/shows were added to Netflix. The earliest date which a movie was added to netflix that is still available today is 2008-01-01. Netflix as a company has been around since 1997 and started their streaming service in 2007. It turns out that "To and From New York" is prized by Netflix.
# https://interestingengineering.com/the-fascinating-history-of-netflix#:~:text=Video%20streaming%20is%20introduced%20in,movies%20on%20their%20personal%20computers.
#
# By the count plot of the release year, we see there is a linear relationship in growth as the years increase. This makes sense as a growing company with a subscription based model, Netflix is inclined to output more Movie/Tv shows to keep members subscribed. Moroever, the countplot shows cummulative titles over the course of years. Therefore, it makes sense that the titles increased by such a large extent over the course of 11 years. Moreover, this agressive strategy was accomplished by getting streaming rights for various titles. For example, in recent times, Netflix bought the show "Cobra Kai", a title originally created by YouTube Originals. Another great example of this is the spanish show "La Casa De Papel", which was critically acclaimed in Spain, but the production was shot down due to low viewership. However, when the the same title was bought by Netflix and made available internationall with the title "Money Heist", **it quickly became the most viewed foreign language title on Netflix**!! (https://www.forbes.com/sites/paultassi/2020/04/22/netflix-reveals-money-heist-has-somehow-drawn-more-viewers-than-tiger-king/?sh=36303bfcf5a1)
#
# This is a great example of how Netflix redirects content in order to match it with audiences they think will view it more!
# ---
# Moving on to the next research question, we can try to look at how directors create content on Netflix. We will try to use the top 5 directors on Netflix in order to simplify our analysis. We were able to extract the most featured directors on Netflix. It turns out that the most frequent director in our data is Raúl Campos, Jan Suter with 18 movies.
t = df['director'].value_counts()
print(t)
sns.countplot(data = df, y = "director",order=pd.value_counts(df['director']).iloc[1:6].index)
# It would be interesting to see the variety of content that these directors have made. We will try to do so by using the column *'rating'*. Rating is an essential column for Netflix, as it allows audiences to check whether the content they are watching is suitable for them based on their age. This becomes even more important, as Netflix can face legal problems for giving incorrect ratings for their titles.
t.head(6)
## To get the top 5 directors on Netflix
# +
Raul = (df.loc[df.director == 'Raúl Campos, Jan Suter', 'rating'].value_counts())
Raboy = (df.loc[df.director == 'Marcus Raboy', 'rating'].value_counts())
Karas = (df.loc[df.director == 'Jay Karas', 'rating'].value_counts())
Molina = (df.loc[df.director == 'Cathy Garcia-Molina', 'rating'].value_counts())
Chapman = (df.loc[df.director == 'Jay Chapman', 'rating'].value_counts())
## Counts and categorizes the various movies the directors have made according to the ratings
# +
Raul.plot.bar(x = "Rating", y = "Frequency")
plt.title('Raúl Campos, Jan Suter range according to rating')
## Plots a barplot to compare Raul Campos, Jan Suter's range of movies according to rating
# -
Raboy.plot.bar(x = "Rating", y = "Frequency")
plt.title('Marcus Raboy range according to rating' )
## Plots a barplot to compare Marcus Raboy's range of movies according to rating
# +
Karas.plot.bar(x = "Rating", y = "Frequency")
plt.title('Jay Karas range according to rating' )
## Plots a barplot to compare Jay Karas's range of movies according to rating
# +
Molina.plot.bar(x = "Rating", y = "Frequency")
plt.title('Cathy Garcia-Molina range according to rating' )
## Plots a barplot to compare Cathy Garcia-Molina's range of movies according to rating
# +
Chapman.plot.bar(x = "Rating", y = "Frequency")
plt.title('Jay Chapman range according to rating' )
## Plots a barplot to compare Jay Chapman's range of movies according to rating
# -
# Based on the graphs above, we can clearly see that the directors tend to specialise in a particular rating. What that essentially means is that directors usually try making movies for a given audience. In the case of the top 5 directors, this happened to be TV-MA. TV-MA is the rating given to Mature Audiences, thus for individuals typically 18 and above. This is consistent with the advent of OTT (Over The Top) platforms, as many countries don't have very well defined laws when it comes to streaming content. Howoever, what is also interesting is that while some of these directors have a clear specialisation, they still create content for other audiences. The next most occuring rating was that of TV-14, which means the content can be viewed by individuals aged 14 and above. It's quite interesting to see that the directors that make content for mature audiences are also making content for teenagers.
#
# We can essentially infer a new insight from these graphs, while directors can clearly target a particular audience, it doesn't prevent them from focusing on creating content for other audiences!!
# ---
# Considering ratings are an integral part of Netflix's service, it's worth exploring further. Let us see the variety of ratings that are available on Netflix.
# +
r = df['rating'].value_counts()
## Created an object that would help determine all the different ratings available and their count
ra = r.to_numpy()
## Because the object r is a series, we converted it to an array so that we can use the array for data visualization
rating_labels = ['TV-MA', 'TV-14', 'TV-PG', 'R', 'PG-13', 'TV-Y', 'TV-Y7', 'PG', 'TV-G', 'NR', 'G', 'Unknown', 'TV-Y7-FV', 'UR', 'NC-17']
## Created labels for all the ratings we found
# +
fig = plt.figure(figsize =(10, 7))
plt.pie(ra, labels = rating_labels)
plt.legend(rating_labels, loc="best")
plt.title('The distribution of ratings across all Netflix titles')
## Created a pie chart to help visualise the ratings and how much space they occupy in the Netflix library
# -
# Now that we can see that TV-MA is the most dominant rating, we can defintely understand why the Top 5 directors made TV-MA movies most frequently. This also makes sense from their revenue model, because even though young children might be a consumer for Netflix, adults (or the actual customer) are mostly likely to pay for their subscriptions. Thus, the pie chart shows the range of consumers for Netflix!!
# We could further explore this by looking at the variety of ratings available in a single country. Let us try to look for the range of ratings in the top 3 countries with most Netflix titles:
# 1. United States
# 2. India
# 3. United Kingdom
# +
US = (df.loc[df.country == 'United States', 'rating'].value_counts())
## Found out the range of ratings available in Netflix US and how much of the library they occupy
US_array = US.to_numpy()
## Converted to an array so that we can use the array for data visualization
US_labels = rating_labels
## Created labels for available ratings for the pie chart
fig = plt.figure(figsize =(10, 7))
plt.pie(US_array, labels = US_labels)
plt.legend(rating_labels, loc="best")
plt.title('The distribution of ratings across all Netflix titles in the United States')
## Created a pie chart to show this distribution
# +
India = (df.loc[df.country == 'India', 'rating'].value_counts())
## Found out the range of ratings available in Netflix India and how much of the library they occupy
I_array = India.to_numpy()
## Converted to an array so that we can use the array for data visualization
I_labels = ['TV-14', 'TV-MA', 'TV-PG', 'TV-Y7', 'TV-G', 'TV-Y','NR', 'PG-13', 'PG', 'R', 'UR', 'TV-Y7-FV']
## Created labels for available ratings for the pie chart
fig = plt.figure(figsize =(10, 7))
plt.pie(I_array, labels = I_labels)
plt.legend(rating_labels, loc="best")
plt.title('The distribution of ratings across all Netflix titles in India')
## Created a pie chart to show this distribution
# +
UK = (df.loc[df.country == 'United Kingdom', 'rating'].value_counts())
## Found out the range of ratings available in Netflix UK and how much of the library they occupy
UK_array = UK.to_numpy()
## Converted to an array so that we can use the array for data visualization
UK_labels = ['TV-MA', 'TV-PG', 'TV-14', 'R', 'TV-G', 'TV-Y', 'PG-13', 'NR', 'TV-Y7', 'PG']
## Created labels for available ratings for the pie chart
fig = plt.figure(figsize =(10, 7))
plt.pie(UK_array, labels = UK_labels)
plt.legend(rating_labels, loc="best")
plt.title('The distribution of ratings across all Netflix titles in the United Kingdom')
## Created a pie chart to show this distribution
# -
# It makes sense that TV-MA is a dominant rating all the 3 counrtires, but we inferred to more insights:
# - India has a higher number of TV-14 titles. This makes a lot of sense, as Indian content has historically relied on creating content that can be viewed by members of the family, or can be watched together with a whole community.
# - The ratings available in a country also depends on the laws in their country, and whether there is strict regulation for violation. The UK has a relativelty relaxed regulation regime, thus ratings there are also more broadly defined!!
# ---
| 60.461538
| 1,229
|
3dd2b2ce7b8f9fd7e21ad9cb0baf521ff7d6512d
|
py
|
python
|
utf-8''C4_W1_Assignment.ipynb
|
snjev310/ML_Misc.
|
['MIT']
|
# -*- coding: utf-8 -*-
# ---
# jupyter:
# jupytext:
# formats: ipynb,py:percent
# text_representation:
# extension: .py
# format_name: percent
# format_version: '1.3'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# %% [markdown]
# # Assignment 1: Neural Machine Translation
#
# Welcome to the first assignment of Course 4. Here, you will build an English-to-German neural machine translation (NMT) model using Long Short-Term Memory (LSTM) networks with attention. Machine translation is an important task in natural language processing and could be useful not only for translating one language to another but also for word sense disambiguation (e.g. determining whether the word "bank" refers to the financial bank, or the land alongside a river). Implementing this using just a Recurrent Neural Network (RNN) with LSTMs can work for short to medium length sentences but can result in vanishing gradients for very long sequences. To solve this, you will be adding an attention mechanism to allow the decoder to access all relevant parts of the input sentence regardless of its length. By completing this assignment, you will:
#
# - learn how to preprocess your training and evaluation data
# - implement an encoder-decoder system with attention
# - understand how attention works
# - build the NMT model from scratch using Trax
# - generate translations using greedy and Minimum Bayes Risk (MBR) decoding
# ## Outline
# - [Part 1: Data Preparation](#1)
# - [1.1 Importing the Data](#1.1)
# - [1.2 Tokenization and Formatting](#1.2)
# - [1.3 tokenize & detokenize helper functions](#1.3)
# - [1.4 Bucketing](#1.4)
# - [1.5 Exploring the data](#1.5)
# - [Part 2: Neural Machine Translation with Attention](#2)
# - [2.1 Attention Overview](#2.1)
# - [2.2 Helper functions](#2.2)
# - [Exercise 01](#ex01)
# - [Exercise 02](#ex02)
# - [Exercise 03](#ex03)
# - [2.3 Implementation Overview](#2.3)
# - [Exercise 04](#ex04)
# - [Part 3: Training](#3)
# - [3.1 TrainTask](#3.1)
# - [Exercise 05](#ex05)
# - [3.2 EvalTask](#3.2)
# - [3.3 Loop](#3.3)
# - [Part 4: Testing](#4)
# - [4.1 Decoding](#4.1)
# - [Exercise 06](#ex06)
# - [Exercise 07](#ex07)
# - [4.2 Minimum Bayes-Risk Decoding](#4.2)
# - [Exercise 08](#ex08)
# - [Exercise 09](#ex09)
# - [Exercise 10](#ex10)
#
# %% [markdown]
# <a name="1"></a>
# # Part 1: Data Preparation
#
# %% [markdown]
# <a name="1.1"></a>
# ## 1.1 Importing the Data
#
# We will first start by importing the packages we will use in this assignment. As in the previous course of this specialization, we will use the [Trax](https://github.com/google/trax) library created and maintained by the [Google Brain team](https://research.google/teams/brain/) to do most of the heavy lifting. It provides submodules to fetch and process the datasets, as well as build and train the model.
# %%
from termcolor import colored
import random
import numpy as np
import trax
from trax import layers as tl
from trax.fastmath import numpy as fastnp
from trax.supervised import training
# !pip list | grep trax
# %% [markdown]
# Next, we will import the dataset we will use to train the model. To meet the storage constraints in this lab environment, we will just use a small dataset from [Opus](http://opus.nlpl.eu/), a growing collection of translated texts from the web. Particularly, we will get an English to German translation subset specified as `opus/medical` which has medical related texts. If storage is not an issue, you can opt to get a larger corpus such as the English to German translation dataset from [ParaCrawl](https://paracrawl.eu/), a large multi-lingual translation dataset created by the European Union. Both of these datasets are available via [Tensorflow Datasets (TFDS)](https://www.tensorflow.org/datasets)
# and you can browse through the other available datasets [here](https://www.tensorflow.org/datasets/catalog/overview). We have downloaded the data for you in the `data/` directory of your workspace. As you'll see below, you can easily access this dataset from TFDS with `trax.data.TFDS`. The result is a python generator function yielding tuples. Use the `keys` argument to select what appears at which position in the tuple. For example, `keys=('en', 'de')` below will return pairs as (English sentence, German sentence).
# %%
# Get generator function for the training set
# This will download the train dataset if no data_dir is specified.
train_stream_fn = trax.data.TFDS('opus/medical',
data_dir='./data/',
keys=('en', 'de'),
eval_holdout_size=0.01, # 1% for eval
train=True)
# Get generator function for the eval set
eval_stream_fn = trax.data.TFDS('opus/medical',
data_dir='./data/',
keys=('en', 'de'),
eval_holdout_size=0.01, # 1% for eval
train=False)
# %% [markdown]
# Notice that TFDS returns a generator *function*, not a generator. This is because in Python, you cannot reset generators so you cannot go back to a previously yielded value. During deep learning training, you use Stochastic Gradient Descent and don't actually need to go back -- but it is sometimes good to be able to do that, and that's where the functions come in. It is actually very common to use generator functions in Python -- e.g., `zip` is a generator function. You can read more about [Python generators](https://book.pythontips.com/en/latest/generators.html) to understand why we use them. Let's print a a sample pair from our train and eval data. Notice that the raw ouput is represented in bytes (denoted by the `b'` prefix) and these will be converted to strings internally in the next steps.
# %%
train_stream = train_stream_fn()
print(colored('train data (en, de) tuple:', 'red'), next(train_stream))
print()
eval_stream = eval_stream_fn()
print(colored('eval data (en, de) tuple:', 'red'), next(eval_stream))
# %% [markdown]
# <a name="1.2"></a>
# ## 1.2 Tokenization and Formatting
#
# Now that we have imported our corpus, we will be preprocessing the sentences into a format that our model can accept. This will be composed of several steps:
#
# **Tokenizing the sentences using subword representations:** As you've learned in the earlier courses of this specialization, we want to represent each sentence as an array of integers instead of strings. For our application, we will use *subword* representations to tokenize our sentences. This is a common technique to avoid out-of-vocabulary words by allowing parts of words to be represented separately. For example, instead of having separate entries in your vocabulary for --"fear", "fearless", "fearsome", "some", and "less"--, you can simply store --"fear", "some", and "less"-- then allow your tokenizer to combine these subwords when needed. This allows it to be more flexible so you won't have to save uncommon words explicitly in your vocabulary (e.g. *stylebender*, *nonce*, etc). Tokenizing is done with the `trax.data.Tokenize()` command and we have provided you the combined subword vocabulary for English and German (i.e. `ende_32k.subword`) saved in the `data` directory. Feel free to open this file to see how the subwords look like.
# %%
# global variables that state the filename and directory of the vocabulary file
VOCAB_FILE = 'ende_32k.subword'
VOCAB_DIR = 'data/'
# Tokenize the dataset.
tokenized_train_stream = trax.data.Tokenize(vocab_file=VOCAB_FILE, vocab_dir=VOCAB_DIR)(train_stream)
tokenized_eval_stream = trax.data.Tokenize(vocab_file=VOCAB_FILE, vocab_dir=VOCAB_DIR)(eval_stream)
# %% [markdown]
# **Append an end-of-sentence token to each sentence:** We will assign a token (i.e. in this case `1`) to mark the end of a sentence. This will be useful in inference/prediction so we'll know that the model has completed the translation.
# %%
# Append EOS at the end of each sentence.
# Integer assigned as end-of-sentence (EOS)
EOS = 1
# generator helper function to append EOS to each sentence
def append_eos(stream):
for (inputs, targets) in stream:
inputs_with_eos = list(inputs) + [EOS]
targets_with_eos = list(targets) + [EOS]
yield np.array(inputs_with_eos), np.array(targets_with_eos)
# append EOS to the train data
tokenized_train_stream = append_eos(tokenized_train_stream)
# append EOS to the eval data
tokenized_eval_stream = append_eos(tokenized_eval_stream)
# %% [markdown]
# **Filter long sentences:** We will place a limit on the number of tokens per sentence to ensure we won't run out of memory. This is done with the `trax.data.FilterByLength()` method and you can see its syntax below.
# %%
# Filter too long sentences to not run out of memory.
# length_keys=[0, 1] means we filter both English and German sentences, so
# both much be not longer that 256 tokens for training / 512 for eval.
filtered_train_stream = trax.data.FilterByLength(
max_length=256, length_keys=[0, 1])(tokenized_train_stream)
filtered_eval_stream = trax.data.FilterByLength(
max_length=512, length_keys=[0, 1])(tokenized_eval_stream)
# print a sample input-target pair of tokenized sentences
train_input, train_target = next(filtered_train_stream)
print(colored(f'Single tokenized example input:', 'red' ), train_input)
print(colored(f'Single tokenized example target:', 'red'), train_target)
# %% [markdown]
# <a name="1.3"></a>
# ## 1.3 tokenize & detokenize helper functions
#
# Given any data set, you have to be able to map words to their indices, and indices to their words. The inputs and outputs to your trax models are usually tensors of numbers where each number corresponds to a word. If you were to process your data manually, you would have to make use of the following:
#
# - <span style='color:blue'> word2Ind: </span> a dictionary mapping the word to its index.
# - <span style='color:blue'> ind2Word:</span> a dictionary mapping the index to its word.
# - <span style='color:blue'> word2Count:</span> a dictionary mapping the word to the number of times it appears.
# - <span style='color:blue'> num_words:</span> total number of words that have appeared.
#
# Since you have already implemented these in previous assignments of the specialization, we will provide you with helper functions that will do this for you. Run the cell below to get the following functions:
#
# - <span style='color:blue'> tokenize(): </span> converts a text sentence to its corresponding token list (i.e. list of indices). Also converts words to subwords (parts of words).
# - <span style='color:blue'> detokenize(): </span> converts a token list to its corresponding sentence (i.e. string).
# %%
# Setup helper functions for tokenizing and detokenizing sentences
def tokenize(input_str, vocab_file=None, vocab_dir=None):
"""Encodes a string to an array of integers
Args:
input_str (str): human-readable string to encode
vocab_file (str): filename of the vocabulary text file
vocab_dir (str): path to the vocabulary file
Returns:
numpy.ndarray: tokenized version of the input string
"""
# Set the encoding of the "end of sentence" as 1
EOS = 1
# Use the trax.data.tokenize method. It takes streams and returns streams,
# we get around it by making a 1-element stream with `iter`.
inputs = next(trax.data.tokenize(iter([input_str]),
vocab_file=vocab_file, vocab_dir=vocab_dir))
# Mark the end of the sentence with EOS
inputs = list(inputs) + [EOS]
# Adding the batch dimension to the front of the shape
batch_inputs = np.reshape(np.array(inputs), [1, -1])
return batch_inputs
def detokenize(integers, vocab_file=None, vocab_dir=None):
"""Decodes an array of integers to a human readable string
Args:
integers (numpy.ndarray): array of integers to decode
vocab_file (str): filename of the vocabulary text file
vocab_dir (str): path to the vocabulary file
Returns:
str: the decoded sentence.
"""
# Remove the dimensions of size 1
integers = list(np.squeeze(integers))
# Set the encoding of the "end of sentence" as 1
EOS = 1
# Remove the EOS to decode only the original tokens
if EOS in integers:
integers = integers[:integers.index(EOS)]
return trax.data.detokenize(integers, vocab_file=vocab_file, vocab_dir=vocab_dir)
# %% [markdown]
# Let's see how we might use these functions:
# %%
# As declared earlier:
# VOCAB_FILE = 'ende_32k.subword'
# VOCAB_DIR = 'data/'
# Detokenize an input-target pair of tokenized sentences
print(colored(f'Single detokenized example input:', 'red'), detokenize(train_input, vocab_file=VOCAB_FILE, vocab_dir=VOCAB_DIR))
print(colored(f'Single detokenized example target:', 'red'), detokenize(train_target, vocab_file=VOCAB_FILE, vocab_dir=VOCAB_DIR))
print()
# Tokenize and detokenize a word that is not explicitly saved in the vocabulary file.
# See how it combines the subwords -- 'hell' and 'o'-- to form the word 'hello'.
print(colored(f"tokenize('hello'): ", 'green'), tokenize('hello', vocab_file=VOCAB_FILE, vocab_dir=VOCAB_DIR))
print(colored(f"detokenize([17332, 140, 1]): ", 'green'), detokenize([17332, 140, 1], vocab_file=VOCAB_FILE, vocab_dir=VOCAB_DIR))
# %% [markdown]
# <a name="1.4"></a>
# ## 1.4 Bucketing
#
# Bucketing the tokenized sentences is an important technique used to speed up training in NLP.
# Here is a
# [nice article describing it in detail](https://medium.com/@rashmi.margani/how-to-speed-up-the-training-of-the-sequence-model-using-bucketing-techniques-9e302b0fd976)
# but the gist is very simple. Our inputs have variable lengths and you want to make these the same when batching groups of sentences together. One way to do that is to pad each sentence to the length of the longest sentence in the dataset. This might lead to some wasted computation though. For example, if there are multiple short sentences with just two tokens, do we want to pad these when the longest sentence is composed of a 100 tokens? Instead of padding with 0s to the maximum length of a sentence each time, we can group our tokenized sentences by length and bucket, as on this image (from the article above):
#
# 
#
# We batch the sentences with similar length together (e.g. the blue sentences in the image above) and only add minimal padding to make them have equal length (usually up to the nearest power of two). This allows to waste less computation when processing padded sequences.
# In Trax, it is implemented in the [bucket_by_length](https://github.com/google/trax/blob/5fb8aa8c5cb86dabb2338938c745996d5d87d996/trax/supervised/inputs.py#L378) function.
# %%
# Bucketing to create streams of batches.
# Buckets are defined in terms of boundaries and batch sizes.
# Batch_sizes[i] determines the batch size for items with length < boundaries[i]
# So below, we'll take a batch of 256 sentences of length < 8, 128 if length is
# between 8 and 16, and so on -- and only 2 if length is over 512.
boundaries = [8, 16, 32, 64, 128, 256, 512]
batch_sizes = [256, 128, 64, 32, 16, 8, 4, 2]
# Create the generators.
train_batch_stream = trax.data.BucketByLength(
boundaries, batch_sizes,
length_keys=[0, 1] # As before: count inputs and targets to length.
)(filtered_train_stream)
eval_batch_stream = trax.data.BucketByLength(
boundaries, batch_sizes,
length_keys=[0, 1] # As before: count inputs and targets to length.
)(filtered_eval_stream)
# Add masking for the padding (0s).
train_batch_stream = trax.data.AddLossWeights(id_to_mask=0)(train_batch_stream)
eval_batch_stream = trax.data.AddLossWeights(id_to_mask=0)(eval_batch_stream)
# %% [markdown]
# <a name="1.5"></a>
# ## 1.5 Exploring the data
#
# We will now be displaying some of our data. You will see that the functions defined above (i.e. `tokenize()` and `detokenize()`) do the same things you have been doing again and again throughout the specialization. We gave these so you can focus more on building the model from scratch. Let us first get the data generator and get one batch of the data.
# %%
input_batch, target_batch, mask_batch = next(train_batch_stream)
# let's see the data type of a batch
print("input_batch data type: ", type(input_batch))
print("target_batch data type: ", type(target_batch))
# let's see the shape of this particular batch (batch length, sentence length)
print("input_batch shape: ", input_batch.shape)
print("target_batch shape: ", target_batch.shape)
# %% [markdown]
# The `input_batch` and `target_batch` are Numpy arrays consisting of tokenized English sentences and German sentences respectively. These tokens will later be used to produce embedding vectors for each word in the sentence (so the embedding for a sentence will be a matrix). The number of sentences in each batch is usually a power of 2 for optimal computer memory usage.
#
# We can now visually inspect some of the data. You can run the cell below several times to shuffle through the sentences. Just to note, while this is a standard data set that is used widely, it does have some known wrong translations. With that, let's pick a random sentence and print its tokenized representation.
# %%
# pick a random index less than the batch size.
index = random.randrange(len(input_batch))
# use the index to grab an entry from the input and target batch
print(colored('THIS IS THE ENGLISH SENTENCE: \n', 'red'), detokenize(input_batch[index], vocab_file=VOCAB_FILE, vocab_dir=VOCAB_DIR), '\n')
print(colored('THIS IS THE TOKENIZED VERSION OF THE ENGLISH SENTENCE: \n ', 'red'), input_batch[index], '\n')
print(colored('THIS IS THE GERMAN TRANSLATION: \n', 'red'), detokenize(target_batch[index], vocab_file=VOCAB_FILE, vocab_dir=VOCAB_DIR), '\n')
print(colored('THIS IS THE TOKENIZED VERSION OF THE GERMAN TRANSLATION: \n', 'red'), target_batch[index], '\n')
# %% [markdown]
# <a name="2"></a>
# # Part 2: Neural Machine Translation with Attention
#
# Now that you have the data generators and have handled the preprocessing, it is time for you to build the model. You will be implementing a neural machine translation model from scratch with attention.
#
# %% [markdown]
# <a name="2.1"></a>
# ## 2.1 Attention Overview
#
# The model we will be building uses an encoder-decoder architecture. This Recurrent Neural Network (RNN) will take in a tokenized version of a sentence in its encoder, then passes it on to the decoder for translation. As mentioned in the lectures, just using a a regular sequence-to-sequence model with LSTMs will work effectively for short to medium sentences but will start to degrade for longer ones. You can picture it like the figure below where all of the context of the input sentence is compressed into one vector that is passed into the decoder block. You can see how this will be an issue for very long sentences (e.g. 100 tokens or more) because the context of the first parts of the input will have very little effect on the final vector passed to the decoder.
#
# <img src='plain_rnn.png'>
#
# Adding an attention layer to this model avoids this problem by giving the decoder access to all parts of the input sentence. To illustrate, let's just use a 4-word input sentence as shown below. Remember that a hidden state is produced at each timestep of the encoder (represented by the orange rectangles). These are all passed to the attention layer and each are given a score given the current activation (i.e. hidden state) of the decoder. For instance, let's consider the figure below where the first prediction "Wie" is already made. To produce the next prediction, the attention layer will first receive all the encoder hidden states (i.e. orange rectangles) as well as the decoder hidden state when producing the word "Wie" (i.e. first green rectangle). Given these information, it will score each of the encoder hidden states to know which one the decoder should focus on to produce the next word. The result of the model training might have learned that it should align to the second encoder hidden state and subsequently assigns a high probability to the word "geht". If we are using greedy decoding, we will output the said word as the next symbol, then restart the process to produce the next word until we reach an end-of-sentence prediction.
#
# <img src='attention_overview.png'>
#
#
# There are different ways to implement attention and the one we'll use for this assignment is the Scaled Dot Product Attention which has the form:
#
# $$Attention(Q, K, V) = softmax(\frac{QK^T}{\sqrt{d_k}})V$$
#
# You will dive deeper into this equation in the next week but for now, you can think of it as computing scores using queries (Q) and keys (K), followed by a multiplication of values (V) to get a context vector at a particular timestep of the decoder. This context vector is fed to the decoder RNN to get a set of probabilities for the next predicted word. The division by square root of the keys dimensionality ($\sqrt{d_k}$) is for improving model performance and you'll also learn more about it next week. For our machine translation application, the encoder activations (i.e. encoder hidden states) will be the keys and values, while the decoder activations (i.e. decoder hidden states) will be the queries.
#
# You will see in the upcoming sections that this complex architecture and mechanism can be implemented with just a few lines of code. Let's get started!
# %% [markdown]
# <a name="2.2"></a>
# ## 2.2 Helper functions
#
# We will first implement a few functions that we will use later on. These will be for the input encoder, pre-attention decoder, and preparation of the queries, keys, values, and mask.
#
# ### 2.2.1 Input encoder
#
# The input encoder runs on the input tokens, creates its embeddings, and feeds it to an LSTM network. This outputs the activations that will be the keys and values for attention. It is a [Serial](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.combinators.Serial) network which uses:
#
# - [tl.Embedding](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.core.Embedding): Converts each token to its vector representation. In this case, it is the the size of the vocabulary by the dimension of the model: `tl.Embedding(vocab_size, d_model)`. `vocab_size` is the number of entries in the given vocabulary. `d_model` is the number of elements in the word embedding.
#
# - [tl.LSTM](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.rnn.LSTM): LSTM layer of size `d_model`. We want to be able to configure how many encoder layers we have so remember to create LSTM layers equal to the number of the `n_encoder_layers` parameter.
#
# <img src = "input_encoder.png">
#
# <a name="ex01"></a>
# ### Exercise 01
#
# **Instructions:** Implement the `input_encoder_fn` function.
# %%
# UNQ_C1
# GRADED FUNCTION
def input_encoder_fn(input_vocab_size, d_model, n_encoder_layers):
""" Input encoder runs on the input sentence and creates
activations that will be the keys and values for attention.
Args:
input_vocab_size: int: vocab size of the input
d_model: int: depth of embedding (n_units in the LSTM cell)
n_encoder_layers: int: number of LSTM layers in the encoder
Returns:
tl.Serial: The input encoder
"""
# create a serial network
input_encoder = tl.Serial(
### START CODE HERE (REPLACE INSTANCES OF `None` WITH YOUR CODE) ###
# create an embedding layer to convert tokens to vectors
tl.Embedding(input_vocab_size, d_model),
# feed the embeddings to the LSTM layers. It is a stack of n_encoder_layers LSTM layers
[tl.LSTM(d_model) for _ in range(n_encoder_layers)]
### END CODE HERE ###
)
return input_encoder
# %% [markdown]
# *Note: To make this notebook more neat, we moved the unit tests to a separate file called `w1_unittest.py`. Feel free to open it from your workspace if needed. We have placed comments in that file to indicate which functions are testing which part of the assignment (e.g. `test_input_encoder_fn()` has the unit tests for UNQ_C1).*
# %%
# BEGIN UNIT TEST
import w1_unittest
w1_unittest.test_input_encoder_fn(input_encoder_fn)
# END UNIT TEST
# %% [markdown]
# ### 2.2.2 Pre-attention decoder
#
# The pre-attention decoder runs on the targets and creates activations that are used as queries in attention. This is a Serial network which is composed of the following:
#
# - [tl.ShiftRight](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.attention.ShiftRight): This pads a token to the beginning of your target tokens (e.g. `[8, 34, 12]` shifted right is `[0, 8, 34, 12]`). This will act like a start-of-sentence token that will be the first input to the decoder. During training, this shift also allows the target tokens to be passed as input to do teacher forcing.
#
# - [tl.Embedding](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.core.Embedding): Like in the previous function, this converts each token to its vector representation. In this case, it is the the size of the vocabulary by the dimension of the model: `tl.Embedding(vocab_size, d_model)`. `vocab_size` is the number of entries in the given vocabulary. `d_model` is the number of elements in the word embedding.
#
# - [tl.LSTM](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.rnn.LSTM): LSTM layer of size `d_model`.
#
# <img src = "pre_attention_decoder.png">
#
# <a name="ex02"></a>
# ### Exercise 02
#
# **Instructions:** Implement the `pre_attention_decoder_fn` function.
#
# %%
# UNQ_C2
# GRADED FUNCTION
def pre_attention_decoder_fn(mode, target_vocab_size, d_model):
""" Pre-attention decoder runs on the targets and creates
activations that are used as queries in attention.
Args:
mode: str: 'train' or 'eval'
target_vocab_size: int: vocab size of the target
d_model: int: depth of embedding (n_units in the LSTM cell)
Returns:
tl.Serial: The pre-attention decoder
"""
# create a serial network
pre_attention_decoder = tl.Serial(
### START CODE HERE (REPLACE INSTANCES OF `None` WITH YOUR CODE) ###
# shift right to insert start-of-sentence token and implement
# teacher forcing during training
tl.ShiftRight(mode=mode),
# run an embedding layer to convert tokens to vectors
tl.Embedding(target_vocab_size,d_model),
# feed to an LSTM layer
tl.LSTM(d_model)
### END CODE HERE ###
)
return pre_attention_decoder
# %%
# BEGIN UNIT TEST
w1_unittest.test_pre_attention_decoder_fn(pre_attention_decoder_fn)
# END UNIT TEST
# %% [markdown]
# ### 2.2.3 Preparing the attention input
#
# This function will prepare the inputs to the attention layer. We want to take in the encoder and pre-attention decoder activations and assign it to the queries, keys, and values. In addition, another output here will be the mask to distinguish real tokens from padding tokens. This mask will be used internally by Trax when computing the softmax so padding tokens will not have an effect on the computated probabilities. From the data preparation steps in Section 1 of this assignment, you should know which tokens in the input correspond to padding.
#
# We have filled the last two lines in composing the mask for you because it includes a concept that will be discussed further next week. This is related to *multiheaded attention* which you can think of right now as computing the attention multiple times to improve the model's predictions. It is required to consider this additional axis in the output so we've included it already but you don't need to analyze it just yet. What's important now is for you to know which should be the queries, keys, and values, as well as to initialize the mask.
#
# <a name="ex03"></a>
# ### Exercise 03
#
# **Instructions:** Implement the `prepare_attention_input` function
#
# %%
# UNQ_C3
# GRADED FUNCTION
def prepare_attention_input(encoder_activations, decoder_activations, inputs):
"""Prepare queries, keys, values and mask for attention.
Args:
encoder_activations fastnp.array(batch_size, padded_input_length, d_model): output from the input encoder
decoder_activations fastnp.array(batch_size, padded_input_length, d_model): output from the pre-attention decoder
inputs fastnp.array(batch_size, padded_input_length): padded input tokens
Returns:
queries, keys, values and mask for attention.
"""
### START CODE HERE (REPLACE INSTANCES OF `None` WITH YOUR CODE) ###
# set the keys and values to the encoder activations
keys = encoder_activations
values = encoder_activations
# set the queries to the decoder activations
queries = decoder_activations
# generate the mask to distinguish real tokens from padding
# hint: inputs is 1 for real tokens and 0 where they are padding
mask = ( inputs !=0 )
### END CODE HERE ###
# add axes to the mask for attention heads and decoder length.
mask = fastnp.reshape(mask, (mask.shape[0], 1, 1, mask.shape[1]))
# broadcast so mask shape is [batch size, attention heads, decoder-len, encoder-len].
# note: for this assignment, attention heads is set to 1.
mask = mask + fastnp.zeros((1, 1, decoder_activations.shape[1], 1))
return queries, keys, values, mask
# %%
# BEGIN UNIT TEST
w1_unittest.test_prepare_attention_input(prepare_attention_input)
# END UNIT TEST
# %% [markdown]
# <a name="2.3"></a>
# ## 2.3 Implementation Overview
#
# We are now ready to implement our sequence-to-sequence model with attention. This will be a Serial network and is illustrated in the diagram below. It shows the layers you'll be using in Trax and you'll see that each step can be implemented quite easily with one line commands. We've placed several links to the documentation for each relevant layer in the discussion after the figure below.
#
# <img src = "NMTModel.png">
# %% [markdown]
# <a name="ex04"></a>
# ### Exercise 04
# **Instructions:** Implement the `NMTAttn` function below to define your machine translation model which uses attention. We have left hyperlinks below pointing to the Trax documentation of the relevant layers. Remember to consult it to get tips on what parameters to pass.
#
# **Step 0:** Prepare the input encoder and pre-attention decoder branches. You have already defined this earlier as helper functions so it's just a matter of calling those functions and assigning it to variables.
#
# **Step 1:** Create a Serial network. This will stack the layers in the next steps one after the other. Like the earlier exercises, you can use [tl.Serial](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.combinators.Serial).
#
# **Step 2:** Make a copy of the input and target tokens. As you see in the diagram above, the input and target tokens will be fed into different layers of the model. You can use [tl.Select](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.combinators.Select) layer to create copies of these tokens. Arrange them as `[input tokens, target tokens, input tokens, target tokens]`.
#
# **Step 3:** Create a parallel branch to feed the input tokens to the `input_encoder` and the target tokens to the `pre_attention_decoder`. You can use [tl.Parallel](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.combinators.Parallel) to create these sublayers in parallel. Remember to pass the variables you defined in Step 0 as parameters to this layer.
#
# **Step 4:** Next, call the `prepare_attention_input` function to convert the encoder and pre-attention decoder activations to a format that the attention layer will accept. You can use [tl.Fn](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.base.Fn) to call this function. Note: Pass the `prepare_attention_input` function as the `f` parameter in `tl.Fn` without any arguments or parenthesis.
#
# **Step 5:** We will now feed the (queries, keys, values, and mask) to the [tl.AttentionQKV](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.attention.AttentionQKV) layer. This computes the scaled dot product attention and outputs the attention weights and mask. Take note that although it is a one liner, this layer is actually composed of a deep network made up of several branches. We'll show the implementation taken [here](https://github.com/google/trax/blob/master/trax/layers/attention.py#L61) to see the different layers used.
#
# ```python
# def AttentionQKV(d_feature, n_heads=1, dropout=0.0, mode='train'):
# """Returns a layer that maps (q, k, v, mask) to (activations, mask).
#
# See `Attention` above for further context/details.
#
# Args:
# d_feature: Depth/dimensionality of feature embedding.
# n_heads: Number of attention heads.
# dropout: Probababilistic rate for internal dropout applied to attention
# activations (based on query-key pairs) before dotting them with values.
# mode: Either 'train' or 'eval'.
# """
# return cb.Serial(
# cb.Parallel(
# core.Dense(d_feature),
# core.Dense(d_feature),
# core.Dense(d_feature),
# ),
# PureAttention( # pylint: disable=no-value-for-parameter
# n_heads=n_heads, dropout=dropout, mode=mode),
# core.Dense(d_feature),
# )
# ```
#
# Having deep layers pose the risk of vanishing gradients during training and we would want to mitigate that. To improve the ability of the network to learn, we can insert a [tl.Residual](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.combinators.Residual) layer to add the output of AttentionQKV with the `queries` input. You can do this in trax by simply nesting the `AttentionQKV` layer inside the `Residual` layer. The library will take care of branching and adding for you.
#
# **Step 6:** We will not need the mask for the model we're building so we can safely drop it. At this point in the network, the signal stack currently has `[attention activations, mask, target tokens]` and you can use [tl.Select](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.combinators.Select) to output just `[attention activations, target tokens]`.
#
# **Step 7:** We can now feed the attention weighted output to the LSTM decoder. We can stack multiple [tl.LSTM](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.rnn.LSTM) layers to improve the output so remember to append LSTMs equal to the number defined by `n_decoder_layers` parameter to the model.
#
# **Step 8:** We want to determine the probabilities of each subword in the vocabulary and you can set this up easily with a [tl.Dense](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.core.Dense) layer by making its size equal to the size of our vocabulary.
#
# **Step 9:** Normalize the output to log probabilities by passing the activations in Step 8 to a [tl.LogSoftmax](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.core.LogSoftmax) layer.
# %%
# UNQ_C4
# GRADED FUNCTION
def NMTAttn(input_vocab_size=33300,
target_vocab_size=33300,
d_model=1024,
n_encoder_layers=2,
n_decoder_layers=2,
n_attention_heads=4,
attention_dropout=0.0,
mode='train'):
"""Returns an LSTM sequence-to-sequence model with attention.
The input to the model is a pair (input tokens, target tokens), e.g.,
an English sentence (tokenized) and its translation into German (tokenized).
Args:
input_vocab_size: int: vocab size of the input
target_vocab_size: int: vocab size of the target
d_model: int: depth of embedding (n_units in the LSTM cell)
n_encoder_layers: int: number of LSTM layers in the encoder
n_decoder_layers: int: number of LSTM layers in the decoder after attention
n_attention_heads: int: number of attention heads
attention_dropout: float, dropout for the attention layer
mode: str: 'train', 'eval' or 'predict', predict mode is for fast inference
Returns:
A LSTM sequence-to-sequence model with attention.
"""
### START CODE HERE (REPLACE INSTANCES OF `None` WITH YOUR CODE) ###
# Step 0: call the helper function to create layers for the input encoder
input_encoder = input_encoder_fn(input_vocab_size, d_model, n_encoder_layers)
# Step 0: call the helper function to create layers for the pre-attention decoder
pre_attention_decoder = pre_attention_decoder_fn(mode, target_vocab_size, d_model)
# Step 1: create a serial network
model = tl.Serial(
# Step 2: copy input tokens and target tokens as they will be needed later.
tl.Select([0,1,0,1]),
# Step 3: run input encoder on the input and pre-attention decoder the target.
tl.Parallel(input_encoder, pre_attention_decoder),
# Step 4: prepare queries, keys, values and mask for attention.
tl.Fn('PrepareAttentionInput', prepare_attention_input, n_out=4),
# Step 5: run the AttentionQKV layer
# nest it inside a Residual layer to add to the pre-attention decoder activations(i.e. queries)
tl.Residual(tl.AttentionQKV(d_model, n_heads=n_attention_heads, dropout=attention_dropout, mode=mode)),
# Step 6: drop attention mask (i.e. index = None
tl.Select([0,2]),
# Step 7: run the rest of the RNN decoder
[tl.LSTM(d_model) for _ in range(n_decoder_layers)],
# Step 8: prepare output by making it the right size
tl.Dense(target_vocab_size),
# Step 9: Log-softmax for output
tl.LogSoftmax()
)
### END CODE HERE
return model
# %%
# BEGIN UNIT TEST
w1_unittest.test_NMTAttn(NMTAttn)
# END UNIT TEST
# %%
# print your model
model = NMTAttn()
print(model)
# %% [markdown]
# **Expected Output:**
#
# ```
# Serial_in2_out2[
# Select[0,1,0,1]_in2_out4
# Parallel_in2_out2[
# Serial[
# Embedding_33300_1024
# LSTM_1024
# LSTM_1024
# ]
# Serial[
# ShiftRight(1)
# Embedding_33300_1024
# LSTM_1024
# ]
# ]
# PrepareAttentionInput_in3_out4
# Serial_in4_out2[
# Branch_in4_out3[
# None
# Serial_in4_out2[
# Parallel_in3_out3[
# Dense_1024
# Dense_1024
# Dense_1024
# ]
# PureAttention_in4_out2
# Dense_1024
# ]
# ]
# Add_in2
# ]
# Select[0,2]_in3_out2
# LSTM_1024
# LSTM_1024
# Dense_33300
# LogSoftmax
# ]
# ```
# %% [markdown]
# <a name="3"></a>
# # Part 3: Training
#
# We will now be training our model in this section. Doing supervised training in Trax is pretty straightforward (short example [here](https://trax-ml.readthedocs.io/en/latest/notebooks/trax_intro.html#Supervised-training)). We will be instantiating three classes for this: `TrainTask`, `EvalTask`, and `Loop`. Let's take a closer look at each of these in the sections below.
#
# %% [markdown]
# <a name="3.1"></a>
# ## 3.1 TrainTask
#
# The [TrainTask](https://trax-ml.readthedocs.io/en/latest/trax.supervised.html#trax.supervised.training.TrainTask) class allows us to define the labeled data to use for training and the feedback mechanisms to compute the loss and update the weights.
#
# <a name="ex05"></a>
# ### Exercise 05
#
# **Instructions:** Instantiate a train task.
# %%
# UNQ_C5
# GRADED
train_task = training.TrainTask(
### START CODE HERE (REPLACE INSTANCES OF `None` WITH YOUR CODE) ###
# use the train batch stream as labeled data
labeled_data= train_batch_stream,
# use the cross entropy loss
loss_layer= tl.CrossEntropyLoss(),
# use the Adam optimizer with learning rate of 0.01
optimizer= trax.optimizers.Adam(0.01),
# use the `trax.lr.warmup_and_rsqrt_decay` as the learning rate schedule
# have 1000 warmup steps with a max value of 0.01
lr_schedule= trax.lr.warmup_and_rsqrt_decay(1000,0.01),
# have a checkpoint every 10 steps
n_steps_per_checkpoint= 10,
### END CODE HERE ###
)
# %%
# BEGIN UNIT TEST
w1_unittest.test_train_task(train_task)
# END UNIT TEST
# %% [markdown]
# <a name="3.2"></a>
# ## 3.2 EvalTask
#
# The [EvalTask](https://trax-ml.readthedocs.io/en/latest/trax.supervised.html#trax.supervised.training.EvalTask) on the other hand allows us to see how the model is doing while training. For our application, we want it to report the cross entropy loss and accuracy.
# %%
eval_task = training.EvalTask(
## use the eval batch stream as labeled data
labeled_data=eval_batch_stream,
## use the cross entropy loss and accuracy as metrics
metrics=[tl.CrossEntropyLoss(), tl.Accuracy()],
)
# %% [markdown]
# <a name="3.3"></a>
# ## 3.3 Loop
#
# The [Loop](https://trax-ml.readthedocs.io/en/latest/trax.supervised.html#trax.supervised.training.Loop) class defines the model we will train as well as the train and eval tasks to execute. Its `run()` method allows us to execute the training for a specified number of steps.
# %%
# define the output directory
output_dir = 'output_dir/'
# remove old model if it exists. restarts training.
# !rm -f ~/output_dir/model.pkl.gz
# define the training loop
training_loop = training.Loop(NMTAttn(mode='train'),
train_task,
eval_tasks=[eval_task],
output_dir=output_dir)
# %%
# NOTE: Execute the training loop. This will take around 8 minutes to complete.
training_loop.run(10)
# %% [markdown]
# <a name="4"></a>
# # Part 4: Testing
#
# We will now be using the model you just trained to translate English sentences to German. We will implement this with two functions: The first allows you to identify the next symbol (i.e. output token). The second one takes care of combining the entire translated string.
#
# We will start by first loading in a pre-trained copy of the model you just coded. Please run the cell below to do just that.
# %%
# instantiate the model we built in eval mode
model = NMTAttn(mode='eval')
# initialize weights from a pre-trained model
model.init_from_file("model.pkl.gz", weights_only=True)
model = tl.Accelerate(model)
# %% [markdown]
# <a name="4.1"></a>
# ## 4.1 Decoding
#
# As discussed in the lectures, there are several ways to get the next token when translating a sentence. For instance, we can just get the most probable token at each step (i.e. greedy decoding) or get a sample from a distribution. We can generalize the implementation of these two approaches by using the `tl.logsoftmax_sample()` method. Let's briefly look at its implementation:
#
# ```python
# def logsoftmax_sample(log_probs, temperature=1.0): # pylint: disable=invalid-name
# """Returns a sample from a log-softmax output, with temperature.
#
# Args:
# log_probs: Logarithms of probabilities (often coming from LogSofmax)
# temperature: For scaling before sampling (1.0 = default, 0.0 = pick argmax)
# """
# # This is equivalent to sampling from a softmax with temperature.
# u = np.random.uniform(low=1e-6, high=1.0 - 1e-6, size=log_probs.shape)
# g = -np.log(-np.log(u))
# return np.argmax(log_probs + g * temperature, axis=-1)
# ```
#
# The key things to take away here are: 1. it gets random samples with the same shape as your input (i.e. `log_probs`), and 2. the amount of "noise" added to the input by these random samples is scaled by a `temperature` setting. You'll notice that setting it to `0` will just make the return statement equal to getting the argmax of `log_probs`. This will come in handy later.
#
# <a name="ex06"></a>
# ### Exercise 06
#
# **Instructions:** Implement the `next_symbol()` function that takes in the `input_tokens` and the `cur_output_tokens`, then return the index of the next word. You can click below for hints in completing this exercise.
#
# <details>
# <summary>
# <font size="3" color="darkgreen"><b>Click Here for Hints</b></font>
# </summary>
# <p>
# <ul>
# <li>To get the next power of two, you can compute <i>2^log_2(token_length + 1)</i> . We add 1 to avoid <i>log(0).</i></li>
# <li>You can use <i>np.ceil()</i> to get the ceiling of a float.</li>
# <li><i>np.log2()</i> will get the logarithm base 2 of a value</li>
# <li><i>int()</i> will cast a value into an integer type</li>
# <li>From the model diagram in part 2, you know that it takes two inputs. You can feed these with this syntax to get the model outputs: <i>model((input1, input2))</i>. It's up to you to determine which variables below to substitute for input1 and input2. Remember also from the diagram that the output has two elements: [log probabilities, target tokens]. You won't need the target tokens so we assigned it to _ below for you. </li>
# <li> The log probabilities output will have the shape: (batch size, decoder length, vocab size). It will contain log probabilities for each token in the <i>cur_output_tokens</i> plus 1 for the start symbol introduced by the ShiftRight in the preattention decoder. For example, if cur_output_tokens is [1, 2, 5], the model will output an array of log probabilities each for tokens 0 (start symbol), 1, 2, and 5. To generate the next symbol, you just want to get the log probabilities associated with the last token (i.e. token 5 at index 3). You can slice the model output at [0, 3, :] to get this. It will be up to you to generalize this for any length of cur_output_tokens </li>
# </ul>
#
# %%
# UNQ_C6
# GRADED FUNCTION
def next_symbol(NMTAttn, input_tokens, cur_output_tokens, temperature):
"""Returns the index of the next token.
Args:
NMTAttn (tl.Serial): An LSTM sequence-to-sequence model with attention.
input_tokens (np.ndarray 1 x n_tokens): tokenized representation of the input sentence
cur_output_tokens (list): tokenized representation of previously translated words
temperature (float): parameter for sampling ranging from 0.0 to 1.0.
0.0: same as argmax, always pick the most probable token
1.0: sampling from the distribution (can sometimes say random things)
Returns:
int: index of the next token in the translated sentence
float: log probability of the next symbol
"""
### START CODE HERE (REPLACE INSTANCES OF `None` WITH YOUR CODE) ###
# set the length of the current output tokens
token_length = len(cur_output_tokens)
# calculate next power of 2 for padding length
padded_length = 2**int(np.ceil(np.log2(token_length + 1)))
# pad cur_output_tokens up to the padded_length
padded = cur_output_tokens + [0]*padded_length
# model expects the output to have an axis for the batch size in front so
# convert `padded` list to a numpy array with shape (x, <padded_length>) where the
# x position is the batch axis. (hint: you can use np.expand_dims() with axis=0 to insert a new axis)
padded_with_batch = np.expand_dims(padded,axis=0)
# get the model prediction. remember to use the `NMAttn` argument defined above.
# hint: the model accepts a tuple as input (e.g. `my_model((input1, input2))`)
output, _ = NMTAttn((input_tokens,padded_with_batch))
# get log probabilities from the last token output
log_probs = output[0,token_length,:]
# get the next symbol by getting a logsoftmax sample (*hint: cast to an int)
symbol = int(tl.logsoftmax_sample(log_probs,temperature))
### END CODE HERE ###
return symbol, float(log_probs[symbol])
# %%
# BEGIN UNIT TEST
w1_unittest.test_next_symbol(next_symbol, model)
# END UNIT TEST
# %% [markdown]
# Now you will implement the `sampling_decode()` function. This will call the `next_symbol()` function above several times until the next output is the end-of-sentence token (i.e. `EOS`). It takes in an input string and returns the translated version of that string.
#
# <a name="ex07"></a>
# ### Exercise 07
#
# **Instructions**: Implement the `sampling_decode()` function.
# %%
# UNQ_C7
# GRADED FUNCTION
def sampling_decode(input_sentence, NMTAttn = None, temperature=0.0, vocab_file=None, vocab_dir=None):
"""Returns the translated sentence.
Args:
input_sentence (str): sentence to translate.
NMTAttn (tl.Serial): An LSTM sequence-to-sequence model with attention.
temperature (float): parameter for sampling ranging from 0.0 to 1.0.
0.0: same as argmax, always pick the most probable token
1.0: sampling from the distribution (can sometimes say random things)
vocab_file (str): filename of the vocabulary
vocab_dir (str): path to the vocabulary file
Returns:
tuple: (list, str, float)
list of int: tokenized version of the translated sentence
float: log probability of the translated sentence
str: the translated sentence
"""
### START CODE HERE (REPLACE INSTANCES OF `None` WITH YOUR CODE) ###
# encode the input sentence
input_tokens = tokenize(input_sentence, vocab_file, vocab_dir)
# initialize the list of output tokens
cur_output_tokens = []
# initialize an integer that represents the current output index
cur_output = 0
# Set the encoding of the "end of sentence" as 1
EOS = 1
# check that the current output is not the end of sentence token
while cur_output != EOS:
# update the current output token by getting the index of the next word (hint: use next_symbol)
cur_output, log_prob = next_symbol(NMTAttn,input_tokens,cur_output_tokens,temperature)
# append the current output token to the list of output tokens
cur_output_tokens.append(cur_output)
# detokenize the output tokens
sentence = detokenize(cur_output_tokens, vocab_file, vocab_dir)
### END CODE HERE ###
return cur_output_tokens, log_prob, sentence
# %%
# Test the function above. Try varying the temperature setting with values from 0 to 1.
# Run it several times with each setting and see how often the output changes.
sampling_decode("I love languages.", model, temperature=0.0, vocab_file=VOCAB_FILE, vocab_dir=VOCAB_DIR)
# %%
# BEGIN UNIT TEST
w1_unittest.test_sampling_decode(sampling_decode, model)
# END UNIT TEST
# %% [markdown]
# We have set a default value of `0` to the temperature setting in our implementation of `sampling_decode()` above. As you may have noticed in the `logsoftmax_sample()` method, this setting will ultimately result in greedy decoding. As mentioned in the lectures, this algorithm generates the translation by getting the most probable word at each step. It gets the argmax of the output array of your model and then returns that index. See the testing function and sample inputs below. You'll notice that the output will remain the same each time you run it.
# %%
def greedy_decode_test(sentence, NMTAttn=None, vocab_file=None, vocab_dir=None):
"""Prints the input and output of our NMTAttn model using greedy decode
Args:
sentence (str): a custom string.
NMTAttn (tl.Serial): An LSTM sequence-to-sequence model with attention.
vocab_file (str): filename of the vocabulary
vocab_dir (str): path to the vocabulary file
Returns:
str: the translated sentence
"""
_,_, translated_sentence = sampling_decode(sentence, NMTAttn, vocab_file=vocab_file, vocab_dir=vocab_dir)
print("English: ", sentence)
print("German: ", translated_sentence)
return translated_sentence
# %%
# put a custom string here
your_sentence = 'I love languages.'
greedy_decode_test(your_sentence, model, vocab_file=VOCAB_FILE, vocab_dir=VOCAB_DIR);
# %%
greedy_decode_test('You are almost done with the assignment!', model, vocab_file=VOCAB_FILE, vocab_dir=VOCAB_DIR);
# %% [markdown]
# <a name="4.2"></a>
# ## 4.2 Minimum Bayes-Risk Decoding
#
# As mentioned in the lectures, getting the most probable token at each step may not necessarily produce the best results. Another approach is to do Minimum Bayes Risk Decoding or MBR. The general steps to implement this are:
#
# 1. take several random samples
# 2. score each sample against all other samples
# 3. select the one with the highest score
#
# You will be building helper functions for these steps in the following sections.
# %% [markdown]
# <a name='4.2.1'></a>
# ### 4.2.1 Generating samples
#
# First, let's build a function to generate several samples. You can use the `sampling_decode()` function you developed earlier to do this easily. We want to record the token list and log probability for each sample as these will be needed in the next step.
# %%
def generate_samples(sentence, n_samples, NMTAttn=None, temperature=0.6, vocab_file=None, vocab_dir=None):
"""Generates samples using sampling_decode()
Args:
sentence (str): sentence to translate.
n_samples (int): number of samples to generate
NMTAttn (tl.Serial): An LSTM sequence-to-sequence model with attention.
temperature (float): parameter for sampling ranging from 0.0 to 1.0.
0.0: same as argmax, always pick the most probable token
1.0: sampling from the distribution (can sometimes say random things)
vocab_file (str): filename of the vocabulary
vocab_dir (str): path to the vocabulary file
Returns:
tuple: (list, list)
list of lists: token list per sample
list of floats: log probability per sample
"""
# define lists to contain samples and probabilities
samples, log_probs = [], []
# run a for loop to generate n samples
for _ in range(n_samples):
# get a sample using the sampling_decode() function
sample, logp, _ = sampling_decode(sentence, NMTAttn, temperature, vocab_file=vocab_file, vocab_dir=vocab_dir)
# append the token list to the samples list
samples.append(sample)
# append the log probability to the log_probs list
log_probs.append(logp)
return samples, log_probs
# %%
# generate 4 samples with the default temperature (0.6)
generate_samples('I love languages.', 4, model, vocab_file=VOCAB_FILE, vocab_dir=VOCAB_DIR)
# %% [markdown]
# ### 4.2.2 Comparing overlaps
#
# Let us now build our functions to compare a sample against another. There are several metrics available as shown in the lectures and you can try experimenting with any one of these. For this assignment, we will be calculating scores for unigram overlaps. One of the more simple metrics is the [Jaccard similarity](https://en.wikipedia.org/wiki/Jaccard_index) which gets the intersection over union of two sets. We've already implemented it below for your perusal.
# %%
def jaccard_similarity(candidate, reference):
"""Returns the Jaccard similarity between two token lists
Args:
candidate (list of int): tokenized version of the candidate translation
reference (list of int): tokenized version of the reference translation
Returns:
float: overlap between the two token lists
"""
# convert the lists to a set to get the unique tokens
can_unigram_set, ref_unigram_set = set(candidate), set(reference)
# get the set of tokens common to both candidate and reference
joint_elems = can_unigram_set.intersection(ref_unigram_set)
# get the set of all tokens found in either candidate or reference
all_elems = can_unigram_set.union(ref_unigram_set)
# divide the number of joint elements by the number of all elements
overlap = len(joint_elems) / len(all_elems)
return overlap
# %%
# let's try using the function. remember the result here and compare with the next function below.
jaccard_similarity([1, 2, 3], [1, 2, 3, 4])
# %% [markdown]
# One of the more commonly used metrics in machine translation is the ROUGE score. For unigrams, this is called ROUGE-1 and as shown in class, you can output the scores for both precision and recall when comparing two samples. To get the final score, you will want to compute the F1-score as given by:
#
# $$score = 2* \frac{(precision * recall)}{(precision + recall)}$$
#
# <a name="ex08"></a>
# ### Exercise 08
#
# **Instructions**: Implement the `rouge1_similarity()` function.
# %%
# UNQ_C8
# GRADED FUNCTION
# for making a frequency table easily
from collections import Counter
def rouge1_similarity(system, reference):
"""Returns the ROUGE-1 score between two token lists
Args:
system (list of int): tokenized version of the system translation
reference (list of int): tokenized version of the reference translation
Returns:
float: overlap between the two token lists
"""
### START CODE HERE (REPLACE INSTANCES OF `None` WITH YOUR CODE) ###
# make a frequency table of the system tokens (hint: use the Counter class)
sys_counter = Counter(system)
# make a frequency table of the reference tokens (hint: use the Counter class)
ref_counter = Counter(reference)
# initialize overlap to 0
overlap = 0
# run a for loop over the sys_counter object (can be treated as a dictionary)
for token in sys_counter:
# lookup the value of the token in the sys_counter dictionary (hint: use the get() method)
token_count_sys = sys_counter.get(token,0)
# lookup the value of the token in the ref_counter dictionary (hint: use the get() method)
token_count_ref = ref_counter.get(token,0)
# update the overlap by getting the smaller number between the two token counts above
overlap += min(token_count_sys,token_count_ref)
# get the precision (i.e. number of overlapping tokens / number of system tokens)
precision = overlap/sum(sys_counter.values())
# get the recall (i.e. number of overlapping tokens / number of reference tokens)
recall = overlap/sum(ref_counter.values())
if precision + recall != 0:
# compute the f1-score
rouge1_score = 2 * (precision*recall)/(precision+recall)
else:
rouge1_score = 0
### END CODE HERE ###
return rouge1_score
# %%
# notice that this produces a different value from the jaccard similarity earlier
rouge1_similarity([1, 2, 3], [1, 2, 3, 4])
# %%
# BEGIN UNIT TEST
w1_unittest.test_rouge1_similarity(rouge1_similarity)
# END UNIT TEST
# %% [markdown]
# ### 4.2.3 Overall score
#
# We will now build a function to generate the overall score for a particular sample. As mentioned earlier, we need to compare each sample with all other samples. For instance, if we generated 30 sentences, we will need to compare sentence 1 to sentences 2 to 30. Then, we compare sentence 2 to sentences 1 and 3 to 30, and so forth. At each step, we get the average score of all comparisons to get the overall score for a particular sample. To illustrate, these will be the steps to generate the scores of a 4-sample list.
#
# 1. Get similarity score between sample 1 and sample 2
# 2. Get similarity score between sample 1 and sample 3
# 3. Get similarity score between sample 1 and sample 4
# 4. Get average score of the first 3 steps. This will be the overall score of sample 1.
# 5. Iterate and repeat until samples 1 to 4 have overall scores.
#
# We will be storing the results in a dictionary for easy lookups.
#
# <a name="ex09"></a>
# ### Exercise 09
#
# **Instructions**: Implement the `average_overlap()` function.
# %%
# UNQ_C9
# GRADED FUNCTION
def average_overlap(similarity_fn, samples, *ignore_params):
"""Returns the arithmetic mean of each candidate sentence in the samples
Args:
similarity_fn (function): similarity function used to compute the overlap
samples (list of lists): tokenized version of the translated sentences
*ignore_params: additional parameters will be ignored
Returns:
dict: scores of each sample
key: index of the sample
value: score of the sample
"""
# initialize dictionary
scores = {}
# run a for loop for each sample
for index_candidate, candidate in enumerate(samples):
### START CODE HERE (REPLACE INSTANCES OF `None` WITH YOUR CODE) ###
# initialize overlap to 0.0
overlap = 0.0
# run a for loop for each sample
for index_sample, sample in enumerate(samples):
# skip if the candidate index is the same as the sample index
if index_candidate == index_sample:
continue
# get the overlap between candidate and sample using the similarity function
sample_overlap = similarity_fn(candidate,sample)
# add the sample overlap to the total overlap
overlap += sample_overlap
# get the score for the candidate by computing the average
score = overlap/index_sample
# save the score in the dictionary. use index as the key.
scores[index_candidate] = score
### END CODE HERE ###
return scores
# %%
average_overlap(jaccard_similarity, [[1, 2, 3], [1, 2, 4], [1, 2, 4, 5]], [0.4, 0.2, 0.5])
# %%
# BEGIN UNIT TEST
w1_unittest.test_average_overlap(average_overlap)
# END UNIT TEST
# %% [markdown]
# In practice, it is also common to see the weighted mean being used to calculate the overall score instead of just the arithmetic mean. We have implemented it below and you can use it in your experiements to see which one will give better results.
# %%
def weighted_avg_overlap(similarity_fn, samples, log_probs):
"""Returns the weighted mean of each candidate sentence in the samples
Args:
samples (list of lists): tokenized version of the translated sentences
log_probs (list of float): log probability of the translated sentences
Returns:
dict: scores of each sample
key: index of the sample
value: score of the sample
"""
# initialize dictionary
scores = {}
# run a for loop for each sample
for index_candidate, candidate in enumerate(samples):
# initialize overlap and weighted sum
overlap, weight_sum = 0.0, 0.0
# run a for loop for each sample
for index_sample, (sample, logp) in enumerate(zip(samples, log_probs)):
# skip if the candidate index is the same as the sample index
if index_candidate == index_sample:
continue
# convert log probability to linear scale
sample_p = float(np.exp(logp))
# update the weighted sum
weight_sum += sample_p
# get the unigram overlap between candidate and sample
sample_overlap = similarity_fn(candidate, sample)
# update the overlap
overlap += sample_p * sample_overlap
# get the score for the candidate
score = overlap / weight_sum
# save the score in the dictionary. use index as the key.
scores[index_candidate] = score
return scores
# %%
weighted_avg_overlap(jaccard_similarity, [[1, 2, 3], [1, 2, 4], [1, 2, 4, 5]], [0.4, 0.2, 0.5])
# %% [markdown]
# ### 4.2.4 Putting it all together
#
# We will now put everything together and develop the `mbr_decode()` function. Please use the helper functions you just developed to complete this. You will want to generate samples, get the score for each sample, get the highest score among all samples, then detokenize this sample to get the translated sentence.
#
# <a name="ex10"></a>
# ### Exercise 10
#
# **Instructions**: Implement the `mbr_overlap()` function.
# %%
# UNQ_C10
# GRADED FUNCTION
def mbr_decode(sentence, n_samples, score_fn, similarity_fn, NMTAttn=None, temperature=0.6, vocab_file=None, vocab_dir=None):
"""Returns the translated sentence using Minimum Bayes Risk decoding
Args:
sentence (str): sentence to translate.
n_samples (int): number of samples to generate
score_fn (function): function that generates the score for each sample
similarity_fn (function): function used to compute the overlap between a pair of samples
NMTAttn (tl.Serial): An LSTM sequence-to-sequence model with attention.
temperature (float): parameter for sampling ranging from 0.0 to 1.0.
0.0: same as argmax, always pick the most probable token
1.0: sampling from the distribution (can sometimes say random things)
vocab_file (str): filename of the vocabulary
vocab_dir (str): path to the vocabulary file
Returns:
str: the translated sentence
"""
### START CODE HERE (REPLACE INSTANCES OF `None` WITH YOUR CODE) ###
# generate samples
samples, log_probs = generate_samples(sentence, n_samples, NMTAttn, temperature, vocab_file, vocab_dir)
# use the scoring function to get a dictionary of scores
# pass in the relevant parameters as shown in the function definition of
# the mean methods you developed earlier
scores = weighted_avg_overlap(similarity_fn, samples, log_probs)
# find the key with the highest score
max_index = max(scores,key=scores.get)
# detokenize the token list associated with the max_index
translated_sentence = detokenize(samples[max_index], vocab_file, vocab_dir)
### END CODE HERE ###
return (translated_sentence, max_index, scores)
# %%
TEMPERATURE = 1.0
# put a custom string here
your_sentence = 'She speaks English and German.'
# %%
mbr_decode(your_sentence, 4, weighted_avg_overlap, jaccard_similarity, model, TEMPERATURE, vocab_file=VOCAB_FILE, vocab_dir=VOCAB_DIR)[0]
# %%
mbr_decode('Congratulations!', 4, average_overlap, rouge1_similarity, model, TEMPERATURE, vocab_file=VOCAB_FILE, vocab_dir=VOCAB_DIR)[0]
# %%
mbr_decode('You have completed the assignment!', 4, average_overlap, rouge1_similarity, model, TEMPERATURE, vocab_file=VOCAB_FILE, vocab_dir=VOCAB_DIR)[0]
# %% [markdown]
# **This unit test take a while to run. Please be patient**
# %%
# BEGIN UNIT TEST
w1_unittest.test_mbr_decode(mbr_decode, model)
# END UNIT TEST
# %% [markdown]
# #### Congratulations! Next week, you'll dive deeper into attention models and study the Transformer architecture. You will build another network but without the recurrent part. It will show that attention is all you need! It should be fun!
| 46.557746
| 1,258
|
584fcc663f96c8aa2ed029d8f406228a379b9ea5
|
py
|
python
|
lectures/Linear Regression.ipynb
|
johnmathews/quant1
|
['CC-BY-4.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# #Linear Regression
# By Evgenia "Jenny" Nitishinskaya and Delaney Granizo-Mackenzie with example algorithms by David Edwards
#
# Part of the Quantopian Lecture Series:
#
# * [www.quantopian.com/lectures](https://www.quantopian.com/lectures)
# * [github.com/quantopian/research_public](https://github.com/quantopian/research_public)
#
# Notebook released under the Creative Commons Attribution 4.0 License.
#
# ---
# Linear regression is a technique that measures the relationship between two variables. If we have an independent variable $X$, and a dependent outcome variable $Y$, linear regression allows us to determine which linear model $Y = \alpha + \beta X$ best explains the data. As an example, let's consider TSLA and SPY. We would like to know how TSLA varies as a function of how SPY varies, so we will take the daily returns of each and regress them against each other.
#
# Python's `statsmodels` library has a built-in linear fit function. Note that this will give a line of best fit; whether or not the relationship it shows is significant is for you to determine. The output will also have some statistics about the model, such as R-squared and the F value, which may help you quantify how good the fit actually is.
# Import libraries
import numpy as np
from statsmodels import regression
import statsmodels.api as sm
import matplotlib.pyplot as plt
import math
# First we'll define a function that performs linear regression and plots the results.
def linreg(X,Y):
# Running the linear regression
X = sm.add_constant(X)
model = regression.linear_model.OLS(Y, X).fit()
a = model.params[0]
b = model.params[1]
X = X[:, 1]
# Return summary of the regression and plot results
X2 = np.linspace(X.min(), X.max(), 100)
Y_hat = X2 * b + a
plt.scatter(X, Y, alpha=0.3) # Plot the raw data
plt.plot(X2, Y_hat, 'r', alpha=0.9); # Add the regression line, colored in red
plt.xlabel('X Value')
plt.ylabel('Y Value')
return model.summary()
# Now we'll get pricing data on TSLA and SPY and perform a regression.
# +
start = '2014-01-01'
end = '2015-01-01'
asset = get_pricing('TSLA', fields='price', start_date=start, end_date=end)
benchmark = get_pricing('SPY', fields='price', start_date=start, end_date=end)
# We have to take the percent changes to get to returns
# Get rid of the first (0th) element because it is NAN
r_a = asset.pct_change()[1:]
r_b = benchmark.pct_change()[1:]
linreg(r_b.values, r_a.values)
# -
# Each point on the above graph represents a day, with the x-coordinate being the return of SPY, and the y-coordinate being the return of TSLA. As we can see, the line of best fit tells us that for every 1% increased return we see from the SPY, we should see an extra 1.92% from TSLA. This is expressed by the parameter $\beta$, which is 1.9271 as estimated. Of course, for decresed return we will also see about double the loss in TSLA, so we haven't gained anything, we are just more volatile.
# ##Linear Regression vs. Correlation
#
# * Linear regression gives us a specific linear model, but is limited to cases of linear dependence.
# * Correlation is general to linear and non-linear dependencies, but doesn't give us an actual model.
# * Both are measures of covariance.
# * Linear regression can give us relationship between Y and many independent variables by making X multidimensional.
# ##Knowing Parameters vs. Estimates
#
# It is very important to keep in mind that all $\alpha$ and $\beta$ parameters estimated by linear regression are just that - estimates. You can never know the underlying true parameters unless you know the physical process producing the data. The paremeters you estimate today may not be the same as the same analysis done including tomorrow's data, and the underlying true parameters may be moving. As such it is very important when doing actual analysis to pay attention to the standard error of the parameter estimates. More material on the standard error will be presented in a later lecture. One way to get a sense of how stable your paremeter estimates are is to estimates them using a rolling window of data and see how much variance there is in the estimates.
#
# ##Ordinary Least Squares
#
# Regression works by optimizing the placement of the line of best fit (or plane in higher dimensions). It does so by defining how bad the fit is using an objective function. In ordinary least squares regression, a very common type and what we use here, the objective function is:
#
# $$\sum_{i=1}^n (Y_i - a - bX_i)^2$$
#
# That is, for each point on the line of best fit, compare it with the real point and take the square of the difference. This function will decrease as we get better paremeter estimates. Regression is a simple case of numerical optimzation that has a closed form solution and does not need any optimizer.
# ##Example case
# Now let's see what happens if we regress two purely random variables.
X = np.random.rand(100)
Y = np.random.rand(100)
linreg(X, Y)
# The above shows a fairly uniform cloud of points. It is important to note that even with 100 samples, the line has a visible slope due to random chance. This is why it is crucial that you use statistical tests and not visualizations to verify your results.
# Now let's make Y dependent on X plus some random noise.
# +
# Generate ys correlated with xs by adding normally-destributed errors
Y = X + 0.2*np.random.randn(100)
linreg(X,Y)
# -
# In a situation like the above, the line of best fit does indeed model the dependent variable Y quite well (with a high $R^2$ value).
# # Evaluating and reporting results
#
# The regression model relies on several assumptions:
# * The independent variable is not random.
# * The variance of the error term is constant across observations. This is important for evaluating the goodness of the fit.
# * The errors are not autocorrelated. The Durbin-Watson statistic detects this; if it is close to 2, there is no autocorrelation.
# * The errors are normally distributed. If this does not hold, we cannot use some of the statistics, such as the F-test.
#
# If we confirm that the necessary assumptions of the regression model are satisfied, we can safely use the statistics reported to analyze the fit. For example, the $R^2$ value tells us the fraction of the total variation of $Y$ that is explained by the model.
#
# When making a prediction based on the model, it's useful to report not only a single value but a confidence interval. The linear regression reports 95% confidence intervals for the regression parameters, and we can visualize what this means using the `seaborn` library, which plots the regression line and highlights the 95% (by default) confidence interval for the regression line:
# +
import seaborn
start = '2014-01-01'
end = '2015-01-01'
asset = get_pricing('TSLA', fields='price', start_date=start, end_date=end)
benchmark = get_pricing('SPY', fields='price', start_date=start, end_date=end)
# We have to take the percent changes to get to returns
# Get rid of the first (0th) element because it is NAN
r_a = asset.pct_change()[1:]
r_b = benchmark.pct_change()[1:]
seaborn.regplot(r_b.values, r_a.values);
# -
# We can also find the standard error of estimate, which measures the standard deviation of the error term $\epsilon$, by getting the `scale` parameter of the model returned by the regression and taking its square root. The formula for standard error of estimate is
# $$ s = \left( \frac{\sum_{i=1}^n \epsilon_i^2}{n-2} \right)^{1/2} $$
#
# This is useful because the variance of the error in our prediction $\hat{Y}$ given the value $X$ is
# $$ s_f^2 = s^2 \left( 1 + \frac{1}{n} + \frac{(X - \mu_X)^2}{(n-1)\sigma_X^2} \right) $$
#
# where $\mu_X$ is the mean of our observations of $X$ and $\sigma_X$ is the standard deviation. Then the 95% confidence interval for the prediction is $\hat{Y} \pm t_cs_f$, where $t_c$ is the critical value of the t-statistic for $n$ samples and a desired 95% confidence.
# ## Mathematical Background
#
# This is a very brief overview of linear regression. For more, please see:
# https://en.wikipedia.org/wiki/Linear_regression
# *This presentation is for informational purposes only and does not constitute an offer to sell, a solicitation to buy, or a recommendation for any security; nor does it constitute an offer to provide investment advisory or other services by Quantopian, Inc. ("Quantopian"). Nothing contained herein constitutes investment advice or offers any opinion with respect to the suitability of any security, and any views expressed herein should not be taken as advice to buy, sell, or hold any security or as an endorsement of any security or company. In preparing the information contained herein, Quantopian, Inc. has not taken into account the investment needs, objectives, and financial circumstances of any particular investor. Any views expressed and data illustrated herein were prepared based upon information, believed to be reliable, available to Quantopian, Inc. at the time of publication. Quantopian makes no guarantees as to their accuracy or completeness. All information is subject to change and may quickly become unreliable for various reasons, including changes in market conditions or economic circumstances.*
| 59.840764
| 1,125
|
e83efe26da1a068089497d20d8eae723eb42e9d8
|
py
|
python
|
predicting-car-prices-with-catboost.ipynb
|
meln-ds/regression-with-catboost
|
['Apache-2.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] toc=true
# <h1>Table of Contents<span class="tocSkip"></span></h1>
# <div class="toc"><ul class="toc-item"><li><span><a href="#Import-libraries" data-toc-modified-id="Import-libraries-1"><span class="toc-item-num">1 </span>Import libraries</a></span></li><li><span><a href="#Locally-defined-functions" data-toc-modified-id="Locally-defined-functions-2"><span class="toc-item-num">2 </span>Locally defined functions</a></span><ul class="toc-item"><li><span><a href="#Metrics" data-toc-modified-id="Metrics-2.1"><span class="toc-item-num">2.1 </span>Metrics</a></span></li><li><span><a href="#Display-functions" data-toc-modified-id="Display-functions-2.2"><span class="toc-item-num">2.2 </span>Display functions</a></span></li><li><span><a href="#Define-features" data-toc-modified-id="Define-features-2.3"><span class="toc-item-num">2.3 </span>Define features</a></span></li></ul></li><li><span><a href="#Global-options" data-toc-modified-id="Global-options-3"><span class="toc-item-num">3 </span>Global options</a></span></li><li><span><a href="#Load--data" data-toc-modified-id="Load--data-4"><span class="toc-item-num">4 </span>Load data</a></span><ul class="toc-item"><li><span><a href="#Training-data" data-toc-modified-id="Training-data-4.1"><span class="toc-item-num">4.1 </span>Training data</a></span></li><li><span><a href="#Out-of-sample-data-(to-predict)" data-toc-modified-id="Out-of-sample-data-(to-predict)-4.2"><span class="toc-item-num">4.2 </span>Out of sample data (to predict)</a></span></li></ul></li><li><span><a href="#Feature-exploration" data-toc-modified-id="Feature-exploration-5"><span class="toc-item-num">5 </span>Feature exploration</a></span><ul class="toc-item"><li><span><a href="#Categorical-features" data-toc-modified-id="Categorical-features-5.1"><span class="toc-item-num">5.1 </span>Categorical features</a></span></li><li><span><a href="#Numerical-features" data-toc-modified-id="Numerical-features-5.2"><span class="toc-item-num">5.2 </span>Numerical features</a></span></li><li><span><a href="#Pair-plot" data-toc-modified-id="Pair-plot-5.3"><span class="toc-item-num">5.3 </span>Pair plot</a></span></li></ul></li><li><span><a href="#Feature-generation" data-toc-modified-id="Feature-generation-6"><span class="toc-item-num">6 </span>Feature generation</a></span></li><li><span><a href="#Feature-selection" data-toc-modified-id="Feature-selection-7"><span class="toc-item-num">7 </span>Feature selection</a></span><ul class="toc-item"><li><span><a href="#Drop-features-(optional)" data-toc-modified-id="Drop-features-(optional)-7.1"><span class="toc-item-num">7.1 </span>Drop features (optional)</a></span></li></ul></li><li><span><a href="#ML-data-preparation" data-toc-modified-id="ML-data-preparation-8"><span class="toc-item-num">8 </span>ML data preparation</a></span><ul class="toc-item"><li><span><a href="#Categorical-feature-encoding" data-toc-modified-id="Categorical-feature-encoding-8.1"><span class="toc-item-num">8.1 </span>Categorical feature encoding</a></span></li><li><span><a href="#Train-test-split" data-toc-modified-id="Train-test-split-8.2"><span class="toc-item-num">8.2 </span>Train test split</a></span></li></ul></li><li><span><a href="#Machine-learning-model" data-toc-modified-id="Machine-learning-model-9"><span class="toc-item-num">9 </span>Machine learning model</a></span><ul class="toc-item"><li><span><a href="#Model-definition" data-toc-modified-id="Model-definition-9.1"><span class="toc-item-num">9.1 </span>Model definition</a></span></li><li><span><a href="#ML-model-training" data-toc-modified-id="ML-model-training-9.2"><span class="toc-item-num">9.2 </span>ML model training</a></span></li></ul></li><li><span><a href="#Model-evaluation" data-toc-modified-id="Model-evaluation-10"><span class="toc-item-num">10 </span>Model evaluation</a></span><ul class="toc-item"><li><span><a href="#Train,-test-predictions" data-toc-modified-id="Train,-test-predictions-10.1"><span class="toc-item-num">10.1 </span>Train, test predictions</a></span></li><li><span><a href="#Regression-coefficients/Feature-importance" data-toc-modified-id="Regression-coefficients/Feature-importance-10.2"><span class="toc-item-num">10.2 </span>Regression coefficients/Feature importance</a></span></li><li><span><a href="#Metrics" data-toc-modified-id="Metrics-10.3"><span class="toc-item-num">10.3 </span>Metrics</a></span></li><li><span><a href="#Model-Performance-plots" data-toc-modified-id="Model-Performance-plots-10.4"><span class="toc-item-num">10.4 </span>Model Performance plots</a></span></li><li><span><a href="#Optionally-retrain-on-the-whole-data-set" data-toc-modified-id="Optionally-retrain-on-the-whole-data-set-10.5"><span class="toc-item-num">10.5 </span>Optionally retrain on the whole data set</a></span></li></ul></li><li><span><a href="#Apply-model-to-OOS-data" data-toc-modified-id="Apply-model-to-OOS-data-11"><span class="toc-item-num">11 </span>Apply model to OOS data</a></span><ul class="toc-item"><li><span><a href="#Subset-to-relevant-columns" data-toc-modified-id="Subset-to-relevant-columns-11.1"><span class="toc-item-num">11.1 </span>Subset to relevant columns</a></span></li><li><span><a href="#Apply-categorical-encoding" data-toc-modified-id="Apply-categorical-encoding-11.2"><span class="toc-item-num">11.2 </span>Apply categorical encoding</a></span></li><li><span><a href="#Apply-model-and-produce-output" data-toc-modified-id="Apply-model-and-produce-output-11.3"><span class="toc-item-num">11.3 </span>Apply model and produce output</a></span></li></ul></li></ul></div>
# -
# # Import libraries
# # Locally defined functions
# +
# this may need to be installed separately with
# # !pip install category-encoders
import category_encoders as ce
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import MinMaxScaler
# python general
import pandas as pd
import numpy as np
from collections import OrderedDict
#scikit learn
import sklearn
from sklearn.base import clone
# model selection
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import KFold
from sklearn.feature_selection import RFE
# ML models
from sklearn import tree
from sklearn.linear_model import LinearRegression
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.ensemble import AdaBoostRegressor
from sklearn.linear_model import Lasso
from sklearn.linear_model import ElasticNet
from sklearn.neighbors import KNeighborsRegressor
import xgboost as xgb
from catboost import CatBoostRegressor
# error metrics
from sklearn.metrics import make_scorer
from sklearn.metrics import mean_absolute_error, mean_squared_error, median_absolute_error
from sklearn.model_selection import cross_val_score
# plotting and display
# %matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib as mpl
from IPython.display import display
pd.options.display.max_columns = None
# widgets and widgets based libraries
import ipywidgets as widgets
from ipywidgets import interact, interactive
# -
# ## Metrics
# +
def rmse(y_true, y_pred):
res = np.sqrt(((y_true - y_pred) ** 2).mean())
return res
def mape(y_true, y_pred):
y_val = np.maximum(np.array(y_true), 1e-8)
return (np.abs(y_true -y_pred)/y_val).mean()
# -
metrics_dict_res = OrderedDict([
('mean_absolute_error', mean_absolute_error),
('median_absolute_error', median_absolute_error),
('root_mean_squared_error', rmse),
('mean abs perc error', mape)
])
def regression_metrics_yin(y_train, y_train_pred, y_test, y_test_pred,
metrics_dict, format_digits=None):
df_results = pd.DataFrame()
for metric, v in metrics_dict.items():
df_results.at[metric, 'train'] = v(y_train, y_train_pred)
df_results.at[metric, 'test'] = v(y_test, y_test_pred)
if format_digits is not None:
df_results = df_results.applymap(('{:,.%df}' % format_digits).format)
return df_results
# ## Display functions
# +
def describe_col(df, col):
display(df[col].describe())
def val_count(df, col):
display(df[col].value_counts())
def show_values(df, col):
print("Number of unique values:", len(df[col].unique()))
return display(df[col].value_counts(dropna=False))
# -
def plot_distribution(df, col, bins=100, figsize=None, xlim=None, font=None, histtype='step'):
if font is not None:
mpl.rc('font', **font)
if figsize is not None:
plt.figure(figsize=figsize)
else:
plt.figure(figsize=(10, 6))
dev = df[col]
dev.plot(kind='hist', bins=bins, density=True, histtype=histtype, color='b', lw=2,alpha=0.99)
print('mean:', dev.mean())
print('median:', dev.median())
if xlim is not None:
plt.xlim(xlim)
return plt.gca()
def plot_feature_importances(model, feature_names=None, n_features=20):
if feature_names is None:
feature_names = range(n_features)
importances = model.feature_importances_
importances_rescaled = 100 * (importances / importances.max())
xlabel = "Relative importance"
sorted_idx = np.argsort(-importances_rescaled)
names_sorted = [feature_names[k] for k in sorted_idx]
importances_sorted = [importances_rescaled[k] for k in sorted_idx]
pos = np.arange(n_features) + 0.5
plt.barh(pos, importances_sorted[:n_features], align='center')
plt.yticks(pos, names_sorted[:n_features])
plt.xlabel(xlabel)
plt.title("Feature importances")
return plt.gca()
def plot_act_vs_pred(y_act, y_pred, scale=1, act_label='actual', pred_label='predicted', figsize=None, xlim=None,
ylim=None, font=None):
if font is not None:
mpl.rc('font', **font)
if figsize is not None:
plt.figure(figsize=figsize)
else:
plt.figure(figsize=(10, 6))
plt.scatter(y_act/scale, y_pred/scale)
x = np.linspace(0, y_act.max()/scale, 10)
plt.plot(x, x)
plt.xlabel(act_label)
plt.ylabel(pred_label)
if xlim is not None:
plt.xlim(xlim)
else:
plt.xlim([0, 1e2])
if ylim is not None:
plt.ylim(ylim)
else:
plt.ylim([0, 1e2])
return plt.gca()
# +
def compute_perc_deviation(y_act, y_pred, absolute=False):
dev = (y_pred - y_act)/y_act * 100
if absolute:
dev = np.abs(dev)
dev.name = 'abs % error'
else:
dev.name = '% error'
return dev
def plot_dev_distribution(y_act, y_pred, absolute=False, bins=100, figsize=None, xlim=None, font=None):
if font is not None:
mpl.rc('font', **font)
if figsize is not None:
plt.figure(figsize=figsize)
else:
plt.figure(figsize=(10, 6))
dev = compute_perc_deviation(y_act, y_pred, absolute=absolute)
dev.plot(kind='hist', bins=bins, density=True)
print('mean % dev:', dev.mean())
print('median % dev:', dev.median())
# plt.vlines(dev.mean(), 0, 0.05)
plt.title('Distribution of errors')
plt.xlabel('% deviation')
if xlim is not None:
plt.xlim(xlim)
else:
plt.xlim([-1e2, 1e2])
return plt.gca()
# -
# ## Define features
# +
categorical_features = [
'Body_Type',
'Driven_Wheels',
'Global_Sales_Sub-Segment',
'Brand',
'Nameplate',
'Transmission',
'Turbo',
'Fuel_Type',
'PropSysDesign',
'Plugin',
'Registration_Type',
'country_name'
]
numeric_features = [
'Generation_Year',
'Length',
'Height',
'Width',
'Engine_KW',
'No_of_Gears',
'Curb_Weight',
'CO2',
'Fuel_cons_combined',
'year'
]
all_numeric_features = list(numeric_features)
all_categorical_features = list(categorical_features)
target = [
'Price_USD'
]
target_name = 'Price_USD'
# -
# # Global options
# +
#ml_model_type = 'Linear Regression'
#ml_model_type = 'Decision Tree'
#ml_model_type = 'Random Forest'
#ml_model_type = 'Gradient Boosting Regressor'
#ml_model_type = 'AdaBoost'
#ml_model_type = 'XGBoost'
ml_model_type = 'CatBoost'
regression_metric = 'mean abs perc error'
do_grid_search_cv = False
scoring_greater_is_better = False # THIS NEEDS TO BE SET CORRECTLY FOR CV GRID SEARCH
do_retrain_total = True
write_predictions_file = True
# relative size of test set
test_size = 0.2
random_state = 33
# -
# # Load data
#
# ## Training data
df = pd.read_csv('/kaggle/input/ihsmarkit-hackathon-june2020/train_data.csv',index_col='vehicle_id')
df['date'] = pd.to_datetime(df['date'])
#g = df['Brand'].value_counts()
#df['Brand'] = np.where(df['Brand'].isin(g.index[g >= 200]), df['Brand'], 'Other')
# basic commands on a dataframe
# df.info()
df.head(5)
# df.shape
# df.head()
# df.tail()
df['country_name'].value_counts()
df.groupby(['year', 'country_name'])['date'].count()
# ## Out of sample data (to predict)
df_oos = pd.read_csv('/kaggle/input/ihsmarkit-hackathon-june2020/oos_data.csv', index_col='vehicle_id')
df_oos['date'] = pd.to_datetime(df_oos['date'])
df_oos['year'] = df_oos['date'].map(lambda d: d.year)
#g_oos = df_oos['Brand'].value_counts()
#df_oos['Brand'] = np.where(df_oos['Brand'].isin(g.index[g >= 200]), df_oos['Brand'], 'Other')
# df_oos.shape
df_oos.head()
df_oos.groupby(['year', 'country_name'])['date'].count()
# # Feature exploration
# ## Categorical features
# unique values, categorical variables
for col in all_categorical_features:
print(col, len(df[col].unique()))
interactive(lambda col: show_values(df, col), col=all_categorical_features)
# ## Numerical features
# summary statistics
df[numeric_features + target].describe()
# +
figsize = (16,12)
sns.set(style='whitegrid', font_scale=2)
bins = 1000
bins = 40
#xlim = [0,100000]
xlim = None
price_mask = df['Price_USD'] < 100000
interactive(lambda col: plot_distribution(df[price_mask], col, bins=bins, xlim=xlim), col=sorted(all_numeric_features + target))
#interactive(lambda col: plot_distribution(df, col, bins=bins, xlim=xlim), col=sorted(all_numeric_features + target))
# -
# ## Pair plot
# this is quite slow
sns.set(style='whitegrid', font_scale=1)
# sns.pairplot(df[numeric_features[:6] + target].iloc[:10000])
#sns.pairplot(df[['Engine_KW'] + target].iloc[:10000])
price_mask = df['Price_USD'] < 100000
df_temp = df[price_mask].copy()
sns.pairplot(df_temp[['Engine_KW'] + target])
# # Feature generation
additional_numeric_features = []
# # Feature selection
#
# You can read about feature selection here
# https://scikit-learn.org/stable/modules/feature_selection.html#
# The dataset is fairly clean with a moderate number of features available. For a start, I did not have to worry too much about missing values in the dataset. A few things I tried out:
# - I tried dropping Nameplate because it has too many categories and when combined with one hot encoding, could make your dataset really sparse. Dropping Nameplate certainly results in superior results when paired with Gradient Boosting algorithms, though in the end I add the feature back in since I decided to use Catboost, which takes care of categorical features really well.
# - I tried setting year and generation year to be categorical instead of numerical, as they should be. However, I did not notice any improvement.
# - There are many highly correlated features which should/could be dropped to reduce the number of dimensions. I have not tried going down the PCA path but would expect an improvement at least for Linear Regression.
# ## Drop features (optional)
# +
features_drop = []
if ml_model_type == 'Linear Regression':
features_drop = categorical_features + numeric_features
features_to_use = ['Engine_KW']
# features_to_use = ['country_name', 'Engine_KW']
for feature in features_to_use:
features_drop.remove(feature)
#else:
#features_drop = categorical_features + numeric_features
#features_to_use = ['Brand', 'country_name', 'Engine_KW']
#features_to_use = ['country_name', 'Engine_KW']
#for feature in features_to_use:
#features_drop.remove(feature)
#features_drop = ['Nameplate']
categorical_features = list(filter(lambda f: f not in features_drop, categorical_features))
numeric_features = list(filter(lambda f: f not in features_drop, numeric_features))
# -
features = categorical_features + numeric_features + additional_numeric_features
model_columns = features + [target_name]
len(model_columns)
# # ML data preparation
#dataframe for further processing
df_proc = df[model_columns].copy()
df_proc.shape
# ## Categorical feature encoding
# I did not use one hot encoding, and instead let Catboost take care of the categorical variables.
# One-hot encoding
#encoder = ce.OneHotEncoder(cols=categorical_features, handle_unknown='value',
#use_cat_names=True)
#encoder.fit(df_proc)
#df_comb_ext = encoder.transform(df_proc)
#features_ext = list(df_comb_ext.columns)
#features_ext.remove(target_name)
# +
#del df_proc
#df_comb_ext.head()
# +
# df_comb_ext.memory_usage(deep=True).sum()/1e9
#features_model
#df_comb_ext.shape
# -
# ## Train test split
#X_train, X_test, y_train, y_test = train_test_split(df_comb_ext[features_ext], df_comb_ext[target_name],
#test_size=test_size, random_state=random_state)
X_train, X_test, y_train, y_test = train_test_split(df_proc[features], df_proc[target_name],
test_size=test_size, random_state=random_state)
print(X_train.shape)
print(X_test.shape)
# Scaling did not seem to improve any performance, which I think could be because I was using a tree-based model, where scaling isn't exactly a deal breaker.
# +
#scaler = MinMaxScaler()
#X_train_scaled = scaler.fit_transform(X_train)
#X_test_scaled = scaler.fit_transform(X_test)
# -
# # Machine learning model
#
# Supervised learning
#
# https://scikit-learn.org/stable/supervised_learning.html
#
# Ensemble methods in scikit learn
#
# https://scikit-learn.org/stable/modules/ensemble.html
#
#
# Decision trees
#
# https://scikit-learn.org/stable/modules/tree.html
#
# Before deciding on a model to use, I try running the dataset through a handful of out of the box sklearn algorithms. Uncomment this cell if you want to check out the results. Gradient Boosting gave me the best result i.e. lowest MSE.
# +
#pipelines = []
#pipelines.append(('ScaledLR', Pipeline([('Scaler', StandardScaler()),('LR',LinearRegression())])))
#pipelines.append(('ScaledEN', Pipeline([('Scaler', StandardScaler()),('EN', ElasticNet())])))
#pipelines.append(('ScaledKNN', Pipeline([('Scaler', StandardScaler()),('KNN', KNeighborsRegressor())])))
#pipelines.append(('ScaledCART', Pipeline([('Scaler', StandardScaler()),('CART', DecisionTreeRegressor())])))
#pipelines.append(('ScaledGBM', Pipeline([('Scaler', StandardScaler()),('GBM', GradientBoostingRegressor())])))
#results = []
#names = []
#for name, model in pipelines:
#cv_results = cross_val_score(model, X_train, y_train, cv=10, scoring='neg_mean_squared_error')
#results.append(cv_results)
#names.append(name)
#msg = "%s: %f (%f)" % (name, cv_results.mean(), cv_results.std())
#print(msg)
# -
# ## Model definition
# +
if ml_model_type == 'Linear Regression':
model_hyper_parameters_dict = OrderedDict(fit_intercept=True, normalize=False)
regressor = LinearRegression(**model_hyper_parameters_dict)
if ml_model_type == 'Decision Tree':
model_hyper_parameters_dict = OrderedDict(max_depth=3, random_state=random_state)
regressor = DecisionTreeRegressor(**model_hyper_parameters_dict)
if ml_model_type == 'Random Forest':
model_hyper_parameters_dict = OrderedDict(n_estimators=10,
max_depth=4,
min_samples_split=2,
max_features='sqrt',
min_samples_leaf=1,
random_state=random_state,
n_jobs=4)
regressor = RandomForestRegressor(**model_hyper_parameters_dict)
if ml_model_type == 'Gradient Boosting Regressor':
model_hyper_parameters_dict = OrderedDict(learning_rate=0.1,
max_depth=6,
subsample=0.8,
max_features=0.2,
n_estimators=200,
random_state=random_state)
regressor = GradientBoostingRegressor(**model_hyper_parameters_dict)
if ml_model_type == 'AdaBoost':
model_hyper_parameters_dict = OrderedDict(n_estimators=180,
random_state=random_state)
regressor = AdaBoostRegressor(**model_hyper_parameters_dict)
if ml_model_type == 'XGBoost':
model_hyper_parameters_dict = OrderedDict(learning_rate=0.01,
colsample_bytree=0.3,
max_depth=3,
subsample=0.8,
n_estimators=1000,
seed=random_state)
regressor = xgb.XGBRegressor(**model_hyper_parameters_dict)
if ml_model_type == 'CatBoost':
model_hyper_parameters_dict = OrderedDict(iterations=4000,
early_stopping_rounds=50,
learning_rate=0.05,
depth=12,
one_hot_max_size=40,
colsample_bylevel=0.5,
bagging_temperature=12,
random_strength=0.7,
reg_lambda=1.0,
eval_metric='RMSE',
logging_level='Silent',
random_seed = random_state)
regressor = CatBoostRegressor(**model_hyper_parameters_dict)
base_regressor = clone(regressor)
if do_grid_search_cv:
scoring = make_scorer(metrics_dict_res[regression_metric], greater_is_better=scoring_greater_is_better)
if ml_model_type == 'Random Forest':
grid_parameters = [{'n_estimators': [10], 'max_depth': [3, 5, 10],
'min_samples_split': [2,4], 'min_samples_leaf': [1]} ]
if ml_model_type == 'XGBoost':
grid_parameters = [{'colsample_bytree':[0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 1.0]}]
if ml_model_type == 'CatBoost':
grid_parameters = [{'learning_rate': [0.1, 0.3, 0.5, 0.8]}]
n_splits = 10
n_jobs = 4
cv_regressor = GridSearchCV(regressor, grid_parameters, cv=n_splits, scoring=scoring, return_train_score=True,
refit=True, n_jobs=n_jobs)
# +
# Use XGBoost API Learner. Comment the whole cell out if you want to use other models
#DM_train = xgb.DMatrix(data=X_train,label=y_train)
#DM_test = xgb.DMatrix(data=X_test,label=y_test)
#params = {"booster":"gblinear", "objective":"reg:linear"}
#xg_reg = xgb.train(params = params, dtrain=DM_train, num_boost_round=5)
# -
# ## ML model training
# +
if do_grid_search_cv:
cv_regressor.fit(X_train, y_train, cat_features=categorical_features)
regressor_best = cv_regressor.best_estimator_
model_hyper_parameters_dict = cv_regressor.best_params_
train_scores = cv_regressor.cv_results_['mean_train_score']
test_scores = cv_regressor.cv_results_['mean_test_score']
test_scores_std = cv_regressor.cv_results_['std_test_score']
cv_results = cv_regressor.cv_results_
elif ml_model_type == 'CatBoost':
regressor.fit(X_train, y_train, cat_features=categorical_features)
else:
regressor.fit(X_train, y_train)
# -
if do_grid_search_cv:
# print(cv_results)
print(model_hyper_parameters_dict)
plt.plot(-train_scores, label='train')
plt.plot(-test_scores, label='test')
plt.xlabel('Parameter set #')
plt.legend()
regressor = regressor_best
# # Model evaluation
# ## Train, test predictions
y_train_pred = regressor.predict(X_train)
y_test_pred = regressor.predict(X_test)
#y_train_pred = xg_reg.predict(DM_train)
#y_test_pred = xg_reg.predict(DM_test)
# ## Regression coefficients/Feature importance
if ml_model_type == 'Linear Regression':
df_reg_coef = (pd.DataFrame(zip(['intercept'] + list(X_train.columns),
[regressor.intercept_] + list(regressor.coef_)))
.rename({0: 'feature', 1: 'coefficient value'}, axis=1))
display(df_reg_coef)
if hasattr(regressor, 'feature_importances_'):
sns.set(style='whitegrid', font_scale=1.5)
plt.figure(figsize=(12,10))
plot_feature_importances(regressor, features_ext, n_features=np.minimum(20, X_train.shape[1]))
# ## Metrics
# +
df_regression_metrics = regression_metrics_yin(y_train, y_train_pred, y_test, y_test_pred,
metrics_dict_res, format_digits=3)
df_output = df_regression_metrics.copy()
df_output.loc['Counts','train'] = len(y_train)
df_output.loc['Counts','test'] = len(y_test)
df_output
# -
# ## Model Performance plots
figsize = (16,10)
xlim = [0, 250]
font={'size': 20}
sns.set(style='whitegrid', font_scale=2.5)
act_label = 'actual price [k$]'
pred_label='predicted price [k$]'
plot_act_vs_pred(y_test, y_test_pred, scale=1000, act_label=act_label, pred_label=pred_label,
figsize=figsize, xlim=xlim, ylim=xlim, font=font)
print()
# +
figsize = (14,8)
xlim = [0, 100]
#xlim = [-100, 100]
# xlim = [-50, 50]
#xlim = [-20, 20]
font={'size': 20}
sns.set(style='whitegrid', font_scale=1.5)
p_error = (y_test_pred - y_test)/y_test *100
df_p_error = pd.DataFrame(p_error.values, columns=['percent_error'])
#display(df_p_error['percent_error'].describe().to_frame())
bins=1000
bins=500
#bins=100
absolute = True
#absolute = False
plot_dev_distribution(y_test, y_test_pred, absolute=absolute, figsize=figsize,
xlim=xlim, bins=bins, font=font)
print()
# -
# ## Optionally retrain on the whole data set
if do_retrain_total:
cv_opt_model = clone(base_regressor.set_params(**model_hyper_parameters_dict))
# train on complete data set
#X_train_full = df_comb_ext[features_ext].copy()
#y_train_full = df_comb_ext[target_name].values
X_train_full = df_proc[features].copy()
y_train_full = df_proc[target_name].values
#cv_opt_model.fit(X_train_full, y_train_full)
cv_opt_model.fit(X_train_full, y_train_full, cat_features=categorical_features)
regressor = cv_opt_model
# # Apply model to OOS data
# +
# df_oos.head()
# -
# ## Subset to relevant columns
df_proc_oos = df_oos[model_columns[:-1]].copy()
df_proc_oos[target_name] = 1
df_proc_oos
df_proc_oos.drop(target_name, axis=1, inplace=True)
# ## Apply categorical encoding
# Again, not needed as I eventually go with Catboost.
# +
#df_comb_ext_oos = encoder.transform(df_proc_oos)
# +
#df_comb_ext_oos.drop(target_name, axis=1, inplace=True)
#df_comb_ext_oos = scaler.fit_transform(df_comb_ext_oos)
# -
# ## Apply model and produce output
#y_oos_pred = regressor.predict(df_comb_ext_oos)
y_oos_pred = regressor.predict(df_proc_oos)
id_col = 'vehicle_id'
df_out = (pd.DataFrame(y_oos_pred, columns=[target_name], index=df_proc_oos.index)
.reset_index()
.rename({'index': id_col}, axis=1))
df_out.head()
df_out.shape
if write_predictions_file:
df_out.to_csv('submission.csv', index=False)
# ## Conclusion
# All in all, I was very impressed with CatBoost's performance. I did not have a chance to try out LightGBM, but comparing to XGBoost and Stochastic Gradient Boosting, CatBoost gave superior results with pretty consistent results between training and testing data, and performance is comparable (albeit I did not track closely).
#
# Next step would be to play around with the hyperparameters tuning -- With the time constraints, I did not have time to try to tinker all the parameters. My three cents:
# - CatBoost deals with categorical features pretty well and I encourage you to make use of that. The one_hot_max_size parameter also allows you to define what is the maximum number of categories. Too many categories will decrease performance of the algorithm.
# - Strike a balance between iterations and learning rate. My suggestion would be to start with a high number of iterations (so you could make use of early stopping) and small learning rate, and then slowly increase learning rate while reducing iterations.
| 38.337302
| 5,856
|
007ffec5f324680c03d36b3bbda088c75d04eb45
|
py
|
python
|
1_mnist.ipynb
|
gmshashank/Pytorch_Lightning
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + colab={"base_uri": "https://localhost:8080/"} id="tYTazKuZVxaw" outputId="5dec9fb0-426e-4801-8546-692923e22ab2"
# !pip install pytorch-lightning
# + id="f0KbdUK_hSxK"
import os
import torch
import torch.nn as nn
from torch.utils.data import DataLoader, random_split
from torchvision import datasets, transforms
# + id="7eg6VIPfhW47"
class MNISTClassifier(nn.Module):
def __init__(self):
super(MNISTClassifier, self).__init__()
# MNIST images are of dimensions(1,28,28)(channel,width,height)
self.layer_1 = nn.Linear(in_features=28 * 28, out_features=128)
self.layer_2 = nn.Linear(in_features=128, out_features=256)
self.layer_3 = nn.Linear(in_features=256, out_features=10)
def forward(self, x):
batch_size, channels, width, height = x.size()
# (b,1,28,28)->(b,1*128*128)
x = x.view(batch_size, -1)
# layer 1
x = self.layer_1(x)
x = torch.relu(x)
# layer 2
x = self.layer_2(x)
x = torch.relu(x)
# layer 3
x = self.layer_3(x)
# probabaility distribution over labels
x = torch.log_softmax(x, dim=1)
return x
# + id="RdO6jyI0hauv"
# Transforms
transform = transforms.Compose(
[transforms.ToTensor(), transforms.Normalize((0.1307), (0.3081))]
)
# + colab={"base_uri": "https://localhost:8080/", "height": 376, "referenced_widgets": ["6fe3e3767e644a4b835828fd38694807", "90cb2a28277243fabf31bf3acc155165", "c5a25a2d09604db49220bae979180dd4", "19e1227d9b5f40659f011e51c3aa0386", "eefb97e928834f1693cc2182349a983a", "2332d1e88b1a4bf4adc4ab08048a53fb", "d7d2c19d15314c7fb61e0148ec69feb6", "e6d743849a3a432e8cac4223f0812c74", "26f055f63f0c448582578fa9ae4ec3f9", "b01bb85707f14d529e4fb7d6eb355945", "f1421288400d4980affd07279f287a9c", "b9ebc85465d0442393c348135bc5d883", "5b711dd881b64fe7bcbac020fd39ab88", "c1f47e70dc9747a0b95c15b07b7f3aab", "89001cd83c6144f6bd33de7834b93bf9", "0a2d8e1ac6d948469ca62dfa59deffdf", "f0268a9111804c8a84edbe09b26c72ce", "c72409f3313b4faca3348bb1ecbfc260", "dc0f90e79d5644859331f0ea16cbb0c1", "f91d1b0fcb7a428a9da5378ae92bb1c3", "75d4bdb2eb134273b21857b3ef33b07a", "f172ddc9c56640cead15783dc80fe947", "4a902e376acf49e5aea50492f7ee0407", "8b1496c78a094dff94630b34eb2d8207", "351ec23594c8486a9bfe102641ed2a4a", "41ac6bcc9d1948358230074db9043e87", "a2073e5f8cf74e39990155d47d1e5252", "7302106525844af29f10e0d7d49f7be7", "82b270b3d4754bd3b796969a0cbf8a4e", "c320ad0736fd467786efb62d5e602e4d", "6674c29b5e9a460eb0a300a0f1c35e6a", "13f1f686420446c19dd0482132c33f4a"]} id="1Jg5XzgwheuS" outputId="73bcffab-cd8d-4395-fc1a-895a7302cf49"
# Training , Validation Data
mnist_train = datasets.MNIST(root=os.getcwd(), train=True, download=True,transform=transform)
mnist_train, mnist_val = random_split(dataset=mnist_train, lengths=[55000, 5000])
# + id="EUnQsCNihgp-"
# Test Data
mnist_test = datasets.MNIST(root=os.getcwd(), train=False, download=True,transform=transform)
# + id="-A7KZ-qGhlWy"
# DataLoaders
mnist_train = DataLoader(dataset=mnist_train, batch_size=64)
mnist_val = DataLoader(dataset=mnist_val, batch_size=64)
mnist_test = DataLoader(dataset=mnist_test, batch_size=64)
# + id="1gIJlKRjhnBR"
# Optimizer
pytorch_model = MNISTClassifier()
optimizer = torch.optim.Adam(params=pytorch_model.parameters(), lr=1e-3)
# + colab={"base_uri": "https://localhost:8080/"} id="Ju5S_FJwhpGd" outputId="971373ba-210e-4e77-a2a9-bfec874fe4ad"
# Loss
def cross_entropy_loss(logits, labels):
return nn.functional.nll_loss(logits, labels)
# + colab={"base_uri": "https://localhost:8080/"} id="r2c3OXufhqul" outputId="082a489a-15bf-4971-e14a-8e30ffcccf78"
# Training Loop
num_epochs = 10
for epoch in range(num_epochs):
print("Epoch: ", epoch)
# Training
for train_batch in mnist_train:
x, y = train_batch
logits = pytorch_model(x)
loss = cross_entropy_loss(logits, y)
print("train loss: ", loss.item())
loss.backward()
optimizer.step()
optimizer.zero_grad()
# Validation
with torch.no_grad():
val_loss = []
for val_batch in mnist_val:
x, y = val_batch
logits = pytorch_model(x)
val_loss.append(cross_entropy_loss(logits, y).item())
val_loss = torch.mean(torch.tensor(val_loss))
print("val_loss: ", val_loss.item())
| 37.720339
| 1,306
|
1aa5c94e95eac2d1f37cca8170f5493dc8c9a159
|
py
|
python
|
Woche 5/2 Inhaltsstoffe Kann Spuren von Intelligenz enthalten.ipynb
|
manfred-l-stark/AMALEA
|
['CC0-1.0', 'CC-BY-4.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# [Nur Colab] Diese Zellen müssen nur auf *Google Colab* ausgeführt werden und installieren Packete und Daten
# !wget -q https://raw.githubusercontent.com/KI-Campus/AMALEA/master/requirements.txt && pip install --quiet -r requirements.txt
# !wget --quiet "https://github.com/KI-Campus/AMALEA/releases/download/data/data.zip" && unzip -q data.zip
# # Inhaltsstoffe: Kann Spuren von Intelligenz enthalten
# ## Faltungsoperation
#
# Wie der Name schon sagt, macht die Faltungsschicht (engl. convolutional layer) Gebrauch von Faltungen. Als kurze Zusammenfassung fasst dieses kleine Widget die Faltung von zwei rechteckigen Funktionen zusammen. Die Faltungsoperation ist durch die folgende Gleichung im kontinuierlichen Bereich gegeben:
#
# \begin{equation*}
# x(t) \ast y(t) = \langle x(t - \tau), y^{\ast}(\tau) \rangle_{\tau} = \int_{-\infty}^{+\infty} x(t -\tau) y(\tau) d \tau
# \end{equation*}
from ipywidgets import interact, interactive, fixed, interact_manual
import numpy as np
from PIL import Image
import matplotlib.pyplot as plt
from scipy import signal
figure_inches = 10
def convolution(tau:tuple, width1:tuple, width2:tuple):
x1 = np.linspace(-3.5, 3.5, num = 1000)
dX = x1[1] - x1[0]
rect1 = np.where(abs(x1) <= width1/2 , 1, 0)
rect2 = np.where(abs(x1- tau)<= width2/2 , 1, 0)
# Convolution of rect1 and rect2
conv = np.convolve(rect1, rect2, 'same') * dX
# Plotting
plt.figure(1, figsize=(16.5,3.5))
plt.plot(x1, rect1, 'b', label = '$rect_{1}$(t)')
plt.plot(x1, rect2, 'r', label = '$rect_{2}$(t- $\\tau$)')
x_gr = x1 - tau
if tau <=0:
index = np.where((np.absolute(x_gr)-np.absolute(tau))<=0.004)
index = index[0][0]
else:
index = np.where(np.absolute(x_gr-tau)<=0.004)
if not index[0].size > 0:
index = [[999]]
index = index[0][0]
plt.plot(x_gr[:index] , conv[:index], 'g', label = '$rect_{1}$ $\\ast$ $rect_{2}$')
plt.axvline(x = tau, color= 'r', dashes = [6,2])
plt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0., prop={'size':15})
plt.ylim(0,np.maximum(np.max(conv),np.max(rect1))+0.1)
plt.xlim(-2.5, 2.5)
plt.grid()
plt.show()
# Interactive Plot
interactive_plot = interactive(convolution, tau = (-2.0, +2.0, 0.25),
width1 = (0.25, 1.75, 0.25),
width2 = (0.25, 1.75, 0.25))
output = interactive_plot.children[-1]
interactive_plot
# Natürlich erfolgt die Berechnung der Faltung nicht im kontinuierlichen Bereich. Daher verwendet numpy die folgende Formel für diskrete Faltung im 1-dimensionalen Raum:
#
# \begin{equation*}
# x_n \ast y_n = \sum_{i = -\infty}^{\infty} x_i \ y_{n-i} = \sum_{i = -\infty}^{\infty} y_{i} \ x_{n-i}
# \end{equation*}
# ### Filter und Faltung in der Bildverarbeitung
#
# Die Faltungsoperation kann auch in zwei Dimensionen angegeben werden:
#
# \begin{equation*}
# x_{mn} \ast \ast \ y_{mn} = \sum_{i = -\infty}^{\infty} \sum_{j = -\infty}^{\infty} x_{ij} \ y_{m-i, n-j} = \sum_{i = -\infty}^{\infty} \sum_{j = -\infty}^{\infty} y_{ij} \ x_{m-i, n-j}
# \end{equation*}
#
# In Bezug auf die Faltung von Bildern im diskreten Bereich ändern sich die Grenzen zu endlichen Werten, die die Größe der Bildform entsprechen.
# Diese Operation kann dann wie folgt visualisiert werden:
#
#
# <img src="images/Faltung1.png" alt="Drawing" style="width: 600px;"/>
# <img src="images/Faltung2.png" alt="Drawing" style="width: 600px;"/>
# <img src="images/Faltung3.png" alt="Drawing" style="width: 600px;"/>
# <img src="images/Faltung4.png" alt="Drawing" style="width: 600px;"/>
#
#
# Beachten Sie, dass die Faltungsschicht den Filter nicht spiegelt, wie in der konventionellen Signalverarbeitung. Von nun an werden Bilder oder räumliche Informationen in 3 Dimensionen definiert durch:
#
# - Height $H$
# - Weight $W$
# - Depth $d$ (channels)
#
# Die diskrete Faltung reduziert die Bilddimensionen wie oben beschrieben. Die folgende Gleichung beschreibt die Reduktion selbst, wobei $K$ die `Kernel`- oder Filterdimensionen, $P$ zusätzliche Werte (meist zur Erhaltung der Ausgangsdimensionen, genannt `Padding`) und $S$ den `Stride` bezeichnet. Es gibt verschiedene Padding-Techniken, z.B. das Hinzufügen von Nullen (Zero Padding). Wenn der Stride größer als 1 ist, überspringt der Kernel bei der Faltung dazwischen liegende Werte.
#
# \begin{align}
# W_{i+1} = \dfrac{(W_{i}-K_{x}+2*P_{x})}{S_x}+1 \\
# H_{i+1} = \dfrac{(H_{i}-K_{y}+2*P_{y})}{S_y}+1
# \end{align}
#
#
# Zunächst beginnen wir mit einem **grauen Bild** namens ascent, wobei W gleich der Breite des Bildes und H gleich der Höhe des Bildes ist. Da dieses Bild ein Graustufenbild ist, ist die Tiefe $d=1$.
# <div class="alert alert-block alert-success">
# <b>Aufgabe 5.2.1:</b>
# Implementieren Sie die Funktion <code>conv</code> welche ein gegebenes Bild <code>image_data</code> mit einem gegebenenen Filter <code>filter_kern</code> filtert. Nehmen Sie an:
#
# * Das Bild liegt entsprechend dem Beispiel (in der folgenden Zelle) als eine Liste von Listen vor
# * Die Tiefe des Bildes ist 1
# </div>
def conv(image_data:list, filter_kern:list)->list:
# STUDENT CODE HERE
#STUDENT CODE until HERE
test_input_data = [[0,0,0,0,0], [0,1,1,1,0], [0,0,2,0,0], [0,3,3,3,0], [0,0,0,0,0], [0,0,0,0,0]]
test_filter = [[0,0], [-1, 1]]
test_result = [[1,0,0,-1],[0,2,-2,0],[3,0,0,-3], [0,0,0,0], [0,0,0,0]]
found = conv(test_input_data, test_filter)
# Die folgende Zeile erzeugt einen Fehler, wenn die Ausgabe der Methode nicht mit der erwarteten übereinstimmt
assert found == test_result
# ### Filtertypen
#
# Bevor wir nun in Richtung praktische Anwendung gehen, schauen wir uns grundlegende Filter an. Außerdem werden wir uns die Effekte der Filter anschauen - hierzu verwenden wir das folgende Bild:
# +
def read_image_as_array(image_path:str, new_size: tuple) -> np.array:
img = Image.open(image_path).convert('L')
img = img.resize(new_size,Image.ANTIALIAS)
return np.array(img)
lama = Image.open('images/lama.png').convert('L')
lama = lama.resize((500,500),Image.ANTIALIAS)
plt.figure(figsize=(figure_inches, figure_inches))
plt.imshow(lama, cmap='gray')
data = np.array(lama)
# -
# #### Identitätsfilter
#
# Der erste Filter entspricht der Identität, d.h. der Wert eines Pixel wird auf genau diesen abgebildet. Um dies zu erreichen wird ein quadratischer Filterkernel benötigt, dessen Größe ungerade ist. Außerdem ist der mittlere Eintrag 1 und alle anderen 0. Ein $3\times 3$-Filterkernel hat somit die Form:
#
# $\left\lbrack\begin{array}{ccc} 0&0&0\\ 0&1&0\\ 0&0&0\end{array}\right\rbrack$
#
# Und nun die angekündigte Anwendung auf das Bild:
plt.figure(figsize=(figure_inches, figure_inches))
filter_kern_id = [[0,0,0],[0,1,0],[0,0,0]]
filtered_data = conv(data, filter_kern_id)
plt.imshow(filtered_data, cmap='gray')
# #### Eckendetektoren
#
# Die nächsten drei Filter ziehlen darauf ab, Ecken im Bild zu finden. Ziel hierbei ist es flächige Bereiche voneinander zu trennen. Die Filter sind oft nach deren Erfinder bzw. Entdecker benannt. In diesem Fall stellt der Sobel2 eine Verbesserung des Sobel1 dar - dieser kann zusätzlich zum horizontalen sowie vertikalen auch im $45^\circ$ Bereich messen.
#
# Roberts: $\left\lbrack\begin{array}{ccc} 1&0&-1\\ 0&0&0\\ -1&0&1 \end{array}\right\rbrack$
#
# Sobel1: $\left\lbrack\begin{array}{ccc} 0&-1&0\\ -1&4&-1\\ 0&-1&0\end{array}\right\rbrack$
#
# Sobel2: $\left\lbrack\begin{array}{ccc} -1&-1&-1\\-1&8&-1\\ -1&-1&-1\end{array}\right\rbrack$
# +
filter_kern_roberts = [[1,0,-1], [0,0,0], [-1,0,1]]
plt.figure(figsize=(figure_inches, figure_inches))
data = read_image_as_array('images/lama.png', (500,500))
filtered_data_e1 = conv(data, filter_kern_roberts)
plt.imshow(filtered_data_e1, cmap='gray')
# +
filter_kern_sobel1 = [[0,-1,0], [-1,4,-1], [0,-1,0]]
plt.figure(figsize=(figure_inches, figure_inches))
data = read_image_as_array('images/lama.png', (500,500))
filtered_data = conv(data, filter_kern_sobel1)
plt.imshow(filtered_data, cmap='gray')
# +
filter_kern_sobel2 = [[-1,-1,-1], [-1,8,-1], [-1,-1,-1]]
plt.figure(figsize=(figure_inches, figure_inches))
data = read_image_as_array('images/lama.png', (500,500))
filtered_data = conv(data, filter_kern_sobel2)
plt.imshow(filtered_data, cmap='gray')
# -
# #### Bildschärfen
#
# Der nächste Filter dient, wie der Name bereits vermuten lässt, dazu, dass Konturen im Bild schärfer werden.
#
# $\left\lbrack\begin{array}{ccc} 0&-1&0\\ -1&5&-1\\ 0&-1&0 \end{array}\right\rbrack$
# +
filter_kern_sharp = [[0,-1,0], [-1,5,-1], [0,-1,0]]
plt.figure(figsize=(figure_inches, figure_inches))
data = read_image_as_array('images/lama.png', (500,500))
filtered_data = conv(data, filter_kern_sharp)
plt.imshow(filtered_data, cmap='gray')
# -
# #### Blur / Unschärfe
#
# Die letzen beiden Filter dienen dazu, das Bild zu glätten. Der erste Filter wird auch als Box-Linear-Filter bezeichnet und ist verhätlinismäßig relativ simple aufgebaut. Der zweite Filter basiert auf einer Gaußverteilung und wird daher als Gauß-Filter bezeichnet.
#
# Box-Linear-Filter: $\frac{1}{9} \left\lbrack\begin{array}{ccc}1&1&1\\ 1&1&1\\ 1&1&1\end{array}\right\rbrack$
#
# Gauß-Filter: $\frac{1}{16} \left\lbrack\begin{array}{ccc}1&2&1\\ 2&4&2\\ 1&2&1\end{array}\right\rbrack$
# +
filter_kern_blf = [[1/9, 1/9, 1/9], [1/9, 1/9, 1/9], [1/9, 1/9, 1/9]]
plt.figure(figsize=(figure_inches, figure_inches))
data = read_image_as_array('images/lama.png', (500,500))
filtered_data = conv(data, filter_kern_blf)
plt.imshow(filtered_data, cmap='gray')
# +
filter_kern_gauss = [[1/16, 2/16, 1/16], [2/16, 4/16, 2/16], [1/16, 2/16, 1/16]]
plt.figure(figsize=(figure_inches, figure_inches))
data = read_image_as_array('images/lama.png', (500,500))
filtered_data = conv(data, filter_kern_gauss)
plt.imshow(filtered_data, cmap='gray')
# -
# ### RGB-Bilder
#
# Farbige Bilder können in der Regel durch RGB-Bilder dargestellt werden, wobei $d$ gleich 3 ist und enthält:
#
# - R (rot),
# - G (grün),
# - B (blau)
#
# Werte für alle Pixel in einem Bild.
# +
lama = Image.open('images/lama.png')
lama = np.array(lama)
fig, ax = plt.subplots(figsize=(figure_inches, figure_inches))
ax.set_title('Lama image 768x1024', fontsize = 15)
ax.imshow(lama, interpolation='nearest')
plt.tight_layout()
# +
# In general deep learning (and in tensorflow) Conv-layers will
# regard all channels and therefore use "cubic" filter
# The filter used here in the example down below is only using d=1 (two - dimensional) of the
# rgb image (therefore red), you can change [:,:,0] to [:,:,1] (green) and [:,:,2] (blue)!
# Try it! :)
prewitt_x = np.array([[-1, 0, 1], [-1, 0, 1], [-1, 0, 1]])
lama_x_prew = signal.convolve2d(lama[:,:,0], prewitt_x, boundary='symm', mode='same')
lama_x_prew = np.absolute(lama_x_prew)
fig, ax = plt.subplots(figsize=(figure_inches, figure_inches))
ax.set_title('Horizontale Ableitung des Lama Bildes', fontsize = 15)
ax.imshow(lama_x_prew, interpolation='nearest', cmap='gray')
plt.tight_layout()
# -
# #### Die Faltungsschicht (engl. Convolutional Layer)
#
# <img src="images/featuremaps.png" alt="Drawing" style="width: 600px;"/>
#
# Eine Faltungsschicht, welche die erste Schicht im Netzwerk sein könnte, ist im Bild oben dargestellt. Ihr Kernel oder Filter mit den Dimensionen $K_x \times K_y \times d$ enthält Gewichte, die während des Trainings aktualisiert werden und auch die Darstellung der Bilder verändern. Eine Aktivierungskarte (engl. activation map) entspricht einer Faltungsoperation mit einem bestimmten Filter und dem zugehörigen Eingangsbild oder den räumlichen Daten der vorherigen Schicht. In den meisten Fällen werden nicht nur ein, sondern mehrere Filter in einer Faltungsschicht gelernt, so dass es mehrere Aktivierungskarten gibt. In diesem speziellen Fall scheint die Ausgabegröße dieser Faltungsschicht im Vergleich zur Eingabegröße größer geworden zu sein. Infolgedessen werden häufig Pooling-Operationen angehängt, um die Daten innerhalb des Netzwerks zu reduzieren. Die nächste Schicht erhält dann wieder räumliche Informationen und verwendet Filter, um die räumlichen Informationen zu extrahieren und zu verändern.
#
#
# **Idea**: _`Spärliche Verbindungen (engl. Sparse Connections)` (nicht vollständig verbundene Schichten wie bei einem MLP) sind als Kernel für große Datenstrukturen gegeben. Die Anzahl der lernbaren Gewichte sinkt!_
#
# Vergleichen wir eine standardmäßige voll verbundene Schicht (engl. fully connected layer) eines MLP mit einer Faltungsschicht für ein reguläres farbiges Bild der Größe $256\times256\times3$:
# - Erstes Hidden Layer in einer voll verbundenen Schicht:
# - Input Neuronen $\rightarrow$ $256*256*3$
# - Beginnen Sie z. B. mit der Hälfte der Neuronen im ersten Hidden Layer $\rightarrow$ $128*256*3$
# - Ergebnisse in Gewichte und Biases $\rightarrow$ $256*256*3*128*256*3 + 128*256*3 = 19.327.451.136$ Parameters
#
#
# - Erste Faltungsschicht in einem faltigen neuronalen Netz: Standard 256 Filter (vernünftige Größe) der Größe $3\times3\times3$
# - Gewichte und Biases $\rightarrow$ $256 * 3 * 3 *3 + 256 = 7.168 $ Parameters
#
# Trotzdem brauchen Faltungen mit räumlichen Blöcken wie in der obigen Abbildung noch Zeit, um verarbeitet zu werden.
# Lokale Informationen werden nur nicht wie globale Abhängigkeiten in Hidden Layers verwendet!
#
# Die **Vorteile** einer Faltungsschicht (`CONV`) gegenüber einer vollverknüpften Schicht sind die folgenden
# - Weniger Parameter für das Training
# - Nutzung der lokalen Strukturen des Bildes
# - Unabhängig von der Position des Merkmals im Bild
#
# **Nachteile** von Faltungsschichten (`CONV`):
# - Informationen müssen räumliche Abhängigkeiten haben (wie bei einem menschlich erkennbaren Bild)
#
# Beim Stapeln mehrerer Faltungsschichten hat ein Kernel der folgenden Faltungsschicht die Form $K_x \times K_y \times d$, wobei $d$ die Anzahl der Kanäle der vorherigen Schicht ist. Die Anzahl der Kanäle ist gegeben durch die Anzahl der verschiedenen Filter, die in der Faltungsschicht verwendet werden. Definiert man also eine Faltungsschicht mit z. B. $nb\_filters=64$, so legt man die dritte Dimension eines Filters in der nächsten Schicht fest. Denn im zweidimensionalen Fall expandiert der Filter immer auf die vorherige Kanaldimension. Betrachtet man CNNs für die Videoanalyse oder für Zeitreihen, so stößt man auf 3-dimensionale Faltungsschichten, die sich nicht nur in den Bilddimensionen bewegen, sondern in einer dritten Dimension (in diesem Fall: Zeit).
# #### Die Poolingsschicht (engl. pooling layer)
#
# <img src="images/maxpool.png" alt="Drawing" style="width: 600px;"/>
#
#
# Quelle: http://cs231n.github.io/convolutional-networks/
#
#
# Die Pooling Schicht ist ein Filter wie alle anderen Filter im neuronalen Faltungsnetzwerk. Allerdings mit der Ausnahme, dass sie ihre Gewichte nicht aktualisiert und eine feste Funktionsoperation durchführt. Die häufigste Pooling-Operation ist das Max-Pooling. Wie der Name schon sagt, wird im Bereich des Kerns nur der Maximalwert weitergegeben. Normalerweise entspricht der Stride den Dimensionen des Kernels. Das Max-Pooling wird nur auf die Höhe und Breite des Bildes angewendet, so dass die Kanaldimensionen nicht betroffen sind. Es wird verwendet, um räumliche Informationen zu reduzieren.
# <div class="alert alert-block alert-success">
# <b>Aufgabe 5.2.2:</b> Implementieren Sie die Funktion <code>max_pool</code> die Maxpooling durchführt. Gegeben ist wieder ein Grauwertbild <code>image_data</code>, d.h. es besitzt nur einen Kanal und Sie können annehmen, dass das Bilder wieder als eine Liste von Listen übergeben wird. Außerdem ist die Größe des Filters <code>filter_size</code> als Tupel und die <code>stride</code> als <code>int</code> gegeben.
# </div>
def max_pool(image_data:list, filter_size:tuple, stride:int)->list:
# STUDENT CODE HERE
# STUDENT CODE until HERE
# +
test_input_data = [[0,0,0,0,0], [0,1,1,1,0], [0,0,2,0,0], [0,3,3,3,0], [0,0,0,0,0], [0,0,0,0,0]]
test_filter_size = (2,2)
stride = 2
test_result = [[1,1],[3,3], [0,0]]
# The folgende Zeile erzeugt einen Fehler, wenn die Ausgabe der Methode nicht mit der erwarteten übereinstimmt
found = max_pool(test_input_data, test_filter_size, stride)
assert found == test_result
# -
# #### ReLU - Schicht oder Aktivierung
# Die "RELU"-Schicht oder Aktivierung verwendet eine elementweise Aktivierungsfunktion auf das Raumvolumen an, wie auf jeden Knoten in einer Hidden Layer. Die Funktion kann als $max(0,x)$ angegeben werden und ist unten dargestellt. Betrachten Sie $\sigma(x)$ als die Aktivierungsfunktion.
# +
import numpy as np
import matplotlib.pyplot as plt
def relu(x:float)->float:
return np.maximum(0,x)
x = np.linspace(-10, 10, num = 1000)
plt.figure(2, figsize=(10,3.5))
plt.plot(x, relu(x), label='ReLU')
plt.title('The ReLU activation')
plt.legend()
plt.xlabel('x')
plt.ylabel('$\sigma(x)$')
plt.tight_layout()
plt.grid()
# -
# ### Zusammenfassung
#
# Die folgende Animation zeigt recht gut, wie ein Faltungsnetzwerk (engl. convolutional network) anhand des `MNIST`-Datensatzes funktioniert.
# Nachdem die Faltungsschichten die Repräsentation der Bilder verändert haben, werden die endgültigen mehrdimensionalen Blöcke in ein langes Array gelegt (die Operation wird "Flattening" genannt) und an voll verbundene Schichten eines neuronalen Netzes weitergeleitet.
#
# [MNIST-CLassification](http://scs.ryerson.ca/~aharley/vis/conv/flat.html)
#
# #### Receptive Field
#
# In der Animation bzw. Simulation von MNIST werden Abhängigkeiten, die als Linien zwischen mehr als zwei Schichten dargestellt werden, nicht abgebildet.
# Dennoch ist es möglich, Beziehungen zwischen beliebigen Schichten innerhalb des Netzes darzustellen. Dadurch ist es möglich, ein gewisses Wissen oder eine Idee über die Anzahl der Faltungsschichten zu erhalten, die für eine Anwendung oder Aufgabe verwendet werden sollten. Betrachten Sie drei übereinander gestapelte Faltungsschichten wie im Bild unten. Ein Wert in der grünen Schicht bezieht sich auf 9 Eingangswerte. Folglich summiert sich ein Wert in der gelben Schicht auf 9 in der grünen Schicht. Ein Eintrag in der gelben Schicht wird also von mehr Werten beeinflusst als die grünen Aktivierungseinträge in Bezug auf das Eingangsbild. Dieser Bereich ist gelb dargestellt und deckt 49 Werte des Eingangsbildes ab. Um die Dimensionen während der Faltungen wie in üblichen CNNs beizubehalten, wurde ein Padding verwendet, um die Dimensionen der Matrix gleich zu halten. Die `Initialmatrix` ist dann von der Größe $7 \times 7$.
#
# <img src="images/ReceptiveField.png" alt="Drawing" style="width: 600px;"/>
#
# Quelle:https://medium.com/mlreview/a-guide-to-receptive-field-arithmetic-for-convolutional-neural-networks-e0f514068807
# <div class="alert alert-block alert-success">
# <b>Frage 5.2.3:</b> Was ist der Hauptunterschied zwischen einer Faltungsschicht (engl. convolutional layer) und einer vollverknüpften Schicht (engl. fully-connected layer) und warum werden überhaupt Filter verwendet?
# </div>
#
# <div class="alert alert-block alert-success">
# <b>Ihre Antwort:</b></div>
#
| 48.032663
| 1,010
|
74994a8fadadfebe1e3954550a3fcfc87f05b34a
|
py
|
python
|
Neural Networks/Exercise 3 - Convolutions/Conv_Augment_Multiclass_Chart_RockPaperScissors).ipynb
|
navajj/Science
|
['Apache-1.1']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] colab_type="text" id="rX8mhOLljYeM"
# ##### Copyright 2019 The TensorFlow Authors.
# + cellView="form" colab_type="code" id="BZSlp3DAjdYf" colab={}
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# + colab_type="code" id="it1c0jCiNCIM" colab={"base_uri": "https://localhost:8080/", "height": 391} outputId="fff711de-66d6-4468-a318-f1db2de13031"
# !wget --no-check-certificate \
# https://storage.googleapis.com/laurencemoroney-blog.appspot.com/rps.zip \
# -O /tmp/rps.zip
# !wget --no-check-certificate \
# https://storage.googleapis.com/laurencemoroney-blog.appspot.com/rps-test-set.zip \
# -O /tmp/rps-test-set.zip
# + colab_type="code" id="PnYP_HhYNVUK" colab={}
import os
import zipfile
local_zip = '/tmp/rps.zip'
zip_ref = zipfile.ZipFile(local_zip, 'r')
zip_ref.extractall('/tmp/')
zip_ref.close()
local_zip = '/tmp/rps-test-set.zip'
zip_ref = zipfile.ZipFile(local_zip, 'r')
zip_ref.extractall('/tmp/')
zip_ref.close()
# + colab_type="code" id="MrxdR83ANgjS" colab={"base_uri": "https://localhost:8080/", "height": 119} outputId="cb874fc5-ef2e-4d96-856f-198827f8c024"
rock_dir = os.path.join('/tmp/rps/rock')
paper_dir = os.path.join('/tmp/rps/paper')
scissors_dir = os.path.join('/tmp/rps/scissors')
print('total training rock images:', len(os.listdir(rock_dir)))
print('total training paper images:', len(os.listdir(paper_dir)))
print('total training scissors images:', len(os.listdir(scissors_dir)))
rock_files = os.listdir(rock_dir)
print(rock_files[:10])
paper_files = os.listdir(paper_dir)
print(paper_files[:10])
scissors_files = os.listdir(scissors_dir)
print(scissors_files[:10])
# + colab_type="code" id="jp9dLel9N9DS" colab={"base_uri": "https://localhost:8080/", "height": 1000} outputId="dbaae03b-1d6a-46d0-c314-fddb20911e78"
# %matplotlib inline
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
pic_index = 2
next_rock = [os.path.join(rock_dir, fname)
for fname in rock_files[pic_index-2:pic_index]]
next_paper = [os.path.join(paper_dir, fname)
for fname in paper_files[pic_index-2:pic_index]]
next_scissors = [os.path.join(scissors_dir, fname)
for fname in scissors_files[pic_index-2:pic_index]]
for i, img_path in enumerate(next_rock+next_paper+next_scissors):
#print(img_path)
img = mpimg.imread(img_path)
plt.imshow(img)
plt.axis('Off')
plt.show()
# + colab_type="code" id="LWTisYLQM1aM" colab={"base_uri": "https://localhost:8080/", "height": 1000} outputId="05b19dd1-3f8f-4c65-8ac1-960dcd7fdfd5"
import tensorflow as tf
import keras_preprocessing
from keras_preprocessing import image
from keras_preprocessing.image import ImageDataGenerator
TRAINING_DIR = "/tmp/rps/"
training_datagen = ImageDataGenerator(
rescale = 1./255,
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
fill_mode='nearest')
VALIDATION_DIR = "/tmp/rps-test-set/"
validation_datagen = ImageDataGenerator(rescale = 1./255)
train_generator = training_datagen.flow_from_directory(
TRAINING_DIR,
target_size=(150,150),
class_mode='categorical',
batch_size=126
)
validation_generator = validation_datagen.flow_from_directory(
VALIDATION_DIR,
target_size=(150,150),
class_mode='categorical',
batch_size=126
)
model = tf.keras.models.Sequential([
# Note the input shape is the desired size of the image 150x150 with 3 bytes color
# This is the first convolution
tf.keras.layers.Conv2D(64, (3,3), activation='relu', input_shape=(150, 150, 3)),
tf.keras.layers.MaxPooling2D(2, 2),
# The second convolution
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
# The third convolution
tf.keras.layers.Conv2D(128, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
# The fourth convolution
tf.keras.layers.Conv2D(128, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
# Flatten the results to feed into a DNN
tf.keras.layers.Flatten(),
tf.keras.layers.Dropout(0.5),
# 512 neuron hidden layer
tf.keras.layers.Dense(512, activation='relu'),
tf.keras.layers.Dense(3, activation='softmax')
])
model.summary()
model.compile(loss = 'categorical_crossentropy', optimizer='rmsprop', metrics=['accuracy'])
history = model.fit(train_generator, epochs=25, steps_per_epoch=20, validation_data = validation_generator, verbose = 1, validation_steps=3)
model.save("rps.h5")
# + colab_type="code" id="aeTRVCr6aosw" colab={"base_uri": "https://localhost:8080/", "height": 298} outputId="88b466d3-fcf0-44d8-ca0a-bdbb64adae04"
import matplotlib.pyplot as plt
acc = history.history['accuracy']
val_acc = history.history['val_accuracy']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'r', label='Training accuracy')
plt.plot(epochs, val_acc, 'b', label='Validation accuracy')
plt.title('Training and validation accuracy')
plt.legend(loc=0)
plt.figure()
plt.show()
# + colab_type="code" id="ZABJp7T3VLCU" colab={"resources": {"http://localhost:8080/nbextensions/google.colab/files.js": {"data": "Ly8gQ29weXJpZ2h0IDIwMTcgR29vZ2xlIExMQwovLwovLyBMaWNlbnNlZCB1bmRlciB0aGUgQXBhY2hlIExpY2Vuc2UsIFZlcnNpb24gMi4wICh0aGUgIkxpY2Vuc2UiKTsKLy8geW91IG1heSBub3QgdXNlIHRoaXMgZmlsZSBleGNlcHQgaW4gY29tcGxpYW5jZSB3aXRoIHRoZSBMaWNlbnNlLgovLyBZb3UgbWF5IG9idGFpbiBhIGNvcHkgb2YgdGhlIExpY2Vuc2UgYXQKLy8KLy8gICAgICBodHRwOi8vd3d3LmFwYWNoZS5vcmcvbGljZW5zZXMvTElDRU5TRS0yLjAKLy8KLy8gVW5sZXNzIHJlcXVpcmVkIGJ5IGFwcGxpY2FibGUgbGF3IG9yIGFncmVlZCB0byBpbiB3cml0aW5nLCBzb2Z0d2FyZQovLyBkaXN0cmlidXRlZCB1bmRlciB0aGUgTGljZW5zZSBpcyBkaXN0cmlidXRlZCBvbiBhbiAiQVMgSVMiIEJBU0lTLAovLyBXSVRIT1VUIFdBUlJBTlRJRVMgT1IgQ09ORElUSU9OUyBPRiBBTlkgS0lORCwgZWl0aGVyIGV4cHJlc3Mgb3IgaW1wbGllZC4KLy8gU2VlIHRoZSBMaWNlbnNlIGZvciB0aGUgc3BlY2lmaWMgbGFuZ3VhZ2UgZ292ZXJuaW5nIHBlcm1pc3Npb25zIGFuZAovLyBsaW1pdGF0aW9ucyB1bmRlciB0aGUgTGljZW5zZS4KCi8qKgogKiBAZmlsZW92ZXJ2aWV3IEhlbHBlcnMgZm9yIGdvb2dsZS5jb2xhYiBQeXRob24gbW9kdWxlLgogKi8KKGZ1bmN0aW9uKHNjb3BlKSB7CmZ1bmN0aW9uIHNwYW4odGV4dCwgc3R5bGVBdHRyaWJ1dGVzID0ge30pIHsKICBjb25zdCBlbGVtZW50ID0gZG9jdW1lbnQuY3JlYXRlRWxlbWVudCgnc3BhbicpOwogIGVsZW1lbnQudGV4dENvbnRlbnQgPSB0ZXh0OwogIGZvciAoY29uc3Qga2V5IG9mIE9iamVjdC5rZXlzKHN0eWxlQXR0cmlidXRlcykpIHsKICAgIGVsZW1lbnQuc3R5bGVba2V5XSA9IHN0eWxlQXR0cmlidXRlc1trZXldOwogIH0KICByZXR1cm4gZWxlbWVudDsKfQoKLy8gTWF4IG51bWJlciBvZiBieXRlcyB3aGljaCB3aWxsIGJlIHVwbG9hZGVkIGF0IGEgdGltZS4KY29uc3QgTUFYX1BBWUxPQURfU0laRSA9IDEwMCAqIDEwMjQ7CgpmdW5jdGlvbiBfdXBsb2FkRmlsZXMoaW5wdXRJZCwgb3V0cHV0SWQpIHsKICBjb25zdCBzdGVwcyA9IHVwbG9hZEZpbGVzU3RlcChpbnB1dElkLCBvdXRwdXRJZCk7CiAgY29uc3Qgb3V0cHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKG91dHB1dElkKTsKICAvLyBDYWNoZSBzdGVwcyBvbiB0aGUgb3V0cHV0RWxlbWVudCB0byBtYWtlIGl0IGF2YWlsYWJsZSBmb3IgdGhlIG5leHQgY2FsbAogIC8vIHRvIHVwbG9hZEZpbGVzQ29udGludWUgZnJvbSBQeXRob24uCiAgb3V0cHV0RWxlbWVudC5zdGVwcyA9IHN0ZXBzOwoKICByZXR1cm4gX3VwbG9hZEZpbGVzQ29udGludWUob3V0cHV0SWQpOwp9CgovLyBUaGlzIGlzIHJvdWdobHkgYW4gYXN5bmMgZ2VuZXJhdG9yIChub3Qgc3VwcG9ydGVkIGluIHRoZSBicm93c2VyIHlldCksCi8vIHdoZXJlIHRoZXJlIGFyZSBtdWx0aXBsZSBhc3luY2hyb25vdXMgc3RlcHMgYW5kIHRoZSBQeXRob24gc2lkZSBpcyBnb2luZwovLyB0byBwb2xsIGZvciBjb21wbGV0aW9uIG9mIGVhY2ggc3RlcC4KLy8gVGhpcyB1c2VzIGEgUHJvbWlzZSB0byBibG9jayB0aGUgcHl0aG9uIHNpZGUgb24gY29tcGxldGlvbiBvZiBlYWNoIHN0ZXAsCi8vIHRoZW4gcGFzc2VzIHRoZSByZXN1bHQgb2YgdGhlIHByZXZpb3VzIHN0ZXAgYXMgdGhlIGlucHV0IHRvIHRoZSBuZXh0IHN0ZXAuCmZ1bmN0aW9uIF91cGxvYWRGaWxlc0NvbnRpbnVlKG91dHB1dElkKSB7CiAgY29uc3Qgb3V0cHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKG91dHB1dElkKTsKICBjb25zdCBzdGVwcyA9IG91dHB1dEVsZW1lbnQuc3RlcHM7CgogIGNvbnN0IG5leHQgPSBzdGVwcy5uZXh0KG91dHB1dEVsZW1lbnQubGFzdFByb21pc2VWYWx1ZSk7CiAgcmV0dXJuIFByb21pc2UucmVzb2x2ZShuZXh0LnZhbHVlLnByb21pc2UpLnRoZW4oKHZhbHVlKSA9PiB7CiAgICAvLyBDYWNoZSB0aGUgbGFzdCBwcm9taXNlIHZhbHVlIHRvIG1ha2UgaXQgYXZhaWxhYmxlIHRvIHRoZSBuZXh0CiAgICAvLyBzdGVwIG9mIHRoZSBnZW5lcmF0b3IuCiAgICBvdXRwdXRFbGVtZW50Lmxhc3RQcm9taXNlVmFsdWUgPSB2YWx1ZTsKICAgIHJldHVybiBuZXh0LnZhbHVlLnJlc3BvbnNlOwogIH0pOwp9CgovKioKICogR2VuZXJhdG9yIGZ1bmN0aW9uIHdoaWNoIGlzIGNhbGxlZCBiZXR3ZWVuIGVhY2ggYXN5bmMgc3RlcCBvZiB0aGUgdXBsb2FkCiAqIHByb2Nlc3MuCiAqIEBwYXJhbSB7c3RyaW5nfSBpbnB1dElkIEVsZW1lbnQgSUQgb2YgdGhlIGlucHV0IGZpbGUgcGlja2VyIGVsZW1lbnQuCiAqIEBwYXJhbSB7c3RyaW5nfSBvdXRwdXRJZCBFbGVtZW50IElEIG9mIHRoZSBvdXRwdXQgZGlzcGxheS4KICogQHJldHVybiB7IUl0ZXJhYmxlPCFPYmplY3Q+fSBJdGVyYWJsZSBvZiBuZXh0IHN0ZXBzLgogKi8KZnVuY3Rpb24qIHVwbG9hZEZpbGVzU3RlcChpbnB1dElkLCBvdXRwdXRJZCkgewogIGNvbnN0IGlucHV0RWxlbWVudCA9IGRvY3VtZW50LmdldEVsZW1lbnRCeUlkKGlucHV0SWQpOwogIGlucHV0RWxlbWVudC5kaXNhYmxlZCA9IGZhbHNlOwoKICBjb25zdCBvdXRwdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQob3V0cHV0SWQpOwogIG91dHB1dEVsZW1lbnQuaW5uZXJIVE1MID0gJyc7CgogIGNvbnN0IHBpY2tlZFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgaW5wdXRFbGVtZW50LmFkZEV2ZW50TGlzdGVuZXIoJ2NoYW5nZScsIChlKSA9PiB7CiAgICAgIHJlc29sdmUoZS50YXJnZXQuZmlsZXMpOwogICAgfSk7CiAgfSk7CgogIGNvbnN0IGNhbmNlbCA9IGRvY3VtZW50LmNyZWF0ZUVsZW1lbnQoJ2J1dHRvbicpOwogIGlucHV0RWxlbWVudC5wYXJlbnRFbGVtZW50LmFwcGVuZENoaWxkKGNhbmNlbCk7CiAgY2FuY2VsLnRleHRDb250ZW50ID0gJ0NhbmNlbCB1cGxvYWQnOwogIGNvbnN0IGNhbmNlbFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgY2FuY2VsLm9uY2xpY2sgPSAoKSA9PiB7CiAgICAgIHJlc29sdmUobnVsbCk7CiAgICB9OwogIH0pOwoKICAvLyBXYWl0IGZvciB0aGUgdXNlciB0byBwaWNrIHRoZSBmaWxlcy4KICBjb25zdCBmaWxlcyA9IHlpZWxkIHsKICAgIHByb21pc2U6IFByb21pc2UucmFjZShbcGlja2VkUHJvbWlzZSwgY2FuY2VsUHJvbWlzZV0pLAogICAgcmVzcG9uc2U6IHsKICAgICAgYWN0aW9uOiAnc3RhcnRpbmcnLAogICAgfQogIH07CgogIGNhbmNlbC5yZW1vdmUoKTsKCiAgLy8gRGlzYWJsZSB0aGUgaW5wdXQgZWxlbWVudCBzaW5jZSBmdXJ0aGVyIHBpY2tzIGFyZSBub3QgYWxsb3dlZC4KICBpbnB1dEVsZW1lbnQuZGlzYWJsZWQgPSB0cnVlOwoKICBpZiAoIWZpbGVzKSB7CiAgICByZXR1cm4gewogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbXBsZXRlJywKICAgICAgfQogICAgfTsKICB9CgogIGZvciAoY29uc3QgZmlsZSBvZiBmaWxlcykgewogICAgY29uc3QgbGkgPSBkb2N1bWVudC5jcmVhdGVFbGVtZW50KCdsaScpOwogICAgbGkuYXBwZW5kKHNwYW4oZmlsZS5uYW1lLCB7Zm9udFdlaWdodDogJ2JvbGQnfSkpOwogICAgbGkuYXBwZW5kKHNwYW4oCiAgICAgICAgYCgke2ZpbGUudHlwZSB8fCAnbi9hJ30pIC0gJHtmaWxlLnNpemV9IGJ5dGVzLCBgICsKICAgICAgICBgbGFzdCBtb2RpZmllZDogJHsKICAgICAgICAgICAgZmlsZS5sYXN0TW9kaWZpZWREYXRlID8gZmlsZS5sYXN0TW9kaWZpZWREYXRlLnRvTG9jYWxlRGF0ZVN0cmluZygpIDoKICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgJ24vYSd9IC0gYCkpOwogICAgY29uc3QgcGVyY2VudCA9IHNwYW4oJzAlIGRvbmUnKTsKICAgIGxpLmFwcGVuZENoaWxkKHBlcmNlbnQpOwoKICAgIG91dHB1dEVsZW1lbnQuYXBwZW5kQ2hpbGQobGkpOwoKICAgIGNvbnN0IGZpbGVEYXRhUHJvbWlzZSA9IG5ldyBQcm9taXNlKChyZXNvbHZlKSA9PiB7CiAgICAgIGNvbnN0IHJlYWRlciA9IG5ldyBGaWxlUmVhZGVyKCk7CiAgICAgIHJlYWRlci5vbmxvYWQgPSAoZSkgPT4gewogICAgICAgIHJlc29sdmUoZS50YXJnZXQucmVzdWx0KTsKICAgICAgfTsKICAgICAgcmVhZGVyLnJlYWRBc0FycmF5QnVmZmVyKGZpbGUpOwogICAgfSk7CiAgICAvLyBXYWl0IGZvciB0aGUgZGF0YSB0byBiZSByZWFkeS4KICAgIGxldCBmaWxlRGF0YSA9IHlpZWxkIHsKICAgICAgcHJvbWlzZTogZmlsZURhdGFQcm9taXNlLAogICAgICByZXNwb25zZTogewogICAgICAgIGFjdGlvbjogJ2NvbnRpbnVlJywKICAgICAgfQogICAgfTsKCiAgICAvLyBVc2UgYSBjaHVua2VkIHNlbmRpbmcgdG8gYXZvaWQgbWVzc2FnZSBzaXplIGxpbWl0cy4gU2VlIGIvNjIxMTU2NjAuCiAgICBsZXQgcG9zaXRpb24gPSAwOwogICAgd2hpbGUgKHBvc2l0aW9uIDwgZmlsZURhdGEuYnl0ZUxlbmd0aCkgewogICAgICBjb25zdCBsZW5ndGggPSBNYXRoLm1pbihmaWxlRGF0YS5ieXRlTGVuZ3RoIC0gcG9zaXRpb24sIE1BWF9QQVlMT0FEX1NJWkUpOwogICAgICBjb25zdCBjaHVuayA9IG5ldyBVaW50OEFycmF5KGZpbGVEYXRhLCBwb3NpdGlvbiwgbGVuZ3RoKTsKICAgICAgcG9zaXRpb24gKz0gbGVuZ3RoOwoKICAgICAgY29uc3QgYmFzZTY0ID0gYnRvYShTdHJpbmcuZnJvbUNoYXJDb2RlLmFwcGx5KG51bGwsIGNodW5rKSk7CiAgICAgIHlpZWxkIHsKICAgICAgICByZXNwb25zZTogewogICAgICAgICAgYWN0aW9uOiAnYXBwZW5kJywKICAgICAgICAgIGZpbGU6IGZpbGUubmFtZSwKICAgICAgICAgIGRhdGE6IGJhc2U2NCwKICAgICAgICB9LAogICAgICB9OwogICAgICBwZXJjZW50LnRleHRDb250ZW50ID0KICAgICAgICAgIGAke01hdGgucm91bmQoKHBvc2l0aW9uIC8gZmlsZURhdGEuYnl0ZUxlbmd0aCkgKiAxMDApfSUgZG9uZWA7CiAgICB9CiAgfQoKICAvLyBBbGwgZG9uZS4KICB5aWVsZCB7CiAgICByZXNwb25zZTogewogICAgICBhY3Rpb246ICdjb21wbGV0ZScsCiAgICB9CiAgfTsKfQoKc2NvcGUuZ29vZ2xlID0gc2NvcGUuZ29vZ2xlIHx8IHt9OwpzY29wZS5nb29nbGUuY29sYWIgPSBzY29wZS5nb29nbGUuY29sYWIgfHwge307CnNjb3BlLmdvb2dsZS5jb2xhYi5fZmlsZXMgPSB7CiAgX3VwbG9hZEZpbGVzLAogIF91cGxvYWRGaWxlc0NvbnRpbnVlLAp9Owp9KShzZWxmKTsK", "ok": true, "headers": [["content-type", "application/javascript"]], "status": 200, "status_text": ""}}, "base_uri": "https://localhost:8080/", "height": 106} outputId="fa665f7e-804c-4265-869d-3ee85c170dd1"
import numpy as np
from google.colab import files
from keras.preprocessing import image
uploaded = files.upload()
for fn in uploaded.keys():
# predicting images
path = fn
img = image.load_img(path, target_size=(150, 150))
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
images = np.vstack([x])
classes = model.predict(images, batch_size=10)
print(fn)
print("paper" if classes[0][0]==1 else "Rock"
if classes[0][1]==1 else "scissors")
| 68.255102
| 7,252
|
006eac1036497e0761a38904b979435b2e7692d0
|
py
|
python
|
docs/source/notebooks/Bayes_factor.ipynb
|
uiuc-arc/pymc3
|
['Apache-2.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Bayes Factors and Marginal Likelihood
# +
import arviz as az
import numpy as np
import pymc3 as pm
from matplotlib import pyplot as plt
from scipy.special import betaln
from scipy.stats import beta
print('Running on PyMC3 v{}'.format(pm.__version__))
# -
az.style.use('arviz-darkgrid')
# The "Bayesian way" to compare models is to compute the _marginal likelihood_ of each model $p(y \mid M_k)$, _i.e._ the probability of the observed data $y$ given the $M_k$ model. This quantity, the marginal likelihood, is just the normalizing constant of Bayes' theorem. We can see this if we write Bayes' theorem and make explicit the fact that all inferences are model-dependant.
#
# $$p (\theta \mid y, M_k ) = \frac{p(y \mid \theta, M_k) p(\theta \mid M_k)}{p( y \mid M_k)}$$
#
# where:
#
# * $y$ is the data
# * $\theta$ the parameters
# * $M_k$ one model out of K competing models
#
#
# Usually when doing inference we do not need to compute this normalizing constant, so in practice we often compute the posterior up to a constant factor, that is:
#
# $$p (\theta \mid y, M_k ) \propto p(y \mid \theta, M_k) p(\theta \mid M_k)$$
#
# However, for model comparison and model averaging the marginal likelihood is an important quantity. Although, it's not the only way to perform these tasks, you can read about model averaging and model selection using alternative methods [here](model_comparison.ipynb), [there](model_averaging.ipynb) and [elsewhere](GLM-model-selection.ipynb).
# ## Bayesian model selection
#
# If our main objective is to choose only one model, the _best_ one, from a set of models we can just choose the one with the largest $p(y \mid M_k)$. This is totally fine if **all models** are assumed to have the same _a priori_ probability. Otherwise, we have to take into account that not all models are equally likely _a priori_ and compute:
#
# $$p(M_k \mid y) \propto p(y \mid M_k) p(M_k)$$
#
# Sometimes the main objective is not to just keep a single model but instead to compare models to determine which ones are more likely and by how much. This can be achieved using Bayes factors:
#
# $$BF = \frac{p(y \mid M_0)}{p(y \mid M_1)}$$
#
# that is, the ratio between the marginal likelihood of two models. The larger the BF the _better_ the model in the numerator ($M_0$ in this example). To ease the interpretation of BFs some authors have proposed tables with levels of *support* or *strength*, just a way to put numbers into words.
#
# * 1-3: anecdotal
# * 3-10: moderate
# * 10-30: strong
# * 30-100: very strong
# * $>$ 100: extreme
#
# Notice that if you get numbers below 1 then the support is for the model in the denominator, tables for those cases are also available. Of course, you can also just take the inverse of the values in the above table or take the inverse of the BF value and you will be OK.
#
# It is very important to remember that these rules are just conventions, simple guides at best. Results should always be put into context of our problems and should be accompanied with enough details so others could evaluate by themselves if they agree with our conclusions. The evidence necessary to make a claim is not the same in particle physics, or a court, or to evacuate a town to prevent hundreds of deaths.
# ## Bayesian model averaging
#
# Instead of choosing one single model from a set of candidate models, model averaging is about getting one meta-model by averaging the candidate models. The Bayesian version of this weights each model by its marginal posterior probability.
#
# $$p(\theta \mid y) = \sum_{k=1}^K p(\theta \mid y, M_k) \; p(M_k \mid y)$$
#
# This is the optimal way to average models if the prior is _correct_ and the _correct_ model is one of the $M_k$ models in our set. Otherwise, _bayesian model averaging_ will asymptotically select the one single model in the set of compared models that is closest in [Kullback-Leibler divergence](https://en.wikipedia.org/wiki/Kullback%E2%80%93Leibler_divergence).
#
# Check this [example](model_averaging.ipynb) as an alternative way to perform model averaging.
# ## Some remarks
#
# Now we will briefly discuss some key facts about the _marginal likelihood_
#
# * The good
# * **Occam Razor included**: Models with more parameters have a larger penalization than models with fewer parameters. The intuitive reason is that the larger the number of parameters the more _spread_ the _prior_ with respect to the likelihood.
#
#
# * The bad
# * Computing the marginal likelihood is, generally, a hard task because it’s an integral of a highly variable function over a high dimensional parameter space. In general this integral needs to be solved numerically using more or less sophisticated methods.
#
# $$p(y \mid M_k) = \int_{\theta_k} p(y \mid \theta_k, M_k) \; p(\theta_k | M_k) \; d\theta_k$$
#
# * The ugly
# * The marginal likelihood depends **sensitively** on the specified prior for the parameters in each model $p(\theta_k \mid M_k)$.
#
# Notice that *the good* and *the ugly* are related. Using the marginal likelihood to compare models is a good idea because a penalization for complex models is already included (thus preventing us from overfitting) and, at the same time, a change in the prior will affect the computations of the marginal likelihood. At first this sounds a little bit silly; we already know that priors affect computations (otherwise we could simply avoid them), but the point here is the word **sensitively**. We are talking about changes in the prior that will keep inference of $\theta$ more or less the same, but could have a big impact in the value of the marginal likelihood.
# ## Computing Bayes factors
#
# The marginal likelihood is generally not available in closed-form except for some restricted models. For this reason many methods have been devised to compute the marginal likelihood and the derived Bayes factors, some of these methods are so simple and [naive](https://radfordneal.wordpress.com/2008/08/17/the-harmonic-mean-of-the-likelihood-worst-monte-carlo-method-ever/) that works very bad in practice. Most of the useful methods have been originally proposed in the field of Statistical Mechanics. This connection is explained because the marginal likelihood is analogous to a central quantity in statistical physics known as the _partition function_ which in turn is closely related to another very important quantity the _free-energy_. Many of the connections between Statistical Mechanics and Bayesian inference are summarized [here](https://arxiv.org/abs/1706.01428).
# ### Using a hierarchical model
#
# Computation of Bayes factors can be framed as a hierarchical model, where the high-level parameter is an index assigned to each model and sampled from a categorical distribution. In other words, we perform inference for two (or more) competing models at the same time and we use a discrete _dummy_ variable that _jumps_ between models. How much time we spend sampling each model is proportional to $p(M_k \mid y)$.
#
# Some common problems when computing Bayes factors this way is that if one model is better than the other, by definition, we will spend more time sampling from it than from the other model. And this could lead to inaccuracies because we will be undersampling the less likely model. Another problem is that the values of the parameters get updated even when the parameters are not used to fit that model. That is, when model 0 is chosen, parameters in model 1 are updated but since they are not used to explain the data, they only get restricted by the prior. If the prior is too vague, it is possible that when we choose model 1, the parameter values are too far away from the previous accepted values and hence the step is rejected. Therefore we end up having a problem with sampling.
#
# In case we find these problems, we can try to improve sampling by implementing two modifications to our model:
#
# * Ideally, we can get a better sampling of both models if they are visited equally, so we can adjust the prior for each model in such a way to favour the less favourable model and disfavour the most favourable one. This will not affect the computation of the Bayes factor because we have to include the priors in the computation.
#
# * Use pseudo priors, as suggested by Kruschke and others. The idea is simple: if the problem is that the parameters drift away unrestricted, when the model they belong to is not selected, then one solution is to try to restrict them artificially, but only when not used! You can find an example of using pseudo priors in a model used by Kruschke in his book and [ported](https://github.com/aloctavodia/Doing_bayesian_data_analysis) to Python/PyMC3.
#
# If you want to learn more about this approach to the computation of the marginal likelihood see [Chapter 12 of Doing Bayesian Data Analysis](http://www.sciencedirect.com/science/book/9780124058880). This chapter also discuss how to use Bayes Factors as a Bayesian alternative to classical hypothesis testing.
# ### Analytically
#
# For some models, like the beta-binomial model (AKA the _coin-flipping_ model) we can compute the marginal likelihood analytically. If we write this model as:
#
# $$\theta \sim Beta(\alpha, \beta)$$
# $$y \sim Bin(n=1, p=\theta)$$
#
# the _marginal likelihood_ will be:
#
# $$p(y) = \binom {n}{h} \frac{B(\alpha + h,\ \beta + n - h)} {B(\alpha, \beta)}$$
#
# where:
#
# * $B$ is the [beta function](https://en.wikipedia.org/wiki/Beta_function) not to get confused with the $Beta$ distribution
# * $n$ is the number of trials
# * $h$ is the number of success
#
# Since we only care about the relative value of the _marginal likelihood_ under two different models (for the same data), we can omit the binomial coefficient $\binom {n}{h}$, thus we can write:
#
# $$p(y) \propto \frac{B(\alpha + h,\ \beta + n - h)} {B(\alpha, \beta)}$$
#
# This expression has been coded in the following cell, but with a twist. We will be using the `betaln` function instead of the `beta` function, this is done to prevent underflow.
def beta_binom(prior, y):
"""
Compute the marginal likelihood, analytically, for a beta-binomial model.
prior : tuple
tuple of alpha and beta parameter for the prior (beta distribution)
y : array
array with "1" and "0" corresponding to the success and fails respectively
"""
alpha, beta = prior
h = np.sum(y)
n = len(y)
p_y = np.exp(betaln(alpha + h, beta+n-h) - betaln(alpha, beta))
return p_y
# Our data for this example consist on 100 "flips of a coin" and the same number of observed "heads" and "tails". We will compare two models one with a uniform prior and one with a _more concentrated_ prior around $\theta = 0.5$
y = np.repeat([1, 0], [50, 50]) # 50 "heads" and 50 "tails"
priors = ((1, 1), (30, 30))
for a, b in priors:
distri = beta(a, b)
x = np.linspace(0, 1, 100)
x_pdf = distri.pdf(x)
plt.plot (x, x_pdf, label=r'$\alpha$ = {:d}, $\beta$ = {:d}'.format(a, b))
plt.yticks([])
plt.xlabel('$\\theta$')
plt.legend()
# The following cell returns the Bayes factor
BF = (beta_binom(priors[1], y) / beta_binom(priors[0], y))
print(round(BF))
# We see that the model with the more concentrated prior $Beta(30, 30)$ has $\approx 5$ times more support than the model with the more extended prior $Beta(1, 1)$. Besides the exact numerical value this should not be surprising since the prior for the most favoured model is concentrated around $\theta = 0.5$ and the data $y$ has equal number of head and tails, consintent with a value of $\theta$ around 0.5.
# ### Sequential Monte Carlo
#
# The [Sequential Monte Carlo](SMC2_gaussians.ipynb) sampler is a method that basically progresses by a series of successive interpolated (or *annealed*) sequences from the prior to the posterior. A nice by-product of this process is that we get an estimation of the marginal likelihood. Actually for numerical reasons the returned value is the marginal log likelihood (this helps to avoid underflow).
# +
n_chains = 1000
models = []
traces = []
for alpha, beta in priors:
with pm.Model() as model:
a = pm.Beta('a', alpha, beta)
yl = pm.Bernoulli('yl', a, observed=y)
trace = pm.sample_smc(1000, random_seed=42)
models.append(model)
traces.append(trace)
# -
BF_smc = np.exp(models[1].marginal_log_likelihood - models[0].marginal_log_likelihood)
print(round(BF_smc))
# As we can see from the previous cell, SMC gives essentially the same answer as the analytical calculation!
#
# The advantage of using SMC is that we can use it to compute the _marginal likelihood_ for a wider range of models as a closed-form expression is no longer needed. The cost we pay for this flexibility is a more expensive computation. We should take into account that for more complex models a more accurate estimation of the _marginal likelihood_ will most likely need a larger number of `draws`. Additionally, a larger number of `n_steps` may help, specially if after stage 1 we notice that SMC uses a number of steps that are close to `n_steps`, i.e. SMC is having trouble to automatically reduce this number.
# ## Bayes factors and inference
#
# In this example we have used Bayes factors to judge which model seems to be better at explaining the data, and we get that one of the models is $\approx 5$ _better_ than the other.
#
# But what about the posterior we get from these models? How different they are?
az.summary(traces[0], var_names='a', kind='stats').round(2)
az.summary(traces[1], var_names='a', kind='stats').round(2)
# We may argue that the results are pretty similar, we have the same mean value for $\theta$, and a slightly wider posterior for `model_0`, as expected since this model has a wider prior. We can also check the posterior predictive distribution to see how similar they are.
_, ax = plt.subplots(figsize=(9, 6))
ppc_0 = pm.sample_posterior_predictive(traces[0], 100, models[0], size=(len(y), 20))
ppc_1 = pm.sample_posterior_predictive(traces[1], 100, models[1], size=(len(y), 20))
for m_0, m_1 in zip(ppc_0['yl'].T, ppc_1['yl'].T):
az.plot_kde(np.mean(m_0, 0), ax=ax, plot_kwargs={'color':'C0'})
az.plot_kde(np.mean(m_1, 0), ax=ax, plot_kwargs={'color':'C1'})
ax.plot([], label='model_0')
ax.plot([], label='model_1')
ax.legend()
ax.set_xlabel('$\\theta$')
ax.set_yticks([]);
# In this example the observed data $y$ is more consistent with `model_1` (because the prior is concentrated around the correct value of $\theta$) than `model_0` (which assigns equal probability to every possible value of $\theta$), and this difference is captured by the Bayes factors. We could say Bayes factors are measuring which model, as a whole, is better, including details of the prior that may be irrelevant for parameter inference. In fact in this example we can also see that it is possible to have two different models, with different Bayes factors, but nevertheless get very similar predictions. The reason is that the data is informative enough to reduce the effect of the prior up to the point of inducing a very similar posterior. As predictions are computed from the posterior we also get very similar predictions. In most scenarios when comparing models what we really care is the predictive accuracy of the models, if two models have similar predictive accuracy we consider both models as similar. To estimate the predictive accuracy we can use tools like WAIC, LOO or cross-validation.
| 69.048889
| 1,106
|
c5255ea0006968bb3094362c870a93d7dcbc5555
|
py
|
python
|
05/CS480_Assignment_5.ipynb
|
ThucNguyen007/cs480student
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3.8.7 64-bit
# name: python387jvsc74a57bd02db524e06e9f5f4ffedc911c917cb75e12dbc923643829bf417064a77eb14d37
# ---
# + [markdown] id="DMrBgGVKWEPJ"
# 
# #Assignment 5
# + id="Ll5BVb2mZS2_"
# In this assignment, we will visualize and explore a CT scan!
# + colab={"base_uri": "https://localhost:8080/"} id="2lscACx4iuKi" outputId="59a97b62-33f7-4ed1-a385-58a597b01f55"
# load numpy and matplotlib
import numpy as np
import matplotlib
# + colab={"base_uri": "https://localhost:8080/"} id="3yT4p3dDmJgI" outputId="f15ca42b-064b-4574-ffc3-991bf5134481"
# we are using pydicom, so lets install it!
# !pip install pydicom
# + [markdown] id="f_SOOwQf78z-"
# **Task 1**: Download and visualize data with SliceDrop! [20 Points]
# + id="vXxbYHfalVQt"
# Please download https://cs480.org/data/ct.zip and extract it on your computer!
# This is a CT scan of an arm in DICOM format.
# + id="wwtBQpTP8QbS"
# 1) Let's explore the data without loading it.
# TODO: Without loading the data, how many slices are there?
# + id="E1Xdz3xw8ZJ6"
# The total of slices are 220. Because there are 220 .dcm inside CT folder
# + id="ZJHks55e8a8K"
# 2) Let's visualize the data with SliceDrop!
# Go to https://slicedrop.com and drag'n'drop all .dcm files into the browser.
# Please use the 2D sliders to show axial, sagittal, and coronal slices in 3D.
# + id="guioyU-e9Q2s"
# TODO Please post a screenshot of SliceDrop's 3D View in the text box below by
# using the Upload image button after double-click.
# + [markdown] id="ZG3wIggWQ_pm"
# 
# + id="iRDRxjS-9LYi"
# + [markdown] id="KbpOUGY9fKLw"
# **Task 2**: Load the data using pydicom as a 3D volume and then reslice it! [35 Points]
# + id="kleZbthp9LcC"
# TODO: Please upload ct.zip using the file panel on the left.
# Then use the following snippet to extract the data.
# + id="JE2DknZUfSG8"
import zipfile
with zipfile.ZipFile('ct.zip', 'r') as zip_ref:
zip_ref.extractall('.')
# + id="wOiAyhP2fSJm"
# 1) Now loop through all the DICOM files and store them in a 3D numpy array.
# Hint: You can either store them in a list first or read the dimensions of a
# single image slice to properly create the 3D numpy array.
# Hint 2: os.listdir(DIR) gives a list of filenames in a directory.
# Hint 2b: This list is not sorted - make sure you sort it.
# Hint 3: The dcmread function loads a single DICOM file.
# Hint 4: You can then use .pixel_array to access the image data.
# + id="qcnvddIGjVOO"
from pydicom import dcmread
import os
from operator import itemgetter
from matplotlib import pyplot as plt
# + id="KN6vNSJpiRf0"
# TODO: YOUR CODE FOR LOADING THE VOLUME AS A 3D NUMPY ARRAY
slices = []
all_files = sorted(os.listdir('ct'))
for i in range(len(all_files)):
files = dcmread('ct' + '/' + all_files[i])
slices.append(files)
imagings_shape = list(slices[0].pixel_array)
imagings_shape.append(len(slices))
data = np.zeros(imagings)
for i, slice in enumerate(slices):
img = slice.pixel_array
data[:, :, i] = img
print(np.shape(data))
# + id="-ASKScaKii8S"
# 2) Now create and show axial, sagittal, and coronal slices from the 3D volume.
# Hint: Please use imshow(XX, cmap='gray') to show the image.
# + id="5pa-UOtUikTK"
# TODO: YOUR CODE FOR AXIAL
axial = plt.imshow(data[:, :, 100], cmap='gray')
# + id="wZwgRylCjB5e"
# TODO: YOUR CODE FOR SAGITTAL
sagittal = plt.imshow(data[:, 100, :], cmap='gray')
# + id="ORHmClyCjDbp"
# TODO: YOUR CODE FOR CORONAL
coronal = plt.imshow(data[100, :, :], cmap='gray')
# + [markdown] id="eqsEUwQKfXnu"
# **Task 3**: Use the Window/Level-technique to visualize the data! [45 Points]
# + id="CkzT93xUmP0G"
# We will now enhance the visualization from above by performing
# Window/Level adjustment.
# Here is one way of doing that:
# vmin = level - window/2
# vmax = level + window/2
# plt.imshow(hu_pixels + rescale, cmap='gray', vmin=vmin, vmax=vmax)
# plt.show()
# + id="dsl2cNGafSMS"
# 1) Please load the Window/Level values from the DICOM file,
# print these values, and then visualize one slice with window/level adjustment.
# Hint: The DICOM header has the following tags.
# (0028, 1050) Window Center
# (0028, 1051) Window Width
# Hint 2: You can use slice[key].value to access DICOM tag values.
# Hint 3: (0028, 1052) Rescale Intercept might be important.
# + id="5ChwNDaznJJb"
# TODO: YOUR CODE
# +
image_slice = dcmread('ct/' + all_files[100])
# Window Center
wind_center = image_slice[0x0028, 0x1050]
# Window width
wind_width = image_slice[0x0028, 0x1051]
# Rescale intercept
rescale_intercept = image_slice[0x0028, 0x1052]
level = image_slice['WindowCenter'].value
window = image_slice['WindowWidth'].value
rescale_intercept = image_slice['RescaleIntercept'].value
vmin = level - window/2
vmax = level + window/2
hu_pixels = image_slice.pixel_array
plt.imshow(hu_pixels + rescale_intercept, cmap='gray', vmin=vmin, vmax=vmax)
plt.show()
# +
level0 = 100
window0 = 100
vmin=level - window/2
vmax=level + window/2
plt.imshow(hu_pixels + rescale, cmap='gray', vmin=vmin, vmax=vmax)
plt.show()
# +
level1 = 70
window1 = 50
vmin=level - window/2
vmax=level + window/2
plt.imshow(hu_pixels + rescale, cmap='gray', vmin=vmin, vmax=vmax)
plt.show()
# +
level = 50
window = 30
vmin=level - window/2
vmax=level + window/2
print("\n vmin \t :", vmin)
print(" vmax \t :", vmax)
plt.imshow(hu_pixels + rescale, cmap='gray', vmin=vmin, vmax=vmax)
plt.show()
# +
level = 30
window = 10
vmin=level - window/2
vmax=level + window/2
print("\n vmin \t :", vmin)
print(" vmax \t :", vmax)
plt.imshow(hu_pixels + rescale, cmap='gray', vmin=vmin, vmax=vmax)
plt.show()
# +
level = 100
window = 200
vmin=level - window/2
vmax=level + window/2
print("\n vmin \t :", vmin)
print(" vmax \t :", vmax)
plt.imshow(hu_pixels + rescale, cmap='gray', vmin=vmin, vmax=vmax)
plt.show()
# + id="_yMGLK2MnVps"
# Which values make sense and why?
# I think the level of value is 100 and window value is 100
# + [markdown] id="1f72yBFNgonn"
# **Bonus**: Create segmentations (label maps) for the volume using thresholding HU! [33 Points]
# + id="suX0YEehgzYq"
# Similar to Window/Level adjustment for visualization, we can threshold
# the volume to highlight the following components using the Hounsfield Units:
# 1) Fat
# 2) Soft Tissue
# 3) Bones
#
# Please create 3 segmentation masks for these structures.
# Then, please visualize each 3 slices per structure to showcase the segmentation.
# Hint: As a reminder, the following code allows thresholding of a numpy array.
# new_mask = imagevolume.copy()
# new_mask[new_mask < XXX] = 0
# Hint2: You might need to cast new_mask to int16 not uint16.
# + id="eiXz11Ytjrcm"
# TODO: YOUR CODE TO SEGMENT FAT
image = (data.copy()).astype(np.int16)
image[data < -70] = 0
image[data > -30] = 0
plt.imshow(image[:, :, 100])
# + id="Rc8QJIyPjuAO"
# TODO: YOUR CODE TO SEGMENT SOFT TISSUE
image = (data.copy()).astype(np.int16)
image[data < 40] = 0
image[data > 20] = 0
plt.imshow(image[100, :, :])
# + id="P3YA8qDhjwLY"
# TODO: YOUR CODE TO SEGMENT BONES
image = (data.copy()).astype(np.int16)
image[data < 1000] = 0
image[data > 20] = 0
plt.imshow(image[100, :, :], cmap = 'gray')
# + id="RVY6FD-Jjz06"
# Are the segmentations good?
# + id="JeVczcqaj3jq"
# TODO: YOUR ANSWER
# my segmentation is not displayed the result as the statement shown.
# + id="JJ6yixJskKqW"
# + id="kWb3h4MKm5t4"
#
# Thank you and Great job!!
#
# _.---._
# .' `.
# :) (:
# \ (@) (@) /
# \ A /
# ) (
# \"""""/
# `._.'
# .=.
# .---._.-.=.-._.---.
# / ':-(_.-: :-._)-:` \
# / /' (__.-: :-.__) `\ \
# / / (___.-` '-.___) \ \
# / / (___.-'^`-.___) \ \
# / / (___.-'=`-.___) \ \
# / / (____.'=`.____) \ \
# / / (___.'=`.___) \ \
# (_.; `---'.=.`---' ;._)
# ;|| __ _.=._ __ ||;
# ;|| ( `.-.=.-.' ) ||;
# ;|| \ `.=.' / ||;
# ;|| \ .=. / ||;
# ;|| .-`.`-._.-'.'-. ||;
# .:::\ ( ,): O O :(, ) /:::.
# |||| ` / /'`--'--'`\ \ ' ||||
# '''' / / \ \ ''''
# / / \ \
# / / \ \
# / / \ \
# / / \ \
# / / \ \
# /.' `.\
# (_)' `(_)
# \\. .//
# \\. .//
# \\. .//
# \\. .//
# \\. .//
# \\. .//
# jgs \\. .//
# ///) (\\\
# ,///' `\\\,
# ///' `\\\
# ""' '""
| 1,436.378125
| 395,180
|
00e9a22f52f38c6441685ac0d75c5cccc3b553f4
|
py
|
python
|
Perceptron/Keras_Iris.ipynb
|
HurleyJames/GoogleColabExercise
|
['CC-BY-4.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + id="eagjilVnnoAw" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="84c20a43-a687-4bd7-ac16-b5e74cb34b83"
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import keras
from keras.models import Sequential
from keras.layers import Dense, Activation
from keras.optimizers import SGD
from sklearn.metrics import classification_report, confusion_matrix
# + id="xDyuSjgBoOXT" colab_type="code" colab={"resources": {"http://localhost:8080/nbextensions/google.colab/files.js": {"data": "Ly8gQ29weXJpZ2h0IDIwMTcgR29vZ2xlIExMQwovLwovLyBMaWNlbnNlZCB1bmRlciB0aGUgQXBhY2hlIExpY2Vuc2UsIFZlcnNpb24gMi4wICh0aGUgIkxpY2Vuc2UiKTsKLy8geW91IG1heSBub3QgdXNlIHRoaXMgZmlsZSBleGNlcHQgaW4gY29tcGxpYW5jZSB3aXRoIHRoZSBMaWNlbnNlLgovLyBZb3UgbWF5IG9idGFpbiBhIGNvcHkgb2YgdGhlIExpY2Vuc2UgYXQKLy8KLy8gICAgICBodHRwOi8vd3d3LmFwYWNoZS5vcmcvbGljZW5zZXMvTElDRU5TRS0yLjAKLy8KLy8gVW5sZXNzIHJlcXVpcmVkIGJ5IGFwcGxpY2FibGUgbGF3IG9yIGFncmVlZCB0byBpbiB3cml0aW5nLCBzb2Z0d2FyZQovLyBkaXN0cmlidXRlZCB1bmRlciB0aGUgTGljZW5zZSBpcyBkaXN0cmlidXRlZCBvbiBhbiAiQVMgSVMiIEJBU0lTLAovLyBXSVRIT1VUIFdBUlJBTlRJRVMgT1IgQ09ORElUSU9OUyBPRiBBTlkgS0lORCwgZWl0aGVyIGV4cHJlc3Mgb3IgaW1wbGllZC4KLy8gU2VlIHRoZSBMaWNlbnNlIGZvciB0aGUgc3BlY2lmaWMgbGFuZ3VhZ2UgZ292ZXJuaW5nIHBlcm1pc3Npb25zIGFuZAovLyBsaW1pdGF0aW9ucyB1bmRlciB0aGUgTGljZW5zZS4KCi8qKgogKiBAZmlsZW92ZXJ2aWV3IEhlbHBlcnMgZm9yIGdvb2dsZS5jb2xhYiBQeXRob24gbW9kdWxlLgogKi8KKGZ1bmN0aW9uKHNjb3BlKSB7CmZ1bmN0aW9uIHNwYW4odGV4dCwgc3R5bGVBdHRyaWJ1dGVzID0ge30pIHsKICBjb25zdCBlbGVtZW50ID0gZG9jdW1lbnQuY3JlYXRlRWxlbWVudCgnc3BhbicpOwogIGVsZW1lbnQudGV4dENvbnRlbnQgPSB0ZXh0OwogIGZvciAoY29uc3Qga2V5IG9mIE9iamVjdC5rZXlzKHN0eWxlQXR0cmlidXRlcykpIHsKICAgIGVsZW1lbnQuc3R5bGVba2V5XSA9IHN0eWxlQXR0cmlidXRlc1trZXldOwogIH0KICByZXR1cm4gZWxlbWVudDsKfQoKLy8gTWF4IG51bWJlciBvZiBieXRlcyB3aGljaCB3aWxsIGJlIHVwbG9hZGVkIGF0IGEgdGltZS4KY29uc3QgTUFYX1BBWUxPQURfU0laRSA9IDEwMCAqIDEwMjQ7Ci8vIE1heCBhbW91bnQgb2YgdGltZSB0byBibG9jayB3YWl0aW5nIGZvciB0aGUgdXNlci4KY29uc3QgRklMRV9DSEFOR0VfVElNRU9VVF9NUyA9IDMwICogMTAwMDsKCmZ1bmN0aW9uIF91cGxvYWRGaWxlcyhpbnB1dElkLCBvdXRwdXRJZCkgewogIGNvbnN0IHN0ZXBzID0gdXBsb2FkRmlsZXNTdGVwKGlucHV0SWQsIG91dHB1dElkKTsKICBjb25zdCBvdXRwdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQob3V0cHV0SWQpOwogIC8vIENhY2hlIHN0ZXBzIG9uIHRoZSBvdXRwdXRFbGVtZW50IHRvIG1ha2UgaXQgYXZhaWxhYmxlIGZvciB0aGUgbmV4dCBjYWxsCiAgLy8gdG8gdXBsb2FkRmlsZXNDb250aW51ZSBmcm9tIFB5dGhvbi4KICBvdXRwdXRFbGVtZW50LnN0ZXBzID0gc3RlcHM7CgogIHJldHVybiBfdXBsb2FkRmlsZXNDb250aW51ZShvdXRwdXRJZCk7Cn0KCi8vIFRoaXMgaXMgcm91Z2hseSBhbiBhc3luYyBnZW5lcmF0b3IgKG5vdCBzdXBwb3J0ZWQgaW4gdGhlIGJyb3dzZXIgeWV0KSwKLy8gd2hlcmUgdGhlcmUgYXJlIG11bHRpcGxlIGFzeW5jaHJvbm91cyBzdGVwcyBhbmQgdGhlIFB5dGhvbiBzaWRlIGlzIGdvaW5nCi8vIHRvIHBvbGwgZm9yIGNvbXBsZXRpb24gb2YgZWFjaCBzdGVwLgovLyBUaGlzIHVzZXMgYSBQcm9taXNlIHRvIGJsb2NrIHRoZSBweXRob24gc2lkZSBvbiBjb21wbGV0aW9uIG9mIGVhY2ggc3RlcCwKLy8gdGhlbiBwYXNzZXMgdGhlIHJlc3VsdCBvZiB0aGUgcHJldmlvdXMgc3RlcCBhcyB0aGUgaW5wdXQgdG8gdGhlIG5leHQgc3RlcC4KZnVuY3Rpb24gX3VwbG9hZEZpbGVzQ29udGludWUob3V0cHV0SWQpIHsKICBjb25zdCBvdXRwdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQob3V0cHV0SWQpOwogIGNvbnN0IHN0ZXBzID0gb3V0cHV0RWxlbWVudC5zdGVwczsKCiAgY29uc3QgbmV4dCA9IHN0ZXBzLm5leHQob3V0cHV0RWxlbWVudC5sYXN0UHJvbWlzZVZhbHVlKTsKICByZXR1cm4gUHJvbWlzZS5yZXNvbHZlKG5leHQudmFsdWUucHJvbWlzZSkudGhlbigodmFsdWUpID0+IHsKICAgIC8vIENhY2hlIHRoZSBsYXN0IHByb21pc2UgdmFsdWUgdG8gbWFrZSBpdCBhdmFpbGFibGUgdG8gdGhlIG5leHQKICAgIC8vIHN0ZXAgb2YgdGhlIGdlbmVyYXRvci4KICAgIG91dHB1dEVsZW1lbnQubGFzdFByb21pc2VWYWx1ZSA9IHZhbHVlOwogICAgcmV0dXJuIG5leHQudmFsdWUucmVzcG9uc2U7CiAgfSk7Cn0KCi8qKgogKiBHZW5lcmF0b3IgZnVuY3Rpb24gd2hpY2ggaXMgY2FsbGVkIGJldHdlZW4gZWFjaCBhc3luYyBzdGVwIG9mIHRoZSB1cGxvYWQKICogcHJvY2Vzcy4KICogQHBhcmFtIHtzdHJpbmd9IGlucHV0SWQgRWxlbWVudCBJRCBvZiB0aGUgaW5wdXQgZmlsZSBwaWNrZXIgZWxlbWVudC4KICogQHBhcmFtIHtzdHJpbmd9IG91dHB1dElkIEVsZW1lbnQgSUQgb2YgdGhlIG91dHB1dCBkaXNwbGF5LgogKiBAcmV0dXJuIHshSXRlcmFibGU8IU9iamVjdD59IEl0ZXJhYmxlIG9mIG5leHQgc3RlcHMuCiAqLwpmdW5jdGlvbiogdXBsb2FkRmlsZXNTdGVwKGlucHV0SWQsIG91dHB1dElkKSB7CiAgY29uc3QgaW5wdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQoaW5wdXRJZCk7CiAgaW5wdXRFbGVtZW50LmRpc2FibGVkID0gZmFsc2U7CgogIGNvbnN0IG91dHB1dEVsZW1lbnQgPSBkb2N1bWVudC5nZXRFbGVtZW50QnlJZChvdXRwdXRJZCk7CiAgb3V0cHV0RWxlbWVudC5pbm5lckhUTUwgPSAnJzsKCiAgY29uc3QgcGlja2VkUHJvbWlzZSA9IG5ldyBQcm9taXNlKChyZXNvbHZlKSA9PiB7CiAgICBpbnB1dEVsZW1lbnQuYWRkRXZlbnRMaXN0ZW5lcignY2hhbmdlJywgKGUpID0+IHsKICAgICAgcmVzb2x2ZShlLnRhcmdldC5maWxlcyk7CiAgICB9KTsKICB9KTsKCiAgY29uc3QgY2FuY2VsID0gZG9jdW1lbnQuY3JlYXRlRWxlbWVudCgnYnV0dG9uJyk7CiAgaW5wdXRFbGVtZW50LnBhcmVudEVsZW1lbnQuYXBwZW5kQ2hpbGQoY2FuY2VsKTsKICBjYW5jZWwudGV4dENvbnRlbnQgPSAnQ2FuY2VsIHVwbG9hZCc7CiAgY29uc3QgY2FuY2VsUHJvbWlzZSA9IG5ldyBQcm9taXNlKChyZXNvbHZlKSA9PiB7CiAgICBjYW5jZWwub25jbGljayA9ICgpID0+IHsKICAgICAgcmVzb2x2ZShudWxsKTsKICAgIH07CiAgfSk7CgogIC8vIENhbmNlbCB1cGxvYWQgaWYgdXNlciBoYXNuJ3QgcGlja2VkIGFueXRoaW5nIGluIHRpbWVvdXQuCiAgY29uc3QgdGltZW91dFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgc2V0VGltZW91dCgoKSA9PiB7CiAgICAgIHJlc29sdmUobnVsbCk7CiAgICB9LCBGSUxFX0NIQU5HRV9USU1FT1VUX01TKTsKICB9KTsKCiAgLy8gV2FpdCBmb3IgdGhlIHVzZXIgdG8gcGljayB0aGUgZmlsZXMuCiAgY29uc3QgZmlsZXMgPSB5aWVsZCB7CiAgICBwcm9taXNlOiBQcm9taXNlLnJhY2UoW3BpY2tlZFByb21pc2UsIHRpbWVvdXRQcm9taXNlLCBjYW5jZWxQcm9taXNlXSksCiAgICByZXNwb25zZTogewogICAgICBhY3Rpb246ICdzdGFydGluZycsCiAgICB9CiAgfTsKCiAgaWYgKCFmaWxlcykgewogICAgcmV0dXJuIHsKICAgICAgcmVzcG9uc2U6IHsKICAgICAgICBhY3Rpb246ICdjb21wbGV0ZScsCiAgICAgIH0KICAgIH07CiAgfQoKICBjYW5jZWwucmVtb3ZlKCk7CgogIC8vIERpc2FibGUgdGhlIGlucHV0IGVsZW1lbnQgc2luY2UgZnVydGhlciBwaWNrcyBhcmUgbm90IGFsbG93ZWQuCiAgaW5wdXRFbGVtZW50LmRpc2FibGVkID0gdHJ1ZTsKCiAgZm9yIChjb25zdCBmaWxlIG9mIGZpbGVzKSB7CiAgICBjb25zdCBsaSA9IGRvY3VtZW50LmNyZWF0ZUVsZW1lbnQoJ2xpJyk7CiAgICBsaS5hcHBlbmQoc3BhbihmaWxlLm5hbWUsIHtmb250V2VpZ2h0OiAnYm9sZCd9KSk7CiAgICBsaS5hcHBlbmQoc3BhbigKICAgICAgICBgKCR7ZmlsZS50eXBlIHx8ICduL2EnfSkgLSAke2ZpbGUuc2l6ZX0gYnl0ZXMsIGAgKwogICAgICAgIGBsYXN0IG1vZGlmaWVkOiAkewogICAgICAgICAgICBmaWxlLmxhc3RNb2RpZmllZERhdGUgPyBmaWxlLmxhc3RNb2RpZmllZERhdGUudG9Mb2NhbGVEYXRlU3RyaW5nKCkgOgogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAnbi9hJ30gLSBgKSk7CiAgICBjb25zdCBwZXJjZW50ID0gc3BhbignMCUgZG9uZScpOwogICAgbGkuYXBwZW5kQ2hpbGQocGVyY2VudCk7CgogICAgb3V0cHV0RWxlbWVudC5hcHBlbmRDaGlsZChsaSk7CgogICAgY29uc3QgZmlsZURhdGFQcm9taXNlID0gbmV3IFByb21pc2UoKHJlc29sdmUpID0+IHsKICAgICAgY29uc3QgcmVhZGVyID0gbmV3IEZpbGVSZWFkZXIoKTsKICAgICAgcmVhZGVyLm9ubG9hZCA9IChlKSA9PiB7CiAgICAgICAgcmVzb2x2ZShlLnRhcmdldC5yZXN1bHQpOwogICAgICB9OwogICAgICByZWFkZXIucmVhZEFzQXJyYXlCdWZmZXIoZmlsZSk7CiAgICB9KTsKICAgIC8vIFdhaXQgZm9yIHRoZSBkYXRhIHRvIGJlIHJlYWR5LgogICAgbGV0IGZpbGVEYXRhID0geWllbGQgewogICAgICBwcm9taXNlOiBmaWxlRGF0YVByb21pc2UsCiAgICAgIHJlc3BvbnNlOiB7CiAgICAgICAgYWN0aW9uOiAnY29udGludWUnLAogICAgICB9CiAgICB9OwoKICAgIC8vIFVzZSBhIGNodW5rZWQgc2VuZGluZyB0byBhdm9pZCBtZXNzYWdlIHNpemUgbGltaXRzLiBTZWUgYi82MjExNTY2MC4KICAgIGxldCBwb3NpdGlvbiA9IDA7CiAgICB3aGlsZSAocG9zaXRpb24gPCBmaWxlRGF0YS5ieXRlTGVuZ3RoKSB7CiAgICAgIGNvbnN0IGxlbmd0aCA9IE1hdGgubWluKGZpbGVEYXRhLmJ5dGVMZW5ndGggLSBwb3NpdGlvbiwgTUFYX1BBWUxPQURfU0laRSk7CiAgICAgIGNvbnN0IGNodW5rID0gbmV3IFVpbnQ4QXJyYXkoZmlsZURhdGEsIHBvc2l0aW9uLCBsZW5ndGgpOwogICAgICBwb3NpdGlvbiArPSBsZW5ndGg7CgogICAgICBjb25zdCBiYXNlNjQgPSBidG9hKFN0cmluZy5mcm9tQ2hhckNvZGUuYXBwbHkobnVsbCwgY2h1bmspKTsKICAgICAgeWllbGQgewogICAgICAgIHJlc3BvbnNlOiB7CiAgICAgICAgICBhY3Rpb246ICdhcHBlbmQnLAogICAgICAgICAgZmlsZTogZmlsZS5uYW1lLAogICAgICAgICAgZGF0YTogYmFzZTY0LAogICAgICAgIH0sCiAgICAgIH07CiAgICAgIHBlcmNlbnQudGV4dENvbnRlbnQgPQogICAgICAgICAgYCR7TWF0aC5yb3VuZCgocG9zaXRpb24gLyBmaWxlRGF0YS5ieXRlTGVuZ3RoKSAqIDEwMCl9JSBkb25lYDsKICAgIH0KICB9CgogIC8vIEFsbCBkb25lLgogIHlpZWxkIHsKICAgIHJlc3BvbnNlOiB7CiAgICAgIGFjdGlvbjogJ2NvbXBsZXRlJywKICAgIH0KICB9Owp9CgpzY29wZS5nb29nbGUgPSBzY29wZS5nb29nbGUgfHwge307CnNjb3BlLmdvb2dsZS5jb2xhYiA9IHNjb3BlLmdvb2dsZS5jb2xhYiB8fCB7fTsKc2NvcGUuZ29vZ2xlLmNvbGFiLl9maWxlcyA9IHsKICBfdXBsb2FkRmlsZXMsCiAgX3VwbG9hZEZpbGVzQ29udGludWUsCn07Cn0pKHNlbGYpOwo=", "ok": true, "headers": [["content-type", "application/javascript"]], "status": 200, "status_text": ""}}, "base_uri": "https://localhost:8080/", "height": 88} outputId="73cf2ad0-12f1-4044-da29-6bc1a91780ef"
from google.colab import files
uploaded = files.upload()
for fn in uploaded.keys():
print('User uploaded file "{name}" with length {length} bytes'.format(
name=fn, length=len(uploaded[fn])))
# + id="6oh1jxfSob0I" colab_type="code" colab={}
df = pd.read_csv("iris.csv")
# + id="BlctFZeVob23" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 204} outputId="7c9d19c8-8eac-4103-b7cf-546925af9c6b"
df.head()
# + id="ZB4Lat0vob6D" colab_type="code" colab={}
inputs_x = df[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']]
inputs_y = df['variety']
# + id="K6KKjcECob-s" colab_type="code" colab={}
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
# + id="zvuQ--EvocBN" colab_type="code" colab={}
inputs_x_scaler = scaler.fit_transform(inputs_x.values)
df_scaler = pd.DataFrame(inputs_x_scaler, index=inputs_x.index, columns=inputs_x.columns)
# + id="mhwERUPAob8h" colab_type="code" colab={}
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(df_scaler.values, inputs_y, test_size=0.2, random_state=42)
# + id="jCpiwOL2qy4i" colab_type="code" colab={}
from sklearn import preprocessing
encoder = preprocessing.LabelEncoder()
y_train = encoder.fit_transform(y_train)
y_test = encoder.fit_transform(y_test)
# + id="NXCw2Minqy7A" colab_type="code" colab={}
y_train = keras.utils.to_categorical(y_train, num_classes=3)
y_test = keras.utils.to_categorical(y_test, num_classes=3)
# + id="SoezPL4rqy9R" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 85} outputId="64a7281c-1a56-471b-983a-857f347db663"
print(X_train.shape)
print(X_test.shape)
print(y_train.shape)
print(y_test.shape)
# + id="hDI9I6Mfqy_9" colab_type="code" colab={}
model = Sequential()
model.add(Dense(100,input_shape=(4,),activation='relu'))
model.add(Dense(3, activation='softmax'))
optimizer = SGD(learning_rate=0.01, momentum=0.9)
model.compile(optimizer=optimizer, loss='categorical_crossentropy', metrics=['accuracy'])
# + id="_PbOfKnAqzCN" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 221} outputId="9b9294cd-8f02-41f6-98a8-cd9954fc2e31"
model.summary()
# + id="DunzzH3_SoC7" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 312} outputId="7bd2c092-ddfd-4a7a-c10c-185e1479c0f0"
from keras.utils import plot_model
plot_model(model, to_file='model_plot.png', show_shapes=True, show_layer_names=True)
from IPython.display import Image
Image(filename='model_plot.png')
# + id="NdPWWA_cqzHC" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 1000} outputId="6a64c790-e8f4-4397-ebe1-cfbb37af1564"
trained_model = model.fit(X_train, y_train, epochs=300, batch_size=32)
# + id="uzHTk-7fqzJg" colab_type="code" colab={}
test_result = model.evaluate(X_test, y_test, verbose=0)
# + id="YZF8-tKVqzFs" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 51} outputId="4569b25b-49c7-4d4d-8c44-9377f825109a"
print("Test score: {}".format(test_result[0]))
print("Test accuracy: {:.2f}".format(test_result[1]))
# + id="g0FTJ5B61MjK" colab_type="code" colab={}
y_pred = model.predict(X_test)
y_test_class = np.argmax(y_test, axis=1)
y_pred_class = np.argmax(y_pred, axis=1)
# + id="I66wnHCpTWLu" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 238} outputId="92988345-87fa-40ae-8b92-b6e71d790329"
print(classification_report(y_test_class,y_pred_class))
print(confusion_matrix(y_test_class,y_pred_class))
# + id="gb1YMYyJ2NqM" colab_type="code" colab={}
df_result = pd.DataFrame.from_dict(trained_model.history)
# + id="BkWdcg-i2WmH" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 282} outputId="3e07c09b-6232-4a52-c8b9-6598bc7c1668"
df_result.plot()
| 106.451327
| 7,663
|
7467cf4c344dc6e78f076e6d7cdb20875619d849
|
py
|
python
|
module3-permutation-boosting/LS_DS_233_assignment.ipynb
|
jrslagle/DS-Unit-2-Applied-Modeling
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="nCc3XZEyG3XV"
# Lambda School Data Science
#
# *Unit 2, Sprint 3, Module 3*
#
# ---
#
#
# # Permutation & Boosting
#
# You will use your portfolio project dataset for all assignments this sprint.
#
# ## Assignment
#
# Complete these tasks for your project, and document your work.
#
# - [ ] If you haven't completed assignment #1, please do so first.
# - [ ] Continue to clean and explore your data. Make exploratory visualizations.
# - [ ] Fit a model. Does it beat your baseline?
# - [ ] Try xgboost.
# - [ ] Get your model's permutation importances.
#
# You should try to complete an initial model today, because the rest of the week, we're making model interpretation visualizations.
#
# But, if you aren't ready to try xgboost and permutation importances with your dataset today, that's okay. You can practice with another dataset instead. You may choose any dataset you've worked with previously.
#
# The data subdirectory includes the Titanic dataset for classification and the NYC apartments dataset for regression. You may want to choose one of these datasets, because example solutions will be available for each.
#
#
# ## Reading
#
# Top recommendations in _**bold italic:**_
#
# #### Permutation Importances
# - _**[Kaggle / Dan Becker: Machine Learning Explainability](https://www.kaggle.com/dansbecker/permutation-importance)**_
# - [Christoph Molnar: Interpretable Machine Learning](https://christophm.github.io/interpretable-ml-book/feature-importance.html)
#
# #### (Default) Feature Importances
# - [Ando Saabas: Selecting good features, Part 3, Random Forests](https://blog.datadive.net/selecting-good-features-part-iii-random-forests/)
# - [Terence Parr, et al: Beware Default Random Forest Importances](https://explained.ai/rf-importance/index.html)
#
# #### Gradient Boosting
# - [A Gentle Introduction to the Gradient Boosting Algorithm for Machine Learning](https://machinelearningmastery.com/gentle-introduction-gradient-boosting-algorithm-machine-learning/)
# - [An Introduction to Statistical Learning](http://www-bcf.usc.edu/~gareth/ISL/ISLR%20Seventh%20Printing.pdf), Chapter 8
# - _**[Gradient Boosting Explained](https://www.gormanalysis.com/blog/gradient-boosting-explained/)**_ — Ben Gorman
# - [Gradient Boosting Explained](http://arogozhnikov.github.io/2016/06/24/gradient_boosting_explained.html) — Alex Rogozhnikov
# - [How to explain gradient boosting](https://explained.ai/gradient-boosting/) — Terence Parr & Jeremy Howard
# + id="qK5wP4yhZoth" executionInfo={"status": "ok", "timestamp": 1607583523909, "user_tz": 420, "elapsed": 8748, "user": {"displayName": "James Slagle", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhHRAEz2_G1d3HzFY6upFC6aFaltoE1eE_4Xy_lQQ=s64", "userId": "05173057312343357198"}}
# %%capture
import sys
# If you're on Colab:
if 'google.colab' in sys.modules:
# DATA_PATH = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-2-Applied-Modeling/master/data/'
# !pip install category_encoders==2.*
# !pip install pandas-profiling==2.*
# !pip install eli5
# If you're working locally:
# else:
# DATA_PATH = '../data/'
# + colab={"base_uri": "https://localhost:8080/", "height": 253} id="FQMPTeJvcCIt" executionInfo={"status": "ok", "timestamp": 1607579114017, "user_tz": 420, "elapsed": 2066, "user": {"displayName": "James Slagle", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhHRAEz2_G1d3HzFY6upFC6aFaltoE1eE_4Xy_lQQ=s64", "userId": "05173057312343357198"}} outputId="bda3972d-3e67-4e22-d9e0-7fa0ce876b15"
# A UCI dataset for predicting crime rates (Unnormalized)
# https://archive.ics.uci.edu/ml/datasets/Communities+and+Crime+Unnormalized
# To extract a list of column names from a website describing a dataset
import re
# column_names = !curl https://archive.ics.uci.edu/ml/datasets/Communities+and+Crime+Unnormalized
# @attribute community numeric
column_names = [ re.search('-- (\S+): ', line) for line in column_names ]
column_names = [ match.group(1) for match in column_names if match ]
column_names = column_names[2:] # 'Creator' and 'Donor' get included from the webpage, but aren't features
# Descriptions for the column names can be found on the main website
# https://archive.ics.uci.edu/ml/datasets/Communities+and+Crime+Unnormalized
import pandas as pd
data_url = 'https://archive.ics.uci.edu/ml/machine-learning-databases/00211/CommViolPredUnnormalizedData.txt'
df = pd.read_csv(data_url, names = column_names, na_values=['?'])
df.head()
# + colab={"base_uri": "https://localhost:8080/", "height": 968, "referenced_widgets": ["672d72f06e0e44edb5143c184bb8224f", "6894d9e0e07641bba42c3b3d62c256a0", "14cb188800ef4a22978c1ee24d67bbcd", "7d63b2fba6ea406e8426b487ced52687", "fe4aec080a634fb0856d7c0fdd572ed9", "965e3d597eea497d992338e59ca20da2", "ad5a7c85ea754afb90c1db5ecae756a6", "9c6fd3c7703045bc9cf21e57307ba1f4", "27a0cc111d9148889ec2002adb2b505c", "d62673d3935b4e6a95130a3697d91067", "24423ba71eb44ecda62f9b12423b84a6", "e7655ab1df774fc9af1023f352432ab8", "c13e452b79fd4251b10315b41074ee2a", "6034175563ce4fc98aa54caac4dc188c", "89791b9049ba4b4a869d1bf563bc4c3f", "f6e5f56d986c41feb11b479d28317311", "1fe85f5649274e4587b439ed6e03f1aa", "516898205c7f4d3996f410ed69bb3127", "a1e20c21e48f4a63b6cd0fac684ce6a8", "ddc612a06d2946b081b74e918471e442", "fd5691fe8f9a4afe9f511d3c05b34dd7", "e5ef5c74cbfb43be8ff6ea525d7dddde", "96e3c35e4a0c435aa70eb6c8bdb7f077", "474bee3f70064710a289fb18f92d64ea", "b0bdbaecd5fd48429a136fee516e260f", "8fecceac83e64f27945f96a0a22be8f5", "518870cf44e142bfa63d16f66ce75e96", "8d684671ef0043318cf9681f00d29d8c", "4d55f01174654652a0d28917fa7c419f", "f10d1b60c11744fdbcebb9fcebcbbe34", "569ddd29f438460d92312b218a240682", "18525db851bc4ee2b5282a6a9fcfc457", "bcb56382154646bd84939eca1576e39c"], "output_embedded_package_id": "1oPH5_O-J2VodcU5RhCmAai_VbxM_Xq8r"} id="PLxs6-rJcL0-" executionInfo={"status": "ok", "timestamp": 1607482874937, "user_tz": 420, "elapsed": 86547, "user": {"displayName": "James Slagle", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhHRAEz2_G1d3HzFY6upFC6aFaltoE1eE_4Xy_lQQ=s64", "userId": "05173057312343357198"}} outputId="638872da-2e42-4856-b19a-ec8a223b806d"
# Get Pandas Profiling Report
from pandas_profiling import ProfileReport
profile = ProfileReport(df, minimal=True).to_notebook_iframe()
profile
# + colab={"base_uri": "https://localhost:8080/", "height": 253} id="LW0gZQrMMQol" executionInfo={"status": "ok", "timestamp": 1607579278133, "user_tz": 420, "elapsed": 663, "user": {"displayName": "James Slagle", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhHRAEz2_G1d3HzFY6upFC6aFaltoE1eE_4Xy_lQQ=s64", "userId": "05173057312343357198"}} outputId="e5faada9-fe24-4dc5-8dcf-ca8594baefa1"
df.head()
# + colab={"base_uri": "https://localhost:8080/"} id="cY1WsKw7cQTo" executionInfo={"status": "ok", "timestamp": 1607582850560, "user_tz": 420, "elapsed": 355, "user": {"displayName": "James Slagle", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhHRAEz2_G1d3HzFY6upFC6aFaltoE1eE_4Xy_lQQ=s64", "userId": "05173057312343357198"}} outputId="5a9292bf-2ba1-4280-9c55-39b17e9306e7"
# Feature Engineering Time!
# Then remove rows where my targets are null
# (turns out models don't like comparing their prediction to null)
good_rows = df[~(df.ViolentCrimesPerPop.isna() | df.nonViolPerPop.isna())]
# The final columns are all various crime rates
features = good_rows[column_names[:129]]
unusable_columns = ['communityname', 'countyCode', 'communityCode', 'fold']
features.drop(columns=unusable_columns, errors='ignore', inplace=True)
# Remove the lemas columns. They 22 are columns on the size and structure of a police department
# but there are only observations for 343 communities, and 1872 missing observations.
lema_columns = [col for col in df.columns if df[col].isna().sum() == 1872]
features.drop(columns=lema_columns, errors='ignore', inplace=True)
# Create a smaller DataFrame (343,22) of just the lemas data
# May use for making better predicitons for these communities
lemas = df[lema_columns]
lemas = lemas[~df.LemasSwornFT.isna()]
# Lets set our targets
y1 = good_rows['ViolentCrimesPerPop']
y2 = good_rows['nonViolPerPop']
# If I want to expand to a true multiclass regression problem
targets = good_rows[column_names[129:]]
print('Features', features.shape)
print('Lemas', lemas.shape)
print('Targets', targets.shape)
print('ViolentCrimesPerPop', y1.shape)
print('nonViolPerPop', y2.shape)
# + colab={"base_uri": "https://localhost:8080/", "height": 231} id="7Nn59aoTLz2A" executionInfo={"status": "error", "timestamp": 1607657468637, "user_tz": 420, "elapsed": 456, "user": {"displayName": "James Slagle", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhHRAEz2_G1d3HzFY6upFC6aFaltoE1eE_4Xy_lQQ=s64", "userId": "05173057312343357198"}} outputId="5fc8a21f-8f3d-409e-fefa-2579509279a1"
# Features
print('Features', features.shape)
display(features.head())
# Lemas
print('\nLemas', lemas.shape)
display(lemas.head())
# Target
print('\nTargets', targets.shape)
display(targets.head())
# + colab={"base_uri": "https://localhost:8080/", "height": 282} id="yILYmqdHe_Ee" executionInfo={"status": "ok", "timestamp": 1607490867403, "user_tz": 420, "elapsed": 672, "user": {"displayName": "James Slagle", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhHRAEz2_G1d3HzFY6upFC6aFaltoE1eE_4Xy_lQQ=s64", "userId": "05173057312343357198"}} outputId="11c1806a-ed9f-48e8-ffe8-1f85ff0db7c0"
# What's the distributions of my targets?
y1.plot.density()
y2.plot.density()
# + id="FbVg5HjzaFgV"
y1.log
# + colab={"base_uri": "https://localhost:8080/", "height": 197} id="cacCkyLRfGVs" executionInfo={"status": "error", "timestamp": 1607657460170, "user_tz": 420, "elapsed": 496, "user": {"displayName": "James Slagle", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhHRAEz2_G1d3HzFY6upFC6aFaltoE1eE_4Xy_lQQ=s64", "userId": "05173057312343357198"}} outputId="f9efeb5a-f55c-42a3-886d-8f231d4aa060"
# Import the train_test_split utility
from sklearn.model_selection import train_test_split
# Create the "remaining" and test datasets
X_train, X_test, y_train, y_test = train_test_split(
features, y1, test_size=0.2, random_state=42)
# + colab={"base_uri": "https://localhost:8080/", "height": 367} id="YkyPxQ3jo1zZ" executionInfo={"status": "error", "timestamp": 1607657415253, "user_tz": 420, "elapsed": 1502, "user": {"displayName": "James Slagle", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhHRAEz2_G1d3HzFY6upFC6aFaltoE1eE_4Xy_lQQ=s64", "userId": "05173057312343357198"}} outputId="6bd8d709-a360-4c47-e82d-c66f7a6ae0d5"
# I iteratively ran this over and over at n_iter=15, and did the following steps
# 1. Take the model parameters from the best 3 models from search.cv_results_
# 2. Adjust param_distributions with the min and max of the top 3 models
# 3. Run again and repeat until the parameters are in a narrow range.
from sklearn.pipeline import make_pipeline
from category_encoders import OrdinalEncoder
from sklearn.impute import SimpleImputer
from sklearn.ensemble import RandomForestRegressor
from scipy.stats import randint, uniform
from sklearn.model_selection import RandomizedSearchCV
from xgboost import XGBClassifier
pipeline = make_pipeline(
OrdinalEncoder(cols=['state']),
SimpleImputer(),
RandomForestRegressor(),
)
# pipeline = make_pipeline(
# OrdinalEncoder(), # cols=['state']
# # SimpleImputer(strategy='median'),
# XGBClassifier(
# n_estimators=100,
# random_state=42,
# n_jobs=-1,
# )
# )
# _, n_features = X_train_val.shape
# depths = np.linspace(2, 2*n_features,dtype='int')
param_distributions = {
'simpleimputer__strategy': ['mean', 'median'],
# 'xgbclassifier__n_estimators': randint(2, 100),
'randomforestregressor__n_estimators': randint(2, 100),
'randomforestregressor__max_depth': randint(2, 80),
# 'randomforestclassifier__max_features': uniform(0, 1),
# 'randomforestclassifier__min_samples_split': randint(2, 500),
# 'randomforestclassifier__min_samples_leaf': randint(1, 500),
# 'randomforestclassifier__bootstrap': [True, False],
# 'randomforestclassifier__ccp_alpha': uniform(0, 1-0),
# 'randomforestclassifier__class_weight': [None],
# 'randomforestclassifier__criterion': ['gini', 'entropy'],
# 'randomforestclassifier__max_leaf_nodes': [None],
# 'randomforestclassifier__max_samples': [None],
# 'randomforestclassifier__min_impurity_decrease': [0.0],
# 'randomforestclassifier__min_impurity_split': [True, False],
# 'randomforestclassifier__min_weight_fraction_leaf': [0.0],
# 'randomforestclassifier__oob_score': [True, False],
# 'randomforestclassifier__verbose': [0],
# 'randomforestclassifier__warm_start': [True, False],
}
# https://scikit-learn.org/stable/modules/generated/sklearn.impute.SimpleImputer.html
# https://contrib.scikit-learn.org/category_encoders/targetencoder.html
# https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html
# If you're on Colab, decrease n_iter & cv parameters
search = RandomizedSearchCV(
pipeline,
param_distributions = param_distributions,
n_iter=20,
cv=5,
scoring='neg_mean_absolute_error',
verbose=10,
return_train_score=True,
n_jobs=-1
)
# https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.RandomizedSearchCV.html
search.fit(X_train, y_train);
best_model = search.best_estimator_
print('Train/Val Accuracy', best_model.score(X_train, y_train))
print(search.best_params_)
# + colab={"base_uri": "https://localhost:8080/"} id="P8WgQwlogxYO" executionInfo={"status": "ok", "timestamp": 1607584686166, "user_tz": 420, "elapsed": 489, "user": {"displayName": "James Slagle", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhHRAEz2_G1d3HzFY6upFC6aFaltoE1eE_4Xy_lQQ=s64", "userId": "05173057312343357198"}} outputId="edd060ab-14b0-44ac-d4b9-606b67425ec1"
y1[:10]
# + colab={"base_uri": "https://localhost:8080/"} id="FkWUXxC1fRq2" executionInfo={"status": "ok", "timestamp": 1607583659710, "user_tz": 420, "elapsed": 22620, "user": {"displayName": "James Slagle", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhHRAEz2_G1d3HzFY6upFC6aFaltoE1eE_4Xy_lQQ=s64", "userId": "05173057312343357198"}} outputId="de2045c3-37da-43af-d918-c344fdcb8638"
import eli5
from eli5.sklearn import PermutationImportance
permuter = PermutationImportance(
best_model,
scoring = 'neg_mean_absolute_error',
n_iter=5,
random_state = 42
)
encoder = OrdinalEncoder(cols=['state'])
X_encoded = encoder.fit_transform(X_train)
permuter.fit(X_encoded, y_train)
# + colab={"base_uri": "https://localhost:8080/"} id="bJ-vGUcofuzz" executionInfo={"status": "ok", "timestamp": 1607584392143, "user_tz": 420, "elapsed": 361, "user": {"displayName": "James Slagle", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhHRAEz2_G1d3HzFY6upFC6aFaltoE1eE_4Xy_lQQ=s64", "userId": "05173057312343357198"}} outputId="7bed52ad-0086-40fb-d70e-a64708801697"
best_model.score(X_test, y_test)
# + colab={"base_uri": "https://localhost:8080/", "height": 1000} id="-Y4uA2SDbsYb" executionInfo={"status": "ok", "timestamp": 1607583739041, "user_tz": 420, "elapsed": 414, "user": {"displayName": "James Slagle", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GhHRAEz2_G1d3HzFY6upFC6aFaltoE1eE_4Xy_lQQ=s64", "userId": "05173057312343357198"}} outputId="33b6136a-984f-4c1d-a65b-118cdfd3d782"
eli5.show_weights(
permuter,
top = None, # no limit
feature_names = X_train.columns.to_list() # must be a list
)
| 57.018248
| 1,680
|
8f8bedbdfe445e374f7c5a181ec195175e7708f7
|
py
|
python
|
tensorflow/tools/docker/notebooks/3_mnist_from_scratch.ipynb
|
fastener/tensorflow
|
['Apache-2.0']
|
# + [markdown] colab_type="text" id="9yupXUk1DKOe"
# # MNIST from scratch
#
# This notebook walks through an example of training a TensorFlow model to do digit classification using the [MNIST data set](http://yann.lecun.com/exdb/mnist/). MNIST is a labeled set of images of handwritten digits.
#
# An example follows.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{}]} colab_type="code" id="sbUKaF8_uDI_" outputId="67a51332-3aea-4c29-8c3d-4752db08ccb3"
from IPython.display import Image
import base64
Image(data=base64.decodestring("iVBORw0KGgoAAAANSUhEUgAAAMYAAABFCAYAAAARv5krAAAYl0lEQVR4Ae3dV4wc1bYG4D3YYJucc8455yCSSIYrBAi4EjriAZHECyAk3rAID1gCIXGRgIvASIQr8UTmgDA5imByPpicTcYGY+yrbx+tOUWpu2e6u7qnZ7qXVFPVVbv2Xutfce+q7hlasmTJktSAXrnn8vR/3/xXmnnadg1aTfxL3/7rwfSPmT+kf/7vf098YRtK+FnaZaf/SS++OjNNathufF9caiT2v/xxqbTGki/SXyM1nODXv/r8+7Tb+r+lnxZNcEFHEG/e3LnpoINXSh/PWzxCy/F9eWjOnDlLrr/++jR16tQakgylqdOWTZOGFqX5C/5IjXNLjdt7/NTvv/+eTjnllLT//vunr776Kl100UVpueWWq8n10lOmpSmTU5o/f0Fa3DDH1ry9p0/++eefaZ999slYYPS0005LK664Yk2eJ02ekqZNnZx+XzA/LfprYgGxePHitOqqq6YZM2akyfPmzUvXXXddHceoic2EOckxDj300CzPggUL0g033NC3OKy00krDer3pppv6FgcBIjvGUkv9u5paZZVVhoHpl4Mvv/wyhfxDQ0NZ7H7EQbacPHny39Tejzj88ccfacqUKRmHEecYf0Nr8GGAQJ8gMHCMPlH0QMzmEBg4RnN4DVr3CQIDx+gTRQ/EbA6BgWM0h9egdZ8g8PeliD4RutfF/Ouvfz9OtZy8aNGiNH/+/GGWl1122XzseYuVNKtqsaI23Ghw0DYCA8doG8JqO+AUG2+8cVq4cGHaY4890vLLL5/WXXfdfI6jvPDCC3lJ8amnnkoezP3000/pl19+GThHtWpIPekYomTxFS7HnkqKjMsss0yGgFE4r62tSBFVJ02aNPyconi9V4/JwzHwT9ZNNtkkeZ6w5ZZbph133DH99ttv6ccff8zXX3nllcRRnHNfv2cNGMQWGRaOrWbUrjsGBRLAA6U4Lhoqw9h2223ztRBq6aWXzsbgvueffz4Lu9NOO2UnYTgrr7xy7tO9nOH111/Pbb744ov0ww8/jAvngAdFMvQDDjggG/0GG2yQX1GZNm1aziCCwzrrrJPl3muvvXKwePnll9M333wzHDCKWPbLMbuAkfISjnvvvXcW/emnn85lqCBqa4a65hiYR/Gk2RNGRlwm3n7ggQfmdrKD9sqJtdZaKxvCnDlz8n3Tp09PXmPYeuutc0SVNQjvnmuvvTa3efzxx9N33303PGZ5rF75DBvvqq233nrp22+/TWeddVbyikpgxCE4vQDhlQUBRfDw2esbs2fPTquvvnqviNN1PuIdJ4GErVx44YUZowsuuCB9+umn6eeff84BspmsWqljhPFDxjGGYx/lDkN33udajCoVlAjRzl4U8LjefRwnPjsXG8OJqKBd8NB1LTU5IHyCd7LJGOYXNoGjFqaGIKtrERDIDKtukfGMH/zRZa1A101+YBF44KfMYzO8VOYYjDWiukiGqc022yyXOUqdzTffPJ/z1ialeqNVxA9gi0wzlOJ5juJlR8JeddVV+ZrIKTq4ZvJp/8EHH+SU+txzz+W2SqmxVFZRplrH5DTRXmGFFdKuu+6azjjjjOzosl5g6D54CQCI4mGjhNQO5occckh2LvLTA6fqJOEnyhU6kNlkZmUuvrtNcFx77bUzhsZWXgoSsm6t4Dsa/tp2DErCmA04HAI4FLjaaqtlBhmnSKiNY4rDtHZFB6jFMMH0RVDH+nCPYxtDCFJnKkniRbDitWjTK3sykQUuMLPn3DZGX8SFnCG/fVyz5zCCBtIHTLshdzif8fERn8cKXxjCNOwCTu3Qf6yqhV4AQokiP489//zzM0DxnQYKwqAtIkko1kQzFFxvaNcJ6u3Pe+65J/cRRvDee+9lA2BInIyRff/997nNO++8k7t0vl2A6vHWynmyiPJ43WKLLbIijz/++LTddtvlTCdzwIWSg9yjxBJ0GN/DDz+c7zv77LOzbEceeWSekwVGgsOsWbNyNo0+qt7DfPvtt8/dmtvIGnPnzk3PPPPMsJ6rHrNef/BBeJA90RprrJEDcNhctMkXR/mnbccwuCjNGTbaaKMc8TBZprITxOdgOvbuKxqGz6LSJ598kseJ9Gi1CYmSv/76a3YyJZWMZJ6Ceskp8EMusihFEAyUmVaa8G2rxTNHIrd733///eH7YeaLNe5xrEzlWNF/HqQDf0Tm+GIbvYdD43MsKAIo/JDgE0G5aFfN8NaWYxiUshikqGYTTUSt0TCkjXsYNqJQQso+rgGa0vX58ccf56hQTtk+48F92rmvlnE1A0on2uKP0Yrw+Nxzzz0zn+ZhjKwRXq6vueaa2TmUiRQfS7SyNeMks9IV9vrvJOl/q622yo4Mfw5Pvm6TMclLdit6shh+YAMnq1E29tEsteUYBgMSgxa5MOAzJZcVXQs4bUR8XxhCHIwzMALCBuCcx5q0tF3u133l8XrRMchFiRYNyMxBKM/5IjZlWVzjULKwACISytIWFsi56aab5mvOKyEikmdAO/iHY+BDCRUZuoPD1e1akECyLseA7d13352DhdKak8Cmlt3U7TSl9p58FwejYK8ncAwKpDTnGDcARbWiAUjHiNEHsITSPlagpEZChcfrZzwSOfBOiQwXLuR3PjAhtwAD08iAMCO/a+5xPTIm3ALjwERf0V+c69QeT7ZujVdLDhgKBrANXAMreMESRkU7rdVPrXNtZ4xIpSLH1VdfnR3j4IMPzkbw2Wefpa+//jovo5188slZsZjArAcvFP3YY4+lSy+9NEdTdTTy0I5xHHfccfm1CH2LtuORKEqmkwVlVU+sBY+IdJRmE0zeeOONnEXuu+++7AhnnnlmWn/99XMJ5brtzTffzHMJx/o555xzkgdb0U8rRtAKrnTYqtG1Ml6teyxInHDCCdlGYByBmG2Z97ChVvFo2zEwbHCRTbqP7EDxPjN2pUBEe86AXAcsg+f10TYMSTvnRM1ulQe1wG/nHEXZZEJZUIYQ5cgWMsEgMgqclFdkdh+MbFFyuddnWMLNfTYkcuuXHlBkpFYNI3dS+mMMfCHHsZWadfUjmQVn8iLywscG21apMscQwR555JEM3KuvvpoZ5LHOmzgjAvBwzFt2/Oijj3Lm4Ayin/MU/eGHH+b2N998c/5MGSaZ44nw7OEd5Rx77LE5+1EehYXxkpes5li2K6+8Mhv8Lrvsko381ltvzcEBfvHQKh5auk9GPvHEE3NJAx+/eKL/HXbYIQcbK3nwN067xAk4s5VHdbvsx0nxrYQeKxJMZAfBA7GlRx99NC9EtCN7JY4RoPBeAHIAyrB3jpHYwqu1d02d7HpZcfqINo5dL7eJMXtxTzk2sgWFM/gcsnCakI2cFOk+523O+Qw7WaeYHYpYRp9xn4BkbPdWSfgJXYYM+ne+2xRj2sdx8EDu8rm4Ntp9pY4RSmb0CIPOAVNGoLA47yU4S2xen37ppZdy9CkLE/3lm8bJHzJbbiavt2Q9p7AkK7oyXAZOLk7gs9c4PJC0AOE8DDyrgJkaWgYQkSPYuAdpWySfteU8HhqKouYq+io6ZfGeZo7xpbT1+jt+jGULfprpq922ePHMBibwjWVq523KVrzBsIzTaMeu1DFi0HI0YyyYtAekY5MltbRyihFJiROBKIYTwMCTWJNubwdQFCXFapK9z96mtbjgs3thFKWnUgjBzNZIya5FOyUcPG36q4LwRgZ6Ix8HtBk3tirGGU0feAkslHfk5PzBh2cXSkvtWqWOOEaRGcoSHdXDMoYn1tK8yaON0ahbCWgFS/vxSnjn5F4ItLeiFAGAzCKc7MDA1OlIjc4pLFKE7FEyxb5ZPNTbtuiv2fvrtddfOFsYXcwj8d8qv/XGq3femLvvvnvOvrIYPPEjG+PDseDbDnXcMXiyiGiyyACOPvrovN95552zV3/++ef5zVveznlEo6CICvG5l/d4JSvHP+qoo7JjKDs4PkVSGPm9HSz9W5rlPEoCQYHjVFXyRGnBOcKA28VOP/qTBWX6YnS2IKB8qYL/enyGHPbKziOOOCLj6sGeslGW8L6Y4ANr2MY99fpsdL7jjmFwkSTSr6gDVCk+tmDQedcJ5LgdwaLPbu7xjJRRNlErSsiQhVHJlOEQoh182o1wRTnharwYs3itnWP9Rd/RD5mLW5yveh/YRhYMjItyBh/wjPat8tEVx6B00RKo5513XpIl7rzzzuwEourMmTOz95uIcyBfTSXYiy++mCOrSFS1klsFrNZ9eGPoJtmeyRx00EE5cpGbIi21XnbZZbkMee2117KMHIKMIVcotVb/vXoOz6I0+URoMlVFcBFE7L1+IjNYIo6v/fo+D3tC+FCR+FHuwNUCgfOtUlccI5hnJMoIBhN1sBICqMoNNaLP3pkiFGciIIBC4HaEbRWk0dyHb3Mp/EY0I6+NsytvyKxsKhpQr8ozGpm1IZ8IbV+PyllGuyh1YBXXOQEcy6R8M5eAHzuxxX3GRvbaCKJ4aRfXrjkG5jEbk00Prxi8SZTJKmc5/PDDc5v99tsvC+hBjWtqStmD0F4Ma1foMvDtfqZMUc3/lYjMSFFW3NS7JtyyoKzSiTocHoFJHMc+MlK7Mta7n9NbATJerbEYvQWIWCVitIyaXrV3nsG7H2Y2GVcbxyj6NX+waKEPmOvbfShwtjhQDDz5Ygt/uuoY+OPtnICDEMBTWsAQUu0NBBsDEgFEWOADAiDaVRERWsCq5i34IRN+TbTJgn8KwzOFuR4KDUXW7Kyik53Ep8w/+RkxWeO5S1EM5wVABguXMGp69dk1x87D0ObdL32GHI5tsDQGHtwbm/Hw4TpnKvNY5Ge0x113DEwT3tIsIdSnDIfxcxJAevCHfE9cXcmotHXfAw88kIFUdgFjLMn4HuZRuh9FExmjRCCnZxRqcPxz8ioUVk9eRhJkPAYHV8ZVFRkjjFSfAtw222yTy2OZ0iv15fHcQ4dKaMcwsBdEEL26RzaIh5+yK7LSBGPno8yOZX+vzRhfXzZ8cRrtyzzkzpr803XHwB8wTJYIRol+VY8zqMMBbP0f+cExE1qTdbU7x3jwwQdzVBYdesExKNiEWx2MfwoOAyCbJ9uRHZvUTcPmsENhGNE4HBKOHKNqZzQu3KNfX9H1nRABQZlbNkpt4SNo4DWIIesDj9qYnwki2giWqol3330348kZLPm7xvi1Pffcc7MzhA3gy/0oeIuxWtmPiWNgNCIFYwcCAa2FA1ikJZz1aeUVsBmge9TyoqGoIqKUFdEKCFXcU0/pHJizVMUnXBiBh6IicdTTzsEOnuZkDE/2rcJI4KMf/TF+0TucwDhkZ+DGL4/nGkPGV/AIC+2RvfP6ZPTI4gu5XNM/Um7RPzuIFyn1zW7wpQ9UHj+fbOHPmDlGCOGBGIeQQfwuq0jnISBQfOHft7JEHN94Q5xF6XLFFVfkyKIEGyuiGAo3r6BIx0imcM6k+6GHHspOEQbcDq+UTl4BwRu7PstUiPEJFsa9/PLL83nXg6d2xnUvoxS5L7744uGyh/wyRpRF9YwSHsHjE088kWWADQeRFThZkTgBstensZG5h4m56oEdcAp9CwTOVUlj6hgECcGBpA6XDazeiLKhVABQAhKB3cNxbEAL4KoEppm+gjf3OMafDf+UW7zeTL/ltqIiAxBMOIIxnLOHgbFsMGQ4InhE0nJfrXw2hnIRD3SFBKmYWDfqE49woFvOzZno3NxM0HDciMjBDsjEBgLTsJHYN+qjmWtj7hjBLKFFQgL7qRz14jHHHJPBcC2M3wRPVDT5ohzZRv0Z16O/sdozAKmdopUH5kftTrzJpl+lk29CcgpLw3BgpMbwwqF/S80pGJ6xO0WM+8Ybbxw2TuOEoTYakwyovB/JKdzDMVQOHvCRzXju890fL11aGhcMqqIxdwwCRkYQDZAaE7lWBhyosQEmQM439MgffDHm0Si8EcuBC0ezcQSZVKYktzFEW+3sfQ4natRvu9eMTS9F7IvHo+m/2fb6LNuCc0WsW+mzHq9j6hgE9YCHp5tkez2EAVjlMOmyUlU2Lis8ygVR0rykyoltPZCaOY9fr32Qp50X6xi7pWCGbsHBvwLgGIcddljGxvcsjOU1GseyiKjJQWydpiqNsBlei85BfhNxeJunVCl31x0jBOMAjJ9jRC3OEERDS7QMI0qQohIYgLSq7FJuMZbi9WZA7kRbvFAWx5Dyy449mjEDG/dyDPW4VSiy2iNvBcCSUdxyyy35OYHrqJUx843j8I/qQpA074BVVdR1x+AIHCIiIGewsqIuds41tSSlOxeOFHuOQ/E+2zPEuFYVKM32U3RMvGy44YbZMTg2B2+GOIXXJcjpR9lkUy/QyZ7GUU8zAD9RCiuR0oQYVv1IMAk7qFL+rjkGg7GZQPLufffdN69QKJtkCAKKjNGu1p7gMgWDYEDRpkpAmu0rnMLehie/RavcI49Sr1ZW0w6V91ac/IsxmdHPB0U5pQ+4+TExDudNUhPufnaKIn7N6m2k9h11jKLRqP+UQJb2eHh4uYjK0LW1D0MpCq0NR4g24RTR/0hCdvM6/m14FtljeTL4D/liedFeO7LYcyh7eMGDY8X16IM8Vp9kWjj2GwWG5IZb2FKVOHTMMTCvDKBgD2Z22223bNynnnpqVrZXBFxjQDZUFJiwIqKHN8qHO+64IxvN/fffn9vG/VWC0UpfeC5uZMEbg/ctM/8SzYOxZ599Nhs4ebSx0ECpcDFvMCdRggkesoQ+zaHU0N4EgAEnue2227JTON+LgaEVDFu5h+w2Wdl33GFkEUIQqYIqdYwwbJGO8q2xOydqUiTFWpJVPzsuUwhlzzFETxlGdFSCqaMB4XwvUzgKWU3AyW4uwFns4QMbilUyxbq8p/4cw3UEB8FDGQUDx/acqB8zRS2dw5qthe3VatPKucocg6JiYu3lP2nfawvekKVITzgJQLH24QTBtPZeE2D89957b27jwZ1IwIm8R2OMWHmJ+3pxTzaK8l+HyMrgTzrppMxqOIEsGoZvz0nsyWiliRMUl2G9aOk6POyLZVUvYtBpniL4wA1m9lVSW46BOQqKpTLK9FnUsxftvW4swssa4dkhCGFCMNfcp08lhM9KKc4h0obgsa8ShHb6Cv5DJnu8IwHB9TB852DkOlzIRV6kXbSVMfQj48BWdhE0TLr1Fe3zQR/+gRMK5yjuq4KjZccQ2SlYjexHmCnSkiLjtsesmlnpQ5naFo1A5GMAHoJxBI709ttv54ygntZWmWEcQMS9VQleRT9kNmfAG0P3HRPGbHnVudg4gEyJOAYiE0wikHAAcxHyxndO4KI/WHEK/Qzo7wjAXfaFNdurikaNtIERRTqmYIYdE2tGEs8hfJ8iFB/3xV67MCjG8NZbb6Unn3wyC+XfDxfnDxFp496qhK6qn5CDA5twK/fIRH5Gb0MMOhxCFgkKjOBoHqKEkmWvueaanG04iTHcP3CKQO0/e3ZhgceP2smqcKyKRuUYlEKhPDL+d5z1c4qVFTDnmBIZMwZ9DiKAzTmvCetPNFR7W7fXXt/KLddqTcyjr17bRybkEF5XiQhPHnMuDlF07MCB3I49l4EDxTrnfsFBJBxQbQSKeGoROqjdurWzIzoGJqRxS2KUf/rpp2flcRDRjRKVCdpFhCwz7rOVKE5z++235/7uuuuuXDq5P5yKEY0np8B3TKb9K1/vLTF0/7MiJtyRPYrq4fx+7R2e7vFDDzDyfx1goPwcUGMEYG/rFI3oGAYW0UUyimQIcRwGzbgpVsZAUTYE065xCtc5GUeSHTyg4kzKs/FKoSBljyhvTz6y2gseZAwlwgI+cNBGtpV9ZRj4BobjFY9O8g0bQcXWaRpxBE5hHuFnJ0XB6dOn56ge2QGDlK2dFSSG4b8kxVzEdSWGVxgYQLzrxJkIGgbTaUE73b9MZ/KNfIMOJpdcckndYZWmFAwv+wgydW/o8wsCK3xnz56dFzx8oxPGtk7QiI5h0FBaeGzRKYIpjDN2ig6lB9OiprmI60qNieIMIXvsQy7yotjH9eI+2hbPDY4bI8D+2JdnWTYY+iwDs78qaUTHEM0sI1pClAVMnqX9ImGQszB6DHoNOLzZNZlGRlEq9JNB9JOsRXvoxDGnsDTudwFUHTNmzMjDqEaU9xYvGgWiZnka0TEo16CeNyCM1SLtwmt5cNEoCOUa5xjQAIFWEGBP5rbKdTRr1qwcfGUMthXVTCt917pnRMdwE6ZiQm0JckADBMYCgWLwtXjTSeq/d5Y7ieag7wmDwMAxJowqB4JUicDAMapEc9DXhEFgcjxcM7vvR4on7bHS1q84WNkpUr/iEL+aOLRw4cIlQCmuIhUBmsjHlpQ9c7EmzjEsN1vd6DeCg8UVT+qRd7b6EQey8wMT+6El8RSu36xhIO8AgQYI9F94bADG4NIAgUDg/wHX+3lgThDIegAAAABJRU5ErkJggg=="), embed=True)
# + [markdown] colab_type="text" id="J0QZYD_HuDJF"
# We're going to be building a model that recognizes these digits as 5, 0, and 4.
#
# # Imports and input data
#
# We'll proceed in steps, beginning with importing and inspecting the MNIST data. This doesn't have anything to do with TensorFlow in particular -- we're just downloading the data archive.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{}]} colab_type="code" executionInfo={"elapsed": 110, "status": "ok", "timestamp": 1446749124399, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="w5vKZqr6CDz9" outputId="794eac6d-a918-4888-e8cf-a8628474d7f1"
import os
import urllib
SOURCE_URL = 'http://yann.lecun.com/exdb/mnist/'
WORK_DIRECTORY = "/tmp/mnist-data"
def maybe_download(filename):
"""A helper to download the data files if not present."""
if not os.path.exists(WORK_DIRECTORY):
os.mkdir(WORK_DIRECTORY)
filepath = os.path.join(WORK_DIRECTORY, filename)
if not os.path.exists(filepath):
filepath, _ = urllib.urlretrieve(SOURCE_URL + filename, filepath)
statinfo = os.stat(filepath)
print 'Succesfully downloaded', filename, statinfo.st_size, 'bytes.'
else:
print 'Already downloaded', filename
return filepath
train_data_filename = maybe_download('train-images-idx3-ubyte.gz')
train_labels_filename = maybe_download('train-labels-idx1-ubyte.gz')
test_data_filename = maybe_download('t10k-images-idx3-ubyte.gz')
test_labels_filename = maybe_download('t10k-labels-idx1-ubyte.gz')
# + [markdown] colab_type="text" id="gCtMhpIoC84F"
# ## Working with the images
#
# Now we have the files, but the format requires a bit of pre-processing before we can work with it. The data is gzipped, requiring us to decompress it. And, each of the images are grayscale-encoded with values from [0, 255]; we'll normalize these to [-0.5, 0.5].
#
# Let's try to unpack the data using the documented format:
#
# [offset] [type] [value] [description]
# 0000 32 bit integer 0x00000803(2051) magic number
# 0004 32 bit integer 60000 number of images
# 0008 32 bit integer 28 number of rows
# 0012 32 bit integer 28 number of columns
# 0016 unsigned byte ?? pixel
# 0017 unsigned byte ?? pixel
# ........
# xxxx unsigned byte ?? pixel
#
# Pixels are organized row-wise. Pixel values are 0 to 255. 0 means background (white), 255 means foreground (black).
#
# We'll start by reading the first image from the test data as a sanity check.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{}]} colab_type="code" executionInfo={"elapsed": 57, "status": "ok", "timestamp": 1446749125010, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="P_3Fm5BpFMDF" outputId="c8e777e0-d891-4eb1-a178-9809f293cc28"
import gzip, binascii, struct, numpy
import matplotlib.pyplot as plt
with gzip.open(test_data_filename) as f:
# Print the header fields.
for field in ['magic number', 'image count', 'rows', 'columns']:
# struct.unpack reads the binary data provided by f.read.
# The format string '>i' decodes a big-endian integer, which
# is the encoding of the data.
print field, struct.unpack('>i', f.read(4))[0]
# Read the first 28x28 set of pixel values.
# Each pixel is one byte, [0, 255], a uint8.
buf = f.read(28 * 28)
image = numpy.frombuffer(buf, dtype=numpy.uint8)
# Print the first few values of image.
print 'First 10 pixels:', image[:10]
# + [markdown] colab_type="text" id="7NXKCQENNRQT"
# The first 10 pixels are all 0 values. Not very interesting, but also unsurprising. We'd expect most of the pixel values to be the background color, 0.
#
# We could print all 28 * 28 values, but what we really need to do to make sure we're reading our data properly is look at an image.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{}]} colab_type="code" executionInfo={"elapsed": 887, "status": "ok", "timestamp": 1446749126640, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="F_5w-cOoNLaG" outputId="77dabc81-e3ee-4fcf-ac72-88038494fb6c"
# %matplotlib inline
# We'll show the image and its pixel value histogram side-by-side.
_, (ax1, ax2) = plt.subplots(1, 2)
# To interpret the values as a 28x28 image, we need to reshape
# the numpy array, which is one dimensional.
ax1.imshow(image.reshape(28, 28), cmap=plt.cm.Greys);
ax2.hist(image, bins=20, range=[0,255]);
# + [markdown] colab_type="text" id="weVoVR-nN0cN"
# The large number of 0 values correspond to the background of the image, another large mass of value 255 is black, and a mix of grayscale transition values in between.
#
# Both the image and histogram look sensible. But, it's good practice when training image models to normalize values to be centered around 0.
#
# We'll do that next. The normalization code is fairly short, and it may be tempting to assume we haven't made mistakes, but we'll double-check by looking at the rendered input and histogram again. Malformed inputs are a surprisingly common source of errors when developing new models.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{}]} colab_type="code" executionInfo={"elapsed": 531, "status": "ok", "timestamp": 1446749126656, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="jc1xCZXHNKVp" outputId="bd45b3dd-438b-41db-ea8f-d202d4a09e63"
# Let's convert the uint8 image to 32 bit floats and rescale
# the values to be centered around 0, between [-0.5, 0.5].
#
# We again plot the image and histogram to check that we
# haven't mangled the data.
scaled = image.astype(numpy.float32)
scaled = (scaled - (255 / 2.0)) / 255
_, (ax1, ax2) = plt.subplots(1, 2)
ax1.imshow(scaled.reshape(28, 28), cmap=plt.cm.Greys);
ax2.hist(scaled, bins=20, range=[-0.5, 0.5]);
# + [markdown] colab_type="text" id="PlqlwkX-O0Hd"
# Great -- we've retained the correct image data while properly rescaling to the range [-0.5, 0.5].
#
# ## Reading the labels
#
# Let's next unpack the test label data. The format here is similar: a magic number followed by a count followed by the labels as `uint8` values. In more detail:
#
# [offset] [type] [value] [description]
# 0000 32 bit integer 0x00000801(2049) magic number (MSB first)
# 0004 32 bit integer 10000 number of items
# 0008 unsigned byte ?? label
# 0009 unsigned byte ?? label
# ........
# xxxx unsigned byte ?? label
#
# As with the image data, let's read the first test set value to sanity check our input path. We'll expect a 7.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{}]} colab_type="code" executionInfo={"elapsed": 90, "status": "ok", "timestamp": 1446749126903, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="d8zv9yZzQOnV" outputId="ad203b2c-f095-4035-e0cd-7869c078da3d"
with gzip.open(test_labels_filename) as f:
# Print the header fields.
for field in ['magic number', 'label count']:
print field, struct.unpack('>i', f.read(4))[0]
print 'First label:', struct.unpack('B', f.read(1))[0]
# + [markdown] colab_type="text" id="zAGrQSXCQtIm"
# Indeed, the first label of the test set is 7.
#
# ## Forming the training, testing, and validation data sets
#
# Now that we understand how to read a single element, we can read a much larger set that we'll use for training, testing, and validation.
#
# ### Image data
#
# The code below is a generalization of our prototyping above that reads the entire test and training data set.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{}]} colab_type="code" executionInfo={"elapsed": 734, "status": "ok", "timestamp": 1446749128718, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="ofFZ5oJeRMDA" outputId="ff2de90b-aed9-4ce5-db8c-9123496186b1"
IMAGE_SIZE = 28
PIXEL_DEPTH = 255
def extract_data(filename, num_images):
"""Extract the images into a 4D tensor [image index, y, x, channels].
For MNIST data, the number of channels is always 1.
Values are rescaled from [0, 255] down to [-0.5, 0.5].
"""
print 'Extracting', filename
with gzip.open(filename) as bytestream:
# Skip the magic number and dimensions; we know these values.
bytestream.read(16)
buf = bytestream.read(IMAGE_SIZE * IMAGE_SIZE * num_images)
data = numpy.frombuffer(buf, dtype=numpy.uint8).astype(numpy.float32)
data = (data - (PIXEL_DEPTH / 2.0)) / PIXEL_DEPTH
data = data.reshape(num_images, IMAGE_SIZE, IMAGE_SIZE, 1)
return data
train_data = extract_data(train_data_filename, 60000)
test_data = extract_data(test_data_filename, 10000)
# + [markdown] colab_type="text" id="0x4rwXxUR96O"
# A crucial difference here is how we `reshape` the array of pixel values. Instead of one image that's 28x28, we now have a set of 60,000 images, each one being 28x28. We also include a number of channels, which for grayscale images as we have here is 1.
#
# Let's make sure we've got the reshaping parameters right by inspecting the dimensions and the first two images. (Again, mangled input is a very common source of errors.)
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{}, {}]} colab_type="code" executionInfo={"elapsed": 400, "status": "ok", "timestamp": 1446749129657, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="0AwSo8mlSja_" outputId="11490c39-7c67-4fe5-982c-ca8278294d96"
print 'Training data shape', train_data.shape
_, (ax1, ax2) = plt.subplots(1, 2)
ax1.imshow(train_data[0].reshape(28, 28), cmap=plt.cm.Greys);
ax2.imshow(train_data[1].reshape(28, 28), cmap=plt.cm.Greys);
# + [markdown] colab_type="text" id="cwBhQ3ouTQcW"
# Looks good. Now we know how to index our full set of training and test images.
# + [markdown] colab_type="text" id="PBCB9aYxRvBi"
# ### Label data
#
# Let's move on to loading the full set of labels. As is typical in classification problems, we'll convert our input labels into a [1-hot](https://en.wikipedia.org/wiki/One-hot) encoding over a length 10 vector corresponding to 10 digits. The vector [0, 1, 0, 0, 0, 0, 0, 0, 0, 0], for example, would correspond to the digit 1.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{"item_id": 1}]} colab_type="code" executionInfo={"elapsed": 191, "status": "ok", "timestamp": 1446749131421, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="9pK1j2WlRwY9" outputId="1ca31655-e14f-405a-b266-6a6c78827af5"
NUM_LABELS = 10
def extract_labels(filename, num_images):
"""Extract the labels into a 1-hot matrix [image index, label index]."""
print 'Extracting', filename
with gzip.open(filename) as bytestream:
# Skip the magic number and count; we know these values.
bytestream.read(8)
buf = bytestream.read(1 * num_images)
labels = numpy.frombuffer(buf, dtype=numpy.uint8)
# Convert to dense 1-hot representation.
return (numpy.arange(NUM_LABELS) == labels[:, None]).astype(numpy.float32)
train_labels = extract_labels(train_labels_filename, 60000)
test_labels = extract_labels(test_labels_filename, 10000)
# + [markdown] colab_type="text" id="hb3Vaq72UUxW"
# As with our image data, we'll double-check that our 1-hot encoding of the first few values matches our expectations.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{"item_id": 1}]} colab_type="code" executionInfo={"elapsed": 127, "status": "ok", "timestamp": 1446749132853, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="uEBID71nUVj1" outputId="3f318310-18dd-49ed-9943-47b4aae7ee69"
print 'Training labels shape', train_labels.shape
print 'First label vector', train_labels[0]
print 'Second label vector', train_labels[1]
# + [markdown] colab_type="text" id="5EwtEhxRUneF"
# The 1-hot encoding looks reasonable.
#
# ### Segmenting data into training, test, and validation
#
# The final step in preparing our data is to split it into three sets: training, test, and validation. This isn't the format of the original data set, so we'll take a small slice of the training data and treat that as our validation set.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{"item_id": 1}]} colab_type="code" executionInfo={"elapsed": 176, "status": "ok", "timestamp": 1446749134110, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="e7aBYBtIVxHE" outputId="bdeae1a8-daff-4743-e594-f1d2229c0f4e"
VALIDATION_SIZE = 5000
validation_data = train_data[:VALIDATION_SIZE, :, :, :]
validation_labels = train_labels[:VALIDATION_SIZE]
train_data = train_data[VALIDATION_SIZE:, :, :, :]
train_labels = train_labels[VALIDATION_SIZE:]
train_size = train_labels.shape[0]
print 'Validation shape', validation_data.shape
print 'Train size', train_size
# + [markdown] colab_type="text" id="1JFhEH8EVj4O"
# # Defining the model
#
# Now that we've prepared our data, we're ready to define our model.
#
# The comments describe the architecture, which fairly typical of models that process image data. The raw input passes through several [convolution](https://en.wikipedia.org/wiki/Convolutional_neural_network#Convolutional_layer) and [max pooling](https://en.wikipedia.org/wiki/Convolutional_neural_network#Pooling_layer) layers with [rectified linear](https://en.wikipedia.org/wiki/Convolutional_neural_network#ReLU_layer) activations before several fully connected layers and a [softmax](https://en.wikipedia.org/wiki/Convolutional_neural_network#Loss_layer) loss for predicting the output class. During training, we use [dropout](https://en.wikipedia.org/wiki/Convolutional_neural_network#Dropout_method).
#
# We'll separate our model definition into three steps:
#
# 1. Defining the variables that will hold the trainable weights.
# 1. Defining the basic model graph structure described above. And,
# 1. Stamping out several copies of the model graph for training, testing, and validation.
#
# We'll start with the variables.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{"item_id": 1}]} colab_type="code" executionInfo={"elapsed": 2081, "status": "ok", "timestamp": 1446749138298, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="Q1VfiAzjzuK8" outputId="f53a39c9-3a52-47ca-d7a3-9f9d84eccf63"
import tensorflow as tf
# We'll bundle groups of examples during training for efficiency.
# This defines the size of the batch.
BATCH_SIZE = 60
# We have only one channel in our grayscale images.
NUM_CHANNELS = 1
# The random seed that defines initialization.
SEED = 42
# This is where training samples and labels are fed to the graph.
# These placeholder nodes will be fed a batch of training data at each
# training step, which we'll write once we define the graph structure.
train_data_node = tf.placeholder(
tf.float32,
shape=(BATCH_SIZE, IMAGE_SIZE, IMAGE_SIZE, NUM_CHANNELS))
train_labels_node = tf.placeholder(tf.float32,
shape=(BATCH_SIZE, NUM_LABELS))
# For the validation and test data, we'll just hold the entire dataset in
# one constant node.
validation_data_node = tf.constant(validation_data)
test_data_node = tf.constant(test_data)
# The variables below hold all the trainable weights. For each, the
# parameter defines how the variables will be initialized.
conv1_weights = tf.Variable(
tf.truncated_normal([5, 5, NUM_CHANNELS, 32], # 5x5 filter, depth 32.
stddev=0.1,
seed=SEED))
conv1_biases = tf.Variable(tf.zeros([32]))
conv2_weights = tf.Variable(
tf.truncated_normal([5, 5, 32, 64],
stddev=0.1,
seed=SEED))
conv2_biases = tf.Variable(tf.constant(0.1, shape=[64]))
fc1_weights = tf.Variable( # fully connected, depth 512.
tf.truncated_normal([IMAGE_SIZE / 4 * IMAGE_SIZE / 4 * 64, 512],
stddev=0.1,
seed=SEED))
fc1_biases = tf.Variable(tf.constant(0.1, shape=[512]))
fc2_weights = tf.Variable(
tf.truncated_normal([512, NUM_LABELS],
stddev=0.1,
seed=SEED))
fc2_biases = tf.Variable(tf.constant(0.1, shape=[NUM_LABELS]))
print 'Done'
# + [markdown] colab_type="text" id="QHB_u04Z4HO6"
# Now that we've defined the variables to be trained, we're ready to wire them together into a TensorFlow graph.
#
# We'll define a helper to do this, `model`, which will return copies of the graph suitable for training and testing. Note the `train` argument, which controls whether or not dropout is used in the hidden layer. (We want to use dropout only during training.)
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{"item_id": 1}]} colab_type="code" executionInfo={"elapsed": 772, "status": "ok", "timestamp": 1446749138306, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="V85_B9QF3uBp" outputId="457d3e49-73ad-4451-c196-421dd4681efc"
def model(data, train=False):
"""The Model definition."""
# 2D convolution, with 'SAME' padding (i.e. the output feature map has
# the same size as the input). Note that {strides} is a 4D array whose
# shape matches the data layout: [image index, y, x, depth].
conv = tf.nn.conv2d(data,
conv1_weights,
strides=[1, 1, 1, 1],
padding='SAME')
# Bias and rectified linear non-linearity.
relu = tf.nn.relu(tf.nn.bias_add(conv, conv1_biases))
# Max pooling. The kernel size spec ksize also follows the layout of
# the data. Here we have a pooling window of 2, and a stride of 2.
pool = tf.nn.max_pool(relu,
ksize=[1, 2, 2, 1],
strides=[1, 2, 2, 1],
padding='SAME')
conv = tf.nn.conv2d(pool,
conv2_weights,
strides=[1, 1, 1, 1],
padding='SAME')
relu = tf.nn.relu(tf.nn.bias_add(conv, conv2_biases))
pool = tf.nn.max_pool(relu,
ksize=[1, 2, 2, 1],
strides=[1, 2, 2, 1],
padding='SAME')
# Reshape the feature map cuboid into a 2D matrix to feed it to the
# fully connected layers.
pool_shape = pool.get_shape().as_list()
reshape = tf.reshape(
pool,
[pool_shape[0], pool_shape[1] * pool_shape[2] * pool_shape[3]])
# Fully connected layer. Note that the '+' operation automatically
# broadcasts the biases.
hidden = tf.nn.relu(tf.matmul(reshape, fc1_weights) + fc1_biases)
# Add a 50% dropout during training only. Dropout also scales
# activations such that no rescaling is needed at evaluation time.
if train:
hidden = tf.nn.dropout(hidden, 0.5, seed=SEED)
return tf.matmul(hidden, fc2_weights) + fc2_biases
print 'Done'
# + [markdown] colab_type="text" id="7bvEtt8C4fLC"
# Having defined the basic structure of the graph, we're ready to stamp out multiple copies for training, testing, and validation.
#
# Here, we'll do some customizations depending on which graph we're constructing. `train_prediction` holds the training graph, for which we use cross-entropy loss and weight regularization. We'll adjust the learning rate during training -- that's handled by the `exponential_decay` operation, which is itself an argument to the `MomentumOptimizer` that performs the actual training.
#
# The vaildation and prediction graphs are much simpler the generate -- we need only create copies of the model with the validation and test inputs and a softmax classifier as the output.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{"item_id": 1}]} colab_type="code" executionInfo={"elapsed": 269, "status": "ok", "timestamp": 1446749139596, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="9pR1EBNT3sCv" outputId="570681b1-f33e-4618-b742-48e12aa58132"
# Training computation: logits + cross-entropy loss.
logits = model(train_data_node, True)
loss = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(
logits, train_labels_node))
# L2 regularization for the fully connected parameters.
regularizers = (tf.nn.l2_loss(fc1_weights) + tf.nn.l2_loss(fc1_biases) +
tf.nn.l2_loss(fc2_weights) + tf.nn.l2_loss(fc2_biases))
# Add the regularization term to the loss.
loss += 5e-4 * regularizers
# Optimizer: set up a variable that's incremented once per batch and
# controls the learning rate decay.
batch = tf.Variable(0)
# Decay once per epoch, using an exponential schedule starting at 0.01.
learning_rate = tf.train.exponential_decay(
0.01, # Base learning rate.
batch * BATCH_SIZE, # Current index into the dataset.
train_size, # Decay step.
0.95, # Decay rate.
staircase=True)
# Use simple momentum for the optimization.
optimizer = tf.train.MomentumOptimizer(learning_rate,
0.9).minimize(loss,
global_step=batch)
# Predictions for the minibatch, validation set and test set.
train_prediction = tf.nn.softmax(logits)
# We'll compute them only once in a while by calling their {eval()} method.
validation_prediction = tf.nn.softmax(model(validation_data_node))
test_prediction = tf.nn.softmax(model(test_data_node))
print 'Done'
# + [markdown] colab_type="text" id="4T21uZJq5UfH"
# # Training and visualizing results
#
# Now that we have the training, test, and validation graphs, we're ready to actually go through the training loop and periodically evaluate loss and error.
#
# All of these operations take place in the context of a session. In Python, we'd write something like:
#
# with tf.Session() as s:
# ...training / test / evaluation loop...
#
# But, here, we'll want to keep the session open so we can poke at values as we work out the details of training. The TensorFlow API includes a function for this, `InteractiveSession`.
#
# We'll start by creating a session and initializing the varibles we defined above.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}} colab_type="code" id="z6Kc5iql6qxV"
# Create a new interactive session that we'll use in
# subsequent code cells.
s = tf.InteractiveSession()
# Use our newly created session as the default for
# subsequent operations.
s.as_default()
# Initialize all the variables we defined above.
tf.initialize_all_variables().run()
# + [markdown] colab_type="text" id="hcG8H-Ka6_mw"
# Now we're ready to perform operations on the graph. Let's start with one round of training. We're going to organize our training steps into batches for efficiency; i.e., training using a small set of examples at each step rather than a single example.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{"item_id": 1}]} colab_type="code" executionInfo={"elapsed": 386, "status": "ok", "timestamp": 1446749389138, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="LYVxeEox71Pg" outputId="9184b5df-009a-4b1b-e312-5be94351351f"
BATCH_SIZE = 60
# Grab the first BATCH_SIZE examples and labels.
batch_data = train_data[:BATCH_SIZE, :, :, :]
batch_labels = train_labels[:BATCH_SIZE]
# This dictionary maps the batch data (as a numpy array) to the
# node in the graph it should be fed to.
feed_dict = {train_data_node: batch_data,
train_labels_node: batch_labels}
# Run the graph and fetch some of the nodes.
_, l, lr, predictions = s.run(
[optimizer, loss, learning_rate, train_prediction],
feed_dict=feed_dict)
print 'Done'
# + [markdown] colab_type="text" id="7bL4-RNm_K-B"
# Let's take a look at the predictions. How did we do? Recall that the output will be probabilities over the possible classes, so let's look at those probabilities.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{"item_id": 1}]} colab_type="code" executionInfo={"elapsed": 160, "status": "ok", "timestamp": 1446749519023, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="2eNitV_4_ZUL" outputId="f1340dd1-255b-4523-bf62-7e3ebb361333"
print predictions[0]
# + [markdown] colab_type="text" id="X5MgraJb_eQZ"
# As expected without training, the predictions are all noise. Let's write a scoring function that picks the class with the maximum probability and compares with the example's label. We'll start by converting the probability vectors returned by the softmax into predictions we can match against the labels.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{"item_id": 1}]} colab_type="code" executionInfo={"elapsed": 220, "status": "ok", "timestamp": 1446750411574, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="wMMlUf5rCKgT" outputId="2c10e96d-52b6-47b0-b6eb-969ad462d46b"
# The highest probability in the first entry.
print 'First prediction', numpy.argmax(predictions[0])
# But, predictions is actually a list of BATCH_SIZE probability vectors.
print predictions.shape
# So, we'll take the highest probability for each vector.
print 'All predictions', numpy.argmax(predictions, 1)
# + [markdown] colab_type="text" id="8pMCIZ3_C2ni"
# Next, we can do the same thing for our labels -- using `argmax` to convert our 1-hot encoding into a digit class.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{"item_id": 1}]} colab_type="code" executionInfo={"elapsed": 232, "status": "ok", "timestamp": 1446750498351, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="kZWp4T0JDDUe" outputId="47b588cd-bc82-45c3-a5d0-8d84dc27a3be"
print 'Batch labels', numpy.argmax(batch_labels, 1)
# + [markdown] colab_type="text" id="bi5Z6whtDiht"
# Now we can compare the predicted and label classes to compute the error rate and confusion matrix for this batch.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{"item_id": 1}, {"item_id": 2}]} colab_type="code" executionInfo={"elapsed": 330, "status": "ok", "timestamp": 1446751307304, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="U4hrLW4CDtQB" outputId="720494a3-cbf9-4687-9d94-e64a33fdd78f"
correct = numpy.sum(numpy.argmax(predictions, 1) == numpy.argmax(batch_labels, 1))
total = predictions.shape[0]
print float(correct) / float(total)
confusions = numpy.zeros([10, 10], numpy.float32)
bundled = zip(numpy.argmax(predictions, 1), numpy.argmax(batch_labels, 1))
for predicted, actual in bundled:
confusions[predicted, actual] += 1
plt.grid(False)
plt.xticks(numpy.arange(NUM_LABELS))
plt.yticks(numpy.arange(NUM_LABELS))
plt.imshow(confusions, cmap=plt.cm.jet, interpolation='nearest');
# + [markdown] colab_type="text" id="iZmx_9DiDXQ3"
# Now let's wrap this up into our scoring function.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{"item_id": 1}]} colab_type="code" executionInfo={"elapsed": 178, "status": "ok", "timestamp": 1446751995007, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="DPJie7bPDaLa" outputId="a06c64ed-f95f-416f-a621-44cccdaba0f8"
def error_rate(predictions, labels):
"""Return the error rate and confusions."""
correct = numpy.sum(numpy.argmax(predictions, 1) == numpy.argmax(labels, 1))
total = predictions.shape[0]
error = 100.0 - (100 * float(correct) / float(total))
confusions = numpy.zeros([10, 10], numpy.float32)
bundled = zip(numpy.argmax(predictions, 1), numpy.argmax(labels, 1))
for predicted, actual in bundled:
confusions[predicted, actual] += 1
return error, confusions
print 'Done'
# + [markdown] colab_type="text" id="sLv22cjeB5Rd"
# We'll need to train for some time to actually see useful predicted values. Let's define a loop that will go through our data. We'll print the loss and error periodically.
#
# Here, we want to iterate over the entire data set rather than just the first batch, so we'll need to slice the data to that end.
#
# (One pass through our training set will take some time on a CPU, so be patient if you are executing this notebook.)
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}} colab_type="code" id="4cgKJrS1_vej"
# Train over the first 1/4th of our training set.
steps = int(train_size / BATCH_SIZE)
for step in xrange(steps):
# Compute the offset of the current minibatch in the data.
# Note that we could use better randomization across epochs.
offset = (step * BATCH_SIZE) % (train_size - BATCH_SIZE)
batch_data = train_data[offset:(offset + BATCH_SIZE), :, :, :]
batch_labels = train_labels[offset:(offset + BATCH_SIZE)]
# This dictionary maps the batch data (as a numpy array) to the
# node in the graph it should be fed to.
feed_dict = {train_data_node: batch_data,
train_labels_node: batch_labels}
# Run the graph and fetch some of the nodes.
_, l, lr, predictions = s.run(
[optimizer, loss, learning_rate, train_prediction],
feed_dict=feed_dict)
# Print out the loss periodically.
if step % 100 == 0:
error, _ = error_rate(predictions, batch_labels)
print 'Step %d of %d' % (step, steps)
print 'Mini-batch loss: %.5f Error: %.5f Learning rate: %.5f' % (l, error, lr)
print 'Validation error: %.1f%%' % error_rate(
validation_prediction.eval(), validation_labels)[0]
# + [markdown] colab_type="text" id="J4LskgGXIDAm"
# The error seems to have gone down. Let's evaluate the results using the test set.
#
# To help identify rare mispredictions, we'll include the raw count of each (prediction, label) pair in the confusion matrix.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{"item_id": 1}, {"item_id": 2}]} colab_type="code" executionInfo={"elapsed": 436, "status": "ok", "timestamp": 1446752934104, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="6Yh1jGFuIKc_" outputId="4e411de4-0fe2-451b-e4ca-8a4854f0db89"
test_error, confusions = error_rate(test_prediction.eval(), test_labels)
print 'Test error: %.1f%%' % test_error
plt.xlabel('Actual')
plt.ylabel('Predicted')
plt.grid(False)
plt.xticks(numpy.arange(NUM_LABELS))
plt.yticks(numpy.arange(NUM_LABELS))
plt.imshow(confusions, cmap=plt.cm.jet, interpolation='nearest');
for i, cas in enumerate(confusions):
for j, count in enumerate(cas):
if count > 0:
xoff = .07 * len(str(count))
plt.text(j-xoff, i+.2, int(count), fontsize=9, color='white')
# + [markdown] colab_type="text" id="yLnS4dGiMwI1"
# We can see here that we're mostly accurate, with some errors you might expect, e.g., '9' is often confused as '4'.
#
# Let's do another sanity check to make sure this matches roughly the distribution of our test set, e.g., it seems like we have fewer '5' values.
# + cellView="both" colab={"autoexec": {"startup": false, "wait_interval": 0}, "output_extras": [{"item_id": 1}]} colab_type="code" executionInfo={"elapsed": 352, "status": "ok", "timestamp": 1446753006584, "user": {"color": "#1FA15D", "displayName": "Michael Piatek", "isAnonymous": false, "isMe": true, "permissionId": "00327059602783983041", "photoUrl": "//lh6.googleusercontent.com/-wKJwK_OPl34/AAAAAAAAAAI/AAAAAAAAAlk/Rh3u6O2Z7ns/s50-c-k-no/photo.jpg", "sessionId": "716a6ad5e180d821", "userId": "106975671469698476657"}, "user_tz": 480} id="x5KOv1AJMgzV" outputId="2acdf737-bab6-408f-8b3c-05fa66d04fe6"
plt.xticks(numpy.arange(NUM_LABELS))
plt.hist(numpy.argmax(test_labels, 1));
# + [markdown] colab_type="text" id="E6DzLSK5M1ju"
# Indeed, we appear to have fewer 5 labels in the test set. So, on the whole, it seems like our model is learning and our early results are sensible.
#
# But, we've only done one round of training. We can greatly improve accuracy by training for longer. To try this out, just re-execute the training cell above.
| 78.455161
| 8,519
|
7b46c72054961e186c869703ea45b362c53edb69
|
py
|
python
|
boston_housing2.ipynb
|
acourtney2015/Udacity-Nano
|
['Apache-2.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# # Machine Learning Engineer Nanodegree
# ## Model Evaluation & Validation
# ## Project 1: Predicting Boston Housing Prices
#
# Welcome to the first project of the Machine Learning Engineer Nanodegree! In this notebook, some template code has already been provided for you, and you will need to implement additional functionality to successfully complete this project. You will not need to modify the included code beyond what is requested. Sections that begin with **'Implementation'** in the header indicate that the following block of code will require additional functionality which you must provide. Instructions will be provided for each section and the specifics of the implementation are marked in the code block with a 'TODO' statement. Please be sure to read the instructions carefully!
#
# In addition to implementing code, there will be questions that you must answer which relate to the project and your implementation. Each section where you will answer a question is preceded by a **'Question X'** header. Carefully read each question and provide thorough answers in the following text boxes that begin with **'Answer:'**. Your project submission will be evaluated based on your answers to each of the questions and the implementation you provide.
#
# >**Note:** Code and Markdown cells can be executed using the **Shift + Enter** keyboard shortcut. In addition, Markdown cells can be edited by typically double-clicking the cell to enter edit mode.
# ## Getting Started
# In this project, you will evaluate the performance and predictive power of a model that has been trained and tested on data collected from homes in suburbs of Boston, Massachusetts. A model trained on this data that is seen as a *good fit* could then be used to make certain predictions about a home — in particular, its monetary value. This model would prove to be invaluable for someone like a real estate agent who could make use of such information on a daily basis.
#
# The dataset for this project originates from the [UCI Machine Learning Repository](https://archive.ics.uci.edu/ml/datasets/Housing). The Boston housing data was collected in 1978 and each of the 506 entries represent aggregated data about 14 features for homes from various suburbs in Boston, Massachusetts. For the purposes of this project, the following preprocessing steps have been made to the dataset:
# - 16 data points have an `'MEDV'` value of 50.0. These data points likely contain **missing or censored values** and have been removed.
# - 1 data point has an `'RM'` value of 8.78. This data point can be considered an **outlier** and has been removed.
# - The features `'RM'`, `'LSTAT'`, `'PTRATIO'`, and `'MEDV'` are essential. The remaining **non-relevant features** have been excluded.
# - The feature `'MEDV'` has been **multiplicatively scaled** to account for 35 years of market inflation.
#
# Run the code cell below to load the Boston housing dataset, along with a few of the necessary Python libraries required for this project. You will know the dataset loaded successfully if the size of the dataset is reported.
# +
# Import libraries necessary for this project
import numpy as np
import pandas as pd
import visuals as vs # Supplementary code
from sklearn.cross_validation import ShuffleSplit
# Pretty display for notebooks
# %matplotlib inline
# Load the Boston housing dataset
data = pd.read_csv('housing.csv')
prices = data['MEDV']
features = data.drop('MEDV', axis = 1)
# Success
print "Boston housing dataset has {} data points with {} variables each.".format(*data.shape)
# -
# ## Data Exploration
# In this first section of this project, you will make a cursory investigation about the Boston housing data and provide your observations. Familiarizing yourself with the data through an explorative process is a fundamental practice to help you better understand and justify your results.
#
# Since the main goal of this project is to construct a working model which has the capability of predicting the value of houses, we will need to separate the dataset into **features** and the **target variable**. The **features**, `'RM'`, `'LSTAT'`, and `'PTRATIO'`, give us quantitative information about each data point. The **target variable**, `'MEDV'`, will be the variable we seek to predict. These are stored in `features` and `prices`, respectively.
# ### Implementation: Calculate Statistics
# For your very first coding implementation, you will calculate descriptive statistics about the Boston housing prices. Since `numpy` has already been imported for you, use this library to perform the necessary calculations. These statistics will be extremely important later on to analyze various prediction results from the constructed model.
#
# In the code cell below, you will need to implement the following:
# - Calculate the minimum, maximum, mean, median, and standard deviation of `'MEDV'`, which is stored in `prices`.
# - Store each calculation in their respective variable.
print prices.head()
print prices.describe()
print np.mean(prices)
# +
# TODO: Minimum price of the data
minimum_price = np.min(prices)
# TODO: Maximum price of the data
maximum_price = np.max(prices)
# TODO: Mean price of the data
mean_price = np.mean(prices)
# TODO: Median price of the data
median_price = np.median(prices)
# TODO: Standard deviation of prices of the data
std_price = np.std(prices)
# Show the calculated statistics
print "Statistics for Boston housing dataset:\n"
print "Minimum price: ${:,.2f}".format(minimum_price)
print "Maximum price: ${:,.2f}".format(maximum_price)
print "Mean price: ${:,.2f}".format(mean_price)
print "Median price ${:,.2f}".format(median_price)
print "Standard deviation of prices: ${:,.2f}".format(std_price)
# -
# ### Question 1 - Feature Observation
# As a reminder, we are using three features from the Boston housing dataset: `'RM'`, `'LSTAT'`, and `'PTRATIO'`. For each data point (neighborhood):
# - `'RM'` is the average number of rooms among homes in the neighborhood.
# - `'LSTAT'` is the percentage of homeowners in the neighborhood considered "lower class" (working poor).
# - `'PTRATIO'` is the ratio of students to teachers in primary and secondary schools in the neighborhood.
#
# _Using your intuition, for each of the three features above, do you think that an increase in the value of that feature would lead to an **increase** in the value of `'MEDV'` or a **decrease** in the value of `'MEDV'`? Justify your answer for each._
# **Hint:** Would you expect a home that has an `'RM'` value of 6 be worth more or less than a home that has an `'RM'` value of 7?
# **Answer: I would think that an increase in the average number of rooms in a home(RM) would lead to an increase in MEDV(prices).Justification the larger the home generally the more rooms although mansions to not fit this line of thought-square footage might be a better predictor; I would predict that an increase in the value of LSTAT would decrease MEDV-jusitification beacuse the lower the salary the less expendible income to take care of yard and home maintenence and the lower the slary the less free hours to work on the home upkeep, (although I personally do not believe this bias is true) and lastly an increase in PTRATIO would lead to a decrease in MEDV-justification- the higher the number of students in the class the less personal attention so parent would want a lower student teacher ratio. A school area that has a loweer student to teacher ratio would be more desirable and higher student to teacher ration would be less desirable**
# ----
#
# ## Developing a Model
# In this second section of the project, you will develop the tools and techniques necessary for a model to make a prediction. Being able to make accurate evaluations of each model's performance through the use of these tools and techniques helps to greatly reinforce the confidence in your predictions.
# ### Implementation: Define a Performance Metric
# It is difficult to measure the quality of a given model without quantifying its performance over training and testing. This is typically done using some type of performance metric, whether it is through calculating some type of error, the goodness of fit, or some other useful measurement. For this project, you will be calculating the [*coefficient of determination*](http://stattrek.com/statistics/dictionary.aspx?definition=coefficient_of_determination), R<sup>2</sup>, to quantify your model's performance. The coefficient of determination for a model is a useful statistic in regression analysis, as it often describes how "good" that model is at making predictions.
#
# The values for R<sup>2</sup> range from 0 to 1, which captures the percentage of squared correlation between the predicted and actual values of the **target variable**. A model with an R<sup>2</sup> of 0 always fails to predict the target variable, whereas a model with an R<sup>2</sup> of 1 perfectly predicts the target variable. Any value between 0 and 1 indicates what percentage of the target variable, using this model, can be explained by the **features**. *A model can be given a negative R<sup>2</sup> as well, which indicates that the model is no better than one that naively predicts the mean of the target variable.*
#
# For the `performance_metric` function in the code cell below, you will need to implement the following:
# - Use `r2_score` from `sklearn.metrics` to perform a performance calculation between `y_true` and `y_predict`.
# - Assign the performance score to the `score` variable.
# TODO: Import 'r2_score'
from sklearn.metrics import r2_score
def performance_metric(y_true, y_predict ):
""" Calculates and returns the performance score between
true and predicted values based on the metric chosen. """
# TODO: Calculate the performance score between 'y_true' and 'y_predict'
score = r2_score(y_true, y_predict)
# Return the score
return score
# ### Question 2 - Goodness of Fit
# Assume that a dataset contains five data points and a model made the following predictions for the target variable:
#
# | True Value | Prediction |
# | :-------------: | :--------: |
# | 3.0 | 2.5 |
# | -0.5 | 0.0 |
# | 2.0 | 2.1 |
# | 7.0 | 7.8 |
# | 4.2 | 5.3 |
# *Would you consider this model to have successfully captured the variation of the target variable? Why or why not?*
#
# Run the code cell below to use the `performance_metric` function and calculate this model's coefficient of determination.
# Calculate the performance of this model
score = performance_metric([3, -0.5, 2, 7, 4.2], [2.5, 0.0, 2.1, 7.8, 5.3])
print "Model has a coefficient of determination, R^2, of {:.3f}.".format(score)
# **Answer:I think the model has reasonably captured the variation in the data because it has fit a rsquared formula which delivers a 0.923 out of 1 or 92% correlation between the actual and predicted values. Possibility there is room to improve this**
# ### Implementation: Shuffle and Split Data
# Your next implementation requires that you take the Boston housing dataset and split the data into training and testing subsets. Typically, the data is also shuffled into a random order when creating the training and testing subsets to remove any bias in the ordering of the dataset.
#
# For the code cell below, you will need to implement the following:
# - Use `train_test_split` from `sklearn.cross_validation` to shuffle and split the `features` and `prices` data into training and testing sets.
# - Split the data into 80% training and 20% testing.
# - Set the `random_state` for `train_test_split` to a value of your choice. This ensures results are consistent.
# - Assign the train and testing splits to `X_train`, `X_test`, `y_train`, and `y_test`.
from sklearn import cross_validation
from sklearn.cross_validation import train_test_split
# +
# TODO: Import 'train_test_split'
# TODO: Shuffle and split the data into training and testing subsets
def shuffle_slpit_data(X, y):
X_train, X_test, y_train, y_test = train_test_split(features, prices, test_size=0.2, random_state=1)
return X_train, y_train, X_test, y_test
# Success
print "Angie, your data has been split"
# -
# ### Question 3 - Training and Testing
# *What is the benefit to splitting a dataset into some ratio of training and testing subsets for a learning algorithm?*
# **Hint:** What could go wrong with not having a way to test your model?
# **Answer: Benefit: Splitting the data into multiple sets allows the alorgorithm to be tested against a set it has not seen before. It tests the robustness of the algorithm against unseen data such as would be encountered in the real world.What could go wrong if you do not have a way to test your model is that the model can learn and perform very well using all the data and the only way that you would know that it actually performs poorly on unseen data is when you go into production against unseen data and it performs poorly. If you had split and tested against the unseen held back data then you would have seen this during model creation. **
# ----
#
# ## Analyzing Model Performance
# In this third section of the project, you'll take a look at several models' learning and testing performances on various subsets of training data. Additionally, you'll investigate one particular algorithm with an increasing `'max_depth'` parameter on the full training set to observe how model complexity affects performance. Graphing your model's performance based on varying criteria can be beneficial in the analysis process, such as visualizing behavior that may not have been apparent from the results alone.
# ### Learning Curves
# The following code cell produces four graphs for a decision tree model with different maximum depths. Each graph visualizes the learning curves of the model for both training and testing as the size of the training set is increased. Note that the shaded region of a learning curve denotes the uncertainty of that curve (measured as the standard deviation). The model is scored on both the training and testing sets using R<sup>2</sup>, the coefficient of determination.
#
# Run the code cell below and use these graphs to answer the following question.
# Produce learning curves for varying training set sizes and maximum depths
vs.ModelLearning(features, prices)
# ### Question 4 - Learning the Data
# *Choose one of the graphs above and state the maximum depth for the model. What happens to the score of the training curve as more training points are added? What about the testing curve? Would having more training points benefit the model?*
# **Hint:** Are the learning curves converging to particular scores?
# **Answer: max_depth = 3 model. As the training points increase the scores goes from one to near 0.8. The testing score increases from just above 0.6 to just below 0.8. It appears having more training points would not benefit max depth 1,3 or 10. In the Max depth 6 graph the final data point score appears to possibly be less than the point before, so this depth could possible perform worse as more training points are added. In max_depth 3 graph it appears the learning curves might be converging towards the score of 0.8. The other graphs also appear to be converging except max_depth = 10 which appears to be more parallel. **
# ### Complexity Curves
# The following code cell produces a graph for a decision tree model that has been trained and validated on the training data using different maximum depths. The graph produces two complexity curves — one for training and one for validation. Similar to the **learning curves**, the shaded regions of both the complexity curves denote the uncertainty in those curves, and the model is scored on both the training and validation sets using the `performance_metric` function.
#
# Run the code cell below and use this graph to answer the following two questions.
vs.ModelComplexity(X_train, y_train)
# ### Question 5 - Bias-Variance Tradeoff
# *When the model is trained with a maximum depth of 1, does the model suffer from high bias or from high variance? How about when the model is trained with a maximum depth of 10? What visual cues in the graph justify your conclusions?*
# **Hint:** How do you know when a model is suffering from high bias or high variance?
# **Answer: I had to lok up the definitions of high bias and high variance from http://www.astroml.org/sklearn_tutorial/practical.html to help answer this question. High bias is underfitting the data. Indicated when both the training and cross-validation errors are very high which means the r2 is low. If this is the case, I can add more features,use a more sophisticated model, use fewer samples or decrease any regularization (penalty terms) that may be in the algorithm. They noted that adding more training data will not help matters if both lines have converged to a relatively high error. High Variance is overfitting. Indicated when the training error is much less than the cross-validation error. If this is the case, adding more training data may not help matters, the training error will climb and the cross validation error will decrease until they begin to converge and both lines tend to converge to a relatively high error. To fix high variance I can use fewer features,use more training samples and/or increase regularization (add penalty terms).
# Looking at the graphs here it appears that when max depth is 1 both the training error is high and the validation error is high (low r2) which indicates high bias (under fitting). When the max depth is 10 the training error is low (high r2) but the validation error is high (lowr2) which indicates high variance. The visual clues are the score points in relationship to the max depth. The shaded uncertainty appears relatively consistant decreasing on the traing score at max depth 10 which makes sense if it is overfitting the data**
# ### Question 6 - Best-Guess Optimal Model
# *Which maximum depth do you think results in a model that best generalizes to unseen data? What intuition lead you to this answer?*
# **Answer: I would guess max depth 4 based upon the graph. It appears that overfitting (high variance) is begining at max depth 6 and the error does not appear improved at max depth 5 as compared to 4 **
# -----
#
# ## Evaluating Model Performance
# In this final section of the project, you will construct a model and make a prediction on the client's feature set using an optimized model from `fit_model`.
# ### Question 7 - Grid Search
# *What is the grid search technique and how it can be applied to optimize a learning algorithm?*
# **Answer: To answer this question I went to http://scikit-learn.org/stable/modules/generated/sklearn.grid_search.GridSearchCV.html and http://scikit-learn.org/stable/modules/generated/sklearn.metrics.make_scorer.html#sklearn.metrics.make_scorer The grid search technique searches over the parameter values to return estimators. Because it can help determine the best parameters for the model it can be useful for parameter tuning**
# ### Question 8 - Cross-Validation
# *What is the k-fold cross-validation training technique? What benefit does this technique provide for grid search when optimizing a model?*
# **Hint:** Much like the reasoning behind having a testing set, what could go wrong with using grid search without a cross-validated set?
# **Answer:I had to research to answer this question. I used https://www.cs.cmu.edu/~schneide/tut5/node42.html to find the definition of the technique: "K-fold cross validation is one way to improve over the holdout method. The data set is divided into k subsets, and the holdout method is repeated k times. Each time, one of the k subsets is used as the test set and the other k-1 subsets are put together to form a training set" It seems to benefit grid search because each data point is in the test set exactly once (and in the training set k-1 times.) which then implies that using grid search without a cross validation set you could overfit or increase the variance **
# ### Implementation: Fitting a Model
# Your final implementation requires that you bring everything together and train a model using the **decision tree algorithm**. To ensure that you are producing an optimized model, you will train the model using the grid search technique to optimize the `'max_depth'` parameter for the decision tree. The `'max_depth'` parameter can be thought of as how many questions the decision tree algorithm is allowed to ask about the data before making a prediction. Decision trees are part of a class of algorithms called *supervised learning algorithms*.
#
# For the `fit_model` function in the code cell below, you will need to implement the following:
# - Use [`DecisionTreeRegressor`](http://scikit-learn.org/stable/modules/generated/sklearn.tree.DecisionTreeRegressor.html) from `sklearn.tree` to create a decision tree regressor object.
# - Assign this object to the `'regressor'` variable.
# - Create a dictionary for `'max_depth'` with the values from 1 to 10, and assign this to the `'params'` variable.
# - Use [`make_scorer`](http://scikit-learn.org/stable/modules/generated/sklearn.metrics.make_scorer.html) from `sklearn.metrics` to create a scoring function object.
# - Pass the `performance_metric` function as a parameter to the object.
# - Assign this scoring function to the `'scoring_fnc'` variable.
# - Use [`GridSearchCV`](http://scikit-learn.org/stable/modules/generated/sklearn.grid_search.GridSearchCV.html) from `sklearn.grid_search` to create a grid search object.
# - Pass the variables `'regressor'`, `'params'`, `'scoring_fnc'`, and `'cv_sets'` as parameters to the object.
# - Assign the `GridSearchCV` object to the `'grid'` variable.
from sklearn import grid_search
from sklearn import tree
from sklearn.metrics import mean_squared_error, make_scorer
from sklearn.grid_search import GridSearchCV
from sklearn.tree import DecisionTreeRegressor
# +
# TODO: Import 'make_scorer', 'DecisionTreeRegressor', and 'GridSearchCV'
def fit_model(X, y):
""" Performs grid search over the 'max_depth' parameter for a
decision tree regressor trained on the input data [X, y]. """
# Create cross-validation sets from the training data
cv_sets = ShuffleSplit(X.shape[0], n_iter = 10, test_size = 0.20, random_state = 0)
# TODO: Create a decision tree regressor object
regressor = DecisionTreeRegressor()
# TODO: Create a dictionary for the parameter 'max_depth' with a range from 1 to 10
params = {'max_depth': (1,2,3,4,5,6,7,8,9,10)}
# TODO: Transform 'performance_metric' into a scoring function using 'make_scorer'
scoring_function = make_scorer(score_func = mean_squared_error, greater_is_better = False)
# TODO: Create the grid search object
grid = GridSearchCV(estimator = regressor,param_grid = params,scoring = scoring_function,cv = cv_sets)
# Fit the grid search object to the data to compute the optimal model
grid = grid.fit(X, y)
# Return the optimal model after fitting the data
return grid.best_estimator_
try:
grid = fit_model(features, prices)
print "Yea, fit a model!", grid
except:
print "Something went wrong with fitting a model."
# -
# ### Making Predictions
# Once a model has been trained on a given set of data, it can now be used to make predictions on new sets of input data. In the case of a *decision tree regressor*, the model has learned *what the best questions to ask about the input data are*, and can respond with a prediction for the **target variable**. You can use these predictions to gain information about data where the value of the target variable is unknown — such as data the model was not trained on.
# ### Question 9 - Optimal Model
# _What maximum depth does the optimal model have? How does this result compare to your guess in **Question 6**?_
#
# Run the code block below to fit the decision tree regressor to the training data and produce an optimal model.
# +
# Fit the training data to the model using grid search
reg = fit_model(X_train, y_train)
# Produce the value for 'max_depth'
print "Parameter 'max_depth' is {} for the optimal model.".format(reg.get_params()['max_depth'])
# -
# **Answer: It choose max depth 5 and I quessed that max depth would be 4 for the optimal model**
# ### Question 10 - Predicting Selling Prices
# Imagine that you were a real estate agent in the Boston area looking to use this model to help price homes owned by your clients that they wish to sell. You have collected the following information from three of your clients:
#
# | Feature | Client 1 | Client 2 | Client 3 |
# | :---: | :---: | :---: | :---: |
# | Total number of rooms in home | 5 rooms | 4 rooms | 8 rooms |
# | Neighborhood poverty level (as %) | 17% | 32% | 3% |
# | Student-teacher ratio of nearby schools | 15-to-1 | 22-to-1 | 12-to-1 |
# *What price would you recommend each client sell his/her home at? Do these prices seem reasonable given the values for the respective features?*
# **Hint:** Use the statistics you calculated in the **Data Exploration** section to help justify your response.
#
# Run the code block below to have your optimized model make predictions for each client's home.
# +
# Produce a matrix for client data
client_data = [[5, 17, 15], # Client 1
[4, 32, 22], # Client 2
[8, 3, 12]] # Client 3
# Show predictions
for i, price in enumerate(reg.predict(client_data)):
print "Predicted selling price for Client {}'s home: ${:,.2f}".format(i+1, price)
# -
# **Answer: The statistics show that the:
# Minimum price: 105,000.00
# Maximum price: 1,024,800.00
# Mean price: 454,342.94
# Median price 438,900.00
# Standard deviation of prices: 165,171.13 so the values appear within reason based upon the statistics of the sales prices the poverty levels and the student teacher ratios. Although client 2 home has an extremely low valuation it would be interesting to see if the current comps maintain that low level over time. To elaborate as requested: client 1 has 5 rooms, a median poverty level and a median student teacher povery level as compared to the other 2 so a predicted selling price of 419,000 appears within the median of the 3 estimates(consistant with the median statistical home price), as is the case for home 2 which has fewer rooms than client 1 but the poverty level is the highest of the 3 and the student teacher ratio is also the highest of the 3 so the lowest valuation matches the the expectation of closest to the minimum price of the 3; also client 3 has 8 rooms, 3% poverty level and the best student teacher ratio which should place it closest to the maximum price of the three. In summation, the features of the clients homes(room.poverty level,student teacher level) which are relatively consistant with anticipated poor valuations and are consistant in their relative comparison to the statistical results also based upon rooms,poverty level and student/teacher ratio. The model appears consistant with the data and features presented. Although one cannot say of this result that these three features are the best predictor of home prices in totality **
# ### Sensitivity
# An optimal model is not necessarily a robust model. Sometimes, a model is either too complex or too simple to sufficiently generalize to new data. Sometimes, a model could use a learning algorithm that is not appropriate for the structure of the data given. Other times, the data itself could be too noisy or contain too few samples to allow a model to adequately capture the target variable — i.e., the model is underfitted. Run the code cell below to run the `fit_model` function ten times with different training and testing sets to see how the prediction for a specific client changes with the data it's trained on.
vs.PredictTrials(features, prices, fit_model, client_data)
# ### Question 11 - Applicability
# *In a few sentences, discuss whether the constructed model should or should not be used in a real-world setting.*
# **Hint:** Some questions to answering:
# - *How relevant today is data that was collected from 1978?*
# - *Are the features present in the data sufficient to describe a home?*
# - *Is the model robust enough to make consistent predictions?*
# - *Would data collected in an urban city like Boston be applicable in a rural city?*
# ***Answer: I don't think the constructed model can be used today in a real world setting. Pricing data from the 1978 would be irrelevant for a pricing prediction today unless the model consistantly updated and trained to keep up with pricing changes dynamically. The features in the data are realatively sufficient to descibe a home but their are variables (features that can make a sugnificant difference in a home price such as a pool or tennis court/putting green/proximity to ocean/view etc. The model appears to have a 69,044.61 difference in price range in its predictions. That is very significant and it would be better to have a model that predicts within a maximum 10,000 range for the real estate market. The data collected in a urban city like boston would have difficulty being applicable in a rural city or in a city like New York where 1 bedroom can be 1 million Or Los Angeles where 2 rooms could be a 5,00 square foot loft. For realestate, I think the model should betrained on data from that individual city if possible to increase accuracy and robustness of the model ***
| 80.895604
| 1,482
|
8ff936ee2d6d178cea151cbb8ac2686158ab533f
|
py
|
python
|
sentiment_classification_imdb_reviews_using_TFHub.ipynb
|
sriran/first-contributions
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/sriran/first-contributions/blob/master/sentiment_classification_imdb_reviews_using_TFHub.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + id="Nff-Zo03ti-v"
import numpy as np
import os
import tensorflow as tf
import tensorflow_datasets as tfds
import tensorflow_hub as hub
import nltk
# + id="JcE7oaOIYDcT" outputId="f8a133eb-2963-4933-91ba-e95c283b4d7d" colab={"base_uri": "https://localhost:8080/", "height": 924, "referenced_widgets": ["e159933ea330488f82130ebf6019c1b9", "1d8e05b954f0408db865e813dac72c13", "8e80445b8720410c8d791d119d966a04", "3c74bf3809e64202a0beb65251e83f36", "88ad6d6bf6114e8fa000ce71dd97efc4", "a828515d034b4ebb9a495e69ee49dd96", "20775160a2714a6087ae56c37bf2bd54", "c79be3dd32a344b0b145312978574ab0", "713c179cf2f045699658621270b5cdd0", "eb8d3f3780504bf9a662b8c7c6db86cb", "68d244a9c6094103ba2c8464a504dd0f", "d8b245361c8145e988a871ecaa23589e", "16686bfc1be64d6098adae5fadaa6185", "267f1b0b36414647a4dd4ee509d59cde", "2ca3d03cb42a4e6db4faa1de17d23a6a", "de65b008e4cf4608a9c6554245ebb710", "ddacb5a9c385477dbc38d492f1cb0b86", "d8c6c79044e24f30a60eec33d01d7d9d", "eafd5d702ba44469b6a67384cac9af54", "0a22d64df0e0435f86ec090e482533ef", "b2a3438d415c4954b92a9fa86abe9228", "72de6439f7194b4cb6182e69114ce13b", "45e53668093f4cf2b67066afa4f71493", "b0b083ce7df64dbf9d2e086532d8f4e1", "fc0245f1c6e44c20ba7e6bbec1d586b4", "6e2446decebb4483ad71de3b65aebe40", "8077c5d5c56d4979b87b073cddd4f72d", "bb34bd5b14cb485b8bf3f0a630209980", "cf7bbdfb79544964a124524a678bbd12", "233d3b167c634dbab99cf18780bf658d", "59fed89f605642639961b852aa609be4", "42a4a8d9c8df486fb3948c55fb57c605", "17915e7a2c654b08874823d126ae7d48", "e0330f16da7c4b3b8656820f820a28cd", "dcbc5a28848a4913b4831228f9d4b391", "7716b2373e6648cf835a551081bcc981", "306f3a35039841b8b297ffa1aa4c0f03", "164b22db53a04d7aac1d0e14dd24429e", "8c2cb509aa20478c8dcc687a071038f8", "fb9c2cd30e9f4c0bb9d1c33650757838", "d80a6fe0f1f147ee9f8c7bc2974c2ad2", "d265d80152a94004b85cbf8f82f734b1", "136eab9dfe2143d5bbaeed15fa16a08d", "c3da9b3e973e4ef9b7c819c5edeec8f2", "766b49da07dc4619889fd327cbfe621f", "10d2f76242984255a1e9b96183cd512d", "120e86a69fd64f6ea2bdbf19e0869ed1", "824465633d69477595d5a99ad28831f9", "6588acb06f6146beadcfc87c198f9ee1", "f47a618c7c804e63820c3d95a6743e48", "b0a4ac0ce792467fa949e0aa508c3d55", "26a17391b14940479feefa297909dc7a", "0a14bbe2e92646b59cc93143c98bc051", "e46a524dad45466fad7558bb9734a008", "9024506c79844ced8c43ec6ab5d42a78", "0fdb391f3232416f9593dd4d7510bb07", "991c9099d10c4212bebfd82f0b771937", "5993a9f0e1864bd18150eed331b0c11d", "7bf04c390e394c70bb06e6bf2cbbfc9b", "cecd755378a04bda82a95aaa1218da81", "5ae1a9d695ed4a3b9d1d965c7044ae41", "a01cdc88828f4aa088a8ca3dd6fcfe14", "d7f7f724d5f04eceb05c47e927589062", "4e9e8efd7f7745a4bd5b26c682f7fd04"]}
(train_examples,test_examples),ds_info=tfds.load('imdb_reviews',split=['train','test'],batch_size=-1,as_supervised=True,with_info=True)
print('info', ds_info)
# + id="3y2RS7FqNPiQ"
train_data,train_labels=tfds.as_numpy(train_examples)
test_data,test_labels=tfds.as_numpy(test_examples)
# + id="-PvUepRRhEZ9" outputId="da705173-0ad0-445e-a2e4-c0a1bf50b195" colab={"base_uri": "https://localhost:8080/", "height": 598}
embedding='https://tfhub.dev/google/nnlm-en-dim50/2'
hub_layer=hub.KerasLayer(embedding,input_shape=[],dtype=tf.string,trainable=True)
hub_layer(train_data[:3])
# + id="EBM6lVNwxtO2" outputId="55a43316-3536-4517-8d25-eaca43b87292" colab={"base_uri": "https://localhost:8080/", "height": 271}
model=tf.keras.Sequential()
model.add(hub_layer)
#model.add(tf.keras.layers.Dropout(0.5))
model.add(tf.keras.layers.Dense(16,activation='relu'))
#model.add(tf.keras.layers.Dropout(0.2))
model.add(tf.keras.layers.Dense(1,activation='sigmoid'))
model.summary()
# + id="dnhJLB3V0b8D"
model.compile(optimizer='adam',
loss=tf.losses.BinaryCrossentropy(from_logits=True),
metrics=['accuracy'])
# + id="FBMPbr1o0gsE"
x_val=train_data[:10000]
p_x_train=train_data[10000:]
y_val=train_labels[:10000]
p_y_train=train_labels[10000:]
# + id="e7M1hVmE2RqF" outputId="233ac216-fd89-4b7e-ee12-30d143a83ca5" colab={"base_uri": "https://localhost:8080/", "height": 35}
print(y_val[:3])
# + id="YpLd0KC303WE" outputId="dd20e8d5-ceb4-4a4b-8177-3ad6f3181388" colab={"base_uri": "https://localhost:8080/", "height": 380}
history = model.fit(p_x_train,
p_y_train,
epochs=10,
batch_size=512,
validation_data=(x_val, y_val)
)
# + id="itW-Ud6h4HK3" outputId="079d519d-1960-4732-a19a-b3e1b0a9dca6" colab={"base_uri": "https://localhost:8080/", "height": 53}
results = model.evaluate(test_data, test_labels)
print(results)
# + id="l4VX0N-Q4gA5" outputId="27dd878b-488b-4a38-ee5c-f1cd95a5b6ed" colab={"base_uri": "https://localhost:8080/", "height": 35}
history_dict = history.history
history_dict.keys()
# + id="-WYmMqIR4k0w" outputId="55896825-44d3-4fd7-8e65-32cd899eb033" colab={"base_uri": "https://localhost:8080/", "height": 573}
import matplotlib.pyplot as plt
acc = history_dict['accuracy']
val_acc = history_dict['val_accuracy']
loss = history_dict['loss']
val_loss = history_dict['val_loss']
epochs = range(1, len(acc) + 1)
# "bo" is for "blue dot"
plt.plot(epochs, loss, 'bo', label='Training loss')
# b is for "solid blue line"
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.show()
plt.clf() # clear figure
plt.plot(epochs, acc, 'bo', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.legend()
plt.show()
| 52.478261
| 2,458
|
8f18f592aa5466dc2401e174c632a78510d2b445
|
py
|
python
|
Modulo3Simat/2_Ajuste de curvas/.ipynb_checkpoints/SubirInterpolacionYAjustes-checkpoint.ipynb
|
IntroCursos/MachingLearning
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] toc="true"
# # Table of Contents
# <div class="toc" style="margin-top: 1em;"><ul class="toc-item" id="toc-level0"><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#Ajuste-de-curvas" data-toc-modified-id="Ajuste-de-curvas-1"><span class="toc-item-num">1 </span>Ajuste de curvas</a></span></li><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#Interpolación" data-toc-modified-id="Interpolación-2"><span class="toc-item-num">2 </span>Interpolación</a></span><ul class="toc-item"><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#Por-ejemplo:" data-toc-modified-id="Por-ejemplo:-2.1"><span class="toc-item-num">2.1 </span>Por ejemplo:</a></span><ul class="toc-item"><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#Interpolación-lineal" data-toc-modified-id="Interpolación-lineal-2.1.1"><span class="toc-item-num">2.1.1 </span>Interpolación lineal</a></span></li><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#Interpolación-polinomial" data-toc-modified-id="Interpolación-polinomial-2.1.2"><span class="toc-item-num">2.1.2 </span>Interpolación polinomial</a></span></li></ul></li><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#Ejercicio-de-Catenaria-de-un-Puente" data-toc-modified-id="Ejercicio-de-Catenaria-de-un-Puente-2.2"><span class="toc-item-num">2.2 </span>Ejercicio de Catenaria de un Puente</a></span><ul class="toc-item"><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#-Resultado:-" data-toc-modified-id="-Resultado:--2.2.1"><span class="toc-item-num">2.2.1 </span><font color="red"> Resultado: </font></a></span></li><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#Función-de-Interpolación-polinómica-(barycentric_interpolate)" data-toc-modified-id="Función-de-Interpolación-polinómica-(barycentric_interpolate)-2.2.2"><span class="toc-item-num">2.2.2 </span>Función de Interpolación polinómica (barycentric_interpolate)</a></span></li><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#Resolviendo-la-Catenaria-del-puente-con-Barycentric" data-toc-modified-id="Resolviendo-la-Catenaria-del-puente-con-Barycentric-2.2.3"><span class="toc-item-num">2.2.3 </span>Resolviendo la Catenaria del puente con Barycentric</a></span></li><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#Resolviendo-el-problema-de-la-temperatura-de-ebullicion-de-la-acetona-$(C_3H_6O)$" data-toc-modified-id="Resolviendo-el-problema-de-la-temperatura-de-ebullicion-de-la-acetona-$(C_3H_6O)$-2.2.4"><span class="toc-item-num">2.2.4 </span>Resolviendo el problema de la temperatura de ebullicion de la acetona $(C_3H_6O)$</a></span></li></ul></li><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#Fenómeno-de-Runge" data-toc-modified-id="Fenómeno-de-Runge-2.3"><span class="toc-item-num">2.3 </span>Fenómeno de Runge</a></span></li><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#Trazadores" data-toc-modified-id="Trazadores-2.4"><span class="toc-item-num">2.4 </span>Trazadores</a></span><ul class="toc-item"><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#Libreria-interp1d" data-toc-modified-id="Libreria-interp1d-2.4.1"><span class="toc-item-num">2.4.1 </span>Libreria interp1d</a></span></li><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#Resolviendo-la-Catenaria-del-puente-con-interp1d" data-toc-modified-id="Resolviendo-la-Catenaria-del-puente-con-interp1d-2.4.2"><span class="toc-item-num">2.4.2 </span>Resolviendo la Catenaria del puente con interp1d</a></span></li></ul></li><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#B-spline" data-toc-modified-id="B-spline-2.5"><span class="toc-item-num">2.5 </span>B-spline</a></span></li></ul></li><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#Ajuste-de-curvas" data-toc-modified-id="Ajuste-de-curvas-3"><span class="toc-item-num">3 </span>Ajuste de curvas</a></span><ul class="toc-item"><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#Ejercicio-de-Catenaria-de-un-Puente" data-toc-modified-id="Ejercicio-de-Catenaria-de-un-Puente-3.1"><span class="toc-item-num">3.1 </span>Ejercicio de Catenaria de un Puente</a></span></li></ul></li><li><span><a href="http://localhost:8888/notebooks/1Jupyter/Iteso_Simulacion2017/Simulaciones/Modulo3Simat/2_Ajuste%20de%20curvas/SubirInterpolacionYAjustes.ipynb#Ejercicio" data-toc-modified-id="Ejercicio-4"><span class="toc-item-num">4 </span>Ejercicio</a></span></li></ul></div>
# -
# # Ajuste de curvas
#
# <p class="importante"> El ajuste de **curvas** consiste en encontrar una curva que contenga una serie de puntos y que posiblemente cumpla una serie de restricciones adicionales.
# </p>
#
# - Veremos una introducción tanto a la interpolación **(cuando se espera un ajuste exacto a determinadas restricciones)**
#
# - Y al ajuste de curvas/análisis de regresión **(cuando se permite una aproximación)**.
#
# <p class="importante">
# $$ \sum_{x=1}^{n}{x+2 -x*2 -x^2}$$
# </p>
# $$ \sum_{x=1}^{n}{x+2 -x*2 -x^2}$$
#
# <b>operaciones comunes</b>
#
# <br>
# Dentro de este sitio podrás tener acceso a gran variedad de <b>recetas</b> de preparacion sencilla,
# aprenderás como realizar <b>operaciones comunes</b> dentro de una cocina y podras plantear tus preguntas
# en nuestro <b>consultorio</b> gourmet donde nuestro cocinero virtual podrá ayudarte con tus proyectos culinarios.
# También te será posible adquirir los <b>libros</b> gastronómicos más populares al precio més accesible...
# <br>
#
#
# $$
# \begin{eqnarray}
# m &=& \ddot{x} + k x + B \dot{x} = 0 \\
# y &=& x^2
# \end{eqnarray}
# $$
# # Interpolación
#
# [comment]: <> (This is a comment, it will not be included)
#
# Se denomina interpolación a la obtención de nuevos puntos partiendo del conocimiento de un conjunto discreto de puntos.
#
#
# ## Por ejemplo:
#
#
# La tabla siguiente presenta la temperatura de ebullicion de la acetona $(C_3H_6O)$ a diferentes presiones.
#
#
#
# |Puntos| 0 |1 |2 |3 |4 |5 |6 |
# |--- |--- |--- |--- |--- |--- |--- |
# |$ T(^{\circ} C)$|56.5 |78.5 |113.0 |144.5 |181.0 |205.0 |214.5 |
# |$P (atm)$|1 |2 |5 |10 |20 |30 |40 |
#
#
# Supóngase que sólo se dispusiera de la segunda tabla
#
#
# |Puntos|0 |1 |2 |3 |
# |---|--- |--- |--- |---
# |$ T(^{\circ} C)$|56.5 |113.0 |181.0 |214.5 |
# |$P (atm)$|1 |5 |20 |40 |
#
# <font color="red">
# Y se desea calcular la temperatura de ebullición de la acetona a 2 atm de presión.</font>
# +
# %matplotlib inline
import numpy as np
import matplotlib.pylab as plt
import matplotlib as mpl
mpl.rc("font",size=12)
To = np.array([56.5,78.5,113.0,144.5,181.0,205.0,214.5])
Po = np.array([1,2,5,10,20,30,40])
T = np.array([56.5,113.0,181.0,214.5])
P = np.array([1,5,20,40])
# +
fig = plt.figure(figsize=(14,6))
plt.subplot(1,2,1)
plt.scatter(Po,To)
plt.xlabel("P(atm)",fontsize=16)
plt.ylabel("$ T(^{\circ} C)$")
plt.title("Tabla original")
plt.grid()
plt.subplot(1,2,2)
plt.scatter(P,T)
plt.xlabel("P(atm)",fontsize=16)
plt.ylabel("$ T(^{\circ} C)$")
plt.title("Tabla reducida")
plt.grid()
# -
# ### Interpolación lineal
# Una forma muy común de resolver este problema es <font color="blue">sustituir los puntos (0) y (1) en la ecuación de la linea recta: $p(x) = a_0 + a_1x$</font> de tal modo que resultan dos ecuaciones con dos incógnitas que son $a_0$ y $a_1$.
#
# <font color="blue">Con la solución del sistema se consigue una aproximación polinomial de primer grado, lo que permite efectuar interpolaciones lineales;</font> es decir, se sustituye el punto (0) en la ecuación de la linea recta y se obtiene
#
# $$56.5 = a_0 + 1a_1 $$
#
# y al sustituir el punto (1)
#
# $$113 = a_0 + 5a_1 $$
#
# <font color="red">sistema que al resolverse da $a_0 = ?$ y $a_1 = ?$</font>
#
# <!-- $a_0 = 42.375$ y $a_1 = 14.125$ -->
#
# Por lo tanto, estos valores generan la ecuación
#
# <!-- ## $$p(x) = 42.375 + 14.125x $$ -->
#
# $$p(x) = a_0 + a_1x $$
#
# La ecuación resultante puede emplearse para aproximar la temperatura cuando la presión es conocida
#
# Al sustituir la presión $x = 2 atm$ se obtiene una temperatura de $70.6^\circ C$. A este proceso se le conoce como interpolación.
#
#
# <font color="red"> Resultado: </font>
#
# <!--
# A = np.array([[1, 1],
# [1, 5]])
# b = np.array([56.5, 113])
#
# x = np.linalg.solve(A, b)
# x
# $a_0 = 42.375$ y $a_1 = 14.125$
# ## $$p(x) = 42.375 + 14.125x $$
# -->
#
# $$p(x) = 42.375 + 14.125x $$
# Utilizando numpy lo podian resolver!!!
plt.plot(P[0:2],T[0:2])
plt.scatter(P[0:2],T[0:2])
plt.scatter(2,70.2,s=80,color="green")
plt.plot([2,2],[50,70.2],color="red")
plt.xlabel("P(atm)")
plt.ylabel("$ T(^{\circ} C)$")
plt.text(P[0],T[0],"$P_0$", fontsize=20,ha='right', va='bottom')
plt.text(P[1],T[1],"$P_1$", fontsize=20,ha='right', va='bottom')
plt.grid()
plt.axis([0,6,50,120])
# ### Interpolación polinomial
#
#
# <font color="blue"> Si se quisiera una aproximación mejor al valor "verdadero" de la temperatura buscada, podrían unirse más puntos de la tabla con una curva suave (sin picos),
# </font>
#
# por ejemplo tres puntos (0),(1) y (2) y graficamente obtener T correspodiente a P=2 atm
#
# Este polinomio es una parábola y tiene la forma general
#
# $$ p_2(x) = a_0 +a_1x + a_2x^2$$
#
# <font color="red">sistema que al resolverse da $a_0 = ?$, $a_1 = ?$ y $a_2=?$</font>
# +
x = np.arange(0,20,.1)
plt.plot(x,p2(x))
plt.scatter(P[0:3],T[0:3])
plt.scatter(2,70.2,s=80,color="green")
plt.plot([2,2],[50,70.2],color="red")
plt.xlabel("P(atm)")
plt.ylabel("$ T(^{\circ} C)$")
plt.title("Parábola")
plt.text(P[0],T[0],"$P_0$", fontsize=20,ha='right', va='bottom')
plt.text(P[1],T[1],"$P_1$", fontsize=20,ha='right', va='bottom')
plt.text(P[2],T[2],"$P_2$", fontsize=20,ha='right', va='bottom')
plt.axis([0,22,50,200])
plt.grid()
# -
p2(2)
# En general, si se desea aproximar una función con **un polinomio de grado $n$, se necesitan $n+1$ puntos**, que sustituidos en la ecuación polinomial de grado $n$ :
#
# $$ p(x) = a_0 +a_1x +a_2x^2 + \ldots + a_nx^n $$
# ## Ejercicio de Catenaria de un Puente
# 
# ### <font color="red"> Resultado: </font>
# +
Poli_Puente = lambda x: a[0] +a[1]*x + a[2]*(x**2)
for i in range(0,16):
print (i,Poli_Puente(i))
# -
# ### Función de Interpolación polinómica (barycentric_interpolate)
from scipy.interpolate import barycentric_interpolate
# ### Resolviendo la Catenaria del puente con Barycentric
# +
Tirante = np.array([0,13,15])
Altura = np.array([20,20,22])
Tirantes_New = np.arange(0,16,1)
y = barycentric_interpolate(Tirante, Altura, Tirantes_New)
plt.plot(Tirante, Altura, "o", Tirantes_New, y, "-")
plt.legend(["data", "Interpolación", "Real"], loc="best")
# -
# ### Resolviendo el problema de la temperatura de ebullicion de la acetona $(C_3H_6O)$
# +
To = np.array([56.5,78.5,113.0,144.5,181.0,205.0,214.5])
Po = np.array([1,2,5,10,20,30,40])
T = np.array([56.5,113.0,181.0,214.5])
P = np.array([1,5,20,40])
x= np.linspace(1,40,num=40 ,endpoint=True)
y =
plt.plot(P, T, "o", x, y, "-", Po, To, "--")
plt.legend(["data", "Interpolación", "Real"], loc="best")
# -
# ## Fenómeno de Runge
#
# Lo primero que vamos a hacer va a ser desterrar la idea de que, sea cual sea el número de puntos que tengamos, podemos construir un polinomio que pase por todos ellos «y que lo haga bien».
#
# Si tenemos N puntos nuestro polinomio tendrá que ser de grado menor o igual que N−1, pero cuando N empieza a ser grande (del orden de 10 o más) a menos que los puntos estén muy cuidadosamente elegidos el polinomio oscilará salvajemente. Esto se conoce como <font color ="red"> fenómeno de Runge </font>.
#
#
# Para ver esto podemos estudiar el clásico ejemplo que dio Runge: tenemos la función
#
# $$f(x) = \frac{1}{1+x^2}$$
#
# veamos qué sucede si la interpolamos en nodos equiespaciados. Para ello vamos a usar la función barycentric_interpolate (según Berrut y Trefethen [II] <font color="blue"> El método de interpolación baricéntrica merece ser conocido como el método estándar de interpolación polinómica»).</font> Esta función recibe tres argumentos:
#
# - una lista de coordenadas x_i de los nodos,
# - una lista de coordenadas y_i de los nodos, y
# - un array x donde evaluar el polinomio interpolante que resulta.
# +
def runge(x):
"""Función de Runge."""
return 1 / (1 + x ** 2)
n=5
xp = np.arange(-n,n+1,1)
fp = runge(xp)
x = np.linspace(-n, n)
y =
plt.plot(xp, fp, "o", x, y, "-", x, runge(x), "--")
plt.legend(["data", "Interpolación", "Real"], loc="best");
# -
# Y no quiero contar nada si escogemos 20 o 100 puntos.
#
# **Existe una forma de mitigar este problema**, que es, como ya hemos dicho, escogiendo los puntos cuidadosamente.
#
# Una de las formas es elegir las raíces de **los polinomios de Chebyshev**, que podemos construir en NumPy usando el módulo polynomial.chebyshev.
#
# Por ejemplo, si queremos como antes 11 nodos tendremos que escoger el polinomio de Chebyshev de grado 11:
# +
from numpy.polynomial import chebyshev
coeffs_cheb = [0] * 11 + [1] # Solo queremos el elemento 11 de la serie
T11 = chebyshev.Chebyshev(coeffs_cheb, [-5, 5])
xp_ch = T11.roots()
# -
# Utilizando estos puntos, la cosa no queda tan mal:
# +
xp = xp_ch
fp = runge(xp)
x = np.linspace(-n, n)
y = barycentric_interpolate(xp, fp, x)
plt.plot(xp, fp, "o", x, y, "-", x, runge(x), "--")
plt.legend(["data", "Interpolación", "Real"], loc="best")
# -
# **Aun así, aún tenemos varios problemas:**
#
# El polinomio sigue oscilando, y esto puede no ser deseable. No siempre podemos escoger los puntos como nosotros queramos. <font color="red"> Por tanto, desde ya vamos a abandonar la idea de usar polinomios y vamos a hablar de trazadores (splines en inglés). </font>
# ## Trazadores
#
# Los trazadores o splines no son más que curvas polinómicas definidas a trozos, normalmente de grado 3 (casi nunca mayor de 5). Al ser cada uno de los trozos de grado pequeño se evita el fenómeno de Runge, y si se empalman los trozos inteligentemente la curva resultante será suave (matemáticamente: diferenciable) hasta cierto punto. Cuando queremos una curva que pase por todos los puntos disponibles un trazador es justamente lo que necesitamos.
#
# El trazador más elemental, el lineal (grado 1), se puede construir rápidamente en NumPy usando <font color="red">np.interp.</font>
#
# El más común, el trazador cúbico (grado 3) se puede construir con la clase <span style="color:#0000FF "> scipy.interpolate.InterpolatedUnivariateSpline.
#
# ### Libreria interp1d
# +
from scipy.interpolate import interp1d
x = np.linspace(0, 10, num=11, endpoint=True)
y = np.cos(-x**2/9.0)
f =
f2 =
xnew = np.linspace(0, 10, num=41, endpoint=True)
plt.plot(x, y, "o", xnew, f(xnew), "-", xnew, f2(xnew), "--")
plt.legend(["data", "linear", "cubic"], loc="best")
plt.show()
# -
# ### Resolviendo la Catenaria del puente con interp1d
#
# ## B-spline
#
# En el subcampo matemático de análisis numérico, una B-spline o Basis spline (o traducido una línea polinómica suave básica), es una función spline que tiene el mínimo soporte con respecto a un determinado grado, suavidad y partición del dominio.
#
# https://es.wikipedia.org/wiki/B-spline
# +
from scipy.interpolate import splrep, splev
n=5
x = np.arange(-n,n+1,1)
y = runge(xp)
tck = splrep(x,y,s=0)
xnew = np.arange(-n,n,.1)
ynew = splev(xnew,tck,der=0)
fig = plt.figure(figsize=(14,6))
plt.plot(x,y,'x',markersize=12)
plt.plot(x,y)
plt.plot(xnew,ynew)
plt.plot(xnew,runge(xnew),"o",alpha=.5) #'b--')
plt.legend(["Nodos",'Linear','Cubic Spline', 'True'])
plt.title('Cubic-spline interpolation')
# -
# +
x = np.arange(0,2*np.pi+np.pi/4,2*np.pi/8)
y = np.sin(x)
tck = splrep(x,y,s=0)
xnew = np.arange(0,2*np.pi,np.pi/50)
ynew = splev(xnew,tck,der=0)
plt.figure()
plt.plot(x,y,'x',xnew,ynew,xnew,np.sin(xnew),x,y,'b')
plt.legend(['Linear','Cubic Spline', 'True'])
plt.axis([-0.05,6.33,-1.05,1.05])
plt.title('Cubic-spline interpolation')
# -
# # Ajuste de curvas
#
#
#
# El ajuste más básico en el que podemos pensar es el ajuste polinómico: buscamos un polinomio que aproxime los datos con el menor error posible.
#
# En ocasiones las cosas son más complicadas que un polinomio (sí, así es la vida).
#
# Pero no pasa nada, porque con la función <font color="blue">scipy.optimize.curve_fit podemos ajustar una serie de datos a cualquier modelo que se nos ocurra, </font> no importa qué tan complicado sea. Sin ir más lejos, tomando el ejemplo de la documentación, supongamos que tenemos unos datos que se ajustan al modelo
#
# $$Ae^{-Bx^2} $$
#
# en Python nuestro modelo será una función que recibirá como primer argumento x y el resto serán los parámetros del mismo:
def func(x, A, B, C):
# """Modelo para nuestros datos."""
return A * np.exp(-B * x ** 2) + C
# Ahora solo necesitamos algunos datos (añadiremos un poco de ruido gaussiano para que tenga más gracia) y podemos probar el ajuste.
#
# La función <font color="blue">scipy.optimize.curve_fit</font> recibe como argumentos:
#
# - el modelo func para los datos,
# - una lista de coordenadas xdata de los puntos, y
# - una lista de coordenadas ydata de los puntos.
#
# Así realizamos el ajuste:
#
# curve_fit
#
# Mínimos cuadrados es una técnica de análisis numérico enmarcada dentro de la optimización matemática, en la que, dados un conjunto de pares ordenados —variable independiente, variable dependiente— y una familia de funciones, se intenta encontrar la función continua, dentro de dicha familia, que mejor se aproxime a los datos (un "mejor ajuste"), de acuerdo con el criterio de mínimo error cuadrático.
#
# curve_fit realiza el ajuste por medio de minimos cuadrados no lineales.
#
# Los Mínimos cuadrados no lineales es la forma de análisis de mínimos cuadrados que se usa para encajar un conjunto de m observaciones con un modelo que es no lineal en n parámetros desconocidos (m > n). Se utiliza en algunas formas de regresión no lineal.
#
# https://es.wikipedia.org/wiki/M%C3%ADnimos_cuadrados_no_lineales
# https://es.wikipedia.org/wiki/M%C3%ADnimos_cuadrados_no_lineales
# https://docs.scipy.org/doc/scipy-0.19.1/reference/generated/scipy.optimize.curve_fit.html
from scipy.optimize import curve_fit
# +
Adat, Bdat, Cdat = 2.5, 1.3, 0.5
xdat = np.linspace(-2, 4, 12)
ydat = func(xdat, Adat, Bdat, Cdat) + 0.2 * np.random.normal(size=len(xdat))
(A, B, C), _ =
#print(A, B, C)
# +
nxdat = np.linspace(-2, 4, 50)
ydatsin =
ydatAjuste =
plt.plot(xdat, ydat,"o", nxdat, ydatsin,"--", nxdat, ydatAjuste)
plt.legend(["Datos", "Sin ruido", "Ajuste"], loc="best")
# -
# ## Ejercicio de Catenaria de un Puente
# # Ejercicio
#
# $$ y = a*e^{b*(x-c)}$$
# +
# curve_fit?
# -
# +
def fun1(x,a):
return a*np.sin(x)
x = np.arange(0,10,.1)
y = 2*np.sin(x)
yruido = y + np.random.normal(0,.2,len(y))
paramOpt, paramCov =
plt.plot(x,yruido,'x',label='Con Ruido')
plt.plot(x,fun1(x,*paramOpt),label='Ajuste')
plt.legend()
# -
# +
from IPython.core.display import HTML
estilo ='''
<style>
@import url(http://fonts.googleapis.com/css?family=ROBOTO|Source+Code+Pro|Montserrat:400,700);
body {
font-family: 'ROBOTO', sans-serif !important;
}
h1 {
color: #0074D9 !important;
font-size: 3rem !important;
}
.importante {
// color: rgb(221, 153, 51) !important;
color: blue !important;
}
table {
color: #3D9970 !important;
align: justify !important;
margin: 0px auto !important;
}
h1, h2, h3, h4, h5, h6 {
font-family: 'ROBOTO', sans-serif !important;
}
p {
font-family: 'ROBOTO', sans-serif !important;
font-size: 1em !important;
line-height: 30px !important;
font-weight: 400 !important;
color: black !important;
}
</style>
'''
HTML(estilo)
# -
td {
color: #F012BE !important;
}
.tablesorter-header-inner{
color: #F012BE !important;
}
| 36.577513
| 6,362
|
58ca30b0969d3ce75a071663e58e374f5de868d1
|
py
|
python
|
Milestone-1-Project/Archive/M1_project_part1_old.ipynb
|
fredgi123/e-commerce-project
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + cell_id="00000-08668a20-ac67-4ffe-9485-220db18020fd" deepnote_cell_type="code"
# %matplotlib inline
import pandas as pd
import numpy as np
import math
import altair as alt
import folium
import matplotlib
import matplotlib.pyplot as plt
from folium import IFrame
from numpy import linalg
import scipy
import branca.colormap as cm
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
import statsmodels.api as sm
import numpy as np
import itertools
# enable correct rendering
alt.renderers.enable('default')
# uses intermediate json files to speed things up
alt.data_transformers.enable('json')
# + cell_id="00001-5aca03cf-2bc2-4199-b8aa-022001875609" deepnote_cell_type="code"
#Importing Main Dataset
df = pd.read_csv('data2.csv', encoding='latin1')
# + cell_id="00002-8c6d5639-2257-401e-b486-aabfcc3e4100" deepnote_cell_type="code"
#Dataframe Cleanup
#Stockcode which contains only digits signifies sale entry
#Hence we will filter out enteries with digit only stockcode
#Carriage did not drop, also look for returns if the qt. is negative and if they have same invoiceid
clean_df = df[df.StockCode.str.contains('^\d', regex=True, na=False)]
# Drop quantities which are negative
clean_df = clean_df[clean_df["Quantity"] >= 0]
#Adding SalesValue column to the Dataframe
clean_df["SalesValue"] = clean_df["Quantity"]*clean_df["UnitPrice"]
#UK Dataframe
uk_df = clean_df[clean_df['Country'] == 'United Kingdom']
#Rest of the World Dataframe
row_df = clean_df.drop(clean_df[clean_df['Country'] == 'United Kingdom'].index)
# + cell_id="00003-36a8cc55-26b4-4db2-848b-7fa96eba954f" deepnote_cell_type="code"
#Grouping sales by customers
customer_df = clean_df.groupby(by=['CustomerID']).agg({'Quantity':['min', 'max', 'sum'], 'UnitPrice':['min', 'max', 'sum']})
print("Number of unique customers = ", customer_df.index.size)
# + cell_id="00004-595aaf49-abf4-4266-ac04-0c8fd754979c" deepnote_cell_type="code"
# Importing Secondary Datasets - Records for year range 2007 to 2011
# HDI could be additional variable
# Smartphone penetration dataset if available
# Credit card
#Purchasing power parity GDP, PPP (constant 2017 international $) | Data (worldbank.org)
gdp_df = pd.read_excel('API_NY.GDP.MKTP.PP.KD_DS2_en_excel_v2_2764839.xls',
sheet_name=0, header=3, usecols="A,B,D,AZ:BD")
#Inflation CPI Consumer price index (2010 = 100) | Data (worldbank.org)
cpi_df = pd.read_excel("API_FP.CPI.TOTL_DS2_en_excel_v2_2765329.xls",
sheet_name=0, header=3, usecols="A,B,D,AZ:BD")
#Debt % versus GDP External debt stocks, long-term (DOD, current US$) | Data (worldbank.org)
#extdebt_df = pd.read_excel("API_DT.DOD.DLXF.CD_DS2_en_excel_v2_2823747.xls",
# sheet_name=0, header=3, usecols="A,B,D,AZ:BD")
#Individuals using the Internet (% of population)
internet_df = pd.read_excel("API_IT.NET.USER.ZS_DS2_en_excel_v2_2764008.xls",
sheet_name=0, header=3, usecols="A,B,D,AZ:BD")
#Exchange rate fluctuation (L5Y) Official exchange rate (LCU per US$, period average) | Data (worldbank.org)
exchrate_df = pd.read_excel("API_PA.NUS.FCRF_DS2_en_excel_v2_2764464.xls",
sheet_name=0, header=3, usecols="A,B,D,AZ:BD")
#Population Population, total | Data (worldbank.org)
pop_df = pd.read_excel("API_SP.POP.TOTL_DS2_en_excel_v2_2764317.xls",
sheet_name=0, header=3, usecols="A,B,D,AZ:BD")
#Merchandise imports Merchandise imports (current US$) | Data (worldbank.org)
merch_df = pd.read_excel("API_TM.VAL.MRCH.CD.WT_DS2_en_excel_v2_2766285.xls",
sheet_name=0, header=3, usecols="A,B,D,AZ:BD")
## new dataset
expendHealth_df = pd.read_excel("Expenditure_on_health.xls",
sheet_name=0, header=3, usecols="A,B,D,AZ:BD")
## new dataset
lifeExpect_df = pd.read_excel("life_expectancy.xls",
sheet_name=0, header=3, usecols="A,B,D,AZ:BD")
## new dataset
PPP_per_capita_df = pd.read_excel("PPP_per_capita.xls",
sheet_name=0, header=3, usecols="A,B,D,AZ:BD")
#CERDI Sea Distance Dataset
seadist_df = pd.read_excel("CERDI-seadistance.xlsx", usecols="A,B,C")
# Cleaning Sea Distance DF to only include entries with UK as the origin
seadist_df = seadist_df[seadist_df["iso1"]=="GBR"]
# + cell_id="00005-32192c21-3c4f-483a-b515-5135a9785a6a" deepnote_cell_type="code"
# Function to normalize country names to code that will be used as a key to combine all datasets
def valeurs(k):
filtered={'United Kingdom': 'GBR',
'France': 'FRA',
'USA': 'USA',
'Belgium': 'BEL',
'Australia': 'AUS',
'EIRE': 'IRL',
'Germany': 'DEU',
'Portugal': 'PRT',
'Japan': 'JPN',
'Denmark': 'DNK',
'Nigeria': 'NGA',
'Netherlands': 'NLD',
'Poland': 'POL',
'Spain': 'ESP',
'Channel Islands': 'CHI',
'Italy': 'ITA',
'Cyprus': 'CYP',
'Greece': 'GRC',
'Norway': 'NOR',
'Austria': 'AUT',
'Sweden': 'SWE',
'United Arab Emirates': 'ARE',
'Finland': 'FIN',
'Switzerland': 'CHE',
'Malta': 'MLT',
'Bahrain': 'BHR',
'Bermuda': 'BMU',
'Hong Kong': 'HKG',
'Singapore': 'SGP',
'Thailand': 'THA',
'Israel': 'ISR',
'Lithuania': 'LTU',
'Lebanon': 'LBN',
'Korea': 'KOR',
'Brazil': 'BRA',
'Canada': 'CAN',
'Iceland': 'ISL'}
try:
x=filtered[k]
except:
x=None
return x
# + cell_id="00006-ad07ce95-2335-4ed9-8cf0-4926f8f6ce74" deepnote_cell_type="code"
# Function to get all the country related information for the analysis task
def countrydf(name, frequency="M"):
# Country specific local dataframe
cdf = clean_df.loc[clean_df["Country"] == name]
# Datetime conversion
cdf["date"] = pd.to_datetime(cdf.InvoiceDate)
# Now lets find cummulative Sales for plot
# First lets group by Date to get transaction total per day
plotdf = cdf.set_index("date").resample(frequency)['SalesValue'].sum()
# Convert series to dataframe
plotdf = plotdf.to_frame()
# Total sales for the country
sales = plotdf["SalesValue"].sum()
# DF that conforms to 13 row format if frequency is set to monthly
if frequency == "M":
plotdf = (plotdf + dummydf).fillna(0)
# Number of unique customers in the country
custcnt = len(cdf["CustomerID"].unique())
# Numpy array of unique stock sold in each country
uniquestock = cdf["StockCode"].unique() #add .tolist() if list output is desired
#Country code
code = valeurs(name)
#GDP, CPI, for year 2011
gdp = gdp_df.loc[gdp_df["Country Code"] == code, "2011"].item()
cpi = cpi_df.loc[cpi_df["Country Code"] == code, "2011"].item()
#exchrate = exchrate_df.loc[exchrate_df["Country Code"] == code, "2011"].item()
pop = pop_df.loc[pop_df["Country Code"] == code, "2011"].item()
merch = merch_df.loc[merch_df["Country Code"] == code, "2011"].item()
internet = internet_df.loc[internet_df["Country Code"] == code, "2011"].item()
life_expect=lifeExpect_df.loc[internet_df["Country Code"] == code, "2011"].item()
expend_health = expendHealth_df.loc[internet_df["Country Code"] == code, "2011"].item()
ppp_capita = PPP_per_capita_df.loc[internet_df["Country Code"] == code, "2011"].item()
#Sea Distance
dist = seadist_df.loc[seadist_df["iso2"]==code, "seadistance"].item()
return {'name':name, 'code':code, 'df':plotdf, 'totalsales':sales, 'customercnt':custcnt,
'uniqueStockID': uniquestock, 'gdp':gdp, 'cpi':cpi, 'population':pop,
'merchsales': merch, 'internet':internet, 'distance':dist, 'expend_health':expend_health,'ppp_cap':ppp_capita,
'life_expect':life_expect}
# Creating dummydf to obtain fixed 13 row plotdf
dummyindex = ['2010-12-31', '2011-01-31', '2011-02-28', '2011-03-31',
'2011-04-30', '2011-05-31', '2011-06-30', '2011-07-31',
'2011-08-31', '2011-09-30', '2011-10-31', '2011-11-30',
'2011-12-31']
dummyvalues = [0,0,0,0,0,0,0,0,0,0,0,0,0]
dummydf = pd.DataFrame({'date':dummyindex, 'SalesValue':dummyvalues})
dummydf = dummydf.set_index('date')
# + cell_id="00007-4f74b957-27c8-4f4c-8c7e-0cb7a622d780" deepnote_cell_type="code"
# Creating Final DF that will be used for regression analysis
# Safe to ignore the SettingWithCopyWarning warning
countries = ['Australia','France', 'USA', 'Belgium', 'EIRE', 'Germany', 'Portugal', 'Japan', 'Denmark', 'Nigeria', \
'Netherlands', 'Poland', 'Spain', 'Italy', 'Cyprus', 'Greece','Norway', 'Austria', 'Sweden', \
'United Arab Emirates', 'Finland', 'Switzerland', 'Malta', 'Bahrain', 'Bermuda', 'Hong Kong', \
'Singapore', 'Thailand', 'Israel', 'Lithuania', 'Lebanon', 'Korea', 'Brazil', 'Canada', 'Iceland']
# Creating list of dictionaries obtained using countrydf function
finallist = [countrydf(country) for country in countries]
# Creating Dataframe from that list
finaldf = pd.DataFrame(finallist)
finaldf.head(3)
liste={'France': 'FRA',
'USA': 'USA',
'Belgium': 'BEL',
'Australia': 'AUS',
'EIRE': 'IRL',
'Germany': 'DEU',
'Portugal': 'PRT',
'Japan': 'JPN',
'Denmark': 'DNK',
'Nigeria': 'NGA',
'Netherlands': 'NLD',
'Poland': 'POL',
'Spain': 'ESP',
'Channel Islands': 'CHI',
'Italy': 'ITA',
'Cyprus': 'CYP',
'Greece': 'GRC',
'Norway': 'NOR',
'Austria': 'AUT',
'Sweden': 'SWE',
'United Arab Emirates': 'ARE',
'Finland': 'FIN',
'Switzerland': 'CHE',
'Unspecified': '',
'Malta': 'MLT',
'Bahrain': 'BHR',
'RSA': '',
'Bermuda': 'BMU',
'Hong Kong': 'HKG',
'Singapore': 'SGP',
'Thailand': 'THA',
'Israel': 'ISR',
'Lithuania': 'LTU',
'West Indies': '',
'Lebanon': 'LBN',
'Korea': 'KOR',
'Brazil': 'BRA',
'Canada': 'CAN',
'Iceland': 'ISL'}
liste=list(liste.keys())
set(countries) ^ set(liste)
# + cell_id="00008-4881859e-e931-42a2-9104-cf0870ebbe6d" deepnote_cell_type="code"
finaldf.columns
# + cell_id="00009-7781d7d3-c98d-41fc-b395-22de1e3e7322" deepnote_cell_type="code"
sales=finaldf.df
sales
# + cell_id="00010-31094ed7-487d-454a-a0b7-78aac2096310" deepnote_cell_type="code"
#Example of how to extract Monthly Sales data for each country
# In this example, we will use Canada and for that code is "CAN"
example = finaldf.loc[finaldf["code"]=='CAN', 'df'].item()
example
# + cell_id="00011-b6d54e9e-6f0b-4d20-83c3-442718806fa5" deepnote_cell_type="code"
# cols=table.columns.difference(['count'])
# cols=['2009-12', '2010-01', '2010-02', '2010-03', '2010-04', '2010-05',
# '2010-06', '2010-07', '2010-08', '2010-09', '2010-10', '2010-11',
# '2010-12']
# + cell_id="00012-741f36d3-0a74-4d60-9972-9cc6fb4d06f4" deepnote_cell_type="code"
##table['count'] = table[table[cols] > 0].count(axis=1)
#table_short=table[table['count']>=7].copy()
# convert negative values to zero
#table_short[table_short < 0] = 0
# table_short.astype(int)
# + cell_id="00013-7a2a0dfb-95af-4ba9-ad89-8f9d3250adb6" deepnote_cell_type="code"
### EXPAND sales by month for each country.
## add a column that
table
# + cell_id="00014-bc3e64bf-6605-4a4c-b4a5-da896ce4c05c" deepnote_cell_type="code"
finaldf.head()
# + cell_id="00015-80cb7f34-9d71-4d1f-8fb9-c0c927377da7" deepnote_cell_type="code"
# + cell_id="00016-b977b553-dcb1-42e1-bd28-272739afe346" deepnote_cell_type="code"
# + cell_id="00017-f1b495b3-dee6-4a44-9ca4-4806b762bf35" deepnote_cell_type="code"
finaldf.columns
# + cell_id="00018-67a08b61-8648-4fe9-9ffb-a483ba6692f3" deepnote_cell_type="code"
# Establishing factors list
liste_factores=['gdp', 'cpi', 'population', 'merchsales', 'internet', 'distance',
'expend_health', 'ppp_cap', 'life_expect']
# liste_factores.append('distance')
print(liste_factores)
# + cell_id="00019-ac78398f-4d6d-45c8-997d-0e7804fafbc3" deepnote_cell_type="code"
da=finaldf[['name','totalsales','gdp', 'cpi', 'population', 'merchsales', 'internet', 'distance',
'expend_health', 'ppp_cap', 'life_expect']]
# + cell_id="00020-b7b40d19-d33d-4666-be4a-029089058b6c" deepnote_cell_type="code"
corr=da[['totalsales','gdp', 'cpi', 'population', 'merchsales', 'internet', 'distance',
'expend_health', 'ppp_cap', 'life_expect']].corr().round(2)
corr.style.background_gradient(cmap='coolwarm')
# + cell_id="00021-2759bff5-d8bc-4348-8f82-28162e091f81" deepnote_cell_type="code"
import seaborn as sns
import altair as alt
## scatter plot or SPLOMS
chart = alt.Chart(da).mark_point(fill='red',fillOpacity=0.6,size=70).encode(
x=alt.X('totalsales:Q',axis=alt.Axis(labelFontSize=12,titleFontSize=14,title="totalsales")),
y=alt.Y('expend_health:Q',axis=alt.Axis(labelFontSize=12,titleFontSize=14,title='expend_health'))
)
chart+chart.transform_regression('totalsales', 'expend_health').mark_line(line=True,fill='black',strokeDash=[1,5],stroke='black',opacity=0.6)
# + cell_id="00022-56d559c3-f7c5-42d5-b4e4-87b5c625e07a" deepnote_cell_type="code"
# Running the algorithm
dico={}
r=0
gagnant=[]
for i in range(1,len(liste_factores)):
# Creating an itertool object with all combinations of factors
liste=(itertools.combinations(liste_factores,i))
for i in liste:
# Creating a string with + to integrate factors into model
element1=' + '.join(list(i))
# Creating string with final parameters for linear regression
a="totalsales ~ {}".format(element1)
# Running the linear regression
model = sm.OLS.from_formula(a, data=da)
result = model.fit()
# Creating a dictionary with all the combinations' adj r squared results
dico[element1]=result.rsquared_adj.round(3)
r1=dico[element1]
# saves best adjusted r-squared and liste of columns
if r1>r:
r=r1
gagnant=element1
# Extracting second best model
second=sorted(dico.items(), key=lambda x: x[1],reverse=True)[:5][1][0]
# Running the model
a="totalsales ~ {}".format(gagnant)
model = sm.OLS.from_formula(a, data=da)
result = model.fit()
# Returns best model according to Adjusted R-squared
result.summary()
# + cell_id="00023-fea52396-994b-474a-b688-6dd37d08d9bb" deepnote_cell_type="code"
# Returns Top 10 models according to adjusted R-squared
ordered_liste=sorted(dico.items(), key=lambda x: x[1],reverse=True)[:10]
ordered_liste
# + cell_id="00024-41bae6b0-2100-46fb-a130-64bc631c1e12" deepnote_cell_type="code"
# Running the model
a="totalsales ~ {}".format(second)
model = sm.OLS.from_formula(a, data=da)
result = model.fit()
# Returns best model according to Adjusted R-squared
result.summary()
# + cell_id="00025-bebfda78-240b-4926-8c48-d218bae56021" deepnote_cell_type="code"
result.params
# + cell_id="00026-ea354bb8-a186-4866-bddc-b5c14ff8811c" deepnote_cell_type="code"
table = pd.pivot_table(df2, values='sales', index=['Code'],
columns=['month_year'], aggfunc=np.sum, fill_value=0)
# + [markdown] tags=[] created_in_deepnote_cell=true deepnote_cell_type="markdown"
# <a style='text-decoration:none;line-height:16px;display:flex;color:#5B5B62;padding:10px;justify-content:end;' href='https://deepnote.com?utm_source=created-in-deepnote-cell&projectId=023a0e03-cc30-4d6d-b448-6d27799cca93' target="_blank">
# <img alt='Created in deepnote.com' style='display:inline;max-height:16px;margin:0px;margin-right:7.5px;' src='data:image/svg+xml;base64,PD94bWwgdmVyc2lvbj0iMS4wIiBlbmNvZGluZz0iVVRGLTgiPz4KPHN2ZyB3aWR0aD0iODBweCIgaGVpZ2h0PSI4MHB4IiB2aWV3Qm94PSIwIDAgODAgODAiIHZlcnNpb249IjEuMSIgeG1sbnM9Imh0dHA6Ly93d3cudzMub3JnLzIwMDAvc3ZnIiB4bWxuczp4bGluaz0iaHR0cDovL3d3dy53My5vcmcvMTk5OS94bGluayI+CiAgICA8IS0tIEdlbmVyYXRvcjogU2tldGNoIDU0LjEgKDc2NDkwKSAtIGh0dHBzOi8vc2tldGNoYXBwLmNvbSAtLT4KICAgIDx0aXRsZT5Hcm91cCAzPC90aXRsZT4KICAgIDxkZXNjPkNyZWF0ZWQgd2l0aCBTa2V0Y2guPC9kZXNjPgogICAgPGcgaWQ9IkxhbmRpbmciIHN0cm9rZT0ibm9uZSIgc3Ryb2tlLXdpZHRoPSIxIiBmaWxsPSJub25lIiBmaWxsLXJ1bGU9ImV2ZW5vZGQiPgogICAgICAgIDxnIGlkPSJBcnRib2FyZCIgdHJhbnNmb3JtPSJ0cmFuc2xhdGUoLTEyMzUuMDAwMDAwLCAtNzkuMDAwMDAwKSI+CiAgICAgICAgICAgIDxnIGlkPSJHcm91cC0zIiB0cmFuc2Zvcm09InRyYW5zbGF0ZSgxMjM1LjAwMDAwMCwgNzkuMDAwMDAwKSI+CiAgICAgICAgICAgICAgICA8cG9seWdvbiBpZD0iUGF0aC0yMCIgZmlsbD0iIzAyNjVCNCIgcG9pbnRzPSIyLjM3NjIzNzYyIDgwIDM4LjA0NzY2NjcgODAgNTcuODIxNzgyMiA3My44MDU3NTkyIDU3LjgyMTc4MjIgMzIuNzU5MjczOSAzOS4xNDAyMjc4IDMxLjY4MzE2ODMiPjwvcG9seWdvbj4KICAgICAgICAgICAgICAgIDxwYXRoIGQ9Ik0zNS4wMDc3MTgsODAgQzQyLjkwNjIwMDcsNzYuNDU0OTM1OCA0Ny41NjQ5MTY3LDcxLjU0MjI2NzEgNDguOTgzODY2LDY1LjI2MTk5MzkgQzUxLjExMjI4OTksNTUuODQxNTg0MiA0MS42NzcxNzk1LDQ5LjIxMjIyODQgMjUuNjIzOTg0Niw0OS4yMTIyMjg0IEMyNS40ODQ5Mjg5LDQ5LjEyNjg0NDggMjkuODI2MTI5Niw0My4yODM4MjQ4IDM4LjY0NzU4NjksMzEuNjgzMTY4MyBMNzIuODcxMjg3MSwzMi41NTQ0MjUgTDY1LjI4MDk3Myw2Ny42NzYzNDIxIEw1MS4xMTIyODk5LDc3LjM3NjE0NCBMMzUuMDA3NzE4LDgwIFoiIGlkPSJQYXRoLTIyIiBmaWxsPSIjMDAyODY4Ij48L3BhdGg+CiAgICAgICAgICAgICAgICA8cGF0aCBkPSJNMCwzNy43MzA0NDA1IEwyNy4xMTQ1MzcsMC4yNTcxMTE0MzYgQzYyLjM3MTUxMjMsLTEuOTkwNzE3MDEgODAsMTAuNTAwMzkyNyA4MCwzNy43MzA0NDA1IEM4MCw2NC45NjA0ODgyIDY0Ljc3NjUwMzgsNzkuMDUwMzQxNCAzNC4zMjk1MTEzLDgwIEM0Ny4wNTUzNDg5LDc3LjU2NzA4MDggNTMuNDE4MjY3Nyw3MC4zMTM2MTAzIDUzLjQxODI2NzcsNTguMjM5NTg4NSBDNTMuNDE4MjY3Nyw0MC4xMjg1NTU3IDM2LjMwMzk1NDQsMzcuNzMwNDQwNSAyNS4yMjc0MTcsMzcuNzMwNDQwNSBDMTcuODQzMDU4NiwzNy43MzA0NDA1IDkuNDMzOTE5NjYsMzcuNzMwNDQwNSAwLDM3LjczMDQ0MDUgWiIgaWQ9IlBhdGgtMTkiIGZpbGw9IiMzNzkzRUYiPjwvcGF0aD4KICAgICAgICAgICAgPC9nPgogICAgICAgIDwvZz4KICAgIDwvZz4KPC9zdmc+' > </img>
# Created in <span style='font-weight:600;margin-left:4px;'>Deepnote</span></a>
| 40.912114
| 2,168
|
3dce731281487d82380c8ea71bf3d5316f40e3e6
|
py
|
python
|
Algorithmic-Trading-in-the-FOREX-Market-master/Models/TensorFlow_LSTM_Keras.ipynb
|
vkumar24/Forx_Algo_Trading
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] colab_type="text" id="w4J2LlgHrd_l"
# **IMPORT LIBRARY**
# + colab={} colab_type="code" id="eOsitRsGrnjr"
import numpy as np
import pandas as pd
import math
import sklearn
import sklearn.preprocessing
import datetime
import seaborn as sns
# + colab={} colab_type="code" id="AlmEwyUbf2I5"
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.pyplot as plt2
import pandas as pd
from pandas import datetime
import math, time
import itertools
from sklearn import preprocessing
import datetime
from sklearn.metrics import mean_squared_error
from math import sqrt
from keras.models import Sequential
from keras.layers.core import Dense, Dropout, Activation
from keras.layers.recurrent import LSTM
from keras.models import load_model
import keras
import pandas_datareader.data as web
import h5py
# + [markdown] colab_type="text" id="jPR8nHH4r0ap"
# **IMPORT DATA**
# + colab={"base_uri": "https://localhost:8080/", "height": 72, "resources": {"http://localhost:8080/nbextensions/google.colab/files.js": {"data": "Ly8gQ29weXJpZ2h0IDIwMTcgR29vZ2xlIExMQwovLwovLyBMaWNlbnNlZCB1bmRlciB0aGUgQXBhY2hlIExpY2Vuc2UsIFZlcnNpb24gMi4wICh0aGUgIkxpY2Vuc2UiKTsKLy8geW91IG1heSBub3QgdXNlIHRoaXMgZmlsZSBleGNlcHQgaW4gY29tcGxpYW5jZSB3aXRoIHRoZSBMaWNlbnNlLgovLyBZb3UgbWF5IG9idGFpbiBhIGNvcHkgb2YgdGhlIExpY2Vuc2UgYXQKLy8KLy8gICAgICBodHRwOi8vd3d3LmFwYWNoZS5vcmcvbGljZW5zZXMvTElDRU5TRS0yLjAKLy8KLy8gVW5sZXNzIHJlcXVpcmVkIGJ5IGFwcGxpY2FibGUgbGF3IG9yIGFncmVlZCB0byBpbiB3cml0aW5nLCBzb2Z0d2FyZQovLyBkaXN0cmlidXRlZCB1bmRlciB0aGUgTGljZW5zZSBpcyBkaXN0cmlidXRlZCBvbiBhbiAiQVMgSVMiIEJBU0lTLAovLyBXSVRIT1VUIFdBUlJBTlRJRVMgT1IgQ09ORElUSU9OUyBPRiBBTlkgS0lORCwgZWl0aGVyIGV4cHJlc3Mgb3IgaW1wbGllZC4KLy8gU2VlIHRoZSBMaWNlbnNlIGZvciB0aGUgc3BlY2lmaWMgbGFuZ3VhZ2UgZ292ZXJuaW5nIHBlcm1pc3Npb25zIGFuZAovLyBsaW1pdGF0aW9ucyB1bmRlciB0aGUgTGljZW5zZS4KCi8qKgogKiBAZmlsZW92ZXJ2aWV3IEhlbHBlcnMgZm9yIGdvb2dsZS5jb2xhYiBQeXRob24gbW9kdWxlLgogKi8KKGZ1bmN0aW9uKHNjb3BlKSB7CmZ1bmN0aW9uIHNwYW4odGV4dCwgc3R5bGVBdHRyaWJ1dGVzID0ge30pIHsKICBjb25zdCBlbGVtZW50ID0gZG9jdW1lbnQuY3JlYXRlRWxlbWVudCgnc3BhbicpOwogIGVsZW1lbnQudGV4dENvbnRlbnQgPSB0ZXh0OwogIGZvciAoY29uc3Qga2V5IG9mIE9iamVjdC5rZXlzKHN0eWxlQXR0cmlidXRlcykpIHsKICAgIGVsZW1lbnQuc3R5bGVba2V5XSA9IHN0eWxlQXR0cmlidXRlc1trZXldOwogIH0KICByZXR1cm4gZWxlbWVudDsKfQoKLy8gTWF4IG51bWJlciBvZiBieXRlcyB3aGljaCB3aWxsIGJlIHVwbG9hZGVkIGF0IGEgdGltZS4KY29uc3QgTUFYX1BBWUxPQURfU0laRSA9IDEwMCAqIDEwMjQ7Ci8vIE1heCBhbW91bnQgb2YgdGltZSB0byBibG9jayB3YWl0aW5nIGZvciB0aGUgdXNlci4KY29uc3QgRklMRV9DSEFOR0VfVElNRU9VVF9NUyA9IDMwICogMTAwMDsKCmZ1bmN0aW9uIF91cGxvYWRGaWxlcyhpbnB1dElkLCBvdXRwdXRJZCkgewogIGNvbnN0IHN0ZXBzID0gdXBsb2FkRmlsZXNTdGVwKGlucHV0SWQsIG91dHB1dElkKTsKICBjb25zdCBvdXRwdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQob3V0cHV0SWQpOwogIC8vIENhY2hlIHN0ZXBzIG9uIHRoZSBvdXRwdXRFbGVtZW50IHRvIG1ha2UgaXQgYXZhaWxhYmxlIGZvciB0aGUgbmV4dCBjYWxsCiAgLy8gdG8gdXBsb2FkRmlsZXNDb250aW51ZSBmcm9tIFB5dGhvbi4KICBvdXRwdXRFbGVtZW50LnN0ZXBzID0gc3RlcHM7CgogIHJldHVybiBfdXBsb2FkRmlsZXNDb250aW51ZShvdXRwdXRJZCk7Cn0KCi8vIFRoaXMgaXMgcm91Z2hseSBhbiBhc3luYyBnZW5lcmF0b3IgKG5vdCBzdXBwb3J0ZWQgaW4gdGhlIGJyb3dzZXIgeWV0KSwKLy8gd2hlcmUgdGhlcmUgYXJlIG11bHRpcGxlIGFzeW5jaHJvbm91cyBzdGVwcyBhbmQgdGhlIFB5dGhvbiBzaWRlIGlzIGdvaW5nCi8vIHRvIHBvbGwgZm9yIGNvbXBsZXRpb24gb2YgZWFjaCBzdGVwLgovLyBUaGlzIHVzZXMgYSBQcm9taXNlIHRvIGJsb2NrIHRoZSBweXRob24gc2lkZSBvbiBjb21wbGV0aW9uIG9mIGVhY2ggc3RlcCwKLy8gdGhlbiBwYXNzZXMgdGhlIHJlc3VsdCBvZiB0aGUgcHJldmlvdXMgc3RlcCBhcyB0aGUgaW5wdXQgdG8gdGhlIG5leHQgc3RlcC4KZnVuY3Rpb24gX3VwbG9hZEZpbGVzQ29udGludWUob3V0cHV0SWQpIHsKICBjb25zdCBvdXRwdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQob3V0cHV0SWQpOwogIGNvbnN0IHN0ZXBzID0gb3V0cHV0RWxlbWVudC5zdGVwczsKCiAgY29uc3QgbmV4dCA9IHN0ZXBzLm5leHQob3V0cHV0RWxlbWVudC5sYXN0UHJvbWlzZVZhbHVlKTsKICByZXR1cm4gUHJvbWlzZS5yZXNvbHZlKG5leHQudmFsdWUucHJvbWlzZSkudGhlbigodmFsdWUpID0+IHsKICAgIC8vIENhY2hlIHRoZSBsYXN0IHByb21pc2UgdmFsdWUgdG8gbWFrZSBpdCBhdmFpbGFibGUgdG8gdGhlIG5leHQKICAgIC8vIHN0ZXAgb2YgdGhlIGdlbmVyYXRvci4KICAgIG91dHB1dEVsZW1lbnQubGFzdFByb21pc2VWYWx1ZSA9IHZhbHVlOwogICAgcmV0dXJuIG5leHQudmFsdWUucmVzcG9uc2U7CiAgfSk7Cn0KCi8qKgogKiBHZW5lcmF0b3IgZnVuY3Rpb24gd2hpY2ggaXMgY2FsbGVkIGJldHdlZW4gZWFjaCBhc3luYyBzdGVwIG9mIHRoZSB1cGxvYWQKICogcHJvY2Vzcy4KICogQHBhcmFtIHtzdHJpbmd9IGlucHV0SWQgRWxlbWVudCBJRCBvZiB0aGUgaW5wdXQgZmlsZSBwaWNrZXIgZWxlbWVudC4KICogQHBhcmFtIHtzdHJpbmd9IG91dHB1dElkIEVsZW1lbnQgSUQgb2YgdGhlIG91dHB1dCBkaXNwbGF5LgogKiBAcmV0dXJuIHshSXRlcmFibGU8IU9iamVjdD59IEl0ZXJhYmxlIG9mIG5leHQgc3RlcHMuCiAqLwpmdW5jdGlvbiogdXBsb2FkRmlsZXNTdGVwKGlucHV0SWQsIG91dHB1dElkKSB7CiAgY29uc3QgaW5wdXRFbGVtZW50ID0gZG9jdW1lbnQuZ2V0RWxlbWVudEJ5SWQoaW5wdXRJZCk7CiAgaW5wdXRFbGVtZW50LmRpc2FibGVkID0gZmFsc2U7CgogIGNvbnN0IG91dHB1dEVsZW1lbnQgPSBkb2N1bWVudC5nZXRFbGVtZW50QnlJZChvdXRwdXRJZCk7CiAgb3V0cHV0RWxlbWVudC5pbm5lckhUTUwgPSAnJzsKCiAgY29uc3QgcGlja2VkUHJvbWlzZSA9IG5ldyBQcm9taXNlKChyZXNvbHZlKSA9PiB7CiAgICBpbnB1dEVsZW1lbnQuYWRkRXZlbnRMaXN0ZW5lcignY2hhbmdlJywgKGUpID0+IHsKICAgICAgcmVzb2x2ZShlLnRhcmdldC5maWxlcyk7CiAgICB9KTsKICB9KTsKCiAgY29uc3QgY2FuY2VsID0gZG9jdW1lbnQuY3JlYXRlRWxlbWVudCgnYnV0dG9uJyk7CiAgaW5wdXRFbGVtZW50LnBhcmVudEVsZW1lbnQuYXBwZW5kQ2hpbGQoY2FuY2VsKTsKICBjYW5jZWwudGV4dENvbnRlbnQgPSAnQ2FuY2VsIHVwbG9hZCc7CiAgY29uc3QgY2FuY2VsUHJvbWlzZSA9IG5ldyBQcm9taXNlKChyZXNvbHZlKSA9PiB7CiAgICBjYW5jZWwub25jbGljayA9ICgpID0+IHsKICAgICAgcmVzb2x2ZShudWxsKTsKICAgIH07CiAgfSk7CgogIC8vIENhbmNlbCB1cGxvYWQgaWYgdXNlciBoYXNuJ3QgcGlja2VkIGFueXRoaW5nIGluIHRpbWVvdXQuCiAgY29uc3QgdGltZW91dFByb21pc2UgPSBuZXcgUHJvbWlzZSgocmVzb2x2ZSkgPT4gewogICAgc2V0VGltZW91dCgoKSA9PiB7CiAgICAgIHJlc29sdmUobnVsbCk7CiAgICB9LCBGSUxFX0NIQU5HRV9USU1FT1VUX01TKTsKICB9KTsKCiAgLy8gV2FpdCBmb3IgdGhlIHVzZXIgdG8gcGljayB0aGUgZmlsZXMuCiAgY29uc3QgZmlsZXMgPSB5aWVsZCB7CiAgICBwcm9taXNlOiBQcm9taXNlLnJhY2UoW3BpY2tlZFByb21pc2UsIHRpbWVvdXRQcm9taXNlLCBjYW5jZWxQcm9taXNlXSksCiAgICByZXNwb25zZTogewogICAgICBhY3Rpb246ICdzdGFydGluZycsCiAgICB9CiAgfTsKCiAgaWYgKCFmaWxlcykgewogICAgcmV0dXJuIHsKICAgICAgcmVzcG9uc2U6IHsKICAgICAgICBhY3Rpb246ICdjb21wbGV0ZScsCiAgICAgIH0KICAgIH07CiAgfQoKICBjYW5jZWwucmVtb3ZlKCk7CgogIC8vIERpc2FibGUgdGhlIGlucHV0IGVsZW1lbnQgc2luY2UgZnVydGhlciBwaWNrcyBhcmUgbm90IGFsbG93ZWQuCiAgaW5wdXRFbGVtZW50LmRpc2FibGVkID0gdHJ1ZTsKCiAgZm9yIChjb25zdCBmaWxlIG9mIGZpbGVzKSB7CiAgICBjb25zdCBsaSA9IGRvY3VtZW50LmNyZWF0ZUVsZW1lbnQoJ2xpJyk7CiAgICBsaS5hcHBlbmQoc3BhbihmaWxlLm5hbWUsIHtmb250V2VpZ2h0OiAnYm9sZCd9KSk7CiAgICBsaS5hcHBlbmQoc3BhbigKICAgICAgICBgKCR7ZmlsZS50eXBlIHx8ICduL2EnfSkgLSAke2ZpbGUuc2l6ZX0gYnl0ZXMsIGAgKwogICAgICAgIGBsYXN0IG1vZGlmaWVkOiAkewogICAgICAgICAgICBmaWxlLmxhc3RNb2RpZmllZERhdGUgPyBmaWxlLmxhc3RNb2RpZmllZERhdGUudG9Mb2NhbGVEYXRlU3RyaW5nKCkgOgogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAnbi9hJ30gLSBgKSk7CiAgICBjb25zdCBwZXJjZW50ID0gc3BhbignMCUgZG9uZScpOwogICAgbGkuYXBwZW5kQ2hpbGQocGVyY2VudCk7CgogICAgb3V0cHV0RWxlbWVudC5hcHBlbmRDaGlsZChsaSk7CgogICAgY29uc3QgZmlsZURhdGFQcm9taXNlID0gbmV3IFByb21pc2UoKHJlc29sdmUpID0+IHsKICAgICAgY29uc3QgcmVhZGVyID0gbmV3IEZpbGVSZWFkZXIoKTsKICAgICAgcmVhZGVyLm9ubG9hZCA9IChlKSA9PiB7CiAgICAgICAgcmVzb2x2ZShlLnRhcmdldC5yZXN1bHQpOwogICAgICB9OwogICAgICByZWFkZXIucmVhZEFzQXJyYXlCdWZmZXIoZmlsZSk7CiAgICB9KTsKICAgIC8vIFdhaXQgZm9yIHRoZSBkYXRhIHRvIGJlIHJlYWR5LgogICAgbGV0IGZpbGVEYXRhID0geWllbGQgewogICAgICBwcm9taXNlOiBmaWxlRGF0YVByb21pc2UsCiAgICAgIHJlc3BvbnNlOiB7CiAgICAgICAgYWN0aW9uOiAnY29udGludWUnLAogICAgICB9CiAgICB9OwoKICAgIC8vIFVzZSBhIGNodW5rZWQgc2VuZGluZyB0byBhdm9pZCBtZXNzYWdlIHNpemUgbGltaXRzLiBTZWUgYi82MjExNTY2MC4KICAgIGxldCBwb3NpdGlvbiA9IDA7CiAgICB3aGlsZSAocG9zaXRpb24gPCBmaWxlRGF0YS5ieXRlTGVuZ3RoKSB7CiAgICAgIGNvbnN0IGxlbmd0aCA9IE1hdGgubWluKGZpbGVEYXRhLmJ5dGVMZW5ndGggLSBwb3NpdGlvbiwgTUFYX1BBWUxPQURfU0laRSk7CiAgICAgIGNvbnN0IGNodW5rID0gbmV3IFVpbnQ4QXJyYXkoZmlsZURhdGEsIHBvc2l0aW9uLCBsZW5ndGgpOwogICAgICBwb3NpdGlvbiArPSBsZW5ndGg7CgogICAgICBjb25zdCBiYXNlNjQgPSBidG9hKFN0cmluZy5mcm9tQ2hhckNvZGUuYXBwbHkobnVsbCwgY2h1bmspKTsKICAgICAgeWllbGQgewogICAgICAgIHJlc3BvbnNlOiB7CiAgICAgICAgICBhY3Rpb246ICdhcHBlbmQnLAogICAgICAgICAgZmlsZTogZmlsZS5uYW1lLAogICAgICAgICAgZGF0YTogYmFzZTY0LAogICAgICAgIH0sCiAgICAgIH07CiAgICAgIHBlcmNlbnQudGV4dENvbnRlbnQgPQogICAgICAgICAgYCR7TWF0aC5yb3VuZCgocG9zaXRpb24gLyBmaWxlRGF0YS5ieXRlTGVuZ3RoKSAqIDEwMCl9JSBkb25lYDsKICAgIH0KICB9CgogIC8vIEFsbCBkb25lLgogIHlpZWxkIHsKICAgIHJlc3BvbnNlOiB7CiAgICAgIGFjdGlvbjogJ2NvbXBsZXRlJywKICAgIH0KICB9Owp9CgpzY29wZS5nb29nbGUgPSBzY29wZS5nb29nbGUgfHwge307CnNjb3BlLmdvb2dsZS5jb2xhYiA9IHNjb3BlLmdvb2dsZS5jb2xhYiB8fCB7fTsKc2NvcGUuZ29vZ2xlLmNvbGFiLl9maWxlcyA9IHsKICBfdXBsb2FkRmlsZXMsCiAgX3VwbG9hZEZpbGVzQ29udGludWUsCn07Cn0pKHNlbGYpOwo=", "headers": [["content-type", "application/javascript"]], "ok": true, "status": 200, "status_text": ""}}} colab_type="code" id="PljmPNQ5f99B" outputId="a03333e3-7d8b-4ec8-9fc6-a8d894955740"
from google.colab import files
uploaded = files.upload()
# + colab={"base_uri": "https://localhost:8080/", "height": 121} colab_type="code" id="CaaSSK9Uf2JF" outputId="1c67c184-1cde-45cd-f8c9-5cf3b34b7ff0"
forex_df = pd.read_csv("Data.csv", index_col = 0)
forex_df.dropna(inplace=True)
forex_df.isnull().sum()
# + colab={"base_uri": "https://localhost:8080/", "height": 233} colab_type="code" id="4EsfbN8OOsrJ" outputId="43cad4bf-636e-4051-8e2b-57ed4489568f"
forex_df.head()
# + colab={"base_uri": "https://localhost:8080/", "height": 294} colab_type="code" id="miXUnjHqxzTS" outputId="9a8cddef-303e-4f30-902c-615731c83eb9"
forex_df.describe()
# + [markdown] colab_type="text" id="4aPbEEm2EOS0"
# **Correlation Plot**
# + colab={"base_uri": "https://localhost:8080/", "height": 365} colab_type="code" id="pJCn-cdGNp3P" outputId="a363542b-0d11-457f-d045-1874a7d616c4"
sns.heatmap(forex_df.corr(),annot= True)
# + [markdown] colab_type="text" id="iB2MKT-eFDgl"
# **Dropping China's Foreign Exchange column**
# + [markdown] colab_type="text" id="g2ZlkTiLPBe6"
# **As DEXCHUS has a corr value of 0.26 we are dropping that column**
# + colab={} colab_type="code" id="oN1QIuUrPVSO"
forex_df.drop('DEXCHUS', axis=1,inplace=True)
# + colab={"base_uri": "https://localhost:8080/", "height": 233} colab_type="code" id="tJSoP7WMPoiX" outputId="edbfdcc2-c10f-4daf-ed8a-c901f30ea01b"
forex_df.head()
# + colab={"base_uri": "https://localhost:8080/", "height": 173} colab_type="code" id="28Va3XDOf2LK" outputId="74fda745-83ad-4e47-9620-1b728789d36e"
forex_df.info()
# + [markdown] colab_type="text" id="HPe_mFAsFQWV"
# **No NULL values in forex_df dataset**
# + [markdown] colab_type="text" id="bEoWn8JOP2sB"
# **Since the range of the columns are at different scales we are NORMALIZING the data**
# + [markdown] colab_type="text" id="HWhanAaZf2LU"
# # Normalization
# + colab={"base_uri": "https://localhost:8080/", "height": 233} colab_type="code" id="-ZqQu78of2LW" outputId="7f6cbb3d-f402-4442-960c-dad9ce8ddfc6"
def normalize_data(df):
min_max_scaler = sklearn.preprocessing.MinMaxScaler()
df['DEXINUS'] = min_max_scaler.fit_transform(df.DEXINUS.values.reshape(-1,1))
df['DEXMXUS'] = min_max_scaler.fit_transform(df.DEXMXUS.values.reshape(-1,1))
df['DEXBZUS'] = min_max_scaler.fit_transform(df.DEXBZUS.values.reshape(-1,1))
df['DEXSZUS'] = min_max_scaler.fit_transform(df.DEXSZUS.values.reshape(-1,1))
return df
forex_df_norm = normalize_data(forex_df)
forex_df_norm.head()
# + [markdown] colab_type="text" id="pNbQ83jbFqAG"
# **Ploting Normalized Forex Market**
# + colab={"base_uri": "https://localhost:8080/", "height": 369} colab_type="code" id="vVEnNdAxsr1t" outputId="a1efd747-2329-4ca5-8f2b-94feb9dcce73"
forex_df_norm[0:].plot()
# + colab={} colab_type="code" id="WictL5-USh0g"
seq_len = 22
d = 0.2
shape = [4, seq_len, 1] # feature, window, output
neurons = [128, 128, 32, 1]
epochs = 100
# + [markdown] colab_type="text" id="AszyLTy0GILu"
# **Train Test Split**
# + [markdown] colab_type="text" id="g0MKS02MGNNc"
# **Converting into arrays for input to the neural network****
# + colab={} colab_type="code" id="nj9LEnlwlJY6"
def load_data(forex, seq_len):
amount_of_features = len(forex.columns)
data = forex.as_matrix()
sequence_length = seq_len + 1 # index starting from 0
result = []
for index in range(len(data) - sequence_length): # maxmimum date = lastmost date - sequence length
result.append(data[index: index + sequence_length]) # index : index + 22days
result = np.array(result)
row = round(0.9 * result.shape[0]) # 90% split
m
train = result[:int(row), :] # 90% date
X_train = train[:, :-1] # all data until day
y_train = train[:, -1][:,-1] # day m + 1 for DEXINUS
X_test = result[int(row):, :-1] # all data until day
y_test = result[int(row):, -1][:,-1] # day m + 1 for DEXINUS
X_train = np.reshape(X_train, (X_train.shape[0], X_train.shape[1], amount_of_features))#
X_test = np.reshape(X_test, (X_test.shape[0], X_test.shape[1], amount_of_features))
return [X_train, y_train, X_test, y_test]
# + [markdown] colab_type="text" id="iVDupf1Rve4I"
# **We are incorporating 4 features(forex makets to predict India's FOREX)**
# + colab={} colab_type="code" id="tRK06ZWAlXRf"
X_train, y_train, X_test, y_test = load_data(forex_df_norm , seq_len)
# + colab={"base_uri": "https://localhost:8080/", "height": 121} colab_type="code" id="_3P_UCDWw4UL" outputId="afe7fadc-833b-4dc5-c1e1-a19ede13737f"
print(X_train.shape[0])#expected number of observations to read each batch
print(X_train.shape[1])# number of time steps
print(X_train.shape[2])#number of features.
print(X_test.shape[0])#expected number of observations to read each batch
print(X_test.shape[1])# number of time steps
print(X_test.shape[2])#number of features.
# + [markdown] colab_type="text" id="yqO10LTkl-Gl"
# **MODEL**
# + colab={} colab_type="code" id="QtEZrR-ul7in"
def build_model2(layers, neurons, d):
model = Sequential()
model.add(LSTM(neurons[0], input_shape=(layers[1], layers[0]), return_sequences=True))
model.add(Dropout(d))
model.add(LSTM(neurons[1], input_shape=(layers[1], layers[0]), return_sequences=False))
model.add(Dropout(d))
model.add(Dense(neurons[2],kernel_initializer="uniform",activation='relu'))
model.add(Dense(neurons[3],kernel_initializer="uniform",activation='linear'))
# model = load_model('my_LSTM_stock_model1000.h5')
# adam = keras.optimizers.Adam(decay=0.2)
model.compile(loss='mse',optimizer='adam', metrics=['accuracy'])
model.summary()
return model
# + colab={"base_uri": "https://localhost:8080/", "height": 346} colab_type="code" id="DbKunRdcg5gz" outputId="cff46d9c-255d-4ebb-dd22-ef48de1f1bc4"
model = build_model2(shape, neurons, d)
# layers = [4, 22, 1]
# + colab={"base_uri": "https://localhost:8080/", "height": 3518} colab_type="code" id="fqLUmM2pTzxy" outputId="9b437961-d277-46e3-c2dd-e38dc8406cfa"
model.fit(
X_train,
y_train,
batch_size=24,
epochs=100,
validation_split=0.1,
verbose=1)
# + colab={} colab_type="code" id="zVJLeQiHCvTj"
def model_score(model, X_train, y_train, X_test, y_test):
trainScore = model.evaluate(X_train, y_train, verbose=0)
print('Train Score: %.5f MSE (%.2f RMSE)' % (trainScore[0], math.sqrt(trainScore[0])))
testScore = model.evaluate(X_test, y_test, verbose=0)
print('Test Score: %.5f MSE (%.2f RMSE)' % (testScore[0], math.sqrt(testScore[0])))
return trainScore[0], testScore[0]
# + colab={"base_uri": "https://localhost:8080/", "height": 69} colab_type="code" id="tHK9wkdHCvZK" outputId="bae0d0a6-9997-4283-dce1-a97d802cdbf7"
model_score(model, X_train, y_train, X_test, y_test)
# + colab={} colab_type="code" id="VyyBM-oziiYY"
def denormalize(forex_df_norm, normalized_value):
forex_df_norm = forex_df_norm['DEXINUS'].values.reshape(-1,1)
normalized_value = normalized_value.reshape(-1,1)
#return df.shape, p.shape
min_max_scaler = preprocessing.MinMaxScaler()
a = min_max_scaler.fit_transform(forex_df_norm)
new = min_max_scaler.inverse_transform(normalized_value)
return new
newp = denormalize(forex_df_norm, p)
newy_test = denormalize(forex_df_norm, y_test)
# + [markdown] colab_type="text" id="rRJRR23BjPH7"
# **Plot**
# + colab={} colab_type="code" id="z8Q80sK3jN4R"
# + colab={"base_uri": "https://localhost:8080/", "height": 348} colab_type="code" id="hoU8qwX6iidX" outputId="a6a34d3a-8580-4ad9-89ca-231723c15509"
import matplotlib.pyplot as plt2
plt2.plot(newp,color='red', label='Prediction')
plt2.plot(newy_test,color='blue', label='Actual')
plt2.legend(loc='best')
plt2.show()
# + colab={} colab_type="code" id="BauL0QyFiicF"
# + [markdown] colab_type="text" id="AFY2z9jd5PGP"
# **END**
# + colab={} colab_type="code" id="L-HUKsd8iiWD"
# + colab={} colab_type="code" id="dZEWQxLu5RO2"
# + colab={} colab_type="code" id="tLRAhNuI5Rgl"
# + colab={} colab_type="code" id="cDU6MZcz5Rt1"
# + colab={} colab_type="code" id="pLiaY4pj5Rrj"
# + colab={} colab_type="code" id="-nJ4TE_V5RoJ"
# + colab={} colab_type="code" id="QWu0QVs_5RmX"
# + colab={} colab_type="code" id="mJiuEz5R5Re1"
# + colab={} colab_type="code" id="qe0o1BZf5Rck"
# + colab={} colab_type="code" id="-tnFFZDS5RZp"
# + colab={} colab_type="code" id="YYr2ySkj5RWr"
# + colab={} colab_type="code" id="1ymtwk4d5RUZ"
# + colab={} colab_type="code" id="kGVAUo8c5RNF"
# + colab={} colab_type="code" id="-hyniJjf5RJ9"
# + colab={} colab_type="code" id="1u_6vZqK5RG8"
# + colab={} colab_type="code" id="pJykfXmDCvXt"
def percentage_difference(model, X_test, y_test):
percentage_diff=[]
p = model.predict(X_test)
for u in range(len(y_test)): # for each data index in test data
pr = p[u][0] # pr = prediction on day u
percentage_diff.append((pr-y_test[u]/pr)*100)
return p
# + colab={} colab_type="code" id="0XhzjLmNVshs"
p = percentage_difference(model, X_test, y_test)
# + colab={} colab_type="code" id="L6FA2VS8CvPs"
def denormalize(forex_df_norm, normalized_value):
stock_name = stock_name['DEXINUS'].values.reshape(-1,1)
normalized_value = normalized_value.reshape(-1,1)
#return df.shape, p.shape
min_max_scaler = preprocessing.MinMaxScaler()
a = min_max_scaler.fit_transform(df)
new = min_max_scaler.inverse_transform(normalized_value)
return new
# + colab={} colab_type="code" id="0oLpvbKiCvOF"
def plot_result(stock_name, normalized_value_p, normalized_value_y_test):
newp = denormalize(stock_name, normalized_value_p)
newy_test = denormalize(stock_name, normalized_value_y_test)
plt2.plot(newp, color='red', label='Prediction')
plt2.plot(newy_test,color='blue', label='Actual')
plt2.legend(loc='best')
plt2.title('The test result for {}'.format(forex_df_norm))
plt2.xlabel('Days')
plt2.ylabel('Adjusted Close')
plt2.show()
# + colab={"base_uri": "https://localhost:8080/", "height": 409} colab_type="code" id="A9nVqLkZCvIP" outputId="38d09e7b-5242-4e75-b888-e45c25ed6175"
plot_result(stock_name, p, y_test)
# + colab={} colab_type="code" id="DgFI5Ckc3hJy"
model.compile(loss='mean_squared_error', optimizer=Adam(lr = 0.0005) , metrics = ['mean_squared_error'])
# + colab={"base_uri": "https://localhost:8080/", "height": 3501} colab_type="code" id="OuXEolkX3hPC" outputId="83c0b790-4362-4a62-ae8d-e08c186e5185"
history = model.fit(X_train, y_train, epochs=100 , batch_size = 25 ,
validation_data = (X_test,y_test))
# + colab={"base_uri": "https://localhost:8080/", "height": 734} colab_type="code" id="A2GmdeLD3hNZ" outputId="6758e15d-3b16-434d-81c2-92c656144893"
import matplotlib.pyplot as plt
plt.plot(history.history['mean_squared_error'])
plt.plot(history.history['val_mean_squared_error'])
plt.title('model mean squared error')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.show()
# summarize history for loss
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.show()
# + colab={"base_uri": "https://localhost:8080/", "height": 69} colab_type="code" id="YFEmUhuF3hH0" outputId="0b895a96-9f29-4989-a514-305a87d04bda"
def model_score(model, X_train, y_train, X_test, y_test):
trainScore = model.evaluate(X_train, y_train, verbose=0)
print('Train Score: %.5f MSE (%.2f RMSE)' % (trainScore[0], math.sqrt(trainScore[0])))
testScore = model.evaluate(X_test, y_test, verbose=0)
print('Test Score: %.5f MSE (%.2f RMSE)' % (testScore[0], math.sqrt(testScore[0])))
return trainScore[0], testScore[0]
model_score(model, X_train, y_train, X_test, y_test)
# + colab={"base_uri": "https://localhost:8080/", "height": 201} colab_type="code" id="icgbmol95pPd" outputId="979ea56d-1fe4-4062-a88c-6768dffa9dfb"
pred = model.predict(X_test)
pred = MinMaxScaler.inverse_transform(pred)
pred[:10]
def denormalize(forex_df_norm, normalized_value):
forex_df_norm = forex_df_norm['DEXINUS'].values.reshape(-1,1)
normalized_value = normalized_value.reshape(-1,1)
#return df.shape, p.shape
min_max_scaler = preprocessing.MinMaxScaler()
a = min_max_scaler.fit_transform(forex_df_norm)
new = min_max_scaler.inverse_transform(normalized_value)
return new
newp = denormalize(forex_df_norm, p)
newy_test = denormalize(forex_df_norm, y_test)
# + colab={} colab_type="code" id="qEaAcO8z5pUf"
def percentage_difference(model, X_test, y_test):
percentage_diff=[]
p = model.predict(X_test)
for u in range(len(y_test)): # for each data index in test data
pr = p[u][0] # pr = prediction on day u
percentage_diff.append((pr-y_test[u]/pr)*100)
return p
# + colab={} colab_type="code" id="XOVrCPO65pZ6"
pred = percentage_difference(model, X_test, y_test)
# + colab={} colab_type="code" id="z_ZurmLk5pYd"
pred = model.predict(testX)
pred = scaler.inverse_transform(pred)
pred[:10]
# + colab={} colab_type="code" id="AhfNCWu35pTH"
# + colab={} colab_type="code" id="ETah0BN45pNq"
# + colab={"base_uri": "https://localhost:8080/", "height": 333} colab_type="code" id="1hADBz5ll7nD" outputId="8eb5ff61-9ab0-460b-8198-7628eb3085e0"
model = build_model2(shape, neurons, d)
# layers = [4, 22, 1]
# + colab={"base_uri": "https://localhost:8080/", "height": 10050} colab_type="code" id="FDjlEWOcl7rs" outputId="838d4ddf-3ed6-4c9a-9608-27df89b86f41"
model.fit(
X_train,
y_train,
batch_size=24,
epochs=epochs,
validation_split=0.1,
verbose=1)
# + colab={} colab_type="code" id="C9YVbTaPl72n"
def percentage_difference(model, X_test, y_test):
percentage_diff=[]
p = model.predict(X_test)
for u in range(len(y_test)): # for each data index in test data
pr = p[u][0] # pr = prediction on day u
percentage_diff.append((pr-y_test[u]/pr)*100)
return p
# + colab={"base_uri": "https://localhost:8080/", "height": 2000} colab_type="code" id="hzca146Sl70e" outputId="e8da787f-4438-4386-8013-5ed38e8e4208"
p = percentage_difference(model, X_test, y_test)
forex_df_norm
# + colab={} colab_type="code" id="s-EwNP7yl7vG"
def denormalize(forex_df_norm, normalized_value):
stock_name = stock_name['DEXINUS'].values.reshape(-1,1)
normalized_value = normalized_value.reshape(-1,1)
#return df.shape, p.shape
min_max_scaler = preprocessing.MinMaxScaler()
a = min_max_scaler.fit_transform(stock_name)
new = min_max_scaler.inverse_transform(normalized_value)
return new
# + colab={} colab_type="code" id="YuNQP40zl7qT"
def plot_result(stock_name, normalized_value_p, normalized_value_y_test):
newp = denormalize(stock_name, normalized_value_p)
newy_test = denormalize(stock_name, normalized_value_y_test)
plt2.plot(newp, color='red', label='Prediction')
plt2.plot(newy_test,color='blue', label='Actual')
plt2.legend(loc='best')
plt2.title('The test result for {}'.format(forex_df_norm))
plt2.xlabel('Days')
plt2.ylabel('DEXINUS')
plt2.show()
# + colab={"base_uri": "https://localhost:8080/", "height": 68} colab_type="code" id="OzhzjHgerQwP" outputId="f3cdebff-5556-421a-f4c5-ac629b475d54"
def model_score(model, X_train, y_train, X_test, y_test):
trainScore = model.evaluate(X_train, y_train, verbose=0)
print('Train Score: %.5f MSE (%.2f RMSE)' % (trainScore[0], math.sqrt(trainScore[0])))
testScore = model.evaluate(X_test, y_test, verbose=0)
print('Test Score: %.5f MSE (%.2f RMSE)' % (testScore[0], math.sqrt(testScore[0])))
return trainScore[0], testScore[0]
model_score(model, X_train, y_train, X_test, y_test)
# + colab={"base_uri": "https://localhost:8080/", "height": 347} colab_type="code" id="zhX_XhK7rQ0i" outputId="fe7245ee-d2a4-4197-abe8-d37cc191c8b9"
import matplotlib.pyplot as plt2
plt2.plot(newp,color='red', label='Prediction')
plt2.plot(newy_test,color='blue', label='Actual')
plt2.legend(loc='best')
plt2.show()
# + [markdown] colab_type="text" id="4igAFhJvB4ug"
# **Saving Model**
# + colab={} colab_type="code" id="ReskBcgfrQuq"
model.save('LSTM_forex_prediction-20170429.h5')
# + [markdown] colab_type="text" id="AH3eSshJCCFz"
# **Hyperparameter Optimization**
# + colab={} colab_type="code" id="cxiubkuQCkhJ"
seq_len = 22
shape = [4, seq_len, 1] # feature, window, output
neurons = [128, 128, 32, 1]
epochs = 300
# + colab={} colab_type="code" id="ZG9eDHQOCO1b"
def quick_measure(forex_df_norm, seq_len, d, shape, neurons, epochs):
df = forex_df_norm
X_train, y_train, X_test, y_test = load_data(df, seq_len)
model = build_model2(shape, neurons, d)
model.fit(X_train, y_train, batch_size=24, epochs=epochs, validation_split=0.1, verbose=1)
# model.save('LSTM_Forex_prediction-20170429.h5')
trainScore, testScore = model_score(model, X_train, y_train, X_test, y_test)
return trainScore, testScore
# + colab={"base_uri": "https://localhost:8080/", "height": 75486} colab_type="code" id="gN-mstHPBJGO" outputId="99b9e575-71f8-488a-90ad-5c8673dda5a7"
dlist = [0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8]
neurons_LSTM = [32, 64, 128, 256, 512, 1024, 2048]
dropout_result = {}
for d in dlist:
trainScore, testScore = quick_measure(forex_df_norm, seq_len, d, shape, neurons, epochs)
dropout_result[d] = testScore
# + colab={} colab_type="code" id="r_aI7m88BJer"
def percentage_difference(model, X_test, y_test):
percentage_diff=[]
p = model.predict(X_test)
for u in range(len(y_test)): # for each data index in test data
pr = p[u][0] # pr = prediction on day u
percentage_diff.append((pr-y_test[u]/pr)*100)
return p
# + colab={} colab_type="code" id="1a02sR1BBJj2"
def model_score(model, X_train, y_train, X_test, y_test):
trainScore = model.evaluate(X_train, y_train, verbose=0)
print('Train Score: %.5f MSE (%.2f RMSE)' % (trainScore[0], math.sqrt(trainScore[0])))
testScore = model.evaluate(X_test, y_test, verbose=0)
print('Test Score: %.5f MSE (%.2f RMSE)' % (testScore[0], math.sqrt(testScore[0])))
return trainScore[0], testScore[0]
model_score(model, X_train, y_train, X_test, y_test)
# + colab={"base_uri": "https://localhost:8080/", "height": 232} colab_type="code" id="teikFNpiBJqo" outputId="66352297-d92b-40a8-8342-751471423ea2"
lists = sorted(seq_len_result.items())
x,y = zip(*lists)
plt.plot(x,y)
plt.title('Finding the best hyperparameter')
plt.xlabel('Days')
plt.ylabel('Mean Square Error')
plt.show()
# + colab={} colab_type="code" id="hwoqXlbCBJio"
# + colab={} colab_type="code" id="5CniBZK9BJdT"
| 45.930272
| 7,663
|
e8b664d9d6c9aca9e3a1d927f4ab798c3fdad4df
|
py
|
python
|
4_LowSignalData.ipynb
|
JoshVarty/MalwareDetection
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python (nlpenv)
# language: python
# name: nlpenv
# ---
# ## Low Signal Data
# Some of the columns appeared to have very little signal so we didn't add them. Let's double check that this is the case.
# +
import pandas as pd
import numpy as np
import fastai
from fastai.tabular import *
from sklearn.metrics import roc_auc_score
import torch
print(fastai.__version__)
# -
path = 'data/'
cont_names = ['Census_TotalPhysicalRAM', 'Census_PrimaryDiskTotalCapacity', 'Census_SystemVolumeTotalCapacity', 'Census_InternalPrimaryDiagonalDisplaySizeInInches', 'Census_InternalPrimaryDisplayResolutionHorizontal', 'Census_InternalPrimaryDisplayResolutionVertical', 'Census_ProcessorCoreCount']
cat_names = ['Wdft_IsGamer', 'OsBuildLab', 'ProductName', 'EngineVersion', 'AppVersion', 'RtpStateBitfield', 'IsSxsPassiveMode', 'DefaultBrowsersIdentifier', 'AVProductsInstalled', 'AVProductsEnabled', 'HasTpm', 'CountryIdentifier', 'OrganizationIdentifier', 'GeoNameIdentifier', 'LocaleEnglishNameIdentifier', 'Platform', 'Processor', 'OsVer', 'OsBuild', 'OsSuite', 'OsPlatformSubRelease', 'SkuEdition', 'IsProtected', 'SMode', 'IeVerIdentifier', 'SmartScreen', 'Firewall', 'UacLuaenable', 'Census_MDC2FormFactor', 'Census_DeviceFamily', 'Census_ProcessorManufacturerIdentifier', 'Census_ProcessorClass', 'Census_PrimaryDiskTypeName', 'Census_HasOpticalDiskDrive', 'Census_ChassisTypeName', 'Census_PowerPlatformRoleName', 'AVProductStatesIdentifier', 'Census_ActivationChannel', 'Census_FirmwareManufacturerIdentifier', 'Census_FlightRing','Census_GenuineStateName','Census_IsAlwaysOnAlwaysConnectedCapable','Census_IsFlightingInternal','Census_IsPenCapable','Census_IsPortableOperatingSystem','Census_IsSecureBootEnabled','Census_IsTouchEnabled','Census_IsVirtualDevice','Census_IsWIMBootEnabled','Census_OEMNameIdentifier','Census_OSBranch','Census_OSBuildNumber','Census_OSEdition','Census_OSInstallLanguageIdentifier','Census_OSUILocaleIdentifier', 'Census_OSWUAutoUpdateOptionsName','Census_ProcessorModelIdentifier','Census_ThresholdOptIn','Census_ThresholdOptIn','Wdft_RegionIdentifier']
useless_columns = ['AutoSampleOptIn', 'Census_InternalBatteryNumberOfCharges', 'Census_InternalBatteryType', 'Census_IsFlightsDisabled', 'Census_OSArchitecture', 'Census_OSSkuName', 'IsBeta', 'PuaMode']
cat_names = cat_names + useless_columns
dep_var = 'HasDetections'
layers = [1000, 500]
batch_size = 512
numRows= 4921483
allNames = cat_names + cont_names + [dep_var]
train_df = pd.read_csv(path + 'train.csv', nrows=numRows, usecols=allNames)
procs = [FillMissing, Categorify, Normalize]
data = (TabularList.from_df(train_df, path=path, cat_names=cat_names, cont_names=cont_names, procs=procs)
.random_split_by_pct(valid_pct=0.01)
.label_from_df(cols=dep_var)
.databunch(bs=batch_size))
#Create new learner, train it and look at losses
learn = tabular_learner(data, layers=layers, ps=[0.1, 0.1], emb_drop=0.04, metrics=[accuracy])
learn.fit_one_cycle(10, 1e-4)
learn.recorder.plot_losses()
#Create new learner, train it and look at losses
learn = tabular_learner(data, layers=layers, ps=[0.1, 0.1], emb_drop=0.04, metrics=[accuracy])
learn.fit_one_cycle(10, 1e-4)
learn.recorder.plot_losses()
#Create new learner, train it and look at losses
learn = tabular_learner(data, layers=layers, ps=[0.1, 0.1], emb_drop=0.04, metrics=[accuracy])
learn.fit_one_cycle(10, 1e-4)
learn.recorder.plot_losses()
| 54.80303
| 1,396
|
d8313baf0f1f73c928738c8ab4588bdd471d5f0f
|
py
|
python
|
tutorials/12-hmm.ipynb
|
spolcyn/brainiak-tutorials
|
['Apache-2.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Hidden Markov Models for Event Segmentation
# [Contributions](#contributions)
#
# If asked to give a quick description of a dinner with friends, you might say something like the following: "First, we met outside the restaurant while waiting for our table. Once we got to our table, we continued talking for a bit before we ordered something to drink and a few appetizers. Later, everyone ordered their dinner. The food arrived after some time and we all began eating. Finally, it was time for dessert and we continued chatting with each other until desert arrived. After this, we split the bill and headed out of the restaurant. We said our goodbyes to each other, while waiting for our taxis, and went home." From this description it is clear that the dinner meeting was composed of stages, or events, that occurred sequentially. Furthermore, these events can be perceived at varying scales. At the longest time scale, the entire dinner could be treated as one event. At smaller time scales, subsets of the meeting such as entering the restaurant, taking off coats, being seated, looking at menus and so on, can be treated as different events. At an even smaller scale, the event of entering the restaurant can be broken up into different sub-events. Regardless of scale, all of these accounts share the property that the event can be represented as a sequence of stages.
#
# The goal of this notebook is to explore methods for finding these “sequence-of-stages” representations in the brain. To accomplish this, we use a machine learning technique called Hidden Markov Modeling (HMM). These models assume that your thoughts progress through a sequence of states, one at a time. You can’t directly observe people’s thoughts, so the states are “hidden” (not directly observable). However, you can directly observe BOLD activity. So the full specification of the HMM is that each “hidden” state (corresponding to thinking about a particular stage of an event) has an observable BOLD correlate (specifically, a spatial pattern across voxels). The goal of HMM analysis is to take the BOLD data timeseries and then infer the sequence of states that the participant’s brain went through, based on the BOLD activity. Note that the broadest formulation of an HMM allows all possible transitions between states (e.g., with three states, some probability of transitioning from S1 to S2, S1 to S3, S2 to S1, S2 to S3, S3 to S1, S3 to S2). However, in the formulation of the HMM used here, we will assume that the participant’s brain can only progress forward between adjacent states (S1 to S2, S2 to S3). This more limited formulation of the HMM is well-suited to situations where we know that events proceed in a stereotyped sequence (e.g., we know that the waiter brings the food after you order the food, not the other way around).
#
# In summary: The HMM analysis used here assumes that the time series of BOLD activity was generated by the participant’s brain progressing through a sequence of states, where each state corresponds to a spatial pattern of BOLD activity. Intuitively, when we do HMM analysis, we are trying to identify when the brain has transitioned from one hidden state to another, by looking at the BOLD time series and estimating when the spatial pattern has switched. By fitting HMM with different numbers of hidden states, we can determine how many states a brain region traverses and identify when the transitions occur. Here we will apply the HMM to fMRI data from watching and recalling a movie (from [Chen et al., 2017](https://www.nature.com/articles/nn.4450)), identifying how different brain regions chunk the movie at different time scales, and examining where in the brain recalling a movie evokes the same sequence of neural patterns as viewing the movie. A full description is given in [Baldassano et al. (2017)](https://doi.org/10.1016/j.neuron.2017.06.041).
#
# <img src="../tutorials/imgs/lab12/hmm_schematics.png" width="800">
#
# A schematic diagram of HMM, from Baldassano et al. (2017).
#
# - (A) Given a set of (unlabeled) time courses from a region of interest, the goal of the event segmentation model is to temporally divide the data into ‘‘events’’ with stable activity patterns, punctuated by ‘‘event boundaries’’ at which activity patterns rapidly transition to a new stable pattern. The number and locations of these event boundaries can then be compared across brain regions or to stimulus annotations.
# - (B) The model can identify event correspondences between datasets (e.g., responses to movie and audio versions of the same narrative) that share the same sequence of event activity patterns, even if the duration of the events is different.
# - (C) The model-identified boundaries can also be used to study processing evoked by event transitions, such as changes in hippocampal activity coupled to event transitions in the cortex.
# - (D) The event segmentation model is implemented as a modified Hidden Markov Model (HMM) in which the latent state $s_t$ for each time point denotes the event to which that time point belongs, starting in event 1 and ending in event $K$. All datapoints during event $k$ are assumed to be exhibit high similarity with an event-specific pattern $m_k$.
#
#
#
# ## Goal of this script
# 1. Understand how HMMs can be useful for analyzing time series.
# 2. Learn how to run HMM in BrainIAK.
# 3. Use HMM to infer events from brain activity.
# 4. Determine the statistical significance from the HMM.
#
# <br>
#
#
# ## Table of Contents
# [0. HMM with toy data](#hmm_toy)
# [1. Detect event boundaries from brain activity](#hmm_brain)
# >[1.1. Data loading](#load_data)
# >[1.2. Formal procedure for fitting HMM](#fit_hmm)
# >>[1.2.1. Inner loop: tune k](#tune_k)
# >>[1.2.2. Outer loop: statistical testing of boundaries](#stats_hmm)
#
# [2. Comparing model boundaries to human-labeled boundaries](#model_to_human)
#
# [3. Aligning movie and recall data](#movie_to_recall)
# >[3.1. Fit HMM on the two datasets](#fit_movie_to_recall)
# >[3.2. Compare the time scale of events between regions](#movie_to_recall_semantic_sensory)
#
#
# #### Exercises
# >[1](#ex1) [2](#ex2) [3](#ex3) [4](#ex4) [5](#ex5) [6](#ex6) [7](#ex7) [8](#ex8)
# >[Novel contribution](#novel)
# +
import warnings
import sys
import os
# import logging
import deepdish as dd
import numpy as np
import brainiak.eventseg.event
import nibabel as nib
from nilearn.input_data import NiftiMasker
import scipy.io
from scipy import stats
from scipy.stats import norm, zscore, pearsonr
from scipy.signal import gaussian, convolve
from sklearn import decomposition
from sklearn.model_selection import LeaveOneOut, KFold
from matplotlib import pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
import matplotlib.patches as patches
import seaborn as sns
# %autosave 5
# %matplotlib inline
sns.set(style = 'white', context='talk', font_scale=1, rc={"lines.linewidth": 2})
if not sys.warnoptions:
warnings.simplefilter("ignore")
from utils import sherlock_h5_data
if not os.path.exists(sherlock_h5_data):
os.makedirs(sherlock_h5_data)
print('Make dir: ', sherlock_h5_data)
else:
print('Data path exists')
from utils import sherlock_dir
# -
# ## 0. HMM with toy data <a id="hmm_toy"></a>
#
# Here we introduce the usage of HMM in BrainIAK with a toy simulation. In particular, we will generate data with a known number $K$ of event boundaries. The following steps are used to run this simulation:
# > 1. Create event labels for each time-point in the toy dataset using `generate_event_labels`.
# > 2. Create data with these event labels using `generate_data`.
# > 3. Check the data matrix to see if we have created events -- time periods where the signal is relatively constant.
# > 4. Segment the signal using the brainiak HMM function: ` brainiak.eventseg.event.EventSegment(K)`.
# > 5. Overlay the results from HMM with the ground truth segments from the data.
# +
def generate_event_labels(T, K, length_std):
event_labels = np.zeros(T, dtype=int)
start_TR = 0
for e in range(K - 1):
length = round(
((T - start_TR) / (K - e)) * (1 + length_std * np.random.randn()))
length = min(max(length, 1), T - start_TR - (K - e))
event_labels[start_TR:(start_TR + length)] = e
start_TR = start_TR + length
event_labels[start_TR:] = K - 1
return event_labels
def generate_data(V, T, event_labels, event_means, noise_std):
simul_data = np.empty((V, T))
for t in range(T):
simul_data[:, t] = stats.multivariate_normal.rvs(
event_means[:, event_labels[t]], cov=noise_std, size=1)
simul_data = stats.zscore(simul_data, axis=1, ddof=1)
return simul_data
# -
# Create and plot simulated data. Imagine the following matrix is the voxel by TR bold activity matrix, averaged across participants.
# +
# Parameters for creating small simulated datasets
V = 10 # number of voxels
K = 10 # number of events
T = 500 # Time points
# Generate the first dataset
np.random.seed(1)
event_means = np.random.randn(V, K)
event_labels = generate_event_labels(T, K, 0.2)
D = generate_data(V, T, event_labels, event_means, 1/4)
# Check the data.
f, ax = plt.subplots(1,1, figsize=(12, 4))
ax.imshow(D, interpolation='nearest', cmap='viridis', aspect='auto')
ax.set_ylabel('Voxels')
ax.set_title('Simulated brain activity')
ax.set_xlabel('Timepoints')
# -
# The goal of the HMM is to identify chunks of time during which activity patterns remain relatively constant. To see if this is a reasonable model for our dataset, we can plot a timepoint-timepoint correlation matrix, showing the similarity between every pair of timepoints in our dataset (averaged over subjects).
f, ax = plt.subplots(1,1, figsize = (10,8))
ax.imshow(np.corrcoef(D.T), cmap='viridis')
title_text = '''
TR-TR correlation matrix
simulated data
'''
ax.set_title(title_text)
ax.set_xlabel('TR')
ax.set_ylabel('TR')
# Calling `brainiak.eventseg.event.EventSegment(k)` fits a hidden markov model with parameter `k`, where `k` is your guess about how many events (or brain states) are in the data. Here we are using the ground truth number of events.
# Find the events in this dataset
hmm_sim = brainiak.eventseg.event.EventSegment(K)
hmm_sim.fit(D.T)
# Another output of the event segmentation fit is the estimated activity pattern for each event: `HMM.event_pat_`.
# +
f, ax = plt.subplots(1,1, figsize=(8, 4))
ax.imshow(hmm_sim.event_pat_.T, cmap='viridis', aspect='auto')
ax.set_title('Estimated brain pattern for each event')
ax.set_ylabel('Event id')
ax.set_xlabel('Voxels')
# -
# One way of visualizing the fit is to mark on the timepoint correlation matrix where the model thinks the borders of the events are. The (soft) event segmentation is in `HMM.segments_[0]`, so we can convert this to hard bounds by taking the argmax.
# +
# plot
f, ax = plt.subplots(1,1, figsize=(12,4))
pred_seg = hmm_sim.segments_[0]
ax.imshow(pred_seg.T, aspect='auto', cmap='viridis')
ax.set_xlabel('Timepoints')
ax.set_ylabel('Event label')
ax.set_title('Predicted event segmentation, by HMM with the ground truth n_events')
f.tight_layout()
# -
# **Exercise 1**:<a id="ex1"></a> What is `hmm_sim.segments_[0]`? If you sum over event ids, you should get 1 for all TRs. Verify this claim. Why is this the case? Hint: reference [BrainIAK documentation](http://brainiak.org/docs/brainiak.eventseg.html).
# The field `HMM.ll_` stores the log likelihood over optimization steps. A description of what is meant by log-likelihood is provided [here](https://towardsdatascience.com/probability-concepts-explained-maximum-likelihood-estimation-c7b4342fdbb1).
#
# In the plot below, the likelihood values closer to zero are larger (e.g., -500 > -900) because they are logs. Also note that the HMM is optimized around 120 iterations in this example.
# +
f, ax = plt.subplots(1,1, figsize=(12, 4))
ax.plot(hmm_sim.ll_)
ax.set_title('Log likelihood')
ax.set_xlabel('EM steps')
sns.despine()
# -
# Here we overlay the predicted event boundaries on top of the TR-TR similarity matrix, which shows HMM is doing a great job recovering the ground truth.
def plot_tt_similarity_matrix(ax, data_matrix, bounds, n_TRs, title_text):
ax.imshow(np.corrcoef(data_matrix.T), cmap='viridis')
ax.set_title(title_text)
ax.set_xlabel('TR')
ax.set_ylabel('TR')
# plot the boundaries
bounds_aug = np.concatenate(([0],bounds,[n_TRs]))
for i in range(len(bounds_aug)-1):
rect = patches.Rectangle(
(bounds_aug[i],bounds_aug[i]),
bounds_aug[i+1]-bounds_aug[i],
bounds_aug[i+1]-bounds_aug[i],
linewidth=2,edgecolor='w',facecolor='none'
)
ax.add_patch(rect)
# +
# extract the boundaries
bounds = np.where(np.diff(np.argmax(pred_seg, axis=1)))[0]
f, ax = plt.subplots(1,1, figsize = (10,8))
title_text = '''
Overlay the predicted event boundaries
on top of the TR-TR correlation matrix
simulated data
'''
plot_tt_similarity_matrix(ax, D, bounds, T, title_text)
f.tight_layout()
# -
# **Exercise 2**: <a id="ex1"></a> Study the effects of over/under-estimating k. Fit HMM with k = 5 then 20. Visualize the resulting boundaries on top of the TR-TR similarity matrices.
# ## 1. Detect event boundaries from brain activity <a id="hmm_brain"></a>
#
# In this section, we will use HMM on a movie dataset. Subjects were watching the first hour of [A Study in Pink](https://en.wikipedia.org/wiki/A_Study_in_Pink), henceforth called the "Sherlock" dataset. Please refer to [Chen et al. (2017)](https://doi.org/10.1038/nn.4450) to learn more about this dataset.
# ### 1.1 Load data <a id="load_data"></a>
#
# Download whole-brain data for 17 subjects. Voxels with low inter-subject correlation (that were not consistently activated across subjects) were removed, and then the data were downsampled into 141 large regions (from a [resting-state atlas](http://www.dpmlab.org/peerj-784.pdf)). After putting this .h5 file into the same directory as this notebook, we can load in the data. In addition to the fMRI data, we have the coordinates of each region, as well as human-labeled boundaries for event boundaries.
# download the data, just need to run these lines once
# !wget https://ndownloader.figshare.com/files/9017983 -O {sherlock_h5_data}/sherlock.h5
# !wget https://ndownloader.figshare.com/files/9055612 -O {sherlock_h5_data}/AG_movie_1recall.h5
D = dd.io.load(os.path.join(sherlock_h5_data, 'sherlock.h5'))
BOLD = D['BOLD']
coords = D['coords']
human_bounds = D['human_bounds']
# **Exercise 3:** <a id="ex3"></a> Inspect the data. What is `BOLD`? Which dimension corresponds to number of subjects (`nSubj`), number of brain regions (`nR`), and number of TRs (`nTR`)? Visualize the data for the 1st subject. What is `coords`? Visualize it with a 3D plot. What is `human_bounds`? Print out this variable and explain the i-th entry.
# When you complete this exercise, these variables must be filled in.
#nR, nTR, nSubj = ?
# **Exercise 4**: <a id="ex4"></a> Compute and visualize the timepoint-timepoint correlation matrix. Is there some block-structure in the correlation matrix? Why?
# ### 1.2 Formal procedure for fitting HMM <a id="fit_hmm"></a>
#
# Here, we introduce some rigorous procedures and relevant statistical tests.
# We need to pick `k` - the assumed number of events when fitting HMM. Then, once we figure out what `k` to use, we need some unseen data to evaluate whether this was a good choice. Therefore we will need to do nested cross-validation. Here is some pseudo code summarizing the procedure:
#
# Given subject 1, subject 2, ... subject N.
#
# > <b>Outer loop</b>: Hold out subject i, for i = 1, 2, ..., N, as the final test subject.
# >> <b>Inner loop</b>: For subject j, for all j $\neq$ i, hold out some subjects as the validation set for parameter tuning and use the rest of the subjects as the training set.
# >>> For all choices of k, fit HMM on the training set, and compute the log likelihood on the validation set.
# >>
# >> Pick the best k (based on the validation set), re-fit the model using subject j, for all j $\neq$ i. And evaluate the the performance of HMM on subject i, the final test subject.
#
#
# The code block below has the nested cv loop (without fitting HMM) to make things more concrete.
# +
# set up the nested cross validation template
n_splits_inner = 4
subj_id_all = np.array([i for i in range(nSubj)])
# set up outer loop loo structure
loo_outer = LeaveOneOut()
loo_outer.get_n_splits(subj_id_all)
for subj_id_train_outer, subj_id_test_outer in loo_outer.split(subj_id_all):
print("Outer:\tTrain:", subj_id_train_outer, "Test:", subj_id_test_outer)
# set up inner loop loo structure
subj_id_all_inner = subj_id_all[subj_id_train_outer]
kf = KFold(n_splits=n_splits_inner)
kf.get_n_splits(subj_id_train_outer)
print('Inner:')
for subj_id_train_inner, subj_id_test_inner in kf.split(subj_id_all_inner):
# inplace update the ids w.r.t. to the inner training set
subj_id_train_inner = subj_id_all_inner[subj_id_train_inner]
subj_id_test_inner = subj_id_all_inner[subj_id_test_inner]
print("-Train:", subj_id_train_inner, "Test:", subj_id_test_inner, ', now try different k...')
print()
# -
# Since the full nested cv procedure is time consuming, we won't do that in this notebook. Instead:
#
# - Inner loop - Split the data in a particular way (train / validation / test), fit HMM with different k and evaluate the log likelihood on some unseen subjects.
#
# - Outer loop - For a fixed k, fit HMM(k) on some training set, and evaluate HMM(k) on the held-out subject.
# #### 1.2.1 Inner loop : tune `k` <a id="tune_k"></a>
#
# For some training set and test set, we can train HMM(`k`) on the training set and evaluate the log likelihood of the trained HMM(`k`) using the test set. For every `k`, we will get the a log likelihood.
#
# Then we can pick the `k`, call it `best_k`, with the largest log likelihood. If we want to evaluate how good this `best_k` is, we need other unseen data.
#
# Here, we pick a particular train-test split and try various `k`.
# **Exercise 5**: <a id="ex5"></a>Estimate a reasonable value for `k`, the number of events. Choose a train - validation - test split. Fit HMM with `k` = 10, 20, ..., 100. Figure out which `k` works the best based on the log likelihood on the held-out subject by plotting log likelihood against all choices of `k`. Use the best `k` to predict event boundaries. Overlay the boundaries on the timepoint-timepoint correlation matrix.
# +
# hold out a subject
subj_id_test = 0
subj_id_val = 1
subj_id_train = [
subj_id for subj_id in range(nSubj)
if subj_id not in [subj_id_test, subj_id_val]
]
BOLD_train = BOLD[:,:,subj_id_train]
BOLD_val = BOLD[:,:,subj_id_val]
BOLD_test = BOLD[:,:,subj_id_test]
print('Whole dataset:\t', np.shape(BOLD))
print('Training set:\t', np.shape(BOLD_train))
print('Tune set:\t', np.shape(BOLD_val))
print('Test set:\t', np.shape(BOLD_test))
print(subj_id_train)
print(subj_id_val)
print(subj_id_test)
# -
# #### 1.2.2 Outer loop: statistical testing of boundaries <a id="stats_hmm"></a>
#
# One way to test whether the model-identified boundaries are consistent across subjects is to fit the model on all but one subject and try to predict something about the held-out subject. There are multiple approaches for doing this (see [here](http://www.dpmlab.org/Neuron17.pdf)), but the simplest is to check whether the model boundaries predict pattern changes in the held-out subject. We therefore measure whether activity patterns 5 TRs apart show a drop in correlation when they are on opposite sides of an event boundary. Here, we pick a `k` and do the full leave-one-subject-out procedure to test the quality of this choice.
#
# For comparison, we generate permuted versions of the model boundaries, in which the distribution of event lengths (the distances between the boundaries) is held constant but the order of the event lengths is shuffled. This should produce a within vs. across boundary difference of zero on average, but the variance of these null boundaries lets us know the interval of chance values and we can see that we are well above chance.
# +
k = 60
w = 5 # window size
nPerm = 1000
within_across = np.zeros((nSubj, nPerm+1))
for left_out in range(nSubj):
# Fit to all but one subject
ev = brainiak.eventseg.event.EventSegment(k)
ev.fit(BOLD[:,:,np.arange(nSubj) != left_out].mean(2).T)
events = np.argmax(ev.segments_[0], axis=1)
# Compute correlations separated by w in time
corrs = np.zeros(nTR-w)
for t in range(nTR-w):
corrs[t] = pearsonr(BOLD[:,t,left_out],BOLD[:,t+w,left_out])[0]
_, event_lengths = np.unique(events, return_counts=True)
# Compute within vs across boundary correlations, for real and permuted bounds
np.random.seed(0)
for p in range(nPerm+1):
within = corrs[events[:-w] == events[w:]].mean()
across = corrs[events[:-w] != events[w:]].mean()
within_across[left_out, p] = within - across
#
perm_lengths = np.random.permutation(event_lengths)
events = np.zeros(nTR, dtype=np.int)
events[np.cumsum(perm_lengths[:-1])] = 1
events = np.cumsum(events)
print('Subj ' + str(left_out+1) + ': within vs across = ' + str(within_across[left_out,0]))
# +
plt.figure(figsize=(2,5))
plt.violinplot(within_across[:,1:].mean(0), showextrema=False)
plt.scatter(1, within_across[:,0].mean(0))
plt.gca().xaxis.set_visible(False)
plt.ylabel('Within vs across boundary correlation')
# -
# **Exercise 6:** <a id="ex6"></a> Is the real within vs. across boundary difference significant with respect to the null distribution of differences? What does this imply?
# ## 2. Comparing model boundaries to human-labeled boundaries <a id="model_to_human"></a>
#
# We can also compare the event boundaries from the model to human-labeled event boundaries. Because there is some ambiguity in both the stimulus and the model about exactly which timepoint the transition occurs at, we will count two boundaries as being a "match" if they are within 3 TRs (4.5 seconds) of each other.
#
# To determine whether the match is statistically significant, we generate permuted versions of the model boundaries as in **Section 1.2.2**. This gives as a null model for comparison: how often will a human and model bound be within 3 TRs of each other by chance?
# +
np.random.seed(0)
event_counts = np.diff(np.concatenate(([0],bounds,[nTR])))
nPerm = 1000
perm_bounds = bounds
threshold = 3
match = np.zeros(nPerm+1)
for p in range(nPerm+1):
for hb in human_bounds:
# check if match
if np.any(np.abs(perm_bounds - hb) <= threshold):
match[p] += 1
match[p] /= len(human_bounds)
perm_counts = np.random.permutation(event_counts)
perm_bounds = np.cumsum(perm_counts)[:-1]
plt.figure(figsize=(2,4))
plt.violinplot(match[1:], showextrema=False)
plt.scatter(1, match[0])
plt.gca().xaxis.set_visible(False)
plt.ylabel('Human-model match')
print('p = ' + str(norm.sf((match[0]-match[1:].mean())/match[1:].std())))
# -
# ## 3. Aligning movie and recall data <a id="movie_to_recall"></a>
#
# A simple model of free recall is that a subject will revisit the same sequence of events experienced during perception, but the lengths of the events will not be identical between perception and recall. We can fit this model by separately estimating event boundaries in movie and recall data, while constraining the event patterns to be the same across the two datasets.
#
# We will now download data consisting of (group-averaged) movie-watching data and free recall data for a single subject. These data are from the angular gyrus. We also have a human-labeled correspondence between the movie and recall (based on the transcript of the subject's verbal recall). The full dataset for all 17 subjects is available [here](https://ndownloader.figshare.com/files/9055471).
# +
D = dd.io.load(os.path.join(sherlock_h5_data,'AG_movie_1recall.h5'))
movie = D['movie']
recall = D['recall']
movie_labels = D['movie_labels']
recall_labels = D['recall_labels']
# -
# **Exercise 7:** <a id="ex7"></a> Inspect the data. Visualize the bold data for movie viewing vs. movie recall (i.e. `movie`, `recall`). Visualize `movie_labels` and `recall_labels`. Compare the number of TRs for the movie viewing data vs. the recall data. Why would they differ, if they do?
# ### 3.1. Fit HMM on the two datasets <a id="fit_movie_to_recall"></a>
#
# We use the same fit function as for a single dataset, but now we pass in both the movie and recall datasets in a list. We assume the two datasets have shared event transitions.
# fit event seg models
k = 25
hmm_ag_mvr = brainiak.eventseg.event.EventSegment(k)
hmm_ag_mvr.fit([movie.T, recall.T])
# This divides the movie and recall into 25 *corresponding* events, with the same 25 event patterns. We can plot the probability that each timepoint is in a particular event.
# +
plt.figure(figsize=(16, 5))
plt.subplot(1,2,1)
plt.imshow(hmm_ag_mvr.segments_[0].T,aspect='auto',origin='lower',cmap='viridis')
plt.xlabel('Movie TRs')
plt.ylabel('Events')
plt.subplot(1,2,2)
plt.imshow(hmm_ag_mvr.segments_[1].T,aspect='auto',origin='lower',cmap='viridis')
plt.xlabel('Recall TRs')
plt.ylabel('Events')
# -
# To get the temporal correspondence between the movie and the recall, we need to find the probability that a movie TR and a recall TR are in the same event (regardless of which event it is):
#
# $$
# \sum_k p(T_M == k) \cdot p(T_R == k)
# $$
# which we can compute by a simple matrix multiplication.
#
# For comparison, we also plot the human-labeled correspondence as white boxes.
# +
f, ax = plt.subplots(1,1, figsize=(10,8))
ax.imshow(
np.dot(hmm_ag_mvr.segments_[1], hmm_ag_mvr.segments_[0].T),
origin='lower',cmap='viridis'
)
# overlay the boundaries
for i in np.unique(movie_labels[movie_labels>0]):
if not np.any(recall_labels==i):
continue
movie_start = np.where(movie_labels==i)[0][0]
movie_end = np.where(movie_labels==i)[0][-1]
recall_start = np.where(recall_labels==i)[0][0]
recall_end = np.where(recall_labels==i)[0][-1]
rect = patches.Rectangle(
(movie_start-0.5, recall_start-0.5),
movie_end - movie_start + 1,
recall_end - recall_start + 1,
linewidth=1,edgecolor='w',facecolor='none'
)
ax.add_patch(rect)
ax.set_xlabel('Movie TRs')
ax.set_ylabel('Recall TRs')
title_text = """
Estimated temporal correspondence, movie vs. recall
human labels overlayed as bounding boxes
ROI = AG
"""
ax.set_title(title_text)
# -
# ### 3.2 Compare the time scale of events between regions. <a id="movie_to_recall_semantic_sensory"></a>
#
# Run HMM on two ROIs that correspond to semantic cognition (posterior medial cortex) and sensory processing (auditory cortex). These regions show differences in their time scale of representation.
# +
def load_labels(subj_id):
label_fname = 'labels_subj%d.mat' % subj_id
labels = scipy.io.loadmat(os.path.join(sherlock_dir, 'labels', label_fname))
recall_times = labels['recall_scenetimes']
movie_times = labels['subj_movie_scenetimes']
return recall_times, movie_times
def read_sherlock_mat_data(fpath):
temp = scipy.io.loadmat(fpath)
return temp['rdata'].T
# -
# **Exercise 8:** <a id="ex8"></a> Fit the HMM to find temporal correspondence between movie vs. recall for both ROIs. Which ROI had a better fit? The code below loads the data for subject 2 (who shows a noticeable ROI difference) but you should extend to all subjects.
# +
subj_id = 2
data_dir_subj = os.path.join(sherlock_dir, 'data_mat', 's%d'%subj_id)
recall_dir = os.path.join(data_dir_subj, 'sherlock_recall')
movie_dir = os.path.join(data_dir_subj, 'sherlock_movie')
# load data
fpath_aud_movie = os.path.join(movie_dir, 'aud_early_sherlock_movie_s%d.mat'%subj_id)
fpath_aud_recall = os.path.join(recall_dir, 'aud_early_sherlock_recall_s%d.mat'%subj_id)
fpath_pmc_movie = os.path.join(movie_dir, 'pmc_nn_sherlock_movie_s%d.mat'%subj_id)
fpath_pmc_recall = os.path.join(recall_dir, 'pmc_nn_sherlock_recall_s%d.mat'%subj_id)
data_aud_movie = read_sherlock_mat_data(fpath_aud_movie)
data_aud_recall = read_sherlock_mat_data(fpath_aud_recall)
data_pmc_movie = read_sherlock_mat_data(fpath_pmc_movie)
data_pmc_recall = read_sherlock_mat_data(fpath_pmc_recall)
# load labels
recall_times, movie_times = load_labels(subj_id)
# -
# **Novel contribution** <a id="novel"></a>: be creative and make one new discovery by adding an analysis, visualization, or optimization.
#
# Here are some ideas:
#
# 1. Use different ROIs and replicate Figure 4 in Baldassano et al. (2017).
# 2. Use RSA to compare all movie viewing patterns vs. all movie recall patterns.
# ### Recommended readings
#
# - Anderson, J. R., Pyke, A. A., & Fincham, J. M. (2016). Hidden stages of cognition revealed in patterns of brain activation. Psychological Science, 27(9), 1215–1226. https://doi.org/10.1177/0956797616654912
#
# - Baldassano, C., Hasson, U., & Norman, K. A. (2018). Representation of real-world event schemas during narrative perception. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 38(45), 9689–9699. https://doi.org/10.1523/JNEUROSCI.0251-18.2018
#
# - Manning, J. R., Fitzpatrick, P. C., & Heusser, A. C. (2018). How is experience transformed into memory? bioRxiv. https://doi.org/10.1101/409987
# ### Contributions <a id="contributions"></a>
#
# - Chris Baldassano developed the initial notebook for
# <a href="https://github.com/brainiak/brainiak/blob/master/examples/eventseg/HiddenMarkovModels.ipynb">brainiak demo</a>.
# - Q add exs; nested cv; tune k; modularize code.
# - M. Kumar added introduction and edited section descriptions.
# - K.A. Norman provided suggestions on the overall content and made edits to this notebook.
| 49.580699
| 1,450
|
3d545c99f0fef65a9f3f1af0f5a13ffee7fa927a
|
py
|
python
|
Datasets/Water/jrc_global_surface_water.ipynb
|
jdgomezmo/gee
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <table class="ee-notebook-buttons" align="left">
# <td><a target="_blank" href="https://github.com/giswqs/earthengine-py-notebooks/tree/master/Datasets/Water/jrc_global_surface_water.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" /> View source on GitHub</a></td>
# <td><a target="_blank" href="https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/Datasets/Water/jrc_global_surface_water.ipynb"><img width=26px src="https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png" />Notebook Viewer</a></td>
# <td><a target="_blank" href="https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/Datasets/Water/jrc_global_surface_water.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" /> Run in Google Colab</a></td>
# </table>
# ## Install Earth Engine API and geemap
# Install the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geemap](https://github.com/giswqs/geemap). The **geemap** Python package is built upon the [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) and [folium](https://github.com/python-visualization/folium) packages and implements several methods for interacting with Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, and `Map.centerObject()`.
# The following script checks if the geemap package has been installed. If not, it will install geemap, which automatically installs its [dependencies](https://github.com/giswqs/geemap#dependencies), including earthengine-api, folium, and ipyleaflet.
#
# **Important note**: A key difference between folium and ipyleaflet is that ipyleaflet is built upon ipywidgets and allows bidirectional communication between the front-end and the backend enabling the use of the map to capture user input, while folium is meant for displaying static data only ([source](https://blog.jupyter.org/interactive-gis-in-jupyter-with-ipyleaflet-52f9657fa7a)). Note that [Google Colab](https://colab.research.google.com/) currently does not support ipyleaflet ([source](https://github.com/googlecolab/colabtools/issues/60#issuecomment-596225619)). Therefore, if you are using geemap with Google Colab, you should use [`import geemap.eefolium`](https://github.com/giswqs/geemap/blob/master/geemap/eefolium.py). If you are using geemap with [binder](https://mybinder.org/) or a local Jupyter notebook server, you can use [`import geemap`](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py), which provides more functionalities for capturing user input (e.g., mouse-clicking and moving).
# +
# Installs geemap package
import subprocess
try:
import geemap
except ImportError:
print('geemap package not installed. Installing ...')
subprocess.check_call(["python", '-m', 'pip', 'install', 'geemap'])
# Checks whether this notebook is running on Google Colab
try:
import google.colab
import geemap.eefolium as geemap
except:
import geemap
# Authenticates and initializes Earth Engine
import ee
try:
ee.Initialize()
except Exception as e:
ee.Authenticate()
ee.Initialize()
# -
# ## Create an interactive map
# The default basemap is `Google Maps`. [Additional basemaps](https://github.com/giswqs/geemap/blob/master/geemap/basemaps.py) can be added using the `Map.add_basemap()` function.
Map = geemap.Map(center=[40,-100], zoom=4)
Map
# ## Add Earth Engine Python script
# Add Earth Engine dataset
dataset = ee.Image('JRC/GSW1_1/GlobalSurfaceWater')
occurrence = dataset.select('occurrence');
occurrenceVis = {'min': 0.0, 'max': 100.0, 'palette': ['ffffff', 'ffbbbb', '0000ff']}
Map.setCenter(59.414, 45.182, 6)
Map.addLayer(occurrence, occurrenceVis, 'Occurrence')
# ## Display Earth Engine data layers
Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.
Map
| 57.319444
| 1,023
|
1a39575000b40cd8ab46cf1fe5a52cdd6923bfca
|
py
|
python
|
project1&2.ipynb
|
sankalpachowdhury/IT-Automation-Projects-
|
['Apache-2.0']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/sankalpachowdhury/IT-Automation-Projects-/blob/master/project1%262.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + id="q_h0TDuKKVf1" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 173} outputId="760d4432-6da7-43b3-a755-e918ddc6206a"
# !python3 -m pip install --upgrade pip
# + id="6NwHLLXMSHJq" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 615} outputId="a73fbf72-a0d0-4046-c3d3-1d764b60c2ee"
# !sudo apt install python3-pil
# + id="DoBqpALdSPjq" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 187} outputId="606e3b63-c602-43da-ef09-1097343b7590"
# !python3 -m pip install --upgrade Pillow
import PIL
# + id="m8Bm-ujYT_Vu" colab_type="code" colab={}
from PIL import Image
im = Image.open("/content/DCGAN.png")
new_im = im.rotate(45)
#new_im = im.resize((640,480))
#new_im.save("example_resized.png")
new_im.rotate(180).resize((640,480)).save("flipped_and_resized.png")
# + id="oaxGyCglWJFr" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 102} outputId="9c5029ec-628b-4313-f965-1f599668d184"
# !curl -c ./cookie -s -L "https://drive.google.com/uc?export=download&id=$11hg55-dKdHN63yJP20dMLAgPJ5oiTOHF" > /dev/null | curl -Lb ./cookie "https://drive.google.com/uc?export=download&confirm=`awk '/download/ {print $NF}' ./cookie`&id=11hg55-dKdHN63yJP20dMLAgPJ5oiTOHF" -o images.zip && sudo rm -rf cookie
# + id="vczd8CVExUR3" colab_type="code" colab={}
# #!/usr/bin/env python3
import os, glob
from PIL import Image
size = 128, 128
for f in glob.glob(os.getcwd()+"/images/ic_*"):
im = Image.open(f).convert('RGB')
print(f)
print(im.format)
new_name = os.path.basename(f)
im.rotate(270).resize((size)).save("/opt/icons/" + new_name, "JPEG")
# + id="y7fJ6n951VUs" colab_type="code" colab={}
#all useful commands
unzip images.zip
# cd images
# ls
pip3 install pillow
chmod +x <script_name>.py
./<script_name>.py
# ls /opt/icons
python3
from PIL import Image
img = Image.open("/opt/icons/ic_edit_location_black_48dp")
img.format, img.size
# + [markdown] id="yON_uc9q65yS" colab_type="text"
# 1. Used APIs
# 2. Used Linax Command shell
# 3. Used python text to convert, fix and resize a image dataset and saving into another path.
# + [markdown] id="5ZWAqINJh4w9" colab_type="text"
# # project 2
# ## Data Serialization
# + id="END8GdBth3tR" colab_type="code" colab={}
import json
with open('people.json', 'w') as people_json:
json.dump(people, people_json, indent=2)
# + id="q6-h1Nl9iU1f" colab_type="code" colab={}
# The Json File
[
{
"name": "Sabrina Green",
"username": "sgreen",
"phone": {
"office": "802-867-5309",
"cell": "802-867-5310"
},
"department": "IT Infrastructure",
"role": "Systems Administrator"
},
{
"name": "Eli Jones",
"username": "ejones",
"phone": {
"office": "684-348-1127"
},
"department": "IT Infrastructure",
"role": "IT Specialist"
},
]
# + id="67r5AS_WibTO" colab_type="code" colab={}
#YAML format
import yaml
with open('people.yaml', 'w') as people_yaml:
yaml.safe_dump(people, people_yaml)
# + id="NsnKVmXxi5ax" colab_type="code" colab={}
#That code will generate a people.yaml file that looks like this:
- department: IT Infrastructure
name: Sabrina Green
phone:
cell: 802-867-5310
office: 802-867-5309
role: Systems Administrator
username: sgreen
- department: IT Infrastructure
name: Eli Jones
phone:
office: 684-348-1127
role: IT Specialist
username: ejones
# + id="274dqwhmtheA" colab_type="code" colab={}
import json
people = [
{
"name": "Sabrina Green",
"username": "sgreen",
"phone": {
"office": "802-867-5309",
"cell": "802-867-5310"
},
"department": "IT Infrastructure",
"role": "Systems Administrator"
},
{
"name": "Eli Jones",
"username": "ejones",
"phone": {
"office": "684-348-1127"
},
"department": "IT Infrastructure",
"role": "IT Specialist"
}
]
with open('people.json', 'w') as people_json:
json.dump(people, people_json, indent=2)
# + [markdown] id="DlW6V6VNuW1C" colab_type="text"
# the dumps() method, which also serializes Python objects, but returns a string instead of writing directly to a file.
# + [markdown] id="UPUNfy6uuvOq" colab_type="text"
# load() method does the inverse of the dump() method. It deserializes JSON from a file into basic Python objects. The loads() method also deserializes JSON into basic Python objects, but parses a string instead of a file.
# + id="tXzujZiwuX4T" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 54} outputId="cfc3fa1b-a0f3-4657-bd4e-9c447c901d9e"
import requests
response = requests.get('https://www.google.com', stream = True)
print(response.raw.read()[:100])
# + id="usb4gLORIHOo" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 35} outputId="f6dd929e-7501-4969-b9f5-00b49681e870"
response.request.headers['Accept-Encoding']
# + id="yJojbv1tIKqp" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 35} outputId="bc8eabc1-614e-4581-915e-8ce28783721f"
response.headers['Content-Encoding']
# + id="XUP0YWVSIfWx" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="e090074c-8a5d-4a96-ebe2-1f3d8384e79e"
# check if response is true or false
response.ok
# + [markdown] id="2GveOiuXI8Ci" colab_type="text"
# [Response Status Codes and Their Meanings](https://www.iana.org/assignments/http-status-codes/http-status-codes.xhtml)
#
# 
#
# + id="49eOGiMYIn-p" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="7efbaacc-3411-4ec1-a1d2-5ccd5864f7e6"
# get response code
response.status_code
# + id="m9euIQ8oJHR7" colab_type="code" colab={}
url = 'https://www.google.com'
response = requests.get(url)
if not response.ok:
raise Exception("GET failed with status code {}".format(response.status_code))
# + [markdown] id="z3OplmpYKNiV" colab_type="text"
# **Raise an HTTPError exception only if the response wasn’t successful**
#
# We can simply write as follows:
# + id="KqikU-icKTdh" colab_type="code" colab={}
response = requests.get(url)
response.raise_for_status()
# + id="cl7HC1KuMEUV" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 35} outputId="3bd44298-ab3a-4b3e-9e1b-f4e08a143549"
p = {"search": "grey kitten",
"max_results": 15}
response = requests.get(url, params=p)
response.request.url
# + id="6VYyhPU_NZUc" colab_type="code" colab={}
response = requests.post("https://example.com/path/to/api", data=p)
# + id="wGacMazxNixO" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 35} outputId="befb0763-b9df-46bf-d409-05cab92ec687"
response.request.url
# + id="c3axAtv3Nqv_" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 35} outputId="d386d391-56f9-427f-f69a-7f269492a0c7"
response.request.body
# + [markdown] id="4yWK8x7nOU-B" colab_type="text"
# **This code creates a dictionary using the data given as parameter and is sent through "requests.post()" method using json = p as a parrameter to the function.**
# + id="IzU61iscN3pe" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="0750c1e8-9b44-440b-d5e2-4d39feef63bb"
response = requests.post("https://example.com/path/to/api", json=p)
response.request.url
response.request.body
# + [markdown] id="IWv5CpmPO6__" colab_type="text"
# #Whats Django?
# + [markdown] id="jpsLnBVrPFs3" colab_type="text"
# Django is a full-stack web framework written in Python.
#
# Web frameworks are commonly split into three basic components: (1) the application code, where you'll add all of your application's logic; (2) the data storage, where you'll configure what data you want to store and how you're storing it; and (3) the web server, where you'll state which pages are served by which logic.
# + [markdown] id="muroGF0KSYax" colab_type="text"
# # **The Project 2 description**
#
# Have to write a script that interacts with a running web service. The web service is part of a company's website and is in charge of storing and displaying the customer reviews of the company.
#
# The reviews are stored in text files in the local disk. My script should open those files, process the information to turn it into the format expected by the web service, then send it to the web service to get stored.
#
#
# + [markdown] id="w9ePyfAfUJ_D" colab_type="text"
# #**Processing Text Files with Python Dictionaries and Uploading to Running Web Service**
# + [markdown] id="uAbn_-KmcG3W" colab_type="text"
# run.py script to request.post() all the text files to server as json and converting the text file as dictionary in python, then converting the python dictionary object to json file and posting it to the live server.
# + [markdown] id="8F11XpvYdRyz" colab_type="text"
# 
#
# + id="iBbtahW9OwB9" colab_type="code" colab={}
# #! /usr/bin/env python3
import os
import requests
# Path to the data
path = "/data/feedback/"
keys = ["title", "name", "date", "feedback"]
folder = os.listdir(path)
for file in folder:
keycount = 0
fb = {}
with open(path + file) as fl:
for line in fl:
value = line.strip()
fb[keys[keycount]] = value
keycount += 1
print(fb)
response = requests.post("http://<IP Address>/feedback/",
json=fb)
print(response.request.body)
print(response.status_code)
# + id="5iA4zDDJcTkA" colab_type="code" colab={}
# + id="Vomy3EoRcS2k" colab_type="code" colab={}
# + id="lcRS1lUGcSS-" colab_type="code" colab={}
| 2,408.813149
| 441,354
|
00783e7d4689a764193c17cc77d236ce8e6e68c6
|
py
|
python
|
concept.ipynb
|
braadbaart/macroeconomics
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: me
# language: python
# name: me
# ---
# ### *One world*
#
# By modeling the relationship between services, trade, and consumption patterns across countries over time that should provide me enough insights to further develop the questions below into actual hypotheses:
#
# * what are the limitations of using money as the principal measure of economic activity (some will be obvious of course, others - hopefully - not so much)?
# * how does the (mis-/non-)valuation of services and the informal economy determine into the economic worldview? and consequently, how to deal with missing data and accurate estimates and/ or valuations of intangible economic activity? and labour pricing in general?
# * what is the impact of the growing share of services (intangibles) in global economic activity on the world economy?
# * what part of the cycle of supply and demand is endogeneous, and what part is exogeneous? this question is closely related to the role of technological development, information diffusion, knowledge / knowhow transfer (education), and the exceedingly dominant role of services in the global economy.
# * could the working models of the **real** economy be improved in the light of the questions posed above?
# * could any of the resulting insights be used to improve the valuation of firms and (e)valuation and remuneration of firm employees?
# * how would the automation of service work through cognitive computing affect the macroeconomic outlook?
# * what is the consumer exposure to macroeconomic policy decisions?
#
# The goal being to build a functioning predictive model of global human economic activity.
#
# *Reminder: continuously perform sanity checks on the completeness and sufficieny of the developed models and systems.*
# ### Research outline
#
# Current technological advances make possible both unprecented economies of scale and scope and the exact matching of supply and demand for labour and skills. These advances would in theory allow firms to significantly reduce overhead on non-productive employees, at the same time creating more meaningful work opportunities for workers. However, given difficulties involved in building up buffers for workers specializing in less in-demand activities, the gig economy currently disproportionally benefits highly skilled workers. Unfortunately, these workers only constitute a small percentage of the total workforce of a country. As a result, the same technologies that enable unprecented economies of scale and scope at the level of the individual worker therefore also work to aggravate existing economic inequalities across the workforce, perpetuating a cycle of privilege and access to education inherent in the socio-economic fabric of our early 21st-century world. Solutions such as universal basic income (UBI) fail to take into account the positive psychological, social, societal and health benifits of individuals participating in and contributing to a community. In the inverse sense, increasing the economic value of the population provides a buffer and leverage against any exclusivist tendencies or policies developed by the most powerful economic and political actors. Where in the last three hundred-fifty years (roughly 1650 CE to 1990 CE) the nation-state had a powerful military incentive to improve the education levels of the workforce and national economy, at the beginning of the 21st century these incentives appear of limited value given the global challenges mankind faces. In fact, the incentivization schemes being maintained at the national level are counterproductive or even damaging when it come to tackling issues such as climate change, epidemic diseases, resource scarcity, poverty reduction, economic development, and the protection of the earth ecosystem. What I argue in this paper is that break is needed from economic the paradigm put forward in it's most clear form in Adam Smith's *The Wealth of Nations* (1776 CE), and the construction of a new social contract, which - in contrast to the nation state - I will refer to as *the human state*.
#
# ##### People
# -----------
#
# Of course merely changing the political and economic discourse won't change the world economic outlook. However, framing the political discourse and greater good along global lines rather than national borders and national interests should - over time - help force politicians and the general public away from the currents of misplaced large-scale bigotry, tribalism, and nepotism still all too common in today's world. Progress as a rallying cry has of course been used and abused throughout history, so a clear definition of what is meant by human development is merited.
#
# ...
#
#
# ##### Technology
# --------------
#
# (innovation, adaptation, education, transference)
#
# ...
#
#
# ##### Trade
# ----------
#
# (including any means of exchange, viz markets, currencies, labour)
#
# ...
#
# #### Policy
# ----------
#
# (roughly speaking, institutions in the broadest sense)
#
# ..
# +
import ipywidgets as widgets
from IPython.display import YouTubeVideo
out = widgets.Output(layout={'border': '1px solid black'})
out.append_stdout('Amartya Sen on demonetization in India')
out.append_display_data(YouTubeVideo('OknuVaSW4M0'))
out
# -
out.clear_output()
# ## Modelling approaches
#
# Short overview of the existing approaches for modelling our global macroeconomic system.
#
# #### Dynamic Stochastic General Equilibrium (DSGE)
#
# - Non-linear approximations of stochastic (Gaussian) processes, for example through vector autoregression (VAR):
#
# $$ \begin{bmatrix} y_{t} \\ x_{t} \end{bmatrix} = A(L) \begin{bmatrix} y_{t-1} \\ x_{t-1} \end{bmatrix} + \begin{bmatrix} u_{yt} \\ u_{xt} \end{bmatrix}$$
#
# with $A(L)$ a matrix polynomial over the lag operator $L$ and $u$ innovations (linear combinations of iid shocks to output $e_{yt}$ and policy $e_{xt}$) on the $t$-th observation (source: Walsh 2010, p. 18). The VAR is system is usually solved by applying least squares over a moving average of the lag operators:
#
# $$y_{t} = \sum\limits_{i=0}^{\infty}a_{1}^{i}u_{yt-i} + \sum\limits_{i=0}^{\infty}a_{1}^{i}a_{2}u_{xt-i-1}$$
#
# #### Deep Neural Network (DNN)
#
# - Nonlinear network representations of sequential processes, for example an RNN with LSTM cells comprising of the following layers and operations:
# * *forget gate layer*: $f_t = \sigma(W_f * [h_{t-1}, x_t] + b_f)$
# * *input gate layer*: $i_t = \sigma(W_i * [h_{t-1}, x_t]+ b_i)$
# * *state update candidate generation op*: $C_t = tanh(W_C * [h_{t-1}, x_t] + b_c)$
# * *internal state update op*: $C_t = f_t * C_{t-1} + i_t * C_t$
# * *output gate layer*: $O_t = \sigma(W_O * [h_{t-1}, x_t] + b_O)$
# * *hidden state output op*: $h_t=O_t * tanh C_t$
#
# where each step in the sequence is modeled in a DNN consisting of a dense layer stacked on to $n$ of the LSTM cells described above. DNNs (either with recurrent or convolutional layers, or a combination of both) have been shown to be powerful enough to allow the modelling of non-linearities and multiple dimensions in time series data.
#
#
# #### Actor-based Computational Economics (ACE)
#
# - Game-theoretical simulations and network models of non-linear complex evolving systems, e.g. using reinforcement learning to optimise stock picks:
# * *environment*:
# * state: market price $s$ of tickers ${a, b, c, ...}$ at time $t$, i.e. ${s_{a,t}, s_{a,t}, s_{a,t}, ...}$
# * *agent*:
# * reward: portfolio value $G = \sum\limits_{t=0}^{t=T} r_t$, the sum (return) of all rewards (increases / decreases in value) of the portfolio of an episode of length $T$.
# * actions: hold, buy $x$, sell $x$ shares of tickers ${a, b, c, ...}$.
# * *policy*: learn a function so as to maximise total reward / return $G$ given the environment and available actions.
# * value function: $V^\pi(s) = E[R|s,\pi]$ the expected value (reward $R$) of a policy $\pi$ given state $s$.
# * state value function: $Q^\pi(s,a) = E[R|s,a,\pi]$ the expected value (reward $R$) of an action $a$ given state $s$ and policy $\pi$.
#
# ## Methodology
#
# The research follows an iterative, step-wise approach in which findings from sub-tasks are used to inform subsequent model-building steps.
#
# At the outset, two steps are identified:
#
# 1. Model the relationship of global economic changes on country-level macroeconomic variables.
# 2. Use the findings in 1) to inform the development of a firm policy simulator that allows local market policy analysis.
# #### Variable under investigation: consumer price inflation
#
# Manipulation of generated country-level policy variables using 1) aggregated (world-level), 2) country-level, and 3) interaction indicator variables as a simulation environment to analyse the impact of government policies, technological change, and commodity prices on consumer price inflation.
| 68.412214
| 2,286
|
57c12856c1402072c60a1dfe696ad3594d4e997e
|
py
|
python
|
Lab_Notebooks/S1_1_Python_Fundamentals.ipynb
|
johannaklahold/2022_ML_Earth_Env_Sci
|
['MIT']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="6waT_XkpBebT"
# # Basic Variables: Numbers and Strings
# + id="eh2mjbN6BpOe"
# comments are anything that comes after the "#" symbol
a = 1 # assign 1 to variable a
b = "hello" # assign "hello" to variable b
# + [markdown] id="v2iA6RH3Bvp1"
# The following identifiers are used as reserved words, or keywords of the language, and cannot be used as ordinary identifiers. They must be spelled exactly as written here:
# + [markdown] id="Fi2nn3tzB3xU"
# ```
# False class finally is return
# None continue for lambda try
# True def from nonlocal while
# and del global not with
# as elif if or yield
# assert else import pass
# break except in raise
# ```
# + [markdown] id="_ZcC-YwUCP5-"
# Additionally, the following a built in functions which are always available in your namespace once you open a Python interpreter
# + [markdown] id="VEa_J6lVCS9D"
# ```
# abs() dict() help() min() setattr() all() dir() hex() next() slice() any()
# divmod() id() object() sorted() ascii() enumerate() input() oct() staticmethod()
# bin() eval() int() open() str() bool() exec() isinstance() ord() sum() bytearray()
# filter() issubclass() pow() super() bytes() float() iter() print() tuple()
# callable() format() len() property() type() chr() frozenset() list() range()
# vars() classmethod() getattr() locals() repr() zip() compile() globals() map()
# reversed() __import__() complex() hasattr() max() round() delattr() hash()
# memoryview() set()
# ```
# + id="kR5n1VLNCgVg"
# how to we see our variables?
print(a)
print(b)
print(a,b)
# + [markdown] id="C1yXwLBlCo48"
# All variables are objects. Every object has a type (class). To find out what type your variables are
# + id="ryTSfvEYCutM"
print(type(a))
print(type(b))
# + id="iFXAA_JjDWNM"
# as a shortcut, iPython notebooks will automatically print whatever is on the last line
type(b)
# + id="YwlGiTAS9HOz"
''' You can make multiline comment using triple quotes. Do note that it does not
really matter if you use single quotes (') or double quotes ("), and different
groups will use different conventions. What is most important is that you're
consistent!
Additionally, you may have noticed that there is a vertical line to the right of
this comment - this is the suggested length limit for each line (80 characters). It's not a hard limit!
But it is suggested that you keep your lines to this length to improve code
readability.'''
'''But this counts as the last line, and will print automatically like before'''
# + id="xuLojJYBDZCB"
# we can check for the type of an object
print(type(a) is int)
print(type(a) is str)
# + id="-bBG9Jwq7SHC"
#f strings allow you to format data easily, but require Python >= 3.6
print(f'The a variable has type {type(a)} and value {a}')
print(f'The b variable has type {type(b)} and value {b}')
# + [markdown] id="ZqIzTDfHDhlA"
# Different objects attributes and methods, which can be accessed via the syntax `variable.method`
#
# IPython will autocomplete if you press `<tab>` to show you the methods available. If you're using Google Colab, you can do the same with `<ctrl> + <space>`
# + id="yTfOLGAlDs1p"
# this returns the method itself
b.capitalize
# + id="GrUqcsqIDwwS"
# this calls the method
b.capitalize()
# there are lots of other methods
# + id="R570VwHCD4Id"
# binary operations act differently on different types of objects
c = 'World'
print(b + c)
print(a + 2)
# + id="Yg_HhkBCGW9I"
print(a + b)
# + [markdown] id="BXqUwjYuEGGg"
# # Math
# + [markdown] id="8UrE--SFEJSb"
# Basic arithmetic and boolean logic is part of the core Python library.
# + id="g0l1x8gmEP43"
# addition / subtraction
1+1-5
# + id="OLv2ZMKrEdSL"
# multiplication
5 * 10
# + id="Dg-qGwZyEhtE"
# division
1/2
# + id="iPeydUeSEjFA"
# that was automatically converted to a float
type(1/2)
# + id="_vE8rLyOEnD3"
# exponentiation
2**4
# + id="3MzrxI3tEo7E"
# rounding
round(9/10)
# + id="dN_vRc8PEr9D"
# built in complex number support
(1+2j) / (3-4j)
# + id="mYap-CwQEvTl"
# logic
True and True
# + id="iZdsFRsyEwmJ"
True and False
# + id="dkv9A8kvE0lR"
True or True
# + id="_hmmntAaE2E-"
(not True) or (not False)
# + [markdown] id="QwzVBDocE4vq"
# # Flow Control
#
# + [markdown] id="5_q3a6nkGyUT"
# ## Conditionals
# + [markdown] id="oBelcHpGE91L"
# The first step to programming & an intro to Python syntax.
# + id="T8aJT1UvFESU"
x = 100
if x > 0:
print('Positive Number')
elif x < 0:
print('Negative Number')
else:
print ('Zero!')
# + id="zsFbWTAqFPjh"
# indentation is MANDATORY
# blocks are closed by indentation level
if x > 0:
print('Positive Number')
if x >= 100:
print('Huge number!')
# + [markdown] id="vbyKoS8kFhlk"
# ## Loops
# + id="iPp59SjhFxxD"
# make a loop
count = 0
while count < 10:
# bad way
# count = count + 1
# better way
count += 1
print(count)
# + [markdown] id="FVah8r57G2-i"
# ## Ranges
# + id="3_Pq8sqVF2Mx"
# use range
for i in range(5):
print(i)
# + [markdown] id="AA48luD5F4LI"
# **Important point**: in Python, we always count from 0!
# + id="7vIMfpbLGEeD"
# what is range?
type(range)
# + id="uB4Ny9YaGIID"
# You can get help in Python by adding ? after an object
# range?
# + id="csQtgDCZGW0h"
# iterate over a list we make up
for pet in ['dog', 'cat', 'fish']:
print(pet, len(pet))
# + [markdown] id="QyqknRFaGYu4"
# What is the thing in brackets? **A list!** Lists are one of the core Python data structures.
# + [markdown] id="HKojPcjYGlW3"
# # Data Structures
# + [markdown] id="u3ByfxgWGorm"
# <a name="Lists"></a>
# ## Lists
# + id="kGKFNMIWJdzF"
l = ['dog', 'cat', 'fish']
type(l)
# + id="vb_1VAGRJgtl"
# list have lots of methods
l.sort()
l
# + id="EbsqKoBgJjM8"
# we can convert a range to a list
r = list(range(5))
r
# + id="oPYGkxiwJngT"
while r:
p = r.pop()
print('p:', p)
print('r:', r)
# + [markdown] id="5DkDjwHvJ5dh"
# There are many different ways to interact with lists. Exploring them is part of the fun of Python.
#
# **list.append(x)** Add an item to the end of the list. Equivalent to `a[len(a):] = [x]`.
#
# **list.extend(L)** Extend the list by appending all the items in the given list. Equivalent to `a[len(a):] = L`.
#
# **list.insert(i, x)** Insert an item at a given position. The first argument is
# the index of the element before which to insert, so `a.insert(0, x)` inserts
# at the front of the list, and `a.insert(len(a), x)` is equivalent to
# `a.append(x)`.
#
# **list.remove(x)** Remove the first item from the list whose value is x. It is an error if there is no such item.
#
# **list.pop([i])** Remove the item at the given position in the list, and return it.
# If no index is specified, `a.pop()` removes and returns the last item in the list.
# (The square brackets around the i in the method signature denote that the
# parameter is optional, not that you should type square brackets at that
# position. You will see this notation frequently in the Python Library Reference.)
#
# **list.clear()** Remove all items from the list. Equivalent to `del a[:]`.
#
# **list.index(x)** Return the index in the list of the first item whose value is x.
# It is an error if there is no such item.
#
# **list.count(x)** Return the number of times x appears in the list.
#
# **list.sort()** Sort the items of the list in place.
#
# **list.reverse()** Reverse the elements of the list in place.
#
# **list.copy()** Return a shallow copy of the list. Equivalent to `a[:]`.
# + [markdown] id="sCiPELD7KzWi"
# Be very careful about how list operations work!
# It's always worth double checking.
# + id="VBB9YHBvLFEI"
# "add" two lists
x = list(range(5))
y = list(range(10,15))
z = x + y
z
# + id="xVHzD9JqLJOi"
# access items from a list
print('first', z[0])
print('last', z[-1])
print('first 3', z[:3])
print('last 3', z[-3:])
print('middle, skipping every other item', z[5:10:2])
# + [markdown] id="3DE2OhaAzFTH"
# **Consider memorizing this syntax!**
# It is central to so much of Python and often proves confusing for users coming from other languages.
# + [markdown] id="WcA5xmUwzIyz"
# In terms of set notation, Python indexing is *left inclusive, right exclusive*. If you remember this, you will never go wrong.
# + id="3tKC5CRZzNr9"
# that means we get an error from the following
N = len(z)
z[N]
# + id="-2_4MU89zfH5"
# this index notation also applies to strings
name = 'Johanna' # Write your full name here
print(name[:4])
# + id="wQnGo7Clzm-Q"
# you can also test for the presence of items in a list
5 in z
# + [markdown] id="ljtMpklWzrEm"
# Lists are not meant for math! They don’t have a datatype.
# + id="VzT89bsTzwDP"
z[4] = 'fish'
z
# + [markdown] id="O8d_n1qJz_-Z"
# Python is full of tricks for iterating and working with lists
# + id="DNQ0OeOj0EhM"
# a cool Python trick: list comprehension
squares = [n**2 for n in range(5)]
squares
# + id="xCZ4Q_u90HDa"
# iterate over two lists together uzing zip
for item1, item2 in zip(x,y):
print('first:', item1, 'second:', item2)
# + [markdown] id="TgqjWKyT0JSv"
# We are almost there. We have the building blocks we need to do basic programming. But Python has some other data structures we need to learn about.
# + [markdown] id="RfEc-2m40PtE"
# ## Tuples
# + [markdown] id="7gIaVYfa0Tsq"
# Tuples are similar to lists, but they are *immutable*—they can’t be extended or modified. What is the point of this? Generally speaking: to pack together inhomogeneous data. Tuples can then be unpacked and distributed by other parts of your code.
# + [markdown] id="-6KdjRmq0dpT"
# Tuples may seem confusing at first, but with time you will come to appreciate them.
# + id="MKOT9XMD0hKw"
# tuples are created with parentheses, or just commas
a = ('Ryan', 33, True)
b = 'Takaya', 25, False
type(b)
# + id="Wp2Y8jz30jVw"
# can be indexed like arrays
print(a[1]) # not the first element!
# + id="XzGBCazt0pHz"
# and they can be unpacked
name, age, status = a
# + [markdown] id="iFMRNBHZ0qtr"
# <a name="Dictionaries"></a>
# ## Dictionaries
# + [markdown] id="pE_jVtju0wzV"
# This is an extremely useful data structure. It maps **keys** to **values**.
#
# Dictionaries are unordered!
# + id="7_3BLE0E05Os"
# different ways to create dictionaries
d = {'name': 'Johanna', 'age': 26} # Write your name and your age here
e = dict(name='Manuel', age=31) # Write someone else's name and their age here
e
# + id="rtlfxEaL2RjY"
# access a value
d['name']
# + [markdown] id="0XL60Qtz2V8I"
# Square brackets `[...]` are Python for “get item” in many different contexts.
# + id="S1T2MRx42acn"
# test for the presence of a key
print('age' in d)
print('height' in e)
# + id="7HuhXXk22cKS"
# try to access a non-existant key
d['height']
# + id="20LVscsW2g5J"
# add a new key
d['height'] = (1,50) # a tuple, e.g. your height in (meters,centimeters)
d
# + id="FouT_hBq2wfA"
# keys don't have to be strings
d[99] = 'ninety nine'
d
# + id="HAgKhynM2yu9"
# iterate over keys
for k in d:
print(k, d[k])
# + id="bJUv_ab-3EGK"
# better way
for key, val in d.items():
print(key, val)
# + [markdown] id="f9nnhgt15fIt"
# # Functions and Classes
# + [markdown] id="vzXIV1G_51VN"
# For longer and more complex tasks, it is important to organize your code into reuseable elements. For example, if you find yourself cutting and pasting the same or similar lines of code over and over, you probably need to define a *function* to encapsulate that code and make it reusable. An important principle in programming in **DRY**: “don’t repeat yourself”. Repetition is tedious and opens you up to errors. Strive for elegance and simplicity in your programs.
# + [markdown] id="kjsvKcre6T36"
# <a name="Functions"></a>
# ## Functions
# + [markdown] id="3Yf3jW616jRD"
# Functions are a central part of advanced python programming. Functions take some inputs (“arguments”) and do something in response. Usually functions return something, but not always.
# + id="9Mnf4o4VIZ-5"
# define a function
def say_hello():
"""Return the word hello."""
return 'Hello'
# + id="ikw5mHhUIfIJ"
# functions are also objects
type(say_hello)
# + id="TI61qJL0Iitl"
# this doesnt call
# say_hello?
# + id="f5WnhuppIo3i"
# this does
say_hello()
# + id="IUKV-PQBIrvu"
# assign the result to something
res = say_hello()
res
# + id="Q6N8OiGPIt_D"
# take some arguments
def say_hello_to(name):
"""Return a greeting to `name`"""
return 'Hello ' + name
# + id="B45GFmD1JU63"
# intended usage
say_hello_to('World')
# + id="dtl2sV1NJWvF"
say_hello_to(10)
# + id="KgyG8PxfJbgR"
# redefine the function
def say_hello_to(name):
"""Return a greeting to `name`"""
return 'Hello ' + str(name)
# + id="XJwrSWFDJcuD"
say_hello_to(10)
# + id="Mnu2O3keJgpO"
# take an optional keyword argument
def say_hello_or_hola(name, french=False):
"""Say hello in multiple languages."""
if french:
greeting = 'Bonjour '
else:
greeting = 'Hello '
return greeting + name
# + id="uJ-y8ccYJ0YP"
print(say_hello_or_hola('')) # Enter your name here
print(say_hello_or_hola('Frédéric', french=True)) # Greet the rector
# + id="Fa-L2UHFKF0i"
# flexible number of arguments
def say_hello_to_everyone(*args):
return ['Bonjour ' + str(a) for a in args]
# + id="Y7eH1WpcKHjA"
say_hello_to_everyone('Niklas', 'Valérie', 'Marie-Élodie') # Greet the deans
# + [markdown] id="uJ6GAy6XLKap"
# ### Pure vs. Impure Functions
# + [markdown] id="mIJByqiRLODp"
# Functions that don’t modify their arguments or produce any other side-effects are called *pure*.
#
# Functions that modify their arguments or cause other actions to occur are called *impure*.
# + [markdown] id="2SGE94AFLXsW"
# Below is an impure function.
# + id="Di5K7iT2Ld85"
def remove_last_from_list(input_list):
input_list.pop()
# + id="GNOLjojELe1W"
names = ['Niklas', 'Valérie', 'Marie-Élodie']
remove_last_from_list(names)
print(names)
remove_last_from_list(names)
print(names)
# + [markdown] id="98OAGfObLqjz"
# We can do something similar with a pure function.
#
# In general, pure functions are safer and more reliable.
# + id="7MY8PcpgLtTt"
def remove_last_from_list_pure(input_list):
new_list = input_list.copy()
new_list.pop()
return new_list
# + id="EMDcZVLiLx_W"
names = ['Niklas', 'Valérie', 'Marie-Élodie']
new_names = remove_last_from_list_pure(names)
print(names)
print(new_names)
# + [markdown] id="hLNtOaDKKt8b"
# We could spend the rest of the day talking about functions, but we have to move on.
# + [markdown] id="gEAn5o8UL8xp"
# ### Namespaces
# + [markdown] id="p6wHhGfqMSsJ"
# In python, a namespace is a mapping between variable names and python object. You can think of it like a dictionary.
#
# The namespace can change depending on where you are in your program. Functions can “see” the variables in the parent namespace, but they can also redefine them in a private scope.
# + id="flPCWOJ1MT3H"
name = 'Johanna' # Enter your name here
# + id="nAiJ-44BL2Tb"
def print_name():
print(name)
def print_name_v2():
name = 'Estelle'
print(name)
print_name()
print_name_v2()
print(name)
# + [markdown] id="GSgKn6oNzaLP"
# It's important that you be aware of the name spaces in your code, specially when dealing with mutable objects.
# + id="0lPV0skBzmga"
friends_list = ['Mario', 'Florence', 'Richard']
pet_tuple = ('Hedwig', 'Na-paw-lyon', 'Cat-hilda')
# + id="qu4UNy2bziet"
def greeter(friends, pets):
print("It's time to say hi to my friends.")
[print(f'Hi {name}! ', end="") for name in friends]
print('\nThese are the names of my pets:')
[print(f'{pet} ', end="") for pet in pets]
print('\n')
def pets_are_friends(friends, pets):
print("I consider both my pets and my friend's pets my friends!")
#add friend's pets
full_pets = pets
full_pets += ('Clifford', 'Crookshanks')
full_friends_list = friends
full_friends_list.extend(full_pets)
print('These are all my friends:')
[print(f'{name} ', end="") for name in full_friends_list]
print('\n')
# + id="KuzrmQOG20zV"
greeter(friends_list, pet_tuple)
pets_are_friends(friends_list, pet_tuple)
greeter(friends_list, pet_tuple)
# + [markdown] id="ImsiXtBiNhWg"
# <a name="Classes"></a>
# ## Classes
# + [markdown] id="2B-qQIb8ONSx"
# We have worked with many different types of python objects so far:
# strings, lists, dictionaries, etc.
# These objects have different attributes and respond in different ways to
# the built-in functions (`len`, etc.)
#
# How can we make our own, custom objects? Answer: by defining classes.
# + [markdown] id="UCZPRuLdObka"
# ### A class to represent a hurricane
# + [markdown] id="JFihKkynnpkZ"
# Let's create our first class below:
# + id="sqs5nuviOnGN"
class Hurricane:
def __init__(self, name):
self.name = name
# + [markdown] id="PfWlZRX5npBY"
# And now let's create an *instance* of that class, `h`:
# + id="ASjmDG4wOp9q"
h = Hurricane('florence')
h
# + [markdown] id="0OZKhsIeOs23"
# Our class only has a single attribute so far, so the instance `h` only has a single attribute as well:
# + id="Df3qyMtXOuXF"
h.name
# + [markdown] id="nwZMRt-ROv_L"
# Let’s add more, along with some input validation:
# + id="__NfaA2tOzBX"
class Hurricane:
def __init__(self, name, category, lon):
self.name = name.upper()
self.category = int(category)
if lon > 180 or lon < -180:
raise ValueError(f'Invalid lon {lon}')
self.lon = lon
# + id="iOMeB_loQlmQ"
h = Hurricane('florence', 4, -46)
h
# + id="wsNidUUXQwHP"
h.name
# + id="RyjkbiLyQ_Og"
h = Hurricane('Ida', 5, 300)
# + [markdown] id="YwNTM1YJRJ1a"
# Now let’s add a custom method:
# + id="ZTprXXYDRMbq"
class Hurricane:
def __init__(self, name, category, lon):
self.name = name.upper()
self.category = int(category)
if lon > 180 or lon < -180:
raise ValueError(f'Invalid lon {lon}')
self.lon = lon
def is_dangerous(self):
return self.category > 1
# + id="7khh5034RReF"
f = Hurricane('florence', 4, -46)
f.is_dangerous()
# + [markdown] id="7qdDLu22RTfM"
# ### Magic / Dunder Methods
# + [markdown] id="Y-KNc9JKRYgp"
# We can implement special methods that begin with double-underscores (i.e. “dunder” methods), which allow us to customize the behavior of our classes. ([Read more here](https://www.python-course.eu/python3_magic_methods.php)). We have already learned one: `__init__`. Let’s implement the `__repr__` method to make our class display something pretty.
#
# + id="u9VRxerKTeHo"
class Hurricane:
def __init__(self, name, category, lon):
self.name = name.upper()
self.category = int(category)
if lon > 180 or lon < -180:
raise ValueError(f'Invalid lon {lon}')
self.lon = lon
def __repr__(self):
return f"<Hurricane {self.name} (cat {self.category})>"
def is_dangerous(self):
return self.category > 1
# + id="zLckKnwsNB2Y"
f = Hurricane('florence', 4, -46)
f
# + [markdown] id="9chcTHjjMe1B"
# # Exercise 1: Creating and Manipulating Simple Data Structures
# + [markdown] id="skoG9IISU14K"
# **Learning Goals**
#
# This assignment will verify that you have the following skills:
#
# * Create lists and dictionaries
# * Iterate through lists, tuples, and dictionaries
# * Index sequences (e.g. lists and tuples)
# * Define functions
# * Use optional keyword arguments in functions
# * Define classes
# * Add a custom method to a class
# + [markdown] id="SD9l8yBjUYcq"
# 
# + [markdown] id="QeijQuaAUgHb"
# Images of the planets in our solar system taken by NASA spacecraft are grouped together to show (from top to bottom) Mercury, Venus, Earth and its moon, Mars, Jupiter, Saturn, Uranus, and Neptune.
# Credits: *NASA*
# + [markdown] id="4rAF7dxIV4sG"
# ## Part I: Lists and Loops
# + [markdown] id="Ro-zgBO8V8z6"
# Hint: *Check out the [Lists subsection](#Lists)*
# + [markdown] id="KhqYJ6XrWFJw"
# **Q1) Create a list with the names of every planet in the solar system (in order)**
#
#
# + id="IsAxsyocWb83"
# Create your list here
planets = ['Mercury','Venus','Earth','Mars','Jupiter','Saturn','Uranus','Neptune']
# + [markdown] id="VtJqCNQuY-WX"
# **Q2) Have Python tell you how many planets there are by examining your list**
# + id="Z6NZf-ujafdT"
# Write your code here
len(planets)
# + [markdown] id="luxRAaIQakvs"
# **Q3) Use slicing to display the first four planets (the rocky planets)**
# + id="iIORvaTRbSeY"
# Write your code here
planets[:4]
# + [markdown] id="tce26Gema6Jc"
# **Q4) Iterate through your planets and print the planet name only if it has an `s` at the end**
# + id="k_SY70I_bCWb"
# Write your code here
for planet in planets:
if planet[-1]=='s': print(planet)
# + [markdown] id="5L6wnk0abhgq"
# ## Part II: Dictionaries
# + [markdown] id="u5T9JrhLc8Y2"
# Hint: *Check out the [Dictionaries subsection](#Dictionaries)*
# + [markdown] id="v0cvw5CpbmOM"
# **Q5) Now create a dictionary that maps each planet name to its mass**
#
# You can use values from this [NASA](https://nssdc.gsfc.nasa.gov/planetary/factsheet/) fact sheet. You can use whatever units you want, but please be consistent.
# + id="9Ij72Bpfb_Dh"
# Write your code here
planets_dict = {'Mercury':0.33, 'Venus':4.87, 'Earth':5.97, 'Mars':0.642, 'Jupiter':1898, 'Saturn':568, 'Uranus':86.6, 'Neptune':102}
# + [markdown] id="cxLpxDMLcDgZ"
# **Q6) Use your dictionary to look up Earth’s mass**
# + id="hbCt0y14cO7f"
# Write your code here
planets_dict['Earth']
# + [markdown] id="uCHeXXiUcRMG"
# **Q7) Loop through the data to create and display a list of planets whose mass is greater than $100 \times 10^{24}$ kg**
# + id="h3Quoxxsdbh9"
# Create the list here
for planet, mass in planets_dict.items():
if mass>100: print(planet)
# + id="R6uOXchRM5Zj"
# Display the list here
# + [markdown] id="4xuQIZY4eAlx"
# **Q8) Now add Pluto to your dictionary**
# + id="G0_cYl6qeNGp"
# Change your dictionary here
planets_dict['Pluto']=0.0130
# + [markdown] id="txSZoB2zeSqR"
# ## Part III: Functions
# + [markdown] id="YrVat4wjqPyW"
# Hint: *Check out the [Functions subsection](#Functions)*
# + [markdown] id="Anyz_CrMebIG"
# **Q9) Write a function to convert from the unit you chose for planetary masses to $M_{Earth}$, the mass of the Earth**
#
# For example, the mass of Jupiter is:
#
# $M_{Jupiter} \approx 1898\times10^{24}kg \approx 318 M_{Earth}$
# + id="wIs-FEisgHtn"
# Write your function here
def convert_mass(mass):
"""Convert mass from 10^24 kg to Earth mass units."""
mass_E = mass/5.97
return mass_E
# + id="E-m-y5Bbhi_u"
# Check that it works for Jupiter
convert_mass(planets_dict['Jupiter'])
# + [markdown] id="wbJzclclhk32"
# **Q10) Now write a single function that can convert from the unit you chose for planetary masses to both $M_{Earth}$ or $M_{Jupiter}$ depending on the keyword argument you specify**
# + id="nW3ajvk1iAM1"
# Write your function here
def convert_mass(mass, unit = 'Earth'):
"""Convert mass from 10^24 kg to Earth (default) or Jupiter mass units."""
if unit == 'Earth': mass_conv = mass/5.97
elif unit == 'Jupiter': mass_conv = mass/1898.
else: print('Invalid unit keyword.')
return mass_conv
# + id="eIX2oONAigX5"
# Check that it works to convert Jupitconvert_mass(planets_dict['Jupiter'])er's mass to $M_{Earth}$
# + id="Jt_v_n4vitnM"
# Bonus: Check that it works to convert Saturn's mass to $M_{Jupiter}$
convert_mass(planets_dict['Saturn'], unit = 'Jupiter')
# + [markdown] id="xB6iqGRni5wq"
# **Q11) Write a function takes one argument (mass in $M_{Jupiter}$) and returns two arguments (mass in $M_{Earth}$ and mass in the unit you chose)**
# + id="Af4iYqy2jmfv"
# Write your function here
def convert_mass_v2(mass):
"""Convert mass from Jupiter mass units to 10^24 kg and Earth mass units."""
mass_kg = mass * 1898.
mass_E = mass_kg/5.97
return mass_kg, mass_E
# + id="uGvaON0UjpEX"
# Check that it works to convert Jupiter's mass to $M_{Earth}$ and to the unit you chose
mass_kg, mass_E = convert_mass_v2(1.)
print(mass_kg, mass_E)
# + id="h-gMPghajw07"
# Bonus: Use the function from Q10 to convert Neptune's mass to $M_{Jupiter}$
# and then the function from Q11 to convert it back to the unit you chose
# Do you get the original value back?
mass = convert_mass(planets_dict['Neptune'], unit = 'Jupiter')
mass_kg, _ = convert_mass_v2(mass)
print(mass_kg)
# + [markdown] id="5s50911okvmB"
# ## Part IV: Classes
# + [markdown] id="aHvWT_RpqVmc"
# Hint: *Check out the [Classes subsection](#Classes)*
# + [markdown] id="GjzqX7Lgk8hJ"
# **Q12) Write a class `planet` with two attributes: `name` and `mass`**
# + id="NevRxMq_m2ta"
# Write your class here
class Planet:
def __init__(self, name, mass):
self.name = name.upper()
self.mass = float(mass)
def __repr__(self):
return f"<Planet {self.name} ({self.mass} * 10^24 kg)>"
# + id="lHOMSMTdnCR9"
# Create a new instance of that class to represent the Earth
e = Planet('Earth', 5.97)
# + id="fqKLa8bNndFy"
# Create a new instance of that class to represent Jupiter
j = Planet('Jupiter', 1898)
# + [markdown] id="CxUgg52cnheI"
# **Q13) Add a custom method `is_light` to the class `planet` that returns `True` if the planet is strictly lighter than Jupiter, and `False` otherwise**
# + id="cNoNKYVZpL6x"
# Change your class here
class Planet:
def __init__(self, name, mass):
self.name = name.upper()
self.mass = float(mass)
def __repr__(self):
return f"<Planet {self.name} ({self.mass} * 10^24 kg)>"
def is_light(self):
return self.mass < 1898
# + id="kONFRjXspOd-"
# Ask Python whether the Earth is lighter than Jupiter
e = Planet('Earth', 5.97)
e.is_light()
# + id="E4vyhiDPpUhL"
# Ask Python whether Jupiter is lighter than itself
j = Planet('Jupiter', 1898)
j.is_light()
# -
| 359.753086
| 324,270
|
e87d2710da09178402e6b621329578f24d612da9
|
py
|
python
|
notebooks/Math Modelling COVID Final Report.ipynb
|
STurner630/mm-major-project-2020
|
['BSD-3-Clause']
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # ZPEM2311 Maths Modelling Project Sophee Turner, Joshua Elphick & Liam Goldsworthy
# ## Research Topic
# How do imposed restrictions impact the future of COVID-19 severity?
#
# Using an SEIR compartmental model, we will be analysing the rates of change among the Susceptible (S), Exposed (E), Infectious (I), and Recovered (R). This analysis will compare the effects of multiple layers of restrictions including masks, quarantine, and travel and social restrictions on the number of people infected with COVID-19 over time. The term restrictions will be used throughout this proposal as a collective term for all of the above imposed restrictions.
# ## Abstract
# This report intends to investigate the impact imposed restrictions have on the severity of COVID-19 infection. It studies the relationship between implementing restrictions and consequential spread. Imposed restrictions may include donning of facemasks, government restrictions on travel through border closures and private gathering limitations, they influence how harshly the population becomes exposed and consequently infected. This report will utilise Victoria as a case study that provides distinct data sets that can be fit into a mathematical model. From this model, data can be extracted through Jupyterlab software using Python coding. The mathematical model that will be used is the epidemiological SEIR model. The way this model works is it segregates the population into four distinct categories, those being Susceptible, Exposed, Infected and Recovered respectively. The rate at which proportions of the population change between these respective states reflects the severity of the spread. This information will be exhibited through various graphs developed on Jupyterlab within this report.
#
# The outcome of the investigation concluded imposed restrictions have a substantial impact on the severity of COVID-19 infection. The data extrapolated through SEIR differential equations demonstrated a 18.83% decrease in population infected and an overall decreased amplitude of the exposed and infected population proportions.
#
# ## Introduction
# Coronavirus disease 2019 (COVID-19) is a highly contagious, respiratory and vascular disease. Common symptoms of this disease include coughing, shortness of breath, lost sense of taste and smell, fatigue and irritated throat. Long term effects of COVID-19 have not yet been defined due to the relatively young age of the disease. It is highly contagious and is most commonly spread through aerosols produced via an infected individual coughing, talking, breathing or sneezing. Government restrictions have been put in place so as to minimise the spread of COVID-19, they address the common forms of spread as mentioned (Victoria State Government, 2020). Through the use of the SEIR with data over a single area (Victoria), we are keeping variables such as population densities, demographic characteristics and infrastructure arrangement constant. This will give a better indication to how imposed restrictions influence the severity.
#
# How do imposed restrictions impact the future of COVID-19 severity? This research question has momentous relevance in the current global pandemic situation. If individuals better understand the impact restrictions have, it will prompt a realisation to the importance abiding fully to such restrictions. Furthermore, the hard evidence that mathematical modelling provides can ensure better governmental decision making on a regional, national and even global level. This justifies the relevance of the question being addressed in the report.
#
# ## Equations SEIR as Defined in our JupyterLab Repository
#
# dSdt= - Beta * S * I
#
# dEdt= ( Beta * S * I) - ( Kappa * E)
#
# dIdt= ( Beta * S * I) -( Gamma * I)
#
# dRdt= Gamma * I
#
# The respective parameters in these equations represent;
#
# = Contraction rate (Beta)
#
# = Mean recovery rate (Gamma)
#
# = Incubation Period (Kappa)
#
# Where,
# S = Susceptible
# E = Exposed
# I = Infected
# R = Recovered
#
# Assumptions,
# = 0.97
# = 14
#
# These equations are slightly different to those presented in the Written Research Proposal submitted earlier. The equations submitted earlier were obtained through online research and belong to authors Godio, Pace and Vergano (2020). The above equations however were deemed appropriate for addressing the research question through some trial runs in Jupyter Lab. These initial runs are illustrated below and demonstrate our differential equations appropriateness:
#
# The below Figures, 1 - 4, have plotted the percentage of the population in each SEIR component against time. The parameter that has been deliberately adjusted to reinforce the appropriateness of the model is parameter Beta () which is the contraction rate. The contraction rate for each of the below figures was 2.5, 2, 1.8 and 1.2 respectively.
from numpy import arange, empty, exp, array, linspace
from scipy.integrate import odeint
from scipy.optimize import minimize
from plotly.offline import init_notebook_mode
from plotly import graph_objs as go
def dSEIRdt(SEIR, t, beta, gamma, kappa):
S, E, I, R = SEIR
dSdt = -beta*S*I
dEdt = beta*S*I-kappa*E
dIdt = beta*S*I-gamma*I
dRdt = gamma*I
return array([dSdt, dEdt, dIdt, dRdt])
# # Basic graph of COVID-19 with no restrictions implemented
# Initial number of infected and recovered individuals, I0 and R0.
# Everyone else, S0, is initially susceptible to infection.
# Initial number of exposed individuals, E0
I0 = 5
S0 = 5926624
R0 = 0
E0 = 0
# Contraction rate, beta, and mean recovery rate, gamma, incubation period (days), kappa.
beta, gamma, kappa = 2.5, .97, 14
#Y axis represents S, E, I and R values
#X axis represents time
t = linspace(0, 30, 30)
SEIR = odeint(dSEIRdt, [1, 0,.00000084, 0], t, (beta, gamma, kappa))
fig = go.Figure()
fig.add_trace(go.Scatter(x=t, y=SEIR[:,0], name='Suseptible'))
fig.add_trace(go.Scatter(x=t, y=SEIR[:,1], name='Exposed'))
fig.add_trace(go.Scatter(x=t, y=SEIR[:,2], name='Infected'))
fig.add_trace(go.Scatter(x=t, y=SEIR[:,3], name='Recovered'))
fig.layout.update(dict(title='SEIR Model - No Restrictions',
xaxis=dict(title='Time (weeks)'),
yaxis=dict(title='Percent of Population x100')))
fig.show('png')
# # Basic graph of COVID-19 with restrictions implemented - marginal effect
# Initial number of infected and recovered individuals, I0 and R0.
# Everyone else, S0, is initially susceptible to infection.
# Initial number of exposed individuals, E0
I0 = 5
S0 = 5926624
R0 = 0
E0 = 0
# Contraction rate, beta, and mean recovery rate, gamma, incubation period, kappa.
beta, gamma, kappa = 2, .97, 14
#Y axis represents S, E, I and R values
#X axis represents time
t = linspace(0, 30, 30)
SEIR = odeint(dSEIRdt, [1, 0,.00000084, 0], t, (beta, gamma, kappa))
fig = go.Figure()
fig.add_trace(go.Scatter(x=t, y=SEIR[:,0], name='Suseptible'))
fig.add_trace(go.Scatter(x=t, y=SEIR[:,1], name='Exposed'))
fig.add_trace(go.Scatter(x=t, y=SEIR[:,2], name='Infected'))
fig.add_trace(go.Scatter(x=t, y=SEIR[:,3], name='Recovered'))
fig.layout.update(dict(title='SEIR Model - Marginal Restrictions',
xaxis=dict(title='Time (weeks)'),
yaxis=dict(title='Percent of Population x100')))
fig.show('png')
# # Basic graph of COVID-19 with restrictions implemented - adequate effect
# Initial number of infected and recovered individuals, I0 and R0.
# Everyone else, S0, is initially susceptible to infection.
# Initial number of exposed individuals, E0
I0 = 5
S0 = 5926624
R0 = 0
E0 = 0
# Contraction rate, beta, and mean recovery rate, gamma, incubation period, kappa.
beta, gamma, kappa = 1.8, .97, 14
#Y axis represents S, E, I and R values
#X axis represents time
t = linspace(0, 40, 40)
SEIR = odeint(dSEIRdt, [1, 0,.00000084, 0], t, (beta, gamma, kappa))
fig = go.Figure()
fig.add_trace(go.Scatter(x=t, y=SEIR[:,0], name='Suseptible'))
fig.add_trace(go.Scatter(x=t, y=SEIR[:,1], name='Exposed'))
fig.add_trace(go.Scatter(x=t, y=SEIR[:,2], name='Infected'))
fig.add_trace(go.Scatter(x=t, y=SEIR[:,3], name='Recovered'))
fig.layout.update(dict(title='SEIR Model - Adequate Restrictions',
xaxis=dict(title='Time (weeks)'),
yaxis=dict(title='Percent of Population x100')))
fig.show('png')
# # Basic graph of COVID-19 with restrictions implemented - great effect
# Initial number of infected and recovered individuals, I0 and R0.
# Everyone else, S0, is initially susceptible to infection.
# Initial number of exposed individuals, E0
I0 = 5
S0 = 5926624
R0 = 0
E0 = 0
# Contraction rate, beta, and mean recovery rate, gamma, incubation period, kappa.
beta, gamma, kappa = 1.2, .97, 14
#Y axis represents S, E, I and R values
#X axis represents time
t = linspace(0, 100, 100)
SEIR = odeint(dSEIRdt, [1, 0,.00000084, 0], t, (beta, gamma, kappa))
fig = go.Figure()
fig.add_trace(go.Scatter(x=t, y=SEIR[:,1], name='Exposed'))
fig.add_trace(go.Scatter(x=t, y=SEIR[:,2], name='Infected'))
fig.layout.update(dict(title='SEIR Model - Strong Restrictions',
xaxis=dict(title='Time (days)'),
yaxis=dict(title='Percent of Population x100')))
fig.show('png')
# ## Observations made are as follows:
#
# S:
# The susceptible population remained susceptible for longer, however was subjected to a far more gradual transition into recovery. The reason for this steadier transition was the far smaller population that had become infected
#
# E:
# The exposed population dropped from around 10% of the population in Figure 1, to 1.3% as evident in Figure 4.
#
# I:
# A drop from 20% of the population being infected to 5% falling into the infected category. Of additional note is that the occurrence of these infections was stretched further in time. The present “flattened curve” means more time and control to the COVID-19 spread will be apparent for the sample population.
#
# R:
# Less of the population is observed to be recovered with increasing (from 90% to 55%), this has occurred due to much less of the population being infected prior.
#
# The above graphs provide a thorough demonstration on how the mathematical model works. The reason for observation in R, being a decreasing recovered population despite reducing contraction rate, is due to the subsequential nature of the SEIR model. Transition through the SEIR stages must occur subsequently, that is in no case is a person able to transition from stage E to R without passing through stage I for example. Therefore, with less people infected, there are less people that can become recovered hence the value drops.
#
# ## Is the Model Appropriate?
#
# The main input that will be adjusted in order to obtain appropriate outputs is the contraction rate (Beta). The output that will be generated is the amount of people exposed and thus infected, this gives information pertaining to the severity of the disease.
#
# The contraction rate is directly dependent on imposed restrictions, thus the model is appropriate for use to address the research question. The above graphs produced outputs that met all expectations that were developed throughout the Written Research Proposal and other prior research. The graphs above are of similar attributes to those of the Reed-Frost SEIR model graphs that analysed the effect of heightened contact rates (Gurarie, 2012), further reinforcing the appropriateness of the chosen model.
#
# With the model described and deemed appropriate, it is now able to be utilised with the COVID-19 Open Data set in order to validate the model for goodness of fit and predictive ability.
#
# ## Model Validation
# For this model we utilized the GoogleCloudPlatform/covid-19-open-data. We specifically used data from Victoria, to validate our data against a state that was suffering from an extreme breakout of infections and decided to implement heavy restrictions to minimise exposure and community transmission.
#
# To enable a comparison, first the data was imported into our jupyter workpage. The imported data sheet is represented in the graph below.
#
import pandas
from plotly import graph_objs as go
data = pandas.read_csv('https://storage.googleapis.com/covid19-open-data/v2/AU_VIC/main.csv')
fig = go.Figure()
fig.add_trace(go.Scatter(x=data['date'], y=data['new_recovered'], name='new_recovered'))
fig.add_trace(go.Scatter(x=data['date'], y=data['new_confirmed'], name='new_confirmed'))
fig.show('png')
# To gain a close comparison as to how the data fit into our model defined previously, we produced a new graph of just exposed and infected trends, this can be seen below.
fig.add_trace(go.Scatter(x=t, y=SEIR[:,1], name='Exposed'))
fig.add_trace(go.Scatter(x=t, y=SEIR[:,2], name='Infected'))
fig.layout.update(dict(title='SEIR Model - Strong Restrictions',
xaxis=dict(title='Time (days)'),
yaxis=dict(title='Percent of Population x100')))
fig.show('png')
# A similarity in the bell-curve tendency of the graphs above can be observed. This provides an initial validation for the fit of our model. Whilst the outline of the graphs are of similar nature, they do have some distinct differences. One key difference is our model sustains a more prolonged time period over which the population spends in the infected category. The real data set demonstrates the peak infection occurred in August, a month on the other side of this peak and the infected could be approximated at 0.
#
# To conduct a full assessment of the validity of our model, it must be fit to the real data set on JupyterLab. By completing this fit, the general tendencies of visual similarity and difference can be observed. Furthermore to visual observation, a RMS function can be conducted that quantitatively indicates the spread of predictions against what actually happens. The RMS provides an indication of error, and is most useful when comparing alternate models to find the most suitable one.
#
#
# Through the attempt to find the goodness of fit and thus validate the predictive ability of our model, we used Fit function to render the below diagram. The blue and red is victorian data, specifically being total recovered and total confirmed respectively. We then have the orange and blue plots, which pertain to our models infected and recovered populations respectively. The comparison is conducted within the time period from 05 July 2020 and 30 September 2020.
#
from numpy import arange, empty, exp, array, linspace
from datetime import date
from plotly.offline import init_notebook_mode
from plotly import graph_objs as go
import pandas
from numpy import zeros, inf
from scipy.integrate import odeint
from scipy.optimize import curve_fit
data = pandas.read_csv('https://storage.googleapis.com/covid19-open-data/v2/AU_VIC/main.csv')
data = data[(data['date'] >= '2020-07-05') & (data['date'] < '2020-09-30')]
def extract_t_from(data):
dates = data['date'].apply(date.fromisoformat).tolist()
return [(d - dates[0]).days for d in dates]
timelist = extract_t_from(data)
def dSEIRdt(SEIR, t):
S, E, I, R = SEIR
dSdt = -beta*S*I
dEdt = beta*S*I-kappa*E
dIdt = beta*S*I-gamma*I
dRdt = gamma*I
return array([dSdt, dEdt, dIdt, dRdt])
def extract_t_from(data):
dates = data['date'].apply(date.fromisoformat).tolist()
return [(d - dates[0]).days for d in dates]
# Initial number of infected and recovered individuals, I0 and R0.
# Everyone else, S0, is initially susceptible to infection.
# Initial number of exposed individuals, E0
I0 = 5
S0 = 30000
R0 = 0
E0 = 0
# Contraction rate, beta, and mean recovery rate, gamma, incubation period (days), kappa.
beta, gamma, kappa = 1.2, .97, 14
t = linspace(0,120,120)
SEIR = odeint(dSEIRdt, [30000, 0,5, 0], t)
fig = go.Figure()
fig.add_trace(go.Scatter(x=data['date'], y=data['total_recovered'], name='total_recovered'))
fig.add_trace(go.Scatter(x=data['date'], y=data['total_confirmed'], name='total_confirmed'))
fig.add_trace(go.Scatter(x=data['date'], y=SEIR[:,0], name='Suseptible'))
fig.add_trace(go.Scatter(x=data['date'], y=SEIR[:,1], name='Exposed'))
fig.add_trace(go.Scatter(x=data['date'], y=SEIR[:,2], name='Infected'))
fig.add_trace(go.Scatter(x=data['date'], y=SEIR[:,3], name='Recovered'))
fig.show('png')
# The graph produced fails to validate the model through the Fit function attempt. This lack of suitability is most likely due to an error in the coding or from incorrect substitution of parameters. Our group explored avenues with different parameters, and alternate approaches to forming the Fit (such as not using String values for the y-axis but rather quantitative figures) however these approaches rendered no more suitable validations. This is unfortunate, as because of the undetermined error we were unable to conduct a Root-Mean-Squared (RMS) function to assess the error produced by the model.
#
# If we were able to use the RMS function, it would grant credibility in the model that extends beyond observed visual comparison
#
# ## Results
# The results obtained from the SEIR model do successfully produce an answer to the proposed research question. The results are divided into answering the research question through the modelling of predicted data, and the comparison of this model to the live data and trends of the COVID-19 spread in Victoria, Australia, in order to validate our findings.
#
#
# _How do imposed restrictions impact infectious rates of COVID-19 in Victoria?_ The SEIR model created using varying contraction rates allows for quantifiable results to manifest. The contraction rate decreases to simulate the increased restrictions imposed on a population and as a result there is a significant decrease in exposed and infected rates in each of the four models.
#
# <b>Contraction Rate</b> - Peak Exposed - Peak Infected
#
# <b>No Restrictions (Beta = 2.5)</b> - 7.04 % - 24.08 %
#
# <b>Marginal Restrictions (Beta = 2)</b> - 4.53 % - 15.85 %
#
# <b>Adequate Restrictions (Beta = 1.8)</b> - 3.38 % - 12.62 %
#
# <b>Strong Restrictions (Beta = 1.2)</b> - 00.13 % - 01.96 %
#
#
# These results show a 5.72% decrease in exposed and 18.83% decrease in infected population between no restrictions and strong restrictions. This indicates that there is a clear decrease in the infected and exposed rates as the contraction rate is decreased. This answers the proposed research question, that as restrictions are increased, the rate of community infection is significantly increased. This indicates that restrictions are effective.
#
# Overall this model was a success, producing results as predicted in the project proposals. It successfully illustrated the effects of restrictions on transmission. As discussed above this was not able to be successfully validated through the comparison to data from Victoria, Australia, however analysis of the Victorian data does still validate this model in a sense. Data from Victoria indicates a similar result to that that this model produced, being that restrictions help to effectively reduce community transmission od COVID-19. Despite the inability to successfully model this data in a graph that would help to validate our findings, the results yielded through the real life implication of heavy restrictions shows the same conclusion that our model found.
#
# ## Conclusions
#
# The intention of this report was to determine the impact imposed restrictions have on the future of COVID-19 severity. Through the development of an appropriate model and utilisation of math modelling our research concluded that severity will reduce significantly if restrictions are imposed. An 18.83% decrease in population infection was found through using differential equations and deliberately varying the parameter pertaining to the contraction rate. The intention of this variation was to mimic the effect of imposed restrictions, the result was influenced by patterns of severity. Our model failed verification against the Victoria data-set but despite this when compared to the raw data visually it is deemed appropriate for concluding imposed restrictions are an effective method of reducing future COVID-19 severity.
#
# ## References
#
# Australian Government Department of Health. (2020, August 03). What you need to know about coronavirus (COVID-19). Retrieved from https://www.health.gov.au/news/health-alerts/novel-coronavirus-2019-ncov-health-alert/what-you-need-to-know-about-coronavirus-covid-19
#
# Davies, N., Eggo, R. M., Kucharski, A. J., Russell, T. W., Liu, Y., & Prem, K. (2020, May). The effect of control strategies to reduce social mixing on outcomes of the COVID-19 epidemic in Wuhan, China: A modelling study. Retrieved from https://www.thelancet.com/action/showPdf?pii=S2468-2667%2820%2930073-6
#
# Godio, A., Pace, F., & Vergnano, A. (2020, May 18). SEIR Modelling of the Italian Epidemic of SARS-CoV-2 Using Computational Swarm Intelligence. Retrieved from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7277829/
# Rachael L. Brown. (2020, June 23). Scientific modelling is steering our response to coronavirus. But what is scientific modelling? Retrieved from https://theconversation.com/scientific-modelling-is-steering-our-response-to-coronavirus-but-what-is-scientific-modelling-135938
#
# Radulescu, A., & Cavanagh, K. (2020, March 26). Management strategies in a SEIR model of COVID 19 community spread. Retrieved August 06, 2020, from https://arxiv.org/pdf/2003.11150.pdf
#
# What you need to know – Version 11. (2020, March 11). Retrieved from https://www.health.gov.au/sites/default/files/documents/2020/03/coronavirus-covid-19-what-you-need-to-know_1.pdf?mc_cid=c24f0a9ced&mc_eid=4c27d1d1ea
#
# Worldometer. (2020). Coronavirus Cases Retrieved from https://www.worldometers.info/coronavirus/
#
# ## Contributions
# Coding primarily conducted by Liam Goldsworthy with member input at various stages. All coding conducted through Liam Goldsworthy's account however, all members had some contributions. The report was completed primarily by Josh Elphick, with some sections being completed by Sophee Turner. Sophee also conducted oversight and proofing of the report and the project overall. Layout and collation conducted by Liam Goldsworthy.
| 63.900568
| 1,108
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.