\nusing Random\n\n# Print without new line\nprint(\"Hello World\")\n\n# Print with new line\nprintln(\"Hello World\")\n\nfor i in range(1, 10, step = 1)\n println(i)\nend\n\nfunction square(a::Int)\n return a*a\nend\n\n# Call the function\nprint(square(6))\n\n# Vectorization \nvec_sq = square.([1, 2, 3, 4])\n\n\n\n# Mapping\nmapped_vec = map(x->square(x)+4, [1,2,3,4])", "meta": {"hexsha": "8f612686de02957873eeec04cb13d31e502732fb", "size": 368, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Study_Session_1/Introduction.jl", "max_stars_repo_name": "khanfarhan10/Study-Group", "max_stars_repo_head_hexsha": "5e5e9c74890a2f31ad9cb67b199db9eac6b6444a", "max_stars_repo_licenses": ["Apache-2.0"], "max_stars_count": 6, "max_stars_repo_stars_event_min_datetime": "2021-01-15T11:06:03.000Z", "max_stars_repo_stars_event_max_datetime": "2021-05-31T20:22:13.000Z", "max_issues_repo_path": "Study_Session_1/Introduction.jl", "max_issues_repo_name": "chaitak-gorai/Study-Group", "max_issues_repo_head_hexsha": "add27a3c6b4d890816ea210b7377f1c5db3c1853", "max_issues_repo_licenses": ["Apache-2.0"], "max_issues_count": 2, "max_issues_repo_issues_event_min_datetime": "2021-01-30T18:32:28.000Z", "max_issues_repo_issues_event_max_datetime": "2021-05-25T18:15:41.000Z", "max_forks_repo_path": "Study_Session_1/Introduction.jl", "max_forks_repo_name": "chaitak-gorai/Study-Group", "max_forks_repo_head_hexsha": "add27a3c6b4d890816ea210b7377f1c5db3c1853", "max_forks_repo_licenses": ["Apache-2.0"], "max_forks_count": 3, "max_forks_repo_forks_event_min_datetime": "2021-01-31T14:11:31.000Z", "max_forks_repo_forks_event_max_datetime": "2021-05-23T11:12:53.000Z", "avg_line_length": 13.6296296296, "max_line_length": 43, "alphanum_fraction": 0.6576086957, "num_tokens": 120, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.38491214448393346, "lm_q2_score": 0.21206881431678098, "lm_q1q2_score": 0.08162786209683726}}
{"text": "### A Pluto.jl notebook ###\n# v0.19.8\n\nusing Markdown\nusing InteractiveUtils\n\n# This Pluto notebook uses @bind for interactivity. When running this notebook outside of Pluto, the following 'mock version' of @bind gives bound variables a default value (instead of an error).\nmacro bind(def, element)\n quote\n local iv = try Base.loaded_modules[Base.PkgId(Base.UUID(\"6e696c72-6542-2067-7265-42206c756150\"), \"AbstractPlutoDingetjes\")].Bonds.initial_value catch; b -> missing; end\n local el = $(esc(element))\n global $(esc(def)) = Core.applicable(Base.get, el) ? Base.get(el) : iv(el)\n el\n end\nend\n\n# \u2554\u2550\u2561 1f0cb625-d1ec-4775-bc75-59d4675d181e\n# \u2560\u2550\u2561 show_logs = false\nbegin\n\tusing Logging\n\tglobal_logger(NullLogger())\n\tdisplay(\"\")\nend\n\n# \u2554\u2550\u2561 25448e4a-345b-4bd6-9cdb-a6a280cfd22c\n# \u2560\u2550\u2561 show_logs = false\n#Set-up packages\nbegin\n\t\n\tusing PlutoUI, DataFrames, HTTP, CSV, Dates, Printf, LaTeXStrings, HypertextLiteral, XLSX\n\t\n\t# gr();\n\t# Plots.GRBackend()\n\n\n\t#Define html elements\n\tnbsp = html\" \" #non-breaking space\n\tvspace = html\"\"\"
\"\"\"\n\tbr = html\"\n\t# \t\n\t# \t
\n\t# \"\"\")\n\n\t\n\t# cellWidth= min(1000, screenWidth*0.9)\n\t# @htl(\"\"\"\n\t# \t\n\t# \"\"\")\n\t\n\n\t#Sets the width of the cells\n\t#begin\n\t#\thtml\"\"\"\"\"\"\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000001\nPLUTO_PROJECT_TOML_CONTENTS = \"\"\"\n[deps]\nPlots = \"91a5bcdd-55d7-5caf-9e0b-520d859cae80\"\nPlutoUI = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\nPrettyTables = \"08abe8d2-0d0c-5749-adfa-8a2ac140af0d\"\n\n[compat]\nPlots = \"~1.25.8\"\nPlutoUI = \"~0.7.34\"\nPrettyTables = \"~1.3.1\"\n\"\"\"\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000002\nPLUTO_MANIFEST_TOML_CONTENTS = \"\"\"\n# This file is machine-generated - editing it directly is not advised\n\njulia_version = \"1.7.2\"\nmanifest_format = \"2.0\"\n\n[[deps.AbstractPlutoDingetjes]]\ndeps = [\"Pkg\"]\ngit-tree-sha1 = \"8eaf9f1b4921132a4cff3f36a1d9ba923b14a481\"\nuuid = \"6e696c72-6542-2067-7265-42206c756150\"\nversion = \"1.1.4\"\n\n[[deps.Adapt]]\ndeps = [\"LinearAlgebra\"]\ngit-tree-sha1 = \"af92965fb30777147966f58acb05da51c5616b5f\"\nuuid = \"79e6a3ab-5dfb-504d-930d-738a2a938a0e\"\nversion = \"3.3.3\"\n\n[[deps.ArgTools]]\nuuid = \"0dad84c5-d112-42e6-8d28-ef12dabb789f\"\n\n[[deps.Artifacts]]\nuuid = \"56f22d72-fd6d-98f1-02f0-08ddc0907c33\"\n\n[[deps.Base64]]\nuuid = \"2a0f44e3-6c83-55bd-87e4-b1978d98bd5f\"\n\n[[deps.Bzip2_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"19a35467a82e236ff51bc17a3a44b69ef35185a2\"\nuuid = \"6e34b625-4abd-537c-b88f-471c36dfa7a0\"\nversion = \"1.0.8+0\"\n\n[[deps.Cairo]]\ndeps = [\"Cairo_jll\", \"Colors\", \"Glib_jll\", \"Graphics\", \"Libdl\", \"Pango_jll\"]\ngit-tree-sha1 = \"d0b3f8b4ad16cb0a2988c6788646a5e6a17b6b1b\"\nuuid = \"159f3aea-2a34-519c-b102-8c37f9878175\"\nversion = \"1.0.5\"\n\n[[deps.Cairo_jll]]\ndeps = [\"Artifacts\", \"Bzip2_jll\", \"Fontconfig_jll\", \"FreeType2_jll\", \"Glib_jll\", \"JLLWrappers\", \"LZO_jll\", \"Libdl\", \"Pixman_jll\", \"Pkg\", \"Xorg_libXext_jll\", \"Xorg_libXrender_jll\", \"Zlib_jll\", \"libpng_jll\"]\ngit-tree-sha1 = \"4b859a208b2397a7a623a03449e4636bdb17bcf2\"\nuuid = \"83423d85-b0ee-5818-9007-b63ccbeb887a\"\nversion = \"1.16.1+1\"\n\n[[deps.ChainRulesCore]]\ndeps = [\"Compat\", \"LinearAlgebra\", \"SparseArrays\"]\ngit-tree-sha1 = \"f9982ef575e19b0e5c7a98c6e75ee496c0f73a93\"\nuuid = \"d360d2e6-b24c-11e9-a2a3-2a2ae2dbcce4\"\nversion = \"1.12.0\"\n\n[[deps.ChangesOfVariables]]\ndeps = [\"ChainRulesCore\", \"LinearAlgebra\", \"Test\"]\ngit-tree-sha1 = \"bf98fa45a0a4cee295de98d4c1462be26345b9a1\"\nuuid = \"9e997f8a-9a97-42d5-a9f1-ce6bfc15e2c0\"\nversion = \"0.1.2\"\n\n[[deps.ColorSchemes]]\ndeps = [\"ColorTypes\", \"Colors\", \"FixedPointNumbers\", \"Luxor\", \"Random\"]\ngit-tree-sha1 = \"5b7d2a8b53c68dfdbce545e957a3b5cc4da80b01\"\nuuid = \"35d6a980-a343-548e-a6ea-1d62b119f2f4\"\nversion = \"3.17.0\"\n\n[[deps.ColorTypes]]\ndeps = [\"FixedPointNumbers\", \"Random\"]\ngit-tree-sha1 = \"024fe24d83e4a5bf5fc80501a314ce0d1aa35597\"\nuuid = \"3da002f7-5984-5a60-b8a6-cbb66c0b333f\"\nversion = \"0.11.0\"\n\n[[deps.Colors]]\ndeps = [\"ColorTypes\", \"FixedPointNumbers\", \"Reexport\"]\ngit-tree-sha1 = \"417b0ed7b8b838aa6ca0a87aadf1bb9eb111ce40\"\nuuid = \"5ae59095-9a9b-59fe-a467-6f913c188581\"\nversion = \"0.12.8\"\n\n[[deps.Compat]]\ndeps = [\"Base64\", \"Dates\", \"DelimitedFiles\", \"Distributed\", \"InteractiveUtils\", \"LibGit2\", \"Libdl\", \"LinearAlgebra\", \"Markdown\", \"Mmap\", \"Pkg\", \"Printf\", \"REPL\", \"Random\", \"SHA\", \"Serialization\", \"SharedArrays\", \"Sockets\", \"SparseArrays\", \"Statistics\", \"Test\", \"UUIDs\", \"Unicode\"]\ngit-tree-sha1 = \"44c37b4636bc54afac5c574d2d02b625349d6582\"\nuuid = \"34da2185-b29b-5c13-b0c7-acf172513d20\"\nversion = \"3.41.0\"\n\n[[deps.CompilerSupportLibraries_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"e66e0078-7015-5450-92f7-15fbd957f2ae\"\n\n[[deps.Contour]]\ndeps = [\"StaticArrays\"]\ngit-tree-sha1 = \"9f02045d934dc030edad45944ea80dbd1f0ebea7\"\nuuid = \"d38c429a-6771-53c6-b99e-75d170b6e991\"\nversion = \"0.5.7\"\n\n[[deps.Crayons]]\ngit-tree-sha1 = \"249fe38abf76d48563e2f4556bebd215aa317e15\"\nuuid = \"a8cc5b0e-0ffa-5ad4-8c14-923d3ee1735f\"\nversion = \"4.1.1\"\n\n[[deps.DataAPI]]\ngit-tree-sha1 = \"cc70b17275652eb47bc9e5f81635981f13cea5c8\"\nuuid = \"9a962f9c-6df0-11e9-0e5d-c546b8b5ee8a\"\nversion = \"1.9.0\"\n\n[[deps.DataStructures]]\ndeps = [\"Compat\", \"InteractiveUtils\", \"OrderedCollections\"]\ngit-tree-sha1 = \"3daef5523dd2e769dad2365274f760ff5f282c7d\"\nuuid = \"864edb3b-99cc-5e75-8d2d-829cb0a9cfe8\"\nversion = \"0.18.11\"\n\n[[deps.DataValueInterfaces]]\ngit-tree-sha1 = \"bfc1187b79289637fa0ef6d4436ebdfe6905cbd6\"\nuuid = \"e2d170a0-9d28-54be-80f0-106bbe20a464\"\nversion = \"1.0.0\"\n\n[[deps.Dates]]\ndeps = [\"Printf\"]\nuuid = \"ade2ca70-3891-5945-98fb-dc099432e06a\"\n\n[[deps.DelimitedFiles]]\ndeps = [\"Mmap\"]\nuuid = \"8bb1440f-4735-579b-a4ab-409b98df4dab\"\n\n[[deps.Distributed]]\ndeps = [\"Random\", \"Serialization\", \"Sockets\"]\nuuid = \"8ba89e20-285c-5b6f-9357-94700520ee1b\"\n\n[[deps.DocStringExtensions]]\ndeps = [\"LibGit2\"]\ngit-tree-sha1 = \"b19534d1895d702889b219c382a6e18010797f0b\"\nuuid = \"ffbed154-4ef7-542d-bbb7-c09d3a79fcae\"\nversion = \"0.8.6\"\n\n[[deps.Downloads]]\ndeps = [\"ArgTools\", \"LibCURL\", \"NetworkOptions\"]\nuuid = \"f43a241f-c20a-4ad4-852c-f6b1247861c6\"\n\n[[deps.EarCut_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"3f3a2501fa7236e9b911e0f7a588c657e822bb6d\"\nuuid = \"5ae413db-bbd1-5e63-b57d-d24a61df00f5\"\nversion = \"2.2.3+0\"\n\n[[deps.Expat_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"ae13fcbc7ab8f16b0856729b050ef0c446aa3492\"\nuuid = \"2e619515-83b5-522b-bb60-26c02a35a201\"\nversion = \"2.4.4+0\"\n\n[[deps.FFMPEG]]\ndeps = [\"FFMPEG_jll\"]\ngit-tree-sha1 = \"b57e3acbe22f8484b4b5ff66a7499717fe1a9cc8\"\nuuid = \"c87230d0-a227-11e9-1b43-d7ebe4e7570a\"\nversion = \"0.4.1\"\n\n[[deps.FFMPEG_jll]]\ndeps = [\"Artifacts\", \"Bzip2_jll\", \"FreeType2_jll\", \"FriBidi_jll\", \"JLLWrappers\", \"LAME_jll\", \"Libdl\", \"Ogg_jll\", \"OpenSSL_jll\", \"Opus_jll\", \"Pkg\", \"Zlib_jll\", \"libass_jll\", \"libfdk_aac_jll\", \"libvorbis_jll\", \"x264_jll\", \"x265_jll\"]\ngit-tree-sha1 = \"d8a578692e3077ac998b50c0217dfd67f21d1e5f\"\nuuid = \"b22a6f82-2f65-5046-a5b2-351ab43fb4e5\"\nversion = \"4.4.0+0\"\n\n[[deps.FileIO]]\ndeps = [\"Pkg\", \"Requires\", \"UUIDs\"]\ngit-tree-sha1 = \"80ced645013a5dbdc52cf70329399c35ce007fae\"\nuuid = \"5789e2e9-d7fb-5bc7-8068-2c6fae9b9549\"\nversion = \"1.13.0\"\n\n[[deps.FixedPointNumbers]]\ndeps = [\"Statistics\"]\ngit-tree-sha1 = \"335bfdceacc84c5cdf16aadc768aa5ddfc5383cc\"\nuuid = \"53c48c17-4a7d-5ca2-90c5-79b7896eea93\"\nversion = \"0.8.4\"\n\n[[deps.Fontconfig_jll]]\ndeps = [\"Artifacts\", \"Bzip2_jll\", \"Expat_jll\", \"FreeType2_jll\", \"JLLWrappers\", \"Libdl\", \"Libuuid_jll\", \"Pkg\", \"Zlib_jll\"]\ngit-tree-sha1 = \"21efd19106a55620a188615da6d3d06cd7f6ee03\"\nuuid = \"a3f928ae-7b40-5064-980b-68af3947d34b\"\nversion = \"2.13.93+0\"\n\n[[deps.Formatting]]\ndeps = [\"Printf\"]\ngit-tree-sha1 = \"8339d61043228fdd3eb658d86c926cb282ae72a8\"\nuuid = \"59287772-0a20-5a39-b81b-1366585eb4c0\"\nversion = \"0.4.2\"\n\n[[deps.FreeType2_jll]]\ndeps = [\"Artifacts\", \"Bzip2_jll\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Zlib_jll\"]\ngit-tree-sha1 = \"87eb71354d8ec1a96d4a7636bd57a7347dde3ef9\"\nuuid = \"d7e528f0-a631-5988-bf34-fe36492bcfd7\"\nversion = \"2.10.4+0\"\n\n[[deps.FriBidi_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"aa31987c2ba8704e23c6c8ba8a4f769d5d7e4f91\"\nuuid = \"559328eb-81f9-559d-9380-de523a88c83c\"\nversion = \"1.0.10+0\"\n\n[[deps.GLFW_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Libglvnd_jll\", \"Pkg\", \"Xorg_libXcursor_jll\", \"Xorg_libXi_jll\", \"Xorg_libXinerama_jll\", \"Xorg_libXrandr_jll\"]\ngit-tree-sha1 = \"51d2dfe8e590fbd74e7a842cf6d13d8a2f45dc01\"\nuuid = \"0656b61e-2033-5cc2-a64a-77c0f6c09b89\"\nversion = \"3.3.6+0\"\n\n[[deps.GR]]\ndeps = [\"Base64\", \"DelimitedFiles\", \"GR_jll\", \"HTTP\", \"JSON\", \"Libdl\", \"LinearAlgebra\", \"Pkg\", \"Printf\", \"Random\", \"RelocatableFolders\", \"Serialization\", \"Sockets\", \"Test\", \"UUIDs\"]\ngit-tree-sha1 = \"4a740db447aae0fbeb3ee730de1afbb14ac798a1\"\nuuid = \"28b8d3ca-fb5f-59d9-8090-bfdbd6d07a71\"\nversion = \"0.63.1\"\n\n[[deps.GR_jll]]\ndeps = [\"Artifacts\", \"Bzip2_jll\", \"Cairo_jll\", \"FFMPEG_jll\", \"Fontconfig_jll\", \"GLFW_jll\", \"JLLWrappers\", \"JpegTurbo_jll\", \"Libdl\", \"Libtiff_jll\", \"Pixman_jll\", \"Pkg\", \"Qt5Base_jll\", \"Zlib_jll\", \"libpng_jll\"]\ngit-tree-sha1 = \"aa22e1ee9e722f1da183eb33370df4c1aeb6c2cd\"\nuuid = \"d2c73de3-f751-5644-a686-071e5b155ba9\"\nversion = \"0.63.1+0\"\n\n[[deps.GeometryBasics]]\ndeps = [\"EarCut_jll\", \"IterTools\", \"LinearAlgebra\", \"StaticArrays\", \"StructArrays\", \"Tables\"]\ngit-tree-sha1 = \"58bcdf5ebc057b085e58d95c138725628dd7453c\"\nuuid = \"5c1252a2-5f33-56bf-86c9-59e7332b4326\"\nversion = \"0.4.1\"\n\n[[deps.Gettext_jll]]\ndeps = [\"Artifacts\", \"CompilerSupportLibraries_jll\", \"JLLWrappers\", \"Libdl\", \"Libiconv_jll\", \"Pkg\", \"XML2_jll\"]\ngit-tree-sha1 = \"9b02998aba7bf074d14de89f9d37ca24a1a0b046\"\nuuid = \"78b55507-aeef-58d4-861c-77aaff3498b1\"\nversion = \"0.21.0+0\"\n\n[[deps.Glib_jll]]\ndeps = [\"Artifacts\", \"Gettext_jll\", \"JLLWrappers\", \"Libdl\", \"Libffi_jll\", \"Libiconv_jll\", \"Libmount_jll\", \"PCRE_jll\", \"Pkg\", \"Zlib_jll\"]\ngit-tree-sha1 = \"a32d672ac2c967f3deb8a81d828afc739c838a06\"\nuuid = \"7746bdde-850d-59dc-9ae8-88ece973131d\"\nversion = \"2.68.3+2\"\n\n[[deps.Graphics]]\ndeps = [\"Colors\", \"LinearAlgebra\", \"NaNMath\"]\ngit-tree-sha1 = \"1c5a84319923bea76fa145d49e93aa4394c73fc2\"\nuuid = \"a2bd30eb-e257-5431-a919-1863eab51364\"\nversion = \"1.1.1\"\n\n[[deps.Graphite2_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"344bf40dcab1073aca04aa0df4fb092f920e4011\"\nuuid = \"3b182d85-2403-5c21-9c21-1e1f0cc25472\"\nversion = \"1.3.14+0\"\n\n[[deps.Grisu]]\ngit-tree-sha1 = \"53bb909d1151e57e2484c3d1b53e19552b887fb2\"\nuuid = \"42e2da0e-8278-4e71-bc24-59509adca0fe\"\nversion = \"1.0.2\"\n\n[[deps.HTTP]]\ndeps = [\"Base64\", \"Dates\", \"IniFile\", \"Logging\", \"MbedTLS\", \"NetworkOptions\", \"Sockets\", \"URIs\"]\ngit-tree-sha1 = \"0fa77022fe4b511826b39c894c90daf5fce3334a\"\nuuid = \"cd3eb016-35fb-5094-929b-558a96fad6f3\"\nversion = \"0.9.17\"\n\n[[deps.HarfBuzz_jll]]\ndeps = [\"Artifacts\", \"Cairo_jll\", \"Fontconfig_jll\", \"FreeType2_jll\", \"Glib_jll\", \"Graphite2_jll\", \"JLLWrappers\", \"Libdl\", \"Libffi_jll\", \"Pkg\"]\ngit-tree-sha1 = \"129acf094d168394e80ee1dc4bc06ec835e510a3\"\nuuid = \"2e76f6c2-a576-52d4-95c1-20adfe4de566\"\nversion = \"2.8.1+1\"\n\n[[deps.Hyperscript]]\ndeps = [\"Test\"]\ngit-tree-sha1 = \"8d511d5b81240fc8e6802386302675bdf47737b9\"\nuuid = \"47d2ed2b-36de-50cf-bf87-49c2cf4b8b91\"\nversion = \"0.0.4\"\n\n[[deps.HypertextLiteral]]\ngit-tree-sha1 = \"2b078b5a615c6c0396c77810d92ee8c6f470d238\"\nuuid = \"ac1192a8-f4b3-4bfe-ba22-af5b92cd3ab2\"\nversion = \"0.9.3\"\n\n[[deps.IOCapture]]\ndeps = [\"Logging\", \"Random\"]\ngit-tree-sha1 = \"f7be53659ab06ddc986428d3a9dcc95f6fa6705a\"\nuuid = \"b5f81e59-6552-4d32-b1f0-c071b021bf89\"\nversion = \"0.2.2\"\n\n[[deps.IniFile]]\ndeps = [\"Test\"]\ngit-tree-sha1 = \"098e4d2c533924c921f9f9847274f2ad89e018b8\"\nuuid = \"83e8ac13-25f8-5344-8a64-a9f2b223428f\"\nversion = \"0.5.0\"\n\n[[deps.InteractiveUtils]]\ndeps = [\"Markdown\"]\nuuid = \"b77e0a4c-d291-57a0-90e8-8db25a27a240\"\n\n[[deps.InverseFunctions]]\ndeps = [\"Test\"]\ngit-tree-sha1 = \"a7254c0acd8e62f1ac75ad24d5db43f5f19f3c65\"\nuuid = \"3587e190-3f89-42d0-90ee-14403ec27112\"\nversion = \"0.1.2\"\n\n[[deps.IrrationalConstants]]\ngit-tree-sha1 = \"7fd44fd4ff43fc60815f8e764c0f352b83c49151\"\nuuid = \"92d709cd-6900-40b7-9082-c6be49f344b6\"\nversion = \"0.1.1\"\n\n[[deps.IterTools]]\ngit-tree-sha1 = \"fa6287a4469f5e048d763df38279ee729fbd44e5\"\nuuid = \"c8e1da08-722c-5040-9ed9-7db0dc04731e\"\nversion = \"1.4.0\"\n\n[[deps.IteratorInterfaceExtensions]]\ngit-tree-sha1 = \"a3f24677c21f5bbe9d2a714f95dcd58337fb2856\"\nuuid = \"82899510-4779-5014-852e-03e436cf321d\"\nversion = \"1.0.0\"\n\n[[deps.JLLWrappers]]\ndeps = [\"Preferences\"]\ngit-tree-sha1 = \"abc9885a7ca2052a736a600f7fa66209f96506e1\"\nuuid = \"692b3bcd-3c85-4b1f-b108-f13ce0eb3210\"\nversion = \"1.4.1\"\n\n[[deps.JSON]]\ndeps = [\"Dates\", \"Mmap\", \"Parsers\", \"Unicode\"]\ngit-tree-sha1 = \"8076680b162ada2a031f707ac7b4953e30667a37\"\nuuid = \"682c06a0-de6a-54ab-a142-c8b1cf79cde6\"\nversion = \"0.21.2\"\n\n[[deps.JpegTurbo_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"b53380851c6e6664204efb2e62cd24fa5c47e4ba\"\nuuid = \"aacddb02-875f-59d6-b918-886e6ef4fbf8\"\nversion = \"2.1.2+0\"\n\n[[deps.Juno]]\ndeps = [\"Base64\", \"Logging\", \"Media\", \"Profile\"]\ngit-tree-sha1 = \"07cb43290a840908a771552911a6274bc6c072c7\"\nuuid = \"e5e0dc1b-0480-54bc-9374-aad01c23163d\"\nversion = \"0.8.4\"\n\n[[deps.LAME_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"f6250b16881adf048549549fba48b1161acdac8c\"\nuuid = \"c1c5ebd0-6772-5130-a774-d5fcae4a789d\"\nversion = \"3.100.1+0\"\n\n[[deps.LZO_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"e5b909bcf985c5e2605737d2ce278ed791b89be6\"\nuuid = \"dd4b983a-f0e5-5f8d-a1b7-129d4a5fb1ac\"\nversion = \"2.10.1+0\"\n\n[[deps.LaTeXStrings]]\ngit-tree-sha1 = \"f2355693d6778a178ade15952b7ac47a4ff97996\"\nuuid = \"b964fa9f-0449-5b57-a5c2-d3ea65f4040f\"\nversion = \"1.3.0\"\n\n[[deps.Latexify]]\ndeps = [\"Formatting\", \"InteractiveUtils\", \"LaTeXStrings\", \"MacroTools\", \"Markdown\", \"Printf\", \"Requires\"]\ngit-tree-sha1 = \"a8f4f279b6fa3c3c4f1adadd78a621b13a506bce\"\nuuid = \"23fbe1c1-3f47-55db-b15f-69d7ec21a316\"\nversion = \"0.15.9\"\n\n[[deps.LibCURL]]\ndeps = [\"LibCURL_jll\", \"MozillaCACerts_jll\"]\nuuid = \"b27032c2-a3e7-50c8-80cd-2d36dbcbfd21\"\n\n[[deps.LibCURL_jll]]\ndeps = [\"Artifacts\", \"LibSSH2_jll\", \"Libdl\", \"MbedTLS_jll\", \"Zlib_jll\", \"nghttp2_jll\"]\nuuid = \"deac9b47-8bc7-5906-a0fe-35ac56dc84c0\"\n\n[[deps.LibGit2]]\ndeps = [\"Base64\", \"NetworkOptions\", \"Printf\", \"SHA\"]\nuuid = \"76f85450-5226-5b5a-8eaa-529ad045b433\"\n\n[[deps.LibSSH2_jll]]\ndeps = [\"Artifacts\", \"Libdl\", \"MbedTLS_jll\"]\nuuid = \"29816b5a-b9ab-546f-933c-edad1886dfa8\"\n\n[[deps.Libdl]]\nuuid = \"8f399da3-3557-5675-b5ff-fb832c97cbdb\"\n\n[[deps.Libffi_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"0b4a5d71f3e5200a7dff793393e09dfc2d874290\"\nuuid = \"e9f186c6-92d2-5b65-8a66-fee21dc1b490\"\nversion = \"3.2.2+1\"\n\n[[deps.Libgcrypt_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Libgpg_error_jll\", \"Pkg\"]\ngit-tree-sha1 = \"64613c82a59c120435c067c2b809fc61cf5166ae\"\nuuid = \"d4300ac3-e22c-5743-9152-c294e39db1e4\"\nversion = \"1.8.7+0\"\n\n[[deps.Libglvnd_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libX11_jll\", \"Xorg_libXext_jll\"]\ngit-tree-sha1 = \"7739f837d6447403596a75d19ed01fd08d6f56bf\"\nuuid = \"7e76a0d4-f3c7-5321-8279-8d96eeed0f29\"\nversion = \"1.3.0+3\"\n\n[[deps.Libgpg_error_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"c333716e46366857753e273ce6a69ee0945a6db9\"\nuuid = \"7add5ba3-2f88-524e-9cd5-f83b8a55f7b8\"\nversion = \"1.42.0+0\"\n\n[[deps.Libiconv_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"42b62845d70a619f063a7da093d995ec8e15e778\"\nuuid = \"94ce4f54-9a6c-5748-9c1c-f9c7231a4531\"\nversion = \"1.16.1+1\"\n\n[[deps.Libmount_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"9c30530bf0effd46e15e0fdcf2b8636e78cbbd73\"\nuuid = \"4b2f31a3-9ecc-558c-b454-b3730dcb73e9\"\nversion = \"2.35.0+0\"\n\n[[deps.Librsvg_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pango_jll\", \"Pkg\", \"gdk_pixbuf_jll\"]\ngit-tree-sha1 = \"25d5e6b4eb3558613ace1c67d6a871420bfca527\"\nuuid = \"925c91fb-5dd6-59dd-8e8c-345e74382d89\"\nversion = \"2.52.4+0\"\n\n[[deps.Libtiff_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"JpegTurbo_jll\", \"Libdl\", \"Pkg\", \"Zlib_jll\", \"Zstd_jll\"]\ngit-tree-sha1 = \"340e257aada13f95f98ee352d316c3bed37c8ab9\"\nuuid = \"89763e89-9b03-5906-acba-b20f662cd828\"\nversion = \"4.3.0+0\"\n\n[[deps.Libuuid_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"7f3efec06033682db852f8b3bc3c1d2b0a0ab066\"\nuuid = \"38a345b3-de98-5d2b-a5d3-14cd9215e700\"\nversion = \"2.36.0+0\"\n\n[[deps.LinearAlgebra]]\ndeps = [\"Libdl\", \"libblastrampoline_jll\"]\nuuid = \"37e2e46d-f89d-539d-b4ee-838fcccc9c8e\"\n\n[[deps.LogExpFunctions]]\ndeps = [\"ChainRulesCore\", \"ChangesOfVariables\", \"DocStringExtensions\", \"InverseFunctions\", \"IrrationalConstants\", \"LinearAlgebra\"]\ngit-tree-sha1 = \"e5718a00af0ab9756305a0392832c8952c7426c1\"\nuuid = \"2ab3a3ac-af41-5b50-aa03-7779005ae688\"\nversion = \"0.3.6\"\n\n[[deps.Logging]]\nuuid = \"56ddb016-857b-54e1-b83d-db4d58db5568\"\n\n[[deps.Luxor]]\ndeps = [\"Base64\", \"Cairo\", \"Colors\", \"Dates\", \"FFMPEG\", \"FileIO\", \"Juno\", \"LaTeXStrings\", \"Random\", \"Requires\", \"Rsvg\"]\ngit-tree-sha1 = \"81a4fd2c618ba952feec85e4236f36c7a5660393\"\nuuid = \"ae8d54c2-7ccd-5906-9d76-62fc9837b5bc\"\nversion = \"3.0.0\"\n\n[[deps.MacroTools]]\ndeps = [\"Markdown\", \"Random\"]\ngit-tree-sha1 = \"3d3e902b31198a27340d0bf00d6ac452866021cf\"\nuuid = \"1914dd2f-81c6-5fcd-8719-6d5c9610ff09\"\nversion = \"0.5.9\"\n\n[[deps.Markdown]]\ndeps = [\"Base64\"]\nuuid = \"d6f4376e-aef5-505a-96c1-9c027394607a\"\n\n[[deps.MbedTLS]]\ndeps = [\"Dates\", \"MbedTLS_jll\", \"Random\", \"Sockets\"]\ngit-tree-sha1 = \"1c38e51c3d08ef2278062ebceade0e46cefc96fe\"\nuuid = \"739be429-bea8-5141-9913-cc70e7f3736d\"\nversion = \"1.0.3\"\n\n[[deps.MbedTLS_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"c8ffd9c3-330d-5841-b78e-0817d7145fa1\"\n\n[[deps.Measures]]\ngit-tree-sha1 = \"e498ddeee6f9fdb4551ce855a46f54dbd900245f\"\nuuid = \"442fdcdd-2543-5da2-b0f3-8c86c306513e\"\nversion = \"0.3.1\"\n\n[[deps.Media]]\ndeps = [\"MacroTools\", \"Test\"]\ngit-tree-sha1 = \"75a54abd10709c01f1b86b84ec225d26e840ed58\"\nuuid = \"e89f7d12-3494-54d1-8411-f7d8b9ae1f27\"\nversion = \"0.5.0\"\n\n[[deps.Missings]]\ndeps = [\"DataAPI\"]\ngit-tree-sha1 = \"bf210ce90b6c9eed32d25dbcae1ebc565df2687f\"\nuuid = \"e1d29d7a-bbdc-5cf2-9ac0-f12de2c33e28\"\nversion = \"1.0.2\"\n\n[[deps.Mmap]]\nuuid = \"a63ad114-7e13-5084-954f-fe012c677804\"\n\n[[deps.MozillaCACerts_jll]]\nuuid = \"14a3606d-f60d-562e-9121-12d972cd8159\"\n\n[[deps.NaNMath]]\ngit-tree-sha1 = \"b086b7ea07f8e38cf122f5016af580881ac914fe\"\nuuid = \"77ba4419-2d1f-58cd-9bb1-8ffee604a2e3\"\nversion = \"0.3.7\"\n\n[[deps.NetworkOptions]]\nuuid = \"ca575930-c2e3-43a9-ace4-1e988b2c1908\"\n\n[[deps.Ogg_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"887579a3eb005446d514ab7aeac5d1d027658b8f\"\nuuid = \"e7412a2a-1a6e-54c0-be00-318e2571c051\"\nversion = \"1.3.5+1\"\n\n[[deps.OpenBLAS_jll]]\ndeps = [\"Artifacts\", \"CompilerSupportLibraries_jll\", \"Libdl\"]\nuuid = \"4536629a-c528-5b80-bd46-f80d51c5b363\"\n\n[[deps.OpenSSL_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"648107615c15d4e09f7eca16307bc821c1f718d8\"\nuuid = \"458c3c95-2e84-50aa-8efc-19380b2a3a95\"\nversion = \"1.1.13+0\"\n\n[[deps.Opus_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"51a08fb14ec28da2ec7a927c4337e4332c2a4720\"\nuuid = \"91d4177d-7536-5919-b921-800302f37372\"\nversion = \"1.3.2+0\"\n\n[[deps.OrderedCollections]]\ngit-tree-sha1 = \"85f8e6578bf1f9ee0d11e7bb1b1456435479d47c\"\nuuid = \"bac558e1-5e72-5ebc-8fee-abe8a469f55d\"\nversion = \"1.4.1\"\n\n[[deps.PCRE_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"b2a7af664e098055a7529ad1a900ded962bca488\"\nuuid = \"2f80f16e-611a-54ab-bc61-aa92de5b98fc\"\nversion = \"8.44.0+0\"\n\n[[deps.Pango_jll]]\ndeps = [\"Artifacts\", \"Cairo_jll\", \"Fontconfig_jll\", \"FreeType2_jll\", \"FriBidi_jll\", \"Glib_jll\", \"HarfBuzz_jll\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"3a121dfbba67c94a5bec9dde613c3d0cbcf3a12b\"\nuuid = \"36c8627f-9965-5494-a995-c6b170f724f3\"\nversion = \"1.50.3+0\"\n\n[[deps.Parsers]]\ndeps = [\"Dates\"]\ngit-tree-sha1 = \"0b5cfbb704034b5b4c1869e36634438a047df065\"\nuuid = \"69de0a69-1ddd-5017-9359-2bf0b02dc9f0\"\nversion = \"2.2.1\"\n\n[[deps.Pixman_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"b4f5d02549a10e20780a24fce72bea96b6329e29\"\nuuid = \"30392449-352a-5448-841d-b1acce4e97dc\"\nversion = \"0.40.1+0\"\n\n[[deps.Pkg]]\ndeps = [\"Artifacts\", \"Dates\", \"Downloads\", \"LibGit2\", \"Libdl\", \"Logging\", \"Markdown\", \"Printf\", \"REPL\", \"Random\", \"SHA\", \"Serialization\", \"TOML\", \"Tar\", \"UUIDs\", \"p7zip_jll\"]\nuuid = \"44cfe95a-1eb2-52ea-b672-e2afdf69b78f\"\n\n[[deps.PlotThemes]]\ndeps = [\"PlotUtils\", \"Requires\", \"Statistics\"]\ngit-tree-sha1 = \"a3a964ce9dc7898193536002a6dd892b1b5a6f1d\"\nuuid = \"ccf2f8ad-2431-5c83-bf29-c5338b663b6a\"\nversion = \"2.0.1\"\n\n[[deps.PlotUtils]]\ndeps = [\"ColorSchemes\", \"Colors\", \"Dates\", \"Printf\", \"Random\", \"Reexport\", \"Statistics\"]\ngit-tree-sha1 = \"6f1b25e8ea06279b5689263cc538f51331d7ca17\"\nuuid = \"995b91a9-d308-5afd-9ec6-746e21dbc043\"\nversion = \"1.1.3\"\n\n[[deps.Plots]]\ndeps = [\"Base64\", \"Contour\", \"Dates\", \"Downloads\", \"FFMPEG\", \"FixedPointNumbers\", \"GR\", \"GeometryBasics\", \"JSON\", \"Latexify\", \"LinearAlgebra\", \"Measures\", \"NaNMath\", \"PlotThemes\", \"PlotUtils\", \"Printf\", \"REPL\", \"Random\", \"RecipesBase\", \"RecipesPipeline\", \"Reexport\", \"Requires\", \"Scratch\", \"Showoff\", \"SparseArrays\", \"Statistics\", \"StatsBase\", \"UUIDs\", \"UnicodeFun\", \"Unzip\"]\ngit-tree-sha1 = \"eb1432ec2b781f70ce2126c277d120554605669a\"\nuuid = \"91a5bcdd-55d7-5caf-9e0b-520d859cae80\"\nversion = \"1.25.8\"\n\n[[deps.PlutoUI]]\ndeps = [\"AbstractPlutoDingetjes\", \"Base64\", \"ColorTypes\", \"Dates\", \"Hyperscript\", \"HypertextLiteral\", \"IOCapture\", \"InteractiveUtils\", \"JSON\", \"Logging\", \"Markdown\", \"Random\", \"Reexport\", \"UUIDs\"]\ngit-tree-sha1 = \"8979e9802b4ac3d58c503a20f2824ad67f9074dd\"\nuuid = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\nversion = \"0.7.34\"\n\n[[deps.Preferences]]\ndeps = [\"TOML\"]\ngit-tree-sha1 = \"2cf929d64681236a2e074ffafb8d568733d2e6af\"\nuuid = \"21216c6a-2e73-6563-6e65-726566657250\"\nversion = \"1.2.3\"\n\n[[deps.PrettyTables]]\ndeps = [\"Crayons\", \"Formatting\", \"Markdown\", \"Reexport\", \"Tables\"]\ngit-tree-sha1 = \"dfb54c4e414caa595a1f2ed759b160f5a3ddcba5\"\nuuid = \"08abe8d2-0d0c-5749-adfa-8a2ac140af0d\"\nversion = \"1.3.1\"\n\n[[deps.Printf]]\ndeps = [\"Unicode\"]\nuuid = \"de0858da-6303-5e67-8744-51eddeeeb8d7\"\n\n[[deps.Profile]]\ndeps = [\"Printf\"]\nuuid = \"9abbd945-dff8-562f-b5e8-e1ebf5ef1b79\"\n\n[[deps.Qt5Base_jll]]\ndeps = [\"Artifacts\", \"CompilerSupportLibraries_jll\", \"Fontconfig_jll\", \"Glib_jll\", \"JLLWrappers\", \"Libdl\", \"Libglvnd_jll\", \"OpenSSL_jll\", \"Pkg\", \"Xorg_libXext_jll\", \"Xorg_libxcb_jll\", \"Xorg_xcb_util_image_jll\", \"Xorg_xcb_util_keysyms_jll\", \"Xorg_xcb_util_renderutil_jll\", \"Xorg_xcb_util_wm_jll\", \"Zlib_jll\", \"xkbcommon_jll\"]\ngit-tree-sha1 = \"ad368663a5e20dbb8d6dc2fddeefe4dae0781ae8\"\nuuid = \"ea2cea3b-5b76-57ae-a6ef-0a8af62496e1\"\nversion = \"5.15.3+0\"\n\n[[deps.REPL]]\ndeps = [\"InteractiveUtils\", \"Markdown\", \"Sockets\", \"Unicode\"]\nuuid = \"3fa0cd96-eef1-5676-8a61-b3b8758bbffb\"\n\n[[deps.Random]]\ndeps = [\"SHA\", \"Serialization\"]\nuuid = \"9a3f8284-a2c9-5f02-9a11-845980a1fd5c\"\n\n[[deps.RecipesBase]]\ngit-tree-sha1 = \"6bf3f380ff52ce0832ddd3a2a7b9538ed1bcca7d\"\nuuid = \"3cdcf5f2-1ef4-517c-9805-6587b60abb01\"\nversion = \"1.2.1\"\n\n[[deps.RecipesPipeline]]\ndeps = [\"Dates\", \"NaNMath\", \"PlotUtils\", \"RecipesBase\"]\ngit-tree-sha1 = \"37c1631cb3cc36a535105e6d5557864c82cd8c2b\"\nuuid = \"01d81517-befc-4cb6-b9ec-a95719d0359c\"\nversion = \"0.5.0\"\n\n[[deps.Reexport]]\ngit-tree-sha1 = \"45e428421666073eab6f2da5c9d310d99bb12f9b\"\nuuid = \"189a3867-3050-52da-a836-e630ba90ab69\"\nversion = \"1.2.2\"\n\n[[deps.RelocatableFolders]]\ndeps = [\"SHA\", \"Scratch\"]\ngit-tree-sha1 = \"cdbd3b1338c72ce29d9584fdbe9e9b70eeb5adca\"\nuuid = \"05181044-ff0b-4ac5-8273-598c1e38db00\"\nversion = \"0.1.3\"\n\n[[deps.Requires]]\ndeps = [\"UUIDs\"]\ngit-tree-sha1 = \"838a3a4188e2ded87a4f9f184b4b0d78a1e91cb7\"\nuuid = \"ae029012-a4dd-5104-9daa-d747884805df\"\nversion = \"1.3.0\"\n\n[[deps.Rsvg]]\ndeps = [\"Cairo\", \"Glib_jll\", \"Librsvg_jll\"]\ngit-tree-sha1 = \"3d3dc66eb46568fb3a5259034bfc752a0eb0c686\"\nuuid = \"c4c386cf-5103-5370-be45-f3a111cca3b8\"\nversion = \"1.0.0\"\n\n[[deps.SHA]]\nuuid = \"ea8e919c-243c-51af-8825-aaa63cd721ce\"\n\n[[deps.Scratch]]\ndeps = [\"Dates\"]\ngit-tree-sha1 = \"0b4b7f1393cff97c33891da2a0bf69c6ed241fda\"\nuuid = \"6c6a2e73-6563-6170-7368-637461726353\"\nversion = \"1.1.0\"\n\n[[deps.Serialization]]\nuuid = \"9e88b42a-f829-5b0c-bbe9-9e923198166b\"\n\n[[deps.SharedArrays]]\ndeps = [\"Distributed\", \"Mmap\", \"Random\", \"Serialization\"]\nuuid = \"1a1011a3-84de-559e-8e89-a11a2f7dc383\"\n\n[[deps.Showoff]]\ndeps = [\"Dates\", \"Grisu\"]\ngit-tree-sha1 = \"91eddf657aca81df9ae6ceb20b959ae5653ad1de\"\nuuid = \"992d4aef-0814-514b-bc4d-f2e9a6c4116f\"\nversion = \"1.0.3\"\n\n[[deps.Sockets]]\nuuid = \"6462fe0b-24de-5631-8697-dd941f90decc\"\n\n[[deps.SortingAlgorithms]]\ndeps = [\"DataStructures\"]\ngit-tree-sha1 = \"b3363d7460f7d098ca0912c69b082f75625d7508\"\nuuid = \"a2af1166-a08f-5f64-846c-94a0d3cef48c\"\nversion = \"1.0.1\"\n\n[[deps.SparseArrays]]\ndeps = [\"LinearAlgebra\", \"Random\"]\nuuid = \"2f01184e-e22b-5df5-ae63-d93ebab69eaf\"\n\n[[deps.StaticArrays]]\ndeps = [\"LinearAlgebra\", \"Random\", \"Statistics\"]\ngit-tree-sha1 = \"a635a9333989a094bddc9f940c04c549cd66afcf\"\nuuid = \"90137ffa-7385-5640-81b9-e52037218182\"\nversion = \"1.3.4\"\n\n[[deps.Statistics]]\ndeps = [\"LinearAlgebra\", \"SparseArrays\"]\nuuid = \"10745b16-79ce-11e8-11f9-7d13ad32a3b2\"\n\n[[deps.StatsAPI]]\ngit-tree-sha1 = \"d88665adc9bcf45903013af0982e2fd05ae3d0a6\"\nuuid = \"82ae8749-77ed-4fe6-ae5f-f523153014b0\"\nversion = \"1.2.0\"\n\n[[deps.StatsBase]]\ndeps = [\"DataAPI\", \"DataStructures\", \"LinearAlgebra\", \"LogExpFunctions\", \"Missings\", \"Printf\", \"Random\", \"SortingAlgorithms\", \"SparseArrays\", \"Statistics\", \"StatsAPI\"]\ngit-tree-sha1 = \"51383f2d367eb3b444c961d485c565e4c0cf4ba0\"\nuuid = \"2913bbd2-ae8a-5f71-8c99-4fb6c76f3a91\"\nversion = \"0.33.14\"\n\n[[deps.StructArrays]]\ndeps = [\"Adapt\", \"DataAPI\", \"StaticArrays\", \"Tables\"]\ngit-tree-sha1 = \"d21f2c564b21a202f4677c0fba5b5ee431058544\"\nuuid = \"09ab397b-f2b6-538f-b94a-2f83cf4a842a\"\nversion = \"0.6.4\"\n\n[[deps.TOML]]\ndeps = [\"Dates\"]\nuuid = \"fa267f1f-6049-4f14-aa54-33bafae1ed76\"\n\n[[deps.TableTraits]]\ndeps = [\"IteratorInterfaceExtensions\"]\ngit-tree-sha1 = \"c06b2f539df1c6efa794486abfb6ed2022561a39\"\nuuid = \"3783bdb8-4a98-5b6b-af9a-565f29a5fe9c\"\nversion = \"1.0.1\"\n\n[[deps.Tables]]\ndeps = [\"DataAPI\", \"DataValueInterfaces\", \"IteratorInterfaceExtensions\", \"LinearAlgebra\", \"TableTraits\", \"Test\"]\ngit-tree-sha1 = \"bb1064c9a84c52e277f1096cf41434b675cd368b\"\nuuid = \"bd369af6-aec1-5ad0-b16a-f7cc5008161c\"\nversion = \"1.6.1\"\n\n[[deps.Tar]]\ndeps = [\"ArgTools\", \"SHA\"]\nuuid = \"a4e569a6-e804-4fa4-b0f3-eef7a1d5b13e\"\n\n[[deps.Test]]\ndeps = [\"InteractiveUtils\", \"Logging\", \"Random\", \"Serialization\"]\nuuid = \"8dfed614-e22c-5e08-85e1-65c5234f0b40\"\n\n[[deps.URIs]]\ngit-tree-sha1 = \"97bbe755a53fe859669cd907f2d96aee8d2c1355\"\nuuid = \"5c2747f8-b7ea-4ff2-ba2e-563bfd36b1d4\"\nversion = \"1.3.0\"\n\n[[deps.UUIDs]]\ndeps = [\"Random\", \"SHA\"]\nuuid = \"cf7118a7-6976-5b1a-9a39-7adc72f591a4\"\n\n[[deps.Unicode]]\nuuid = \"4ec0a83e-493e-50e2-b9ac-8f72acf5a8f5\"\n\n[[deps.UnicodeFun]]\ndeps = [\"REPL\"]\ngit-tree-sha1 = \"53915e50200959667e78a92a418594b428dffddf\"\nuuid = \"1cfade01-22cf-5700-b092-accc4b62d6e1\"\nversion = \"0.4.1\"\n\n[[deps.Unzip]]\ngit-tree-sha1 = \"34db80951901073501137bdbc3d5a8e7bbd06670\"\nuuid = \"41fe7b60-77ed-43a1-b4f0-825fd5a5650d\"\nversion = \"0.1.2\"\n\n[[deps.Wayland_jll]]\ndeps = [\"Artifacts\", \"Expat_jll\", \"JLLWrappers\", \"Libdl\", \"Libffi_jll\", \"Pkg\", \"XML2_jll\"]\ngit-tree-sha1 = \"3e61f0b86f90dacb0bc0e73a0c5a83f6a8636e23\"\nuuid = \"a2964d1f-97da-50d4-b82a-358c7fce9d89\"\nversion = \"1.19.0+0\"\n\n[[deps.Wayland_protocols_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"66d72dc6fcc86352f01676e8f0f698562e60510f\"\nuuid = \"2381bf8a-dfd0-557d-9999-79630e7b1b91\"\nversion = \"1.23.0+0\"\n\n[[deps.XML2_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Libiconv_jll\", \"Pkg\", \"Zlib_jll\"]\ngit-tree-sha1 = \"1acf5bdf07aa0907e0a37d3718bb88d4b687b74a\"\nuuid = \"02c8fc9c-b97f-50b9-bbe4-9be30ff0a78a\"\nversion = \"2.9.12+0\"\n\n[[deps.XSLT_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Libgcrypt_jll\", \"Libgpg_error_jll\", \"Libiconv_jll\", \"Pkg\", \"XML2_jll\", \"Zlib_jll\"]\ngit-tree-sha1 = \"91844873c4085240b95e795f692c4cec4d805f8a\"\nuuid = \"aed1982a-8fda-507f-9586-7b0439959a61\"\nversion = \"1.1.34+0\"\n\n[[deps.Xorg_libX11_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libxcb_jll\", \"Xorg_xtrans_jll\"]\ngit-tree-sha1 = \"5be649d550f3f4b95308bf0183b82e2582876527\"\nuuid = \"4f6342f7-b3d2-589e-9d20-edeb45f2b2bc\"\nversion = \"1.6.9+4\"\n\n[[deps.Xorg_libXau_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"4e490d5c960c314f33885790ed410ff3a94ce67e\"\nuuid = \"0c0b7dd1-d40b-584c-a123-a41640f87eec\"\nversion = \"1.0.9+4\"\n\n[[deps.Xorg_libXcursor_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libXfixes_jll\", \"Xorg_libXrender_jll\"]\ngit-tree-sha1 = \"12e0eb3bc634fa2080c1c37fccf56f7c22989afd\"\nuuid = \"935fb764-8cf2-53bf-bb30-45bb1f8bf724\"\nversion = \"1.2.0+4\"\n\n[[deps.Xorg_libXdmcp_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"4fe47bd2247248125c428978740e18a681372dd4\"\nuuid = \"a3789734-cfe1-5b06-b2d0-1dd0d9d62d05\"\nversion = \"1.1.3+4\"\n\n[[deps.Xorg_libXext_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libX11_jll\"]\ngit-tree-sha1 = \"b7c0aa8c376b31e4852b360222848637f481f8c3\"\nuuid = \"1082639a-0dae-5f34-9b06-72781eeb8cb3\"\nversion = \"1.3.4+4\"\n\n[[deps.Xorg_libXfixes_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libX11_jll\"]\ngit-tree-sha1 = \"0e0dc7431e7a0587559f9294aeec269471c991a4\"\nuuid = \"d091e8ba-531a-589c-9de9-94069b037ed8\"\nversion = \"5.0.3+4\"\n\n[[deps.Xorg_libXi_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libXext_jll\", \"Xorg_libXfixes_jll\"]\ngit-tree-sha1 = \"89b52bc2160aadc84d707093930ef0bffa641246\"\nuuid = \"a51aa0fd-4e3c-5386-b890-e753decda492\"\nversion = \"1.7.10+4\"\n\n[[deps.Xorg_libXinerama_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libXext_jll\"]\ngit-tree-sha1 = \"26be8b1c342929259317d8b9f7b53bf2bb73b123\"\nuuid = \"d1454406-59df-5ea1-beac-c340f2130bc3\"\nversion = \"1.1.4+4\"\n\n[[deps.Xorg_libXrandr_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libXext_jll\", \"Xorg_libXrender_jll\"]\ngit-tree-sha1 = \"34cea83cb726fb58f325887bf0612c6b3fb17631\"\nuuid = \"ec84b674-ba8e-5d96-8ba1-2a689ba10484\"\nversion = \"1.5.2+4\"\n\n[[deps.Xorg_libXrender_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libX11_jll\"]\ngit-tree-sha1 = \"19560f30fd49f4d4efbe7002a1037f8c43d43b96\"\nuuid = \"ea2f1a96-1ddc-540d-b46f-429655e07cfa\"\nversion = \"0.9.10+4\"\n\n[[deps.Xorg_libpthread_stubs_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"6783737e45d3c59a4a4c4091f5f88cdcf0908cbb\"\nuuid = \"14d82f49-176c-5ed1-bb49-ad3f5cbd8c74\"\nversion = \"0.1.0+3\"\n\n[[deps.Xorg_libxcb_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"XSLT_jll\", \"Xorg_libXau_jll\", \"Xorg_libXdmcp_jll\", \"Xorg_libpthread_stubs_jll\"]\ngit-tree-sha1 = \"daf17f441228e7a3833846cd048892861cff16d6\"\nuuid = \"c7cfdc94-dc32-55de-ac96-5a1b8d977c5b\"\nversion = \"1.13.0+3\"\n\n[[deps.Xorg_libxkbfile_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libX11_jll\"]\ngit-tree-sha1 = \"926af861744212db0eb001d9e40b5d16292080b2\"\nuuid = \"cc61e674-0454-545c-8b26-ed2c68acab7a\"\nversion = \"1.1.0+4\"\n\n[[deps.Xorg_xcb_util_image_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_xcb_util_jll\"]\ngit-tree-sha1 = \"0fab0a40349ba1cba2c1da699243396ff8e94b97\"\nuuid = \"12413925-8142-5f55-bb0e-6d7ca50bb09b\"\nversion = \"0.4.0+1\"\n\n[[deps.Xorg_xcb_util_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libxcb_jll\"]\ngit-tree-sha1 = \"e7fd7b2881fa2eaa72717420894d3938177862d1\"\nuuid = \"2def613f-5ad1-5310-b15b-b15d46f528f5\"\nversion = \"0.4.0+1\"\n\n[[deps.Xorg_xcb_util_keysyms_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_xcb_util_jll\"]\ngit-tree-sha1 = \"d1151e2c45a544f32441a567d1690e701ec89b00\"\nuuid = \"975044d2-76e6-5fbe-bf08-97ce7c6574c7\"\nversion = \"0.4.0+1\"\n\n[[deps.Xorg_xcb_util_renderutil_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_xcb_util_jll\"]\ngit-tree-sha1 = \"dfd7a8f38d4613b6a575253b3174dd991ca6183e\"\nuuid = \"0d47668e-0667-5a69-a72c-f761630bfb7e\"\nversion = \"0.3.9+1\"\n\n[[deps.Xorg_xcb_util_wm_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_xcb_util_jll\"]\ngit-tree-sha1 = \"e78d10aab01a4a154142c5006ed44fd9e8e31b67\"\nuuid = \"c22f9ab0-d5fe-5066-847c-f4bb1cd4e361\"\nversion = \"0.4.1+1\"\n\n[[deps.Xorg_xkbcomp_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libxkbfile_jll\"]\ngit-tree-sha1 = \"4bcbf660f6c2e714f87e960a171b119d06ee163b\"\nuuid = \"35661453-b289-5fab-8a00-3d9160c6a3a4\"\nversion = \"1.4.2+4\"\n\n[[deps.Xorg_xkeyboard_config_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_xkbcomp_jll\"]\ngit-tree-sha1 = \"5c8424f8a67c3f2209646d4425f3d415fee5931d\"\nuuid = \"33bec58e-1273-512f-9401-5d533626f822\"\nversion = \"2.27.0+4\"\n\n[[deps.Xorg_xtrans_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"79c31e7844f6ecf779705fbc12146eb190b7d845\"\nuuid = \"c5fb5394-a638-5e4d-96e5-b29de1b5cf10\"\nversion = \"1.4.0+3\"\n\n[[deps.Zlib_jll]]\ndeps = [\"Libdl\"]\nuuid = \"83775a58-1f1d-513f-b197-d71354ab007a\"\n\n[[deps.Zstd_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"e45044cd873ded54b6a5bac0eb5c971392cf1927\"\nuuid = \"3161d3a3-bdf6-5164-811a-617609db77b4\"\nversion = \"1.5.2+0\"\n\n[[deps.gdk_pixbuf_jll]]\ndeps = [\"Artifacts\", \"Glib_jll\", \"JLLWrappers\", \"JpegTurbo_jll\", \"Libdl\", \"Libtiff_jll\", \"Pkg\", \"Xorg_libX11_jll\", \"libpng_jll\"]\ngit-tree-sha1 = \"c23323cd30d60941f8c68419a70905d9bdd92808\"\nuuid = \"da03df04-f53b-5353-a52f-6a8b0620ced0\"\nversion = \"2.42.6+1\"\n\n[[deps.libass_jll]]\ndeps = [\"Artifacts\", \"Bzip2_jll\", \"FreeType2_jll\", \"FriBidi_jll\", \"HarfBuzz_jll\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Zlib_jll\"]\ngit-tree-sha1 = \"5982a94fcba20f02f42ace44b9894ee2b140fe47\"\nuuid = \"0ac62f75-1d6f-5e53-bd7c-93b484bb37c0\"\nversion = \"0.15.1+0\"\n\n[[deps.libblastrampoline_jll]]\ndeps = [\"Artifacts\", \"Libdl\", \"OpenBLAS_jll\"]\nuuid = \"8e850b90-86db-534c-a0d3-1478176c7d93\"\n\n[[deps.libfdk_aac_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"daacc84a041563f965be61859a36e17c4e4fcd55\"\nuuid = \"f638f0a6-7fb0-5443-88ba-1cc74229b280\"\nversion = \"2.0.2+0\"\n\n[[deps.libpng_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Zlib_jll\"]\ngit-tree-sha1 = \"94d180a6d2b5e55e447e2d27a29ed04fe79eb30c\"\nuuid = \"b53b4c65-9356-5827-b1ea-8c7a1a84506f\"\nversion = \"1.6.38+0\"\n\n[[deps.libvorbis_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Ogg_jll\", \"Pkg\"]\ngit-tree-sha1 = \"b910cb81ef3fe6e78bf6acee440bda86fd6ae00c\"\nuuid = \"f27f6e37-5d2b-51aa-960f-b287f2bc3b7a\"\nversion = \"1.3.7+1\"\n\n[[deps.nghttp2_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"8e850ede-7688-5339-a07c-302acd2aaf8d\"\n\n[[deps.p7zip_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"3f19e933-33d8-53b3-aaab-bd5110c3b7a0\"\n\n[[deps.x264_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"4fea590b89e6ec504593146bf8b988b2c00922b2\"\nuuid = \"1270edf5-f2f9-52d2-97e9-ab00b5d0237a\"\nversion = \"2021.5.5+0\"\n\n[[deps.x265_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"ee567a171cce03570d77ad3a43e90218e38937a9\"\nuuid = \"dfaa095f-4041-5dcd-9319-2fabd8486b76\"\nversion = \"3.5.0+0\"\n\n[[deps.xkbcommon_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Wayland_jll\", \"Wayland_protocols_jll\", \"Xorg_libxcb_jll\", \"Xorg_xkeyboard_config_jll\"]\ngit-tree-sha1 = \"ece2350174195bb31de1a63bea3a41ae1aa593b6\"\nuuid = \"d8fb68d0-12a3-5cfd-a85a-d49703b185fd\"\nversion = \"0.9.1+5\"\n\"\"\"\n\n# \u2554\u2550\u2561 Cell order:\n# \u255f\u25008ba6a00a-7a53-4dcc-af4d-cb85aa9b4d68\n# \u255f\u25008869f117-1fc6-4cc5-9803-b8ec72a109a6\n# \u255f\u250017724757-604e-4c77-a42b-238ba121e88f\n# \u255f\u2500431ccdf0-93a9-4b3c-9576-8854ba2f1fad\n# \u255f\u25007efaf27c-8e0a-40f9-ac28-af90357450a3\n# \u255f\u250055ea8324-f36d-40bd-99e3-92e7fcbafca9\n# \u255f\u25003502da52-7d2b-4c79-82ce-7424d756cd9b\n# \u255f\u2500c392c8a9-361d-47e4-933e-2e51b793e069\n# \u255f\u250091345d04-f502-4e90-935e-a4dc250244db\n# \u255f\u2500719784fe-3ef6-4e53-ad52-82140e3d0b5e\n# \u255f\u25001d4d6059-5b79-43cd-ac17-ef2aa057ecad\n# \u255f\u2500d46ced14-a5d3-45a9-9d43-38dc7879b0c7\n# \u255f\u2500d3568c5d-16bf-4698-9891-0be65d62b36c\n# \u255f\u250055d4ab48-9589-4fd9-ac06-3338fdb418c4\n# \u255f\u250054370424-3add-4f93-91b0-e136166842ae\n# \u2560\u2550451ae3a0-8068-4747-95a7-d31955808f29\n# \u2560\u255006a90381-b4cb-4d32-af0d-2f084364129c\n# \u2560\u2550b6890de0-8923-11ec-3552-3113bdd53f86\n# \u255f\u250000000000-0000-0000-0000-000000000001\n# \u255f\u250000000000-0000-0000-0000-000000000002\n", "meta": {"hexsha": "7319b22a8e4288dfb1d8848c9238eb6392cff3a3", "size": 53910, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "lectures/Lecture-6-5440-7770-S2022.jl", "max_stars_repo_name": "mavisbrown/CHEME-5440-7770-Cornell-Spring-2022", "max_stars_repo_head_hexsha": "6bbfa8f58f75df4bc8d2146ae768a816a1273a46", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 2, "max_stars_repo_stars_event_min_datetime": "2022-02-07T23:40:03.000Z", "max_stars_repo_stars_event_max_datetime": "2022-02-14T19:06:26.000Z", "max_issues_repo_path": "lectures/Lecture-6-5440-7770-S2022.jl", "max_issues_repo_name": "mavisbrown/CHEME-5440-7770-Cornell-Spring-2022", "max_issues_repo_head_hexsha": "6bbfa8f58f75df4bc8d2146ae768a816a1273a46", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "lectures/Lecture-6-5440-7770-S2022.jl", "max_forks_repo_name": "mavisbrown/CHEME-5440-7770-Cornell-Spring-2022", "max_forks_repo_head_hexsha": "6bbfa8f58f75df4bc8d2146ae768a816a1273a46", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 12, "max_forks_repo_forks_event_min_datetime": "2022-02-03T15:15:33.000Z", "max_forks_repo_forks_event_max_datetime": "2022-03-08T22:46:36.000Z", "avg_line_length": 37.0006863418, "max_line_length": 742, "alphanum_fraction": 0.7253941755, "num_tokens": 22209, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4532618480153861, "lm_q2_score": 0.17553807793655438, "lm_q1q2_score": 0.07956471360259151}}
{"text": "using Test \nusing clima_demo \n\n@testset \"My test\" begin \n\t@test 1 + 1 == 2\nend\n", "meta": {"hexsha": "02a7dfc9b7cd27b64fb622d9b476d51ebab07f1e", "size": 79, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/runtests.jl", "max_stars_repo_name": "costachris/clima_demo", "max_stars_repo_head_hexsha": "0b132414dd6effe035bcdcdf6a045af42e51436b", "max_stars_repo_licenses": ["Apache-2.0"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "test/runtests.jl", "max_issues_repo_name": "costachris/clima_demo", "max_issues_repo_head_hexsha": "0b132414dd6effe035bcdcdf6a045af42e51436b", "max_issues_repo_licenses": ["Apache-2.0"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "test/runtests.jl", "max_forks_repo_name": "costachris/clima_demo", "max_forks_repo_head_hexsha": "0b132414dd6effe035bcdcdf6a045af42e51436b", "max_forks_repo_licenses": ["Apache-2.0"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 11.2857142857, "max_line_length": 25, "alphanum_fraction": 0.6582278481, "num_tokens": 29, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4960938294709195, "lm_q2_score": 0.1602660403494035, "lm_q1q2_score": 0.07950699369107649}}
{"text": "# NOTE: All tests of this file can be run with any number of processes.\r\n# Nearly all of the functionality can however be verified with one single process\r\n# (thanks to the usage of periodic boundaries in most of the full halo update tests).\r\n\r\npush!(LOAD_PATH, \"../src\")\r\nusing Test\r\nusing ImplicitGlobalGrid; GG = ImplicitGlobalGrid\r\nimport MPI\r\nusing CUDA\r\nimport ImplicitGlobalGrid: @require, longnameof\r\n\r\ntest_gpu = CUDA.functional()\r\n\r\nif test_gpu\r\n\tglobal cuzeros = CUDA.zeros\r\n\tglobal allocators = [zeros, cuzeros]\r\n\tglobal ArrayConstructors = [Array, CuArray]\r\nelse\r\n\tglobal cuzeros = nothing # To enable statements like: if zeros==cuzeros\r\n\tglobal allocators = [zeros]\r\n\tglobal ArrayConstructors = [Array]\r\nend\r\n\r\n\r\n## Test setup\r\nMPI.Init();\r\nnprocs = MPI.Comm_size(MPI.COMM_WORLD); # NOTE: these tests can run with any number of processes.\r\nndims_mpi = GG.NDIMS_MPI;\r\nnneighbors_per_dim = GG.NNEIGHBORS_PER_DIM; # Should be 2 (one left and one right neighbor).\r\nnx = 7;\r\nny = 5;\r\nnz = 6;\r\ndx = 1.0\r\ndy = 1.0\r\ndz = 1.0\r\n\r\n@testset \"$(basename(@__FILE__)) (processes: $nprocs)\" begin\r\n\t@testset \"1. argument check (allocator: $(longnameof(zeros)))\" for zeros in allocators\r\n\t\tinit_global_grid(nx, ny, nz, quiet=true, init_MPI=false);\r\n\t\tP = zeros(nx, ny, nz );\r\n\t\tSxz = zeros(nx-2,ny-1,nz-2);\r\n\t\tA = zeros(nx-1,ny+2,nz+1);\r\n\t A2 = A;\n\t\tZ = zeros(ComplexF64, nx-1,ny+2,nz+1);\n\t\tZ2 = Z;\r\n\t @test_throws ErrorException update_halo!(P, Sxz, A) # Error: Sxz has no halo.\r\n\t @test_throws ErrorException update_halo!(P, Sxz, A, Sxz) # Error: Sxz and Sxz have no halo.\r\n\t @test_throws ErrorException update_halo!(P, A, A) # Error: A is given twice.\r\n\t @test_throws ErrorException update_halo!(P, A, A2) # Error: A2 is duplicate of A (an alias; it points to the same memory).\r\n\t @test_throws ErrorException update_halo!(P, A, A, A2) # Error: the second A and A2 are duplicates of the first A.\n\t\t@test_throws ErrorException update_halo!(Z, Z2) # Error: Z2 is duplicate of Z (an alias; it points to the same memory).\n\t\t@test_throws ErrorException update_halo!(Z, P) # Error: P is of different type than Z.\n\t\t@test_throws ErrorException update_halo!(Z, P, A) # Error: P and A are of different type than Z.\r\n\t\tfinalize_global_grid(finalize_MPI=false);\r\n\tend;\r\n\r\n\t@testset \"2. buffer allocation (allocator: $(longnameof(zeros)))\" for zeros in allocators\r\n\t\tinit_global_grid(nx, ny, nz, periodx=1, periody=1, periodz=1, quiet=true, init_MPI=false);\r\n\t\tP = zeros(nx, ny, nz );\r\n\t\tA = zeros(nx-1,ny+2,nz+1);\r\n\t\tB = zeros(Float32, nx+1, ny+2, nz+3);\r\n\t\tC = zeros(Float32, nx+1, ny+1, nz+1);\n\t\tZ = zeros(ComplexF16, nx, ny, nz );\n\t\tY = zeros(ComplexF16, nx-1, ny+2, nz+1);\r\n\t @testset \"free buffers\" begin\r\n\t @require GG.get_sendbufs_raw() === nothing\r\n\t @require GG.get_recvbufs_raw() === nothing\r\n\t GG.allocate_bufs(P);\r\n\t @require GG.get_sendbufs_raw() !== nothing\r\n\t @require GG.get_recvbufs_raw() !== nothing\r\n\t GG.free_update_halo_buffers();\r\n\t @test GG.get_sendbufs_raw() === nothing\r\n\t @test GG.get_recvbufs_raw() === nothing\r\n\t end;\r\n\t @testset \"allocate single\" begin\r\n\t GG.free_update_halo_buffers();\r\n\t GG.allocate_bufs(P);\r\n\t for bufs_raw in [GG.get_sendbufs_raw(), GG.get_recvbufs_raw()]\r\n\t @test length(bufs_raw) == 1 # 1 array\r\n\t @test length(bufs_raw[1]) == nneighbors_per_dim # 2 neighbors per dimension\r\n\t for n = 1:nneighbors_per_dim\r\n\t @test length(bufs_raw[1][n]) >= prod(sort([size(P)...])[2:end]) # required length: max halo elements in any of the dimensions\r\n\t end\r\n\t end\r\n\t end;\n\t\t@testset \"allocate single (Complex)\" begin\n\t GG.free_update_halo_buffers();\n\t GG.allocate_bufs(Z);\n\t for bufs_raw in [GG.get_sendbufs_raw(), GG.get_recvbufs_raw()]\n\t @test length(bufs_raw) == 1 # 1 array\n\t @test length(bufs_raw[1]) == nneighbors_per_dim # 2 neighbors per dimension\n\t for n = 1:nneighbors_per_dim\n\t @test length(bufs_raw[1][n]) >= prod(sort([size(Z)...])[2:end]) # required length: max halo elements in any of the dimensions\n\t end\n\t end\n\t end;\r\n\t @testset \"keep 1st, allocate 2nd\" begin\r\n\t GG.free_update_halo_buffers();\r\n\t GG.allocate_bufs(P);\r\n\t GG.allocate_bufs(A, P);\r\n\t for bufs_raw in [GG.get_sendbufs_raw(), GG.get_recvbufs_raw()]\r\n\t @test length(bufs_raw) == 2 # 2 arrays\r\n\t @test length(bufs_raw[1]) == nneighbors_per_dim # 2 neighbors per dimension\r\n\t @test length(bufs_raw[2]) == nneighbors_per_dim # 2 neighbors per dimension\r\n\t for n = 1:nneighbors_per_dim\r\n\t @test length(bufs_raw[1][n]) >= prod(sort([size(A)...])[2:end]) # required length: max halo elements in any of the dimensions\r\n\t @test length(bufs_raw[2][n]) >= prod(sort([size(P)...])[2:end]) # ...\r\n\t end\r\n\t end\r\n\t end;\n\t\t@testset \"keep 1st, allocate 2nd (Complex)\" begin\n\t GG.free_update_halo_buffers();\n\t GG.allocate_bufs(Z);\n\t GG.allocate_bufs(Y, Z);\n\t for bufs_raw in [GG.get_sendbufs_raw(), GG.get_recvbufs_raw()]\n\t @test length(bufs_raw) == 2 # 2 arrays\n\t @test length(bufs_raw[1]) == nneighbors_per_dim # 2 neighbors per dimension\n\t @test length(bufs_raw[2]) == nneighbors_per_dim # 2 neighbors per dimension\n\t for n = 1:nneighbors_per_dim\n\t @test length(bufs_raw[1][n]) >= prod(sort([size(Y)...])[2:end]) # required length: max halo elements in any of the dimensions\n\t @test length(bufs_raw[2][n]) >= prod(sort([size(Z)...])[2:end]) # ...\n\t end\n\t end\n\t end;\r\n\t @testset \"reinterpret (no allocation)\" begin\r\n\t GG.free_update_halo_buffers();\r\n\t GG.allocate_bufs(A, P);\r\n\t GG.allocate_bufs(B, C); # The new arrays contain Float32 (A, and P were Float64); B and C have a halo with more elements than A and P had, but they require less space in memory\r\n\t for bufs_raw in [GG.get_sendbufs_raw(), GG.get_recvbufs_raw()]\r\n\t @test length(bufs_raw) == 2 # Still 2 arrays: B, C (even though they are different then before: was A and P)\r\n\t @test length(bufs_raw[1]) == nneighbors_per_dim # 2 neighbors per dimension\r\n\t @test length(bufs_raw[2]) == nneighbors_per_dim # 2 neighbors per dimension\r\n\t for n = 1:nneighbors_per_dim\r\n\t @test length(bufs_raw[1][n]) >= prod(sort([size(B)...])[2:end]) # required length: max halo elements in any of the dimensions\r\n\t @test length(bufs_raw[2][n]) >= prod(sort([size(C)...])[2:end]) # ...\r\n\t end\r\n\t @test all([eltype(bufs_raw[i][n]) == Float32 for i=1:length(bufs_raw), n=1:nneighbors_per_dim])\r\n\t end\r\n\t end;\n\t\t@testset \"reinterpret (no allocation) (Complex)\" begin\n\t GG.free_update_halo_buffers();\n\t GG.allocate_bufs(A, P);\n\t GG.allocate_bufs(Y, Z); # The new arrays contain Float32 (A, and P were Float64); B and C have a halo with more elements than A and P had, but they require less space in memory\n\t for bufs_raw in [GG.get_sendbufs_raw(), GG.get_recvbufs_raw()]\n\t @test length(bufs_raw) == 2 # Still 2 arrays: B, C (even though they are different then before: was A and P)\n\t @test length(bufs_raw[1]) == nneighbors_per_dim # 2 neighbors per dimension\n\t @test length(bufs_raw[2]) == nneighbors_per_dim # 2 neighbors per dimension\n\t for n = 1:nneighbors_per_dim\n\t @test length(bufs_raw[1][n]) >= prod(sort([size(Y)...])[2:end]) # required length: max halo elements in any of the dimensions\n\t @test length(bufs_raw[2][n]) >= prod(sort([size(Z)...])[2:end]) # ...\n\t end\n\t @test all([eltype(bufs_raw[i][n]) == ComplexF16 for i=1:length(bufs_raw), n=1:nneighbors_per_dim])\n\t end\n\t end;\r\n\t @testset \"(cu)sendbuf / (cu)recvbuf\" begin\r\n\t\t\tsendbuf, recvbuf = (GG.sendbuf, GG.recvbuf);\r\n\t\t\t@static if (test_gpu && zeros == cuzeros) sendbuf, recvbuf = (GG.cusendbuf, GG.curecvbuf); end\r\n\t GG.free_update_halo_buffers();\r\n\t GG.allocate_bufs(A, P);\r\n\t for dim = 1:ndims(A), n = 1:nneighbors_per_dim\r\n\t @test all(size(sendbuf(n,dim,1,A)) .== size(A)[1:ndims(A).!=dim])\r\n\t @test all(size(recvbuf(n,dim,1,A)) .== size(A)[1:ndims(A).!=dim])\r\n\t end\r\n\t for dim = 1:ndims(P), n = 1:nneighbors_per_dim\r\n\t @test all(size(sendbuf(n,dim,2,P)) .== size(P)[1:ndims(P).!=dim])\r\n\t @test all(size(recvbuf(n,dim,2,P)) .== size(P)[1:ndims(P).!=dim])\r\n\t end\r\n\t end;\n\t\t@testset \"(cu)sendbuf / (cu)recvbuf (Complex)\" begin\n\t\t\tsendbuf, recvbuf = (GG.sendbuf, GG.recvbuf);\n\t\t\t@static if (test_gpu && zeros == cuzeros) sendbuf, recvbuf = (GG.cusendbuf, GG.curecvbuf); end\n\t GG.free_update_halo_buffers();\n\t GG.allocate_bufs(Y, Z);\n\t for dim = 1:ndims(Y), n = 1:nneighbors_per_dim\n\t @test all(size(sendbuf(n,dim,1,Y)) .== size(Y)[1:ndims(Y).!=dim])\n\t @test all(size(recvbuf(n,dim,1,Y)) .== size(Y)[1:ndims(Y).!=dim])\n\t end\n\t for dim = 1:ndims(Z), n = 1:nneighbors_per_dim\n\t @test all(size(sendbuf(n,dim,2,Z)) .== size(Z)[1:ndims(Z).!=dim])\n\t @test all(size(recvbuf(n,dim,2,Z)) .== size(Z)[1:ndims(Z).!=dim])\n\t end\n\t end;\r\n\t\tfinalize_global_grid(finalize_MPI=false);\r\n\tend;\r\n\r\n\t@testset \"3. data transfer components\" begin\r\n\t\t@testset \"iwrite_sendbufs! / iread_recvbufs!\" begin\r\n\t\t\t@testset \"sendranges / recvranges (allocator: $(longnameof(zeros)))\" for zeros in allocators\r\n\t\t\t\tinit_global_grid(nx, ny, nz, periodx=1, periody=1, periodz=1, overlapz=3, quiet=true, init_MPI=false);\r\n\t\t\t\tP = zeros(nx, ny, nz );\r\n\t\t\t\tA = zeros(nx-1,ny+2,nz+1);\r\n\t\t\t\t@test GG.sendranges(1, 1, P) == [ 2:2, 1:size(P,2), 1:size(P,3)]\r\n\t\t\t\t@test GG.sendranges(2, 1, P) == [size(P,1)-1:size(P,1)-1, 1:size(P,2), 1:size(P,3)]\r\n\t\t\t\t@test GG.sendranges(1, 2, P) == [ 1:size(P,1), 2:2, 1:size(P,3)]\r\n\t\t\t\t@test GG.sendranges(2, 2, P) == [ 1:size(P,1), size(P,2)-1:size(P,2)-1, 1:size(P,3)]\r\n\t\t\t\t@test GG.sendranges(1, 3, P) == [ 1:size(P,1), 1:size(P,2), 3:3]\r\n\t\t\t\t@test GG.sendranges(2, 3, P) == [ 1:size(P,1), 1:size(P,2), size(P,3)-2:size(P,3)-2]\r\n\t\t\t\t@test GG.recvranges(1, 1, P) == [ 1:1, 1:size(P,2), 1:size(P,3)]\r\n\t\t\t\t@test GG.recvranges(2, 1, P) == [ size(P,1):size(P,1), 1:size(P,2), 1:size(P,3)]\r\n\t\t\t\t@test GG.recvranges(1, 2, P) == [ 1:size(P,1), 1:1, 1:size(P,3)]\r\n\t\t\t\t@test GG.recvranges(2, 2, P) == [ 1:size(P,1), size(P,2):size(P,2), 1:size(P,3)]\r\n\t\t\t\t@test GG.recvranges(1, 3, P) == [ 1:size(P,1), 1:size(P,2), 1:1]\r\n\t\t\t\t@test GG.recvranges(2, 3, P) == [ 1:size(P,1), 1:size(P,2), size(P,3):size(P,3)]\r\n\t\t\t\t@test_throws ErrorException GG.sendranges(1, 1, A)\r\n\t\t\t\t@test_throws ErrorException GG.sendranges(2, 1, A)\r\n\t\t\t\t@test GG.sendranges(1, 2, A) == [ 1:size(A,1), 4:4, 1:size(A,3)]\r\n\t\t\t\t@test GG.sendranges(2, 2, A) == [ 1:size(A,1), size(A,2)-3:size(A,2)-3, 1:size(A,3)]\r\n\t\t\t\t@test GG.sendranges(1, 3, A) == [ 1:size(A,1), 1:size(A,2), 4:4]\r\n\t\t\t\t@test GG.sendranges(2, 3, A) == [ 1:size(A,1), 1:size(A,2), size(A,3)-3:size(A,3)-3]\r\n\t\t\t\t@test_throws ErrorException GG.recvranges(1, 1, A)\r\n\t\t\t\t@test_throws ErrorException GG.recvranges(2, 1, A)\r\n\t\t\t\t@test GG.recvranges(1, 2, A) == [ 1:size(A,1), 1:1, 1:size(A,3)]\r\n\t\t\t\t@test GG.recvranges(2, 2, A) == [ 1:size(A,1), size(A,2):size(A,2), 1:size(A,3)]\r\n\t\t\t\t@test GG.recvranges(1, 3, A) == [ 1:size(A,1), 1:size(A,2), 1:1]\r\n\t\t\t\t@test GG.recvranges(2, 3, A) == [ 1:size(A,1), 1:size(A,2), size(A,3):size(A,3)]\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\r\n\t\t\t@testset \"write_h2h! / read_h2h!\" begin\r\n\t\t\t\tinit_global_grid(nx, ny, nz, quiet=true, init_MPI=false);\r\n\t\t\t\tP = zeros(nx, ny, nz );\r\n\t\t\t\tP .= [iz*1e2 + iy*1e1 + ix for ix=1:size(P,1), iy=1:size(P,2), iz=1:size(P,3)];\r\n\t\t\t\tP2 = zeros(size(P));\r\n\t\t\t\t# (dim=1)\r\n\t\t\t\tbuf = zeros(size(P,2), size(P,3));\r\n\t\t\t\tranges = [2:2, 1:size(P,2), 1:size(P,3)];\r\n\t\t\t\tGG.write_h2h!(buf, P, ranges, 1);\r\n\t\t\t\t@test all(buf[:] .== P[ranges[1],ranges[2],ranges[3]][:])\r\n\t\t\t\tGG.read_h2h!(buf, P2, ranges, 1);\r\n\t\t\t\t@test all(buf[:] .== P2[ranges[1],ranges[2],ranges[3]][:])\r\n\t\t\t\t# (dim=2)\r\n\t\t\t\tbuf = zeros(size(P,1), size(P,3));\r\n\t\t\t\tranges = [1:size(P,1), 3:3, 1:size(P,3)];\r\n\t\t\t\tGG.write_h2h!(buf, P, ranges, 2);\r\n\t\t\t\t@test all(buf[:] .== P[ranges[1],ranges[2],ranges[3]][:])\r\n\t\t\t\tGG.read_h2h!(buf, P2, ranges, 2);\r\n\t\t\t\t@test all(buf[:] .== P2[ranges[1],ranges[2],ranges[3]][:])\r\n\t\t\t\t# (dim=3)\r\n\t\t\t\tbuf = zeros(size(P,1), size(P,2));\r\n\t\t\t\tranges = [1:size(P,1), 1:size(P,2), 4:4];\r\n\t\t\t\tGG.write_h2h!(buf, P, ranges, 3);\r\n\t\t\t\t@test all(buf[:] .== P[ranges[1],ranges[2],ranges[3]][:])\r\n\t\t\t\tGG.read_h2h!(buf, P2, ranges, 3);\r\n\t\t\t\t@test all(buf[:] .== P2[ranges[1],ranges[2],ranges[3]][:])\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\r\n\t\t\t@static if test_gpu\r\n\t\t\t\t@testset \"write_d2x! / write_d2h_async! / read_x2d! / read_h2d_async!\" begin\r\n\t\t\t\t\tinit_global_grid(nx, ny, nz, quiet=true, init_MPI=false);\r\n\t\t\t\t\tP = zeros(nx, ny, nz );\r\n\t\t\t\t\tP .= [iz*1e2 + iy*1e1 + ix for ix=1:size(P,1), iy=1:size(P,2), iz=1:size(P,3)];\r\n\t\t\t\t\tP = CuArray(P);\r\n\t\t\t\t\t# (dim=1)\r\n\t\t\t\t\tdim = 1;\r\n\t\t\t\t\tP2 = cuzeros(eltype(P),size(P));\r\n\t\t\t\t\tbuf = zeros(size(P,2), size(P,3));\r\n\t\t\t\t\tbuf_d, buf_h = GG.register(buf);\r\n\t\t\t\t\tranges = [2:2, 1:size(P,2), 1:size(P,3)];\r\n\t\t\t\t\tnthreads = (1, 1, 1);\r\n\t halosize = [r[end] - r[1] + 1 for r in ranges];\r\n\t\t\t\t\tnblocks = Tuple(ceil.(Int, halosize./nthreads));\r\n\t @cuda blocks=nblocks threads=nthreads GG.write_d2x!(buf_d, P, ranges[1], ranges[2], ranges[3], dim); CUDA.synchronize();\r\n\t\t\t\t\t@test all(buf[:] .== Array(P[ranges[1],ranges[2],ranges[3]][:]))\r\n\t\t\t\t\t@cuda blocks=nblocks threads=nthreads GG.read_x2d!(buf_d, P2, ranges[1], ranges[2], ranges[3], dim); CUDA.synchronize();\r\n\t\t\t\t\t@test all(buf[:] .== Array(P2[ranges[1],ranges[2],ranges[3]][:]))\r\n\t\t\t\t\tbuf .= 0.0;\r\n\t\t\t\t\tP2 .= 0.0;\r\n\t\t\t\t\tcustream = stream();\r\n\t\t\t\t\tGG.write_d2h_async!(buf, P, ranges, dim, custream); CUDA.synchronize();\r\n\t\t\t\t\t@test all(buf[:] .== Array(P[ranges[1],ranges[2],ranges[3]][:]))\r\n\t\t\t\t\tGG.read_h2d_async!(buf, P2, ranges, dim, custream); CUDA.synchronize();\r\n\t\t\t\t\t@test all(buf[:] .== Array(P2[ranges[1],ranges[2],ranges[3]][:]))\r\n\t\t\t\t\tCUDA.Mem.unregister(buf_h);\r\n\t\t\t\t\t# (dim=2)\r\n\t\t\t\t\tdim = 2;\r\n\t\t\t\t\tP2 = cuzeros(eltype(P),size(P));\r\n\t\t\t\t\tbuf = zeros(size(P,1), size(P,3));\r\n\t\t\t\t\tbuf_d, buf_h = GG.register(buf);\r\n\t\t\t\t\tranges = [1:size(P,1), 3:3, 1:size(P,3)];\r\n\t\t\t\t\tnthreads = (1, 1, 1);\r\n\t\t\t\t\thalosize = [r[end] - r[1] + 1 for r in ranges];\r\n\t\t\t\t\tnblocks = Tuple(ceil.(Int, halosize./nthreads));\r\n\t\t\t\t\t@cuda blocks=nblocks threads=nthreads GG.write_d2x!(buf_d, P, ranges[1], ranges[2], ranges[3], dim); CUDA.synchronize();\r\n\t\t\t\t\t@test all(buf[:] .== Array(P[ranges[1],ranges[2],ranges[3]][:]))\r\n\t\t\t\t\t@cuda blocks=nblocks threads=nthreads GG.read_x2d!(buf_d, P2, ranges[1], ranges[2], ranges[3], dim); CUDA.synchronize();\r\n\t\t\t\t\t@test all(buf[:] .== Array(P2[ranges[1],ranges[2],ranges[3]][:]))\r\n\t\t\t\t\tbuf .= 0.0;\r\n\t\t\t\t\tP2 .= 0.0;\r\n\t\t\t\t\tcustream = stream();\r\n\t\t\t\t\tGG.write_d2h_async!(buf, P, ranges, dim, custream); CUDA.synchronize();\r\n\t\t\t\t\t@test all(buf[:] .== Array(P[ranges[1],ranges[2],ranges[3]][:]))\r\n\t\t\t\t\tGG.read_h2d_async!(buf, P2, ranges, dim, custream); CUDA.synchronize();\r\n\t\t\t\t\t@test all(buf[:] .== Array(P2[ranges[1],ranges[2],ranges[3]][:]))\r\n\t\t\t\t\tCUDA.Mem.unregister(buf_h);\r\n\t\t\t\t\t# (dim=3)\r\n\t\t\t\t\tdim = 3\r\n\t\t\t\t\tP2 = cuzeros(eltype(P),size(P));\r\n\t\t\t\t\tbuf = zeros(size(P,1), size(P,2));\r\n\t\t\t\t\tbuf_d, buf_h = GG.register(buf);\r\n\t\t\t\t\tranges = [1:size(P,1), 1:size(P,2), 4:4];\r\n\t\t\t\t\tnthreads = (1, 1, 1);\r\n\t\t\t\t\thalosize = [r[end] - r[1] + 1 for r in ranges];\r\n\t\t\t\t\tnblocks = Tuple(ceil.(Int, halosize./nthreads));\r\n\t\t\t\t\t@cuda blocks=nblocks threads=nthreads GG.write_d2x!(buf_d, P, ranges[1], ranges[2], ranges[3], dim); CUDA.synchronize();\r\n\t\t\t\t\t@test all(buf[:] .== Array(P[ranges[1],ranges[2],ranges[3]][:]))\r\n\t\t\t\t\t@cuda blocks=nblocks threads=nthreads GG.read_x2d!(buf_d, P2, ranges[1], ranges[2], ranges[3], dim); CUDA.synchronize();\r\n\t\t\t\t\t@test all(buf[:] .== Array(P2[ranges[1],ranges[2],ranges[3]][:]))\r\n\t\t\t\t\tbuf .= 0.0;\r\n\t\t\t\t\tP2 .= 0.0;\r\n\t\t\t\t\tcustream = stream();\r\n\t\t\t\t\tGG.write_d2h_async!(buf, P, ranges, dim, custream); CUDA.synchronize();\r\n\t\t\t\t\t@test all(buf[:] .== Array(P[ranges[1],ranges[2],ranges[3]][:]))\r\n\t\t\t\t\tGG.read_h2d_async!(buf, P2, ranges, dim, custream); CUDA.synchronize();\r\n\t\t\t\t\t@test all(buf[:] .== Array(P2[ranges[1],ranges[2],ranges[3]][:]))\r\n\t\t\t\t\tCUDA.Mem.unregister(buf_h);\r\n\t\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\t\tend;\r\n\t\t\tend\r\n\t\t\t@testset \"iwrite_sendbufs! (allocator: $(longnameof(zeros)))\" for zeros in allocators\r\n\t\t\t\tinit_global_grid(nx, ny, nz, periodx=1, periody=1, periodz=1, overlapz=3, quiet=true, init_MPI=false);\r\n\t\t\t\tP = zeros(nx, ny, nz );\r\n\t\t\t\tA = zeros(nx-1,ny+2,nz+1);\r\n\t\t\t\tif zeros == cuzeros\r\n\t\t\t\t\tP = CuArray([iz*1e2 + iy*1e1 + ix for ix=1:size(P,1), iy=1:size(P,2), iz=1:size(P,3)]);\r\n\t\t\t\t\tA = CuArray([iz*1e2 + iy*1e1 + ix for ix=1:size(A,1), iy=1:size(A,2), iz=1:size(A,3)]);\r\n\t\t\t\telse\r\n\t\t\t\t\tP .= [iz*1e2 + iy*1e1 + ix for ix=1:size(P,1), iy=1:size(P,2), iz=1:size(P,3)];\r\n\t\t\t\t\tA .= [iz*1e2 + iy*1e1 + ix for ix=1:size(A,1), iy=1:size(A,2), iz=1:size(A,3)];\r\n\t\t\t\tend\r\n\t\t\t\tGG.allocate_bufs(P, A);\r\n\t\t\t\tif (zeros == cuzeros)\r\n\t\t\t\t\tGG.allocate_custreams(P, A);\r\n\t\t\t\telse\r\n\t\t\t\t\tGG.allocate_tasks(P, A);\r\n\t\t\t\tend\r\n\t\t\t\tdim = 1\r\n\t\t\t\tn = 1\r\n\t\t\t\tGG.iwrite_sendbufs!(n, dim, P, 1);\r\n\t\t\t\tGG.iwrite_sendbufs!(n, dim, A, 2);\r\n\t\t\t\tGG.wait_iwrite(n, P, 1);\r\n\t\t\t\tGG.wait_iwrite(n, A, 2);\r\n\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t@test all(GG.cusendbuf_flat(n,dim,1,P) .== CuArray(P[2,:,:][:]))\r\n\t\t\t\t\t@test all(GG.cusendbuf_flat(n,dim,2,A) .== 0.0)\r\n\t\t\t\telse\r\n\t\t\t\t\t@test all(GG.sendbuf_flat(n,dim,1,P) .== Array(P[2,:,:][:]))\r\n\t\t\t\t\t@test all(GG.sendbuf_flat(n,dim,2,A) .== 0.0)\r\n\t\t\t\tend\r\n\t\t\t\tn = 2\r\n\t\t\t\tGG.iwrite_sendbufs!(n, dim, P, 1);\r\n\t\t\t\tGG.iwrite_sendbufs!(n, dim, A, 2);\r\n\t\t\t\tGG.wait_iwrite(n, P, 1);\r\n\t\t\t\tGG.wait_iwrite(n, A, 2);\r\n\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t@test all(GG.cusendbuf_flat(n,dim,1,P) .== CuArray(P[end-1,:,:][:]))\r\n\t\t\t\t\t@test all(GG.cusendbuf_flat(n,dim,2,A) .== 0.0)\r\n\t\t\t\telse\r\n\t\t\t\t\t@test all(GG.sendbuf_flat(n,dim,1,P) .== Array(P[end-1,:,:][:]))\r\n\t\t\t\t\t@test all(GG.sendbuf_flat(n,dim,2,A) .== 0.0)\r\n\t\t\t\tend\r\n\t\t\t\tdim = 2\r\n\t\t\t\tn = 1\r\n\t\t\t\tGG.iwrite_sendbufs!(n, dim, P, 1);\r\n\t\t\t\tGG.iwrite_sendbufs!(n, dim, A, 2);\r\n\t\t\t\tGG.wait_iwrite(n, P, 1);\r\n\t\t\t\tGG.wait_iwrite(n, A, 2);\r\n\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t@test all(GG.cusendbuf_flat(n,dim,1,P) .== CuArray(P[:,2,:][:]))\r\n\t\t\t\t\t@test all(GG.cusendbuf_flat(n,dim,2,A) .== CuArray(A[:,4,:][:]))\r\n\t\t\t\telse\r\n\t\t\t\t\t@test all(GG.sendbuf_flat(n,dim,1,P) .== Array(P[:,2,:][:]))\r\n\t\t\t\t\t@test all(GG.sendbuf_flat(n,dim,2,A) .== Array(A[:,4,:][:]))\r\n\t\t\t\tend\r\n\t\t\t\tn = 2\r\n\t\t\t\tGG.iwrite_sendbufs!(n, dim, P, 1);\r\n\t\t\t\tGG.iwrite_sendbufs!(n, dim, A, 2);\r\n\t\t\t\tGG.wait_iwrite(n, P, 1);\r\n\t\t\t\tGG.wait_iwrite(n, A, 2);\r\n\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t@test all(GG.cusendbuf_flat(n,dim,1,P) .== CuArray(P[:,end-1,:][:]))\r\n\t\t\t\t\t@test all(GG.cusendbuf_flat(n,dim,2,A) .== CuArray(A[:,end-3,:][:]))\r\n\t\t\t\telse\r\n\t\t\t\t\t@test all(GG.sendbuf_flat(n,dim,1,P) .== Array(P[:,end-1,:][:]))\r\n\t\t\t\t\t@test all(GG.sendbuf_flat(n,dim,2,A) .== Array(A[:,end-3,:][:]))\r\n\t\t\t\tend\r\n\t\t\t\tdim = 3\r\n\t\t\t\tn = 1\r\n\t\t\t\tGG.iwrite_sendbufs!(n, dim, P, 1);\r\n\t\t\t\tGG.iwrite_sendbufs!(n, dim, A, 2);\r\n\t\t\t\tGG.wait_iwrite(n, P, 1);\r\n\t\t\t\tGG.wait_iwrite(n, A, 2);\r\n\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t@test all(GG.cusendbuf_flat(n,dim,1,P) .== CuArray(P[:,:,3][:]))\r\n\t\t\t\t\t@test all(GG.cusendbuf_flat(n,dim,2,A) .== CuArray(A[:,:,4][:]))\r\n\t\t\t\telse\r\n\t\t\t\t\t@test all(GG.sendbuf_flat(n,dim,1,P) .== Array(P[:,:,3][:]))\r\n\t\t\t\t\t@test all(GG.sendbuf_flat(n,dim,2,A) .== Array(A[:,:,4][:]))\r\n\t\t\t\tend\r\n\t\t\t\tn = 2\r\n\t\t\t\tGG.iwrite_sendbufs!(n, dim, P, 1);\r\n\t\t\t\tGG.iwrite_sendbufs!(n, dim, A, 2);\r\n\t\t\t\tGG.wait_iwrite(n, P, 1);\r\n\t\t\t\tGG.wait_iwrite(n, A, 2);\r\n\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t@test all(GG.cusendbuf_flat(n,dim,1,P) .== CuArray(P[:,:,end-2][:]))\r\n\t\t\t\t\t@test all(GG.cusendbuf_flat(n,dim,2,A) .== CuArray(A[:,:,end-3][:]))\r\n\t\t\t\telse\r\n\t\t\t\t\t@test all(GG.sendbuf_flat(n,dim,1,P) .== Array(P[:,:,end-2][:]))\r\n\t\t\t\t\t@test all(GG.sendbuf_flat(n,dim,2,A) .== Array(A[:,:,end-3][:]))\r\n\t\t\t\tend\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\r\n\t\t\t@testset \"iread_recvbufs! (allocator: $(longnameof(zeros)))\" for zeros in allocators\r\n\t\t\t\tinit_global_grid(nx, ny, nz, periodx=1, periody=1, periodz=1, overlapz=3, quiet=true, init_MPI=false);\r\n\t\t\t\tP = zeros(nx, ny, nz );\r\n\t\t\t\tA = zeros(nx-1,ny+2,nz+1);\r\n\t\t\t\tGG.allocate_bufs(P, A);\r\n\t\t\t\tif (zeros == cuzeros)\r\n\t\t\t\t\tGG.allocate_custreams(P, A);\r\n\t\t\t\telse\r\n\t\t\t\t\tGG.allocate_tasks(P, A);\r\n\t\t\t\tend\r\n\t\t\t\tdim = 1\r\n\t\t\t\tfor n = 1:nneighbors_per_dim\r\n\t\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t\tGG.curecvbuf_flat(n,dim,1,P) .= dim*1e2 + n*1e1 + 1;\r\n\t\t\t\t\t\tGG.curecvbuf_flat(n,dim,2,A) .= dim*1e2 + n*1e1 + 2;\r\n\t\t\t\t\telse\r\n\t\t\t\t\t\tGG.recvbuf_flat(n,dim,1,P) .= dim*1e2 + n*1e1 + 1;\r\n\t\t\t\t\t\tGG.recvbuf_flat(n,dim,2,A) .= dim*1e2 + n*1e1 + 2;\r\n\t\t\t\t\tend\r\n\t\t\t\tend\r\n\t\t\t\tn = 1\r\n\t\t\t\tGG.iread_recvbufs!(n, dim, P, 1);\r\n\t\t\t\tGG.iread_recvbufs!(n, dim, A, 2);\r\n\t\t\t\tGG.wait_iread(n, P, 1);\r\n\t\t\t\tGG.wait_iread(n, A, 2);\r\n\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t@test all(GG.curecvbuf_flat(n,dim,1,P) .== CuArray(P[1,:,:][:]))\r\n\t\t\t\t\t@test all( 0.0 .== CuArray(A[1,:,:][:]))\r\n\t\t\t\telse\r\n\t\t\t\t\t@test all(GG.recvbuf_flat(n,dim,1,P) .== Array(P[1,:,:][:]))\r\n\t\t\t\t\t@test all( 0.0 .== Array(A[1,:,:][:]))\r\n\t\t\t\tend\r\n\t\t\t\tn = 2\r\n\t\t\t\tGG.iread_recvbufs!(n, dim, P, 1);\r\n\t\t\t\tGG.iread_recvbufs!(n, dim, A, 2);\r\n\t\t\t\tGG.wait_iread(n, P, 1);\r\n\t\t\t\tGG.wait_iread(n, A, 2);\r\n\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t@test all(GG.curecvbuf_flat(n,dim,1,P) .== CuArray(P[end,:,:][:]))\r\n\t\t\t\t\t@test all( 0.0 .== CuArray(A[end,:,:][:]))\r\n\t\t\t\telse\r\n\t\t\t\t\t@test all(GG.recvbuf_flat(n,dim,1,P) .== Array(P[end,:,:][:]))\r\n\t\t\t\t\t@test all( 0.0 .== Array(A[end,:,:][:]))\r\n\t\t\t\tend\r\n\t\t\t\tdim = 2\r\n\t\t\t\tfor n = 1:nneighbors_per_dim\r\n\t\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t\tGG.curecvbuf_flat(n,dim,1,P) .= dim*1e2 + n*1e1 + 1;\r\n\t\t\t\t\t\tGG.curecvbuf_flat(n,dim,2,A) .= dim*1e2 + n*1e1 + 2;\r\n\t\t\t\t\telse\r\n\t\t\t\t\t\tGG.recvbuf_flat(n,dim,1,P) .= dim*1e2 + n*1e1 + 1;\r\n\t\t\t\t\t\tGG.recvbuf_flat(n,dim,2,A) .= dim*1e2 + n*1e1 + 2;\r\n\t\t\t\t\tend\r\n\t\t\t\tend\r\n\t\t\t\tn = 1\r\n\t\t\t\tGG.iread_recvbufs!(n, dim, P, 1);\r\n\t\t\t\tGG.iread_recvbufs!(n, dim, A, 2);\r\n\t\t\t\tGG.wait_iread(n, P, 1);\r\n\t\t\t\tGG.wait_iread(n, A, 2);\r\n\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t@test all(GG.curecvbuf_flat(n,dim,1,P) .== CuArray(P[:,1,:][:]))\r\n\t\t\t\t\t@test all(GG.curecvbuf_flat(n,dim,2,A) .== CuArray(A[:,1,:][:]))\r\n\t\t\t\telse\r\n\t\t\t\t\t@test all(GG.recvbuf_flat(n,dim,1,P) .== Array(P[:,1,:][:]))\r\n\t\t\t\t\t@test all(GG.recvbuf_flat(n,dim,2,A) .== Array(A[:,1,:][:]))\r\n\t\t\t\tend\r\n\t\t\t\tn = 2\r\n\t\t\t\tGG.iread_recvbufs!(n, dim, P, 1);\r\n\t\t\t\tGG.iread_recvbufs!(n, dim, A, 2);\r\n\t\t\t\tGG.wait_iread(n, P, 1);\r\n\t\t\t\tGG.wait_iread(n, A, 2);\r\n\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t@test all(GG.curecvbuf_flat(n,dim,1,P) .== CuArray(P[:,end,:][:]))\r\n\t\t\t\t\t@test all(GG.curecvbuf_flat(n,dim,2,A) .== CuArray(A[:,end,:][:]))\r\n\t\t\t\telse\r\n\t\t\t\t\t@test all(GG.recvbuf_flat(n,dim,1,P) .== Array(P[:,end,:][:]))\r\n\t\t\t\t\t@test all(GG.recvbuf_flat(n,dim,2,A) .== Array(A[:,end,:][:]))\r\n\t\t\t\tend\r\n\t\t\t\tdim = 3\r\n\t\t\t\tfor n = 1:nneighbors_per_dim\r\n\t\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t\tGG.curecvbuf_flat(n,dim,1,P) .= dim*1e2 + n*1e1 + 1;\r\n\t\t\t\t\t\tGG.curecvbuf_flat(n,dim,2,A) .= dim*1e2 + n*1e1 + 2;\r\n\t\t\t\t\telse\r\n\t\t\t\t\t\tGG.recvbuf_flat(n,dim,1,P) .= dim*1e2 + n*1e1 + 1;\r\n\t\t\t\t\t\tGG.recvbuf_flat(n,dim,2,A) .= dim*1e2 + n*1e1 + 2;\r\n\t\t\t\t\tend\r\n\t\t\t\tend\r\n\t\t\t\tn = 1\r\n\t\t\t\tGG.iread_recvbufs!(n, dim, P, 1);\r\n\t\t\t\tGG.iread_recvbufs!(n, dim, A, 2);\r\n\t\t\t\tGG.wait_iread(n, P, 1);\r\n\t\t\t\tGG.wait_iread(n, A, 2);\r\n\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t@test all(GG.curecvbuf_flat(n,dim,1,P) .== CuArray(P[:,:,1][:]))\r\n\t\t\t\t\t@test all(GG.curecvbuf_flat(n,dim,2,A) .== CuArray(A[:,:,1][:]))\r\n\t\t\t\telse\r\n\t\t\t\t\t@test all(GG.recvbuf_flat(n,dim,1,P) .== Array(P[:,:,1][:]))\r\n\t\t\t\t\t@test all(GG.recvbuf_flat(n,dim,2,A) .== Array(A[:,:,1][:]))\r\n\t\t\t\tend\r\n\t\t\t\tn = 2\r\n\t\t\t\tGG.iread_recvbufs!(n, dim, P, 1);\r\n\t\t\t\tGG.iread_recvbufs!(n, dim, A, 2);\r\n\t\t\t\tGG.wait_iread(n, P, 1);\r\n\t\t\t\tGG.wait_iread(n, A, 2);\r\n\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t@test all(GG.curecvbuf_flat(n,dim,1,P) .== CuArray(P[:,:,end][:]))\r\n\t\t\t\t\t@test all(GG.curecvbuf_flat(n,dim,2,A) .== CuArray(A[:,:,end][:]))\r\n\t\t\t\telse\r\n\t\t\t\t\t@test all(GG.recvbuf_flat(n,dim,1,P) .== Array(P[:,:,end][:]))\r\n\t\t\t\t\t@test all(GG.recvbuf_flat(n,dim,2,A) .== Array(A[:,:,end][:]))\r\n\t\t\t\tend\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\r\n\t\t\tif (nprocs==1)\r\n\t\t\t\t@testset \"sendrecv_halo_local (allocator: $(longnameof(zeros)))\" for zeros in allocators\r\n\t\t\t\t\tinit_global_grid(nx, ny, nz, periodx=1, periody=1, periodz=1, overlapz=3, quiet=true, init_MPI=false);\r\n\t\t\t\t\tP = zeros(nx, ny, nz );\r\n\t\t\t\t\tA = zeros(nx-1,ny+2,nz+1);\r\n\t\t\t\t\tGG.allocate_bufs(P, A);\r\n\t\t\t\t\tdim = 1\r\n\t\t\t\t\tfor n = 1:nneighbors_per_dim\r\n\t\t\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t\t\tGG.cusendbuf_flat(n,dim,1,P) .= dim*1e2 + n*1e1 + 1;\r\n\t\t\t\t\t\t\tGG.cusendbuf_flat(n,dim,2,A) .= dim*1e2 + n*1e1 + 2;\r\n\t\t\t\t\t\telse\r\n\t\t\t\t\t\t\tGG.sendbuf_flat(n,dim,1,P) .= dim*1e2 + n*1e1 + 1;\r\n\t\t\t\t\t\t\tGG.sendbuf_flat(n,dim,2,A) .= dim*1e2 + n*1e1 + 2;\r\n\t\t\t\t\t\tend\r\n\t\t\t\t\tend\r\n\t\t\t\t\tfor n = 1:nneighbors_per_dim\r\n\t\t\t\t\t\tGG.sendrecv_halo_local(n, dim, P, 1);\r\n\t\t\t\t\t\tGG.sendrecv_halo_local(n, dim, A, 2);\r\n\t\t\t\t\tend\r\n\t\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t\t@test all(GG.curecvbuf_flat(1,dim,1,P) .== GG.cusendbuf_flat(2,dim,1,P));\r\n\t\t\t\t\t\t@test all(GG.curecvbuf_flat(1,dim,2,A) .== 0.0); # There is no halo (ol(dim,A) < 2).\r\n\t\t\t\t\t\t@test all(GG.curecvbuf_flat(2,dim,1,P) .== GG.cusendbuf_flat(1,dim,1,P));\r\n\t\t\t\t\t\t@test all(GG.curecvbuf_flat(2,dim,2,A) .== 0.0); # There is no halo (ol(dim,A) < 2).\r\n\t\t\t\t\telse\r\n\t\t\t\t\t\t@test all(GG.recvbuf_flat(1,dim,1,P) .== GG.sendbuf_flat(2,dim,1,P));\r\n\t\t\t\t\t\t@test all(GG.recvbuf_flat(1,dim,2,A) .== 0.0); # There is no halo (ol(dim,A) < 2).\r\n\t\t\t\t\t\t@test all(GG.recvbuf_flat(2,dim,1,P) .== GG.sendbuf_flat(1,dim,1,P));\r\n\t\t\t\t\t\t@test all(GG.recvbuf_flat(2,dim,2,A) .== 0.0); # There is no halo (ol(dim,A) < 2).\r\n\t\t\t\t\tend\r\n\t\t\t\t\tdim = 2\r\n\t\t\t\t\tfor n = 1:nneighbors_per_dim\r\n\t\t\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t\t\tGG.cusendbuf_flat(n,dim,1,P) .= dim*1e2 + n*1e1 + 1;\r\n\t\t\t\t\t\t\tGG.cusendbuf_flat(n,dim,2,A) .= dim*1e2 + n*1e1 + 2;\r\n\t\t\t\t\t\telse\r\n\t\t\t\t\t\t\tGG.sendbuf_flat(n,dim,1,P) .= dim*1e2 + n*1e1 + 1;\r\n\t\t\t\t\t\t\tGG.sendbuf_flat(n,dim,2,A) .= dim*1e2 + n*1e1 + 2;\r\n\t\t\t\t\t\tend\r\n\t\t\t\t\tend\r\n\t\t\t\t\tfor n = 1:nneighbors_per_dim\r\n\t\t\t\t\t\tGG.sendrecv_halo_local(n, dim, P, 1);\r\n\t\t\t\t\t\tGG.sendrecv_halo_local(n, dim, A, 2);\r\n\t\t\t\t\tend\r\n\t\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t\t@test all(GG.curecvbuf_flat(1,dim,1,P) .== GG.cusendbuf_flat(2,dim,1,P));\r\n\t\t\t\t\t\t@test all(GG.curecvbuf_flat(1,dim,2,A) .== GG.cusendbuf_flat(2,dim,2,A));\r\n\t\t\t\t\t\t@test all(GG.curecvbuf_flat(2,dim,1,P) .== GG.cusendbuf_flat(1,dim,1,P));\r\n\t\t\t\t\t\t@test all(GG.curecvbuf_flat(2,dim,2,A) .== GG.cusendbuf_flat(1,dim,2,A));\r\n\t\t\t\t\telse\r\n\t\t\t\t\t\t@test all(GG.recvbuf_flat(1,dim,1,P) .== GG.sendbuf_flat(2,dim,1,P));\r\n\t\t\t\t\t\t@test all(GG.recvbuf_flat(1,dim,2,A) .== GG.sendbuf_flat(2,dim,2,A));\r\n\t\t\t\t\t\t@test all(GG.recvbuf_flat(2,dim,1,P) .== GG.sendbuf_flat(1,dim,1,P));\r\n\t\t\t\t\t\t@test all(GG.recvbuf_flat(2,dim,2,A) .== GG.sendbuf_flat(1,dim,2,A));\r\n\t\t\t\t\tend\r\n\t\t\t\t\tdim = 3\r\n\t\t\t\t\tfor n = 1:nneighbors_per_dim\r\n\t\t\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t\t\tGG.cusendbuf_flat(n,dim,1,P) .= dim*1e2 + n*1e1 + 1;\r\n\t\t\t\t\t\t\tGG.cusendbuf_flat(n,dim,2,A) .= dim*1e2 + n*1e1 + 2;\r\n\t\t\t\t\t\telse\r\n\t\t\t\t\t\t\tGG.sendbuf_flat(n,dim,1,P) .= dim*1e2 + n*1e1 + 1;\r\n\t\t\t\t\t\t\tGG.sendbuf_flat(n,dim,2,A) .= dim*1e2 + n*1e1 + 2;\r\n\t\t\t\t\t\tend\r\n\t\t\t\t\tend\r\n\t\t\t\t\tfor n = 1:nneighbors_per_dim\r\n\t\t\t\t\t\tGG.sendrecv_halo_local(n, dim, P, 1);\r\n\t\t\t\t\t\tGG.sendrecv_halo_local(n, dim, A, 2);\r\n\t\t\t\t\tend\r\n\t\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t\t@test all(GG.curecvbuf_flat(1,dim,1,P) .== GG.cusendbuf_flat(2,dim,1,P));\r\n\t\t\t\t\t\t@test all(GG.curecvbuf_flat(1,dim,2,A) .== GG.cusendbuf_flat(2,dim,2,A));\r\n\t\t\t\t\t\t@test all(GG.curecvbuf_flat(2,dim,1,P) .== GG.cusendbuf_flat(1,dim,1,P));\r\n\t\t\t\t\t\t@test all(GG.curecvbuf_flat(2,dim,2,A) .== GG.cusendbuf_flat(1,dim,2,A));\r\n\t\t\t\t\telse\r\n\t\t\t\t\t\t@test all(GG.recvbuf_flat(1,dim,1,P) .== GG.sendbuf_flat(2,dim,1,P));\r\n\t\t\t\t\t\t@test all(GG.recvbuf_flat(1,dim,2,A) .== GG.sendbuf_flat(2,dim,2,A));\r\n\t\t\t\t\t\t@test all(GG.recvbuf_flat(2,dim,1,P) .== GG.sendbuf_flat(1,dim,1,P));\r\n\t\t\t\t\t\t@test all(GG.recvbuf_flat(2,dim,2,A) .== GG.sendbuf_flat(1,dim,2,A));\r\n\t\t\t\t\tend\r\n\t\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\t\tend\r\n\t\t\tend\r\n\t\tend;\r\n\t\tif (nprocs>1)\r\n\t\t\t@testset \"irecv_halo! / isend_halo (allocator: $(longnameof(zeros)))\" for zeros in allocators\r\n\t\t\t\tme, dims, nprocs, coords, comm = init_global_grid(nx, ny, nz, dimy=1, dimz=1, periodx=1, quiet=true, init_MPI=false);\r\n\t\t\t\tP = zeros(nx,ny,nz);\r\n\t\t\t\tA = zeros(nx,ny,nz);\r\n\t\t\t\tdim = 1;\r\n\t\t\t\tGG.allocate_bufs(P, A);\r\n\t\t\t\tfor n = 1:nneighbors_per_dim\r\n\t\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t\tGG.cusendbuf(n,dim,1,P) .= 9.0;\r\n\t\t\t\t\t\tGG.curecvbuf(n,dim,1,P) .= 0;\r\n\t\t\t\t\t\tGG.cusendbuf(n,dim,2,A) .= 9.0;\r\n\t\t\t\t\t\tGG.curecvbuf(n,dim,2,A) .= 0;\r\n\t\t\t\t\telse\r\n\t\t\t\t\t\tGG.sendbuf(n,dim,1,P) .= 9.0;\r\n\t\t\t\t\t\tGG.recvbuf(n,dim,1,P) .= 0;\r\n\t\t\t\t\t\tGG.sendbuf(n,dim,2,A) .= 9.0;\r\n\t\t\t\t\t\tGG.recvbuf(n,dim,2,A) .= 0;\r\n\t\t\t\t\tend\r\n\t\t\t\tend\r\n\t\t\t\treqs = fill(MPI.REQUEST_NULL, 2, nneighbors_per_dim, 2);\r\n\t\t\t\tfor n = 1:nneighbors_per_dim\r\n\t reqs[1,n,1] = GG.irecv_halo!(n, dim, P, 1);\r\n\t\t\t\t\treqs[2,n,1] = GG.irecv_halo!(n, dim, A, 2);\r\n\t reqs[1,n,2] = GG.isend_halo(n, dim, P, 1);\r\n\t\t\t\t\treqs[2,n,2] = GG.isend_halo(n, dim, A, 2);\r\n\t end\r\n\t\t\t\t@test all(reqs .!= [MPI.REQUEST_NULL])\r\n\t MPI.Waitall!(reqs[:]);\r\n\t\t\t\tfor n = 1:nneighbors_per_dim\r\n\t\t\t\t\tif zeros == cuzeros && GG.cudaaware_MPI(dim)\r\n\t\t\t\t\t\t@test all(GG.curecvbuf(n,dim,1,P) .== 9.0)\r\n\t\t\t\t\t\t@test all(GG.curecvbuf(n,dim,2,A) .== 9.0)\r\n\t\t\t\t\telse\r\n\t\t\t\t\t\t@test all(GG.recvbuf(n,dim,1,P) .== 9.0)\r\n\t\t\t\t\t\t@test all(GG.recvbuf(n,dim,2,A) .== 9.0)\r\n\t\t\t\t\tend\r\n\t\t\t\tend\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\r\n\t\tend\r\n\tend;\r\n\r\n\t# (Backup field filled with encoded coordinates and set boundary to zeros; then update halo and compare with backuped field; it should be the same again, except for the boundaries that are not halos)\r\n\t@testset \"4. halo update (allocator: $(longnameof(Array)))\" for Array in ArrayConstructors\r\n\t\t@testset \"basic grid (default: periodic)\" begin\r\n\t\t\t@testset \"1D\" begin\r\n\t\t \tinit_global_grid(nx, 1, 1, periodx=1, quiet=true, init_MPI=false);\r\n\t\t\t\tP = zeros(nx);\r\n\t\t\t\tP .= [x_g(ix,dx,P) for ix=1:size(P,1)];\r\n\t\t\t\tP_ref = copy(P);\r\n\t\t\t\tP[[1, end]] .= 0.0;\r\n\t\t\t\tP = Array(P);\r\n\t\t\t\tP_ref = Array(P_ref);\r\n\t\t\t\t@require !all(P .== P_ref)\r\n\t\t\t\tupdate_halo!(P);\r\n\t\t\t\t@test all(P .== P_ref)\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\r\n\t\t\t@testset \"2D\" begin\r\n\t\t\t\tinit_global_grid(nx, ny, 1, periodx=1, periody=1, quiet=true, init_MPI=false);\r\n\t\t\t\tP = zeros(nx, ny);\r\n\t\t\t\tP .= [y_g(iy,dy,P)*1e1 + x_g(ix,dx,P) for ix=1:size(P,1), iy=1:size(P,2)];\r\n\t\t\t\tP_ref = copy(P);\r\n\t\t\t\tP[[1, end], :] .= 0.0;\r\n\t\t\t\tP[ :,[1, end]] .= 0.0;\r\n\t\t\t\tP = Array(P);\r\n\t\t\t\tP_ref = Array(P_ref);\r\n\t\t\t\t@require !all(P .== P_ref)\r\n\t\t\t\tupdate_halo!(P);\r\n\t\t\t\t@test all(P .== P_ref)\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\r\n\t\t\t@testset \"3D\" begin\r\n\t\t\t\tinit_global_grid(nx, ny, nz, periodx=1, periody=1, periodz=1, quiet=true, init_MPI=false);\r\n\t\t\t\tP = zeros(nx, ny, nz);\r\n\t\t\t\tP .= [z_g(iz,dz,P)*1e2 + y_g(iy,dy,P)*1e1 + x_g(ix,dx,P) for ix=1:size(P,1), iy=1:size(P,2), iz=1:size(P,3)];\r\n\t\t\t\tP_ref = copy(P);\r\n\t\t\t\tP[[1, end], :, :] .= 0.0;\r\n\t\t\t\tP[ :,[1, end], :] .= 0.0;\r\n\t\t\t\tP[ :, :,[1, end]] .= 0.0;\r\n\t\t\t\tP = Array(P);\r\n\t\t\t\tP_ref = Array(P_ref);\r\n\t\t\t\t@require !all(P .== P_ref)\r\n\t\t\t\tupdate_halo!(P);\r\n\t\t\t\t@test all(P .== P_ref)\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\r\n\t\t\t@testset \"3D (non-default overlap)\" begin\r\n\t\t\t\tinit_global_grid(nx, ny, nz, periodx=1, periody=1, periodz=1, overlapx=4, overlapz=3, quiet=true, init_MPI=false);\r\n\t\t\t\tP = zeros(nx, ny, nz);\r\n\t\t\t\tP .= [z_g(iz,dz,P)*1e2 + y_g(iy,dy,P)*1e1 + x_g(ix,dx,P) for ix=1:size(P,1), iy=1:size(P,2), iz=1:size(P,3)];\r\n\t\t\t\tP_ref = copy(P);\r\n\t\t\t\tP[[1, end], :, :] .= 0.0;\r\n\t\t\t\tP[ :,[1, end], :] .= 0.0;\r\n\t\t\t\tP[ :, :,[1, end]] .= 0.0;\r\n\t\t\t\tP = Array(P);\r\n\t\t\t\tP_ref = Array(P_ref);\r\n\t\t\t\t@require !all(P .== P_ref)\r\n\t\t\t\tupdate_halo!(P);\r\n\t\t\t\t@test all(P .== P_ref)\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\r\n\t\t\t@testset \"3D (not periodic)\" begin\r\n\t\t\t\tme, dims, nprocs, coords = init_global_grid(nx, ny, nz, quiet=true, init_MPI=false);\r\n\t\t\t\tP = zeros(nx, ny, nz);\r\n\t\t\t\tP .= [z_g(iz,dz,P)*1e2 + y_g(iy,dy,P)*1e1 + x_g(ix,dx,P) for ix=1:size(P,1), iy=1:size(P,2), iz=1:size(P,3)];\r\n\t\t\t\tP_ref = copy(P);\r\n\t\t\t\tP[[1, end], :, :] .= 0.0;\r\n\t\t\t\tP[ :,[1, end], :] .= 0.0;\r\n\t\t\t\tP[ :, :,[1, end]] .= 0.0;\r\n\t\t\t\tP = Array(P);\r\n\t\t\t\tP_ref = Array(P_ref);\r\n\t\t\t\t@require !all(P .== P_ref)\r\n\t\t\t\tupdate_halo!(P);\r\n\t\t\t\t@test all(P[2:end-1,2:end-1,2:end-1] .== P_ref[2:end-1,2:end-1,2:end-1])\r\n\t\t\t\tif (coords[1] == 0) @test all(P[ 1, :, :] .== 0.0); else @test all(P[ 1,2:end-1,2:end-1] .== P_ref[ 1,2:end-1,2:end-1]); end # Verifcation of corner values would be cumbersome here; it is already sufficiently covered in the periodic tests.\r\n\t\t\t\tif (coords[1] == dims[1]-1) @test all(P[end, :, :] .== 0.0); else @test all(P[ end,2:end-1,2:end-1] .== P_ref[ end,2:end-1,2:end-1]); end\r\n\t\t\t\tif (coords[2] == 0) @test all(P[ :, 1, :] .== 0.0); else @test all(P[2:end-1, 1,2:end-1] .== P_ref[2:end-1, 1,2:end-1]); end\r\n\t\t\t\tif (coords[2] == dims[2]-1) @test all(P[ :,end, :] .== 0.0); else @test all(P[2:end-1, end,2:end-1] .== P_ref[2:end-1, end,2:end-1]); end\r\n\t\t\t\tif (coords[3] == 0) @test all(P[ :, :, 1] .== 0.0); else @test all(P[2:end-1,2:end-1, 1] .== P_ref[2:end-1,2:end-1, 1]); end\r\n\t\t\t\tif (coords[3] == dims[3]-1) @test all(P[ :, :,end] .== 0.0); else @test all(P[2:end-1,2:end-1, end] .== P_ref[2:end-1,2:end-1, end]); end\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\r\n\t\tend;\r\n\t\t@testset \"staggered grid (default: periodic)\" begin\r\n\t\t\t@testset \"1D\" begin\r\n\t\t\t\tinit_global_grid(nx, 1, 1, periodx=1, quiet=true, init_MPI=false);\r\n\t\t\t\tVx = zeros(nx+1);\r\n\t\t\t\tVx .= [x_g(ix,dx,Vx) for ix=1:size(Vx,1)];\r\n\t\t\t\tVx_ref = copy(Vx);\r\n\t\t\t\tVx[[1, end]] .= 0.0;\r\n\t\t\t\tVx = Array(Vx);\r\n\t\t\t\tVx_ref = Array(Vx_ref);\r\n\t\t\t\t@require !all(Vx .== Vx_ref)\r\n\t\t\t\tupdate_halo!(Vx);\r\n\t\t\t\t@test all(Vx .== Vx_ref)\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\r\n\t\t\t@testset \"2D\" begin\r\n\t\t\t\tinit_global_grid(nx, ny, 1, periodx=1, periody=1, quiet=true, init_MPI=false);\r\n\t\t\t\tVy = zeros(nx,ny+1);\r\n\t\t\t\tVy .= [y_g(iy,dy,Vy)*1e1 + x_g(ix,dx,Vy) for ix=1:size(Vy,1), iy=1:size(Vy,2)];\r\n\t\t\t\tVy_ref = copy(Vy);\r\n\t\t\t\tVy[[1, end], :] .= 0.0;\r\n\t\t\t\tVy[ :,[1, end]] .= 0.0;\r\n\t\t\t\tVy = Array(Vy);\r\n\t\t\t\tVy_ref = Array(Vy_ref);\r\n\t\t\t\t@require !all(Vy .== Vy_ref)\r\n\t\t\t\tupdate_halo!(Vy);\r\n\t\t\t\t@test all(Vy .== Vy_ref)\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\r\n\t\t\t@testset \"3D\" begin\r\n\t\t\t\tinit_global_grid(nx, ny, nz, periodx=1, periody=1, periodz=1, quiet=true, init_MPI=false);\r\n\t\t\t\tVz = zeros(nx,ny,nz+1);\r\n\t\t\t\tVz .= [z_g(iz,dz,Vz)*1e2 + y_g(iy,dy,Vz)*1e1 + x_g(ix,dx,Vz) for ix=1:size(Vz,1), iy=1:size(Vz,2), iz=1:size(Vz,3)];\r\n\t\t\t\tVz_ref = copy(Vz);\r\n\t\t\t\tVz[[1, end], :, :] .= 0.0;\r\n\t\t\t\tVz[ :,[1, end], :] .= 0.0;\r\n\t\t\t\tVz[ :, :,[1, end]] .= 0.0;\r\n\t\t\t\tVz = Array(Vz);\r\n\t\t\t\tVz_ref = Array(Vz_ref);\r\n\t\t\t\t@require !all(Vz .== Vz_ref)\r\n\t\t\t\tupdate_halo!(Vz);\r\n\t\t\t\t@test all(Vz .== Vz_ref)\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\r\n\t\t\t@testset \"3D (non-default overlap)\" begin\r\n\t\t\t\tinit_global_grid(nx, ny, nz, periodx=1, periody=1, periodz=1, overlapx=3, overlapz=3, quiet=true, init_MPI=false);\r\n\t\t\t\tVx = zeros(nx+1,ny,nz);\r\n\t\t\t\tVx .= [z_g(iz,dz,Vx)*1e2 + y_g(iy,dy,Vx)*1e1 + x_g(ix,dx,Vx) for ix=1:size(Vx,1), iy=1:size(Vx,2), iz=1:size(Vx,3)];\r\n\t\t\t\tVx_ref = copy(Vx);\r\n\t\t\t\tVx[[1, end], :, :] .= 0.0;\r\n\t\t\t\tVx[ :,[1, end], :] .= 0.0;\r\n\t\t\t\tVx[ :, :,[1, end]] .= 0.0;\r\n\t\t\t\tVx = Array(Vx);\r\n\t\t\t\tVx_ref = Array(Vx_ref);\r\n\t\t\t\t@require !all(Vx .== Vx_ref)\r\n\t\t\t\tupdate_halo!(Vx);\r\n\t\t\t\t@test all(Vx .== Vx_ref)\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\r\n\t\t\t@testset \"3D (not periodic)\" begin\r\n\t\t\t\tme, dims, nprocs, coords = init_global_grid(nx, ny, nz, quiet=true, init_MPI=false);\r\n\t\t\t\tVz = zeros(nx,ny,nz+1);\r\n\t\t\t\tVz .= [z_g(iz,dz,Vz)*1e2 + y_g(iy,dy,Vz)*1e1 + x_g(ix,dx,Vz) for ix=1:size(Vz,1), iy=1:size(Vz,2), iz=1:size(Vz,3)];\r\n\t\t\t\tVz_ref = copy(Vz);\r\n\t\t\t\tVz[[1, end], :, :] .= 0.0;\r\n\t\t\t\tVz[ :,[1, end], :] .= 0.0;\r\n\t\t\t\tVz[ :, :,[1, end]] .= 0.0;\r\n\t\t\t\tVz = Array(Vz);\r\n\t\t\t\tVz_ref = Array(Vz_ref);\r\n\t\t\t\t@require !all(Vz .== Vz_ref)\r\n\t\t\t\tupdate_halo!(Vz);\r\n\t\t\t\t@test all(Vz[2:end-1,2:end-1,2:end-1] .== Vz_ref[2:end-1,2:end-1,2:end-1])\r\n\t\t\t\tif (coords[1] == 0) @test all(Vz[ 1, :, :] .== 0.0); else @test all(Vz[ 1,2:end-1,2:end-1] .== Vz_ref[ 1,2:end-1,2:end-1]); end # Verifcation of corner values would be cumbersome here; it is already sufficiently covered in the periodic tests.\r\n\t\t\t\tif (coords[1] == dims[1]-1) @test all(Vz[end, :, :] .== 0.0); else @test all(Vz[ end,2:end-1,2:end-1] .== Vz_ref[ end,2:end-1,2:end-1]); end\r\n\t\t\t\tif (coords[2] == 0) @test all(Vz[ :, 1, :] .== 0.0); else @test all(Vz[2:end-1, 1,2:end-1] .== Vz_ref[2:end-1, 1,2:end-1]); end\r\n\t\t\t\tif (coords[2] == dims[2]-1) @test all(Vz[ :,end, :] .== 0.0); else @test all(Vz[2:end-1, end,2:end-1] .== Vz_ref[2:end-1, end,2:end-1]); end\r\n\t\t\t\tif (coords[3] == 0) @test all(Vz[ :, :, 1] .== 0.0); else @test all(Vz[2:end-1,2:end-1, 1] .== Vz_ref[2:end-1,2:end-1, 1]); end\r\n\t\t\t\tif (coords[3] == dims[3]-1) @test all(Vz[ :, :,end] .== 0.0); else @test all(Vz[2:end-1,2:end-1, end] .== Vz_ref[2:end-1,2:end-1, end]); end\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\r\n\t\t\t@testset \"2D (no halo in one dim)\" begin\r\n\t\t\t\tinit_global_grid(nx, ny, 1, periodx=1, periody=1, quiet=true, init_MPI=false);\r\n\t\t\t\tA = zeros(nx-1,ny+2);\r\n\t\t\t\tA .= [y_g(iy,dy,A)*1e1 + x_g(ix,dx,A) for ix=1:size(A,1), iy=1:size(A,2)];\r\n\t\t\t\tA_ref = copy(A);\r\n\t\t\t\tA[[1, end], :] .= 0.0;\r\n\t\t\t\tA[ :,[1, end]] .= 0.0;\r\n\t\t\t\tA = Array(A);\r\n\t\t\t\tA_ref = Array(A_ref);\r\n\t\t\t\t@require !all(A .== A_ref)\r\n\t\t\t\tupdate_halo!(A);\r\n\t\t\t\t@test all(A[2:end-1,:] .== A_ref[2:end-1,:])\r\n\t\t\t\t@test all(A[[1, end],:] .== 0.0)\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\r\n\t\t\t@testset \"3D (no halo in one dim)\" begin\r\n\t\t\t\tinit_global_grid(nx, ny, nz, periodx=1, periody=1, periodz=1, quiet=true, init_MPI=false);\r\n\t\t\t\tA = zeros(nx+2,ny-1,nz+1);\r\n\t\t\t\tA .= [z_g(iz,dz,A)*1e2 + y_g(iy,dy,A)*1e1 + x_g(ix,dx,A) for ix=1:size(A,1), iy=1:size(A,2), iz=1:size(A,3)];\r\n\t\t\t\tA_ref = copy(A);\r\n\t\t\t\tA[[1, end], :, :] .= 0.0;\r\n\t\t\t\tA[ :,[1, end], :] .= 0.0;\r\n\t\t\t\tA[ :, :,[1, end]] .= 0.0;\r\n\t\t\t\tA = Array(A);\r\n\t\t\t\tA_ref = Array(A_ref);\r\n\t\t\t\t@require !all(A .== A_ref)\r\n\t\t\t\tupdate_halo!(A);\r\n\t\t\t\t@test all(A[:,2:end-1,:] .== A_ref[:,2:end-1,:])\r\n\t\t\t\t@test all(A[:,[1, end],:] .== 0.0)\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\n\t\t\t@testset \"3D (Complex)\" begin\n\t\t\t\tinit_global_grid(nx, ny, nz, periodx=1, periody=1, periodz=1, quiet=true, init_MPI=false);\n\t\t\t\tVz = zeros(ComplexF16,nx,ny,nz+1);\n\t\t\t\tVz .= [(1+im)*(z_g(iz,dz,Vz)*1e2 + y_g(iy,dy,Vz)*1e1 + x_g(ix,dx,Vz)) for ix=1:size(Vz,1), iy=1:size(Vz,2), iz=1:size(Vz,3)];\n\t\t\t\tVz_ref = copy(Vz);\n\t\t\t\tVz[[1, end], :, :] .= 0.0;\n\t\t\t\tVz[ :,[1, end], :] .= 0.0;\n\t\t\t\tVz[ :, :,[1, end]] .= 0.0;\n\t\t\t\tVz = Array(Vz);\n\t\t\t\tVz_ref = Array(Vz_ref);\n\t\t\t\t@require !all(Vz .== Vz_ref)\n\t\t\t\tupdate_halo!(Vz);\n\t\t\t\t@test all(Vz .== Vz_ref)\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\n\t\t\tend;\r\n\t\t\t# @testset \"3D (changing datatype)\" begin\r\n\t\t\t# \tinit_global_grid(nx, ny, nz, periodx=1, periody=1, periodz=1, quiet=true, init_MPI=false);\r\n\t\t\t# \tVz = zeros(nx,ny,nz+1);\r\n\t\t\t# \tVz .= [z_g(iz,dz,Vz)*1e2 + y_g(iy,dy,Vz)*1e1 + x_g(ix,dx,Vz) for ix=1:size(Vz,1), iy=1:size(Vz,2), iz=1:size(Vz,3)];\r\n\t\t\t# \tVz_ref = copy(Vz);\r\n\t\t\t# \tVx = zeros(Float32,nx+1,ny,nz);\r\n\t\t\t# \tVx .= [z_g(iz,dz,Vx)*1e2 + y_g(iy,dy,Vx)*1e1 + x_g(ix,dx,Vx) for ix=1:size(Vx,1), iy=1:size(Vx,2), iz=1:size(Vx,3)];\r\n\t\t\t# \tVx_ref = copy(Vx);\r\n\t\t\t# \tVz[[1, end], :, :] .= 0.0;\r\n\t\t\t# \tVz[ :,[1, end], :] .= 0.0;\r\n\t\t\t# \tVz[ :, :,[1, end]] .= 0.0;\r\n\t\t\t# \tVz = Array(Vz);\r\n\t\t\t# \tVz_ref = Array(Vz_ref);\r\n\t\t\t# \t@require !all(Vz .== Vz_ref)\r\n\t\t\t# \tupdate_halo!(Vz);\r\n\t\t\t# \t@test all(Vz .== Vz_ref)\r\n\t\t\t# \tVx[[1, end], :, :] .= 0.0;\r\n\t\t\t# \tVx[ :,[1, end], :] .= 0.0;\r\n\t\t\t# \tVx[ :, :,[1, end]] .= 0.0;\r\n\t\t\t# \tVx = Array(Vx);\r\n\t\t\t# \tVx_ref = Array(Vx_ref);\r\n\t\t\t# \t@require !all(Vx .== Vx_ref)\r\n\t\t\t# \tupdate_halo!(Vx);\r\n\t\t\t# \t@test all(Vx .== Vx_ref)\r\n\t\t\t# \t#TODO: added for GPU - quick fix:\r\n\t\t\t# \tVz = zeros(nx,ny,nz+1);\r\n\t\t\t# \tVz .= [z_g(iz,dz,Vz)*1e2 + y_g(iy,dy,Vz)*1e1 + x_g(ix,dx,Vz) for ix=1:size(Vz,1), iy=1:size(Vz,2), iz=1:size(Vz,3)];\r\n\t\t\t# \tVz_ref = copy(Vz);\r\n\t\t\t# \tVz[[1, end], :, :] .= 0.0;\r\n\t\t\t# \tVz[ :,[1, end], :] .= 0.0;\r\n\t\t\t# \tVz[ :, :,[1, end]] .= 0.0;\r\n\t\t\t# \tVz = Array(Vz);\r\n\t\t\t# \tVz_ref = Array(Vz_ref);\r\n\t\t\t# \t@require !all(Vz .== Vz_ref)\r\n\t\t\t# \tupdate_halo!(Vz);\r\n\t\t\t# \t@test all(Vz .== Vz_ref)\r\n\t\t\t# \tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\t# end;\n\t\t\t# @testset \"3D (changing datatype) (Complex)\" begin\n\t\t\t# \tinit_global_grid(nx, ny, nz, periodx=1, periody=1, periodz=1, quiet=true, init_MPI=false);\n\t\t\t# \tVz = zeros(nx,ny,nz+1);\n\t\t\t# \tVz .= [z_g(iz,dz,Vz)*1e2 + y_g(iy,dy,Vz)*1e1 + x_g(ix,dx,Vz) for ix=1:size(Vz,1), iy=1:size(Vz,2), iz=1:size(Vz,3)];\n\t\t\t# \tVz_ref = copy(Vz);\n\t\t\t# \tVx = zeros(ComplexF64,nx+1,ny,nz);\n\t\t\t# \tVx .= [(1+im)*(z_g(iz,dz,Vx)*1e2 + y_g(iy,dy,Vx)*1e1 + x_g(ix,dx,Vx)) for ix=1:size(Vx,1), iy=1:size(Vx,2), iz=1:size(Vx,3)];\n\t\t\t# \tVx_ref = copy(Vx);\n\t\t\t# \tVz[[1, end], :, :] .= 0.0;\n\t\t\t# \tVz[ :,[1, end], :] .= 0.0;\n\t\t\t# \tVz[ :, :,[1, end]] .= 0.0;\n\t\t\t# \tVz = Array(Vz);\n\t\t\t# \tVz_ref = Array(Vz_ref);\n\t\t\t# \t@require !all(Vz .== Vz_ref)\n\t\t\t# \tupdate_halo!(Vz);\n\t\t\t# \t@test all(Vz .== Vz_ref)\n\t\t\t# \tVx[[1, end], :, :] .= 0.0;\n\t\t\t# \tVx[ :,[1, end], :] .= 0.0;\n\t\t\t# \tVx[ :, :,[1, end]] .= 0.0;\n\t\t\t# \tVx = Array(Vx);\n\t\t\t# \tVx_ref = Array(Vx_ref);\n\t\t\t# \t@require !all(Vx .== Vx_ref)\n\t\t\t# \tupdate_halo!(Vx);\n\t\t\t# \t@test all(Vx .== Vx_ref)\n\t\t\t# \t#TODO: added for GPU - quick fix:\n\t\t\t# \tVz = zeros(nx,ny,nz+1);\n\t\t\t# \tVz .= [z_g(iz,dz,Vz)*1e2 + y_g(iy,dy,Vz)*1e1 + x_g(ix,dx,Vz) for ix=1:size(Vz,1), iy=1:size(Vz,2), iz=1:size(Vz,3)];\n\t\t\t# \tVz_ref = copy(Vz);\n\t\t\t# \tVz[[1, end], :, :] .= 0.0;\n\t\t\t# \tVz[ :,[1, end], :] .= 0.0;\n\t\t\t# \tVz[ :, :,[1, end]] .= 0.0;\n\t\t\t# \tVz = Array(Vz);\n\t\t\t# \tVz_ref = Array(Vz_ref);\n\t\t\t# \t@require !all(Vz .== Vz_ref)\n\t\t\t# \tupdate_halo!(Vz);\n\t\t\t# \t@test all(Vz .== Vz_ref)\n\t\t\t# \tfinalize_global_grid(finalize_MPI=false);\n\t\t\t# end;\r\n\t\t\t@testset \"3D (two fields simultaneously)\" begin\r\n\t\t\t\tinit_global_grid(nx, ny, nz, periodx=1, periody=1, periodz=1, quiet=true, init_MPI=false);\r\n\t\t\t\tVz = zeros(nx,ny,nz+1);\r\n\t\t\t\tVz .= [z_g(iz,dz,Vz)*1e2 + y_g(iy,dy,Vz)*1e1 + x_g(ix,dx,Vz) for ix=1:size(Vz,1), iy=1:size(Vz,2), iz=1:size(Vz,3)];\r\n\t\t\t\tVz_ref = copy(Vz);\r\n\t\t\t\tVx = zeros(nx+1,ny,nz);\r\n\t\t\t\tVx .= [z_g(iz,dz,Vx)*1e2 + y_g(iy,dy,Vx)*1e1 + x_g(ix,dx,Vx) for ix=1:size(Vx,1), iy=1:size(Vx,2), iz=1:size(Vx,3)];\r\n\t\t\t\tVx_ref = copy(Vx);\r\n\t\t\t\tVz[[1, end], :, :] .= 0.0;\r\n\t\t\t\tVz[ :,[1, end], :] .= 0.0;\r\n\t\t\t\tVz[ :, :,[1, end]] .= 0.0;\r\n\t\t\t\tVx[[1, end], :, :] .= 0.0;\r\n\t\t\t\tVx[ :,[1, end], :] .= 0.0;\r\n\t\t\t\tVx[ :, :,[1, end]] .= 0.0;\r\n\t\t\t\tVz = Array(Vz);\r\n\t\t\t\tVz_ref = Array(Vz_ref);\r\n\t\t\t\tVx = Array(Vx);\r\n\t\t\t\tVx_ref = Array(Vx_ref);\r\n\t\t\t\t@require !all(Vz .== Vz_ref)\r\n\t\t\t\t@require !all(Vx .== Vx_ref)\r\n\t\t\t\tupdate_halo!(Vz, Vx);\r\n\t\t\t\t@test all(Vz .== Vz_ref)\r\n\t\t\t\t@test all(Vx .== Vx_ref)\r\n\t\t\t\tfinalize_global_grid(finalize_MPI=false);\r\n\t\t\tend;\r\n\t\tend;\r\n\tend;\r\nend;\r\n\r\n## Test tear down\r\nMPI.Finalize()\r\n", "meta": {"hexsha": "80f01c6f8374d40aa50a728fab533a9243df7e73", "size": 46804, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/test_update_halo.jl", "max_stars_repo_name": "luraess/ImplicitGlobalGrid.jl", "max_stars_repo_head_hexsha": "fc3c5be93b1280a9286d90a03fc8ffe892f9ec72", "max_stars_repo_licenses": ["BSD-3-Clause"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "test/test_update_halo.jl", "max_issues_repo_name": "luraess/ImplicitGlobalGrid.jl", "max_issues_repo_head_hexsha": "fc3c5be93b1280a9286d90a03fc8ffe892f9ec72", "max_issues_repo_licenses": ["BSD-3-Clause"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "test/test_update_halo.jl", "max_forks_repo_name": "luraess/ImplicitGlobalGrid.jl", "max_forks_repo_head_hexsha": "fc3c5be93b1280a9286d90a03fc8ffe892f9ec72", "max_forks_repo_licenses": ["BSD-3-Clause"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 48.3512396694, "max_line_length": 269, "alphanum_fraction": 0.5304888471, "num_tokens": 17094, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.43014734858584286, "lm_q2_score": 0.1847675084034608, "lm_q1q2_score": 0.0794772538445611}}
{"text": "x = 1; y = 2\n\nif (x < y)\n println(\"x($x) is less than y($y)\")\nelseif (x > y)\n println(\"x($x) is greater than y($y)\")\nelse\n println(\"x($x) is equal to y($y)\")\nend\n\n# Leaky Variable Scope\n\nif (x < y)\n relation = \"x($x) is less than y($y)\"\nelseif (x > y)\n relation = \"x($x) is greater than y($y)\"\nelse\n relation = \"x($x) is equal to y($y)\"\nend\n\nprintln(relation)\n\n# Ternary Operator\n# - can be nested\n\nz = x == y ? \"x($x) is equal to y($y)\" : \"x($x) is not equal to y($y)\"", "meta": {"hexsha": "b121062de541ec2a83f98c5f4f7833e30ba03bcf", "size": 475, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "julia-programming/6-control-flow/conditional-evaluation.jl", "max_stars_repo_name": "cadamsmith/julia-programming", "max_stars_repo_head_hexsha": "56435144a2775f5c5e75b3eec9023983caa86d3d", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "julia-programming/6-control-flow/conditional-evaluation.jl", "max_issues_repo_name": "cadamsmith/julia-programming", "max_issues_repo_head_hexsha": "56435144a2775f5c5e75b3eec9023983caa86d3d", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "julia-programming/6-control-flow/conditional-evaluation.jl", "max_forks_repo_name": "cadamsmith/julia-programming", "max_forks_repo_head_hexsha": "56435144a2775f5c5e75b3eec9023983caa86d3d", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 18.2692307692, "max_line_length": 70, "alphanum_fraction": 0.56, "num_tokens": 183, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.3345894279828469, "lm_q2_score": 0.23651622568252118, "lm_q1q2_score": 0.07913582865977668}}
{"text": "### A Pluto.jl notebook ###\n# v0.17.7\n\nusing Markdown\nusing InteractiveUtils\n\n# \u2554\u2550\u2561 3f9f4948-74c3-11ec-2daf-51719517f805\n# Import packages\nbegin\n\t# Parse HTML\n\tusing Gumbo\n\n\t# CSS selector for parsed HTML\n\tusing Cascadia\n\n\t# Pluto Notebook utilities\n\tusing PlutoUI\n\n\t# Work with tabular data (similar to pandas in Python)\n\tusing DataFrames\n\n\t# General purpose linear algebra\n\tusing LinearAlgebra\n\n\t# Working with Dates and DateTimes\n\tusing Dates\n\n\t# Make plots for data visualization\n\tusing Plots\nend\n\n# \u2554\u2550\u2561 2398b4e5-5920-442e-980b-f75359ba73ae\nmd\"\"\"\n# Assignment 1\n\n- Name: Eric Nguyen\n- TUID: 915786865\n\"\"\"\n\n# \u2554\u2550\u2561 97d595d3-3c44-4112-a7a8-4efe4c1ba943\nmd\"\"\"\n# Problem 1\n\nThe goal is to provide better data about the top 50 solar flares recorded so far than those shown by [SpaceWeatherLive.com](https://www.spaceweatherlive.com/en/solar-activity/top-50-solar-flares).\nUse [this messy NASA data](http://cdaw.gsfc.nasa.gov/CME_list/radio/waves_type2.html) to add more features for the top 50 solar flares.\nYou need to scrape this information directly from each HTML page.\nYou can read here more about [Solar Flares](https://en.wikipedia.org/wiki/Solar_flare), [coronal mass ejections](https://www.spaceweatherlive.com/en/help/what-is-a-coronal-mass-ejection-cme), and [the solar flare alphabet soup](http://spaceweather.com/glossary/flareclasses.html).\n\"\"\"\n\n# \u2554\u2550\u2561 62ba405d-4d1a-42b7-bacc-1350bf90564e\nmd\"## Part 1: Data scraping and preparation\"\n\n# \u2554\u2550\u2561 d6c88a68-c221-417b-bb78-06938fe97407\nmd\"\"\"\n### Task 1: Scrape your competitor's data\n\nScrape data for the top 50 solar flares shown in [SpaceWeatherLive.com](https://www.spaceweatherlive.com/en/solar-activity/top-50-solar-flares).\n\"\"\"\n\n# \u2554\u2550\u2561 151282f9-0a35-435c-9cba-e7736efccb3f\nswldf() = let\n\t# Make request to website for HTML data using cookies to gain authorization for web scraping\n\tdata = read(`curl 'https://www.spaceweatherlive.com/en/solar-activity/top-50-solar-flares.html' \\\n -H 'authority: www.spaceweatherlive.com' \\\n -H 'cache-control: max-age=0' \\\n -H 'sec-ch-ua: \" Not;A Brand\";v=\"99\", \"Google Chrome\";v=\"97\", \"Chromium\";v=\"97\"' \\\n -H 'sec-ch-ua-mobile: ?0' \\\n -H 'sec-ch-ua-platform: \"macOS\"' \\\n -H 'upgrade-insecure-requests: 1' \\\n -H 'user-agent: Mozilla/5.0 (Macintosh; Intel Mac OS X 10_15_7) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/97.0.4692.71 Safari/537.36' \\\n -H 'accept: text/html,application/xhtml+xml,application/xml;q=0.9,image/avif,image/webp,image/apng,*/*;q=0.8,application/signed-exchange;v=b3;q=0.9' \\\n -H 'sec-fetch-site: same-origin' \\\n -H 'sec-fetch-mode: navigate' \\\n -H 'sec-fetch-user: ?1' \\\n -H 'sec-fetch-dest: document' \\\n -H 'referer: https://www.spaceweatherlive.com/en/solar-activity/top-50-solar-flares.html' \\\n -H 'accept-language: en-US,en;q=0.9' \\\n -H 'cookie: CookieScriptConsent={\"action\":\"accept\",\"categories\":\"[\\\"targeting\\\",\\\"performance\\\"]\",\"CMP\":\"CPSyPX3PSyPX3F2ADBENBdCsAP_AAH_AAAAAH6Nf_X__bX9j-_59__t0eY1f9_7_v-wzjhfdt-8N2P_X_L0X42E7PF36pq4KuR4Eu3LBIQNlHOHUTUmw6okVrTPsak2Mr7NKJ7LEmnMZe2dYGHtfn91TuZKY7_78_9fz3_-v_v___9f3r-3_3__59X---_e_V399zLv9_____9nA_YAkw1L4ALsSxwZJo0qhRAhCsJDoBQAUUAwtE1hAwuCnZWAR6ggYAITUBGBECDEFGLAIABAIAkIiAkAPBAIgCIBAACAFSAhAARMAgsALAwCAAUA0LECKAIQJCDI4KjlMCAiRaKCeysASi72NMIQyiwAoFH9FRgIlCCBYGQkLBzGAAAAA.YAAAAAAAAAAA\",\"key\":\"cd200ab5-5ddb-4df2-8057-ad9de673f1e6\"}; _gid=GA1.2.1779020148.1642128786; SPSI=e3d46e0ee55ab21c983b674967abeb21; SPSE=QtZcE7RE5wJ43mJgik/6IiEWjaVvJvUBWREba0C084quUcv8c2WF9rgKyHChd+zcp+4hVlkLs6x5OJPlBXX6HA==; spcsrf=2db0574ff9f3e8b9fbc45b8804c9f061; sp_lit=/BKwhMbFX5iEuAmhsZlBwA==; UTGv2=h47aacd40fcd612bc33976e2edcd25116129; adOtr=643eeedS055; PRLST=ua/Ry/Ai; _ga_25HHLX0QL3=GS1.1.1642218626.6.1.1642219616.0; _ga=GA1.2.737209106.1642128785' \\\n --compressed`, String)\n\n\t# Parse HTML data using CSS selectors\n\tpage = parsehtml(data)\n\ttable = first(eachmatch(sel\".table-responsive-md > table\", page.root))\n\tthead = first(eachmatch(sel\"thead\", table))\n\ttbody = first(eachmatch(sel\"tbody\", table))\n\n\t# Use reshape() to convert list of to 1x8-Matrix\n\t# Vertically concatenate each Matrix to form 50x8 Matrix\n\tdatarows = reduce(vcat, [reshape([Gumbo.text(td.children[1]) for td \u2208 tr.children], (1, :)) for tr \u2208 eachmatch(sel\"tr\", tbody)])\n\n\t# Put rows of data into DataFrame\n\tdf = DataFrame(datarows, [:rank, :x_class, :date, :region, :start_time, :max_time, :end_time, :movie])\nend\n\n# \u2554\u2550\u2561 8f922beb-d685-4790-a08b-0d24810c417d\nmd\"#### DataFrame for top 50 solar flares from SpaceWeatherLive.com\"\n\n# \u2554\u2550\u2561 c93284d7-0cb2-4991-a44e-a30e53c29a93\nswldf()\n\n# \u2554\u2550\u2561 99a21f98-bd1a-4bc4-a619-668e824bb788\nmd\"### Task 2: Tidy the top 50 solar flare data\"\n\n# \u2554\u2550\u2561 6fa4452d-ddcd-4df7-996a-ba75ae21ba6e\ntidy_swldf() = let\n\tdf = swldf()\n\n\t# Remove '+' from x_class\n\tdf[!,:x_class][last.(df[!,:x_class]) .== '+'] = chop.(df[!,:x_class][last.(df[!,:x_class]) .== '+'])\n\n\t# Drop last column of DataFrame\n\tselect!(df, Not(:movie))\n\n\t# Set missing values to missing (if they exist)\n\tallowmissing!(df, :region)\n\tdf[!,:region][df[!,:region] .== \"-\"] .= missing\n\t\n\t# Convert rank to integer\n\tdf.rank = parse.(Int, df.rank)\n\n\t# Convert dates and times to DateTime\n\tdf[!,:date] = Date.(df[:, :date], \"yyyy/mm/dd\")\n\tdf[!,:start_time] = DateTime.(df[:, :date], Time.(df[:, :start_time]))\n\tdf[!,:max_time] = DateTime.(df[:, :date], Time.(df[:, :max_time]))\n\tdf[!,:end_time] = DateTime.(df[:, :date], Time.(df[:, :end_time]))\n\n\t# Drop date columns\n\tselect!(df, Not(:date))\n\n\t# Rename columns\n\trename!(df, :start_time => :start_datetime)\n\trename!(df, :max_time => :max_datetime)\n\trename!(df, :end_time => :end_datetime)\n\n\t# Reorder columns\n\tdf[!, [:rank, :x_class, :start_datetime, :max_datetime, :end_datetime, :region]]\nend\n\n# \u2554\u2550\u2561 e2c62cf7-9c2e-48d7-97a3-0990328e6ec5\nmd\"#### Tidied DataFrame for top 50 solar flares from SpaceWeatherLive.com\"\n\n# \u2554\u2550\u2561 ff744877-012a-4edf-bdf9-a9e29744cff3\ntidy_swldf()\n\n# \u2554\u2550\u2561 d5bf8c5a-229e-4a2f-bbff-6d1ef6fcf9c6\nmd\"\"\"\n### Task 3: Scrape the NASA data\n\nScrape the [NASA data](http://cdaw.gsfc.nasa.gov/CME_list/radio/waves_type2.html) to get additional features about the solar flares using the [table format description](http://cdaw.gsfc.nasa.gov/CME_list/radio/waves_type2_description.htm).\n\"\"\"\n\n# \u2554\u2550\u2561 467168a3-33d0-46e4-acdc-3aea85fd2ff5\nnasadf() = let\n\t# Make request to website for HTML data\n\tdata = read(`curl https://cdaw.gsfc.nasa.gov/CME_list/radio/waves_type2.html`, String)\n\n\t# Parse HTML data using CSS selectors\n\tpage = parsehtml(data)\n\tpre = Gumbo.text(first(eachmatch(sel\"pre\", page.root)))\n\tdata = reduce(vcat, [reshape(split(row)[1:14], (1,:)) for row \u2208 split(pre, '\\n')[12:end-1]])\n\n\t# Put data into DataFrame\n\tdf = DataFrame(data, [:start_date, :start_time, :end_date, :end_time, :start_frequency, :end_frequency, :flare_location, :flare_region, :flare_classification, :cme_date, :cme_time, :cme_angle, :cme_width, :cme_speed])\nend\n\n# \u2554\u2550\u2561 c7e4ee98-8c09-4eb5-aa9d-df7a8e2b51d7\nmd\"#### DataFrame of solar flare data from NASA\"\n\n# \u2554\u2550\u2561 9f986c37-8f68-4348-a587-1cc18f924fee\nnasadf()\n\n# \u2554\u2550\u2561 fd3f32e1-29b6-4766-b1a4-60ce09c9e8f9\nmd\"### Task 4: Tidy the NASA table\"\n\n# \u2554\u2550\u2561 cc46809e-3436-4748-a29a-2a82cc197d66\ntidy_nasadf() = let\n\tdf = nasadf()\n\tallowmissing!(df)\n\n\t# Replace missing data with 'missing'\n\tdf[!,:start_frequency][df[!,:start_frequency] .== \"????\"] .= missing\n\tdf[!,:end_frequency][df[!,:end_frequency] .== \"????\"] .= missing\n\tdf[!,:flare_location][df[!,:flare_location] .== \"------\"] .= missing\n\tdf[!,:flare_region][df[!,:flare_region] .== \"-----\"] .= missing\n\tdf[!,:flare_classification][df[!,:flare_classification] .== \"----\"] .= missing\n\tdf[!,:cme_date][df[!,:cme_date] .== \"--/--\"] .= missing\n\tdf[!,:cme_time][df[!,:cme_time] .== \"--:--\"] .= missing\n\tdf[!,:cme_angle][df[!,:cme_angle] .== \"----\"] .= missing\n\tdf[!,:cme_width][df[!,:cme_width] .== \"----\" .|| df[!,:cme_width] .== \"---\"] .= missing\n\tdf[!,:cme_speed][df[!,:cme_speed] .== \"----\"] .= missing\n\n\t# Create is_halo column and set Halo angles to missing\n\tdf.is_halo = df[!,:cme_angle] .== \"Halo\"\n\tdf[!,:cme_angle][coalesce.(df.is_halo, false)] .= missing\n\n\t# Indicate lower bound and strip non-numeric parts of width\n\tdf.width_lower_bound = occursin.(\">\", coalesce.(df[!,:cme_width], \"\"))\n\tdf[!,:cme_width][df.width_lower_bound] = replace.(df[!,:cme_width][df.width_lower_bound], \">\" => \"\")\n\tdf[!,:cme_width] = replace.(coalesce.(df[!,:cme_width], \"-1\"), \"h\" => \"\")\n\t\n\t# Convert dates and times to DateTime\n\tdf[!,:start_date] = Date.(df[:, :start_date], \"yyyy/mm/dd\")\n\tdf[!,:end_date] = Date.(string.(year.(df[!,:start_date])) .* \"/\" .* df[:, :end_date], \"yyyy/mm/dd\")\n\t\n\tdf[!,:start_time] = DateTime.(df[:, :start_date], Time.(df[:, :start_time]))\n\tdf[!,:end_time][df[!,:end_time] .== \"24:00\"] .= \"23:59\" # Fix invalid time format\n\tdf[!,:end_time] = DateTime.(df[:, :end_date], Time.(df[:, :end_time]))\n\t\n\tdf[!,:cme_date] = Date.(coalesce.(string.(year.(df[!,:start_date])) .* \"/\" .* df[!,:cme_date], \"0\"), \"yyyy/mm/dd\")\n\tallowmissing!(df, :cme_date)\n\tdf[!,:cme_date][year.(df[!,:cme_date]) .== 0] .= missing\n\t\n\tdf[!,:cme_time] = DateTime.(coalesce.(df[:, :cme_date], Date(0)), Time.(coalesce.(df[:, :cme_time], \"0\")))\n\tallowmissing!(df, :cme_time)\n\tdf[!,:cme_time][year.(df[!,:cme_time]) .== 0] .= missing\n\n\t# Parse int columns\n\tfor col \u2208 [:start_frequency, :end_frequency, :cme_angle, :cme_width, :cme_speed]\n\t\tdf[!,col] .= parse.(Int, coalesce.(df[!,col], \"-1\"))\n\t\tallowmissing!(df, col)\n\t\tdf[!,col][df[!,col] .== -1] .= missing\n\tend\n\t\n\t# Drop date columns\n\tselect!(df, Not(:start_date))\n\tselect!(df, Not(:end_date))\n\tselect!(df, Not(:cme_date))\n\t\n\t# Rename columns\n\trename!(df, :start_time => :start_datetime)\n\trename!(df, :end_time => :end_datetime)\n\trename!(df, :cme_time => :cme_datetime)\n\trename!(df, :cme_angle => :cpa)\n\trename!(df, :cme_width => :width)\n\trename!(df, :cme_speed => :speed)\n\n\tdf\nend\n\n# \u2554\u2550\u2561 8710210a-2dcf-4d63-a274-4ab44445a17f\nmd\"#### Tidied DataFrame of solar flare data from NASA\"\n\n# \u2554\u2550\u2561 12328375-5e07-49c1-83da-bdae88fcc8d4\ntidy_nasadf()\n\n# \u2554\u2550\u2561 1e97263b-5fa9-497b-b610-7f3ad30b5520\nmd\"## Part 2: Analysis\"\n\n# \u2554\u2550\u2561 238002f4-c32b-41bc-8342-ac238828f74a\nmd\"### Task 5: Replication\"\n\n# \u2554\u2550\u2561 3156a7d1-d1a3-414e-8794-c895ed08f3b0\nreplicate_nasadf() = let\n\tdf = tidy_nasadf()\n\n\t# Sort DataFrame by flare classification in descending order\n\tdf = sort(subset(dropmissing(df, :flare_classification), :flare_classification => x -> occursin.('X', x)), order(:flare_classification, by = x -> parse(Float64, x[2:end])), rev=true)\n\t\n\t# Rename columns\n\trename!(df, :cme_datetime => :max_datetime)\n\trename!(df, :flare_classification => :x_class)\n\trename!(df, :flare_region => :region)\n\n\t# Truncate regions\n\tdf[!,:region][length.(coalesce.(df[!,:region], \"\")) .> 4] .= chop.(df[!,:region][length.(coalesce.(df[!,:region], \"\")) .> 4], head = 1, tail = 0)\n\n\t# Remove trailing dots from x_class\n\tdf[!,:x_class][last.(df[!,:x_class]) .== '.'] .= chop.(df[!,:x_class][last.(df[!,:x_class]) .== '.'])\n\n\t# Reorder the columns\n\tdf[!, [:x_class, :start_datetime, :max_datetime, :end_datetime, :region]]\nend\n\n# \u2554\u2550\u2561 b35a3d34-628e-470d-96b2-076a1e3d5ba5\nmd\"#### DataFrame replication for top 50 solar flares using NASA data\"\n\n# \u2554\u2550\u2561 6048f590-30e0-41ea-bcb9-a283d700668b\nreplicate_nasadf()[1:50,:]\n\n# \u2554\u2550\u2561 ec522eda-7c21-4691-863e-4b9adf1414b3\nmd\"\"\"\n**Task 5 result.** Using the NASA table, I cannot produce an exact replicate due to discrepancies between the tables, e.g., some of the dates/times are different between the tables, and there are some records in the tables that don't exist in the other table.\n\"\"\"\n\n# \u2554\u2550\u2561 0509b052-02c7-4cf5-9c1f-fa5cf584360c\nmd\"### Task 6: Integration\"\n\n# \u2554\u2550\u2561 9a30c019-fc18-492c-b147-14c234c8ff12\nintegrated_nasadf() = let\n\tdf = replicate_nasadf()\n\tdf_t50 = tidy_swldf()\n\n\t# Find most similar row from SpaceWeatherLive.com\n\tsimilarities = [\n\t\tfindmin([\n\t\t\tsqrt.(\n\t\t\t\t# Compare x_class\n\t\t\t\t(parse.(Float64, chop.(row[:x_class], head = 1, tail = 0)) - parse.(Float64, chop.(t50row[:x_class], head = 1, tail = 0))).^2 +\n\n\t\t\t\t# Compare start_datetime\n\t\t\t\tDates.value.(Dates.Minute.(row[:start_datetime] - t50row[:start_datetime])).^2 +\n\n\t\t\t\t# Compare max_datetime\n\t\t\t\tDates.value.(Dates.Minute.(coalesce.(row[:max_datetime], DateTime(0)) - t50row[:max_datetime])).^2 .*\n\n\t\t\t\t# Ignore missing max_datetimes\n\t\t\t\tDates.value.(Dates.Minute.(coalesce.(row[:max_datetime], DateTime(0)))) +\n\n\t\t\t\t# Compare end_datetime\n\t\t\t\tDates.value.(Dates.Minute.(row[:end_datetime] - t50row[:end_datetime])).^2 +\n\n\t\t\t\t# Compare region\n\t\t\t\t(parse.(Int, coalesce.(row[:region], \"0\")) - parse.(Int, t50row[:region])).^2 .*\n\n\t\t\t\t# Ignore missing regions\n\t\t\t\tparse.(Int, coalesce.(row[:region], \"0\"))\n\t\t\t) for t50row \u2208 eachrow(df_t50)]) for row \u2208 eachrow(df)\n\t]\n\n\t# Add rank column\n\tinsertcols!(df, 1, :rank => [similarities[x][2] for x \u2208 1:length(similarities)])\n\tallowmissing!(df, :rank)\n\tbuf = Set()\n\tmin_xclass = minimum(parse.(Float64, chop.(df_t50[!,:x_class], head = 1, tail = 0)))\n\tfor row \u2208 eachrow(df)\n\t\tif row[:rank] \u2208 buf || parse(Float64, chop(row[:x_class], head = 1, tail = 0)) < min_xclass\n\t\t\trow[:rank] = missing\n\t\telse\n\t\t\tpush!(buf, row[:rank])\n\t\tend\n\tend\n\n\t# Remove rows with missing rank\n\tdf = df[(!ismissing).(df[!,:rank]), :]\n\t\n\t# Sort rows by rank\n\tsort(df)\nend\n\n# \u2554\u2550\u2561 074eb18b-05eb-456e-89c7-39b7b122e14e\nmd\"#### Integrated DataFrame of top 50 solar flares from NASA\"\n\n# \u2554\u2550\u2561 53a2adeb-4d89-41e5-8a38-bb0abb2466eb\nintegrated_nasadf()\n\n# \u2554\u2550\u2561 f9ac8b88-2383-4ef2-a9b9-01435654ebf8\nmd\"### Task 7: Attributes visualization\"\n\n# \u2554\u2550\u2561 e389aaa7-82c8-4569-ace4-d13e2e3fb3ec\ndataviz(feature) = let\n\tdf = tidy_nasadf()\n\tdf_t50 = integrated_nasadf()\n\tt50 = filter(row -> row[:start_datetime] \u2208 df_t50[!,:start_datetime], df)\n\tnott50 = filter(row -> row[:start_datetime] \u2209 df_t50[!,:start_datetime], df)\n\tscatter(nott50[!,:start_datetime], nott50[!,feature], label=\"Other\")\n\tscatter!(t50[!,:start_datetime], t50[!,feature], label=\"Top 50\", marker=:star, markersize=8)\n\txlabel!(\"Starting DateTime\")\nend\n\n# \u2554\u2550\u2561 c4311bd3-6cd0-4bf9-8210-8b6555d3175f\nlet\n\tdataviz(:start_frequency)\n\tylabel!(\"Starting frequencies (kHz)\")\n\ttitle!(\"Starting frequencies over time\")\nend\n\n# \u2554\u2550\u2561 8f8db2f8-bf75-4828-862c-2b361eb33692\nlet\n\tdataviz(:end_frequency)\n\tylabel!(\"Ending frequencies (kHz)\")\n\ttitle!(\"Ending frequencies over time\")\nend\n\n# \u2554\u2550\u2561 8e6d61eb-923d-47c3-9306-237529a60672\nlet\n\tdataviz(:width)\n\tylabel!(\"CME width in the sky plane (degrees)\")\n\ttitle!(\"CME width over time\")\nend\n\n# \u2554\u2550\u2561 b5eb2437-a13e-4b69-a378-e977ecf0fba7\nlet\n\tdataviz(:speed)\n\txlabel!(\"Starting DateTime\")\n\tylabel!(\"CME Speed in the sky plane (km/s)\")\n\ttitle!(\"CME Speeds over time\")\nend\n\n# \u2554\u2550\u2561 55ec5071-e9a5-4acf-9e15-9947cc9be2b5\nmd\"### Task 8: Attributes comparison\"\n\n# \u2554\u2550\u2561 046bd1e5-e6c8-46b4-bace-11fe8ef95b50\nlet\n\tdf = tidy_nasadf()\n\tdf_t50 = integrated_nasadf()\n\tt50 = filter(row -> row[:start_datetime] \u2208 df_t50[!,:start_datetime], df)\n\tnott50 = filter(row -> row[:start_datetime] \u2209 df_t50[!,:start_datetime], df)\n\tt50ratio = sum(coalesce.(t50[!,:is_halo], false)) / length(t50[!,:is_halo])\n\tnott50ratio = sum(coalesce.(nott50[!,:is_halo], false)) / length(nott50[!,:is_halo])\n\tbar([\"Top 50\", \"Other\"], [t50ratio, nott50ratio], color = [theme_palette(:auto)[1], theme_palette(:auto)[2]], legend = false)\n\tylabel!(\"Ratio of Halo CMEs vs. Number of CMEs\")\n\ttitle!(\"Comparision of ratios of Halo CMEs between the\\n top 50 solar flares and the rest of the solar flares\")\nend\n\n# \u2554\u2550\u2561 e6c5c88c-1d13-4785-bdea-6048cc905dcc\nmd\"### Task 9: Events distribution\"\n\n# \u2554\u2550\u2561 d1ffbd03-3454-4e95-8c8f-3466a0d3ecbc\nlet\n\tdf = tidy_nasadf()\n\tdf_t50 = integrated_nasadf()\n\tt50 = filter(row -> row[:start_datetime] \u2208 df_t50[!,:start_datetime], df)\n\tnott50 = filter(row -> row[:start_datetime] \u2209 df_t50[!,:start_datetime], df)\n\n\t# Plot the number of years instead of months to make it easier to visualize\n\tnumyears = length(df[!,:start_datetime][1]:Year(1):df[!,:start_datetime][end])\n\th = histogram(nott50[!,:start_datetime], label=\"Other\", nbins = numyears)\n\thistogram!(h, t50[!,:start_datetime], label=\"Top 50\", nbins = numyears)\n\tplot(h, xticks = [df[!,:start_datetime][1], df[!,:start_datetime][end\u00f72], df[!,:start_datetime][end]], xformatter = x -> Date(Dates.epochms2datetime(x)))\n\ttitle!(\"Number of solar flares per month\")\nend\n\n# \u2554\u2550\u2561 bd510480-984e-44ef-ac45-2fbfa909eb9e\nmd\"There appears to be a cluster of strong flares between the years 2001-2003 however there are no other clusters apparent in the data.\"\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000001\nPLUTO_PROJECT_TOML_CONTENTS = \"\"\"\n[deps]\nCascadia = \"54eefc05-d75b-58de-a785-1a3403f0919f\"\nDataFrames = \"a93c6f00-e57d-5684-b7b6-d8193f3e46c0\"\nDates = \"ade2ca70-3891-5945-98fb-dc099432e06a\"\nGumbo = \"708ec375-b3d6-5a57-a7ce-8257bf98657a\"\nLinearAlgebra = \"37e2e46d-f89d-539d-b4ee-838fcccc9c8e\"\nPlots = \"91a5bcdd-55d7-5caf-9e0b-520d859cae80\"\nPlutoUI = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\n\n[compat]\nCascadia = \"~1.0.1\"\nDataFrames = \"~1.3.1\"\nGumbo = \"~0.8.0\"\nPlots = \"~1.25.6\"\nPlutoUI = \"~0.7.29\"\n\"\"\"\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000002\nPLUTO_MANIFEST_TOML_CONTENTS = \"\"\"\n# This file is machine-generated - editing it directly is not advised\n\njulia_version = \"1.7.1\"\nmanifest_format = \"2.0\"\n\n[[deps.AbstractPlutoDingetjes]]\ndeps = [\"Pkg\"]\ngit-tree-sha1 = \"8eaf9f1b4921132a4cff3f36a1d9ba923b14a481\"\nuuid = \"6e696c72-6542-2067-7265-42206c756150\"\nversion = \"1.1.4\"\n\n[[deps.AbstractTrees]]\ngit-tree-sha1 = \"03e0550477d86222521d254b741d470ba17ea0b5\"\nuuid = \"1520ce14-60c1-5f80-bbc7-55ef81b5835c\"\nversion = \"0.3.4\"\n\n[[deps.Adapt]]\ndeps = [\"LinearAlgebra\"]\ngit-tree-sha1 = \"af92965fb30777147966f58acb05da51c5616b5f\"\nuuid = \"79e6a3ab-5dfb-504d-930d-738a2a938a0e\"\nversion = \"3.3.3\"\n\n[[deps.ArgTools]]\nuuid = \"0dad84c5-d112-42e6-8d28-ef12dabb789f\"\n\n[[deps.Artifacts]]\nuuid = \"56f22d72-fd6d-98f1-02f0-08ddc0907c33\"\n\n[[deps.Base64]]\nuuid = \"2a0f44e3-6c83-55bd-87e4-b1978d98bd5f\"\n\n[[deps.Bzip2_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"19a35467a82e236ff51bc17a3a44b69ef35185a2\"\nuuid = \"6e34b625-4abd-537c-b88f-471c36dfa7a0\"\nversion = \"1.0.8+0\"\n\n[[deps.Cairo_jll]]\ndeps = [\"Artifacts\", \"Bzip2_jll\", \"Fontconfig_jll\", \"FreeType2_jll\", \"Glib_jll\", \"JLLWrappers\", \"LZO_jll\", \"Libdl\", \"Pixman_jll\", \"Pkg\", \"Xorg_libXext_jll\", \"Xorg_libXrender_jll\", \"Zlib_jll\", \"libpng_jll\"]\ngit-tree-sha1 = \"4b859a208b2397a7a623a03449e4636bdb17bcf2\"\nuuid = \"83423d85-b0ee-5818-9007-b63ccbeb887a\"\nversion = \"1.16.1+1\"\n\n[[deps.Cascadia]]\ndeps = [\"AbstractTrees\", \"Gumbo\"]\ngit-tree-sha1 = \"95629728197821d21a41778d0e0a49bc2d58ab9b\"\nuuid = \"54eefc05-d75b-58de-a785-1a3403f0919f\"\nversion = \"1.0.1\"\n\n[[deps.ChainRulesCore]]\ndeps = [\"Compat\", \"LinearAlgebra\", \"SparseArrays\"]\ngit-tree-sha1 = \"926870acb6cbcf029396f2f2de030282b6bc1941\"\nuuid = \"d360d2e6-b24c-11e9-a2a3-2a2ae2dbcce4\"\nversion = \"1.11.4\"\n\n[[deps.ChangesOfVariables]]\ndeps = [\"ChainRulesCore\", \"LinearAlgebra\", \"Test\"]\ngit-tree-sha1 = \"bf98fa45a0a4cee295de98d4c1462be26345b9a1\"\nuuid = \"9e997f8a-9a97-42d5-a9f1-ce6bfc15e2c0\"\nversion = \"0.1.2\"\n\n[[deps.ColorSchemes]]\ndeps = [\"ColorTypes\", \"Colors\", \"FixedPointNumbers\", \"Random\"]\ngit-tree-sha1 = \"6b6f04f93710c71550ec7e16b650c1b9a612d0b6\"\nuuid = \"35d6a980-a343-548e-a6ea-1d62b119f2f4\"\nversion = \"3.16.0\"\n\n[[deps.ColorTypes]]\ndeps = [\"FixedPointNumbers\", \"Random\"]\ngit-tree-sha1 = \"024fe24d83e4a5bf5fc80501a314ce0d1aa35597\"\nuuid = \"3da002f7-5984-5a60-b8a6-cbb66c0b333f\"\nversion = \"0.11.0\"\n\n[[deps.Colors]]\ndeps = [\"ColorTypes\", \"FixedPointNumbers\", \"Reexport\"]\ngit-tree-sha1 = \"417b0ed7b8b838aa6ca0a87aadf1bb9eb111ce40\"\nuuid = \"5ae59095-9a9b-59fe-a467-6f913c188581\"\nversion = \"0.12.8\"\n\n[[deps.Compat]]\ndeps = [\"Base64\", \"Dates\", \"DelimitedFiles\", \"Distributed\", \"InteractiveUtils\", \"LibGit2\", \"Libdl\", \"LinearAlgebra\", \"Markdown\", \"Mmap\", \"Pkg\", \"Printf\", \"REPL\", \"Random\", \"SHA\", \"Serialization\", \"SharedArrays\", \"Sockets\", \"SparseArrays\", \"Statistics\", \"Test\", \"UUIDs\", \"Unicode\"]\ngit-tree-sha1 = \"44c37b4636bc54afac5c574d2d02b625349d6582\"\nuuid = \"34da2185-b29b-5c13-b0c7-acf172513d20\"\nversion = \"3.41.0\"\n\n[[deps.CompilerSupportLibraries_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"e66e0078-7015-5450-92f7-15fbd957f2ae\"\n\n[[deps.Contour]]\ndeps = [\"StaticArrays\"]\ngit-tree-sha1 = \"9f02045d934dc030edad45944ea80dbd1f0ebea7\"\nuuid = \"d38c429a-6771-53c6-b99e-75d170b6e991\"\nversion = \"0.5.7\"\n\n[[deps.Crayons]]\ngit-tree-sha1 = \"b618084b49e78985ffa8422f32b9838e397b9fc2\"\nuuid = \"a8cc5b0e-0ffa-5ad4-8c14-923d3ee1735f\"\nversion = \"4.1.0\"\n\n[[deps.DataAPI]]\ngit-tree-sha1 = \"cc70b17275652eb47bc9e5f81635981f13cea5c8\"\nuuid = \"9a962f9c-6df0-11e9-0e5d-c546b8b5ee8a\"\nversion = \"1.9.0\"\n\n[[deps.DataFrames]]\ndeps = [\"Compat\", \"DataAPI\", \"Future\", \"InvertedIndices\", \"IteratorInterfaceExtensions\", \"LinearAlgebra\", \"Markdown\", \"Missings\", \"PooledArrays\", \"PrettyTables\", \"Printf\", \"REPL\", \"Reexport\", \"SortingAlgorithms\", \"Statistics\", \"TableTraits\", \"Tables\", \"Unicode\"]\ngit-tree-sha1 = \"cfdfef912b7f93e4b848e80b9befdf9e331bc05a\"\nuuid = \"a93c6f00-e57d-5684-b7b6-d8193f3e46c0\"\nversion = \"1.3.1\"\n\n[[deps.DataStructures]]\ndeps = [\"Compat\", \"InteractiveUtils\", \"OrderedCollections\"]\ngit-tree-sha1 = \"3daef5523dd2e769dad2365274f760ff5f282c7d\"\nuuid = \"864edb3b-99cc-5e75-8d2d-829cb0a9cfe8\"\nversion = \"0.18.11\"\n\n[[deps.DataValueInterfaces]]\ngit-tree-sha1 = \"bfc1187b79289637fa0ef6d4436ebdfe6905cbd6\"\nuuid = \"e2d170a0-9d28-54be-80f0-106bbe20a464\"\nversion = \"1.0.0\"\n\n[[deps.Dates]]\ndeps = [\"Printf\"]\nuuid = \"ade2ca70-3891-5945-98fb-dc099432e06a\"\n\n[[deps.DelimitedFiles]]\ndeps = [\"Mmap\"]\nuuid = \"8bb1440f-4735-579b-a4ab-409b98df4dab\"\n\n[[deps.Distributed]]\ndeps = [\"Random\", \"Serialization\", \"Sockets\"]\nuuid = \"8ba89e20-285c-5b6f-9357-94700520ee1b\"\n\n[[deps.DocStringExtensions]]\ndeps = [\"LibGit2\"]\ngit-tree-sha1 = \"b19534d1895d702889b219c382a6e18010797f0b\"\nuuid = \"ffbed154-4ef7-542d-bbb7-c09d3a79fcae\"\nversion = \"0.8.6\"\n\n[[deps.Downloads]]\ndeps = [\"ArgTools\", \"LibCURL\", \"NetworkOptions\"]\nuuid = \"f43a241f-c20a-4ad4-852c-f6b1247861c6\"\n\n[[deps.EarCut_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"3f3a2501fa7236e9b911e0f7a588c657e822bb6d\"\nuuid = \"5ae413db-bbd1-5e63-b57d-d24a61df00f5\"\nversion = \"2.2.3+0\"\n\n[[deps.Expat_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"b3bfd02e98aedfa5cf885665493c5598c350cd2f\"\nuuid = \"2e619515-83b5-522b-bb60-26c02a35a201\"\nversion = \"2.2.10+0\"\n\n[[deps.FFMPEG]]\ndeps = [\"FFMPEG_jll\"]\ngit-tree-sha1 = \"b57e3acbe22f8484b4b5ff66a7499717fe1a9cc8\"\nuuid = \"c87230d0-a227-11e9-1b43-d7ebe4e7570a\"\nversion = \"0.4.1\"\n\n[[deps.FFMPEG_jll]]\ndeps = [\"Artifacts\", \"Bzip2_jll\", \"FreeType2_jll\", \"FriBidi_jll\", \"JLLWrappers\", \"LAME_jll\", \"Libdl\", \"Ogg_jll\", \"OpenSSL_jll\", \"Opus_jll\", \"Pkg\", \"Zlib_jll\", \"libass_jll\", \"libfdk_aac_jll\", \"libvorbis_jll\", \"x264_jll\", \"x265_jll\"]\ngit-tree-sha1 = \"d8a578692e3077ac998b50c0217dfd67f21d1e5f\"\nuuid = \"b22a6f82-2f65-5046-a5b2-351ab43fb4e5\"\nversion = \"4.4.0+0\"\n\n[[deps.FixedPointNumbers]]\ndeps = [\"Statistics\"]\ngit-tree-sha1 = \"335bfdceacc84c5cdf16aadc768aa5ddfc5383cc\"\nuuid = \"53c48c17-4a7d-5ca2-90c5-79b7896eea93\"\nversion = \"0.8.4\"\n\n[[deps.Fontconfig_jll]]\ndeps = [\"Artifacts\", \"Bzip2_jll\", \"Expat_jll\", \"FreeType2_jll\", \"JLLWrappers\", \"Libdl\", \"Libuuid_jll\", \"Pkg\", \"Zlib_jll\"]\ngit-tree-sha1 = \"21efd19106a55620a188615da6d3d06cd7f6ee03\"\nuuid = \"a3f928ae-7b40-5064-980b-68af3947d34b\"\nversion = \"2.13.93+0\"\n\n[[deps.Formatting]]\ndeps = [\"Printf\"]\ngit-tree-sha1 = \"8339d61043228fdd3eb658d86c926cb282ae72a8\"\nuuid = \"59287772-0a20-5a39-b81b-1366585eb4c0\"\nversion = \"0.4.2\"\n\n[[deps.FreeType2_jll]]\ndeps = [\"Artifacts\", \"Bzip2_jll\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Zlib_jll\"]\ngit-tree-sha1 = \"87eb71354d8ec1a96d4a7636bd57a7347dde3ef9\"\nuuid = \"d7e528f0-a631-5988-bf34-fe36492bcfd7\"\nversion = \"2.10.4+0\"\n\n[[deps.FriBidi_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"aa31987c2ba8704e23c6c8ba8a4f769d5d7e4f91\"\nuuid = \"559328eb-81f9-559d-9380-de523a88c83c\"\nversion = \"1.0.10+0\"\n\n[[deps.Future]]\ndeps = [\"Random\"]\nuuid = \"9fa8497b-333b-5362-9e8d-4d0656e87820\"\n\n[[deps.GLFW_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Libglvnd_jll\", \"Pkg\", \"Xorg_libXcursor_jll\", \"Xorg_libXi_jll\", \"Xorg_libXinerama_jll\", \"Xorg_libXrandr_jll\"]\ngit-tree-sha1 = \"0c603255764a1fa0b61752d2bec14cfbd18f7fe8\"\nuuid = \"0656b61e-2033-5cc2-a64a-77c0f6c09b89\"\nversion = \"3.3.5+1\"\n\n[[deps.GR]]\ndeps = [\"Base64\", \"DelimitedFiles\", \"GR_jll\", \"HTTP\", \"JSON\", \"Libdl\", \"LinearAlgebra\", \"Pkg\", \"Printf\", \"Random\", \"RelocatableFolders\", \"Serialization\", \"Sockets\", \"Test\", \"UUIDs\"]\ngit-tree-sha1 = \"4a740db447aae0fbeb3ee730de1afbb14ac798a1\"\nuuid = \"28b8d3ca-fb5f-59d9-8090-bfdbd6d07a71\"\nversion = \"0.63.1\"\n\n[[deps.GR_jll]]\ndeps = [\"Artifacts\", \"Bzip2_jll\", \"Cairo_jll\", \"FFMPEG_jll\", \"Fontconfig_jll\", \"GLFW_jll\", \"JLLWrappers\", \"JpegTurbo_jll\", \"Libdl\", \"Libtiff_jll\", \"Pixman_jll\", \"Pkg\", \"Qt5Base_jll\", \"Zlib_jll\", \"libpng_jll\"]\ngit-tree-sha1 = \"aa22e1ee9e722f1da183eb33370df4c1aeb6c2cd\"\nuuid = \"d2c73de3-f751-5644-a686-071e5b155ba9\"\nversion = \"0.63.1+0\"\n\n[[deps.GeometryBasics]]\ndeps = [\"EarCut_jll\", \"IterTools\", \"LinearAlgebra\", \"StaticArrays\", \"StructArrays\", \"Tables\"]\ngit-tree-sha1 = \"58bcdf5ebc057b085e58d95c138725628dd7453c\"\nuuid = \"5c1252a2-5f33-56bf-86c9-59e7332b4326\"\nversion = \"0.4.1\"\n\n[[deps.Gettext_jll]]\ndeps = [\"Artifacts\", \"CompilerSupportLibraries_jll\", \"JLLWrappers\", \"Libdl\", \"Libiconv_jll\", \"Pkg\", \"XML2_jll\"]\ngit-tree-sha1 = \"9b02998aba7bf074d14de89f9d37ca24a1a0b046\"\nuuid = \"78b55507-aeef-58d4-861c-77aaff3498b1\"\nversion = \"0.21.0+0\"\n\n[[deps.Glib_jll]]\ndeps = [\"Artifacts\", \"Gettext_jll\", \"JLLWrappers\", \"Libdl\", \"Libffi_jll\", \"Libiconv_jll\", \"Libmount_jll\", \"PCRE_jll\", \"Pkg\", \"Zlib_jll\"]\ngit-tree-sha1 = \"a32d672ac2c967f3deb8a81d828afc739c838a06\"\nuuid = \"7746bdde-850d-59dc-9ae8-88ece973131d\"\nversion = \"2.68.3+2\"\n\n[[deps.Graphite2_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"344bf40dcab1073aca04aa0df4fb092f920e4011\"\nuuid = \"3b182d85-2403-5c21-9c21-1e1f0cc25472\"\nversion = \"1.3.14+0\"\n\n[[deps.Grisu]]\ngit-tree-sha1 = \"53bb909d1151e57e2484c3d1b53e19552b887fb2\"\nuuid = \"42e2da0e-8278-4e71-bc24-59509adca0fe\"\nversion = \"1.0.2\"\n\n[[deps.Gumbo]]\ndeps = [\"AbstractTrees\", \"Gumbo_jll\", \"Libdl\"]\ngit-tree-sha1 = \"e711d08d896018037d6ff0ad4ebe675ca67119d4\"\nuuid = \"708ec375-b3d6-5a57-a7ce-8257bf98657a\"\nversion = \"0.8.0\"\n\n[[deps.Gumbo_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"29070dee9df18d9565276d68a596854b1764aa38\"\nuuid = \"528830af-5a63-567c-a44a-034ed33b8444\"\nversion = \"0.10.2+0\"\n\n[[deps.HTTP]]\ndeps = [\"Base64\", \"Dates\", \"IniFile\", \"Logging\", \"MbedTLS\", \"NetworkOptions\", \"Sockets\", \"URIs\"]\ngit-tree-sha1 = \"0fa77022fe4b511826b39c894c90daf5fce3334a\"\nuuid = \"cd3eb016-35fb-5094-929b-558a96fad6f3\"\nversion = \"0.9.17\"\n\n[[deps.HarfBuzz_jll]]\ndeps = [\"Artifacts\", \"Cairo_jll\", \"Fontconfig_jll\", \"FreeType2_jll\", \"Glib_jll\", \"Graphite2_jll\", \"JLLWrappers\", \"Libdl\", \"Libffi_jll\", \"Pkg\"]\ngit-tree-sha1 = \"129acf094d168394e80ee1dc4bc06ec835e510a3\"\nuuid = \"2e76f6c2-a576-52d4-95c1-20adfe4de566\"\nversion = \"2.8.1+1\"\n\n[[deps.Hyperscript]]\ndeps = [\"Test\"]\ngit-tree-sha1 = \"8d511d5b81240fc8e6802386302675bdf47737b9\"\nuuid = \"47d2ed2b-36de-50cf-bf87-49c2cf4b8b91\"\nversion = \"0.0.4\"\n\n[[deps.HypertextLiteral]]\ngit-tree-sha1 = \"2b078b5a615c6c0396c77810d92ee8c6f470d238\"\nuuid = \"ac1192a8-f4b3-4bfe-ba22-af5b92cd3ab2\"\nversion = \"0.9.3\"\n\n[[deps.IOCapture]]\ndeps = [\"Logging\", \"Random\"]\ngit-tree-sha1 = \"f7be53659ab06ddc986428d3a9dcc95f6fa6705a\"\nuuid = \"b5f81e59-6552-4d32-b1f0-c071b021bf89\"\nversion = \"0.2.2\"\n\n[[deps.IniFile]]\ndeps = [\"Test\"]\ngit-tree-sha1 = \"098e4d2c533924c921f9f9847274f2ad89e018b8\"\nuuid = \"83e8ac13-25f8-5344-8a64-a9f2b223428f\"\nversion = \"0.5.0\"\n\n[[deps.InteractiveUtils]]\ndeps = [\"Markdown\"]\nuuid = \"b77e0a4c-d291-57a0-90e8-8db25a27a240\"\n\n[[deps.InverseFunctions]]\ndeps = [\"Test\"]\ngit-tree-sha1 = \"a7254c0acd8e62f1ac75ad24d5db43f5f19f3c65\"\nuuid = \"3587e190-3f89-42d0-90ee-14403ec27112\"\nversion = \"0.1.2\"\n\n[[deps.InvertedIndices]]\ngit-tree-sha1 = \"bee5f1ef5bf65df56bdd2e40447590b272a5471f\"\nuuid = \"41ab1584-1d38-5bbf-9106-f11c6c58b48f\"\nversion = \"1.1.0\"\n\n[[deps.IrrationalConstants]]\ngit-tree-sha1 = \"7fd44fd4ff43fc60815f8e764c0f352b83c49151\"\nuuid = \"92d709cd-6900-40b7-9082-c6be49f344b6\"\nversion = \"0.1.1\"\n\n[[deps.IterTools]]\ngit-tree-sha1 = \"fa6287a4469f5e048d763df38279ee729fbd44e5\"\nuuid = \"c8e1da08-722c-5040-9ed9-7db0dc04731e\"\nversion = \"1.4.0\"\n\n[[deps.IteratorInterfaceExtensions]]\ngit-tree-sha1 = \"a3f24677c21f5bbe9d2a714f95dcd58337fb2856\"\nuuid = \"82899510-4779-5014-852e-03e436cf321d\"\nversion = \"1.0.0\"\n\n[[deps.JLLWrappers]]\ndeps = [\"Preferences\"]\ngit-tree-sha1 = \"22df5b96feef82434b07327e2d3c770a9b21e023\"\nuuid = \"692b3bcd-3c85-4b1f-b108-f13ce0eb3210\"\nversion = \"1.4.0\"\n\n[[deps.JSON]]\ndeps = [\"Dates\", \"Mmap\", \"Parsers\", \"Unicode\"]\ngit-tree-sha1 = \"8076680b162ada2a031f707ac7b4953e30667a37\"\nuuid = \"682c06a0-de6a-54ab-a142-c8b1cf79cde6\"\nversion = \"0.21.2\"\n\n[[deps.JpegTurbo_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"d735490ac75c5cb9f1b00d8b5509c11984dc6943\"\nuuid = \"aacddb02-875f-59d6-b918-886e6ef4fbf8\"\nversion = \"2.1.0+0\"\n\n[[deps.LAME_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"f6250b16881adf048549549fba48b1161acdac8c\"\nuuid = \"c1c5ebd0-6772-5130-a774-d5fcae4a789d\"\nversion = \"3.100.1+0\"\n\n[[deps.LZO_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"e5b909bcf985c5e2605737d2ce278ed791b89be6\"\nuuid = \"dd4b983a-f0e5-5f8d-a1b7-129d4a5fb1ac\"\nversion = \"2.10.1+0\"\n\n[[deps.LaTeXStrings]]\ngit-tree-sha1 = \"f2355693d6778a178ade15952b7ac47a4ff97996\"\nuuid = \"b964fa9f-0449-5b57-a5c2-d3ea65f4040f\"\nversion = \"1.3.0\"\n\n[[deps.Latexify]]\ndeps = [\"Formatting\", \"InteractiveUtils\", \"LaTeXStrings\", \"MacroTools\", \"Markdown\", \"Printf\", \"Requires\"]\ngit-tree-sha1 = \"a8f4f279b6fa3c3c4f1adadd78a621b13a506bce\"\nuuid = \"23fbe1c1-3f47-55db-b15f-69d7ec21a316\"\nversion = \"0.15.9\"\n\n[[deps.LibCURL]]\ndeps = [\"LibCURL_jll\", \"MozillaCACerts_jll\"]\nuuid = \"b27032c2-a3e7-50c8-80cd-2d36dbcbfd21\"\n\n[[deps.LibCURL_jll]]\ndeps = [\"Artifacts\", \"LibSSH2_jll\", \"Libdl\", \"MbedTLS_jll\", \"Zlib_jll\", \"nghttp2_jll\"]\nuuid = \"deac9b47-8bc7-5906-a0fe-35ac56dc84c0\"\n\n[[deps.LibGit2]]\ndeps = [\"Base64\", \"NetworkOptions\", \"Printf\", \"SHA\"]\nuuid = \"76f85450-5226-5b5a-8eaa-529ad045b433\"\n\n[[deps.LibSSH2_jll]]\ndeps = [\"Artifacts\", \"Libdl\", \"MbedTLS_jll\"]\nuuid = \"29816b5a-b9ab-546f-933c-edad1886dfa8\"\n\n[[deps.Libdl]]\nuuid = \"8f399da3-3557-5675-b5ff-fb832c97cbdb\"\n\n[[deps.Libffi_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"0b4a5d71f3e5200a7dff793393e09dfc2d874290\"\nuuid = \"e9f186c6-92d2-5b65-8a66-fee21dc1b490\"\nversion = \"3.2.2+1\"\n\n[[deps.Libgcrypt_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Libgpg_error_jll\", \"Pkg\"]\ngit-tree-sha1 = \"64613c82a59c120435c067c2b809fc61cf5166ae\"\nuuid = \"d4300ac3-e22c-5743-9152-c294e39db1e4\"\nversion = \"1.8.7+0\"\n\n[[deps.Libglvnd_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libX11_jll\", \"Xorg_libXext_jll\"]\ngit-tree-sha1 = \"7739f837d6447403596a75d19ed01fd08d6f56bf\"\nuuid = \"7e76a0d4-f3c7-5321-8279-8d96eeed0f29\"\nversion = \"1.3.0+3\"\n\n[[deps.Libgpg_error_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"c333716e46366857753e273ce6a69ee0945a6db9\"\nuuid = \"7add5ba3-2f88-524e-9cd5-f83b8a55f7b8\"\nversion = \"1.42.0+0\"\n\n[[deps.Libiconv_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"42b62845d70a619f063a7da093d995ec8e15e778\"\nuuid = \"94ce4f54-9a6c-5748-9c1c-f9c7231a4531\"\nversion = \"1.16.1+1\"\n\n[[deps.Libmount_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"9c30530bf0effd46e15e0fdcf2b8636e78cbbd73\"\nuuid = \"4b2f31a3-9ecc-558c-b454-b3730dcb73e9\"\nversion = \"2.35.0+0\"\n\n[[deps.Libtiff_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"JpegTurbo_jll\", \"Libdl\", \"Pkg\", \"Zlib_jll\", \"Zstd_jll\"]\ngit-tree-sha1 = \"340e257aada13f95f98ee352d316c3bed37c8ab9\"\nuuid = \"89763e89-9b03-5906-acba-b20f662cd828\"\nversion = \"4.3.0+0\"\n\n[[deps.Libuuid_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"7f3efec06033682db852f8b3bc3c1d2b0a0ab066\"\nuuid = \"38a345b3-de98-5d2b-a5d3-14cd9215e700\"\nversion = \"2.36.0+0\"\n\n[[deps.LinearAlgebra]]\ndeps = [\"Libdl\", \"libblastrampoline_jll\"]\nuuid = \"37e2e46d-f89d-539d-b4ee-838fcccc9c8e\"\n\n[[deps.LogExpFunctions]]\ndeps = [\"ChainRulesCore\", \"ChangesOfVariables\", \"DocStringExtensions\", \"InverseFunctions\", \"IrrationalConstants\", \"LinearAlgebra\"]\ngit-tree-sha1 = \"e5718a00af0ab9756305a0392832c8952c7426c1\"\nuuid = \"2ab3a3ac-af41-5b50-aa03-7779005ae688\"\nversion = \"0.3.6\"\n\n[[deps.Logging]]\nuuid = \"56ddb016-857b-54e1-b83d-db4d58db5568\"\n\n[[deps.MacroTools]]\ndeps = [\"Markdown\", \"Random\"]\ngit-tree-sha1 = \"3d3e902b31198a27340d0bf00d6ac452866021cf\"\nuuid = \"1914dd2f-81c6-5fcd-8719-6d5c9610ff09\"\nversion = \"0.5.9\"\n\n[[deps.Markdown]]\ndeps = [\"Base64\"]\nuuid = \"d6f4376e-aef5-505a-96c1-9c027394607a\"\n\n[[deps.MbedTLS]]\ndeps = [\"Dates\", \"MbedTLS_jll\", \"Random\", \"Sockets\"]\ngit-tree-sha1 = \"1c38e51c3d08ef2278062ebceade0e46cefc96fe\"\nuuid = \"739be429-bea8-5141-9913-cc70e7f3736d\"\nversion = \"1.0.3\"\n\n[[deps.MbedTLS_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"c8ffd9c3-330d-5841-b78e-0817d7145fa1\"\n\n[[deps.Measures]]\ngit-tree-sha1 = \"e498ddeee6f9fdb4551ce855a46f54dbd900245f\"\nuuid = \"442fdcdd-2543-5da2-b0f3-8c86c306513e\"\nversion = \"0.3.1\"\n\n[[deps.Missings]]\ndeps = [\"DataAPI\"]\ngit-tree-sha1 = \"bf210ce90b6c9eed32d25dbcae1ebc565df2687f\"\nuuid = \"e1d29d7a-bbdc-5cf2-9ac0-f12de2c33e28\"\nversion = \"1.0.2\"\n\n[[deps.Mmap]]\nuuid = \"a63ad114-7e13-5084-954f-fe012c677804\"\n\n[[deps.MozillaCACerts_jll]]\nuuid = \"14a3606d-f60d-562e-9121-12d972cd8159\"\n\n[[deps.NaNMath]]\ngit-tree-sha1 = \"f755f36b19a5116bb580de457cda0c140153f283\"\nuuid = \"77ba4419-2d1f-58cd-9bb1-8ffee604a2e3\"\nversion = \"0.3.6\"\n\n[[deps.NetworkOptions]]\nuuid = \"ca575930-c2e3-43a9-ace4-1e988b2c1908\"\n\n[[deps.Ogg_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"887579a3eb005446d514ab7aeac5d1d027658b8f\"\nuuid = \"e7412a2a-1a6e-54c0-be00-318e2571c051\"\nversion = \"1.3.5+1\"\n\n[[deps.OpenBLAS_jll]]\ndeps = [\"Artifacts\", \"CompilerSupportLibraries_jll\", \"Libdl\"]\nuuid = \"4536629a-c528-5b80-bd46-f80d51c5b363\"\n\n[[deps.OpenSSL_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"648107615c15d4e09f7eca16307bc821c1f718d8\"\nuuid = \"458c3c95-2e84-50aa-8efc-19380b2a3a95\"\nversion = \"1.1.13+0\"\n\n[[deps.Opus_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"51a08fb14ec28da2ec7a927c4337e4332c2a4720\"\nuuid = \"91d4177d-7536-5919-b921-800302f37372\"\nversion = \"1.3.2+0\"\n\n[[deps.OrderedCollections]]\ngit-tree-sha1 = \"85f8e6578bf1f9ee0d11e7bb1b1456435479d47c\"\nuuid = \"bac558e1-5e72-5ebc-8fee-abe8a469f55d\"\nversion = \"1.4.1\"\n\n[[deps.PCRE_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"b2a7af664e098055a7529ad1a900ded962bca488\"\nuuid = \"2f80f16e-611a-54ab-bc61-aa92de5b98fc\"\nversion = \"8.44.0+0\"\n\n[[deps.Parsers]]\ndeps = [\"Dates\"]\ngit-tree-sha1 = \"d7fa6237da8004be601e19bd6666083056649918\"\nuuid = \"69de0a69-1ddd-5017-9359-2bf0b02dc9f0\"\nversion = \"2.1.3\"\n\n[[deps.Pixman_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"b4f5d02549a10e20780a24fce72bea96b6329e29\"\nuuid = \"30392449-352a-5448-841d-b1acce4e97dc\"\nversion = \"0.40.1+0\"\n\n[[deps.Pkg]]\ndeps = [\"Artifacts\", \"Dates\", \"Downloads\", \"LibGit2\", \"Libdl\", \"Logging\", \"Markdown\", \"Printf\", \"REPL\", \"Random\", \"SHA\", \"Serialization\", \"TOML\", \"Tar\", \"UUIDs\", \"p7zip_jll\"]\nuuid = \"44cfe95a-1eb2-52ea-b672-e2afdf69b78f\"\n\n[[deps.PlotThemes]]\ndeps = [\"PlotUtils\", \"Requires\", \"Statistics\"]\ngit-tree-sha1 = \"a3a964ce9dc7898193536002a6dd892b1b5a6f1d\"\nuuid = \"ccf2f8ad-2431-5c83-bf29-c5338b663b6a\"\nversion = \"2.0.1\"\n\n[[deps.PlotUtils]]\ndeps = [\"ColorSchemes\", \"Colors\", \"Dates\", \"Printf\", \"Random\", \"Reexport\", \"Statistics\"]\ngit-tree-sha1 = \"68604313ed59f0408313228ba09e79252e4b2da8\"\nuuid = \"995b91a9-d308-5afd-9ec6-746e21dbc043\"\nversion = \"1.1.2\"\n\n[[deps.Plots]]\ndeps = [\"Base64\", \"Contour\", \"Dates\", \"Downloads\", \"FFMPEG\", \"FixedPointNumbers\", \"GR\", \"GeometryBasics\", \"JSON\", \"Latexify\", \"LinearAlgebra\", \"Measures\", \"NaNMath\", \"PlotThemes\", \"PlotUtils\", \"Printf\", \"REPL\", \"Random\", \"RecipesBase\", \"RecipesPipeline\", \"Reexport\", \"Requires\", \"Scratch\", \"Showoff\", \"SparseArrays\", \"Statistics\", \"StatsBase\", \"UUIDs\", \"UnicodeFun\", \"Unzip\"]\ngit-tree-sha1 = \"db7393a80d0e5bef70f2b518990835541917a544\"\nuuid = \"91a5bcdd-55d7-5caf-9e0b-520d859cae80\"\nversion = \"1.25.6\"\n\n[[deps.PlutoUI]]\ndeps = [\"AbstractPlutoDingetjes\", \"Base64\", \"ColorTypes\", \"Dates\", \"Hyperscript\", \"HypertextLiteral\", \"IOCapture\", \"InteractiveUtils\", \"JSON\", \"Logging\", \"Markdown\", \"Random\", \"Reexport\", \"UUIDs\"]\ngit-tree-sha1 = \"7711172ace7c40dc8449b7aed9d2d6f1cf56a5bd\"\nuuid = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\nversion = \"0.7.29\"\n\n[[deps.PooledArrays]]\ndeps = [\"DataAPI\", \"Future\"]\ngit-tree-sha1 = \"db3a23166af8aebf4db5ef87ac5b00d36eb771e2\"\nuuid = \"2dfb63ee-cc39-5dd5-95bd-886bf059d720\"\nversion = \"1.4.0\"\n\n[[deps.Preferences]]\ndeps = [\"TOML\"]\ngit-tree-sha1 = \"2cf929d64681236a2e074ffafb8d568733d2e6af\"\nuuid = \"21216c6a-2e73-6563-6e65-726566657250\"\nversion = \"1.2.3\"\n\n[[deps.PrettyTables]]\ndeps = [\"Crayons\", \"Formatting\", \"Markdown\", \"Reexport\", \"Tables\"]\ngit-tree-sha1 = \"dfb54c4e414caa595a1f2ed759b160f5a3ddcba5\"\nuuid = \"08abe8d2-0d0c-5749-adfa-8a2ac140af0d\"\nversion = \"1.3.1\"\n\n[[deps.Printf]]\ndeps = [\"Unicode\"]\nuuid = \"de0858da-6303-5e67-8744-51eddeeeb8d7\"\n\n[[deps.Qt5Base_jll]]\ndeps = [\"Artifacts\", \"CompilerSupportLibraries_jll\", \"Fontconfig_jll\", \"Glib_jll\", \"JLLWrappers\", \"Libdl\", \"Libglvnd_jll\", \"OpenSSL_jll\", \"Pkg\", \"Xorg_libXext_jll\", \"Xorg_libxcb_jll\", \"Xorg_xcb_util_image_jll\", \"Xorg_xcb_util_keysyms_jll\", \"Xorg_xcb_util_renderutil_jll\", \"Xorg_xcb_util_wm_jll\", \"Zlib_jll\", \"xkbcommon_jll\"]\ngit-tree-sha1 = \"ad368663a5e20dbb8d6dc2fddeefe4dae0781ae8\"\nuuid = \"ea2cea3b-5b76-57ae-a6ef-0a8af62496e1\"\nversion = \"5.15.3+0\"\n\n[[deps.REPL]]\ndeps = [\"InteractiveUtils\", \"Markdown\", \"Sockets\", \"Unicode\"]\nuuid = \"3fa0cd96-eef1-5676-8a61-b3b8758bbffb\"\n\n[[deps.Random]]\ndeps = [\"SHA\", \"Serialization\"]\nuuid = \"9a3f8284-a2c9-5f02-9a11-845980a1fd5c\"\n\n[[deps.RecipesBase]]\ngit-tree-sha1 = \"6bf3f380ff52ce0832ddd3a2a7b9538ed1bcca7d\"\nuuid = \"3cdcf5f2-1ef4-517c-9805-6587b60abb01\"\nversion = \"1.2.1\"\n\n[[deps.RecipesPipeline]]\ndeps = [\"Dates\", \"NaNMath\", \"PlotUtils\", \"RecipesBase\"]\ngit-tree-sha1 = \"37c1631cb3cc36a535105e6d5557864c82cd8c2b\"\nuuid = \"01d81517-befc-4cb6-b9ec-a95719d0359c\"\nversion = \"0.5.0\"\n\n[[deps.Reexport]]\ngit-tree-sha1 = \"45e428421666073eab6f2da5c9d310d99bb12f9b\"\nuuid = \"189a3867-3050-52da-a836-e630ba90ab69\"\nversion = \"1.2.2\"\n\n[[deps.RelocatableFolders]]\ndeps = [\"SHA\", \"Scratch\"]\ngit-tree-sha1 = \"cdbd3b1338c72ce29d9584fdbe9e9b70eeb5adca\"\nuuid = \"05181044-ff0b-4ac5-8273-598c1e38db00\"\nversion = \"0.1.3\"\n\n[[deps.Requires]]\ndeps = [\"UUIDs\"]\ngit-tree-sha1 = \"838a3a4188e2ded87a4f9f184b4b0d78a1e91cb7\"\nuuid = \"ae029012-a4dd-5104-9daa-d747884805df\"\nversion = \"1.3.0\"\n\n[[deps.SHA]]\nuuid = \"ea8e919c-243c-51af-8825-aaa63cd721ce\"\n\n[[deps.Scratch]]\ndeps = [\"Dates\"]\ngit-tree-sha1 = \"0b4b7f1393cff97c33891da2a0bf69c6ed241fda\"\nuuid = \"6c6a2e73-6563-6170-7368-637461726353\"\nversion = \"1.1.0\"\n\n[[deps.Serialization]]\nuuid = \"9e88b42a-f829-5b0c-bbe9-9e923198166b\"\n\n[[deps.SharedArrays]]\ndeps = [\"Distributed\", \"Mmap\", \"Random\", \"Serialization\"]\nuuid = \"1a1011a3-84de-559e-8e89-a11a2f7dc383\"\n\n[[deps.Showoff]]\ndeps = [\"Dates\", \"Grisu\"]\ngit-tree-sha1 = \"91eddf657aca81df9ae6ceb20b959ae5653ad1de\"\nuuid = \"992d4aef-0814-514b-bc4d-f2e9a6c4116f\"\nversion = \"1.0.3\"\n\n[[deps.Sockets]]\nuuid = \"6462fe0b-24de-5631-8697-dd941f90decc\"\n\n[[deps.SortingAlgorithms]]\ndeps = [\"DataStructures\"]\ngit-tree-sha1 = \"b3363d7460f7d098ca0912c69b082f75625d7508\"\nuuid = \"a2af1166-a08f-5f64-846c-94a0d3cef48c\"\nversion = \"1.0.1\"\n\n[[deps.SparseArrays]]\ndeps = [\"LinearAlgebra\", \"Random\"]\nuuid = \"2f01184e-e22b-5df5-ae63-d93ebab69eaf\"\n\n[[deps.StaticArrays]]\ndeps = [\"LinearAlgebra\", \"Random\", \"Statistics\"]\ngit-tree-sha1 = \"2ae4fe21e97cd13efd857462c1869b73c9f61be3\"\nuuid = \"90137ffa-7385-5640-81b9-e52037218182\"\nversion = \"1.3.2\"\n\n[[deps.Statistics]]\ndeps = [\"LinearAlgebra\", \"SparseArrays\"]\nuuid = \"10745b16-79ce-11e8-11f9-7d13ad32a3b2\"\n\n[[deps.StatsAPI]]\ngit-tree-sha1 = \"d88665adc9bcf45903013af0982e2fd05ae3d0a6\"\nuuid = \"82ae8749-77ed-4fe6-ae5f-f523153014b0\"\nversion = \"1.2.0\"\n\n[[deps.StatsBase]]\ndeps = [\"DataAPI\", \"DataStructures\", \"LinearAlgebra\", \"LogExpFunctions\", \"Missings\", \"Printf\", \"Random\", \"SortingAlgorithms\", \"SparseArrays\", \"Statistics\", \"StatsAPI\"]\ngit-tree-sha1 = \"51383f2d367eb3b444c961d485c565e4c0cf4ba0\"\nuuid = \"2913bbd2-ae8a-5f71-8c99-4fb6c76f3a91\"\nversion = \"0.33.14\"\n\n[[deps.StructArrays]]\ndeps = [\"Adapt\", \"DataAPI\", \"StaticArrays\", \"Tables\"]\ngit-tree-sha1 = \"2ce41e0d042c60ecd131e9fb7154a3bfadbf50d3\"\nuuid = \"09ab397b-f2b6-538f-b94a-2f83cf4a842a\"\nversion = \"0.6.3\"\n\n[[deps.TOML]]\ndeps = [\"Dates\"]\nuuid = \"fa267f1f-6049-4f14-aa54-33bafae1ed76\"\n\n[[deps.TableTraits]]\ndeps = [\"IteratorInterfaceExtensions\"]\ngit-tree-sha1 = \"c06b2f539df1c6efa794486abfb6ed2022561a39\"\nuuid = \"3783bdb8-4a98-5b6b-af9a-565f29a5fe9c\"\nversion = \"1.0.1\"\n\n[[deps.Tables]]\ndeps = [\"DataAPI\", \"DataValueInterfaces\", \"IteratorInterfaceExtensions\", \"LinearAlgebra\", \"TableTraits\", \"Test\"]\ngit-tree-sha1 = \"bb1064c9a84c52e277f1096cf41434b675cd368b\"\nuuid = \"bd369af6-aec1-5ad0-b16a-f7cc5008161c\"\nversion = \"1.6.1\"\n\n[[deps.Tar]]\ndeps = [\"ArgTools\", \"SHA\"]\nuuid = \"a4e569a6-e804-4fa4-b0f3-eef7a1d5b13e\"\n\n[[deps.Test]]\ndeps = [\"InteractiveUtils\", \"Logging\", \"Random\", \"Serialization\"]\nuuid = \"8dfed614-e22c-5e08-85e1-65c5234f0b40\"\n\n[[deps.URIs]]\ngit-tree-sha1 = \"97bbe755a53fe859669cd907f2d96aee8d2c1355\"\nuuid = \"5c2747f8-b7ea-4ff2-ba2e-563bfd36b1d4\"\nversion = \"1.3.0\"\n\n[[deps.UUIDs]]\ndeps = [\"Random\", \"SHA\"]\nuuid = \"cf7118a7-6976-5b1a-9a39-7adc72f591a4\"\n\n[[deps.Unicode]]\nuuid = \"4ec0a83e-493e-50e2-b9ac-8f72acf5a8f5\"\n\n[[deps.UnicodeFun]]\ndeps = [\"REPL\"]\ngit-tree-sha1 = \"53915e50200959667e78a92a418594b428dffddf\"\nuuid = \"1cfade01-22cf-5700-b092-accc4b62d6e1\"\nversion = \"0.4.1\"\n\n[[deps.Unzip]]\ngit-tree-sha1 = \"34db80951901073501137bdbc3d5a8e7bbd06670\"\nuuid = \"41fe7b60-77ed-43a1-b4f0-825fd5a5650d\"\nversion = \"0.1.2\"\n\n[[deps.Wayland_jll]]\ndeps = [\"Artifacts\", \"Expat_jll\", \"JLLWrappers\", \"Libdl\", \"Libffi_jll\", \"Pkg\", \"XML2_jll\"]\ngit-tree-sha1 = \"3e61f0b86f90dacb0bc0e73a0c5a83f6a8636e23\"\nuuid = \"a2964d1f-97da-50d4-b82a-358c7fce9d89\"\nversion = \"1.19.0+0\"\n\n[[deps.Wayland_protocols_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"66d72dc6fcc86352f01676e8f0f698562e60510f\"\nuuid = \"2381bf8a-dfd0-557d-9999-79630e7b1b91\"\nversion = \"1.23.0+0\"\n\n[[deps.XML2_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Libiconv_jll\", \"Pkg\", \"Zlib_jll\"]\ngit-tree-sha1 = \"1acf5bdf07aa0907e0a37d3718bb88d4b687b74a\"\nuuid = \"02c8fc9c-b97f-50b9-bbe4-9be30ff0a78a\"\nversion = \"2.9.12+0\"\n\n[[deps.XSLT_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Libgcrypt_jll\", \"Libgpg_error_jll\", \"Libiconv_jll\", \"Pkg\", \"XML2_jll\", \"Zlib_jll\"]\ngit-tree-sha1 = \"91844873c4085240b95e795f692c4cec4d805f8a\"\nuuid = \"aed1982a-8fda-507f-9586-7b0439959a61\"\nversion = \"1.1.34+0\"\n\n[[deps.Xorg_libX11_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libxcb_jll\", \"Xorg_xtrans_jll\"]\ngit-tree-sha1 = \"5be649d550f3f4b95308bf0183b82e2582876527\"\nuuid = \"4f6342f7-b3d2-589e-9d20-edeb45f2b2bc\"\nversion = \"1.6.9+4\"\n\n[[deps.Xorg_libXau_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"4e490d5c960c314f33885790ed410ff3a94ce67e\"\nuuid = \"0c0b7dd1-d40b-584c-a123-a41640f87eec\"\nversion = \"1.0.9+4\"\n\n[[deps.Xorg_libXcursor_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libXfixes_jll\", \"Xorg_libXrender_jll\"]\ngit-tree-sha1 = \"12e0eb3bc634fa2080c1c37fccf56f7c22989afd\"\nuuid = \"935fb764-8cf2-53bf-bb30-45bb1f8bf724\"\nversion = \"1.2.0+4\"\n\n[[deps.Xorg_libXdmcp_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"4fe47bd2247248125c428978740e18a681372dd4\"\nuuid = \"a3789734-cfe1-5b06-b2d0-1dd0d9d62d05\"\nversion = \"1.1.3+4\"\n\n[[deps.Xorg_libXext_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libX11_jll\"]\ngit-tree-sha1 = \"b7c0aa8c376b31e4852b360222848637f481f8c3\"\nuuid = \"1082639a-0dae-5f34-9b06-72781eeb8cb3\"\nversion = \"1.3.4+4\"\n\n[[deps.Xorg_libXfixes_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libX11_jll\"]\ngit-tree-sha1 = \"0e0dc7431e7a0587559f9294aeec269471c991a4\"\nuuid = \"d091e8ba-531a-589c-9de9-94069b037ed8\"\nversion = \"5.0.3+4\"\n\n[[deps.Xorg_libXi_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libXext_jll\", \"Xorg_libXfixes_jll\"]\ngit-tree-sha1 = \"89b52bc2160aadc84d707093930ef0bffa641246\"\nuuid = \"a51aa0fd-4e3c-5386-b890-e753decda492\"\nversion = \"1.7.10+4\"\n\n[[deps.Xorg_libXinerama_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libXext_jll\"]\ngit-tree-sha1 = \"26be8b1c342929259317d8b9f7b53bf2bb73b123\"\nuuid = \"d1454406-59df-5ea1-beac-c340f2130bc3\"\nversion = \"1.1.4+4\"\n\n[[deps.Xorg_libXrandr_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libXext_jll\", \"Xorg_libXrender_jll\"]\ngit-tree-sha1 = \"34cea83cb726fb58f325887bf0612c6b3fb17631\"\nuuid = \"ec84b674-ba8e-5d96-8ba1-2a689ba10484\"\nversion = \"1.5.2+4\"\n\n[[deps.Xorg_libXrender_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libX11_jll\"]\ngit-tree-sha1 = \"19560f30fd49f4d4efbe7002a1037f8c43d43b96\"\nuuid = \"ea2f1a96-1ddc-540d-b46f-429655e07cfa\"\nversion = \"0.9.10+4\"\n\n[[deps.Xorg_libpthread_stubs_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"6783737e45d3c59a4a4c4091f5f88cdcf0908cbb\"\nuuid = \"14d82f49-176c-5ed1-bb49-ad3f5cbd8c74\"\nversion = \"0.1.0+3\"\n\n[[deps.Xorg_libxcb_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"XSLT_jll\", \"Xorg_libXau_jll\", \"Xorg_libXdmcp_jll\", \"Xorg_libpthread_stubs_jll\"]\ngit-tree-sha1 = \"daf17f441228e7a3833846cd048892861cff16d6\"\nuuid = \"c7cfdc94-dc32-55de-ac96-5a1b8d977c5b\"\nversion = \"1.13.0+3\"\n\n[[deps.Xorg_libxkbfile_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libX11_jll\"]\ngit-tree-sha1 = \"926af861744212db0eb001d9e40b5d16292080b2\"\nuuid = \"cc61e674-0454-545c-8b26-ed2c68acab7a\"\nversion = \"1.1.0+4\"\n\n[[deps.Xorg_xcb_util_image_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_xcb_util_jll\"]\ngit-tree-sha1 = \"0fab0a40349ba1cba2c1da699243396ff8e94b97\"\nuuid = \"12413925-8142-5f55-bb0e-6d7ca50bb09b\"\nversion = \"0.4.0+1\"\n\n[[deps.Xorg_xcb_util_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libxcb_jll\"]\ngit-tree-sha1 = \"e7fd7b2881fa2eaa72717420894d3938177862d1\"\nuuid = \"2def613f-5ad1-5310-b15b-b15d46f528f5\"\nversion = \"0.4.0+1\"\n\n[[deps.Xorg_xcb_util_keysyms_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_xcb_util_jll\"]\ngit-tree-sha1 = \"d1151e2c45a544f32441a567d1690e701ec89b00\"\nuuid = \"975044d2-76e6-5fbe-bf08-97ce7c6574c7\"\nversion = \"0.4.0+1\"\n\n[[deps.Xorg_xcb_util_renderutil_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_xcb_util_jll\"]\ngit-tree-sha1 = \"dfd7a8f38d4613b6a575253b3174dd991ca6183e\"\nuuid = \"0d47668e-0667-5a69-a72c-f761630bfb7e\"\nversion = \"0.3.9+1\"\n\n[[deps.Xorg_xcb_util_wm_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_xcb_util_jll\"]\ngit-tree-sha1 = \"e78d10aab01a4a154142c5006ed44fd9e8e31b67\"\nuuid = \"c22f9ab0-d5fe-5066-847c-f4bb1cd4e361\"\nversion = \"0.4.1+1\"\n\n[[deps.Xorg_xkbcomp_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_libxkbfile_jll\"]\ngit-tree-sha1 = \"4bcbf660f6c2e714f87e960a171b119d06ee163b\"\nuuid = \"35661453-b289-5fab-8a00-3d9160c6a3a4\"\nversion = \"1.4.2+4\"\n\n[[deps.Xorg_xkeyboard_config_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Xorg_xkbcomp_jll\"]\ngit-tree-sha1 = \"5c8424f8a67c3f2209646d4425f3d415fee5931d\"\nuuid = \"33bec58e-1273-512f-9401-5d533626f822\"\nversion = \"2.27.0+4\"\n\n[[deps.Xorg_xtrans_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"79c31e7844f6ecf779705fbc12146eb190b7d845\"\nuuid = \"c5fb5394-a638-5e4d-96e5-b29de1b5cf10\"\nversion = \"1.4.0+3\"\n\n[[deps.Zlib_jll]]\ndeps = [\"Libdl\"]\nuuid = \"83775a58-1f1d-513f-b197-d71354ab007a\"\n\n[[deps.Zstd_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"cc4bf3fdde8b7e3e9fa0351bdeedba1cf3b7f6e6\"\nuuid = \"3161d3a3-bdf6-5164-811a-617609db77b4\"\nversion = \"1.5.0+0\"\n\n[[deps.libass_jll]]\ndeps = [\"Artifacts\", \"Bzip2_jll\", \"FreeType2_jll\", \"FriBidi_jll\", \"HarfBuzz_jll\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Zlib_jll\"]\ngit-tree-sha1 = \"5982a94fcba20f02f42ace44b9894ee2b140fe47\"\nuuid = \"0ac62f75-1d6f-5e53-bd7c-93b484bb37c0\"\nversion = \"0.15.1+0\"\n\n[[deps.libblastrampoline_jll]]\ndeps = [\"Artifacts\", \"Libdl\", \"OpenBLAS_jll\"]\nuuid = \"8e850b90-86db-534c-a0d3-1478176c7d93\"\n\n[[deps.libfdk_aac_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"daacc84a041563f965be61859a36e17c4e4fcd55\"\nuuid = \"f638f0a6-7fb0-5443-88ba-1cc74229b280\"\nversion = \"2.0.2+0\"\n\n[[deps.libpng_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Zlib_jll\"]\ngit-tree-sha1 = \"94d180a6d2b5e55e447e2d27a29ed04fe79eb30c\"\nuuid = \"b53b4c65-9356-5827-b1ea-8c7a1a84506f\"\nversion = \"1.6.38+0\"\n\n[[deps.libvorbis_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Ogg_jll\", \"Pkg\"]\ngit-tree-sha1 = \"b910cb81ef3fe6e78bf6acee440bda86fd6ae00c\"\nuuid = \"f27f6e37-5d2b-51aa-960f-b287f2bc3b7a\"\nversion = \"1.3.7+1\"\n\n[[deps.nghttp2_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"8e850ede-7688-5339-a07c-302acd2aaf8d\"\n\n[[deps.p7zip_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"3f19e933-33d8-53b3-aaab-bd5110c3b7a0\"\n\n[[deps.x264_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"4fea590b89e6ec504593146bf8b988b2c00922b2\"\nuuid = \"1270edf5-f2f9-52d2-97e9-ab00b5d0237a\"\nversion = \"2021.5.5+0\"\n\n[[deps.x265_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"ee567a171cce03570d77ad3a43e90218e38937a9\"\nuuid = \"dfaa095f-4041-5dcd-9319-2fabd8486b76\"\nversion = \"3.5.0+0\"\n\n[[deps.xkbcommon_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\", \"Wayland_jll\", \"Wayland_protocols_jll\", \"Xorg_libxcb_jll\", \"Xorg_xkeyboard_config_jll\"]\ngit-tree-sha1 = \"ece2350174195bb31de1a63bea3a41ae1aa593b6\"\nuuid = \"d8fb68d0-12a3-5cfd-a85a-d49703b185fd\"\nversion = \"0.9.1+5\"\n\"\"\"\n\n# \u2554\u2550\u2561 Cell order:\n# \u255f\u25002398b4e5-5920-442e-980b-f75359ba73ae\n# \u2560\u25503f9f4948-74c3-11ec-2daf-51719517f805\n# \u255f\u250097d595d3-3c44-4112-a7a8-4efe4c1ba943\n# \u255f\u250062ba405d-4d1a-42b7-bacc-1350bf90564e\n# \u255f\u2500d6c88a68-c221-417b-bb78-06938fe97407\n# \u2560\u2550151282f9-0a35-435c-9cba-e7736efccb3f\n# \u255f\u25008f922beb-d685-4790-a08b-0d24810c417d\n# \u2560\u2550c93284d7-0cb2-4991-a44e-a30e53c29a93\n# \u255f\u250099a21f98-bd1a-4bc4-a619-668e824bb788\n# \u2560\u25506fa4452d-ddcd-4df7-996a-ba75ae21ba6e\n# \u255f\u2500e2c62cf7-9c2e-48d7-97a3-0990328e6ec5\n# \u2560\u2550ff744877-012a-4edf-bdf9-a9e29744cff3\n# \u255f\u2500d5bf8c5a-229e-4a2f-bbff-6d1ef6fcf9c6\n# \u2560\u2550467168a3-33d0-46e4-acdc-3aea85fd2ff5\n# \u255f\u2500c7e4ee98-8c09-4eb5-aa9d-df7a8e2b51d7\n# \u2560\u25509f986c37-8f68-4348-a587-1cc18f924fee\n# \u255f\u2500fd3f32e1-29b6-4766-b1a4-60ce09c9e8f9\n# \u2560\u2550cc46809e-3436-4748-a29a-2a82cc197d66\n# \u255f\u25008710210a-2dcf-4d63-a274-4ab44445a17f\n# \u2560\u255012328375-5e07-49c1-83da-bdae88fcc8d4\n# \u255f\u25001e97263b-5fa9-497b-b610-7f3ad30b5520\n# \u255f\u2500238002f4-c32b-41bc-8342-ac238828f74a\n# \u2560\u25503156a7d1-d1a3-414e-8794-c895ed08f3b0\n# \u255f\u2500b35a3d34-628e-470d-96b2-076a1e3d5ba5\n# \u2560\u25506048f590-30e0-41ea-bcb9-a283d700668b\n# \u255f\u2500ec522eda-7c21-4691-863e-4b9adf1414b3\n# \u255f\u25000509b052-02c7-4cf5-9c1f-fa5cf584360c\n# \u2560\u25509a30c019-fc18-492c-b147-14c234c8ff12\n# \u255f\u2500074eb18b-05eb-456e-89c7-39b7b122e14e\n# \u2560\u255053a2adeb-4d89-41e5-8a38-bb0abb2466eb\n# \u255f\u2500f9ac8b88-2383-4ef2-a9b9-01435654ebf8\n# \u2560\u2550e389aaa7-82c8-4569-ace4-d13e2e3fb3ec\n# \u2560\u2550c4311bd3-6cd0-4bf9-8210-8b6555d3175f\n# \u2560\u25508f8db2f8-bf75-4828-862c-2b361eb33692\n# \u2560\u25508e6d61eb-923d-47c3-9306-237529a60672\n# \u2560\u2550b5eb2437-a13e-4b69-a378-e977ecf0fba7\n# \u255f\u250055ec5071-e9a5-4acf-9e15-9947cc9be2b5\n# \u2560\u2550046bd1e5-e6c8-46b4-bace-11fe8ef95b50\n# \u255f\u2500e6c5c88c-1d13-4785-bdea-6048cc905dcc\n# \u2560\u2550d1ffbd03-3454-4e95-8c8f-3466a0d3ecbc\n# \u255f\u2500bd510480-984e-44ef-ac45-2fbfa909eb9e\n# \u255f\u250000000000-0000-0000-0000-000000000001\n# \u255f\u250000000000-0000-0000-0000-000000000002\n", "meta": {"hexsha": "4a50b24f90ddbe57e42d8e0169ced6fe502f1010", "size": 51663, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "assignment-01.jl", "max_stars_repo_name": "airicbear/CIS4523", "max_stars_repo_head_hexsha": "8c7433eb4a359f72255fc9a356cfaa8b10fbeb81", "max_stars_repo_licenses": ["Unlicense"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "assignment-01.jl", "max_issues_repo_name": "airicbear/CIS4523", "max_issues_repo_head_hexsha": "8c7433eb4a359f72255fc9a356cfaa8b10fbeb81", "max_issues_repo_licenses": ["Unlicense"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "assignment-01.jl", "max_forks_repo_name": "airicbear/CIS4523", "max_forks_repo_head_hexsha": "8c7433eb4a359f72255fc9a356cfaa8b10fbeb81", "max_forks_repo_licenses": ["Unlicense"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 35.777700831, "max_line_length": 957, "alphanum_fraction": 0.7248707973, "num_tokens": 22672, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.5, "lm_q2_score": 0.1581743527484317, "lm_q1q2_score": 0.07908717637421585}}
{"text": "### A Pluto.jl notebook ###\n# v0.12.21\n\nusing Markdown\nusing InteractiveUtils\n\n# \u2554\u2550\u2561 b0dff76e-8726-11eb-31c8-ab5878dd9ed4\nbegin\n\tusing PlutoUI\n\tusing NativeSVG\nend\n\n# \u2554\u2550\u2561 bef082a6-8726-11eb-088f-5ffda6b048e7\nmd\"\"\"# Tuples\n\nThis chapter presents one more built-in type, the tuple, and then shows how arrays, dictionaries, and tuples work together. It also introduces a useful feature for variable-length argument arrays, the gather and scatter operators.\"\"\"\n\n# \u2554\u2550\u2561 d10a4cb8-8726-11eb-27e2-b55de2487e54\nmd\"\"\"## Tuples Are Immutable\n\nA *tuple* is a sequence of values. The values can be of any type, and they are indexed by integers, so in that respect tuples are a lot like arrays. The important difference is that tuples are immutable and that each element can have its own type.\n\nSyntactically, a tuple is a comma-separated list of values:\n\n```julia\njulia> t = 'a', 'b', 'c', 'd', 'e' \n('a', 'b', 'c', 'd', 'e')\n```\n\nAlthough it is not necessary, it is common to enclose tuples in parentheses:\n\n```julia\njulia> t = ('a', 'b', 'c', 'd', 'e') \n('a', 'b', 'c', 'd', 'e')\n```\n\nTo create a tuple with a single element, you have to include a final comma:\n\n```julia\njulia> t1 = ('a',) \n('a',)\njulia> typeof(t1) \nTuple{Char}\n```\n\n!!! danger\n A value in parentheses without comma is not a tuple:\n \n ```julia\n julia> t2 = ('a')\n 'a': ASCII/Unicode U+0061 (category Ll: Letter, lowercase) \n julia> typeof(t2) \n Char\n ```\n\nAnother way to create a tuple is using the built-in function `tuple`. With no argument, it creates an empty tuple:\n\n```julia\njulia> tuple() \n()\n```\n\nIf multiple arguments are provided, the result is a tuple with the given arguments:\n\n```julia\njulia> t3 = tuple(1, 'a', pi) \n(1, 'a', \u03c0 = 3.1415926535897...)\n```\n\nBecause `tuple` is the name of a built-in function, you should avoid using it as a variable name.\n\nMost array operators also work on tuples. The bracket operator indexes an element:\n\n```julia\njulia> t = ('a', 'b', 'c', 'd', 'e'); julia> t[1]\n'a': ASCII/Unicode U+0061 (category Ll: Letter, lowercase)\n```\n\nAnd the slice operator selects a range of elements:\n\n```julia\njulia> t[2:4] \n('b', 'c', 'd')\n```\n\nBut if you try to modify one of the elements of the tuple, you get an error:\n\n```julia\njulia> t[1] = 'A'\nERROR: MethodError: no method matching setindex!(::NTuple{5,Char},::Char, ::Int64)\n```\n\nBecause tuples are immutable, you can\u2019t modify the elements.\n\nThe relational operators work with tuples and other sequences. Julia starts by comparing the first element from each sequence. If they are equal, it goes on to the next elements, and so on until it finds elements that differ. Subsequent elements are not considered (even if they are really big):\n\n```julia\njulia>(0, 1, 2) < (0, 3, 4)\ntrue\njulia> (0, 1, 2000000) < (0, 3, 4) \ntrue\n```\n\"\"\"\n\n# \u2554\u2550\u2561 c7ffd72e-8727-11eb-0f6d-fb8e9395741c\nmd\"\"\"## Tuple Assignment\n\nIt is often useful to swap the values of two variables. With conventional assignments, you have to use a temporary variable. For example, to swap `a` and `b`:\n\n```julia\ntemp=a \na=b \nb=temp\n```\n\nThis solution is cumbersome; *tuple assignment* is more elegant:\n\n```julia\na, b = b, a\n```\n\nThe left side is a tuple of variables; the right side is a tuple of expressions. Each value is assigned to its respective variable. All the expressions on the right side are evaluated before any of the assignments.\n\nThe number of variables on the left has to be fewer than the number of values on the right:\n\n```julia\njulia> (a, b) = (1, 2, 3)\n(1, 2, 3)\njulia>a, b, c= 1, 2\nERROR: BoundsError: attempt to access (1, 2) at index [3]\n```\n\nMore generally, the right side can be any kind of sequence (string, array, or tuple). For example, to split an email address into a username and a domain, you could write:\n\n```julia\njulia> addr = \"julius.caesar@rome\" \n\"julius.caesar@rome\"\njulia> uname, domain = split(addr, '@')\n2-element Array{SubString{String},1}:\n \"julius.caesar\"\n \"rome\"\n```\n\nThe return value from `split` is an array with two elements; the first element is assigned to `uname`, the second to `domain`:\n\n```julia\njulia> uname \n\"julius.caesar\" \njulia> domain \n\"rome\"\n```\n\"\"\"\n\n# \u2554\u2550\u2561 574b6fb0-8728-11eb-0eca-51b778a97602\nmd\"\"\"## Tuples as Return Values\n\nStrictly speaking, a function can only return one value, but if the value is a tuple, the effect is the same as returning multiple values. For example, if you want to divide two integers and compute the quotient and remainder, it is inefficient to compute `x \u00f7 y` and then `x % y`. It is better to compute them both at the same time.\nThe built-in function divrem takes two arguments and returns a tuple of two values, the quotient and remainder. You can store the result as a tuple:\n\n```julia\njulia> t = divrem(7, 3) \n(2, 1)\n```\n\nOr use tuple assignment to store the elements separately:\n\n```julia\njulia> q, r = divrem(7, 3);\njulia> @show q r; \nq = 2\nr = 1\n```\n\nHere is an example of a function that returns a tuple:\n\n```julia\nminmax(t) = minimum(t), maximum(t)\n```\n\n`maximum` and `minimum` are built-in functions that find the largest and smallest ele\u2010 ments of a sequence. `minmax` computes both and returns a tuple of two values. The built-in function `extrema` is more efficient.\n\"\"\"\n\n# \u2554\u2550\u2561 a5a354be-8728-11eb-2ab3-4f994c1939c1\nmd\"\"\"## Variable-Length Argument Tuples\n\nFunctions can take a variable number of arguments. A parameter name that ends with `...` *gathers* arguments into a tuple. For example, `printall` takes any number of arguments and prints them:\n\n```julia\nprintall(args...) = println(args)\n```\n\nThe gather parameter can have any name you like, but `args` is conventional. Here\u2019s how the function works:\n\n```julia\njulia> printall(1, 2.0, '3') \n(1, 2.0, '3')\n```\n\nThe complement of gather is *scatter*. If you have a sequence of values and you want to pass it to a function as multiple arguments, you can use the `...` operator. For example, `divrem` takes exactly two arguments; it doesn\u2019t work with a tuple:\n\n```julia\njulia> t = (7, 3); \n\njulia> divrem(t)\nERROR: MethodError: no method matching divrem(::Tuple{Int64,Int64})\n```\n\nBut if you scatter the tuple, it works:\n\n```julia\njulia> divrem(t...) \n(2, 1)\n```\n\nMany of the built-in functions use variable-length argument tuples. For example, `max` and `min` can take any number of arguments:\n\n```julia\njulia> max(1, 2, 3) \n3\n```\n\nBut sum does not: \n\n```julia\njulia> sum(1, 2, 3)\nERROR: MethodError: no method matching sum(::Int64, ::Int64, ::Int64)\n```\n\nIn the Julia world, gather is often called \u201cslurp\u201d and scatter \u201csplat.\u201d\n\"\"\"\n\n# \u2554\u2550\u2561 19ee0db4-8729-11eb-3ca7-737de22181ff\nmd\"\"\"#### Exercise 12-1\n\nWrite a function called `sumall` that takes any number of arguments and returns their sum.\"\"\"\n\n# \u2554\u2550\u2561 3d55da4a-8729-11eb-3d2f-37fb30c62793\nmd\"\"\"## Arrays and Tuples\n\n`zip` is a built-in function that takes two or more sequences and returns a collection of tuples where each tuple contains one element from each sequence. The name of the function refers to a zipper, which joins and interleaves two rows of teeth.\n\nThis example zips a string and an array:\n\n```julia\njulia> s = \"abc\"; \n\njulia> t = [1, 2, 3];\njulia> zip(s, t) \nBase.Iterators.Zip{Tuple{String,Array{Int64,1}}}((\"abc\", [1, 2, 3]))\n```\n\nThe result is a *zip object* that knows how to iterate through the pairs. The most common use of `zip` is in a `for` loop:\n\n```julia\njulia> for pair in zip(s, t) \n\t \t println(pair)\n\t end\n('a', 1)\n('b', 2)\n('c', 3)\n```\n\nA zip object is a kind of *iterator*, which is any object that iterates through a sequence. Iterators are similar to arrays in some ways, but unlike arrays, you can\u2019t use an index to select an element from an iterator.\n\nIf you want to use array operators and functions, you can use a zip object to make an array:\n\n```julia\njulia> collect(zip(s, t))\n3-element Array{Tuple{Char,Int64},1}:\n ('a', 1)\n ('b', 2)\n ('c', 3)\n```\n\nThe result is an array of tuples; in this example, each tuple contains a character from the string and the corresponding element from the array.\n\nIf the sequences are not the same length, the result has the length of the shorter one:\n\n```julia\njulia> collect(zip(\"Anne\", \"Elk\")) \n3-element Array{Tuple{Char,Char},1}:\n ('A', 'E')\n ('n', 'l')\n ('n', 'k')\n```\n\nYou can use tuple assignment in a `for` loop to traverse an array of tuples: \n\n```julia\njulia> t = [('a', 1), ('b', 2), ('c', 3)];\n\njulia> for (letter, number) in t \n println(number, \" \", letter)\n end\n1a 2b 3c\n```\n\nEach time through the loop, Julia selects the next tuple in the array and assigns the elements to `letter` and `number`. The parentheses around (letter, number) are compulsory.\n\nIf you combine `zip`, `for`, and tuple assignment, you get a useful idiom for traversing two (or more) sequences at the same time. For example, `hasmatch` takes two sequences, `t1` and `t2`, and returns true if there is an index `i` such that `t1[i] == t2[i]`:\n\n```julia\nfunction hasmatch(t1, t2) \n\tfor (x, y) in zip(t1, t2)\n\t\tif x == y \n\t\t\treturn true\n\t\tend \n\tend\n\tfalse \nend\n```\n\nIf you need to traverse the elements of a sequence and their indices, you can use the built-in function `enumerate`:\n\n```julia\njulia> for (index, element) in enumerate(\"abc\") \n println(index, \" \", element)\n end\n1 a \n2 b \n3 c\n```\n\nThe result from `enumerate` is an enumerate object, which iterates a sequence of pairs; each pair contains an index (starting from 1) and an element from the given sequence.\n\"\"\"\n\n# \u2554\u2550\u2561 221a3360-872a-11eb-2ead-8d1358efe435\nmd\"\"\"## Dictionaries and Tuples\n\nDictionaries can be used as iterators that iterate the key-value pairs. You can use a dictionary in a `for` loop like this:\n\n```julia\njulia> d = Dict('a'=>1, 'b'=>2, 'c'=>3);\n\njulia> for (key, value) in d \n println(key, \" \", value)\n end\na 1 \nc 3 \nb 2\n```\n\nAs you should expect from a dictionary, the items are in no particular order.\n\nGoing in the other direction, you can use an array of tuples to initialize a new dictionary:\n\n```julia\njulia> t = [('a', 1), ('c', 3), ('b', 2)];\n\njulia> d = Dict(t) \nDict{Char,Int64} with 3 entries:\n 'a' => 1\n 'c' => 3\n 'b' => 2\n```\n\nCombining `Dict` with `zip` yields a concise way to create a dictionary:\n\n```julia\njulia> d = Dict(zip(\"abc\", 1:3)) \nDict{Char,Int64} with 3 entries:\n 'a' => 1\n 'c' => 3\n 'b' => 2\n```\n\nIt is common to use tuples as keys in dictionaries. For example, a telephone directory might map from *last-name*, *first-name* pairs to telephone numbers. Assuming that we have defined `last`, `first`, and `number`, we could write:\n\n```julia\ndirectory[last, first] = number\n```\n\nThe expression in brackets is a tuple. We could use tuple assignment to traverse this\ndictionary:\n\n```julia\nfor ((last, first), number) in directory \n\tprintln(first, \" \", last, \" \", number)\nend\n```\n\nThis loop traverses the key-value pairs in `directory`, which are tuples. It assigns the elements of the key in each tuple to `last` and `first`, and the value to `number`, then prints the name and corresponding telephone number.\n\nThere are two ways to represent tuples in a state diagram. The more detailed version shows the indices and elements just as they appear in an array. For example, the tuple `(\"Cleese\", \"John\")` would appear as in Figure 12-1.\n\"\"\"\n\n# \u2554\u2550\u2561 ce6beec2-872a-11eb-3b39-5531bfc637cb\nDrawing(width=720, height=70) do\n\tdefs() do\n marker(id=\"arrow\", markerWidth=\"10\", markerHeight=\"10\", refX=\"0\", refY=\"3\", orient=\"auto\", markerUnits=\"strokeWidth\") do\n \t\tpath(d=\"M0,0 L0,6 L9,3 z\", fill=\"black\")\n\t\tend\n\tend\n\trect(x=285, y=10, width=150, height=50, fill=\"rgb(242, 242, 242)\", stroke=\"black\")\n\ttext(x=295, y=30, font_family=\"JuliaMono, monospace\", font_size=\"0.85rem\", font_weight=600) do \n\t\tstr(\"1\") \n\tend\n\tline(x1=315, y1=25, x2=355, y2=25, stroke=\"black\", marker_end=\"url(#arrow)\")\n\ttext(x=375, y=30, font_family=\"JuliaMono, monospace\", font_size=\"0.85rem\", font_weight=600) do \n\t\tstr(\"Cleese\") \n\tend\n\ttext(x=295, y=50, font_family=\"JuliaMono, monospace\", font_size=\"0.85rem\", font_weight=600) do \n\t\tstr(\"2\") \n\tend\n\tline(x1=315, y1=45, x2=355, y2=45, stroke=\"black\", marker_end=\"url(#arrow)\")\n\ttext(x=375, y=50, font_family=\"JuliaMono, monospace\", font_size=\"0.85rem\", font_weight=600) do \n\t\tstr(\"John\") \n\tend\nend\n\n# \u2554\u2550\u2561 bf393bf4-872b-11eb-0233-cbb084e3ad3f\nmd\"*Figure 12-1. State diagram.*\"\n\n# \u2554\u2550\u2561 c580c4b4-872b-11eb-1aaf-b51e08f9f26c\nmd\"\"\"But in a larger diagram you might want to leave out the details. For example, a diagram of the telephone directory might appear as in Figure 12-2.\"\"\"\n\n# \u2554\u2550\u2561 c608335e-872b-11eb-278e-51296de15fbc\nDrawing(width=720, height=150) do\n\tdefs() do\n marker(id=\"arrow\", markerWidth=\"10\", markerHeight=\"10\", refX=\"0\", refY=\"3\", orient=\"auto\", markerUnits=\"strokeWidth\") do\n \t\tpath(d=\"M0,0 L0,6 L9,3 z\", fill=\"black\")\n\t\tend\n\tend\n\trect(x=175, y=10, width=370, height=130, fill=\"rgb(242, 242, 242)\", stroke=\"black\")\n\ttext(x=355, y=30, font_family=\"JuliaMono, monospace\", font_size=\"0.85rem\", font_weight=600, text_anchor=\"end\") do \n\t\tstr(\"(\\\"Cleese\\\", \\\"John\\\")\") \n\tend\n\tline(x1=365, y1=25, x2=405, y2=25, stroke=\"black\", marker_end=\"url(#arrow)\")\n\ttext(x=425, y=30, font_family=\"JuliaMono, monospace\", font_size=\"0.85rem\", font_weight=600) do \n\t\tstr(\"08700 100 222\") \n\tend\n\ttext(x=355, y=50, font_family=\"JuliaMono, monospace\", font_size=\"0.85rem\", font_weight=600, text_anchor=\"end\") do \n\t\tstr(\"(\\\"Chapman\\\", \\\"Graham\\\")\") \n\tend\n\tline(x1=365, y1=45, x2=405, y2=45, stroke=\"black\", marker_end=\"url(#arrow)\")\n\ttext(x=425, y=50, font_family=\"JuliaMono, monospace\", font_size=\"0.85rem\", font_weight=600) do \n\t\tstr(\"08700 100 222\") \n\tend\n\ttext(x=355, y=70, font_family=\"JuliaMono, monospace\", font_size=\"0.85rem\", font_weight=600, text_anchor=\"end\") do \n\t\tstr(\"(\\\"Idle\\\", \\\"Eric\\\")\") \n\tend\n\tline(x1=365, y1=65, x2=405, y2=65, stroke=\"black\", marker_end=\"url(#arrow)\")\n\ttext(x=425, y=70, font_family=\"JuliaMono, monospace\", font_size=\"0.85rem\", font_weight=600) do \n\t\tstr(\"08700 100 222\") \n\tend\n\ttext(x=355, y=90, font_family=\"JuliaMono, monospace\", font_size=\"0.85rem\", font_weight=600, text_anchor=\"end\") do \n\t\tstr(\"(\\\"Gilliam\\\", \\\"Terry\\\")\") \n\tend\n\tline(x1=365, y1=85, x2=405, y2=85, stroke=\"black\", marker_end=\"url(#arrow)\")\n\ttext(x=425, y=90, font_family=\"JuliaMono, monospace\", font_size=\"0.85rem\", font_weight=600) do \n\t\tstr(\"08700 100 222\") \n\tend\n\ttext(x=355, y=110, font_family=\"JuliaMono, monospace\", font_size=\"0.85rem\", font_weight=600, text_anchor=\"end\") do \n\t\tstr(\"(\\\"Jones\\\", \\\"Terry\\\")\") \n\tend\n\tline(x1=365, y1=105, x2=405, y2=105, stroke=\"black\", marker_end=\"url(#arrow)\")\n\ttext(x=425, y=110, font_family=\"JuliaMono, monospace\", font_size=\"0.85rem\", font_weight=600) do \n\t\tstr(\"08700 100 222\") \n\tend\n\ttext(x=355, y=130, font_family=\"JuliaMono, monospace\", font_size=\"0.85rem\", font_weight=600, text_anchor=\"end\") do \n\t\tstr(\"(\\\"Palin\\\", \\\"Michael\\\")\") \n\tend\n\tline(x1=365, y1=125, x2=405, y2=125, stroke=\"black\", marker_end=\"url(#arrow)\")\n\ttext(x=425, y=130, font_family=\"JuliaMono, monospace\", font_size=\"0.85rem\", font_weight=600) do \n\t\tstr(\"08700 100 222\") \n\tend\nend\n\n# \u2554\u2550\u2561 1f4b8a1e-872d-11eb-20b1-014ddea498de\nmd\"*Figure 12-2. State diagram.*\"\n\n# \u2554\u2550\u2561 286fe504-872d-11eb-06d3-e1c2682eff9c\nmd\"\"\"Here the tuples are shown using Julia syntax as a graphical shorthand. The telephone number in the diagram is the complaints line for the BBC, so please don\u2019t call it.\"\"\"\n\n# \u2554\u2550\u2561 3260e720-872d-11eb-065c-93bbb15621b9\nmd\"\"\"## Sequences of Sequences\n\nI have focused on arrays of tuples, but almost all of the examples in this chapter also work with arrays of arrays, tuples of tuples, and tuples of arrays. To avoid enumerating the possible combinations, it is sometimes easier to talk about sequences of sequences.\n\nIn many contexts, the different kinds of sequences (strings, arrays, and tuples) can be used interchangeably. So how should you choose one over the others?\n\nTo start with the obvious, strings are more limited than other sequences because the elements have to be characters. They are also immutable. If you need the ability to change the characters in a string (as opposed to creating a new string), you might want to use an array of characters instead.\n\nArrays are more common than tuples, mostly because they are mutable. But there are a few cases where you might prefer tuples:\n\n* In some contexts, like a `return` statement, it is syntactically simpler to create a tuple than an array.\n* If you are passing a sequence as an argument to a function, using tuples reduces the potential for unexpected behavior due to aliasing.\n* For performance reasons. The compiler can specialize on the type.\n\nBecause tuples are immutable, they don\u2019t provide functions like `sort!` and `reverse!`, which modify existing arrays. But Julia provides the built-in functions `sort`, which takes an array and returns a new array with the same elements in sorted order, and `reverse`, which takes any sequence and returns a sequence of the same type in reverse order.\n\"\"\"\n\n# \u2554\u2550\u2561 6cd09676-872d-11eb-0ffd-3fd772eb7db4\nmd\"\"\"## Debugging\n\nArrays, dictionaries, and tuples are examples of *data structures*; in this chapter we are starting to see compound data structures, like arrays of tuples, or dictionaries that contain tuples as keys and arrays as values. Compound data structures are useful, but they are prone to what I call *shape errors*; that is, errors caused when a data structure has the wrong type, size, or structure. For example, if you are expecting an array with one integer and I give you a plain old integer (not in an array), it won\u2019t work.\n\nJulia allows you to attach a type to elements of a sequence. How this is done is detailed in Lecture 17. Specifying the type eliminates a lot of shape errors.\n\"\"\"\n\n# \u2554\u2550\u2561 96a213d2-872d-11eb-25e4-fdd44a16150d\nmd\"\"\"## Glossary\n\n*tuple*:\nAn immutable sequence of elements where every element can have its own type.\n\n*tuple assignment*:\nAn assignment with a sequence on the right side and a tuple of variables on the left. The right side is evaluated and then its elements are assigned to the variables on the left.\n\n*gather*:\nThe operation of assembling a variable-length argument tuple.\n\n*scatter*:\nThe operation of treating a sequence as a list of arguments.\n\n*zip object*:\nThe result of calling a built-in function zip, an object that iterates through a sequence of tuples.\n\n*iterator*:\nAn object that can iterate through a sequence, but that does not provide array operators and functions.\n\n*data structure*:\nA collection of related values, often organized in arrays, dictionaries, tuples, etc.\n\n*shape error*:\nAn error caused because a value has the wrong shape; that is, the wrong type or size.\n\"\"\"\n\n# \u2554\u2550\u2561 e7f038d6-872d-11eb-28a3-6307d3e72e85\nmd\"\"\"## Exercises \n\n#### Exercise 12-2\n\nWrite a function called `mostfrequent` that takes a string and prints the letters in decreasing order of frequency. Find text samples from several different languages and see how letter frequency varies between languages. Compare your results with the tables at [https://en.wikipedia.org/wiki/Letter_frequencies](https://en.wikipedia.org/wiki/Letter_frequencies).\n\"\"\"\n\n# \u2554\u2550\u2561 05a7a222-872e-11eb-3de9-c73ab5da92a0\nmd\"\"\"#### Exercise 12-3\n\nMore anagrams!\n\n1. Write a program that reads a word list from a file (see \u201cReading Word Lists\u201d) and prints all the sets of words that are anagrams. Here is an example of what the output might look like:\n \n ```julia\n [\"deltas\", \"desalt\", \"lasted\", \"salted\", \"slated\", \"staled\"]\n [\"retainers\", \"ternaries\"]\n [\"generating\", \"greatening\"]\n [\"resmelts\", \"smelters\", \"termless\"]\n ```\n \n !!! tip\n You might want to build a dictionary that maps from a collection of letters to an array of words that can be spelled with those letters. The question is, how can you represent the col\u2010 lection of letters in a way that can be used as a key?\n\n2. Modify the previous program so that it prints the longest array of anagrams first, followed by the second longest, and so on.\n\n3. In Scrabble a \u201cbingo\u201d is when you play all seven tiles in your rack, along with a letter on the board, to form an eight-letter word. What collection of eight letters forms the most possible bingos?\n\"\"\"\n\n# \u2554\u2550\u2561 586908a2-872e-11eb-3919-f96ba66ee674\nmd\"\"\"#### Exercise 12-4\n\nTwo words form a \u201cmetathesis pair\u201d if you can transform one into the other by swap\u2010 ping two letters; for example, \u201cconverse\u201d and \u201cconserve.\u201d Write a program that finds all of the metathesis pairs in *words.txt*.\n\n!!! tip\n Don\u2019t test all pairs of words, and don\u2019t test all possible swaps.\n\n*Credit*: This exercise is inspired by an example at [http://puzzlers.org](http://puzzlers.org).\n\"\"\"\n\n# \u2554\u2550\u2561 8a7b57dc-872e-11eb-189c-7f2d7cb2e695\nmd\"\"\"#### Exercise 12-5\n\nHere\u2019s another Car Talk Puzzler:\n\n> What is the longest English word, that remains a valid English word, as you remove its letters one at a time?\n> \n> Now, letters can be removed from either end, or the middle, but you can\u2019t rearrange any of the letters. Every time you drop a letter, you wind up with another English word. If you do that, you\u2019re eventually going to wind up with one letter and that too is going to be an English word\u2014one that\u2019s found in the dictionary. I want to know what\u2019s the longest word. What\u2019s the word, and how many letters does it have?\n> \n> I\u2019m going to give you a little modest example: Sprite. Ok? You start off with sprite, you take a letter off, one from the interior of the word, take the r away, and we\u2019re left with the word spite, then we take the e off the end, we\u2019re left with spit, we take the s off, we\u2019re left with pit, it, and I.\n\nWrite a program to find all words that can be reduced in this way, and then find the longest one.\n\n!!! tip\n This exercise is a little more challenging than most, so here are some suggestions:\n \n 1. You might want to write a function that takes a word and com\u2010 putes an array of all the words that can be formed by removing one letter. These are the \u201cchildren\u201d of the word.\n \n 2. Recursively, a word is reducible if any of its children are reducible. As a base case, you can consider the empty string reducible.\n \n 3. The word list I provided, *words.txt*, doesn\u2019t contain singleletter words. So, you might want to add \u201cI,\u201d \u201ca,\u201d and the empty string.\n \n 4. To improve the performance of your program, you might want to memoize the words that are known to be reducible.\n\"\"\"\n\n# \u2554\u2550\u2561 Cell order:\n# \u255f\u2500b0dff76e-8726-11eb-31c8-ab5878dd9ed4\n# \u255f\u2500bef082a6-8726-11eb-088f-5ffda6b048e7\n# \u255f\u2500d10a4cb8-8726-11eb-27e2-b55de2487e54\n# \u255f\u2500c7ffd72e-8727-11eb-0f6d-fb8e9395741c\n# \u255f\u2500574b6fb0-8728-11eb-0eca-51b778a97602\n# \u255f\u2500a5a354be-8728-11eb-2ab3-4f994c1939c1\n# \u255f\u250019ee0db4-8729-11eb-3ca7-737de22181ff\n# \u255f\u25003d55da4a-8729-11eb-3d2f-37fb30c62793\n# \u255f\u2500221a3360-872a-11eb-2ead-8d1358efe435\n# \u255f\u2500ce6beec2-872a-11eb-3b39-5531bfc637cb\n# \u255f\u2500bf393bf4-872b-11eb-0233-cbb084e3ad3f\n# \u255f\u2500c580c4b4-872b-11eb-1aaf-b51e08f9f26c\n# \u255f\u2500c608335e-872b-11eb-278e-51296de15fbc\n# \u255f\u25001f4b8a1e-872d-11eb-20b1-014ddea498de\n# \u255f\u2500286fe504-872d-11eb-06d3-e1c2682eff9c\n# \u255f\u25003260e720-872d-11eb-065c-93bbb15621b9\n# \u255f\u25006cd09676-872d-11eb-0ffd-3fd772eb7db4\n# \u255f\u250096a213d2-872d-11eb-25e4-fdd44a16150d\n# \u255f\u2500e7f038d6-872d-11eb-28a3-6307d3e72e85\n# \u255f\u250005a7a222-872e-11eb-3de9-c73ab5da92a0\n# \u255f\u2500586908a2-872e-11eb-3919-f96ba66ee674\n# \u255f\u25008a7b57dc-872e-11eb-189c-7f2d7cb2e695\n", "meta": {"hexsha": "892cdcbcd468c70d0181886bc0613eddffff9466", "size": 23195, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Lectures/Lecture12.jl", "max_stars_repo_name": "BenLauwens/ES123", "max_stars_repo_head_hexsha": "8531db895a8d4555e63219d79d4dcc527a685458", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 12, "max_stars_repo_stars_event_min_datetime": "2018-03-07T18:04:42.000Z", "max_stars_repo_stars_event_max_datetime": "2021-02-10T15:07:23.000Z", "max_issues_repo_path": "Lectures/Lecture12.jl", "max_issues_repo_name": "BenLauwens/ES123", "max_issues_repo_head_hexsha": "8531db895a8d4555e63219d79d4dcc527a685458", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Lectures/Lecture12.jl", "max_forks_repo_name": "BenLauwens/ES123", "max_forks_repo_head_hexsha": "8531db895a8d4555e63219d79d4dcc527a685458", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 2, "max_forks_repo_forks_event_min_datetime": "2018-08-12T05:29:31.000Z", "max_forks_repo_forks_event_max_datetime": "2020-01-02T21:38:27.000Z", "avg_line_length": 37.5323624595, "max_line_length": 522, "alphanum_fraction": 0.7102392757, "num_tokens": 7269, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.3007455664065234, "lm_q2_score": 0.26284183159693775, "lm_q1q2_score": 0.07904851551894908}}
{"text": "\n#=\nCreated on Saturday 26 December 2020\nLast update: Monday 27 September 2021\n\n@author: Bram De Jaegher\nbram.de.jaegher@gmail.com\n=#\n\n# --- Types --- #\n\nabstract type AbstractQuestion end\nabstract type AbstractDifficulty end\nabstract type AbstractQuestionBlock end\nstruct NoDiff <: AbstractDifficulty end\nstruct Easy <: AbstractDifficulty end\nstruct Intermediate <: AbstractDifficulty end\nstruct Hard <: AbstractDifficulty end\n\n\"\"\"\nDefines a question.\n\n# Arguments\n\n- `description` (md_str): a markdown string to be displayed above the validators\n- `validators` (Array{Bool}): an array of booleans with the tests the answer to the question should solve.\n- `status` (md_str): a markdown string used to change/display the question state (correct, missing, incorrect).\n\tthe default value is missing and should probably not be changed.\n\nDifficulty types\n------\n- `T \u2208 [NoDiff, Easy, Intermediate, Hard]` \n- At this point purely easthetic since the difficulty is only used as display\n\"\"\"\nmutable struct Question{T<:AbstractDifficulty} <: AbstractQuestion \n\tdescription::Markdown.MD\n\tvalidators::Any\n\tstatus::Markdown.MD\n\tdifficulty::T\n\n\tQuestion{T}(;description=md\"\", \n\t\t\t\t\t\tvalidators=[], \n\t\t\t\t\t\tstatus=md\"\") where {T<:AbstractDifficulty} = new{T}(description, validators, status, T())\nend\n\n\"\"\"\nDefines a Question block.\n\n# Arguments\n- `title` (md_str): a markdown string to be displayed as title\n- `description` (md_str): the general descriptions, this is diplayed directly below the title.\n- `hints` (Array{md_str}): an array of markdown strings for hint admonitions but this can me any markdown and will be displayed in the \\\"hints\\\" section.\n- `questions` (Array{AbstractQuestion}): an array of questions. Currently exactly one mandatory question is expected but 0-\u221e optional questions can be defined.\n\n# Examples\n```julia\nq\u2081 = Question(;\n\tdescription=md\\\"\\\"\\\"\n\tComplete the function `myclamp(x)` that clamps a number `x` between 0 and 1.\n\n\tOpen assignments always return `missing`. \n\t\\\"\\\"\\\",\n\tvalidators= @safe[myclamp(-1)==0, myclamp(0.3)==0.3, myclamp(1.1)==1.0]\n)\n\nq\u2082 = Question{Easy}(;\n\tdescription=md\\\"\\\"\\\"\n\tTry to make the clamping also work for arrays.\n\t\\\"\\\"\\\",\n\tvalidators= @safe[myclamp([2.0, 0.3])==[1.0, 0.3]]\n)\n\nq\u2083 = Question{Intermediate}(;\n\tdescription=md\\\"\\\"\\\"\n\tThis is an intermediate question. Surely you can complete this\n\t\\\"\\\"\\\",\n\tvalidators= @safe[true]\n)\n\nq\u2084 = Question{Hard}(;\n\tdescription=md\\\"\\\"\\\"\n\tI admit, this one is definitely harder\n\t\\\"\\\"\\\",\n\tvalidators= @safe[false]\n)\n\nqb = QuestionBlock(;\ntitle=md\"### Question 1.0: What a crazy exercise\",\ndescription=md\\\"\\\"\\\"\n\tSome additional general kind off description and all.\n\tAnything markdowny. Just make sure to use the triple accolades.\n\t\n\t\\\"\\\"\\\",\nquestions = [q\u2081, q\u2082, q\u2083, q\u2084],\nhints=[\thint(md\"Have you tried this?\"),\n\t\thint(md\"Have you tried switching it on and off again?\")]\n);\n```\n\"\"\"\nmutable struct QuestionBlock <: AbstractQuestionBlock\n\ttitle::Markdown.MD\n\tdescription::Markdown.MD\n\thints::Array{Markdown.MD}\n\tquestions::Array{T} where {T<:AbstractQuestion}\n\n\tQuestionBlock(;title=md\"\",\n\t\t\t\t\t\t\t\t\tdescription=md\"\",\n\t\t\t\t\t\t\t\t\thints = Markdown.MD[],\n\t\t\t\t\t\t\t\t\tquestions = [Question()]) = new(title, description, hints, questions) \nend\n\n# --- Rendering --- # \nBase.show(io::IO, ::MIME\"text/html\", q::AbstractQuestionBlock) = print(io::IO, tohtml(q))\n\nfunction tohtml(q::QuestionBlock)\n\t\n\thint_string = \"\"\n\tif length(q.hints) > 0\n\t\thint_string = \"Hints:
\"\n\t\tfor hint in q.hints\n\t\t\thint_string *= \"\" * html(hint) * \"
\"\n\t\tend\n\tend\n\t\n\tN_mandatory = sum(isa.(q.questions,Question))\n\tstate_string = \"\"\n\tfor index in 1:N_mandatory\n\t\tif q.questions[index].description !== \"\"\n\t\t\tstate_string *= \" $(html(q.questions[index].description))
\"\n\t\tend\n\t\tstate_string *= \" $(html(q.questions[index].status))
\"\n\tend\n\n\tout = \"\"\"\n\t\t\n\t\t\t$(html(q.title))\n\t\t\t
$(html(q.description))
\n\t\t\t$state_string\n\t\t\t\n\t\t\t$hint_string\n\t\t
© Pascal, April 2021
\")\n\"\"\"\n\n# \u2554\u2550\u2561 6bcfba84-98ab-4d26-b98c-d86cb65dc277\nPlutoUI.TableOfContents(indent=true, depth=4, aside=true)\n\n# \u2554\u2550\u2561 f88a1737-7a84-4103-82c8-1920bae6b2e8\nmd\"\"\"\n### What is data wrangling?\n\nTake raw data (and/or unorganized data) and turn them (data as plural of datum) into something useful.\n\nA possible data wrangling is as follows:\n\n\n read | $\\Rightarrow$ | clean | $\\Rightarrow$ | Tidy \n --- | --- | --- | --- | --- \n | | $\\Uparrow$ | | $\\Downarrow$ \n | | **Visualize** | $\\Leftarrow$ | **Analyze** \n\n\n**Julia ecosystem**:\n\nRead | Clean, Tidy, Analyze | Visualize\n--- | --- | --- |\nCSV | DataFrames| Plots, StatsPlots\nParquet | DataFramesMeta | Makie, StatsMakie\nSASLib | Query| Gadfly\nReadStat | JuliaDB, JuliaDBMeta | Gaston\nJSON, JSON2/3 | | VegaLite\nLightXML, EzXML | | UnicodePlots\nXLSX, ExcelReader, Taro | | \nPDFIO | |\n\"\"\"\n\n# \u2554\u2550\u2561 02ac758d-3706-4bf4-8a36-83aff4422d0e\nmd\"\"\"\n### Read\n\nUtilize automated tools to read data into memory as cleanly as possible.\n\n`CSV.jl`:\n - Skip leading/trailing rows\n - Parse missing values e.g \"NA\"\n - Parse date/time values\n - Normalize columns\n - Select/drop columns on read\n - User specified column types\n - Auto-delimiter detection\n - Automatic pooled string columns\n - Parse booleans\n - Transpose data\n\"\"\"\n\n# \u2554\u2550\u2561 fb1f4efc-6b89-48a6-95d5-8e3661edf53d\nmd\"\"\"\n### Clean\n\n - Delete unwanted columns\n - Delete rows with missing data\n - Rename columns\n - Fix column data type\n - ...\n\nExamples\n```julia\ndescribe(df) # take a quick peek\n\nselect!(df, 1:5) # keep \"good\" columns / delete unwanted ones \nselect!(df, Not([\"garbaggecol1\", \"garbaggecol2\"]))\n\n\nrename!(df, \"Region Name\" => :region_name)\n\ndropmissing!(df) # Remove rows with missing data\ndropmissing!(df, Between(:x3, :x5))\n```\n\n\"\"\"\n\n# \u2554\u2550\u2561 f22c3d88-e0e6-4d1d-92d4-c99f80aa4733\nmd\"\"\"\n### Tidy Data\n\n - Each variable forms a *column*\n - Each observation forms a *row*\n - Each type of observational unit forms a *table*\n\n\n**Tidy Recipes, examples:**\n\n```julia\n# Stack all date columns into rows\n# The column names are stored ion a new column \"Date\"\nsdf = stack(df, Not(:Region_name); variable_name=:Date)\nsdf = stack(df, Not(1); variable_name=:Date)\nsdf = stack(df, 2:5; variable_name=:Date)\n\n# Date column is a CategoricalArray type. Make it Date type\nsdf.Date = [Date(get(x)) for x \u2208 sdf.Date]\n# or maybe using broadcast:\nsdf.Date .= get.(sdf.Data) .|> Date\n\n# Hoe to turn it back to a wide format?\ndf2 = unstack(sdf, :date, :value)\n\n# It may introduce Union{Missing, T} column type. Let's fix it.\ndisallowmissing!(df2, 2:5)\n```\n\"\"\"\n\n# \u2554\u2550\u2561 2dc07426-ad94-46a0-8da2-68d687ad87c2\nmd\"\"\"\n### Analyze\n\nGet some insights from data. \n\n**Analuyze Recipe**:\n\n```julia\nselct(df, :region_name, 4:5) # select by column\n\nfilter(:region_name => ==(\"Abilene, TX\"), df) # note df is the 2nd arg here\nfilter(\"2020-01-31\" => >(400_000), df)\n\nsort(df, :region_name)\nsort(df, :region_name, rev=true)\n\n# Transform means adding new columns - many more ways of using Transform\ntransform(df, :region_name => ByRow(length) => :region_name_len)\n\n# Group\ngroupby(sdf, :Date)\n\n# Summarize the grouped data:\ncombine(groupby(sdf, :Date), :value => mean => :avg)\n\n# Using Pipe.jl to build a transformation pipeline\n@pipe sdf |>\n groupby(_, :Date) |>\n\tcombine(_, :value => mean => :avg)\n\ncountry = DataFrame(Name=[\"United States\"])\n\n# Joining data is easy\ninnerjoin(df, country, on=[:region_name => :name])\nleftjoin(df, country, on=[:region_name => :name])\nrightjoin(df, country, on=[:region_name => :name])\nouterjoin(df, country, on=[:region_name => :name])\n\n# Also\nsemijoin(df, country, on=[:region_name => :name])\nantijoin(df, country, on=[:region_name => :name])\n```\n\"\"\"\n\n# \u2554\u2550\u2561 cb11e3e6-f385-44db-b739-cf056a7f8841\nmd\"\"\"\n### Visualize\n\nGain more insight by looking at the data in a graphical form.\n\n\n**Visualize Recipes:**\n\n`Plots.jl` | `StatsPlots.jl`\n--- | ---\n`plot` | `groupedbar`\n`scatter` | `corrplot`\n`histogram` | `marginhist`\n`heatmap` | `boxplot`\n`bar` | `violin`\n... | ...\n\n\"\"\"\n\n# \u2554\u2550\u2561 944f1843-1802-45b7-a505-8bb32ad09902\nmd\"\"\"\n## Let's practice\n\n### Tidying\n\"\"\"\n\n# \u2554\u2550\u2561 f9d0761d-a859-404a-812f-f2a424c7b191\nbegin\n\tdf = CSV.File(\"data/youth_suicide.csv\", header=true) |> DataFrame;\n\tsize(df)\nend\n\n# \u2554\u2550\u2561 6790c878-d494-4247-ab1a-d5122d3d845b\nfirst(df, 5)\n\n# \u2554\u2550\u2561 ff4cc1c3-5967-4614-82eb-cc8771e962dc\nnames(df)\n\n# \u2554\u2550\u2561 5315e6b5-344e-42e7-a9a7-b53cfaf2b5b5\nwith_terminal() do\n\tdisplaytable(names(df), index=true)\nend\n\n# \u2554\u2550\u2561 8083c3d3-51f6-437e-ba12-38f8b5814e1e\ndescribe(df, :eltype, :nmissing, :first => first)\n\n# \u2554\u2550\u2561 e70b9da8-34f9-4193-a86e-5e7dc9ed23aa\n## turn colums 2 to 19 to row (observations)\nsdf = stack(df, 2:19; variable_name=:type_year);\n\n# \u2554\u2550\u2561 e783b82c-052e-49d3-87c8-70142b990d5f\nsize(sdf)\n\n# \u2554\u2550\u2561 e9d507d7-df67-4bd4-b80b-02385b2f9be5\nbegin\n\t## Now spread content of column `:type_year` into 2 separate columns\n\tsdf[!, :ctype] .= split.(sdf.type_year, \" \") .|> a -> getindex(a, 1)\n\tsdf[!, :year] .= split.(sdf.type_year, \" \") .|> a -> getindex(a, 2)\n\tselect!(sdf, Not(:type_year))\n\tfirst(sdf, 3)\nend\n\n# \u2554\u2550\u2561 723dc0d9-96c4-43c0-81fd-611209e10106\n## Let's check if the Total type really contains the total\n## 1 - Pick a county\nsdf[sdf.County .== \"Yakima\", :]\n\n# \u2554\u2550\u2561 0031eb09-8e5f-47fe-b04d-1600fad8f994\n## Remove the total row which is redundant\n\nsdf\u2081 = @pipe sdf |>\n\tfilter(:ctype => \u2260(\"Total\"), _) |>\n\tfilter(:year => \u2260(\"(2008-2012)\"), _);\n\n# \u2554\u2550\u2561 f8ee2398-2b69-4133-b2d6-11a2c12b9d27\nsdf\u2081[sdf\u2081.County .== \"Yakima\", :]\n\n# \u2554\u2550\u2561 c52c8ce7-c68b-4b7b-b507-5a813d32cf93\ndescribe(sdf\u2081)\n\n# \u2554\u2550\u2561 4b14a6ad-6dc5-4319-95bf-007e84ae6877\nunique(sdf\u2081.year)\n## Now we want integer instead of string and we want to get rid of the parentheses.\n\n# \u2554\u2550\u2561 54a8cfd0-5daa-40e3-8a0e-12413cc36e83\n## mutate column year from String to Int\nsdf\u2081[!, :year] .= replace.(sdf\u2081.year, r\"[\\(\\)]\" => \"\") |> a -> parse.(Int, a)\n\n# \u2554\u2550\u2561 3378787b-c9cf-4d47-a119-2a35abc3b0b4\ndescribe(sdf\u2081, :eltype, :min, :max, :nunique, :nmissing)\n\n# \u2554\u2550\u2561 53aa180c-7260-46cf-a2a3-4a49e0c30844\nbegin\n\t# Male and Female variables can be put back as columns now\n\tdf\u2081 = unstack(sdf\u2081, :ctype, :value)\n\tdf\u2081[df\u2081.County .== \"Yakima\", :]\nend\n\n# \u2554\u2550\u2561 56d8dcc1-8b76-4332-bbe4-f7081e178300\nmd\"\"\"\n### Visualize\n\"\"\"\n\n# \u2554\u2550\u2561 73a99bae-e8a6-44d1-af74-9778e17df7b9\n# Plot King county's yearly rate\nfilter(:County => ==(\"King\"), df\u2081)\n\n# \u2554\u2550\u2561 5eaa88b0-95e4-4410-bd99-cb9ad40fdb79\n# bar chart\n@pipe df\u2081 |>\n\tfilter(:County => ==(\"King\"), _) |>\n\tbar(_.year, _.Male, \n title = \"King County Youth Suicides (Male)\",\n legend = :none,\n size = (450, 300))\n\n# \u2554\u2550\u2561 5548eaac-d390-4cb1-bf49-0b8985c1f0f0\nbegin\n\t# Prepare to plot both Male and Female together\n\tdf_stacked = stack(df\u2081, 3:4; variable_name = \"gender\");\n\tnames(df_stacked)\nend\n\n# \u2554\u2550\u2561 bb947d94-62f3-45f6-a0d1-9ed596e289c8\n@pipe df_stacked |>\n\tfilter(:County => ==(\"King\"), _) |>\n\t groupedbar( # StatsPlots.jl\n\t\t_.year, # x-axis \n\t _.value; # y-axis\n group = _.gender,\n bar_position = :stack,\n bar_width = 0.7,\n title = \"King County Suicides\",\n size = (450, 300),\n legend = :topleft)\n\n# \u2554\u2550\u2561 5fadd975-cbcf-4a0e-b493-0382875354ba\n@pipe df\u2081 |>\n groupby(_, :County)\n\n# \u2554\u2550\u2561 a92c1c18-39bd-4dcf-81a7-f2f62577e156\n# 5 year totals\n@pipe df\u2081 |>\n groupby(_, :County) |>\n combine(_, :Female => sum, :Male => sum)\n\n# \u2554\u2550\u2561 2c21d894-7e57-4f92-b08d-a4d527f5c25a\n# 5 year totals with male/female combined\n@pipe df\u2081 |>\n groupby(_, :County) |>\n combine(_, :Female => sum, :Male => sum) |>\n select(_, :County, [:Female_sum, :Male_sum] => ByRow(+) => :Total)\n\n# \u2554\u2550\u2561 72aeaa39-6529-4fc3-ba86-f2b113a5a075\nlet \n data\u2081 = @pipe df\u2081 |>\n groupby(_, :year) |>\n combine(_, :Female => sum => :Female, :Male => sum => :Male) |>\n stack(_, 2:3; variable_name = :gender, value_name = :suicides) \n\t\n plot(data\u2081.year, data\u2081.suicides; groups = data\u2081.gender,\n title = \"Youth Suicides Trend\",\n legend = :topleft,\n linewidth = 3,\n size = (450, 300))\nend\n\n# \u2554\u2550\u2561 a7802df0-6819-4650-a539-ccbb99333e92\nlet\n data\u2082 = @pipe df\u2081 |>\n select(_, :County, :year, [:Female, :Male] => ByRow(+) => :total) |>\n unstack(_, :year, :total)\n\t\n values = Matrix(data\u2082[:, 2:end])\n\t\n StatsPlots.heatmap(2008:2012, data\u2082.County, values;\n title = \"Youth Suicide Heatmap\",\n xticks = :all,\n yticks = :all,\n size = (400, 600))\nend\n\n# \u2554\u2550\u2561 8018394c-a123-43c4-aaaf-3d942284e182\nhtml\"\"\"\n\n\"\"\"\n\n# \u2554\u2550\u2561 Cell order:\n# \u255f\u2500578530b0-9b50-11eb-07e1-5db7ceb1b1c7\n# \u2560\u2550000c1c07-fd7a-4e08-9e36-3f0aa6069309\n# \u255f\u25006bcfba84-98ab-4d26-b98c-d86cb65dc277\n# \u255f\u2500f88a1737-7a84-4103-82c8-1920bae6b2e8\n# \u255f\u250002ac758d-3706-4bf4-8a36-83aff4422d0e\n# \u255f\u2500fb1f4efc-6b89-48a6-95d5-8e3661edf53d\n# \u255f\u2500f22c3d88-e0e6-4d1d-92d4-c99f80aa4733\n# \u255f\u25002dc07426-ad94-46a0-8da2-68d687ad87c2\n# \u255f\u2500cb11e3e6-f385-44db-b739-cf056a7f8841\n# \u255f\u2500944f1843-1802-45b7-a505-8bb32ad09902\n# \u2560\u2550f9d0761d-a859-404a-812f-f2a424c7b191\n# \u2560\u25506790c878-d494-4247-ab1a-d5122d3d845b\n# \u2560\u2550ff4cc1c3-5967-4614-82eb-cc8771e962dc\n# \u2560\u25505315e6b5-344e-42e7-a9a7-b53cfaf2b5b5\n# \u2560\u25508083c3d3-51f6-437e-ba12-38f8b5814e1e\n# \u2560\u2550e70b9da8-34f9-4193-a86e-5e7dc9ed23aa\n# \u2560\u2550e783b82c-052e-49d3-87c8-70142b990d5f\n# \u2560\u2550e9d507d7-df67-4bd4-b80b-02385b2f9be5\n# \u2560\u2550723dc0d9-96c4-43c0-81fd-611209e10106\n# \u2560\u25500031eb09-8e5f-47fe-b04d-1600fad8f994\n# \u2560\u2550f8ee2398-2b69-4133-b2d6-11a2c12b9d27\n# \u2560\u2550c52c8ce7-c68b-4b7b-b507-5a813d32cf93\n# \u2560\u25504b14a6ad-6dc5-4319-95bf-007e84ae6877\n# \u2560\u255054a8cfd0-5daa-40e3-8a0e-12413cc36e83\n# \u2560\u25503378787b-c9cf-4d47-a119-2a35abc3b0b4\n# \u2560\u255053aa180c-7260-46cf-a2a3-4a49e0c30844\n# \u255f\u250056d8dcc1-8b76-4332-bbe4-f7081e178300\n# \u2560\u255073a99bae-e8a6-44d1-af74-9778e17df7b9\n# \u2560\u25505eaa88b0-95e4-4410-bd99-cb9ad40fdb79\n# \u2560\u25505548eaac-d390-4cb1-bf49-0b8985c1f0f0\n# \u2560\u2550bb947d94-62f3-45f6-a0d1-9ed596e289c8\n# \u2560\u25505fadd975-cbcf-4a0e-b493-0382875354ba\n# \u2560\u2550a92c1c18-39bd-4dcf-81a7-f2f62577e156\n# \u2560\u25502c21d894-7e57-4f92-b08d-a4d527f5c25a\n# \u2560\u255072aeaa39-6529-4fc3-ba86-f2b113a5a075\n# \u2560\u2550a7802df0-6819-4650-a539-ccbb99333e92\n# \u255f\u25008018394c-a123-43c4-aaaf-3d942284e182\n", "meta": {"hexsha": "8d03f9af8158aa606c21b62885711debbd32118d", "size": 10747, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Julia_Investigation/data_wrangling_with_julia.jl", "max_stars_repo_name": "pascal-p/julia-notebooks", "max_stars_repo_head_hexsha": "568c884c8b0de8ce34a84e8d1ce5fb6994cf32b8", "max_stars_repo_licenses": ["BSD-3-Clause"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2021-08-01T20:34:56.000Z", "max_stars_repo_stars_event_max_datetime": "2021-08-01T20:34:56.000Z", "max_issues_repo_path": "Julia_Investigation/data_wrangling_with_julia.jl", "max_issues_repo_name": "pascal-p/julia-notebooks", "max_issues_repo_head_hexsha": "568c884c8b0de8ce34a84e8d1ce5fb6994cf32b8", "max_issues_repo_licenses": ["BSD-3-Clause"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Julia_Investigation/data_wrangling_with_julia.jl", "max_forks_repo_name": "pascal-p/julia-notebooks", "max_forks_repo_head_hexsha": "568c884c8b0de8ce34a84e8d1ce5fb6994cf32b8", "max_forks_repo_licenses": ["BSD-3-Clause"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2022-01-10T09:03:18.000Z", "max_forks_repo_forks_event_max_datetime": "2022-01-10T09:03:18.000Z", "avg_line_length": 25.8341346154, "max_line_length": 102, "alphanum_fraction": 0.6655810924, "num_tokens": 4524, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.403566839388498, "lm_q2_score": 0.19436782035217448, "lm_q1q2_score": 0.07844040693835844}}
{"text": "using HTTP, JSON, DataFrames, CSV\n\n\n\"\"\"\nReturns total num of escaped chars and lines for any cell, including any outputs\n\"\"\"\nfunction get_cell_counts_shared(cell)\n char_total, lines_total = 0, 0\n for line in cell[\"source\"]\n char_total += length(String(line))\n end\n\n # Count outputs if they exist\n if haskey(cell, \"outputs\")\n char_outputs, lines_outputs = get_counts_outputs(cell[\"outputs\"])\n char_total += char_outputs\n lines_total += lines_outputs\n end\n\n lines_total += length(cell[\"source\"])\n return char_total, lines_total\nend\n\n\n\"\"\"\nReturns number of escaped chars and lines from an array of outputs. It considers only the following:\n - The `text` portion of an `stdout` output\n - A `text/plain` portion of a `data` output\n\"\"\"\nfunction get_counts_outputs(outputs)\n char_total, line_total = 0, 0\n\n # Loops through lines, checks if keys exist, add to counts if they do\n for output in outputs\n if haskey(output, \"name\") && output[\"name\"] == \"stdout\"\n for line in output[\"text\"]\n char_total += length(line)\n end\n line_total += length(output[\"text\"])\n elseif haskey(output, \"data\") && haskey(output[\"data\"], \"text/plain\")\n for line in output[\"data\"][\"text/plain\"]\n char_total += length(line)\n end\n line_total += length(output[\"data\"][\"text/plain\"])\n end\n end\n return char_total, line_total\nend\n\n\"\"\"\nReturns array of counts for #, ##, ###, and #### (respectively) in a cell.\n\"\"\"\nfunction get_cell_counts_markdown(cell)\n count = zeros(Int32, 4)\n for line in cell[\"source\"]\n # Loops through each index of the above array, also using it to count the number of # in a row\n for i in eachindex(count)\n # Considers a # surrounded by non-#es a match. Also takes into account BoL and EoL.\n count[i] += length(collect(m.match for m in eachmatch(Regex(\"^#{$i}[^#]|[^#]#{$i}[^#]|[^#]#{$i}^\"), String(line))))\n end\n end\n return count\nend\n\n\"\"\"\nReturns array of counts for `return`, `for`, `if`, and `using` (respectively) in a cell\n\"\"\"\nfunction get_cell_counts_code(cell)\n matches = [\"return\", \"for\", \"if\", \"using\"]\n count = zeros(Int32, 4)\n for line in cell[\"source\"]\n # Loop through array of matches, add each match to the count\n for i in eachindex(matches)\n count[i] += length(collect(m.match for m in eachmatch(Regex(matches[i]), String(line))))\n end\n end\n return count\nend\n\n\"\"\"\nReturns a formatted, processed row representing a markdown cell\n\"\"\"\nfunction process_cell_markdown(cell_number, markdown_cell)\n return [cell_number, get_cell_counts_shared(markdown_cell)..., get_cell_counts_markdown(markdown_cell)...]\nend\n\n\"\"\"\nReturns a formatted, processed row representing a code cell\n\"\"\"\nfunction process_cell_code(cell_number, code_cell)\n return [cell_number, get_cell_counts_shared(code_cell)..., get_cell_counts_code(code_cell)...]\nend\n\n\"\"\"\nIterates through all given cells, formats the markdown and code cells into two separate DataFrames, returns them.\n\"\"\"\nfunction process_cells_all(all_cells)\n markdown_df = DataFrame(cell_number = Int[], character_count = Int[], line_count = Int[], one_hash = Int[], two_hash = Int[], three_hash = Int[], four_hash = Int[])\n code_df = DataFrame(cell_number = Int[], character_count = Int[], line_count = Int[], returns = Int[], fors = Int[], ifs = Int[], usings = Int[])\n for i in 1:lastindex(all_cells)\n cell = all_cells[i]\n if cell[\"cell_type\"] == \"markdown\"\n push!(markdown_df, process_cell_markdown(i, cell))\n elseif cell[\"cell_type\"] == \"code\"\n push!(code_df, process_cell_code(i, cell))\n end\n end\n return markdown_df, code_df\nend\n\n\n# Basic request - no need to pull out into a function\n\nr = HTTP.request(\"GET\", \"https://raw.githubusercontent.com/yoninazarathy/ProgrammingCourse-with-Julia-SimulationAnalysisAndLearningSystems/main/practicals_jupyter/practical_B_julia_essentials.ipynb\")\nnotebook_json = JSON.parse(String(r.body))\n\n\n# Print the summary (with the help of an occasional inline)\n\nprintln(\"Total number of cells: \" * string(length(notebook_json[\"cells\"])))\n\ncode_cells_count = count(cell->(cell[\"cell_type\"] == \"code\"), notebook_json[\"cells\"])\nprintln(\"Number of code cells: \" * string(code_cells_count))\n\nprintln(\"Number of markdown cells: \" * string(count(cell->(cell[\"cell_type\"] == \"markdown\"), notebook_json[\"cells\"])))\n\ncode_cells_nooutput_count = count(cell->(cell[\"cell_type\"] == \"code\" && length(cell[\"outputs\"]) == 0), notebook_json[\"cells\"])\nprintln(\"Number of code cells w/o output: \" * string(code_cells_nooutput_count))\nprintln(\"Number of code cells w/ output: \" * string(code_cells_count - code_cells_nooutput_count))\n\n\n\"\"\"\nQuick function that takes a list of cells and iterates through their outputs, returning a somewhat comprehensive list of keys\n\"\"\"\nfunction total_keys_in_outputs(cells)\n unique_keys = Set()\n for cell in cells\n if haskey(cell, \"outputs\")\n for output in cell[\"outputs\"]\n union!(unique_keys, Set(keys(output)))\n for key in keys(output)\n if isa(output[key], Dict)\n union!(unique_keys, Set(keys(output[key])))\n end\n end\n end\n end\n end\n return unique_keys\nend\n\n\n# It always worried me whether I missed an edge case in the outputs - so, this is my chosen 'extra summary'\n\nprintln(\"Number of unique keys present in output fields: \" * string(length(total_keys_in_outputs(notebook_json[\"cells\"]))))\n\n# Process cells, chuck em into the csvs with proper formatting\n\nmarkdown_df, code_df = process_cells_all(notebook_json[\"cells\"])\n\nCSV.write(\"markdown_summary.csv\", markdown_df; header = [\"cell_number\", \"character_count\", \"line_count\", \"#\", \"##\", \"###\", \"####\"])\n\nCSV.write(\"code_summary.csv\", code_df; header = [\"cell_number\", \"character_count\", \"line_count\", \"return\", \"for\", \"if\", \"using\"])\n\n\n# Read the files back, then print them out with decent formatting\nmarkdown_csv, code_csv = CSV.File(\"markdown_summary.csv\"; limit=4, header=0) |> DataFrame, CSV.File(\"code_summary.csv\"; limit=4, header=0) |> DataFrame\nprintln(markdown_csv)\nprintln(code_csv)", "meta": {"hexsha": "00de7977c4f1ff5280392e0df4073cedb2eece1d", "size": 6337, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "q2/q2_solution.jl", "max_stars_repo_name": "XxX-Daniil-underscore-Zaikin-XxX/Daniil-Zaikin-2504-2021-HW2", "max_stars_repo_head_hexsha": "bda2cc752f2dd1df1d4982865fe090e05e846dd6", "max_stars_repo_licenses": ["Unlicense"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "q2/q2_solution.jl", "max_issues_repo_name": "XxX-Daniil-underscore-Zaikin-XxX/Daniil-Zaikin-2504-2021-HW2", "max_issues_repo_head_hexsha": "bda2cc752f2dd1df1d4982865fe090e05e846dd6", "max_issues_repo_licenses": ["Unlicense"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "q2/q2_solution.jl", "max_forks_repo_name": "XxX-Daniil-underscore-Zaikin-XxX/Daniil-Zaikin-2504-2021-HW2", "max_forks_repo_head_hexsha": "bda2cc752f2dd1df1d4982865fe090e05e846dd6", "max_forks_repo_licenses": ["Unlicense"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 37.4970414201, "max_line_length": 199, "alphanum_fraction": 0.6684551049, "num_tokens": 1501, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.28140560742914383, "lm_q2_score": 0.27825678771720325, "lm_q1q2_score": 0.0783030203688419}}
{"text": "## formatting conveniences\n\n\"markdown can leave wrapping p's\"\nfunction strip_p(txt)\n if occursin(r\"^\", txt)\n txt = replace(replace(txt, r\"^
\" => \"\"), r\"
$\" => \"\")\n end\n txt\nend\n\nfunction md(x)\n out = sprint(io -> show(io, \"text/html\", Markdown.parse(string(x))))\n strip_p(out)\nend\n\nmarkdown_to_latex(x) = sprint(io -> show(io, \"text/latex\", Markdown.parse(x)))\n\n\"\"\"\nHide output and input, but execute cell.\n\nExamples\n```\n2 + 2\nInvisible()\n```\n\"\"\"\nmutable struct Invisible\nend\n\n\"\"\"\nShow output as HTML\n\nExamples\n```\nHTMLoutput(\"em \")\n```\n\n\"\"\"\nmutable struct HTMLoutput\n x\nend\nBase.show(io::IO, ::MIME\"text/plain\", x::HTMLoutput) = print(io, \"\"\"$(x.x)
\"\"\")\nBase.show(io::IO, ::MIME\"text/html\", x::HTMLoutput) = print(io, x.x)\nBase.show(io::IO, ::MIME\"text/latex\", x::HTMLoutput) = println(io, \"...unable to display raw html...\")\n\n\n\n\"\"\"\nShow as input, but do not execute.\nExamples:\n```\nVerbatim(\"This will print, but not be executed\")\n```\n\"\"\"\nmutable struct Verbatim\n x\nend\nBase.show(io::IO, ::MIME\"text/plain\", x::Verbatim) = print(io, \"\"\"$(x.x) \"\"\")\nBase.show(io::IO, ::MIME\"text/html\", x::Verbatim) = print(io, x.x)\nBase.show(io::IO, ::MIME\"text/latex\", x::Verbatim) = print(io, \"\\verb@$(markdown_to_latex(x.x))@\")\n\n\n\"\"\"\nHide input, but show output\n\nExamples\n```\nx = 2 + 2\nOutputonly(x)\n```\n\"\"\"\nmutable struct Outputonly\n x\nend\n\n\n\"\"\"\n JSXGraph(f; [ID], [CLASS], [WIDTH], [HEIGHT]\n\nShow jsxgraph commands contained in file `f`.\n\"\"\"\nfunction JSXGraph(f, caption=\"JSXGraph Demo\"; ID=\"jsxgraph\", CLASS=\"jsxgraph\", WIDTH=600, HEIGHT=400)\n JSXGRAPH(read(f, String),\n markdown(caption),\n ID, CLASS, WIDTH, HEIGHT)\nend\n\nmutable struct JSXGRAPH\n FILE_CONTENTS\n CAPTION\n ID\n CLASS\n WIDTH\n HEIGHT\nend\n\n#\n\n\n## XXX Put in centered\njsxgraph_tpl = Mustache.mt\"\"\"\n\n\n\n\n\n\n\n\n\"\"\"\n\nBase.show(io::IO, ::MIME\"text/html\", x::JSXGRAPH) = Mustache.render(io, jsxgraph_tpl, x)\nBase.show(io::IO, x::JSXGRAPH) = print(io, \"JSXGraph unavailable\")\n\n## Bootstrap things\nabstract type Bootstrap end\nBase.show(io::IO, ::MIME\"text/html\", x::Bootstrap) = print(io, \"\"\"$(x.x)\"\"\")\nBase.show(io::IO, ::MIME\"text/latex\", x::Bootstrap) = print(io, \"\"\"XXX BOOTSTRAP $(markdown_to_latex(x.x))\"\"\")\n\nmutable struct Alert <: Bootstrap\n x\n d::Dict\nend\n\n### An alert\nfunction alert(txt; kwargs...)\n d = Dict()\n for (k,v) in kwargs\n d[k] = v\n end\n Alert(txt, d)\nend\n\nwarning(txt; kwargs...) = alert(txt; class=\"warning\", kwargs...)\nnote(txt; kwargs...) = alert(txt; class=\"info\", kwargs...)\n\n\nfunction Base.show(io::IO, ::MIME\"text/html\", x::Alert)\n cls = haskey(x.d,:class) ? x.d[:class] : \"success\"\n txt = sprint(io -> show(io, \"text/html\", Markdown.parse(x.x)))\n tpl = \"\"\"$txt
\"\"\"\n\n print(io, tpl)\nend\n\nfunction Base.show(io::IO, ::MIME\"text/latex\", x::Alert)\n println(io, \"\"\"\n\\\\begin{mdframed}\n$(markdown_to_latex(x.x))\n\\\\end{mdframed}\n\"\"\")\nend\n\n\n\n\nmutable struct Example <: Bootstrap\n x\n d::Dict\nend\n\n## use nm=\"name\" to pass along name\nfunction example(txt; kwargs...)\n d = Dict()\n for (k,v) in kwargs\n d[k] = v\n end\n Example(txt, d)\nend\n\n\nfunction Base.show(io::IO, ::MIME\"text/html\", x::Example)\n nm = haskey(x.d,:nm) ? \" $(x.d[:nm]) \" : \"\"\n txt = sprint(io -> show(io, \"text/html\", Markdown.parse(x.x)))\n tpl = \"\"\"\n\n\nexample: $nm$txt\n\n
\n\n\"\"\"\n\n print(io, tpl)\nend\n\nfunction Base.show(io::IO, ::MIME\"text/latex\", x::Example)\n println(io, \"\"\"Example $(markdown_to_latex(x.x))\"\"\")\nend\n\n\nmutable struct Popup <: Bootstrap\n x\n title\n icon\n label\nend\n\n\"\"\"\n\nCreate a button to toggle the display of more detail.\n\nCan modify text, title, icon and label (for the button)\n\nThe text, title, and label can use Markdown.\n\nLaTeX markup does not work, as MathJax rendering is not supported in the popup.\n\n\"\"\"\npopup(x; title=\" \", icon=\"star\", label=\"click me\") = Popup(x, title, icon, label)\n\n\npopup_html_tpl = mt\"\"\"\n\n \n\"\"\"\n# issue with formatted content\nfunction Base.show(io::IO, ::MIME\"text/html\", x::Popup)\n d = Dict()\n d[\"title\"] = x.title #sprint(io -> show(io, \"text/html\", Markdown.parse(x.title)))\n d[\"icon\"] = x.icon\n label = x.label #sprint(io -> show(io, \"text/html\", Markdown.parse(x.label)))\n d[\"button_label\"] = label\n d[\"body\"] = x.x #sprint(io -> show(io, \"text/html\", Markdown.parse(x.x)))\n Mustache.render(io, popup_html_tpl, d)\nend\n\nfunction Base.show(io::IO, ::MIME\"text/latex\", x::Popup)\n println(io, \"\"\"\n\\\\begin{quotation}\n$(markdown_to_latex(x.x))\n\\\\end{quotation}\n\"\"\")\nend\n\n\"\"\"\n\nWay to convert rectangular gird of values into a table\n\n\"\"\"\nmutable struct Table <: Bootstrap\n x\nend\ntable(x) = Table(x)\n\ntable_html_tpl=mt\"\"\"\n\n\n
\n{{{:nms}}}\n{{{:body}}}\n
\n
$(join(map(string, vals), \" \")) \"\n for j in 1:n\n val = Base.invokelatest(getindex, x.x, i, j)\n if ismissing(val)\n val = \".\"\n end\n bdy = bdy * \"$(md(val)) \"\n end\n bdy = bdy * \" \\n\"\n end\n d[:body] = bdy\n Mustache.render(io, table_html_tpl, d)\nend\n\nfunction df_to_table(df, label=\"label\", caption=\"caption\")\n nc = size(df, 2)\n perc = string(round(1/nc, digits=2))\n fmt = \"l\" * repeat(\"p{$perc\\\\textwidth}\", nc-1)\n header = join(string.(names(df)), \" & \")\n row = join([\"{{{:$x}}}\" for x in map(string, names(df))], \" & \")\n\ntpl=\"\"\"\n\\\\begin{table}[!ht]\n \\\\centering\n \\\\begin{tabular}{$fmt}\n $header\\\\\\\\\n \\\\midrule\\\\\\\\\n{{#:DF}} $row\\\\\\\\\n{{/:DF}}\n \\\\bottomrule\n \\\\end{tabular}\n \\\\label{tab:$label}\n\n\\\\end{table}\n\"\"\"\n\n Mustache.render(tpl, DF=df)\nend\n\nfunction Base.show(io::IO, ::MIME\"text/latex\", x::Table)\n d = markdown_to_latex.(x.x)\n println(io, df_to_table(d))\nend\n\n\n\nmutable struct NamedTable <: Bootstrap\ndata\nrownames\ncolnames\nend\n\nfunction Base.show(io::IO, ::MIME\"text/html\", x::NamedTable)\n vals = x.data\n cnames = x.colnames\n rnames = x.rownames\n d = Dict()\n d[:nms] = \"$(join(map(string, cnames), \" \")) \", rnames[i],\" \")\n for j in 1:n\n val = Base.invokelatest(getindex, x.data, i, j)\n print(buf, \"\", md(val), \" \")\n end\n println(buf, \"\n#==\n==#\nfooter_html_tpl = \"\"\"\n
\n \n
\n\"\"\"\n", "meta": {"hexsha": "07bebc6157182568c5e39bfbad2b6255d409014b", "size": 10199, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/WeaveSupport/formatting.jl", "max_stars_repo_name": "jverzani/CalculusWithJulia.jl", "max_stars_repo_head_hexsha": "6ee5135e82c11a1f83b024556be55ad6cbf2622d", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 31, "max_stars_repo_stars_event_min_datetime": "2019-08-29T02:00:11.000Z", "max_stars_repo_stars_event_max_datetime": "2022-02-20T11:15:12.000Z", "max_issues_repo_path": "src/WeaveSupport/formatting.jl", "max_issues_repo_name": "jverzani/CalculusWithJulia.jl", "max_issues_repo_head_hexsha": "6ee5135e82c11a1f83b024556be55ad6cbf2622d", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 16, "max_issues_repo_issues_event_min_datetime": "2020-12-03T15:00:01.000Z", "max_issues_repo_issues_event_max_datetime": "2022-03-11T00:57:57.000Z", "max_forks_repo_path": "src/WeaveSupport/formatting.jl", "max_forks_repo_name": "jverzani/CalculusWithJulia.jl", "max_forks_repo_head_hexsha": "6ee5135e82c11a1f83b024556be55ad6cbf2622d", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 10, "max_forks_repo_forks_event_min_datetime": "2020-01-07T10:53:24.000Z", "max_forks_repo_forks_event_max_datetime": "2021-09-15T06:08:38.000Z", "avg_line_length": 21.6539278132, "max_line_length": 112, "alphanum_fraction": 0.6101578586, "num_tokens": 3056, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.2942149597859341, "lm_q2_score": 0.2658804847339313, "lm_q1q2_score": 0.07822601612385825}}
{"text": "# ------------------------------------------------------------\n# We can use a barrier function\nusing BenchmarkTools\n\nfunction double_sum(data)\n total = 0\n for v in data\n total += 2 * v\n end\n return total\nend\n\nfunction double_sum_of_random_data(n)\n data = random_data(n)\n return double_sum(data)\nend\n\n@btime double_sum_of_random_data(100000);\n@btime double_sum_of_random_data(100001);\n\n#=\njulia> @btime double_sum_of_random_data(100000);\n 245.044 \u03bcs (2 allocations: 781.33 KiB)\n\njulia> @btime double_sum_of_random_data(100001);\n 180.454 \u03bcs (2 allocations: 781.39 KiB)\n\n=#\n\n# Another potential issue is the accumulator.\n\n@code_warntype double_sum(rand(Int, 3));\n@code_warntype double_sum(rand(Float64, 3));\n# -- red text for the Float64 call against the `total` variable\n#=\njulia> @code_warntype double_sum(rand(Int, 3));\nVariables\n #self#::Core.Compiler.Const(double_sum, false)\n data::Array{Int64,1}\n total::Int64\n @_4::Union{Nothing, Tuple{Int64,Int64}}\n v::Int64\n\nBody::Int64\n1 \u2500 (total = 0)\n\u2502 %2 = data::Array{Int64,1}\n\u2502 (@_4 = Base.iterate(%2))\n\u2502 %4 = (@_4 === nothing)::Bool\n\u2502 %5 = Base.not_int(%4)::Bool\n\u2514\u2500\u2500 goto #4 if not %5\n2 \u2504 %7 = @_4::Tuple{Int64,Int64}::Tuple{Int64,Int64}\n\u2502 (v = Core.getfield(%7, 1))\n\u2502 %9 = Core.getfield(%7, 2)::Int64\n\u2502 %10 = total::Int64\n\u2502 %11 = (2 * v)::Int64\n\u2502 (total = %10 + %11)\n\u2502 (@_4 = Base.iterate(%2, %9))\n\u2502 %14 = (@_4 === nothing)::Bool\n\u2502 %15 = Base.not_int(%14)::Bool\n\u2514\u2500\u2500 goto #4 if not %15\n3 \u2500 goto #2\n4 \u2504 return total\n\njulia> @code_warntype double_sum(rand(Float64, 3));\nVariables\n #self#::Core.Compiler.Const(double_sum, false)\n data::Array{Float64,1}\n total::Union{Float64, Int64}\n @_4::Union{Nothing, Tuple{Float64,Int64}}\n v::Float64\n\nBody::Union{Float64, Int64}\n1 \u2500 (total = 0)\n\u2502 %2 = data::Array{Float64,1}\n\u2502 (@_4 = Base.iterate(%2))\n\u2502 %4 = (@_4 === nothing)::Bool\n\u2502 %5 = Base.not_int(%4)::Bool\n\u2514\u2500\u2500 goto #4 if not %5\n2 \u2504 %7 = @_4::Tuple{Float64,Int64}::Tuple{Float64,Int64}\n\u2502 (v = Core.getfield(%7, 1))\n\u2502 %9 = Core.getfield(%7, 2)::Int64\n\u2502 %10 = total::Union{Float64, Int64}\n\u2502 %11 = (2 * v)::Float64\n\u2502 (total = %10 + %11)\n\u2502 (@_4 = Base.iterate(%2, %9))\n\u2502 %14 = (@_4 === nothing)::Bool\n\u2502 %15 = Base.not_int(%14)::Bool\n\u2514\u2500\u2500 goto #4 if not %15\n3 \u2500 goto #2\n4 \u2504 return total\n\n=#\n\n# There are better ways to write it.\n\n# Fix 1 - use zero function to match type\nfunction double_sum(data)\n total = zero(eltype(data))\n for v in data\n total += 2 * v\n end\n return total\nend\n\n# check again - no more red text in the output\n@code_warntype double_sum(rand(Int, 3));\n@code_warntype double_sum(rand(Float64, 3));\n\n# Fix 2 - leverage type parameter, more flexible\nfunction double_sum(data::AbstractVector{T}) where {T <: Number}\n total = zero(T)\n for v in data\n total += v\n end\n return total\nend\n\n# check again - no more red text in the output\n@code_warntype double_sum(rand(Int, 3));\n@code_warntype double_sum(rand(Float64, 3));\n\n# -----\n# using zero(s), one(s), and similar\n\n#=\njulia> zero(Int)\n0\n\njulia> zeros(Float64, 5)\n5-element Array{Float64,1}:\n 0.0\n 0.0\n 0.0\n 0.0\n 0.0\n\njulia> zeros(Float64, 5)^C\n\njulia> one(UInt8)\n0x01\n\njulia> ones(UInt8, 5)\n5-element Array{UInt8,1}:\n 0x01\n 0x01\n 0x01\n 0x01\n 0x01\n\njulia> A = rand(3,4)\n3\u00d74 Array{Float64,2}:\n 0.275294 0.333407 0.380679 0.680078\n 0.795881 0.297336 0.478146 0.377958\n 0.534937 0.0791554 0.916814 0.210469\n\njulia> B = similar(A)\n3\u00d74 Array{Float64,2}:\n 9.88131e-324 4.94066e-324 4.94066e-324 4.94066e-324\n 9.88131e-324 4.94066e-324 4.94066e-324 0.0 \n 4.94066e-324 4.94066e-324 4.94066e-324 2.32833e-314\n\njulia> zeros(axes(A))\n3\u00d74 Array{Float64,2}:\n 0.0 0.0 0.0 0.0\n 0.0 0.0 0.0 0.0\n 0.0 0.0 0.0 0.0\n\n=#", "meta": {"hexsha": "edb17e4b47ebcf90ebae58183f749416ac3c453a", "size": 3870, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Chapter06/BarrierFunctionPattern/2_barrier_function.jl", "max_stars_repo_name": "Moelf/Hands-on-Design-Patterns-and-Best-Practices-with-Julia", "max_stars_repo_head_hexsha": "ee95448d05c63db8de3a5e9e27c9351cb6cc0e88", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 167, "max_stars_repo_stars_event_min_datetime": "2020-02-07T14:38:43.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-28T09:33:00.000Z", "max_issues_repo_path": "Chapter06/BarrierFunctionPattern/2_barrier_function.jl", "max_issues_repo_name": "Moelf/Hands-on-Design-Patterns-and-Best-Practices-with-Julia", "max_issues_repo_head_hexsha": "ee95448d05c63db8de3a5e9e27c9351cb6cc0e88", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Chapter06/BarrierFunctionPattern/2_barrier_function.jl", "max_forks_repo_name": "Moelf/Hands-on-Design-Patterns-and-Best-Practices-with-Julia", "max_forks_repo_head_hexsha": "ee95448d05c63db8de3a5e9e27c9351cb6cc0e88", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 33, "max_forks_repo_forks_event_min_datetime": "2020-02-14T05:17:17.000Z", "max_forks_repo_forks_event_max_datetime": "2022-01-24T07:38:59.000Z", "avg_line_length": 23.0357142857, "max_line_length": 64, "alphanum_fraction": 0.6072351421, "num_tokens": 1485, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4339814648038986, "lm_q2_score": 0.1801066618860355, "lm_q1q2_score": 0.07816295294624218}}
{"text": "### A Pluto.jl notebook ###\n# v0.16.4\n\nusing Markdown\nusing InteractiveUtils\n\n# This Pluto notebook uses @bind for interactivity. When running this notebook outside of Pluto, the following 'mock version' of @bind gives bound variables a default value (instead of an error).\nmacro bind(def, element)\n quote\n local el = $(esc(element))\n global $(esc(def)) = Core.applicable(Base.get, el) ? Base.get(el) : missing\n el\n end\nend\n\n# \u2554\u2550\u2561 e4dca908-1016-11ec-0b7f-e9d01bd4e1b2\nmd\"\"\"\n# Solution Explorer\nThis is an interactive visualization of by-county solutions. Solutions\nare taken from the `out` directory in the repository root, and can be\nproduced by executing `m-core/sweep.m` in MatLab.\n\n*Note:* currently, only county-level solutions are supported. Solutions aggregated\nto a different level will not be displayed.\n\n---\nAuthors: Kara Ignatenko, 2021;\n\t\t Yaroslav Salii, 2021\n\n\nChangelog:\n- 2021-10-09 v.1.0: First complete version\n- 2021-10-24 v.1.1: Add control effort choropleth \n\n\"\"\"\n\n# \u2554\u2550\u2561 3389dc98-b90f-4c6a-8c96-31840b0516ea\n# a workaround to allow dynamically reloaded includes\n# --refresh this cell to reload the included files\nmodule includes\n\tinclude(\"viz-ui.jl\")\n\tinclude(\"vega-specs.jl\")\n\tinclude(\"exporter.jl\")\nend\n\n# \u2554\u2550\u2561 e598c897-61bf-4bc2-91db-e225362f5606\n# Start off by importing all the stuff\nbegin\n\tusing VegaLite,VegaDatasets\n\tusing DataFrames, CSV, Statistics, Revise\n\tusing PlutoUI, HypertextLiteral\n\t\n\tUI = includes.VizUI\n\tSpecs = includes.VegaSpecs\n\tExp = includes.Exporter\n\t\t\n\tthisPath = splitpath(@__DIR__)\n\tprojRoot = thisPath[1:findfirst(isequal(\"epi-net-m\"), thisPath)]\n\t\n\tconst iDir = joinpath(projRoot...,\"out\") #here be the simulator's outputs\n\t\nend\n\n# \u2554\u2550\u2561 0d909765-a87d-4a3c-bb9f-ff80e463992a\n#TODO: file selector based on the contents of /out\nbegin\n\t#get the list of csv files in ../out\n\toutDir = readdir(iDir)\n\tcsvs = filter(endswith(\".csv\"), outDir)\n\t#get the list of unique solution names (prefixes)\n\tsolNames = map(fname -> split(fname, \"-\")[1], csvs) |> unique\n\tfilter!(contains(\"cty\"), solNames)\n\t\n\t#create a
to pick from the prefixes \n\tpicker = @bind slnName Select(solNames)\n\t\n\tmd\"\"\"\n\tSelect solution: $(picker)\n\t\"\"\"\nend\n\n# \u2554\u2550\u2561 ced90367-407c-4b67-8276-68edc17933d3\n\"\"\"\nRead the solutions into dataframes and stuff them into a named tuple \n:f,:f0 are *fractional* per-node per-day s/z/r\n:a,:a0 are *absolute* per-node per-day S/Z/R \n:avgc are per-day average infected fraction z, zNull, and average optimal control effort u\n\"\"\"\nfunction rdSolutions(iName, slnDir)\n\tslnSuffs = [\"-frac.csv\",\"-frac0.csv\",\"-abs.csv\",\"-abs0.csv\",\"-avg.csv\"]\n slns= map( p -> CSV.read(p,DataFrame), (joinpath(iDir,slnName * suff) for suff in slnSuffs))\n return NamedTuple([:f,:f0,:a,:a0, :avgc] .=> slns) \nend\n\n# \u2554\u2550\u2561 6a2ca31f-716c-48f4-8e1a-70e3c033358b\ntry\n\tss = rdSolutions(slnName, iDir)\n\t\n\tglobal a0Median = ss.a0.Z180 |> median\n\tglobal a0Max = ss.a0.Z180 |> maximum\n\t\n\t#get rid of irrelevant (for now) data by only selecting the Z values\n\ta_clean = select(ss.a, \"id\", r\"^Z\")\n\ta0_clean = select(ss.a0, \"id\", r\"^Z\")\n\tf_clean = select(ss.f, \"id\", r\"^u\")\n\t\n\t#toss everything into one dataframe to simplify rendering\n\t#for the null case, add _NULL to the column names\n\tglobal sol = innerjoin(a_clean, a0_clean, on=\"id\", renamecols=(\"\" => \"_NULL\"))\n\t#add *control effort* data; f_clean should be fine too\n\tglobal sol2 = innerjoin(sol, f_clean, on=\"id\") \n\t\n\t\n\t#\"-avg.csv\" has 3 columns: z_avg; zNull_avg; u_avg, and per-day rows. \n\t#insert day numbers\n\tinsertcols!(ss.avgc,1,:day => 0:(nrow(ss.avgc)-1))\n\tglobal long_avgc = stack(ss.avgc,[:z_avg,:zNull_avg,:u_avg],variable_name=:symbol)\n\t\t\t\n\tmd\"Solution files read successfully!\"\ncatch err\n\tmd\"**Error while reading solution files:** $(err)\"\nend\n\n# \u2554\u2550\u2561 dcabd381-1316-449b-9ebf-68410ee6e3fd\n@bind day UI.dayPicker(0, 180, [1, 10])\n\n# \u2554\u2550\u2561 cc533197-dbe3-41b6-8478-64f82da52ba8\nmd\"Day selected: $day\"\n\n# \u2554\u2550\u2561 8398bb71-aa8e-42b6-b3e3-c08ca57ee5b0\nbegin\n\tplt_u = Specs.pltCtrlByCty(sol2,day)\n\tUI.vegaEmbed(plt_u)\nend\n\n# \u2554\u2550\u2561 a6a64f62-316a-48f5-bd5c-0eebff53022d\nbegin\n\tpltZ = Specs.pltZOptVsNullByCty(sol, day, a0Median)\n\tUI.vegaEmbed(pltZ)\nend\n\n# \u2554\u2550\u2561 f4bf1143-c315-4521-b19b-06bf0373828a\nbegin\n\tpltAvgZandU = Specs.pltAvgInfdCtrl(long_avgc)\n\tUI.vegaEmbed(pltAvgZandU)\nend\n\n# \u2554\u2550\u2561 86ddda30-351d-41f6-94b2-4c84dea08448\nbegin \n\tsel = @bind format Select([\"svg\", \"pdf\"])\n\tcbx = @bind exportMap UI.booleanButton(\"Export map\")\n\tcbx2 = @bind exportPlot UI.booleanButton(\"Export avg. plot\")\n\tfigDir = Exp.figDir \n\t@htl(\"\"\"Select export format: $sel $cbx $cbx2 \u2013 Will be saved to $figDir\"\"\")\nend\n\n# \u2554\u2550\u2561 bfd9753c-323a-4a82-b5e6-057e7519ed6c\nif exportMap\n\tExp.savePlot(slnName * \"_day$(day)_\", pltZ, format)\nend\n\n# \u2554\u2550\u2561 ff5a5927-6bc6-4b05-b0e8-e0bfdc623440\nif exportPlot\n\tExp.savePlot(slnName * \"_avgZ_\", pltAvgZandU, format)\nend\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000001\nPLUTO_PROJECT_TOML_CONTENTS = \"\"\"\n[deps]\nCSV = \"336ed68f-0bac-5ca0-87d4-7b16caf5d00b\"\nDataFrames = \"a93c6f00-e57d-5684-b7b6-d8193f3e46c0\"\nHypertextLiteral = \"ac1192a8-f4b3-4bfe-ba22-af5b92cd3ab2\"\nPlutoUI = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\nRevise = \"295af30f-e4ad-537b-8983-00126c2a3abe\"\nStatistics = \"10745b16-79ce-11e8-11f9-7d13ad32a3b2\"\nVegaDatasets = \"0ae4a718-28b7-58ec-9efb-cded64d6d5b4\"\nVegaLite = \"112f6efa-9a02-5b7d-90c0-432ed331239a\"\n\n[compat]\nCSV = \"~0.8.5\"\nDataFrames = \"~1.2.2\"\nHypertextLiteral = \"~0.9.0\"\nPlutoUI = \"~0.7.9\"\nRevise = \"~3.1.19\"\nVegaDatasets = \"~2.1.1\"\nVegaLite = \"~2.6.0\"\n\"\"\"\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000002\nPLUTO_MANIFEST_TOML_CONTENTS = \"\"\"\n# This file is machine-generated - editing it directly is not advised\n\n[[ArgTools]]\nuuid = \"0dad84c5-d112-42e6-8d28-ef12dabb789f\"\n\n[[Artifacts]]\nuuid = \"56f22d72-fd6d-98f1-02f0-08ddc0907c33\"\n\n[[Base64]]\nuuid = \"2a0f44e3-6c83-55bd-87e4-b1978d98bd5f\"\n\n[[CSV]]\ndeps = [\"Dates\", \"Mmap\", \"Parsers\", \"PooledArrays\", \"SentinelArrays\", \"Tables\", \"Unicode\"]\ngit-tree-sha1 = \"b83aa3f513be680454437a0eee21001607e5d983\"\nuuid = \"336ed68f-0bac-5ca0-87d4-7b16caf5d00b\"\nversion = \"0.8.5\"\n\n[[ChainRulesCore]]\ndeps = [\"Compat\", \"LinearAlgebra\", \"SparseArrays\"]\ngit-tree-sha1 = \"30ee06de5ff870b45c78f529a6b093b3323256a3\"\nuuid = \"d360d2e6-b24c-11e9-a2a3-2a2ae2dbcce4\"\nversion = \"1.3.1\"\n\n[[CodeTracking]]\ndeps = [\"InteractiveUtils\", \"UUIDs\"]\ngit-tree-sha1 = \"9aa8a5ebb6b5bf469a7e0e2b5202cf6f8c291104\"\nuuid = \"da1fd8a2-8d9e-5ec2-8556-3022fb5608a2\"\nversion = \"1.0.6\"\n\n[[CodecZlib]]\ndeps = [\"TranscodingStreams\", \"Zlib_jll\"]\ngit-tree-sha1 = \"ded953804d019afa9a3f98981d99b33e3db7b6da\"\nuuid = \"944b1d66-785c-5afd-91f1-9de20f533193\"\nversion = \"0.7.0\"\n\n[[Compat]]\ndeps = [\"Base64\", \"Dates\", \"DelimitedFiles\", \"Distributed\", \"InteractiveUtils\", \"LibGit2\", \"Libdl\", \"LinearAlgebra\", \"Markdown\", \"Mmap\", \"Pkg\", \"Printf\", \"REPL\", \"Random\", \"SHA\", \"Serialization\", \"SharedArrays\", \"Sockets\", \"SparseArrays\", \"Statistics\", \"Test\", \"UUIDs\", \"Unicode\"]\ngit-tree-sha1 = \"727e463cfebd0c7b999bbf3e9e7e16f254b94193\"\nuuid = \"34da2185-b29b-5c13-b0c7-acf172513d20\"\nversion = \"3.34.0\"\n\n[[CompilerSupportLibraries_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"e66e0078-7015-5450-92f7-15fbd957f2ae\"\n\n[[ConstructionBase]]\ndeps = [\"LinearAlgebra\"]\ngit-tree-sha1 = \"f74e9d5388b8620b4cee35d4c5a618dd4dc547f4\"\nuuid = \"187b0558-2788-49d3-abe0-74a17ed4e7c9\"\nversion = \"1.3.0\"\n\n[[Crayons]]\ngit-tree-sha1 = \"3f71217b538d7aaee0b69ab47d9b7724ca8afa0d\"\nuuid = \"a8cc5b0e-0ffa-5ad4-8c14-923d3ee1735f\"\nversion = \"4.0.4\"\n\n[[DataAPI]]\ngit-tree-sha1 = \"bec2532f8adb82005476c141ec23e921fc20971b\"\nuuid = \"9a962f9c-6df0-11e9-0e5d-c546b8b5ee8a\"\nversion = \"1.8.0\"\n\n[[DataFrames]]\ndeps = [\"Compat\", \"DataAPI\", \"Future\", \"InvertedIndices\", \"IteratorInterfaceExtensions\", \"LinearAlgebra\", \"Markdown\", \"Missings\", \"PooledArrays\", \"PrettyTables\", \"Printf\", \"REPL\", \"Reexport\", \"SortingAlgorithms\", \"Statistics\", \"TableTraits\", \"Tables\", \"Unicode\"]\ngit-tree-sha1 = \"d785f42445b63fc86caa08bb9a9351008be9b765\"\nuuid = \"a93c6f00-e57d-5684-b7b6-d8193f3e46c0\"\nversion = \"1.2.2\"\n\n[[DataStructures]]\ndeps = [\"Compat\", \"InteractiveUtils\", \"OrderedCollections\"]\ngit-tree-sha1 = \"7d9d316f04214f7efdbb6398d545446e246eff02\"\nuuid = \"864edb3b-99cc-5e75-8d2d-829cb0a9cfe8\"\nversion = \"0.18.10\"\n\n[[DataValueInterfaces]]\ngit-tree-sha1 = \"bfc1187b79289637fa0ef6d4436ebdfe6905cbd6\"\nuuid = \"e2d170a0-9d28-54be-80f0-106bbe20a464\"\nversion = \"1.0.0\"\n\n[[DataValues]]\ndeps = [\"DataValueInterfaces\", \"Dates\"]\ngit-tree-sha1 = \"d88a19299eba280a6d062e135a43f00323ae70bf\"\nuuid = \"e7dc6d0d-1eca-5fa6-8ad6-5aecde8b7ea5\"\nversion = \"0.4.13\"\n\n[[Dates]]\ndeps = [\"Printf\"]\nuuid = \"ade2ca70-3891-5945-98fb-dc099432e06a\"\n\n[[DelimitedFiles]]\ndeps = [\"Mmap\"]\nuuid = \"8bb1440f-4735-579b-a4ab-409b98df4dab\"\n\n[[Distributed]]\ndeps = [\"Random\", \"Serialization\", \"Sockets\"]\nuuid = \"8ba89e20-285c-5b6f-9357-94700520ee1b\"\n\n[[DocStringExtensions]]\ndeps = [\"LibGit2\"]\ngit-tree-sha1 = \"a32185f5428d3986f47c2ab78b1f216d5e6cc96f\"\nuuid = \"ffbed154-4ef7-542d-bbb7-c09d3a79fcae\"\nversion = \"0.8.5\"\n\n[[DoubleFloats]]\ndeps = [\"GenericLinearAlgebra\", \"LinearAlgebra\", \"Polynomials\", \"Printf\", \"Quadmath\", \"Random\", \"Requires\", \"SpecialFunctions\"]\ngit-tree-sha1 = \"1c962cf7e75c09a5f1fbf504df7d6a06447a1129\"\nuuid = \"497a8b3b-efae-58df-a0af-a86822472b78\"\nversion = \"1.1.23\"\n\n[[Downloads]]\ndeps = [\"ArgTools\", \"LibCURL\", \"NetworkOptions\"]\nuuid = \"f43a241f-c20a-4ad4-852c-f6b1247861c6\"\n\n[[ExprTools]]\ngit-tree-sha1 = \"b7e3d17636b348f005f11040025ae8c6f645fe92\"\nuuid = \"e2ba6199-217a-4e67-a87a-7c52f15ade04\"\nversion = \"0.1.6\"\n\n[[FileIO]]\ndeps = [\"Pkg\", \"Requires\", \"UUIDs\"]\ngit-tree-sha1 = \"937c29268e405b6808d958a9ac41bfe1a31b08e7\"\nuuid = \"5789e2e9-d7fb-5bc7-8068-2c6fae9b9549\"\nversion = \"1.11.0\"\n\n[[FilePaths]]\ndeps = [\"FilePathsBase\", \"MacroTools\", \"Reexport\", \"Requires\"]\ngit-tree-sha1 = \"919d9412dbf53a2e6fe74af62a73ceed0bce0629\"\nuuid = \"8fc22ac5-c921-52a6-82fd-178b2807b824\"\nversion = \"0.8.3\"\n\n[[FilePathsBase]]\ndeps = [\"Dates\", \"Mmap\", \"Printf\", \"Test\", \"UUIDs\"]\ngit-tree-sha1 = \"0f5e8d0cb91a6386ba47bd1527b240bd5725fbae\"\nuuid = \"48062228-2e41-5def-b9a4-89aafe57970f\"\nversion = \"0.9.10\"\n\n[[FileWatching]]\nuuid = \"7b1f6079-737a-58dc-b8bc-7a2ca5c1b5ee\"\n\n[[Formatting]]\ndeps = [\"Printf\"]\ngit-tree-sha1 = \"8339d61043228fdd3eb658d86c926cb282ae72a8\"\nuuid = \"59287772-0a20-5a39-b81b-1366585eb4c0\"\nversion = \"0.4.2\"\n\n[[Future]]\ndeps = [\"Random\"]\nuuid = \"9fa8497b-333b-5362-9e8d-4d0656e87820\"\n\n[[GenericLinearAlgebra]]\ndeps = [\"LinearAlgebra\", \"Printf\", \"Random\"]\ngit-tree-sha1 = \"ff291c1827030ffaacaf53e3c83ed92d4d5e6fb6\"\nuuid = \"14197337-ba66-59df-a3e3-ca00e7dcff7a\"\nversion = \"0.2.5\"\n\n[[HTTP]]\ndeps = [\"Base64\", \"Dates\", \"IniFile\", \"MbedTLS\", \"Sockets\"]\ngit-tree-sha1 = \"c7ec02c4c6a039a98a15f955462cd7aea5df4508\"\nuuid = \"cd3eb016-35fb-5094-929b-558a96fad6f3\"\nversion = \"0.8.19\"\n\n[[HypertextLiteral]]\ngit-tree-sha1 = \"72053798e1be56026b81d4e2682dbe58922e5ec9\"\nuuid = \"ac1192a8-f4b3-4bfe-ba22-af5b92cd3ab2\"\nversion = \"0.9.0\"\n\n[[IniFile]]\ndeps = [\"Test\"]\ngit-tree-sha1 = \"098e4d2c533924c921f9f9847274f2ad89e018b8\"\nuuid = \"83e8ac13-25f8-5344-8a64-a9f2b223428f\"\nversion = \"0.5.0\"\n\n[[InteractiveUtils]]\ndeps = [\"Markdown\"]\nuuid = \"b77e0a4c-d291-57a0-90e8-8db25a27a240\"\n\n[[Intervals]]\ndeps = [\"Dates\", \"Printf\", \"RecipesBase\", \"Serialization\", \"TimeZones\"]\ngit-tree-sha1 = \"323a38ed1952d30586d0fe03412cde9399d3618b\"\nuuid = \"d8418881-c3e1-53bb-8760-2df7ec849ed5\"\nversion = \"1.5.0\"\n\n[[InvertedIndices]]\ngit-tree-sha1 = \"bee5f1ef5bf65df56bdd2e40447590b272a5471f\"\nuuid = \"41ab1584-1d38-5bbf-9106-f11c6c58b48f\"\nversion = \"1.1.0\"\n\n[[IrrationalConstants]]\ngit-tree-sha1 = \"f76424439413893a832026ca355fe273e93bce94\"\nuuid = \"92d709cd-6900-40b7-9082-c6be49f344b6\"\nversion = \"0.1.0\"\n\n[[IterableTables]]\ndeps = [\"DataValues\", \"IteratorInterfaceExtensions\", \"Requires\", \"TableTraits\", \"TableTraitsUtils\"]\ngit-tree-sha1 = \"70300b876b2cebde43ebc0df42bc8c94a144e1b4\"\nuuid = \"1c8ee90f-4401-5389-894e-7a04a3dc0f4d\"\nversion = \"1.0.0\"\n\n[[IteratorInterfaceExtensions]]\ngit-tree-sha1 = \"a3f24677c21f5bbe9d2a714f95dcd58337fb2856\"\nuuid = \"82899510-4779-5014-852e-03e436cf321d\"\nversion = \"1.0.0\"\n\n[[JLLWrappers]]\ndeps = [\"Preferences\"]\ngit-tree-sha1 = \"642a199af8b68253517b80bd3bfd17eb4e84df6e\"\nuuid = \"692b3bcd-3c85-4b1f-b108-f13ce0eb3210\"\nversion = \"1.3.0\"\n\n[[JSON]]\ndeps = [\"Dates\", \"Mmap\", \"Parsers\", \"Unicode\"]\ngit-tree-sha1 = \"8076680b162ada2a031f707ac7b4953e30667a37\"\nuuid = \"682c06a0-de6a-54ab-a142-c8b1cf79cde6\"\nversion = \"0.21.2\"\n\n[[JSONSchema]]\ndeps = [\"HTTP\", \"JSON\", \"ZipFile\"]\ngit-tree-sha1 = \"b84ab8139afde82c7c65ba2b792fe12e01dd7307\"\nuuid = \"7d188eb4-7ad8-530c-ae41-71a32a6d4692\"\nversion = \"0.3.3\"\n\n[[JuliaInterpreter]]\ndeps = [\"CodeTracking\", \"InteractiveUtils\", \"Random\", \"UUIDs\"]\ngit-tree-sha1 = \"e273807f38074f033d94207a201e6e827d8417db\"\nuuid = \"aa1ae85d-cabe-5617-a682-6adf51b2e16a\"\nversion = \"0.8.21\"\n\n[[LazyArtifacts]]\ndeps = [\"Artifacts\", \"Pkg\"]\nuuid = \"4af54fe1-eca0-43a8-85a7-787d91b784e3\"\n\n[[LibCURL]]\ndeps = [\"LibCURL_jll\", \"MozillaCACerts_jll\"]\nuuid = \"b27032c2-a3e7-50c8-80cd-2d36dbcbfd21\"\n\n[[LibCURL_jll]]\ndeps = [\"Artifacts\", \"LibSSH2_jll\", \"Libdl\", \"MbedTLS_jll\", \"Zlib_jll\", \"nghttp2_jll\"]\nuuid = \"deac9b47-8bc7-5906-a0fe-35ac56dc84c0\"\n\n[[LibGit2]]\ndeps = [\"Base64\", \"NetworkOptions\", \"Printf\", \"SHA\"]\nuuid = \"76f85450-5226-5b5a-8eaa-529ad045b433\"\n\n[[LibSSH2_jll]]\ndeps = [\"Artifacts\", \"Libdl\", \"MbedTLS_jll\"]\nuuid = \"29816b5a-b9ab-546f-933c-edad1886dfa8\"\n\n[[Libdl]]\nuuid = \"8f399da3-3557-5675-b5ff-fb832c97cbdb\"\n\n[[LinearAlgebra]]\ndeps = [\"Libdl\"]\nuuid = \"37e2e46d-f89d-539d-b4ee-838fcccc9c8e\"\n\n[[LogExpFunctions]]\ndeps = [\"ChainRulesCore\", \"DocStringExtensions\", \"IrrationalConstants\", \"LinearAlgebra\"]\ngit-tree-sha1 = \"86197a8ecb06e222d66797b0c2d2f0cc7b69e42b\"\nuuid = \"2ab3a3ac-af41-5b50-aa03-7779005ae688\"\nversion = \"0.3.2\"\n\n[[Logging]]\nuuid = \"56ddb016-857b-54e1-b83d-db4d58db5568\"\n\n[[LoweredCodeUtils]]\ndeps = [\"JuliaInterpreter\"]\ngit-tree-sha1 = \"491a883c4fef1103077a7f648961adbf9c8dd933\"\nuuid = \"6f1432cf-f94c-5a45-995e-cdbf5db27b0b\"\nversion = \"2.1.2\"\n\n[[MacroTools]]\ndeps = [\"Markdown\", \"Random\"]\ngit-tree-sha1 = \"0fb723cd8c45858c22169b2e42269e53271a6df7\"\nuuid = \"1914dd2f-81c6-5fcd-8719-6d5c9610ff09\"\nversion = \"0.5.7\"\n\n[[Markdown]]\ndeps = [\"Base64\"]\nuuid = \"d6f4376e-aef5-505a-96c1-9c027394607a\"\n\n[[MbedTLS]]\ndeps = [\"Dates\", \"MbedTLS_jll\", \"Random\", \"Sockets\"]\ngit-tree-sha1 = \"1c38e51c3d08ef2278062ebceade0e46cefc96fe\"\nuuid = \"739be429-bea8-5141-9913-cc70e7f3736d\"\nversion = \"1.0.3\"\n\n[[MbedTLS_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"c8ffd9c3-330d-5841-b78e-0817d7145fa1\"\n\n[[Missings]]\ndeps = [\"DataAPI\"]\ngit-tree-sha1 = \"2ca267b08821e86c5ef4376cffed98a46c2cb205\"\nuuid = \"e1d29d7a-bbdc-5cf2-9ac0-f12de2c33e28\"\nversion = \"1.0.1\"\n\n[[Mmap]]\nuuid = \"a63ad114-7e13-5084-954f-fe012c677804\"\n\n[[Mocking]]\ndeps = [\"ExprTools\"]\ngit-tree-sha1 = \"748f6e1e4de814b101911e64cc12d83a6af66782\"\nuuid = \"78c3b35d-d492-501b-9361-3d52fe80e533\"\nversion = \"0.7.2\"\n\n[[MozillaCACerts_jll]]\nuuid = \"14a3606d-f60d-562e-9121-12d972cd8159\"\n\n[[MutableArithmetics]]\ndeps = [\"LinearAlgebra\", \"SparseArrays\", \"Test\"]\ngit-tree-sha1 = \"3927848ccebcc165952dc0d9ac9aa274a87bfe01\"\nuuid = \"d8a4904e-b15c-11e9-3269-09a3773c0cb0\"\nversion = \"0.2.20\"\n\n[[NetworkOptions]]\nuuid = \"ca575930-c2e3-43a9-ace4-1e988b2c1908\"\n\n[[NodeJS]]\ndeps = [\"Pkg\"]\ngit-tree-sha1 = \"905224bbdd4b555c69bb964514cfa387616f0d3a\"\nuuid = \"2bd173c7-0d6d-553b-b6af-13a54713934c\"\nversion = \"1.3.0\"\n\n[[Nullables]]\ngit-tree-sha1 = \"8f87854cc8f3685a60689d8edecaa29d2251979b\"\nuuid = \"4d1e1d77-625e-5b40-9113-a560ec7a8ecd\"\nversion = \"1.0.0\"\n\n[[OpenSpecFun_jll]]\ndeps = [\"Artifacts\", \"CompilerSupportLibraries_jll\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"13652491f6856acfd2db29360e1bbcd4565d04f1\"\nuuid = \"efe28fd5-8261-553b-a9e1-b2916fc3738e\"\nversion = \"0.5.5+0\"\n\n[[OrderedCollections]]\ngit-tree-sha1 = \"85f8e6578bf1f9ee0d11e7bb1b1456435479d47c\"\nuuid = \"bac558e1-5e72-5ebc-8fee-abe8a469f55d\"\nversion = \"1.4.1\"\n\n[[Parsers]]\ndeps = [\"Dates\"]\ngit-tree-sha1 = \"bfd7d8c7fd87f04543810d9cbd3995972236ba1b\"\nuuid = \"69de0a69-1ddd-5017-9359-2bf0b02dc9f0\"\nversion = \"1.1.2\"\n\n[[Pkg]]\ndeps = [\"Artifacts\", \"Dates\", \"Downloads\", \"LibGit2\", \"Libdl\", \"Logging\", \"Markdown\", \"Printf\", \"REPL\", \"Random\", \"SHA\", \"Serialization\", \"TOML\", \"Tar\", \"UUIDs\", \"p7zip_jll\"]\nuuid = \"44cfe95a-1eb2-52ea-b672-e2afdf69b78f\"\n\n[[PlutoUI]]\ndeps = [\"Base64\", \"Dates\", \"InteractiveUtils\", \"JSON\", \"Logging\", \"Markdown\", \"Random\", \"Reexport\", \"Suppressor\"]\ngit-tree-sha1 = \"44e225d5837e2a2345e69a1d1e01ac2443ff9fcb\"\nuuid = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\nversion = \"0.7.9\"\n\n[[Polynomials]]\ndeps = [\"Intervals\", \"LinearAlgebra\", \"MutableArithmetics\", \"RecipesBase\"]\ngit-tree-sha1 = \"0bbfdcd8cda81b8144de4be8a67f5717e959a005\"\nuuid = \"f27b6e38-b328-58d1-80ce-0feddd5e7a45\"\nversion = \"2.0.14\"\n\n[[PooledArrays]]\ndeps = [\"DataAPI\", \"Future\"]\ngit-tree-sha1 = \"a193d6ad9c45ada72c14b731a318bedd3c2f00cf\"\nuuid = \"2dfb63ee-cc39-5dd5-95bd-886bf059d720\"\nversion = \"1.3.0\"\n\n[[Preferences]]\ndeps = [\"TOML\"]\ngit-tree-sha1 = \"00cfd92944ca9c760982747e9a1d0d5d86ab1e5a\"\nuuid = \"21216c6a-2e73-6563-6e65-726566657250\"\nversion = \"1.2.2\"\n\n[[PrettyTables]]\ndeps = [\"Crayons\", \"Formatting\", \"Markdown\", \"Reexport\", \"Tables\"]\ngit-tree-sha1 = \"0d1245a357cc61c8cd61934c07447aa569ff22e6\"\nuuid = \"08abe8d2-0d0c-5749-adfa-8a2ac140af0d\"\nversion = \"1.1.0\"\n\n[[Printf]]\ndeps = [\"Unicode\"]\nuuid = \"de0858da-6303-5e67-8744-51eddeeeb8d7\"\n\n[[Quadmath]]\ndeps = [\"Printf\", \"Random\", \"Requires\"]\ngit-tree-sha1 = \"5a8f74af8eae654086a1d058b4ec94ff192e3de0\"\nuuid = \"be4d8f0f-7fa4-5f49-b795-2f01399ab2dd\"\nversion = \"0.5.5\"\n\n[[REPL]]\ndeps = [\"InteractiveUtils\", \"Markdown\", \"Sockets\", \"Unicode\"]\nuuid = \"3fa0cd96-eef1-5676-8a61-b3b8758bbffb\"\n\n[[Random]]\ndeps = [\"Serialization\"]\nuuid = \"9a3f8284-a2c9-5f02-9a11-845980a1fd5c\"\n\n[[RecipesBase]]\ngit-tree-sha1 = \"44a75aa7a527910ee3d1751d1f0e4148698add9e\"\nuuid = \"3cdcf5f2-1ef4-517c-9805-6587b60abb01\"\nversion = \"1.1.2\"\n\n[[Reexport]]\ngit-tree-sha1 = \"45e428421666073eab6f2da5c9d310d99bb12f9b\"\nuuid = \"189a3867-3050-52da-a836-e630ba90ab69\"\nversion = \"1.2.2\"\n\n[[Requires]]\ndeps = [\"UUIDs\"]\ngit-tree-sha1 = \"4036a3bd08ac7e968e27c203d45f5fff15020621\"\nuuid = \"ae029012-a4dd-5104-9daa-d747884805df\"\nversion = \"1.1.3\"\n\n[[Revise]]\ndeps = [\"CodeTracking\", \"Distributed\", \"FileWatching\", \"JuliaInterpreter\", \"LibGit2\", \"LoweredCodeUtils\", \"OrderedCollections\", \"Pkg\", \"REPL\", \"Requires\", \"UUIDs\", \"Unicode\"]\ngit-tree-sha1 = \"1947d2d75463bd86d87eaba7265b0721598dd803\"\nuuid = \"295af30f-e4ad-537b-8983-00126c2a3abe\"\nversion = \"3.1.19\"\n\n[[SHA]]\nuuid = \"ea8e919c-243c-51af-8825-aaa63cd721ce\"\n\n[[SentinelArrays]]\ndeps = [\"Dates\", \"Random\"]\ngit-tree-sha1 = \"54f37736d8934a12a200edea2f9206b03bdf3159\"\nuuid = \"91c51154-3ec4-41a3-a24f-3f23e20d615c\"\nversion = \"1.3.7\"\n\n[[Serialization]]\nuuid = \"9e88b42a-f829-5b0c-bbe9-9e923198166b\"\n\n[[Setfield]]\ndeps = [\"ConstructionBase\", \"Future\", \"MacroTools\", \"Requires\"]\ngit-tree-sha1 = \"fca29e68c5062722b5b4435594c3d1ba557072a3\"\nuuid = \"efcf1570-3423-57d1-acb7-fd33fddbac46\"\nversion = \"0.7.1\"\n\n[[SharedArrays]]\ndeps = [\"Distributed\", \"Mmap\", \"Random\", \"Serialization\"]\nuuid = \"1a1011a3-84de-559e-8e89-a11a2f7dc383\"\n\n[[Sockets]]\nuuid = \"6462fe0b-24de-5631-8697-dd941f90decc\"\n\n[[SortingAlgorithms]]\ndeps = [\"DataStructures\"]\ngit-tree-sha1 = \"b3363d7460f7d098ca0912c69b082f75625d7508\"\nuuid = \"a2af1166-a08f-5f64-846c-94a0d3cef48c\"\nversion = \"1.0.1\"\n\n[[SparseArrays]]\ndeps = [\"LinearAlgebra\", \"Random\"]\nuuid = \"2f01184e-e22b-5df5-ae63-d93ebab69eaf\"\n\n[[SpecialFunctions]]\ndeps = [\"ChainRulesCore\", \"LogExpFunctions\", \"OpenSpecFun_jll\"]\ngit-tree-sha1 = \"a322a9493e49c5f3a10b50df3aedaf1cdb3244b7\"\nuuid = \"276daf66-3868-5448-9aa4-cd146d93841b\"\nversion = \"1.6.1\"\n\n[[Statistics]]\ndeps = [\"LinearAlgebra\", \"SparseArrays\"]\nuuid = \"10745b16-79ce-11e8-11f9-7d13ad32a3b2\"\n\n[[Suppressor]]\ngit-tree-sha1 = \"a819d77f31f83e5792a76081eee1ea6342ab8787\"\nuuid = \"fd094767-a336-5f1f-9728-57cf17d0bbfb\"\nversion = \"0.2.0\"\n\n[[TOML]]\ndeps = [\"Dates\"]\nuuid = \"fa267f1f-6049-4f14-aa54-33bafae1ed76\"\n\n[[TableShowUtils]]\ndeps = [\"DataValues\", \"Dates\", \"JSON\", \"Markdown\", \"Test\"]\ngit-tree-sha1 = \"14c54e1e96431fb87f0d2f5983f090f1b9d06457\"\nuuid = \"5e66a065-1f0a-5976-b372-e0b8c017ca10\"\nversion = \"0.2.5\"\n\n[[TableTraits]]\ndeps = [\"IteratorInterfaceExtensions\"]\ngit-tree-sha1 = \"c06b2f539df1c6efa794486abfb6ed2022561a39\"\nuuid = \"3783bdb8-4a98-5b6b-af9a-565f29a5fe9c\"\nversion = \"1.0.1\"\n\n[[TableTraitsUtils]]\ndeps = [\"DataValues\", \"IteratorInterfaceExtensions\", \"Missings\", \"TableTraits\"]\ngit-tree-sha1 = \"78fecfe140d7abb480b53a44f3f85b6aa373c293\"\nuuid = \"382cd787-c1b6-5bf2-a167-d5b971a19bda\"\nversion = \"1.0.2\"\n\n[[Tables]]\ndeps = [\"DataAPI\", \"DataValueInterfaces\", \"IteratorInterfaceExtensions\", \"LinearAlgebra\", \"TableTraits\", \"Test\"]\ngit-tree-sha1 = \"d0c690d37c73aeb5ca063056283fde5585a41710\"\nuuid = \"bd369af6-aec1-5ad0-b16a-f7cc5008161c\"\nversion = \"1.5.0\"\n\n[[Tar]]\ndeps = [\"ArgTools\", \"SHA\"]\nuuid = \"a4e569a6-e804-4fa4-b0f3-eef7a1d5b13e\"\n\n[[Test]]\ndeps = [\"InteractiveUtils\", \"Logging\", \"Random\", \"Serialization\"]\nuuid = \"8dfed614-e22c-5e08-85e1-65c5234f0b40\"\n\n[[TextParse]]\ndeps = [\"CodecZlib\", \"DataStructures\", \"Dates\", \"DoubleFloats\", \"Mmap\", \"Nullables\", \"WeakRefStrings\"]\ngit-tree-sha1 = \"eb1f4fb185c8644faa2d18d14c72f2c24412415f\"\nuuid = \"e0df1984-e451-5cb5-8b61-797a481e67e3\"\nversion = \"1.0.2\"\n\n[[TimeZones]]\ndeps = [\"Dates\", \"Future\", \"LazyArtifacts\", \"Mocking\", \"Pkg\", \"Printf\", \"RecipesBase\", \"Serialization\", \"Unicode\"]\ngit-tree-sha1 = \"6c9040665b2da00d30143261aea22c7427aada1c\"\nuuid = \"f269a46b-ccf7-5d73-abea-4c690281aa53\"\nversion = \"1.5.7\"\n\n[[TranscodingStreams]]\ndeps = [\"Random\", \"Test\"]\ngit-tree-sha1 = \"216b95ea110b5972db65aa90f88d8d89dcb8851c\"\nuuid = \"3bb67fe8-82b1-5028-8e26-92a6c54297fa\"\nversion = \"0.9.6\"\n\n[[URIParser]]\ndeps = [\"Unicode\"]\ngit-tree-sha1 = \"53a9f49546b8d2dd2e688d216421d050c9a31d0d\"\nuuid = \"30578b45-9adc-5946-b283-645ec420af67\"\nversion = \"0.4.1\"\n\n[[UUIDs]]\ndeps = [\"Random\", \"SHA\"]\nuuid = \"cf7118a7-6976-5b1a-9a39-7adc72f591a4\"\n\n[[Unicode]]\nuuid = \"4ec0a83e-493e-50e2-b9ac-8f72acf5a8f5\"\n\n[[Vega]]\ndeps = [\"DataStructures\", \"DataValues\", \"Dates\", \"FileIO\", \"FilePaths\", \"IteratorInterfaceExtensions\", \"JSON\", \"JSONSchema\", \"MacroTools\", \"NodeJS\", \"Pkg\", \"REPL\", \"Random\", \"Setfield\", \"TableTraits\", \"TableTraitsUtils\", \"URIParser\"]\ngit-tree-sha1 = \"43f83d3119a868874d18da6bca0f4b5b6aae53f7\"\nuuid = \"239c3e63-733f-47ad-beb7-a12fde22c578\"\nversion = \"2.3.0\"\n\n[[VegaDatasets]]\ndeps = [\"DataStructures\", \"DataValues\", \"FilePaths\", \"IterableTables\", \"IteratorInterfaceExtensions\", \"JSON\", \"TableShowUtils\", \"TableTraits\", \"TableTraitsUtils\", \"TextParse\"]\ngit-tree-sha1 = \"c997c7217f37205c5795de8c797f8f8531890f1d\"\nuuid = \"0ae4a718-28b7-58ec-9efb-cded64d6d5b4\"\nversion = \"2.1.1\"\n\n[[VegaLite]]\ndeps = [\"Base64\", \"DataStructures\", \"DataValues\", \"Dates\", \"FileIO\", \"FilePaths\", \"IteratorInterfaceExtensions\", \"JSON\", \"MacroTools\", \"NodeJS\", \"Pkg\", \"REPL\", \"Random\", \"TableTraits\", \"TableTraitsUtils\", \"URIParser\", \"Vega\"]\ngit-tree-sha1 = \"3e23f28af36da21bfb4acef08b144f92ad205660\"\nuuid = \"112f6efa-9a02-5b7d-90c0-432ed331239a\"\nversion = \"2.6.0\"\n\n[[WeakRefStrings]]\ndeps = [\"DataAPI\", \"Parsers\", \"Random\", \"Test\"]\ngit-tree-sha1 = \"9ef95db08bf767499a74586bcbd4b5df30c19b9f\"\nuuid = \"ea10d353-3f73-51f8-a26c-33c1cb351aa5\"\nversion = \"1.1.0\"\n\n[[ZipFile]]\ndeps = [\"Libdl\", \"Printf\", \"Zlib_jll\"]\ngit-tree-sha1 = \"c3a5637e27e914a7a445b8d0ad063d701931e9f7\"\nuuid = \"a5390f91-8eb1-5f08-bee0-b1d1ffed6cea\"\nversion = \"0.9.3\"\n\n[[Zlib_jll]]\ndeps = [\"Libdl\"]\nuuid = \"83775a58-1f1d-513f-b197-d71354ab007a\"\n\n[[nghttp2_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"8e850ede-7688-5339-a07c-302acd2aaf8d\"\n\n[[p7zip_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"3f19e933-33d8-53b3-aaab-bd5110c3b7a0\"\n\"\"\"\n\n# \u2554\u2550\u2561 Cell order:\n# \u255f\u2500e4dca908-1016-11ec-0b7f-e9d01bd4e1b2\n# \u2560\u25503389dc98-b90f-4c6a-8c96-31840b0516ea\n# \u255f\u2500e598c897-61bf-4bc2-91db-e225362f5606\n# \u2560\u25500d909765-a87d-4a3c-bb9f-ff80e463992a\n# \u2560\u2550ced90367-407c-4b67-8276-68edc17933d3\n# \u2560\u25506a2ca31f-716c-48f4-8e1a-70e3c033358b\n# \u2560\u2550cc533197-dbe3-41b6-8478-64f82da52ba8\n# \u2560\u2550dcabd381-1316-449b-9ebf-68410ee6e3fd\n# \u2560\u25508398bb71-aa8e-42b6-b3e3-c08ca57ee5b0\n# \u2560\u2550a6a64f62-316a-48f5-bd5c-0eebff53022d\n# \u255f\u2500f4bf1143-c315-4521-b19b-06bf0373828a\n# \u2560\u255086ddda30-351d-41f6-94b2-4c84dea08448\n# \u2560\u2550bfd9753c-323a-4a82-b5e6-057e7519ed6c\n# \u2560\u2550ff5a5927-6bc6-4b05-b0e8-e0bfdc623440\n# \u255f\u250000000000-0000-0000-0000-000000000001\n# \u255f\u250000000000-0000-0000-0000-000000000002\n", "meta": {"hexsha": "ac94c4dd1580397e6550538249b13f75b899dc95", "size": 24188, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "tools/viz/solution-explorer.jl", "max_stars_repo_name": "yvs314/epi-net-m", "max_stars_repo_head_hexsha": "d4ddc68612247e420ae5d6c1bcd2cf488ce3d8bf", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "tools/viz/solution-explorer.jl", "max_issues_repo_name": "yvs314/epi-net-m", "max_issues_repo_head_hexsha": "d4ddc68612247e420ae5d6c1bcd2cf488ce3d8bf", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 6, "max_issues_repo_issues_event_min_datetime": "2021-08-30T22:55:22.000Z", "max_issues_repo_issues_event_max_datetime": "2021-11-18T20:01:24.000Z", "max_forks_repo_path": "tools/viz/solution-explorer.jl", "max_forks_repo_name": "yvs314/epi-net-m", "max_forks_repo_head_hexsha": "d4ddc68612247e420ae5d6c1bcd2cf488ce3d8bf", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 30.8127388535, "max_line_length": 280, "alphanum_fraction": 0.7327187035, "num_tokens": 10545, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.46879062662624377, "lm_q2_score": 0.16667540468797667, "lm_q1q2_score": 0.07813586740685935}}
{"text": "# The following test cases and comments are adapted from http://brainfuck.org/tests.b\n\nusing Test\nif !@isdefined(BrainFuck)\n include(joinpath(@__DIR__, \"..\", \"src\", \"BrainFuck.jl\"))\nend\noutput = IOBuffer()\n\n# Here are some little programs for testing brainfuck implementations.\n\n@testset \"IO test\" begin\n # This is for testing i/o; give it a return followed by an EOF. (Try it both\n # with file input \u2014 a file consisting only of one blank line \u2014 and with\n # keyboard input, i.e. hit return and then ctrl-d (Unix) or ctrl-z (Windows).)\n # It should give two lines of output; the two lines should be identical, and\n # should be lined up one over the other. If that doesn't happen, ten is not\n # coming through as newline on output.\n # The content of the lines tells how input is being processed; each line\n # should be two uppercase letters.\n # Anything with O in it means newline is not coming through as ten on input.\n # LK means newline input is working fine, and EOF leaves the cell unchanged\n # (which I recommend).\n # LB means newline input is working fine, and EOF translates as 0.\n # LA means newline input is working fine, and EOF translates as -1.\n # Anything else is fairly unexpected.\n iotest = \">,>+++++++++,>+++++++++++[<++++++<++++++<+>>>-]<<.>.<<-.>.>.<<.\"\n\n BrainFuck.interpret(iotest, input = \"\\n\", output = output)\n @test String(take!(output)) == \"LK\\nLK\\n\"\n\n BrainFuck.interpret(iotest, input = open(joinpath(@__DIR__, \"empty.txt\")), output = output)\n @test String(take!(output)) == \"LK\\nLK\\n\"\nend\n\n@testset \"Memory size\" begin\n memorysizetest = \"++++[>++++++<-]>[>+++++>+++++++<<-]>>++++<[[>[[>>+<<-]<]>>>-]>-[>+>+<<-]>]+++++[>+++++++<<++>-]>.<<.\"\n # Goes to cell 30000 and reports from there with a #. (Verifies that the\n # array is big enough.)\n BrainFuck.interpret(memorysizetest, output = output)\n @test String(take!(output)) == \"#\\n\"\n\n BrainFuck.compile(memorysizetest)(output = output)\n @test String(take!(output)) == \"#\\n\"\nend\n\n@testset \"Several obscure tests\" begin\n # Tests for several obscure problems. Should output an H.\n obscuretest = \"\"\"[]++++++++++[>>+>+>++++++[<<+<+++>>>-]<<<<-]\n \"A*\\$\";?@![#>>+<<]>[>>]<<<<[>++<[-]]>.>.\"\"\"\n @test BrainFuck.interpret(obscuretest, output = String) == \"H\\n\"\n @test BrainFuck.compile(obscuretest)(output = String) == \"H\\n\"\nend\n\n@testset \"Unmatched brackets\" begin\n # Should ideally give error message \"unmatched [\" or the like, and not give\n # any output. Not essential.\n unmatchedleft = \"+++++[>+++++++>++<<-]>.>.[\"\n @test_throws ArgumentError BrainFuck.interpret(unmatchedleft)\n @test_throws ArgumentError BrainFuck.compile(unmatchedleft)\n\n # Should ideally give error message \"unmatched ]\" or the like, and not give\n # any output. Not essential.\n unmatchedright = \"+++++[>+++++++>++<<-]>.>.][\"\n @test_throws ArgumentError BrainFuck.interpret(unmatchedright)\n @test_throws ArgumentError BrainFuck.compile(unmatchedright)\nend\n\n@testset \"Deep brackets\" begin\n # [Daniel Cristofani's] pathological program rot13.b is good for testing the\n # response to deep brackets; the input \"~mlk zyx\" should produce the output \"~zyx mlk\".\n rot13 = \"\"\"\n ,\n [>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-\n [>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-\n [>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-\n [>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-\n [>++++++++++++++<-\n [>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-\n [>>+++++[<----->-]<<-\n [>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-\n [>++++++++++++++<-\n [>+<-[>+<-[>+<-[>+<-[>+<-\n [>++++++++++++++<-\n [>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-\n [>>+++++[<----->-]<<-\n [>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-[>+<-\n [>++++++++++++++<-\n [>+<-]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]\n ]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]]>.[-]<,]\n \"\"\"\n\n @test BrainFuck.interpret(rot13, input = \"~mlk zyx\", output = String) == \"~zyx mlk\"\n @test BrainFuck.compile(rot13)(input = \"~mlk zyx\", output = String) == \"~zyx mlk\"\nend\n", "meta": {"hexsha": "82dbb8e681b2c78f361e7cc513c8dac81dbfcbb5", "size": 4430, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/Brainfuck/test/BrainFuck.jl", "max_stars_repo_name": "stellartux/Esolangs", "max_stars_repo_head_hexsha": "5ea59ad63c3d4089f39166ab71ef1d768eeefad2", "max_stars_repo_licenses": ["Unlicense"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/Brainfuck/test/BrainFuck.jl", "max_issues_repo_name": "stellartux/Esolangs", "max_issues_repo_head_hexsha": "5ea59ad63c3d4089f39166ab71ef1d768eeefad2", "max_issues_repo_licenses": ["Unlicense"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/Brainfuck/test/BrainFuck.jl", "max_forks_repo_name": "stellartux/Esolangs", "max_forks_repo_head_hexsha": "5ea59ad63c3d4089f39166ab71ef1d768eeefad2", "max_forks_repo_licenses": ["Unlicense"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 46.6315789474, "max_line_length": 123, "alphanum_fraction": 0.4909706546, "num_tokens": 1456, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4726834766204328, "lm_q2_score": 0.16451646494473965, "lm_q1q2_score": 0.0777642146113831}}
{"text": "# Additional tests of the to_dot function\n\n# This adds tests where:\n# 1) there are isolated vertices\n# 2) there are vertex attributes to be shown in .dot\n# 3) the graph verifies implements_edge_list(.) == true\n# and implements_vertex_map(.) == true\n\n\n# These functions help with dealing with the fact that in two equivalent\n# renderings of the same graph one may obtain equivalent .dots with permuted\n# lines. This results of the fact that no ordering of vertices is implied by\n# the code\n# ```for vtx in vertices(graph) .... end```. For similar reasons,\n# attributes are extracted of a Dict in arbitrary order.\n#\n# So:\n# 1) lines are sorted (result is independent of order in vertices and edges)\n# 2) attributes are checksummed (with an order independent checksum (not a good\n# one)). This is not a precise verification of attributes, but good enough\n# for testing\n\n# NOTE: if to_dot is modified to emit lines with .dot comments, these tests\n# mail fail erroneously....\n\n\ncomRX = Base.compile(r\"^[^\\[]+\\[([^\\[]+)\\]\\h*$\"x)\nfunction rewriteAttrs(a::AbstractString)\n m = match(comRX,a)\n if m!=nothing\n attrs = m.captures[1]\n offset= m.offsets[1]\n chksum= mod(reduce(+,\n map(x->convert(Int,x),collect(attrs)), init=0 ), 25)\n ch = convert(Char, convert(Int,'a') - 1 + chksum)\n a[1:offset-1] * \"$ch\" * a[ offset+length(attrs) : end ]\n else\n return a\n end\nend\n\nfunction check_same_dot(a::AbstractString,b::AbstractString)\n sa=sort( map( rewriteAttrs, split( a, \"\\n\")))\n sb=sort( map( rewriteAttrs, split( b, \"\\n\")))\n la = map(rewriteAttrs,sa)\n lb = map(rewriteAttrs,sb)\n return la==lb\nend\n\n\n\nmodule testDOT1\n\nusing Graphs\nusing Test\n\n\n###########\n# test dot output for graphs for which\n# true == implements_edge_list && implements_vertex_map\n# and no vertex attributes\n###########\n\n### 1) graph without node attributes\nsgd = simple_graph(3)\n\n@test @show implements_edge_list(sgd)==true\n@test @show implements_vertex_map(sgd)==true\n\nadd_vertex!(sgd)\n\ndot1=to_dot(sgd)\nprintln(dot1)\n@test Main.check_same_dot(dot1,\"digraph graphname {\\n1\\n2\\n3\\n4\\n}\\n\")\n\n### 2) graph without node attributes but with some edges\nadd_edge!(sgd,1,3)\nadd_edge!(sgd,3,1)\nadd_edge!(sgd,2,3)\n\ndot2=to_dot(sgd)\nprintln(dot2)\n@test Main.check_same_dot(dot2,\n \"digraph graphname {\\n1\\n2\\n3\\n4\\n1 -> 3\\n3 -> 1\\n2 -> 3\\n}\\n\")\n\n\nend # module testDOT1\n\nmodule testDOT2\n\nusing Graphs\nusing Test\n\n###########\n# test dot output for graphs for which\n# true == implements_edge_list && implements_vertex_map\n# and vertex attributes\n###########\n\nstruct MyVtxType\n name::AbstractString\nend\n\nimport Graphs.attributes\nfunction Graphs.attributes(vtx::MyVtxType,g::G) where {G<:AbstractGraph}\n rd = Graphs.AttributeDict()\n rd[\"label\"]=vtx.name\n rd[\"color\"]=\"bisque\"\n rd\nend\n\n### 3) directed graph with node attributes and some disconnected vertices\n\nag = Graphs.graph( map( MyVtxType,[ \"a\", \"b\", \"c\",\"d\"]), Graphs.Edge{MyVtxType}[],\n is_directed=true)\n\nvl = ag.vertices\n\nadd_edge!(ag, vl[1], vl[3] )\nadd_edge!(ag, vl[3], vl[1] )\nadd_edge!(ag, vl[2], vl[3])\n\n@test @show implements_edge_list(ag)==true\n@test @show implements_vertex_map(ag)==true\n\ndot3 = to_dot( ag )\nprintln(dot3)\n\ncompDot3 = \"digraph graphname {\\n1\\t[\\\"label\\\"=\\\"a\\\",\\\"color\\\"=\\\"bisque\\\"]\\n2\\t[\\\"label\\\"=\\\"b\\\",\\\"color\\\"=\\\"bisque\\\"]\\n3\\t[\\\"label\\\"=\\\"c\\\",\\\"color\\\"=\\\"bisque\\\"]\\n4\\t[\\\"label\\\"=\\\"d\\\",\\\"color\\\"=\\\"bisque\\\"]\\n1 -> 3\\n3 -> 1\\n2 -> 3\\n}\\n\"\n\n@test Main.check_same_dot(dot3,compDot3)\n\n### 4) undirected graph with node attributes and some disconnected vertices\n\n\nagu = Graphs.graph( map( MyVtxType,[ \"a\", \"b\", \"c\",\"d\"]), Graphs.Edge{MyVtxType}[],\n is_directed=false)\n\nvl = agu.vertices\n\nadd_edge!(agu, vl[1], vl[3] )\nadd_edge!(agu, vl[2], vl[3])\n\ndot4=to_dot(agu)\nprintln(dot4)\n\n@test @show implements_edge_list(agu)==true\n@test @show implements_vertex_map(agu)==true\n\n@test Main.check_same_dot(dot4,\"graph graphname {\\n1\\t[\\\"label\\\"=\\\"a\\\",\\\"color\\\"=\\\"bisque\\\"]\\n2\\t[\\\"label\\\"=\\\"b\\\",\\\"color\\\"=\\\"bisque\\\"]\\n3\\t[\\\"label\\\"=\\\"c\\\",\\\"color\\\"=\\\"bisque\\\"]\\n4\\t[\\\"label\\\"=\\\"d\\\",\\\"color\\\"=\\\"bisque\\\"]\\n1 -- 3\\n2 -- 3\\n}\\n\"\n)\n\nend # module testDOT2\n", "meta": {"hexsha": "86a7e9cdcdf0b22072efd1e8742a5ff41079a4cb", "size": 4339, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/dot2.jl", "max_stars_repo_name": "UnofficialJuliaMirrorSnapshots/Graphs.jl-86223c79-3864-5bf0-83f7-82e725a168b6", "max_stars_repo_head_hexsha": "b76b9914178a417085681167643af510547e8a0a", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 41, "max_stars_repo_stars_event_min_datetime": "2018-08-19T13:26:30.000Z", "max_stars_repo_stars_event_max_datetime": "2021-10-07T08:17:35.000Z", "max_issues_repo_path": "test/dot2.jl", "max_issues_repo_name": "JuliaAttic/OldGraphs.jl", "max_issues_repo_head_hexsha": "39a9c6efacc190ac79db47dd4dd951657058d2c0", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 5, "max_issues_repo_issues_event_min_datetime": "2019-02-20T22:05:43.000Z", "max_issues_repo_issues_event_max_datetime": "2019-05-24T15:20:41.000Z", "max_forks_repo_path": "test/dot2.jl", "max_forks_repo_name": "JuliaAttic/OldGraphs.jl", "max_forks_repo_head_hexsha": "39a9c6efacc190ac79db47dd4dd951657058d2c0", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 6, "max_forks_repo_forks_event_min_datetime": "2019-02-12T18:37:11.000Z", "max_forks_repo_forks_event_max_datetime": "2020-07-03T14:09:36.000Z", "avg_line_length": 28.1753246753, "max_line_length": 243, "alphanum_fraction": 0.6383959438, "num_tokens": 1281, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4532618480153861, "lm_q2_score": 0.17106118533966355, "lm_q1q2_score": 0.07753550899075838}}
{"text": "#%%\nprintln(\"hello\")\nprintln(\"Julia\")\n#%%\n\n#%%\nnum1=22\nnum2=22.35\nprintln(\"n1: \",num1,\" \",typeof(num1))\nprintln(\"n2: \",num2,\" \",typeof(num2))\nd=num1/num2\nprintln(d)\nstrA=\"myString\"\nprintln(typeof(strA))\n\nnum3=convert(Float64,num1)\nprintln(\"n3: \",num3,\" \",typeof(num1))\n\n#%%\n\n#%%\nfunction funcA(x)\n return x*5\nend\nprintln(funcA(2))\nprintln(funcA([2,22]))\n#%%\n", "meta": {"hexsha": "dedd1582f44c7c1d701d5e8923b1a8526db8268a", "size": 361, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "first.jl", "max_stars_repo_name": "urmi-21/hello-julia", "max_stars_repo_head_hexsha": "08b635acc2f09e0225fefd62f05cf0db70ab5945", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "first.jl", "max_issues_repo_name": "urmi-21/hello-julia", "max_issues_repo_head_hexsha": "08b635acc2f09e0225fefd62f05cf0db70ab5945", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "first.jl", "max_forks_repo_name": "urmi-21/hello-julia", "max_forks_repo_head_hexsha": "08b635acc2f09e0225fefd62f05cf0db70ab5945", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 12.8928571429, "max_line_length": 37, "alphanum_fraction": 0.6315789474, "num_tokens": 132, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4610167793123159, "lm_q2_score": 0.16667541089705434, "lm_q1q2_score": 0.07684016112231687}}
{"text": "# ------------------------------------------------------------------------------------------\n# # Strings\n#\n# Topics:\n# 1. How to get a string\n# 2. String interpolation\n# 3. String concatenation\n# ------------------------------------------------------------------------------------------\n\n# ------------------------------------------------------------------------------------------\n# ## How to get a string\n#\n# Enclose your characters in \" \" or \"\"\" \"\"\"!\n# ------------------------------------------------------------------------------------------\n\ns1 = \"I am a string.\"\n\ns2 = \"\"\"I am also a string. \"\"\"\n\n# ------------------------------------------------------------------------------------------\n# There are a couple functional differences between strings enclosed in single and triple\n# quotes. in end`:\n\nfor i in 1:5\n println(i)\nend\n\n\n# !!! info\n# Ranges are constructed as `start:stop`, or `start:step:stop`.\n\n#-\n\nfor i in [1.2, 2.3, 3.4, 4.5, 5.6]\n println(i)\nend\n\n# This for-each loop also works with dictionaries:\n\nfor (key, value) in Dict(\"A\" => 1, \"B\" => 2.5, \"D\" => 2 - 3im)\n println(\"$(key): $(value)\")\nend\n\n# Note that in contrast to vector languages like Matlab and R, loops do not\n# result in a significant performance degradation in Julia.\n\n# ## Control Flow\n\n# Julia control flow is similar to Matlab, using the keywords\n# `if-elseif-else-end`, and the logical operators `||` and `&&` for **or** and\n# **and** respectively:\n\nfor i in 0:3:15\n if i < 5\n println(\"$(i) is less than 5\")\n elseif i < 10\n println(\"$(i) is less than 10\")\n else\n if i == 10\n println(\"the value is 10\")\n else\n println(\"$(i) is bigger than 10\")\n end\n end\nend\n\n# ## Comprehensions\n\n# Similar to languages like Haskell and Python, Julia supports the use of simple\n# loops in the construction of arrays and dictionaries, called comprehenions.\n#\n# A list of increasing integers:\n\n[i for i in 1:5]\n\n# Matrices can be built by including multiple indices:\n\n[i * j for i in 1:5, j in 5:10]\n\n# Conditional statements can be used to filter out some values:\n\n[i for i in 1:10 if i % 2 == 1]\n\n# A similar syntax can be used for building dictionaries:\n\nDict(\"$(i)\" => i for i in 1:10 if i % 2 == 1)\n\n# ## Functions\n\n# A simple function is defined as follows:\n\nfunction print_hello()\n println(\"hello\")\nend\nprint_hello()\n\n# Arguments can be added to a function:\n\nfunction print_it(x)\n println(x)\nend\nprint_it(\"hello\")\nprint_it(1.234)\nprint_it(:my_id)\n\n# Optional keyword arguments are also possible:\n\nfunction print_it(x; prefix = \"value:\")\n println(\"$(prefix) $(x)\")\nend\nprint_it(1.234)\nprint_it(1.234, prefix = \"val:\")\n\n# The keyword `return` is used to specify the return values of a function:\n\nfunction mult(x; y = 2.0)\n return x * y\nend\n\nmult(4.0)\n\n#-\n\nmult(4.0, y = 5.0)\n\n# ### Anonymous functions\n\n# The syntax `input -> output` creates an anonymous function. These are most\n# useful when passed to other functions. For example:\n\nf = x -> x^2\nf(2)\n\n#-\n\nmap(x -> x^2, 1:4)\n\n# ### Type parameters\n\n# We can constrain the inputs to a function using type parameters, which are\n# `::` followed by the type of the input we want. For example:\n\nfunction foo(x::Int)\n return x^2\nend\n\nfunction foo(x::Float64)\n return exp(x)\nend\n\nfunction foo(x::Number)\n return x + 1\nend\n\n@show foo(2)\n@show foo(2.0)\n@show foo(1 + 1im)\nnothing #hide\n\n# But what happens if we call `foo` with something we haven't defined it for?\n\ntry #hide\nfoo([1, 2, 3])\ncatch err; showerror(stdout, err) end #hide\n\n# We get a dreaded `MethodError`! A `MethodError` means that you passed a\n# function something that didn't match the type that it was expecting. In this\n# case, the error message says that it doesn't know how to handle an\n# `Array{Int64, 1}`, but it does know how to handle `Float64`, `Int64`, and\n# `Number`.\n#\n# !!! tip\n# Read the \"Closest candidates\" part of the error message carefully to get a\n# hint as to what was expected.\n\n# ### Broadcasting\n\n# In the example above, we didn't define what to do if `f` was passed an\n# `Array`. Luckily, Julia provides a convienient syntax for mapping `f`\n# element-wise over arrays! Just add a `.` between the name of the function and\n# the opening `(`. This works for _any_ function, including functions with\n# multiple arguments. For example:\n\nf.([1, 2, 3])\n\n# !!! tip\n# Get a `MethodError` when calling a function that takes an `Array`? Try\n# broadcasting it!\n\n# ## Mutable vs immutable objects\n\n# Some types in Julia are *mutable*, which means you can change the values\n# inside them. A good example is an array. You can modify the contents of an\n# array without having to make a new array.\n\n# In contrast, types like `Float64` are *immutable*. You can't modify the\n# contents of a `Float64`.\n\n# This is something to be aware of when passing types into functions. For\n# example:\n\nfunction mutability_example(mutable_type::Vector{Int}, immutable_type::Int)\n mutable_type[1] += 1\n immutable_type += 1\n return\nend\n\nmutable_type = [1, 2, 3]\nimmutable_type = 1\n\nmutability_example(mutable_type, immutable_type)\n\nprintln(\"mutable_type: $(mutable_type)\")\nprintln(\"immutable_type: $(immutable_type)\")\n\n# Because `Vector{Int}` is a mutable type, modifying the variable inside the\n# function changed the value outside of the function. In constrast, the change\n# to `immutable_type` didn't modify the value outside the function.\n\n# You can check mutability with the `isimmutable` function:\n\nisimmutable([1, 2, 3])\n\n#-\n\nisimmutable(1)\n\n# ## The package manager\n\n# ### Installing packages\n\n# No matter how wonderful Julia's base language is, at some point you will want\n# to use an extension package. Some of these are built-in, for example random\n# number generation is available in the `Random` package in the standard\n# library. These packages are loaded with the commands `using` and `import`.\n\nusing Random # The equivalent of Python's `from Random import *`\nimport Random # The equivalent of Python's `import Random`\n\nRandom.seed!(33)\n\n[rand() for i in 1:10]\n\n# The Package Manager is used to install packages that are not part of Julia's\n# standard library.\n\n# For example the following can be used to install JuMP,\n# ```julia\n# using Pkg\n# Pkg.add(\"JuMP\")\n# ```\n\n# For a complete list of registed Julia packages see the package listing at\n# [JuliaHub](https://juliahub.com).\n\n# From time to you may wish to use a Julia package that is not registered. In\n# this case a git repository URL can be used to install the package.\n# ```julia\n# using Pkg\n# Pkg.add(\"https://github.com/user-name/MyPackage.jl.git\")\n# ```\n\n# ### Package environments\n\n# By default, `Pkg.add` will add packages to Julia's global environment.\n# However, Julia also has built-in support for virtual environments.\n\n# Activate a virtual environment with:\n# ```julia\n# import Pkg; Pkg.activate(\"/path/to/environment\")\n# ```\n\n# You can see what packages are installed in the current environment with\n# `Pkg.status()`.\n\n# !!! tip\n# We _strongly_ recommend you create a Pkg environment for each project\n# that you create in Julia, and add only the packages that you need, instead\n# of adding lots of packages to the global environment. The [Pkg manager documentation](https://julialang.github.io/Pkg.jl/v1/environments/)\n# has more information on this topic.\n", "meta": {"hexsha": "7af139a58f5a649eaa7dc30aabf6341a649b5b5d", "size": 16499, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "lang/Julia/an_introduction_to_julia.jl", "max_stars_repo_name": "ethansaxenian/RosettaDecode", "max_stars_repo_head_hexsha": "8ea1a42a5f792280b50193ad47545d14ee371fb7", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "lang/Julia/an_introduction_to_julia.jl", "max_issues_repo_name": "ethansaxenian/RosettaDecode", "max_issues_repo_head_hexsha": "8ea1a42a5f792280b50193ad47545d14ee371fb7", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "lang/Julia/an_introduction_to_julia.jl", "max_forks_repo_name": "ethansaxenian/RosettaDecode", "max_forks_repo_head_hexsha": "8ea1a42a5f792280b50193ad47545d14ee371fb7", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 27.8699324324, "max_line_length": 144, "alphanum_fraction": 0.672889266, "num_tokens": 4510, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.44939263446475963, "lm_q2_score": 0.16667539847889923, "lm_q1q2_score": 0.07490269642289611}}
{"text": "# Type Hierarchy Exercise\n#\n# Demonstration of multiple dispatch as opposed to conditional formatting\n\n#module LJtypeHierarchy\n\nabstract type AbstractPerson end\n\nstruct Person <: AbstractPerson\n name::String\nend\n\nstruct Student <: AbstractPerson\n name::String\n grade::Int\nend\n\nstruct GroupLeader <: AbstractPerson\n name::String\n group::String\nend\n\n#end\n\nprintln(\"Hello World\")\n\nadam = Person(\"adam\")\nprintln(\"$(adam.name) is a Person\")\ncameron = Student(\"cameron\", 95)\njohn = GroupLeader(\"John\", \"CDAS\")\n\nperson_info(p::Person) = println(\"$(p.name) is a Person\")\nperson_info(p::Student) = println(\"$(p.name) is a Student with grade $(p.grade)\")\nperson_info(p::GroupLeader) = println(\"$(p.name) is a GroupLeader of $(p.group)\")\n\npeople = (adam, cameron, john)\n\n\nfor person in people\n person_info(person)\nend\n", "meta": {"hexsha": "d6245ac5690a1406c696e70f4cad2e7253dd9311", "size": 825, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "TypeHierarchy.jl", "max_stars_repo_name": "AdamVStephen/learning-julia", "max_stars_repo_head_hexsha": "442e0dfab5c3e224927e1ca96516af87d8e81319", "max_stars_repo_licenses": ["CC0-1.0"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "TypeHierarchy.jl", "max_issues_repo_name": "AdamVStephen/learning-julia", "max_issues_repo_head_hexsha": "442e0dfab5c3e224927e1ca96516af87d8e81319", "max_issues_repo_licenses": ["CC0-1.0"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "TypeHierarchy.jl", "max_forks_repo_name": "AdamVStephen/learning-julia", "max_forks_repo_head_hexsha": "442e0dfab5c3e224927e1ca96516af87d8e81319", "max_forks_repo_licenses": ["CC0-1.0"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 19.6428571429, "max_line_length": 81, "alphanum_fraction": 0.7151515152, "num_tokens": 211, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4843800842769843, "lm_q2_score": 0.15405755880753266, "lm_q1q2_score": 0.07462241331869914}}
{"text": "### A Pluto.jl notebook ###\n# v0.17.1\n\nusing Markdown\nusing InteractiveUtils\n\n# This Pluto notebook uses @bind for interactivity. When running this notebook outside of Pluto, the following 'mock version' of @bind gives bound variables a default value (instead of an error).\nmacro bind(def, element)\n quote\n local iv = try Base.loaded_modules[Base.PkgId(Base.UUID(\"6e696c72-6542-2067-7265-42206c756150\"), \"AbstractPlutoDingetjes\")].Bonds.initial_value catch; b -> missing; end\n local el = $(esc(element))\n global $(esc(def)) = Core.applicable(Base.get, el) ? Base.get(el) : iv(el)\n el\n end\nend\n\n# \u2554\u2550\u2561 75fe8fd0-2563-11ec-30ef-d15cdc3d4e63\nusing PlutoUI\n\n# \u2554\u2550\u2561 d4b5618f-f1a7-4483-b738-fb94e1e2bbbe\nhtml\"\"\"\n \ud83d\udcbb L\u00f3gica de Programa\u00e7\u00e3o por Franco Naghetini \u00e9 licenciado sob CC BY 4.0
\n\"\"\"\n\n# \u2554\u2550\u2561 deee1735-37ee-4c97-a4df-8fcb61e95d6a\nPlutoUI.TableOfContents(aside=true, title=\"Sum\u00e1rio\",\n\t\t\t\t\t\tindent=true, depth=2)\n\n# \u2554\u2550\u2561 488dcacd-109d-41f2-b904-3d17193e6190\nmd\"\"\"\n\n\"\"\"\n\n# \u2554\u2550\u2561 6ca3e113-02db-4cef-ad9e-3941ac7d7a6d\nmd\"\"\"\n# \ud83d\udcbb L\u00f3gica de Programa\u00e7\u00e3o\n\nO ato de **programar** pode ser entendido como uma forma de se comunicar com as m\u00e1quinas a partir de um conjunto de instru\u00e7\u00f5es n\u00e3o amb\u00edguas com o intuito de se realizar uma determinada tarefa. Essa comunica\u00e7\u00e3o com as m\u00e1quinas \u00e9 realizada por meio das **linguagens de programa\u00e7\u00e3o**, como Python, R, C e Julia.\n\nNesse contexto, a **l\u00f3gica de programa\u00e7\u00e3o** \u00e9 de suma import\u00e2ncia, uma vez que ela define a estrutura das instru\u00e7\u00f5es dadas \u00e0s m\u00e1quinas, ou seja, define a forma que voc\u00ea se comunica com as m\u00e1quinas. Se quer aprender mais sobre a import\u00e2ncia da l\u00f3gica de programa\u00e7\u00e3o assista [este v\u00eddeo](https://www.youtube.com/watch?v=l7lIPXij85I) do canal Programa\u00e7\u00e3o Din\u00e2mica.\n\nNeste m\u00f3dulo, estudaremos alguns conceitos b\u00e1sicos de l\u00f3gica de programa\u00e7\u00e3o. Para isso, utilizaremos [Julia](https://julialang.org/), uma linguagem de programa\u00e7\u00e3o moderna e de prop\u00f3sito geral que possui diversos recursos favor\u00e1veis ao desenvolvimento de rotinas geoestat\u00edsticas.\n\n> A linguagem \u00e9 *simples* como Python e *r\u00e1pida* como C \ud83d\ude80\n\nA seguir, ser\u00e3o apresentados alguns conceitos b\u00e1sicos de l\u00f3gica de programa\u00e7\u00e3o, al\u00e9m de uma introdu\u00e7\u00e3o sobre os recursos interativos da linguagem Julia.\n\"\"\"\n\n# \u2554\u2550\u2561 14db5525-7c0a-433b-a23c-088db728f46b\nmd\"\"\"\n>##### \ud83d\udcda Sobre\n>- Voc\u00ea pode exportar este notebook como PDF ou HTML est\u00e1tico. Para isso, clique no \u00edcone \ud83d\udd3a\ud83d\udd34, localizado no canto superior direito da pagina. Entretanto, ambos os formatos n\u00e3o s\u00e3o compat\u00edveis com os recursos interativos do notebook.\n>- Caso deseje executar alguma c\u00e9lula do notebook, clique no \u00edcone \u25b6\ufe0f, localizado no canto inferior direito da c\u00e9lula.\n>- Algumas c\u00e9lulas encontram-se ocultadas (e.g. c\u00e9lulas que geram os plots). Voc\u00ea pode clicar no \u00edcone \ud83d\udc41\ufe0f, localizado no canto superior esquerdo da c\u00e9lula, para ocult\u00e1-la ou exib\u00ed-la.\n>- A explica\u00e7\u00e3o das c\u00e9lulas que geram os plots est\u00e1 fora do escopo deste notebook. Entretanto, a sintaxe \u00e9 bem intuitiva e pode ser facilmente compreendida!\n>- Voc\u00ea pode ainda clicar no \u00edcone `...`, no canto superior direito de uma c\u00e9lula, para exclu\u00ed-la do notebook.\n>- Algumas c\u00e9lulas deste notebook encontram-se encapsuladas pela express\u00e3o `md\"...\"` (e.g. esta c\u00e9lula). Essas s\u00e3o c\u00e9lulas de texto chamadas de *markdown*. Caso deseje aprender um pouco mais sobre a linguagem *markdown*, clique [aqui](https://docs.pipz.com/central-de-ajuda/learning-center/guia-basico-de-markdown#open).\n>- No Pluto, todos os pacotes devem ser importados/baixados na primeira c\u00e9lula do notebook. Clique no \u00edcone \ud83d\udc41\ufe0f para exibir essa c\u00e9lula ou consulte a se\u00e7\u00e3o *Pacotes utilizados* deste notebook para saber mais informa\u00e7\u00f5es sobre os pacotes.\n>- Utilize a macro ` @which` para verificar a qual pacote uma determinada fun\u00e7\u00e3o pertence.\n>- Voc\u00ea pode utilizar este notebook da forma que quiser, basta referenciar [este link](https://github.com/fnaghetini/intro-to-geostats). Consulte a [licen\u00e7a] (https://github.com/fnaghetini/intro-to-geostats/blob/main/LICENSE) para saber mais detalhes.\n>- Para mais informa\u00e7\u00f5es acesse o [README](https://github.com/fnaghetini/intro-to-geostats/blob/main/README.md) do projeto \ud83d\ude80\n\"\"\"\n\n# \u2554\u2550\u2561 c111da75-d294-4def-93fb-56953c3585ad\nmd\"\"\"\n\n## 1. Vari\u00e1veis\n\nAs **vari\u00e1veis** podem ser entendidas como partes da mem\u00f3ria (ou \"caixas\") onde se armazena **valores** de diferentes **tipos** para serem posteriormente processados. Cada vari\u00e1vel \u00e9 identificada/rotulada por um **nome**.\n\nAs vari\u00e1veis podem ser **num\u00e9ricas**...\n\n\"\"\"\n\n# \u2554\u2550\u2561 7ceb9c27-0310-4f97-8349-91286c1f9235\nidade = 24\n\n# \u2554\u2550\u2561 218bb8c3-729b-43c7-9a53-d3b579cf2d21\naltura = 1.58\n\n# \u2554\u2550\u2561 3b3e6955-1573-4620-8109-d80b71cf0708\nmd\" As vari\u00e1veis podem armazenar **cadeias de caracteres**... \"\n\n# \u2554\u2550\u2561 f2e6adcd-9095-496f-a8e3-88069537015c\nnome = \"Camila\"\n\n# \u2554\u2550\u2561 870cbb76-b0c3-4d14-be22-86e0d34ebe58\nfrase = \"Ol\u00e1, mundo! \ud83c\udf0e\"\n\n# \u2554\u2550\u2561 55a2c622-0f9f-45db-b534-55873c0759d4\nmd\"\"\"\n> \u26a0\ufe0f As cadeias de caracteres (**strings**) devem ser encapsuladas por \u00e1spas duplas. Emojis tamb\u00e9m s\u00e3o considerados cadeias de caracteres! Clique [aqui](https://getemoji.com/) para copiar emojis e colar nas c\u00e9lulas deste notebook.\n\"\"\"\n\n# \u2554\u2550\u2561 449c91c3-7f03-4ffd-95e7-02cdb58323fd\nmd\"\"\"\nUm recurso muito utilizado e presente em diversas linguagens de programa\u00e7\u00e3o \u00e9 a **interpola\u00e7\u00e3o de strings**. Esse procedimento utiliza o valor de uma vari\u00e1vel dentro de uma string, de modo que a string interpolada atua como um \"template\". Em Julia, usamos o s\u00edmbolo `$` para interpolar strings.\n\nA c\u00e9lula abaixo mostra um exemplo de interpola\u00e7\u00e3o de string, utilizando as vari\u00e1veis j\u00e1 criadas `nome`, `idade` e `altura`...\n\"\"\"\n\n# \u2554\u2550\u2561 8d463f50-d694-41eb-bbad-c40f93471852\n\"Ol\u00e1! Meu nome \u00e9 $nome, tenho $idade anos e tenho $altura m de altura.\"\n\n# \u2554\u2550\u2561 4d31b989-7a7d-43ca-a1f8-831db58f1c8e\nmd\"\"\"\n\nPodemos utilizar *s\u00edmbolos matem\u00e1ticos* como nomes das vari\u00e1veis. Para inserir o s\u00edmbolo $\\alpha$, por exemplo, digita-se `\\alpha` + `TAB`. No caso do s\u00edmbolo $\\gamma$, digita-se `\\gamma` + `TAB`.\n\nCique [aqui](https://docs.julialang.org/en/v1/manual/unicode-input/) para acessar a lista completa de s\u00edmbolos dispon\u00edveis.\n\n\"\"\"\n\n# \u2554\u2550\u2561 b0d01238-1213-45c3-9bef-a443afc6b87c\n\u03b2 = 2.0\n\n# \u2554\u2550\u2561 2cce4c5e-7890-4c6c-bd7c-eecc8437bef3\n\u03f5 = 0.5\n\n# \u2554\u2550\u2561 1b454935-c858-4817-928d-da116011d7b0\n\u03b2 + \u03f5\n\n# \u2554\u2550\u2561 96ced3bd-69e7-490c-b8be-b06a30049a31\n\u03c0\n\n# \u2554\u2550\u2561 3de2d16f-8849-4462-9ef3-0cc1830285c6\nmd\"\"\"\n> \u26a0\ufe0f Perceba que o s\u00edmbolo $\u03c0$ j\u00e1 possui um valor associado na linguagem Julia.\n\"\"\"\n\n# \u2554\u2550\u2561 d28119c3-6800-430d-9ffe-d6f25e8e5c2a\nmd\"\"\"\n## 2. Fun\u00e7\u00f5es\n\nAs **fun\u00e7\u00f5es**, no mundo da computa\u00e7\u00e3o, s\u00e3o similares \u00e0s fun\u00e7\u00f5es matem\u00e1ticas. De forma geral, uma fun\u00e7\u00e3o recebe valores de **entrada**, processa-os e retorna valores de **sa\u00edda**.\n\nEm Julia, existem, essencialmente, tr\u00eas maneiras distintas de se construir fun\u00e7\u00f5es:\n\n\"\"\"\n\n# \u2554\u2550\u2561 bb495116-8ff9-4a66-9b66-6f6fe990c5d9\nf(\u03c7) = 5\u03c7 + 25\n\n# \u2554\u2550\u2561 a63d4a04-0ceb-4e4f-90ba-30dc798e1b48\nfunction areatriangulo(b,h)\n\tA = (b * h) / 2\n\treturn A\nend\n\n# \u2554\u2550\u2561 9cf1d023-1421-4adb-af35-64aa6a8ddb53\nmd\"\"\"\n> \u26a0\ufe0f O comando `return` foi utilizado aqui apenas por fins did\u00e1ticos. Entretanto, poder\u00edamos omiti-lo.\n\"\"\"\n\n# \u2554\u2550\u2561 5b168a45-b5a2-4415-80f4-e2347c21730a\ndiagonal = l -> l * \u221a2\n\n# \u2554\u2550\u2561 1a1ac9a0-b8f0-46d1-9d89-21b6bc0e6aec\nf(2) + areatriangulo(10,4) / diagonal(1)\n\n# \u2554\u2550\u2561 784426f8-ac63-4555-adb2-cbfb9b37fdda\nmd\"\"\"\n#### \ud83d\udd8a\ufe0f Exerc\u00edcio\n\nCrie uma fun\u00e7\u00e3o `recurso` que retorne a tonelagem em metal contido de um dep\u00f3sito de volume `V`, densidade m\u00e9dia `\u03c1` e teor m\u00e9dio `T` (em %).\n\n\"\"\"\n\n# \u2554\u2550\u2561 ffc5e222-8ae8-4314-a616-e4588d7808fe\nrecurso(V, \u03c1, T) = missing\n\n# \u2554\u2550\u2561 5dc76f3b-badf-4951-9eb4-f61ca66dfb98\nmd\"\"\"\n## 3. Cole\u00e7\u00f5es\n\nAs **cole\u00e7\u00f5es** s\u00e3o conjuntos de elementos, normalmente do mesmo tipo, mas n\u00e3o necessariamente. S\u00e3o exemplos *vetores*, *matrizes* e *tensores*.\n\nOs **vetores** podem ser escritos como...\n\n\"\"\"\n\n# \u2554\u2550\u2561 d349ac4d-44bb-4505-a6c4-3096d8c48ba0\nvetor1 = [1,2,3,4,5,6,7,8,9,10]\n\n# \u2554\u2550\u2561 9e181c4a-a759-4f92-8444-6a23387a20b9\nvetor_2 = collect(10:20)\n\n# \u2554\u2550\u2561 d39f503a-158e-4774-851e-d786014b8f2b\nmd\" As **matrizes** podem ser escritas como... \"\n\n# \u2554\u2550\u2561 b5387324-df28-422d-8e74-4fefa8054e7f\nA = [1 2\n\t 3 4]\n\n# \u2554\u2550\u2561 570f62db-edd7-429d-a084-2686f4b6923c\nB = [1 2 3; 4 5 6; 7 8 9]\n\n# \u2554\u2550\u2561 7e8a4e33-0e9c-45f5-b8bc-e2adece0806d\nmd\" Os **tensores** podem ser escritos como... \"\n\n# \u2554\u2550\u2561 204bcedc-4123-4845-bcac-493ce3544582\ntensor_de_zeros = zeros(5,5,2)\n\n# \u2554\u2550\u2561 92b0cb5f-41a1-420f-8b81-3b967db8138f\ntensor_de_uns = ones(2,2,3)\n\n# \u2554\u2550\u2561 7471120d-6e00-46be-be11-649c8cdbb32c\nmd\"\"\"\nUma opera\u00e7\u00e3o muito utilizada \u00e9 o **fatiamento**, que visa obter apenas certo(s) elemento(s) de uma cole\u00e7\u00e3o. O fatiamento de vetores, por exemplo, \u00e9 realizado a partir da seguinte sintaxe:\n\n```julia\nvetor[\u00edndice inicial:\u00edndice final]\n```\n\nObserve o vetor `v\u2081`...\n\n\"\"\"\n\n# \u2554\u2550\u2561 b31b883d-5a69-4e04-b8b5-cedca81dcf9a\nv\u2081 = [10,20,30,40,50,60,70,80,90,100]\n\n# \u2554\u2550\u2561 c7f94dcd-64a2-4e97-b38d-363dc6f9f757\nmd\" O primeiro elemento pode ser obtido a partir do fatiamento de `v\u2081`...\"\n\n# \u2554\u2550\u2561 2a25a884-44cf-4460-ab23-02e728201f90\nv\u2081[1]\n\n# \u2554\u2550\u2561 e127bdf8-2a53-4534-a628-1463a5735f41\nmd\"\"\"\n\n> \u26a0\ufe0f Assim como R e diferentemente de Python ou JavaScript, a numera\u00e7\u00e3o do \u00edndice se inicia em **1** na linguagem Julia.\n\nOs tr\u00eas primeiros elementos de `v\u2081` podem ser obtidos como...\n\n\"\"\"\n\n# \u2554\u2550\u2561 0806feb3-33eb-402c-92d2-af0b153834ba\nv\u2081[1:3]\n\n# \u2554\u2550\u2561 1a5f467b-7244-4587-b01c-61fd22bfbb9b\nv\u2081[begin:3]\n\n# \u2554\u2550\u2561 4b1ec558-5f94-417a-aa3c-d13a0039a6a0\nmd\" Os dois \u00faltimos elementos de `v\u2081` podem ser obtidos como...\"\n\n# \u2554\u2550\u2561 6bee210a-11cc-40c4-bfb0-d14a6b2495b4\nv\u2081[9:10]\n\n# \u2554\u2550\u2561 10cc97dd-a390-4d4a-9644-20b654ff66bb\nv\u2081[end-1:end]\n\n# \u2554\u2550\u2561 04738501-16fa-43e1-b640-47682c155bcb\nmd\"\"\"\n\n#### \ud83d\udd8a\ufe0f Exerc\u00edcio\n\nFatie o vetor `v\u2082`, de modo que apenas os elementos da terceira posi\u00e7\u00e3o em diante sejam retornados. A fatia resultante deve ser armazenada na vari\u00e1vel `v\u2083`.\n\n\"\"\"\n\n# \u2554\u2550\u2561 f9317c0c-2389-43a8-8b91-f8b2f278e258\nbegin\n\tv\u2082 = collect(10:15)\n\tv\u2083 = missing\nend\n\n# \u2554\u2550\u2561 8253149c-e1b0-4e3f-9f5b-909621dba86e\nmd\"\"\"\n## 4. Condicionais\n\nAs **estruturas condicionais** s\u00e3o utilizadas em situa\u00e7\u00f5es em que se deseja executar algum trecho de c\u00f3digo apenas quando uma **condi\u00e7\u00e3o** \u00e9 satisfeita:\n\n```julia\nif condi\u00e7\u00e3o\n\tcomando_1\nelse\n\tcomando_2\nend\n```\n\nO exemplo abaixo mostra uma fun\u00e7\u00e3o `\u00e9granitoide` que utiliza estruturas condicionais. **Se** o conte\u00fado em quartzo for menor que 20%, a fun\u00e7\u00e3o retorna a frase \"N\u00e3o \u00e9 granitoide \ud83d\ude10\". **Caso contr\u00e1rio**, a frase \"\u00c9 granitoide!\" \u00e9 retornada.\n\n\"\"\"\n\n# \u2554\u2550\u2561 4a778055-7ade-4275-aa87-95c121b31232\nfunction \u00e9granitoide(quartzo)\n\tif quartzo < 20\n \t\trochaignea = \"N\u00e3o \u00e9 granitoide \ud83d\ude10\"\n\telse\n \t\trochaignea = \"\u00c9 granitoide!\"\n\tend\n\t\n\treturn rochaignea\nend\n\n# \u2554\u2550\u2561 ca5453f5-1768-4f59-b09d-e40bb4981622\n\u00e9granitoide(30)\n\n# \u2554\u2550\u2561 217945a3-919b-4c5f-afb0-6eaadf4e9022\n\u00e9granitoide(5)\n\n# \u2554\u2550\u2561 1f0abfbe-49cd-4529-8f4d-65f8286b6f0e\nmd\"\"\"\n\n#### \ud83d\udd8a\ufe0f Exerc\u00edcio\n\nAgora, crie uma fun\u00e7\u00e3o `tiporocha` que recebe o nome de uma rocha e retorna o seu tipo:\n\n* \"gabro\" \u2192 \"\u00edgnea\"\n* \"gnaisse\" \u2192 \"metam\u00f3rfica\"\n* \"ritmito\" \u2192 \"sedimentar\"\n\n\"\"\"\n\n# \u2554\u2550\u2561 6fcc1433-c348-4825-8503-782304ed508b\nfunction tiporocha(rocha)\n\tmissing\nend\n\n# \u2554\u2550\u2561 3cfbddfb-be59-4019-965f-13bdbac2295a\nmd\"\"\"\n\n## 5. La\u00e7os de repeti\u00e7\u00e3o\n\nOs **la\u00e7os de repeti\u00e7\u00e3o** s\u00e3o utilizados quando se deseja repetir determinado trecho do algoritmo m\u00faltiplas vezes. O n\u00famero de repeti\u00e7\u00f5es pode ser indeterminado, mas necessariamente finito (*Dauricio, 2015*).\n\nUma das repeti\u00e7\u00f5es mais utilizadas \u00e9 o `for`, que apresenta a seguinte sintaxe:\n\n```julia\nfor elemento in vetor\n\tcomando\nend\n```\n\nO comando `for` tamb\u00e9m pode ser utilizado para criar vetores/listas, a partir das chamadas **list comprehensions**. A sintaxe \u00e9 definida da seguinte maneira:\n\n```julia\n[comando for elemento in vetor]\n```\n\nO exemplo abaixo mostra a utiliza\u00e7\u00e3o do recurso *list comprehension* para a cria\u00e7\u00e3o de um vetor constitu\u00eddo por pot\u00eancias de 2:\n\n\"\"\"\n\n# \u2554\u2550\u2561 abc428c6-b762-47bc-b5ac-7abedd1a7021\n[2 ^ num for num in 1:10]\n\n# \u2554\u2550\u2561 bc8fa2b9-0803-40e7-af91-2196a11aae44\nmd\"\"\"\n\n#### \ud83d\udd8a\ufe0f Exerc\u00edcio\n\nEscreva a fun\u00e7\u00e3o `raiz` que retorna a ra\u00edz quadrada de todos os elementos do vetor `v\u2084`.\n\n\"\"\"\n\n# \u2554\u2550\u2561 729eaff4-7aa1-4db1-a04f-57216175b029\nraiz(v\u2084) = missing\n\n# \u2554\u2550\u2561 5eb8ff96-2adf-465d-bf8c-bd4e64fa6342\nmd\"\"\"\nPodemos ainda utilizar a sintaxe *list comprehension* para criarmos uma tabuada em apenas uma linha!\n\"\"\"\n\n# \u2554\u2550\u2561 0d420048-c4f9-4149-b25b-149ef77f3264\n[linha * coluna for linha in 1:10, coluna in 1:10]\n\n# \u2554\u2550\u2561 f8f272e0-17ca-47e5-8dd0-577eb6322c90\nmd\"\"\"\n\n## 6. Interatividade\n\nO ambiente [Pluto](https://github.com/fonsp/Pluto.jl) e o pacote [PlutoUI](https://github.com/fonsp/PlutoUI.jl) proporciona uma s\u00e9rie de recursos interativos a partir da macro `@bind`. Esses recursos permitem que o valor de uma vari\u00e1vel seja alterado de acordo com a intera\u00e7\u00e3o entre o usu\u00e1rio e o notebook.\n\nO recurso **Slider** funciona como uma barra de deslizamento para vari\u00e1veis que assumem valores cont\u00ednuos. A sua sintaxe \u00e9 dada por:\n\n```julia\n@bind nome_var Slider(inicio:passo:fim)\n```\n\nO *slider* abaixo mostra o teor de Cu (%) em uma amostra...\n\n\"\"\"\n\n# \u2554\u2550\u2561 e137bf5e-d795-49ac-b641-b5f7364cfac9\n@bind CU Slider(0.05:0.01:3.00, default=1.0, show_value=true)\n\n# \u2554\u2550\u2561 fae5e284-66e7-4599-9905-f0d5e64387ea\nmd\" A amostra apresenta $CU % de Cu\"\n\n# \u2554\u2550\u2561 9ee680a7-2866-4f1b-84af-0fdb737246ad\nmd\"\"\"\nUm outro elemento interativo \u00e9 o **TextField**, que se refere a uma caixa de texto. Sua sintaxe \u00e9 definida como:\n\n```julia\n@bind nome_var TextField()\n```\n\n\"\"\"\n\n# \u2554\u2550\u2561 6c7e7d3e-d544-4b59-9504-e28fe2cc3619\nmd\"\"\"\n\n#### \ud83d\udd8a\ufe0f Exerc\u00edcio\n\nCom seus conhecimentos geol\u00f3gicos, tente identificar qual rocha \u00e9 exibida na Figura 01. Escreva o nome da rocha (apenas com letras min\u00fasculas) na caixa de texto abaixo.\n\n\"\"\"\n\n# \u2554\u2550\u2561 2fd0708e-b225-4f8c-8f4f-b46305e06366\nmd\"\"\"\n\n\n_**Figura 01:** Identifique a rocha acima. Extra\u00eddo de [sandatlas.org](https://www.sandatlas.org/mylonite/)._\n\"\"\"\n\n# \u2554\u2550\u2561 98a32d2d-7173-406d-92cf-dfa847d47c49\nmd\" Rocha: $(@bind rocha TextField())\"\n\n# \u2554\u2550\u2561 df1f77ac-bbf5-4d42-91bf-e634ebee4bb9\nmd\"\"\" Podemos tamb\u00e9m utilizar o recurso interativo **Select** que, por sua vez, atua como uma lista suspensa. Esse comando possui a seguinte sintaxe:\n\n```julia\n@bind nome_var Select([\"elem_1\", \"elem_2\", ..., \"elem_n\"])\n```\n\n\"\"\"\n\n# \u2554\u2550\u2561 ba7ce05c-cae6-4eec-9760-e4e253986061\nmd\"\"\"\n\n#### \ud83d\udd8a\ufe0f Exerc\u00edcio\n\nNa lista suspensa seguir, selecione a op\u00e7\u00e3o que corresponda \u00e0 correta subclasse do silicato exibido na Figura 02.\n\n\"\"\"\n\n# \u2554\u2550\u2561 b844b981-2922-4056-811d-8c61b904d996\nmd\"\"\"\n\n\n_**Figura 02:** Identifique a subclasse do silicato acima. Extra\u00eddo de: [geology.com](https://geology.com/minerals/titanite.shtml)._\n\"\"\"\n\n# \u2554\u2550\u2561 87832d6c-525d-4060-a1e8-d06a611b5dfe\nmd\"\"\" Subclasse: $(@bind silicato Select([\"Tectossilicato\",\"Filossilicato\",\n\t\t\t\t\t\t\t\t\t\t \"Inossilicato\",\"Ciclossilicato\",\n\t\t\t\t\t\t\t\t\t\t \"Sorossilicato\",\"Nesossilicato\"]))\n\"\"\"\n\n# \u2554\u2550\u2561 a93dd751-a6b3-454d-9f70-b864fdbbd968\nmd\"\"\"\n## Refer\u00eancias\n\n*Dauricio, J. S. **Algoritmos e L\u00f3gica de Programa\u00e7\u00e3o**. Londrina: Editora e Distribuidora Educacional S.A., 2015.*\n\"\"\"\n\n# \u2554\u2550\u2561 8f866ee4-ce44-4d42-be56-7764168c1c71\nmd\"\"\"\n## Recursos adicionais\n\nAbaixo, s\u00e3o listados alguns recursos complementares a este notebook:\n\n> [Documenta\u00e7\u00e3o da Linguagem Julia](https://docs.julialang.org/en/v1/)\n\n> [YouTube - Julia Language](https://www.youtube.com/c/TheJuliaLanguage)\n\n> [YouTube - Programa\u00e7\u00e3o Din\u00e2mica](https://www.youtube.com/playlist?list=PL5TJqBvpXQv4TO4Y_JZExBCBzNXDLYUCj)\n\"\"\"\n\n# \u2554\u2550\u2561 ce463e41-8cb2-46cb-9656-1503997a883e\nmd\"\"\"\n## Pacotes utilizados\n\nOs seguintes pacotes foram utilizados neste notebook:\n\n| Pacote | Descri\u00e7\u00e3o |\n|:---------------------------------------------:|:-------------------:|\n|[PlutoUI](https://github.com/fonsp/PlutoUI.jl) | Widgets interativos |\n\n\"\"\"\n\n# \u2554\u2550\u2561 bb899ab7-b3d8-493f-aaa6-85a4710a6690\nbegin\n\thint(text) = Markdown.MD(Markdown.Admonition(\"hint\", \"Dica\", [text]))\n\n\talmost(text) = Markdown.MD(Markdown.Admonition(\"warning\", \"Quase l\u00e1!\", [text]))\n\n\tstill_missing(text=md\"Troque `missing` pela sua resposta.\") = Markdown.MD(Markdown.Admonition(\"warning\", \"Aqui vamos n\u00f3s!\", [text]))\n\n\tkeep_working(text=md\"A resposta n\u00e3o est\u00e1 correta \ud83d\ude14\") = Markdown.MD(Markdown.Admonition(\"danger\", \"N\u00e3o desanime, voc\u00ea est\u00e1 quase l\u00e1!\", [text]))\n\n\tyays = [md\"Fant\u00e1stico!\", md\"\u00d3timo!\", md\"Yay \u2764\", md\"Legal! \ud83c\udf89\", md\"Muito bem!\", md\"Bom trabalho!\", md\"Voc\u00ea conseguiu a resposta certa!\", md\"Vamos seguir para pr\u00f3xima se\u00e7\u00e3o.\"]\n\n\tcorrect(text=rand(yays)) = Markdown.MD(Markdown.Admonition(\"correct\", \"Certa resposta!\", [text]))\n\n\tnot_defined(variable_name) = Markdown.MD(Markdown.Admonition(\"danger\", \"Opa!\", [md\"Tenha certeza que definiu uma vari\u00e1vel chamada **$(Markdown.Code(string(variable_name)))**\"]))\nend;\n\n# \u2554\u2550\u2561 a4f3b825-6806-4c64-bbe0-6d7383015d68\nbegin\n\ts1 = false\n\t_rec = recurso(12500000,2.7,5)\n\tif ismissing(_rec)\n\t\tstill_missing()\n\telseif _rec \u2248 (12500000 * 2.7 * 5) / 100\n\t\ts1 = true\n\t\tcorrect()\n\telseif _rec isa Number\n\t\talmost(md\"A f\u00f3rmula n\u00e3o est\u00e1 certa...\")\n\telse\n\t\tkeep_working()\n\tend\nend\n\n# \u2554\u2550\u2561 3ccac0bc-9f78-4ece-988b-ba832811e538\nhint(md\"Algu\u00e9m me contou que a f\u00f3rmula \u00e9 $\\frac{V\u03c1T}{100}$...\")\n\n# \u2554\u2550\u2561 a84b90c1-fa85-499a-9fa4-ac1f33ed8469\nbegin\n\ts2 = false\n\tif ismissing(v\u2083)\n\t\tstill_missing()\n\telseif v\u2083 == v\u2082[3:end]\n\t\ts2 = true\n\t\tcorrect()\n\telse\n\t\tkeep_working()\n\tend\nend\n\n# \u2554\u2550\u2561 e2c42c9e-abbb-4a33-844d-ef0f0c9b4939\nhint(md\"Utilize `end` como \u00edndice final.\")\n\n# \u2554\u2550\u2561 33b81b24-ca33-4dd3-a85e-0371ca8623e8\nbegin\n\ts3 = false\n\t_rcktype = tiporocha.([\"gabro\",\"gnaisse\",\"ritmito\"])\n\tif all(ismissing.(_rcktype))\n\t\tstill_missing()\n\telseif all(_rcktype .== [\"\u00edgnea\",\"metam\u00f3rfica\",\"sedimentar\"])\n\t\ts3 = true\n\t\tcorrect()\n\telseif _rcktype \u2286 [\"\u00edgnea\",\"metam\u00f3rfica\",\"sedimentar\"]\n\t\talmost(md\"A resposta n\u00e3o est\u00e1 100% correta...\")\n\telse\n\t\tkeep_working()\n\tend\nend\n\n# \u2554\u2550\u2561 e462ca14-b38a-46b0-8faa-2261a2cfc150\nhint(md\"Basta escrever uma sequ\u00eancia de `if rocha == \\\"gabro\\\" return \\\"\u00edgnea\\\" end`\")\n\n# \u2554\u2550\u2561 7d12857d-d767-49e5-b7de-11e696f1d84a\nbegin\n\ts4 = false\n\t_sqrt = raiz([1 4; 9 16])\n\tif ismissing(_sqrt)\n\t\tstill_missing()\n\telseif _sqrt == [1 2; 3 4]\n\t\ts4 = true\n\t\tcorrect()\n\telseif _sqrt == [1, 2, 3, 4]\n\t\talmost(md\"Tente usar a dica!\")\n\telse\n\t\tkeep_working()\n\tend\nend\n\n# \u2554\u2550\u2561 a9f386e1-8c6e-40e7-ba67-b3792c50f87e\nhint(md\"A nota\u00e7\u00e3o list comprehension `[f(x) for v in v\u2084]` pode ser bem \u00fatil!\")\n\n# \u2554\u2550\u2561 f1080b76-7564-4419-884f-70a49bfe9d57\nbegin\n\ts5 = false\n\tif rocha == \"\"\n\t\talmost(md\"Identifique a rocha!\")\n\telseif rocha == \"milonito\"\n\t\ts5 = true\n\t\tcorrect()\n\telseif rocha == \"protomilonito\" || rocha == \"ultramilonito\"\n\t\talmost(md\"\"\"A porcentagem de matriz \"triturada\" \u00e9 de 50% a 90%.\"\"\")\n\telse\n\t\tkeep_working()\n\tend\nend\n\n# \u2554\u2550\u2561 844171ee-dfc3-4985-a6a1-7aa7986d1ac4\nhint(md\"Essa rocha \u00e9 um t\u00edpico produto de recristaliza\u00e7\u00e3o din\u00e2mica!\")\n\n# \u2554\u2550\u2561 8b0df84d-a112-49ec-9f7b-c6e5efd45c48\nbegin\n\ts6 = false\n\tif silicato == \"Nesossilicato\"\n\t\ts6 = true\n\t\tcorrect()\n\telse\n\t\tkeep_working()\n\tend\nend\n\n# \u2554\u2550\u2561 55298aee-618b-4704-98ff-e81fdbddd9a7\nhint(md\"A f\u00f3rmula qu\u00edmica deste silicato \u00e9 `CaTiO(SiO\u2084)`.\")\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000001\nPLUTO_PROJECT_TOML_CONTENTS = \"\"\"\n[deps]\nPlutoUI = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\n\n[compat]\nPlutoUI = \"~0.7.14\"\n\"\"\"\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000002\nPLUTO_MANIFEST_TOML_CONTENTS = \"\"\"\n# This file is machine-generated - editing it directly is not advised\n\n[[Base64]]\nuuid = \"2a0f44e3-6c83-55bd-87e4-b1978d98bd5f\"\n\n[[Dates]]\ndeps = [\"Printf\"]\nuuid = \"ade2ca70-3891-5945-98fb-dc099432e06a\"\n\n[[HypertextLiteral]]\ngit-tree-sha1 = \"72053798e1be56026b81d4e2682dbe58922e5ec9\"\nuuid = \"ac1192a8-f4b3-4bfe-ba22-af5b92cd3ab2\"\nversion = \"0.9.0\"\n\n[[IOCapture]]\ndeps = [\"Logging\", \"Random\"]\ngit-tree-sha1 = \"f7be53659ab06ddc986428d3a9dcc95f6fa6705a\"\nuuid = \"b5f81e59-6552-4d32-b1f0-c071b021bf89\"\nversion = \"0.2.2\"\n\n[[InteractiveUtils]]\ndeps = [\"Markdown\"]\nuuid = \"b77e0a4c-d291-57a0-90e8-8db25a27a240\"\n\n[[JSON]]\ndeps = [\"Dates\", \"Mmap\", \"Parsers\", \"Unicode\"]\ngit-tree-sha1 = \"8076680b162ada2a031f707ac7b4953e30667a37\"\nuuid = \"682c06a0-de6a-54ab-a142-c8b1cf79cde6\"\nversion = \"0.21.2\"\n\n[[Logging]]\nuuid = \"56ddb016-857b-54e1-b83d-db4d58db5568\"\n\n[[Markdown]]\ndeps = [\"Base64\"]\nuuid = \"d6f4376e-aef5-505a-96c1-9c027394607a\"\n\n[[Mmap]]\nuuid = \"a63ad114-7e13-5084-954f-fe012c677804\"\n\n[[Parsers]]\ndeps = [\"Dates\"]\ngit-tree-sha1 = \"a8709b968a1ea6abc2dc1967cb1db6ac9a00dfb6\"\nuuid = \"69de0a69-1ddd-5017-9359-2bf0b02dc9f0\"\nversion = \"2.0.5\"\n\n[[PlutoUI]]\ndeps = [\"Base64\", \"Dates\", \"HypertextLiteral\", \"IOCapture\", \"InteractiveUtils\", \"JSON\", \"Logging\", \"Markdown\", \"Random\", \"Reexport\", \"UUIDs\"]\ngit-tree-sha1 = \"d1fb76655a95bf6ea4348d7197b22e889a4375f4\"\nuuid = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\nversion = \"0.7.14\"\n\n[[Printf]]\ndeps = [\"Unicode\"]\nuuid = \"de0858da-6303-5e67-8744-51eddeeeb8d7\"\n\n[[Random]]\ndeps = [\"Serialization\"]\nuuid = \"9a3f8284-a2c9-5f02-9a11-845980a1fd5c\"\n\n[[Reexport]]\ngit-tree-sha1 = \"45e428421666073eab6f2da5c9d310d99bb12f9b\"\nuuid = \"189a3867-3050-52da-a836-e630ba90ab69\"\nversion = \"1.2.2\"\n\n[[SHA]]\nuuid = \"ea8e919c-243c-51af-8825-aaa63cd721ce\"\n\n[[Serialization]]\nuuid = \"9e88b42a-f829-5b0c-bbe9-9e923198166b\"\n\n[[UUIDs]]\ndeps = [\"Random\", \"SHA\"]\nuuid = \"cf7118a7-6976-5b1a-9a39-7adc72f591a4\"\n\n[[Unicode]]\nuuid = \"4ec0a83e-493e-50e2-b9ac-8f72acf5a8f5\"\n\"\"\"\n\n# \u2554\u2550\u2561 Cell order:\n# \u255f\u250075fe8fd0-2563-11ec-30ef-d15cdc3d4e63\n# \u255f\u2500d4b5618f-f1a7-4483-b738-fb94e1e2bbbe\n# \u255f\u2500deee1735-37ee-4c97-a4df-8fcb61e95d6a\n# \u255f\u2500488dcacd-109d-41f2-b904-3d17193e6190\n# \u255f\u25006ca3e113-02db-4cef-ad9e-3941ac7d7a6d\n# \u255f\u250014db5525-7c0a-433b-a23c-088db728f46b\n# \u255f\u2500c111da75-d294-4def-93fb-56953c3585ad\n# \u2560\u25507ceb9c27-0310-4f97-8349-91286c1f9235\n# \u2560\u2550218bb8c3-729b-43c7-9a53-d3b579cf2d21\n# \u255f\u25003b3e6955-1573-4620-8109-d80b71cf0708\n# \u2560\u2550f2e6adcd-9095-496f-a8e3-88069537015c\n# \u2560\u2550870cbb76-b0c3-4d14-be22-86e0d34ebe58\n# \u255f\u250055a2c622-0f9f-45db-b534-55873c0759d4\n# \u255f\u2500449c91c3-7f03-4ffd-95e7-02cdb58323fd\n# \u2560\u25508d463f50-d694-41eb-bbad-c40f93471852\n# \u255f\u25004d31b989-7a7d-43ca-a1f8-831db58f1c8e\n# \u2560\u2550b0d01238-1213-45c3-9bef-a443afc6b87c\n# \u2560\u25502cce4c5e-7890-4c6c-bd7c-eecc8437bef3\n# \u2560\u25501b454935-c858-4817-928d-da116011d7b0\n# \u2560\u255096ced3bd-69e7-490c-b8be-b06a30049a31\n# \u255f\u25003de2d16f-8849-4462-9ef3-0cc1830285c6\n# \u255f\u2500d28119c3-6800-430d-9ffe-d6f25e8e5c2a\n# \u2560\u2550bb495116-8ff9-4a66-9b66-6f6fe990c5d9\n# \u2560\u2550a63d4a04-0ceb-4e4f-90ba-30dc798e1b48\n# \u255f\u25009cf1d023-1421-4adb-af35-64aa6a8ddb53\n# \u2560\u25505b168a45-b5a2-4415-80f4-e2347c21730a\n# \u2560\u25501a1ac9a0-b8f0-46d1-9d89-21b6bc0e6aec\n# \u255f\u2500784426f8-ac63-4555-adb2-cbfb9b37fdda\n# \u2560\u2550ffc5e222-8ae8-4314-a616-e4588d7808fe\n# \u255f\u2500a4f3b825-6806-4c64-bbe0-6d7383015d68\n# \u255f\u25003ccac0bc-9f78-4ece-988b-ba832811e538\n# \u255f\u25005dc76f3b-badf-4951-9eb4-f61ca66dfb98\n# \u2560\u2550d349ac4d-44bb-4505-a6c4-3096d8c48ba0\n# \u2560\u25509e181c4a-a759-4f92-8444-6a23387a20b9\n# \u255f\u2500d39f503a-158e-4774-851e-d786014b8f2b\n# \u2560\u2550b5387324-df28-422d-8e74-4fefa8054e7f\n# \u2560\u2550570f62db-edd7-429d-a084-2686f4b6923c\n# \u255f\u25007e8a4e33-0e9c-45f5-b8bc-e2adece0806d\n# \u2560\u2550204bcedc-4123-4845-bcac-493ce3544582\n# \u2560\u255092b0cb5f-41a1-420f-8b81-3b967db8138f\n# \u255f\u25007471120d-6e00-46be-be11-649c8cdbb32c\n# \u2560\u2550b31b883d-5a69-4e04-b8b5-cedca81dcf9a\n# \u255f\u2500c7f94dcd-64a2-4e97-b38d-363dc6f9f757\n# \u2560\u25502a25a884-44cf-4460-ab23-02e728201f90\n# \u255f\u2500e127bdf8-2a53-4534-a628-1463a5735f41\n# \u2560\u25500806feb3-33eb-402c-92d2-af0b153834ba\n# \u2560\u25501a5f467b-7244-4587-b01c-61fd22bfbb9b\n# \u255f\u25004b1ec558-5f94-417a-aa3c-d13a0039a6a0\n# \u2560\u25506bee210a-11cc-40c4-bfb0-d14a6b2495b4\n# \u2560\u255010cc97dd-a390-4d4a-9644-20b654ff66bb\n# \u255f\u250004738501-16fa-43e1-b640-47682c155bcb\n# \u2560\u2550f9317c0c-2389-43a8-8b91-f8b2f278e258\n# \u255f\u2500a84b90c1-fa85-499a-9fa4-ac1f33ed8469\n# \u255f\u2500e2c42c9e-abbb-4a33-844d-ef0f0c9b4939\n# \u255f\u25008253149c-e1b0-4e3f-9f5b-909621dba86e\n# \u2560\u25504a778055-7ade-4275-aa87-95c121b31232\n# \u2560\u2550ca5453f5-1768-4f59-b09d-e40bb4981622\n# \u2560\u2550217945a3-919b-4c5f-afb0-6eaadf4e9022\n# \u255f\u25001f0abfbe-49cd-4529-8f4d-65f8286b6f0e\n# \u2560\u25506fcc1433-c348-4825-8503-782304ed508b\n# \u255f\u250033b81b24-ca33-4dd3-a85e-0371ca8623e8\n# \u255f\u2500e462ca14-b38a-46b0-8faa-2261a2cfc150\n# \u255f\u25003cfbddfb-be59-4019-965f-13bdbac2295a\n# \u2560\u2550abc428c6-b762-47bc-b5ac-7abedd1a7021\n# \u255f\u2500bc8fa2b9-0803-40e7-af91-2196a11aae44\n# \u2560\u2550729eaff4-7aa1-4db1-a04f-57216175b029\n# \u255f\u25007d12857d-d767-49e5-b7de-11e696f1d84a\n# \u255f\u2500a9f386e1-8c6e-40e7-ba67-b3792c50f87e\n# \u255f\u25005eb8ff96-2adf-465d-bf8c-bd4e64fa6342\n# \u2560\u25500d420048-c4f9-4149-b25b-149ef77f3264\n# \u255f\u2500f8f272e0-17ca-47e5-8dd0-577eb6322c90\n# \u255f\u2500e137bf5e-d795-49ac-b641-b5f7364cfac9\n# \u255f\u2500fae5e284-66e7-4599-9905-f0d5e64387ea\n# \u255f\u25009ee680a7-2866-4f1b-84af-0fdb737246ad\n# \u255f\u25006c7e7d3e-d544-4b59-9504-e28fe2cc3619\n# \u255f\u25002fd0708e-b225-4f8c-8f4f-b46305e06366\n# \u255f\u250098a32d2d-7173-406d-92cf-dfa847d47c49\n# \u255f\u2500f1080b76-7564-4419-884f-70a49bfe9d57\n# \u255f\u2500844171ee-dfc3-4985-a6a1-7aa7986d1ac4\n# \u255f\u2500df1f77ac-bbf5-4d42-91bf-e634ebee4bb9\n# \u255f\u2500ba7ce05c-cae6-4eec-9760-e4e253986061\n# \u255f\u2500b844b981-2922-4056-811d-8c61b904d996\n# \u255f\u250087832d6c-525d-4060-a1e8-d06a611b5dfe\n# \u255f\u25008b0df84d-a112-49ec-9f7b-c6e5efd45c48\n# \u255f\u250055298aee-618b-4704-98ff-e81fdbddd9a7\n# \u255f\u2500a93dd751-a6b3-454d-9f70-b864fdbbd968\n# \u255f\u25008f866ee4-ce44-4d42-be56-7764168c1c71\n# \u255f\u2500ce463e41-8cb2-46cb-9656-1503997a883e\n# \u255f\u2500bb899ab7-b3d8-493f-aaa6-85a4710a6690\n# \u255f\u250000000000-0000-0000-0000-000000000001\n# \u255f\u250000000000-0000-0000-0000-000000000002\n", "meta": {"hexsha": "b2e5507a77a43c502d703a7b340757e08e7d1331", "size": 25804, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "1-logica_de_programacao.jl", "max_stars_repo_name": "fnaghetini/intro-to-geostats", "max_stars_repo_head_hexsha": "6f82ddb50f6b0aa6c86d0ef2095b1ca3791d0420", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 10, "max_stars_repo_stars_event_min_datetime": "2021-10-31T12:49:19.000Z", "max_stars_repo_stars_event_max_datetime": "2021-11-24T11:39:45.000Z", "max_issues_repo_path": "1-logica_de_programacao.jl", "max_issues_repo_name": "fnaghetini/intro-to-geostats", "max_issues_repo_head_hexsha": "6f82ddb50f6b0aa6c86d0ef2095b1ca3791d0420", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "1-logica_de_programacao.jl", "max_forks_repo_name": "fnaghetini/intro-to-geostats", "max_forks_repo_head_hexsha": "6f82ddb50f6b0aa6c86d0ef2095b1ca3791d0420", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 2, "max_forks_repo_forks_event_min_datetime": "2021-11-08T23:14:35.000Z", "max_forks_repo_forks_event_max_datetime": "2021-11-22T19:42:08.000Z", "avg_line_length": 32.7878017789, "max_line_length": 760, "alphanum_fraction": 0.7298093319, "num_tokens": 11125, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.22270014914315836, "lm_q2_score": 0.334589441253186, "lm_q1q2_score": 0.07451311846881055}}
{"text": "# ---\n# title: 139. Word Break\n# id: problem139\n# author: zhwang\n# date: 2022-02-18\n# difficulty: Medium\n# categories: Dynamic Programming\n# link: , Expr(:tuple, plist...), Expr(:call, f1, alist...)))\n # if f has non-scalar output, sum it\n isbits(y) || (f2=f; f=(x...)->toscalar(f2(x...)))\n return (f,fargs...)\nend\n\nfunction randin(range, dims...; eps=EPS)\n if isa(range, UnitRange{Int64})\n rand(range, dims...)\n elseif range==(-Inf,Inf)\n randn(dims...)\n elseif range==(0,Inf)\n eps-log(rand(dims...))\n elseif range==(1,Inf)\n eps+1-log(rand(dims...))\n elseif range==(-1,Inf)\n eps-1-log(rand(dims...))\n elseif range==(-1,1)\n (1-eps)*(2rand(dims...)-1)\n elseif range==(0,1)\n eps+(1-2eps)*rand(dims...)\n elseif range==(0,2)\n eps+2*(1-eps)*rand(dims...)\n elseif range==(-Inf,-1,1,Inf)\n x = sec(randn(dims...))\n sign(x)*eps + x\n else\n error(\"Unknown range $range\")\n end\nend\n\nfunction addtest1(f,r) # unary\n addtest(f,randin(r))\n addtest(f,randin(r,2))\nend\n\nfunction addtest2(f,r1,r2=r1) # binary\n addtest(f,randin(r1),randin(r2))\n addtest(f,randin(r1),randin(r2,2))\n addtest(f,randin(r1,2),randin(r2))\n addtest(f,randin(r1,2),randin(r2,2))\nend\n\nfunction addtest3(f,r1,r2=r1) # broadcasting\n addtest2(f,r1,r2)\n addtest(f,randin(r1,2),randin(r2,2,2))\n addtest(f,randin(r1,2,2),randin(r2,2))\n addtest(f,randin(r1,1,2),randin(r2,2,2))\n addtest(f,randin(r1,2,2),randin(r2,1,2))\nend\n\n\n# EPS, RTOL, ATOL = 1e-4, 1e-4, 1e-6\nEPS, RTOL, ATOL = 1e-4, 1e-2, 1e-4\n\n# TODO: do sampling or random direction for large args\n\"\"\"\ncheck_grads(fun, args...) checks the computed gradients for fun(args)\ncomparing them with numeric approximations.\n\"\"\"\nfunction check_grads(fun, args...; eps=EPS, rtol=RTOL, atol=ATOL, fname=fun)\n @dbgutil((:check_grads,fname,:args,args...))\n isempty(args) && error(\"No args given\")\n exact = ntuple(i->grad(fun,i)(args...), length(args))\n numeric = nd(fun, args...; eps=eps)\n @dbgutil((:check_grads,fname,:exact,exact,:numeric,numeric))\n same = isequivalent(exact, numeric; rtol=rtol, atol=atol)\n same || warn((:check_grads,fname,:args,args,:exact,exact,:numeric,numeric))\n return same\nend\n\nfunction nd(f, args...; eps=EPS)\n @dbgutil((:nd,f,args..., :eps, eps))\n unary_f = x->f(x...)\n unary_nd(unary_f, args, eps)\nend\n\nunary_nd(f, x::Tuple, eps) = ntuple(i->unary_nd(indexed_function(f, x, i), x[i], eps), length(x))\nunary_nd(f, x::Associative, eps) = (a=similar(x); for(k,v) in x; a[k] = unary_nd(indexed_function(f, x, k), v, eps); end; a)\nunary_nd(f, x::AbstractArray, eps) = reshape(eltype(x)[unary_nd(indexed_function(f, x, i), v, eps) for (i,v) in enumerate(x)], size(x))\nunary_nd(f, x::Complex, eps) = ((f(x + eps/2) - f(x - eps/2)) / eps - im*(f(x + im*eps/2) - f(x - im*eps/2)) / eps)\nunary_nd(f, x::Real, eps) = ((f(x + eps/2) - f(x - eps/2)) / eps)\n\nfunction indexed_function(fun, arg, index)\n function partial_function(x)\n if isa(arg, Tuple)\n local_arg = (arg[1:index-1]..., x, arg[index+1:end]...)\n else\n local_arg = copy(arg); local_arg[index] = x\n end\n return fun(local_arg)\n end\n return partial_function\nend\n\n# isequivalent uses isapprox for Number and AbstractArray{T<:Number}\nisequivalent(x::Number,y::Number; o...)=isapprox(x,y;o...)\nisequivalent{T<:Number,S<:Number}(x::AbstractArray{T},y::AbstractArray{S}; o...)=isapprox(x,y;o...)\n\n# isequivalent extends to Tuple, Associative, and other Arrays, comparing elementwise\nisequivalent(x::Tuple, y::Tuple; o...)=(length(x)==length(y) && all(i->isequivalent(x[i],y[i];o...), 1:length(x)))\nisequivalent(x::AbstractArray, y::AbstractArray; o...)=(length(x)==length(y) && all(i->isequivalent(x[i],y[i];o...), 1:length(x)))\nisequivalent(x::Associative, y::Associative; o...)=all(k->isequivalent(get(x,k,nothing),get(y,k,nothing);o...), unique([keys(x)...,keys(y)...]))\n\n# isequivalent treats `nothing` as equivalent to zero or zero array.\nisequivalent(x::Number,z::Void; o...)=isequivalent(z,x;o...)\nisequivalent{T<:Number}(x::AbstractArray{T},z::Void; o...)=isequivalent(z,x;o...)\nisequivalent(z::Void,x::Number; o...)=isapprox(zero(x),x;o...)\nisequivalent{T<:Number}(z::Void,x::AbstractArray{T}; rtol::Real=Base.rtoldefault(T), atol::Real=0, norm::Function=vecnorm) = (norm(x) <= atol/(1-rtol)) # Modified from: linalg/generic.jl:522\n\n# The way broadcasting works in Julia:\n# y = f(x...) where f is a broadcasting operation.\n# size(y) = broadcast_shape(x...)\n# ndims(y) = max ndims(x)\n# size(y,i) = max size(x,i)\n# size(x,i) = 1 or size(y,i) for all x and i<=ndims(x)\n# if ndims(x) < ndims(y) the extra dimensions of x are treated as 1\n\nfunction unbroadcast(x, dx)\n if size(x)==size(dx)\n return dx\n elseif isa(getval(x),Number)\n return sum(dx)\n else\n d = []\n for i=1:ndims(dx)\n size(x,i) == size(dx,i) && continue\n size(x,i) != 1 && throw(DimensionMismatch())\n push!(d,i)\n end\n length(d)==1 && (d=d[1])\n return sum(dx, d)\n end\nend\n\nfunction toscalar(xv; rng=MersenneTwister())\n x = getval(xv)\n isa(x,Number) && return xv\n isa(x,OneHot) && (x = full(x))\n idx = isa(x,Tuple) ? (1:length(x)) : eachindex(x)\n s = 0\n for i in idx\n s += xv[i] * rand(rng)\n end\n return s\nend\n\n# sumvalues sums values of dictionaries, otherwise acts like sum:\n\nsumvalues(x)=sum(x)\nsumvalues(x::Associative)=sum(values(x))\n@primitive sumvalues(x::Associative),ds fillvalues(ds,x)\nfillvalues(v,x)=(y=similar(x);for k in keys(x); y[k]=v; end; y)\n@primitive fillvalues(v,x),dxv sumvalues(dxv) nothing\naddtest(sumvalues, Dict(1=>1.,2=>2.))\naddtest(fillvalues, 0., Dict(1=>1.,2=>2.,3=>3.))\n\n# This needs more work:\n# @primitive values(x),dy Dict(map((a,b)->(a=>b), keys(x), dy))\n\n# It gets tiresome to write `Type{Grad{1}}` after a while, here are\n# some convenient aliases:\n\ntypealias D1 Type{Grad{1}}\ntypealias D2 Type{Grad{2}}\nif !isdefined(:Dn)\ntypealias Dn{N} Type{Grad{N}}\nend\n\n# Pretty print for debugging:\n_dbg(x)=x # extend to define short printable representations\n_dbg(x::Tuple)=map(_dbg,x)\n_dbg(x::Node)=\"N$(id2(x))_$(id2(x.value))\"\n_dbg(x::Value)=\"V$(id2(x))_$(_dbg(x.value))\"\n_dbg(x::Tape)=\"T$(join([id2(x),map(id2,x)...],'_'))\"\n_dbg(x::AbstractArray)=\"A$(join([id2(x),size(x)...],'_'))\"\nid2(x)=Int(object_id(x)%1000)\n\nBase.show(io::IO, n::Value) = print(io, _dbg(n))\nBase.show(io::IO, n::Node) = print(io, _dbg(n))\nBase.show(io::IO, n::Tape) = print(io, _dbg(n))\n\n", "meta": {"hexsha": "0435f0b644a12776798e13cc91373690f8e95114", "size": 15794, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/util.jl", "max_stars_repo_name": "JuliaPackageMirrors/AutoGrad.jl", "max_stars_repo_head_hexsha": "3775730f69e07a37a998cd4489ee7c7bcbc3c4a5", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/util.jl", "max_issues_repo_name": "JuliaPackageMirrors/AutoGrad.jl", "max_issues_repo_head_hexsha": "3775730f69e07a37a998cd4489ee7c7bcbc3c4a5", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/util.jl", "max_forks_repo_name": "JuliaPackageMirrors/AutoGrad.jl", "max_forks_repo_head_hexsha": "3775730f69e07a37a998cd4489ee7c7bcbc3c4a5", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 32.9041666667, "max_line_length": 190, "alphanum_fraction": 0.5919336457, "num_tokens": 4864, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.49609382947091946, "lm_q2_score": 0.14608725262486594, "lm_q1q2_score": 0.07247298459155538}}
{"text": "# read_samples\n\n\"\"\"\n\nRead sample output files created by StanSample.jl.\n\nThis method is added to StanRun's read_sample function.\n\n### Method\n```julia\nread_samples(model::SampleModel; start=1)\n```\n\n### Required arguments\n```julia\n* `model` : SampleModel\n```\n\n### Optional arguments\n```julia\n* `start=1` : First draw saved in MCMCHains.Chains object\n```\n\n\"\"\"\nfunction read_samples(model::SampleModel; start=1)\n\n local a3d, monitors, index, idx, indvec, ftype, noofsamples\n \n output_base = model.output_base\n name_base =\"_chain\"\n n_samples = model.method. num_samples \n n_chains = StanBase.get_n_chains(model)\n \n # Handle save_warmup\n start = model.method.save_warmup ? model.method.num_warmup+1 : start\n \n # a3d will contain the samples such that a3d[s, i, c] where\n\n # s: num_samples\n # i: variables (from cmdstan .csv file)\n # c: n_chains\n\n # Read .csv files created by each chain\n \n for i in 1:n_chains\n if isfile(output_base*name_base*\"_$(i).csv\")\n #noofsamples = 0\n instream = open(output_base*name_base*\"_$(i).csv\")\n #\n # Skip initial set of commented lines, e.g. containing cmdstan version info, etc.\n #\n skipchars(isspace, instream, linecomment='#')\n #\n # First non-comment line contains names of variables\n #\n line = Unicode.normalize(readline(instream), newline2lf=true)\n idx = split(strip(line), \",\")\n index = [idx[k] for k in 1:length(idx)] \n indvec = 1:length(index)\n \n if i == 1\n a3d = fill(0.0, n_samples, length(indvec), n_chains)\n end\n \n #println(size(a3d))\n skipchars(isspace, instream, linecomment='#')\n for j in 1:n_samples\n skipchars(isspace, instream, linecomment='#')\n line = Unicode.normalize(readline(instream), newline2lf=true)\n if eof(instream) && length(line) < 2\n close(instream)\n break\n else\n flds = parse.(Float64, split(strip(line), \",\"))\n flds = reshape(flds[indvec], 1, length(indvec))\n a3d[j,:,i] = flds\n end\n end # read in samples\n end # read in next file\n end # read in file for each chain\n \n cnames = convert.(String, idx[indvec])\n chns = convert_a3d(a3d, cnames, Val(:mcmcchains); start=start)\n\nend # end of read_samples\n\nfunction read_samples(model::StanModel; chain=1)\n read_samples(default_output_base(model)*\"_chain_$(chain).csv\")\nend\n\n\"\"\"\n\n# convert_a3d\n\nConvert the output file created by cmdstan to the shape of choice.\n\n### Method\n```julia\nconvert_a3d(a3d_array, cnames, ::Val{Symbol}; start=1)\n```\n### Required arguments\n```julia\n* `a3d_array::Array{Float64, 3},` : Read in from output files created by cmdstan \n* `cnames::Vector{AbstractString}` : Monitored variable names\n```\n\n### Optional arguments\n```julia\n* `::Val{Symbol}` : Output format\n* `::start=1` : First draw for MCMCChains.Chains\n```\nMethod called is based on the output_format defined in the stanmodel, e.g.:\n\n stanmodel = Stanmodel(`num_samples`=1200, thin=2, name=\"bernoulli\", \n model=bernoullimodel, `output_format`=:mcmcchains);\n\nCurrent formats supported are:\n\n1. :array (a3d_array format, the default for CmdStan)\n2. :namedarray (NamedArrays object)\n3. :dataframe (DataFrames object)\n4. :mambachains (Mamba.Chains object)\n5. :mcmcchains (TuringLang/MCMCChains.Chains object)\n\nOptions 3 through 5 are respectively provided by the packages StanDataFrames, \nStanMamba, StanMCMCChains and StanMCMCChains.\n```\n\n### Return values\n```julia\n* `res` : Draws converted to the specified format.\n```\n\"\"\"\nconvert_a3d(a3d_array, cnames, ::Val{:array}; start=1) = a3d_array\n\nconvert_a3d(a3d_array, cnames, ::Val{:namedarray}; start=1) = \n [NamedArray(a3d_array[:,:,i], (collect(1:size(a3d_array, 1)), Symbol.(cnames))) \n for i in 1:size(a3d_array, 3)]\n\nfunction convert_a3d(a3d_array, cnames, ::Val{:mcmcchains}; start=1)\n pi = filter(p -> length(p) > 2 && p[end-1:end] == \"__\", cnames)\n p = filter(p -> !(p in pi), cnames)\n\n MCMCChains.Chains(a3d_array,\n cnames,\n Dict(\n :parameters => p,\n :internals => pi\n );\n start=start\n )\nend\n", "meta": {"hexsha": "a980c7a09e180a4579d9fd47f0a7f5fd3a7f5e26", "size": 4259, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/stansamples/read_samples.jl", "max_stars_repo_name": "UnofficialJuliaMirror/StanSample.jl-c1514b29-d3a0-5178-b312-660c88baa699", "max_stars_repo_head_hexsha": "768894f98284a1840f01fd9c6c51c5247bae2ad5", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/stansamples/read_samples.jl", "max_issues_repo_name": "UnofficialJuliaMirror/StanSample.jl-c1514b29-d3a0-5178-b312-660c88baa699", "max_issues_repo_head_hexsha": "768894f98284a1840f01fd9c6c51c5247bae2ad5", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/stansamples/read_samples.jl", "max_forks_repo_name": "UnofficialJuliaMirror/StanSample.jl-c1514b29-d3a0-5178-b312-660c88baa699", "max_forks_repo_head_hexsha": "768894f98284a1840f01fd9c6c51c5247bae2ad5", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 27.8366013072, "max_line_length": 120, "alphanum_fraction": 0.6386475699, "num_tokens": 1231, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4765796510636759, "lm_q2_score": 0.15203223778010538, "lm_q1q2_score": 0.07245547083167243}}
{"text": "function truncate(y, negtrunc, postrunc)\r\n\r\n# Truncate input if its too big or too small\r\n if (y < negtrunc) # If y is less than the value negtrunc\r\n truncy = negtrunc;\r\n elseif (y > postrunc)\r\n truncy= postrunc; # If y is greater than the value postrunc\r\n else\r\n truncy = y;\r\n end \r\n\r\n return truncy \r\n\r\nend\r\n\r\n\r\n ", "meta": {"hexsha": "88bee2b3323d52096f5cb6f597b689c0d35d52ec", "size": 394, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "LifecycleCostaDias/v5_julia/code_numericTools/truncate.jl", "max_stars_repo_name": "floswald/ucl-econ-julia", "max_stars_repo_head_hexsha": "c0b9077382d4245fb1276ae2f517cc9372259c25", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2017-08-18T00:50:53.000Z", "max_stars_repo_stars_event_max_datetime": "2017-08-18T00:50:53.000Z", "max_issues_repo_path": "LifecycleCostaDias/v5_julia/code_numericTools/truncate.jl", "max_issues_repo_name": "floswald/ucl-econ-julia", "max_issues_repo_head_hexsha": "c0b9077382d4245fb1276ae2f517cc9372259c25", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 3, "max_issues_repo_issues_event_min_datetime": "2015-05-01T13:10:23.000Z", "max_issues_repo_issues_event_max_datetime": "2015-05-14T08:44:31.000Z", "max_forks_repo_path": "LifecycleCostaDias/v5_julia/code_numericTools/truncate.jl", "max_forks_repo_name": "floswald/ucl-econ-julia", "max_forks_repo_head_hexsha": "c0b9077382d4245fb1276ae2f517cc9372259c25", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 12, "max_forks_repo_forks_event_min_datetime": "2015-04-25T11:54:42.000Z", "max_forks_repo_forks_event_max_datetime": "2022-02-10T01:23:04.000Z", "avg_line_length": 23.1764705882, "max_line_length": 67, "alphanum_fraction": 0.5355329949, "num_tokens": 103, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.40733340004593027, "lm_q2_score": 0.17781086729958678, "lm_q1q2_score": 0.0724283051422564}}
{"text": "## Could tidy up this HTML to make it look nicer\nhtml_templates = Dict()\n\n# thumbs up/down don't show in my editor\ngrading_partial = \"\"\"\n if(correct) {\n msgBox.innerHTML = \" \ud83d\udc4d {{#:CORRECT}}{{{:CORRECT}}}{{/:CORRECT}}{{^:CORRECT}}Correct{{/:CORRECT}}
\";\n var explanation = document.getElementById(\"explanation_{{:ID}}\")\n if (explanation != null) {\n explanation.style.display = \"none\";\n }\n } else {\n msgBox.innerHTML = \"\ud83d\udc4e {{#:INCORRECT}}{{{:INCORRECT}}}{{/:INCORRECT}}{{^:INCORRECT}}Incorrect{{/:INCORRECT}}
\";\n var explanation = document.getElementById(\"explanation_{{:ID}}\")\n if (explanation != null) {\n explanation.style.display = \"block\";\n }\n }\n\"\"\"\n\n## Basic question\n## has label and hint option.\n## Hint is put with label when present; otherwise, it appears at bottom of form.\n## this is overridden with input widget in how show method is called\nhtml_templates[\"question_tpl\"] = mt\"\"\"\n\n\n\n\"\"\"\n\nhtml_templates[\"input_grading_script\"] = jmt\"\"\"\ndocument.getElementById(\"{{:ID}}\").addEventListener(\"change\", function() {\n var correct = {{{:CORRECT_ANSWER}}};\n var msgBox = document.getElementById('{{:ID}}_message');\n $(grading_partial)\n});\n\"\"\"\n\n##\nhtml_templates[\"inputq_form\"] = mt\"\"\"\n\n \n \n {{#:UNITS}}{{{:UNITS}}}{{/:UNITS}}{{#:HINT}} \ud83c\udf81 {{/:HINT}}\n \n
\n\"\"\"\n\n## Multiple choice (one of many)\n## XXX add {{INLINE}}\nhtml_templates[\"Radioq\"] = mt\"\"\"\n{{#:ITEMS}}\n\n \n \n {{{:LABEL}}}\n \n \n \n
\n{{/:ITEMS}}\n\"\"\"\n\nhtml_templates[\"radio_grading_script\"] = \"\"\"\ndocument.querySelectorAll('input[name=\"radio_{{:ID}}\"]').forEach(function(rb) {\nrb.addEventListener(\"change\", function() {\n var correct = rb.value == {{:CORRECT_ANSWER}};\n var msgBox = document.getElementById('{{:ID}}_message');\n $(grading_partial)\n})});\n\"\"\"\n## ----\n\nhtml_templates[\"Buttonq\"] = mt\"\"\"\n\n {{#:BUTTONS}}\n \n {{{:TEXT}}\n \n {{/:BUTTONS}}\n
\n\n\"\"\"\n\n\n\nhtml_templates[\"Multiq\"] = mt\"\"\"\n{{#:ITEMS}}\n\n \n \n {{{:LABEL}}}\n \n \n \n
\n{{/:ITEMS}}\n\"\"\"\n\nhtml_templates[\"multi_grading_script\"] = \"\"\"\ndocument.querySelectorAll('input[name=\"check_{{:ID}}\"]').forEach(function(rb) {\nrb.addEventListener(\"change\", function() {\n var choice_buttons = document.getElementsByName(\"check_{{:ID}}\");\n var selected = [];\n for (var i=0; i < choice_buttons.length; i++) {\n if (choice_buttons[i].checked) {\n selected.push(i+1)\n }\n }\n var a = selected;\n var b = {{{:CORRECT_ANSWER}}};\n // https://stackoverflow.com/questions/7837456/how-to-compare-arrays-in-javascript\n var correct = (a.length === b.length && a.find((v,i) => v !== b[i]) === undefined)\n var msgBox = document.getElementById('{{:ID}}_message');\n $(grading_partial)\n})});\n\"\"\"\n\nhtml_templates[\"MultiButtonq\"] = mt\"\"\"\n\n {{#:BUTTONS}}\n
\n {{{:TEXT}}\n \n {{/:BUTTONS}}\n\n
\n DONE\n \n\n\n\"\"\"\n\nhtml_templates[\"multi_button_grading_script\"] = \"\"\"\n\n// toggle button select\ndocument.querySelectorAll('[id^=\"button_{{:ID}}_\"]').forEach(function(btn) {\n btn.addEventListener(\"click\", function(btn) {\n\tvar unclicked = (this.value == \"unclicked\")\n\tif (unclicked) {\n this.style.background = \"{{{:SELECTED_COLOR}}}\";\n\t this.value = \"clicked\";\n this.setAttribute(\"aria-pressed\", \"true\");\n\t} else {\n\t this.style.background = null;\n\t this.value=\"unclicked\";\n this.setAttribute(\"aria-pressed\", \"false\");\n\t}\n });\n})\n\n// grade question\ndocument.querySelector('[id^=\"button_{{:ID}}-done\"]').addEventListener(\"click\", function() {\n this.disabled = true;\n var multi_choice_buttons = document.querySelectorAll('[id^=\"button_{{:ID}}_\"]');\n var selected = [];\n var correct = {{{:CORRECT_ANSWER}}};\n for (var i=0; i < multi_choice_buttons.length; i++) {\n\tvar btn = multi_choice_buttons[i];\n btn.disabled = true;\n\tvar btn_txt = btn.innerHTML;\n\tif (multi_choice_buttons[i].value == \"clicked\") {\n\t selected.push(i+1)\n\t if (correct.indexOf(i+1) < 0) {\n\t\tbtn.innerHTML = \"{{{:INCORRECT_flag}}}\" + btn_txt\n\t } else {\n\t\tbtn.innerHTML = \"{{{:CORRECT_flag}}}\" + btn_txt\n btn.style.fontSize = \"1.1rem\";\n btn.style.borderRadius = \"12px\";\n }\n\t} else {\n\t if (correct.indexOf(i+1) < 0) {\n\t\tbtn.innerHTML = \"{{{:INCORRECT_flag}}}\" + btn_txt\n\t } else {\n\t\tbtn.innerHTML = \"{{{:CORRECT_flag}}}\" + btn_txt\n btn.style.fontSize = \"1.1rem\";\n btn.style.borderRadius = \"12px\";\n\t }\n\t}\n }\n var a = selected;\n var b = {{{:CORRECT_ANSWER}}}\n // https://stackoverflow.com/questions/7837456/how-to-compare-arrays-in-javascript\n var correct = (a.length === b.length && a.find((v,i) => v !== b[i]) === undefined)\n var msgBox = document.getElementById('{{:ID}}_message');\n $(grading_partial)\n})\n\"\"\"\n\n## ----\n\nhtml_templates[\"Matchq\"] = mt\"\"\"\n
\n{{#:ITEMS}}\n\n \n {{{:QUESTION}}} \n \n \n \n {{{:BLANK}}} \n {{#:ANSWER_CHOICES}}\n {{{:LABEL}}} \n {{/:ANSWER_CHOICES}}\n \n \n \n{{/:ITEMS}}\n
\n\"\"\"\n\nhtml_templates[\"matchq_grading_script\"] = \"\"\"\n function callback(element, iterator) {\n\t element.addEventListener(\"change\", function() {\n\t var a = [];\n\t var selectors = document.querySelectorAll('[id ^= \"select_{{:ID}}\"]');\n\t Array.prototype.forEach.call(selectors, function (element, iterator) {\n\t\t a.push(element.value);\n\t })\n var b = {{{:CORRECT_ANSWER}}};\n // https://stackoverflow.com/questions/7837456/how-to-compare-arrays-in-javascript\n var correct = (a.length === b.length && a.find((v,i) => v !== b[i]) === undefined)\n var msgBox = document.getElementById('{{:ID}}_message');\n $(grading_partial)\n\t })\n }\n Array.prototype.forEach.call(document.querySelectorAll('[id ^= \"select_{{:ID}}\"]'), callback);\n\"\"\"\n\n\n## ----\n\nhtml_templates[\"fill_in_blank_select\"] = \"\"\"\n
\n {{{:BLANK}}} \n {{#:ANSWER_CHOICES}}\n {{{:LABEL}}} \n {{/:ANSWER_CHOICES}}\n \n\"\"\"\n\nhtml_templates[\"fill_in_blank_input\"] = \"\"\"\n
\n\"\"\"\n\n## -------\nhtml_templates[\"hotspot\"] = \"\"\"\n
\n
\n
\n\"\"\"\n\nhtml_templates[\"hotspot_grading_script\"] = \"\"\"\n document.getElementById(\"hotspot_{{{:ID}}}\").addEventListener(\"click\", function(e) {\n var u = e.offsetX;\n var v = e.offsetY;\n var w = this.offsetWidth;\n var h = this.offsetHeight\n\n var x = u/w;\n var y = (h-v)/h\n\n var correct = {{{:CORRECT_ANSWER}}}\n var msgBox = document.getElementById('{{:ID}}_message');\n $(grading_partial)\n\n })\n\"\"\"\n\n## -----\n## -------\nhtml_templates[\"plotlylight_grading_script\"] = \"\"\"\ndocument.getElementById(\"{{{:ID}}}\").on(\"plotly_click\", function(e) {\n var x = e.points[0].x\n var y = e.points[0].y\n\n var correct = {{{:CORRECT_ANSWER}}}\n var msgBox = document.getElementById('{{:ID}}_message');\n $(grading_partial)\n\n })\n\"\"\"\n", "meta": {"hexsha": "da0602fac88335ed15367bcaabdd18a43a2db9eb", "size": 10981, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/html_templates.jl", "max_stars_repo_name": "jverzani/QuizQuestions.jl", "max_stars_repo_head_hexsha": "afe6e2c96f92dedabf89fcc41710843891a326f3", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 6, "max_stars_repo_stars_event_min_datetime": "2022-02-02T03:37:09.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-29T13:49:11.000Z", "max_issues_repo_path": "src/html_templates.jl", "max_issues_repo_name": "jverzani/QuizQuestions.jl", "max_issues_repo_head_hexsha": "afe6e2c96f92dedabf89fcc41710843891a326f3", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 5, "max_issues_repo_issues_event_min_datetime": "2022-02-03T06:54:28.000Z", "max_issues_repo_issues_event_max_datetime": "2022-03-30T14:59:50.000Z", "max_forks_repo_path": "src/html_templates.jl", "max_forks_repo_name": "jverzani/QuizQuestions.jl", "max_forks_repo_head_hexsha": "afe6e2c96f92dedabf89fcc41710843891a326f3", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 32.4881656805, "max_line_length": 231, "alphanum_fraction": 0.5840998088, "num_tokens": 2970, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.2598256379609837, "lm_q2_score": 0.2782567937024021, "lm_q1q2_score": 0.07229824894070444}}
{"text": "## Exercise 9-3\n# Write a function named avoids that takes a word and a string of forbidden letters, and that returns true if the word doesn\u2019t use any of the forbidden letters.\nprintln(\"Ans:\\n[Part 1]\")\n\nfunction avoids(string, forbidden_letters)\n for letter in forbidden_letters\n if letter \u2208 string\n return false\n end\n end\n return true\nend\n\nprintln(avoids(\"hello world\", \"hwe\"))\nprintln(avoids(\"printf\", \"xyz\"))\n\n# Modify your program to prompt the user to enter a string of forbidden letters and then print the number of words that don\u2019t contain any of them. Can you find a combination of 5 forbidden letters that excludes the smallest number of words?\nprintln(\"[Part 2]\")\n\nprint(\"Please Enter forbidden letters: \")\nforbidden_letters = strip(readline())\n\ncount = 0\nfor line in eachline(\"words.txt\")\n if avoids(line, forbidden_letters)\n global count += 1\n end\nend\n\nprintln(\"There are total of $count letters which avoids the forbidden letters.\")\n\nprintln(\"End.\")\n", "meta": {"hexsha": "36c64c82f1c795d791809b4f3add4df7fa7e6644", "size": 1011, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Chapter9/ex3.jl", "max_stars_repo_name": "yashppawar/ThinkJuliaExercises.jl", "max_stars_repo_head_hexsha": "72145b969dc51ebcac413a10004175ebc63cd5c2", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2022-02-13T14:11:30.000Z", "max_stars_repo_stars_event_max_datetime": "2022-02-13T14:11:30.000Z", "max_issues_repo_path": "Chapter9/ex3.jl", "max_issues_repo_name": "yashppawar/ThinkJuliaExercises.jl", "max_issues_repo_head_hexsha": "72145b969dc51ebcac413a10004175ebc63cd5c2", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Chapter9/ex3.jl", "max_forks_repo_name": "yashppawar/ThinkJuliaExercises.jl", "max_forks_repo_head_hexsha": "72145b969dc51ebcac413a10004175ebc63cd5c2", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 30.6363636364, "max_line_length": 240, "alphanum_fraction": 0.7210682493, "num_tokens": 231, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.34510525748676846, "lm_q2_score": 0.2094696714602651, "lm_q1q2_score": 0.07228908490496358}}
{"text": "\n## Page(\"SOme blurb\", (q1,q2,q3, ...);\n#DBSubject=\"Calculus\",\n#KEYWORDS=\"limits\",\n#AuthorText=\"John Verzani\"\n#AuthorText2=\"Joseph Maher\"\n#)\nraw\"\"\"\n Page(intro, questions; context=\"\", meta...)\n\n\nCreate a page which prints as a `pg` file.\n\n* `intro` may be marked up using modified markdown \n* `questions` is a tuple of questions or `QUESTIONS` object\n* `context` optional value to create page context. Typical usage: `context=\"Interval\"`\n* `answer_context`: Dictionary of context for all answers on the page, e.g \n```\nanswer_context=Dict(:operators=>Dict(:undefine=>\"'+','-','*','/','^'\"),\n :functions=>Dict(:disable=>\"'All'\"),\n :constants=>Dict(:remove=>\"\\\"e\\\"\")\n )\n```\nThe context dictionary above, is aliased as `numbers_only`.\n\n* `meta` for page meta data.\n\nIf `ENV[\"BRANDING\"]` is set, it will be printed on each page generated.\n\nExample:\n\n```\nusing JuliaWeBWorK\nmeta=(AuthorText=\"Julia\", Institution=\"JuliaAcademy\", Question=\"1\")\nintro = raw\"# Problem 1\"\nq1 = numericq(\"What is ``{{:a1}} + {{:a2}}``?\", (x,y)->x+y, (1:5, 1:5))\np = Page(intro, (q1,); meta...) \n# open(\"mynew.pg\",\"w\") do io\n# print(io, p)\n# end\n```\n\"\"\"\nstruct Page\n intro\n questions\n meta_information\n context\n answer_context\n function Page(intro, questions; context=\"\",answer_context=\"\", kwargs...)\n new(intro, questions, Dict(kwargs...), context, answer_context)\n end\nend\n\n# utility function\nfunction kv(io::IO, v,d, ops=[])\n if isa(d, Dict)\n if(v != nothing)\n push!(ops, v)\n for (vv, dd) in d\n kv(io, vv, dd, ops)\n end\n else\n for (kk,vv) in d\n kv(io, kk, vv)\n end\n end\n else\n println(io, \"\"\"Context()->$(join(string.(ops), \"->\"))->$(v)($(d));\\n\"\"\")\n end\nend\n\n\n\"\"\"\n numbers_only\n\nDictionary to pass to `answer_context` to turn off WeBWorK's simplification pass.\nThere is no means to turn this off per problem, only per page.\n\"\"\"\nnumbers_only = Dict(:operators=>Dict(:undefine=>\"'+','-','*','/','^'\"),\n :functions=>Dict(:disable=>\"'All'\"),\n :constants=>Dict(:remove=>\"\\\"e\\\"\")\n )\n\nfunction Base.show(io::IO, p::Page)\n\n for (k,v) in p.meta_information\n println(io, \"## $k('$v')\")\n end\n \n \n println(io, raw\"\"\"\nDOCUMENT();\n\nloadMacros(\"PG.pl\",\"PGbasicmacros.pl\",\"PGanswermacros.pl\");\nloadMacros(\"PGstandard.pl\");\nloadMacros(\"MathObjects.pl\");\nloadMacros(\"Parser.pl\");\nloadMacros(\"AnswerFormatHelp.pl\");\nloadMacros(\"parserRadioButtons.pl\");\nloadMacros(\"PGchoicemacros.pl\");\nloadMacros(\"PGessaymacros.pl\");\n##loadMacros(\"PGML.pl\");\n##loadMacros(\"PGcourse.pl\");\n\nContext()->{format}{number} = \"%.16g\";\nContext()->variables->add(y=>'Real', z=>'Real', t=>'Real', u=>'Real', m=>'Real', n=>'Real');\nContext()->flags->set(ignoreEndpointTypes=> 1);\n\nmy %seen; # hat tip to https://perlmaven.com/unique-values-in-an-array-in-perl; filter Context->strings->add\n$seen{\"yes\"} = 1; $seen{\"no\"}=1;$seen{\"true\"} = 1; $seen{\"false\"}=1;\nContext()->strings->add(qq(yes)=>{},qq(no)=>{},qq(true)=>{},qq(false)=>{});\n$ATSYMS = qw\"@syms\"; \n\n\"\"\")\n\n println(io, \"TEXT(beginproblem());\")\n\n ## add in space for popup (called by imagelink)\n println(io, raw\"\"\"\nHEADER_TEXT(<
\n \n \n\"\"\")\n\n ## print javascript headers, as needed\n for T in unique(typeof.(p.questions))\n print(io, javascript_headers(T))\n end\n \n println(io, raw\"\"\" \nEOF\n\"\"\")\n ## add in somee missing formatting styles\n println(io,\"\"\"\n\\$BBLOCKQUOTE = MODES(\nHTML=>\"\",\nTeX =>\"\"\n);\n\n\\$EBLOCKQUOTE = MODES(\nHTML=>\" \",\nTeX=>\"\"\n);\n\n\\$ADMONITION = MODES(\nHTML=>\"☆ \",\nTeX=>\"\\\\(\\\\bigwhitestar)\");\n\"\"\")\n\n\n if length(p.context) > 0\n println(io, \"Context(\\\"$(p.context)\\\");\")\n end\n\n println(io, \"\\n## ---------- create answer values ----------\\n\")\n for q in p.questions\n print(io, create_answer(q))\n println(io, \"\")\n end\n\n println(io, \"\\n## ---------- show questions ----------\\n\")\n \n\n println(io, raw\"\"\"$branding_ = <<\"END_BRANDING\";\"\"\")\n println(io, get(ENV, \"BRANDING\", \"\"))\n println(io, \"\"\"END_BRANDING\"\"\")\n println(io, raw\"\"\"$branding = MODES(HTML=>$branding_, TeX=>\"[nothing to see]\");\"\"\")\n \n println(io, \"BEGIN_TEXT\\n\")\n println(io, raw\"\"\"$branding\"\"\")\n\n #intro = replace(p.intro, \"\\\\\" => \"\\\\\\\\\") # had this, replaced with\n intro = Mustache.render(p.intro)\n print(io, escape_string(intro))\n\n println(io, \"\\n\\n\\$HR\\$PAR\\n\")\n \n for q in p.questions\n print(io, show_question(q))\n println(io,\"\\$PAR\\n\")\n end\n\n println(io, \"END_TEXT\")\n\n println(io, \"\\n## ---------- show answers ----------\\n\")\n\n ops = []\n if length(p.answer_context) > 0\n kv(io, nothing, p.answer_context)\n end\n\n for q in p.questions\n println(io, show_answer(q))\n println(io,\"\")\n end\n\n ## Solutions go at end (when added)\n soln_io = IOBuffer()\n for q in p.questions\n print(soln_io, show_solution(q))\n end\n solns = String(take!(soln_io))\n\n if length(solns) > 0\n \n println(io, \"#***************************************** Solution: \")\n println(io, \"\"\"\nContext()->texStrings;\nSOLUTION(EV3(<<\"END_SOLUTION\"));\n\"\"\")\n println(io, escape_string(solns)) \n\n println(io, \"\"\"\nEND_SOLUTION\nContext()->normalStrings;\n\"\"\")\n end\n\n\n println(io, \"ENDDOCUMENT();\")\n\nend\n\n\"\"\"\n PAGE(SCRIPTNAME)\n\nWrite a page to a file name based on the value of `SCRIPTNAME`. Returns an anonymous function \nwhich can be called repeatedly to write a page with a filename based on `SCRIPTNAME`.\n\nThis is designed to be used as `PAGE = JuliaWeBWorK.PAGE(@__FILE__)`. Then from one script file\nseveral related `pg` files can be generated. This might be useful for authoring exams\nwhere it is a good practice to have many separate problems and not one big one with many\nparts.\n\n```\nusing JuliaWeBWorK\nPAGE = write_page(@__FILE__)\n\nq = numericq(raw\"What is \\\\({{:a1}} + {{:a2}}\\\\)?\", (a,b) -> a+b, (1:4, 2:5))\nPAGE(\"Addition\", (q,)) # writes to SCRIPT_BASE_NAME-1.pg\n\nq = numericq(raw\"What is \\\\({{:a1}} - {{:a2}}\\\\)?\", (a,b) -> a-b, (1:4, 2:5))\nPAGE(\"subtraction\", (q,)) # writes to SCRIPT_BASE_NAME-2.pg\n\nq = numericq(raw\"What is \\\\({{:a1}} * {{:a2}}\\\\)?\", (a,b) -> a*b, (1:4, 2:5))\nPAGE(\"multiplication\", (q,)) # writes to SCRIPT_BASE_NAME-3.pg\n```\n\n\n\"\"\"\nfunction PAGE(SCRIPTNAME)\n base_nm = replace(SCRIPTNAME, r\".jl$\" => \"\")\n ctr = Ref(1)\n # return an anonymous function for printing a page\n # to the numbered .pg file.\n (args...;kwargs...) -> begin\n fname = base_nm * \"-$(ctr[]).pg\"\n ctr[] += 1\n open(fname, \"w\") do io\n show(io, Page(args..., kwargs...))\n end\n end\nend\n\n\n### ------ CONVENIENCES ------------\n\n\n# simple struct to hold questions\n# * can be passed to `page`\n# * the call method calls `push!(q, x)` so that questions can be piped into t\n# these objects, as in `numericq(...) |> qs`\nstruct Questions\n qs\nend\nQUESTIONS() = Questions(Any[])\nBase.iterate(q::Questions) = iterate(q.qs)\nBase.iterate(q::Questions, st) = iterate(q.qs, st)\nBase.length(q::Questions) = length(q.qs)\nBase.push!(q::Questions, x) = push!(q.qs, x)\n## returning `x` allows use like `u = randomizer(...) |> qs`\n(q::Questions)(x) = (push!(q, x); x)\n\n\n## ----\n\"return iterator over the letters `(a)`, `(b)`, ... Calling function increments letters\"\nfunction LETTERS()\n letters = [\"(a)\", \"(b)\", \"(c)\", \"(d)\", \"(e)\", \"(f)\", \"(g)\", \"(h)\", \"(i)\", \"(j)\", \"(k)\", \"(l)\", \"(m)\", \"(n)\", \"(o)\", \"(p)\", \"(q)\", \"(r)\", \"(s)\", \"(t)\", \"(u)\", \"(v)\", \"(w)\", \"(x)\", \"(y)\", \"(z)\"]\n idx = Base.Ref(1)\n () -> begin\n idx[] = idx[] + 1\n letters[idx[]-1]\n end\nend\n\n### ------ HACKS ------------\n\n## A total hack to print `@syms` in a block\n## First \\{\\} are expanded, then $... and @... are substituted\n## so we can't generalize through the \\{...\\} phase\nATSYMS = \"\\$ATSYMS\"; export ATSYMS\n\n# https://github.com/JuliaLang/julia/blob/master/stdlib/InteractiveUtils/src/clipboard.jl\nfunction mac_clipboard(p)\n open(pipeline(`pbcopy`, stderr=stderr), \"w\") do io\n show(io, p)\n end\nend\nexport mac_clipboard\n\n \n", "meta": {"hexsha": "ecea2b03d19993496e4e559eaddb918f3d1d87c3", "size": 8652, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/page.jl", "max_stars_repo_name": "mth229/JuliaWeBWorK.jl", "max_stars_repo_head_hexsha": "10a04eeaa45c29158e8a6e6613a1b4d6472c8913", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2021-05-25T01:43:39.000Z", "max_stars_repo_stars_event_max_datetime": "2021-05-25T01:43:39.000Z", "max_issues_repo_path": "src/page.jl", "max_issues_repo_name": "mth229/JuliaWeBWorK.jl", "max_issues_repo_head_hexsha": "10a04eeaa45c29158e8a6e6613a1b4d6472c8913", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/page.jl", "max_forks_repo_name": "mth229/JuliaWeBWorK.jl", "max_forks_repo_head_hexsha": "10a04eeaa45c29158e8a6e6613a1b4d6472c8913", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 26.953271028, "max_line_length": 196, "alphanum_fraction": 0.5670365233, "num_tokens": 2506, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.3486451488696663, "lm_q2_score": 0.20689403903542758, "lm_q1q2_score": 0.0721326030397532}}
{"text": "# ---\n# jupyter:\n# jupytext:\n# formats: ipynb,jl:hydrogen\n# text_representation:\n# extension: .jl\n# format_name: hydrogen\n# format_version: '1.3'\n# jupytext_version: 1.11.2\n# kernelspec:\n# display_name: Julia 1.6.2\n# language: julia\n# name: julia-1.6\n# ---\n\n# %% [markdown]\n# # Attempt to use ConcreteStructs.jl with Parameters.jl\n#\n# * Gen Kuroki\n# * 2021-09-06\n# * https://github.com/jonniedie/ConcreteStructs.jl/issues/4#issuecomment-913998030\n#\n# I have tried to make ConcreteStructs.jl and Parameters.jl work together well.\n#\n# __Conclusion:__ It is possible to do so by making the following two changes.\n#\n# * Change `@concrete` not creating the inner constructor, so that the `Foo{__T_a, __T_b, __T_c}(a, b, c)`-type default constructor will be defined.\n# * Change `@with_kw` expanding macros in the argument, [like `Base.@kwdef`](https://github.com/JuliaLang/julia/blob/4931faa34a8a1c98b39fb52ed4eb277729120128/base/util.jl#L455).\n#\n# Then `@concrete` works well with `@with_kw` and more completely with `Base.@kwdef`.\n#\n# See below for details.\n\n# %%\nVERSION\n\n# %%\nusing ConcreteStructs\nusing Parameters\n\nmacro macroexpand_rmln(code)\n :(macroexpand($__module__, $(QuoteNode(code)), recursive=true) |>\n Base.remove_linenums!)\nend\n\n# %% [markdown]\n# ## Plain struct\n\n# %%\nstruct Foo{A, B, C}\n a::A\n b::B\n c::C\nend\n\n# %%\nmethods(Foo)\n\n# %%\nmethods(Foo{1,2,3})\n\n# %% [markdown]\n# The default constructor `Foo{A, B, C}(a, b, c)` is defined.\n\n# %% [markdown]\n# ## @concrete struct\n#\n# `@concrete` removes the `Foo_concrete{__T_a, __T_b, __T_c}(a, b, c)`-type default constructor.\n\n# %%\n@concrete struct Foo_concrete\n a\n b\n c\nend\n\n# %%\nmethods(Foo_concrete)\n\n# %%\nmethods(Foo_concrete{1,2,3})\n\n# %% [markdown]\n# `Foo_concrete{__T_a, __T_b, __T_c}(a, b, c)` is not defined because only the inner constructor `Foo_concrete(a::__T_a, b::__T_b, c::__T_c)` is defined.\n\n# %%\n@macroexpand_rmln @concrete struct Foo_concrete\n a\n b\n c\nend\n\n# %% [markdown]\n# ## Base.@kwdef concrete struct\n\n# %%\nBase.@kwdef @concrete struct Foo_kwdef_concrete\n a = 1\n b = 2.0\n c = \"three\"\nend\n\n# %%\nFoo_kwdef_concrete()\n\n# %%\nFoo_kwdef_concrete{Int, Float64, String}(a = 4, b = 5.0, c = \"six\")\n\n# %% [markdown]\n# The reason for this error is that `Foo_kwdef_concrete{__T_a, __T_b, __T_c}(a, b, c)` is not defined.\n\n# %%\nmethods(Foo_kwdef_concrete)\n\n# %%\nmethods(Foo_kwdef_concrete{1,2,3})\n\n# %%\n@macroexpand_rmln Base.@kwdef @concrete struct Foo_kwdef_concrete\n a\n b\n c\nend\n\n# %% [markdown]\n# ## @with_kw @concrete struct causes error\n\n# %%\n@with_kw @concrete struct Foo_with_kw_concrete\n a = 1\n b = 2.0\n c = \"three\"\nend\n\n# %% [markdown]\n# [The first line of `Base.@kwdef`](https://github.com/JuliaLang/julia/blob/4931faa34a8a1c98b39fb52ed4eb277729120128/base/util.jl#L455) expands macros in the argument expression:\n#\n# ```julia\n# expr = macroexpand(__module__, expr) # to expand @static\n# ```\n#\n# This is what makes `Base.@kwdef @concrete struct` possible. Make a similar change to `Parameters.@with_kw`.\n\n# %% [markdown]\n# ## Change @with_kw expanding macros in the argument\n\n# %%\n@eval Parameters macro with_kw(typedef)\n typedef = macroexpand(__module__, typedef) # inserted\n return esc(with_kw(typedef, __module__, true))\nend\n\n# %%\n@with_kw @concrete struct Foo_with_kw_concrete\n a = 1\n b = 2.0\n c = \"three\"\nend\n\n# %% [markdown]\n# Okay, it seems to have worked. But...\n\n# %%\nmethods(Foo_with_kw_concrete)\n\n# %%\nmethods(Foo_with_kw_concrete{1,2,3})\n\n# %%\nFoo_with_kw_concrete()\n\n# %% [markdown]\n# The reason for this error is that `Foo_with_kw_concrete{__T_a, __T_b, __T_c}(a, b, c)` is not defined again.\n\n# %%\n@macroexpand_rmln @with_kw @concrete struct Foo_with_kw_concrete\n a = 1\n b = 2.0\n c = \"three\"\nend\n\n# %% [markdown]\n# ## Change `@concrete` not creating the inner constructor\n#\n# Change `ConcreteStructs._concretize(expr)` not creating the inner constructor.\n\n# %%\n@eval ConcreteStructs function _concretize(expr)\n expr isa Expr && expr.head == :struct || error(\"Invalid usage of @concrete\")\n \n is_mutable = expr.args[1]\n struct_name, type_params, super = _parse_head(expr.args[2])\n line_tuples = _parse_line.(expr.args[3].args)\n lines = first.(line_tuples)\n type_params_full = (type_params..., filter(x -> x!==nothing, last.(line_tuples))...)\n\n struct_type = if length(type_params_full) == 0\n struct_name\n else\n Expr(:curly, struct_name, type_params_full...)\n end\n\n head = Expr(:(<:), struct_type, super)\n # constructor_expr = _make_constructor(struct_name, type_params, type_params_full, lines)\n # body = Expr(:block, lines..., constructor_expr)\n body = Expr(:block, lines...)\n struct_expr = Expr(:struct, is_mutable, head, body)\n \n return struct_expr, struct_name, type_params\nend\n\n# %%\n@macroexpand_rmln @concrete struct Bar_concrete\n a = 1\n b = 2.0\n c = \"three\"\nend\n\n# %% [markdown]\n# The inner constructor has been deleted.\n\n# %% [markdown]\n# ## @concrete works well with @with_kw\n\n# %%\n@with_kw @concrete struct Bar_with_kw_concrete\n a = 1\n b = 2.0\n c = \"three\"\nend\n\n# %%\nmethods(Bar_with_kw_concrete)\n\n# %%\nmethods(Bar_with_kw_concrete{1,2,3})\n\n# %% [markdown]\n# The default constructor `Bar_with_kw_concrete{__T_a, __T_b, __T_c}(a, b, c)` is implicitly defined.\n\n# %%\nBar_with_kw_concrete()\n\n# %%\nBar_with_kw_concrete(c = '3')\n\n# %%\nBar_with_kw_concrete{Int, Float64, String}(a = 4, b = 5.0, c = \"six\")\n\n# %%\nBar_with_kw_concrete(4, 5.0, \"six\")\n\n# %% [markdown]\n# `ConcreteStructs.@concrete` works well with `Parameters.@with_kw`!\n\n# %%\n@macroexpand_rmln @with_kw @concrete struct Bar_with_kw_concrete\n a = 1\n b = 2.0\n c = \"three\"\nend\n\n# %% [markdown]\n# ## @concrete works well more completely with Base.@kwdef\n\n# %%\nBase.@kwdef @concrete struct Bar_kwdef_concrete\n a = 1\n b = 2.0\n c = \"three\"\nend\n\n# %%\nBar_kwdef_concrete()\n\n# %%\nBar_kwdef_concrete(c = '3')\n\n# %%\nBar_kwdef_concrete{Int, Float64, String}(a = 4, b = 5.0, c = \"six\") # not error\n\n# %%\nBar_kwdef_concrete(4, 5.0, \"six\")\n\n# %% [markdown]\n# `ConcreteStructs.@concrete` works more completely well with `Base.@kwdef`!\n\n# %%\n@macroexpand_rmln Base.@kwdef @concrete struct Bar_kwdef_concrete\n a = 1\n b = 2.0\n c = \"three\"\nend\n\n# %%\n", "meta": {"hexsha": "f5761427bf96370df735019b53680e567ae8bf25", "size": 6342, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "0019/ConcreteStructs.jl with Parameters.jl.jl", "max_stars_repo_name": "genkuroki/public", "max_stars_repo_head_hexsha": "339ea5dfd424492a6b21d1df299e52d48902de18", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 10, "max_stars_repo_stars_event_min_datetime": "2021-06-06T00:33:49.000Z", "max_stars_repo_stars_event_max_datetime": "2022-01-24T06:56:08.000Z", "max_issues_repo_path": "0019/ConcreteStructs.jl with Parameters.jl.jl", "max_issues_repo_name": "genkuroki/public", "max_issues_repo_head_hexsha": "339ea5dfd424492a6b21d1df299e52d48902de18", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "0019/ConcreteStructs.jl with Parameters.jl.jl", "max_forks_repo_name": "genkuroki/public", "max_forks_repo_head_hexsha": "339ea5dfd424492a6b21d1df299e52d48902de18", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 3, "max_forks_repo_forks_event_min_datetime": "2021-08-02T11:58:34.000Z", "max_forks_repo_forks_event_max_datetime": "2021-12-11T11:46:05.000Z", "avg_line_length": 21.4256756757, "max_line_length": 178, "alphanum_fraction": 0.6641438032, "num_tokens": 2040, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.3849121585956185, "lm_q2_score": 0.18713267989575072, "lm_q1q2_score": 0.07202964376245631}}
{"text": "# This file is a part of AstroLib.jl. License is MIT \"Expat\".\n# Copyright (C) 2016 Mos\u00e8 Giordano.\n\n\"\"\"\n ymd2dn(date) -> number_of_days\n\n### Purpose ###\n\nConvert from a date to day of the year.\n\n### Explanation ###\n\nReturns the day of the year for `date` with January 1st being day 1.\n\n### Arguments ###\n\n* `date`: the date with `Date` type. Can be a single date or an array of dates.\n\n### Output ###\n\nThe day of the year for the given `date`. If `date` is an array, returns an\narray of days.\n\n### Example ###\n\nFind the days of the year for March 5 in the years 2015 and 2016 (this is a leap\nyear).\n\n``` julia\nymd2dn([Date(2015, 3, 5), Date(2016, 3, 5)])\n# => 2-element Array{Int64,1}:\n# 64\n# 65\n```\n\n### Note ###\n\n`ydn2md` converts from year and day number of year to a date.\n\"\"\"\nconst ymd2dn = Dates.dayofyear\n", "meta": {"hexsha": "f45ee60992da8e48da35a2a919d3fce0aa5696d9", "size": 824, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/ymd2dn.jl", "max_stars_repo_name": "JuliaPackageMirrors/AstroLib.jl", "max_stars_repo_head_hexsha": "d56c7307efa7e784554c1ee806664af51b82a052", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/ymd2dn.jl", "max_issues_repo_name": "JuliaPackageMirrors/AstroLib.jl", "max_issues_repo_head_hexsha": "d56c7307efa7e784554c1ee806664af51b82a052", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/ymd2dn.jl", "max_forks_repo_name": "JuliaPackageMirrors/AstroLib.jl", "max_forks_repo_head_hexsha": "d56c7307efa7e784554c1ee806664af51b82a052", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 20.0975609756, "max_line_length": 80, "alphanum_fraction": 0.6444174757, "num_tokens": 265, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4378234844434674, "lm_q2_score": 0.1645164608483867, "lm_q1q2_score": 0.07202917013694794}}
{"text": "\"\"\"\nFunction replaces the negative values in a datacube with missing. \n\"\"\"\nfunction replace_negative(datacube)\n for i in 1:prod(size(datacube))\n if ismissing(datacube[i]) \n continue\n elseif datacube[i] < 0\n datacube[i] = missing\n else\n continue\n end\n end\n return datacube\nend\n", "meta": {"hexsha": "43845804e2d12d5570257c894a36b18f34e0f5a1", "size": 350, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/data_cleaning/replace_negative.jl", "max_stars_repo_name": "JuliaPlanet/NighttimeLights.jl", "max_stars_repo_head_hexsha": "100641b875e7af3c4eaa9e9daacd8e06e8940458", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 8, "max_stars_repo_stars_event_min_datetime": "2021-08-01T02:22:59.000Z", "max_stars_repo_stars_event_max_datetime": "2022-01-16T03:19:42.000Z", "max_issues_repo_path": "src/data_cleaning/replace_negative.jl", "max_issues_repo_name": "xKDR/NighttimeLights.jl", "max_issues_repo_head_hexsha": "3b3015c8c8d45372cde9d5a9bd76feae6a1588c1", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/data_cleaning/replace_negative.jl", "max_forks_repo_name": "xKDR/NighttimeLights.jl", "max_forks_repo_head_hexsha": "3b3015c8c8d45372cde9d5a9bd76feae6a1588c1", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2021-08-09T10:57:53.000Z", "max_forks_repo_forks_event_max_datetime": "2021-08-09T10:57:53.000Z", "avg_line_length": 21.875, "max_line_length": 66, "alphanum_fraction": 0.5857142857, "num_tokens": 82, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.44939263446475963, "lm_q2_score": 0.16026603633858202, "lm_q1q2_score": 0.07202237628542027}}
{"text": "#=\nP04 (*) Find the number of elements of a list.\n=#\n\nfunction my_length(coll)\n # With built-in `length` function\n length(coll)\nend\n\nfunction len_with_count(coll, count)\n # With tail recursion\n if isempty(coll)\n count\n else\n len_with_count(coll[2:end], count + 1)\n end\nend\n\nfunction my_length_2(coll)\n # Counting from 0\n len_with_count(coll, 0)\nend\n\n@assert my_length(()) == 0\n@assert my_length((:a, :b, :c)) == 3\n@assert my_length((1, 2, 3, 4)) == 4\n@assert my_length(((:a, :b), (:c, :d), (:e, :f))) == 3\n\n@assert my_length_2(()) == 0\n@assert my_length_2((:a, :b, :c)) == 3\n@assert my_length_2((1, 2, 3, 4)) == 4\n@assert my_length_2(((:a, :b), (:c, :d), (:e, :f))) == 3\n\nprintln(\"Tests passed: JL-04.jl\")\n", "meta": {"hexsha": "e9ab1f5491ee08672660e2b46fb81c8949d06ea2", "size": 744, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "JL-04.jl", "max_stars_repo_name": "microamp/jl-99", "max_stars_repo_head_hexsha": "5d49a7e1617394e6cbc06f1a94fe6230b3025d73", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "JL-04.jl", "max_issues_repo_name": "microamp/jl-99", "max_issues_repo_head_hexsha": "5d49a7e1617394e6cbc06f1a94fe6230b3025d73", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "JL-04.jl", "max_forks_repo_name": "microamp/jl-99", "max_forks_repo_head_hexsha": "5d49a7e1617394e6cbc06f1a94fe6230b3025d73", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 21.2571428571, "max_line_length": 56, "alphanum_fraction": 0.5913978495, "num_tokens": 276, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4493926492132671, "lm_q2_score": 0.16026602831693942, "lm_q1q2_score": 0.07202237504423789}}
{"text": "# a() is some operation\na() = sum(i for i in 1:100000)\n\n# b is the Task of \nb = Task(a)\n\n# shows the current task\ncurrent_task()\n\n# Query if the specified is done\nprintln(\"Is the task done yet?:\", istaskdone(b))\n\nschedule(b)\n\nyield()\n\nresult = istaskdone(b)\nprintln(\"Is the task done?:\", result)", "meta": {"hexsha": "bfd5072bbb44202708de6f697c9571854cae2583", "size": 295, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "intro_to_tasks.jl", "max_stars_repo_name": "ryanorsinger/JuliaPlayground", "max_stars_repo_head_hexsha": "11f67480cf82b42ba05bcd57bdb776dbf95196e0", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "intro_to_tasks.jl", "max_issues_repo_name": "ryanorsinger/JuliaPlayground", "max_issues_repo_head_hexsha": "11f67480cf82b42ba05bcd57bdb776dbf95196e0", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "intro_to_tasks.jl", "max_forks_repo_name": "ryanorsinger/JuliaPlayground", "max_forks_repo_head_hexsha": "11f67480cf82b42ba05bcd57bdb776dbf95196e0", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 16.3888888889, "max_line_length": 48, "alphanum_fraction": 0.6745762712, "num_tokens": 86, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.44939263446475963, "lm_q2_score": 0.16026603032235007, "lm_q1q2_score": 0.07202237358176995}}
{"text": "#========================================================================================#\n#\tLaboratory 7\n#\n# Encapsulation and software design.\n#\n# Author: Niall Palfreyman, 06/02/2022\n#========================================================================================#\n[\n\tActivity(\n\t\t\"\"\"\n\t\tIn this laboratory we look at the issue of encapsulation - a super-important topic in\n\t\tmodern software engineering. The point is that if everyone is able to change the\n\t\tvalue of important variables, our code will be EXTREMELY hard for people to understand\n\t\tand debug. I have seen one firm come to bankruptcy because of this problem. How does it\n\t\tcome about?\n\n\t\tIn a moment, you will discover that the variables inside a module or function have LOCAL\n\t\tSCOPE - that is, they are only visible and available inside that function, and not in\n\t\tthe GLOBAL SCOPE outside the function.\n\n\t\tSometimes, we are tempted to pass data from one function to another by storing that data\n\t\tin global variables, but this ALWAYS brings with it the danger that someone might change\n\t\tthat data by accident - particularly because it is often very difficult for users of our\n\t\tprogram to notice that we are using global variables to pass the data!\n\n\t\tA large part of the science of informatics concerns how to pass data in ways that\n\t\tprotect that data from being changed by accident, so that is what we will investigate\n\t\tin this laboratory.\n\n\t\tIn our first experiment, enter the following code, then tell me the value of paula:\n\n\t\tlinus = [5,4,3,2,1]; paula = 5\n\t\t\"\"\",\n\t\t\"\",\n\t\tx -> x==5\n\t),\n\tActivity(\n\t\t\"\"\"\n\t\tNow enter the following function:\n\n\t\tfunction change_paula()\n\t\t\tpaula = 7\n\t\t\tpaula\n\t\tend\n\n\t\tThen call the function change_paula() and tell me the value you get back:\n\t\t\"\"\",\n\t\t\"Your result may (not) surprise you, depending on how you think about scoping rules\",\n\t\tx -> x==7\n\t),\n\tActivity(\n\t\t\"\"\"\n\t\tYour result shows us that the variable paula has two different meanings: the meaning in\n\t\tthe GLOBAL scope outside the function change_paula, and the meaning inside the LOCAL\n\t\tscope of change_paula.\n\n\t\tNow tell me the current value of paula:\n\t\t\"\"\",\n\t\t\"Ask Julia for the value of paula\",\n\t\tx -> x==5\n\t),\n\tActivity(\n\t\t\"\"\"\n\t\tAha! So although we can change the value of LOCAL paula inside the function change_paula(),\n\t\tthis does not affect the value of GLOBAL paula. In fact, there exist two different variables\n\t\tnamed paula: the GLOBAL variable containing the value 5, and a LOCAL variable containing\n\t\tthe value 7. When change_paula() ends, the variables in its local scope are thrown away,\n\t\tand the LOCAL paula disappears.\n\n\t\tIf we REALLY want to change the global value of paula, we can do so by redefining the\n\t\tfunction change_paula():\n\n\t\tfunction change_paula()\n\t\t\tglobal paula = 7\n\t\t\tpaula\n\t\tend\n\n\t\tNOTE: Doing this is a Very Bad Idea! Tell me the value of paula now:\n\t\t\"\"\",\n\t\t\"\",\n\t\tx -> x==7\n\t),\n\tActivity(\n\t\t\"\"\"\n\t\tSo Julia does allow us to make use of global values inside a local scope, but it forces us\n\t\tto announce this by using the keyword \"global\".\n\n\t\tThere is a further issue here. Enter this code:\n\n\t\tfunction change_linus()\n\t\t\tlinus[3] = 7\n\t\t\tlinus\n\t\tend\n\n\t\tAgain, the return value tells us that we are able to change the value of a local variable\n\t\tnamed linus. But now tell me the value of the GLOBAL variable named linus:\n\t\t\"\"\",\n\t\t\"\",\n\t\tx -> x==[5,4,7,2,1]\n\t),\n\tActivity(\n\t\t\"\"\"\n\t\tThe point here is that linus is a Vector that refers to its contents (5,4,3,2,1). Julia\n\t\tdoes not allow us to change the value of linus, however it DOES allow us to change the\n\t\tCONTENTS that linus refers to. So global variables are still unsafe! What are we to do?\n\t\tThe solution is this:\n\n\t\t\tALWAYS encapsulate (i.e.: wrap/hide) EVERYTHING you do inside a MODULE!\n\n\t\tModules offer a very effective of preventing our variables and code from being changed by\n\t\tother programmers. Let's see how to do this. Enter the following code:\n\n\t\tmodule MyModule\n\t\t\tfunction change_paula1()\n\t\t\t\tglobal paula = 9\n\t\t\t\tpaula\n\t\t\tend\n\t\tend\n\n\t\tNow call MyModule.change_paula1(), then tell me the value of paula afterwards:\n\t\t\"\"\",\n\t\t\"\",\n\t\tx -> x==7\n\t),\n\tActivity(\n\t\t\"\"\"\n\t\tOK, so putting change_paula1() inside the module MyModule means it cannot interfere\n\t\twith the value of our global variable paula. But wait! We know that we can load\n\t\tmodules into global scope by means of the keyword \"using\": will that make it possible\n\t\tfor users change paula's value by accident? Load the module MyModule now:\n\n\t\tusing .MyModule\n\n\t\tRepeat the previous experiment - what is the value of paula afterwards?\n\t\t\"\"\",\n\t\t\"\",\n\t\tx -> x==7\n\t),\n\tActivity(\n\t\t\"\"\"\n\t\tGreat! Now that we know how to hide variables and functions inside a module, we can do\n\t\tsome real live software development! In the next laboratory, we will develop a genetic\n\t\talgorithm (GA), and GAs need to work with arrays of random values. However, generating\n\t\trandom numbers is very time-expensive, so in this laboratory we will develop a software\n\t\ttool called a Casino that will later help us generate arrays of random values very quickly.\n\n\t\tThere is just one thing we need to do first. Our modules can get quite complex, so we\n\t\twill build them up step-by-step within the Ingolstadt filesystem. If you look in the\n\t\tdirectory Ingolstadt/Development/Casinos, you will find a file named Casinos.jl. At\n\t\tpresent, this is just a dummy file that doesn't do anything except create a module named\n\t\tCasinos that contains the single variable test. Notice that the module Casinos is prefixed\n\t\tby a triple-quoted multi-line string that describes its purpose.\n\t\t\n\t\tWhenever you start writing a new module, always start the same way I have here - creating\n\t\ta very simple file that you can gradually expand into a complex program. We call this\n\t\tstyle of programming AGILE programming, because it is a good way to get started quickly\n\t\ton the job of programming a new module.\n\n\t\tTell me the value that I have assigned to test inside the Casinos module:\n\t\t\"\"\",\n\t\t\"The program statement is in line 11 of Casinos.jl\",\n\t\tx -> x==5\n\t),\n\tActivity(\n\t\t\"\"\"\n\t\tNow we'll test the Casinos module by reading and parsing the file Casinos.jl. Enter the\n\t\tfollowing line from inside the Ingolstadt folder:\n\n\t\tinclude(\"src/Development/Casinos/Casinos.jl\")\n\n\t\tNow you can test the Casinos module by telling me the answer to the following line of code:\n\n\t\tCasinos.test\n\t\t\"\"\",\n\t\t\"\",\n\t\tx -> x==5\n\t),\n\tActivity(\n\t\t\"\"\"\n\t\tBingo! Our new module works! Now we can develop our Casinos module in the file Casinos.jl.\n\t\tWe start by setting up a USE-CASE for the module - that is, we sketch out how we will want\n\t\tto use the module when it is finished. Replace your module definition in the file Casinos.jl\n\t\tby the following code, then move on to the next activity:\n\n\t\t\tmodule Casinos\n\n\t\t\t#--------------------------------------------------------------------------------------\n\t\t\t\\\"\"\"\n\t\t\t\tunittest()\n\n\t\t\tUnit-test the Casinos module.\n\t\t\t\\\"\"\"\n\t\t\tfunction unittest()\n\t\t\t\tprintln(\"\\\\n============ Unit test Casinos: ===============\")\n\t\t\t\tprintln(\"Casino deck of randomness 2 for matrix withdrawals up to size (2x3):\")\n\t\t\t\tcasino = Casino(2,3,2)\n\t\t\t\tdisplay( casino.deck)\n\t\t\t\tprintln()\n\t\t\t\n\t\t\t\tprintln(\"Draw several (2x3) matrices from the casino:\")\n\t\t\t\tdisplay( draw( casino,2,3)); println()\n\t\t\t\tdisplay( draw( casino,2,3)); println()\n\t\t\t\tdisplay( draw( casino,2,3)); println()\n\t\t\t\n\t\t\t\tprintln(\"Finally, reshuffle the casino and redisplay its deck:\")\n\t\t\t\tshuffle!(casino)\n\t\t\t\tdisplay(casino.deck)\n\t\t\tend\n\n\t\t\tend # of Casinos\n\t\t\"\"\",\n\t\t\"Note: you can't compile or run Casinos yet - just enter this code and move on\",\n\t\tx -> true\n\t),\n\tActivity(\n\t\t\"\"\"\n\t\tYou can test your new module by reincluding Casinos.jl:\n\n\t\tinclude(\"src/Development/Casinos/Casinos.jl\")\n\n\t\tIf you enter Casinos.unittest() at the Julia prompt, you will see that the command\n\t\truns, but throws lots of errors (exceptions). We'll now start fixing those errors...\n\n\t\tFirst, comment out the lines of unittest() that are causing problems. Insert the\n\t\tmulti-line comment marker #= at the beginning of the third line of unittest() so that\n\t\tit looks like this:\n\t\t\t\n\t\t#=\tcasino = Casino(2,3,2)\n\n\t\tNext, close this multi-line comment by inserting the marker =# to the right of the final\n\t\tline of unittest():\n\n\t\t\tdisplay(casino.deck) =#\n\n\t\tReinclude Casinos.jl. Now you should be able to run Casino.unittest() without errors.\n\t\tTell me the last six characters that you see in the output:\n\t\t\"\"\",\n\t\t\"You should get two lines of output, and the final characters specify a matrix size\",\n\t\tx -> x == \"(2x3):\"\n\t),\n\tActivity(\n\t\t\"\"\"\n\t\tTo start developing our new module, we'll define the type Casino. Insert the following\n\t\tcode AFTER the \"module\" line and BEFORE the comment box before unittest(), and check that\n\t\teverything still includes and runs properly:\n\n\t\tusing Random\n\n\t\t#-----------------------------------------------------------------------------------------\n\t\t# Module types:\n\t\t\n\t\t\\\"\"\"\n\t\t\tCasino\n\t\t\n\t\tA Casino can return arrays of random numbers in the range [0,1), up to a maximum number of rows\n\t\t(maxrows), and a maximum number of columns (maxcols). It also contains a deck of prepared\n\t\trandom numbers from which it draws the arrays.\n\t\t\\\"\"\"\n\t\tstruct Casino\n\t\t\tmaxrows::Int\t\t\t\t\t\t\t# Maximum number of drawable rows\n\t\t\tmaxcols::Int\t\t\t\t\t\t\t# Maximum number of drawable columns\n\t\t\trandomness::Int\t\t\t\t\t\t\t# How randomised will our withdrawals be?\n\t\t\tdeck::Matrix\t\t\t\t\t\t\t# Repository of random numbers in [0,1)\n\t\t\n\t\t\t\"The one and only constructor\"\n\t\t\tfunction Casino(maxrows::Int,maxcols::Int,randomness::Int=5)\n\t\t\t\tnew(\n\t\t\t\t\tmaxrows, maxcols, randomness,\n\t\t\t\t\trand((maxrows+1)*randomness,(maxcols+1)*randomness)\n\t\t\t\t)\n\t\t\tend\n\t\tend\n\t\t\n\t\t#-----------------------------------------------------------------------------------------\n\t\t# Module methods:\n\t\t\"\"\",\n\t\t\"Remember your code won't do anything new yet: just getting it to run is your first goal!\",\n\t\tx -> true\n\t),\n\tActivity(\n\t\t\"\"\"\n\t\tRight, now let's activate the new code. Remove the open-comment marker from the third line\n\t\tof unittest() and insert it instead at the beginning of the 7th line:\n\n\t\t#=\tprintln(\"Draw several (2x3) matrices from the casino:\")\n\n\t\tThis reveals the lines 3-6. Test that your program can now correctly create and display a\n\t\tCasino. Now tell me the size of the Casino deck, and think about why I have designed this\n\t\tsize to depend on the constructor argument randomness:\n\t\t\"\"\",\n\t\t\"\",\n\t\tx -> x == (6,8)\n\t),\n\tActivity(\n\t\t\"\"\"\n\t\tDid you work out why the size of the deck depends on randomness? The idea is that we\n\t\twant to draw random matricess from the Casino like drawing cards from a shuffled deck of\n\t\tcards, and that is only possible if the deck contains more cards than we actually need.\n\n\t\tNow we'll implement the draw() method. First reveal the next four lines of unittest() and\n\t\tinsert the following dummy code immediately after the \"Module methods:\" comment. Now test\n\t\tthis dummy version of draw to make sure it is robust before moving on:\n\n\t\t\\\"\"\"\n\t\t\tdraw( casino, nrows, ncols)\n\t\t\n\t\tDraw the required number of rows and columns from the casino deck, first ensuring that the\n\t\tdeck is large enough to support the withdrawal.\n\t\t\\\"\"\"\n\t\tfunction draw( casino::Casino, nrows::Int, ncols::Int)\n\t\t\tif nrows > casino.maxrows || ncols > casino.maxcols\n\t\t\t\t# Repository is too small - throw exception:\n\t\t\t\terror( \"Requested withdrawal is too large\")\n\t\t\tend\n\t\t\n\t\t\t# Choose random offsets and strides for drawing a matrix of size (nrows x ncols) from\n\t\t\t# the deck, assuming that it is big enough to support the withdrawal:\n\t\t\tones(nrows,ncols)\n\t\tend\n\n\t\t\"\"\",\n\t\t\"\",\n\t\tx -> true\n\t),\n\tActivity(\n\t\t\"\"\"\n\t\tNow comes the cool part of the code. Notice how we have made absolutely sure that by the\n\t\ttime we get to the dummy line \"ones(nrows,ncols)\", we can rely on the values nrows and\n\t\tncols being small enough to be able to draw our new random matrix. Now replace this\n\t\tdummy line by the following code and test it:\n\n\t\t\tdeckrows, deckcols = size(casino.deck)\n\t\t\toffset_r = rand( 1 : (deckrows-nrows))\n\t\t\tstride_r = (nrows <= 1) ? 1 :\n\t\t\t\t\t\t\trand( 1 : (deckrows-offset_r) \u00f7 (nrows-1))\n\t\t\toffset_c = rand( 1 : (deckcols-ncols))\n\t\t\tstride_c = (ncols <= 1) ? 1 :\n\t\t\t\t\t\t\trand( 1 : (deckcols-offset_c) \u00f7 (ncols-1))\n\t\t\n\t\t\t# Return a randomly chosen table of slices from the deck:\n\t\t\tcasino.deck[\n\t\t\t\t(offset_r : stride_r : (offset_r + (nrows-1)*stride_r)),\n\t\t\t\t(offset_c : stride_c : (offset_c + (ncols-1)*stride_c))\n\t\t\t]\n\t\t\"\"\",\n\t\t\"\",\n\t\tx -> true\n\t),\n\tActivity(\n\t\t\"\"\"\n\t\tFinally, we'll now give Casinos the functionality of shuffling. We want users to be\n\t\table to shuffle the contents of the deck to give new random values. Reveal all lines\n\t\tof code in unittest(), insert the command \"using Random\" at the beginning of the Casinos\n\t\tmodule - immediately after the line \"module Casinos\", then insert the following code after\n\t\tdraw() in Casinos. Then test the module again:\n\n\t\t#-----------------------------------------------------------------------------------------\n\t\t\\\"\"\"\n\t\t\tshuffle!( casino)\n\n\t\tReassign random values in the deck.\n\t\t\\\"\"\"\n\t\tfunction shuffle!( casino::Casino)\n\t\t\trand!( casino.deck)\n\t\tend\n\n\t\t\"\"\",\n\t\t\"\",\n\t\tx -> true\n\t),\n\tActivity(\n\t\t\"\"\"\n\t\tCongratulations! You have written your first Julia module! You can test its functionality\n\t\tfor yourself by doing something like this at the Julia prompt:\n\n\t\tinclude(\"src/Development/Casinos/Casinos.jl\")\n\t\tcasino = Casinos.Casino(3,3,5)\n\t\tCasinos.draw(casino,3,3)\n\n\t\tMake sure you also test error cases like this:\n\n\t\tCasinos.draw(casino,7,9)\n\t\t\"\"\",\n\t\t\"\",\n\t\tx -> true\n\t),\n\tActivity(\n\t\t\"\"\"\n\t\tThere is just one small thing we can do to make life easier for users of the Casinos\n\t\tmodule: We shall expose the new functionality. After all, it's a pain to have to write\n\t\t\"Casinos.\" in front of every command. To avoid this, insert the following lines between\n\t\tthe line \"module Casinos\" and the line \"using Random\":\n\n\t\t# Externally callable methods of Casinos\n\t\texport Casino, draw, shuffle!\n\t\t\n\t\tNow reinclude Casinos.jl and repeat your tests from the previous activity, but this time\n\t\tfirst enter \"using .Casinos\" at the Julia prompt. You should now be able to call all the\n\t\tCasinos functionality without typing \"Casinos.\" in front of everything. :)\n\t\t\"\"\",\n\t\t\"\",\n\t\tx -> true\n\t),\n]", "meta": {"hexsha": "19bd363b4e63f4e27506a2cfa9687aab4f4c4da4", "size": 14246, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Labs/INLab007.jl", "max_stars_repo_name": "NiallPalfreyman/Ingolstadt.jl", "max_stars_repo_head_hexsha": "c8b92b4b2bd8659d2584f153efe70dd7e99c5e9c", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2022-03-25T09:00:42.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-25T09:00:42.000Z", "max_issues_repo_path": "Labs/INLab007.jl", "max_issues_repo_name": "NiallPalfreyman/Ingolstadt.jl", "max_issues_repo_head_hexsha": "c8b92b4b2bd8659d2584f153efe70dd7e99c5e9c", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Labs/INLab007.jl", "max_forks_repo_name": "NiallPalfreyman/Ingolstadt.jl", "max_forks_repo_head_hexsha": "c8b92b4b2bd8659d2584f153efe70dd7e99c5e9c", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 3, "max_forks_repo_forks_event_min_datetime": "2022-03-18T14:20:14.000Z", "max_forks_repo_forks_event_max_datetime": "2022-03-25T09:00:45.000Z", "avg_line_length": 35.2623762376, "max_line_length": 97, "alphanum_fraction": 0.6796995648, "num_tokens": 3697, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.3311197264277872, "lm_q2_score": 0.21733752104706247, "lm_q1q2_score": 0.07196474051159676}}
{"text": "##Scoping Exercises\n\n# Scoping Example\n\nx=5; y=7; #Defined globally\nfunction scopeTest(z)\n x += z #Changes global value\n y = Vector{Float64}(1) #Declares a variable, local scope\n y[1] = 2\n return x + y + z\nend\n\n# Caution Example\n\nfunction f1()\n @parallel for i = 1:100\n var = 10\n if var < 100\n var = var + 1\n end\n end\n var = 100 + 10\nend\nf1()\nfunction f2()\n @parallel for i = 1:100\n var = 10\n if var < 100\n var = var + 1\n end\n end\nend\nf2()\n", "meta": {"hexsha": "cd9b7a3659d72f8b04e632f8327ede40aa630373", "size": 477, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "ExampleCode/scoping.jl", "max_stars_repo_name": "jla524/IntroToJulia", "max_stars_repo_head_hexsha": "2301ed94f1459893dcc67f67fc9b65df8d45d0ee", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 251, "max_stars_repo_stars_event_min_datetime": "2016-05-17T06:47:27.000Z", "max_stars_repo_stars_event_max_datetime": "2022-01-30T16:07:03.000Z", "max_issues_repo_path": "ExampleCode/scoping.jl", "max_issues_repo_name": "jla524/IntroToJulia", "max_issues_repo_head_hexsha": "2301ed94f1459893dcc67f67fc9b65df8d45d0ee", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 50, "max_issues_repo_issues_event_min_datetime": "2016-10-25T16:11:42.000Z", "max_issues_repo_issues_event_max_datetime": "2021-10-02T12:08:06.000Z", "max_forks_repo_path": "ExampleCode/scoping.jl", "max_forks_repo_name": "jla524/IntroToJulia", "max_forks_repo_head_hexsha": "2301ed94f1459893dcc67f67fc9b65df8d45d0ee", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 98, "max_forks_repo_forks_event_min_datetime": "2016-05-24T16:44:39.000Z", "max_forks_repo_forks_event_max_datetime": "2022-01-19T18:13:08.000Z", "avg_line_length": 14.0294117647, "max_line_length": 58, "alphanum_fraction": 0.5870020964, "num_tokens": 182, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4416730056646256, "lm_q2_score": 0.16238003058646674, "lm_q1q2_score": 0.0717188761690386}}
{"text": "using Pkg\nPkg.add(\"JuMP\")\nPkg.add(\"StructArrays\")\nPkg.add(\"DataFrames\")\nPkg.add(\"XLSX\")\nPkg.add(\"StatsBase\")\nPkg.add(\"Query\")\nPkg.add(\"Gadfly\")\nPkg.add(\"Cairo\")\nPkg.add(\"Fontconfig\")\nPkg.add(\"CSV\")\nPkg.add(\"JLD\")\n\n# these packages need their software installed at the following locations\nENV[\"CPLEX_STUDIO_BINARIES\"] = \"/opt/ibm/ILOG/CPLEX_Studio1210/cplex/bin/x86-64_linux/\"\nPkg.add(\"CPLEX\")\nENV[\"GUROBI_HOME\"] = \"/opt/gurobi911/linux64/\"\nPkg.add(\"Gurobi\")\n", "meta": {"hexsha": "2906614077cc552f3ad47fe7c228ae1f6b41df7d", "size": 458, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "setup.jl", "max_stars_repo_name": "Dibillilia/JuliaOptHeuristics", "max_stars_repo_head_hexsha": "e7acc803d8037969b98534cccba87b99028a0f5b", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "setup.jl", "max_issues_repo_name": "Dibillilia/JuliaOptHeuristics", "max_issues_repo_head_hexsha": "e7acc803d8037969b98534cccba87b99028a0f5b", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "setup.jl", "max_forks_repo_name": "Dibillilia/JuliaOptHeuristics", "max_forks_repo_head_hexsha": "e7acc803d8037969b98534cccba87b99028a0f5b", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 24.1052631579, "max_line_length": 87, "alphanum_fraction": 0.7248908297, "num_tokens": 159, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4843800842769843, "lm_q2_score": 0.14804719051380194, "lm_q1q2_score": 0.07171111061804614}}
{"text": "using Pkg\n\nPkg.update()\n\nPkg.add(\"Plots\")\nPkg.add(\"DataFrames\")\nPkg.add(\"CSV\")\n\nPkg.build(\"Plots\")\nPkg.build(\"DataFrames\")\nPkg.build(\"CSV\")", "meta": {"hexsha": "df26774360cfc184b622cb33d98882202f7593c1", "size": 139, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "setup.jl", "max_stars_repo_name": "yaozhenghangma/Julia-notebook", "max_stars_repo_head_hexsha": "e659053295e0b6b780405057b7e387a0d47379d1", "max_stars_repo_licenses": ["Apache-2.0"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "setup.jl", "max_issues_repo_name": "yaozhenghangma/Julia-notebook", "max_issues_repo_head_hexsha": "e659053295e0b6b780405057b7e387a0d47379d1", "max_issues_repo_licenses": ["Apache-2.0"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "setup.jl", "max_forks_repo_name": "yaozhenghangma/Julia-notebook", "max_forks_repo_head_hexsha": "e659053295e0b6b780405057b7e387a0d47379d1", "max_forks_repo_licenses": ["Apache-2.0"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 12.6363636364, "max_line_length": 23, "alphanum_fraction": 0.6834532374, "num_tokens": 48, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4455295350395727, "lm_q2_score": 0.16026603433317135, "lm_q1q2_score": 0.07140325175909402}}
{"text": "using BenchmarkTools\r\n\r\nfunction my_append()\r\n\r\n v = []\r\n\r\n for i in 1:10000\r\n push!(v, 5)\r\n end\r\nend\r\n\r\nfunction my_prealloc()\r\n\r\n v = Vector{Int64}(undef, 10000)\r\n\r\n for i in 1:10000\r\n v[i] = 5\r\n end\r\nend\r\n\r\n# @benchmark my_append()\r\n@benchmark my_prealloc()", "meta": {"hexsha": "e74711d740444af080635ab5a7df4204f7d5f831", "size": 292, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "julia-learning-src/advance/02c-benchmarking.jl", "max_stars_repo_name": "nunesmelo/djs-office-hours", "max_stars_repo_head_hexsha": "aa5cc3dfe3072c5608bf25f5dab27cbfa3664713", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 11, "max_stars_repo_stars_event_min_datetime": "2021-03-27T14:23:15.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-26T14:50:37.000Z", "max_issues_repo_path": "julia-learning-src/advance/02c-benchmarking.jl", "max_issues_repo_name": "nunesmelo/djs-office-hours", "max_issues_repo_head_hexsha": "aa5cc3dfe3072c5608bf25f5dab27cbfa3664713", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "julia-learning-src/advance/02c-benchmarking.jl", "max_forks_repo_name": "nunesmelo/djs-office-hours", "max_forks_repo_head_hexsha": "aa5cc3dfe3072c5608bf25f5dab27cbfa3664713", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 6, "max_forks_repo_forks_event_min_datetime": "2021-03-29T20:01:24.000Z", "max_forks_repo_forks_event_max_datetime": "2022-03-08T14:33:47.000Z", "avg_line_length": 13.2727272727, "max_line_length": 36, "alphanum_fraction": 0.551369863, "num_tokens": 89, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4455295350395727, "lm_q2_score": 0.1602660323277607, "lm_q1q2_score": 0.07140325086562435}}
{"text": "\"\"\"\n BostonHousing(; as_df = true, dir = nothing)\n\nThe classical Boston Housing tabular dataset.\n\nSources:\n (a) Origin: This dataset was taken from the StatLib library which is\n maintained at Carnegie Mellon University.\n (b) Creator: Harrison, D. and Rubinfeld, D.L. 'Hedonic prices and the \n demand for clean air', J. Environ. Economics & Management,\n vol.5, 81-102, 1978.\n (c) Date: July 7, 1993\n\nNumber of Instances: 506\n\nNumber of Attributes: 13 continuous attributes (including target\n attribute \"MEDV\"), 1 binary-valued attribute.\n\n# Arguments\n\n$ARGUMENTS_SUPERVISED_TABLE\n\n# Fields\n\n$FIELDS_SUPERVISED_TABLE\n\n# Methods\n\n$METHODS_SUPERVISED_TABLE\n\n# Examples\n \n```julia-repl\njulia> using MLDatasets: BostonHousing\n\njulia> dataset = BostonHousing()\nBostonHousing:\n metadata => Dict{String, Any} with 5 entries\n features => 506\u00d713 DataFrame\n targets => 506\u00d71 DataFrame\n dataframe => 506\u00d714 DataFrame\n\n\njulia> dataset[1:5][1]\n5\u00d713 DataFrame\n Row \u2502 CRIM ZN INDUS CHAS NOX RM AGE DIS RAD TAX PTRATIO B LSTAT \n \u2502 Float64 Float64 Float64 Int64 Float64 Float64 Float64 Float64 Int64 Int64 Float64 Float64 Float64 \n\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\n 1 \u2502 0.00632 18.0 2.31 0 0.538 6.575 65.2 4.09 1 296 15.3 396.9 4.98\n 2 \u2502 0.02731 0.0 7.07 0 0.469 6.421 78.9 4.9671 2 242 17.8 396.9 9.14\n 3 \u2502 0.02729 0.0 7.07 0 0.469 7.185 61.1 4.9671 2 242 17.8 392.83 4.03\n 4 \u2502 0.03237 0.0 2.18 0 0.458 6.998 45.8 6.0622 3 222 18.7 394.63 2.94\n 5 \u2502 0.06905 0.0 2.18 0 0.458 7.147 54.2 6.0622 3 222 18.7 396.9 5.33\n\njulia> dataset[1:5][2]\n5\u00d71 DataFrame\nRow \u2502 MEDV \n \u2502 Float64 \n\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\n 1 \u2502 24.0\n 2 \u2502 21.6\n 3 \u2502 34.7\n 4 \u2502 33.4\n 5 \u2502 36.2 \n\njulia> X, y = BostonHousing(as_df=false)[]\n([0.00632 0.02731 \u2026 0.10959 0.04741; 18.0 0.0 \u2026 0.0 0.0; \u2026 ; 396.9 396.9 \u2026 393.45 396.9; 4.98 9.14 \u2026 6.48 7.88], [24.0 21.6 \u2026 22.0 11.9])\n```\n\"\"\"\nstruct BostonHousing <: SupervisedDataset\n metadata::Dict{String, Any}\n features\n targets\n dataframe\nend\n\nfunction BostonHousing(; as_df = true, dir = nothing)\n @assert dir === nothing \"custom `dir` is not supported at the moment.\"\n path = joinpath(@__DIR__, \"..\", \"..\", \"..\", \"data\", \"boston_housing.csv\")\n df = read_csv(path)\n features = df[!, Not(:MEDV)]\n targets = df[!, [:MEDV]]\n \n metadata = Dict{String, Any}()\n metadata[\"path\"] = path\n metadata[\"feature_names\"] = names(features)\n metadata[\"target_names\"] = names(targets)\n metadata[\"n_observations\"] = size(targets, 1)\n metadata[\"description\"] = BOSTONHOUSING_DESCR\n \n if !as_df \n features = df_to_matrix(features) \n targets = df_to_matrix(targets)\n df = nothing\n end\n \n return BostonHousing(metadata, features, targets, df)\nend\n\nconst BOSTONHOUSING_DESCR = \"\"\"\nThe Boston Housing Dataset.\n\nSources:\n (a) Origin: This dataset was taken from the StatLib library which is\n maintained at Carnegie Mellon University.\n (b) Creator: Harrison, D. and Rubinfeld, D.L. 'Hedonic prices and the \n demand for clean air', J. Environ. Economics & Management,\n vol.5, 81-102, 1978.\n (c) Date: July 7, 1993\n\nNumber of Instances: 506\n\nNumber of Attributes: 13 continuous attributes (including target\n attribute \"MEDV\"), 1 binary-valued attribute.\n\nFeatures:\n\n 1. CRIM per capita crime rate by town\n 2. ZN proportion of residential land zoned for lots over 25,000 sq.ft.\n 3. INDUS proportion of non-retail business acres per town\n 4. CHAS Charles River dummy variable (= 1 if tract bounds river; 0 otherwise)\n 5. NOX nitric oxides concentration (parts per 10 million)\n 6. RM average number of rooms per dwelling\n 7. AGE proportion of owner-occupied units built prior to 1940\n 8. DIS weighted distances to five Boston employment centres\n 9. RAD index of accessibility to radial highways\n 10. TAX full-value property-tax rate per 10,000 dollars\n 11. PTRATIO pupil-teacher ratio by town\n 12. B 1000(Bk - 0.63)^2 where Bk is the proportion of blacks by town\n 13. LSTAT % lower status of the population\n \nTarget:\n \n 14. MEDV Median value of owner-occupied homes in 1000's of dollars \n\nNote: Variable #14 seems to be censored at 50.00 (corresponding to a median price of \\\\\\$50,000); \nCensoring is suggested by the fact that the highest median price of exactly \\\\\\$50,000 is reported in 16 cases, \nwhile 15 cases have prices between \\\\\\$40,000 and \\\\\\$50,000, with prices rounded to the nearest hundred. \nHarrison and Rubinfeld do not mention any censoring.\n\nThe data file stored in this repo is a copy of the UCI ML housing dataset. \nhttps://archive.ics.uci.edu/ml/machine-learning-databases/housing/\n\"\"\"\n\n# Deprecated in v0.6,\nfunction Base.getproperty(::Type{BostonHousing}, s::Symbol)\n if s == :features\n @warn \"BostonHousing.features() is deprecated, use `BostonHousing().features` instead.\"\n return () -> BostonHousing(as_df=false).features\n elseif s == :targets\n @warn \"BostonHousing.targets() is deprecated, use `BostonHousing().targets` instead.\"\n return () -> BostonHousing(as_df=false).targets\n elseif s == :feature_names\n @warn \"BostonHousing.feature_names() is deprecated, use `BostonHousing().feature_names` instead.\"\n return () -> lowercase.(BostonHousing().metadata[\"feature_names\"])\n else\n return getfield(BostonHousing, s)\n end\nend\n", "meta": {"hexsha": "811db30b0cb7fd4eb7451158d97def591e190a6d", "size": 5961, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/datasets/misc/boston_housing.jl", "max_stars_repo_name": "soham-chitnis10/MLDatasets.jl", "max_stars_repo_head_hexsha": "795ad8d1be3695024091e4ca8c0162397d0b525b", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/datasets/misc/boston_housing.jl", "max_issues_repo_name": "soham-chitnis10/MLDatasets.jl", "max_issues_repo_head_hexsha": "795ad8d1be3695024091e4ca8c0162397d0b525b", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/datasets/misc/boston_housing.jl", "max_forks_repo_name": "soham-chitnis10/MLDatasets.jl", "max_forks_repo_head_hexsha": "795ad8d1be3695024091e4ca8c0162397d0b525b", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 37.25625, "max_line_length": 137, "alphanum_fraction": 0.6129843986, "num_tokens": 1835, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4455295350395727, "lm_q2_score": 0.16026602631152884, "lm_q1q2_score": 0.07140324818521537}}
{"text": "using Pkg\nPkg.activate(pwd())\n\n# # Tuples\n\nt = (1, 2.0, \"3\")\nt = 1, 2.0, \"3\"\n\n#+\n\ntypeof(t)\n\n#+\n\nt[1] # the first element\nt[end] # the last element\nt[1:2] # the first two elements\n\n#+\n\na, b, c = t\nprintln(\"The values stored in the tuple are: $a, $b and $c\")\n\n# ### Exercise:\n# Create a tuple that contains the first four letters of the alphabet (these letters should\n# be of type `String`). Then unpack this tuple into four variables `a`, `b`, `c` and `d`.\n# \n# ---\n# ### Solution:\n\n\n\n# ---\n# \n# # Named Tuples\n\nt = (a = 1, b = 2.0, c = \"3\")\n\n#+\n\na = 1;\nb = 2.0;\nc = \"3\";\n\nt = (; a, b, c)\n\n#+\n\nt[1] # the first element\nt[end] # the last element\nt[1:2] # error\n\n#+\n\nt.a\nt.c\na, b, c = t\n\nprintln(\"The values stored in the tuple are: a = $a, b = $b\")", "meta": {"hexsha": "cc9a94e7d34b23fc1d5f13a348ae4b4b36bee7b4", "size": 747, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "lecture_02/02-tuples.jl", "max_stars_repo_name": "JuliaTeachingCTU/Julia-for-Optimization-and-Learning-Scripts", "max_stars_repo_head_hexsha": "8e00299449736e4ccf47c247aa9d80f99a7e5b92", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "lecture_02/02-tuples.jl", "max_issues_repo_name": "JuliaTeachingCTU/Julia-for-Optimization-and-Learning-Scripts", "max_issues_repo_head_hexsha": "8e00299449736e4ccf47c247aa9d80f99a7e5b92", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "lecture_02/02-tuples.jl", "max_forks_repo_name": "JuliaTeachingCTU/Julia-for-Optimization-and-Learning-Scripts", "max_forks_repo_head_hexsha": "8e00299449736e4ccf47c247aa9d80f99a7e5b92", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 12.6610169492, "max_line_length": 91, "alphanum_fraction": 0.5542168675, "num_tokens": 284, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4455295350395727, "lm_q2_score": 0.1602660263115288, "lm_q1q2_score": 0.07140324818521536}}
{"text": "# Code for Video 1.6: \r\n\r\nt1 = (108, 3.14,\"Julia\",'\u03b1') # (108,3.14,\"Julia\",'\u03b1')\r\ntypeof(t1) # Tuple{Int64,Float64,ASCIIString,Char}\r\n\r\na, b, c, d = t1 # (108,3.14,\"Julia\",'\u03b1')\r\n\r\na # 108\r\nb # 3.14\r\nc # \"Julia\"\r\nd # '\u03b1'\r\n\r\na, b = b, a # (3.14,108)\r\nprintln(\"a is now $a, b is now $b\") \r\n# a is now 3.14, b is now 108\r\n\r\nt1[3] # \"Julia\"\r\n\r\nfor e in t1\r\n print(e, \" - \")\r\nend\r\n# 108 - 3.14 - Julia - \u03b1 -\r\n\r\n# t1[4] = '\u03b2'\r\n# ERROR: MethodError: `setindex!` has no method matching setindex!(::Tuple{Int64,F\r\n# loat64,ASCIIString,Char}, ::Char, ::Int64)\r\n\r\ns = Set(Int64[11, 14, 13, 7, 14, 11])\r\n# Set([7,14,13,11])\r\ns2 = Set(Any[11, 3.14, \"Julia\", '\u03b1'])\r\n# Set(Any[3.14,'\u03b1',\"Julia\",11])\r\nintersect(s, s2) # Set([11])\r\n\r\ndiris = Dict(\"I. setosa\" => 50,\r\n \"I. versicolor\" => 50,\r\n \"I. virginica\" => 50)\r\n# Dict{ASCIIString,Int64} with 3 entries:\r\n# \"I. virginica\" => 50\r\n# \"I. versicolor\" => 50\r\n# \"I. setosa\" => 50\r\n\r\ndiriss = Dict( :setosa => 50,\r\n :versicolor => 50,\r\n :virginica => 50)\r\n# Dict{Symbol,Int64} with 3 entries:\r\n# :versicolor => 50\r\n# :virginica => 50\r\n# :setosa => 50\r\n\r\ndiris[\"I. setosa\"] # 50\r\ndiriss[:setosa] # 50\r\n\r\ndiriss[:setosa] = 53\r\ndiriss\r\n# Dict{Symbol,Int64} with 3 entries:\r\n# :versicolor => 50\r\n# :virginica => 50\r\n# :setosa => 53\r\n\r\nhaskey(diris, \"I. setosa\") # true\r\n\r\nkeys(diris)\r\n# Base.KeyIterator for a Dict{ASCIIString,Int64} with 3 entries. Keys:\r\n# \"I. virginica\"\r\n# \"I. versicolor\"\r\n# \"I. setosa\"\r\n\r\nvalues(diris)\r\n# Base.ValueIterator for a Dict{ASCIIString,Int64} with 3 entries. Values:\r\n# 50\r\n# 50\r\n# 50\r\n\r\nfor k in keys(diris)\r\n print(k, \" - \")\r\nend\r\n# I. virginica - I. versicolor - I. setosa -\r\n\r\nfor (k, v) in diris\r\n println(\"$k has $v measurements\")\r\nend\r\n# I. virginica has 50 measurements\r\n# I. versicolor has 50 measurements\r\n# I. setosa has 50 measurements", "meta": {"hexsha": "3786d7813247652dc46717975623b444804bb4e0", "size": 1911, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Section 1/1_6.jl", "max_stars_repo_name": "lytemar/Julia-for-Data-Science-Video", "max_stars_repo_head_hexsha": "e7cb2427b10979d4be0f1e00be1cc1090f4da736", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 4, "max_stars_repo_stars_event_min_datetime": "2019-09-01T15:05:48.000Z", "max_stars_repo_stars_event_max_datetime": "2021-11-14T14:09:53.000Z", "max_issues_repo_path": "Section 1/1_6.jl", "max_issues_repo_name": "lytemar/Julia-for-Data-Science-Video", "max_issues_repo_head_hexsha": "e7cb2427b10979d4be0f1e00be1cc1090f4da736", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Section 1/1_6.jl", "max_forks_repo_name": "lytemar/Julia-for-Data-Science-Video", "max_forks_repo_head_hexsha": "e7cb2427b10979d4be0f1e00be1cc1090f4da736", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 4, "max_forks_repo_forks_event_min_datetime": "2019-09-01T15:05:49.000Z", "max_forks_repo_forks_event_max_datetime": "2021-06-08T15:00:22.000Z", "avg_line_length": 22.75, "max_line_length": 83, "alphanum_fraction": 0.5510204082, "num_tokens": 745, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4455295203152604, "lm_q2_score": 0.16026603032235004, "lm_q1q2_score": 0.07140324761234759}}
{"text": "using LibFoo\nusing Test\n\n@test 1 == 1", "meta": {"hexsha": "2b474685fb611353f31efb7afad6160515ecaa70", "size": 37, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "stdlib/Pkg/test/test_packages/BigProject/test/LibFoo.jl/test/runtests.jl", "max_stars_repo_name": "ninjin/julia", "max_stars_repo_head_hexsha": "2d589cca94c502a696fc5e234835560e28b9efd3", "max_stars_repo_licenses": ["Zlib"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "stdlib/Pkg/test/test_packages/BigProject/test/LibFoo.jl/test/runtests.jl", "max_issues_repo_name": "ninjin/julia", "max_issues_repo_head_hexsha": "2d589cca94c502a696fc5e234835560e28b9efd3", "max_issues_repo_licenses": ["Zlib"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "stdlib/Pkg/test/test_packages/BigProject/test/LibFoo.jl/test/runtests.jl", "max_forks_repo_name": "ninjin/julia", "max_forks_repo_head_hexsha": "2d589cca94c502a696fc5e234835560e28b9efd3", "max_forks_repo_licenses": ["Zlib"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 9.25, "max_line_length": 12, "alphanum_fraction": 0.7027027027, "num_tokens": 13, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.3960681520167196, "lm_q2_score": 0.18010665968560707, "lm_q1q2_score": 0.0713345118675826}}
{"text": "### A Pluto.jl notebook ###\n# v0.17.3\n\nusing Markdown\nusing InteractiveUtils\n\n# \u2554\u2550\u2561 c730d000-5ed6-11ec-36b6-39741ccad6c7\nbegin\n\tusing Pkg; Pkg.activate(@__DIR__); Pkg.instantiate()\n\tPkg.precompile()\n\n\tusing PlutoUI\n\tusing BenchmarkTools\n\tusing DataFrames\n\tusing DelimitedFiles\n\tusing CSV\n\tusing XLSX\n\tusing Downloads\n\tusing JLD, NPZ, MAT, RData\n\tusing RCall\nend;\n\n# \u2554\u2550\u2561 62c5d4f2-9776-43ae-8125-0d429d6cde42\nPlutoUI.TableOfContents(aside=true, indent=true, depth=3)\n\n# \u2554\u2550\u2561 34f92fb1-6c1e-4db6-b795-8a526007ea92\nhtml\"\"\"\n\n\t
\n\"\"\"\n\n# \u2554\u2550\u2561 13a10dac-8c7e-4e72-9fa9-d58ca8dd0202\nmd\"\"\"\n# \ud83d\udcbe Data\n\nBeing able to easily load and process data is a crucial task that can make any data science more pleasant. In this notebook, we will cover most common types often encountered in data science tasks, and we will be using this data throughout the rest of this tutorial.\n\n[YouTube link](https://www.youtube.com/watch?v=iG1dZBaxS-U)\n\"\"\"\n\n# \u2554\u2550\u2561 5100df3f-c5d3-4346-bcd9-8afc92bc559c\nmd\"\"\"\n## Get some data\n\nIn Julia, it's pretty easy to dowload a file from the web using the `download` function. But also, you can use your favorite command line commad to download files by easily switching from Julia via the `;` key. Let's try both.\n\n**Note:** `download` depends on external tools such as curl, wget or fetch. So you must have one of these.\n\"\"\"\n\n# \u2554\u2550\u2561 abe62085-b630-4816-af11-b39133892849\ndata\u2081 = Downloads.download(\"https://raw.githubusercontent.com/nassarhuda/easy_data/master/programming_languages.csv\", \"programminglanguages.csv\")\n\n# \u2554\u2550\u2561 51c909b1-1d37-458f-acd6-68f4041bc92f\nmd\"\"\"\n## Read your data from text files\n\nThe key question here is to load data from files such as `csv` files, `xlsx` files, or just raw text files. We will go over some Julia packages that will allow us to read such files very easily.\n\"\"\"\n\n# \u2554\u2550\u2561 6483858c-2e58-4930-b4e9-76b47032eac4\nmd\"\"\"\n### DelimitedFiles.jl\n\nLet's start with the package `DelimitedFiles` which is in the standard library. When loaded, the file is stored as a `Matrix`.\n\n> **readdlm**(source, \n> delim::AbstractChar, \n> T::Type, \n> eol::AbstractChar; \n> header=false, \n> skipstart=0, \n> skipblanks=true, \n> use_mmap, \n> quotes=true, \n> dims, \n> comments=false, \n> comment_char='#')\n\"\"\"\n\n# \u2554\u2550\u2561 89a5a316-5058-4ff2-8d71-fc832a84c58d\nbegin\n\tdata\u2082, header = readdlm(\"programming_languages.csv\",',';header=true)\n\theader\nend\n\n# \u2554\u2550\u2561 853a4e90-ac39-4d73-9953-c7b70e8f78ef\ndata\u2082\n\n# \u2554\u2550\u2561 3a22c018-2ca7-41df-9ee4-91dc8b64d940\nmd\"\"\"\n> **Note:** the output table is a Matrix (2D Array).\n\nWe can use the function `writedlm`, from `DelimitedFiles.jl` package, to write to a text file...\n\"\"\"\n\n# \u2554\u2550\u2561 b4fcdcf9-793f-4d4a-a5fb-950108cb8188\nwritedlm(\"output/programminglanguages_dlm.txt\", data\u2082, '-')\n\n# \u2554\u2550\u2561 cfa37c7c-810c-4095-8887-262dee4a9d04\nmd\"\"\"\n### CSV.jl\n\nA more powerful package to use here is the `CSV.jl` package. By default, this package imports the data to a `DataFrame`, which can have several advantages as we will see below.\n\n_In general, `CSV.jl` is the recommended way to load CSVs in Julia_. Only use `DelimitedFiles.jl` when you have a more complicated file where you want to specify several things.\n\"\"\"\n\n# \u2554\u2550\u2561 4b651b5a-e562-46ef-a3b5-8ee85b5a61ed\ndf = CSV.read(\"programming_languages.csv\", DataFrame)\n\n# \u2554\u2550\u2561 11471bbf-b7b4-4b6e-90cb-9e9ff47bbe4a\nmd\"\"\"\nWe can use the `typeof` function to make sure that the output is a DataFrame.\n\"\"\"\n\n# \u2554\u2550\u2561 2652dc01-d80d-46d9-ae7a-9afa534e88cb\ntypeof(df)\n\n# \u2554\u2550\u2561 b2f35e6b-8c19-4729-87a3-292858bfbcb5\nmd\"\"\"\nWe can use the macro `@btime`, from `BenchMarkTools.jl`, to prove that `CSV.jl` is faster than `DelimitedFiles.jl`...\n\n**Note:** Check Julia REPL to see the processing time.\n\"\"\"\n\n# \u2554\u2550\u2561 e1210609-a216-4d54-be63-04f10c5c15bc\n@btime df\u2081, h\u2081 = readdlm(\"programming_languages.csv\",',';header=true);\n\n# \u2554\u2550\u2561 008b635f-c4e5-475a-83af-4bc3fc722344\n@btime df\u2082 = CSV.read(\"programming_languages.csv\", DataFrame);\n\n# \u2554\u2550\u2561 934c8549-c86c-45e6-a9f5-56297c73513d\nmd\"\"\"\nWe can use the function `write`, from `CSV.jl` package, to write to a text file...\n\n**Note:** if we want to write a Matrix, we could first convert it into a DataFrame (using the `DataFrame` function) and, then, write it using the `write` function.\n\"\"\"\n\n# \u2554\u2550\u2561 92708ece-3620-4cf5-b09d-098002044623\nCSV.write(\"output/programminglanguages_CSV.csv\", df)\n\n# \u2554\u2550\u2561 cac4f1b5-f9e3-469e-9576-553a1cf1b363\nmd\"\"\"\n### Some DataFrame Operations\n\nThere are different ways to slice a DataFrame. Suppose that we want just the first 10 rows of a DataFrame...\n\"\"\"\n\n# \u2554\u2550\u2561 3d378ec4-659f-4032-a5b7-6d6b6d402c2f\ndf[1:10,:]\n\n# \u2554\u2550\u2561 e50ae64c-ff64-4597-bf75-d7c921d23225\nmd\"\"\"\nSuppose that we want all the instances of a given feature (column)...\n\"\"\"\n\n# \u2554\u2550\u2561 d9ea3574-2ffb-4fdb-9ced-db1dc669ecd9\ndf[!,\"year\"]\n\n# \u2554\u2550\u2561 09ff6d8b-15f2-4d8e-a775-dee357a417e4\ndf.year\n\n# \u2554\u2550\u2561 843a668f-7b08-4bce-b02a-3153d0eb7d0d\ndf[!,\"year\"] == df.year\n\n# \u2554\u2550\u2561 159a35f4-09fd-47bd-9935-88668a7e7f27\ntypeof(df.year)\n\n# \u2554\u2550\u2561 494ebede-80d0-430e-91aa-0a9050a1bb29\nmd\"\"\"\nWe can use the `names` function to get an array of all column names...\n\"\"\"\n\n# \u2554\u2550\u2561 00d12274-3057-4a52-8400-0c8c3c358e6b\nnames(df)\n\n# \u2554\u2550\u2561 fede3ce2-da03-4ff9-a13e-7ce55ec05bf6\nmd\"\"\"\nWe can use the `describe` function to check some data stats...\n\"\"\"\n\n# \u2554\u2550\u2561 74783bbb-3c9d-4e15-bc54-b3d8e7c2d230\ndescribe(df)\n\n# \u2554\u2550\u2561 e4b3a444-c487-4fe8-8048-873b8a3affb7\nmd\"\"\"\nLet's create two DataFrames...\n\"\"\"\n\n# \u2554\u2550\u2561 0becba29-d5ff-4595-86ab-6b0575674047\nbegin\n\tfoods = [\"Apple\",\"Cucumber\",\"Tomato\",\"Banana\"]\n\tcalories = [105,47,22,105]\n\n\tdf_cal = DataFrame(Item=foods, Calories=calories)\nend\n\n# \u2554\u2550\u2561 9d522bdc-d066-40ac-84c4-47287d9e07e0\nbegin\n\tprices = [0.85,1.6,0.8,0.6]\n\n\tdf_price = DataFrame(Item=foods, Price=prices)\nend\n\n# \u2554\u2550\u2561 79de9741-c913-4c00-8833-4f1b68f80ea5\nmd\"\"\"\nNow, we can use the `innerjoin` function to join both DataFrames on `Item` column...\n\"\"\"\n\n# \u2554\u2550\u2561 fd724d83-41bb-422c-860e-2bbeff17e0dc\ndf_comb = innerjoin(df_cal, df_price, on=:Item)\n\n# \u2554\u2550\u2561 51301eeb-709a-4ea9-a1a5-d7967f8dd55d\nmd\"\"\"\n### XLSX.jl\n\nAnother type of files that we may often need to read is XLSX files. Let's try to read this type of file using the `XLSX.jl` package...\n\n> **readdata**(filepath, sheetname, cellrange)\n\"\"\"\n\n# \u2554\u2550\u2561 64e7e90b-cd7d-4569-a715-4f27f1787e40\nxlsx\u2081 = XLSX.readdata(\"data/zillow_data_download_april2020.xlsx\",\n \t\t\t \t \"Sale_counts_city\",\n \t\t\t \t \"A1:F9\")\n\n# \u2554\u2550\u2561 e5f2c69b-fbd8-4048-aaed-792a64a4d66c\nmd\"\"\"\nIf you don't want to specify cell ranges, you can use the `readtable` function. However, this will take a little longer...\n\n> **readtable**(filepath, sheetname)\n\"\"\"\n\n# \u2554\u2550\u2561 26047637-1b77-4203-b705-e34f74137295\nxlsx\u2082 = XLSX.readtable(\"data/zillow_data_download_april2020.xlsx\",\n\t\t\t\t\t \"Sale_counts_city\")\n\n# \u2554\u2550\u2561 0f3d505c-a575-419f-8155-fd071f510714\ntypeof(xlsx\u2082)\n\n# \u2554\u2550\u2561 c2f1eab2-862a-4eb8-b233-71567cdc9963\nmd\"\"\"\nHere, `xlsx\u2082` is a tuple of two items. The first item is an vector of vectors where each vector corresponds to a column in the excel file...\n\"\"\"\n\n# \u2554\u2550\u2561 3a924b3e-3dbe-49b6-9e5e-5a48b0ffd385\nxlsx\u2082[1]\n\n# \u2554\u2550\u2561 58bad348-0586-4d0f-8d54-7c3e1a7a2c40\nmd\"\"\"\nWe can access the first column by typing...\n\"\"\"\n\n# \u2554\u2550\u2561 036de982-36ad-431a-8445-517178626911\nxlsx\u2082[1][1]\n\n# \u2554\u2550\u2561 4ead9039-f4c2-4f82-bf1e-2146718014a3\nmd\"\"\"\nWe can access the third column first 10 elements...\n\"\"\"\n\n# \u2554\u2550\u2561 bb04dc6c-e62e-4b2c-a247-ebe4836d8013\nxlsx\u2082[1][3][begin:10]\n\n# \u2554\u2550\u2561 d9550872-fa7c-4ebd-9a4d-828182c8e49d\nmd\"\"\"\nOn the other hand, the second is the header with the column names...\n\"\"\"\n\n# \u2554\u2550\u2561 e01a65ab-394a-4718-b9ae-ca2bb30e20b5\nxlsx\u2082[2]\n\n# \u2554\u2550\u2561 1eb26b3c-8a5a-4648-bb73-f8291b656edc\nmd\"\"\"\nWe could check the names of the last 10 columns...\n\"\"\"\n\n# \u2554\u2550\u2561 cb027055-2cf6-4f74-b265-9d01c23c2967\nxlsx\u2082[2][end-9:end]\n\n# \u2554\u2550\u2561 12dccfc3-e976-46e6-81b0-dc51b48309f4\nmd\"\"\"\nAnd we can easily store this data in a `DataFrame`. We can use the \"splat\" operator to unwrap these arrays and pass them to the DataFrame constructor...\n\"\"\"\n\n# \u2554\u2550\u2561 403ac783-9c02-4fd1-8506-afe0af36a14b\ndf_xlsx = DataFrame(xlsx\u2082...)\n# df_xlsx = DataFrame(xlsx\u2082[1], xlsx\u2082[2])\n\n# \u2554\u2550\u2561 36d8898e-980c-4f54-bbff-277ded2229ea\nmd\"\"\"\n**Note:** the first argument is the actual data and the second one comprises the column names.\n\nWe can also easily write data to an XLSX file (takes several amount of time)...\n\"\"\"\n\n# \u2554\u2550\u2561 8d769d53-1902-4a09-8228-ce60174b22bb\n# XLSX.writetable(\"output/writefile_using_XLSX.xlsx\",xlsx\u2082[1],xlsx\u2082[2])\n\n# \u2554\u2550\u2561 bc29d2b5-e6c2-4b45-8b79-cbb012659be1\nmd\"\"\"\n## Importing your data\n\nOften, the data you want to import is not stored in plain text, and you might want to import different kinds of types. Here we will go over importing `jld`, `npz`, `rda`, and `mat` files. Hopefully, these four will capture the types from four common programming languages used in Data Science (Julia, Python, R and Matlab).\n\nWe will use a toy example here of a very small matrix. But the same syntax will hold for bigger files.\n\"\"\"\n\n# \u2554\u2550\u2561 7c2fef9c-4e9a-4637-bc8e-6636af5f98ae\nmd\"\"\"\n### Julia Data (JLD)\n\nJLD, for which files conventionally have the extension `.jld`, is a widely-used format for data storage with the Julia programming language.\n\nJLD is a specific \"dialect\" of HDF5, a cross-platform, multi-language data storage format most frequently used for scientific data. By comparison with \"plain\" HDF5, JLD files automatically add attributes and naming conventions to preserve type information for each object.\n\"\"\"\n\n# \u2554\u2550\u2561 8f51aeb5-3e75-4e5a-8c8f-ed0201eb8ce0\n# load JLD file\njld_data = JLD.load(\"data/mytempdata.jld\")\n\n# \u2554\u2550\u2561 879ac394-8598-4399-957f-1280ecd99478\n# see the data\njld_data[\"tempdata\"]\n\n# \u2554\u2550\u2561 0d27e3b3-79e3-424d-bc6b-94252900f7fb\n# save JLD file\nsave(\"output/mywrite.jld\", \"A\", jld_data)\n\n# \u2554\u2550\u2561 13053d54-8680-41e2-b416-8a7bc50f6c92\n# JLD type\ntypeof(jld_data)\n\n# \u2554\u2550\u2561 e0f49c77-0c0d-4740-9227-5619d14748b6\nmd\"\"\"\n### Numpy Data (NPZ)\n\nSeveral arrays into a single file in uncompressed Numpy `.npz` format.\n\"\"\"\n\n# \u2554\u2550\u2561 f4ddf1bf-0cbf-47c5-bb5f-f8abf0fb8ac2\n# load NPZ file\nnpz_data = npzread(\"data/mytempdata.npz\")\n\n# \u2554\u2550\u2561 a979107e-f2cf-42da-aefa-5cb249bc7073\n# save NPZ file\nnpzwrite(\"output/mywrite.npz\", npz_data)\n\n# \u2554\u2550\u2561 edd2093e-f84a-4563-acb7-219f2bd07399\n# NPZ type\ntypeof(npz_data)\n\n# \u2554\u2550\u2561 be91882d-fe73-40d0-8e75-30c2e8ddd564\nmd\"\"\"\n### R Data (RDA)\n\nThe RData format (usually with extension `.rdata` or `.rda`) is a format designed for use with R, a system for statistical computation and related graphics, for storing a complete R workspace or selected \"objects\" from a workspace in a form that can be loaded back by R.\n\"\"\"\n\n# \u2554\u2550\u2561 1f3092e8-3f3a-4d29-a9f5-e64ef96c13a8\n# load RDA file\nR_data = RData.load(\"data/mytempdata.rda\")\n\n# \u2554\u2550\u2561 cf9a1b00-79dd-4305-93ad-69f229319ce2\n# see the data\nR_data[\"tempdata\"]\n\n# \u2554\u2550\u2561 1b49419d-18cd-4617-9442-15080e3ef588\nbegin\n\t# save RDA file\n\t@rput R_data\n\tR\"save(R_data, file=\\\"output/mywrite.rda\\\")\"\nend\n\n# \u2554\u2550\u2561 c3bce6db-d503-4a68-b83f-278c6d603f07\nmd\"\"\"\n**Note:** we must use the `RCall.jl` package to save `.rda` files.\n\"\"\"\n\n# \u2554\u2550\u2561 7131ac1c-984e-4ad9-a1cd-b1d46a4f240e\n# RDA type\ntypeof(R_data)\n\n# \u2554\u2550\u2561 4f70b5df-0026-43cd-b769-d4ca50145fbd\nmd\"\"\"\n### MATLAB\u00ae Data (MAT)\n\nMAT-files are binary MATLAB\u00ae `.mat` files that store workspace variables.\n\"\"\"\n\n# \u2554\u2550\u2561 0c1201b9-6acb-4206-a806-e09e04c6a65e\n# load MAT file\nmatlab_data = matread(\"data/mytempdata.mat\")\n\n# \u2554\u2550\u2561 cc555ffc-0eea-4e35-b0d1-36d3a734f9f2\n# see the data\nmatlab_data[\"tempdata\"]\n\n# \u2554\u2550\u2561 5516dd98-3a87-4801-a62e-e91a95e4923a\n# save MAT file\nmatwrite(\"output/mywrite.mat\", matlab_data)\n\n# \u2554\u2550\u2561 53be8986-1f06-45a5-9303-5018a3ddf06f\n# MAT type\ntypeof(matlab_data)\n\n# \u2554\u2550\u2561 ecefa934-49a8-4abf-880d-d91d3f9f9d05\nmd\"\"\"\nAll files, when loaded, are `Dict`. The only exception is NPZ data, which is loaded as an `Matrix`.\n\"\"\"\n\n# \u2554\u2550\u2561 01be19ba-2aaa-4746-af5d-8ce7f1cf3a55\nmd\"\"\"\n## Processing different types of data\n\nWe will mainly cover `Vector` (`Matrix` included), `DataFrame`, and `Dict`. Let's bring back our programming languages dataset `data\u2082` and start playing it the matrix it's stored in.\n\"\"\"\n\n# \u2554\u2550\u2561 3a6f9dc0-2186-440a-9d78-5da6acabcdae\nmd\"\"\"\n### Matrix\n\"\"\"\n\n# \u2554\u2550\u2561 4f9dca90-9f1a-4c0e-a510-fa640d109b0d\ndata\u2082\n\n# \u2554\u2550\u2561 57b8265f-be5c-4f05-91c6-6c4fec4f0a6b\nmd\"\"\"\nHere are some quick questions we might want to ask about this simple data.\n\n> **Q1:** Which year was was a given language invented?\n\"\"\"\n\n# \u2554\u2550\u2561 157e9e3d-2e10-418e-997e-0178c14d7b3d\nfunction year_created_mtx(data, language::String)\n loc = findfirst(data[:,2] .== language)\n !isnothing(loc) && return data[loc,1]\n\terror(\"Error: Language not found!\")\nend\n\n# \u2554\u2550\u2561 e7bd695e-c7d6-4b38-8520-7d915e740015\nmd\"\"\"\n**Note:** this function return the year just when `loc` local variable is not `nothing`. Otherwise, it returns an error message.\n\"\"\"\n\n# \u2554\u2550\u2561 c9ead4e0-aa01-43bc-8107-72d8ec4c073b\nbegin\n\tjulia_year1 = year_created_mtx(data\u2082, \"Julia\")\n\n\t\"Julia was created in $julia_year1.\"\nend\n\n# \u2554\u2550\u2561 92b4a2d9-b271-42b3-97d1-647300a9439a\nbegin\n\tcobol_year1 = year_created_mtx(data\u2082, \"COBOL\")\n\n\t\"COBOL was created in $cobol_year1.\"\nend\n\n# \u2554\u2550\u2561 01a73335-69b9-4297-9910-a441848fe2c5\nmd\"\"\"\n> **Q2:** How many languages were created in a given year?\n\"\"\"\n\n# \u2554\u2550\u2561 fbf69873-af41-428c-9bba-d63cdb20149b\nfunction langs_per_year_mtx(data, year::Int64)\n\tcount = length(findall(data[:,1] .== year))\n\n\treturn count\nend\n\n# \u2554\u2550\u2561 1ea19d7e-5748-4559-b2d2-fd4f4d449516\nbegin\n\tlangs_1988\u2081 = langs_per_year_mtx(data\u2082, 1988)\n\n\t\"In 1988, $langs_1988\u2081 language(s) was/were created.\"\nend\n\n# \u2554\u2550\u2561 2592f7cd-95ec-493c-93e1-bbedf433367f\nbegin\n\tlangs_2006\u2081 = langs_per_year_mtx(data\u2082, 2006)\n\n\t\"In 2006, $langs_2006\u2081 language(s) was/were created.\"\nend\n\n# \u2554\u2550\u2561 12121f45-8a7b-4709-bc85-b2fde7313a21\nmd\"\"\"\n### DataFrame\n\nNow let's try to store this data in a DataFrame...\n\"\"\"\n\n# \u2554\u2550\u2561 d455f80e-c5b5-4b1a-9667-86fade5ee3d3\n# anonymous column names\ndata\u2083 = DataFrame(data\u2082, :auto)\n\n# \u2554\u2550\u2561 42c72fde-8331-4e40-b761-c50d0e2e8486\n# specifying column names\ndata\u2084 = DataFrame(Year = data\u2082[:,1], Language = data\u2082[:,2])\n\n# \u2554\u2550\u2561 201a8534-91ee-4ac9-a120-51b29f196c23\n# specifying both column names and data types\ndata\u2085 = DataFrame(Year = Int.(data\u2082[:,1]), Language = string.(data\u2082[:,2]))\n\n# \u2554\u2550\u2561 0b14e9fc-62e6-4ad7-ab67-eb36e340749c\nmd\"\"\"\n> **Q1:** Which year was was a given language invented?\n\"\"\"\n\n# \u2554\u2550\u2561 679b2b83-07f9-40bd-bdd1-89e7233bfade\nfunction year_created_df(df,language::String)\n loc = findfirst(df.Language .== language)\n\t!isnothing(loc) && return df.Year[loc]\n\terror(\"Error: Language not found!\")\nend\n\n# \u2554\u2550\u2561 f4738476-8b05-4dbc-a15e-6b3d59c90c50\nmd\"\"\"\n**Note:** this function return the year just when `loc` local variable is not `nothing`. Otherwise, it returns an error message.\n\"\"\"\n\n# \u2554\u2550\u2561 c6b170be-6576-480e-b2ba-65d3b853871a\nbegin\n\tjulia_year2 = year_created_df(data\u2085, \"Julia\")\n\n\t\"Julia was created in $julia_year2.\"\nend\n\n# \u2554\u2550\u2561 9977fdaa-db30-43a5-bd58-14087c2163e4\nbegin\n\tcobol_year2 = year_created_df(data\u2085, \"COBOL\")\n\n\t\"COBOL was created in $cobol_year2.\"\nend\n\n# \u2554\u2550\u2561 a137ebd7-9db4-4c29-a694-b1acad5214a3\nmd\"\"\"\n> **Q2:** How many languages were created in a given year?\n\"\"\"\n\n# \u2554\u2550\u2561 2cc2938b-58d5-4c27-bdab-dddf22231bc8\nfunction langs_per_year_df(df, year::Int64)\n count = length(findall(df.Year.==year))\n return count\nend\n\n# \u2554\u2550\u2561 ae7a1714-2245-4d5a-89bb-10b3adff787a\nbegin\n\tlangs_1988\u2082 = langs_per_year_mtx(data\u2085, 1988)\n\n\t\"In 1988, $langs_1988\u2082 language(s) was/were created.\"\nend\n\n# \u2554\u2550\u2561 ce4ac86d-5e06-4c3e-93d6-ebdee46439bd\nbegin\n\tlangs_2006\u2082 = langs_per_year_mtx(data\u2085, 2006)\n\n\t\"In 2006, $langs_2006\u2082 language(s) was/were created.\"\nend\n\n# \u2554\u2550\u2561 515b7479-97d8-464b-8245-1034f3cd8bdc\nmd\"\"\"\n### Dictionary\n\nNext, we'll use dictionaries. A quick way to create a dictionary is with the `Dict()` command. But this creates a dictionary without types. Here, we will specify the types of this dictionary.\n\"\"\"\n\n# \u2554\u2550\u2561 3b08a2c8-546b-4a6c-8ae0-2a1e64578af8\n# dict from a list of tuples\nDict([(\"A\", 1), (\"B\", 2)])\n\n# \u2554\u2550\u2561 21551a38-57fc-42c2-81a2-b314bb06250b\nmd\"\"\"\n**Note:** the key type is `String`, while the value type is `Int64`.\n\"\"\"\n\n# \u2554\u2550\u2561 6da1f5a7-56a2-4a7a-abfc-25ec3f44f892\n# dictionary from a list of tuples\nDict([(\"A\", 1), (\"B\", 2), (\"C\", [1,2,3])])\n\n# \u2554\u2550\u2561 43ad8297-d94c-45a0-941f-ab0d5bfd2ddb\nmd\"\"\"\n**Note:** the key type is `String`, while the value type is `Any`, since we have integers and arrays as values.\n\"\"\"\n\n# \u2554\u2550\u2561 9390d98f-94eb-48bb-ba9c-e6b3f67f762f\n# empty dict\nDict()\n\n# \u2554\u2550\u2561 30718df7-5085-4d0d-8a86-7b4a08ff20b2\n# empty dict, specifying that keys are Integers and values are Vectors of Strings\ndict\u2081 = Dict{Integer, Vector{String}}()\n\n# \u2554\u2550\u2561 f77a5189-19b4-4f99-b394-72190253912f\n# appending a key-value pair to dict\ndict\u2081[2012] = [\"Julia\", \"Programming\", \"Language\"]\n\n# \u2554\u2550\u2561 50ba8f3b-5104-4d4f-85ea-831d4afe9164\n# updated dict\ndict\u2081\n\n# \u2554\u2550\u2561 bb814af7-62e3-4744-955f-31b557540057\nmd\"\"\"\nNow, let's populate the dictionary with years as keys and vectors that hold all the programming languages created in each year as their values. Even though this looks like more work, we often need to do it just once.\n\"\"\"\n\n# \u2554\u2550\u2561 9193df28-7ed1-4853-a97b-aa5a1232e07b\nbegin\n\tlang_dict = Dict{Integer, Vector{String}}()\n\n\tfor i in 1:size(data\u2082,1)\n\t\tyear, lang = data\u2082[i,:]\n\t\tif year \u2208 keys(lang_dict)\n\t\t\tlang_dict[year] = push!(lang_dict[year], lang)\n\t\telse\n\t\t\tlang_dict[year] = [lang]\n\t\tend\n\tend\nend\n\n# \u2554\u2550\u2561 c5aca233-9a26-4800-9b8d-7f0f03852626\nlang_dict\n\n# \u2554\u2550\u2561 7b2d5919-9629-4ca9-8195-f26183734044\nmd\"\"\"\n> **Note:** there is an `enumerate()` method in Julia which is useful when you need not only the values `x` over which you are iterating, but also the number of iterations (index) so far.\n**Syntax:**\n```julia\nfor (index, value) in enumerate(df.column)\n```\n\"\"\"\n\n# \u2554\u2550\u2561 d8054919-1554-4c73-a4a2-09383ee09224\nmd\"\"\"\nWe can check the size of the dictionary...\n\"\"\"\n\n# \u2554\u2550\u2561 7d57f6cc-ab44-4658-a73e-92c873111155\nlength(keys(lang_dict))\n\n# \u2554\u2550\u2561 a322a0e7-3984-4c00-95a5-7b23fc6ec06f\nmd\"\"\"\nOr we can check the size of the vector made up by unique `Year` values...\n\"\"\"\n\n# \u2554\u2550\u2561 e0a28251-a7d6-4c17-b49e-84ffc91c60b6\nlength(unique(data\u2082[:,1]))\n\n# \u2554\u2550\u2561 9073e71f-b922-44a4-a470-5cb081588fcd\nmd\"\"\"\n> **Q1:** Which year was was a given language invented?\n\"\"\"\n\n# \u2554\u2550\u2561 f9325e7b-8df8-448a-98b2-9ea8add93da6\nfunction year_created_dict(dict,language::String)\n keys_vec = collect(keys(dict))\n lookup = map(keyid -> findfirst(dict[keyid] .== language), keys_vec)\n \n return keys_vec[findfirst((!isnothing).(lookup))]\nend\n\n# \u2554\u2550\u2561 c33df74d-dd81-4efa-81d6-a5deb4685f1d\nbegin\n\tjulia_year3 = year_created_dict(lang_dict, \"Julia\")\n\n\t\"Julia was created in $julia_year3.\"\nend\n\n# \u2554\u2550\u2561 a08ad2fd-5d3c-4e76-a059-0608de3e36aa\nmd\"\"\"\n> **Q2:** How many languages were created in a given year?\n\"\"\"\n\n# \u2554\u2550\u2561 fb6df54d-0765-4493-b889-17707ff0cf14\nlangs_per_year_dict(dict, year::Int64) = length(dict[year])\n\n# \u2554\u2550\u2561 d5e60574-5afd-498c-aedd-1f5ffc4c6bd7\nbegin\n\tlangs_1988\u2083 = langs_per_year_dict(lang_dict, 1988)\n\n\t\"In 1988, $langs_1988\u2083 language(s) was/were created.\"\nend\n\n# \u2554\u2550\u2561 ee8bcf97-9efa-47f0-a82e-cfaa2efd68e6\nmd\"\"\"\n## Missing data\n\nLet's remove the first year value from our data (Matrix) and, after that, create a DataFrame...\n\"\"\"\n\n# \u2554\u2550\u2561 b3927e97-7516-4a80-bcc1-49614cf67166\nbegin\n\tdata\u2082[1,1] = missing\n\n\tdata\u2086 = DataFrame(Year=data\u2082[:,1], Language=data\u2082[:,2])\nend\n\n# \u2554\u2550\u2561 dc7ed277-b044-464e-8ad3-998a83b00d1b\nmd\"\"\"\nWe can now use the `dropmissing!` function to remove (inplace) the line with missing value...\n\"\"\"\n\n# \u2554\u2550\u2561 e2b6b920-2e05-4168-af16-07e9611d895d\ndropmissing!(data\u2086)\n\n# \u2554\u2550\u2561 Cell order:\n# \u255f\u2500c730d000-5ed6-11ec-36b6-39741ccad6c7\n# \u255f\u250062c5d4f2-9776-43ae-8125-0d429d6cde42\n# \u255f\u250034f92fb1-6c1e-4db6-b795-8a526007ea92\n# \u255f\u250013a10dac-8c7e-4e72-9fa9-d58ca8dd0202\n# \u255f\u25005100df3f-c5d3-4346-bcd9-8afc92bc559c\n# \u2560\u2550abe62085-b630-4816-af11-b39133892849\n# \u255f\u250051c909b1-1d37-458f-acd6-68f4041bc92f\n# \u255f\u25006483858c-2e58-4930-b4e9-76b47032eac4\n# \u2560\u255089a5a316-5058-4ff2-8d71-fc832a84c58d\n# \u2560\u2550853a4e90-ac39-4d73-9953-c7b70e8f78ef\n# \u255f\u25003a22c018-2ca7-41df-9ee4-91dc8b64d940\n# \u2560\u2550b4fcdcf9-793f-4d4a-a5fb-950108cb8188\n# \u255f\u2500cfa37c7c-810c-4095-8887-262dee4a9d04\n# \u2560\u25504b651b5a-e562-46ef-a3b5-8ee85b5a61ed\n# \u255f\u250011471bbf-b7b4-4b6e-90cb-9e9ff47bbe4a\n# \u2560\u25502652dc01-d80d-46d9-ae7a-9afa534e88cb\n# \u255f\u2500b2f35e6b-8c19-4729-87a3-292858bfbcb5\n# \u2560\u2550e1210609-a216-4d54-be63-04f10c5c15bc\n# \u2560\u2550008b635f-c4e5-475a-83af-4bc3fc722344\n# \u255f\u2500934c8549-c86c-45e6-a9f5-56297c73513d\n# \u2560\u255092708ece-3620-4cf5-b09d-098002044623\n# \u255f\u2500cac4f1b5-f9e3-469e-9576-553a1cf1b363\n# \u2560\u25503d378ec4-659f-4032-a5b7-6d6b6d402c2f\n# \u255f\u2500e50ae64c-ff64-4597-bf75-d7c921d23225\n# \u2560\u2550d9ea3574-2ffb-4fdb-9ced-db1dc669ecd9\n# \u2560\u255009ff6d8b-15f2-4d8e-a775-dee357a417e4\n# \u2560\u2550843a668f-7b08-4bce-b02a-3153d0eb7d0d\n# \u2560\u2550159a35f4-09fd-47bd-9935-88668a7e7f27\n# \u255f\u2500494ebede-80d0-430e-91aa-0a9050a1bb29\n# \u2560\u255000d12274-3057-4a52-8400-0c8c3c358e6b\n# \u255f\u2500fede3ce2-da03-4ff9-a13e-7ce55ec05bf6\n# \u2560\u255074783bbb-3c9d-4e15-bc54-b3d8e7c2d230\n# \u255f\u2500e4b3a444-c487-4fe8-8048-873b8a3affb7\n# \u2560\u25500becba29-d5ff-4595-86ab-6b0575674047\n# \u2560\u25509d522bdc-d066-40ac-84c4-47287d9e07e0\n# \u255f\u250079de9741-c913-4c00-8833-4f1b68f80ea5\n# \u2560\u2550fd724d83-41bb-422c-860e-2bbeff17e0dc\n# \u255f\u250051301eeb-709a-4ea9-a1a5-d7967f8dd55d\n# \u2560\u255064e7e90b-cd7d-4569-a715-4f27f1787e40\n# \u255f\u2500e5f2c69b-fbd8-4048-aaed-792a64a4d66c\n# \u2560\u255026047637-1b77-4203-b705-e34f74137295\n# \u2560\u25500f3d505c-a575-419f-8155-fd071f510714\n# \u255f\u2500c2f1eab2-862a-4eb8-b233-71567cdc9963\n# \u2560\u25503a924b3e-3dbe-49b6-9e5e-5a48b0ffd385\n# \u255f\u250058bad348-0586-4d0f-8d54-7c3e1a7a2c40\n# \u2560\u2550036de982-36ad-431a-8445-517178626911\n# \u255f\u25004ead9039-f4c2-4f82-bf1e-2146718014a3\n# \u2560\u2550bb04dc6c-e62e-4b2c-a247-ebe4836d8013\n# \u255f\u2500d9550872-fa7c-4ebd-9a4d-828182c8e49d\n# \u2560\u2550e01a65ab-394a-4718-b9ae-ca2bb30e20b5\n# \u255f\u25001eb26b3c-8a5a-4648-bb73-f8291b656edc\n# \u2560\u2550cb027055-2cf6-4f74-b265-9d01c23c2967\n# \u255f\u250012dccfc3-e976-46e6-81b0-dc51b48309f4\n# \u2560\u2550403ac783-9c02-4fd1-8506-afe0af36a14b\n# \u255f\u250036d8898e-980c-4f54-bbff-277ded2229ea\n# \u2560\u25508d769d53-1902-4a09-8228-ce60174b22bb\n# \u255f\u2500bc29d2b5-e6c2-4b45-8b79-cbb012659be1\n# \u255f\u25007c2fef9c-4e9a-4637-bc8e-6636af5f98ae\n# \u2560\u25508f51aeb5-3e75-4e5a-8c8f-ed0201eb8ce0\n# \u2560\u2550879ac394-8598-4399-957f-1280ecd99478\n# \u2560\u25500d27e3b3-79e3-424d-bc6b-94252900f7fb\n# \u2560\u255013053d54-8680-41e2-b416-8a7bc50f6c92\n# \u255f\u2500e0f49c77-0c0d-4740-9227-5619d14748b6\n# \u2560\u2550f4ddf1bf-0cbf-47c5-bb5f-f8abf0fb8ac2\n# \u2560\u2550a979107e-f2cf-42da-aefa-5cb249bc7073\n# \u2560\u2550edd2093e-f84a-4563-acb7-219f2bd07399\n# \u255f\u2500be91882d-fe73-40d0-8e75-30c2e8ddd564\n# \u2560\u25501f3092e8-3f3a-4d29-a9f5-e64ef96c13a8\n# \u2560\u2550cf9a1b00-79dd-4305-93ad-69f229319ce2\n# \u2560\u25501b49419d-18cd-4617-9442-15080e3ef588\n# \u255f\u2500c3bce6db-d503-4a68-b83f-278c6d603f07\n# \u2560\u25507131ac1c-984e-4ad9-a1cd-b1d46a4f240e\n# \u255f\u25004f70b5df-0026-43cd-b769-d4ca50145fbd\n# \u2560\u25500c1201b9-6acb-4206-a806-e09e04c6a65e\n# \u2560\u2550cc555ffc-0eea-4e35-b0d1-36d3a734f9f2\n# \u2560\u25505516dd98-3a87-4801-a62e-e91a95e4923a\n# \u2560\u255053be8986-1f06-45a5-9303-5018a3ddf06f\n# \u255f\u2500ecefa934-49a8-4abf-880d-d91d3f9f9d05\n# \u255f\u250001be19ba-2aaa-4746-af5d-8ce7f1cf3a55\n# \u255f\u25003a6f9dc0-2186-440a-9d78-5da6acabcdae\n# \u2560\u25504f9dca90-9f1a-4c0e-a510-fa640d109b0d\n# \u255f\u250057b8265f-be5c-4f05-91c6-6c4fec4f0a6b\n# \u2560\u2550157e9e3d-2e10-418e-997e-0178c14d7b3d\n# \u255f\u2500e7bd695e-c7d6-4b38-8520-7d915e740015\n# \u2560\u2550c9ead4e0-aa01-43bc-8107-72d8ec4c073b\n# \u2560\u255092b4a2d9-b271-42b3-97d1-647300a9439a\n# \u255f\u250001a73335-69b9-4297-9910-a441848fe2c5\n# \u2560\u2550fbf69873-af41-428c-9bba-d63cdb20149b\n# \u2560\u25501ea19d7e-5748-4559-b2d2-fd4f4d449516\n# \u2560\u25502592f7cd-95ec-493c-93e1-bbedf433367f\n# \u255f\u250012121f45-8a7b-4709-bc85-b2fde7313a21\n# \u2560\u2550d455f80e-c5b5-4b1a-9667-86fade5ee3d3\n# \u2560\u255042c72fde-8331-4e40-b761-c50d0e2e8486\n# \u2560\u2550201a8534-91ee-4ac9-a120-51b29f196c23\n# \u255f\u25000b14e9fc-62e6-4ad7-ab67-eb36e340749c\n# \u2560\u2550679b2b83-07f9-40bd-bdd1-89e7233bfade\n# \u255f\u2500f4738476-8b05-4dbc-a15e-6b3d59c90c50\n# \u2560\u2550c6b170be-6576-480e-b2ba-65d3b853871a\n# \u2560\u25509977fdaa-db30-43a5-bd58-14087c2163e4\n# \u255f\u2500a137ebd7-9db4-4c29-a694-b1acad5214a3\n# \u2560\u25502cc2938b-58d5-4c27-bdab-dddf22231bc8\n# \u2560\u2550ae7a1714-2245-4d5a-89bb-10b3adff787a\n# \u2560\u2550ce4ac86d-5e06-4c3e-93d6-ebdee46439bd\n# \u255f\u2500515b7479-97d8-464b-8245-1034f3cd8bdc\n# \u2560\u25503b08a2c8-546b-4a6c-8ae0-2a1e64578af8\n# \u2560\u255021551a38-57fc-42c2-81a2-b314bb06250b\n# \u2560\u25506da1f5a7-56a2-4a7a-abfc-25ec3f44f892\n# \u255f\u250043ad8297-d94c-45a0-941f-ab0d5bfd2ddb\n# \u2560\u25509390d98f-94eb-48bb-ba9c-e6b3f67f762f\n# \u2560\u255030718df7-5085-4d0d-8a86-7b4a08ff20b2\n# \u2560\u2550f77a5189-19b4-4f99-b394-72190253912f\n# \u2560\u255050ba8f3b-5104-4d4f-85ea-831d4afe9164\n# \u255f\u2500bb814af7-62e3-4744-955f-31b557540057\n# \u2560\u25509193df28-7ed1-4853-a97b-aa5a1232e07b\n# \u2560\u2550c5aca233-9a26-4800-9b8d-7f0f03852626\n# \u255f\u25007b2d5919-9629-4ca9-8195-f26183734044\n# \u255f\u2500d8054919-1554-4c73-a4a2-09383ee09224\n# \u2560\u25507d57f6cc-ab44-4658-a73e-92c873111155\n# \u255f\u2500a322a0e7-3984-4c00-95a5-7b23fc6ec06f\n# \u2560\u2550e0a28251-a7d6-4c17-b49e-84ffc91c60b6\n# \u255f\u25009073e71f-b922-44a4-a470-5cb081588fcd\n# \u2560\u2550f9325e7b-8df8-448a-98b2-9ea8add93da6\n# \u2560\u2550c33df74d-dd81-4efa-81d6-a5deb4685f1d\n# \u255f\u2500a08ad2fd-5d3c-4e76-a059-0608de3e36aa\n# \u2560\u2550fb6df54d-0765-4493-b889-17707ff0cf14\n# \u2560\u2550d5e60574-5afd-498c-aedd-1f5ffc4c6bd7\n# \u255f\u2500ee8bcf97-9efa-47f0-a82e-cfaa2efd68e6\n# \u2560\u2550b3927e97-7516-4a80-bcc1-49614cf67166\n# \u255f\u2500dc7ed277-b044-464e-8ad3-998a83b00d1b\n# \u2560\u2550e2b6b920-2e05-4168-af16-07e9611d895d\n", "meta": {"hexsha": "c46a5ac323833890ed7e7d1aaf24c7030bd7d04e", "size": 24674, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "01_Data.jl", "max_stars_repo_name": "fnaghetini/DataScience", "max_stars_repo_head_hexsha": "5cd1c2a9c72aacc8a67800c74e394fb0e2045f2d", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "01_Data.jl", "max_issues_repo_name": "fnaghetini/DataScience", "max_issues_repo_head_hexsha": "5cd1c2a9c72aacc8a67800c74e394fb0e2045f2d", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "01_Data.jl", "max_forks_repo_name": "fnaghetini/DataScience", "max_forks_repo_head_hexsha": "5cd1c2a9c72aacc8a67800c74e394fb0e2045f2d", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 29.4439140811, "max_line_length": 323, "alphanum_fraction": 0.731944557, "num_tokens": 11321, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.32423541204073586, "lm_q2_score": 0.22000708951749934, "lm_q1q2_score": 0.07133408932158945}}
{"text": "module MINLPTests\n\nusing JuMP\nusing Test\n\n###\n### Default tolerances that are used in the tests.\n###\n\n# Absolute tolerance when checking the objective value.\nconst OPT_TOL = 1e-6\n\n# Absolute tolerance when checking the primal solution value.\nconst PRIMAL_TOL = 1e-6\n\n# Absolue tolerance when checking the dual solution value.\nconst DUAL_TOL = 1e-6\n\n###\n### Default expected status codes for different types of problems and solvers.\n###\n\n# We only distinguish between feasible and infeasible problems now.\n@enum ProblemTypeCode FEASIBLE_PROBLEM INFEASIBLE_PROBLEM\n\n# Target status codes for local solvers:\nconst TERMINATION_TARGET_LOCAL = Dict(\n FEASIBLE_PROBLEM => JuMP.MOI.LOCALLY_SOLVED,\n INFEASIBLE_PROBLEM => JuMP.MOI.LOCALLY_INFEASIBLE,\n)\nconst PRIMAL_TARGET_LOCAL = Dict(\n FEASIBLE_PROBLEM => JuMP.MOI.FEASIBLE_POINT,\n INFEASIBLE_PROBLEM => JuMP.MOI.INFEASIBLE_POINT,\n)\n\n# Target status codes for global solvers:\nconst TERMINATION_TARGET_GLOBAL = Dict(\n FEASIBLE_PROBLEM => JuMP.MOI.OPTIMAL,\n INFEASIBLE_PROBLEM => JuMP.MOI.INFEASIBLE,\n)\nconst PRIMAL_TARGET_GLOBAL = Dict(\n FEASIBLE_PROBLEM => JuMP.MOI.FEASIBLE_POINT,\n INFEASIBLE_PROBLEM => JuMP.MOI.NO_SOLUTION,\n)\n\n###\n### Helper functions for the tests.\n###\n\nfunction check_status(\n model,\n problem_type::ProblemTypeCode,\n termination_target = TERMINATION_TARGET_LOCAL,\n primal_target = PRIMAL_TARGET_LOCAL,\n)\n @test JuMP.termination_status(model) == termination_target[problem_type]\n @test JuMP.primal_status(model) == primal_target[problem_type]\nend\n\nfunction check_objective(model, solution; tol = OPT_TOL)\n if !isnan(tol)\n @test isapprox(JuMP.objective_value(model), solution, atol = tol)\n end\nend\n\nfunction check_solution(variables, solutions; tol = PRIMAL_TOL)\n if !isnan(tol)\n @assert length(variables) == length(solutions)\n for (variable, solution) in zip(variables, solutions)\n @test isapprox(JuMP.value(variable), solution, atol = tol)\n end\n end\nend\n\nfunction check_dual(constraints, solutions; tol = DUAL_TOL)\n if !isnan(tol)\n @assert length(constraints) == length(solutions)\n for (constraint, solution) in zip(constraints, solutions)\n @test isapprox(JuMP.dual(constraint), solution, atol = tol)\n end\n end\nend\n\n###\n### Loop through and include every model function.\n###\n\nfor directory in [\"nlp\", \"nlp-cvx\", \"nlp-mi\"]\n files = readdir(joinpath(@__DIR__, directory))\n for file_name in filter(f -> endswith(f, \".jl\"), files)\n include(joinpath(@__DIR__, directory, file_name))\n end\nend\n\n\"\"\"\n test_directory(\n directory,\n optimizer;\n debug::Bool = false,\n exclude = String[],\n include = String[],\n objective_tol = OPT_TOL,\n primal_tol = PRIMAL_TOL,\n dual_tol = DUAL_TOL,\n termination_target = TERMINATION_TARGET_LOCAL,\n primal_target = PRIMAL_TARGET_LOCAL,\n )\n\nTest all of the files in `directory` using `optimizer`.\n\nIf `debug`, print the name of the file befor running it.\n\nUse `exclude` and `include` to run a subset of the files in a directory.\n\nUse the remaining args to control tolerances and status targets.\n\n## Example\n\nTest all but nlp_001_010:\n```julia\ntest_directory(\"nlp\", optimizer; exclude = [\"001_010\"])\n```\n\nTest only nlp_001_010:\n```julia\ntest_directory(\"nlp\", optimizer; include = [\"001_010\"])\n```\n\"\"\"\nfunction test_directory(\n directory,\n optimizer;\n debug::Bool = false,\n exclude = String[],\n include = String[],\n objective_tol = OPT_TOL,\n primal_tol = PRIMAL_TOL,\n dual_tol = DUAL_TOL,\n termination_target = TERMINATION_TARGET_LOCAL,\n primal_target = PRIMAL_TARGET_LOCAL,\n)\n @testset \"$(directory)\" begin\n models = _list_of_models(directory, exclude, include)\n @testset \"$(model_name)\" for model_name in models\n if debug\n println(\"Running $(model_name)\")\n end\n getfield(MINLPTests, model_name)(\n optimizer,\n objective_tol,\n primal_tol,\n dual_tol,\n termination_target,\n primal_target,\n )\n end\n end\nend\n\nfunction _list_of_models(\n directory,\n exclude::Vector{String},\n include::Vector{String},\n)\n dir = replace(directory, \"-\" => \"_\")\n if length(include) > 0\n return [Symbol(\"$(dir)_$(i)\") for i in include]\n else\n models = Symbol[]\n for file in readdir(joinpath(@__DIR__, directory))\n if !endswith(file, \".jl\")\n continue\n end\n file = replace(file, \".jl\" => \"\")\n if file in exclude\n continue\n end\n push!(models, Symbol(\"$(dir)_$(file)\"))\n end\n return models\n end\nend\n\n###\n### Helper functions to test a subset of models.\n###\n\nfunction test_nlp(\n optimizer;\n debug::Bool = false,\n exclude = String[],\n objective_tol = OPT_TOL,\n primal_tol = PRIMAL_TOL,\n dual_tol = DUAL_TOL,\n termination_target = TERMINATION_TARGET_LOCAL,\n primal_target = PRIMAL_TARGET_LOCAL,\n)\n return test_directory(\n \"nlp\",\n optimizer;\n debug = debug,\n exclude = exclude,\n objective_tol = objective_tol,\n primal_tol = primal_tol,\n dual_tol = dual_tol,\n termination_target = termination_target,\n primal_target = primal_target,\n )\nend\n\nfunction test_nlp_cvx(\n optimizer;\n debug::Bool = false,\n exclude = String[],\n objective_tol = OPT_TOL,\n primal_tol = PRIMAL_TOL,\n dual_tol = DUAL_TOL,\n termination_target = TERMINATION_TARGET_LOCAL,\n primal_target = PRIMAL_TARGET_LOCAL,\n)\n return test_directory(\n \"nlp-cvx\",\n optimizer;\n debug = debug,\n exclude = exclude,\n objective_tol = objective_tol,\n primal_tol = primal_tol,\n dual_tol = dual_tol,\n termination_target = termination_target,\n primal_target = primal_target,\n )\nend\n\nfunction test_nlp_mi(\n optimizer;\n debug::Bool = false,\n exclude = String[],\n objective_tol = OPT_TOL,\n primal_tol = PRIMAL_TOL,\n dual_tol = DUAL_TOL,\n termination_target = TERMINATION_TARGET_LOCAL,\n primal_target = PRIMAL_TARGET_LOCAL,\n)\n return test_directory(\n \"nlp-mi\",\n optimizer;\n debug = debug,\n exclude = exclude,\n objective_tol = objective_tol,\n primal_tol = primal_tol,\n dual_tol = dual_tol,\n termination_target = termination_target,\n primal_target = primal_target,\n )\nend\n\n### Tests that haven't been updated.\n\ninclude(\"nlp-mi-cvx/tests.jl\")\ninclude(\"poly/tests.jl\")\ninclude(\"poly-cvx/tests.jl\")\ninclude(\"poly-mi/tests.jl\")\ninclude(\"poly-mi-cvx/tests.jl\")\n\nend\n", "meta": {"hexsha": "f74d5b33bbb3f7745f0f41c3edfe083921144872", "size": 6805, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/MINLPTests.jl", "max_stars_repo_name": "jump-dev/MINLPTests.jl", "max_stars_repo_head_hexsha": "9dc2b751de7470dcb4d9a726e11339a8e0bf745c", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 3, "max_stars_repo_stars_event_min_datetime": "2020-07-04T22:02:43.000Z", "max_stars_repo_stars_event_max_datetime": "2021-04-16T15:49:34.000Z", "max_issues_repo_path": "src/MINLPTests.jl", "max_issues_repo_name": "jump-dev/MINLPTests.jl", "max_issues_repo_head_hexsha": "9dc2b751de7470dcb4d9a726e11339a8e0bf745c", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 14, "max_issues_repo_issues_event_min_datetime": "2020-06-14T16:44:08.000Z", "max_issues_repo_issues_event_max_datetime": "2022-03-27T01:32:27.000Z", "max_forks_repo_path": "src/MINLPTests.jl", "max_forks_repo_name": "jump-dev/MINLPTests.jl", "max_forks_repo_head_hexsha": "9dc2b751de7470dcb4d9a726e11339a8e0bf745c", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 2, "max_forks_repo_forks_event_min_datetime": "2020-07-24T16:22:09.000Z", "max_forks_repo_forks_event_max_datetime": "2020-07-25T02:23:53.000Z", "avg_line_length": 25.679245283, "max_line_length": 78, "alphanum_fraction": 0.6537839824, "num_tokens": 1640, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.5, "lm_q2_score": 0.14223189864583877, "lm_q1q2_score": 0.07111594932291938}}
{"text": "\"\"\"\r\n# Get the categories for an economic data series in the FRED database.\r\n\r\n## Arguments\r\n\r\n - `symbol` : String specifying the name (id) of the time series.\r\n\r\n## Examples\r\n\r\n```jldoctests\r\njulia> get_category(\"GDPC1\")\r\njulia> get_category(\"FEDFUNDS\")\r\njulia> get_category(\"T10Y2Y\")\r\n```\r\n\"\"\"\r\nfunction get_category(symbol::String)\r\n\r\n url = \"https://api.stlouisfed.org/fred/series/categories\"\r\n parameters = Dict(\"api_key\" => ENV[\"API_FRED\"],\r\n \"file_type\" => \"json\",\r\n \"series_id\" => symbol)\r\n\r\n response = HTTP.request(\"GET\", url; query = parameters)\r\n body = JSON.parse(String(response.body))[\"categories\"][1][\"name\"]\r\n\r\n return(body)\r\nend\r\n", "meta": {"hexsha": "250b9bf60c0b1327313caec24cc085e731c16a88", "size": 709, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/get_category.jl", "max_stars_repo_name": "JuliaTagBot/FredApi.jl", "max_stars_repo_head_hexsha": "6e997585368bb316fe505b5c7ad55c52c6cbb22f", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 6, "max_stars_repo_stars_event_min_datetime": "2020-02-15T19:18:40.000Z", "max_stars_repo_stars_event_max_datetime": "2021-12-16T07:37:12.000Z", "max_issues_repo_path": "src/get_category.jl", "max_issues_repo_name": "JuliaTagBot/FredApi.jl", "max_issues_repo_head_hexsha": "6e997585368bb316fe505b5c7ad55c52c6cbb22f", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 5, "max_issues_repo_issues_event_min_datetime": "2020-08-25T21:43:03.000Z", "max_issues_repo_issues_event_max_datetime": "2021-12-31T03:54:17.000Z", "max_forks_repo_path": "src/get_category.jl", "max_forks_repo_name": "JuliaTagBot/FredApi.jl", "max_forks_repo_head_hexsha": "6e997585368bb316fe505b5c7ad55c52c6cbb22f", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 7, "max_forks_repo_forks_event_min_datetime": "2020-02-08T10:43:07.000Z", "max_forks_repo_forks_event_max_datetime": "2021-12-07T10:14:34.000Z", "avg_line_length": 25.3214285714, "max_line_length": 71, "alphanum_fraction": 0.6121297602, "num_tokens": 176, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.44939263446475963, "lm_q2_score": 0.1581743527484317, "lm_q1q2_score": 0.07108238908637592}}
{"text": "const ColKey = Union{Symbol,AbstractString}\n\n\"\"\"\n coerce(A, specs...; tight=false, verbosity=1)\n\nGiven a table `A`, return a copy of `A`, ensuring that the element\nscitypes of the columns match the new specification, `specs`. There\nare three valid specifiations:\n\n(i) one or more `column_name=>Scitype` pairs:\n\n coerce(X, col1=>Sciyype1, col2=>Scitype2, ... ; verbosity=1)\n\n(ii) one or more `OldScitype=>NewScitype` pairs (`OldScitype` covering\nboth the `OldScitype` and `Union{Missing,OldScitype}` cases):\n\n coerce(X, OldScitype1=>NewSciyype1, OldScitype2=>NewScitype2, ... ; verbosity=1)\n\n(iii) a dictionary of scientific types keyed on column names:\n\n coerce(X, d::AbstractDict{<:ColKey, <:Type}; verbosity=1)\n\nwhere `ColKey = Union{Symbol,AbstractString}`.\n\n### Examples\n\nSpecifiying `column_name=>Scitype` pairs:\n\n```\nusing CategoricalArrays, DataFrames, Tables\nX = DataFrame(name=[\"Siri\", \"Robo\", \"Alexa\", \"Cortana\"],\n height=[152, missing, 148, 163],\n rating=[1, 5, 2, 1])\nXc = coerce(X, :name=>Multiclass, :height=>Continuous, :rating=>OrderedFactor)\nschema(Xc).scitypes # (Multiclass, Continuous, OrderedFactor)\n```\n\nSpecifying `OldScitype=>NewScitype` pairs:\n\n```\nX = (x = [1, 2, 3],\n y = rand(3),\n z = [10, 20, 30])\nXc = coerce(X, Count=>Continuous)\nschema(Xfixed).scitypes # (Continuous, Continuous, Continuous)\n```\n\"\"\"\ncoerce(X, a...; kw...) = coerce(Val(ST.trait(X)), X, a...; kw...)\n\n# Non tabular data is not supported\ncoerce(::Val{:other}, X, a...; kw...) =\n throw(CoercionError(\"`coerce` is undefined for non-tabular data.\"))\n\n\n_bad_dictionary() = throw(ArgumentError(\n \"A dictionary specifying a scitype conversion \"*\n \"must have type `AbstractDict{<:ColKey, <:Type}`. It's keys must \"*\n \"be column names and its values be scientific types. \"*\n \"E.g., `Dict(:cats=>Continuous, :dogs=>Textual`. \"))\ncoerce(::Val{:table}, X, types_dict::AbstractDict; kw...) =\n _bad_dictionary()\n\n_bad_specs() =\n throw(ArgumentError(\n \"Invalid `specs` in `coerce(X, specs...; kwargs...)`. \"*\n \"Valid `specs` are: (i) one or more pairs of \"*\n \"the form `column_name=>Scitype`; (ii) one or more pairs \"*\n \"of the from `OldScitype=>NewScitype`; or (iii) a \"*\n \"dictionary of scientific \"*\n \"types keyed on column names. \"))\ncoerce(::Val{:table}, X, specs...; kw...) = _bad_specs()\n\nfunction coerce(::Val{:table},\n X,\n types_dict::AbstractDict{<:ColKey, <:Type};\n kw...)\n isempty(types_dict) && return X\n names = schema(X).names\n X_ct = Tables.columntable(X)\n ct_new = (_coerce_col(X_ct, col, types_dict; kw...) for col in names)\n return Tables.materializer(X)(NamedTuple{names}(ct_new))\nend\n\n# -------------------------------------------------------------\n# utilities for coerce\n\nstruct CoercionError <: Exception\n m::String\nend\n\nfunction _coerce_col(Xcol,\n name,\n types_dict::AbstractDict{Symbol, <:Type};\n kw...)\n y = Tables.getcolumn(Xcol, name)\n if haskey(types_dict, name)\n coerce_type = types_dict[name]\n return coerce(y, coerce_type; kw...)\n end\n return y\nend\n\n# -------------------------------------------------------------\n# alternative ways to do coercion, both for coerce and coerce!\n\n# The following extends the two methods so that a mixture of\n# symbol=>type and type=>type pairs can be specified in place of a\n# dictionary:\n\nfeature_scitype_pairs(p::Pair{<:ColKey,<:Type}, X) = [Symbol(first(p)) => last(p), ]\nfunction feature_scitype_pairs(p::Pair{<:Type,<:Type}, X)\n from_scitype = first(p)\n to_scitype = last(p)\n sch = schema(X)\n ret = Pair{Symbol,Type}[]\n for j in eachindex(sch.names)\n if sch.scitypes[j] <: Union{Missing,from_scitype}\n push!(ret, Pair(sch.names[j], to_scitype))\n end\n end\n return ret\nend\n\nfor c in (:coerce, :coerce!)\n ex = quote\n function $c(::Val{:table},\n X,\n mixed_pairs::Pair{<:Union{<:ColKey,<:Type},<:Type}...;\n kw...)\n components = map(p -> feature_scitype_pairs(p, X), mixed_pairs)\n pairs = vcat(components...)\n\n # must construct dictionary by hand to check no conflicts:\n scitype_given_feature = Dict{Symbol,Type}()\n for p in pairs\n feature = first(p)\n if haskey(scitype_given_feature, feature)\n throw(ArgumentError(\"`coerce` argments cannot be \"*\n \"resolved to determined a \"*\n \"*unique* scitype for each \"*\n \"feature. \"))\n else\n scitype_given_feature[feature] = last(p)\n end\n end\n\n return $c(X, scitype_given_feature; kw...)\n end\n end\n eval(ex)\nend\n\n# -------------------------------------------------------------\n# In place coercion\n\n\"\"\"\ncoerce!(X, ...)\n\nSame as [`ScientificTypes.coerce`](@ref) except it does the modification in\nplace provided `X` supports in-place modification (at the moment, only the\nDataFrame! does). An error is thrown otherwise. The arguments are the same as\n`coerce`.\n\n\"\"\"\ncoerce!(X, a...; kw...) = begin\n coerce!(Val(ST.trait(X)), X, a...; kw...)\nend\n\ncoerce!(::Val{:other}, X, a...; kw...) =\n throw(CoercionError(\"`coerce!` is undefined for non-tabular data.\"))\n\ncoerce!(::Val{:table}, X, types_dict::AbstractDict; kw...) =\n _bad_dictionary()\n\ncoerce!(::Val{:table}, X, specs...; kw...) = _bad_specs()\n\nfunction coerce!(::Val{:table},\n X,\n types_dict::AbstractDict{<:ColKey, <:Type};\n kw...)\n # DataFrame --> coerce_df!\n if is_type(X, :DataFrames, :DataFrame)\n return coerce_df!(X, types_dict; kw...)\n end\n # Everything else\n throw(ArgumentError(\"In place coercion not supported for $(typeof(X)).\" *\n \"Try `coerce` instead.\"))\nend\n\n# -------------------------------------------------------------\n# utilities for coerce!\n\n\"\"\"\n coerce_df!(df, pairs...; kw...)\n\nIn place coercion for a dataframe.(Unexported method)\n\"\"\"\nfunction coerce_df!(df, tdict::AbstractDict{<:ColKey, <:Type}; kw...)\n names = schema(df).names\n for name in names\n name in keys(tdict) || continue\n coerce_type = tdict[name]\n df[!, name] = coerce(df[!, name], coerce_type; kw...)\n end\n return df\nend\n\n\n\"\"\"\n is_type(X, spkg, stype)\n\nCheck that an object `X` is of a given type that may be defined in a package\nthat is not loaded in the current environment.\nAs an example say `DataFrames` is not loaded in the current environment, a\nfunction from some package could still return a DataFrame in which case it\ncan be checked with\n\n```\nis_type(X, :DataFrames, :DataFrame)\n```\n\"\"\"\nfunction is_type(X, spkg::Symbol, stype::Symbol)\n # If the package is loaded, then it will just be `stype`\n # otherwise it will be `spkg.stype`\n rx = Regex(\"^($spkg\\\\.)?$stype\")\n return ifelse(match(rx, \"$(typeof(X))\") === nothing, false, true)\nend\n", "meta": {"hexsha": "00935d6ae3ec02fc3ade14d9f04904036bc08eee", "size": 7208, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/coerce.jl", "max_stars_repo_name": "alan-turing-institute/ScientificTypes.jl", "max_stars_repo_head_hexsha": "cecd1eb3bc0c22c25e7777073ce1347cf20871ab", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 43, "max_stars_repo_stars_event_min_datetime": "2019-08-02T05:52:08.000Z", "max_stars_repo_stars_event_max_datetime": "2021-05-18T08:27:13.000Z", "max_issues_repo_path": "src/coerce.jl", "max_issues_repo_name": "alan-turing-institute/ScientificTypes.jl", "max_issues_repo_head_hexsha": "cecd1eb3bc0c22c25e7777073ce1347cf20871ab", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 110, "max_issues_repo_issues_event_min_datetime": "2019-08-01T21:50:19.000Z", "max_issues_repo_issues_event_max_datetime": "2021-06-12T13:05:19.000Z", "max_forks_repo_path": "src/coerce.jl", "max_forks_repo_name": "alan-turing-institute/ScientificTypes.jl", "max_forks_repo_head_hexsha": "cecd1eb3bc0c22c25e7777073ce1347cf20871ab", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 9, "max_forks_repo_forks_event_min_datetime": "2020-01-01T23:25:02.000Z", "max_forks_repo_forks_event_max_datetime": "2020-11-29T01:39:33.000Z", "avg_line_length": 31.6140350877, "max_line_length": 84, "alphanum_fraction": 0.5828246393, "num_tokens": 1900, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.41489884579676883, "lm_q2_score": 0.17106119379155804, "lm_q1q2_score": 0.07097309186473483}}
{"text": "# # Stuff I Like About The Julia Programming Language\n\n# `git_club 2020-06-19` - Hannes\n\n# [View this notebook online](https://nbviewer.jupyter.org/github/Hasnep/stuff-i-like-about-julia/blob/master/stuff-i-like-about-julia.ipynb)\n\n# > Julia's unofficial tagline is \"Looks like Python, feels like Lisp, runs like Fortran.\"\n# > I've been learning Julia for slightly less than a year now, and I'd like to share some of the things I've enjoyed about it.\n# > I'll give an overview of the language's main features with code examples and discuss whether it really is the future scientific computing.\n\n# Technically:\n# > Julia is a high level, JIT compiled, dynamic language designed with multiple dispatch, automatic differentiation and metaprogramming.\n\n# In practice:\n# > Julia finds a balance between being fast and easy to use with lots of features you'll miss when you use another language.\n\n# ## History\n\n# - Julia was started in 2009 by Jeff Bezanson, Stefan Karpinski, Viral B. Shah, and Alan Edelman\n# - Announced in 2012\n# - Version 1.0 was released in 2018\n# - Now on version 1.4, heading for 1.5 soon\n\n# ## Julia is easy to use\n\n# Let's write a function from scratch to sum an array of numbers, first in Python:\n# ```python\n# def my_sum(array):\n# \"\"\"\n# Sum a list.\n# \"\"\"\n# total = 0\n# for x in array:\n# total += x\n# return total\n# ```\n\n# Then in Julia:\n\n\"\"\"\nSum an array.\n\"\"\"\nfunction my_sum(array)\n total = 0\n for x in array\n total += x\n end\n return total\nend\n\n# This syntax will be familliar if you've used Python.\n \nmy_sum(1:10)\n\n# Benchmarks for this function in different languages:\n# - C: ~10ms\n# - Python: ~500ms\n# - Julia: ~10ms\n# > Source: [An Introduction to Julia (Beginner Level) | SciPy 2018 Tutorial | Jane Herriman, Sacha Verweij](https://www.youtube.com/watch?v=b5xvVyzUnXI)\n\n# ## It's fast\n\n# Julia was designed to solve the \"two language problem\", where researchers use a slower, high-level language for research and then have to use a slower, low-level language once they hit a bottleneck or for production.\n# Julia is both languages at the same time!\n\n# Some benchmarks (from the Julia website) of different languages relative to C.\n# \n# > Source: https://julialang.org/benchmarks/\n\n# These benchmarks try to compare implementations of the same algorithm.\n# Julia is comparable to compiled languages like Rust, Go, Fortran, etc. and is sometimes faster than C.\n# Python is sometimes 100x slower than C.\n# R is a bit slower than that.\n\n# How is it so fast?\n\n# Generally languages fall into two categories:\n\n# - Compiled languages like C or Rust compile all the code before you run\n# - Interpreted languages like Python or R don't compile\n\n# Julia uses a _Just-In-Time_ (JIT) compiler which compiles a function the first time it is called.\n\n# In my opinion, speed doesn't matter all that much\n\n# - JIT compilation takes a short while\n# - My time is more valuable than the computer's time\n\n# Now, back to some code!\n\n# ## Dynamically typed\n\n# Let's write a function to calculate the $n$th Fibonacci number.\n# We want the input `n` to only allow integers and we want to specify the output should only be integers.\n\n\"\"\"\nCalculate the nth Fibonacci number.\n\"\"\"\nfunction fib(n::Integer)::Integer\n if n < 2\n return n\n else\n return fib(n - 1) + fib(n - 2)\n end\nend\n\n# If we try to run the function with a non-integer input\n\n# ```julia\n# julia> fib(0.5)\n# ```\n\n# Julia will throw an error:\n\n# ```\n# ERROR: MethodError: no method matching fib(::Float64)\n# Closest candidates are:\n# fib(::Integer)\n# ```\n\nfib(20)\n\n# There is also a one-line function notation that is useful for short functions\n\nshort_fib(n::Integer)::Integer = n < 2 ? n : short_fib(n - 1) + short_fib(n - 2)\n\nshort_fib(20)\n\n# ## It's free!\n\n# Julia is MIT licenced.\n\n# ## Broadcasting\n\n# In Python, most functions accept one element.\n# Running this code:\n# ```python\n# >>> import math\n# >>> math.sin([1, 2, 3])\n# ```\n# will give this error:\n# ```\n# TypeError: must be real number, not list\n# ```\n# Pythonistas would probably use a list comprehension:\n# ```python\n# >>> [math.sin(x) for x in [1, 2, 3]]\n# ```\n# or a map:\n# ```python\n# >>> map(math.sin, [1, 2, 3])\n# ```\n# ```\n# [0.8414709848078965, 0.9092974268256817, 0.1411200080598672]\n# ```\n\n# In R, most functions are vectorised\n# ```R\n# > sin(c(1, 2, 3))\n# ```\n# ```\n# [1] 0.8414710 0.9092974 0.1411200\n# ```\n\n# In Julia, functions are not vectorised, for example, this line:\n# ```julia\n# julia> sin([1, 2, 3])\n# ```\n# gives this error:\n# ```\n# ERROR: MethodError: no method matching sin(::Array{Int64,1})\n# ```\n\n# Using the broadcast operator, the funciton is applied elementwise!\n\nsin.([1, 2, 3])\n\n# Broadcasting even works for user functions\n \nfib.(1:10)\n\n# And for operators:\n\n## What are the first 10 square numbers?\n(1:10).^2\n\n# This is powerful, but sometimes tricky syntax.\n# For example, adding a dot makes the length function broadcast over the array:\n\nlength(split(\"How many words are in this sentence?\"))\n# +\nlength.(split(\"How many characters are each of these words?\"))\n\n# ## Useful Unicode characters\n\n# Julia lets you type lots of symbols using LaTeX-y abbreviations, e.g. type `\\alpha` and press tab for \u03b1 or `\\sqrt` and tab for \u221a.\n\n\u03b1 = 37\n\n# The square root symbol is an abbreviation for the `sqrt()` function:\n\n\u221a\u03b1\n\n# Some constants like \u03c0 for pi and \u212f for Euler's constant are predefined:\n\n## Use the approximate equals sign \u2248 because this calculation is not exact\n\u212f^(im * \u03c0) \u2248 -1\n \n# You can define your own operators using either built in functions:\n\nconst \u2282 = issubset\n[2, 5] \u2282 [1, 2, 3, 4, 5]\n\n# or user functions:\n\nconst \u2211 = my_sum\n\u2211(1:10)\n\n# Because Julia supports almost any symbol you can type as a variable name, that includes emojis!\n\n\ud83d\udd25 = 10\n\ud83d\udc36 = 20\n\ud83c\udf2d = 30\n\ud83d\udd25 + \ud83d\udc36 == \ud83c\udf2d\n\n# ## Julia is (mostly) written in Julia\n\n# If you can read Julia code, you can also read Julia's source code to understand what it does.\n# I looked at the most recent PR as an example:\n\ntensor(A::AbstractArray, B::AbstractArray) = [a * b for a in A, b in B]\nconst \u2297 = tensor\n#-\n[1, 2] \u2297 [3, 4, 5]\n\n# Python's numpy is fast because it's mostly written in C/C++ (51.4%), but if you want to do something that numpy can't do, you need to either use C++ or write slower Python code.\n\n# This bridges the gap between a Julia user and a Julia developer.\n# For every user of Julia, there's another possible contributer.\n\n# ## Multiple dispatch\n\n# As an example, let's build a small Julia package based on [Measurements.jl](https://github.com/JuliaPhysics/Measurements.jl/).\n\n\"\"\"\nA number with some error.\n\"\"\"\nstruct Uncertain <: Real\n val::Real\n err::Real\nend\n\n# Define the standard gravity on earth.\n\ng = Uncertain(9.8, 0.1)\n\n# Wouldn't it be nicer to show an uncertain number with the plus/minus symbol?\n# Let's write a show function that dispatches on the Uncertain type:\n\nBase.show(io::IO, x::Uncertain) = print(io, \"$(x.val) \u2213 $(x.err)\")\n\ng\n\n# Let's define the plus/minus operator to create `Uncertain` numbers:\n\n\u2213(a::Real, b::Real) = Uncertain(a, b)\nmy_height = 190 \u2213 1\n\n# How do you add two uncertain measurements?\n\nmy_brothers_height = 175 \u2213 2\n\n# ```julia\n# julia> my_height + my_brothers_height\n# ```\n# ```\n# ERROR: + not defined for Uncertain\n# ```\n\n# $$\n# Q = a + b \\\\\n# {\\delta Q} = \\sqrt{(\\delta a)^2 + (\\delta b)^2}\n# $$\n\nBase.:+(a::Uncertain, b::Uncertain) = (a.val + b.val) \u2213 sqrt(a.err^2 + b.err^2)\nmy_height + my_brothers_height\n\n# Similar for subtraction.\n\nBase.:-(a::Uncertain, b::Uncertain) = (a.val - b.val) \u2213 sqrt(a.err^2 + b.err^2)\nmy_height - my_brothers_height\n\n# Slightly more complicated for multiplcation and division.\n\n\nfunction Base.:*(a::Uncertain, b::Uncertain) \n total_value = a.val * b.val\n total_error = total_value * sqrt((a.err / a.val)^2 + (b.err / b.val)^2)\n return Uncertain(total_value, total_error)\nend\n\nfunction Base.:/(a::Uncertain, b::Uncertain) \n total_value = a.val / b.val\n total_error = total_value * sqrt((a.err / a.val)^2 + (b.err / b.val)^2)\n return Uncertain(total_value, total_error)\nend\n#-\nmy_height * my_brothers_height\n#-\nmy_height / my_brothers_height\n\n# Finally powers, again the exact formula is not important.\n\nBase.:^(a::Uncertain, b::Real) = (a.val^b) \u2213 (abs(b) * a.val^(b - 1) * a.err)\nmy_height^2.0\n\n# Finally, we need to tell Julia what to do if we give it an `Uncertain` number and some other number in the same operation.\n\n# ```julia\n# julia> 1 + g\n# ```\n# ```\n# ERROR: promotion of types Int64 and Uncertain failed to change any arguments\n# ```\n\n# We want to convert both numbers to our `Uncertain` type:\n\nBase.promote_rule(::Type{Uncertain}, ::Type{T}) where T <: Real = Uncertain\n\n# Coverting a real number to our Uncertain type just means we add an error of 0: \n\nBase.convert(::Type{Uncertain}, x::Real) = Uncertain(x, 0)\n\n# When Julia wants to convert an Uncertain number it doesn't need to do anything :\n\nBase.convert(::Type{Uncertain}, x::Uncertain) = x\n#-\n1 + g\n\n# Now we can solve a simple problem, how much time would it take if I dropped a ball from my height and my brother caught it at his height?\n\n# Solve for t:\n# $$\n# t = \\frac{\\sqrt{2 a s + u^2} - u}{a} = \\frac{\\sqrt{2 g (h_1 - h_2)}}{g}\n# $$\n\nt = ((2 * g * (my_height - my_brothers_height))^0.5) / g\n\n# Let's use the actual Measurements.jl package\n\nusing Measurements: Measurement, \u00b1\n\n# Let's solve the same problem as before and make sure we get the same result:\n\ng = 9.8 \u00b1 0.1\nmy_height = 190 \u00b1 1\nmy_brothers_height = 175 \u00b1 2\n\nt = (2 * g * (my_height - my_brothers_height))^0.5 / g\n\n# This next part is inspired by a JuliaCon talk called [_The Unreasonable Effectiveness of Multiple Dispatch_](https://www.youtube.com/watch?v=kc9HwsxE1OY) by one of the Julia co-founders, Stefan Karpinski.\n\n# All the \"methods\" are outside the class definition, that can include being in a completely different package.\n\n# Let's solve the same problem using differential equations!\n\n# Calculate the position of the ball between time $t = 0$ and $t = 3$\n# $$\n# v = \\frac{ds}{dt}\n# $$\n# $$\n# f(t) = -g t = - (9.8 \\pm 0.1) t\n# $$\n# With initial conditions $s_0 = 190 \\pm 1$\n\nusing DifferentialEquations\n#-\nf(s, p, t) = -g * t\ns\u2080 = my_height\ntime_span = (0.0, 3.0)\nproblem = ODEProblem(f, s\u2080, time_span)\n#-\nsolution = solve(problem, Tsit5(), saveat=0.1)\n#-\nusing Plots\n#-\nplot(\n solution.t,\n solution.u,\n title=\"Solution to the ODE\",\n xaxis=\"Time (t) in seconds\",\n yaxis=\"Displacement s(t) in metres\"\n)\n\n# ## Interoperability\n\n# We can import any python module using the PyCall package:\n\nusing PyCall\n#-\nmath = pyimport(\"math\")\n#-\nmath.sqrt(100)\n\n# We can mix Python functions and variables with Julia code:\n\nmath.sin.([\u03c0, math.pi, 2\u03c0, 2 * math.pi])\n\n# We can define Python functions that call Julia funcitons\n\npy\"\"\"\ndef pyfib(n, fun):\n if n < 2:\n return n\n else:\n return fun(n - 1, pyfib) + fun(n - 2, pyfib)\n\"\"\"\n#-\nfunction jlfib(n, fun)\n if n < 2\n return n\n else\n return fun(n - 1, jlfib) + fun(n - 2, jlfib)\n end\nend\n#-\njlfib(20, py\"pyfib\")\n\n# If you have some code written in another language (like Python), there can be a smooth transition to using Julia.\n\n# # Summary\n\n# - Julia looks a bit like Python\n# - It's fast (with some caveats)\n# - It's free!\n# - It's written in Julia\n# - It has powerful features like:\n# - Broadcasting syntax\n# - Unicode variables\n# - Multiple dispatch\n# - Good interoperability\n\n# Other stuff I haven't mentioned:\n# - Automatic differentiation (Swift also has this now)\n# - Metaprogramming\n# - Data science stuff (The first two letters of Jupyter Notebooks are named after Julia!)\n", "meta": {"hexsha": "f7192404a272cba43f68d01a4b9a00041950a72e", "size": 11755, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/stuff-i-like-about-julia.jl", "max_stars_repo_name": "Hasnep/stuff-i-like-about-julia", "max_stars_repo_head_hexsha": "47e92d4889ac9a9e18213066f66dcb84d7476b41", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/stuff-i-like-about-julia.jl", "max_issues_repo_name": "Hasnep/stuff-i-like-about-julia", "max_issues_repo_head_hexsha": "47e92d4889ac9a9e18213066f66dcb84d7476b41", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/stuff-i-like-about-julia.jl", "max_forks_repo_name": "Hasnep/stuff-i-like-about-julia", "max_forks_repo_head_hexsha": "47e92d4889ac9a9e18213066f66dcb84d7476b41", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 26.2975391499, "max_line_length": 218, "alphanum_fraction": 0.6801361123, "num_tokens": 3432, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4186969238628498, "lm_q2_score": 0.1688569500503904, "lm_q1q2_score": 0.07069988555896134}}
{"text": "# Copyright (c) 2019 Arpit Bhatia and contributors #src\n# #src\n# Permission is hereby granted, free of charge, to any person obtaining a copy #src\n# of this software and associated documentation files (the \"Software\"), to deal #src\n# in the Software without restriction, including without limitation the rights #src\n# to use, copy, modify, merge, publish, distribute, sublicense, and/or sell #src\n# copies of the Software, and to permit persons to whom the Software is #src\n# furnished to do so, subject to the following conditions: #src\n# #src\n# The above copyright notice and this permission notice shall be included in all #src\n# copies or substantial portions of the Software. #src\n# #src\n# THE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR #src\n# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, #src\n# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE #src\n# AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER #src\n# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, #src\n# OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE #src\n# SOFTWARE. #src\n\n# # Solvers and Solutions\n\n# **Originally Contributed by**: Arpit Bhatia\n\n# The purpose of this part of the tutorial is to introduce you to solvers and\n# how to use them with JuMP. We'll also learn what to do with a problem after\n# the solver has finished optimizing it.\n\n# ## What is a Solver?\n\n# A solver is a software package that incorporates algorithms for finding\n# solutions to one or more classes of problem. For example, GLPK, which we used\n# in the previous tutorials is a solver for linear programming (LP) and mixed\n# integer programming (MIP) problems. It incorporates algorithms such as the\n# simplex method, interior-point method etc. JuMP currently supports a number of\n# open-source and commercial solvers which can be viewed in the [Supported-solvers](@ref)\n# table.\n\n# ## What is MathOptInterface?\n\n# Each mathematical optimization solver API has its own concepts and data\n# structures for representing optimization models and obtaining results.\n# However, it is often desirable to represent an instance of an optimization\n# problem at a higher level so that it is easy to try using different solvers.\n\n# MathOptInterface (MOI) is an abstraction layer designed to provide an\n# interface to mathematical optimization solvers so that users do not need to\n# understand multiple solver-specific APIs. MOI can be used directly, or through\n# a higher-level modeling interface like JuMP.\n\n# Note that JuMP re-exports MathOptInterface and you can use the shortcut `MOI`\n# to refer to MathOptInterface in your code.\n\n# ## Constructing a model\n\n# JuMP models can be created in three different modes: `AUTOMATIC`, `MANUAL` and\n# `DIRECT`. We'll use the following LP to illustrate them.\n\n# ```math\n# \\begin{aligned}\n# & \\max_{x,y} & x + 2y \\\\\n# & \\;\\;\\text{s.t.} & x + y &\\leq 1 \\\\\n# & & 0\\leq x, y &\\leq 1 \\\\\n# \\end{aligned}\n# ```\n\nusing JuMP\nusing GLPK\n\n# ### `AUTOMATIC` Mode\n\n# #### With Optimizer\n\n# This is the easiest method to use a solver in JuMP. In order to do so, we\n# simply set the solver inside the Model constructor.\n\nmodel_auto = Model(GLPK.Optimizer)\n@variable(model_auto, 0 <= x <= 1)\n@variable(model_auto, 0 <= y <= 1)\n@constraint(model_auto, x + y <= 1)\n@objective(model_auto, Max, x + 2y)\noptimize!(model_auto)\nobjective_value(model_auto)\n\n# #### No Optimizer (at first)\n\n# It is also possible to create a JuMP model with no optimizer attached. After\n# the model object is initialized empty and all its variables, constraints and\n# objective are set, then we can attach the solver at `optimize!` time.\n\nmodel_auto_no = Model()\n@variable(model_auto_no, 0 <= x <= 1)\n@variable(model_auto_no, 0 <= y <= 1)\n@constraint(model_auto_no, x + y <= 1)\n@objective(model_auto_no, Max, x + 2y)\nset_optimizer(model_auto_no, GLPK.Optimizer)\noptimize!(model_auto_no)\nobjective_value(model_auto_no)\n\n# Note that we can also enforce the automatic mode by passing\n# `caching_mode = MOIU.AUTOMATIC` in the Model function call.\n\n# ### `MANUAL` Mode\n\n# This mode is similar to the `AUTOMATIC` mode, but there are less protections\n# from the user getting errors from the solver API. On the other side, nothing\n# happens silently, which might give the user more control. It requires\n# attaching the solver before the solve step using the `MOIU.attach_optimizer()`\n# function.\n\nmodel_manual = Model(GLPK.Optimizer, caching_mode = MOIU.MANUAL)\n@variable(model_manual, 0 <= x <= 1)\n@variable(model_manual, 0 <= y <= 1)\n@constraint(model_manual, x + y <= 1)\n@objective(model_manual, Max, x + 2y)\nMOIU.attach_optimizer(model_manual)\noptimize!(model_manual)\nobjective_value(model_manual)\n\n# ### `DIRECT` Mode\n\n# Some solvers are able to handle the problem data directly. This is common for\n# LP/MIP solver but not very common for open-source conic solvers. In this case\n# we do not set a optimizer, we set a backend which is more generic and is able\n# to hold data and not only solving a model.\n\nmodel_direct = direct_model(GLPK.Optimizer())\n@variable(model_direct, 0 <= x <= 1)\n@variable(model_direct, 0 <= y <= 1)\n@constraint(model_direct, x + y <= 1)\n@objective(model_direct, Max, x + 2y)\noptimize!(model_direct)\nobjective_value(model_direct)\n\n# ### Solver Options\n\n# Many of the solvers also allow options to be passed in. However, these options\n# are solver-specific. To find out the various options available, please check\n# out the individual solver packages. Some examples for the GLPK solver are\n# given below.\n\nusing GLPK\n\n# To turn off printing (i.e. silence the solver),\n\nmodel = Model(optimizer_with_attributes(GLPK.Optimizer, \"msg_lev\" => 0));\n\n# To increase the maximum number of simplex iterations:\n\nmodel = Model(optimizer_with_attributes(GLPK.Optimizer, \"it_lim\" => 10_000));\n\n# To set the solution timeout limit (in milliseconds):\n\nmodel = Model(optimizer_with_attributes(GLPK.Optimizer, \"tm_lim\" => 5_000));\n\n# ## How to querying the solution\n\n# So far we have seen all the elements and constructs related to writing a JuMP\n# optimization model. In this section we reach the point of what to do with a\n# solved problem. JuMP follows closely the concepts defined in MathOptInterface\n# to answer user questions about a finished call to `optimize!(model)`. The\n# three main steps in querying a solution are given below. We'll use the model\n# we created in `AUTOMATIC` mode with an optimizer attached in this section.\n\n# ### The termination status\n\n# Termination statuses are meant to explain the reason why the optimizer stopped\n# executing in the most recent call to `optimize!`.\n\ntermination_status(model_auto)\n\n# You can view the different termination status codes by referring to the docs\n# or though checking the possible types using the below command.\n\ndisplay(typeof(MOI.OPTIMAL))\n\n# ### The primal and dual status\n\n# These statuses indicate what kind of result is available to be queried from\n# the model. It's possible that no result is available to be queried. We shall\n# discuss more on the dual status and solutions in the Duality tutorial.\n\nprimal_status(model_auto)\n\n#-\n\ndual_status(model_auto)\n\n# As we saw before, the result (solution) status codes can be viewed directly\n# from Julia.\n\ndisplay(typeof(MOI.FEASIBLE_POINT))\n\n# ### Getting the primal solution\n\n# Provided the primal status is not `MOI.NO_SOLUTION`, we can inspect the\n# solution values and optimal cost.\n\nvalue(x)\n\n#-\n\nvalue(y)\n\n#-\n\nobjective_value(model_auto)\n\n# Since it is possible that no solution is available to be queried from the\n# model, calls to [`value`](@ref) may throw errors. Hence, it is recommended to\n# check for the presence of solutions.\n\nmodel_no_solution = Model(GLPK.Optimizer)\n@variable(model_no_solution, 0 <= x <= 1)\n@variable(model_no_solution, 0 <= y <= 1)\n@constraint(model_no_solution, x + y >= 3)\n@objective(model_no_solution, Max, x + 2y)\n\noptimize!(model_no_solution)\n\ntry #hide\nif termination_status(model_no_solution) == MOI.OPTIMAL\n optimal_solution = value(x)\n optimal_objective = objective_value(model_no_solution)\nelseif termination_status(model_no_solution) == MOI.TIME_LIMIT && has_values(model_no_solution)\n suboptimal_solution = value(x)\n suboptimal_objective = objective_value(model_no_solution)\nelse\n error(\"The model was not solved correctly.\")\nend\ncatch err; showerror(stderr, err); end #hide\n", "meta": {"hexsha": "0bec0f5928d0847cdf694a9ae0cdcfa3c870d383", "size": 8887, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "lang/Julia/solvers_and_solutions.jl", "max_stars_repo_name": "ethansaxenian/RosettaDecode", "max_stars_repo_head_hexsha": "8ea1a42a5f792280b50193ad47545d14ee371fb7", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "lang/Julia/solvers_and_solutions.jl", "max_issues_repo_name": "ethansaxenian/RosettaDecode", "max_issues_repo_head_hexsha": "8ea1a42a5f792280b50193ad47545d14ee371fb7", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "lang/Julia/solvers_and_solutions.jl", "max_forks_repo_name": "ethansaxenian/RosettaDecode", "max_forks_repo_head_hexsha": "8ea1a42a5f792280b50193ad47545d14ee371fb7", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 38.3060344828, "max_line_length": 95, "alphanum_fraction": 0.7168898391, "num_tokens": 2092, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4186969093556867, "lm_q2_score": 0.1688569500503904, "lm_q1q2_score": 0.07069988310932603}}
{"text": "## Exercise 5-1\n## As an exercise, draw a stack diagram for printn called with s = \"Hello\" and n = 2. Then write a function called do_n that takes a function object and a number, n, as arguments, and that calls the given function n times.\nprintln(\"Ans: \")\n\nprintln(\" n --> 2\")\nprintln(\" n --> 1\")\nprintln(\" n --> 0\")\n\nfunction printhello()\n println(\"Hello\")\nend \n\nfunction do_n(func, n)\n if n <= 0\n return\n end\n\n func()\n do_n(func, n - 1)\nend \n\ndo_n(printhello, 3)\n\nprintln(\"End.\")\n", "meta": {"hexsha": "ebae24f20e36268239abd25aac9620ac9a3e9c23", "size": 504, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Chapter5/ex1.jl", "max_stars_repo_name": "yashppawar/ThinkJuliaExercises.jl", "max_stars_repo_head_hexsha": "72145b969dc51ebcac413a10004175ebc63cd5c2", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2022-02-13T14:11:30.000Z", "max_stars_repo_stars_event_max_datetime": "2022-02-13T14:11:30.000Z", "max_issues_repo_path": "Chapter5/ex1.jl", "max_issues_repo_name": "yashppawar/ThinkJuliaExercises.jl", "max_issues_repo_head_hexsha": "72145b969dc51ebcac413a10004175ebc63cd5c2", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Chapter5/ex1.jl", "max_forks_repo_name": "yashppawar/ThinkJuliaExercises.jl", "max_forks_repo_head_hexsha": "72145b969dc51ebcac413a10004175ebc63cd5c2", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 20.16, "max_line_length": 222, "alphanum_fraction": 0.6329365079, "num_tokens": 148, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.27825680567280014, "lm_q2_score": 0.25386101261427363, "lm_q1q2_score": 0.0706385544549102}}
{"text": "\"\"\"\n\tternary(cmd0::String=\"\", arg1=[]; kwargs...)\n\nreads (x,y) pairs and generates PostScript code that will plot lines,\npolygons, or symbols at those locations on a map.\n\nFull option list at [`ternary`](http://gmt.soest.hawaii.edu/doc/latest/ternary.html)\n\nParameters\n----------\n\n- **A** : **straight_lines** : -- Str -- \n\n By default, geographic line segments are drawn as great circle arcs. To draw them as straight\n lines, use the -A flag.\n [`-A`](http://gmt.soest.hawaii.edu/doc/latest/ternary.html#a)\n- $(GMT.opt_J)\n- $(GMT.opt_R)\n- $(GMT.opt_B)\n- $(GMT.opt_C)\n- **G** : **fill** : **markerfacecolor** : **MarkerFaceColor** : -- Str --\n\n Select color or pattern for filling of symbols or polygons. BUT WARN: the alias 'fill' will set the\n color of polygons OR symbols but not the two together. If your plot has polygons and symbols, use\n 'fill' for the polygons and 'markerfacecolor' for filling the symbols. Same applyies for W bellow\n [`-G`](http://gmt.soest.hawaii.edu/doc/latest/ternary.html#g)\n- **L** : **labels** : -- Str -- Flags = a/b/c\n\n Set the labels for the three diagram vertices [none]. \n [`-L`](http://gmt.soest.hawaii.edu/doc/latest/ternary.html#l)\n- **M** : **no_plot** : -- Bool or [] --\n\n Do no plotting. Instead, convert the input (a,b,c[,*z*]) records to Cartesian (x,y,[,*z*]) records,\n where x, y are normalized coordinates on the triangle (i.e., 0-1 in xand 0-sqrt(3)/2 in y).\n [`-M`](http://gmt.soest.hawaii.edu/doc/latest/ternary.html#m)\n- **N** : **no_clip** : -- Bool or [] --\n\n Do NOT clip symbols that fall outside map border \n [`-N`](http://gmt.soest.hawaii.edu/doc/latest/ternary.html#n)\n- $(GMT.opt_P)\n- **S** : **symbol** : **marker** : **Marker** : -- Str --\n\n Plot symbols (including vectors, pie slices, fronts, decorated or quoted lines). \n [`-S`](http://gmt.soest.hawaii.edu/doc/latest/ternary.html#s)\n Alternatively select a sub-set of symbols using the aliases: **marker** or **Marker** and values:\n\n + **-**, **x_dash**\n + **+**, **plus**\n + **a**, *, **star**\n + **c**, **circle**\n + **d**, **diamond**\n + **g**, **octagon**\n + **h**, **hexagon**\n + **i**, **v**, **inverted_tri**\n + **n**, **pentagon**\n + **p**, **.**, **point**\n + **r**, **rectangle**\n + **s**, **square**\n + **t**, **^**, **triangle**\n + **x**, **cross**\n + **y**, **y_dash**\n- **W** : **line_attrib** : **markeredgecolor** : **MarkerEdgeColor** : -- Str --\n\n Set pen attributes for lines or the outline of symbols\n [`-W`](http://gmt.soest.hawaii.edu/doc/latest/ternary.html#w)\n WARNING: the pen attributes will set the pen of polygons OR symbols but not the two together.\n If your plot has polygons and symbols, use **W** or **line_attribs** for the polygons and\n **markeredgecolor** or **MarkerEdgeColor** for filling the symbols. Similar to S above.\n- $(GMT.opt_U)\n- $(GMT.opt_V)\n- $(GMT.opt_X)\n- $(GMT.opt_Y)\n- **axis** : **aspect** : -- Str --\n When equal to \"equal\" makes a square plot.\n- $(GMT.opt_a)\n- $(GMT.opt_bi)\n- $(GMT.opt_di)\n- $(GMT.opt_e)\n- $(GMT.opt_f)\n- $(GMT.opt_g)\n- $(GMT.opt_h)\n- $(GMT.opt_i)\n- $(GMT.opt_p)\n- $(GMT.opt_t)\n- $(GMT.opt_swap_xy)\n\"\"\"\nfunction ternary(cmd0::String=\"\", arg1=[]; caller=[], K=false, O=false, first=true, kwargs...)\n\n\targ2 = []\t\t# May be needed if GMTcpt type is sent in via C\n\tN_args = isempty_(arg1) ? 0 : 1\n\n\t((isempty(cmd0) && isempty_(arg1) || occursin(\" -\", cmd0)) && return monolitic(\"ternary\", cmd0, arg1)\t# Monolitic mode\n\n\td = KW(kwargs)\n\toutput, opt_T, fname_ext = fname_out(d)\t\t# OUTPUT may have been an extension only\n\n\topt_J = \" -JX12c/10.4c\" # Equilateral triangle\n\tfor sym in [:axis :aspect]\n\t\tif (haskey(d, sym))\n\t\t\tif (d[sym] == \"equal\")\t\t\t\t# Need also a 'tight' option\n\t\t\t\topt_J = \" -JX12c\"\n\t\t\tend\n\t\t\tbreak\n\t\tend\n\tend\n\tcmd, opt_B, opt_J, opt_R = parse_BJR(d, \"\", caller, O, opt_J)\n\tcmd = parse_UVXY(cmd, d)\n\tcmd, = parse_a(cmd, d)\n\tcmd, opt_bi = parse_bi(cmd, d)\n\tcmd, opt_di = parse_di(cmd, d)\n\tcmd, = parse_e(cmd, d)\n\tcmd, = parse_f(cmd, d)\n\tcmd, = parse_g(cmd, d)\n\tcmd, = parse_h(cmd, d)\n\tcmd, opt_i = parse_i(cmd, d)\n\tcmd, = parse_p(cmd, d)\n\tcmd, = parse_t(cmd, d)\n\tcmd, = parse_swap_xy(cmd, d)\n\tcmd = parse_params(cmd, d)\n\n\tcmd, K, O, opt_B = set_KO(cmd, opt_B, first, K, O)\t\t# Set the K O dance\n\n\t# If file name sent in, read it and compute a tight -R if this was not provided \n\tcmd, arg1, = read_data(d, cmd0, cmd, arg1, opt_R, opt_i, opt_bi, opt_di, false)\n\n\tcmd, arg1, arg2, = add_opt_cpt(d, cmd, [:C :color :cmap], 'C', N_args, arg1, arg2)\n\n\tcmd = add_opt(cmd, 'L', d, [:L :straight_lines])\n\tcmd = add_opt(cmd, 'M', d, [:M :offset])\n\tcmd = add_opt(cmd, 'N', d, [:N :error_bars])\n\n\tcmd = add_opt(cmd, 'G', d, [:G :fill])\n\topt_Gsymb = \"\"\t\t\t# Filling color for symbols\n\tfor sym in [:G :markerfacecolor :MarkerFaceColor]\n\t\tif (haskey(d, sym))\n\t\t\topt_Gsymb = \" -G\" * arg2str(d[sym])\n\t\t\tbreak\n\t\tend\n\tend\n\n\topt_Wmarker = \"\"\n\tfor sym in [:markeredgecolor :MarkerEdgeColor]\n\t\tif (haskey(d, sym))\n\t\t\topt_Wmarker = \"0.5p,\" * arg2str(d[sym])\t\t# 0.25p is so thin\n\t\t\tbreak\n\t\tend\n\tend\n\n\tcmd = add_opt(cmd, 'L', d, [:L :labels])\n\tcmd = add_opt(cmd, 'M', d, [:M :no_plot])\n\tcmd = add_opt(cmd, 'N', d, [:N :no_clip])\n\n\topt_W = \"\"\n\tpen = build_pen(d)\t\t\t\t\t\t# Either a full pen string or empty (\"\")\n\tif (!isempty(pen))\n\t\topt_W = \" -W\" * pen\n\telse\n\t\tfor sym in [:W :line_attrib]\n\t\t\tif (haskey(d, sym))\n\t\t\t\tif (isa(d[sym], Tuple))\t\t# Like this it can hold the pen, not extended atts\n\t\t\t\t\topt_W = \" -W\" * parse_pen(d[sym])\n\t\t\t\telse\n\t\t\t\t\topt_W = \" -W\" * arg2str(d[sym])\n\t\t\t\tend\n\t\t\t\tbreak\n\t\t\tend\n\t\tend\n\tend\n\n\topt_S = \"\"\n\tfor sym in [:S :symbol]\n\t\tif (haskey(d, sym))\n\t\t\topt_S = \" -S\" * arg2str(d[sym])\n\t\t\tbreak\n\t\tend\n\tend\n\tif (isempty(opt_S))\t\t\t# OK, no symbol given via the -S option. So fish in aliases\n\t\tmarca = \"\"\n\t\tfor sym in [:marker :Marker]\n\t\t\tif (haskey(d, sym))\n\t\t\t\tt = d[sym]\n\t\t\t\tif (isa(t, Symbol))\tt = string(t)\tend\n\t\t\t\tif (t == \"-\" || t == \"x-dash\") marca = \"-\"\n\t\t\t\telseif (t == \"+\" || t == \"plus\") marca = \"+\"\n\t\t\t\telseif (t == \"a\" || t == \"*\" || t == \"star\") marca = \"a\"\n\t\t\t\telseif (t == \"c\" || t == \"circle\") marca = \"c\"\n\t\t\t\telseif (t == \"d\" || t == \"diamond\") marca = \"d\"\n\t\t\t\telseif (t == \"g\" || t == \"octagon\") marca = \"g\"\n\t\t\t\telseif (t == \"h\" || t == \"hexagon\") marca = \"h\"\n\t\t\t\telseif (t == \"i\" || t == \"v\" || t == \"inverted_tri\") marca = \"i\"\n\t\t\t\telseif (t == \"n\" || t == \"pentagon\") marca = \"n\"\n\t\t\t\telseif (t == \"p\" || t == \".\" || t == \"point\") marca = \"p\"\n\t\t\t\telseif (t == \"r\" || t == \"rectangle\") marca = \"r\"\n\t\t\t\telseif (t == \"s\" || t == \"square\") marca = \"s\"\n\t\t\t\telseif (t == \"t\" || t == \"^\" || t == \"triangle\") marca = \"t\"\n\t\t\t\telseif (t == \"x\" || t == \"cross\") marca = \"x\"\n\t\t\t\telseif (is3D && (t == \"u\" || t == \"cube\")) marca = \"u\"\n\t\t\t\telseif (t == \"y\" || t == \"y-dash\") marca = \"y\"\n\t\t\t\tend\n\t\t\t\tbreak\n\t\t\tend\n\t\tend\n\t\tif (!isempty(marca))\n\t\t\tdone = false\n\t\t\tfor sym in [:markersize :MarkerSize :size]\n\t\t\t\tif (haskey(d, sym))\n\t\t\t\t\tmarca = marca * arg2str(d[sym])\n\t\t\t\t\tdone = true\n\t\t\t\t\tbreak\n\t\t\t\tend\n\t\t\tend\n\t\t\tif (!done) marca = marca * \"8p\" end\t\t\t# Default to 8p\n\t\tend\n\t\tif (!isempty(marca)) opt_S = \" -S\" * marca end\n\tend\n\n\tif (!isempty(opt_S))\t\t\t# \n\t\topt_ML = \"\"\n\t\tfor sym in [:markerline :MarkerLine]\n\t\t\tif (haskey(d, sym))\n\t\t\t\tif (isa(d[sym], Tuple))\t# Like this it can hold the pen, not extended atts\n\t\t\t\t\topt_ML = \" -W\" * parse_pen(d[sym])\n\t\t\t\telse\n\t\t\t\t\topt_ML = \" -W\" * arg2str(d[sym])\n\t\t\t\tend\n\t\t\t\tif (!isempty(opt_Wmarker))\n\t\t\t\t\topt_Wmarker = \"\"\n\t\t\t\t\t@warn(\"markerline overrides markeredgecolor\")\n\t\t\t\tend\n\t\t\t\tbreak\n\t\t\tend\n\t\tend\n\t\tif (!isempty(opt_W) && !isempty(opt_ML))\n\t\t\t@warn(\"You cannot use both markeredgecolor and W or line_attrib keys.\")\n\t\tend\n\tend\n\n\tif (!isempty(opt_W) && isempty(opt_S)) \t\t\t# We have a line/polygon request\n\t\tcmd = [finish_PS(d, cmd * opt_W, output, K, O)]\n\telseif (isempty(opt_W) && !isempty(opt_S))\t\t# We have a symbol request\n\t\tif (!isempty(opt_Wmarker) && isempty(opt_W))\n\t\t\topt_Gsymb = opt_Gsymb * \" -W\" * opt_Wmarker\t# Piggy back in this option string\n\t\tend\n\t\tif (!isempty(opt_ML))\t\t\t\t\t\t# If we have a symbol outline pen\n\t\t\tcmd = cmd * opt_ML\n\t\tend\n\t\tcmd = [finish_PS(d, cmd * opt_S * opt_Gsymb, output, K, O)]\n\telseif (!isempty(opt_W) && !isempty(opt_S))\t\t# We have both line/polygon and a symbol\n\t\t# that is not a vector (because Vector width is set by -W)\n\t\tif (opt_S[4] == 'v' || opt_S[4] == 'V' || opt_S[4] == '=')\n\t\t\tcmd = [finish_PS(d, cmd * opt_W * opt_S * opt_Gsymb, output, K, O)]\n\t\telse\n\t\t\tif (!isempty(opt_Wmarker))\n\t\t\t\topt_Wmarker = \" -W\" * opt_Wmarker\t# Set Symbol edge color \n\t\t\tend\n\t\t\tcmd1 = cmd * opt_W\n\t\t\tcmd2 = replace(cmd, opt_B => \"\") * opt_S * opt_Gsymb * opt_Wmarker\t# Don't repeat option -B\n\t\t\tif (!isempty(opt_ML))\t\t\t\t\t# If we have a symbol outline pen\n\t\t\t\tcmd1 = cmd1 * opt_ML\n\t\t\tend\n\t\t\tcmd = [finish_PS(d, cmd1, output, true, O)\n\t\t\t finish_PS(d, cmd2, output, K, true)]\n\t\tend\n\telseif (!isempty(opt_S) && !isempty(opt_ML))\t\t# We have a symbol outline pen\n\t\tcmd = [finish_PS(d, cmd * opt_ML * opt_S * opt_Gsymb, output, K, O)]\n\telse\n\t\tcmd = [finish_PS(d, cmd, output, K, O)]\n\tend\n\n\treturn finish_PS_module(d, cmd, \"\", output, fname_ext, opt_T, K, \"ternary\", arg1, arg2)\nend\n\n# ---------------------------------------------------------------------------------------------------\nternary!(cmd0::String=\"\", arg1=[]; caller=[], K=true, O=true, first=false, kw...) =\n\tternary(cmd0, arg1; caller=caller, K=K, O=O, first=first, kw...)\nternary!(arg1=[]; caller=[], K=true, O=true, first=false, kw...) =\n\tternary(\"\", arg1; caller=caller, K=K, O=O, first=first, kw...)\n\npsternary = ternary # Aliases\npsternary! = ternary! # Aliases", "meta": {"hexsha": "3c343a658485d0287b78d0a4a0cad992066425a3", "size": 9699, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/psternary.jl", "max_stars_repo_name": "JuliaDocsForks/GMT.jl", "max_stars_repo_head_hexsha": "53d9c030559fe3f079f3b662ce262e7ed682d357", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/psternary.jl", "max_issues_repo_name": "JuliaDocsForks/GMT.jl", "max_issues_repo_head_hexsha": "53d9c030559fe3f079f3b662ce262e7ed682d357", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/psternary.jl", "max_forks_repo_name": "JuliaDocsForks/GMT.jl", "max_forks_repo_head_hexsha": "53d9c030559fe3f079f3b662ce262e7ed682d357", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 34.6392857143, "max_line_length": 119, "alphanum_fraction": 0.5732549747, "num_tokens": 3209, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.49609382947091946, "lm_q2_score": 0.14223190046381004, "lm_q1q2_score": 0.07056036817401817}}
{"text": "### A Pluto.jl notebook ###\n# v0.17.1\n\nusing Markdown\nusing InteractiveUtils\n\n# \u2554\u2550\u2561 c50a52e2-30fa-11ec-00e9-4f4a81ed30ed\nusing CSV, DataFrames\n\n# \u2554\u2550\u2561 9ba9e100-6929-4312-8cd8-75de0cb05a3c\niris = CSV.File(\"iris_dataset.csv\") |> DataFrame\n\n# \u2554\u2550\u2561 b1a1e7fc-58de-45ef-b6c4-355f5e30b7ef\ndescribe(iris)\n\n# \u2554\u2550\u2561 63668d67-5d58-415d-9fc9-6a891e26ddbd\nnrow(iris)\n\n# \u2554\u2550\u2561 5346265b-2829-4e49-afc7-a016db72db17\nunique(iris.Species)\n\n# \u2554\u2550\u2561 148805ea-c9d6-4bdf-ac79-b9bea8d24f4c\nspecies = groupby(iris, :Species)\n\n# \u2554\u2550\u2561 7b9ee6d2-95b1-4400-a21d-313b0a97b229\ncombine(species, nrow)\n\n# \u2554\u2550\u2561 9f30a14b-eded-4829-bd2a-6b85548ba6b8\ncombine(species, :SepalLength => minimum => :MinSepalLength)\n\n# \u2554\u2550\u2561 6e73a20f-860e-4b2e-8dc0-64f1b93f6fa1\ncolnames = names(iris)\n\n# \u2554\u2550\u2561 27013be8-56eb-4455-9d2a-53c475d6147a\nnumeric_columns = colnames[1:4]\n\n# \u2554\u2550\u2561 4f193ca1-f0a6-472d-a9bf-6deb725c7bb6\ntransform(iris, numeric_columns .=> (x -> 10x) .=> numeric_columns)\n\n# \u2554\u2550\u2561 432548a4-67b2-45c9-a0bd-fb3099568f15\nbegin\n\tiris[!, :Sample] = 1:nrow(iris)\n\tlong = stack(iris, Not([:Species, :Sample]))\nend\n\n# \u2554\u2550\u2561 b94f9bc0-2e63-4cd5-b4b5-b64c41d79270\nbegin\n\tlong.Dimension = [\n\t\treplace(x, r\"Sepal|Petal\" => \"\") \n\t\tfor x in long.variable\n\t\t]\n\tlong.Structure = [\n\t\treplace(x, r\"Length|Width\" => \"\") \n\t\tfor x in long.variable\n\t\t]\n\tselect!(long, Not(:variable))\n\tunstack(long, :Dimension, :value)\nend\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000001\nPLUTO_PROJECT_TOML_CONTENTS = \"\"\"\n[deps]\nCSV = \"336ed68f-0bac-5ca0-87d4-7b16caf5d00b\"\nDataFrames = \"a93c6f00-e57d-5684-b7b6-d8193f3e46c0\"\n\n[compat]\nCSV = \"~0.9.10\"\nDataFrames = \"~1.2.2\"\n\"\"\"\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000002\nPLUTO_MANIFEST_TOML_CONTENTS = \"\"\"\n# This file is machine-generated - editing it directly is not advised\n\n[[ArgTools]]\nuuid = \"0dad84c5-d112-42e6-8d28-ef12dabb789f\"\n\n[[Artifacts]]\nuuid = \"56f22d72-fd6d-98f1-02f0-08ddc0907c33\"\n\n[[Base64]]\nuuid = \"2a0f44e3-6c83-55bd-87e4-b1978d98bd5f\"\n\n[[CSV]]\ndeps = [\"CodecZlib\", \"Dates\", \"FilePathsBase\", \"InlineStrings\", \"Mmap\", \"Parsers\", \"PooledArrays\", \"SentinelArrays\", \"Tables\", \"Unicode\", \"WeakRefStrings\"]\ngit-tree-sha1 = \"74147e877531d7c172f70b492995bc2b5ca3a843\"\nuuid = \"336ed68f-0bac-5ca0-87d4-7b16caf5d00b\"\nversion = \"0.9.10\"\n\n[[CodecZlib]]\ndeps = [\"TranscodingStreams\", \"Zlib_jll\"]\ngit-tree-sha1 = \"ded953804d019afa9a3f98981d99b33e3db7b6da\"\nuuid = \"944b1d66-785c-5afd-91f1-9de20f533193\"\nversion = \"0.7.0\"\n\n[[Compat]]\ndeps = [\"Base64\", \"Dates\", \"DelimitedFiles\", \"Distributed\", \"InteractiveUtils\", \"LibGit2\", \"Libdl\", \"LinearAlgebra\", \"Markdown\", \"Mmap\", \"Pkg\", \"Printf\", \"REPL\", \"Random\", \"SHA\", \"Serialization\", \"SharedArrays\", \"Sockets\", \"SparseArrays\", \"Statistics\", \"Test\", \"UUIDs\", \"Unicode\"]\ngit-tree-sha1 = \"dce3e3fea680869eaa0b774b2e8343e9ff442313\"\nuuid = \"34da2185-b29b-5c13-b0c7-acf172513d20\"\nversion = \"3.40.0\"\n\n[[Crayons]]\ngit-tree-sha1 = \"3f71217b538d7aaee0b69ab47d9b7724ca8afa0d\"\nuuid = \"a8cc5b0e-0ffa-5ad4-8c14-923d3ee1735f\"\nversion = \"4.0.4\"\n\n[[DataAPI]]\ngit-tree-sha1 = \"cc70b17275652eb47bc9e5f81635981f13cea5c8\"\nuuid = \"9a962f9c-6df0-11e9-0e5d-c546b8b5ee8a\"\nversion = \"1.9.0\"\n\n[[DataFrames]]\ndeps = [\"Compat\", \"DataAPI\", \"Future\", \"InvertedIndices\", \"IteratorInterfaceExtensions\", \"LinearAlgebra\", \"Markdown\", \"Missings\", \"PooledArrays\", \"PrettyTables\", \"Printf\", \"REPL\", \"Reexport\", \"SortingAlgorithms\", \"Statistics\", \"TableTraits\", \"Tables\", \"Unicode\"]\ngit-tree-sha1 = \"d785f42445b63fc86caa08bb9a9351008be9b765\"\nuuid = \"a93c6f00-e57d-5684-b7b6-d8193f3e46c0\"\nversion = \"1.2.2\"\n\n[[DataStructures]]\ndeps = [\"Compat\", \"InteractiveUtils\", \"OrderedCollections\"]\ngit-tree-sha1 = \"7d9d316f04214f7efdbb6398d545446e246eff02\"\nuuid = \"864edb3b-99cc-5e75-8d2d-829cb0a9cfe8\"\nversion = \"0.18.10\"\n\n[[DataValueInterfaces]]\ngit-tree-sha1 = \"bfc1187b79289637fa0ef6d4436ebdfe6905cbd6\"\nuuid = \"e2d170a0-9d28-54be-80f0-106bbe20a464\"\nversion = \"1.0.0\"\n\n[[Dates]]\ndeps = [\"Printf\"]\nuuid = \"ade2ca70-3891-5945-98fb-dc099432e06a\"\n\n[[DelimitedFiles]]\ndeps = [\"Mmap\"]\nuuid = \"8bb1440f-4735-579b-a4ab-409b98df4dab\"\n\n[[Distributed]]\ndeps = [\"Random\", \"Serialization\", \"Sockets\"]\nuuid = \"8ba89e20-285c-5b6f-9357-94700520ee1b\"\n\n[[Downloads]]\ndeps = [\"ArgTools\", \"LibCURL\", \"NetworkOptions\"]\nuuid = \"f43a241f-c20a-4ad4-852c-f6b1247861c6\"\n\n[[FilePathsBase]]\ndeps = [\"Dates\", \"Mmap\", \"Printf\", \"Test\", \"UUIDs\"]\ngit-tree-sha1 = \"d962b5a47b6d191dbcd8ae0db841bc70a05a3f5b\"\nuuid = \"48062228-2e41-5def-b9a4-89aafe57970f\"\nversion = \"0.9.13\"\n\n[[Formatting]]\ndeps = [\"Printf\"]\ngit-tree-sha1 = \"8339d61043228fdd3eb658d86c926cb282ae72a8\"\nuuid = \"59287772-0a20-5a39-b81b-1366585eb4c0\"\nversion = \"0.4.2\"\n\n[[Future]]\ndeps = [\"Random\"]\nuuid = \"9fa8497b-333b-5362-9e8d-4d0656e87820\"\n\n[[InlineStrings]]\ndeps = [\"Parsers\"]\ngit-tree-sha1 = \"19cb49649f8c41de7fea32d089d37de917b553da\"\nuuid = \"842dd82b-1e85-43dc-bf29-5d0ee9dffc48\"\nversion = \"1.0.1\"\n\n[[InteractiveUtils]]\ndeps = [\"Markdown\"]\nuuid = \"b77e0a4c-d291-57a0-90e8-8db25a27a240\"\n\n[[InvertedIndices]]\ngit-tree-sha1 = \"bee5f1ef5bf65df56bdd2e40447590b272a5471f\"\nuuid = \"41ab1584-1d38-5bbf-9106-f11c6c58b48f\"\nversion = \"1.1.0\"\n\n[[IteratorInterfaceExtensions]]\ngit-tree-sha1 = \"a3f24677c21f5bbe9d2a714f95dcd58337fb2856\"\nuuid = \"82899510-4779-5014-852e-03e436cf321d\"\nversion = \"1.0.0\"\n\n[[LibCURL]]\ndeps = [\"LibCURL_jll\", \"MozillaCACerts_jll\"]\nuuid = \"b27032c2-a3e7-50c8-80cd-2d36dbcbfd21\"\n\n[[LibCURL_jll]]\ndeps = [\"Artifacts\", \"LibSSH2_jll\", \"Libdl\", \"MbedTLS_jll\", \"Zlib_jll\", \"nghttp2_jll\"]\nuuid = \"deac9b47-8bc7-5906-a0fe-35ac56dc84c0\"\n\n[[LibGit2]]\ndeps = [\"Base64\", \"NetworkOptions\", \"Printf\", \"SHA\"]\nuuid = \"76f85450-5226-5b5a-8eaa-529ad045b433\"\n\n[[LibSSH2_jll]]\ndeps = [\"Artifacts\", \"Libdl\", \"MbedTLS_jll\"]\nuuid = \"29816b5a-b9ab-546f-933c-edad1886dfa8\"\n\n[[Libdl]]\nuuid = \"8f399da3-3557-5675-b5ff-fb832c97cbdb\"\n\n[[LinearAlgebra]]\ndeps = [\"Libdl\"]\nuuid = \"37e2e46d-f89d-539d-b4ee-838fcccc9c8e\"\n\n[[Logging]]\nuuid = \"56ddb016-857b-54e1-b83d-db4d58db5568\"\n\n[[Markdown]]\ndeps = [\"Base64\"]\nuuid = \"d6f4376e-aef5-505a-96c1-9c027394607a\"\n\n[[MbedTLS_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"c8ffd9c3-330d-5841-b78e-0817d7145fa1\"\n\n[[Missings]]\ndeps = [\"DataAPI\"]\ngit-tree-sha1 = \"bf210ce90b6c9eed32d25dbcae1ebc565df2687f\"\nuuid = \"e1d29d7a-bbdc-5cf2-9ac0-f12de2c33e28\"\nversion = \"1.0.2\"\n\n[[Mmap]]\nuuid = \"a63ad114-7e13-5084-954f-fe012c677804\"\n\n[[MozillaCACerts_jll]]\nuuid = \"14a3606d-f60d-562e-9121-12d972cd8159\"\n\n[[NetworkOptions]]\nuuid = \"ca575930-c2e3-43a9-ace4-1e988b2c1908\"\n\n[[OrderedCollections]]\ngit-tree-sha1 = \"85f8e6578bf1f9ee0d11e7bb1b1456435479d47c\"\nuuid = \"bac558e1-5e72-5ebc-8fee-abe8a469f55d\"\nversion = \"1.4.1\"\n\n[[Parsers]]\ndeps = [\"Dates\"]\ngit-tree-sha1 = \"ae4bbcadb2906ccc085cf52ac286dc1377dceccc\"\nuuid = \"69de0a69-1ddd-5017-9359-2bf0b02dc9f0\"\nversion = \"2.1.2\"\n\n[[Pkg]]\ndeps = [\"Artifacts\", \"Dates\", \"Downloads\", \"LibGit2\", \"Libdl\", \"Logging\", \"Markdown\", \"Printf\", \"REPL\", \"Random\", \"SHA\", \"Serialization\", \"TOML\", \"Tar\", \"UUIDs\", \"p7zip_jll\"]\nuuid = \"44cfe95a-1eb2-52ea-b672-e2afdf69b78f\"\n\n[[PooledArrays]]\ndeps = [\"DataAPI\", \"Future\"]\ngit-tree-sha1 = \"a193d6ad9c45ada72c14b731a318bedd3c2f00cf\"\nuuid = \"2dfb63ee-cc39-5dd5-95bd-886bf059d720\"\nversion = \"1.3.0\"\n\n[[PrettyTables]]\ndeps = [\"Crayons\", \"Formatting\", \"Markdown\", \"Reexport\", \"Tables\"]\ngit-tree-sha1 = \"d940010be611ee9d67064fe559edbb305f8cc0eb\"\nuuid = \"08abe8d2-0d0c-5749-adfa-8a2ac140af0d\"\nversion = \"1.2.3\"\n\n[[Printf]]\ndeps = [\"Unicode\"]\nuuid = \"de0858da-6303-5e67-8744-51eddeeeb8d7\"\n\n[[REPL]]\ndeps = [\"InteractiveUtils\", \"Markdown\", \"Sockets\", \"Unicode\"]\nuuid = \"3fa0cd96-eef1-5676-8a61-b3b8758bbffb\"\n\n[[Random]]\ndeps = [\"Serialization\"]\nuuid = \"9a3f8284-a2c9-5f02-9a11-845980a1fd5c\"\n\n[[Reexport]]\ngit-tree-sha1 = \"45e428421666073eab6f2da5c9d310d99bb12f9b\"\nuuid = \"189a3867-3050-52da-a836-e630ba90ab69\"\nversion = \"1.2.2\"\n\n[[SHA]]\nuuid = \"ea8e919c-243c-51af-8825-aaa63cd721ce\"\n\n[[SentinelArrays]]\ndeps = [\"Dates\", \"Random\"]\ngit-tree-sha1 = \"f45b34656397a1f6e729901dc9ef679610bd12b5\"\nuuid = \"91c51154-3ec4-41a3-a24f-3f23e20d615c\"\nversion = \"1.3.8\"\n\n[[Serialization]]\nuuid = \"9e88b42a-f829-5b0c-bbe9-9e923198166b\"\n\n[[SharedArrays]]\ndeps = [\"Distributed\", \"Mmap\", \"Random\", \"Serialization\"]\nuuid = \"1a1011a3-84de-559e-8e89-a11a2f7dc383\"\n\n[[Sockets]]\nuuid = \"6462fe0b-24de-5631-8697-dd941f90decc\"\n\n[[SortingAlgorithms]]\ndeps = [\"DataStructures\"]\ngit-tree-sha1 = \"b3363d7460f7d098ca0912c69b082f75625d7508\"\nuuid = \"a2af1166-a08f-5f64-846c-94a0d3cef48c\"\nversion = \"1.0.1\"\n\n[[SparseArrays]]\ndeps = [\"LinearAlgebra\", \"Random\"]\nuuid = \"2f01184e-e22b-5df5-ae63-d93ebab69eaf\"\n\n[[Statistics]]\ndeps = [\"LinearAlgebra\", \"SparseArrays\"]\nuuid = \"10745b16-79ce-11e8-11f9-7d13ad32a3b2\"\n\n[[TOML]]\ndeps = [\"Dates\"]\nuuid = \"fa267f1f-6049-4f14-aa54-33bafae1ed76\"\n\n[[TableTraits]]\ndeps = [\"IteratorInterfaceExtensions\"]\ngit-tree-sha1 = \"c06b2f539df1c6efa794486abfb6ed2022561a39\"\nuuid = \"3783bdb8-4a98-5b6b-af9a-565f29a5fe9c\"\nversion = \"1.0.1\"\n\n[[Tables]]\ndeps = [\"DataAPI\", \"DataValueInterfaces\", \"IteratorInterfaceExtensions\", \"LinearAlgebra\", \"TableTraits\", \"Test\"]\ngit-tree-sha1 = \"fed34d0e71b91734bf0a7e10eb1bb05296ddbcd0\"\nuuid = \"bd369af6-aec1-5ad0-b16a-f7cc5008161c\"\nversion = \"1.6.0\"\n\n[[Tar]]\ndeps = [\"ArgTools\", \"SHA\"]\nuuid = \"a4e569a6-e804-4fa4-b0f3-eef7a1d5b13e\"\n\n[[Test]]\ndeps = [\"InteractiveUtils\", \"Logging\", \"Random\", \"Serialization\"]\nuuid = \"8dfed614-e22c-5e08-85e1-65c5234f0b40\"\n\n[[TranscodingStreams]]\ndeps = [\"Random\", \"Test\"]\ngit-tree-sha1 = \"216b95ea110b5972db65aa90f88d8d89dcb8851c\"\nuuid = \"3bb67fe8-82b1-5028-8e26-92a6c54297fa\"\nversion = \"0.9.6\"\n\n[[UUIDs]]\ndeps = [\"Random\", \"SHA\"]\nuuid = \"cf7118a7-6976-5b1a-9a39-7adc72f591a4\"\n\n[[Unicode]]\nuuid = \"4ec0a83e-493e-50e2-b9ac-8f72acf5a8f5\"\n\n[[WeakRefStrings]]\ndeps = [\"DataAPI\", \"InlineStrings\", \"Parsers\"]\ngit-tree-sha1 = \"c69f9da3ff2f4f02e811c3323c22e5dfcb584cfa\"\nuuid = \"ea10d353-3f73-51f8-a26c-33c1cb351aa5\"\nversion = \"1.4.1\"\n\n[[Zlib_jll]]\ndeps = [\"Libdl\"]\nuuid = \"83775a58-1f1d-513f-b197-d71354ab007a\"\n\n[[nghttp2_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"8e850ede-7688-5339-a07c-302acd2aaf8d\"\n\n[[p7zip_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"3f19e933-33d8-53b3-aaab-bd5110c3b7a0\"\n\"\"\"\n\n# \u2554\u2550\u2561 Cell order:\n# \u2560\u2550c50a52e2-30fa-11ec-00e9-4f4a81ed30ed\n# \u2560\u25509ba9e100-6929-4312-8cd8-75de0cb05a3c\n# \u2560\u2550b1a1e7fc-58de-45ef-b6c4-355f5e30b7ef\n# \u2560\u255063668d67-5d58-415d-9fc9-6a891e26ddbd\n# \u2560\u25505346265b-2829-4e49-afc7-a016db72db17\n# \u2560\u2550148805ea-c9d6-4bdf-ac79-b9bea8d24f4c\n# \u2560\u25507b9ee6d2-95b1-4400-a21d-313b0a97b229\n# \u2560\u25509f30a14b-eded-4829-bd2a-6b85548ba6b8\n# \u2560\u25506e73a20f-860e-4b2e-8dc0-64f1b93f6fa1\n# \u2560\u255027013be8-56eb-4455-9d2a-53c475d6147a\n# \u2560\u25504f193ca1-f0a6-472d-a9bf-6deb725c7bb6\n# \u2560\u2550432548a4-67b2-45c9-a0bd-fb3099568f15\n# \u2560\u2550b94f9bc0-2e63-4cd5-b4b5-b64c41d79270\n# \u255f\u250000000000-0000-0000-0000-000000000001\n# \u255f\u250000000000-0000-0000-0000-000000000002\n", "meta": {"hexsha": "0d4e1459b4fb6742ab7029d552a9b14fd57011de", "size": 10638, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Chapter05/describe_iris_dataframe.jl", "max_stars_repo_name": "PacktPublishing/Interactive-Visualization-with-Julia", "max_stars_repo_head_hexsha": "6f9b2f0798784892b7d6965dd56fb2ef42d1012a", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2021-10-04T16:28:23.000Z", "max_stars_repo_stars_event_max_datetime": "2021-10-04T16:28:23.000Z", "max_issues_repo_path": "Chapter05/describe_iris_dataframe.jl", "max_issues_repo_name": "PacktPublishing/Interactive-Visualization-with-Julia", "max_issues_repo_head_hexsha": "6f9b2f0798784892b7d6965dd56fb2ef42d1012a", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Chapter05/describe_iris_dataframe.jl", "max_forks_repo_name": "PacktPublishing/Interactive-Visualization-with-Julia", "max_forks_repo_head_hexsha": "6f9b2f0798784892b7d6965dd56fb2ef42d1012a", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 28.0686015831, "max_line_length": 280, "alphanum_fraction": 0.7280503854, "num_tokens": 4967, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.43782349911420193, "lm_q2_score": 0.16026603032235007, "lm_q1q2_score": 0.0701682341848741}}
{"text": "@testset \"Basic Type coercion tests\" begin\n X = (x=10:10:44, y=1:4, z=collect(\"abcd\"))\n\n @test_throws ScientificTypes.CoercionError coerce(X, :x=>Float64)\n @test_throws ScientificTypes.CoercionError coerce(X, :x=>Textual)\n\n types = Dict(:x => Continuous, :z => Multiclass)\n X_coerced = coerce(X, types)\n @test X_coerced == coerce(X, :x => Continuous, :z => Multiclass)\n @test scitype_union(X_coerced.x) === Continuous\n @test scitype_union(X_coerced.z) <: Multiclass\n @test !X_coerced.z.pool.ordered\n @test_throws MethodError coerce([\"a\", \"b\", \"c\"], Count)\n\n y = collect(Float64, 1:5)\n y_coerced = coerce(y, Count)\n @test scitype_union(y_coerced) === Count\n @test y_coerced == y\n y = [1//2, 3//4, 6//5]\n y_coerced = coerce(y, Continuous)\n @test scitype_union(y_coerced) === Continuous\n @test y_coerced \u2248 y\n X_coerced = coerce(X, Dict(:z => OrderedFactor))\n @test X_coerced == coerce(X, :z => OrderedFactor)\n @test X_coerced.x === X.x\n @test scitype_union(X_coerced.z) <: OrderedFactor\n @test X_coerced.z.pool.ordered\n\n # Check no-op coercion\n y = rand(Float64, 5)\n @test coerce(y, Continuous) === y\n y = rand(Float32, 5)\n @test coerce(y, Continuous) === y\n y = rand(BigFloat, 5)\n @test coerce(y, Continuous) === y\n y = rand(Int, 5)\n @test coerce(y, Count) === y\n y = big.(y)\n @test coerce(y, Count) === y\n y = rand(UInt32, 5)\n @test coerce(y, Count) === y\n X_coerced = coerce(X, Dict{Symbol, Type}())\n @test X_coerced.x === X.x\n @test X_coerced.z === X.z\n z = categorical(X.z)\n @test coerce(z, Multiclass) === z\n z = categorical(X.z, compress=true, ordered=false)\n @test coerce(z, Multiclass) === z\n z = categorical(X.z, compress=true, ordered=true)\n @test coerce(z, OrderedFactor) === z\n\n # missing values\n y_coerced = @test_logs(\n (:info, r\"Trying to coerce from `Union{Missing,\"),\n coerce([4, 7, missing], Continuous))\n @test ismissing(y_coerced == [4.0, 7.0, missing])\n @test scitype_union(y_coerced) === Union{Missing,Continuous}\n y_coerced = @test_logs(\n (:info, r\"Trying to coerce from `Any\"),\n coerce(Any[4, 7.0, missing], Continuous))\n @test ismissing(y_coerced == [4.0, 7.0, missing])\n @test scitype_union(y_coerced) === Union{Missing,Continuous}\n y_coerced = @test_logs(\n (:info, r\"Trying to coerce from `Union{Missing,\"),\n coerce([4.0, 7.0, missing], Count))\n @test ismissing(y_coerced == [4, 7, missing])\n @test scitype_union(y_coerced) === Union{Missing,Count}\n y_coerced = @test_logs(\n (:info, r\"Trying to coerce from `Any\"),\n coerce(Any[4, 7.0, missing], Count))\n @test ismissing(y_coerced == [4, 7, missing])\n @test scitype_union(y_coerced) === Union{Missing,Count}\n @test scitype_union(@test_logs(\n (:info, r\"Trying to coerce from `Union{Missing,\"),\n coerce(['x', 'y', missing], Multiclass))) ===\n Union{Missing, Multiclass{2}}\n @test scitype_union(@test_logs(\n (:info, r\"Trying to coerce from `Union{Missing,\"),\n coerce(['x', 'y', missing], OrderedFactor))) ===\n Union{Missing, OrderedFactor{2}}\n # non-missing Any vectors\n @test coerce(Any[4, 7], Continuous) == [4.0, 7.0]\n @test coerce(Any[4.0, 7.0], Continuous) == [4, 7]\n\n # Finite conversions:\n @test scitype_union(coerce(['x', 'y'], Finite)) === Multiclass{2}\n @test scitype_union(@test_logs(\n (:info, r\"Trying to coerce from `Union{Missing,\"),\n coerce(['x', 'y', missing], Finite))) === Union{Missing, Multiclass{2}}\n\n # More finite conversions (to check resolution of #48):\n y = categorical([1, 2, 3, missing]) # unordered\n yc = coerce(y, Union{Missing,OrderedFactor})\n @test isordered(yc)\n @test yc[1].pool.ordered\n @test scitype(yc) == Vec{Union{Missing, OrderedFactor{3}}}\n @test scitype_union(yc) == Union{Missing, OrderedFactor{3}}\n @test scitype_union(y) == Union{Missing, Multiclass{3}}\n\n # tests fix for issue https://github.com/JuliaAI/ScientificTypes.jl/issues/161\n X = (x=10:10:44, y=1:4, z=collect(\"abcd\"))\n Xc = coerce(X, :x => Continuous, \"y\" => Continuous)\n @test scitype_union(Xc.x) === Continuous\n @test scitype_union(Xc.y) === Continuous\nend\n\n# issue #62 (ScientficTypes)\n@testset \"coersion of Type=>Type\" begin\n X = (x=[1,2,1,2,5,1,0,7],\n y=[0,1,0,1,0,1,0,1],\n z=['a','b','a','b','a','a',missing,missing])\n Xc = coerce(X, :y=>OrderedFactor)\n Xc = coerce(Xc, Count=>Continuous)\n @test elscitype(Xc.x) == Continuous\n @test elscitype(Xc.y) == OrderedFactor{2}\n Xc = coerce(Xc, OrderedFactor=>Count)\n @test elscitype(Xc.y) == Count\n Xc = coerce(Xc, :z=>Multiclass, verbosity=0)\n Xc = coerce(Xc, Multiclass=>OrderedFactor, verbosity=0)\n @test elscitype(Xc.z) == Union{Missing,OrderedFactor{2}}\n Xc = coerce(X, Count=>Continuous, Unknown=>Multiclass, verbosity=0)\n @test elscitype(Xc.x) == Continuous\n @test elscitype(Xc.y) == Continuous\n @test elscitype(Xc.z) == Union{Missing, Multiclass{2}}\nend\n\n# issue #13\n@testset \"coersion of mixture of Type=>Type and Symbol=>Type\" begin\n X = (x=10:10:44, y=1:4, z=collect(\"abcd\"), w=[\"a\", \"b\", \"c\", missing])\n @test ScientificTypes.feature_scitype_pairs(:x => Continuous, X) ==\n [:x => Continuous, ]\n @test ScientificTypes.feature_scitype_pairs(Count => Continuous, X) ==\n [:x => Union{Continuous}, :y=> Union{Continuous}]\n X1 = coerce(X, :z => Multiclass, Count=>Continuous)\n @test schema(X1).scitypes ==\n (Continuous, Continuous, Multiclass{4}, Union{Missing,Textual})\n X2 = coerce(X, :z => Multiclass, :w=>Multiclass, Count=>Continuous,\n verbosity=0)\n @test schema(X2).scitypes ==\n (Continuous, Continuous, Multiclass{4}, Union{Missing,Multiclass{3}})\n @test_throws ArgumentError coerce(X, Count => Continuous, :x=>Multiclass)\nend\n\n@testset \"coerce!\" begin\n df = DataFrame((x=ones(Int,5), y=ones(5)))\n @test scitype(df) == Table{Union{AbstractArray{Continuous,1},\n AbstractArray{Count,1}}}\n coerce!(df, :x=>Continuous)\n @test scitype(df) == Table{AbstractArray{Continuous,1}}\n \n df = DataFrame((\n x=ones(Int, 50),\n y=ones(50),\n z=collect(\"abbabaacbcabbabaacbbcccbccbbabaaacacbcabcbccaabaaa\")\n ))\n @test scitype(df) == Table{Union{AbstractArray{Continuous,1},\n AbstractArray{Count,1},\n AbstractArray{Unknown,1}}}\n \n coerce!(df, autotype(df, :few_to_finite))\n @test scitype(df) == Table{Union{AbstractArray{Multiclass{3},1},\n AbstractArray{OrderedFactor{1},1}}}\n \n @test_throws ScientificTypes.CoercionError coerce!(randn(5, 5))\n\n df2 = DataFrame(x=[1,2,3,4],\n y=[\"a\",\"b\",\"c\",\"a\"],\n z = Union{Missing,Int}[10, 20, 30, 40])\n coerce!(df2, Textual=>Finite)\n coerce!(df2, Union{Missing, Count}=>Count, tight=true) # issue #166\n @test scitype(df2) == Table{Union{AbstractArray{Count,1},\n AbstractArray{Multiclass{3},1} }}\n \n # issue #9 (coerce Text => Num)\n df3 = DataFrame(x=[\"1\",\"2\",\"3\"], y=[2,3,4])\n coerce!(df3, Textual=>Count)\n @test scitype(df3) == Table{AbstractArray{Count,1}}\n \n x = [1,2,3,4]\n @test_throws ScientificTypes.CoercionError coerce!(x, Continuous)\n\nend\n", "meta": {"hexsha": "15e7ec4dc7a09adae3fdef58304e3bcfce7520ae", "size": 7465, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/coerce.jl", "max_stars_repo_name": "felixcremer/ScientificTypes.jl", "max_stars_repo_head_hexsha": "593c73f5a7c47a52d474bbbe08ea8865bf0dd625", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 45, "max_stars_repo_stars_event_min_datetime": "2021-06-17T16:58:25.000Z", "max_stars_repo_stars_event_max_datetime": "2022-02-17T16:24:37.000Z", "max_issues_repo_path": "test/coerce.jl", "max_issues_repo_name": "felixcremer/ScientificTypes.jl", "max_issues_repo_head_hexsha": "593c73f5a7c47a52d474bbbe08ea8865bf0dd625", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 46, "max_issues_repo_issues_event_min_datetime": "2021-06-14T00:19:06.000Z", "max_issues_repo_issues_event_max_datetime": "2022-03-28T01:47:27.000Z", "max_forks_repo_path": "test/coerce.jl", "max_forks_repo_name": "felixcremer/ScientificTypes.jl", "max_forks_repo_head_hexsha": "593c73f5a7c47a52d474bbbe08ea8865bf0dd625", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 5, "max_forks_repo_forks_event_min_datetime": "2021-06-17T16:26:57.000Z", "max_forks_repo_forks_event_max_datetime": "2022-03-09T15:57:25.000Z", "avg_line_length": 40.7923497268, "max_line_length": 82, "alphanum_fraction": 0.609912927, "num_tokens": 2372, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.5, "lm_q2_score": 0.14033624589467186, "lm_q1q2_score": 0.07016812294733593}}
{"text": "using Random\nexport customop,\nxavier_init,\nload_op_and_grad,\nload_op,\nuse_gpu,\ninstall,\nload_system_op,\nregister,\ndebug,\ndoctor,\nnnuq,\ncompile,\nlist_physical_devices,\nMCMCSimple,\nsimulate,\ndiagnose,\nget_placement,\ntimestamp,\nload_library,\nsleep_for,\nget_library_symbols\n\n\"\"\"\n xavier_init(size, dtype=Float64)\n\nReturns a matrix of size `size` and its values are from Xavier initialization. \n\"\"\"\nfunction xavier_init(size, dtype=Float64)\n in_dim = size[1]\n xavier_stddev = 1. / sqrt(in_dim / 2.)\n return randn(dtype, size...)*xavier_stddev\nend\n\n############### custom operators ##################\n\"\"\"\n cmake(DIR::String=\"..\"; CMAKE_ARGS::Union{Array{String}, String} = \"\")\n\nThe built-in Cmake command for building C/C++ libraries. If extra Cmake arguments are needed, please specify it through `CMAKE_ARGS`.\n\n# Example \n```\nADCME.cmake(CMAKE_ARGS=[\"SHARED=YES\", \"STAITC=NO\"])\n```\n\nThe executed command might be:\n```\n/home/darve/kailaix/.julia/adcme/bin/cmake -G Ninja -DCMAKE_MAKE_PROGRAM=/home/darve/kailaix/.julia/adcme/bin/ninja -DJULIA=/home/darve/kailaix/julia-1.3.1/bin/julia -DCMAKE_C_COMPILER=/home/darve/kailaix/.julia/adcme/bin/x86_64-conda_cos6-linux-gnu-gcc -DCMAKE_CXX_COMPILER=/home/darve/kailaix/.julia/adcme/bin/x86_64-conda_cos6-linux-gnu-g++ SHARED=YES STATIC=NO ..\n```\n\"\"\"\nfunction cmake(DIR::String=\"..\"; CMAKE_ARGS::Union{Array{String}, String} = \"\")\n DIR = abspath(DIR)\n ENV_ = copy(ENV)\n LD_PATH = Sys.iswindows() ? \"PATH\" : \"LD_LIBRARY_PATH\"\n if haskey(ENV_, LD_PATH)\n ENV_[LD_PATH] = ENV[LD_PATH]*\":$LIBDIR\"\n else\n ENV_[LD_PATH] = LIBDIR\n end\n\n if has_mpi(false)\n ENV_[\"MPI_INCLUDE_PATH\"], ENV_[\"MPI_C_LIBRARIES\"] = get_mpi()\n end\n\n if Sys.iswindows()\n if !haskey(ENV_, \"VS150COMNTOOLS\")\n # @warn \"VS150COMNTOOLS is not set, default to /c/Program Files (x86)/Microsoft Visual Studio/2017/Community/Common7/Tools\" maxlog=1\n ENV_[\"VS150COMNTOOLS\"] = \"/c/Program Files (x86)/Microsoft Visual Studio/2017/Community/Common7/Tools\"\n end\n # @info \"Do remember to add ADD_DEFINITIONS(-DNOMINMAX) to your CMakeLists.txt\" maxlog=1\n run(setenv(`$CMAKE -G\"Visual Studio 15\" -DJULIA=\"$(joinpath(Sys.BINDIR, \"julia\"))\" -A x64 $CMAKE_ARGS $DIR`, ENV_)) # very important, x64\n else\n run(setenv(`$CMAKE -G Ninja -DCMAKE_MAKE_PROGRAM=$NINJA -DJULIA=\"$(joinpath(Sys.BINDIR, \"julia\"))\" -DCMAKE_C_COMPILER=$CC -DCMAKE_CXX_COMPILER=$CXX $CMAKE_ARGS $DIR`, ENV_))\n end\nend\n\nfunction make()\n ENV_ = copy(ENV)\n LD_PATH = Sys.iswindows() ? \"PATH\" : \"LD_LIBRARY_PATH\"\n if haskey(ENV_, LD_PATH)\n ENV_[LD_PATH] = ENV[LD_PATH]*\":$LIBDIR\"\n else\n ENV_[LD_PATH] = LIBDIR\n end\n if Sys.iswindows()\n sln_file = filter(x->endswith(x, \".sln\"), readdir())\n if length(sln_file)==0\n error(\"No .sln file found. Did you run `ADCME.cmake()`?\")\n elseif length(sln_file)>1\n error(\"More than 1 .sln file found. Check your program.\")\n else\n sln_file = sln_file[1]\n end \n run(`cmd /c $CMAKE --build . -j --target ALL_BUILD --config Release`)\n else\n if haskey(ENV, \"TRAVIS_BRANCH\")\n run(setenv(`$NINJA -j1`, ENV_))\n else\n run(setenv(`$NINJA -j20`, ENV_))\n end\n end\nend\n\n\"\"\"\n make_library(Libdir::String)\n\nMake shared library in `Libdir`. The structure of the source codes files are \n\n```\n- Libdir \n - *.cpp \n - *.h \n - CMakeLists\n - build (Optional)\n```\n\"\"\"\nfunction make_library(Libdir::String)\n if !isdir(Libdir)\n error(\"$Libdir is not a valid directory.\")\n end\n PWD = pwd()\n cd(Libdir)\n if !isdir(\"build\")\n mkdir(\"build\")\n end\n cd(\"build\")\n if !isfile(\"Makefile\")\n ADCME.cmake()\n end\n ADCME.make()\n cd(PWD)\nend\n\nload_op_dict = Dict{Tuple{String, String}, PyObject}()\nload_op_grad_dict = Dict{Tuple{String, String}, PyObject}()\n\n@doc \"\"\"\n load_op(oplibpath::Union{PyObject, String}, opname::String; verbose::Union{Missing, Bool} = missing)\n\nLoads the operator `opname` from library `oplibpath`.\n\"\"\"\nfunction load_op(oplibpath::Union{PyObject, String}, opname::String; verbose::Union{Missing, Bool} = missing)\n verbose = coalesce(verbose, options.customop.verbose)\n oplibpath = get_library(oplibpath)\n if haskey(load_op_dict, (oplibpath,opname))\n return load_op_dict[(oplibpath,opname)]\n end\n s = getproperty(load_library(oplibpath), opname)\n load_op_dict[(oplibpath,opname)] = s\n verbose && printstyled(\"Load library operator: $oplibpath ==> $opname\\n\", color=:green)\n return s\nend\n\n@doc \"\"\"\n load_op_and_grad(oplibpath::Union{PyObject, String}, opname::String; multiple::Bool=false)\n\nLoads the operator `opname` from library `oplibpath`; gradients are also imported. \nIf `multiple` is true, the operator is assumed to have multiple outputs. \n\"\"\"\nfunction load_op_and_grad(oplibpath::Union{PyObject, String}, opname::String; \n multiple::Bool=false, verbose::Union{Missing, Bool} = missing)\n verbose = coalesce(verbose, options.customop.verbose)\n is_system_op = oplibpath == LIBADCME\n\n if isa(oplibpath, String)\n if Sys.iswindows()\n a, b = splitdir(oplibpath)\n if length(b) >=3 && b[1:3]==\"lib\"\n b = b[4:end]\n end\n oplibpath = joinpath(a, b)\n end\n if splitext(oplibpath)[2]==\"\"\n oplibpath = oplibpath * (Sys.islinux() ? \n \".so\" : Sys.isapple() ? \".dylib\" : \".dll\")\n end\n oplibpath = abspath(oplibpath)\n if haskey(load_op_grad_dict, (oplibpath,opname))\n return load_op_grad_dict[(oplibpath,opname)]\n end\n if !isfile(oplibpath)\n error(\"Library $oplibpath does not exist.\")\n end\n end\n\n opname_grad = opname*\"_grad\"\n fn_name = opname*randstring(8)\n try\n if is_system_op\npy\"\"\"\nimport tensorflow as tf\nlib$$fn_name = $libadcme\n\"\"\"\n elseif isa(oplibpath, String)\npy\"\"\"\nimport tensorflow as tf\nlib$$fn_name = tf.load_op_library($oplibpath)\n\"\"\"\n else \npy\"\"\"\nimport tensorflow as tf\nlib$$fn_name = $oplibpath\n\"\"\"\n end\nif !multiple\npy\"\"\"\n@tf.custom_gradient\ndef $$fn_name(*args):\n u = lib$$fn_name.$$opname(*args)\n def grad(dy):\n return lib$$fn_name.$$opname_grad(dy, u, *args)\n return u, grad\n\"\"\"\nelse\npy\"\"\"\n@tf.custom_gradient\ndef $$fn_name(*args):\n u = lib$$fn_name.$$opname(*args)\n def grad(*dy):\n dy = [y for y in dy if y is not None and y.dtype in [tf.float64, tf.float32]] # only float64 and float32 can backpropagate gradients\n return lib$$fn_name.$$opname_grad(*dy, *u, *args)\n return u, grad\n\"\"\"\nend\ncatch(e)\n printstyled(\"Failed load $oplibpath or its symbols. Error Message from the TensorFlow backend\\n$(string(e))\\n\", color=:red)\n Libdl.dlopen(oplibpath)\nend\n s = py\"$$fn_name\"\n if isa(oplibpath, String)\n load_op_grad_dict[(oplibpath,opname)] = s\n verbose && printstyled(\"Load library operator (with gradient, multiple outputs = $multiple): $oplibpath ==> $opname\\n\", color=:green)\n end\n return s\nend\n\n\n\"\"\"\n load_system_op(opname::String, grad::Bool=true; multiple::Bool=false)\n\nLoads custom operator from CustomOps directory (shipped with ADCME instead of TensorFlow)\nFor example \n```\ns = \"SparseOperator\"\noplib = \"libSO\"\ngrad = true\n```\nthis will direct Julia to find library `CustomOps/SparseOperator/libSO.dylib` on MACOSX\n\"\"\"\nfunction load_system_op(opname::String, grad::Bool=true; multiple::Bool=false)\n if !isfile(LIBADCME)\n @info \"$LIBADCME does not exist. Precompiling...\"\n ADCME.precompile()\n end\n if grad\n load_op_and_grad(LIBADCME, opname; multiple=multiple, verbose=false)\n else\n load_op(LIBADCME, opname, verbose=false)\n end\nend\n\n\"\"\"\n compile(s::String; force::Bool=false)\n\nCompiles the library given by path `deps/s`. If `force` is false, `compile` first check whether \nthe binary product exists. If the binary product exists, return 2. Otherwise, `compile` tries to \ncompile the binary product, and returns 0 if successful; it return 1 otherwise. \n\"\"\"\nfunction compile(s::String; force::Bool=false, customdir::Bool = false)\n PWD = pwd()\n dir = s \n if !customdir\n dir = joinpath(joinpath(\"$(@__DIR__)\", \"../deps/CustomOps\"), s)\n end\n if !isdir(dir)\n @warn(\"Folder for the operator $s does not exist: $dir\")\n return 1\n end\n cd(dir)\n \n local surfix \n if Sys.isapple()\n surfix = \".dylib\"\n elseif Sys.islinux()\n surfix = \".so\"\n elseif Sys.iswindows()\n surfix = \".dll\"\n end\n if !force && isdir(\"build\") # check if product exists \n files = readdir(\"build\")\n if any([endswith(x, surfix) for x in files])\n @warn(\"The binary product exists.\")\n cd(PWD)\n return 2\n end\n end\n rm(\"build\",force=true,recursive=true)\n mkdir(\"build\")\n cd(\"build\")\n try\n cmake()\n make()\n cd(PWD)\n return 0\n catch e \n error(\"Compilation error: $e\")\n cd(PWD)\n return 1\n end\nend\n\n\"\"\"\n precompile(force::Bool=false)\n\nPrecompile the built-in custom operators. \n\"\"\"\nfunction Base.:precompile(force::Bool=false)\n PWD = pwd()\n cd(\"$(@__DIR__)/../deps/CustomOps\")\n if force\n try\n rm(\"build\", force=true, recursive=true)\n catch \n error(\"\"\"Failed to remove build directory. Follow the following steps and try again:\n1. Quit ALL julia processes that use ADCME;\n2. Remove $(joinpath(pwd(), \"build\")) manually.\"\"\")\n end\n end\n change_directory(\"build\")\n require_cmakecache() do \n if Sys.isapple()\n try\n ADCME.cmake()\n catch \n @info \"Use system clang...\"\n mv(joinpath(BINDIR, \"clang\"), joinpath(BINDIR, \"clang_original\"))\n mv(joinpath(BINDIR, \"clang++\"), joinpath(BINDIR, \"clang++_original\"))\n symlink(\"/usr/bin/clang\",joinpath(BINDIR, \"clang\"))\n symlink(\"/usr/bin/clang++\",joinpath(BINDIR, \"clang++\"))\n ADCME.cmake()\n end\n else\n ADCME.cmake()\n end\n end\n require_library(\"adcme\") do \n ADCME.make()\n end\n cd(PWD)\n global libadcme = tf.load_op_library(LIBADCME)\nend\n\n\"\"\"\n compile()\n\nCompile a custom operator in the current directory. A `CMakeLists.txt` must be present. \n\"\"\"\nfunction compile()\n PWD = pwd()\n if !isfile(\"CMakeLists.txt\")\n error(SystemError(\"No CMakeLists.txt in the current directory found.\"))\n end\n if !isdir(\"build\")\n mkdir(\"build\")\n end\n cd(\"build\")\n try \n cmake()\n make()\n catch e\n @warn \"Compiling failed: $e\"\n finally\n cd(PWD)\n end\nend\n\n\n\"\"\"\n customop(;with_mpi::Bool = false)\n\nCreate a new custom operator. Typically users call `customop` twice: the first call generates a `customop.txt`, \nusers edit the content in the file; the second all generates C++ source code, CMakeLists.txt, and gradtest.jl from `customop.txt`.\n\n# Example\n```julia-repl\njulia> customop() # create an editable `customop.txt` file\n[ Info: Edit custom_op.txt for custom operators\njulia> customop() # after editing `customop.txt`, call it again to generate interface files.\n```\n\n# Options \n- `with_mpi`: Whether the custom operator uses MPI\n\"\"\"\nfunction customop(;with_mpi::Bool = false)\n # install_custom_op_dependency()\n py_dir = \"$(@__DIR__)/../deps/CustomOpsTemplate\"\n if !(\"custom_op.txt\" in readdir(\".\"))\n cp(\"$(py_dir)/custom_op.example\", \"custom_op.txt\")\n @info \"Edit custom_op.txt for custom operators\"\n return\n else\n python = PyCall.python\n with_mpi = with_mpi ? 1 : 0\n run(`$python $(py_dir)/customop.py custom_op.txt $py_dir $with_mpi`)\n end\n nothing\nend\n\n\n\nfunction use_gpu(i::Union{Nothing,Int64}=nothing)\n if length(CUDA_INC)==0\n error(\"\"\"ADCME is not built against GPU. Set ENV[\"GPU\"]=1 and rebuild GPU.\"\"\")\n end\n dl = pyimport(\"tensorflow.python.client.device_lib\")\n if !isnothing(i) && i>=1\n i = join(collect(0:i-1),',') \n ENV[\"CUDA_VISIBLE_DEVICES\"] = i \n elseif !isnothing(i) && i==0\n ENV[\"CUDA_VISIBLE_DEVICES\"] = \"\"\n end\n local_device_protos = dl.list_local_devices()\n return [x.name for x in local_device_protos if x.device_type == \"GPU\"]\nend\n\nfunction list_physical_devices(cpu_or_gpu::String = \"all\")\n dl = pyimport(\"tensorflow.python.client.device_lib\")\n local_device_protos = dl.list_local_devices()\n CPU = [x.name for x in local_device_protos if x.device_type == \"CPU\"]\n GPU = [x.name for x in local_device_protos if x.device_type == \"GPU\"]\n if cpu_or_gpu == \"all\"\n return [CPU;GPU]\n elseif cpu_or_gpu == \"GPU\"\n return GPU \n elseif cpu_or_gpu == \"CPU\"\n return CPU \n else\n error(ArgumentError(\"$cpu_or_gpu is not a valid input. Expected: all, CPU, or GPU\"))\n end\nend\n\n\n\"\"\"\n install(s::String; force::Bool = false, islocal::Bool = false)\n\nInstall a custom operator from a URL, a directory (when `islocal` is true), or a string. In any of the three case, \n`install` copy the folder to $(abspath(joinpath(LIBADCME, \"../../Plugin\"))). \nWhen `s` is a string, `s` is converted to \n\nhttps://github.com/ADCMEMarket/\n\"\"\"\nfunction install(s::String; force::Bool = false, islocal::Bool = false)\n if !islocal && !startswith(s, \"http\")\n s = \"https://github.com/ADCMEMarket/\"*s\n end \n _, name = splitdir(s)\n if force \n rm(joinpath(LIBPLUGIN, name), force=true, recursive=true)\n elseif isdir(joinpath(LIBPLUGIN, name)) && (\"build\" in readdir(joinpath(LIBPLUGIN, name)))\n return _plugin_lib(joinpath(LIBPLUGIN, name))\n end\n try\n LibGit2.clone(s, joinpath(LIBPLUGIN, name))\n catch\n LibGit2.clone(\"git://$(s[9:end]).git\", joinpath(LIBPLUGIN, name))\n end\n PWD = pwd()\n cd(joinpath(LIBPLUGIN, name))\n if isfile(joinpath(LIBPLUGIN, name, \"build.jl\"))\n include(joinpath(LIBPLUGIN, name, \"build.jl\"))\n end\n cmakelists = String(read(\"CMakeLists.txt\"))\n if !occursin(\"cmake_minimum_required\", cmakelists)\n cmakelists = replace(String(read(joinpath(LIBPLUGIN, \"CMakeLists.txt\"))), \"[INSTRUCTION]\"=>cmakelists)\n end\n open(\"CMakeLists.txt\", \"w\") do io \n write(io, cmakelists)\n end\n mkdir(\"build\")\n cd(\"build\")\n ADCME.cmake()\n ADCME.make()\n cd(PWD) \n return _plugin_lib(joinpath(LIBPLUGIN, name))\nend\n\nfunction _plugin_lib(D)\n files = readdir(joinpath(D, \"build\"))\n dylib = filter(x->endswith(x, \".$dlext\"), files)\n if length(dylib)==0\n error(SystemError(\"No dynamic library found.\"))\n elseif length(dylib)>1\n error(SystemError(\"More then one dynamic libraries found.\"))\n else\n return joinpath(D, \"build\", dylib[1])\n end\nend\n\n@doc raw\"\"\"\n register(forward::Function, backward::Function; multiple::Bool=false)\n\nRegister a function `forward` with back-propagated gradients rule `backward` to the backward. \n\u2218 `forward`: it takes $n$ inputs and outputs $m$ tensors. When $m>1$, the keyword `multiple` must be true. \n\u2218 `backward`: it takes $\\tilde m$ top gradients from float/double output tensors of `forward`, $m$ outputs of the `forward`, \n and $n$ inputs of the `forward`. `backward` outputs $n$ gradients for each input of `forward`. When input $i$ of\n `forward` is not float/double, `backward` should return `nothing` for the corresponding gradients. \n \n# Example \n```julia\nforward = x->log(1+exp(x))\nbackward = (dy, y, x)->dy*(1-1/(1+y))\nf = register(forward, backward)\n```\n\"\"\"\nfunction register(forward::Function, backward::Function; multiple::Bool=false)\n fn_name = \"customgrad_\"*randstring(8)\n if !multiple\npy\"\"\"\nimport tensorflow as tf\n@tf.custom_gradient\ndef $$fn_name(*args):\n u = $forward(*args)\n def grad(dy):\n return $backward(dy, u, *args)\n return u, grad\n\"\"\"\n else\npy\"\"\"\nimport tensorflow as tf\n@tf.custom_gradient\ndef $$fn_name(*args):\n u = forward_$$fn_name(*args)\n def grad(*dy):\n dy = [y for y in dy if y is not None and y.dtype in [tf.float64, tf.float32]] # only float64 and float32 can backpropagate gradients\n return backward_$$fn_name(*dy, *u, *args)\n return u, grad\n\"\"\"\n end\n return py\"$$fn_name\"\nend\n\n\"\"\"\n debug(sess::PyObject, o::PyObject)\n\nIn the case a session run yields an error from the TensorFlow backend, this function can help print the exact error. \nFor example, you might encounter `InvalidArgumentError()` with no detailed error information, and this function can be useful for debugging.\n\"\"\"\nfunction debug(sess::PyObject, o::PyObject)\npy\"\"\"\nimport tensorflow as tf\nimport traceback\ntry:\n $sess.run($o)\nexcept Exception:\n print(traceback.format_exc())\n\"\"\"\nend\n\n\"\"\"\n debug(libfile::String = \"\")\n\nLoading custom operator shared library. If the loading fails, detailed error message is printed.\n\"\"\"\nfunction debug(libfile::String = \"\")\n if libfile==\"\"\n libfile = LIBADCME\n end\npy\"\"\"\nimport tensorflow as tf\nimport traceback\ntry:\n tf.load_op_library($libfile)\nexcept Exception:\n print(traceback.format_exc())\n\"\"\"\nend\n\n\"\"\"\n doctor()\n\nReports health of the current installed ADCME package. If some components are broken, possible fix is proposed.\n\"\"\"\nfunction doctor()\n function yes(name)\n printstyled(\"[\u2714\ufe0f] $name\\n\", color=:green, bold=true)\n end\n function no(name, diagnose, instruction)\n printstyled(\"[\u2718] $name\\n\", color=:red, bold=true)\n printstyled(\"\\n[Reason]\\n\", color=:magenta)\n printstyled(\"$diagnose\\n\\n\", color=:blue)\n printstyled(\"\\n[Instruction]\\n\", color=:magenta)\n printstyled(\"$instruction\\n\\n\", color=:blue)\n end\n\n c = true \n if VERSION>=v\"1.4\" && VERSION=6) && (tf.__version__[1:6]==\"1.15.0\")\n if c \n yes(\"TensorFlow version\")\n else\n no(\"TensorFlow version\", \n\"\"\"Your TensorFlow version is $(tf.__version__). The TensorFlow version shipped with ADCME is 1.15.0.\"\"\",\n\"\"\"Set ENV[\"FORCE_REINSTALL_ADCME\"] = 1 and rebuild ADCME\njulia> ENV[\"FORCE_REINSTALL_ADCME\"] = 1\njulia> ]\npkg> build ADCME\"\"\")\n end \n\n\n\n c = (PyCall.python==ADCME.PYTHON)\n if c \n yes(\"Python executable file\")\n else\n no(\"Python executable file\", \n\"\"\"PyCall Python path $(PyCall.python) does not match the ADCME-compatible Python $(ADCME.PYTHON)\"\"\",\n\"\"\"Rebuild PyCall with a compatible Python version:\n\nusing Pkg\nENV[\"PYTHON\"] = \"$(ADCME.PYTHON)\"\nPkg.build(\"PyCall\")\n\"\"\")\n end \n\n c = true \n try \n if Sys.iswindows()\n run(`cmd /c where julia`)\n else \n read(`which julia`)\n end\n catch\n c = false\n end\n\n if c \n yes(\"Julia path\")\n else\n no(\"Julia path (Optional)\", \n\"\"\"`julia` outputs nothing. This will break custom operator compilation.\"\"\",\n\"\"\"Add your julia binary path to your environment path, e.g. (Unix systems) \n\nexport PATH=$(Sys.BINDIR):\\$PATH\n\nFor convenience, you can add the above line to your `~/.bashrc` (Linux) or `~/.bash_profile` (Apple).\nFor Windows, you need to add it to system environment.\"\"\")\n end\n\n c = Sys.iswindows() ?\n haskey(ENV, \"PATH\") && occursin(ADCME.LIBDIR, ENV[\"PATH\"]) :\n haskey(ENV, \"LD_LIBRARY_PATH\") && occursin(ADCME.LIBDIR, ENV[\"LD_LIBRARY_PATH\"])\n if c \n yes(\"Dynamic library path\")\n else\n no(\"Dynamic library path (Optional)\", \n\"\"\"$(ADCME.LIBDIR) is not in LD_LIBRARY_PATH. This MAY break custom operator compilation. However, in most cases, ADCME automatic fixes this problem for you.\"\"\",\n\"\"\"Add your dynamic library path path to your environment path, e.g. (Unix systems) \n\nexport LD_LIBRARY_PATH=$(ADCME.LIBDIR):\\$LD_LIBRARY_PATH\n\nFor convenience, you can add the above line to your `~/.bashrc` (Linux or Apple).\nFor Windows, you need to add it to PATH instead of LD_LIBRARY_PATH.\"\"\")\n end\n\n c = Sys.WORD_SIZE==64\n if c \n yes(\"Memory Address Length = 64\")\n else \n no(\"Memory Address Length\",\n\"\"\"Your memory address length is $(Sys.WORD_SIZE). ADCME is only tested against 64-bit machine.\"\"\",\n\"\"\"If you do not need custom operators, then it's fine. Otherwise you need to switch to a 64-bit machine\"\"\")\n end\n\n if Sys.iswindows()\n c = isfile(ADCME.MAKE*\".exe\") && occursin(\"15\", (ADCME.MAKE)) && occursin(\"2017\", ADCME.MAKE)\n if c \n yes(\"C Compiler\")\n else\n no(\"C Compiler\", \n\"\"\"You specified that the C compiler for custom operators is \n$(ADCME.MAKE)\nHowever, one of the following requirements is not met: \n1*. The file you specified $(ADCME.MAKE*\".exe\") does not exist.\n2**. (Optional) For compatibility, we suggest you use Microsoft Visual Studio 2017 (Version number: 15).\n\n* The path is actually not needed in compilation, but we raise such an issue here in case you obtain some compilation errors in the future.\n\n* We check the version by looking for \"15\" and \"2017\" in the path specification. If you are sure your compiler is correct, you can ignore this message. \"\"\",\n\"\"\"Manually edit $(abspath(joinpath(splitdir(pathof(ADCME))[1], \"../deps/deps.jl\"))) and modify `MAKE` to be the correct compiler.\"\"\")\n end \n\n end\n \n \n\n c = haskey(ENV, \"PATH\") && occursin(ADCME.BINDIR, ENV[\"PATH\"])\n if c \n yes(\"Binaries path\")\n else\n no(\"Binaries path\", \n\"\"\"$(ADCME.BINDIR) is not in PATH. This path contains compatible tools such as a GCC compiler, `cmake`, `make`, or any other tools you want to use directly from terminal.\nHowever, setting the path is NOT a requirement, and ADCME works totally fine without any action.\"\"\",\n\"\"\"(Optional) Add your binary path to your environment path, e.g. (Unix systems) \n\nexport PATH=$(ADCME.BINDIR):\\$PATH\n\nFor convenience, you can add the above line to your `~/.bashrc` (Linux) or `~/.bash_profile` (Apple).\nFor Windows, you need to add it to system environment.\"\"\")\n end\n\n if length(ADCME.CUDA_INC)>0\n c = Sys.iswindows() ?\n haskey(ENV, \"PATH\") && occursin(ADCME.LIBCUDA, ENV[\"PATH\"]) :\n haskey(ENV, \"LD_LIBRARY_PATH\") && occursin(ADCME.LIBCUDA, ENV[\"LD_LIBRARY_PATH\"])\n if c \n yes(\"CUDA LD_LIBRARY_PATH\")\n else\n no(\"CUDA LD_LIBRARY_PATH\", \n \"\"\"$(ADCME.LIBCUDA) is not in LD_LIBRARY_PATH. This path contains compatible tools such as a GCC compiler, `cmake`, `make`, etc.\"\"\",\n \"\"\"The fix is OPTIONAL.\n Add your binary path to your environment path, e.g. (Unix systems) \n \n export LD_LIBRARY_PATH=$(ADCME.LIBCUDA):\\$LD_LIBRARY_PATH\n \n For convenience, you can add the above line to your `~/.bashrc` (Linux or Apple).\n For Windows, you need to add it to PATH instead of LD_LIBRARY_PATH.\"\"\")\n end\n\n try \n if !Sys.iswindows()\n Libdl.dlpath(\"libcuda\")\n Libdl.dlpath(\"libcudnn\")\n Libdl.dlpath(\"libcublas\")\n else\n Libdl.dlpath(\"cudart64_100\")\n Libdl.dlpath(\"cudnn64_7\")\n Libdl.dlpath(\"cublas64_100\")\n end\n yes(\"CUDA Shared Library\")\n catch\n no(\"CUDA Shared Library\", \n \"\"\"libcuda, libcudnn, and (or) libcublas can not be loaded.\"\"\",\n \"\"\"If you intend to use GPU, this fix is mandatory. Make sure cudatoolkit and cudnn libraries can be found in\n$(ADCME.LIBCUDA)\nand `nvcc` is in your path.\"\"\")\n end\n\n c = isdir(ADCME.CUDA_INC) && \"cuda.h\" in readdir(ADCME.CUDA_INC)\n if c \n yes(\"CUDA Header Files\")\n\n if !isfile(joinpath(ADCME.TF_INC, \"third_party/gpus/cuda/include/cuda_fp16.h\"))\n println(\"Fixing third_party/gpus/cuda/include...\")\n if !ispath(joinpath(ADCME.TF_INC, \"third_party/gpus/cuda/\"))\n mkpath(joinpath(ADCME.TF_INC, \"third_party/gpus/cuda/\"))\n end\n rm(joinpath(ADCME.TF_INC, \"third_party/gpus/cuda/include/\"), force=true, recursive=true)\n symlink(ADCME.CUDA_INC, joinpath(ADCME.TF_INC, \"third_party/gpus/cuda/include\"))\n end\n\n else \n no(\"CUDA Header Files\",\n \"\"\"Cuda include library does not exist or `cuda.h` is missing.\"\"\",\n \"\"\"It might be possible that your cuda include library is located somewhere else other than $(ADCME.CUDA_INC). Fix the dependency file.\"\"\")\n end\n else\n no(\"GPU Support (Optional)\", \n \"\"\"ADCME is not compiled against GPU.\"\"\",\n \"\"\"If you intend to use GPU, set ENV[\"GPU\"] = 1 and then rebuild ADCME.\"\"\")\n end\n\n \n depsfile = abspath(joinpath(@__DIR__, \"../deps/deps.jl\"))\n println(\"============================================================\\n$(depsfile)\\n============================================================\")\n println(read(depsfile, String))\n\n @assert isdir(ADCME.BINDIR)\n @assert isdir(ADCME.LIBDIR)\n @assert isdir(ADCME.TF_INC)\n @assert isdir(ADCME.PREFIXDIR)\n if !Sys.iswindows()\n @assert isfile(ADCME.CC) || islink(ADCME.CC)\n @assert isfile(ADCME.CXX) || islink(ADCME.CXX)\n @assert isfile(ADCME.NINJA)\n else \n @assert isfile(ADCME.MAKE)\n end\n @assert isfile(ADCME.CMAKE)\n @assert isfile(ADCME.TF_LIB_FILE) || islink(ADCME.TF_LIB_FILE)\n @assert isdir(ADCME.INCDIR)\nend\n\n\n\"\"\"\n test_gpu()\n\nTests the GPU ultilities\n\"\"\"\nfunction test_gpu()\n PWD = pwd()\n run(`which nvcc`)\n cd(\"$(@__DIR__)/../examples/gpu_custom_op\")\n mkdir(\"build\")\n cd(\"build\")\n ADCME.cmake()\n ADCME.make()\n cd(\"..\")\n include(\"gputest.jl\")\n cd(PWD)\nend\n\n\n@doc raw\"\"\"\n nnuq(H::Array{Float64,2}, invR::Union{Float64, Array{Float64,2}}, invQ::Union{Float64, Array{Float64,2}})\n\nReturns the variance matrix for the Baysian inversion. \n\nThe negative log likelihood function is\n\n$$l(s) =\\frac{1}{2} (y-h(s))^T R^{-1} (y-h(s)) + \\frac{1}{2} s^T Q^{-1} s$$\n\nThe covariance matrix is computed by first linearizing $h(s)$\n\n$$h(s)\\approx h(s_0) + \\nabla h(s_0) (s-s_0)$$\n\nand then computing the second order derivative\n\n$$V = \\left(\\frac{\\partial^2 l}{\\partial s^T\\partial s}\\right)^{-1} = (H^T R^{-1} H + Q^{-1})^{-1}$$\n\nNote the result is independent of $s_0$, $y_0$, and only depends on $\\nabla h(s_0)$\n\"\"\"\nfunction nnuq(H::Array{Float64,2}, invR::Union{Float64, Array{Float64,2}}, invQ::Union{Float64, Array{Float64,2}})\n if isa(invQ, Float64)\n invQ = invQ * I \n end\n \u03a3 = inv(H' * invR * H + invQ)\n (\u03a3 + \u03a3')/2\nend\n\n\n\nmutable struct MCMCSimple\n logf::Function\n proposal::Function\n \u03b80::Array{Float64, 1}\n ub::Float64\n lb::Float64 \n L::Array{Float64, 1}\n AC::Array{Float64, 1}\nend\n\n@doc raw\"\"\"\n MCMCSimple(obs::Array{Float64, 1}, h::Function, \n \u03c3::Float64, \u03b80::Array{Float64,1}, lb::Float64, ub::Float64)\n\nA very simple yet useful interface for MCMC simulation in many scientific computing problems. \n\n- `obs`: Observations\n- `h`: Forward computation function\n- `\u03c3`: Noise standard deviation for the observed data \n- `ub`, `lb`: upper and lower bound\n- `\u03b80`: Initial guess \n\nThe mathematical model is \n\n```math\ny_{obs} = h(\\theta)\n```\n\nand we have a hard constraint `lb\\leq \\theta \\leq ub`. \n\"\"\"\nfunction MCMCSimple(obs::Array{Float64, 1}, h::Function, \n \u03c3::Float64, \u03b80::Array{Float64,1}, lb::Float64, ub::Float64,\n \u03b4::Union{Missing, Float64}=missing)\n \u03c4 = (ub-lb)/6\n \u03b4 = coalesce(\u03b4, (ub-lb)/100)\n function logf(x)\n -sum((obs - h(x)).^2/2\u03c3^2) - sum((x-\u03b80).^2)/2\u03c4^2\n end\n function proposal(x)\n x + (rand(length(x)) .- 0.5)*2 * \u03b4\n end\n MCMCSimple(logf, proposal, \u03b80, ub, lb, zeros(0), zeros(0))\nend\n\nfunction simulate(ms::MCMCSimple, N::Int64, burnin::Union{Int64, Missing} = missing)\n burnin = coalesce(burnin, Int64(round(N*0.2)))\n sim = zeros(N, length(ms.\u03b80))\n sim[1,:] = ms.\u03b80\n L = zeros(N)\n AC = ones(N)\n AC[1] = NaN\n L[1] = ms.logf(ms.\u03b80)\n k = 1\n for i = 2:N \n sim[i,:], L[i], k_ = _MCMCSimple_simulate(ms, sim[i-1,:])\n k += k_ \n AC[i] = k/i\n end\n ms.AC = AC\n ms.L = L \n return sim\nend\n\nfunction diagnose(ms::MCMCSimple)\n if !isdefined(Main, :PyPlot)\n error(\"Package PyPlot.jl must be imported in the main module using `import PyPlot` or `using PyPlot`\")\n end\n Main.PyPlot.figure(figsize = (10, 4))\n Main.PyPlot.subplot(121)\n Main.PyPlot.title(\"Acceptance Rate\")\n Main.PyPlot.plot(ms.AC)\n Main.PyPlot.ylim(0,1.05)\n Main.PyPlot.subplot(122)\n Main.PyPlot.title(\"Log Likelihood\")\n Main.PyPlot.plot(ms.L)\nend\n\nfunction _MCMCSimple_simulate(ms::MCMCSimple, x::Array{Float64})\n local x_star\n while true\n x_star = ms.proposal(x)\n if all(x_star.<=ms.ub) && all(x_star.>=ms.lb)\n break\n end\n end\n \u0394 = ms.logf(x_star) - ms.logf(x)\n if log(rand())<\u0394\n return x_star, ms.logf(x_star), 1\n else \n return x, ms.logf(x), 0\n end\nend\n\n\n\n\"\"\"\n get_placement()\n\nReturns the operation placements.\n\"\"\"\nfunction get_placement()\n sess = Session(config=tf.ConfigProto(log_device_placement=true))\n originalSTDOUT = stdout\n (outRead, outWrite) = redirect_stdout()\n init(sess)\n close(outWrite)\n data = readavailable(outRead)\n close(outRead)\n redirect_stdout(originalSTDOUT)\n lines = split(String(data), '\\n')[1:end-1]\nend\n\n\"\"\"\n sleep_for(t::Union{PyObject, <:Real})\n\nSleeps for `t` seconds. \n\"\"\"\nfunction sleep_for(t::Union{PyObject, <:Real})\n sleep_for_ = load_system_op(\"sleep_for\", false)\n sleep_for_(convert_to_tensor(t, dtype=Float64))\nend\n\n\"\"\"\n timestamp(deps::Union{PyObject, <:Real, Missing}=missing)\n\nThese functions are usually used with [`bind`](@ref) for profiling. \nNote the timing is not very accurate in a multithreaded environment.\n\n- `deps`: `deps` is always executed before returning the timestamp.\n# Example \n```julia\na = constant(3.0)\nt0 = timestamp(a)\nsleep_time = sleep_for(a)\nt1 = timestamp(sleep_time)\nsess = Session(); init(sess)\nt0_, t1_ = run(sess, [t0, t1])\ntime = t1_ - t0_\n```\n\"\"\"\nfunction timestamp(deps::Union{PyObject, <:Real, Missing}=missing)\n deps = coalesce(deps, 0.0)\n deps = convert_to_tensor(deps, dtype = Float64)\n timer_ = load_system_op(\"timer\", false)\n timer_(deps)\nend\n\n\"\"\"\n load_library(filename::String)\n\nLoad custom operator libraries. If used with \n\"\"\"\nfunction load_library(filename::String)\n filename = get_library(filename)\n keyname = \"custom_op_library_\"*filename\n if haskey(STORAGE, keyname)\n return STORAGE[keyname]\n end\n if !isfile(filename)\n error(\"File $filename not found. If you intend to load a library by absolute path, try `tf.load_op_library(filename)`.\")\n end\n try\n STORAGE[keyname] = tf.load_op_library(filename)\n catch e \n error(\"Failed to load library: $filename. Original error message:\\n$e\")\n end\n return STORAGE[keyname] \nend\n\n\"\"\"\n get_library_symbols(file::Union{String, PyObject})\n\nReturns the symbols in the custom op library `file`.\n\"\"\"\nfunction get_library_symbols(files::Union{String, PyObject})\n if isa(files, String)\n files = load_library(files)\n end\n files = keys(files)\n filter(x->islowercase(String(x)[1]) && String(x)!=\"tf_export\" && !(occursin(\"fallback\", String(x))) && String(x)!=\"deprecated_endpoints\", files)\nend\n\n\nfunction Base.:NamedTuple(df::PyObject)\n names = tuple(Symbol.(df.index.tolist())...)\n vals = tuple(df.values...)\n NamedTuple{names}(vals)\nend", "meta": {"hexsha": "f0edd25210b4f9549c64c558720ade3ee2fa3dbc", "size": 31973, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/extra.jl", "max_stars_repo_name": "ziyiyin97/ADCME.jl", "max_stars_repo_head_hexsha": "1c9b2c1ae63059d79a5a6a7b86eee64796868755", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/extra.jl", "max_issues_repo_name": "ziyiyin97/ADCME.jl", "max_issues_repo_head_hexsha": "1c9b2c1ae63059d79a5a6a7b86eee64796868755", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/extra.jl", "max_forks_repo_name": "ziyiyin97/ADCME.jl", "max_forks_repo_head_hexsha": "1c9b2c1ae63059d79a5a6a7b86eee64796868755", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 30.7432692308, "max_line_length": 367, "alphanum_fraction": 0.6344102837, "num_tokens": 8986, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4263215925474903, "lm_q2_score": 0.16451645470385745, "lm_q1q2_score": 0.07013691696961556}}
{"text": "module errors\ninclude(\"_errors.jl\")\n\nimport Base: InterpreterIP, show_method_candidates, ExceptionStack\n\nimport Term:\n theme, highlight, reshape_text, read_file_lines, load_code_and_highlight, split_lines\nimport Term.style: apply_style\nimport ..panel: Panel, TextBox\nimport ..renderables: RenderableText\nimport ..layout: hLine, Spacer\nimport ..consoles: Console\nimport ..measure: Measure\n\nexport install_stacktrace\n\nconst ErrorsExplanations = Dict(\n ArgumentError => \"The parameters to a function call do not match a valid signature.\",\n AssertionError => \"comes up when an assertion's check fails (e.g., `@assert 1==2`)\",\n BoundsError => \"comes up when trying to acces a container at invalid position (e.g., a string a='abcd' with 4 characters cannot be accessed as a[5]).\",\n DimensionMismatch => \"comes up when trying to perform an operation on objects which don't have matching dimensionality (e.g., summing matrixes of different size).\",\n DivideError => \"comes up when attempting integer division with 0 as denominator. [blue]2/0=Inf[/blue] is okay, but [orange1]div(2, )[/orange1] will give an error\",\n DomainError => \"comes up when the argument to a function is outside its domain (e.g., \u221a(-1))\",\n ErrorException => \"is a generic error type\",\n KeyError => \"comes up when attempting to access a non-existing [blue]Dict[/blue] key.\",\n InexactError => \"comes up when a type cannot exactly be converted to another (e.g. Int(2.5) cannot convert Float64 to Int64, but Int(round(2.5)) will work)\",\n LoadError => \"occurs when another comes up while evaluating 'include', 'require' or 'using' statements\",\n MethodError => \"comes up when to method can be found with a given name and for a given set of argument types.\",\n StackOverflowError => \"usually comes up when functions call each other recursively.\",\n TypeError => \"is a type assertion failure, or calling an intrinsic function with an incorrect argument type.\",\n UndefKeywordError => \"comes up when a function has a keyword argument with no default value and no value is passed to a function call\",\n UndefVarError => \"comes up when a variable is used which is either not defined, or, which is not visible in the current variables scope (e.g.: variable defined in function A and used in function B)\",\n)\n\n_width() = min(Console(stderr).width, 100)\n\n# ----------------------- error type specific messages ----------------------- #\n\n# ! ARGUMENT ERROR\nfunction error_message(io::IO, er::ArgumentError)\n return er.msg, \"\"\nend\n\n# ! ASSERTION ERROR\nfunction error_message(io::IO, er::AssertionError)\n return er.msg, \"\"\nend\n\n# ! BOUNDS ERROR\nfunction error_message(io::IO, er::BoundsError)\n # @info \"bounds error\" er fieldnames(typeof(er))\n main_msg = \"Attempted to access $(_highlight_with_type(er.a)) at index $(_highlight_with_type(er.i))\"\n\n additional_msg = \"\"\n\n if isdefined(er, :a)\n if er.a isa AbstractString\n nunits = ncodeunits(er.a)\n additional_msg = \"S\\ntring has $nunits codeunits, $(length(er.a)) characters.\"\n end\n else\n additional_msg = \"\\n[red]Variable is not defined!.[/red]\"\n end\n return main_msg, additional_msg\nend\n\n# ! Domain ERROR\nfunction error_message(io::IO, er::DomainError)\n # @info \"err exceprion\" er fieldnames(DomainError) er.val\n # msg = split(er.msg, \" around \")[1]\n return er.msg, \"\\nThe invalid value is: $(_highlight_with_type(er.val)).\"\nend\n\n# ! DimensionMismatch\nfunction error_message(io::IO, er::DimensionMismatch)\n return _highlight_numbers(er.msg), \"\"\nend\n\n# ! DivideError\nfunction error_message(io::IO, er::DivideError)\n return \"Attempted integer division by [blue]0[/blue]\", \"\"\nend\n\n# ! EXCEPTION ERROR\nfunction error_message(io::IO, er::ErrorException)\n # @info \"err exceprion\" er fieldnames(ErrorException) er.msg\n msg = split(er.msg, \" around \")[1]\n return msg, \"\"\nend\n\n# ! KeyError\nfunction error_message(io::IO, er::KeyError)\n # @info \"err KeyError\" er fieldnames(KeyError)\n msg = \"Key $(_highlight_with_type(er.key)) not found!\"\n return msg, \"\"\nend\n\n# ! InexactError\nfunction error_message(io::IO, er::InexactError)\n # @info \"load error message\" fieldnames(InexactError)\n msg = \"Cannot convert $(_highlight_with_type(er.val)) to type [$(theme.type)]$(er.T)[/$(theme.type)]\"\n subm = \"\\nConversion error in function: $(_highlight(er.func))\"\n return msg, subm\nend\n\n# ! LoadError\nfunction error_message(io::IO, er::LoadError)\n # @info \"load error message\" fieldnames(LoadError)\n msg = \"At [grey62 underline]$(er.file)[/grey62 underline] line [bold]$(er.line)\"\n subm = \"The cause is an error of type: [bright_red]$(string(typeof(er.error)))\"\n return msg, subm\nend\n\n# ! METHOD ERROR\n_method_regexes = [r\"!Matched+[:a-zA-Z]*\\{+[a-zA-Z\\s \\,]*\\}\", r\"!Matched+[:a-zA-Z]*\"]\nfunction error_message(io::IO, er::MethodError; kwargs...)\n # get main error message\n _args = join([string(ar) * _highlight(typeof(ar)) for ar in er.args], \"\\n \")\n fn_name = \"$(_highlight(string(er.f)))\"\n main_line = \"No method matching $fn_name with aguments:\\n \" * _args\n\n # get recomended candidates\n _candidates = split(sprint(show_method_candidates, er; context = io), \"\\n\")\n candidates::Vector{String} = []\n\n for can in _candidates[3:(end - 1)]\n fun, file = split(can, \" at \")\n name, args = split(fun, \"(\"; limit = 2)\n # name = \"[red]$name[/red]\"\n\n for regex in _method_regexes\n for match in collect(eachmatch(regex, args))\n args = replace(\n args, match.match => \"[dim red]$(match.match[9:end])[/dim red]\"\n )\n end\n end\n\n file, lineno = split(file, \":\")\n\n # println(RenderableText(name, \"red\"))\n push!(candidates, fn_name * \"(\" * args)\n push!(candidates, \"[dim]$file [bold dim](line: $lineno)[/bold dim][/dim]\\n\")\n end\n candidates =\n length(candidates) == 0 ? [\"[dim]no candidate method found[/dim]\"] : candidates\n\n return main_line * \"\\n\",\n Panel(\n \"\\n\" * join(candidates, \"\\n\");\n width = _width() - 10,\n title = \"closest candidates\",\n title_style = \"yellow\",\n style = \"blue dim\",\n )\nend\n\n# ! StackOverflowError\nfunction error_message(io::IO, er::StackOverflowError)\n return \"Stack overflow error: too many function calls.\", \"\"\nend\n\n# ! TYPE ERROR\nfunction error_message(io::IO, er::TypeError)\n # @info \"type err\" er fieldnames(typeof(er)) er.func er.context er.expected er.got\n # var = string(er.var)\n msg = \"In `[$(theme.emphasis_light) italic]$(er.func)` > `$(er.context)[/$(theme.emphasis_light) italic]` got\"\n msg *= \" [orange1 bold]$(er.got)[/orange1 bold][$(theme.type)](::$(typeof(er.got)))[/$(theme.type)] but expected argument of type\"\n msg *= \" [$(theme.type)]::$(er.expected)[/$(theme.type)]\"\n return msg, \"\"\nend\n\n# ! UndefKeywordError\nfunction error_message(io::IO, er::UndefKeywordError)\n # @info \"UndefKeywordError\" er er.var typeof(er.var) fieldnames(typeof(er.var))\n var = string(er.var)\n return \"Undefined function keyword argument: '$(_highlight(er.var))'.\", \"\"\nend\n\n# ! UNDEFVAR ERROR\nfunction error_message(io::IO, er::UndefVarError)\n # @info \"undef var error\" er er.var typeof(er.var)\n var = string(er.var)\n return \"Undefined variable '$(_highlight(er.var))'.\", \"\"\nend\n\n# ! STRING INDEX ERROR\nfunction error_message(io::IO, er::StringIndexError)\n # @info er typeof(er) fieldnames(typeof(er)) \n m1 = \"attempted to access a String at index $(er.index)\\n\"\n return m1, \"\"\nend\n\n# ! catch all other errors\nfunction error_message(io::IO, er)\n @debug \"Error message type doesnt have a specialized method!\" er typeof(er) fieldnames(\n typeof(er)\n )\n if hasfield(typeof(er), :error)\n # @info \"nested error\" typeof(er.error)\n m1, m2 = error_message(io, er.error)\n msg = \"\\n[bold red]LoadError:[/bold red]\\n\" * m1\n else\n msg = if hasfield(typeof(er), :msg)\n er.msg\n else\n \"no message for error of type $(typeof(er)), sorry.\"\n end\n m2 = \"\"\n end\n return msg, m2\nend\n\n# ---------------------------------------------------------------------------- #\n# INSTALL STACKTRACE #\n# ---------------------------------------------------------------------------- #\nfunction install_stacktrace()\n @eval begin\n\n # ---------------------------- handle load errors ---------------------------- #\n\n function Base.showerror(io::IO, er::LoadError, bt; backtrace = true)\n print(\"\\n\")\n println(hLine(_width(), \"[bold red]LoadError[/bold red]\"; style = \"dim red\"))\n Base.display_error(io, er, bt)\n\n return Base.showerror(io, er.error, bt; backtrace = true)\n end\n\n \"\"\"\n prints a line to mark te start of the error followed\n by the error's stack trace\n \"\"\"\n\n function Base.showerror(io::IO, er, bt; backtrace = true)\n ename = string(typeof(er))\n print(hLine(_width(), \"[bold red]$ename[/bold red]\"; style = \"dim red\"))\n\n try\n stack = style_backtrace(io, bt)\n print(stack)\n catch stack_error\n @warn \"failed to generate stack trace\" er stack_error\n println.(style_stacktrace_simple(bt))\n end\n end\n\n # # ------------------ handle all other errors (no backtrace) ------------------ #\n \"\"\"\n Re-define Base module function. Prints a nicely formatted error message.\n \"\"\"\n \n function Base.display_error(io::IO, er, bt)\n try\n err, err_msg = style_error(io, er)\n println(err / err_msg)\n catch styling_error\n @error \"Failed to generate styled error message\" styling_error stacktrace()\n\n println(apply_style(\"Original error: [bright_red]$(string(er))\"))\n end\n end\n end\nend\nend\n", "meta": {"hexsha": "adaaec683262f1c53eb255ee8e3e403586868703", "size": 10083, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/errors.jl", "max_stars_repo_name": "thazhemadam/Term.jl", "max_stars_repo_head_hexsha": "f69284b823f8952e445f96066413807c79743925", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/errors.jl", "max_issues_repo_name": "thazhemadam/Term.jl", "max_issues_repo_head_hexsha": "f69284b823f8952e445f96066413807c79743925", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/errors.jl", "max_forks_repo_name": "thazhemadam/Term.jl", "max_forks_repo_head_hexsha": "f69284b823f8952e445f96066413807c79743925", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 37.9060150376, "max_line_length": 203, "alphanum_fraction": 0.619756025, "num_tokens": 2505, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4610167941228965, "lm_q2_score": 0.15203223778010544, "lm_q1q2_score": 0.07008941486471411}}
{"text": "function fix(a)\r\n if a>0\r\n return floor(a)\r\n elseif a<0\r\n return ceil(a)\r\n else\r\n return a\r\n end\r\nend \r\n", "meta": {"hexsha": "87f4cb76918d410ee5e52666a72d3858da2182eb", "size": 137, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "fix.jl", "max_stars_repo_name": "umbertosaetti/J-F16", "max_stars_repo_head_hexsha": "d17c15ea94f149494094ba76f27b640c8a200202", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 5, "max_stars_repo_stars_event_min_datetime": "2022-01-03T21:16:03.000Z", "max_stars_repo_stars_event_max_datetime": "2022-01-10T07:54:32.000Z", "max_issues_repo_path": "fix.jl", "max_issues_repo_name": "umbertosaetti/J-F16", "max_issues_repo_head_hexsha": "d17c15ea94f149494094ba76f27b640c8a200202", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "fix.jl", "max_forks_repo_name": "umbertosaetti/J-F16", "max_forks_repo_head_hexsha": "d17c15ea94f149494094ba76f27b640c8a200202", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 13.7, "max_line_length": 24, "alphanum_fraction": 0.4671532847, "num_tokens": 38, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.48438008427698437, "lm_q2_score": 0.1441488530327406, "lm_q1q2_score": 0.06982283358042952}}
{"text": "## formatting conveniences\n\n\"markdown can leave wrapping p's\"\nfunction strip_p(txt)\n if occursin(r\"^\", txt)\n txt = replace(replace(txt, r\"^
\" => \"\"), r\"
$\" => \"\")\n end\n txt\nend\n\nfunction md(x)\n out = sprint(io -> show(io, \"text/html\", Markdown.parse(string(x))))\n strip_p(out)\nend\n\nmarkdown_to_latex(x) = sprint(io -> show(io, \"text/latex\", Markdown.parse(x)))\n\n\"\"\"\nHide output and input, but execute cell.\n\nExamples\n```\n2 + 2\nInvisible()\n```\n\"\"\"\nmutable struct Invisible\nend\n\n\"\"\"\nShow output as HTML\n\nExamples\n```\nHTMLoutput(\"em \")\n```\n\n\"\"\"\nmutable struct HTMLoutput\n x\nend\nBase.show(io::IO, ::MIME\"text/plain\", x::HTMLoutput) = print(io, \"\"\"$(x.x)
\"\"\")\nBase.show(io::IO, ::MIME\"text/html\", x::HTMLoutput) = print(io, x.x)\nBase.show(io::IO, ::MIME\"text/latex\", x::HTMLoutput) = println(io, \"...unable to display raw html...\")\n\n\n\n\"\"\"\nShow as input, but do not execute.\nExamples:\n```\nVerbatim(\"This will print, but not be executed\")\n```\n\"\"\"\nmutable struct Verbatim\n x\nend\nBase.show(io::IO, ::MIME\"text/plain\", x::Verbatim) = print(io, \"\"\"$(x.x) \"\"\")\nBase.show(io::IO, ::MIME\"text/html\", x::Verbatim) = print(io, x.x)\nBase.show(io::IO, ::MIME\"text/latex\", x::Verbatim) = print(io, \"\\verb@$(markdown_to_latex(x.x))@\")\n\n\n\"\"\"\nHide input, but show output\n\nExamples\n```\nx = 2 + 2\nOutputonly(x)\n```\n\"\"\"\nmutable struct Outputonly\n x\nend\n\n## Bootstrap things\nabstract type Bootstrap end\nBase.show(io::IO, ::MIME\"text/html\", x::Bootstrap) = print(io, \"\"\"$(x.x)\"\"\")\nBase.show(io::IO, ::MIME\"text/latex\", x::Bootstrap) = print(io, \"\"\"XXX BOOTSTRAP $(markdown_to_latex(x.x))\"\"\")\n\nmutable struct Alert <: Bootstrap\n x\n d::Dict\nend\n\n### An alert\nfunction alert(txt; kwargs...)\n d = Dict()\n for (k,v) in kwargs\n d[k] = v\n end\n Alert(txt, d)\nend\n\nwarning(txt; kwargs...) = alert(txt; class=\"warning\", kwargs...)\nnote(txt; kwargs...) = alert(txt; class=\"info\", kwargs...)\n\n\nfunction Base.show(io::IO, ::MIME\"text/html\", x::Alert)\n cls = haskey(x.d,:class) ? x.d[:class] : \"success\"\n txt = sprint(io -> show(io, \"text/html\", Markdown.parse(x.x)))\n tpl = \"\"\"\n\n\n$txt\n\n
\n\"\"\"\n\n print(io, tpl)\nend\n\nfunction Base.show(io::IO, ::MIME\"text/latex\", x::Alert)\n println(io, \"\"\"\n\\\\begin{mdframed}\n$(markdown_to_latex(x.x))\n\\\\end{mdframed}\n\"\"\")\nend\n \n\n\n\nmutable struct Example <: Bootstrap\n x\n d::Dict\nend\n\n## use nm=\"name\" to pass along name\nfunction example(txt; kwargs...)\n d = Dict()\n for (k,v) in kwargs\n d[k] = v\n end\n Example(txt, d)\nend\n\n\nfunction Base.show(io::IO, ::MIME\"text/html\", x::Example)\n nm = haskey(x.d,:nm) ? \" $(x.d[:nm]) \" : \"\"\n txt = sprint(io -> show(io, \"text/html\", Markdown.parse(x.x)))\n tpl = \"\"\"\n\n\nexample: $nm$txt\n\n
\n\n\"\"\"\n\n print(io, tpl)\nend\n\nfunction Base.show(io::IO, ::MIME\"text/latex\", x::Example)\n println(io, \"\"\"Example $(markdown_to_latex(x.x))\"\"\")\nend\n\n\nmutable struct Popup <: Bootstrap\n x\n title\n icon\n label\nend\n\n\"\"\"\n\nCreate a button to toggle the display of more detail.\n\nCan modify text, title, icon and label (for the button)\n\nThe text, title, and label can use Markdown.\n\nLaTeX markup does not work, as MathJax rendering is not supported in the popup.\n\n\"\"\"\npopup(x; title=\" \", icon=\"share-alt\", label=\" \") = Popup(x, title, icon, label)\n\npopup_html_tpl=mt\"\"\"\n\\\n \\\n\"\"\"\n\nfunction Base.show(io::IO, ::MIME\"text/html\", x::Popup)\n d = Dict()\n d[\"title\"] = sprint(io -> show(io, \"text/html\", Markdown.parse(x.title)))\n d[\"icon\"] = x.icon\n label = sprint(io -> show(io, \"text/html\", Markdown.parse(x.label)))\n d[\"button_label\"] = strip_p(label)\n d[\"body\"] = sprint(io -> show(io, \"text/html\", Markdown.parse(x.x)))\n println(d)\n Mustache.render(io, popup_html_tpl, d)\nend\n\nfunction Base.show(io::IO, ::MIME\"text/latex\", x::Popup)\n println(io, \"\"\"\n\\\\begin{quotation}\n$(markdown_to_latex(x.x))\n\\\\end{quotation}\n\"\"\")\nend\n\n\"\"\"\n\nWay to convert rectangular gird of values into a table\n\n\"\"\"\nmutable struct Table <: Bootstrap\n x\nend\ntable(x) = Table(x)\n\ntable_html_tpl=mt\"\"\"\n\n\n
\n{{{:nms}}}\n{{{:body}}}\n
\n
$(join(map(string, vals), \" \")) \"\n for j in 1:n\n val = Base.invokelatest(getindex, x.x, i, j)\n if ismissing(val)\n val = \".\"\n end\n bdy = bdy * \"$(md(val)) \"\n end\n bdy = bdy * \" \\n\"\n end\n d[:body] = bdy\n Mustache.render(io, table_html_tpl, d)\nend\n\nfunction df_to_table(df, label=\"label\", caption=\"caption\")\n nc = size(df, 2)\n perc = string(round(1/nc, digits=2))\n fmt = \"l\" * repeat(\"p{$perc\\\\textwidth}\", nc-1)\n header = join(string.(names(df)), \" & \")\n row = join([\"{{{:$x}}}\" for x in map(string, names(df))], \" & \")\n\ntpl=\"\"\"\n\\\\begin{table}[!ht]\n \\\\centering\n \\\\begin{tabular}{$fmt}\n $header\\\\\\\\\n \\\\midrule\\\\\\\\\n{{#:DF}} $row\\\\\\\\\n{{/:DF}} \n \\\\bottomrule\n \\\\end{tabular}\n \\\\label{tab:$label}\n\n\\\\end{table}\n\"\"\"\n\n Mustache.render(tpl, DF=df)\nend\n\nfunction Base.show(io::IO, ::MIME\"text/latex\", x::Table)\n d = markdown_to_latex.(x.x)\n println(io, df_to_table(d))\nend\n \n\n\nmutable struct NamedTable <: Bootstrap\ndata\nrownames\ncolnames\nend\n\nfunction Base.show(io::IO, ::MIME\"text/html\", x::NamedTable)\n vals = x.data\n cnames = x.colnames\n rnames = x.rownames\n d = Dict()\n d[:nms] = \"$(join(map(string, cnames), \" \")) \", rnames[i],\" \")\n for j in 1:n\n val = Base.invokelatest(getindex, x.data, i, j)\n print(buf, \"\", md(val), \" \")\n end\n println(buf, \"\"\n)=#\n", "meta": {"hexsha": "1d701970f87b181d5989da757ae803f34ba50cb5", "size": 992, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "docs/make.jl", "max_stars_repo_name": "fatteneder/SymPy.jl", "max_stars_repo_head_hexsha": "8b25bb9d0941443404de1b3a79205fee32080f6c", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "docs/make.jl", "max_issues_repo_name": "fatteneder/SymPy.jl", "max_issues_repo_head_hexsha": "8b25bb9d0941443404de1b3a79205fee32080f6c", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "docs/make.jl", "max_forks_repo_name": "fatteneder/SymPy.jl", "max_forks_repo_head_hexsha": "8b25bb9d0941443404de1b3a79205fee32080f6c", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 27.5555555556, "max_line_length": 73, "alphanum_fraction": 0.627016129, "num_tokens": 256, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.44167300566462553, "lm_q2_score": 0.15610489744545744, "lm_q1q2_score": 0.06894731925370332}}
{"text": "# -----------------------------------------------------------------\n# scitype function (generic)\n\n\"\"\"\n scitype(X)\n\nThe scientific type (interpretation) of `X`, as distinct from its\nmachine type, as specified by the active convention.\n\n### Examples from the MLJ convention\n\n```\njulia> using ScientificTypes # or `using MLJ`\njulia> scitype(3.14)\nContinuous\n\njulia> scitype([1, 2, 3, missing])\nAbstractArray{Union{Missing, Count},1}\n\njulia> scitype((5, \"beige\"))\nTuple{Count, Textual}\n\njulia> using CategoricalArrays\njulia> X = (gender = categorical(['M', 'M', 'F', 'M', 'F']),\n ndevices = [1, 3, 2, 3, 2])\njulia> scitype(X)\nTable{Union{AbstractArray{Count,1}, AbstractArray{Multiclass{2},1}}}\n```\n\nThe specific behavior of `scitype` is governed by the active\nconvention, as returned by `ScientificTypesBase.convention()`. The\n[ScientificTypes.jl\ndocumentation](https://alan-turing-institute.github.io/ScientificTypes.jl/dev/)\ndetails the convention demonstrated above.\n\n\"\"\"\nscitype(X; kw...) = scitype(X, convention(); kw...)\nscitype(X, C; kw...) = scitype(X, C, Val(trait(X)); kw...)\n\nscitype(X, C, ::Val{:other}; kw...) = Unknown\nscitype(::Missing; kw...) = Missing\nscitype(::Nothing; kw...) = Nothing\n\nscitype(t::Tuple, ::Convention; kw...) = Tuple{scitype.(t; kw...)...}\n\n# -----------------------------------------------------------------\n# convenience methods for scitype over unions\n\n\"\"\"\n scitype_union(A)\n\nReturn the type union, over all elements `x` generated by the iterable `A`,\nof `scitype(x)`. See also [`scitype`](@ref).\n\"\"\"\nfunction scitype_union(A)\n isempty(A) && return scitype(eltype(A))\n reduce((a,b)->Union{a,b}, (scitype(el) for el in A))\nend\n\n\n# -----------------------------------------------------------------\n# Scitype for arrays\n\n\"\"\"\n Scitype(T, C)\n\nMethod for implementers of a convention `C` to enable speed-up of\nscitype evaluations for arrays.\n\nIn general, one cannot infer the scitype of an object of type\n`AbstractArray{T, N}` from the machine type `T` alone.\n\nNevertheless, for some *restricted* machine types `U`, the statement\n`type(X) == AbstractArray{T, N}` for some `T<:U` already allows one\ndeduce that `scitype(X) = AbstractArray{S,N}`, where `S` is determined\nby `U` alone. This is the case in the *MLJ* convention, for example,\nif `U = Integer`, in which case `S = Count`.\n\nSuch shortcuts are specified as follows:\n\n```\nScitype(::Type{<:U}, ::C) = S\n```\n\nwhich incurs a considerable speed-up in the computation of `scitype(X)`.\nThere is also a speed-up for the case that `T <: Union{U, Missing}`.\n\nFor example, in the *MLJ* convention, one has\n\n```\nScitype(::Type{<:Integer}, ::MLJ) = Count\n```\n\"\"\"\nfunction Scitype end\n\nScitype(::Type, ::Convention) = Unknown\n\n# to distinguish between Any type and Union{T,Missing} for some more\n# specialised `T`, we define the Any case explicitly\nScitype(::Type{Any}, ::Convention) = Unknown\n\n# for the case Union{T,Missing} we return Union{S,Missing} with S\n# the scientific type corresponding to T\nScitype(::Type{Union{T,Missing}}, C::Convention) where T =\n Union{Missing,Scitype(T, C)}\n\n# for the case Missing, we return Missing\nScitype(::Type{Missing}, C::Convention) = Missing\n\nScitype(::Type{Nothing}, C::Convention) = Nothing\n\n# Broadcasting over arrays\n\nscitype(A::Arr{T}, C::Convention, ::Val{:other}; kw...) where T =\n arr_scitype(A, C, Scitype(T, C); kw...)\n\n\"\"\"\n arr_scitype(A, C, S; tight)\n\nReturn the scitype associated with an array `A` of type `{T,N}` assuming an\nexplicit `Scitype` correspondance exist mapping `T` to `S`.\nIf `tight=true` and `T>:Missing` then the function checks whether there are\n\"true missing values\", otherwise it constructs a \"tight copy\" of the array\nwithout a `Union{Missing,S}` type.\n\"\"\"\nfunction arr_scitype(A::Arr{T,N}, C::Convention, S::Type;\n tight::Bool=false) where {T,N}\n # no explicit scitype available\n S === Unknown && return Arr{scitype_union(A),N}\n # otherwise return `Arr{S,N}` or `Arr{Union{Missing,S},N}`\n if T >: Missing\n if tight\n has_missings = findfirst(ismissing, A) !== nothing\n !has_missings && return Arr{nonmissing(S),N}\n end\n return Arr{Union{S,Missing},N}\n end\n return Arr{S,N}\nend\n\n\"\"\"\n elscitype(A)\n\nReturn the element scientific type of an abstract array `A`. By definition, if\n`scitype(A) = AbstractArray{S,N}`, then `elscitype(A) = S`.\n\"\"\"\nelscitype(X::Arr; kw...) = scitype(X; kw...) |> _get_elst\n\n_get_elst(st::Type{Arr{T,N}}) where {T,N} = T\n", "meta": {"hexsha": "84400d56b75640e16ffdcf9bb5d25db404c554ea", "size": 4542, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/scitype.jl", "max_stars_repo_name": "juliohm/ScientificTypesBase.jl", "max_stars_repo_head_hexsha": "f52047c09da2b0e437515bdd58f0e865c32e06c6", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/scitype.jl", "max_issues_repo_name": "juliohm/ScientificTypesBase.jl", "max_issues_repo_head_hexsha": "f52047c09da2b0e437515bdd58f0e865c32e06c6", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/scitype.jl", "max_forks_repo_name": "juliohm/ScientificTypesBase.jl", "max_forks_repo_head_hexsha": "f52047c09da2b0e437515bdd58f0e865c32e06c6", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 30.28, "max_line_length": 79, "alphanum_fraction": 0.6402465874, "num_tokens": 1298, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4532618627863437, "lm_q2_score": 0.15203223970113983, "lm_q1q2_score": 0.06891041617051856}}
{"text": "module NumberSuffix\n\n export SuffixIt\n\n function SuffixIt(num)::String\n if (typeof(num)!=Int64)\n throw(TypeError(num,\"cannot suffix this type of argument\",Int64))\n end\n if (num<0)\n throw(DomainError(x, \"argument must be nonnegative\"))\n end\n suffixedNumber=string(num)\n lastDigit=num%10\n if (lastDigit==0 || lastDigit>=4)\n suffixedNumber=suffixedNumber*\"th\"\n elseif (lastDigit==1)\n suffixedNumber=suffixedNumber*\"st\"\n elseif (lastDigit==2)\n suffixedNumber=suffixedNumber*\"nd\"\n else\n suffixedNumber=suffixedNumber*\"rd\"\n end\n return suffixedNumber\n end\n\nend", "meta": {"hexsha": "938b293ba0704c46cb4174fac1b58eb76204b259", "size": 713, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/NumberSuffix.jl", "max_stars_repo_name": "eulerkochy/NumberSuffix.jl", "max_stars_repo_head_hexsha": "5ed924ae59701c9dfcec6e70d1829ea3448df286", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/NumberSuffix.jl", "max_issues_repo_name": "eulerkochy/NumberSuffix.jl", "max_issues_repo_head_hexsha": "5ed924ae59701c9dfcec6e70d1829ea3448df286", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/NumberSuffix.jl", "max_forks_repo_name": "eulerkochy/NumberSuffix.jl", "max_forks_repo_head_hexsha": "5ed924ae59701c9dfcec6e70d1829ea3448df286", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 27.4230769231, "max_line_length": 77, "alphanum_fraction": 0.5932678822, "num_tokens": 174, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.48438008427698437, "lm_q2_score": 0.14223188773801163, "lm_q1q2_score": 0.06889429376941265}}
{"text": "\"\"\"\n Imputor\n\nAn imputor stores information about imputing values in `AbstractArray`s and `Tables.table`s.\nNew imputation methods are expected to subtype `Imputor` and, at minimum,\nimplement the `_impute!(data::AbstractArrays, imp::)` method.\n\nWhile fallback `impute` and `impute!` methods are provided to extend your `_impute!` methods to\nn-dimensional arrays and tables, you can always override these methods to change the\nbehaviour as necessary.\n\"\"\"\nabstract type Imputor end\n\n#=\nThese default methods are required because @auto_hash_equals doesn't\nplay nice with Base.@kwdef\n=#\nfunction Base.hash(imp::T, h::UInt) where T <: Imputor\n h = hash(Symbol(T), h)\n\n for f in fieldnames(T)\n h = hash(getfield(imp, f), h)\n end\n\n return h\nend\n\nfunction Base.:(==)(a::T, b::T) where T <: Imputor\n result = true\n\n for f in fieldnames(T)\n if !isequal(getfield(a, f), getfield(b, f))\n result = false\n break\n end\n end\n\n return result\nend\n\n\"\"\"\n impute(data::T, imp; kwargs...) -> T\n\nReturns a new copy of the `data` with the missing data imputed by the imputor `imp`.\nFor matrices and tables, data is imputed one variable/column at a time.\nIf this is not the desired behaviour then you should overload this method or specify a different `dims` value.\n\n# Arguments\n* `data`: the data to be impute\n* `imp::Imputor`: the Imputor method to use\n\n# Returns\n* the input `data` with values imputed\n\n# Example\n```jldoctest\njulia> using Impute: Interpolate, impute\n\njulia> v = [1.0, 2.0, missing, missing, 5.0]\n5-element Vector{Union{Missing, Float64}}:\n 1.0\n 2.0\n missing\n missing\n 5.0\n\njulia> impute(v, Interpolate())\n5-element Vector{Union{Missing, Float64}}:\n 1.0\n 2.0\n 3.0\n 4.0\n 5.0\n```\n\"\"\"\nfunction impute(data, imp::Imputor; kwargs...)\n # NOTE: We don't use a return type declaration here because `trycopy` isn't guaranteed\n # to return the same type passed in. For example, subarrays and subdataframes will\n # return a regular array or dataframe.\n return impute!(trycopy(data), imp; kwargs...)\nend\n\n\"\"\"\n impute!(data::A, imp; dims=:, kwargs...) -> A\n\nImpute the `missing` values in the array `data` using the imputor `imp`.\nOptionally, you can specify the dimension to impute along.\n\n# Arguments\n* `data::AbstractArray{Union{T, Missing}}`: the data to be impute along dimensions `dims`\n* `imp::Imputor`: the Imputor method to use\n\n# Keyword Arguments\n* `dims=:`: The dimension to impute along. `:rows` and `:cols` are also supported for matrices.\n\n# Returns\n* `AbstractArray{Union{T, Missing}}`: the input `data` with values imputed\n\n# NOTES\n1. Matrices have a deprecated `dims=2` special case as `dims=:` is a breaking change\n2. Mutation isn't guaranteed for all array types, hence we return the result\n3. `eachslice` is used internally which requires Julia 1.1\n\n# Example\n```jldoctest\njulia> using Impute: Interpolate, impute!\n\njulia> M = [1.0 2.0 missing missing 5.0; 1.1 2.2 3.3 missing 5.5]\n2\u00d75 Matrix{Union{Missing, Float64}}:\n 1.0 2.0 missing missing 5.0\n 1.1 2.2 3.3 missing 5.5\n\njulia> impute!(M, Interpolate(); dims=1)\n2\u00d75 Matrix{Union{Missing, Float64}}:\n 1.0 2.0 3.0 4.0 5.0\n 1.1 2.2 3.3 4.4 5.5\n\njulia> M\n2\u00d75 Matrix{Union{Missing, Float64}}:\n 1.0 2.0 3.0 4.0 5.0\n 1.1 2.2 3.3 4.4 5.5\n```\n\"\"\"\nfunction impute!(\n data::A, imp::Imputor; dims=:, kwargs...\n)::A where A <: AbstractArray{Union{T, Missing}} where T\n dims === Colon() && return _impute!(data, imp; kwargs...)\n\n for x in eachslice(data; dims=dims)\n _impute!(x, imp; kwargs...)\n end\n\n return data\nend\n\n\nfunction impute!(\n data::M, imp::Imputor; dims=nothing, kwargs...\n)::M where M <: AbstractMatrix{Union{T, Missing}} where T\n dims === Colon() && return _impute!(data, imp; kwargs...)\n # We're calling our `dim` function to throw a depwarn if `dims === nothing`\n d = dim(data, dims)\n\n for x in eachslice(data; dims=d)\n _impute!(x, imp; kwargs...)\n end\n\n return data\nend\n\nimpute!(data::AbstractMatrix{Missing}, imp::Imputor; kwargs...) = data\n\n\"\"\"\n impute!(data::T, imp; kwargs...) -> T where T <: AbstractVector{<:NamedTuple}\n\nSpecial case rowtables which are arrays, but we want to fallback to the tables method.\n\"\"\"\nfunction impute!(data::T, imp::Imputor)::T where T <: AbstractVector{<:NamedTuple}\n return materializer(data)(impute!(Tables.columns(data), imp))\nend\n\n\"\"\"\n impute!(data::AbstractArray, imp) -> data\n\n\nJust returns the `data` when the array doesn't contain `missing`s\n\"\"\"\nimpute!(data::AbstractArray, imp::Imputor; kwargs...) = disallowmissing(data)\n\n\"\"\"\n impute!(data::AbstractArray{Missing}, imp) -> data\n\nJust return the `data` when the array only contains `missing`s\n\"\"\"\nimpute!(data::AbstractArray{Missing}, imp::Imputor; kwargs...) = data\n\n\n\"\"\"\n impute!(table, imp; cols=nothing) -> table\n\nImputes the data in a table by imputing the values 1 column at a time;\nif this is not the desired behaviour custom imputor methods should overload this method.\n\n# Arguments\n* `imp::Imputor`: the Imputor method to use\n* `table`: the data to impute\n\n# Keyword Arguments\n* `cols`: The columns to impute along (default is to impute all columns)\n\n# Returns\n* the input `data` with values imputed\n\n# Example\n```jldoctest\njulia> using DataFrames; using Impute: Interpolate, impute\n\n\njulia> df = DataFrame(:a => [1.0, 2.0, missing, missing, 5.0], :b => [1.1, 2.2, 3.3, missing, 5.5])\n5\u00d72 DataFrame\n Row \u2502 a b\n \u2502 Float64? Float64?\n\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\n 1 \u2502 1.0 1.1\n 2 \u2502 2.0 2.2\n 3 \u2502 missing 3.3\n 4 \u2502 missing missing\n 5 \u2502 5.0 5.5\n\njulia> impute(df, Interpolate())\n5\u00d72 DataFrame\n Row \u2502 a b\n \u2502 Float64? Float64?\n\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\n 1 \u2502 1.0 1.1\n 2 \u2502 2.0 2.2\n 3 \u2502 3.0 3.3\n 4 \u2502 4.0 4.4\n 5 \u2502 5.0 5.5\n```\n\"\"\"\nfunction impute!(table::T, imp::Imputor; cols=nothing)::T where T\n # TODO: We could probably handle iterators of tables here\n istable(table) || throw(MethodError(impute!, (table, imp)))\n\n # Extract a columns iterator that we should be able to use to mutate the data.\n # NOTE: Mutation is not guaranteed for all table types, but it avoid copying the data\n columntable = Tables.columns(table)\n\n cnames = cols === nothing ? propertynames(columntable) : cols\n for cname in cnames\n impute!(getproperty(columntable, cname), imp)\n end\n\n return table\nend\n\nfiles = [\n \"interp.jl\",\n \"knn.jl\",\n \"locf.jl\",\n \"nocb.jl\",\n \"replace.jl\",\n \"srs.jl\",\n \"substitute.jl\",\n \"svd.jl\",\n]\n\nfor file in files\n include(joinpath(\"imputors\", file))\nend\n", "meta": {"hexsha": "838cd9703ae7515a1c036140cb5f6b4ce96da1d2", "size": 6712, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/imputors.jl", "max_stars_repo_name": "pitmonticone/Impute.jl", "max_stars_repo_head_hexsha": "bd2e1f2c62a7b9d29cf25cb0bd2d5290cc569d07", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 57, "max_stars_repo_stars_event_min_datetime": "2017-05-18T22:52:17.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-29T17:09:14.000Z", "max_issues_repo_path": "src/imputors.jl", "max_issues_repo_name": "pitmonticone/Impute.jl", "max_issues_repo_head_hexsha": "bd2e1f2c62a7b9d29cf25cb0bd2d5290cc569d07", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 103, "max_issues_repo_issues_event_min_datetime": "2017-04-11T23:08:16.000Z", "max_issues_repo_issues_event_max_datetime": "2022-01-23T00:04:56.000Z", "max_forks_repo_path": "src/imputors.jl", "max_forks_repo_name": "pitmonticone/Impute.jl", "max_forks_repo_head_hexsha": "bd2e1f2c62a7b9d29cf25cb0bd2d5290cc569d07", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 10, "max_forks_repo_forks_event_min_datetime": "2017-12-21T17:12:10.000Z", "max_forks_repo_forks_event_max_datetime": "2020-12-12T07:20:56.000Z", "avg_line_length": 26.6349206349, "max_line_length": 110, "alphanum_fraction": 0.647794994, "num_tokens": 2129, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.49609382947091946, "lm_q2_score": 0.13846179412408713, "lm_q1q2_score": 0.06869004168243244}}
{"text": "Pkg.update()\n\n# Install required Julia packages\nPkg.add(\"SpecialFunctions\")\nPkg.add(\"FastGaussQuadrature\")\nPkg.add(\"GSL\")\nPkg.add(\"IterativeSolvers\")\nPkg.add(\"LinearMaps\")\nPkg.add(\"PyPlot\")\nPkg.add(\"SymPy\")\nPkg.add(\"PyCall\")\nPkg.add(\"LowRankApprox\")\nPkg.add(\"MATLAB\")\nPkg.add(\"NullableArrays\")\nPkg.add(\"Revise\")\nPkg.add(\"FMMLIB2D\")\nPkg.add(\"FINUFFT\")\nPkg.add(\"LaTeXStrings\")\nPkg.add(\"JLD\")\nPkg.add(\"Glob\")\nPkg.add(\"HDF5\")\nPkg.add(\"FileIO\")\nPkg.add(\"Humanize\")\nPkg.add(\"Interpolations\")\n", "meta": {"hexsha": "3ba48d43274fd40673211784425bc21298a924fa", "size": 486, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "julia/setup.jl", "max_stars_repo_name": "askhamwhat/inse-fiem-2d", "max_stars_repo_head_hexsha": "06e1f8610b35da67900e1acef1004a0456a696c1", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 9, "max_stars_repo_stars_event_min_datetime": "2019-08-20T12:53:24.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-16T00:15:10.000Z", "max_issues_repo_path": "julia/setup.jl", "max_issues_repo_name": "askhamwhat/inse-fiem-2d", "max_issues_repo_head_hexsha": "06e1f8610b35da67900e1acef1004a0456a696c1", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "julia/setup.jl", "max_forks_repo_name": "askhamwhat/inse-fiem-2d", "max_forks_repo_head_hexsha": "06e1f8610b35da67900e1acef1004a0456a696c1", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 2, "max_forks_repo_forks_event_min_datetime": "2020-03-28T18:44:17.000Z", "max_forks_repo_forks_event_max_datetime": "2020-12-14T04:17:59.000Z", "avg_line_length": 19.44, "max_line_length": 33, "alphanum_fraction": 0.7181069959, "num_tokens": 171, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.45713671682749485, "lm_q2_score": 0.1500288186418291, "lm_q1q2_score": 0.0685836815834334}}
{"text": "using Test\nusing MPI\n\nif get(ENV,\"JULIA_MPI_TEST_ARRAYTYPE\",\"\") == \"CuArray\"\n import CUDA\n ArrayType = CUDA.CuArray\nelse\n ArrayType = Array\nend\n\nMPI.Init()\n\ncomm = MPI.COMM_WORLD\nsize = MPI.Comm_size(comm)\nrank = MPI.Comm_rank(comm)\n\ndst = mod(rank+1, size)\nsrc = mod(rank-1, size)\n\nN = 32\n\nsend_mesg = ArrayType{Float64}(undef,N)\nrecv_mesg = ArrayType{Float64}(undef,N)\nrecv_mesg_expected = ArrayType{Float64}(undef,N)\n\nfill!(send_mesg, Float64(rank))\nfill!(recv_mesg_expected, Float64(src))\n\nrreq = MPI.Irecv!(recv_mesg, src, src+32, comm)\nsreq = MPI.Isend(send_mesg, dst, rank+32, comm)\n\nstats = MPI.Waitall!([sreq, rreq])\n@test rreq isa MPI.Request\n@test sreq isa MPI.Request\n@test MPI.Get_source(stats[2]) == src\n@test MPI.Get_tag(stats[2]) == src+32\n@test recv_mesg == recv_mesg_expected\n\n(done, stats) = MPI.Testall!([sreq, rreq])\n@test done\n\nif rank == 0\n MPI.send(send_mesg, dst, rank+32, comm)\n recv_mesg = recv_mesg_expected\nelseif rank == size-1\n (recv_mesg, _) = MPI.recv(src, src+32, comm)\nelse\n (recv_mesg, _) = MPI.recv(src, src+32, comm)\n MPI.send(send_mesg, dst, rank+32, comm)\nend\n@test recv_mesg == recv_mesg_expected\n\n\nif rank == 0\n MPI.Send(Float64(rank), dst, rank+32, comm)\n recv_val = Float64(src)\nelseif rank == size-1\n (recv_val, _) = MPI.Recv(Float64, src, src+32, comm)\nelse\n (recv_val, _) = MPI.Recv(Float64, src, src+32, comm)\n MPI.Send(Float64(rank), dst, rank+32, comm)\nend\n@test recv_val == Float64(src)\n\n\nrreq = MPI.Irecv!(recv_mesg, src, src+32, comm)\nsreq = MPI.Isend(send_mesg, dst, rank+32, comm)\n\nreq_arr = [sreq,rreq]\ninds = MPI.Waitsome(req_arr)\nfor i in inds\n @test MPI.Test(req_arr[i])\nend\n\nrreq = MPI.Irecv!(recv_mesg, src, src+32, comm)\nMPI.Cancel!(rreq)\nMPI.Wait(rreq)\n@test rreq.buffer == nothing\n\nGC.gc()\n\n# ---------------------\n# MPI_Sendrecv function\n# ---------------------\n#\n# send datatype\n# ---------------------\n# We test this function by executing a left shift of the leftmost element in a 1D\n# cartesian topology with periodic boundary conditions.\n#\n# Assume we have two processors, the data will look like this\n# proc 0 | proc 1\n# 0 0 0 | 1 1 1\n#\n# After the shift the data will contain\n# proc 0 | proc 1\n# 0 0 1 | 1 1 0\n#\n# init data\ncomm_rank = MPI.Comm_rank(comm)\ncomm_size = MPI.Comm_size(comm)\na = Float64[comm_rank, comm_rank, comm_rank]\n\n# construct cartesian communicator with 1D topology\ncomm_cart = MPI.Cart_create(comm, 1, Cint[comm_size], Cint[1], false)\n\n# get source and dest ranks using Cart_shift\nsrc_rank, dest_rank = MPI.Cart_shift(comm_cart, 0, -1)\n\n# execute left shift using subarrays\nMPI.Sendrecv!(@view(a[1]), dest_rank, 0,\n @view(a[3]), src_rank, 0, comm_cart)\n\n@test a == [comm_rank, comm_rank, (comm_rank+1) % comm_size]\n\n# send elements from a buffer\n# ---------------------------\na = Float64[comm_rank, comm_rank, comm_rank]\nb = Float64[ -1, -1, -1]\nMPI.Sendrecv!(@view(a[1:2]), dest_rank, 1,\n @view(b[1:2]), src_rank, 1, comm_cart)\n\n@test b == [(comm_rank+1) % comm_size, (comm_rank+1) % comm_size, -1]\n\n# send entire buffer\n# ---------------------------\na = Float64[comm_rank, comm_rank, comm_rank]\nb = Float64[ -1, -1, -1]\nMPI.Sendrecv!(a, dest_rank, 2,\n b, src_rank, 2, comm_cart)\n\n@test b == [(comm_rank+1) % comm_size, (comm_rank+1) % comm_size, (comm_rank+1) % comm_size]\n\n@test MPI.Waitall!(MPI.Request[]) == MPI.Status[]\n\nMPI.Finalize()\n# @test MPI.Finalized()\n", "meta": {"hexsha": "21971cd66de5ab04caed24d45d68b38a928034bd", "size": 3489, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/test_sendrecv.jl", "max_stars_repo_name": "mikegros/MPI.jl", "max_stars_repo_head_hexsha": "3941405967b16884de159c69dd5168e205373535", "max_stars_repo_licenses": ["Unlicense"], "max_stars_count": 255, "max_stars_repo_stars_event_min_datetime": "2015-01-08T03:24:13.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-28T18:27:43.000Z", "max_issues_repo_path": "test/test_sendrecv.jl", "max_issues_repo_name": "mikegros/MPI.jl", "max_issues_repo_head_hexsha": "3941405967b16884de159c69dd5168e205373535", "max_issues_repo_licenses": ["Unlicense"], "max_issues_count": 397, "max_issues_repo_issues_event_min_datetime": "2015-01-06T06:50:36.000Z", "max_issues_repo_issues_event_max_datetime": "2022-03-13T23:24:49.000Z", "max_forks_repo_path": "test/test_sendrecv.jl", "max_forks_repo_name": "mikegros/MPI.jl", "max_forks_repo_head_hexsha": "3941405967b16884de159c69dd5168e205373535", "max_forks_repo_licenses": ["Unlicense"], "max_forks_count": 118, "max_forks_repo_forks_event_min_datetime": "2015-02-07T04:16:58.000Z", "max_forks_repo_forks_event_max_datetime": "2022-03-13T14:58:55.000Z", "avg_line_length": 25.2826086957, "max_line_length": 92, "alphanum_fraction": 0.6494697621, "num_tokens": 1106, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.48828339529583464, "lm_q2_score": 0.1403362476923775, "lm_q1q2_score": 0.06852385950631133}}
{"text": "module MVDA\n\nusing DataFrames: copy, copyto!\nusing DataDeps, CSV, DataFrames, CodecZlib\nusing Parameters, Printf, MLDataUtils, ProgressMeter\nusing LinearAlgebra, Random, Statistics, StatsBase, StableRNGs\nusing KernelFunctions\nusing DelimitedFiles, Plots\nusing Polyester, MLDataUtils\n\nimport Base: show, iterate\n\n##### DATA #####\n\n#=\nUses DataDeps to download data as needed.\nInspired by UCIData.jl: https://github.com/JackDunnNZ/UCIData.jl\n=#\n\nconst DATA_DIR = joinpath(@__DIR__, \"data\")\n\n\"\"\"\n`list_datasets()`\n\nList available datasets in MVDA.\n\"\"\"\nlist_datasets() = map(x -> splitext(x)[1], readdir(DATA_DIR))\n\nfunction __init__()\n for dataset in list_datasets()\n include(joinpath(DATA_DIR, dataset * \".jl\"))\n end\nend\n\n\"\"\"\n`dataset(str)`\n\nLoad a dataset named `str`, if available. Returns data as a `DataFrame` where\nthe first column contains labels/targets and the remaining columns correspond to\ndistinct features.\n\"\"\"\nfunction dataset(str)\n # Locate dataset file.\n dataset_path = @datadep_str str\n file = readdir(dataset_path)\n index = findfirst(x -> occursin(\"data.\", x), file)\n if index isa Int\n dataset_file = joinpath(dataset_path, file[index])\n else # is this unreachable?\n error(\"Failed to locate a data.* file in $(dataset_path)\")\n end\n \n # Read dataset file as a DataFrame.\n df = if splitext(dataset_file)[2] == \".csv\"\n CSV.read(dataset_file, DataFrame)\n else # assume .csv.gz\n open(GzipDecompressorStream, dataset_file, \"r\") do stream\n CSV.read(stream, DataFrame)\n end\n end\n return df\nend\n\n\"\"\"\nProcess the dataset located at the given `path`.\n\nThis is an extra step to give fine-grain control in generating files with DataDeps.jl.\n\"\"\"\nfunction process_dataset(path::AbstractString; header=false, missingstrings=\"\", kwargs...)\n input_df = CSV.read(path, DataFrame, header=header, missingstrings=missingstrings)\n process_dataset(input_df; kwargs...)\n rm(path)\nend\n\n\"\"\"\nFinal step in processing the given dataset `input_df`.\n\nThis standardizes cached files that live in ~/.julia/datadeps so that labels/targets appear in first column\nfollowed by features in the remaining columns.\nWe also check for uniqueness in features.\n\"\"\"\nfunction process_dataset(input_df::DataFrame;\n target_index=-1,\n feature_indices=1:0,\n ext=\".csv\")\n # Build output DataFrame.\n output_df = DataFrame()\n output_df.target = input_df[!, target_index]\n output_df = hcat(output_df, input_df[!, feature_indices], makeunique=true)\n output_cols = [ :target; [Symbol(\"x\", n) for n in eachindex(feature_indices)] ]\n rename!(output_df, output_cols)\n dropmissing!(output_df)\n \n # Write to disk.\n output_path = \"data\" * ext\n if ext == \".csv\"\n CSV.write(output_path, output_df, delim=',', writeheader=true)\n elseif ext == \".csv.gz\"\n open(GzipCompressorStream, output_path, \"w\") do stream\n CSV.write(stream, output_df, delim=\",\", writeheader=true)\n end\n else\n error(\"Unknown file extension option '$(ext)'\")\n end\nend\n\n##### IMPLEMENTATION #####\n\ninclude(\"problem.jl\")\ninclude(\"utilities.jl\")\ninclude(\"projections.jl\")\ninclude(\"simulation.jl\")\n\nabstract type AbstractMMAlg end\n\ninclude(joinpath(\"algorithms\", \"SD.jl\"))\ninclude(joinpath(\"algorithms\", \"MMSVD.jl\"))\ninclude(joinpath(\"algorithms\", \"CyclicVDA.jl\"))\n\nconst DEFAULT_ANNEALING = geometric_progression\nconst DEFAULT_CALLBACK = __do_nothing_callback__\nconst DEFAULT_SCORE_FUNCTION = prediction_error\n\n\"\"\"\n fit(algorithm, problem, \u03f5, s; kwargs...)\n\nSolve optimization problem at sparsity level `s` using a deadzone of size `\u03f5`.\n\nThe solution is obtained via a proximal distance `algorithm` that gradually anneals parameter estimates\ntoward the target sparsity set.\n\"\"\"\nfunction fit(algorithm::AbstractMMAlg, problem::MVDAProblem, \u03f5::Real, s::Real; kwargs...)\n extras = __mm_init__(algorithm, problem, nothing) # initialize extra data structures\n MVDA.fit!(algorithm, problem, \u03f5, s, extras, (true,false,); kwargs...)\nend\n\n\"\"\"\n fit!(algorithm, problem, \u03f5, s, [extras], [update_extras]; kwargs...)\n\nSame as `fit_MVDA(algorithm, problem, \u03f5, s)`, but with preallocated data structures in `extras`.\n\n!!! Note\n The caller should specify whether to update data structures depending on `s` and `\u03c1` using `update_extras[1]` and `update_extras[2]`, respectively.\n\n Convergence is determined based on the rule `dist < dtol || abs(dist - old) < rtol * (1 + old)`, where `dist` is the squared distance and `dtol` and `rtol` are tolerance parameters.\n\n!!! Tip\n The `extras` argument can be constructed using `extras = __mm_init__(algorithm, problem, nothing)`.\n\n# Keyword Arguments\n\n- `nouter`: The number of outer iterations; i.e. the maximum number of `\u03c1` values to use in annealing (default=`100`).\n- `dtol`: An absolute tolerance parameter for the squared distance (default=`1e-6`).\n- `rtol`: A relative tolerance parameter for the squared distance (default=`1e-6`).\n- `rho_init`: The initial value for `\u03c1` (default=1.0).\n- `rho_max`: The maximum value for `\u03c1` (default=1e8).\n- `rhof`: A function `rhof(\u03c1, iter, rho_max)` used to determine the next value for `\u03c1` in the annealing sequence. The default multiplies `\u03c1` by `1.2`.\n- `verbose`: Print convergence information (default=`false`).\n- `cb`: A callback function for extending functionality.\n\nSee also: [`MVDA.anneal!`](@ref) for additional keyword arguments applied at the annealing step.\n\"\"\"\nfunction fit!(algorithm::AbstractMMAlg, problem::MVDAProblem, \u03f5::Real, s::Real,\n extras=nothing,\n update_extras::NTuple{2,Bool}=(true,false,);\n nouter::Int=100,\n dtol::Real=1e-6,\n rtol::Real=1e-6,\n rho_init::Real=1.0,\n rho_max::Real=1e8,\n rhof::Function=DEFAULT_ANNEALING,\n verbose::Bool=false,\n cb::Function=DEFAULT_CALLBACK,\n kwargs...)\n # Check for missing data structures.\n if extras isa Nothing\n error(\"Detected missing data structures for algorithm $(algorithm).\")\n end\n\n # Get problem info and extra data structures.\n @unpack intercept, coeff, coeff_prev, proj = problem\n @unpack projection = extras\n \n # Fix model size(s).\n k = sparsity_to_k(problem, s)\n\n # Initialize \u03c1 and iteration count.\n \u03c1, iters = rho_init, 0\n\n # Update data structures due to hyperparameters.\n update_extras[1] && __mm_update_sparsity__(algorithm, problem, \u03f5, \u03c1, k, extras)\n update_extras[2] && __mm_update_rho__(algorithm, problem, \u03f5, \u03c1, k, extras)\n\n # Check initial values for loss, objective, distance, and norm of gradient.\n apply_projection(projection, problem, k)\n init_result = __evaluate_objective__(problem, \u03f5, \u03c1, extras)\n result = SubproblemResult(0, init_result)\n cb(0, problem, \u03f5, \u03c1, k, result)\n old = sqrt(result.distance)\n\n for iter in 1:nouter\n # Solve minimization problem for fixed rho.\n verbose && print(\"\\n\",iter,\" \u03c1 = \",\u03c1)\n result = MVDA.anneal!(algorithm, problem, \u03f5, \u03c1, s, extras, (false,true,); verbose=verbose, cb=cb, kwargs...)\n\n # Update total iteration count.\n iters += result.iters\n\n cb(iter, problem, \u03f5, \u03c1, k, result)\n\n # Check for convergence to constrained solution.\n dist = sqrt(result.distance)\n if dist < dtol || abs(dist - old) < rtol * (1 + old)\n break\n else\n old = dist\n end\n \n # Update according to annealing schedule.\n \u03c1 = ifelse(iter < nouter, rhof(\u03c1, iter, rho_max), \u03c1)\n end\n \n # Project solution to the constraint set.\n apply_projection(projection, problem, k)\n loss, obj, dist, gradsq = __evaluate_objective__(problem, \u03f5, \u03c1, extras)\n\n if verbose\n print(\"\\n\\niters = \", iters)\n print(\"\\n\u2211\u1d62 max{0, |y\u1d62-B\u1d40x\u1d62|\u2082-\u03f5}\u00b2 = \", loss)\n print(\"\\nobjective = \", obj)\n print(\"\\ndistance = \", sqrt(dist))\n println(\"\\n|gradient| = \", sqrt(gradsq))\n end\n\n return SubproblemResult(iters, loss, obj, dist, gradsq)\nend\n\n\"\"\"\n anneal(algorithm, problem, \u03f5, \u03c1, s; kwargs...)\n\nSolve the `\u03c1`-penalized optimization problem at sparsity level `s` with deadzone `\u03f5`.\n\"\"\"\nfunction anneal(algorithm::AbstractMMAlg, problem::MVDAProblem, \u03f5::Real, \u03c1::Real, s::Real; kwargs...)\n extras = __mm_init__(algorithm, problem, nothing)\n MVDA.anneal!(algorithm, problem, \u03f5, \u03c1, s, extras, (true,true,); kwargs...)\nend\n\n\"\"\"\n anneal!(algorithm, problem, \u03f5, \u03c1, s, [extras], [update_extras]; kwargs...)\n\nSame as `anneal(algorithm, problem, \u03f5, \u03c1, s)`, but with preallocated data structures in `extras`.\n\n!!! Note\n The caller should specify whether to update data structures depending on `s` and `\u03c1` using `update_extras[1]` and `update_extras[2]`, respectively.\n\n Convergence is determined based on the rule `gradsq < gtol`, where `gradsq` is squared Euclidean norm of the gradient and `gtol` is a tolerance parameter.\n\n!!! Tip\n The `extras` argument can be constructed using `extras = __mm_init__(algorithm, problem, nothing)`.\n\n# Keyword Arguments\n\n- `ninner`: The maximum number of iterations (default=`10^4`).\n- `gtol`: An absoluate tolerance parameter on the squared Euclidean norm of the gradient (default=`1e-6`).\n- `nesterov_threshold`: The number of early iterations before applying Nesterov acceleration (default=`10`).\n- `verbose`: Print convergence information (default=`false`).\n- `cb`: A callback function for extending functionality.\n\"\"\"\nfunction anneal!(algorithm::AbstractMMAlg, problem::MVDAProblem, \u03f5::Real, \u03c1::Real, s::Real,\n extras=nothing,\n update_extras::NTuple{2,Bool}=(true,true);\n ninner::Int=10^4,\n gtol::Real=1e-6,\n nesterov_threshold::Int=10,\n verbose::Bool=false,\n cb::Function=DEFAULT_CALLBACK,\n kwargs...\n )\n # Check for missing data structures.\n if extras isa Nothing\n error(\"Detected missing data structures for algorithm $(algorithm).\")\n end\n\n # Get problem info and extra data structures.\n @unpack intercept, coeff, coeff_prev, proj = problem\n @unpack projection = extras\n\n # Fix model size(s).\n k = sparsity_to_k(problem, s)\n\n # Update data structures due to hyperparameters.\n update_extras[1] && __mm_update_sparsity__(algorithm, problem, \u03f5, \u03c1, k, extras)\n update_extras[2] && __mm_update_rho__(algorithm, problem, \u03f5, \u03c1, k, extras)\n\n # Check initial values for loss, objective, distance, and norm of gradient.\n apply_projection(projection, problem, k)\n result = __evaluate_objective__(problem, \u03f5, \u03c1, extras)\n cb(0, problem, \u03f5, \u03c1, k, result)\n old = result.objective\n\n if sqrt(result.gradient) < gtol\n return SubproblemResult(0, result)\n end\n\n # Use previous estimates in case of warm start.\n copyto!(coeff.all, coeff_prev.all)\n\n # Initialize iteration counts.\n iters = 0\n nesterov_iter = 1\n verbose && @printf(\"\\n%-5s\\t%-8s\\t%-8s\\t%-8s\\t%-8s\", \"iter.\", \"loss\", \"objective\", \"distance\", \"|gradient|\")\n for iter in 1:ninner\n iters += 1\n\n # Apply the algorithm map to minimize the quadratic surrogate.\n __mm_iterate__(algorithm, problem, \u03f5, \u03c1, k, extras)\n\n # Update loss, objective, distance, and gradient.\n apply_projection(projection, problem, k)\n result = __evaluate_objective__(problem, \u03f5, \u03c1, extras)\n\n cb(iter, problem, \u03f5, \u03c1, k, result)\n\n if verbose\n @printf(\"\\n%4d\\t%4.3e\\t%4.3e\\t%4.3e\\t%4.3e\", iter, result.loss, result.objective, sqrt(result.distance), sqrt(result.gradient))\n end\n\n # Assess convergence.\n obj = result.objective\n gradsq = sqrt(result.gradient)\n if gradsq < gtol\n break\n elseif iter < ninner\n needs_reset = iter < nesterov_threshold || obj > old\n nesterov_iter = __apply_nesterov__!(coeff.all, coeff_prev.all, nesterov_iter, needs_reset)\n old = obj\n end\n end\n # Save parameter estimates in case of warm start.\n copyto!(coeff_prev.all, coeff.all)\n\n return SubproblemResult(iters, result)\nend\n\n\"\"\"\n cv(algorithm, problem, grids; [at], [kwargs...])\n\nSplit data in `problem` into cross-validation and a test sets, then run cross-validation over the `grids`.\n\n# Keyword Arguments\n\n- `at`: A value between `0` and `1` indicating the proportion of samples/instances used for cross-validation, with remaining samples used for a test set (default=`0.8`).\n\nSee also: [`MVDA.cv(algorithm::AbstractMMAlg, problem::MVDAProblem, grids::Tuple{E,S}, dataset_split::Tuple{Any,Any})`](@ref)\n\"\"\"\nfunction cv(algorithm::AbstractMMAlg, problem::MVDAProblem, grids::Tuple{E,S}; at::Real=0.8, kwargs...) where {E,S}\n # Split data into cross-validation and test sets.\n @unpack p, Y, X, intercept = problem\n dataset_split = splitobs((Y, view(X, :, 1:p)), at=at, obsdim=1)\n MVDA.cv(algorithm, problem, grids, dataset_split; kwargs...)\nend\n\n\"\"\"\n cv(algorithm, problem, grids, dataset_split; [kwargs...])\n\nRun k-fold cross-validation over hyperparameters `(\u03f5, s)` for deadzone radius and sparsity level, respectively.\n\nThe given `problem` should enter with initial model parameters in `problem.coeff.all`.\nHyperparameters are specified in `grids = (\u03f5_grid, s_grid)`, and data subsets are given as `dataset_split = (cv_set, test_set)`.\n\n# Keyword Arguments\n\n- `nfolds`: The number of folds to run in cross-validation.\n- `scoref`: A function that evaluates a classifier over training, validation, and testing sets (default uses misclassification error).\n- `show_progress`: Toggles progress bar.\n\nAdditional arguments are propagated to `fit` and `anneal`. See also [`MVDA.fit`](@ref) and [`MVDA.anneal`](@ref).\n\"\"\"\nfunction cv(algorithm::AbstractMMAlg, problem::MVDAProblem, grids::Tuple{E,S}, dataset_split::Tuple{Any,Any};\n lambda::Real=1e-3,\n maxiter::Int=10^4,\n tol::Real=1e-4,\n nfolds::Int=5,\n scoref::Function=DEFAULT_SCORE_FUNCTION,\n cb::Function=DEFAULT_CALLBACK,\n show_progress::Bool=true,\n kwargs...) where {E,S}\n # Initialize the output.\n cv_set, test_set = dataset_split\n \u03f5_grid, s_grid = grids\n n\u03f5, ns = length(\u03f5_grid), length(s_grid)\n alloc_score_arrays(a, b, c) = [Matrix{Float64}(undef, a, b) for _ in 1:c]\n result = (;\n train=alloc_score_arrays(ns, n\u03f5, nfolds),\n validation=alloc_score_arrays(ns, n\u03f5, nfolds),\n test=alloc_score_arrays(ns, n\u03f5, nfolds),\n time=alloc_score_arrays(ns, n\u03f5, nfolds),\n )\n\n # Run cross-validation.\n if show_progress\n progress_bar = Progress(nfolds * n\u03f5 * ns, 1, \"Running CV w/ $(algorithm)... \")\n end\n\n for (k, fold) in enumerate(kfolds(cv_set, k=nfolds, obsdim=1))\n # Retrieve the training set and validation set.\n # TODO: Does this guarantee copies?\n train_set, validation_set = fold\n train_Y, train_X = getobs(train_set, obsdim=1)\n val_Y, val_X = getobs(validation_set, obsdim=1)\n test_Y, test_X = getobs(test_set, obsdim=1)\n \n # Standardize ALL data based on the training set.\n F = StatsBase.fit(ZScoreTransform, train_X, dims=1)\n has_nan = any(isnan, F.scale) || any(isnan, F.mean)\n has_inf = any(isinf, F.scale) || any(isinf, F.mean)\n has_zero = any(iszero, F.scale)\n if has_nan\n error(\"Detected NaN in z-score.\")\n elseif has_inf\n error(\"Detected Inf in z-score.\")\n elseif has_zero\n for idx in eachindex(F.scale)\n x = F.scale[idx]\n F.scale[idx] = ifelse(iszero(x), one(x), x)\n end\n end\n\n foreach(X -> StatsBase.transform!(F, X), (train_X, val_X, test_X))\n \n # Create a problem object for the training set.\n train_idx, _ = parentindices(train_set[1])\n train_problem = change_data(problem, train_Y, train_X)\n extras = __mm_init__(algorithm, train_problem, nothing)\n\n for (j, \u03f5) in enumerate(\u03f5_grid)\n # Set initial model parameters.\n set_initial_coefficients!(train_problem, problem, train_idx)\n \n for (i, s) in enumerate(s_grid)\n # Obtain solution as function of (\u03f5, s).\n if s != 0.0\n result.time[k][i,j] = @elapsed MVDA.fit!(algorithm, train_problem, \u03f5, s, extras, (true, false,);\n cb=cb, kwargs...\n )\n else# s == 0\n result.time[k][i,j] = @elapsed MVDA.init!(algorithm, train_problem, \u03f5, lambda, extras;\n maxiter=maxiter, gtol=tol, nesterov_threshold=0,\n )\n end\n copyto!(train_problem.coeff.all, train_problem.proj.all)\n\n # Evaluate the solution.\n r = scoref(train_problem, (train_Y, train_X), (val_Y, val_X), (test_Y, test_X))\n for (arr, val) in zip(result, r)\n arr[k][i,j] = val\n end\n\n # Update the progress bar.\n if show_progress\n spercent = string(round(100*s, digits=6), '%')\n next!(progress_bar, showvalues=[(:fold, k), (:sparsity, spercent), (:\u03f5, \u03f5)])\n end\n end\n end\n end\n\n return result\nend\n\nfunction cv_estimation(algorithm::AbstractMMAlg, problem::MVDAProblem, grids::Tuple{E,S}; at::Real=0.8, kwargs...) where {E,S}\n # Split data into cross-validation and test sets.\n @unpack p, Y, X, intercept = problem\n dataset_split = splitobs((Y, view(X, :, 1:p)), at=at, obsdim=1)\n MVDA.cv_estimation(algorithm, problem, grids, dataset_split; kwargs...)\nend\n\nfunction cv_estimation(algorithm::AbstractMMAlg, problem::MVDAProblem, grids::Tuple{E,S}, dataset_split::Tuple{Any,Any};\n nreplicates::Int=10,\n show_progress::Bool=true,\n rng::AbstractRNG=StableRNG(1903),\n kwargs...) where {E,S}\n # Retrieve subsets and create index set into cross-validation set.\n cv_set, test_set = dataset_split\n\n if show_progress\n progress_bar = Progress(nreplicates, 1, \"Running CV w/ $(algorithm)... \")\n end\n\n # Replicate CV procedure several times.\n replicate = NamedTuple[]\n for r in 1:nreplicates\n # Shuffle cross-validation data.\n cv_shuffled = shuffleobs(cv_set, obsdim=1, rng=rng)\n\n # Run k-fold cross-validation and store results.\n result = MVDA.cv(algorithm, problem, grids, (cv_shuffled, test_set); show_progress=false, kwargs...)\n push!(replicate, result)\n\n # Update the progress bar.\n if show_progress\n next!(progress_bar, showvalues=[(:replicate, r),])\n end\n end\n\n return replicate\nend\n\n\"\"\"\n```init!(algorithm, problem, \u03f5, \u03bb, [_extras_]; [maxiter=10^3], [gtol=1e-6], [nesterov_threshold=10], [verbose=false])```\n\nInitialize a `problem` with its `\u03bb`-regularized solution.\n\"\"\"\nfunction init!(algorithm::AbstractMMAlg, problem::MVDAProblem, \u03f5, \u03bb, _extras_=nothing;\n maxiter::Int=10^3,\n gtol::Real=1e-6,\n nesterov_threshold::Int=10,\n verbose::Bool=false,\n )\n # Check for missing data structures.\n extras = __mm_init__(algorithm, problem, _extras_)\n\n # Get problem info and extra data structures.\n @unpack coeff, coeff_prev, proj = problem\n\n # Update data structures due to hyperparameters.\n __mm_update_lambda__(algorithm, problem, \u03f5, \u03bb, extras)\n\n # Initialize coefficients.\n randn!(coeff.all)\n copyto!(coeff_prev.all, coeff.all)\n\n # Check initial values for loss, objective, distance, and norm of gradient.\n result = __evaluate_reg_objective__(problem, \u03f5, \u03bb, extras)\n old = result.objective\n\n if sqrt(result.gradient) < gtol\n return SubproblemResult(0, result)\n end\n\n # Initialize iteration counts.\n iters = 0\n nesterov_iter = 1\n verbose && @printf(\"\\n%-5s\\t%-8s\\t%-8s\\t%-8s\", \"iter.\", \"loss\", \"objective\", \"|gradient|\")\n for iter in 1:maxiter\n iters += 1\n\n # Apply the algorithm map to minimize the quadratic surrogate.\n __reg_iterate__(algorithm, problem, \u03f5, \u03bb, extras)\n\n # Update loss, objective, and gradient.\n result = __evaluate_reg_objective__(problem, \u03f5, \u03bb, extras)\n\n if verbose\n @printf(\"\\n%4d\\t%4.3e\\t%4.3e\\t%4.3e\", iter, result.loss, result.objective, sqrt(result.gradient))\n end\n\n # Assess convergence.\n obj = result.objective\n gradsq = sqrt(result.gradient)\n if gradsq < gtol\n break\n elseif iter < maxiter\n needs_reset = iter < nesterov_threshold || obj > old\n nesterov_iter = __apply_nesterov__!(coeff.all, coeff_prev.all, nesterov_iter, needs_reset)\n old = obj\n end\n end\n # Save parameter estimates in case of warm start.\n copyto!(coeff_prev.all, coeff.all)\n copyto!(proj.all, coeff.all)\n\n return SubproblemResult(iters, result)\nend\n\n# function fit_MVDA(algorithm::CyclicVDA, problem, \u03f5, \u03b4, \u03bb\u2081, \u03bb\u2082;\n# niter::Int=10^3,\n# atol=1e-4,\n# )\n# @unpack Y, X, res, coeff = problem\n# # \u03b4 = 1 / 20\n# # \u03f5 = 1//2 * sqrt(2*c/(c-1))\n# n, p, c = probdims(problem)\n# \u03bc\u2081 = n * \u03bb\u2081\n# \u03bc\u2082 = n * \u03bb\u2082\n\n# # initialize residuals\n# mul!(res.main.all, X, coeff.all)\n# axpby!(1.0, Y, -1.0, res.main.all)\n# extras = nothing\n\n# full_objective, _, _ = fetch_objective(problem, p+1, 1, \u03f5, \u03b4, \u03bb\u2081, \u03bb\u2082)\n# penalty1 = 0.0\n# penalty2 = 0.0\n# for j in 1:p # does not include intercept here\n# \u03b2 = view(problem.coeff.all, j, :)\n# penalty1 = penalty1 + \u03bc\u2081 * norm(\u03b2, 1)\n# penalty2 = penalty2 + \u03bc\u2082 * norm(\u03b2, 2)\n# end\n# full_objective = full_objective + penalty2 + penalty1\n\n# iters = 0\n# for iter in 1:niter\n# iters += 1\n\n# __mm_iterate__(algorithm, problem, \u03f5, \u03b4, \u03bb\u2081, \u03bb\u2082, extras)\n# loss, _, _ = fetch_objective(problem, p+1, 1, \u03f5, \u03b4, \u03bb\u2081, \u03bb\u2082)\n# penalty1 = 0.0\n# penalty2 = 0.0\n# for j in 1:p # does not include intercept here\n# \u03b2 = view(problem.coeff.all, j, :)\n# penalty1 = penalty1 + \u03bc\u2081 * norm(\u03b2, 1)\n# penalty2 = penalty2 + \u03bc\u2082 * norm(\u03b2, 2)\n# end\n# objective = loss + penalty2 + penalty1\n\n# if objective > full_objective error(\"Descent failure\") end\n\n# if full_objective - objective < atol break end\n# full_objective = objective\n# end\n\n# copyto!(problem.proj.all, coeff.all)\n\n# return full_objective, penalty1, penalty2\n# end\n\nexport IterationResult, SubproblemResult\nexport MVDAProblem, SD, MMSVD # CyclicVDA\n\nend\n", "meta": {"hexsha": "4f8e89d8e42c605c000876c71d5943d53ad34b93", "size": 22496, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/MVDA.jl", "max_stars_repo_name": "alanderos91/SparseMVDA", "max_stars_repo_head_hexsha": "a2ade5627b3d05fb8346ee8f7f342b312011f6b4", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/MVDA.jl", "max_issues_repo_name": "alanderos91/SparseMVDA", "max_issues_repo_head_hexsha": "a2ade5627b3d05fb8346ee8f7f342b312011f6b4", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/MVDA.jl", "max_forks_repo_name": "alanderos91/SparseMVDA", "max_forks_repo_head_hexsha": "a2ade5627b3d05fb8346ee8f7f342b312011f6b4", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 35.5949367089, "max_line_length": 185, "alphanum_fraction": 0.650737909, "num_tokens": 6107, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4882833952958347, "lm_q2_score": 0.14033624589467186, "lm_q1q2_score": 0.06852385862852152}}
{"text": "# As with the first half of this assignment, `learning_julia1_exercises.jl`,\n# fill in the missing implementation and pass the `@test`s.\n#\n# If you're writing your code within the `@testset`, the functions will not be available outside it.\n# (A `@testset` defines an enclosed scope that doesn't \"pollute\" the outside.)\n# You may find this makes it harder to develop and debug your code.\n# Remember that in VS Code you can select a range of lines and hit Ctrl-Enter to evaluate it,\n# and that may make it easier to experiment in VS Code's Julia REPL.\n#\n# We're also extending your usage of Test by introducing `@test_throws`.\n# Read its help to see what it does. When you pass argument(s) to the exception type, you check that, e.g.,\n# a particular message will be passed to the user.\n# Starting in Julia 1.8 it will be possible to check the error message without being concerned about\n# the exception type. (https://github.com/JuliaLang/julia/pull/41888)\n\nusing Test\n\n@testset \"Learning Julia 2\" begin\n # Problem 1 [Adapted from ThinkJulia, exercise 3-2]:\n # Create a function `rightjustify` that prints a string with the right number of leading spaces\n # so that the final character aligns with a chosen column.\n # I've given you the signature of one method; also create another method that supplies `stdout`\n # as the default `io` choice. See the style guide for more info about recommended best\n # practices for ordering arguments for Julia functions:\n # https://docs.julialang.org/en/v1/manual/style-guide/#Write-functions-with-argument-ordering-similar-to-Julia-Base\n function rightjustify(io::IO, str::AbstractString, col::Integer = 70)\n # fill me in!\n end\n # also define a method that doesn't have `io` supplied\n\n # here are my tests (don't modify this code)\n checkstr(ostr, refstr, len) = @test length(ostr) == len && endswith(ostr, refstr) && strip(ostr) == refstr\n for str in (\"short\", \"kind of medium\", \"getting longer, but not too huge\")\n checkstr(sprint(rightjustify, str), str, 70)\n checkstr(sprint(rightjustify, str, 80), str, 80)\n end\n mktemp() do fname, io\n str = \"send me to stdout!\"\n redirect_stdout(io) do\n rightjustify(str)\n end\n seek(io, 0)\n checkstr(chomp(read(io, String)), str, 70)\n end\n\n\n # Problem 2: adapted from https://exercism.org/tracks/julia/exercises/robot-simulator\n # (Exercism is a site offering programming exercises in many different languages, with a nice\n # framework for in-browser coding, automated testing, progress-monitoring, and even mentoring.)\n # The version of this problem on Exercism encourages you to develop a custom type, which is great,\n # but here we're going to treat it as an exercise in control-flow.\n # If you wish, you can put everything in a single function.\n #\n # A robot starts at a location `(x, y)`, where both `x` and `y` are integers, facing in a specified direction\n # ('N'=North, 'S'=South, 'E'=East, 'W'=West). `x` is the East-West axis and `y` the North-South axis.\n # You also supply a string, where each character of the string is one of the following:\n # - 'A': advance one step in the direction it's facing\n # - 'R': turn right\n # - 'L': turn left\n # When the robot turns, it pivots without changing its location. For example, \"RAALAL\" means:\n # 1. turn right\n # 2. advance two spaces\n # 3. turn left\n # 4. advance one space\n # 5. turn left\n #\n # Your code should return both the final position and the direction the robot is facing.\n # See if you can come up with a more efficient approach than a huge nest of `if...else...end`\n # statements. My solution is less than 20 lines of code.\n # Hint: some of the better solutions might use `%` or `mod1` (read their help strings).\n\n function robot_command(cmd, (x, y), dir) # argument destructuring syntax: https://docs.julialang.org/en/v1/manual/functions/#Argument-destructuring\n # you write the body\n end\n\n # Single steps (these are deliberately written out in long form to avoid giving too much of the solution away)\n @test robot_command(\"R\", (0, 0), 'N') == ((0, 0), 'E')\n @test robot_command(\"R\", (0, 0), 'E') == ((0, 0), 'S')\n @test robot_command(\"R\", (0, 0), 'S') == ((0, 0), 'W')\n @test robot_command(\"R\", (0, 0), 'W') == ((0, 0), 'N')\n @test robot_command(\"L\", (0, 0), 'N') == ((0, 0), 'W')\n @test robot_command(\"L\", (0, 0), 'E') == ((0, 0), 'N')\n @test robot_command(\"L\", (0, 0), 'S') == ((0, 0), 'E')\n @test robot_command(\"L\", (0, 0), 'W') == ((0, 0), 'S')\n @test robot_command(\"A\", (0, 0), 'N') == ((0, 1), 'N')\n @test robot_command(\"A\", (0, 0), 'E') == ((1, 0), 'E')\n @test robot_command(\"A\", (0, 0), 'S') == ((0, -1), 'S')\n @test robot_command(\"A\", (0, 0), 'W') == ((-1, 0), 'W')\n @test_throws ErrorException(\"robot command K not recognized\") robot_command(\"K\", (0, 0), 'W')\n # Sequences\n for x in (7, 22), y in (3, -7)\n @test robot_command(\"RAALAL\", (x, y), 'N') == ((x+2, y+1), 'W')\n @test robot_command(\"ARAAALA\", (x, y), 'W') == ((x-2, y+3), 'W')\n for dir in ('N', 'E', 'S', 'W')\n @test robot_command(\"ALALALAL\", (x, y), dir) == ((x, y), dir)\n end\n end\n\n\n # Problem 3: introduction to structures\n # Note that when you redefine structures, you may have to shut down your Julia session and restart it.\n # (Julia won't let you change an existing `struct` definition.)\n # a) create a new subtype of `Integer` called `MyInt`, one that stores an `Int` internally\n\n @test MyInt(3) isa Integer\n @test !ismutable(MyInt(3))\n\n # b) extend `Base`'s `==` so that you can compare `MyInt` against any other `Integer`s\n # You can either `import Base: ==` and write `==(a, b)` methods, or write your new\n # methods using module qualification, `Base.:(==)(a, b)`. The `:` is needed only for operators;\n # when extending other functions, you don't need `:`.\n\n @test MyInt(3) == 3\n @test 2 == MyInt(2)\n @test MyInt(1) != MyInt(2) # this will follow automatically if you define `==`\n\n # c) extend `+` so that `MyInt` \"wraps around\" at 5: `MyInt(5) + 1` return `MyInt(1)` and then it starts counting up again.\n\n @test MyInt(1) + 1 === MyInt(2)\n @test MyInt(2) + 1 === MyInt(3)\n @test MyInt(3) + 1 === MyInt(4)\n @test MyInt(4) + 1 === MyInt(5)\n @test MyInt(5) + 1 === MyInt(1)\n @test MyInt(4) + 2 === MyInt(1)\n @test MyInt(3) + 10 === MyInt(3)\n\n # d) also extend the `typemax` \"trait\" for `MyInt`. Read the docs for `typemax`, and then do `@edit typemax(UInt8)`\n # to see how such methods are implemented. Mimic that definition.\n\n @test typemax(MyInt) === MyInt(5)\n @test typemax(MyInt(2)) === MyInt(5) # note you didn't have to write a method specifically for this test\n @test typemax(UInt8) === 0xff # just to make sure it's not broken\n\n # Problem 4: encapsulating data in objects & functions\n # a) Create a parametric `struct` representing a unit Gaussian in 1d: if `g isa Gaussian`, then\n # - `g.\u03bc` should return the mean (center) of the Gaussian, a scalar of type `T`\n # - `g.\u03c3` should return the standard deviation, a scalar also of type `T`\n # Also, `T` should only be allowed to be a subtype of `Real`\n\n g = Gaussian(1.0, 0.5)\n @test g isa Gaussian{Float64}\n @test g.\u03bc === 1.0\n @test g.\u03c3 === 0.5\n g = Gaussian(2, 5)\n @test g isa Gaussian{Int}\n @test g.\u03bc === 2\n @test g.\u03c3 === 5\n @test_throws TypeError Gaussian{String}\n\n # b) make objects of type Gaussian callable (review https://docs.julialang.org/en/v1/manual/methods/#Function-like-objects\n # as needed), so that `g(x)` returns `exp(-(x - \u03bc)^2/(2*\u03c3^2))`.\n\n g = Gaussian(1.0, 0.5)\n @test g(2) \u2248 0.1353352832366127\n @test g(2.0) == g(2)\n @test g(1.25) \u2248 0.8824969025845955\n g = Gaussian(2, 5)\n @test g(-3) \u2248 0.6065306597126334\n\n # c) now do the same thing with a *closure*, a function that wraps data. Here's a demo:\n \"\"\"\n f = adder(x)\n\n Return a function `f` that adds `x` to any number.\n \"\"\"\n function adder(x)\n return y::Number -> x+y\n end\n f = adder(3)\n @test f isa Function\n @test f(2) == 5\n @test_throws MethodError f(\"hello\") # f isn't defined on strings because of the `y::Number`\n\n # Another way you could have written the above is\n # function adder(x)\n # f = function(y::Number)\n # return x + y\n # end\n # return f\n # end\n # If that's clearer to you, feel free to use that form.\n\n # Now you write one: `f = gaussian_closure(\u03bc, \u03c3)` should return a function `f` such that\n # `f(x) == exp(-(x - \u03bc)^2/(2*\u03c3^2))`. Enforce the fact that both `\u03bc` and `\u03c3` must be `Real`,\n # as must `x`, but don't require them to be the same type.\n\n f = gaussian_closure(2, 5)\n @test f isa Function\n @test f(-3) \u2248 0.6065306597126334\n @test_throws MethodError f(\"hello\")\n @test_throws MethodError gaussian_closure(\"hello\", 5)\n @test_throws MethodError gaussian_closure(2, [5])\nend\n", "meta": {"hexsha": "9fac243edbf66bba40a15d945bc8a262ff97fcc7", "size": 9145, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "homeworks/learning_julia2_exercises.jl", "max_stars_repo_name": "HsupoLeng/AdvancedScientificComputing", "max_stars_repo_head_hexsha": "035f1e08a606bc514e07a6adfcde465b755bf588", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "homeworks/learning_julia2_exercises.jl", "max_issues_repo_name": "HsupoLeng/AdvancedScientificComputing", "max_issues_repo_head_hexsha": "035f1e08a606bc514e07a6adfcde465b755bf588", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "homeworks/learning_julia2_exercises.jl", "max_forks_repo_name": "HsupoLeng/AdvancedScientificComputing", "max_forks_repo_head_hexsha": "035f1e08a606bc514e07a6adfcde465b755bf588", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 47.1391752577, "max_line_length": 152, "alphanum_fraction": 0.6276653909, "num_tokens": 2821, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.3522017956470284, "lm_q2_score": 0.1943678016853152, "lm_q1q2_score": 0.06845668876953354}}
{"text": "# Note: Functions surrounded by a comment blocks are there because `Vararg` is still allocating.\n# When Vararg is fast enough, they can simply be removed.\n\n\n#######################\n# UndefBlocksInitializer #\n#######################\n\n\"\"\"\n UndefBlocksInitializer\n\nSingleton type used in block array initialization, indicating the\narray-constructor-caller would like an uninitialized block array. See also\nundef_blocks (@ref), an alias for UndefBlocksInitializer().\n\nExamples\n\n\u2261\u2261\u2261\u2261\u2261\u2261\u2261\u2261\u2261\u2261\n```julia\njulia> BlockArray(undef_blocks, Matrix{Float32}, [1,2], [3,2])\n2\u00d72-blocked 3\u00d75 BlockArray{Float32,2}:\n #undef #undef #undef \u2502 #undef #undef\n \u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\n #undef #undef #undef \u2502 #undef #undef\n #undef #undef #undef \u2502 #undef #undef\n ```\n\"\"\"\nstruct UndefBlocksInitializer end\n\n\"\"\"\n undef_blocks\n\nAlias for UndefBlocksInitializer(), which constructs an instance of the singleton\ntype UndefBlocksInitializer (@ref), used in block array initialization to indicate the\narray-constructor-caller would like an uninitialized block array.\n\nExamples\n\n\u2261\u2261\u2261\u2261\u2261\u2261\u2261\u2261\u2261\u2261\n```julia\njulia> BlockArray(undef_blocks, Matrix{Float32}, [1,2], [3,2])\n2\u00d72-blocked 3\u00d75 BlockArray{Float32,2}:\n #undef #undef #undef \u2502 #undef #undef\n ------------------------\u253c----------------\n #undef #undef #undef \u2502 #undef #undef\n #undef #undef #undef \u2502 #undef #undef\n ```\n\"\"\"\nconst undef_blocks = UndefBlocksInitializer()\n\n##############\n# BlockArray #\n##############\n\nfunction _BlockArray end\n\n\"\"\"\n BlockArray{T, N, R<:AbstractArray{<:AbstractArray{T,N},N}, BS<:AbstractBlockSizes{N}} <: AbstractBlockArray{T, N}\n\nA `BlockArray` is an array where each block is stored contiguously. This means that insertions and retrieval of blocks\ncan be very fast and non allocating since no copying of data is needed.\n\nIn the type definition, `R` defines the array type that holds the blocks, for example `Matrix{Matrix{Float64}}`.\n\"\"\"\nstruct BlockArray{T, N, R <: AbstractArray{<:AbstractArray{T,N},N}, BS<:AbstractBlockSizes{N}} <: AbstractBlockArray{T, N}\n blocks::R\n block_sizes::BS\n\n global function _BlockArray(blocks::R, block_sizes::BS) where {T, N, R<:AbstractArray{<:AbstractArray{T,N},N}, BS<:AbstractBlockSizes{N}}\n new{T, N, R, BS}(blocks, block_sizes)\n end\nend\n\n# Auxilary outer constructors\nfunction _BlockArray(blocks::R, block_sizes::Vararg{AbstractVector{Int}, N}) where {T, N, R<:AbstractArray{<:AbstractArray{T,N},N}}\n return _BlockArray(blocks, BlockSizes(block_sizes...))\nend\n\nfunction _BlockArray(blocks::R, block_sizes::BS) where {T, N, R<:AbstractArray{<:AbstractArray{V,N} where V,N}, BS<:AbstractBlockSizes{N}}\n _BlockArray(convert(AbstractArray{AbstractArray{mapreduce(eltype,promote_type,blocks),N},N}, blocks), block_sizes)\nend\n\nconst BlockMatrix{T, R <: AbstractMatrix{<:AbstractMatrix{T}}} = BlockArray{T, 2, R}\nconst BlockVector{T, R <: AbstractVector{<:AbstractVector{T}}} = BlockArray{T, 1, R}\nconst BlockVecOrMat{T, R} = Union{BlockMatrix{T, R}, BlockVector{T, R}}\n\n################\n# Constructors #\n################\n\n@inline _BlockArray(::Type{R}, block_sizes::Vararg{AbstractVector{Int}, N}) where {T, N, R<:AbstractArray{<:AbstractArray{T,N},N}} =\n _BlockArray(R, BlockSizes(block_sizes...))\n\nfunction _BlockArray(::Type{R}, block_sizes::BlockSizes{N}) where {T, N, R<:AbstractArray{<:AbstractArray{T,N},N}}\n n_blocks = nblocks(block_sizes)\n blocks = R(undef, n_blocks)\n _BlockArray(blocks, block_sizes)\nend\n\n@inline undef_blocks_BlockArray(::Type{R}, block_sizes::Vararg{AbstractVector{Int}, N}) where {T, N, R<:AbstractArray{<:AbstractArray{T,N},N}} =\n _BlockArray(R, block_sizes...)\n\n\"\"\"\nConstructs a `BlockArray` with uninitialized blocks from a block type `R` with sizes defind by `block_sizes`.\n\n```jldoctest; setup = quote using BlockArrays end\njulia> BlockArray(undef_blocks, Matrix{Float64}, [1,3], [2,2])\n2\u00d72-blocked 4\u00d74 BlockArray{Float64,2}:\n #undef \u2502 #undef #undef #undef \u2502\n --------\u253c--------------------------\u253c\n #undef \u2502 #undef #undef #undef \u2502\n #undef \u2502 #undef #undef #undef \u2502\n --------\u253c--------------------------\u253c\n #undef \u2502 #undef #undef #undef \u2502\n```\n\"\"\"\n@inline BlockArray(::UndefBlocksInitializer, ::Type{R}, block_sizes::Vararg{AbstractVector{Int}, N}) where {T, N, R<:AbstractArray{T,N}} =\n undef_blocks_BlockArray(Array{R,N}, block_sizes...)\n\n@inline BlockArray{T}(::UndefBlocksInitializer, block_sizes::Vararg{AbstractVector{Int}, N}) where {T, N} =\n BlockArray(undef_blocks, Array{T,N}, block_sizes...)\n\n@inline BlockArray{T,N}(::UndefBlocksInitializer, block_sizes::Vararg{AbstractVector{Int}, N}) where {T, N} =\n BlockArray(undef_blocks, Array{T,N}, block_sizes...)\n\n@inline BlockArray{T,N,R}(::UndefBlocksInitializer, block_sizes::Vararg{AbstractVector{Int}, N}) where {T, N, R<:AbstractArray{<:AbstractArray{T,N},N}} =\n undef_blocks_BlockArray(R, block_sizes...)\n\n\n@generated function initialized_blocks_BlockArray(::Type{R}, block_sizes::BlockSizes{N}) where R<:AbstractArray{V,N} where {T,N,V<:AbstractArray{T,N}}\n return quote\n block_arr = _BlockArray(R, block_sizes)\n @nloops $N i i->(1:nblocks(block_sizes, i)) begin\n block_index = @ntuple $N i\n setblock!(block_arr, similar(V, blocksize(block_sizes, block_index)), block_index...)\n end\n\n return block_arr\n end\nend\n\n\ninitialized_blocks_BlockArray(::Type{R}, block_sizes::Vararg{AbstractVector{Int}, N}) where {T, N, R<:AbstractArray{<:AbstractArray{T,N},N}} =\n initialized_blocks_BlockArray(R, BlockSizes(block_sizes...))\n\n@inline BlockArray{T}(::UndefInitializer, block_sizes::BlockSizes{N}) where {T, N} =\n initialized_blocks_BlockArray(Array{Array{T,N},N}, block_sizes)\n\n@inline BlockArray{T, N}(::UndefInitializer, block_sizes::BlockSizes{N}) where {T, N} =\n initialized_blocks_BlockArray(Array{Array{T,N},N}, block_sizes)\n\n@inline BlockArray{T, N, R}(::UndefInitializer, block_sizes::BlockSizes{N}) where {T, N, R<:AbstractArray{<:AbstractArray{T,N},N}} =\n initialized_blocks_BlockArray(R, block_sizes)\n\n@inline BlockArray{T}(::UndefInitializer, block_sizes::Vararg{AbstractVector{Int}, N}) where {T, N} =\n initialized_blocks_BlockArray(Array{Array{T,N},N}, block_sizes...)\n\n@inline BlockArray{T, N}(::UndefInitializer, block_sizes::Vararg{AbstractVector{Int}, N}) where {T, N} =\n initialized_blocks_BlockArray(Array{Array{T,N},N}, block_sizes...)\n\n@inline BlockArray{T, N, R}(::UndefInitializer, block_sizes::Vararg{AbstractVector{Int}, N}) where {T, N, R<:AbstractArray{<:AbstractArray{T,N},N}} =\n initialized_blocks_BlockArray(R, block_sizes...)\n\nfunction BlockArray(arr::AbstractArray{T, N}, block_sizes::Vararg{AbstractVector{Int}, N}) where {T,N}\n for i in 1:N\n if sum(block_sizes[i]) != size(arr, i)\n throw(DimensionMismatch(\"block size for dimension $i: $(block_sizes[i]) does not sum to the array size: $(size(arr, i))\"))\n end\n end\n BlockArray(arr, BlockSizes(block_sizes...))\nend\n\n@generated function BlockArray(arr::AbstractArray{T, N}, block_sizes::BlockSizes{N}) where {T,N}\n return quote\n block_arr = _BlockArray(Array{typeof(arr),N}, block_sizes)\n @nloops $N i i->(1:nblocks(block_sizes, i)) begin\n block_index = @ntuple $N i\n indices = globalrange(block_sizes, block_index)\n setblock!(block_arr, arr[indices...], block_index...)\n end\n\n return block_arr\n end\nend\n\nBlockVector(blocks::AbstractVector, block_sizes::AbstractBlockSizes{1}) = BlockArray(blocks, block_sizes)\nBlockVector(blocks::AbstractVector, block_sizes::AbstractVector{Int}) = BlockArray(blocks, block_sizes)\nBlockMatrix(blocks::AbstractMatrix, block_sizes::AbstractBlockSizes{2}) = BlockArray(blocks, block_sizes)\nBlockMatrix(blocks::AbstractMatrix, block_sizes::Vararg{AbstractVector{Int},2}) = BlockArray(blocks, block_sizes...)\n\n\"\"\"\n mortar(blocks::AbstractArray)\n mortar(blocks::AbstractArray{R, N}, sizes_1, sizes_2, ..., sizes_N)\n mortar(blocks::AbstractArray{R, N}, block_sizes::BlockSizes{N})\n\nConstruct a `BlockArray` from `blocks`. `block_sizes` is computed from\n`blocks` if it is not given.\n\n# Examples\n```jldoctest; setup = quote using BlockArrays end\njulia> blocks = permutedims(reshape([\n 1ones(1, 3), 2ones(1, 2),\n 3ones(2, 3), 4ones(2, 2),\n ], (2, 2)))\n2\u00d72 Array{Array{Float64,2},2}:\n [1.0 1.0 1.0] [2.0 2.0]\n [3.0 3.0 3.0; 3.0 3.0 3.0] [4.0 4.0; 4.0 4.0]\n\njulia> mortar(blocks)\n2\u00d72-blocked 3\u00d75 BlockArray{Float64,2}:\n 1.0 1.0 1.0 \u2502 2.0 2.0\n \u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\n 3.0 3.0 3.0 \u2502 4.0 4.0\n 3.0 3.0 3.0 \u2502 4.0 4.0\n\njulia> ans == mortar(\n (1ones(1, 3), 2ones(1, 2)),\n (3ones(2, 3), 4ones(2, 2)),\n )\ntrue\n```\n\"\"\"\nmortar(blocks::AbstractArray{R, N}, block_sizes::AbstractBlockSizes{N}) where {R, N} =\n _BlockArray(blocks, block_sizes)\n\nmortar(blocks::AbstractArray{R, N}, block_sizes::Vararg{AbstractVector{Int}, N}) where {R, N} =\n _BlockArray(blocks, block_sizes...)\n\nmortar(blocks::AbstractArray) = mortar(blocks, sizes_from_blocks(blocks))\n\nsizes_from_blocks(blocks) = sizes_from_blocks(blocks, axes(blocks)) #\u00a0allow overriding on axes\n\nfunction sizes_from_blocks(blocks::AbstractArray{<:Any, N}, _) where N\n if length(blocks) == 0\n return zeros.(Int, size(blocks))\n end\n if !all(b -> ndims(b) == N, blocks)\n error(\"All blocks must have ndims consistent with ndims = $N of `blocks` array.\")\n end\n fullsizes = map!(size, Array{NTuple{N,Int}, N}(undef, size(blocks)), blocks)\n block_sizes = ntuple(ndims(blocks)) do i\n [s[i] for s in view(fullsizes, ntuple(j -> j == i ? (:) : 1, ndims(blocks))...)]\n end\n checksizes(fullsizes, block_sizes)\n return BlockSizes(block_sizes...)\nend\n\ngetsizes(block_sizes, block_index) = getindex.(block_sizes, block_index)\n\n@generated function checksizes(fullsizes::Array{NTuple{N,Int}, N}, block_sizes::NTuple{N,Vector{Int}}) where N\n quote\n @nloops $N i fullsizes begin\n block_index = @ntuple $N i\n if fullsizes[block_index...] != getsizes(block_sizes, block_index)\n error(\"size(blocks[\", strip(repr(block_index), ['(', ')']),\n \"]) (= \", fullsizes[block_index...],\n \") is incompatible with expected size: \",\n getsizes(block_sizes, block_index))\n end\n end\n return fullsizes\n end\nend\n\n\"\"\"\n mortar((block_11, ..., block_1m), ... (block_n1, ..., block_nm))\n\nConstruct a `BlockMatrix` with `n * m` blocks. Each `block_ij` must be an\n`AbstractMatrix`.\n\"\"\"\nmortar(rows::Vararg{NTuple{M, AbstractMatrix}}) where M =\n mortar(permutedims(reshape(\n foldl(append!, rows, init=eltype(eltype(rows))[]),\n M, length(rows))))\n\n# Convert AbstractArrays that conform to block array interface\nconvert(::Type{BlockArray{T,N,R}}, A::BlockArray{T,N,R}) where {T,N,R} = A\nconvert(::Type{BlockArray{T,N}}, A::BlockArray{T,N}) where {T,N} = A\nconvert(::Type{BlockArray{T}}, A::BlockArray{T}) where {T} = A\nconvert(::Type{BlockArray}, A::BlockArray) = A\n\nBlockArray{T, N}(A::AbstractArray{T2, N}) where {T,T2,N} =\n BlockArray(Array{T, N}(A), blocksizes(A))\nBlockArray{T1}(A::AbstractArray{T2, N}) where {T1,T2,N} = BlockArray{T1, N}(A)\nBlockArray(A::AbstractArray{T, N}) where {T,N} = BlockArray{T, N}(A)\n\nconvert(::Type{BlockArray{T, N}}, A::AbstractArray{T2, N}) where {T,T2,N} =\n BlockArray(convert(Array{T, N}, A), blocksizes(A))\nconvert(::Type{BlockArray{T1}}, A::AbstractArray{T2, N}) where {T1,T2,N} =\n convert(BlockArray{T1, N}, A)\nconvert(::Type{BlockArray}, A::AbstractArray{T, N}) where {T,N} =\n convert(BlockArray{T, N}, A)\n\ncopy(A::BlockArray) = _BlockArray(copy.(A.blocks), copy(A.block_sizes))\n\n################################\n# AbstractBlockArray Interface #\n################################\n@inline blocksizes(block_array::BlockArray) = block_array.block_sizes\n\n@inline function getblock(block_arr::BlockArray{T,N}, block::Vararg{Integer, N}) where {T,N}\n @boundscheck blockcheckbounds(block_arr, block...)\n block_arr.blocks[block...]\nend\n\n@inline function Base.getindex(block_arr::BlockArray{T,N}, blockindex::BlockIndex{N}) where {T,N}\n @boundscheck blockcheckbounds(block_arr, Block(blockindex.I))\n @inbounds block = getblock(block_arr, blockindex.I...)\n @boundscheck checkbounds(block, blockindex.\u03b1...)\n @inbounds v = block[blockindex.\u03b1...]\n return v\nend\n\n\n###########################\n# AbstractArray Interface #\n###########################\n\n@inline function Base.similar(block_array::BlockArray{T,N}, ::Type{T2}) where {T,N,T2}\n _BlockArray(similar(block_array.blocks, Array{T2, N}), copy(blocksizes(block_array)))\nend\n\n@inline function Base.getindex(block_arr::BlockArray{T, N}, i::Vararg{Integer, N}) where {T,N}\n @boundscheck checkbounds(block_arr, i...)\n @inbounds v = block_arr[global2blockindex(blocksizes(block_arr), i)]\n return v\nend\n\n@inline function Base.setindex!(block_arr::BlockArray{T, N}, v, i::Vararg{Integer, N}) where {T,N}\n @boundscheck checkbounds(block_arr, i...)\n @inbounds block_arr[global2blockindex(blocksizes(block_arr), i)] = v\n return block_arr\nend\n\n############\n# Indexing #\n############\n\nfunction _check_setblock!(block_arr::BlockArray{T, N}, v, block::NTuple{N, Integer}) where {T,N}\n for i in 1:N\n if size(v, i) != blocksize(block_arr, i, block[i])\n throw(DimensionMismatch(string(\"tried to assign $(size(v)) array to \", blocksize(block_arr, block), \" block\")))\n end\n end\nend\n\n\n@inline function setblock!(block_arr::BlockArray{T, N}, v, block::Vararg{Integer, N}) where {T,N}\n @boundscheck blockcheckbounds(block_arr, block...)\n @boundscheck _check_setblock!(block_arr, v, block)\n @inbounds block_arr.blocks[block...] = v\n return block_arr\nend\n\n@propagate_inbounds function Base.setindex!(block_array::BlockArray{T, N}, v, block_index::BlockIndex{N}) where {T,N}\n getblock(block_array, block_index.I...)[block_index.\u03b1...] = v\nend\n\nBase.dataids(arr::BlockArray) = (dataids(arr.blocks)..., dataids(arr.block_sizes)...)\n# This is not entirely valid. In principle, we have to concatenate\n# all dataids of all blocks. However, it makes `dataids` non-inferable.\n\n########\n# Misc #\n########\n\n@generated function Base.Array(block_array::BlockArray{T, N, R}) where {T,N,R}\n # TODO: This will fail for empty block array\n return quote\n block_sizes = blocksizes(block_array)\n arr = similar(block_array.blocks[1], size(block_array)...)\n @nloops $N i i->(1:nblocks(block_sizes, i)) begin\n block_index = @ntuple $N i\n indices = globalrange(block_sizes, block_index)\n arr[indices...] = getblock(block_array, block_index...)\n end\n\n return arr\n end\nend\n\n@generated function copyto!(block_array::BlockArray{T, N, R}, arr::R) where {T,N,R <: AbstractArray}\n return quote\n block_sizes = blocksizes(block_array)\n\n @nloops $N i i->(1:nblocks(block_sizes, i)) begin\n block_index = @ntuple $N i\n indices = globalrange(block_sizes, block_index)\n copyto!(getblock(block_array, block_index...), arr[indices...])\n end\n\n return block_array\n end\nend\n\nfunction Base.fill!(block_array::BlockArray, v)\n for block in block_array.blocks\n fill!(block, v)\n end\n block_array\nend\n\nfunction lmul!(\u03b1::Number, block_array::BlockArray)\n for block in block_array.blocks\n lmul!(\u03b1, block)\n end\n block_array\nend\n\nfunction rmul!(block_array::BlockArray, \u03b1::Number)\n for block in block_array.blocks\n rmul!(block, \u03b1)\n end\n block_array\nend\n", "meta": {"hexsha": "a728dde7f6d8d732dab9f96f5492d15811a9b8a1", "size": 15764, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/blockarray.jl", "max_stars_repo_name": "UnofficialJuliaMirrorSnapshots/BlockArrays.jl-8e7c35d0-a365-5155-bbbb-fb81a777f24e", "max_stars_repo_head_hexsha": "5812573bf3ee4b5797ba631e6b7a8b02c0f9d3d5", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2018-12-30T10:45:16.000Z", "max_stars_repo_stars_event_max_datetime": "2018-12-30T10:45:16.000Z", "max_issues_repo_path": "src/blockarray.jl", "max_issues_repo_name": "UnofficialJuliaMirrorSnapshots/BlockArrays.jl-8e7c35d0-a365-5155-bbbb-fb81a777f24e", "max_issues_repo_head_hexsha": "5812573bf3ee4b5797ba631e6b7a8b02c0f9d3d5", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/blockarray.jl", "max_forks_repo_name": "UnofficialJuliaMirrorSnapshots/BlockArrays.jl-8e7c35d0-a365-5155-bbbb-fb81a777f24e", "max_forks_repo_head_hexsha": "5812573bf3ee4b5797ba631e6b7a8b02c0f9d3d5", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 37.8942307692, "max_line_length": 153, "alphanum_fraction": 0.6609997463, "num_tokens": 4526, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.48047869292635403, "lm_q2_score": 0.14223188773801163, "lm_q1q2_score": 0.06833939151280775}}
{"text": "for p in (\"Knet\",\"AutoGrad\",\"ArgParse\",\"Compat\")\n Pkg.installed(p) == nothing && Pkg.add(p)\nend\n\n\"\"\"\ncharlm.jl: Knet8 version (c) Emre Yolcu, Deniz Yuret, 2016\n\nThis example implements an LSTM network for training and testing\ncharacter-level language models inspired by [\"The Unreasonable\nEffectiveness of Recurrent Neural\nNetworks\"](http://karpathy.github.io/2015/05/21/rnn-effectiveness) from\nthe Andrej Karpathy blog. The model can be trained with different\ngenres of text, and can be used to generate original text in the same\nstyle.\n\nExample usage:\n\n* `julia charlm.jl`: trains a model using its own code.\n\n* `julia charlm.jl --data foo.txt`: uses foo.txt to train instead.\n\n* `julia charlm.jl --data foo.txt bar.txt`: uses foo.txt for training\n and bar.txt for validation. Any number of files can be specified,\n the first two will be used for training and validation, the rest for\n testing.\n\n* `julia charlm.jl --best foo.jld --save bar.jld`: saves the best\n model (according to validation set) to foo.jld, last model to\n bar.jld.\n\n* `julia charlm.jl --load foo.jld --generate 1000`: generates 1000\n characters from the model in foo.jld.\n\n* `julia charlm.jl --help`: describes all available options.\n \n\"\"\"\nmodule CharLM\n\nusing Knet,AutoGrad,ArgParse,Compat\n\nfunction main(args=ARGS)\n s = ArgParseSettings()\n s.description=\"charlm.jl (c) Emre Yolcu, Deniz Yuret, 2016. Character level language model based on http://karpathy.github.io/2015/05/21/rnn-effectiveness.\"\n s.exc_handler=ArgParse.debug_handler\n @add_arg_table s begin\n (\"--datafiles\"; nargs='+'; help=\"If provided, use first file for training, second for dev, others for test.\")\n (\"--loadfile\"; help=\"Initialize model from file\")\n (\"--savefile\"; help=\"Save final model to file\")\n (\"--bestfile\"; help=\"Save best model to file\")\n (\"--generate\"; arg_type=Int; default=0; help=\"If non-zero generate given number of characters.\")\n (\"--hidden\"; nargs='+'; arg_type=Int; default=[256]; help=\"Sizes of one or more LSTM layers.\")\n (\"--embed\"; arg_type=Int; default=256; help=\"Size of the embedding vector.\")\n (\"--epochs\"; arg_type=Int; default=3; help=\"Number of epochs for training.\")\n (\"--batchsize\"; arg_type=Int; default=128; help=\"Number of sequences to train on in parallel.\")\n (\"--seqlength\"; arg_type=Int; default=100; help=\"Number of steps to unroll the network for.\")\n (\"--decay\"; arg_type=Float64; default=0.9; help=\"Learning rate decay.\")\n (\"--lr\"; arg_type=Float64; default=4.0; help=\"Initial learning rate.\")\n (\"--gclip\"; arg_type=Float64; default=3.0; help=\"Value to clip the gradient norm at.\")\n (\"--winit\"; arg_type=Float64; default=0.3; help=\"Initial weights set to winit*randn().\")\n (\"--gcheck\"; arg_type=Int; default=0; help=\"Check N random gradients.\")\n (\"--seed\"; arg_type=Int; default=-1; help=\"Random number seed.\")\n (\"--atype\"; default=(gpu()>=0 ? \"KnetArray{Float32}\" : \"Array{Float32}\"); help=\"array type: Array for cpu, KnetArray for gpu\")\n (\"--fast\"; action=:store_true; help=\"skip loss printing for faster run\")\n #TODO (\"--dropout\"; arg_type=Float64; default=0.0; help=\"Dropout probability.\")\n end\n println(s.description)\n isa(args, AbstractString) && (args=split(args))\n o = parse_args(args, s; as_symbols=true)\n println(\"opts=\",[(k,v) for (k,v) in o]...)\n o[:seed] > 0 && srand(o[:seed])\n o[:atype] = eval(parse(o[:atype]))\n if any(f->(o[f]!=nothing), (:loadfile, :savefile, :bestfile))\n Pkg.installed(\"JLD\")==nothing && Pkg.add(\"JLD\") # error(\"Please Pkg.add(\\\"JLD\\\") to load or save files.\")\n eval(Expr(:using,:JLD))\n end\n\n # we initialize a model from loadfile, train using datafiles (both optional).\n # if the user specifies neither, train a model using the charlm.jl source code.\n isempty(o[:datafiles]) && o[:loadfile]==nothing && push!(o[:datafiles],@__FILE__) # shakespeare()\n\n # read text and report lengths\n text = map((@compat readstring), o[:datafiles])\n !isempty(text) && info(\"Chars read: $(map((f,c)->(basename(f),length(c)),o[:datafiles],text))\")\n\n # vocab (char_to_index) comes from the initial model if there is one, otherwise from the datafiles.\n # if there is an initial model make sure the data has no new vocab\n if o[:loadfile]==nothing\n vocab = Dict{Char,Int}()\n for t in text, c in t; get!(vocab, c, 1+length(vocab)); end\n model = initweights(o[:atype], o[:hidden], length(vocab), o[:embed], o[:winit])\n else\n info(\"Loading model from $(o[:loadfile])\")\n vocab = load(o[:loadfile], \"vocab\") \n for t in text, c in t; haskey(vocab, c) || error(\"Unknown char $c\"); end\n model = map(p->convert(o[:atype],p), load(o[:loadfile], \"model\"))\n end\n info(\"$(length(vocab)) unique chars.\")\n if !isempty(text)\n train!(model, text, vocab, o)\n end\n if o[:savefile] != nothing\n info(\"Saving last model to $(o[:savefile])\")\n save(o[:savefile], \"model\", model, \"vocab\", vocab)\n end\n if o[:generate] > 0\n state = initstate(o[:atype],o[:hidden],1)\n generate(model, state, vocab, o[:generate])\n end\nend\n\n\nfunction train!(model, text, vocab, o)\n s0 = initstate(o[:atype], o[:hidden], o[:batchsize])\n data = map(t->minibatch(t, vocab, o[:batchsize]), text)\n lr = o[:lr]\n if o[:fast]\n @time (for epoch=1:o[:epochs]\n train1(model, copy(s0), data[1]; slen=o[:seqlength], lr=lr, gclip=o[:gclip])\n end; gpu()>=0 && Knet.cudaDeviceSynchronize())\n return\n end\n losses = map(d->loss(model,copy(s0),d), data)\n println((:epoch,0,:loss,losses...))\n devset = ifelse(length(data) > 1, 2, 1)\n devlast = devbest = losses[devset]\n for epoch=1:o[:epochs]\n @time train1(model, copy(s0), data[1]; slen=o[:seqlength], lr=lr, gclip=o[:gclip])\n @time losses = map(d->loss(model,copy(s0),d), data)\n println((:epoch,epoch,:loss,losses...))\n if o[:gcheck] > 0\n gradcheck(loss, model, copy(s0), data[1], 1:o[:seqlength]; gcheck=o[:gcheck])\n end\n devloss = losses[devset]\n if devloss < devbest\n devbest = devloss\n if o[:bestfile] != nothing\n info(\"Saving best model to $(o[:bestfile])\")\n save(o[:bestfile], \"model\", model, \"vocab\", vocab)\n end\n end\n if devloss > devlast\n lr *= o[:decay]\n info(\"New learning rate: $lr\")\n end\n devlast = devloss\n end\nend \n\n\n# sequence[t]: input token at time t\n# state is modified in place\nfunction train1(param, state, sequence; slen=100, lr=1.0, gclip=0.0)\n for t = 1:slen:length(sequence)-slen\n range = t:t+slen-1\n gloss = lossgradient(param, state, sequence, range)\n gscale = lr\n if gclip > 0\n gnorm = sqrt(mapreduce(sumabs2, +, 0, gloss))\n if gnorm > gclip\n gscale *= gclip / gnorm\n end\n end\n for k in 1:length(param)\n # param[k] -= gscale * gloss[k]\n axpy!(-gscale, gloss[k], param[k])\n end\n isa(state,Vector{Any}) || error(\"State should not be Boxed.\")\n # The following is needed in case AutoGrad boxes state values during gradient calculation\n for i = 1:length(state)\n state[i] = AutoGrad.getval(state[i])\n end\n end\nend\n\n# param[2k-1,2k]: weight and bias for the k'th lstm layer\n# param[end-2]: embedding matrix\n# param[end-1,end]: weight and bias for final prediction\nfunction initweights(atype, hidden, vocab, embed, winit)\n param = Array(Any, 2*length(hidden)+3)\n input = embed\n for k = 1:length(hidden)\n param[2k-1] = winit*randn(input+hidden[k], 4*hidden[k])\n param[2k] = zeros(1, 4*hidden[k])\n param[2k][1:hidden[k]] = 1 # forget gate bias\n input = hidden[k]\n end\n param[end-2] = winit*randn(vocab,embed)\n param[end-1] = winit*randn(hidden[end],vocab)\n param[end] = zeros(1,vocab)\n return map(p->convert(atype,p), param)\nend\n\n# state[2k-1,2k]: hidden and cell for the k'th lstm layer\nfunction initstate(atype, hidden, batchsize)\n state = Array(Any, 2*length(hidden))\n for k = 1:length(hidden)\n state[2k-1] = zeros(batchsize,hidden[k])\n state[2k] = zeros(batchsize,hidden[k])\n end\n return map(s->convert(atype,s), state)\nend\n\nfunction lstm(weight,bias,hidden,cell,input)\n gates = hcat(input,hidden) * weight .+ bias\n hsize = size(hidden,2)\n forget = sigm(gates[:,1:hsize])\n ingate = sigm(gates[:,1+hsize:2hsize])\n outgate = sigm(gates[:,1+2hsize:3hsize])\n change = tanh(gates[:,1+3hsize:end])\n cell = cell .* forget + ingate .* change\n hidden = outgate .* tanh(cell)\n return (hidden,cell)\nend\n\n# s[2k-1,2k]: hidden and cell for the k'th lstm layer\n# w[2k-1,2k]: weight and bias for k'th lstm layer\n# w[end-2]: embedding matrix\n# w[end-1,end]: weight and bias for final prediction\n# state is modified in place\nfunction predict(w, s, x)\n x = x * w[end-2]\n for i = 1:2:length(s)\n (s[i],s[i+1]) = lstm(w[i],w[i+1],s[i],s[i+1],x)\n x = s[i]\n end\n return x * w[end-1] .+ w[end]\nend\n\n# sequence[t]: input token at time t\n# state is modified in place\nfunction loss(param,state,sequence,range=1:length(sequence)-1)\n total = 0.0; count = 0\n atype = typeof(AutoGrad.getval(param[1]))\n input = convert(atype,sequence[first(range)])\n for t in range\n ypred = predict(param,state,input)\n ynorm = logp(ypred,2) # ypred .- log(sum(exp(ypred),2))\n ygold = convert(atype,sequence[t+1])\n total += sum(ygold .* ynorm)\n count += size(ygold,1)\n input = ygold\n end\n return -total / count\nend\n\nlossgradient = grad(loss)\n\nfunction generate(param, state, vocab, nchar)\n index_to_char = Array(Char, length(vocab))\n for (k,v) in vocab; index_to_char[v] = k; end\n input = oftype(param[1], zeros(1,length(vocab)))\n index = 1\n for t in 1:nchar\n ypred = predict(param,state,input)\n input[index] = 0\n index = sample(exp(logp(ypred)))\n print(index_to_char[index])\n input[index] = 1\n end\n println()\nend\n\nfunction sample(p)\n p = convert(Array,p)\n r = rand()\n for c = 1:length(p)\n r -= p[c]\n r < 0 && return c\n end\nend\n\nfunction shakespeare()\n file = Knet.dir(\"data\",\"100.txt\")\n if !isfile(file)\n info(\"Downloading 'The Complete Works of William Shakespeare'\")\n url = \"http://www.gutenberg.org/files/100/100.txt\"\n download(url,file)\n end\n return file\nend\n\nfunction minibatch(chars, char_to_index, batch_size)\n nbatch = div(length(chars), batch_size)\n vocab_size = length(char_to_index)\n data = [ falses(batch_size, vocab_size) for i=1:nbatch ] # using BitArrays\n cidx = 0\n for c in chars # safest way to iterate over utf-8 text\n idata = 1 + cidx % nbatch\n row = 1 + div(cidx, nbatch)\n row > batch_size && break\n col = char_to_index[c]\n data[idata][row,col] = 1\n cidx += 1\n end\n return data\nend\n\n# To be able to load/save KnetArrays:\nif Pkg.installed(\"JLD\") != nothing\n import JLD: writeas, readas\n type KnetJLD; a::Array; end\n writeas(c::KnetArray) = KnetJLD(Array(c))\n readas(d::KnetJLD) = KnetArray(d.a)\nend\n\n# This allows both non-interactive (shell command) and interactive calls like:\n# $ julia charlm.jl --epochs 10\n# julia> CharLM.main(\"--epochs 10\")\n!isinteractive() && !isdefined(Core.Main,:load_only) && main(ARGS)\n\nend # module\n\n# Note: 10.txt used in the sample runs below was generated using\n# head -10000 100.txt > 10.txt\n# where 100.txt is the file downloaded by shakespeare().\n\n\n\n### SAMPLE RUN 74a2e6c+ Mon Sep 19 14:03:10 EEST 2016\n### Implemented multi-layer. Removed the keepstate option fixing it to true.\n### Note that winit default changed so I specify it below for comparison.\n### The slight difference is due to keepstate.\n\n# julia> CharLM.main(\"--data 10.txt --winit 0.3 --fast\")\n# charlm.jl (c) Emre Yolcu, Deniz Yuret, 2016. Character level language model based on http://karpathy.github.io/2015/05/21/rnn-effectiveness.\n# opts=(:lr,4.0)(:atype,\"KnetArray{Float32}\")(:winit,0.3)(:savefile,nothing)(:loadfile,nothing)(:generate,0)(:bestfile,nothing)(:gclip,3.0)(:hidden,[256])(:epochs,3)(:decay,0.9)(:gcheck,0)(:seqlength,100)(:seed,42)(:embed,256)(:batchsize,128)(:datafiles,Any[\"10.txt\"])(:fast,true)\n# INFO: Chars read: [(\"10.txt\",425808)]\n# INFO: 87 unique chars.\n# 1.406687 seconds (1.74 M allocations: 196.799 MB, 2.49% gc time)\n# (:epoch,0,:loss,6.1075509900258)\n# 4.002638 seconds (6.12 M allocations: 374.188 MB, 2.41% gc time)\n# 3.990772 seconds (6.11 M allocations: 374.129 MB, 2.44% gc time)\n# 4.006249 seconds (6.12 M allocations: 374.240 MB, 2.53% gc time)\n# 1.405878 seconds (1.75 M allocations: 197.059 MB, 2.56% gc time)\n# (:epoch,3,:loss,1.8713183968407767)\n\n\n\n### SAMPLE RUN 4ce58d1+ Fri Sep 16 12:24:00 EEST 2016\n### Transposed everything so getindex does not need to copy\n\n# charlm.jl (c) Emre Yolcu, Deniz Yuret, 2016. Character level language model based on http://karpathy.github.io/2015/05/21/rnn-effectiveness.\n# opts=(:keepstate,false)(:lr,4.0)(:atype,\"KnetArray{Float32}\")(:winit,0.3)(:savefile,nothing)(:loadfile,nothing)(:generate,0)(:bestfile,nothing)(:gclip,3.0)(:embedding,256)(:hidden,256)(:epochs,3)(:decay,0.9)(:gcheck,0)(:seqlength,100)(:seed,42)(:batchsize,128)(:datafiles,Any[\"data/10.txt\"])(:fast,true)\n# INFO: Chars read: [(\"10.txt\",425808)]\n# INFO: 87 unique chars.\n# 1.388652 seconds (1.73 M allocations: 196.686 MB, 2.04% gc time)\n# (:epoch,0,:loss,6.1075509900258)\n# 3.940298 seconds (6.06 M allocations: 373.166 MB, 2.06% gc time)\n# 3.935995 seconds (6.06 M allocations: 373.244 MB, 2.07% gc time)\n# 3.934983 seconds (6.06 M allocations: 373.245 MB, 2.08% gc time)\n# 1.390374 seconds (1.73 M allocations: 196.820 MB, 2.11% gc time)\n# (:epoch,3,:loss,1.860654126576015)\n\n\n\n### SAMPLE RUN 31136d5+ Wed Sep 14 17:51:44 EEST 2016: using vcat(x,h) and vcat(w...)\n### optimized learning parameters: winit=0.3, lr=4.0, gclip=3.0\n\n# charlm.jl (c) Emre Yolcu, Deniz Yuret, 2016. Character level language model based on http://karpathy.github.io/2015/05/21/rnn-effectiveness.\n# opts=(:keepstate,false)(:lr,4.0)(:atype,\"KnetArray{Float32}\")(:winit,0.3)(:savefile,nothing)(:loadfile,nothing)(:generate,0)(:bestfile,nothing)(:gclip,3.0)(:embedding,256)(:hidden,256)(:epochs,3)(:decay,0.9)(:gcheck,0)(:seqlength,100)(:seed,42)(:batchsize,128)(:datafiles,Any[\"10.txt\"])(:fast,true)\n# INFO: Chars read: [(\"10.txt\",425808)]\n# INFO: 87 unique chars.\n# 1.596959 seconds (1.79 M allocations: 197.432 MB, 2.56% gc time)\n# (:epoch,0,:loss,5.541976199042528)\n# 4.421566 seconds (6.23 M allocations: 375.843 MB, 2.54% gc time)\n# 4.418540 seconds (6.25 M allocations: 376.058 MB, 2.51% gc time)\n# 4.402317 seconds (6.26 M allocations: 376.297 MB, 2.66% gc time)\n# 1.594677 seconds (1.81 M allocations: 197.737 MB, 2.64% gc time)\n# (:epoch,3,:loss,1.8484550957572192)\n\n\n### SAMPLE RUN 80503e7+ Wed Sep 14 17:35:36 EEST 2016: using vcat(x,h)\n#\n# charlm.jl (c) Emre Yolcu, Deniz Yuret, 2016. Character level language model based on http://karpathy.github.io/2015/05/21/rnn-effectiveness.\n# opts=(:keepstate,false)(:lr,1.0)(:atype,\"KnetArray{Float32}\")(:savefile,nothing)(:loadfile,nothing)(:generate,0)(:bestfile,nothing)(:embedding,256)(:gclip,5.0)(:hidden,256)(:epochs,3)(:decay,0.9)(:gcheck,0)(:seqlength,100)(:seed,42)(:batchsize,128)(:datafiles,Any[\"10.txt\"])(:fast,true)\n# INFO: Chars read: [(\"10.txt\",425808)]\n# INFO: 87 unique chars.\n# 1.930180 seconds (1.95 M allocations: 213.741 MB, 1.82% gc time)\n# (:epoch,0,:loss,4.462641664662756)\n# 4.968101 seconds (7.47 M allocations: 454.259 MB, 2.24% gc time)\n# 4.963733 seconds (7.47 M allocations: 454.363 MB, 2.26% gc time)\n# 4.967413 seconds (7.45 M allocations: 454.024 MB, 2.14% gc time)\n# 1.945658 seconds (1.98 M allocations: 214.183 MB, 2.02% gc time)\n# (:epoch,3,:loss,3.2389672966290237)\n\n\n### SAMPLE RUN 65f57ff+ Wed Sep 14 10:02:30 EEST 2016: separate x, h, w, b\n#\n# charlm.jl (c) Emre Yolcu, Deniz Yuret, 2016. Character level language model based on http://karpathy.github.io/2015/05/21/rnn-effectiveness.\n# opts=(:keepstate,false)(:lr,1.0)(:atype,\"KnetArray{Float32}\")(:savefile,nothing)(:loadfile,nothing)(:generate,0)(:bestfile,nothing)(:embedding,256)(:gclip,5.0)(:hidden,256)(:epochs,3)(:decay,0.9)(:gcheck,0)(:seqlength,100)(:seed,42)(:batchsize,128)(:datafiles,Any[\"10.txt\"])(:fast,true)\n# INFO: Chars read: [(\"10.txt\",425808)]\n# INFO: 87 unique chars.\n# 2.156358 seconds (2.31 M allocations: 237.913 MB, 2.30% gc time)\n# (:epoch,0,:loss,4.465127425659868)\n# 6.287736 seconds (9.54 M allocations: 574.703 MB, 2.84% gc time)\n# 6.272144 seconds (9.54 M allocations: 574.633 MB, 2.80% gc time)\n# 6.277462 seconds (9.54 M allocations: 574.637 MB, 2.86% gc time)\n# 2.165516 seconds (2.34 M allocations: 238.323 MB, 2.56% gc time)\n# (:epoch,3,:loss,3.226540256084356)\n\n\n### SAMPLE OUTPUT (with head -10000 100.txt): first version\n# julia> CharLM.main(\"--gpu --data 10.txt\")\n# opts=(:lr,1.0)(:savefile,nothing)(:loadfile,nothing)(:dropout,0.0)(:generate,0)(:bestfile,nothing)(:embedding,256)(:gclip,5.0)(:hidden,256)(:epochs,10)(:nlayer,1)(:decay,0.9)(:gpu,true)(:seqlength,100)(:seed,42)(:batchsize,128)(:datafiles,Any[\"10.txt\"])\n# INFO: Chars read: [(\"10.txt\",425808)]\n# INFO: 87 unique chars.\n# (0,4.465127425659868)\n# 2.182693 seconds (2.36 M allocations: 240.394 MB, 1.74% gc time)\n# 7.861079 seconds (10.12 M allocations: 601.311 MB, 1.84% gc time)\n# (1,3.3244698245543285)\n# 2.159062 seconds (2.35 M allocations: 239.217 MB, 1.80% gc time)\n# 6.200085 seconds (9.55 M allocations: 575.573 MB, 2.28% gc time)\n# (2,3.24593908969621)\n# 2.352389 seconds (2.34 M allocations: 239.381 MB, 1.51% gc time)\n# 6.211946 seconds (9.55 M allocations: 575.568 MB, 2.21% gc time)\n# (3,3.226540256084356)\n\n", "meta": {"hexsha": "27c71fa1752b535324012bb0bf877522f1b15adf", "size": 18100, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "examples/charlm.jl", "max_stars_repo_name": "enzotarta/Knet.jl", "max_stars_repo_head_hexsha": "4330f3fec812ccbbfeabbcf6aa63450ca743f9c2", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "examples/charlm.jl", "max_issues_repo_name": "enzotarta/Knet.jl", "max_issues_repo_head_hexsha": "4330f3fec812ccbbfeabbcf6aa63450ca743f9c2", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "examples/charlm.jl", "max_forks_repo_name": "enzotarta/Knet.jl", "max_forks_repo_head_hexsha": "4330f3fec812ccbbfeabbcf6aa63450ca743f9c2", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 43.3014354067, "max_line_length": 305, "alphanum_fraction": 0.6443093923, "num_tokens": 5976, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.5, "lm_q2_score": 0.13660839354130014, "lm_q1q2_score": 0.06830419677065007}}
{"text": "# ---\n# title: 236. Lowest Common Ancestor of a Binary Tree\n# id: problem236\n# author: zhwang\n# date: 2022-02-10\n# difficulty: Medium\n# categories: Tree\n# link: Student name, Student name, Student name ... Student name\nSmith School of Chemical and Biomolecular Engineering, Cornell University, Ithaca NY 14850
\n\"\"\"\n\n# \u2554\u2550\u2561 87a183bc-3857-4189-8103-18c46ff3245d\nmd\"\"\"\n#### Build the stoichiometric array\n\"\"\"\n\n# \u2554\u2550\u2561 5338451e-3c4b-4030-bbbb-42eaf4209a89\nbegin\n\n\t# fill me in\n\t# ...\n\t\nend\n\n# \u2554\u2550\u2561 6970dab5-16bd-4898-b88d-723cb1b3d89e\nmd\"\"\"\n#### Convex analysis: extreme pathways\n\"\"\"\n\n# \u2554\u2550\u2561 97b0763d-dcab-4afa-b660-52e18b3d523f\nbegin\n\n\t# fill me in\n\t# ...\n\t\nend\n\n# \u2554\u2550\u2561 b473b17e-3bf5-4b6c-af24-fe57b5a7e7e9\nmd\"\"\"\n#### Metabolite connectivity array (MCA)\n\"\"\"\n\n# \u2554\u2550\u2561 999ae1fd-5341-4f66-9db2-dec53fa0cd49\n\n\n# \u2554\u2550\u2561 b7e5d1a6-57ed-4d09-a039-a4bd12386367\nmd\"\"\"\n#### Reaction connectivity array (RCA)\n\"\"\"\n\n# \u2554\u2550\u2561 4520fc6e-7305-487e-924d-af22406e6d45\nbegin\n\n\t# fill me in ...\nend\n\n# \u2554\u2550\u2561 267865de-1b5c-4579-861b-c6c46beb4739\nfunction ingredients(path::String)\n\t\n\t# this is from the Julia source code (evalfile in base/loading.jl)\n\t# but with the modification that it returns the module instead of the last object\n\tname = Symbol(\"lib\")\n\tm = Module(name)\n\tCore.eval(m,\n Expr(:toplevel,\n :(eval(x) = $(Expr(:core, :eval))($name, x)),\n :(include(x) = $(Expr(:top, :include))($name, x)),\n :(include(mapexpr::Function, x) = $(Expr(:top, :include))(mapexpr, $name, x)),\n :(include($path))))\n\tm\nend\n\n# \u2554\u2550\u2561 67f5db98-88d0-11ec-27ac-b57538a166f4\nbegin\n\t# import some packages -\n\tusing PlutoUI\n\tusing PrettyTables\n\tusing LinearAlgebra\n\t\n\t# setup paths -\n\tconst _PATH_TO_NOTEBOOK = pwd()\n\tconst _PATH_TO_SRC = joinpath(_PATH_TO_NOTEBOOK,\"src\")\n\n\t# load the PS2 code lib -\n\tlib = ingredients(joinpath(_PATH_TO_SRC, \"Include.jl\"));\n\n\t# return -\n\tnothing\nend\n\n# \u2554\u2550\u2561 ab2bcfd5-3ba7-4388-8a3c-2cb95fba989a\nhtml\"\"\"\n\"\"\"\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000001\nPLUTO_PROJECT_TOML_CONTENTS = \"\"\"\n[deps]\nLinearAlgebra = \"37e2e46d-f89d-539d-b4ee-838fcccc9c8e\"\nPlutoUI = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\nPrettyTables = \"08abe8d2-0d0c-5749-adfa-8a2ac140af0d\"\n\n[compat]\nPlutoUI = \"~0.7.34\"\nPrettyTables = \"~1.3.1\"\n\"\"\"\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000002\nPLUTO_MANIFEST_TOML_CONTENTS = \"\"\"\n# This file is machine-generated - editing it directly is not advised\n\njulia_version = \"1.7.1\"\nmanifest_format = \"2.0\"\n\n[[deps.AbstractPlutoDingetjes]]\ndeps = [\"Pkg\"]\ngit-tree-sha1 = \"8eaf9f1b4921132a4cff3f36a1d9ba923b14a481\"\nuuid = \"6e696c72-6542-2067-7265-42206c756150\"\nversion = \"1.1.4\"\n\n[[deps.ArgTools]]\nuuid = \"0dad84c5-d112-42e6-8d28-ef12dabb789f\"\n\n[[deps.Artifacts]]\nuuid = \"56f22d72-fd6d-98f1-02f0-08ddc0907c33\"\n\n[[deps.Base64]]\nuuid = \"2a0f44e3-6c83-55bd-87e4-b1978d98bd5f\"\n\n[[deps.ColorTypes]]\ndeps = [\"FixedPointNumbers\", \"Random\"]\ngit-tree-sha1 = \"024fe24d83e4a5bf5fc80501a314ce0d1aa35597\"\nuuid = \"3da002f7-5984-5a60-b8a6-cbb66c0b333f\"\nversion = \"0.11.0\"\n\n[[deps.CompilerSupportLibraries_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"e66e0078-7015-5450-92f7-15fbd957f2ae\"\n\n[[deps.Crayons]]\ngit-tree-sha1 = \"249fe38abf76d48563e2f4556bebd215aa317e15\"\nuuid = \"a8cc5b0e-0ffa-5ad4-8c14-923d3ee1735f\"\nversion = \"4.1.1\"\n\n[[deps.DataAPI]]\ngit-tree-sha1 = \"cc70b17275652eb47bc9e5f81635981f13cea5c8\"\nuuid = \"9a962f9c-6df0-11e9-0e5d-c546b8b5ee8a\"\nversion = \"1.9.0\"\n\n[[deps.DataValueInterfaces]]\ngit-tree-sha1 = \"bfc1187b79289637fa0ef6d4436ebdfe6905cbd6\"\nuuid = \"e2d170a0-9d28-54be-80f0-106bbe20a464\"\nversion = \"1.0.0\"\n\n[[deps.Dates]]\ndeps = [\"Printf\"]\nuuid = \"ade2ca70-3891-5945-98fb-dc099432e06a\"\n\n[[deps.Downloads]]\ndeps = [\"ArgTools\", \"LibCURL\", \"NetworkOptions\"]\nuuid = \"f43a241f-c20a-4ad4-852c-f6b1247861c6\"\n\n[[deps.FixedPointNumbers]]\ndeps = [\"Statistics\"]\ngit-tree-sha1 = \"335bfdceacc84c5cdf16aadc768aa5ddfc5383cc\"\nuuid = \"53c48c17-4a7d-5ca2-90c5-79b7896eea93\"\nversion = \"0.8.4\"\n\n[[deps.Formatting]]\ndeps = [\"Printf\"]\ngit-tree-sha1 = \"8339d61043228fdd3eb658d86c926cb282ae72a8\"\nuuid = \"59287772-0a20-5a39-b81b-1366585eb4c0\"\nversion = \"0.4.2\"\n\n[[deps.Hyperscript]]\ndeps = [\"Test\"]\ngit-tree-sha1 = \"8d511d5b81240fc8e6802386302675bdf47737b9\"\nuuid = \"47d2ed2b-36de-50cf-bf87-49c2cf4b8b91\"\nversion = \"0.0.4\"\n\n[[deps.HypertextLiteral]]\ngit-tree-sha1 = \"2b078b5a615c6c0396c77810d92ee8c6f470d238\"\nuuid = \"ac1192a8-f4b3-4bfe-ba22-af5b92cd3ab2\"\nversion = \"0.9.3\"\n\n[[deps.IOCapture]]\ndeps = [\"Logging\", \"Random\"]\ngit-tree-sha1 = \"f7be53659ab06ddc986428d3a9dcc95f6fa6705a\"\nuuid = \"b5f81e59-6552-4d32-b1f0-c071b021bf89\"\nversion = \"0.2.2\"\n\n[[deps.InteractiveUtils]]\ndeps = [\"Markdown\"]\nuuid = \"b77e0a4c-d291-57a0-90e8-8db25a27a240\"\n\n[[deps.IteratorInterfaceExtensions]]\ngit-tree-sha1 = \"a3f24677c21f5bbe9d2a714f95dcd58337fb2856\"\nuuid = \"82899510-4779-5014-852e-03e436cf321d\"\nversion = \"1.0.0\"\n\n[[deps.JSON]]\ndeps = [\"Dates\", \"Mmap\", \"Parsers\", \"Unicode\"]\ngit-tree-sha1 = \"8076680b162ada2a031f707ac7b4953e30667a37\"\nuuid = \"682c06a0-de6a-54ab-a142-c8b1cf79cde6\"\nversion = \"0.21.2\"\n\n[[deps.LibCURL]]\ndeps = [\"LibCURL_jll\", \"MozillaCACerts_jll\"]\nuuid = \"b27032c2-a3e7-50c8-80cd-2d36dbcbfd21\"\n\n[[deps.LibCURL_jll]]\ndeps = [\"Artifacts\", \"LibSSH2_jll\", \"Libdl\", \"MbedTLS_jll\", \"Zlib_jll\", \"nghttp2_jll\"]\nuuid = \"deac9b47-8bc7-5906-a0fe-35ac56dc84c0\"\n\n[[deps.LibGit2]]\ndeps = [\"Base64\", \"NetworkOptions\", \"Printf\", \"SHA\"]\nuuid = \"76f85450-5226-5b5a-8eaa-529ad045b433\"\n\n[[deps.LibSSH2_jll]]\ndeps = [\"Artifacts\", \"Libdl\", \"MbedTLS_jll\"]\nuuid = \"29816b5a-b9ab-546f-933c-edad1886dfa8\"\n\n[[deps.Libdl]]\nuuid = \"8f399da3-3557-5675-b5ff-fb832c97cbdb\"\n\n[[deps.LinearAlgebra]]\ndeps = [\"Libdl\", \"libblastrampoline_jll\"]\nuuid = \"37e2e46d-f89d-539d-b4ee-838fcccc9c8e\"\n\n[[deps.Logging]]\nuuid = \"56ddb016-857b-54e1-b83d-db4d58db5568\"\n\n[[deps.Markdown]]\ndeps = [\"Base64\"]\nuuid = \"d6f4376e-aef5-505a-96c1-9c027394607a\"\n\n[[deps.MbedTLS_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"c8ffd9c3-330d-5841-b78e-0817d7145fa1\"\n\n[[deps.Mmap]]\nuuid = \"a63ad114-7e13-5084-954f-fe012c677804\"\n\n[[deps.MozillaCACerts_jll]]\nuuid = \"14a3606d-f60d-562e-9121-12d972cd8159\"\n\n[[deps.NetworkOptions]]\nuuid = \"ca575930-c2e3-43a9-ace4-1e988b2c1908\"\n\n[[deps.OpenBLAS_jll]]\ndeps = [\"Artifacts\", \"CompilerSupportLibraries_jll\", \"Libdl\"]\nuuid = \"4536629a-c528-5b80-bd46-f80d51c5b363\"\n\n[[deps.Parsers]]\ndeps = [\"Dates\"]\ngit-tree-sha1 = \"0b5cfbb704034b5b4c1869e36634438a047df065\"\nuuid = \"69de0a69-1ddd-5017-9359-2bf0b02dc9f0\"\nversion = \"2.2.1\"\n\n[[deps.Pkg]]\ndeps = [\"Artifacts\", \"Dates\", \"Downloads\", \"LibGit2\", \"Libdl\", \"Logging\", \"Markdown\", \"Printf\", \"REPL\", \"Random\", \"SHA\", \"Serialization\", \"TOML\", \"Tar\", \"UUIDs\", \"p7zip_jll\"]\nuuid = \"44cfe95a-1eb2-52ea-b672-e2afdf69b78f\"\n\n[[deps.PlutoUI]]\ndeps = [\"AbstractPlutoDingetjes\", \"Base64\", \"ColorTypes\", \"Dates\", \"Hyperscript\", \"HypertextLiteral\", \"IOCapture\", \"InteractiveUtils\", \"JSON\", \"Logging\", \"Markdown\", \"Random\", \"Reexport\", \"UUIDs\"]\ngit-tree-sha1 = \"8979e9802b4ac3d58c503a20f2824ad67f9074dd\"\nuuid = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\nversion = \"0.7.34\"\n\n[[deps.PrettyTables]]\ndeps = [\"Crayons\", \"Formatting\", \"Markdown\", \"Reexport\", \"Tables\"]\ngit-tree-sha1 = \"dfb54c4e414caa595a1f2ed759b160f5a3ddcba5\"\nuuid = \"08abe8d2-0d0c-5749-adfa-8a2ac140af0d\"\nversion = \"1.3.1\"\n\n[[deps.Printf]]\ndeps = [\"Unicode\"]\nuuid = \"de0858da-6303-5e67-8744-51eddeeeb8d7\"\n\n[[deps.REPL]]\ndeps = [\"InteractiveUtils\", \"Markdown\", \"Sockets\", \"Unicode\"]\nuuid = \"3fa0cd96-eef1-5676-8a61-b3b8758bbffb\"\n\n[[deps.Random]]\ndeps = [\"SHA\", \"Serialization\"]\nuuid = \"9a3f8284-a2c9-5f02-9a11-845980a1fd5c\"\n\n[[deps.Reexport]]\ngit-tree-sha1 = \"45e428421666073eab6f2da5c9d310d99bb12f9b\"\nuuid = \"189a3867-3050-52da-a836-e630ba90ab69\"\nversion = \"1.2.2\"\n\n[[deps.SHA]]\nuuid = \"ea8e919c-243c-51af-8825-aaa63cd721ce\"\n\n[[deps.Serialization]]\nuuid = \"9e88b42a-f829-5b0c-bbe9-9e923198166b\"\n\n[[deps.Sockets]]\nuuid = \"6462fe0b-24de-5631-8697-dd941f90decc\"\n\n[[deps.SparseArrays]]\ndeps = [\"LinearAlgebra\", \"Random\"]\nuuid = \"2f01184e-e22b-5df5-ae63-d93ebab69eaf\"\n\n[[deps.Statistics]]\ndeps = [\"LinearAlgebra\", \"SparseArrays\"]\nuuid = \"10745b16-79ce-11e8-11f9-7d13ad32a3b2\"\n\n[[deps.TOML]]\ndeps = [\"Dates\"]\nuuid = \"fa267f1f-6049-4f14-aa54-33bafae1ed76\"\n\n[[deps.TableTraits]]\ndeps = [\"IteratorInterfaceExtensions\"]\ngit-tree-sha1 = \"c06b2f539df1c6efa794486abfb6ed2022561a39\"\nuuid = \"3783bdb8-4a98-5b6b-af9a-565f29a5fe9c\"\nversion = \"1.0.1\"\n\n[[deps.Tables]]\ndeps = [\"DataAPI\", \"DataValueInterfaces\", \"IteratorInterfaceExtensions\", \"LinearAlgebra\", \"TableTraits\", \"Test\"]\ngit-tree-sha1 = \"bb1064c9a84c52e277f1096cf41434b675cd368b\"\nuuid = \"bd369af6-aec1-5ad0-b16a-f7cc5008161c\"\nversion = \"1.6.1\"\n\n[[deps.Tar]]\ndeps = [\"ArgTools\", \"SHA\"]\nuuid = \"a4e569a6-e804-4fa4-b0f3-eef7a1d5b13e\"\n\n[[deps.Test]]\ndeps = [\"InteractiveUtils\", \"Logging\", \"Random\", \"Serialization\"]\nuuid = \"8dfed614-e22c-5e08-85e1-65c5234f0b40\"\n\n[[deps.UUIDs]]\ndeps = [\"Random\", \"SHA\"]\nuuid = \"cf7118a7-6976-5b1a-9a39-7adc72f591a4\"\n\n[[deps.Unicode]]\nuuid = \"4ec0a83e-493e-50e2-b9ac-8f72acf5a8f5\"\n\n[[deps.Zlib_jll]]\ndeps = [\"Libdl\"]\nuuid = \"83775a58-1f1d-513f-b197-d71354ab007a\"\n\n[[deps.libblastrampoline_jll]]\ndeps = [\"Artifacts\", \"Libdl\", \"OpenBLAS_jll\"]\nuuid = \"8e850b90-86db-534c-a0d3-1478176c7d93\"\n\n[[deps.nghttp2_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"8e850ede-7688-5339-a07c-302acd2aaf8d\"\n\n[[deps.p7zip_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"3f19e933-33d8-53b3-aaab-bd5110c3b7a0\"\n\"\"\"\n\n# \u2554\u2550\u2561 Cell order:\n# \u255f\u25006b1ad54f-61e4-490d-9032-7a557e8dc82f\n# \u255f\u25007057c8e4-9e94-4a28-a885-07f5c96ebe39\n# \u255f\u250087a183bc-3857-4189-8103-18c46ff3245d\n# \u2560\u25505338451e-3c4b-4030-bbbb-42eaf4209a89\n# \u2560\u25506970dab5-16bd-4898-b88d-723cb1b3d89e\n# \u2560\u255097b0763d-dcab-4afa-b660-52e18b3d523f\n# \u255f\u2500b473b17e-3bf5-4b6c-af24-fe57b5a7e7e9\n# \u2560\u2550999ae1fd-5341-4f66-9db2-dec53fa0cd49\n# \u255f\u2500b7e5d1a6-57ed-4d09-a039-a4bd12386367\n# \u2560\u25504520fc6e-7305-487e-924d-af22406e6d45\n# \u2560\u255067f5db98-88d0-11ec-27ac-b57538a166f4\n# \u2560\u2550267865de-1b5c-4579-861b-c6c46beb4739\n# \u255f\u2500ab2bcfd5-3ba7-4388-8a3c-2cb95fba989a\n# \u255f\u250000000000-0000-0000-0000-000000000001\n# \u255f\u250000000000-0000-0000-0000-000000000002\n", "meta": {"hexsha": "f2abe9e5e66d014f9fab15b2f157a6d780dbb3a1", "size": 10245, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "problem_sets/PS2/PS2-5440-7770-Template.jl", "max_stars_repo_name": "za-gao/CHEME-5440-7770-Cornell-Spring-2022", "max_stars_repo_head_hexsha": "ad8c9a10c28bf893c9fa11f23561d37ed4883831", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "problem_sets/PS2/PS2-5440-7770-Template.jl", "max_issues_repo_name": "za-gao/CHEME-5440-7770-Cornell-Spring-2022", "max_issues_repo_head_hexsha": "ad8c9a10c28bf893c9fa11f23561d37ed4883831", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "problem_sets/PS2/PS2-5440-7770-Template.jl", "max_forks_repo_name": "za-gao/CHEME-5440-7770-Cornell-Spring-2022", "max_forks_repo_head_hexsha": "ad8c9a10c28bf893c9fa11f23561d37ed4883831", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 26.2020460358, "max_line_length": 196, "alphanum_fraction": 0.7190824793, "num_tokens": 4570, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.39233681595684605, "lm_q2_score": 0.1732882080640581, "lm_q1q2_score": 0.06798734379472}}
{"text": "abstract type Question end\n\nmutable struct Stringq <: Question\n re::Regex\n label\n hint\nend\n\n\"\"\"\n stringq(re::Regex; label=\"\", hint=\"\")\n\nMatch string answer with regular expression\n\nArguments:\n\n* `re`: a regular expression for grading\n* `label`: optional label for the form element\n* `hint`: optional plain-text hint that can be seen on hover\n\n\"\"\"\nstringq(re::Regex; label=\"\", hint=\"\") = Stringq(re, label, hint)\n\n\n##\nmutable struct Numericq <: Question\n val\n tol\n units\n label\n hint\nend\n\n\"\"\"\n numericq(value, tol=1e-3; label=\"\", hint=\"\", units=\"\")\n\nMatch a numeric answer\n\nArguments:\n\n* `value`: the numeric answer\n* `tol`: ``|answer - value| \\\\le tol`` is used to determine correctness\n* `label`: optional label for the form element\n* `hint`: optional plain-text hint that can be seen on hover\n* `units`: a string indicating expected units.\n\n\"\"\"\nfunction numericq(val, tol=1e-3, args...;\n label=\"\",\n hint=\"\", units::AbstractString=\"\")\n\n Numericq(val, tol, units, label, hint)\nend\n\nnumericq(val::Int; kwargs...) = numericq(val, 0; kwargs...)\n\n\n\n##\nmutable struct Radioq <: Question\n choices\n answer\n values\n labels\n label\n hint\n inline\nend\n\n\"\"\"\n radioq(choices, answer; label=\"\", hint=\"\", keep_order=false)\n\nMultiple choice question (one of several)\n\nArguments:\n\n* `choices`: indexable collection of choices. As seen in the example, choices can be formatted with markdown.\n\n* `answer::Int`: index of correct choice\n\n* `keep_order::Boolean`: if `true` keeps display order of choices, otherwise they are shuffled.\n\n* `inline::Bool`: hint to render inline (or not) if supported\n\n* `label`: optional label for the form element\n\n* `hint`: optional plain-text hint that can be seen on hover\n\nExample:\n\n```\nchoices = [\"beta\", raw\"``\\\\beta``\", \"`beta`\"]\nanswer = 2\nradioq(choices, answer; hint=\"Which is the Greek symbol?\")\n```\n\n\"\"\"\nfunction radioq(choices, answer::Integer;\n label=\"\", hint=\"\", inline::Bool=(hint!=\"\"),\n keep_order::Bool=false)\n inds = collect(1:length(choices))\n values = copy(inds)\n labels = choices\n !keep_order && shuffle!(inds)\n\n Radioq(choices[inds], findfirst(isequal(answer), inds),\n values, labels[inds], label, hint, inline)\nend\n\n\n\n\n##\nmutable struct Multiq <: Question\n choices\n answer\n values\n labels\n label\n hint\n inline\nend\n\n\"\"\"\n multiq(choices, answers; label=\"\", hint=\"\", keep_order=false)\n\nMultiple choice question (one *or more* of several)\n\nArguments:\n\n* `choices`: indexable collection of choices. As seen in the example, choices can be formatted with markdown.\n\n* `answers::Vector{Int}`: index of correct choice(s)\n\n* `keep_order::Boolean`: if `true` keeps display order of choices, otherwise they are shuffled.\n\n* `inline::Bool`: hint to render inline (or not) if supported\n\n* `label`: optional label for the form element\n\n* `hint`: optional plain-text hint that can be seen on hover\n\nExample:\n\n```\nchoices = [\"pear\", \"tomato\", \"banana\"]\nanswers = [1,3]\nmultiplecq(choices, answers; hint=\"not the red one!\")\n```\n\n\"\"\"\nfunction multiq(choices, answers;\n label=\"\", hint=\"\", inline::Bool=(hint!=\"\"),\n keep_order::Bool=false)\n inds = collect(1:length(choices))\n values = copy(inds)\n labels = choices\n !keep_order && shuffle!(inds)\n\n Multiq(choices[inds], findall(in(answers), inds),\n values, labels[inds], label, hint, inline)\nend\n\n\n\"\"\"\n booleanq(ans; [label, hint])\nTrue of false questions:\n\nExample:\n\n```\nbooleanq(true; label=\"Does it hurt...\")\n```\n\n\"\"\"\nfunction booleanq(ans::Bool;\n labels::Vector=[\"true\", \"false\"],\n label=\"\", hint::AbstractString=\"\",\n inline::Bool=true)\n choices = labels[1:2]\n ans = 2 - ans\n radioq(choices, ans;\n label=label, hint=hint, inline=inline, keep_order=true)\nend\n\n\"\"\"\n yesnoq(ans)\n\nBoolean question with `yes` or `no` labels.\n\nExamples:\n\n```\nyesnoq(\"yes\")\nyesnoq(true)\n```\n\n\"\"\"\nyesnoq(ans::AbstractString, args...; kwargs...) = radioq([\"Yes\", \"No\"], ans == \"yes\" ? 1 : 2, args...; keep_order=true, kwargs...)\nyesnoq(ans::Bool, args...; kwargs...) = yesnoq(ans ? \"yes\" : \"no\", args...;kwargs...)\n", "meta": {"hexsha": "e9e18f326d46cc7c73746c878fc597893db289f6", "size": 4257, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/question_types.jl", "max_stars_repo_name": "sylvaticus/QuizQuestions.jl", "max_stars_repo_head_hexsha": "27b35af704429264b44c1fe2a653ab7fb0ffa9c5", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/question_types.jl", "max_issues_repo_name": "sylvaticus/QuizQuestions.jl", "max_issues_repo_head_hexsha": "27b35af704429264b44c1fe2a653ab7fb0ffa9c5", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/question_types.jl", "max_forks_repo_name": "sylvaticus/QuizQuestions.jl", "max_forks_repo_head_hexsha": "27b35af704429264b44c1fe2a653ab7fb0ffa9c5", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 21.285, "max_line_length": 130, "alphanum_fraction": 0.6328400282, "num_tokens": 1076, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.476579651063676, "lm_q2_score": 0.14223189137395395, "lm_q1q2_score": 0.06778482516112565}}
{"text": "module JuliennedArrays\n\nimport Base: axes, getindex, setindex!, size\nusing Base: promote_op, @pure, @propagate_inbounds, tail\n\nmap_unrolled(call, variables::Tuple{}) = ()\nmap_unrolled(call, variables) =\n call(first(variables)), map_unrolled(call, tail(variables))...\n\nmap_unrolled(call, variables1::Tuple{}, variables2::Tuple{}) = ()\nmap_unrolled(call, variables1, variables2) =\n call(first(variables1), first(variables2)),\n map_unrolled(call, tail(variables1), tail(variables2))...\n\nis_in(needle::Needle, straw1::Needle, straws...) where {Needle} = True()\nis_in(needle, straw1, straws...) = is_in(needle, straws...)\nis_in(needle) = False()\n\nin_unrolled(straws, needle1, needles...) =\n is_in(needle1, straws...),\n in_unrolled(straws, needles...)...\nin_unrolled(straws) = ()\n\n@pure as_vals(them::Int...) = map(Val, them)\n\nabstract type TypedBool end\n\"\"\"\n struct True\n\nTyped `true`\n\"\"\"\nstruct True <: TypedBool end\n\"\"\"\n struct False\n\nTyped `false`\n\"\"\"\nstruct False <: TypedBool end\n\n@inline untyped(::True) = true\n@inline untyped(::False) = false\n\nnot(::False) = True()\nnot(::True) = False()\n\nexport True\nexport False\n\ngetindex_unrolled(into::Tuple{}, switches::Tuple{}) = ()\nfunction getindex_unrolled(into, switches)\n next = getindex_unrolled(tail(into), tail(switches))\n if untyped(first(switches))\n (first(into), next...)\n else\n next\n end\nend\n\nsetindex_unrolled(old::Tuple{}, something, ::Tuple{}) = ()\nsetindex_unrolled(old, new, switches) =\n if untyped(first(switches))\n first(new),\n setindex_unrolled(tail(old), tail(new), tail(switches))...\n else\n first(old),\n setindex_unrolled(tail(old), new, tail(switches))...\n end\n\nstruct Slices{Item, Dimensions, Whole, Alongs} <: AbstractArray{Item, Dimensions}\n whole::Whole\n alongs::Alongs\nend\nSlices{Item, Dimensions}(whole::Whole, alongs::Alongs) where {Item, Dimensions, Whole, Alongs} =\n Slices{Item, Dimensions, Whole, Alongs}(whole, alongs)\n\naxes(slices::Slices) =\n getindex_unrolled(axes(slices.whole), map_unrolled(not, slices.alongs))\nsize(slices::Slices) = map_unrolled(length, axes(slices))\n\nslice_index(slices, indices) = setindex_unrolled(\n axes(slices.whole),\n indices,\n map_unrolled(not, slices.alongs)\n)\n@propagate_inbounds getindex(slices::Slices, indices::Int...) =\n view(slices.whole, slice_index(slices, indices)...)\n@propagate_inbounds setindex!(slices::Slices, value, indices::Int...) =\n slices.whole[slice_index(slices, indices)...] = value\n\naxis_or_1(switch, axis) =\n if untyped(switch)\n axis\n else\n 1\n end\n\"\"\"\n Slices(whole, alongs::TypedBool...)\n\nSlice `whole` into `view`s.\n\n`alongs`, made of [`True`](@ref) and [`False`](@ref) objects, shows which dimensions will be replaced with `:` when slicing.\n\n```jldoctest\njulia> using JuliennedArrays\n\njulia> whole = [1 2; 3 4];\n\njulia> slices = Slices(whole, False(), True())\n2-element Slices{SubArray{Int64,1,Array{Int64,2},Tuple{Int64,Base.OneTo{Int64}},true},1,Array{Int64,2},Tuple{False,True}}:\n [1, 2]\n [3, 4]\n\njulia> slices[1] == whole[1, :]\ntrue\n\njulia> slices[1] = [2, 1];\n\njulia> whole\n2\u00d72 Array{Int64,2}:\n 2 1\n 3 4\n\njulia> larger = rand(5, 5, 5);\n\njulia> larger_slices = Slices(larger, True(), False(), False());\n\njulia> size(first(larger_slices))\n(5,)\n```\n\"\"\"\nSlices(whole::AbstractArray, alongs::TypedBool...) =\n Slices{\n typeof(@inbounds view(\n whole,\n map_unrolled(axis_or_1, alongs, axes(whole))...\n )),\n length(getindex_unrolled(alongs, map_unrolled(not, alongs)))\n }(whole, alongs)\n\n\"\"\"\n Slices(whole, alongs::Int...)\n\nAlternative syntax: `alongs` is which dimensions will be replaced with `:` when slicing.\n\n```jldoctest\njulia> using JuliennedArrays\n\njulia> input = reshape(1:8, 2, 2, 2)\n2\u00d72\u00d72 reshape(::UnitRange{Int64}, 2, 2, 2) with eltype Int64:\n[:, :, 1] =\n 1 3\n 2 4\n\n[:, :, 2] =\n 5 7\n 6 8\n\njulia> Slices(input, 1, 3)\n2-element Slices{SubArray{Int64,2,Base.ReshapedArray{Int64,3,UnitRange{Int64},Tuple{}},Tuple{Base.OneTo{Int64},Int64,Base.OneTo{Int64}},false},1,Base.ReshapedArray{Int64,3,UnitRange{Int64},Tuple{}},Tuple{True,False,True}}:\n [1 5; 2 6]\n [3 7; 4 8]\n```\n\"\"\"\nSlices(whole::AbstractArray{Item, NumberOfDimensions}, alongs::Int...) where {Item, NumberOfDimensions} =\n Slices(whole, in_unrolled(\n as_vals(alongs...),\n ntuple(Val, NumberOfDimensions)...\n )...)\nexport Slices\n\nstruct Align{Item, Dimensions, Sliced, Alongs} <: AbstractArray{Item, Dimensions}\n slices::Sliced\n alongs::Alongs\nend\nAlign{Item, Dimensions}(slices::Sliced, alongs::Alongs) where {Item, Dimensions, Sliced, Alongs} =\n Align{Item, Dimensions, Sliced, Alongs}(slices, alongs)\n\naxes(aligned::Align) = setindex_unrolled(\n setindex_unrolled(\n aligned.alongs,\n axes(aligned.slices),\n map_unrolled(not, aligned.alongs)\n ),\n axes(first(aligned.slices)),\n aligned.alongs\n)\nsize(aligned::Align) = map_unrolled(length, axes(aligned))\n\nsplit_indices(aligned, indices) =\n getindex_unrolled(indices, map_unrolled(not, aligned.alongs)),\n getindex_unrolled(indices, aligned.alongs)\n@propagate_inbounds function getindex(aligned::Align, indices::Int...)\n outer, inner = split_indices(aligned, indices)\n aligned.slices[outer...][inner...]\nend\n@propagate_inbounds function setindex!(aligned::Align, value, indices::Int...)\n outer, inner = split_indices(aligned, indices)\n aligned.slices[outer...][inner...] = value\nend\n\n\"\"\"\n Align(slices, alongs::TypedBool...)\n\n`Align` an array of arrays, all with the same size.\n\n`alongs`, made of [`True`](@ref) and [`False`](@ref) objects, shows which dimensions will be taken up by the inner arrays. Inverse of [`Slices`](@ref).\n\n```jldoctest\njulia> using JuliennedArrays\n\njulia> slices = [[1, 2], [3, 4]];\n\njulia> aligned = Align(slices, False(), True())\n2\u00d72 Align{Int64,2,Array{Array{Int64,1},1},Tuple{False,True}}:\n 1 2\n 3 4\n\njulia> aligned[1, :] == slices[1]\ntrue\n\njulia> aligned[1, 1] = 0;\n\njulia> slices\n2-element Array{Array{Int64,1},1}:\n [0, 2]\n [3, 4]\n```\n\"\"\"\nAlign(slices::AbstractArray{<:AbstractArray{Item, InnerDimensions}, OuterDimensions}, alongs::TypedBool...) where {Item, InnerDimensions, OuterDimensions} =\n Align{Item, OuterDimensions + InnerDimensions}(slices, alongs)\nexport Align\n\n\"\"\"\n Along(slices, alongs::Int...)\n\nAlternative syntax: `alongs` is which dimensions will be taken up by the inner arrays.\n\n```jldoctest\njulia> using JuliennedArrays\n\njulia> input = reshape(1:8, 2, 2, 2)\n2\u00d72\u00d72 reshape(::UnitRange{Int64}, 2, 2, 2) with eltype Int64:\n[:, :, 1] =\n 1 3\n 2 4\n\n[:, :, 2] =\n 5 7\n 6 8\n\njulia> slices = collect(Slices(input, 1, 3))\n2-element Array{SubArray{Int64,2,Base.ReshapedArray{Int64,3,UnitRange{Int64},Tuple{}},Tuple{Base.OneTo{Int64},Int64,Base.OneTo{Int64}},false},1}:\n [1 5; 2 6]\n [3 7; 4 8]\n\njulia> Align(slices, 1, 3)\n2\u00d72\u00d72 Align{Int64,3,Array{SubArray{Int64,2,Base.ReshapedArray{Int64,3,UnitRange{Int64},Tuple{}},Tuple{Base.OneTo{Int64},Int64,Base.OneTo{Int64}},false},1},Tuple{True,False,True}}:\n[:, :, 1] =\n 1 3\n 2 4\n\n[:, :, 2] =\n 5 7\n 6 8\n```\n\"\"\"\nAlign(slices::AbstractArray{<:AbstractArray{Item, InnerDimensions}, OuterDimensions}, alongs::Int...) where {Item, InnerDimensions, OuterDimensions} =\n Align(slices, in_unrolled(\n as_vals(alongs...),\n ntuple(Val, InnerDimensions + OuterDimensions)...\n )...)\n\nend\n", "meta": {"hexsha": "3b5b7981449f889d47f078a2e26ffe119e6f3b6b", "size": 7415, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/JuliennedArrays.jl", "max_stars_repo_name": "UnofficialJuliaMirrorSnapshots/JuliennedArrays.jl-5cadff95-7770-533d-a838-a1bf817ee6e0", "max_stars_repo_head_hexsha": "8dc49be38ce5d85c0adb92e31917ccd6c5ac3827", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/JuliennedArrays.jl", "max_issues_repo_name": "UnofficialJuliaMirrorSnapshots/JuliennedArrays.jl-5cadff95-7770-533d-a838-a1bf817ee6e0", "max_issues_repo_head_hexsha": "8dc49be38ce5d85c0adb92e31917ccd6c5ac3827", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/JuliennedArrays.jl", "max_forks_repo_name": "UnofficialJuliaMirrorSnapshots/JuliennedArrays.jl-5cadff95-7770-533d-a838-a1bf817ee6e0", "max_forks_repo_head_hexsha": "8dc49be38ce5d85c0adb92e31917ccd6c5ac3827", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 27.0620437956, "max_line_length": 222, "alphanum_fraction": 0.6737693864, "num_tokens": 2349, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.480478678047907, "lm_q2_score": 0.14033624949008322, "lm_q1q2_score": 0.06742857563719645}}
{"text": "a = let\n i=3\n i+=5\n i # the value returned from the computation\nend\n\na\n\nb = let i=5\n i+=42\n i\nend\n\nc = let i=10\n i+=42\n i\nend\n\nc\n\ni\n\nd = begin\n i=41\n i+=1\n i\nend\n\n\ni\nd\n\nconst C = 299792458 # m / s, this is an Int\n\nC = 300000000 # change the value of C\n\nC = 2.998 * 1e8 #change the type of C, not permitted\n\n#%%\nmodule ScopeTestModule\nexport a1\na1 = 25\nb1 = 42\nend # end of module\n#%%\nusing .ScopeTestModule\n\na1\nb1\n\nScopeTestModule.b1=26\n", "meta": {"hexsha": "2d0b12a274f923167272e3d53434cbe8bae27bb0", "size": 467, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "lesson-scope/scope.jl", "max_stars_repo_name": "JeffreySarnoff/techytok-examples", "max_stars_repo_head_hexsha": "5c34c9fc0660da1a69f9e3959465ed80659c30cd", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 9, "max_stars_repo_stars_event_min_datetime": "2019-10-28T11:21:11.000Z", "max_stars_repo_stars_event_max_datetime": "2021-07-14T09:06:49.000Z", "max_issues_repo_path": "lesson-scope/scope.jl", "max_issues_repo_name": "JeffreySarnoff/techytok-examples", "max_issues_repo_head_hexsha": "5c34c9fc0660da1a69f9e3959465ed80659c30cd", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "lesson-scope/scope.jl", "max_forks_repo_name": "JeffreySarnoff/techytok-examples", "max_forks_repo_head_hexsha": "5c34c9fc0660da1a69f9e3959465ed80659c30cd", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 7, "max_forks_repo_forks_event_min_datetime": "2020-01-20T23:53:35.000Z", "max_forks_repo_forks_event_max_datetime": "2022-01-10T20:51:24.000Z", "avg_line_length": 8.9807692308, "max_line_length": 52, "alphanum_fraction": 0.5952890792, "num_tokens": 191, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.5, "lm_q2_score": 0.13477592958647097, "lm_q1q2_score": 0.06738796479323549}}
{"text": "#------------------------------------------------------------------------------\n\"\"\"\n printmat([fh::IO],x;width=10,prec=3,NoPrinting=false,StringFmt=\"\")\n\nPrint all elements of matrix with predefined formatting.\n\n# Input\n- `fh::IO`: (optional) file handle. If not supplied, prints to screen\n- `x::Array`: string, date or array to print\n- `width::Int`: (keyword) scalar, minimum width of printed cells\n- `prec::Int`: (keyword) scalar, precision of printed cells\n- `NoPrinting::Bool`: (keyword) bool, true: no printing, just return formatted string\n- `StringFmt::String`: (keyword) string, \"\", \"html\", \"csv\"\n\n# Output\n- str (if NoPrinting) string, (otherwise nothing)\n\n# Examples\n```\nx = [11 12;21 22]\nprintmat(x)\n```\n```\nx = Any[1 \"ab\"; Date(2018,10,7) 3.14]\nprintmat(x,width=20)\n```\nCan also call as\n```\nopt = Dict(:width=>10,:prec=>3,:NoPrinting=>false,:StringFmt=>\"\")\nprintmat(x;opt...) #notice , and ...\n```\n(not all keywords are needed)\n\n# Requires\n- Dates\n- fmtNumPs\n\n# To do\n\n\nPaul.Soderlind@unisg.ch\n\n\"\"\"\nfunction printmat(fh::IO,x;width=10,prec=3,NoPrinting=false,StringFmt=\"\")\n\n if isa(x,Union{String,Date,DateTime,Missing}) #eg. a single Date\n str = string(lpad(x,width),\"\\n\")\n if NoPrinting\n return str\n else\n print(fh,str,\"\\n\")\n return nothing\n end\n elseif isa(x,Nothing)\n return nothing\n end\n\n if ndims(x) > 2\n @warn(\"more than 2 dimensions\")\n return nothing\n end\n\n (m,n) = (size(x,1),size(x,2))\n\n iob = IOBuffer()\n for i = 1:m #loop over lines\n for j = 1:n-1 #loop over columns 1:n-1\n writeElementPs(iob,x,i,j,width,prec,StringFmt)\n end\n if StringFmt == \"csv\" #last (n) column\n writeElementPs(iob,x,i,n,width,prec,\"\") #no , at end of line\n else\n writeElementPs(iob,x,i,n,width,prec,StringFmt)\n end\n write(iob,\"\\n\") #newline\n end\n str = String(take!(iob))\n\n if NoPrinting #no printing, just return str\n return str\n else #print, return nothing\n print(fh,str,\"\\n\")\n return nothing\n end\n\nend\n #when fh is not supplied: printing to screen\nprintmat(x;width=10,prec=3,NoPrinting=false,StringFmt=\"\") = printmat(stdout::IO,\n x,width=width,prec=prec,NoPrinting=NoPrinting,StringFmt=StringFmt)\n#------------------------------------------------------------------------------\n\n\n#------------------------------------------------------------------------------\n\"\"\"\n printTable([fh::IO],x,colNames=[],rowNames=[];\n width=10,prec=3,NoPrinting=false,StringFmt=\"\",cell00=\"\")\n\nPrint formatted table with row names (1st column) column names (1st row),\nand data matrix (the rest).\n\n\n# Input\n- `fh::IO`: (optional) file handle. If not supplied, prints to screen\n- `x::Array`: (of numbers, dates, strings, ...) to print\n- `colNames::Array`: of strings with column headers\n- `rowNames::Array`: of strings with row labels\n- `width::Int`: (keyword) scalar, minimum width of printed cells. [10]\n- `prec::Int`: (keyword) scalar, precision of printed cells. [3]\n- `NoPrinting::Bool`: (keyword) bool, true: no printing, just return formatted string [false]\n- `StringFmt::String`: (keyword) string, \"\", \"html\", \"csv\"\n- `cell00::String`: (keyword) string, for row 0, column 0\n\n# Output\n- `str::String`: (if NoPrinting) string, (otherwise nothing)\n\n# Example\n```\nxA = [1 \"ab\" \"abc\"; \"ccc\" 3.14 missing]\nprintTable(xA,colNames,[\"1\";\"4\"],width=12,prec=2)\n```\nCan also call as\n```\nopt = Dict(:width=>10,:prec=>3,:NoPrinting=>false,:StringFmt=>\"\",:cell00=>\"\")\nprintTable(x;opt...) #notice , and ...\n```\n(not all keywords are needed)\n\n# Requires\n- Dates\n- printmat\n\n\"\"\"\nfunction printTable(fh::IO,x,colNames=[],rowNames=[];\n width=10,prec=3,NoPrinting=false,StringFmt=\"\",cell00=\"\")\n\n isempty(x) && return nothing #do nothing is isempty(x)\n\n (m,n) = (size(x,1),size(x,2))\n\n if isempty(rowNames) #create row names \"r1\"\n rowNames = [string(\"r\",i) for i = 1:m]\n end\n if isempty(colNames) #create column names \"c1\"\n colNames = [string(\"c\",i) for i = 1:n]\n end\n\n rNamesWidth = maximum([length(rowNames[i]) for i = 1:length(rowNames)]) #max length of rowNames\n rNamesWidth = max(rNamesWidth,length(cell00))\n\n iob = IOBuffer()\n if StringFmt == \"html\" #print column names\n write(iob,\"\",lpad(cell00,rNamesWidth),\" \")\n for i = 1:n\n write(iob,\"\",lpad(colNames[i],width),\" \")\n end\n write(iob,\"\",rpad(rowNames[i],rNamesWidth),\" \",strLR,\" \")\n elseif StringFmt == \"csv\"\n strLR = string(strLR,\",\")\n end\n\n return strLR\n\nend\n#------------------------------------------------------------------------------\n\n\n#------------------------------------------------------------------------------\nfunction printblue(x...)\n foreach(z->printstyled(z,color=:blue,bold=true),x)\n print(\"\\n\")\nend\nfunction printred(x...)\n foreach(z->printstyled(z,color=:red,bold=true),x)\n print(\"\\n\")\nend\nfunction printmagenta(x...)\n foreach(z->printstyled(z,color=:magenta,bold=true),x)\n print(\"\\n\")\nend\nfunction printyellow(x...)\n foreach(z->printstyled(z,color=:yellow,bold=true),x)\n print(\"\\n\")\nend\n#------------------------------------------------------------------------------\n\n\n#------------------------------------------------------------------------------\nfunction printwhere(txt)\n println(@__FILE__,\" \",@__LINE__,\" \",txt)\nend\n#------------------------------------------------------------------------------\n\n", "meta": {"hexsha": "d051d982cde474679b6f3d6bdaa7a41caf1ae812", "size": 13859, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "printmat.jl", "max_stars_repo_name": "snowdj/JuliaTutorial", "max_stars_repo_head_hexsha": "35a0c370b248101a4f05e2bccdffdcbe9f3ea9ef", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 20, "max_stars_repo_stars_event_min_datetime": "2016-01-15T21:13:48.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-18T21:21:18.000Z", "max_issues_repo_path": "printmat.jl", "max_issues_repo_name": "fmyilmaz/JuliaTutorial", "max_issues_repo_head_hexsha": "b77a113835f47f430bd6b458731d2cec06da69b6", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 1, "max_issues_repo_issues_event_min_datetime": "2016-03-05T13:07:59.000Z", "max_issues_repo_issues_event_max_datetime": "2016-03-07T11:11:21.000Z", "max_forks_repo_path": "printmat.jl", "max_forks_repo_name": "fmyilmaz/JuliaTutorial", "max_forks_repo_head_hexsha": "b77a113835f47f430bd6b458731d2cec06da69b6", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 13, "max_forks_repo_forks_event_min_datetime": "2016-03-05T13:01:10.000Z", "max_forks_repo_forks_event_max_datetime": "2021-09-07T19:44:57.000Z", "avg_line_length": 32.919239905, "max_line_length": 116, "alphanum_fraction": 0.5212497294, "num_tokens": 3582, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.43014734858584286, "lm_q2_score": 0.15610489155639232, "lm_q1q2_score": 0.06714810520426269}}
{"text": "using PkgBenchmark\r\nresults = benchmarkpkg(\"TaylorSeries\")\r\nshow(results)\r\n\r\n#=\r\n# specify tag and uncommit to benchmark versus prior tagged version\r\ntag =\r\nresults = judge(\"TaylorSeries\", tag)\r\nshow(results)\r\n=#\r\n\r\nexport_markdown(\"results.md\", results)\r\n", "meta": {"hexsha": "c074a5399bf95d5a4b4ec103631e2d20aa0b475c", "size": 256, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "benchmark/run_benchmark.jl", "max_stars_repo_name": "mewilhel/TaylorSeries.jl", "max_stars_repo_head_hexsha": "e169d1f20217d345b8ff24ec9124b01b95b32953", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "benchmark/run_benchmark.jl", "max_issues_repo_name": "mewilhel/TaylorSeries.jl", "max_issues_repo_head_hexsha": "e169d1f20217d345b8ff24ec9124b01b95b32953", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "benchmark/run_benchmark.jl", "max_forks_repo_name": "mewilhel/TaylorSeries.jl", "max_forks_repo_head_hexsha": "e169d1f20217d345b8ff24ec9124b01b95b32953", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 19.6923076923, "max_line_length": 68, "alphanum_fraction": 0.73046875, "num_tokens": 56, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4532618480153861, "lm_q2_score": 0.14804719615221756, "lm_q1q2_score": 0.06710414572145049}}
{"text": "## Exercise 7-1\n## Rewrite the function printn from Recursion using iteration instead of recursion.\nprintln(\"Ans: \")\n\nfunction printn(s, n::Int)\n while n > 0\n println(s)\n n -= 1\n end\nend \n\nprintn(\"Hello iteration!\", 4)\n\nprintln(\"End.\")\n", "meta": {"hexsha": "d04f5565fff29320737666650a2e1e5f6dcb2ace", "size": 256, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Chapter7/ex1.jl", "max_stars_repo_name": "yashppawar/ThinkJuliaExercises.jl", "max_stars_repo_head_hexsha": "72145b969dc51ebcac413a10004175ebc63cd5c2", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2022-02-13T14:11:30.000Z", "max_stars_repo_stars_event_max_datetime": "2022-02-13T14:11:30.000Z", "max_issues_repo_path": "Chapter7/ex1.jl", "max_issues_repo_name": "yashppawar/ThinkJuliaExercises.jl", "max_issues_repo_head_hexsha": "72145b969dc51ebcac413a10004175ebc63cd5c2", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Chapter7/ex1.jl", "max_forks_repo_name": "yashppawar/ThinkJuliaExercises.jl", "max_forks_repo_head_hexsha": "72145b969dc51ebcac413a10004175ebc63cd5c2", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 17.0666666667, "max_line_length": 83, "alphanum_fraction": 0.6328125, "num_tokens": 71, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.22270013882530884, "lm_q2_score": 0.30074559147596, "lm_q1q2_score": 0.0669760849727959}}
{"text": "### A Pluto.jl notebook ###\n# v0.19.8\n\nusing Markdown\nusing InteractiveUtils\n\n# This Pluto notebook uses @bind for interactivity. When running this notebook outside of Pluto, the following 'mock version' of @bind gives bound variables a default value (instead of an error).\nmacro bind(def, element)\n quote\n local iv = try Base.loaded_modules[Base.PkgId(Base.UUID(\"6e696c72-6542-2067-7265-42206c756150\"), \"AbstractPlutoDingetjes\")].Bonds.initial_value catch; b -> missing; end\n local el = $(esc(element))\n global $(esc(def)) = Core.applicable(Base.get, el) ? Base.get(el) : iv(el)\n el\n end\nend\n\n# \u2554\u2550\u2561 2efa05e7-b130-4a95-9eab-d8bb9d16a29b\n# \u2560\u2550\u2561 show_logs = false\nbegin\n\tusing Logging\n\tglobal_logger(NullLogger())\n\tdisplay(\"\")\nend\n\n# \u2554\u2550\u2561 c3e429e4-e7e9-4db6-852c-906630f909a4\n# \u2560\u2550\u2561 show_logs = false\n#Set-up packages\nbegin\n\t\n\tusing DataFrames, HTTP, CSV, Dates, Plots, PlutoUI, Printf, LaTeXStrings, HypertextLiteral, XLSX\n\t\n\tgr();\n\tPlots.GRBackend()\n\n\n\t#Define html elements\n\tnbsp = html\" \" #non-breaking space\n\tvspace = html\"\"\"
\"\"\"\n\tbr = html\"\n\t# \t\n\t# \t
\n\t# \"\"\")\n\n\t\n\t# cellWidth= min(1000, screenWidth*0.9)\n\t# @htl(\"\"\"\n\t# \t\n\t# \"\"\")\n\t\n\n\t#Sets the width of the cells\n\t#begin\n\t#\thtml\"\"\"\n\t# \"\"\")\n\t\n\n\t#Sets the width of the cells\n\t#begin\n\t#\thtml\"\"\"\n\t\"\"\")\n\n# \u2554\u2550\u2561 c763ed72-82c9-445c-a8f7-a0c40982e4d9\n@skip_as_script toc()\n\n# \u2554\u2550\u2561 955705f9-c90d-495d-86b4-5f3b5bc9fc8e\nbegin\n\tstruct Slider\n\t\txs\n\tend\n\t\n\tBase.get(s::Slider) = first(s.xs)\n\t\n\tBase.show(io::IO, m::MIME\"text/html\", s::Slider) = show(io, m, @htl(\"\n\t\t$(\n\tmap(lines) do l\n\t\tkeys = [Symbol(m.match) for m in eachmatch(r, l)]\n\t\trest = split(l, r; keepempty=true)\n\t\t\n\t\tresult = vcat((\n\t\t\t[(isempty(r) ? @htl(\"\") : preish(r)), embed_display(d[k].x)]\n\t\t\tfor (r,k) in zip(rest, keys)\n\t\t\t)...)\n\t\t\n\t\tpush!(result, preish(last(rest)))\n\t\t\n\t\t@htl(\"$(result) \")\n\tend\n\t)\n\t\t \"\"\")\n\tshow(io, m, h)\nend\n\n# \u2554\u2550\u2561 ef6fc423-f1b1-4dcb-a059-276121391bc6\nprettycolors(e) = Markdown.MD([Markdown.Code(\"julia\", expr_to_str(e))])\n\n# \u2554\u2550\u2561 0f31dd2e-0331-4d4c-8db5-9ce188cd3730\n@skip_as_script [lens_to_getter(:source, [FieldLens(:x), PropertyLens(:y)])] .|> prettycolors\n\n# \u2554\u2550\u2561 cecba3e6-98e8-408a-97dd-96b67c4f42cf\n@skip_as_script [lens_to_setter(:dest, [FieldLens(:x), PropertyLens(:y)], :value)] .|> prettycolors\n\n# \u2554\u2550\u2561 7e6c2162-97e9-4835-b650-52c9723c327f\nmd\"## Utils\"\n\n# \u2554\u2550\u2561 1ac164c8-88fc-4a87-a194-60ef616fb399\nflatmap(args...) = vcat(map(args...)...)\n\n# \u2554\u2550\u2561 1c1b64b1-107e-4d43-9ce2-569c3034017e\nfunction expr_lenses_for_quoted(e::Expr, code_loweredish_with_lenses)::Frames\n\tif e.head == :$\n\t\tframes = code_loweredish_with_lenses(e.args[1])\n\t\tapply_lens_to_frames([FieldLens(:args), PropertyLens(1)], frames)\n\telse\n\t\targument_frames = flatmap(enumerate(e.args)) do (i, arg)\n\t\t\tframes = expr_lenses_for_quoted(arg, code_loweredish_with_lenses)\n\t\t\tapply_lens_to_frames([FieldLens(:args), PropertyLens(i)], frames)\n\t\tend\n\tend\nend\n\n# \u2554\u2550\u2561 ce90612e-ffc1-4e30-9d89-531f11fd75eb\n\"\"\"\n code_loweredish_with_lenses(e::Expr)::Vector{ExprWithLens}\n\nTransforms an expression into a set of list of expressions that would be executed one after eachother. It gives every expression a lens referencing where it is inside the original expression. This way you can execute each expr, and then put the result in the expression to create the step-by-step execution.\n\nIt doesn't \"dedupe\" the expressions, so when you run the last expression in the list (which will just be the original expression), it doesn't take advantage of any previously run expressions in the same list. You'll have to do that later, manually.\n\nIn PlutoTest this is done with [TODO](@ref)\n\"\"\"\nfunction code_loweredish_with_lenses(e::Expr)::Frames\n\tif e.head == :kw\n\t\tframes = code_loweredish_with_lenses(e.args[2])\n\t\tapply_lens_to_frames([FieldLens(:args), PropertyLens(2)], frames)\n\telseif e.head == :(=)\n\t\t# This was getting closer, but the whole thing still is quite hard...\n\t\t# so for now any assignment just throws :D\n\t\t# We need quite some smartness in build_step_by_step_blocks.\n\t\tthrow(CantStepifyThisYetException(e))\n\t\t\n\t\tframes = code_loweredish_with_lenses(e.args[2])\n\t\tlens = [FieldLens(:args), PropertyLens(2)]\n\t\t[\n\t\t\tapply_lens_to_frames(lens, frames)...,\n\t\t\tExprWithLens(expr=e.args[2], lens=lens, expr_to_show=e),\n\t\t\t# It now adds the whole `x = ...` expression as well,\n\t\t\t# which doesn't look that good in the output...\n\t\t\t# But we'll have to live with it for now\n\t\t]\n\telseif e.head == :parameters\n\t\tflatmap(enumerate(e.args)) do (i, arg)\n\t\t\tframes = code_loweredish_with_lenses(arg)\n\t\t\tapply_lens_to_frames([FieldLens(:args), PropertyLens(i)], frames)\n\t\tend\n\telseif e.head == :quote\n\t\tframes = expr_lenses_for_quoted(e.args[1], code_loweredish_with_lenses)\n\t\targument_frames = apply_lens_to_frames(\n\t\t\t[FieldLens(:args), PropertyLens(1)],\n\t\t\tframes\n\t\t)\n\t\t[argument_frames..., ExprWithLens(expr=e, lens=[])]\n\telseif e.head == :...\n\t\tframes = code_loweredish_with_lenses(e.args[1])\n\t\tapply_lens_to_frames([FieldLens(:args), PropertyLens(1)], frames)\n\telseif e.head == :macrocall || e.head == :ref\n\t\t[ExprWithLens(expr=e, lens=[])]\n\telseif e.head == :call\n\t\t# With calls we don't want to dive into the callee if it is just a symbol\n\t\t# (This would expand everything like x == y to #function(:==)(x,y) which\n\t\t# is definitely not what we want)\n\t\tpossibly_callee_frames = if e.args[begin] isa Symbol\n\t\t\t[]\n\t\telse\n\t\t\tframes = code_loweredish_with_lenses(e.args[begin])\n\t\t\tapply_lens_to_frames([FieldLens(:args), PropertyLens(firstindex(e.args))], frames)\n\t\tend\n\t\t\n\t\targument_frames = flatmap(enumerate(e.args[begin+1:end])) do (i, arg)\n\t\t\tframes = code_loweredish_with_lenses(arg)\n\t\t\t# @info \"III\" i arg\n\t\t\tapply_lens_to_frames([FieldLens(:args), PropertyLens(i+1)], frames)\n\t\tend\n\t\t\n\t\t[possibly_callee_frames..., argument_frames..., ExprWithLens(expr=e, lens=[])]\n\n\telseif (\n\t\te.head == :begin ||\n\t\te.head == :block ||\n\t\te.head == :vect ||\n\t\te.head == :string ||\n\t\te.head == :. ||\n\t\te.head == :tuple ||\n\t\te.head == :let\n\t)\n\t\targument_frames = flatmap(enumerate(e.args)) do (i, arg)\n\t\t\tframes = code_loweredish_with_lenses(arg)\n\t\t\tapply_lens_to_frames([FieldLens(:args), PropertyLens(i)], frames)\n\t\tend\n\t\t\n\t\t[argument_frames..., ExprWithLens(expr=e, lens=[])]\n\telseif (\n\t\te.head == :if ||\n\t\te.head == :elseif ||\n\t\te.head == :else ||\n\t\te.head == :&& ||\n\t\te.head == :|| ||\n\t\te.head == :try ||\n\t\te.head == :catch ||\n\t\te.head == :finally\n\t)\n\t\tif ERROR_ON_UNKNOWN_EXPRESSION_TYPE\n\t\t\tthrow(CantStepifyThisYetException(e))\n\t\telse\n\t\t\t[ExprWithLens(expr=e, lens=[])]\n\t\tend\n\telse\n\t\tif ERROR_ON_UNKNOWN_EXPRESSION_TYPE\n\t\t\tthrow(CantStepifyThisYetException(e))\n\t\telse\n\t\t\t[ExprWithLens(expr=e, lens=[])]\n\t\tend\n\tend\nend;\n\n# \u2554\u2550\u2561 e1c306e3-0a47-4149-a9fb-ec7ab380fa11\n\"\"\"\n\tstep_by_step(expr::Expr)\n\nThe preparing for step-by-step testing happens in two steps itself.\n\nFirst there is [`code_loweredish_with_lenses`](@ref) which takes an expression and splits it up in [`ExprWithLens`](@ref)s. These are separate expressions with a lens specifying where in the original expression the result should be placed.\n\nSecond part is combining all those expressions into a block that gradually executes those separate parts, and at each step saves the whole expression to be shown. That's what [`build_step_by_step_blocks`](@ref) is for.\n\nThis functions combines these two. This is the main function used inside the test macro. The reason it is a separate function and not a macro on its own, is because macro hygiene is weird... \n\"\"\"\nfunction step_by_step(expr)\t\n\tlowered = code_loweredish_with_lenses(expr)\n\texpr_ref_lens = gensym(\"expr_ref\")\n\tsteps_lens = gensym(\"steps\")\n\t\n\tquote\n\t\tbegin\n\t\t\ttry\n\t\t\t\texpr = $(QuoteNode(expr))\n\t\t\t\t$steps_lens = Any[expr]\n\t\t\t\t$expr_ref_lens = Ref{Any}(expr)\n\t\t\t\t$(build_step_by_step_blocks(lowered;\n\t\t\t\t\texpr_ref_lens=expr_ref_lens,\n\t\t\t\t\tsteps_lens=steps_lens,\n\t\t\t\t)...)\n\t\t\t\t$steps_lens\n\t\t\tcatch error\n\t\t\t\terror.steps\n\t\t\tend\n\t\tend\n\tend\nend\n\n# \u2554\u2550\u2561 b6e8a170-12cc-4d97-905d-274e2609bfd8\nfunction test(expr, extra_args...)\n\tstep_by_step\n\tTest.test_expr!(\"\", expr, extra_args...)\n\t\t\n\tquote\n\t\tlocal expr_raw = $(QuoteNode(expr))\n\t\ttry\t\t\t\n\t\t\tsteps = $(step_by_step(expr))\n\n\t\t\tresult = unwrap_computed(last(steps))\n\t\t\t\n\t\t\tif result === true\n\t\t\t\tCorrectCall(expr_raw, steps)\n\t\t\telse\n\t\t\t\tWrongCall(expr_raw, steps)\n\t\t\tend\n\t\tcatch error\n\t\t\tif error isa PartialEvaluatedException\n\t\t\t\tErrorCall(\n\t\t\t\t\texpr=expr_raw,\n\t\t\t\t\tsteps=error.steps,\n\t\t\t\t\terror=error.error,\n\t\t\t\t)\n\t\t\telse\n\t\t\t\trethrow(error)\n\t\t\tend\n\t\tend\n\tend\nend\n\n# \u2554\u2550\u2561 d7dc79e6-1f58-4414-aeef-667bdb0dd200\nmacro pretty_step_by_step(e)\n\tquote\n\t\tresulting_expressions = try\n\t\t\t$(step_by_step(e))\n\t\tcatch error\n\t\t\tif error isa PartialEvaluatedException\n\t\t\t\terror.steps\n\t\t\telse\n\t\t\t\trethrow(error)\n\t\t\tend\n\t\tend \n\t\t\n\t\tresulting_expressions .|> prettycolors\n\tend\nend\n\n# \u2554\u2550\u2561 ba4a5762-33da-40e6-94fa-cca9befc6d5a\nexample_equals = @skip_as_script let\n\t@pretty_step_by_step sqrt(sqrt(16)) == 4\nend\n\n# \u2554\u2550\u2561 9101631b-81ca-4c7c-94da-81d9e106df78\nexample_call_spread = @skip_as_script let\n\t@pretty_step_by_step max([1,2,3]...) != min([1,2,3]...)\nend\n\n# \u2554\u2550\u2561 3f0e5a49-5eec-42cd-bd2c-254b277840bf\nexample_show_variable_value = @skip_as_script let\n\tx = [1,2,3]\n\t@pretty_step_by_step x == [1,2,3]\nend\n\n# \u2554\u2550\u2561 fc26d26a-a9a5-4646-b85b-12eac66d96cb\nexample_show_thrown_error = @skip_as_script let\n\t@pretty_step_by_step sqrt(sqrt(16) - 5)\nend\n\n# \u2554\u2550\u2561 312ef6a6-55aa-4913-9416-15e79b4e3362\nexample_with_nested_macro = @skip_as_script let\n\t@pretty_step_by_step @return_one() + 2 == 3\nend\n\n# \u2554\u2550\u2561 1aa319c8-5e1d-4dd9-ae22-ad99e46e7b4d\nexample_keyword_arguments = @skip_as_script let\n\t@pretty_step_by_step round(sqrt(2), digits=Int(sqrt(16)))\nend\n\n# \u2554\u2550\u2561 605d2481-23be-4ad9-82c9-e375b7be8669\n# Seems very similar to `example_keyword_arguments`, but this one\n# has a `;`, which makes a liiiitle bit different AST\nexample_keyword_arguments_explicit = @skip_as_script let\n\t@pretty_step_by_step round(sqrt(2); digits=Int(sqrt(16)))\nend\n\n# \u2554\u2550\u2561 60a398c9-9fe8-4b90-b863-1568183641d9\nexample_returned_function = @skip_as_script let\n\tfunction_that_returns_function = () -> function X() 10 end\n\t@pretty_step_by_step function_that_returns_function()() == 10\nend\n\n# \u2554\u2550\u2561 a661e172-6afb-42ff-bd43-bb5b787ee5ed\nmacro eval_step_by_step(e)\n\tstep_by_step(e)\nend\n\n# \u2554\u2550\u2561 b4b317d7-bed1-489c-9650-8d336e330689\nrs = @skip_as_script @eval_step_by_step(begin\n\t\t(1+2) + (7-6)\n\t\tplot(2000 .+ 30 .* rand(2+2))\n\t\t4+5\n\t\tsqrt(sqrt(sqrt(5)))\n\tend) .|> SlottedDisplay\n\n# \u2554\u2550\u2561 93ed973f-daf6-408b-9d4b-d53495418610\n@skip_as_script @bind rindex Slider(eachindex(rs))\n\n# \u2554\u2550\u2561 dea898a0-1904-4d09-ad0b-6915008fe946\n@skip_as_script rs[rindex]\n\n# \u2554\u2550\u2561 b0ab9327-8240-4d34-bdd9-3f8f5117bb29\nstruct PlutoStylesheet\n\tcode\nend\n\n# \u2554\u2550\u2561 8a2e8348-49cf-4855-b5b3-cdee33e5ed67\n# const pluto_test_css = PlutoStylesheet(\"\"\"\npluto_test_css = PlutoStylesheet(\"\"\"\npt-dot {\n\tflex: 0 0 auto;\n\tbackground: grey;\n\twidth: 1em;\n\theight: 1em;\n\tbottom: -.1em;\n\tborder-radius: 100%;\n\tmargin-right: .7em;\n\tdisplay: block;\n\tposition: relative;\n\tcursor: pointer;\n}\n\npt-dot.floating {\n\tposition: fixed;\n\tz-index: 60;\n\tvisibility: hidden;\n\ttransition: transform linear 240ms;\n\topacity: .8;\n}\n.show-top-float > pt-dot.floating.top,\n.show-bottom-float > pt-dot.floating.bottom {\n\tvisibility: visible;\n}\n\npt-dot.floating.top {\n\ttop: 5px;\n}\npt-dot.floating.bottom {\n\tbottom: 5px;\n}\n\n\n.fail > pt-dot {\n\tbackground: #f75d5d;\n\n}\n.pass > pt-dot {\n\tbackground: #56a038;\n}\n\n@keyframes fadeout {\n 0% { opacity: 1;}\n 100% { opacity: 0; pointer-events: none;}\n}\n\n\n.pass > pt-dot.floating {\n animation: fadeout 2s;\n\n\tanimation-fill-mode: both;\n\tanimation-delay: 2s;\n\n\t/*opacity: 0.4;*/\n}\n\n\n.pluto-test {\n\tfont-family: \"JuliaMono\", monospace;\n\tfont-size: 0.75rem;\n\twhite-space: normal;\n\tpadding: 4px;\n\n\tmin-height: 25px;\n}\n\n\n.pluto-test.pass {\n\tcolor: rgba(0, 0, 0, 0.5);\n}\n\n@media (prefers-color-scheme: dark) {\n\t.pluto-test.pass {\n\t\tcolor: rgba(200, 200, 200, 0.5);\n\t}\n}\n\n.pluto-test.fail {\nbackground: linear-gradient(90deg, #ff2e2e14, transparent);\nborder-radius: 7px;\n}\n\n\n.pluto-test>.arg_result {\n\tflex: 0 0 auto;\n}\n\n.pluto-test>.arg_result>div,\n.pluto-test>.arg_result>div>pluto-display>div {\n\tdisplay: inline-flex;\n}\n\n\n.pluto-test>.comma {\n\tmargin-right: .5em;\n}\n\n.pluto-test.call>code {\n\tpadding: 0px;\n}\n\n.pluto-test.call.infix-operator>div {\n\toverflow-x: auto;\n}\n\n.pluto-test {\n\tdisplay: flex;\n\talign-items: baseline;\n}\n\n.pluto-test.call.infix-operator>.fname {\n\tmargin: 0px .6em;\n\t/*color: darkred;*/\n}\n\n\n/* expanding */\n\n\n.pluto-test:not(.expanded) {\n\tcursor: pointer;\n}\n\n.pluto-test:not(.expanded) > p-frame-viewer > p-frame-controls {\n\tdisplay: none;\n\t\n}\n\n.pluto-test.expanded > p-frame-viewer {\n max-width: 100%;\n}\n.pluto-test.expanded > p-frame-viewer > p-frames > slotted-code > line-like {\n\tflex-wrap: wrap;\n}\n.pluto-test.expanded > p-frame-viewer > p-frames > slotted-code > line-like > pluto-display[mime=\"application/vnd.pluto.tree+object\"] {\n\t/*flex-basis: 100%;*/\n}\n\"\"\")\n\n# \u2554\u2550\u2561 42671258-07a0-4015-8f47-4b3032595f08\n# const frames_css = PlutoStylesheet(\"\"\"\nframes_css = PlutoStylesheet(\"\"\"\np-frame-viewer {\n\tdisplay: inline-flex;\n\tflex-direction: column;\n}\np-frames,\np-frame-controls {\n\tdisplay: inline-flex;\n}\n\"\"\")\n\n# \u2554\u2550\u2561 e968fc57-d850-4e2d-9410-8777d03b7b3c\nfunction frames(fs::Vector; startframe::Union{Nothing,Int}=nothing)\n\tl = length(fs)\n\t\n\tstartframe = if isnothing(startframe)\n\t\tl > 2 ? l - 1 : l\n\telse\n\t\tstartframe\n\tend\n\t\n\t@htl(\"\"\"\n\t\t\n\t\t$(fs)\n\t\t \n\t\t\n\t\t\n\t\t\t \n\t\t\n\t\t\n\t\t\n\t\n\t\t\n\t \"\"\")\nend\n\n# \u2554\u2550\u2561 74c19786-1ba7-4865-a993-590a779ae564\n@skip_as_script frames(rs)\n\n# \u2554\u2550\u2561 8480d0d7-bdf7-468d-9344-5b789e33921c\n# const slotted_code_css = PlutoStylesheet(\"\"\"\nslotted_code_css = PlutoStylesheet(\"\"\"\nslotted-code {\n\tfont-family: \"JuliaMono\", monospace;\n\tfont-size: .75rem;\n\tdisplay: flex;\n\tflex-direction: column;\n}\npre-ish {\n\twhite-space: pre;\n}\n\nline-like {\n\tdisplay: flex;\n\talign-items: baseline;\n}\n\"\"\")\n\n# \u2554\u2550\u2561 b273d3d3-648f-4d34-94e7-e49277d4ba29\nwith_slotted_css(x) = @htl(\"\"\"\n\t$(x)\n\t\n\t\"\"\")\n\n# \u2554\u2550\u2561 326f7661-3482-4bf2-a97b-57cc7ac60ee2\nmacro visual_debug(expr)\n\tframes\n\tSlottedDisplay\n\tvar\"@eval_step_by_step\"\n\twith_slotted_css\n\tquote\n\t\t@eval_step_by_step($(expr)) .|> SlottedDisplay |> frames |> with_slotted_css\n\tend\nend\n\n# \u2554\u2550\u2561 a2cbb0c3-23b9-4091-9ca7-5ba96e85e3a3\n@skip_as_script @visual_debug begin\n\t(1+2) + (7-6)\n\tplot(2000 .+ 30 .* rand(2+2))\n\t4+5\n\tsqrt(sqrt(sqrt(5)))\n\tmd\"#### Wow\"\nend\n\n# \u2554\u2550\u2561 1e619ca9-e00f-46d0-b327-85b33929787f\nfunction Base.show(io::IO, mime::MIME\"text/html\", stylesheet::PlutoStylesheet)\n\t# show(io, mime, md\"`\n\t\t\n\t\t\n\t\t\"\"\")\n\t\tBase.show(io, m, result)\n\tend\n\n\tmd\"\"\"\n\t```julia\n\tfunction Base.show(io::IO, m::MIME\"text/html\", call::Union{WrongCall,CorrectCall,ErrorCall})\n\t```\n\t\"\"\"\nend\n\n# \u2554\u2550\u2561 a4a067b5-8b4b-4846-b986-0417d83cba48\nmacro test(main_expr, expr...)\n\tshow_for_test_result_should_be_defined_before_test_macro\n\t\n\tsource = QuoteNode(__source__)\n\torig_expr = QuoteNode(main_expr)\n\t\n\tquote\n\t\tpluto_result, julia_test_result = try\n\t\t\tresult = $(test(main_expr, expr...))\n\t\t\t(result, Test.Returned(result isa CorrectCall, \"\", $(source)))\n\t\tcatch err\t\n\t\t\trethrow(err)\n\t\t\t(err, Test.Threw(err, Base.catch_stack(), $(source)))\n\t\tend\n\n\t\ttry\n\t\t\tTest.do_test(julia_test_result, $(orig_expr))\n\t\tcatch; end\n\t\t\n\t\tpluto_result\n\tend\nend\n\n# \u2554\u2550\u2561 73d74146-8f60-4388-aaba-0dfe4215cb5d\n@skip_as_script @test sqrt(20-11) == 3\n\n# \u2554\u2550\u2561 71b22e76-2b50-4d16-85f6-9dad0415630e\n@skip_as_script @test iseven(123 + 7^3)\n\n# \u2554\u2550\u2561 6762ed72-f422-43a9-a782-de78f739c0ae\n@skip_as_script @test 4+4 \u2208 [1:7...]\n\n# \u2554\u2550\u2561 f77275b9-90aa-4e07-a608-981b5df727af\n@skip_as_script @test is_good_boy(first(friends))\n\n# \u2554\u2550\u2561 de7e71e2-e5e9-4f05-a61d-658f01a3e937\n@skip_as_script @test is_bad_boy(first(friends))\n\n# \u2554\u2550\u2561 37529063-8ee9-46a6-85cc-94db292da541\n@skip_as_script @test sqrt(sqrt(16)) == sqrt(2)\n\n# \u2554\u2550\u2561 89f78031-1c54-468b-9ab8-7410c51df10e\nexport @test\n\n# \u2554\u2550\u2561 97eb4444-a22c-47f2-9247-3bce6d7e179e\nexample_longer_fn_name = @skip_as_script begin\n\t@test Test.Pass(:symbol, 10, 10, 10) isa Test.Pass\nend\n\n# \u2554\u2550\u2561 24f2eb92-5fd7-429b-b2ea-a987195c6edb\nexample_cant_stepify_assignments = @skip_as_script let\n\t@test @return_error(begin\n\t\t@expand_at_runtime @test begin\n\t\t\tx = 1 + 3\n\t\t\tx\n\t\tend\n\tend).error isa CantStepifyThisYetException\nend\n\n# \u2554\u2550\u2561 bedc3586-6b85-4de2-9ea1-79d842db6b56\nexample_cant_stepify_try_catch = @skip_as_script let\n\t@test @return_error(begin\n\t\t@expand_at_runtime @test try\n\t\t\t0 / 0\n\t\tcatch error\n\t\t\t\"Oops\"\n\t\tend\n\tend).error isa CantStepifyThisYetException\nend\n\n# \u2554\u2550\u2561 42f34453-9935-4e50-a62b-9dcf31d72601\nexample_cant_stepify_if_else = @skip_as_script let\n\t@test @return_error(begin\n\t\t@expand_at_runtime @test if 4 > 5\n\t\t\t\"Yeahhh\"\n\t\telse\n\t\t\t\"Nohhhh\"\n\t\tend\n\tend).error isa CantStepifyThisYetException\nend\n\n# \u2554\u2550\u2561 1b869c21-b8cd-4aaa-91a9-370c1b7a3d32\n@skip_as_script @test quote\n\tbegin begin begin 123 end end end\nend |> remove_linenums |> remove_singleline_blocks === 123\n\n# \u2554\u2550\u2561 Cell order:\n# \u255f\u2500ab02837b-79ec-40d7-bff1-c1d2dd7362ef\n# \u2560\u255073d74146-8f60-4388-aaba-0dfe4215cb5d\n# \u2560\u255071b22e76-2b50-4d16-85f6-9dad0415630e\n# \u2560\u25506762ed72-f422-43a9-a782-de78f739c0ae\n# \u2560\u2550f77275b9-90aa-4e07-a608-981b5df727af\n# \u2560\u2550de7e71e2-e5e9-4f05-a61d-658f01a3e937\n# \u2560\u255037529063-8ee9-46a6-85cc-94db292da541\n# \u255f\u250056347b7e-5007-45f8-8f6d-8ac8cc719637\n# \u255f\u2500bf2abe01-6ae0-4066-8704-12f64e04511b\n# \u255f\u2500e46cf3e0-aa15-4c17-a925-3e9fc5109d54\n# \u255f\u25005b70aaf1-9623-4f55-b055-4263ed8be31d\n# \u255f\u2500fd8428a3-9fa3-471a-8b2d-5bbb8fdb3137\n# \u255f\u2500191f1f04-18d4-485b-af8b-a2f073b7043b\n# \u255f\u2500ec1fd70a-d92a-4688-98b2-135879f07141\n# \u2560\u255089f78031-1c54-468b-9ab8-7410c51df10e\n# \u2560\u2550cf314b21-3f4f-4637-b1ce-ec1d5d5af966\n# \u2560\u255078704300-0531-4f8e-8aa5-3f588fbdd190\n# \u2560\u25509129342b-f560-4901-81a2-56e3f8641521\n# \u2560\u2550c763ed72-82c9-445c-a8f7-a0c40982e4d9\n# \u255f\u25008a2e8348-49cf-4855-b5b3-cdee33e5ed67\n# \u255f\u250042671258-07a0-4015-8f47-4b3032595f08\n# \u255f\u25000d70962a-3880-4dee-a439-35068d019f5a\n# \u2560\u2550113cc425-e224-4f77-bfbd-ef4eb1d1ed70\n# \u2560\u25506188f559-bcab-4da6-84b2-a3fe522a5c3c\n# \u2560\u2550c24b46ce-bcbb-4dc9-8a59-b5b1bd2cd617\n# \u2560\u25505041085e-a406-4ed4-ab82-84d8f126cf0f\n# \u2560\u255003ccd498-83c3-41bb-84d7-625adabd7aee\n# \u2560\u25501bcf8bd1-c8a3-49a1-9791-d813aa856399\n# \u2560\u255014c525a1-eca1-466b-8e63-3a90d7d7111c\n# \u255f\u2500a2efc968-246c-40c2-b285-2ec94b185a44\n# \u255f\u2500e1c306e3-0a47-4149-a9fb-ec7ab380fa11\n# \u2560\u2550b6e8a170-12cc-4d97-905d-274e2609bfd8\n# \u2560\u2550a4a067b5-8b4b-4846-b986-0417d83cba48\n# \u255f\u25009c3f6eab-b1c3-4607-add8-d6d7e468c11a\n# \u255f\u2500dbfbcc16-c740-436c-bbf0-fee16b0a20c5\n# \u2560\u2550d97987a0-bdc0-46ed-a6a5-f35c1ce961dc\n# \u2560\u2550a6709e08-964d-46ea-9813-2c70a834824b\n# \u255f\u25007c2bab29-8609-4575-b2ca-7feb34915645\n# \u2560\u255069bfb438-7ecf-4f9b-8bc4-51e07aa46ef1\n# \u255f\u2500838b5904-1de2-4d9f-8f3c-a93ec224d40e\n# \u2560\u2550a3c41025-2f4a-4f9c-8577-72e4b7abbb98\n# \u2560\u25503e79ff61-6532-4879-9402-86473aa7d960\n# \u2560\u2550275c5f57-623d-439f-b09d-f7c745e0bed6\n# \u2560\u255010803c0d-d0a5-45c5-b7ef-9659e441df69\n# \u255f\u2500b056a99d-5b13-47ba-a199-d788410e3c99\n# \u255f\u25005b093e83-78c1-4187-b406-56e79800e1be\n# \u2560\u2550a461f1fd-b5a5-4ae3-a47c-067a6081fb24\n# \u2560\u255038e54516-cdf4-4c1d-815b-68e1e7a7f6f7\n# \u2560\u2550411271a6-4236-45e2-ab34-f26410108821\n# \u255f\u2500f9c81ab1-556c-4d81-bee8-2897c20e324d\n# \u255f\u2500a392d2d6-5a16-4383-b0ef-5003aa2de9fa\n# \u255f\u2500ae95b691-f54b-4bf5-b17b-3e5bd1edf75e\n# \u255f\u250012119016-fa61-4d38-8c58-821ea435df7d\n# \u2560\u25509bed78b6-5a8f-44ce-ab66-cab685daf264\n# \u255f\u250074929fa6-d1f7-41cd-ab55-48f35d5fbf28\n# \u255f\u2500f1ede628-d158-4296-befd-3eaa87cdad27\n# \u2560\u25502f6e353d-2cdc-46d6-9727-01b0a6167ca0\n# \u2560\u255017dea9e5-84ea-4476-a318-cc475043c83b\n# \u255f\u25005e66e59b-fdb8-4373-b231-097b0227dc5c\n# \u2560\u2550c47252b9-8869-4878-b9bf-7eeb7ed17c9a\n# \u2560\u2550227129bc-4415-4240-ad55-815bde65a5a1\n# \u2560\u2550ce90612e-ffc1-4e30-9d89-531f11fd75eb\n# \u255f\u25000a3f5c6c-6e1b-458c-bf91-523a0b639b41\n# \u255f\u250043fe89d7-f33e-4dfa-853e-327e981feb1e\n# \u255f\u2500fc000550-3053-483e-bc41-6aed22c3999c\n# \u255f\u25003f11ca4c-dd06-47c9-92e2-cb97c18a06db\n# \u255f\u2500b155d336-f746-4c82-8206-ab1a49cedea8\n# \u255f\u2500f9b2a11d-8c4e-47a5-9d93-38025fae9a95\n# \u255f\u2500221aa13b-aa25-4145-8076-da77432364bb\n# \u255f\u25002a514f2f-79c8-4b0d-be8a-170f3386d5d5\n# \u255f\u25009fb4d52d-77f2-4032-a769-6d5e60be43bf\n# \u255f\u25001c1b64b1-107e-4d43-9ce2-569c3034017e\n# \u255f\u2500cade56ad-312e-40cf-bcda-11480ce27852\n# \u255f\u2500810470b8-0a6c-48b8-aba2-a2058b8d9f59\n# \u255f\u2500a29d5277-e97a-4cca-8e31-8037f9cfdd80\n# \u255f\u25004f7aac13-9e49-4b2b-8d78-53f583f6130a\n# \u255f\u2500cc7102e1-6af0-43bb-8cf0-43e2cec210e3\n# \u255f\u2500f5d9a4c5-300f-4dae-8507-346ec0b74632\n# \u255f\u2500ec6f1b07-d026-45ca-996d-be7693664cd7\n# \u255f\u2500dadf1c50-6588-4345-a240-69a72336c7cd\n# \u255f\u2500d384e3fc-b207-48ce-bc7b-1b47a14b1581\n# \u255f\u2500d7dc79e6-1f58-4414-aeef-667bdb0dd200\n# \u255f\u2500a661e172-6afb-42ff-bd43-bb5b787ee5ed\n# \u255f\u2500a6e8c835-f209-445a-9f43-cdf2ecfd1b57\n# \u255f\u25005759b2cc-1e96-4069-ae42-bc159c7cf5fb\n# \u255f\u2500ba4a5762-33da-40e6-94fa-cca9befc6d5a\n# \u255f\u25009101631b-81ca-4c7c-94da-81d9e106df78\n# \u255f\u25003f0e5a49-5eec-42cd-bd2c-254b277840bf\n# \u255f\u2500fc26d26a-a9a5-4646-b85b-12eac66d96cb\n# \u255f\u250094ebb761-21fb-4015-acb3-26310b19b0dc\n# \u255f\u2500312ef6a6-55aa-4913-9416-15e79b4e3362\n# \u2560\u255097eb4444-a22c-47f2-9247-3bce6d7e179e\n# \u255f\u2500716d9ddc-18dc-4973-924e-e5ebf9161ff6\n# \u255f\u25001aa319c8-5e1d-4dd9-ae22-ad99e46e7b4d\n# \u255f\u2500605d2481-23be-4ad9-82c9-e375b7be8669\n# \u255f\u2500bc08755d-721f-403e-af95-36494b8fb7bc\n# \u255f\u2500586826a5-d667-4035-9796-bd2db61498d6\n# \u255f\u2500a8fd09d1-c5ca-47f3-8fb3-32d8aeef3e59\n# \u255f\u250024f2eb92-5fd7-429b-b2ea-a987195c6edb\n# \u2560\u2550bedc3586-6b85-4de2-9ea1-79d842db6b56\n# \u2560\u255042f34453-9935-4e50-a62b-9dcf31d72601\n# \u255f\u2500de94f2b5-96ae-4936-870f-7639a39fd40d\n# \u255f\u250060a398c9-9fe8-4b90-b863-1568183641d9\n# \u255f\u250021d4560e-721f-4ed4-9db7-86a8151ab22c\n# \u255f\u250099afc7f4-727c-4277-8311-f2ffa94830ae\n# \u255f\u25004956526a-daf9-43c9-bff3-ff2446016e2e\n# \u255f\u250084ff6a23-c134-4910-b630-a7ad45f3bf29\n# \u255f\u2500318363d0-6d9e-4144-b478-b775f437edaf\n# \u255f\u250067fd07b7-340b-4e24-bc06-e4c85b186872\n# \u255f\u2500c6d5597c-d505-4125-88c4-10415934d2a4\n# \u255f\u2500872b4877-30dd-4a92-a3c8-69eb50675dcb\n# \u255f\u2500c877c109-db16-468c-8f3c-8294db859d6d\n# \u2560\u2550ab0a19b8-cf7c-4c4f-802a-f85eef81fc02\n# \u255f\u25008480d0d7-bdf7-468d-9344-5b789e33921c\n# \u2560\u25506f5ba692-4b6a-405a-8cd3-1a8f9cc06611\n# \u255f\u2500b4b317d7-bed1-489c-9650-8d336e330689\n# \u2560\u255093ed973f-daf6-408b-9d4b-d53495418610\n# \u2560\u2550dea898a0-1904-4d09-ad0b-6915008fe946\n# \u255f\u2500b5763c10-e11c-4389-b6fc-421d2c9682f1\n# \u255f\u250074c19786-1ba7-4865-a993-590a779ae564\n# \u2560\u2550e968fc57-d850-4e2d-9410-8777d03b7b3c\n# \u255f\u25003d5abd58-02ab-4b91-a7a3-d9068d4df017\n# \u255f\u2500326f7661-3482-4bf2-a97b-57cc7ac60ee2\n# \u255f\u2500b273d3d3-648f-4d34-94e7-e49277d4ba29\n# \u2560\u2550a2cbb0c3-23b9-4091-9ca7-5ba96e85e3a3\n# \u255f\u2500f9ed2487-a7f6-4ce9-b673-f8a298cd5fc3\n# \u255f\u250020166ec9-7084-4d58-8b19-3aa51cc8f2c6\n# \u255f\u25000f31dd2e-0331-4d4c-8db5-9ce188cd3730\n# \u255f\u2500cecba3e6-98e8-408a-97dd-96b67c4f42cf\n# \u2560\u25501633fe05-cb51-4032-b6b6-f23db72bbd49\n# \u2560\u25507c312943-c48b-40e7-a499-227f7ff8aa59\n# \u2560\u2550a0207e25-0398-4104-8c0f-a8fbd9fe1d53\n# \u255f\u25007d14b79c-74e5-4986-80b7-de7cd7d48670\n# \u255f\u25005950488e-2008-48d8-9095-7f9421df191e\n# \u255f\u250077cc33a3-c2bc-4f2d-ba88-e3693ec79b0c\n# \u255f\u25005a3a0f63-dcce-49c9-84fd-a6317184820f\n# \u255f\u250035f63c4e-3583-4ea8-a057-31f18f8a09d6\n# \u255f\u2500ef59d0f0-0f02-4089-a49d-53fb0427c3a0\n# \u255f\u250035b2770e-1db6-4327-bf86-c27a4b61dbd3\n# \u255f\u250022640a2f-ea38-4517-a4f3-7a65e60ffebe\n# \u255f\u2500d414f840-4952-4de5-a565-7fdc81a94817\n# \u255f\u250064bf02a4-4fe3-424d-ae6e-5906c3395278\n# \u255f\u2500f3916810-1911-48bd-936b-776206fcad54\n# \u255f\u2500122c27a5-a6e8-45ef-a968-b9b4b3f9ad09\n# \u255f\u25009126f47d-cbc7-411f-93bd-8684ba06c9e9\n# \u255f\u2500955705f9-c90d-495d-86b4-5f3b5bc9fc8e\n# \u255f\u2500187c3005-cd43-45a0-8cbd-bc96b9cb39da\n# \u255f\u25006c0156a9-7281-4326-9e1f-989efa73bb7b\n# \u255f\u25008d3df0c0-eb48-4dae-97a8-8c01f0b0a34b\n# \u255f\u2500ef6fc423-f1b1-4dcb-a059-276121391bc6\n# \u255f\u2500b765dbfe-4e58-4bb9-b1d6-aa4378d4e9c9\n# \u255f\u25001b869c21-b8cd-4aaa-91a9-370c1b7a3d32\n# \u255f\u2500dbd41240-9fc4-4e25-8b25-2b68afa679f2\n# \u255f\u25007cc07d1b-7757-4428-8028-dc892bf05f2f\n# \u255f\u2500e0837338-e657-4bdc-ae91-1de9224da78d\n# \u255f\u250064df4678-0721-4911-8289-fb18f55e6657\n# \u255f\u250058845ff9-821b-45d4-b5ec-96e1949bb277\n# \u255f\u25004d5f44e4-85e9-4985-9b76-73be5e097186\n# \u255f\u2500dd495e00-d74d-47d4-a5d5-422fb147ec3b\n# \u255f\u2500e414c28a-8111-4821-ab25-21aff8289d26\n# \u255f\u2500c13e0f00-d3c4-4f1d-9531-84ed480c81f3\n# \u255f\u25007e6c2162-97e9-4835-b650-52c9723c327f\n# \u2560\u25501ac164c8-88fc-4a87-a194-60ef616fb399\n# \u2560\u2550b0ab9327-8240-4d34-bdd9-3f8f5117bb29\n# \u255f\u25001e619ca9-e00f-46d0-b327-85b33929787f\n", "meta": {"hexsha": "72e5f2bc25b38ac87989c9ffe758d8f01cdca856", "size": 52772, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/notebook.jl", "max_stars_repo_name": "simeonschaub/PlutoTest.jl", "max_stars_repo_head_hexsha": "6d0200e0ae44b4225beb7973db189b7824cc7b9e", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/notebook.jl", "max_issues_repo_name": "simeonschaub/PlutoTest.jl", "max_issues_repo_head_hexsha": "6d0200e0ae44b4225beb7973db189b7824cc7b9e", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/notebook.jl", "max_forks_repo_name": "simeonschaub/PlutoTest.jl", "max_forks_repo_head_hexsha": "6d0200e0ae44b4225beb7973db189b7824cc7b9e", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 27.4282744283, "max_line_length": 311, "alphanum_fraction": 0.7115515804, "num_tokens": 20741, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4455295497638851, "lm_q2_score": 0.14033624769237754, "lm_q1q2_score": 0.06252394524993803}}
{"text": "### A Pluto.jl notebook ###\n# v0.16.0\n\nusing Markdown\nusing InteractiveUtils\n\n# \u2554\u2550\u2561 42e620aa-5f4c-11eb-2ebf-85814cf720e7\nbegin \n\tusing Pkg; Pkg.activate(\".\") # using the env in /notebooks/day2\n\tusing PlutoUI\n\tusing DSJulia\nend\n\n# \u2554\u2550\u2561 122cffca-5fdc-11eb-3555-b39b818f1116\nlet \n\tusing JuMP\n\tusing GLPK\nend\n\n# \u2554\u2550\u2561 0963185c-5fdc-11eb-0eed-89d514850353\nusing BioSequences\n\n# \u2554\u2550\u2561 24806108-5fdc-11eb-2f19-bb09f836f893\nusing Sockets\n\n# \u2554\u2550\u2561 4621c212-5fc7-11eb-2c9d-ad577506420e\nusing Plots\n\n# \u2554\u2550\u2561 b2c1cef8-5fe5-11eb-20c7-134432196893\nusing Distributed\n\n# \u2554\u2550\u2561 31c1e25e-5e53-11eb-2467-9153d30962d5\nmd\"\"\"\n# Metaprogramming\n\nThe strongest legacy of [Lisp](https://en.wikipedia.org/wiki/Lisp_(programming_language)) in the Julia language is its metaprogramming support. Like Lisp,\nJulia represents its own code as a data structure of the language itself. Since code is represented\nby objects that can be created and manipulated from within the language, it is possible for a\nprogram to transform and generate its own code. This allows sophisticated code generation without\nextra build steps, and also allows true Lisp-style macros operating at the level of abstract syntax trees.\nBecause all data types and\ncode in Julia are represented by Julia data structures, powerful reflection\ncapabilities are available to explore the internals of a program and its types just like any other\ndata.\n\"\"\"\n\n# \u2554\u2550\u2561 4ab33c0e-5e53-11eb-2e63-2dd6f06de3ba\nmd\"\"\"\n## Program representation\n\nEvery Julia program starts its life as a string:\n\"\"\"\n\n# \u2554\u2550\u2561 e76d8f04-5e53-11eb-26df-db496622642d\nprog = \"1 + 1\"\n\n# \u2554\u2550\u2561 f361d734-5e53-11eb-3957-61bd0a370b5a\nmd\"\"\"\n**What happens next?**\n\nThe next step is to parse each string\ninto an object called an expression, represented by the Julia type `Expr`. \nParsing means taking the input (in this case, a string) and building a data structure \u2013 often some kind of parse tree, [abstract syntax tree](https://en.wikipedia.org/wiki/Abstract_syntax_tree) or other hierarchical structure, giving a structural representation of the input while checking for correct syntax. \n\"\"\"\n\n# \u2554\u2550\u2561 15aaa5b6-5e54-11eb-067f-9dcd096940b6\nex1 = Meta.parse(prog)\n\n# \u2554\u2550\u2561 34b6a71e-5e54-11eb-0324-4bffbe85c813\ntypeof(ex1)\n\n# \u2554\u2550\u2561 22770a30-5e54-11eb-28db-1359ab1f402f\nmd\"\"\"\n`Expr` objects contains two parts:\n\n * a `Symbol` identifying the kind of expression. A symbol is an [interned string](https://en.wikipedia.org/wiki/String_interning)\n identifier (string interning is a method of storing only one copy of each distinct string value, which must be immutable).\n\n\"\"\"\n\n# \u2554\u2550\u2561 41f9633c-5e54-11eb-323b-e9703c674b0f\nex1.head\n\n# \u2554\u2550\u2561 48104478-5e54-11eb-2f23-c98c4ccbb763\nmd\"\"\"\n * the expression arguments, which may be symbols, other expressions, or literal values:\n\"\"\"\n\n# \u2554\u2550\u2561 538e2054-5e54-11eb-2b9c-451637e1d8ee\nex1.args\n\n# \u2554\u2550\u2561 6e3beaf8-5e54-11eb-154b-95f5887efcb0\nmd\"\"\"\nExpressions may also be constructed directly in [prefix notation](https://en.wikipedia.org/wiki/Polish_notation) (= Polish notation, a mathematical notation in which operators precede their operands, i.e. `+ 1 1` instead of `1 + 1` with infix notation):\n\"\"\"\n\n# \u2554\u2550\u2561 7de376a8-5e54-11eb-0356-9b74225b671e\nex2 = Expr(:call, :+, 1, 1)\n\n# \u2554\u2550\u2561 825b45a6-5e54-11eb-27e6-f3cca8a2f67e\nmd\"\"\"\nThe two expressions constructed above \u2013 by parsing and by direct construction \u2013 are equivalent:\n\"\"\"\n\n# \u2554\u2550\u2561 8b23cac8-5e54-11eb-2424-5972ea3d2e21\nex1 == ex2\n\n# \u2554\u2550\u2561 8ecd49ba-5e54-11eb-24b6-a328043b7a07\nmd\"\"\"\n**The key point here is that Julia code is internally represented as a data structure that is accessible\nfrom the language itself.**\n\nThe `dump` function provides indented and annotated display of `Expr` objects:\n\"\"\"\n\n# \u2554\u2550\u2561 985a57f2-5e54-11eb-1f2f-bf23014c6171\n@terminal dump(ex2)\n\n# \u2554\u2550\u2561 d66ea502-5e54-11eb-2694-4766a05aed38\nmd\"\"\"\n### Symbols\n\nThe `:` character has two syntactic purposes in Julia. The first form creates a [`Symbol`](@ref),\nan [interned string](https://en.wikipedia.org/wiki/String_interning) used as one building-block\nof expressions:\n\"\"\"\n\n# \u2554\u2550\u2561 db1e9936-5e54-11eb-376f-8ddea65d976c\ns = :foo\n\n# \u2554\u2550\u2561 e4f50fda-5e54-11eb-2210-6b8eb531d898\ntypeof(s)\n\n# \u2554\u2550\u2561 ea891b6e-5e54-11eb-24c9-357a8a65d1c0\nmd\"\"\"\nThe `Symbol` constructor takes any number of arguments and creates a new symbol by concatenating\ntheir string representations together:\n\"\"\"\n\n\n# \u2554\u2550\u2561 efe79c46-5e54-11eb-23c9-3107ea94e5b9\n:foo == Symbol(\"foo\")\n\n# \u2554\u2550\u2561 f5fac50e-5e54-11eb-2ff9-cf7e30a4a8ad\nSymbol(\"func\", 10)\n\n# \u2554\u2550\u2561 fe35b31e-5e54-11eb-258e-29913f1e8723\nSymbol(:var, '_', \"sym\")\n\n# \u2554\u2550\u2561 0464f1be-5e55-11eb-34df-ef1217012663\nmd\"\"\"Note that to use `:` syntax, the symbol's name must be a valid identifier.\nOtherwise the `Symbol(str)` constructor must be used.\n\nIn the context of an expression, symbols are used to indicate access to variables; when an expression\nis evaluated, a symbol is replaced with the value bound to that symbol in the appropriate **scope**.\n\"\"\"\n\n# \u2554\u2550\u2561 abfc3392-5e5a-11eb-149d-ad3dd2755524\nmd\"\"\"\n### Quoting\n\nThe second syntactic purpose of the `:` character is to create expression objects without using\nthe explicit `Expr` constructor. This is referred to as *quoting*. The `:` character, followed\nby paired parentheses around a single statement of Julia code, produces an `Expr` object based\non the enclosed code. Here is an example of the short form used to quote an arithmetic expression:\n\"\"\"\n\n# \u2554\u2550\u2561 917a579c-5e5a-11eb-3fe6-cb0dcad5e4e6\nex = :(a + b * c + 1)\n\n# \u2554\u2550\u2561 82d3220a-5e5a-11eb-3c49-d5e7afb1af2a\ntypeof(ex)\n\n# \u2554\u2550\u2561 67954702-5e5a-11eb-1ab1-e9a285ee208d\nex.args\n\n# \u2554\u2550\u2561 2ae6ac9c-5e5a-11eb-060c-b3eae1df0493\nmd\"\"\"\n### Interpolation\n\nDirect construction of `Expr` objects with value arguments is powerful, but `Expr` constructors\ncan be tedious compared to \"normal\" Julia syntax. As an alternative, Julia allows *interpolation* of\nliterals or expressions into quoted expressions. Interpolation is indicated by a prefix `$`.\n\nIn this example, the value of variable `a` is interpolated:\n\"\"\"\n\n# \u2554\u2550\u2561 5ef3d532-5e5a-11eb-30b3-a569e0bd736e\na = 1\n\n# \u2554\u2550\u2561 3bb1f892-5e5a-11eb-0946-e5e977a4e5b2\n:($a + b)\n\n# \u2554\u2550\u2561 5763cca0-5e5a-11eb-1341-3d9bfb47fb58\nmd\"\"\"\nInterpolating into an unquoted expression is not supported and will cause a compile-time error:\n\"\"\"\n\n# \u2554\u2550\u2561 e6a3902a-5e58-11eb-2bd2-4bc779bcebe7\n$a + b\n\n# \u2554\u2550\u2561 11991248-5e55-11eb-2748-992f6fe48620\nmd\"\"\"\nThe use of `$` for expression interpolation is intentionally reminiscent of string interpolation and command interpolation. Expression interpolation allows convenient, readable programmatic construction of complex Julia expressions.\n\"\"\"\n\n# \u2554\u2550\u2561 1c016bec-5e5d-11eb-3687-931da82f8c04\nmd\"\"\"\n\n### Splatting interpolation\n\nNotice that the `$` interpolation syntax allows inserting only a single expression into an\nenclosing expression.\nOccasionally, you have an array of expressions and need them all to become arguments of\nthe surrounding expression.\nThis can be done with the syntax `$(args...)`.\n\nFor example, the following code generates a function call where the number of arguments is\ndetermined programmatically:\n\"\"\"\n\n# \u2554\u2550\u2561 552ea3bc-5e5d-11eb-0c97-e1e6db526df3\nargs = [:x, :y, :z]\n\n# \u2554\u2550\u2561 629c4f9a-5e5d-11eb-2731-bf7457755829\n:(f(1, $(args...)))\n\n# \u2554\u2550\u2561 66c113d0-5e5d-11eb-0aab-b3fa7f98eae5\nmd\"\"\"\n### Evaluating expressions\n\nGiven an expression object, one can cause Julia to evaluate (execute) it at global scope using `eval`\n\"\"\"\n\n# \u2554\u2550\u2561 6fc2fb9c-5e5d-11eb-0f21-6524555a9c42\n:(1 + 2)\n\n# \u2554\u2550\u2561 6dab1d58-5e5d-11eb-3381-a1796f8d14c5\neval(:(1 + 2))\n\n# \u2554\u2550\u2561 b833d8ba-5e5d-11eb-3311-899efb15c1b0\nmd\"\"\"\n## Macros\n\nNow that we have an understanding of the basic concepts of code representation in Julia, we can introduce the core concept of this notebook: macros. \nMacros provide a method to include generated code in the final body of a program. A macro maps\na tuple of arguments to a returned *expression*, and the resulting expression is compiled directly\nrather than requiring a runtime `eval` call. Macro arguments may include expressions,\nliteral values, and symbols. \n\nIn the following examples, we will show that macros allow us to\n1. modify code before it runs\n2. elegantly add new features or syntax\n3. process strings at compile time instead of runtime\n\n### Basics\n\nHere is an extraordinarily simple macro:\n\"\"\"\n\n# \u2554\u2550\u2561 169c6aa2-5e5e-11eb-1d84-e7e3b4641e10\nmacro sayhello()\n\treturn :( println(\"Hello, world!\") )\nend\n\n# \u2554\u2550\u2561 ae1525ea-5e5e-11eb-3906-593143776559\n@terminal @sayhello\n\n# \u2554\u2550\u2561 c40cebe6-5e5e-11eb-1f7f-37134b7a449f\nmd\"\"\"\nMacros have a dedicated character in Julia's syntax: the `@` (at-sign), followed by the unique\nname declared in a `macro NAME ... end` block. In this example, the compiler will replace all\ninstances of `@sayhello` with:\n\"\"\"\n\n# \u2554\u2550\u2561 f5ab41f4-5e5f-11eb-02f7-efcb3e05e616\n:( println(\"Hello, world!\") )\n\n# \u2554\u2550\u2561 0001a53a-5e60-11eb-18df-d7e9e7dcf43f\nmd\"\"\"\nWhen `@sayhello` is entered in the REPL, the expression executes immediately, thus we only see the evaluation result:\n\"\"\"\n\n# \u2554\u2550\u2561 bda7fd66-5e5e-11eb-1e06-eb67a0178e14\n@terminal @sayhello()\n\n# \u2554\u2550\u2561 1707320e-5e60-11eb-3f33-798c81663488\nmd\"\"\"Now, consider a slightly more complex macro:\"\"\"\n\n# \u2554\u2550\u2561 1fdf9600-5e60-11eb-30d3-b167811ae47f\nmacro sayhello(name)\n\treturn :( println(\"Hello, \", $name) )\nend\n\n# \u2554\u2550\u2561 2dcacdfc-5e60-11eb-3d04-9f2ffb46d0fe\nmd\"\"\"\nThis macro takes one argument: `name`. When `@sayhello` is encountered, the quoted expression is *expanded* to interpolate the value of the argument into the final expression:\n\"\"\"\n\n# \u2554\u2550\u2561 36fc5076-5e60-11eb-04be-bd9f542b1ff3\n@terminal @sayhello(\"Mr. Bond\")\n\n# \u2554\u2550\u2561 0c787198-61a0-11eb-1e30-fbcb5a624076\n@terminal @sayhello \"Mr. Bond\"\n\n# \u2554\u2550\u2561 4a732eb8-5e60-11eb-197b-0729d0f1be15\nmd\"\"\"\nWe can view the quoted return expression using the function `macroexpand` or the macro `@macroexpand` (**important note:**\nthis is an extremely useful tool for debugging macros).\nWe can see that the `\"Mr. Bond\"` literal has been interpolated into the expression.\"\"\"\n\n# \u2554\u2550\u2561 536addec-5e61-11eb-1a9d-d7f2c190d6c2\n@macroexpand @sayhello(\"Mr. Bond\")\n\n# \u2554\u2550\u2561 855412b2-5e61-11eb-3aba-bb862489414e\nmd\"\"\"\n\n### Hold on: why macros?\n\nWe have already seen a function `f(::Expr...) -> Expr` in a previous section. In fact, `macroexpand` is also such a function. So, why do macros exist?\n\nMacros are necessary because they execute when code is parsed, therefore, macros allow the programmer\nto generate and include fragments of customized code *before* the full program is run. To illustrate\nthe difference, consider the following example.\n\nNote that in this case, you need to look at your REPL to see the output. A macro definition is not allowed in the local scope, so we cannot wrap this with our `@terminal` macro.\n\"\"\"\n\n# \u2554\u2550\u2561 cabb3cdc-5e62-11eb-2479-bb8337c05292\nmacro twostep(arg)\n\tprintln(\"I execute at parse time. The argument is: \", arg)\n\tstr1 = \"I execute at runtime. \"\n\tstr2 = \"The argument is: \"\n\tmessage = str1 * str2\n\treturn :(println($message, $arg))\nend\n\n# \u2554\u2550\u2561 f33f0cfe-5f52-11eb-30e0-93c4b899aaa7\nmd\"\"\"Note that the computation of message is compiled away if we expand the macro!\"\"\"\n\n# \u2554\u2550\u2561 01feeca0-5e63-11eb-11a0-1b66a92127d1\nex_twostep = @macroexpand @twostep :(1, 2, 3)\n\n# \u2554\u2550\u2561 f1969888-5e65-11eb-18a3-879d2f87b447\n@terminal dump(ex_twostep)\n\n# \u2554\u2550\u2561 0613f620-5e66-11eb-08d9-01dd3a403321\nmd\"\"\"\n### Macro invocation\n\nMacros are invoked with the following general syntax:\n\n```julia\n@name expr1 expr2 ...\n@name(expr1, expr2, ...)\n```\n\n\"\"\"\n\n# \u2554\u2550\u2561 29a2c0ee-5e66-11eb-2194-6f1c3cef563a\nmd\"\"\"\nNote the distinguishing `@` before the macro name and the lack of commas between the argument\nexpressions in the first form, and the lack of whitespace after `@name` in the second form. The\ntwo styles should not be mixed. For example, the following syntax is different from the examples\nabove; it passes the tuple `(expr1, expr2, ...)` as one argument to the macro:\n\n```julia\n@name (expr1, expr2, ...)\n```\n\"\"\"\n\n# \u2554\u2550\u2561 a1e86478-5e66-11eb-2ef9-6d460e707013\nmd\"\"\"\n### Building an advanced macro\n\nHere is a simplified definition of Julia's `@assert` macro, which checks if an expression is true:\n\n(`... ? ... : ...` is the ternary if-else operator seen in `01-basics.jl` )\n\"\"\"\n\n# \u2554\u2550\u2561 e61381aa-5e66-11eb-3347-5d26b61e6c17\nmacro assert(ex)\n\treturn :( $ex ? nothing : throw(AssertionError($(string(ex)))) )\nend\n\n# \u2554\u2550\u2561 a43490de-5e67-11eb-253e-f7455ade3ffe\nmd\"\"\"This macro can be used like this:\"\"\"\n\n# \u2554\u2550\u2561 c79ad04c-5e67-11eb-1fd9-bf7ff40f0862\nmd\"\"\"\nIn place of the written syntax, the macro call is expanded at parse time to its returned result. This is equivalent to writing:\"\"\"\n\n# \u2554\u2550\u2561 f34e9020-5e67-11eb-1987-e3e6a0969705\n1 == 1.0 ? nothing : throw(AssertionError(\"1 == 1.0\"))\n\n# \u2554\u2550\u2561 f81955fe-5e67-11eb-11b9-5f9fd0096a09\n1 == 0 ? nothing : throw(AssertionError(\"1 == 0\"))\n\n# \u2554\u2550\u2561 fb772ef4-5e67-11eb-218e-330ba087bcc8\nmd\"\"\"\nThat is, in the first call, the expression `:(1 == 1.0)` is spliced into the test condition slot,\nwhile the value of `string(:(1 == 1.0))` is spliced into the assertion message slot. The entire\nexpression, thus constructed, is placed into the syntax tree where the `@assert` macro call occurs.\nThen at execution time, if the test expression evaluates to true, then `nothing` is returned,\nwhereas if the test is false, an error is raised indicating the asserted expression that was false.\nNotice that it would not be possible to write this as a function, since only the *value* of the\ncondition is available and it would be impossible to display the expression that computed it in\nthe error message.\n\nThe actual definition of `@assert` in Julia Base is more complicated. It allows the\nuser to optionally specify their own error message, instead of just printing the failed expression.\nJust like in functions with a variable number of arguments, this is specified with an ellipses\nfollowing the last argument:\n\"\"\"\n\n# \u2554\u2550\u2561 2df2baa8-5e68-11eb-2f28-9968b7ecbcd5\nmacro assert(ex, msgs...)\n\tmsg_body = isempty(msgs) ? ex : msgs[1]\n\tmsg = string(msg_body)\n\treturn :($ex ? nothing : throw(AssertionError($msg)))\nend\n\n# \u2554\u2550\u2561 b0dfd0dc-5e67-11eb-19cf-5970a2e80bfa\n@assert 1 == 1.0\n\n# \u2554\u2550\u2561 c42d2d6a-5e67-11eb-1564-2dcb0e91fb19\n@assert 1 == 0\n\n# \u2554\u2550\u2561 376fe8e4-5e68-11eb-1d7d-c9b1945b133f\nmd\"\"\"\nNow `@assert` has two modes of operation depending upon the number of arguments it receives!\nIf there is only one argument, the tuple of expressions captured by `msgs` will be empty and it\nwill behave the same as the simpler definition above with only one argument. But now if the user specifies a second argument,\nit is printed in the message body instead of the failing expression. You can inspect the result\nof a macro expansion with `@macroexpand`.\n\"\"\"\n\n# \u2554\u2550\u2561 4750ce04-5e68-11eb-237e-3fed9eb1f4c5\n@macroexpand @assert a == b\n\n# \u2554\u2550\u2561 4dce26dc-5e68-11eb-1ff7-8974f93e3cb8\n@macroexpand @assert a==b \"a should equal b!\"\n\n# \u2554\u2550\u2561 558920ca-5e68-11eb-3533-d5d596e3884d\nmd\"\"\"\n\nThere is yet another case that the actual `@assert` macro handles: what if, in addition to printing\n\"a should equal b,\" we wanted to print their values? One might naively try to use string interpolation\nin the custom message, e.g., `@assert a==b \"a ($a) should equal b ($b)!\"`, but this won't work\nas expected with the above macro. Can you see why? Recall from string interpolation that an interpolated string is rewritten to a call to `string`. Compare:\n\"\"\"\n\n# \u2554\u2550\u2561 6d69106a-5e68-11eb-0d50-e17b4db5db74\ntypeof(:(\"a should equal b\"))\n\n# \u2554\u2550\u2561 715464cc-5e68-11eb-3e1a-977db04a7db6\ntypeof(:(\"a ($a) should equal b ($b)!\"))\n\n# \u2554\u2550\u2561 7588018e-5e68-11eb-3be2-b97ce124d6bf\n@terminal dump(:(\"a ($a) should equal b ($b)!\"))\n\n# \u2554\u2550\u2561 84771eaa-5e68-11eb-0684-cf1a3118d10b\nmd\"\"\"\nSo now instead of getting a plain string in `msg_body`, the macro is receiving a full expression that will need to be evaluated in order to display as expected. This can be spliced directly into the returned expression as an argument to the `string` call; see `error.jl` for the complete implementation.\n\nThe `@assert` macro makes great use of splicing into quoted expressions to simplify the manipulation of expressions inside the macro body.\n\"\"\"\n\n# \u2554\u2550\u2561 43a19ace-5e6b-11eb-005b-b7d2d9a46878\nmd\"\"\"\n### Macros and dispatch\n\nMacros, just like Julia functions, are generic. This means they can also have multiple method definitions, thanks to multiple dispatch:\n\"\"\"\n\n# \u2554\u2550\u2561 508167c4-5e6b-11eb-2587-b967b79cf74c\nmacro m end\n\n# \u2554\u2550\u2561 59f86d98-5e6b-11eb-3259-7312501b70bf\nmacro m(args...)\n\t\"$(length(args)) arguments\"\nend\n\n# \u2554\u2550\u2561 5edf55e2-5e6b-11eb-383e-5965ae694c4c\nmacro m(x,y)\n \t\"Two arguments\"\nend\n\n# \u2554\u2550\u2561 3dc3aa10-5e6c-11eb-3338-7777b8cb97a5\nmd\"\"\"\nHowever one should keep in mind, that macro dispatch is based on the types of AST\nthat are handed to the macro, not the types that the AST evaluates to at runtime:\"\"\"\n\n# \u2554\u2550\u2561 34565ba8-5e6c-11eb-2a27-2ff13394bcc8\nmacro m(::Int)\n\t\"An Integer\"\nend\n\n# \u2554\u2550\u2561 65c94e9c-5e6b-11eb-38a8-4d60c692222b\n@m \"asdl\"\n\n# \u2554\u2550\u2561 42fc5284-5e6c-11eb-0edb-9555a320ec55\n@m 1 2\n\n# \u2554\u2550\u2561 02e4bf64-5e6d-11eb-36b5-fb57c87bff81\n@m 3\n\n# \u2554\u2550\u2561 07f8cc34-5e6d-11eb-36e4-b56b2b92a2e2\nx = 2\n\n# \u2554\u2550\u2561 0bbfce80-5e6d-11eb-07f6-b71086357384\n@m x\n\n# \u2554\u2550\u2561 0e00bff6-5e6d-11eb-342d-bfded7ef8a96\nmd\"\"\"\n## Code Generation\n\nWhen a significant amount of repetitive boilerplate code is required, it is common to generate\nit programmatically to avoid redundancy. In most languages, this requires an extra build step,\nand a separate program to generate the repetitive code. In Julia, expression interpolation and `eval` allow such code generation to take place in the normal course of program execution.\nFor example, consider the following custom type\n\"\"\"\n\n# \u2554\u2550\u2561 36805374-5e6d-11eb-205c-0524096bdf27\nstruct MyNumber\n x::Float64\nend\n\n# \u2554\u2550\u2561 4056c9bc-5e6d-11eb-377e-238ef8633c74\nmd\"\"\"for which we want to add a number of methods to. We can do this programmatically in the following loop:\"\"\"\n\n# \u2554\u2550\u2561 49807d8c-5e6d-11eb-0170-f1419837d958\nfor op = (:sin, :cos, :tan, :log, :exp, :log)\n @eval Base.$op(a::MyNumber) = MyNumber($op(a.x))\nend\n\n# \u2554\u2550\u2561 4ef3f280-5e6d-11eb-2020-b7cd9e6b9c03\nmd\"\"\"and we can now use those functions with our custom type:\"\"\"\n\n# \u2554\u2550\u2561 53f36f36-5e6d-11eb-3f06-6de0eff4d173\ny = MyNumber(\u03c0)\n\n# \u2554\u2550\u2561 3428d7b8-5e6d-11eb-32bc-af1df0579e60\nsin(y)\n\n# \u2554\u2550\u2561 18e3753a-5f55-11eb-0a11-47ca6abe9186\nmd\"This will not work since we only have defined `log` and not `log10`.\"\n\n# \u2554\u2550\u2561 2f4fe6ce-5e6c-11eb-1744-2f9534c8b6ba\nlog10(y)\n\n# \u2554\u2550\u2561 cae2546a-5e6b-11eb-3fc2-69f9a15107cf\nmd\"\"\"In this manner, Julia acts as its own preprocessor, and allows code generation from inside the language.\"\"\"\n\n# \u2554\u2550\u2561 0ac5096e-61a1-11eb-24ba-13bc0a3377c6\n\n\n# \u2554\u2550\u2561 efc1357a-61a0-11eb-20f5-a15338080e4c\nmd\"### Example: domain specific languages\"\n\n# \u2554\u2550\u2561 4d9de330-5f55-11eb-0d89-eb81e6c9ffab\nmd\"\nCode generation can for instance be used to simplify the creation of a mathematical optimisation problem. In this case, we we'll use the `JuMP` package.\n`JuMP` ('Julia for Mathematical Programming') is an open-source modeling language that is embedded in Julia. It allows users to formulate various classes of optimization problems (linear, mixed-integer, quadratic, conic quadratic, semidefinite, and nonlinear) with easy-to-read code. `JuMP` also makes advanced optimization techniques easily accessible from a high-level language. \n\nAs a dummy example, let us consider the following linear programming problem:\n\n``\\max_{x,y}\\,\\,x + 2y``\n\n``\\text{s.t.}``\n\n``x + y \\leq 1``\n\n``0\\leq x, y \\leq 1``\n\nWhich can be transcribed into a `JuMP` model as:\n\"\n\n# \u2554\u2550\u2561 92458f54-5f57-11eb-179f-c749ce06f7e2\nlet\n\tmodel = Model(GLPK.Optimizer)\n\t@variable(model, 0 <= x <= 1)\n\t@variable(model, 0 <= y <= 1)\n\t@constraint(model, x + y <= 1)\n\t@objective(model, Max, x + 2y)\n\toptimize!(model)\n\tvalue(x), value(y), objective_value(model)\nend\n\n# \u2554\u2550\u2561 d1222dcc-5f57-11eb-2c87-77a16ea8e65d\nmd\"\nWithout the macros, the code would be more difficult to read. As an example, check the `macroexpansion` of `@constraint` to see the bunch of code that is generated behind the scenes:\n\"\n\n# \u2554\u2550\u2561 0b1f3542-5f58-11eb-1610-75df3e2a4a76\nlet\n\tmodel = Model(GLPK.Optimizer)\n\t@macroexpand @constraint(model, x + y <= 1)\nend\n\n# \u2554\u2550\u2561 57f243ec-5fe3-11eb-04d9-d776ccba4c0f\nmd\"Finally, this section on code generation ends with two more examples:\n1. a macro to repeat a certain expression n-times\n2. a macro to repeat a certain expression until a condition is met\n\nAre these macros the best way to tackle the problems at hand? Maybe not, but they do give a nice illustration of how the code generation works.\n\"\n\n# \u2554\u2550\u2561 211cf9ca-5fe3-11eb-2596-71b1848753d7\nmacro dotimes(n, body)\n quote\n for i = 1:$(esc(n))\n $(esc(body))\n end\n end\nend\n\n# \u2554\u2550\u2561 35ccaee2-5fe3-11eb-3bd0-e355b0fd3f71\n@macroexpand @dotimes 3 println(\"hi there\")\n\n# \u2554\u2550\u2561 434669d2-5fe3-11eb-1f4a-27edf1f9e7f6\nmacro until(condition, block)\n quote\n while true\n $(esc(block))\n if $(esc(condition))\n break\n end\n end\n end\nend\n\n# \u2554\u2550\u2561 9612965e-5fe3-11eb-172a-59cc73a26ab6\nPlutoUI.with_terminal() do\n\ti = 0\n\t@until i == 10 begin\n\t\ti += 1\n\t\tprintln(i) \n\tend\nend\n\n# \u2554\u2550\u2561 3c2cce20-5fe3-11eb-32e3-f542f9a94a6e\n@macroexpand @until j == 10 begin\n\tj += 1\n\tprintln(j) \nend\n\n# \u2554\u2550\u2561 10a8532a-5e60-11eb-3331-974e708cb39d\nmd\"\"\"\n## Non-Standard String Literals\n\nString literals prefixed by an identifier are called non-standard string literals, and can have different semantics than un-prefixed string literals. For example:\n\n\n * `r\"^\\s*(?:#|$)\"` produces a regular expression object rather than a string\n * `b\"DATA\\xff\\u2200\"` is a byte array literal for `[68,65,84,65,255,226,136,128]`.\n\nPerhaps surprisingly, these behaviors are not hard-coded into the Julia parser or compiler. Instead,\nthey are custom behaviors provided by a general mechanism that anyone can use: prefixed string\nliterals are parsed as calls to specially-named macros. For example, the regular expression macro\nis just the following:\n```julia\nmacro r_str(p)\n Regex(p)\nend\n```\n\"\"\"\n\n# \u2554\u2550\u2561 d8a9ad46-5e7a-11eb-1cce-3d4bc49cd332\nmd\"\"\"\nThat's all. This macro says that the literal contents of the string literal `r\"^\\s*(?:#|$)\"` should\nbe passed to the `@r_str` macro and the result of that expansion should be placed in the syntax\ntree where the string literal occurs. In other words, the expression `r\"^\\s*(?:#|$)\"` is equivalent\nto placing the following object directly into the syntax tree:\n\"\"\"\n\n# \u2554\u2550\u2561 e85b2d3c-5e7a-11eb-3b89-e11bc274f7cf\nRegex(\"^\\\\s*(?:#|\\$)\")\n\n# \u2554\u2550\u2561 ec73cd70-5e7a-11eb-0285-6b1fabd1289d\nmd\"\"\"\nNot only is the string literal form shorter and far more convenient, but it is also more efficient:\nsince the regular expression is compiled, which takes time, and the `Regex` object is actually created *when the code is compiled*,\nthe compilation occurs only once, rather than every time the code is executed. Consider if the\nregular expression occurs in a loop:\n\"\"\"\n\n# \u2554\u2550\u2561 f1d24ba2-5e7a-11eb-010a-f543cfc306b5\nPlutoUI.with_terminal() do\n\tfor line \u2208 [\"first\", \"sec#nd\", \"third\"]\n\t\tm = match(r\"#\", line)\n\t\tif m === nothing\n\t\t\tprintln(\"nothing found\")\n\t\telse\n\t\t\tprintln(\"found something\")\n\t\tend\n\tend\nend\n\n# \u2554\u2550\u2561 f838762c-5e7a-11eb-000a-e963fab5e97d\nmd\"\"\"\nSince the regular expression `r\"^\\s*(?:#|$)\"` is compiled and inserted into the syntax tree when\nthis code is parsed, the expression is only compiled once instead of each time the loop is executed.\nIn order to accomplish this without macros, one would have to write this loop like this:\n\"\"\"\n\n# \u2554\u2550\u2561 149c1c12-5e7b-11eb-297d-6da57f45891b\nPlutoUI.with_terminal() do\n\tre = Regex(\"#\")\n\tfor line \u2208 [\"first\", \"sec#nd\", \"third\"]\n\t\tm = match(re, line)\n\t\tif m === nothing\n\t\t\tprintln(\"nothing found\")\n\t\telse\n\t\t\tprintln(\"found something\")\n\t\tend\n\tend\nend\n\n# \u2554\u2550\u2561 780da60e-5e7c-11eb-0c1a-55be73da188f\nmd\"\"\"\nMoreover, if the compiler could not determine that the regex object was constant over all loops,\ncertain optimizations might not be possible, making this version still less efficient than the\nmore convenient literal form above. Of course, there are still situations where the non-literal\nform is more convenient: if one needs to interpolate a variable into the regular expression, one\nmust take this more verbose approach. In the vast majority of use cases, however, regular expressions\nare not constructed based on run-time data. In this majority of cases, the ability to write regular\nexpressions as compile-time values is invaluable.\n\"\"\"\n\n# \u2554\u2550\u2561 90a672d0-5f5c-11eb-3a62-1b034cd41e67\nmd\"\nDesigning your own custom string literals can be done as such:\n\"\n\n# \u2554\u2550\u2561 b4104d12-5e7c-11eb-3e22-a78b74526129\nmacro foo_str(str, flag)\n # do stuff\n\tstr, flag\nend\n\n# \u2554\u2550\u2561 20393e86-5e7d-11eb-18e3-613890472903\nfoo\"this is the string\"theflag\n\n# \u2554\u2550\u2561 5069df16-5e7d-11eb-217d-6b740e9b3559\nmd\"\"\"\nThe first example of a custom macro can be found in every notebook in this course! It is the markdown string literal, which allows the usage of Markdown markup language to prettify these lectures!\n\n```julia\nmd\"I am a Markdown string with glorious **formatting capabilities**.\"\n```\nI am a Markdown string with glorious *formatting* **capabilities**.\n\"\"\"\n\n# \u2554\u2550\u2561 1a50ff0e-5e7d-11eb-2fc4-cd5c12015751\nmd\"The second example is from the `BioSequences.jl` package. In that bioinformatics package, you can define sequences e.g. DNA and RNA as string literals:\n\"\n\n# \u2554\u2550\u2561 0664eb22-5e7d-11eb-07a6-ed35143bf03f\ndna\"ACGT\"\n\n# \u2554\u2550\u2561 a965bd70-5e7c-11eb-13dd-5fe83950af11\nmd\"Repetition and concatenation can be performed pretty straightforward:\"\n\n# \u2554\u2550\u2561 fb0c59fa-5f61-11eb-1ab7-bf7b80746aa2\nrepeat(dna\"TTAGGG\", 10)\n\n# \u2554\u2550\u2561 a157dafc-5e7c-11eb-105d-4db3ee9dbec6\ndna = dna\"ACGT\" * dna\"TGCAA\"\n\n# \u2554\u2550\u2561 0ec9887c-5f5e-11eb-29e5-931881dcb222\nmd\"Other typical string operations such as pushing new values work as you would expect\"\n\n# \u2554\u2550\u2561 1d4aca5a-5f5e-11eb-23c3-d1ddef4e6ee6\npush!(dna, DNA_A)\n\n# \u2554\u2550\u2561 c4c2137a-5f5d-11eb-1e06-8f5e2b1f1c0e\nmd\"There exist methods to convert a DNA sequence to its RNA equivalent:\"\n\n# \u2554\u2550\u2561 e13fcbbe-5f5d-11eb-2074-fdbc945861e1\nrna = convert(LongRNASeq, dna)\n\n# \u2554\u2550\u2561 2c1308f4-5f5e-11eb-058f-79cbf7dd23c6\nmd\"Note that altough the printout of the DNA and RNA object is different because of different nucleotides. The information content is the same, and as such this statement is true:\"\n\n# \u2554\u2550\u2561 fdf46c92-5f5d-11eb-345b-e145c9c2874c\ndna.data === rna.data\n\n# \u2554\u2550\u2561 37da420a-5f61-11eb-3688-09e6188165e3\nmd\"Remember before that we mentioned that you can add a flag to a string literal? In `BioSequences.jl` this has a use case. \n\nIf you have a function that generates a sequence, and you want it to create a new sequence each time it is called, then you can add a flag to the end of the sequence literal to dictate behaviour: A flag of 's' means 'static': the sequence will be allocated before code is run, as is the default behaviour. However providing 'd' flag changes the behaviour: 'd' means 'dynamic': the sequence will be allocated whilst the code is running, and not before. So to change foo so as it creates a new sequence each time it is called, simply add the 'd' flag to the sequence literal:\"\n\n# \u2554\u2550\u2561 d9f18616-5e8d-11eb-1b1a-3bcc749fb467\nfunction getdna_dynamic()\n\ts = dna\"CTT\"d # 'd' flag appended to the string literal.\n\tpush!(s, DNA_A)\nend\n\n# \u2554\u2550\u2561 ea3da4dc-5e8d-11eb-0910-091b926aea58\ngetdna_dynamic()\n\n# \u2554\u2550\u2561 f13dd22a-5e8d-11eb-1def-137de66123ba\ngetdna_dynamic()\n\n# \u2554\u2550\u2561 be11ec9a-5f61-11eb-374a-c96489ea582d\nmd\"Output of `getdna_dynamic()` stays the same!\"\n\n# \u2554\u2550\u2561 f44d757e-5e8d-11eb-18f1-719d3c7b8688\nfunction getdna_static()\n\ts = dna\"CTT\"s # 's' flag appended to the string literal.\n\tpush!(s, DNA_A)\nend\n\n# \u2554\u2550\u2561 fa310fdc-5e8d-11eb-024b-7ded3e213112\ngetdna_static()\n\n# \u2554\u2550\u2561 1acb02d4-5e8e-11eb-232c-f756e92c3f97\ngetdna_static()\n\n# \u2554\u2550\u2561 2dcb4600-5e8e-11eb-093e-59eb79f0cf20\nmd\"\"\"\nBe careful when you are using sequence literals inside of functions, and inside the bodies of things like for loops. And if you use them and are unsure, use the 's' and 'd' flags to ensure the behaviour you get is the behaviour you intend.\n\"\"\"\n\n# \u2554\u2550\u2561 2e2601b6-5e94-11eb-3613-eb5fee19b6b7\nmd\"\n## Overview of some interesting macros\n\"\n\n# \u2554\u2550\u2561 311a4666-5fc5-11eb-2cc0-ef66e5be96e3\nmd\"check if an expression is true\"\n\n# \u2554\u2550\u2561 4622422a-5fc5-11eb-20f2-9bfe466f8f30\n@assert true == true\n\n# \u2554\u2550\u2561 5ba5d6b6-5fc5-11eb-256b-73a954a5db68\nmd\"Integers and floating point numbers with arbitrary precision. This macro exists because promoting a floating point number to a `BigFloat` will keep the approximation error of `Float64`.\"\n\n# \u2554\u2550\u2561 a47ab60e-5fc5-11eb-368a-d76fc6ee640d\nbig\"0.1\"\n\n# \u2554\u2550\u2561 a99546c2-5fc5-11eb-0902-71cb283543c8\n@big_str \"0.1\"\n\n# \u2554\u2550\u2561 af2c7d4e-5fc5-11eb-30d4-a367d2c0db23\nbig(0.1)\n\n# \u2554\u2550\u2561 4a335466-5fc6-11eb-2273-312fd3928616\nmd\"There exists a lot more unique string literals than we have shown here, such as html strings, ip address literals, etc.\"\n\n# \u2554\u2550\u2561 17f771dc-5fc6-11eb-3b83-ed51f87194e0\nhtml\"\"\"1 println("hello world!" )\n
\")\n\t\tprint(io, \"
\")\n\tend\nend\n\n# \u2554\u2550\u2561 353b54bf-2cd7-4ece-840c-01d9b121dbbd\nThing()\n\n# \u2554\u2550\u2561 f268819d-e75b-4818-8779-b719625b5b4c\nmd\"# Putting this all Together\"\n\n# \u2554\u2550\u2561 5b5e3de9-8413-4ab9-854d-69832e677c1d\nbegin\n\tdata = randn(100)\n\tnobs_ui = @bind nobs Slider(10:10:100)\n\tseries_ui = @bind series Select([\"scatter\", \"line\", \"bar\"])\n\t\n\tmd\"\"\"\n\tThis is a small UI for editing the elements of a plot.\n\n\t- It's neat!\n\n\t$nobs_ui $series_ui\n\t\"\"\"\nend\n\n# \u2554\u2550\u2561 784f192c-0830-44fa-bf09-bcc0752961c0\nplot(data[1:nobs], seriestype=Symbol(series), title=\"Nobs: $nobs\")\n\n# \u2554\u2550\u2561 Cell order:\n# \u255f\u2500361b5f40-6c24-46b2-a389-f819e3481f71\n# \u2560\u2550dd18192a-a85c-11eb-202d-914944b9ef89\n# \u2560\u2550d88098cf-aa9a-424b-8682-77aad6901795\n# \u2560\u255071eac437-61c2-42b4-86a1-4a8b73b67f85\n# \u255f\u2500798d0410-8615-4164-935b-1419e1ee61b9\n# \u255f\u2500c77a4e1e-a8be-41c8-aaff-9b066184c24c\n# \u255f\u250013b03cc2-f2cf-40d6-8b08-bedc7a0acc95\n# \u2560\u2550e762440f-bfc4-411e-a63b-d61fc9ffc4ae\n# \u2560\u255094e52639-6072-4649-a7b9-33efa2791918\n# \u255f\u2500ccac4170-071a-4dd8-b87c-c2cb23f61819\n# \u2560\u255055593255-a4ca-49df-9f6e-12411e9fe9d2\n# \u2560\u255055518562-76b8-4969-bc03-efe3bd138e68\n# \u2560\u25506d676090-70e7-4ef8-b888-ea0b7a3c0491\n# \u255f\u2500b7aa9aa5-49d6-4baa-962b-f65dc748b9f2\n# \u2560\u255039af1a40-21e9-4e02-8193-b33ffd77ec98\n# \u2560\u2550353b54bf-2cd7-4ece-840c-01d9b121dbbd\n# \u255f\u2500f268819d-e75b-4818-8779-b719625b5b4c\n# \u2560\u2550f46bcfa1-445d-4928-abd0-a551cef1e36f\n# \u2560\u25505b5e3de9-8413-4ab9-854d-69832e677c1d\n# \u2560\u2550784f192c-0830-44fa-bf09-bcc0752961c0\n", "meta": {"hexsha": "8d948daa63c3a8d33e725a0e5d377c57134bdb1b", "size": 3154, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "pluto_notebooks/first-steps-5-pluto.jl", "max_stars_repo_name": "joshday/JuliaForDataScience.jl", "max_stars_repo_head_hexsha": "0c330d65ac43733e6820e5c698cb882b28fe4db5", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 9, "max_stars_repo_stars_event_min_datetime": "2018-08-22T15:21:42.000Z", "max_stars_repo_stars_event_max_datetime": "2022-01-26T04:19:53.000Z", "max_issues_repo_path": "pluto_notebooks/first-steps-5-pluto.jl", "max_issues_repo_name": "joshday/JuliaForDataScience.jl", "max_issues_repo_head_hexsha": "0c330d65ac43733e6820e5c698cb882b28fe4db5", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 1, "max_issues_repo_issues_event_min_datetime": "2018-09-29T01:11:07.000Z", "max_issues_repo_issues_event_max_datetime": "2018-10-03T02:00:22.000Z", "max_forks_repo_path": "pluto_notebooks/first-steps-5-pluto.jl", "max_forks_repo_name": "joshday/JuliaForDataScience.jl", "max_forks_repo_head_hexsha": "0c330d65ac43733e6820e5c698cb882b28fe4db5", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 3, "max_forks_repo_forks_event_min_datetime": "2018-11-21T00:52:10.000Z", "max_forks_repo_forks_event_max_datetime": "2020-02-08T11:05:43.000Z", "avg_line_length": 24.640625, "max_line_length": 195, "alphanum_fraction": 0.7067216233, "num_tokens": 1566, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.42632159254749036, "lm_q2_score": 0.1441488511943849, "lm_q1q2_score": 0.061453767805081376}}
{"text": "\n@inline function _to_cartesian(a, i::CanonicalInt)\n @inbounds(CartesianIndices(ntuple(dim -> indices(a, dim), Val(ndims(a))))[i])\nend\n@inline function _to_linear(a, i::Tuple{CanonicalInt,Vararg{CanonicalInt}})\n _strides2int(offsets(a), size_to_strides(size(a), static(1)), i) + static(1)\nend\n\n\"\"\"\n is_splat_index(::Type{T}) -> StaticBool\n \nReturns `static(true)` if `T` is a type that splats across multiple dimensions. \n\"\"\"\nis_splat_index(@nospecialize(x)) = is_splat_index(typeof(x))\nis_splat_index(::Type{T}) where {T} = static(false)\n_is_splat(::Type{I}, i::StaticInt) where {I} = is_splat_index(_get_tuple(I, i))\n\n\"\"\"\n ndims_index(::Type{I}) -> StaticInt\n\nReturns the number of dimension that an instance of `I` maps to when indexing. For example,\n`CartesianIndex{3}` maps to 3 dimensions. If this method is not explicitly defined, then `1`\nis returned.\n\n\"\"\"\nndims_index(@nospecialize(i)) = ndims_index(typeof(i))\nndims_index(::Type{I}) where {I} = static(1)\nndims_index(::Type{<:AbstractCartesianIndex{N}}) where {N} = static(N)\nndims_index(::Type{<:AbstractArray{T}}) where {T} = ndims_index(T)\nndims_index(::Type{<:AbstractArray{Bool,N}}) where {N} = static(N)\nndims_index(::Type{<:LogicalIndex{<:Any,<:AbstractArray{Bool,N}}}) where {N} = static(N)\n_ndims_index(::Type{I}, i::StaticInt) where {I} = ndims_index(_get_tuple(I, i))\n\n\"\"\"\n to_indices(A, I::Tuple) -> Tuple\n\nConverts the tuple of indexing arguments, `I`, into an appropriate form for indexing into `A`.\nTypically, each index should be an `Int`, `StaticInt`, a collection with values of `Int`, or a collection with values of `CartesianIndex`\nThis is accomplished in three steps after the initial call to `to_indices`:\n\n# Extended help\n\nThis implementation differs from that of `Base.to_indices` in the following ways:\n\n* `to_indices(A, I)` never results in recursive processing of `I` through\n `to_indices(A, axes(A), I)`. This is avoided through the use of an internal `@generated`\n method that aligns calls of `to_indices` and `to_index` based on the return values of\n `ndims_index`. This is beneficial because the compiler currently does not optimize away\n the increased time spent recursing through\n each additional argument that needs converting. For example:\n ```julia\n julia> x = rand(4,4,4,4,4,4,4,4,4,4);\n\n julia> inds1 = (1, 2, 1, 2, 1, 2, 1, 2, 1, 2);\n\n julia> inds2 = (1, CartesianIndex(1, 2), 1, CartesianIndex(1, 2), 1, CartesianIndex(1, 2), 1);\n\n julia> inds3 = (fill(true, 4, 4), 2, fill(true, 4, 4), 2, 1, fill(true, 4, 4), 1);\n\n julia> @btime Base.to_indices(\\$x, \\$inds2)\n 1.105 \u03bcs (12 allocations: 672 bytes)\n (1, 1, 2, 1, 1, 2, 1, 1, 2, 1)\n\n julia> @btime ArrayInterface.to_indices(\\$x, \\$inds2)\n 0.041 ns (0 allocations: 0 bytes)\n (1, 1, 2, 1, 1, 2, 1, 1, 2, 1)\n\n julia> @btime Base.to_indices(\\$x, \\$inds3);\n 340.629 ns (14 allocations: 768 bytes)\n\n julia> @btime ArrayInterface.to_indices(\\$x, \\$inds3);\n 11.614 ns (0 allocations: 0 bytes)\n\n ```\n* Recursing through `to_indices(A, axes, I::Tuple{I1,Vararg{Any}})` is intended to provide\n context for processing `I1`. However, this doesn't tell use how many dimensions are\n consumed by what is in `Vararg{Any}`. Using `ndims_index` to directly align the axes of\n `A` with each value in `I` ensures that a `CartesiaIndex{3}` at the tail of `I` isn't\n incorrectly assumed to only consume one dimension.\n* `Base.to_indices` may fail to infer the returned type. This is the case for `inds2` and\n `inds3` in the first bullet on Julia 1.6.4.\n* Specializing by dispatch through method definitions like this:\n `to_indices(::ArrayType, ::Tuple{AxisType,Vararg{Any}}, ::Tuple{::IndexType,Vararg{Any}})`\n require an excessive number of hand written methods to avoid ambiguities. Furthermore, if\n `AxisType` is wrapping another axis that should have unique behavior, then unique parametric \n types need to also be explicitly defined.\n* `to_index(axes(A, dim), index)` is called, as opposed to `Base.to_index(A, index)`. The\n `IndexStyle` of the resulting axis is used to allow indirect dispatch on nested axis types\n within `to_index`.\n\"\"\"\nto_indices(A, ::Tuple{}) = ()\n@inline function to_indices(a::A, inds::I) where {A,I}\n _to_indices(\n a,\n inds,\n IndexStyle(A),\n static(ndims(A)),\n eachop(_ndims_index, nstatic(Val(known_length(I))), I),\n eachop(_is_splat, nstatic(Val(known_length(I))), I)\n )\nend\n@generated function _to_indices(A, inds::I, ::S, ::StaticInt{N}, ::NDI, ::IS) where {I,S,N,NDI,IS}\n cnt = zeros(Int, known_length(NDI))\n splat_position = 0\n remaining = N\n for i in 1:known_length(NDI)\n ndi = known(NDI.parameters[i])\n splat = known(IS.parameters[i])\n if splat && splat_position === 0\n splat_position = i\n else\n remaining -= ndi\n cnt[i] = ndi\n end\n end\n if splat_position !== 0\n cnt[splat_position] = max(0, remaining)\n else\n # if there are additional trailing dimensions not consumed by the index then we have\n # to assume it's linear indexing or that these are trailing dimensions.\n cnt[end] += max(0, remaining)\n end\n\n t = Expr(:tuple)\n dim = 0\n for i in 1:known_length(NDI)\n if i === known_length(NDI) && S <: IndexLinear\n ICall = :LinearIndices\n else\n ICall = :CartesianIndices\n end\n c = cnt[i]\n iexpr = :(@inbounds(getfield(inds, $i))::$(I.parameters[i]))\n if dim === N\n push!(t.args, :(to_index($(ICall)(()), $iexpr)))\n elseif c === 1\n dim += 1\n push!(t.args, :(to_index(@inbounds(getfield(axs, $dim)), $iexpr)))\n else\n subaxs = Expr(:tuple)\n for _ in 1:c\n if dim < N\n dim += 1\n push!(subaxs.args, :(@inbounds(getfield(axs, $dim))))\n end\n end\n push!(t.args, :(to_index($(ICall)($subaxs), $iexpr)))\n end\n end\n Expr(:block,\n Expr(:meta, :inline),\n Expr(:(=), :axs, :(lazy_axes(A))),\n :(_flatten_tuples($t))\n )\nend\n@generated function _flatten_tuples(inds::I) where {I}\n t = Expr(:tuple)\n for i in 1:known_length(I)\n p = I.parameters[i]\n if p <: Tuple\n for j in 1:known_length(p)\n push!(t.args, :(@inbounds(getfield(getfield(inds, $i), $j))))\n end\n else\n push!(t.args, :(@inbounds(getfield(inds, $i))))\n end\n end\n t\nend\n\n\"\"\"\n to_index([::IndexStyle, ]axis, arg) -> index\n\nConvert the argument `arg` that was originally passed to `getindex` for the dimension\ncorresponding to `axis` into a form for native indexing (`Int`, Vector{Int}, etc.). New\naxis types with unique behavior should use an `IndexStyle` trait:\n\n```julia\nto_index(axis::MyAxisType, arg) = to_index(IndexStyle(axis), axis, arg)\nto_index(::MyIndexStyle, axis, arg) = ...\n```\n\"\"\"\nto_index(x, i::Slice) = i\nto_index(x, ::Colon) = indices(x)\n# logical indexing\nto_index(x, i::AbstractArray{Bool}) = LogicalIndex(i)\nto_index(x::LinearIndices, i::AbstractArray{Bool}) = LogicalIndex{Int}(i)\n# cartesian indexing\n@inline to_index(x, i::CartesianIndices{0}) = i\n@inline to_index(x, i::CartesianIndices) = axes(i)\n@inline to_index(x, i::CartesianIndex) = Tuple(i)\n@inline to_index(x, i::NDIndex) = Tuple(i)\n@inline to_index(x, i::AbstractArray{<:AbstractCartesianIndex}) = i\n# integer indexing\nto_index(x, i::AbstractArray{<:Integer}) = i\nto_index(x, @nospecialize(i::StaticInt)) = i\nto_index(x, i::Integer) = Int(i)\n@inline to_index(x, i) = to_index(IndexStyle(x), x, i)\nfunction to_index(S::IndexStyle, x, i)\n throw(ArgumentError(\n \"invalid index: $S does not support indices of type $(typeof(i)) for instances of type $(typeof(x)).\"\n ))\nend\n\n\"\"\"\n unsafe_reconstruct(A, data; kwargs...)\n\nReconstruct `A` given the values in `data`. New methods using `unsafe_reconstruct`\nshould only dispatch on `A`.\n\"\"\"\nfunction unsafe_reconstruct(axis::OneTo, data; kwargs...)\n if axis === data\n return axis\n else\n return OneTo(data)\n end\nend\nfunction unsafe_reconstruct(axis::UnitRange, data; kwargs...)\n if axis === data\n return axis\n else\n return UnitRange(first(data), last(data))\n end\nend\nfunction unsafe_reconstruct(axis::OptionallyStaticUnitRange, data; kwargs...)\n if axis === data\n return axis\n else\n return OptionallyStaticUnitRange(static_first(data), static_last(data))\n end\nend\nfunction unsafe_reconstruct(A::AbstractUnitRange, data; kwargs...)\n return static_first(data):static_last(data)\nend\n\n\"\"\"\n to_axes(A, inds) -> Tuple\n\nConstruct new axes given the corresponding `inds` constructed after\n`to_indices(A, args) -> inds`. This method iterates through each pair of axes and\nindices calling [`to_axis`](@ref).\n\"\"\"\n@inline function to_axes(A, inds::Tuple)\n if ndims(A) === 1\n return (to_axis(axes(A, 1), first(inds)),)\n elseif length(inds) === 1\n return (to_axis(eachindex(IndexLinear(), A), first(inds)),)\n else\n return to_axes(A, axes(A), inds)\n end\nend\n# drop this dimension\nto_axes(A, a::Tuple, i::Tuple{<:Integer,Vararg{Any}}) = to_axes(A, tail(a), tail(i))\nto_axes(A, a::Tuple, i::Tuple{I,Vararg{Any}}) where {I} = _to_axes(ndims_index(I), A, a, i)\nfunction _to_axes(::StaticInt{1}, A, axs::Tuple, inds::Tuple)\n return (to_axis(first(axs), first(inds)), to_axes(A, tail(axs), tail(inds))...)\nend\n@propagate_inbounds function _to_axes(::StaticInt{N}, A, axs::Tuple, inds::Tuple) where {N}\n axes_front, axes_tail = Base.IteratorsMD.split(axs, Val(N))\n if IndexStyle(A) === IndexLinear()\n axis = to_axis(LinearIndices(axes_front), getfield(inds, 1))\n else\n axis = to_axis(CartesianIndices(axes_front), getfield(inds, 1))\n end\n return (axis, to_axes(A, axes_tail, tail(inds))...)\nend\nto_axes(A, ::Tuple{Ax,Vararg{Any}}, ::Tuple{}) where {Ax} = ()\nto_axes(A, ::Tuple{}, ::Tuple{}) = ()\n\n\"\"\"\n to_axis(old_axis, index) -> new_axis\n\nConstruct an `new_axis` for a newly constructed array that corresponds to the\npreviously executed `to_index(old_axis, arg) -> index`. `to_axis` assumes that\n`index` has already been confirmed to be in bounds. The underlying indices of\n`new_axis` begins at one and extends the length of `index` (i.e., one-based indexing).\n\"\"\"\n@inline function to_axis(axis, inds)\n if !can_change_size(axis) &&\n (known_length(inds) !== nothing && known_length(axis) === known_length(inds))\n return axis\n else\n return to_axis(IndexStyle(axis), axis, inds)\n end\nend\n\n# don't need to check size b/c slice means it's the entire axis\n@inline function to_axis(axis, inds::Slice)\n if can_change_size(axis)\n return copy(axis)\n else\n return axis\n end\nend\nto_axis(S::IndexLinear, axis, inds) = StaticInt(1):static_length(inds)\n\n\"\"\"\n ArrayInterface.getindex(A, args...)\n\nRetrieve the value(s) stored at the given key or index within a collection. Creating\nanother instance of `ArrayInterface.getindex` should only be done by overloading `A`.\nChanging indexing based on a given argument from `args` should be done through,\n[`to_index`](@ref), or [`to_axis`](@ref).\n\"\"\"\nfunction getindex(A, args...)\n inds = to_indices(A, args)\n @boundscheck checkbounds(A, inds...)\n unsafe_getindex(A, inds...)\nend\nfunction getindex(A; kwargs...)\n inds = to_indices(A, order_named_inds(dimnames(A), values(kwargs)))\n @boundscheck checkbounds(A, inds...)\n unsafe_getindex(A, inds...)\nend\n@propagate_inbounds getindex(x::Tuple, i::Int) = getfield(x, i)\n@propagate_inbounds getindex(x::Tuple, ::StaticInt{i}) where {i} = getfield(x, i)\n\n## unsafe_getindex ##\nfunction unsafe_getindex(a::A) where {A}\n parent_type(A) <: A && throw(MethodError(unsafe_getindex, (A,)))\n unsafe_getindex(parent(a))\nend\n\n# TODO Need to manage index transformations between nested layers of arrays\nfunction unsafe_getindex(a::A, i::CanonicalInt) where {A}\n if IndexStyle(A) === IndexLinear()\n parent_type(A) <: A && throw(MethodError(unsafe_getindex, (A, i)))\n return unsafe_getindex(parent(a), i)\n else\n return unsafe_getindex(a, _to_cartesian(a, i)...)\n end\nend\nfunction unsafe_getindex(a::A, i::CanonicalInt, ii::Vararg{CanonicalInt}) where {A}\n if IndexStyle(A) === IndexLinear()\n return unsafe_getindex(a, _to_linear(a, (i, ii...)))\n else\n parent_type(A) <: A && throw(MethodError(unsafe_getindex, (A, i)))\n return unsafe_getindex(parent(a), i, ii...)\n end\nend\n\nunsafe_getindex(a, i::Vararg{Any}) = unsafe_get_collection(a, i)\n\nunsafe_getindex(A::Array) = Base.arrayref(false, A, 1)\nunsafe_getindex(A::Array, i::CanonicalInt) = Base.arrayref(false, A, Int(i))\n@inline function unsafe_getindex(A::Array, i::CanonicalInt, ii::Vararg{CanonicalInt})\n unsafe_getindex(A, _to_linear(A, (i, ii...)))\nend\n\nunsafe_getindex(A::LinearIndices, i::CanonicalInt) = Int(i)\nunsafe_getindex(A::CartesianIndices, i::CanonicalInt, ii::Vararg{CanonicalInt}) = CartesianIndex(i, ii...)\nunsafe_getindex(A::CartesianIndices, i::CanonicalInt) = @inbounds(A[i])\n\nunsafe_getindex(A::ReshapedArray, i::CanonicalInt) = @inbounds(parent(A)[i])\nfunction unsafe_getindex(A::ReshapedArray, i::CanonicalInt, ii::Vararg{CanonicalInt})\n @inbounds(parent(A)[_to_linear(A, (i, ii...))])\nend\n\nunsafe_getindex(A::SubArray, i::CanonicalInt) = @inbounds(A[i])\nunsafe_getindex(A::SubArray, i::CanonicalInt, ii::Vararg{CanonicalInt}) = @inbounds(A[i, ii...])\n\n# This is based on Base._unsafe_getindex from https://github.com/JuliaLang/julia/blob/c5ede45829bf8eb09f2145bfd6f089459d77b2b1/base/multidimensional.jl#L755.\n#=\n unsafe_get_collection(A, inds)\n\nReturns a collection of `A` given `inds`. `inds` is assumed to have been bounds-checked.\n=#\nfunction unsafe_get_collection(A, inds)\n axs = to_axes(A, inds)\n dest = similar(A, axs)\n if map(Base.unsafe_length, axes(dest)) == map(Base.unsafe_length, axs)\n Base._unsafe_getindex!(dest, A, inds...)\n else\n Base.throw_checksize_error(dest, axs)\n end\n return dest\nend\n_ints2range(x::Integer) = x:x\n_ints2range(x::AbstractRange) = x\n@inline function unsafe_get_collection(A::CartesianIndices{N}, inds) where {N}\n if (length(inds) === 1 && N > 1) || stride_preserving_index(typeof(inds)) === False()\n return Base._getindex(IndexStyle(A), A, inds...)\n else\n return CartesianIndices(to_axes(A, _ints2range.(inds)))\n end\nend\n@inline function unsafe_get_collection(A::LinearIndices{N}, inds) where {N}\n if length(inds) === 1 && isone(_ndims_index(typeof(inds), static(1)))\n return @inbounds(eachindex(A)[first(inds)])\n elseif stride_preserving_index(typeof(inds)) === True()\n return LinearIndices(to_axes(A, _ints2range.(inds)))\n else\n return Base._getindex(IndexStyle(A), A, inds...)\n end\nend\n\n\"\"\"\n ArrayInterface.setindex!(A, args...)\n\nStore the given values at the given key or index within a collection.\n\"\"\"\n@propagate_inbounds function setindex!(A, val, args...)\n can_setindex(A) || error(\"Instance of type $(typeof(A)) are not mutable and cannot change elements after construction.\")\n inds = to_indices(A, args)\n @boundscheck checkbounds(A, inds...)\n unsafe_setindex!(A, val, inds...)\nend\n@propagate_inbounds function setindex!(A, val; kwargs...)\n can_setindex(A) || error(\"Instance of type $(typeof(A)) are not mutable and cannot change elements after construction.\")\n inds = to_indices(A, order_named_inds(dimnames(A), values(kwargs)))\n @boundscheck checkbounds(A, inds...)\n unsafe_setindex!(A, val, inds...)\nend\n\n## unsafe_setindex! ##\nfunction unsafe_setindex!(a::A, v) where {A}\n parent_type(A) <: A && throw(MethodError(unsafe_setindex!, (A, v)))\n return unsafe_setindex!(parent(a), v)\nend\n# TODO Need to manage index transformations between nested layers of arrays\nfunction unsafe_setindex!(a::A, v, i::CanonicalInt) where {A}\n if IndexStyle(A) === IndexLinear()\n parent_type(A) <: A && throw(MethodError(unsafe_setindex!, (A, v, i)))\n return unsafe_setindex!(parent(a), v, i)\n else\n return unsafe_setindex!(a, v, _to_cartesian(a, i)...)\n end\nend\nfunction unsafe_setindex!(a::A, v, i::CanonicalInt, ii::Vararg{CanonicalInt}) where {A}\n if IndexStyle(A) === IndexLinear()\n return unsafe_setindex!(a, v, _to_linear(a, (i, ii...)))\n else\n parent_type(A) <: A && throw(MethodError(unsafe_setindex!, (A, v, i, ii...)))\n return unsafe_setindex!(parent(a), v, i, ii...)\n end\nend\n\n\nfunction unsafe_setindex!(A::Array{T}, v) where {T}\n Base.arrayset(false, A, convert(T, v)::T, 1)\nend\nfunction unsafe_setindex!(A::Array{T}, v, i::CanonicalInt) where {T}\n return Base.arrayset(false, A, convert(T, v)::T, Int(i))\nend\n\nunsafe_setindex!(a, v, i::Vararg{Any}) = unsafe_set_collection!(a, v, i)\n\n# This is based on Base._unsafe_setindex!.\n#=\n unsafe_set_collection!(A, val, inds)\n\nSets `inds` of `A` to `val`. `inds` is assumed to have been bounds-checked.\n=#\nunsafe_set_collection!(A, v, i) = Base._unsafe_setindex!(IndexStyle(A), A, v, i...)\n", "meta": {"hexsha": "3e29b2c98bcb78c8df34caf532905d9a2115a7aa", "size": 17224, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/indexing.jl", "max_stars_repo_name": "N5N3/ArrayInterface.jl", "max_stars_repo_head_hexsha": "fedb318e9d55fba85f0e9c08b0180a44ae4540a1", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 55, "max_stars_repo_stars_event_min_datetime": "2021-03-09T23:44:15.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-19T04:54:06.000Z", "max_issues_repo_path": "src/indexing.jl", "max_issues_repo_name": "N5N3/ArrayInterface.jl", "max_issues_repo_head_hexsha": "fedb318e9d55fba85f0e9c08b0180a44ae4540a1", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 129, "max_issues_repo_issues_event_min_datetime": "2021-03-02T07:24:09.000Z", "max_issues_repo_issues_event_max_datetime": "2022-03-31T12:42:14.000Z", "max_forks_repo_path": "src/indexing.jl", "max_forks_repo_name": "N5N3/ArrayInterface.jl", "max_forks_repo_head_hexsha": "fedb318e9d55fba85f0e9c08b0180a44ae4540a1", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 8, "max_forks_repo_forks_event_min_datetime": "2021-05-17T04:20:13.000Z", "max_forks_repo_forks_event_max_datetime": "2022-02-23T10:32:42.000Z", "avg_line_length": 37.8549450549, "max_line_length": 157, "alphanum_fraction": 0.6655829076, "num_tokens": 4948, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.5, "lm_q2_score": 0.12252322052837361, "lm_q1q2_score": 0.061261610264186805}}
{"text": "\nimport Base: ==\n==(x,y,z,ws...) = x==y && ==(y,z,ws...)\n\n\n\"\"\" \nReturn the type's fields as a tuple\n\"\"\"\n@generated fieldvalues(x) = Expr(:tuple, (:(x.$f) for f=fieldnames(x))...)\n@generated fields(x) = Expr(:tuple, (:($f=x.$f) for f=fieldnames(x))...)\nfirstfield(x) = first(fieldvalues(x))\n\n\n\n\"\"\"\nRewrites `@! x = f(args...)` to `x = f!(x,args...)`\n\nSpecial cases for `*` and `\\\\` forward to `mul!` and `ldiv!`, respectively.\n\"\"\"\nmacro !(ex)\n if @capture(ex, x_ = f_(args__; kwargs_...))\n esc(:($(Symbol(string(f,\"!\")))($x,$(args...); $kwargs...)))\n elseif @capture(ex, x_ = f_(args__))\n if f == :*\n f = :mul\n elseif f==:\\\n f = :ldiv\n end\n esc(:($x = $(Symbol(string(f,\"!\")))($x,$(args...))::typeof($x))) # ::typeof part helps inference sometimes\n else\n error(\"Usage: @! x = f(...)\")\n end\nend\n\n\nnan2zero(x::T) where {T} = !isfinite(x) ? zero(T) : x\nnan2zero(x::Diagonal{T}) where {T} = Diagonal{T}(nan2zero.(x.diag))\nnan2inf(x::T) where {T} = !isfinite(x) ? T(Inf) : x\n\n\n\"\"\" Return a tuple with the expression repeated n times \"\"\"\nmacro repeated(ex,n)\n :(tuple($(repeated(esc(ex),n)...)))\nend\n\n\"\"\" \nPack some variables in a dictionary \n\n```julia\n> x = 3\n> y = 4\n> @dictpack x y z=>5\nDict(:x=>3,:y=>4,:z=>5)\n```\n\"\"\"\nmacro dictpack(exs...)\n kv(ex::Symbol) = :($(QuoteNode(ex))=>$(esc(ex)))\n kv(ex) = isexpr(ex,:call) && ex.args[1]==:(=>) ? :($(QuoteNode(ex.args[2]))=>$(esc(ex.args[3]))) : error()\n :(Dict($((kv(ex) for ex=exs)...)))\nend\n\n\"\"\" \nPack some variables into a NamedTuple. E.g.:\n\n```julia\n> x = 3\n> y = 4\n> @namedtuple(x, y, z=5)\n(x=3,y=4,z=5)\n```\n\"\"\"\nmacro namedtuple(exs...)\n if length(exs)==1 && isexpr(exs[1],:tuple)\n exs = exs[1].args\n end\n kv(ex::Symbol) = :($(esc(ex))=$(esc(ex)))\n kv(ex) = isexpr(ex,:(=)) ? :($(esc(ex.args[1]))=$(esc(ex.args[2]))) : error()\n Expr(:tuple, (kv(ex) for ex=exs)...)\nend\n\n\n\n# these allow pinv and sqrt of SMatrices of Diagonals to work correctly, which\n# we use for the T-E block of the covariance. hopefully some of this can be cut\n# down on in the futue with some PRs into StaticArrays.\npermutedims(A::SMatrix{2,2}) = @SMatrix[A[1] A[3]; A[2] A[4]]\nfunction sqrt(A::SMatrix{2,2,<:Diagonal})\n a,b,c,d = A\n s = @. sqrt(a*d-b*c)\n t = pinv(@. sqrt(a+d+2s))\n @SMatrix[t*(a+s) t*b; t*c t*(d+s)]\nend\nfunction pinv(A::SMatrix{2,2,<:Diagonal})\n a,b,c,d = A\n idet = pinv(@. a*d-b*c)\n @SMatrix[d*idet -(b*idet); -(c*idet) a*idet]\nend\n\n\n# some usefule tuple manipulation functions:\n\n# see: https://discourse.julialang.org/t/efficient-tuple-concatenation/5398/10\n# and https://github.com/JuliaLang/julia/issues/27988\n@inline tuplejoin(x) = x\n@inline tuplejoin(x, y) = (x..., y...)\n@inline tuplejoin(x, y, z...) = (x..., tuplejoin(y, z...)...)\n\n# see https://discourse.julialang.org/t/any-way-to-make-this-one-liner-type-stable/10636/2\nusing Base: tuple_type_cons, tuple_type_head, tuple_type_tail, first, tail\nmap_tupleargs(f,::Type{T}) where {T<:Tuple} = \n (f(tuple_type_head(T)), map_tupleargs(f,tuple_type_tail(T))...)\nmap_tupleargs(f,::Type{T},::Type{S}) where {T<:Tuple,S<:Tuple} = \n (f(tuple_type_head(T),tuple_type_head(S)), map_tupleargs(f,tuple_type_tail(T),tuple_type_tail(S))...)\nmap_tupleargs(f,::Type{T},s::Tuple) where {T<:Tuple} = \n (f(tuple_type_head(T),first(s)), map_tupleargs(f,tuple_type_tail(T),tail(s))...)\nmap_tupleargs(f,::Type{<:Tuple{}}...) = ()\nmap_tupleargs(f,::Type{<:Tuple{}},::Tuple) = ()\n\n\n# returns the base parametric type with all type parameters stripped out\nbasetype(::Type{T}) where {T} = T.name.wrapper\n@generated function basetype(t::UnionAll)\n unwrap_expr(s::UnionAll, t=:t) = unwrap_expr(s.body, :($t.body))\n unwrap_expr(::DataType, t) = t\n :($(unwrap_expr(t.parameters[1])).name.wrapper)\nend\n\n\nfunction ensuresame(args...)\n @assert all(args .== Ref(args[1]))\n args[1]\nend\n\n\ntuple_type_len(::Type{<:NTuple{N,Any}}) where {N} = N\n\n\nensure1d(x::Union{Tuple,AbstractArray}) = x\nensure1d(x) = (x,)\n\n\n# see https://discourse.julialang.org/t/dispatching-on-the-result-of-unwrap-unionall-seems-weird/25677\n# for why we need this\n# to use, just decorate the custom show_datatype with it, and make sure the args\n# are named `io` and `t`.\nmacro show_datatype(ex)\n def = splitdef(ex)\n def[:body] = quote\n isconcretetype(t) ? $(def[:body]) : invoke(Base.show_datatype, Tuple{IO,DataType}, io, t)\n end\n esc(combinedef(def))\nend\n\n\n\n\"\"\"\n # symmetric in any of its final arguments except for bar:\n @sym_memo foo(bar, @sym(args...)) = \n # symmetric in (i,j), but not baz\n @sym_memo foo(baz, @sym(i, j)) = \n \nThe `@sym_memo` macro should be applied to a definition of a function\nwhich is symmetric in some of its arguments. The arguments in which its\nsymmetric are specified by being wrapping them in @sym, and they must come at\nthe very end. The resulting function will be memoized and permutations of the\narguments which are equal due to symmetry will only be computed once.\n\"\"\"\nmacro sym_memo(funcdef)\n \n \n sfuncdef = splitdef(funcdef)\n \n asymargs = sfuncdef[:args][1:end-1]\n symargs = collect(@match sfuncdef[:args][end] begin\n Expr(:macrocall, [head, _, ex...]), if head==Symbol(\"@sym\") end => ex\n _ => error(\"final argument(s) should be marked @sym\")\n end)\n sfuncdef[:args] = [asymargs..., symargs...]\n \n sfuncdef[:body] = quote\n symargs = [$(symargs...)]\n sorted_symargs = sort(symargs)\n if symargs==sorted_symargs\n $((sfuncdef[:body]))\n else\n $(sfuncdef[:name])($(asymargs...), sorted_symargs...)\n end\n end\n \n esc(:(@memoize $(combinedef(sfuncdef))))\n \nend\n\n\n@doc doc\"\"\"\n```\n@subst sum(x*$(y+1) for x=1:2)\n```\n \nbecomes\n\n```\nlet tmp=(y+1)\n sum(x*tmp for x=1:2)\nend\n```\n\nto aid in writing clear/succinct code that doesn't recompute things\nunnecessarily.\n\"\"\"\nmacro subst(ex)\n \n subs = []\n ex = postwalk(ex) do x\n if isexpr(x, Symbol(raw\"$\"))\n var = gensym()\n push!(subs, :($(esc(var))=$(esc(x.args[1]))))\n var\n else\n x\n end\n end\n \n quote\n let $(subs...)\n $(esc(ex))\n end\n end\n\nend\n\n\n\"\"\"\n @invokelatest expr...\n \nRewrites all non-broadcasted function calls anywhere within an expression to use\n`Base.invokelatest`. This means functions can be called that have a newer world\nage, at the price of making things non-inferrable.\n\"\"\"\nmacro invokelatest(ex)\n function walk(x)\n if isdef(x)\n x.args[2:end] .= map(walk, x.args[2:end])\n x\n elseif @capture(x, f_(args__; kwargs__)) && !startswith(string(f),'.')\n :(Base.invokelatest($f, $(map(walk,args)...); $(map(walk,kwargs)...)))\n elseif @capture(x, f_(args__)) && !startswith(string(f),'.')\n :(Base.invokelatest($f, $(map(walk,args)...)))\n elseif isexpr(x)\n x.args .= map(walk, x.args)\n x\n else\n x\n end\n end\n esc(walk(ex))\nend\n\n\n\"\"\"\n @ondemand(Package.function)(args...; kwargs...)\n @ondemand(Package.Submodule.function)(args...; kwargs...)\n\nJust like calling `Package.function` or `Package.Submodule.function`, but\n`Package` will be loaded on-demand if it is not already loaded. The call is no\nlonger inferrable.\n\"\"\"\nmacro ondemand(ex)\n get_root_package(x) = @capture(x, a_.b_) ? get_root_package(a) : x\n quote\n @eval import $(get_root_package(ex))\n (args...; kwargs...) -> Base.invokelatest($(esc(ex)), args...; kwargs...)\n end\nend\n\nfunction tmap(f, args...)\n @static if nthreads()==1 || VERSIONThreads.@spawn(f(args...)),args...))\n end\nend\n\n\nget_kwarg_names(func::Function) = Vector{Symbol}(kwarg_decl(first(methods(func)), typeof(methods(func).mt.kwsorter)))\nkwarg_decl(m::Method,kw::DataType) = VERSION<=v\"1.3.999\" ? Base.kwarg_decl(m,kw) : Base.kwarg_decl(m)\n\n# maps a function recursively across all arguments of a Broadcasted expression,\n# using the function `broadcasted` to reconstruct the `Broadcasted` object at\n# each point.\nmap_bc_args(f, bc::Broadcasted) = broadcasted(bc.f, map(arg->map_bc_args(f, arg), bc.args)...)\nmap_bc_args(f, arg) = f(arg)\n\n\n# adapting a closure adapts the captured variables\n# this could probably be a PR into Adapt.jl\n@generated function adapt_structure(to, f::F) where {F<:Function}\n if fieldcount(F) == 0\n :f\n else\n quote\n captured_vars = $(Expr(:tuple, (:(adapt(to, f.$x)) for x=fieldnames(F))...))\n $(Expr(:new, :($(F.name.wrapper){map(typeof,captured_vars)...}), (:(captured_vars[$i]) for i=1:fieldcount(F))...))\n end\n end\nend\n\nadapt_structure(to, d::Dict) = Dict(k => adapt(to, v) for (k,v) in d)\n", "meta": {"hexsha": "6686a32728ee89a3fe09be8a04006b3f724c5513", "size": 8880, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/util.jl", "max_stars_repo_name": "JuliaTagBot/CMBLensing.jl", "max_stars_repo_head_hexsha": "59913ff7a889587e706f3cf634c1e6b379bc1574", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/util.jl", "max_issues_repo_name": "JuliaTagBot/CMBLensing.jl", "max_issues_repo_head_hexsha": "59913ff7a889587e706f3cf634c1e6b379bc1574", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/util.jl", "max_forks_repo_name": "JuliaTagBot/CMBLensing.jl", "max_forks_repo_head_hexsha": "59913ff7a889587e706f3cf634c1e6b379bc1574", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 28.7378640777, "max_line_length": 126, "alphanum_fraction": 0.6025900901, "num_tokens": 2744, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.5, "lm_q2_score": 0.12252320290590249, "lm_q1q2_score": 0.061261601452951245}}
{"text": "import Pkg; Pkg.add(Pkg.PackageSpec(url=\"https://github.com/JuliaComputing/JuliaAcademyData.jl\"))\nusing JuliaAcademyData; activate(\"Parallel_Computing\")\n\n# # GPUs\n#\n# The graphics processor in your computer is _itself_ like a mini-computer highly\n# tailored for massively and embarassingly parallel operations (like computing how light will bounce\n# off of every point on a 3D mesh of triangles).\n#\n# Of course, recently their utility in other applications has become more clear\n# and thus the GPGPU was born.\n#\n# Just like how we needed to send data to other processes, we need to send our\n# data to the GPU to do computations there.\n\n#-\n\n# ## How is a GPU different from a CPU?\n#\n# This is what a typical consumer CPU looks like:\n#\n# 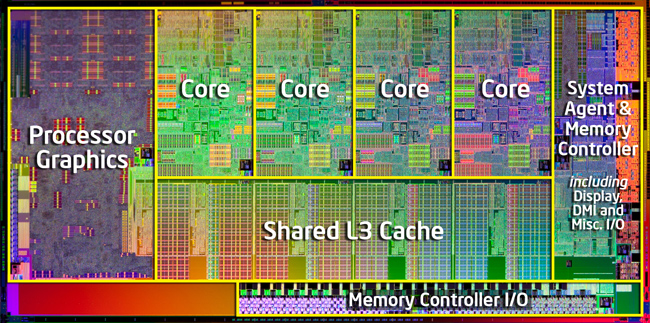\n#\n# And this is what a GPU looks like:\n#\n# 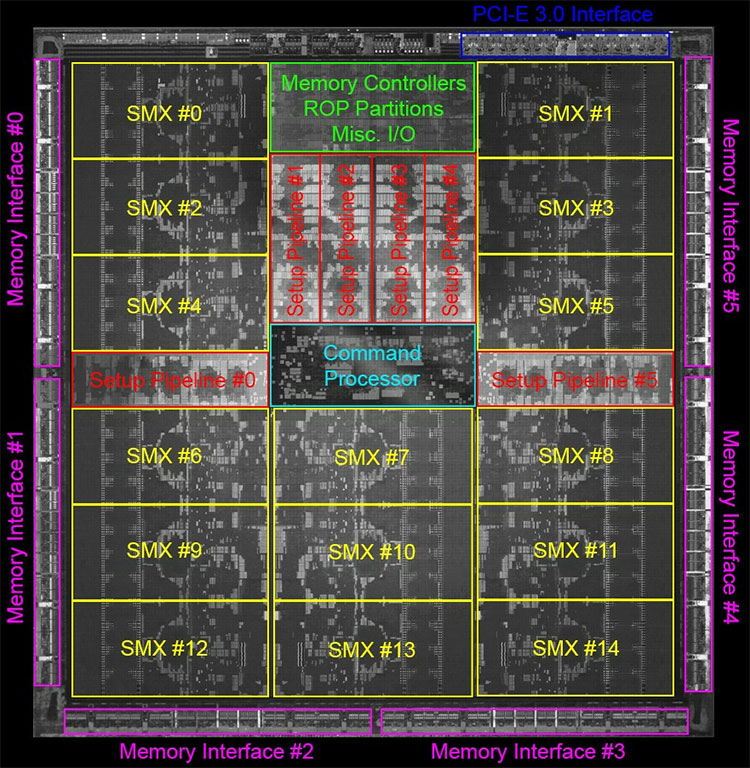\n#\n# Each SMX isn't just one \"core\", each is a _streaming multiprocessor_ capable of running hundreds of threads simultaneously itself. There are so many threads, in fact, that you reason about them in groups of 32 \u2014\u00a0called a \"warp.\" No, no [that warp](https://www.google.com/search?tbm=isch&q=warp&tbs=imgo:1), [this one](https://www.google.com/search?tbm=isch&q=warp%20weaving&tbs=imgo:1).\n#\n# The card above supports up to 6 warps per multiprocessor, with 32 threads each, times 15 multiprocessors... 2880 threads at a time!\n#\n# Also note the memory interfaces.\n#\n# --------------\n#\n# Each thread is relatively limited \u2014\u00a0and a warp is almost like a SIMD unit that supports branching. Except it's still only executing one instruction even after a branch:\n#\n# 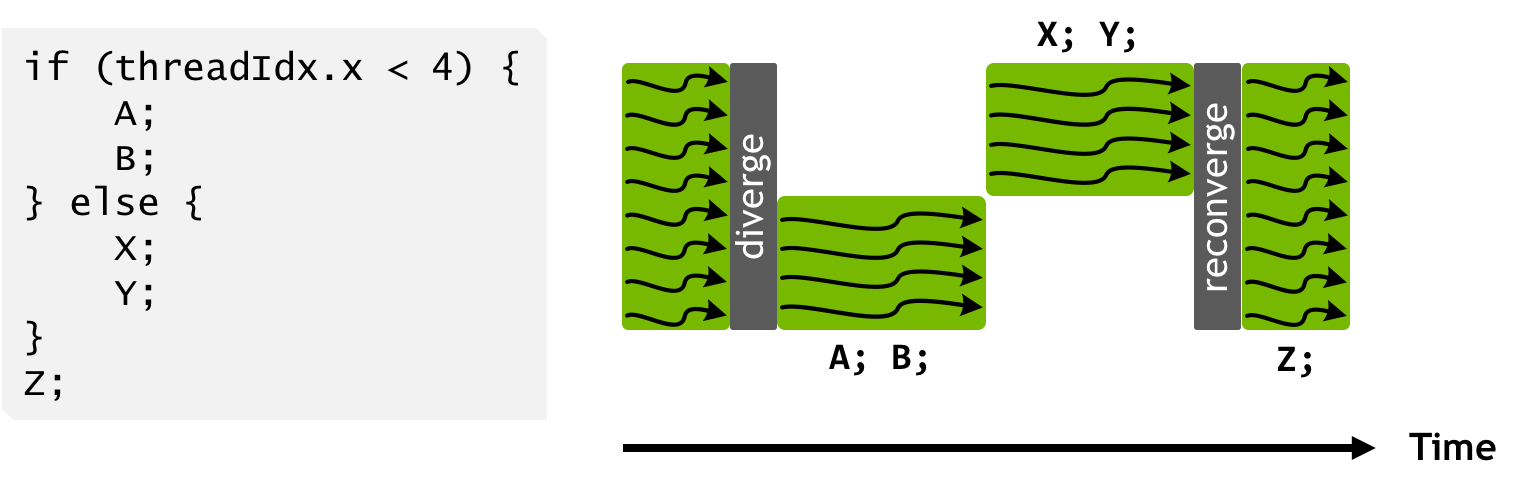\n\n#-\n\n# You can inspect the installed GPUs with nvidia-smi:\n\n#nb ;nvidia-smi\n#jl run(`nvidia-smi`)\n\n# ## Example\n#\n# The deep learning MNIST example: https://fluxml.ai/experiments/mnist/\n#\n# This is how it looks on the CPU:\n\nusing Flux, Flux.Data.MNIST, Statistics\nusing Flux: onehotbatch, onecold, crossentropy, throttle\nusing Base.Iterators: repeated, partition\n\nimgs = MNIST.images()\nlabels = onehotbatch(MNIST.labels(), 0:9)\n\n## Partition into batches of size 32\ntrain = [(cat(float.(imgs[i])..., dims = 4), labels[:,i])\n for i in partition(1:60_000, 32)]\n## Prepare test set (first 1,000 images)\ntX = cat(float.(MNIST.images(:test)[1:1000])..., dims = 4)\ntY = onehotbatch(MNIST.labels(:test)[1:1000], 0:9)\n\nm = Chain(\n Conv((3, 3), 1=>32, relu),\n Conv((3, 3), 32=>32, relu),\n x -> maxpool(x, (2,2)),\n Conv((3, 3), 32=>16, relu),\n x -> maxpool(x, (2,2)),\n Conv((3, 3), 16=>10, relu),\n x -> reshape(x, :, size(x, 4)),\n Dense(90, 10), softmax)\n\nloss(x, y) = crossentropy(m(x), y)\naccuracy(x, y) = mean(onecold(m(x)) .== onecold(y))\n## opt = ADAM() # <-- Move Flux.params(m) here!\n## @time Flux.train!(loss, Flux.params(m), train[1:10], opt, cb = () -> @show(accuracy(tX, tY)))\nopt = ADAM(Flux.params(m), ) # <-- Move Flux.params(m) here!\nFlux.train!(loss, train[1:1], opt, cb = () -> @show(accuracy(tX, tY)))\n@time Flux.train!(loss, train[1:10], opt, cb = () -> @show(accuracy(tX, tY)))\n\n# Now let's re-do it on a GPU. \"All\" it takes is moving the data there with `gpu`!\n\ninclude(datapath(\"scripts/fixupCUDNN.jl\")) # JuliaBox uses an old version of CuArrays; this backports a fix for it\ngputrain = gpu.(train[1:10])\ngpum = gpu(m)\ngputX = gpu(tX)\ngputY = gpu(tY)\ngpuloss(x, y) = crossentropy(gpum(x), y)\ngpuaccuracy(x, y) = mean(onecold(gpum(x)) .== onecold(y))\ngpuopt = ADAM(Flux.params(gpum), )\nFlux.train!(gpuloss, gpu.(train[1:1]), gpuopt, cb = () -> @show(gpuaccuracy(gputX, gputY)))\n@time Flux.train!(gpuloss, gputrain, gpuopt, cb = () -> @show(gpuaccuracy(gputX, gputY)))\n\n# ## Defining your own GPU kernels\n#\n# So that's leveraging Flux's ability to work with GPU arrays \u2014 which is magical\n# and awesome \u2014\u00a0but you don't always have a library to lean on like that.\n# How might you define your own GPU kernel?\n#\n# Recall the monte carlo pi example:\n\nfunction serialpi(n)\n inside = 0\n for i in 1:n\n x, y = rand(), rand()\n inside += (x^2 + y^2 <= 1)\n end\n return 4 * inside / n\nend\n\n# How could we express this on the GPU?\n\nusing CuArrays.CURAND\nfunction findpi_gpu(n)\n 4 * sum(curand(Float64, n).^2 .+ curand(Float64, n).^2 .<= 1) / n\nend\nfindpi_gpu(10_000_000)\n\n#-\n\nusing BenchmarkTools\n@btime findpi_gpu(10_000_000)\n@btime serialpi(10_000_000)\n\n# That leans on broadcast to build the GPU kernel \u2014 and is creating three arrays\n# in the process \u2014 but it's still much faster than our serial pi from before.\n\n#-\n\n# In general, using CuArrays and broadcast is one of the best ways to just\n# get everything to work. If you really want to get your hands dirty, you\n# can use [CUDAnative.jl](https://github.com/JuliaGPU/CUDAnative.jl) to manually specify exactly how everything works,\n# but be forewarned, it's not for the [faint at heart](https://github.com/JuliaGPU/CUDAnative.jl/blob/master/examples/reduce/reduce.jl)! (If you've done CUDA\n# programming in C or C++, it's very similar.)\n\n", "meta": {"hexsha": "5ae17cf07eefd7dda6aa6e793aca1034f8612f84", "size": 5302, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Courses/Parallel_Computing/080 GPUs.jl", "max_stars_repo_name": "fercarozzi/JuliaAcademyMaterials", "max_stars_repo_head_hexsha": "4c7501d42e698379050fd6e6d469f3f84428cdcd", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 45, "max_stars_repo_stars_event_min_datetime": "2020-02-13T00:50:27.000Z", "max_stars_repo_stars_event_max_datetime": "2021-12-05T07:57:22.000Z", "max_issues_repo_path": "Courses/Parallel_Computing/080 GPUs.jl", "max_issues_repo_name": "fercarozzi/JuliaAcademyMaterials", "max_issues_repo_head_hexsha": "4c7501d42e698379050fd6e6d469f3f84428cdcd", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 52, "max_issues_repo_issues_event_min_datetime": "2019-10-30T16:22:28.000Z", "max_issues_repo_issues_event_max_datetime": "2020-01-26T20:02:43.000Z", "max_forks_repo_path": "Courses/Parallel_Computing/080 GPUs.jl", "max_forks_repo_name": "fercarozzi/JuliaAcademyMaterials", "max_forks_repo_head_hexsha": "4c7501d42e698379050fd6e6d469f3f84428cdcd", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 27, "max_forks_repo_forks_event_min_datetime": "2020-02-26T11:33:28.000Z", "max_forks_repo_forks_event_max_datetime": "2022-02-25T22:34:53.000Z", "avg_line_length": 37.8714285714, "max_line_length": 390, "alphanum_fraction": 0.6938890985, "num_tokens": 1602, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.49218813572079556, "lm_q2_score": 0.12421300997306337, "lm_q1q2_score": 0.06113616981091064}}
{"text": "#=\nExample of using a breakpoint (in this case an infiltration point) to debug a\nfunction contained in this file.\n=#\n\nusing Infiltrator\n\nfunction calcs(x, y)\n a = x + y^2\n @infiltrate\n b = (a + 3) / x^2\n return b\nend\n\nz = calcs(2, 9)\nprintln(\"z is $z\")\n", "meta": {"hexsha": "05ab004eaa9d16d6a1a17a0c170ef0abb7f395d1", "size": 265, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "examples/debugging.jl", "max_stars_repo_name": "wigging/julia-computing", "max_stars_repo_head_hexsha": "927ee1667d5aa88a9f634fb4135a58d1244613eb", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "examples/debugging.jl", "max_issues_repo_name": "wigging/julia-computing", "max_issues_repo_head_hexsha": "927ee1667d5aa88a9f634fb4135a58d1244613eb", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "examples/debugging.jl", "max_forks_repo_name": "wigging/julia-computing", "max_forks_repo_head_hexsha": "927ee1667d5aa88a9f634fb4135a58d1244613eb", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 15.5882352941, "max_line_length": 77, "alphanum_fraction": 0.6339622642, "num_tokens": 91, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.40733340004593027, "lm_q2_score": 0.1500288243424251, "lm_q1q2_score": 0.061111751124293644}}
{"text": "mutable struct Person\n age::Int64\n\n function Person(age::Int64) \n this = new()\n if age < 0\n println(\"Age is not valid\")\n this.age = 0\n return this\n else\n this.age = age\n return this \n end\n end\nend\n\nfunction yearPasses(self::Person)\n self.age += 1\nend\n\nfunction amIOld(self::Person)\n if self.age < 13\n println(\"You are young.\")\n elseif self.age < 18\n println(\"You are a teenager.\")\n else\n println(\"You are old.\")\n end\nend\n\n# a = Person(111)\n# yearPasses(a)\n# println(a.age)\n# amIOld(a)\n", "meta": {"hexsha": "09f5098e3aa8d82d285790d3ddff67fadba9e594", "size": 614, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Hackerrank/30 Days of Code/Julia/day 04.jl", "max_stars_repo_name": "Next-Gen-UI/Code-Dynamics", "max_stars_repo_head_hexsha": "a9b9d5e3f27e870b3e030c75a1060d88292de01c", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "Hackerrank/30 Days of Code/Julia/day 04.jl", "max_issues_repo_name": "Next-Gen-UI/Code-Dynamics", "max_issues_repo_head_hexsha": "a9b9d5e3f27e870b3e030c75a1060d88292de01c", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Hackerrank/30 Days of Code/Julia/day 04.jl", "max_forks_repo_name": "Next-Gen-UI/Code-Dynamics", "max_forks_repo_head_hexsha": "a9b9d5e3f27e870b3e030c75a1060d88292de01c", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 17.5428571429, "max_line_length": 39, "alphanum_fraction": 0.5325732899, "num_tokens": 166, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4532618480153861, "lm_q2_score": 0.134775915685312, "lm_q1q2_score": 0.06108878061149038}}
{"text": "export Queue\n\nmutable struct Queue{T}\n data::Vector{T}\n Queue{T}(d) where T = new(T[d])\nend\n\nfunction Base.show(io::IO, q::Queue)\n print(io, \"Queue($(q.data))\")\nend\n\nfunction Base.pop!(q::Queue)\n popfirst!(q.data)\nend\n\nfunction Base.push!(q::Queue, d)\n push!(q.data, d)\nend\n", "meta": {"hexsha": "fea62a22d3ec5a1c64afce75a859e62f2515df57", "size": 284, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/queue.jl", "max_stars_repo_name": "yuehhua/DataStructure101", "max_stars_repo_head_hexsha": "64a3de64295c133bb7b7fcf114090c01d49521f3", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 5, "max_stars_repo_stars_event_min_datetime": "2020-04-16T17:39:11.000Z", "max_stars_repo_stars_event_max_datetime": "2021-11-05T12:30:13.000Z", "max_issues_repo_path": "src/queue.jl", "max_issues_repo_name": "yuehhua/DataStructure101", "max_issues_repo_head_hexsha": "64a3de64295c133bb7b7fcf114090c01d49521f3", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 19, "max_issues_repo_issues_event_min_datetime": "2020-03-02T07:54:58.000Z", "max_issues_repo_issues_event_max_datetime": "2021-04-12T06:17:55.000Z", "max_forks_repo_path": "src/queue.jl", "max_forks_repo_name": "yuehhua/DataStructure101", "max_forks_repo_head_hexsha": "64a3de64295c133bb7b7fcf114090c01d49521f3", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2020-04-21T09:05:07.000Z", "max_forks_repo_forks_event_max_datetime": "2020-04-21T09:05:07.000Z", "avg_line_length": 14.9473684211, "max_line_length": 36, "alphanum_fraction": 0.6373239437, "num_tokens": 90, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.3522017956470284, "lm_q2_score": 0.1732882037945951, "lm_q1q2_score": 0.061032416540904605}}
{"text": "# ---\n# title: 144. Binary Tree Preorder Traversal\n# id: problem144\n# author: Tian Jun\n# date: 2020-10-31\n# difficulty: Medium\n# categories: Stack, Tree\n# link: \n", "meta": {"hexsha": "d8dc606c487b3c94a5635d5399fb5e29f660c0f5", "size": 10245, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "notebooks/04_tuning/notebook.jl", "max_stars_repo_name": "roland-KA/MLJTutorial.jl", "max_stars_repo_head_hexsha": "00d2294dbe458504f7e2d200b1ed84097b9ff3e7", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 14, "max_stars_repo_stars_event_min_datetime": "2021-11-02T10:46:22.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-25T23:36:21.000Z", "max_issues_repo_path": "notebooks/04_tuning/notebook.jl", "max_issues_repo_name": "roland-KA/MLJTutorial.jl", "max_issues_repo_head_hexsha": "00d2294dbe458504f7e2d200b1ed84097b9ff3e7", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 23, "max_issues_repo_issues_event_min_datetime": "2021-11-11T14:36:35.000Z", "max_issues_repo_issues_event_max_datetime": "2022-02-14T04:31:54.000Z", "max_forks_repo_path": "notebooks/04_tuning/notebook.jl", "max_forks_repo_name": "roland-KA/MLJTutorial.jl", "max_forks_repo_head_hexsha": "00d2294dbe458504f7e2d200b1ed84097b9ff3e7", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 6, "max_forks_repo_forks_event_min_datetime": "2021-11-11T12:08:43.000Z", "max_forks_repo_forks_event_max_datetime": "2021-12-30T14:27:30.000Z", "avg_line_length": 34.4949494949, "max_line_length": 172, "alphanum_fraction": 0.6979014153, "num_tokens": 2654, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.3738758227716966, "lm_q2_score": 0.16238003666671086, "lm_q1q2_score": 0.06070996981046479}}
{"text": "using InstantiateFromURL\nactivate_github(\"QuantEcon/QuantEconLectureAllPackages\", tag = \"v0.9.5\");\n", "meta": {"hexsha": "840593ddcb997a5bb1c968b5a6c81eb1a872635e", "size": 99, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "rst_files/_static/includes/alldeps_no_using.jl", "max_stars_repo_name": "aderdzyan/QuantEcon-lecture-source-jl", "max_stars_repo_head_hexsha": "2a2b79ac6544d0de58abcb588026ff35a022246b", "max_stars_repo_licenses": ["BSD-3-Clause"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "rst_files/_static/includes/alldeps_no_using.jl", "max_issues_repo_name": "aderdzyan/QuantEcon-lecture-source-jl", "max_issues_repo_head_hexsha": "2a2b79ac6544d0de58abcb588026ff35a022246b", "max_issues_repo_licenses": ["BSD-3-Clause"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "rst_files/_static/includes/alldeps_no_using.jl", "max_forks_repo_name": "aderdzyan/QuantEcon-lecture-source-jl", "max_forks_repo_head_hexsha": "2a2b79ac6544d0de58abcb588026ff35a022246b", "max_forks_repo_licenses": ["BSD-3-Clause"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 33.0, "max_line_length": 73, "alphanum_fraction": 0.8181818182, "num_tokens": 31, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.35936414516010196, "lm_q2_score": 0.16885695841556153, "lm_q1q2_score": 0.060681136515343154}}
{"text": "# ------------------------------------------------------------------------------------------\n# # Julia for Data Science - Data\n# Prepared by [@nassarhuda](https://github.com/nassarhuda)! \ud83d\ude03\n#\n# In the next few notebooks, we will discuss why *Julia* is the tool you want to use for\n# your data science applications.\n#\n# We will cover the following:\n# 1. Reading and writing files\n# 1. DataFrames\n# 1. RDatasets\n# 1. FileIO\n# 1. File types\n#\n#\n# ### Data: Build a strong relationship with your data.\n# Every data science task has one main ingredient, the _data_! Most likely, you want to use\n# your data to learn something new. But before the _new_ part, what about the data you\n# already have? Let's make sure you can **read** it, **store** it, and **understand** it\n# before you start using it.\n#\n# Julia makes this step really easy with data structures and packages to process the data,\n# as well as existing functions that are readily usable on your data.\n#\n# The goal of this first part is get you acquainted with some Julia's tools to manage your\n# data.\n# ------------------------------------------------------------------------------------------\n\n# ------------------------------------------------------------------------------------------\n# **Reading and writing to files is really easy in Julia.** `).\n\n# The new model behaves like any other transformer:\n\nmach = machine(pipe, X) |> fit!;\nXsmall = transform(mach, X)\nschema(Xsmall)\n\n# Want to combine this pre-processing with ridge regression?\n\nrgs = @load RidgeRegressor pkg=MLJLinearModels\npipe2 = @pipeline encoder reducer rgs\n\n# Now our pipeline is a supervised model, instead of a transformer,\n# whose performance we can evaluate:\n\nmach = machine(pipe2, X, y) |> fit!\nevaluate!(mach, measure=mae, resampling=Holdout()) # CV(nfolds=6) is default\n\n\n# ### Training of composite models is \"smart\"\n\n# Now notice what happens if we train on all the data, then change a\n# regressor hyper-parameter and retrain:\n\nfit!(mach)\n\n#-\n\npipe2.ridge_regressor.lambda = 0.1\nfit!(mach)\n\n# Second time only the ridge regressor is retrained!\n\n# Mutate a hyper-parameter of the `PCA` model and every model except\n# the `ContinuousEncoder` (which comes before it will be retrained):\n\npipe2.pca.pratio = 0.9999\nfit!(mach)\n\n\n# ### Inspecting composite models\n\n# The dot syntax used above to change the values of *nested*\n# hyper-parameters is also useful when inspecting the learned\n# parameters and report generated when training a composite model:\n\nfitted_params(mach).ridge_regressor\n\n#-\n\nreport(mach).pca\n\n\n# ### Incorporating target transformations\n\n# Next, suppose that instead of using the raw `:price` as the\n# training target, we want to use the log-price (a common practice in\n# dealing with house price data). However, suppose that we still want\n# to report final *predictions* on the original linear scale (and use\n# these for evaluation purposes). Then we supply appropriate functions\n# to key-word arguments `target` and `inverse`.\n\n# First we'll overload `log` and `exp` for broadcasting:\nBase.log(v::AbstractArray) = log.(v)\nBase.exp(v::AbstractArray) = exp.(v)\n\n# Now for the new pipeline:\n\npipe3 = @pipeline encoder reducer rgs target=log inverse=exp\nmach = machine(pipe3, X, y)\nevaluate!(mach, measure=mae)\n\n# MLJ will also allow you to insert *learned* target\n# transformations. For example, we might want to apply\n# `Standardizer()` to the target, to standarize it, or\n# `UnivariateBoxCoxTransformer()` to make it look Gaussian. Then\n# instead of specifying a *function* for `target`, we specify a\n# unsupervised *model* (or model type). One does not specify `inverse`\n# because only models implementing `inverse_transform` are\n# allowed.\n\n# Let's see which of these two options results in a better outcome:\n\nbox = UnivariateBoxCoxTransformer(n=20)\nstand = Standardizer()\n\npipe4 = @pipeline encoder reducer rgs target=box\nmach = machine(pipe4, X, y)\nevaluate!(mach, measure=mae)\n\n#-\n\npipe4.target = stand\nevaluate!(mach, measure=mae)\n\n\n# ### Resources for Part 3\n\n# - From the MLJ manual:\n# - [Transformers and other unsupervised models](https://alan-turing-institute.github.io/MLJ.jl/dev/transformers/)\n# - [Linear pipelines](https://alan-turing-institute.github.io/MLJ.jl/dev/composing_models/#Linear-pipelines-1)\n# - From Data Science Tutorials:\n# - [Composing models](https://alan-turing-institute.github.io/DataScienceTutorials.jl/getting-started/composing-models/)\n\n\n# ### Exercises for Part 3\n\n# #### Exercise 7\n\n# Consider again the Horse Colic classification problem considered in\n# Exercise 6, but with all features, `Finite` and `Infinite`:\n\ny, X = unpack(horse, ==(:outcome), name -> true);\nschema(X)\n\n# (a) Define a pipeline that:\n# - uses `Standardizer` to ensure that features that are already\n# continuous are centred at zero and have unit variance\n# - re-encodes the full set of features as `Continuous`, using\n# `ContinuousEncoder`\n# - uses the `KMeans` clustering model from `Clustering.jl`\n# to reduce the dimension of the feature space to `k=10`.\n# - trains a `EvoTreeClassifier` (a gradient tree boosting\n# algorithm in `EvoTrees.jl`) on the reduced data, using\n# `nrounds=50` and default values for the other\n# hyper-parameters\n\n# (b) Evaluate the pipeline on all data, using 6-fold cross-validation\n# and `cross_entropy` loss.\n\n# ☆(c) Plot a learning curve which examines the effect on this loss\n# as the tree booster parameter `max_depth` varies from 2 to 10.\n\n# Presentation mode \"\n\n# \u2554\u2550\u2561 5c402564-e0e1-11eb-105b-951cfa19cacd\nmd\"\"\"\n# Julia for Statistics and Data Science\n\n- *Cecile Ane*, *Douglas Bates* & *Claudia Solis-Lemus* all with U. of Wisconsin - Madison\n\n1. What is **Julia**?\n - How does it differ from other languages, particularly **R** and **Python**, used for data science?\n - How do I get started?\n2. A deep dive into a shallow function - how does compilation work?\n3. Motivating example -- `MixedModels.jl` compared to R's `lme4`\n4. Some recommended packages for Statistics and Data Science \n5. `RCall.jl` and `PyCall.jl`: to allow you to build on your current skills\n\"\"\"\n\n# \u2554\u2550\u2561 7e8e3903-a76d-4310-b5df-8508fdd7dba3\nmd\"\"\"\n## What is **Julia**?\n\n- an Open Source language designed for \"technical computing\". The motivation is best explained in [this blog post](https://julialang.org/blog/2012/02/why-we-created-julia/) by its creators.\n- development of the [language itself](https://github.com/JuliaLang/julia) and most packages, e.g. [DataFrames](https://github.com/JuliaData/DataFrames.jl), takes place on github.\n- a few things to notice about [github.com/JuliaLang/julia](https://github.com/JuliaLang/julia)\n - nearly 1100 contributors\n - over 40,000 issues or pull requests\n - over 50,000 commits\n - the most common coding language used in the `julia` repository is Julia\n- installation is easiest using binary downloads from [julialang.org](https://julialang.org/downloads)\n\"\"\"\n\n# \u2554\u2550\u2561 dc65b3ed-2acf-4bf5-be2e-77dde963a7f1\nmd\"\"\"\n## Package Ecosystem\n- Julia packages are public git repositories (usually on `github.com` or `gitlab.com`) listed in the [General Registry](https://github.com/JuliaRegistries/General)\n- see the [Packages tab at julialang.org](https://julialang.org/packages) for links to package exploration tools. I prefer the one at [JuliaHub](https://juliahub.com/ui/packages)\n + when browsing Julia packages visit the repository and look at the Languages bar on the right hand side. Most packages are 100% Julia code. As [Erik Engheim wrote](https://medium.com/codex/is-julia-really-fast-12cd7caef96b), \"it's turtles all the way down\".\n + [this blog posting](https://julialang.org/blog/2021/08/general-survey/) provides some analysis of the package registry contents\n- setting version numbers according to [semantic versioning](https://semver.org/) is strongly encouraged\n- often packages are maintained by organizations on github.com, e.g. DataFrames.jl belongs to the JuliaData organization.\n\"\"\"\n\n# \u2554\u2550\u2561 bb892d27-63f6-4267-bfcc-758e743c31d2\nmd\"\"\"\n## Ways to use Julia\n1. Start the App and use the Read-Eval-Print-Loop (REPL) **[demo this]**\n - the REPL has name completion and other conveniences\n - the REPL has \"modes\"\n + standard Julia input\n + help mode\n + package manager mode\n + shell mode\n + some packages, e.g. `RCall`, enable their own modes\n2. An Integrated Development Environment (IDE)\n - [Visual Studio Code](https://code.visualstudio.com/) (with the Julia extension) is the most widely used\n3. [Jupyter notebooks](https://jupyter.org) using the [IJulia package](https://github.com/JuliaLang/IJulia.jl)\n4. [Pluto](https://github.com/fonsp/Pluto.jl) notebooks, which we are using here.\n - (+) notebooks are self-contained with regard to packages needed and versions thereof\n - (+) notebooks are reactive - changes in one place are propagated to others\n - (-) reactivity sometimes requires awkward code phrasing\n - (-) output displayed is the result of `show` on the value of a block, which sometimes misses things that would normally be displayed\n\"\"\"\n\n# \u2554\u2550\u2561 864522f6-a214-4397-91cb-8d4d670e7638\nmd\"\"\"\n## Why Julia?\n- allows you to bypass the **two language problem**\n + high-level, *dynamic* languages (R, Python, Matlab/Octave, etc.) allow for ease of use and high productivity\n + often code in these languages is unable to give the performance required for high-throughput, production uses\n + the usual solution is to re-write the performance-critical parts in a low-level, *static* language (C, C++, Fortran) and integrate that with the higher-level code\n- language and support system are carefully crafted to provide **both** high level tools and high performance\n- recently developed language with the benefit of hindsight and modern tools.\n\"\"\"\n\n# \u2554\u2550\u2561 d78b2581-31cd-4a08-9c8f-59aedc5c3086\nmd\"\"\"\n## Why not Julia?\n- recently developed language\n + ecosystem is not as mature as R, Python, etc.\n + don't have time to learn a new language and support system because this proposal is due Friday\n\"\"\"\n\n# \u2554\u2550\u2561 0aab07a1-893e-4c8b-ae8d-198146058d2e\nmd\"\"\"\n## In what ways is **Julia** similar to **R**?\n\n- Algorithms are expressed as **functions**\n- Functions are *generic* - different *methods* can be defined for argument *signatures*\n * in **R** a function must be explicitly designated as a generic function\n * **R** has different systems of generic functions and methods (S3 and S4)\n - S3 method dispatch is on the first argument's class only (single dispatch)\n - S4 allows for **multiple dispatch**\n- In Julia all functions are generic and all functions use multiple dispatch\n\"\"\"\n\n# \u2554\u2550\u2561 85fcacb1-7ad2-4fb8-9c87-28cfaa0f1404\nmd\"\"\"\n## What makes Julia fast?\n- The first time a particular method is invoked, it is compiled using a *Just In Time* compiler based on [**LLVM**](https://llvm.org) - the low-level virtual machine\n * good news - the actual execution is fast and subsequent calls are fast\n * bad news - the first call can be slow because of the compilation overhead\n + known informally as the \"Time To First Plot\" problem\n- Ease of programming and ease of compilation are often antithetical goals\n * ease of programming emphasizes generality provided by languages with dynamic types\n * ease of compilation requires very specific, usually static, types b/c that's what the processor works with\n- In Julia methods are often defined for general, abstract argument types but compilation takes place for specific, concrete types.\n\nTime for a deep dive (notebook named `2compilation.jl`)\n\"\"\"\n\n# \u2554\u2550\u2561 Cell order:\n# \u255f\u25008b92a6cf-cd6a-4364-9f59-598bcf206ef1\n# \u255f\u25005c402564-e0e1-11eb-105b-951cfa19cacd\n# \u255f\u25007e8e3903-a76d-4310-b5df-8508fdd7dba3\n# \u255f\u2500dc65b3ed-2acf-4bf5-be2e-77dde963a7f1\n# \u255f\u2500bb892d27-63f6-4267-bfcc-758e743c31d2\n# \u255f\u2500864522f6-a214-4397-91cb-8d4d670e7638\n# \u255f\u2500d78b2581-31cd-4a08-9c8f-59aedc5c3086\n# \u255f\u25000aab07a1-893e-4c8b-ae8d-198146058d2e\n# \u255f\u250085fcacb1-7ad2-4fb8-9c87-28cfaa0f1404\n", "meta": {"hexsha": "1c0906e155b4b5a027a053f39979ecf4bf342b56", "size": 6667, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "notebooks/1intro.jl", "max_stars_repo_name": "palday/JSM2021", "max_stars_repo_head_hexsha": "bc7cd1232f10bd23a33686b346643557e2ada91d", "max_stars_repo_licenses": ["CC0-1.0"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "notebooks/1intro.jl", "max_issues_repo_name": "palday/JSM2021", "max_issues_repo_head_hexsha": "bc7cd1232f10bd23a33686b346643557e2ada91d", "max_issues_repo_licenses": ["CC0-1.0"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "notebooks/1intro.jl", "max_forks_repo_name": "palday/JSM2021", "max_forks_repo_head_hexsha": "bc7cd1232f10bd23a33686b346643557e2ada91d", "max_forks_repo_licenses": ["CC0-1.0"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 52.0859375, "max_line_length": 264, "alphanum_fraction": 0.7364631768, "num_tokens": 1953, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.1919327864472368, "lm_q2_score": 0.3140505385717077, "lm_q1q2_score": 0.060276594953323284}}
{"text": "#**************************************************************************************\n# Input_Interface.jl\n# =============== part of the GeoEfficiency.jl package.\n#\n# all the input either from the console or from the csv files to the package is handled by some function here.\n#\n#**************************************************************************************\n\n#------------------ consts - globals - imports ----------------------------\n\nusing Compat\nusing Compat.MathConstants\nusing Compat.DelimitedFiles\nimport Compat: @info, @warn, @error\n\n@compat isconst(@__MODULE__, :dataFolder ) \t||\tconst dataFolder = string(@__MODULE__)\n@compat isconst(@__MODULE__, :dataDir )\t\t||\tconst dataDir = joinpath(homedir(), dataFolder); \t\nmkpath(dataDir)\n\nconst Detectors = \"Detectors.csv\";\nconst srcHeights = \"srcHeights.csv\";\nconst srcRhos = \"srcRhos.csv\";\nconst srcRadii = \"srcRadii.csv\";\nconst srcLengths = \"srcLengths.csv\";\n\n\n@enum SrcType srcUnknown=-1 srcPoint=0 srcLine=1 srcDisk=2 srcVolume=3 srcNotPoint =4\nglobal srcType = srcUnknown\n\n#------------------ typeofSrc --------------------------------------\n\n\"\"\"\n typeofSrc()::SrcType\n\nreturn the current value of the global `GeoEfficiency.srcType`.\n\n\"\"\"\ntypeofSrc()::SrcType = srcType # srcType !== SrcType, but\n\n\"\"\"\n typeofSrc(x::Int)::SrcType\n\nset and return the value of the global `GeoEfficiency.srcType` corresponding to `x`.\n\n * srcUnknown = -1 also any negative integer treated as so, \n * srcPoint = 0, \n * srcLine = 1, \n * srcDisk = 2, \n * srcVolume = 3, \n * srcNotPoint = 4 also any greater than 4 integer treated as so.\n\n\"\"\"\nfunction typeofSrc(x::Int)::SrcType\n\tglobal srcType = if x < 0\n\t\t\t\t\tSrcType(-1)\n\t\t\t\telseif x > 4\n\t\t\t\t\tSrcType(4)\n\t\t\t\telse\n\t\t\t\t\tSrcType(x)\n\t\t\t\tend\nend #function\n\n\n#------------------------ setSrcToPoint ---------------------------\n\n\"\"\"\n setSrcToPoint()::Bool\n\nreturn whether the source type is a point or not.\n\"\"\"\nsetSrcToPoint()::Bool = srcType === srcPoint\n\n\"\"\"\n\n setSrcToPoint(yes::Bool)::Bool\n\nreturn whether the source type is a point or not after setting `srcType` to ``srcPoint`` if \n`yes` = ``true`` else if `yes` = ``false`` setting it to ``srcNotPoint`` if it was not already \nset to other non-point type (``srcDisk``, ``srcLine``, ``srcVolume``).\n\n!!! note\n * The user can use this function to change the source type any time.\n * The source type is set the fist time asked for source.\n\n**see also:** [`typeofSrc(::Int)`](@ref).\n\n\"\"\"\nfunction setSrcToPoint(yes::Bool)::Bool\n\tglobal srcType = if yes \n\t\t\t\t\t\tsrcPoint\n\t\t\t\t\telseif srcType in [srcUnknown, srcPoint] \n\t\t\t\t\t\tsrcNotPoint\n\t\t\t\t\telse\n\t\t\t\t\t\tsrcType \n\t\t\t\t\tend\n\treturn srcType === srcPoint\nend\n\n\"\"\"\n\tsetSrcToPoint(prompt::AbstractString)::Bool\n\nreturn whether the source type is a point or not. only prompt the user to set the source \ntype if it were not already set before. \n\n**see also:** [`typeofSrc(::Int)`](@ref), [`setSrcToPoint(::Bool)`](@ref).\n\n\"\"\"\nsetSrcToPoint(prompt::AbstractString)::Bool = srcType != srcUnknown ?\tsetSrcToPoint() :\n\t\t\t\t\t\t\t\t\t\t\tsetSrcToPoint(input(prompt) |> lowercase != \"n\" )\n\n\n#---------------------------- input ---------------------------------\n\n\"\"\" # UnExported\n\n input(prompt::AbstractString = \"? \", incolor::Symbol = :green)\n\nreturn a string delimited by new line excluding the new line. prompt the user with the massage `prompt` defaults to `? `. \n`incolor` specify the prompt text color, default to ``green``.\n\n\"\"\"\nfunction input(prompt::AbstractString = \"? \", incolor::Symbol = :green)\n printstyled(prompt, color=incolor); chomp(readline())\nend # function\n\n\n#---------------------------- getfloat -----------------------------------\n\n\"\"\"# UnExported\n\n\tgetfloat(prompt::AbstractString = \"? \", from::Real = 0.0, to::Real = Inf; value::AbstractString=\"nothing\")::Float64\n\nprompts the user with the massage `prompt` defaults to `? ` to input a numerical expression \nevaluate to a numerical value and asserts that the value is in the semi open interval [`from`, `to`[\nbefore returning it as a `Float64`.\n\n!!! note\n * a blank input (i.e just a return) is considered as being ``0.0``.\n * input from the `console` can be numerical expression not just a number.\n * All ``5/2``, ``5//2``, ``exp(2)``, ``pi``, ``1E-2``, ``5.2/3``, ``sin(1)``, ``pi/2/3`` \n are valid mathematical expressions.\n * the key word argument `value` , if provided the function will not ask for input from the \n `console` and take it as if it where inputted from the `console` [``for test propose mainly``].\n\n# Examples\n```\njulia> getfloat(\"input a number:\", value=\"3\")\n3.0\n\njulia> getfloat(\"input a number:\", value=\"\")\n0.0\n\njulia> getfloat(\"input a number:\", value=\"5/2\")\n2.5\n\njulia> getfloat(\"input a number:\", value=\"5//2\")\n2.5\n\njulia> getfloat(\"input a number:\", value=\"pi\")\n3.141592653589793\n```\n\n\"\"\"\nfunction getfloat(prompt::AbstractString = \"? \", from::Real = 0.0, to::Real = Inf; value::AbstractString=\"nothing\") ::Float64\n\ttry\n\t\t\"nothing\" == value ? value = input(prompt) : nothing\n\t\t\"\" == value && return 0.0\t\t# just pressing return is interapted as <0.0>\n\t\tval::Float64 = Meta.parse(value) |> eval |> float\n\t\t@assert from <= val < to\n\t\treturn val\n\n catch err\n if isa(err, AssertionError) \n\t\t\t@warn(\"\"\"input `$value` evaluated to be outside the semi open interval [$from, $to[,\n\t\t\t\\n Please: provide an adequate value\"\"\")\n else \n\t\t\t@warn(\"\"\"input `$value` cannot be parsed to a valid numerical value!,\n\t\t\t\\n Please: provide a valid expression\"\"\")\n end #if \n \n return getfloat(prompt, from, to)\n\n end #try\nend\t\n\n\n#---------------------------- detector_info_from_csvFile ------------------------------\n\n\"\"\"# UnExported\n\n\t detector_info_from_csvFile(detectors::AbstractString = Detectors, \n datadir::AbstractString = dataDir)\nreturn a vector{Detector} based on information in the file of name `detectors` found in the \ndirectory `datadir`.\n\n!!! note\n * if no path is given the second argument `datadir` is default to ``$(dataDir)`` as set by \n the constant ``dataDir``. \n * if no file name is specified the name of the predefined file ``$Detectors`` as set by \n the constant ``Detectors``. \n * the no argument method is the most useful; other methods are mainly for ``test propose``.\n\n\"\"\"\nfunction detector_info_from_csvFile(detectors::AbstractString = Detectors, \n datadir::AbstractString = dataDir)\n detector_info_array::Matrix{Float64} = Matrix{Float64}(undef, 0, 0)\n @info(\"opening '$(detectors)'......\")\n try\n detector_info_array = readdlm(joinpath(datadir, detectors), ',', header=true)[1];\n return getDetectors(detector_info_array)\n\t\t\n catch err\n if isa(err, SystemError) \n\t\t @error(\"detector_info_from_csvFile: Some thing went wrong, may be the file '$(joinpath( datadir, detectors))' can't be found\")\n\t\tend\n rethrow()\n\n end #try\n\nend #function\n\n\n#---------------------------- read_from_csvFile --------------------------------\n\n\"\"\"# UnExported\n\n\tread_from_csvFile(csv_data::AbstractString, \n datadir::AbstractString = dataDir)::Vector{Float64}\n\nreturn Vector{Float64} based on data in csv file named `csv_data`. directory `datadir` point to \nwhere the file is located default to ``$(dataDir)`` as set by the constant `dataDir`.\n\n\"\"\"\nfunction read_from_csvFile(csv_data::AbstractString, datadir::AbstractString = datadir)::Vector{Float64}\n\t@info(\"Opening `$(csv_data)`......\")\n\ttry\n\t\tindata = readdlm(joinpath(datadir, csv_data), ',', header=true)[1][:,1]\n\t\treturn float(indata ) |> sort;\n\n\tcatch err\n\t if isa(err, SystemError) \n\t\t @error(\"Some thing went wrong, may be `$(csv_data)` can't be found in `$(datadir)`\")\n\t\t\n\t\telse\n\t\t #println(err)\n\t\t\n\t\tend\t\t\n\t\treturn Float64[0.0]\n\n\tend #try\nend #function\n\n\n#--------------------------- read_batch_info ------------------------------------\n\n\"\"\"# UnExported\n\n\tread_batch_info()\n\nread `detectors` and `sources` parameters from the predefined csv files.\n\nReturn a tuple\n\t (detectors_array,\n\t\tsrcHeights_array,\n\t\tsrcRhos_array,\n\t\tsrcRadii_array,\n\t\tsrcLengths_array,\n\t\tGeoEfficiency_isPoint)\n\n\"\"\"\nread_batch_info() = read_batch_info(datadir,\n detectors, \n\t\t\t\t\t\t\t\t srcHeights,\n\t\t\t\t\t\t\t\t srcRhos,\n\t\t\t\t\t\t\t\t srcRadii,\n\t\t\t\t\t\t\t\t srcLengths)\n\n\n\"\"\"# UnExported\n\n\tread_batch_info(datadir::AbstractString,\n detectors::AbstractString, \n srcHeights::AbstractString,\n srcRhos::AbstractString,\n srcRadii::AbstractString,\n srcLengths::AbstractString)\n\nread `detectors` and `sources` parameters from the location given in the argument list.\n\nReturn a tuple\n\n\t (detectors_array,\n\t\tsrcHeights_array,\n\t\tsrcRhos_array,\n\t\tsrcRadii_array,\n\t\tsrcLengths_array,\n\t\tisPoint)\n\n\"\"\"\t\t\t\t\t\t\t\t \nfunction read_batch_info(datadir::AbstractString,\n detectors::AbstractString, \n\t\t\t\t\t srcHeights::AbstractString,\n\t\t\t\t\t srcRhos::AbstractString,\n\t\t\t\t\t srcRadii::AbstractString,\n\t\t\t\t\t srcLengths::AbstractString)\n\n\t@info(\"Starting the Batch Mode ....\")\n\tisPoint = setSrcToPoint(\"\\n Is it a point source {Y|n} ?\")\n\n\t@info(\"Reading data from `CSV files` at $datadir .....\")\n\tdetectors_array ::Vector{RadiationDetector} = try detector_info_from_csvFile(detectors, datadir); catch err; getDetectors(); end\n\tsrcHeights_array::Vector{Float64} = read_from_csvFile(srcHeights, datadir)\n\tsrcRhos_array ::Vector{Float64} = [0.0]\n\tsrcRadii_array ::Vector{Float64} = [0.0]\n\tsrcLengths_array::Vector{Float64} = [0.0]\n\n\tfunction batchfailure(err::AbstractString)\n\t\t@warn(err, \", transfer to direct data input via the `console`......\")\n\t\tsleep(3); src = source()\n\t\tsrcHeights_array, srcRhos_array, srcRadii_array , srcLengths_array = \n\t\t[src[1].Height] , [src[1].Rho] , [src[2]] , [src[3]]\n\t\treturn nothing\n\tend #function\n\n\tif srcHeights_array == [0.0]\n\t\tbatchfailure(\"`$(srcHeights)` is not found in `$(datadir)`)\")\n\n\telseif isPoint\n\t\tsrcRhos_array =\tread_from_csvFile(srcRhos, datadir)\n\n\telse\n\t\tsrcRadii_array = read_from_csvFile(srcRadii, datadir)\n\t\tif srcRadii_array == [0.0]\n\t\t\tbatchfailure(\"`$(srcRadii)` is not found in `$(datadir)`)\")\n\n\t\telse\n\t\t\tsrcLengths_array = read_from_csvFile(srcLengths, datadir)\n\n\t\tend #if\n\tend #if\n\t#println(\"\\n Results log\\n=============\")\n\treturn (\n\t\tdetectors_array,\n\t\tsrcHeights_array,\n\t\tsrcRhos_array,\n\t\tsrcRadii_array,\n\t\tsrcLengths_array,\n\t\tisPoint,\n\t\t)\nend #function\n\n\n#------------------------- getDetectors -------------------------------------\n\n\"\"\"\n\n getDetectors(detectors_array::Vector{<:Detector} = Detector[])\n\nreturn the `detectors_array` extended by the entered detectors and sorted according to the \ndetector volume. \nprompt the user to input detector parameters from the `console`.\n\n!!! note\n If no array received in the input an empty array will be created to receive the converted detectors.\n\n\"\"\"\nfunction getDetectors(detectors_array::Vector{<:RadiationDetector} = RadiationDetector[])\n\tVector{RadiationDetector}(detectors_array); @info(\"Please, input the detector information via the console\")\n\twhile(true)\n\t\ttry push!(detectors_array, RadiationDetector()); catch err\tprintln(err); @warn(\"Please: Enter a New Detector\"); continue; end\n\t\tlowercase(input(\n\t\t\t\"\"\"\\n\n \t - To add a new detector press return\\n\n \t - To quit press 'q'|'Q' then return\\n\n\t\t\t\\n\\t your Choice: \"\"\", :blue)) == \"q\" && break\n\tend #while\n\treturn detectors_array |> sort\nend #function\n\n\n\"\"\"\n\n\tgetDetectors(detector_info_array::Matrix{<:Real}, detectors_array::Vector{<:Detector} = Detector[] ;console_FB=true) \n\nreturn `detectors_array`, after extending it with the successfully converted detectors. while, \nattempt to convert detectors from the information in `detector_info_array`. \n\n!!! note\n if `console_FB` argument is set to true , the function will call `getDetectors()` to take input\n from the `console` if the `detector_info_array` is empty or contain no numerical element.\n\n\"\"\"\nfunction getDetectors(detector_info_array::Matrix{<:Real}, detectors_array::Vector{<:Detector} = Detector[] ; console_FB=true) \n\t\n\tif isempty(detector_info_array) \n\t\tif console_FB \n\t\t\t@info(\"The new detectors information may entred via the console\")\n\t\t\treturn getDetectors(detectors_array)\n\t\telse\t\n\t\t \terror(\"getDetectors: Empty `detector_info_array`\")\n\t\tend\n\t\t\n\telse\n\t\tVector{RadiationDetector}(detectors_array)\n\t\tfor i_th_line = 1:size(detector_info_array)[1]\n\t\t\ttry push!(detectors_array, RadiationDetector((detector_info_array[i_th_line,:])...)) end #try\n\t\tend #for\n\n\t\treturn detectors_array |> sort\n\tend #if\nend #function\n", "meta": {"hexsha": "cf4d3bb2116eaf36804beb8709faa3a7f0ea0209", "size": 12725, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/Input_Interface.jl", "max_stars_repo_name": "GeoEfficiency/GeoEfficiency.github.io", "max_stars_repo_head_hexsha": "4d87b43cd81685d1589eb23b7d0e73ff5f8ad026", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2020-09-18T14:29:17.000Z", "max_stars_repo_stars_event_max_datetime": "2020-09-18T14:29:17.000Z", "max_issues_repo_path": "src/Input_Interface.jl", "max_issues_repo_name": "GeoEfficiency/GeoEfficiency.github.io", "max_issues_repo_head_hexsha": "4d87b43cd81685d1589eb23b7d0e73ff5f8ad026", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/Input_Interface.jl", "max_forks_repo_name": "GeoEfficiency/GeoEfficiency.github.io", "max_forks_repo_head_hexsha": "4d87b43cd81685d1589eb23b7d0e73ff5f8ad026", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2019-04-13T20:07:53.000Z", "max_forks_repo_forks_event_max_datetime": "2019-04-13T20:07:53.000Z", "avg_line_length": 30.8859223301, "max_line_length": 132, "alphanum_fraction": 0.6375638507, "num_tokens": 3266, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4532618627863437, "lm_q2_score": 0.13296423504419935, "lm_q1q2_score": 0.06026761686009504}}
{"text": "#\n# rubberindex.jl -\n#\n# Rubber-index for Julia arrays.\n#\n\n\"\"\"\n colons(n)\n\nyields a `n`-tuple of colons `:` (a.k.a. `Colon()`).\n\nWhen `n` is known at compile time, it is faster to call:\n\n colons(Val(n))\n\nThis method is suitable to extract sub-arrays of build views when some kind of\nrubber index is needed. For instance:\n\n slice(A::AbstractArray{T,N}, i::Integer) where {T,N} =\n A[colons(Val(N-1))..., i]\n\ndefines a function that returns the `i`-th slice of `A` assuming index `i`\nrefers the last index of `A`. Using the rubber-index `..`, a shorter\ndefinition is:\n\n slice(A::AbstractArray, i) = A[.., i]\n\nwhich is also able to deal with multiple trailing indices if `i` is a\n`CartesianIndex`.\n\nSee also: `..`, [`RubberIndex`](@ref).\n\n\"\"\"\ncolons(n::Integer) =\n (n == 0 ? () :\n n == 1 ? (:,) :\n n == 2 ? (:,:,) :\n n == 3 ? (:,:,:,) :\n n == 4 ? (:,:,:,:,) :\n n == 5 ? (:,:,:,:,:,) :\n n == 6 ? (:,:,:,:,:,:,) :\n n == 7 ? (:,:,:,:,:,:,:,) :\n n == 8 ? (:,:,:,:,:,:,:,:,) :\n n == 9 ? (:,:,:,:,:,:,:,:,:,) :\n n == 10 ? (:,:,:,:,:,:,:,:,:,:,) :\n _colons(n))\n\nfunction _colons(n::Integer)\n n \u2265 0 || bad_ndims(n)\n return ([Colon() for i in 1:n]...,)\nend\n\n@noinline bad_ndims(n::Integer) =\n throw(ArgumentError(string(\"number of dimensions should be \u2265 0, got \", n)))\n\ncolons(v::Val{N}) where {N} = ntuple(colon, v)\n\n# Just yields a colon whatever the argument.\ncolon(x) = Colon()\n\n\"\"\"\n\n`RubberIndex` is the singleron type that represents any number of indices. The\nconstant `..` is defined as `RubberIndex()` and can be used in array indexation\nto left and/or right justify the other indices. For instance, assuming `A` is\na `3\u00d74\u00d75\u00d76` array, then all the following equalities hold:\n\n A[..] == A[:,:,:,:]\n A[..,3] == A[:,:,:,3]\n A[2,..] == A[2,:,:,:]\n A[..,2:4,5] == A[:,:,2:4,5]\n A[2:3,..,1,2:4] == A[2:3,:,1,2:4]\n\nAs you can see, the advantage of the rubber index `..` is that it automatically\nexpands as the number of colons needed to have the correct number of indices.\nThe expressions are also more readable.\n\nThe rubber index may also be used for setting values. For instance:\n\n A[..] .= 1 # to fill A with ones\n A[..,3] = A[..,2] # to copy A[:,:,:,2] in A[:,:,:,3]\n A[..,3] .= A[..,2] # idem but faster\n A[2,..] = A[3,..] # to copy A[3,:,:,:] in A[2,:,:,:]\n A[..,2:4,5] .= 7 # to set all elements in A[:,:,2:4,5] to 7\n\nLeading/trailing indices may be specified as Cartesian indices (of type\n`CartesianIndex`).\n\n!!! warning\n There are two known limitations:\n 1. The `end` reserved word can only be used in intervals specified *before*\n the rubber index but not *after*. This limitation is due to the Julia\n parser cannot be avoided.\n 2. At most 9 indices can be specified before the rubber index. This\n can be extended by editing the source code.\n\nSee also: [`colons`](@ref).\n\n\"\"\" RubberIndex\n\nstruct RubberIndex end\n\nconst .. = RubberIndex()\nconst \u2026 = .. # FIXME: should be deprecated\n\n# Quickly get the tuple inside a Cartesian index.\nto_tuple(I::CartesianIndex) = I.I\n\n# Grow tuple of colons until tuple of axes is empty.\n@inline growcolons(colons, inds::Tuple{}) = colons\n@inline growcolons(colons, inds::Tuple) = growcolons((colons..., :), tail(inds))\n\n# Drop as many colons as there are specified indices.\n@inline dropcolons(colons, I::Tuple{}) = colons\n@inline dropcolons(colons::Tuple{}, ::Tuple{}) = ()\n@noinline dropcolons(colons::Tuple{}, I::Tuple) =\n throw(ArgumentError(\"too many indices specifed\"))\n@inline dropcolons(colons, I::Tuple) =\n dropcolons(tail(colons), tail(I))\n@inline dropcolons(colons, I::Tuple{CartesianIndex, Vararg}) =\n dropcolons(dropcolons(colons, to_tuple(I[1])), tail(I))\n@noinline dropcolons(colons, I::Tuple{RubberIndex, Vararg}) =\n throw(ArgumentError(\"more than one rubber index specified\"))\n\n@inline function to_indices(A, inds, I::Tuple{RubberIndex, Vararg})\n # Align the remaining indices to the tail of the `inds`. First\n # `growcolons` is called to build a tuple of as many as colons as the\n # number of remaining axes. Second, `dropcolons` is used to drop as\n # many colons as the number of specified indices (taking into account\n # Cartesian indices).\n colons = dropcolons(growcolons((), inds), tail(I))\n to_indices(A, inds, (colons..., tail(I)...))\nend\n\n# avoid copying if indexing with .. alone, see\n# https://github.com/JuliaDiffEq/OrdinaryDiffEq.jl/issues/214\n@inline Base.getindex(A::AbstractArray, ::RubberIndex) = A\n\n# The following is needed to allow for statements like `A[..] .+= expr` to\n# work properly.\ndotview(A::AbstractArray{T,N}, ::RubberIndex) where {T,N} =\n dotview(A, ntuple(colon, Val{N}())...)\n", "meta": {"hexsha": "e6a5e428bd5a39a835e3adf29c2b11970aecbfba", "size": 4784, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/rubberindex.jl", "max_stars_repo_name": "emmt/ArrayTools.jl", "max_stars_repo_head_hexsha": "f4f8ba53bcd4b5cbada93e314e82ec4a518fcbd2", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 8, "max_stars_repo_stars_event_min_datetime": "2019-10-28T12:12:54.000Z", "max_stars_repo_stars_event_max_datetime": "2021-02-16T06:03:15.000Z", "max_issues_repo_path": "src/rubberindex.jl", "max_issues_repo_name": "emmt/ArrayTools.jl", "max_issues_repo_head_hexsha": "f4f8ba53bcd4b5cbada93e314e82ec4a518fcbd2", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 2, "max_issues_repo_issues_event_min_datetime": "2020-11-08T00:25:27.000Z", "max_issues_repo_issues_event_max_datetime": "2021-12-03T10:15:44.000Z", "max_forks_repo_path": "src/rubberindex.jl", "max_forks_repo_name": "emmt/ArrayTools.jl", "max_forks_repo_head_hexsha": "f4f8ba53bcd4b5cbada93e314e82ec4a518fcbd2", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 33.4545454545, "max_line_length": 80, "alphanum_fraction": 0.6151755853, "num_tokens": 1459, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4843800842769843, "lm_q2_score": 0.12421301321508334, "lm_q1q2_score": 0.06016630980942023}}
{"text": "\"\"\"\n sign_cadlag(x::N)::N where {N<:Real}\n\nThis function works like the sign function but is ``1`` for input ``0``.\n\n### Input\n\n- `x` -- real scalar\n\n### Output\n\n``1`` if ``x \u2265 0``, ``-1`` otherwise.\n\n### Notes\n\nThis is the sign function right-continuous at zero (see\n[c\u00e0dl\u00e0g function](https://en.wikipedia.org/wiki/C%C3%A0dl%C3%A0g)).\nIt can be used with vector-valued arguments via the dot operator.\n\n### Examples\n\n```jldoctest\njulia> LazySets.sign_cadlag.([-0.6, 1.3, 0.0])\n3-element Array{Float64,1}:\n -1.0\n 1.0\n 1.0\n```\n\"\"\"\nfunction sign_cadlag(x::N)::N where {N<:Real}\n return x < zero(x) ? -one(x) : one(x)\nend\n\n\"\"\"\n substitute(substitution::Dict{Int, T}, x::AbstractVector{T}) where {T}\n\nApply a substitution to a given vector.\n\n### Input\n\n- `substitution` -- substitution (a mapping from an index to a new value)\n- `x` -- vector\n\n### Output\n\nA fresh vector corresponding to `x` after `substitution` was applied.\n\"\"\"\nfunction substitute(substitution::Dict{Int, T}, x::AbstractVector{T}) where {T}\n return substitute!(substitution, copy(x))\nend\n\n\"\"\"\n substitute!(substitution::Dict{Int, T}, x::AbstractVector{T}) where {T}\n\nApply a substitution to a given vector.\n\n### Input\n\n- `substitution` -- substitution (a mapping from an index to a new value)\n- `x` -- vector (modified in this function)\n\n### Output\n\nThe same (but see the Notes below) vector `x` but after `substitution` was\napplied.\n\n### Notes\n\nThe vector `x` is modified in-place if it has type `Vector` or `SparseVector`.\nOtherwise, we first create a new `Vector` from it.\n\"\"\"\nfunction substitute!(substitution::Dict{Int, T}, x::AbstractVector{T}) where {T}\n return substitute!(Vector(x), substitution)\nend\n\nfunction substitute!(substitution::Dict{Int, T},\n x::Union{Vector{T}, SparseVector{T}}) where {T}\n for (index, value) in substitution\n x[index] = value\n end\n return x\nend\n\n\"\"\"\n reseed(rng::AbstractRNG, seed::Union{Int, Nothing})::AbstractRNG\n\nReset the RNG seed if the seed argument is a number.\n\n### Input\n\n- `rng` -- random number generator\n- `seed` -- seed for reseeding\n\n### Output\n\nThe input RNG if the seed is `nothing`, and a reseeded RNG otherwise.\n\"\"\"\nfunction reseed(rng::AbstractRNG, seed::Union{Int, Nothing})::AbstractRNG\n if seed != nothing\n return Random.seed!(rng, seed)\n end\n return rng\nend\n\n\"\"\"\n StrictlyIncreasingIndices\n\nIterator over the vectors of `m` strictly increasing indices from 1 to `n`.\n\n### Fields\n\n- `n` -- size of the index domain\n- `m` -- number of indices to choose (resp. length of the vectors)\n\n### Notes\n\nThe vectors are modified in-place.\n\nThe iterator ranges over ``\\\\binom{n}{m}`` (`n` choose `m`) possible vectors.\n\nThis implementation results in a lexicographic order with the last index growing\nfirst.\n\n### Examples\n\n```jldoctest\njulia> for v in LazySets.StrictlyIncreasingIndices(4, 2)\n println(v)\n end\n[1, 2]\n[1, 3]\n[1, 4]\n[2, 3]\n[2, 4]\n[3, 4]\n```\n\"\"\"\nstruct StrictlyIncreasingIndices\n n::Int\n m::Int\n\n function StrictlyIncreasingIndices(n::Int, m::Int)\n @assert n >= m > 0 \"require n >= m > 0\"\n new(n, m)\n end\nend\n\nBase.eltype(::Type{StrictlyIncreasingIndices}) = Vector{Int}\nBase.length(sii::StrictlyIncreasingIndices) = binomial(sii.n, sii.m)\n\n# initialization\nfunction Base.iterate(sii::StrictlyIncreasingIndices)\n v = [1:sii.m;]\n return (v, v)\nend\n\n# normal iteration\nfunction Base.iterate(sii::StrictlyIncreasingIndices, state::AbstractVector{Int})\n v = state\n i = sii.m\n diff = sii.n\n if i == diff\n return nothing\n end\n while v[i] == diff\n i -= 1\n diff -= 1\n end\n # update vector\n v[i] += 1\n for j in i+1:sii.m\n v[j] = v[j-1] + 1\n end\n # detect termination: first index has maximum value\n if i == 1 && v[1] == (sii.n - sii.m + 1)\n return (v, nothing)\n end\n return (v, v)\nend\n\n# termination\nfunction Base.iterate(sii::StrictlyIncreasingIndices, state::Nothing)\n return nothing\nend\n\n\"\"\"\n subtypes(interface, concrete::Bool)\n\nReturn the concrete subtypes of a given interface.\n\n### Input\n\n- `interface` -- an abstract type, usually a set interface\n- `concrete` -- if `true`, seek further the inner abstract subtypes of the given\n interface, otherwise return only the direct subtypes of `interface`\n\n### Output\n\nA list with the subtypes of the abstract type `interface`, sorted alphabetically.\n\n### Examples\n\nConsider the `AbstractPolytope` interface. If we include the abstract subtypes\nof this interface,\n\n```jldoctest subtypes\njulia> using LazySets: subtypes\n\njulia> subtypes(AbstractPolytope, false)\n4-element Array{Any,1}:\n AbstractCentrallySymmetricPolytope\n AbstractPolygon\n HPolytope\n VPolytope\n```\n\nWe can use this function to obtain the concrete subtypes of\n`AbstractCentrallySymmetricPolytope` and `AbstractPolygon` (further until all\nconcrete types are obtained), using the `concrete` flag:\n\n```jldoctest subtypes\njulia> subtypes(AbstractPolytope, true)\n14-element Array{Type,1}:\n Ball1\n BallInf\n HPolygon\n HPolygonOpt\n HPolytope\n Hyperrectangle\n Interval\n LineSegment\n Singleton\n SymmetricIntervalHull\n VPolygon\n VPolytope\n ZeroSet\n Zonotope\n```\n\"\"\"\nfunction subtypes(interface, concrete::Bool)\n\n subtypes_to_test = subtypes(interface)\n\n # do not seek the concrete subtypes further\n if !concrete\n return sort(subtypes_to_test, by=string)\n end\n\n result = Vector{Type}()\n i = 0\n while i < length(subtypes_to_test)\n i += 1\n subtype = subtypes_to_test[i]\n new_subtypes = subtypes(subtype)\n if isempty(new_subtypes)\n # base type found\n push!(result, subtype)\n else\n # yet another interface layer\n append!(subtypes_to_test, new_subtypes)\n end\n end\n return sort(result, by=string)\nend\n", "meta": {"hexsha": "5c7ad124f08a3de1c20226ea3b4816242c30597c", "size": 5887, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/helper_functions.jl", "max_stars_repo_name": "ueliwechsler/LazySets.jl", "max_stars_repo_head_hexsha": "9bc60f06aea88026656a17a07a7ed2c337386033", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/helper_functions.jl", "max_issues_repo_name": "ueliwechsler/LazySets.jl", "max_issues_repo_head_hexsha": "9bc60f06aea88026656a17a07a7ed2c337386033", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/helper_functions.jl", "max_forks_repo_name": "ueliwechsler/LazySets.jl", "max_forks_repo_head_hexsha": "9bc60f06aea88026656a17a07a7ed2c337386033", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 22.2150943396, "max_line_length": 84, "alphanum_fraction": 0.6675726176, "num_tokens": 1655, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4455295350395727, "lm_q2_score": 0.1347759226358913, "lm_q1q2_score": 0.060046654146498075}}
{"text": "# \nfunction printsum(a)\n println(summary(a), \": \", repr(a))\nend\n# \nfor i in 1:5\n print(i, \", \")\nend\n#> 1, 2, 3, 4, 5,\n# In loop definitions \"in\" is equivilent to \"=\" \n# (AFAIK, the two are interchangable in this context)\nfor i = 1:5\n print(i, \", \")\nend\nprintln() #> 1, 2, 3, 4, 5,\n\n# arrays can also be looped over directly:\na1 = [1,2,3,4]\nfor i in a1\n print(i, \", \")\nend\nprintln() #> 1, 2, 3, 4,\n\n# **continue** and **break** work in the same way as python\na2 = collect(1:20)\nfor i in a2\n if i % 2 != 0\n continue\n end\n print(i, \", \")\n if i >= 8\n break\n end\nend\nprintln() #> 2, 4, 6, 8,\n\n# if the array is being manipulated during evaluation a while loop shoud be used\n# [pop](https://docs.julialang.org/en/v1/base/collections/#Base.pop!-Tuple{Any,Any,Any}) removes the last element from an array\nwhile !isempty(a1)\n print(pop!(a1), \", \")\nend\nprintln() #> 4, 3, 2, 1,\n\nd1 = Dict(1=>\"one\", 2=>\"two\", 3=>\"three\")\n# dicts may be looped through using the keys function:\nfor k in sort(collect(keys(d1)))\n print(k, \": \", d1[k], \", \")\nend\nprintln() #> 1: one, 2: two, 3: three,\n\n# like python [enumerate](https://docs.julialang.org/en/v1/base/iterators/#Base.Iterators.enumerate) can be used to get both the index and value in a loop\na3 = [\"one\", \"two\", \"three\"]\nfor (i, v) in enumerate(a3)\n print(i, \": \", v, \", \")\nend\nprintln() #> 1: one, 2: two, 3: three,\n\n# (note enumerate starts from 1 since Julia arrays are 1 indexed unlike python)\n\n# [map](https://docs.julialang.org/en/v1/base/collections/#Base.map) works as you might expect performing the given function on each member of \n# an array or iter much like comprehensions\na4 = map((x) -> x^2, [1, 2, 3, 7])\nprint(a4) \nprintln() #> [1, 4, 9, 49]\n", "meta": {"hexsha": "742ffbf53af21979bf6a6ec78983b141fda079a9", "size": 1758, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/loops_map.jl", "max_stars_repo_name": "talgatomarov/JuliaByExample", "max_stars_repo_head_hexsha": "9fc5516b7f6d03bd2f340b7a9bb52a06fedb7458", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 281, "max_stars_repo_stars_event_min_datetime": "2015-02-01T01:18:56.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-19T19:51:35.000Z", "max_issues_repo_path": "src/loops_map.jl", "max_issues_repo_name": "talgatomarov/JuliaByExample", "max_issues_repo_head_hexsha": "9fc5516b7f6d03bd2f340b7a9bb52a06fedb7458", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 16, "max_issues_repo_issues_event_min_datetime": "2015-04-10T04:23:38.000Z", "max_issues_repo_issues_event_max_datetime": "2021-09-23T12:32:59.000Z", "max_forks_repo_path": "src/loops_map.jl", "max_forks_repo_name": "talgatomarov/JuliaByExample", "max_forks_repo_head_hexsha": "9fc5516b7f6d03bd2f340b7a9bb52a06fedb7458", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 101, "max_forks_repo_forks_event_min_datetime": "2015-03-24T20:45:23.000Z", "max_forks_repo_forks_event_max_datetime": "2022-02-09T23:24:46.000Z", "avg_line_length": 27.0461538462, "max_line_length": 154, "alphanum_fraction": 0.6177474403, "num_tokens": 627, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.45713670203584295, "lm_q2_score": 0.13117323564948066, "lm_q1q2_score": 0.059964100340174055}}
{"text": "#\n# Using LabelledArrays source code and types to \n# \"pass through\" all properties to an underlying\n# LArray or SLArray. This allows us to write \n# a new abstract type that *functions*\n# like a LabelledArray type!\n#\n# All code in this file was copied, and modified \n# from LabelledArrays.jl source code. Their license is shown below. \n# All credit goes to LabelledArrays.jl developers.\n#\n\nconst __LABELLED_ARRAYS_LICENSE = \"\"\"\nThe LabelledArrays.jl package is licensed under the MIT \"Expat\" License:\n\n> Copyright (c) 2017: Christopher Rackauckas.\n>\n>\n> Permission is hereby granted, free of charge, to any person obtaining a copy\n>\n> of this software and associated documentation files (the \"Software\"), to deal\n>\n> in the Software without restriction, including without limitation the rights\n>\n> to use, copy, modify, merge, publish, distribute, sublicense, and/or sell\n>\n> copies of the Software, and to permit persons to whom the Software is\n>\n> furnished to do so, subject to the following conditions:\n>\n>\n>\n> The above copyright notice and this permission notice shall be included in all\n>\n> copies or substantial portions of the Software.\n>\n>\n>\n> THE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\n>\n> IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,\n>\n> FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE\n>\n> AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER\n>\n> LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,\n>\n> OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE\n>\n> SOFTWARE.\n>\n>\n\"\"\"\n\nconst __LABELLED_ARRAYS_CREDITS = \"\"\"\nThis source code which provides this functionality was copied directly from LabelledArrays.jl source code. The LabelledArrays.jl license text is provided in Julia's **Extended Help** section (accessible via `@doc`, or `??` in Julia's REPL).\n\n# Extended Help\n\n__LabelledArrays.jl License__\n\n$__LABELLED_ARRAYS_LICENSE\n\"\"\"\n\n\"\"\"\n$(TYPEDEF)\n\nA supertype for types that *function* like \n`LabelledArray.LArray` or `LabelledArray.SLArray` \ninstances, but are under a new type tree. This is\nused in `GeneralAstrodynamics` for parameterizing \nastrodynamics state vectors by physical units.\n\n!!! note\n All subtypes __must__ have only one field:\n a `LabelledArrays.LArray` or `LabelledArrays.SLArray`\n field called `__rawdata`. All methods on this abstract\n type require this field to be called `__rawdata`!\n\nNearly all code which acts on this type is copied \nand / or modified from LabelledArrays.jl source code.\nAll credit goes to LabelledArrays.jl developers. \nThe LabelledArrays.jl LICENSE file is provided \nin this docstring under Julia's __Extended Help__\ndocstring section.\n\n# Extended help\n\n__LabelledArrays.jl License__\n\n$__LABELLED_ARRAYS_LICENSE\n\"\"\"\nabstract type ParameterizedLabelledArray{F,N,T,L} <: DenseArray{F,N} end\n\n\"\"\"\n$(SIGNATURES)\n\nReturns dot-accessible property names for a `ParameterizedLabelledArray`.\n\"\"\"\nBase.propertynames(::ParameterizedLabelledArray{F,N,T}) where {F,N,T} = T isa NamedTuple ? keys(T) : T\n\n\"\"\"\n$(SIGNATURES)\n\nOverrides `Base.getproperty` for all `ParameterizedLabelledArray` instances.\n\n$__LABELLED_ARRAYS_CREDITS\n\"\"\"\nBase.@propagate_inbounds function Base.getproperty(x::ParameterizedLabelledArray, s::Symbol)\n if s \u2208 propertynames(x)\n return getproperty(getfield(x, :__rawdata), s)\n end\n return getfield(x, s) # will throw an error if s is not :__rawdata!\nend\n\n\"\"\"\n$(SIGNATURES)\n\nSets indices of a `ParameterizedLabelledArray` via label.\n\n$__LABELLED_ARRAYS_CREDITS\n\"\"\"\nBase.@propagate_inbounds function Base.setproperty!(x::ParameterizedLabelledArray, s::Symbol,y)\n if s \u2208 propertynames(x)\n return setproperty!(getfield(x, :__rawdata), s, y)\n end\n setfield!(x, s, y)\nend\n\n\"\"\"\n$(SIGNATURES)\n\nOverrides `similar` for `ParameterizedLabelledArray` instances.\n\n$__LABELLED_ARRAYS_CREDITS\n\"\"\"\nfunction Base.similar(x::ParameterizedLabelledArray, ::Type{S}, dims::NTuple{N,Int}) where {S,N}\n tmp = similar(x.__rawdata, S, dims)\n return typeof(x)(tmp)\nend\n\n\"\"\"\n$(SIGNATURES)\n\nShallow copies a `ParameterizedLabelledArray`.\n\n$__LABELLED_ARRAYS_CREDITS\n\"\"\"\nBase.copy(x::ParameterizedLabelledArray) = typeof(x)(copy(getfield(x,:__rawdata)))\n\n\"\"\"\n$(SIGNATURES)\n\nDeep copies a `ParameterizedLabelledArray`.\n\n$__LABELLED_ARRAYS_CREDITS\n\"\"\"\nBase.deepcopy(x::ParameterizedLabelledArray) = typeof(x)(deepcopy(getfield(x, :__rawdata)))\n\n\"\"\"\n$(SIGNATURES)\n\nCopies one `ParameterizedLabelledArray` to another.\n\n$__LABELLED_ARRAYS_CREDITS\n\"\"\"\nBase.copyto!(x::C,y::C) where C <: ParameterizedLabelledArray = copyto!(getfield(x,:__rawdata),getfield(y,:__rawdata))\n\n\"\"\"\n$(SIGNATURES)\n\nProvides `unsafe_convert` for `ParameterizedLabelledArray` types for use with LAPACK.\n\n$__LABELLED_ARRAYS_CREDITS\n\"\"\"\nBase.unsafe_convert(::Type{Ptr{T}}, a::ParameterizedLabelledArray{F}) where {T, F} = Base.unsafe_convert(Ptr{T}, getfield(a,:__rawdata))\n\n\"\"\"\n$(SIGNATURES)\n\nConverts the underlying floating point type for a `ParameterizedLabelledArray`.\n\n$__LABELLED_ARRAYS_CREDITS\n\"\"\"\nBase.convert(::Type{T}, x) where {T<:ParameterizedLabelledArray} = T(x)\n\n\"\"\"\n$(SIGNATURES)\n\nConverts the underlying floating point type for a `ParameterizedLabelledArray`.\n\n$__LABELLED_ARRAYS_CREDITS\n\"\"\"\nBase.convert(::Type{T}, x::T) where {T<:ParameterizedLabelledArray} = x\n\n\"\"\"\n$(SIGNATURES)\n\nConverts the underlying floating point type for a `ParameterizedLabelledArray`.\n\n$__LABELLED_ARRAYS_CREDITS\n\"\"\"\nBase.convert(::Type{<:Array},x::ParameterizedLabelledArray) = convert(Array, getfield(x,:__rawdata))\n\n\"\"\"\n$(SIGNATURES)\n\nReshapes a `ParameterizedLabelledArray`.\n\n$__LABELLED_ARRAYS_CREDITS\n\"\"\"\nArrayInterface.restructure(x::ParameterizedLabelledArray{F1}, y::ParameterizedLabelledArray{F2}) where {F1, F2} = reshape(y, size(x)...)\n\n\"\"\"\n$(SIGNATURES)\n\nImplements `dataids` for a `ParameterizedLabelledArray` instance.\n\n$__LABELLED_ARRAYS_CREDITS\n\"\"\"\nBase.dataids(A::ParameterizedLabelledArray) = Base.dataids(A.__rawdata)\n\n\"\"\"\n$(SIGNATURES)\n\nReturns the index of the `ParameterizedLabelledArray`.\n\n$__LABELLED_ARRAYS_CREDITS\n\"\"\"\nBase.getindex(state::ParameterizedLabelledArray, args...) = Base.getindex(state.__rawdata, args...)\n\n\"\"\"\n$(SIGNATURES)\n\nSets the index of the `ParameterizedLabelledArray`.\n\n$__LABELLED_ARRAYS_CREDITS\n\"\"\"\nBase.setindex!(state::ParameterizedLabelledArray, args...) = Base.setindex!(state.__rawdata, args...)\n\n\"\"\"\n$(SIGNATURES)\n\nReturns the memory stride for any `ParameterizedLabelledArray`.\n\n$__LABELLED_ARRAYS_CREDITS\n\"\"\"\nBase.elsize(::ParameterizedLabelledArray{F}) where F = sizeof(F)\n", "meta": {"hexsha": "d3c8e662778d7b666d85cb008f8e0a8bac975fb9", "size": 6644, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/States/Common/ParameterizedLabelledArrays.jl", "max_stars_repo_name": "pbouffard/GeneralAstrodynamics.jl", "max_stars_repo_head_hexsha": "80f175a5b3c6dac2140e645b016d39f131ecea05", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 13, "max_stars_repo_stars_event_min_datetime": "2021-05-25T00:32:05.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-22T23:31:08.000Z", "max_issues_repo_path": "src/States/Common/ParameterizedLabelledArrays.jl", "max_issues_repo_name": "pbouffard/GeneralAstrodynamics.jl", "max_issues_repo_head_hexsha": "80f175a5b3c6dac2140e645b016d39f131ecea05", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 16, "max_issues_repo_issues_event_min_datetime": "2020-10-11T16:07:53.000Z", "max_issues_repo_issues_event_max_datetime": "2021-04-10T12:28:56.000Z", "max_forks_repo_path": "src/States/Common/ParameterizedLabelledArrays.jl", "max_forks_repo_name": "cadojo/UnitfulAstrodynamics.jl", "max_forks_repo_head_hexsha": "c24888c1500450ea420169af4ca54dbd5bc99092", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2021-08-28T12:13:09.000Z", "max_forks_repo_forks_event_max_datetime": "2021-08-28T12:13:09.000Z", "avg_line_length": 26.576, "max_line_length": 240, "alphanum_fraction": 0.757525587, "num_tokens": 1727, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4960938294709195, "lm_q2_score": 0.12085324357297283, "lm_q1q2_score": 0.059954548408097885}}
{"text": "function pattern(n::Integer)::String\n if n < 1\n return \"\"\n end\n output = Char[]\n sizehint!(output,n^2)\n # only convert one time the integers to strings\n numbers = Array{String}(undef,n)\n map!( string, numbers, 1:n )\n # generate the pattern\n for last = 1:n\n for idx = n:-1:last\n append!(output, numbers[idx] )\n end\n push!(output, '\\n' )\n end\n # end-1 because no \\n at the end\n String( output[1:end-1] )\nend", "meta": {"hexsha": "78e934200092175c75c6e219195cf763ef35f847", "size": 446, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "7_kyu/Complete_The_Pattern_2.jl", "max_stars_repo_name": "UlrichBerntien/Codewars-Katas", "max_stars_repo_head_hexsha": "bbd025e67aa352d313564d3862db19fffa39f552", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "7_kyu/Complete_The_Pattern_2.jl", "max_issues_repo_name": "UlrichBerntien/Codewars-Katas", "max_issues_repo_head_hexsha": "bbd025e67aa352d313564d3862db19fffa39f552", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "7_kyu/Complete_The_Pattern_2.jl", "max_forks_repo_name": "UlrichBerntien/Codewars-Katas", "max_forks_repo_head_hexsha": "bbd025e67aa352d313564d3862db19fffa39f552", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 23.4736842105, "max_line_length": 49, "alphanum_fraction": 0.600896861, "num_tokens": 139, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4960938294709195, "lm_q2_score": 0.12085323249047067, "lm_q1q2_score": 0.059954542910136945}}
{"text": "println(\"Hello, World!\")\n\n# This is a comment\n\n#= This is a multi-line comment\nThe second line of the comment =#\n\n# println (\"this is not allowed\")\nprintln(\"This is allowed.\")\nprintln(\"This is also allowed.\");\n\nprintln(1 + 2)\n1 + 2 # Will not print in script\n\n# Installing, updating and removing packages\nusing Pkg # Alternatively, enter pkg mode in REPL with ]\nPkg.add(\"Example\")\nPkg.status()\nPkg.update(\"Example\") # Pkg.update() updates all packages\nPkg.rm(\"Example\") # removes from environment, does not delete files\n\n# Environments\n# In pkg mode:\n# activate tutorial\n# st\n# add Example\n# activate # goes back to default environment", "meta": {"hexsha": "c73c73fb0f6ba78233bac5248427c754f4937470", "size": 635, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/intro.jl", "max_stars_repo_name": "EnzioKam/learning_julia", "max_stars_repo_head_hexsha": "a673e2d721d734333c79bcc4cf04f678ed378bdb", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/intro.jl", "max_issues_repo_name": "EnzioKam/learning_julia", "max_issues_repo_head_hexsha": "a673e2d721d734333c79bcc4cf04f678ed378bdb", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/intro.jl", "max_forks_repo_name": "EnzioKam/learning_julia", "max_forks_repo_head_hexsha": "a673e2d721d734333c79bcc4cf04f678ed378bdb", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 23.5185185185, "max_line_length": 67, "alphanum_fraction": 0.7228346457, "num_tokens": 157, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.1602660403494035, "lm_q2_score": 0.37387583672470853, "lm_q1q2_score": 0.05991959993418913}}
{"text": "## Exercise 4-10\n## Write an appropriately general set of functions that can draw shapes as in Turtle pies.\n## https://benlauwens.github.io/ThinkJulia.jl/latest/book.html#fig04-3\nprintln(\"Ans: \")\n\nprintln(\"End.\")\n", "meta": {"hexsha": "9561a74f1b512e533cf77f8a3189c92ce76cc484", "size": 213, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Chapter4/ex10.jl", "max_stars_repo_name": "yashppawar/ThinkJuliaExercises.jl", "max_stars_repo_head_hexsha": "72145b969dc51ebcac413a10004175ebc63cd5c2", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2022-02-13T14:11:30.000Z", "max_stars_repo_stars_event_max_datetime": "2022-02-13T14:11:30.000Z", "max_issues_repo_path": "Chapter4/ex10.jl", "max_issues_repo_name": "yashppawar/ThinkJuliaExercises.jl", "max_issues_repo_head_hexsha": "72145b969dc51ebcac413a10004175ebc63cd5c2", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Chapter4/ex10.jl", "max_forks_repo_name": "yashppawar/ThinkJuliaExercises.jl", "max_forks_repo_head_hexsha": "72145b969dc51ebcac413a10004175ebc63cd5c2", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 30.4285714286, "max_line_length": 90, "alphanum_fraction": 0.7417840376, "num_tokens": 58, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4263215925474903, "lm_q2_score": 0.1403362422992606, "lm_q1q2_score": 0.059828370309151245}}
{"text": "import MathOptInterface\nusing Test\n\nconst MOI = MathOptInterface\nconst MOIU = MOI.Utilities\nconst MOF = MOI.FileFormats.MOF\n\nconst TEST_MOF_FILE = \"test.mof.json\"\n\n@test sprint(show, MOF.Model()) == \"A MathOptFormat Model\"\n\ninclude(\"nonlinear.jl\")\n\nstruct UnsupportedSet <: MOI.AbstractSet end\nstruct UnsupportedFunction <: MOI.AbstractFunction end\n\nfunction test_model_equality(model_string, variables, constraints; suffix=\"\")\n model = MOF.Model(validate = true)\n MOIU.loadfromstring!(model, model_string)\n MOI.write_to_file(model, TEST_MOF_FILE * suffix)\n model_2 = MOF.Model()\n MOI.read_from_file(model_2, TEST_MOF_FILE * suffix)\n MOIU.test_models_equal(model, model_2, variables, constraints)\n MOF.validate(TEST_MOF_FILE * suffix)\nend\n\n@testset \"Error handling: read_from_file\" begin\n failing_models_dir = joinpath(@__DIR__, \"failing_models\")\n\n @testset \"Non-empty model\" begin\n model = MOF.Model(warn=true)\n MOI.add_variable(model)\n @test !MOI.is_empty(model)\n exception = ErrorException(\n \"Cannot read model from file as destination model is not empty.\")\n @test_throws exception MOI.read_from_file(\n model, joinpath(@__DIR__, \"empty_model.mof.json\"))\n options = MOF.get_options(model)\n @test options.warn\n MOI.empty!(model)\n @test MOI.is_empty(model)\n MOI.read_from_file(\n model, joinpath(@__DIR__, \"empty_model.mof.json\"))\n options2 = MOF.get_options(model)\n @test options2.warn\n end\n\n @testset \"$(filename)\" for filename in filter(\n f -> endswith(f, \".mof.json\"), readdir(failing_models_dir))\n @test_throws Exception MOI.read_from_file(MOF.Model(),\n joinpath(failing_models_dir, filename))\n end\nend\n@testset \"Names\" begin\n @testset \"Blank variable name\" begin\n model = MOF.Model()\n variable_index = MOI.add_variable(model)\n @test_throws Exception MOF.moi_to_object(variable_index, model)\n MOI.FileFormats.create_unique_names(model, warn=true)\n @test MOF.moi_to_object(variable_index, model) ==\n MOF.OrderedObject(\"name\" => \"x1\")\n end\n @testset \"Duplicate variable name\" begin\n model = MOF.Model()\n x = MOI.add_variable(model)\n MOI.set(model, MOI.VariableName(), x, \"x\")\n y = MOI.add_variable(model)\n MOI.set(model, MOI.VariableName(), y, \"x\")\n @test MOF.moi_to_object(x, model) == MOF.OrderedObject(\"name\" => \"x\")\n @test MOF.moi_to_object(y, model) == MOF.OrderedObject(\"name\" => \"x\")\n MOI.FileFormats.create_unique_names(model, warn=true)\n @test MOF.moi_to_object(x, model) == MOF.OrderedObject(\"name\" => \"x\")\n @test MOF.moi_to_object(y, model) == MOF.OrderedObject(\"name\" => \"x_1\")\n end\n @testset \"Blank constraint name\" begin\n model = MOF.Model()\n x = MOI.add_variable(model)\n MOI.set(model, MOI.VariableName(), x, \"x\")\n c = MOI.add_constraint(model, MOI.SingleVariable(x), MOI.ZeroOne())\n name_map = Dict(x => \"x\")\n MOI.FileFormats.create_unique_names(model, warn=true)\n @test MOF.moi_to_object(c, model, name_map)[\"name\"] == \"c1\"\n end\n @testset \"Duplicate constraint name\" begin\n model = MOF.Model()\n x = MOI.add_variable(model)\n MOI.set(model, MOI.VariableName(), x, \"x\")\n c1 = MOI.add_constraint(model, MOI.SingleVariable(x), MOI.LessThan(1.0))\n c2 = MOI.add_constraint(model, MOI.SingleVariable(x), MOI.GreaterThan(0.0))\n MOI.set(model, MOI.ConstraintName(), c1, \"c\")\n MOI.set(model, MOI.ConstraintName(), c2, \"c\")\n name_map = Dict(x => \"x\")\n @test MOF.moi_to_object(c1, model, name_map)[\"name\"] == \"c\"\n @test MOF.moi_to_object(c2, model, name_map)[\"name\"] == \"c\"\n MOI.FileFormats.create_unique_names(model, warn=true)\n @test MOF.moi_to_object(c1, model, name_map)[\"name\"] == \"c_1\"\n @test MOF.moi_to_object(c2, model, name_map)[\"name\"] == \"c\"\n end\nend\n@testset \"round trips\" begin\n @testset \"Empty model\" begin\n model = MOF.Model(validate = true)\n MOI.write_to_file(model, TEST_MOF_FILE)\n model_2 = MOF.Model(validate = true)\n MOI.read_from_file(model_2, TEST_MOF_FILE)\n MOIU.test_models_equal(model, model_2, String[], String[])\n end\n @testset \"FEASIBILITY_SENSE\" begin\n model = MOF.Model(validate = true)\n x = MOI.add_variable(model)\n MOI.set(model, MOI.VariableName(), x, \"x\")\n MOI.set(model, MOI.ObjectiveSense(), MOI.FEASIBILITY_SENSE)\n MOI.write_to_file(model, TEST_MOF_FILE)\n model_2 = MOF.Model(validate = true)\n MOI.read_from_file(model_2, TEST_MOF_FILE)\n MOIU.test_models_equal(model, model_2, [\"x\"], String[])\n end\n @testset \"Empty function term\" begin\n model = MOF.Model(validate = true)\n x = MOI.add_variable(model)\n MOI.set(model, MOI.VariableName(), x, \"x\")\n c = MOI.add_constraint(model,\n MOI.ScalarAffineFunction(MOI.ScalarAffineTerm{Float64}[], 0.0),\n MOI.GreaterThan(1.0)\n )\n MOI.set(model, MOI.ConstraintName(), c, \"c\")\n MOI.write_to_file(model, TEST_MOF_FILE)\n model_2 = MOF.Model(validate = true)\n MOI.read_from_file(model_2, TEST_MOF_FILE)\n MOIU.test_models_equal(model, model_2, [\"x\"], [\"c\"])\n end\n @testset \"min objective\" begin\n test_model_equality(\"\"\"\n variables: x\n minobjective: x\n \"\"\", [\"x\"], String[])\n end\n @testset \"max objective\" begin\n test_model_equality(\"\"\"\n variables: x\n maxobjective: x\n \"\"\", [\"x\"], String[], suffix=\".gz\")\n end\n @testset \"min scalaraffine\" begin\n test_model_equality(\"\"\"\n variables: x\n minobjective: 1.2x + 0.5\n \"\"\", [\"x\"], String[])\n end\n @testset \"max scalaraffine\" begin\n test_model_equality(\"\"\"\n variables: x\n maxobjective: 1.2x + 0.5\n \"\"\", [\"x\"], String[], suffix=\".gz\")\n end\n @testset \"singlevariable-in-lower\" begin\n test_model_equality(\"\"\"\n variables: x\n minobjective: 1.2x + 0.5\n c1: x >= 1.0\n \"\"\", [\"x\"], [\"c1\"])\n end\n @testset \"singlevariable-in-upper\" begin\n test_model_equality(\"\"\"\n variables: x\n maxobjective: 1.2x + 0.5\n c1: x <= 1.0\n \"\"\", [\"x\"], [\"c1\"], suffix=\".gz\")\n end\n @testset \"singlevariable-in-interval\" begin\n test_model_equality(\"\"\"\n variables: x\n minobjective: 1.2x + 0.5\n c1: x in Interval(1.0, 2.0)\n \"\"\", [\"x\"], [\"c1\"])\n end\n @testset \"singlevariable-in-equalto\" begin\n test_model_equality(\"\"\"\n variables: x\n minobjective: 1.2x + 0.5\n c1: x == 1.0\n \"\"\", [\"x\"], [\"c1\"])\n end\n @testset \"singlevariable-in-zeroone\" begin\n test_model_equality(\"\"\"\n variables: x\n minobjective: 1.2x + 0.5\n c1: x in ZeroOne()\n \"\"\", [\"x\"], [\"c1\"])\n end\n @testset \"singlevariable-in-integer\" begin\n test_model_equality(\"\"\"\n variables: x\n minobjective: 1.2x + 0.5\n c1: x in Integer()\n \"\"\", [\"x\"], [\"c1\"])\n end\n @testset \"singlevariable-in-Semicontinuous\" begin\n test_model_equality(\"\"\"\n variables: x\n minobjective: 1.2x + 0.5\n c1: x in Semicontinuous(1.0, 2.0)\n \"\"\", [\"x\"], [\"c1\"])\n end\n @testset \"singlevariable-in-Semiinteger\" begin\n test_model_equality(\"\"\"\n variables: x\n minobjective: 1.2x + 0.5\n c1: x in Semiinteger(1.0, 2.0)\n \"\"\", [\"x\"], [\"c1\"])\n end\n @testset \"scalarquadratic-objective\" begin\n test_model_equality(\"\"\"\n variables: x\n minobjective: 1.0*x*x + -2.0x + 1.0\n \"\"\", [\"x\"], String[])\n end\n @testset \"SOS1\" begin\n test_model_equality(\"\"\"\n variables: x, y, z\n minobjective: x\n c1: [x, y, z] in SOS1([1.0, 2.0, 3.0])\n \"\"\", [\"x\", \"y\", \"z\"], [\"c1\"])\n end\n @testset \"SOS2\" begin\n test_model_equality(\"\"\"\n variables: x, y, z\n minobjective: x\n c1: [x, y, z] in SOS2([1.0, 2.0, 3.0])\n \"\"\", [\"x\", \"y\", \"z\"], [\"c1\"])\n end\n @testset \"Reals\" begin\n test_model_equality(\"\"\"\n variables: x, y, z\n minobjective: x\n c1: [x, y, z] in Reals(3)\n \"\"\", [\"x\", \"y\", \"z\"], [\"c1\"])\n end\n @testset \"Zeros\" begin\n test_model_equality(\"\"\"\n variables: x, y, z\n minobjective: x\n c1: [x, y, z] in Zeros(3)\n \"\"\", [\"x\", \"y\", \"z\"], [\"c1\"])\n end\n @testset \"Nonnegatives\" begin\n test_model_equality(\"\"\"\n variables: x, y, z\n minobjective: x\n c1: [x, y, z] in Nonnegatives(3)\n \"\"\", [\"x\", \"y\", \"z\"], [\"c1\"])\n end\n @testset \"Nonpositives\" begin\n test_model_equality(\"\"\"\n variables: x, y, z\n minobjective: x\n c1: [x, y, z] in Nonpositives(3)\n \"\"\", [\"x\", \"y\", \"z\"], [\"c1\"])\n end\n @testset \"PowerCone\" begin\n test_model_equality(\"\"\"\n variables: x, y, z\n minobjective: x\n c1: [x, y, z] in PowerCone(2.0)\n \"\"\", [\"x\", \"y\", \"z\"], [\"c1\"])\n end\n @testset \"DualPowerCone\" begin\n test_model_equality(\"\"\"\n variables: x, y, z\n minobjective: x\n c1: [x, y, z] in DualPowerCone(0.5)\n \"\"\", [\"x\", \"y\", \"z\"], [\"c1\"])\n end\n @testset \"GeometricMeanCone\" begin\n test_model_equality(\"\"\"\n variables: x, y, z\n minobjective: x\n c1: [x, y, z] in GeometricMeanCone(3)\n \"\"\", [\"x\", \"y\", \"z\"], [\"c1\"])\n end\n @testset \"vectoraffine-in-zeros\" begin\n test_model_equality(\"\"\"\n variables: x, y\n minobjective: x\n c1: [1.0x + -3.0, 2.0y + -4.0] in Zeros(2)\n \"\"\", [\"x\", \"y\"], [\"c1\"])\n end\n @testset \"vectorquadratic-in-nonnegatives\" begin\n test_model_equality(\"\"\"\n variables: x, y\n minobjective: x\n c1: [1.0*x*x + -2.0x + 1.0, 2.0y + -4.0] in Nonnegatives(2)\n \"\"\", [\"x\", \"y\"], [\"c1\"])\n end\n @testset \"ExponentialCone\" begin\n test_model_equality(\"\"\"\n variables: x, y, z\n minobjective: x\n c1: [x, y, z] in ExponentialCone()\n \"\"\", [\"x\", \"y\", \"z\"], [\"c1\"])\n end\n @testset \"DualExponentialCone\" begin\n test_model_equality(\"\"\"\n variables: x, y, z\n minobjective: x\n c1: [x, y, z] in DualExponentialCone()\n \"\"\", [\"x\", \"y\", \"z\"], [\"c1\"])\n end\n @testset \"SecondOrderCone\" begin\n test_model_equality(\"\"\"\n variables: x, y, z\n minobjective: x\n c1: [x, y, z] in SecondOrderCone(3)\n \"\"\", [\"x\", \"y\", \"z\"], [\"c1\"])\n end\n @testset \"RotatedSecondOrderCone\" begin\n test_model_equality(\"\"\"\n variables: x, y, z\n minobjective: x\n c1: [x, y, z] in RotatedSecondOrderCone(3)\n \"\"\", [\"x\", \"y\", \"z\"], [\"c1\"])\n end\n @testset \"PositiveSemidefiniteConeTriangle\" begin\n test_model_equality(\"\"\"\n variables: x1, x2, x3\n minobjective: x1\n c1: [x1, x2, x3] in PositiveSemidefiniteConeTriangle(2)\n \"\"\", [\"x1\", \"x2\", \"x3\"], [\"c1\"])\n end\n @testset \"PositiveSemidefiniteConeSquare\" begin\n test_model_equality(\"\"\"\n variables: x1, x2, x3, x4\n minobjective: x1\n c1: [x1, x2, x3, x4] in PositiveSemidefiniteConeSquare(2)\n \"\"\", [\"x1\", \"x2\", \"x3\", \"x4\"], [\"c1\"])\n end\n @testset \"LogDetConeTriangle\" begin\n test_model_equality(\"\"\"\n variables: t, u, x1, x2, x3\n minobjective: x1\n c1: [t, u, x1, x2, x3] in LogDetConeTriangle(2)\n \"\"\", [\"t\", \"u\", \"x1\", \"x2\", \"x3\"], [\"c1\"])\n end\n @testset \"LogDetConeSquare\" begin\n test_model_equality(\"\"\"\n variables: t, u, x1, x2, x3, x4\n minobjective: x1\n c1: [t, u, x1, x2, x3, x4] in LogDetConeSquare(2)\n \"\"\", [\"t\", \"u\", \"x1\", \"x2\", \"x3\", \"x4\"], [\"c1\"])\n end\n @testset \"RootDetConeTriangle\" begin\n test_model_equality(\"\"\"\n variables: t, x1, x2, x3\n minobjective: x1\n c1: [t, x1, x2, x3] in RootDetConeTriangle(2)\n \"\"\", [\"t\", \"x1\", \"x2\", \"x3\"], [\"c1\"])\n end\n @testset \"RootDetConeSquare\" begin\n test_model_equality(\"\"\"\n variables: t, x1, x2, x3, x4\n minobjective: x1\n c1: [t, x1, x2, x3, x4] in RootDetConeSquare(2)\n \"\"\", [\"t\", \"x1\", \"x2\", \"x3\", \"x4\"], [\"c1\"])\n end\n @testset \"IndicatorSet\" begin\n test_model_equality(\"\"\"\n variables: x, y\n minobjective: x\n c1: [x, y] in IndicatorSet{ACTIVATE_ON_ONE}(GreaterThan(1.0))\n c2: x >= 0.0\n \"\"\", [\"x\", \"y\"], [\"c1\", \"c2\"])\n\n test_model_equality(\"\"\"\n variables: x, y\n minobjective: x\n c1: [x, y] in IndicatorSet{ACTIVATE_ON_ZERO}(GreaterThan(1.0))\n c2: x >= 0.0\n \"\"\", [\"x\", \"y\"], [\"c1\", \"c2\"])\n end\n @testset \"NormOneCone\" begin\n test_model_equality(\"\"\"\n variables: x, y\n minobjective: x\n c1: [x, y] in NormOneCone(2)\n c2: x >= 0.0\n \"\"\", [\"x\", \"y\"], [\"c1\", \"c2\"])\n end\n @testset \"NormInfinityCone\" begin\n test_model_equality(\"\"\"\n variables: x, y\n minobjective: x\n c1: [x, y] in NormInfinityCone(2)\n c2: x >= 0.0\n \"\"\", [\"x\", \"y\"], [\"c1\", \"c2\"])\n end\n @testset \"RelativeEntropyCone\" begin\n test_model_equality(\"\"\"\n variables: x, y, z\n minobjective: x\n c1: [x, y, z] in RelativeEntropyCone(3)\n c2: x >= 0.0\n \"\"\", [\"x\", \"y\", \"z\"], [\"c1\", \"c2\"])\n end\n @testset \"NormSpectralCone\" begin\n test_model_equality(\"\"\"\n variables: x, y, z\n minobjective: x\n c1: [x, y, z] in NormSpectralCone(1, 2)\n \"\"\", [\"x\", \"y\", \"z\"], [\"c1\"])\n end\n @testset \"NormNuclearCone\" begin\n test_model_equality(\"\"\"\n variables: x, y, z\n minobjective: x\n c1: [x, y, z] in NormNuclearCone(1, 2)\n \"\"\", [\"x\", \"y\", \"z\"], [\"c1\"])\n end\n # Clean up\n sleep(1.0) # allow time for unlink to happen\n rm(TEST_MOF_FILE, force=true)\n rm(TEST_MOF_FILE * \".gz\", force=true)\nend\n", "meta": {"hexsha": "f7b0637c15611f38918f57fc9d71790a92320927", "size": 14702, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/FileFormats/MOF/MOF.jl", "max_stars_repo_name": "egbuck/MathOptInterface.jl", "max_stars_repo_head_hexsha": "a95e7d68adb2e60e40dd8c0bffe4fdbcfde24590", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "test/FileFormats/MOF/MOF.jl", "max_issues_repo_name": "egbuck/MathOptInterface.jl", "max_issues_repo_head_hexsha": "a95e7d68adb2e60e40dd8c0bffe4fdbcfde24590", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "test/FileFormats/MOF/MOF.jl", "max_forks_repo_name": "egbuck/MathOptInterface.jl", "max_forks_repo_head_hexsha": "a95e7d68adb2e60e40dd8c0bffe4fdbcfde24590", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 34.756501182, "max_line_length": 83, "alphanum_fraction": 0.5340089784, "num_tokens": 4531, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4882833952958347, "lm_q2_score": 0.12252320771203078, "lm_q1q2_score": 0.05982604786416719}}
{"text": "using Distributed\n\nusing Test\n\n@everywhere begin\n using MLJBase\n using MLJTuning\n using ..Models\n import ComputationalResources: CPU1, CPUProcesses, CPUThreads\nend\n\nusing Random\nRandom.seed!(1234*myid())\nusing .TestUtilities\n\nN = 30\nx1 = rand(N);\nx2 = rand(N);\nx3 = rand(N);\nX = (x1=x1, x2=x2, x3=x3);\ny = 2*x1 .+ 5*x2 .- 3*x3 .+ 0.4*rand(N);\n\nm(K) = KNNRegressor(K=K)\nr = [m(K) for K in 13:-1:2]\n\n# TODO: replace the above with the line below and post an issue on\n# the failure (a bug in Distributed, I reckon):\n# r = (m(K) for K in 13:-1:2)\n\n@testset \"constructor\" begin\n @test_throws(MLJTuning.ERR_SPECIFY_RANGE,\n TunedModel(model=first(r), tuning=Grid(), measure=rms))\n @test_throws(MLJTuning.ERR_SPECIFY_RANGE,\n TunedModel(model=first(r), measure=rms))\n @test_throws(MLJTuning.ERR_BOTH_DISALLOWED,\n TunedModel(model=first(r),\n models=r, tuning=Explicit(), measure=rms))\n tm = TunedModel(models=r, tuning=Explicit(), measure=rms)\n @test tm.tuning isa Explicit && tm.range ==r && tm.model == first(r)\n @test input_scitype(tm) == Unknown\n @test TunedModel(models=r, measure=rms) == tm\n @test_logs (:info, r\"No measure\") @test TunedModel(models=r) == tm\n\n @test_throws(MLJTuning.ERR_SPECIFY_MODEL,\n TunedModel(range=r, measure=rms))\n @test_throws(MLJTuning.ERR_MODEL_TYPE,\n TunedModel(model=42, tuning=Grid(),\n range=r, measure=rms))\n @test_logs (:info, MLJTuning.INFO_MODEL_IGNORED) tm =\n TunedModel(model=42, tuning=Explicit(), range=r, measure=rms)\n @test_logs (:info, r\"No measure\") tm =\n TunedModel(model=first(r), range=r)\n @test_throws(MLJTuning.ERR_SPECIFY_RANGE_OR_MODELS,\n TunedModel(tuning=Explicit(), measure=rms))\n @test_throws(MLJTuning.ERR_NEED_EXPLICIT,\n TunedModel(models=r, tuning=Grid()))\n tm = TunedModel(model=first(r), range=r, measure=rms)\n @test tm.tuning isa Grid\n @test input_scitype(tm) == Table(Continuous)\nend\n\nresults = [(evaluate(model, X, y,\n resampling=CV(nfolds=2),\n measure=rms,\n verbosity=0,)).measurement[1] for model in r]\n\n@testset \"measure compatibility check\" begin\n tm = TunedModel(model=first(r), tuning=Explicit(),\n range=r, resampling=CV(nfolds=2),\n measures=cross_entropy)\n @test_logs((:error, r\"Problem\"),\n (:info, r\"\"),\n (:info, r\"\"),\n @test_throws ArgumentError fit(tm, 0, X, y))\nend\n\n@testset_accelerated \"basic fit (CPU1)\" accel begin\n printstyled(\"\\n Testing progressmeter basic fit with $(accel) and CPU1 resampling \\n\", color=:bold)\n best_index = argmin(results)\n tm = TunedModel(model=first(r), tuning=Explicit(),\n range=r, resampling=CV(nfolds=2),\n measures=[rms, l1], acceleration=accel)\n verbosity = accel isa CPU1 ? 2 : 1\n fitresult, meta_state, _report = fit(tm, verbosity, X, y);\n history, _, state = meta_state;\n results2 = map(event -> event.measurement[1], history)\n @test results2 \u2248 results\n @test fitresult.model == collect(r)[best_index]\n @test _report.best_model == collect(r)[best_index]\n @test _report.history[5] == MLJTuning.delete(history[5], :metadata)\n\n # training_losses:\n losses = training_losses(tm, _report)\n @test all(eachindex(losses)) do i\n minimum(results[1:i]) == losses[i]\n end\n @test MLJBase.iteration_parameter(tm) == :n\n\nend\n\n@static if VERSION >= v\"1.3.0-DEV.573\"\n@testset_accelerated \"Basic fit (CPUThreads)\" accel begin\n printstyled(\"\\n Testing progressmeter basic fit with $(accel) and CPUThreads resampling \\n\", color=:bold)\n tm = TunedModel(model=first(r), tuning=Explicit(),\n range=r, resampling=CV(nfolds=2),\n measures=[rms, l1], acceleration= CPUThreads(),\n acceleration_resampling=accel)\n fitresult, meta_state, report = fit(tm, 1, X, y);\n history, _, state = meta_state;\n results3 = map(event -> event.measurement[1], history)\n @test results3 \u2248 results\nend\nend\n@testset_accelerated \"Basic fit (CPUProcesses)\" accel begin\n printstyled(\"\\n Testing progressmeter basic fit with $(accel) and CPUProcesses resampling \\n\", color=:bold)\n best_index = argmin(results)\n tm = TunedModel(model=first(r), tuning=Explicit(),\n range=r, resampling=CV(nfolds=2),\n measures=[rms, l1], acceleration=CPUProcesses(),\n acceleration_resampling=accel)\n fitresult, meta_state, report = fit(tm, 1, X, y);\n history, _, state = meta_state;\n results4 = map(event -> event.measurement[1], history)\n @test results4 \u2248 results\nend\n\n@testset_accelerated(\"under/over supply of models\", accel, begin\n tm = TunedModel(model=first(r), tuning=Explicit(),\n range=r, measures=[rms, l1],\n acceleration=accel,\n resampling=CV(nfolds=2),\n n=4)\n mach = machine(tm, X, y)\n fit!(mach, verbosity=0)\n history = MLJBase.report(mach).history\n @test map(event -> event.measurement[1], history) \u2248 results[1:4]\n\n tm.n += 2\n @test_logs((:info, r\"^Updating\"),\n (:info, r\"^Attempting to add 2.*to 6\"),\n fit!(mach, verbosity=1))\n history = MLJBase.report(mach).history\n @test map(event -> event.measurement[1], history) \u2248 results[1:6]\n\n tm.n=100\n @test_logs (:info, r\"Only 12\") fit!(mach, verbosity=0)\n history = MLJBase.report(mach).history\n @test map(event -> event.measurement[1], history) \u2248 results\nend)\n\n@everywhere begin\n\n # variation of the Explicit strategy that annotates the models\n # with metadata\n mutable struct MockExplicit <: MLJTuning.TuningStrategy end\n\n annotate(model) = (model, params(model)[1])\n\n _length(x) = length(x)\n _length(::Nothing) = 0\n MLJTuning.models(tuning::MockExplicit,\n model,\n history,\n state,\n n_remaining,\n verbosity) =\n annotate.(state)[_length(history) + 1:end], state\n\n function default_n(tuning::Explicit, range)\n try\n length(range)\n catch MethodError\n DEFAULT_N\n end\n\n end\n\nend\n\n@testset_accelerated(\"passing of model metadata\", accel,\n begin\n tm = TunedModel(model=first(r), tuning=MockExplicit(),\n range=r, resampling=CV(nfolds=2),\n measures=[rms, l1], acceleration=accel)\n fitresult, meta_state, report = fit(tm, 0, X, y);\n history, _, state = meta_state;\n @test all(history) do event\n event.metadata == event.model.K\n end\n end)\n\n\n\n@testset \"data caching\" begin\n X = (x1= ones(5),);\n y = coerce(collect(\"abcaa\"), Multiclass);\n\n m(b) = ConstantClassifier(testing=true, bogus=b)\n r = [m(b) for b in 1:5]\n\n tuned_model = TunedModel(model=first(r), tuning=Explicit(),\n range=r, resampling=Holdout(fraction_train=0.8),\n measure=log_loss, cache=true)\n\n # There is reformatting and resampling of X and y for training of\n # first model, and then resampling on the prediction side only\n # thereafter. Then, finally, for training on all supplied data, there\n # is reformatting and resampling again:\n @test_logs(\n (:info, \"reformatting X, y\"), # fit 1\n (:info, \"resampling X, y\"), # fit 1\n (:info, \"resampling X\"), # predict 1\n (:info, \"resampling X\"), # predict 2\n (:info, \"resampling X\"), # predict 3\n (:info, \"resampling X\"), # predict 4\n (:info, \"resampling X\"), # predict 5\n (:info, \"reformatting X, y\"), # fit on all data\n (:info, \"resampling X, y\"), # fit one all data\n fit(tuned_model, 0, X, y)\n );\n\n # Otherwise, resampling and reformatting happen for every model\n # trained, and we get a reformat on the prediction side every\n # time:\n tuned_model.cache = false\n @test_logs(\n (:info, \"reformatting X, y\"), # fit 1\n (:info, \"resampling X, y\"), # fit 1\n (:info, \"reformatting X\"), # predict 1\n (:info, \"reformatting X, y\"), # fit 2\n (:info, \"resampling X, y\"), # fit 2\n (:info, \"reformatting X\"), # predict 2\n (:info, \"reformatting X, y\"), # fit 3\n (:info, \"resampling X, y\"), # fit 3\n (:info, \"reformatting X\"), # predict 3\n (:info, \"reformatting X, y\"), # fit 4\n (:info, \"resampling X, y\"), # fit 4\n (:info, \"reformatting X\"), # predict 4\n (:info, \"reformatting X, y\"), # fit 5\n (:info, \"resampling X, y\"), # fit 5\n (:info, \"reformatting X\"), # predict 5\n (:info, \"reformatting X, y\"), # fit on all data\n (:info, \"resampling X, y\"), # fit on all data\n fit(tuned_model, 0, X, y)\n );\nend\n\n@testset \"issue #128\" begin\n X, y = make_regression(10, 2)\n dtc = DecisionTreeRegressor()\n r = range(dtc, :max_depth, lower=1, upper=50);\n\n tmodel = TunedModel(model=dtc, ranges=[r, ],\n tuning=Grid(resolution=50),\n measure=mae,\n n=48);\n mach = machine(tmodel, X, y)\n fit!(mach, verbosity=0);\n\n @test length(report(mach).history) == 48\n\n tmodel.n = 49\n fit!(mach, verbosity=0);\n\n @test length(report(mach).history) == 49\nend\n\n@testset_accelerated \"Resampling reproducibility\" accel begin\n X, y = make_regression(100, 2)\n dcr = DeterministicConstantRegressor()\n\n # Hold out reproducibility\n homodel = TunedModel(tuning=Explicit(),\n models=fill(dcr, 10),\n resampling=Holdout(rng=StableRNG(1234)),\n acceleration_resampling=accel,\n measure=mae)\n homach = machine(homodel, X, y)\n fit!(homach, verbosity=0);\n horep = report(homach)\n measurements = getproperty.(horep.history, :measurement)\n @test all(==(measurements[1]), measurements)\n\n # Cross-validation reproducibility\n cvmodel = TunedModel(tuning=Explicit(),\n models=fill(dcr, 10),\n resampling=CV(nfolds=5, rng=StableRNG(1234)),\n acceleration_resampling=accel,\n measure=mae)\n cvmach = machine(cvmodel, X, y)\n fit!(cvmach, verbosity=0);\n cvrep = report(cvmach)\n per_folds = getproperty.(cvrep.history, :per_fold)\n @test all(==(per_folds[1]), per_folds)\nend\n\n@testset \"deterministic metrics for probabilistic models\" begin\n\n # https://github.com/JuliaAI/MLJBase.jl/pull/599 allows mix of\n # deterministic and probabilistic metrics:\n X, y = MLJBase.make_blobs()\n model = DecisionTreeClassifier()\n range = MLJBase.range(model, :max_depth, values=[1,2])\n tmodel = TunedModel(model=model,\n range=range,\n measures=[MisclassificationRate(),\n LogLoss()])\n mach = machine(tmodel, X, y)\n @test_logs fit!(mach, verbosity=0)\n\nend\n\ntrue\n", "meta": {"hexsha": "849329f577cdeb225c55c1db75802e2ed6b97c51", "size": 11452, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/tuned_models.jl", "max_stars_repo_name": "alan-turing-institute/MLJTuning.jl", "max_stars_repo_head_hexsha": "b058e8df63ebbd67ef8df823480a42cfd45462b7", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 41, "max_stars_repo_stars_event_min_datetime": "2020-01-28T04:27:57.000Z", "max_stars_repo_stars_event_max_datetime": "2021-06-11T05:56:59.000Z", "max_issues_repo_path": "test/tuned_models.jl", "max_issues_repo_name": "alan-turing-institute/MLJTuning.jl", "max_issues_repo_head_hexsha": "b058e8df63ebbd67ef8df823480a42cfd45462b7", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 64, "max_issues_repo_issues_event_min_datetime": "2020-02-03T21:26:17.000Z", "max_issues_repo_issues_event_max_datetime": "2021-06-11T03:49:59.000Z", "max_forks_repo_path": "test/tuned_models.jl", "max_forks_repo_name": "alan-turing-institute/MLJTuning.jl", "max_forks_repo_head_hexsha": "b058e8df63ebbd67ef8df823480a42cfd45462b7", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 8, "max_forks_repo_forks_event_min_datetime": "2020-02-08T11:38:35.000Z", "max_forks_repo_forks_event_max_datetime": "2021-05-29T10:25:21.000Z", "avg_line_length": 36.4713375796, "max_line_length": 111, "alphanum_fraction": 0.5832169053, "num_tokens": 3081, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4882833952958347, "lm_q2_score": 0.12252320450794524, "lm_q1q2_score": 0.05982604629966542}}
{"text": "module ResizableArraysTests\n\nusing Test\nusing ResizableArrays\nusing ResizableArrays: checkdimension, checkdimensions, _same_elements\nusing Base: unsafe_convert, elsize\n\n# FIXME: used @generated\nslice(A::AbstractArray{<:Any,2}, I) = A[:,I]\nslice(A::AbstractArray{<:Any,3}, I) = A[:,:,I]\nslice(A::AbstractArray{<:Any,4}, I) = A[:,:,:,I]\nslice(A::AbstractArray{<:Any,5}, I) = A[:,:,:,:,I]\n\nsum_v1(iter::AbstractArray) = (s = zero(eltype(iter));\n for x in iter; s += x; end;\n return s)\nsum_v2(iter::AbstractArray) = (s = zero(eltype(iter));\n @inbounds for x in iter; s += x; end;\n return s)\nsum_v3(iter::AbstractArray) = (s = zero(eltype(iter));\n @inbounds @simd for x in iter; s += x; end;\n return s)\n\n@testset \"Basic methods\" begin\n @testset \"Utilities\" begin\n @test checkdimension(Bool, \u03c0) == false\n @test checkdimensions(Bool, ()) == true\n @test checkdimensions(Bool, (1,)) == true\n @test checkdimensions(Bool, (1,2,0)) == true\n @test checkdimensions(Bool, (1,-2,0)) == false\n @test_throws ErrorException checkdimensions((1,-2,0))\n @test isgrowable(\u03c0) == false\n @test isgrowable((1,2,3)) == false\n @test isgrowable([1,2,3]) == true\n for T in (Int16, Float32, Tuple{Float64,Float64})\n @test elsize(ResizableArray{T,3,Vector{T}}) == sizeof(T)\n end\n\n # Make sure all variants of _same_elements are tested.\n A = randn(3,4)\n indexstyles = (IndexLinear(), IndexCartesian())\n for indexstyle1 in indexstyles, indexstyle2 in indexstyles\n @test _same_elements(indexstyle1, A, indexstyle2, A, length(A))\n end\n\n end\n @testset \"Dimensions: $dims\" for dims in ((), (3,), (2,3), (2,3,4))\n altdims = map(UInt, dims) # used later\n N = length(dims)\n if N > 0\n A = rand(dims...)\n T = eltype(A)\n else\n T = Float64\n A = Array{T}(undef, dims)\n A[1] = rand(dims...)\n end\n B = ResizableArray{T}(undef, size(A))\n @test isgrowable(B) == (N > 0)\n @test IndexStyle(typeof(B)) == IndexLinear()\n @test eltype(B) == eltype(A)\n @test elsize(B) == elsize(A)\n @test sizeof(B) == sizeof(A)\n @test ndims(B) == ndims(A) == N\n @test size(B) == size(A)\n @test all(d -> size(B,d) == size(A,d), 1:(N+2))\n @test axes(B) == axes(A)\n @test Base.axes1(B) == axes(B,1)\n @test length(B) == length(A) == prod(dims)\n @test maxlength(B) == length(B)\n @test all(d -> axes(B,d) == axes(A,d), 1:(N+2))\n @test unsafe_convert(Ptr{T}, B) == unsafe_convert(Ptr{T}, parent(B))\n @test pointer(B) == pointer(parent(B))\n @test pointer(B, 2) == pointer(parent(B), 2)\n copyto!(B, A)\n @test all(i -> A[i] == B[i], 1:length(A))\n @test all(i -> A[i] == B[i], CartesianIndices(A))\n @test all(i -> A[i] == parent(B)[i], 1:length(A))\n @test A == B\n for i in eachindex(B)\n B[i] = rand()\n end\n copyto!(A, B)\n @test A == B\n if N > 0\n # Extend array B.\n tmpdims = collect(size(B))\n tmpdims[end] += 1\n resize!(B, tmpdims...)\n for i in length(A)+1:length(B); B[i] = 0; end\n @test maxlength(B) == length(B) == prod(tmpdims)\n @test A != B\n @test B != A\n @test all(i -> B[i] == A[i], 1:length(A))\n C = view(B, axes(A)...)\n @test C != B && B != C\n @test C == A && A == C\n # Shrink array B.\n oldmaxlen = maxlength(B)\n resize!(B, dims)\n @test B == A\n @test C == B && B == C\n @test maxlength(B) == oldmaxlen\n # Use copy to make a fresh resizable copy\n C = copy(ResizableArray, B)\n @test C == B\n @test pointer(C) != pointer(B)\n @test maxlength(C) == length(C)\n C = copy(ResizableArray{T}, B)\n @test C == B\n @test pointer(C) != pointer(B)\n @test maxlength(C) == length(C)\n C = copy(ResizableArray{T,N}, B)\n @test C == B\n @test pointer(C) != pointer(B)\n @test maxlength(C) == length(C)\n shrink!(B)\n @test B == A\n @test maxlength(B) == length(B)\n end\n\n # Check errors.\n @test_throws BoundsError B[0]\n @test_throws BoundsError B[length(B) + 1]\n @test_throws ErrorException resize!(B, (dims..., 5))\n @test_throws DimensionMismatch ResizableArray{T,N+1}(A)\n @test_throws DimensionMismatch copy(ResizableArray{T,N+1}, A)\n @test_throws ErrorException ResizableArray{T,N,Vector{Char}}(A)\n @test_throws ErrorException copy(ResizableArray{T,N,Vector{Char}}, A)\n\n # Make a copy of A using a resizable array.\n C = copyto!(similar(ResizableArray{T}, axes(A)), A)\n @test C == A\n\n # Check equality for a different list of dimensions.\n C = rand(7)\n @test (B == C) == false\n @test (C == B) == false\n @test (B == ResizableArray(C)) == false\n @test (ResizableArray(C) == B) == false\n\n # Check various constructors and custom buffer\n # (do not splat dimensions if N=0).\n buf = Vector{T}(undef, length(A))\n for arg in (undef, buf), sz in (dims, altdims)\n if isa(arg, Vector)\n # No parameters.\n C = copyto!(ResizableArray(arg, sz), A)\n @test eltype(C) == eltype(A) && C == A\n if N > 0\n C = copyto!(ResizableArray(arg, sz...), A)\n @test eltype(C) == eltype(A) && C == A\n end\n end\n # Parameter {T}.\n C = copyto!(ResizableArray{T}(arg, sz), A)\n @test eltype(C) == eltype(A) && C == A\n if N > 0\n C = copyto!(ResizableArray{T}(arg, sz...), A)\n @test eltype(C) == eltype(A) && C == A\n end\n # Parameters {T,N}.\n C = copyto!(ResizableArray{T,N}(arg, sz), A)\n if N > 0\n @test eltype(C) == eltype(A) && C == A\n C = copyto!(ResizableArray{T,N}(arg, sz...), A)\n end\n end\n\n # Use constructor to convert array.\n C = ResizableArray{T,N}()\n resize!(C, dims)\n copyto!(C, A)\n @test eltype(C) == eltype(A) && C == A\n\n # Use constructor to convert ordinary array.\n C = ResizableArray(A)\n @test eltype(C) == eltype(A) && C == A\n C = ResizableArray{T}(A)\n @test eltype(C) == eltype(A) && C == A\n C = ResizableArray{T,N}(A)\n @test eltype(C) == eltype(A) && C == A\n\n # Use convert to convert ordinary array.\n C = convert(ResizableArray, A)\n @test eltype(C) == eltype(A) && C == A\n C = convert(ResizableArray{T}, A)\n @test eltype(C) == eltype(A) && C == A\n C = convert(ResizableArray{T,N}, A)\n @test eltype(C) == eltype(A) && C == A\n\n # Use convert to convert resizable array, result should be identical.\n for pass in 1:4\n C = (pass == 1 ? convert(ResizableArray, B) :\n pass == 2 ? convert(ResizableArray{T}, B) :\n pass == 3 ? convert(ResizableArray{T,N}, B) :\n pass == 4 ? convert(ResizableArray{T,N,Vector}, B) : nothing)\n @test eltype(C) == eltype(B)\n @test C == B\n @test pointer(C) == pointer(B)\n end\n # Use convert to convert resizable array, result should be different.\n B = ResizableArray{Int16,N}(undef, dims)\n for i in 1:length(B); B[i] = i; end\n for pass in 1:3\n C = (pass == 1 ? convert(ResizableArray{Int32}, B) :\n pass == 2 ? convert(ResizableArray{Int32,N}, B) :\n pass == 3 ? convert(ResizableArray{Int32,N,Vector}, B) :\n nothing)\n @test eltype(C) == Int32\n @test elsize(C) == sizeof(eltype(C))\n @test sizeof(C) == elsize(C)*length(C)\n @test C == B\n @test pointer(C) != pointer(B)\n end\n end\nend\n\n@testset \"Queue methods\" begin\n T = Float32\n @testset \"Dimensions: $dims\" for dims in ((3,), (2,3), (2,3,4))\n N = length(dims)\n m = 5\n altdims = map(UInt16, dims) # used later\n extdims = (dims..., m)\n extaltdims = (altdims..., Int16(m))\n A = rand(T, dims..., m)\n B = rand(T, dims)\n C = rand(T, dims)\n R = ResizableArray(A)\n append!(R, B)\n @test slice(R, 1:m) == A\n @test slice(R, m+1) == B\n prepend!(R, C)\n @test slice(R, 1) == C\n @test slice(R, 2:m+1) == A\n @test slice(R, m+2) == B\n @test length(extdims) == length(extaltdims) == N+1\n for hint in (prod(extdims), extdims, extaltdims)\n R = sizehint!(ResizableArray{T}(undef, dims..., 0), hint...)\n for k in 1:m\n if isodd(k)\n append!(R, B)\n else\n prepend!(R, C)\n end\n end\n @test slice(R, 1) == C\n @test slice(R, m) == B\n end\n end\nend\n\n@testset \"Iterations\" begin\n T = Float64\n @testset \"Dimensions: $dims\" for dims in ((3,), (2,3), (2,3,4))\n N = length(dims)\n A = rand(T, dims)\n B = ResizableArray(A)\n val = sum(A)\n @test val \u2248 sum(B)\n @test val \u2248 sum_v1(B)\n @test val \u2248 sum_v2(B)\n @test val \u2248 sum_v3(B)\n end\nend\n\nend # module\n", "meta": {"hexsha": "fce77deb7d38cb05a32a9e7de939cafa4dffcc61", "size": 9885, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/runtests.jl", "max_stars_repo_name": "JuliaTagBot/ResizableArrays.jl", "max_stars_repo_head_hexsha": "c2d29fbab36e687dce951d7e7ededc254c4ee783", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "test/runtests.jl", "max_issues_repo_name": "JuliaTagBot/ResizableArrays.jl", "max_issues_repo_head_hexsha": "c2d29fbab36e687dce951d7e7ededc254c4ee783", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "test/runtests.jl", "max_forks_repo_name": "JuliaTagBot/ResizableArrays.jl", "max_forks_repo_head_hexsha": "c2d29fbab36e687dce951d7e7ededc254c4ee783", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 36.7472118959, "max_line_length": 78, "alphanum_fraction": 0.4838644411, "num_tokens": 2802, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.44939264921326705, "lm_q2_score": 0.13296422645481662, "lm_q1q2_score": 0.05975314597712281}}
{"text": "using MPI\n\nfunction do_hello()\n comm = MPI.COMM_WORLD\n rk = MPI.Comm_rank(comm)\n sz = (MPI.Comm_size(comm))\n println(\"Hello world, I'm $(rk) of $(sz)\")\n MPI.Barrier(comm)\nend\n\nfunction main()\n MPI.Init()\n do_hello()\n MPI.Finalize()\nend\n\nmain()\n", "meta": {"hexsha": "9c88316f6752cec0f64cdd832b8fba2e273194fa", "size": 268, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "messagePassingInterfaceExer/hello.jl", "max_stars_repo_name": "terasakisatoshi/juliaExer", "max_stars_repo_head_hexsha": "e3c2195f39de858915a3dcd47684eccbb7ecb552", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 2, "max_stars_repo_stars_event_min_datetime": "2020-05-02T01:24:20.000Z", "max_stars_repo_stars_event_max_datetime": "2020-10-04T12:03:25.000Z", "max_issues_repo_path": "messagePassingInterfaceExer/hello.jl", "max_issues_repo_name": "terasakisatoshi/juliaExer", "max_issues_repo_head_hexsha": "e3c2195f39de858915a3dcd47684eccbb7ecb552", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "messagePassingInterfaceExer/hello.jl", "max_forks_repo_name": "terasakisatoshi/juliaExer", "max_forks_repo_head_hexsha": "e3c2195f39de858915a3dcd47684eccbb7ecb552", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 14.8888888889, "max_line_length": 46, "alphanum_fraction": 0.6119402985, "num_tokens": 76, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.40356685373537454, "lm_q2_score": 0.14804720179063333, "lm_q1q2_score": 0.059746943430972}}
{"text": "# Lies! Lies and Slander! Supposedly ccache doesn't make things fail, but\n# WHAT DO WE HAVE?! Literal proof that this is not the case. This whole thing\n# fails to do the proper symbol renaming with `ccache`, which is TERRIBLE.\nENV[\"BINARYBUILDER_USE_CCACHE\"] = \"false\"\nusing BinaryBuilder\n\n# Collection of sources required to build Arpack\nname = \"Arpack\"\nversion = v\"3.7.0\"\nsources = [\n \"https://github.com/opencollab/arpack-ng.git\" =>\n \"a580f6fc103c4bba1630ddd957b0b5f58dec284c\",\n]\n\n\n# Bash recipe for building across all platforms\nscript = raw\"\"\"\ncd $WORKSPACE/srcdir\nmkdir arpack-build\ncd arpack-build\n\n# arpack tests require finding libgfortran when linking with C linkers,\n# and gcc doesn't automatically add that search path. So we do it for it.\nEXE_LINK_FLAGS=\"${LDFLAGS}\"\nif [[ ${target} != *darwin* ]]; then\n EXE_LINK_FLAGS=\"${EXE_LINK_FLAGS} -Wl,-rpath-link,/opt/${target}/${target}/lib -Wl,-rpath-link,/opt/${target}/${target}/lib64\"\nfi\n\n# Symbols that have float32, float64, complexf32, and complexf64 support\nSDCZ_SYMBOLS=\"axpy copy gemv geqr2 lacpy lahqr lanhs larnv lartg \\\n lascl laset scal trevc trmm trsen gbmv gbtrf gbtrs \\\n gttrf gttrs pttrf pttrs\"\n\n# All symbols that have float32/float64 support (including the SDCZ_SYMBOLS above)\nSD_SYMBOLS=\"${SDCZ_SYMBOLS} dot ger labad laev2 lamch lanst lanv2 \\\n lapy2 larf larfg lasr nrm2 orm2r rot steqr swap\"\n\n# All symbols that have complexf32/complexf64 support (including the SDCZ_SYMBOLS above)\nCZ_SYMBOLS=\"${SDCZ_SYMBOLS} dotc geru unm2r\"\n\n# Add in (s|d)*_64 symbol remappings:\nfor sym in ${SD_SYMBOLS}; do\n SYMBOL_DEFS=\"${SYMBOL_DEFS} -Ds${sym}=s${sym}_64 -Dd${sym}=d${sym}_64\"\ndone\n\n# Add in (c|z)*_64 symbol remappings:\nfor sym in ${CZ_SYMBOLS}; do\n SYMBOL_DEFS=\"${SYMBOL_DEFS} -Dc${sym}=c${sym}_64 -Dz${sym}=z${sym}_64\"\ndone\n\n# Add one-off symbol mappings; things that don't fit into any other bucket:\nfor sym in scnrm2 dznrm2 csscal zdscal dgetrf dgetrs; do\n SYMBOL_DEFS=\"${SYMBOL_DEFS} -D${sym}=${sym}_64\"\ndone\n\n# Set up not only lowercase symbol remappings, but uppercase as well:\nSYMBOL_DEFS=\"${SYMBOL_DEFS} ${SYMBOL_DEFS^^}\"\n\nFFLAGS=\"${FFLAGS} -O2 -fPIC -ffixed-line-length-none -cpp\"\nLIBOPENBLAS=openblas\nif [[ ${nbits} == 64 ]]; then\n LIBOPENBLAS=openblas64_\n FFLAGS=\"${FFLAGS} -fdefault-integer-8 ${SYMBOL_DEFS}\"\nfi\n\ncmake ../arpack-ng -DCMAKE_INSTALL_PREFIX=\"$prefix\" -DCMAKE_TOOLCHAIN_FILE=\"/opt/$target/$target.toolchain\" -DBUILD_SHARED_LIBS=ON -DBLAS_LIBRARIES=\"-L$prefix/lib -l${LIBOPENBLAS}\" -DLAPACK_LIBRARIES=\"-L$prefix/lib -l${LIBOPENBLAS}\" -DCMAKE_Fortran_FLAGS=\"${FFLAGS}\" -DCMAKE_EXE_LINKER_FLAGS=\"${EXE_LINK_FLAGS}\"\n\nmake -j${nproc} VERBOSE=1\nmake install VERBOSE=1\n\n# For now, we'll have to adjust the name of the OpenBLAS library on macOS.\n# Eventually, this should be fixed upstream\nif [[ ${target} == \"x86_64-apple-darwin14\" ]]; then\n echo \"-- Modifying library name for OpenBLAS\"\n install_name_tool -change libopenblas64_.0.3.0.dev.dylib libopenblas64_.dylib ${prefix}/lib/libarpack.2.1.0.dylib\nfi\n\"\"\"\n\n# These are the platforms we will build for by default, unless further\n# platforms are passed in on the command line. We enable the full\n# combinatorial explosion of GCC versions because this package most\n# definitely links against libgfortran.\nplatforms = expand_gcc_versions(supported_platforms())\n\n# The products that we will ensure are always built\nproducts(prefix) = [\n LibraryProduct(prefix, \"libarpack\", :libarpack)\n]\n\n# Dependencies that must be installed before this package can be built\ndependencies = [\n \"https://github.com/JuliaLinearAlgebra/OpenBLASBuilder/releases/download/v0.3.0-3/build_OpenBLAS.v0.3.0.jl\"\n]\n\n# Build the tarballs, and possibly a `build.jl` as well.\nbuild_tarballs(ARGS, name, version, sources, script, platforms, products, dependencies)\n\n", "meta": {"hexsha": "f434774eba67fde3f10ec8c8b6f303931b2d34a8", "size": 3850, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "build_tarballs.jl", "max_stars_repo_name": "daviehh/ArpackBuilder", "max_stars_repo_head_hexsha": "e5e96a12af42513f66ffb04066941002c77dc190", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "build_tarballs.jl", "max_issues_repo_name": "daviehh/ArpackBuilder", "max_issues_repo_head_hexsha": "e5e96a12af42513f66ffb04066941002c77dc190", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "build_tarballs.jl", "max_forks_repo_name": "daviehh/ArpackBuilder", "max_forks_repo_head_hexsha": "e5e96a12af42513f66ffb04066941002c77dc190", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 39.2857142857, "max_line_length": 311, "alphanum_fraction": 0.7381818182, "num_tokens": 1173, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.5, "lm_q2_score": 0.11920292202211755, "lm_q1q2_score": 0.05960146101105877}}
{"text": "### A Pluto.jl notebook ###\n# v0.17.1\n\nusing Markdown\nusing InteractiveUtils\n\n# \u2554\u2550\u2561 88d2a9b8-d8e4-47a2-b1de-369d8fb44c20\nusing PlutoUI; PlutoUI.TableOfContents(title = \"Benchmarking Procesos\")\n\n# \u2554\u2550\u2561 07ddd182-470c-11ec-3704-93171dbc510e\nbegin\n\tusing CSV, DataFrames, DataFramesMeta, Dates, IndexedTables\n\tusing FileIO, JLD2, JuliaDB, JuliaDBMeta, Statistics, StatsBase \n\tusing Plots, StatsPlots\n\t#cd(raw\"\\\\srvfs02\\Departamento de Investigacion Economica\\Departamento\\BD Internacional\\Remesas\")\n\tcd(raw\"D:\\Departamento\\BD Internacional\\Remesas\")\n\t#include(\"functions/fn_all.jl\");\n\tPlots.plotly()\n\t#ENV[\"COLUMNS\"]=1000\n\t#ENV[\"LINES\"] = 60\n\t\n\t## Cargar base desde archivo, como dataframe\n\t#directorio = \"//srvfs02/Departamento de Investigacion Economica/Departamento/BD Internacional/Remesas/\"\n\tdirectorio = \"D:/Departamento/BD Internacional/Remesas/\"\nend\n\n# \u2554\u2550\u2561 d1d0d4b2-92c1-4faa-8e42-7b7241fa1cc2\nbegin\n\t# 801197710224\n\tx = round.(rand(5)*10^12)#00000000000)\n\tusing Formatting\n\tformat.(x)\n\t#x\n\tround.(x./10^11)\nend\n\n# \u2554\u2550\u2561 454637cc-589c-459d-93fd-7da3ba6fd67c\nmd\"# Leer CSVs\"\n\n# \u2554\u2550\u2561 53266a68-82e6-4024-af9d-8cefe89399f9\nglobal function mergeCSV(fecha_proceso)\n\tn1 = Dates.now()\n\tsearchdir(path, key) = Base.filter(\n\t\tx -> Base.contains(x, key), Base.readdir(path))\n\tkey = fecha_proceso * \".csv\"\n\tpath = \"csv/\"\n\tcsv = searchdir(path, key)\n\tNArchivo_csv = Base.size(csv)[1];\n\n\t# Importar archivos, seleccionar y renombrar variables\n\t#path * csv[1]\n\tbases_csv = []\n\n\tfor i in 1:NArchivo_csv\n\t\tBase.push!(bases_csv,\n\t\t\tCSV.read(path * csv[i],\n\t\t\t\tdelim = ',',\n\t\t\t\tDataFrames.DataFrame))\n\t\tDataFrames.rename!(bases_csv[i],\n\t\t\t:Fecha_Contable => :Fecha,\n\t\t\t:Departamento => :DeptoTransacc,\n\t\t\t:Municipio => :MunicTransacc,\n\t\t\t:RTN_Identidad => :ID)\n\t\tbases_csv[i] = DataFramesMeta.select(bases_csv[i],\n\t\t\t:Fecha,\n\t\t\t#:Agente_Cambiario,\n\t\t\t#:Tipo_Transaccion,\n\t\t\t#:Actividad_Economica,\n\t\t\t#:Medio_Pago,\n\t\t\t:DeptoTransacc,\n\t\t\t:MunicTransacc,\n\t\t\t:Tasa_Cambio,\n\t\t\t:Monto,\n\t\t\t:ID,\n\t\t\t:Tipo_Cliente);\n\tend\n\n\t# Unir en una sola tabla\n\tdatabase = Base.copy(bases_csv[1])\n\tfor i in 2:NArchivo_csv\n\t\tdatabase = Base.vcat(\n\t\t\tdatabase, bases_csv[i])\n\tend\n\tdatabase\nend\n\n# \u2554\u2550\u2561 4a5b02b5-83d5-46b9-88ec-41f4d0df54d1\ndatabase = mergeCSV(\"2021\")\n\n# \u2554\u2550\u2561 ae2c7408-7ca2-4166-98be-f9ba3d8476ed\nmd\"# Cambiar Formatos\"\n\n# \u2554\u2550\u2561 38b7bde3-7c34-4930-8582-bfd067a05c09\nmd\"## Fecha\"\n\n# \u2554\u2550\u2561 cd85e83f-f380-480a-9c78-5fdbbba068ab\nbegin\n\tdatabase[!, :Fechas] .= Dates.Date(\"1-1-1\")\n\tdatabase[!, :Days] .= 0\n database[!, :Weeks] .= 0\n database[!, :Months] .= 0\n database[!, :Years] .= 0\n\tdatabase\nend\n\n# \u2554\u2550\u2561 5cb827be-5ad3-49a3-b920-b1df8a8cb861\n\n\n# \u2554\u2550\u2561 df699c60-d076-4319-9d95-13ea2afe955c\n\n\n# \u2554\u2550\u2561 fbd5cb7c-e722-4a2e-84ef-4c6702134234\n# Benchmarking\n#=\n@time database[!, :Fechas] = Dates.Date.(\n\t\tdatabase[!, :Fecha],\n\t\tDates.DateFormat(\"d/m/Y H:M:S\"))\n@time for i in 1:Base.size(database)[1]\n\tdatabase[i, :Fechas] = Dates.Date.(\n\t\tdatabase[i, :Fecha],\n\t\tDates.DateFormat(\"d/m/Y H:M:S\"))\n\tend\n=#\n\n# \u2554\u2550\u2561 e5552459-e2ad-4f44-9760-757ea37fa233\n@time for i in 1:Base.size(database)[1]\n\tdatabase[i, :Fechas] = Dates.Date.(\n\t\tdatabase[i, :Fecha],\n\t\tDates.DateFormat(\"d/m/Y H:M:S\"))\nend\n\n# \u2554\u2550\u2561 50ca420b-2a11-4366-9541-555585eeb5ea\n\n\n# \u2554\u2550\u2561 b45cf1f6-c70c-450b-a8e4-29f7eb44e791\ndatabase\n\n# \u2554\u2550\u2561 e3c415b1-0964-4337-9f9f-c1a9616a8b5d\n\n\n# \u2554\u2550\u2561 029a721d-4a55-4cc0-8898-61bdfb80c10c\nmd\"## Depurar IDs, solo enteros\"\n\n# \u2554\u2550\u2561 7ffa2874-0690-4587-91db-f0e55570bcb1\nglobal function StrToInt(x)\n\ttry\n\t\tBase.parse.(Int64, x)\n\tcatch\n\t\treturn 0\n\tend\nend\n\n# \u2554\u2550\u2561 11a22c40-1da4-4f9c-a650-000230546bfa\nmd\"## Tipo de Cliente\"\n\n# \u2554\u2550\u2561 3e06e476-3c9d-42c4-b167-c2ce926b2c3c\nbegin\n\tdatabase[!, :Tipo_Clientes] .= 0\n\tfor i in 1:Base.size(database)[1]\n\t\tif database[i, :Tipo_Cliente] == \"Natural Residente\"\n\t\t\tdatabase[i, :Tipo_Clientes] = 1\n\t\telse\n\t\t\tdatabase[i, :Tipo_Clientes] = 0\n\t\tend\n\tend\nend\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000001\nPLUTO_PROJECT_TOML_CONTENTS = \"\"\"\n[deps]\nCSV = \"336ed68f-0bac-5ca0-87d4-7b16caf5d00b\"\nDataFrames = \"a93c6f00-e57d-5684-b7b6-d8193f3e46c0\"\nDataFramesMeta = \"1313f7d8-7da2-5740-9ea0-a2ca25f37964\"\nDates = \"ade2ca70-3891-5945-98fb-dc099432e06a\"\nFileIO = \"5789e2e9-d7fb-5bc7-8068-2c6fae9b9549\"\nFormatting = \"59287772-0a20-5a39-b81b-1366585eb4c0\"\nIndexedTables = \"6deec6e2-d858-57c5-ab9b-e6ca5bd20e43\"\nJLD2 = \"033835bb-8acc-5ee8-8aae-3f567f8a3819\"\nJuliaDB = \"a93385a2-3734-596a-9a66-3cfbb77141e6\"\nJuliaDBMeta = \"2c06ca41-a429-545c-b8f0-5ca7dd64ba19\"\nPlots = \"91a5bcdd-55d7-5caf-9e0b-520d859cae80\"\nPlutoUI = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\nStatistics = \"10745b16-79ce-11e8-11f9-7d13ad32a3b2\"\nStatsBase = \"2913bbd2-ae8a-5f71-8c99-4fb6c76f3a91\"\nStatsPlots = \"f3b207a7-027a-5e70-b257-86293d7955fd\"\n\n[compat]\nCSV = \"~0.8.5\"\nDataFrames = \"~0.22.7\"\nDataFramesMeta = \"~0.8.0\"\nFileIO = \"~1.11.2\"\nFormatting = \"~0.4.2\"\nIndexedTables = \"~0.8.1\"\nJLD2 = \"~0.4.3\"\nJuliaDB = \"~0.9.0\"\nJuliaDBMeta = \"~0.4.0\"\nPlots = \"~0.28.4\"\nPlutoUI = \"~0.7.1\"\nStatsBase = \"~0.32.2\"\nStatsPlots = \"~0.14.5\"\n\"\"\"\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000002\nPLUTO_MANIFEST_TOML_CONTENTS = \"\"\"\n# This file is machine-generated - editing it directly is not advised\n\n[[AbstractFFTs]]\ndeps = [\"LinearAlgebra\"]\ngit-tree-sha1 = \"485ee0867925449198280d4af84bdb46a2a404d0\"\nuuid = \"621f4979-c628-5d54-868e-fcf4e3e8185c\"\nversion = \"1.0.1\"\n\n[[Adapt]]\ndeps = [\"LinearAlgebra\"]\ngit-tree-sha1 = \"84918055d15b3114ede17ac6a7182f68870c16f7\"\nuuid = \"79e6a3ab-5dfb-504d-930d-738a2a938a0e\"\nversion = \"3.3.1\"\n\n[[ArgTools]]\nuuid = \"0dad84c5-d112-42e6-8d28-ef12dabb789f\"\n\n[[Arpack]]\ndeps = [\"Arpack_jll\", \"Libdl\", \"LinearAlgebra\"]\ngit-tree-sha1 = \"2ff92b71ba1747c5fdd541f8fc87736d82f40ec9\"\nuuid = \"7d9fca2a-8960-54d3-9f78-7d1dccf2cb97\"\nversion = \"0.4.0\"\n\n[[Arpack_jll]]\ndeps = [\"Libdl\", \"OpenBLAS_jll\", \"Pkg\"]\ngit-tree-sha1 = \"e214a9b9bd1b4e1b4f15b22c0994862b66af7ff7\"\nuuid = \"68821587-b530-5797-8361-c406ea357684\"\nversion = \"3.5.0+3\"\n\n[[Artifacts]]\nuuid = \"56f22d72-fd6d-98f1-02f0-08ddc0907c33\"\n\n[[AxisAlgorithms]]\ndeps = [\"LinearAlgebra\", \"Random\", \"SparseArrays\", \"WoodburyMatrices\"]\ngit-tree-sha1 = \"66771c8d21c8ff5e3a93379480a2307ac36863f7\"\nuuid = \"13072b0f-2c55-5437-9ae7-d433b7a33950\"\nversion = \"1.0.1\"\n\n[[Base64]]\nuuid = \"2a0f44e3-6c83-55bd-87e4-b1978d98bd5f\"\n\n[[BinaryProvider]]\ndeps = [\"Libdl\", \"Logging\", \"SHA\"]\ngit-tree-sha1 = \"ecdec412a9abc8db54c0efc5548c64dfce072058\"\nuuid = \"b99e7846-7c00-51b0-8f62-c81ae34c0232\"\nversion = \"0.5.10\"\n\n[[CSV]]\ndeps = [\"Dates\", \"Mmap\", \"Parsers\", \"PooledArrays\", \"SentinelArrays\", \"Tables\", \"Unicode\"]\ngit-tree-sha1 = \"b83aa3f513be680454437a0eee21001607e5d983\"\nuuid = \"336ed68f-0bac-5ca0-87d4-7b16caf5d00b\"\nversion = \"0.8.5\"\n\n[[Calculus]]\ndeps = [\"LinearAlgebra\"]\ngit-tree-sha1 = \"f641eb0a4f00c343bbc32346e1217b86f3ce9dad\"\nuuid = \"49dc2e85-a5d0-5ad3-a950-438e2897f1b9\"\nversion = \"0.5.1\"\n\n[[CategoricalArrays]]\ndeps = [\"DataAPI\", \"Future\", \"JSON\", \"Missings\", \"Printf\", \"Statistics\", \"StructTypes\", \"Unicode\"]\ngit-tree-sha1 = \"18d7f3e82c1a80dd38c16453b8fd3f0a7db92f23\"\nuuid = \"324d7699-5711-5eae-9e2f-1d82baa6b597\"\nversion = \"0.9.7\"\n\n[[Chain]]\ngit-tree-sha1 = \"cac464e71767e8a04ceee82a889ca56502795705\"\nuuid = \"8be319e6-bccf-4806-a6f7-6fae938471bc\"\nversion = \"0.4.8\"\n\n[[ChainRulesCore]]\ndeps = [\"Compat\", \"LinearAlgebra\", \"SparseArrays\"]\ngit-tree-sha1 = \"f885e7e7c124f8c92650d61b9477b9ac2ee607dd\"\nuuid = \"d360d2e6-b24c-11e9-a2a3-2a2ae2dbcce4\"\nversion = \"1.11.1\"\n\n[[ChangesOfVariables]]\ndeps = [\"LinearAlgebra\", \"Test\"]\ngit-tree-sha1 = \"9a1d594397670492219635b35a3d830b04730d62\"\nuuid = \"9e997f8a-9a97-42d5-a9f1-ce6bfc15e2c0\"\nversion = \"0.1.1\"\n\n[[Clustering]]\ndeps = [\"Distances\", \"LinearAlgebra\", \"NearestNeighbors\", \"Printf\", \"SparseArrays\", \"Statistics\", \"StatsBase\"]\ngit-tree-sha1 = \"75479b7df4167267d75294d14b58244695beb2ac\"\nuuid = \"aaaa29a8-35af-508c-8bc3-b662a17a0fe5\"\nversion = \"0.14.2\"\n\n[[CodecZlib]]\ndeps = [\"BinaryProvider\", \"Libdl\", \"TranscodingStreams\"]\ngit-tree-sha1 = \"05916673a2627dd91b4969ff8ba6941bc85a960e\"\nuuid = \"944b1d66-785c-5afd-91f1-9de20f533193\"\nversion = \"0.6.0\"\n\n[[ColorTypes]]\ndeps = [\"FixedPointNumbers\", \"Random\"]\ngit-tree-sha1 = \"b9de8dc6106e09c79f3f776c27c62360d30e5eb8\"\nuuid = \"3da002f7-5984-5a60-b8a6-cbb66c0b333f\"\nversion = \"0.9.1\"\n\n[[Colors]]\ndeps = [\"ColorTypes\", \"FixedPointNumbers\", \"InteractiveUtils\", \"Printf\", \"Reexport\"]\ngit-tree-sha1 = \"177d8b959d3c103a6d57574c38ee79c81059c31b\"\nuuid = \"5ae59095-9a9b-59fe-a467-6f913c188581\"\nversion = \"0.11.2\"\n\n[[CommonSubexpressions]]\ndeps = [\"MacroTools\", \"Test\"]\ngit-tree-sha1 = \"7b8a93dba8af7e3b42fecabf646260105ac373f7\"\nuuid = \"bbf7d656-a473-5ed7-a52c-81e309532950\"\nversion = \"0.3.0\"\n\n[[Compat]]\ndeps = [\"Base64\", \"Dates\", \"DelimitedFiles\", \"Distributed\", \"InteractiveUtils\", \"LibGit2\", \"Libdl\", \"LinearAlgebra\", \"Markdown\", \"Mmap\", \"Pkg\", \"Printf\", \"REPL\", \"Random\", \"SHA\", \"Serialization\", \"SharedArrays\", \"Sockets\", \"SparseArrays\", \"Statistics\", \"Test\", \"UUIDs\", \"Unicode\"]\ngit-tree-sha1 = \"dce3e3fea680869eaa0b774b2e8343e9ff442313\"\nuuid = \"34da2185-b29b-5c13-b0c7-acf172513d20\"\nversion = \"3.40.0\"\n\n[[CompilerSupportLibraries_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"e66e0078-7015-5450-92f7-15fbd957f2ae\"\n\n[[Contour]]\ndeps = [\"StaticArrays\"]\ngit-tree-sha1 = \"9f02045d934dc030edad45944ea80dbd1f0ebea7\"\nuuid = \"d38c429a-6771-53c6-b99e-75d170b6e991\"\nversion = \"0.5.7\"\n\n[[Crayons]]\ngit-tree-sha1 = \"3f71217b538d7aaee0b69ab47d9b7724ca8afa0d\"\nuuid = \"a8cc5b0e-0ffa-5ad4-8c14-923d3ee1735f\"\nversion = \"4.0.4\"\n\n[[Dagger]]\ndeps = [\"Distributed\", \"LinearAlgebra\", \"MemPool\", \"Profile\", \"Random\", \"Serialization\", \"SharedArrays\", \"SparseArrays\", \"Statistics\", \"StatsBase\", \"Test\"]\ngit-tree-sha1 = \"39dd9351c6637a71d1925e26e49adcd8a25ebf0b\"\nuuid = \"d58978e5-989f-55fb-8d15-ea34adc7bf54\"\nversion = \"0.8.0\"\n\n[[DataAPI]]\ngit-tree-sha1 = \"cc70b17275652eb47bc9e5f81635981f13cea5c8\"\nuuid = \"9a962f9c-6df0-11e9-0e5d-c546b8b5ee8a\"\nversion = \"1.9.0\"\n\n[[DataFrames]]\ndeps = [\"CategoricalArrays\", \"Compat\", \"DataAPI\", \"Future\", \"InvertedIndices\", \"IteratorInterfaceExtensions\", \"LinearAlgebra\", \"Markdown\", \"Missings\", \"PooledArrays\", \"PrettyTables\", \"Printf\", \"REPL\", \"Reexport\", \"SortingAlgorithms\", \"Statistics\", \"TableTraits\", \"Tables\", \"Unicode\"]\ngit-tree-sha1 = \"d50972453ef464ddcebdf489d11885468b7b83a3\"\nuuid = \"a93c6f00-e57d-5684-b7b6-d8193f3e46c0\"\nversion = \"0.22.7\"\n\n[[DataFramesMeta]]\ndeps = [\"Chain\", \"DataFrames\", \"MacroTools\", \"Reexport\"]\ngit-tree-sha1 = \"55be907a531471de062f321147006c04b8c1e75f\"\nuuid = \"1313f7d8-7da2-5740-9ea0-a2ca25f37964\"\nversion = \"0.8.0\"\n\n[[DataStructures]]\ndeps = [\"InteractiveUtils\", \"OrderedCollections\"]\ngit-tree-sha1 = \"88d48e133e6d3dd68183309877eac74393daa7eb\"\nuuid = \"864edb3b-99cc-5e75-8d2d-829cb0a9cfe8\"\nversion = \"0.17.20\"\n\n[[DataValueInterfaces]]\ngit-tree-sha1 = \"bfc1187b79289637fa0ef6d4436ebdfe6905cbd6\"\nuuid = \"e2d170a0-9d28-54be-80f0-106bbe20a464\"\nversion = \"1.0.0\"\n\n[[DataValues]]\ndeps = [\"DataValueInterfaces\", \"Dates\"]\ngit-tree-sha1 = \"d88a19299eba280a6d062e135a43f00323ae70bf\"\nuuid = \"e7dc6d0d-1eca-5fa6-8ad6-5aecde8b7ea5\"\nversion = \"0.4.13\"\n\n[[Dates]]\ndeps = [\"Printf\"]\nuuid = \"ade2ca70-3891-5945-98fb-dc099432e06a\"\n\n[[DelimitedFiles]]\ndeps = [\"Mmap\"]\nuuid = \"8bb1440f-4735-579b-a4ab-409b98df4dab\"\n\n[[DiffEqDiffTools]]\ndeps = [\"LinearAlgebra\", \"SparseArrays\", \"StaticArrays\"]\ngit-tree-sha1 = \"b992345a39b4d9681342ae795a8dacc100730182\"\nuuid = \"01453d9d-ee7c-5054-8395-0335cb756afa\"\nversion = \"0.14.0\"\n\n[[DiffResults]]\ndeps = [\"StaticArrays\"]\ngit-tree-sha1 = \"c18e98cba888c6c25d1c3b048e4b3380ca956805\"\nuuid = \"163ba53b-c6d8-5494-b064-1a9d43ac40c5\"\nversion = \"1.0.3\"\n\n[[DiffRules]]\ndeps = [\"LogExpFunctions\", \"NaNMath\", \"Random\", \"SpecialFunctions\"]\ngit-tree-sha1 = \"3287dacf67c3652d3fed09f4c12c187ae4dbb89a\"\nuuid = \"b552c78f-8df3-52c6-915a-8e097449b14b\"\nversion = \"1.4.0\"\n\n[[Distances]]\ndeps = [\"LinearAlgebra\", \"Statistics\", \"StatsAPI\"]\ngit-tree-sha1 = \"837c83e5574582e07662bbbba733964ff7c26b9d\"\nuuid = \"b4f34e82-e78d-54a5-968a-f98e89d6e8f7\"\nversion = \"0.10.6\"\n\n[[Distributed]]\ndeps = [\"Random\", \"Serialization\", \"Sockets\"]\nuuid = \"8ba89e20-285c-5b6f-9357-94700520ee1b\"\n\n[[Distributions]]\ndeps = [\"FillArrays\", \"LinearAlgebra\", \"PDMats\", \"Printf\", \"QuadGK\", \"Random\", \"SpecialFunctions\", \"Statistics\", \"StatsBase\", \"StatsFuns\"]\ngit-tree-sha1 = \"9c41285c57c6e0d73a21ed4b65f6eec34805f937\"\nuuid = \"31c24e10-a181-5473-b8eb-7969acd0382f\"\nversion = \"0.23.8\"\n\n[[DocStringExtensions]]\ndeps = [\"LibGit2\"]\ngit-tree-sha1 = \"b19534d1895d702889b219c382a6e18010797f0b\"\nuuid = \"ffbed154-4ef7-542d-bbb7-c09d3a79fcae\"\nversion = \"0.8.6\"\n\n[[DoubleFloats]]\ndeps = [\"GenericLinearAlgebra\", \"LinearAlgebra\", \"Polynomials\", \"Printf\", \"Quadmath\", \"Random\", \"Requires\", \"SpecialFunctions\"]\ngit-tree-sha1 = \"79ce76892ef75c65b210fc531891e2427152d4fd\"\nuuid = \"497a8b3b-efae-58df-a0af-a86822472b78\"\nversion = \"1.1.24\"\n\n[[Downloads]]\ndeps = [\"ArgTools\", \"LibCURL\", \"NetworkOptions\"]\nuuid = \"f43a241f-c20a-4ad4-852c-f6b1247861c6\"\n\n[[ExprTools]]\ngit-tree-sha1 = \"b7e3d17636b348f005f11040025ae8c6f645fe92\"\nuuid = \"e2ba6199-217a-4e67-a87a-7c52f15ade04\"\nversion = \"0.1.6\"\n\n[[FFMPEG]]\ndeps = [\"BinaryProvider\", \"Libdl\"]\ngit-tree-sha1 = \"9143266ba77d3313a4cf61d8333a1970e8c5d8b6\"\nuuid = \"c87230d0-a227-11e9-1b43-d7ebe4e7570a\"\nversion = \"0.2.4\"\n\n[[FFTW]]\ndeps = [\"AbstractFFTs\", \"FFTW_jll\", \"LinearAlgebra\", \"MKL_jll\", \"Preferences\", \"Reexport\"]\ngit-tree-sha1 = \"463cb335fa22c4ebacfd1faba5fde14edb80d96c\"\nuuid = \"7a1cc6ca-52ef-59f5-83cd-3a7055c09341\"\nversion = \"1.4.5\"\n\n[[FFTW_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"c6033cc3892d0ef5bb9cd29b7f2f0331ea5184ea\"\nuuid = \"f5851436-0d7a-5f13-b9de-f02708fd171a\"\nversion = \"3.3.10+0\"\n\n[[FileIO]]\ndeps = [\"Pkg\", \"Requires\", \"UUIDs\"]\ngit-tree-sha1 = \"2db648b6712831ecb333eae76dbfd1c156ca13bb\"\nuuid = \"5789e2e9-d7fb-5bc7-8068-2c6fae9b9549\"\nversion = \"1.11.2\"\n\n[[FillArrays]]\ndeps = [\"LinearAlgebra\", \"Random\", \"SparseArrays\"]\ngit-tree-sha1 = \"4863cbb7910079369e258dee4add9d06ead5063a\"\nuuid = \"1a297f60-69ca-5386-bcde-b61e274b549b\"\nversion = \"0.8.14\"\n\n[[FixedPointNumbers]]\ngit-tree-sha1 = \"d14a6fa5890ea3a7e5dcab6811114f132fec2b4b\"\nuuid = \"53c48c17-4a7d-5ca2-90c5-79b7896eea93\"\nversion = \"0.6.1\"\n\n[[Formatting]]\ndeps = [\"Printf\"]\ngit-tree-sha1 = \"8339d61043228fdd3eb658d86c926cb282ae72a8\"\nuuid = \"59287772-0a20-5a39-b81b-1366585eb4c0\"\nversion = \"0.4.2\"\n\n[[ForwardDiff]]\ndeps = [\"CommonSubexpressions\", \"DiffResults\", \"DiffRules\", \"LinearAlgebra\", \"LogExpFunctions\", \"NaNMath\", \"Preferences\", \"Printf\", \"Random\", \"SpecialFunctions\", \"StaticArrays\"]\ngit-tree-sha1 = \"6406b5112809c08b1baa5703ad274e1dded0652f\"\nuuid = \"f6369f11-7733-5829-9624-2563aa707210\"\nversion = \"0.10.23\"\n\n[[Future]]\ndeps = [\"Random\"]\nuuid = \"9fa8497b-333b-5362-9e8d-4d0656e87820\"\n\n[[GR]]\ndeps = [\"Base64\", \"DelimitedFiles\", \"LinearAlgebra\", \"Printf\", \"Random\", \"Serialization\", \"Sockets\", \"Test\"]\ngit-tree-sha1 = \"c690c2ab22ac9ee323d9966deae61a089362b25c\"\nuuid = \"28b8d3ca-fb5f-59d9-8090-bfdbd6d07a71\"\nversion = \"0.44.0\"\n\n[[GenericLinearAlgebra]]\ndeps = [\"LinearAlgebra\", \"Printf\", \"Random\"]\ngit-tree-sha1 = \"ac44f4f51ffee9ff1ea50bd3fbb5677ea568d33d\"\nuuid = \"14197337-ba66-59df-a3e3-ca00e7dcff7a\"\nversion = \"0.2.7\"\n\n[[GeometryTypes]]\ndeps = [\"ColorTypes\", \"FixedPointNumbers\", \"LinearAlgebra\", \"StaticArrays\"]\ngit-tree-sha1 = \"78f0ce9d01993b637a8f28d84537d75dc0ce8eef\"\nuuid = \"4d00f742-c7ba-57c2-abde-4428a4b178cb\"\nversion = \"0.7.10\"\n\n[[Glob]]\ngit-tree-sha1 = \"4df9f7e06108728ebf00a0a11edee4b29a482bb2\"\nuuid = \"c27321d9-0574-5035-807b-f59d2c89b15c\"\nversion = \"1.3.0\"\n\n[[Grisu]]\ngit-tree-sha1 = \"53bb909d1151e57e2484c3d1b53e19552b887fb2\"\nuuid = \"42e2da0e-8278-4e71-bc24-59509adca0fe\"\nversion = \"1.0.2\"\n\n[[IndexedTables]]\ndeps = [\"DataValues\", \"Dates\", \"IteratorInterfaceExtensions\", \"LinearAlgebra\", \"OnlineStats\", \"PooledArrays\", \"Random\", \"Serialization\", \"SparseArrays\", \"Statistics\", \"TableTraits\", \"TableTraitsUtils\", \"Test\", \"WeakRefStrings\"]\ngit-tree-sha1 = \"cb4406eeed8729f8179940d39e6dec5598971341\"\nuuid = \"6deec6e2-d858-57c5-ab9b-e6ca5bd20e43\"\nversion = \"0.8.1\"\n\n[[IntelOpenMP_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"d979e54b71da82f3a65b62553da4fc3d18c9004c\"\nuuid = \"1d5cc7b8-4909-519e-a0f8-d0f5ad9712d0\"\nversion = \"2018.0.3+2\"\n\n[[InteractiveUtils]]\ndeps = [\"Markdown\"]\nuuid = \"b77e0a4c-d291-57a0-90e8-8db25a27a240\"\n\n[[Interpolations]]\ndeps = [\"AxisAlgorithms\", \"LinearAlgebra\", \"OffsetArrays\", \"Random\", \"Ratios\", \"SharedArrays\", \"SparseArrays\", \"StaticArrays\", \"WoodburyMatrices\"]\ngit-tree-sha1 = \"2b7d4e9be8b74f03115e64cf36ed2f48ae83d946\"\nuuid = \"a98d9a8b-a2ab-59e6-89dd-64a1c18fca59\"\nversion = \"0.12.10\"\n\n[[Intervals]]\ndeps = [\"Dates\", \"Printf\", \"RecipesBase\", \"Serialization\", \"TimeZones\"]\ngit-tree-sha1 = \"323a38ed1952d30586d0fe03412cde9399d3618b\"\nuuid = \"d8418881-c3e1-53bb-8760-2df7ec849ed5\"\nversion = \"1.5.0\"\n\n[[InverseFunctions]]\ndeps = [\"Test\"]\ngit-tree-sha1 = \"a7254c0acd8e62f1ac75ad24d5db43f5f19f3c65\"\nuuid = \"3587e190-3f89-42d0-90ee-14403ec27112\"\nversion = \"0.1.2\"\n\n[[InvertedIndices]]\ngit-tree-sha1 = \"bee5f1ef5bf65df56bdd2e40447590b272a5471f\"\nuuid = \"41ab1584-1d38-5bbf-9106-f11c6c58b48f\"\nversion = \"1.1.0\"\n\n[[IrrationalConstants]]\ngit-tree-sha1 = \"7fd44fd4ff43fc60815f8e764c0f352b83c49151\"\nuuid = \"92d709cd-6900-40b7-9082-c6be49f344b6\"\nversion = \"0.1.1\"\n\n[[IterTools]]\ngit-tree-sha1 = \"05110a2ab1fc5f932622ffea2a003221f4782c18\"\nuuid = \"c8e1da08-722c-5040-9ed9-7db0dc04731e\"\nversion = \"1.3.0\"\n\n[[IteratorInterfaceExtensions]]\ngit-tree-sha1 = \"a3f24677c21f5bbe9d2a714f95dcd58337fb2856\"\nuuid = \"82899510-4779-5014-852e-03e436cf321d\"\nversion = \"1.0.0\"\n\n[[JLD2]]\ndeps = [\"CodecZlib\", \"DataStructures\", \"MacroTools\", \"Mmap\", \"Pkg\", \"Printf\", \"Requires\", \"UUIDs\"]\ngit-tree-sha1 = \"9f2f2f24e60305feb6ae293a617ddf06f429efc3\"\nuuid = \"033835bb-8acc-5ee8-8aae-3f567f8a3819\"\nversion = \"0.4.3\"\n\n[[JLLWrappers]]\ndeps = [\"Preferences\"]\ngit-tree-sha1 = \"642a199af8b68253517b80bd3bfd17eb4e84df6e\"\nuuid = \"692b3bcd-3c85-4b1f-b108-f13ce0eb3210\"\nversion = \"1.3.0\"\n\n[[JSON]]\ndeps = [\"Dates\", \"Mmap\", \"Parsers\", \"Unicode\"]\ngit-tree-sha1 = \"8076680b162ada2a031f707ac7b4953e30667a37\"\nuuid = \"682c06a0-de6a-54ab-a142-c8b1cf79cde6\"\nversion = \"0.21.2\"\n\n[[JuliaDB]]\ndeps = [\"Dagger\", \"DataValues\", \"Dates\", \"Distributed\", \"Glob\", \"IndexedTables\", \"MemPool\", \"Nullables\", \"OnlineStats\", \"OnlineStatsBase\", \"PooledArrays\", \"Printf\", \"Random\", \"RecipesBase\", \"Serialization\", \"Statistics\", \"StatsBase\", \"Test\", \"TextParse\", \"WeakRefStrings\"]\ngit-tree-sha1 = \"83cfc7566e079721ab9c2b19efa6a26bc5ba9d20\"\nuuid = \"a93385a2-3734-596a-9a66-3cfbb77141e6\"\nversion = \"0.9.0\"\n\n[[JuliaDBMeta]]\ndeps = [\"Dagger\", \"Distributed\", \"IndexedTables\", \"IterTools\", \"JuliaDB\", \"MacroTools\", \"Reexport\", \"Test\"]\ngit-tree-sha1 = \"ca0c4c156d4a3b8055dcabd36eacb3d7469b8583\"\nuuid = \"2c06ca41-a429-545c-b8f0-5ca7dd64ba19\"\nversion = \"0.4.0\"\n\n[[KernelDensity]]\ndeps = [\"Distributions\", \"FFTW\", \"Interpolations\", \"Optim\", \"StatsBase\", \"Test\"]\ngit-tree-sha1 = \"c1048817fe5711f699abc8fabd47b1ac6ba4db04\"\nuuid = \"5ab0869b-81aa-558d-bb23-cbf5423bbe9b\"\nversion = \"0.5.1\"\n\n[[LazyArtifacts]]\ndeps = [\"Artifacts\", \"Pkg\"]\nuuid = \"4af54fe1-eca0-43a8-85a7-787d91b784e3\"\n\n[[LearnBase]]\ndeps = [\"LinearAlgebra\", \"SparseArrays\", \"StatsBase\", \"Test\"]\ngit-tree-sha1 = \"c4b5da6d68517f46f70ed5157b28336b56cd2ff3\"\nuuid = \"7f8f8fb0-2700-5f03-b4bd-41f8cfc144b6\"\nversion = \"0.2.2\"\n\n[[LibCURL]]\ndeps = [\"LibCURL_jll\", \"MozillaCACerts_jll\"]\nuuid = \"b27032c2-a3e7-50c8-80cd-2d36dbcbfd21\"\n\n[[LibCURL_jll]]\ndeps = [\"Artifacts\", \"LibSSH2_jll\", \"Libdl\", \"MbedTLS_jll\", \"Zlib_jll\", \"nghttp2_jll\"]\nuuid = \"deac9b47-8bc7-5906-a0fe-35ac56dc84c0\"\n\n[[LibGit2]]\ndeps = [\"Base64\", \"NetworkOptions\", \"Printf\", \"SHA\"]\nuuid = \"76f85450-5226-5b5a-8eaa-529ad045b433\"\n\n[[LibSSH2_jll]]\ndeps = [\"Artifacts\", \"Libdl\", \"MbedTLS_jll\"]\nuuid = \"29816b5a-b9ab-546f-933c-edad1886dfa8\"\n\n[[Libdl]]\nuuid = \"8f399da3-3557-5675-b5ff-fb832c97cbdb\"\n\n[[LineSearches]]\ndeps = [\"LinearAlgebra\", \"NLSolversBase\", \"NaNMath\", \"Parameters\", \"Printf\"]\ngit-tree-sha1 = \"f27132e551e959b3667d8c93eae90973225032dd\"\nuuid = \"d3d80556-e9d4-5f37-9878-2ab0fcc64255\"\nversion = \"7.1.1\"\n\n[[LinearAlgebra]]\ndeps = [\"Libdl\"]\nuuid = \"37e2e46d-f89d-539d-b4ee-838fcccc9c8e\"\n\n[[LogExpFunctions]]\ndeps = [\"ChainRulesCore\", \"ChangesOfVariables\", \"DocStringExtensions\", \"InverseFunctions\", \"IrrationalConstants\", \"LinearAlgebra\"]\ngit-tree-sha1 = \"be9eef9f9d78cecb6f262f3c10da151a6c5ab827\"\nuuid = \"2ab3a3ac-af41-5b50-aa03-7779005ae688\"\nversion = \"0.3.5\"\n\n[[Logging]]\nuuid = \"56ddb016-857b-54e1-b83d-db4d58db5568\"\n\n[[LossFunctions]]\ndeps = [\"InteractiveUtils\", \"LearnBase\", \"Markdown\", \"Random\", \"RecipesBase\", \"SparseArrays\", \"Statistics\", \"StatsBase\", \"Test\"]\ngit-tree-sha1 = \"08d87fec43e7d335811dfae5b55dbfc5690e915b\"\nuuid = \"30fc2ffe-d236-52d8-8643-a9d8f7c094a7\"\nversion = \"0.5.1\"\n\n[[MKL_jll]]\ndeps = [\"Artifacts\", \"IntelOpenMP_jll\", \"JLLWrappers\", \"LazyArtifacts\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"5455aef09b40e5020e1520f551fa3135040d4ed0\"\nuuid = \"856f044c-d86e-5d09-b602-aeab76dc8ba7\"\nversion = \"2021.1.1+2\"\n\n[[MacroTools]]\ndeps = [\"Markdown\", \"Random\"]\ngit-tree-sha1 = \"3d3e902b31198a27340d0bf00d6ac452866021cf\"\nuuid = \"1914dd2f-81c6-5fcd-8719-6d5c9610ff09\"\nversion = \"0.5.9\"\n\n[[Markdown]]\ndeps = [\"Base64\"]\nuuid = \"d6f4376e-aef5-505a-96c1-9c027394607a\"\n\n[[MbedTLS_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"c8ffd9c3-330d-5841-b78e-0817d7145fa1\"\n\n[[Measures]]\ngit-tree-sha1 = \"e498ddeee6f9fdb4551ce855a46f54dbd900245f\"\nuuid = \"442fdcdd-2543-5da2-b0f3-8c86c306513e\"\nversion = \"0.3.1\"\n\n[[MemPool]]\ndeps = [\"DataStructures\", \"Distributed\", \"Mmap\", \"Random\", \"Serialization\", \"Sockets\", \"Test\"]\ngit-tree-sha1 = \"d52799152697059353a8eac1000d32ba8d92aa25\"\nuuid = \"f9f48841-c794-520a-933b-121f7ba6ed94\"\nversion = \"0.2.0\"\n\n[[Missings]]\ndeps = [\"DataAPI\"]\ngit-tree-sha1 = \"f8c673ccc215eb50fcadb285f522420e29e69e1c\"\nuuid = \"e1d29d7a-bbdc-5cf2-9ac0-f12de2c33e28\"\nversion = \"0.4.5\"\n\n[[Mmap]]\nuuid = \"a63ad114-7e13-5084-954f-fe012c677804\"\n\n[[Mocking]]\ndeps = [\"Compat\", \"ExprTools\"]\ngit-tree-sha1 = \"29714d0a7a8083bba8427a4fbfb00a540c681ce7\"\nuuid = \"78c3b35d-d492-501b-9361-3d52fe80e533\"\nversion = \"0.7.3\"\n\n[[MozillaCACerts_jll]]\nuuid = \"14a3606d-f60d-562e-9121-12d972cd8159\"\n\n[[MultivariateStats]]\ndeps = [\"Arpack\", \"LinearAlgebra\", \"SparseArrays\", \"Statistics\", \"StatsBase\"]\ngit-tree-sha1 = \"352fae519b447bf52e6de627b89f448bcd469e4e\"\nuuid = \"6f286f6a-111f-5878-ab1e-185364afe411\"\nversion = \"0.7.0\"\n\n[[MutableArithmetics]]\ndeps = [\"LinearAlgebra\", \"SparseArrays\", \"Test\"]\ngit-tree-sha1 = \"7bb6853d9afec54019c1397c6eb610b9b9a19525\"\nuuid = \"d8a4904e-b15c-11e9-3269-09a3773c0cb0\"\nversion = \"0.3.1\"\n\n[[NLSolversBase]]\ndeps = [\"Calculus\", \"DiffEqDiffTools\", \"DiffResults\", \"Distributed\", \"ForwardDiff\"]\ngit-tree-sha1 = \"f1b8ed89fa332f410cfc7c937682eb4d0b361521\"\nuuid = \"d41bc354-129a-5804-8e4c-c37616107c6c\"\nversion = \"7.5.0\"\n\n[[NaNMath]]\ngit-tree-sha1 = \"bfe47e760d60b82b66b61d2d44128b62e3a369fb\"\nuuid = \"77ba4419-2d1f-58cd-9bb1-8ffee604a2e3\"\nversion = \"0.3.5\"\n\n[[NearestNeighbors]]\ndeps = [\"Distances\", \"StaticArrays\"]\ngit-tree-sha1 = \"16baacfdc8758bc374882566c9187e785e85c2f0\"\nuuid = \"b8a86587-4115-5ab1-83bc-aa920d37bbce\"\nversion = \"0.4.9\"\n\n[[NetworkOptions]]\nuuid = \"ca575930-c2e3-43a9-ace4-1e988b2c1908\"\n\n[[Nullables]]\ngit-tree-sha1 = \"8f87854cc8f3685a60689d8edecaa29d2251979b\"\nuuid = \"4d1e1d77-625e-5b40-9113-a560ec7a8ecd\"\nversion = \"1.0.0\"\n\n[[Observables]]\ngit-tree-sha1 = \"3469ef96607a6b9a1e89e54e6f23401073ed3126\"\nuuid = \"510215fc-4207-5dde-b226-833fc4488ee2\"\nversion = \"0.3.3\"\n\n[[OffsetArrays]]\ndeps = [\"Adapt\"]\ngit-tree-sha1 = \"043017e0bdeff61cfbb7afeb558ab29536bbb5ed\"\nuuid = \"6fe1bfb0-de20-5000-8ca7-80f57d26f881\"\nversion = \"1.10.8\"\n\n[[OnlineStats]]\ndeps = [\"DataStructures\", \"Dates\", \"LearnBase\", \"LinearAlgebra\", \"LossFunctions\", \"OnlineStatsBase\", \"PenaltyFunctions\", \"Random\", \"RecipesBase\", \"Reexport\", \"Statistics\", \"StatsBase\", \"SweepOperator\", \"Test\"]\ngit-tree-sha1 = \"a783fe25075b9d09d6dd2ba67dafd374d835acfa\"\nuuid = \"a15396b6-48d5-5d58-9928-6d29437db91e\"\nversion = \"0.23.0\"\n\n[[OnlineStatsBase]]\ndeps = [\"Dates\", \"LearnBase\", \"Statistics\", \"Test\"]\ngit-tree-sha1 = \"2d62fb8d9b1b407f9f80e99f4a63f32dea90423c\"\nuuid = \"925886fa-5bf2-5e8e-b522-a9147a512338\"\nversion = \"0.10.2\"\n\n[[OpenBLAS_jll]]\ndeps = [\"Artifacts\", \"CompilerSupportLibraries_jll\", \"Libdl\"]\nuuid = \"4536629a-c528-5b80-bd46-f80d51c5b363\"\n\n[[OpenSpecFun_jll]]\ndeps = [\"Artifacts\", \"CompilerSupportLibraries_jll\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"13652491f6856acfd2db29360e1bbcd4565d04f1\"\nuuid = \"efe28fd5-8261-553b-a9e1-b2916fc3738e\"\nversion = \"0.5.5+0\"\n\n[[Optim]]\ndeps = [\"Compat\", \"FillArrays\", \"LineSearches\", \"LinearAlgebra\", \"NLSolversBase\", \"NaNMath\", \"Parameters\", \"PositiveFactorizations\", \"Printf\", \"SparseArrays\", \"StatsBase\"]\ngit-tree-sha1 = \"c05aa6b694d426df87ff493306c1c5b4b215e148\"\nuuid = \"429524aa-4258-5aef-a3af-852621145aeb\"\nversion = \"0.22.0\"\n\n[[OrderedCollections]]\ngit-tree-sha1 = \"85f8e6578bf1f9ee0d11e7bb1b1456435479d47c\"\nuuid = \"bac558e1-5e72-5ebc-8fee-abe8a469f55d\"\nversion = \"1.4.1\"\n\n[[PDMats]]\ndeps = [\"LinearAlgebra\", \"SparseArrays\", \"SuiteSparse\", \"Test\"]\ngit-tree-sha1 = \"95a4038d1011dfdbde7cecd2ad0ac411e53ab1bc\"\nuuid = \"90014a1f-27ba-587c-ab20-58faa44d9150\"\nversion = \"0.10.1\"\n\n[[Parameters]]\ndeps = [\"OrderedCollections\", \"UnPack\"]\ngit-tree-sha1 = \"34c0e9ad262e5f7fc75b10a9952ca7692cfc5fbe\"\nuuid = \"d96e819e-fc66-5662-9728-84c9c7592b0a\"\nversion = \"0.12.3\"\n\n[[Parsers]]\ndeps = [\"Dates\"]\ngit-tree-sha1 = \"bfd7d8c7fd87f04543810d9cbd3995972236ba1b\"\nuuid = \"69de0a69-1ddd-5017-9359-2bf0b02dc9f0\"\nversion = \"1.1.2\"\n\n[[PenaltyFunctions]]\ndeps = [\"InteractiveUtils\", \"LearnBase\", \"LinearAlgebra\", \"RecipesBase\", \"Reexport\", \"Test\"]\ngit-tree-sha1 = \"b0baaa5218ca0ffd6a8ae37ef0b58e0df688ac8b\"\nuuid = \"06bb1623-fdd5-5ca2-a01c-88eae3ea319e\"\nversion = \"0.1.2\"\n\n[[Pkg]]\ndeps = [\"Artifacts\", \"Dates\", \"Downloads\", \"LibGit2\", \"Libdl\", \"Logging\", \"Markdown\", \"Printf\", \"REPL\", \"Random\", \"SHA\", \"Serialization\", \"TOML\", \"Tar\", \"UUIDs\", \"p7zip_jll\"]\nuuid = \"44cfe95a-1eb2-52ea-b672-e2afdf69b78f\"\n\n[[PlotThemes]]\ndeps = [\"PlotUtils\", \"Requires\", \"Statistics\"]\ngit-tree-sha1 = \"87a4ea7f8c350d87d3a8ca9052663b633c0b2722\"\nuuid = \"ccf2f8ad-2431-5c83-bf29-c5338b663b6a\"\nversion = \"1.0.3\"\n\n[[PlotUtils]]\ndeps = [\"Colors\", \"Dates\", \"Printf\", \"Random\", \"Reexport\"]\ngit-tree-sha1 = \"51e742162c97d35f714f9611619db6975e19384b\"\nuuid = \"995b91a9-d308-5afd-9ec6-746e21dbc043\"\nversion = \"0.6.5\"\n\n[[Plots]]\ndeps = [\"Base64\", \"Contour\", \"Dates\", \"FFMPEG\", \"FixedPointNumbers\", \"GR\", \"GeometryTypes\", \"JSON\", \"LinearAlgebra\", \"Measures\", \"NaNMath\", \"Pkg\", \"PlotThemes\", \"PlotUtils\", \"Printf\", \"REPL\", \"Random\", \"RecipesBase\", \"Reexport\", \"Requires\", \"Showoff\", \"SparseArrays\", \"Statistics\", \"StatsBase\", \"UUIDs\"]\ngit-tree-sha1 = \"efbe466a790d7e8a5c4b5ee1601c0c8edc99780b\"\nuuid = \"91a5bcdd-55d7-5caf-9e0b-520d859cae80\"\nversion = \"0.28.4\"\n\n[[PlutoUI]]\ndeps = [\"Base64\", \"Dates\", \"InteractiveUtils\", \"Logging\", \"Markdown\", \"Random\", \"Suppressor\"]\ngit-tree-sha1 = \"45ce174d36d3931cd4e37a47f93e07d1455f038d\"\nuuid = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\nversion = \"0.7.1\"\n\n[[Polynomials]]\ndeps = [\"Intervals\", \"LinearAlgebra\", \"MutableArithmetics\", \"RecipesBase\"]\ngit-tree-sha1 = \"79bcbb379205f1c62913fa9ebecb413c7a35f8b0\"\nuuid = \"f27b6e38-b328-58d1-80ce-0feddd5e7a45\"\nversion = \"2.0.18\"\n\n[[PooledArrays]]\ndeps = [\"DataAPI\"]\ngit-tree-sha1 = \"b1333d4eced1826e15adbdf01a4ecaccca9d353c\"\nuuid = \"2dfb63ee-cc39-5dd5-95bd-886bf059d720\"\nversion = \"0.5.3\"\n\n[[PositiveFactorizations]]\ndeps = [\"LinearAlgebra\"]\ngit-tree-sha1 = \"17275485f373e6673f7e7f97051f703ed5b15b20\"\nuuid = \"85a6dd25-e78a-55b7-8502-1745935b8125\"\nversion = \"0.2.4\"\n\n[[Preferences]]\ndeps = [\"TOML\"]\ngit-tree-sha1 = \"00cfd92944ca9c760982747e9a1d0d5d86ab1e5a\"\nuuid = \"21216c6a-2e73-6563-6e65-726566657250\"\nversion = \"1.2.2\"\n\n[[PrettyTables]]\ndeps = [\"Crayons\", \"Formatting\", \"Markdown\", \"Reexport\", \"Tables\"]\ngit-tree-sha1 = \"574a6b3ea95f04e8757c0280bb9c29f1a5e35138\"\nuuid = \"08abe8d2-0d0c-5749-adfa-8a2ac140af0d\"\nversion = \"0.11.1\"\n\n[[Printf]]\ndeps = [\"Unicode\"]\nuuid = \"de0858da-6303-5e67-8744-51eddeeeb8d7\"\n\n[[Profile]]\ndeps = [\"Printf\"]\nuuid = \"9abbd945-dff8-562f-b5e8-e1ebf5ef1b79\"\n\n[[QuadGK]]\ndeps = [\"DataStructures\", \"LinearAlgebra\"]\ngit-tree-sha1 = \"78aadffb3efd2155af139781b8a8df1ef279ea39\"\nuuid = \"1fd47b50-473d-5c70-9696-f719f8f3bcdc\"\nversion = \"2.4.2\"\n\n[[Quadmath]]\ndeps = [\"Printf\", \"Random\", \"Requires\"]\ngit-tree-sha1 = \"5a8f74af8eae654086a1d058b4ec94ff192e3de0\"\nuuid = \"be4d8f0f-7fa4-5f49-b795-2f01399ab2dd\"\nversion = \"0.5.5\"\n\n[[REPL]]\ndeps = [\"InteractiveUtils\", \"Markdown\", \"Sockets\", \"Unicode\"]\nuuid = \"3fa0cd96-eef1-5676-8a61-b3b8758bbffb\"\n\n[[Random]]\ndeps = [\"Serialization\"]\nuuid = \"9a3f8284-a2c9-5f02-9a11-845980a1fd5c\"\n\n[[Ratios]]\ndeps = [\"Requires\"]\ngit-tree-sha1 = \"01d341f502250e81f6fec0afe662aa861392a3aa\"\nuuid = \"c84ed2f1-dad5-54f0-aa8e-dbefe2724439\"\nversion = \"0.4.2\"\n\n[[RecipesBase]]\ngit-tree-sha1 = \"7bdce29bc9b2f5660a6e5e64d64d91ec941f6aa2\"\nuuid = \"3cdcf5f2-1ef4-517c-9805-6587b60abb01\"\nversion = \"0.7.0\"\n\n[[Reexport]]\ndeps = [\"Pkg\"]\ngit-tree-sha1 = \"7b1d07f411bc8ddb7977ec7f377b97b158514fe0\"\nuuid = \"189a3867-3050-52da-a836-e630ba90ab69\"\nversion = \"0.2.0\"\n\n[[Requires]]\ndeps = [\"UUIDs\"]\ngit-tree-sha1 = \"4036a3bd08ac7e968e27c203d45f5fff15020621\"\nuuid = \"ae029012-a4dd-5104-9daa-d747884805df\"\nversion = \"1.1.3\"\n\n[[Rmath]]\ndeps = [\"Random\", \"Rmath_jll\"]\ngit-tree-sha1 = \"bf3188feca147ce108c76ad82c2792c57abe7b1f\"\nuuid = \"79098fc4-a85e-5d69-aa6a-4863f24498fa\"\nversion = \"0.7.0\"\n\n[[Rmath_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"68db32dff12bb6127bac73c209881191bf0efbb7\"\nuuid = \"f50d1b31-88e8-58de-be2c-1cc44531875f\"\nversion = \"0.3.0+0\"\n\n[[SHA]]\nuuid = \"ea8e919c-243c-51af-8825-aaa63cd721ce\"\n\n[[SentinelArrays]]\ndeps = [\"Dates\", \"Random\"]\ngit-tree-sha1 = \"f45b34656397a1f6e729901dc9ef679610bd12b5\"\nuuid = \"91c51154-3ec4-41a3-a24f-3f23e20d615c\"\nversion = \"1.3.8\"\n\n[[Serialization]]\nuuid = \"9e88b42a-f829-5b0c-bbe9-9e923198166b\"\n\n[[SharedArrays]]\ndeps = [\"Distributed\", \"Mmap\", \"Random\", \"Serialization\"]\nuuid = \"1a1011a3-84de-559e-8e89-a11a2f7dc383\"\n\n[[Showoff]]\ndeps = [\"Dates\", \"Grisu\"]\ngit-tree-sha1 = \"ee010d8f103468309b8afac4abb9be2e18ff1182\"\nuuid = \"992d4aef-0814-514b-bc4d-f2e9a6c4116f\"\nversion = \"0.3.2\"\n\n[[Sockets]]\nuuid = \"6462fe0b-24de-5631-8697-dd941f90decc\"\n\n[[SortingAlgorithms]]\ndeps = [\"DataStructures\", \"Random\", \"Test\"]\ngit-tree-sha1 = \"03f5898c9959f8115e30bc7226ada7d0df554ddd\"\nuuid = \"a2af1166-a08f-5f64-846c-94a0d3cef48c\"\nversion = \"0.3.1\"\n\n[[SparseArrays]]\ndeps = [\"LinearAlgebra\", \"Random\"]\nuuid = \"2f01184e-e22b-5df5-ae63-d93ebab69eaf\"\n\n[[SpecialFunctions]]\ndeps = [\"OpenSpecFun_jll\"]\ngit-tree-sha1 = \"d8d8b8a9f4119829410ecd706da4cc8594a1e020\"\nuuid = \"276daf66-3868-5448-9aa4-cd146d93841b\"\nversion = \"0.10.3\"\n\n[[StaticArrays]]\ndeps = [\"LinearAlgebra\", \"Random\", \"Statistics\"]\ngit-tree-sha1 = \"da4cf579416c81994afd6322365d00916c79b8ae\"\nuuid = \"90137ffa-7385-5640-81b9-e52037218182\"\nversion = \"0.12.5\"\n\n[[Statistics]]\ndeps = [\"LinearAlgebra\", \"SparseArrays\"]\nuuid = \"10745b16-79ce-11e8-11f9-7d13ad32a3b2\"\n\n[[StatsAPI]]\ngit-tree-sha1 = \"1958272568dc176a1d881acb797beb909c785510\"\nuuid = \"82ae8749-77ed-4fe6-ae5f-f523153014b0\"\nversion = \"1.0.0\"\n\n[[StatsBase]]\ndeps = [\"DataAPI\", \"DataStructures\", \"LinearAlgebra\", \"Missings\", \"Printf\", \"Random\", \"SortingAlgorithms\", \"SparseArrays\", \"Statistics\"]\ngit-tree-sha1 = \"19bfcb46245f69ff4013b3df3b977a289852c3a1\"\nuuid = \"2913bbd2-ae8a-5f71-8c99-4fb6c76f3a91\"\nversion = \"0.32.2\"\n\n[[StatsFuns]]\ndeps = [\"Rmath\", \"SpecialFunctions\"]\ngit-tree-sha1 = \"ced55fd4bae008a8ea12508314e725df61f0ba45\"\nuuid = \"4c63d2b9-4356-54db-8cca-17b64c39e42c\"\nversion = \"0.9.7\"\n\n[[StatsPlots]]\ndeps = [\"Clustering\", \"DataStructures\", \"DataValues\", \"Distributions\", \"Interpolations\", \"KernelDensity\", \"MultivariateStats\", \"Observables\", \"Plots\", \"RecipesBase\", \"Reexport\", \"StatsBase\", \"TableOperations\", \"Tables\", \"Widgets\"]\ngit-tree-sha1 = \"388e6588a1e09e763b0f38c498aeb350b5f76828\"\nuuid = \"f3b207a7-027a-5e70-b257-86293d7955fd\"\nversion = \"0.14.5\"\n\n[[StructTypes]]\ndeps = [\"Dates\", \"UUIDs\"]\ngit-tree-sha1 = \"d24a825a95a6d98c385001212dc9020d609f2d4f\"\nuuid = \"856f2bd8-1eba-4b0a-8007-ebc267875bd4\"\nversion = \"1.8.1\"\n\n[[SuiteSparse]]\ndeps = [\"Libdl\", \"LinearAlgebra\", \"Serialization\", \"SparseArrays\"]\nuuid = \"4607b0f0-06f3-5cda-b6b1-a6196a1729e9\"\n\n[[Suppressor]]\ngit-tree-sha1 = \"a819d77f31f83e5792a76081eee1ea6342ab8787\"\nuuid = \"fd094767-a336-5f1f-9728-57cf17d0bbfb\"\nversion = \"0.2.0\"\n\n[[SweepOperator]]\ndeps = [\"LinearAlgebra\", \"Test\"]\ngit-tree-sha1 = \"2039aaa96f7b21634a1b3246cb2699dd1d26d473\"\nuuid = \"7522ee7d-7047-56d0-94d9-4bc626e7058d\"\nversion = \"0.2.0\"\n\n[[TOML]]\ndeps = [\"Dates\"]\nuuid = \"fa267f1f-6049-4f14-aa54-33bafae1ed76\"\n\n[[TableOperations]]\ndeps = [\"Tables\", \"Test\"]\ngit-tree-sha1 = \"208630a14884abd110a8f8008b0882f0d0f5632c\"\nuuid = \"ab02a1b2-a7df-11e8-156e-fb1833f50b87\"\nversion = \"0.2.1\"\n\n[[TableTraits]]\ndeps = [\"IteratorInterfaceExtensions\"]\ngit-tree-sha1 = \"c06b2f539df1c6efa794486abfb6ed2022561a39\"\nuuid = \"3783bdb8-4a98-5b6b-af9a-565f29a5fe9c\"\nversion = \"1.0.1\"\n\n[[TableTraitsUtils]]\ndeps = [\"DataValues\", \"IteratorInterfaceExtensions\", \"Missings\", \"TableTraits\"]\ngit-tree-sha1 = \"78fecfe140d7abb480b53a44f3f85b6aa373c293\"\nuuid = \"382cd787-c1b6-5bf2-a167-d5b971a19bda\"\nversion = \"1.0.2\"\n\n[[Tables]]\ndeps = [\"DataAPI\", \"DataValueInterfaces\", \"IteratorInterfaceExtensions\", \"LinearAlgebra\", \"TableTraits\", \"Test\"]\ngit-tree-sha1 = \"fed34d0e71b91734bf0a7e10eb1bb05296ddbcd0\"\nuuid = \"bd369af6-aec1-5ad0-b16a-f7cc5008161c\"\nversion = \"1.6.0\"\n\n[[Tar]]\ndeps = [\"ArgTools\", \"SHA\"]\nuuid = \"a4e569a6-e804-4fa4-b0f3-eef7a1d5b13e\"\n\n[[Test]]\ndeps = [\"InteractiveUtils\", \"Logging\", \"Random\", \"Serialization\"]\nuuid = \"8dfed614-e22c-5e08-85e1-65c5234f0b40\"\n\n[[TextParse]]\ndeps = [\"CodecZlib\", \"DataStructures\", \"Dates\", \"DoubleFloats\", \"Mmap\", \"Nullables\", \"PooledArrays\", \"WeakRefStrings\"]\ngit-tree-sha1 = \"26b43d6746b52cca13c4cdef90f89652273b413e\"\nuuid = \"e0df1984-e451-5cb5-8b61-797a481e67e3\"\nversion = \"0.9.1\"\n\n[[TimeZones]]\ndeps = [\"Dates\", \"Future\", \"LazyArtifacts\", \"Mocking\", \"Pkg\", \"Printf\", \"RecipesBase\", \"Serialization\", \"Unicode\"]\ngit-tree-sha1 = \"a5688ffdbd849a98503c6650effe79fe89a41252\"\nuuid = \"f269a46b-ccf7-5d73-abea-4c690281aa53\"\nversion = \"1.5.9\"\n\n[[TranscodingStreams]]\ndeps = [\"Random\", \"Test\"]\ngit-tree-sha1 = \"216b95ea110b5972db65aa90f88d8d89dcb8851c\"\nuuid = \"3bb67fe8-82b1-5028-8e26-92a6c54297fa\"\nversion = \"0.9.6\"\n\n[[UUIDs]]\ndeps = [\"Random\", \"SHA\"]\nuuid = \"cf7118a7-6976-5b1a-9a39-7adc72f591a4\"\n\n[[UnPack]]\ngit-tree-sha1 = \"387c1f73762231e86e0c9c5443ce3b4a0a9a0c2b\"\nuuid = \"3a884ed6-31ef-47d7-9d2a-63182c4928ed\"\nversion = \"1.0.2\"\n\n[[Unicode]]\nuuid = \"4ec0a83e-493e-50e2-b9ac-8f72acf5a8f5\"\n\n[[WeakRefStrings]]\ndeps = [\"Missings\", \"Random\", \"Test\"]\ngit-tree-sha1 = \"960639a12ffd223ee463e93392aeb260fa325566\"\nuuid = \"ea10d353-3f73-51f8-a26c-33c1cb351aa5\"\nversion = \"0.5.8\"\n\n[[Widgets]]\ndeps = [\"Colors\", \"Dates\", \"Observables\", \"OrderedCollections\"]\ngit-tree-sha1 = \"80661f59d28714632132c73779f8becc19a113f2\"\nuuid = \"cc8bc4a8-27d6-5769-a93b-9d913e69aa62\"\nversion = \"0.6.4\"\n\n[[WoodburyMatrices]]\ndeps = [\"LinearAlgebra\", \"SparseArrays\"]\ngit-tree-sha1 = \"de67fa59e33ad156a590055375a30b23c40299d3\"\nuuid = \"efce3f68-66dc-5838-9240-27a6d6f5f9b6\"\nversion = \"0.5.5\"\n\n[[Zlib_jll]]\ndeps = [\"Libdl\"]\nuuid = \"83775a58-1f1d-513f-b197-d71354ab007a\"\n\n[[nghttp2_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"8e850ede-7688-5339-a07c-302acd2aaf8d\"\n\n[[p7zip_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"3f19e933-33d8-53b3-aaab-bd5110c3b7a0\"\n\"\"\"\n\n# \u2554\u2550\u2561 Cell order:\n# \u2560\u255088d2a9b8-d8e4-47a2-b1de-369d8fb44c20\n# \u2560\u255007ddd182-470c-11ec-3704-93171dbc510e\n# \u2560\u2550d1d0d4b2-92c1-4faa-8e42-7b7241fa1cc2\n# \u255f\u2500454637cc-589c-459d-93fd-7da3ba6fd67c\n# \u2560\u255053266a68-82e6-4024-af9d-8cefe89399f9\n# \u2560\u25504a5b02b5-83d5-46b9-88ec-41f4d0df54d1\n# \u255f\u2500ae2c7408-7ca2-4166-98be-f9ba3d8476ed\n# \u255f\u250038b7bde3-7c34-4930-8582-bfd067a05c09\n# \u2560\u2550cd85e83f-f380-480a-9c78-5fdbbba068ab\n# \u2560\u25505cb827be-5ad3-49a3-b920-b1df8a8cb861\n# \u2560\u2550df699c60-d076-4319-9d95-13ea2afe955c\n# \u2560\u2550fbd5cb7c-e722-4a2e-84ef-4c6702134234\n# \u2560\u2550e5552459-e2ad-4f44-9760-757ea37fa233\n# \u2560\u255050ca420b-2a11-4366-9541-555585eeb5ea\n# \u2560\u2550b45cf1f6-c70c-450b-a8e4-29f7eb44e791\n# \u2560\u2550e3c415b1-0964-4337-9f9f-c1a9616a8b5d\n# \u255f\u2500029a721d-4a55-4cc0-8898-61bdfb80c10c\n# \u2560\u25507ffa2874-0690-4587-91db-f0e55570bcb1\n# \u255f\u250011a22c40-1da4-4f9c-a650-000230546bfa\n# \u2560\u25503e06e476-3c9d-42c4-b167-c2ce926b2c3c\n# \u255f\u250000000000-0000-0000-0000-000000000001\n# \u255f\u250000000000-0000-0000-0000-000000000002\n", "meta": {"hexsha": "fa346b9f94c19d229ee0c7f39fb1d336f2a583d2", "size": 36196, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Benchmarking_Julia.jl", "max_stars_repo_name": "ElvisCasco/Remesas", "max_stars_repo_head_hexsha": "511165eaee92d77f976d23ed57e46264df225a3c", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "Benchmarking_Julia.jl", "max_issues_repo_name": "ElvisCasco/Remesas", "max_issues_repo_head_hexsha": "511165eaee92d77f976d23ed57e46264df225a3c", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Benchmarking_Julia.jl", "max_forks_repo_name": "ElvisCasco/Remesas", "max_forks_repo_head_hexsha": "511165eaee92d77f976d23ed57e46264df225a3c", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 31.3656845754, "max_line_length": 303, "alphanum_fraction": 0.735274616, "num_tokens": 16216, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.5, "lm_q2_score": 0.11920292045759122, "lm_q1q2_score": 0.05960146022879561}}
{"text": "using SafeTestsets\n# TODO add description to the tests\n\n## ===-===-===-===-===-===-===-===-\n@safetestset \"MatrixProcessing tests\" begin\n include(\"matrixProcessing_tests.jl\")\nend\n\n## ===-===-===-===-===-===-===-===-\n@safetestset \"MatrixOrganization tests\" begin\n include(\"matrixOrganisation_tests.jl\")\nend\n\n## ===-===-===-===-===-===-===-===-\n@safetestset \"BettiCurves tests\" begin\n include(\"bettiCurves_tests.jl\")\nend\n\n## ===-===-===-===-===-===-===-===-\n", "meta": {"hexsha": "32c6b80f005862807c1ddf0032e7a27c669d42e7", "size": 464, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/runtests.jl", "max_stars_repo_name": "edd26/TopologyPreprocessing.jl", "max_stars_repo_head_hexsha": "273497114e8adf84244d3b24b0155b3ae813b84d", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2021-01-08T02:01:57.000Z", "max_stars_repo_stars_event_max_datetime": "2021-01-08T02:01:57.000Z", "max_issues_repo_path": "test/runtests.jl", "max_issues_repo_name": "edd26/TopologyPreprocessing", "max_issues_repo_head_hexsha": "12948c0ff3885f8fc2fa6b98d46b8d1c599715fa", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 5, "max_issues_repo_issues_event_min_datetime": "2020-11-30T16:38:47.000Z", "max_issues_repo_issues_event_max_datetime": "2022-02-10T15:10:02.000Z", "max_forks_repo_path": "test/runtests.jl", "max_forks_repo_name": "edd26/TopologyPreprocessing", "max_forks_repo_head_hexsha": "12948c0ff3885f8fc2fa6b98d46b8d1c599715fa", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 23.2, "max_line_length": 45, "alphanum_fraction": 0.5431034483, "num_tokens": 152, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.5, "lm_q2_score": 0.11920291107043361, "lm_q1q2_score": 0.059601455535216806}}
{"text": "\"\"\"\nRead CSV-formatted posterior output.\n\n# API\n\nSee [`StanCSV.read_chain`](@ref) and [`StanCSV.matching_files`], neither is exported.\n\n# File format\n\nPosterior samples are supposed to be available in a CSV file which follows the following\nconventions:\n\n1. Whitespace and content after `#` is ignored. No fields are quoted or escaped. If a line\nhas a `#`, it cannot have content.\n\n2. The first non-ignored line contains a comma-separated list of variable names, of the\nformat `var[.i1.i2.\u2026]` where optional indexes follow dots. Indexes for the same variable are\ncontiguous, and have a column-major layout.\n\n3. Subsequent lines contain the same number of comma-separated floating-point values.\n\n`cmdstan` outputs posterior samples in this format (diagnostics are ignored for now), hence\nthe name.\n\"\"\"\nmodule StanCSV\n\nusing ArgCheck: @argcheck\nusing DocStringExtensions: FUNCTIONNAME, SIGNATURES\nimport ..Chains\n\n####\n#### Header parsing building blocks\n####\n\n\"\"\"\n$(SIGNATURES)\n\nParse a Stan variable name as `name::Symbol => indexes::Tuple`.\n\"\"\"\nfunction parse_variable_name(variable_name::AbstractString)\n parts = split(variable_name, '.')\n @argcheck !isempty(first(parts))\n name = Symbol(strip(first(parts)))\n index = tuple(parse.(Ref(Int), parts[2:end])...)\n name => index\nend\n\n\"\"\"\n$(SIGNATURES)\n\nGiven a vector of `name => index` pairs and a starting index `i`, which should be `1`s, find\nthe last index for this variable (ie the dimensions) and check that that intermediate\nindices are contiguous (in column major ordering).\n\nReturn `name`, `dimensions`, and the position for the continuation.\n\"\"\"\nfunction collapse_contiguous_dimensions(names_indexes, i)\n name, index = names_indexes[i]\n @argcheck all(index .== 1) \"Indexes for $(name) don't start with ones.\"\n n = length(names_indexes)\n j = i + 1\n while j \u2264 n\n if names_indexes[j][1] \u2260 name\n break\n else\n j += 1\n end\n end\n dimensions = names_indexes[j - 1][2]\n indexes = CartesianIndices(dimensions)\n for (k, ix) in zip(i:(j-1), CartesianIndices(dimensions))\n index_k = names_indexes[k][2]\n @argcheck CartesianIndex(index_k) == ix \"Non-contiguous index $(k) for $(name).\"\n end\n name, dimensions, j\nend\n\n\"\"\"\n$(SIGNATURES)\n\nParse a vector of `names => indexes` to a column schema.\n\"\"\"\nfunction parse_schema(names_indexes)\n n = length(names_indexes)\n i = 1\n schema = Pair{Symbol,Tuple{Vararg{Int}}}[]\n while i \u2264 n\n name, dimensions, next_i = collapse_contiguous_dimensions(names_indexes, i)\n @argcheck all(s -> first(schema) \u2260 name, schema) \"Duplicate name $(name) in column $(i).\"\n push!(schema, name => dimensions)\n i = next_i\n end\n Chains.IndexSchema(NamedTuple{Tuple(first.(schema))}(last.(schema)))\nend\n\n####\n#### CSV file reading.\n####\n\n\"\"\"\n$(SIGNATURES)\n\nTest if `line` is a comment line.\n\"\"\"\nis_comment_line(line) = occursin(r\"^ *#\", line)\n\n\"\"\"\n$(SIGNATURES)\n\nFind the first non-comment line and read it as a `Chains.IndexSchema`. When there is no\nsuch line, throw an `EOFError`.\n\"\"\"\nfunction read_schema(io::IO)\n while true # will throw an EOFError if not fount\n line = readline(io; keep = false)\n if !is_comment_line(line)\n return parse_schema(parse_variable_name.(split(line, ',')))\n end\n end\nend\n\n\"\"\"\n$(SIGNATURES)\n\nRead `nfields` fields from `io`, parse as `Float64`, and push into `buffer`, returning\n`true`. Fewer fields than `nfields` results in an error.\n\nIf a line starts with a `'#'`, do no parsing, return `false`.\n\"\"\"\nfunction read_csv_line!(buffer, io::IO, nfields::Integer)\n line = readline(io; keep = false)\n is_comment_line(line) && return false\n pos = 1\n last_pos = lastindex(line)\n for _ in 1:nfields\n @argcheck pos \u2264 last_pos \"Fewer than $(nfields) fields in line.\"\n delim_pos = something(findnext(isequal(','), line, pos), last_pos + 1)\n push!(buffer, parse(Float64, SubString(line, pos, delim_pos - 1)))\n pos = delim_pos + 1\n end\n true\nend\n\nfunction read_csv_flat_data!(buffer, io, nfields)\n row_count = 0\n while !eof(io)\n row_count += read_csv_line!(buffer, io, nfields)\n end\n row_count\nend\n\nfunction csv_buffer_to_array(buffer, row_count, dims)\n n = length(dims)\n permutedims(reshape(buffer, dims..., row_count), vcat([1 + n], 1:n))\nend\n\nfunction read_chain(io::IO)\n sch = read_schema(io)\n nfields = length(sch)\n buffer = Vector{Float64}()\n row_count = read_csv_flat_data!(buffer, io, nfields)\n sample_matrix = collect(permutedims(reshape(buffer, nfields, row_count)))\n Chains.Chain(sch, sample_matrix)\nend\n\n\"\"\"\n$(SIGNATURES)\n\nRead data from a CSV file, that uses the output format from Stan.\n\nReturn a vector of `name => array` pairs, where `name::Symbol` is the variable name, and the\n`array` is the data for that variable.\n\nConsecutive columns for the same variable name with `n`-dimensional indexes are assembled\ninto an `n+1` dimensional array, with index `var[row, i1, i2, \u2026]` containing the results\nfor `var.i1.i2.\u2026` in the given `row`.\n\"\"\"\nread_chain(filename::AbstractString) = open(read_chain, filename, \"r\")\n\n\"\"\"\n$(SIGNATURES)\n\nReturn a vector `id::Int => filename` where `filename` is `prefix_.csv` and the path\nis a file. Prefix should be a path, eg\n\n```julia\n$(FUNCTIONNAME)(\"/tmp/samples_\")\n```\n\nwill return `[1 => \"/tmp/samples_1.csv\", 2 => \"/tmp/samples_2.csv\", \u2026]` if the files exist.\n\nRecommended use: obtaining all the filenames in a directory.\n\"\"\"\nfunction matching_files(prefix::AbstractString)\n dir = dirname(prefix)\n base = basename(prefix)\n pattern = Regex(\"^\" * base * raw\"(\\d+)\\.csv$\")\n ids_files = Pair{Int,String}[]\n for filename in readdir(dir)\n m = match(pattern, filename)\n p = joinpath(dir, filename)\n if m \u2262 nothing && isfile(p)\n push!(ids_files, parse(Int, m.captures[1]) => p)\n end\n end\n @argcheck allunique(first.(ids_files)) \"Non-unique file ids, perhaps because of 0-padding?\"\n sort!(ids_files, by = first)\n ids_files\nend\n\nend\n", "meta": {"hexsha": "ede427bc43a19f7c87fcd05a0448470da954a3f9", "size": 6123, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/stan_csv.jl", "max_stars_repo_name": "JuliaTagBot/MCMCStorage.jl", "max_stars_repo_head_hexsha": "69497470ed71c00ee975e6a1d243c4f2f83ca49e", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2019-06-14T01:37:19.000Z", "max_stars_repo_stars_event_max_datetime": "2019-06-14T01:37:19.000Z", "max_issues_repo_path": "src/stan_csv.jl", "max_issues_repo_name": "JuliaTagBot/MCMCStorage.jl", "max_issues_repo_head_hexsha": "69497470ed71c00ee975e6a1d243c4f2f83ca49e", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/stan_csv.jl", "max_forks_repo_name": "JuliaTagBot/MCMCStorage.jl", "max_forks_repo_head_hexsha": "69497470ed71c00ee975e6a1d243c4f2f83ca49e", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2020-02-08T11:48:39.000Z", "max_forks_repo_forks_event_max_datetime": "2020-02-08T11:48:39.000Z", "avg_line_length": 28.8820754717, "max_line_length": 97, "alphanum_fraction": 0.6731994121, "num_tokens": 1604, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.44167300566462553, "lm_q2_score": 0.13477591568531197, "lm_q1q2_score": 0.05952688377193389}}
{"text": "# Bogumi\u0142 Kami\u0144ski, 2022\n\n# Codes for chapter 12\n\n# Codes for section 12.1\n\n# Code for listing 12.1\n\nimport Downloads\nusing SHA\ngit_zip = \"git_web_ml.zip\"\nif !isfile(git_zip)\n Downloads.download(\"https://snap.stanford.edu/data/\" *\n \"git_web_ml.zip\",\n git_zip)\nend\nisfile(git_zip)\nopen(sha256, git_zip) == [0x56, 0xc0, 0xc1, 0xc2,\n 0xc4, 0x60, 0xdc, 0x4c,\n 0x7b, 0xf8, 0x93, 0x57,\n 0xb1, 0xfe, 0xc0, 0x20,\n 0xf4, 0x5e, 0x2e, 0xce,\n 0xba, 0xb8, 0x1d, 0x13,\n 0x1d, 0x07, 0x3b, 0x10,\n 0xe2, 0x8e, 0xc0, 0x31]\n\n# Code for opening a zip archive\n\nimport ZipFile\ngit_archive = ZipFile.Reader(git_zip)\n\n# Code for listing 12.2\n\nfunction ingest_to_df(archive::ZipFile.Reader, filename::AbstractString)\n idx = only(findall(x -> x.name == filename, archive.files))\n return CSV.read(read(archive.files[idx]), DataFrame)\nend\n\n# Code for working with zip archive\n\ngit_archive.files\n\ngit_archive.files[2].name\n\nfindall(x -> x.name == \"git_web_ml/musae_git_edges.csv\", git_archive.files)\nfindall(x -> x.name == \"\", git_archive.files)\n\nonly(findall(x -> x.name == \"git_web_ml/musae_git_edges.csv\", git_archive.files))\nonly(findall(x -> x.name == \"\", git_archive.files))\n\n# Code for listing 12.3\n\nusing CSV\nusing DataFrames\nedges_df = ingest_to_df(git_archive, \"git_web_ml/musae_git_edges.csv\");\nclasses_df = ingest_to_df(git_archive, \"git_web_ml/musae_git_target.csv\");\nclose(git_archive)\nsummary(edges_df)\ndescribe(edges_df, :min, :max, :mean, :nmissing, :eltype)\nsummary(classes_df)\ndescribe(classes_df, :min, :max, :mean, :nmissing, :eltype)\n\n# Code for updating data frame columns using broadcasting\n\nedges_df .+= 1\nclasses_df.id .+= 1\n\n# Code for examples of data frame broadcasting\n\ndf = DataFrame(a=1:3, b=[4, missing, 5])\ndf .^ 2\ncoalesce.(df, 0)\ndf .+ [10, 11, 12]\n\n# Code for checking the order of :id column in a data frame\n\nclasses_df.id == axes(classes_df, 1)\n\n# Code for the difference between ! and : in broadcasting assignment\n\ndf = DataFrame(a=1:3, b=1:3)\ndf[!, :a] .= \"x\"\ndf[:, :b] .= \"x\"\ndf\n\n# Code for the difference between ! and : in assignment\n\ndf = DataFrame(a=1:3, b=1:3, c=1:3)\ndf[!, :a] = [\"x\", \"y\", \"z\"]\ndf[:, :b] = [\"x\", \"y\", \"z\"]\ndf[:, :c] = [11, 12, 13]\ndf\n\n# Codes for section 12.2\n\n# Code for listing 12.4\n\nusing Graphs\ngh = SimpleGraph(nrow(classes_df))\nfor (src, dst) in eachrow(edges_df)\n add_edge!(gh, src, dst)\nend\ngh\nne(gh)\nnv(gh)\n\n# Code for iterator destruction in iteration specification\n\nmat = [1 2; 3 4; 5 6]\nfor (x1, x2) in eachrow(mat)\n @show x1, x2\nend\n\n# Code for getting degrees of nodes in the graph\n\ndegree(gh)\n\n# Code for adding a column to a data frame\n\nclasses_df.deg = degree(gh)\n\n# Code for the difference between ! and : when adding a column\n\ndf = DataFrame()\nx = [1, 2, 3]\ndf[!, :x1] = x\ndf[:, :x2] = x\ndf\ndf.x1 === x\ndf.x2 === x\ndf.x2 == x\n\n# Code for creating a column using broadcasting\n\ndf.x3 .= 1\ndf\n\n# Code for edge iterator of a graph\n\nedges(gh)\n\ne1 = first(edges(gh))\ndump(e1)\ne1.src\ne1.dst\n\n# Code for listing 12.5\n\nfunction deg_class(gh, class)\n deg_ml = zeros(Int, length(class))\n deg_web = zeros(Int, length(class))\n for edge in edges(gh)\n a, b = edge.src, edge.dst\n if class[b] == 1\n deg_ml[a] += 1\n else\n deg_web[a] += 1\n end\n if class[a] == 1\n deg_ml[b] += 1\n else\n deg_web[b] += 1\n end\n end\n return (deg_ml, deg_web)\nend\n\n# Code for computing machine learning and web neighbors for gh graph\n\nclasses_df.deg_ml, classes_df.deg_web =\ndeg_class(gh, classes_df.ml_target)\n\n# Code for checking type stability of deg_class function\n\n@time deg_class(gh, classes_df.ml_target);\n@code_warntype deg_class(gh, classes_df.ml_target)\n\n# Code for checking the classes_df summary statistics\n\ndescribe(classes_df, :min, :max, :mean, :std)\n\n# Code for average degree of node in the graph\n\n2 * ne(gh) / nv(gh)\n\n# Code for checking correctness of computations\n\nclasses_df.deg_ml + classes_df.deg_web == classes_df.deg\n\n# Code for showing that DataFrames.jl checks consistency of stored objects\n\ndf = DataFrame(a=1, b=11)\npush!(df.a, 2)\ndf\n\n# Codes for section 12.3\n\n# Code for computing groupwise means of columns\n\nusing Statistics\nfor type in [0, 1], col in [\"deg_ml\", \"deg_web\"]\n println((type, col, mean(classes_df[classes_df.ml_target .== type, col])))\nend\n\ngdf = groupby(classes_df, :ml_target)\n\ncombine(gdf,\n :deg_ml => mean => :mean_deg_ml,\n :deg_web => mean => :mean_deg_web)\n\nusing DataFramesMeta\n@combine(gdf,\n :mean_deg_ml = mean(:deg_ml),\n :mean_deg_web = mean(:deg_web))\n\n# Code for simple plotting of relationship between developer degree and type\n\nusing Plots\nscatter(classes_df.deg_ml, classes_df.deg_web;\n color=[x == 1 ? \"black\" : \"gray\" for x in classes_df.ml_target],\n xlabel=\"degree ml\", ylabel=\"degree web\", labels=false)\n\n# Code for aggregation of degree data\n\nagg_df = combine(groupby(classes_df, [:deg_ml, :deg_web]),\n :ml_target => (x -> 1 - mean(x)) => :web_mean)\n\n# Code for comparison how Julia parses expressions\n\n:ml_target => (x -> 1 - mean(x)) => :web_mean\n:ml_target => x -> 1 - mean(x) => :web_mean\n\n# Code for aggregation using DataFramesMeta.jl\n\n@combine(groupby(classes_df, [:deg_ml, :deg_web]),\n :web_mean = 1 - mean(:ml_target))\n\n# Code for getting summary information about the aggregated data frame\n\ndescribe(agg_df)\n\n# Code for log1p function\n\nlog1p(0)\n\n# Code for listing 12.6\n\nfunction gen_ticks(maxv)\n max2 = round(Int, log2(maxv))\n tick = [0; 2 .^ (0:max2)]\n return (log1p.(tick), tick)\nend\n\nlog1pjitter(x) = log1p(x) - 0.05 + rand() / 10\n\nusing Random\nRandom.seed!(1234);\nscatter(log1pjitter.(agg_df.deg_ml),\n log1pjitter.(agg_df.deg_web);\n zcolor=agg_df.web_mean,\n xlabel=\"degree ml\", ylabel=\"degree web\",\n markersize=2, markerstrokewidth=0, markeralpha=0.8,\n legend=:topleft, labels=\"fraction web\",\n xticks=gen_ticks(maximum(classes_df.deg_ml)),\n yticks=gen_ticks(maximum(classes_df.deg_web)))\n\n# Code for fitting logistic regression model\n\nusing GLM\nglm(@formula(ml_target~log1p(deg_ml)+log1p(deg_web)), classes_df, Binomial(), LogitLink())\n\n# Code for inspecting @formula result\n\n@formula(ml_target~log1p(deg_ml)+log1p(deg_web))\n\n# Code for inserting columns to a data frame\n\ndf = DataFrame(x=1:2)\ninsertcols!(df, :y => 4:5)\ninsertcols!(df, :y => 4:5)\ninsertcols!(df, :z => 1)\n\ninsertcols!(df, 1, :a => 0)\ninsertcols!(df, :x, :pre_x => 2)\ninsertcols!(df, :x, :post_x => 3, after=true)\n", "meta": {"hexsha": "98878f4c9392367bfe44016f3c66ee99300a3f49", "size": 6745, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "ch12.jl", "max_stars_repo_name": "Mo-Gul/JuliaForDataAnalysis", "max_stars_repo_head_hexsha": "28af42c8fb30bb56569281c9d2978ad76069e3b8", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "ch12.jl", "max_issues_repo_name": "Mo-Gul/JuliaForDataAnalysis", "max_issues_repo_head_hexsha": "28af42c8fb30bb56569281c9d2978ad76069e3b8", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "ch12.jl", "max_forks_repo_name": "Mo-Gul/JuliaForDataAnalysis", "max_forks_repo_head_hexsha": "28af42c8fb30bb56569281c9d2978ad76069e3b8", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 23.5839160839, "max_line_length": 90, "alphanum_fraction": 0.6532246108, "num_tokens": 2083, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4726834766204328, "lm_q2_score": 0.1259227615549033, "lm_q1q2_score": 0.059521608717417476}}
{"text": "ENV[\"JUPYTER\"]=\"/home/idies/miniconda3/bin/jupyter\"\nusing Pkg\nPkg.add(\"IJulia\")\nPkg.add(\"Plots\")\nPkg.add(\"PyPlot\")\nPkg.add(\"PyCall\")\n", "meta": {"hexsha": "647557dd60b08894d9582dd2b0814a268e7ad9a0", "size": 133, "ext": "jl", "lang": "Julia", "max_stars_repo_path": ".refactored/oceanography/2.0/julia-setup.jl", "max_stars_repo_name": "sciserver/sciserver-base-images", "max_stars_repo_head_hexsha": "bde00a4d28b4e7e42e2dd3240c0a2bab5d88bf88", "max_stars_repo_licenses": ["Apache-2.0"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2021-06-28T21:57:35.000Z", "max_stars_repo_stars_event_max_datetime": "2021-06-28T21:57:35.000Z", "max_issues_repo_path": ".refactored/sciserver-julia/julia-setup.jl", "max_issues_repo_name": "sciserver/sciserver-base-images", "max_issues_repo_head_hexsha": "bde00a4d28b4e7e42e2dd3240c0a2bab5d88bf88", "max_issues_repo_licenses": ["Apache-2.0"], "max_issues_count": 7, "max_issues_repo_issues_event_min_datetime": "2020-04-01T20:34:53.000Z", "max_issues_repo_issues_event_max_datetime": "2021-12-20T17:17:22.000Z", "max_forks_repo_path": ".refactored/oceanography/2.1/julia-setup.jl", "max_forks_repo_name": "sciserver/sciserver-base-images", "max_forks_repo_head_hexsha": "bde00a4d28b4e7e42e2dd3240c0a2bab5d88bf88", "max_forks_repo_licenses": ["Apache-2.0"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 19.0, "max_line_length": 51, "alphanum_fraction": 0.7067669173, "num_tokens": 52, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4532618480153861, "lm_q2_score": 0.13117323225300487, "lm_q1q2_score": 0.059455821661148434}}
{"text": "\r\n# ============================================================================ #\r\n# Variable\r\n# ============================================================================ #\r\n\r\n\r\nmutable struct Variable <: AbstractVariable\r\n name::String\r\n value::Float64\r\n\r\n function Variable(name, value = 0.0)\r\n return new(string(name), value)\r\n end\r\nend\r\n\r\nmacro variable(varname, value = default_variable_value())\r\n lhs = esc(varname)\r\n rhs = :( Variable( $(QuoteNode(varname)), $value) )\r\n ex = :($(lhs) = $(rhs))\r\n\r\n return ex\r\nend\r\n\r\nfunction evaluate(x::Variable)\r\n return x.value\r\nend\r\n\r\nfunction evaluate(x::A) where {A <: AbstractArray{<: Variable}}\r\n s = similar(x)\r\n for ii in eachindex(x)\r\n s[ii] = x[ii].value\r\n end\r\n return s\r\nend\r\n\r\nfunction to_string(x::Variable)\r\n return x.name\r\nend\r\n\r\nfunction clone(x::Variable)\r\n y = Variable(x.name, x.value)\r\n return y\r\nend\r\n\r\n\r\n", "meta": {"hexsha": "0055d8991c8a5417a1bbc6268580b80618644682", "size": 943, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "symbolics/src/variable.jl", "max_stars_repo_name": "HomoModelicus/julia", "max_stars_repo_head_hexsha": "26be81348032ccd2728046193ce627c823a3804b", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "symbolics/src/variable.jl", "max_issues_repo_name": "HomoModelicus/julia", "max_issues_repo_head_hexsha": "26be81348032ccd2728046193ce627c823a3804b", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "symbolics/src/variable.jl", "max_forks_repo_name": "HomoModelicus/julia", "max_forks_repo_head_hexsha": "26be81348032ccd2728046193ce627c823a3804b", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 20.5, "max_line_length": 81, "alphanum_fraction": 0.4899257688, "num_tokens": 210, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4843800842769844, "lm_q2_score": 0.12252322533450247, "lm_q1q2_score": 0.05934781021341426}}
{"text": "\nstruct RandomStrategy{G <: Game} <: Strategy end\n\nfunction BoardGames.getmove(board, s::RandomStrategy)\n rand(getmoves(board))\nend\n\nfunction BoardGames.getvarsnames(s::RandomStrategy)\n return String[]\nend\n\nfunction BoardGames.getvalues(s::RandomStrategy)\n return ()\nend\n\nBoardGames.name(::RandomStrategy) = \"Random Selection\"\n\nfunction Base.copy(s::RandomStrategy)\n return s\nend", "meta": {"hexsha": "86b60f8c5e46c3a87f457ba587c0c2ff5900049c", "size": 391, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/RandomStrategy.jl", "max_stars_repo_name": "antcap96/BoardGamesStrategies.jl", "max_stars_repo_head_hexsha": "b7fda322d76726ef778eaf663a759f02c38de78a", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2021-05-12T09:22:10.000Z", "max_stars_repo_stars_event_max_datetime": "2021-05-12T09:22:10.000Z", "max_issues_repo_path": "src/RandomStrategy.jl", "max_issues_repo_name": "antcap96/BoardGamesStrategies.jl", "max_issues_repo_head_hexsha": "b7fda322d76726ef778eaf663a759f02c38de78a", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/RandomStrategy.jl", "max_forks_repo_name": "antcap96/BoardGamesStrategies.jl", "max_forks_repo_head_hexsha": "b7fda322d76726ef778eaf663a759f02c38de78a", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 19.55, "max_line_length": 54, "alphanum_fraction": 0.7570332481, "num_tokens": 91, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4649015713733884, "lm_q2_score": 0.12765263361856616, "lm_q1q2_score": 0.059345909959222835}}
{"text": "### A Pluto.jl notebook ###\n# v0.14.1\n\nusing Markdown\nusing InteractiveUtils\n\n# \u2554\u2550\u2561 de098278-8e74-11eb-13e3-49c1b6e06e7d\nusing Pkg; Pkg.activate(\"MLJ_env\", shared=true)\n\n# \u2554\u2550\u2561 ac463e7a-8b59-11eb-229e-db560e17c5f5\nbegin\n\tusing Test\n\tusing PlutoUI\nend\n\n# \u2554\u2550\u2561 8c80e072-8b59-11eb-3c21-a18fe43c4536\nmd\"\"\"\n## Databases and SQL\n\nref. from book **\"Data Science from Scratch\"**, Chap 24\n\n$(html\"© Pascal, April 2021
\")\n\"\"\"\n\n# \u2554\u2550\u2561 e7373726-8b59-11eb-2a2b-b5138e4f5268\nPlutoUI.TableOfContents(indent=true, depth=4, aside=true)\n\n# \u2554\u2550\u2561 f5ee64b2-8b59-11eb-2751-0778efd589cd\nhtml\"\"\"\n\n\"\"\"\n\n# \u2554\u2550\u2561 81290d1c-8ce2-11eb-3340-337957fd81b7\nhtml\"\"\"\n pkey is given by id(self)\n\t row_ids = filter(p -> p.first == id(self), row)\n\t if length(row_ids) == 0\n\t\tpk_id = gen_pkey_value(self)\n\t\trow = [row..., id(self) => pk_id]\n\t else\n\t\tid_val = row_ids[1][2] ## value assoc with this pkey\n\t\trow_already_inserted(self, id_val) && (return nothing)\n\t end\n\t push!(self.rows, Dict(row...))\n\tend\n\n\tfunction insert(self::Table, row::Vector{Any})\n\t \"\"\"\n\t row is something like: [9, \"Foo\", 2 ] - just pure values\n\t \"\"\"\n\t length(row) \u2260 length(self.types) && id(self) \u2208 map(p -> p[1], row) &&\n\t\tthrow(ArgumentError(\"Mismatch with expected number of columns \"))\n\t #\n\t check_value_dtype(self, row)\n\t ## row[1] is the value assoc. with pkey\n\t row_already_inserted(self, row[1]) && (return nothing)\n\t push!(self.rows, Dict(zip(self.columns, row)))\n\tend\n\n\tfunction insert(self::Table, row::Row)\n\t \"\"\"\n\t row is a Dict, like: insert(Users, Dict(:name => \"Foo\", :num_friends => 5))\n\t \"\"\"\n\t length(row) \u2260 length(self.types) && haskey(row, id(self)) &&\n\t\tthrow(ArgumentError(\"Mismatch with expected number of columns\"))\n\t check_value_dtype(self, row)\n\t if haskey(row, id(self))\n\t\trow_already_inserted(self, row[id(self)]) && (return nothing)\n\t else\n\t\trow[id(self)] = gen_pkey_value(self)\n\t end\n\t push!(self.rows, row)\n\tend\n\n\tinsert(self::Table, rows::Vector{T}) where T <: GRow = insert.(Ref(self), rows)\n\n\tfunction coltype(self::Table, colname::Symbol)::UDT\n\t ix = findfirst(col -> col == colname, self.columns)\n\t ix === nothing && throw(ArgumentError(\"column $(colname) inexistent\"))\n\t self.types[ix]\n\tend\n\n\t# function Base.show(io::IO, self::Table)\n\t# \ts = \"num. records: $(length(self)):\\n\"\n\t# \tfor rd \u2208 self.rows\n\t# \t\ts\u2081 = []\n\t# \t\tfor (k, v) \u2208 rd\n\t# \t\t\tpush!(s\u2081, \"$(k): $(v)\")\n\t# \t\tend\n\t# \t\ts = string(s, \" <\", join(s\u2081, \", \"), \">\\n\")\n\t# \tend\n\t# \tprint(io, \"$(s)\")\n\t# end\n\n\n\t##\n\t## Internal checkers\n\t##\n\n\tfunction check_value_dtype(self::Table, values::Vector{DT}) where DT <: Any\n\t dtypes = self.types\n\t for (v, dt) \u2208 zip(values, dtypes)\n\t\t!(typeof(v) <: dt) && v !== nothing &&\n\t\t throw(\"Expected type for <$(v)>: $(dt), got: $(typeof(v))\")\n\t end\n\tend\n\n\n\tfunction check_value_dtype(self::Table,\n\t\t\t\t\t\t\t row::Union{Row, Vector{Pair{Symbol, DT}}}) where DT <: Any\n\t (cols, types) = self.columns, self.types\n\t col_type = Dict(zip(cols, types))\n\t for (k, v) \u2208 row\n\t\t(haskey(col_type, k) && typeof(v) <: col_type[k]) ||\n\t\t throw(\"Expected type/2: $(col_type[k]), got: $(typeof(v))\")\n\t end\n\tend\n\n\tfunction row_already_inserted(self::Table, id_val::Any)::Bool\n\t\tid(self) === nothing && return false ## no pkey => ignore check\n\t\t#\n\t\tres = filter(r -> r[id(self)] == id_val, self.rows)\n\t\tlength(res) > 0 ## row already inserted id > 0\n\tend\n\nend\n\n# \u2554\u2550\u2561 d146a8ac-92cc-11eb-29b1-bb065a468477\nmd\"\"\"\nInsert using an array/vector:\n\"\"\"\n\n# \u2554\u2550\u2561 13a0940e-8ce4-11eb-231c-f331a607203c\nbegin\n\tinsert(Users, [0, \"Hero\", 0])\n\t@assert length(Users) \u2265 1\nend\n\n# \u2554\u2550\u2561 e3458d34-92cc-11eb-2426-79f229abd908\nmd\"\"\"\nInsert using a vector of pairs:\n\"\"\"\n\n# \u2554\u2550\u2561 96c2a668-9290-11eb-074d-19ffeb68eba6\nbegin\n\tinsert(Users, [id(Users) => 1, :name => \"Dunn\", :num_friends => 2])\n\t@assert length(Users) \u2265 2\nend\n\n# \u2554\u2550\u2561 f1357972-92cc-11eb-1f73-43aae5a43aa5\nmd\"\"\"\nInsert using a dictionary\n\"\"\"\n\n# \u2554\u2550\u2561 5a3a8fdc-929d-11eb-25e3-e309dffdb455\nbegin\n\t## Insert with a Dict\n\tinsert(Users, Dict(id(Users) => 2, :name => \"Sue\", :num_friends => 3))\n\t@assert length(Users) \u2265 3\nend\n\n# \u2554\u2550\u2561 0292e48e-92cd-11eb-1882-81a2faaad136\nmd\"\"\"\nInsert using a dictionary, no primary key value specified:\n\"\"\"\n\n# \u2554\u2550\u2561 8545d17c-929e-11eb-0989-ffd4749bbb25\nbegin\n\t## Insert with a Dict no pkey/1\n\tinsert(Users, Dict(:name => \"Ayumi\", :num_friends => 5))\n\t@assert length(Users) \u2265 4\nend\n\n# \u2554\u2550\u2561 95d2b19e-92a4-11eb-0f91-e343c9317983\nbegin\n\t## Insert with a Dict no pkey/2\n\tinsert(Users, [:name => \"PasMas\", :num_friends => 5])\n\t@assert length(Users) \u2265 5\nend\n\n# \u2554\u2550\u2561 8a945abc-012f-4ace-a67a-b7e0f520346b\nbegin\n\t## Insert with a Dict no pkey/2 + null value (encoded as nothing)\n\t# insert(Users,\n\t# \t[:name => \"Foobar\", :num_friends => nothing])\n\tinsert(Users,\n\t\t[:name => \"Foobar\", :num_friends => nothing, :user_id => 17])\nend\n\n# \u2554\u2550\u2561 28ba0bf6-92cd-11eb-0669-75eadb768518\nmd\"\"\"\nInsert using a collection (vector of rows):\n\"\"\"\n\n# \u2554\u2550\u2561 d4562f7e-9290-11eb-2d8c-952f2e0edfed\nbegin\n\t## Insert a collection (Vector of) of Records \n\tinsert(Users, [\n\t\t[5, \"Chi\", 3],\n\t\t[6, \"Thor\", 3],\n\t\t[7, \"Clive\", 2],\n\t\t[8, \"Devin\", 2],\n\t\t[9, \"Kate\", 2],\n\t\t[10, \"Kaze\", nothing]\n\t])\n\t@assert length(Users) \u2265 10\nend\n\n# \u2554\u2550\u2561 f877c3e6-929c-11eb-00cd-c15568f99627\nUsers\n\n# \u2554\u2550\u2561 d5657a88-92c4-11eb-2a4f-c7dcaa97bcf6\nbegin\n\t@test_throws ArgumentError coltype(Users, :foobar)\n\t@test coltype(Users, :num_friends) == U_IN\n\t@test coltype(Users, :name) == U_SN\nend\n\n# \u2554\u2550\u2561 80ddf0f4-8ce2-11eb-3046-331119a0dc9b\nhtml\"\"\"\nop))), TimeArray{Float64,2}) == true\n @test isa(convert(cl.>op), TimeArray{Float64,1}) == true\n @test isa(convert(merge(cl.op)), TimeArray{Float64,2}) == true\n end\nend\n\n\n@testset \"index by integer works with both 1d and 2d time array\" begin\n @testset \"1d time array\" begin\n @test cl[1].timestamp == [Date(2000,1,3)]\n @test cl[1].values == [111.94]\n @test cl[1].colnames == [\"Close\"]\n @test cl[1].meta == \"AAPL\"\n end\n\n @testset \"2d time array\" begin\n @test ohlc[1].timestamp == [Date(2000,1,3)]\n @test ohlc[1].values == [104.88 112.5 101.69 111.94]\n @test ohlc[1].colnames == [\"Open\", \"High\", \"Low\",\"Close\"]\n @test ohlc[1].meta == \"AAPL\"\n end\nend\n\n\n@testset \"ordered collection methods\" begin\n @testset \"iterator protocol is valid\" begin\n @test !isempty(op)\n @test isempty(op[op .< 0])\n @test start(op) == 1\n @test next(op, 1) == ((op.timestamp[1], op.values[1,:]), 2)\n @test done(op, length(op)+1) == true\n end\n\n @testset \"end keyword returns correct index\" begin\n @test ohlc[end].timestamp[1] == ohlc.timestamp[end]\n end\n\n @testset \"getindex on single Int and Date\" begin\n @test ohlc[1].timestamp == [Date(2000,1,3)]\n @test ohlc[Date(2000,1,3)].timestamp == [Date(2000,1,3)]\n end\n\n @testset \"getindex on array of Int and Date\" begin\n @test ohlc[[1,10]].timestamp == [Date(2000,1,3), Date(2000,1,14)]\n @test ohlc[[Date(2000,1,3),Date(2000,1,14)]].timestamp == [Date(2000,1,3), Date(2000,1,14)]\n end\n\n @testset \"getindex on range of Int and Date\" begin\n @test ohlc[1:2].timestamp == [Date(2000,1,3), Date(2000,1,4)]\n @test ohlc[1:2:4].timestamp == [Date(2000,1,3), Date(2000,1,5)]\n @test ohlc[Int8(1):Int8(2):Int8(4)].timestamp == [Date(2000,1,3), Date(2000,1,5)]\n @test ohlc[Date(2000,1,3):Day(1):Date(2000,1,4)].timestamp == [Date(2000,1,3), Date(2000,1,4)]\n end\n\n @testset \"getindex on range of DateTime when only Date is in timestamp\" begin\n @test_throws(\n MethodError,\n ohlc[DateTime(2000,1,3,0,0,0)])\n @test_throws(\n MethodError,\n ohlc[[DateTime(2000,1,3,0,0,0),DateTime(2000,1,14,0,0,0)]])\n @test_throws(\n MethodError,\n ohlc[DateTime(2000,1,3,0,0,0):Day(1):DateTime(2000,1,4,0,0,0)])\n end\n\n @testset \"getindex on range of Date\" begin\n @test length(cl[Date(2000,1,1):Date(2001,12,31)]) == 500\n end\n\n @testset \"getindex on single column name\" begin\n @test size(ohlc[\"Open\"].values, 2) == 1\n @test size(ohlc[\"Open\"][Date(2000,1,3):Day(1):Date(2000,1,14)].values, 1) == 10\n end\n\n @testset \"getindex on multiple column name\" begin\n @test ohlc[\"Open\", \"Close\"].values[1] == 104.88\n @test ohlc[\"Open\", \"Close\"].values[2] == 108.25\n @test ohlc[\"Open\", \"Close\"].values[501] == 111.94\n end\n\n @testset \"getindex on 1d returns 1d object\" begin\n @test isa(cl[1], TimeArray{Float64,1}) == true\n @test isa(cl[1:2], TimeArray{Float64,1}) == true\n end\n\n @testset \"getindex on a 1d Boolean TimeArray returns appropriate rows\" begin\n @test ohlc[op .> cl][2].values == ohlc[4].values\n @test ohlc[op[300:end] .> cl][2].timestamp == ohlc[303].timestamp\n # MethodError, Bool must be 1D-TimeArray\n @test_throws MethodError ohlc[merge(op.>cl, op. GLPK.Optimizer())\ncreate_baseline(my_suite, \"glpk_master\"; directory = \"/tmp\", verbose = true)\n```\n\"\"\"\nfunction create_baseline(\n suite::BenchmarkTools.BenchmarkGroup,\n name::String;\n directory::String = \"\",\n kwargs...,\n)\n tune!(suite)\n BenchmarkTools.save(\n joinpath(directory, name * \"_params.json\"),\n params(suite),\n )\n results = run(suite; kwargs...)\n BenchmarkTools.save(joinpath(directory, name * \"_baseline.json\"), results)\n return\nend\n\n\"\"\"\n compare_against_baseline(\n suite, name::String; directory::String = \"\",\n report_filename::String = \"report.txt\"\n )\n\nRun all benchmarks in `suite` and compare against files called `name` in\n`directory` that were created by a call to `create_baseline`.\n\nA report summarizing the comparison is written to `report_filename` in\n`directory`.\n\nExtra `kwargs` are based to `BenchmarkTools.run`.\n\n### Examples\n\n```julia\nmy_suite = suite(() -> GLPK.Optimizer())\ncompare_against_baseline(\n my_suite, \"glpk_master\"; directory = \"/tmp\", verbose = true\n)\n```\n\"\"\"\nfunction compare_against_baseline(\n suite::BenchmarkTools.BenchmarkGroup,\n name::String;\n directory::String = \"\",\n report_filename::String = \"report.txt\",\n kwargs...,\n)\n params_filename = joinpath(directory, name * \"_params.json\")\n baseline_filename = joinpath(directory, name * \"_baseline.json\")\n if !isfile(params_filename) || !isfile(baseline_filename)\n error(\"You create a baseline with `create_baseline` first.\")\n end\n loadparams!(\n suite,\n BenchmarkTools.load(params_filename)[1],\n :evals,\n :samples,\n )\n new_results = run(suite; kwargs...)\n old_results = BenchmarkTools.load(baseline_filename)[1]\n open(joinpath(directory, report_filename), \"w\") do io\n println(stdout, \"\\n========== Results ==========\")\n println(io, \"\\n========== Results ==========\")\n for key in keys(new_results)\n judgement = judge(\n BenchmarkTools.median(new_results[key]),\n BenchmarkTools.median(old_results[key]),\n )\n println(stdout, \"\\n\", key)\n println(io, \"\\n\", key)\n show(stdout, MIME\"text/plain\"(), judgement)\n show(io, MIME\"text/plain\"(), judgement)\n end\n end\n return\nend\n\n###\n### Benchmarks\n###\n\nmacro add_benchmark(f)\n name = f.args[1].args[1]\n return quote\n $(esc(f))\n BENCHMARKS[String($(Base.Meta.quot(name)))] = $(esc(name))\n end\nend\n\n@add_benchmark function add_variable(new_model)\n model = new_model()\n for i in 1:10_000\n MOI.add_variable(model)\n end\n return model\nend\n\n@add_benchmark function add_variables(new_model)\n model = new_model()\n MOI.add_variables(model, 10_000)\n return model\nend\n\n@add_benchmark function add_variable_constraint(new_model)\n model = new_model()\n x = MOI.add_variables(model, 10_000)\n for (i, xi) in enumerate(x)\n MOI.add_constraint(model, MOI.SingleVariable(xi), MOI.LessThan(1.0 * i))\n end\n return model\nend\n\n@add_benchmark function add_variable_constraints(new_model)\n model = new_model()\n x = MOI.add_variables(model, 10_000)\n MOI.add_constraints(\n model,\n MOI.SingleVariable.(x),\n MOI.LessThan.(1.0:10_000.0),\n )\n return model\nend\n\n@add_benchmark function delete_variable(new_model)\n model = new_model()\n x = MOI.add_variables(model, 1_000)\n MOI.add_constraint.(model, MOI.SingleVariable.(x), Ref(MOI.LessThan(1.0)))\n MOI.delete.(model, x)\n return model\nend\n\n@add_benchmark function delete_variable_constraint(new_model)\n model = new_model()\n x = MOI.add_variables(model, 1_000)\n cons =\n MOI.add_constraint.(\n model,\n MOI.SingleVariable.(x),\n Ref(MOI.LessThan(1.0)),\n )\n for con in cons\n MOI.delete(model, con)\n end\n cons =\n MOI.add_constraint.(\n model,\n MOI.SingleVariable.(x),\n Ref(MOI.LessThan(1.0)),\n )\n MOI.set(model, MOI.ObjectiveSense(), MOI.MAX_SENSE)\n MOI.set(\n model,\n MOI.ObjectiveFunction{MOI.ScalarAffineFunction{Float64}}(),\n MOI.ScalarAffineFunction(MOI.ScalarAffineTerm.(1.0, x), 0.0),\n )\n MOI.optimize!(model)\n for con in cons\n MOI.delete(model, con)\n end\n return model\nend\n\n@add_benchmark function add_constraint(new_model)\n model = new_model()\n index = MOI.add_variables(model, 10_000)\n for (i, x) in enumerate(index)\n MOI.add_constraint(\n model,\n MOI.ScalarAffineFunction([MOI.ScalarAffineTerm(1.0, x)], 0.0),\n MOI.LessThan(1.0 * i),\n )\n end\n return model\nend\n\n@add_benchmark function add_constraints(new_model)\n model = new_model()\n x = MOI.add_variables(model, 10_000)\n MOI.add_constraints(\n model,\n [\n MOI.ScalarAffineFunction([MOI.ScalarAffineTerm(1.0, xi)], 0.0) for\n xi in x\n ],\n MOI.LessThan.(1:1.0:10_000),\n )\n return model\nend\n\n@add_benchmark function delete_constraint(new_model)\n model = new_model()\n index = MOI.add_variables(model, 1_000)\n cons = Vector{\n MOI.ConstraintIndex{\n MOI.ScalarAffineFunction{Float64},\n MOI.LessThan{Float64},\n },\n }(\n undef,\n 1_000,\n )\n for (i, x) in enumerate(index)\n cons[i] = MOI.add_constraint(\n model,\n MOI.ScalarAffineFunction([MOI.ScalarAffineTerm(1.0, x)], 0.0),\n MOI.LessThan(1.0 * i),\n )\n end\n for con in cons\n MOI.delete(model, con)\n end\n return model\nend\n\n@add_benchmark function copy_model(new_model)\n model = new_model()\n index = MOI.add_variables(model, 1_000)\n cons = Vector{\n MOI.ConstraintIndex{\n MOI.ScalarAffineFunction{Float64},\n MOI.LessThan{Float64},\n },\n }(\n undef,\n 1_000,\n )\n for (i, x) in enumerate(index)\n cons[i] = MOI.add_constraint(\n model,\n MOI.ScalarAffineFunction([MOI.ScalarAffineTerm(1.0, x)], 0.0),\n MOI.LessThan(1.0 * i),\n )\n end\n\n model2 = new_model()\n MOI.copy_to(model2, model)\n # MOI.copy_to(model2, model, filter_constraints=(x) -> x in cons[1:500])\n\n return model2\nend\n\nend\n", "meta": {"hexsha": "bb2e7eb178a8363c41582bec5c65c9634323ca19", "size": 7457, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/Benchmarks/Benchmarks.jl", "max_stars_repo_name": "guimarqu/MathOptInterface.jl", "max_stars_repo_head_hexsha": "cc1814d8d02d6708a307e4e12abcf81f379d2373", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/Benchmarks/Benchmarks.jl", "max_issues_repo_name": "guimarqu/MathOptInterface.jl", "max_issues_repo_head_hexsha": "cc1814d8d02d6708a307e4e12abcf81f379d2373", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/Benchmarks/Benchmarks.jl", "max_forks_repo_name": "guimarqu/MathOptInterface.jl", "max_forks_repo_head_hexsha": "cc1814d8d02d6708a307e4e12abcf81f379d2373", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 25.3639455782, "max_line_length": 80, "alphanum_fraction": 0.6223682446, "num_tokens": 1953, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.3106943959796865, "lm_q2_score": 0.18952109819626, "lm_q1q2_score": 0.05888314312949386}}
{"text": "\"\"\"\nRun a coarse thermal bubble simulation and save the output to NetCDF at the\n10th time step. Then read back the output and test that it matches the model's\nstate.\n\"\"\"\nfunction run_thermal_bubble_netcdf_tests(arch)\n Nx, Ny, Nz = 16, 16, 16\n Lx, Ly, Lz = 100, 100, 100\n \u0394t = 6\n\n grid = RegularCartesianGrid(size=(Nx, Ny, Nz), length=(Lx, Ly, Lz))\n closure = ConstantIsotropicDiffusivity(\u03bd=4e-2, \u03ba=4e-2)\n model = Model(architecture=arch, grid=grid, closure=closure)\n\n # Add a cube-shaped warm temperature anomaly that takes up the middle 50%\n # of the domain volume.\n i1, i2 = round(Int, Nx/4), round(Int, 3Nx/4)\n j1, j2 = round(Int, Ny/4), round(Int, 3Ny/4)\n k1, k2 = round(Int, Nz/4), round(Int, 3Nz/4)\n model.tracers.T.data[i1:i2, j1:j2, k1:k2] .+= 0.01\n\n outputs = Dict(\"v\"=>model.velocities.v,\n \"u\"=>model.velocities.u,\n \"w\"=>model.velocities.w,\n \"T\"=>model.tracers.T,\n \"S\"=>model.tracers.S)\n nc_writer = NetCDFOutputWriter(model, outputs,\n filename=\"dumptest.nc\",\n frequency=10)\n push!(model.output_writers, nc_writer)\n\n xC_slice = 1:10\n xF_slice = 2:11\n yC_slice = 10:15\n yF_slice = 1\n zC_slice = 10\n zF_slice = 9:11\n nc_sliced_writer = NetCDFOutputWriter(model, outputs,\n filename=\"dumptestsliced.nc\",\n frequency=10,\n xC=xC_slice, xF=xF_slice, yC=yC_slice,\n yF=yF_slice, zC=zC_slice, zF=zF_slice)\n push!(model.output_writers, nc_sliced_writer)\n\n time_step!(model, 10, \u0394t)\n\n close(nc_writer)\n close(nc_sliced_writer)\n\n u = read_output(nc_writer, \"u\")\n v = read_output(nc_writer, \"v\")\n w = read_output(nc_writer, \"w\")\n T = read_output(nc_writer, \"T\")\n S = read_output(nc_writer, \"S\")\n \n @test all(u .\u2248 Array(interiorparent(model.velocities.u)))\n @test all(v .\u2248 Array(interiorparent(model.velocities.v)))\n @test all(w .\u2248 Array(interiorparent(model.velocities.w)))\n @test all(T .\u2248 Array(interiorparent(model.tracers.T)))\n @test all(S .\u2248 Array(interiorparent(model.tracers.S)))\n\n u_sliced = read_output(nc_sliced_writer, \"u\")\n v_sliced = read_output(nc_sliced_writer, \"v\")\n w_sliced = read_output(nc_sliced_writer, \"w\")\n T_sliced = read_output(nc_sliced_writer, \"T\")\n S_sliced = read_output(nc_sliced_writer, \"S\")\n\n @test all(u_sliced .\u2248 Array(interiorparent(model.velocities.u))[xF_slice, yC_slice, zC_slice])\n @test all(v_sliced .\u2248 Array(interiorparent(model.velocities.v))[xC_slice, yF_slice, zC_slice])\n @test all(w_sliced .\u2248 Array(interiorparent(model.velocities.w))[xC_slice, yC_slice, zF_slice])\n @test all(T_sliced .\u2248 Array(interiorparent(model.tracers.T))[xC_slice, yC_slice, zC_slice])\n @test all(S_sliced .\u2248 Array(interiorparent(model.tracers.S))[xC_slice, yC_slice, zC_slice])\nend\n\nfunction run_jld2_file_splitting_tests(arch)\n model = Model(grid=RegularCartesianGrid(size=(16, 16, 16), length=(1, 1, 1)))\n\n u(model) = Array(model.velocities.u.data.parent)\n fields = Dict(:u => u)\n\n function fake_bc_init(file, model)\n file[\"boundary_conditions/fake\"] = \u03c0\n end\n\n ow = JLD2OutputWriter(model, fields; dir=\".\", prefix=\"test\", frequency=1,\n init=fake_bc_init, including=[:grid],\n max_filesize=200KiB, force=true)\n push!(model.output_writers, ow)\n\n # 531 KiB of output will be written which should get split into 3 files.\n time_step!(model, 10, 1)\n\n # Test that files has been split according to size as expected.\n @test filesize(\"test_part1.jld2\") > 200KiB\n @test filesize(\"test_part2.jld2\") > 200KiB\n @test filesize(\"test_part3.jld2\") < 200KiB\n\n for n in string.(1:3)\n filename = \"test_part\" * n * \".jld2\"\n jldopen(filename, \"r\") do file\n # Test to make sure all files contain structs from `including`.\n @test file[\"grid/Nx\"] == 16\n\n # Test to make sure all files contain info from `init` function.\n @test file[\"boundary_conditions/fake\"] == \u03c0\n end\n\n # Leave test directory clean.\n rm(filename)\n end\nend\n\n\"\"\"\nRun two coarse rising thermal bubble simulations and make sure that when\nrestarting from a checkpoint, the restarted simulation matches the non-restarted\nsimulation numerically.\n\"\"\"\nfunction run_thermal_bubble_checkpointer_tests(arch)\n Nx, Ny, Nz = 16, 16, 16\n Lx, Ly, Lz = 100, 100, 100\n \u0394t = 6\n\n grid = RegularCartesianGrid(size=(Nx, Ny, Nz), length=(Lx, Ly, Lz))\n closure = ConstantIsotropicDiffusivity(\u03bd=4e-2, \u03ba=4e-2)\n true_model = Model(architecture=arch, grid=grid, closure=closure)\n\n # Add a cube-shaped warm temperature anomaly that takes up the middle 50%\n # of the domain volume.\n i1, i2 = round(Int, Nx/4), round(Int, 3Nx/4)\n j1, j2 = round(Int, Ny/4), round(Int, 3Ny/4)\n k1, k2 = round(Int, Nz/4), round(Int, 3Nz/4)\n true_model.tracers.T.data[i1:i2, j1:j2, k1:k2] .+= 0.01\n\n checkpointed_model = deepcopy(true_model)\n\n time_step!(true_model, 9, \u0394t)\n\n checkpointer = Checkpointer(checkpointed_model; frequency=5, force=true)\n push!(checkpointed_model.output_writers, checkpointer)\n\n # Checkpoint should be saved as \"checkpoint5.jld\" after the 5th iteration.\n time_step!(checkpointed_model, 5, \u0394t)\n\n # Remove all knowledge of the checkpointed model.\n checkpointed_model = nothing\n\n restored_model = restore_from_checkpoint(\"checkpoint5.jld2\")\n\n time_step!(restored_model, 4, \u0394t; init_with_euler=false)\n\n rm(\"checkpoint0.jld2\", force=true)\n rm(\"checkpoint5.jld2\", force=true)\n\n # Now the true_model and restored_model should be identical.\n @test all(restored_model.velocities.u.data .\u2248 true_model.velocities.u.data)\n @test all(restored_model.velocities.v.data .\u2248 true_model.velocities.v.data)\n @test all(restored_model.velocities.w.data .\u2248 true_model.velocities.w.data)\n @test all(restored_model.tracers.T.data .\u2248 true_model.tracers.T.data)\n @test all(restored_model.tracers.S.data .\u2248 true_model.tracers.S.data)\n @test all(restored_model.timestepper.G\u207f.u.data .\u2248 true_model.timestepper.G\u207f.u.data)\n @test all(restored_model.timestepper.G\u207f.v.data .\u2248 true_model.timestepper.G\u207f.v.data)\n @test all(restored_model.timestepper.G\u207f.w.data .\u2248 true_model.timestepper.G\u207f.w.data)\n @test all(restored_model.timestepper.G\u207f.T.data .\u2248 true_model.timestepper.G\u207f.T.data)\n @test all(restored_model.timestepper.G\u207f.S.data .\u2248 true_model.timestepper.G\u207f.S.data)\nend\n\n@testset \"Output writers\" begin\n println(\"Testing output writers...\")\n\n for arch in archs\n @testset \"NetCDF [$(typeof(arch))]\" begin\n println(\" Testing NetCDF output writer [$(typeof(arch))]...\")\n run_thermal_bubble_netcdf_tests(arch)\n end\n\n @testset \"JLD2 [$(typeof(arch))]\" begin\n println(\" Testing JLD2 output writer [$(typeof(arch))]...\")\n run_jld2_file_splitting_tests(arch)\n end\n\n @testset \"Checkpointer [$(typeof(arch))]\" begin\n println(\" Testing Checkpointer [$(typeof(arch))]...\")\n run_thermal_bubble_checkpointer_tests(arch)\n end\n end\nend\n", "meta": {"hexsha": "f962578180514080f3cfc21a27640dec2aacd344", "size": 7406, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/test_output_writers.jl", "max_stars_repo_name": "UnofficialJuliaMirror/Oceananigans.jl-9e8cae18-63c1-5223-a75c-80ca9d6e9a09", "max_stars_repo_head_hexsha": "d753151ca11649756cd6452faf443306002cac79", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "test/test_output_writers.jl", "max_issues_repo_name": "UnofficialJuliaMirror/Oceananigans.jl-9e8cae18-63c1-5223-a75c-80ca9d6e9a09", "max_issues_repo_head_hexsha": "d753151ca11649756cd6452faf443306002cac79", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "test/test_output_writers.jl", "max_forks_repo_name": "UnofficialJuliaMirror/Oceananigans.jl-9e8cae18-63c1-5223-a75c-80ca9d6e9a09", "max_forks_repo_head_hexsha": "d753151ca11649756cd6452faf443306002cac79", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 39.6042780749, "max_line_length": 98, "alphanum_fraction": 0.6473129895, "num_tokens": 2139, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.48047867804790706, "lm_q2_score": 0.1225232237324595, "lm_q1q2_score": 0.05886979656914009}}
{"text": "function insert_element(values)\n push!(values, -10)\nend\n\nlist = [1,2,3]\ninsert_element(list)\nprintln(list)\n\nf(x,y) = x^3 + x*y - y\ng(a, b = 5) = a + b\nk(x; a1=1, a2=2) = x * (a1+a2)\n\nprintln(f(3,2))\n\nprintln(\"g(3) + f(1,2) = $( g(3) + f(1,2) )\")\nprintln(\"k(2) = $(k(2))\")\n\ncubes = map(x->x^3, [1,2,3,5])\nprintln(\"$cubes\")\n\nprintln(\"$([1:3])\")\n\na = split(\"A,B,C,D\",\",\")\nprintln(\"typeof(a): $(typeof(a))\")\nprintln(\"a: $(a)\")\n\nstr::String = \"This is a string\"\nprintln(str)\n", "meta": {"hexsha": "e957baa8b33b8920e0411caacefa4c11b568c5ba", "size": 471, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "ch3.jl", "max_stars_repo_name": "ykyang/org.allnix.julia", "max_stars_repo_head_hexsha": "58933a5848dec81c53d591b4163e9a70df62ddd8", "max_stars_repo_licenses": ["Apache-2.0"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "ch3.jl", "max_issues_repo_name": "ykyang/org.allnix.julia", "max_issues_repo_head_hexsha": "58933a5848dec81c53d591b4163e9a70df62ddd8", "max_issues_repo_licenses": ["Apache-2.0"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "ch3.jl", "max_forks_repo_name": "ykyang/org.allnix.julia", "max_forks_repo_head_hexsha": "58933a5848dec81c53d591b4163e9a70df62ddd8", "max_forks_repo_licenses": ["Apache-2.0"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 16.2413793103, "max_line_length": 45, "alphanum_fraction": 0.5350318471, "num_tokens": 201, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.480478678047907, "lm_q2_score": 0.12252320931407355, "lm_q1q2_score": 0.05886978964141307}}
{"text": "\"\"\"\n ispangram(input)\n\nReturn `true` if `input` contains every alphabetic character (case insensitive).\n\n\"\"\"\nfunction ispangram(input)\n input_low = lowercase(input)\n lower_alpha = 'a':'z'\n\n return all(\n [(c in input_low) for c in lower_alpha]) \n\nend\n\n", "meta": {"hexsha": "fe6ac098d55b9f2426a1da1f2dfadc4a9745b65a", "size": 273, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "julia/pangram/pangram.jl", "max_stars_repo_name": "jfcliang/jl-exercism-solutions", "max_stars_repo_head_hexsha": "8e8c2ee48dbea3175bcd0d0f2071de945014ce0f", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "julia/pangram/pangram.jl", "max_issues_repo_name": "jfcliang/jl-exercism-solutions", "max_issues_repo_head_hexsha": "8e8c2ee48dbea3175bcd0d0f2071de945014ce0f", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "julia/pangram/pangram.jl", "max_forks_repo_name": "jfcliang/jl-exercism-solutions", "max_forks_repo_head_hexsha": "8e8c2ee48dbea3175bcd0d0f2071de945014ce0f", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 17.0625, "max_line_length": 80, "alphanum_fraction": 0.6483516484, "num_tokens": 67, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.480478678047907, "lm_q2_score": 0.12252320450794524, "lm_q1q2_score": 0.05886978733217089}}
{"text": "function sum_values(x,y)\n return x+y\nend", "meta": {"hexsha": "9fb6c616a38e25b77ab16d1ce5082fe211dbe830", "size": 43, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/simple_function.jl", "max_stars_repo_name": "jeff788/JuliaPackageTest", "max_stars_repo_head_hexsha": "bc3e40fa840343fdf9d11239e5e701d4c9bec0b3", "max_stars_repo_licenses": ["BSD-3-Clause"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/simple_function.jl", "max_issues_repo_name": "jeff788/JuliaPackageTest", "max_issues_repo_head_hexsha": "bc3e40fa840343fdf9d11239e5e701d4c9bec0b3", "max_issues_repo_licenses": ["BSD-3-Clause"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/simple_function.jl", "max_forks_repo_name": "jeff788/JuliaPackageTest", "max_forks_repo_head_hexsha": "bc3e40fa840343fdf9d11239e5e701d4c9bec0b3", "max_forks_repo_licenses": ["BSD-3-Clause"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 14.3333333333, "max_line_length": 24, "alphanum_fraction": 0.7209302326, "num_tokens": 12, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.5, "lm_q2_score": 0.1175721304535499, "lm_q1q2_score": 0.05878606522677495}}
{"text": "#==============================================================================\n Generate DataFrame of Column Names and Type from a DataFrame\n\n author: mahiki@users.noreply.github.com\n=============================================================================#\n\nusing DataFrames, Dates\n\n\"\"\"\n printschema(df::DataFrame)\n\nReturn a DataFrame of ordered columns and column type for input dataFrame df\n\n# Examples\n```jldoctest\njulia> df = DataFrame(a = 1:2, b = [1.0, \u03c0], c = [Date(\"20191002\", \"yyyymmdd\"), Date(\"21120101\", \"yyyymmdd\")], d = [1//2, missing]);\njulia> printschema(df)\n4\u00d72 DataFrame\n Row \u2502 variable value\n \u2502 String Type\n\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\n 1 \u2502 a Int64\n 2 \u2502 b Float64\n 3 \u2502 c Date\n 4 \u2502 d Union{Missing, Rational{Int64}}\n```\n\"\"\"\nfunction printschema(df::DataFrame)\n dfcols = mapcols(eltype, df)\n dfcolnames = names(dfcols)\n stack(dfcols, dfcolnames)\nend\n\n\n", "meta": {"hexsha": "9c85af7ca5c1705ff0d38d793f2940f78f5e0cb1", "size": 965, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/printschema.jl", "max_stars_repo_name": "mahiki/DesertIslandDisk", "max_stars_repo_head_hexsha": "f8e3e31c0123322807863749cdd2bb5198adb889", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2021-09-07T04:15:35.000Z", "max_stars_repo_stars_event_max_datetime": "2021-09-07T04:15:35.000Z", "max_issues_repo_path": "src/printschema.jl", "max_issues_repo_name": "mahiki/DesertIslandDisk", "max_issues_repo_head_hexsha": "f8e3e31c0123322807863749cdd2bb5198adb889", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 3, "max_issues_repo_issues_event_min_datetime": "2021-10-05T01:10:46.000Z", "max_issues_repo_issues_event_max_datetime": "2022-01-20T08:04:06.000Z", "max_forks_repo_path": "src/printschema.jl", "max_forks_repo_name": "mahiki/DesertIslandDisk", "max_forks_repo_head_hexsha": "f8e3e31c0123322807863749cdd2bb5198adb889", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 27.5714285714, "max_line_length": 132, "alphanum_fraction": 0.4984455959, "num_tokens": 244, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4921881357207956, "lm_q2_score": 0.11920292515117027, "lm_q1q2_score": 0.058670265502620035}}
{"text": "\"Your optional docstring here\"\nfunction distance(a, b)\n if length(a) != length(b)\n throw((a, b), ArgumentError(\"Unequal strand lengths!\"))\n end\n hamming_distance = 0\n\n for i in 1:length(a)\n if a[i] != b[i]\n hamming_distance += 1\n end\n end\n\n return hamming_distance\nend\n", "meta": {"hexsha": "e1946059c608682072d365c033b16f953075b1de", "size": 319, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "julia/hamming/hamming.jl", "max_stars_repo_name": "jfcliang/jl-exercism-solutions", "max_stars_repo_head_hexsha": "8e8c2ee48dbea3175bcd0d0f2071de945014ce0f", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "julia/hamming/hamming.jl", "max_issues_repo_name": "jfcliang/jl-exercism-solutions", "max_issues_repo_head_hexsha": "8e8c2ee48dbea3175bcd0d0f2071de945014ce0f", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "julia/hamming/hamming.jl", "max_forks_repo_name": "jfcliang/jl-exercism-solutions", "max_forks_repo_head_hexsha": "8e8c2ee48dbea3175bcd0d0f2071de945014ce0f", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 19.9375, "max_line_length": 63, "alphanum_fraction": 0.5862068966, "num_tokens": 88, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.49218813572079556, "lm_q2_score": 0.11920291107043361, "lm_q1q2_score": 0.0586702585722485}}
{"text": "module GnuplotSimple\n\nusing Gnuplot\nimport Gnuplot: PlotElement, DatasetText\n\nexport plot\n\n\n\n\n\"\"\"\n`plot(x,Y; kwargs...)`\n\nA simple plot recipe for `Gnuplot.jl`.\n\nRequired: vector-like `x` and vector or matrix-like `Y`.\n\nAll keyword arguments are optional:\n\n`title`, `xlab`, `ylab`, `key` should be strings.\n\n`grid`, `scatter`, `xlog`, `ylog`, `mono` are boolean.\n\n`xr` and `yr` are like `[0,1]`. Use `NaN` for unbounded.\n\n`labels`, `linecolor`, `linetype` should be vectors of strings (at least as many as columns in `Y`).\n\n`linewidth` should be a vector of numbers.\n\n\n\n\n```julia\nusing Gnuplot, GnuplotSimple\nx = 0:0.05:2\ny = x.^(1:3)'\n@gp plot(x,y; title=\"My Title\",xlab=\"My x\",ylab=\"My y\",xr=[0,1],labels=[\"y1\",\"y2\",\"y3\"])\n```\n\"\"\"\nfunction plot(x,Y; grid::Bool=true, scatter::Bool=false, title=nothing, xlab=nothing, ylab=nothing, key=nothing, labels=nothing, xr=[NaN,NaN], yr=[NaN,NaN],\n linetype=nothing, linewidth=nothing, linecolor=nothing, xlog::Bool=false, ylog::Bool=false, mono::Bool=false)\n # assertions and input processing\n @assert !isempty(x)\n @assert !isempty(Y)\n Lx = length(x)\n Ly, Ny = size(Y)\n dimsx = length(size(x))\n dimsY = length(size(Y))\n @assert dimsx == 1 \"x must be 1D\"\n @assert dimsY <= 2 \"Y must be 1D or 2D\"\n @assert Lx == Ly \"x and Y (or its columns) must have equal number of observations\"\n \n ptype = ifelse(scatter,\"p\",\"l\")\n \n # plot setup\n out = Vector{Gnuplot.PlotElement}()\n grid && push!(out,PlotElement(cmds=[\"set grid\"]))\n mono && push!(out,PlotElement(cmds=[\"set monochrome\"]))\n !isnothing(title) && push!(out, PlotElement(;tit=title))\n !isnothing(xlab) && push!(out, PlotElement(;xlabel=xlab))\n !isnothing(ylab) && push!(out, PlotElement(;ylabel=ylab))\n !isnothing(key) && push!(out, PlotElement(;key=key))\n !isnothing(xr) && push!(out, PlotElement(;xrange=xr))\n !isnothing(yr) && push!(out, PlotElement(;yrange=yr))\n push!(out, PlotElement(;xlog=xlog,ylog=ylog))\n\n # do plotting\n for i in 1:Ny\n plotstr = \"w $ptype\"\n if !isnothing(labels)\n if length(labels) >= i\n thislab = labels[i]\n plotstr = string(plotstr,\" t '$thislab'\")\n end\n end\n if !isnothing(linetype)\n if length(linetype) >= i\n thislt = linetype[i]\n plotstr = string(plotstr,\" $thislt\")\n end\n end\n if !isnothing(linewidth)\n if length(linewidth) >= i\n thislw = linewidth[i]\n plotstr = string(plotstr,\" lw $thislw\")\n end\n end\n if !isnothing(linecolor)\n if length(linecolor) >= i\n thiscolor = linecolor[i]\n plotstr = string(plotstr,\" lc rgb '$thiscolor'\")\n end\n end\n \n push!(out, PlotElement(data=DatasetText(x, Y[:,i]), plot=plotstr))\n end\n return out\nend\n\n\n\n\nend # module\n", "meta": {"hexsha": "b4e7a8944da099ab93c20e75d9d4602fbbe9ac69", "size": 2950, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/GnuplotSimple.jl", "max_stars_repo_name": "tbeason/GnuplotSimple.jl", "max_stars_repo_head_hexsha": "a9a2134de21b0ea5754c95620cd8d6b0ecf7b96c", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/GnuplotSimple.jl", "max_issues_repo_name": "tbeason/GnuplotSimple.jl", "max_issues_repo_head_hexsha": "a9a2134de21b0ea5754c95620cd8d6b0ecf7b96c", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/GnuplotSimple.jl", "max_forks_repo_name": "tbeason/GnuplotSimple.jl", "max_forks_repo_head_hexsha": "a9a2134de21b0ea5754c95620cd8d6b0ecf7b96c", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 28.3653846154, "max_line_length": 156, "alphanum_fraction": 0.5894915254, "num_tokens": 865, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4843800842769843, "lm_q2_score": 0.12085323249047068, "lm_q1q2_score": 0.05853889893888017}}
{"text": "\nusing Latexify: Latexify\n\n# Recipe to hook into Latexify.jl's `latexify` function for `Sym`s.\n# We do not use SymPy's `latex` function here since it behaves a bit different\n# from Latexify.jl. Thus, we let Latexify.jl handle everything for the sake of\n# consistency.\n# For example, SymPy doesn't print a multiplication symbol by default while\n# Latexify uses a `\"\\\\cdot\"` (unless the keyword argument `cdot=false` is set).\nLatexify.@latexrecipe function _(x::Sym)\n return string(x)\nend\n", "meta": {"hexsha": "35184a80caf6ac879c6eecea848cf75195712ce8", "size": 488, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/latexify_recipe.jl", "max_stars_repo_name": "simeonschaub/SymPy.jl", "max_stars_repo_head_hexsha": "937f3c209df413b47007413ddce3c750df8a4deb", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 179, "max_stars_repo_stars_event_min_datetime": "2016-09-09T07:38:56.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-26T23:14:33.000Z", "max_issues_repo_path": "src/latexify_recipe.jl", "max_issues_repo_name": "simeonschaub/SymPy.jl", "max_issues_repo_head_hexsha": "937f3c209df413b47007413ddce3c750df8a4deb", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 256, "max_issues_repo_issues_event_min_datetime": "2016-09-08T13:37:01.000Z", "max_issues_repo_issues_event_max_datetime": "2022-03-01T12:26:16.000Z", "max_forks_repo_path": "src/latexify_recipe.jl", "max_forks_repo_name": "simeonschaub/SymPy.jl", "max_forks_repo_head_hexsha": "937f3c209df413b47007413ddce3c750df8a4deb", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 68, "max_forks_repo_forks_event_min_datetime": "2016-11-07T20:26:42.000Z", "max_forks_repo_forks_event_max_datetime": "2022-02-15T03:16:24.000Z", "avg_line_length": 37.5384615385, "max_line_length": 79, "alphanum_fraction": 0.7520491803, "num_tokens": 135, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.48438008427698437, "lm_q2_score": 0.12085322774082714, "lm_q1q2_score": 0.058538896638247434}}
{"text": "\n# # Introduction to [PowerSystems.jl](https://github.com/NREL-SIIP/PowerSystems.jl)\n#\n\n# **Originally Contributed by**: Clayton Barrows and Jose Daniel Lara\n\n# ## Introduction\n\n# This notebook is intended to show a power system data specification framework that exploits the\n# capabilities of Julia to improve performance and allow modelers to develop modular software\n# to create problems with different complexities and enable large scale analysis. The\n# [PowerSystems documentation](https://nrel-siip.github.io/PowerSystems.jl/stable/) is also\n# an excellent resource.\n#\n# ### Objective\n# PowerSystems.jl provides a type specification for bulk power system data.\n# The objective is to exploit Julia's integration of dynamic types to enable efficient data\n# handling and enable functional dispatch in modeling and analysis applications\n# As explained in Julia's documentation:\n#\n# \"Julia\u2019s type system is dynamic, but gains some of the advantages of static type systems\n# by making it possible to indicate that certain values are of specific types. This can be\n# of great assistance in generating efficient code, but even more significantly, it allows\n# method dispatch on the types of function arguments to be deeply integrated with the language.\"\n#\n# For more details on Julia types, refer to the [documentation](https://docs.julialang.org/en/v1/)\n#\n#\n# ## Environment and packages\n#\n# PowerSystems.jl relies on a framework for data handling established in\n# [InfrastructureSystems.jl](https://github.com/NREL-SIIP/InfrastructureSystems.jl).\n# Users of PowerSystems.jl should not need to interact directly with InfrastructureSystems.jl.\n# However, it's worth recognizing that InfrastructureSystems provides much of the back end\n# code for managing and accessing data, especially time series data.\n#\n# The examples in this notebook depend upon Julia 1.5 and a specific set of package releases\n# as defined in the `Manifest.toml`.\nusing Pkg\nPkg.status()\n\nusing SIIPExamples;\nusing PowerSystems;\nusing D3TypeTrees;\nIS = PowerSystems.IS\n\n# ## Types in PowerSystems\n# PowerSystems.jl provides a type hierarchy for specifying power system data. Data that\n# describes infrastructure components is held in `struct`s. For example, a `Bus` is defined\n# as follows with fields for the parameters required to describe a bus (along with an\n# `internal` field used by InfrastructureSystems to improve the efficiency of handling data).\n\nprint_struct(Bus)\n\n# ### Type Hierarchy\n# PowerSystems is intended to organize data containers by the behavior of the devices that\n# the data represents. To that end, a type hierarchy has been defined with several levels of\n# abstract types starting with `InfrastructureSystemsType`. There are a bunch of subtypes of\n# `InfrastructureSystemsType`, but the important ones to know about are:\n# - `Component`: includes all elements of power system data\n# - `Topology`: includes non physical elements describing network connectivity\n# - `Service`: includes descriptions of system requirements (other than energy balance)\n# - `Device`: includes descriptions of all the physical devices in a power system\n# - `InfrastructureSystems.DeviceParameter`: includes structs that hold data describing the\n# dynamic, or economic capabilities of `Device`.\n# - `TimeSeriesData`: Includes all time series types\n# - `time series`: includes structs to define time series of forecasted data where multiple\n# values can represent each time stamp\n# - `StaticTimeSeries`: includes structs to define time series with a single value for each\n# time stamp\n# - `System`: collects all of the `Component`s\n#\n# *The following trees are made with [D3TypeTrees](https://github.com/claytonpbarrows/D3TypeTrees.jl),\n# nodes that represent Structs will show the Fields in the hoverover tooltip.*\n\n# TypeTree(InfrastructureSystemsType)\n\n# ### `TimeSeriesData`\n# [_Read the Docs!_](https://nrel-siip.github.io/PowerSystems.jl/stable/modeler_guide/time_series/)\n# Every `Component` has a `time_series_container::InfrastructureSystems.TimeSeriesContainer`\n# field. `TimeSeriesData` are used to hold time series information that describes the\n# temporally dependent data of fields within the same struct. For example, the\n# `ThermalStandard.time_series_container` field can\n# describe other fields in the struct (`available`, `activepower`, `reactivepower`).\n\n# `TimeSeriesData`s themselves can take the form of the following:\nTypeTree(TimeSeriesData)\n\n# In each case, the time series contains fields for `scaling_factor_multiplier` and `data`\n# to identify the details of th `Component` field that the time series describes, and the\n# time series `data`. For example: we commonly want to use a time series to\n# describe the maximum active power capability of a renewable generator. In this case, we\n# can create a `SingleTimeSeries` with a `TimeArray` and an accessor function to the\n# maximum active power field in the struct describing the generator. In this way, we can\n# store a scaling factor time series that will get multiplied by the maximum active power\n# rather than the magnitudes of the maximum active power time series.\n\nprint_struct(Deterministic)\n\n# Examples of how to create and add time series to system can be found in the\n# [Add Time Series Example](../PowerSystems.jl Examples/add_forecasts.ipynb)\n\n# ### System\n# The `System` object collects all of the individual components into a single struct along\n# with some metadata about the system itself (e.g. `base_power`)\n\nprint_struct(System)\n\n# ## Basic example\n# PowerSystems contains a few basic data files (mostly for testing and demonstration).\n\nBASE_DIR = abspath(joinpath(dirname(Base.find_package(\"PowerSystems\")), \"..\"))\ninclude(joinpath(BASE_DIR, \"test\", \"data_5bus_pu.jl\")) #.jl file containing 5-bus system data\nnodes_5 = nodes5() # function to create 5-bus buses\n\n# ### Create a `System`\n\nsys = System(\n 100.0,\n nodes_5,\n vcat(thermal_generators5(nodes_5), renewable_generators5(nodes_5)),\n loads5(nodes_5),\n branches5(nodes_5),\n)\n\n# ### Accessing `System` Data\n# PowerSystems provides functional interfaces to all data. The following examples outline\n# the intended approach to accessing data expressed using PowerSystems.\n\n# PowerSystems enforces unique `name` fields between components of a particular concrete type.\n# So, in order to retrieve a specific component, the user must specify the type of the component\n# along with the name and system\n\n# #### Accessing components\n@show get_component(Bus, sys, \"nodeA\")\n@show get_component(Line, sys, \"1\")\n\n# Similarly, you can access all the components of a particular type: *note: the return type\n# of get_components is a `FlattenIteratorWrapper`, so call `collect` to get an `Array`\n\nget_components(Bus, sys) |> collect\n\n# `get_components` also works on abstract types:\n\nget_components(Branch, sys) |> collect\n\n# The fields within a component can be accessed using the `get_*` functions:\n# *It's highly recommended that users avoid using the `.` to access fields since we make no\n# guarantees on the stability field names and locations. We do however promise to keep the\n# accessor functions stable.*\n\nbus1 = get_component(Bus, sys, \"nodeA\")\n@show get_name(bus1);\n@show get_magnitude(bus1);\n\n# #### Accessing `TimeSeries`\n\n# First we need to add some time series to the `System`\nloads = collect(get_components(PowerLoad, sys))\nfor (l, ts) in zip(loads, load_timeseries_DA[2])\n add_time_series!(\n sys,\n l,\n Deterministic(\n \"activepower\",\n Dict(TimeSeries.timestamp(load_timeseries_DA[2][1])[1] => ts),\n ),\n )\nend\n\n# If we want to access a specific time series for a specific component, we need to specify:\n# - time series type\n# - `component`\n# - initial_time\n# - label\n#\n# We can find the initial time of all the time series in the system:\nget_forecast_initial_times(sys)\n\n# We can find the names of all time series attached to a component:\nts_names = get_time_series_names(Deterministic, loads[1])\n\n# We can access a specific time series for a specific component:\nta = get_time_series_array(Deterministic, loads[1], ts_names[1])\n\n# Or, we can just get the values of the time series:\nts = get_time_series_values(Deterministic, loads[1], ts_names[1])\n", "meta": {"hexsha": "127cad9cf8bf519df736e526132e8b3c0e97b30b", "size": 8255, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "script/2_PowerSystems_examples/PowerSystems_intro.jl", "max_stars_repo_name": "raphaelsaavedra/SIIPExamples.jl", "max_stars_repo_head_hexsha": "d7304c84fe5382db2ff4c20f058bc1e5d01cae8c", "max_stars_repo_licenses": ["BSD-3-Clause"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "script/2_PowerSystems_examples/PowerSystems_intro.jl", "max_issues_repo_name": "raphaelsaavedra/SIIPExamples.jl", "max_issues_repo_head_hexsha": "d7304c84fe5382db2ff4c20f058bc1e5d01cae8c", "max_issues_repo_licenses": ["BSD-3-Clause"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "script/2_PowerSystems_examples/PowerSystems_intro.jl", "max_forks_repo_name": "raphaelsaavedra/SIIPExamples.jl", "max_forks_repo_head_hexsha": "d7304c84fe5382db2ff4c20f058bc1e5d01cae8c", "max_forks_repo_licenses": ["BSD-3-Clause"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 43.9095744681, "max_line_length": 102, "alphanum_fraction": 0.766444579, "num_tokens": 1885, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.48438008427698437, "lm_q2_score": 0.1208532261576127, "lm_q1q2_score": 0.058538895871369886}}
{"text": "using Documenter\n\nmakedocs(\n sitename = \"\ud83c\udfa2 Control Theory\",\n format = Documenter.LaTeX(),\n authors = \"Joe Carpinelli\",\n pages = [\n \"Frontmatter\" => [\n \"Welcome!\" => \"index.md\",\n \"Chapter 1: Introduction\" => \"Chapter 1: Introduction.md\"\n ],\n \"Topic 1: Dynamics\" => [\n \"Chapter 2: General Dynamics\" => \"Chapter 2: General Dynamics.md\",\n \"Chapter 3: Flight Dynamics\" => \"Chapter 3: Flight Dynamics.md\",\n \"Chapter 4: Linear Dynamics\" => \"Chapter 4: Linear Dynamics.md\",\n \"Chapter 5: Equilibrium Points\" => \"Chapter 5: Equilibrium Points.md\",\n \"Chapter 6: Linearization\" => \"Chapter 6: Linearization.md\"\n ],\n \"Topic 2: Systems\" => [],\n \"Topic 3: Controls\" => [],\n \"Topic 4: Analysis\" => []\n ]\n)", "meta": {"hexsha": "28a7a10352924b0a9084368e43ed4652c229026b", "size": 810, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "docs/make_pdf.jl", "max_stars_repo_name": "cadojo/controls", "max_stars_repo_head_hexsha": "d128521e5277a2d77323362420bb3401b04159ca", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 2, "max_stars_repo_stars_event_min_datetime": "2021-06-29T01:35:03.000Z", "max_stars_repo_stars_event_max_datetime": "2021-06-29T04:04:56.000Z", "max_issues_repo_path": "docs/make_pdf.jl", "max_issues_repo_name": "cadojo/exploring-control-theory", "max_issues_repo_head_hexsha": "eaa8c4653c3e77c6f3272a18630984f1b33097b3", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "docs/make_pdf.jl", "max_forks_repo_name": "cadojo/exploring-control-theory", "max_forks_repo_head_hexsha": "eaa8c4653c3e77c6f3272a18630984f1b33097b3", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 35.2173913043, "max_line_length": 78, "alphanum_fraction": 0.5530864198, "num_tokens": 218, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.3557749071749625, "lm_q2_score": 0.16451646904109266, "lm_q1q2_score": 0.05853083150184733}}
{"text": "import Base.BaseDocs: @kw_str\n\n\"\"\"\n**\u6b22\u8fce\u6765\u5230 Julia $(string(VERSION)).** \u5b8c\u6574\u7684\u4e2d\u6587\u624b\u518c\u53ef\u4ee5\u5728\u8fd9\u91cc\u627e\u5230\n\n https://docs.juliacn.com/\n\n\u66f4\u591a\u4e2d\u6587\u8d44\u6599\u548c\u6559\u7a0b\uff0c\u4e5f\u8bf7\u5173\u6ce8 Julia \u4e2d\u6587\u793e\u533a\n\n https://cn.julialang.org\n\n\u65b0\u624b\u8bf7\u53c2\u8003\u4e2d\u6587 discourse \u4e0a\u7684\u65b0\u624b\u6307\u5f15\n\n https://discourse.juliacn.com/t/topic/159\n\n\u8f93\u5165 `?`\uff0c \u7136\u540e\u8f93\u5165\u4f60\u60f3\u8981\u67e5\u770b\u5e2e\u52a9\u6587\u6863\u7684\u51fd\u6570\u6216\u8005\u5b8f\u540d\u79f0\u5c31\u53ef\u4ee5\u67e5\u770b\u5b83\u4eec\u7684\u6587\u6863\u3002\u4f8b\u5982 `?cos`, \u6216\u8005 `?@time` \u7136\u540e\u6309\u56de\u8f66\u952e\u5373\u53ef\u3002\n\n\u5728 REPL \u4e2d\u8f93\u5165 `ENV[\"REPL_LOCALE\"]=\"\"` \u5c06\u6062\u590d\u82f1\u6587\u6a21\u5f0f\u3002\u518d\u6b21\u56de\u5230\u4e2d\u6587\u6a21\u5f0f\u8bf7\u8f93\u5165 `ENV[\"REPL_LOCALE\"]=\"zh_CN\"`\u3002\n\"\"\"\nkw\"help\", kw\"?\", kw\"julia\", kw\"\"\n\n\n\"\"\"\n using\n\n`using Foo` \u5c06\u4f1a\u52a0\u8f7d\u4e00\u4e2a\u540d\u4e3a `Foo` \u7684\u6a21\u5757\uff08module\uff09\u6216\u8005\u4e00\u4e2a\u5305\uff0c\u7136\u540e\u5176 [`export`](@ref) \u7684\u540d\u79f0\u5c06\u53ef\u4ee5\u76f4\u63a5\u4f7f\u7528\u3002\u4e0d\u8bba\u662f\u5426\u88ab `export`\uff0c\u540d\u79f0\u90fd\u53ef\u4ee5\u901a\u8fc7\u70b9\u6765\u8bbf\u95ee\uff08\u4f8b\u5982\uff0c\u8f93\u5165 `Foo.foo` \u6765\u8bbf\u95ee\u5230 `foo`\uff09\u3002\u67e5\u770b[\u624b\u518c\u4e2d\u5173\u4e8e\u6a21\u5757\u7684\u90e8\u5206](@ref modules)\u4ee5\u83b7\u53d6\u66f4\u591a\u7ec6\u8282\u3002\n\"\"\"\nkw\"using\"\n\n\"\"\"\n import\n\n`import Foo` \u5c06\u4f1a\u52a0\u8f7d\u4e00\u4e2a\u540d\u4e3a `Foo` \u7684\u6a21\u5757\uff08module\uff09\u6216\u8005\u4e00\u4e2a\u5305\u3002`Foo` \u6a21\u5757\u4e2d\u7684\u540d\u79f0\u53ef\u4ee5\u901a\u8fc7\u70b9\u6765\u8bbf\u95ee\u5230\uff08\u4f8b\u5982\uff0c\u8f93\u5165 `Foo.foo` \u53ef\u4ee5\u83b7\u53d6\u5230 `foo`\uff09\u3002\u67e5\u770b[\u624b\u518c\u4e2d\u5173\u4e8e\u6a21\u5757\u7684\u90e8\u5206](@ref modules)\u4ee5\u83b7\u53d6\u66f4\u591a\u7ec6\u8282\u3002\n\"\"\"\nkw\"import\"\n\n\"\"\"\n export\n\n`export` \u88ab\u7528\u6765\u5728\u6a21\u5757\u4e2d\u544a\u8bc9Julia\u54ea\u4e9b\u51fd\u6570\u6216\u8005\u540d\u5b57\u53ef\u4ee5\u7531\u7528\u6237\u4f7f\u7528\u3002\u4f8b\u5982 `export foo` \u5c06\u5728 [`using`](@ref) \u8fd9\u4e2a module \u7684\u65f6\u5019\u4f7f\u5f97 `foo`\u53ef\u4ee5\u76f4\u63a5\u88ab\u8bbf\u95ee\u5230\u3002\u67e5\u770b[\u624b\u518c\u4e2d\u5173\u4e8e\u6a21\u5757\u7684\u90e8\u5206](@ref modules)\u4ee5\u83b7\u53d6\u66f4\u591a\u7ec6\u8282\u3002\n\"\"\"\nkw\"export\"\n\n\"\"\"\n abstract type\n\n`abstract type` \u58f0\u660e\u6765\u4e00\u4e2a\u4e0d\u80fd\u5b9e\u4f8b\u5316\u7684\u7c7b\u578b\uff0c\u5b83\u5c06\u4ec5\u4ec5\u4f5c\u4e3a\u7c7b\u578b\u56fe\u4e2d\u7684\u4e00\u4e2a\u8282\u70b9\u5b58\u5728\uff0c\u4ece\u800c\u80fd\u591f\u63cf\u8ff0\u4e00\u7cfb\u5217\u76f8\u4e92\u5173\u8054\u7684\u5177\u4f53\u7c7b\u578b\uff08concrete type\uff09\uff1a\u8fd9\u4e9b\u5177\u4f53\u7c7b\u578b\u90fd\u662f\u62bd\u8c61\u7c7b\u578b\u7684\u5b50\u8282\u70b9\u3002\u62bd\u8c61\u7c7b\u578b\u5728\u6982\u5ff5\u4e0a\u4f7f\u5f97 Julia \u7684\u7c7b\u578b\u7cfb\u7edf\u4e0d\u4ec5\u4ec5\u662f\u4e00\u7cfb\u5217\u5bf9\u8c61\u7684\u96c6\u5408\u3002\u4f8b\u5982\uff1a\n\n```julia\nabstract type Number end\nabstract type Real <: Number end\n```\n\n[`Number`](@ref) \u6ca1\u6709\u7236\u8282\u70b9\uff08\u7236\u7c7b\u578b\uff09, \u800c [`Real`](@ref) \u662f `Number` \u7684\u4e00\u4e2a\u62bd\u8c61\u5b50\u7c7b\u578b\u3002\n\"\"\"\nkw\"abstract type\"\n\n\"\"\"\n module\n\n`module` \u4f1a\u58f0\u660e\u4e00\u4e2a `Module` \u7c7b\u578b\u7684\u5b9e\u4f8b\u7528\u4e8e\u63cf\u8ff0\u4e00\u4e2a\u72ec\u7acb\u7684\u53d8\u91cf\u540d\u7a7a\u95f4\u3002\u5728\u4e00\u4e2a\u6a21\u5757\uff08module\uff09\u91cc\uff0c\u4f60\u53ef\u4ee5\u63a7\u5236\u6765\u81ea\u4e8e\u5176\u5b83\u6a21\u5757\u7684\u540d\u5b57\u662f\u5426\u53ef\u89c1\uff08\u901a\u8fc7\u8f7d\u5165\uff0cimport\uff09\uff0c\u4f60\u4e5f\u53ef\u4ee5\u51b3\u5b9a\u4f60\u7684\u540d\u5b57\u6709\u54ea\u4e9b\u662f\u53ef\u4ee5\u516c\u5f00\u7684\uff08\u901a\u8fc7\u66b4\u9732\uff0cexport\uff09\u3002\u6a21\u5757\u4f7f\u5f97\u4f60\u5728\u5728\u521b\u5efa\u4e0a\u5c42\u5b9a\u4e49\u65f6\u65e0\u9700\u62c5\u5fc3\u547d\u540d\u51b2\u7a81\u3002\u67e5\u770b[\u624b\u518c\u4e2d\u5173\u4e8e\u6a21\u5757\u7684\u90e8\u5206](@ref modules)\u4ee5\u83b7\u53d6\u66f4\u591a\u7ec6\u8282\u3002\n\n# \u4f8b\u5b50\n```julia\nmodule Foo\nimport Base.show\nexport MyType, foo\n\nstruct MyType\n x\nend\n\nbar(x) = 2x\nfoo(a::MyType) = bar(a.x) + 1\nshow(io::IO, a::MyType) = print(io, \"MyType \\$(a.x)\")\nend\n```\n\"\"\"\nkw\"module\"\n\n\"\"\"\n baremodule\n\n`baremodule` \u5c06\u58f0\u660e\u4e00\u4e2a\u4e0d\u5305\u542b `using Base` \u6216\u8005 `eval` \u5b9a\u4e49\u7684\u6a21\u5757\u3002\u4f46\u662f\u5b83\u5c06\u4ecd\u7136\u8f7d\u5165 `Core` \u6a21\u5757\u3002\n\"\"\"\nkw\"baremodule\"\n\n\"\"\"\n primitive type\n\n`primitive type` \u58f0\u660e\u4e86\u4e00\u4e2a\u5176\u6570\u636e\u4ec5\u4ec5\u7531\u4e00\u7cfb\u5217\u4e8c\u8fdb\u5236\u6570\u8868\u793a\u7684\u5177\u4f53\u7c7b\u578b\u3002\u6bd4\u8f83\u5e38\u89c1\u7684\u4f8b\u5b50\u662f\u6574\u6570\u7c7b\u578b\u548c\u6d6e\u70b9\u7c7b\u578b\u3002\u4e0b\u9762\u662f\u4e00\u4e9b\u5185\u7f6e\u7684\u539f\u59cb\u7c7b\u578b\uff08primitive type\uff09\uff1a\n\n```julia\nprimitive type Char 32 end\nprimitive type Bool <: Integer 8 end\n```\n\n\u540d\u79f0\u540e\u9762\u7684\u6570\u5b57\u8868\u8fbe\u4e86\u8fd9\u4e2a\u7c7b\u578b\u5b58\u50a8\u6240\u9700\u7684\u6bd4\u7279\u6570\u76ee\u3002\u76ee\u524d\u8fd9\u4e2a\u6570\u5b57\u8981\u6c42\u662f 8 bit \u7684\u500d\u6570\u3002[`Bool`](@ref) \u7c7b\u578b\u7684\u58f0\u660e\u5c55\u793a\u4e86\u4e00\u4e2a\u539f\u59cb\u7c7b\u578b\u5982\u4f55\u9009\u62e9\u6210\u4e3a\u53e6\u4e00\u4e2a\u7c7b\u578b\u7684\u5b50\u7c7b\u578b\u3002\n\"\"\"\nkw\"primitive type\"\n\n\n\"\"\"\n macro\n\n`macro` \u5b9a\u4e49\u4e86\u4e00\u79cd\u4f1a\u5c06\u751f\u6210\u7684\u4ee3\u7801\u5305\u542b\u5728\u6700\u7ec8\u7a0b\u5e8f\u4f53\u4e2d\u7684\u65b9\u6cd5\uff0c\u8fd9\u79f0\u4e4b\u4e3a\u5b8f\u3002\u4e00\u4e2a\u5b8f\u5c06\u4e00\u7cfb\u5217\u8f93\u5165\u6620\u5c04\u5230\u4e00\u4e2a\u8868\u8fbe\u5f0f\uff0c\u7136\u540e\u6240\u8fd4\u56de\u7684\u8868\u8fbe\u5f0f\u5c06\u4f1a\u88ab\u76f4\u63a5\u8fdb\u884c\u7f16\u8bd1\u800c\u4e0d\u9700\u8981\u5728\u8fd0\u884c\u65f6\u8c03\u7528 `eval` \u51fd\u6570\u3002\u5b8f\u7684\u8f93\u5165\u53ef\u4ee5\u5305\u62ec\u8868\u8fbe\u5f0f\u3001\u5b57\u9762\u91cf\u548c\u7b26\u53f7\u3002\u4f8b\u5982\uff1a\n\n# \u4f8b\u5b50\n\n```jldoctest\njulia> macro sayhello(name)\n return :( println(\"Hello, \", \\$name, \"!\") )\n end\n@sayhello (macro with 1 method)\n\njulia> @sayhello \"\u5c0f\u660e\"\nHello, \u5c0f\u660e!\n```\n\"\"\"\nkw\"macro\"\n\n\"\"\"\n local\n\n`local`\u5c06\u4f1a\u5b9a\u4e49\u4e00\u4e2a\u65b0\u7684\u5c40\u90e8\u53d8\u91cf\u3002\n\n\u67e5\u770b[\u624b\u518c\uff1a\u53d8\u91cf\u4f5c\u7528\u57df](@ref scope-of-variables)\u4ee5\u83b7\u53d6\u66f4\u8be6\u7ec6\u7684\u4fe1\u606f\u3002\n\n# \u4f8b\u5b50\n```jldoctest\njulia> function foo(n)\n x = 0\n for i = 1:n\n local x # introduce a loop-local x\n x = i\n end\n x\n end\nfoo (generic function with 1 method)\n\njulia> foo(10)\n0\n```\n\"\"\"\nkw\"local\"\n\n\"\"\"\n global\n\n`global x` \u5c06\u4f1a\u4f7f\u5f97\u5f53\u524d\u4f5c\u7528\u57df\u548c\u5f53\u524d\u4f5c\u7528\u6240\u5305\u542b\u7684\u4f5c\u7528\u57df\u91cc\u7684 `x` \u6307\u5411\u540d\u4e3a `x` \u7684\u5168\u5c40\u53d8\u91cf\u3002\u67e5\u770b[\u624b\u518c\uff1a\u53d8\u91cf\u4f5c\u7528\u57df](@ref scope-of-variables)\u4ee5\u83b7\u53d6\u66f4\u591a\u4fe1\u606f\u3002\n\n# \u4f8b\u5b50\n```jldoctest\njulia> z = 3\n3\n\njulia> function foo()\n global z = 6 # use the z variable defined outside foo\n end\nfoo (generic function with 1 method)\n\njulia> foo()\n6\n\njulia> z\n6\n```\n\"\"\"\nkw\"global\"\n\n\"\"\"\n let\n\n`let` \u4f1a\u5728\u6bcf\u6b21\u88ab\u8fd0\u884c\u65f6\u58f0\u660e\u4e00\u4e2a\u65b0\u7684\u53d8\u91cf\u7ed1\u5b9a\u3002\u8fd9\u4e2a\u65b0\u7684\u53d8\u91cf\u7ed1\u5b9a\u5c06\u62e5\u6709\u4e00\u4e2a\u65b0\u7684\u5730\u5740\u3002\u8fd9\u91cc\u7684\u4e0d\u540c\u53ea\u6709\u5f53\u53d8\u91cf\u901a\u8fc7\u95ed\u5305\u751f\u5b58\u5728\u5b83\u4eec\u7684\u4f5c\u7528\u57df\u5916\u65f6\u624d\u4f1a\u663e\u73b0\u3002`let` \u8bed\u6cd5\u63a5\u53d7\u9017\u53f7\u5206\u5272\u7684\u4e00\u7cfb\u5217\u8d4b\u503c\u8bed\u53e5\u548c\u53d8\u91cf\u540d\uff1a\n\n```julia\nlet var1 = value1, var2, var3 = value3\n code\nend\n```\n\n\u8fd9\u4e9b\u8d4b\u503c\u8bed\u53e5\u662f\u6309\u7167\u987a\u5e8f\u6c42\u503c\u7684\uff0c\u7b49\u53f7\u53f3\u8fb9\u7684\u8868\u8fbe\u5f0f\u5c06\u4f1a\u9996\u5148\u6c42\u503c\uff0c\u7136\u540e\u624d\u7ed1\u5b9a\u7ed9\u5de6\u8fb9\u7684\u53d8\u91cf\u3002\u56e0\u6b64\u8fd9\u4f7f\u5f97 `let x = x` \u8fd9\u6837\u7684\u8868\u8fbe\u5f0f\u6709\u610f\u4e49\uff0c\u56e0\u4e3a\u8fd9\u4e24\u4e2a `x` \u53d8\u91cf\u5c06\u5177\u6709\u4e0d\u540c\u7684\u5730\u5740\u3002\n\"\"\"\nkw\"let\"\n\n\"\"\"\n quote\n\n`quote` \u4f1a\u5c06\u5176\u5305\u542b\u7684\u4ee3\u7801\u6269\u53d8\u6210\u4e00\u4e2a\u591a\u91cd\u7684\u8868\u8fbe\u5f0f\u5bf9\u8c61\uff0c\u800c\u65e0\u9700\u663e\u793a\u8c03\u7528 `Expr` \u7684\u6784\u9020\u5668\u3002\u8fd9\u79f0\u4e4b\u4e3a\u5f15\u7528\uff0c\u6bd4\u5982\u8bf4\n\n```julia\nex = quote\n x = 1\n y = 2\n x + y\nend\n```\n\n\u548c\u5176\u5b83\u5f15\u7528\u65b9\u5f0f\u4e0d\u540c\u7684\u662f\uff0c`:( ... )`\u5f62\u5f0f\u7684\u5f15\u7528\uff08\u88ab\u5305\u542b\u65f6\uff09\u5c06\u4f1a\u5728\u8868\u8fbe\u5f0f\u6811\u91cc\u5f15\u5165\u4e00\u4e2a\u5728\u64cd\u4f5c\u8868\u8fbe\u5f0f\u6811\u65f6\u5fc5\u987b\u8981\u8003\u8651\u7684 `QuoteNode` \u5143\u7d20\u3002\u800c\u5728\u5176\u5b83\u573a\u666f\u4e0b\uff0c`:( ... )`\u548c `quote .. end` \u4ee3\u7801\u5757\u662f\u88ab\u540c\u7b49\u5bf9\u5f85\u7684\u3002\n\"\"\"\nkw\"quote\"\n\n\n\"\"\"\n '\n\n\u5384\u7c73\u7b97\u7b26\uff08\u5171\u8f6d\u8f6c\u7f6e\uff09\uff0c\u53c2\u89c1 [`adjoint`](@ref)\n\n# \u4f8b\u5b50\n```jldoctest\njulia> A = [1.0 -2.0im; 4.0im 2.0]\n2\u00d72 Array{Complex{Float64},2}:\n 1.0+0.0im -0.0-2.0im\n 0.0+4.0im 2.0+0.0im\n\njulia> A'\n2\u00d72 Array{Complex{Float64},2}:\n 1.0-0.0im 0.0-4.0im\n -0.0+2.0im 2.0-0.0im\n```\n\"\"\"\nkw\"'\"\n\n\"\"\"\n const\n\n`const` \u88ab\u7528\u6765\u58f0\u660e\u5e38\u6570\u5168\u5c40\u53d8\u91cf\u3002\u5728\u5927\u90e8\u5206\uff08\u5c24\u5176\u662f\u6027\u80fd\u654f\u611f\u7684\u4ee3\u7801\uff09\u5168\u5c40\u53d8\u91cf\u5e94\u5f53\u88ab\u58f0\u660e\u4e3a\u5e38\u6570\u3002\n\n```julia\nconst x = 5\n```\n\n\u53ef\u4ee5\u4f7f\u7528\u5355\u4e2a `const` \u58f0\u660e\u591a\u4e2a\u5e38\u6570\u53d8\u91cf\u3002\n\n```julia\nconst y, z = 7, 11\n```\n\n\u6ce8\u610f `const` \u53ea\u4f1a\u4f5c\u7528\u4e8e\u4e00\u4e2a `=` \u64cd\u4f5c\uff0c\u56e0\u6b64 `const x = y = 1` \u58f0\u660e\u4e86 `x` \u662f\u5e38\u6570\uff0c\u800c `y` \u4e0d\u662f\u3002\u5728\u53e6\u4e00\u65b9\u9762\uff0c`const x = const y = 1`\u58f0\u660e\u4e86 `x` \u548c `y` \u90fd\u662f\u5e38\u6570\u3002\n\n\u6ce8\u610f\u300c\u5e38\u6570\u6027\u8d28\u300d\u5e76\u4e0d\u4f1a\u5f3a\u5236\u5bb9\u5668\u5185\u90e8\u53d8\u6210\u5e38\u6570\uff0c\u6240\u4ee5\u5982\u679c `x` \u662f\u4e00\u4e2a\u6570\u7ec4\u6216\u8005\u5b57\u5178\uff08\u4e3e\u4f8b\u6765\u8bb2\uff09\u4f60\u4ecd\u7136\u53ef\u4ee5\u7ed9\u5b83\u4eec\u6dfb\u52a0\u6216\u8005\u5220\u9664\u5143\u7d20\u3002\n\n\u4e25\u683c\u6765\u8bb2\uff0c\u4f60\u751a\u81f3\u53ef\u4ee5\u91cd\u65b0\u5b9a\u4e49 `const`\uff08\u5e38\u6570\uff09\u53d8\u91cf\uff0c\u5c3d\u7ba1\u8fd9\u5c06\u4f1a\u8ba9\u7f16\u8bd1\u5668\u4ea7\u751f\u4e00\u4e2a\u8b66\u544a\u3002\u552f\u4e00\u4e25\u683c\u7684\u8981\u6c42\u662f\u8fd9\u4e2a\u53d8\u91cf\u7684**\u7c7b\u578b**\u4e0d\u80fd\u6539\u53d8\uff0c\u8fd9\u4e5f\u662f\u4e3a\u4ec0\u4e48\u5e38\u6570\u53d8\u91cf\u4f1a\u6bd4\u4e00\u822c\u7684\u5168\u5c40\u53d8\u91cf\u66f4\u5feb\u7684\u539f\u56e0\u3002\n\"\"\"\nkw\"const\"\n\n\"\"\"\n function\n\n\u51fd\u6570\u7531 `function` \u5173\u952e\u8bcd\u5b9a\u4e49\uff1a\n\n```julia\nfunction add(a, b)\n return a + b\nend\n```\n\n\u6216\u8005\u662f\u66f4\u77ed\u7684\u5f62\u5f0f\uff1a\n\n```julia\nadd(a, b) = a + b\n```\n\n[`return`](@ref) \u5173\u952e\u8bcd\u7684\u4f7f\u7528\u65b9\u6cd5\u548c\u5176\u5b83\u8bed\u8a00\u5b8c\u5168\u4e00\u6837\uff0c\u4f46\u662f\u5e38\u5e38\u662f\u4e0d\u4f7f\u7528\u7684\u3002\u4e00\u4e2a\u6ca1\u6709\u663e\u793a\u58f0\u660e `return` \u7684\u51fd\u6570\u5c06\u8fd4\u56de\u51fd\u6570\u4f53\u6700\u540e\u4e00\u4e2a\u8868\u8fbe\u5f0f\u3002\n\"\"\"\nkw\"function\"\n\n\"\"\"\n return\n\n`return` \u53ef\u4ee5\u7528\u6765\u5728\u51fd\u6570\u4f53\u4e2d\u7acb\u5373\u9000\u51fa\u5e76\u8fd4\u56de\u7ed9\u5b9a\u503c\uff0c\u4f8b\u5982\n\n```julia\nfunction compare(a, b)\n a == b && return \"equal to\"\n a < b ? \"less than\" : \"greater than\"\nend\n```\n\n\u901a\u5e38\uff0c\u4f60\u53ef\u4ee5\u5728\u51fd\u6570\u4f53\u7684\u4efb\u610f\u4f4d\u7f6e\u653e\u7f6e `return` \u8bed\u53e5\uff0c\u5305\u62ec\u5728\u591a\u5c42\u5d4c\u5957\u7684\u5faa\u73af\u548c\u6761\u4ef6\u8868\u8fbe\u5f0f\u4e2d\uff0c\u4f46\u8981\u6ce8\u610f `do` \u5757\u3002\u4f8b\u5982\uff1a\n\n```julia\nfunction test1(xs)\n for x in xs\n iseven(x) && return 2x\n end\nend\n\nfunction test2(xs)\n map(xs) do x\n iseven(x) && return 2x\n x\n end\nend\n```\n\n\u5728\u7b2c\u4e00\u4e2a\u4f8b\u5b50\u4e2d\uff0creturn \u4e00\u78b0\u5230\u5076\u6570\u5c31\u8df3\u51fa\u5305\u542b\u5b83\u7684\u51fd\u6570\uff0c\u56e0\u6b64 `test1([5,6,7])` \u8fd4\u56de `12`\u3002\n\n\u4f60\u53ef\u80fd\u5e0c\u671b\u7b2c\u4e8c\u4e2a\u4f8b\u5b50\u7684\u884c\u4e3a\u4e0e\u6b64\u76f8\u540c\uff0c\u4f46\u5b9e\u9645\u4e0a\uff0c\u8fd9\u91cc\u7684 `return` \u53ea\u4f1a\u8df3\u51fa\uff08\u5728 `do` \u5757\u4e2d\u7684\uff09*\u5185\u90e8*\u51fd\u6570\u5e76\u628a\u503c\u8fd4\u56de\u7ed9 `map`\u3002\u4e8e\u662f\uff0c`test2([5,6,7])` \u8fd4\u56de `[5,12,7]`\u3002\n\"\"\"\nkw\"return\"\n\n# \u4eff\u7167 https://docs.juliacn.com/latest/manual/control-flow/#man-conditional-evaluation-1\n\"\"\"\n if/elseif/else\n\n`if`/`elseif`/`else` \u6267\u884c\u6761\u4ef6\u8868\u8fbe\u5f0f\uff08Conditional evaluation\uff09\u53ef\u4ee5\u6839\u636e\u5e03\u5c14\u8868\u8fbe\u5f0f\u7684\u503c\uff0c\u8ba9\u90e8\u5206\u4ee3\u7801\u88ab\u6267\u884c\u6216\u8005\u4e0d\u88ab\u6267\u884c\u3002\u4e0b\u9762\u662f\u5bf9 `if`-`elseif`-`else` \u6761\u4ef6\u8bed\u6cd5\u7684\u5206\u6790\uff1a\n\n```julia\nif x < y\n println(\"x is less than y\")\nelseif x > y\n println(\"x is greater than y\")\nelse\n println(\"x is equal to y\")\nend\n```\n\n\u5982\u679c\u8868\u8fbe\u5f0f `x < y` \u662f `true`\uff0c\u90a3\u4e48\u5bf9\u5e94\u7684\u4ee3\u7801\u5757\u4f1a\u88ab\u6267\u884c\uff1b\u5426\u5219\u5224\u65ad\u6761\u4ef6\u8868\u8fbe\u5f0f `x > y`\uff0c\u5982\u679c\u5b83\u662f `true`\uff0c\u5219\u6267\u884c\u5bf9\u5e94\u7684\u4ee3\u7801\u5757\uff1b\u5982\u679c\u6ca1\u6709\u8868\u8fbe\u5f0f\u662f true\uff0c\u5219\u6267\u884c `else` \u4ee3\u7801\u5757\u3002`elseif` \u548c `else` \u4ee3\u7801\u5757\u662f\u53ef\u9009\u7684\uff0c\u5e76\u4e14\u53ef\u4ee5\u4f7f\u7528\u4efb\u610f\u591a\u4e2a `elseif` \u4ee3\u7801\u5757\u3002\n\"\"\"\nkw\"if\", kw\"elseif\", kw\"else\"\n\n\"\"\"\n for\n\n`for` \u5faa\u73af\u901a\u8fc7\u8fed\u4ee3\u4e00\u7cfb\u5217\u503c\u6765\u91cd\u590d\u8ba1\u7b97\u5faa\u73af\u4f53\u3002\n\n# \u4f8b\u5b50\n```jldoctest\njulia> for i in [1, 4, 0]\n println(i)\n end\n1\n4\n0\n```\n\"\"\"\nkw\"for\"\n\n# \u4eff\u7167 https://docs.juliacn.com/latest/manual/control-flow/#man-loops-1\n\"\"\"\n while\n\n`while` \u5faa\u73af\u4f1a\u91cd\u590d\u6267\u884c\u6761\u4ef6\u8868\u8fbe\u5f0f\uff0c\u5e76\u5728\u8be5\u8868\u8fbe\u5f0f\u4e3a `true` \u65f6\u7ee7\u7eed\u6267\u884c while \u5faa\u73af\u7684\u4e3b\u4f53\u90e8\u5206\u3002\u5f53 while \u5faa\u73af\u7b2c\u4e00\u6b21\u6267\u884c\u65f6\uff0c\u5982\u679c\u6761\u4ef6\u8868\u8fbe\u5f0f\u4e3a false\uff0c\u90a3\u4e48\u4e3b\u4f53\u4ee3\u7801\u5c31\u4e00\u6b21\u4e5f\u4e0d\u4f1a\u88ab\u6267\u884c\u3002\n\n# \u4f8b\u5b50\njldoctest\njulia> i = 1\n1\n\njulia> while i < 5\n println(i)\n global i += 1\n end\n1\n2\n3\n4\n```\n\"\"\"\nkw\"while\"\n\n\"\"\"\n end\n\n`end` \u6807\u8bb0\u4e00\u4e2a\u8868\u8fbe\u5f0f\u5757\u7684\u7ed3\u675f\uff0c\u4f8b\u5982 [`module`](@ref)\u3001[`struct`](@ref)\u3001[`mutable struct`](@ref)\u3001[`begin`](@ref)\u3001[`let`](@ref)\u3001[`for`](@ref) \u7b49\u3002`end` \u5728\u7d22\u5f15\u6570\u7ec4\u65f6\u4e5f\u53ef\u4ee5\u7528\u6765\u8868\u793a\u7ef4\u5ea6\u7684\u6700\u540e\u4e00\u4e2a\u7d22\u5f15\u3002\n\n# \u4f8b\u5b50\n```jldoctest\njulia> A = [1 2; 3 4]\n2\u00d72 Array{Int64,2}:\n 1 2\n 3 4\n\njulia> A[end, :]\n2-element Array{Int64,1}:\n 3\n 4\n```\n\"\"\"\nkw\"end\"\n\n# \u4eff\u7167 https://docs.juliacn.com/latest/manual/control-flow/#try/catch-%E8%AF%AD%E5%8F%A5-1\n\"\"\"\n try/catch\n\n`try/catch` \u8bed\u53e5\u53ef\u4ee5\u7528\u6765\u6355\u83b7 `Exception`\uff0c\u5e76\u8fdb\u884c\u5f02\u5e38\u5904\u7406\u3002\u4f8b\u5982\uff0c\u4e00\u4e2a\u81ea\u5b9a\u4e49\u7684\u5e73\u65b9\u6839\u51fd\u6570\u53ef\u4ee5\u901a\u8fc7 `Exception` \u6765\u5b9e\u73b0\u81ea\u52a8\u6309\u9700\u8c03\u7528\u6c42\u89e3\u5b9e\u6570\u6216\u8005\u590d\u6570\u5e73\u65b9\u6839\u7684\u65b9\u6cd5\uff1a\n\n```julia\nf(x) = try\n sqrt(x)\ncatch\n sqrt(complex(x, 0))\nend\n```\n\n`try/catch` \u8bed\u53e5\u5141\u8bb8\u4fdd\u5b58 `Exception` \u5230\u4e00\u4e2a\u53d8\u91cf\u4e2d\uff0c\u4f8b\u5982 `catch y`\u3002\n\n`try/catch` \u7ec4\u4ef6\u7684\u5f3a\u5927\u4e4b\u5904\u5728\u4e8e\u80fd\u591f\u5c06\u9ad8\u5ea6\u5d4c\u5957\u7684\u8ba1\u7b97\u7acb\u523b\u89e3\u8026\u6210\u66f4\u9ad8\u5c42\u6b21\u5730\u8c03\u7528\u51fd\u6570\u3002\n\"\"\"\nkw\"try\", kw\"catch\"\n\n# \u4eff\u7167 https://docs.juliacn.com/latest/manual/control-flow/#finally-%E5%AD%90%E5%8F%A5-1\n\"\"\"\n finally\n\n\u65e0\u8bba\u4ee3\u7801\u5757\u662f\u5982\u4f55\u9000\u51fa\u7684\uff0c\u90fd\u8ba9\u4ee3\u7801\u5757\u5728\u9000\u51fa\u65f6\u8fd0\u884c\u67d0\u6bb5\u4ee3\u7801\u3002\u8fd9\u91cc\u662f\u4e00\u4e2a\u786e\u4fdd\u4e00\u4e2a\u6253\u5f00\u7684\u6587\u4ef6\u88ab\u5173\u95ed\u7684\u4f8b\u5b50\uff1a\n\n```julia\nf = open(\"file\")\ntry\n operate_on_file(f)\nfinally\n close(f)\nend\n```\n\n\u5f53\u63a7\u5236\u6d41\u79bb\u5f00 `try` \u4ee3\u7801\u5757\uff08\u4f8b\u5982\uff0c\u9047\u5230 `return`\uff0c\u6216\u8005\u6b63\u5e38\u7ed3\u675f\uff09\uff0c`close(f)` \u5c31\u4f1a\u88ab\u6267\u884c\u3002\u5982\u679c `try` \u4ee3\u7801\u5757\u7531\u4e8e\u5f02\u5e38\u9000\u51fa\uff0c\u8fd9\u4e2a\u5f02\u5e38\u4f1a\u7ee7\u7eed\u4f20\u9012\u3002`catch` \u4ee3\u7801\u5757\u53ef\u4ee5\u548c `try` \u8fd8\u6709 `finally` \u914d\u5408\u4f7f\u7528\u3002\u8fd9\u65f6 `finally` \u4ee3\u7801\u5757\u4f1a\u5728 `catch` \u5904\u7406\u9519\u8bef\u4e4b\u540e\u624d\u8fd0\u884c\u3002\n\"\"\"\nkw\"finally\"\n\n\"\"\"\n break\n\n\u7acb\u5373\u8df3\u51fa\u5f53\u524d\u5faa\u73af\u3002\n\n# \u4f8b\u5b50\n```jldoctest\njulia> i = 0\n0\n\njulia> while true\n global i += 1\n i > 5 && break\n println(i)\n end\n1\n2\n3\n4\n5\n```\n\"\"\"\nkw\"break\"\n\n\"\"\"\n continue\n\n\u8df3\u8fc7\u5f53\u524d\u5faa\u73af\u8fed\u4ee3\u7684\u5269\u4f59\u90e8\u5206\u3002\n\n# \u4f8b\u5b50\n```jldoctest\njulia> for i = 1:6\n iseven(i) && continue\n println(i)\n end\n1\n3\n5\n```\n\"\"\"\nkw\"continue\"\n\n\"\"\"\n do\n\n\u521b\u5efa\u4e00\u4e2a\u533f\u540d\u51fd\u6570\u3002\u4f8b\u5982\uff1a\n\n```julia\nmap(1:10) do x\n 2x\nend\n```\n\n\u7b49\u4ef7\u4e8e `map(x->2x, 1:10)`\u3002\n\n\u50cf\u8fd9\u6837\u4fbf\u53ef\u4f7f\u7528\u591a\u4e2a\u53c2\u6570\uff1a\n\n```julia\nmap(1:10, 11:20) do x, y\n x + y\nend\n```\n\"\"\"\nkw\"do\"\n\n\"\"\"\n ...\n\n\u300csplat\u300d\u8fd0\u7b97\u7b26 `...` \u8868\u793a\u53c2\u6570\u5e8f\u5217\u3002`...` \u53ef\u4ee5\u5728\u51fd\u6570\u5b9a\u4e49\u4e2d\u7528\u6765\u8868\u793a\u8be5\u51fd\u6570\u63a5\u53d7\u4efb\u610f\u6570\u91cf\u7684\u53c2\u6570\u3002`...` \u4e5f\u53ef\u4ee5\u7528\u6765\u5c06\u51fd\u6570\u4f5c\u7528\u4e8e\u53c2\u6570\u5e8f\u5217\u3002\n\n# \u4f8b\u5b50\n```jldoctest\njulia> add(xs...) = reduce(+, xs)\nadd (generic function with 1 method)\n\njulia> add(1, 2, 3, 4, 5)\n15\n\njulia> add([1, 2, 3]...)\n6\n\njulia> add(7, 1:100..., 1000:1100...)\n111107\n```\n\"\"\"\nkw\"...\"\n\n\"\"\"\n ;\n\n`;` \u5728 Julia \u4e2d\u5177\u6709\u4e0e\u8bb8\u591a\u7c7b C \u8bed\u8a00\u76f8\u4f3c\u7684\u4f5c\u7528\uff0c\u7528\u4e8e\u5206\u9694\u524d\u4e00\u4e2a\u8bed\u53e5\u7684\u7ed3\u5c3e\u3002`;` \u5728\u6362\u884c\u4e2d\u4e0d\u662f\u5fc5\u8981\u7684\uff0c\u4f46\u53ef\u4ee5\u7528\u4e8e\u5728\u5355\u884c\u4e2d\u5206\u9694\u8bed\u53e5\u6216\u8005\u5c06\u591a\u4e2a\u8868\u8fbe\u5f0f\u8fde\u63a5\u4e3a\u5355\u4e2a\u8868\u8fbe\u5f0f\u3002`;` \u4e5f\u7528\u4e8e\u6291\u5236 REPL \u548c\u7c7b\u4f3c\u754c\u9762\u4e2d\u7684\u8f93\u51fa\u6253\u5370\u3002\n\n# \u4f8b\u5b50\n```julia\njulia> function foo()\n x = \"Hello, \"; x *= \"World!\"\n return x\n end\nfoo (generic function with 1 method)\n\njulia> bar() = (x = \"Hello, Mars!\"; return x)\nbar (generic function with 1 method)\n\njulia> foo();\n\njulia> bar()\n\"Hello, Mars!\"\n```\n\"\"\"\nkw\";\"\n\n\"\"\"\n x && y\n\n\u77ed\u8def\u5e03\u5c14 AND\u3002\n\"\"\"\nkw\"&&\"\n\n\"\"\"\n x || y\n\n\u77ed\u8def\u5e03\u5c14 OR\u3002\n\"\"\"\nkw\"||\"\n\n\"\"\"\n ccall((function_name, library), returntype, (argtype1, ...), argvalue1, ...)\n ccall(function_name, returntype, (argtype1, ...), argvalue1, ...)\n ccall(function_pointer, returntype, (argtype1, ...), argvalue1, ...)\n\n\u8c03\u7528\u7531 C \u5bfc\u51fa\u7684\u5171\u4eab\u5e93\u91cc\u7684\u51fd\u6570\uff0c\u8be5\u51fd\u6570\u7531\u5143\u7ec4 `(function_name, library)` \u6307\u5b9a\uff0c\u5176\u4e2d\u7684\u6bcf\u4e2a\u7ec4\u4ef6\u90fd\u662f\u5b57\u7b26\u4e32\u6216\u7b26\u53f7\u3002\u82e5\u4e0d\u6307\u5b9a\u5e93\uff0c\u8fd8\u53ef\u4ee5\u4f7f\u7528 `function_name` \u7684\u7b26\u53f7\u6216\u5b57\u7b26\u4e32\uff0c\u5b83\u4f1a\u5728\u5f53\u524d\u8fdb\u7a0b\u4e2d\u89e3\u6790\u3002\u53e6\u5916\uff0c`ccall` \u4e5f\u53ef\u7528\u4e8e\u8c03\u7528\u51fd\u6570\u6307\u9488 `function_pointer`\uff0c\u6bd4\u5982 `dlsym` \u8fd4\u56de\u7684\u51fd\u6570\u6307\u9488\u3002\n\n\u8bf7\u6ce8\u610f\u53c2\u6570\u7c7b\u578b\u5143\u7ec4\u5fc5\u987b\u662f\u5b57\u9762\u4e0a\u7684\u5143\u7ec4\uff0c\u800c\u4e0d\u662f\u5143\u7ec4\u7c7b\u578b\u7684\u53d8\u91cf\u6216\u8868\u8fbe\u5f0f\u3002\n\n\u901a\u8fc7\u81ea\u52a8\u63d2\u5165\u5bf9 `unsafe_convert(argtype, cconvert(argtype, argvalue))` \u7684\u8c03\u7528\uff0c\u6bcf\u4e2a\u4f20\u7ed9 `ccall` \u7684 `argvalue` \u5c06\u88ab\u7c7b\u578b\u8f6c\u6362\u4e3a\u5bf9\u5e94\u7684 `argtype`\u3002\uff08\u6709\u5173\u7684\u8be6\u7ec6\u4fe1\u606f\uff0c\u8bf7\u53c2\u9605 [`unsafe_convert`](@ref Base.unsafe_convert) \u548c [`cconvert`](@ref Base.cconvert) \u7684\u6587\u6863\u3002\uff09\u5728\u5927\u591a\u6570\u60c5\u51b5\u4e0b\uff0c\u8fd9\u53ea\u4f1a\u7b80\u5355\u5730\u8c03\u7528 `convert(argtype, argvalue)`\u3002\n\"\"\"\nkw\"ccall\"\n\n\"\"\"\n begin\n\n`begin...end` \u8868\u793a\u4e00\u4e2a\u4ee3\u7801\u5757\u3002\n\n```julia\nbegin\n println(\"Hello, \")\n println(\"World!\")\nend\n```\n\n\u901a\u5e38\uff0c`begin` \u4e0d\u4f1a\u662f\u5fc5\u9700\u7684\uff0c\u56e0\u4e3a\u8bf8\u5982 [`function`](@ref) and [`let`](@ref) \u4e4b\u7c7b\u7684\u5173\u952e\u5b57\u4f1a\u9690\u5f0f\u5730\u5f00\u59cb\u4ee3\u7801\u5757\u3002\u53e6\u8bf7\u53c2\u9605 [`;`](@ref)\u3002\n\"\"\"\nkw\"begin\"\n\n\"\"\"\n struct\n\nstruct \u662f Julia \u4e2d\u6700\u5e38\u7528\u7684\u6570\u636e\u7c7b\u578b\uff0c\u7531\u540d\u79f0\u548c\u4e00\u7ec4\u5b57\u6bb5\u6307\u5b9a\u3002\n\n```julia\nstruct Point\n x\n y\nend\n```\n\n\u53ef\u5bf9\u5b57\u6bb5\u65bd\u52a0\u7c7b\u578b\u9650\u5236\uff0c\u8be5\u9650\u5236\u4e5f\u53ef\u88ab\u53c2\u6570\u5316\uff1a\n\n```julia\n struct Point{X}\n x::X\n y::Float64\n end\n```\n\nstruct \u53ef\u4ee5\u901a\u8fc7 `<:` \u8bed\u6cd5\u58f0\u660e\u4e00\u4e2a\u62bd\u8c61\u8d85\u7c7b\u578b\uff1a\nA struct can also declare an abstract super type via `<:` syntax:\n\n```julia\nstruct Point <: AbstractPoint\n x\n y\nend\n```\n\n`struct` \u9ed8\u8ba4\u662f\u4e0d\u53ef\u53d8\u7684\uff1b\u8fd9\u4e9b\u7c7b\u578b\u7684\u5b9e\u4f8b\u5728\u6784\u9020\u540e\u4e0d\u80fd\u88ab\u4fee\u6539\u3002\u5982\u9700\u4fee\u6539\u5b9e\u4f8b\uff0c\u8bf7\u4f7f\u7528 [`mutable struct`](@ref) \u6765\u58f0\u660e\u4e00\u4e2a\u53ef\u4ee5\u4fee\u6539\u5176\u5b9e\u4f8b\u7684\u7c7b\u578b\u3002\n\n\u6709\u5173\u66f4\u591a\u7ec6\u8282\uff0c\u6bd4\u5982\u600e\u4e48\u5b9a\u4e49\u6784\u9020\u51fd\u6570\uff0c\u8bf7\u53c2\u9605\u624b\u518c\u7684 [\u590d\u5408\u7c7b\u578b](@ref) \u7ae0\u8282\u3002\n\"\"\"\nkw\"struct\"\n\n\"\"\"\n mutable struct\n\n`mutable struct` \u7c7b\u4f3c\u4e8e [`struct`](@ref)\uff0c\u4f46\u53e6\u5916\u5141\u8bb8\u5728\u6784\u9020\u540e\u8bbe\u7f6e\u7c7b\u578b\u7684\u5b57\u6bb5\u3002\u6709\u5173\u8be6\u7ec6\u4fe1\u606f\uff0c\u8bf7\u53c2\u9605 [\u590d\u5408\u7c7b\u578b](@ref)\u3002\n\"\"\"\nkw\"mutable struct\"\n\n\"\"\"\n new\n\n\u4ec5\u5728\u5185\u90e8\u6784\u9020\u51fd\u6570\u4e2d\u53ef\u7528\u7684\u7279\u6b8a\u51fd\u6570\uff0c\u7528\u6765\u521b\u5efa\u8be5\u7c7b\u578b\u7684\u5bf9\u8c61\u3002\u6709\u5173\u66f4\u591a\u4fe1\u606f\uff0c\u8bf7\u53c2\u9605\u624b\u518c\u7684 [\u5185\u90e8\u6784\u9020\u65b9\u6cd5](@ref) \u7ae0\u8282\u3002\n\"\"\"\nkw\"new\"\n\n\"\"\"\n where\n\n`where` \u5173\u952e\u5b57\u521b\u5efa\u4e00\u4e2a\u7c7b\u578b\uff0c\u8be5\u7c7b\u578b\u662f\u5176\u4ed6\u7c7b\u578b\u5728\u4e00\u4e9b\u53d8\u91cf\u4e0a\u6240\u6709\u503c\u7684\u8fed\u4ee3\u5e76\u96c6\u3002\u4f8b\u5982 `Vector{T} where T<:Real` \u5305\u542b\u6240\u6709\u5143\u7d20\u7c7b\u578b\u662f\u67d0\u79cd `Real` \u7684 [`Vector`](@ref)\u3002\n\n\u5982\u679c\u7701\u7565\uff0c\u53d8\u91cf\u4e0a\u754c\u9ed8\u8ba4\u4e3a `Any`\uff1a\n\n```julia\nVector{T} where T # short for `where T<:Any`\n```\n\n\u53d8\u91cf\u4e5f\u53ef\u4ee5\u5177\u6709\u4e0b\u754c\uff1a\n\n```julia\nVector{T} where T>:Int\nVector{T} where Int<:T<:Real\n```\n\n\u5d4c\u5957\u7684 `where` \u4e5f\u6709\u7b80\u6d01\u7684\u8bed\u6cd5\u3002\u4f8b\u5982\uff0c\u8fd9\u884c\u4ee3\u7801\uff1a\n\n```julia\nPair{T, S} where S<:Array{T} where T<:Number\n```\n\n\u53ef\u4ee5\u7f29\u5199\u4e3a\uff1a\n\n```julia\nPair{T, S} where {T<:Number, S<:Array{T}}\n```\n\n\u8fd9\u79cd\u5f62\u5f0f\u5e38\u89c1\u4e8e\u65b9\u6cd5\u7b7e\u540d\uff1a\n\n\u8bf7\u6ce8\u610f\uff0c\u5728\u8fd9\u79cd\u5f62\u5f0f\u4e2d\uff0c\u6700\u5916\u5c42\u53d8\u91cf\u5217\u5728\u6700\u524d\u9762\u3002\u8fd9\u4e0e\u4f7f\u7528\u8bed\u6cd5 `T{p1, p2, ...}` \u5c06\u7c7b\u578b\u300c\u4f5c\u7528\u300d\u4e8e\u53c2\u6570\u503c\u65f6\u6240\u66ff\u6362\u53d8\u91cf\u7684\u6b21\u5e8f\u76f8\u5339\u914d\u3002\n\"\"\"\nkw\"where\"\n\n\"\"\"\n ans\n\n\u4e00\u4e2a\u5f15\u7528\u6700\u540e\u4e00\u6b21\u8ba1\u7b97\u7ed3\u679c\u7684\u53d8\u91cf\uff0c\u5728\u4ea4\u4e92\u5f0f\u63d0\u793a\u7b26\u4e2d\u4f1a\u81ea\u52a8\u8bbe\u7f6e\u3002\n\"\"\"\nkw\"ans\"\n\n\"\"\"\n Union{}\n\n`Union{}`\uff0c\u5373\u7a7a\u7684\u7c7b\u578b [`Union`](@ref)\uff0c\u662f\u6ca1\u6709\u503c\u7684\u7c7b\u578b\u3002\u4e5f\u5c31\u662f\u8bf4\uff0c\u5b83\u5177\u6709\u51b3\u5b9a\u6027\u6027\u8d28\uff1a\u5bf9\u4e8e\u4efb\u4f55 `x`\uff0c`isa(x, Union{}) == false`\u3002`Base.Bottom` \u88ab\u5b9a\u4e49\u4e3a\u5176\u522b\u540d\uff0c`Union{}` \u7684\u7c7b\u578b\u662f `Core.TypeofBottom`\u3002\n\n# \u4f8b\u5b50\n```jldoctest\njulia> isa(nothing, Union{})\nfalse\n```\n\"\"\"\nkw\"Union{}\", Base.Bottom\n\n\"\"\"\n ::\n\n`::` \u8fd0\u7b97\u7b26\u7528\u4e8e\u7c7b\u578b\u58f0\u660e\uff0c\u5728\u7a0b\u5e8f\u4e2d\u53ef\u88ab\u9644\u52a0\u5230\u8868\u8fbe\u5f0f\u548c\u53d8\u91cf\u540e\u3002\u8be6\u89c1\u624b\u518c\u7684 [\u7c7b\u578b\u58f0\u660e](@ref) \u7ae0\u8282\n\n\u5728\u7c7b\u578b\u58f0\u660e\u5916\uff0c`::` \u7528\u4e8e\u65ad\u8a00\u7a0b\u5e8f\u4e2d\u7684\u8868\u8fbe\u5f0f\u548c\u53d8\u91cf\u5177\u6709\u7ed9\u5b9a\u7c7b\u578b\u3002\n\n# \u4f8b\u5b50\n```jldoctest\njulia> (1+2)::AbstractFloat\nERROR: TypeError: typeassert: expected AbstractFloat, got Int64\n\njulia> (1+2)::Int\n3\n```\n\"\"\"\nkw\"::\"\n\n\"\"\"\nJulia \u7684\u57fa\u7840\u5e93\u3002\n\"\"\"\nkw\"Base\"\n", "meta": {"hexsha": "5b5abdfbfeebcba59678d56501022aa5209333d6", "size": 11505, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/basedocs.jl", "max_stars_repo_name": "UnofficialJuliaMirror/JuliaZH.jl-652e05fd-ed22-5b6c-bf99-44e63a676e5f", "max_stars_repo_head_hexsha": "78c38dadab009de38e34f12af89eff240bb9a724", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 504, "max_stars_repo_stars_event_min_datetime": "2018-07-19T03:38:47.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-31T04:55:11.000Z", "max_issues_repo_path": "src/basedocs.jl", "max_issues_repo_name": "UnofficialJuliaMirror/JuliaZH.jl-652e05fd-ed22-5b6c-bf99-44e63a676e5f", "max_issues_repo_head_hexsha": "78c38dadab009de38e34f12af89eff240bb9a724", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 78, "max_issues_repo_issues_event_min_datetime": "2018-07-18T16:21:06.000Z", "max_issues_repo_issues_event_max_datetime": "2022-01-04T14:44:51.000Z", "max_forks_repo_path": "src/basedocs.jl", "max_forks_repo_name": "UnofficialJuliaMirror/JuliaZH.jl-652e05fd-ed22-5b6c-bf99-44e63a676e5f", "max_forks_repo_head_hexsha": "78c38dadab009de38e34f12af89eff240bb9a724", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 114, "max_forks_repo_forks_event_min_datetime": "2018-07-18T19:24:19.000Z", "max_forks_repo_forks_event_max_datetime": "2022-03-06T08:23:42.000Z", "avg_line_length": 16.3191489362, "max_line_length": 258, "alphanum_fraction": 0.6392003477, "num_tokens": 6183, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.32082130082460697, "lm_q2_score": 0.18242552825126704, "lm_q1q2_score": 0.05852599527718758}}
{"text": "mutable struct Point\n x::Int64\n y::Float64\n meta\nend\np = Point(0, 0.0, \"Origin\")\np.x # \u8bbf\u95ee\u57df\np.meta = 2 # \u6539\u53d8\u57df\u7684\u503c\np.x = 1.5 # \u9519\u8bef\uff0c\u6570\u636e\u7c7b\u578b\u4e0d\u5339\u914d\np.z = 1 # \u9519\u8bef\uff0c\u6ca1\u6709\u8fd9\u4e2a\u57df\nfieldnames(Point) # \u83b7\u53d6\u6240\u6709\u7684\u57df\u540d\n", "meta": {"hexsha": "5f9ad620262639b6644531df5c35dbffadecb934", "size": 222, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/struct.jl", "max_stars_repo_name": "zhaiyusci/Julia-Notes", "max_stars_repo_head_hexsha": "b8b6992af2a04d9eff5612f3d673f7e1e705a034", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/struct.jl", "max_issues_repo_name": "zhaiyusci/Julia-Notes", "max_issues_repo_head_hexsha": "b8b6992af2a04d9eff5612f3d673f7e1e705a034", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 1, "max_issues_repo_issues_event_min_datetime": "2021-04-23T13:40:13.000Z", "max_issues_repo_issues_event_max_datetime": "2021-04-23T13:40:29.000Z", "max_forks_repo_path": "src/struct.jl", "max_forks_repo_name": "zhaiyusci/Julia-Notes", "max_forks_repo_head_hexsha": "b8b6992af2a04d9eff5612f3d673f7e1e705a034", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 18.5, "max_line_length": 30, "alphanum_fraction": 0.518018018, "num_tokens": 109, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.42250463481418826, "lm_q2_score": 0.13846179590164853, "lm_q1q2_score": 0.058500750513142685}}
{"text": "\"\"\"\n grdimage(cmd0::String=\"\", arg1=[], arg2=[], arg3=[]; kwargs...)\n\nProduces a gray-shaded (or colored) map by plotting rectangles centered on each grid node and assigning them a gray-shade (or color) based on the z-value.\n\nFull option list at [`grdimage`](http://gmt.soest.hawaii.edu/doc/latest/grdimage.html)\n\nParameters\n----------\n\n- **A** : **img_out** : **image_out** : -- Str --\n\n Save an image in a raster format instead of PostScript.\n [`-A`](http://gmt.soest.hawaii.edu/doc/latest/grdimage.html#a)\n- $(GMT.opt_J)\n- $(GMT.opt_B)\n- $(GMT.opt_C)\n- **D** : **img_in** : **image_in** : -- Str or [] --\n\n Specifies that the grid supplied is an image file to be read via GDAL.\n [`-D`](http://gmt.soest.hawaii.edu/doc/latest/grdimage.html#d)\n- **E** : **dpi** : -- Int or [] -- \n\n Sets the resolution of the projected grid that will be created.\n [`-E`](http://gmt.soest.hawaii.edu/doc/latest/grdimage.html#e)\n- **G** : -- Str or Int --\n\n [`-G`](http://gmt.soest.hawaii.edu/doc/latest/grdimage.html#g)\n- **I** : **shade** : **intensity** : **intensfile** : -- Str or GMTgrid --\n\n Gives the name of a grid file or GMTgrid with intensities in the (-1,+1) range,\n or a grdgradient shading flags.\n [`-I`](http://gmt.soest.hawaii.edu/doc/latest/grdimage.html#i)\n- **M** : **monochrome** : -- Bool or [] --\n\n Force conversion to monochrome image using the (television) YIQ transformation.\n [`-M`](http://gmt.soest.hawaii.edu/doc/latest/grdimage.html#m)\n- **N** : **noclip** : -- Bool or [] --\n\n Do not clip the image at the map boundary.\n [`-N`](http://gmt.soest.hawaii.edu/doc/latest/grdimage.html#n)\n- $(GMT.opt_P)\n- **Q** : **nan_t** : **nan_alpha** : -- Bool or [] --\n\n Make grid nodes with z = NaN transparent, using the colormasking feature in PostScript Level 3.\n- $(GMT.opt_R)\n- $(GMT.opt_U)\n- $(GMT.opt_V)\n- $(GMT.opt_X)\n- $(GMT.opt_Y)\n- $(GMT.opt_f)\n- $(GMT.opt_n)\n- $(GMT.opt_p)\n- $(GMT.opt_t)\n\"\"\"\nfunction grdimage(cmd0::String=\"\", arg1=[], arg2=[], arg3=[], arg4=[]; K=false, O=false, first=true, kwargs...)\n\n\tlength(kwargs) == 0 && occursin(\" -\", cmd0) && return monolitic(\"grdimage\", cmd0, arg1)\t# Speedy mode\n\n\td = KW(kwargs)\n\toutput, opt_T, fname_ext = fname_out(d)\t\t# OUTPUT may have been an extension only\n\n\tcmd, opt_B, = parse_BJR(d, \"\", \"\", O, \" -JX12c/0\")\n\tcmd = parse_UVXY(cmd, d)\n\tcmd, = parse_f(cmd, d)\n\tcmd, = parse_n(cmd, d)\n\tcmd, = parse_p(cmd, d)\n\tcmd, = parse_t(cmd, d)\n\tcmd = parse_params(cmd, d)\n\n\tcmd, K, O, = set_KO(cmd, opt_B, first, K, O)\t\t\t# Set the K O dance\n\n\tcmd = add_opt(cmd, 'A', d, [:A :img_out :image_out])\n\tcmd = add_opt(cmd, 'D', d, [:D :img_in :image_in])\n\tcmd = add_opt(cmd, 'E', d, [:E :dpi])\n\tcmd = add_opt(cmd, 'G', d, [:G])\n\tcmd = add_opt(cmd, 'M', d, [:M :monochrome])\n\tcmd = add_opt(cmd, 'N', d, [:N :noclip])\n\tcmd = add_opt(cmd, 'Q', d, [:Q :nan_t :nan_alpha])\n\n\tcmd, got_fname, arg1 = find_data(d, cmd0, cmd, 1, arg1)\t\t# Find how data was transmitted\n\tif (got_fname == 0 && isempty_(arg1))\t\t\t# Than it must be using the three r,g,b grids\n\t\tcmd, got_fname, arg1, arg2, arg3 = find_data(d, cmd0, cmd, 3, arg1, arg2, arg3)\n\t\tif (got_fname == 0 && isempty_(arg1))\n\t\t\terror(\"No input data to use in grdimage.\")\n\t\tend\n\tend\n\n\tif (isa(arg1, Array{<:Number}))\n\t\targ1 = mat2grid(arg1)\n\t\tif (!isempty_(arg2) && isa(arg2, Array{<:Number})) arg2 = mat2grid(arg2) end\n\t\tif (!isempty_(arg3) && isa(arg3, Array{<:Number})) arg3 = mat2grid(arg3) end\n\tend\n\n\tN_used = got_fname == 0 ? 1 : 0\t\t# To know whether a cpt will go to arg1 or arg2\n\tcmd, arg1, arg2, = add_opt_cpt(d, cmd, [:C :color :cmap], 'C', N_used, arg1, arg2)\n\tif (!isempty_(arg3) && occursin(\"-C\", cmd))\t\t# This lieves out the case when the r,g,b were sent as a text.\n\t\terror(\"Cannot use the three R,G,B grids and a color table.\")\n\tend\n\n\tfor sym in [:I :shade :intensity :intensfile]\n\t\tif (haskey(d, sym))\n\t\t\tif (!isa(d[sym], GMTgrid))\t\t# Uff, simple. Either a file name or a -A type modifier\n\t\t\t\tcmd = cmd * \" -I\" * arg2str(d[sym])\n\t\t\telse\n\t\t\t\tcmd, N = put_in_slot(cmd, d[sym], 'I', [arg1, arg2, arg3, arg4])\n\t\t\t\tif (N == 1) arg1 = d[sym]\n\t\t\t\telseif (N == 2) arg2 = d[sym]\n\t\t\t\telseif (N == 3) arg3 = d[sym]\n\t\t\t\telseif (N == 4) arg4 = d[sym]\n\t\t\t\tend\n\t\t\tend\n\t\t\tbreak\n\t\tend\n\tend\n\n\tcmd = finish_PS(d, cmd, output, K, O)\n return finish_PS_module(d, cmd, \"\", output, fname_ext, opt_T, K, \"grdimage\", arg1, arg2, arg3, arg4)\nend\n\n# ---------------------------------------------------------------------------------------------------\ngrdimage!(cmd0::String=\"\", arg1=[], arg2=[], arg3=[], arg4=[]; K=true, O=true, first=false, kw...) =\n\tgrdimage(cmd0, arg1, arg2, arg3, arg4; K=true, O=true, first=false, kw...) \n\ngrdimage(arg1, cmd0::String=\"\", arg2=[], arg3=[], arg4=[]; K=false, O=false, first=true, kw...) =\n\tgrdimage(cmd0, arg1, arg2, arg3, arg4; K=K, O=O, first=first, kw...)\n\ngrdimage!(arg1, cmd0::String=\"\", arg2=[], arg3=[], arg4=[]; K=true, O=true, first=false, kw...) =\n\tgrdimage(cmd0, arg1, arg2, arg3, arg4; K=K, O=O, first=first, kw...)", "meta": {"hexsha": "7d25c406b86728a3b3611f2255269d9502c4d558", "size": 5001, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/grdimage.jl", "max_stars_repo_name": "JuliaDocsForks/GMT.jl", "max_stars_repo_head_hexsha": "53d9c030559fe3f079f3b662ce262e7ed682d357", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/grdimage.jl", "max_issues_repo_name": "JuliaDocsForks/GMT.jl", "max_issues_repo_head_hexsha": "53d9c030559fe3f079f3b662ce262e7ed682d357", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/grdimage.jl", "max_forks_repo_name": "JuliaDocsForks/GMT.jl", "max_forks_repo_head_hexsha": "53d9c030559fe3f079f3b662ce262e7ed682d357", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 38.7674418605, "max_line_length": 154, "alphanum_fraction": 0.5970805839, "num_tokens": 1674, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.411110869232168, "lm_q2_score": 0.1422318950098963, "lm_q1q2_score": 0.058473077990056924}}
{"text": "\"\"\"\nThrow an `AssertionError` if conditions like `op(exp1, exp2)` are `false`, where `op` is a conditional infix operator.\n\n# Examples\n\n```\njulia> a = 3; b = 4;\njulia> @assert_op a == b\nERROR: AssertionError: 3 == 4\n\njulia> @assert_op a + 3 > b + 4\nERROR: AssertionError: 6 > 8\n```\n\"\"\"\nmacro assert_op(expr)\n assert_op(expr)\nend\n\nfunction assert_op(expr::Expr)\n # Only special case expressions of the form `expr1 == expr2`\n if length(expr.args) == 3 && expr.head == :call\n return assert_op(expr.args[1], expr.args[2], expr.args[3])\n else\n return :(@assert $(expr))\n end\nend\n\nfunction assert_op(op, exp1, exp2)\n return :(\n if !$op($(esc(exp1)), $(esc(exp2)))\n val1 = $(esc(exp1))\n val2 = $(esc(exp2))\n op_str = $(esc(op))\n throw(AssertionError(\"$val1 $op_str $val2\"))\n end\n )\nend\n", "meta": {"hexsha": "28e5ac96fc4ed9095de1037d170e890a45f4defb", "size": 872, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/utils/assert_op.jl", "max_stars_repo_name": "m-bossart/InfrastructureSystems.jl", "max_stars_repo_head_hexsha": "1879ffa82715c2ec1446d3781818972120d2bfaf", "max_stars_repo_licenses": ["BSD-3-Clause"], "max_stars_count": 25, "max_stars_repo_stars_event_min_datetime": "2020-04-25T20:30:52.000Z", "max_stars_repo_stars_event_max_datetime": "2022-01-25T01:01:09.000Z", "max_issues_repo_path": "src/utils/assert_op.jl", "max_issues_repo_name": "m-bossart/InfrastructureSystems.jl", "max_issues_repo_head_hexsha": "1879ffa82715c2ec1446d3781818972120d2bfaf", "max_issues_repo_licenses": ["BSD-3-Clause"], "max_issues_count": 196, "max_issues_repo_issues_event_min_datetime": "2020-04-01T21:36:00.000Z", "max_issues_repo_issues_event_max_datetime": "2022-03-22T03:22:18.000Z", "max_forks_repo_path": "src/utils/assert_op.jl", "max_forks_repo_name": "m-bossart/InfrastructureSystems.jl", "max_forks_repo_head_hexsha": "1879ffa82715c2ec1446d3781818972120d2bfaf", "max_forks_repo_licenses": ["BSD-3-Clause"], "max_forks_count": 13, "max_forks_repo_forks_event_min_datetime": "2020-07-15T16:32:36.000Z", "max_forks_repo_forks_event_max_datetime": "2022-03-29T16:09:30.000Z", "avg_line_length": 22.9473684211, "max_line_length": 118, "alphanum_fraction": 0.5802752294, "num_tokens": 264, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.3998116407397951, "lm_q2_score": 0.14608724704829565, "lm_q1q2_score": 0.05840738193353887}}
{"text": "a = b = 0\n\nn = parse(Int, readline())\nfor i in 1:n\n sa, sb = split(readline())\n if sa < sb\n global b += 3\n elseif sa > sb\n global a += 3\n else\n a += 1\n b += 1\n end\nend\nprintln(a, \" \", b)\n\n\n", "meta": {"hexsha": "5fad9d6618a8637d539806e1df1e072f5eaa9cd4", "size": 232, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "ITP1/ITP1_9/ITP1_9_C.jl", "max_stars_repo_name": "goropikari/AizuOnlineJudge_solution_julia", "max_stars_repo_head_hexsha": "4c135f2359b421024800cbabd6efb458345075b0", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "ITP1/ITP1_9/ITP1_9_C.jl", "max_issues_repo_name": "goropikari/AizuOnlineJudge_solution_julia", "max_issues_repo_head_hexsha": "4c135f2359b421024800cbabd6efb458345075b0", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "ITP1/ITP1_9/ITP1_9_C.jl", "max_forks_repo_name": "goropikari/AizuOnlineJudge_solution_julia", "max_forks_repo_head_hexsha": "4c135f2359b421024800cbabd6efb458345075b0", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 12.8888888889, "max_line_length": 30, "alphanum_fraction": 0.4310344828, "num_tokens": 83, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.476579651063676, "lm_q2_score": 0.1225232173242878, "lm_q1q2_score": 0.05839207215960802}}
{"text": "# This file was generated, do not modify it. # hide\nusing RDatasets, DataFrames\nboston = dataset(\"MASS\", \"Boston\")\nfirst(boston, 3)", "meta": {"hexsha": "fa011cb368200b648b0dd9f49388b90b3059d730", "size": 131, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "_assets/pages/isl/lab-3/code/ex2.jl", "max_stars_repo_name": "giordano/DataScienceTutorials.jl", "max_stars_repo_head_hexsha": "8284298842e0d77061cf8ee767d0899fb7d051ff", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 29, "max_stars_repo_stars_event_min_datetime": "2021-08-09T11:35:53.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-07T06:20:43.000Z", "max_issues_repo_path": "_assets/pages/isl/lab-3/code/ex2.jl", "max_issues_repo_name": "giordano/DataScienceTutorials.jl", "max_issues_repo_head_hexsha": "8284298842e0d77061cf8ee767d0899fb7d051ff", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 56, "max_issues_repo_issues_event_min_datetime": "2019-10-22T00:06:41.000Z", "max_issues_repo_issues_event_max_datetime": "2020-05-21T14:38:09.000Z", "max_forks_repo_path": "_assets/pages/isl/lab-3/code/ex2.jl", "max_forks_repo_name": "giordano/DataScienceTutorials.jl", "max_forks_repo_head_hexsha": "8284298842e0d77061cf8ee767d0899fb7d051ff", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 9, "max_forks_repo_forks_event_min_datetime": "2019-11-20T16:25:04.000Z", "max_forks_repo_forks_event_max_datetime": "2020-05-05T11:55:15.000Z", "avg_line_length": 32.75, "max_line_length": 51, "alphanum_fraction": 0.7328244275, "num_tokens": 39, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4571367168274948, "lm_q2_score": 0.12765263527792128, "lm_q1q2_score": 0.05835470658532657}}
{"text": "using Test\nusing Mimi\n\n@defcomp ArgTester begin\n varA = Variable(index=[time])\n parA = Parameter()\n \n function run_timestep(p, v, d, t)\n v.varA[t] = p.parA\n end\nend\n\nm = Model()\n\n# trying to run model with no components\nset_dimension!(m, :time, 1:10)\n@test_throws ErrorException run(m)\n", "meta": {"hexsha": "5b211a924b16c54e2f4cc2c3d0642d85851b5cec", "size": 295, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/test_num_components.jl", "max_stars_repo_name": "arnavgautam/Mimi.jl", "max_stars_repo_head_hexsha": "7c6c43bf721166b76b997dd6fd4d167c57225e33", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "test/test_num_components.jl", "max_issues_repo_name": "arnavgautam/Mimi.jl", "max_issues_repo_head_hexsha": "7c6c43bf721166b76b997dd6fd4d167c57225e33", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "test/test_num_components.jl", "max_forks_repo_name": "arnavgautam/Mimi.jl", "max_forks_repo_head_hexsha": "7c6c43bf721166b76b997dd6fd4d167c57225e33", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 16.3888888889, "max_line_length": 40, "alphanum_fraction": 0.6847457627, "num_tokens": 93, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.45713671682749474, "lm_q2_score": 0.12765261702501565, "lm_q1q2_score": 0.058354698241253215}}
{"text": "module Example\n\nexport func\n\n\"\"\"\n func(x)\n\nReturns double the number `x` plus `1`.\n\nHere's an equation:\n\n``\\\\frac{n!}{k!(n - k)!} = \\\\binom{n}{k}``\n\n\"\"\"\nfunc(x) = 2x + 1\n\nend\n", "meta": {"hexsha": "2cba04500b4dc95f0556e1ed0de0d6e84788a6ff", "size": 178, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/Example.jl", "max_stars_repo_name": "jorgepz/Julia_Docs_Testbed", "max_stars_repo_head_hexsha": "f1271de3a024e194f5161b28cb9e7e7d11fca238", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/Example.jl", "max_issues_repo_name": "jorgepz/Julia_Docs_Testbed", "max_issues_repo_head_hexsha": "f1271de3a024e194f5161b28cb9e7e7d11fca238", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 3, "max_issues_repo_issues_event_min_datetime": "2021-06-23T14:34:23.000Z", "max_issues_repo_issues_event_max_datetime": "2021-06-24T20:35:58.000Z", "max_forks_repo_path": "src/Example.jl", "max_forks_repo_name": "jorgepz/Julia_Docs_Testbed", "max_forks_repo_head_hexsha": "f1271de3a024e194f5161b28cb9e7e7d11fca238", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 9.8888888889, "max_line_length": 42, "alphanum_fraction": 0.5449438202, "num_tokens": 63, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4960938294709195, "lm_q2_score": 0.11757213663746784, "lm_q1q2_score": 0.05832681150355962}}
{"text": "# # GridapMakie\n#\n# [](https://gridap.github.io/GridapMakie.jl/stable)\n# [](https://gridap.github.io/GridapMakie.jl/dev)\n# [](https://github.com/gridap/GridapMakie.jl/actions)\n# [](https://codecov.io/gh/gridap/GridapMakie.jl)\n\n# ## Overview\n\n# The visualization of numerical results is an important part of finite element (FE) computations. However, before the inception of GridapMakie.jl, the \n# only approach available to data visualization of [Gridap.jl](https://github.com/gridap/Gridap.jl) computations was to write simulation \n# data to data files (e.g., in vtu format) for later visualization with, e.g., Paraview or VisIt. From the idea of visually inspecting \n# data from Julia code directly or to manipulate it with packages of the Julia \n# open-source package ecosystem, [GridapMakie.jl](https://github.com/gridap/GridapMakie.jl) is born. As a part of the Google Summer of \n# Code 2021 program, GridapMakie adopts [Makie.jl](https://github.com/JuliaPlots/Makie.jl) as a second visualization back-end for \n# Gridap.jl simulations. This package is thought as a built-in tool to assess the user in their FE calculations with a smoother workflow \n# in a highly intuitive API.\n#\n# ## Installation\n\n# According to Makie's guidelines, it is enough to install one of its backends, e.g. GLMakie. Additionally, Gridap provides the plot objects\n# to be visualized and `FileIO` allows to save the figures plotted. \n\n# ```julia\n# julia> ] \n# pkg> add Gridap, GridapMakie, GLMakie, FileIO\n# ```\n#\n# ## Examples\n\n# First things first, we shall be using the three packages as well as `FileIO`.\n# We may as well create directories to store downloaded meshes and output files\n\nusing Gridap, GridapMakie, GLMakie\nusing FileIO\nmkdir(\"models\")\nmkdir(\"images\")\n\n# ### 2D Plots\n\n# Then, let us consider a simple, 2D simplexified cartesian triangulation \u03a9\n\ndomain = (0, 1, 0, 1)\ncell_nums = (10, 10)\nmodel = CartesianDiscreteModel(domain, cell_nums) |> simplexify\n\u03a9 = Triangulation(model)\n\n\n# The visualization of the vertices, edges, and faces of \u03a9 can be achieved as follows\n\nfig = plot(\u03a9)\nwireframe!(\u03a9, color=:black, linewidth=2)\nscatter!(\u03a9, marker=:star8, markersize=20, color=:blue)\nsave(\"images/2d_Fig1.png\", fig)\n# \n#
\n\n# We now consider a FE function `uh` constructed with Gridap\n\nreffe = ReferenceFE(lagrangian, Float64, 1)\nV = FESpace(model, reffe)\nuh = interpolate(x->sin(\u03c0*(x[1]+x[2])), V)\n\n# and plot it over \u03a9, adding a colorbar\n\nfig, _ , plt = plot(\u03a9, uh)\nColorbar(fig[1,2], plt)\nsave(\"images/2d_Fig11.png\", fig)\n# \n#
\n\n# On the other hand, we may as well plot cell values\n\ncelldata = \u03c0*rand(num_cells(\u03a9)) .-1\nfig, _ , plt = plot(\u03a9, color=celldata, colormap=:heat)\nColorbar(fig[2,1], plt, vertical=false)\nsave(\"images/2d_Fig13.png\", fig)\n# \n#
\n\n# If we are only interested in the boundary of \u03a9, namely \u0393\n\n\u0393 = BoundaryTriangulation(model)\nfig, _ , plt = plot(\u0393, uh, colormap=:algae, linewidth=10)\nColorbar(fig[1,2], plt)\nsave(\"images/2d_Fig111.png\", fig)\n# \n#
\n\n# ### 3D Plots\n\n# In addition to the 2D plots, GridapMakie is able to handle more complex geometries. For example, \n# take the mesh from the [first Gridap tutorial](https://gridap.github.io/Tutorials/stable/pages/t001_poisson/#Tutorial-1:-Poisson-equation-1),\n# which can be downloaded using\n\nurl = \"https://github.com/gridap/GridapMakie.jl/raw/d5d74190e68bd310483fead8a4154235a61815c5/_readme/model.json\"\ndownload(url,\"models/model.json\")\n\n# Therefore, we may as well visualize such mesh\n\nmodel = DiscreteModelFromFile(\"models/model.json\")\n\u03a9 = Triangulation(model)\n\u2202\u03a9 = BoundaryTriangulation(model)\nfig = plot(\u03a9, shading=true)\nwireframe!(\u2202\u03a9, color=:black)\nsave(\"images/3d_Fig1.png\", fig)\n# \n#
\n\nv(x) = sin(\u03c0*(x[1]+x[2]+x[3]))\nfig, ax, plt = plot(\u03a9, v, shading=true)\nColorbar(fig[1,2], plt)\nsave(\"images/3d_Fig2.png\", fig)\n# \n#
\n\n# we can even plot functions in certain subdomains, e.g.\n\n\u0393 = BoundaryTriangulation(model, tags=[\"square\", \"triangle\", \"circle\"])\nfig = plot(\u0393, v, colormap=:rainbow, shading=true)\nwireframe!(\u2202\u03a9, linewidth=0.5, color=:gray)\nsave(\"images/3d_Fig3.png\", fig)\n# \n#
\n\n# ### Animations and interactivity\n\n# Finally, by using Makie [Observables](https://makie.juliaplots.org/stable/interaction/nodes.html), we\n# can create animations or interactive plots. For example, if the nodal field has a time dependence\n\nt = Observable(0.0)\nu = lift(t) do t\n x->sin(\u03c0*(x[1]+x[2]+x[3]))*cos(\u03c0*t)\nend\nfig = plot(\u03a9, u, colormap=:rainbow, shading=true, colorrange=(-1,1))\nwireframe!(\u2202\u03a9, color=:black, linewidth=0.5)\nframerate = 30\ntimestamps = range(0, 2, step=1/framerate)\nrecord(fig, \"images/animation.gif\", timestamps; framerate=framerate) do this_t\n t[] = this_t\nend\n# \n#
", "meta": {"hexsha": "728a2c53f58777eff1075d7ddf33a2f29539ffe5", "size": 5561, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "_readme/README.jl", "max_stars_repo_name": "gridap/GridapMakie.jl", "max_stars_repo_head_hexsha": "524f3a4f8b63a4a9ac9b965865fc4bb3170ae98c", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 26, "max_stars_repo_stars_event_min_datetime": "2020-08-11T12:13:28.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-25T01:22:36.000Z", "max_issues_repo_path": "_readme/README.jl", "max_issues_repo_name": "gridap/GridapMakie.jl", "max_issues_repo_head_hexsha": "524f3a4f8b63a4a9ac9b965865fc4bb3170ae98c", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 28, "max_issues_repo_issues_event_min_datetime": "2020-08-12T11:02:12.000Z", "max_issues_repo_issues_event_max_datetime": "2022-03-01T11:42:36.000Z", "max_forks_repo_path": "_readme/README.jl", "max_forks_repo_name": "gridap/GridapMakie.jl", "max_forks_repo_head_hexsha": "524f3a4f8b63a4a9ac9b965865fc4bb3170ae98c", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 36.8278145695, "max_line_length": 152, "alphanum_fraction": 0.7187556195, "num_tokens": 1767, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.46879062662624377, "lm_q2_score": 0.12421300186801369, "lm_q1q2_score": 0.05822989098083293}}
{"text": "f(x, y...) = y\n@assert f(1, 2, 3) == (2, 3)\n\ng(x; y...) = y\n@assert g(1, a=2, b=5) == [(:a,2),(:b,5)]", "meta": {"hexsha": "104f8cb1ac1189ed5bda8a000e89afab8824441b", "size": 101, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "julia/parameter_packing_and_unpacking.jl", "max_stars_repo_name": "BBK-DCSIS-Programming-Paradigms-2018-19/code", "max_stars_repo_head_hexsha": "fbf4e45899b7555fc3b12fcb1fefa172ef415060", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 2, "max_stars_repo_stars_event_min_datetime": "2018-10-17T18:35:39.000Z", "max_stars_repo_stars_event_max_datetime": "2018-10-27T13:18:08.000Z", "max_issues_repo_path": "julia/parameter_packing_and_unpacking.jl", "max_issues_repo_name": "BBK-DCSIS-Programming-Paradigms-2018-19/code", "max_issues_repo_head_hexsha": "fbf4e45899b7555fc3b12fcb1fefa172ef415060", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "julia/parameter_packing_and_unpacking.jl", "max_forks_repo_name": "BBK-DCSIS-Programming-Paradigms-2018-19/code", "max_forks_repo_head_hexsha": "fbf4e45899b7555fc3b12fcb1fefa172ef415060", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2018-11-28T21:50:43.000Z", "max_forks_repo_forks_event_max_datetime": "2018-11-28T21:50:43.000Z", "avg_line_length": 20.2, "max_line_length": 41, "alphanum_fraction": 0.3564356436, "num_tokens": 61, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.46879062662624377, "lm_q2_score": 0.12421299700498412, "lm_q1q2_score": 0.05822988870109024}}
{"text": "abstract type BinaryNode{T} <: AbstractNode{T} end\n\n### Interface\n\n# Return the node's value.\ngetvalue(n::BinaryNode) = nothing\n\n# Set the node's value.\nsetvalue!(n::BinaryNode) = nothing\n\n# Return node's left child.\nleft(n::BinaryNode) = nothing\n\n# Set the node' left child.\nsetleft!(n::BinaryNode, node) = nothing\n\n# Return node's left child.\nright(n::BinaryNode) = nothing\n\n# Set the node' left child.\nsetright!(n::BinaryNode, node) = nothing\n\n# Check if node is a leaf.\nisleaf(n::BinaryNode) = nothing\n\n# generic traversal and others\n# function traversal() end\n\n# preorder, inorder, postorder, levelorder\n", "meta": {"hexsha": "f271bd4c3da0a0f4136edb847e2e9df54ffb6d68", "size": 609, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/tree/abstract_binarynode.jl", "max_stars_repo_name": "hesseltuinhof/DataStructures.jl", "max_stars_repo_head_hexsha": "50c630bfb9b0eb43329bfd92148e1ca8b3fec9b1", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/tree/abstract_binarynode.jl", "max_issues_repo_name": "hesseltuinhof/DataStructures.jl", "max_issues_repo_head_hexsha": "50c630bfb9b0eb43329bfd92148e1ca8b3fec9b1", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/tree/abstract_binarynode.jl", "max_forks_repo_name": "hesseltuinhof/DataStructures.jl", "max_forks_repo_head_hexsha": "50c630bfb9b0eb43329bfd92148e1ca8b3fec9b1", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 20.3, "max_line_length": 50, "alphanum_fraction": 0.7192118227, "num_tokens": 166, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.414898860266261, "lm_q2_score": 0.1403362494900832, "lm_q1q2_score": 0.05822534996747717}}
{"text": "#### Elementwise manipulations (scaling/clamping/type conversion) ####\n\n# This file exists primarily to handle conversions for display and\n# saving to disk. Both of these operations require UFixed-valued\n# elements, but with display we always want to convert to 8-bit\n# whereas saving can handle 16-bit.\n# We also can't trust that user images are clamped properly.\n# Finally, this supports adjustable contrast limits.\n\n# Structure of MapInfo subtype definitions:\n# - type definition\n# - constructors for scalars\n# - constructors for AbstractArrays\n# - similar (syntax: similar(mapi, ToType, FromType))\n# - implementation of map() for scalars\n# - implementation of map() for AbstractArrays\n# map(mapi::MapInfo{T}, x) should return an object of type T (for x not an array)\n# map1(mapi::MapInfo{T}, x) is designed to allow T<:Color to work on\n# scalars x::Fractional\n\n\n# Dispatch-based elementwise manipulations\n\"\"\"\n`MapInfo{T}` is an abstract type that encompasses objects designed to\nperform intensity or color transformations on pixels. For example,\nbefore displaying an image in a window, you might need to adjust the\ncontrast settings; `MapInfo` objects provide a means to describe these\ntransformations without calculating them immediately. This delayed\nexecution can be useful in many contexts. For example, if you want to\ndisplay a movie, it would be quite wasteful to have to first transform\nthe entire movie; instead, `MapInfo` objects allow one to specify a\ntransformation to be performed on-the-fly as particular frames are\ndisplayed.\n\nYou can create your own custom `MapInfo` objects. For example, given a\ngrayscale image, you could color \"saturated\" pixels red using\n\n```jl\nimmutable ColorSaturated{C<:AbstractRGB} <: MapInfo{C}\nend\n\nBase.map{C}(::ColorSaturated{C}, val::Union{Number,Gray}) = ifelse(val == 1, C(1,0,0), C(val,val,val))\n\nimgc = map(ColorSaturated{RGB{U8}}(), img)\n```\n\nFor pre-defined types see `MapNone`, `BitShift`, `ClampMinMax`, `ScaleMinMax`,\n`ScaleAutoMinMax`, and `ScaleSigned`.\n\"\"\"\nabstract MapInfo{T}\neltype{T}(mapi::MapInfo{T}) = T\n\n\n## MapNone\n\"`MapNone(T)` is a `MapInfo` object that converts `x` to have type `T`.\"\nimmutable MapNone{T} <: MapInfo{T}; end\n\n# Constructors\nMapNone{T}(::Type{T}) = MapNone{T}()\nMapNone{T}(val::T) = MapNone{T}()\nMapNone{T}(A::AbstractArray{T}) = MapNone{T}()\n\nsimilar{T}(mapi::MapNone, ::Type{T}, ::Type) = MapNone{T}()\n\n# Implementation\nmap{T}(mapi::MapNone{T}, val::Union{Number,Colorant}) = convert(T, val)\nmap1(mapi::Union{MapNone{RGB24}, MapNone{ARGB32}}, b::Bool) = ifelse(b, 0xffuf8, 0x00uf8)\nmap1(mapi::Union{MapNone{RGB24},MapNone{ARGB32}}, val::Fractional) = convert(UFixed8, val)\nmap1{CT<:Colorant}(mapi::MapNone{CT}, val::Fractional) = convert(eltype(CT), val)\n\nmap{T<:Colorant}(mapi::MapNone{T}, img::AbstractImageIndexed{T}) = convert(Image{T}, img)\nmap{C<:Colorant}(mapi::MapNone{C}, img::AbstractImageDirect{C}) = img # ambiguity resolution\nmap{T}(mapi::MapNone{T}, img::AbstractArray{T}) = img\n\n\n## BitShift\n\"\"\"\n`BitShift{T,N}` performs a \"saturating rightward bit-shift\" operation.\nIt is particularly useful in converting high bit-depth images to 8-bit\nimages for the purpose of display. For example,\n\n```\nmap(BitShift(UFixed8, 8), 0xa2d5uf16) === 0xa2uf8\n```\n\nconverts a `UFixed16` to the corresponding `UFixed8` by discarding the\nleast significant byte. However,\n\n```\nmap(BitShift(UFixed8, 7), 0xa2d5uf16) == 0xffuf8\n```\n\nbecause `0xa2d5>>7 == 0x0145 > typemax(UInt8)`.\n\nWhen applicable, the main advantage of using `BitShift` rather than\n`MapNone` or `ScaleMinMax` is speed.\n\"\"\"\nimmutable BitShift{T,N} <: MapInfo{T} end\nBitShift{T}(::Type{T}, n::Int) = BitShift{T,n}() # note that this is not type-stable\n\nsimilar{S,T,N}(mapi::BitShift{S,N}, ::Type{T}, ::Type) = BitShift{T,N}()\n\n# Implementation\nimmutable BS{N} end\n_map{T<:Unsigned,N}(::Type{T}, ::Type{BS{N}}, val::Unsigned) = (v = val>>>N; tm = oftype(val, typemax(T)); convert(T, ifelse(v > tm, tm, v)))\n_map{T<:UFixed,N}(::Type{T}, ::Type{BS{N}}, val::UFixed) = reinterpret(T, _map(FixedPointNumbers.rawtype(T), BS{N}, reinterpret(val)))\nmap{T<:Real,N}(mapi::BitShift{T,N}, val::Real) = _map(T, BS{N}, val)\nmap{T<:Real,N}(mapi::BitShift{Gray{T},N}, val::Gray) = Gray(_map(T, BS{N}, val.val))\nmap1{N}(mapi::Union{BitShift{RGB24,N},BitShift{ARGB32,N}}, val::Unsigned) = _map(UInt8, BS{N}, val)\nmap1{N}(mapi::Union{BitShift{RGB24,N},BitShift{ARGB32,N}}, val::UFixed) = _map(UFixed8, BS{N}, val)\nmap1{CT<:Colorant,N}(mapi::BitShift{CT,N}, val::UFixed) = _map(eltype(CT), BS{N}, val)\n\n\n## Clamp types\n# The Clamp types just enforce bounds, but do not scale or offset\n\n# Types and constructors\nabstract AbstractClamp{T} <: MapInfo{T}\n\"\"\"\n`ClampMin(T, minvalue)` is a `MapInfo` object that clamps pixel values\nto be greater than or equal to `minvalue` before converting to type `T`.\n\nSee also: `ClampMax`, `ClampMinMax`.\n\"\"\"\nimmutable ClampMin{T,From} <: AbstractClamp{T}\n min::From\nend\nClampMin{T,From}(::Type{T}, min::From) = ClampMin{T,From}(min)\nClampMin{T}(min::T) = ClampMin{T,T}(min)\n\"\"\"\n`ClampMax(T, maxvalue)` is a `MapInfo` object that clamps pixel values\nto be less than or equal to `maxvalue` before converting to type `T`.\n\nSee also: `ClampMin`, `ClampMinMax`.\n\"\"\"\nimmutable ClampMax{T,From} <: AbstractClamp{T}\n max::From\nend\nClampMax{T,From}(::Type{T}, max::From) = ClampMax{T,From}(max)\nClampMax{T}(max::T) = ClampMax{T,T}(max)\nimmutable ClampMinMax{T,From} <: AbstractClamp{T}\n min::From\n max::From\nend\n\"\"\"\n`ClampMinMax(T, minvalue, maxvalue)` is a `MapInfo` object that clamps\npixel values to be between `minvalue` and `maxvalue` before converting\nto type `T`.\n\nSee also: `ClampMin`, `ClampMax`, and `Clamp`.\n\"\"\"\nClampMinMax{T,From}(::Type{T}, min::From, max::From) = ClampMinMax{T,From}(min,max)\nClampMinMax{T}(min::T, max::T) = ClampMinMax{T,T}(min,max)\n\"\"\"\n`Clamp(C)` is a `MapInfo` object that clamps color values to be within\ngamut. For example,\n\n```\nmap(Clamp(RGB{U8}), RGB(1.2, -0.4, 0.6)) === RGB{U8}(1, 0, 0.6)\n```\n\"\"\"\nimmutable Clamp{T} <: AbstractClamp{T} end\nClamp{T}(::Type{T}) = Clamp{T}()\n\nsimilar{T,F}(mapi::ClampMin, ::Type{T}, ::Type{F}) = ClampMin{T,F}(convert(F, mapi.min))\nsimilar{T,F}(mapi::ClampMax, ::Type{T}, ::Type{F}) = ClampMax{T,F}(convert(F, mapi.max))\nsimilar{T,F}(mapi::ClampMinMax, ::Type{T}, ::Type{F}) = ClampMin{T,F}(convert(F, mapi.min), convert(F, mapi.max))\nsimilar{T,F}(mapi::Clamp, ::Type{T}, ::Type{F}) = Clamp{T}()\n\n# Implementation\nmap{T<:Real,F<:Real}(mapi::ClampMin{T,F}, val::F) = convert(T, max(val, mapi.min))\nmap{T<:Real,F<:Real}(mapi::ClampMax{T,F}, val::F) = convert(T, min(val, mapi.max))\nmap{T<:Real,F<:Real}(mapi::ClampMinMax{T,F}, val::F) = convert(T,min(max(val, mapi.min), mapi.max))\nmap{T<:Fractional,F<:Real}(mapi::ClampMin{Gray{T},F}, val::F) = convert(Gray{T}, max(val, mapi.min))\nmap{T<:Fractional,F<:Real}(mapi::ClampMax{Gray{T},F}, val::F) = convert(Gray{T}, min(val, mapi.max))\nmap{T<:Fractional,F<:Real}(mapi::ClampMinMax{Gray{T},F}, val::F) = convert(Gray{T},min(max(val, mapi.min), mapi.max))\nmap{T<:Fractional,F<:Fractional}(mapi::ClampMin{Gray{T},F}, val::Gray{F}) = convert(Gray{T}, max(val, mapi.min))\nmap{T<:Fractional,F<:Fractional}(mapi::ClampMax{Gray{T},F}, val::Gray{F}) = convert(Gray{T}, min(val, mapi.max))\nmap{T<:Fractional,F<:Fractional}(mapi::ClampMinMax{Gray{T},F}, val::Gray{F}) = convert(Gray{T},min(max(val, mapi.min), mapi.max))\nmap{T<:Fractional,F<:Fractional}(mapi::ClampMin{Gray{T},Gray{F}}, val::Gray{F}) = convert(Gray{T}, max(val, mapi.min))\nmap{T<:Fractional,F<:Fractional}(mapi::ClampMax{Gray{T},Gray{F}}, val::Gray{F}) = convert(Gray{T}, min(val, mapi.max))\nmap{T<:Fractional,F<:Fractional}(mapi::ClampMinMax{Gray{T},Gray{F}}, val::Gray{F}) = convert(Gray{T},min(max(val, mapi.min), mapi.max))\nmap1{T<:Union{RGB24,ARGB32},F<:Fractional}(mapi::ClampMin{T,F}, val::F) = convert(UFixed8, max(val, mapi.min))\nmap1{T<:Union{RGB24,ARGB32},F<:Fractional}(mapi::ClampMax{T,F}, val::F) = convert(UFixed8, min(val, mapi.max))\nmap1{T<:Union{RGB24,ARGB32},F<:Fractional}(mapi::ClampMinMax{T,F}, val::F) = convert(UFixed8,min(max(val, mapi.min), mapi.max))\nmap1{CT<:Colorant,F<:Fractional}(mapi::ClampMin{CT,F}, val::F) = convert(eltype(CT), max(val, mapi.min))\nmap1{CT<:Colorant,F<:Fractional}(mapi::ClampMax{CT,F}, val::F) = convert(eltype(CT), min(val, mapi.max))\nmap1{CT<:Colorant,F<:Fractional}(mapi::ClampMinMax{CT,F}, val::F) = convert(eltype(CT), min(max(val, mapi.min), mapi.max))\n\nmap{To<:Real}(::Clamp{To}, val::Real) = clamp01(To, val)\nmap{To<:Real}(::Clamp{Gray{To}}, val::AbstractGray) = Gray(clamp01(To, val.val))\nmap{To<:Real}(::Clamp{Gray{To}}, val::Real) = Gray(clamp01(To, val))\nmap1{CT<:AbstractRGB}(::Clamp{CT}, val::Real) = clamp01(eltype(CT), val)\nmap1{P<:TransparentRGB}(::Clamp{P}, val::Real) = clamp01(eltype(P), val)\n\n# Also available as a stand-alone function\nclamp01{T}(::Type{T}, x::Real) = convert(T, min(max(x, zero(x)), one(x)))\nclamp01(x::Real) = clamp01(typeof(x), x)\nclamp01(x::Colorant) = clamp01(typeof(x), x)\nclamp01{Cdest<:AbstractRGB }(::Type{Cdest}, x::AbstractRGB) = (To = eltype(Cdest);\n Cdest(clamp01(To, red(x)), clamp01(To, green(x)), clamp01(To, blue(x))))\nclamp01{Pdest<:TransparentRGB}(::Type{Pdest}, x::TransparentRGB) = (To = eltype(Pdest);\n Pdest(clamp01(To, red(x)), clamp01(To, green(x)), clamp01(To, blue(x)), clamp01(To, alpha(x))))\n\n# clamp is generic for any colorspace; this version does the right thing for any RGB type\nclamp(x::Union{AbstractRGB, TransparentRGB}) = clamp01(x)\n\n## ScaleMinMax\n\"\"\"\n`ScaleMinMax(T, min, max, [scalefactor])` is a `MapInfo` object that\nclamps the image at the specified `min`/`max` values, subtracts the\n`min` value, scales the result by multiplying by `scalefactor`, and\nfinally converts to type `T`. If `scalefactor` is not specified, it\ndefaults to scaling the interval `[min,max]` to `[0,1]`.\n\nAlternative constructors include `ScaleMinMax(T, img)` for which\n`min`, `max`, and `scalefactor` are computed from the minimum and\nmaximum values found in `img`.\n\nSee also: `ScaleMinMaxNaN`, `ScaleAutoMinMax`, `MapNone`, `BitShift`.\n\"\"\"\nimmutable ScaleMinMax{To,From,S<:AbstractFloat} <: MapInfo{To}\n min::From\n max::From\n s::S\n\n function ScaleMinMax(min, max, s)\n min >= max && error(\"min must be smaller than max\")\n new(min, max, s)\n end\nend\n\nScaleMinMax{To,From}(::Type{To}, min::From, max::From, s::AbstractFloat) = ScaleMinMax{To,From,typeof(s)}(min, max, s)\nScaleMinMax{To<:Union{Fractional,Colorant},From}(::Type{To}, mn::From, mx::From) = ScaleMinMax(To, mn, mx, 1.0f0/(convert(Float32, mx)-convert(Float32, mn)))\n\n# ScaleMinMax constructors that take AbstractArray input\nScaleMinMax{To,From<:Real}(::Type{To}, img::AbstractArray{From}, mn::Real, mx::Real) = ScaleMinMax(To, convert(From,mn), convert(From,mx), 1.0f0/(convert(Float32, convert(From, mx))-convert(Float32,convert(From, mn))))\nScaleMinMax{To,From<:Real}(::Type{To}, img::AbstractArray{Gray{From}}, mn::Real, mx::Real) = ScaleMinMax(To, convert(From,mn), convert(From,mx), 1.0f0/(convert(Float32, convert(From,mx))-convert(Float32, convert(From,mn))))\nScaleMinMax{To,From<:Real,R<:Real}(::Type{To}, img::AbstractArray{From}, mn::Gray{R}, mx::Gray{R}) = ScaleMinMax(To, convert(From,mn.val), convert(From,mx.val), 1.0f0/(convert(Float32, convert(From,mx.val))-convert(Float32, convert(From,mn.val))))\nScaleMinMax{To,From<:Real,R<:Real}(::Type{To}, img::AbstractArray{Gray{From}}, mn::Gray{R}, mx::Gray{R}) = ScaleMinMax(To, convert(From,mn.val), convert(From,mx.val), 1.0f0/(convert(Float32, convert(From,mx.val))-convert(Float32, convert(From,mn.val))))\nScaleMinMax{To}(::Type{To}, img::AbstractArray) = ScaleMinMax(To, img, minfinite(img), maxfinite(img))\nScaleMinMax{To,CV<:AbstractRGB}(::Type{To}, img::AbstractArray{CV}) = (imgr = reinterpret(eltype(CV), img); ScaleMinMax(To, minfinite(imgr), maxfinite(imgr)))\n\nsimilar{T,F,To,From,S}(mapi::ScaleMinMax{To,From,S}, ::Type{T}, ::Type{F}) = ScaleMinMax{T,F,S}(convert(F,mapi.min), convert(F.mapi.max), mapi.s)\n\n# Implementation\nfunction map{To<:Union{Real,AbstractGray},From<:Union{Real,AbstractGray}}(mapi::ScaleMinMax{To,From}, val::From)\n g = gray(val)\n t = ifelse(g < mapi.min, zero(From), ifelse(g > mapi.max, mapi.max-mapi.min, g-mapi.min))\n convert(To, mapi.s*t)\nend\nfunction map{To<:Union{Real,AbstractGray},From<:Union{Real,AbstractGray}}(mapi::ScaleMinMax{To,From}, val::Union{Real,Colorant})\n map(mapi, convert(From, val))\nend\nfunction map1{To<:Union{RGB24,ARGB32},From<:Real}(mapi::ScaleMinMax{To,From}, val::From)\n t = ifelse(val < mapi.min, zero(From), ifelse(val > mapi.max, mapi.max-mapi.min, val-mapi.min))\n convert(UFixed8, mapi.s*t)\nend\nfunction map1{To<:Colorant,From<:Real}(mapi::ScaleMinMax{To,From}, val::From)\n t = ifelse(val < mapi.min, zero(From), ifelse(val > mapi.max, mapi.max-mapi.min, val-mapi.min))\n convert(eltype(To), mapi.s*t)\nend\nfunction map1{To<:Union{RGB24,ARGB32},From<:Real}(mapi::ScaleMinMax{To,From}, val::Union{Real,Colorant})\n map1(mapi, convert(From, val))\nend\nfunction map1{To<:Colorant,From<:Real}(mapi::ScaleMinMax{To,From}, val::Union{Real,Colorant})\n map1(mapi, convert(From, val))\nend\n\n## ScaleSigned\n\"\"\"\n`ScaleSigned(T, scalefactor)` is a `MapInfo` object designed for\nvisualization of images where the pixel's sign has special meaning.\nIt multiplies the pixel value by `scalefactor`, then clamps to the\ninterval `[-1,1]`. If `T` is a floating-point type, it stays in this\nrepresentation. If `T` is an `AbstractRGB`, then it is encoded as a\nmagenta (positive)/green (negative) image, with the intensity of the\ncolor proportional to the clamped absolute value.\n\"\"\"\nimmutable ScaleSigned{T, S<:AbstractFloat} <: MapInfo{T}\n s::S\nend\nScaleSigned{T}(::Type{T}, s::AbstractFloat) = ScaleSigned{T, typeof(s)}(s)\n\nScaleSigned{T}(::Type{T}, img::AbstractArray) = ScaleSigned(T, 1.0f0/maxabsfinite(img))\nScaleSigned(img::AbstractArray) = ScaleSigned(Float32, img)\n\nsimilar{T,To,S}(mapi::ScaleSigned{To,S}, ::Type{T}, ::Type) = ScaleSigned{T,S}(mapi.s)\n\nmap{T}(mapi::ScaleSigned{T}, val::Real) = convert(T, clamppm(mapi.s*val))\nfunction map{C<:AbstractRGB}(mapi::ScaleSigned{C}, val::Real)\n x = clamppm(mapi.s*val)\n g = UFixed8(abs(x))\n ifelse(x >= 0, C(g, zero(UFixed8), g), C(zero(UFixed8), g, zero(UFixed8)))\nend\n\nclamppm(x::Real) = ifelse(x >= 0, min(x, one(x)), max(x, -one(x)))\n\n## ScaleAutoMinMax\n# Works only on whole arrays, not values\n\"\"\"\n`ScaleAutoMinMax(T)` constructs a `MapInfo` object that causes images\nto be dynamically scaled to their specific min/max values, using the\nsame algorithm for `ScaleMinMax`. When displaying a movie, the min/max\nwill be recalculated for each frame, so this can result in\ninconsistent contrast scaling.\n\"\"\"\nimmutable ScaleAutoMinMax{T} <: MapInfo{T} end\nScaleAutoMinMax{T}(::Type{T}) = ScaleAutoMinMax{T}()\nScaleAutoMinMax() = ScaleAutoMinMax{UFixed8}()\n\nsimilar{T}(mapi::ScaleAutoMinMax, ::Type{T}, ::Type) = ScaleAutoMinMax{T}()\n\n## NaN-nulling mapping\n\"\"\"\n`ScaleMinMaxNaN(smm)` constructs a `MapInfo` object from a\n`ScaleMinMax` object `smm`, with the additional property that `NaN`\nvalues map to zero.\n\nSee also: `ScaleMinMax`.\n\"\"\"\nimmutable ScaleMinMaxNaN{To,From,S} <: MapInfo{To}\n smm::ScaleMinMax{To,From,S}\nend\n\n\"\"\"\n`Clamp01NaN(T)` or `Clamp01NaN(img)` constructs a `MapInfo` object\nthat clamps grayscale or color pixels to the interval `[0,1]`, sending\n`NaN` pixels to zero.\n\"\"\"\nimmutable Clamp01NaN{T} <: MapInfo{T} end\n\nClamp01NaN{T}(A::AbstractArray{T}) = Clamp01NaN{T}()\n\n# Implementation\nsimilar{T,F,To,From,S}(mapi::ScaleMinMaxNaN{To,From,S}, ::Type{T}, ::Type{F}) = ScaleMinMaxNaN{T,F,S}(similar(mapi.smm, T, F))\nsimilar{T}(mapi::Clamp01NaN, ::Type{T}, ::Type) = Clamp01NaN{T}()\n\nBase.map{To}(smmn::ScaleMinMaxNaN{To}, g::Number) = isnan(g) ? zero(To) : map(smmn.smm, g)\nBase.map{To}(smmn::ScaleMinMaxNaN{To}, g::Gray) = isnan(g) ? zero(To) : map(smmn.smm, g)\n\nfunction Base.map{T<:RGB}(::Clamp01NaN{T}, c::AbstractRGB)\n r, g, b = red(c), green(c), blue(c)\n if isnan(r) || isnan(g) || isnan(b)\n return T(0,0,0)\n end\n T(clamp(r, 0, 1), clamp(g, 0, 1), clamp(b, 0, 1))\nend\nfunction Base.map{T<:Union{Fractional,Gray}}(::Clamp01NaN{T}, c::Union{Fractional,AbstractGray})\n g = gray(c)\n if isnan(g)\n return T(0)\n end\n T(clamp(g, 0, 1))\nend\n\n# Conversions to RGB{T}, RGBA{T}, RGB24, ARGB32,\n# for grayscale, AbstractRGB, and abstract ARGB inputs.\n# This essentially \"vectorizes\" map over a single pixel's color channels using map1\nfor SI in (MapInfo, AbstractClamp)\n for ST in subtypes(SI)\n ST.abstract && continue\n ST == ScaleSigned && continue # ScaleSigned gives an RGB from a scalar, so don't \"vectorize\" it\n @eval begin\n # Grayscale and GrayAlpha inputs\n map(mapi::$ST{RGB24}, g::Gray) = map(mapi, g.val)\n map(mapi::$ST{RGB24}, g::Real) = (x = map1(mapi, g); convert(RGB24, RGB{UFixed8}(x,x,x)))\n function map(mapi::$ST{RGB24}, g::AbstractFloat)\n if isfinite(g)\n x = map1(mapi, g)\n convert(RGB24, RGB{UFixed8}(x,x,x))\n else\n RGB24(0)\n end\n end\n map{G<:Gray}(mapi::$ST{RGB24}, g::TransparentColor{G}) = map(mapi, gray(g))\n map(mapi::$ST{ARGB32}, g::Gray) = map(mapi, g.val)\n function map(mapi::$ST{ARGB32}, g::Real)\n x = map1(mapi, g)\n convert(ARGB32, ARGB{UFixed8}(x,x,x,0xffuf8))\n end\n function map{G<:Gray}(mapi::$ST{ARGB32}, g::TransparentColor{G})\n x = map1(mapi, gray(g))\n convert(ARGB32, ARGB{UFixed8}(x,x,x,map1(mapi, g.alpha)))\n end\n end\n for O in (:RGB, :BGR)\n @eval begin\n map{T}(mapi::$ST{$O{T}}, g::Gray) = map(mapi, g.val)\n function map{T}(mapi::$ST{$O{T}}, g::Real)\n x = map1(mapi, g)\n $O{T}(x,x,x)\n end\n end\n end\n for OA in (:RGBA, :ARGB, :BGRA)\n exAlphaGray = ST == MapNone ? :nothing : quote\n function map{T,G<:Gray}(mapi::$ST{$OA{T}}, g::TransparentColor{G})\n x = map1(mapi, gray(g))\n $OA{T}(x,x,x,map1(mapi, g.alpha))\n end # avoids an ambiguity warning with MapNone definitions\n end\n @eval begin\n map{T}(mapi::$ST{$OA{T}}, g::Gray) = map(mapi, g.val)\n function map{T}(mapi::$ST{$OA{T}}, g::Real)\n x = map1(mapi, g)\n $OA{T}(x,x,x)\n end\n $exAlphaGray\n end\n end\n @eval begin\n # AbstractRGB and abstract ARGB inputs\n map(mapi::$ST{RGB24}, rgb::AbstractRGB) =\n convert(RGB24, RGB{UFixed8}(map1(mapi, red(rgb)), map1(mapi, green(rgb)), map1(mapi, blue(rgb))))\n map{C<:AbstractRGB, TC}(mapi::$ST{RGB24}, argb::TransparentColor{C,TC}) =\n convert(RGB24, RGB{UFixed8}(map1(mapi, red(argb)), map1(mapi, green(argb)),\n map1(mapi, blue(argb))))\n map{C<:AbstractRGB, TC}(mapi::$ST{ARGB32}, argb::TransparentColor{C,TC}) =\n convert(ARGB32, ARGB{UFixed8}(map1(mapi, red(argb)), map1(mapi, green(argb)),\n map1(mapi, blue(argb)), map1(mapi, alpha(argb))))\n map(mapi::$ST{ARGB32}, rgb::AbstractRGB) =\n convert(ARGB32, ARGB{UFixed8}(map1(mapi, red(rgb)), map1(mapi, green(rgb)), map1(mapi, blue(rgb))))\n end\n for O in (:RGB, :BGR)\n @eval begin\n map{T}(mapi::$ST{$O{T}}, rgb::AbstractRGB) =\n $O{T}(map1(mapi, red(rgb)), map1(mapi, green(rgb)), map1(mapi, blue(rgb)))\n map{T,C<:AbstractRGB, TC}(mapi::$ST{$O{T}}, argb::TransparentColor{C,TC}) =\n $O{T}(map1(mapi, red(argb)), map1(mapi, green(argb)), map1(mapi, blue(argb)))\n end\n end\n for OA in (:RGBA, :ARGB, :BGRA)\n @eval begin\n map{T, C<:AbstractRGB, TC}(mapi::$ST{$OA{T}}, argb::TransparentColor{C,TC}) =\n $OA{T}(map1(mapi, red(argb)), map1(mapi, green(argb)),\n map1(mapi, blue(argb)), map1(mapi, alpha(argb)))\n map{T}(mapi::$ST{$OA{T}}, argb::ARGB32) = map(mapi, convert(RGBA{UFixed8}, argb))\n map{T}(mapi::$ST{$OA{T}}, rgb::AbstractRGB) =\n $OA{T}(map1(mapi, red(rgb)), map1(mapi, green(rgb)), map1(mapi, blue(rgb)))\n map{T}(mapi::$ST{$OA{T}}, rgb::RGB24) = map(mapi, convert(RGB{UFixed8}, argb))\n end\n end\n end\nend\n\n# # Apply to any Colorant\n# map(f::Callable, x::Color) = f(x)\n# map(mapi, x::Color) = map(mapi, convert(RGB, x))\n# map{C<:Color, TC}(f::Callable, x::TransparentColor{C, TC}) = f(convert(ARGB, x))\n# map{C<:Color, TC}(mapi, x::TransparentColor{C, TC}) = map(mapi, convert(ARGB, x))\n\n## Fallback definitions of map() for array types\n\nfunction map{T}(mapi::MapInfo{T}, img::AbstractArray)\n out = similar(img, T)\n map!(mapi, out, img)\nend\n\nmap{C<:Colorant,R<:Real}(mapi::MapNone{C}, img::AbstractImageDirect{R}) = mapcd(mapi, img) # ambiguity resolution\nmap{C<:Colorant,R<:Real}(mapi::MapInfo{C}, img::AbstractImageDirect{R}) = mapcd(mapi, img)\nfunction mapcd{C<:Colorant,R<:Real}(mapi::MapInfo{C}, img::AbstractImageDirect{R})\n # For this case we have to check whether color is defined along an array axis\n cd = colordim(img)\n if cd > 0\n dims = setdiff(1:ndims(img), cd)\n out = similar(img, C, size(img)[dims])\n map!(mapi, out, img, TypeConst{cd})\n else\n out = similar(img, C)\n map!(mapi, out, img)\n end\n out # note this isn't type-stable\nend\n\nfunction map{T<:Colorant}(mapi::MapInfo{T}, img::AbstractImageIndexed)\n out = Image(Array(T, size(img)), properties(img))\n map!(mapi, out, img)\nend\n\nmap!{T,T1,T2,N}(mapi::MapInfo{T1}, out::AbstractArray{T,N}, img::AbstractArray{T2,N}) =\n _map_a!(mapi, out, img)\nfunction _map_a!{T,T1,T2,N}(mapi::MapInfo{T1}, out::AbstractArray{T,N}, img::AbstractArray{T2,N})\n mi = take(mapi, img)\n dimg = data(img)\n dout = data(out)\n size(dout) == size(dimg) || throw(DimensionMismatch())\n for I in eachindex(dout, dimg)\n @inbounds dout[I] = map(mi, dimg[I])\n end\n out\nend\n\ntake(mapi::MapInfo, img::AbstractArray) = mapi\ntake{T}(mapi::ScaleAutoMinMax{T}, img::AbstractArray) = ScaleMinMax(T, img)\n\n# Indexed images (colormaps)\nmap!{T,T1,N}(mapi::MapInfo{T}, out::AbstractArray{T,N}, img::AbstractImageIndexed{T1,N}) =\n _mapindx!(mapi, out, img)\nfunction _mapindx!{T,T1,N}(mapi::MapInfo{T}, out::AbstractArray{T,N}, img::AbstractImageIndexed{T1,N})\n dimg = data(img)\n dout = data(out)\n cmap = map(mapi, img.cmap)\n for I in eachindex(dout, dimg)\n @inbounds dout[I] = cmap[dimg[I]]\n end\n out\nend\n\n# For when color is encoded along dimension CD\n# NC is the number of color channels\n# This is a very flexible implementation: color can be stored along any dimension, and it handles conversions to\n# many different colorspace representations.\nfor (CT, NC) in ((Union{AbstractRGB,RGB24}, 3), (Union{RGBA,ARGB,ARGB32}, 4), (Union{AGray,GrayA,AGray32}, 2))\n for N = 1:4\n N1 = N+1\n @eval begin\nfunction map!{T<:$CT,T1,T2,CD}(mapi::MapInfo{T}, out::AbstractArray{T1,$N}, img::AbstractArray{T2,$N1}, ::Type{TypeConst{CD}})\n mi = take(mapi, img)\n dimg = data(img)\n dout = data(out)\n # Set up the index along the color axis\n # We really only need dimension CD, but this will suffice\n @nexprs $NC k->(@nexprs $N1 d->(j_k_d = k))\n # Loop over all the elements in the output, performing the conversion on each color component\n @nloops $N i dout d->(d(j_k_d = i_d)) : (@nexprs $NC k->(j_k_{d+1} = i_d))) begin\n @inbounds @nref($N, dout, i) = @ncall $NC T k->(map1(mi, @nref($N1, dimg, j_k)))\n end\n out\nend\n end\n end\nend\n\n\n#### MapInfo defaults\n# Each \"client\" can define its own methods. \"clients\" include UFixed,\n# RGB24/ARGB32, and ImageMagick\n\nconst bitshiftto8 = ((UFixed10, 2), (UFixed12, 4), (UFixed14, 6), (UFixed16, 8))\n\n# typealias GrayType{T<:Fractional} Union{T, Gray{T}}\ntypealias GrayArray{T<:Fractional} Union{AbstractArray{T}, AbstractArray{Gray{T}}}\n# note, though, that we need to override for AbstractImage in case the\n# \"colorspace\" property is defined differently\n\n# mapinfo{T<:Union{Real,Colorant}}(::Type{T}, img::AbstractArray{T}) = MapNone(img)\n\"\"\"\n`mapi = mapinf(T, img)` returns a `MapInfo` object that is deemed\nappropriate for converting pixels of `img` to be of type `T`. `T` can\neither be a specific type (e.g., `RGB24`), or you can specify an\nabstract type like `Clamp` and it will return one of the `Clamp`\nfamily of `MapInfo` objects.\n\nYou can define your own rules for `mapinfo`. For example, the\n`ImageMagick` package defines methods for how pixels values should be\nconverted before saving images to disk.\n\"\"\"\nmapinfo{T<:UFixed}(::Type{T}, img::AbstractArray{T}) = MapNone(img)\nmapinfo{T<:AbstractFloat}(::Type{T}, img::AbstractArray{T}) = MapNone(img)\n\n# Grayscale methods\nmapinfo(::Type{UFixed8}, img::GrayArray{UFixed8}) = MapNone{UFixed8}()\nmapinfo(::Type{Gray{UFixed8}}, img::GrayArray{UFixed8}) = MapNone{Gray{UFixed8}}()\nmapinfo(::Type{GrayA{UFixed8}}, img::AbstractArray{GrayA{UFixed8}}) = MapNone{GrayA{UFixed8}}()\nfor (T,n) in bitshiftto8\n @eval mapinfo(::Type{UFixed8}, img::GrayArray{$T}) = BitShift{UFixed8,$n}()\n @eval mapinfo(::Type{Gray{UFixed8}}, img::GrayArray{$T}) = BitShift{Gray{UFixed8},$n}()\n @eval mapinfo(::Type{GrayA{UFixed8}}, img::AbstractArray{GrayA{$T}}) = BitShift{GrayA{UFixed8},$n}()\nend\nmapinfo{T<:UFixed,F<:AbstractFloat}(::Type{T}, img::GrayArray{F}) = ClampMinMax(T, zero(F), one(F))\nmapinfo{T<:UFixed,F<:AbstractFloat}(::Type{Gray{T}}, img::GrayArray{F}) = ClampMinMax(Gray{T}, zero(F), one(F))\nmapinfo{T<:AbstractFloat, R<:Real}(::Type{T}, img::AbstractArray{R}) = MapNone(T)\n\nmapinfo(::Type{RGB24}, img::Union{AbstractArray{Bool}, BitArray}) = MapNone{RGB24}()\nmapinfo(::Type{ARGB32}, img::Union{AbstractArray{Bool}, BitArray}) = MapNone{ARGB32}()\nmapinfo{F<:Fractional}(::Type{RGB24}, img::GrayArray{F}) = ClampMinMax(RGB24, zero(F), one(F))\nmapinfo{F<:Fractional}(::Type{ARGB32}, img::AbstractArray{F}) = ClampMinMax(ARGB32, zero(F), one(F))\n\n# Color->Color methods\nmapinfo(::Type{RGB{UFixed8}}, img) = MapNone{RGB{UFixed8}}()\nmapinfo(::Type{RGBA{UFixed8}}, img) = MapNone{RGBA{UFixed8}}()\nfor (T,n) in bitshiftto8\n @eval mapinfo(::Type{RGB{UFixed8}}, img::AbstractArray{RGB{$T}}) = BitShift{RGB{UFixed8},$n}()\n @eval mapinfo(::Type{RGBA{UFixed8}}, img::AbstractArray{RGBA{$T}}) = BitShift{RGBA{UFixed8},$n}()\nend\nmapinfo{F<:Fractional}(::Type{RGB{UFixed8}}, img::AbstractArray{RGB{F}}) = Clamp(RGB{UFixed8})\nmapinfo{F<:Fractional}(::Type{RGBA{UFixed8}}, img::AbstractArray{RGBA{F}}) = Clamp(RGBA{UFixed8})\n\n\n\n# Color->RGB24/ARGB32\nmapinfo(::Type{RGB24}, img::AbstractArray{RGB24}) = MapNone{RGB24}()\nmapinfo(::Type{ARGB32}, img::AbstractArray{ARGB32}) = MapNone{ARGB32}()\nfor C in tuple(subtypes(AbstractRGB)..., Gray)\n C == RGB24 && continue\n @eval mapinfo(::Type{RGB24}, img::AbstractArray{$C{UFixed8}}) = MapNone{RGB24}()\n @eval mapinfo(::Type{ARGB32}, img::AbstractArray{$C{UFixed8}}) = MapNone{ARGB32}()\n for (T, n) in bitshiftto8\n @eval mapinfo(::Type{RGB24}, img::AbstractArray{$C{$T}}) = BitShift{RGB24, $n}()\n @eval mapinfo(::Type{ARGB32}, img::AbstractArray{$C{$T}}) = BitShift{ARGB32, $n}()\n end\n @eval mapinfo{F<:AbstractFloat}(::Type{RGB24}, img::AbstractArray{$C{F}}) = ClampMinMax(RGB24, zero(F), one(F))\n @eval mapinfo{F<:AbstractFloat}(::Type{ARGB32}, img::AbstractArray{$C{F}}) = ClampMinMax(ARGB32, zero(F), one(F))\n for AC in subtypes(TransparentColor)\n length(AC.parameters) == 2 || continue\n @eval mapinfo(::Type{ARGB32}, img::AbstractArray{$AC{$C{UFixed8},UFixed8}}) = MapNone{ARGB32}()\n @eval mapinfo(::Type{RGB24}, img::AbstractArray{$AC{$C{UFixed8},UFixed8}}) = MapNone{RGB24}()\n for (T, n) in bitshiftto8\n @eval mapinfo(::Type{ARGB32}, img::AbstractArray{$AC{$C{$T},$T}}) = BitShift{ARGB32, $n}()\n @eval mapinfo(::Type{RGB24}, img::AbstractArray{$AC{$C{$T},$T}}) = BitShift{RGB24, $n}()\n end\n @eval mapinfo{F<:AbstractFloat}(::Type{ARGB32}, img::AbstractArray{$AC{$C{F},F}}) = ClampMinMax(ARGB32, zero(F), one(F))\n @eval mapinfo{F<:AbstractFloat}(::Type{RGB24}, img::AbstractArray{$AC{$C{F},F}}) = ClampMinMax(RGB24, zero(F), one(F))\n end\nend\n\nmapinfo{CT<:Colorant}(::Type{RGB24}, img::AbstractArray{CT}) = MapNone{RGB24}()\nmapinfo{CT<:Colorant}(::Type{ARGB32}, img::AbstractArray{CT}) = MapNone{ARGB32}()\n\n\n# UInt32 conversions will use ARGB32 for images that have an alpha channel,\n# and RGB24 when not\nmapinfo{CV<:Union{Fractional,Color,AbstractGray}}(::Type{UInt32}, img::AbstractArray{CV}) = mapinfo(RGB24, img)\nmapinfo{CV<:TransparentColor}(::Type{UInt32}, img::AbstractArray{CV}) = mapinfo(ARGB32, img)\nmapinfo(::Type{UInt32}, img::Union{AbstractArray{Bool},BitArray}) = mapinfo(RGB24, img)\nmapinfo(::Type{UInt32}, img::AbstractArray{UInt32}) = MapNone{UInt32}()\n\n\n# Clamping mapinfo client. Converts to RGB and uses UFixed, clamping\n# floating-point values to [0,1].\nmapinfo{T<:UFixed}(::Type{Clamp}, img::AbstractArray{T}) = MapNone{T}()\nmapinfo{T<:AbstractFloat}(::Type{Clamp}, img::AbstractArray{T}) = ClampMinMax(UFixed8, zero(T), one(T))\nlet handled = Set()\nfor ACV in (Color, AbstractRGB)\n for CV in subtypes(ACV)\n (length(CV.parameters) == 1 && !(CV.abstract)) || continue\n CVnew = CV<:AbstractGray ? Gray : RGB\n @eval mapinfo{T<:UFixed}(::Type{Clamp}, img::AbstractArray{$CV{T}}) = MapNone{$CVnew{T}}()\n @eval mapinfo{CV<:$CV}(::Type{Clamp}, img::AbstractArray{CV}) = Clamp{$CVnew{UFixed8}}()\n CVnew = CV<:AbstractGray ? Gray : BGR\n AC, CA = alphacolor(CV), coloralpha(CV)\n if AC in handled\n continue\n end\n push!(handled, AC)\n ACnew, CAnew = alphacolor(CVnew), coloralpha(CVnew)\n @eval begin\n mapinfo{T<:UFixed}(::Type{Clamp}, img::AbstractArray{$AC{T}}) = MapNone{$ACnew{T}}()\n mapinfo{P<:$AC}(::Type{Clamp}, img::AbstractArray{P}) = Clamp{$ACnew{UFixed8}}()\n mapinfo{T<:UFixed}(::Type{Clamp}, img::AbstractArray{$CA{T}}) = MapNone{$CAnew{T}}()\n mapinfo{P<:$CA}(::Type{Clamp}, img::AbstractArray{P}) = Clamp{$CAnew{UFixed8}}()\n end\n end\nend\nend\nmapinfo(::Type{Clamp}, img::AbstractArray{RGB24}) = MapNone{RGB{UFixed8}}()\nmapinfo(::Type{Clamp}, img::AbstractArray{ARGB32}) = MapNone{BGRA{UFixed8}}()\n\n\n# Backwards-compatibility\nuint32color(img) = map(mapinfo(UInt32, img), img)\nuint32color!(buf, img::AbstractArray) = map!(mapinfo(UInt32, img), buf, img)\nuint32color!(buf, img::AbstractArray, mi::MapInfo) = map!(mi, buf, img)\nuint32color!{T,N}(buf::Array{UInt32,N}, img::AbstractImageDirect{T,N}) =\n map!(mapinfo(UInt32, img), buf, img)\nuint32color!{T,N,N1}(buf::Array{UInt32,N}, img::AbstractImageDirect{T,N1}) =\n map!(mapinfo(UInt32, img), buf, img, TypeConst{colordim(img)})\nuint32color!{T,N}(buf::Array{UInt32,N}, img::AbstractImageDirect{T,N}, mi::MapInfo) =\n map!(mi, buf, img)\nuint32color!{T,N,N1}(buf::Array{UInt32,N}, img::AbstractImageDirect{T,N1}, mi::MapInfo) =\n map!(mi, buf, img, TypeConst{colordim(img)})\n\n\"\"\"\n```\nimgsc = sc(img)\nimgsc = sc(img, min, max)\n```\n\nApplies default or specified `ScaleMinMax` mapping to the image.\n\"\"\"\nsc(img::AbstractArray) = map(ScaleMinMax(UFixed8, img), img)\nsc(img::AbstractArray, mn::Real, mx::Real) = map(ScaleMinMax(UFixed8, img, mn, mx), img)\n\nfor (fn,T) in ((:float32, Float32), (:float64, Float64), (:ufixed8, UFixed8),\n (:ufixed10, UFixed10), (:ufixed12, UFixed12), (:ufixed14, UFixed14),\n (:ufixed16, UFixed16))\n @eval begin\n function $fn{C<:Colorant}(A::AbstractArray{C})\n newC = eval(C.name.name){$T}\n convert(Array{newC}, A)\n end\n $fn{C<:Colorant}(img::AbstractImage{C}) = shareproperties(img, $fn(data(img)))\n end\nend\n\n\nufixedsc{T<:UFixed}(::Type{T}, img::AbstractImageDirect) = map(mapinfo(T, img), img)\nufixed8sc(img::AbstractImageDirect) = ufixedsc(UFixed8, img)\n", "meta": {"hexsha": "1b65412f52fb34d3e52492e151f6e6277d3b9542", "size": 32574, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/map.jl", "max_stars_repo_name": "rsrock/Images.jl", "max_stars_repo_head_hexsha": "8e4192a04c45be0f93f8b13b189249ed93f394c1", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/map.jl", "max_issues_repo_name": "rsrock/Images.jl", "max_issues_repo_head_hexsha": "8e4192a04c45be0f93f8b13b189249ed93f394c1", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/map.jl", "max_forks_repo_name": "rsrock/Images.jl", "max_forks_repo_head_hexsha": "8e4192a04c45be0f93f8b13b189249ed93f394c1", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 46.5342857143, "max_line_length": 253, "alphanum_fraction": 0.6518388899, "num_tokens": 10646, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.48828341018881344, "lm_q2_score": 0.11920292045759122, "lm_q1q2_score": 0.058204808505498515}}
{"text": "# constructing:\nstart_time = time()\n# long computation\ntime_elapsed = time() - start_time\nprintln(\"Time elapsed: $time_elapsed\") #> 0.0009999275207519531\n\nusing Dates\nd = Date(2018,9,1) #> 2018-09-01\ndt = DateTime(2018,9,1,12,30,59,1) #> 2018-09-01T12:30:59.001\n# accessors:\nyear(d)\nmonth(d)\nweek(d)\nday(d)\n# functions:\nisleapyear(d)\ndayofyear(d)\nmonthname(d)\ndaysinmonth(d)", "meta": {"hexsha": "bafcbbf291048c68729065dc29248880375d8211", "size": 375, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Chapter02/dates.jl", "max_stars_repo_name": "tavoludra1/Julia1.0", "max_stars_repo_head_hexsha": "92dc1e3955393394f10f538dceded45facd1cff6", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 26, "max_stars_repo_stars_event_min_datetime": "2018-09-29T03:07:28.000Z", "max_stars_repo_stars_event_max_datetime": "2021-11-08T13:12:33.000Z", "max_issues_repo_path": "Chapter02/dates.jl", "max_issues_repo_name": "tavoludra1/Julia1.0", "max_issues_repo_head_hexsha": "92dc1e3955393394f10f538dceded45facd1cff6", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Chapter02/dates.jl", "max_forks_repo_name": "tavoludra1/Julia1.0", "max_forks_repo_head_hexsha": "92dc1e3955393394f10f538dceded45facd1cff6", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 11, "max_forks_repo_forks_event_min_datetime": "2018-09-16T05:55:20.000Z", "max_forks_repo_forks_event_max_datetime": "2021-06-08T14:59:54.000Z", "avg_line_length": 19.7368421053, "max_line_length": 64, "alphanum_fraction": 0.7146666667, "num_tokens": 143, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4804786780479071, "lm_q2_score": 0.12085324357297283, "lm_q1q2_score": 0.05806740670974372}}
{"text": "module CytofImg\n\nimport Cytof5\nusing Plots\npyplot()\nusing StatPlots\nusing Distributions # std, mean, quantile\nusing RCall # TODO: GET RID OF THIS DEPENDENCY!\n\nstartupMsg = \"\"\"\nTo scale font size globally for plots (for presentations mostly), do this:\n```\njulia> Plots.scalefontsizes(2) # scales all texts by factor of 2\n\n# To reset\njulia> Plots.reset_defaults()\n```\n\"\"\"\nprintln(startupMsg)\n\nfunction getPosterior(sym::Symbol, monitor)\n return [ m[sym] for m in monitor]\nend\n\n\n\"\"\"\nturn on legend with `legend=true`\n\nNote that xlabel / ylabel can be supplied by either:\n\n```\njulia> xlabel!(\"my xlab\")\n# OR\njulia> plotZ(Z, xlabel=\"my xlab\")\n```\n\n\"\"\"\nfunction plotZ(Z::Matrix{T}; kw...) where {T <: Number}\n # FIXME: doesn't work for Z having either 1 row or 1 column!\n J, K = size(Z)\n if J < 2 || K < 2\n println(\"heatmap is not supported for single row / column matrices\")\n return\n end\n\n img = heatmap(Z, c=:Greys, legend=:none, border=true, bordercolor=:lightgrey; kw...)\n if J > 1\n hline!((2:J) .- .5, c=:lightgrey)\n end\n if K > 1\n vline!((2:K) .- .5, c=:lightgrey)\n end\n\n return img\nend\n\nfunction plotY_heatmap(y::Matrix{T}; clim=(-5, 5), c=:pu_or) where {T <: Number}\n img = heatmap(y, c=c, clim=clim, background_color_inside=:black, bordercolor=:transparent);\n return img\nend\n\nfunction plotYZ(y::Matrix{V}, Z::Matrix{T}; ycolor=:balance, clim=(-5, 5), heights=[.7, .3], kw...) where {T <: Number, V <: Number}\n # TODO: change proportions\n heatY = plotY_heatmap(y, c=ycolor, clim=clim)\n heatZ = plotZ(Matrix(Z'))\n img = plot(heatY, heatZ, layout=grid(2, 1, heights=heights); kw...);\n return img\nend\n\nfunction annotateHeatmap(mat::Matrix{T}; digits=2, textsize=6) where T\n rows, cols = size(mat)\n for c in 1:cols\n for r in 1:rows\n annotate!([(float(c), float(r),\n text(\"$(round(mat[r,c], digits=digits))\", textsize))])\n end\n end\nend\n\nfunction plotPost(x::Vector{T}; a::Float64=0.05, q_digits::Int=3, sd_digits::Int=3, add=false,\n trace::Bool=true, useDensity::Bool=true, showAccRate::Bool=true,\n parent=1, offset=1, traceFont=font(16), kw...) where {T <: Number}\n denColor = (0, .99, :steelblue)\n if useDensity\n if add\n img = StatPlots.density!(x, label=\"\", grid=false, yaxis=false,\n linecolor=:transparent,\n fill=denColor, bordercolor=:transparent; kw...)\n else\n img = StatPlots.density(x, label=\"\", grid=false, bordercolor=:transparent, yaxis=false,\n fill=denColor, linecolor=:transparent; kw...)\n end\n else\n if add\n img = histogram!(x, label=\"\", normed=true, grid=false,\n bordercolor=:white, yaxis=false,\n linecolor=:transparent, c=:steelblue; kw...)\n else\n img = histogram(x, label=\"\", normed=true, grid=false,\n bordercolor=:white, yaxis=false,\n linecolor=:transparent, c=:steelblue; kw...)\n end\n end\n\n\n if trace\n accRate = length(unique(x)) / length(x)\n\t\ttraceTitle = showAccRate ? \"acc: $(Int(round(accRate*100)))%\" : \"\"\n # x y W H\n plot!(x, inset_subplots=(parent, bbox(0, .1, .3, .3, :top, :right)), subplot=parent+offset,\n axis=false, legend=false, title=traceTitle,\n c=:grey70, grid=false, titlefont=traceFont, linewidths=.5)\n\n end\n\n xmean = mean(x)\n xsd = std(x)\n q = quantile(x, [a/2, 1-a/2])\n\n xticks!(round.([q; xmean], digits=q_digits), axiscolor=:transparent; kw...)\n #vline!(q, c=:red, linewidths=1, line=:dot, label=\"95% CI\", legend=:best; kw...)\n #vline!([xmean], c=:red, linewidths=2, legend=:best,\n # label=\"Mean: $(round(xmean, digits=q_digits))\"; kw...)\n #hline!([0], label=\"SD: $(round(xsd, digits=sd_digits))\", \n # linewidth=0, bgcolor_legend=:transparent; kw...)\n vline!(q, c=:red, label=\"\", linewidths=1, line=:dot; kw...)\n vline!([xmean], c=:red, label=\"\", linewidths=1; kw...)\n hline!([0], linewidth=0, label=\"\", bgcolor_legend=:transparent; kw...)\n\n # TODO:\n # Option for traceplot\n\n return img\nend\n\nfunction plotPosts(X::Matrix{T}; a::Float64=0.05, q_digits::Int=3,\n sd_digits::Int=3, cor_digits::Int=3, titles=:none,\n detail=false, hist2d::Bool=false, trace::Bool=true,\n traceFont=font(16), kw...) where {T <: Number}\n N = size(X, 2)\n\n if titles == :none\n titles = fill(\"\", N)\n end\n\n img = plot(;layout=(N, N))\n counter = 0\n offset = 0\n for r in 1:N\n for c in 1:N\n counter += 1\n\n if r == c\n offset += 1\n plotPost(X[:, r], a=a, q_digits=q_digits, sd_digits=sd_digits,\n subplot=counter, add=true, title=titles[c], trace=trace,\n parent=counter, offset=N*N-counter+offset,\n traceFont=traceFont; kw...)\n elseif r < c\n if hist2d\n if detail\n histogram2d!(X[:, c], X[:,r], subplot=counter, colorbar=:none, grid=false)\n else\n histogram2d!(X[:, c], X[:,r], subplot=counter, colorbar=:none, grid=false,\n axis=false)\n end\n else\n plot!(X[:, c], X[:,r], subplot=counter, c=:grey70, grid=false,\n axis=false, legend=false, linewidths=.5)\n end\n else # r > c\n corXrc = cor(X[:, r], X[:, c])\n annotate!([(0.5, 0.5, text(\"r=$(round(corXrc, digits=cor_digits))\", \n Int(ceil(abs(corXrc) * 5))+5, :center))],\n subplot=counter, axis=false, grid=false)\n end\n end\n end\n\n return img\nend\n\nfunction postProbMiss(b0, b1, i::Int;\n y::Vector{Float64}=collect(-10:.1:10),\n credibility::Float64=.95)\n\n N, I = size(b0)\n @assert size(b0) == size(b1)\n \n len_y = length(y)\n alpha = 1 - credibility\n p_lower = alpha / 2\n p_upper = 1 - alpha / 2\n\n pmiss = hcat([Cytof5.Model.prob_miss.(yi, b0[:,i], b1[:,i]) for yi in y]...)\n pmiss_mean = vec(mean(pmiss, dims=1))\n pmiss_lower = [ quantile(pmiss[:, col], p_lower) for col in 1:len_y ]\n pmiss_upper = [ quantile(pmiss[:, col], p_upper) for col in 1:len_y ]\n\n return (pmiss_mean, pmiss_lower, pmiss_upper, y)\nend\n\nfunction pairwiseAlloc(Z, W, i)\n J, K = size(Z)\n A = zeros(J, J)\n for r in 1:J\n for c in 1:J\n for k in 1:K\n if Z[r, k] == 1 && Z[c, k] == 1\n A[r, c] += W[i, k]\n A[c, r] += W[i, k]\n end\n end\n end\n end\n\n return A\nend # pairwiseAlloc\n\nfunction estimate_ZWi_index(monitor, i)\n As = [pairwiseAlloc(m[:Z], m[:W], i) for m in monitor]\n\n Amean = mean(As)\n mse = [ mean((A - Amean) .^ 2) for A in As]\n\n return argmin(mse)\nend\n\nfunction countmap(x::Vector{T}) where {T <: Number}\n out = Dict{Int, T}()\n for xi in x\n if xi in keys(out)\n out[xi] += 1\n else\n out[xi] = 1\n end\n end\n return out\nend\n\nfunction reorder_lam_i(lam_i::Vector{T}, W_i::Vector{V}) where {T <: Number, V <: Number}\n N = length(lam_i)\n new_lam_i = Vector{Int}(undef, N)\n K = length(W_i)\n ord = sortperm(W_i)\n\n for k in 1:K\n new_lam_i[lam_i .== ord[k]] .= k\n end\n\n return new_lam_i\nend\n\n# TODO: Implement the threshold\nfunction yZ_inspect(monitor, y::Vector{Matrix{Float64}},\n i::Int; thresh=0.7, ycolor=:balance,\n addLines::Bool=true, marker_names = missing,\n clim=(-5,5), W_digits=1, kw...)\n idx_best = estimate_ZWi_index(monitor, i)\n Zi = monitor[idx_best][:Z]\n Wi = monitor[idx_best][:W][i, :]\n lami = monitor[idx_best][:lam][i]\n new_lami = reorder_lam_i(lami, Wi)\n ord = sortperm(new_lami)\n K = length(Wi)\n J = size(y[i], 2)\n\n W_order = sortperm(Wi)\n\n cumProb = 0.0; k = K\n for w in reverse(Wi[W_order])\n cumProb += w\n if cumProb > thresh\n break\n end\n k -= 1\n end\n W_order = W_order[k:end]\n\n plotYZ(y[i][ord, :], Zi[:, W_order], clim=clim, ycolor=ycolor)\n\n # TODO: cell types on left, percentage on right\n W_val = map(x->\"$(x)%\", round.(Wi[W_order] * 100, digits=W_digits))\n #plot!(yticks=(1:K, W_order), subplot=2)\n plot!(yticks=(1:length(W_order), W_val), subplot=2, ymirror=true)\n\n # add marker names if provided\n if !ismissing(marker_names)\n plot!(xticks=(1:J, marker_names), subplot=1, rotation=90)\n end\n\n if addLines\n groups = cumsum(sort(collect(values(countmap(new_lami)))))\n hline!(groups, c=:yellow, subplot=1, linewidths=1, legend=false)\n end\nend\n\nfunction postSummary(x::Matrix{T}; alpha::Float64=.05) where {T <: Number}\n K = size(x, 2)\n xmean = mean(x, dims=1)\n xsd = std(x, dims=1)\n xqLower = [ quantile(x[:, k], alpha / 2) for k in 1:K ]\n xqLower = reshape(xqLower, 1, K)\n xqUpper = [ quantile(x[:, k], 1- alpha / 2) for k in 1:K ]\n xqUpper = reshape(xqUpper, 1, K)\n xmedian = median(x, dims=1)\n\n summaryMatrix = Matrix([xmean; xsd; xqLower; xmedian; xqUpper]')\n println(\"mean sd lower upper median\")\n return summaryMatrix\nend\n\n# TODO: GET RID OF THIS!\nR\"\"\"\nari <- function(x, y) {\n x = as.vector(x)\n y = as.vector(y)\n n = length(x)\n stopifnot(n == length(y))\n tab = table(x, y)\n index = sum(choose(tab, 2))\n rowSumsChoose2.sum = sum(choose(rowSums(tab), 2))\n colSumsChoose2.sum = sum(choose(colSums(tab), 2))\n expected.index = rowSumsChoose2.sum * colSumsChoose2.sum/choose(n,\n 2)\n max.index = mean(c(rowSumsChoose2.sum, colSumsChoose2.sum))\n (index - expected.index)/(max.index - expected.index)\n}\n\"\"\"\nari = R\"ari\"\n\nfunction confusion(x::Vector{T}, y::Vector{T}) where {T <: Number}\n @assert length(y) == length(x)\n n = length(x)\n confusionMat = zeros(Int, n, n)\n for i in 1:n\n for j in 1:n\n if x[i] == y[j]\n confusionMat[i, j] += 1\n end\n end\n end\n return confusionMat\nend\n\n#=\nfunction ari(x::Vector{T}, y::Vector{T}) where {T <: Number}\n @assert length(y) == length(x)\n n = length(x)\n tab = confusion(x, y)\n # TODO: https://en.wikipedia.org/wiki/Rand_index\nend\n=#\n\n\n\nend\n", "meta": {"hexsha": "3cbd4455640f9599264567e332f935aca157706b", "size": 9970, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "sims/sim_study/CytofImg.jl", "max_stars_repo_name": "luiarthur/cytof5", "max_stars_repo_head_hexsha": "6b4df5e9fd94bfd586e96579b8c618fdf6f913ed", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2020-01-30T21:56:52.000Z", "max_stars_repo_stars_event_max_datetime": "2020-01-30T21:56:52.000Z", "max_issues_repo_path": "sims/sim_study/CytofImg.jl", "max_issues_repo_name": "luiarthur/cytof5", "max_issues_repo_head_hexsha": "6b4df5e9fd94bfd586e96579b8c618fdf6f913ed", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 27, "max_issues_repo_issues_event_min_datetime": "2018-12-20T18:22:25.000Z", "max_issues_repo_issues_event_max_datetime": "2021-02-24T03:13:32.000Z", "max_forks_repo_path": "sims/sim_study/CytofImg.jl", "max_forks_repo_name": "luiarthur/cytof5", "max_forks_repo_head_hexsha": "6b4df5e9fd94bfd586e96579b8c618fdf6f913ed", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 27.8491620112, "max_line_length": 132, "alphanum_fraction": 0.5870611836, "num_tokens": 3179, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.480478678047907, "lm_q2_score": 0.12085323090725615, "lm_q1q2_score": 0.05806740062413689}}
{"text": "using InteractiveUtils # 0.7 \u4e4b\u540e\u9700\u8981\u8c03\u7528\u8fd9\u4e2a\u6807\u51c6\u5e93\n\nfunction view_tree(T, depth=0)\n println(\" \"^depth, T)\n for each in subtypes(T)\n view_tree(each, depth+1)\n end\nend\n\nabstract type AbstractAnimal end\n\nabstract type AbstractBird <: AbstractAnimal end\nabstract type AbstractDog <: AbstractAnimal end\nabstract type AbstractCat <: AbstractAnimal end\n\nstruct Sparrow <: AbstractBird end\nstruct Kitty <: AbstractCat end\nstruct Snoope <: AbstractDog end\n\nview_tree(AbstractAnimal)", "meta": {"hexsha": "fa30f48a6d02b50a556194a59b92b453cf23cf2d", "size": 480, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "julia/005_abstrac.jl", "max_stars_repo_name": "liushooter/OneDayOneCommit", "max_stars_repo_head_hexsha": "87dc037fcb21c9cd91723c282d1b618bef3e0414", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "julia/005_abstrac.jl", "max_issues_repo_name": "liushooter/OneDayOneCommit", "max_issues_repo_head_hexsha": "87dc037fcb21c9cd91723c282d1b618bef3e0414", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "julia/005_abstrac.jl", "max_forks_repo_name": "liushooter/OneDayOneCommit", "max_forks_repo_head_hexsha": "87dc037fcb21c9cd91723c282d1b618bef3e0414", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 24.0, "max_line_length": 48, "alphanum_fraction": 0.7625, "num_tokens": 122, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.48047867804790706, "lm_q2_score": 0.12085322457439822, "lm_q1q2_score": 0.05806739758133369}}
{"text": "using Interact, Blink, Colors\nexport interactive_orbitdiagram, scaleod\n\n\"\"\"\n controlwindow(D, n, Ttr, density, i)\nCreate an Electron control window for the orbit diagram interactive application.\n\n```julia\nreturn n, Ttr, density, i, \u25a2update, \u25a2back, \u25a2reset, \u03b1,\n \u2b1cpmin, \u2b1cpmax, \u2b1cumin, \u2b1cumax\n```\n\nAll returned values are `Observable`s.\nTheir value corresponds to the one chosen in the Electron window.\nItems with `\u25a2` are buttons and with `\u2b1c` are the boxes with limits.\n\"\"\"\nfunction controlwindow(D, n, Ttr, density, i)\n n = Interact.textbox(string(n); value = n, label = \"n\")\n Ttr = Interact.textbox(string(Ttr); value = Ttr, label = \"Ttr\")\n density = Interact.textbox(string(density); value = density, label = \"density\")\n\n i = Interact.dropdown(OrderedDict(string(j) => j for j in 1:D); label = \"variable\")\n \u25a2update = Interact.button(\"update\")\n \u25a2back = Interact.button(\"\u2190 back\")\n \u25a2reset = Interact.button(\"reset\")\n\n \u03b1 = Interact.slider(0:0.001:1; value = 0.1, label = \"\u03b1 (transparency)\")\n\n # Limit boxes\n \u2b1cpmin = Interact.textbox(; value = 0.0, label = \"pmin\")\n \u2b1cpmax = Interact.textbox(; value = 1.0, label = \"pmax\")\n \u2b1cumin = Interact.textbox(; value = 0.0, label = \"umin\")\n \u2b1cumax = Interact.textbox(; value = 1.0, label = \"umax\")\n\n w = Window(Dict(:height => 400, :title => \"Orbit Diagram controls\"))\n s = 5em\n body!(w, Interact.vbox(\n \u03b1,\n Interact.hbox(n, Ttr, density),\n Interact.hbox(i, hskip(s), \u25a2update, hskip(s), \u25a2back, hskip(s), \u25a2reset),\n Interact.hline(),\n Interact.hbox(\u2b1cpmin, \u2b1cpmax),\n Interact.hbox(\u2b1cumin, \u2b1cumax)\n )\n )\n\n return n, Ttr, density, i, \u25a2update, \u25a2back, \u25a2reset, observe(\u03b1),\n \u2b1cpmin, \u2b1cpmax, \u2b1cumin, \u2b1cumax\nend\n\n\n\n\"\"\"\n interactive_orbitdiagram(ds::DiscreteDynamicalSystem,\n i::Int, p_index, p_min, p_max;\n density = 500, u0 = get_state(ds), Ttr = 200, n = 500,\n parname = \"p\"\n )\n\nOpen an interactive application for exploring orbit diagrams (ODs) of\ndiscrete systems. The functionality works for _any_ discrete system.\n\nOnce initialized it opens a Makie plot window and an Electron control window.\n\n## Interaction\nBy using the Electron window you are able to update all parameters of the OD\ninteractively (like e.g. `n` or `Ttr`). You have to press `update` after changing\nthese parameters. You can even decide which variable to get the OD for,\nby choosing one of the variables from the wheel (again, press `update` afterwards).\n\nIn the Makie window you can interactively zoom into the OD. Click\nand drag with the left mouse button to select a region in the OD. This region is then\n**re-computed** at a higher resolution (i.e. we don't \"just zoom\").\n\nBack in the Electron window, you can press `reset` to bring the OD in the original\nstate (and variable). Pressing `back` will go back through the history of your exploration\nHistory is stored when any change happens (besides transparency).\n\n## Accessing the data\nWhat is plotted on the application window is a _true_ orbit diagram, not a plotting\nshorthand. This means that all data are obtainable and usable directly.\nInternally we always scale the orbit diagram to [0,1]\u00b2 (to allow `Float64` precision\neven though plotting is `Float32`-based). This however means that it is\nnecessary to transform the data in real scale. This is done through the function\n[`scaleod`](@ref) which accepts the 5 arguments returned from the current function:\n```\nod, pmin, pmax, umin, umax = interactive_orbitdiagram(...)\nps, us = scaleod(od, pmin, pmax, umin, umax)\n```\n\"\"\"\nfunction interactive_orbitdiagram(ds::DiscreteDynamicalSystem,\n i::Int, p_index, p_min, p_max;\n density = 1000, u0 = get_state(ds), Ttr = 200, n = 500,\n parname = \"p\"\n )\n\n # UI elements\n n, Ttr, density, i, \u25a2update, \u25a2back, \u25a2reset, \u03b1, \u2b1cpmin, \u2b1cpmax, \u2b1cumin, \u2b1cumax =\n controlwindow(dimension(ds), n, Ttr, density, i)\n\n # Initial Orbit diagram data\n integ = integrator(ds, u0)\n pmin, pmax = p_min, p_max\n odinit, xmin, xmax = minimal_normalized_od(integ, i[], p_index, pmin, pmax, density[], n[], Ttr[], u0)\n od_node = Observable(odinit)\n densityinit = density[]; ninit = n[]; Ttrinit = Ttr[]\n\n # History stores the variable index and true diagram limits\n history = [(i[], pmin, pmax, xmin, xmax, n[], Ttr[], density[])]\n update_controls!(history[end], i, n, Ttr, density, \u2b1cpmin, \u2b1cpmax, \u2b1cumin, \u2b1cumax)\n\n color = lift(a -> RGBA(0,0,0,a), \u03b1)\n scplot = Scene(resolution = (1200, 800))\n scatter!(scplot, od_node, markersize = 0.008, color = color)\n\n scplot[Axis][:ticks][:ranges] = (collect(0:0.25:1), collect(0:0.25:1))\n scplot[Axis][:ticks][:labels] =\n ([\"pmin\", \" \", \" \", \" \", \"pmax\"], [\"umin\", \" \", \" \", \" \", \"umax\"])\n scplot[Axis][:names][:axisnames] = (parname, \"u\"*subscript(i[]))\n\n display(scplot)\n rect = select_rectangle(scplot)\n\n # Uppon interactively selecting a rectangle, with value `r` (in [0,1]\u00b2)\n on(rect) do r\n spmin, sxmin = r.origin\n spmax, sxmax = r.origin + r.widths\n # Convert p,x to true values\n j, ppmin, ppmax, pxmin, pxmax = history[end]\n pdif = ppmax - ppmin; xdif = pxmax - pxmin\n pmin = spmin*pdif + ppmin\n pmax = spmax*pdif + ppmin\n xmin = sxmin*xdif + pxmin\n xmax = sxmax*xdif + pxmin\n\n od_node[] = minimal_normalized_od(\n integ, j, p_index, pmin, pmax,\n density[], n[], Ttr[], u0, xmin, xmax\n )\n # update history and controls\n push!(history, (j, pmin, pmax, xmin, xmax, n[], Ttr[], density[]))\n update_controls!(history[end], i, n, Ttr, density, \u2b1cpmin, \u2b1cpmax, \u2b1cumin, \u2b1cumax)\n end\n\n # Upon hitting the update button\n on(\u25a2update) do clicks\n j, pmin, pmax, xmin, xmax, m, T, d = history[end]\n # Check if there was any change:\n if !(\u2b1cpmin[] == pmin && \u2b1cpmax[] == pmax && \u2b1cumin[] == xmin && \u2b1cumax[] == xmax &&\n j == i[] && m == n[] && T == Ttr[] && d == density[])\n\n pmin, pmax, xmin, xmax = \u2b1cpmin[], \u2b1cpmax[], \u2b1cumin[], \u2b1cumax[]\n j, m, T, d = i[], n[], Ttr[], density[]\n\n od_node[] = minimal_normalized_od(\n integ, j, p_index, pmin, pmax, density[], n[], Ttr[], u0, xmin, xmax\n )\n # Update history and controls\n push!(history, (j, pmin, pmax, xmin, xmax, m, T, d))\n update_controls!(history[end], i, n, Ttr, density, \u2b1cpmin, \u2b1cpmax, \u2b1cumin, \u2b1cumax)\n scplot[Axis][:names][:axisnames] = (parname, \"u\"*subscript(i[]))\n end\n end\n\n # Upon hitting the \"reset\" button\n on(\u25a2reset) do clicks\n if length(history) > 1\n deleteat!(history, 2:length(history))\n j, pmin, pmax, xmin, xmax, m, T, d = history[end]\n od_node[] = odinit\n # Update controls/labels\n update_controls!(history[end], i, n, Ttr, density, \u2b1cpmin, \u2b1cpmax, \u2b1cumin, \u2b1cumax)\n scplot[Axis][:names][:axisnames] = (parname, \"u\"*subscript(j))\n end\n end\n\n # Upon hitting the \"back\" button\n on(\u25a2back) do clicks\n if length(history) > 1\n pop!(history)\n j, pmin, pmax, xmin, xmax, m, T, d = history[end]\n od_node[] = minimal_normalized_od(\n integ, j, p_index, pmin, pmax, d, m, T, u0, xmin, xmax\n )\n # Update limits in textboxes\n update_controls!(history[end], i, n, Ttr, density, \u2b1cpmin, \u2b1cpmax, \u2b1cumin, \u2b1cumax)\n scplot[Axis][:names][:axisnames] = (parname, \"u\"*subscript(j))\n end\n end\n\n display(scplot)\n return od_node, \u2b1cpmin, \u2b1cpmax, \u2b1cumin, \u2b1cumax\nend\n\n\n\"\"\"\n minimal_normalized_od(integ, i, p_index, pmin, pmax,\n density, n, Ttr, u0)\n minimal_normalized_od(integ, i, p_index, pmin, pmax,\n density, n, Ttr, u0, xmin, xmax)\n\nCompute and return a minimal and normalized orbit diagram (OD).\n\nAll points are stored in a single vector of `Point2f0` to ensure fastest possible\nplotting. In addition all numbers are scaled to [0, 1]. This allows us to have\n64-bit precision while display is only 32-bit!\n\nThe version with `xmin, xmax` only keeps points with limits between the\nreal `xmin, xmax` (in the normal units of the dynamical system).\n\"\"\"\nfunction minimal_normalized_od(integ, i, p_index, pmin, pmax,\n density::Int, n::Int, Ttr::Int, u0)\n\n pvalues = range(pmin, stop = pmax, length = density)\n pdif = pmax - pmin\n od = Vector{Point2f0}() # make this pre-allocated\n xmin = eltype(integ.u)(Inf); xmax = eltype(integ.u)(-Inf)\n #= @inbounds =# for (j, p) in enumerate(pvalues)\n pp = (p - pmin)/pdif # p to plot, in [0, 1]\n DynamicalSystemsBase.reinit!(integ, u0)\n integ.p[p_index] = p\n DynamicalSystemsBase.step!(integ, Ttr)\n for z in 1:n\n DynamicalSystemsBase.step!(integ)\n x = integ.u[i]\n push!(od, Point2f0(pp, integ.u[i]))\n # update limits\n if x < xmin\n xmin = x\n elseif x > xmax\n xmax = x\n end\n end\n end\n # normalize x values to [0, 1]\n xdif = xmax - xmin\n #= @inbounds =# for j in eachindex(od)\n x = od[j][2]; p = od[j][1]\n od[j] = Point2f0(p, (x - xmin)/xdif)\n end\n return od, xmin, xmax\nend\n\nfunction minimal_normalized_od(integ, i, p_index, pmin, pmax,\n density::Int, n::Int, Ttr::Int, u0, xmin, xmax)\n\n pvalues = range(pmin, stop = pmax, length = density)\n pdif = pmax - pmin; xdif = xmax - xmin\n od = Vector{Point2f0}()\n #= @inbounds =# for p in pvalues\n pp = (p - pmin)/pdif # p to plot, in [0, 1]\n DynamicalSystemsBase.reinit!(integ, u0)\n integ.p[p_index] = p\n DynamicalSystemsBase.step!(integ, Ttr)\n for z in 1:n\n DynamicalSystemsBase.step!(integ)\n x = integ.u[i]\n if xmin \u2264 x \u2264 xmax\n push!(od, Point2f0(pp, (integ.u[i] - xmin)/xdif))\n end\n end\n end\n return od\nend\n\nfunction update_controls!(h, i, n, Ttr, density, \u2b1cpmin, \u2b1cpmax, \u2b1cumin, \u2b1cumax)\n j, pmin, pmax, xmin, xmax, m, T, d = h\n i[] = j; n[] = m; Ttr[] = T; density[] = d\n \u2b1cpmin[] = pmin; \u2b1cpmax[] = pmax\n \u2b1cumin[] = xmin; \u2b1cumax[] = xmax\n return\nend\n\n\"\"\"\n scaleod(od, pmin, pmax, umin, umax) -> ps, us\nGiven the return values of [`interactive_orbitdiagram`](@ref), produce\norbit diagram data scaled correctly in data units. Return the data as a vector of\nparameter values and a vector of corresponding variable values.\n\"\"\"\nfunction scaleod(od, pmin, pmax, umin, umax)\n oddata = od[]; L = length(oddata);\n T = promote_type(typeof(umin[]), Float32)\n ps = zeros(T, L); us = copy(ps)\n udif = umax[] - umin[]; um = umin[]\n pdif = pmax[] - pmin[]; pm = pmin[]\n @inbounds for i \u2208 1:length(oddata)\n p, u = oddata[i]\n ps[i] = pm + pdif*p; us[i] = um + udif*u\n end\n return ps, us\nend\n\n# TODO: Use `\u03b1` directly after Simon's fix, no need for observe(\u03b1)\n# TODO: Add exit button that closes both windows\n# TODO: Make marker GLMakie.FastPixel()\n# TODO: Allow initial state to be a function of paramter (define function `get_u(f, p)`)\n", "meta": {"hexsha": "c8967a0f1b002ab3ba1a3f499bd0b71a5471f423", "size": 11250, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/orbitdiagram.jl", "max_stars_repo_name": "UnofficialJuliaMirrorSnapshots/InteractiveChaos.jl-81850811-659e-51ab-a7c2-5dd848ff4aa4", "max_stars_repo_head_hexsha": "e1beca1584272a389408cec2f05655105194bc68", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/orbitdiagram.jl", "max_issues_repo_name": "UnofficialJuliaMirrorSnapshots/InteractiveChaos.jl-81850811-659e-51ab-a7c2-5dd848ff4aa4", "max_issues_repo_head_hexsha": "e1beca1584272a389408cec2f05655105194bc68", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/orbitdiagram.jl", "max_forks_repo_name": "UnofficialJuliaMirrorSnapshots/InteractiveChaos.jl-81850811-659e-51ab-a7c2-5dd848ff4aa4", "max_forks_repo_head_hexsha": "e1beca1584272a389408cec2f05655105194bc68", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 37.8787878788, "max_line_length": 106, "alphanum_fraction": 0.6023111111, "num_tokens": 3566, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.5, "lm_q2_score": 0.11596071978154186, "lm_q1q2_score": 0.05798035989077093}}
{"text": "module SyntaxTree\n\n# This file is part of SyntaxTree.jl. It is licensed under the MIT license\n# Copyright (C) 2018 Michael Reed\n\nexport linefilter!, linefilter, callcount, genfun, @genfun\n\nif VERSION < v\"0.7.0\"\n linefilter(expr) = linefilter!(expr)\nelse\n @deprecate linefilter(expr) linefilter!(expr)\nend\n\n\"\"\"\n linefilter!(::Expr)\n\nRecursively filters out `:LineNumberNode` from `Expr` objects.\n\"\"\"\n@noinline function linefilter!(expr::Expr)\n total = length(expr.args)\n i = 0\n while i < total\n i += 1\n if expr.args[i] |> typeof == Expr\n if expr.args[i].head == :line\n deleteat!(expr.args,i)\n total -= 1\n i -= 1\n else\n expr.args[i] = linefilter!(expr.args[i])\n end\n elseif expr.args[i] |> typeof == LineNumberNode\n if expr.head == :macrocall\n expr.args[i] = nothing\n else\n deleteat!(expr.args,i)\n total -= 1\n i -= 1\n end\n end\n end\n return expr\nend\n\n\"\"\"\n sub(T::DataType,expr::Expr)\n\nMake a substitution to convert numerical values inside an `Expr` to type `T`.\n\"\"\"\n@noinline function sub(T::DataType,expr)\n if typeof(expr) == Expr\n ixpr = deepcopy(expr)\n if ixpr.head == :call && ixpr.args[1] == :^\n ixpr.args[2] = sub(T,ixpr.args[2])\n if typeof(ixpr.args[3]) == Expr\n ixpr.args[3] = sub(T,ixpr.args[3])\n end\n elseif ixpr.head == :macrocall &&\n ixpr.args[1] \u2208 [Symbol(\"@int128_str\"), Symbol(\"@big_str\")]\n return convert(T,eval(ixpr))\n else\n for a \u2208 1:length(ixpr.args)\n ixpr.args[a] = sub(T,ixpr.args[a])\n end\n end\n return ixpr\n elseif typeof(expr) <: Number\n return convert(T,expr)\n end\n return expr\nend\n\n\"\"\"\n SyntaxTree.abs(expr)\n\nApply `abs` to the expression recursively.\n\"\"\"\n@noinline function abs(expr)\n if typeof(expr) == Expr\n ixpr = deepcopy(expr)\n if ixpr.head == :call && ixpr.args[1] == :^\n ixpr.args[2] = abs(ixpr.args[2])\n if typeof(ixpr.args[3]) == Expr\n ixpr.args[3] = abs(ixpr.args[3])\n end\n elseif ixpr.head == :macrocall &&\n ixpr.args[1] \u2208 [Symbol(\"@int128_str\"), Symbol(\"@big_str\")]\n val = VERSION < v\"0.7\" ? (ixpr.args[1],) : (ixpr.args[1],nothing)\n rep = ('-',\"\")\n return Expr(:macrocall,val...,replace(ixpr.args[end],(VERSION < v\"0.7\" ? rep : (Pair(rep...),))...))\n else\n ixpr.head == :call && ixpr.args[1] == :- && (ixpr.args[1] = :+)\n for a \u2208 1:length(ixpr.args)\n ixpr.args[a] = abs(ixpr.args[a])\n end\n end\n return ixpr\n elseif typeof(expr) <: Number\n return Base.abs(expr)\n end\n return expr\nend\n\n\"\"\"\n alg(expr,f=:(1+\u03f5))\n\nRecursively insert a machine epsilon bound (1+\u03f5) per call in `expr`.\n\"\"\"\n@noinline function alg(expr,f=:(1+\u03f5))\n if typeof(expr) == Expr\n ixpr = deepcopy(expr)\n if ixpr.head == :call\n ixpr.args[2:end] = alg.(ixpr.args[2:end],Ref(f))\n ixpr = Expr(:call,:*,f,ixpr)\n end\n return ixpr\n else\n return expr\n end\nend\n\n\"\"\"\n @genfun(expr, args)\n\nReturns an anonymous function based on the given `expr` and `args`.\n\n```Julia\njulia> @genfun x^2+y^2 x y\n```\n\"\"\"\nmacro genfun(expr,args...); :(($(args...),)->$expr) end\n\n\"\"\"\n genfun(expr, args)\n\nReturns an anonymous function based on the given `expr` and `args`.\n\n```Julia\njulia> genfun(:(x^2+y^2),[:x,:y])\njulia> genfun(:(x^2+y^2),(:x,:y))\njulia> genfun(:(x^2+y^2),:x,:y)\n```\n\"\"\"\ngenfun(expr,args::Union{Vector,Tuple}) = eval(:(($(args...),)->$expr))\ngenfun(expr,args::Symbol...) = genfun(expr,args)\n\n\"\"\"\n @genlatest(expr, args)\n\nReturns an invokelatest function based on the given `expr` and `args`.\n\n```Julia\njulia> @genlatest x^2+y^2 x y\n```\n\"\"\"\nmacro genlatest(expr,args,gs = gensym())\n eval(Expr(:function,Expr(:call,gs,args.args...),expr))\n :($(Expr(:tuple,args.args...))->Base.invokelatest($gs,$(args.args...)))\nend\n\n\"\"\"\n genlatest(expr, args)\n\nReturns an invokelatest function based on the given `expr` and `args`.\n\n```Julia\njulia> genlatest(:(x^2+y^2),[:x,:y])\njulia> genlatest(:(x^2+y^2),(:x,:y))\njulia> genlatest(:(x^2+y^2),:x,:y)\n```\n\"\"\"\nfunction genlatest(expr,args::T,gs=gensym()) where T<:Union{Vector,Tuple}\n eval(Expr(:function,Expr(:call,gs,args...),expr))\n if length(args) == 0\n ()->Base.invokelatest(eval(gs))\n elseif length(args) == 1\n (a)->Base.invokelatest(eval(gs),a)\n elseif length(args) == 2\n (a,b)->Base.invokelatest(eval(gs),a,b)\n elseif length(args) == 3\n (a,b,c)->Base.invokelatest(eval(gs),a,b,c)\n elseif length(args) == 4\n (a,b,c,d)->Base.invokelatest(eval(gs),a,b,c,d)\n elseif length(args) == 5\n (a,b,c,d,e)->Base.invokelatest(eval(gs),a,b,c,d,e)\n elseif length(args) == 6\n (a,b,c,d,e,f)->Base.invokelatest(eval(gs),a,b,c,d,e,f)\n elseif length(args) == 7\n (a,b,c,d,e,f,g)->Base.invokelatest(eval(gs),a,b,c,d,e,f,g)\n end\nend\ngenlatest(expr,arg,gs=gensym()) = genlatest(expr,(arg,),gs)\n\n\"\"\"\n callcount(expr)\n\nReturns a count of the `call` operations in `expr`.\n\"\"\"\n@noinline function callcount(expr)\n c = 0\n if typeof(expr) == Expr\n expr.head == :call && (c += 1)\n c += sum(callcount.(expr.args))\n end\n return c\nend\n\ninclude(\"exprval.jl\")\n\nend # module\n", "meta": {"hexsha": "b7b26da27a458456163a8b78bb7f4ab45ca61073", "size": 5605, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/SyntaxTree.jl", "max_stars_repo_name": "UnofficialJuliaMirrorSnapshots/SyntaxTree.jl-a4af3ec5-f8ac-5fed-a759-c2e80b4d74cb", "max_stars_repo_head_hexsha": "631d8141e9ae509952d6c6b6477082d0b541e47f", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 13, "max_stars_repo_stars_event_min_datetime": "2018-10-06T10:07:20.000Z", "max_stars_repo_stars_event_max_datetime": "2022-02-08T14:12:52.000Z", "max_issues_repo_path": "src/SyntaxTree.jl", "max_issues_repo_name": "UnofficialJuliaMirrorSnapshots/SyntaxTree.jl-a4af3ec5-f8ac-5fed-a759-c2e80b4d74cb", "max_issues_repo_head_hexsha": "631d8141e9ae509952d6c6b6477082d0b541e47f", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/SyntaxTree.jl", "max_forks_repo_name": "UnofficialJuliaMirrorSnapshots/SyntaxTree.jl-a4af3ec5-f8ac-5fed-a759-c2e80b4d74cb", "max_forks_repo_head_hexsha": "631d8141e9ae509952d6c6b6477082d0b541e47f", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2020-02-18T19:45:09.000Z", "max_forks_repo_forks_event_max_datetime": "2020-02-18T19:45:09.000Z", "avg_line_length": 26.5639810427, "max_line_length": 116, "alphanum_fraction": 0.5482604817, "num_tokens": 1682, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.5, "lm_q2_score": 0.11596070756094144, "lm_q1q2_score": 0.05798035378047072}}
{"text": "using Test\nusing DifferentialAlgebra\n\n@test 1+1==2\n", "meta": {"hexsha": "9d8ddf14ed121accc5164f4c6648357aff5ec58d", "size": 51, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/runtests.jl", "max_stars_repo_name": "Stopa42/DifferentialAlgebra.jl", "max_stars_repo_head_hexsha": "4938f848903836867438dba2d70c7a295e67ffde", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "test/runtests.jl", "max_issues_repo_name": "Stopa42/DifferentialAlgebra.jl", "max_issues_repo_head_hexsha": "4938f848903836867438dba2d70c7a295e67ffde", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "test/runtests.jl", "max_forks_repo_name": "Stopa42/DifferentialAlgebra.jl", "max_forks_repo_head_hexsha": "4938f848903836867438dba2d70c7a295e67ffde", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 10.2, "max_line_length": 25, "alphanum_fraction": 0.7843137255, "num_tokens": 16, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.41869689484852374, "lm_q2_score": 0.13846179412408713, "lm_q1q2_score": 0.05797352325491085}}
{"text": "\"\"\"\nTools for generating Julia packages for _educational_ purposes.\n\n# Extended Help\n\n## Overview \n\nHave you ever wanted to write a note-set, or a textbook, or a \nseries of homework assignments with tools within the Julia \necosystem? Your instincts are correct! Mediums like \n`Documenter.jl` websites, `Pluto.jl` notebooks, and `Markdown.jl` \nand PDF formats each have their own strengths, and all can help \nyou write informative, and educational content. \n\nWouldn't it be nice if there was a tool which allowed you to \nwrite _one_ file, and generate content within all of the following \nmediums automatically? Well, it already exists, and it's called \n`Literate.jl`.\n\nAlright, but wouldn't it be nice if there was a tool to help you \ncreate packages with structures that `Literate.jl` can benefit\nfrom? That's where `EducationalPkg` comes in! The following \nuse cases are a few examples that might benefit from \n`EducationalPkg`.\n\n1. Writing a note-set which is compiled with `Documenter.jl`\n2. Developing a bunch of homework assignments with `Pluto.jl`\n3. Showing several investigations, or experiments that you \nwant accessible in a variety of mediums\n\n## Usage\n\n```julia\njulia> import EducationalPkg\nedu} # enter `}` in the REPL to enter `EducationalPkg` mode!\nedu} generate FutureNobelPrize\n```\n\n\"\"\"\nmodule EducationalPkg\n\nimport Pkg\n\nusing Pluto\nusing Literate\nusing Documenter\nusing DocStringExtensions\n\ninclude(joinpath(\"EduREPL\", \"EduREPL.jl\"))\n\nmodule EduREPLInit\n using ..EduREPL\n function __init__()\n if isdefined(Base, :active_repl)\n EduREPL.runEduREPL()\n end\n end\nend\n\nend # module\n", "meta": {"hexsha": "f9037210ff146ddb27433ced02f36a7f202b0256", "size": 1630, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/EducationalPkg.jl", "max_stars_repo_name": "cadojo/EducationalPkg.jl", "max_stars_repo_head_hexsha": "2c7acd114dd758b42090f17bf396d63f195482da", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/EducationalPkg.jl", "max_issues_repo_name": "cadojo/EducationalPkg.jl", "max_issues_repo_head_hexsha": "2c7acd114dd758b42090f17bf396d63f195482da", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/EducationalPkg.jl", "max_forks_repo_name": "cadojo/EducationalPkg.jl", "max_forks_repo_head_hexsha": "2c7acd114dd758b42090f17bf396d63f195482da", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 26.7213114754, "max_line_length": 67, "alphanum_fraction": 0.7558282209, "num_tokens": 413, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.23370634623958195, "lm_q2_score": 0.24798742624020279, "lm_q1q2_score": 0.057956235299955625}}
{"text": "\n# rewrite.jl - expression pattern matching and rewriting.\n#\n# The general idea behind functions in this file is to provide easy means\n# of finding specific pieces of expressions and using them elsewhere.\n# At the time of writing it is used in 2 parts of this package:\n#\n# * for applyging derivatives, e.g. `x^n` ==> `n*x^(n-1)`\n# * for expression simplification, e.g. `1 * x` ==> `x`\n#\n# Pieces of expression are matched to so-called placeholders - symbols that either\n# start with `_` (e.g. `_x`) or are passed via `phs` paraneter, or set globally\n# using `set_default_placeholders(::Set{Symbol})`. Using list of placeholders\n# instead of _-prefixed names is convenient when writing a lot of transformation\n# rules where using underscores creates unnecessary noise.\n\n\nconst Symbolic = Union{Expr, Symbol}\nconst Numeric = Union{Number, Array}\n\n## pattern matching\n\nconst DEFAULT_PHS = [Set{Symbol}()]\nset_default_placeholders(set::Set{Symbol}) = (DEFAULT_PHS[1] = set)\n\nisplaceholder(x, phs) = false\nisplaceholder(x::Symbol, phs) = (startswith(string(x), \"_\")\n || in(x, phs))\n# isplaceholder(ex::Expr, phs) = ex.head == :... && isplaceholder(ex.args[1], phs)\n\nfunction find_key(d::Dict{K, V}, val) where {K,V}\n r = nothing\n for (k,v) in d\n if v == val\n r = k\n break\n end\n end\n return r\nend\n\n\nfunction matchex!(m::Dict{Symbol,Any}, p::QuoteNode, x::QuoteNode;\n opts...)\n return matchex!(m, p.value, x.value)\nend\n\n\nfunction matchex!(m::Dict{Symbol,Any}, ps::Vector, xs::Vector; opts...)\n opts = to_dict(opts)\n phs = get(opts, :phs, Set([]))\n length(ps) <= length(xs) || return false\n for i in eachindex(ps)\n if isa(ps[i], Expr) && ps[i].head == :... && isplaceholder(ps[i].args[1], phs)\n p = ps[i].args[1]\n haskey(m, p) && m[p] != xs[i] && m[p] != xs[i:end] && return false\n # TODO: add something here?\n m[p] = xs[i:end]\n return true\n else\n matchex!(m, ps[i], xs[i]; opts...) || return false\n end\n end\n # matched everything, didn't encounter dots expression\n return length(ps) == length(xs)\nend\n\n\nfunction matchex!(m::Dict{Symbol,Any}, p, x; phs=DEFAULT_PHS[1], allow_ex=true, exact=false)\n allow_ex = exact ? false : allow_ex # override allow_ex=false if exact==true\n if isplaceholder(p, phs)\n if haskey(m, p) && m[p] != x\n # different bindings to the same pattern, treat as no match\n return false\n elseif !allow_ex && isa(x, Expr)\n # x is expression, but matching to expression is not allowed, treat as no match\n return false\n elseif exact\n k = find_key(m, x)\n if k != p\n return false\n else\n m[p] = x\n return true\n end\n else \n m[p] = x\n return true\n end\n elseif isa(p, Expr) && isa(x, Expr)\n return (matchex!(m, p.head, x.head) &&\n matchex!(m, p.args, x.args; phs=phs, allow_ex=allow_ex, exact=exact))\n else\n return p == x\n end\nend\n\n\n\"\"\"\nMatch expression `ex` to a pattern `pat`, return nullable dictionary of matched\nsymbols or rpatpressions.\nExample:\n\n```\nex = :(u ^ v)\npat = :(_x ^ _n)\nmatchex(pat, ex)\n# ==> Union{ Dict{Symbol,Any}(:_n=>:v,:_x=>:u), Void }\n```\n\nNOTE: two symbols match if they are equal or symbol in pattern is a placeholder.\nPlaceholder is any symbol that starts with '_'. It's also possible to pass\nlist of placeholder names (not necessarily starting wiht '_') via `phs` parameter:\n\n```\nex = :(u ^ v)\npat = :(x ^ n)\nmatchex(pat, ex; phs=Set([:x, :n]))\n# ==> Union{ Dict{Symbol,Any}(:n=>:v,:x=>:u), Void } \n```\n\nSeveral elements may be matched using `...` expression, e.g.:\n\n```\nex = :(A[i, j, k])\npat = :(x[I...])\nmatchex(pat, ex; phs=Set([:x, :I]))\n# ==> Union{ Dict(:x=>:A, :I=>[:i,:j,:k]), Void }\n```\n\nOptional parameters:\n\n * phs::Set{Symbol} = DEFAULT_PHS[1]\n A set of placeholder symbols\n * allow_ex::Boolean = true\n Allow matchinng of symbol pattern to an expression. Example:\n\n matchex(:(_x + 1), :(a*b + 1); allow_ex=true) # ==> matches\n matchex(:(_x + 1), :(a*b + 1); allow_ex=false) # ==> doesn't match\n * exact::Boolean = false\n Allow matching of the same expression to different keys\n\n matchex(:(_x + _y), :(a + a); exact=false) # ==> matches\n matchex(:(_x = _y), :(a + a); exact=true) # ==> doesn't match\n\n\"\"\"\nfunction matchex(pat, ex; opts...)\n m = Dict{Symbol,Any}()\n res = matchex!(m, pat, ex; opts...)\n if res\n return m\n else\n return nothing\n end\nend\n\n\"\"\"\nCheck if expression matches pattern. See `matchex()` for details.\n\"\"\"\nfunction matchingex(pat, ex; opts...)\n return matchex(pat, ex; opts...) != nothing\nend\n\n\n## find rpatpression\n\nfunction findex!(res::Vector, pat, ex; phs=DEFAULT_PHS[1])\n if matchingex(pat, ex; phs=phs)\n push!(res, ex)\n elseif expr_like(ex)\n for arg in ex.args\n findex!(res, pat, arg; phs=phs)\n end\n end\nend\n\n\n\"\"\"\nFind sub-expressions matching a pattern. Example:\n\n ex = :(a * f(x) + b * f(y))\n pat = :(f(_))\n findex(pat, ex) # ==> [:(f(x)), :(f(y))]\n\n\"\"\"\nfunction findex(pat, ex; phs=DEFAULT_PHS[1])\n res = Any[]\n findex!(res, pat, ex; phs=phs)\n return res\nend\n\n\n## substitution\n\n\"\"\"\nGiven a list of expression arguments, flatten the dotted ones. Example:\n\n args = [:foo, :([a, b, c]...)]\n flatten_dots(args)\n # ==> [:foo, :a, :b, :c]\n\"\"\"\nfunction flatten_dots(args::Vector)\n new_args = Vector{Any}()\n for arg in args\n if isa(arg, Expr) && arg.head == :... && isa(arg.args[1], AbstractArray)\n for x in arg.args[1]\n push!(new_args, x)\n end\n else\n push!(new_args, arg)\n end\n end\n return new_args\nend\n\n\n\"\"\"\nSubstitute symbols in `ex` according to substitute table `st`.\nExample:\n\n ex = :(x ^ n)\n subs(ex, x=2) # ==> :(2 ^ n)\n\nalternatively:\n\n subs(ex, Dict(:x => 2)) # ==> :(2 ^ n)\n\nIf `ex` contains a :(xs...) argument and `st` contains an array-valued\nsabstitute for it, the substitute will be flattened:\n\n ex = :(foo(xs...))\n subs(ex, Dict(:xs => [:a, :b, :c]))\n # ==> :(foo(a, b, c))\n\"\"\"\nfunction subs(ex::Expr, st::Dict)\n if haskey(st, ex)\n return st[ex]\n # elseif ex.head == :... && haskey(st, ex.args[1]) \n else\n new_args = [subs(arg, st) for arg in ex.args]\n new_args = flatten_dots(new_args)\n return Expr(ex.head, new_args...)\n end\nend\n\n\nfunction subs(s::Symbol, st::Dict)\n return haskey(st, s) ? st[s] : s\nend\n\nsubs(q::QuoteNode, st::Dict) = QuoteNode(subs(q.value, st))\nsubs(x::Any, st::Dict) = x\nsubs(ex; st...) = subs(ex, to_dict(st))\n\n\n## remove rpatpression\n\n\"\"\"\nRemove rpatpression conforming to a pattern.\nExample:\n\n ex = :(x * (m == n))\n pat = :(_i == _j)\n ex = without(ex, pat) # ==> :x\n\"\"\"\nfunction without(ex::Expr, pat; phs=DEFAULT_PHS[1])\n new_args_without = [without(arg, pat; phs=phs) for arg in ex.args]\n new_args = filter(arg -> !matchingex(pat, arg; phs=phs), new_args_without)\n if ex.head == :call && length(new_args) == 2 &&\n (ex.args[1] == :+ || ex.args[1] == :*)\n # pop argument of now-single-valued operation\n # TODO: make more general, e.g. handle (x - y) with x removed\n return new_args[2]\n elseif ex.head == :call && length(new_args) == 1 && ex.args[1] == :*\n return 1.0\n elseif ex.head == :call && length(new_args) == 1 && ex.args[1] == :+\n return 0.0\n else\n return Expr(ex.head, new_args...) |> simplify\n end\nend\n\nwithout(x, pat; phs=DEFAULT_PHS[1]) = x\n\n\n## rewriting\n\n\"\"\"\nrewrite(ex, pat, rpat)\n\nRewrite expression `ex` according to a transform from pattern `pat`\nto a substituting expression `rpat`.\nExample (derivative of x^n):\n\n ex = :(u ^ v)\n pat = :(_x ^ _n)\n rpat = :(_n * _x ^ (_n - 1))\n rewrite(ex, pat, rpat) # ==> :(v * u ^ (v - 1))\n\"\"\"\nfunction rewrite(ex::Symbolic, pat::Symbolic, rpat::Any; opts...)\n st = matchex(pat, ex; opts...)\n if st == nothing\n error(\"Expression $ex doesn't match pattern $pat\")\n else\n return subs(rpat, st)\n end\nend\n\n\n\"\"\"\nSame as rewrite, but returns Union{Expr, Void} and doesn't throw an error\nwhen expression doesn't match pattern\n\"\"\"\nfunction tryrewrite(ex::Symbolic, pat::Symbolic, rpat::Any; opts...)\n st = matchex(pat, ex; opts...)\n if st == nothing\n return nothing\n else\n return subs(rpat, st)\n end\nend\n\n\n\n\"\"\"\nrewrite_all(ex, rules)\n\nRecursively rewrite an expression according to a list of rules like [pat => rpat]\nExample:\n\n ex = :(foo(bar(foo(A))))\n rules = [:(foo(x)) => :(quux(x)),\n :(bar(x)) => :(baz(x))]\n rewrite_all(ex, rules; phs=[:x])\n # ==> :(quux(baz(quux(A))))\n\"\"\"\nfunction rewrite_all(ex::Symbolic, rules; opts...)\n new_ex = ex\n if isa(ex, Expr)\n new_args = [rewrite_all(arg, rules; opts...)\n for arg in ex.args]\n new_ex = Expr(ex.head, new_args...)\n end\n for (pat, rpat) in rules\n if matchingex(pat, new_ex; opts...)\n new_ex = rewrite(new_ex, pat, rpat; opts...)\n end\n end\n return new_ex\nend\n\n\n\"\"\"\nrewrite_all(ex, pat, rpat)\n\nRecursively rewrite all occurrences of a pattern in an expression.\nExample:\n\n ex = :(foo(bar(foo(A))))\n pat = :(foo(x))\n rpat = :(quux(x))\n rewrite_all(ex, pat, rpat; phs=[:x])\n # ==> :(quux(bar(quux(A))))\n\"\"\"\nfunction rewrite_all(ex::Symbolic, pat::Symbolic, rpat; opts...)\n return rewrite_all(ex, [pat => rpat]; opts...)\nend\n\nrewrite_all(x, pat, rpat; opts...) = x\nrewrite_all(x, rules; opts...) = x\n", "meta": {"hexsha": "79e84b140423170521c6bd64ac8e1b49bfc2ea0f", "size": 9850, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/rewrite.jl", "max_stars_repo_name": "UnofficialJuliaMirror/Espresso.jl-6912e4f1-e036-58b0-9138-08d1e6358ea9", "max_stars_repo_head_hexsha": "3fc95cdf87ec8f7c7180723b1047715eb9918605", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 53, "max_stars_repo_stars_event_min_datetime": "2016-08-15T12:25:32.000Z", "max_stars_repo_stars_event_max_datetime": "2022-01-24T00:19:00.000Z", "max_issues_repo_path": "src/rewrite.jl", "max_issues_repo_name": "dfdx/Hydra.jl", "max_issues_repo_head_hexsha": "5362d8cef68d0f0f91a34834dc7095ad7ab024f6", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 25, "max_issues_repo_issues_event_min_datetime": "2016-08-14T15:41:56.000Z", "max_issues_repo_issues_event_max_datetime": "2020-10-08T21:02:28.000Z", "max_forks_repo_path": "src/rewrite.jl", "max_forks_repo_name": "dfdx/Hydra.jl", "max_forks_repo_head_hexsha": "5362d8cef68d0f0f91a34834dc7095ad7ab024f6", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 12, "max_forks_repo_forks_event_min_datetime": "2017-01-20T07:14:13.000Z", "max_forks_repo_forks_event_max_datetime": "2021-08-03T12:32:22.000Z", "avg_line_length": 26.5498652291, "max_line_length": 92, "alphanum_fraction": 0.5726903553, "num_tokens": 2873, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4073334000459302, "lm_q2_score": 0.1422318986458388, "lm_q1q2_score": 0.05793580287039765}}
{"text": "## CPItree.jl - Tipos y m\u00e9todos para operar la estructura jer\u00e1rquica del IPC\n\n## ----------------------------------------------------------------------------\n# Main type definitions\n# ----------------------------------------------------------------------------\n\n\"\"\"\n Item{T}(code::String, name::String, weight::T<:AbstractFloat)\n\nRepresenta un gasto b\u00e1sico en la estructura de nodos del IPC. Es el nivel m\u00e1s\nbajo de la estructura. Almacena el c\u00f3digo del gasto b\u00e1sico, su nombre o\ndescripci\u00f3n y su ponderaci\u00f3n en el IPC. Los datos de este nodo deben estar\ndisponibles en alg\u00fan [`FullCPIBase`](@ref). \n\nPosee los campos: \n- `code`: que almacena el c\u00f3digo del gasto b\u00e1sico como un `String`.\n- `name`: que almacena el nombre del gasto b\u00e1sico como un `String`.\n- `weight::T`: que almacena la ponderaci\u00f3n del gasto b\u00e1sico como un valor\n flotante de tipo `T`.\n\nAunque este nodo ser\u00e1 usualmente creado de manera autom\u00e1tica por m\u00e9todos como\n[`CPITree`](@ref), se pueden crear estructuras jer\u00e1rquicas manualmente. Por\nejemplo, para crear un nodo del nivel inferior: \n```julia-repl\njulia> Item(\"_011101\", \"Item A\", 7.352945f0)\nItem{Float32}(\"_011101\", \"Item A\", 7.352945f0)\n```\n\nVer tambi\u00e9n: [`Group`](@ref), [`CPITree`](@ref).\n\"\"\"\nstruct Item{T}\n code::String\n name::String\n weight::T\nend\n\n\"\"\"\n Group{S,T}\n\n Group(code, name, children::Vector{S}) where S\n Group(code, name, children...) \n\nRepresenta un nodo de agrupaci\u00f3n de cualquier nivel en la estructura de nodos\ndel IPC. Puede almacenar gastos b\u00e1sicos u otros grupos de mayor jerarqu\u00eda.\nAlmacena el c\u00f3digo del grupo, su nombre o descripci\u00f3n y su ponderaci\u00f3n en el\nIPC. \n\nPosee los campos: \n- `code`: almacena el c\u00f3digo del grupo como un `String`.\n- `name`: almacena el nombre del grupo como un `String`.\n- `weight::T`: almacena la ponderaci\u00f3n del grupo como un valor flotante de tipo\n `T`.\n- `children::Vector{S}`: almacena el vector de nodos \"hijos\" de la estructura.\n Por ejemplo, este vector podr\u00eda ser un vector de elementos `Item` para agrupar\n un conjunto de gastos b\u00e1sicos.\n\nAunque este nodo ser\u00e1 usualmente creado de manera autom\u00e1tica por m\u00e9todos como\n[`CPITree`](@ref), se pueden crear estructuras jer\u00e1rquicas manualmente. Por\nejemplo, para crear un grupo: \n```julia-repl\njulia> a = Item(\"_011201\", \"Item B\", 6.7442417f0)\nItem{Float32}(\"_011201\", \"Item B\", 6.7442417f0)\n\njulia> b = Item(\"_011202\", \"Item C\", 7.394718f0)\nItem{Float32}(\"_011202\", \"Item C\", 7.394718f0)\n\njulia> g = Group(\"_0112\", \"Subgr._0112\", a, b)\nGroup{Item{Float32}, Float32}\n_0112: Subgr._0112 [14.13896] \n\u251c\u2500 _011201: Item B [6.7442417]\n\u2514\u2500 _011202: Item C [7.394718]\n```\n\nVer tambi\u00e9n: [`Item`](@ref), [`CPITree`](@ref).\n\"\"\"\nstruct Group{S,T}\n code::String\n name::String\n weight::T\n children::Vector{S}\n\n function Group(code, name, children...)\n sum_weights = sum(child.weight for child in children)\n T = eltype(sum_weights)\n S = eltype(children)\n new{S,T}(code, name, sum_weights, S[children...])\n end\n Group(code, name, children::Vector{S}) where {S} = Group(code, name, children...)\nend\n\n# Redefine methods for getting children\nchildren(::Item) = ()\nchildren(g::Group) = g.children\n\n# Redefine how to print a node in the print_tree function \nprintnode(io::IO, g::Item) = print(io, g.code * \": \" * g.name * \" [\" * string(g.weight) * \"] \")\nprintnode(io::IO, g::Group) = print(io, g.code * \": \" * g.name * \" [\" * string(g.weight) * \"] \")\n\n# How to show a Group\nfunction Base.show(io::IO, g::Group)\n println(io, typeof(g)) \n print_tree(io, g)\nend\n\n\n## ----------------------------------------------------------------------------\n# CPI tree functions \n# ----------------------------------------------------------------------------\n\n\"\"\"\n cpi_tree_nodes(codes::Vector{<:AbstractString}; \n characters::(NTuple{N, Int} where N), depth::Int=1, chars::Int=characters[depth], prefix::AbstractString=\"\", \n base::FullCPIBase, \n group_names::Vector{<:AbstractString}, \n group_codes::Vector{<:AbstractString})\n\nConstruye y devuelve una lista de nodos a partir de la lista de c\u00f3digos `codes`\no de la especificaci\u00f3n jer\u00e1rquica de caracteres en `characters`. Los nombres y\nlas ponderaciones del nivel inferior (nivel de gasto b\u00e1sico) son\nobtenidas de la estructura `base`. Se debe proveer el vector de c\u00f3digos y\nnombres de todas las jerarqu\u00edas superiores en la estructura de c\u00f3digos en los\nvectores `group_names` y `group_codes`.\n\nEsta funci\u00f3n permite crear \u00fanicamente la estructura jer\u00e1rquica de nodos. Para construir de manera autom\u00e1tica una estructura del IPC, se recomienda utilizar preferentemente [`CPITree`](@ref).\n\nVea tambi\u00e9n: [`CPITree`](@ref).\n\"\"\"\nfunction cpi_tree_nodes(codes::Vector{<:AbstractString}; \n characters::(NTuple{N, Int} where N), depth::Int=1, chars::Int=characters[depth], prefix::AbstractString=\"\", \n base::FullCPIBase, \n group_names::Vector{<:AbstractString}, \n group_codes::Vector{<:AbstractString})\n \n # Get available starting codes \n available = filter(code -> startswith(code, prefix), codes)\n \n # Base case: \n # If code length is the last available then we reached a leaf node\n if chars == last(characters)\n # With available codes construct leaf CPI nodes\n children = map(available) do code\n # Find the code index \n icode = findfirst(==(code), codes)\n Item(code, base.names[icode], base.w[icode])\n end\n return children\n end\n\n # Get possible prefixes values from the available list. Possible prefixes\n # are the ones from the beginning of the string to the number of chars,\n # which depends on the depth we are on.\n possibles = unique(getindex.(available, Ref(1:chars)))\n\n # For each available code, call the function itself with the next downward hierarchy\n groups = map(possibles) do prefixcode\n # Go get the children in the next downward level\n children = cpi_tree_nodes(codes; \n characters, depth=depth+1, prefix=prefixcode,\n base, group_names, group_codes\n )\n # Get the group name \n @debug \"Prefix code:\" prefixcode\n i = findfirst(==(prefixcode), group_codes)\n if i === nothing \n @warn \"C\u00f3digo de grupo para $(prefixcode) no encontrado en `group_codes`. Utilizando nombre gen\u00e9rico.\"\n gname = \"Group: $prefixcode\"\n else\n gname = group_names[i]\n end\n # With the children create a group \n group = Group(prefixcode, gname, children)\n group\n end\n\n # Return the group for the upward parents\n return groups\nend\n\n\"\"\"\n get_cpi_tree(; \n base::FullCPIBase, \n group_names::Vector{<:AbstractString}, \n group_codes::Vector{<:AbstractString}, \n characters::(NTuple{N, Int} where N),\n upperlevel_code = \"_0\", \n upperlevel_name = \"IPC\")\n\nFunci\u00f3n superior para obtener estructura jer\u00e1rquica del IPC. Devuelve el nodo\nsuperior del \u00e1rbol jer\u00e1rquico. Utiliza la funci\u00f3n de m\u00e1s bajo nivel\n`cpi_tree_nodes` para construir los nodos del nivel m\u00e1s alto y hacia abajo en la\nestructura jer\u00e1rquica. Se debe proveer el vector de c\u00f3digos y nombres de todas\nlas jerarqu\u00edas superiores en la estructura de c\u00f3digos en los vectores\n`group_names` y `group_codes`. \n\nEsta funci\u00f3n permite crear \u00fanicamente la estructura jer\u00e1rquica de nodos. Para construir de manera autom\u00e1tica una estructura del IPC, se recomienda utilizar preferentemente [`CPITree`](@ref).\n\nVea tambi\u00e9n: [`CPITree`](@ref).\n\"\"\"\nfunction get_cpi_tree(; \n base::FullCPIBase, \n group_names::Vector{<:AbstractString}, \n group_codes::Vector{<:AbstractString}, \n characters::(NTuple{N, Int} where N),\n upperlevel_code = \"_0\", \n upperlevel_name = \"IPC\")\n\n length(characters) >= 2 || throw(ArgumentError(\"`characters` debe ser una tupla con al menos dos valores\"))\n for i in 2:length(characters)\n characters[i] > characters[i-1] || throw(ArgumentError(\"Valores en `characters` deben ser ascendentes hasta el largo de los c\u00f3digos en base\"))\n end\n # Get the codes list from the FullCPIBase object\n codes = base.codes\n @debug \"Codes:\" codes\n\n # Call lower level tree building function\n upper_nodes = cpi_tree_nodes(codes; characters, base, group_names, group_codes)\n \n # Build upper level tree node\n tree = Group(upperlevel_code, upperlevel_name, upper_nodes)\n tree\nend\n\n\n## Build functions to find the inner tree of a given code\nfunction find_tree(code, tree::Item) \n code == tree.code && return tree \n nothing\nend\n\nfunction find_tree(code, tree::Group)\n # Most basic case: the code is the same as the tree in which to search \n code == tree.code && return tree\n\n # Search in the tree's nodes \n for child in tree.children\n # If code searched is one of the children, return the child\n code == child.code && return child\n\n # Look if the code starts the same as the child's code, i.e the code\n # contains the child's code. If so, find in the inner tree and break out\n # of this level's search\n contains(code, child.code) && return find_tree(code, child)\n end\n\n # Code not found at any level\n nothing\nend\n\n# Redefine getindex to search for specific nodes within the tree\nBase.getindex(tree::Union{Item, Group}, code::AbstractString) = find_tree(code, tree)\n\n\n\n\n## ----------------------------------------------------------------------------\n# Functions to compute any code's price index from the lower level data\n# ----------------------------------------------------------------------------\n\n# Basic case: compute index of an Item, which is stored in the `base` structure\nfunction compute_index(good::Item, base::FullCPIBase)\n i = findfirst(==(good.code), base.codes)\n # Maybe add a warning here if code not found\n base.ipc[:, i]\nend\n\n# Recursive function to compute inner price indexes for groups\nfunction compute_index(group, base::FullCPIBase)\n # Get the indexes of the children. At the lowest level the dispatch will\n # select compute_index(::Item, ::FullCPIBase) to return the Goods indices\n ipcs = mapreduce(c -> compute_index(c, base), hcat, group.children)\n\n # If there exists only one good in the group, that is the group's index\n size(ipcs, 2) == 1 && return ipcs\n \n # Get the weights\n weights = map(c -> c.weight, group.children) \n\n # Return normalized sum product \n ipcs * weights / sum(weights)\nend\n\n# Edge case called when searching for a node that doest not exist. For example,\n# if called compute_index(tree[\"_0101101\"], base) and node with code \"_0101101\"\n# does not exist, returns nothing and raises a warning\nfunction compute_index(::Nothing, ::FullCPIBase)\n @warn \"Nodo no disponible en la estructura\"\n nothing \nend\n\n\n## ----------------------------------------------------------------------------\n# In-place functions to compute any code's price index from the lower level data\n# ----------------------------------------------------------------------------\n\nfunction compute_index!(cache::Dict, good::Item, base::FullCPIBase)\n i = findfirst(==(good.code), base.codes)\n cache[good.code] = base.ipc[:, i] # save a copy in the cache\n cache[good.code]\nend\n\nfunction compute_index!(cache::Dict, group::Group, base::FullCPIBase)\n # If code is available in cache, just return it\n group.code in keys(cache) && return cache[group.code]\n\n # Else, compute the index and store it \n # Get the indexes of the children. At the lowest level the dispatch will\n # select compute_index(::Item, ::FullCPIBase) to return the Goods indices\n ipcs = mapreduce(c -> compute_index!(cache, c, base), hcat, group.children)\n\n # If there exists only one good in the group, that is the group's index\n if size(ipcs, 2) == 1 \n cache[group.code] = ipcs \n return ipcs\n end\n \n # Get the weights\n weights = map(c -> c.weight, group.children) \n\n # Store normalized sum product \n cache[group.code] = ipcs * weights / sum(weights)\n cache[group.code]\nend\n\nfunction compute_index!(::Dict, ::Nothing, ::FullCPIBase)\n @warn \"Nodo no disponible en la estructura\"\n nothing \nend\n\n\n## ----------------------------------------------------------------------------\n# CPITree: estructura contenedora del \u00e1rbol jer\u00e1rquico y los datos necesarios\n# para computar cualquier nodo en la estructura del IPC.\n# ----------------------------------------------------------------------------\n\n\"\"\"\n CPITree\n\n CPITree(base::FullCPIBase, tree::Union{Group, Item}, group_names::Vector{String}, group_codes::Vector{String})\n CPITree(; base::FullCPIBase, groupsdf::DataFrame, characters::(NTuple{N, Int} where N), upperlevel_code = \"_0\", upperlevel_name = \"IPC\")\n\nContenedor envolvente de un \u00e1rbol jer\u00e1rquico del IPC y los datos necesarios de\nlos gastos b\u00e1sicos para computar cualquier jerarqu\u00eda dentro del \u00e1rbol. Permite\nvisualizar y explorar la composici\u00f3n del IPC de un pa\u00eds, as\u00ed como computar los\n\u00edndices de precios de las diferentes jerarqu\u00edas de la estructura del IPC. Est\u00e1\ncompuesto por: \n- Un objeto `base`, de tipo [`FullCPIBase`](@ref), el cual almacena las series\n de tiempo de los \u00edndices de los gastos b\u00e1sicos. \n- Un objeto `tree` que contiene la estructura de nodos del IPC.\n- El vector de nombres `group_names` de los grupos del \u00e1rbol `tree`.\n- El vector de c\u00f3digos `group_codes` de los grupos del \u00e1rbol `tree`.\n\nEl constructor simple requiere una estructura jer\u00e1rquica de nodos como la\ndevuelta por [`get_cpi_tree`](@ref). Al utilizar el constructor con `groupsdf` y\n`characters`, se construye autom\u00e1ticamente un `CPITree` utilizando los c\u00f3digos\ncomo indicadores de la estructura jer\u00e1rquica. Los c\u00f3digos de los gastos b\u00e1sicos\ncontenidos en `base` describen c\u00f3mo se agrupan las jerarqu\u00edas. Por ejemplo, el\nsiguiente `FullCPIBase` contiene 10 gastos b\u00e1sicos: \n```julia-repl\njulia> base\nFullCPIBase{Float32, Float32}: 36 per\u00edodos \u00d7 10 gastos b\u00e1sicos Jan-01-Dec-03\n\u250c\u2500\u2500\u2500\u2500\u2500\u252c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u252c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u252c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u252c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u252c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u252c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u252c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u252c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u252c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u252c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u252c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2510\n\u2502 Row \u2502 Dates \u2502 _011101 \u2502 _011201 \u2502 _011202 \u2502 _021101 \u2502 _022101 \u2502 _031101 \u2502 _041101 \u2502 _041201 \u2502 _041202 \u2502 _041301 \u2502\n\u2502 \u2502 \u2502 Item A \u2502 Item B \u2502 Item C \u2502 Item D \u2502 Item E \u2502 Item F \u2502 Item G \u2502 Item H \u2502 Item I \u2502 Item J \u2502\n\u2502 \u2502 \u2502 11.8947 \u2502 2.39931 \u2502 7.80836 \u2502 1.58585 \u2502 20.141 \u2502 12.0526 \u2502 9.56901 \u2502 14.6256 \u2502 15.0276 \u2502 4.89602 \u2502\n\u251c\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2524\n\u2502 1 \u2502 2001-01-01 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 101.13 \u2502 100.00 \u2502 100.00 \u2502 100.51 \u2502 100.00 \u2502\n\u2502 2 \u2502 2001-02-01 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 102.28 \u2502 100.00 \u2502 100.00 \u2502 101.02 \u2502 100.00 \u2502\n\u2502 3 \u2502 2001-03-01 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 103.44 \u2502 100.00 \u2502 100.00 \u2502 101.53 \u2502 100.00 \u2502\n\u2502 4 \u2502 2001-04-01 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 104.61 \u2502 100.00 \u2502 100.00 \u2502 102.05 \u2502 100.00 \u2502\n\u2502 5 \u2502 2001-05-01 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 105.79 \u2502 100.00 \u2502 100.00 \u2502 102.56 \u2502 100.00 \u2502\n\u2502 \u22ee \u2502 \u22ee \u2502 \u22ee \u2502 \u22ee \u2502 \u22ee \u2502 \u22ee \u2502 \u22ee \u2502 \u22ee \u2502 \u22ee \u2502 \u22ee \u2502 \u22ee \u2502 \u22ee \u2502\n\u2502 32 \u2502 2003-08-01 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 143.39 \u2502 100.00 \u2502 100.00 \u2502 117.59 \u2502 100.00 \u2502\n\u2502 33 \u2502 2003-09-01 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 145.02 \u2502 100.00 \u2502 100.00 \u2502 118.19 \u2502 100.00 \u2502\n\u2502 34 \u2502 2003-10-01 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 146.66 \u2502 100.00 \u2502 100.00 \u2502 118.79 \u2502 100.00 \u2502\n\u2502 35 \u2502 2003-11-01 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 148.32 \u2502 100.00 \u2502 100.00 \u2502 119.39 \u2502 100.00 \u2502\n\u2502 36 \u2502 2003-12-01 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 100.00 \u2502 150.00 \u2502 100.00 \u2502 100.00 \u2502 120.00 \u2502 100.00 \u2502\n\u2514\u2500\u2500\u2500\u2500\u2500\u2534\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2534\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2534\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2534\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2534\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2534\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2534\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2534\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2534\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2534\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2534\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2518\n 26 rows omitted\n```\n\nEn este ejemplo, el argumento debe especificarse como `characters = (3, 4, 5,\n7)`, pues los c\u00f3digos indican las jerarqu\u00edas en el IPC de manera siguiente: \n- Los primeros 3 caracteres indican la jerarqu\u00eda de *divisi\u00f3n de gasto*. \n- El siguiente caracter indica la jerarqu\u00eda de *agrupaci\u00f3n de gasto*. \n- El siguiente caracter indica la jerarqu\u00eda de *subgrupo de gasto*. \n- Los siguientes 2 caracteres indican el *n\u00famero de gasto b\u00e1sico dentro de su\n grupo*. \n\nPor su parte, el DataFrame `groupsdf` debe tener la estructura m\u00ednima siguiente: \n- La primera columna debe ser de tipo `String` y contiene los c\u00f3digos de los\n grupos disponibles en la estructura del IPC. \n- La segunda columna debe ser de tipo `String` y contiene las descripciones o\nnombres de los grupos disponibles en la estructura del IPC. Por ejemplo, el\nDataFrame `groupsdf` puede verse de esta forma: \n```\n17\u00d72 DataFrame\n Row \u2502 code description \n \u2502 String String \n\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\n 1 \u2502 _01 Div._01\n 2 \u2502 _02 Div._02\n 3 \u2502 _03 Div._03\n 4 \u2502 _04 Div._04\n 5 \u2502 _011 Agr._011\n 6 \u2502 _021 Agr._021\n 7 \u2502 _022 Agr._022\n 8 \u2502 _031 Agr._031\n 9 \u2502 _041 Agr._041\n 10 \u2502 _0111 Subgr._0111\n 11 \u2502 _0112 Subgr._0112\n 12 \u2502 _0211 Subgr._0211\n 13 \u2502 _0221 Subgr._0221\n 14 \u2502 _0311 Subgr._0311\n 15 \u2502 _0411 Subgr._0411\n 16 \u2502 _0412 Subgr._0412\n 17 \u2502 _0413 Subgr._0413\n```\n\nPor ejemplo, al construir un `CPITree` con la estructura de c\u00f3digos indicada anteriormente y el DataFrame de ejemplo, el \u00e1rbol del IPC se puede ver de esta forma: \n```julia-repl\njulia> tree = CPITree(; base, groupsdf, characters=(3,4,5,7))\nCPITree{Group{Group{Group{Group{Item{Float32}, Float32}, Float32}, Float32}, Float32}} con datos\n\u2514\u2500\u2192 FullCPIBase{Float32, Float32}: 36 per\u00edodos \u00d7 10 gastos b\u00e1sicos Jan-01-Dec-03\n_0: IPC [100.0]\n\u251c\u2500 _01: Div._01 [21.491905]\n\u2502 \u2514\u2500 _011: Agr._011 [21.491905]\n\u2502 \u251c\u2500 _0111: Subgr._0111 [7.352945]\n\u2502 \u2502 \u2514\u2500 _011101: Item A [7.352945]\n\u2502 \u2514\u2500 _0112: Subgr._0112 [14.13896]\n\u2502 \u251c\u2500 _011201: Item B [6.7442417]\n\u2502 \u2514\u2500 _011202: Item C [7.394718]\n\u251c\u2500 _02: Div._02 [3.0530455]\n\u2502 \u251c\u2500 _021: Agr._021 [1.1036392]\n\u2502 \u2502 \u2514\u2500 _0211: Subgr._0211 [1.1036392]\n\u2502 \u2502 \u2514\u2500 _021101: Item D [1.1036392]\n\u2502 \u2514\u2500 _022: Agr._022 [1.9494063]\n\u2502 \u2514\u2500 _0221: Subgr._0221 [1.9494063]\n\u2502 \u2514\u2500 _022101: Item E [1.9494063]\n\u251c\u2500 _03: Div._03 [11.68543]\n\u2502 \u2514\u2500 _031: Agr._031 [11.68543]\n\u2502 \u2514\u2500 _0311: Subgr._0311 [11.68543]\n\u2502 \u2514\u2500 _031101: Item F [11.68543]\n\u2514\u2500 _04: Div._04 [63.769615]\n \u2514\u2500 _041: Agr._041 [63.769615]\n \u251c\u2500 _0411: Subgr._0411 [16.103952]\n \u2502 \u2514\u2500 _041101: Item G [16.103952]\n \u251c\u2500 _0412: Subgr._0412 [28.824577]\n \u2502 \u251c\u2500 _041201: Item H [11.367162]\n \u2502 \u2514\u2500 _041202: Item I [17.457417]\n \u2514\u2500 _0413: Subgr._0413 [18.841085]\n \u2514\u2500 _041301: Item J [18.841085]\n```\n\"\"\"\nstruct CPITree{G}\n base::FullCPIBase\n tree::G\n group_names::Vector{String}\n group_codes::Vector{String}\n function CPITree(base::FullCPIBase, tree::Union{Group, Item}, group_names::Vector{String}, group_codes::Vector{String})\n G = typeof(tree)\n new{G}(base, tree, group_names, group_codes)\n end\nend\n\nfunction CPITree(; \n base::FullCPIBase, \n groupsdf::DataFrame, \n characters::(NTuple{N, Int} where N),\n upperlevel_code = \"_0\", \n upperlevel_name = \"IPC\")\n\n # Obtener c\u00f3digos y nombres de los grupos en las jerarqu\u00edas\n group_codes = convert.(String, groupsdf[!, 1])\n group_names = groupsdf[!, 2]\n\n tree = get_cpi_tree(;\n base, \n characters,\n group_names, \n group_codes,\n upperlevel_code, \n upperlevel_name\n )\n\n CPITree(base, tree, group_names, group_codes)\nend\n\n\nfunction Base.show(io::IO, cpitree::CPITree)\n println(io, typeof(cpitree), \" con datos\")\n println(io, \"\u2514\u2500\u2192 \", sprint(summary, cpitree.base)) \n print_tree(io, cpitree.tree)\nend\n\n\"\"\"\n Base.getindex(cpitree::CPITree, code::AbstractString)\n\nEste m\u00e9todo se utiliza para indexar el \u00e1rbol jer\u00e1rquico `cpitree` y obtener una\nestructura similar cuyo nodo superior sea el nodo con el c\u00f3digo provisto `code`. Por ejemplo, si tenemos el siguiente \u00e1rbol: \n```julia-repl\njulia> tree\nCPITree{Group{Group{Group{Group{Item{Float32}, Float32}, Float32}, Float32}, Float32}} con datos\n\u2514\u2500\u2192 FullCPIBase{Float32, Float32}: 36 per\u00edodos \u00d7 10 gastos b\u00e1sicos Jan-01-Dec-03\n_0: IPC [100.0]\n\u251c\u2500 _01: Div._01 [21.491905]\n\u2502 \u2514\u2500 _011: Agr._011 [21.491905]\n\u2502 \u251c\u2500 _0111: Subgr._0111 [7.352945]\n\u2502 \u2502 \u2514\u2500 _011101: Item A [7.352945]\n\u2502 \u2514\u2500 _0112: Subgr._0112 [14.13896]\n\u2502 \u251c\u2500 _011201: Item B [6.7442417]\n\u2502 \u2514\u2500 _011202: Item C [7.394718]\n\u251c\u2500 _02: Div._02 [3.0530455]\n\u2502 \u251c\u2500 _021: Agr._021 [1.1036392] \n\u2502 \u2502 \u2514\u2500 _0211: Subgr._0211 [1.1036392]\n\u2502 \u2502 \u2514\u2500 _021101: Item D [1.1036392]\n\u2502 \u2514\u2500 _022: Agr._022 [1.9494063]\n\u2502 \u2514\u2500 _0221: Subgr._0221 [1.9494063]\n\u2502 \u2514\u2500 _022101: Item E [1.9494063]\n\u251c\u2500 _03: Div._03 [11.68543]\n\u2502 \u2514\u2500 _031: Agr._031 [11.68543]\n\u2502 \u2514\u2500 _0311: Subgr._0311 [11.68543]\n\u2502 \u2514\u2500 _031101: Item F [11.68543]\n\u2514\u2500 _04: Div._04 [63.769615]\n \u2514\u2500 _041: Agr._041 [63.769615]\n \u251c\u2500 _0411: Subgr._0411 [16.103952]\n \u2502 \u2514\u2500 _041101: Item G [16.103952]\n \u251c\u2500 _0412: Subgr._0412 [28.824577]\n \u2502 \u251c\u2500 _041201: Item H [11.367162]\n \u2502 \u2514\u2500 _041202: Item I [17.457417]\n \u2514\u2500 _0413: Subgr._0413 [18.841085]\n \u2514\u2500 _041301: Item J [18.841085]\n```\n\nAl indexar por un c\u00f3digo, como `_041`, obtenemos una estructura similar a partir de ese nodo:\n```julia-repl\njulia> tree[\"_041\"]\nCPITree{Group{Group{Item{Float32}, Float32}, Float32}} con datos\n\u2514\u2500\u2192 FullCPIBase{Float32, Float32}: 36 per\u00edodos \u00d7 10 gastos b\u00e1sicos Jan-01-Dec-03\n_041: Agr._041 [63.769615]\n\u251c\u2500 _0411: Subgr._0411 [16.103952]\n\u2502 \u2514\u2500 _041101: Item G [16.103952] \n\u251c\u2500 _0412: Subgr._0412 [28.824577]\n\u2502 \u251c\u2500 _041201: Item H [11.367162]\n\u2502 \u2514\u2500 _041202: Item I [17.457417]\n\u2514\u2500 _0413: Subgr._0413 [18.841085]\n \u2514\u2500 _041301: Item J [18.841085]\n```\n\"\"\"\nfunction Base.getindex(cpitree::CPITree, code::AbstractString) \n node = cpitree.tree[code]\n node === nothing && return nothing\n CPITree(cpitree.base, node, cpitree.group_names, cpitree.group_codes)\nend\n\n\"\"\"\n compute_index(cpitree::CPITree [, code::AbstractString])\n\nPermite computar el \u00edndice de precios de la jerarqu\u00eda provista en `code`. Si se\nomite `code`, se computa la jerarqu\u00eda padre de la estructura `cpitree`. Si el\nnodo no se encuentra en el \u00e1rbol, devuelve `nothing`.\n\n```julia-repl\njulia> tree\nCPITree{Group{Group{Group{Group{Item{Float32}, Float32}, Float32}, Float32}, Float32}} con datos\n\u2514\u2500\u2192 FullCPIBase{Float32, Float32}: 36 per\u00edodos \u00d7 10 gastos b\u00e1sicos Jan-01-Dec-03\n_0: IPC [100.0]\n\u251c\u2500 _01: Div._01 [21.491905]\n\u2502 \u2514\u2500 _011: Agr._011 [21.491905]\n\u2502 \u251c\u2500 _0111: Subgr._0111 [7.352945]\n\u2502 \u2502 \u2514\u2500 _011101: Item A [7.352945]\n\u2502 \u2514\u2500 _0112: Subgr._0112 [14.13896]\n\u2502 \u251c\u2500 _011201: Item B [6.7442417]\n\u2502 \u2514\u2500 _011202: Item C [7.394718]\n\u251c\u2500 _02: Div._02 [3.0530455]\n\u2502 \u251c\u2500 _021: Agr._021 [1.1036392]\n\u2502 \u2502 \u2514\u2500 _0211: Subgr._0211 [1.1036392]\n\u2502 \u2502 \u2514\u2500 _021101: Item D [1.1036392]\n\u2502 \u2514\u2500 _022: Agr._022 [1.9494063]\n\u2502 \u2514\u2500 _0221: Subgr._0221 [1.9494063]\n\u2502 \u2514\u2500 _022101: Item E [1.9494063]\n\u251c\u2500 _03: Div._03 [11.68543]\n\u2502 \u2514\u2500 _031: Agr._031 [11.68543]\n\u2502 \u2514\u2500 _0311: Subgr._0311 [11.68543]\n\u2502 \u2514\u2500 _031101: Item F [11.68543]\n\u2514\u2500 _04: Div._04 [63.769615]\n \u2514\u2500 _041: Agr._041 [63.769615]\n \u251c\u2500 _0411: Subgr._0411 [16.103952]\n \u2502 \u2514\u2500 _041101: Item G [16.103952]\n \u251c\u2500 _0412: Subgr._0412 [28.824577]\n \u2502 \u251c\u2500 _041201: Item H [11.367162]\n \u2502 \u2514\u2500 _041202: Item I [17.457417]\n \u2514\u2500 _0413: Subgr._0413 [18.841085]\n \u2514\u2500 _041301: Item J [18.841085]\n\njulia> compute_index(tree, \"_041\")\n36-element Vector{Float32}:\n 100.13899\n 100.2787\n 100.41912\n 100.560234\n 100.70207\n 100.84464\n 100.98792\n \u22ee\n 104.65377\n 104.81638\n 104.97984\n 105.14412\n 105.309235\n 105.47519\n\njulia> compute_index(tree[\"_041\"])\n36-element Vector{Float32}:\n 100.13899\n 100.2787\n 100.41912\n 100.560234\n 100.70207\n 100.84464\n 100.98792\n \u22ee\n 104.65377\n 104.81638\n 104.97984\n 105.14412\n 105.309235\n 105.47519\n\njulia> a = tree[\"_041302\"]\n\njulia> a === nothing\ntrue\n```\n\"\"\"\nfunction compute_index(cpitree::CPITree, code::AbstractString)\n node = cpitree.tree[code]\n node === nothing && return nothing \n compute_index(node, cpitree.base)\nend\n\n# When single argument, compute the top level node\nfunction compute_index(cpitree::CPITree)\n node = cpitree.tree\n compute_index(node, cpitree.base)\nend\n\n\nchildren(cpitree::CPITree) = children(cpitree.tree)\n\n", "meta": {"hexsha": "1f7a7c62d79591819a6c12faf9a4db010b1e4d0b", "size": 24677, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/tree/CPItree.jl", "max_stars_repo_name": "DIE-BG/CPIDataBase.jl", "max_stars_repo_head_hexsha": "bc541a4217d4ec30c0959238b38a494548798f39", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/tree/CPItree.jl", "max_issues_repo_name": "DIE-BG/CPIDataBase.jl", "max_issues_repo_head_hexsha": "bc541a4217d4ec30c0959238b38a494548798f39", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 5, "max_issues_repo_issues_event_min_datetime": "2022-02-10T03:41:00.000Z", "max_issues_repo_issues_event_max_datetime": "2022-02-24T02:04:02.000Z", "max_forks_repo_path": "src/tree/CPItree.jl", "max_forks_repo_name": "DIE-BG/CPIDataBase.jl", "max_forks_repo_head_hexsha": "bc541a4217d4ec30c0959238b38a494548798f39", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 37.9646153846, "max_line_length": 190, "alphanum_fraction": 0.6274263484, "num_tokens": 8096, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.44167300566462553, "lm_q2_score": 0.13117322546005342, "lm_q1q2_score": 0.057935672751665376}}
{"text": "#\n# ResizableArrays.jl --\n#\n# Implement arrays which are resizable.\n#\n\nmodule ResizableArrays\n\nexport\n ResizableArray,\n ResizableMatrix,\n ResizableVector,\n isgrowable,\n maxlength,\n shrink!\n\nusing Base: elsize, tail, OneTo, throw_boundserror, @propagate_inbounds\n\n\"\"\"\n\n```julia\nResizableArray{T}(undef, dims)\n```\n\nyields a resizable array with uninitialized elements of type `T` and dimensions\n`dims`. Dimensions may be a tuple of integers or a a list of integers. The\nnumber `N` of dimensions may be explicitly specified:\n\n```julia\nResizableArray{T,N}(undef, dims)\n```\n\nTo create an empty resizable array of given rank and element type, call:\n\n```julia\nResizableArray{T,N}()\n```\n\nThe dimensions of a resizable array `A` may be changed by calling\n`resize!(A,dims)` with `dims` the new dimensions. The number of dimensions\nmust remain unchanged but the length of the array may change. Depending on the\ntype of the object backing the storage of the array, it may be possible or not\nto augment the number of elements of the array. When array elements are stored\nin a regular Julia vector, the number of element can always be augmented.\nChanging only the last dimension of a resizable array preserves its contents.\n\nResizable arrays are designed to re-use storage if possible to avoid calling\nthe garbage collector. This may be useful for real-time applications. As a\nconsequence, the storage used by a resizable array `A` can only grow unless\n`skrink!(A)` is called to reduce the storage to the minimum. The call\n`copy(ResizableArray,A)` yields a copy of `A` which is a resizable array.\n\nTo improve performances, call `sizehint!(A,n)` to indicate the minimum number\nof elements to preallocate for `A` (`n` can be a number of elements or array\ndimensions).\n\nThe `ResizableArray` constructor and the `convert` method can be used to to\nconvert an array `A` to a resizable array:\n\n```julia\nResizableArray(A)\nconvert(ResizableArray, A)\n```\n\nElement type `T` and number of dimensions `N` may be specified:\n\n```julia\nResizableArray{T[,N]}(A)\nconvert(ResizableArray{T[,N]}, A)\n```\n\n`N` must match `ndims(A)` but `T` may be different from `eltype(A)`. If\npossible, the `convert` method returns the input array while the\n`ResizableArray` constructor always returns a new instance.\n\nThe default storage for the elements of a resizable array is provided by a\nregular Julia vector. To use an object `buf` to store the elements of a\nresizable array, use one of the following:\n\n```julia\nA = ResizableArray(buf, dims)\nA = ResizableArray{T}(buf, dims)\nA = ResizableArray{T,N}(buf, dims)\n```\n\nThe buffer `buf` must store its elements contiguously using linear indexing\nstyle with 1-based indices and have element type `T`, that is\n`IndexStyle(typeof(buf))` and `eltype(buf)` must yield `IndexLinear()` and `T`\nrespectively. The methods, `IndexStyle`, `eltype`, `length`, `getindex` and\n`setindex!` must be applicable for the type of `buf`. If the method `resize!`\nis applicable for `buf`, the number of elements of `A` can be augmented;\notherwise the maximum number of elements of `A` is `length(buf)`.\n\n!!! warning\n When explictely providing a resizable buffer `buf` for backing the\n storage of a resizable array `A`, you have the responsibility to make\n sure that the same buffer is not resized elsewhere. Otherwise a\n segmentation fault may occur because `A` might assume a wrong buffer\n size. To avoid this, the best is to make sure that only `A` owns `buf`\n and only `A` manages its size. In the current implementation, the size\n of the internal buffer is never reduced so the same buffer may be\n safely shared by different resizable arrays.\n\n\"\"\"\nmutable struct ResizableArray{T,N,B} <: DenseArray{T,N}\n len::Int\n dims::NTuple{N,Int}\n vals::B\n # Inner constructor for provided storage buffer.\n function ResizableArray{T,N}(buf::B,\n dims::NTuple{N,Int}) where {T,N,B}\n eltype(B) === T || error(\"buffer has a different element type\")\n IndexStyle(B) === IndexLinear() ||\n error(\"buffer must have linear indexing style\")\n checkdimensions(dims)\n len = prod(dims)\n length(buf) \u2265 len || error(\"buffer is too small\")\n return new{T,N,B}(len, dims, buf)\n end\n # Inner constructor using regular Julia's vector to store elements.\n function ResizableArray{T,N}(::UndefInitializer,\n dims::NTuple{N,Int}) where {T,N}\n checkdimensions(dims)\n len = prod(dims)\n buf = Vector{T}(undef, len)\n return new{T,N,Vector{T}}(len, dims, buf)\n end\n\nend\n\n# Calling the `ResizableArray` constructor always creates a new instance.\n\nResizableArray(arg, dims::Integer...) =\n ResizableArray(arg, dims)\nResizableArray(arg, dims::Tuple{Vararg{Integer}}) =\n ResizableArray(arg, map(Int, dims))\nResizableArray(buf::B, dims::NTuple{N,Int}) where {N,B} =\n ResizableArray{eltype(B),N}(buf, dims)\n\nResizableArray{T}(arg, dims::Integer...) where {T} =\n ResizableArray{T}(arg, dims)\nResizableArray{T}(arg, dims::Tuple{Vararg{Integer}}) where {T} =\n ResizableArray{T}(arg, map(Int, dims))\nResizableArray{T}(arg, dims::NTuple{N,Int}) where {T,N} =\n ResizableArray{T,N}(arg, dims)\n\nResizableArray{T,N}(arg, dims::Integer...) where {T,N} =\n ResizableArray{T,N}(arg, dims)\nfunction ResizableArray{T,N}(arg, dims::Tuple{Vararg{Integer}}) where {T,N}\n length(dims) == N || _throw_mismatching_number_of_dimensions()\n return ResizableArray{T,N}(arg, map(Int, dims))\nend\n\nResizableArray{T,N,B}(A::AbstractArray) where {T,N,B} =\n (Vector{T} <: B ? ResizableArray{T,N}(A) :\n _throw_invalid_buffer_type(B,T))\nResizableArray{T,N}(A::AbstractArray{<:Any,M}) where {T,N,M} =\n (M == N ? copyto!(ResizableArray{T,N}(undef, size(A)), A) :\n _throw_mismatching_number_of_dimensions())\nResizableArray{T}(A::AbstractArray) where {T} =\n copyto!(ResizableArray{T}(undef, size(A)), A)\nResizableArray(A::AbstractArray{T}) where {T} =\n copyto!(ResizableArray{T}(undef, size(A)), A)\n\n# Constructor for, initially empty, workspace of given rank and element type.\nResizableArray{T,N}() where {T,N} =\n ResizableArray{T,N}(undef, ntuple(i -> 0, Val(N)))\n\n@noinline _throw_mismatching_number_of_dimensions() =\n throw(DimensionMismatch(\"mismatching number of dimensions\"))\n\n# TypeError is more appropriate but we want a specific error message.\n@noinline _throw_invalid_buffer_type(::Type{B},::Type{T}) where {B,T} =\n throw(ErrorException(\"invalid buffer type $B (must be \u2265 Vector{$T})\"))\n\n# Make a resizable copy.\nBase.copy(::Type{ResizableArray}, A::AbstractArray) =\n ResizableArray(A)\nBase.copy(::Type{ResizableArray{T}}, A::AbstractArray) where {T} =\n ResizableArray{T}(A)\nBase.copy(::Type{ResizableArray{T,N}}, A::AbstractArray) where {T,N} =\n ResizableArray{T,N}(A)\nBase.copy(::Type{ResizableArray{T,N,B}}, A::AbstractArray) where {T,N,B} =\n ResizableArray{T,N,B}(A)\n\n# Unlike the `ResizableArray` constructor, calling the `convert` method avoids\n# creating a new instance if possible.\nBase.convert(::Type{ResizableArray{T,N,B}}, A::ResizableArray{T,N,C}) where {T,N,B,C<:B} = A\nBase.convert(::Type{ResizableArray{T,N}}, A::ResizableArray{T,N}) where {T,N} = A\nBase.convert(::Type{ResizableArray{T}}, A::ResizableArray{T}) where {T} = A\nBase.convert(::Type{ResizableArray}, A::ResizableArray) = A\nBase.convert(::Type{ResizableArray{T,N,B}}, A::AbstractArray) where {T,N,B} =\n ResizableArray{T,N,B}(A)\nBase.convert(::Type{ResizableArray{T,N}}, A::AbstractArray) where {T,N} =\n ResizableArray{T,N}(A)\nBase.convert(::Type{ResizableArray{T}}, A::AbstractArray) where {T} =\n ResizableArray{T}(A)\nBase.convert(::Type{ResizableArray}, A::AbstractArray) =\n ResizableArray(A)\n\n\"\"\"\n```julia\nResizableVector{T}\n```\n\nSupertype for one-dimensional resizable arrays with elements of type `T`.\nAlias for [`ResizableArray{T,1}`](@ref).\n\n\"\"\"\nconst ResizableVector{T,B} = ResizableArray{T,1,B}\n\n\"\"\"\n```julia\nResizableMatrix{T}\n```\n\nSupertype for two-dimensional resizable arrays with elements of type `T`.\nAlias for [`ResizableArray{T,2}`](@ref).\n\n\"\"\"\nconst ResizableMatrix{T,B} = ResizableArray{T,2,B}\n\n\"\"\"\n```julia\ncheckdimension(Bool, dim) -> boolean\n```\n\nyields whether `dim` is a valid dimension length (that is a nonnegative\ninteger).\n\n\"\"\"\n@inline checkdimension(::Type{Bool}, dim::Integer) = (dim \u2265 0)\n@inline checkdimension(::Type{Bool}, dim) = false\n\n\"\"\"\n```julia\ncheckdimensions(Bool, dims) -> boolean\n```\n\nyields whether `dims` is a valid list of dimensions.\n\n```julia\ncheckdimensions(dims)\n```\nthrows an error if `dims` is not a valid list of dimensions.\n\n\"\"\"\n@inline checkdimensions(::Type{Bool}, dims::Tuple) =\n checkdimension(Bool, dims[1]) & checkdimensions(Bool, tail(dims))\n@inline checkdimensions(::Type{Bool}, ::Tuple{}) = true\n@inline checkdimensions(dims::Tuple) =\n checkdimensions(Bool, dims) || throw_invalid_dimensions()\n\n@noinline throw_invalid_dimensions() =\n error(\"invalid dimension(s)\")\n\n\"\"\"\n```julia\nisgrowable(x) -> boolean\n```\n\nyields whether `x` is a growable object, that is its size can be augmented.\n\n\"\"\"\nisgrowable(A::ResizableArray) = isgrowable(A.vals)\nisgrowable(A::ResizableArray{T,0}) where {T} = false\nisgrowable(::Vector) = true\nisgrowable(::Any) = false\n\n\"\"\"\n```julia\nmaxlength(A)\n```\n\nyields the maximum number of elements which can be stored in resizable\narray `A` without resizing its internal buffer.\n\nSee also: [`ResizableArray`](@ref).\n\n\"\"\"\nmaxlength(A::ResizableArray) = length(A.vals)\n\nBase.length(A::ResizableArray) = A.len\nBase.size(A::ResizableArray) = A.dims\nBase.size(A::ResizableArray{T,N}, d::Integer) where {T,N} =\n (d > N ? 1 : d \u2265 1 ? A.dims[d] : error(\"out of range dimension\"))\nBase.axes(A::ResizableArray) = map(OneTo, A.dims)\nBase.axes(A::ResizableArray, d::Integer) = Base.OneTo(size(A, d))\n@inline Base.axes1(A::ResizableArray{<:Any,0}) = OneTo(1)\n@inline Base.axes1(A::ResizableArray) = OneTo(A.dims[1])\nBase.IndexStyle(::Type{<:ResizableArray}) = IndexLinear()\nBase.parent(A::ResizableArray) = A.vals\nBase.similar(::Type{ResizableArray{T}}, dims::NTuple{N,Int}) where {T,N} =\n ResizableArray{T,N}(undef, dims)\n\n# Make sizeof() return the number of bytes of the actual contents.\nBase.elsize(::Type{ResizableArray{T,N,B}}) where {T,N,B} = elsize(B)\nBase.sizeof(A::ResizableArray) = elsize(A)*length(A)\n\n# Make ResizableArray's efficient iterators.\n@inline Base.iterate(A::ResizableArray, i=1) =\n ((i % UInt) - 1 < length(A) ? (@inbounds A[i], i + 1) : nothing)\n\n# Equality is false if rank is different.\nBase.:(==)(::ResizableArray, ::AbstractArray) = false\nBase.:(==)(::AbstractArray, ::ResizableArray) = false\nBase.:(==)(::ResizableArray, ::ResizableArray) = false\n\nBase.:(==)(A::ResizableVector{<:Any}, B::ResizableVector{<:Any}) =\n (length(A) == length(B) && _same_elements(A.vals, B.vals, length(A)))\n\nBase.:(==)(A::ResizableArray{<:Any,N}, B::ResizableArray{<:Any,N}) where {N} =\n (size(A) == size(B) && _same_elements(A.vals, B.vals, length(A)))\n\nBase.:(==)(A::ResizableVector{<:Any}, B::Vector{<:Any}) =\n (length(A) == length(B) && _same_elements(A.vals, B, length(A)))\nBase.:(==)(A::Vector{<:Any}, B::ResizableVector{<:Any}) =\n (length(A) == length(B) && _same_elements(A, B.vals, length(A)))\n\nBase.:(==)(A::ResizableArray{<:Any,N}, B::Array{<:Any,N}) where {N} =\n (size(A) == size(B) && _same_elements(A.vals, B, length(A)))\nBase.:(==)(A::Array{<:Any,N}, B::ResizableArray{<:Any,N}) where {N} =\n (size(A) == size(B) && _same_elements(A, B.vals, length(A)))\n\nBase.:(==)(A::ResizableArray{<:Any,N}, B::AbstractArray{<:Any,N}) where {N} =\n (axes(A) == axes(B) && _same_elements(A.vals, B, length(A)))\nBase.:(==)(A::AbstractArray{<:Any,N}, B::ResizableArray{<:Any,N}) where {N} =\n (axes(A) == axes(B) && _same_elements(A, B.vals, length(A)))\n\n# Yields whether the `n` first elements of `A` and `B` are identical. This\n# method is unsafe as it assumes that `A` and `B` have at least `n` elements.\n_same_elements(A, B, n::Integer) =\n _same_elements(IndexStyle(A), A, IndexStyle(B), B, Int(n))\n\nfunction _same_elements(::IndexLinear, A, ::IndexLinear, B, n::Int)\n @inbounds for i in Base.OneTo(n)\n A[i] == B[i] || return false\n end\n return true\nend\n\nfunction _same_elements(::IndexLinear, A, ::IndexStyle, B, n::Int)\n i = 0\n @inbounds for j in eachindex(B)\n (i += 1) \u2264 n || return true\n A[i] == B[j] || return false\n end\n return (i \u2265 n)\nend\n\nfunction _same_elements(::IndexStyle, A, ::IndexLinear, B, n::Int)\n j = 0\n @inbounds for i in eachindex(A)\n (j += 1) \u2264 n || return true\n A[i] == B[j] || return false\n end\n return (j \u2265 n)\nend\n\nfunction _same_elements(::IndexStyle, A, ::IndexStyle, B, n::Int)\n # this case should never occur\n j = 0\n @inbounds for i in eachindex(A,B)\n (j += 1) \u2264 n || return true\n A[i] == B[i] || return false\n end\n return (j \u2265 n)\nend\n\nBase.resize!(A::ResizableArray, dims::Integer...) = resize!(A, dims)\nfunction Base.resize!(A::ResizableArray, dims::Tuple{Vararg{Integer}})\n length(dims) == ndims(A) ||\n error(\"changing the number of dimensions is not allowed\")\n return resize!(A, map(Int, dims))\nend\nfunction Base.resize!(A::ResizableArray{T,N}, dims::NTuple{N,Int}) where {T,N}\n if dims != size(A)\n checkdimensions(dims)\n newlen = prod(dims)\n newlen > length(A.vals) && resize!(A.vals, newlen)\n A.dims = dims\n A.len = newlen\n end\n return A\nend\n\nBase.sizehint!(A::ResizableArray, dims::Integer...) = sizehint!(A, dims)\nfunction Base.sizehint!(A::ResizableArray, dims::Tuple{Vararg{Integer}})\n length(dims) == ndims(A) ||\n error(\"changing the number of dimensions is not allowed\")\n checkdimensions(dims)\n len = prod(dims)\n len > maxlength(A) && sizehint!(parent(A), len)\n return A\nend\nfunction Base.sizehint!(A::ResizableArray, len::Integer)\n len \u2265 0 || throw(ArgumentError(\"number of elements must be nonnegative\"))\n len > maxlength(A) && sizehint!(parent(A), len)\n return A\nend\n\n\"\"\"\n\n```julia\nshrink!(A) -> A\n```\n\nshrinks as much as possible the storage of resizable array `A` and returns `A`.\nCall `copy(ResizableArray,A)` to make a copy of `A` which is a resizable array\nwith skrinked storage.\n\n\"\"\"\nfunction shrink!(A::ResizableArray)\n length(A) < length(A.vals) && resize!(A.vals, length(A))\n return A\nend\n\n\"\"\"\n\n```julia\ngrow!(A, B, prepend=false) -> A\n```\n\ngrows resizable array `A` with the elements of `B` and returns `A`. If\n`prepend` is `true`, the elements of `B` are inserted before those of `A`;\notherwise, the elements of `B` are appended after those of `A`. By default,\n`prepend` is `false`.\n\nAssuming `A` has `N` dimensions, array `B` may have `N` or `N-1` dimensions.\nThe `N-1` leading dimensions of `A` and `B` must be identical and are the\nleading dimensions of the result. If `B` has the same number of dimensions as\n`A`, the last dimension of the result is the sum of the last dimensions of `A`\nand `B`; otherwise, the last dimension of the result is one plus the last\ndimension of `A`.\n\nDepending on argument `prepend`, calling the `grow!` method is equivalent to\ncalling `append!` or `prepend!` methods.\n\nSee also [`ResizableArray`](@ref).\n\n\"\"\"\nfunction grow!(A::ResizableArray{<:Any,N},\n B::AbstractArray{<:Any,M}, prepend::Bool=false) where {N,M}\n N - 1 \u2264 M \u2264 N || throw(DimensionMismatch(\"invalid number of dimensions\"))\n indA = axes(A)\n indB = axes(B)\n @inbounds for d in 1:N-1\n indA[d] == indB[d] ||\n throw(DimensionMismatch(\"leading dimensions must be identical\"))\n end\n dimN = length(indA[N]) + (M == N ? length(indB[N]) : 1)\n lenA = length(A)\n lenB = length(B)\n minlen = lenA + lenB\n buf = A.vals\n length(buf) \u2265 minlen || resize!(buf, minlen)\n if prepend\n copyto!(buf, lenB + 1, buf, 1, lenA)\n copyto!(buf, 1, B, 1, lenB)\n else\n copyto!(buf, lenA + 1, B, 1, lenB)\n end\n A.len = lenA + lenB\n A.dims = ntuple(d -> (d < N ? A.dims[d] : dimN), Val(N))\n return A\nend\n\nBase.append!(dst::ResizableArray, src::AbstractArray) =\n grow!(dst, src, false)\n\nBase.prepend!(dst::ResizableArray, src::AbstractArray) =\n grow!(dst, src, true)\n\n@inline @propagate_inbounds Base.getindex(A::ResizableArray, i::Int) =\n (@boundscheck checkbounds(A, i);\n @inbounds getindex(A.vals, i))\n\n@inline @propagate_inbounds Base.setindex!(A::ResizableArray, x, i::Int) =\n (@boundscheck checkbounds(A, i);\n @inbounds setindex!(A.vals, x, i))\n\n@inline Base.checkbounds(::Type{Bool}, A::ResizableArray, i::Int) =\n (i % UInt) - 1 < length(A)\n\nBase.copyto!(dst::ResizableArray{T}, src::Array{T}) where {T} =\n copyto!(dst, 1, src, 1, length(src))\nBase.copyto!(dst::Array{T}, src::ResizableArray{T}) where {T} =\n copyto!(dst, 1, src, 1, length(src))\nBase.copyto!(dst::ResizableArray{T}, src::ResizableArray{T}) where {T} =\n copyto!(dst, 1, src, 1, length(src))\n\nfunction Base.copyto!(dst::ResizableArray{T}, doff::Integer,\n src::Array{T}, soff::Integer, n::Integer) where {T}\n if n != 0\n checkcopyto(length(dst), doff, length(src), soff, n)\n unsafe_copyto!(dst.vals, doff, src, soff, n)\n end\n return dst\nend\n\nfunction Base.copyto!(dst::Array{T}, doff::Integer,\n src::ResizableArray{T}, soff::Integer,\n n::Integer) where {T}\n if n != 0\n checkcopyto(length(dst), doff, length(src), soff, n)\n unsafe_copyto!(dst, doff, src.vals, soff, n)\n end\n return dst\nend\n\nfunction Base.copyto!(dst::ResizableArray{T}, doff::Integer,\n src::ResizableArray{T}, soff::Integer,\n n::Integer) where {T}\n if n != 0\n checkcopyto(length(dst), doff, length(src), soff, n)\n unsafe_copyto!(dst.vals, doff, src.vals, soff, n)\n end\n return dst\nend\n\n@inline function checkcopyto(dlen::Integer, doff::Integer,\n slen::Integer, soff::Integer, n::Integer)\n @noinline throw_invalid_length() =\n throw(ArgumentError(\"number of elements to copy must be nonnegative\"))\n n \u2265 0 || throw_invalid_length()\n (doff > 0 && doff - 1 + n \u2264 dlen &&\n soff > 0 && soff - 1 + n \u2264 slen) || throw(BoundsError())\nend\n\nBase.unsafe_convert(::Type{Ptr{T}}, A::ResizableArray{T}) where {T} =\n Base.unsafe_convert(Ptr{T}, A.vals)\n\nBase.pointer(A::ResizableArray) = pointer(A.vals)\nBase.pointer(A::ResizableArray, i::Integer) = pointer(A.vals, i)\n\nend # module\n", "meta": {"hexsha": "1fb640696321114ac4372e3129168b0689d6e82f", "size": 18591, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/ResizableArrays.jl", "max_stars_repo_name": "JuliaTagBot/ResizableArrays.jl", "max_stars_repo_head_hexsha": "c2d29fbab36e687dce951d7e7ededc254c4ee783", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/ResizableArrays.jl", "max_issues_repo_name": "JuliaTagBot/ResizableArrays.jl", "max_issues_repo_head_hexsha": "c2d29fbab36e687dce951d7e7ededc254c4ee783", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/ResizableArrays.jl", "max_forks_repo_name": "JuliaTagBot/ResizableArrays.jl", "max_forks_repo_head_hexsha": "c2d29fbab36e687dce951d7e7ededc254c4ee783", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 34.6201117318, "max_line_length": 92, "alphanum_fraction": 0.6662363509, "num_tokens": 5321, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.44167300566462553, "lm_q2_score": 0.13117321527062692, "lm_q1q2_score": 0.05793566825127075}}
{"text": "\"\"\"\n scatterplot(x, y; kwargs...)\n\nDescription\n============\n\nDraws the given points on a new canvas.\n\nThe first (optional) vector `x` should contain the horizontal\npositions for all the points. The second vector `y` should then\ncontain the corresponding vertical positions respectively. This\nmeans that the two vectors must be of the same length and\nordering.\n\nUsage\n======\n\n scatterplot([x], y; name = \"\", marker = :pixel, title = \"\", xlabel = \"\", ylabel = \"\", labels = true, border = :solid, margin = 3, padding = 1, color = :auto, xlim = (0, 0), ylim = (0, 0), canvas = BrailleCanvas, grid = true)\n\nArguments\n==========\n\n- **`x`** : Optional. The horizontal position for each point.\n If omitted, the axes of `y` will be used as `x`.\n\n- **`y`** : The vertical position for each point.\n\n- **`name`** : Annotation of the current drawing to be displayed on the right\n\n- **`marker`** : Choose a marker from $(keys(MARKERS)), a `Char`, a unit length `String` or a vector of these\n\n$DOC_PLOT_PARAMS\n\n- **`height`** : Number of character rows that should be used for plotting.\n\n- **`xlim`** : Plotting range for the x axis.\n `(0, 0)` stands for automatic.\n\n- **`ylim`** : Plotting range for the y axis.\n `(0, 0)` stands for automatic.\n\n- **`canvas`** : The type of canvas that should be used for drawing.\n\n- **`grid`** : If `true`, draws grid-lines at the origin.\n\nReturns\n========\n\nA plot object of type `Plot{T<:Canvas}`\n\nAuthor(s)\n==========\n\n- Christof Stocker (Github: https://github.com/Evizero)\n\nExamples\n=========\n\n```julia-repl\njulia> scatterplot(randn(50), randn(50), title = \"My Scatterplot\")\n My Scatterplot\n \u250c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2510\n 3 \u2502\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2846\u2800\u2800\u2800\u2800\u2800\u2880\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2502\n \u2502\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2847\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2502\n \u2502\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2847\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2810\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2502\n \u2502\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2810\u2800\u2847\u2800\u2800\u2800\u2800\u2800\u2800\u2820\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2502\n \u2502\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2821\u2800\u2800\u2800\u2800\u2800\u2847\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2840\u2801\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2502\n \u2502\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2808\u2800\u2800\u2801\u2800\u2847\u2800\u2800\u2800\u2800\u2800\u2800\u2810\u2800\u2800\u2800\u2800\u2800\u2800\u2802\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2502\n \u2502\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2810\u2804\u2800\u2800\u2800\u2800\u2840\u2800\u2802\u2847\u2800\u2800\u2800\u2802\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2502\n \u2502\u2824\u2824\u2824\u2824\u2824\u2824\u2834\u2824\u2824\u2824\u282c\u2824\u282c\u2824\u2824\u2824\u2867\u28a5\u2864\u2824\u2824\u2864\u2824\u2824\u2824\u282c\u2824\u2824\u2824\u2824\u2824\u2824\u2824\u2824\u2864\u2824\u2824\u2824\u2824\u2804\u2502\n \u2502\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2801\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2857\u2810\u2828\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2502\n \u2502\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2810\u2800\u2800\u2800\u2800\u2847\u2800\u2810\u2800\u2828\u2800\u2800\u2802\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2880\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2502\n \u2502\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2820\u2800\u2804\u2800\u2800\u2800\u2800\u2800\u2847\u2800\u2800\u2800\u2810\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2502\n \u2502\u2800\u2800\u2800\u2800\u2801\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2801\u2802\u2830\u2847\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2502\n \u2502\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2847\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2502\n \u2502\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2847\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2502\n -3 \u2502\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2847\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2800\u2502\n \u2514\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2518\n -2 3\n```\n\nSee also\n=========\n\n[`Plot`](@ref), [`lineplot`](@ref), [`stairs`](@ref),\n[`BrailleCanvas`](@ref), [`BlockCanvas`](@ref),\n[`AsciiCanvas`](@ref), [`DotCanvas`](@ref)\n\"\"\"\nfunction scatterplot(\n x::AbstractVector, y::AbstractVector;\n canvas::Type = BrailleCanvas, color::Union{UserColorType,AbstractVector} = :auto,\n marker::Union{MarkerType,AbstractVector} = :pixel, name = \"\", kw...\n)\n new_plot = Plot(x, y, canvas; kw...)\n scatterplot!(new_plot, x, y; color = color, name = name, marker = marker)\nend\n\nscatterplot(y::AbstractVector; kw...) = scatterplot(axes(y, 1), y; kw...)\n\nfunction scatterplot!(\n plot::Plot{<:Canvas}, x::AbstractVector, y::AbstractVector;\n color::Union{UserColorType,AbstractVector} = :auto,\n marker::Union{MarkerType,AbstractVector} = :pixel, name = \"\"\n)\n color = (color == :auto) ? next_color!(plot) : color\n name == \"\" || label!(plot, :r, string(name), color isa AbstractVector ? color[1] : color)\n if marker \u2208 (:pixel, :auto)\n points!(plot, x, y, color)\n else\n for (xi, yi, mi, ci) in zip(x, y, iterable(marker), iterable(color))\n annotate!(plot, xi, yi, char_marker(mi); color=ci)\n end\n end\n plot\nend\n\nscatterplot!(plot::Plot{<:Canvas}, y::AbstractVector; kw...) = scatterplot!(\n plot, axes(y, 1), y; kw...\n)\n", "meta": {"hexsha": "68fd25bd329897078ae559372d794a96f1b3c303", "size": 3840, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/interface/scatterplot.jl", "max_stars_repo_name": "Mechachleopteryx/UnicodePlots.jl", "max_stars_repo_head_hexsha": "6584706d886744f5609d40ad25c4f1c2e507f919", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 844, "max_stars_repo_stars_event_min_datetime": "2015-08-15T05:22:06.000Z", "max_stars_repo_stars_event_max_datetime": "2021-09-01T11:13:32.000Z", "max_issues_repo_path": "src/interface/scatterplot.jl", "max_issues_repo_name": "Mechachleopteryx/UnicodePlots.jl", "max_issues_repo_head_hexsha": "6584706d886744f5609d40ad25c4f1c2e507f919", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 140, "max_issues_repo_issues_event_min_datetime": "2015-08-16T15:45:45.000Z", "max_issues_repo_issues_event_max_datetime": "2021-09-01T11:22:54.000Z", "max_forks_repo_path": "src/interface/scatterplot.jl", "max_forks_repo_name": "Mechachleopteryx/UnicodePlots.jl", "max_forks_repo_head_hexsha": "6584706d886744f5609d40ad25c4f1c2e507f919", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 59, "max_forks_repo_forks_event_min_datetime": "2015-08-15T02:31:13.000Z", "max_forks_repo_forks_event_max_datetime": "2021-08-16T10:30:02.000Z", "avg_line_length": 32.0, "max_line_length": 228, "alphanum_fraction": 0.48671875, "num_tokens": 1452, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4726834766204329, "lm_q2_score": 0.12252320450794524, "lm_q1q2_score": 0.05791469427349186}}
{"text": "function is_leap_year(year)\n # https://docs.julialang.org/en/v1/manual/control-flow/#Short-Circuit-Evaluation\n return year % 4 == 0 && (year % 100 != 0 || year % 400 == 0)\nend\n", "meta": {"hexsha": "510e4a7ce5da13c94033471a1a1d0e5272056f71", "size": 182, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "exercism/leap.jl", "max_stars_repo_name": "joaopalmeiro/learning-julia", "max_stars_repo_head_hexsha": "12fc7abb712ad94fe1dbf8f02af8a383bfb65750", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "exercism/leap.jl", "max_issues_repo_name": "joaopalmeiro/learning-julia", "max_issues_repo_head_hexsha": "12fc7abb712ad94fe1dbf8f02af8a383bfb65750", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 4, "max_issues_repo_issues_event_min_datetime": "2022-01-07T00:39:37.000Z", "max_issues_repo_issues_event_max_datetime": "2022-01-11T08:46:14.000Z", "max_forks_repo_path": "exercism/leap.jl", "max_forks_repo_name": "joaopalmeiro/learning-julia", "max_forks_repo_head_hexsha": "12fc7abb712ad94fe1dbf8f02af8a383bfb65750", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 36.4, "max_line_length": 84, "alphanum_fraction": 0.6538461538, "num_tokens": 59, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4726834766204328, "lm_q2_score": 0.12252320450794522, "lm_q1q2_score": 0.057914694273491836}}
{"text": "### A Pluto.jl notebook ###\n# v0.17.2\n\nusing Markdown\nusing InteractiveUtils\n\n# This Pluto notebook uses @bind for interactivity. When running this notebook outside of Pluto, the following 'mock version' of @bind gives bound variables a default value (instead of an error).\nmacro bind(def, element)\n quote\n local iv = try Base.loaded_modules[Base.PkgId(Base.UUID(\"6e696c72-6542-2067-7265-42206c756150\"), \"AbstractPlutoDingetjes\")].Bonds.initial_value catch; b -> missing; end\n local el = $(esc(element))\n global $(esc(def)) = Core.applicable(Base.get, el) ? Base.get(el) : iv(el)\n el\n end\nend\n\n# \u2554\u2550\u2561 6c1969e0-02f5-11eb-3fa2-09931a63b1ac\nbegin\n using PoreMatMod, PlutoUI, Bio3DView\n HOME = joinpath(homedir(), \".PoreMatModGO\")\n\trc[:paths][:crystals] = HOME\n\trc[:paths][:moieties] = HOME\n\tif !isdir(HOME)\n\t\tmkdir(HOME)\n\tend\n\tcd(HOME)\nmd\"\"\"\n# \ud83d\udca0 PoreMatModGO \ud83d\ude80\n\nThis notebook interactively substitutes moieties within a crystal using a modified implementation of Ullmann's algorithm to perform substructure searches and applying singular value decomposition to align fragments of the generated materials. Read the docs [here](SimonEnsemble.github.io/PoreMatMod.jl).\n\nSee the original publication on PoreMatMod.jl here: [(article)](https://doi.org/10.33774/chemrxiv-2021-vx5r3) [(GitHub)](https://github.com/SimonEnsemble/PoreMatMod.jl)\n\"\"\"\nend\n\n# \u2554\u2550\u2561 5dc43a20-10b8-11eb-26dc-7fb98e9aeb1a\nmd\"\"\"\nAdrian Henle, [Simon Ensemble](http://simonensemble.github.io), 2021\n\n$(Resource(\"https://simonensemble.github.io/osu_logo.jpg\", :width => 250))\n\"\"\"\n\n# \u2554\u2550\u2561 90696d20-10b7-11eb-20b5-6174faeaf613\n@bind load_inputs Button(\"Reset\")\n\n# \u2554\u2550\u2561 50269ffe-02ef-11eb-0614-f11975d991fe\nbegin load_inputs\n # input fields: query, replacement, xtal\n md\"\"\"\n ##### Input Files\n\n Parent Crystal $(@bind parent_crystal FilePicker())\n\n Search Moiety $(@bind search_moiety FilePicker())\n\n Replace Moiety $(@bind replace_moiety FilePicker())\n \"\"\"\nend\n\n# \u2554\u2550\u2561 33b1fb50-0f73-11eb-2ab2-9d2cb6c5a533\n# write file input strings to files in temp directory\nbegin\n # dict for tracking load status of inputs\n isloaded = Dict([:replacement => false, :query => false, :parent => false])\n # replacement loader\n if !isnothing(replace_moiety)\n write(\"replacement.xyz\", replace_moiety[\"data\"])\n replacement = moiety(\"replacement.xyz\")\n isloaded[:replacement] = true\n end\n # query loader\n if !isnothing(search_moiety)\n write(\"query.xyz\", search_moiety[\"data\"])\n query = moiety(\"query.xyz\")\n isloaded[:query] = true\n end\n # xtal loader\n if !isnothing(parent_crystal)\n write(\"parent.cif\", parent_crystal[\"data\"])\n xtal = Crystal(\"parent.cif\", check_overlap=false)\n Xtals.strip_numbers_from_atom_labels!(xtal)\n infer_bonds!(xtal, true)\n isloaded[:parent] = true\n end\n # run search and display terminal message\n if isloaded[:query] && isloaded[:parent]\n search = query \u2208 xtal\n with_terminal() do\n @info \"Search Results\" isomorphisms=nb_isomorphisms(search) locations=nb_locations(search)\n end\n end\nend\n\n# \u2554\u2550\u2561 415e9210-0f71-11eb-15c8-e7484b5be309\n# choose replacement type\nif all(values(isloaded))\n md\"\"\"\n ### Find/Replace Options\n\n Mode $(@bind replace_mode Select([\"\", \"random replacement at each location\", \"random replacement at n random locations\", \"random replacement at specific locations\", \"specific replacements\"]))\n \"\"\"\nend\n\n# \u2554\u2550\u2561 3997c4d0-0f75-11eb-2976-c161879b8d0c\n# options populated w/ conditional logic based on mode selection\nif all(values(isloaded))\n\tlocal output = nothing\n\tx = [\"$(x)\" for x in 1:nb_locations(search)]\n\tif replace_mode == \"random replacement at each location\"\n\t\toutput = nothing\n\telseif replace_mode == \"random replacement at n random locations\"\n\t\toutput = md\"Number of locations $(@bind nb_loc Slider(1:nb_locations(search)))\"\n\telseif replace_mode == \"random replacement at specific locations\"\n\t\toutput = md\"Locations $(@bind loc MultiSelect(x))\"\n\telseif replace_mode == \"specific replacements\"\n\t\toutput = md\"\"\"\n\tLocations $(@bind loc MultiSelect(x))\n\n\tOrientations $(@bind ori TextField())\n\t\"\"\"\n\tend\n\toutput\nend\n\n# \u2554\u2550\u2561 69edca20-0f94-11eb-13ba-334438ca2406\nif all(values(isloaded))\n\tnew_xtal_flag = true\n\tif replace_mode == \"random replacement at each location\"\n\t\tnew_xtal = substructure_replace(search, replacement)\n\telseif replace_mode == \"random replacement at n random locations\" && nb_loc > 0\n\t\tnew_xtal = substructure_replace(search, replacement, nb_loc=nb_loc)\n\telseif replace_mode == \"random replacement at specific locations\" && loc \u2260 []\n\t\tnew_xtal = substructure_replace(search, replacement, loc=[parse(Int, x) for x in loc])\n\telseif replace_mode == \"specific replacements\"\n\t\tif loc \u2260 [] && ori \u2260 \"\" && length(loc) == length(split(ori, \",\"))\n\t\t\tnew_xtal = substructure_replace(search, replacement,\n\t\t\t\tloc=[parse(Int, x) for x in loc],\n\t\t\t\tori=[parse(Int, x) for x in split(ori, \",\")])\n\t\telse\n\t\t\tnew_xtal_flag = false\n\t\tend\n\telse\n\t\tnew_xtal_flag = false\n\tend\n\tif new_xtal_flag\n\t\twith_terminal() do\n\t\t\tif replace_mode == \"random replacement at each location\"\n\t\t\t\t@info replace_mode new_xtal\n\t\t\telseif replace_mode == \"random replacement at n random locations\"\n\t\t\t\t@info replace_mode nb_loc new_xtal\n\t\t\telseif replace_mode == \"random replacement at specific locations\"\n\t\t\t\t@info replace_mode loc new_xtal\n\t\t\telseif replace_mode == \"specific replacements\"\n\t\t\t\t@info replace_mode loc ori new_xtal\n\t\t\tend\n\t\tend\n\tend\nend;\n\n# \u2554\u2550\u2561 5918f770-103d-11eb-0537-81036bd3e675\nif all(values(isloaded)) && new_xtal_flag\n\twrite_cif(new_xtal, \"crystal.cif\")\n\twrite_xyz(new_xtal, \"atoms.xyz\")\n\twrite_vtk(new_xtal.box, \"unit_cell.vtk\")\n\twrite_bond_information(new_xtal, \"bonds.vtk\")\n\tno_pb = deepcopy(new_xtal)\n\tdrop_cross_pb_bonds!(no_pb)\n\twrite_mol2(new_xtal, filename=\"crystal.mol2\")\n\twrite_mol2(no_pb, filename=\"view.mol2\")\n\tviewfile(\"view.mol2\", \"mol2\", vtkcell=\"unit_cell.vtk\", axes=Axes(4, 0.25))\nend\n\n# \u2554\u2550\u2561 31832e30-1054-11eb-24ed-219fd3e236a1\nif all(values(isloaded)) && new_xtal_flag\n download_cif = DownloadButton(read(\"crystal.cif\"), \"crystal.cif\")\n download_box = DownloadButton(read(\"unit_cell.vtk\"), \"unit_cell.vtk\")\n download_xyz = DownloadButton(read(\"atoms.xyz\"), \"atoms.xyz\")\n download_bonds = DownloadButton(read(\"bonds.vtk\"), \"bonds.vtk\")\n\tdownload_mol2 = DownloadButton(read(\"crystal.mol2\"), \"crystal.mol2\")\nmd\"\"\"\n### Output Files\nComplete Crystal $download_mol2 $download_cif\n\nComponents $download_xyz $download_bonds $download_box\n\"\"\"\nend\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000001\nPLUTO_PROJECT_TOML_CONTENTS = \"\"\"\n[deps]\nBio3DView = \"99c8bb3a-9d13-5280-9740-b4880ed9c598\"\nPlutoUI = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\nPoreMatMod = \"2de0d7f0-0963-4438-8bc8-7e7ffe3dc69a\"\n\n[compat]\nBio3DView = \"~0.1.3\"\nPlutoUI = \"~0.7.14\"\nPoreMatMod = \"~0.2.0\"\n\"\"\"\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000002\nPLUTO_MANIFEST_TOML_CONTENTS = \"\"\"\n# This file is machine-generated - editing it directly is not advised\n\n[[ANSIColoredPrinters]]\ngit-tree-sha1 = \"574baf8110975760d391c710b6341da1afa48d8c\"\nuuid = \"a4c015fc-c6ff-483c-b24f-f7ea428134e9\"\nversion = \"0.0.1\"\n\n[[AbstractPlutoDingetjes]]\ndeps = [\"Pkg\"]\ngit-tree-sha1 = \"abb72771fd8895a7ebd83d5632dc4b989b022b5b\"\nuuid = \"6e696c72-6542-2067-7265-42206c756150\"\nversion = \"1.1.2\"\n\n[[ArgTools]]\nuuid = \"0dad84c5-d112-42e6-8d28-ef12dabb789f\"\n\n[[ArnoldiMethod]]\ndeps = [\"LinearAlgebra\", \"Random\", \"StaticArrays\"]\ngit-tree-sha1 = \"62e51b39331de8911e4a7ff6f5aaf38a5f4cc0ae\"\nuuid = \"ec485272-7323-5ecc-a04f-4719b315124d\"\nversion = \"0.2.0\"\n\n[[Artifacts]]\nuuid = \"56f22d72-fd6d-98f1-02f0-08ddc0907c33\"\n\n[[Base64]]\nuuid = \"2a0f44e3-6c83-55bd-87e4-b1978d98bd5f\"\n\n[[BinaryProvider]]\ndeps = [\"Libdl\", \"Logging\", \"SHA\"]\ngit-tree-sha1 = \"ecdec412a9abc8db54c0efc5548c64dfce072058\"\nuuid = \"b99e7846-7c00-51b0-8f62-c81ae34c0232\"\nversion = \"0.5.10\"\n\n[[Bio3DView]]\ndeps = [\"Requires\"]\ngit-tree-sha1 = \"7f472efd9b6af772307dd017f9deeff2a243754f\"\nuuid = \"99c8bb3a-9d13-5280-9740-b4880ed9c598\"\nversion = \"0.1.3\"\n\n[[CSV]]\ndeps = [\"Dates\", \"Mmap\", \"Parsers\", \"PooledArrays\", \"SentinelArrays\", \"Tables\", \"Unicode\"]\ngit-tree-sha1 = \"b83aa3f513be680454437a0eee21001607e5d983\"\nuuid = \"336ed68f-0bac-5ca0-87d4-7b16caf5d00b\"\nversion = \"0.8.5\"\n\n[[CategoricalArrays]]\ndeps = [\"DataAPI\", \"Future\", \"JSON\", \"Missings\", \"Printf\", \"Statistics\", \"StructTypes\", \"Unicode\"]\ngit-tree-sha1 = \"18d7f3e82c1a80dd38c16453b8fd3f0a7db92f23\"\nuuid = \"324d7699-5711-5eae-9e2f-1d82baa6b597\"\nversion = \"0.9.7\"\n\n[[ChainRulesCore]]\ndeps = [\"Compat\", \"LinearAlgebra\", \"SparseArrays\"]\ngit-tree-sha1 = \"f885e7e7c124f8c92650d61b9477b9ac2ee607dd\"\nuuid = \"d360d2e6-b24c-11e9-a2a3-2a2ae2dbcce4\"\nversion = \"1.11.1\"\n\n[[ChangesOfVariables]]\ndeps = [\"LinearAlgebra\", \"Test\"]\ngit-tree-sha1 = \"9a1d594397670492219635b35a3d830b04730d62\"\nuuid = \"9e997f8a-9a97-42d5-a9f1-ce6bfc15e2c0\"\nversion = \"0.1.1\"\n\n[[Compat]]\ndeps = [\"Base64\", \"Dates\", \"DelimitedFiles\", \"Distributed\", \"InteractiveUtils\", \"LibGit2\", \"Libdl\", \"LinearAlgebra\", \"Markdown\", \"Mmap\", \"Pkg\", \"Printf\", \"REPL\", \"Random\", \"SHA\", \"Serialization\", \"SharedArrays\", \"Sockets\", \"SparseArrays\", \"Statistics\", \"Test\", \"UUIDs\", \"Unicode\"]\ngit-tree-sha1 = \"dce3e3fea680869eaa0b774b2e8343e9ff442313\"\nuuid = \"34da2185-b29b-5c13-b0c7-acf172513d20\"\nversion = \"3.40.0\"\n\n[[Conda]]\ndeps = [\"JSON\", \"VersionParsing\"]\ngit-tree-sha1 = \"299304989a5e6473d985212c28928899c74e9421\"\nuuid = \"8f4d0f93-b110-5947-807f-2305c1781a2d\"\nversion = \"1.5.2\"\n\n[[Configurations]]\ndeps = [\"ExproniconLite\", \"OrderedCollections\", \"TOML\"]\ngit-tree-sha1 = \"79e812c535bb9780ba00f3acba526bde5652eb13\"\nuuid = \"5218b696-f38b-4ac9-8b61-a12ec717816d\"\nversion = \"0.16.6\"\n\n[[Crayons]]\ngit-tree-sha1 = \"3f71217b538d7aaee0b69ab47d9b7724ca8afa0d\"\nuuid = \"a8cc5b0e-0ffa-5ad4-8c14-923d3ee1735f\"\nversion = \"4.0.4\"\n\n[[DataAPI]]\ngit-tree-sha1 = \"cc70b17275652eb47bc9e5f81635981f13cea5c8\"\nuuid = \"9a962f9c-6df0-11e9-0e5d-c546b8b5ee8a\"\nversion = \"1.9.0\"\n\n[[DataFrames]]\ndeps = [\"CategoricalArrays\", \"Compat\", \"DataAPI\", \"Future\", \"InvertedIndices\", \"IteratorInterfaceExtensions\", \"LinearAlgebra\", \"Markdown\", \"Missings\", \"PooledArrays\", \"PrettyTables\", \"Printf\", \"REPL\", \"Reexport\", \"SortingAlgorithms\", \"Statistics\", \"TableTraits\", \"Tables\", \"Unicode\"]\ngit-tree-sha1 = \"d50972453ef464ddcebdf489d11885468b7b83a3\"\nuuid = \"a93c6f00-e57d-5684-b7b6-d8193f3e46c0\"\nversion = \"0.22.7\"\n\n[[DataStructures]]\ndeps = [\"Compat\", \"InteractiveUtils\", \"OrderedCollections\"]\ngit-tree-sha1 = \"7d9d316f04214f7efdbb6398d545446e246eff02\"\nuuid = \"864edb3b-99cc-5e75-8d2d-829cb0a9cfe8\"\nversion = \"0.18.10\"\n\n[[DataValueInterfaces]]\ngit-tree-sha1 = \"bfc1187b79289637fa0ef6d4436ebdfe6905cbd6\"\nuuid = \"e2d170a0-9d28-54be-80f0-106bbe20a464\"\nversion = \"1.0.0\"\n\n[[Dates]]\ndeps = [\"Printf\"]\nuuid = \"ade2ca70-3891-5945-98fb-dc099432e06a\"\n\n[[DelimitedFiles]]\ndeps = [\"Mmap\"]\nuuid = \"8bb1440f-4735-579b-a4ab-409b98df4dab\"\n\n[[Distributed]]\ndeps = [\"Random\", \"Serialization\", \"Sockets\"]\nuuid = \"8ba89e20-285c-5b6f-9357-94700520ee1b\"\n\n[[DocStringExtensions]]\ndeps = [\"LibGit2\"]\ngit-tree-sha1 = \"b19534d1895d702889b219c382a6e18010797f0b\"\nuuid = \"ffbed154-4ef7-542d-bbb7-c09d3a79fcae\"\nversion = \"0.8.6\"\n\n[[Documenter]]\ndeps = [\"ANSIColoredPrinters\", \"Base64\", \"Dates\", \"DocStringExtensions\", \"IOCapture\", \"InteractiveUtils\", \"JSON\", \"LibGit2\", \"Logging\", \"Markdown\", \"REPL\", \"Test\", \"Unicode\"]\ngit-tree-sha1 = \"f425293f7e0acaf9144de6d731772de156676233\"\nuuid = \"e30172f5-a6a5-5a46-863b-614d45cd2de4\"\nversion = \"0.27.10\"\n\n[[Downloads]]\ndeps = [\"ArgTools\", \"LibCURL\", \"NetworkOptions\"]\nuuid = \"f43a241f-c20a-4ad4-852c-f6b1247861c6\"\n\n[[ExproniconLite]]\ngit-tree-sha1 = \"8b08cc88844e4d01db5a2405a08e9178e19e479e\"\nuuid = \"55351af7-c7e9-48d6-89ff-24e801d99491\"\nversion = \"0.6.13\"\n\n[[FIGlet]]\ndeps = [\"BinaryProvider\", \"Pkg\"]\ngit-tree-sha1 = \"bfc6b52f75b4720581e3e49ae786da6764e65b6a\"\nuuid = \"3064a664-84fe-4d92-92c7-ed492f3d8fae\"\nversion = \"0.2.1\"\n\n[[FileIO]]\ndeps = [\"Pkg\", \"Requires\", \"UUIDs\"]\ngit-tree-sha1 = \"2db648b6712831ecb333eae76dbfd1c156ca13bb\"\nuuid = \"5789e2e9-d7fb-5bc7-8068-2c6fae9b9549\"\nversion = \"1.11.2\"\n\n[[FileWatching]]\nuuid = \"7b1f6079-737a-58dc-b8bc-7a2ca5c1b5ee\"\n\n[[Formatting]]\ndeps = [\"Printf\"]\ngit-tree-sha1 = \"8339d61043228fdd3eb658d86c926cb282ae72a8\"\nuuid = \"59287772-0a20-5a39-b81b-1366585eb4c0\"\nversion = \"0.4.2\"\n\n[[FromFile]]\ngit-tree-sha1 = \"81e918d0ed5978fcdacd06b7c64c0c5074c4d55a\"\nuuid = \"ff7dd447-1dcb-4ce3-b8ac-22a812192de7\"\nversion = \"0.1.2\"\n\n[[Future]]\ndeps = [\"Random\"]\nuuid = \"9fa8497b-333b-5362-9e8d-4d0656e87820\"\n\n[[FuzzyCompletions]]\ndeps = [\"REPL\"]\ngit-tree-sha1 = \"2cc2791b324e8ed387a91d7226d17be754e9de61\"\nuuid = \"fb4132e2-a121-4a70-b8a1-d5b831dcdcc2\"\nversion = \"0.4.3\"\n\n[[GitHubActions]]\ndeps = [\"JSON\", \"Logging\"]\ngit-tree-sha1 = \"56e01ec63d13e1cf015d9ff586156eae3cc7cd6f\"\nuuid = \"6b79fd1a-b13a-48ab-b6b0-aaee1fee41df\"\nversion = \"0.1.4\"\n\n[[Graphs]]\ndeps = [\"ArnoldiMethod\", \"DataStructures\", \"Distributed\", \"Inflate\", \"LinearAlgebra\", \"Random\", \"SharedArrays\", \"SimpleTraits\", \"SparseArrays\", \"Statistics\"]\ngit-tree-sha1 = \"92243c07e786ea3458532e199eb3feee0e7e08eb\"\nuuid = \"86223c79-3864-5bf0-83f7-82e725a168b6\"\nversion = \"1.4.1\"\n\n[[HTTP]]\ndeps = [\"Base64\", \"Dates\", \"IniFile\", \"Logging\", \"MbedTLS\", \"NetworkOptions\", \"Sockets\", \"URIs\"]\ngit-tree-sha1 = \"0fa77022fe4b511826b39c894c90daf5fce3334a\"\nuuid = \"cd3eb016-35fb-5094-929b-558a96fad6f3\"\nversion = \"0.9.17\"\n\n[[Hyperscript]]\ndeps = [\"Test\"]\ngit-tree-sha1 = \"8d511d5b81240fc8e6802386302675bdf47737b9\"\nuuid = \"47d2ed2b-36de-50cf-bf87-49c2cf4b8b91\"\nversion = \"0.0.4\"\n\n[[HypertextLiteral]]\ngit-tree-sha1 = \"2b078b5a615c6c0396c77810d92ee8c6f470d238\"\nuuid = \"ac1192a8-f4b3-4bfe-ba22-af5b92cd3ab2\"\nversion = \"0.9.3\"\n\n[[IOCapture]]\ndeps = [\"Logging\", \"Random\"]\ngit-tree-sha1 = \"f7be53659ab06ddc986428d3a9dcc95f6fa6705a\"\nuuid = \"b5f81e59-6552-4d32-b1f0-c071b021bf89\"\nversion = \"0.2.2\"\n\n[[Inflate]]\ngit-tree-sha1 = \"f5fc07d4e706b84f72d54eedcc1c13d92fb0871c\"\nuuid = \"d25df0c9-e2be-5dd7-82c8-3ad0b3e990b9\"\nversion = \"0.1.2\"\n\n[[IniFile]]\ndeps = [\"Test\"]\ngit-tree-sha1 = \"098e4d2c533924c921f9f9847274f2ad89e018b8\"\nuuid = \"83e8ac13-25f8-5344-8a64-a9f2b223428f\"\nversion = \"0.5.0\"\n\n[[InteractiveUtils]]\ndeps = [\"Markdown\"]\nuuid = \"b77e0a4c-d291-57a0-90e8-8db25a27a240\"\n\n[[InverseFunctions]]\ndeps = [\"Test\"]\ngit-tree-sha1 = \"a7254c0acd8e62f1ac75ad24d5db43f5f19f3c65\"\nuuid = \"3587e190-3f89-42d0-90ee-14403ec27112\"\nversion = \"0.1.2\"\n\n[[InvertedIndices]]\ngit-tree-sha1 = \"bee5f1ef5bf65df56bdd2e40447590b272a5471f\"\nuuid = \"41ab1584-1d38-5bbf-9106-f11c6c58b48f\"\nversion = \"1.1.0\"\n\n[[IrrationalConstants]]\ngit-tree-sha1 = \"7fd44fd4ff43fc60815f8e764c0f352b83c49151\"\nuuid = \"92d709cd-6900-40b7-9082-c6be49f344b6\"\nversion = \"0.1.1\"\n\n[[IteratorInterfaceExtensions]]\ngit-tree-sha1 = \"a3f24677c21f5bbe9d2a714f95dcd58337fb2856\"\nuuid = \"82899510-4779-5014-852e-03e436cf321d\"\nversion = \"1.0.0\"\n\n[[JLD2]]\ndeps = [\"DataStructures\", \"FileIO\", \"MacroTools\", \"Mmap\", \"Pkg\", \"Printf\", \"Reexport\", \"TranscodingStreams\", \"UUIDs\"]\ngit-tree-sha1 = \"46b7834ec8165c541b0b5d1c8ba63ec940723ffb\"\nuuid = \"033835bb-8acc-5ee8-8aae-3f567f8a3819\"\nversion = \"0.4.15\"\n\n[[JSON]]\ndeps = [\"Dates\", \"Mmap\", \"Parsers\", \"Unicode\"]\ngit-tree-sha1 = \"8076680b162ada2a031f707ac7b4953e30667a37\"\nuuid = \"682c06a0-de6a-54ab-a142-c8b1cf79cde6\"\nversion = \"0.21.2\"\n\n[[LibCURL]]\ndeps = [\"LibCURL_jll\", \"MozillaCACerts_jll\"]\nuuid = \"b27032c2-a3e7-50c8-80cd-2d36dbcbfd21\"\n\n[[LibCURL_jll]]\ndeps = [\"Artifacts\", \"LibSSH2_jll\", \"Libdl\", \"MbedTLS_jll\", \"Zlib_jll\", \"nghttp2_jll\"]\nuuid = \"deac9b47-8bc7-5906-a0fe-35ac56dc84c0\"\n\n[[LibGit2]]\ndeps = [\"Base64\", \"NetworkOptions\", \"Printf\", \"SHA\"]\nuuid = \"76f85450-5226-5b5a-8eaa-529ad045b433\"\n\n[[LibSSH2_jll]]\ndeps = [\"Artifacts\", \"Libdl\", \"MbedTLS_jll\"]\nuuid = \"29816b5a-b9ab-546f-933c-edad1886dfa8\"\n\n[[Libdl]]\nuuid = \"8f399da3-3557-5675-b5ff-fb832c97cbdb\"\n\n[[LinearAlgebra]]\ndeps = [\"Libdl\"]\nuuid = \"37e2e46d-f89d-539d-b4ee-838fcccc9c8e\"\n\n[[LogExpFunctions]]\ndeps = [\"ChainRulesCore\", \"ChangesOfVariables\", \"DocStringExtensions\", \"InverseFunctions\", \"IrrationalConstants\", \"LinearAlgebra\"]\ngit-tree-sha1 = \"be9eef9f9d78cecb6f262f3c10da151a6c5ab827\"\nuuid = \"2ab3a3ac-af41-5b50-aa03-7779005ae688\"\nversion = \"0.3.5\"\n\n[[Logging]]\nuuid = \"56ddb016-857b-54e1-b83d-db4d58db5568\"\n\n[[MacroTools]]\ndeps = [\"Markdown\", \"Random\"]\ngit-tree-sha1 = \"3d3e902b31198a27340d0bf00d6ac452866021cf\"\nuuid = \"1914dd2f-81c6-5fcd-8719-6d5c9610ff09\"\nversion = \"0.5.9\"\n\n[[Markdown]]\ndeps = [\"Base64\"]\nuuid = \"d6f4376e-aef5-505a-96c1-9c027394607a\"\n\n[[MbedTLS]]\ndeps = [\"Dates\", \"MbedTLS_jll\", \"Random\", \"Sockets\"]\ngit-tree-sha1 = \"1c38e51c3d08ef2278062ebceade0e46cefc96fe\"\nuuid = \"739be429-bea8-5141-9913-cc70e7f3736d\"\nversion = \"1.0.3\"\n\n[[MbedTLS_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"c8ffd9c3-330d-5841-b78e-0817d7145fa1\"\n\n[[MetaGraphs]]\ndeps = [\"Graphs\", \"JLD2\", \"Random\"]\ngit-tree-sha1 = \"2af69ff3c024d13bde52b34a2a7d6887d4e7b438\"\nuuid = \"626554b9-1ddb-594c-aa3c-2596fe9399a5\"\nversion = \"0.7.1\"\n\n[[Missings]]\ndeps = [\"DataAPI\"]\ngit-tree-sha1 = \"f8c673ccc215eb50fcadb285f522420e29e69e1c\"\nuuid = \"e1d29d7a-bbdc-5cf2-9ac0-f12de2c33e28\"\nversion = \"0.4.5\"\n\n[[Mmap]]\nuuid = \"a63ad114-7e13-5084-954f-fe012c677804\"\n\n[[MozillaCACerts_jll]]\nuuid = \"14a3606d-f60d-562e-9121-12d972cd8159\"\n\n[[MsgPack]]\ndeps = [\"Serialization\"]\ngit-tree-sha1 = \"a8cbf066b54d793b9a48c5daa5d586cf2b5bd43d\"\nuuid = \"99f44e22-a591-53d1-9472-aa23ef4bd671\"\nversion = \"1.1.0\"\n\n[[NetworkOptions]]\nuuid = \"ca575930-c2e3-43a9-ace4-1e988b2c1908\"\n\n[[OrderedCollections]]\ngit-tree-sha1 = \"85f8e6578bf1f9ee0d11e7bb1b1456435479d47c\"\nuuid = \"bac558e1-5e72-5ebc-8fee-abe8a469f55d\"\nversion = \"1.4.1\"\n\n[[Parsers]]\ndeps = [\"Dates\"]\ngit-tree-sha1 = \"bfd7d8c7fd87f04543810d9cbd3995972236ba1b\"\nuuid = \"69de0a69-1ddd-5017-9359-2bf0b02dc9f0\"\nversion = \"1.1.2\"\n\n[[Pkg]]\ndeps = [\"Artifacts\", \"Dates\", \"Downloads\", \"LibGit2\", \"Libdl\", \"Logging\", \"Markdown\", \"Printf\", \"REPL\", \"Random\", \"SHA\", \"Serialization\", \"TOML\", \"Tar\", \"UUIDs\", \"p7zip_jll\"]\nuuid = \"44cfe95a-1eb2-52ea-b672-e2afdf69b78f\"\n\n[[Pluto]]\ndeps = [\"Base64\", \"Configurations\", \"Dates\", \"Distributed\", \"FileWatching\", \"FuzzyCompletions\", \"HTTP\", \"InteractiveUtils\", \"Logging\", \"Markdown\", \"MsgPack\", \"Pkg\", \"REPL\", \"Sockets\", \"TableIOInterface\", \"Tables\", \"UUIDs\"]\ngit-tree-sha1 = \"a5b3fee95de0c0a324bab53a03911395936d15d9\"\nuuid = \"c3e4b0f8-55cb-11ea-2926-15256bba5781\"\nversion = \"0.17.2\"\n\n[[PlutoSliderServer]]\ndeps = [\"Base64\", \"Configurations\", \"Distributed\", \"FromFile\", \"GitHubActions\", \"HTTP\", \"Logging\", \"Pkg\", \"Pluto\", \"SHA\", \"Sockets\", \"TOML\", \"UUIDs\"]\ngit-tree-sha1 = \"ed9660bb2c9eee9d389601bd80a10cee3dd64f0b\"\nuuid = \"2fc8631c-6f24-4c5b-bca7-cbb509c42db4\"\nversion = \"0.2.7\"\n\n[[PlutoUI]]\ndeps = [\"AbstractPlutoDingetjes\", \"Base64\", \"Dates\", \"Hyperscript\", \"HypertextLiteral\", \"IOCapture\", \"InteractiveUtils\", \"JSON\", \"Logging\", \"Markdown\", \"Random\", \"Reexport\", \"UUIDs\"]\ngit-tree-sha1 = \"b68904528fd538f1cb6a3fbc44d2abdc498f9e8e\"\nuuid = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\nversion = \"0.7.21\"\n\n[[PooledArrays]]\ndeps = [\"DataAPI\", \"Future\"]\ngit-tree-sha1 = \"db3a23166af8aebf4db5ef87ac5b00d36eb771e2\"\nuuid = \"2dfb63ee-cc39-5dd5-95bd-886bf059d720\"\nversion = \"1.4.0\"\n\n[[PoreMatMod]]\ndeps = [\"Bio3DView\", \"CSV\", \"DataFrames\", \"FIGlet\", \"Graphs\", \"LinearAlgebra\", \"MetaGraphs\", \"PlutoSliderServer\", \"PlutoUI\", \"Reexport\", \"StatsBase\", \"Xtals\"]\ngit-tree-sha1 = \"e4b40bedba7a4aadefe93b52b809de10539bc915\"\nuuid = \"2de0d7f0-0963-4438-8bc8-7e7ffe3dc69a\"\nversion = \"0.2.6\"\n\n[[PrettyTables]]\ndeps = [\"Crayons\", \"Formatting\", \"Markdown\", \"Reexport\", \"Tables\"]\ngit-tree-sha1 = \"574a6b3ea95f04e8757c0280bb9c29f1a5e35138\"\nuuid = \"08abe8d2-0d0c-5749-adfa-8a2ac140af0d\"\nversion = \"0.11.1\"\n\n[[Printf]]\ndeps = [\"Unicode\"]\nuuid = \"de0858da-6303-5e67-8744-51eddeeeb8d7\"\n\n[[PyCall]]\ndeps = [\"Conda\", \"Dates\", \"Libdl\", \"LinearAlgebra\", \"MacroTools\", \"Serialization\", \"VersionParsing\"]\ngit-tree-sha1 = \"4ba3651d33ef76e24fef6a598b63ffd1c5e1cd17\"\nuuid = \"438e738f-606a-5dbb-bf0a-cddfbfd45ab0\"\nversion = \"1.92.5\"\n\n[[REPL]]\ndeps = [\"InteractiveUtils\", \"Markdown\", \"Sockets\", \"Unicode\"]\nuuid = \"3fa0cd96-eef1-5676-8a61-b3b8758bbffb\"\n\n[[Random]]\ndeps = [\"Serialization\"]\nuuid = \"9a3f8284-a2c9-5f02-9a11-845980a1fd5c\"\n\n[[Reexport]]\ngit-tree-sha1 = \"45e428421666073eab6f2da5c9d310d99bb12f9b\"\nuuid = \"189a3867-3050-52da-a836-e630ba90ab69\"\nversion = \"1.2.2\"\n\n[[Requires]]\ndeps = [\"UUIDs\"]\ngit-tree-sha1 = \"4036a3bd08ac7e968e27c203d45f5fff15020621\"\nuuid = \"ae029012-a4dd-5104-9daa-d747884805df\"\nversion = \"1.1.3\"\n\n[[SHA]]\nuuid = \"ea8e919c-243c-51af-8825-aaa63cd721ce\"\n\n[[SentinelArrays]]\ndeps = [\"Dates\", \"Random\"]\ngit-tree-sha1 = \"f45b34656397a1f6e729901dc9ef679610bd12b5\"\nuuid = \"91c51154-3ec4-41a3-a24f-3f23e20d615c\"\nversion = \"1.3.8\"\n\n[[Serialization]]\nuuid = \"9e88b42a-f829-5b0c-bbe9-9e923198166b\"\n\n[[SharedArrays]]\ndeps = [\"Distributed\", \"Mmap\", \"Random\", \"Serialization\"]\nuuid = \"1a1011a3-84de-559e-8e89-a11a2f7dc383\"\n\n[[SimpleTraits]]\ndeps = [\"InteractiveUtils\", \"MacroTools\"]\ngit-tree-sha1 = \"5d7e3f4e11935503d3ecaf7186eac40602e7d231\"\nuuid = \"699a6c99-e7fa-54fc-8d76-47d257e15c1d\"\nversion = \"0.9.4\"\n\n[[Sockets]]\nuuid = \"6462fe0b-24de-5631-8697-dd941f90decc\"\n\n[[SortingAlgorithms]]\ndeps = [\"DataStructures\", \"Random\", \"Test\"]\ngit-tree-sha1 = \"03f5898c9959f8115e30bc7226ada7d0df554ddd\"\nuuid = \"a2af1166-a08f-5f64-846c-94a0d3cef48c\"\nversion = \"0.3.1\"\n\n[[SparseArrays]]\ndeps = [\"LinearAlgebra\", \"Random\"]\nuuid = \"2f01184e-e22b-5df5-ae63-d93ebab69eaf\"\n\n[[StaticArrays]]\ndeps = [\"LinearAlgebra\", \"Random\", \"Statistics\"]\ngit-tree-sha1 = \"3c76dde64d03699e074ac02eb2e8ba8254d428da\"\nuuid = \"90137ffa-7385-5640-81b9-e52037218182\"\nversion = \"1.2.13\"\n\n[[Statistics]]\ndeps = [\"LinearAlgebra\", \"SparseArrays\"]\nuuid = \"10745b16-79ce-11e8-11f9-7d13ad32a3b2\"\n\n[[StatsAPI]]\ngit-tree-sha1 = \"0f2aa8e32d511f758a2ce49208181f7733a0936a\"\nuuid = \"82ae8749-77ed-4fe6-ae5f-f523153014b0\"\nversion = \"1.1.0\"\n\n[[StatsBase]]\ndeps = [\"DataAPI\", \"DataStructures\", \"LinearAlgebra\", \"LogExpFunctions\", \"Missings\", \"Printf\", \"Random\", \"SortingAlgorithms\", \"SparseArrays\", \"Statistics\", \"StatsAPI\"]\ngit-tree-sha1 = \"2bb0cb32026a66037360606510fca5984ccc6b75\"\nuuid = \"2913bbd2-ae8a-5f71-8c99-4fb6c76f3a91\"\nversion = \"0.33.13\"\n\n[[StructTypes]]\ndeps = [\"Dates\", \"UUIDs\"]\ngit-tree-sha1 = \"d24a825a95a6d98c385001212dc9020d609f2d4f\"\nuuid = \"856f2bd8-1eba-4b0a-8007-ebc267875bd4\"\nversion = \"1.8.1\"\n\n[[TOML]]\ndeps = [\"Dates\"]\nuuid = \"fa267f1f-6049-4f14-aa54-33bafae1ed76\"\n\n[[TableIOInterface]]\ngit-tree-sha1 = \"9a0d3ab8afd14f33a35af7391491ff3104401a35\"\nuuid = \"d1efa939-5518-4425-949f-ab857e148477\"\nversion = \"0.1.6\"\n\n[[TableTraits]]\ndeps = [\"IteratorInterfaceExtensions\"]\ngit-tree-sha1 = \"c06b2f539df1c6efa794486abfb6ed2022561a39\"\nuuid = \"3783bdb8-4a98-5b6b-af9a-565f29a5fe9c\"\nversion = \"1.0.1\"\n\n[[Tables]]\ndeps = [\"DataAPI\", \"DataValueInterfaces\", \"IteratorInterfaceExtensions\", \"LinearAlgebra\", \"TableTraits\", \"Test\"]\ngit-tree-sha1 = \"fed34d0e71b91734bf0a7e10eb1bb05296ddbcd0\"\nuuid = \"bd369af6-aec1-5ad0-b16a-f7cc5008161c\"\nversion = \"1.6.0\"\n\n[[Tar]]\ndeps = [\"ArgTools\", \"SHA\"]\nuuid = \"a4e569a6-e804-4fa4-b0f3-eef7a1d5b13e\"\n\n[[Test]]\ndeps = [\"InteractiveUtils\", \"Logging\", \"Random\", \"Serialization\"]\nuuid = \"8dfed614-e22c-5e08-85e1-65c5234f0b40\"\n\n[[TranscodingStreams]]\ndeps = [\"Random\", \"Test\"]\ngit-tree-sha1 = \"216b95ea110b5972db65aa90f88d8d89dcb8851c\"\nuuid = \"3bb67fe8-82b1-5028-8e26-92a6c54297fa\"\nversion = \"0.9.6\"\n\n[[URIs]]\ngit-tree-sha1 = \"97bbe755a53fe859669cd907f2d96aee8d2c1355\"\nuuid = \"5c2747f8-b7ea-4ff2-ba2e-563bfd36b1d4\"\nversion = \"1.3.0\"\n\n[[UUIDs]]\ndeps = [\"Random\", \"SHA\"]\nuuid = \"cf7118a7-6976-5b1a-9a39-7adc72f591a4\"\n\n[[Unicode]]\nuuid = \"4ec0a83e-493e-50e2-b9ac-8f72acf5a8f5\"\n\n[[VersionParsing]]\ngit-tree-sha1 = \"e575cf85535c7c3292b4d89d89cc29e8c3098e47\"\nuuid = \"81def892-9a0e-5fdd-b105-ffc91e053289\"\nversion = \"1.2.1\"\n\n[[Xtals]]\ndeps = [\"Bio3DView\", \"CSV\", \"DataFrames\", \"Documenter\", \"FIGlet\", \"Graphs\", \"JLD2\", \"LinearAlgebra\", \"Logging\", \"MetaGraphs\", \"Printf\", \"PyCall\", \"UUIDs\"]\ngit-tree-sha1 = \"3147503cd35c4f2b3744fe36301c7de3efee98c5\"\nuuid = \"ede5f01d-793e-4c47-9885-c447d1f18d6d\"\nversion = \"0.3.9\"\n\n[[Zlib_jll]]\ndeps = [\"Libdl\"]\nuuid = \"83775a58-1f1d-513f-b197-d71354ab007a\"\n\n[[nghttp2_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"8e850ede-7688-5339-a07c-302acd2aaf8d\"\n\n[[p7zip_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"3f19e933-33d8-53b3-aaab-bd5110c3b7a0\"\n\"\"\"\n\n# \u2554\u2550\u2561 Cell order:\n# \u255f\u25006c1969e0-02f5-11eb-3fa2-09931a63b1ac\n# \u255f\u250050269ffe-02ef-11eb-0614-f11975d991fe\n# \u255f\u250033b1fb50-0f73-11eb-2ab2-9d2cb6c5a533\n# \u255f\u2500415e9210-0f71-11eb-15c8-e7484b5be309\n# \u255f\u25003997c4d0-0f75-11eb-2976-c161879b8d0c\n# \u255f\u250069edca20-0f94-11eb-13ba-334438ca2406\n# \u255f\u25005918f770-103d-11eb-0537-81036bd3e675\n# \u255f\u250031832e30-1054-11eb-24ed-219fd3e236a1\n# \u255f\u25005dc43a20-10b8-11eb-26dc-7fb98e9aeb1a\n# \u255f\u250090696d20-10b7-11eb-20b5-6174faeaf613\n# \u255f\u250000000000-0000-0000-0000-000000000001\n# \u255f\u250000000000-0000-0000-0000-000000000002\n", "meta": {"hexsha": "a4d1c13bea5627c2532e315d6d5ec0edc1ca2a2b", "size": 24746, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/PoreMatModGO.jl", "max_stars_repo_name": "eahenle/PoreMatMod.jl", "max_stars_repo_head_hexsha": "e23345111708563f82d7e92d771af3f1dc578f8b", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 4, "max_stars_repo_stars_event_min_datetime": "2021-09-04T11:47:47.000Z", "max_stars_repo_stars_event_max_datetime": "2022-01-19T01:03:18.000Z", "max_issues_repo_path": "src/PoreMatModGO.jl", "max_issues_repo_name": "eahenle/PoreMatMod.jl", "max_issues_repo_head_hexsha": "e23345111708563f82d7e92d771af3f1dc578f8b", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 41, "max_issues_repo_issues_event_min_datetime": "2021-09-06T18:04:41.000Z", "max_issues_repo_issues_event_max_datetime": "2022-03-30T19:13:39.000Z", "max_forks_repo_path": "src/PoreMatModGO.jl", "max_forks_repo_name": "eahenle/PoreMatMod.jl", "max_forks_repo_head_hexsha": "e23345111708563f82d7e92d771af3f1dc578f8b", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 2, "max_forks_repo_forks_event_min_datetime": "2022-02-08T19:41:50.000Z", "max_forks_repo_forks_event_max_datetime": "2022-02-20T06:08:50.000Z", "avg_line_length": 32.1794538362, "max_line_length": 303, "alphanum_fraction": 0.7291683504, "num_tokens": 10232, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.45326184801538616, "lm_q2_score": 0.1276526369372765, "lm_q1q2_score": 0.05786007012222709}}
{"text": "export AbstractPolynomial\n\nconst SymbolLike = Union{AbstractString,Char,Symbol}\n\n\"\"\"\n AbstractPolynomial{T}\n\nAn abstract container for various polynomials. \n\n# Properties\n- `coeffs` - The coefficients of the polynomial\n- `var` - The indeterminate of the polynomial\n\"\"\"\nabstract type AbstractPolynomial{T} end\n\n# We want \u27d2(P{\u03b1\u2026,T}) = P{\u03b1\u2026}; this default\n# works for most cases\n\u27d2(P::Type{<:AbstractPolynomial}) = constructorof(P)\n\n# convert `as` into polynomial of type P based on instance, inheriting variable\n# (and for LaurentPolynomial the offset)\n_convert(p::P, as) where {P <: AbstractPolynomial} = \u27d2(P)(as, p.var)\n\n\"\"\"\n Polynomials.@register(name)\n\nGiven a polynomial with `name`, creates some common convenience constructors and conversions to minimize code required for implementation of a new polynomial type.\n\n# Example\n```julia\nstruct MyPolynomial{T} <: AbstractPolynomial{T} end\n\nPolynomials.@register MyPolynomial\n```\n\n# Implementations\nThis will implement simple self-conversions like `convert(::Type{MyPoly}, p::MyPoly) = p` and creates two promote rules. The first allows promotion between two types (e.g. `promote(Polynomial, ChebyshevT)`) and the second allows promotion between parametrized types (e.g. `promote(Polynomial{T}, Polynomial{S})`). \n\nFor constructors, it implements the shortcut for `MyPoly(...) = MyPoly{T}(...)`, singleton constructor `MyPoly(x::Number, ...)`, conversion constructor `MyPoly{T}(n::S, ...)`, and `variable` alternative `MyPoly(var=:x)`.\n\"\"\"\nmacro register(name)\n poly = esc(name)\n quote\n Base.convert(::Type{P}, p::P) where {P<:$poly} = p\n Base.convert(P::Type{<:$poly}, p::$poly{T}) where {T} = P(coeffs(p), p.var)\n Base.promote(p::P, q::Q) where {T, P <:$poly{T}, Q <: $poly{T}} = p,q\n Base.promote_rule(::Type{<:$poly{T}}, ::Type{<:$poly{S}}) where {T,S} =\n $poly{promote_type(T, S)}\n Base.promote_rule(::Type{<:$poly{T}}, ::Type{S}) where {T,S<:Number} =\n $poly{promote_type(T, S)}\n $poly(coeffs::AbstractVector{T}, var::SymbolLike = :x) where {T} =\n $poly{T}(coeffs, Symbol(var))\n $poly{T}(x::AbstractVector{S}, var::SymbolLike = :x) where {T,S<:Number} =\n $poly(T.(x), Symbol(var))\n function $poly(coeffs::G, var::SymbolLike=:x) where {G}\n !Base.isiterable(G) && throw(ArgumentError(\"coeffs is not iterable\"))\n $poly(collect(coeffs), var)\n end\n $poly{T}(n::S, var::SymbolLike = :x) where {T, S<:Number} =\n n * one($poly{T}, Symbol(var))\n $poly(n::S, var::SymbolLike = :x) where {S <: Number} = n * one($poly{S}, Symbol(var))\n $poly{T}(var::SymbolLike=:x) where {T} = variable($poly{T}, Symbol(var))\n $poly(var::SymbolLike=:x) = variable($poly, Symbol(var))\n end\nend\n\n\nmacro registerN(name, params...)\n poly = esc(name)\n \u03b1s = tuple(esc.(params)...)\n quote\n Base.convert(::Type{P}, q::Q) where {$(\u03b1s...),T, P<:$poly{$(\u03b1s...),T}, Q <: $poly{$(\u03b1s...),T}} = q\n Base.convert(::Type{$poly{$(\u03b1s...)}}, q::Q) where {$(\u03b1s...),T, Q <: $poly{$(\u03b1s...),T}} = q \n Base.promote(p::P, q::Q) where {$(\u03b1s...),T, P <:$poly{$(\u03b1s...),T}, Q <: $poly{$(\u03b1s...),T}} = p,q\n Base.promote_rule(::Type{<:$poly{$(\u03b1s...),T}}, ::Type{<:$poly{$(\u03b1s...),S}}) where {$(\u03b1s...),T,S} =\n $poly{$(\u03b1s...),promote_type(T, S)}\n Base.promote_rule(::Type{<:$poly{$(\u03b1s...),T}}, ::Type{S}) where {$(\u03b1s...),T,S<:Number} = \n $poly{$(\u03b1s...),promote_type(T,S)}\n\n function $poly{$(\u03b1s...),T}(x::AbstractVector{S}, var::SymbolLike = :x) where {$(\u03b1s...),T,S}\n $poly{$(\u03b1s...),T}(T.(x), Symbol(var))\n end\n $poly{$(\u03b1s...)}(coeffs::AbstractVector{T}, var::SymbolLike=:x) where {$(\u03b1s...),T} =\n $poly{$(\u03b1s...),T}(coeffs, Symbol(var))\n $poly{$(\u03b1s...),T}(n::Number, var::SymbolLike = :x) where {$(\u03b1s...),T} = n*one($poly{$(\u03b1s...),T}, Symbol(var))\n $poly{$(\u03b1s...)}(n::Number, var::SymbolLike = :x) where {$(\u03b1s...)} = n*one($poly{$(\u03b1s...)}, Symbol(var))\n $poly{$(\u03b1s...),T}(var::SymbolLike=:x) where {$(\u03b1s...), T} = variable($poly{$(\u03b1s...),T}, Symbol(var))\n $poly{$(\u03b1s...)}(var::SymbolLike=:x) where {$(\u03b1s...)} = variable($poly{$(\u03b1s...)}, Symbol(var))\n end\nend\n\n\n# deprecated. If desired, replace with @registerN type parameters... macro\n# Macros to register POLY{\u03b1, T} and POLY{\u03b1, \u03b2, T}\nmacro register1(name)\n @warn \"@register1 is deprecated use @registerN\"\n poly = esc(name)\n quote\n Base.convert(::Type{P}, p::P) where {P<:$poly} = p\n Base.promote(p::P, q::Q) where {\u03b1,T, P <:$poly{\u03b1,T}, Q <: $poly{\u03b1,T}} = p,q\n Base.promote_rule(::Type{<:$poly{\u03b1,T}}, ::Type{<:$poly{\u03b1,S}}) where {\u03b1,T,S} =\n $poly{\u03b1,promote_type(T, S)}\n Base.promote_rule(::Type{<:$poly{\u03b1,T}}, ::Type{S}) where {\u03b1,T,S<:Number} = \n $poly{\u03b1,promote_type(T,S)}\n function $poly{\u03b1,T}(x::AbstractVector{S}, var::SymbolLike = :x) where {\u03b1,T,S}\n $poly{\u03b1,T}(T.(x), Symbol(var))\n end\n $poly{\u03b1}(coeffs::AbstractVector{T}, var::SymbolLike=:x) where {\u03b1,T} =\n $poly{\u03b1,T}(coeffs, Symbol(var))\n $poly{\u03b1,T}(n::Number, var::SymbolLike = :x) where {\u03b1,T} = n*one($poly{\u03b1,T}, Symbol(var))\n $poly{\u03b1}(n::Number, var::SymbolLike = :x) where {\u03b1} = n*one($poly{\u03b1}, Symbol(var))\n $poly{\u03b1,T}(var::SymbolLike=:x) where {\u03b1, T} = variable($poly{\u03b1,T}, Symbol(var))\n $poly{\u03b1}(var::SymbolLike=:x) where {\u03b1} = variable($poly{\u03b1}, Symbol(var))\n end\nend\n\n\n# Macro to register POLY{\u03b1, \u03b2, T}\nmacro register2(name)\n @warn \"@register2 is deprecated use @registerN\"\n poly = esc(name)\n quote\n Base.convert(::Type{P}, p::P) where {P<:$poly} = p\n Base.promote(p::P, q::Q) where {\u03b1,\u03b2,T, P <:$poly{\u03b1,\u03b2,T}, Q <: $poly{\u03b1,\u03b2,T}} = p,q \n Base.promote_rule(::Type{<:$poly{\u03b1,\u03b2,T}}, ::Type{<:$poly{\u03b1,\u03b2,S}}) where {\u03b1,\u03b2,T,S} =\n $poly{\u03b1,\u03b2,promote_type(T, S)}\n Base.promote_rule(::Type{<:$poly{\u03b1,\u03b2,T}}, ::Type{S}) where {\u03b1,\u03b2,T,S<:Number} =\n $poly{\u03b1,\u03b2,promote_type(T, S)}\n $poly{\u03b1,\u03b2}(coeffs::AbstractVector{T}, var::SymbolLike = :x) where {\u03b1,\u03b2,T} =\n $poly{\u03b1,\u03b2,T}(coeffs, Symbol(var))\n $poly{\u03b1,\u03b2,T}(x::AbstractVector{S}, var::SymbolLike = :x) where {\u03b1,\u03b2,T,S<:Number} = $poly{\u03b1,\u03b2,T}(T.(x), var)\n $poly{\u03b1,\u03b2,T}(n::Number, var::SymbolLike = :x) where {\u03b1,\u03b2,T} = n*one($poly{\u03b1,\u03b2,T}, Symbol(var))\n $poly{\u03b1,\u03b2}(n::Number, va::SymbolLiker = :x) where {\u03b1,\u03b2} = n*one($poly{\u03b1,\u03b2}, Symbol(var))\n $poly{\u03b1,\u03b2,T}(var::SymbolLike=:x) where {\u03b1,\u03b2, T} = variable($poly{\u03b1,\u03b2,T}, Symbol(var))\n $poly{\u03b1,\u03b2}(var::SymbolLike=:x) where {\u03b1,\u03b2} = variable($poly{\u03b1,\u03b2}, Symbol(var))\n end\nend\n\n", "meta": {"hexsha": "4bb0a1007d1f9c215d013ba81ccc25831b17c6ce", "size": 6746, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/abstract.jl", "max_stars_repo_name": "michakraus/Polynomials.jl", "max_stars_repo_head_hexsha": "8b6a3d282d8f67cfca219ec91aa4076257a16ff0", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/abstract.jl", "max_issues_repo_name": "michakraus/Polynomials.jl", "max_issues_repo_head_hexsha": "8b6a3d282d8f67cfca219ec91aa4076257a16ff0", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 1, "max_issues_repo_issues_event_min_datetime": "2017-01-09T18:43:34.000Z", "max_issues_repo_issues_event_max_datetime": "2017-01-11T11:18:49.000Z", "max_forks_repo_path": "src/abstract.jl", "max_forks_repo_name": "michakraus/Polynomials.jl", "max_forks_repo_head_hexsha": "8b6a3d282d8f67cfca219ec91aa4076257a16ff0", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 48.5323741007, "max_line_length": 314, "alphanum_fraction": 0.5677438482, "num_tokens": 2265, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.3702253925955866, "lm_q2_score": 0.15610489940847916, "lm_q1q2_score": 0.05779399766959875}}
{"text": "\"\"\"\n\tgrdvector(cmd0::String=\"\", arg1=nothing, arg2=nothing, kwargs...)\n\nTakes two 2-D grid files which represents the x- and y-components of a vector field and produces\na vector field plot by drawing vectors with orientation and length according to the information\nin the files. Alternatively, polar coordinate r, theta grids may be given instead.\n\nFull option list at [`grdvector`]($(GMTdoc)grdvector.html)\n\nParameters\n----------\n\n- **A** | **polar** :: [Type => Bool] \n\n The grid contain polar (r, theta) components instead of Cartesian (x, y) [Default is Cartesian components].\n ($(GMTdoc)grdvector.html#a)\n- $(GMT.opt_B)\n- $(GMT.opt_C)\n- **G** | **fill** :: [Type => Str | Number]\n\n Sets color or shade for vector interiors [Default is no fill].\n ($(GMTdoc)grdvector.html#g)\n- **I** | **inc** :: [Type => Sytr | Number]\t``Arg=[x]dx[/dy]``\n\n Only plot vectors at nodes every x_inc, y_inc apart (must be multiples of original grid spacing).\n ($(GMTdoc)grdvector.html#i)\n- **N** | **noclip** | **no_clip** :: [Type => Bool]\n\n Do NOT clip symbols that fall outside map border \n ($(GMTdoc)grdvector.html#n)\n- **Q** | **vec** | **vector** | **arrow** :: [Type => Str]\n\n Modify vector parameters. For vector heads, append vector head size [Default is 0, i.e., stick-plot].\n ($(GMTdoc)grdvector.html#q)\n- $(GMT.opt_P)\n- $(GMT.opt_R)\n- **S** | **vec_scale** :: [Type => Str | Number]\t\t``Arg = [i|l]scale[unit]``\n\n Sets scale for vector plot length in data units per plot distance measurement unit [1].\n ($(GMTdoc)grdvector.html#s)\n- **T** | **sign_scale** :: [Type => Bool]\n\n Means the azimuths of Cartesian data sets should be adjusted according to the signs of the\n scales in the x- and y-directions [Leave alone].\n ($(GMTdoc)grdvector.html#t)\n- $(GMT.opt_U)\n- $(GMT.opt_V)\n- **W** | **pen** :: [Type => Str | Number]\n\n Sets the attributes for the particular line.\n ($(GMTdoc)grdvector.html#w)\n- $(GMT.opt_X)\n- $(GMT.opt_Y)\n- **Z** | **azimuth** :: [Type => Bool]\n\n The theta grid provided contains azimuths rather than directions (implies -A).\n ($(GMTdoc)grdvector.html#z)\n- $(GMT.opt_V)\n- $(GMT.opt_f)\n\"\"\"\nfunction grdvector(cmd0::String=\"\", arg1=nothing, arg2=nothing; first=true, kwargs...)\n\n\tlength(kwargs) == 0 && return monolitic(\"grdvector\", cmd0, arg1, arg2)\n\n\td, K, O = init_module(first, kwargs...)\t\t# Also checks if the user wants ONLY the HELP mode\n\n\tcmd::String = parse_BJR(d, \"\", \"\", O, \" -JX12c/0\")[1]\n\tcmd = parse_common_opts(d, cmd, [:I :UVXY :f :p :t :params], first)[1]\n\tcmd = parse_these_opts(cmd, d, [[:A :polar], [:N :noclip :no_clip], [:S :vec_scale], [:T :sign_scale], [:Z :azimuth]])\n\n # Check case in which the two grids were transmitted by name. \n (cmd0 != \"\" && isa(arg1, String)) && (cmd0 *= \" \" * arg1; arg1 = nothing)\n\n\tcmd, got_fname, arg1 = find_data(d, cmd0, cmd, arg1)\t# Find how data was transmitted\n\n\tN_used = got_fname == 0 ? 1 : 0\t\t# To know whether a cpt will go to arg1 or arg2\n\tcmd, arg1, arg2, = add_opt_cpt(d, cmd, CPTaliases, 'C', N_used, arg1, arg2)\n\topt_Q = parse_Q_grdvec(d, [:Q :vec :vector :arrow])\n\t!occursin(\" -G\", opt_Q) && (cmd = add_opt_fill(cmd, d, [:G :fill], 'G'))\t# If fill not passed in arrow, try from regular option\n\tcmd *= add_opt_pen(d, [:W :pen], \"W\", true)\t\t\t\t\t\t\t\t\t# TRUE to also seek (lw,lc,ls)\n\t(!occursin(\" -C\", cmd) && !occursin(\" -W\", cmd) && !occursin(\" -G\", opt_Q)) && (cmd *= \" -W0.5\")\t# If still nothing, set -W.\n\t(opt_Q != \"\") && (cmd *= opt_Q)\n\n return finish_PS_module(d, \"grdvector \" * cmd, \"\", K, O, true, arg1, arg2)\nend\n\n# ---------------------------------------------------------------------------------------------------\nfunction parse_Q_grdvec(d::Dict, symbs::Array{<:Symbol})::String\n\t(show_kwargs[1]) && return print_kwarg_opts(symbs, \"NamedTuple | String\")\n\tcmd::String = \"\"\n if ((val = find_in_dict(d, symbs)[1]) !== nothing)\n\t\tif (isa(val, String)) cmd *= \" -Q\" * val\t\t# An hard core GMT string directly with options\n\t\telse cmd *= \" -Q\" * vector_attrib(val)\n\t\tend\n\t\tif ((ind = findfirst(\"+g\", cmd)) !== nothing) # -Q0.4+e+gred+n0.4+pcyan+h0\n\t\t\tcmd *= \" -G\" * split(cmd[ind[1]+2:end], \"+\")[1]\t# Add a -G (does the same) to have at least one of the -G, -W, -C\n\t\tend\n\tend\n\treturn cmd\nend\n\n# ---------------------------------------------------------------------------------------------------\ngrdvector(arg1, arg2=nothing, cmd0::String=\"\"; kw...) = grdvector(cmd0, arg1, arg2; first=true, kw...)\ngrdvector!(cmd0::String=\"\", arg1=nothing, arg2=nothing; kw...) = grdvector(cmd0, arg1, arg2; first=false, kw...)\ngrdvector!(arg1, arg2=nothing, cmd0::String=\"\"; kw...) = grdvector(cmd0, arg1, arg2; first=false, kw...)\n", "meta": {"hexsha": "35595f5feefa89a8b9f2685659a235840f5c0e4b", "size": 4689, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/grdvector.jl", "max_stars_repo_name": "stevengj/GMT.jl", "max_stars_repo_head_hexsha": "40f8ac4b2e352369adf98c8e2a7864e454599e18", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/grdvector.jl", "max_issues_repo_name": "stevengj/GMT.jl", "max_issues_repo_head_hexsha": "40f8ac4b2e352369adf98c8e2a7864e454599e18", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/grdvector.jl", "max_forks_repo_name": "stevengj/GMT.jl", "max_forks_repo_head_hexsha": "40f8ac4b2e352369adf98c8e2a7864e454599e18", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 44.2358490566, "max_line_length": 128, "alphanum_fraction": 0.5986351034, "num_tokens": 1442, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.3849121303722487, "lm_q2_score": 0.15002882814282253, "lm_q1q2_score": 0.0577479158577058}}
{"text": "\"\"\"\n Clock{T}\n\nManages a simulation's time information.\n\"\"\"\nmutable struct Clock{T}\n time::T # current simulation time\n dt::T # simulation timestep\n stop_time::T # simulation end time\nend\n\ntick!(clock::Clock) = (clock.time += clock.dt)\n\nstop_time_exceeded(clock::Clock) = (clock.time >= clock.stop_time)\n", "meta": {"hexsha": "22f7cdc3630d1db65afa2a360b8998458bdecaad", "size": 337, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/CoupledSimulations/clock.jl", "max_stars_repo_name": "CliMA/CouplerMachine", "max_stars_repo_head_hexsha": "34768de8dbca05b8c03c5c24626dea6c14404b03", "max_stars_repo_licenses": ["Apache-2.0"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2022-03-05T07:09:01.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-05T07:09:01.000Z", "max_issues_repo_path": "src/CoupledSimulations/clock.jl", "max_issues_repo_name": "CliMA/CouplerMachine", "max_issues_repo_head_hexsha": "34768de8dbca05b8c03c5c24626dea6c14404b03", "max_issues_repo_licenses": ["Apache-2.0"], "max_issues_count": 20, "max_issues_repo_issues_event_min_datetime": "2021-04-01T13:23:06.000Z", "max_issues_repo_issues_event_max_datetime": "2021-11-02T04:15:01.000Z", "max_forks_repo_path": "src/CoupledSimulations/clock.jl", "max_forks_repo_name": "CliMA/ClimaCoupler.jl", "max_forks_repo_head_hexsha": "d21349895a9bdaf9e192b534977deff9bb05385b", "max_forks_repo_licenses": ["Apache-2.0"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2020-11-05T16:29:27.000Z", "max_forks_repo_forks_event_max_datetime": "2020-11-05T16:29:27.000Z", "avg_line_length": 22.4666666667, "max_line_length": 66, "alphanum_fraction": 0.6439169139, "num_tokens": 82, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.48438008427698437, "lm_q2_score": 0.11920292358664392, "lm_q1q2_score": 0.05773952217296151}}
{"text": "#=\nMain generic API\n\n@author : Spencer Lyon \n@date : 2015-04-13 11:29:12\n\n=#\n\n# ----------- #\n# Manager API #\n# ----------- #\n\"\"\"\n`verbose(m::IterationManager) -> Bool`\n\nSpecifies if the iteration manager should be verbose and print status updates.\n\nChecks the `verbose` field of the manager and returns it. If the field\ndoesn't exist, the default is false.\n\n\"\"\"\nverbose(m::IterationManager) = isdefined(m, :verbose) ? m.verbose : false\n\n\"\"\"\n`prefix(m::IterationManager) -> String`\n\nDefines the prefix that should be applied to all printed messages\n\nChecks the `prefix` field of the manager and returns it. If the field\ndoesn't exist, the default is `\"\"`.\n\n\"\"\"\nprefix(m::IterationManager) = isdefined(m, :print_prefix) ? m.print_prefix : \"\"\n\n\"\"\"\n`print_now(m::IterationManager, n::Int) -> Bool`\n\nSpecifies if the iteration manager should print on the `n`th iteration\n\"\"\"\nfunction print_now(mgr::IterationManager, n::Int)\n if verbose(mgr)\n n == 1 && return true\n isdefined(mgr, :print_skip) ? n % mgr.print_skip == 0 : true\n else\n false\n end\nend\n\n# fallback in case x is not an Int\nprint_now(mgr::IterationManager, x::Any) = verbose(mgr)\n\n# --------- #\n# State API #\n# --------- #\n\n# default methods for the `by` argument of `update!`\ndefault_by(x::T, y::T) where T <: AbstractArray = maximum(abs.(x .- y))\ndefault_by(x::T, y::T) where T <: Number = abs(x - y)\n\nfunction default_by(x::S, y::T) where S where T\n msg = \"default_by not implemented for types x::$S, y::$T\"\n throw(ArgumentError(msg))\nend\n\nnum_iter(::IterationState) = nothing\nLinearAlgebra.norm(::IterationState) = Inf\n\n\"\"\"\n`default_by(x, y)`\n\nGives the default comparison `by` argument to `managed_iteration` based on the\ntypes of x and y\n\"\"\" default_by\n\ndisplay_iter(istate::IterationState, prefix=\"\") =\n display_iter(stdout, istate, prefix)\n\ndisplay_iter(io::IO, istate::T, prefix=\"\") where T <: IterationState =\n error(\"`display_iter` must be implemented directly by type $T\")\n\n\"\"\"\n display_iter([io::IO=stdout], st::IterationState, [prefix::String])\n\nA `display` method for an iteration state. Must be implemented by all concrete\nsubtypes of `IterationState`\n\"\"\"\ndisplay_iter\n\n# --------------------------- #\n# Combining Manager and State #\n# --------------------------- #\n\n\"\"\"\n finished(mgr::IterationManager, istate::IterationState)::Bool\n\nGiven a manager and a state, determine if the iterations have finished and\nshould thus be terminated\n\"\"\"\nfinished\n\n# ------------- #\n# default hooks #\n# ------------- #\n\"\"\"\n pre_hook(mgr::IterationManager, istate::IterationState)\n\nCalled before iterations begin.\n\nFor the default manager and state this is used to print the column headers for\nverbose output printing\n\"\"\"\npre_hook(mgr::IterationManager, istate::IterationState) =\n verbose(mgr) && display_iter(istate, prefix(mgr))\n\nprint_now(mgr::IterationManager, istate::IterationState) =\n print_now(mgr, num_iter(istate))\n\n\"\"\"\n iter_hook(mgr::IterationManager, istate::IterationState)\n\nCalled after every iteration.\n\nFor the default manager and state this is used to print updates on the\niterations if `verbose(mgr) == true`.\n\"\"\"\niter_hook(mgr::IterationManager, istate::IterationState) =\n print_now(mgr, istate) && display_iter(istate, prefix(mgr))\n\n\"\"\"\n post_hook(mgr::IterationManager, istate::IterationState)\n\nCalled after iterations have finished\n\nFor the default manager this is used to print a warning if the maximum number\nof iterations was exceeded.\n\"\"\"\nfunction post_hook(mgr::IterationManager, istate::IterationState)\n if !(isdefined(mgr, :maxiter))\n return nothing\n end\n if num_iter(istate) >= mgr.maxiter\n @warn \"Maximum iterations exceeded. Algorithm may not have converged\"\n end\n nothing\nend\n\nfunction managed_iteration(\n f::Base.Callable,\n mgr::IterationManager,\n istate::IterationState{T};\n by=default_by\n ) where T\n pre_hook(mgr, istate)\n\n while !(finished(mgr, istate))\n v = f(istate.prev)::T # if we don't get a T back it should be an error\n update!(istate, v; by=by)\n iter_hook(mgr, istate)\n end\n\n post_hook(mgr, istate)\n istate\nend\n\n\"\"\"\n managed_iteration!(\n f!::Base.Callable,\n mgr::IterationManager,\n dest::T,\n istate::IterationState{T};\n by::Base.Callable=default_by\n ) where T <: AbstractArray\n\nGiven a function with signature `f!(dest::T, src::T)`, run a non-allocating\nversion of managed_iteration where each step calls the mutating function `f!`\nthat fills its first argument based on the value of the second argument. All\nother behavior is equivalent to other forms of `managed_iteration`\n\n\"\"\"\nfunction managed_iteration!(\n f!::Base.Callable,\n mgr::IterationManager,\n dest::T,\n istate::IterationState{T};\n by::Base.Callable=default_by\n ) where T <: AbstractArray\n pre_hook(mgr, istate)\n\n while !(finished(mgr, istate))\n f!(dest, istate.prev)\n update!(istate, dest; by=by)\n iter_hook(mgr, istate)\n end\n\n post_hook(mgr, istate)\n istate\nend\n\n# kwarg version to create default manger/state\nfunction managed_iteration(\n f::Base.Callable,\n init;\n tol::Float64=NaN,\n maxiter::Int=typemax(Int),\n by::Base.Callable=default_by,\n verbose::Bool=true,\n print_skip::Int=div(maxiter, 5)\n )\n mgr = DefaultManager(tol, maxiter, verbose, print_skip)\n istate = DefaultState(init)\n managed_iteration(f, mgr, istate; by=by)\nend\n", "meta": {"hexsha": "d3246e4d15ac12d69525c2695087533a0214e5ce", "size": 5564, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/api.jl", "max_stars_repo_name": "UnofficialJuliaMirror/IterationManagers.jl-69680d3f-cf1d-5315-a21f-791b8226f004", "max_stars_repo_head_hexsha": "7d210fbcd7b52b7d987bccee4ce7fe89d2490c01", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 10, "max_stars_repo_stars_event_min_datetime": "2015-04-15T08:04:58.000Z", "max_stars_repo_stars_event_max_datetime": "2016-11-28T09:15:17.000Z", "max_issues_repo_path": "src/api.jl", "max_issues_repo_name": "UnofficialJuliaMirror/IterationManagers.jl-69680d3f-cf1d-5315-a21f-791b8226f004", "max_issues_repo_head_hexsha": "7d210fbcd7b52b7d987bccee4ce7fe89d2490c01", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 5, "max_issues_repo_issues_event_min_datetime": "2015-04-24T21:34:54.000Z", "max_issues_repo_issues_event_max_datetime": "2016-08-20T19:28:19.000Z", "max_forks_repo_path": "src/api.jl", "max_forks_repo_name": "UnofficialJuliaMirror/IterationManagers.jl-69680d3f-cf1d-5315-a21f-791b8226f004", "max_forks_repo_head_hexsha": "7d210fbcd7b52b7d987bccee4ce7fe89d2490c01", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 4, "max_forks_repo_forks_event_min_datetime": "2017-05-16T06:01:20.000Z", "max_forks_repo_forks_event_max_datetime": "2020-08-01T10:14:31.000Z", "avg_line_length": 26.2452830189, "max_line_length": 79, "alphanum_fraction": 0.6703810208, "num_tokens": 1413, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.43398147944527615, "lm_q2_score": 0.13296425050508953, "lm_q1q2_score": 0.057704022147531064}}
{"text": "### A Pluto.jl notebook ###\n# v0.15.1\n\nusing Markdown\nusing InteractiveUtils\n\n# This Pluto notebook uses @bind for interactivity. When running this notebook outside of Pluto, the following 'mock version' of @bind gives bound variables a default value (instead of an error).\nmacro bind(def, element)\n quote\n local el = $(esc(element))\n global $(esc(def)) = Core.applicable(Base.get, el) ? Base.get(el) : missing\n el\n end\nend\n\n# \u2554\u2550\u2561 ac87ced2-986f-4e6c-80b2-104b25c171c2\nbegin\n\tusing CSV, JLD2, FITSIO, FileIO# , HDF5\n\tusing DataFrames #, Query\n\tusing PyCall\n\tusing PlutoUI, PlutoTeachingTools\n\teval(Meta.parse(code_for_check_type_funcs))\nend\n\n# \u2554\u2550\u2561 e0e60e09-3808-4d6b-a773-6ba59c02f517\nmd\"\"\"\n# Astro 528, Lab 3, Exercise 2\n# Benchmarking File I/O &\n# (and calling Python packages)\n\"\"\"\n\n# \u2554\u2550\u2561 8c1d81ab-d215-4972-afeb-7e00bf6063c2\nmd\"\"\"\nFor many applications, its important that we be able to read input data from a file and/or to write our outputes to files so they can be reused later. Disk access is typically much slower than accessing system memory. Therefore, disk access can easily become the limiting factor for a project. In this set of exercises, you'll see examples of how to perform basic file I/O. In the process, you'll also see how to import Python packages, call python functions and access data computed by python functions.\n\nYou'll be provided with most of the code you need, so that you can focus on comparing how much disk space and time is required by different file formats. Near the end of the lab, you'll be asked to think about when each type of file format would be a good choice for you to use in your research projects.\n\"\"\"\n\n# \u2554\u2550\u2561 1607eac9-e76f-4d1f-a9ce-981ce3be9bea\nmd\"\"\"\n### Download some data\nFirst, we're going to download some data from the web. Julia has a built in `download` function that can be handy for this. It relies on your system having some utilities already installed (e.g., `curl`, `wget` or `fetch`). If you run this on a local system and run into trouble, then you can leave the cell below, and manually download the file to the data subdirectory.\n\"\"\"\n\n# \u2554\u2550\u2561 f27e1e8f-15eb-4754-a94c-7f37c54b871e\nbegin \n\tpath = basename(pwd())==\"test\" ? \"../data/\" : \"data/\"\n\tif !isdir(path) mkdir(path) end # make sure there's a data directory\n\turl = \"https://exoplanetarchive.ipac.caltech.edu/data/KeplerData/Simulated/kplr_dr25_inj1_plti.txt\"\n\tfilename_ipac = joinpath(path,basename(url)) # extract the filename and prepend \"data/\"\n\ttime_to_download = NaN\n\tif !isfile(filename_ipac) # skip downloading if file already exists\n\t time_to_download = @elapsed download(url,filename_ipac)\n\tend\nend\n\n# \u2554\u2550\u2561 80f02c3a-6751-48df-92ec-13f5c7d8c71e\nif !isnan(time_to_download)\n\tmd\"Downloading the file took $time_to_download seconds.\"\nend\n\n# \u2554\u2550\u2561 624c9038-3008-4e78-a149-60796dacf9c0\nmd\"\"\"\nPreviously, everything you needed for an assignment was included in a GitHub repository. So why did I make you download the file?\n\nNotice the size of the file. Git is great for tracking source code, but it wasn't really designed for working with large files (especially large _binary_ files). Since we're not going to be editing it, we'll simply download it once. Besides, it's useful to know how to download a file from within a julia script and to compare the time required to download the file from the internet to the time required to read the file from disk.\n\"\"\"\n\n# \u2554\u2550\u2561 2eb255d9-707d-4224-a0ce-fe90a1c69722\nmd\"\"\"\n## Calling AstroPy to read data in unusual formats\n\nHere I've picked a file containing the results of applying the pipeline for NASA's Kepler mission to [simulated data](https://exoplanetarchive.ipac.caltech.edu/docs/KeplerSimulated.html) in which the signals of simulated \"planet's\" have been injected into actual Kepler data. This data set is the basis for computing the efficiency of the Kepler pipeline at detecting real planets. This dataset has played an important role in enabling astronomers to estimate the occurrence rates of planets around other stars. \nFor documentation of its contents, you could read [its documentation](https://exoplanetarchive.ipac.caltech.edu/docs/KSCI-19110-001.pdf). However, that's not necessary for this lab. For now, we'll just do some basic manipulations of the file, and the details of its contents aren't important. That said, it is important to know the _file format_. \n\nThis data file that we downloaded is in [IPAC format](https://irsa.ipac.caltech.edu/applications/DDGEN/Doc/ipac_tbl.html). \nIt would be tedious to learn the details of every file format that is used in astronomy, let alone to write our own code to read them. Fortunately, there are packages that can read the most common file formats. Sometimes astronomers use specialized/less common file formats which aren't always implemented in native Julia. Reading such files is an example of something that [astropy](http://docs.astropy.org) is particularly good for. It provides a function [`astropy.io.ascii.read`](http://docs.astropy.org/en/stable/io/ascii/) that will read a file in IPAC for us. \nSince astropy is written in Python, we import the [`PyCall`](https://github.com/stevengj/PyCall.jl) package, so we can import python packages and call python functions from Julia. \nSince reading a file from disk is typically limited by the rate of getting data from disk, rather than compute speed, it's usually not a problem that Python isn't particularly fast, when it's comes to reading files.\n\"\"\"\n\n# \u2554\u2550\u2561 8571ccaf-5fbc-4593-82db-ead073a4074f\nmd\"\"\"\nAt the bottom of this notebook, I've run `using PyCall`. Here, we'll import the required python module using `pyimport`. \n\"\"\"\n\n# \u2554\u2550\u2561 442604d0-b490-404b-8b1c-7e1c01cebad7\nastropy_io_ascii = pyimport(\"astropy.io.ascii\")\n\n# \u2554\u2550\u2561 70f5fe63-fea1-4afc-b03d-f113b8bd621e\nmd\"\"\"\nNow, we can review the documentation for [`astropy.io.ascii.read`](http://docs.astropy.org/en/stable/io/ascii/) and call that function load the data in our input file. Let's use `@time`, so we can compare the time required to read various formats.\n\"\"\"\n\n# \u2554\u2550\u2561 8cd5a772-a3aa-4166-92e6-39436d0d2278\nwith_terminal() do \n\t@time astropy_io_ascii.read(filename_ipac, format=\"ipac\", fast_reader=false)\nend\n\n# \u2554\u2550\u2561 074b00ce-dae6-4e4f-8673-49be73085327\nmd\"\"\"\nOn the plus side, the data was read in. However, what is the type of the data that astropy read for us and stored into `data_from_astropy`? Let's check.\n\"\"\"\n\n# \u2554\u2550\u2561 ced3ecc9-94ea-408f-bdef-d34802166663\nbegin\n\ttime_to_read_with_astropy = @elapsed data_from_astropy = astropy_io_ascii.read(filename_ipac, format=\"ipac\", fast_reader=false)\n\ttypeof(data_from_astropy)\nend\n\n# \u2554\u2550\u2561 c62adfb6-6ef8-45fd-992f-07bca59f82cd\nmd\"\"\"\nSince Python is a weakly-typed language, it's type is `PyObject`. That can contain most anything! \nThat flexibility can be convenient, but it is also one of the reasons that Python is not a good language for high-performance computing. To enable Julia to work efficiently with the data, we'll want Julia to know what type the data is and store a list of strictly-typed columns into a the data into a `DataFrame`.\nThe PyCall package provides an interface for For accessing data from PyObjects. Because of the weak typing issue, the syntax and details can be a bit confusing. If you're not intending to call Python for your class project, then there's no reason to worry about that these details. \n\"\"\"\n\n# \u2554\u2550\u2561 03edae54-4368-4b81-8713-17f93bbb9ed9\nprotip(md\"\"\"\nThis list is just for students who are curious about accessing Python data and methods from Julia.\n\t\n - Given o::PyObject, o.attribute in Julia is equivalent to o.attribute in Python, with automatic type conversion. To get an attribute as a PyObject without type conversion, do o.\"attribute\" instead. The keys(o::PyObject) function returns an array of the available attribute symbols.\n\n - Given o::PyObject, get(o, key) is equivalent to o[key] in Python, with automatic type conversion. To get as a PyObject without type conversion, do get(o, PyObject, key), or more generally get(o, SomeType, key). You can also supply a default value to use if the key is not found by get(o, key, default) or get(o, SomeType, key, default). Similarly, set!(o, key, val) is equivalent to o[key] = val in Python, and delete!(o, key) is equivalent to del o[key] in Python. For one or more integer indices, o[i] in Julia is equivalent to o[i-1] in Python.\n\n - You can call an o::PyObject via o(args...) just like in Python (assuming that the object is callable in Python). The explicit pycall form is still useful in Julia if you want to specify the return type.\n\n - pystr(o) and pyrepr(o) are analogous to str and repr in Python, respectively.\n\n - There's more information about accessing data in PyObjects (and other types to contain Python data) in the [PyCall documentation](https://github.com/JuliaPy/PyCall.jl#types). (The above info is copied directly from there.)\n\"\"\")\n\n# \u2554\u2550\u2561 e0b82e6c-043c-4e66-a2a4-ae0f3e4e5404\nmd\"\"\"\nFor now, you can get a [`Dict` or \"Dictionary\"](https://docs.julialang.org/en/v1/base/collections/index.html#Dictionaries-1) by using `data_from_astropy.columns`. The dictionary consists of a set of _keys_ (in this case strings), where each key is associated with a _value_ (in this case a Vector or 1-d array). \nOften times, the data you want to work with can be represented as a table. For efficiency's sake, it's usually best to represent these as a bunch of `Vector`'s, each containing one columns of data. Using a `Dict` allows you to give the columns names (instead of just numbers, so you're less likely to access the wrong column) and allows each column to have a different type (again useful for Julia to optimize your code). \n\"\"\"\n\n# \u2554\u2550\u2561 aa62f1e5-6af5-43f6-be11-665ed25986c0\ndict_from_astropy = data_from_astropy.columns\n\n# \u2554\u2550\u2561 540468dc-3f9f-457b-9b44-efb3ec036166\nmd\"\"\"\nMost of the columns can be automatically converted to an array with a known Julia type. For example, `data_from_astropy.columns[\"KIC_ID\"]` returns data as an `Array{Int64,1}`. However, value associated with `TCE_ID` is some `PyObject` that can't be automatically reinterpretted as a Julia array. \n\"\"\"\n\n# \u2554\u2550\u2561 21e3adbb-0176-4963-bdfc-0ba2422af4bf\ntypeof(dict_from_astropy[\"KIC_ID\"])\n\n# \u2554\u2550\u2561 d27aef3b-bfe7-466a-9580-93fc53dcef18\ntypeof(dict_from_astropy[\"TCE_ID\"])\n\n# \u2554\u2550\u2561 02f638d9-ec2f-4072-9fb3-e6fabac1b1e6\nmd\"\"\"Since many `KIC_ID`'s don't have an associated `TCE_ID`, there are many missing entries. Python is trying to store a list with lots of empty entries efficiently, but PyCall doesn't (yet?) know how to deal with this `MaskedColumn` for us. Working with the data when Julia can't know its type would be very inefficient. Therefore, we want to create an array of Strings that allows Julia to represent this data more efficiently. Technically, it will be an array where each element is either a `String` or a `missing`. It took a little tinkering, but eventually, I figured out how to extract that data into an efficient Julia object.\n\"\"\"\n\n# \u2554\u2550\u2561 481c5b7a-4660-4954-84f3-29d33bd73f3d\nbegin\n\tTCE_ID_list = map(x -> x != nothing ? x : missing, data_from_astropy.columns[\"TCE_ID\"].data.tolist())\n\tTCE_ID_list_type = typeof(TCE_ID_list)\n\tTCE_ID_list\nend\n\n# \u2554\u2550\u2561 aba84742-a927-4ce5-9f7c-6e535db1ee47\nprotip(md\"Note that the type of `TCE_ID_list` is a $TCE_ID_list_type. Julia is able to store and compute on vectors with missing data very efficiently thanks to its rich type system.\")\n\n# \u2554\u2550\u2561 aa7c4dfe-a3d4-448b-a559-1bc7b338a1dc\nmd\"\"\"\nNow let's replace the value associated with TCE_ID with this list.\n\"\"\"\n\n# \u2554\u2550\u2561 a97f9c8e-dbd3-4613-9c6e-c471340ea2d6\ndict_from_astropy[\"TCE_ID\"] = TCE_ID_list;\n\n# \u2554\u2550\u2561 7c7430a0-d16e-485b-bbd9-7f231fee853a\nmd\"\"\"\nIf we just wantted to access the data, then we could use the data stored in the columns as a dictionary. \nHowever, a dictionary doesn't guarentee anything about the relationship of the value of different keys.\nFor example, in a table, each column should have the same number of rows. Therefore, we'll switch from representing the data as a dictionary and start using a `DataFrame`, a type provided by the [DataFrames.jl package](https://github.com/JuliaData/DataFrames.jl). A `DataFrame` can be thought of as a table, where the data for each column is stored as an array. A `DataFrame` also provides some additional features to allow easy and efficient access and manipulation of the table that will come in useful later. \n\"\"\"\n\n# \u2554\u2550\u2561 cebe52b3-386c-4d31-8497-c19b6c742577\nbegin\n\t# First, we'll create and copy a small `DataFrame`, so the functions get compiled before we start timing things.\n\tsmall_dict = Dict(:a=>[1,2],:b=>[3.0,4.0],:c=>[\"hello\",missing])\n\tsmall_df1 = DataFrame(small_dict)\n\tsmall_df2 = DataFrame(small_dict, copycols=false)\nend;\n\n# \u2554\u2550\u2561 3d09d38a-ca72-43f2-bd85-17fc0e05a645\nmd\"\"\"\nWe'll use our existing dictionary to initialize a `DataFrame`. \n\"\"\"\n\n# \u2554\u2550\u2561 9257c879-d96f-4d2b-994e-d542617b0c65\nwith_terminal() do\n\tsmall_dict # makes sure already compiled code for creating small DataFrame\n\t@time df_tmp = DataFrame(dict_from_astropy)\nend\n\n# \u2554\u2550\u2561 307a7a29-afa6-4c4d-88ae-4f757aeba892\nmd\"\"\"\n2a. Look at how much memory was allocated during this line of code. \nDo you think it involved allocating and copying lots of data from the dictionary?\n\n\"\"\"\n\n# \u2554\u2550\u2561 afdc442a-2931-4129-824e-98431e1d8be2\nresponse_2a = missing\n\n# \u2554\u2550\u2561 c42deea3-acf1-491a-97bd-fdc472917584\ndisplay_msg_if_fail(check_type_isa(:response_2a,response_2a,[AbstractString,Markdown.MD]))\n\n# \u2554\u2550\u2561 7c1f4a27-dfd7-428a-97ef-eac1784e8d6f\nmd\"\"\"By default `DataFrames` prioritizes _safe operations_ over efficient ones. However, it also provides options for more efficient operations for times when users want to invest a little more time to get good performance. For example, we can tell it not to copy the data, using `copycols=false`).\"\"\"\n\n# \u2554\u2550\u2561 97111ccc-8a16-4e7d-adf7-0dcd90524be3\nwith_terminal() do\n\t\t@time df = DataFrame(dict_from_astropy, copycols=false)\nend\n\n# \u2554\u2550\u2561 f45668b1-1cbd-48e6-9224-d469826e093b\nmd\"\"\"\n2b. Look at how much memory was allocated the second time. \nDid Julia make a new copy of all of the data? How did the time required compare to the first attempt?\n\"\"\"\n\n# \u2554\u2550\u2561 52beb148-6214-4a03-a0cf-6c2a6da77d40\nresponse_2b = missing\n\n# \u2554\u2550\u2561 1ecbd400-0157-45ce-b1b8-db764012ba5e\ndisplay_msg_if_fail(check_type_isa(:response_2b,response_2b,[AbstractString,Markdown.MD]))\n\n# \u2554\u2550\u2561 191ba96e-2573-4bc1-a352-46a66e0a5c4f\nmd\"\"\"\n## Writing a CSV file\n\nThe IPAC format allows for significant metadata, but reading not-so-common file formats can be annoying. \nLet's say that we'd like to write the data to a [CSV file](https://en.wikipedia.org/wiki/Comma-separated_values), so that it's easier for other programs to read in. Using the CSV package, we can read a CSV file into a DataFrame (or several other tabular data structures) and write CSV files from a DataFrame with code like the following.\n\"\"\"\n\n# \u2554\u2550\u2561 82a757ad-566d-4c1d-8b3d-366ffd980fb4\nbegin\n\t# Read & write a small test file so compilation time not included below\n\tn_small_df = 1024\n\tsmall_csv_filename = \"random_numbers.csv\"\n\tsmall_df = DataFrame(a=rand(n_small_df),b=rand(n_small_df) ) \n\tCSV.write(joinpath(path,small_csv_filename),small_df) \n\tsmall_df_from_csv = CSV.read(joinpath(path,small_csv_filename),DataFrame)\nend;\n\n# \u2554\u2550\u2561 3e550b71-4750-460b-be18-911a848a8f49\nmd\"\"\"\nNow, let's write the DataFrame from our IPAC file to a CSV file. We'll investigate how the filesize and time to read files compares.\n\"\"\"\n\n# \u2554\u2550\u2561 b0c3875b-ef07-4e92-a0a8-55f42b266c6b\nbegin\n\tfilename_csv = replace(filename_ipac, \".txt\" => \".csv\") \n\tdf = DataFrame(dict_from_astropy, copycols=false)\n\twith_terminal() do\n\t\t@time CSV.write(filename_csv,df)\n\tend\nend\n\n# \u2554\u2550\u2561 58e021c6-500f-40d8-a388-a5732bc808b3\nmd\"\"\"Next, we'll compare the filesizes for the IPAC file and the CSV file containing the same data. \n\n2c. Which do you expect will be larger? Why?\nOnce you've made a prediction, mouse over the hint box below to see the sizes of the two files. If you were suprsised, try to explain what happened.\"\"\"\n\n# \u2554\u2550\u2561 36bfd224-a13b-438c-9106-382c15a6a1d2\nresponse_2c = missing\n\n# \u2554\u2550\u2561 7f4c3ab8-041e-4ef0-bb2e-320f5293cdde\ndisplay_msg_if_fail(check_type_isa(:response_2c,response_2c,[AbstractString,Markdown.MD]))\n\n# \u2554\u2550\u2561 195c2df4-7e1c-49f8-871f-f76b985dab6d\nbegin\n\tipac_filesize = filesize(filename_ipac) /1024^2\n\tcsv_filesize = filesize(filename_csv) /1024^2\n\thint(md\"The IPAC file size is $ipac_filesize MB versus $csv_filesize MB for the CSV.\")\nend\n\n# \u2554\u2550\u2561 32124f8a-f5cf-41d1-97a9-ce1d05145fde\nmd\"\"\"\n\n## Reading CSV files\n\n2d. Think back to how long it took to read in the file in IPAC format ($time_to_read_with_astropy seconds). How long do you think it will take to read in the same data once it's been stored in CSV format?\n\"\"\"\n\n# \u2554\u2550\u2561 507e1516-5433-49eb-831d-32426f30895e\nresponse_2d = missing\n\n# \u2554\u2550\u2561 eac67cc9-754b-4f7d-add8-93900a1b5b49\ndisplay_msg_if_fail(check_type_isa(:response_2d,response_2d,[AbstractString,Markdown.MD]))\n\n# \u2554\u2550\u2561 64224c6b-c5a0-44f2-b2a0-7f77759cb848\nmd\"\"\"\nNow, try reading it in and see. \n\"\"\"\n\n# \u2554\u2550\u2561 73c06bb1-be49-46bd-b7f1-c45cc56af7b4\nmd\"\"\"\nI'm ready to benchmark reading a CSV file. $(@bind ready_read_csv CheckBox()) \n$(@bind go_read_csv Button(\"Rerun the benchmarks.\"))\n\"\"\"\n\n# \u2554\u2550\u2561 ebfaa677-829b-4bf9-bdbb-19f3c87dd3a4\nif ready_read_csv\n\tgo_read_csv\n\ttime_read_csv = @elapsed df_csv = CSV.read(filename_csv, DataFrame)\n\tmd\"It took $time_read_csv seconds to read the CSV file.\"\nend\n\n# \u2554\u2550\u2561 945f5a55-3026-4497-9ece-8af878c87788\nmd\"\"\"\n2e. How did the time required to read the data in CSV format compare to the time to read the data in IPAC format? If you were suprsised, try to explain what happened.\n\"\"\"\n\n# \u2554\u2550\u2561 57397ee4-9efc-48b3-b640-d2b7a10da633\nresponse_2e = missing\n\n# \u2554\u2550\u2561 8059a6a3-384a-4344-8a23-650ee0be10c2\ndisplay_msg_if_fail(check_type_isa(:response_2e,response_2e,[AbstractString,Markdown.MD]))\n\n# \u2554\u2550\u2561 f5b93929-2c59-4360-8c41-97a1324ba455\nmd\"2f. How do you think the sizes of the files in the two formats will compare? Once you've made your prediction, mouse over the hint box. If you were suprsised, try to explain what happened.\"\n\n# \u2554\u2550\u2561 122196fa-45ca-4031-85eb-4afd4782de9e\nresponse_2f = missing\n\n# \u2554\u2550\u2561 e9dc1456-616b-4e4b-b209-9f6ba4c48607\ndisplay_msg_if_fail(check_type_isa(:response_2f,response_2f,[AbstractString,Markdown.MD]))\n\n# \u2554\u2550\u2561 f15d37a7-d962-4da0-977f-76729a3313be\nhint(md\"The IPAC file size is $ipac_filesize MB versus $csv_filesize MB for the CSV.\")\n\n# \u2554\u2550\u2561 28f195c4-4f61-4873-85d6-b4e3aaa3660f\nmd\"\"\"\n## Binary formats: HDF5/JLD2\n\nThere are numerous binary file formats that one could use. Here, we'll try using JLD2 which is a subset of the [HDF5](https://www.hdfgroup.org/solutions/hdf5/) file format. This means that when [Julia's JLD2 package](https://github.com/JuliaIO/JLD2.jl) writes jld2 files, they can be read by other programs that can read HDF5 files. However, a generic HDF5 file is not a valid JLD2 file. If you want to read a HDF5 file, then you can use Julia's [HDF5.jl package](https://github.com/JuliaIO/HDF5.jl). The [FileIO.jl](https://github.com/JuliaIO/FileIO.jl) package provides a common interface for reading and writing from multiple file formats, including these and several others.\n\nAs before, we'll call each function once using a small DataFrame, just so they get compiled before we benchmark them.\n\"\"\"\n\n# \u2554\u2550\u2561 3837e439-250b-4577-b0d7-93352ec19f6e\nbegin\n\tfilename_jld2_small = replace(small_csv_filename, \".csv\" => \".jld2\") \n\t@save joinpath(path,filename_jld2_small) small_df \n\tsmall_df_from_jld2 = load(joinpath(path,filename_jld2_small), \"small_df\")\nend;\n\n# \u2554\u2550\u2561 df9b701f-d314-4512-b2ea-1f6ae015166c\nresponse_2g = missing\n\n# \u2554\u2550\u2561 377f7527-a338-4526-bb24-9766c635e719\ndisplay_msg_if_fail(check_type_isa(:response_2g,response_2g,[AbstractString,Markdown.MD]))\n\n# \u2554\u2550\u2561 25f24754-16d4-4343-b54a-cf8ea1358ce9\nbegin \n\tsmall_jld2_filesize = filesize(joinpath(path,filename_jld2_small))/1024\n\tsmall_csv_filesize = filesize(joinpath(path,small_csv_filename))/1024\n\thint(md\"For the small random data set, the JLD2 file's size is $small_jld2_filesize KB versus $small_csv_filesize KB for the CSV file.\")\nend\n\n# \u2554\u2550\u2561 8255b5a9-cbe6-4604-bedd-0e366f311096\nmd\"\"\"\nLet's check the filesizes for the JLD2 file to the CSV file. \nThe small CSV file we created is $small_jld2_filesize KB. \n\n2g. How large would you guess the JLD2 file will be? Once you've made your prediction, mouse over the hint box. If you were suprised, try to explain what happened.\n\"\"\"\n\n# \u2554\u2550\u2561 570fd826-23fd-46ee-bdb4-58fb0c45719a\nmd\"\"\"\nNow let's time how long it takes to save the data from IPAC into a JLD2 file.\n\"\"\"\n\n# \u2554\u2550\u2561 691410bb-0472-4800-a90d-29ddf947de3e\nbegin\n\tfilename_jld2 = replace(filename_ipac, \".txt\" => \".jld2\") \n\twith_terminal() do \n\t\t@time @save filename_jld2 df\n\tend\nend\n\n# \u2554\u2550\u2561 8cbb1c90-bd94-44b5-80b6-81d38f3e6252\nmd\"2h. How long do you think it will take to load the data from the JLD2 file? \"\n\n# \u2554\u2550\u2561 c3065acf-6205-455f-ba74-ca51f3f6761b\nresponse_2h = missing\n\n# \u2554\u2550\u2561 9c392be9-1505-40bc-a290-68085a1c2700\ndisplay_msg_if_fail(check_type_isa(:response_2h,response_2h,[AbstractString,Markdown.MD]))\n\n# \u2554\u2550\u2561 d738bdcc-5f83-4dfa-a17f-e9ea23db2986\nmd\"\"\"\nNow time how long it takes to load the data from the JLD2 file.\n\"\"\"\n\n# \u2554\u2550\u2561 d55ce157-099c-4c1b-94db-62918f04e5fe\nmd\"\"\"\nI'm ready to benchmark reading a JLD2 file. $(@bind ready_read_jld2 CheckBox()) \n$(@bind go_read_jld2 Button(\"Rerun the benchmarks.\"))\n\"\"\"\n\n# \u2554\u2550\u2561 206f464b-55fe-46aa-85b7-8f0246a0aaad\nif ready_read_jld2\n\tgo_read_jld2\n\ttime_read_jld2 = @elapsed df_jld2_read_back_in = load(filename_jld2, \"df\")\n\tmd\"It took $time_read_jld2 seconds to read the JLD2 file.\"\nend\n\n# \u2554\u2550\u2561 6f72d1b2-63f6-4301-8272-bb2d6d2d049e\nmd\"\"\"\n2i. How does the time required to read and write the JLD2 file compare to the time required to read the IPAC and CSV formatted files? If you were suprised, try to explain what happend.\n\"\"\"\n\n# \u2554\u2550\u2561 01201c37-0b79-46b1-a001-e716f5b3ba67\nresponse_2i = missing\n\n# \u2554\u2550\u2561 e3d37bc1-a119-4add-a111-899ee0caea05\ndisplay_msg_if_fail(check_type_isa(:response_2i,response_2i,[AbstractString,Markdown.MD]))\n\n# \u2554\u2550\u2561 6dff3f21-fac0-42e1-910a-f969a231374f\nmd\"\"\"\nNext, we'll compare the filesizes for the JLD2 file to the CSV file. \n\n2j. How large would you guess the JLD2 file will be? Once you've made your prediction, mouse over the hint box. If you were suprised, try to explain what happened.\n\"\"\"\n\n# \u2554\u2550\u2561 b26b8253-e6cd-49f4-81c5-2a3c2963a37c\nresponse_2j = missing\n\n# \u2554\u2550\u2561 e5dc123f-1596-4311-9398-f0cfe80a5342\ndisplay_msg_if_fail(check_type_isa(:response_2j,response_2j,[AbstractString,Markdown.MD]))\n\n# \u2554\u2550\u2561 7b9d26c2-899e-45e9-b664-39d5f1adfe3f\nbegin \n\tjld2_filesize = filesize(filename_jld2) / 1024^2;\n\thint(md\"The JLD2 file size is $jld2_filesize MB versus $csv_filesize MB for the CSV.\")\nend\n\n# \u2554\u2550\u2561 fc01d57f-c90b-4231-96be-ddd48656d55e\nmd\"\"\"\n## Flexible Format: FITS\n\nAstronomers often use the [FITS file format](https://en.wikipedia.org/wiki/FITS). Like [HDF\n5](https://www.hdfgroup.org/solutions/hdf5/), it's a very flexible (e.g., it can store both text and binary data) and thus complicated file\n format. \nTherefore, most languages call a common [FITSIO library written in C](https://heasarc.gsfc.nasa.gov/fitsio/), rather than implementing code themselves. Indeed, that's what [Julia's FITSIO.jl package](https://github.com/JuliaAstro/FITSIO.jl) does.\n\nUnfortunately, the FITSIO package isn't as polished as the others. It expects a `Dict` rather than a `DataFrame`, and it can't handle missing values. So I've provided some helper functions at the bottom of the notebook. Also, FITS files have complicated headers, so I'll provide a function to read all the tabular data from a simple FITS file. As usual, we'll use each function once, so that Julia compiles them before we start timing.\n\"\"\"\n\n# \u2554\u2550\u2561 3b232365-f2fe-4edb-a39f-3e37c8cbb666\nmd\"Now we can time how long it takes to write and read the data as FITS files.\"\n\n# \u2554\u2550\u2561 568862b3-6fce-426a-a9e2-e558adf3932a\nmd\"\"\"\nI'm ready to benchmark reading a FITS file. $(@bind ready_read_fits CheckBox()) \n$(@bind go_read_fits Button(\"Rerun the benchmarks.\"))\n\"\"\"\n\n# \u2554\u2550\u2561 ae79ee89-d788-4612-8b1d-fc22d85c7744\nmd\"2k. How do the read/write times and file sizes for FITS compare to CSV and JLD2?\"\n\n# \u2554\u2550\u2561 3745c237-ba48-4f2c-959e-441484244764\nresponse_2k = missing\n\n# \u2554\u2550\u2561 6d061411-6f19-4119-aefb-cc380198b0ce\ndisplay_msg_if_fail(check_type_isa(:response_2k,response_2k,[AbstractString,Markdown.MD]))\n\n# \u2554\u2550\u2561 8def87d2-f10b-4a82-b353-a6477eeead9b\nmd\"## Implications for your project\"\n\n# \u2554\u2550\u2561 c74b3105-f480-4688-b85f-3e7dff70da3b\nmd\"\"\"\n2l. How does the time required to read any of the above formats compare to the time required to download the files ($time_to_download seconds)? \nWill your project need to transfer large files over the internet? If so, very roughly how large do you expect they will be? \n\"\"\"\n\n# \u2554\u2550\u2561 55438c09-1d94-4ff7-90c3-0cc6064a091e\nresponse_2l = missing\n\n# \u2554\u2550\u2561 bcc796c9-db11-4a09-a5f9-215127ac0938\ndisplay_msg_if_fail(check_type_isa(:response_2l,response_2l,[AbstractString,Markdown.MD]))\n\n# \u2554\u2550\u2561 92b2ecdd-9491-4f16-8cb9-bacbcf180280\nmd\"\"\"\n2m. Will your project need to read in any input files? If so, what format are they in? \n\nI assume everyone's project code will write at least some output files? Very roughly, how large do you expect that they'll be? What format(s) would be a good choice for your project?\"\"\"\n\n# \u2554\u2550\u2561 83216915-cdcf-4f0f-829f-a5ff4c4b8da0\nresponse_2m = missing\n\n# \u2554\u2550\u2561 452872f9-2009-4733-b2d3-28f262ae19b7\ndisplay_msg_if_fail(check_type_isa(:response_2m,response_2m,[AbstractString,Markdown.MD]))\n\n# \u2554\u2550\u2561 29415ddc-e002-4f56-a169-95f7b1c36be9\nmd\"# Helper Functions\"\n\n# \u2554\u2550\u2561 fb23d6c6-b812-4fe1-b224-0014bedbd43f\nChooseDisplayMode()\n\n# \u2554\u2550\u2561 1e53aa10-dff6-40d5-89e2-da194ffc2052\nTableOfContents()\n\n# \u2554\u2550\u2561 14cca8ce-cc61-4fae-b871-21c3fd23d0ea\n\"Convert a DataFrame to a Dict, replacing missing values with 0 or an empty string.\"\nfunction convert_dataframe_to_dict_remove_missing(df::DataFrame)\n d = Dict(map(n->\"$n\"=> # create a dictionary\n ( any(ismissing.(df[!,n])) ? # if column contains a missing\n map(x-> !ismissing(x) ? # search for missings\n x : # leave non-missing values alone\n ( (eltype(df[!,n]) <: Number) ? zero(eltype(n)) : \"\")\n , df[!,n]) # but replace missing with 0 or \"\"\n : df[!,n] ), # if nothing is missing, just use column as is\n names(df) )) \nend\n\n# \u2554\u2550\u2561 28fc8de4-749b-4093-b32f-c398f8d27d3d\n\"Write a DataFrame to a FITS file, replacing missing values with 0 or an empty string.\"\nfunction write_dataframe_as_fits(filename::String, df::DataFrame)\n try \n dict = convert_dataframe_to_dict_remove_missing(df) \n fits_file = FITS(filename,\"w\")\n write(fits_file, dict )\n close(fits_file)\n catch\n @warn(\"There was a problem writing a dataframe to \" * filename * \".\")\n end\nend\n\n\n# \u2554\u2550\u2561 57b422e5-0ad0-4674-bdd3-a8358bc7aaeb\n\n\"Read the columns of the first table from a FITS file into a Dict\"\nfunction read_fits_tables(filename::String)\n dict = Dict{String,Any}()\n fits_file = FITS(filename,\"r\")\n # fits_file[1] is image data, we're interested in the table\n @assert length(fits_file) >= 2\n header = read_header(fits_file[2])\n for i in 1:length(header)\n c = get_comment(header,i)\n if !occursin(\"label for field\",c)\n continue\n end\n h = header[i]\n @assert typeof(h) == String\n try \n dict[h] = read(fits_file[2],h)\n catch\n @warn \"# Problem reading table column \" * h * \".\"\n end\n end\n close(fits_file)\n return dict\nend\n\n\n# \u2554\u2550\u2561 e05f16d6-eb50-49a9-bf14-95d63c9da7ff\nbegin\n\tfilename_fits_small = replace(small_csv_filename, \".csv\" => \".fits\") \n\twrite_dataframe_as_fits(joinpath(path,filename_fits_small),small_df)\n\tread_fits_tables(joinpath(path,filename_fits_small))\nend;\n\n# \u2554\u2550\u2561 1992595f-9976-4b18-bf9c-df8a73d30dc8\nbegin\n\tfilename_fits_small # make sure have already compiled functions\n\tfilename_fits = replace(filename_ipac, \".txt\" => \".fits\") \n\twith_terminal() do \n\t\t@time write_dataframe_as_fits(filename_fits,df)\n\tend\nend\n\n# \u2554\u2550\u2561 90d5244e-17be-4601-b922-8c254f1248bf\nbegin\n\tfits_filesize = filesize(filename_fits) /1024^2\n\thint(md\"The FITS file size is $fits_filesize MB versus $jld2_filesize MB for the JLD2 and $csv_filesize MB for the CSV.\")\nend\n\n# \u2554\u2550\u2561 52f9edd0-79b0-4a9a-9930-3a05d3aa2447\nif ready_read_fits\n\tgo_read_fits\n\ttime_read_fits = @elapsed read_fits_tables(filename_fits)\n\tmd\"It took $time_read_fits seconds to read the FITS file.\"\nend\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000001\nPLUTO_PROJECT_TOML_CONTENTS = \"\"\"\n[deps]\nCSV = \"336ed68f-0bac-5ca0-87d4-7b16caf5d00b\"\nDataFrames = \"a93c6f00-e57d-5684-b7b6-d8193f3e46c0\"\nFITSIO = \"525bcba6-941b-5504-bd06-fd0dc1a4d2eb\"\nFileIO = \"5789e2e9-d7fb-5bc7-8068-2c6fae9b9549\"\nJLD2 = \"033835bb-8acc-5ee8-8aae-3f567f8a3819\"\nPlutoTeachingTools = \"661c6b06-c737-4d37-b85c-46df65de6f69\"\nPlutoUI = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\nPyCall = \"438e738f-606a-5dbb-bf0a-cddfbfd45ab0\"\n\n[compat]\nCSV = \"~0.8.5\"\nDataFrames = \"~1.2.2\"\nFITSIO = \"~0.16.7\"\nFileIO = \"~1.10.1\"\nJLD2 = \"~0.4.13\"\nPlutoTeachingTools = \"~0.1.3\"\nPlutoUI = \"~0.7.9\"\nPyCall = \"~1.92.3\"\n\"\"\"\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000002\nPLUTO_MANIFEST_TOML_CONTENTS = \"\"\"\n# This file is machine-generated - editing it directly is not advised\n\n[[ArgTools]]\nuuid = \"0dad84c5-d112-42e6-8d28-ef12dabb789f\"\n\n[[Artifacts]]\nuuid = \"56f22d72-fd6d-98f1-02f0-08ddc0907c33\"\n\n[[Base64]]\nuuid = \"2a0f44e3-6c83-55bd-87e4-b1978d98bd5f\"\n\n[[CFITSIO]]\ndeps = [\"CFITSIO_jll\"]\ngit-tree-sha1 = \"c860f5545064216f86aa3365ec186ce7ced6a935\"\nuuid = \"3b1b4be9-1499-4b22-8d78-7db3344d1961\"\nversion = \"1.3.0\"\n\n[[CFITSIO_jll]]\ndeps = [\"Artifacts\", \"JLLWrappers\", \"LibCURL_jll\", \"Libdl\", \"Pkg\"]\ngit-tree-sha1 = \"2fabb5fc48d185d104ca7ed7444b475705993447\"\nuuid = \"b3e40c51-02ae-5482-8a39-3ace5868dcf4\"\nversion = \"3.49.1+0\"\n\n[[CSV]]\ndeps = [\"Dates\", \"Mmap\", \"Parsers\", \"PooledArrays\", \"SentinelArrays\", \"Tables\", \"Unicode\"]\ngit-tree-sha1 = \"b83aa3f513be680454437a0eee21001607e5d983\"\nuuid = \"336ed68f-0bac-5ca0-87d4-7b16caf5d00b\"\nversion = \"0.8.5\"\n\n[[Compat]]\ndeps = [\"Base64\", \"Dates\", \"DelimitedFiles\", \"Distributed\", \"InteractiveUtils\", \"LibGit2\", \"Libdl\", \"LinearAlgebra\", \"Markdown\", \"Mmap\", \"Pkg\", \"Printf\", \"REPL\", \"Random\", \"SHA\", \"Serialization\", \"SharedArrays\", \"Sockets\", \"SparseArrays\", \"Statistics\", \"Test\", \"UUIDs\", \"Unicode\"]\ngit-tree-sha1 = \"727e463cfebd0c7b999bbf3e9e7e16f254b94193\"\nuuid = \"34da2185-b29b-5c13-b0c7-acf172513d20\"\nversion = \"3.34.0\"\n\n[[Conda]]\ndeps = [\"JSON\", \"VersionParsing\"]\ngit-tree-sha1 = \"299304989a5e6473d985212c28928899c74e9421\"\nuuid = \"8f4d0f93-b110-5947-807f-2305c1781a2d\"\nversion = \"1.5.2\"\n\n[[Crayons]]\ngit-tree-sha1 = \"3f71217b538d7aaee0b69ab47d9b7724ca8afa0d\"\nuuid = \"a8cc5b0e-0ffa-5ad4-8c14-923d3ee1735f\"\nversion = \"4.0.4\"\n\n[[DataAPI]]\ngit-tree-sha1 = \"ee400abb2298bd13bfc3df1c412ed228061a2385\"\nuuid = \"9a962f9c-6df0-11e9-0e5d-c546b8b5ee8a\"\nversion = \"1.7.0\"\n\n[[DataFrames]]\ndeps = [\"Compat\", \"DataAPI\", \"Future\", \"InvertedIndices\", \"IteratorInterfaceExtensions\", \"LinearAlgebra\", \"Markdown\", \"Missings\", \"PooledArrays\", \"PrettyTables\", \"Printf\", \"REPL\", \"Reexport\", \"SortingAlgorithms\", \"Statistics\", \"TableTraits\", \"Tables\", \"Unicode\"]\ngit-tree-sha1 = \"d785f42445b63fc86caa08bb9a9351008be9b765\"\nuuid = \"a93c6f00-e57d-5684-b7b6-d8193f3e46c0\"\nversion = \"1.2.2\"\n\n[[DataStructures]]\ndeps = [\"Compat\", \"InteractiveUtils\", \"OrderedCollections\"]\ngit-tree-sha1 = \"7d9d316f04214f7efdbb6398d545446e246eff02\"\nuuid = \"864edb3b-99cc-5e75-8d2d-829cb0a9cfe8\"\nversion = \"0.18.10\"\n\n[[DataValueInterfaces]]\ngit-tree-sha1 = \"bfc1187b79289637fa0ef6d4436ebdfe6905cbd6\"\nuuid = \"e2d170a0-9d28-54be-80f0-106bbe20a464\"\nversion = \"1.0.0\"\n\n[[Dates]]\ndeps = [\"Printf\"]\nuuid = \"ade2ca70-3891-5945-98fb-dc099432e06a\"\n\n[[DelimitedFiles]]\ndeps = [\"Mmap\"]\nuuid = \"8bb1440f-4735-579b-a4ab-409b98df4dab\"\n\n[[Distributed]]\ndeps = [\"Random\", \"Serialization\", \"Sockets\"]\nuuid = \"8ba89e20-285c-5b6f-9357-94700520ee1b\"\n\n[[Downloads]]\ndeps = [\"ArgTools\", \"LibCURL\", \"NetworkOptions\"]\nuuid = \"f43a241f-c20a-4ad4-852c-f6b1247861c6\"\n\n[[FITSIO]]\ndeps = [\"CFITSIO\", \"Printf\", \"Reexport\", \"Tables\"]\ngit-tree-sha1 = \"85b66005c9d16d3d27c9d06fd3be5e25074128da\"\nuuid = \"525bcba6-941b-5504-bd06-fd0dc1a4d2eb\"\nversion = \"0.16.7\"\n\n[[FileIO]]\ndeps = [\"Pkg\", \"Requires\", \"UUIDs\"]\ngit-tree-sha1 = \"256d8e6188f3f1ebfa1a5d17e072a0efafa8c5bf\"\nuuid = \"5789e2e9-d7fb-5bc7-8068-2c6fae9b9549\"\nversion = \"1.10.1\"\n\n[[Formatting]]\ndeps = [\"Printf\"]\ngit-tree-sha1 = \"8339d61043228fdd3eb658d86c926cb282ae72a8\"\nuuid = \"59287772-0a20-5a39-b81b-1366585eb4c0\"\nversion = \"0.4.2\"\n\n[[Future]]\ndeps = [\"Random\"]\nuuid = \"9fa8497b-333b-5362-9e8d-4d0656e87820\"\n\n[[InteractiveUtils]]\ndeps = [\"Markdown\"]\nuuid = \"b77e0a4c-d291-57a0-90e8-8db25a27a240\"\n\n[[InvertedIndices]]\ndeps = [\"Test\"]\ngit-tree-sha1 = \"15732c475062348b0165684ffe28e85ea8396afc\"\nuuid = \"41ab1584-1d38-5bbf-9106-f11c6c58b48f\"\nversion = \"1.0.0\"\n\n[[IteratorInterfaceExtensions]]\ngit-tree-sha1 = \"a3f24677c21f5bbe9d2a714f95dcd58337fb2856\"\nuuid = \"82899510-4779-5014-852e-03e436cf321d\"\nversion = \"1.0.0\"\n\n[[JLD2]]\ndeps = [\"DataStructures\", \"FileIO\", \"MacroTools\", \"Mmap\", \"Pkg\", \"Printf\", \"Reexport\", \"TranscodingStreams\", \"UUIDs\"]\ngit-tree-sha1 = \"59ee430ac5dc87bc3eec833cc2a37853425750b4\"\nuuid = \"033835bb-8acc-5ee8-8aae-3f567f8a3819\"\nversion = \"0.4.13\"\n\n[[JLLWrappers]]\ndeps = [\"Preferences\"]\ngit-tree-sha1 = \"642a199af8b68253517b80bd3bfd17eb4e84df6e\"\nuuid = \"692b3bcd-3c85-4b1f-b108-f13ce0eb3210\"\nversion = \"1.3.0\"\n\n[[JSON]]\ndeps = [\"Dates\", \"Mmap\", \"Parsers\", \"Unicode\"]\ngit-tree-sha1 = \"8076680b162ada2a031f707ac7b4953e30667a37\"\nuuid = \"682c06a0-de6a-54ab-a142-c8b1cf79cde6\"\nversion = \"0.21.2\"\n\n[[LaTeXStrings]]\ngit-tree-sha1 = \"c7f1c695e06c01b95a67f0cd1d34994f3e7db104\"\nuuid = \"b964fa9f-0449-5b57-a5c2-d3ea65f4040f\"\nversion = \"1.2.1\"\n\n[[LibCURL]]\ndeps = [\"LibCURL_jll\", \"MozillaCACerts_jll\"]\nuuid = \"b27032c2-a3e7-50c8-80cd-2d36dbcbfd21\"\n\n[[LibCURL_jll]]\ndeps = [\"Artifacts\", \"LibSSH2_jll\", \"Libdl\", \"MbedTLS_jll\", \"Zlib_jll\", \"nghttp2_jll\"]\nuuid = \"deac9b47-8bc7-5906-a0fe-35ac56dc84c0\"\n\n[[LibGit2]]\ndeps = [\"Base64\", \"NetworkOptions\", \"Printf\", \"SHA\"]\nuuid = \"76f85450-5226-5b5a-8eaa-529ad045b433\"\n\n[[LibSSH2_jll]]\ndeps = [\"Artifacts\", \"Libdl\", \"MbedTLS_jll\"]\nuuid = \"29816b5a-b9ab-546f-933c-edad1886dfa8\"\n\n[[Libdl]]\nuuid = \"8f399da3-3557-5675-b5ff-fb832c97cbdb\"\n\n[[LinearAlgebra]]\ndeps = [\"Libdl\"]\nuuid = \"37e2e46d-f89d-539d-b4ee-838fcccc9c8e\"\n\n[[Logging]]\nuuid = \"56ddb016-857b-54e1-b83d-db4d58db5568\"\n\n[[MacroTools]]\ndeps = [\"Markdown\", \"Random\"]\ngit-tree-sha1 = \"0fb723cd8c45858c22169b2e42269e53271a6df7\"\nuuid = \"1914dd2f-81c6-5fcd-8719-6d5c9610ff09\"\nversion = \"0.5.7\"\n\n[[Markdown]]\ndeps = [\"Base64\"]\nuuid = \"d6f4376e-aef5-505a-96c1-9c027394607a\"\n\n[[MbedTLS_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"c8ffd9c3-330d-5841-b78e-0817d7145fa1\"\n\n[[Missings]]\ndeps = [\"DataAPI\"]\ngit-tree-sha1 = \"2ca267b08821e86c5ef4376cffed98a46c2cb205\"\nuuid = \"e1d29d7a-bbdc-5cf2-9ac0-f12de2c33e28\"\nversion = \"1.0.1\"\n\n[[Mmap]]\nuuid = \"a63ad114-7e13-5084-954f-fe012c677804\"\n\n[[MozillaCACerts_jll]]\nuuid = \"14a3606d-f60d-562e-9121-12d972cd8159\"\n\n[[NetworkOptions]]\nuuid = \"ca575930-c2e3-43a9-ace4-1e988b2c1908\"\n\n[[OrderedCollections]]\ngit-tree-sha1 = \"85f8e6578bf1f9ee0d11e7bb1b1456435479d47c\"\nuuid = \"bac558e1-5e72-5ebc-8fee-abe8a469f55d\"\nversion = \"1.4.1\"\n\n[[Parsers]]\ndeps = [\"Dates\"]\ngit-tree-sha1 = \"bfd7d8c7fd87f04543810d9cbd3995972236ba1b\"\nuuid = \"69de0a69-1ddd-5017-9359-2bf0b02dc9f0\"\nversion = \"1.1.2\"\n\n[[Pkg]]\ndeps = [\"Artifacts\", \"Dates\", \"Downloads\", \"LibGit2\", \"Libdl\", \"Logging\", \"Markdown\", \"Printf\", \"REPL\", \"Random\", \"SHA\", \"Serialization\", \"TOML\", \"Tar\", \"UUIDs\", \"p7zip_jll\"]\nuuid = \"44cfe95a-1eb2-52ea-b672-e2afdf69b78f\"\n\n[[PlutoTeachingTools]]\ndeps = [\"LaTeXStrings\", \"Markdown\", \"PlutoUI\", \"Random\"]\ngit-tree-sha1 = \"e2b63ee022e0b20f43fcd15cda3a9047f449e3b4\"\nuuid = \"661c6b06-c737-4d37-b85c-46df65de6f69\"\nversion = \"0.1.4\"\n\n[[PlutoUI]]\ndeps = [\"Base64\", \"Dates\", \"InteractiveUtils\", \"JSON\", \"Logging\", \"Markdown\", \"Random\", \"Reexport\", \"Suppressor\"]\ngit-tree-sha1 = \"44e225d5837e2a2345e69a1d1e01ac2443ff9fcb\"\nuuid = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\nversion = \"0.7.9\"\n\n[[PooledArrays]]\ndeps = [\"DataAPI\", \"Future\"]\ngit-tree-sha1 = \"cde4ce9d6f33219465b55162811d8de8139c0414\"\nuuid = \"2dfb63ee-cc39-5dd5-95bd-886bf059d720\"\nversion = \"1.2.1\"\n\n[[Preferences]]\ndeps = [\"TOML\"]\ngit-tree-sha1 = \"00cfd92944ca9c760982747e9a1d0d5d86ab1e5a\"\nuuid = \"21216c6a-2e73-6563-6e65-726566657250\"\nversion = \"1.2.2\"\n\n[[PrettyTables]]\ndeps = [\"Crayons\", \"Formatting\", \"Markdown\", \"Reexport\", \"Tables\"]\ngit-tree-sha1 = \"0d1245a357cc61c8cd61934c07447aa569ff22e6\"\nuuid = \"08abe8d2-0d0c-5749-adfa-8a2ac140af0d\"\nversion = \"1.1.0\"\n\n[[Printf]]\ndeps = [\"Unicode\"]\nuuid = \"de0858da-6303-5e67-8744-51eddeeeb8d7\"\n\n[[PyCall]]\ndeps = [\"Conda\", \"Dates\", \"Libdl\", \"LinearAlgebra\", \"MacroTools\", \"Serialization\", \"VersionParsing\"]\ngit-tree-sha1 = \"169bb8ea6b1b143c5cf57df6d34d022a7b60c6db\"\nuuid = \"438e738f-606a-5dbb-bf0a-cddfbfd45ab0\"\nversion = \"1.92.3\"\n\n[[REPL]]\ndeps = [\"InteractiveUtils\", \"Markdown\", \"Sockets\", \"Unicode\"]\nuuid = \"3fa0cd96-eef1-5676-8a61-b3b8758bbffb\"\n\n[[Random]]\ndeps = [\"Serialization\"]\nuuid = \"9a3f8284-a2c9-5f02-9a11-845980a1fd5c\"\n\n[[Reexport]]\ngit-tree-sha1 = \"5f6c21241f0f655da3952fd60aa18477cf96c220\"\nuuid = \"189a3867-3050-52da-a836-e630ba90ab69\"\nversion = \"1.1.0\"\n\n[[Requires]]\ndeps = [\"UUIDs\"]\ngit-tree-sha1 = \"4036a3bd08ac7e968e27c203d45f5fff15020621\"\nuuid = \"ae029012-a4dd-5104-9daa-d747884805df\"\nversion = \"1.1.3\"\n\n[[SHA]]\nuuid = \"ea8e919c-243c-51af-8825-aaa63cd721ce\"\n\n[[SentinelArrays]]\ndeps = [\"Dates\", \"Random\"]\ngit-tree-sha1 = \"54f37736d8934a12a200edea2f9206b03bdf3159\"\nuuid = \"91c51154-3ec4-41a3-a24f-3f23e20d615c\"\nversion = \"1.3.7\"\n\n[[Serialization]]\nuuid = \"9e88b42a-f829-5b0c-bbe9-9e923198166b\"\n\n[[SharedArrays]]\ndeps = [\"Distributed\", \"Mmap\", \"Random\", \"Serialization\"]\nuuid = \"1a1011a3-84de-559e-8e89-a11a2f7dc383\"\n\n[[Sockets]]\nuuid = \"6462fe0b-24de-5631-8697-dd941f90decc\"\n\n[[SortingAlgorithms]]\ndeps = [\"DataStructures\"]\ngit-tree-sha1 = \"b3363d7460f7d098ca0912c69b082f75625d7508\"\nuuid = \"a2af1166-a08f-5f64-846c-94a0d3cef48c\"\nversion = \"1.0.1\"\n\n[[SparseArrays]]\ndeps = [\"LinearAlgebra\", \"Random\"]\nuuid = \"2f01184e-e22b-5df5-ae63-d93ebab69eaf\"\n\n[[Statistics]]\ndeps = [\"LinearAlgebra\", \"SparseArrays\"]\nuuid = \"10745b16-79ce-11e8-11f9-7d13ad32a3b2\"\n\n[[Suppressor]]\ngit-tree-sha1 = \"a819d77f31f83e5792a76081eee1ea6342ab8787\"\nuuid = \"fd094767-a336-5f1f-9728-57cf17d0bbfb\"\nversion = \"0.2.0\"\n\n[[TOML]]\ndeps = [\"Dates\"]\nuuid = \"fa267f1f-6049-4f14-aa54-33bafae1ed76\"\n\n[[TableTraits]]\ndeps = [\"IteratorInterfaceExtensions\"]\ngit-tree-sha1 = \"c06b2f539df1c6efa794486abfb6ed2022561a39\"\nuuid = \"3783bdb8-4a98-5b6b-af9a-565f29a5fe9c\"\nversion = \"1.0.1\"\n\n[[Tables]]\ndeps = [\"DataAPI\", \"DataValueInterfaces\", \"IteratorInterfaceExtensions\", \"LinearAlgebra\", \"TableTraits\", \"Test\"]\ngit-tree-sha1 = \"d0c690d37c73aeb5ca063056283fde5585a41710\"\nuuid = \"bd369af6-aec1-5ad0-b16a-f7cc5008161c\"\nversion = \"1.5.0\"\n\n[[Tar]]\ndeps = [\"ArgTools\", \"SHA\"]\nuuid = \"a4e569a6-e804-4fa4-b0f3-eef7a1d5b13e\"\n\n[[Test]]\ndeps = [\"InteractiveUtils\", \"Logging\", \"Random\", \"Serialization\"]\nuuid = \"8dfed614-e22c-5e08-85e1-65c5234f0b40\"\n\n[[TranscodingStreams]]\ndeps = [\"Random\", \"Test\"]\ngit-tree-sha1 = \"216b95ea110b5972db65aa90f88d8d89dcb8851c\"\nuuid = \"3bb67fe8-82b1-5028-8e26-92a6c54297fa\"\nversion = \"0.9.6\"\n\n[[UUIDs]]\ndeps = [\"Random\", \"SHA\"]\nuuid = \"cf7118a7-6976-5b1a-9a39-7adc72f591a4\"\n\n[[Unicode]]\nuuid = \"4ec0a83e-493e-50e2-b9ac-8f72acf5a8f5\"\n\n[[VersionParsing]]\ngit-tree-sha1 = \"80229be1f670524750d905f8fc8148e5a8c4537f\"\nuuid = \"81def892-9a0e-5fdd-b105-ffc91e053289\"\nversion = \"1.2.0\"\n\n[[Zlib_jll]]\ndeps = [\"Libdl\"]\nuuid = \"83775a58-1f1d-513f-b197-d71354ab007a\"\n\n[[nghttp2_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"8e850ede-7688-5339-a07c-302acd2aaf8d\"\n\n[[p7zip_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"3f19e933-33d8-53b3-aaab-bd5110c3b7a0\"\n\"\"\"\n\n# \u2554\u2550\u2561 Cell order:\n# \u255f\u2500e0e60e09-3808-4d6b-a773-6ba59c02f517\n# \u255f\u25008c1d81ab-d215-4972-afeb-7e00bf6063c2\n# \u255f\u25001607eac9-e76f-4d1f-a9ce-981ce3be9bea\n# \u2560\u2550f27e1e8f-15eb-4754-a94c-7f37c54b871e\n# \u2560\u255080f02c3a-6751-48df-92ec-13f5c7d8c71e\n# \u255f\u2500624c9038-3008-4e78-a149-60796dacf9c0\n# \u255f\u25002eb255d9-707d-4224-a0ce-fe90a1c69722\n# \u255f\u25008571ccaf-5fbc-4593-82db-ead073a4074f\n# \u2560\u2550442604d0-b490-404b-8b1c-7e1c01cebad7\n# \u255f\u250070f5fe63-fea1-4afc-b03d-f113b8bd621e\n# \u2560\u25508cd5a772-a3aa-4166-92e6-39436d0d2278\n# \u255f\u2500074b00ce-dae6-4e4f-8673-49be73085327\n# \u2560\u2550ced3ecc9-94ea-408f-bdef-d34802166663\n# \u255f\u2500c62adfb6-6ef8-45fd-992f-07bca59f82cd\n# \u255f\u250003edae54-4368-4b81-8713-17f93bbb9ed9\n# \u255f\u2500e0b82e6c-043c-4e66-a2a4-ae0f3e4e5404\n# \u2560\u2550aa62f1e5-6af5-43f6-be11-665ed25986c0\n# \u255f\u2500540468dc-3f9f-457b-9b44-efb3ec036166\n# \u2560\u255021e3adbb-0176-4963-bdfc-0ba2422af4bf\n# \u2560\u2550d27aef3b-bfe7-466a-9580-93fc53dcef18\n# \u255f\u250002f638d9-ec2f-4072-9fb3-e6fabac1b1e6\n# \u2560\u2550481c5b7a-4660-4954-84f3-29d33bd73f3d\n# \u255f\u2500aba84742-a927-4ce5-9f7c-6e535db1ee47\n# \u255f\u2500aa7c4dfe-a3d4-448b-a559-1bc7b338a1dc\n# \u2560\u2550a97f9c8e-dbd3-4613-9c6e-c471340ea2d6\n# \u255f\u25007c7430a0-d16e-485b-bbd9-7f231fee853a\n# \u255f\u2500cebe52b3-386c-4d31-8497-c19b6c742577\n# \u255f\u25003d09d38a-ca72-43f2-bd85-17fc0e05a645\n# \u2560\u25509257c879-d96f-4d2b-994e-d542617b0c65\n# \u255f\u2500307a7a29-afa6-4c4d-88ae-4f757aeba892\n# \u2560\u2550afdc442a-2931-4129-824e-98431e1d8be2\n# \u255f\u2500c42deea3-acf1-491a-97bd-fdc472917584\n# \u255f\u25007c1f4a27-dfd7-428a-97ef-eac1784e8d6f\n# \u2560\u255097111ccc-8a16-4e7d-adf7-0dcd90524be3\n# \u255f\u2500f45668b1-1cbd-48e6-9224-d469826e093b\n# \u2560\u255052beb148-6214-4a03-a0cf-6c2a6da77d40\n# \u255f\u25001ecbd400-0157-45ce-b1b8-db764012ba5e\n# \u255f\u2500191ba96e-2573-4bc1-a352-46a66e0a5c4f\n# \u2560\u255082a757ad-566d-4c1d-8b3d-366ffd980fb4\n# \u255f\u25003e550b71-4750-460b-be18-911a848a8f49\n# \u2560\u2550b0c3875b-ef07-4e92-a0a8-55f42b266c6b\n# \u255f\u250058e021c6-500f-40d8-a388-a5732bc808b3\n# \u2560\u255036bfd224-a13b-438c-9106-382c15a6a1d2\n# \u255f\u25007f4c3ab8-041e-4ef0-bb2e-320f5293cdde\n# \u255f\u2500195c2df4-7e1c-49f8-871f-f76b985dab6d\n# \u255f\u250032124f8a-f5cf-41d1-97a9-ce1d05145fde\n# \u2560\u2550507e1516-5433-49eb-831d-32426f30895e\n# \u255f\u2500eac67cc9-754b-4f7d-add8-93900a1b5b49\n# \u255f\u250064224c6b-c5a0-44f2-b2a0-7f77759cb848\n# \u255f\u250073c06bb1-be49-46bd-b7f1-c45cc56af7b4\n# \u255f\u2500ebfaa677-829b-4bf9-bdbb-19f3c87dd3a4\n# \u255f\u2500945f5a55-3026-4497-9ece-8af878c87788\n# \u2560\u255057397ee4-9efc-48b3-b640-d2b7a10da633\n# \u255f\u25008059a6a3-384a-4344-8a23-650ee0be10c2\n# \u255f\u2500f5b93929-2c59-4360-8c41-97a1324ba455\n# \u2560\u2550122196fa-45ca-4031-85eb-4afd4782de9e\n# \u255f\u2500e9dc1456-616b-4e4b-b209-9f6ba4c48607\n# \u255f\u2500f15d37a7-d962-4da0-977f-76729a3313be\n# \u255f\u250028f195c4-4f61-4873-85d6-b4e3aaa3660f\n# \u2560\u25503837e439-250b-4577-b0d7-93352ec19f6e\n# \u255f\u25008255b5a9-cbe6-4604-bedd-0e366f311096\n# \u2560\u2550df9b701f-d314-4512-b2ea-1f6ae015166c\n# \u255f\u2500377f7527-a338-4526-bb24-9766c635e719\n# \u255f\u250025f24754-16d4-4343-b54a-cf8ea1358ce9\n# \u255f\u2500570fd826-23fd-46ee-bdb4-58fb0c45719a\n# \u2560\u2550691410bb-0472-4800-a90d-29ddf947de3e\n# \u255f\u25008cbb1c90-bd94-44b5-80b6-81d38f3e6252\n# \u2560\u2550c3065acf-6205-455f-ba74-ca51f3f6761b\n# \u255f\u25009c392be9-1505-40bc-a290-68085a1c2700\n# \u255f\u2500d738bdcc-5f83-4dfa-a17f-e9ea23db2986\n# \u255f\u2500d55ce157-099c-4c1b-94db-62918f04e5fe\n# \u255f\u2500206f464b-55fe-46aa-85b7-8f0246a0aaad\n# \u255f\u25006f72d1b2-63f6-4301-8272-bb2d6d2d049e\n# \u2560\u255001201c37-0b79-46b1-a001-e716f5b3ba67\n# \u255f\u2500e3d37bc1-a119-4add-a111-899ee0caea05\n# \u255f\u25006dff3f21-fac0-42e1-910a-f969a231374f\n# \u2560\u2550b26b8253-e6cd-49f4-81c5-2a3c2963a37c\n# \u255f\u2500e5dc123f-1596-4311-9398-f0cfe80a5342\n# \u255f\u25007b9d26c2-899e-45e9-b664-39d5f1adfe3f\n# \u255f\u2500fc01d57f-c90b-4231-96be-ddd48656d55e\n# \u2560\u2550e05f16d6-eb50-49a9-bf14-95d63c9da7ff\n# \u255f\u25003b232365-f2fe-4edb-a39f-3e37c8cbb666\n# \u2560\u25501992595f-9976-4b18-bf9c-df8a73d30dc8\n# \u255f\u2500568862b3-6fce-426a-a9e2-e558adf3932a\n# \u255f\u250052f9edd0-79b0-4a9a-9930-3a05d3aa2447\n# \u255f\u250090d5244e-17be-4601-b922-8c254f1248bf\n# \u255f\u2500ae79ee89-d788-4612-8b1d-fc22d85c7744\n# \u2560\u25503745c237-ba48-4f2c-959e-441484244764\n# \u255f\u25006d061411-6f19-4119-aefb-cc380198b0ce\n# \u255f\u25008def87d2-f10b-4a82-b353-a6477eeead9b\n# \u255f\u2500c74b3105-f480-4688-b85f-3e7dff70da3b\n# \u2560\u255055438c09-1d94-4ff7-90c3-0cc6064a091e\n# \u255f\u2500bcc796c9-db11-4a09-a5f9-215127ac0938\n# \u255f\u250092b2ecdd-9491-4f16-8cb9-bacbcf180280\n# \u2560\u255083216915-cdcf-4f0f-829f-a5ff4c4b8da0\n# \u255f\u2500452872f9-2009-4733-b2d3-28f262ae19b7\n# \u255f\u250029415ddc-e002-4f56-a169-95f7b1c36be9\n# \u255f\u2500fb23d6c6-b812-4fe1-b224-0014bedbd43f\n# \u2560\u25501e53aa10-dff6-40d5-89e2-da194ffc2052\n# \u2560\u2550ac87ced2-986f-4e6c-80b2-104b25c171c2\n# \u2560\u255014cca8ce-cc61-4fae-b871-21c3fd23d0ea\n# \u2560\u255028fc8de4-749b-4093-b32f-c398f8d27d3d\n# \u2560\u255057b422e5-0ad0-4674-bdd3-a8358bc7aaeb\n# \u255f\u250000000000-0000-0000-0000-000000000001\n# \u255f\u250000000000-0000-0000-0000-000000000002\n", "meta": {"hexsha": "9f1deb054894d9474c6a7f29abbb30367ac1ac72", "size": 44325, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "ex2.jl", "max_stars_repo_name": "PsuAstro528/lab3-start", "max_stars_repo_head_hexsha": "adc592c7a3d00a2402ebe1943ab4a09c8d8cabfa", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2019-11-20T16:12:26.000Z", "max_stars_repo_stars_event_max_datetime": "2019-11-20T16:12:26.000Z", "max_issues_repo_path": "ex2.jl", "max_issues_repo_name": "PsuAstro528/lab3-start", "max_issues_repo_head_hexsha": "adc592c7a3d00a2402ebe1943ab4a09c8d8cabfa", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "ex2.jl", "max_forks_repo_name": "PsuAstro528/lab3-start", "max_forks_repo_head_hexsha": "adc592c7a3d00a2402ebe1943ab4a09c8d8cabfa", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 40.5535224154, "max_line_length": 683, "alphanum_fraction": 0.7465313029, "num_tokens": 17288, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.34158249943831703, "lm_q2_score": 0.16885695214168317, "lm_q1q2_score": 0.05767857976009242}}
{"text": "module IOcdf\n\n\"\"\"\n module EmpiricalCDFS.IOcdf\n\nBinary IO functions for `EmpiricalCDFs`.\n\nTypes: `CDFfile`\n\nFunctions: `save`, `save`, `readcdf`, `readcdfinfo`, `header`, `version`, `getcdf`, `data`\n\n`CDFfile` contains an `AbstractEmpiricalCDF` as a field. Many functions on the latter are forwarded to\nthe former.\n\"\"\"\nIOcdf\n\nexport CDFfile, save, readcdf, readcdfinfo, header, version, getcdf, data\n\nimport Statistics\nusing ..EmpiricalCDFs: AbstractEmpiricalCDF, EmpiricalCDF, EmpiricalCDFHi\n\nimport ..EmpiricalCDFs: data, linprint, logprint, getindex, getcdfindex, counts\n\ninclude(\"readstring.jl\")\n\n#### CDFfile type\n\n@enum CDFTYPE EmpiricalCDFtype=1 EmpiricalCDFHitype=2\n\nconst CDFfileVersion = v\"0.0.2\"\n\n\"\"\"\n CDFfile(cdf::AbstractEmpiricalCDF, header=\"\")\n\n struct CDFfile{T <: AbstractEmpiricalCDF}\n cdf::T\n header::String\n end\n\nBinary data file for `AbstractEmpiricalCDF`\n\nThe file format is\n\n- Identifying string\n- `n::Int64` number of bytes in the header string\n- `s::String` The header string\n- `t::Int64` Type of `AbstractEmpiricalCDF`, 1 or 2. 1 for `EmpiricalCDF`, 2 for `EmpiricalCDFHi`.\n- `lowreject::Float64` the lower cutoff, only for `EmpiricalCDFHi`.\n- `npts::Int64` number of data points in the CDF\n- `npts` data points of type `Float64`\n\"\"\"\nstruct CDFfile{T <: AbstractEmpiricalCDF}\n cdf::T\n header::String\n vn::VersionNumber\nend\n\nCDFfile(cdf) = CDFfile(cdf,\"\",CDFfileVersion)\n\ndata(cdff::CDFfile) = data(cdff.cdf)\n\nfunction Base.show(io::IO, cdff::CDFfile)\n print(io, string(typeof(cdff)), \"(\")\n show(io,cdff.cdf)\n print(io, \",\")\n print(io, \"v\", cdff.vn)\n print(io, \")\")\n nothing\nend\n\n\"\"\"\n header::String = header(cdff::CDFfile)\n\nReturn the header from `cdff`.\n\"\"\"\nheader(cdff::CDFfile) = cdff.header\n\n\"\"\"\n cdf::AbstractEmpiricalCDF = getcdf(cdff::CDFfile)\n\nReturn the CDF from `cdff`.\n\"\"\"\ngetcdf(cdff::CDFfile) = cdff.cdf\n\n\"\"\"\n version(cdff::CDFfile)\n\nReturn the version number of the file format.\n\"\"\"\nversion(cdff::CDFfile) = cdff.vn\n\nfor f in (:sort!, :push!, :append!, :getindex, :length, :size, :firstindex, :lastindex, :rand, :minimum, :maximum)\n @eval Base.$(f)(cdff::CDFfile, args...) = $(f)(cdff.cdf, args...)\nend\n\nfor f in (:mean, :std, :quantile )\n @eval Statistics.$(f)(cdff::CDFfile, args...) = $(f)(cdff.cdf, args...)\nend\n\nfor f in (:getcdfindex, :counts)\n# :mle, :KSstatistic, :mleKS, :scanmle)\n @eval $(f)(cdff::CDFfile, args...) = $(f)(cdff.cdf, args...)\nend\n\nfor f in (:linprint, :logprint )\n @eval $(f)(ioorfile, cdff::CDFfile, args...) = $(f)(ioorfile, cdff.cdf, args...)\nend\n\n(cdff::CDFfile)(x::Real) = getcdf(cdff)(x)\n(cdff::CDFfile)(v::AbstractArray) = getcdf(cdff)(v)\n\n####\n\nfunction make_CDFfile_version_string(v)\n nchars = 100\n s = \"CDFfile \" * string(v)\n s = s * \" \"^(nchars-length(s))\nend\n\nmake_CDFfile_version_string() = make_CDFfile_version_string(CDFfileVersion)\n\nfunction read_CDFfile_version_string(io::IO)\n nchars = 100\n local s\n try\n s = readstring(io,nchars)\n catch\n error(\"Failed reading identifying string and version number from CDFfile\")\n end\n (filetype,versionstring) = split(s)\n if filetype != \"CDFfile\"\n error(\"Failed reading version number from CDFfile\")\n end\n local vn\n try\n vn = VersionNumber(versionstring)\n catch\n error(\"Unable to parse version number string '$versionstring'\")\n end\n vn\nend\n\n#### Writing CDFfile and CDF\n\nfunction save(io::IO,cdff::CDFfile)\n write(io, make_CDFfile_version_string())\n write(io,sizeof(cdff.header))\n write(io,cdff.header)\n save(io,cdff.cdf)\nend\n\nfunction save(io::IO,cdf::EmpiricalCDF)\n write(io, Int64(EmpiricalCDFtype))\n _writedata(io,cdf)\nend\n\nfunction save(io::IO,cdf::EmpiricalCDFHi)\n write(io, Int64(EmpiricalCDFHitype))\n write(io, convert(Float64, cdf.lowreject))\n _writedata(io, cdf)\nend\n\nfunction _writedata(io::IO,cdf::AbstractEmpiricalCDF)\n write(io, convert(Int64, length(cdf)))\n for x in cdf.xdata\n write(io,x)\n end\nend\n\n\"\"\"\n save(fn::String, cdf::AbstractEmpiricalCDF, header::String=\"\")\n\nwrite `cdf` to file `fn` in a fast binary format.\n\"\"\"\nfunction save(fn::String, cdf::AbstractEmpiricalCDF, header::String=\"\")\n io = open(fn,\"w\")\n save(io, CDFfile(cdf,header,CDFfileVersion))\n close(io)\nend\n\n#save(fn::String, cdf::AbstractEmpiricalCDF) = save(fn,cdf,\"\")\n\n#### Reading CDFfile and CDF\n\nfunction readcdfdata(io::IO, cdf::AbstractEmpiricalCDF)\n npts = read(io,Int64)\n resize!(cdf.xdata,npts)\n@inbounds for i in 1:npts\n x = read(io,Float64)\n cdf.xdata[i] = x\n end\nend\n\nfunction readcdf(io::IO)\n info = readcdfinfo(io)\n (vn,header) = (info.vn,info.header)\n cdf = typeof(info) == CDFInfo ? EmpiricalCDF() : EmpiricalCDFHi(info.lowreject)\n readcdfdata(io,cdf)\n CDFfile(cdf,header,vn)\nend\n\nstruct CDFInfo\n vn::VersionNumber\n header::String\n npts::Int\nend\n\nfunction Base.show(io::IO, i::CDFInfo)\n print(io,typeof(i),\"(\")\n print(io,\"length(header)=\", length(i.header))\n print(io,\",vn=v\", i.vn)\n print(io,\",npts=\",i.npts,\")\")\nend\n\nstruct CDFHiInfo\n vn::VersionNumber\n header::String\n npts::Int\n lowreject::Float64\nend\n\nfunction Base.show(io::IO, i::CDFHiInfo)\n print(io,typeof(i),\"(\")\n print(io,\"length(header)=\", length(i.header))\n print(io,\",vn=v\", i.vn)\n print(io,\",npts=\",i.npts)\n print(io,\",lowreject=\",i.lowreject,\")\")\nend\n\nfunction readcdfinfo(io::IO)\n vn = read_CDFfile_version_string(io)\n header = readlengthandstring(io)\n cdftype = CDFTYPE(read(io,Int64))\n local lowreject\n if cdftype == EmpiricalCDFtype\n npts = peektype(io,Int64)\n CDFInfo(vn,header,npts)\n elseif cdftype == EmpiricalCDFHitype\n lowreject = read(io,Float64)\n npts = peektype(io,Int64)\n CDFHiInfo(vn,header,npts,lowreject)\n else\n error(\"Uknown cdf type \", cdftype)\n end\nend\n\n\"\"\"\n readcdfinfo(fn::String)\n\nReturn an object containing information about the cdf saved in the binary\nfile `fn`. The data itself is not read.\n\"\"\"\nfunction readcdfinfo(fn::String)\n io = open(fn,\"r\")\n info = readcdfinfo(io)\n close(io)\n info\nend\n\n\"\"\"\n readcdf(fn::String)\n\nRead an empirical CDF from file `fn`. Return an object\n`cdff` of type `CDFfile`. The header is in field `header`.\nThe cdf is in in field `cdf`.\n\"\"\"\nfunction readcdf(fn::String)\n io = open(fn,\"r\")\n cdffile = readcdf(io)\n close(io)\n cdffile\nend\n\nend # module IO\n", "meta": {"hexsha": "24eb0382ad7f0b11dc21f939decb82ff4a574f64", "size": 6495, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/IOcdf.jl", "max_stars_repo_name": "jlapeyre/EmpiricalCDFs.jl", "max_stars_repo_head_hexsha": "37c68e4f382486c56f67e8e1b1b4f00c907f6ed0", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 26, "max_stars_repo_stars_event_min_datetime": "2018-05-09T16:49:10.000Z", "max_stars_repo_stars_event_max_datetime": "2022-02-08T07:46:20.000Z", "max_issues_repo_path": "src/IOcdf.jl", "max_issues_repo_name": "jlapeyre/EmpiricalCDFs.jl", "max_issues_repo_head_hexsha": "37c68e4f382486c56f67e8e1b1b4f00c907f6ed0", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 9, "max_issues_repo_issues_event_min_datetime": "2018-12-13T08:43:47.000Z", "max_issues_repo_issues_event_max_datetime": "2020-02-21T08:46:00.000Z", "max_forks_repo_path": "src/IOcdf.jl", "max_forks_repo_name": "jlapeyre/EmpiricalCDFs.jl", "max_forks_repo_head_hexsha": "37c68e4f382486c56f67e8e1b1b4f00c907f6ed0", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 4, "max_forks_repo_forks_event_min_datetime": "2018-05-21T17:48:03.000Z", "max_forks_repo_forks_event_max_datetime": "2021-08-03T12:54:17.000Z", "avg_line_length": 23.6181818182, "max_line_length": 114, "alphanum_fraction": 0.6635873749, "num_tokens": 2001, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.47657965106367595, "lm_q2_score": 0.120853219824755, "lm_q1q2_score": 0.05759618533400346}}
{"text": "\"\"\"\n @add_kwonly function_definition\n\nDefine keyword-only version of the `function_definition`.\n\n @add_kwonly function f(x; y=1)\n ...\n end\n\nexpands to:\n\n function f(x; y=1)\n ...\n end\n function f(; x = error(\"No argument x\"), y=1)\n ...\n end\n\"\"\"\nmacro add_kwonly(ex)\n esc(add_kwonly(ex))\nend\n\nadd_kwonly(ex::Expr) = add_kwonly(Val{ex.head}, ex)\n\nfunction add_kwonly(::Type{<: Val}, ex)\n error(\"add_only does not work with expression $(ex.head)\")\nend\n\nfunction add_kwonly(::Union{Type{Val{:function}},\n Type{Val{:(=)}}}, ex::Expr)\n body = ex.args[2:end] # function body\n default_call = ex.args[1] # e.g., :(f(a, b=2; c=3))\n kwonly_call = add_kwonly(default_call)\n if kwonly_call === nothing\n return ex\n end\n\n return quote\n begin\n $ex\n $(Expr(ex.head, kwonly_call, body...))\n end\n end\nend\n\nfunction add_kwonly(::Type{Val{:where}}, ex::Expr)\n default_call = ex.args[1]\n rest = ex.args[2:end]\n kwonly_call = add_kwonly(default_call)\n if kwonly_call === nothing\n return nothing\n end\n return Expr(:where, kwonly_call, rest...)\nend\n\nfunction add_kwonly(::Type{Val{:call}}, default_call::Expr)\n # default_call is, e.g., :(f(a, b=2; c=3))\n funcname = default_call.args[1] # e.g., :f\n required = [] # required positional arguments; e.g., [:a]\n optional = [] # optional positional arguments; e.g., [:(b=2)]\n default_kwargs = []\n for arg in default_call.args[2:end]\n if isa(arg, Symbol)\n push!(required, arg)\n elseif arg.head == :(::)\n push!(required, arg)\n elseif arg.head == :kw\n push!(optional, arg)\n elseif arg.head == :parameters\n @assert default_kwargs == [] # can I have :parameters twice?\n default_kwargs = arg.args\n else\n error(\"Not expecting to see: $arg\")\n end\n end\n if isempty(required) && isempty(optional)\n # If the function is already keyword-only, do nothing:\n return nothing\n end\n if isempty(required)\n # It's not clear what should be done. Let's not support it at\n # the moment:\n error(\"At least one positional mandatory argument is required.\")\n end\n\n kwonly_kwargs = Expr(:parameters, [\n Expr(:kw, pa, :(error($(\"No argument $pa\"))))\n for pa in required\n ]..., optional..., default_kwargs...)\n kwonly_call = Expr(:call, funcname, kwonly_kwargs)\n # e.g., :(f(; a=error(...), b=error(...), c=1, d=2))\n\n return kwonly_call\nend\n\nfunction num_types_in_tuple(sig)\n length(sig.parameters)\nend\n\nfunction num_types_in_tuple(sig::UnionAll)\n length(Base.unwrap_unionall(sig).parameters)\nend\n\nfunction numargs(f)\n typ = Tuple{Any, Val{:analytic}, Vararg}\n typ2 = Tuple{Any, Type{Val{:analytic}}, Vararg} # This one is required for overloaded types\n typ3 = Tuple{Any, Val{:jac}, Vararg}\n typ4 = Tuple{Any, Type{Val{:jac}}, Vararg} # This one is required for overloaded types\n typ5 = Tuple{Any, Val{:tgrad}, Vararg}\n typ6 = Tuple{Any, Type{Val{:tgrad}}, Vararg} # This one is required for overloaded types\n numparam = maximum([(m.sig<:typ || m.sig<:typ2 || m.sig<:typ3 || m.sig<:typ4 || m.sig<:typ5 || m.sig<:typ6) ? 0 : num_types_in_tuple(m.sig) for m in methods(f)])\n return (numparam-1) #-1 in v0.5 since it adds f as the first parameter\nend\n\nfunction isinplace(f,inplace_param_number)\n numargs(f)>=inplace_param_number\nend\n\n### Default Linsolve\n\n# Try to be as smart as possible\n# lu! if Matrix\n# lu if sparse\n# gmres if operator\n\nmutable struct DefaultLinSolve\n A\n iterable\nend\nDefaultLinSolve() = DefaultLinSolve(nothing, nothing)\n\nfunction (p::DefaultLinSolve)(x,A,b,update_matrix=false;tol=nothing, kwargs...)\n if p.iterable isa Vector && eltype(p.iterable) <: LinearAlgebra.BlasInt # `iterable` here is the pivoting vector\n F = LU{eltype(A)}(A, p.iterable, zero(LinearAlgebra.BlasInt))\n ldiv!(x, F, b)\n return nothing\n end\n if update_matrix\n if typeof(A) <: Matrix\n blasvendor = BLAS.vendor()\n # if the user doesn't use OpenBLAS, we assume that is a better BLAS\n # implementation like MKL\n #\n # RecursiveFactorization seems to be consistantly winning below 100\n # https://discourse.julialang.org/t/ann-recursivefactorization-jl/39213\n if ArrayInterface.can_setindex(x) && (size(A,1) <= 100 || ((blasvendor === :openblas || blasvendor === :openblas64) && size(A,1) <= 500))\n p.A = RecursiveFactorization.lu!(A)\n else\n p.A = lu!(A)\n end\n elseif typeof(A) <: Tridiagonal\n p.A = lu!(A)\n elseif typeof(A) <: Union{SymTridiagonal}\n p.A = ldlt!(A)\n elseif typeof(A) <: Union{Symmetric,Hermitian}\n p.A = bunchkaufman!(A)\n elseif typeof(A) <: SparseMatrixCSC\n p.A = lu(A)\n elseif ArrayInterface.isstructured(A)\n p.A = factorize(A)\n elseif !(typeof(A) <: AbstractDiffEqOperator)\n # Most likely QR is the one that is overloaded\n # Works on things like CuArrays\n p.A = qr(A)\n end\n end\n\n if typeof(A) <: Union{Matrix,SymTridiagonal,Tridiagonal,Symmetric,Hermitian} # No 2-arg form for SparseArrays!\n x .= b\n ldiv!(p.A,x)\n # Missing a little bit of efficiency in a rare case\n #elseif typeof(A) <: DiffEqArrayOperator\n # ldiv!(x,p.A,b)\n elseif ArrayInterface.isstructured(A) || A isa SparseMatrixCSC\n ldiv!(x,p.A,b)\n elseif typeof(A) <: AbstractDiffEqOperator\n # No good starting guess, so guess zero\n if p.iterable === nothing\n p.iterable = IterativeSolvers.gmres_iterable!(x,A,b;initially_zero=true,restart=5,maxiter=5,tol=1e-16,kwargs...)\n p.iterable.reltol = tol\n end\n x .= false\n iter = p.iterable\n purge_history!(iter, x, b)\n\n for residual in iter\n end\n else\n ldiv!(x,p.A,b)\n end\n return nothing\nend\n\nfunction (p::DefaultLinSolve)(::Type{Val{:init}},f,u0_prototype)\n DefaultLinSolve()\nend\n\nconst DEFAULT_LINSOLVE = DefaultLinSolve()\n\n@inline UNITLESS_ABS2(x) = real(abs2(x))\n@inline DEFAULT_NORM(u::Union{AbstractFloat,Complex}) = @fastmath abs(u)\n@inline DEFAULT_NORM(u::Array{T}) where T<:Union{AbstractFloat,Complex} =\n sqrt(real(sum(abs2,u)) / length(u))\n@inline DEFAULT_NORM(u::StaticArray{T}) where T<:Union{AbstractFloat,Complex} =\n sqrt(real(sum(abs2,u)) / length(u))\n@inline DEFAULT_NORM(u::RecursiveArrayTools.AbstractVectorOfArray) =\n sum(sqrt(real(sum(UNITLESS_ABS2,_u)) / length(_u)) for _u in u.u)\n@inline DEFAULT_NORM(u::AbstractArray) = sqrt(real(sum(UNITLESS_ABS2,u)) / length(u))\n@inline DEFAULT_NORM(u) = norm(u)\n\n\"\"\"\n prevfloat_tdir(x, x0, x1)\n\nMove `x` one floating point towards x0.\n\"\"\"\nfunction prevfloat_tdir(x, x0, x1)\n x1 > x0 ? prevfloat(x) : nextfloat(x)\nend\n\nfunction nextfloat_tdir(x, x0, x1)\n x1 > x0 ? nextfloat(x) : prevfloat(x)\nend\n\nfunction max_tdir(a, b, x0, x1)\n x1 > x0 ? max(a, b) : min(a, b)\nend\n\nalg_autodiff(alg::AbstractNewtonAlgorithm{CS,AD}) where {CS,AD} = AD\nalg_autodiff(alg) = false\n\n\"\"\"\n value_derivative(f, x)\n\nCompute `f(x), d/dx f(x)` in the most efficient way.\n\"\"\"\nfunction value_derivative(f::F, x::R) where {F,R}\n T = typeof(ForwardDiff.Tag(f, R))\n out = f(ForwardDiff.Dual{T}(x, one(x)))\n ForwardDiff.value(out), ForwardDiff.extract_derivative(T, out)\nend\n\nvalue(x) = x\nvalue(x::Dual) = ForwardDiff.value(x)\nvalue(x::AbstractArray{<:Dual}) = map(ForwardDiff.value, x)\n", "meta": {"hexsha": "df2ae74b913403957c0244a69ca67087bfdeb94b", "size": 7356, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/utils.jl", "max_stars_repo_name": "utkarsh530/NonlinearSolve.jl", "max_stars_repo_head_hexsha": "f232c8f06a991cbc1849659017ee6daa414d5bbc", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/utils.jl", "max_issues_repo_name": "utkarsh530/NonlinearSolve.jl", "max_issues_repo_head_hexsha": "f232c8f06a991cbc1849659017ee6daa414d5bbc", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/utils.jl", "max_forks_repo_name": "utkarsh530/NonlinearSolve.jl", "max_forks_repo_head_hexsha": "f232c8f06a991cbc1849659017ee6daa414d5bbc", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 30.0244897959, "max_line_length": 163, "alphanum_fraction": 0.6500815661, "num_tokens": 2196, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.34510525748676846, "lm_q2_score": 0.1666754088273618, "lm_q1q2_score": 0.05752055988007909}}
{"text": "\"\"\"\n\thistogram(cmd0::String=\"\", arg1=[]; kwargs...)\n\nReads file and examines the first data column to calculate histogram parameters based on the bin-width provided.\n\nFull option list at [`pshistogram`](http://gmt.soest.hawaii.edu/doc/latest/pshistogram.html)\n\nParameters\n----------\n\n- $(GMT.opt_J)\n- **W** : **bin** : **width** : -- Number or Str --\n\n\tSets the bin width used for histogram calculations.\n\t[`-W`](http://gmt.soest.hawaii.edu/doc/latest/pshistogram.html#w)\n- **A** : **horizontal** : -- Bool or [] --\n\n\tPlot the histogram horizontally from x = 0 [Default is vertically from y = 0].\n\t[`-A`](http://gmt.soest.hawaii.edu/doc/latest/pshistogram.html#a)\n- $(GMT.opt_B)\n- **C** : **color** : -- Str or GMTcpt --\n\n\tGive a CPT. The mid x-value for each bar is used to look-up the bar color.\n\t[`-C`](http://gmt.soest.hawaii.edu/doc/latest/pshistogram.html#c)\n- **D** : **annot** : **annotate** : -- Str or [] --\n\n\tAnnotate each bar with the count it represents.\n\t[`-D`](http://gmt.soest.hawaii.edu/doc/latest/pshistogram.html#d)\n- **F** : **center** : -- Bool or [] --\n\n\tCenter bin on each value. [Default is left edge].\n\t[`-F`](http://gmt.soest.hawaii.edu/doc/latest/pshistogram.html#f)\n- **G** : **fill** : -- Number or Str --\n\n\tSelect filling of bars [if no G, L or C set G=100].\n\t[`-G`](http://gmt.soest.hawaii.edu/doc/latest/pshistogram.html#g)\n- **I** : **inquire** : -- Bool or [] --\n\n\tInquire about min/max x and y after binning.\n\t[`-I`](http://gmt.soest.hawaii.edu/doc/latest/pshistogram.html#i)\n- **L** : **labels** : -- Str or [] --\n\n\tDraw bar outline using the specified pen thickness [if no G, L or C set L=0.5].\n\t[`-L`](http://gmt.soest.hawaii.edu/doc/latest/pshistogram.html#l)\n- **N** : **normal** : -- Str --\n\n\tDraw the equivalent normal distribution; append desired pen [0.5p,black].\n\t[`-N`](http://gmt.soest.hawaii.edu/doc/latest/pshistogram.html#n)\n- $(GMT.opt_P)\n- **Q** : **alpha** : -- Number or [] --\n\n\tSets the confidence level used to determine if the mean resultant is significant.\n\t[`-Q`](http://gmt.soest.hawaii.edu/doc/latest/pshistogram.html#q)\n- **R** : **region** : -- Str --\n\n\tSpecifies the \u2018region\u2019 of interest in (r,azimuth) space. r0 is 0, r1 is max length in units.\n\t[`-R`](http://gmt.soest.hawaii.edu/doc/latest/pshistogram.html#r)\n- **S** : **stairs** : -- Str or number --\n\n\tDraws a stairs-step diagram which does not include the internal bars of the default histogram.\n\t[`-S`](http://gmt.soest.hawaii.edu/doc/latest/pshistogram.html#s)\n- **Z** : **kind** : -- Number or Str --\n\n\tChoose between 6 types of histograms.\n\t[`-Z`](http://gmt.soest.hawaii.edu/doc/latest/pshistogram.html#z)\n- $(GMT.opt_U)\n- $(GMT.opt_V)\n- $(GMT.opt_X)\n- $(GMT.opt_Y)\n- $(GMT.opt_bi)\n- $(GMT.opt_di)\n- $(GMT.opt_e)\n- $(GMT.opt_h)\n- $(GMT.opt_i)\n- $(GMT.opt_p)\n- $(GMT.opt_t)\n- $(GMT.opt_swap_xy)\n\"\"\"\nfunction histogram(cmd0::String=\"\", arg1=[]; caller=[], K=false, O=false, first=true, kwargs...)\n\n\targ2 = []\t\t# May be needed if GMTcpt type is sent in via C\n\tN_args = isempty_(arg1) ? 0 : 1\n\n\tlength(kwargs) == 0 && return monolitic(\"pshistogram\", cmd0, arg1)\t# Speedy mode\n\n\td = KW(kwargs)\n\toutput, opt_T, fname_ext = fname_out(d)\t\t# OUTPUT may have been an extension only\n\n\tcmd, opt_B, opt_J, opt_R = parse_BJR(d, \"\", caller, O, \" -JX12c/12c\")\n\tcmd, = parse_JZ(cmd, d)\n\tcmd = parse_UVXY(cmd, d)\n\tcmd, opt_bi = parse_bi(cmd, d)\n\tcmd, opt_di = parse_di(cmd, d)\n\tcmd, = parse_e(cmd, d)\n\tcmd, = parse_h(cmd, d)\n\tcmd, opt_i = parse_i(cmd, d)\n\tcmd, = parse_p(cmd, d)\n\tcmd, = parse_t(cmd, d)\n\tcmd, = parse_swap_xy(cmd, d)\n\tcmd = parse_params(cmd, d)\n\n\tcmd, K, O, opt_B = set_KO(cmd, opt_B, first, K, O)\t\t# Set the K O dance\n\n\t# If file name sent in, read it and compute a tight -R if this was not provided\n\tif (isempty(opt_R)) opt_R = \" \" end\t\t# So it doesn't try to find the -R in next call\n\tcmd, arg1, opt_R, opt_i = read_data(d, cmd0, cmd, arg1, opt_R, opt_i, opt_bi, opt_di)\n\n\tcmd, arg1, arg2, = add_opt_cpt(d, cmd, [:C :color :cmap], 'C', N_args, arg1, arg2)\n\n\tcmd = add_opt(cmd, 'A', d, [:A :horizontal])\n\tcmd = add_opt(cmd, 'D', d, [:D :annot :annotate])\n\tcmd = add_opt(cmd, 'F', d, [:F :center])\n\tcmd = add_opt(cmd, 'G', d, [:G :fill])\n\tcmd = add_opt(cmd, 'I', d, [:I :inquire])\n\topt_L = opt_pen(d, 'L', [:L :pen])\n\tcmd = add_opt(cmd, 'Q', d, [:Q :cumulative])\n\tcmd = add_opt(cmd, 'S', d, [:S :stairs])\n\tcmd = add_opt(cmd, 'W', d, [:W :bin :width])\n\tcmd = add_opt(cmd, 'Z', d, [:Z :kind])\n\tif (isempty(opt_L) && !occursin(\"-G\", cmd) && !occursin(\"-C\", cmd))\t\t# If no -L, -G or -C set these defaults\n\t\tcmd = cmd * \" -G150\" * \" -L0.5p\"\n\telseif (!isempty(opt_L))\n\t\tcmd = cmd * opt_L\n\tend\n\n\tfor symb in [:N :normal]\n\t\tif (haskey(d, symb))\n\t\t\tif (isa(d[symb], Number)) cmd = @sprintf(\"%s -N%d\", cmd, d[symb])\n\t\t\telseif (isa(d[symb], String)) cmd = cmd * \" -N\" * d[symb]\n\t\t\telseif (isa(d[symb], Tuple)) cmd = cmd * \" -N\" * parse_arg_and_pen(d[symb])\n\t\t\tend\n\t\t\tbreak\n\t\tend\n\tend\n\n\tcmd = finish_PS(d, cmd, output, K, O)\n\treturn finish_PS_module(d, cmd, \"\", output, fname_ext, opt_T, K, \"pshistogram\", arg1, arg2)\nend\n\n# ---------------------------------------------------------------------------------------------------\nhistogram!(cmd0::String=\"\", arg1=[]; caller=[], K=true, O=true, first=false, kw...) =\n\thistogram(cmd0, arg1; caller=caller, K=K, O=O, first=first, kw...)\n\nhistogram(arg1=[]; caller=[], K=false, O=false, first=true, kw...) =\n\thistogram(\"\", arg1; caller=caller, K=K, O=O, first=first, kw...)\n\nhistogram!(arg1=[]; caller=[], K=true, O=true, first=false, kw...) =\n\thistogram(\"\", arg1; caller=caller, K=K, O=O, first=first, kw...)\n\npshistogram = histogram\t\t\t# Alias\npshistogram! = histogram!\t\t\t# Alias", "meta": {"hexsha": "fc14c550e1440f29fb972931b729d82ec825c71d", "size": 5646, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/pshistogram.jl", "max_stars_repo_name": "JuliaDocsForks/GMT.jl", "max_stars_repo_head_hexsha": "53d9c030559fe3f079f3b662ce262e7ed682d357", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/pshistogram.jl", "max_issues_repo_name": "JuliaDocsForks/GMT.jl", "max_issues_repo_head_hexsha": "53d9c030559fe3f079f3b662ce262e7ed682d357", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/pshistogram.jl", "max_forks_repo_name": "JuliaDocsForks/GMT.jl", "max_forks_repo_head_hexsha": "53d9c030559fe3f079f3b662ce262e7ed682d357", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 37.3907284768, "max_line_length": 112, "alphanum_fraction": 0.6126461211, "num_tokens": 1817, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.43782349911420193, "lm_q2_score": 0.1311732373477186, "lm_q1q2_score": 0.057430725765715875}}
{"text": "# Julia docs: https://docs.julialang.org/en/latest/manual/variables-and-scoping/\n\nx = 42\n\nfunction f()\n y = x + 3 # equivalent to `local y = x + 3`\n @show y\nend\n\nf()\n\ny\n\n# x is global whereas y is local (to the function f)\n\n# we can create a global variables from a local scope\n# but we must be explicit about it.\nfunction g()\n global y = x + 3\n @show y\nend\n\ng()\n\ny\n\n\n# we ALWAYS need to be explicit when writing to globals from a local scope:\nfunction h()\n x = x + 3\n @show x\nend\n\nh()\n\nfunction h2()\n global x = x + 3\n @show x\nend\n\nh2()\n\n# This fact can lead to subtle somewhat unintuitive errors:\na = 0\nfor i in 1:10\n a += 1\nend\n\n\n\n# Note that if we do not use global scope, everything is simple and intuitive:\nfunction k()\n a = 0\n for i in 1:10\n a += 1\n end\n a\nend\n\nk()\n\n\n\n# Variables are inherited from non-global scope\nfunction nested()\n function inner()\n # Try with and without \"local\"\n j = 2\n k = j + 1\n end\n j = 0\n k = 0\n return inner(), j, k\nend\n\nnested()\n\n\n\nfunction inner()\n j = 2 # Try with and without \"local\"\n k = j + 1\nend\n\nfunction nested()\n j = 0\n k = 0\n return inner(), j, k\nend\n\nnested()\n", "meta": {"hexsha": "f8330fd964739db3a16d9739b7854815f0b7e99a", "size": 1204, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "backup/scoping.jl", "max_stars_repo_name": "de-souza/JuliaOulu20", "max_stars_repo_head_hexsha": "f9685a8b769a66d8f1145d7428bb4894918830b0", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 98, "max_stars_repo_stars_event_min_datetime": "2019-07-26T20:02:31.000Z", "max_stars_repo_stars_event_max_datetime": "2021-08-06T08:12:15.000Z", "max_issues_repo_path": "backup/scoping.jl", "max_issues_repo_name": "de-souza/JuliaOulu20", "max_issues_repo_head_hexsha": "f9685a8b769a66d8f1145d7428bb4894918830b0", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 5, "max_issues_repo_issues_event_min_datetime": "2019-07-25T14:24:54.000Z", "max_issues_repo_issues_event_max_datetime": "2019-10-25T17:37:37.000Z", "max_forks_repo_path": "backup/scoping.jl", "max_forks_repo_name": "de-souza/JuliaOulu20", "max_forks_repo_head_hexsha": "f9685a8b769a66d8f1145d7428bb4894918830b0", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 25, "max_forks_repo_forks_event_min_datetime": "2020-02-11T08:28:07.000Z", "max_forks_repo_forks_event_max_datetime": "2021-06-18T06:21:50.000Z", "avg_line_length": 13.0869565217, "max_line_length": 80, "alphanum_fraction": 0.5905315615, "num_tokens": 379, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4378235137849365, "lm_q2_score": 0.13117322546005344, "lm_q1q2_score": 0.057430722485424295}}
{"text": "\"\"\"\natleast2d \\\\\n\nFor formatting the dimensions of the random variables to at least 2 dimensions \\\\\nArguments: random_variable \\\\\nReturns: A 2-d version of the variable \\\\\n\"\"\"\nfunction atleast2d(random_variable::Array)\n is_one_d = 1 == ndims(random_variable)\n return is_one_d ? reshape(random_variable, size(random_variable)[1], 1) : random_variable\nend\n", "meta": {"hexsha": "3a2fa21f7a0910553404554a522b64420434f93b", "size": 361, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/atleast2d.jl", "max_stars_repo_name": "thomasjdelaney/Jentropy.jl", "max_stars_repo_head_hexsha": "7d991182ea3deb1643907edda77b02782da8b933", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2021-06-05T18:19:27.000Z", "max_stars_repo_stars_event_max_datetime": "2021-06-05T18:19:27.000Z", "max_issues_repo_path": "src/atleast2d.jl", "max_issues_repo_name": "thomasjdelaney/Jentropy.jl", "max_issues_repo_head_hexsha": "7d991182ea3deb1643907edda77b02782da8b933", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/atleast2d.jl", "max_forks_repo_name": "thomasjdelaney/Jentropy.jl", "max_forks_repo_head_hexsha": "7d991182ea3deb1643907edda77b02782da8b933", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 30.0833333333, "max_line_length": 91, "alphanum_fraction": 0.7479224377, "num_tokens": 89, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.48828339529583464, "lm_q2_score": 0.11757214127540647, "lm_q1q2_score": 0.05740852433415701}}
{"text": "# # how to print\n# # assigning variables\n# # syntax for basic math\n\n# println(\"Print to Standard output\")\n# 123\n\n# # the \"ans\" variable holds the last expression in memory\n\n# # basic math operations\n# sum = 3 + 8\n# difference = 10 - 3\n# product = 20 * 3\n# power = 10^2\n# println(product)\n\ns1 = \"this is a string\"\ns2 = \"\"\" this is another string\"\"\"\n\n# typeof(string)\n\n# typeof('a')\n# # typeof('aa')\n\n# # String Interpolation\n# # dollar sign to throw variables into a string\n# a = 5\n# print(\"The value of a is $a\")\n# println()\n# b = 3\n# print(\"The sum of a and b is $(a + b)\")\n\n# String concatenation\n# Method 1, use the string function\nstring(\"con\", \"cat\", \"e\", \"nate\")\nstring(\"con\", 3, \"cat\", 3, \"e\", 3, \"nate\")\n\na = 1\nb = 2\n# Method 2 of string concatenation\n\"$a$b\"\n\n\n\n", "meta": {"hexsha": "9a00b5a629779d2955ac810a303d16aab1ebcd23", "size": 770, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "playground.jl", "max_stars_repo_name": "ryanorsinger/JuliaPlayground", "max_stars_repo_head_hexsha": "11f67480cf82b42ba05bcd57bdb776dbf95196e0", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "playground.jl", "max_issues_repo_name": "ryanorsinger/JuliaPlayground", "max_issues_repo_head_hexsha": "11f67480cf82b42ba05bcd57bdb776dbf95196e0", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "playground.jl", "max_forks_repo_name": "ryanorsinger/JuliaPlayground", "max_forks_repo_head_hexsha": "11f67480cf82b42ba05bcd57bdb776dbf95196e0", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 17.1111111111, "max_line_length": 58, "alphanum_fraction": 0.6207792208, "num_tokens": 255, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4035668537353746, "lm_q2_score": 0.14223189682786755, "lm_q1q2_score": 0.05740007910363691}}
{"text": "# x, y without type, this is a generic function\nfunction f(x, y)\n x + y\nend\n\nprintln(f(1, 2))\n# 3\ng = f # g is f\nprintln(g(1, 2))\n\nfunction with_return()\n return \"I have return\"\nend\nprintln(with_return())\n\nfunction hypot(x, y)\n x = abs(x)\n y = abs(y)\n if x > y\n r = y/x\n return x*sqrt(1+r*r)\n end\n if y == 0\n return zero(x)\n end\n r = x/y\n return y*sqrt(1+r*r)\nend\n\nprintln(hypot(3, 4))\n# 5\n\nprintln(+(1, 2, 3) == (1 + 2 + 3))\n# operator is function\n", "meta": {"hexsha": "7fe6dfe41b340aaaa3339ebc717189feda5dd2f1", "size": 500, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "julia/basic/function.jl", "max_stars_repo_name": "dannypsnl/languages-learn", "max_stars_repo_head_hexsha": "2351a235bd55e720394111237d41f65482eb89ec", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 2, "max_stars_repo_stars_event_min_datetime": "2018-01-04T00:47:25.000Z", "max_stars_repo_stars_event_max_datetime": "2018-01-12T08:07:50.000Z", "max_issues_repo_path": "julia/basic/function.jl", "max_issues_repo_name": "dannypsnl/languages-learn", "max_issues_repo_head_hexsha": "2351a235bd55e720394111237d41f65482eb89ec", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 1, "max_issues_repo_issues_event_min_datetime": "2018-01-08T14:45:55.000Z", "max_issues_repo_issues_event_max_datetime": "2018-01-09T05:02:09.000Z", "max_forks_repo_path": "julia/basic/function.jl", "max_forks_repo_name": "dannypsnl/languages-learn", "max_forks_repo_head_hexsha": "2351a235bd55e720394111237d41f65482eb89ec", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 14.2857142857, "max_line_length": 47, "alphanum_fraction": 0.536, "num_tokens": 188, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.44939264921326716, "lm_q2_score": 0.12765261204695083, "lm_q1q2_score": 0.05736614550677266}}
{"text": "#=\nSet\n=#\n\nusing SimpleDataStructures\n\nss = SimpleSet{Int}()\npush!(ss, 1)\n1 in ss\n2 in ss\npush!(ss, 2)\n2 in ss\n\ndelete!(ss, 1)\ndelete!(ss, 2)\n\nSimpleSet([1,2,3,1,2,3])\n", "meta": {"hexsha": "cdc9e9e580cdc089cec778a133c7c79ec0d8ad3c", "size": 168, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "examples/set.jl", "max_stars_repo_name": "harryscholes/SimpleDataStructures.jl", "max_stars_repo_head_hexsha": "bce776a723aa1026c37246b128203afab2d2424c", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "examples/set.jl", "max_issues_repo_name": "harryscholes/SimpleDataStructures.jl", "max_issues_repo_head_hexsha": "bce776a723aa1026c37246b128203afab2d2424c", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "examples/set.jl", "max_forks_repo_name": "harryscholes/SimpleDataStructures.jl", "max_forks_repo_head_hexsha": "bce776a723aa1026c37246b128203afab2d2424c", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 9.3333333333, "max_line_length": 26, "alphanum_fraction": 0.619047619, "num_tokens": 74, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.48047867804790706, "lm_q2_score": 0.11920291419948609, "lm_q1q2_score": 0.057274458634027164}}
{"text": "module TestMOIWrapper\n\nusing Test\nusing MathOptInterface\nimport Clp\n\nconst MOI = MathOptInterface\n\nfunction runtests()\n for name in names(@__MODULE__; all = true)\n if startswith(\"$(name)\", \"test_\")\n @testset \"$(name)\" begin\n getfield(@__MODULE__, name)()\n end\n end\n end\nend\n\nfunction test_SolverName()\n @test MOI.get(Clp.Optimizer(), MOI.SolverName()) == \"Clp\"\n return\nend\n\nfunction test_supports_default_copy_to()\n @test !MOI.supports_incremental_interface(Clp.Optimizer())\n return\nend\n\nfunction test_runtests()\n model = MOI.Bridges.full_bridge_optimizer(\n MOI.Utilities.CachingOptimizer(\n MOI.Utilities.UniversalFallback(MOI.Utilities.Model{Float64}()),\n Clp.Optimizer(),\n ),\n Float64,\n )\n MOI.set(model, MOI.Silent(), true)\n MOI.Test.runtests(\n model,\n MOI.Test.Config(\n exclude = Any[MOI.DualObjectiveValue, MOI.ObjectiveBound],\n ),\n exclude = [\n # Unable to prove infeasibility\n \"test_conic_NormInfinityCone_INFEASIBLE\",\n \"test_conic_NormOneCone_INFEASIBLE\",\n ],\n )\n return\nend\n\nfunction test_Nonexistant_unbounded_ray()\n model = MOI.Utilities.CachingOptimizer(\n MOI.Utilities.UniversalFallback(MOI.Utilities.Model{Float64}()),\n Clp.Optimizer(),\n )\n MOI.set(model, MOI.Silent(), true)\n x = MOI.add_variables(model, 5)\n MOI.set(\n model,\n MOI.ObjectiveFunction{MOI.ScalarAffineFunction{Float64}}(),\n MOI.ScalarAffineFunction(MOI.ScalarAffineTerm.(1.0, x), 0.0),\n )\n MOI.set(model, MOI.ObjectiveSense(), MOI.MAX_SENSE)\n MOI.optimize!(model)\n status = MOI.get(model, MOI.TerminationStatus())\n @test status == MOI.DUAL_INFEASIBLE\n return\nend\n\nfunction test_RawOptimizerAttribute()\n model = Clp.Optimizer()\n MOI.set(model, MOI.RawOptimizerAttribute(\"LogLevel\"), 1)\n @test MOI.get(model, MOI.RawOptimizerAttribute(\"LogLevel\")) == 1\n MOI.set(model, MOI.RawOptimizerAttribute(\"LogLevel\"), 2)\n @test MOI.get(model, MOI.RawOptimizerAttribute(\"LogLevel\")) == 2\n\n MOI.set(model, MOI.RawOptimizerAttribute(\"SolveType\"), 1)\n @test MOI.get(model, MOI.RawOptimizerAttribute(\"SolveType\")) == 1\n MOI.set(model, MOI.RawOptimizerAttribute(\"SolveType\"), 4)\n @test MOI.get(model, MOI.RawOptimizerAttribute(\"SolveType\")) == 4\n\n MOI.set(model, MOI.RawOptimizerAttribute(\"PresolveType\"), 1)\n @test MOI.get(model, MOI.RawOptimizerAttribute(\"PresolveType\")) == 1\n MOI.set(model, MOI.RawOptimizerAttribute(\"PresolveType\"), 0)\n @test MOI.get(model, MOI.RawOptimizerAttribute(\"PresolveType\")) == 0\n return\nend\n\nfunction test_All_parameters()\n model = Clp.Optimizer()\n param = MOI.RawOptimizerAttribute(\"NotAnOption\")\n @test !MOI.supports(model, param)\n @test_throws MOI.UnsupportedAttribute(param) MOI.get(model, param)\n @test_throws MOI.UnsupportedAttribute(param) MOI.set(model, param, false)\n for key in Clp.SUPPORTED_PARAMETERS\n @test MOI.supports(model, MOI.RawOptimizerAttribute(key))\n value = MOI.get(model, MOI.RawOptimizerAttribute(key))\n MOI.set(model, MOI.RawOptimizerAttribute(key), value)\n @test MOI.get(model, MOI.RawOptimizerAttribute(key)) == value\n end\n return\nend\n\nfunction test_copy_to_bug()\n model = MOI.Utilities.Model{Float64}()\n x = MOI.add_variable(model)\n con = [\n MOI.add_constraint(\n model,\n MOI.ScalarAffineFunction([MOI.ScalarAffineTerm(1.0, x)], 0.0),\n MOI.EqualTo(1.0),\n ) for i in 1:2\n ]\n clp = Clp.Optimizer()\n index_map = MOI.copy_to(clp, model)\n @test index_map[con[1]] != index_map[con[2]]\n return\nend\n\nfunction test_options_after_empty!()\n model = Clp.Optimizer()\n @test MOI.get(model, MOI.Silent()) == false\n MOI.set(model, MOI.Silent(), true)\n @test MOI.get(model, MOI.Silent()) == true\n MOI.empty!(model)\n @test MOI.get(model, MOI.Silent()) == true\n return\nend\n\nend # module TestMOIWrapper\n\nTestMOIWrapper.runtests()\n", "meta": {"hexsha": "52534dab065cd46bd1e709a4161b542d98de1134", "size": 4095, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/MOI_wrapper.jl", "max_stars_repo_name": "jump-dev/Clp.jl", "max_stars_repo_head_hexsha": "eb6ec27e1207593d98671dbdc2b429d3f2961d47", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 10, "max_stars_repo_stars_event_min_datetime": "2020-08-29T16:56:20.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-04T23:36:55.000Z", "max_issues_repo_path": "test/MOI_wrapper.jl", "max_issues_repo_name": "jump-dev/Clp.jl", "max_issues_repo_head_hexsha": "eb6ec27e1207593d98671dbdc2b429d3f2961d47", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 42, "max_issues_repo_issues_event_min_datetime": "2020-06-13T16:30:11.000Z", "max_issues_repo_issues_event_max_datetime": "2022-03-03T03:37:09.000Z", "max_forks_repo_path": "test/MOI_wrapper.jl", "max_forks_repo_name": "jump-dev/Clp.jl", "max_forks_repo_head_hexsha": "eb6ec27e1207593d98671dbdc2b429d3f2961d47", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 5, "max_forks_repo_forks_event_min_datetime": "2020-10-09T12:10:56.000Z", "max_forks_repo_forks_event_max_datetime": "2021-10-08T02:12:14.000Z", "avg_line_length": 30.5597014925, "max_line_length": 77, "alphanum_fraction": 0.6625152625, "num_tokens": 1148, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4610167793123159, "lm_q2_score": 0.12421300348902362, "lm_q1q2_score": 0.05726427881721912}}
{"text": "### A Pluto.jl notebook ###\n# v0.14.4\n\nusing Markdown\nusing InteractiveUtils\n\n# \u2554\u2550\u2561 1d460401-7c48-4718-914e-ba3fc9f7950a\nbegin\n import Pkg\n # activate a clean environment\n Pkg.activate(mktempdir())\n\n Pkg.add([\n Pkg.PackageSpec(name=\"PlutoUI\"),\n Pkg.PackageSpec(name=\"Unitful\"),\n # ... keep adding your packages\n ])\n\n using PlutoUI\n\tusing Unitful\n\t\n\tfunction Image(filepath::String)\n\t\t@assert !isempty(filepath)\n\t\tPlutoUI.LocalResource(joinpath(split(@__FILE__, '#')[1] * \".assets\", filepath))\n\tend\nend;\n\n# \u2554\u2550\u2561 baaa0f44-a9d8-11eb-0d0d-53b78cd74315\nmd\"# Physics II Final Exam Review\n\n- PHYS 1062 Section 003 (Spring 2021)\n- Instructor: Zbigniew Dziembowski\n- Temple University\"\n\n# \u2554\u2550\u2561 c565da05-80ca-4b72-af73-d5da2f1c2abc\nmd\"### Information about the exam\n\n- Please print out and bring to the exam two copies of the IRDEA worksheet.\n The worksheet is available for download in the *Final exam* Module.\n It includes a one sectence honesty statement which you have to sign.\n\n- The exam will have two parts:\n - A quiz consisting of 25 multiple choice questions covering **units 5 through 12**.\n The quiz setting will be such that there will be only one attempt given.\n You will see one question at a time and the questions will be locked after answering.\n The quiz will be available on Canvas.\n - 2 word problems covering **units 9-12**.\n These problems, where you are expected to show your explanations, must be solved using the IRDEA worksheet.\n The solutions must be uploaded for grading on Canvas (like the in-class problem solutions).\n\n- Final exam is worth 234 pts (125pts/quiz and 54.5 pts/problem)\n\n- Extra Credit\n - 4 pts extra credit per problem will be given, if a solution is submitted on time\n and on the IRDEA worksheet with signed the one sentence honesty statement.\n\n- Penalties\n - 4 pts penalty per problem will be applied if a solution is uploaded to Canvas less than 10 mins LATE.\n - 8 pts penalty per problem if uploaded to Canvas 10-20 mins LATE.\n - Solution is not graded if it is uploaded to Canvas more than 20 mins. LATE.\n\n- Do not wait to the last moment. Upload each solution once you completed it!\n\n- After the exam, we reserve the right to ask anyone to work out a few new problems with us to spot check the exam inconsistency.\n\"\n\n# \u2554\u2550\u2561 a1c99c25-e72e-4c4f-9ee9-855dad795483\nmd\"### How to review and prepare for the final exam\n\n1. Review the lecture notes for Unit 5 through 12.\n Especially, go through the quick-check questions in the posted lecture notes.\n Then take the practice quiz to be posted on Canvas by Sunday, April 25th.\n\n\n2. The contents of the two word-problem section of the exam will be limited to the contents of Units 9 through 12.\n\n To prepare for this section of the exam, redo, using a copy of the RIDEA worksheet, all exercises and problems in the DT and PSW homework for Unit 9 through 12.\n Use the wording of the assignments as in the back of Chapters 29, 30, 31 and 34, respectively.\n Check your answers against the answers to those exercises and problems as given in the file posted on Canvas (\\\"DT-PSW list for Unit XX with answers\\\") in the module **Final exam.**\"\n\n# \u2554\u2550\u2561 c5cf2733-8d62-4f97-9c6c-97c6bce90b02\nmd\"### Electricity and Magnetism formulas\"\n\n# \u2554\u2550\u2561 7abb1db1-d637-446c-9e55-3457db3db17e\nmd\"$$\\begin{gather*}\n\\vec{E} = k \\frac{q}{r^2} \\hat{r}; \\qquad \\vec{F} = q\\vec{E} \\\\\nV_B - V_A = -\\int_A^B \\vec{E} \\cdot d\\vec{r} \\\\\nV = k \\frac{q}{r}; \\qquad U = qV = k\\frac{q_1q_2}{r} \\\\\nC \\equiv \\frac{Q}{\\Delta V_C} = \\frac{\\epsilon_0 A}{d} \\\\\nU = \\frac{1}{2} \\frac{Q^2}{C} \\\\\nI = \\frac{dQ}{dt} \\qquad J = v_d e n_e \\\\\nI = \\frac{\\Delta V}{R} \\qquad R = \\rho \\frac{L}{A} \\qquad P = I \\mathcal{E} \\\\\n\\frac{1}{C_{\\text{eq}}} = \\sum \\frac{1}{C_i} \\qquad C_{\\text{eq}} = \\sum C_i \\\\\nR_{\\text{eq}} = \\sum R_i \\qquad \\frac{1}{R_{\\text{eq}}} = \\sum \\frac{1}{R_i} \\\\\n\\vec{F} = I\\vec{L} \\times \\vec{B} \\qquad \\vec{F} = q\\vec{v} \\times \\vec{B} \\\\\nr = \\frac{mv}{qB} \\qquad f = \\frac{qB}{2\\pi m} \\\\\nF = \\frac{\\mu_0}{2\\pi} \\frac{I_1 I_2}{d} L\n\\end{gather*}$$\"\n\n# \u2554\u2550\u2561 be1d2a6e-8c24-4a1e-b84b-aabfad8e6135\nmd\"$$\\begin{gather*}\n\\tau = NIAB \\sin{\\theta} \\\\\nB = \\frac{\\mu_0}{2\\pi} \\frac{I}{r} \\qquad B = \\frac{\\mu_0 I a^2}{2(x^2 + a^2)^{3/2}} \\\\\n\\vec{B} = \\frac{\\mu_0}{2\\pi} \\frac{\\vec{\\mu}}{z^3} \\qquad B = \\frac{\\mu_0 IN}{L} \\\\\n\\Phi_{\\text{m}} = \\int \\vec{B} \\cdot d\\vec{A} \\qquad \\mathcal{E} = \\left|\\frac{d\\Phi_{\\text{m}}}{dt}\\right| \\\\\nE_{\\text{inside}} = \\frac{r}{2} \\left|\\frac{dB}{dt}\\right| \\qquad V_S = \\left(\\frac{N_S}{N_P}\\right) V_P \\\\\nU_L = \\frac{1}{2} LI^2 \\qquad L_{\\text{sol}} = \\frac{\\mu_0 N^2 A}{\\ell} \\\\\n\\mathcal{E} = NB \\omega A \\sin{\\omega t} \\\\\nI = \\frac{P}{A} = S_{\\text{avg}} = \\frac{1}{2c\\mu_0} {E_0}^2 = \\frac{c\\epsilon_0}{2} {E_0}^2 \\\\\nk = \\frac{2\\pi}{\\lambda} \\qquad \\omega = 2\\pi f \\qquad f\\lambda = \\frac{\\omega}{k} = v \\\\\nv = \\frac{\\omega}{k} = \\frac{1}{\\sqrt{\\epsilon_0 \\mu_0}} \\qquad E = \\frac{\\omega}{k} B = cB\n\\end{gather*}$$\"\n\n# \u2554\u2550\u2561 6c9182b4-2bf5-40a8-8a7e-ecfb1f7e59f3\nmd\"#### Relevant constants and conversion factors\n\n$$\\begin{gather*}\nk \\approx 9.0 \\times 10^9 \\text{ N} \\cdot \\text{m}^2 / \\text{C}^2 \\\\\n\\mu_0 = 4\\pi \\times 10^{-7} \\text{ T} \\cdot \\text{m} / \\text{A} \\\\\n\\epsilon_0 = \\frac{1}{4\\pi k} = 8.85 \\times 10^{-12} \\text{ C}^2 / \\text{N}\\cdot\\text{m}^2 \\\\\n1 \\text{ eV} = 1.602 \\times 10^{-19} \\text{ J} \\\\\ne = 1.60 \\times 10^{-19} \\text{ C} \\qquad c = 3.00 \\times 10^8 \\text{ m/c} \\\\\nm_e = 9.11 \\times 10^{-31} \\text{ kg} \\qquad m_p = 1.67 \\times 10^{-27} \\text{ kg}\n\\end{gather*}$$\n\"\n\n# \u2554\u2550\u2561 b670ab02-1fe7-4204-9747-237348707d97\nbegin\n\tk = 9.0e9u\"N*m^2/C^2\"\n\t\u03bc0 = 1.26e-6u\"T*m/A\"\n\t\u03f50 = 8.85e-12u\"C^2/(N*m^2)\"\n\te = 1.60e-19u\"C\"\n\tc = 3.00e8u\"m/s\"\n\tme = 9.11e-31u\"kg\"\n\tmp = 1.67e-27u\"kg\"\n\t\n\t# Other constants\n\tB_earth = 50e-6u\"T\" # Earth's magnetic field\n\tg = 9.8u\"m/s^2\"\nend;\n\n# \u2554\u2550\u2561 3240dda1-8d4c-4551-8878-8c2ddd9f12d1\nmd\"### Ray Optics\n| Medium | $n$ |\n| :--- | :--- |\n| Vacuum | 1.00 exact |\n| Air (actual) | 1.0003 |\n| Air (accepted) | 1.00 |\n| Water | 1.33 |\n| Ethyl alcohol | 1.36 |\n| Oil | 1.46 |\n| Glass (typical) | 1.50 |\n| Polystyrene plastic | 1.59 |\n| Cubic zirconia | 2.18 |\n| Diamond | 2.41 |\n| Silicon (infrared) | 3.50 |\n\n$$\\begin{gather*}\nn = \\frac{c}{v} \\qquad n_1 \\sin{\\theta_1} = n_2 \\sin_{\\theta_2} \\\\\n\\frac{1}{s} + \\frac{1}{s'} = \\frac{1}{f} \\\\\nM = \\frac{h'}{h} = -\\frac{s'}{s} \\\\\n|f| = \\frac{R}{2}\n\\end{gather*}$$\n\"\n\n# \u2554\u2550\u2561 8a56ceb5-dbd9-4950-bbfe-50b56c682487\nmd\"# Notebook environment\"\n\n# \u2554\u2550\u2561 98b7f219-578b-4717-8dfa-5f22f86c04b2\nmd\"# Topics covered\n\n- Unit 5: Chapters 22 and 23---Electric Charges and Forces, The Electric Field\n- Unit 6: Chapters 25 and 26---The Electric Potential, Potential and Field\n- Unit 7: Chapter 27---Current and Resistance\n- Unit 8: Chapter 28---Fundamentals of Circuits\n- Unit 9: Chapter 29---The Magnetic Field\n- Unit 10: Chapter 30---Electromagnetic Induction\n- Unit 11: Chapter 31---Electromagnetic Fields and Waves\n- Unit 12: Chapter 34---Ray Optics\"\n\n# \u2554\u2550\u2561 f6b01ecd-9629-422c-800e-a41512fa9933\nmd\"# QuickChecks\"\n\n# \u2554\u2550\u2561 81d41ebd-9c50-49a1-b77d-1f6c600477e7\nmd\"## Unit 5 The Electric Charge, Force and Field\"\n\n# \u2554\u2550\u2561 067e90ac-b370-4c53-b1d8-8ee65189b036\nmd\"### Unit 5 Part 1 QuickCheck #1\n\nWhich is the direction of the net force on the charge at the lower left?\"\n\n# \u2554\u2550\u2561 179020bb-048a-4040-b517-0c081a5b68c0\nImage(\"Unit5/P1QC1.png\")\n\n# \u2554\u2550\u2561 8065e0db-4aa8-447f-a130-49feedcddad7\nmd\"**Answer:** B, down to the left\"\n\n# \u2554\u2550\u2561 5745f4ea-219d-4e60-b269-9e08f010836a\nmd\"### Unit 5 Part 2 QuickCheck #1\n\nWhat is the direction of the net electric field at the dot?\"\n\n# \u2554\u2550\u2561 885d48fa-4a05-4468-8492-0cae4bfb7b26\nImage(\"Unit5/P2QC1.png\")\n\n# \u2554\u2550\u2561 2d690e4b-3d09-4907-99de-5f0ef4662b7e\nmd\"**Answer:** D, to the right\"\n\n# \u2554\u2550\u2561 eae4eef0-1701-406f-966d-736a2b2381f5\nmd\"### Unit 5 Part 2 QuickCheck #2\n\nAn electron is in the plane that bisects a dipole.\nWhat is the direction of the electric force on the electron?\"\n\n# \u2554\u2550\u2561 9ca8f846-5d5b-4b28-85b6-90c5f9c068b6\nImage(\"Unit5/P2QC2.png\")\n\n# \u2554\u2550\u2561 04912a55-5871-4917-8c95-7f03bd4abf1d\nmd\"**Answer:** A, up because the electron goes towards the proton\"\n\n# \u2554\u2550\u2561 42f03b7e-9fec-40bc-8d22-e594a22ab54e\nmd\"### Unit 5 Part 3 QuickCheck #1\n\nThree points inside a parallel-plate capacitor are marked.\nWhich is true?\n\nA. $E_1 > E_2 > E_3$\n\nB. $E_1 < E_2 < E_3$\n\nC. $E_1 = E_2 = E_3$\n\nD. $E_1 = E_3 > E_2$\n\"\n\n# \u2554\u2550\u2561 34b75779-b546-4628-ac0a-1d8fedc231eb\nImage(\"Unit5/P3QC1.png\")\n\n# \u2554\u2550\u2561 fb93dd3c-de36-4087-9870-8365161f7d1f\nmd\"**Answer:**\nC. $E_1 = E_2 = E_3$\"\n\n# \u2554\u2550\u2561 ab703047-2e82-4c13-a9d9-c97ffd42af67\nmd\"## Unit 6 The Electric Potential, and Capacitors\"\n\n# \u2554\u2550\u2561 253d5697-9088-424f-aa2e-c5df2c7675bf\nmd\"### Unit 6 Part 2 QuickCheck #1\n\nAt the midpoint between these two equal but opposite charges,\n\nA. $\\vec{E} = \\vec{0}; \\;V = 0$\n\nB. $\\vec{E} = \\vec{0}; \\;V > 0$\n\nC. $\\vec{E} = \\vec{0}; \\;V < 0$\n\nD. $\\vec{E}$ points right; $V = 0$\n\nE. $\\vec{E}$ points left; $V = 0$\n\"\n\n# \u2554\u2550\u2561 fcdb7ea5-6203-4e74-b746-4470052b5f99\nImage(\"Unit6/P2QC1.png\")\n\n# \u2554\u2550\u2561 5db75338-4b19-4037-942d-979856662d6c\nmd\"**Answer:** D. $\\vec{E}$ points right; $V = 0$\n\n**Explanation:** $V = \\frac{k q_1}{r} + \\frac{k q_2}{r} = 0$\"\n\n# \u2554\u2550\u2561 74378daa-7994-403e-9c41-86bae271a74d\nmd\"### Unit 6 QuickCheck #2 (Exercise 25.6)\nWhat is the electrostatic energy of the group of charges in figure below?\n\nA. positive\n\nB. negative\n\nC. zero\n\"\n\n# \u2554\u2550\u2561 af3ab252-16c6-4e6f-bdc6-9a01fede5a30\nImage(\"Unit6/P2QC2.png\")\n\n# \u2554\u2550\u2561 64bbc192-28dd-4db4-8dab-1cde2a031bc5\nmd\"**Answer:** C. zero\n\n**Explanation:**\n\n$\\begin{gather*}\nU = \\;? \\\\\nU = k \\sum_{i 1 \\text{ W})$.\"\n\n# \u2554\u2550\u2561 060fd1a0-5548-4eb4-b4c2-f2db077e0d50\nmd\"### Unit 8 Part 1 QuickCheck #2\n\nWhich bulb is brighter?\n\nA. The 60 W bulb\n\nB. The 100 W bulb\n\nC. Their brightnesses are the same.\n\nD. There's not enough information to tell.\"\n\n# \u2554\u2550\u2561 8eb2caa8-5341-40a7-8ae5-aad8e44c1af1\nImage(\"Unit8/P1QC2.png\")\n\n# \u2554\u2550\u2561 860bbf33-9025-44ee-aa26-7217f0bc7562\nmd\"**Answer:** A. The 60 W bulb\n\n**Explanation:**\n\n$\\begin{gather*}\nV = 120 \\text{ V} \\\\\nP_{60} = 60 \\text{ W} \\\\\nP_{100} = 100 \\text{ W} \\\\\nP_R = \\frac{V^2}{R} \\implies R = \\frac{(\\Delta V_R)^2}{P_R} \\\\\nR_{60} = \\frac{V^2}{60} \\;\\Omega \\\\\nR_{100} = \\frac{V^2}{100} \\;\\Omega \\\\\nV = IR \\\\\nR_{60} > R_{100} \\implies V_{60} > V_{100}\n\\end{gather*}$\"\n\n# \u2554\u2550\u2561 28acdfbe-1020-4921-8c6c-0e38d0732d78\nmd\"### Unit 8 Part 2 QuickCheck #1\n\nThe battery current $I$ is\n\nA. 3 A\n\nB. 2 A\n\nC. 1 A\n\nD. 2/3 A\n\nE. 1/2 A\"\n\n# \u2554\u2550\u2561 e06efb86-5366-49c3-acb6-f9b209bf5d81\nImage(\"Unit8/P2QC1.png\")\n\n# \u2554\u2550\u2561 6c9233eb-058a-4e8a-9c9b-22964e210dd1\nmd\"**Answer:** 2/3 A\n\n**Explanation:** Use Ohm's Law $V = IR$\"\n\n# \u2554\u2550\u2561 c83063bd-0bba-47e7-b869-e169d6e9c03e\nmd\"### Unit 8 Part 2 QuickCheck #2\n\nWhat does the (ideal) ammeter read?\n\nA. 6 A\n\nB. 3 A\n\nC. 2 A\n\nD. Some other value\n\nE. Nothing because this will fry the meter.\"\n\n# \u2554\u2550\u2561 c9102584-2196-4483-b79f-60089ad9e1b4\nImage(\"Unit8/P2QC2.png\")\n\n# \u2554\u2550\u2561 b30d503e-ab6c-4515-9a68-81327795aa6c\nmd\"**Answer:** E. Nothing because this will fry the meter.\n\n**Explanation:** Since the ammeter is ideal, its resistance $R = 0 \\implies I = \\lim_{R \\to 0} \\frac{V}{R} = \\infty$\"\n\n# \u2554\u2550\u2561 85e2b232-6a09-4621-ab97-e2345b676999\nmd\"### Unit 8 Part 2 QuickCheck #3\n\nThe battery current $I$ is\n\nA. 3 A\n\nB. 2 A\n\nC. 1 A\n\nD. 2/3 A\n\nE. 1/2 A\"\n\n# \u2554\u2550\u2561 69bdb37e-2f06-4c78-a5bc-c3fce3cf6be8\nImage(\"Unit8/P2QC3.png\")\n\n# \u2554\u2550\u2561 f9ba57aa-4113-40e0-af24-8d7066baef4c\nmd\"**Answer:** 3 A\n\n**Explanation:** The problem involves resistors in parallel and Ohm's Law.\n\n$\\begin{gather*}\nI = \\;? \\\\\nV = 12 \\text{ V} \\\\\n\\frac{1}{R} = \\frac{1}{12} + \\frac{1}{6} = \\frac{3}{12} \\implies R = 4 \\\\\nV = IR \\implies I = \\frac{V}{R} = \\frac{12}{4} = 3 \\text{ A}\n\\end{gather*}$\n\"\n\n# \u2554\u2550\u2561 cdc4c352-a21c-4d88-8a6e-28da5974e893\nmd\"### Unit 8 Part 2 QuickCheck #4\n\nWhat does the (ideal) voltmeter read?\n\nA. 6 V\n\nB. 3 V\n\nC. 2 V\n\nD. Some other value\n\nE. Nothing because this will fry the meter.\n\"\n\n# \u2554\u2550\u2561 bc7ebdc7-3614-43ad-bb1c-695f535c339b\nImage(\"Unit8/P2QC4.png\")\n\n# \u2554\u2550\u2561 3badd910-d0be-435d-8b61-751d2fc46984\nmd\"**Answer:** A. 6 V\n\n**Answer:** An ideal voltmeter will have very high resistance to measure the voltage of the circuit.\"\n\n# \u2554\u2550\u2561 f5871103-8943-4085-8ec8-f7c19155262c\nmd\"## Unit 9 Magnetism: Force and Field\"\n\n# \u2554\u2550\u2561 64256f0e-f61a-41d9-9f07-f53f12b81d9f\nmd\"### Unit 9 Part 1 QuickCheck #1\n\nA long straight wire extends into and out of the screen.\nThe current in the wire is\n\nA. Into the screen.\n\nB. Out of the screen.\n\nC. There is no current in the wire.\n\nD. Not enough info to tell the direction.\"\n\n# \u2554\u2550\u2561 56988b0d-40f0-485c-bfd1-468431e2f906\nImage(\"Unit9/P1QC1.png\")\n\n# \u2554\u2550\u2561 baf581de-7077-4c25-a5df-b5e82490b113\nmd\"**Answer:** Out of the screen.\n\n**Explanation:** Use the right-hand rule along the direction of the magnetic field.\"\n\n# \u2554\u2550\u2561 11c087b0-7823-41ac-8034-e66875bcaec1\nmd\"### Unit 9 Part 1 QuickCheck #2\n\nWhat is the direction of the magnetic field at the position of the dot?\n\nA. Into the screen\n\nB. Out of the screen\n\nC. Up\n\nD. Down\n\nE. Left\"\n\n# \u2554\u2550\u2561 ad23f43f-c8ac-48fd-9b21-901658710684\nImage(\"Unit9/P1QC2.png\")\n\n# \u2554\u2550\u2561 a59c5606-b60e-44c8-a7e3-b84cd89b322c\nmd\"**Answer:** C. Up\n\n**Explanation:** Use the right-hand rule for the cross product $\\vec{v} \\times \\hat{r}$ where $\\hat{r}$ is the distance between the two points and determine the direction of the magnetic field at the given point.\"\n\n# \u2554\u2550\u2561 69065726-b456-4ab1-b8a6-22e638304388\nmd\"### Unit 9 Part 2 QuickCheck #1\n\nCompared to the magnetic field at point A, the magnetic field at point B is\n\nA. Half as strong, same direction.\n\nB. Half as strong, opposite direction.\n\nC. One-quarter as strong, same direction.\n\nD. One-quarter as strong, opposite direction.\n\nE. Can't compare without knowing $I$.\"\n\n# \u2554\u2550\u2561 47e713c1-0741-4135-ad0c-8b7ad003c141\nImage(\"Unit9/P2QC1.png\")\n\n# \u2554\u2550\u2561 45baa2c9-e2a9-44fc-a6a3-9e1b149d3ec6\nmd\"**Answer:** B. Half as strong, opposite direction.\n\n**Explanation:** The magnetic field of a current carrying wire is\n\n$B = \\frac{\\mu_0}{2\\pi} \\frac{I}{r}$\"\n\n# \u2554\u2550\u2561 a44466e8-0954-4738-9836-e7d445d41a91\nmd\"### Unit 9 Part 3 QuickCheck #1\n\nWhat is the current direction in this loop?\nAnd where is the north magnetic pole of this current loop?\n\nA. Current CW; north pole on top\n\nB. Current CW; north pole on bottom\n\nC. Current CCW; north pole on top\n\nD. Current CCW; north pole on bottom\"\n\n# \u2554\u2550\u2561 13f7bece-837b-473e-bb86-df55d3f5dbdf\nImage(\"Unit9/P3QC1.png\")\n\n# \u2554\u2550\u2561 6917fdb5-0538-4995-9ac5-14d7fc92b903\nmd\"**Answer:** B. Current CW; north pole on bottom\n\n**Explanation:** Use the right-hand rule.\"\n\n# \u2554\u2550\u2561 018a2522-f201-44a7-8f63-d67411970a90\nmd\"### Unit 9 Part 5 QuickCheck #1\n\nThe direction of the magnetic force on the proton is\n\nA. To the right.\n\nB. To the left.\n\nC. Into the screen.\n\nD. Out of the screen.\n\nE. The magnetic force is zero.\"\n\n# \u2554\u2550\u2561 ebe19088-8374-441c-9c1b-c00194737814\nImage(\"Unit9/P5QC1.png\")\n\n# \u2554\u2550\u2561 30625a27-4961-4eff-be88-27a351a3db12\nmd\"**Answer:** D. Out of the screen.\n\n**Explanation:** Use the right-hand rule for $\\vec{v} \\times \\vec{B}$\"\n\n# \u2554\u2550\u2561 66bd1832-dd76-43fa-ab08-018c8e726885\nmd\"### Unit 9 Part 5 QuickCheck #2\n\nThe direction of the magnetic force on the electron is\n\nA. Upward.\n\nB. Downward.\n\nC. Into the screen.\n\nD. Out of the screen.\n\nE. The magnetic force is zero.\"\n\n# \u2554\u2550\u2561 08c4b0d3-a431-46dd-a0ae-3bd5a59b81e3\nImage(\"Unit9/P5QC2.png\")\n\n# \u2554\u2550\u2561 eff73afd-3ded-49d2-9411-ec11933fb31f\nmd\"**Answer:** E. The magnetic force is zero.\n\n**Explanation:** Use the right-hand rule for $\\vec{v} \\times \\vec{B}$\"\n\n# \u2554\u2550\u2561 6e2b45e2-67bd-4512-9b3c-6f25dc75824f\nmd\"### Unit 9 Part 6 QuickCheck #1\n\nThe horizontal wire can be levitated---held up against the force of gravity---if the current in the wire is\n\nA. Right to left.\n\nB. Left to right.\n\nC. It can't be done with this magnetic field.\"\n\n# \u2554\u2550\u2561 9cf0faf9-ea42-45da-9788-975cfecdca37\nImage(\"Unit9/P6QC1.png\")\n\n# \u2554\u2550\u2561 67aca9a5-da3f-4a2c-9159-2dfb922217f8\nmd\"**Answer:** B. Left to right\n\n**Explanation:** Use the right-hand rule for $\\vec{v} \\times \\vec{B}$\"\n\n# \u2554\u2550\u2561 37aeddcf-e1cf-40a0-925e-20d600f24845\nmd\"### Unit 9 Part 6 QuickCheck #2\n\nIf released from rest, the current loop will\n\nA. Move upward.\n\nB. Move downward.\n\nC. Rotate clockwise.\n\nD. Rotate counterclockwise.\n\nE. Do something not listed here.\"\n\n# \u2554\u2550\u2561 5fb3d735-8df5-4e69-aa47-ff467dee9047\nImage(\"Unit9/P6QC2.png\")\n\n# \u2554\u2550\u2561 c4f15be9-ba69-4b9f-9b5a-407a975c2fbe\nmd\"**Answer:** D. Rotate counterclockwise.\n\n**Explanation:** Use the right-hand rule for $\\vec{v} \\times \\vec{B}$ on both ends of the current loop to determine the direction of each end.\"\n\n# \u2554\u2550\u2561 2c330d4d-8fa2-4c3f-a5b8-8a3ac19fd075\nmd\"## Unit 10 Electromagnetic Induction\"\n\n# \u2554\u2550\u2561 a2fbf237-3b84-4331-be33-e13c0f726838\nmd\"### Unit 10 Part 2 QuickCheck #1\n\nAn induced current flows clockwise as the metal bar is pushed to the right.\nThe original magnetic field points\n\nA. Up.\n\nB. Down.\n\nC. Into the screen.\n\nD. Out of the screen.\n\nE. To the right.\"\n\n# \u2554\u2550\u2561 84cdc818-8a8c-4888-b3bd-12399e60a739\nImage(\"Unit10/P2QC1.png\")\n\n# \u2554\u2550\u2561 b759a10f-6110-4e66-8121-e1c8bf24efee\nmd\"**Answer:** C. Into the screen\n\n**Explanation:** Apply Lenz's Law for direction of current induced in a loop.\"\n\n# \u2554\u2550\u2561 9c245886-b491-4adf-9ccd-0f8d20326949\nmd\"Unit 10 Part 2 QuickCheck #2\n\nThe current in the straight wire is decreasing.\nWhich is true?\n\nA. There is a clockwise induced current in the loop.\n\nB. There is a counterclockwise induced current in the loop.\n\nC. There is no induced current in the loop.\"\n\n# \u2554\u2550\u2561 c352a601-03e9-4f47-9d8e-2e7e806a7433\nImage(\"Unit10/P2QC2.png\")\n\n# \u2554\u2550\u2561 d085be3a-89ec-4672-a47d-f5f2c020885b\nmd\"**Answer:** A. There is a clockwise induced current in the loop.\n\n**Explanation:** Apply Lenz's law for direction of current induced in a loop for decreasing magnetic flux. To oppose the decrease (pointing out of the screen), the magnetic field of the induced current must point into the screen.\"\n\n# \u2554\u2550\u2561 3d7c7ea7-24a4-4acc-8238-e71118fef4f1\nmd\"## Unit 11 EM Fields and Waves\"\n\n# \u2554\u2550\u2561 27a07162-335c-4d3a-9771-2a3793490631\nmd\"### Unit 11 Part 1 QuickCheck #1\"\n\n# \u2554\u2550\u2561 04beeb35-c9b8-4aca-9afa-6d9cefaacc62\nImage(\"Unit11/P1QC1.png\")\n\n# \u2554\u2550\u2561 2f72f311-4bb9-4459-a1ee-3a3b5339b13f\nmd\"**Answer**: A.\"\n\n# \u2554\u2550\u2561 082f1121-811b-4404-b561-6ce9ca6a05ce\nmd\"### Unit 11 Part 1 QuickCheck #2\n\nIn which direction is this electro-magnetic wave traveling?\n\nA. Up\n\nB. Down\n\nC. Into the screen\n\nD. Out of the screen\n\nE. These are not allowable fields for an electromagnetic wave\"\n\n# \u2554\u2550\u2561 4584596e-c66f-45b3-ad3d-3042f13a1eca\nImage(\"Unit11/P1QC2.png\")\n\n# \u2554\u2550\u2561 99a92b4f-0a27-4681-a372-e6db84db033a\nmd\"**Answer:** A. Up\n\n**Explanation:** Use the right-hand rule for $\\vec{E} \\times \\vec{B}$ to determine the direction of the electro-magnetic wave.\"\n\n# \u2554\u2550\u2561 3a92f0e7-983c-433d-a83d-7c4e1a52a206\nmd\"### Unit 11 Part 2 QuickCheck #1\"\n\n# \u2554\u2550\u2561 ae445aa3-dcd9-4d97-87f7-7be0a2287cf3\nImage(\"Unit11/P2QC1.png\")\n\n# \u2554\u2550\u2561 6c521a07-2a6b-4ce8-a772-68b511bddf50\nmd\"**Answer:** A.\n\n**Explanation:** Use the right-hand rule where $\\vec{E} \\times \\vec{B}$ determines the direction of the wave.\"\n\n# \u2554\u2550\u2561 90a8d78d-cdf9-4485-ba31-1364e2c920ae\nmd\"## Unit 12 Ray Optics\"\n\n# \u2554\u2550\u2561 1f9d99d8-d613-4e62-9d46-5ebc61b029d6\nmd\"### Unit 12 Part 1 QuickCheck #1\n\nA laser beam passing from medium 1 to medium 2 is refracted as shown.\nWhich is true?\n\nA. $n_1 < n_2$\n\nB. $n_1 > n_2$\n\nC. There's not enough information to compare $n_1$ and $n_2$.\"\n\n# \u2554\u2550\u2561 141ea713-8276-4799-8e9c-f46f14e30c44\nImage(\"Unit12/P1QC1.png\")\n\n# \u2554\u2550\u2561 59508d51-2fab-4cab-816c-6d0c88979af8\nmd\"**Answer:** B. $n_1 > n_2$\n\n**Explanation:** The light bends away from the normal.\"\n\n# \u2554\u2550\u2561 685e68c4-2be2-4fc7-b22f-9dc78b231236\nmd\"### Unit 12 Part 2 QuickCheck #1\n\nIn an optical setup consisting of a single mirror and an object we measure the magnification factor $m = +3$.\nWe can conclude that the image is:\n\nA. Inverted and virtual\n\nB. Inverted and real\n\nC. Upright and virtual\n\nD. Upright and real\"\n\n# \u2554\u2550\u2561 f0d975f2-dc73-476a-9dec-6c7939511066\nmd\"**Answer:** Upright and virtual\n\n**Explanation:** The mirror must be concave to produce an enlarged image. The image must be upright since the magnification is positive. The image is only upright with the condition that the object's distance from the mirror is less than the focal length, i.e., $s < f$. The image will form behind the mirror so it will be virtual.\"\n\n# \u2554\u2550\u2561 d1a284e5-e3d9-4d9c-b350-f6a9f9d4bf23\nmd\"### Unit 12 Part 3 QuickCheck #1\n\nIn an optical setup consisting of a single lens and an object we measure the magnification factor $m = -0.5$.\nWe can conclude that object in this setup is:\n\nA. Between lens and focal point\n\nB. Between focal point and point of twice the focal length\n\nC. Exactly at point of twice the focal length\n\nD. At distance greater than twice the focal length\"\n\n# \u2554\u2550\u2561 a76b95fb-bf1e-4fea-ae0b-927017a78d1b\nmd\"**Answer:** D. At distance greater than twice the focal length\n\n**Explanation:** The image must be inverted, so a converging lens must be used. Use two rays to determine the height of the image: one passing through the lens bending towards the focal point and another passing through the center of the lens. Notice that the image will only get smaller for distances larger than $2f$.\"\n\n# \u2554\u2550\u2561 368b2131-d30c-42ae-9550-b51073e7592b\nmd\"# Final Practice Quiz\"\n\n# \u2554\u2550\u2561 0acf5486-3702-4e8a-889e-871916d2ecf3\nmd\"### Question 1\n\nWhich is the direction of the net force on the charge at the lower left?\n\n- A\n\n- D\n\n- B\n\n- C\n\n- E. None of these.\"\n\n# \u2554\u2550\u2561 bfef7588-5c90-4a37-967e-660eb3e5efcd\nImage(\"Unit5/P1QC1.png\")\n\n# \u2554\u2550\u2561 86fa33b9-17da-4322-b195-fdc99687c518\nmd\"**Answer:** B\n\n**Explanation:** Apply the principle of superposition.\"\n\n# \u2554\u2550\u2561 9fdb361f-cb53-4270-a59c-6b967da468ea\nmd\"### Question 2\n\nWhich is the direction of the net force on the charge at the top?\n\n- A\n\n- D\n\n- C\n\n- E. None of these.\n\n- B\"\n\n# \u2554\u2550\u2561 4c54fa63-06a1-4519-8104-aea78a2bbfbe\nImage(\"Unit5/P2QC1.png\")\n\n# \u2554\u2550\u2561 a68a80e3-b74e-4dbf-93ce-2892bc6dafd5\nmd\"**Answer:** D\n\n**Explanation:** The positive charge pushes, the negative charge pulls. Their vertical forces cancel each other out.\"\n\n# \u2554\u2550\u2561 cda191c4-afb2-4512-a930-171f5757a094\nmd\"### Question 3\n\nA uniform electric field, with a magnitude of 500 V/m, is directed parallel to the +x axis.\nIf the potential at x = 5.0 m is 2500 V, what is the potential at x = 2.0 m?\n\n- 1000V\n\n- 2000V\n\n- 4000V\n\n- 500V\"\n\n# \u2554\u2550\u2561 c721d98c-3586-450c-a22f-a4ff048d12c6\nmd\"\n##### Incorrect\n**Answer:** 1000V\"\n\n# \u2554\u2550\u2561 b55d0b05-3713-46ea-819e-d180e662a47e\nmd\"##### Correct answer\n4000V\n\n**Explanation:**\nThe electric field's magnitude $E$ is the rate at which the potential $U$ is decreasing.\nThen the potential at 2.0 m must be greater than the potential at 5.0 m.\n\n$\\begin{gather*}\nE = 500 \\text{ V/m} \\\\\nU_1 = 2500 \\text{ V} \\\\\nx_1 = 5.0 \\text{ m} \\\\\nx_0 = 2.0 \\text{ m} \\\\\nU_2 = U_1 + E(x_1 - x_0) = 2500 + 500(3) = 4000 \\text{ V}\n\\end{gather*}$\"\n\n# \u2554\u2550\u2561 16f0dd9e-f5bb-4173-a07c-7feba801837f\nlet\n\tE = 500\n\tU1 = 2500\n\tx1 = 5.0\n\tx0 = 2.0\n\tU2 = U1 + E * (x1 - x0)\nend\n\n# \u2554\u2550\u2561 b87d87a8-da10-4af4-94af-868947f45d02\nmd\"### Question 4\n\nA parallel-plate capacitor is connected to a battery and becomes fully charged.\nThe capacitor is then disconnected, and the separation between the plates is increased in such a way that no charge leaks off.\nThe energy stored in this capacitor has\n\n- increased.\n\n- become zero.\n\n- decreased.\n\n- not changed.\"\n\n# \u2554\u2550\u2561 e664b06c-2924-423c-9f1b-52174fca7cc3\nmd\"**Answer:** increased.\n\n**Explanation:** $C = \\frac{\\epsilon_0 A}{d}$\"\n\n# \u2554\u2550\u2561 7ec6edf3-7b6d-4ea2-8a4f-86dd2178eb91\nmd\"### Question 5\n\nThe plates of a parallel-plate capacitor are maintained with constant voltage by a battery as they are pulled apart.\nWhat happens to the strength of the electric field during this process?\n\n- It remains constant\n\n- cannot be determined from the information given\n\n- It decreases.\n\n- It increases.\n\n- not changed.\"\n\n# \u2554\u2550\u2561 6de0da98-8dfe-496d-adc8-82c5c9af499e\nmd\"**Answer:** It decreases.\n\n**Explanation:** $C = \\frac{\\epsilon_0 A}{d}$\"\n\n# \u2554\u2550\u2561 d518d932-52d5-45de-aaf4-d94dfaf3eb64\nmd\"### Question 6\n\nConsider two copper wires.\nOne has twice the cross-sectional area of the other.\nHow do the resistances of these two wires compare?\n\n- The thicker wire has twice the resistance of the shorter\n\n- Both wires have the same resistance\n\n- none of the given answers\n\n- The thicker wire has half the resistance of the shorter wire.\"\n\n# \u2554\u2550\u2561 570a8d6f-b108-4bba-81a4-8adffe79cea5\nmd\"**Answer:** The thicker wire has half the resistance of the shorter wire.\n\n**Explanation:** $R = \\rho \\frac{L}{A}$\"\n\n# \u2554\u2550\u2561 e275bc47-c73c-4cf5-bbdb-bebee2fb08af\nmd\"### Question 7\n\nConsider two copper wires.\nOne has twice the length of the other.\nHow do the resistances of these two wires compare?\n\n- The longer wire has twice the resistance of the shorter wire.\n\n- The longer wire has half the resistance of the shorter wire.\n\n- Both wires have the same resistance.\n\n- none of the given answers\"\n\n# \u2554\u2550\u2561 07adb0ac-6cb2-40ed-ad5f-659a6f58303b\nmd\"\n##### Incorrect\n**Answer:** Both wires have the same resistance\"\n\n# \u2554\u2550\u2561 276749db-45c7-43b9-a746-e48a6bdede2e\nmd\"##### Correct answer\n\nThe longer wire has twice the resistance of the shorter wire.\n\n**Explanation:**\nRefer to the formula sheet to find that the resistance of a wire is\n\n$R = \\rho \\frac{L}{A}$\n\"\n\n# \u2554\u2550\u2561 4689568d-ba6d-4954-86ba-3fcadb52f957\nmd\"### Question 8\n\nIf the voltage across a circuit of constant resistance is doubled, the power dissipated by that circuit will\n\n- decrease to one fourth.\n\n- decrease to one half.\n\n- double.\n\n- quadruple.\"\n\n# \u2554\u2550\u2561 9a0b5bac-f167-4150-beaa-3c770dbc8cac\nmd\"**Answer:** quadruple.\n\n**Explanation:** $P = \\frac{V^2}{R}$\"\n\n# \u2554\u2550\u2561 7094b76b-519e-4427-91fe-88d33304fef1\nmd\"### Question 9\n\nIf the resistance in a constant voltage circuit is doubled, the power dissipated by that circuit will\n\n- decrease to one-fourth its original value.\n\n- increase by a factor of four.\n\n- decrease to one-half its original value.\n\n- increase by a factor of two.\"\n\n# \u2554\u2550\u2561 82836166-8d43-4c15-a2d8-ac451a1d20ab\nmd\"**Answer:** decrease to one-half its original value.\n\n**Explanation:** $P = \\frac{V^2}{R}$\"\n\n# \u2554\u2550\u2561 06389e81-54a5-46df-b7a6-fd11b44c5406\nmd\"### Question 10\n\nIf the resistance in a circuit with constant current flowing is doubled, the power dissipated by that circuit will\n\n- decrease to one fourth.\n\n- quadruple\n\n- decrease to one half.\n\n- double\"\n\n# \u2554\u2550\u2561 5b87298f-9b97-4f7c-aab5-b3d740f9cab0\nmd\"**Answer:** double\n\n**Explanation:** $P = I^2 R$\"\n\n# \u2554\u2550\u2561 980aa663-98cd-42be-9b92-238a452f537a\nmd\"#### Question 11\n\nConsider three identical resistors, each of resistance R.\nThe maximum power each can dissipate is P.\nThree of the resistors are connected in series, and a fourth is connected in parallel with these three.\nWhat is the maximum power this network can dissipate?\n\n- 2P\n\n- 3P\n\n- 3P/2\n\n- 4P/3\"\n\n# \u2554\u2550\u2561 44d12feb-10cf-48b2-929a-8e6ff39a9496\nmd\"**Answer:** 4P/3\"\n\n# \u2554\u2550\u2561 9f4b96a0-2b5e-47d0-8694-1c1d2144fd01\nmd\"### Question 12\n\nA charged particle is observed traveling in a circular path in a uniform magnetic field.\nIf the particle had been traveling twice as fast, the radius of the circular path would be\n\n- four times the original radius.\n\n- twice the original radius.\n\n- one-fourth the original radius.\n\n- one-half the original radius.\"\n\n# \u2554\u2550\u2561 2434b3b5-2da8-4c1b-8bf2-522fc75b5cd1\nmd\"**Answer:** one-half the original radius.\n\n**Explanation:** $2B = \\frac{\\mu_0}{2\\pi} \\frac{I}{r} \\implies r \\propto \\frac{1}{2}$\"\n\n# \u2554\u2550\u2561 72bd9e28-5048-48f2-a110-f5feaa67caee\nmd\"### Question 13\n\nAt a particular instant, a proton moves eastward at speed V in a uniform magnetic field that is directed straight downward.\nThe magnetic force that acts on it is\n\n- directed to the north.\n\n- directed upward.\n\n- zero\n\n- directed to the south.\"\n\n# \u2554\u2550\u2561 a8dd4343-6913-4e19-bc12-23e89c9e6236\nmd\"**Answer:** directed to the north.\"\n\n# \u2554\u2550\u2561 239d8478-4571-4155-90e3-95ae1deb4939\nmd\"### Question 14\n\nAn electron moving along the +x axis enters a region where there is a uniform magnetic field in the +y direction.\nWhat is the direction of the magnetic force on the electron? (+x to the right, +y up, and +z out of the page.)\n\n- -z direction\n\n- -x direction\n\n- -y direction\n\n- +z direction\"\n\n# \u2554\u2550\u2561 a748ef26-adc8-4fe5-ad2b-8c8b1d79b54d\nmd\"**Answer:** -z direction\"\n\n# \u2554\u2550\u2561 f584bd06-21b6-41c2-b729-84b77acd2e2f\nmd\"### Question 15\n\nWhat is the current direction in this loop?\nAnd where is the north magnetic pole of this current loop?\n\n- Current CW; north pole on top\n\n- Current CW; north pole on bottom\n\n- Current CCW; north pole on bottom\n\n- Current CCW; north pole on top\"\n\n# \u2554\u2550\u2561 0fbb0fa0-6c8e-497f-856f-2f3f8f2ce735\nImage(\"Unit9/P3QC1.png\")\n\n# \u2554\u2550\u2561 4e172839-92b8-411a-ab77-3f8238d3a5e1\nmd\"**Answer:** Current CW; north pole on bottom\"\n\n# \u2554\u2550\u2561 51cf30e3-8fdc-4b62-a1a6-9252ed608dc9\nmd\"### Question 16\n\nA long, straight wire extends into and out of the screen.\nThe current in the wire is\n\n- Into the screen\n\n- Not enough info to tell the direction.\n\n- There is no current in the wire.\n\n- Out of the screen.\"\n\n# \u2554\u2550\u2561 1183767b-3d27-49c0-91ba-fd65e96cf773\nImage(\"Unit9/P1QC1.png\")\n\n# \u2554\u2550\u2561 9086f546-aa48-438e-b682-b0f27b2bff5c\nmd\"**Answer:** Out of the screen.\"\n\n# \u2554\u2550\u2561 d78a8b7b-4ea8-45ad-9d8d-331a090170a4\nmd\"### Question 17\n\nSolenoid 2 has twice the diameter, twice the length, and twice as many turns as solenoid 1.\nHow does the field B\u2082 at the center of solenoid 2 compare to B\u2081 at the center of solenoid 1?\n\n- B2 = 4B1\n\n- B2 = B1\n\n- B2 = B1/2\n\n- B2 = 2B1\n\n- B2 = B1/4\"\n\n# \u2554\u2550\u2561 b7181523-93b0-4865-9c91-9ee965124bfa\nImage(\"Quiz/Q17.png\")\n\n# \u2554\u2550\u2561 fe0fb4f9-6d8f-48e7-9809-71c55035ba75\nmd\"**Answer:** B2 = B1\"\n\n# \u2554\u2550\u2561 39fe00bb-0eb1-4f31-8073-e17186da71e7\nmd\"### Question 18\n\nWhere is the north magnetic pole of this current loop?\n\n- Right side\n\n- Top side\n\n- Left side\n\n- Bottom side\"\n\n# \u2554\u2550\u2561 818d7074-6ce6-4746-a15e-a101ae653ec9\nImage(\"Quiz/Q18.png\")\n\n# \u2554\u2550\u2561 da06ac7d-67f3-483c-aad0-98529cf44a28\nmd\"**Answer:** Bottom side\"\n\n# \u2554\u2550\u2561 eae1f535-37ac-48dd-8f2c-65b88852fa43\nmd\"### Question 19\n\nWhat is the induced current direction in the loop?\n\n- In at the top, out at the bottom.\n\n- Out at the top, in at the bottom.\"\n\n# \u2554\u2550\u2561 d0224ffa-a5ef-465c-89fe-bc293e4d46ad\nImage(\"Quiz/Q19.png\")\n\n# \u2554\u2550\u2561 4ff2da18-3d84-4cc8-9588-9fcc017aa4db\nmd\"\n##### Incorrect\n**Answer:** Out at the top, in at the bottom.\"\n\n# \u2554\u2550\u2561 73709c95-3e75-48b9-9465-b80684920db7\nmd\"##### Correct answer\n\nIn at the top, out at the bottom.\n\n**Explanation:**\nBecome familiar with Lenz's Law.\nThe change in magnetic flux is increasing, so the induced current's direction is switched.\n\"\n\n# \u2554\u2550\u2561 6036b824-7567-444a-8bad-a0472aeb549e\nmd\"### Question 20\n\nThe bar magnet is pushed toward the center of a wire loop.\nWhich is true?\n\n- There is a clockwise induced current in the loop.\n\n- There is no induced current in the loop.\n\n- There is a counterclockwise induced current in the loop.\"\n\n# \u2554\u2550\u2561 9a0f2ce9-6ac6-4c9a-a1dc-dc2e93187247\nImage(\"Quiz/Q20.png\")\n\n# \u2554\u2550\u2561 3bd6fefb-1475-4044-b637-f2358469d38f\nmd\"**Answer:** There is a clockwise induced current in the loop.\"\n\n# \u2554\u2550\u2561 87b9e579-9e52-4d85-8c33-8438c548184a\nmd\"### Question 21\n\nThe current in the straight wire is decreasing.\nWhich is true?\n\n- There is no induced current in the loop.\n\n- There is a counterclockwise induced current in the loop.\n\n- There is a clockwise induced current in the loop.\"\n\n# \u2554\u2550\u2561 8633003f-0de3-466a-974a-9087eaea00ec\nImage(\"Unit10/P2QC2.png\")\n\n# \u2554\u2550\u2561 ffa99665-c461-43ee-9939-e6027e53fad8\nmd\"**Answer:** There is a clockwise induced current in the loop.\"\n\n# \u2554\u2550\u2561 81deea62-431f-404e-9df4-7536baa0cabd\nmd\"### Question 22\n\nThe out of the page magnetic field in the circular region is decreasing.\nWhich is the induced electric field?\n\n- D\n\n- B\n\n- C\n\n- A\"\n\n# \u2554\u2550\u2561 cb6cc92b-2d9a-4ee7-9e82-a175c0d1202f\nImage(\"Quiz/Q22.png\")\n\n# \u2554\u2550\u2561 16e94b95-66bd-4b73-8185-e564eb2d6789\nmd\"\n##### Incorrect\n**Answer:** A.\"\n\n# \u2554\u2550\u2561 6516bc36-7df5-4522-90e1-57ddbf01bd9d\nmd\"##### Correct answer\n\nB.\n\n**Explanation:** Use the right-hand rule into the screen. The electric field is induced to oppose the direction of the increasing magnetic field.\"\n\n# \u2554\u2550\u2561 aab4689d-9e64-486c-8913-99972148d7f5\nmd\"### Question 23\n\nAn electromagnetic wave is traveling to the east.\nAt one instant at a given point its E vector points straight up.\nWhat is the direction of its B vector?\n\n- down\n\n- east\n\n- south\n\n- north\"\n\n# \u2554\u2550\u2561 b7a99f37-aeb3-4123-aa05-dadfca72f06e\nmd\"\n##### Incorrect\n**Answer:** north\"\n\n# \u2554\u2550\u2561 c6563fbe-7118-4ddc-a922-9e9327b46bc5\nmd\"##### Correct answer\n\nsouth\n\n**Explanation:** Apply the right-hand rule where $\\vec{E} \\times \\vec{B}$ determines the direction of the electromagnetic wave.\"\n\n# \u2554\u2550\u2561 79f4dc36-9ea9-4f22-9e0a-23f7e7bc139e\nmd\"### Question 24\n\nA lens creates an image as shown.\nIn this situation, the object distance $s$ is\n\n- Larger than the focal length f\n\n- Larger than 2f\n\n- Equal to the focal length f.\n\n- Larger than the focal length f but smaller than 2f\"\n\n# \u2554\u2550\u2561 80c5e3e4-61bd-46cd-aa7c-1f6b098269a6\nImage(\"Quiz/Q24.png\")\n\n# \u2554\u2550\u2561 cd63ab55-26fe-4015-b171-0a1073eee06d\nmd\"**Answer:** Larger than the focal length f but smaller than 2f\"\n\n# \u2554\u2550\u2561 f08902fd-e2b8-47bc-b6fe-ffee0c0b5267\nmd\"### Question 25\n\nA lens creates an image as shown.\nIn this situation, the image distance $s'$ is\n\n- Larger than the focal length f.\n\n- Smaller than focal length f.\n\n- Equal to the focal length f.\"\n\n# \u2554\u2550\u2561 ed2b1790-224a-442e-9971-dc8f5313dbcc\nImage(\"Quiz/Q24.png\")\n\n# \u2554\u2550\u2561 c32eba5a-fc0e-4eb3-b179-3c4ef4dbc000\nmd\"**Answer:** Larger than the focal length f.\"\n\n# \u2554\u2550\u2561 574b6d09-5147-404b-a0b7-3a7f6c88e251\nmd\"### Question 26\n\nYou see an upright, magnified image of your face when you look into magnifying \\\"cosmetic mirror\\\". The image is located\n\n- Behind the mirror's surface.\n\n- In front of the mirror's surface.\n\n- Only in your mind because it's a virtual image\n\n- On the mirror's surface.\"\n\n# \u2554\u2550\u2561 6a8c8c80-3833-42ef-b5e2-b36385d2a456\nmd\"**Answer:** Behind the mirror's surface\"\n\n# \u2554\u2550\u2561 3d9f7f6a-f6d4-4566-80ca-61b791cde540\nmd\"### Question 27\n\nIn an optical setup consisting of a single concave mirror and an object we measure the magnification factor M=-3.\nWe can conclude that this image is:\n\n- Inverted and virtual\n\n- Upright and virtual\n\n- Upright and real\n\n- Inverted and real\"\n\n# \u2554\u2550\u2561 e25ab1b9-ead0-48cd-8bb4-8611cba660dc\nmd\"**Answer:** Inverted and real\"\n\n# \u2554\u2550\u2561 e18a7261-ef49-4a4d-a961-4d4977554e1d\nmd\"### Question 28\n\nIn an optical setup consisting of a single lens and an object we measure the magnification factor M = -1.0.\nWe can conclude that object in this setup is:\n\n- At distance greater than twice the focal length\n\n- Exactly at point of twice the focal length\n\n- Between focal point and point of twice the focal length\n\n- Between lens and focal point\"\n\n# \u2554\u2550\u2561 2c9665eb-2e88-4405-8eb6-4804253a34dd\nmd\"**Answer:** Exactly at point of twice the focal length\"\n\n# \u2554\u2550\u2561 bb10b8d6-d5e3-415f-bda7-9f26cd52f3a6\nmd\"## Diagnostic Test 5\"\n\n# \u2554\u2550\u2561 ff1c72bf-e271-46df-a288-8619a9f0a2e4\nmd\"### Conceptual Question 22.14\n\nCharges **A** and **B** in the figure are equal. Each charge exerts a force on the other of magnitude $F$. Suppose the charge of **B** is increased by a factor of 4, but everything else is unchanged.\"\n\n# \u2554\u2550\u2561 c40cfee1-3a46-4b05-ad7e-e2a8126f56cd\nImage(\"Unit5/DTP1.png\")\n\n# \u2554\u2550\u2561 c24e62c5-08e4-465c-b8cb-80328a8df16f\nmd\"#### Part A\n\nIn terms of $F$, what is the magnitude of the force on **A**?\n\n**Express your answer in terms of $F$.**\"\n\n# \u2554\u2550\u2561 19d8e693-2fa4-4311-8fab-40c636d19169\nmd\"**Answer:** $F_A = 4F$\"\n\n# \u2554\u2550\u2561 c810aee0-42df-4e43-9a0c-5b3a37e0c892\nmd\"#### Part B\n\nIn terms of $F$, what is the magnitude of the force on **B**?\n\n**Express your answer in terms of $F$.**\"\n\n# \u2554\u2550\u2561 058a4e62-655b-4e86-8ac9-c14c9f303153\nmd\"**Answer:** $F_B = 4F$\"\n\n# \u2554\u2550\u2561 9594aa75-3585-4a4b-8f16-0e23e999268d\nmd\"### Conceptual Question 22.15\n\nThe electric force on a charged particle in an electric field is $F$.\"\n\n# \u2554\u2550\u2561 4379e2f3-d536-40c1-bff6-dbd652c496ff\nmd\"#### Part A\n\nWhat will be the force if the particle's charge is tripled and the electric field strength is halved?\n\n**Give your answer in terms of F**\"\n\n# \u2554\u2550\u2561 746b5d5b-5fde-4270-9e52-129f21ac722b\nmd\"**Answer:** $F' = \\frac{3}{2}F$\n\n**Explanation:** $\\vec{F} = q\\vec{E}$\"\n\n# \u2554\u2550\u2561 40616231-34a0-4308-ac97-05ef93da2c86\nmd\"### Problem 22.17 - Enhanced - with Feedback\n\nYou may want to review (Pages 613 - 616).\"\n\n# \u2554\u2550\u2561 f8c5aaf6-7cbb-4a87-b08c-fbea27e0658a\nImage(\"Unit5/DTP3.png\")\n\n# \u2554\u2550\u2561 65c05232-36bb-4df8-8146-4e8237e11758\nmd\"#### Part A\n\nWhat is the magnitude of the net electric force on charge A in the figure (Figure 1)?\nAssume that $q_1$ = 1.2 nC and $q_2$ = 5.2 nC.\n\n**Express your answer to two significant figures and include the appropriate units.**\"\n\n# \u2554\u2550\u2561 ae4eb3d7-cb85-4655-9e12-fdbb1e99af1b\nmd\"**Answer:**\n\n**Explanation:** Use $F = \\frac{k q_1 q_2}{r}$\n\n$\\begin{gather*}\nq_1 = 1.2 \\text{ nC} = 1.2 \\times 10^{-9} \\text{ C} \\\\\nq_2 = 5.2 \\text{ nC} = 5.2 \\times 10^{-9} \\text{ C} \\\\\nr = 1 \\text{ cm} = 0.01 \\text{ m} \\\\\n|F| = \\left|\\frac{kq_1q_1}{r^2} - \\frac{kq_1q_2}{(2r)^2}\\right| = 1.1 \\times 10^{-5} \\text{ N}\n\\end{gather*}$\"\n\n# \u2554\u2550\u2561 9319bfbb-97c1-44f4-b605-16ee995476c3\nlet\n\tq1 = 1.2e-9\n\tq2 = 5.2e-9\n\tr = 1 / 100\n\tk = 9e9\n\tF = k*q1*q1 / r^2 - k*q1*q2/(2r)^2\nend\n\n# \u2554\u2550\u2561 ef5e8506-4a02-42a1-950d-4a53aecd42d7\nmd\"#### Part B\n\nWhat is the direction of the net electric force on charge A?\n\n- right\n\n- left\n\n- up\n\n- down\"\n\n# \u2554\u2550\u2561 9f5e3964-3c67-41c8-b194-3dcdd6d485c2\nmd\"**Answer:** left\n\n**Explanation:** Compare the two forces being exerted on charge A.\"\n\n# \u2554\u2550\u2561 5fe3b6d7-2277-4c43-8e21-7cd4203433cc\nlet\n\tq1 = 1.2e-9\n\tq2 = 5.2e-9\n\tr = 1 / 100\n\tk = 9e9\n\tFB = k*q1*q1 / r^2\n\tFA = -k*q1*q2 / (2r)^2\n\tFB, FA\nend\n\n# \u2554\u2550\u2561 c50bcc01-6ab1-43d6-bdb0-4e9e17b524c0\nmd\"## Diagnostic Test 9 (Chapter 29)\"\n\n# \u2554\u2550\u2561 4830439e-3665-44c1-88c4-2b63da020890\nmd\"### Problem 29.5\"\n\n# \u2554\u2550\u2561 dde0edeb-b85c-4bca-b5e9-a75255dc50af\nImage(\"Unit9/DTP1.png\")\n\n# \u2554\u2550\u2561 1120ad44-3519-4a0e-b464-d3666470ca35\nmd\"#### Part A\n\nWhat is the magnetic field at the position of the dot in the figure (Figure 1)?\nGive your answer as a vector.\n\nExpress your answer in terms of the unit vectors $\\hat{i}$, $\\hat{j}$, and $\\hat{k}$. Use the 'unit vector' button to denote unit vectors in your answer.\"\n\n# \u2554\u2550\u2561 fefd36c1-8607-4825-b72b-4d4f0f357523\nmd\"**Answer:** \n$\\begin{gather*}\nB = \\frac{\\mu_0}{4\\pi} \\frac{qv\\sin{\\theta}}{r^2}\n\\end{gather*}$\"\n\n# \u2554\u2550\u2561 ffb0edda-d3b5-4154-9df0-95758f3f3f12\nlet\n\tv = 2e7u\"m/s\"\n\t\u03b8 = 45u\"\u00b0\"\n\tq = e\n\tx = (2/100)u\"m\"\n\ty = (2/100)u\"m\"\n\t\n\t# B = ?\n\t# Magnetic field of a moving charge\n\t# B = (\u03bc0 / 4\u03c0) * (qvsin(\u03b8) / r^2)\n\t\n\t# r = ?\n\t# Distance formula\n\tr = sqrt(x^2 + y^2)u\"m\"\n\t\n\tB = (\u03bc0 / 4\u03c0) * (q*v*sin(\u03b8))/(r^2)\nend\n\n# \u2554\u2550\u2561 c45dde4e-d60f-4543-a2ee-d061f45bc848\nmd\"### Problem 29.9\n\nAlthough the evidence is weak, there has been concern in recent years over possible health effects from the magnetic fields generated by electric transmission lines. A typical high-voltage transmission line is 20 m above the ground and carries a 200 A current at a potential of 110 kV.\"\n\n# \u2554\u2550\u2561 0bcd7967-9648-4f80-8b8b-7a1459f963b7\nmd\"#### Part A\n\nWhat is the magnetic field strength on the ground directly under such a transmission line?\n\n**Express your answer with the appropriate units.**\"\n\n# \u2554\u2550\u2561 ee17d3a7-a845-4b0d-991f-0d1546bd89c8\nmd\"**Answer:**\n$B = \\frac{\\mu_0}{2\\pi} \\frac{I}{r}$\"\n\n# \u2554\u2550\u2561 0497b136-6780-4e31-9066-5cab808fa9f3\nlet\n\tr = 20u\"m\"\n\tI = 200u\"A\"\n\t\n\t# B = ?\n\t# Magnetic field of a wire\n\tB = (\u03bc0 / 2\u03c0) * (I / r)\nend\n\n# \u2554\u2550\u2561 a57d36a2-c69a-4a4b-b623-e7067527a8d6\nmd\"#### Part B\n\nWhat percentage is this of the earth's magnetic field of 50 \u03bcT?\n\n$\\frac{B_{\\text{wire}}}{B_{\\text{earth}}}$\"\n\n# \u2554\u2550\u2561 42774779-7c00-46c6-af46-29e6bfa5b362\nlet\n\t(r, I) = 20u\"m\", 200u\"A\"\n\tB_wire = (\u03bc0 / 2\u03c0) * (I / r)\n\t\n\t(B_wire / B_earth) * 100\nend\n\n# \u2554\u2550\u2561 924acf6e-5ada-4a64-ad51-1ff5ce84bb5e\nmd\"# Notice\n\nFrom this point on, I will no longer write out each problem since it takes too long and my exam is coming up very soon.\"\n\n# \u2554\u2550\u2561 7bb28d6d-6d18-4209-b48e-5e87eedec74b\nmd\"### Problem 29.14\"\n\n# \u2554\u2550\u2561 d878a54c-a6e3-4fa7-81c9-5b045781f353\nlet\n\tI = 12u\"A\"\n\tr = (2/100)u\"m\"\n\t\n\t# B = ?\n\t# Magnetic field of a wire + superposition principle\n\t# B = (\u03bc0 / 2\u03c0) * (I / r)\n\t\n\tB1a = (\u03bc0 / 2\u03c0) * (I / r)\n\tB2a = -(\u03bc0 / 2\u03c0) * (I / 3r)\n\tB1b = (\u03bc0 / 2\u03c0) * (I / r)\n\tB2b = (\u03bc0 / 2\u03c0) * (I / r)\n\tB1c = -(\u03bc0 / 2\u03c0) * (I / 3r)\n\tB2c = (\u03bc0 / 2\u03c0) * (I / r)\n\t\n\tB1a + B2a, (B1a, B2a), B1b + B2b, B1c + B2c, (B1c, B2c)\nend\n\n# \u2554\u2550\u2561 862df6ac-1091-48b6-93a3-6cf64aa27dde\nmd\"### Problem 29.46\"\n\n# \u2554\u2550\u2561 294246d2-c696-4f46-b746-d5f40234eb59\nlet\n\tI = 4.0u\"A\"\n\tr1 = (1 / 100)u\"m\"\n\tr2 = (2.0 / 100)u\"m\"\n\ts1 = 2\u03c0 * r1\n\ts2 = 2\u03c0 * r2\n\t\n\t# B = ?\n\t# Magnetic field of a loop + superposition principle\n\t# B = (\u03bc0 / 2\u03c0) * (I / r)\n\t\n\tB1 = (\u03bc0 / 4\u03c0) * (I / r1)\n\tB2 = (\u03bc0 / 4\u03c0) * (I / r2)\n\t\n\tB1 + B2 # Incorrect!\nend\n\n# \u2554\u2550\u2561 9a3ac337-de34-41cb-96cf-7c2dd7cd47e3\nmd\"### Problem 29.44\"\n\n# \u2554\u2550\u2561 5bfab755-ed80-4318-ba3e-5774c89fff9f\nmd\"$B = \\frac{\\mu_0 IR^2}{2(z^2 + R^2)^{3/2}} = \\frac{\\mu_0 I}{4R}$\n\n$(z^2 + R^2)^{3/2} = 2R^3$\n\n$z^2 + R^2 = 1.59R^2$\n\n$z^2 = 0.59R^2$\n\n$z = 0.77$\"\n\n# \u2554\u2550\u2561 06f1e08c-95b3-430d-86a1-3de041fe1506\nlet\n\t# z = ?\n\t# Magnetic field of a circular loop\n\t# B = (\u03bc0 I R^2) / (2(z^2 + R^2)^(3/2))\n\t\n\t# Compare to center of loop, z = 0\n\t# B = (\u03bc0 I) / 2R\n\t#\n\t# Half it\n\t# B = (\u03bc0 I) / 4R\n\t\n\tsqrt(2^(2/3) - 1)\nend\n\n# \u2554\u2550\u2561 64d28e04-f67f-4b98-afd6-9c16a15f2277\nmd\"### Problem 29.29\"\n\n# \u2554\u2550\u2561 a502e763-da48-4811-ab9b-6d2075c94758\nlet\n\tB = 3.0000u\"T\"\n\tu = 1.6605e-27u\"kg\"\n\tmO2 = 2 * 15.995 * u\n\tmN2 = 2 * 14.003 * u\n\tmCO = 12.000 * u + 15.995 * u\n\tq = 1.6022e-19u\"C\"\n\t\n\t# f = ?\n\t# Frequency of cyclotron\n\t# f = qB / (2\u03c0 * m)\n\t\n\tfO2 = q*B / (2\u03c0 * mO2)\n\tfN2 = q*B / (2\u03c0 * mN2)\n\tfCO = q*B / (2\u03c0 * mCO)\n\tfO2, fN2, fCO # Hz\nend\n\n# \u2554\u2550\u2561 634882f3-baf9-4e98-a7c2-03c08314322d\nmd\"### Problem 29.30\"\n\n# \u2554\u2550\u2561 0a95a636-be2d-43cc-ac71-707c88d5f117\nlet\n\tv = (0.1c)u\"m/s\"\n\td = (42 / 100)u\"m\"\n\tr = (d / 2)u\"m\"\n\tq = e\n\tB = (mp * v) / (q * r)\nend\n\n# \u2554\u2550\u2561 9fa6bc2f-f99f-4d4b-91fa-695d04f0e83b\nmd\"### Problem 29.33\"\n\n# \u2554\u2550\u2561 2b72b700-5b08-4744-b29e-2714ec33580a\nlet\n\tm = (2 / 1000)u\"kg\"\n\tI = 1.6u\"A\"\n\td = (12 / 100)u\"m\"\n\tB = m*g / (I*d)\nend\n\n# \u2554\u2550\u2561 998d6c8b-3af1-4549-b633-13f4977e9afe\nmd\"### Problem 29.34\"\n\n# \u2554\u2550\u2561 a18585ce-d13b-4581-9c49-65c54bc3436f\nlet\n\tL = 10 / 100\n\td = 5 / 1000\n\tF = 7.2e-5\n\tV = 9\n\tR1 = 2\n\tI1 = V / R1\n\tI2 = F * 2\u03c0 * d / (\u03bc0 * I1 * L)\n\tR2 = V / I2\nend\n\n# \u2554\u2550\u2561 d8f53b47-1267-426b-a6d6-15b397dc8d9e\nmd\"### Problem 29.38\"\n\n# \u2554\u2550\u2561 f6840354-2325-470c-8fd1-a3246f236550\nlet\n\ts = (5.50 / 100)u\"m\"\n\tI = (510 / 1000)u\"A\"\n\tB = 1.60u\"T\"\n\t\u03b8 = 30u\"\u00b0\"\n\t\u03c4 = I*s^2*B*sin(\u03b8)\nend\n\n# \u2554\u2550\u2561 dc205f5d-0512-4429-8f26-f60e9f07115b\nmd\"## PSW 9 (Chapter 29)\"\n\n# \u2554\u2550\u2561 d6e4e26e-cb79-4aa1-8d97-1f2f238aeb10\nmd\"### Problem 29.15\"\n\n# \u2554\u2550\u2561 9f8323d5-1480-490a-b088-be9d69bf6e00\nlet\n\td = (6.0 / 1000)u\"m\"\n\t\u2130 = 60u\"V\"\n\tR100 = 100u\"\u03a9\"\n\tR30 = 30u\"\u03a9\"\n\tR20 = 20u\"\u03a9\"\n\tR = (1/R100 + 1/(R30+R20))^-1\n\tI = \u2130 / R\n\tI1 = \u2130 / R100\n\tI2 = \u2130 / (R30 + R20)\n\tB = (\u03bc0 / 2\u03c0) * (I2 / d)\nend\n\n# \u2554\u2550\u2561 4a0ecf24-5fc7-4586-9313-3c2fc33ea91c\nmd\"### Problem 29.37\"\n\n# \u2554\u2550\u2561 c494b302-bf92-44b5-abf2-59956da672d5\nlet\n\tl = 1u\"m\"\n\tm = (58 / 1000)u\"kg\"\n\tr = (4 / 100)u\"m\"\n\t\u03b8 = 60u\"\u00b0\"\n\t# 2F = mg\n\t# \u27f9 2IlB sin(\u03b8) = mg\n\t# \u27f9 \u03bc0 * I^2 sin(\u03b8) / \u03c0r = mg\n\t# \u27f9 I = sqrt(mg * \u03c0r / (\u03bc0 * sin(\u03b8)))\n\tI = sqrt(m*g * \u03c0*r / (\u03bc0 * sin(\u03b8)))\nend\n\n# \u2554\u2550\u2561 c8d2f67e-33ec-4ecd-b7f4-2037c1ac91ec\nmd\"### Problem 29.50\"\n\n# \u2554\u2550\u2561 6bc0b95d-be29-42ba-93ca-bb1bc910256e\nlet\n\td = (2 / 100)u\"m\"\n\tL = (8 / 100)u\"m\"\n\tB = 0.10u\"T\"\n\tI = 1.6u\"A\"\n\tN = B * L / (\u03bc0 * I)\nend\n\n# \u2554\u2550\u2561 971c6622-d701-4142-addf-55c2fb8007bd\nmd\"### Problem 29.52\"\n\n# \u2554\u2550\u2561 beedbeca-0477-4162-9c9e-05aa9cfd907f\nmd\"\n$B = \\frac{\\mu_0 I a^2}{2(x^2 + a^2)^{3/2}}$\"\n\n# \u2554\u2550\u2561 63619364-64ad-44aa-bbc5-1a847358c892\nlet\n\td = (18 / 100)u\"m\"\n\tr = (d / 2)\n\tB = 6e-12u\"T\"\n\tx = r\n\tI = (B * 2 * (x^2 + r^2)^(3/2)) / (\u03bc0 * r^2)\n\tI = (B * 2 * (r^2 + r^2)^(3/2)) / (\u03bc0 * r^2)\n\tI = (B * 2 * (2r^2)^(3/2)) / (\u03bc0 * r^2)\n\tI = (B * 2 * (sqrt(2) * r)^(3)) / (\u03bc0 * r^2)\nend\n\n# \u2554\u2550\u2561 d05ed603-19ce-4c7d-b67d-78e7258378d5\nmd\"### Problem 29.73\"\n\n# \u2554\u2550\u2561 21c3aacf-abb7-4f65-8681-440ee3c13341\nlet\n\tm = 4.0u\"kg\"\n\ts = 4.0u\"m\"\n\tI = 25u\"A\"\n\t\u03b8 = 25u\"\u00b0\"\n\t\n\t# Force acts on center of object\n\t# \u03c4 = Fd cos(\u03b8)\n\t# \u27f9 mg (s / 2) cos(\u03b8) = Is^2B sin(90-\u03b8)\n\t# \u27f9 B = mg cos(\u03b8) / (2Is sin(90-\u03b8))\n\tB = m*g*cos(\u03b8) / (2*I*s*sin(90u\"\u00b0\"-\u03b8))\nend\n\n# \u2554\u2550\u2561 651147e8-f7f8-4b87-9793-21e9b82a7c91\nmd\"### Problem 29.65\"\n\n# \u2554\u2550\u2561 f30306c0-0006-4a6f-873a-07fadcdc5d2d\nlet\n\tB = u\"T\"(29.0u\"mT\")\n\tv = 4.9e6u\"m/s\"\n\t\u03b8 = 30u\"\u00b0\"\n\tvpp = v*cos(\u03b8)\n\tvpl = v*sin(\u03b8)\n\tq = e\n\tr = me*(vpp) / (q*B)\n\t\n\t\u0394t = 2\u03c0*r / vpp\n\tp = vpl * \u0394t\nend\n\n# \u2554\u2550\u2561 bfec2cfd-b400-4e73-8a40-7b3665209a5d\nmd\"## Diagnostic Test 10 (Chapter 30)\"\n\n# \u2554\u2550\u2561 1b65d5b3-ffb4-4c4c-a071-24cf1cfdd0c4\nmd\"### Problem 30.4\"\n\n# \u2554\u2550\u2561 6a010c2a-f8e6-44db-84ff-8e9a9593a29e\nlet\n\ta = u\"m\"(30u\"cm\")\n\tB1 = 2.0u\"T\"\n\tB2 = 1.0u\"T\"\n\t\u03a61 = a^2 * B1\n\t\u03a62 = a^2 * B2\n\t\u03a61 - \u03a62\nend\n\n# \u2554\u2550\u2561 e5254b57-0930-4083-b8d5-b37ae9292052\nmd\"### Problem 30.5\"\n\n# \u2554\u2550\u2561 d7fd0a6d-2df6-4614-abf0-72971f07220b\nlet\n\tA = u\"m^2\"((5*10)u\"cm^2\")\n\tB = 5.50e-2u\"T\"\n\t\u03b8 = 45u\"\u00b0\"\n\t\u03a61 = A*B*cos(\u03b8)\n\t\u03a62 = A*B*cos(\u03b8)\n\t\u03a61 + \u03a62\nend\n\n# \u2554\u2550\u2561 e6375fb6-ae08-4442-b7c5-26953bd1cd95\nmd\"### Problem 30.8\"\n\n# \u2554\u2550\u2561 adcf50c9-8c45-46fa-aca3-10a5c660f80b\nlet\n\tds = u\"m\"(2.6u\"cm\")\n\tdl = u\"m\"(7.0u\"cm\")\n\tA = 1/4 * \u03c0 * ds^2\n\tB = 0.16u\"T\"\n\t\u03a61 = \u03a62 = A*B\nend\n\n# \u2554\u2550\u2561 549d2a7f-d307-4c96-9df5-6f936842cc59\nmd\"### Problem 30.11\"\n\n# \u2554\u2550\u2561 4cd51b45-2e77-41a2-9691-2d5e1fc01d3f\nlet\n\ts = u\"m\"(20u\"cm\")\n\tB = 0.30u\"T\"\n\t\u03b8 = 60u\"\u00b0\"\n\tb = s/2\n\th = s*sin(\u03b8)\n\tA = b*h / 2\n\t\u03a6 = A*B\nend\n\n# \u2554\u2550\u2561 3945a727-df1d-4717-b9d1-3b14f9e07adf\nmd\"### Problem 30.12\n\n- Yes, clockwise. Use Lenz's law.\"\n\n# \u2554\u2550\u2561 9c8a76a8-7caa-4d0d-9608-4e98d3955677\nmd\"### Problem 30.13\"\n\n# \u2554\u2550\u2561 bf938d16-4f96-409a-b3e7-39f6873ca275\nlet\n\tB = 0.50u\"T\"\n\tv = 55u\"m/s\"\n\tR = 0.70u\"\u03a9\"\n\tl = u\"m\"(5.0u\"cm\")\n\t\u2130 = v*l*B\n\tI = \u2130 / R\nend\n\n# \u2554\u2550\u2561 e4ecd474-4d48-4414-93b4-6eb029da45eb\nmd\"### Problem 30.15\"\n\n# \u2554\u2550\u2561 b798ffc4-bc44-4add-885f-96b88cf66960\nlet\n\tR = 0.30u\"\u03a9\"\n\tI = u\"A\"(150u\"mA\")\n\ts = u\"m\"(8.0u\"cm\")\n\tE = I*R\n\tA = s^2\n\t# E = d\u03a6/dt = A |dB/dt|\n\tdBdt = E/A\nend\n\n# \u2554\u2550\u2561 1d2210ec-00a9-4960-8686-85df92dae42d\nmd\"### Problem 30.22\"\n\n# \u2554\u2550\u2561 d2162fdf-dc4b-4138-8cc8-6ac803d82566\nlet\n\tV1 = 13000u\"V\"\n\tf = 60u\"Hz\"\n\tV2 = 120u\"V\"\n\tN2 = 150\n\tN1 = N2*V1/V2\n\t\n\tI2 = 220u\"A\"\n\t# I1V1 = I2V2 \u27f9 I1 = I2V2/V1\n\tI1 = I2*V2/V1\n\t\n\tN1, I1\nend\n\n# \u2554\u2550\u2561 57086fd7-66cb-4a72-b589-dc5fa3c8c8c2\nmd\"### Problem 30.26\"\n\n# \u2554\u2550\u2561 a7972df8-734e-4108-86b6-c0a5d0ede63d\nlet\n\tL = (100 / 1000)u\"H\"\n\tRw = 5.0u\"\u03a9\"\n\t\u2130 = 9u\"V\"\n\tRi = 3.0u\"\u03a9\"\n\tR = Rw + Ri\n\tI = \u2130/R\n\tUL = 1/2 * L * I^2\nend\n\n# \u2554\u2550\u2561 465bc5f9-d40c-4227-8962-da6746b1dcc4\nmd\"### Problem 30.27\"\n\n# \u2554\u2550\u2561 2817d2a7-fb3f-4986-adb9-3fb36a66d1b9\nlet\n\td = u\"m\"(3.00u\"cm\")\n\tA = 1/4 * \u03c0 * d^2\n\t\u2113 = u\"m\"(14.0u\"cm\")\n\tN = 200\n\tI = 0.750u\"A\"\n\tLsol = \u03bc0 * N^2 * A / \u2113\n\tUL = 1/2 * Lsol * I^2\nend\n\n# \u2554\u2550\u2561 6f906af6-1379-42a3-b24d-dd5d257241c9\nmd\"### Problem 30.19\"\n\n# \u2554\u2550\u2561 b94bf1ef-fb85-4e1a-92f8-3f47e143f9e6\nlet\n\t# F = ma = qE \u27f9 a = qE/m\n\tdBdt = 0.7u\"T/s\"\n\tra = u\"m\"(1.0u\"cm\")\n\trb = u\"m\"(0.0u\"cm\")\n\trc = u\"m\"(1.0u\"cm\")\n\trd = u\"m\"(2.0u\"cm\")\n\tq = e\n\tEa = ra/2 * dBdt\n\tEb = rb/2 * dBdt\n\tEc = rc/2 * dBdt\n\tEd = rd/2 * dBdt\n\taa = q*Ea / mp # upwards\n\tab = q*Eb / mp # a = 0\n\tac = q*Ec / mp # downwards\n\tad = q*Ed / mp # downwards\n\taa,ab,ac,ad\nend\n\n# \u2554\u2550\u2561 1568e950-6ae3-4eee-8c28-8db36795a929\nmd\"### Problem 30.20\"\n\n# \u2554\u2550\u2561 66cb890a-45af-4faf-8ecd-02df1bcdf3e6\nlet\n\td = u\"m\"(5.0u\"cm\")\n\tB = 2.0u\"T\"\n\tdBdt = 3.40u\"T/s\"\n\tr = u\"m\"(1.60u\"cm\")\n\tE = r/2 * dBdt\nend\n\n# \u2554\u2550\u2561 22fc3d80-341b-4902-89c1-c1a587c28a08\nmd\"## PSW 10 (Chapter 30)\"\n\n# \u2554\u2550\u2561 63b35422-6697-4a1f-8d60-55c977a87360\nmd\"### Problem 30.39\"\n\n# \u2554\u2550\u2561 d410ae9c-d876-4ac2-9d7d-44154cc24325\nlet\n\tN = 100\n\tdc = u\"m\"(5.0u\"cm\")\n\tdw = u\"m\"(0.50u\"mm\")\n\trc = dc / 2\n\tI = 4.0u\"A\"\n\t\u03c1 = 1.7e-8u\"\u03a9\"\n\n\t# |dB/dt| = ?\n\t# Faraday's Law\n\t# E = N|d\u03a6/dt| = NA |dB/dt| \u27f9 |dB/dt| = E/NA\n\t\n\tAw = 1/4 * \u03c0 * dw^2\n\tL = dw * N\n\tR = N * \u03c1 * (2\u03c0*rc) / Aw\n\tE = I * R\n\t\n\tA = 1/4 * \u03c0 * dc^2\n\t\n\tdBdt = E/(N*A)\nend\n\n# \u2554\u2550\u2561 e0a50b78-31a1-45ee-be74-2c6d8b5f13b8\nmd\"### Problem 30.49\"\n\n# \u2554\u2550\u2561 77c5d223-6de7-454f-b6d3-2895600e2b1a\nlet\n\td = u\"m\"(18u\"cm\")\n\tN = 120\n\tf = 60u\"Hz\"\n\t\u2130 = 350u\"V\"\n\t\n\t# B = ?\n\t# Electromotive force of a generator\n\t# \u2130 = NB\u03c9A sin\u03c9t \u27f9 B = \u2130/(N\u03c9Asin(\u03c9t)) = \u2130/(N\u03c9A) since peak of sin = 1\n\t\n\t\u03c9 = 2\u03c0 * f\n\tA = 1/4 * \u03c0 * d^2\n\tB = \u2130/(N*\u03c9*A)\nend\n\n# \u2554\u2550\u2561 85dee8d0-d364-4744-a5ac-5459f7c1a95c\nmd\"### Problem 30.50\"\n\n# \u2554\u2550\u2561 91d1441c-8d8b-4652-b089-5d06d191d21d\nlet\n\tNc = 40\n\tdc = u\"m\"(4.0u\"cm\")\n\tR = 0.40u\"\u03a9\"\n\t\n\tds = u\"m\"(4.0u\"cm\")\n\tLs = u\"m\"(20u\"cm\")\n\tNs = 200\n\t\n\tf = 60u\"Hz\"\n\tI = 0.20u\"A\"\n\t\n\t# I0 = ?\n\t# Magnetic field of a solenoid\n\t# Bsol = \u03bc0 INs / Ls = \u03bc0 (I0 sin(2\u03c0ft)) Ns / Ls\n\t\n\t# Magnetic flux through coil\n\t# \u03a6m = Nc ABsol = Nc A (\u03bc0 (I0 sin(2\u03c0ft)) Ns / Ls)\n\t\n\t# Induced emf in coil\n\t# \u2130 = -d\u03a6/dt = Nc A (2\u03c0f \u03bc0 (I0 cos(2\u03c0ft)) Ns / Ls)\n\t# I = \u2130/R = Nc A (2\u03c0f \u03bc0 (I0 cos(2\u03c0ft)) Ns / Ls) / R\n\t\n\t# Solve for I0\n\t# I0 = (I * Ls * R) / (Nc A 2\u03c0f \u03bc0 Ns)\n\t\n\tA = 1/4 * \u03c0 * ds^2\n\tI0 = I*Ls*R / (Nc*A*2\u03c0*f*\u03bc0*Ns) # The correct answer is 6.0 A.\nend\n\n# \u2554\u2550\u2561 2e69b5bd-0b56-4bf2-a338-b4d373dde517\nmd\"### Problem 30.65\"\n\n# \u2554\u2550\u2561 e65a8d69-d925-4de2-9edd-c33669b756a8\nlet\n\tA = 6.3e-2u\"m^2\"\n\tB = 5.3u\"T\"\n\t\u2130 = 9e-2u\"V\"\n\t\n\t# \u0394t = ?\n\t# Faraday's Law\n\t# \u2130 = \u0394\u03a6m/\u0394t \u27f9 \u0394t = \u0394\u03a6m/\u2130\n\t\n\t# \u0394\u03a6m = ?\n\t# Magnetic flux equation\n\t\u0394\u03a6m = A*B\n\t\n\t\u0394t = \u0394\u03a6m/\u2130\nend\n\n# \u2554\u2550\u2561 e0d4b7ca-578b-4d25-a9e6-81baafa497b0\nmd\"### Problem 30.80\"\n\n# \u2554\u2550\u2561 f44da9f7-87fa-4a44-864b-11e54553b3b0\nlet\n\tR = 2.4e-2u\"\u03a9\"\n\tI = 15u\"A\"\n\tr = u\"m\"(2.0u\"cm\")\n\tl = u\"m\"(4.0u\"cm\")\n\tx = u\"m\"(1.0u\"cm\")\n\tv = 10u\"m/s\"\n\t\n\t# I_loop = ?\n\t# Ohm's Law\n\t# \u2130 = IR \u27f9 I = \u2130/R\n\t\n\t# \u2130 = ?\n\t# Faraday's Law\n\t# \u2130 = d\u03a6/dt = d/dt(AB) = d/dt(xlB) = vlB = vl(B_r - B_rx)\n\t\n\t# B_r = ?\n\t# Magnetic field of a wire\n\tB_r = (\u03bc0 / 2\u03c0) * (I / r)\n\t\n\t# B_rx = ?\n\t# Magnetic field of a wire\n\tB_rx = (\u03bc0 / 2\u03c0) * (I / (r+x))\n\t\n\t\u2130 = v*l*(B_r - B_rx)\n\t\n\tI = \u2130/R\nend\n\n# \u2554\u2550\u2561 ca39044c-f352-4fb9-a8be-355f4a01a17d\nmd\"### Problem 30.85\"\n\n# \u2554\u2550\u2561 092aa2fc-ddc6-4729-911c-aa4e21ad7bd9\nlet\n\td = u\"m\"(2.1u\"cm\")\n\tN = 1200\n\tL = 1u\"m\"\n\tr = u\"m\"(0.60u\"cm\")\n\t\u2130 = 5.4e-4u\"V/m\"\n\t\n\t# dI/dt = ?\n\t# Magnetic field of a solenoid\n\t# dB/dt = (\u03bc0 N / L) (dI/dt) \u27f9 dI/dt = (dB/dt) / (\u03bc0 N / L)\n\t\n\t# dB/dt = ?\n\t# Faraday's Law of induction\n\t# \u2130 = r/2 (dB/dt) \u27f9 dB/dt = 2\u2130 / r\n\t\n\tdBdt = 2\u2130 / r\n\t\n\tdIdt = dBdt / (\u03bc0 * N / L)\nend\n\n# \u2554\u2550\u2561 3ece80ba-1bde-48a9-93ff-d6b9778b43e2\nmd\"## Diagnostic Test 11 (Chapter 31)\"\n\n# \u2554\u2550\u2561 07586a45-e8b7-466d-99b5-5b1fc20b32eb\nmd\"### Conceptual Question 31.7\"\n\n# \u2554\u2550\u2561 88053dfe-70e5-498a-bfd1-cb7eb33fad96\nlet\n\t# Part A\n\t# Out of the page\n\t\n\t# Part B\n\t# Upward\nend\n\n# \u2554\u2550\u2561 4c49f6cf-0f0e-408c-acde-ad77faa9903c\nmd\"### Problem 31.19\"\n\n# \u2554\u2550\u2561 da63cb45-9b93-4423-8fd7-0b6aae9e4a29\nlet\n\td = u\"m\"(2.0u\"mm\")\n\tP = u\"W\"(1.6u\"mW\")\n\t\n\t# E0 = ?\n\t# Intensity of electromagnetic wave\n\t# P/A = 1/2c\u03bc0 E0^2\n\t# \u27f9 E0 = sqrt((P/A) * 2c\u03bc0)\n\t\n\t# A = ?\n\tA = 1/4 * \u03c0 * d^2\n\t\n\tE0 = sqrt((P/A) * 2c*\u03bc0)\n\t\n\t# B0 = ?\n\t# EM Field strength relationship\n\t# E0 = cB0\n\t# \u27f9 B0 = E0/c\n\tB0 = E0/c\n\t\n\tE0, B0\nend\n\n# \u2554\u2550\u2561 472fc923-c527-47e4-9ea2-e2a762c08f62\nmd\"### Problem 31.21\"\n\n# \u2554\u2550\u2561 916addb5-4a54-422d-b9c9-5b24c3f16e3f\nlet\n\tP = u\"W\"(160u\"MW\")\n\td = u\"m\"(1.6u\"\u03bcm\")\n\t\n\t# E0 = ?\n\t# Intensity of electromagnetic wave\n\t# P/A = 1/2c\u03bc0 E0^2\n\t# \u27f9 E0 = sqrt((P/A) * 2c\u03bc0)\n\t\n\t# A = ?\n\tA = 1/4 * \u03c0 * d^2\n\t\n\tE0 = sqrt((P/A) * 2c*\u03bc0)\n\t\n\t# Ehydrogen = ?\n\t# Electric field produced by a point charge\n\tq = e\n\tr = u\"m\"(0.053u\"nm\")\n\tEhydrogen = k*q/r^2\n\t\n\tE0, E0 / Ehydrogen\nend\n\n# \u2554\u2550\u2561 bfafde7f-8d61-41fb-be8c-d3bf12404fab\nmd\"### Problem 31.22\"\n\n# \u2554\u2550\u2561 892661d3-046f-4df9-9269-89f26ca79b4c\nlet\n\tP = 1000u\"W\"\n\t\u03bb = u\"m\"(10u\"\u03bcm\")\n\td = u\"m\"(3.0u\"mm\")\n\t\n\t# F = ?\n\t# Radiation force due to beam of light\n\tF = P / c\nend\n\n# \u2554\u2550\u2561 04898c9a-5eca-4d1f-bd19-aec0d16f51d8\nmd\"### Conceptual Question 31.10\"\n\n# \u2554\u2550\u2561 8f1b5656-c3e6-4490-ba16-fbeb3f9c574e\nlet\n\t\u03b8 = Dict(\n\t\t:a => 90u\"\u00b0\",\n\t\t:b => 45u\"\u00b0\",\n\t\t:c => 30u\"\u00b0\",\n\t\t:d => 0u\"\u00b0\",\n\t\t:e => 30u\"\u00b0\"\n\t)\n\t\n\tsort(collect(\u03b8), by=x->(cos(x[2])^2), rev=true)\nend\n\n# \u2554\u2550\u2561 705a1476-eb5d-4f76-af51-4cf3bb61d36d\nmd\"## PSW 11 (Chapter 31)\"\n\n# \u2554\u2550\u2561 ff9db5b4-9875-4279-8957-4cfd3245d6f7\nmd\"### Problem 31.47\"\n\n# \u2554\u2550\u2561 61c3c8a1-4a00-4c1f-8c82-6594bcdfb006\nlet\n\tr = u\"m\"(4.5e9u\"km\")\n\tP = 21u\"W\"\n\t\n\t# I = ?\n\t# Intensity of point source\n\t# I = P/4\u03c0r^2\n\tI = P/(4\u03c0*r^2)\n\t\n\t# E0 = ?\n\t# Intensity of electromagnetic wave\n\t# I = 1/2c\u03bc0 E0^2\n\t# \u27f9 E0 = sqrt(2Ic\u03bc0)\n\tE0 = sqrt(2I*c*\u03bc0)\n\t\n\tI, E0\nend\n\n# \u2554\u2550\u2561 241d99b8-c672-40f3-8b9d-854413a57630\nmd\"### Problem 31.59\"\n\n# \u2554\u2550\u2561 d40d40e9-e141-4161-8cc7-403d30e74304\nlet\n\ts = u\"m\"(10u\"cm\")\n\tE0 = u\"V/m\"(11u\"kV/m\")\n\t\u0394T = 45\n\t\n\t# t = ?\n\t# 0.85P = E/t\n\t# \u27f9 t = E/0.85P\n\t\n\t# P = ?\n\t# Intensity of electromagnetic wave\n\t# P/A = 1/2c\u03bc0 E0^2\n\t# \u27f9 P = A/2c\u03bc0 E0^2\n\t\n\tbegin\n\t\t# A = ?\n\t\t# Area of one side of the cube\n\t\tA = s^2\n\n\t\tP = A/(2c*\u03bc0) * E0^2\n\tend\n\t\n\t# E = ?\n\t# Specific heat of water\n\t# E = Q = mc\u0394T = \u03c1Vc\u0394T\n\t\n\tbegin\n\t\t# \u03c1 = ?\n\t\t# Density of water\n\t\t\u03c1 = 1000u\"kg/m^3\"\n\n\t\t# V = ?\n\t\t# Volume of water\n\t\tV = s^3\n\n\t\t# c = ?\n\t\t# Specific heat capacity of water\n\t\tcw = 4190u\"J/(kg*K)\"\n\t\n\t\tE = Q = \u03c1*V*cw*\u0394T\n\tend\n\t\n\tt = E/0.85P\nend\n\n# \u2554\u2550\u2561 df7ed866-611b-4b64-94fd-b5876ed39336\nmd\"### Problem 31.60\"\n\n# \u2554\u2550\u2561 c74ce47c-697f-4fa6-9d27-bef932bd2d54\nlet\n\tm = 80u\"kg\"\n\t\u0394x = 5.0u\"m\"\n\tP = 1000u\"W\"\n\tt0 = u\"s\"(1.0u\"hr\")\n\t\n\t# t = ?\n\t# Initial time plus extra time\n\t# t = t0 + t1\n\t\n\tbegin\n\t\t# t1 = ?\n\t\t# Change in position over time\n\t\t# v = \u0394x1/t1 \u27f9 t = \u0394x1/v\n\t\t\n\t\tbegin\n\t\t\t# \u0394x1 = ?\n\t\t\t# Change in position after 1.0hr\n\t\t\t# \u0394x1 = \u0394x - \u0394x1\n\n\t\t\tbegin\n\t\t\t\t# \u0394x0 = ?\n\t\t\t\t# Kinematic equation\n\t\t\t\t# \u0394x0 = 1/2 a(t0)^2\n\t\t\t\t\n\t\t\t\tbegin\n\t\t\t\t\t# a = ?\n\t\t\t\t\t# Newton's second law\n\t\t\t\t\t# F = ma \u27f9 a = F/m\n\t\t\t\t\t\n\t\t\t\t\tbegin\n\t\t\t\t\t\t# F = ?\n\t\t\t\t\t\t# Force due to beam of light\n\t\t\t\t\t\t# F = P/c\n\t\t\t\t\t\t\n\t\t\t\t\t\tF = P/c\n\t\t\t\t\tend\n\t\t\t\t\t\n\t\t\t\t\ta = F/m\n\t\t\t\tend\n\t\t\t\t\n\t\t\t\t\u0394x0 = 1/2 * a * t0^2\n\t\t\tend\n\t\t\t\n\t\t\t\u0394x1 = \u0394x - \u0394x0\n\t\tend\n\t\t\n\t\tbegin\n\t\t\t# v = ?\n\t\t\t# Kinematic equation\n\t\t\t# v = at0\n\t\t\t\n\t\t\tv = a*t0\n\t\tend\n\t\t\n\t\tt1 = \u0394x1/v\n\tend\n\t\n\tt = t0 + t1\n\tu\"hr\"(t)\nend\n\n# \u2554\u2550\u2561 69e73591-3bdf-4e31-9ce9-4c1f44e207a8\nmd\"### Problem 31.37\"\n\n# \u2554\u2550\u2561 3388d0f4-ace3-4322-89ba-a06ba0840adf\nlet\n\tI = 5.0u\"A\"\n\td = u\"m\"(1.0u\"cm\")\n\tR = d / 2\n\tr = u\"m\"(1.8u\"mm\")\n\t\n\t# B = ?\n\t# Magnetic field of a wire\n\tB1 = (\u03bc0 / 2\u03c0) * (I / r)\n\t\n\t# B = ?\n\t# Biot-Savart Law\n\tB2 = (\u03bc0 / 2\u03c0) * (r / R^2) * I\n\t\n\tB1, B2\nend\n\n# \u2554\u2550\u2561 aa2e1f09-bb0e-452d-b277-32ed87f1d805\nmd\"## Diagnostic Test 12 (Chapter 34)\"\n\n# \u2554\u2550\u2561 49522fe9-e9d9-433c-92a2-072d60e1576e\nmd\"### Problem 34.40\"\n\n# \u2554\u2550\u2561 e9661551-68aa-41fc-be08-e88b8444e4f6\nlet\n\ts = 30.0u\"cm\"\n\tf = -22.0u\"cm\"\n\t\n\t# s\u2032 = ?\n\t# Mirror equation\n\t# 1/s + 1/s\u2032 = 1/f\n\t# \u27f9 s\u2032f + sf = ss\u2032\n\t# \u27f9 s\u2032(f - s) = -sf\n\t# \u27f9 s\u2032 = -sf/(f - s)\n\ts\u2032 = -s*f/(f - s)\nend\n\n# \u2554\u2550\u2561 8d7d25f5-f27e-4b86-a845-15e9a3dd2f72\nmd\"### Problem 34.41\"\n\n# \u2554\u2550\u2561 3548c0a7-1de7-4534-a930-58d80e9efad7\nlet\n\th = 1.3u\"cm\"\n\ts = 18.0u\"cm\"\n\tf = 56.0u\"cm\"\n\t\n\t# s\u2032 = ?\n\t# Mirror equation\n\t# 1/s + 1/s\u2032 = 1/f\n\t# \u27f9 s\u2032f + sf = ss\u2032\n\t# \u27f9 s\u2032(f - s) = -sf\n\t# \u27f9 s\u2032 = -sf/(f - s)\n\ts\u2032 = -s*f/(f - s)\n\t\n\t# h\u2032 = ?\n\t# Magnification equation\n\t# M = h\u2032/h = -s\u2032/s\n\t# \u27f9 h\u2032 = -s\u2032/s h\n\th\u2032 = -s\u2032/s * h\n\t\n\ts\u2032, h\u2032\nend\n\n# \u2554\u2550\u2561 b550aa41-be86-4336-91cd-816565045da5\nmd\"### Problem 34.33\"\n\n# \u2554\u2550\u2561 0fe25f6d-0132-4fa4-a1b8-a03963fd1ee7\nlet\n\th = 2.2u\"cm\"\n\ts = 36.0u\"cm\"\n\tf = 18.0u\"cm\"\n\t\n\t# s\u2032 = ?\n\t# Lens equation\n\t# 1/s + 1/s\u2032 = 1/f\n\t# \u27f9 s\u2032f + sf = ss\u2032\n\t# \u27f9 s\u2032(f - s) = -sf\n\t# \u27f9 s\u2032 = -sf/(f - s)\n\ts\u2032 = -s*f/(f - s)\n\t\n\t# h\u2032 = ?\n\t# Magnification equation\n\t# M = h\u2032/h = -s\u2032/s\n\t# \u27f9 h\u2032 = -s\u2032/s h\n\th\u2032 = -s\u2032/s * h\n\t\n\ts\u2032, abs(h\u2032)\nend\n\n# \u2554\u2550\u2561 a6e0b275-276f-4c0b-b548-4a4c283a6330\nmd\"### Problem 34.16\"\n\n# \u2554\u2550\u2561 f0a5c1d8-0de4-4455-b6e7-73b0426cd04c\nlet\n\t# d = ?\n\t# Transparent hemisphere\n\t# d = R / n\nend\n\n# \u2554\u2550\u2561 ceccbe3d-5c9a-47bf-a168-18154f30729b\nmd\"### Problem 34.11\"\n\n# \u2554\u2550\u2561 6f920972-e2ab-4e62-855e-9bac912831df\nlet\n\ty = 1.0u\"cm\"\n\t\u03b81 = 69u\"\u00b0\"\n\tn1 = 1\n\tn2 = 1.33\n\tn3 = 1.50\n\t\n\t# \u03b82 = ?\n\t# Snell's Law\n\t# n1 sin(\u03b81) = n2 sin(\u03b82)\n\t# \u27f9 \u03b82 = asin(n1 sin(\u03b81) / n2)\n\t\u03b82 = asind(n1 * sind(\u03b81) / n2)\n\t\n\t# \u03b83 = ?\n\t# Snell's Law\n\t# n2 sin(\u03b82) = n3 sin(\u03b83)\n\t# \u27f9 \u03b83 = asin(n2 sin(\u03b82) / n3)\n\t\u03b83 = asind(n2 * sind(\u03b82) / n3)\nend\n\n# \u2554\u2550\u2561 3be6b392-ea30-4015-a718-41f43797626e\nmd\"### Problem 34.7\"\n\n# \u2554\u2550\u2561 1c600f48-8f4d-4841-bfcf-14e805e284b8\nlet\n\tb = 5.0u\"m\"\n\th = 3.0u\"m\"\n\tmid = h / 2\n\t\n\t# \u03d5 = ?\n\t# Trigonometry\n\t# \u03d5 = atan(h/x)\n\t\n\tbegin\n\t\t# x = ?\n\t\t# Trigonometry relationship\n\t\t# tan(\u03d5) = h/x = mid / (b - x)\n\t\t# \u27f9 (b - x) h = x mid\n\t\t# \u27f9 bh - xh = x mid\n\t\t# \u27f9 bh = x(mid + h)\n\t\t# \u27f9 x = bh / (mid + h)\n\t\t\n\t\tx = b*h / (mid + h)\n\tend\n\t\n\t\u03d5 = atand(h/x)\nend\n\n# \u2554\u2550\u2561 f4730b14-2902-457d-a5f8-7e9140926b1e\nmd\"### Problem 34.14\"\n\n# \u2554\u2550\u2561 2fbca1ac-29e5-4f5b-8cac-faf6b48045eb\nlet\n\tn1 = 1.60\n\tn2 = 1.45\n\t\n\t# \u03b8c = ?\n\t# Critical angle\n\t\u03b8c = asind(n2/n1)u\"\u00b0\"\n\t\n\t\u03b8max = 90u\"\u00b0\" - \u03b8c\nend\n\n# \u2554\u2550\u2561 c9cae5ed-a959-4c20-a4a6-21b6962f7fc5\nmd\"### Problem 34.37\"\n\n# \u2554\u2550\u2561 53439784-13c7-4f08-949d-e60cd41d4494\nlet\n\th = 2.2u\"cm\"\n\ts = 15u\"cm\"\n\tf = -23u\"cm\"\n\t\n\t# s\u2032 = ?\n\t# Lens equation\n\t# 1/s + 1/s\u2032 = 1/f\n\t# \u27f9 s\u2032f + sf = ss\u2032\n\t# \u27f9 s\u2032(f - s) = -sf\n\t# \u27f9 s\u2032 = -sf/(f - s)\n\ts\u2032 = -s*f/(f - s)\n\t\n\t# h\u2032 = ?\n\t# Magnification equation\n\t# M = h\u2032/h = -s\u2032/s\n\t# \u27f9 h\u2032 = -s\u2032/s h\n\th\u2032 = -s\u2032/s * h\n\t\n\ts\u2032, abs(h\u2032)\nend\n\n# \u2554\u2550\u2561 e98b7caa-8df6-4292-93f5-f364ddeda2a3\nmd\"### Problem 34.5\"\n\n# \u2554\u2550\u2561 692de57c-99d1-439c-ab59-945db21dfe47\nlet\n\t(Ax, Ay) = (10u\"cm\", 5u\"cm\")\n\t(Bx, By) = (15u\"cm\", 15u\"cm\")\n\t\n\t# d = ?\n\t# Initial displacement\n\t# d = d0 + y\n\t\n\td0 = Ay\n\t\n\tbegin\n\t\t# y = ?\n\t\t# Use trigonometry relationship\n\t\t# tan(\u03d5) = (By - Ay)/y = By/((By - Ay) - y)\n\t\t# let dy = By - Ay\n\t\t# \u27f9 dy (dy - y) = By * y\n\t\t# \u27f9 dy^2 - dy y = By * y\n\t\t# \u27f9 dy^2 = y * (By + dy)\n\t\t# \u27f9 y = dy^2 / (By + dy)\n\t\t\n\t\tdy = By - Ay\n\t\ty = dy^2 / (By + dy)\n\tend\n\t\n\td = d0 + y\nend\n\n# \u2554\u2550\u2561 0efd04be-af2c-45e4-9aaa-b3fe5fb9c7d0\nmd\"### Problem 34.34\"\n\n# \u2554\u2550\u2561 da59c2ff-871e-4dee-9ec8-467ed823f11a\nlet\n\th = 1.1u\"cm\"\n\ts = 87.0u\"cm\"\n\tf = 31.0u\"cm\"\n\t\n\t# s\u2032 = ?\n\t# Lens equation\n\t# 1/s + 1/s\u2032 = 1/f\n\t# \u27f9 s\u2032f + sf = ss\u2032\n\t# \u27f9 s\u2032(f - s) = -sf\n\t# \u27f9 s\u2032 = -sf/(f - s)\n\ts\u2032 = -s*f/(f - s)\n\t\n\t# h\u2032 = ?\n\t# Magnification equation\n\t# M = h\u2032/h = -s\u2032/s\n\t# \u27f9 h\u2032 = -s\u2032/s h\n\th\u2032 = -s\u2032/s * h\n\t\n\ts\u2032, abs(h\u2032)\nend\n\n# \u2554\u2550\u2561 b520e5e2-702d-4131-9d10-3b12b04bda7e\nmd\"## PSW 12 (Chapter 34)\"\n\n# \u2554\u2550\u2561 c843abfd-f681-4850-83c0-f80241b3b122\nmd\"### Problem 34.52\"\n\n# \u2554\u2550\u2561 210917a3-6257-4723-a0ee-5bc04dd87884\nlet\n\tx1 = 2.1u\"m\"\n\ty1 = 1.0u\"m\"\n\ty2 = 3.0u\"m\"\n\tn1 = 1.00\n\tn2 = 1.33\n\t\n\t# d = ?\n\t# d = x1 + x2\n\t\n\tbegin\n\t\t# x2 = ?\n\t\t# Trigonometry\n\t\t# tan(\u03b82) = x2/y2 \u27f9 x2 = y2 tan(\u03b82)\n\t\t\n\t\tbegin\n\t\t\t# \u03b82 = ?\n\t\t\t# Snell's Law\n\t\t\t# n1 sin(\u03b81) = n2 sin(\u03b82)\n\t\t\t# \u27f9 \u03b82 = asin(n1 sin(\u03b81) / n2)\n\n\t\t\tbegin\n\t\t\t\t# \u03b81 = ?\n\t\t\t\t# Trigonometry\n\t\t\t\t# tan(\u03b81) = x1/y1 \u27f9 \u03b81 = atan(x1/y1)\n\t\t\t\t\u03b81 = atand(x1/y1)\n\t\t\tend\n\t\t\t\n\t\t\t\u03b82 = asind(n1 * sind(\u03b81) / n2)\n\t\tend\n\t\t\n\t\tx2 = y2 * tand(\u03b82)\n\tend\n\t\n\td = x1 + x2\nend\n\n# \u2554\u2550\u2561 ad1e49db-25cf-425a-88dd-47422330e85a\nmd\"### Problem 34.53\"\n\n# \u2554\u2550\u2561 b51230db-b12f-4c92-a140-79002eb2c1e0\nlet\n\ty = 1.90u\"m\"\n\tn1 = 1.00\n\ts = u\"m\"(36.0u\"cm\")\n\ts\u2032 = u\"m\"(46.0u\"cm\")\n\tx = s\u2032 - s\n\t\n\t# n2 = ?\n\t# Snell's law\n\t# n1 sin(\u03b81) = n2 sin(\u03b82)\n\t# \u27f9 n1 = n2 sin(\u03b82)\n\t# \u27f9 n2 = n1 / sin(\u03b82)\n\t\n\tbegin\n\t\t# \u03b82 = ?\n\t\t# Trigonometry\n\t\t# tan(\u03b82) = x/y \u27f9 \u03b82 = atan(x/y)\n\t\t\n\t\t\u03b82 = atand(x/y)\n\tend\n\t\n\tn2 = n1 / sind(\u03b82) # Incorrect!\nend\n\n# \u2554\u2550\u2561 9bf8ac4d-5c33-4dd5-a2d2-5ae1b03ebcd2\nmd\"### Problem 34.63\"\n\n# \u2554\u2550\u2561 8799b871-5a4c-4663-afcc-35e58ccf502e\nlet\n\tM = 1.5\n\ts = 1.8u\"cm\"\n\t\n\t# f = ?\n\t# Mirror equation\n\t# 1/s + 1/s\u2032 = 1/f\n\t# \u27f9 s\u2032f + sf = ss\u2032\n\t# \u27f9 f = ss\u2032 / (s\u2032 + s)\n\n\tbegin\n\t\t# s\u2032 = ?\n\t\t# Magnification equation\n\t\t# M = -s\u2032/s\n\t\t# \u27f9 s\u2032 = -M * s\n\t\t\n\t\ts\u2032 = -M * s\n\tend\n\t\n\tf = s*s\u2032 / (s\u2032 + s)\nend\n\n# \u2554\u2550\u2561 9e90dab3-6ba7-4298-889d-047bac5b5059\nmd\"### Problem 34.76\"\n\n# \u2554\u2550\u2561 9df87359-662d-46d0-8983-d45c2798a93c\nlet\n\tM = 3\n\tr = 30u\"cm\"\n\tf = r / 2\n\t\n\t# s = ?\n\t# Mirror equation\n\t# 1/s + 1/s\u2032 = 1/f\n\t# \u27f9 1/s + 1/3s = 1/f\n\t# \u27f9 3f - f = 3s\n\t# \u27f9 2f = 3s\n\t# \u27f9 s = 2f/3\n\t\n\ts = 2f/3\nend\n\n# \u2554\u2550\u2561 a47cc5f3-9375-4128-bac4-da8bd689b46c\nmd\"### Problem 34.69\"\n\n# \u2554\u2550\u2561 ed90dde8-3230-4fe9-87c2-615aab4269bb\nlet\n\tr = 2.6u\"m\"\n\tM = -2\n\t\n\t# f = ?\n\t# Mirror equation\n\t# 1/s + 1/s\u2032 = 1/f\n\t# \u27f9 s\u2032f + sf = ss\u2032\n\t# \u27f9 f = ss\u2032 / (s\u2032 + s)\n\t\n\t# s = ?\n\t# Magnification equation\n\t# M = -s\u2032/s = -(r - s)/s\n\t# \u27f9 sM = s - r\n\t# \u27f9 s = r / (1 - M)\n\ts = r / (1 - M)\n\t\n\t# s\u2032 = ?\n\t# s + s\u2032 = r\n\t# \u27f9 s\u2032 = r - s\n\ts\u2032 = r - s\n\t\n\tf = s*s\u2032 / (s\u2032 + s)\n\t\n\tf, s\nend\n\n# \u2554\u2550\u2561 14f633eb-43f6-44b3-8558-6871f318d908\nmd\"### Problem 34.73\"\n\n# \u2554\u2550\u2561 bd8ebbf4-7a57-4216-9690-f7c407fcfc58\nlet\n\ts = 13u\"cm\"\n\tM = 2\n\t\n\t# s\u2032 = ?\n\t# Magnification equation\n\t# M = -s\u2032/s \u27f9 s\u2032 = -M*s\n\ts\u2032 = -M * s\nend\n\n# \u2554\u2550\u2561 Cell order:\n# \u255f\u2500baaa0f44-a9d8-11eb-0d0d-53b78cd74315\n# \u255f\u2500c565da05-80ca-4b72-af73-d5da2f1c2abc\n# \u255f\u2500a1c99c25-e72e-4c4f-9ee9-855dad795483\n# \u255f\u2500c5cf2733-8d62-4f97-9c6c-97c6bce90b02\n# \u255f\u25007abb1db1-d637-446c-9e55-3457db3db17e\n# \u255f\u2500be1d2a6e-8c24-4a1e-b84b-aabfad8e6135\n# \u255f\u25006c9182b4-2bf5-40a8-8a7e-ecfb1f7e59f3\n# \u2560\u2550b670ab02-1fe7-4204-9747-237348707d97\n# \u255f\u25003240dda1-8d4c-4551-8878-8c2ddd9f12d1\n# \u255f\u25008a56ceb5-dbd9-4950-bbfe-50b56c682487\n# \u2560\u25501d460401-7c48-4718-914e-ba3fc9f7950a\n# \u255f\u250098b7f219-578b-4717-8dfa-5f22f86c04b2\n# \u255f\u2500f6b01ecd-9629-422c-800e-a41512fa9933\n# \u255f\u250081d41ebd-9c50-49a1-b77d-1f6c600477e7\n# \u255f\u2500067e90ac-b370-4c53-b1d8-8ee65189b036\n# \u255f\u2500179020bb-048a-4040-b517-0c081a5b68c0\n# \u255f\u25008065e0db-4aa8-447f-a130-49feedcddad7\n# \u255f\u25005745f4ea-219d-4e60-b269-9e08f010836a\n# \u255f\u2500885d48fa-4a05-4468-8492-0cae4bfb7b26\n# \u255f\u25002d690e4b-3d09-4907-99de-5f0ef4662b7e\n# \u255f\u2500eae4eef0-1701-406f-966d-736a2b2381f5\n# \u255f\u25009ca8f846-5d5b-4b28-85b6-90c5f9c068b6\n# \u255f\u250004912a55-5871-4917-8c95-7f03bd4abf1d\n# \u255f\u250042f03b7e-9fec-40bc-8d22-e594a22ab54e\n# \u255f\u250034b75779-b546-4628-ac0a-1d8fedc231eb\n# \u255f\u2500fb93dd3c-de36-4087-9870-8365161f7d1f\n# \u255f\u2500ab703047-2e82-4c13-a9d9-c97ffd42af67\n# \u255f\u2500253d5697-9088-424f-aa2e-c5df2c7675bf\n# \u255f\u2500fcdb7ea5-6203-4e74-b746-4470052b5f99\n# \u255f\u25005db75338-4b19-4037-942d-979856662d6c\n# \u255f\u250074378daa-7994-403e-9c41-86bae271a74d\n# \u255f\u2500af3ab252-16c6-4e6f-bdc6-9a01fede5a30\n# \u255f\u250064bbc192-28dd-4db4-8dab-1cde2a031bc5\n# \u255f\u2500185600d3-131d-491d-869d-06a865280a5c\n# \u255f\u25005fd137c7-a14e-4a53-9c9f-39f0992b1e43\n# \u255f\u25008a9474e5-4b73-4a3e-9573-fa0ca066f9de\n# \u255f\u2500d8c9b9e2-eaa1-40e5-9899-f60d8fb9dddb\n# \u255f\u2500f6000dca-7d5a-4bb6-93ec-3de97124fe51\n# \u255f\u250005c12afa-6ce0-4ab6-aac9-bb6a4f7a20e8\n# \u255f\u2500f30d8566-72fd-4658-a4cb-fcd2ac33b063\n# \u255f\u2500b6605cff-3e3c-40ed-b003-547a908a89f3\n# \u255f\u2500c53a0513-5a65-453b-8833-f4be54af3e0c\n# \u255f\u25006be1aa76-6cf2-4e57-bde4-7effcc453073\n# \u255f\u25008acec63d-de21-4698-9e51-579c16d2e398\n# \u255f\u25001e73fe74-4f6d-4137-b771-2cfac7cf6332\n# \u255f\u2500762087fa-ffcf-4691-8c67-6837555bbfb0\n# \u255f\u2500d6dba861-4f6e-4aad-9b8c-1e18ee2639af\n# \u255f\u25006cf18ee6-6b70-466f-9e23-32368cb35865\n# \u255f\u2500c5ee0bee-d2db-4daa-bbcf-5ac9f30b2f12\n# \u255f\u250024a53a41-96a8-4bab-a8cf-1f3cd6ad30ff\n# \u255f\u2500060fd1a0-5548-4eb4-b4c2-f2db077e0d50\n# \u255f\u25008eb2caa8-5341-40a7-8ae5-aad8e44c1af1\n# \u255f\u2500860bbf33-9025-44ee-aa26-7217f0bc7562\n# \u255f\u250028acdfbe-1020-4921-8c6c-0e38d0732d78\n# \u255f\u2500e06efb86-5366-49c3-acb6-f9b209bf5d81\n# \u255f\u25006c9233eb-058a-4e8a-9c9b-22964e210dd1\n# \u255f\u2500c83063bd-0bba-47e7-b869-e169d6e9c03e\n# \u255f\u2500c9102584-2196-4483-b79f-60089ad9e1b4\n# \u255f\u2500b30d503e-ab6c-4515-9a68-81327795aa6c\n# \u255f\u250085e2b232-6a09-4621-ab97-e2345b676999\n# \u255f\u250069bdb37e-2f06-4c78-a5bc-c3fce3cf6be8\n# \u255f\u2500f9ba57aa-4113-40e0-af24-8d7066baef4c\n# \u255f\u2500cdc4c352-a21c-4d88-8a6e-28da5974e893\n# \u255f\u2500bc7ebdc7-3614-43ad-bb1c-695f535c339b\n# \u255f\u25003badd910-d0be-435d-8b61-751d2fc46984\n# \u255f\u2500f5871103-8943-4085-8ec8-f7c19155262c\n# \u255f\u250064256f0e-f61a-41d9-9f07-f53f12b81d9f\n# \u255f\u250056988b0d-40f0-485c-bfd1-468431e2f906\n# \u255f\u2500baf581de-7077-4c25-a5df-b5e82490b113\n# \u255f\u250011c087b0-7823-41ac-8034-e66875bcaec1\n# \u255f\u2500ad23f43f-c8ac-48fd-9b21-901658710684\n# \u255f\u2500a59c5606-b60e-44c8-a7e3-b84cd89b322c\n# \u255f\u250069065726-b456-4ab1-b8a6-22e638304388\n# \u255f\u250047e713c1-0741-4135-ad0c-8b7ad003c141\n# \u255f\u250045baa2c9-e2a9-44fc-a6a3-9e1b149d3ec6\n# \u255f\u2500a44466e8-0954-4738-9836-e7d445d41a91\n# \u255f\u250013f7bece-837b-473e-bb86-df55d3f5dbdf\n# \u255f\u25006917fdb5-0538-4995-9ac5-14d7fc92b903\n# \u255f\u2500018a2522-f201-44a7-8f63-d67411970a90\n# \u255f\u2500ebe19088-8374-441c-9c1b-c00194737814\n# \u255f\u250030625a27-4961-4eff-be88-27a351a3db12\n# \u255f\u250066bd1832-dd76-43fa-ab08-018c8e726885\n# \u255f\u250008c4b0d3-a431-46dd-a0ae-3bd5a59b81e3\n# \u255f\u2500eff73afd-3ded-49d2-9411-ec11933fb31f\n# \u255f\u25006e2b45e2-67bd-4512-9b3c-6f25dc75824f\n# \u255f\u25009cf0faf9-ea42-45da-9788-975cfecdca37\n# \u255f\u250067aca9a5-da3f-4a2c-9159-2dfb922217f8\n# \u255f\u250037aeddcf-e1cf-40a0-925e-20d600f24845\n# \u255f\u25005fb3d735-8df5-4e69-aa47-ff467dee9047\n# \u255f\u2500c4f15be9-ba69-4b9f-9b5a-407a975c2fbe\n# \u255f\u25002c330d4d-8fa2-4c3f-a5b8-8a3ac19fd075\n# \u255f\u2500a2fbf237-3b84-4331-be33-e13c0f726838\n# \u255f\u250084cdc818-8a8c-4888-b3bd-12399e60a739\n# \u255f\u2500b759a10f-6110-4e66-8121-e1c8bf24efee\n# \u255f\u25009c245886-b491-4adf-9ccd-0f8d20326949\n# \u255f\u2500c352a601-03e9-4f47-9d8e-2e7e806a7433\n# \u255f\u2500d085be3a-89ec-4672-a47d-f5f2c020885b\n# \u255f\u25003d7c7ea7-24a4-4acc-8238-e71118fef4f1\n# \u255f\u250027a07162-335c-4d3a-9771-2a3793490631\n# \u255f\u250004beeb35-c9b8-4aca-9afa-6d9cefaacc62\n# \u255f\u25002f72f311-4bb9-4459-a1ee-3a3b5339b13f\n# \u255f\u2500082f1121-811b-4404-b561-6ce9ca6a05ce\n# \u255f\u25004584596e-c66f-45b3-ad3d-3042f13a1eca\n# \u255f\u250099a92b4f-0a27-4681-a372-e6db84db033a\n# \u255f\u25003a92f0e7-983c-433d-a83d-7c4e1a52a206\n# \u255f\u2500ae445aa3-dcd9-4d97-87f7-7be0a2287cf3\n# \u255f\u25006c521a07-2a6b-4ce8-a772-68b511bddf50\n# \u255f\u250090a8d78d-cdf9-4485-ba31-1364e2c920ae\n# \u255f\u25001f9d99d8-d613-4e62-9d46-5ebc61b029d6\n# \u255f\u2500141ea713-8276-4799-8e9c-f46f14e30c44\n# \u255f\u250059508d51-2fab-4cab-816c-6d0c88979af8\n# \u255f\u2500685e68c4-2be2-4fc7-b22f-9dc78b231236\n# \u255f\u2500f0d975f2-dc73-476a-9dec-6c7939511066\n# \u255f\u2500d1a284e5-e3d9-4d9c-b350-f6a9f9d4bf23\n# \u255f\u2500a76b95fb-bf1e-4fea-ae0b-927017a78d1b\n# \u255f\u2500368b2131-d30c-42ae-9550-b51073e7592b\n# \u255f\u25000acf5486-3702-4e8a-889e-871916d2ecf3\n# \u255f\u2500bfef7588-5c90-4a37-967e-660eb3e5efcd\n# \u255f\u250086fa33b9-17da-4322-b195-fdc99687c518\n# \u255f\u25009fdb361f-cb53-4270-a59c-6b967da468ea\n# \u255f\u25004c54fa63-06a1-4519-8104-aea78a2bbfbe\n# \u255f\u2500a68a80e3-b74e-4dbf-93ce-2892bc6dafd5\n# \u255f\u2500cda191c4-afb2-4512-a930-171f5757a094\n# \u255f\u2500c721d98c-3586-450c-a22f-a4ff048d12c6\n# \u255f\u2500b55d0b05-3713-46ea-819e-d180e662a47e\n# \u2560\u255016f0dd9e-f5bb-4173-a07c-7feba801837f\n# \u255f\u2500b87d87a8-da10-4af4-94af-868947f45d02\n# \u255f\u2500e664b06c-2924-423c-9f1b-52174fca7cc3\n# \u255f\u25007ec6edf3-7b6d-4ea2-8a4f-86dd2178eb91\n# \u255f\u25006de0da98-8dfe-496d-adc8-82c5c9af499e\n# \u255f\u2500d518d932-52d5-45de-aaf4-d94dfaf3eb64\n# \u255f\u2500570a8d6f-b108-4bba-81a4-8adffe79cea5\n# \u255f\u2500e275bc47-c73c-4cf5-bbdb-bebee2fb08af\n# \u255f\u250007adb0ac-6cb2-40ed-ad5f-659a6f58303b\n# \u255f\u2500276749db-45c7-43b9-a746-e48a6bdede2e\n# \u255f\u25004689568d-ba6d-4954-86ba-3fcadb52f957\n# \u255f\u25009a0b5bac-f167-4150-beaa-3c770dbc8cac\n# \u255f\u25007094b76b-519e-4427-91fe-88d33304fef1\n# \u255f\u250082836166-8d43-4c15-a2d8-ac451a1d20ab\n# \u255f\u250006389e81-54a5-46df-b7a6-fd11b44c5406\n# \u255f\u25005b87298f-9b97-4f7c-aab5-b3d740f9cab0\n# \u255f\u2500980aa663-98cd-42be-9b92-238a452f537a\n# \u255f\u250044d12feb-10cf-48b2-929a-8e6ff39a9496\n# \u255f\u25009f4b96a0-2b5e-47d0-8694-1c1d2144fd01\n# \u255f\u25002434b3b5-2da8-4c1b-8bf2-522fc75b5cd1\n# \u255f\u250072bd9e28-5048-48f2-a110-f5feaa67caee\n# \u255f\u2500a8dd4343-6913-4e19-bc12-23e89c9e6236\n# \u255f\u2500239d8478-4571-4155-90e3-95ae1deb4939\n# \u255f\u2500a748ef26-adc8-4fe5-ad2b-8c8b1d79b54d\n# \u255f\u2500f584bd06-21b6-41c2-b729-84b77acd2e2f\n# \u255f\u25000fbb0fa0-6c8e-497f-856f-2f3f8f2ce735\n# \u255f\u25004e172839-92b8-411a-ab77-3f8238d3a5e1\n# \u255f\u250051cf30e3-8fdc-4b62-a1a6-9252ed608dc9\n# \u255f\u25001183767b-3d27-49c0-91ba-fd65e96cf773\n# \u255f\u25009086f546-aa48-438e-b682-b0f27b2bff5c\n# \u255f\u2500d78a8b7b-4ea8-45ad-9d8d-331a090170a4\n# \u255f\u2500b7181523-93b0-4865-9c91-9ee965124bfa\n# \u255f\u2500fe0fb4f9-6d8f-48e7-9809-71c55035ba75\n# \u255f\u250039fe00bb-0eb1-4f31-8073-e17186da71e7\n# \u255f\u2500818d7074-6ce6-4746-a15e-a101ae653ec9\n# \u255f\u2500da06ac7d-67f3-483c-aad0-98529cf44a28\n# \u255f\u2500eae1f535-37ac-48dd-8f2c-65b88852fa43\n# \u255f\u2500d0224ffa-a5ef-465c-89fe-bc293e4d46ad\n# \u255f\u25004ff2da18-3d84-4cc8-9588-9fcc017aa4db\n# \u255f\u250073709c95-3e75-48b9-9465-b80684920db7\n# \u255f\u25006036b824-7567-444a-8bad-a0472aeb549e\n# \u255f\u25009a0f2ce9-6ac6-4c9a-a1dc-dc2e93187247\n# \u255f\u25003bd6fefb-1475-4044-b637-f2358469d38f\n# \u255f\u250087b9e579-9e52-4d85-8c33-8438c548184a\n# \u255f\u25008633003f-0de3-466a-974a-9087eaea00ec\n# \u255f\u2500ffa99665-c461-43ee-9939-e6027e53fad8\n# \u255f\u250081deea62-431f-404e-9df4-7536baa0cabd\n# \u255f\u2500cb6cc92b-2d9a-4ee7-9e82-a175c0d1202f\n# \u255f\u250016e94b95-66bd-4b73-8185-e564eb2d6789\n# \u255f\u25006516bc36-7df5-4522-90e1-57ddbf01bd9d\n# \u255f\u2500aab4689d-9e64-486c-8913-99972148d7f5\n# \u255f\u2500b7a99f37-aeb3-4123-aa05-dadfca72f06e\n# \u255f\u2500c6563fbe-7118-4ddc-a922-9e9327b46bc5\n# \u255f\u250079f4dc36-9ea9-4f22-9e0a-23f7e7bc139e\n# \u255f\u250080c5e3e4-61bd-46cd-aa7c-1f6b098269a6\n# \u255f\u2500cd63ab55-26fe-4015-b171-0a1073eee06d\n# \u255f\u2500f08902fd-e2b8-47bc-b6fe-ffee0c0b5267\n# \u255f\u2500ed2b1790-224a-442e-9971-dc8f5313dbcc\n# \u255f\u2500c32eba5a-fc0e-4eb3-b179-3c4ef4dbc000\n# \u255f\u2500574b6d09-5147-404b-a0b7-3a7f6c88e251\n# \u255f\u25006a8c8c80-3833-42ef-b5e2-b36385d2a456\n# \u255f\u25003d9f7f6a-f6d4-4566-80ca-61b791cde540\n# \u255f\u2500e25ab1b9-ead0-48cd-8bb4-8611cba660dc\n# \u255f\u2500e18a7261-ef49-4a4d-a961-4d4977554e1d\n# \u255f\u25002c9665eb-2e88-4405-8eb6-4804253a34dd\n# \u255f\u2500bb10b8d6-d5e3-415f-bda7-9f26cd52f3a6\n# \u255f\u2500ff1c72bf-e271-46df-a288-8619a9f0a2e4\n# \u255f\u2500c40cfee1-3a46-4b05-ad7e-e2a8126f56cd\n# \u255f\u2500c24e62c5-08e4-465c-b8cb-80328a8df16f\n# \u255f\u250019d8e693-2fa4-4311-8fab-40c636d19169\n# \u255f\u2500c810aee0-42df-4e43-9a0c-5b3a37e0c892\n# \u255f\u2500058a4e62-655b-4e86-8ac9-c14c9f303153\n# \u255f\u25009594aa75-3585-4a4b-8f16-0e23e999268d\n# \u255f\u25004379e2f3-d536-40c1-bff6-dbd652c496ff\n# \u255f\u2500746b5d5b-5fde-4270-9e52-129f21ac722b\n# \u255f\u250040616231-34a0-4308-ac97-05ef93da2c86\n# \u255f\u2500f8c5aaf6-7cbb-4a87-b08c-fbea27e0658a\n# \u255f\u250065c05232-36bb-4df8-8146-4e8237e11758\n# \u255f\u2500ae4eb3d7-cb85-4655-9e12-fdbb1e99af1b\n# \u2560\u25509319bfbb-97c1-44f4-b605-16ee995476c3\n# \u255f\u2500ef5e8506-4a02-42a1-950d-4a53aecd42d7\n# \u255f\u25009f5e3964-3c67-41c8-b194-3dcdd6d485c2\n# \u2560\u25505fe3b6d7-2277-4c43-8e21-7cd4203433cc\n# \u255f\u2500c50bcc01-6ab1-43d6-bdb0-4e9e17b524c0\n# \u255f\u25004830439e-3665-44c1-88c4-2b63da020890\n# \u255f\u2500dde0edeb-b85c-4bca-b5e9-a75255dc50af\n# \u255f\u25001120ad44-3519-4a0e-b464-d3666470ca35\n# \u255f\u2500fefd36c1-8607-4825-b72b-4d4f0f357523\n# \u2560\u2550ffb0edda-d3b5-4154-9df0-95758f3f3f12\n# \u255f\u2500c45dde4e-d60f-4543-a2ee-d061f45bc848\n# \u255f\u25000bcd7967-9648-4f80-8b8b-7a1459f963b7\n# \u255f\u2500ee17d3a7-a845-4b0d-991f-0d1546bd89c8\n# \u2560\u25500497b136-6780-4e31-9066-5cab808fa9f3\n# \u255f\u2500a57d36a2-c69a-4a4b-b623-e7067527a8d6\n# \u2560\u255042774779-7c00-46c6-af46-29e6bfa5b362\n# \u255f\u2500924acf6e-5ada-4a64-ad51-1ff5ce84bb5e\n# \u255f\u25007bb28d6d-6d18-4209-b48e-5e87eedec74b\n# \u2560\u2550d878a54c-a6e3-4fa7-81c9-5b045781f353\n# \u255f\u2500862df6ac-1091-48b6-93a3-6cf64aa27dde\n# \u2560\u2550294246d2-c696-4f46-b746-d5f40234eb59\n# \u255f\u25009a3ac337-de34-41cb-96cf-7c2dd7cd47e3\n# \u255f\u25005bfab755-ed80-4318-ba3e-5774c89fff9f\n# \u2560\u255006f1e08c-95b3-430d-86a1-3de041fe1506\n# \u255f\u250064d28e04-f67f-4b98-afd6-9c16a15f2277\n# \u2560\u2550a502e763-da48-4811-ab9b-6d2075c94758\n# \u255f\u2500634882f3-baf9-4e98-a7c2-03c08314322d\n# \u2560\u25500a95a636-be2d-43cc-ac71-707c88d5f117\n# \u255f\u25009fa6bc2f-f99f-4d4b-91fa-695d04f0e83b\n# \u2560\u25502b72b700-5b08-4744-b29e-2714ec33580a\n# \u255f\u2500998d6c8b-3af1-4549-b633-13f4977e9afe\n# \u2560\u2550a18585ce-d13b-4581-9c49-65c54bc3436f\n# \u255f\u2500d8f53b47-1267-426b-a6d6-15b397dc8d9e\n# \u2560\u2550f6840354-2325-470c-8fd1-a3246f236550\n# \u255f\u2500dc205f5d-0512-4429-8f26-f60e9f07115b\n# \u255f\u2500d6e4e26e-cb79-4aa1-8d97-1f2f238aeb10\n# \u2560\u25509f8323d5-1480-490a-b088-be9d69bf6e00\n# \u255f\u25004a0ecf24-5fc7-4586-9313-3c2fc33ea91c\n# \u2560\u2550c494b302-bf92-44b5-abf2-59956da672d5\n# \u255f\u2500c8d2f67e-33ec-4ecd-b7f4-2037c1ac91ec\n# \u2560\u25506bc0b95d-be29-42ba-93ca-bb1bc910256e\n# \u255f\u2500971c6622-d701-4142-addf-55c2fb8007bd\n# \u255f\u2500beedbeca-0477-4162-9c9e-05aa9cfd907f\n# \u2560\u255063619364-64ad-44aa-bbc5-1a847358c892\n# \u255f\u2500d05ed603-19ce-4c7d-b67d-78e7258378d5\n# \u2560\u255021c3aacf-abb7-4f65-8681-440ee3c13341\n# \u255f\u2500651147e8-f7f8-4b87-9793-21e9b82a7c91\n# \u2560\u2550f30306c0-0006-4a6f-873a-07fadcdc5d2d\n# \u255f\u2500bfec2cfd-b400-4e73-8a40-7b3665209a5d\n# \u255f\u25001b65d5b3-ffb4-4c4c-a071-24cf1cfdd0c4\n# \u2560\u25506a010c2a-f8e6-44db-84ff-8e9a9593a29e\n# \u255f\u2500e5254b57-0930-4083-b8d5-b37ae9292052\n# \u2560\u2550d7fd0a6d-2df6-4614-abf0-72971f07220b\n# \u255f\u2500e6375fb6-ae08-4442-b7c5-26953bd1cd95\n# \u2560\u2550adcf50c9-8c45-46fa-aca3-10a5c660f80b\n# \u255f\u2500549d2a7f-d307-4c96-9df5-6f936842cc59\n# \u2560\u25504cd51b45-2e77-41a2-9691-2d5e1fc01d3f\n# \u255f\u25003945a727-df1d-4717-b9d1-3b14f9e07adf\n# \u255f\u25009c8a76a8-7caa-4d0d-9608-4e98d3955677\n# \u2560\u2550bf938d16-4f96-409a-b3e7-39f6873ca275\n# \u255f\u2500e4ecd474-4d48-4414-93b4-6eb029da45eb\n# \u2560\u2550b798ffc4-bc44-4add-885f-96b88cf66960\n# \u255f\u25001d2210ec-00a9-4960-8686-85df92dae42d\n# \u2560\u2550d2162fdf-dc4b-4138-8cc8-6ac803d82566\n# \u255f\u250057086fd7-66cb-4a72-b589-dc5fa3c8c8c2\n# \u2560\u2550a7972df8-734e-4108-86b6-c0a5d0ede63d\n# \u255f\u2500465bc5f9-d40c-4227-8962-da6746b1dcc4\n# \u2560\u25502817d2a7-fb3f-4986-adb9-3fb36a66d1b9\n# \u255f\u25006f906af6-1379-42a3-b24d-dd5d257241c9\n# \u2560\u2550b94bf1ef-fb85-4e1a-92f8-3f47e143f9e6\n# \u255f\u25001568e950-6ae3-4eee-8c28-8db36795a929\n# \u2560\u255066cb890a-45af-4faf-8ecd-02df1bcdf3e6\n# \u255f\u250022fc3d80-341b-4902-89c1-c1a587c28a08\n# \u255f\u250063b35422-6697-4a1f-8d60-55c977a87360\n# \u2560\u2550d410ae9c-d876-4ac2-9d7d-44154cc24325\n# \u255f\u2500e0a50b78-31a1-45ee-be74-2c6d8b5f13b8\n# \u2560\u255077c5d223-6de7-454f-b6d3-2895600e2b1a\n# \u255f\u250085dee8d0-d364-4744-a5ac-5459f7c1a95c\n# \u2560\u255091d1441c-8d8b-4652-b089-5d06d191d21d\n# \u255f\u25002e69b5bd-0b56-4bf2-a338-b4d373dde517\n# \u2560\u2550e65a8d69-d925-4de2-9edd-c33669b756a8\n# \u255f\u2500e0d4b7ca-578b-4d25-a9e6-81baafa497b0\n# \u2560\u2550f44da9f7-87fa-4a44-864b-11e54553b3b0\n# \u255f\u2500ca39044c-f352-4fb9-a8be-355f4a01a17d\n# \u2560\u2550092aa2fc-ddc6-4729-911c-aa4e21ad7bd9\n# \u255f\u25003ece80ba-1bde-48a9-93ff-d6b9778b43e2\n# \u255f\u250007586a45-e8b7-466d-99b5-5b1fc20b32eb\n# \u2560\u255088053dfe-70e5-498a-bfd1-cb7eb33fad96\n# \u255f\u25004c49f6cf-0f0e-408c-acde-ad77faa9903c\n# \u2560\u2550da63cb45-9b93-4423-8fd7-0b6aae9e4a29\n# \u255f\u2500472fc923-c527-47e4-9ea2-e2a762c08f62\n# \u2560\u2550916addb5-4a54-422d-b9c9-5b24c3f16e3f\n# \u255f\u2500bfafde7f-8d61-41fb-be8c-d3bf12404fab\n# \u2560\u2550892661d3-046f-4df9-9269-89f26ca79b4c\n# \u255f\u250004898c9a-5eca-4d1f-bd19-aec0d16f51d8\n# \u2560\u25508f1b5656-c3e6-4490-ba16-fbeb3f9c574e\n# \u255f\u2500705a1476-eb5d-4f76-af51-4cf3bb61d36d\n# \u255f\u2500ff9db5b4-9875-4279-8957-4cfd3245d6f7\n# \u2560\u255061c3c8a1-4a00-4c1f-8c82-6594bcdfb006\n# \u255f\u2500241d99b8-c672-40f3-8b9d-854413a57630\n# \u2560\u2550d40d40e9-e141-4161-8cc7-403d30e74304\n# \u255f\u2500df7ed866-611b-4b64-94fd-b5876ed39336\n# \u2560\u2550c74ce47c-697f-4fa6-9d27-bef932bd2d54\n# \u255f\u250069e73591-3bdf-4e31-9ce9-4c1f44e207a8\n# \u2560\u25503388d0f4-ace3-4322-89ba-a06ba0840adf\n# \u255f\u2500aa2e1f09-bb0e-452d-b277-32ed87f1d805\n# \u255f\u250049522fe9-e9d9-433c-92a2-072d60e1576e\n# \u2560\u2550e9661551-68aa-41fc-be08-e88b8444e4f6\n# \u255f\u25008d7d25f5-f27e-4b86-a845-15e9a3dd2f72\n# \u2560\u25503548c0a7-1de7-4534-a930-58d80e9efad7\n# \u255f\u2500b550aa41-be86-4336-91cd-816565045da5\n# \u2560\u25500fe25f6d-0132-4fa4-a1b8-a03963fd1ee7\n# \u255f\u2500a6e0b275-276f-4c0b-b548-4a4c283a6330\n# \u2560\u2550f0a5c1d8-0de4-4455-b6e7-73b0426cd04c\n# \u255f\u2500ceccbe3d-5c9a-47bf-a168-18154f30729b\n# \u2560\u25506f920972-e2ab-4e62-855e-9bac912831df\n# \u255f\u25003be6b392-ea30-4015-a718-41f43797626e\n# \u2560\u25501c600f48-8f4d-4841-bfcf-14e805e284b8\n# \u255f\u2500f4730b14-2902-457d-a5f8-7e9140926b1e\n# \u2560\u25502fbca1ac-29e5-4f5b-8cac-faf6b48045eb\n# \u255f\u2500c9cae5ed-a959-4c20-a4a6-21b6962f7fc5\n# \u2560\u255053439784-13c7-4f08-949d-e60cd41d4494\n# \u255f\u2500e98b7caa-8df6-4292-93f5-f364ddeda2a3\n# \u2560\u2550692de57c-99d1-439c-ab59-945db21dfe47\n# \u2560\u25500efd04be-af2c-45e4-9aaa-b3fe5fb9c7d0\n# \u2560\u2550da59c2ff-871e-4dee-9ec8-467ed823f11a\n# \u255f\u2500b520e5e2-702d-4131-9d10-3b12b04bda7e\n# \u255f\u2500c843abfd-f681-4850-83c0-f80241b3b122\n# \u2560\u2550210917a3-6257-4723-a0ee-5bc04dd87884\n# \u255f\u2500ad1e49db-25cf-425a-88dd-47422330e85a\n# \u2560\u2550b51230db-b12f-4c92-a140-79002eb2c1e0\n# \u255f\u25009bf8ac4d-5c33-4dd5-a2d2-5ae1b03ebcd2\n# \u2560\u25508799b871-5a4c-4663-afcc-35e58ccf502e\n# \u255f\u25009e90dab3-6ba7-4298-889d-047bac5b5059\n# \u2560\u25509df87359-662d-46d0-8983-d45c2798a93c\n# \u255f\u2500a47cc5f3-9375-4128-bac4-da8bd689b46c\n# \u2560\u2550ed90dde8-3230-4fe9-87c2-615aab4269bb\n# \u255f\u250014f633eb-43f6-44b3-8558-6871f318d908\n# \u2560\u2550bd8ebbf4-7a57-4216-9690-f7c407fcfc58\n", "meta": {"hexsha": "54516ac2dc40c83c2180e67cc7c4c51641a39ce3", "size": 77041, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "PHYS1062FinalsReview.jl", "max_stars_repo_name": "airicbear/finals-review", "max_stars_repo_head_hexsha": "4c3fafe0fde2d656b28baebce516b2b22a494067", "max_stars_repo_licenses": ["Unlicense"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "PHYS1062FinalsReview.jl", "max_issues_repo_name": "airicbear/finals-review", "max_issues_repo_head_hexsha": "4c3fafe0fde2d656b28baebce516b2b22a494067", "max_issues_repo_licenses": ["Unlicense"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "PHYS1062FinalsReview.jl", "max_forks_repo_name": "airicbear/finals-review", "max_forks_repo_head_hexsha": "4c3fafe0fde2d656b28baebce516b2b22a494067", "max_forks_repo_licenses": ["Unlicense"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 23.3953841482, "max_line_length": 332, "alphanum_fraction": 0.660868888, "num_tokens": 38833, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.43014736319616964, "lm_q2_score": 0.13296424535145931, "lm_q1q2_score": 0.05719421953729878}}
{"text": "# Before running this, please make sure to activate and instantiate the environment\n# corresponding to [this `Project.toml`](https://raw.githubusercontent.com/alan-turing-institute/MLJTutorials/master/Project.toml) and [this `Manifest.toml`](https://raw.githubusercontent.com/alan-turing-institute/MLJTutorials/master/Manifest.toml)\n# so that you get an environment which matches the one used to generate the tutorials:\n#\n# ```julia\n# cd(\"MLJTutorials\") # cd to folder with the *.toml\n# using Pkg; Pkg.activate(\".\"); Pkg.instantiate()\n# ```\n\n# ## Machine type vs Scientific Type## ### Why make a distinction?## When analysing data, it is important to distinguish between## * *how the data is encoded* (e.g. `Int`), and# * *how the data should be interpreted* (e.g. a class label, a count, ...)## How the data is encoded will be referred to as the **machine type** whereas how the data should be interpreted will be referred to as the **scientific type** (or `scitype`).## In some cases, this may be un-ambiguous, for instance if you have a vector of floating point values, this should usually be interpreted as a continuous feature (e.g.: weights, speeds, temperatures, ...).## In many other cases however, there may be ambiguities, we list a few examples below:## * A vector of `Int` e.g. `[1, 2, ...]` which should be interpreted as categorical labels,# * A vector of `Int` e.g. `[1, 2, ...]` which should be interpreted as count data,# * A vector of `String` e.g. `[\"High\", \"Low\", \"High\", ...]` which should be interpreted as ordered categorical labels,# * A vector of `String` e.g. `[\"John\", \"Maria\", ...]` which should not interpreted as informative data,# * A vector of floating points `[1.5, 1.5, -2.3, -2.3]` which should be interpreted as categorical data (e.g. the few possible values of some setting), etc.## ### The Scientific Types## The package [ScientificTypes.jl](https://github.com/alan-turing-institute/ScientificTypes.jl) defines a barebone type hierarchy which can be used to indicate how a particular feature should be interpreted; in particular:## ```plaintext# Found# \u251c\u2500 Known# \u2502 \u251c\u2500 Textual# \u2502 \u251c\u2500 Finite# \u2502 \u2502 \u251c\u2500 Multiclass# \u2502 \u2502 \u2514\u2500 OrderedFactor# \u2502 \u2514\u2500 Infinite# \u2502 \u251c\u2500 Continuous# \u2502 \u2514\u2500 Count# \u2514\u2500 Unknown# ```## A *scientific type convention* is a specific implementation indicating how machine types can be related to scientific types. It may also provide helper functions to convert data to a given scitype.## The convention used in MLJ is implemented in [MLJScientificTypes.jl](https://github.com/alan-turing-institute/ScientificTypes.jl).# This is what we will use throughout; you never need to use ScientificTypes.jl# unless you intend to implement your own scientific type convention.## ### Inspecting the scitype## The `schema` function\nusing RDatasets, MLJScientificTypes\nboston = dataset(\"MASS\", \"Boston\")\nsch = schema(boston)\n\n# In this cases, most of the variables have a (machine) type `Float64` and# their default interpretation is `Continuous`.# There is also `:Chas`, `:Rad` and `:Tax` that have a (machine) type `Int64`# and their default interpretation is `Count`.## While the interpretation as `Continuous` is usually fine, the interpretation# as `Count` needs a bit more attention.# For instance note that:\nunique(boston.Chas)\n\n# so even though it's got a machine type of `Int64` and consequently a# default interpretation of `Count`, it would be more appropriate to interpret# it as an `OrderedFactor`.## ### Changing the scitype## In order to re-specify the scitype(s) of feature(s) in a dataset, you can# use the `coerce` function and specify pairs of variable name and scientific# type:\nboston2 = coerce(boston, :Chas => OrderedFactor);\n\n# the effect of this is to convert the `:Chas` column to an ordered categorical# vector:\neltype(boston2.Chas)\n\n# corresponding to the `OrderedFactor` scitype:\nelscitype(boston2.Chas)\n\n# You can also specify multiple pairs in one shot with `coerce`:\nboston3 = coerce(boston, :Chas => OrderedFactor, :Rad => OrderedFactor);\n\n# ### String and Unknown## If a feature in your dataset has String elements, then the default scitype# is `Textual`; you can either choose to drop such columns or to coerce them# to categorical:\nfeature = [\"AA\", \"BB\", \"AA\", \"AA\", \"BB\"]\nelscitype(feature)\n\n# which you can coerce:\nfeature2 = coerce(feature, Multiclass)\nelscitype(feature2)\n\n# ## Tips and tricks## ### Type to Type coercion## In some cases you will want to reinterpret all features currently# interpreted as some scitype `S1` into some other scitype `S2`.# An example is if some features are currently interpreted as `Count` because# their original type was `Int` but you want to consider all such as# `Continuous`:\ndata = select(boston, [:Rad, :Tax])\nschema(data)\n\n# let's coerce from `Count` to `Continuous`:\ndata2 = coerce(data, Count => Continuous)\nschema(data2)\n\n# ### Autotype## A last useful tool is `autotype` which allows you to specify *rules* to# define the interpretation of features automatically.# You can code your own rules but there are three useful ones that are pre-# coded:## * the `:few_to_finite` rule which checks how many unique entries are present# in a vector and if there are \"few\" suggests a categorical type,# * the `:discrete_to_continuous` rule converts `Integer` or `Count` to# `Continuous`# * the `:string_to_multiclass` which returns `Multiclass` for any string-like# column.## For instance:\nboston3 = coerce(boston, autotype(boston, :few_to_finite))\nschema(boston3)\n\n# You can also specify multiple rules, see [the docs](https://alan-turing-institute.github.io/MLJScientificTypes.jl/stable/#Automatic-type-conversion-for-tabular-data-1) for more information.\n# This file was generated using Literate.jl, https://github.com/fredrikekre/Literate.jl\n\n", "meta": {"hexsha": "a4446a680ba3860d051df3f1e5f238579e4f7aa9", "size": 5792, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "__site/generated/scripts/D0-scitype.jl", "max_stars_repo_name": "ven-k/MLJTutorials", "max_stars_repo_head_hexsha": "42151c8a96ad701aeaf763d53c8b7c6689eb6e8d", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "__site/generated/scripts/D0-scitype.jl", "max_issues_repo_name": "ven-k/MLJTutorials", "max_issues_repo_head_hexsha": "42151c8a96ad701aeaf763d53c8b7c6689eb6e8d", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "__site/generated/scripts/D0-scitype.jl", "max_forks_repo_name": "ven-k/MLJTutorials", "max_forks_repo_head_hexsha": "42151c8a96ad701aeaf763d53c8b7c6689eb6e8d", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 109.2830188679, "max_line_length": 2245, "alphanum_fraction": 0.7353245856, "num_tokens": 1495, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.43014733397551624, "lm_q2_score": 0.1329642333263228, "lm_q1q2_score": 0.05719421047941624}}
{"text": "module FranklinTheorems\n\nimport Franklin\nexport lx_fakebiblabel, lx_newcounter, lx_stepcounter, lx_arabic\n\n\nmodule ExportConfigPath\n\n\texport css_path, config_path\n\n\t\"\"\"\n\tReturns a path to FranklinTheorem's default CSS file. Copy this file to the Franklin `\\\\_css\\\\` directory.\n\t\"\"\"\n\tfunction css_path()\n\t\treturn normpath(joinpath(@__FILE__, \"..\", \"FranklinTheorems.css\"))\n\tend\n\n\t\"\"\"\n\tReturns a path to FranklinTheorem's markdown file, full of LaTeX-style definitions.\n\tBy default, the theorem, lemma, and definition blocks are defined.\n\tIf a list of strings is given, then the associated counters and environments are created.\n\tSince this is dynamic, because of the optional parameters, this returns a path to a tempfile.\n\t\"\"\"\n\tfunction config_path()\n\t\tconfig_path([])\n\tend\n\tfunction config_path(theorem_class_names)\n\t\tall_class_names = [[\"theorem\", \"lemma\", \"definition\"]; theorem_class_names]\n\t\tbaseMarkdownText = read(normpath(joinpath(@__FILE__, \"..\", \"FranklinTheorems.md\")), String)\n\n\t\tf = tempname()\n\t\twrite(f, baseMarkdownText * \"\\n\" * make_custom_environments(all_class_names))\n\t\treturn f\n\tend\n\n\t\"\"\"\n\tReturns a path to a temporary file that includes Franklin-Markdown code for a custom amstheorem-style environment.\n\t\"\"\"\n\tfunction make_custom_environments(theorem_class_names)\n\t\t\n\t\tenv_string = \"\"\n\t\tenable_string = \"\\\\newcommand{\\\\enabletheorems}{\"\n\n\t\tfor i=1:length(theorem_class_names)\n\t\t\tdefn_name = theorem_class_names[i]\n\t\t\tdefn_titlecase = titlecase(defn_name)\n\t\t\tdefn_body = \"\"\"\n\\\\newcommand{\\\\$(defn_name)ref}[1]{\\\\cite{!#1}}\n\n\\\\newenvironment{$(defn_name)}[2]{\n\t\\\\stepcounter{Num$(defn_name)}\n\t\\\\begin{thmBlock}{$(defn_titlecase) \\\\arabic{Num$(defn_name)}}{#1}{$(defn_titlecase)}{ \\\\fakebiblabel{!#2 @ $(defn_titlecase) \\\\arabic{Num$(defn_name)}} }\n}{\n\t\\\\end{thmBlock}\n}\n\t\t\t\"\"\"\n\t\t\tenv_string = env_string * \"\\n\" * defn_body\n\t\t\tenable_string = enable_string * \"\\n\" * \"\\\\newcounter{Num$(defn_name)}\"\n\t\tend\n\n\t\tenable_string = enable_string * \"\\n}\"\n\n\t\treturn env_string * \"\\n\" * enable_string\n\tend\n\nend\n\nusing .ExportConfigPath\n\n\n\n\"\"\"\nSpecialized code for automatic numbering by abuse of the biblabel function.\n\n- Given an input of form `\\\\fakebiblable{@ Legible Name}`, this returns the string `\"Legible Name\"`\n- Given an input of form `\\\\fakebiblabel{goob @ Legible Name}`, this runs `\\\\biblabel{goob}{Legible Name}`, so that Franklin's referencing system keeps track of `goob` as a name that points to `\"Legible Name\"`, and the function returns the string `\"goob\"`.\n\nThis `@` symbol approach is a hack to allow a single optional input variable to the function.\nThe `@` symbol was chosen because of its relevance to LaTeX.\n\nFurther, the string `\"Legible Name\"` is always parsed by Franklin, which has two notable consequences\n1. Other lx_functions like `\\\\arabic{counter}` can be used in the full name\n2. Markdown syntaxing like `_italics_` will be parsed into `italics`, which breaks the references\n\nSo, while `Legible Name` can reference counters, it should not have any markdown styling.\n\"\"\"\nfunction lx_fakebiblabel(lxc, _)\n\targs = Franklin.content(lxc.braces[1]) |> strip |> split\n\n\tnvars = length(args)\n\n\tif nvars < 2\n\t\terror(\"Bad fakebiblabel input: Less than 2 inputs were given. Input was:\\n\\\\fakebiblablel{\" * join(args, \" \") * \"}\")\n\tend\n\n\tif args[1] == \"@\"\n\t\treturn Franklin.fd2html(join(args[2:end], \" \"), internal=true, nop=true)\n\telseif args[2] == \"@\"\n\t\tref_name = args[1]\n\t\tlegible_name = Franklin.fd2html(join(args[3:end], \" \"), internal=true, nop=true)\n\t\tFranklin.fd2html(\"\\\\biblabel{\" * ref_name * \"}{\" * legible_name * \"}\", internal=true)\n\t\treturn ref_name\n\telse\n\t\terror(\"Bad fakebiblabel input: @ was not the second input. Input was:\\n\\\\fakebiblablel{\" * join(args, \" \") * \"}\")\n\tend\nend\n\n\n# Module for the counters. By making this into a module, this hides\n# access to the counters dictionary. If you need a new way to interact\n# with the system of counters, impliment a new method in this module.\nmodule LxCounters\n\t\n\t# Used for latex-style parsing\n\tusing FranklinUtils\n\n\texport lx_newcounter, lx_stepcounter, lx_arabic\n\n\t# The core dictionary that stores all the things we want to count.\n\tcounters = Dict{String,Int64}()\n\n\t\"\"\"\n\tCall `\\\\newcounter{name}` to create a new counter. Returns an empty string.\n\t\"\"\"\n\tfunction lx_newcounter(lxc, _)\n\t\targs, kwargs = lxargs(lxc)\n\n\t\tname = string(args[1])\n\n\t\tdelete!(counters, name)\n\t\tget!(counters, name, 0)\n\t\treturn \"\"\n\tend\n\n\t\"\"\"\n\tCall `\\\\stepcounter{name}` to incriment the value of a counter by 1. Returns an empty string.\n\t\"\"\"\n\tfunction lx_stepcounter(lxc, _)\n\t\targs, kwargs = lxargs(lxc)\n\n\t\tname = string(args[1])\n\n\t\tcounters[name] = counters[name] + 1\n\t\treturn \"\"\n\tend\n\n\t\"\"\"\n\tCall `\\\\arabic{name}` to return the value of a counter, as an arabic number.\n\t\"\"\"\n\tfunction lx_arabic(lxc, _)\n\t\targs, kwargs = lxargs(lxc)\n\n\t\tname = string(args[1])\n\n\t\treturn string(counters[name])\n\tend\n\nend\n\nusing .LxCounters\n\nend # Module\n", "meta": {"hexsha": "ee13e7c9d446d7bac995e9c099887daec8ee1b35", "size": 4911, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/FranklinTheorems.jl", "max_stars_repo_name": "Shuvomoy/FranklinTheorems.jl", "max_stars_repo_head_hexsha": "febec5a2e5c980d4912f15ce7fae92f00fad54b9", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 4, "max_stars_repo_stars_event_min_datetime": "2021-02-17T15:55:00.000Z", "max_stars_repo_stars_event_max_datetime": "2021-11-20T21:38:40.000Z", "max_issues_repo_path": "src/FranklinTheorems.jl", "max_issues_repo_name": "Shuvomoy/FranklinTheorems.jl", "max_issues_repo_head_hexsha": "febec5a2e5c980d4912f15ce7fae92f00fad54b9", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/FranklinTheorems.jl", "max_forks_repo_name": "Shuvomoy/FranklinTheorems.jl", "max_forks_repo_head_hexsha": "febec5a2e5c980d4912f15ce7fae92f00fad54b9", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2021-11-20T21:38:42.000Z", "max_forks_repo_forks_event_max_datetime": "2021-11-20T21:38:42.000Z", "avg_line_length": 30.1288343558, "max_line_length": 256, "alphanum_fraction": 0.7151293016, "num_tokens": 1336, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.43014733397551624, "lm_q2_score": 0.13296422989056964, "lm_q1q2_score": 0.05719420900153618}}
{"text": "# conditional loop\ni = 0\nwhile i <= 5 \n print(i)\n print(\",\")\n global i += 1\nend\nprint(i)\n", "meta": {"hexsha": "768550b444204cb0a4b9a16f35632fdfa82d1ad8", "size": 101, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "demo/while_loop.jl", "max_stars_repo_name": "elucian/julia", "max_stars_repo_head_hexsha": "653dc99961b6e27c2a3910c2594ba1a94df84063", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "demo/while_loop.jl", "max_issues_repo_name": "elucian/julia", "max_issues_repo_head_hexsha": "653dc99961b6e27c2a3910c2594ba1a94df84063", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "demo/while_loop.jl", "max_forks_repo_name": "elucian/julia", "max_forks_repo_head_hexsha": "653dc99961b6e27c2a3910c2594ba1a94df84063", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 11.2222222222, "max_line_length": 18, "alphanum_fraction": 0.5148514851, "num_tokens": 34, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4073334000459302, "lm_q2_score": 0.1403362440969662, "lm_q1q2_score": 0.05716363945769285}}
{"text": "module MyFunRep\n\n# Write your package code here.\n\"\"\"\nA simple add method for testing\n ```julia> my_fun1(2,3)\n ```\n\"\"\"\nmy_fun1(x,y) = x+y \n\n\"\"\"\nA sample function as an experiment\n ```julia> my_fun2(1,2,3)\n\"\"\"\nfunction my_fun2(x,y,z)\n return x+y+z\nend\n\n\"\"\"\n func(x)\nReturns double the number `x` plus `1`.\n```julia-repl\njulia > func(2)\n```\n\"\"\"\nfunc(x) = 2x + 1\n\n\"\"\"\n func2(x,y)\nReturns something else double the number `x` plus `1`.\n\"\"\"\nfunc2(x,y) = 2x + 1 + 3y\n\nexport my_fun1, my_fun2, func,func2\n\nend\n", "meta": {"hexsha": "8e22f7465308d54c8a246ec1802424e60da397fe", "size": 520, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/MyFunRep.jl", "max_stars_repo_name": "geekymode/MyFunRep.jl", "max_stars_repo_head_hexsha": "a128a6a52ca8b33510c0975ab8aeeb289273b0ab", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/MyFunRep.jl", "max_issues_repo_name": "geekymode/MyFunRep.jl", "max_issues_repo_head_hexsha": "a128a6a52ca8b33510c0975ab8aeeb289273b0ab", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/MyFunRep.jl", "max_forks_repo_name": "geekymode/MyFunRep.jl", "max_forks_repo_head_hexsha": "a128a6a52ca8b33510c0975ab8aeeb289273b0ab", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 14.0540540541, "max_line_length": 54, "alphanum_fraction": 0.6057692308, "num_tokens": 183, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.47268347662043286, "lm_q2_score": 0.1208532340736852, "lm_q1q2_score": 0.05712532684277248}}
{"text": "# saveTable(\"test.txt\", \"\", [[1 2]; [3 4]])\n# saveTable(\"test.txt\", \"\", rand(2, 2))\n\nusing DelimitedFiles\n\nfunction saveTable(outfile, tag, outmatrix)\n open(outfile, \"a\") do file\n write(file, \"$(tag)\\n\")\n writedlm(file, outmatrix, \"\\t\")\n end\nend\n", "meta": {"hexsha": "fc559d5ff3226e2595494f8086284abf38c8649d", "size": 298, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "docs/programs/saveTable.jl", "max_stars_repo_name": "kylebarron/tablefill", "max_stars_repo_head_hexsha": "5f56a54c63939e03538d7530f5a64ede2b2455be", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 8, "max_stars_repo_stars_event_min_datetime": "2017-12-25T20:07:26.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-21T01:28:00.000Z", "max_issues_repo_path": "docs/programs/saveTable.jl", "max_issues_repo_name": "kylebarron/tablefill", "max_issues_repo_head_hexsha": "5f56a54c63939e03538d7530f5a64ede2b2455be", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 8, "max_issues_repo_issues_event_min_datetime": "2018-07-05T21:52:46.000Z", "max_issues_repo_issues_event_max_datetime": "2021-10-23T23:56:36.000Z", "max_forks_repo_path": "docs/programs/saveTable.jl", "max_forks_repo_name": "kylebarron/tablefill", "max_forks_repo_head_hexsha": "5f56a54c63939e03538d7530f5a64ede2b2455be", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 2, "max_forks_repo_forks_event_min_datetime": "2017-08-06T16:29:52.000Z", "max_forks_repo_forks_event_max_datetime": "2020-04-08T22:00:29.000Z", "avg_line_length": 24.8333333333, "max_line_length": 59, "alphanum_fraction": 0.610738255, "num_tokens": 96, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4726834766204329, "lm_q2_score": 0.12085322140796939, "lm_q1q2_score": 0.0571253208558979}}
{"text": "\"\"\"\nThis is a Julia translation of the schema expected by typescript.\n\nCalling [`to_json`](@ref) on a SemagramSchema should give a schema that\ntypescript can understand.\n\nThere is also a macro here for creating a SemagramSchema: [`@semagramschema`](@ref),\nwhich makes writing down a schema much more convenient.\n\"\"\"\nmodule Schema\n\nexport AttributeType, Numeric, Stringlike,\n BoxProperties, PortProperties, WireProperties,\n SemagramSchema, BoxDesc, PortDesc, WireDesc, DataDesc,\n @semagramschema\n \nimport ..Muesli: to_json, from_json\nusing ..Muesli\nusing ..SVG\nusing Catlab.Present, Catlab.Theories\nusing MLStyle\n\n@enum AttributeType begin\n Numeric\n Stringlike\nend\n\nto_json(x::AttributeType) = @match x begin\n Numeric => \"Numeric\"\n Stringlike => \"Stringlike\"\nend\n\nfunction from_json(d::Any, ::Type{AttributeType})\n @match d begin\n \"Numeric\" => Numeric\n \"Stringlike\" => Stringlike\n end\nend\n\nstruct BoxProperties\n weights::Vector{Tuple{AttributeType, Symbol}}\n shape::String\n label::Union{Symbol,Nothing}\nend\n\nto_json(x::BoxProperties) = generic_to_json(x)\nfrom_json(d::Dict{String,<:Any}, ::Type{BoxProperties}) = generic_from_json(d, BoxProperties)\n\nstruct PortProperties\n weights::Vector{Tuple{AttributeType, Symbol}}\n box::Symbol\n box_map::Symbol\n style::String\nend\n\nto_json(x::PortProperties) = generic_to_json(x)\nfrom_json(d::Dict{String,<:Any}, ::Type{PortProperties}) = generic_from_json(d, PortProperties)\n\nstruct WireProperties\n weights::Vector{Tuple{AttributeType, Symbol}}\n src::Tuple{String, Symbol}\n src_map::Symbol\n tgt::Tuple{String, Symbol}\n tgt_map::Symbol\n style::String\nend\n\nto_json(x::WireProperties) = generic_to_json(x)\nfrom_json(d::Dict{String,<:Any}, ::Type{WireProperties}) = generic_from_json(d, WireProperties)\n\nstruct SemagramSchema\n box_types::Dict{Symbol, BoxProperties}\n port_types::Dict{Symbol, PortProperties}\n wire_types::Dict{Symbol, WireProperties}\nend\n\nto_json(x::SemagramSchema) = generic_to_json(x)\nfrom_json(d::Dict{String, <:Any}, ::Type{SemagramSchema}) = generic_from_json(d, SemagramSchema)\n\nstruct BoxDesc\n name::Symbol\n shape::SVGNode\n label::Union{Symbol,Nothing}\n function BoxDesc(name,shape=Circle,label=nothing)\n new(name,shape, label)\n end\nend\n\nstruct PortDesc\n name::Symbol\n box_map::Symbol\n style::String\n function PortDesc(name,box_map,style=\"Circular\")\n new(name,box_map,style)\n end\nend\n\nstruct WireDesc\n name::Symbol\n src_map::Symbol\n tgt_map::Symbol\n style::String\n function WireDesc(name,src_map,tgt_map,style=\"DefaultWire\")\n new(name,src_map,tgt_map,style)\n end\nend\n\nstruct DataDesc\n name::Symbol\n type::AttributeType\nend\n\nconst EntityDesc = Union{BoxDesc, PortDesc, WireDesc, DataDesc}\n\nfunction pres_to_semagramschema(p::Presentation, descs::Array)\n ws = SemagramSchema(Dict(),Dict(),Dict())\n datadescs = filter(desc -> typeof(desc) <: DataDesc, descs)\n datatypes = Dict(map(desc -> desc.name => desc.type, datadescs)...)\n function weights(ob::Symbol)\n attrs = filter(attr -> nameof(dom(attr)) == ob, p.generators[:Attr])\n map(attr -> (get(datatypes, nameof(codom(attr)), Stringlike), nameof(attr)), attrs)\n end\n for desc in descs\n ob = desc.name\n if typeof(desc)<:BoxDesc\n ws.box_types[ob] = BoxProperties(weights(ob),write_svg(desc.shape),desc.label)\n elseif typeof(desc)<:PortDesc\n box = nameof(codom(p[desc.box_map]))\n ws.port_types[ob] = PortProperties(weights(ob),box,desc.box_map,desc.style)\n elseif typeof(desc)<:WireDesc\n src = nameof(codom(p[desc.src_map]))\n src_type = if src \u2208 keys(ws.box_types)\n \"Box\"\n else\n \"Port\"\n end\n tgt = nameof(codom(p[desc.tgt_map]))\n tgt_type = if src \u2208 keys(ws.box_types)\n \"Box\"\n else\n \"Port\"\n end\n ws.wire_types[ob] = WireProperties(\n weights(ob),\n (src_type, src), desc.src_map,\n (tgt_type, tgt), desc.tgt_map,\n desc.style\n )\n end\n end\n ws\nend\n\nq(x) = Expr(:quote,x)\n\n\"\"\"\nSee the examples/general documentation for how to use this.\n\"\"\"\nmacro semagramschema(head,body)\n descs = @match body begin\n Expr(:block,lines...) => begin\n map(lines) do line\n @match line begin\n Expr(:macrocall, mname, _, desc, args...) => begin\n (name,morphs) = @match desc begin\n name::Symbol => (name,[])\n Expr(:call, name, args...) => (name,args)\n end\n constructor = if mname == Symbol(\"@box\")\n BoxDesc\n elseif mname == Symbol(\"@port\")\n PortDesc\n elseif mname == Symbol(\"@wire\")\n WireDesc\n elseif mname == Symbol(\"@data\")\n DataDesc\n end\n Expr(:call, constructor, q(name), q.(morphs)..., args...)\n end\n _ => missing\n end\n end\n end\n _ => error(\"the body must be a block\")\n end\n descs = filter(x -> !ismissing(x), descs)\n name, pres = @match head begin\n Expr(:call, name, pres) => (name, pres)\n _ => error(\"the head must be a name and presentation\")\n end\n :($(esc(name)) = pres_to_semagramschema($(esc(pres)), $(esc(Expr(:vect, descs...)))))\nend\n\nend\n", "meta": {"hexsha": "dae47028dfc468b21d246d71f843595c48dd1a82", "size": 5172, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/Schema.jl", "max_stars_repo_name": "bosonbaas/Semagrams.jl", "max_stars_repo_head_hexsha": "8e2cab4548650ad26bda44290843276927cca14d", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/Schema.jl", "max_issues_repo_name": "bosonbaas/Semagrams.jl", "max_issues_repo_head_hexsha": "8e2cab4548650ad26bda44290843276927cca14d", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/Schema.jl", "max_forks_repo_name": "bosonbaas/Semagrams.jl", "max_forks_repo_head_hexsha": "8e2cab4548650ad26bda44290843276927cca14d", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 26.5230769231, "max_line_length": 96, "alphanum_fraction": 0.6664733179, "num_tokens": 1405, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.49218813572079556, "lm_q2_score": 0.11596072894699293, "lm_q1q2_score": 0.05707449499724494}}
{"text": "using Documenter, NumericalAnalysis\n\n\nmakedocs(\n modules = [NumericalAnalysis],\n clean = false,\n sitename = \"NumericalAnalysis.jl\",\n pages = Any[\n \"Home\" => \"index.md\",\n \"Functions\" => Any[\n \"Basic function\" => \"m/basic.md\",\n \"SEq in one variable\" => \"m/SEq1.md\",\n \"Interpolation and the Lagrange Polynomial\" => \"m/Polynomial.md\",\n \"Numerical Differentiation and Integration\" => \"m/NCalculus.md\",\n ],\n ],\n)\n", "meta": {"hexsha": "1e19ab3095ca2858acb69e1f3184ab073dfb89cd", "size": 487, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "docs/make.jl", "max_stars_repo_name": "ZhouZhuofei/NumericalAnalysis.jl", "max_stars_repo_head_hexsha": "1e4926d6968fa72cc6ba102ad04052a77044351a", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "docs/make.jl", "max_issues_repo_name": "ZhouZhuofei/NumericalAnalysis.jl", "max_issues_repo_head_hexsha": "1e4926d6968fa72cc6ba102ad04052a77044351a", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 11, "max_issues_repo_issues_event_min_datetime": "2020-09-24T17:58:58.000Z", "max_issues_repo_issues_event_max_datetime": "2020-12-11T00:37:24.000Z", "max_forks_repo_path": "docs/make.jl", "max_forks_repo_name": "ZhouZhuofei/NumericalAnalysis.jl", "max_forks_repo_head_hexsha": "1e4926d6968fa72cc6ba102ad04052a77044351a", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 27.0555555556, "max_line_length": 77, "alphanum_fraction": 0.5667351129, "num_tokens": 125, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.49218813572079556, "lm_q2_score": 0.11596072436426731, "lm_q1q2_score": 0.05707449274168176}}
{"text": "#######################################################################################################################################################################################################\n#\n# Changes to the function\n# General\n# 2021-Dec-24: migrate the function from PkgUtility to NetcdfIO\n# 2022-Jan-28: remove the complicated funtion to create var and dim at the same time\n# 2022-Jan-28: add global attributes to the generated file\n#\n#######################################################################################################################################################################################################\n\"\"\"\nNCDatasets.jl does not have a convenient function (1 line command) to save dataset as a file. Thus, we provide a few methods as supplements:\n\n$(METHODLIST)\n\n\"\"\"\nfunction save_nc! end\n\n\n\"\"\"\nTo save the code and effort to redefine the common attributes like latitude, longitude, and cycle index, we provide a shortcut method that handles these within the function:\n\n save_nc!(file::String, var_name::String, var_data::Array{T,N}, var_attribute::Dict{String,String}; compress::Int = 4, growable::Bool = false) where {T<:Union{AbstractFloat,Int,String},N}\n\nSave the 1D, 2D, or 3D data as netcdf file, given\n- `file` Path to save the dataset\n- `var_name` Variable name for the data in the NC file\n- `var_data` Data to save\n- `var_attribute` Variable attributes for the data, such as unit and long name\n- `compress` Compression level fro NetCDF, default is 4\n- `growable` If true, make index growable, default is false\n\n---\n# Examples\n```julia\n# generate data to write into NC file\ndata1 = rand(12) .+ 273.15;\ndata2 = rand(36,18) .+ 273.15;\ndata3 = rand(36,18,12) .+ 273.15;\n\n# save data as NC files (2D and 3D)\nsave_nc!(\"data1.nc\", \"data1\", data1, Dict(\"description\" => \"Random temperature\", \"unit\" => \"K\"));\nsave_nc!(\"data2.nc\", \"data2\", data2, Dict(\"description\" => \"Random temperature\", \"unit\" => \"K\"));\nsave_nc!(\"data3.nc\", \"data3\", data3, Dict(\"description\" => \"Random temperature\", \"unit\" => \"K\"));\n```\n\"\"\"\nsave_nc!(file::String, var_name::String, var_data::Array{T,N}, var_attribute::Dict{String,String}; compress::Int = 4, growable::Bool = false) where {T<:Union{AbstractFloat,Int,String},N} = (\n @assert 1 <= N <= 3 \"Variable must be a 1D, 2D, or 3D dataset!\";\n @assert 0 <= compress <= 9 \"Compression rate must be within 0 to 9\";\n\n # create the file\n _dset = Dataset(file, \"c\");\n\n # global title attribute\n for (_title,_notes) in ATTR_ABOUT\n _dset.attrib[_title] = _notes;\n end;\n\n # the case if the dimension is 1D\n if N==1\n _n_ind = (growable ? Inf : length(var_data));\n _inds = collect(eachindex(var_data));\n add_nc_dim!(_dset, \"ind\", _n_ind);\n append_nc!(_dset, \"ind\", _inds, ATTR_CYC, [\"ind\"]; compress=compress);\n append_nc!(_dset, var_name, var_data, var_attribute, [\"ind\"]; compress=compress);\n\n close(_dset);\n\n return nothing\n end;\n\n # if the dimension is 2D or 3D\n _n_lon = size(var_data, 1);\n _n_lat = size(var_data, 2);\n _res_lon = 360 / _n_lon;\n _res_lat = 180 / _n_lat;\n _lons = collect(_res_lon/2:_res_lon:360) .- 180;\n _lats = collect(_res_lat/2:_res_lat:180) .- 90;\n add_nc_dim!(_dset, \"lon\", _n_lon);\n add_nc_dim!(_dset, \"lat\", _n_lat);\n append_nc!(_dset, \"lon\", _lons, ATTR_LON, [\"lon\"]; compress=compress);\n append_nc!(_dset, \"lat\", _lats, ATTR_LAT, [\"lat\"]; compress=compress);\n\n if N==2\n append_nc!(_dset, var_name, var_data, var_attribute, [\"lon\", \"lat\"]; compress=compress);\n elseif N==3\n _n_ind = (growable ? Inf : size(var_data,3));\n _inds = collect(1:_n_ind);\n add_nc_dim!(_dset, \"ind\", _n_ind);\n append_nc!(_dset, \"ind\", _inds, ATTR_CYC, [\"ind\"]; compress=compress);\n append_nc!(_dset, var_name, var_data, var_attribute, [\"lon\", \"lat\", \"ind\"]; compress=compress);\n end;\n\n close(_dset);\n\n return nothing\n);\n\n\n\"\"\"\nThis method saves DataFrame as a NetCDF file to save more space (compared to a CSV file).\n\n save_nc!(file::String, df::DataFrame, var_names::Vector{String}, var_attributes::Vector{Dict{String,String}}; compress::Int = 4, growable::Bool = false)\n\nSave DataFrame to NetCDF, given\n- `file` Path to save the data\n- `df` DataFrame to save\n- `var_names` The label of data in DataFrame to save\n- `var_attributes` Variable attributes for the data to save\n- `compress` Compression level fro NetCDF, default is 4\n- `growable` If true, make index growable, default is false\n\n---\n# Examples\n```julia\ndf = DataFrame();\ndf[!,\"A\"] = rand(5);\ndf[!,\"B\"] = rand(5);\ndf[!,\"C\"] = rand(5);\nsave_nc!(\"dataf.nc\", df, [\"A\",\"B\"], [Dict(\"A\" => \"Attribute A\"), Dict(\"B\" => \"Attribute B\")]);\n```\n\"\"\"\nsave_nc!(file::String, df::DataFrame, var_names::Vector{String}, var_attributes::Vector{Dict{String,String}}; compress::Int = 4, growable::Bool = false) = (\n @assert 0 <= compress <= 9 \"Compression rate must be within 0 to 9\";\n @assert length(var_names) == length(var_attributes) \"Variable name and attributes lengths must match!\";\n\n # create the file\n _dset = Dataset(file, \"c\");\n\n # global title attribute\n for (_title,_notes) in ATTR_ABOUT\n _dset.attrib[_title] = _notes;\n end;\n\n # define dimension related variables\n _n_ind = (growable ? Inf : size(df)[1]);\n _inds = collect(1:_n_ind);\n\n # save the variables\n add_nc_dim!(_dset, \"ind\", _n_ind);\n append_nc!(_dset, \"ind\", _inds, ATTR_CYC, [\"ind\"]; compress=compress);\n for _i in eachindex(var_names)\n append_nc!(_dset, var_names[_i], df[:, var_names[_i]], var_attributes[_i], [\"ind\"]; compress = compress);\n end;\n\n close(_dset);\n\n return nothing\n);\n\n\n\"\"\"\nThis method is a simplified version of the method above, namely when users do not want to define the attributes.\n\n save_nc!(file::String, df::DataFrame; compress::Int = 4, growable::Bool = false)\n\nSave DataFrame to NetCDF, given\n- `file` Path to save the data\n- `df` DataFrame to save\n- `notes` Global attributes (notes)\n- `compress` Compression level fro NetCDF, default is 4\n- `growable` If true, make index growable, default is false\n\n---\n# Examples\n```julia\ndf = DataFrame();\ndf[!,\"A\"] = rand(5);\ndf[!,\"B\"] = rand(5);\ndf[!,\"C\"] = rand(5);\nsave_nc!(\"test.nc\", df);\n```\n\"\"\"\nsave_nc!(file::String, df::DataFrame; compress::Int = 4, growable::Bool = false) = (\n _var_names = names(df);\n _var_attrs = [Dict{String,String}(_vn => _vn) for _vn in _var_names];\n\n save_nc!(file, df, _var_names, _var_attrs; compress=compress, growable = growable);\n\n return nothing\n);\n", "meta": {"hexsha": "d0bb9a8f4769a616395d5e8243c066024c070178", "size": 6626, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/save.jl", "max_stars_repo_name": "Yujie-W/NetcdfIO.jl", "max_stars_repo_head_hexsha": "015b1b8bd26739721a0c6a0fb0fd9dad9b8ea8ca", "max_stars_repo_licenses": ["Apache-2.0"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2021-12-30T22:36:35.000Z", "max_stars_repo_stars_event_max_datetime": "2021-12-30T22:36:35.000Z", "max_issues_repo_path": "src/save.jl", "max_issues_repo_name": "Yujie-W/NetcdfIO.jl", "max_issues_repo_head_hexsha": "015b1b8bd26739721a0c6a0fb0fd9dad9b8ea8ca", "max_issues_repo_licenses": ["Apache-2.0"], "max_issues_count": 10, "max_issues_repo_issues_event_min_datetime": "2021-12-29T06:59:22.000Z", "max_issues_repo_issues_event_max_datetime": "2022-03-12T00:59:43.000Z", "max_forks_repo_path": "src/save.jl", "max_forks_repo_name": "Yujie-W/NetcdfIO.jl", "max_forks_repo_head_hexsha": "015b1b8bd26739721a0c6a0fb0fd9dad9b8ea8ca", "max_forks_repo_licenses": ["Apache-2.0"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 36.6077348066, "max_line_length": 199, "alphanum_fraction": 0.6193782071, "num_tokens": 1894, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4921881357207956, "lm_q2_score": 0.11596071978154188, "lm_q1q2_score": 0.057074490486118686}}
{"text": "module WeaveSupport\n## Modified from\n## https://github.com/SciML/SciMLTutorials.jl/blob/master/src/SciMLTutorials.jl\n\nimport Base64: base64encode\nusing Random\n\nusing Weave\n\nusing Mustache\nimport Markdown\nusing JSON\nusing Reexport\n@reexport using LaTeXStrings\n#using SymPy\n#function Base.show(io::IO, ::MIME\"text/latex\", x::SymPy.SymbolicObject)\n# print(io, SymPy.as_markdown(sympy.latex(x)))\n#end\n\n\nusing Pkg\n\ninclude(\"formatting.jl\")\ninclude(\"bootstrap.jl\")\ninclude(\"questions.jl\")\ninclude(\"show-methods.jl\")\ninclude(\"toc.jl\")\n\n#import Plots\n# just show body, not standalone\n#function Plots._show(io::IO, ::MIME\"text/html\", plt::Plots.Plot{Plots.PlotlyBackend})\n# write(io, Plots.html_body(plt))\n#end\n\n\n## we have jmd files that convert to html files\n## using a specialized template\n\nconst repo_directory = joinpath(@__DIR__,\"..\", \"..\")\nconst cssfile = joinpath(@__DIR__, \"..\", \"..\", \"templates\", \"skeleton_css.css\")\nconst htmlfile = joinpath(@__DIR__,\"..\", \"..\", \"templates\", \"bootstrap.tpl\")\nconst latexfile = joinpath(@__DIR__, \"..\", \"..\", \"templates\", \"julia_tex.tpl\")\n\nfunction weave_file(folder, file; build_list=(:script,:html,:pdf,:github,:notebook), force=false, kwargs...)\n\n\n jmddir = joinpath(repo_directory,\"CwJ\",folder)\n tmp = joinpath(jmddir, file)\n bnm = replace(basename(tmp), r\".jmd$\" => \"\")\n\n\n if !force\n testfile = joinpath(repo_directory, \"html\", folder, bnm*\".html\")\n if isfile(testfile) && (mtime(testfile) >= mtime(tmp))\n return\n end\n end\n\n Pkg.activate(dirname(tmp))\n Pkg.instantiate()\n args = Dict{Symbol,String}(:folder=>folder,:file=>file)\n\n if :script \u2208 build_list\n println(\"Building Script\")\n dir = joinpath(repo_directory,\"script\",folder)\n isdir(dir) || mkpath(dir)\n args[:doctype] = \"script\"\n tangle(tmp;out_path=dir)\n end\n\n if :html \u2208 build_list\n println(\"Building HTML\")\n dir = joinpath(repo_directory,\"html\",folder)\n isdir(dir) || mkpath(dir)\n\n figdir = joinpath(jmddir,\"figures\")\n htmlfigdir = joinpath(dir, \"figures\")\n\n if isdir(figdir)\n isdir(htmlfigdir) && rm(htmlfigdir, recursive=true)\n cp(figdir, htmlfigdir)\n end\n\n Weave.set_chunk_defaults!(:wrap=>false)\n args[:doctype] = \"html\"\n #weave(tmp,doctype = \"md2html\",out_path=dir,args=args; fig_ext=\".svg\", css=cssfile, kwargs...)\n weave(tmp,doctype = \"md2html\", out_path=dir,args=args; fig_ext=\".svg\",\n template=htmlfile,\n fig_path=tempdir(),\n kwargs...)\n\n # clean up\n isdir(htmlfigdir) && rm(htmlfigdir, recursive=true)\n\n end\n\n if :pdf \u2208 build_list\n\n eval(quote using Tectonic end) # load Tectonic; wierd testing error\n\n println(\"Building PDF\")\n dir = joinpath(repo_directory,\"pdf\",folder)\n isdir(dir) || mkpath(dir)\n\n fig_path = \"_figures_\" * bnm\n figdir = joinpath(jmddir,\"figures\")\n texfigdir = joinpath(dir, \"figures\")\n\n if isdir(figdir)\n isdir(texfigdir) && rm(texfigdir, recursive=true)\n cp(figdir, texfigdir)\n end\n\n args[:doctype] = \"pdf\"\n try\n weave(tmp,doctype=\"md2tex\",out_path=dir,args=args;\n template=latexfile,\n fig_path=fig_path,\n kwargs...)\n\n texfile = joinpath(dir, bnm * \".tex\")\n Base.invokelatest(Tectonic.tectonic, bin -> run(`$bin $texfile`))\n\n\n # clean up\n for ext in (\".tex\",)\n f = joinpath(dir, bnm * ext)\n isfile(f) && rm(f)\n end\n\n catch ex\n @warn \"PDF generation failed\" exception=(ex, catch_backtrace())\n\n end\n\n isdir(texfigdir) && rm(texfigdir, recursive=true)\n isdir(joinpath(dir,fig_path)) && rm(joinpath(dir,fig_path), recursive=true)\n for ext in (\".aux\", \".log\", \".out\")\n f = joinpath(dir, bnm * ext)\n isfile(f) && rm(f)\n end\n\n end\n\n if :github \u2208 build_list\n println(\"Building Github Markdown\")\n dir = joinpath(repo_directory,\"markdown\",folder)\n isdir(dir) || mkpath(dir)\n args[:doctype] = \"github\"\n weave(tmp,doctype = \"github\",out_path=dir, args=args;\n fig_path=tempdir(),\n kwargs...)\n end\n\n if :notebook \u2208 build_list\n println(\"Building Notebook\")\n dir = joinpath(repo_directory,\"notebook\",folder)\n isdir(dir) || mkpath(dir)\n args[:doctype] = \"notebook\"\n Weave.convert_doc(tmp,joinpath(dir,file[1:end-4]*\".ipynb\"))\n end\nend\n\n\"\"\"\n weave_all(; force=false, build_list=(:script,:html,:pdf,:github,:notebook))\n\nRun `weave` on all source files.\n\n* `force`: by default, only run `weave` on files with `html` file older than the source file in `CwJ`\n* `build_list`: list of output types to be built. The default is all types\n\nThe files will be built as subdirectories in the package directory. This is returned by `pathof(CalculusWithJulia)`.\n\n\"\"\"\nfunction weave_all(;force=false, build_list=(:script,:html,:pdf,:github,:notebook))\n for folder in readdir(joinpath(repo_directory,\"CwJ\"))\n folder == \"test.jmd\" && continue\n weave_folder(folder; force=force, build_list=build_list)\n end\nend\n\nfunction weave_folder(folder; force=false, build_list=(:script,:html,:pdf,:github,:notebook))\n !isnothing(match(r\"\\.ico$\", folder)) && return nothing\n for file in readdir(joinpath(repo_directory,\"CwJ\",folder))\n !occursin(r\".jmd$\", basename(file)) && continue\n println(\"Building $(joinpath(folder,file))\")\n try\n weave_file(folder,file; force=force, build_list=build_list)\n catch\n end\n end\nend\n\n\n\n\n\nmacro q_str(x)\n \"`$x`\"\nend\n\nexport mmd\nexport @q_str\nexport ImageFile, Verbatim, Invisible, Outputonly, HTMLonly, JSXGraph\nexport alert, warning, note\nexport example, popup, table\nexport gif_to_data\nexport numericq, radioq, booleanq, yesnoq, shortq, longq, multiq\n\n\nend # module\n", "meta": {"hexsha": "b23a3e896382d7b73523b8dfe9b24f291a21d10f", "size": 6058, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/WeaveSupport/WeaveSupport.jl", "max_stars_repo_name": "jverzani/CalculusWithJulia.jl", "max_stars_repo_head_hexsha": "6ee5135e82c11a1f83b024556be55ad6cbf2622d", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 31, "max_stars_repo_stars_event_min_datetime": "2019-08-29T02:00:11.000Z", "max_stars_repo_stars_event_max_datetime": "2022-02-20T11:15:12.000Z", "max_issues_repo_path": "src/WeaveSupport/WeaveSupport.jl", "max_issues_repo_name": "jverzani/CalculusWithJulia.jl", "max_issues_repo_head_hexsha": "6ee5135e82c11a1f83b024556be55ad6cbf2622d", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 16, "max_issues_repo_issues_event_min_datetime": "2020-12-03T15:00:01.000Z", "max_issues_repo_issues_event_max_datetime": "2022-03-11T00:57:57.000Z", "max_forks_repo_path": "src/WeaveSupport/WeaveSupport.jl", "max_forks_repo_name": "jverzani/CalculusWithJulia.jl", "max_forks_repo_head_hexsha": "6ee5135e82c11a1f83b024556be55ad6cbf2622d", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 10, "max_forks_repo_forks_event_min_datetime": "2020-01-07T10:53:24.000Z", "max_forks_repo_forks_event_max_datetime": "2021-09-15T06:08:38.000Z", "avg_line_length": 28.5754716981, "max_line_length": 116, "alphanum_fraction": 0.6223175966, "num_tokens": 1617, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.37022540649291935, "lm_q2_score": 0.15405756851741473, "lm_q1q2_score": 0.057036025927670646}}
{"text": "d1 = Dict(1 => 4.2, 2 => 5.3) \n# Dict{Int64,Float64} with 2 entries:\n# 2 => 5.3\n# 1 => 4.2\nd1 = Dict{Int64,Float64}(1 => 4.2, 2 => 5.3)\nd1 = [1 => 4.2, 2 => 5.3]\n# 2-element Array{Pair{Int64,Float64},1}:\n# 1 => 4.2\n# 2 => 5.3\n\nd2 = Dict{Any,Any}(\"a\"=>1, (2,3)=>true) \n\nd3 = Dict(:A => 100, :B => 200)\n# Dict{Symbol,Int64} with 2 entries:\n# :A => 100\n# :B => 200\nd3 = Dict{Symbol,Int64}(:A => 100, :B => 200)\nd3[:B] #> 200\n# d3[:Z] #> ERROR: KeyError: key :Z not found\nget(d3, :Z, 999) #> 999\n\nd3[:A] = 150 #> d3 is now [:A => 150, :B => 200]\nd3[:C] = 300 #> d3 is now [:A => 150, :B => 200, :C => 300]\nlength(d3) #> 3\n# d3[\"CVO\"] = 500 #> ERROR: KeyError: key \"CVO\" not found \n# d3[:CVO] = \"Julia\" #> ERROR: KeyError: key \"CVO\" not found\n\ndmus = Dict{Symbol,String}(:first_name => \"Louis\", :surname => \"Armstrong\", :occupation => \"musician\", \n \t\t :date_of_birth => \"4/8/1901\")\n# Dict{Symbol,String} with 4 entries:\n# :date_of_birth => \"4/8/1901\"\n# :occupation => \"musician\"\n# :surname => \"Armstrong\"\n# :first_name => \"Louis\"\n\nhaskey(d3, :Z) #> false\nhaskey(d3, :B) #> true\n\nd4 = Dict() #> Dict{Any,Any} with 0 entries\nd4[\"lang\"] = \"Julia\"\n\nd5 = Dict{Float64, Int64}() #> Dict{Float64,Int64} with 0 entries\n# d5[\"c\"] = 6 #> ERROR: MethodError: `convert` has no method matching convert(::Type{Float64}, ::ASCIIString)\n\nd3 = Dict(:A => 100, :B => 200) \ndelete!(d3, :B) #> Dict{Symbol,Int64} with 1 entry: :A => 100\n\nd3 = Dict(:A => 100, :B => 200) \nki = keys(d3) #> KeyIterator\nfor k in ki\n println(k)\nend #> A B\n:A in ki #> true\n:Z in ki #> false\n\ncollect(keys(d3)) #> 2-element Array{Symbol,1}:\n# :A\n# :B\nvi = values(d3) #> ValueIterator\nfor v in values(d3)\n println(v)\nend #> 100 200\n\nkeys1 = [\"J.S. Bach\", \"Woody Allen\", \"Barack Obama\"]\nvalues1 = [ 1685, 1935, 1961]\nd5 = Dict(zip(keys1, values1))\n#>\n# Dict{String,Int64} with 3 entries:\n# \"J.S. Bach\" => 1685\n# \"Woody Allen\" => 1935\n# \"Barack Obama\" => 1961\n\nfor (k, v) in d5\n println(\"$k was born in $v\")\nend\nfor p in d5\n println(\"$(p[1]) was born in $(p[2])\")\nend\n#J.S. Bach was born in 1685\n#Barack Obama was born in 1961\n#Woody Allen was born in 1935\n\ncapitals = Dict{String, String}(\"France\"=> \"Paris\", \"China\"=>\"Beijing\")\n# Dict{String,String} with 2 entries:\n# \"China\" => \"Beijing\"\n# \"France\" => \"Paris\"\n\n# neat tricks:\ndict = Dict(\"a\" => 1, \"b\" => 2, \"c\" => 3) \narrkey = [key for (key, value) in dict] #> 3-element Array{String,1}: \"a\" \"b\" \"c\"\n# same as collect(keys(dict)) \narrval = [value for (key, value) in dict] #> 3-element Array{Int64,1}: 1 2 3\n# same as collect(values(dict))\n", "meta": {"hexsha": "8af62d133f6617bfa038f46f44d14085e296989c", "size": 2603, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Chapter05/dicts.jl", "max_stars_repo_name": "tavoludra1/Julia1.0", "max_stars_repo_head_hexsha": "92dc1e3955393394f10f538dceded45facd1cff6", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 26, "max_stars_repo_stars_event_min_datetime": "2018-09-29T03:07:28.000Z", "max_stars_repo_stars_event_max_datetime": "2021-11-08T13:12:33.000Z", "max_issues_repo_path": "Chapter05/dicts.jl", "max_issues_repo_name": "tavoludra1/Julia1.0", "max_issues_repo_head_hexsha": "92dc1e3955393394f10f538dceded45facd1cff6", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Chapter05/dicts.jl", "max_forks_repo_name": "tavoludra1/Julia1.0", "max_forks_repo_head_hexsha": "92dc1e3955393394f10f538dceded45facd1cff6", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 11, "max_forks_repo_forks_event_min_datetime": "2018-09-16T05:55:20.000Z", "max_forks_repo_forks_event_max_datetime": "2021-06-08T14:59:54.000Z", "avg_line_length": 27.6914893617, "max_line_length": 109, "alphanum_fraction": 0.5747214752, "num_tokens": 1106, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.3849121444839335, "lm_q2_score": 0.1480472055495773, "lm_q1q2_score": 0.05698516737294149}}
{"text": "using Oceananigans.Models: AbstractModel\nusing Oceananigans.Utils: prettykeys\n\nmutable struct NaNChecker{F}\n fields :: F\n erroring :: Bool\nend\n\nNaNChecker(fields) = NaNChecker(fields, false) # default\n\nfunction Base.summary(nc::NaNChecker)\n fieldnames = prettykeys(nc.fields)\n if nc.erroring\n return \"Erroring NaNChecker for $fieldnames\"\n else\n return \"NaNChecker for $fieldnames\"\n end\nend\n\nBase.show(io, nc::NaNChecker) = print(io, summary(nc))\n\n\"\"\"\n NaNChecker(; fields, erroring=false)\n\nReturn a `NaNChecker`, which sets `sim.running=false` if a `NaN` is detected\nin any member of `fields` when `NaNChecker(sim)` is called. `fields` should be\na container with key-value pairs like a dictionary or `NamedTuple`.\n\nIf `erroring=true`, the `NaNChecker` will throw an error on NaN detection.\n\"\"\"\nNaNChecker(; fields, erroring=false) = NaNChecker(fields, erroring)\n\nhasnan(field) = any(isnan, parent(field)) \nhasnan(model::AbstractModel) = hasnan(first(fields(model)))\n\nfunction (nc::NaNChecker)(simulation)\n for (name, field) in pairs(nc.fields)\n if hasnan(field)\n simulation.running = false\n\n if nc.erroring\n clock = simulation.model.clock\n error(\"time = $(clock.time), iteration = $(clock.iteration): NaN found in field $name. Aborting simulation.\")\n else\n @info \"NaN found in field $name. Stopping simulation.\"\n end\n end\n end\n return nothing\nend\n\n\"\"\"\n erroring_NaNChecker!(simulation)\n\nToggles `simulation`'s `NaNChecker` to throw an error when a `NaN` is detected.\n\"\"\"\nfunction erroring_NaNChecker!(simulation)\n simulation.callbacks[:nan_checker].func.erroring = true\n return nothing\nend\n\n", "meta": {"hexsha": "fb120a7cf19eeb7b5fdefde285a649632a04d818", "size": 1747, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/Simulations/nan_checker.jl", "max_stars_repo_name": "johnryantaylor/Oceananigans.jl", "max_stars_repo_head_hexsha": "b769cc4f42f2d531133fb6359f197633efc975c7", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 239, "max_stars_repo_stars_event_min_datetime": "2019-03-05T03:46:44.000Z", "max_stars_repo_stars_event_max_datetime": "2020-05-04T21:53:14.000Z", "max_issues_repo_path": "src/Simulations/nan_checker.jl", "max_issues_repo_name": "johnryantaylor/Oceananigans.jl", "max_issues_repo_head_hexsha": "b769cc4f42f2d531133fb6359f197633efc975c7", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 654, "max_issues_repo_issues_event_min_datetime": "2019-03-02T02:20:29.000Z", "max_issues_repo_issues_event_max_datetime": "2020-05-02T00:13:53.000Z", "max_forks_repo_path": "src/Simulations/nan_checker.jl", "max_forks_repo_name": "ali-ramadhan/OceanDispatch.jl", "max_forks_repo_head_hexsha": "65b8851d37052e90ca4a3e0c4a1c20398b0ee09a", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 45, "max_forks_repo_forks_event_min_datetime": "2019-03-05T18:25:16.000Z", "max_forks_repo_forks_event_max_datetime": "2020-05-04T08:04:25.000Z", "avg_line_length": 28.1774193548, "max_line_length": 125, "alphanum_fraction": 0.6823125358, "num_tokens": 421, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4649015713733885, "lm_q2_score": 0.12252322533450248, "lm_q1q2_score": 0.056961239987745965}}
{"text": "LATEXIFY(x) = md\"$(latexify(x))\"\n\nLATEXIFY(s::AbstractString, x) = md\"$(s) $(latexify(x))\"\n\nL = LATEXIFY\n\nexport LATEXIFY, L", "meta": {"hexsha": "0c2f71ddb91277b1f5f582368050e88a0e5f86a1", "size": 124, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/pluto_helpers.jl", "max_stars_repo_name": "PtFEM/NumericalMethodsforEngineers.jl", "max_stars_repo_head_hexsha": "e4a997a14adbb86b7efe1586962df39eb9285ebb", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 5, "max_stars_repo_stars_event_min_datetime": "2018-07-23T18:12:52.000Z", "max_stars_repo_stars_event_max_datetime": "2020-11-25T03:32:45.000Z", "max_issues_repo_path": "src/pluto_helpers.jl", "max_issues_repo_name": "PtFEM/NumericalMethodsforEngineers.jl", "max_issues_repo_head_hexsha": "e4a997a14adbb86b7efe1586962df39eb9285ebb", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 8, "max_issues_repo_issues_event_min_datetime": "2018-07-23T21:46:36.000Z", "max_issues_repo_issues_event_max_datetime": "2021-04-27T23:14:46.000Z", "max_forks_repo_path": "src/pluto_helpers.jl", "max_forks_repo_name": "PtFEM/NumericalMethodsforEngineers.jl", "max_forks_repo_head_hexsha": "e4a997a14adbb86b7efe1586962df39eb9285ebb", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 5, "max_forks_repo_forks_event_min_datetime": "2018-10-27T14:13:34.000Z", "max_forks_repo_forks_event_max_datetime": "2021-11-20T18:54:06.000Z", "avg_line_length": 17.7142857143, "max_line_length": 56, "alphanum_fraction": 0.6451612903, "num_tokens": 52, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4649015713733885, "lm_q2_score": 0.12252320450794522, "lm_q1q2_score": 0.056961230305446774}}
{"text": "### A Pluto.jl notebook ###\n# v0.17.2\n\nusing Markdown\nusing InteractiveUtils\n\n# \u2554\u2550\u2561 227de887-4284-46fb-83e2-22d33830c9cf\nusing LaTeXStrings\n\n# \u2554\u2550\u2561 1eb8d032-dbeb-4544-8bcd-e512f19d6cd5\n\u03b8 = round(1/3, digits=2)\n\n# \u2554\u2550\u2561 ec80fb74-5aa2-11ec-0e31-bb7b0d8fe4f7\nmd\"\"\"\n``\\theta`` = $(round (\u03b8, digits = 2))\n\"\"\"\n\n# \u2554\u2550\u2561 76f42d5d-7b83-4d11-a52b-261ad8b55567\nmd\"\"\"\n``\\theta`` = $\u03b8\n\"\"\"\n\n# \u2554\u2550\u2561 18fbc708-96ea-4a2a-b631-442295d59b86\nmd\"\"\"\n\u03b8 = $(round (\u03b8, digits = 2))\n\"\"\"\n\n# \u2554\u2550\u2561 147eb4f3-bbf7-49d4-b7db-2c017878ff21\n\u03b8\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000001\nPLUTO_PROJECT_TOML_CONTENTS = \"\"\"\n[deps]\nLaTeXStrings = \"b964fa9f-0449-5b57-a5c2-d3ea65f4040f\"\n\n[compat]\nLaTeXStrings = \"~1.3.0\"\n\"\"\"\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000002\nPLUTO_MANIFEST_TOML_CONTENTS = \"\"\"\n# This file is machine-generated - editing it directly is not advised\n\njulia_version = \"1.7.0\"\nmanifest_format = \"2.0\"\n\n[[deps.LaTeXStrings]]\ngit-tree-sha1 = \"f2355693d6778a178ade15952b7ac47a4ff97996\"\nuuid = \"b964fa9f-0449-5b57-a5c2-d3ea65f4040f\"\nversion = \"1.3.0\"\n\"\"\"\n\n# \u2554\u2550\u2561 Cell order:\n# \u2560\u2550227de887-4284-46fb-83e2-22d33830c9cf\n# \u2560\u25501eb8d032-dbeb-4544-8bcd-e512f19d6cd5\n# \u2560\u2550ec80fb74-5aa2-11ec-0e31-bb7b0d8fe4f7\n# \u2560\u255076f42d5d-7b83-4d11-a52b-261ad8b55567\n# \u255f\u250018fbc708-96ea-4a2a-b631-442295d59b86\n# \u2560\u2550147eb4f3-bbf7-49d4-b7db-2c017878ff21\n# \u255f\u250000000000-0000-0000-0000-000000000001\n# \u255f\u250000000000-0000-0000-0000-000000000002\n", "meta": {"hexsha": "fd6c3886c1d882deee8730a6497559329e7ef409", "size": 1372, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "notebooks/intros/latex.jl", "max_stars_repo_name": "StatisticalRethinkingJulia/SR2StanPluto.jl", "max_stars_repo_head_hexsha": "6eea864f4ca098320d7bc295c6254304c86d7d19", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 2, "max_stars_repo_stars_event_min_datetime": "2021-12-17T01:36:29.000Z", "max_stars_repo_stars_event_max_datetime": "2021-12-20T00:23:37.000Z", "max_issues_repo_path": "notebooks/intros/latex.jl", "max_issues_repo_name": "StatisticalRethinkingJulia/SR2StanPluto.jl", "max_issues_repo_head_hexsha": "6eea864f4ca098320d7bc295c6254304c86d7d19", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "notebooks/intros/latex.jl", "max_forks_repo_name": "StatisticalRethinkingJulia/SR2StanPluto.jl", "max_forks_repo_head_hexsha": "6eea864f4ca098320d7bc295c6254304c86d7d19", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2022-02-21T21:01:50.000Z", "max_forks_repo_forks_event_max_datetime": "2022-02-21T21:01:50.000Z", "avg_line_length": 22.1290322581, "max_line_length": 69, "alphanum_fraction": 0.7128279883, "num_tokens": 710, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.48438006939036565, "lm_q2_score": 0.11757213354550883, "lm_q1q2_score": 0.056949598205146904}}
{"text": "\"\"\"\n TestPackage.jl\n\nHallo!\n\"\"\"\nmodule TestPackage\n\nexport plusTwo\n\n\"\"\"\n plusTwo(x)\n\nSum the numeric \"2\" to whatever it receives as input\n\nA more detailed explanation can go here, although I guess it is not needed in this case\n\n# Arguments\n* `x`: The amount to which we want to add 2\n\n# Notes\n* Notes can go here\n\n# Examples\n```julia\njulia> five = plusTwo(3)\n5\n```\n\"\"\"\nplusTwo(x) = x + 2\n\nend\n", "meta": {"hexsha": "34ea6e8cc76f32786ec4be5b8ce8892e45baff6e", "size": 399, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/TestPackage.jl", "max_stars_repo_name": "matthiasbaitsch/TestPackage.jl", "max_stars_repo_head_hexsha": "e6ac91e1f60e18715a758f7d2eb2e256a90cdaee", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/TestPackage.jl", "max_issues_repo_name": "matthiasbaitsch/TestPackage.jl", "max_issues_repo_head_hexsha": "e6ac91e1f60e18715a758f7d2eb2e256a90cdaee", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/TestPackage.jl", "max_forks_repo_name": "matthiasbaitsch/TestPackage.jl", "max_forks_repo_head_hexsha": "e6ac91e1f60e18715a758f7d2eb2e256a90cdaee", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 12.46875, "max_line_length": 87, "alphanum_fraction": 0.671679198, "num_tokens": 117, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.42250463481418826, "lm_q2_score": 0.134775927848826, "lm_q1q2_score": 0.056943454177511614}}
{"text": "\"\"\"\n\nConstruct a csv file containing data for all instances\n\"\"\"\n\n@everywhere ROOT = pwd()\n@everywhere include(joinpath(ROOT,\"..\",\"src_PowSysMod\", \"PowSysMod_body.jl\"))\n@everywhere include(\"para_fct.jl\"))\n\n\n# NOTE: Launch in console using 'julia -p nbparathreads compute_GOC_all_scenario.jl'\n# where the nb of cores is likely a good value for nbparathreads\n\ninstances_path = joinpath(\"instances\", \"GOC\")\nfolders = readdir(instances_path)\n\n##empty dictionaries which will contain data to export in csv\ninstances_nb_scenario_data = Dict{String,Int64}()\ninstances_nb_contingency_data = Dict{String,Int64}()\ninstances_nb_bus_data = Dict{String,Int64}()\ninstances_nb_links_data = Dict{String,Int64}()\ninstances_nb_violated_constraints = Dict{String, Float64}()\ninstances_nb_cc_Qgen_active = Dict{String, Float64}()\n\n\n##loop on instances to get data\nepsilon = 1e-3\nfor folder in folders\n instances_nb_scenario_data[folder] = get_nb_scenarios(instances_path,folder)\n scenario = readdir(joinpath(instances_path, folder))[1]\n instances_nb_contingency_data[folder] = get_nb_contingencies(folder,scenario)\n instances_nb_bus_data[folder] = get_nb_bus(folder, scenario)\n instances_nb_links_data[folder] = get_nb_links(folder,scenario)\n instances_nb_cc_Qgen_active[folder] = mean(get_nb_cc_reactive_power_active(folder, scenario, epsilon) for scenario in readdir(joinpath(instances_path, folder)) if scenario !=\"scorepara.csv\")\n #instances_nb_violated_constraints[folder] = mean(get_feasibility(folder, scenario, epsilon) for scenario in readdir(joinpath(instances_path, folder)))\nend\n##create csv file\ntouch(\"GOCdata.csv\")\nf = open(\"GOCdata.csv\", \"w\")\nwrite(f, \"Instance;Nb_scenarios;Nb_contingencies;Nb_bus;Nb_links;Mean nb of violated constraints by GOC solution at $(epsilon);Mean nb of reactive Qgen contingency constraints at $(epsilon)\\n\")\nfor (folder, nb_scenarios) in instances_nb_scenario_data\n nb_cont = instances_nb_contingency_data[folder]\n nb_bus = instances_nb_bus_data[folder]\n nb_links = instances_nb_links_data[folder]\n # nb_vc = instances_nb_violated_constraints[folder]\n nb_vc = \"NONE\"\n nb_cc_act = instances_nb_cc_Qgen_active[folder]\n write(f, \"$(folder);$(nb_scenarios);$(nb_cont);$(nb_bus);$(nb_links);$(nb_vc);$(nb_cc_act)\\n\")\nend\nclose(f)\n\n\n\n##functions to get specific data\n\"\"\"\n get_nb_scenarios(instances_path,folder)\n\nReturns the number of scenarios for an instance `folder` in `instances_path`.\n\"\"\"\nfunction get_nb_scenarios(instances_path,folder)\n folder_path = joinpath(instances_path, folder)\n scenarios = sort(filter(x->!ismatch(r\"\\.\", x), readdir(folder_path)), by=x->parse(split(x, \"_\")[2]))\n nb_scenarios = length(scenarios)\n return nb_scenarios\nend\n\n\"\"\"\n get_nb_bus(folder, scenario)\n\nReturns the number of bus for a `scenario` in instance `folder`.\n\"\"\"\nfunction get_nb_bus(folder, scenario)\n power_data = getpowerdata([folder, scenario])\n try\n nb_bus = Int(power_data[\"totalbus\"])\n return nb_bus\n catch\n warn(\"power_data coming from .mat file does not have key totalbus\")\n return 0.0\n end\nend\n\n\"\"\"\n get_nb_contingencies(folder,scenario)\n\nReturns the number of contingencies for a `scenario` in instance `folder`.\n\"\"\"\nfunction get_nb_contingencies(folder,scenario)\n contingency_data = getcontingencydata([folder, scenario])\n nb_contingency = size(contingency_data,1)\n return nb_contingency\nend\n\n\"\"\"\n get_nb_links(folder, scenario)\n\nReturns the number of links for a `scenario` in instance `folder`.\n\"\"\"\nfunction get_nb_links(folder, scenario)\n power_data = getpowerdata([folder, scenario])\n try\n nb_links = Int(power_data[\"totalbranch\"])\n return nb_links\n catch\n warn(\"power_data coming from .mat file does not have key totalbranch\")\n return 0.0\n end\nend\n\n\"\"\"\n get_feasibility(folder, scenario, epsilon)\n\nReturns the number of violated constraints at `epsilon` (except balance constraints) for a `scenario` in instance `folder`.\n\"\"\"\nfunction get_feasibility(folder, scenario, epsilon)\n try\n violations = test_feasibility_GOC([folder, scenario],epsilon)\n nb_violated_constraints = 0\n for (scenario, dict_infeas) in violations\n nb_violated_constraints += length(dict_infeas)\n end\n return nb_violated_constraints\n\n catch\n return -1.0\n end\nend\n\n\n\"\"\"\n get_nb_cc_reactive_power_active(folder, scenario,epsilon)\n\nReturns the number of active constraints of type contingency Qgen coupling constraints at `epsilon` for a `scenario` in instance `folder`.\n\"\"\"\nfunction get_nb_cc_reactive_power_active(folder, scenario,epsilon)\n try\n return nb_active_constraints_in_scenario_cc_Qgen([folder, scenario], epsilon)\n catch\n println([folder, scenario])\n return -1.0\n end\nend\n", "meta": {"hexsha": "278c30b7b5a80e1d11f0079cd3c3b813147c1cbe", "size": 4835, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "dev/collect_data_instances.jl", "max_stars_repo_name": "Gimaju/GOC", "max_stars_repo_head_hexsha": "daaee3ff9a0c8d21a739597ce0a7172213f43924", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "dev/collect_data_instances.jl", "max_issues_repo_name": "Gimaju/GOC", "max_issues_repo_head_hexsha": "daaee3ff9a0c8d21a739597ce0a7172213f43924", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "dev/collect_data_instances.jl", "max_forks_repo_name": "Gimaju/GOC", "max_forks_repo_head_hexsha": "daaee3ff9a0c8d21a739597ce0a7172213f43924", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 33.8111888112, "max_line_length": 194, "alphanum_fraction": 0.7420889349, "num_tokens": 1191, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.42250463481418826, "lm_q2_score": 0.13477592089824647, "lm_q1q2_score": 0.05694345124085955}}
{"text": "\nusing Distributed\nif nprocs() == 1\n addprocs(3)\nend\n# Number of logical CPU cores available in the system.\n# println(\"CPU cores: \", Sys.CPU_CORES)\nSys.cpu_summary()\n# Number of available processes.\n# println(\"nprocs: \", nprocs())\n# println(\"nworkers: \", nworkers())\nprintln(\"nprocs: \", nprocs())\nprintln(\"nworkers: \", nworkers())\n\nn = 20000000\n# Ordinary for loop.\nlet\n global sum1\n sum1 = 0\n @time for i = 1:n\n sum1 += Int(rand(Bool));\n end\nend\n\n# Parallel for loop\n# @time @parallel (+) for i = 1:20000\n@time sum2 = @distributed (+) for i = 1:n\n Int(rand(Bool));\nend\n\n# Predefined function.\n@time sum3 =sum(rand(0:1, n))\n\n# For a not so small amount of work:\n# n = 200000000;\n# n = 20000;\n# @time @parallel (+) for i = 1:n\n@time sum4 = @distributed (+) for i = 1:n\n Int(rand(Bool))\nend\n\n# println(sum1)\n# println(sum2)\n# println(sum3)\n# println(sum4)\n\n# DO NOT run sum(rand(0:1, n)) or the ordinary for loop with 'n': You may run\n# out of memory.\n", "meta": {"hexsha": "f313033b9d38bf9d399f2e6989323e9f0c9aa493", "size": 979, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "devel/dev_parallel_example.jl", "max_stars_repo_name": "mjirik/LarSurf.jl", "max_stars_repo_head_hexsha": "de2eaec62dfe8c63e7d621bc973aa01d8de019c6", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 2, "max_stars_repo_stars_event_min_datetime": "2019-09-17T22:56:08.000Z", "max_stars_repo_stars_event_max_datetime": "2020-01-04T09:50:42.000Z", "max_issues_repo_path": "devel/dev_parallel_example.jl", "max_issues_repo_name": "mjirik/lario3d.jl", "max_issues_repo_head_hexsha": "de2eaec62dfe8c63e7d621bc973aa01d8de019c6", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 1, "max_issues_repo_issues_event_min_datetime": "2019-11-16T15:47:22.000Z", "max_issues_repo_issues_event_max_datetime": "2019-11-18T17:43:46.000Z", "max_forks_repo_path": "devel/dev_parallel_example.jl", "max_forks_repo_name": "mjirik/lario3d.jl", "max_forks_repo_head_hexsha": "de2eaec62dfe8c63e7d621bc973aa01d8de019c6", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2021-03-05T15:01:47.000Z", "max_forks_repo_forks_event_max_datetime": "2021-03-05T15:01:47.000Z", "avg_line_length": 19.9795918367, "max_line_length": 77, "alphanum_fraction": 0.6353421859, "num_tokens": 320, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4225046348141882, "lm_q2_score": 0.13477591568531197, "lm_q1q2_score": 0.05694344903837056}}
{"text": "\"\"\"\n count_nucleotides(strand)\n\nThe frequency of each nucleotide within `strand` as a dictionary.\n\nInvalid strands raise a `DomainError`.\n\n\"\"\"\nfunction count_nucleotides(strand)\n dna = Dict('A'=>0,'C'=>0,'G'=>0,'T'=>0)\n\n for i in strand\n try\n dna[i] += 1\n catch\n throw(DomainError(\"Invalid Key\"))\n end\n end\n\n return dna\nend\n", "meta": {"hexsha": "58f163174d5fe042cd900c7aed7938344f93046f", "size": 350, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "julia/nucleotide-count/nucleotide-count.jl", "max_stars_repo_name": "res0nat0r/exercism", "max_stars_repo_head_hexsha": "b3f7df7cefee52cecb932af7ca80280060bfdaf4", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "julia/nucleotide-count/nucleotide-count.jl", "max_issues_repo_name": "res0nat0r/exercism", "max_issues_repo_head_hexsha": "b3f7df7cefee52cecb932af7ca80280060bfdaf4", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "julia/nucleotide-count/nucleotide-count.jl", "max_forks_repo_name": "res0nat0r/exercism", "max_forks_repo_head_hexsha": "b3f7df7cefee52cecb932af7ca80280060bfdaf4", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 15.9090909091, "max_line_length": 65, "alphanum_fraction": 0.6342857143, "num_tokens": 104, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4455295350395727, "lm_q2_score": 0.1276526236624356, "lm_q1q2_score": 0.05687301406690649}}
{"text": "struct Config{T<:Real}\n atol::T # absolute tolerance for ...\n rtol::T # relative tolerance for ...\n solve::Bool # optimize and test result\n query_number_of_constraints::Bool # can get `MOI.NumberOfConstraints` attribute\n query::Bool # can get objective function, and constraint functions, and constraint sets\n modify_lhs::Bool # can modify function of a constraint\n duals::Bool # test dual solutions\n dual_objective_value::Bool # test `DualObjectiveValue`\n infeas_certificates::Bool # check for primal or dual infeasibility certificates when appropriate\n # The expected \"optimal\" status returned by the solver. Either\n # MOI.OPTIMAL or MOI.LOCALLY_SOLVED.\n optimal_status::MOI.TerminationStatusCode\n basis::Bool # can get variable and constraint basis status\n\n \"\"\"\n Config{T}(;\n atol::Real = Base.rtoldefault(T),\n rtol::Real = Base.rtoldefault(T),\n solve::Bool = true,\n query_number_of_constraints::Bool = true,\n query::Bool = true,\n modify_lhs::Bool = true,\n duals::Bool = true,\n dual_objective_value::Bool = duals,\n infeas_certificates::Bool = true,\n optimal_status = MOI.OPTIMAL,\n basis::Bool = false,\n )\n\n Return an object that is used to configure various tests.\n\n ## Keywords\n\n * `atol::Real = Base.rtoldefault(T)`: Control the absolute tolerance used\n when comparing solutions.\n * `rtol::Real = Base.rtoldefault(T)`: Control the relative tolerance used\n when comparing solutions.\n * `solve::Bool = true`: Set to `false` to skip tests requiring a call to\n [`MOI.optimize!`](@ref)\n * `query_number_of_constraints::Bool = true`: Set to `false` to skip tests\n requiring a call to [`MOI.NumberOfConstraints`](@ref).\n * `query::Bool = true`: Set to `false` to skip tests requiring a call to\n [`MOI.get`](@ref) for [`MOI.ConstraintFunction`](@ref) and\n [`MOI.ConstraintSet`](@ref)\n * `modify_lhs::Bool = true`:\n * `duals::Bool = true`: Set to `false` to skip tests querying\n [`MOI.ConstraintDual`](@ref).\n * `dual_objective_value::Bool = duals`: Set to `false` to skip tests\n querying [`MOI.DualObjectiveValue`](@ref).\n * `infeas_certificates::Bool = true`: Set to `false` to skip tests querying\n primal and dual infeasibility certificates.\n * `optimal_status = MOI.OPTIMAL`: Set to `MOI.LOCALLY_SOLVED` if the solver\n cannot prove global optimality.\n * `basis::Bool = false`: Set to `true` if the solver supports\n [`MOI.ConstraintBasisStatus`](@ref) and [`MOI.VariableBasisStatus`](@ref).\n \"\"\"\n function Config{T}(;\n atol::Real = Base.rtoldefault(T),\n rtol::Real = Base.rtoldefault(T),\n solve::Bool = true,\n query_number_of_constraints::Bool = true,\n query::Bool = true,\n modify_lhs::Bool = true,\n duals::Bool = true,\n dual_objective_value::Bool = duals,\n infeas_certificates::Bool = true,\n optimal_status = MOI.OPTIMAL,\n basis::Bool = false,\n ) where {T<:Real}\n return new(\n atol,\n rtol,\n solve,\n query_number_of_constraints,\n query,\n modify_lhs,\n duals,\n dual_objective_value,\n infeas_certificates,\n optimal_status,\n basis,\n )\n end\n Config(; kwargs...) = Config{Float64}(; kwargs...)\nend\n\n@deprecate TestConfig Config\n\n\"\"\"\n @moitestset setname subsets\n\nDefines a function `setnametest(model, config, exclude)` that runs the tests defined in the dictionary `setnametests`\nwith the model `model` and config `config` except the tests whose dictionary key is in `exclude`.\nIf `subsets` is `true` then each test runs in fact multiple tests hence the `exclude` argument is passed\nas it can also contains test to be excluded from these subsets of tests.\n\"\"\"\nmacro moitestset(setname, subsets = false)\n testname = Symbol(string(setname) * \"test\")\n testdict = Symbol(string(testname) * \"s\")\n if subsets\n runtest = :(f(model, config, exclude))\n else\n runtest = :(f(model, config))\n end\n return esc(\n :(\n function $testname(\n model::$MOI.ModelLike,\n config::$MOI.DeprecatedTest.Config,\n exclude::Vector{String} = String[],\n )\n for (name, f) in $testdict\n if name in exclude\n continue\n end\n @testset \"$name\" begin\n $runtest\n end\n end\n end\n ),\n )\nend\n", "meta": {"hexsha": "84d0cfdfb6b03b99566ad1750dcff96be3ea6419", "size": 4745, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/DeprecatedTest/config.jl", "max_stars_repo_name": "manuelbb-upb/MathOptInterface.jl", "max_stars_repo_head_hexsha": "54b6bcb723acb2b2d79584e2f27ea56fd4c7777c", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 132, "max_stars_repo_stars_event_min_datetime": "2020-06-20T00:45:49.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-28T22:06:34.000Z", "max_issues_repo_path": "src/DeprecatedTest/config.jl", "max_issues_repo_name": "manuelbb-upb/MathOptInterface.jl", "max_issues_repo_head_hexsha": "54b6bcb723acb2b2d79584e2f27ea56fd4c7777c", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 460, "max_issues_repo_issues_event_min_datetime": "2020-06-08T14:12:55.000Z", "max_issues_repo_issues_event_max_datetime": "2022-03-31T19:03:39.000Z", "max_forks_repo_path": "src/DeprecatedTest/config.jl", "max_forks_repo_name": "manuelbb-upb/MathOptInterface.jl", "max_forks_repo_head_hexsha": "54b6bcb723acb2b2d79584e2f27ea56fd4c7777c", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 28, "max_forks_repo_forks_event_min_datetime": "2020-06-08T01:45:29.000Z", "max_forks_repo_forks_event_max_datetime": "2022-03-23T10:36:25.000Z", "avg_line_length": 37.96, "max_line_length": 117, "alphanum_fraction": 0.6037934668, "num_tokens": 1133, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.3998116264369279, "lm_q2_score": 0.14223190046381004, "lm_q1q2_score": 0.05686596745565113}}
{"text": "using Test\n\n@testset \"Create tests for your project\" begin\n\n@testset \"Testing linear algebra results\" begin\n @test 1 == 1\nend;\n\nend; # ex1.jl\n\n", "meta": {"hexsha": "d20b338115a8389022dfa2759e8c5d0eca3576cf", "size": 145, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/test1.jl", "max_stars_repo_name": "PsuAstro528/project-template", "max_stars_repo_head_hexsha": "2397a484cbb09f60f632d6a725682ca6faaa9fcf", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2019-11-20T16:12:35.000Z", "max_stars_repo_stars_event_max_datetime": "2019-11-20T16:12:35.000Z", "max_issues_repo_path": "test/test1.jl", "max_issues_repo_name": "PsuAstro528/project-template", "max_issues_repo_head_hexsha": "2397a484cbb09f60f632d6a725682ca6faaa9fcf", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "test/test1.jl", "max_forks_repo_name": "PsuAstro528/project-template", "max_forks_repo_head_hexsha": "2397a484cbb09f60f632d6a725682ca6faaa9fcf", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 13.1818181818, "max_line_length": 47, "alphanum_fraction": 0.7034482759, "num_tokens": 43, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4765796510636759, "lm_q2_score": 0.11920292515117027, "lm_q1q2_score": 0.0568096884743142}}
{"text": "# Bogumi\u0142 Kami\u0144ski, 2019-03-25\n\nurl = \"https://projecteuler.net/project/resources/p096_sudoku.txt\"\nfilename = \"p096_sudoku.txt\"\nif !isfile(filename) # download only if needed\n println(\"File not found. Fetching from a remote location ...\")\n try\n # this uses OS tools and might fail if they are not found\n download(url, filename)\n catch\n # thus we provide a fallback\n using HTTP\n r = HTTP.get(url)\n write(filename, r.body)\n end\nend\n", "meta": {"hexsha": "6e138bc2f9d21c04b67eb2086fc7cf3bddf78b69", "size": 484, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "7. Sudoku/getproblems.jl", "max_stars_repo_name": "bkamins/UEP-Workshop-20190405", "max_stars_repo_head_hexsha": "7fcbb371c3c2efd862ce58752cb1a6cef4300fea", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 5, "max_stars_repo_stars_event_min_datetime": "2019-03-25T20:10:03.000Z", "max_stars_repo_stars_event_max_datetime": "2021-03-12T01:24:01.000Z", "max_issues_repo_path": "7. Sudoku/getproblems.jl", "max_issues_repo_name": "bkamins/UEP-Workshop-20190405", "max_issues_repo_head_hexsha": "7fcbb371c3c2efd862ce58752cb1a6cef4300fea", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "7. Sudoku/getproblems.jl", "max_forks_repo_name": "bkamins/UEP-Workshop-20190405", "max_forks_repo_head_hexsha": "7fcbb371c3c2efd862ce58752cb1a6cef4300fea", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 4, "max_forks_repo_forks_event_min_datetime": "2019-04-03T19:31:29.000Z", "max_forks_repo_forks_event_max_datetime": "2019-08-09T06:11:08.000Z", "avg_line_length": 28.4705882353, "max_line_length": 66, "alphanum_fraction": 0.6549586777, "num_tokens": 127, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.476579651063676, "lm_q2_score": 0.1192029188930649, "lm_q1q2_score": 0.05680968549182854}}
{"text": "const MYID = myid()\nconst OTHERIDS = filter(id-> id != MYID, procs())[rand(1:(nprocs()-1))]\n\nfunction check_leaks()\n if length(DistributedArrays.refs) > 0\n sleep(0.1) # allow time for any cleanup to complete and test again\n length(DistributedArrays.refs) > 0 && warn(\"Probable leak of \", length(DistributedArrays.refs), \" darrays\")\n end\nend\n\n@testset \"test distribute\" begin\n A = rand(1:100, (100,100))\n\n @testset \"test default distribute\" begin\n DA = distribute(A)\n @test length(procs(DA)) == nworkers()\n @test sum(DA) == sum(A)\n close(DA)\n end\n\n @testset \"test distribute with procs arguments\" begin\n DA = distribute(A, procs = procs())\n @test length(procs(DA)) == nprocs()\n @test sum(DA) == sum(A)\n close(DA)\n end\n\n @testset \"test distribute with procs and dist arguments\" begin\n DA = distribute(A, procs = [1, 2], dist = [1,2])\n @test size(procs(DA)) == (1,2)\n @test sum(DA) == sum(A)\n close(DA)\n end\n\n @testset \"Create darray with unconventional distribution and distibute like it\" begin\n block = 10\n Y = nworkers() * block\n X = nworkers() * block\n remote_parts = map(workers()) do wid\n remotecall(rand, wid, block, Y)\n end\n DA1 = DArray(reshape(remote_parts, (length(remote_parts), 1)))\n A = rand(X, Y)\n DA2 = distribute(A, DA1)\n\n @test size(DA1) == size(DA2)\n\n close(DA1)\n close(DA2)\n end\nend\n\ncheck_leaks()\n\n@testset \"test DArray equality\" begin\n D = drand((200,200), [MYID, OTHERIDS])\n DC = copy(D)\n\n @testset \"test isequal(::DArray, ::DArray)\" begin\n @test D == DC\n end\n\n @testset \"test copy(::DArray) does a copy of each localpart\" begin\n @spawnat OTHERIDS localpart(DC)[1] = 0\n @test fetch(@spawnat OTHERIDS localpart(D)[1] != 0)\n end\n\n close(D)\n close(DC)\nend\n\ncheck_leaks()\n\n@testset \"test DArray similar\" begin\n D = drand((200,200), [MYID, OTHERIDS])\n DS = similar(D,Float16)\n\n @testset \"test eltype of a similar\" begin\n @test eltype(DS) == Float16\n end\n\n @testset \"test dims of a similar\" begin\n @test size(D) == size(DS)\n end\n close(D)\n close(DS)\nend\n\ncheck_leaks()\n\n@testset \"test DArray reshape\" begin\n D = drand((200,200), [MYID, OTHERIDS])\n\n @testset \"Test error-throwing in reshape\" begin\n @test_throws DimensionMismatch reshape(D,(100,100))\n end\n\n DR = reshape(D,(100,400))\n @testset \"Test reshape\" begin\n @test size(DR) == (100,400)\n end\n close(D)\nend\n\ncheck_leaks()\n\n@testset \"test @DArray comprehension constructor\" begin\n\n @testset \"test valid use of @DArray\" begin\n D = @DArray [i+j for i=1:10, j=1:10]\n @test D == [i+j for i=1:10, j=1:10]\n close(D)\n end\n\n @testset \"test invalid use of @DArray\" begin\n @test_throws ArgumentError eval(:((@DArray [1,2,3,4])))\n end\nend\n\ncheck_leaks()\n\n@testset \"test DArray / Array conversion\" begin\n D = drand((200,200), [MYID, OTHERIDS])\n\n @testset \"test convert(::Array, ::(Sub)DArray)\" begin\n S = convert(Matrix{Float64}, D[1:150, 1:150])\n A = convert(Matrix{Float64}, D)\n\n @test A[1:150,1:150] == S\n D2 = convert(DArray{Float64,2,Matrix{Float64}}, A)\n @test D2 == D\n @test fetch(@spawnat MYID localpart(D)[1,1]) == D[1,1]\n @test fetch(@spawnat OTHERIDS localpart(D)[1,1]) == D[1,101]\n close(D2)\n\n S2 = convert(Vector{Float64}, D[4, 23:176])\n @test A[4, 23:176] == S2\n\n S3 = convert(Vector{Float64}, D[23:176, 197])\n @test A[23:176, 197] == S3\n\n S4 = zeros(4)\n setindex!(S4, D[3:4, 99:100], :)\n @test S4 == vec(D[3:4, 99:100])\n @test S4 == vec(A[3:4, 99:100])\n\n S5 = zeros(2,2)\n setindex!(S5, D[1,1:4], :, 1:2)\n @test vec(S5) == D[1, 1:4]\n @test vec(S5) == A[1, 1:4]\n end\n close(D)\nend\n\ncheck_leaks()\n\n@testset \"copy!\" begin\n D1 = dzeros((10,10))\n r1 = remotecall_wait(() -> randn(3,10), workers()[1])\n r2 = remotecall_wait(() -> randn(7,10), workers()[2])\n D2 = DArray(reshape([r1; r2], 2, 1))\n copy!(D2, D1)\n @test D1 == D2\n close(D1)\n close(D2)\nend\n\ncheck_leaks()\n\n@testset \"test DArray reduce\" begin\n D = DArray(id->fill(myid(), map(length,id)), (10,10), [MYID, OTHERIDS])\n\n @testset \"test reduce\" begin\n @test reduce(+, D) == ((50*MYID) + (50*OTHERIDS))\n end\n\n @testset \"test map / reduce\" begin\n D2 = map(x->1, D)\n @test reduce(+, D2) == 100\n close(D2)\n end\n\n @testset \"test map! / reduce\" begin\n map!(x->1, D)\n @test reduce(+, D) == 100\n end\n close(D)\nend\n\ncheck_leaks()\n\n@testset \"test scale\" begin\n A = randn(100,100)\n DA = distribute(A)\n @test scale!(DA, 2) == scale!(A, 2)\n close(DA)\nend\n\ncheck_leaks()\n\n@testset \"test scale!(b, A)\" begin\n A = randn(100, 100)\n b = randn(100)\n DA = distribute(A)\n @test scale!(b, A) == scale!(b, DA)\n close(DA)\n A = randn(100, 100)\n b = randn(100)\n DA = distribute(A)\n @test scale!(A, b) == scale!(DA, b)\n close(DA)\nend\n\ncheck_leaks()\n\n@testset \"test mapreduce on DArrays\" begin\n for _ = 1:25, f = [x -> Int128(2x), x -> Int128(x^2), x -> Int128(x^2 + 2x - 1)], opt = [+, *]\n A = rand(1:5, rand(2:30))\n DA = distribute(A)\n @test mapreduce(f, opt, DA) - mapreduce(f, opt, A) == 0\n close(DA)\n end\nend\n\ncheck_leaks()\n\n@testset \"test mapreducedim on DArrays\" begin\n D = DArray(I->fill(myid(), map(length,I)), (73,73), [MYID, OTHERIDS])\n D2 = map(x->1, D)\n @test mapreducedim(t -> t*t, +, D2, 1) == mapreducedim(t -> t*t, +, convert(Array, D2), 1)\n @test mapreducedim(t -> t*t, +, D2, 2) == mapreducedim(t -> t*t, +, convert(Array, D2), 2)\n @test mapreducedim(t -> t*t, +, D2, (1,2)) == mapreducedim(t -> t*t, +, convert(Array, D2), (1,2))\n\n # Test non-regularly chunked DArrays\n r1 = DistributedArrays.remotecall(() -> sprandn(3, 10, 0.1), workers()[1])\n r2 = DistributedArrays.remotecall(() -> sprandn(7, 10, 0.1), workers()[2])\n D = DArray(reshape([r1; r2], (2,1)))\n @test Array(sum(D, 2)) == sum(Array(D), 2)\n\n # close(D)\n # close(D2)\n darray_closeall() # temp created by the mapreduce above\nend\n\ncheck_leaks()\n\n@testset \"test mapreducdim, reducedim on DArrays\" begin\n dims = (20,20,20)\n DA = drandn(dims)\n A = convert(Array, DA)\n\n @testset \"dimension $dms\" for dms in (1, 2, 3, (1,2), (1,3), (2,3), (1,2,3))\n @test mapreducedim(t -> t*t, +, A, dms) \u2248 mapreducedim(t -> t*t, +, DA, dms)\n @test mapreducedim(t -> t*t, +, A, dms, 1.0) \u2248 mapreducedim(t -> t*t, +, DA, dms, 1.0)\n @test reducedim(*, A, dms) \u2248 reducedim(*, DA, dms)\n @test reducedim(*, A, dms, 2.0) \u2248 reducedim(*, DA, dms, 2.0)\n end\n close(DA)\n darray_closeall() # temp created by the mapreduce above\nend\n\ncheck_leaks()\n\n@testset \"test statistical functions on DArrays\" begin\n dims = (20,20,20)\n DA = drandn(dims)\n A = convert(Array, DA)\n\n @testset \"test $f for dimension $dms\" for f in (mean, ), dms in (1, 2, 3, (1,2), (1,3), (2,3), (1,2,3))\n # std is pending implementation\n @test f(DA,dms) \u2248 f(A,dms)\n end\n\n close(DA)\n darray_closeall() # temporaries created above\nend\n\ncheck_leaks()\n\n@testset \"test sum on DArrays\" begin\n A = randn(100,100)\n DA = distribute(A)\n\n # sum either throws an ArgumentError or a CompositeException of ArgumentErrors\n try\n sum(DA, -1)\n catch err\n if isa(err, CompositeException)\n @test !isempty(err.exceptions)\n for excep in err.exceptions\n # Unpack the remote exception\n orig_err = excep.ex.captured.ex\n @test isa(orig_err, ArgumentError)\n end\n else\n @test isa(err, ArgumentError)\n end\n end\n try\n sum(DA, 0)\n catch err\n if isa(err, CompositeException)\n @test !isempty(err.exceptions)\n for excep in err.exceptions\n # Unpack the remote exception\n orig_err = excep.ex.captured.ex\n @test isa(orig_err, ArgumentError)\n end\n else\n @test isa(err, ArgumentError)\n end\n end\n\n @test sum(DA) \u2248 sum(A)\n @test sum(DA,1) \u2248 sum(A,1)\n @test sum(DA,2) \u2248 sum(A,2)\n @test sum(DA,3) \u2248 sum(A,3)\n close(DA)\n darray_closeall() # temporaries created above\nend\n\ncheck_leaks()\n\n@testset \"test size on DArrays\" begin\n\n A = randn(100,100)\n DA = distribute(A)\n\n @test_throws BoundsError size(DA, 0)\n @test size(DA,1) == size(A,1)\n @test size(DA,2) == size(A,2)\n @test size(DA,3) == size(A,3)\n close(DA)\nend\n\ncheck_leaks()\n\n# test length / endof\n@testset \"test collections API\" begin\n A = randn(23,23)\n DA = distribute(A)\n\n @testset \"test length\" begin\n @test length(DA) == length(A)\n end\n\n @testset \"test endof\" begin\n @test endof(DA) == endof(A)\n end\n close(DA)\nend\n\ncheck_leaks()\n\n@testset \"test max / min / sum\" begin\n a = map(x->Int(round(rand() * 100)) - 50, Array(Int, 100,1000))\n d = distribute(a)\n\n @test sum(d) == sum(a)\n @test maximum(d) == maximum(a)\n @test minimum(d) == minimum(a)\n @test maxabs(d) == maxabs(a)\n @test minabs(d) == minabs(a)\n @test sumabs(d) == sumabs(a)\n @test sumabs2(d) == sumabs2(a)\n close(d)\nend\n\ncheck_leaks()\n\n@testset \"test all / any\" begin\n a = map(x->Int(round(rand() * 100)) - 50, Array(Int, 100,1000))\n a = [true for i in 1:100]\n d = distribute(a)\n\n @test all(d)\n @test any(d)\n\n close(d)\n\n a[50] = false\n d = distribute(a)\n @test !all(d)\n @test any(d)\n\n close(d)\n\n a = [false for i in 1:100]\n d = distribute(a)\n @test !all(d)\n @test !any(d)\n\n close(d)\n\n d = dones(10,10)\n @test !all(x-> x>1.0, d)\n @test all(x-> x>0.0, d)\n\n close(d)\n\n a = ones(10,10)\n a[10] = 2.0\n d = distribute(a)\n @test any(x-> x == 1.0, d)\n @test any(x-> x == 2.0, d)\n @test !any(x-> x == 3.0, d)\n\n close(d)\nend\n\ncheck_leaks()\n\n@testset \"test count\" begin\n a = ones(10,10)\n a[10] = 2.0\n d = distribute(a)\n\n @test count(x-> x == 2.0, d) == 1\n @test count(x-> x == 1.0, d) == 99\n @test count(x-> x == 0.0, d) == 0\n\n close(d)\nend\n\ncheck_leaks()\n\n@testset \"test prod\" begin\n a = fill(2, 10);\n d = distribute(a);\n @test prod(d) == 2^10\n\n close(d)\nend\n\ncheck_leaks()\n\n@testset \"test zeros\" begin\n @testset \"1D dzeros default element type\" begin\n A = dzeros(10)\n @test A == zeros(10)\n @test eltype(A) == Float64\n @test size(A) == (10,)\n close(A)\n end\n\n @testset \"1D dzeros with specified element type\" begin\n A = dzeros(Int, 10)\n @test A == zeros(10)\n @test eltype(A) == Int\n @test size(A) == (10,)\n close(A)\n end\n\n @testset \"2D dzeros default element type, Dims constuctor\" begin\n A = dzeros((10,10))\n @test A == zeros((10,10))\n @test eltype(A) == Float64\n @test size(A) == (10,10)\n close(A)\n end\n\n @testset \"2D dzeros specified element type, Dims constructor\" begin\n A = dzeros(Int, (10,10))\n @test A == zeros(Int, (10,10))\n @test eltype(A) == Int\n @test size(A) == (10,10)\n close(A)\n end\n\n @testset \"2D dzeros, default element type\" begin\n A = dzeros(10,10)\n @test A == zeros(10,10)\n @test eltype(A) == Float64\n @test size(A) == (10,10)\n close(A)\n end\n\n @testset \"2D dzeros, specified element type\" begin\n A = dzeros(Int, 10, 10)\n @test A == zeros(Int, 10, 10)\n @test eltype(A) == Int\n @test size(A) == (10,10)\n close(A)\n end\nend\n\ncheck_leaks()\n\n@testset \"test dones\" begin\n @testset \"1D dones default element type\" begin\n A = dones(10)\n @test A == ones(10)\n @test eltype(A) == Float64\n @test size(A) == (10,)\n close(A)\n end\n\n @testset \"1D dones with specified element type\" begin\n A = dones(Int, 10)\n @test eltype(A) == Int\n @test size(A) == (10,)\n close(A)\n end\n\n @testset \"2D dones default element type, Dims constuctor\" begin\n A = dones((10,10))\n @test A == ones((10,10))\n @test eltype(A) == Float64\n @test size(A) == (10,10)\n close(A)\n end\n\n @testset \"2D dones specified element type, Dims constructor\" begin\n A = dones(Int, (10,10))\n @test A == ones(Int, (10,10))\n @test eltype(A) == Int\n @test size(A) == (10,10)\n close(A)\n end\n\n @testset \"2D dones, default element type\" begin\n A = dones(10,10)\n @test A == ones(10,10)\n @test eltype(A) == Float64\n @test size(A) == (10,10)\n close(A)\n end\n\n @testset \"2D dones, specified element type\" begin\n A = dones(Int, 10, 10)\n @test A == ones(Int, 10, 10)\n @test eltype(A) == Int\n @test size(A) == (10,10)\n close(A)\n end\nend\n\ncheck_leaks()\n\n@testset \"test drand\" begin\n @testset \"1D drand\" begin\n A = drand(100)\n @test eltype(A) == Float64\n @test size(A) == (100,)\n @test all(x-> x >= 0.0 && x <= 1.0, A)\n close(A)\n end\n\n @testset \"1D drand, specified element type\" begin\n A = drand(Int, 100)\n @test eltype(A) == Int\n @test size(A) == (100,)\n close(A)\n end\n\n @testset \"2D drand, Dims constructor\" begin\n A = drand((50,50))\n @test eltype(A) == Float64\n @test size(A) == (50,50)\n @test all(x-> x >= 0.0 && x <= 1.0, A)\n close(A)\n end\n\n @testset \"2D drand\" begin\n A = drand(100,100)\n @test eltype(A) == Float64\n @test size(A) == (100,100)\n @test all(x-> x >= 0.0 && x <= 1.0, A)\n close(A)\n end\n\n @testset \"2D drand, Dims constructor, specified element type\" begin\n A = drand(Int, (100,100))\n @test eltype(A) == Int\n @test size(A) == (100,100)\n close(A)\n end\n\n @testset \"2D drand, specified element type\" begin\n A = drand(Int, 100, 100)\n @test eltype(A) == Int\n @test size(A) == (100,100)\n close(A)\n end\nend\n\ncheck_leaks()\n\n@testset \"test randn\" begin\n @testset \"1D drandn\" begin\n A = drandn(100)\n @test eltype(A) == Float64\n @test size(A) == (100,)\n close(A)\n end\n\n @testset \"2D drandn, Dims constructor\" begin\n A = drandn((50,50))\n @test eltype(A) == Float64\n @test size(A) == (50,50)\n close(A)\n end\n\n @testset \"2D drandn\" begin\n A = drandn(100,100)\n @test eltype(A) == Float64\n @test size(A) == (100,100)\n close(A)\n end\nend\n\ncheck_leaks()\n\n@testset \"test c/transpose\" begin\n @testset \"test ctranspose real\" begin\n A = drand(Float64, 100, 200)\n @test A' == Array(A)'\n close(A)\n end\n @testset \"test ctranspose complex\" begin\n A = drand(Complex128, 200, 100)\n @test A' == Array(A)'\n close(A)\n end\n @testset \"test transpose real\" begin\n A = drand(Float64, 200, 100)\n @test A.' == Array(A).'\n close(A)\n end\n @testset \"test ctranspose complex\" begin\n A = drand(Complex128, 100, 200)\n @test A.' == Array(A).'\n close(A)\n end\n\n darray_closeall() # close the temporaries created above\nend\n\ncheck_leaks()\n\n@testset \"test convert from subdarray\" begin\n a = drand(20, 20);\n\n s = view(a, 1:5, 5:8)\n @test isa(s, SubDArray)\n @test s == convert(DArray, s)\n\n s = view(a, 6:5, 5:8)\n @test isa(s, SubDArray)\n @test s == convert(DArray, s)\n close(a)\n darray_closeall() # close the temporaries created above\nend\n\ncheck_leaks()\n\n@testset \"test scalar math\" begin\n a = drand(20, 20);\n b = convert(Array, a)\n @testset \"$f\" for f in (-, abs, abs2, acos, acosd, acot,\n acotd, acsch, angle, asech, asin,\n asind, asinh, atan, atand, atanh,\n big, cbrt, ceil, cis, complex, conj,\n cos, cosc, cosd, cosh, cospi, cot,\n cotd, coth, csc, cscd, csch, dawson,\n deg2rad, digamma, erf, erfc, erfcinv,\n erfcx, erfi, erfinv, exp, exp10, exp2,\n expm1, exponent, float, floor, gamma, imag,\n invdigamma, isfinite, isinf, isnan, lfact,\n lgamma, log, log10, log1p, log2, rad2deg, real,\n sec, secd, sech, sign, sin, sinc, sind,\n sinh, sinpi, sqrt, tan, tand, tanh, trigamma)\n @test f.(a) == f.(b)\n end\n a = a + 1\n b = b + 1\n @testset \"$f\" for f in (asec, asecd, acosh, acsc, acscd, acoth)\n @test f.(a) == f.(b)\n end\n close(a)\n darray_closeall() # close the temporaries created above\nend\n\ncheck_leaks()\n\n@testset \"test mapslices\" begin\n A = randn(5,5,5)\n D = distribute(A, procs = workers(), dist = [1, 1, min(nworkers(), 5)])\n @test mapslices(svdvals, D, (1,2)) \u2248 mapslices(svdvals, A, (1,2))\n @test mapslices(svdvals, D, (1,3)) \u2248 mapslices(svdvals, A, (1,3))\n @test mapslices(svdvals, D, (2,3)) \u2248 mapslices(svdvals, A, (2,3))\n @test mapslices(sort, D, (1,)) \u2248 mapslices(sort, A, (1,))\n @test mapslices(sort, D, (2,)) \u2248 mapslices(sort, A, (2,))\n @test mapslices(sort, D, (3,)) \u2248 mapslices(sort, A, (3,))\n\n # issue #3613\n B = mapslices(sum, dones(Float64, (2,3,4), workers(), [1,1,min(nworkers(),4)]), [1,2])\n @test size(B) == (1,1,4)\n @test all(B.==6)\n\n # issue #5141\n C1 = mapslices(x-> maximum(-x), D, [])\n @test C1 == -D\n\n # issue #5177\n c = dones(Float64, (2,3,4,5), workers(), [1,1,1,min(nworkers(),5)])\n m1 = mapslices(x-> ones(2,3), c, [1,2])\n m2 = mapslices(x-> ones(2,4), c, [1,3])\n m3 = mapslices(x-> ones(3,4), c, [2,3])\n @test size(m1) == size(m2) == size(m3) == size(c)\n\n n1 = mapslices(x-> ones(6), c, [1,2])\n n2 = mapslices(x-> ones(6), c, [1,3])\n n3 = mapslices(x-> ones(6), c, [2,3])\n n1a = mapslices(x-> ones(1,6), c, [1,2])\n n2a = mapslices(x-> ones(1,6), c, [1,3])\n n3a = mapslices(x-> ones(1,6), c, [2,3])\n @test (size(n1a) == (1,6,4,5) && size(n2a) == (1,3,6,5) && size(n3a) == (2,1,6,5))\n @test (size(n1) == (6,1,4,5) && size(n2) == (6,3,1,5) && size(n3) == (2,6,1,5))\n close(D)\n close(c)\n darray_closeall() # close the temporaries created above\nend\n\ncheck_leaks()\n\n@testset \"test scalar ops\" begin\n a = drand(20,20)\n b = convert(Array, a)\n c = drand(20,20)\n d = convert(Array, c)\n\n @testset \"$f\" for f in (+, -, .+, .-, .*, ./, .%, div, mod)\n x = rand()\n @test f(a, x) == f(b, x)\n @test f(x, a) == f(x, b)\n @test f(a, c) == f(b, d)\n end\n\n close(a)\n close(c)\n\n a = dones(Int, 20, 20)\n b = convert(Array, a)\n @testset \"$f\" for f in (.<<, .>>)\n @test f(a, 2) == f(b, 2)\n @test f(2, a) == f(2, b)\n @test f(a, a) == f(b, b)\n end\n\n @testset \"$f\" for f in (rem,)\n x = rand()\n @test f(a, x) == f(b, x)\n end\n close(a)\n close(c)\n darray_closeall() # close the temporaries created above\nend\n\ncheck_leaks()\n\n@testset \"test broadcast ops\" begin\n wrkrs = workers()\n nwrkrs = length(wrkrs)\n nrows = 20 * nwrkrs\n ncols = 10 * nwrkrs\n a = drand((nrows,ncols), wrkrs, (1, nwrkrs))\n m = mean(a, 1)\n c = a .- m\n d = convert(Array, a) .- convert(Array, m)\n @test c == d\n darray_closeall()\nend\n\ncheck_leaks()\n\n@testset \"test matrix multiplication\" begin\n A = drandn(20,20)\n b = drandn(20)\n B = drandn(20,20)\n\n @test norm(convert(Array, A*b) - convert(Array, A)*convert(Array, b), Inf) < sqrt(eps())\n @test norm(convert(Array, A*B) - convert(Array, A)*convert(Array, B), Inf) < sqrt(eps())\n @test norm(convert(Array, A'*b) - convert(Array, A)'*convert(Array, b), Inf) < sqrt(eps())\n @test norm(convert(Array, A'*B) - convert(Array, A)'*convert(Array, B), Inf) < sqrt(eps())\n close(A)\n close(b)\n close(B)\n darray_closeall() # close the temporaries created above\nend\n\ncheck_leaks()\n\n@testset \"test norm\" begin\n x = drandn(20)\n\n @test abs(norm(x) - norm(convert(Array, x))) < sqrt(eps())\n @test abs(norm(x, 1) - norm(convert(Array, x), 1)) < sqrt(eps())\n @test abs(norm(x, 2) - norm(convert(Array, x), 2)) < sqrt(eps())\n @test abs(norm(x, Inf) - norm(convert(Array, x), Inf)) < sqrt(eps())\n close(x)\nend\n\ncheck_leaks()\n\n@testset \"test axpy!\" begin\n x = drandn(20)\n y = drandn(20)\n\n @test norm(convert(Array, LinAlg.axpy!(2.0, x, copy(y))) - LinAlg.axpy!(2.0, convert(Array, x), convert(Array, y))) < sqrt(eps())\n @test_throws DimensionMismatch LinAlg.axpy!(2.0, x, zeros(length(x) + 1))\n close(x)\n close(y)\n darray_closeall() # close the temporaries created above\nend\n\ncheck_leaks()\n\n@testset \"test ppeval\" begin\n A = drandn((10, 10, nworkers()), workers(), [1, 1, nworkers()])\n B = drandn((10, nworkers()), workers(), [1, nworkers()])\n\n R = zeros(10, nworkers())\n for i = 1:nworkers()\n R[:, i] = convert(Array, A)[:, :, i]*convert(Array, B)[:, i]\n end\n @test convert(Array, ppeval(*, A, B)) \u2248 R\n @test sum(ppeval(eigvals, A)) \u2248 sum(ppeval(eigvals, A, eye(10, 10)))\n close(A)\n close(B)\n darray_closeall() # close the temporaries created above\nend\n\ncheck_leaks()\n\n@testset \"test nnz\" begin\n A = sprandn(10, 10, 0.5)\n @test nnz(distribute(A)) == nnz(A)\nend\n\n@testset \"test matmatmul\" begin\n A = drandn(30, 30)\n B = drandn(30, 20)\n a = convert(Array, A)\n b = convert(Array, B)\n\n AB = A * B\n AtB = A.' * B\n AcB = A' * B\n\n ab = a * b\n atb = a.' * b\n acb = a' * b\n\n @test AB \u2248 ab\n @test AtB \u2248 atb\n @test AcB \u2248 acb\n darray_closeall() # close the temporaries created above\nend\n\n@testset \"sort, T = $T\" for i in 0:6, T in [Int, Float64]\n d = DistributedArrays.drand(T, 10^i)\n @testset \"sample = $sample\" for sample in Any[true, false, (minimum(d),maximum(d)), rand(T, 10^i>512 ? 512 : 10^i)]\n d2 = DistributedArrays.sort(d; sample=sample)\n\n @test length(d) == length(d2)\n @test sort(convert(Array, d)) == convert(Array, d2)\n end\n darray_closeall() # close the temporaries created above\nend\n\ncheck_leaks()\n\ndarray_closeall()\n\n@testset \"test for any leaks\" begin\n sleep(1.0) # allow time for any cleanup to complete\n allrefszero = Bool[remotecall_fetch(()->length(DistributedArrays.refs) == 0, p) for p in procs()]\n @test all(allrefszero)\n\n allregistrieszero = Bool[remotecall_fetch(()->length(DistributedArrays.registry) == 0, p) for p in procs()]\n @test all(allregistrieszero)\nend\n", "meta": {"hexsha": "a08ae03481288098475ecaf9596db5106cda8b77", "size": 22763, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/darray.jl", "max_stars_repo_name": "JuliaPackageMirrors/DistributedArrays.jl", "max_stars_repo_head_hexsha": "9018902e7c323b39a02d1e515aa283fbb1f913f1", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "test/darray.jl", "max_issues_repo_name": "JuliaPackageMirrors/DistributedArrays.jl", "max_issues_repo_head_hexsha": "9018902e7c323b39a02d1e515aa283fbb1f913f1", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "test/darray.jl", "max_forks_repo_name": "JuliaPackageMirrors/DistributedArrays.jl", "max_forks_repo_head_hexsha": "9018902e7c323b39a02d1e515aa283fbb1f913f1", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 25.75, "max_line_length": 133, "alphanum_fraction": 0.546896279, "num_tokens": 7887, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.45713671682749474, "lm_q2_score": 0.12421301969912356, "lm_q1q2_score": 0.056782332012486274}}
{"text": "@testset \"Primitives\" begin\n\nusing Base.Test\nusing DataValueArrays\n\nn = rand(1:5)\nsiz = [ rand(2:5) for i in n ]\nA = rand(siz...)\nM = rand(Bool, siz...)\nX = DataValueArray(A, M)\ni = rand(1:length(X))\n@test values(X, i) == X.values[i]\n@test isnull(X, i) == X.isnull[i]\nI = [ rand(1:size(X,i)) for i in 1:n ]\n@test values(X, I...) == X.values[I...]\n@test isnull(X, I...) == X.isnull[I...]\n\n# ----- test Base.similar, Base.size ----------------------------------------#\n\nx = DataValueArray(Int, (5, 2))\n@test isa(x, DataValueMatrix{Int})\n@test size(x) === (5, 2)\n\ny = similar(x, DataValue{Int}, (3, 3))\n@test isa(y, DataValueMatrix{Int})\n@test size(y) === (3, 3)\n\nz = similar(x, DataValue{Int}, (2,))\n@test isa(z, DataValueVector{Int})\n@test size(z) === (2, )\n\n# test common use-patterns for 'similar'\ndv = DataValueArray(Int, 2)\ndm = DataValueArray(Int, 2, 2)\ndt = DataValueArray(Int, 2, 2, 2)\n\nsimilar(dv)\nsimilar(dm)\nsimilar(dt)\n\nsimilar(dv, 2)\nsimilar(dm, 2, 2)\nsimilar(dt, 2, 2, 2)\n\n# ----- test Base.copy/Base.copy! --------------------------------------------#\n\n#copy\nx = DataValueArray([1, 2, nothing], Int, Void)\ny = DataValueArray([3, nothing, 5], Int, Void)\n@test isequal(copy(x), x)\n@test isequal(copy!(y, x), x)\n\n\n# copy!\nfunction nonbits(dv)\n ret = similar(dv, Integer)\n for i = 1:length(dv)\n if !dv.isnull[i]\n ret[i] = dv[i]\n end\n end\n ret\nend\nset1 = Any[DataValueArray([1, nothing, 3], Int, Void),\n DataValueArray([nothing, 5], Int, Void),\n DataValueArray([1, 2, 3, 4, 5], Int, Void),\n DataValueArray(Int[]),\n DataValueArray([nothing, 5, 3], Int, Void),\n DataValueArray([1, 5, 3], Int, Void)]\nset2 = map(nonbits, set1)\n\nfor (dest, src, bigsrc, emptysrc, res1, res2) in Any[set1, set2]\n @test isequal(copy!(copy(dest), src), res1)\n @test isequal(copy!(copy(dest), 1, src), res1)\n\n @test isequal(copy!(copy(dest), 2, src, 2), res2)\n @test isequal(copy!(copy(dest), 2, src, 2, 1), res2)\n\n @test isequal(copy!(copy(dest), 99, src, 99, 0), dest)\n\n @test isequal(copy!(copy(dest), 1, emptysrc), dest)\n @test_throws BoundsError copy!(dest, 1, emptysrc, 1)\n\n for idx in [0, 4]\n @test_throws BoundsError copy!(dest, idx, src)\n @test_throws BoundsError copy!(dest, idx, src, 1)\n @test_throws BoundsError copy!(dest, idx, src, 1, 1)\n @test_throws BoundsError copy!(dest, 1, src, idx)\n @test_throws BoundsError copy!(dest, 1, src, idx, 1)\n end\n\n @test_throws ArgumentError copy!(dest, 1, src, 1, -1)\n\n @test_throws BoundsError copy!(dest, bigsrc)\n\n @test_throws BoundsError copy!(dest, 3, src)\n @test_throws BoundsError copy!(dest, 3, src, 1)\n @test_throws BoundsError copy!(dest, 3, src, 1, 2)\n @test_throws BoundsError copy!(dest, 1, src, 2, 2)\nend\n\n# ----- test Base.fill! ------------------------------------------------------#\n\nX = DataValueArray(Int, 10, 2)\nfill!(X, DataValue(10))\nY = DataValueArray(Float64, 10)\nfill!(Y, rand(Float64))\n\n@test X.values == fill(10, 10, 2)\n@test isequal(X.isnull, fill(false, 10, 2))\n@test isequal(Y.isnull, fill(false, 10))\n\nfill!(X, DataValue())\n@test isequal(X.isnull, fill(true, 10, 2))\n\n# ----- test Base.deepcopy ---------------------------------------------------#\n\nY1 = deepcopy(Y)\n@test isequal(Y1, Y)\n@assert !(Y === Y1)\n\n# ----- test Base.resize! ----------------------------------------------------#\n\nresize!(Y1, 20)\n@test Y1.values[1:10] == Y.values[1:10]\n@test Y1.isnull[1:10] == Y.isnull[1:10]\n@test Y1.isnull[11:20] == fill(true, 10)\n\nresize!(Y1, 5)\n@test Y1.values[1:5] == Y.values[1:5]\n@test Y1.isnull[1:5] == Y.isnull[1:5]\n\n# ----- test Base.reshape ----------------------------------------------------#\n\nY1 = reshape(copy(Y), length(Y), 1)\n@test size(Y1) == (length(Y), 1)\n@test all(i->isequal(Y1[i], Y[i]), 1:length(Y))\nY2 = reshape(Y1, 1, length(Y1))\n@test size(Y2) == (1, length(Y1))\n@test all(i->isequal(Y1[i], Y2[i]), 1:length(Y2))\n# Test that arrays share the same data\nY2.values[1] += 1\nY2.isnull[2] = true\n@test all(i->isequal(Y1[i], Y2[i]), 1:length(Y2))\n\n# ----- test Base.ndims ------------------------------------------------------#\n\nfor n in 1:4\n @test ndims(DataValueArray(Int, collect(1:n)...)) == n\nend\n\n# ----- test Base.length -----------------------------------------------------#\n\n@test length(DataValueArray(Int, 10)) == 10\n@test length(DataValueArray(Int, 5, 5)) == 25\n@test length(DataValueArray(Int, (3, 3, 3))) == 27\n\n# ----- test Base.endof ------------------------------------------------------#\n\n@test endof(DataValueArray(collect(1:10))) == 10\n@test endof(DataValueArray([1, 2, nothing, 4, nothing])) == 5\n\n# ----- test Base.find -------------------------------------------------------#\n\nz = DataValueArray(rand(Bool, 10))\n@test find(z) == find(z.values)\n\nz = DataValueArray([false, true, false, true, false, true])\n@test isequal(find(z), [2, 4, 6])\n\n# ----- test dropnull --------------------------------------------------------#\n\n# dropnull(X::DataValueVector)\nz = DataValueArray([1, 2, 3, 'a', 5, 'b', 7, 'c'], Int, Char)\n@test dropnull(z) == [1, 2, 3, 5, 7]\n\n# dropnull(X::AbstractVector)\nA = Any[1, 2, 3, DataValue(), 5, DataValue(), 7, DataValue()]\n@test dropnull(A) == [1, 2, 3, 5, 7]\n\n# dropnull(X::AbstractVector{<:DataValue})\nB = [1, 2, 3, DataValue(), 5, DataValue(), 7, DataValue()]\n@test dropnull(B) == [1, 2, 3, 5, 7]\n# assert dropnull returns copy for !(DataValue <: eltype(X))\nnullfree = [1, 2, 3, 4]\nreturned_copy = dropnull(nullfree)\n@test nullfree == returned_copy && !(nullfree === returned_copy)\n\n# ----- test dropnull! -------------------------------------------------------#\n\n# for each, assert returned values are unwrapped and inplace change\n# dropnull!(X::DataValueVector)\n@test dropnull!(z) == [1, 2, 3, 5, 7]\n@test isequal(z, DataValueArray([1, 2, 3, 5, 7]))\n\n# dropnull!(X::AbstractVector)\n@test dropnull!(A) == [1, 2, 3, 5, 7]\n@test isequal(A, Any[1, 2, 3, 5, 7])\n\n# dropnull!(X::AbstractVector{<:DataValue})\n@test dropnull!(B) == [1, 2, 3, 5, 7]\n@test isequal(B, DataValue[1, 2, 3, 5, 7])\n\n# when no nulls present, dropnull! returns input vector\nreturned_view = dropnull!(nullfree)\n@test nullfree == returned_view && nullfree === returned_view\n\n# test that dropnull! returns unwrapped values when DataValues are present\nX = [false, 1, :c, \"string\", DataValue(\"I am not null\"), DataValue()]\n@test !any(x -> isa(x, DataValue), dropnull!(X))\n@test any(x -> isa(x, DataValue), X)\nY = Any[false, 1, :c, \"string\", DataValue(\"I am not null\"), DataValue()]\n@test !any(x -> isa(x, DataValue), dropnull!(Y))\n@test any(x -> isa(x, DataValue), Y)\n\n# ----- test any(isnull, X) --------------------------------------------------#\n\n# any(isnull, X::DataValueArray)\nz = DataValueArray([1, 2, 3, 'a', 5, 'b', 7, 'c'], Int, Char)\n@test any(isnull, z) == true\n@test any(isnull, dropnull(z)) == false\nz = DataValueArray(Int, 10)\n@test any(isnull, z) == true\n\n# any(isnull, A::AbstractArray)\nA2 = [DataValue(1), DataValue(2), DataValue(3)]\n@test any(isnull, A2) == false\npush!(A2, DataValue{Int}())\n@test any(isnull, A2) == true\n\n# any(isnull, xs::NTuple)\n@test any(isnull, (DataValue(1), DataValue(2))) == false\n@test any(isnull, (DataValue{Int}(), DataValue(1), 3, 6)) == true\n\n# any(isnull, S::SubArray{T, N, U<:DataValueArray})\nA = rand(10, 3, 3)\nM = rand(Bool, 10, 3, 3)\nX = DataValueArray(A, M)\ni, j = rand(1:3), rand(1:3)\nS = view(X, :, i, j)\n\n@test any(isnull, S) == any(isnull, X[:, i, j])\nX = DataValueArray(A)\nS = view(X, :, i, j)\n@test any(isnull, S) == false\n\n\n# ----- test all(isnull, X) --------------------------------------------------#\n\n# all(isnull, X::DataValueArray)\nz = DataValueArray(Int, 10)\n@test all(isnull, z) == true\nz[1] = 10\n@test all(isnull, z) == false\n\n# all(isnull, X::AbstractArray{<:DataValue})\n@test all(isnull, DataValue{Int}[DataValue(), DataValue()]) == true\n@test all(isnull, DataValue{Int}[DataValue(1), DataValue()]) == false\n\n# all(isnull, X::Any)\n@test all(isnull, Any[DataValue(), DataValue()]) == true\n@test all(isnull, [1, 2]) == false\n@test all(isnull, 1:3) == false\n\n# ----- test Base.isnan ------------------------------------------------------#\n\nx = DataValueArray([1, 2, NaN, 4, 5, NaN, Inf, nothing], Float64, Void)\n_x = isnan(x)\n@test isequal(_x, DataValueArray([false, false, true, false,\n false, true, false, nothing], Bool, Void))\n@test _x.isnull[8] == true\n\n# ----- test Base.isfinite ---------------------------------------------------#\n\n_x = isfinite(x)\n@test isequal(_x, DataValueArray([true, true, false, true,\n true, false, false, nothing], Bool, Void))\n@test _x.isnull[8] == true\n\n# ----- test conversion methods ----------------------------------------------#\n\nu = DataValueArray(collect(1:10))\nv = DataValueArray(Int, 4, 4)\nfill!(v, 4)\nw = DataValueArray(['a', 'b', 'c', 'd', 'e', 'f', nothing], Char, Void)\nx = DataValueArray([(i, j, k) for i in 1:10, j in 1:10, k in 1:10])\ny = DataValueArray([2, 4, 6, 8, 10])\nz = DataValueArray([i*j for i in 1:10, j in 1:10])\n_z = DataValueArray(reshape(collect(1:100), 10, 10),\n convert(Array{Bool},\n reshape([mod(j, 2) for i in 1:10, j in 1:10],\n (10, 10))))\n_x = DataValueArray([false, true, false, nothing, false, true, nothing],\n Bool, Void)\na = [i*j*k for i in 1:2, j in 1:2, k in 1:2]\nb = collect(1:10)\nc = [i for i in 1:10, j in 1:10]\n\ne = convert(DataValueArray, a)\nf = convert(DataValueArray{Float64}, b)\ng = convert(DataValueArray, c)\nh = convert(DataValueArray{Float64}, g)\n\n@test_throws NullException convert(Array{Char, 1}, w)\n@test convert(Array{Char, 1},\n DataValueArray(dropnull(w))) == ['a', 'b', 'c', 'd', 'e', 'f']\n@test_throws NullException convert(Array{Char}, w)\n@test convert(Array{Float64}, u) == Float64[1, 2, 3, 4, 5, 6, 7, 8, 9, 10]\n@test convert(Vector{Float64}, y) == Float64[2, 4, 6, 8, 10]\n@test convert(Matrix{Float64}, z) == Float64[i*j for i in 1:10, j in 1:10]\n@test_throws NullException convert(Array, w)\n@test convert(Array, v) == [4 4 4 4; 4 4 4 4; 4 4 4 4; 4 4 4 4]\n@test convert(Array, z) == [i*j for i in 1:10, j in 1:10]\n@test convert(Array, u) == [1, 2, 3, 4, 5, 6, 7, 8, 9, 10]\n@test convert(Array, _x, false) == [false, true, false, false,\n false, true, false]\n@test convert(Array{Int, 1}, _x, 0) == [0, 1, 0, 0, 0, 1, 0]\n@test convert(Vector, _x, false) == [false, true, false, false,\n false, true, false]\n@test sum(convert(Matrix, _z, 0)) == 2775\n@test isequal(e[:, :, 1], DataValueArray([1 2; 2 4]))\n@test isequal(f, DataValueArray(Float64[i for i in 1:10]))\n@test isa(g, DataValueArray{Int, 2})\n@test isa(h, DataValueArray{Float64, 2})\n\n# Base.convert{T}(::Type{Vector}, X::DataValueVector{T})\nX = DataValueArray([1, 2, 3, 4, 5])\n@test convert(Vector, X) == [1, 2, 3, 4, 5]\npush!(X, DataValue())\n@test_throws NullException convert(Vector, X)\n\n# Base.convert{T}(::Type{Matrix}, X::DataValueMatrix{T})\nY = DataValueArray([1 2; 3 4; 5 6; 7 8; 9 10])\n@test convert(Matrix, Y) == [1 2; 3 4; 5 6; 7 8; 9 10]\nZ = DataValueArray([1 2; 3 4; 5 6; 7 8; 9 nothing], Int, Void)\n@test_throws NullException convert(Matrix, Z)\n\n# float(X::DataValueArray)\nA = rand(Int, 20)\nM = rand(Bool, 20)\nX = DataValueArray(A, M)\n@test isequal(float(X), DataValueArray(float(A), M))\n\n# ----- test Base.hash (julia/base/hashing.jl:5) -----------------------------#\n\n# Omitted for now, pending investigation into DataValueArray-specific\n# method.\n# TODO: reinstate testing once decision whether or not to implement\n# DataValueArray-specific hash method is reached.\n\n# ----- test unique (julia/base/set.jl:107) ----------------------------------#\n\nx = DataValueArray([1, nothing, -2, 1, nothing, 4], Int, Void)\n@assert isequal(unique(x), DataValueArray([1, nothing, -2, 4], Int, Void))\n@assert isequal(unique(reverse(x)),\n DataValueArray([4, nothing, 1, -2], Int, Void))\n\n\n\nend\n", "meta": {"hexsha": "c62f6d74c7325c9f428edf3cf199f77bcfae14e8", "size": 12038, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/primitives.jl", "max_stars_repo_name": "davidanthoff/DataArrays2.jl", "max_stars_repo_head_hexsha": "75c0a234076f977f16a931a6c9e1020a82457db0", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "test/primitives.jl", "max_issues_repo_name": "davidanthoff/DataArrays2.jl", "max_issues_repo_head_hexsha": "75c0a234076f977f16a931a6c9e1020a82457db0", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "test/primitives.jl", "max_forks_repo_name": "davidanthoff/DataArrays2.jl", "max_forks_repo_head_hexsha": "75c0a234076f977f16a931a6c9e1020a82457db0", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 33.3462603878, "max_line_length": 79, "alphanum_fraction": 0.5647948164, "num_tokens": 3996, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.46879062662624377, "lm_q2_score": 0.12085324198975819, "lm_q1q2_score": 0.05665486704219182}}
{"text": "module LP\n\nimport ..FileFormats\nimport VecMathOptInterface\nconst MOI = VecMathOptInterface\n\n# Julia 1.6 removes Grisu from Base. Previously, we went\n# print_shortest(io, x) = Base.Grisu.print_shortest(io, x)\n# To avoid adding Grisu as a dependency, use the following printing heuristic.\n# TODO(odow): consider printing 1.0 as 1.0 instead of 1, i.e., without the\n# rounding branch.\nfunction print_shortest(io::IO, x::Real)\n x_int = round(Int, x)\n if isapprox(x, x_int)\n print(io, x_int)\n else\n print(io, x)\n end\n return\nend\n\nMOI.Utilities.@model(\n Model,\n (MOI.ZeroOne, MOI.Integer),\n (MOI.EqualTo, MOI.GreaterThan, MOI.LessThan, MOI.Interval),\n (),\n (),\n (),\n (MOI.ScalarAffineFunction,),\n (),\n ()\n)\nfunction MOI.supports(\n ::Model{T},\n ::MOI.ObjectiveFunction{<:MOI.ScalarQuadraticFunction{T}},\n) where {T}\n return false\nend\n\nstruct Options\n maximum_length::Int\n warn::Bool\nend\n\nfunction get_options(m::Model)\n default_options = Options(255, false)\n return get(m.ext, :LP_OPTIONS, default_options)\nend\n\n\"\"\"\n Model(; kwargs...)\n\nCreate an empty instance of FileFormats.LP.Model.\n\nKeyword arguments are:\n\n - `maximum_length::Int=255`: the maximum length for the name of a variable.\n lp_solve 5.0 allows only 16 characters, while CPLEX 12.5+ allow 255.\n\n - `warn::Bool=false`: print a warning when variables or constraints are renamed.\n\"\"\"\nfunction Model(; maximum_length::Int = 255, warn::Bool = false)\n model = Model{Float64}()\n options = Options(maximum_length, warn)\n model.ext[:LP_OPTIONS] = options\n return model\nend\n\nfunction Base.show(io::IO, ::Model)\n print(io, \"A .LP-file model\")\n return\nend\n\n# ==============================================================================\n#\n# Base.write\n#\n# ==============================================================================\n\nconst START_REG = r\"^([\\.0-9eE])\"\nconst NAME_REG = r\"([^a-zA-Z0-9\\!\\\"\\#\\$\\%\\&\\(\\)\\/\\,\\.\\;\\?\\@\\_\\`\\'\\{\\}\\|\\~])\"\n\nfunction write_function(\n io::IO,\n model::Model,\n func::MOI.VariableIndex,\n variable_names::Dict{MOI.VariableIndex,String},\n)\n print(io, variable_names[func])\n return\nend\n\nfunction write_function(\n io::IO,\n model::Model,\n func::MOI.ScalarAffineFunction{Float64},\n variable_names::Dict{MOI.VariableIndex,String},\n)\n is_first_item = true\n if !(func.constant \u2248 0.0)\n print_shortest(io, func.constant)\n is_first_item = false\n end\n for term in func.terms\n if !(term.coefficient \u2248 0.0)\n if is_first_item\n print_shortest(io, term.coefficient)\n is_first_item = false\n else\n print(io, term.coefficient < 0 ? \" - \" : \" + \")\n print_shortest(io, abs(term.coefficient))\n end\n\n print(io, \" \", variable_names[term.variable])\n end\n end\n return\nend\n\nfunction write_constraint_suffix(io::IO, set::MOI.LessThan)\n print(io, \" <= \")\n print_shortest(io, set.upper)\n println(io)\n return\nend\n\nfunction write_constraint_suffix(io::IO, set::MOI.GreaterThan)\n print(io, \" >= \")\n print_shortest(io, set.lower)\n println(io)\n return\nend\n\nfunction write_constraint_suffix(io::IO, set::MOI.EqualTo)\n print(io, \" = \")\n print_shortest(io, set.value)\n println(io)\n return\nend\n\nfunction write_constraint_suffix(io::IO, set::MOI.Interval)\n print(io, \" <= \")\n print_shortest(io, set.upper)\n println(io)\n return\nend\n\nfunction write_constraint_prefix(io::IO, set::MOI.Interval)\n print_shortest(io, set.lower)\n print(io, \" <= \")\n return\nend\n\nwrite_constraint_prefix(io::IO, set) = nothing\n\nfunction write_constraint(\n io::IO,\n model::Model,\n index::MOI.ConstraintIndex,\n variable_names::Dict{MOI.VariableIndex,String};\n write_name::Bool = true,\n)\n func = MOI.get(model, MOI.ConstraintFunction(), index)\n set = MOI.get(model, MOI.ConstraintSet(), index)\n if write_name\n print(io, MOI.get(model, MOI.ConstraintName(), index), \": \")\n end\n write_constraint_prefix(io, set)\n write_function(io, model, func, variable_names)\n return write_constraint_suffix(io, set)\nend\n\nconst SCALAR_SETS = (\n MOI.LessThan{Float64},\n MOI.GreaterThan{Float64},\n MOI.EqualTo{Float64},\n MOI.Interval{Float64},\n)\n\nfunction write_sense(io::IO, model::Model)\n if MOI.get(model, MOI.ObjectiveSense()) == MOI.MAX_SENSE\n println(io, \"maximize\")\n else\n println(io, \"minimize\")\n end\n return\nend\n\nfunction write_objective(\n io::IO,\n model::Model,\n variable_names::Dict{MOI.VariableIndex,String},\n)\n print(io, \"obj: \")\n obj_func_type = MOI.get(model, MOI.ObjectiveFunctionType())\n obj_func = MOI.get(model, MOI.ObjectiveFunction{obj_func_type}())\n write_function(io, model, obj_func, variable_names)\n println(io)\n return\nend\n\n\"\"\"\n Base.write(io::IO, model::FileFormats.LP.Model)\n\nWrite `model` to `io` in the LP file format.\n\"\"\"\nfunction Base.write(io::IO, model::Model)\n options = get_options(model)\n FileFormats.create_unique_names(\n model,\n warn = options.warn,\n replacements = [\n s -> match(START_REG, s) !== nothing ? \"_\" * s : s,\n s -> replace(s, NAME_REG => \"_\"),\n s -> s[1:min(length(s), options.maximum_length)],\n ],\n )\n variable_names = Dict{MOI.VariableIndex,String}(\n index => MOI.get(model, MOI.VariableName(), index) for\n index in MOI.get(model, MOI.ListOfVariableIndices())\n )\n write_sense(io, model)\n write_objective(io, model, variable_names)\n println(io, \"subject to\")\n for S in SCALAR_SETS\n for index in MOI.get(\n model,\n MOI.ListOfConstraintIndices{MOI.ScalarAffineFunction{Float64},S}(),\n )\n write_constraint(\n io,\n model,\n index,\n variable_names;\n write_name = true,\n )\n end\n end\n println(io, \"Bounds\")\n free_variables = Set(keys(variable_names))\n for S in SCALAR_SETS\n for index in\n MOI.get(model, MOI.ListOfConstraintIndices{MOI.VariableIndex,S}())\n delete!(free_variables, MOI.VariableIndex(index.value))\n write_constraint(\n io,\n model,\n index,\n variable_names;\n write_name = false,\n )\n end\n end\n for index in MOI.get(\n model,\n MOI.ListOfConstraintIndices{MOI.VariableIndex,MOI.ZeroOne}(),\n )\n delete!(free_variables, MOI.VariableIndex(index.value))\n end\n for variable in sort(collect(free_variables), by = x -> x.value)\n println(io, variable_names[variable], \" free\")\n end\n for (S, str_S) in [(MOI.Integer, \"General\"), (MOI.ZeroOne, \"Binary\")]\n indices =\n MOI.get(model, MOI.ListOfConstraintIndices{MOI.VariableIndex,S}())\n if length(indices) > 0\n println(io, str_S)\n for index in indices\n write_function(\n io,\n model,\n MOI.get(model, MOI.ConstraintFunction(), index),\n variable_names,\n )\n println(io)\n end\n end\n end\n println(io, \"End\")\n return\nend\n\n# ==============================================================================\n#\n# Base.read!\n#\n# ==============================================================================\n\nfunction Base.read!(io::IO, model::Model)\n return error(\"read! is not implemented for LP files.\")\nend\n\nend\n", "meta": {"hexsha": "edefc6c13a54e231ae8c822e8eee94460c7c3652", "size": 7668, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/FileFormats/LP/LP.jl", "max_stars_repo_name": "manuelbb-upb/MathOptInterface.jl", "max_stars_repo_head_hexsha": "54b6bcb723acb2b2d79584e2f27ea56fd4c7777c", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/FileFormats/LP/LP.jl", "max_issues_repo_name": "manuelbb-upb/MathOptInterface.jl", "max_issues_repo_head_hexsha": "54b6bcb723acb2b2d79584e2f27ea56fd4c7777c", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/FileFormats/LP/LP.jl", "max_forks_repo_name": "manuelbb-upb/MathOptInterface.jl", "max_forks_repo_head_hexsha": "54b6bcb723acb2b2d79584e2f27ea56fd4c7777c", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 26.0816326531, "max_line_length": 81, "alphanum_fraction": 0.5850286907, "num_tokens": 1917, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4687906266262437, "lm_q2_score": 0.12085324198975818, "lm_q1q2_score": 0.0566548670421918}}
{"text": "module JuliaBlackBoard\n\nusing CommonMark\nusing Mustache\nusing Base64\n\n# for LaTeX\nusing Tectonic\nusing ImageMagick_jll\nusing TtH_jll\n\n\n# Exports\nexport question\nexport MC, MA, TF, ESS, ORD, MAT, FIB, FIB_PLUS, FIL, NUM, SR, OP, JUMBLED_SENTENCE #, QUIZ_BOWL\nexport Plot, File, Tikz, LaTeX, preview, LaTeX\u2032\nexport lquestion, mdquestion # tentative\nexport @ltx_str, @mdltx_str, @MT_str, @mt_str # reexoprt\n\n\ninclude(\"mustache-additions.jl\")\ninclude(\"questions.jl\")\n\nend\n", "meta": {"hexsha": "6e5b91b31c995521f4fb8871f41786c529b003eb", "size": 468, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/JuliaBlackBoard.jl", "max_stars_repo_name": "mth229/JuliaBlackBoard.jl", "max_stars_repo_head_hexsha": "66188b1aca1eb5ab8268a97dc51283019fda542c", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2020-11-10T04:33:36.000Z", "max_stars_repo_stars_event_max_datetime": "2020-11-10T04:33:36.000Z", "max_issues_repo_path": "src/JuliaBlackBoard.jl", "max_issues_repo_name": "mth229/JuliaBlackBoard.jl", "max_issues_repo_head_hexsha": "66188b1aca1eb5ab8268a97dc51283019fda542c", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/JuliaBlackBoard.jl", "max_forks_repo_name": "mth229/JuliaBlackBoard.jl", "max_forks_repo_head_hexsha": "66188b1aca1eb5ab8268a97dc51283019fda542c", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2020-11-10T04:30:39.000Z", "max_forks_repo_forks_event_max_datetime": "2020-11-10T04:30:39.000Z", "avg_line_length": 18.72, "max_line_length": 96, "alphanum_fraction": 0.7670940171, "num_tokens": 153, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.46879062662624377, "lm_q2_score": 0.12085322932404165, "lm_q1q2_score": 0.056654861104622624}}
{"text": "module data_loader\nusing CSV, DataFrames, Random\n\nfunction read_csv(file_name)\n df = CSV.read(\"data_proj_414.csv\";\n delim=\",\",\n types=Dict(\"Potter\"=>Bool,\n \"Weasley\"=>Bool,\n \"Granger\"=>Bool))\n df = DataFrame(df)\n return df\nend\n\nfunction data_split(df, proportion=0.9)\n n = nrow(df)\n idx = shuffle(1:n)\n train_idx = view(idx, 1:floor(Int, proportion*n))\n test_idx = view(idx, (floor(Int, proportion*n)+1):n)\n train_df = df[train_idx, :]\n test_df = df[test_idx, :]\n return train_df, test_df\nend\n\nfunction save_csv(df, file_name)\n CSV.write(file_name, df, delim=\"\\t\")\nend\nend # data_loader\n", "meta": {"hexsha": "c102b1caf489b42aee4797859592ce6568d2757f", "size": 633, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/data_loader.jl", "max_stars_repo_name": "JackSnowWolf/VE414_final_project", "max_stars_repo_head_hexsha": "b766b84886f41d79ff9a18e4e0f676037d1124a9", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/data_loader.jl", "max_issues_repo_name": "JackSnowWolf/VE414_final_project", "max_issues_repo_head_hexsha": "b766b84886f41d79ff9a18e4e0f676037d1124a9", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/data_loader.jl", "max_forks_repo_name": "JackSnowWolf/VE414_final_project", "max_forks_repo_head_hexsha": "b766b84886f41d79ff9a18e4e0f676037d1124a9", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2019-09-02T00:56:44.000Z", "max_forks_repo_forks_event_max_datetime": "2019-09-02T00:56:44.000Z", "avg_line_length": 22.6071428571, "max_line_length": 56, "alphanum_fraction": 0.6524486572, "num_tokens": 186, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.44939263446475963, "lm_q2_score": 0.1259227582746744, "lm_q1q2_score": 0.05658876008012503}}
{"text": "\"\"\"\nCore functionality\n\n## Basic usage\n\nThe primary export of the package is `Mat{T,N}`, which is the Julia type for\n`cv::Mat`. `Mat{T,N}` is designed to be a subtype of `AbstractArray{T,N}`. It\nhas element type (`T`) and dimension (`N`) as type parameters, whereas\n`cv::Mat` doesn't. Note that matrix construction interface is different between\nJulia and C++. `cv::Mat(3,3,CV_8U)` in C++ can be translated in Julia as\n`Mat{UInt8}(3,3)`.\n\n### Create uninitialized matrix:\n\n```julia\nusing CVCore\n\njulia> A = Mat{Float64}(3,3)\n3\u00d73 CVCore.Mat{Float64,2}:\n 3.39519e-313 4.94066e-324 2.122e-314\n NaN 1.72723e-77 -2.32036e77\n 1.97626e-323 0.0 0.0\n```\n\n### Create matrix filled with Scalar (`cv::Scalar`)\n\n```julia\njulia> A = Mat{Float64}(3,3,Scalar(1))\n3\u00d73 CVCore.Mat{Float64,2}:\n 1.0 1.0 1.0\n 1.0 1.0 1.0\n 1.0 1.0 1.0\n```\n\n### Matirx operations\n\n```julia\njulia> A * A\nCVCore.MatExpr{Float64,2}\n3\u00d73 CVCore.Mat{Float64,2}:\n 3.0 3.0 3.0\n 3.0 3.0 3.0\n 3.0 3.0 3.0\n\njulia> A + A\nCVCore.MatExpr{Float64,2}\n3\u00d73 CVCore.Mat{Float64,2}:\n 2.0 2.0 2.0\n 2.0 2.0 2.0\n 2.0 2.0 2.0\n\njulia> A - A\nCVCore.MatExpr{Float64,2}\n3\u00d73 CVCore.Mat{Float64,2}:\n 0.0 0.0 0.0\n 0.0 0.0 0.0\n 0.0 0.0 0.0\n```\n\n### Create multi-channel matrix\n\n```julia\njulia> A = Mat{Float64}(3,3,3,Scalar(1,2,3))\n3\u00d73\u00d73 CVCore.Mat{Float64,3}:\n[:, :, 1] =\n 1.0 1.0 1.0\n 1.0 1.0 1.0\n 1.0 1.0 1.0\n\n[:, :, 2] =\n 2.0 2.0 2.0\n 2.0 2.0 2.0\n 2.0 2.0 2.0\n\n[:, :, 3] =\n 3.0 3.0 3.0\n 3.0 3.0 3.0\n 3.0 3.0 3.0\n```\n\n### Conversion between `Mat{T,N}` and `Array{T,N}`\n\n```julia\njulia> B = Array(A)\n3\u00d73 Array{Float64,2}:\n 1.0 1.0 1.0\n 1.0 1.0 1.0\n 1.0 1.0 1.0\n\njulia> C = Mat(B)\n3\u00d73 CVCore.Mat{Float64,2}:\n 1.0 1.0 1.0\n 1.0 1.0 1.0\n 1.0 1.0 1.0\n```\n\nNote that `Mat(B)` where `B` is an array shares the underlying data.\n\n\"\"\"\nmodule CVCore\n\nexport AbstractCvMat, MatExpr, Mat, UMat, depth, channels, flags, dims, rows,\n cols, clone, total, isContinuous, elemSize, Scalar, TermCriteria,\n convertScaleAbs!, convertScaleAbs, addWeighted!, addWeighted\n\n#=\nNaming convention:\n 1. Cxx types should have prefix cv: e.g. cv::Mat -> cvMat\n 2. Julia types should not have prefix cv: e.g. cv::Mat -> Mat\n=#\n\nusing LibOpenCV\nusing Cxx\n\n\"\"\"\nReturns a refrence of the interal structure\n\"\"\"\nfunction handle end\n\ninclude(\"macros.jl\")\ninclude(\"const.jl\")\n\nimport Base: convert, eltype, size\n\n### Cxx types ###\n\ncvScalar_{T} = cxxt\"cv::Scalar_<$T>\"\n\nconst cvScalar = cxxt\"cv::Scalar\"\ncvScalar(v) = icxx\"return cv::Scalar($v);\"\n\nconst AbstractCvScalar = Union{cvScalar, cvScalar_}\n\ncvPoint_{T} = cxxt\"cv::Point_<$T>\"\ncvPoint_{T}(x, y) where {T} = icxx\"cv::Point_<$T>($x, $y);\"\neltype(p::cvPoint_{T}) where {T} = T\n\ncvPoint3_{T} = cxxt\"cv::Point3_<$T>\"\ncvPoint3_{T}(x, y, z) where {T} = icxx\"cv::Point3_<$T>($x, $y, $z);\"\neltype(p::cvPoint3_{T}) where {T} = T\n\nconst cvPoint = cxxt\"cv::Point\"\ncvPoint(x, y) = icxx\"cv::Point($x, $y);\"\n\nconst AbstractCvPoint = Union{cvPoint, cvPoint_}\n\ncvSize_{T} = cxxt\"cv::Size_<$T>\"\ncvSize_{T}(x, y) where {T} = icxx\"cv::Size_<$T>($x, $y);\"\neltype(s::cvSize_{T}) where {T} = T\n\nconst cvSize = cxxt\"cv::Size\"\ncvSize(x, y) = icxx\"cv::Size($x, $y);\"\n\nconst AbstractCvSize = Union{cvSize, cvSize_}\n\nheight(s::AbstractCvSize) = Int(icxx\"$s.height;\")\nwidth(s::AbstractCvSize) = Int(icxx\"$s.width;\")\narea(s::AbstractCvSize) = Int(icxx\"$s.area();\")\n\n\"\"\"Determine julia type from the depth of cv::Mat\n\"\"\"\nfunction jltype(depth)\n if depth == CV_8U\n return UInt8\n elseif depth == CV_8S\n return Int8\n elseif depth == CV_16U\n return UInt16\n elseif depth == CV_16S\n return Int16\n elseif depth == CV_32S\n return Int32\n elseif depth == CV_32F\n return Float32\n elseif depth == CV_64F\n return Float64\n else\n error(\"This shouldn't happen\")\n end\nend\n\n\"\"\"Determine cv::Mat depth from Julia type\n\"\"\"\nfunction cvdepth(T)\n if T == UInt8\n return CV_8U\n elseif T == Int8\n return CV_8S\n elseif T == UInt16\n return CV_16U\n elseif T == Int16\n return CV_16S\n elseif T == Int32\n return CV_32S\n elseif T == Float32\n return CV_32F\n elseif T == Float64\n return CV_64F\n else\n error(\"$T: not supported in cv::Mat\")\n end\nend\n\nmat_depth(flags) = flags & CV_MAT_DEPTH_MASK\nmat_channel(flags) = (flags & CV_MAT_CN_MASK) >> CV_CN_SHIFT + 1\nmaketype(depth, cn) = mat_depth(depth) + ((cn-1) << CV_CN_SHIFT)\n\n### Scalar ###\n\n# TODO: need to be subtype of cv::Vec\nconst Scalar = cvScalar_\nScalar{T}(s1=01,s2=0,s3=0,s4=0) where {T} =\n icxx\"return cv::Scalar_<$T>($s1,$s2,$s3,$s4);\"\nScalar(s1=0,s2=0,s3=0,s4=0) = Scalar{Float64}(s1,s2,s3,s4)\neltype(s::Scalar{T}) where {T} = T\n\n### cv::TermCriteria ###\n\nconst TERM_CRITERIA_COUNT = icxx\"cv::TermCriteria::COUNT;\"\nconst TERM_CRITERIA_MAX_ITER = icxx\"cv::TermCriteria::MAX_ITER;\"\nconst TERM_CRITERIA_EPS = icxx\"cv::TermCriteria::EPS;\"\n\nconst TermCriteria = cxxt\"cv::TermCriteria\"\nTermCriteria(typ, maxCount, epsilon) =\n icxx\"cv::TermCriteria($typ, $maxCount, $epsilon);\"\n\ninclude(\"mat.jl\")\n\n@gencxxf(convertScaleAbs!(src::AbstractCvMat, dst::AbstractCvMat,\n alpha=1,beta=0), \"cv::convertScaleAbs\")\nfunction convertScaleAbs(src::AbstractCvMat, alpha=1, beta=0)\n dst = similar_empty(src)\n convertScaleAbs!(src, dst, alpha, beta)\n dst\nend\n\n@gencxxf(addWeighted!(src1::AbstractCvMat, alpha, src2::AbstractCvMat, beta,\n gamma, dst::AbstractCvMat, dtype=-1), \"cv::addWeighted\")\nfunction addWeighted(src1::AbstractCvMat, alpha, src2::AbstractCvMat, beta,\n gamma, dtype=-1)\n dst = similar_empty(src)\n addWeighted!(src1, alpha, src2, beta, gamma, dst, dtype)\n dst\nend\n\nend # module\n", "meta": {"hexsha": "51c706d0aed61bdc4d3771ea507d441e0409bd72", "size": 5715, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/CVCore.jl", "max_stars_repo_name": "r9y9/CVCore.jl", "max_stars_repo_head_hexsha": "d57d9f0ba5e13a25bdb85ad4bfcacf63437b9234", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2016-09-04T04:15:15.000Z", "max_stars_repo_stars_event_max_datetime": "2016-09-04T04:15:15.000Z", "max_issues_repo_path": "src/CVCore.jl", "max_issues_repo_name": "r9y9/CVCore.jl", "max_issues_repo_head_hexsha": "d57d9f0ba5e13a25bdb85ad4bfcacf63437b9234", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 3, "max_issues_repo_issues_event_min_datetime": "2016-06-22T08:02:34.000Z", "max_issues_repo_issues_event_max_datetime": "2020-08-09T11:11:28.000Z", "max_forks_repo_path": "src/CVCore.jl", "max_forks_repo_name": "r9y9/CVCore.jl", "max_forks_repo_head_hexsha": "d57d9f0ba5e13a25bdb85ad4bfcacf63437b9234", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 2, "max_forks_repo_forks_event_min_datetime": "2017-04-14T23:56:07.000Z", "max_forks_repo_forks_event_max_datetime": "2021-10-01T18:04:06.000Z", "avg_line_length": 23.1376518219, "max_line_length": 79, "alphanum_fraction": 0.6365704287, "num_tokens": 2270, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.48047867804790706, "lm_q2_score": 0.11757212736159103, "lm_q1q2_score": 0.056490900329977424}}
{"text": "\"\"\"\n ArcEager()\n\nArc-Eager transition system for dependency parsing.\n\n# Transitions\n\n| Transition | Definition |\n|:----------- |:--------------------------------------------- |\n| LeftArc(l) | (\u03c3\\\\|s, b\\\\|\u03b2, A) \u2192 (\u03c3, b\\\\|\u03b2, A \u222a (b, l, s)) |\n| RightArc(l) | (\u03c3\\\\|s, b\\\\|\u03b2, A) \u2192 (\u03c3, b\\\\|\u03b2, A \u222a (b, l, s)) |\n| Reduce | (\u03c3\\\\|s, \u03b2, A) \u2192 (\u03c3, \u03b2, A) |\n| Shift\t | (\u03c3, b\\\\|\u03b2, A) \u2192 (\u03c3\\\\|b, \u03b2, A) |\n\n# Preconditions\n\n| Transition | Condition |\n|:----------- |:-------------------------------- |\n| LeftArc(l) | \u00ac[s = 0], \u00ac\u2203k\u2203l'[(k, l', i) \u03f5 A] |\n| RightArc(l) | \u00ac\u2203k\u2203l'[(k, l', j) \u03f5 A] |\n| Reduce | \u2203k\u2203l[(k, l, i) \u03f5 A] |\n\n# References\n\n[Nivre 2003](http://stp.lingfil.uu.se/~nivre/docs/iwpt03.pdf), [Nivre 2008](https://www.aclweb.org/anthology/J08-4003.pdf).\n\"\"\"\nstruct ArcEager <: AbstractTransitionSystem end\n\ninitconfig(::ArcEager, graph::DependencyTree) =\n ArcEagerConfig(graph)\n\ntransition_space(::ArcEager, labels=[]) =\n isempty(labels) ? [LeftArc(), RightArc(), Reduce(), Shift()] :\n [LeftArc.(labels)..., RightArc.(labels)..., Reduce(), Shift()]\n\nprojective_only(::ArcEager) = true\n\nstruct ArcEagerConfig <: AbstractParserConfiguration\n stack::Vector{Int}\n buffer::Vector{Int}\n A::Vector{Token}\nend\n\nArcEagerConfig(sentence) = stack_buffer_config(ArcEagerConfig, sentence)\n\nbuffer(cfg::ArcEagerConfig) = cfg.buffer\nstack(cfg::ArcEagerConfig) = cfg.stack\ntokens(cfg::ArcEagerConfig) = cfg.A\n\nfunction apply_transition(f, cfg::ArcEagerConfig, a...; k...)\n \u03c3, \u03b2, A = f(cfg.stack, cfg.buffer, cfg.A, a...; k...)\n return ArcEagerConfig(\u03c3, \u03b2, A)\nend\n\nleftarc(cfg::ArcEagerConfig, args...; kwargs...) =\n apply_transition(leftarc_reduce, cfg, args...; kwargs...)\n\nrightarc(cfg::ArcEagerConfig, args...; kwargs...) =\n apply_transition(rightarc_shift, cfg, args...; kwargs...)\n\nreduce(cfg::ArcEagerConfig) = apply_transition(reduce, cfg)\n\nshift(cfg::ArcEagerConfig) = apply_transition(shift, cfg)\n\nisfinal(cfg::ArcEagerConfig) = all(has_head, cfg.A)\n\nhas_head(cfg::ArcEagerConfig, k) = has_head(token(cfg, k))\n\n\"\"\"\n static_oracle(cfg::ArcEagerConfig, gold, arc=untyped)\n\nDefault static oracle function for arc-eager dependency parsing.\n\nSee [Goldberg & Nivre 2012](https://www.aclweb.org/anthology/C12-1059.pdf).\n(Also called Arc-Eager-Reduce in [Qi & Manning 2017](https://nlp.stanford.edu/pubs/qi2017arcswift.pdf)).\n\"\"\"\nfunction static_oracle(cfg::ArcEagerConfig, gold, arc=untyped)\n if stacklength(cfg) >= 1\n (\u03c3, s) = popstack(cfg)\n if bufferlength(cfg) >= 1\n (b, \u03b2) = shiftbuffer(cfg)\n has_arc(gold, b, s) && return LeftArc(arc(gold[s])...)\n has_arc(gold, s, b) && return RightArc(arc(gold[b])...)\n end\n if all(k -> k > 0 && has_head(cfg, k), [s ; deps(gold, s)])\n return Reduce()\n end\n end\n return Shift()\nend\n\n\"\"\"\n static_oracle_prefer_shift(cfg::ArcEagerConfig, tree, arc=untyped)\n\nStatic oracle for arc-eager dependency parsing. Similar to the\n\"regular\" static oracle, but always Shift when ambiguity is present.\n\nSee [Qi & Manning 2017](https://nlp.stanford.edu/pubs/qi2017arcswift.pdf).\n\"\"\"\nfunction static_oracle_prefer_shift(cfg::ArcEagerConfig, tree, arc=untyped)\n l = i -> arc(token(tree, i))\n gold_arc = (a, b) -> has_arc(tree, a, b)\n (\u03c3, s), (b, \u03b2) = popstack(cfg), shiftbuffer(cfg)\n gold_arc(b, s) && return LeftArc(l(s)...)\n gold_arc(s, b) && return RightArc(l(b)...)\n must_reduce = false\n for k in stack(cfg)\n if gold_arc(k, b) || gold_arc(b, k)\n must_reduce = true\n break\n elseif has_head(token(cfg, k), -1)\n break\n end\n end\n has_right_children = any(k -> s in rightdeps(tree, k), buffer(cfg))\n if !must_reduce || s > 0 && !has_head(cfg, s) || has_right_children\n return Shift()\n else\n return Reduce()\n end\nend\n\n\"\"\"\n dynamic_oracle(cfg::ArgEagerConfig, tree, arc=untyped)\n\nDynamic oracle function for arc-eager parsing.\n\nFor details, see [Goldberg & Nivre 2012](https://aclweb.org/anthology/C12-1059).\n\"\"\"\ndynamic_oracle(cfg::ArcEagerConfig, tree, arc=untyped) =\n filter(t -> cost(t, cfg, tree) == 0, possible_transitions(cfg, tree, arc))\n\n# see figure 2 in goldberg & nivre 2012 \"a dynamic oracle...\"\npossible_transitions(cfg::ArcEagerConfig, graph::DependencyTree, arc=untyped) =\n possible_transitions(cfg, arc)\n\nfunction possible_transitions(cfg::ArcEagerConfig, arc=untyped)\n ts = TransitionOperator[]\n if is_possible(LeftArc(), cfg)\n s = last(stack(cfg))\n push!(ts, LeftArc(arc(token(cfg, s))...))\n end\n if is_possible(RightArc(), cfg)\n b = first(buffer(cfg))\n push!(ts, RightArc(arc(token(cfg, b))...))\n end\n is_possible(Reduce(), cfg) && push!(ts, Reduce())\n is_possible(Shift(), cfg) && push!(ts, Shift())\n return ts\nend\n\nfunction cost(t::LeftArc, cfg::ArcEagerConfig, gold)\n # left arc cost: num of arcs (k,l',s), (s,l',k) s.t. k \u03f5 \u03b2\n \u03c3, s = popstack(cfg)\n b, \u03b2 = shiftbuffer(cfg)\n if has_arc(gold, b, s)\n 0\n else\n count(k -> has_arc(gold, k, s) || has_arc(gold, s, k), \u03b2)\n end\nend\n\nfunction cost(t::RightArc, cfg::ArcEagerConfig, gold)\n # right arc cost: num of gold arcs (k,l',b), s.t. k \u03f5 \u03c3 or k \u03f5 \u03b2,\n # plus num of gold arcs (b,l',k) s.t. k \u03f5 \u03c3\n \u03c3, s = popstack(cfg)\n b, \u03b2 = shiftbuffer(cfg)\n if has_arc(gold, s, b)\n 0\n else\n count(k -> has_arc(gold, k, b), [\u03c3 ; \u03b2]) + count(k -> has_arc(gold, b, k), \u03c3)\n end\nend\n\nfunction cost(t::Reduce, cfg::ArcEagerConfig, gold)\n # num of gold arcs (s,l',k) s.t. k \u03f5 b|\u03b2\n \u03c3, s = popstack(cfg)\n count(k -> has_arc(gold, s, k), buffer(cfg))\nend\n\nfunction cost(t::Shift, cfg::ArcEagerConfig, gold)\n # num of gold arcs (k,l',b), (b,l',k) s.t. k \u03f5 s|\u03c3\n b, \u03b2 = shiftbuffer(cfg)\n count(k -> has_arc(gold, k, b) || has_arc(gold, b, k), stack(cfg))\nend\n\nfunction is_possible(::LeftArc, cfg::ArcEagerConfig)\n if length(cfg.stack) > 0 && length(cfg.buffer) > 0\n s = last(stack(cfg))\n return s != 0 && !has_head(token(cfg, s))\n end\n return false\nend\n\nfunction is_possible(::RightArc, cfg::ArcEagerConfig)\n return length(cfg.stack) > 0 && length(cfg.buffer) > 0 && \n !has_head(token(cfg, first(buffer(cfg))))\nend\n\nis_possible(::Reduce, cfg::ArcEagerConfig) =\n stacklength(cfg) > 0 && has_head(token(cfg, last(stack(cfg))))\n\nis_possible(::Shift, cfg::ArcEagerConfig) = bufferlength(cfg) > 0\n\n==(cfg1::ArcEagerConfig, cfg2::ArcEagerConfig) =\n cfg1.stack == cfg2.stack && cfg1.buffer == cfg2.buffer && cfg1.A == cfg2.A\n\nBase.getindex(cfg::ArcEagerConfig, i) = token(cfg, i)\n", "meta": {"hexsha": "78791e5cc5d79bf5a99f92295f67d32d62526c0a", "size": 6781, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/transition_parsing/systems/arc_eager.jl", "max_stars_repo_name": "dellison/DependencyTrees.jl", "max_stars_repo_head_hexsha": "d5889ac8ce64f00b500bf3485507e7ab94375b3c", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 9, "max_stars_repo_stars_event_min_datetime": "2019-03-13T02:07:49.000Z", "max_stars_repo_stars_event_max_datetime": "2021-11-07T21:30:21.000Z", "max_issues_repo_path": "src/transition_parsing/systems/arc_eager.jl", "max_issues_repo_name": "dellison/DependencyTrees.jl", "max_issues_repo_head_hexsha": "d5889ac8ce64f00b500bf3485507e7ab94375b3c", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 6, "max_issues_repo_issues_event_min_datetime": "2018-09-09T22:43:46.000Z", "max_issues_repo_issues_event_max_datetime": "2020-09-19T17:10:31.000Z", "max_forks_repo_path": "src/transition_parsing/systems/arc_eager.jl", "max_forks_repo_name": "dellison/DependencyTrees.jl", "max_forks_repo_head_hexsha": "d5889ac8ce64f00b500bf3485507e7ab94375b3c", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 5, "max_forks_repo_forks_event_min_datetime": "2019-07-25T04:22:52.000Z", "max_forks_repo_forks_event_max_datetime": "2021-08-03T12:25:41.000Z", "avg_line_length": 32.4449760766, "max_line_length": 123, "alphanum_fraction": 0.602860935, "num_tokens": 2098, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.46101677931231594, "lm_q2_score": 0.1225232125181592, "lm_q1q2_score": 0.056485256826120184}}
{"text": "# !!! this script needs Julia v 0.4 or higher !!!\n# constructing:\nstart_time = time()\n# long computation\ntime_elapsed = time() - start_time\nprintln(\"Time elapsed: $time_elapsed\") #> 0.0010001659393310547\n\nd = Date(2014,9,1) #> 2014-09-01\ndt = DateTime(2014,9,1,12,30,59,1) #> 2014-09-01T12:30:59.001\n# accessors:\nDates.year(d)\nDates.month(d)\nDates.week(d)\nDates.day(d)\n# functions:\nDates.isleapyear(d)\nDates.dayofyear(d)\nDates.monthname(d)\nDates.daysinmonth(d)\n", "meta": {"hexsha": "de9a17600628a8fe09a3ff1d03e32f18743d5448", "size": 461, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Module 1/Chapter02/dates.jl", "max_stars_repo_name": "PacktPublishing/Julia-High-Performance-Programming", "max_stars_repo_head_hexsha": "861d655d163d8b87bb05478bfd255735b9263d60", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 22, "max_stars_repo_stars_event_min_datetime": "2017-02-12T15:36:27.000Z", "max_stars_repo_stars_event_max_datetime": "2022-02-28T03:30:39.000Z", "max_issues_repo_path": "Module 1/Chapter02/dates.jl", "max_issues_repo_name": "PacktPublishing/Julia-High-Performance-Programming", "max_issues_repo_head_hexsha": "861d655d163d8b87bb05478bfd255735b9263d60", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Module 1/Chapter02/dates.jl", "max_forks_repo_name": "PacktPublishing/Julia-High-Performance-Programming", "max_forks_repo_head_hexsha": "861d655d163d8b87bb05478bfd255735b9263d60", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 14, "max_forks_repo_forks_event_min_datetime": "2017-02-10T16:19:46.000Z", "max_forks_repo_forks_event_max_datetime": "2021-09-07T11:46:44.000Z", "avg_line_length": 23.05, "max_line_length": 63, "alphanum_fraction": 0.7136659436, "num_tokens": 163, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.46101677931231594, "lm_q2_score": 0.12252321251815919, "lm_q1q2_score": 0.05648525682612018}}
{"text": "\nfunction f1(x, y)\n m = 0\n n = 2\n\n if x > 10\n m = x - 10\n println(\"OK\")\n else\n m = 3 * x\n end\n\n while y < 3\n y += 1\n n = 2 * y\n end\n\n d = m + n\n return d\nend\n\nres = f1(1, 1) # should be 9\nif res < 10\n println(\"Here1\")\nelse\n println(\"Here2\")\nend\n\ns = [1,2,3,4,5,6]\n\nfor num in s\n k = num + 1\n println(k)\nend", "meta": {"hexsha": "77a562906e4021d7a8a80e3e3b9fc42ecbaa258c", "size": 334, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/blocks/blocks1.jl", "max_stars_repo_name": "judy-vscode/judy", "max_stars_repo_head_hexsha": "9bc57e1905aeba0d9a4d0b1f208bd6cfb3d9db1c", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 18, "max_stars_repo_stars_event_min_datetime": "2019-01-17T05:54:07.000Z", "max_stars_repo_stars_event_max_datetime": "2021-11-28T17:46:08.000Z", "max_issues_repo_path": "test/blocks/blocks1.jl", "max_issues_repo_name": "judy-vscode/judy", "max_issues_repo_head_hexsha": "9bc57e1905aeba0d9a4d0b1f208bd6cfb3d9db1c", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 7, "max_issues_repo_issues_event_min_datetime": "2018-12-25T11:20:16.000Z", "max_issues_repo_issues_event_max_datetime": "2021-01-29T18:49:04.000Z", "max_forks_repo_path": "test/blocks/blocks1.jl", "max_forks_repo_name": "judy-vscode/judy", "max_forks_repo_head_hexsha": "9bc57e1905aeba0d9a4d0b1f208bd6cfb3d9db1c", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 8, "max_forks_repo_forks_event_min_datetime": "2019-01-17T05:53:53.000Z", "max_forks_repo_forks_event_max_datetime": "2021-11-28T17:46:09.000Z", "avg_line_length": 9.8235294118, "max_line_length": 28, "alphanum_fraction": 0.4730538922, "num_tokens": 162, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.46101676450173545, "lm_q2_score": 0.1225232029059025, "lm_q1q2_score": 0.056485250580068806}}
{"text": "\"\"\"\n AbstractMillNode\n\nSupertype for any structure representing a data node.\n\"\"\"\nabstract type AbstractMillNode end\n\n\"\"\"\n AbstractProductNode <: AbstractMillNode\n\nSupertype for any structure representing a data node implementing a Cartesian product of data in subtrees.\n\"\"\"\nabstract type AbstractProductNode <: AbstractMillNode end\n\n\"\"\"\n AbstractBagNode <: AbstractMillNode\n\nSupertype for any data node structure representing a multi-instance learning problem.\n\"\"\"\nabstract type AbstractBagNode <: AbstractMillNode end\n\n\"\"\"\n Mill.data(n::AbstractMillNode)\n\nReturn data stored in node `n`.\n\n# Examples\n```jldoctest\njulia> Mill.data(ArrayNode([1 2; 3 4], \"metadata\"))\n2\u00d72 Matrix{Int64}:\n 1 2\n 3 4\n\njulia> Mill.data(BagNode(ArrayNode([1 2; 3 4]), bags([1:3, 4:4]), \"metadata\"))\n2\u00d72 ArrayNode{Matrix{Int64}, Nothing}:\n 1 2\n 3 4\n```\n\nSee also: [`Mill.metadata`](@ref)\n\"\"\"\ndata(n::AbstractMillNode) = getfield(n, :data)\n\n\"\"\"\n Mill.metadata(n::AbstractMillNode)\n\nReturn metadata stored in node `n`.\n\n# Examples\n```jldoctest\njulia> Mill.metadata(ArrayNode([1 2; 3 4], \"metadata\"))\n\"metadata\"\n\njulia> Mill.metadata(BagNode(ArrayNode([1 2; 3 4]), bags([1:3, 4:4]), \"metadata\"))\n\"metadata\"\n```\n\nSee also: [`Mill.data`](@ref)\n\"\"\"\nmetadata(x::AbstractMillNode) = getfield(x, :metadata)\n\n\"\"\"\n catobs(ns...)\n\nMerge multiple nodes storing samples (observations) into one suitably promoting in the process if possible.\n\nSimilar to `Base.cat` but concatenates along the abstract \\\"axis\\\" where samples are stored.\n\nIn case of repeated calls with varying number of arguments or argument types, use `reduce(catobs, [ns...])`\nto save compilation time.\n\n# Examples\n```jldoctest\njulia> catobs(ArrayNode(zeros(2, 2)), ArrayNode([1 2; 3 4]))\n2\u00d74 ArrayNode{Matrix{Float64}, Nothing}:\n 0.0 0.0 1.0 2.0\n 0.0 0.0 3.0 4.0\n\njulia> n = ProductNode((t1=ArrayNode(randn(2, 3)), t2=BagNode(ArrayNode(randn(3, 8)), bags([1:3, 4:5, 6:8]))))\nProductNode with 3 obs\n \u251c\u2500\u2500 t1: ArrayNode(2\u00d73 Array with Float64 elements)\n \u2514\u2500\u2500 t2: BagNode with 3 obs\n \u2514\u2500\u2500 ArrayNode(3\u00d78 Array with Float64 elements)\n\njulia> catobs(n[1], n[3])\nProductNode with 2 obs\n \u251c\u2500\u2500 t1: ArrayNode(2\u00d72 Array with Float64 elements)\n \u2514\u2500\u2500 t2: BagNode with 2 obs\n \u2514\u2500\u2500 ArrayNode(3\u00d76 Array with Float64 elements)\n```\n\nSee also: [`Mill.subset`](@ref).\n\"\"\"\nfunction catobs end\n\n\"\"\"\n subset(n, i)\n\nExtract a subset `i` of samples (observations) stored in node `n`.\n\nSimilar to `Base.getindex` or `MLDataPattern.getobs` but defined for all `Mill.jl` compatible data as well.\n\n# Examples\n```jldoctest\njulia> Mill.subset(ArrayNode(NGramMatrix([\"Hello\", \"world\"])), 2)\n2053\u00d71 ArrayNode{NGramMatrix{String, Vector{String}, Int64}, Nothing}:\n \"world\"\n\njulia> Mill.subset(BagNode(ArrayNode(randn(2, 8)), [1:2, 3:3, 4:7, 8:8]), 1:3)\nBagNode with 3 obs\n \u2514\u2500\u2500 ArrayNode(2\u00d77 Array with Float64 elements)\n```\n\nSee also: [`catobs`](@ref).\n\"\"\"\nfunction subset end\n\n\"\"\"\n removeinstances(n::AbstractBagNode, mask)\n\nRemove instances from `n` using `mask` and remap bag indices accordingly.\n\n# Examples\n```jldoctest\njulia> b1 = BagNode(ArrayNode([1 2 3; 4 5 6]), bags([1:2, 0:-1, 3:3]))\nBagNode with 3 obs\n \u2514\u2500\u2500 ArrayNode(2\u00d73 Array with Int64 elements)\n\njulia> b2 = removeinstances(b1, [false, true, true])\nBagNode with 3 obs\n \u2514\u2500\u2500 ArrayNode(2\u00d72 Array with Int64 elements)\n\njulia> b2.data\n2\u00d72 ArrayNode{Matrix{Int64}, Nothing}:\n 2 3\n 5 6\n\njulia> b2.bags\nAlignedBags{Int64}(UnitRange{Int64}[1:1, 0:-1, 2:2])\n```\n\"\"\"\nfunction removeinstances end\n\n\"\"\"\n dropmeta(n:AbstractMillNode)\n\nDrop metadata stored in data node `n`.\n\n# Examples\n```jldoctest\njulia> n1 = ArrayNode(NGramMatrix([\"foo\", \"bar\"]), [\"metafoo\", \"metabar\"])\n2053\u00d72 ArrayNode{NGramMatrix{String, Vector{String}, Int64}, Vector{String}}:\n \"foo\"\n \"bar\"\n\njulia> n2 = dropmeta(n1)\n2053\u00d72 ArrayNode{NGramMatrix{String, Vector{String}, Int64}, Nothing}:\n \"foo\"\n \"bar\"\n\njulia> isnothing(Mill.metadata(n2))\ntrue\n```\n\nSee also: [`Mill.metadata`](@ref).\n\"\"\"\nfunction dropmeta end\n\n\"\"\"\n mapdata(f, x)\n\nRecursively apply `f` to data in all leaves of `x`.\n\n# Examples\n```jldoctest\njulia> n1 = ProductNode((a=ArrayNode(zeros(2,2)), b=ArrayNode(ones(2,2))))\nProductNode with 2 obs\n \u251c\u2500\u2500 a: ArrayNode(2\u00d72 Array with Float64 elements)\n \u2514\u2500\u2500 b: ArrayNode(2\u00d72 Array with Float64 elements)\n\njulia> n2 = Mill.mapdata(x -> x .+ 1, n1)\nProductNode with 2 obs\n \u251c\u2500\u2500 a: ArrayNode(2\u00d72 Array with Float64 elements)\n \u2514\u2500\u2500 b: ArrayNode(2\u00d72 Array with Float64 elements)\n\njulia> Mill.data(n2).a\n2\u00d72 ArrayNode{Matrix{Float64}, Nothing}:\n 1.0 1.0\n 1.0 1.0\n\njulia> Mill.data(n2).b\n2\u00d72 ArrayNode{Matrix{Float64}, Nothing}:\n 2.0 2.0\n 2.0 2.0\n```\n\"\"\"\nmapdata(f, x) = f(x)\n\n# functions to make datanodes compatible with getindex and with MLDataPattern\nBase.getindex(x::T, i::BitArray{1}) where T <: AbstractMillNode = x[findall(i)]\nBase.getindex(x::T, i::Vector{Bool}) where T <: AbstractMillNode = x[findall(i)]\nBase.getindex(x::AbstractMillNode, i::Int) = x[i:i]\nBase.lastindex(ds::AbstractMillNode) = nobs(ds)\nMLDataPattern.getobs(x::AbstractMillNode, i) = x[i]\nMLDataPattern.getobs(x::AbstractMillNode, i, ::ObsDim.Undefined) = x[i]\nMLDataPattern.getobs(x::AbstractMillNode, i, ::ObsDim.Last) = x[i]\n\nsubset(x::AbstractMatrix, i) = x[:, i]\nsubset(x::AbstractVector, i) = x[i]\nsubset(x::AbstractMillNode, i) = x[i]\nsubset(x::DataFrame, i) = x[i, :]\nsubset(::Missing, i) = missing\nsubset(::Nothing, i) = nothing\nsubset(xs::Union{Tuple, NamedTuple}, i) = map(x -> x[i], xs)\n\ninclude(\"arraynode.jl\")\n\nStatsBase.nobs(::Missing) = nothing\n\ninclude(\"bagnode.jl\")\ninclude(\"weighted_bagnode.jl\")\nStatsBase.nobs(a::AbstractBagNode) = length(a.bags)\nStatsBase.nobs(a::AbstractBagNode, ::Type{ObsDim.Last}) = nobs(a)\nBase.ndims(x::AbstractBagNode) = Colon()\n\ninclude(\"productnode.jl\")\n\nBase.ndims(x::AbstractProductNode) = Colon()\nStatsBase.nobs(a::AbstractProductNode) = nobs(a.data[1], ObsDim.Last)\nStatsBase.nobs(a::AbstractProductNode, ::Type{ObsDim.Last}) = nobs(a)\n\ninclude(\"lazynode.jl\")\n\ncatobs(as::Maybe{ArrayNode}...) = reduce(catobs, collect(as))\ncatobs(as::Maybe{BagNode}...) = reduce(catobs, collect(as))\ncatobs(as::Maybe{WeightedBagNode}...) = reduce(catobs, collect(as))\ncatobs(as::Maybe{ProductNode}...) = reduce(catobs, collect(as))\ncatobs(as::Maybe{LazyNode}...) = reduce(catobs, collect(as))\n\nBase.cat(as::AbstractMillNode...; dims=:) = catobs(as...)\n\n# reduction of common datatypes the way we like it\nBase.reduce(::typeof(catobs), as::Vector{<:DataFrame}) = reduce(vcat, as)\nBase.reduce(::typeof(catobs), as::Vector{<:AbstractMatrix}) = reduce(hcat, as)\nBase.reduce(::typeof(catobs), as::Vector{<:AbstractVector}) = reduce(vcat, as)\nBase.reduce(::typeof(catobs), as::Vector{Missing}) = missing\nBase.reduce(::typeof(catobs), as::Vector{Nothing}) = nothing\nBase.reduce(::typeof(catobs), as::Vector{Union{Missing, Nothing}}) = nothing\n\nfunction Base.reduce(::typeof(catobs), as::Vector{Maybe{T}}) where T <: AbstractMillNode\n reduce(catobs, skipmissing(as) |> collect)\nend\n\nfunction Base.show(io::IO, @nospecialize(n::AbstractMillNode))\n print(io, nameof(typeof(n)))\n if !get(io, :compact, false)\n _show_data(IOContext(io, :compact => true), n)\n if !(n isa ArrayNode)\n print(io, \" with \", nobs(n), \" obs\")\n end\n end\nend\n\n_show_data(io, n::LazyNode{Name}) where {Name} = print(io, \"{\", Name, \"}\")\n_show_data(io, _) = print(io)\n", "meta": {"hexsha": "967054245812077f1a7b5d35de9a365d02121055", "size": 7373, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/datanodes/datanode.jl", "max_stars_repo_name": "tlauli/Mill.jl", "max_stars_repo_head_hexsha": "a76adf8f91c55a12c193be10921085bf4fd44a4e", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/datanodes/datanode.jl", "max_issues_repo_name": "tlauli/Mill.jl", "max_issues_repo_head_hexsha": "a76adf8f91c55a12c193be10921085bf4fd44a4e", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/datanodes/datanode.jl", "max_forks_repo_name": "tlauli/Mill.jl", "max_forks_repo_head_hexsha": "a76adf8f91c55a12c193be10921085bf4fd44a4e", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 27.7180451128, "max_line_length": 110, "alphanum_fraction": 0.6879153669, "num_tokens": 2367, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4073334000459302, "lm_q2_score": 0.1384617799035966, "lm_q1q2_score": 0.05640010758454325}}
{"text": "function merge_sorted_arrays(x::Vector{T}, y::Vector{T})::Vector{T} where {T}\n x_len=length(x)\n y_len=length(y)\n a,b=1,1\n i = 1\n merged_array=Vector{T}(undef, x_len + y_len)\n @inbounds while (a <= x_len) && (b <= y_len)\n if x[a] < y[b]\n merged_array[i] = x[a]\n a += 1\n i += 1\n else\n merged_array[i] = y[b]\n b += 1\n i += 1\n end\n end\n while a <= x_len\n @inbounds merged_array[i] = x[a]\n a += 1\n i += 1\n end\n while b <= y_len\n @inbounds merged_array[i] = y[b]\n b += 1\n i += 1\n end\n return merged_array\nend", "meta": {"hexsha": "fdb0c15ab9c5ff7314c0aea9c4dc815d595e423d", "size": 667, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/merge_sort.jl", "max_stars_repo_name": "Dinesh-Adhithya-H/ReadCoverageDistributions.jl", "max_stars_repo_head_hexsha": "21b059b08b6ca915fa5cda45c9928c8a17d5eb0b", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/merge_sort.jl", "max_issues_repo_name": "Dinesh-Adhithya-H/ReadCoverageDistributions.jl", "max_issues_repo_head_hexsha": "21b059b08b6ca915fa5cda45c9928c8a17d5eb0b", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 4, "max_issues_repo_issues_event_min_datetime": "2021-07-04T08:46:07.000Z", "max_issues_repo_issues_event_max_datetime": "2021-12-17T15:53:29.000Z", "max_forks_repo_path": "src/merge_sort.jl", "max_forks_repo_name": "Dinesh-Adhithya-H/ReadCoverageDistributions.jl", "max_forks_repo_head_hexsha": "21b059b08b6ca915fa5cda45c9928c8a17d5eb0b", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 23.0, "max_line_length": 77, "alphanum_fraction": 0.4527736132, "num_tokens": 215, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.5, "lm_q2_score": 0.11279540926528922, "lm_q1q2_score": 0.05639770463264461}}
{"text": "using PlotUtils\nusing Test\nusing Statistics: mean\nusing Dates\nusing Random\nusing StableRNGs\n\nrng = StableRNG(42)\n\n# TODO: real tests\n\n# ----------------------\n# colors\n\nconst C = RGBA{Float64}\nconst C0 = RGBA{PlotUtils.Colors.N0f8}\n\n@testset \"colors\" begin\n\n @test plot_color(nothing) == C(0, 0, 0, 0)\n @test plot_color(false) == C(0, 0, 0, 0)\n @test_throws ErrorException plot_color(true)\n\n @test plot_color(:red) == parse(C, :red)\n @test plot_color(\"red\") == parse(C, \"red\")\n @test_throws ErrorException plot_color(\"notacolor\")\n\n @test plot_color(colorant\"red\") == C(1, 0, 0, 1)\n\n grad = cgrad()\n @test typeof(grad) == PlotUtils.ContinuousColorGradient\n @test plot_color(grad) === grad\n\n grad = cgrad([:red, \"blue\"])\n @test color_list(grad) == C[colorant\"red\", colorant\"blue\"]\n @test grad.values == collect(range(0, stop = 1, length = 2))\n\n grad = cgrad([:red, \"blue\"], alpha = 0.5)\n @test C0.(color_list(grad)) == C0[C(1, 0, 0, 0.5), C(0, 0, 1, 0.5)]\n @test grad.values == collect(range(0, stop = 1, length = 2))\n\n grad = cgrad([:red,:blue], [0,0.1,1])\n @test length(color_list(grad)) == 3\n @test grad.values == [0,0.1,1]\n\n cs = plot_color(rand(rng, 10))\n @test typeof(cs) == Vector{C}\n @test length(cs) == 10\n\n cs = plot_color(rand(rng, 4, 4))\n @test typeof(cs) == Matrix{C}\n @test length(cs) == 16\n @test size(cs) == (4, 4)\n\n\n cs = plot_color(rand(rng, 10), 0.5)\n @test typeof(cs) == Vector{C}\n @test length(cs) == 10\n for c in cs\n @test alpha(c) == 0.5\n end\n\n cs = plot_color(rand(rng, 4, 4), 0.5)\n @test typeof(cs) == Matrix{C}\n @test length(cs) == 16\n @test size(cs) == (4, 4)\n for c in cs\n @test alpha(c) == 0.5\n end\nend\n\n# ----------------------\n# gradients\n\n@testset \"gradients\" begin\n grad = cgrad(:inferno)\n @test length(grad) == 256\n @test RGB(grad.colors[1]) == RGB(0.001462, 0.000466, 0.013866)\n @test RGB(grad.colors[end]) == RGB(0.988362, 0.998364, 0.644924)\nend\n\n# ----------------------\n# ticks\n\n# Copied from Plots.is_uniformly_spaced to avoid dependency on recent version\n# on Plots which is not used on Travis.\nfunction is_uniformly_spaced(v; tol = 1e-6)\n dv = diff(v)\n maximum(dv) - minimum(dv) < tol * mean(abs.(dv))\nend\n\n@testset \"ticks\" begin\n @test optimize_ticks(-1, 2) == ([-1.0,0.0,1.0,2.0], -1.0, 2.0)\n dt1, dt2 = Dates.value(DateTime(2000)), Dates.value(DateTime(2100))\n @test optimize_datetime_ticks(dt1, dt2) == (\n [63113990400000, 63902908800000, 64691827200000, 65480745600000],\n [\"2001-01-01\", \"2026-01-01\", \"2051-01-01\", \"2076-01-01\"])\n\n @testset \"small range\" begin\n @testset \"small range $x, $(i)\u03f5\" for x in exp10.(-12:12), i in -5:5\n y = x + i * eps(x)\n x, y = minmax(x, y)\n ticks = PlotUtils.optimize_ticks(x, y)[1]\n @test issorted(ticks)\n @test all(x .<= ticks .<= y)\n # Fails:\n # @test allunique(ticks)\n end\n end\n\n function test_ticks(x, y, ticks)\n @test issorted(ticks)\n @test all(x .<= ticks .<= y)\n if x < y\n @test length(ticks) >= 2\n @test is_uniformly_spaced(ticks)\n end\n end\n\n @testset \"fixed ranges\" begin\n @testset \"fixed range $x..$y\" for (x, y) in [(2, 14),(14, 25),(16, 36),(57, 69)]\n test_ticks(x, y, optimize_ticks(x, y)[1])\n test_ticks(-y, -x, optimize_ticks(-y, -x)[1])\n end\n end\n\n @testset \"random ranges\" begin\n r = [minmax(rand(rng, -100:100, 2)...) .* 10.0^i for _ = 1:10, i = -5:5]\n @testset \"random range $x..$y\" for (x, y) in r\n test_ticks(x, y, optimize_ticks(x, y)[1])\n end\n end\n\n # issue 86\n let x = -1.0, y = 13.0\n test_ticks(x, y, optimize_ticks(x, y, k_min = 4, k_max = 8)[1])\n end\n\n @testset \"digits\" begin\n @testset \"digits $((10^n) - 1)*10^$i\" for n in 1:9, i in -9:9\n y0 = 10^n\n x0 = y0 - 1\n x, y = (x0, y0) .* 10.0^i\n ticks = optimize_ticks(x, y)[1]\n test_ticks(x, y, ticks)\n end\n end\nend\n\n# ----------------------\n# adapted grid\n\n@testset \"adapted grid\" begin\n f = sin\n int = (0, \u03c0)\n xs, fs = adapted_grid(f, int)\n l = length(xs) - 1\n for i in 1:l\n for \u03bb in 0:0.1:1\n # test that `f` is well approximated by a line\n # in the interval `(xs[i], xs[i+1])`\n x = \u03bb * xs[i] + (1 - \u03bb) * xs[i + 1]\n y = \u03bb * fs[i] + (1 - \u03bb) * fs[i + 1]\n @test y \u2248 f(x) atol = 1e-2\n end\n end\n\n int = (2, 2)\n xs, fs = adapted_grid(f, int)\n @test xs == [2]\n @test fs == [f(2)]\n\n int = (2, 1)\n @test_throws ArgumentError adapted_grid(f, int)\nend\n\n@testset \"zscale\" begin\n bkg = 30 .* randn(rng, 8192) .+ 1000\n data = bkg .+ 100 .* randn(rng, 8192) .+ 2500\n defects = rand(rng, CartesianIndices(bkg), 500)\n data[defects] .= rand(rng, [0, 1e7], 500)\n cmin, cmax = zscale(data)\n # values calculated using IRAF\n @test cmin \u2248 2841.586 atol = 1e-3\n @test cmax \u2248 4171.648 atol = 1e-3\n @test cmin > minimum(data)\n @test cmax < maximum(data)\n\n data = vcat(1:100)\n cmin, cmax = zscale(data)\n @test cmin == 1\n @test cmax == 100\n\n # Make sure output is finite\n data = vcat(0:999, NaN)\n cmin, cmax = zscale(data)\n @test cmin == 0\n @test cmax == 999\nend\n", "meta": {"hexsha": "2a598a9c81e3f5a97ea68877d5928584f71bdbb1", "size": 5440, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/runtests.jl", "max_stars_repo_name": "t-bltg/PlotUtils.jl", "max_stars_repo_head_hexsha": "4007370cd3c3b9a6cdc62871687dac21364190ff", "max_stars_repo_licenses": ["Apache-2.0", "CC0-1.0"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "test/runtests.jl", "max_issues_repo_name": "t-bltg/PlotUtils.jl", "max_issues_repo_head_hexsha": "4007370cd3c3b9a6cdc62871687dac21364190ff", "max_issues_repo_licenses": ["Apache-2.0", "CC0-1.0"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "test/runtests.jl", "max_forks_repo_name": "t-bltg/PlotUtils.jl", "max_forks_repo_head_hexsha": "4007370cd3c3b9a6cdc62871687dac21364190ff", "max_forks_repo_licenses": ["Apache-2.0", "CC0-1.0"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 27.2, "max_line_length": 88, "alphanum_fraction": 0.5380514706, "num_tokens": 1947, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.5, "lm_q2_score": 0.11279539882690352, "lm_q1q2_score": 0.05639769941345176}}
{"text": "import Base: ==, length, in, eltype;\n\nexport Problem, InstrumentedProblem,\n actions, get_result, goal_test, path_cost, value,\n format_instrumented_results,\n Node, expand, child_node, solution, path, ==,\n search,\n GAState, mate, mutate,\n tree_search, graph_search,\n breadth_first_tree_search, depth_first_tree_search, depth_first_graph_search,\n breadth_first_search, best_first_graph_search, uniform_cost_search,\n recursive_dls, depth_limited_search, iterative_deepening_search,\n greedy_best_first_graph_search,\n Graph, make_undirected, connect_nodes, get_linked_nodes, get_nodes,\n UndirectedGraph, RandomGraph,\n GraphProblem,\n astar_search, recursive_best_first_search,\n hill_climbing, exp_schedule, simulated_annealing,\n or_search, and_search, and_or_graph_search,\n OnlineDFSAgentProgram, update_state, execute,\n OnlineSearchProblem, LRTAStarAgentProgram,\n learning_realtime_astar_cost,\n genetic_search, genetic_algorithm,\n NQueensProblem, conflict, conflicted,\n random_boggle, print_boggle, boggle_neighbors, int_sqrt,\n WordList, lookup, length, in,\n BoggleFinder, set_board, find, words, score,\n boggle_hill_climbing, mutate_boggle,\n execute_searcher, compare_searchers, beautify_node;\n\n#=\n\n Problem is a abstract problem that contains a initial state and goal state.\n\n=#\nmutable struct Problem <: AbstractProblem\n initial::String\n goal::Union{Nothing, String}\n\n function Problem(initial_state::String; goal_state::Union{Nothing, String}=nothing)\n return new(initial_state, goal_state);\n end\nend\n\n\"\"\"\n actions(ap::T, state::String) where {T <: AbstractProblem}\n\nReturn an array of possible actions that can be executed in the given state 'state'.\n\"\"\"\nfunction actions(ap::T, state::String) where {T <: AbstractProblem}\n println(\"actions() is not implemented yet for \", typeof(ap), \"!\");\n nothing;\nend\n\n\"\"\"\n get_result(ap::T, state::String, action::String) where {T <: AbstractProblem}\n\nReturn the resulting state from executing the given action 'action' in the given state 'state'.\n\"\"\"\nfunction get_result(ap::T, state::String, action::String) where {T <: AbstractProblem}\n println(\"get_result() is not implemented yet for \", typeof(ap), \"!\");\n nothing;\nend\n\n\"\"\"\n goal_test(ap::T, state::String) where {T <: AbstractProblem}\n\nReturn a boolean value representing whether the given state 'state' is a goal state in the given\nproblem 'ap'.\n\"\"\"\nfunction goal_test(ap::T, state::String) where {T <: AbstractProblem}\n return ap.goal == state;\nend\n\n\"\"\"\n path_cost(ap::T, cost::Float64, state1::String, action::String, state2::String) where {T <: AbstractProblem}\n path_cost(ap::T, cost::Float64, state1::AbstractVector, action::Int64, state2::AbstractVector) where {T <: AbstractProblem}\n\nReturn the cost of a solution path arriving at 'state2' from 'state1' with the given action 'action' and\ncost 'cost' to arrive at 'state1'. The default path_cost() method costs 1 for every step in a path.\n\"\"\"\nfunction path_cost(ap::T, cost::Float64, state1::String, action::String, state2::String) where {T <: AbstractProblem}\n return cost + 1;\nend\n\nfunction path_cost(ap::T, cost::Float64, state1::AbstractVector, action::Int64, state2::AbstractVector) where {T <: AbstractProblem}\n return cost + 1;\nend\n\n\"\"\"\n value(ap::T, state::String) where {T <: AbstractProblem}\n\nReturn a value for the given state 'state' in the given problem 'ap'.\n\nThis value is used in optimization problems such as hill climbing or simulated annealing.\n\"\"\"\nfunction value(ap::T, state::String) where {T <: AbstractProblem}\n println(\"value() is not implemented yet for \", typeof(ap), \"!\");\n nothing;\nend\n\n#=\n\n InstrumentedProblem is a AbstractProblem implementation that wraps another AbstractProblem\n\n implementation and tracks the number of function calls made. This problem is used in\n\n compare_searchers() and execute_searcher().\n\n=#\nmutable struct InstrumentedProblem <: AbstractProblem\n problem::AbstractProblem\n actions::Int64\n results::Int64\n goal_tests::Int64\n found # can be any DataType, but check for Nothing DataType later\n\n function InstrumentedProblem(ap::T) where {T <: AbstractProblem}\n return new(ap, Int64(0), Int64(0), Int64(0), nothing);\n end\nend\n\nfunction actions(ap::InstrumentedProblem, state::AbstractVector)\n ap.actions = ap.actions + 1;\n return actions(ap.problem, state);\nend\n\nfunction actions(ap::InstrumentedProblem, state::String)\n ap.actions = ap.actions + 1;\n return actions(ap.problem, state);\nend\n\nfunction get_result(ap::InstrumentedProblem, state::String, action::String)\n ap.results = ap.results + 1;\n return get_result(ap.problem, state, action);\nend\n\nfunction get_result(ap::InstrumentedProblem, state::AbstractVector, action::Int64)\n ap.results = ap.results + 1;\n return get_result(ap.problem, state, action);\nend\n\nfunction goal_test(ap::InstrumentedProblem, state::String)\n ap.goal_tests = ap.goal_tests + 1;\n local result::Bool = goal_test(ap.problem, state);\n if (result)\n ap.found = state;\n end\n return result;\nend\n\nfunction goal_test(ap::InstrumentedProblem, state::AbstractVector)\n ap.goal_tests = ap.goal_tests + 1;\n local result::Bool = goal_test(ap.problem, state);\n if (result)\n ap.found = state;\n end\n return result;\nend\n\nfunction path_cost(ap::InstrumentedProblem, cost::Float64, state1::String, action::String, state2::String)\n return path_cost(ap.problem, cost, state1, action, state2);\nend\n\nfunction path_cost(ap::InstrumentedProblem, cost::Float64, state1::AbstractVector, action::Int64, state2::AbstractVector)\n return path_cost(ap.problem, cost, state1, action, state2);\nend\n\nfunction value(ap::InstrumentedProblem, state::String)\n return value(ap.problem, state);\nend\n\nfunction value(ap::InstrumentedProblem, state::AbstractVector)\n return value(ap.problem, state);\nend\n\nfunction format_instrumented_results(ap::InstrumentedProblem)\n return @sprintf(\"<%4d/%4d/%4d/%s>\", ap.actions, ap.goal_tests, ap.results, string(ap.found));\nend\n\n# A node should not exist without a state.\nmutable struct Node{T}\n state::T\n path_cost::Float64\n depth::UInt32\n action::Union{Nothing, String, Int64, Tuple}\n parent::Union{Nothing, Node}\n f::Float64\n\n function Node{T}(state::T; parent::Union{Nothing, Node}=nothing, action::Union{Nothing, String, Int64, Tuple}=nothing, path_cost::Float64=0.0, f::Union{Nothing, Float64}=nothing) where T\n nn = new(state, path_cost, UInt32(0), action, parent);\n if (typeof(parent) <: Node)\n nn.depth = UInt32(parent.depth + 1);\n end\n if (typeof(f) <: Float64)\n nn.f = f;\n end\n return nn;\n end\nend\n\n\"\"\"\n expand(n::Node, ap::T) where {T <: AbstractProblem}\n\nReturn an array of nodes reachable by 1 step from the given node 'n' in the problem 'ap'.\n\"\"\"\nfunction expand(n::Node, ap::T) where {T <: AbstractProblem}\n return collect(child_node(n, ap, act) for act in actions(ap, n.state));\nend\n\n\"\"\"\n child_node(n::Node, ap::T, action::String) where {T <: AbstractProblem}\n\nReturn a child node for the given node 'n' in problem 'ap' after executing the action 'action' (Fig. 3.10).\n\"\"\"\nfunction child_node(n::Node, ap::T, action::String) where {T <: AbstractProblem}\n local next_node = get_result(ap, n.state, action);\n return Node{typeof(next_node)}(next_node, parent=n, action=action, path_cost=path_cost(ap, n.path_cost, n.state, action, next_node));\nend\n\nfunction child_node(n::Node, ap::T, action::Int64) where {T <: AbstractProblem}\n local next_node = get_result(ap, n.state, action);\n return Node{typeof(next_node)}(next_node, parent=n, action=action, path_cost=path_cost(ap, n.path_cost, n.state, action, next_node));\nend\n\nfunction child_node(n::Node, ap::T, action::Tuple) where {T <: AbstractProblem}\n local next_node = get_result(ap, n.state, action);\n return Node{typeof(next_node)}(next_node, parent=n, action=action, path_cost=path_cost(ap, n.path_cost, n.state, action, next_node));\nend\n\n\"\"\"\n solution(n::Node)\n\nReturn an array of actions to get from the root node of node 'n' to the given node 'n'.\n\"\"\"\nfunction solution(n::Node)\n local path_sequence = path(n);\n return [node.action for node in path_sequence[2:length(path_sequence)]];\nend\n\n\"\"\"\n path(n::Node)\n\nReturn the path between the root node of node 'n' to the given node 'n' as an array of nodes.\n\"\"\"\nfunction path(n::Node)\n local node = n;\n local path_back = [];\n while true\n push!(path_back, node);\n if (!(node.parent === nothing))\n node = node.parent;\n else\n # The root node does not have a parent node.\n break;\n end\n end\n path_back = reverse(path_back);\n return path_back;\nend\n\nfunction ==(n1::Node, n2::Node)\n return (n1.state == n2.state);\nend\n\n#=\n\n SimpleProblemSolvingAgentProgram is a abstract problem solving agent (Fig. 3.1).\n\n=#\nmutable struct SimpleProblemSolvingAgentProgram <: AgentProgram\n state::Union{Nothing, String}\n goal::Union{Nothing, String}\n seq::Array{String, 1}\n problem::Union{Nothing, Problem}\n\n function SimpleProblemSolvingAgentProgram(;initial_state::Union{Nothing, String}=nothing)\n return new(initial_state, nothing, Array{String, 1}(), nothing);\n end\nend\n\nfunction execute(spsap::SimpleProblemSolvingAgentProgram, percept::Tuple{Any, Any})\n spsap.state = update_state(spsap, spsap.state, percept);\n if (length(spsap.seq) == 0)\n spsap.goal = formulate_problem(spsap, spsap.state);\n spsap.problem = forumate_problem(spsap, spsap.state, spsap.goal);\n spsap.seq = search(spsap, spsap.problem);\n if (length(spsap.seq) == 0)\n return Nothing;\n end\n end\n local action = popfirst!(spsap.seq);\n return action;\nend\n\nfunction update_state(spsap::SimpleProblemSolvingAgentProgram, state::String, percept::Tuple{Any, Any})\n println(\"update_state() is not implemented yet for \", typeof(spsap), \"!\");\n nothing;\nend\n\nfunction formulate_goal(spsap::SimpleProblemSolvingAgentProgram, state::String)\n println(\"formulate_goal() is not implemented yet for \", typeof(spsap), \"!\");\n nothing;\nend\n\nfunction formulate_problem(spsap::SimpleProblemSolvingAgentProgram, state::String, goal::String)\n println(\"formulate_problem() is not implemented yet for \", typeof(spsap), \"!\");\n nothing;\nend\n\nfunction search(spsap::SimpleProblemSolvingAgentProgram, problem::T) where {T <: AbstractProblem}\n println(\"search() is not implemented yet for \", typeof(spsap), \"!\");\n nothing;\nend\n\nstruct GAState\n genes::Array{Any, 1}\n\n function GAState(genes::Array{Any, 1})\n return new(Array{Any,1}(deepcopy(genes)));\n end\nend\n\nfunction mate(ga_state::T, other::T) where {T <: GAState}\n local c = rand(RandomDeviceInstance, range(1, stop=length(ga_state.genes)));\n local new_ga_state = deepcopy(ga_state[1:c]);\n for element in other.genes[(c + 1):length(other.genes)]\n push!(new_ga_state, element);\n end\n return new_ga_state;\nend\n\nfunction mutate(ga_state::T) where {T <: GAState}\n println(\"mutate() is not implemented yet for \", typeof(ga_state), \"!\");\n nothing;\nend\n\n\"\"\"\n tree_search{T1 <: AbstractProblem, T2 <: Queue}(problem::T1, frontier::T2)\n\nSearch the given problem by using the general tree search algorithm (Fig. 3.7) and return the node solution.\n\"\"\"\nfunction tree_search(problem::T1, frontier::T2) where {T1 <: AbstractProblem, T2 <: Queue}\n push!(frontier, Node{typeof(problem.initial)}(problem.initial));\n while (length(frontier) != 0)\n local node = pop!(frontier);\n if (goal_test(problem, node.state))\n return node;\n end\n extend!(frontier, expand(node, problem));\n end\n return nothing;\nend\n\nfunction tree_search(problem::InstrumentedProblem, frontier::T) where {T <: Queue}\n push!(frontier, Node{typeof(problem.problem.initial)}(problem.problem.initial));\n while (length(frontier) != 0)\n local node = pop!(frontier);\n if (goal_test(problem, node.state))\n return node;\n end\n extend!(frontier, expand(node, problem));\n end\n return nothing;\nend\n\n\"\"\"\n graph_search{T1 <: AbstractProblem, T2 <: Queue}(problem::T1, frontier::T2)\n\nSearch the given problem by using the general graph search algorithm (Fig. 3.7) and return the node solution.\n\nThe uniform cost algorithm (Fig. 3.14) should be used when the frontier is a priority queue.\n\"\"\"\nfunction graph_search(problem::T1, frontier::T2) where {T1 <: AbstractProblem, T2 <: Queue}\n local explored::Set;\n if (typeof(problem.initial) <: Tuple)\n explored = Set{NTuple}();\n else\n explored = Set{typeof(problem.initial)}();\n end\n push!(frontier, Node{typeof(problem.initial)}(problem.initial));\n while (length(frontier) != 0)\n local node = pop!(frontier);\n if (goal_test(problem, node.state))\n return node;\n end\n push!(explored, node.state);\n extend!(frontier, collect(child_node for child_node in expand(node, problem)\n if (!(child_node.state in explored) && !(child_node in frontier))));\n end\n return nothing;\nend\n\nfunction graph_search(problem::InstrumentedProblem, frontier::T) where {T <: Queue}\n local explored::Set;\n if (typeof(problem.problem.initial) <: Tuple)\n explored = Set{NTuple}();\n else\n explored = Set{typeof(problem.problem.initial)}();\n end\n push!(frontier, Node{typeof(problem.problem.initial)}(problem.problem.initial));\n while (length(frontier) != 0)\n local node = pop!(frontier);\n if (goal_test(problem, node.state))\n return node;\n end\n push!(explored, node.state);\n extend!(frontier, collect(child_node for child_node in expand(node, problem)\n if (!(child_node.state in explored) && !(child_node in frontier))));\n end\n return nothing;\nend\n\n\"\"\"\n breadth_first_tree_search(problem::T) where {T <: AbstractProblem}\n\nSearch the shallowest nodes in the search tree first.\n\"\"\"\nfunction breadth_first_tree_search(problem::T) where {T <: AbstractProblem}\n return tree_search(problem, FIFOQueue());\nend\n\n\"\"\"\n depth_first_tree_search(problem::T) where {T <: AbstractProblem}\n\nSearch the deepest nodes in the search tree first.\n\"\"\"\nfunction depth_first_tree_search(problem::T) where {T <: AbstractProblem}\n return tree_search(problem, Stack());\nend\n\n\"\"\"\n depth_first_graph_search(problem::T) where {T <: AbstractProblem}\n\nSearch the deepest nodes in the search tree first.\n\"\"\"\nfunction depth_first_graph_search(problem::T) where {T <: AbstractProblem}\n return graph_search(problem, Stack());\nend\n\n\"\"\"\n breadth_first_search(problem::T) where {T <: AbstractProblem}\n breadth_first_search(problem::InstrumentedProblem)\n\nReturn a solution by using the breadth-first search algorithm (Fig. 3.11)\non the given problem 'problem'. Otherwise, return 'nothing' on failure.\n\"\"\"\nfunction breadth_first_search(problem::T) where {T <: AbstractProblem}\n local node = Node{typeof(problem.initial)}(problem.initial);\n if (goal_test(problem, node.state))\n return node;\n end\n local frontier = FIFOQueue();\n push!(frontier, node);\n local explored = Set{String}();\n while (length(frontier) != 0)\n node = pop!(frontier);\n push!(explored, node.state);\n for child_node in expand(node, problem)\n if (!(child_node.state in explored) && !(child_node in frontier))\n if (goal_test(problem, child_node.state))\n return child_node;\n end\n push!(frontier, child_node);\n end\n end\n end\n return nothing;\nend\n\nfunction breadth_first_search(problem::InstrumentedProblem)\n local node = Node{typeof(problem.problem.initial)}(problem.problem.initial);\n if (goal_test(problem, node.state))\n return node;\n end\n local frontier = FIFOQueue();\n push!(frontier, node);\n local explored = Set{String}();\n while (length(frontier) != 0)\n node = pop!(frontier);\n push!(explored, node.state);\n for child_node in expand(node, problem)\n if (!(child_node.state in explored) && !(child_node in frontier))\n if (goal_test(problem, child_node.state))\n return child_node;\n end\n push!(frontier, child_node);\n end\n end\n end\n return nothing;\nend\n\n\"\"\"\n best_first_graph_search(problem::T, f::Function) where {T <: AbstractProblem}\n\nSearch the nodes in the given problem 'problem' by visiting the nodes with the lowest\nscores returned by f(). If f() is a heuristics estimate function to the goal state, then\nthis function becomes greedy best first search. If f() is a function that gets the node's\ndepth, then this function becomes breadth-first search.\n\nReturns a solution if found, otherwise returns 'nothing' on failure.\n\nThis function uses f as a Function, because using f as an MemoizedFunction exhibits unusual\nbehavior when relying on MemoizedFunction by producing unexpected results.\n\"\"\"\nfunction best_first_graph_search(problem::T, f::Function) where {T <: AbstractProblem}\n local node = Node{typeof(problem.initial)}(problem.initial);\n if (goal_test(problem, node.state))\n return node;\n end\n local frontier = PQueue();\n push!(frontier, node, f);\n local explored = Set{typeof(problem.initial)}();\n while (length(frontier) != 0)\n node = pop!(frontier);\n if (goal_test(problem, node.state))\n return node;\n end\n push!(explored, node.state);\n for child_node in expand(node, problem)\n if (!(child_node.state in explored) &&\n !(child_node in collect(getindex(x, 2) for x in frontier.array)))\n push!(frontier, child_node, f);\n elseif (child_node in [getindex(x, 2) for x in frontier.array])\n # Recall that Nodes can share the same state and different values for other fields.\n local existing_node = pop!(collect(getindex(x, 2)\n for x in frontier.array\n if (getindex(x, 2) == child_node)));\n if (f(child_node) < f(existing_node))\n delete!(frontier, existing_node);\n push!(frontier, child_node, f);\n end\n end\n end\n end\n return nothing;\nend\n\n\n\"\"\"\n uniform_cost_search(problem::T) where {T <: AbstractProblem}\n\nSearch the given problem by using the uniform cost algorithm (Fig. 3.14) and return the node solution.\n\nsolution() can be used on the node solution to reconstruct the path taken to the solution.\n\"\"\"\nfunction uniform_cost_search(problem::T) where {T <: AbstractProblem}\n return best_first_graph_search(problem, (function(n::Node)return n.path_cost;end));\nend\n\nfunction recursive_dls(node::Node, problem::T, limit::Int64) where {T <: AbstractProblem}\n if (goal_test(problem, node.state))\n return node;\n elseif (node.depth == limit)\n return \"cutoff\";\n else\n local cutoff_occurred = false;\n for child_node in expand(node, problem)\n local result = recursive_dls(child_node, problem, limit);\n if (result == \"cutoff\")\n cutoff_occurred = true;\n elseif (!(typeof(result) <: Nothing))\n return result;\n end\n end\n return if_(cutoff_occurred, \"cutoff\", nothing);\n end\nend;\n\n\"\"\"\n depth_limited_search(problem::T; limit::Int64) where {T <: AbstractProblem}\n\nSearch the given problem by using the depth limited tree search algorithm (Fig. 3.17)\nand return the node solution if a solution was found. Otherwise, this function returns 'nothing'.\n\nsolution() can be used on the node solution to reconstruct the path taken to the solution.\n\"\"\"\nfunction depth_limited_search(problem::T; limit::Int64=50) where {T <: AbstractProblem}\n return recursive_dls(Node{typeof(problem.initial)}(problem.initial), problem, limit);\nend\n\nfunction depth_limited_search(problem::InstrumentedProblem; limit::Int64=50)\n return recursive_dls(Node{typeof(problem.problem.initial)}(problem.problem.initial), problem, limit);\nend\n\n\"\"\"\n iterative_deepening_search(problem::T) where {T <: AbstractProblem}\n\nSearch the given problem by using the iterative deepening search algorithm (Fig. 3.18)\nand return the node solution if a solution was found. Otherwise, this function returns 'nothing'.\n\nsolution() can be used on the node solution to reconstruct the path taken to the solution.\n\"\"\"\nfunction iterative_deepening_search(problem::T) where {T <: AbstractProblem}\n for depth in 1:typemax(Int64)\n local result = depth_limited_search(problem, limit=depth)\n if (result != \"cutoff\")\n return result;\n end\n end\n return nothing;\nend\n\nconst greedy_best_first_graph_search = best_first_graph_search;\n\n#=\n\n Graph is a graph that consists of nodes (vertices) and edges (links).\n\n The Graph constructor uses the keyword 'directed' to specify if the graph\n\n is directed or undirected.\n\n\n For an example Graph instance:\n\n Graph(dict=Dict([(\"A\", Dict([(\"B\", 1), (\"C\", 2)]))]))\n\n The example Graph is a directed graph with 3 vertices (\"A\", \"B\", and \"C\")\n\n and link \"A\"=>\"B\" (length 1) and \"A\"=>\"C\" (length 2).\n\n=#\nstruct Graph{N}\n dict::Dict{N, Any}\n locations::Dict{N, Tuple{Any, Any}}\n directed::Bool\n\n function Graph{N}(;dict::Union{Nothing, Dict{N, }}=nothing, locations::Union{Nothing, Dict{N, Tuple{Any, Any}}}=nothing, directed::Bool=true) where N\n local ng::Graph;\n if ((typeof(dict) <: Nothing) && (typeof(locations) <: Nothing))\n ng = new(Dict{Any, Any}(), Dict{Any, Tuple{Any, Any}}(), Bool(directed));\n elseif (typeof(locations) <: Nothing)\n ng = new(Dict{eltype(dict.keys), Any}(dict), Dict{Any, Tuple{Any, Any}}(), Bool(directed));\n else\n ng = new(Dict{eltype(dict.keys), Any}(dict), Dict{eltype(locations.keys), Tuple{Any, Any}}(locations), Bool(directed));\n end\n if (!ng.directed)\n make_undirected(ng);\n end\n return ng;\n end\n\n function Graph{N}(graph::Graph{N}) where N\n return new(Dict{Any, Any}(graph.dict), Dict{String, Tuple{Any, Any}}(graph.locations), Bool(graph.directed));\n end\nend\n\neltype(::Type{<:Graph{T}}) where {T} = T\n\nfunction make_undirected(graph::Graph)\n for location_A in keys(graph.dict)\n for (location_B, d) in graph.dict[location_A]\n connect_nodes(graph, location_B, location_A, distance=d);\n end\n end\nend\n\n\"\"\"\n connect_nodes(graph::Graph{N}, A::N, B::N; distance::Int64=Int64(1)) where N\n\nAdd a link between Node 'A' to Node 'B'. If the graph is undirected, then add\nthe inverse link from Node 'B' to Node 'A'.\n\"\"\"\nfunction connect_nodes(graph::Graph{N}, A::N, B::N; distance::Int64=Int64(1)) where N\n get!(graph.dict, A, Dict{String, Int64}())[B]=distance;\n if (!graph.directed)\n get!(graph.dict, B, Dict{String, Int64}())[A]=distance;\n end\n nothing;\nend\n\n\"\"\"\n get_linked_nodes(graph::Graph{N}, a::N; b::Union{Nothing, N}=nothing) where N\n\nReturn a dictionary of nodes and their distances if the 'b' keyword is not given.\nOtherwise, return the distance between 'a' and 'b'.\n\"\"\"\nfunction get_linked_nodes(graph::Graph{N}, a::N; b::Union{Nothing, N}=nothing) where N\n local linked = get!(graph.dict, a, Dict{Any, Any}());\n if (typeof(b) <: Nothing)\n return linked;\n else\n return get(linked, b, nothing);\n end\nend\n\nfunction get_nodes(graph::Graph)\n return collect(keys(graph.dict));\nend\n\n\"\"\"\n UndirectedGraph(dict::Dict{T, }, locations::Dict{T, Tuple{Any, Any}}) where T\n UndirectedGraph()\n\nReturn an undirected graph from the given dictionary of links 'dict' and dictionary \nof locations 'locations' if given.\n\"\"\"\nfunction UndirectedGraph(dict::Dict{T, }, locations::Dict{T, Tuple{Any, Any}}) where T\n return Graph{eltype(dict.keys)}(dict=dict, locations=locations, directed=false);\nend\n\nfunction UndirectedGraph()\n return Graph{Any}(directed=false);\nend\n\n\"\"\"\n RandomGraph()\n\nReturn a random graph with the specified nodes and number of links.\n\"\"\"\nfunction RandomGraph(;nodes::UnitRange=1:10,\n min_links::Int64=2,\n width::Int64=400,\n height::Int64=300,\n curvature::Function=(function()\n return (0.4*rand(RandomDeviceInstance)) + 1.1;\n end))\n local g = UndirectedGraph();\n for node in nodes\n g.locations[node] = Tuple((rand(RandomDeviceInstance, 1:width), rand(RandomDeviceInstance, 1:height)));\n end\n for i in 1:min_link\n for node in nodes\n if (get_linked_nodes(g, node) < min_links)\n local here = g.locations[node];\n local neighbor = argmin(nodes, (function(n, ; graph::Graph=g, current_node::Node=node, current_location::Tuple=here)\n if (n == current_node || get_linked_nodes(graph, current_node, n) != nothing)\n return Inf;\n end\n return distance(g.locations[n], current_location);\n end));\n local d = distance(g.locations[neighbor], here) * curvature();\n connect(g, node, neighbor, Int64(floor(d)));\n end\n end\n end\n return g;\nend\n\n#=\n\n GraphProblem is the problem of searching a graph from one node to another node.\n\n=#\nstruct GraphProblem <: AbstractProblem\n initial::String\n goal::String\n graph::Graph\n h::MemoizedFunction\n\n\n function GraphProblem(initial_state::String, goal_state::String, graph::Graph)\n return new(initial_state, goal_state, Graph{eltype(graph)}(graph), MemoizedFunction(initial_to_goal_distance));\n end\nend\n\nfunction actions(gp::GraphProblem, loc::String)\n return collect(keys(get_linked_nodes(gp.graph,loc)));\nend\n\nfunction get_result(gp::GraphProblem, state::String, action::String)\n return action;\nend\n\nfunction path_cost(gp::GraphProblem, current_cost::Float64, location_A::String, action::String, location_B::String)\n local AB_distance::Float64;\n if (haskey(gp.graph.dict, location_A) && haskey(gp.graph.dict[location_A], location_B))\n AB_distance= Float64(get_linked_nodes(gp.graph,location_A, b=location_B));\n else\n AB_distance = Float64(Inf);\n end\n return current_cost + AB_distance;\nend\n\n\"\"\"\n initial_to_goal_distance(gp::GraphProblem, n::Node)\n\nCompute the straight line distance between the initial state and goal state.\n\"\"\"\nfunction initial_to_goal_distance(gp::GraphProblem, n::Node)\n local locations = gp.graph.locations;\n if (isempty(locations))\n return Inf;\n else\n return Float64(floor(distance(locations[n.state], locations[gp.goal])));\n end\nend\n\nfunction initial_to_goal_distance(gp::InstrumentedProblem, n::Node)\n local locations = gp.problem.graph.locations;\n if (isempty(locations))\n return Inf;\n else\n return Float64(floor(distance(locations[n.state], locations[gp.problem.goal])));\n end\nend\n\n\"\"\"\n astar_search(problem::GraphProblem; h::Union{Nothing, Function}=nothing)\n\nApply the A* search (best-first graph search with f(n)=g(n)+h(n)) to the given problem 'problem'.\nIf the 'h' keyword is not used, this function uses the function problem.h.\n\nThis function uses mh as a Function, because using mh as an MemoizedFunction exhibits unusual\nbehavior when relying on MemoizedFunction by producing unexpected results.\n\"\"\"\nfunction astar_search(problem::GraphProblem; h::Union{Nothing, Function}=nothing)\n local mh::Function;\n if (!(typeof(h) <: Nothing))\n mh = h;\n else\n mh = problem.h.f;\n end\n return best_first_graph_search(problem,\n (function(node::Node; h::Function=mh, prob::GraphProblem=problem)\n return node.path_cost + h(prob, node);\n end));\nend\n\n\"\"\"\n RBFS(problem::T1, node::T2, flmt::Float64, h::MemoizedFunction) where {T1 <: AbstractProblem, T2 <: Node}\n\nRecursively calls RBFS() with a new 'flmt' value and returns its solution to recursive_best_first_search().\n\"\"\"\nfunction RBFS(problem::T1, node::T2, flmt::Float64, h::MemoizedFunction) where {T1 <: AbstractProblem, T2 <: Node}\n if (goal_test(problem, node.state))\n return node, 0.0;\n end\n local successors = expand(node, problem);\n if (length(successors) == 0);\n return node, Inf;\n end\n for successor in successors\n successor.f = max(successor.path_cost + eval_memoized_function(h, problem, successor), node.f);\n end\n while (true)\n sort!(successors, lt=(function(n1::Node, n2::Node)return isless(n1.f, n2.f);end));\n local best::Node = successors[1];\n if (best.f > flmt)\n return nothing, best.f;\n end\n local alternative::Float64;\n if (length(successors) > 1)\n alternative = successors[1].f;\n else\n alternative = Inf;\n end\n result, best.f = RBFS(problem, best, min(flmt, alternative), h);\n if (!(result === nothing))\n return result, best.f;\n end\n end\nend\n\n\"\"\"\n recursive_best_first_search(problem::T; h::Union{Nothing, MemoizedFunction}) where {T <: AbstractProblem}\n\nSearch the given problem by using the recursive best first search algorithm (Fig. 3.26)\nand return the node solution.\n\nsolution() can be used on the node solution to reconstruct the path taken to the solution.\n\"\"\"\nfunction recursive_best_first_search(problem::T; h::Union{Nothing, MemoizedFunction}=nothing) where {T <: AbstractProblem}\n local mh::MemoizedFunction; #memoized h(n) function\n if (!(typeof(h) <: Nothing))\n mh = MemoizedFunction(h);\n else\n mh = problem.h;\n end\n\n local node = Node{typeof(problem.initial)}(problem.initial);\n node.f = eval_memoized_function(mh, problem, node);\n result, bestf = RBFS(problem, node, Inf, mh);\n return result;\nend\n\nfunction recursive_best_first_search(problem::InstrumentedProblem; h::Union{Nothing, MemoizedFunction}=nothing)\n local mh::MemoizedFunction; #memoized h(n) function\n if (!(typeof(h) <: Nothing))\n mh = MemoizedFunction(h);\n else\n mh = problem.problem.h;\n end\n\n local node = Node{typeof(problem.problem.initial)}(problem.problem.initial);\n node.f = eval_memoized_function(mh, problem, node);\n result, bestf = RBFS(problem, node, Inf, mh);\n return result;\nend\n\n\"\"\"\n hill_climbing(problem::T) where {T <: AbstractProblem}\n\nReturn a state that is a local maximum for the given problem 'problem' by using\nthe hill-climbing search algorithm (Fig. 4.2) on the initial state of the problem.\n\"\"\"\nfunction hill_climbing(problem::T) where {T <: AbstractProblem}\n local current_node = Node{typeof(problem.initial)}(problem.initial);\n while (true)\n local neighbors = expand(current_node, problem);\n if (length(neighbors) == 0)\n break;\n end\n local neighbor = argmax_random_tie(neighbors,\n (function(n::Node,; p::AbstractProblem=problem)\n return value(p, n.state);\n end));\n if (value(problem, neighbor.state) <= value(problem, current_node.state))\n break;\n end\n current_node = neighbor;\n end\n return current_node.state;\nend\n\n\"\"\"\n exp_schedule(;kvar::Int64=20, delta::Float64=0.005, lmt::Int64=100)\n\nReturn a scheduled time for simulated annealing.\n\"\"\"\nfunction exp_schedule(;kvar::Int64=20, delta::Float64=0.005, lmt::Int64=100)\n return (function(t::Real; k=kvar, d=delta, limit=lmt)\n return if_((t < limit), (k * exp(-d * t)), 0);\n end);\nend\n\n\"\"\"\n simulated_annealing(problem::T; schedule::Function=exp_schedule()) where {T <: AbstractProblem}\n\nReturn the solution node by applying the simulated annealing algorithm (Fig. 4.5) on the given\nproblem 'problem' and schedule function 'schedule'. If a solution node can't be found,\nthis function returns 'nothing' on failure.\n\"\"\"\nfunction simulated_annealing(problem::T; schedule::Function=exp_schedule()) where {T <: AbstractProblem}\n local current_node = Node{typeof(problem.initial)}(problem.initial);\n for t in 0:(typemax(Int64) - 1)\n local temperature::Float64 = schedule(t);\n if (temperature == 0)\n return current_node;\n end\n local neighbors = expand(current_node, problem);\n if (length(neighbors) == 0)\n return current_node;\n end\n local next_node = rand(RandomDeviceInstance, neighbors);\n delta_e = value(problem, next_node.state) - value(problem, current_node.state);\n if ((delta_e > 0) || (exp(delta_e/temperature) > rand(RandomDeviceInstance)))\n current_node = next_node;\n end\n end\n return nothing;\nend\n\n#=\n\n and_search() and or_search() are used by and_or_graph_search().\n\n=#\nfunction or_search(problem::T, state::AbstractVector, path::AbstractVector) where {T <: AbstractProblem}\n if (goal_test(problem, state))\n return [];\n end\n if (state in path)\n return nothing;\n end\n for action in actions(problem, state)\n local plan = and_search(get_result(problem, state, action), vcat(path, [state,]));\n if (plan != nothing)\n return [action, plan];\n end\n end\n return nothing;\nend\n\nfunction or_search(problem::T, state::String, path::AbstractVector) where {T <: AbstractProblem}\n if (goal_test(problem, state))\n return [];\n end\n if (state in path)\n return nothing;\n end\n for action in actions(problem, state)\n local plan = and_search(problem, get_result(problem, state, action), vcat(path, [state,]));\n if (plan != nothing)\n return [action, plan];\n end\n end\n return nothing;\nend\n\nfunction and_search(problem::T, states::AbstractVector, path::AbstractVector) where {T <: AbstractVector}\n local plan = Dict{Any, Any}();\n for state in states\n plan[state] = or_search(problem, state, path);\n if (plan[state] == nothing)\n return nothing;\n end\n end\n return plan;\nend\n\n\"\"\"\n and_or_graph_search(problem::T) where {T <: AbstractProblem}\n\nReturn a conditional plan by using the algorithm for searching and-or graphs (Fig. 4.11)\non the given problem 'problem'. This function returns 'nothing' on failure.\n\"\"\"\nfunction and_or_graph_search(problem::T) where {T <: AbstractProblem}\n return or_search(problem, problem.initial, []);\nend\n\n#=\n\n OnlineDFSAgentProgram is a online depth first search agent (Fig. 4.21)\n\n implementation of AgentProgram.\n\n=#\nmutable struct OnlineDFSAgentProgram <: AgentProgram\n result::Dict\n untried::Dict\n unbacktracked::Dict\n state::Union{Nothing, String}\n action::Union{Nothing, String}\n problem::AbstractProblem\n\n function OnlineDFSAgentProgram(problem::T) where {T <: AbstractProblem}\n return new(Dict(), Dict(), Dict(), nothing, nothing, problem);\n end\nend\n\nfunction update_state(odfsap::OnlineDFSAgentProgram, percept::String)\n return percept;\nend\n\nfunction execute(odfsap::OnlineDFSAgentProgram, percept::String)\n local s_prime::String = update_state(odfsap, percept);\n if (goal_test(odfsap.problem, s_prime))\n odfsap.action = nothing;\n else\n if (!(s_prime in keys(odfsap.untried)))\n odfsap.untried[s_prime] = actions(odfsap.problem, s_prime);\n end\n if (!(odfsap.state === nothing))\n if (haskey(odfsap.result, (odfsap.state, odfsap.action)))\n if (s_prime != odfsap.result[(odfsap.state, odfsap.action)])\n odfsap.result[(odfsap.state, odfsap.action)] = s_prime;\n pushfirst!(odfsap.unbacktracked[s_prime], odfsap.state);\n end\n else\n if (s_prime != [])\n odfsap.result[(odfsap.state, odfsap.action)] = s_prime;\n pushfirst!(odfsap.unbacktracked[s_prime], odfsap.state);\n end\n end\n end\n if (length(odfsap.untried[s_prime]) == 0)\n if (length(odfsap.unbacktracked[s_prime]) == 0)\n odfsap.action = nothing;\n else\n first_item = popfirst!(odfsap.unbacktracked[s_prime]);\n for (state, b) in keys(odfsap.result)\n if (odfsap.result[(state, b)] == first_item)\n odfsap.action = b;\n break;\n end\n end\n end\n else\n odfsap.action = popfirst!(odfsap.untried[s_prime]);\n end\n end\n odfsap.state = s_prime;\n return odfsap.action;\nend\n\n#=\n\n OnlineSearchProblem is a AbstractProblem implementation of a online search problem\n\n that can be solved by a online search agent.\n\n=#\nstruct OnlineSearchProblem <: AbstractProblem\n initial::String\n goal::String\n graph::Graph\n least_costs::Dict\n h::Function\n\n function OnlineSearchProblem(initial::String, goal::String, graph::Graph, least_costs::Dict)\n return new(initial, goal, graph, least_costs, online_search_least_cost);\n end\nend\n\nfunction actions(osp::OnlineSearchProblem, state::String)\n return collect(keys(osp.graph.dict[state]));\nend\n\nfunction get_result(osp::OnlineSearchProblem, state::String, action::String)\n return osp.graph.dict[state][action];\nend\n\nfunction online_search_least_cost(osp::OnlineSearchProblem, state::String)\n return osp.least_costs[state];\nend\n\nfunction path_cost(osp::OnlineSearchProblem, state1::String, action::String, state2::String)\n return 1;\nend\n\nfunction goal_test(osp::OnlineSearchProblem, state::String)\n if (state == osp.goal)\n return true;\n else\n return false;\n end\nend\n\n#=\n\n LRTAStarAgentProgram is an AgentProgram implementation of LRTA*-Agent (Fig. 4.24).\n\n The 'result' field is not necessary as the given problem contains the results table.\n\n=#\nmutable struct LRTAStarAgentProgram <: AgentProgram\n H::Dict\n state::Union{Nothing, String}\n action::Union{Nothing, String}\n problem::AbstractProblem\n\n function LRTAStarAgentProgram(problem::T) where {T <: AbstractProblem}\n return new(Dict(), nothing, nothing, problem);\n end\nend\n\nfunction learning_realtime_astar_cost(lrtaap::LRTAStarAgentProgram, state::String, action::String, s_prime::String, H::Dict)\n if (haskey(lrtaap.H, s_prime))\n return path_cost(lrtaap.problem, state, action, s_prime) + lrtaap.H[s_prime];\n else\n return path_cost(lrtaap.problem, state, action, s_prime) + lrtaap.problem.h(lrtaap.problem, s_prime);\n end\nend\n\n\"\"\"\n execute(lrtaap::LRTAStarAgentProgram, s_prime::String)\n\nReturn an action given the percept 's_prime' and by using the LRTA*-Agent\nprogram (Fig. 4.24). If the current state of the agent is at the goal\nstate, return 'nothing'.\n\"\"\"\nfunction execute(lrtaap::LRTAStarAgentProgram, s_prime::String)\n if (goal_test(lrtaap.problem, s_prime))\n lrtaap.action = nothing;\n return nothing;\n else\n if (!haskey(lrtaap.H, s_prime))\n lrtaap.H[s_prime] = lrtaap.problem.h(lrtaap.problem, s_prime);\n end\n if (!(lrtaap.state === nothing))\n lrtaap.H[lrtaap.state] = reduce(min, learning_realtime_astar_cost(lrtaap,\n lrtaap.state,\n b,\n get_result(lrtaap.problem, lrtaap.state, b),\n lrtaap.H)\n for b in actions(lrtaap.problem, lrtaap.state));\n end\n lrtaap.action = argmin(actions(lrtaap.problem, s_prime),\n (function(b::String)\n return learning_realtime_astar_cost(lrtaap,\n s_prime,\n b,\n get_result(lrtaap.problem, s_prime, b),\n lrtaap.H);\n end));\n lrtaap.state = s_prime;\n return lrtaap.action;\n end\nend\n\nfunction genetic_search(problem::T; ngen::Int64=1000, pmut::Float64=0.1, n::Int64=20) where {T <: AbstractProblem}\n local s = problem.initial;\n local states = [result(s, action) for action in actions(problem, s)];\n shuffle!(RandomDeviceInstance, states);\n if (length(states) < n)\n n = length(states);\n end\n return genetic_algorithm(states[1:n], value, ngen=ngen, pmut=pmut);\nend\n\nfunction genetic_algorithm(population::T, fitness::Function; ngen::Int64=1000, pmut::Float64=0.1) where {T <: AbstractVector}\n for i in 1:ngen\n local new_population = Array{Any, 1}();\n for j in 1:length(population)\n local fitnesses = map(fitness, population);\n p1, p2 = weighted_sample_with_replacement(population, fitnesses, 2);\n local child = mate(p1, p2);\n if (rand(RandomDeviceInstance) < pmut)\n mutate(child);\n end\n push!(new_population, child);\n end\n population = new_population;\n end\n return argmax(population, fitness);\nend\n\n# Simplified road map of Romania example (Fig. 3.2)\nromania = UndirectedGraph(Dict(\n Pair(\"A\", Dict(\"Z\"=>75, \"S\"=>140, \"T\"=>118)),\n Pair(\"B\", Dict(\"U\"=>85, \"P\"=>101, \"G\"=>90, \"F\"=>211)),\n Pair(\"C\", Dict(\"D\"=>120, \"R\"=>146, \"P\"=>138)),\n Pair(\"D\", Dict(\"M\"=>75)),\n Pair(\"E\", Dict(\"H\"=>86)),\n Pair(\"F\", Dict(\"S\"=>99)),\n Pair(\"H\", Dict(\"U\"=>98)),\n Pair(\"I\", Dict(\"V\"=>92, \"N\"=>87)),\n Pair(\"L\", Dict(\"T\"=>111, \"M\"=>70)),\n Pair(\"O\", Dict(\"Z\"=>71, \"S\"=>151)),\n Pair(\"P\", Dict(\"R\"=>97)),\n Pair(\"R\", Dict(\"S\"=>80)),\n Pair(\"U\", Dict(\"V\"=>142)),\n ),\n Dict{String, Tuple{Any, Any}}(\n \"A\"=>( 91, 492), \"B\"=>(400, 327), \"C\"=>(253, 288), \"D\"=>(165, 299),\n \"E\"=>(562, 293), \"F\"=>(305, 449), \"G\"=>(375, 270), \"H\"=>(534, 350),\n \"I\"=>(473, 506), \"L\"=>(165, 379), \"M\"=>(168, 339), \"N\"=>(406, 537),\n \"O\"=>(131, 571), \"P\"=>(320, 368), \"R\"=>(233, 410), \"S\"=>(207, 457),\n \"T\"=>( 94, 410), \"U\"=>(456, 350), \"V\"=>(509, 444), \"Z\"=>(108, 531),\n )\n );\n\n# One-dimensional state space example (Fig. 4.23)\none_dim_state_space = Graph{String}(dict=Dict{String, Dict{String, String}}([Pair(\"State_1\", Dict([Pair(\"Right\", \"State_2\")])),\n Pair(\"State_2\", Dict([Pair(\"Right\", \"State_3\"),\n Pair(\"Left\", \"State_1\")])),\n Pair(\"State_3\", Dict([Pair(\"Right\", \"State_4\"),\n Pair(\"Left\", \"State_2\")])),\n Pair(\"State_4\", Dict([Pair(\"Right\", \"State_5\"),\n Pair(\"Left\", \"State_3\")])),\n Pair(\"State_5\", Dict([Pair(\"Right\", \"State_6\"),\n Pair(\"Left\", \"State_4\")])),\n Pair(\"State_6\", Dict([Pair(\"Left\", \"State_5\")]))]));\n\none_dim_state_space_least_costs = Dict([Pair(\"State_1\", 8),\n Pair(\"State_2\", 9),\n Pair(\"State_3\", 2),\n Pair(\"State_4\", 2),\n Pair(\"State_5\", 4),\n Pair(\"State_6\", 3)]);\n\n# Principal states and territories of Australia example (Fig. 6.1)\naustralia = UndirectedGraph(Dict(\n Pair(\"T\", Dict()),\n Pair(\"SA\", Dict(\"WA\"=>1, \"NT\"=>1, \"Q\"=>1, \"NSW\"=>1, \"V\"=>1)),\n Pair(\"NT\", Dict(\"WA\"=>1, \"Q\"=>1)),\n Pair(\"NSW\", Dict(\"Q\"=>1, \"V\"=>1)),\n ),\n Dict{String, Tuple{Any, Any}}(\"WA\"=>(120, 24), \"NT\"=>(135, 20), \"SA\"=>(135, 30),\n \"Q\"=>(145, 20), \"NSW\"=>(145, 32), \"T\"=>(145, 42), \"V\"=>(145, 37),\n )\n );\n\n#=\n\n NQueensProblem is the problem of placing 'N' non-attacking queens on a 'N'x'N' chess board.\n\n Each state is represented as an 'N' element array where the value of 'r' at index 'c' implies\n\n that a queen occupies the position at row 'r' and column 'c'. The columns are values are filled\n\n from left to right.\n\n=#\nstruct NQueensProblem <: AbstractProblem\n N::Int64\n initial::Array{Union{Nothing, Int64}, 1}\n\n function NQueensProblem(n::Int64)\n return new(n, fill(nothing, n));\n end\nend\n\nfunction actions(problem::NQueensProblem, state::AbstractVector)\n if (!(state[length(state)] === nothing))\n return Array{Any, 1}([]);\n else\n local col = utils.null_index(state);\n return collect(row for row in 1:problem.N if (!conflicted(problem, state, row, col)));\n end\nend\n\nfunction conflict(problem::NQueensProblem, row1::Int64, col1::Int64, row2::Int64, col2::Int64)\n return ((row1 == row2) ||\n (col1 == col2) ||\n (row1 - col1 == row2 - col2) ||\n (row1 + col1 == row2 + col2));\nend\n\nfunction conflict(problem::NQueensProblem, row1::Int64, col1::Int64, row2::Nothing, col2::Int64)\n error(\"conflict(): 'row2' is not initialized!\");\nend\n\nfunction conflicted(problem::NQueensProblem, state::AbstractVector, row::Int64, col::Int64)\n return any(conflict(problem, row, col, state[i], i) for i in 1:(col-1));\nend\n\nfunction get_result(problem::NQueensProblem, state::AbstractVector, row::Int64)\n local col = utils.null_index(state);\n local new_result = deepcopy(state);\n new_result[col] = row;\n return new_result;\nend\n\nfunction goal_test(problem::NQueensProblem, state::AbstractVector)\n if ((state[length(state)] === nothing))\n return false;\n end\n return !any(conflicted(problem, state, state[col], col) for col in 1:length(state));\nend\n\ncapital_case_alphabet = \"ABCDEFGHIJKLMNOPQRSTUVWXYZ\";\n\ncubes16 = Array{String, 1}([\"FORIXB\", \"MOQABJ\", \"GURILW\", \"SETUPL\",\n \"CMPDAE\", \"ACITAO\", \"SLCRAE\", \"ROMASH\",\n \"NODESW\", \"HEFIYE\", \"ONUDTK\", \"TEVIGN\",\n \"ANEDVZ\", \"PINESH\", \"ABILYT\", \"GKYLEU\"]);\n\nfunction random_boggle(;n::Int64=4)\n local cubes = collect(cubes16[(i % 16) + 1] for i in 0:((n * n) - 1));\n shuffle!(RandomDeviceInstance, cubes);\n return map((function(array::String)\n return rand(RandomDeviceInstance, collect(array));\n end), cubes);\nend\n\nboyan_best = collect(\"RSTCSDEIAEGNLRPEATESMSSID\");\n\nfunction print_boggle(board::Array{Char, 1})\n local nn::Int64 = length(board);\n local n::Int64 = int_sqrt(nn);\n local board_str::String = \"\";\n for i in 0:(nn - 1)\n if ((i % n == 0) && (i > 0))\n board_str = board_str * \"\\n\";\n end\n if (board[i + 1] == 'Q')\n board_str = board_str * \"Qu \";\n else\n board_str = board_str * String([board[i + 1]]) * \" \";\n end\n end\n print(board_str);\n nothing;\nend\n\nfunction boggle_neighbors(nn::Int64; cache::Dict=Dict{Any, Any}())\n if haskey(cache, nn)\n return cache[nn];\n end\n local n::Int64 = int_sqrt(nn)\n local neighbors::AbstractVector = Array{Any, 1}(undef, nn);\n for i in 0:(nn - 1)\n neighbors[i + 1] = Array{Int64, 1}([]);\n on_top::Bool = (i < n);\n on_bottom::Bool = (i >= (nn - n));\n on_left::Bool = (i % n == 0);\n on_right::Bool = ((i + 1) % n == 0);\n if (!on_top)\n push!(neighbors[i + 1], (i + 1) - n);\n if (!on_left)\n push!(neighbors[i + 1], (i + 1) - n - 1);\n end\n if (!on_right)\n push!(neighbors[i + 1], (i + 1) - n + 1);\n end\n end\n if (!on_bottom)\n push!(neighbors[i + 1], (i + 1) + n);\n if (!on_left)\n push!(neighbors[i + 1], (i + 1) + n - 1);\n end\n if (!on_right)\n push!(neighbors[i + 1], (i + 1) + n + 1);\n end\n end\n if (!on_left)\n push!(neighbors[i + 1], (i + 1) - 1);\n end\n if (!on_right)\n push!(neighbors[i + 1], (i + 1) + 1);\n end\n end\n cache[nn] = neighbors;\n return neighbors;\nend\n\nfunction int_sqrt(n::Number)\n return Int64(sqrt(n));\nend\n\n#=\n\n WordList contains an array of words.\n\n=#\nstruct WordList\n words::Array{String, 1}\n bounds::Dict{Char, Tuple{Any, Any}}\n\n function WordList(filename::String; min_len::Int64=3)\n local wlba = read(filename);\n local wls = uppercase(String(wlba));\n local wlsa = sort(map(strip, split(wls, '\\n')));\n local wlsa_filtered = collect(s for s in wlsa if (length(s) >= min_len));\n nwl::WordList = new(wlsa_filtered, Dict{Char, Tuple{Any, Any}}());\n for c in capital_case_alphabet\n fc::Char = c + 1; #following character\n nwl.bounds[c] = (searchsorted(nwl.words, String([c]), 1, length(nwl.words), Base.Order.Forward).stop + 1,\n searchsorted(nwl.words, String([fc]), 1, length(nwl.words), Base.Order.Forward).stop + 1);\n end\n return nwl;\n end\nend\n\nfunction lookup(wl::WordList, prefix::String; lo::Int64=1, hi::Union{Nothing, Int64}=nothing)\n local words = wl.words;\n if (typeof(hi) <: Nothing)\n hi = length(words);\n end\n local i::Int64 = searchsorted(words, prefix, lo, hi, Base.Order.Forward).start;\n\n #'i' is only larger than length of words when the returned index is not in WordList.\n if (i <= length(words) && startswith(words[i], prefix))\n return i, (words[i] == prefix);\n else\n return nothing, false;\n end\nend\n\nlength(wl::WordList) = length(wl.words);\n\nin(prefix::String, wl::WordList) = getindex(lookup(wl, prefix), 2);\n\n#=\n\n BoggleFinder contains the words found on the Boggle board\n\n and the array of possible words for the Boggle board.\n\n=#\nmutable struct BoggleFinder\n wordlist::WordList\n scores::AbstractVector\n found::Dict\n board::AbstractVector\n neighbors::AbstractVector\n\n function BoggleFinder(;board::Union{Nothing, AbstractVector}=nothing, fn::Union{Nothing, String}=nothing)\n local wlfn::String;\n if (typeof(fn) <: Nothing)\n if (is_windows())\n wlfn = \"..\\\\aima-data\\\\EN-text\\\\wordlist.txt\";\n elseif (is_apple() || is_unix())\n wlfn = \"../aima-data/EN-text/wordlist.txt\";\n end\n else\n wlfn = fn;\n end\n nbf = new(WordList(wlfn),\n vcat([0, 0, 0, 0, 1, 2, 3, 5], fill(11, 100)),\n Dict{Any, Any}())\n if (!(typeof(board) <: Nothing))\n set_board(nbf, board=board);\n end\n return nbf;\n end\nend\n\nfunction set_board(bf::BoggleFinder; board::Union{Nothing, AbstractVector}=nothing)\n if (typeof(board) <: Nothing)\n board = random_boggle();\n end\n bf.board = board;\n bf.neighbors = boggle_neighbors(length(board));\n bf.found = Dict{Any, Any}();\n for i in 1:length(board)\n lo::Int64, hi::Int64 = bf.wordlist.bounds[board[i]];\n find(bf, lo, hi, i, [], \"\");\n end\n return bf;\nend\n\nfunction find(bf::BoggleFinder, lo::Int64, hi::Int64, i::Int64, visited::AbstractVector, prefix::String)\n if i in visited\n return nothing;\n end\n wordpos, is_word::Bool = lookup(bf.wordlist, prefix, lo=lo, hi=hi);\n if (!(typeof(wordpos) <: Nothing))\n if (is_word)\n bf.found[prefix] = true;\n end\n push!(visited, i);\n local c = bf.board[i];\n if (c == 'Q')\n c = \"QU\";\n else\n c = String([c]);\n end\n prefix = prefix * c;\n for j in bf.neighbors[i]\n find(bf, wordpos, hi, j, visited, prefix);\n end\n pop!(visited);\n end\n return nothing;\nend\n\nfunction words(bf::BoggleFinder)\n return collect(keys(bf.found));\nend\n\nfunction score(bf::BoggleFinder)\n return sum(collect(scores[len(w)] for w in words(bf)));\nend\n\nlength(bf::BoggleFinder) = length(bf.found);\n\n\"\"\"\n boggle_hill_climbing(;board::Union{Nothing, AbstractVector}=nothing, ntimes::Int64=100, verbose::Bool=true)\n\nSolve the inverse Boggle by using hill climbing (initially use a random Boggle board and changing it).\n\nReturn the best Boggle board and its length.\n\"\"\"\nfunction boggle_hill_climbing(;board::Union{Nothing, AbstractVector}=nothing, ntimes::Int64=100, verbose::Bool=true)\n finder = BoggleFinder();\n if (typeof(board) <: Nothing)\n board = random_boggle();\n end\n local best_length::Int64 = length(set_board(finder, board=board));\n for t in 1:ntimes\n i, old_char = mutate_boggle(board);\n local new_length::Int64 = length(set_board(finder, board=board));\n if (new_length > best_length)\n best_length = new_length;\n if (verbose)\n println(best_length, \" \", t, \" \", board);\n end\n else\n board[i] = old_char;\n end\n end\n if (verbose)\n print_boggle(board);\n end\n return board, best_length;\nend\n\nfunction mutate_boggle(board::AbstractArray)\n local i::Int64 = rand(RandomDeviceInstance, 1:length(board));\n local old_char::Char = board[i];\n board[i] = rand(RandomDeviceInstance, collect(rand(RandomDeviceInstance, cubes16)));\n return i, old_char;\nend\n\nfunction execute_searcher(searcher::Function, problem::T) where {T <: AbstractProblem}\n local p = InstrumentedProblem(problem);\n searcher(p);\n return p;\nend\n\nfunction compare_searchers(problems::Array{T, 1},\n header::Array{String, 1};\n searchers::Array{Function, 1}=[breadth_first_tree_search,\n breadth_first_search,\n depth_first_graph_search,\n iterative_deepening_search,\n depth_limited_search,\n recursive_best_first_search]) where {T <: AbstractProblem}\n local table = vcat(permutedims(hcat(header), [2, 1]), \n hcat(map(string, searchers), \n permutedims(reduce(hcat,\n collect(\n collect(format_instrumented_results(execute_searcher(s, p)) for p in problems)\n for s in searchers)),\n [2,1])));\n return table;\nend\n\nfunction beautify_node(n::Node)\n return @sprintf(\"%s%s%s\", \"\");\nend\n\n", "meta": {"hexsha": "da70a9fa6585ca56f7a10e3135fddbc1a18c2723", "size": 56665, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "search.jl", "max_stars_repo_name": "mikhail-j/aima-julia-aimacode-fork", "max_stars_repo_head_hexsha": "07ea7acd534266fd2b10ee0d4aeb847a88225ac0", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 131, "max_stars_repo_stars_event_min_datetime": "2016-07-23T14:31:27.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-29T07:49:47.000Z", "max_issues_repo_path": "search.jl", "max_issues_repo_name": "mikhail-j/aima-julia-aimacode-fork", "max_issues_repo_head_hexsha": "07ea7acd534266fd2b10ee0d4aeb847a88225ac0", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 21, "max_issues_repo_issues_event_min_datetime": "2017-03-30T19:04:57.000Z", "max_issues_repo_issues_event_max_datetime": "2021-08-08T16:35:26.000Z", "max_forks_repo_path": "search.jl", "max_forks_repo_name": "mikhail-j/aima-julia-aimacode-fork", "max_forks_repo_head_hexsha": "07ea7acd534266fd2b10ee0d4aeb847a88225ac0", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 65, "max_forks_repo_forks_event_min_datetime": "2016-09-24T17:30:46.000Z", "max_forks_repo_forks_event_max_datetime": "2022-03-29T07:49:50.000Z", "avg_line_length": 35.4599499374, "max_line_length": 190, "alphanum_fraction": 0.6105179564, "num_tokens": 13881, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4416730056646256, "lm_q2_score": 0.1276526269811457, "lm_q1q2_score": 0.05638071943974791}}
{"text": "# test for unify.jl\r\nusing Test\r\n#include(\"utils.jl\") # for isinvar()\r\n#include(\"unify.jl\")\r\n\r\n#==\r\nidea:\r\nunify([:x,:y], \"P(x,y)\", \"P(x,f(x))\")\r\nunify([:x,:y], :(P(x,y)), :(P(x,f(x))))\r\n\r\n:(P(x,y)) == Meta.Meta.parse(\"P(x,y)\")\r\nyoucan readline(from console) and Meta.parse it to get the Expr\r\n==#\r\n\r\n@testset \"unify0 find a pair unifiable\" begin\r\n @test unify0([], :P, :P) ==()\r\n @test unify0([:x], :x, :x) ==()\r\n @test unify0([:x], :x, :a) ==(:x, :a)\r\n @test unify0([:x], :a, :x) ==(:x, :a)\r\n\r\n @test_throws ICMP unify0([:x], :P, :Q)\r\n @test_throws ICMP unify0([], :P, :Q)\r\n\r\n @test unify0([:x], :(P(x)), :(P(a))) == (:x, :a)\r\n @test unify0([:x], :(P(x)), :(P(f(a)))) == (:x,:(f(a)))\r\n @test unify0([:x], :(P(f(a))), :(P(f(x)))) == (:x,:a)\r\n @test unify0([:x], :(P(f(a))), :(P(x))) == (:x,:(f(a)))\r\n\r\n @test_throws ICMP unify0([:x],:(P(g(a))),:(P(f(x))))\r\n @test_throws ICMP unify0([:x],:(P(f(a))),:(P(f(b))))\r\n @test_throws ICMP unify0([:x],:(P(a)), :(P(f(b)))) \r\nend\r\n\r\n@testset \"unify1\" begin\r\n @test unify1([], :(f()), :(f()), []) == []\r\n @test unify1([:x], :x, :x, [:x]) == [:x]\r\n @test unify1([:x], :x, :a, [:x]) == [:a]\r\n @test unify1([:x], :x, :x, [:x]) == [:x]\r\n\r\n @test unify1([:x], :(f(x)), :(f(x)), [:x]) == [:x]\r\n @test unify1([:x], :(f(x)), :(f(a)), [:x]) == [:a]\r\n @test unify1([:x], :(f(x)), :(f(x)), [:b]) == [:b]\r\n\r\n @test_throws ICMP unify1([:x], :(f(x)), :(f(a)), [:b])\r\n\r\n @test unify1([:x,:y], :x , :(f(a)), [:x,:y]) == [:(f(a)), :y]\r\n @test unify1([:x,:y], :x , :(f(a)), [:x,:a]) == [:(f(a)), :a]\r\n\r\n @test unify1([:x,:y,:z], :a , :z, [:y,:z,:z]) == [:a,:a,:a]\r\n @test unify1([:x,:y,:z], :z , :a, [:y,:z,:z]) == [:a,:a,:a]\r\n\r\n @test unify1([:x,:y,:z], :z , :(f(a)), [:y,:z,:z]) == [:(f(a)),:(f(a)),:(f(a))]\r\n\r\n @test unify1([:x,:y], :x , :(f(a)), [:x,:x]) == [:(f(a)), :(f(a))]\r\n\r\n @test unify1([:x,:y,:z],:x,:(f(a)),[:y,:z,:z]) == [:(f(a)),:(f(a)),:(f(a))]\r\n @test unify1([:x,:y,:z],:z,:(f(a)),[:y,:z,:z]) == [:(f(a)),:(f(a)),:(f(a))]\r\n @test unify1([:x,:y,:z,:w,:u],:z,:(f(a)),[:y,:z,:w,:u,:u]) == [:(f(a)),:(f(a)),:(f(a)),:(f(a)),:(f(a))]\r\n\r\n @test unify1([:x,:y], :x , :(f(a)), [:y,:y]) == [:(f(a)), :(f(a))]\r\n\r\n @test unify1([:x,:y], :(f(x)), :(f(x)), [:x,:a]) == [:x,:a]\r\n @test unify1([:x,:y], :(f(x)), :(f(b)), [:x,:a]) == [:b,:a]\r\n\r\n @test unify1([:x,:y], :(f(b)), :(f(x)), [:x,:a]) == [:b,:a]\r\n @test unify1([:x,:y], :(f(g(b))), :(f(x)), [:x,:a]) == [:(g(b)),:a]\r\n\r\n @test unify1([:x,:y], :(f(x)), :(f(y)), [:b,:y]) == [:b,:b]\r\n\r\n @test unify1([:x,:y], :(f(g(b,y))), :(f(x)), [:x,:(h(a))]) == [:(g(b,y)),:(h(a))]\r\n @test unify1([:x,:y], :x, :p, [:y,:y]) == [:p,:p]\r\n\r\n @test unify1([:x,:y], :x, :y, [:p,:y]) == [:p,:p]\r\n\r\nend\r\n\r\n@testset \"unify\" begin\r\n @test_throws ICMP unify([],:(P()),:(Q()))\r\n\r\n @test unify([:x],:(),:())== [:x]\r\n @test unify([:x],:(P()),:(P())) == [:x]\r\n @test_throws ICMP unify([:x],:(P()),:(Q()))\r\n\r\n\r\n @test unify([:x],:(P(x)), :(P(x))) == [:x]\r\n @test unify([:y],:(P(x)), :(P(x))) == [:y]\r\n\r\n @test unify([:x],:(P(x)), :(P(y))) == [:y]\r\n @test unify([:y],:(P(x)), :(P(y))) == [:x]\r\n\r\n @testset \"with number\" begin\r\n @test unify([], :(f(2)), :(f(2))) == []\r\n @test unify([:x], :(f(x)), :(f(2))) == [2]\r\n @test unify([:x], :(f(3)), :(f(x))) == [3]\r\n @test unify([:x,:y], :(f(3,y)), :(f(x,23))) == [3,23]\r\n end\r\n# in this case, x and y are constants\r\n @test_throws ICMP unify([:z],:(P(x)), :(P(y)))\r\n\r\n @test unify([],:(P(a)), :(P(a))) == []\r\n @test_throws ICMP unify([],:(P(a)), :(P(b)))\r\n\r\n (\u03c3=unify([:x,:y],:(P(x)),:(P(y))); @test \u03c3==[:x,:x] || \u03c3==[:y,:y])\r\n\r\n @test unify([:x,:y],:(P(x)),:(P(a))) == [:a,:y]\r\n @test unify([:x,:y],:(P(f(x))),:(P(f(a)))) == [:a,:y]\r\n\r\n @test unify([:x,:y],:(P(x)),:(P(f(a)))) == [:(f(a)),:y]\r\n (\u03c3 = unify([:x,:y],:(P(x,f(a,x))),:(P(y,f(a,y)))); @test \u03c3 == [:y,:y] || \u03c3 == [:y,:y])\r\n\r\n @test unify([:x,:y],:(P(x,f(x))),:(P(a,f(y)))) == [:a,:a]\r\n @test unify([:x,:y],:(P(x,y)),:(P(y,f(a)))) == [:(f(a)),:(f(a))]\r\n\r\n @test unify([:x,:y], :(f(g(b,y),h(a))), :(f(x,y))) == [:(g(b,h(a))),:(h(a))]\r\n\r\n @testset \"fixed point of subst\" begin\r\n @test fp_subst([:x,:y],[:x,:y]) == [:x,:y]\r\n @test fp_subst([:x,:y],[:a,:x]) == [:a,:a]\r\n\r\n @test fp_subst([:x,:y],[:x,:x]) == [:x,:x]\r\n\r\n @test fp_subst([:x,:y,:z],[:a,:(f(x,z)),:b]) == [:a,:(f(a,b)),:b]\r\n @test fp_subst([:x,:y,:z,:w],[:x,:(f(g(z,w),h(w))),:(k(w)),:(m(a))]) == [:x,:(f(g(k(m(a)),m(a)),h(m(a)))),:(k(m(a))),:(m(a))]\r\n\r\n @test fp_subst([:x,:y,:z,:u],[:(f(y)),:(g(z)),:(h(u)),:(k(a))]) == [:(f(g(h(k(a))))),:(g(h(k(a)))),:(h(k(a))),:(k(a))]\r\n\r\n @test unify([:x,:y,:z,:u],:(P(x,y,z,u)),:(P(f(y),g(z),h(u),k(a)))) == [:(f(g(h(k(a))))),:(g(h(k(a)))),:(h(k(a))),:(k(a))]\r\n\r\n\r\n @testset \"loop check for fixed point of subst\" begin\r\n @test_throws Loop fp_subst([:x,:y],[:(f(y)),:x])\r\n @test_throws Loop fp_subst([:x,:y,:z],[:a,:(f(x,z)),:(g(y))]) == [:a,:(f(a,b)),:b]\r\n @test_throws Loop fp_subst([:x,:y,:z,:w],[:a,:(f(x,z)),:(g(w)),:(h(z))]) == [:a,:(f(a,b)),:b]\r\n end\r\n end\r\n\r\n @testset \"unify intervention\" begin\r\n @test unify([:x,:y],:(P(f(x),x)),:(P(y,a))) == [:a,:(f(a))]\r\n\r\n @test unify([:x,:y,:w],:(P(f(w,x),w,x)),:(P(y,h(b),g(a)))) == [:(g(a)),:(f(h(b),g(a))),:(h(b))]\r\n\r\n @test unify([:x,:y,:w,:u,:z,:n],:(P(f(w,z),w,g(n),z)),:(P(y,h(b),u,h(u)))) == [:x,:(f(h(b),h(g(n)))),:(h(b)),:(g(n)),:(h(g(n))),:n]\r\n\r\n @test unify([:x,:y,:z,:w,:u,:n,:v],:((P(f(x),y,h(y,z),z,k(n)))),:((P(w,g(w),u,q(v),v)))) == [:x,:(g(f(x))),:(q(k(n))),:(f(x)),:(h(g(f(x)),q(k(n)))),:n,:(k(n))]\r\n\r\n @test unify([:x,:y,:z,:w,:u,:n,:v],:((P(f(y,z),y,h(z),z,k(n)))),:((P(w,g(u,v),u,q(v),v)))) == [:x,:(g(h(q(k(n))),k(n))),:(q(k(n))),:(f(g(h(q(k(n))),k(n)),q(k(n)))),:(h(q(k(n)))),:n,:(k(n))]\r\n\r\n end\r\n\r\n @test unify([:x,:y], :(f(a=x,b=x)), :(f(a=p,b=y))) == [:p, :p]\r\n\r\n @testset \"unify loop test fail\" begin\r\n @test_throws Loop unify([:x,:y],:(P(x)),:(P(f(x))))\r\n @test_throws Loop unify([:x,:y],:(P(x,y)),:(P(y,f(y))))\r\n end\r\n\r\nend\r\n\r\n", "meta": {"hexsha": "d2312c02de5a56845c86cb6b473c463f0afa5a82", "size": 5897, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Prover/testunify.jl", "max_stars_repo_name": "sazare/cheapViews", "max_stars_repo_head_hexsha": "b235bafa85e5734c0aaa303d480cb7f4f97ab949", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "Prover/testunify.jl", "max_issues_repo_name": "sazare/cheapViews", "max_issues_repo_head_hexsha": "b235bafa85e5734c0aaa303d480cb7f4f97ab949", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Prover/testunify.jl", "max_forks_repo_name": "sazare/cheapViews", "max_forks_repo_head_hexsha": "b235bafa85e5734c0aaa303d480cb7f4f97ab949", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 37.0880503145, "max_line_length": 192, "alphanum_fraction": 0.3823978294, "num_tokens": 2607, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.49218813572079556, "lm_q2_score": 0.11436853825632723, "lm_q1q2_score": 0.05629083762949418}}
{"text": "using OREnvironment\nusing ORInterface\nusing Test\n\n@testset \"Building Status\" begin\n numConstraints = 5;\n status = OREnvironment.constructStatus(numConstraints);\n # conditions after creation\n @test OREnvironment.is_feasible(status) == false;\n @test OREnvironment.is_optimal(status) == false;\n @test OREnvironment.get_objfunction(status) == 0.0;\n for i in 1:numConstraints\n @test OREnvironment.get_constraint_consumption(status, i) == 0.0;\n end\n # conditions after modification\n OREnvironment.set_feasible!(status, true);\n OREnvironment.set_optimal!(status, true);\n OREnvironment.set_objfunction!(status, 16.0);\n OREnvironment.set_constraint_consumption!(status, 1, 15.0);\n @test OREnvironment.is_feasible(status) == true;\n @test OREnvironment.is_optimal(status) == true;\n @test OREnvironment.get_objfunction(status) == 16.0;\n @test OREnvironment.get_constraint_consumption(status, 1) == 15.0;\nend\n\n@testset \"is_first_status_better\" begin\n numConstraints = 5;\n s1 = OREnvironment.constructStatus(numConstraints);\n s2 = OREnvironment.constructStatus(numConstraints);\n OREnvironment.set_objfunction!(s1, 34.0);\n OREnvironment.set_objfunction!(s2, 3.0);\n\n @testset \"feasibility not required\" begin\n feasibility = false;\n @test OREnvironment.is_first_status_better(s1,s2,:max, feasibility) == true;\n @test OREnvironment.is_first_status_better(s1,s2,:min, feasibility) == false;\n OREnvironment.set_objfunction!(s2, 34.0);\n @test OREnvironment.is_first_status_better(s1,s2,:max, feasibility) == false;\n @test OREnvironment.is_first_status_better(s2,s1,:max, feasibility) == false;\n @test OREnvironment.is_first_status_better(s1,s2,:min, feasibility) == false;\n @test OREnvironment.is_first_status_better(s2,s1,:min, feasibility) == false;\n\n OREnvironment.set_objfunction!(s1, 34.0);\n OREnvironment.set_objfunction!(s2, 3.0);\n OREnvironment.set_feasible!(s1, true);\n OREnvironment.set_feasible!(s2, false);\n @test OREnvironment.is_first_status_better(s1,s2,:max, feasibility) == true;\n @test OREnvironment.is_first_status_better(s1,s2,:min, feasibility) == false;\n OREnvironment.set_objfunction!(s2, 34.0);\n @test OREnvironment.is_first_status_better(s1,s2,:max, feasibility) == false;\n @test OREnvironment.is_first_status_better(s2,s1,:max, feasibility) == false;\n @test OREnvironment.is_first_status_better(s1,s2,:min, feasibility) == false;\n @test OREnvironment.is_first_status_better(s2,s1,:min, feasibility) == false;\n\n OREnvironment.set_objfunction!(s1, 34.0);\n OREnvironment.set_objfunction!(s2, 3.0);\n OREnvironment.set_feasible!(s1, false);\n OREnvironment.set_feasible!(s2, true);\n @test OREnvironment.is_first_status_better(s1,s2,:max, feasibility) == true;\n @test OREnvironment.is_first_status_better(s1,s2,:min, feasibility) == false;\n OREnvironment.set_objfunction!(s2, 34.0);\n @test OREnvironment.is_first_status_better(s1,s2,:max, feasibility) == false;\n @test OREnvironment.is_first_status_better(s2,s1,:max, feasibility) == false;\n @test OREnvironment.is_first_status_better(s1,s2,:min, feasibility) == false;\n @test OREnvironment.is_first_status_better(s2,s1,:min, feasibility) == false;\n\n OREnvironment.set_objfunction!(s1, 34.0);\n OREnvironment.set_objfunction!(s2, 3.0);\n OREnvironment.set_feasible!(s1, true);\n OREnvironment.set_feasible!(s2, true);\n @test OREnvironment.is_first_status_better(s1,s2,:max, feasibility) == true;\n @test OREnvironment.is_first_status_better(s1,s2,:min, feasibility) == false;\n OREnvironment.set_objfunction!(s2, 34.0);\n @test OREnvironment.is_first_status_better(s1,s2,:max, feasibility) == false;\n @test OREnvironment.is_first_status_better(s2,s1,:max, feasibility) == false;\n @test OREnvironment.is_first_status_better(s1,s2,:min, feasibility) == false;\n @test OREnvironment.is_first_status_better(s2,s1,:min, feasibility) == false;\n end\n\n @testset \"feasibility required\" begin\n feasibility = true;\n OREnvironment.set_objfunction!(s1, 34.0);\n OREnvironment.set_objfunction!(s2, 3.0)\n OREnvironment.set_feasible!(s1, false);\n OREnvironment.set_feasible!(s2, false);\n @test OREnvironment.is_first_status_better(s1,s2,:max, feasibility) == true;\n @test OREnvironment.is_first_status_better(s1,s2,:min, feasibility) == false;\n OREnvironment.set_objfunction!(s2, 34.0);\n @test OREnvironment.is_first_status_better(s1,s2,:max, feasibility) == false;\n @test OREnvironment.is_first_status_better(s2,s1,:max, feasibility) == false;\n @test OREnvironment.is_first_status_better(s1,s2,:min, feasibility) == false;\n @test OREnvironment.is_first_status_better(s2,s1,:min, feasibility) == false;\n\n OREnvironment.set_objfunction!(s1, 34.0);\n OREnvironment.set_objfunction!(s2, 3.0);\n OREnvironment.set_feasible!(s1, true);\n OREnvironment.set_feasible!(s2, false);\n @test OREnvironment.is_first_status_better(s1,s2,:max, feasibility) == true;\n @test OREnvironment.is_first_status_better(s1,s2,:min, feasibility) == true;\n OREnvironment.set_objfunction!(s2, 34.0);\n @test OREnvironment.is_first_status_better(s1,s2,:max, feasibility) == true;\n @test OREnvironment.is_first_status_better(s2,s1,:max, feasibility) == false;\n @test OREnvironment.is_first_status_better(s1,s2,:min, feasibility) == true;\n @test OREnvironment.is_first_status_better(s2,s1,:min, feasibility) == false;\n\n OREnvironment.set_objfunction!(s1, 34.0);\n OREnvironment.set_objfunction!(s2, 3.0);\n OREnvironment.set_feasible!(s1, false);\n OREnvironment.set_feasible!(s2, true);\n @test OREnvironment.is_first_status_better(s1,s2,:max, feasibility) == false;\n @test OREnvironment.is_first_status_better(s1,s2,:min, feasibility) == false;\n OREnvironment.set_objfunction!(s2, 34.0);\n @test OREnvironment.is_first_status_better(s1,s2,:max, feasibility) == false;\n @test OREnvironment.is_first_status_better(s2,s1,:max, feasibility) == true;\n @test OREnvironment.is_first_status_better(s1,s2,:min, feasibility) == false;\n @test OREnvironment.is_first_status_better(s2,s1,:min, feasibility) == true;\n\n OREnvironment.set_objfunction!(s1, 34.0);\n OREnvironment.set_objfunction!(s2, 3.0);\n OREnvironment.set_feasible!(s1, true);\n OREnvironment.set_feasible!(s2, true);\n @test OREnvironment.is_first_status_better(s1,s2,:max, feasibility) == true;\n @test OREnvironment.is_first_status_better(s1,s2,:min, feasibility) == false;\n OREnvironment.set_objfunction!(s2, 34.0);\n @test OREnvironment.is_first_status_better(s1,s2,:max, feasibility) == false;\n @test OREnvironment.is_first_status_better(s2,s1,:max, feasibility) == false;\n @test OREnvironment.is_first_status_better(s1,s2,:min, feasibility) == false;\n @test OREnvironment.is_first_status_better(s2,s1,:min, feasibility) == false;\n end\nend\n\n@testset \"Worst Value\" begin\n @test OREnvironment.worst_value(:max) == -Inf;\n @test OREnvironment.worst_value(:min) == Inf;\nend\n\n@testset \"update_status\" begin\n numConstraints = 5;\n # first status\n s1 = OREnvironment.constructStatus(numConstraints);\n OREnvironment.set_objfunction!(s1, 34.0);\n OREnvironment.set_feasible!(s1, true);\n OREnvironment.set_optimal!(s1, true);\n OREnvironment.set_constraint_consumption!(s1, 1, 1.2);\n OREnvironment.set_constraint_consumption!(s1, 2, 2.2);\n OREnvironment.set_constraint_consumption!(s1, 3, 3.2);\n OREnvironment.set_constraint_consumption!(s1, 4, 4.2);\n OREnvironment.set_constraint_consumption!(s1, 5, 5.2);\n\n @test OREnvironment.is_feasible(s1) == true;\n @test OREnvironment.is_optimal(s1) == true;\n @test OREnvironment.get_objfunction(s1) == 34.0;\n @test OREnvironment.get_constraint_consumption(s1, 1) == 1.2;\n @test OREnvironment.get_constraint_consumption(s1, 2) == 2.2;\n @test OREnvironment.get_constraint_consumption(s1, 3) == 3.2;\n @test OREnvironment.get_constraint_consumption(s1, 4) == 4.2;\n @test OREnvironment.get_constraint_consumption(s1, 5) == 5.2;\n\n # second status\n s2 = OREnvironment.constructStatus(numConstraints);\n @test OREnvironment.is_feasible(s2) == false;\n @test OREnvironment.is_optimal(s2) == false;\n @test OREnvironment.get_objfunction(s2) == 0.0;\n for i in 1:numConstraints\n @test OREnvironment.get_constraint_consumption(s2, i) == 0.0;\n end\n\n # update\n OREnvironment.update_status!(s2, s1);\n\n @test OREnvironment.is_feasible(s1) == true;\n @test OREnvironment.is_optimal(s1) == true;\n @test OREnvironment.get_objfunction(s1) == 34.0;\n @test OREnvironment.get_constraint_consumption(s1, 1) == 1.2;\n @test OREnvironment.get_constraint_consumption(s1, 2) == 2.2;\n @test OREnvironment.get_constraint_consumption(s1, 3) == 3.2;\n @test OREnvironment.get_constraint_consumption(s1, 4) == 4.2;\n @test OREnvironment.get_constraint_consumption(s1, 5) == 5.2;\n\n @test OREnvironment.is_feasible(s2) == true;\n @test OREnvironment.is_optimal(s2) == true;\n @test OREnvironment.get_objfunction(s2) == 34.0;\n @test OREnvironment.get_constraint_consumption(s2, 1) == 1.2;\n @test OREnvironment.get_constraint_consumption(s2, 2) == 2.2;\n @test OREnvironment.get_constraint_consumption(s2, 3) == 3.2;\n @test OREnvironment.get_constraint_consumption(s2, 4) == 4.2;\n @test OREnvironment.get_constraint_consumption(s2, 5) == 5.2;\nend\n", "meta": {"hexsha": "7e192c4c669af2b8c3315a68919d34aa3d2ba190", "size": 9798, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/Status.jl", "max_stars_repo_name": "DavidGarHeredia/OREnvironment.jl", "max_stars_repo_head_hexsha": "d3bc3c3bdbb0e282ff09ab4bab5a74fcf372b45f", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "test/Status.jl", "max_issues_repo_name": "DavidGarHeredia/OREnvironment.jl", "max_issues_repo_head_hexsha": "d3bc3c3bdbb0e282ff09ab4bab5a74fcf372b45f", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 4, "max_issues_repo_issues_event_min_datetime": "2021-07-24T13:31:34.000Z", "max_issues_repo_issues_event_max_datetime": "2021-09-24T17:33:36.000Z", "max_forks_repo_path": "test/Status.jl", "max_forks_repo_name": "DavidGarHeredia/OREnvironment.jl", "max_forks_repo_head_hexsha": "d3bc3c3bdbb0e282ff09ab4bab5a74fcf372b45f", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 51.8412698413, "max_line_length": 85, "alphanum_fraction": 0.7081036946, "num_tokens": 2978, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4921881357207955, "lm_q2_score": 0.11436853523769844, "lm_q1q2_score": 0.0562908361437609}}
{"text": "using Pkg\n\n# Pkg.add(\"BinaryBuilder\")\nPkg.add(\"IJulia\")\nPkg.add(\"NamedArrays\")\nPkg.add(\"Feather\")\nPkg.add(\"DataFrames\")\nPkg.add(\"Conda\")\n# Problems with Plots > GR > GR_jll > Qt5Base:\n# - dylib not found\n# - if renamed 'binary' to .dylib >> issue with _NSAppearanceNameDarkAqua not found (this is only avail. on 10.14!))\n# cf. here: https://discourse.julialang.org/t/problem-with-qt-dependency-of-plots-jl/58765\n# cf. here: https://github.com/jheinen/GR.jl/issues/392\n# v1.1.2 is causing the dylib not found error (and subsequently the _NSAppearanceNameDarkAqua when renaming the framework binaries to dylib)\n# v1.1.1 is causing the _NSAppearanceNameDarkAqua not found error \nENV[\"JULIA_GR_PROVIDER\"] = \"GR\" \nPkg.add(Pkg.PackageSpec(;name=\"Plots\", version=\"1.1.0\"))\n\nPkg.add(\"WebIO\")\nPkg.add(\"Interact\")\n\nusing Plots\nusing IJulia\nusing WebIO\n\n# precompile the packages ? \nPkg.precompile()\n\n", "meta": {"hexsha": "63d9a2c7d3721e85516db7f0936c20cd589353bf", "size": 890, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "jlab_server-dist/julia/depot_init.jl", "max_stars_repo_name": "benbenz/jupyterlab-econ-desktop", "max_stars_repo_head_hexsha": "78ed9c8c6d84f520bb7edcfb5b2603f68447eb10", "max_stars_repo_licenses": ["BSD-3-Clause"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "jlab_server-dist/julia/depot_init.jl", "max_issues_repo_name": "benbenz/jupyterlab-econ-desktop", "max_issues_repo_head_hexsha": "78ed9c8c6d84f520bb7edcfb5b2603f68447eb10", "max_issues_repo_licenses": ["BSD-3-Clause"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "jlab_server-dist/julia/depot_init.jl", "max_forks_repo_name": "benbenz/jupyterlab-econ-desktop", "max_forks_repo_head_hexsha": "78ed9c8c6d84f520bb7edcfb5b2603f68447eb10", "max_forks_repo_licenses": ["BSD-3-Clause"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 30.6896551724, "max_line_length": 140, "alphanum_fraction": 0.7382022472, "num_tokens": 286, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4921881357207955, "lm_q2_score": 0.11436853372838407, "lm_q1q2_score": 0.056290835400894274}}
{"text": "n = parse(Int64, readline())\n\nfor i in 1:n\n S = readline()\n\n len = length(S)\n\n odd = \"\"\n even = \"\"\n\n for j in 1:2:len\n odd = odd*S[j]\n end\n for j in 2:2:len\n even = even*S[j]\n end\n println(odd,\" \",even)\n # println(even)\nend", "meta": {"hexsha": "0d19299bf11196a18bcd8f379eed13262991562d", "size": 267, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Hackerrank/30 Days of Code/Julia/day 06.jl", "max_stars_repo_name": "Next-Gen-UI/Code-Dynamics", "max_stars_repo_head_hexsha": "a9b9d5e3f27e870b3e030c75a1060d88292de01c", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "Hackerrank/30 Days of Code/Julia/day 06.jl", "max_issues_repo_name": "Next-Gen-UI/Code-Dynamics", "max_issues_repo_head_hexsha": "a9b9d5e3f27e870b3e030c75a1060d88292de01c", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Hackerrank/30 Days of Code/Julia/day 06.jl", "max_forks_repo_name": "Next-Gen-UI/Code-Dynamics", "max_forks_repo_head_hexsha": "a9b9d5e3f27e870b3e030c75a1060d88292de01c", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 14.0526315789, "max_line_length": 28, "alphanum_fraction": 0.4756554307, "num_tokens": 89, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.42250463481418826, "lm_q2_score": 0.1329642436335826, "lm_q1q2_score": 0.05617800919975158}}
{"text": "# This file was generated, do not modify it. # hide\ndescribe(data)\n\n#\u00a0The describe() function shows that there are several features with missing values.", "meta": {"hexsha": "d432a2da1a5f7cfc9658e43d36a006030a540f3a", "size": 152, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "__site/assets/data/processing/code/ex6.jl", "max_stars_repo_name": "giordano/DataScienceTutorials.jl", "max_stars_repo_head_hexsha": "8284298842e0d77061cf8ee767d0899fb7d051ff", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 29, "max_stars_repo_stars_event_min_datetime": "2021-08-09T11:35:53.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-07T06:20:43.000Z", "max_issues_repo_path": "__site/assets/data/processing/code/ex6.jl", "max_issues_repo_name": "giordano/DataScienceTutorials.jl", "max_issues_repo_head_hexsha": "8284298842e0d77061cf8ee767d0899fb7d051ff", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 56, "max_issues_repo_issues_event_min_datetime": "2019-10-22T00:06:41.000Z", "max_issues_repo_issues_event_max_datetime": "2020-05-21T14:38:09.000Z", "max_forks_repo_path": "__site/assets/data/processing/code/ex6.jl", "max_forks_repo_name": "giordano/DataScienceTutorials.jl", "max_forks_repo_head_hexsha": "8284298842e0d77061cf8ee767d0899fb7d051ff", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 9, "max_forks_repo_forks_event_min_datetime": "2019-11-20T16:25:04.000Z", "max_forks_repo_forks_event_max_datetime": "2020-05-05T11:55:15.000Z", "avg_line_length": 38.0, "max_line_length": 84, "alphanum_fraction": 0.7697368421, "num_tokens": 33, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.42250462027098473, "lm_q2_score": 0.1329642333263228, "lm_q1q2_score": 0.056178002911160624}}
{"text": "### A Pluto.jl notebook ###\n# v0.18.0\n\nusing Markdown\nusing InteractiveUtils\n\n# This Pluto notebook uses @bind for interactivity. When running this notebook outside of Pluto, the following 'mock version' of @bind gives bound variables a default value (instead of an error).\nmacro bind(def, element)\n quote\n local iv = try Base.loaded_modules[Base.PkgId(Base.UUID(\"6e696c72-6542-2067-7265-42206c756150\"), \"AbstractPlutoDingetjes\")].Bonds.initial_value catch; b -> missing; end\n local el = $(esc(element))\n global $(esc(def)) = Core.applicable(Base.get, el) ? Base.get(el) : iv(el)\n el\n end\nend\n\n# \u2554\u2550\u2561 cab5e5da-8609-11ec-2b2c-2d98fe66bb99\nbegin\n import Pkg\n\tENV[\"JULIA_MARGO_LOAD_PYPLOT\"] = \"no thank you\"\n Pkg.activate(mktempdir())\n Pkg.add([\n Pkg.PackageSpec(name=\"Plots\", version=\"1\"),\n Pkg.PackageSpec(name=\"ClimateMARGO\", version=v\"0.3.2\"),\n Pkg.PackageSpec(name=\"PlutoUI\", version=\"0.7\"),\n Pkg.PackageSpec(name=\"HypertextLiteral\", version=\"0.9\"),\n ])\n\t\n\tusing Plots\n\tusing Plots.Colors\n\tusing ClimateMARGO\n\tusing ClimateMARGO.Models\n\tusing ClimateMARGO.Optimization\n\tusing ClimateMARGO.Diagnostics\n\tusing PlutoUI\n using HypertextLiteral\n\t\n\tPlots.default(linewidth=5)\nend;\n\n# \u2554\u2550\u2561 cc8614a3-dd69-42d4-9954-44885bac93eb\nmd\"## Exploring uncertainty in climate forcing, feedbacks, and ocean heat uptake.\n\n*by Henri F. Drake*\n\"\n\n# \u2554\u2550\u2561 84ff8ba3-fd8a-4bc9-ab4e-7f763f98d34d\nmd\"##### Anthropogenic forcing\"\n\n# \u2554\u2550\u2561 8037ac4e-5c70-4aa7-9982-7442baaa83a0\nmd\"##### The climate response\"\n\n# \u2554\u2550\u2561 4575f6fd-d1ec-4fd1-bd56-268fab59a533\n\n\n# \u2554\u2550\u2561 7ea69497-8b83-45f9-93d3-bbce4c3c10b2\nreveal_box = @bind reveal CheckBox(default=false);\n\n# \u2554\u2550\u2561 71975b7f-e8b7-4097-b573-8e383bb83868\nif reveal\nmd\"The analytical scalings are:\n\n$\\text{TCR}\\propto \\frac{a}{\\lambda + \\kappa}, \\quad\\quad\\quad \\text{ECS} \\propto \\frac{a}{\\lambda}, \\quad\\quad\\quad \\tau = \\frac{C_{D}}{\\lambda}\\,\\frac{\\lambda + \\kappa}{\\kappa}.$\"\nelse\nmd\"Reveal solution: $(reveal_box)\"\nend\n\n# \u2554\u2550\u2561 8b17a78f-f3d8-4777-becd-01de58974de3\n\n\n# \u2554\u2550\u2561 56e32b2b-8830-4f2c-97ea-7bc5820dca1b\n\u03c1_slider = @bind \u03c1 Slider(0.:0.5:3, default=3, show_value=true);\n\n# \u2554\u2550\u2561 7e63eaf5-6841-4f2d-9388-427d9905fd2e\ndo_optimize_box = @bind do_optimize CheckBox(default=false);\n\n# \u2554\u2550\u2561 a442349e-f6ba-4841-93a6-d7ea7235371a\nif reveal\nmd\"#### Exercise 2: policy solutions\nExplore the sensivity of ''optimal'' policy solutions. Check the box: $(do_optimize_box)\n\"\nend\n\n# \u2554\u2550\u2561 053db221-7f7f-4358-915f-bcfb9f6b6783\nif do_optimize\n\tmd\"*Note the emissions reductions (and carbon dioxide removal) and corresponding warming reductions above!*\"\nend\n\n# \u2554\u2550\u2561 2f4a79c4-5455-4b8d-9a85-1aac1cdad3f7\nif do_optimize\n\tmd\"#### Exercise 3: Intergenerational (in)equity\n\nYou may have guessed the ''optimal'' level of warming depends strongly on economic preferences, e.g. how much the model values the welfare of future generations.\n \nHow does the ''optimal'' policy response change as we decrease the ''discount rate'' towards intergenerational equity?\n\n $(\u03c1_slider) % per year\n\t\"\nend\n\n# \u2554\u2550\u2561 42a58da0-bffc-4fd1-ab7b-0a90b0664e4e\nbegin\n\tif do_optimize\n\t\tlast_year = 2800\n\telse\n\t\tlast_year = 4000\n\tend\n\tend_year_slider = @bind end_year Slider(2100:50:last_year, default=2200, show_value=true);\nend;\n\n# \u2554\u2550\u2561 97e66e74-7f38-4811-8d11-f69df3e5ce49\nmd\"\n#### Exercise 1: transient and equilibrium global warming\nLengthen the simulation's time horizon and graphically explore the scaling of the following emergent properties with the four key parameters above:\n\nA) the Transient Climate Response (TCR), i.e. the warming in 2150 when forcing stabilizes\n\nB) the Equilibrium Climate Sensivity (ECS), i.e. the warming when temperatures have equilibrated with\n\nC) the timescale $\\tau_{D}$ of the equilibration\n\n$(end_year_slider)\n\"\n\n# \u2554\u2550\u2561 56b88523-aa8e-42d7-8637-17b0a0c6fd22\n\n\n# \u2554\u2550\u2561 b72a5d5f-1806-4ae0-95e1-d3b1e4b845ce\n\n\n# \u2554\u2550\u2561 4fee292d-9cf1-40ec-b96d-0eacb0135cae\nmd\"# Appendix\"\n\n# \u2554\u2550\u2561 1a3b7318-d0a8-424a-96db-1deb03da716b\ncmip5_std = Dict(\"a\"=>0.9*log(2.), \"\u03bb\"=>0.31, \"\u03ba\"=>0.18, \"Cd\"=>62.)\n\n# \u2554\u2550\u2561 8742ea1c-54af-4a7d-8f41-1566a71c4d94\nblob(el, color = \"blue\") = @htl(\"\"\"$(el)
\"\"\");\n\n# \u2554\u2550\u2561 24d098eb-a548-4c80-935b-0e9bec229105\nfunction default_parameters(years)::ClimateModelParameters\n\tresult = deepcopy(ClimateMARGO.IO.included_configurations[\"default\"])\n\tresult.domain = years isa Domain ? years : Domain(step(years), first(years), last(years))\n\tresult.economics.baseline_emissions = ramp_emissions(result.domain)\n result.economics.extra_CO\u2082 = zeros(size(result.economics.baseline_emissions))\n\treturn result\nend;\n\n# \u2554\u2550\u2561 226ae32f-36d4-4563-8af4-1aa9d16702c6\nbegin\n\t# Create default parameter values\n\tyears = 2020:6.0:end_year;\n\tdefault_params = default_parameters(years);\n\tdefault_params.physics.a = 6.9*log(2.);\n\tphys = default_params.physics\nend\n\n# \u2554\u2550\u2561 c834b860-866d-4f21-b51a-4fa301d17348\na_slider = @bind a Slider(\n\tround(phys.a-2*cmip5_std[\"a\"], digits=2)\n\t:round(4*cmip5_std[\"a\"]/100., digits=2)\n\t:round(phys.a+2*cmip5_std[\"a\"], digits=2),\n\tdefault=phys.a, show_value=true\n);\n\n# \u2554\u2550\u2561 27c8bca8-55f9-4289-a839-cebbbf696169\n\u03bb_slider = @bind \u03bb Slider(\n\tround(phys.B-2*cmip5_std[\"\u03bb\"], digits=2)\n\t:round(4*cmip5_std[\"\u03bb\"]/100., digits=2)\n\t:round(phys.B+2*cmip5_std[\"\u03bb\"], digits=2),\n\tdefault=phys.B, show_value=true\n);\n\n# \u2554\u2550\u2561 672287f1-f1bd-4899-9d7e-29a6a5646f10\n\u03ba_slider = @bind \u03ba Slider(\n\tround(phys.\u03ba-2*cmip5_std[\"\u03ba\"], digits=2)\n\t:round(4*cmip5_std[\"\u03ba\"]/100., digits=2)\n\t:round(phys.\u03ba+2*cmip5_std[\"\u03ba\"], digits=2),\n\tdefault=phys.\u03ba, show_value=true\n);\n\n# \u2554\u2550\u2561 58441503-2998-472e-b466-5d4c832f9042\nCd_slider = @bind Cd Slider(\n\tmax(Int(round(phys.Cd-2*cmip5_std[\"Cd\"], digits=0)), 20)\n\t:Int(round(4*cmip5_std[\"Cd\"]/100., digits=0))\n\t:Int(round(phys.Cd+2*cmip5_std[\"Cd\"], digits=0)),\n\tdefault=phys.Cd, show_value=true\n);\n\n# \u2554\u2550\u2561 9c7d7e6c-d2d9-4754-b2e3-72c3b9e0ec06\nlet\n\tblob(\n\t\t@htl(\"\"\"\n\tEnergy Balance Model Parameters \n\tDefaults are CMIP5 multi-model mean; ranges are \u00b12\u03c3. \n\t\n\t\n\t\n\tValue \n\t \n\t\n\t\t\n\n\t\tCO2 forcing parameter, a [W m\u207b\u00b2] \n\t\t$(a_slider) \n\t\t \t\n\n\t\t\n\t\tFeedback parameter, \u03bb [W m\u207b\u00b2 K\u207b\u00b9] \n\t\t$(\u03bb_slider) \n\t\t\n\n\t\t\n\t\t Heat uptake rate, \u03ba [W m\u207b\u00b2 K\u207b\u00b9] \n\t\t$(\u03ba_slider) \n\t\t \n\t\t\n\t\t\n\t\t Deep ocean heat capacity, C\u2080 [W yr m\u207b\u00b2 K\u207b\u00b9] \n\t\t$(Cd_slider) \n\t\t \n\t\t\n\t\t\n\t \n\t
\n\t\t\"\"\"),\n\t\t\"#c0ecff33\"\n\t)\nend\n\n# \u2554\u2550\u2561 8c1ddae9-a9dd-4556-9027-4f701a064604\nbegin\n\t# Overwrite defaults with slider values\n\tparams = deepcopy(default_params)\n\tparams.economics.\u03c1 = \u03c1/100. + params.economics.\u03b3;\n\tparams.economics.\u03b2 = 0.003;\n\tparams.physics.T0 = 1.0;\n\tparams.physics.a = a;\n\tparams.physics.B = \u03bb;\n\tparams.physics.\u03ba = \u03ba;\n\tparams.physics.Cd = Cd;\n\tm = ClimateModel(params);\n\tmax_deployment = Dict(\"mitigate\"=>1., \"remove\"=>0.5, \"geoeng\"=>0., \"adapt\"=>0.)\n\tif do_optimize\n\t\topt = optimize_controls!(\n\t\t\tm, obj_option=\"net_benefit\", max_deployment=max_deployment\n\t\t);\n\tend\n\tm;\nend\n\n# \u2554\u2550\u2561 1285f653-a4a8-4c5c-b395-2f3ffd24811d\nbegin\n\tp_emit = plot(t(m), effective_emissions(m), color=:grey, linewidth=3, alpha=0.5, label=\"baseline fossil growth\")\n\tif do_optimize\n\t\tplot!(p_emit, t(m), effective_emissions(m, M=true, R=true), linewidth=5, color=:deepskyblue2, label=\"''optimal'' policy\")\n\tend\n\tplot!(p_emit, t(m), 0. *t(m), linestyle=:dash, color=:black, alpha=0.3, linewidth = 2.5, label=\"\")\n\tplot!(p_emit, ylim=(-5., 15.), xlim=(t(m)[1], t(m)[end]))\n\tplot!(p_emit, ylabel=\"emissions [ppm/year]\", xlabel=\"year\")\n\tplot!(p_emit, size=(700, 225), margins=2.75Plots.Measures.mm)\nend\n\n# \u2554\u2550\u2561 13b0c090-ff52-4eca-98a5-1306f859c58f\nbegin\n\tp = plot(t(m), T(m), color=:grey, linewidth=3, alpha=0.5, label=\"baseline fossil growth\")\n\tif do_optimize\n\t\tplot!(t(m), T(m, M=true, R=true), linewidth=5, color=:deepskyblue2, label=\"''optimal'' policy\")\n\tend\n\tif minimum(T(m, M=true, R=true)) < 0.\n\t\tplot!(ylim=(-2, max(8, maximum(T(m, M=true, R=true)))), xlim=(t(m)[1], t(m)[end]))\n\telse\n\t\tplot!(ylim=(0, max(8, maximum(T(m, M=true, R=true)))), xlim=(t(m)[1], t(m)[end]))\n\tend\n\tplot!(ylabel=\"warming (relative to P-I)\", xlabel=\"year\")\n\tplot!(yticks=[-1., 0., 1., 1.5, 2., 2.5, 3., 4., 5., 6., 7., 8., 9, 10.])\n\tplot!(size=(700, 250), margins=2.75Plots.Measures.mm)\n\tplot!(legend=:topleft)\nend\n\n# \u2554\u2550\u2561 10befe5b-9e7c-40a6-9079-1ab5a380888b\nif do_optimize\n\tp_discount = plot(\n\t\tt(m),\n\t\t(1 .- (m.economics.\u03c1 - m.economics.\u03b3)).^((t(m) .- t(m)[1])) * 100., label=\"\"\n\t)\n\tplot!(xlabel=\"year\", ylabel=\"discount factor [%]\", ylim = (0, 100))\nend\n\n# \u2554\u2550\u2561 Cell order:\n# \u255f\u2500cc8614a3-dd69-42d4-9954-44885bac93eb\n# \u255f\u250084ff8ba3-fd8a-4bc9-ab4e-7f763f98d34d\n# \u255f\u25001285f653-a4a8-4c5c-b395-2f3ffd24811d\n# \u255f\u25008037ac4e-5c70-4aa7-9982-7442baaa83a0\n# \u255f\u250013b0c090-ff52-4eca-98a5-1306f859c58f\n# \u255f\u25009c7d7e6c-d2d9-4754-b2e3-72c3b9e0ec06\n# \u255f\u250097e66e74-7f38-4811-8d11-f69df3e5ce49\n# \u255f\u25004575f6fd-d1ec-4fd1-bd56-268fab59a533\n# \u255f\u250071975b7f-e8b7-4097-b573-8e383bb83868\n# \u255f\u25007ea69497-8b83-45f9-93d3-bbce4c3c10b2\n# \u255f\u2500a442349e-f6ba-4841-93a6-d7ea7235371a\n# \u255f\u2500053db221-7f7f-4358-915f-bcfb9f6b6783\n# \u255f\u25008b17a78f-f3d8-4777-becd-01de58974de3\n# \u255f\u25002f4a79c4-5455-4b8d-9a85-1aac1cdad3f7\n# \u255f\u250010befe5b-9e7c-40a6-9079-1ab5a380888b\n# \u255f\u250042a58da0-bffc-4fd1-ab7b-0a90b0664e4e\n# \u255f\u250056e32b2b-8830-4f2c-97ea-7bc5820dca1b\n# \u255f\u25007e63eaf5-6841-4f2d-9388-427d9905fd2e\n# \u255f\u250056b88523-aa8e-42d7-8637-17b0a0c6fd22\n# \u255f\u2500b72a5d5f-1806-4ae0-95e1-d3b1e4b845ce\n# \u255f\u25004fee292d-9cf1-40ec-b96d-0eacb0135cae\n# \u2560\u2550cab5e5da-8609-11ec-2b2c-2d98fe66bb99\n# \u2560\u2550226ae32f-36d4-4563-8af4-1aa9d16702c6\n# \u2560\u25508c1ddae9-a9dd-4556-9027-4f701a064604\n# \u2560\u25501a3b7318-d0a8-424a-96db-1deb03da716b\n# \u255f\u25008742ea1c-54af-4a7d-8f41-1566a71c4d94\n# \u255f\u2500c834b860-866d-4f21-b51a-4fa301d17348\n# \u255f\u250027c8bca8-55f9-4289-a839-cebbbf696169\n# \u255f\u2500672287f1-f1bd-4899-9d7e-29a6a5646f10\n# \u255f\u250058441503-2998-472e-b466-5d4c832f9042\n# \u255f\u250024d098eb-a548-4c80-935b-0e9bec229105\n", "meta": {"hexsha": "6a9cd632985e651978f614e002af0ce93fad2413", "size": 10045, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "ebm.jl", "max_stars_repo_name": "ClimateMARGO/interactive", "max_stars_repo_head_hexsha": "9351c5d33e4a17f0272bd397f7bb429ab50c1ecd", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 5, "max_stars_repo_stars_event_min_datetime": "2021-09-24T18:15:49.000Z", "max_stars_repo_stars_event_max_datetime": "2022-02-07T21:21:09.000Z", "max_issues_repo_path": "ebm.jl", "max_issues_repo_name": "ClimateMARGO/interactive", "max_issues_repo_head_hexsha": "9351c5d33e4a17f0272bd397f7bb429ab50c1ecd", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 8, "max_issues_repo_issues_event_min_datetime": "2022-02-03T13:31:45.000Z", "max_issues_repo_issues_event_max_datetime": "2022-02-11T20:47:51.000Z", "max_forks_repo_path": "ebm.jl", "max_forks_repo_name": "ClimateMARGO/interactive", "max_forks_repo_head_hexsha": "9351c5d33e4a17f0272bd397f7bb429ab50c1ecd", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 30.0748502994, "max_line_length": 195, "alphanum_fraction": 0.7014435042, "num_tokens": 4246, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.48438008427698437, "lm_q2_score": 0.1159607258918425, "lm_q1q2_score": 0.05616906618031096}}
{"text": "## RESAMPLING STRATEGIES\n\nabstract type ResamplingStrategy <: MLJType end\nshow_as_constructed(::Type{<:ResamplingStrategy}) = true\n\n# resampling strategies are `==` if they have the same type and their\n# field values are `==`:\nfunction ==(s1::S, s2::S) where S <: ResamplingStrategy\n return all(getfield(s1, fld) == getfield(s2, fld) for fld in fieldnames(S))\nend\n\n# fallbacks:\ntrain_test_pairs(s::ResamplingStrategy, rows, X, y, w) =\n train_test_pairs(s, rows, X, y)\ntrain_test_pairs(s::ResamplingStrategy, rows, X, y) =\n train_test_pairs(s, rows, y)\ntrain_test_pairs(s::ResamplingStrategy, rows, y) =\n train_test_pairs(s, rows)\n\n\n# Helper to interpret rng, shuffle in case either is `nothing` or if\n# `rng` is an integer:\nfunction shuffle_and_rng(shuffle, rng)\n if rng isa Integer\n rng = MersenneTwister(rng)\n end\n\n if shuffle === nothing\n shuffle = ifelse(rng===nothing, false, true)\n end\n\n if rng === nothing\n rng = Random.GLOBAL_RNG\n end\n\n return shuffle, rng\nend\n\n\"\"\"\n holdout = Holdout(; fraction_train=0.7,\n shuffle=nothing,\n rng=nothing)\n\nHoldout resampling strategy, for use in `evaluate!`, `evaluate` and in tuning.\n\n train_test_pairs(holdout, rows)\n\nReturns the pair `[(train, test)]`, where `train` and `test` are\nvectors such that `rows=vcat(train, test)` and\n`length(train)/length(rows)` is approximatey equal to fraction_train`.\n\nPre-shuffling of `rows` is controlled by `rng` and `shuffle`. If `rng`\nis an integer, then the `Holdout` keyword constructor resets it to\n`MersenneTwister(rng)`. Otherwise some `AbstractRNG` object is\nexpected.\n\nIf `rng` is left unspecified, `rng` is reset to `Random.GLOBAL_RNG`,\nin which case rows are only pre-shuffled if `shuffle=true` is\nspecified.\n\n\"\"\"\nstruct Holdout <: ResamplingStrategy\n fraction_train::Float64\n shuffle::Bool\n rng::Union{Int,AbstractRNG}\n\n function Holdout(fraction_train, shuffle, rng)\n 0 < fraction_train < 1 ||\n error(\"`fraction_train` must be between 0 and 1.\")\n return new(fraction_train, shuffle, rng)\n end\nend\n\n# Keyword Constructor\nHoldout(; fraction_train::Float64=0.7, shuffle=nothing, rng=nothing) =\n Holdout(fraction_train, shuffle_and_rng(shuffle, rng)...)\n\nfunction train_test_pairs(holdout::Holdout, rows)\n\n train, test = partition(rows, holdout.fraction_train,\n shuffle=holdout.shuffle, rng=holdout.rng)\n return [(train, test),]\n\nend\n\n\n\"\"\"\n cv = CV(; nfolds=6, shuffle=nothing, rng=nothing)\n\nCross-validation resampling strategy, for use in `evaluate!`,\n`evaluate` and tuning.\n\n train_test_pairs(cv, rows)\n\nReturns an `nfolds`-length iterator of `(train, test)` pairs of\nvectors (row indices), where each `train` and `test` is a sub-vector\nof `rows`. The `test` vectors are mutually exclusive and exhaust\n`rows`. Each `train` vector is the complement of the corresponding\n`test` vector. With no row pre-shuffling, the order of `rows` is\npreserved, in the sense that `rows` coincides precisely with the\nconcatenation of the `test` vectors, in the order they are\ngenerated. All but the last `test` vector have equal length.\n\nPre-shuffling of `rows` is controlled by `rng` and `shuffle`. If `rng`\nis an integer, then the `CV` keyword constructor resets it to\n`MersenneTwister(rng)`. Otherwise some `AbstractRNG` object is\nexpected.\n\nIf `rng` is left unspecified, `rng` is reset to `Random.GLOBAL_RNG`,\nin which case rows are only pre-shuffled if `shuffle=true` is\nexplicitly specified.\n\n\"\"\"\nstruct CV <: ResamplingStrategy\n nfolds::Int\n shuffle::Bool\n rng::Union{Int,AbstractRNG}\n function CV(nfolds, shuffle, rng)\n nfolds > 1 || error(\"Must have nfolds > 1. \")\n return new(nfolds, shuffle, rng)\n end\nend\n\n# Constructor with keywords\nCV(; nfolds::Int=6, shuffle=nothing, rng=nothing) =\n CV(nfolds, shuffle_and_rng(shuffle, rng)...)\n\nfunction train_test_pairs(cv::CV, rows)\n\n n_observations = length(rows)\n nfolds = cv.nfolds\n\n if cv.shuffle\n rows=shuffle!(cv.rng, collect(rows))\n end\n\n # number of observations per fold\n k = floor(Int, n_observations/nfolds)\n k > 0 || error(\"Inusufficient data for $nfolds-fold cross-validation.\\n\"*\n \"Try reducing nfolds. \")\n\n # define the (trainrows, testrows) pairs:\n firsts = 1:k:((nfolds - 1)*k + 1) # itr of first `test` rows index\n seconds = k:k:(nfolds*k) # itr of last `test` rows index\n\n ret = map(1:nfolds) do k\n f = firsts[k]\n s = seconds[k]\n k < nfolds || (s = n_observations)\n return (vcat(rows[1:(f - 1)], rows[(s + 1):end]), # trainrows\n rows[f:s]) # testrows\n end\n\n return ret\nend\n\n\"\"\"\n stratified_cv = StratifiedCV(; nfolds=6,\n shuffle=false,\n rng=Random.GLOBAL_RNG)\n\nStratified cross-validation resampling strategy, for use in\n`evaluate!`, `evaluate` and in tuning. Applies only to classification\nproblems (`OrderedFactor` or `Multiclass` targets).\n\n train_test_pairs(stratified_cv, rows, y)\n\nReturns an `nfolds`-length iterator of `(train, test)` pairs of\nvectors (row indices) where each `train` and `test` is a sub-vector of\n`rows`. The `test` vectors are mutually exclusive and exhaust\n`rows`. Each `train` vector is the complement of the corresponding\n`test` vector.\n\nUnlike regular cross-validation, the distribution of the levels of the\ntarget `y` corresponding to each `train` and `test` is constrained, as\nfar as possible, to replicate that of `y[rows]` as a whole.\n\nSpecifically, the data is split into a number of groups on which `y`\nis constant, and each individual group is resampled according to the\nordinary cross-validation strategy `CV(nfolds=nfolds)`. To obtain the\nfinal `(train, test)` pairs of row indices, the per-group pairs are\ncollated in such a way that each collated `train` and `test` respects\nthe original order of `rows` (after shuffling, if `shuffle=true`).\n\nPre-shuffling of `rows` is controlled by `rng` and `shuffle`. If `rng`\nis an integer, then the `StratifedCV` keyword constructor resets it to\n`MersenneTwister(rng)`. Otherwise some `AbstractRNG` object is\nexpected.\n\nIf `rng` is left unspecified, `rng` is reset to `Random.GLOBAL_RNG`,\nin which case rows are only pre-shuffled if `shuffle=true` is\nexplicitly specified.\n\n\"\"\"\nstruct StratifiedCV <: ResamplingStrategy\n nfolds::Int\n shuffle::Bool\n rng::Union{Int,AbstractRNG}\n function StratifiedCV(nfolds, shuffle, rng)\n nfolds > 1 || error(\"Must have nfolds > 1. \")\n return new(nfolds, shuffle, rng)\n end\nend\n\n# Constructor with keywords\nStratifiedCV(; nfolds::Int=6, shuffle=nothing, rng=nothing) =\n StratifiedCV(nfolds, shuffle_and_rng(shuffle, rng)...)\n\nfunction train_test_pairs(stratified_cv::StratifiedCV, rows, X, y)\n\n n_observations = length(rows)\n nfolds = stratified_cv.nfolds\n\n if stratified_cv.shuffle\n rows=shuffle!(stratified_cv.rng, collect(rows))\n end\n\n st = scitype(y)\n st <: AbstractArray{<:Finite} ||\n error(\"Supplied target has scitpye $st but stratified \"*\n \"cross-validation applies only to classification problems. \")\n\n\n freq_given_level = countmap(y[rows])\n minimum(values(freq_given_level)) >= nfolds ||\n error(\"The number of observations for which the target takes on a \"*\n \"given class must, for each class, exceed `nfolds`. Try \"*\n \"reducing `nfolds`. \")\n\n levels_seen = keys(freq_given_level) |> collect\n\n cv = CV(nfolds=nfolds)\n\n # the target is constant on each stratum, a subset of `rows`:\n class_rows = [rows[y[rows] .== c] for c in levels_seen]\n\n # get the cv train/test pairs for each level:\n train_test_pairs_per_level = (train_test_pairs(cv, class_rows[m])\n for m in eachindex(levels_seen))\n\n # just the train rows in each level:\n trains_per_level = map(x -> first.(x),\n train_test_pairs_per_level)\n\n # just the test rows in each level:\n tests_per_level = map(x -> last.(x),\n train_test_pairs_per_level)\n\n # for each fold, concatenate the train rows over levels:\n trains_per_fold = map(x->vcat(x...), zip(trains_per_level...))\n\n # for each fold, concatenate the test rows over levels:\n tests_per_fold = map(x->vcat(x...), zip(tests_per_level...))\n\n # restore ordering specified by rows:\n trains_per_fold = map(trains_per_fold) do train\n filter(in(train), rows)\n end\n tests_per_fold = map(tests_per_fold) do test\n filter(in(test), rows)\n end\n\n # re-assemble:\n return zip(trains_per_fold, tests_per_fold) |> collect\n\nend\n\n\n## EVALUATION TYPE\n\nconst PerformanceEvaluation = NamedTuple{(:measure, :measurement,\n :per_fold, :per_observation)}\n# pretty printing:\nround3(x) = round(x, sigdigits=3)\n_short(v::Vector{<:Real}) = MLJBase.short_string(v)\n_short(v::Vector) = string(\"[\", join(_short.(v), \", \"), \"]\")\n_short(::Missing) = missing\n\nfunction Base.show(io::IO, ::MIME\"text/plain\", e::PerformanceEvaluation)\n data = hcat(e.measure, round3.(e.measurement),\n [round3.(v) for v in e.per_fold])\n header = [\"_.measure\", \"_.measurement\", \"_.per_fold\"]\n PrettyTables.pretty_table(io, data, header;\n header_crayon=Crayon(bold=false),\n alignment=:l)\n println(io, \"_.per_observation = $(_short(e.per_observation))\")\nend\n\nfunction Base.show(io::IO, e::PerformanceEvaluation)\n summary = Tuple(round3.(e.measurement))\n print(io, \"PerformanceEvaluation$summary\")\nend\n\n\n## EVALUATION METHODS\n\nfunction _check_measure(model, measure, y, operation, override)\n\n override && (return nothing)\n\n T = scitype(y)\n\n T == Unknown && (return nothing)\n target_scitype(measure) == Unknown && (return nothing)\n prediction_type(measure) == :unknown && (return nothing)\n\n avoid = \"\\nTo override measure checks, set check_measure=false. \"\n\n T <: target_scitype(measure) ||\n throw(ArgumentError(\n \"\\nscitype of target = $T but target_scitype($measure) = \"*\n \"$(target_scitype(measure)).\"*avoid))\n\n if model isa Probabilistic\n if operation == predict\n if prediction_type(measure) != :probabilistic\n suggestion = \"\"\n if target_scitype(measure) <: Finite\n suggestion = \"\\nPerhaps you want to set operation=\"*\n \"predict_mode. \"\n elseif target_scitype(measure) <: Continuous\n suggestion = \"\\nPerhaps you want to set operation=\"*\n \"predict_mean or operation=predict_median. \"\n else\n suggestion = \"\"\n end\n throw(ArgumentError(\n \"\\n$model <: Probabilistic but prediction_type($measure) = \"*\n \":$(prediction_type(measure)). \"*suggestion*avoid))\n end\n end\n end\n\n model isa Deterministic && prediction_type(measure) != :deterministic &&\n throw(ArgumentError(\"$model <: Deterministic but \"*\n \"prediction_type($measure) =\"*\n \":$(prediction_type(measure)).\"*avoid))\n\n return nothing\n\nend\n\nfunction _process_weights_measures(weights, measures, mach,\n operation, verbosity, check_measure)\n\n if measures === nothing\n candidate = default_measure(mach.model)\n candidate === nothing && error(\"You need to specify measure=... \")\n _measures = [candidate, ]\n elseif !(measures isa AbstractVector)\n _measures = [measures, ]\n else\n _measures = measures\n end\n\n y = mach.args[2]\n\n [ _check_measure(mach.model, m, y, operation, !check_measure)\n for m in _measures ]\n\n if weights != nothing\n weights isa AbstractVector{<:Real} ||\n throw(ArgumentError(\"`weights` must be a `Real` vector.\"))\n length(weights) == nrows(y) ||\n throw(DimensionMismatch(\"`weights` and target \"*\n \"have different lengths. \"))\n _weights = weights\n elseif length(mach.args) == 3\n verbosity < 1 ||\n @info \"Passing machine sample weights to any supported measures. \"\n _weights = mach.args[3]\n else\n _weights = weights\n end\n\n return _weights, _measures\n\nend\n\n\"\"\"\n evaluate!(mach,\n resampling=CV(),\n measure=nothing,\n weights=nothing,\n operation=predict,\n n = 1,\n acceleration=default_resource(),\n force=false,\n verbosity=1)\n\nEstimate the performance of a machine `mach` wrapping a supervised\nmodel in data, using the specified `resampling` strategy (defaulting\nto 6-fold cross-validation) and `measure`, which can be a single\nmeasure or vector.\n\nDo `subtypes(MLJ.ResamplingStrategy)` to obtain a list of available\nresampling strategies. If `resampling` is not an object of type\n`MLJ.ResamplingStrategy`, then a vector of pairs (of the form\n`(train_rows, test_rows)` is expected. For example, setting\n\n resampling = [(1:100), (101:200)),\n (101:200), (1:100)]\n\ngives two-fold cross-validation using the first 200 rows of data.\n\nThe resampling strategy is applied repeatedly if `repeats > 1`. For\n`resampling = CV(nfolds=5)`, for example, this generates a total of\n`5n` test folds for evaluation and subsequent aggregation.\n\nIf `resampling isa MLJ.ResamplingStrategy` then one may optionally\nrestrict the data used in evaluation by specifying `rows`.\n\nAn optional `weights` vector may be passed for measures that support\nsample weights (`MLJ.supports_weights(measure) == true`), which is\nignored by those that don't.\n\n*Important:* If `mach` already wraps sample weights `w` (as in `mach =\nmachine(model, X, y, w)`) then these weights, which are used for\n*training*, are automatically passed to the measures for\nevaluation. However, for evaluation purposes, any `weights` specified\nas a keyword argument will take precedence over `w`.\n\nUser-defined measures are supported; see the manual for details.\n\nIf no measure is specified, then `default_measure(mach.model)` is\nused, unless this default is `nothing` and an error is thrown.\n\nThe `acceleration` keyword argument is used to specify the compute resource (a\nsubtype of `ComputationalResources.AbstractResource`) that will be used to\naccelerate/parallelize the resampling operation.\n\nAlthough evaluate! is mutating, `mach.model` and `mach.args` are\nuntouched.\n\n### Return value\n\nA property-accessible object of type `PerformanceEvaluation` with\nthese properties:\n\n- `measure`: the vector of specified measures\n\n- `measurements`: the corresponding measurements, aggregated across the\n test folds using the aggregation method defined for each measure (do\n `aggregation(measure)` to inspect)\n\n- `per_fold`: a vector of vectors of individual test fold evaluations\n (one vector per measure)\n\n- `per_observation`: a vector of vectors of individual observation\n evaluations of those measures for which\n `reports_each_observation(measure)` is true, which is otherwise\n reported `missing`.\n\nSee also [`evaluate`](@ref)\n\n\"\"\"\nfunction evaluate!(mach::Machine{<:Supervised};\n resampling=CV(),\n measures=nothing, measure=measures, weights=nothing,\n operation=predict, acceleration=default_resource(),\n rows=nothing, repeats=1, force=false,\n check_measure=true, verbosity=1)\n\n # this method just checks validity of options, preprocess the\n # weights and measures, and dispatches a strategy-specific\n # `evaluate!`\n\n repeats > 0 || error(\"Need n > 0. \")\n\n if resampling isa TrainTestPairs\n if rows !== nothing\n error(\"You cannot specify `rows` unless `resampling \"*\n \"isa MLJ.ResamplingStrategy` is true. \")\n end\n if repeats != 1 && verbosity > 0\n @warn \"repeats > 1 not supported unless \"*\n \"`resampling<:ResamplingStrategy. \"\n end\n end\n\n _weights, _measures =\n _process_weights_measures(weights, measure, mach,\n operation, verbosity, check_measure)\n\n if verbosity >= 0 && weights !== nothing\n unsupported = filter(_measures) do m\n !supports_weights(m)\n end\n if !isempty(unsupported)\n unsupported_as_string = string(unsupported[1])\n unsupported_as_string *=\n reduce(*, [string(\", \", m) for m in unsupported[2:end]])\n @warn \"Sample weights ignored in evaluations of the following\"*\n \" measures, as unsupported: \\n$unsupported_as_string \"\n end\n end\n\n evaluate!(mach, resampling, _weights, rows, verbosity, repeats,\n _measures, operation, acceleration, force)\n\nend\n\n\"\"\"\n evaluate(model, X, y; measure=nothing, options...)\n evaluate(model, X, y, w; measure=nothing, options...)\n\nEvaluate the performance of a supervised model `model` on input data\n`X` and target `y`, optionally specifying sample weights `w` for\ntraining, where supported. The same weights are passed to measures\nthat support sample weights, unless this behaviour is overridden by\nexplicitly specifying the option `weights=...`.\n\nSee the machine version `evaluate!` for the complete list of options.\n\n\"\"\"\nevaluate(model::Supervised, args...; kwargs...) =\n evaluate!(machine(model, args...); kwargs...)\n\nconst AbstractRow = Union{AbstractVector{<:Integer}, Colon}\nconst TrainTestPair = Tuple{AbstractRow,AbstractRow}\nconst TrainTestPairs = AbstractVector{<:TrainTestPair}\n\nfunction _evaluate!(func::Function, res::CPU1, nfolds, verbosity)\n p = Progress(nfolds + 1, dt=0, desc=\"Evaluating over $nfolds folds: \",\n barglyphs=BarGlyphs(\"[=> ]\"), barlen=25, color=:yellow)\n verbosity > 0 && next!(p)\n return reduce(vcat, (func(k, p, verbosity) for k in 1:nfolds))\nend\nfunction _evaluate!(func::Function, res::CPUProcesses, nfolds, verbosity)\n # TODO: use pmap here ?:\n return @distributed vcat for k in 1:nfolds\n func(k)\n end\nend\n@static if VERSION >= v\"1.3.0-DEV.573\"\n function _evaluate!(func::Function, res::CPUThreads, nfolds, verbosity)\n task_vec = [Threads.@spawn func(k) for k in 1:nfolds]\n return reduce(vcat, fetch.(task_vec))\n end\nend\n\n# Evaluation when resampling is a TrainTestPairs (core evaluator):\nfunction evaluate!(mach::Machine, resampling, weights,\n rows, verbosity, repeats,\n measures, operation, acceleration, force)\n\n # Note: `rows` and `n` are ignored here\n\n resampling isa TrainTestPairs ||\n error(\"`resampling` must be an \"*\n \"`MLJ.ResamplingStrategy` or tuple of pairs \"*\n \"of the form `(train_rows, test_rows)`\")\n\n X = mach.args[1]\n y = mach.args[2]\n\n nfolds = length(resampling)\n\n nmeasures = length(measures)\n\n function get_measurements(k)\n train, test = resampling[k]\n fit!(mach; rows=train, verbosity=verbosity-1, force=force)\n Xtest = selectrows(X, test)\n ytest = selectrows(y, test)\n if weights == nothing\n wtest = nothing\n else\n wtest = weights[test]\n end\n yhat = operation(mach, Xtest)\n return [value(m, yhat, Xtest, ytest, wtest)\n for m in measures]\n end\n function get_measurements(k, p, verbosity) # p = progress meter\n ret = get_measurements(k)\n verbosity > 0 && next!(p)\n return ret\n end\n\n measurements_flat = if acceleration isa CPUProcesses\n ## TODO: progress meter for distributed case\n if verbosity > 0\n @info \"Distributing cross-validation computation \" *\n \"among $(nworkers()) workers.\"\n end\n end\n measurements_flat =\n _evaluate!(get_measurements, acceleration, nfolds, verbosity)\n\n # in the following rows=folds, columns=measures:\n measurements_matrix = permutedims(\n reshape(measurements_flat, (nmeasures, nfolds)))\n\n # measurements for each observation:\n per_observation = map(1:nmeasures) do k\n m = measures[k]\n if reports_each_observation(m)\n [measurements_matrix[:,k]...]\n else\n missing\n end\n end\n\n # measurements for each fold:\n per_fold = map(1:nmeasures) do k\n m = measures[k]\n if reports_each_observation(m)\n broadcast(MLJBase.aggregate, per_observation[k], [m,])\n else\n [measurements_matrix[:,k]...]\n end\n end\n\n # overall aggregates:\n per_measure = map(1:nmeasures) do k\n m = measures[k]\n MLJBase.aggregate(per_fold[k], m)\n end\n\n ret = (measure=measures,\n measurement=per_measure,\n per_fold=per_fold,\n per_observation=per_observation)\n\n return ret\n\nend\n\nfunction actual_rows(rows, N, verbosity)\n unspecified_rows = (rows === nothing)\n _rows = unspecified_rows ? (1:N) : rows\n unspecified_rows || @info \"Creating subsamples from a subset of all rows. \"\n return _rows\nend\n\n# Evaluation when resampling is a ResamplingStrategy:\nfunction evaluate!(mach::Machine, resampling::ResamplingStrategy,\n weights, rows, verbosity, repeats, args...)\n\n y = mach.args[2]\n _rows = actual_rows(rows, length(y), verbosity)\n\n repeated_train_test_pairs =\n vcat([train_test_pairs(resampling, _rows, mach.args...)\n for i in 1:repeats]...)\n\n return evaluate!(mach::Machine,\n repeated_train_test_pairs,\n weights, nothing, verbosity, repeats, args...)\n\nend\n\n\n## RESAMPLER - A MODEL WRAPPER WITH `evaluate` OPERATION\n\n\"\"\"\n resampler = Resampler(model=ConstantRegressor(),\n resampling=CV(),\n measure=nothing,\n weights=nothing,\n operation=predict,\n repeats = 1,\n acceleration=default_resource(),\n check_measure=true)\n\nResampling model wrapper, used internally by the `fit` method of\n`TunedModel` instances. See [`evaluate!](@ref) for options. Not\nintended for general use.\n\nGiven a machine `mach = machine(resampler, args...)` one obtains a\nperformance evaluation of the specified `model`, performed according\nto the prescribed `resampling` strategy and other parameters, using\ndata `args...`, by calling `fit!(mach)` followed by\n`evaluate(mach)`. The advantage over using `evaluate(model, X, y)` is\nthat the latter call always calls `fit` on the `model` but\n`fit!(mach)` only calls `update` after the first call.\n\nThe sample `weights` are passed to the specified performance\nmeasures that support weights for evaluation.\n\n*Important:* If `weights` are left unspecified, then any weight vector\n`w` used in constructing the resampler machine, as in\n`resampler_machine = machine(resampler, X, y, w)` (which is then used\nin *training* the model) will also be used in evaluation.\n\n\"\"\"\nmutable struct Resampler{S,M<:Supervised} <: Supervised\n model::M\n resampling::S # resampling strategy\n measure\n weights::Union{Nothing,AbstractVector{<:Real}}\n operation\n acceleration::AbstractResource\n check_measure::Bool\n repeats::Int\nend\n\nMLJBase.is_wrapper(::Type{<:Resampler}) = true\nMLJBase.supports_weights(::Type{<:Resampler{<:Any,M}}) where M =\n supports_weights(M)\nMLJBase.is_pure_julia(::Type{<:Resampler}) = true\n\nfunction MLJBase.clean!(resampler::Resampler)\n warning = \"\"\n if resampler.measure === nothing\n measure = default_measure(resampler.model)\n if measure === nothing\n error(\"No default measure known for $(resampler.model). \"*\n \"You must specify measure=... \")\n else\n warning *= \"No `measure` specified. \"*\n \"Setting `measure=$measure`. \"\n end\n end\n return warning\nend\n\nfunction Resampler(; model=ConstantRegressor(), resampling=CV(),\n measure=nothing, weights=nothing, operation=predict,\n acceleration=default_resource(), check_measure=true, repeats=1)\n\n resampler = Resampler(model, resampling, measure, weights, operation,\n acceleration, check_measure, repeats)\n message = MLJBase.clean!(resampler)\n isempty(message) || @warn message\n\n return resampler\n\nend\n\nfunction MLJBase.fit(resampler::Resampler, verbosity::Int, args...)\n\n mach = machine(resampler.model, args...)\n\n weights, measures =\n _process_weights_measures(resampler.weights, resampler.measure,\n mach, resampler.operation,\n verbosity, resampler.check_measure)\n\n fitresult = evaluate!(mach, resampler.resampling,\n weights, nothing, verbosity - 1, resampler.repeats,\n measures, resampler.operation,\n resampler.acceleration, false)\n\n cache = (mach, deepcopy(resampler.resampling))\n report = NamedTuple()\n\n return fitresult, cache, report\n\nend\n\n# in special case of holdout, we can reuse the underlying model's\n# machine, provided the training_fraction has not changed:\nfunction MLJBase.update(resampler::Resampler{Holdout},\n verbosity::Int, fitresult, cache, args...)\n\n old_mach, old_resampling = cache\n\n if old_resampling.fraction_train == resampler.resampling.fraction_train\n mach = old_mach\n else\n mach = machine(resampler.model, args...)\n cache = (mach, deepcopy(resampler.resampling))\n end\n\n weights, measures =\n _process_weights_measures(resampler.weights, resampler.measure,\n mach, resampler.operation,\n verbosity, resampler.check_measure)\n mach.model = resampler.model\n fitresult = evaluate!(mach, resampler.resampling,\n weights, nothing, verbosity - 1, resampler.repeats,\n measures, resampler.operation,\n resampler.acceleration, false)\n\n\n report = NamedTuple\n\n return fitresult, cache, report\n\nend\n\nMLJBase.input_scitype(::Type{<:Resampler{S,M}}) where {S,M} =\n MLJBase.input_scitype(M)\nMLJBase.target_scitype(::Type{<:Resampler{S,M}}) where {S,M} =\n MLJBase.target_scitype(M)\nMLJBase.package_name(::Type{<:Resampler}) = \"MLJBase\"\n\nMLJBase.load_path(::Type{<:Resampler}) = \"MLJBase.Resampler\"\n\nevaluate(resampler::Resampler, fitresult) = fitresult\n\nfunction evaluate(machine::AbstractMachine{<:Resampler})\n if isdefined(machine, :fitresult)\n return evaluate(machine.model, machine.fitresult)\n else\n throw(error(\"$machine has not been trained.\"))\n end\nend\n\n", "meta": {"hexsha": "7ee7571cf0ecb257dd4d3910d0f8a3f02e202608", "size": 27063, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/resampling.jl", "max_stars_repo_name": "darenasc/MLJBase.jl", "max_stars_repo_head_hexsha": "88d70061e2823528026998c2019d25b885dc37e4", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/resampling.jl", "max_issues_repo_name": "darenasc/MLJBase.jl", "max_issues_repo_head_hexsha": "88d70061e2823528026998c2019d25b885dc37e4", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/resampling.jl", "max_forks_repo_name": "darenasc/MLJBase.jl", "max_forks_repo_head_hexsha": "88d70061e2823528026998c2019d25b885dc37e4", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 33.7443890274, "max_line_length": 80, "alphanum_fraction": 0.6503344049, "num_tokens": 6509, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4843800842769844, "lm_q2_score": 0.11596071825396675, "lm_q1q2_score": 0.05616906248067606}}
{"text": "module Queues\n\nexport Queue, enqueue!, dequeue!, peek\n\ninclude(\"LinkedLists.jl\")\nusing .LinkedLists\n\nstruct Queue{T}\n list::LinkedList{T}\nend\n\nQueue(t::Type{T}) where {T} = Queue{T}(LinkedList{T}())\nQueue{T}() where {T} = Queue{T}(LinkedList{T}())\nQueue() = Queue{Any}(LinkedList{Any}())\n\nfunction enqueue!(q::Queue{T}, elem::T) where {T}\n pushfirst!(q.list, Node(elem))\n q\nend\n\nfunction dequeue!(q::Queue{T}) where {T}\n isempty(q) && throw(ArgumentError(\"Queue must not be empty\"))\n pop!(q.list).data\nend\n\nfunction peek(q::Queue{T}) where {T}\n isempty(q) && throw(ArgumentError(\"Queue must not be empty\"))\n (q.list[end]).data\nend\n\nfunction Base.length(q::Queue{T}) where {T}\n length(q.list)\nend\n\nfunction Base.isempty(q::Queue{T}) where {T}\n length(q) == 0\nend\n\nfunction Base.empty!(q::Queue{T}) where {T}\n empty!(q.list)\n\n q\nend\n\nfunction Base.eltype(q::Queue{T}) where {T}\n eltype(q.list)\nend\n\nfunction Base.iterate(q::Queue{T}, state::Union{Queue,Nothing} = q) where {T}\n (state == nothing || isempty(q)) && return nothing\n dequeue!(q), q\nend\n\nend\n", "meta": {"hexsha": "72a244b9a4d15b0ee283916a2a27c0e9c770e8f5", "size": 1066, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Chapter 6/Queues.jl", "max_stars_repo_name": "lytemar/Hands-On-Data-Structures-and-Algorithms-with-Julia", "max_stars_repo_head_hexsha": "c3a0dfbc6dba61fc8e8d5062a69bdafa62f78543", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 13, "max_stars_repo_stars_event_min_datetime": "2019-08-14T03:23:55.000Z", "max_stars_repo_stars_event_max_datetime": "2021-10-04T06:36:56.000Z", "max_issues_repo_path": "Chapter 6/Queues.jl", "max_issues_repo_name": "lytemar/Hands-On-Data-Structures-and-Algorithms-with-Julia", "max_issues_repo_head_hexsha": "c3a0dfbc6dba61fc8e8d5062a69bdafa62f78543", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Chapter 6/Queues.jl", "max_forks_repo_name": "lytemar/Hands-On-Data-Structures-and-Algorithms-with-Julia", "max_forks_repo_head_hexsha": "c3a0dfbc6dba61fc8e8d5062a69bdafa62f78543", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 7, "max_forks_repo_forks_event_min_datetime": "2019-11-15T16:43:35.000Z", "max_forks_repo_forks_event_max_datetime": "2021-10-06T07:48:52.000Z", "avg_line_length": 19.3818181818, "max_line_length": 77, "alphanum_fraction": 0.6641651032, "num_tokens": 324, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.47657965106367595, "lm_q2_score": 0.11757213199952937, "lm_q1q2_score": 0.05603248564314816}}
{"text": "#=\nQueue\n=#\n\nusing SimpleDataStructures\n\nsq = SimpleQueue{Int}()\n\nenqueue!(sq, 1)\nenqueue!(sq, 2)\ndequeue!(sq)\nenqueue!(sq, 3)\nenqueue!(sq, 4)\ndequeue!(sq)\ndequeue!(sq)\ndequeue!(sq)\ndequeue!(sq)\n", "meta": {"hexsha": "589259b07381f15fd381b42c23dd0375ca61b047", "size": 195, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "examples/queue.jl", "max_stars_repo_name": "harryscholes/SimpleDataStructures.jl", "max_stars_repo_head_hexsha": "bce776a723aa1026c37246b128203afab2d2424c", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "examples/queue.jl", "max_issues_repo_name": "harryscholes/SimpleDataStructures.jl", "max_issues_repo_head_hexsha": "bce776a723aa1026c37246b128203afab2d2424c", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "examples/queue.jl", "max_forks_repo_name": "harryscholes/SimpleDataStructures.jl", "max_forks_repo_head_hexsha": "bce776a723aa1026c37246b128203afab2d2424c", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 10.8333333333, "max_line_length": 26, "alphanum_fraction": 0.6717948718, "num_tokens": 72, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4571367168274948, "lm_q2_score": 0.12252321732428778, "lm_q1q2_score": 0.05600986130276655}}
{"text": "# This file was generated, do not modify it. # hide\nusing ScientificTypes\ndata = coerce(boston, :Tax=>Continuous, :Rad=>Continuous);", "meta": {"hexsha": "84d3d0f6747da8991f07a7abf5d84af4e1f86f27", "size": 132, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "assets/pages/isl/lab-3/code/ex4.jl", "max_stars_repo_name": "vdayanand/MLJTutorials", "max_stars_repo_head_hexsha": "702effdcfadb44fb7c4fe61355f0c481595c95bf", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "assets/pages/isl/lab-3/code/ex4.jl", "max_issues_repo_name": "vdayanand/MLJTutorials", "max_issues_repo_head_hexsha": "702effdcfadb44fb7c4fe61355f0c481595c95bf", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "assets/pages/isl/lab-3/code/ex4.jl", "max_forks_repo_name": "vdayanand/MLJTutorials", "max_forks_repo_head_hexsha": "702effdcfadb44fb7c4fe61355f0c481595c95bf", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 44.0, "max_line_length": 58, "alphanum_fraction": 0.75, "num_tokens": 34, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.49609382947091957, "lm_q2_score": 0.11279540926528922, "lm_q1q2_score": 0.055957106529156975}}
{"text": "# See learn_Plotly.jl for more examples\r\n# http://juliaplots.org/PlotlyJS.jl/stable/\r\n\r\nusing PlotlyJS, HTTP, CSV\r\nimport JSON\r\nimport DataFrames\r\n#import RDatasets\r\nimport WebIO\r\n#import UUIDs\r\nimport DefaultApplication\r\n\r\n\"\"\"\r\n dataset(dir, name)\r\n\r\nReplace the dataset function from RDatasets. It stopped working.\r\n\"\"\"\r\nfunction dataset(dir, name)\r\n url = \"https://raw.githubusercontent.com/vincentarelbundock/Rdatasets/master/csv/$(dir)/$(name).csv\"\r\n df = urldownload(url)\r\n\r\n return df\r\nend\r\n\r\nfunction urldownload(url)\r\n body = HTTP.get(url).body\r\n csv = CSV.File(body)\r\n df = DataFrames.DataFrame(csv)\r\n\r\n return df\r\nend\r\n\r\n#\r\n# Building Blocks\r\n#\r\n\r\n\"\"\"\r\n\r\nhttp://juliaplots.org/PlotlyJS.jl/stable/building_traces_layouts/\r\n\"\"\"\r\nfunction sec_Traces()\r\n # http://juliaplots.org/PlotlyJS.jl/stable/building_traces_layouts/\r\n t = PlotlyJS.scatter(\r\n x=[1,2,3,4,5],\r\n y=[1,6,3,6,1],\r\n mode=\"markers+text\",\r\n name=\"Team A\",\r\n text=[\"A1\", \"A2\", \"A3\", \"A4\", \"A5\"],\r\n textposition=\"top center\",\r\n textfont_family=\"Raleway, sans-serif\", # _ for nested attributes\r\n marker_size=12\r\n )\r\n #print(JSON.json(t,2))\r\n\r\n # Access attributes\r\n @show getindex(t, :text)\r\n @show t[:text]\r\n @show t[\"textfont.family\"]\r\n @show t[\"textfont_family\"]\r\n\r\n # Setting attributes\r\n t[\"marker_color\"] =\"red\"\r\n t[\"line_width\"] = 5\r\n\r\n return t\r\nend\r\n\r\nfunction sec_Layouts()\r\n l = Layout(\r\n title=\"Penguins\",\r\n xaxis_range=[0, 42.0],\r\n xaxis_title=\"fish\",\r\n xaxis_showgrid=true,\r\n yaxis_title=\"Weight\",\r\n yaxis_showgrid=true,\r\n legend_x=0.7,\r\n legend_y=1.15\r\n )\r\n \r\n # Using attr()\r\n l = Layout(\r\n title=\"Penguins\",\r\n xaxis=attr(range=[0, 42.0], title=\"fish\", showgrid=true),\r\n yaxis=attr(title=\"Weight\", showgrid=true),\r\n legend_x=0.7,\r\n legend_y=1.15\r\n )\r\n return l\r\nend\r\n\r\nfunction sec_DataFrames()\r\n iris = RDatasets.dataset(\"datasets\", \"iris\")\r\n trace = scatter(\r\n iris,\r\n x=:SepalLength,\r\n y=:SepalWidth,\r\n marker_color=:red\r\n )\r\n\r\n # TODO: group\r\n return iris\r\nend\r\n\r\n#\r\n# Putting it Together\r\n#\r\n\r\n\"\"\"\r\n\r\nhttp://juliaplots.org/PlotlyJS.jl/stable/syncplots/#Plot\r\n\"\"\"\r\nfunction sec_Plot()\r\n iris = dataset(\"datasets\", \"iris\")\r\n layout = Layout(title=\"Iris\", width=800, height=600)\r\n plot = Plot(iris, x=Symbol(\"Sepal.Length\"), y=Symbol(\"Sepal.Width\"), layout,\r\n mode=\"markers\", marker_size=8,\r\n group=:Species\r\n )\r\n @info typeof(plot)\r\n\r\n return plot\r\nend\r\n#display(sec_Plot())\r\n\r\nfunction sec_SyncPlots1()\r\n p = sec_Plot()\r\n sp = plot(p)\r\n display(sp)\r\n\r\n # Call back\r\n # only works when launch from console\r\n obs = WebIO.on(sp[\"hover\"]) do data\r\n println(\"\\nYou hovered over $data\")\r\n end\r\nend\r\n\r\nfunction sec_SyncPlots2()\r\n p = plot(rand(10,4))\r\n display(p)\r\n WebIO.on(p[\"hover\"]) do data\r\n println(\"\\nYou hovered over: $data\")\r\n end\r\nend\r\n\r\nfunction sec_SyncPlots3()\r\n colors = [fill(\"red\", 10), fill(\"blue\", 10)]\r\n symbols = [fill(\"circle\", 10), fill(\"circle\", 10)]\r\n\r\n ys = [rand(10), rand(10)]\r\n # list comprehension\r\n scatters = [\r\n scatter(y=y, marker=attr(color=c, symbol=s, size=15), line_color=c[1])\r\n for (y,c,s) in zip(ys, colors, symbols)\r\n ]\r\n p = plot(scatters)\r\n display(p)\r\n \r\n # Callback\r\n WebIO.on(p[\"click\"]) do data\r\n colors = (fill(\"red\", 10), fill(\"blue\", 10))\r\n symbols = (fill(\"circle\", 10), fill(\"circle\", 10))\r\n for point in data[\"points\"]\r\n # zero-offset -> unit-offset\r\n colors[point[\"curveNumber\"] + 1][point[\"pointIndex\"] + 1] = \"gold\"\r\n symbols[point[\"curveNumber\"] + 1][point[\"pointIndex\"] + 1] = \"star\"\r\n end\r\n restyle!(p, marker_color=colors, marker_symbol=symbols)\r\n end\r\n\r\n nothing\r\nend\r\n\r\n\r\n#\r\n# Styles\r\n#\r\n\r\nfunction sec_Cyclers()\r\n style = Style(global_trace=attr(\r\n marker_color=Cycler([\"green\",\"red\"]),\r\n line_color=\"blue\"\r\n )\r\n )\r\n #p = plot(rand(10,3), style=style)\r\n p = plot(\r\n [\r\n scatter(y=rand(4)),\r\n scatter(y=rand(4), marker_color=\"black\"),\r\n scatter(y=rand(4)),\r\n scatter(y=rand(4)),\r\n ],\r\n style=style\r\n ) \r\n display(p)\r\nend\r\n\r\n#\r\n# 3D\r\n# \r\nusing PlotlyJS, DataFrames, Colors, Distributions, LinearAlgebra\r\n\r\nfunction random_line()\r\n n = 400\r\n rw() = cumsum(randn(n))\r\n trace1 = scatter3d(;x=rw(),y=rw(), z=rw(), mode=\"lines\",\r\n marker=attr(color=\"#1f77b4\", size=12, symbol=\"circle\",\r\n line=attr(color=\"rgb(0,0,0)\", width=0)),\r\n line=attr(color=\"#1f77b4\", width=1))\r\n trace2 = scatter3d(;x=rw(),y=rw(), z=rw(), mode=\"lines\",\r\n marker=attr(color=\"#9467bd\", size=12, symbol=\"circle\",\r\n line=attr(color=\"rgb(0,0,0)\", width=0)),\r\n line=attr(color=\"rgb(44, 160, 44)\", width=1))\r\n trace3 = scatter3d(;x=rw(),y=rw(), z=rw(), mode=\"lines\",\r\n marker=attr(color=\"#bcbd22\", size=12, symbol=\"circle\",\r\n line=attr(color=\"rgb(0,0,0)\", width=0)),\r\n line=attr(color=\"#bcbd22\", width=1))\r\n layout = Layout(autosize=false, width=500, height=500,\r\n margin=attr(l=0, r=0, b=0, t=65))\r\n plot([trace1, trace2, trace3], layout)\r\nend\r\n#display(random_line())\r\n\r\nfunction topo_surface()\r\n z = Vector[[27.80985, 49.61936, 83.08067, 116.6632, 130.414, 150.7206, 220.1871,\r\n 156.1536, 148.6416, 203.7845, 206.0386, 107.1618, 68.36975, 45.3359,\r\n 49.96142, 21.89279, 17.02552, 11.74317, 14.75226, 13.6671, 5.677561,\r\n 3.31234, 1.156517, -0.147662],\r\n [27.71966, 48.55022, 65.21374, 95.27666, 116.9964, 133.9056, 152.3412,\r\n 151.934, 160.1139, 179.5327, 147.6184, 170.3943, 121.8194, 52.58537,\r\n 33.08871, 38.40972, 44.24843, 69.5786, 4.019351, 3.050024, 3.039719,\r\n 2.996142, 2.967954, 1.999594],\r\n [30.4267, 33.47752, 44.80953, 62.47495, 77.43523, 104.2153, 102.7393, 137.0004,\r\n 186.0706, 219.3173, 181.7615, 120.9154, 143.1835, 82.40501, 48.47132,\r\n 74.71461, 60.0909, 7.073525, 6.089851, 6.53745, 6.666096, 7.306965, 5.73684,\r\n 3.625628],\r\n [16.66549, 30.1086, 39.96952, 44.12225, 59.57512, 77.56929, 106.8925,\r\n 166.5539, 175.2381, 185.2815, 154.5056, 83.0433, 62.61732, 62.33167,\r\n 60.55916, 55.92124, 15.17284, 8.248324, 36.68087, 61.93413, 20.26867,\r\n 68.58819, 46.49812, 0.2360095],\r\n [8.815617, 18.3516, 8.658275, 27.5859, 48.62691, 60.18013, 91.3286,\r\n 145.7109, 116.0653, 106.2662, 68.69447, 53.10596, 37.92797, 47.95942,\r\n 47.42691, 69.20731, 44.95468, 29.17197, 17.91674, 16.25515, 14.65559,\r\n 17.26048, 31.22245, 46.71704],\r\n [6.628881, 10.41339, 24.81939, 26.08952, 30.1605, 52.30802, 64.71007,\r\n 76.30823, 84.63686, 99.4324, 62.52132, 46.81647, 55.76606, 82.4099,\r\n 140.2647, 81.26501, 56.45756, 30.42164, 17.28782, 8.302431, 2.981626,\r\n 2.698536, 5.886086, 5.268358],\r\n [21.83975, 6.63927, 18.97085, 32.89204, 43.15014, 62.86014, 104.6657,\r\n 130.2294, 114.8494, 106.9873, 61.89647, 55.55682, 86.80986, 89.27802,\r\n 122.4221, 123.9698, 109.0952, 98.41956, 77.61374, 32.49031, 14.67344,\r\n 7.370775, 0.03711011, 0.6423392],\r\n [53.34303, 26.79797, 6.63927, 10.88787, 17.2044, 56.18116, 79.70141,\r\n 90.8453, 98.27675, 80.87243, 74.7931, 75.54661, 73.4373, 74.11694, 68.1749,\r\n 46.24076, 39.93857, 31.21653, 36.88335, 40.02525, 117.4297, 12.70328,\r\n 1.729771, 0],\r\n [25.66785, 63.05717, 22.1414, 17.074, 41.74483, 60.27227, 81.42432, 114.444,\r\n 102.3234, 101.7878, 111.031, 119.2309, 114.0777, 110.5296, 59.19355,\r\n 42.47175, 14.63598, 6.944074, 6.944075, 27.74936, 0, 0, 0.09449376, 0.07732264],\r\n [12.827, 69.20554, 46.76293, 13.96517, 33.88744, 61.82613, 84.74799,\r\n 121.122, 145.2741, 153.1797, 204.786, 227.9242, 236.3038, 228.3655,\r\n 79.34425, 25.93483, 6.944074, 6.944074, 6.944075, 7.553681, 0, 0, 0, 0],\r\n [0, 68.66396, 59.0435, 33.35762, 47.45282, 57.8355, 78.91689, 107.8275,\r\n 168.0053, 130.9597, 212.5541, 165.8122, 210.2429, 181.1713, 189.7617,\r\n 137.3378, 84.65395, 8.677168, 6.956576, 8.468093, 0, 0, 0, 0],\r\n [0, 95.17499, 80.03818, 59.89862, 39.58476, 50.28058, 63.81641, 80.61302,\r\n 66.37824, 198.7651, 244.3467, 294.2474, 264.3517, 176.4082, 60.21857,\r\n 77.41475, 53.16981, 56.16393, 6.949235, 7.531059, 3.780177, 0, 0, 0],\r\n [0, 134.9879, 130.3696, 96.86325, 75.70494, 58.86466, 57.20374, 55.18837,\r\n 78.128, 108.5582, 154.3774, 319.1686, 372.8826, 275.4655, 130.2632, 54.93822,\r\n 25.49719, 8.047439, 8.084393, 5.115252, 5.678269, 0, 0, 0],\r\n [0, 48.08919, 142.5558, 140.3777, 154.7261, 87.9361, 58.11092, 52.83869,\r\n 67.14822, 83.66798, 118.9242, 150.0681, 272.9709, 341.1366, 238.664, 190.2,\r\n 116.8943, 91.48672, 14.0157, 42.29277, 5.115252, 0, 0, 0],\r\n [0, 54.1941, 146.3839, 99.48143, 96.19411, 102.9473, 76.14089, 57.7844,\r\n 47.0402, 64.36799, 84.23767, 162.7181, 121.3275, 213.1646, 328.482,\r\n 285.4489, 283.8319, 212.815, 164.549, 92.29631, 7.244015, 1.167, 0, 0],\r\n [0, 6.919659, 195.1709, 132.5253, 135.2341, 89.85069, 89.45549, 60.29967,\r\n 50.33806, 39.17583, 59.06854, 74.52159, 84.93402, 187.1219, 123.9673,\r\n 103.7027, 128.986, 165.1283, 249.7054, 95.39966, 10.00284, 2.39255, 0, 0],\r\n [0, 21.73871, 123.1339, 176.7414, 158.2698, 137.235, 105.3089, 86.63255, 53.11591,\r\n 29.03865, 30.40539, 39.04902, 49.23405, 63.27853, 111.4215, 101.1956,\r\n 40.00962, 59.84565, 74.51253, 17.06316, 2.435141, 2.287471, -0.0003636982, 0],\r\n [0, 0, 62.04672, 136.3122, 201.7952, 168.1343, 95.2046, 58.90624, 46.94091,\r\n 49.27053, 37.10416, 17.97011, 30.93697, 33.39257, 44.03077, 55.64542,\r\n 78.22423, 14.42782, 9.954997, 7.768213, 13.0254, 21.73166, 2.156372,\r\n 0.5317867],\r\n [0, 0, 79.62993, 139.6978, 173.167, 192.8718, 196.3499, 144.6611, 106.5424,\r\n 57.16653, 41.16107, 32.12764, 13.8566, 10.91772, 12.07177, 22.38254,\r\n 24.72105, 6.803666, 4.200841, 16.46857, 15.70744, 33.96221, 7.575688,\r\n -0.04880907],\r\n [0, 0, 33.2664, 57.53643, 167.2241, 196.4833, 194.7966, 182.1884, 119.6961,\r\n 73.02113, 48.36549, 33.74652, 26.2379, 16.3578, 6.811293, 6.63927, 6.639271,\r\n 8.468093, 6.194273, 3.591233, 3.81486, 8.600739, 5.21889, 0],\r\n [0, 0, 29.77937, 54.97282, 144.7995, 207.4904, 165.3432, 171.4047, 174.9216,\r\n 100.2733, 61.46441, 50.19171, 26.08209, 17.18218, 8.468093, 6.63927,\r\n 6.334467, 6.334467, 5.666687, 4.272203, 0, 0, 0, 0],\r\n [0, 0, 31.409, 132.7418, 185.5796, 121.8299, 185.3841, 160.6566, 116.1478,\r\n 118.1078, 141.7946, 65.56351, 48.84066, 23.13864, 18.12932, 10.28531,\r\n 6.029663, 6.044627, 5.694764, 3.739085, 3.896037, 0, 0, 0],\r\n [0, 0, 19.58994, 42.30355, 96.26777, 187.1207, 179.6626, 221.3898, 154.2617,\r\n 142.1604, 148.5737, 67.17937, 40.69044, 39.74512, 26.10166, 14.48469,\r\n 8.65873, 3.896037, 3.571392, 3.896037, 3.896037, 3.896037, 1.077756, 0],\r\n [0.001229679, 3.008948, 5.909858, 33.50574, 104.3341, 152.2165, 198.1988,\r\n 191.841, 228.7349, 168.1041, 144.2759, 110.7436, 57.65214, 42.63504,\r\n 27.91891, 15.41052, 8.056102, 3.90283, 3.879774, 3.936718, 3.968634,\r\n 0.1236256, 3.985531, -0.1835741],\r\n [0, 5.626141, 7.676256, 63.16226, 45.99762, 79.56688, 227.311, 203.9287,\r\n 172.5618, 177.1462, 140.4554, 123.9905, 110.346, 65.12319, 34.31887,\r\n 24.5278, 9.561069, 3.334991, 5.590495, 5.487353, 5.909499, 5.868994,\r\n 5.833817, 3.568177]]\r\n trace = surface(z=z)\r\n layout = Layout(title=\"Mt. Bruno Elevation\", autosize=false, width=500,\r\n height=500, margin=attr(l=65, r=50, b=65, t=90))\r\n plot(trace, layout)\r\nend\r\n#display(topo_surface())\r\nfunction multiple_surface()\r\n z1 = Vector[[8.83, 8.89, 8.81, 8.87, 8.9, 8.87],\r\n [8.89, 8.94, 8.85, 8.94, 8.96, 8.92],\r\n [8.84, 8.9, 8.82, 8.92, 8.93, 8.91],\r\n [8.79, 8.85, 8.79, 8.9, 8.94, 8.92],\r\n [8.79, 8.88, 8.81, 8.9, 8.95, 8.92],\r\n [8.8, 8.82, 8.78, 8.91, 8.94, 8.92],\r\n [8.75, 8.78, 8.77, 8.91, 8.95, 8.92],\r\n [8.8, 8.8, 8.77, 8.91, 8.95, 8.94],\r\n [8.74, 8.81, 8.76, 8.93, 8.98, 8.99],\r\n [8.89, 8.99, 8.92, 9.1, 9.13, 9.11],\r\n [8.97, 8.97, 8.91, 9.09, 9.11, 9.11],\r\n [9.04, 9.08, 9.05, 9.25, 9.28, 9.27],\r\n [9, 9.01, 9, 9.2, 9.23, 9.2],\r\n [8.99, 8.99, 8.98, 9.18, 9.2, 9.19],\r\n [8.93, 8.97, 8.97, 9.18, 9.2, 9.18]]\r\n z2 = map(x->x.+1, z1)\r\n z3 = map(x->x.-1, z1)\r\n trace1 = surface(z=z1, colorscale=\"Viridis\")\r\n trace2 = surface(z=z2, showscale=false, opacity=0.9, colorscale=\"Viridis\")\r\n trace3 = surface(z=z3, showscale=false, opacity=0.9, colorscale=\"Viridis\")\r\n plot([trace1, trace2, trace3])\r\nend\r\n#display(multiple_surface())\r\n\r\nfunction clustering_alpha_shapes()\r\n # load data\r\n iris = dataset(\"datasets\", \"iris\")\r\n nms = unique(iris[!,:Species])\r\n colors = [RGB(0.89, 0.1, 0.1), RGB(0.21, 0.50, 0.72), RGB(0.28, 0.68, 0.3)]\r\n\r\n data = GenericTrace[]\r\n\r\n for (i, nm) in enumerate(nms)\r\n df = iris[iris[!,:Species] .== nm, :]\r\n x=df[!,Symbol(\"Sepal.Length\")]\r\n y=df[!,Symbol(\"Sepal.Width\")]\r\n z=df[!,Symbol(\"Petal.Length\")]\r\n trace = scatter3d(;name=nm, mode=\"markers\",\r\n marker_size=3, marker_color=colors[i], marker_line_width=0,\r\n x=x, y=y, z=z)\r\n push!(data, trace)\r\n\r\n cluster = mesh3d(;color=colors[i], opacity=0.3, x=x, y=y, z=z)\r\n push!(data, cluster)\r\n end\r\n\r\n # notice the nested attrs to create complex JSON objects\r\n layout = Layout(width=800, height=550, autosize=false, title=\"Iris dataset\",\r\n scene=attr(xaxis=attr(gridcolor=\"rgb(255, 255, 255)\",\r\n zerolinecolor=\"rgb(255, 255, 255)\",\r\n showbackground=true,\r\n backgroundcolor=\"rgb(230, 230,230)\"),\r\n yaxis=attr(gridcolor=\"rgb(255, 255, 255)\",\r\n zerolinecolor=\"rgb(255, 255, 255)\",\r\n showbackground=true,\r\n backgroundcolor=\"rgb(230, 230,230)\"),\r\n zaxis=attr(gridcolor=\"rgb(255, 255, 255)\",\r\n zerolinecolor=\"rgb(255, 255, 255)\",\r\n showbackground=true,\r\n backgroundcolor=\"rgb(230, 230,230)\"),\r\n aspectratio=attr(x=1, y=1, z=0.7),\r\n aspectmode = \"manual\"))\r\n\r\n # Need to use Plot to work with Dash \r\n # return plot(data, layout)\r\n return Plot(data, layout)\r\nend\r\n#display(clustering_alpha_shapes())\r\n\r\nfunction scatter_3d()\r\n \u03a3 = fill(0.5, 3, 3) + Diagonal([0.5, 0.5, 0.5])\r\n obs1 = rand(MvNormal(zeros(3), \u03a3), 200)'\r\n obs2 = rand(MvNormal(zeros(3), 0.5\u03a3), 100)'\r\n\r\n trace1 = scatter3d(;x=obs1[:, 1], y=obs1[:, 2], z=obs1[:, 3],\r\n mode=\"markers\", opacity=0.8,\r\n marker_size=12, marker_line_width=0.5,\r\n marker_line_color=\"rgba(217, 217, 217, 0.14)\")\r\n\r\n trace2 = scatter3d(;x=obs2[:, 1], y=obs2[:, 2], z=obs2[:, 3],\r\n mode=\"markers\", opacity=0.9,\r\n marker=attr(color=\"rgb(127, 127, 127)\",\r\n symbol=\"circle\", line_width=1.0,\r\n line_color=\"rgb(204, 204, 204)\"))\r\n\r\n layout = Layout(margin=attr(l=0, r=0, t=0, b=0))\r\n\r\n return Plot([trace1, trace2], layout)\r\nend\r\n#display(scatter_3d())\r\n\r\nfunction trisurf()\r\n facecolor = repeat([\r\n \t\"rgb(50, 200, 200)\",\r\n \t\"rgb(100, 200, 255)\",\r\n \t\"rgb(150, 200, 115)\",\r\n \t\"rgb(200, 200, 50)\",\r\n \t\"rgb(230, 200, 10)\",\r\n \t\"rgb(255, 140, 0)\"\r\n ], inner=[2])\r\n\r\n t = mesh3d(\r\n x=[0, 0, 1, 1, 0, 0, 1, 1],\r\n y=[0, 1, 1, 0, 0, 1, 1, 0],\r\n z=[0, 0, 0, 0, 1, 1, 1, 1],\r\n i=[7, 0, 0, 0, 4, 4, 2, 6, 4, 0, 3, 7],\r\n j=[3, 4, 1, 2, 5, 6, 5, 5, 0, 1, 2, 2],\r\n k=[0, 7, 2, 3, 6, 7, 1, 2, 5, 5, 7, 6],\r\n facecolor=facecolor)\r\n\r\n plot(t)\r\nend\r\n#display(trisurf())\r\n\r\nfunction meshcube()\r\n t = mesh3d(\r\n x=[0, 0, 1, 1, 0, 0, 1, 1],\r\n y=[0, 1, 1, 0, 0, 1, 1, 0],\r\n z=[0, 0, 0, 0, 1, 1, 1, 1],\r\n i=[7, 0, 0, 0, 4, 4, 6, 6, 4, 0, 3, 2],\r\n j=[3, 4, 1, 2, 5, 6, 5, 2, 0, 1, 6, 3],\r\n k=[0, 7, 2, 3, 6, 7, 1, 1, 5, 5, 7, 6],\r\n intensity=range(0, stop=1, length=8),\r\n colorscale=[\r\n [0, \"rgb(255, 0, 255)\"],\r\n [0.5, \"rgb(0, 255, 0)\"],\r\n [1, \"rgb(0, 0, 255)\"]\r\n ]\r\n )\r\n plot(t)\r\nend\r\n#display(meshcube())\r\n\r\n#\r\n# Contour\r\n#\r\nfunction contour1()\r\n \r\nend\r\n\r\n\r\n\r\n#\r\n# Line and Scatter\r\n#\r\nusing PlotlyJS, DataFrames, CSV, Dates\r\nfunction linescatter1()\r\n l = Layout(\r\n #title=\"Penguins\",\r\n #xaxis=attr(range=[0, 42.0], title=\"fish\", showgrid=true),\r\n #yaxis=attr(title=\"Weight\", showgrid=true),\r\n legend=attr(x=0.1, y=0.1)\r\n )\r\n trace1 = scatter(;x=1:4, y=[10, 15, 13, 17], mode=\"markers\")\r\n trace2 = scatter(;x=2:5, y=[16, 5, 11, 9], mode=\"lines\")\r\n trace3 = scatter(;x=1:4, y=[12, 9, 15, 12], mode=\"lines+markers\")\r\n #p = Plot([trace1, trace2, trace3], l)\r\n plot([trace1, trace2, trace3], l)\r\nend\r\n#display(linescatter1())\r\n\r\nfunction linescatter2()\r\n trace1 = scatter(;x=1:5, y=[1, 6, 3, 6, 1],\r\n mode=\"markers\", name=\"Team A\",\r\n text=[\"A-1\", \"A-2\", \"A-3\", \"A-4\", \"A-5\"],\r\n marker_size=12)\r\n\r\n trace2 = scatter(;x=1:5+0.5, y=[4, 1, 7, 1, 4],\r\n mode=\"markers\", name= \"Team B\",\r\n text=[\"B-a\", \"B-b\", \"B-c\", \"B-d\", \"B-e\"])\r\n # setting marker.size this way is _equivalent_ to what we did for trace1\r\n trace2[\"marker\"] = Dict(:size => 12)\r\n\r\n data = [trace1, trace2]\r\n layout = Layout(;title=\"Data Labels Hover\", xaxis_range=[0.75, 5.25],\r\n yaxis_range=[0, 8])\r\n plot(data, layout)\r\nend\r\n#display(linescatter2())\r\n\r\nfunction linescatter3()\r\n trace1 = scatter(;x=1:5, y=[1, 6, 3, 6, 1],\r\n mode=\"markers+text\", name=\"Team A\",\r\n textposition=\"top center\",\r\n text=[\"A-1\", \"A-2\", \"A-3\", \"A-4\", \"A-5\"],\r\n marker_size=12, textfont_family=\"Raleway, sans-serif\")\r\n\r\n trace2 = scatter(;x=1:5+0.5, y=[4, 1, 7, 1, 4],\r\n mode=\"markers+text\", name= \"Team B\",\r\n textposition=\"bottom center\",\r\n text= [\"B-a\", \"B-b\", \"B-c\", \"B-d\", \"B-e\"],\r\n marker_size=12, textfont_family=\"Times New Roman\")\r\n\r\n data = [trace1, trace2]\r\n\r\n layout = Layout(;title=\"Data Labels on the Plot\", xaxis_range=[0.75, 5.25],\r\n yaxis_range=[0, 8], legend_y=0.5, legend_yref=\"paper\",\r\n legend=attr(family=\"Arial, sans-serif\", size=20,\r\n color=\"grey\"))\r\n plot(data, layout)\r\nend\r\n#display(linescatter3())\r\n\r\nfunction linescatter4()\r\n trace1 = scatter(;y=fill(5, 40), mode=\"markers\", marker_size=40,\r\n marker_color=0:39)\r\n layout = Layout(title=\"Scatter Plot with a Color Dimension\")\r\n plot(trace1, layout)\r\nend\r\n#display(linescatter4())\r\n\r\nfunction linescatter5()\r\n\r\n country = [\"Switzerland (2011)\", \"Chile (2013)\", \"Japan (2014)\",\r\n \"United States (2012)\", \"Slovenia (2014)\", \"Canada (2011)\",\r\n \"Poland (2010)\", \"Estonia (2015)\", \"Luxembourg (2013)\",\r\n \"Portugal (2011)\"]\r\n\r\n votingPop = [40, 45.7, 52, 53.6, 54.1, 54.2, 54.5, 54.7, 55.1, 56.6]\r\n regVoters = [49.1, 42, 52.7, 84.3, 51.7, 61.1, 55.3, 64.2, 91.1, 58.9]\r\n\r\n # notice use of `attr` function to make nested attributes\r\n trace1 = scatter(;x=votingPop, y=country, mode=\"markers\",\r\n name=\"Percent of estimated voting age population\",\r\n marker=attr(color=\"rgba(156, 165, 196, 0.95)\",\r\n line_color=\"rgba(156, 165, 196, 1.0)\",\r\n line_width=1, size=16, symbol=\"circle\"))\r\n\r\n trace2 = scatter(;x=regVoters, y=country, mode=\"markers\",\r\n name=\"Percent of estimated registered voters\")\r\n # also could have set the marker props above by using a dict\r\n trace2[\"marker\"] = Dict(:color => \"rgba(204, 204, 204, 0.95)\",\r\n :line => Dict(:color=> \"rgba(217, 217, 217, 1.0)\",\r\n :width=> 1),\r\n :symbol => \"circle\",\r\n :size => 16)\r\n\r\n data = [trace1, trace2]\r\n layout = Layout(Dict{Symbol,Any}(:paper_bgcolor => \"rgb(254, 247, 234)\",\r\n :plot_bgcolor => \"rgb(254, 247, 234)\");\r\n title=\"Votes cast for ten lowest voting age population in OECD countries\",\r\n width=600, height=600, hovermode=\"closest\",\r\n margin=Dict(:l => 140, :r => 40, :b => 50, :t => 80),\r\n xaxis=attr(showgrid=false, \r\n showline=true,\r\n linecolor=\"rgb(102, 102, 102)\",\r\n titlefont_color=\"rgb(204, 204, 204)\",\r\n tickfont_color=\"rgb(102, 102, 102)\",\r\n autotick=false, dtick=10, ticks=\"outside\",\r\n tickcolor=\"rgb(102, 102, 102)\"),\r\n legend=attr(font_size=10, yanchor=\"middle\",\r\n xanchor=\"right\"),\r\n )\r\n\r\n return Plot(data, layout)\r\nend\r\n#display(linescatter5())\r\n\r\nfunction linescatter6()\r\n trace1 = scatter(;x=[52698, 43117], y=[53, 31],\r\n mode=\"markers\",\r\n name=\"North America\",\r\n text=[\"United States\", \"Canada\"],\r\n marker=attr(color=\"rgb(164, 194, 244)\", size=12,\r\n line=attr(color=\"white\", width=0.5))\r\n )\r\n\r\n trace2 = scatter(;x=[39317, 37236, 35650, 30066, 29570, 27159, 23557, 21046, 18007],\r\n y=[33, 20, 13, 19, 27, 19, 49, 44, 38],\r\n mode=\"markers\", name=\"Europe\",\r\n marker_size=12, marker_color=\"rgb(255, 217, 102)\",\r\n text=[\"Germany\", \"Britain\", \"France\", \"Spain\", \"Italy\",\r\n \"Czech Rep.\", \"Greece\", \"Poland\", \"Portugal\"])\r\n\r\n trace3 = scatter(;x=[42952, 37037, 33106, 17478, 9813, 5253, 4692, 3899],\r\n y=[23, 42, 54, 89, 14, 99, 93, 70],\r\n mode=\"markers\",\r\n name=\"Asia/Pacific\",\r\n marker_size=12, marker_color=\"rgb(234, 153, 153)\",\r\n text=[\"Australia\", \"Japan\", \"South Korea\", \"Malaysia\",\r\n \"China\", \"Indonesia\", \"Philippines\", \"India\"])\r\n\r\n trace4 = scatter(;x=[19097, 18601, 15595, 13546, 12026, 7434, 5419],\r\n y=[43, 47, 56, 80, 86, 93, 80],\r\n mode=\"markers\", name=\"Latin America\",\r\n marker_size=12, marker_color=\"rgb(142, 124, 195)\",\r\n text=[\"Chile\", \"Argentina\", \"Mexico\", \"Venezuela\",\r\n \"Venezuela\", \"El Salvador\", \"Bolivia\"])\r\n\r\n data = [trace1, trace2, trace3, trace4]\r\n\r\n layout = Layout(;title=\"Quarter 1 Growth\",\r\n xaxis=attr(title=\"GDP per Capital\", showgrid=false, zeroline=false),\r\n yaxis=attr(title=\"Percent\", zeroline=false))\r\n\r\n plot(data, layout)\r\nend\r\n#display(linescatter6())\r\n\r\n\r\nfunction dumbell()\r\n # reference: https://plot.ly/r/dumbbell-plots/\r\n # read Data into dataframe\r\n nm = tempname()\r\n url = \"https://raw.githubusercontent.com/plotly/datasets/master/school_earnings.csv\"\r\n download(url, nm)\r\n df = CSV.read(nm, DataFrame)\r\n rm(nm)\r\n\r\n # sort dataframe by male earnings\r\n df = sort(df, :Men, rev=false)\r\n\r\n men = scatter(;y=df[!,:School], x=df[!,:Men], mode=\"markers\", name=\"Men\",\r\n marker=attr(color=\"blue\", size=12))\r\n women = scatter(;y=df[!,:School], x=df[!,:Women], mode=\"markers\", name=\"Women\",\r\n marker=attr(color=\"pink\", size=12))\r\n\r\n lines = map(eachrow(df)) do r\r\n scatter(y=fill(r[:School], 2), x=[r[:Women], r[:Men]], mode=\"lines\",\r\n name=r[:School], showlegend=false, line_color=\"gray\")\r\n end\r\n\r\n data = Base.typed_vcat(GenericTrace, men, women, lines)\r\n layout = Layout(width=650, height=650, margin_l=100, yaxis_title=\"School\",\r\n xaxis_title=\"Annual Salary (thousands)\",\r\n title=\"Gender earnings disparity\")\r\n\r\n plot(data, layout)\r\nend\r\n#display(dumbell())\r\n\r\nfunction errorbars1()\r\n trace1 = scatter(;x=vcat(1:10, 10:-1:1),\r\n y=vcat(2:11, 9:-1:0),\r\n fill=\"tozerox\",\r\n fillcolor=\"rgba(0, 100, 80, 0.2)\",\r\n line_color=\"transparent\",\r\n name=\"Fair\",\r\n showlegend=false)\r\n\r\n trace2 = scatter(;x=vcat(1:10, 10:-1:1),\r\n y=[5.5, 3.0, 5.5, 8.0, 6.0, 3.0, 8.0, 5.0, 6.0, 5.5, 4.75,\r\n 5.0, 4.0, 7.0, 2.0, 4.0, 7.0, 4.4, 2.0, 4.5],\r\n fill=\"tozerox\",\r\n fillcolor=\"rgba(0, 176, 246, 0.2)\",\r\n line_color=\"transparent\",\r\n name=\"Premium\",\r\n showlegend=false)\r\n\r\n trace3 = scatter(;x=vcat(1:10, 10:-1:1),\r\n y=[11.0, 9.0, 7.0, 5.0, 3.0, 1.0, 3.0, 5.0, 3.0, 1.0,\r\n -1.0, 1.0, 3.0, 1.0, -0.5, 1.0, 3.0, 5.0, 7.0, 9.],\r\n fill=\"tozerox\",\r\n fillcolor=\"rgba(231, 107, 243, 0.2)\",\r\n line_color=\"transparent\",\r\n name=\"Fair\",\r\n showlegend=false)\r\n\r\n trace4 = scatter(;x=1:10, y=1:10,\r\n line_color=\"rgb(00, 100, 80)\",\r\n mode=\"lines\",\r\n name=\"Fair\")\r\n\r\n trace5 = scatter(;x=1:10,\r\n y=[5.0, 2.5, 5.0, 7.5, 5.0, 2.5, 7.5, 4.5, 5.5, 5.],\r\n line_color=\"rgb(0, 176, 246)\",\r\n mode=\"lines\",\r\n name=\"Premium\")\r\n\r\n trace6 = scatter(;x=1:10, y=vcat(10:-2:0, [2, 4,2, 0]),\r\n line_color=\"rgb(231, 107, 243)\",\r\n mode=\"lines\",\r\n name=\"Ideal\")\r\n data = [trace1, trace2, trace3, trace4, trace5, trace6]\r\n layout = Layout(;paper_bgcolor=\"rgb(255, 255, 255)\",\r\n plot_bgcolor=\"rgb(229, 229, 229)\",\r\n\r\n xaxis=attr(gridcolor=\"rgb(255, 255, 255)\",\r\n range=[1, 10],\r\n showgrid=true,\r\n showline=false,\r\n showticklabels=true,\r\n tickcolor=\"rgb(127, 127, 127)\",\r\n ticks=\"outside\",\r\n zeroline=false),\r\n\r\n yaxis=attr(gridcolor=\"rgb(255, 255, 255)\",\r\n showgrid=true,\r\n showline=false,\r\n showticklabels=true,\r\n tickcolor=\"rgb(127, 127, 127)\",\r\n ticks=\"outside\",\r\n zeroline=false))\r\n\r\n plot(data, layout)\r\nend\r\n#display(errorbars1())\r\n\r\n#t = sec_Traces()\r\n#l = sec_Layouts()\r\n#iris = sec_DataFrames()\r\n#p = sec_Plot()\r\n#display(plot(p))\r\n\r\n# Open in Pane when run from VS Code\r\n# Open in Electron when run from console\r\n#sec_SyncPlots()\r\n\r\n# Open in default browser\r\n#savefig(p, \"plotlyjs.html\")\r\n#savefig(p, \"plotlyjs.png\")\r\n#DefaultApplication.open(\"plotlyjs.html\")\r\n\r\n#sec_Cyclers()\r\n\r\nnothing\r\n", "meta": {"hexsha": "a7bc2380dac83fd0dccdac0c0808b441a932f4d9", "size": 29616, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "learn/learn_PlotlyJS.jl", "max_stars_repo_name": "ykyang/org.allnix.julia", "max_stars_repo_head_hexsha": "58933a5848dec81c53d591b4163e9a70df62ddd8", "max_stars_repo_licenses": ["Apache-2.0"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "learn/learn_PlotlyJS.jl", "max_issues_repo_name": "ykyang/org.allnix.julia", "max_issues_repo_head_hexsha": "58933a5848dec81c53d591b4163e9a70df62ddd8", "max_issues_repo_licenses": ["Apache-2.0"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "learn/learn_PlotlyJS.jl", "max_forks_repo_name": "ykyang/org.allnix.julia", "max_forks_repo_head_hexsha": "58933a5848dec81c53d591b4163e9a70df62ddd8", "max_forks_repo_licenses": ["Apache-2.0"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 40.4038199181, "max_line_length": 105, "alphanum_fraction": 0.4929092382, "num_tokens": 10343, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.42632159254749025, "lm_q2_score": 0.13117321527062692, "lm_q1q2_score": 0.055921974033748434}}
{"text": "module LP\n\nimport ..MathOptFormat\n\nimport MathOptInterface\nconst MOI = MathOptInterface\n\nMOI.Utilities.@model(InnerModel,\n (MOI.ZeroOne, MOI.Integer),\n (MOI.EqualTo, MOI.GreaterThan, MOI.LessThan, MOI.Interval),\n (),\n (),\n (),\n (MOI.ScalarAffineFunction,),\n (),\n ()\n)\n\nstruct Options\n maximum_length::Int\n warn::Bool\n warn_once::Bool\n warned_start::Set{Char}\n warned_illegal::Set{Char}\nend\n\nfunction get_options(m::InnerModel)\n default_options = Options(255, false, false, Set{Char}(), Set{Char}())\n return get(m.ext, :LP_OPTIONS, default_options)\nend\n\n\"\"\"\n Model(; kwargs...)\n\nCreate an empty instance of MathOptFormat.LP.Model.\n\nKeyword arguments are:\n\n - `maximum_length::Int=255`: the maximum length for the name of a variable.\n lp_solve 5.0 allows only 16 characters, while CPLEX 12.5+ allow 255.\n\n - `warn::Bool=false`: print a warning when variables or constraints are renamed.\n\n - `warn_once::Bool=false`: print a warning when variables or constraints are\n renamed, but only once per kind of replacement (e.g., once per illegal\n character).\n\"\"\"\nfunction Model(;\n maximum_length::Int = 255, warn::Bool = false, warn_once::Bool = false\n)\n model = InnerModel{Float64}()\n options = Options(maximum_length, warn, warn_once, Set{Char}(), Set{Char}())\n model.ext[:LP_OPTIONS] = options\n return model\nend\n\nfunction Base.show(io::IO, ::InnerModel)\n print(io, \"A .LP-file model\")\n return\nend\n\n# ==============================================================================\n#\n# MOI.write_to_file\n#\n# ==============================================================================\n\n\nconst START_REG = r\"^([\\.0-9eE])\"\nconst NAME_REG = r\"([^a-zA-Z0-9\\!\\\"\\#\\$\\%\\&\\(\\)\\/\\,\\.\\;\\?\\@\\_\\`\\'\\{\\}\\|\\~])\"\n\nfunction sanitized_name(name::String, options::Options)\n m = match(START_REG, name)\n if m !== nothing\n plural = length(m.match) > 1\n\n if options.warn || (options.warn_once && !(m.match[1] in options.warned_start))\n @warn(\"Name $(name) cannot start with a period, a number, e, or E. \" *\n \"Prepending an underscore to name.\")\n push!(options.warned_start, m.match[1])\n end\n\n return sanitized_name(\"_\" * name, options)\n end\n\n m = match(NAME_REG, name)\n if m !== nothing\n plural = length(m.match) > 1\n\n if options.warn || (options.warn_once && !(m.match[1] in options.warned_illegal))\n @warn(\"Name $(name) contains $(ifelse(plural, \"\", \"an \"))\" *\n \"illegal character$(ifelse(plural, \"s\", \"\")): \" *\n \"\\\"$(m.match)\\\". Removing the offending \" *\n \"character$(ifelse(plural, \"s\", \"\")) from name.\")\n push!(options.warned_illegal, m.match[1])\n end\n\n return sanitized_name(replace(name, NAME_REG => s\"_\"), options)\n end\n\n # Truncate at the end to fit as many characters as possible.\n if length(name) > options.maximum_length\n @warn(\"Name $(name) too long (length: $(length(name)); \" *\n \"maximum: $(options.maximum_length)). Truncating.\")\n return sanitized_name(String(name[1:options.maximum_length]), options)\n end\n\n return name\nend\n\nfunction write_function(io::IO, model::InnerModel, func::MOI.SingleVariable, sanitized_names::Dict{MOI.VariableIndex, String})\n print(io, sanitized_names[func.variable])\n return\nend\n\nfunction write_function(io::IO, model::InnerModel, func::MOI.ScalarAffineFunction{Float64}, sanitized_names::Dict{MOI.VariableIndex, String})\n is_first_item = true\n if !(func.constant \u2248 0.0)\n Base.Grisu.print_shortest(io, func.constant)\n is_first_item = false\n end\n for term in func.terms\n if !(term.coefficient \u2248 0.0)\n if is_first_item\n Base.Grisu.print_shortest(io, term.coefficient)\n is_first_item = false\n else\n print(io, term.coefficient < 0 ? \" - \" : \" + \")\n Base.Grisu.print_shortest(io, abs(term.coefficient))\n end\n\n print(io, \" \", sanitized_names[term.variable_index])\n end\n end\n return\nend\n\nfunction write_constraint_suffix(io::IO, set::MOI.LessThan)\n print(io, \" <= \", )\n Base.Grisu.print_shortest(io, set.upper)\n println(io)\n return\nend\n\nfunction write_constraint_suffix(io::IO, set::MOI.GreaterThan)\n print(io, \" >= \", )\n Base.Grisu.print_shortest(io, set.lower)\n println(io)\n return\nend\n\nfunction write_constraint_suffix(io::IO, set::MOI.EqualTo)\n print(io, \" = \", )\n Base.Grisu.print_shortest(io, set.value)\n println(io)\n return\nend\n\nfunction write_constraint_suffix(io::IO, set::MOI.Interval)\n print(io, \" <= \", )\n Base.Grisu.print_shortest(io, set.upper)\n println(io)\n return\nend\n\nfunction write_constraint_prefix(io::IO, set::MOI.Interval)\n Base.Grisu.print_shortest(io, set.lower)\n print(io, \" <= \")\n return\nend\n\nwrite_constraint_prefix(io::IO, set) = nothing\n\nfunction write_constraint(io::IO, model::InnerModel, index, sanitized_names::Dict{MOI.VariableIndex, String}; write_name::Bool = true)\n func = MOI.get(model, MOI.ConstraintFunction(), index)\n set = MOI.get(model, MOI.ConstraintSet(), index)\n if write_name\n print(io, MOI.get(model, MOI.ConstraintName(), index), \": \")\n end\n write_constraint_prefix(io, set)\n write_function(io, model, func, sanitized_names)\n write_constraint_suffix(io, set)\nend\n\nconst SCALAR_SETS = (\n MOI.LessThan{Float64}, MOI.GreaterThan{Float64}, MOI.EqualTo{Float64},\n MOI.Interval{Float64}\n)\n\nfunction write_sense(io::IO, model::InnerModel)\n if MOI.get(model, MOI.ObjectiveSense()) == MOI.MAX_SENSE\n println(io, \"maximize\")\n else\n println(io, \"minimize\")\n end\n return\nend\n\nfunction write_objective(io::IO, model::InnerModel, sanitized_names::Dict{MOI.VariableIndex, String})\n print(io, \"obj: \")\n obj_func_type = MOI.get(model, MOI.ObjectiveFunctionType())\n obj_func = MOI.get(model, MOI.ObjectiveFunction{obj_func_type}())\n write_function(io, model, obj_func, sanitized_names)\n println(io)\n return\nend\n\nfunction MOI.write_to_file(model::InnerModel, io::IO)\n options = get_options(model)\n max_length = options.maximum_length\n # Ensure each variable has a unique name that does not infringe LP constraints.\n MathOptFormat.create_unique_names(model, warn = options.warn)\n sanitized_names = Dict{MOI.VariableIndex, String}()\n sanitized_names_set = Set{String}()\n for v in MOI.get(model, MOI.ListOfVariableIndices())\n proposed_sanitized_name = sanitized_name(MOI.get(model, MOI.VariableName(), v), options)\n # In case of duplicate names after sanitization, add a number at the end.\n if proposed_sanitized_name in sanitized_names_set\n # If the name is already too long, make some space for the suffix.\n if length(proposed_sanitized_name) >= max_length\n proposed_sanitized_name = String(proposed_sanitized_name[1:max_length - 2])\n end\n i = 1\n while proposed_sanitized_name * '_' * string(i) in sanitized_names_set\n i += 1\n\n # If the maximum length constraint would be broken with the *next* i,\n # truncate a bit more the proposed_sanitized_name.\n if length(proposed_sanitized_name * '_' * string(i)) > max_length\n proposed_sanitized_name = String(proposed_sanitized_name[1:length(proposed_sanitized_name) - 1])\n end\n end\n proposed_sanitized_name *= '_' * string(i)\n end\n push!(sanitized_names_set, proposed_sanitized_name)\n sanitized_names[v] = proposed_sanitized_name\n end\n\n write_sense(io, model)\n write_objective(io, model, sanitized_names)\n println(io, \"subject to\")\n for S in SCALAR_SETS\n for index in MOI.get(model, MOI.ListOfConstraintIndices{MOI.ScalarAffineFunction{Float64}, S}())\n write_constraint(io, model, index, sanitized_names; write_name = true)\n end\n end\n\n println(io, \"Bounds\")\n for S in SCALAR_SETS\n for index in MOI.get(model, MOI.ListOfConstraintIndices{MOI.SingleVariable, S}())\n write_constraint(io, model, index, sanitized_names; write_name = false)\n end\n end\n\n for (S, str_S) in [(MOI.Integer, \"General\"), (MOI.ZeroOne, \"Binary\")]\n indices = MOI.get(model, MOI.ListOfConstraintIndices{MOI.SingleVariable, S}())\n if length(indices) > 0\n println(io, str_S)\n for index in indices\n write_function(io, model, MOI.get(model, MOI.ConstraintFunction(), index), sanitized_names)\n println(io)\n end\n end\n end\n\n println(io, \"End\")\n\n return\nend\n\n# ==============================================================================\n#\n# MOI.read_from_to_file\n#\n# ==============================================================================\n\nfunction MOI.read_from_file(model::InnerModel, io::IO)\n error(\"Read from file is not implemented for LP files.\")\nend\n\nend\n", "meta": {"hexsha": "f2f01a1e0f4efa35e5f9deb7f4f61b529248bf6e", "size": 9134, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/LP/LP.jl", "max_stars_repo_name": "josepalos/MathOptFormat.jl", "max_stars_repo_head_hexsha": "04c41af0a81a867d3cd78beafd2617cdd033456e", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/LP/LP.jl", "max_issues_repo_name": "josepalos/MathOptFormat.jl", "max_issues_repo_head_hexsha": "04c41af0a81a867d3cd78beafd2617cdd033456e", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/LP/LP.jl", "max_forks_repo_name": "josepalos/MathOptFormat.jl", "max_forks_repo_head_hexsha": "04c41af0a81a867d3cd78beafd2617cdd033456e", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 32.390070922, "max_line_length": 141, "alphanum_fraction": 0.624589446, "num_tokens": 2273, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.41489883132727684, "lm_q2_score": 0.1347759191606016, "lm_q1q2_score": 0.055918371350793146}}
{"text": "## Exercise 4-9\n## Write an appropriately general set of functions that can draw flowers as in Turtle flowers.\n## https://benlauwens.github.io/ThinkJulia.jl/latest/book.html#fig04-2\nprintln(\"Ans: \")\n\nprintln(\"End.\")\n", "meta": {"hexsha": "15f483bc576e54194d8587511a352e6bdb63108a", "size": 216, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Chapter4/ex9.jl", "max_stars_repo_name": "yashppawar/ThinkJuliaExercises.jl", "max_stars_repo_head_hexsha": "72145b969dc51ebcac413a10004175ebc63cd5c2", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2022-02-13T14:11:30.000Z", "max_stars_repo_stars_event_max_datetime": "2022-02-13T14:11:30.000Z", "max_issues_repo_path": "Chapter4/ex9.jl", "max_issues_repo_name": "yashppawar/ThinkJuliaExercises.jl", "max_issues_repo_head_hexsha": "72145b969dc51ebcac413a10004175ebc63cd5c2", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Chapter4/ex9.jl", "max_forks_repo_name": "yashppawar/ThinkJuliaExercises.jl", "max_forks_repo_head_hexsha": "72145b969dc51ebcac413a10004175ebc63cd5c2", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 30.8571428571, "max_line_length": 94, "alphanum_fraction": 0.7453703704, "num_tokens": 58, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4148988457967689, "lm_q2_score": 0.13477590699708825, "lm_q1q2_score": 0.05591836825430458}}
{"text": "\"\"\"\n NOCB <: Imputor\n\nFills in missing data using the Next Observation Carried Backward (NOCB) approach.\n\"\"\"\nstruct NOCB <: Imputor end\n\n\"\"\"\n impute!(imp::NOCB, ctx::Context, data::AbstractVector)\n\nIterates backwards through the `data` and fills missing data with the next\nexisting observation.\n\nWARNING: missing elements at the tail of the array may not be imputed if there is no\nexisting observation to carry backward. As a result, this method does not guarantee\nthat all missing values will be imputed.\n\n# Usage\n```\n\n```\n\"\"\"\nfunction impute!(imp::NOCB, ctx::Context, data::AbstractVector)\n end_idx = findlast(ctx, data) - 1\n for i in end_idx:-1:1\n if ismissing(ctx, data[i])\n data[i] = data[i+1]\n end\n end\n\n return data\nend\n", "meta": {"hexsha": "dd7c914460bffb226a3b098bb8fbb21feab3de04", "size": 770, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/imputors/nocb.jl", "max_stars_repo_name": "nalimilan/Impute.jl", "max_stars_repo_head_hexsha": "73e78d1a18abbad7679745f085aea2fa136f15bd", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/imputors/nocb.jl", "max_issues_repo_name": "nalimilan/Impute.jl", "max_issues_repo_head_hexsha": "73e78d1a18abbad7679745f085aea2fa136f15bd", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/imputors/nocb.jl", "max_forks_repo_name": "nalimilan/Impute.jl", "max_forks_repo_head_hexsha": "73e78d1a18abbad7679745f085aea2fa136f15bd", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 23.3333333333, "max_line_length": 84, "alphanum_fraction": 0.6883116883, "num_tokens": 206, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.46879064146934857, "lm_q2_score": 0.1192029235866439, "lm_q1q2_score": 0.05588121501320453}}
{"text": "using BasicPOMCP\nusing Test\nusing POMDPs\nusing POMDPModels\nusing NBInclude\nusing D3Trees\nusing Random\nusing POMDPSimulators\nusing POMDPModelTools\nusing POMDPTesting\nusing POMDPLinter: @requirements_info, @show_requirements, requirements_info\nusing ParticleFilters: n_particles, particles, particle, weights, weighted_particles, weight_sum, weight\n\nimport POMDPs:\n\ttransition,\n\tobservation,\n reward,\n discount,\n\tinitialstate,\n\tupdater,\n\tstates,\n\tactions,\n\tobservations\n\nstruct ConstObsPOMDP <: POMDP{Bool, Symbol, Bool} end\nupdater(problem::ConstObsPOMDP) = DiscreteUpdater(problem)\ninitialstate(::ConstObsPOMDP) = BoolDistribution(0.0)\ntransition(p::ConstObsPOMDP, s::Bool, a::Symbol) = BoolDistribution(0.0)\nobservation(p::ConstObsPOMDP, a::Symbol, sp::Bool) = BoolDistribution(1.0)\nreward(p::ConstObsPOMDP, s::Bool, a::Symbol, sp::Bool) = 1.\ndiscount(p::ConstObsPOMDP) = 0.9\nstates(p::ConstObsPOMDP) = (true, false)\nactions(p::ConstObsPOMDP) = (:the_only_action,)\nobservations(p::ConstObsPOMDP) = (true, false)\n\n@testset \"POMDPTesting\" begin\n\tpomdp = BabyPOMDP()\n\ttest_solver(POMCPSolver(), BabyPOMDP())\nend;\n\n@testset \"type stability\" begin\n\tpomdp = BabyPOMDP()\n\tsolver = POMCPSolver(rng = MersenneTwister(1))\n\tplanner = solve(solver, pomdp)\n\tb = initialstate(pomdp)\n tree = BasicPOMCP.POMCPTree(pomdp, b, solver.tree_queries)\n node = BasicPOMCP.POMCPObsNode(tree, 1)\n\n r = @inferred BasicPOMCP.simulate(planner, rand(MersenneTwister(1), initialstate(pomdp)), node, 20)\nend;\n\n@testset \"belief dependent actions\" begin\n\tpomdp = ConstObsPOMDP()\n\tfunction POMDPs.actions(m::ConstObsPOMDP, b::LeafNodeBelief)\n\t\t@test currentobs(b) == true\n @test history(b)[end].o == true\n @test history(b)[end].a == :the_only_action\n @test mean(b) == 0.0\n @test mode(b) == 0.0\n @test only(support(b)) == false\n @test pdf(b, false) == 1.0\n @test pdf(b, true) == 0.0\n @test rand(b) == false\n @test n_particles(b) == 1\n @test only(particles(b)) == false\n @test only(weights(b)) == 1.0\n @test only(weighted_particles(b)) == (false => 1.0)\n @test weight_sum(b) == 1.0\n @test weight(b, 1) == 1.0 \n @test particle(b, 1) == false\n\n # old type name - this can be removed when upgrading versions\n @test b isa AOHistoryBelief\n return actions(m)\n\tend\n\n\tsolver = POMCPSolver(rng = MersenneTwister(1))\n\tplanner = solve(solver, pomdp)\n\tb = initialstate(pomdp)\n tree = BasicPOMCP.POMCPTree(pomdp, b, solver.tree_queries)\n node = BasicPOMCP.POMCPObsNode(tree, 1)\n\n @inferred BasicPOMCP.simulate(planner, rand(MersenneTwister(1), initialstate(pomdp)), node, 20)\nend;\n\n@testset \"simulation\" begin\n\tpomdp = BabyPOMDP()\n\tsolver = POMCPSolver(rng = MersenneTwister(1))\n\tplanner = solve(solver, pomdp)\n solver = POMCPSolver(max_time=0.1, tree_queries=typemax(Int), rng = MersenneTwister(1))\n planner = solve(solver, pomdp)\n\tb = initialstate(pomdp)\n\n a, info = action_info(planner, b)\n println(\"time below should be about 0.1 seconds\")\n etime = @elapsed a, info = action_info(planner, b)\n @show etime\n @test etime < 0.2\n @show info[:search_time_us]\n\n sim = HistoryRecorder(max_steps=10)\n simulate(sim, pomdp, planner, updater(pomdp))\nend;\n\n@testset \"d3t\" begin\n\tpomdp = BabyPOMDP()\n solver = POMCPSolver(max_time=0.1, tree_queries=typemax(Int), rng = MersenneTwister(1))\n planner = solve(solver, pomdp)\n\tb = initialstate(pomdp)\n a, info = action_info(planner, b, tree_in_info=true)\n\n d3t = D3Tree(info[:tree], title=\"test tree\")\n # inchrome(d3t)\n show(stdout, MIME(\"text/plain\"), d3t)\n\n\n solver = POMCPSolver(max_time=0.1, tree_queries=typemax(Int), rng=MersenneTwister(1), tree_in_info=true)\n planner = solve(solver, pomdp)\n a, info = action_info(planner, b)\n\n d3t = D3Tree(info[:tree], title=\"test tree (tree_in_info solver option)\")\nend;\n\n@testset \"Minimal_Example\" begin\n @nbinclude(joinpath(dirname(@__FILE__), \"..\", \"notebooks\", \"Minimal_Example.ipynb\"))\nend;\n\n@testset \"consistency\" begin\n # test consistency when rng is specified\n pomdp = BabyPOMDP()\n solver = POMCPSolver(rng = MersenneTwister(1))\n planner = solve(solver, pomdp)\n hist1 = simulate(HistoryRecorder(max_steps=1000, rng=MersenneTwister(3)), pomdp, planner)\n\n solver = POMCPSolver(rng = MersenneTwister(1))\n planner = solve(solver, pomdp)\n hist2 = simulate(HistoryRecorder(max_steps=1000, rng=MersenneTwister(3)), pomdp, planner)\n\n @test discounted_reward(hist1) == discounted_reward(hist2)\nend;\n\n@testset \"requires\" begin\n # REQUIREMENTS\n solver = POMCPSolver()\n pomdp = TigerPOMDP()\n\n println(\"============== @requirements_info with only solver:\")\n requirements_info(solver)\n println(\"============== @requirements_info with solver and pomdp:\")\n requirements_info(solver, pomdp)\n @show_requirements POMDPs.solve(solver, pomdp)\nend;\n\n@testset \"errors\" begin\n struct TerminalPOMDP <: POMDP{Int, Int, Int} end\n POMDPs.isterminal(::TerminalPOMDP, s) = true\n POMDPs.actions(::TerminalPOMDP) = [1,2,3]\n\n solver = POMCPSolver()\n planner = solve(solver, TerminalPOMDP())\n @test_throws AllSamplesTerminal action(planner, Deterministic(1))\n let ex = nothing\n try\n action(planner, Deterministic(1))\n catch ex\n end\n\n @test sprint(showerror, ex) == \"\"\"\n Planner failed to choose an action because all states sampled from the belief were terminal.\n\n To see the belief, catch this exception as ex and see ex.belief.\n\n To specify an action for this case, use the default_action solver parameter.\n \"\"\"\n end\nend\n", "meta": {"hexsha": "71758530ffa95364dc35ebf5e6d93de9b487e015", "size": 5681, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/runtests.jl", "max_stars_repo_name": "WhiffleFish/BasicPOMCP.jl", "max_stars_repo_head_hexsha": "de923f282ccde51335057d78c14c8a61af61d3fd", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 25, "max_stars_repo_stars_event_min_datetime": "2017-10-09T04:54:27.000Z", "max_stars_repo_stars_event_max_datetime": "2021-09-20T21:24:33.000Z", "max_issues_repo_path": "test/runtests.jl", "max_issues_repo_name": "WhiffleFish/BasicPOMCP.jl", "max_issues_repo_head_hexsha": "de923f282ccde51335057d78c14c8a61af61d3fd", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 24, "max_issues_repo_issues_event_min_datetime": "2017-09-11T19:32:34.000Z", "max_issues_repo_issues_event_max_datetime": "2021-08-11T21:08:21.000Z", "max_forks_repo_path": "test/runtests.jl", "max_forks_repo_name": "WhiffleFish/BasicPOMCP.jl", "max_forks_repo_head_hexsha": "de923f282ccde51335057d78c14c8a61af61d3fd", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 13, "max_forks_repo_forks_event_min_datetime": "2018-02-28T01:50:41.000Z", "max_forks_repo_forks_event_max_datetime": "2021-09-20T21:27:55.000Z", "avg_line_length": 32.0960451977, "max_line_length": 108, "alphanum_fraction": 0.6884351347, "num_tokens": 1730, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4882833952958347, "lm_q2_score": 0.11436853372838406, "lm_q1q2_score": 0.05584425596390156}}
{"text": "@testset \"model.jl\" begin\r\n @testset \"lastVar\" begin\r\n trailer = SeaPearl.Trailer()\r\n model = SeaPearl.CPModel(trailer)\r\n\r\n x = SeaPearl.IntVar(2, 6, \"x\", trailer)\r\n y = SeaPearl.IntVar(2, 6, \"y\", trailer)\r\n \r\n SeaPearl.addVariable!(model, x)\r\n SeaPearl.addVariable!(model, y)\r\n\r\n valueselection = SeaPearl.BasicHeuristic()\r\n @test isnothing(model.statistics.lastVar)\r\n v = valueselection(SeaPearl.DecisionPhase, model, x)\r\n @test model.statistics.lastVar.id==\"x\"\r\n SeaPearl.assign!(x, v)\r\n v = valueselection(SeaPearl.DecisionPhase, model, y)\r\n @test model.statistics.lastVar.id==\"y\"\r\n SeaPearl.assign!(y, v)\r\n\r\n end\r\n\r\n @testset \"addVariable!()\" begin\r\n trailer = SeaPearl.Trailer()\r\n x = SeaPearl.IntVar(2, 6, \"x\", trailer)\r\n y = SeaPearl.IntVar(2, 6, \"y\", trailer)\r\n\r\n model = SeaPearl.CPModel(trailer)\r\n\r\n SeaPearl.addVariable!(model, x)\r\n SeaPearl.addVariable!(model, y)\r\n\r\n @test length(model.variables) == 2\r\n @test SeaPearl.branchable_variables(model) == Dict{String, SeaPearl.AbstractVar}([\"x\" => x, \"y\" => y])\r\n\r\n z = SeaPearl.IntVar(2, 6, \"y\", trailer)\r\n\r\n @test_throws AssertionError SeaPearl.addVariable!(model, z)\r\n\r\n # Not branching\r\n trailer = SeaPearl.Trailer()\r\n x = SeaPearl.IntVar(2, 6, \"x\", trailer)\r\n y = SeaPearl.IntVar(2, 6, \"y\", trailer)\r\n\r\n model = SeaPearl.CPModel(trailer)\r\n\r\n SeaPearl.addVariable!(model, x; branchable=false)\r\n SeaPearl.addVariable!(model, y)\r\n\r\n @test length(model.variables) == 2\r\n @test SeaPearl.branchable_variables(model) == Dict{String, SeaPearl.AbstractVar}([\"y\" => y])\r\n\r\n # Trying to branch on Set variable\r\n trailer = SeaPearl.Trailer()\r\n y = SeaPearl.IntSetVar(2, 6, \"y\", trailer)\r\n\r\n model = SeaPearl.CPModel(trailer)\r\n\r\n @test_throws AssertionError SeaPearl.addVariable!(model, y)\r\n end\r\n\r\n @testset \"merge!()\" begin\r\n test1 = SeaPearl.CPModification(\"x\" => [2, 3, 4],\"z\" => [11, 12, 13, 14, 15],\"y\" => [7, 8])\r\n test2 = SeaPearl.CPModification(\"x\" => [5],\"y\" => [7, 8])\r\n\r\n SeaPearl.merge!(test1, test2)\r\n\r\n @test test1 == SeaPearl.CPModification(\"x\" => [2, 3, 4, 5],\"z\" => [11, 12, 13, 14, 15],\"y\" => [7, 8, 7, 8])\r\n end\r\n\r\n @testset \"addToPrunedDomains!()\" begin\r\n test1 = SeaPearl.CPModification(\"x\" => [2, 3, 4],\"z\" => [11, 12, 13, 14, 15],\"y\" => [7, 8])\r\n trailer = SeaPearl.Trailer()\r\n x = SeaPearl.IntVar(2, 6, \"x\", trailer)\r\n t = SeaPearl.IntVar(2, 6, \"t\", trailer)\r\n b = SeaPearl.BoolVar(\"b\", trailer)\r\n\r\n SeaPearl.addToPrunedDomains!(test1, x, [5, 6])\r\n\r\n @test test1 == SeaPearl.CPModification(\"x\" => [2, 3, 4, 5, 6],\"z\" => [11, 12, 13, 14, 15],\"y\" => [7, 8])\r\n\r\n SeaPearl.addToPrunedDomains!(test1, t, [5, 6])\r\n\r\n @test test1 == SeaPearl.CPModification(\"x\" => [2, 3, 4, 5, 6],\"z\" => [11, 12, 13, 14, 15],\"y\" => [7, 8], \"t\" => [5, 6])\r\n\r\n SeaPearl.addToPrunedDomains!(test1, b, [true])\r\n\r\n @test test1 == SeaPearl.CPModification(\"x\" => [2, 3, 4, 5, 6],\"z\" => [11, 12, 13, 14, 15],\"y\" => [7, 8], \"t\" => [5, 6], \"b\" => [true])\r\n\r\n end\r\n\r\n @testset \"solutionFound()\" begin\r\n trailer = SeaPearl.Trailer()\r\n x = SeaPearl.IntVar(2, 6, \"x\", trailer)\r\n y = SeaPearl.IntVar(2, 6, \"y\", trailer)\r\n\r\n model = SeaPearl.CPModel(trailer)\r\n\r\n SeaPearl.addVariable!(model, x)\r\n SeaPearl.addVariable!(model, y)\r\n\r\n @test !SeaPearl.solutionFound(model)\r\n\r\n constraint = SeaPearl.EqualConstant(x, 3, trailer)\r\n constraint2 = SeaPearl.Equal(x, y, trailer)\r\n SeaPearl.addConstraint!(model, constraint)\r\n SeaPearl.addConstraint!(model, constraint2)\r\n\r\n SeaPearl.fixPoint!(model)\r\n\r\n @test SeaPearl.solutionFound(model)\r\n end\r\n\r\n @testset \"triggerFoundSolution!()\" begin\r\n trailer = SeaPearl.Trailer()\r\n x = SeaPearl.IntVar(2, 2, \"x\", trailer)\r\n y = SeaPearl.IntVar(3, 3, \"y\", trailer)\r\n\r\n model = SeaPearl.CPModel(trailer)\r\n\r\n SeaPearl.addVariable!(model, x)\r\n SeaPearl.addVariable!(model, y)\r\n SeaPearl.addObjective!(model, y)\r\n\r\n SeaPearl.triggerFoundSolution!(model)\r\n\r\n @test length(model.statistics.solutions) == 1\r\n @test model.statistics.solutions[1] == Dict(\"x\" => 2,\"y\" => 3)\r\n @test model.statistics.nodevisitedpersolution[1] == 0\r\n @test model.objectiveBound == 2\r\n @test model.statistics.numberOfSolutions == 1\r\n @test model.statistics.objectives[1] == 3\r\n @test model.statistics.numberOfInfeasibleSolutions == 0\r\n @test model.statistics.numberOfSolutionsBeforeRestart == 1\r\n @test model.statistics.numberOfInfeasibleSolutionsBeforeRestart == 0\r\n @test model.statistics.numberOfNodesBeforeRestart == 0\r\n end\r\n\r\n @testset \"tightenObjective!()\" begin\r\n trailer = SeaPearl.Trailer()\r\n x = SeaPearl.IntVar(2, 2, \"x\", trailer)\r\n y = SeaPearl.IntVar(3, 3, \"y\", trailer)\r\n\r\n model = SeaPearl.CPModel(trailer)\r\n\r\n SeaPearl.addVariable!(model, x)\r\n SeaPearl.addVariable!(model, y)\r\n SeaPearl.addObjective!(model, y)\r\n\r\n @test isnothing(model.objectiveBound)\r\n\r\n\r\n SeaPearl.tightenObjective!(model)\r\n\r\n @test model.objectiveBound == 2\r\n end\r\n\r\n @testset \"belowLimits()\" begin\r\n trailer = SeaPearl.Trailer()\r\n model = SeaPearl.CPModel(trailer)\r\n\r\n model.statistics.numberOfNodes = 1500\r\n model.statistics.numberOfSolutions = 15\r\n SeaPearl.tic()\r\n @test SeaPearl.belowLimits(model)\r\n\r\n model.limit.numberOfNodes = 1501\r\n model.limit.numberOfSolutions = 16\r\n model.limit.searchingTime = 1\r\n SeaPearl.tic()\r\n sleep(0.1)\r\n @test SeaPearl.belowLimits(model)\r\n\r\n model.statistics.numberOfNodes = 1501\r\n @test !SeaPearl.belowLimits(model)\r\n\r\n model.statistics.numberOfNodes = 1500\r\n model.statistics.numberOfSolutions = 16\r\n @test !SeaPearl.belowLimits(model)\r\n\r\n SeaPearl.tic()\r\n sleep(1)\r\n @test !SeaPearl.belowLimits(model)\r\n end\r\n\r\n @testset \"belowNodeLimit()\" begin\r\n trailer = SeaPearl.Trailer()\r\n model = SeaPearl.CPModel(trailer)\r\n\r\n model.statistics.numberOfNodes = 1500\r\n\r\n @test SeaPearl.belowNodeLimit(model)\r\n\r\n model.limit.numberOfNodes = 1501\r\n @test SeaPearl.belowNodeLimit(model)\r\n\r\n model.statistics.numberOfNodes = 1501\r\n @test !SeaPearl.belowNodeLimit(model)\r\n end\r\n\r\n @testset \"belowSolutionLimits()\" begin\r\n trailer = SeaPearl.Trailer()\r\n model = SeaPearl.CPModel(trailer)\r\n\r\n model.statistics.numberOfSolutions = 15\r\n\r\n @test SeaPearl.belowSolutionLimit(model)\r\n\r\n model.limit.numberOfSolutions = 16\r\n @test SeaPearl.belowSolutionLimit(model)\r\n\r\n model.statistics.numberOfSolutions = 16\r\n @test !SeaPearl.belowSolutionLimit(model)\r\n end\r\n\r\n @testset \"belowTimeLimits()\" begin\r\n trailer = SeaPearl.Trailer()\r\n model = SeaPearl.CPModel(trailer)\r\n\r\n SeaPearl.tic()\r\n sleep(0.1)\r\n @test SeaPearl.belowTimeLimit(model)\r\n\r\n model.limit.searchingTime = 1\r\n SeaPearl.tic()\r\n sleep(0.1)\r\n\r\n @test SeaPearl.belowTimeLimit(model)\r\n\r\n model.limit.searchingTime = 0\r\n SeaPearl.tic()\r\n sleep(0.1)\r\n @test !SeaPearl.belowTimeLimit(model)\r\n end\r\n\r\n @testset \"Base.isempty()\" begin\r\n trailer = SeaPearl.Trailer()\r\n model = SeaPearl.CPModel(trailer)\r\n\r\n @test isempty(model)\r\n\r\n x = SeaPearl.IntVar(2, 2, \"x\", trailer)\r\n SeaPearl.addVariable!(model, x)\r\n\r\n @test !isempty(model)\r\n end\r\n\r\n @testset \"Base.empty!()\" begin\r\n\r\n trailer = SeaPearl.Trailer()\r\n model = SeaPearl.CPModel(trailer)\r\n\r\n x = SeaPearl.IntVar(2, 2, \"x\", trailer)\r\n SeaPearl.addVariable!(model, x)\r\n\r\n empty!(model)\r\n\r\n @test isempty(model)\r\n end\r\n\r\n @testset \"reset_model!()\" begin\r\n trailer = SeaPearl.Trailer()\r\n model = SeaPearl.CPModel(trailer)\r\n\r\n x = SeaPearl.IntVar(2, 5, \"x\", trailer)\r\n SeaPearl.addVariable!(model, x)\r\n SeaPearl.addObjective!(model, x)\r\n\r\n SeaPearl.assign!(x, 3)\r\n SeaPearl.fixPoint!(model)\r\n SeaPearl.triggerFoundSolution!(model)\r\n\r\n @test SeaPearl.length(x.domain) == 1\r\n @test model.objectiveBound == 2\r\n SeaPearl.reset_model!(model)\r\n @test SeaPearl.length(x.domain) == 4\r\n @test isnothing(model.objectiveBound)\r\n end\r\n @testset \"restart_search\" begin\r\n\r\n trailer = SeaPearl.Trailer()\r\n model = SeaPearl.CPModel(trailer)\r\n\r\n x = SeaPearl.IntVar(1, 2, \"x\", trailer)\r\n y = SeaPearl.IntVar(1, 2, \"y\", trailer)\r\n z = SeaPearl.IntVar(1, 2, \"z\", trailer)\r\n\r\n SeaPearl.addVariable!(model, x)\r\n SeaPearl.addVariable!(model, y)\r\n SeaPearl.addVariable!(model, z)\r\n SeaPearl.addConstraint!(model, SeaPearl.NotEqual(x, y, trailer))\r\n SeaPearl.addConstraint!(model, SeaPearl.NotEqual(y, z, trailer))\r\n SeaPearl.addConstraint!(model, SeaPearl.NotEqual(x, z, trailer))\r\n\r\n SeaPearl.search!(model, SeaPearl.DFSearch(), SeaPearl.MinDomainVariableSelection(), SeaPearl.BasicHeuristic())\r\n @test model.statistics.numberOfInfeasibleSolutionsBeforeRestart == 2\r\n @test model.statistics.numberOfNodesBeforeRestart == 3\r\n @test model.statistics.numberOfSolutionsBeforeRestart == 0\r\n\r\n SeaPearl.restart_search!(model)\r\n\r\n @test model.statistics.numberOfInfeasibleSolutionsBeforeRestart == 0\r\n @test model.statistics.numberOfNodesBeforeRestart == 0\r\n @test model.statistics.numberOfSolutionsBeforeRestart == 0\r\n end\r\n\r\n @testset \"addKnownObjective!\" begin\r\n trailer = SeaPearl.Trailer()\r\n model = SeaPearl.CPModel(trailer)\r\n\r\n SeaPearl.addKnownObjective!(model, 1)\r\n @test model.knownObjective == 1\r\n\r\n end\r\n @testset \"triggerInfeasible!\" begin\r\n\r\n trailer = SeaPearl.Trailer()\r\n model = SeaPearl.CPModel(trailer)\r\n\r\n x = SeaPearl.IntVar(1, 2, \"x\", trailer)\r\n y = SeaPearl.IntVar(1, 2, \"y\", trailer)\r\n z = SeaPearl.IntVar(1, 2, \"z\", trailer)\r\n\r\n SeaPearl.addVariable!(model, x)\r\n SeaPearl.addVariable!(model, y)\r\n SeaPearl.addVariable!(model, z)\r\n\r\n SeaPearl.addConstraint!(model, SeaPearl.NotEqual(x, y, trailer))\r\n SeaPearl.addConstraint!(model, SeaPearl.NotEqual(y, z, trailer))\r\n\r\n\r\n @test model.statistics.infeasibleStatusPerVariable[\"x\"] == 1\r\n @test model.statistics.infeasibleStatusPerVariable[\"y\"] == 2\r\n @test model.statistics.infeasibleStatusPerVariable[\"z\"] == 1\r\n\r\n constraint = model.constraints[1]\r\n SeaPearl.triggerInfeasible!(constraint, model)\r\n @test model.statistics.infeasibleStatusPerVariable[\"x\"] == 2\r\n @test model.statistics.infeasibleStatusPerVariable[\"y\"] == 3\r\n\r\n end\r\nend\r\n", "meta": {"hexsha": "609a988b64c22b3483f3c7e61fcad8d3fad80775", "size": 11286, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/CP/core/model.jl", "max_stars_repo_name": "pitmonticone/SeaPearl.jl", "max_stars_repo_head_hexsha": "0c0ca5ec5cce81515acd202ea2d87c985c0c3fea", "max_stars_repo_licenses": ["BSD-3-Clause"], "max_stars_count": 44, "max_stars_repo_stars_event_min_datetime": "2021-04-20T16:29:52.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-31T07:17:03.000Z", "max_issues_repo_path": "test/CP/core/model.jl", "max_issues_repo_name": "pitmonticone/SeaPearl.jl", "max_issues_repo_head_hexsha": "0c0ca5ec5cce81515acd202ea2d87c985c0c3fea", "max_issues_repo_licenses": ["BSD-3-Clause"], "max_issues_count": 65, "max_issues_repo_issues_event_min_datetime": "2021-04-23T17:20:56.000Z", "max_issues_repo_issues_event_max_datetime": "2022-03-22T23:42:24.000Z", "max_forks_repo_path": "test/CP/core/model.jl", "max_forks_repo_name": "pitmonticone/SeaPearl.jl", "max_forks_repo_head_hexsha": "0c0ca5ec5cce81515acd202ea2d87c985c0c3fea", "max_forks_repo_licenses": ["BSD-3-Clause"], "max_forks_count": 6, "max_forks_repo_forks_event_min_datetime": "2021-05-10T23:32:49.000Z", "max_forks_repo_forks_event_max_datetime": "2022-02-15T02:44:34.000Z", "avg_line_length": 32.9037900875, "max_line_length": 143, "alphanum_fraction": 0.5972000709, "num_tokens": 3209, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.48828339529583464, "lm_q2_score": 0.11436853070975538, "lm_q1q2_score": 0.05584425448995529}}
{"text": "##### model_diagnostics.jl\n# Created by Alexander Dowling\n\n###############\n##### Exported functions for model diagnostics\n\nfunction round_to_bounds!(m::Model, epsilon::Float64=1.0E-6, f=STDOUT)\n\t# This function rounds each element of the solution vector to the closest bound if\n\t# it is within epsilon.\n\t# Why is this necessary? Some solvers (e.g., IPOPT) give solutions that are slightly\n\t# infeasible or violate bounds. This is problematic in a meta-algorithm with calls\n\t# to other solvers.\n\t\n\tx = get_x(m, :Unsolved)\n\t\n\tup = m.colUpper\n\tlo = m.colLower\n\t\n\tfor i = 1:length(x)\n\t\n\t\tv = Variable(m, i)\n\t\n\t\tif(up[i] < lo[i])\n\t\t\tprintln(f,\"Warning: Upper bound (\",up[i],\") is smaller than lower bound (\",lo[i],\") for variable \",v)\n\t\tend\n\t\n\t\tx_ = NaN\n\t\n\t\tif(x[i] > up[i])\n\t\t\tx_ = up[i]\n\t\telseif(x[i] < lo[i])\n\t\t\tx_ = lo[i]\n\t\telseif(up[i] - x[i] < epsilon)\n\t\t\tx_ = up[i]\n\t\telseif(x[i] - lo[i] < epsilon)\n\t\t\tx_ = lo[i]\n\t\tend\n\t\t\n\t\tif(!isnan(x_))\n\t\t\tsetvalue(v, x_)\n\t\tend\n\t\n\tend\n\t\n\treturn nothing\n\nend\n\nfunction printVariableDiagnostics(m::Model, epsilon::Float64=1.0E-6, f=STDOUT, status::Symbol=:Unknown)\n\tx = get_x(m, status)\n\n\tprintln(f,\"Uninitialized Variables: \")\n\tfor i = 1:length(x)\n\t\tif(isnan(x[i]))\n\t\t\tv = Variable(m, i)\n\t\t\t#println(\"x[\",i,\"] = \",v)\n\t\t\tprintln(f,v)\n\t\tend\n\tend\n\tprintln(f,\" \")\n\n\tprintln(f,\"Variable Violating Bounds: \")\n\tup = m.colUpper\n\tlo = m.colLower\n\n\tfor i = 1:length(x)\n\t\tviol_up = x[i] - up[i] > epsilon\n\t\tviol_lo = x[i] - lo[i] < -epsilon\n\t\tif(viol_up || viol_lo)\n\n\t\t\tv = Variable(m, i)\n\t\t\tprint(f,v, \" = \",x[i], \", bounds: [ \")\n\n\t\t\tif(viol_lo)\n\t\t\t\tprint_with_color(:red, f, string(lo[i]))\n\t\t\telse\n\t\t\t\tprint(f, string(lo[i]))\n\t\t\tend\n\n\t\t\tprint(f, \" , \")\n\n\t\t\tif(viol_up)\n\t\t\t\tprint_with_color(:green, f, string(up[i]))\n\t\t\telse\n\t\t\t\tprint(f, string(up[i]))\n\t\t\tend\n\n\t\t\tprintln(f, \" ]\")\n\t\tend\n\tend\n\n\treturn nothing\n\nend\n\nfunction printInfeasibleEquations(m2::Model, eqns, f=STDOUT, status::Symbol=:Unknown)\n\n\tdd = DegenData(m2, status)\n\treturn printInfeasibleEquations(m2, dd, eqns, f)\n\nend\n\nfunction printInfeasibleEquations(m2::Model, dd::DegenData, eqns, f=STDOUT)\n\n\tprintln(f,\"Checking infeasibility for specified equations: \")\n\n\tr = reshape(min(dd.g - dd.gLB, 0) + max(dd.g - dd.gUB, 0),length(dd.g))\n\n\tfor i = eqns\n\t\tprint(f, \"r[\",string(i),\"] = \")\n\t\tprint_with_color(:green,f,string(r[i]))\n\t\tprint(f,\"\\n\")\n\n\t\tprintEquation(m2, dd, i, f)\n\n\t\tprintln(f, \" \")\n\n\tend\n\n\treturn nothing\n\nend\n\nfunction printInfeasibleEquations(m2::Model, epsilon::Float64, f=STDOUT, status::Symbol=:Unknown)\n\n\tdd = DegenData(m2, f, status)\n\treturn printInfeasibleEquations(m2, dd, epsilon, f)\nend\n\nfunction printInfeasibleEquations(m2::Model, dd::DegenData, epsilon::Float64, f=STDOUT)\n\n\tprintln(f,\"Infeasible equations: \")\n\n\t#=\n\tprintln(\"size(dd.g) = \",size(dd.g))\n\tprintln(\"size(dd.gLB) = \",size(dd.gLB))\n\tprintln(\"size(dd.gUB) = \",size(dd.gUB))\n\t=#\n\n\tr = reshape(min(dd.g - dd.gLB, 0) + max(dd.g - dd.gUB, 0),length(dd.g))\n\n\tk = sortperm(abs(r))\n\n\tfor j = 1:length(k)\n\n\t\ti = k[j]\n\n\t\tif(abs(r[i]) >= epsilon || isnan(r[i]))\n\t\t\tprint(f,\"r[\",string(i),\"] = \")\n\t\t\tprint_with_color(:green,f,string(r[i]))\n\t\t\tprint(f,\"\\n\")\n\n\t\t\tprintEquation(m2, dd, i,f)\n\n\t\t\tprintVariablesInEquation(m2, dd, i, true, true, f)\n\n\t\t\tprintln(f,\" \")\n\n\t\tend\n\n\tend\n\n\treturn nothing\n\nend\n\nfunction printInactiveEquations(m2::Model, epsilon::Float64=1E-6, f=STDOUT)\n\n\tdd = DegenData(m2, f)\n\treturn printInactiveEquations(m2, dd, epsilon, f)\n\nend\n\nfunction printInactiveEquations(m2::Model, dd::DegenData, epsilon::Float64=1E-6, f=STDOUT)\n\n\tprintln(f,\"Inactive Equations: \")\n\n\tinactive = ((dd.gUB - dd.g) .> epsilon) & ((dd.g - dd.gLB) .> epsilon)\n\n\tfor i = 1:length(dd.g)\n\t\tif(((dd.gUB[i] - dd.g[i]) .> epsilon) & ((dd.g[i] - dd.gLB[i]) .> epsilon))\n\t\t\tprintEquation(m2, dd, i, f)\n\t\tend\n\tend\n\n\treturn nothing\n\nend\n\nfunction checkVarBounds(m::Model, f=STDOUT)\n\n\tlow = m.colLower\n\tup = m.colUpper\n\tval = m.colVal\n\n\tn = length(val)\n\n\tfor i = 1:n\n\t\tif(up[i] < low[i] || val[i] < low[i] || val[i] > up[i])\n\n\t\t\tprint(f,getname(m,i),\":\\t\\t\",\"Lower: \")\n\n\t\t\tif(up[i] < low[i] || val[i] < low[i])\n\t\t\t\tprint_with_color(:red,f,string(low[i]))\n\t\t\telse\n\t\t\t\tprint(f,low[i])\n\t\t\tend\n\n\t\t\tprint(f,\"\\t Upper: \")\n\n\t\t\tif(up[i] < low[i] || val[i] > up[i])\n\t\t\t\tprint_with_color(:blue,f,string(up[i]))\n\t\t\telse\n\t\t\t\tprint(f,up[i])\n\t\t\tend\n\n\t\t\tprint(f,\"\\t Value: \")\n\n\t\t\tif(val[i] < low[i] || val[i] > up[i])\n\t\t\t\tprint_with_color(:green,f,string(val[i]))\n\t\t\telse\n\t\t\t\tprint(f,val[i])\n\t\t\tend\n\n\t\t\tprint(f,\"\\n\")\n\t\tend\n\tend\n\n\treturn nothing\n\nend\n\nfunction printBound(m::Model, i::Int64, epsiActive::Float64, f=STDOUT)\n\n\tv = Variable(m, i)\n\n\tup = m.colUpper[i]\n\tlo = m.colLower[i]\n\tval = m.colVal[i]\n\n\tif(abs(val-lo) < epsiActive)\n\t\tprint(f,string(lo),\" <= \")\n\tend\n\n\tprint(getname(v))\n\n\tif(abs(val-up) < epsiActive)\n\t\tprint(f,\" <= \",string(up))\n\tend\n\n\tprint(f,\"\\n\")\n\n\treturn nothing\n\nend\n\nfunction printRows(m::Model, rows::Int64, f=STDOUT; lite::Bool=false)\n\treturn printRows(m, Array([rows]), f, lite)\nend\n\nfunction printRows(m::Model, rows::Array{Int64,1}, f=STDOUT; lite::Bool=false)\n\n\tdd = DegenData(m, f)\n\treturn printRows(m, dd, rows, f, lite)\n\nend\n\nfunction printRows(m::Model, dd::DegenData, rows::Int64, f=STDOUT; lite::Bool=false)\n\treturn printRows(m, dd, Array([rows]), f, lite=lite)\nend\n\nfunction printRows(m::Model, dd::DegenData, rows::Array{Int64,1}, f=STDOUT; lite::Bool=false)\n\n\tif(lite && length(rows) == 1)\n\t\n\t\tr = rows[1]\n\t\t\n\t\tif(r <= dd.nConstr)\n\t\t\tprintEquation(m, dd, rows[1], f)\n\t\telse\n\t\t\tprintBound(m, r - dd.nConstr, 1E-6, f)\n\t\tend\n\telse\n\n\t\tfor i = 1:length(rows)\n\t\t\tr = rows[i]\n\t\t\t\n\t\t\tprintln(f,\"**********************************\")\n\t\t\t\n\t\t\tif(r <= dd.nConstr)\n\t\t\t\t\t\n\t\t\t\tprintln(f,\"Constraint \",string(r),\"...\")\n\t\t\t\tprintEquation(m, dd, r, f)\n\t\t\t\tprintln(f,string(dd.gLB[r]),\" <= \", string(dd.g[r]), \" <= \",string(dd.gUB[r]))\n\n\t\t\t\tprintln(f,\" \")\n\t\t\t\tprintln(f,\"Involved Variables:\")\n\t\t\t\tk = find(dd.iR .== r)\n\n\t\t\t\tfor j in unique(dd.jC[k])\n\t\t\t\t\tv = Variable(m,j)\n\t\t\t\t\tprint(f,getname(v),\" \\t\")\n\t\t\t\t\tprintln(f,getlowerbound(v),\" <= \",getvalue(v),\" <= \",getupperbound(v))\n\t\t\t\tend\n\t\t\t\n\t\t\telse\n\t\t\t\tv = r - dd.nConstr\n\t\t\t\tprintln(f,\"Bound for variable \",v)\n\t\t\t\tprintBounds(m, v, 1E-6, f)\n\t\t\t\t\n\t\t\tend\n\n\t\t\tprintln(f,\" \")\n\n\t\tend\n\tend\n\n\n\treturn nothing\n\nend\n\nfunction checkEquationScaling(m::Model, status::Symbol=:Unknown, f=STDOUT)\n\n\tdd = DegenData(m, status)\n\t\n\tcheckJacobianScaling(m, dd, f)\n\n\treturn nothing\n\nend", "meta": {"hexsha": "7e8638430307dc46d37877fe3c6e677961767f0c", "size": 6373, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/model_diagnostics.jl", "max_stars_repo_name": "adowling2/DegeneracyHunter", "max_stars_repo_head_hexsha": "44b569c9504819be8f07544c3e5968ddbdf9cc86", "max_stars_repo_licenses": ["Apache-2.0"], "max_stars_count": 13, "max_stars_repo_stars_event_min_datetime": "2016-09-06T18:15:26.000Z", "max_stars_repo_stars_event_max_datetime": "2020-11-04T14:10:38.000Z", "max_issues_repo_path": "src/model_diagnostics.jl", "max_issues_repo_name": "adowling2/DegeneracyHunter", "max_issues_repo_head_hexsha": "44b569c9504819be8f07544c3e5968ddbdf9cc86", "max_issues_repo_licenses": ["Apache-2.0"], "max_issues_count": 5, "max_issues_repo_issues_event_min_datetime": "2016-09-02T00:32:50.000Z", "max_issues_repo_issues_event_max_datetime": "2021-02-26T12:12:28.000Z", "max_forks_repo_path": "src/model_diagnostics.jl", "max_forks_repo_name": "adowling2/DegeneracyHunter", "max_forks_repo_head_hexsha": "44b569c9504819be8f07544c3e5968ddbdf9cc86", "max_forks_repo_licenses": ["Apache-2.0"], "max_forks_count": 4, "max_forks_repo_forks_event_min_datetime": "2017-04-13T13:07:45.000Z", "max_forks_repo_forks_event_max_datetime": "2020-12-27T18:15:06.000Z", "avg_line_length": 19.253776435, "max_line_length": 104, "alphanum_fraction": 0.6107013965, "num_tokens": 2076, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4882833952958347, "lm_q2_score": 0.11436852316318397, "lm_q1q2_score": 0.055844250805089785}}
{"text": "#=\n\n Reading input\n\n=#\nfunction read_input()\n\n pass=[]\n policy=[]\n letter=[]\n open(\"./input\", \"r\") do f\n while ! eof(f)\n s::String = readline(f)\n entry = split(s,\" \")\n push!(pass,entry[3])\n push!(letter,entry[2][1])\n\n policy_split = split(entry[1],\"-\")\n policy_tuple = (parse(Int32,policy_split[1]), \n parse(Int32,policy_split[2]))\n push!(policy,policy_tuple)\n end\n end\n\n return pass, policy, letter\nend\n\ninput = read_input()\npass = input[1]\npolicy = input[2]\nletter = input[3]\n\n\n\n\n#=\n\n--- Day 2: Password Philosophy ---\n\nYour flight departs in a few days from the coastal airport; the easiest way down to \nthe coast from here is via toboggan.\n\nThe shopkeeper at the North Pole Toboggan Rental Shop is having a bad day. \"Something's \nwrong with our computers; we can't log in!\" You ask if you can take a look.\n\nTheir password database seems to be a little corrupted: some of the passwords wouldn't \nhave been allowed by the Official Toboggan Corporate Policy that was in effect when they \nwere chosen.\n\nTo try to debug the problem, they have created a list (your puzzle input) of passwords \n(according to the corrupted database) and the corporate policy when that password was set.\n\nFor example, suppose you have the following list:\n\n1-3 a: abcde\n1-3 b: cdefg\n2-9 c: ccccccccc\n\nEach line gives the password policy and then the password. The password policy indicates \nthe lowest and highest number of times a given letter must appear for the password to be \nvalid. For example, 1-3 a means that the password must contain a at least 1 time and at \nmost 3 times.\n\nIn the above example, 2 passwords are valid. The middle password, cdefg, is not; it contains \nno instances of b, but needs at least 1. The first and third passwords are valid: they contain \none a or nine c, both within the limits of their respective policies.\n\nHow many passwords are valid according to their policies?\n\n=#\n\n\nfunction is_valid(policy, letter, password)\n letter_count = count(==(letter), password)\n if letter_countpolicy[2]\n return false\n else\n return true\n end\nend\n\nfunction count_valid_passwords(policy, letter, pass)\n count_valid = 0\n for i in 1:size(pass,1)\n if is_valid(policy[i], letter[i], pass[i])\n count_valid+=1\n end\n end\n return count_valid\nend\n\nprintln(size(policy,1),\", \",size(letter,1), \", \", size(pass,1))\nprintln(\"Valid passwords found: \", count_valid_passwords(policy, letter, pass))\n\n\n#=\n--- Part Two ---\n\nWhile it appears you validated the passwords correctly, they don't seem to be \nwhat the Official Toboggan Corporate Authentication System is expecting.\n\nThe shopkeeper suddenly realizes that he just accidentally explained the password \npolicy rules from his old job at the sled rental place down the street! The Official \nToboggan Corporate Policy actually works a little differently.\n\nEach policy actually describes two positions in the password, where 1 means the \nfirst character, 2 means the second character, and so on. (Be careful; Toboggan \nCorporate Policies have no concept of \"index zero\"!) Exactly one of these positions \nmust contain the given letter. Other occurrences of the letter are irrelevant for \nthe purposes of policy enforcement.\n\nGiven the same example list from above:\n\n 1-3 a: abcde is valid: position 1 contains a and position 3 does not.\n 1-3 b: cdefg is invalid: neither position 1 nor position 3 contains b.\n 2-9 c: ccccccccc is invalid: both position 2 and position 9 contain c.\n\nHow many passwords are valid according to the new interpretation of the policies?\n=#\n\nfunction is_valid(policy, letter, password)\n in_fst_pos = (password[policy[1]] == letter)\n in_snd_pos = (password[policy[2]] == letter)\n\n if in_fst_pos\n if in_snd_pos\n return false\n else\n return true\n end\n elseif in_snd_pos\n return true\n else\n return false\n end\nend\n\nfunction count_valid_passwords(policy, letter, pass)\n count_valid = 0\n for i in 1:size(pass,1)\n if is_valid(policy[i], letter[i], pass[i])\n count_valid+=1\n end\n end\n return count_valid\nend\n\nprintln(\"Valid passwords found with second policy: \", count_valid_passwords(policy, letter, pass))", "meta": {"hexsha": "79dab5720a6ef06ce8e079d844719f8adcb50880", "size": 4432, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Day2/password_philosophy.jl", "max_stars_repo_name": "colombelli/AoC2020-julia", "max_stars_repo_head_hexsha": "ae857741dccad0dc4f7da3636b3c7dd6fe00bd3d", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "Day2/password_philosophy.jl", "max_issues_repo_name": "colombelli/AoC2020-julia", "max_issues_repo_head_hexsha": "ae857741dccad0dc4f7da3636b3c7dd6fe00bd3d", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Day2/password_philosophy.jl", "max_forks_repo_name": "colombelli/AoC2020-julia", "max_forks_repo_head_hexsha": "ae857741dccad0dc4f7da3636b3c7dd6fe00bd3d", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 29.5466666667, "max_line_length": 98, "alphanum_fraction": 0.6949458484, "num_tokens": 1065, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4882833952958346, "lm_q2_score": 0.11436851561661297, "lm_q1q2_score": 0.05584424712022446}}
{"text": "@testset \"1089.duplicate-zeros.jl\" begin\n arr = [1,0,2,3,0,4,5,0]\n duplicate_zeros!(arr)\n @test arr == [1,0,0,2,3,0,0,4]\n\n arr2 = [1,2,3]\n duplicate_zeros!(arr2)\n @test arr2 == [1,2,3]\nend\n", "meta": {"hexsha": "e273f8b33ffd9bcb6bb3b722bd517ec2c75d612e", "size": 207, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/problems/1089.duplicate-zeros.jl", "max_stars_repo_name": "Moelf/LeetCode.jl", "max_stars_repo_head_hexsha": "c5a37331356060b743bc850c5255d6695d30fb57", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "test/problems/1089.duplicate-zeros.jl", "max_issues_repo_name": "Moelf/LeetCode.jl", "max_issues_repo_head_hexsha": "c5a37331356060b743bc850c5255d6695d30fb57", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "test/problems/1089.duplicate-zeros.jl", "max_forks_repo_name": "Moelf/LeetCode.jl", "max_forks_repo_head_hexsha": "c5a37331356060b743bc850c5255d6695d30fb57", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 20.7, "max_line_length": 40, "alphanum_fraction": 0.5603864734, "num_tokens": 95, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4610167793123159, "lm_q2_score": 0.12085323090725614, "lm_q1q2_score": 0.05571536728235085}}
{"text": "### A Pluto.jl notebook ###\n# v0.18.2\n\nusing Markdown\nusing InteractiveUtils\n\n# \u2554\u2550\u2561 c3530026-5a87-4447-88f5-fc744cb48ff2\nbegin\n\tusing PlutoUI \nend\n\n# \u2554\u2550\u2561 2c895e14-e71a-435d-86d4-c1b101877b3c\nmd\"\"\"\n### The End of the Beginning: Whole-Cell Metabolic Engineering\n\nIn the first lecture, we introduced metabolic engineering. Today, we'll close the loop at put together all we learned. Metabolic engineering is the practice of optimizing genetic and regulatory processes within cells to increase the cell's production of a desired small molecule or protein product of interest. \n\nMetabolic engineers manipulate the biochemical networks cells use to convert raw materials into molecules necessary for the cell's survival. Metabolic engineering specifically seeks to:\n\n1. Mathematically model biochemical networks, calculate the yield (product divided by substrate) of valuable products and identify parts of the network that constrain the production of these products of interest. \n1. Use genetic engineering techniques to modify the biochemical network to relieve constraints limiting production. Metabolic engineers can then model the modified network to calculate the new product yield and identify new constraints (back to 1).\n\nIn this (the final lecture of Part I of the course), we'll:\n\n* Incorporate the costs of transcription and translation into the flux balance analysis problem\n* Introduce a workaround (often used in practice) to account for the metabolic cost of gene expression\n* Close the loop: using a metabolic model and flux balance to optimize glycan production in _E.coli_. \n\n\"\"\"\n\n# \u2554\u2550\u2561 ce5c8e9c-7652-4c3e-98a7-98e3567dc695\nmd\"\"\"\n### Integrating the Cost and Logic of Gene Expression With Flux Balance Analysis\n\nThe [Allen and Palsson study](https://pubmed.ncbi.nlm.nih.gov/12453446/) gave us a roadmap of integrating the cost of gene expression with the metabolic operation of the cell. Further, [Vilkhovoy et al.](https://pubmed.ncbi.nlm.nih.gov/29944340/) showed how we could do this in a cell-free system. However, how do you account for gene expression in metabolic models at the whole-genome scale?\n\n* [Thiele I, Jamshidi N, Fleming RM, Palsson B\u00d8. Genome-scale reconstruction of Escherichia coli's transcriptional and translational machinery: a knowledge base, its mathematical formulation, and its functional characterization. PLoS Comput Biol. 2009;5(3):e1000312. doi:10.1371/journal.pcbi.1000312](https://www.ncbi.nlm.nih.gov/labs/pmc/articles/PMC2648898/?report=classic)\n* [Lerman JA, Hyduke DR, Latif H, Portnoy VA, Lewis NE, Orth JD, Schrimpe-Rutledge AC, Smith RD, Adkins JN, Zengler K, Palsson BO. In silico method for modeling metabolism and gene product expression at genome scale. Nat Commun. 2012 Jul 3;3:929. doi: 10.1038/ncomms1928. PMID: 22760628; PMCID: PMC3827721.](https://pubmed.ncbi.nlm.nih.gov/22760628/)\n\"\"\"\n\n# \u2554\u2550\u2561 bfd134fe-b311-4768-a53b-2c5c4c1b121a\nmd\"\"\"\n##### Aside: A question of timescales\nOne open question is whether we need to solve the gene expression and flux estimation problem simultaneously. The time scale of enzyme activity (and hence metabolic flux) is much faster than gene expression. Why? \n\n* The $k_{cat}$ for [phosphofructokinase-2 (pfk) in _E.coli_ is 9240 s$^{-1}$](https://bionumbers.hms.harvard.edu/bionumber.aspx?id=104955&ver=7&trm=kcat+pfk+in+E.+coli&org=). Thus, pfk has a characteristic time scale of $\\tau\\sim{k_{cat}}^{-1}$ or about 1$\\times$10$^{-4}$ s.\n* The $k_{cat}$ for RNAP polymerase is $k_{cat}$=$e_{x}L^{-1}$. If we assume [$e_{x}\\sim{35}$ nt/s](https://bionumbers.hms.harvard.edu/bionumber.aspx?id=111871&ver=1&trm=elongation+rate+RNAP+E.coli+&org=) and average read length [$L\\sim{924}$ nt](https://bionumbers.hms.harvard.edu/bionumber.aspx?id=111922&ver=3&trm=avarge+gene+length+in+E.+coli&org=), this gives $k_{cat}\\sim$ 0.038 s$^{-1}$ or a timescale of $\\tau\\sim$26.4 s.\n\nThus, the time scale of metabolic flux (e.g., the timescale of enzyme activity) is approximately 5-orders of magnitude faster than gene expression. Therefore, gene expression seems constant from the perspective of metabolic flux; hence, we treat these scales separately in traditional models. \n\"\"\"\n\n# \u2554\u2550\u2561 8041cb6c-b80f-4e81-8d1d-5a2e9942d809\nmd\"\"\"\n##### Aside: How do you account for macromolecular synthesis without the E/ME formulation?\nThe E/ME formulation of Palsson and coworkers is detailed and requires more time investment than perhaps you're willing to give in a metabolic engineering application. How do you account for the energy and resource cost of producing enzymes (and all the other machinery) required to make a cell?\n\nThe traditional way metabolic engineers address this question is to treat cell mass formation as just another reaction, e.g., the growth reaction $\\mu$: \n\n$$\\left\\{precursors\\right\\}~\\rightarrow~cell$$\n\nwhere each cellmass precursor has some stoichiometric coefficient $\\sigma_{i,\\mu}$ which describes how much of precursor $i$ is consumed (or produced) during cellmass formation. This formulation leads to species material balances of the form (assuming cellmass specific units, in a constant volume batch culture):\n\n$$\\frac{dx_{i}}{dt} = \\sum_{j=1}^{\\mathcal{R}}\\sigma_{ij}\\hat{r}_{j} - \\left(x_{i}-\\sigma_{i,\\mu}\\right)\\mu\\qquad{i=1,2,\\dots,\\mathcal{M}}$$\n\nAt steady state, for example in a flux balance analysis problem, these balances form the species constraints:\n\n$$\\sum_{j=1}^{\\mathcal{R}}\\sigma_{ij}\\hat{r}_{j} - \\left(x_{i}-\\sigma_{i,\\mu}\\right)\\mu = 0\\qquad{i=1,2,\\dots,\\mathcal{M}}$$\n\nPalsson and coworkers gave an example cell mass reaction for _E.coli_ in:\n\n* [Orth JD, Fleming RM, Palsson B\u00d8. Reconstruction and Use of Microbial Metabolic Networks: the Core Escherichia coli Metabolic Model as an Educational Guide. EcoSal Plus. 2010 Sep;4(1). doi: 10.1128/ecosalplus.10.2.1. PMID: 26443778.](https://pubmed.ncbi.nlm.nih.gov/26443778/)\n\nA recent paper on sampling uncertain biomass stoichiometric coefficients:\n\n* [Dinh et al. (2022) Quantifying the propagation of parametric uncertainty on flux balance analysis. Metabolic Engineering, 69:26-39; https://doi.org/10.1016/j.ymben.2021.10.012](https://www.sciencedirect.com/science/article/pii/S1096717621001634)\n\n\"\"\"\n\n# \u2554\u2550\u2561 06c3eafa-10b3-4a47-9a06-39f6abd29db3\nmd\"\"\"\n### Using flux balance analysis to improve metabolic performance\nUltimately a metabolic engineer uses a model to improve the performance of a production host or cell-free system. There is a huge variety of design approaches to improve metabolic function; let's look at a recent one from Cornell:\n\n* [Wayman JA, Glasscock C, Mansell TJ, DeLisa MP, Varner JD. Improving designer glycan production in Escherichia coli through model-guided metabolic engineering. Metab Eng Commun. 2019;9:e00088. Published 2019 Mar 29. doi:10.1016/j.mec.2019.e00088](https://www.ncbi.nlm.nih.gov/labs/pmc/articles/PMC6454127/)\n\n\"\"\"\n\n# \u2554\u2550\u2561 b15b540b-8f13-4f21-b2da-510d34143b33\nmd\"\"\"\n### Summary and conclusions\n\nToday, we:\n\n* Incorporated the costs of transcription and translation into the flux balance analysis problem\n* Introduced a workaround (often used in practice) to account for the metabolic cost of gene expression\n* Closed the loop: using a metabolic model and flux balance to optimize glycan production in _E.coli_. \n\"\"\"\n\n# \u2554\u2550\u2561 a818128b-d642-49e0-8c7c-db36b37ea882\nmd\"\"\"\n### Alas, if we just had some more time, it would have been cool to talk about:\n\n##### So many interesting tools:\n\n* [Lewis NE, Nagarajan H, Palsson BO. Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods. Nat Rev Microbiol. 2012 Feb 27;10(4):291-305. doi: 10.1038/nrmicro2737. PMID: 22367118; PMCID: PMC3536058.](https://pubmed.ncbi.nlm.nih.gov/22367118/)\n\n##### Better constraints give better flux estimates:\n\n* [Buescher JM, Antoniewicz MR, Boros LG, et al. A roadmap for interpreting (13)C metabolite labeling patterns from cells. Curr Opin Biotechnol. 2015;34:189-201. doi:10.1016/j.copbio.2015.02.003](https://www.ncbi.nlm.nih.gov/labs/pmc/articles/PMC4552607/)\n\n##### Flux balance analysis prediction of mutant response is not correct:\n\n* [Segr\u00e8 D, Vitkup D, Church GM. Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci U S A. 2002 Nov 12;99(23):15112-7. doi: 10.1073/pnas.232349399. Epub 2002 Nov 1. PMID: 12415116; PMCID: PMC137552.](https://pubmed.ncbi.nlm.nih.gov/12415116/)\n\n##### Molecular crowding plays a role in metabolism (non-representative random sample):\n\n\n* [Beg QK, Vazquez A, Ernst J, de Menezes MA, Bar-Joseph Z, Barab\u00e1si AL, Oltvai ZN. Intracellular crowding defines the mode and sequence of substrate uptake by Escherichia coli and constrains its metabolic activity. Proc Natl Acad Sci U S A. 2007 Jul 31;104(31):12663-8. doi: 10.1073/pnas.0609845104. Epub 2007 Jul 24. PMID: 17652176; PMCID: PMC1937523.](https://pubmed.ncbi.nlm.nih.gov/17652176/)'\n* [Conrado RJ, Mansell TJ, Varner JD, DeLisa MP. Stochastic reaction-diffusion simulation of enzyme compartmentalization reveals improved catalytic efficiency for a synthetic metabolic pathway. Metab Eng. 2007 Jul;9(4):355-63. doi: 10.1016/j.ymben.2007.05.002. Epub 2007 May 26. PMID: 17601761.](https://pubmed.ncbi.nlm.nih.gov/17601761/)\n* [Klumpp S, Scott M, Pedersen S, Hwa T. Molecular crowding limits translation and cell growth. Proc Natl Acad Sci U S A. 2013 Oct 15;110(42):16754-9. doi: 10.1073/pnas.1310377110. Epub 2013 Sep 30. PMID: 24082144; PMCID: PMC3801028.](https://pubmed.ncbi.nlm.nih.gov/22760628/)\n\n\"\"\"\n\n# \u2554\u2550\u2561 84d8a1c4-483c-4cc2-bdd0-7c88d26fd319\nTableOfContents(title=\"\ud83d\udcda Lecture Outline\", indent=true, depth=5, aside=true)\n\n# \u2554\u2550\u2561 7988263a-a5f5-11ec-3836-150105d04264\nhtml\"\"\"\n\"\"\"\n\n# \u2554\u2550\u2561 dee24b55-a5c5-4a47-9e1c-88ea0cf1cd61\nhtml\"\"\"\n\"\"\"\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000001\nPLUTO_PROJECT_TOML_CONTENTS = \"\"\"\n[deps]\nPlutoUI = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\n\n[compat]\nPlutoUI = \"~0.7.37\"\n\"\"\"\n\n# \u2554\u2550\u2561 00000000-0000-0000-0000-000000000002\nPLUTO_MANIFEST_TOML_CONTENTS = \"\"\"\n# This file is machine-generated - editing it directly is not advised\n\njulia_version = \"1.7.2\"\nmanifest_format = \"2.0\"\n\n[[deps.AbstractPlutoDingetjes]]\ndeps = [\"Pkg\"]\ngit-tree-sha1 = \"8eaf9f1b4921132a4cff3f36a1d9ba923b14a481\"\nuuid = \"6e696c72-6542-2067-7265-42206c756150\"\nversion = \"1.1.4\"\n\n[[deps.ArgTools]]\nuuid = \"0dad84c5-d112-42e6-8d28-ef12dabb789f\"\n\n[[deps.Artifacts]]\nuuid = \"56f22d72-fd6d-98f1-02f0-08ddc0907c33\"\n\n[[deps.Base64]]\nuuid = \"2a0f44e3-6c83-55bd-87e4-b1978d98bd5f\"\n\n[[deps.ColorTypes]]\ndeps = [\"FixedPointNumbers\", \"Random\"]\ngit-tree-sha1 = \"024fe24d83e4a5bf5fc80501a314ce0d1aa35597\"\nuuid = \"3da002f7-5984-5a60-b8a6-cbb66c0b333f\"\nversion = \"0.11.0\"\n\n[[deps.CompilerSupportLibraries_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"e66e0078-7015-5450-92f7-15fbd957f2ae\"\n\n[[deps.Dates]]\ndeps = [\"Printf\"]\nuuid = \"ade2ca70-3891-5945-98fb-dc099432e06a\"\n\n[[deps.Downloads]]\ndeps = [\"ArgTools\", \"LibCURL\", \"NetworkOptions\"]\nuuid = \"f43a241f-c20a-4ad4-852c-f6b1247861c6\"\n\n[[deps.FixedPointNumbers]]\ndeps = [\"Statistics\"]\ngit-tree-sha1 = \"335bfdceacc84c5cdf16aadc768aa5ddfc5383cc\"\nuuid = \"53c48c17-4a7d-5ca2-90c5-79b7896eea93\"\nversion = \"0.8.4\"\n\n[[deps.Hyperscript]]\ndeps = [\"Test\"]\ngit-tree-sha1 = \"8d511d5b81240fc8e6802386302675bdf47737b9\"\nuuid = \"47d2ed2b-36de-50cf-bf87-49c2cf4b8b91\"\nversion = \"0.0.4\"\n\n[[deps.HypertextLiteral]]\ngit-tree-sha1 = \"2b078b5a615c6c0396c77810d92ee8c6f470d238\"\nuuid = \"ac1192a8-f4b3-4bfe-ba22-af5b92cd3ab2\"\nversion = \"0.9.3\"\n\n[[deps.IOCapture]]\ndeps = [\"Logging\", \"Random\"]\ngit-tree-sha1 = \"f7be53659ab06ddc986428d3a9dcc95f6fa6705a\"\nuuid = \"b5f81e59-6552-4d32-b1f0-c071b021bf89\"\nversion = \"0.2.2\"\n\n[[deps.InteractiveUtils]]\ndeps = [\"Markdown\"]\nuuid = \"b77e0a4c-d291-57a0-90e8-8db25a27a240\"\n\n[[deps.JSON]]\ndeps = [\"Dates\", \"Mmap\", \"Parsers\", \"Unicode\"]\ngit-tree-sha1 = \"3c837543ddb02250ef42f4738347454f95079d4e\"\nuuid = \"682c06a0-de6a-54ab-a142-c8b1cf79cde6\"\nversion = \"0.21.3\"\n\n[[deps.LibCURL]]\ndeps = [\"LibCURL_jll\", \"MozillaCACerts_jll\"]\nuuid = \"b27032c2-a3e7-50c8-80cd-2d36dbcbfd21\"\n\n[[deps.LibCURL_jll]]\ndeps = [\"Artifacts\", \"LibSSH2_jll\", \"Libdl\", \"MbedTLS_jll\", \"Zlib_jll\", \"nghttp2_jll\"]\nuuid = \"deac9b47-8bc7-5906-a0fe-35ac56dc84c0\"\n\n[[deps.LibGit2]]\ndeps = [\"Base64\", \"NetworkOptions\", \"Printf\", \"SHA\"]\nuuid = \"76f85450-5226-5b5a-8eaa-529ad045b433\"\n\n[[deps.LibSSH2_jll]]\ndeps = [\"Artifacts\", \"Libdl\", \"MbedTLS_jll\"]\nuuid = \"29816b5a-b9ab-546f-933c-edad1886dfa8\"\n\n[[deps.Libdl]]\nuuid = \"8f399da3-3557-5675-b5ff-fb832c97cbdb\"\n\n[[deps.LinearAlgebra]]\ndeps = [\"Libdl\", \"libblastrampoline_jll\"]\nuuid = \"37e2e46d-f89d-539d-b4ee-838fcccc9c8e\"\n\n[[deps.Logging]]\nuuid = \"56ddb016-857b-54e1-b83d-db4d58db5568\"\n\n[[deps.Markdown]]\ndeps = [\"Base64\"]\nuuid = \"d6f4376e-aef5-505a-96c1-9c027394607a\"\n\n[[deps.MbedTLS_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"c8ffd9c3-330d-5841-b78e-0817d7145fa1\"\n\n[[deps.Mmap]]\nuuid = \"a63ad114-7e13-5084-954f-fe012c677804\"\n\n[[deps.MozillaCACerts_jll]]\nuuid = \"14a3606d-f60d-562e-9121-12d972cd8159\"\n\n[[deps.NetworkOptions]]\nuuid = \"ca575930-c2e3-43a9-ace4-1e988b2c1908\"\n\n[[deps.OpenBLAS_jll]]\ndeps = [\"Artifacts\", \"CompilerSupportLibraries_jll\", \"Libdl\"]\nuuid = \"4536629a-c528-5b80-bd46-f80d51c5b363\"\n\n[[deps.Parsers]]\ndeps = [\"Dates\"]\ngit-tree-sha1 = \"85b5da0fa43588c75bb1ff986493443f821c70b7\"\nuuid = \"69de0a69-1ddd-5017-9359-2bf0b02dc9f0\"\nversion = \"2.2.3\"\n\n[[deps.Pkg]]\ndeps = [\"Artifacts\", \"Dates\", \"Downloads\", \"LibGit2\", \"Libdl\", \"Logging\", \"Markdown\", \"Printf\", \"REPL\", \"Random\", \"SHA\", \"Serialization\", \"TOML\", \"Tar\", \"UUIDs\", \"p7zip_jll\"]\nuuid = \"44cfe95a-1eb2-52ea-b672-e2afdf69b78f\"\n\n[[deps.PlutoUI]]\ndeps = [\"AbstractPlutoDingetjes\", \"Base64\", \"ColorTypes\", \"Dates\", \"Hyperscript\", \"HypertextLiteral\", \"IOCapture\", \"InteractiveUtils\", \"JSON\", \"Logging\", \"Markdown\", \"Random\", \"Reexport\", \"UUIDs\"]\ngit-tree-sha1 = \"bf0a1121af131d9974241ba53f601211e9303a9e\"\nuuid = \"7f904dfe-b85e-4ff6-b463-dae2292396a8\"\nversion = \"0.7.37\"\n\n[[deps.Printf]]\ndeps = [\"Unicode\"]\nuuid = \"de0858da-6303-5e67-8744-51eddeeeb8d7\"\n\n[[deps.REPL]]\ndeps = [\"InteractiveUtils\", \"Markdown\", \"Sockets\", \"Unicode\"]\nuuid = \"3fa0cd96-eef1-5676-8a61-b3b8758bbffb\"\n\n[[deps.Random]]\ndeps = [\"SHA\", \"Serialization\"]\nuuid = \"9a3f8284-a2c9-5f02-9a11-845980a1fd5c\"\n\n[[deps.Reexport]]\ngit-tree-sha1 = \"45e428421666073eab6f2da5c9d310d99bb12f9b\"\nuuid = \"189a3867-3050-52da-a836-e630ba90ab69\"\nversion = \"1.2.2\"\n\n[[deps.SHA]]\nuuid = \"ea8e919c-243c-51af-8825-aaa63cd721ce\"\n\n[[deps.Serialization]]\nuuid = \"9e88b42a-f829-5b0c-bbe9-9e923198166b\"\n\n[[deps.Sockets]]\nuuid = \"6462fe0b-24de-5631-8697-dd941f90decc\"\n\n[[deps.SparseArrays]]\ndeps = [\"LinearAlgebra\", \"Random\"]\nuuid = \"2f01184e-e22b-5df5-ae63-d93ebab69eaf\"\n\n[[deps.Statistics]]\ndeps = [\"LinearAlgebra\", \"SparseArrays\"]\nuuid = \"10745b16-79ce-11e8-11f9-7d13ad32a3b2\"\n\n[[deps.TOML]]\ndeps = [\"Dates\"]\nuuid = \"fa267f1f-6049-4f14-aa54-33bafae1ed76\"\n\n[[deps.Tar]]\ndeps = [\"ArgTools\", \"SHA\"]\nuuid = \"a4e569a6-e804-4fa4-b0f3-eef7a1d5b13e\"\n\n[[deps.Test]]\ndeps = [\"InteractiveUtils\", \"Logging\", \"Random\", \"Serialization\"]\nuuid = \"8dfed614-e22c-5e08-85e1-65c5234f0b40\"\n\n[[deps.UUIDs]]\ndeps = [\"Random\", \"SHA\"]\nuuid = \"cf7118a7-6976-5b1a-9a39-7adc72f591a4\"\n\n[[deps.Unicode]]\nuuid = \"4ec0a83e-493e-50e2-b9ac-8f72acf5a8f5\"\n\n[[deps.Zlib_jll]]\ndeps = [\"Libdl\"]\nuuid = \"83775a58-1f1d-513f-b197-d71354ab007a\"\n\n[[deps.libblastrampoline_jll]]\ndeps = [\"Artifacts\", \"Libdl\", \"OpenBLAS_jll\"]\nuuid = \"8e850b90-86db-534c-a0d3-1478176c7d93\"\n\n[[deps.nghttp2_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"8e850ede-7688-5339-a07c-302acd2aaf8d\"\n\n[[deps.p7zip_jll]]\ndeps = [\"Artifacts\", \"Libdl\"]\nuuid = \"3f19e933-33d8-53b3-aaab-bd5110c3b7a0\"\n\"\"\"\n\n# \u2554\u2550\u2561 Cell order:\n# \u255f\u25002c895e14-e71a-435d-86d4-c1b101877b3c\n# \u255f\u2500ce5c8e9c-7652-4c3e-98a7-98e3567dc695\n# \u255f\u2500bfd134fe-b311-4768-a53b-2c5c4c1b121a\n# \u255f\u25008041cb6c-b80f-4e81-8d1d-5a2e9942d809\n# \u255f\u250006c3eafa-10b3-4a47-9a06-39f6abd29db3\n# \u255f\u2500b15b540b-8f13-4f21-b2da-510d34143b33\n# \u255f\u2500a818128b-d642-49e0-8c7c-db36b37ea882\n# \u2560\u255084d8a1c4-483c-4cc2-bdd0-7c88d26fd319\n# \u2560\u2550c3530026-5a87-4447-88f5-fc744cb48ff2\n# \u255f\u25007988263a-a5f5-11ec-3836-150105d04264\n# \u255f\u2500dee24b55-a5c5-4a47-9e1c-88ea0cf1cd61\n# \u255f\u250000000000-0000-0000-0000-000000000001\n# \u255f\u250000000000-0000-0000-0000-000000000002\n", "meta": {"hexsha": "22c3a7a3f1c4f2a529ab0b83f1db20314e2a40a9", "size": 17419, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "lectures/Lecture-13-5440-7770-S2022.jl", "max_stars_repo_name": "varnerlab/CHEME-5440-7770-Cornell-Spring-2022", "max_stars_repo_head_hexsha": "f580d12cb9d08657b6e7488280e5bf8391bb8530", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 2, "max_stars_repo_stars_event_min_datetime": "2022-02-07T23:40:03.000Z", "max_stars_repo_stars_event_max_datetime": "2022-02-14T19:06:26.000Z", "max_issues_repo_path": "lectures/Lecture-13-5440-7770-S2022.jl", "max_issues_repo_name": "varnerlab/CHEME-5440-7770-Cornell-Spring-2022", "max_issues_repo_head_hexsha": "f580d12cb9d08657b6e7488280e5bf8391bb8530", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "lectures/Lecture-13-5440-7770-S2022.jl", "max_forks_repo_name": "varnerlab/CHEME-5440-7770-Cornell-Spring-2022", "max_forks_repo_head_hexsha": "f580d12cb9d08657b6e7488280e5bf8391bb8530", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 12, "max_forks_repo_forks_event_min_datetime": "2022-02-03T15:15:33.000Z", "max_forks_repo_forks_event_max_datetime": "2022-03-08T22:46:36.000Z", "avg_line_length": 41.6722488038, "max_line_length": 429, "alphanum_fraction": 0.7331649348, "num_tokens": 6142, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.3522017684487511, "lm_q2_score": 0.15817435274843167, "lm_q1q2_score": 0.05570928676123421}}
{"text": "using PMI\nusing Test\n\n@test PMI.initialized() == false\n\nPMI.init()\n\n@test PMI.initialized()\n\nnranks = PMI.get_size()\nrank = PMI.get_rank()\n\n@test nranks >= 1\n@test 0 <= rank < nranks\n\nPMI.barrier()\n\n@test PMI.get_clique_size() >= 1\n@test rank \u2208 PMI.get_clique_ranks()\n\nPMI.finalize()", "meta": {"hexsha": "d39a3742e3c2bdef169ffe01b5a4df68e7319b7d", "size": 283, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/test_setup.jl", "max_stars_repo_name": "JuliaParallel/PMI.jl", "max_stars_repo_head_hexsha": "85c04b987d6f53bedbeb6d63b1b93344199aaae3", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2021-08-13T11:25:10.000Z", "max_stars_repo_stars_event_max_datetime": "2021-08-13T11:25:10.000Z", "max_issues_repo_path": "test/test_setup.jl", "max_issues_repo_name": "JuliaParallel/PMI.jl", "max_issues_repo_head_hexsha": "85c04b987d6f53bedbeb6d63b1b93344199aaae3", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 1, "max_issues_repo_issues_event_min_datetime": "2020-11-30T02:37:28.000Z", "max_issues_repo_issues_event_max_datetime": "2020-11-30T02:37:28.000Z", "max_forks_repo_path": "test/test_setup.jl", "max_forks_repo_name": "JuliaParallel/PMI.jl", "max_forks_repo_head_hexsha": "85c04b987d6f53bedbeb6d63b1b93344199aaae3", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 13.4761904762, "max_line_length": 35, "alphanum_fraction": 0.6855123675, "num_tokens": 90, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.43014734858584286, "lm_q2_score": 0.12940274166401952, "lm_q1q2_score": 0.055662246226516776}}
{"text": "n = parse(Int64, readline())\nA = map((x) -> parse(Int64,x) , split(readline(), \" \"))\n\nfor i in n:-1:1\n print(A[i],\" \")\nend\nprintln()", "meta": {"hexsha": "3b86ce39f779f8de1acf5df924d0c7915c3c1e1d", "size": 135, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Hackerrank/30 Days of Code/Julia/day 07.jl", "max_stars_repo_name": "Next-Gen-UI/Code-Dynamics", "max_stars_repo_head_hexsha": "a9b9d5e3f27e870b3e030c75a1060d88292de01c", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "Hackerrank/30 Days of Code/Julia/day 07.jl", "max_issues_repo_name": "Next-Gen-UI/Code-Dynamics", "max_issues_repo_head_hexsha": "a9b9d5e3f27e870b3e030c75a1060d88292de01c", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Hackerrank/30 Days of Code/Julia/day 07.jl", "max_forks_repo_name": "Next-Gen-UI/Code-Dynamics", "max_forks_repo_head_hexsha": "a9b9d5e3f27e870b3e030c75a1060d88292de01c", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 19.2857142857, "max_line_length": 55, "alphanum_fraction": 0.5481481481, "num_tokens": 47, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4301473631961697, "lm_q2_score": 0.12940273494909915, "lm_q1q2_score": 0.055662245228727834}}
{"text": "macro import_basic_huge()\n :(\n using ComplexVisual:\n CV_Error, cv_error,\n CV_TranslateByOffset, CV_MultiplyByFactor,\n CV_CyclicValue, cv_set_value!,\n cv_create_angle_cross_test, cv_half,\n CV_AttachType, cv_north, cv_south, cv_east, cv_west\n )\nend\n\n# {{{ Error Handling\nstruct CV_Error <: Exception\n msg :: String;\nend\ncv_error(messages::String...) = throw(CV_Error(join(messages)))\n\ncv_error(something...) = cv_error(\n (@sprintf(\"%s\", thing) for thing in something)...)\n# }}}\n\n# {{{ helper functions for showing concrete datatypes with their fields\n\n\"\"\"\nreturn fieldnames of a Datatype. Put the `parent_...` fields at the end.\n\"\"\"\nfunction cv_sorted_fieldnames(t::DataType)\n fnames = fieldnames(t)\n return tuple(\n filter(x -> !contains(string(x), \"parent_\"), fnames)...,\n filter(x -> contains(string(x), \"parent_\"), fnames)...)\nend\n\nfunction cv_show_value_replacements(value)\n if value isa Cairo.CairoSurfaceImage || value isa Cairo.CairoContext\n value = value.ptr\n end\n if value isa CV_LineSegments\n value = string(length(value)) * \" CV_LineSegments\"\n end\n return value\nend\n\n\"\"\"\nimplementation for `show(io, obj)` for ComplexVisual-objects with\n`typeof(obj) isa DataType`\n\"\"\"\nfunction cv_show_impl(io::IO, obj)\n t = typeof(obj)::DataType\n print(io, string(t.name.name), '(')\n first = true\n fnames = cv_sorted_fieldnames(t)\n for name_sym in fnames\n value = getfield(obj, name_sym)\n vtype = typeof(value)\n value = cv_show_value_replacements(value)\n if !first\n print(io, \", \")\n end\n print(io, string(name_sym), \": \")\n if contains(string(vtype.name.name), '#')\n show(io, MIME(\"text/plain\"), value)\n else\n show(io, value)\n end\n first = false\n end\n print(io, ')')\n return nothing\nend\n\n\"\"\"\nimplementation for `show(io, mime, obj)` for ComplexVisual-objects with\n`typeof(obj) isa DataType` and where `mime` is \"text/plain\".\n\"\"\"\nfunction cv_show_impl(io::IO, m::MIME{Symbol(\"text/plain\")}, obj)\n t = typeof(obj)\n outer_indent = (get(io, :cv_indent, \"\")::AbstractString)\n indent = outer_indent * \" \"\n iio = IOContext(io, :cv_indent => indent)\n println(io, string(t.name.name), '(')\n fnames = cv_sorted_fieldnames(t)\n for name_sym in fnames\n print(io, indent, string(name_sym), \": \")\n show(iio, m, cv_show_value_replacements(getfield(obj, name_sym)))\n println(io)\n end\n print(io, outer_indent, ')')\n return nothing\nend\n# }}} \n\n# {{{ Dynamic helpers\n\"\"\"\nA mutable struct with one (mutable) `value` (i.e. degree of freedom).\nThe struct is callable. When called, the input is translated by the\ngiven `value`.\n\"\"\"\nmutable struct CV_TranslateByOffset{N<:Number} # {{{\n value :: N\n\n function CV_TranslateByOffset(::Type{T}) where {T<:Number}\n return new{T}(zero(T))\n end\nend\nfunction (tbp::CV_TranslateByOffset{N})(z::N) where {N}\n return z + tbp.value\nend # }}}\n\n\"\"\"\nA mutable struct with one `factor` as degree of freedom.\nThe struct is callable. When called, the input is multiplied by\nthe given `factor.\n\nIf the factor is a complex number then this can be used to code\na scaling and rotation operator.\n\"\"\"\nmutable struct CV_MultiplyByFactor{N<:Number} # {{{\n factor :: N\n function CV_MultiplyByFactor(::Type{T}) where {T<:Number}\n return new{T}(one(T))\n end\nend\nfunction (mbf::CV_MultiplyByFactor{N})(z::N) where {N}\n return mbf.factor * z\nend\n# }}}\n\n\"\"\"\nA mutable struct with an integer `value` in the interval [1, maxvalue].\nThe struct is callable. When called, the value is increased by 1 or \nset to 1 if maxvalue was reached.\n\"\"\"\nmutable struct CV_CyclicValue{maxV} # {{{\n value :: Int\n\n function CV_CyclicValue(maxV::Int)\n maxV < 1 && cv_error(\"CV_CyclicValue: maxV must be >= 1\")\n return new{maxV}(1)\n end\nend\nfunction (cv::CV_CyclicValue{maxV})() where {maxV}\n cv.value = (cv.value == maxV) ? 1 : cv.value + 1\n return nothing\nend\n\nfunction cv_set_value!(cv::CV_CyclicValue{maxV}, new_value::Int) where {maxV}\n if !(1 \u2264 new_value \u2264 maxV)\n cv_error(\"cv_set_value: new_value must be in [1, \", string(maxV),\n \"]; but found: \", string(new_value))\n end\n cv.value = new_value\n return nothing\nend\n\n\n# }}}\n\n# }}}\n\n# {{{ Helper functions\n\"\"\"\ncreates a test, which checks if a line segment [z,w] intersects\nthe line segemnt [rmin*exp(i\u03d5), rmax*exp(i\u03d5)].\n\"\"\"\nfunction cv_create_angle_cross_test(\u03d5::Real, rmin::Real, rmax::Real; \u03b4=100eps())\n em\u03d5, rmin, rmax = exp(-Float64(\u03d5)*1im), Float64(rmin), Float64(rmax)\n\n return (z, w) -> begin\n hz = z*em\u03d5\n rhz, ihz = real(hz), imag(hz)\n z == w && return (rmin \u2264 rhz \u2264 rmax) && (abs(ihz) \u2264 \u03b4)\n\n hw = w*em\u03d5\n rhw, ihw = real(hw), imag(hw)\n if (abs(ihz) \u2264 \u03b4) && (abs(ihw) \u2264 \u03b4)\n # both are very close to the cross-boundary\n return (rmin \u2264 rhz \u2264 rmax) || (rmin \u2264 rhw \u2264 rmax)\n else\n sign(ihz)*sign(ihw) != -1 && return false\n r = (rhz - rhw + rhw*ihz - rhz*ihw)/(ihz - ihw)\n return rmin \u2264 r \u2264 rmax\n end\n end\nend\n\ncv_half(x::N) where {N<:Integer} = x \u00f7 N(2)\ncv_half(x::N) where {N<:AbstractFloat} = x/2\n\n# }}}\n\n# {{{ Directions/Locations \nconst cv_north, CV_northT = Val(:north), Val{:north}\nconst cv_south, CV_southT = Val(:south), Val{:south}\nconst cv_east, CV_eastT = Val(:east), Val{:east}\nconst cv_west, CV_westT = Val(:west), Val{:west}\nconst CV_AttachType = Union{CV_northT, CV_southT, CV_eastT, CV_westT}\n# }}}\n\n# vim:syn=julia:cc=79:fdm=marker:sw=4:ts=4:\n", "meta": {"hexsha": "986677de7e747d7c1378aed2fb2c4710f6b4a221", "size": 5719, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/basic.jl", "max_stars_repo_name": "April-Hannah-Lena/ComplexVisual.jl", "max_stars_repo_head_hexsha": "954defe63191cc507793ffccfb748fb646de4b07", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 3, "max_stars_repo_stars_event_min_datetime": "2021-05-05T12:21:41.000Z", "max_stars_repo_stars_event_max_datetime": "2021-06-19T14:10:13.000Z", "max_issues_repo_path": "src/basic.jl", "max_issues_repo_name": "April-Hannah-Lena/ComplexVisual.jl", "max_issues_repo_head_hexsha": "954defe63191cc507793ffccfb748fb646de4b07", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 3, "max_issues_repo_issues_event_min_datetime": "2021-05-11T16:17:42.000Z", "max_issues_repo_issues_event_max_datetime": "2021-08-11T11:59:11.000Z", "max_forks_repo_path": "src/basic.jl", "max_forks_repo_name": "April-Hannah-Lena/ComplexVisual.jl", "max_forks_repo_head_hexsha": "954defe63191cc507793ffccfb748fb646de4b07", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 2, "max_forks_repo_forks_event_min_datetime": "2021-05-06T10:16:41.000Z", "max_forks_repo_forks_event_max_datetime": "2021-06-02T15:35:10.000Z", "avg_line_length": 28.4527363184, "max_line_length": 80, "alphanum_fraction": 0.6254589963, "num_tokens": 1672, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4416729909662417, "lm_q2_score": 0.12592275499444552, "lm_q1q2_score": 0.055616679829106}}
{"text": "lines = readlines(\"input.txt\")\nre = r\"(\\w+)\"\n\nfunction main()\n uniq = 0\n for line = lines\n input, output = split(line, '|')\n signals = [m.match for m = eachmatch(re, output)]\n for signal = signals\n if length(signal) == 2 # one\n uniq += 1\n elseif length(signal) == 3 # seven\n uniq += 1\n elseif length(signal) == 4 # four\n uniq += 1\n elseif length(signal) == 7 # eight\n uniq += 1\n end\n end\n end\n println(uniq)\nend\n\nmain()\n", "meta": {"hexsha": "0fc53dacd876220913b50681eba3bd97502a40fb", "size": 575, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "08/8a.jl", "max_stars_repo_name": "dominikbayerl/aoc-2021", "max_stars_repo_head_hexsha": "2fb3a61216b6b34255e9bffc86d84cf7ac7f319a", "max_stars_repo_licenses": ["Unlicense"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "08/8a.jl", "max_issues_repo_name": "dominikbayerl/aoc-2021", "max_issues_repo_head_hexsha": "2fb3a61216b6b34255e9bffc86d84cf7ac7f319a", "max_issues_repo_licenses": ["Unlicense"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "08/8a.jl", "max_forks_repo_name": "dominikbayerl/aoc-2021", "max_forks_repo_head_hexsha": "2fb3a61216b6b34255e9bffc86d84cf7ac7f319a", "max_forks_repo_licenses": ["Unlicense"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 23.0, "max_line_length": 57, "alphanum_fraction": 0.4539130435, "num_tokens": 148, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.49218813572079556, "lm_q2_score": 0.11279541373888335, "lm_q1q2_score": 0.05551656440599681}}
{"text": "using Markdown\nusing Latexify\n\narr = [\"x/(y-1)\", 1.0, 3//2, :(x-y), :symb]\n\nM = vcat(hcat(arr...), hcat(arr...))\nhead = [\"col$i\" for i in 1:size(M, 2)]\nside = [\"row$i\" for i in 1:size(M, 1)]\n\n@test mdtable(arr) == Markdown.md\"\n| $\\frac{x}{y - 1}$ |\n| -----------------:|\n| $1.0$ |\n| $\\frac{3}{2}$ |\n| $x - y$ |\n| $symb$ |\n\"\n\n@test mdtable(arr; head = [\"head\"]) == Markdown.md\"\n| head |\n| -----------------:|\n| $\\frac{x}{y - 1}$ |\n| $1.0$ |\n| $\\frac{3}{2}$ |\n| $x - y$ |\n| $symb$ |\n\"\n\n@test mdtable(arr; head = [\"head\"], side=1:length(arr)) == Markdown.md\"\n| \u2218 | head |\n| ---:| -----------------:|\n| 1 | $\\frac{x}{y - 1}$ |\n| 2 | $1.0$ |\n| 3 | $\\frac{3}{2}$ |\n| 4 | $x - y$ |\n| 5 | $symb$ |\n\"\n\n@test mdtable(arr; head = [\"head\"], side=1:length(arr)+1) == Markdown.md\"\n| 1 | head |\n| ---:| -----------------:|\n| 2 | $\\frac{x}{y - 1}$ |\n| 3 | $1.0$ |\n| 4 | $\\frac{3}{2}$ |\n| 5 | $x - y$ |\n| 6 | $symb$ |\n\"\n\n@test mdtable(arr, arr) == Markdown.md\"\n| $\\frac{x}{y - 1}$ | $\\frac{x}{y - 1}$ |\n| -----------------:| -----------------:|\n| $1.0$ | $1.0$ |\n| $\\frac{3}{2}$ | $\\frac{3}{2}$ |\n| $x - y$ | $x - y$ |\n| $symb$ | $symb$ |\n\"\n\n@test mdtable(arr, arr; head = [\"col1\", \"col2\"]) == Markdown.md\"\n| col1 | col2 |\n| -----------------:| -----------------:|\n| $\\frac{x}{y - 1}$ | $\\frac{x}{y - 1}$ |\n| $1.0$ | $1.0$ |\n| $\\frac{3}{2}$ | $\\frac{3}{2}$ |\n| $x - y$ | $x - y$ |\n| $symb$ | $symb$ |\n\"\n\n@test mdtable(M) == Markdown.md\"\n| $\\frac{x}{y - 1}$ | $1.0$ | $\\frac{3}{2}$ | $x - y$ | $symb$ |\n| -----------------:| -----:| -------------:| -------:| ------:|\n| $\\frac{x}{y - 1}$ | $1.0$ | $\\frac{3}{2}$ | $x - y$ | $symb$ |\n\"\n\n@test mdtable(M, head=head) == Markdown.md\"\n| col1 | col2 | col3 | col4 | col5 |\n| -----------------:| -----:| -------------:| -------:| ------:|\n| $\\frac{x}{y - 1}$ | $1.0$ | $\\frac{3}{2}$ | $x - y$ | $symb$ |\n| $\\frac{x}{y - 1}$ | $1.0$ | $\\frac{3}{2}$ | $x - y$ | $symb$ |\n\"\n\n@test mdtable(M, head=head, side=side) == Markdown.md\"\n| \u2218 | col1 | col2 | col3 | col4 | col5 |\n| ----:| -----------------:| -----:| -------------:| -------:| ------:|\n| row1 | $\\frac{x}{y - 1}$ | $1.0$ | $\\frac{3}{2}$ | $x - y$ | $symb$ |\n| row2 | $\\frac{x}{y - 1}$ | $1.0$ | $\\frac{3}{2}$ | $x - y$ | $symb$ |\n\"\n\n@test mdtable(M, head=side, side=head, transpose=true) == Markdown.md\"\n| \u2218 | row1 | row2 |\n| ----:| -----------------:| -----------------:|\n| col1 | $\\frac{x}{y - 1}$ | $\\frac{x}{y - 1}$ |\n| col2 | $1.0$ | $1.0$ |\n| col3 | $\\frac{3}{2}$ | $\\frac{3}{2}$ |\n| col4 | $x - y$ | $x - y$ |\n| col5 | $symb$ | $symb$ |\n\"\n\n\nm = [\"one_two_tree\"; \"four_five_six\"; \"seven_eight\"]\n@test latexify(m; env=:mdtable, latex=false, escape_underscores=true) == Markdown.md\"\n| one\\_two\\_tree |\n| -------------:|\n| four\\_five\\_six |\n| seven_eight |\n\"\n\n\nusing DataFrames\nd = DataFrame(A = 11:13, B = [:X, :Y, :Z])\n\n\n@test latexify(d; env=:mdtable, side=1:3) == Markdown.md\"\n| \u2218 | A | B |\n| ---:| ----:| ---:|\n| 1 | $11$ | $X$ |\n| 2 | $12$ | $Y$ |\n| 3 | $13$ | $Z$ |\n\"\n\n@test latexify(d; env=:mdtable) == Markdown.md\"\n| A | B |\n| ----:| ---:|\n| $11$ | $X$ |\n| $12$ | $Y$ |\n| $13$ | $Z$ |\n\"\n\n\n\n@test latexify(((1.0, 2), (3, 4)); env=:mdtable) == Markdown.md\"\n| $1.0$ | $3$ |\n| -----:| ---:|\n| $2$ | $4$ |\n\"\n\n# @test_throws MethodError mdtable(M; bad_kwarg=\"should error\")\n", "meta": {"hexsha": "cc5ed499b636c0794c6d7c3e21d1e611e10d6807", "size": 3829, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/mdtable_test.jl", "max_stars_repo_name": "EuklidAlexandria/Latexify.jl", "max_stars_repo_head_hexsha": "20371f5c891345fbfce4064a0ef226a8bc51f14d", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 2, "max_stars_repo_stars_event_min_datetime": "2020-07-19T01:42:16.000Z", "max_stars_repo_stars_event_max_datetime": "2021-04-01T12:38:30.000Z", "max_issues_repo_path": "test/mdtable_test.jl", "max_issues_repo_name": "EuklidAlexandria/Latexify.jl", "max_issues_repo_head_hexsha": "20371f5c891345fbfce4064a0ef226a8bc51f14d", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "test/mdtable_test.jl", "max_forks_repo_name": "EuklidAlexandria/Latexify.jl", "max_forks_repo_head_hexsha": "20371f5c891345fbfce4064a0ef226a8bc51f14d", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 27.9489051095, "max_line_length": 85, "alphanum_fraction": 0.3298511361, "num_tokens": 1448, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.46490157137338844, "lm_q2_score": 0.11920292045759123, "lm_q1q2_score": 0.055417625033031194}}
{"text": "\"\"\"\n# Description\n\nGiven a tuple of symbols, return a `Dict{Symbol, Int}`` mapping each symbol to\nits index in the input tuple.\n\n# Arguments\n\n1. `column_names::NTuple{N, Symbol} where N`: A tuple of symbols.\n\n# Return Values\n\n1. `index::Dict{Symbol, Int}`: A dictionary mapping symbols to their indices in\n the input tuple.\n\n# Examples\n\n```\njulia> index_column_names(((:a, false), (:b, false)))\nDict{ColumnName,Int64} with 2 entries:\n (:a, false) => 1\n (:b, false) => 2\n```\n\"\"\"\nfunction index_column_names(column_names::NTuple{N, ColumnName}) where N\n Dict{ColumnName, Int}(column_names .=> 1:length(column_names))\nend\n", "meta": {"hexsha": "6310792f66a5e1412954eb2644bd761f63e275d3", "size": 625, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/query/expression_operations/index_column_names.jl", "max_stars_repo_name": "johnmyleswhite/Volcanito.jl", "max_stars_repo_head_hexsha": "d182ea3715016e3ef215cd54e8737ef422132350", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 28, "max_stars_repo_stars_event_min_datetime": "2020-08-28T14:55:25.000Z", "max_stars_repo_stars_event_max_datetime": "2022-02-21T09:54:01.000Z", "max_issues_repo_path": "src/query/expression_operations/index_column_names.jl", "max_issues_repo_name": "johnmyleswhite/Volcanito.jl", "max_issues_repo_head_hexsha": "d182ea3715016e3ef215cd54e8737ef422132350", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 29, "max_issues_repo_issues_event_min_datetime": "2020-08-28T16:24:08.000Z", "max_issues_repo_issues_event_max_datetime": "2020-09-29T16:26:01.000Z", "max_forks_repo_path": "src/query/expression_operations/index_column_names.jl", "max_forks_repo_name": "johnmyleswhite/Volcanito.jl", "max_forks_repo_head_hexsha": "d182ea3715016e3ef215cd54e8737ef422132350", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 4, "max_forks_repo_forks_event_min_datetime": "2020-09-06T14:21:02.000Z", "max_forks_repo_forks_event_max_datetime": "2022-02-12T10:14:07.000Z", "avg_line_length": 22.3214285714, "max_line_length": 79, "alphanum_fraction": 0.6944, "num_tokens": 180, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.48438008427698437, "lm_q2_score": 0.11436853221906972, "lm_q1q2_score": 0.05539783927490799}}
{"text": "\"\"\"\n parserl(T, io)\n\nReads a line from `io` and parses the result to type `T`. Typically used for single-value lines.\n\"\"\"\nparserl(T, io) = parse(T, readline(io))\n\n\n\"\"\"\n parsesrl(T, io; sep = \",\", headskip = 0, tailskip = 0)\n\nReads a line from `io`, splits the line over `sep`. Use keywords `headskip` and `tailskip` to optionally exclude the head and tail of the string before splitting.\n\n#Usage\njulia> parsesrl(Int, io) # read \"1,0,1\"\n\n\"\"\"\nfunction parsesrl(T, io; sep = \",\", headskip = 0, tailskip = 0)\n l = readline(io)\n i = 1 + headskip\n j = length(l) - tailskip\n l = l[i:j]\n parse.(T, split(l, sep))\nend", "meta": {"hexsha": "d8a2f0e095defbe1784630a767a6ab4a6cc5e3de", "size": 629, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/io/utilities.jl", "max_stars_repo_name": "Michiel-VL/GCSPET.jl", "max_stars_repo_head_hexsha": "1b288f878c85e988f8ba8ed7791dc6f60e011a4b", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/io/utilities.jl", "max_issues_repo_name": "Michiel-VL/GCSPET.jl", "max_issues_repo_head_hexsha": "1b288f878c85e988f8ba8ed7791dc6f60e011a4b", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/io/utilities.jl", "max_forks_repo_name": "Michiel-VL/GCSPET.jl", "max_forks_repo_head_hexsha": "1b288f878c85e988f8ba8ed7791dc6f60e011a4b", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 26.2083333333, "max_line_length": 162, "alphanum_fraction": 0.6279809221, "num_tokens": 195, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4843800842769844, "lm_q2_score": 0.11436852618181248, "lm_q1q2_score": 0.05539783635058083}}
{"text": "name = \"Jacob\"\nnum = 42\n\nif true\n println(\"True!\")\nelseif false\n println(\"False!\")\nelse\n println(\"WTF?!\")\nend\n\nfor x in 1:10\n y = x^2\n println(\"$(x) squared is $(y)\")\nend\n\nfunction area(width, height)\n return width * height\nend\n\nprintln(area(12, 34))\n", "meta": {"hexsha": "b82a907f82abe227cb4daa092e86abc7dec7b8d5", "size": 269, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "Julia/mail.jl", "max_stars_repo_name": "jgphilpott/babel", "max_stars_repo_head_hexsha": "47d81262ed3fb7a05302a8986782b99762f393a8", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2021-12-10T14:53:57.000Z", "max_stars_repo_stars_event_max_datetime": "2021-12-10T14:53:57.000Z", "max_issues_repo_path": "Julia/mail.jl", "max_issues_repo_name": "jgphilpott/babel", "max_issues_repo_head_hexsha": "47d81262ed3fb7a05302a8986782b99762f393a8", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "Julia/mail.jl", "max_forks_repo_name": "jgphilpott/babel", "max_forks_repo_head_hexsha": "47d81262ed3fb7a05302a8986782b99762f393a8", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 12.2272727273, "max_line_length": 35, "alphanum_fraction": 0.6022304833, "num_tokens": 87, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.38861802670584894, "lm_q2_score": 0.1422318931919251, "lm_q1q2_score": 0.055273877666883006}}
{"text": "### A Pluto.jl notebook ###\n# v0.18.0\n\nusing Markdown\nusing InteractiveUtils\n\n# \u2554\u2550\u2561 aff9313b-2fd8-46ca-b399-47fa21b324c5\nbegin\n import Pkg\n Pkg.activate(Base.current_project())\n Pkg.instantiate()\nend\n\n# \u2554\u2550\u2561 bcca4c48-89be-11ec-3c89-3189fd524f08\nmd\"\"\"\n# Template-matching in Julia using Pluto.jl\n\n## Introduction\n\nJulia is a general programming language suited for scientific computing applications. Moreover, Julia is a dynamic, high-level language: executing a program written in Julia does not require the user to compile it beforehand, as for C or Fortran, and its syntax is similar to MATLAB or Python. Nonetheless, its performance is close to ones of static languages.\n\nPluto is a Julia library for writing notebooks similar (but better in many respect) to Jupyter notebooks. In this notebook I will work out an example and very briefly introduce Julia and Pluto. The goal is to template match some signals from acoustic measurements of hydraulic fracture growth performed under triaxial confinement in laboratory (more informations [here](https://www.researchgate.net/profile/Seyyedmaalek-Momeni/publication/352780078_Combining_active_and_passive_acoustic_methods_to_image_hydraulic_fracture_growth_in_laboratory_experiments/links/60d87837a6fdccb745ea29dd/Combining-active-and-passive-acoustic-methods-to-image-hydraulic-fracture-growth-in-laboratory-experiments.pdf)). \n\"\"\"\n\n# \u2554\u2550\u2561 11ba1299-05e6-46c1-8716-ac06d5fc46a5\nmd\"\"\"\n## Julia and Pluto.jl\n\n### Pluto.jl\n\nA Pluto notebook is a sequence of cells. Each cell contain some Julia code, which can be executed. In Julia, every statement is an expression and hence evaluates to a value. Pluto converts that value to an HTML element and shows it. What you are reading is the output of a cell containing just a string, which returns the string itself [^1]. Pluto is able to parse the content of cells and detect dependencies between one cell and another. When the user evaluates a cell, for example changing the value of a variable, Pluto will re-evaluate all the cells that depend on that one and in the right order.\n\n\nNotice that Pluto requires one statement per cell, hence if you want to put more than one statement in a single cell you need to wrap them between the keywords `begin` and `end`, which are used to chain statements. The resulting block will evaluate to the value of the last statement.\n\nFor example\n\n```julia\nbegin\n\tx = 1\n\ty = x + 1\n\t\"hello world\"\n\t2 * y\nend\n```\n\nevaluates to 4.\n\n[^1]: Actually, the value that is rendered as the piece of text you are reading is not a string, but an object whose type is defined in the Markdown standard library. You can create these object prefixing the string literals with `md`. In this way, the content will be formatted using Markdown. Otherwise you can use plain strings.\n\"\"\"\n\n# \u2554\u2550\u2561 6dca0719-eeb3-42d5-8585-f89a77e579b3\nmd\"\"\"\n### Defining Functions in Julia\n\nJulia does not have methods attached to objects and accessible using the dot notation like in Python or C++. The main way of organizing code in Julia is by writing functions, and eventually grouping them into modules.\n\nFunction are defined using the `function` keyword. For example, the following will multiply the first two arguments and add the product to the third.\n\n```julia\nfunction f(x, y, z)\n\tx * y + z\nend\n```\n\nTh `return` keyword is implied in the last statement of a function definition, but can be supplyied if one wants to.\n\nNotice that the first time a function is called, Julia compiles on the fly that function. This is called just in time compilation or JIT. Hence the execution time of a function call change drastically between the first and subsequent calls. This is something to keep in mind when writing Julia code. There are some subtleties related to JIT compilation and the types of the arguments of a function, but we will talk about that in the next section.\n\"\"\"\n\n# \u2554\u2550\u2561 364a442a-17ef-455d-9608-aecd3126db59\nmd\"\"\"\n### Arrays and other types in Julia\n\nIn theory, one might write Julia code without thinking or caring about types, like in Python. However, it is practically impossibile to write and debug efficient code without some knowledge of the topic. One of the most important things to know is that the compiler can produce optimized machine code only for type-stable functions, where a function is called type-stabile if the type of its output is determined only from the types of its arguments (and not from their values). \n\nThe most used type in scientific computing is probably the array, i.e. a data type that represent indexed objects of the same type and contiguous in memory. Julia has a rich type system that includes arrays. In particular, arrays are an example of parametric types in Julia, i.e. types that depends on other types. The arguments of a dependent type are surrounded by braces, so for example `Array{Float64, 1}` is the type of one-dimensional arrays of 64-bit floating points numbers. To produce an exemplar of this type, you have to call its constructor. The constructor is a function with the same name of the type. In the case of one-dimensional array (or vector) constructors, they take two arguments, the value with which filling the array and the number of elements, so for example `Array{Float64, 1}(1.0, 10)` will be a ten element vector filled with the number 1.0. Array indexing in Julia is similar to Python with one important exceptions, in Julia one starts to count from 1 instead of 0, as you would do in Fortran or Matlab. Also, the last element is indicated with the keyword `end` and you can apply a function that acts on scalars element-wise on arrays posfixing a dot to its name in the function call, as in the function definition below (really, the dot notation is used for broadcasting, a powerful concept that should be familiar to Numpy user).\n\"\"\"\n\n# \u2554\u2550\u2561 Cell order:\n# \u2560\u2550aff9313b-2fd8-46ca-b399-47fa21b324c5\n# \u255f\u2500bcca4c48-89be-11ec-3c89-3189fd524f08\n# \u255f\u250011ba1299-05e6-46c1-8716-ac06d5fc46a5\n# \u255f\u25006dca0719-eeb3-42d5-8585-f89a77e579b3\n# \u255f\u2500364a442a-17ef-455d-9608-aecd3126db59\n", "meta": {"hexsha": "fd33329fa4a98d644634a1c38b57a988d22b9b82", "size": 6030, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "notebooks/0_julia_intro.jl", "max_stars_repo_name": "stefanocampanella/TemplateMatching.jl", "max_stars_repo_head_hexsha": "6a016534630a257f97012d8f73586b11d15747ab", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "notebooks/0_julia_intro.jl", "max_issues_repo_name": "stefanocampanella/TemplateMatching.jl", "max_issues_repo_head_hexsha": "6a016534630a257f97012d8f73586b11d15747ab", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 1, "max_issues_repo_issues_event_min_datetime": "2022-02-08T09:45:59.000Z", "max_issues_repo_issues_event_max_datetime": "2022-02-08T09:45:59.000Z", "max_forks_repo_path": "notebooks/0_julia_intro.jl", "max_forks_repo_name": "stefanocampanella/TemplateMatching.jl", "max_forks_repo_head_hexsha": "6a016534630a257f97012d8f73586b11d15747ab", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 70.1162790698, "max_line_length": 1364, "alphanum_fraction": 0.7842454395, "num_tokens": 1517, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.3886180267058489, "lm_q2_score": 0.14223188773801163, "lm_q1q2_score": 0.0552738755473939}}
{"text": "module TestDescribe\n\nusing Test\nusing RLEVectors\n\n@testset begin\n\n x = RLEVector([4, 5, 6], [3, 6, 9])\n\n # nrun\n @test nrun(x) == 3\n\n # length\n @test length(x) == 9\n\n # size\n @test size(x) == (9,)\n @test size(x, 1) == 9\n @test size(x, 2) == (9, 1)\n\n # isempty\n @test isempty(x) == false\n @test isempty(RLEVector(Int[], Int[])) == true\n\n # ==\n @test x == x\n\n # isequal\n @test isequal(x, x)\n\n # eltype, endtype\n @test endtype(x) == Int64\n @test eltype(x) == Int64\n\n\nend # testset\n\nend # module\n", "meta": {"hexsha": "5f5d6653c1cd9cf12ceed4d3226392b7d4ac9f89", "size": 549, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/test_describe.jl", "max_stars_repo_name": "phaverty/RleVectors.jl", "max_stars_repo_head_hexsha": "52fffeb9ce6549441771fd62997f387826415963", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 29, "max_stars_repo_stars_event_min_datetime": "2015-10-28T01:45:09.000Z", "max_stars_repo_stars_event_max_datetime": "2021-10-10T07:52:55.000Z", "max_issues_repo_path": "test/test_describe.jl", "max_issues_repo_name": "phaverty/RleVectors.jl", "max_issues_repo_head_hexsha": "52fffeb9ce6549441771fd62997f387826415963", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 43, "max_issues_repo_issues_event_min_datetime": "2016-06-02T08:38:37.000Z", "max_issues_repo_issues_event_max_datetime": "2022-02-28T21:07:27.000Z", "max_forks_repo_path": "test/test_describe.jl", "max_forks_repo_name": "phaverty/RleVectors.jl", "max_forks_repo_head_hexsha": "52fffeb9ce6549441771fd62997f387826415963", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 11, "max_forks_repo_forks_event_min_datetime": "2015-11-20T23:22:31.000Z", "max_forks_repo_forks_event_max_datetime": "2021-05-23T22:45:35.000Z", "avg_line_length": 14.0769230769, "max_line_length": 50, "alphanum_fraction": 0.5173041894, "num_tokens": 204, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.47657965106367595, "lm_q2_score": 0.11596071672639166, "lm_q1q2_score": 0.055264517914557505}}
{"text": "using Dates\nusing Statistics\nusing Test\n\nusing MarketData\n\nusing TimeSeries\n\n\n@testset \"meta\" begin\n\n\n@testset \"construction with and without meta field\" begin\n nometa = TimeArray(timestamp(cl), values(cl), colnames(cl))\n\n @testset \"default meta field to nothing\" begin\n @test meta(nometa) == nothing\n end\n\n @testset \"allow objects in meta field\" begin\n @test meta(mdata) == \"Apple\"\n end\nend\n\n\n@testset \"get index operations preserve meta\" begin\n @testset \"index by integer row\" begin\n @test meta(mdata[1]) == \"Apple\"\n end\n\n @testset \"index by integer range\" begin\n @test meta(mdata[1:2]) == \"Apple\"\n end\n\n @testset \"index by column name\" begin\n @test meta(mdata[:Close]) == \"Apple\"\n end\n\n @testset \"index by date range\" begin\n @test meta(mdata[[Date(2000,1,3), Date(2000,1,14)]]) == \"Apple\"\n end\nend\n\n\n@testset \"split operations preserve meta\" begin\n @testset \"when\" begin\n @test meta(when(mdata, dayofweek, 1)) == \"Apple\"\n end\n\n @testset \"from\" begin\n @test meta(from(mdata, Date(2000,1,1))) == \"Apple\"\n end\n\n @testset \"to\" begin\n @test meta(to(mdata, Date(2000,1,1))) == \"Apple\"\n end\nend\n\n\n@testset \"apply operations preserve meta\" begin\n @testset \"lag\" begin\n @test meta(lag(mdata)) == \"Apple\"\n end\n\n @testset \"lead\" begin\n @test meta(lead(mdata)) == \"Apple\"\n end\n\n @testset \"percentchange\" begin\n @test meta(percentchange(mdata)) == \"Apple\"\n end\n\n @testset \"moving\" begin\n @test meta(moving(mean,mdata,10)) == \"Apple\"\n end\n\n @testset \"upto\" begin\n @test meta(upto(sum, mdata)) == \"Apple\"\n end\nend\n\n\n@testset \"combine operations preserve meta\" begin\n @testset \"merge when both have identical meta\" begin\n @test meta(merge(cl, op)) == \"AAPL\"\n @test meta(merge(cl, op, :left)) == \"AAPL\"\n @test meta(merge(cl, op, :right)) == \"AAPL\"\n @test meta(merge(cl, op, :outer)) == \"AAPL\"\n end\n\n @testset \"merged meta field value concatenates when both objects' meta field values are strings\" begin\n @test meta(merge(mdata, cl)) == \"Apple_AAPL\"\n @test meta(merge(mdata, cl, :left)) == \"Apple_AAPL\"\n @test meta(merge(mdata, cl, :right)) == \"Apple_AAPL\"\n @test meta(merge(mdata, cl, :outer)) == \"Apple_AAPL\"\n end\n\n @testset \"merge when supplied with meta\" begin\n @test meta(merge(mdata, mdata, meta=47)) == 47\n @test meta(merge(mdata, mdata, :left, meta=47)) == 47\n @test meta(merge(mdata, mdata, :right, meta=47)) == 47\n @test meta(merge(mdata, mdata, :outer, meta=47)) == 47\n @test meta(merge(mdata, cl, meta=47)) == 47\n @test meta(merge(mdata, cl, :left, meta=47)) == 47\n @test meta(merge(mdata, cl, :right, meta=47)) == 47\n @test meta(merge(mdata, cl, :outer, meta=47)) == 47\n end\n\n @testset \"merged meta field value for disparate types in meta field defaults to Void\" begin\n @test meta(merge(mdata, merge(cl, op, meta=47))) == nothing\n @test meta(merge(mdata, merge(cl, op, meta=47), :left)) == nothing\n @test meta(merge(mdata, merge(cl, op, meta=47), :right)) == nothing\n @test meta(merge(mdata, merge(cl, op, meta=47), :outer)) == nothing\n end\n\n @testset \"collapse\" begin\n @test meta(collapse(mdata, week, first)) == \"Apple\"\n end\nend\n\n\n@testset \"basecall operations preserve meta\" begin\n @testset \"basecall\" begin\n @test meta(basecall(mdata, cumsum)) == \"Apple\"\n end\nend\n\n\n@testset \"mathematical and comparison operations preserve meta\" begin\n @testset \".+\" begin\n @test meta(mdata .+ mdata) == \"Apple\"\n @test meta(mdata .+ cl) == nothing\n end\n\n @testset \".<\" begin\n @test meta(mdata .< mdata) == \"Apple\"\n @test meta(mdata .< cl) == nothing\n end\nend\n\n\n@testset \"readwrite accepts meta argument\" begin\n @testset \"Apple is present\" begin\n @test meta(mdata) == \"Apple\"\n end\nend\n\n\nend # @testset \"meta\"\n", "meta": {"hexsha": "9314096f41953b0d6414f1000a6040580da8e9a4", "size": 4086, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/meta.jl", "max_stars_repo_name": "bovine3dom/TimeSeries.jl", "max_stars_repo_head_hexsha": "49859163fcf5ebe46ce0a8a07645a1edf4cf94db", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "test/meta.jl", "max_issues_repo_name": "bovine3dom/TimeSeries.jl", "max_issues_repo_head_hexsha": "49859163fcf5ebe46ce0a8a07645a1edf4cf94db", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "test/meta.jl", "max_forks_repo_name": "bovine3dom/TimeSeries.jl", "max_forks_repo_head_hexsha": "49859163fcf5ebe46ce0a8a07645a1edf4cf94db", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 27.24, "max_line_length": 106, "alphanum_fraction": 0.5971610377, "num_tokens": 1245, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4571367168274948, "lm_q2_score": 0.1208532261576127, "lm_q1q2_score": 0.05524644702370178}}
{"text": "# Copyright (c) 2013: Clp.jl contributors\n#\n# Use of this source code is governed by an MIT-style license that can be found\n# in the LICENSE.md file or at https://opensource.org/licenses/MIT.\n\nmodule TestMOIWrapper\n\nusing Test\nusing MathOptInterface\nimport Clp\n\nconst MOI = MathOptInterface\n\nfunction runtests()\n for name in names(@__MODULE__; all = true)\n if startswith(\"$(name)\", \"test_\")\n @testset \"$(name)\" begin\n getfield(@__MODULE__, name)()\n end\n end\n end\nend\n\nfunction test_SolverName()\n @test MOI.get(Clp.Optimizer(), MOI.SolverName()) == \"Clp\"\n return\nend\n\nfunction test_supports_default_copy_to()\n @test !MOI.supports_incremental_interface(Clp.Optimizer())\n return\nend\n\nfunction test_runtests()\n # This is what JuMP would construct\n model = MOI.Utilities.CachingOptimizer(\n MOI.Utilities.UniversalFallback(MOI.Utilities.Model{Float64}()),\n MOI.instantiate(Clp.Optimizer; with_bridge_type = Float64),\n )\n @test model.optimizer.model.model_cache isa\n MOI.Utilities.UniversalFallback{Clp.OptimizerCache}\n # `Variable.ZerosBridge` makes dual needed by some tests fail.\n MOI.Bridges.remove_bridge(\n model.optimizer,\n MathOptInterface.Bridges.Variable.ZerosBridge{Float64},\n )\n MOI.set(model, MOI.Silent(), true)\n MOI.Test.runtests(\n model,\n MOI.Test.Config(\n exclude = Any[MOI.DualObjectiveValue, MOI.ObjectiveBound],\n ),\n exclude = [\n # Unable to prove infeasibility\n \"test_conic_NormInfinityCone_INFEASIBLE\",\n \"test_conic_NormOneCone_INFEASIBLE\",\n ],\n )\n return\nend\n\nfunction test_Nonexistant_unbounded_ray()\n inner = Clp.Optimizer()\n model =\n MOI.Utilities.CachingOptimizer(MOI.default_cache(inner, Float64), inner)\n MOI.set(model, MOI.Silent(), true)\n x = MOI.add_variables(model, 5)\n MOI.set(\n model,\n MOI.ObjectiveFunction{MOI.ScalarAffineFunction{Float64}}(),\n MOI.ScalarAffineFunction(MOI.ScalarAffineTerm.(1.0, x), 0.0),\n )\n MOI.set(model, MOI.ObjectiveSense(), MOI.MAX_SENSE)\n MOI.optimize!(model)\n status = MOI.get(model, MOI.TerminationStatus())\n @test status == MOI.DUAL_INFEASIBLE\n return\nend\n\nfunction test_RawOptimizerAttribute()\n model = Clp.Optimizer()\n MOI.set(model, MOI.RawOptimizerAttribute(\"LogLevel\"), 1)\n @test MOI.get(model, MOI.RawOptimizerAttribute(\"LogLevel\")) == 1\n MOI.set(model, MOI.RawOptimizerAttribute(\"LogLevel\"), 2)\n @test MOI.get(model, MOI.RawOptimizerAttribute(\"LogLevel\")) == 2\n\n MOI.set(model, MOI.RawOptimizerAttribute(\"SolveType\"), 1)\n @test MOI.get(model, MOI.RawOptimizerAttribute(\"SolveType\")) == 1\n MOI.set(model, MOI.RawOptimizerAttribute(\"SolveType\"), 4)\n @test MOI.get(model, MOI.RawOptimizerAttribute(\"SolveType\")) == 4\n\n MOI.set(model, MOI.RawOptimizerAttribute(\"PresolveType\"), 1)\n @test MOI.get(model, MOI.RawOptimizerAttribute(\"PresolveType\")) == 1\n MOI.set(model, MOI.RawOptimizerAttribute(\"PresolveType\"), 0)\n @test MOI.get(model, MOI.RawOptimizerAttribute(\"PresolveType\")) == 0\n return\nend\n\nfunction test_All_parameters()\n model = Clp.Optimizer()\n param = MOI.RawOptimizerAttribute(\"NotAnOption\")\n @test !MOI.supports(model, param)\n @test_throws MOI.UnsupportedAttribute(param) MOI.get(model, param)\n @test_throws MOI.UnsupportedAttribute(param) MOI.set(model, param, false)\n for key in Clp.SUPPORTED_PARAMETERS\n @test MOI.supports(model, MOI.RawOptimizerAttribute(key))\n value = MOI.get(model, MOI.RawOptimizerAttribute(key))\n MOI.set(model, MOI.RawOptimizerAttribute(key), value)\n @test MOI.get(model, MOI.RawOptimizerAttribute(key)) == value\n end\n return\nend\n\nfunction test_copy_to_bug()\n model = MOI.Utilities.Model{Float64}()\n x = MOI.add_variable(model)\n con = [\n MOI.add_constraint(\n model,\n MOI.ScalarAffineFunction([MOI.ScalarAffineTerm(1.0, x)], 0.0),\n MOI.EqualTo(1.0),\n ) for i in 1:2\n ]\n clp = Clp.Optimizer()\n index_map = MOI.copy_to(clp, model)\n @test index_map[con[1]] != index_map[con[2]]\n return\nend\n\nfunction test_options_after_empty!()\n model = Clp.Optimizer()\n @test MOI.get(model, MOI.Silent()) == false\n MOI.set(model, MOI.Silent(), true)\n @test MOI.get(model, MOI.Silent()) == true\n MOI.empty!(model)\n @test MOI.get(model, MOI.Silent()) == true\n return\nend\n\nend # module TestMOIWrapper\n\nTestMOIWrapper.runtests()\n", "meta": {"hexsha": "d82c1f57b40b1b2d45cb5a8d4381889780d6a11f", "size": 4568, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/MOI_wrapper.jl", "max_stars_repo_name": "JuliaOpt/Clp.jl", "max_stars_repo_head_hexsha": "8efff608c2b9e2af5cc0d87fd829b16d7e3683d4", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 27, "max_stars_repo_stars_event_min_datetime": "2016-07-12T14:14:24.000Z", "max_stars_repo_stars_event_max_datetime": "2020-04-25T19:29:36.000Z", "max_issues_repo_path": "test/MOI_wrapper.jl", "max_issues_repo_name": "JuliaOpt/Clp.jl", "max_issues_repo_head_hexsha": "8efff608c2b9e2af5cc0d87fd829b16d7e3683d4", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 69, "max_issues_repo_issues_event_min_datetime": "2015-06-04T09:36:49.000Z", "max_issues_repo_issues_event_max_datetime": "2020-06-05T23:30:28.000Z", "max_forks_repo_path": "test/MOI_wrapper.jl", "max_forks_repo_name": "JuliaOpt/Clp.jl", "max_forks_repo_head_hexsha": "8efff608c2b9e2af5cc0d87fd829b16d7e3683d4", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 29, "max_forks_repo_forks_event_min_datetime": "2015-06-04T08:10:12.000Z", "max_forks_repo_forks_event_max_datetime": "2020-04-29T14:06:45.000Z", "avg_line_length": 31.9440559441, "max_line_length": 80, "alphanum_fraction": 0.6757880911, "num_tokens": 1268, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4571367168274948, "lm_q2_score": 0.12085322457439823, "lm_q1q2_score": 0.05524644629995632}}
{"text": "module SearchablePDFs\n\nusing Pkg\nusing Pkg.Artifacts\nusing Random\n\nusing ProgressMeter\nusing Scratch\nusing CSV\nusing Comonicon\n\nusing Poppler_jll\nusing unpaper_jll\nusing Tesseract_jll\n\nexport ocr\n\n#####\n##### Utilities\n#####\n\nfunction argument_error(msg; exception=isinteractive())\n if exception\n throw(ArgumentError(msg))\n else\n printstyled(\"ERROR:\"; bold=true, color=:red)\n printstyled(\" \", msg, \"\\n\"; color=:red)\n exit(1)\n end\nend\n\nfunction require_extension(path, ext; exception=isinteractive())\n _ext = splitext(path)[2]\n _ext == ext ||\n argument_error(\"Expected $path to have file extension `$ext`; got `$(_ext)`\";\n exception)\n return nothing\nend\n\nfunction require_no_file(path; exception=isinteractive())\n isfile(path) && argument_error(\"File already exists at `$(path)`!\"; exception)\n return nothing\nend\n\n# For now we will hardcode this choice of training data\nfunction get_data_path()\n return artifact\"tessdata_fast\"\nend\n\n# a place to store intermediate files; we could use temporary directories,\n# but I've occasionally run into permissions issues there\n# so I'd prefer to use a more local location. This also means that we are\n# more in charge of the cleanup, which can be good for debugging.\nfunction get_scratch_dir(pdf)\n return joinpath(@get_scratch!(\"pdf_tmps\"),\n splitext(basename(pdf))[1] * \"_\" * string(randstring(10)))\nend\n\n# https://discourse.julialang.org/t/collecting-all-output-from-shell-commands/15592/7\nfunction run_and_collect_logs(cmd::Cmd)\n out = Pipe()\n err = Pipe()\n process = run(pipeline(cmd; stdout=out, stderr=err); wait=false)\n close(out.in)\n close(err.in)\n\n stdout = @async String(read(out))\n stderr = @async String(read(err))\n wait(process)\n return (stdout=fetch(stdout), stderr=fetch(stderr), code=process.exitcode)\nend\n\n# There's gotta be a better way...\nfunction num_pages(pdf)\n result = read(`$(Poppler_jll.pdfinfo()) $pdf`, String)\n m = match(r\"Pages\\:\\s*([0-9]*)\", result)\n return parse(Int, m.captures[1])\nend\n\n#####\n##### Step 1: Extract images\n#####\n\n# Use Poppler to extract the image\nfunction get_images(pdf, page_range::UnitRange{Int}, tmp, total_pages)\n logs = run_and_collect_logs(`$(Poppler_jll.pdftoppm()) -f $(first(page_range)) -l $(last(page_range)) $pdf -tiff -forcenum $(tmp)/page`)\n @debug \"`pdftoppm`\" logs\n paths = [joinpath(tmp, string(\"page-\", lpad(page, ndigits(total_pages), '0'), \".tif\"))\n for page in page_range]\n return paths, (; binary=\"pdftoppm\", logs...)\nend\n\n# Clean up an image with unpaper\nfunction unpaper(img)\n img_base, img_ext = splitext(img)\n img_unpaper = img_base * \"_unpaper\" * img_ext\n logs = run_and_collect_logs(`$(unpaper_jll.unpaper()) $img $img_unpaper`)\n return (; img_unpaper, logs=(; binary=\"unpaper\", logs...))\nend\n\n#####\n##### Step 2: Use tesseract to generate a one-page searchable PDF from an image\n#####\n\nfunction make_pdf(img; tesseract_nthreads)\n data_path = get_data_path() * \"/\"\n img_base, img_ext = splitext(img)\n output = img_base\n tesseract = addenv(Tesseract_jll.tesseract(), \"OMP_THREAD_LIMIT\" => tesseract_nthreads)\n cmd = `$tesseract -l eng+equ --tessdata-dir $data_path $img $output -c tessedit_create_pdf=1`\n @debug \"Tesseracting!\" img\n logs = run_and_collect_logs(cmd)\n @debug logs\n return (; pdf=output * \".pdf\", logs=(; binary=\"tesseract\", logs...))\nend\n\n#####\n##### Step 3: collect all the PDFs into one with `pdfunite`\n#####\n\nfunction unite_pdfs(pdfs, output)\n logs = run_and_collect_logs(`$(Poppler_jll.pdfunite()) $pdfs $output`)\n return (; binary=\"pdfunite\", logs...)\nend\n\n# unites all the pdfs in `pdfs`, `max_files` at a time.\n# I ran into \"too many open files\" errors otherwise\n# (which seems weird... maybe ? It was on MacOS)\nfunction unite_many_pdfs!(unite_progress_meter, all_logs, tmp, pdfs, output_path;\n max_files_per_unite=100)\n isdir(tmp) || mkdir(tmp)\n\n output_paths = map(enumerate(Iterators.partition(pdfs, max_files_per_unite))) do (i,\n current_pdfs)\n current_output_path = joinpath(tmp, string(\"section_\", i, \".pdf\"))\n unite_logs = unite_pdfs(current_pdfs, current_output_path)\n put!(all_logs, (; page=missing, unite_logs...))\n next!(unite_progress_meter; step=length(current_pdfs))\n return current_output_path\n end\n\n unite_logs = unite_pdfs(output_paths, output_path)\n put!(all_logs, (; page=missing, unite_logs...))\n\n next!(unite_progress_meter; step=length(output_paths))\n return nothing\nend\n\n#####\n##### Apply steps 1 -- 3\n#####\n\n\"\"\"\n ocr(pdf, output_path=string(splitext(pdf)[1], \"_OCR\", \".pdf\"); apply_unpaper=false,\n ntasks=Sys.CPU_THREADS - 1, tesseract_nthreads=1, pages=num_pages(pdf),\n cleanup_after=true, cleanup_at_exit=true, tmp=get_scratch_dir(pdf),\n verbose=true)\n \nReads in a PDF located at `pdf`, uses Tesseract to OCR each page and combines the results into a pdf located `output_path`.\n\nKeyword arguments:\n\n* `ntasks`: how many parallel tasks to use for launching `tesseract` and `pdftoppm`.\n* `tesseract_nthreads`: how many threads to direct Tesseract to use\n* `apply_unpaper`: whether or not to apply `unpaper` to try to improve the image quality\n* `tmp`: a directory to store intermediate files. This directory is deleted at the end of the function if `cleanup_after` is set to `true`, and when the Julia session is ended if `cleanup_at_exit` is set to `true`.\n* `pages=nothing`: the number of pages of the PDF to process; the default of `nothing` indicates all pages in the PDF. It can help in debugging to set this to something small.\n* `verbose`: show a progress bar for each step of the process.\n\nSet `ENV[\"JULIA_DEBUG\"] = SearchablePDFs` to see (many) debug messages.\n\"\"\"\nfunction ocr(pdf, output_path=string(splitext(pdf)[1], \"_OCR\", \".pdf\"); apply_unpaper=false,\n ntasks=Sys.CPU_THREADS - 1, tesseract_nthreads=1, pages=nothing,\n cleanup_after=true, cleanup_at_exit=true, tmp=get_scratch_dir(pdf),\n verbose=true, force=false, max_files_per_unite=100)\n isfile(pdf) || argument_error(\"Input file not found at `$pdf`\"; exception=true)\n force || require_no_file(output_path; exception=true)\n require_extension(pdf, \".pdf\"; exception=true)\n require_extension(output_path, \".pdf\"; exception=true)\n\n total_pages = num_pages(pdf)\n\n if pages === nothing\n pages = total_pages\n elseif pages > total_pages\n argument_error(\"`pages` must be less than the total number of pages ($(total_pages))\";\n exception=true)\n end\n\n mkpath(tmp)\n if cleanup_at_exit\n atexit(() -> rm(tmp; force=true, recursive=true))\n end\n\n @debug \"Found file\" pdf pages tmp\n\n all_logs = Channel{@NamedTuple{page::Union{Int,UnitRange{Int},Missing},binary::String,\n stdout::String,stderr::String,code::Int}}(Inf)\n\n @debug \"Generating images...\"\n imag_prog = Progress(pages; desc=\"(1/3) Extracting images: \", enabled=verbose)\n\n img_paths_grps = asyncmap(Iterators.partition(1:pages, 20); ntasks) do page_range\n paths, pdftoppm_logs = get_images(pdf, page_range, tmp, total_pages)\n put!(all_logs, (; page=page_range, pdftoppm_logs...))\n next!(imag_prog; step=length(page_range))\n return paths\n end\n\n img_paths = reduce(vcat, img_paths_grps)\n\n @debug \"Finished generating images. Starting tesseracting...\"\n ocr_prog = Progress(pages; desc=\"(2/3) OCRing: \", enabled=verbose)\n pdfs = asyncmap(enumerate(img_paths); ntasks) do (page, img)\n @debug \"img\" page img\n if apply_unpaper\n img, unpaper_logs = unpaper(img)\n put!(all_logs, (; page, unpaper_logs...))\n end\n pdf, tesseract_logs = make_pdf(img; tesseract_nthreads)\n put!(all_logs, (; page, tesseract_logs...))\n next!(ocr_prog)\n return pdf\n end\n @debug \"Finished processing pages. Uniting...\"\n unite_dir = joinpath(tmp, \"unite\")\n unite_progress_meter = Progress(pages + cld(pages, max_files_per_unite) + 1;\n desc=\"(3/3) Collecting pages: \", enabled=verbose)\n unite_many_pdfs!(unite_progress_meter, all_logs, unite_dir, pdfs, output_path;\n max_files_per_unite)\n @debug \"Done uniting pdfs\"\n if cleanup_after\n @debug \"Cleaning up\"\n rm(tmp; recursive=true, force=true)\n end\n @debug \"Done\"\n if verbose\n isfile(output_path) || @error \"File was not generated, check the logs!\"\n end\n close(all_logs)\n return (; output_path, logs=collect(all_logs), tmp)\nend\n\n#####\n##### CLI interface\n#####\n\n\"\"\"\nCreate a searchable version of a PDF.\n\"\"\"\n@main function searchable(input_pdf::String,\n output_path::String=string(splitext(input_pdf)[1], \"_OCR\",\n \".pdf\"); apply_unpaper::Bool=false,\n ntasks::Int=Sys.CPU_THREADS - 1, tesseract_nthreads::Int=1,\n keep_intermediates::Bool=false,\n tmp::String=get_scratch_dir(input_pdf), quiet::Bool=false,\n logfile::Union{Nothing,String}=nothing, force::Bool=false)\n # some of these are redundant with checks inside `ocr`; that's because we want to do them before the \"Starting to ocr\" message,\n # and we want them to exit if they fail in a non-interactive context, instead of printing a stacktracee.\n isfile(input_pdf) || argument_error(\"Input file not found at `$(input_pdf)`\")\n force || require_no_file(output_path)\n require_extension(input_pdf, \".pdf\")\n require_extension(output_path, \".pdf\")\n if logfile !== nothing\n force || require_no_file(logfile)\n require_extension(logfile, \".csv\")\n end\n verbose = !quiet\n verbose &&\n println(\"Starting to ocr `$(input_pdf)`; result will be located at `$(output_path)`.\")\n result = ocr(input_pdf, output_path; apply_unpaper, ntasks, tesseract_nthreads,\n cleanup_after=!keep_intermediates, cleanup_at_exit=!keep_intermediates,\n tmp, verbose)\n verbose && println(\"\\nOutput is located at `$(output_path)`.\")\n if keep_intermediates && verbose\n println(\"Intermediate files located at `$tmp`.\")\n end\n if logfile !== nothing\n verbose && println(\"Writing logs...\")\n CSV.write(logfile, result.logs)\n verbose && println(\"Logs written to `$(logfile)`.\")\n end\n\n return result\nend\n\nend # module\n", "meta": {"hexsha": "a35a69f702d1af965132be471a559f7b5adf0011", "size": 10785, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/SearchablePDFs.jl", "max_stars_repo_name": "ericphanson/PDFSandwich.jl", "max_stars_repo_head_hexsha": "90ba0ebcb6ceb8e8d599c78ed46b8c3d59c23dd5", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 14, "max_stars_repo_stars_event_min_datetime": "2021-05-26T20:41:40.000Z", "max_stars_repo_stars_event_max_datetime": "2022-01-24T20:39:08.000Z", "max_issues_repo_path": "src/SearchablePDFs.jl", "max_issues_repo_name": "ericphanson/PDFSandwich.jl", "max_issues_repo_head_hexsha": "90ba0ebcb6ceb8e8d599c78ed46b8c3d59c23dd5", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 7, "max_issues_repo_issues_event_min_datetime": "2021-03-13T03:13:43.000Z", "max_issues_repo_issues_event_max_datetime": "2021-12-02T01:01:36.000Z", "max_forks_repo_path": "src/SearchablePDFs.jl", "max_forks_repo_name": "ericphanson/PDFSandwich.jl", "max_forks_repo_head_hexsha": "90ba0ebcb6ceb8e8d599c78ed46b8c3d59c23dd5", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2021-05-26T20:21:09.000Z", "max_forks_repo_forks_event_max_datetime": "2021-05-26T20:21:09.000Z", "avg_line_length": 37.4479166667, "max_line_length": 214, "alphanum_fraction": 0.6577654149, "num_tokens": 2678, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.33111973962899144, "lm_q2_score": 0.16667540675766923, "lm_q1q2_score": 0.05518951728815567}}
{"text": "using Base.Dates\nusing Base.Test\n\nusing MarketData\n\nusing TimeSeries\n\n\n@testset \"meta\" begin\n\n\n@testset \"construction with and without meta field\" begin\n nometa = TimeArray(cl.timestamp, cl.values, cl.colnames)\n\n @testset \"default meta field to nothing\" begin\n @test nometa.meta == nothing\n end\n\n @testset \"allow objects in meta field\" begin\n @test mdata.meta == \"Apple\"\n end\nend\n\n\n@testset \"get index operations preserve meta\" begin\n @testset \"index by integer row\" begin\n @test mdata[1].meta == \"Apple\"\n end\n\n @testset \"index by integer range\" begin\n @test mdata[1:2].meta == \"Apple\"\n end\n\n @testset \"index by column name\" begin\n @test mdata[\"Close\"].meta == \"Apple\"\n end\n\n @testset \"index by date range\" begin\n @test mdata[[Date(2000,1,3), Date(2000,1,14)]].meta == \"Apple\"\n end\nend\n\n\n@testset \"split operations preserve meta\" begin\n @testset \"when\" begin\n @test when(mdata, dayofweek, 1).meta == \"Apple\"\n end\n\n @testset \"from\" begin\n @test from(mdata, Date(2000,1,1)).meta == \"Apple\"\n end\n\n @testset \"to\" begin\n @test to(mdata, Date(2000,1,1)).meta == \"Apple\"\n end\nend\n\n\n@testset \"apply operations preserve meta\" begin\n @testset \"lag\" begin\n @test lag(mdata).meta == \"Apple\"\n end\n\n @testset \"lead\" begin\n @test lead(mdata).meta == \"Apple\"\n end\n\n @testset \"percentchange\" begin\n @test percentchange(mdata).meta == \"Apple\"\n end\n\n @testset \"moving\" begin\n @test moving(mdata,mean,10).meta == \"Apple\"\n end\n\n @testset \"upto\" begin\n @test upto(mdata,sum).meta == \"Apple\"\n end\nend\n\n\n@testset \"combine operations preserve meta\" begin\n @testset \"merge when both have identical meta\" begin\n @test merge(cl, op).meta == \"AAPL\"\n @test merge(cl, op, :left).meta == \"AAPL\"\n @test merge(cl, op, :right).meta == \"AAPL\"\n @test merge(cl, op, :outer).meta == \"AAPL\"\n end\n\n @testset \"merged meta field value concatenates when both objects' meta field values are strings\" begin\n @test merge(mdata, cl).meta == \"Apple_AAPL\"\n @test merge(mdata, cl, :left).meta == \"Apple_AAPL\"\n @test merge(mdata, cl, :right).meta == \"Apple_AAPL\"\n @test merge(mdata, cl, :outer).meta == \"Apple_AAPL\"\n end\n\n @testset \"merge when supplied with meta\" begin\n @test merge(mdata, mdata, meta=47).meta == 47\n @test merge(mdata, mdata, :left, meta=47).meta == 47\n @test merge(mdata, mdata, :right, meta=47).meta == 47\n @test merge(mdata, mdata, :outer, meta=47).meta == 47\n @test merge(mdata, cl, meta=47).meta == 47\n @test merge(mdata, cl, :left, meta=47).meta == 47\n @test merge(mdata, cl, :right, meta=47).meta == 47\n @test merge(mdata, cl, :outer, meta=47).meta == 47\n end\n\n @testset \"merged meta field value for disparate types in meta field defaults to Void\" begin\n @test merge(mdata, merge(cl, op, meta=47)).meta == Void\n @test merge(mdata, merge(cl, op, meta=47), :left).meta == Void\n @test merge(mdata, merge(cl, op, meta=47), :right).meta == Void\n @test merge(mdata, merge(cl, op, meta=47), :outer).meta == Void\n end\n\n @testset \"collapse\" begin\n @test collapse(mdata, week, first).meta == \"Apple\"\n end\nend\n\n\n@testset \"basecall operations preserve meta\" begin\n @testset \"basecall\" begin\n @test basecall(mdata, cumsum).meta == \"Apple\"\n end\nend\n\n\n@testset \"mathematical and comparison operations preserve meta\" begin\n @testset \".+\" begin\n @test (mdata .+ mdata).meta == \"Apple\"\n @test (mdata .+ cl).meta == Void\n end\n\n @testset \".<\" begin\n @test (mdata .< mdata).meta == \"Apple\"\n @test (mdata .< cl).meta == Void\n end\nend\n\n\n@testset \"readwrite accepts meta argument\" begin\n @testset \"Apple is present\" begin\n @test mdata.meta == \"Apple\"\n end\nend\n\n\nend # @testset \"meta\"\n", "meta": {"hexsha": "56740d02d9f61d70495e6854af8da5af38b44b39", "size": 4019, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/meta.jl", "max_stars_repo_name": "BenjaminBorn/TimeSeries.jl", "max_stars_repo_head_hexsha": "c38509ea8427afd74cc140e0f7f6c7ca0cb39c06", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "test/meta.jl", "max_issues_repo_name": "BenjaminBorn/TimeSeries.jl", "max_issues_repo_head_hexsha": "c38509ea8427afd74cc140e0f7f6c7ca0cb39c06", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "test/meta.jl", "max_forks_repo_name": "BenjaminBorn/TimeSeries.jl", "max_forks_repo_head_hexsha": "c38509ea8427afd74cc140e0f7f6c7ca0cb39c06", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2021-09-16T22:57:15.000Z", "max_forks_repo_forks_event_max_datetime": "2021-09-16T22:57:15.000Z", "avg_line_length": 26.9731543624, "max_line_length": 106, "alphanum_fraction": 0.6008957452, "num_tokens": 1216, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4960938294709195, "lm_q2_score": 0.11124119472172637, "lm_q1q2_score": 0.055186070284421476}}
{"text": "\"\"\"\n# Summary statistics\n\n```julia\ndescribe(ts::TS)\n```\n\nCompute summary statistics of `ts`. The output is a `DataFrame`\ncontaining standard statistics along with number of missing values and\ndata types of columns.\n\n# Examples\n```jldoctest; setup = :(using TSx, DataFrames, Dates, Random, Statistics)\njulia> using Random;\njulia> random(x) = rand(MersenneTwister(123), x...);\njulia> ts = TS(random(([1, 2, 3, 4, missing], 10)))\njulia> describe(ts)\n2\u00d77 DataFrame\n Row \u2502 variable mean min median max nmissing eltype\n \u2502 Symbol Float64 Int64 Float64 Int64 Int64 Type\n\u2500\u2500\u2500\u2500\u2500\u253c\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\n 1 \u2502 Index 5.5 1 5.5 10 0 Int64\n 2 \u2502 x1 2.75 2 3.0 4 2 Union{Missing, Int64}\n\n```\n\"\"\"\nfunction describe(io::IO, ts::TS)\n DataFrames.describe(ts.coredata)\nend\nTSx.describe(ts::TS) = TSx.describe(stdout, ts)\n\n\nfunction Base.show(io::IO, ts::TS)\n println(\"(\", TSx.nrow(ts), \" x \", TSx.ncol(ts), \") TS with \", eltype(index(ts)), \" Index\")\n println(\"\")\n DataFrames.show(ts.coredata, show_row_number=false, summary=false)\n return nothing\nend\nBase.show(ts::TS) = show(stdout, ts)\n\n\nfunction Base.summary(io::IO, ts::TS)\n println(\"(\", nr(ts), \" x \", nc(ts), \") TS\")\nend\n\n\"\"\"\n# Size methods\n\n```julia\nnrow(ts::TS)\nnr(ts::TS)\n```\n\nReturn the number of rows of `ts`. `nr` is an alias for `nrow`.\n\n# Examples\n```jldoctest; setup = :(using TSx, DataFrames, Dates, Random, Statistics)\njulia> ts = TS(collect(1:10))\njulia> TSx.nrow(ts)\n10\n```\n\"\"\"\nfunction nrow(ts::TS)\n DataFrames.size(ts.coredata)[1]\nend\n# alias\nnr = TSx.nrow\n\nfunction Base.lastindex(ts::TS)\n lastindex(index(ts))\nend\n\nfunction Base.length(ts::TS)\n TSx.nrow(ts)\nend\n\n# Number of columns\n\"\"\"\n# Size methods\n\n```julia\nncol(ts::TS)\n```\n\nReturn the number of columns of `ts`. `nc` is an alias for `ncol`.\n\n# Examples\n```jldoctest; setup = :(using TSx, DataFrames, Dates, Random, Statistics)\njulia> using Random;\n\njulia> random(x) = rand(MersenneTwister(123), x);\n\njulia> TSx.ncol(TS([random(100) random(100) random(100)]))\n3\n\njulia> nc(TS([random(100) random(100) random(100)]))\n3\n```\n\"\"\"\nfunction ncol(ts::TS)\n DataFrames.size(ts.coredata)[2] - 1\nend\n# alias\nnc = TSx.ncol\n\n# Size of\n\"\"\"\n# Size methods\n```julia\nsize(ts::TS)\n```\n\nReturn the number of rows and columns of `ts` as a tuple.\n\n# Examples\n```jldoctest; setup = :(using TSx, DataFrames, Dates, Random, Statistics)\njulia> TSx.size(TS([collect(1:100) collect(1:100) collect(1:100)]))\n(100, 3)\n```\n\"\"\"\nfunction size(ts::TS)\n nr = TSx.nrow(ts)\n nc = TSx.ncol(ts)\n (nr, nc)\nend\n\n# Return index column\n\"\"\"\n# Index column\n\n```julia\nindex(ts::TS)\n```\n\nReturn the index vector from the `coredata` DataFrame.\n\n# Examples\n\n```jldoctest; setup = :(using TSx, DataFrames, Dates, Random, Statistics)\njulia> using Random;\n\njulia> random(x) = rand(MersenneTwister(123), x);\n\njulia> ts = TS(random(10), Date(\"2022-02-01\"):Month(1):Date(\"2022-02-01\")+Month(9));\n\n\njulia> show(ts)\n(10 x 1) TS with Dates.Date Index\n\n Index x1\n Date Float64\n\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\n 2022-02-01 0.768448\n 2022-03-01 0.940515\n 2022-04-01 0.673959\n 2022-05-01 0.395453\n 2022-06-01 0.313244\n 2022-07-01 0.662555\n 2022-08-01 0.586022\n 2022-09-01 0.0521332\n 2022-10-01 0.26864\n 2022-11-01 0.108871\n\njulia> index(ts)\n10-element Vector{Date}:\n 2022-02-01\n 2022-03-01\n 2022-04-01\n 2022-05-01\n 2022-06-01\n 2022-07-01\n 2022-08-01\n 2022-09-01\n 2022-10-01\n 2022-11-01\n\njulia> eltype(index(ts))\nDate\n```\n\"\"\"\nfunction index(ts::TS)\n ts.coredata[!, :Index]\nend\n\n\"\"\"\n# Column names\n```julia\nnames(ts::TS)\n```\n\nReturn a `Vector{String}` containing column names of the TS object,\nexcludes index.\n\n# Examples\n```jldoctest; setup = :(using TSx, DataFrames, Dates, Random, Statistics)\njulia> names(TS([1:10 11:20]))\n2-element Vector{String}:\n \"x1\"\n \"x2\"\n```\n\"\"\"\n \nfunction names(ts::TS)\n names(ts.coredata[!, Not(:Index)])\nend\n\n\n\"\"\"\n# First Row\n```julia\nfirst(ts::TS)\n```\n\nReturns the first row of `ts` as a TS object.\n\n# Examples\n```jldoctest; setup = :(using TSx, DataFrames, Dates, Random)\njulia> first(TS(1:10))\n(10 x 1) TS with Dates.Date Index\n\n Index x1\n Date Float64\n\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\n 2022-02-01 0.768448\n\n```\n\"\"\"\nfunction Base.first(ts::TS)\n TS(Base.first(ts.coredata,1))\nend\n\n\n\"\"\"\n# Head\n```julia\nhead(ts::TS, n::Int = 10)\n```\nReturns the first `n` rows of `ts`.\n\n# Examples\n```jldoctest; setup = :(using TSx, DataFrames, Dates, Random)\njulia> head(TS(1:100))\n(10 x 1) TS with Int64 Index\n\n Index x1 \n Int64 Int64 \n\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\n 1 1\n 2 2\n 3 3\n 4 4\n 5 5\n 6 6\n 7 7\n 8 8\n 9 9\n 10 10\n```\n\"\"\"\nfunction head(ts::TS, n::Int = 10)\n TS(Base.first(ts.coredata, n))\nend\n\n\n\"\"\"\n# Tail\n```julia\ntail(ts::TS, n::Int = 10)\n```\n\nReturns the last `n` rows of `ts`.\n\n```jldoctest; setup = :(using TSx, DataFrames, Dates, Random)\njulia> tail(TS(1:100))\n(10 x 1) TS with Int64 Index\n\n Index x1 \n Int64 Int64 \n\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\u2500\n 91 91\n 92 92\n 93 93\n 94 94\n 95 95\n 96 96\n 97 97\n 98 98\n 99 99\n 100 100\n```\n\"\"\"\nfunction tail(ts::TS, n::Int = 10)\n TS(DataFrames.last(ts.coredata, n))\nend\n", "meta": {"hexsha": "7a951f24de87cc2681ce62d8bfec31f8b0766c13", "size": 5341, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/utils.jl", "max_stars_repo_name": "xKDR/TSx.jl", "max_stars_repo_head_hexsha": "82189a7475159b3c1ded8448c548e79b0e14b25b", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/utils.jl", "max_issues_repo_name": "xKDR/TSx.jl", "max_issues_repo_head_hexsha": "82189a7475159b3c1ded8448c548e79b0e14b25b", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/utils.jl", "max_forks_repo_name": "xKDR/TSx.jl", "max_forks_repo_head_hexsha": "82189a7475159b3c1ded8448c548e79b0e14b25b", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 18.1050847458, "max_line_length": 94, "alphanum_fraction": 0.5948324284, "num_tokens": 1891, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.42632159254749036, "lm_q2_score": 0.1294027332703691, "lm_q1q2_score": 0.05516717932782187}}
{"text": "# script to install required packages\n\nmodule Setup\n using Pkg\n\n packages = [\n :PackageCompiler, :PyCall, :PyPlot,\n :Plots, :DataFrames, :CSV, :Test,\n :StatsPlots, :Statistics, :FreqTables, \n :NamedArrays, :Distributions, :LinearAlgebra,\n :StatsBase, :PDMats, :Combinatorics, :SpecialFunctions\n ]\n\n function install_packages()\n for pkg in packages\n Pkg.add(String(pkg))\n end \n end\n\n function update_packages()\n for pkg in packages\n Pkg.update(String(pkg))\n end \n end\nend", "meta": {"hexsha": "1642540b0c57938fd19d6981afc19ebc39754312", "size": 590, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "setup.jl", "max_stars_repo_name": "ShisatoYano/JuliaAutonomy", "max_stars_repo_head_hexsha": "d1643add4ab9625996fafeac23fc03f25eedff12", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 17, "max_stars_repo_stars_event_min_datetime": "2021-03-10T12:43:52.000Z", "max_stars_repo_stars_event_max_datetime": "2021-11-01T16:40:19.000Z", "max_issues_repo_path": "setup.jl", "max_issues_repo_name": "ShisatoYano/JuliaAutonomy", "max_issues_repo_head_hexsha": "d1643add4ab9625996fafeac23fc03f25eedff12", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "setup.jl", "max_forks_repo_name": "ShisatoYano/JuliaAutonomy", "max_forks_repo_head_hexsha": "d1643add4ab9625996fafeac23fc03f25eedff12", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 2, "max_forks_repo_forks_event_min_datetime": "2021-08-14T02:46:28.000Z", "max_forks_repo_forks_event_max_datetime": "2021-11-07T09:19:40.000Z", "avg_line_length": 23.6, "max_line_length": 62, "alphanum_fraction": 0.5847457627, "num_tokens": 146, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.37754066879814546, "lm_q2_score": 0.14608725076600915, "lm_q1q2_score": 0.05515387835708148}}
{"text": "\"\"\"\nDifferenceLists, (c) 2018, Bill Burdick (William R. Burdick Jr.)\nMIT Licensed (see LICENSE file).\n\"\"\"\n\nmodule DifferenceLists\n\nexport DL, dl, concat, push, pushfirst, todl, dlconcat\n\n\"\"\"\n DL(func)\n\nGiven function `func`, construct a difference list.\n\nDifference lists are highly efficient, immutable, concatenate and prepend in constant time, and iterate in time N.\n\n# Examples\n```jldoctest\njulia> [x for x = dl(1, 2, 3)]\n3-element Array{Int64,1}:\n 1\n 2\n 3\n```\n\"\"\"\nstruct DL\n func\nend\n\n\"\"\"\n dl()::DL\n dl(items...)::DL\n\nConstruct a difference list of `items`.\n\n# Examples\n```jldoctest\njulia> dl()\ndl()\n\njulia> dl(1)\ndl(1)\n\njulia> dl(1, 2, 3)\ndl(1, 2, 3)\n\njulia> dl(1, dl(2, 3), 4)\ndl(1, dl(2, 3), 4)\n```\n\"\"\"\ndl() = DL(last -> last)\ndl(items...) = todl(items)\n\n\"\"\"\n todl(items)\n\nCreate a difference list from something you can iterate over\n\n# Examples\n```jldoctest\njulia> todl([1, 2, 3])\ndl(1, 2, 3)\n```\n\"\"\"\ntodl(items) = DL(last -> nextFor(items, iterate(items), last))\ntodl(dl::DL) = dl\n\n\"\"\"\n nextFor(items, state, last)\n\nCompute the next iteration value for an embedded collection.\n\"\"\"\nnextFor(items, ::Nothing, last) = last\nnextFor(items, (item, state), last) = item, (items, state, last)\n\n\"\"\"\n push(item, dl::DL)\n\nPush an item onto the end of a difference list.\n\n# Examples\n```jldoctest\njulia> push(2, push(1, dl(7, 8, 9)))\ndl(7, 8, 9, 1, 2)\n```\n\"\"\"\npush(dl::DL, items...) = concat(dl, todl(items))\n\n\"\"\"\n pushfirst(item, dl::DL)\n\nPush an item onto the front of a difference list.\n\n# Examples\n```jldoctest\njulia> pushfirst(1, pushfirst(2, dl(7, 8, 9)))\ndl(1, 2, 7, 8, 9)\n```\n\"\"\"\npushfirst(dl::DL, items...) = concat(todl(items), dl)\n\n\"\"\"\n concat(lists::DL...)::DL\n\nConcatenate difference lists in constant time\n\nSee also: [`dl`](@ref)\n\n# Examples\n```jldoctest\njulia> concat(dl(1, 2), dl(3, 4))\ndl(1, 2, 3, 4)\n\njulia> concat(dl(1), dl(2))\ndl(1, 2)\n```\n\"\"\"\nconcat(lists::DL...) = DL(last -> foldr((x, y) -> x.func(y), lists, init=last))\ndlconcat(lists...) = DL(last -> foldr((x, y) -> x.func(y), map(todl, lists), init=last))\n\n\"\"\"\n (a::DL)(lists::DL...)::DL\n\nA difference list itself can be used as shorthand for concat.\n\nSee also: [`dl`](@ref), [`concat`](@ref)\n\n# Examples\n```jldoctest\njulia> dl(1, 2)(dl(3, 4), dl(5, 6, 7))\ndl(1, 2, 3, 4, 5, 6, 7)\n```\n\"\"\"\n(a::DL)(lists...) = concat(a, map(todl, lists)...)\n\n# Iteration support\nBase.iterate(d::DL) = d.func(nothing)\nBase.iterate(::DL, cur::Tuple{Any, Any}) = cur\nBase.iterate(::DL, (items, state, last)::Tuple{Any, Any, Any}) = nextFor(items, iterate(items, state), last)\nBase.iterate(::DL, ::Nothing) = nothing\nBase.IteratorSize(::DL) = Base.SizeUnknown()\n\n# value display support\nBase.show(io::IO, dl::DL) = print(io, \"dl(\", join([sprint(show, x) for x = dl], \", \"), \")\")\n\nend # module\n", "meta": {"hexsha": "47651b107a307e60008a00db576db50c2ce234d4", "size": 2775, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/DifferenceLists.jl", "max_stars_repo_name": "UnofficialJuliaMirror/DifferenceLists.jl-6951a811-51c6-5b2e-b13c-b6857b9511c8", "max_stars_repo_head_hexsha": "db485d4c59969dafb2c6cd5b8fcb4d5329c8d1d1", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/DifferenceLists.jl", "max_issues_repo_name": "UnofficialJuliaMirror/DifferenceLists.jl-6951a811-51c6-5b2e-b13c-b6857b9511c8", "max_issues_repo_head_hexsha": "db485d4c59969dafb2c6cd5b8fcb4d5329c8d1d1", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 1, "max_issues_repo_issues_event_min_datetime": "2018-09-06T14:42:00.000Z", "max_issues_repo_issues_event_max_datetime": "2018-09-07T07:18:01.000Z", "max_forks_repo_path": "src/DifferenceLists.jl", "max_forks_repo_name": "UnofficialJuliaMirror/DifferenceLists.jl-6951a811-51c6-5b2e-b13c-b6857b9511c8", "max_forks_repo_head_hexsha": "db485d4c59969dafb2c6cd5b8fcb4d5329c8d1d1", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2021-08-03T12:27:13.000Z", "max_forks_repo_forks_event_max_datetime": "2021-08-03T12:27:13.000Z", "avg_line_length": 18.8775510204, "max_line_length": 114, "alphanum_fraction": 0.6154954955, "num_tokens": 979, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.43782351378493656, "lm_q2_score": 0.12592276811536138, "lm_q1q2_score": 0.05513194880179329}}
{"text": "# ------------------------------------------------------------------------------------------\n# # Packages\n#\n# Julia has over 2000 registered packages, making packages a huge part of the Julia\n# ecosystem.\n#\n# Even so, the package ecosystem still has some growing to do. Notably, we have first class\n# function calls to other languages, providing excellent foreign function interfaces. We\n# can easily call into python or R, for example, with `PyCall` or `Rcall`.\n#\n# This means that you don't have to wait until the Julia ecosystem is fully mature, and that\n# moving to Julia doesn't mean you have to give up your favorite package/library from\n# another language!\n#\n# To see all available packages, check out\n#\n# https://pkg.julialang.org/\n# or\n# https://juliaobserver.com/\n#\n# For now, let's learn how to use a package.\n# ------------------------------------------------------------------------------------------\n\n# ------------------------------------------------------------------------------------------\n# The first time you use a package on a given Julia installation, you need to use the\n# package manager to explicitly add it:\n# ------------------------------------------------------------------------------------------\n\nusing Pkg\nPkg.add(\"Example\")\n\n# ------------------------------------------------------------------------------------------\n# Every time you use Julia (start a new session at the REPL, or open a notebook for the\n# first time, for example), you load the package with the `using` keyword\n# ------------------------------------------------------------------------------------------\n\nusing Example\n\n# ------------------------------------------------------------------------------------------\n# In the source code of `Example.jl` at\n# https://github.com/JuliaLang/Example.jl/blob/master/src/Example.jl\n# we see the following function declared\n#\n# ```\n# hello(who::String) = \"Hello, $who\"\n# ```\n#\n# Having loaded `Example`, we should now be able to call `hello`\n# ------------------------------------------------------------------------------------------\n\nhello(\"it's me. I was wondering if after all these years you'd like to meet.\")\n\n# ------------------------------------------------------------------------------------------\n# Now let's play with the Colors package\n# ------------------------------------------------------------------------------------------\n\nPkg.add(\"Colors\")\n\nusing Colors\n\n# ------------------------------------------------------------------------------------------\n# Let's create a palette of 100 different colors\n# ------------------------------------------------------------------------------------------\n\npalette = distinguishable_colors(100)\n\n# ------------------------------------------------------------------------------------------\n# and then we can create a randomly checkered matrix using the `rand` command\n# ------------------------------------------------------------------------------------------\n\nrand(palette, 3, 3)\n\n# ------------------------------------------------------------------------------------------\n# In the next notebook, we'll use a new package to plot datasets.\n# ------------------------------------------------------------------------------------------\n\n# ------------------------------------------------------------------------------------------\n# ### Exercises\n#\n# #### 7.1\n# Load the Primes package (source code at https://github.com/JuliaMath/Primes.jl).\n# ------------------------------------------------------------------------------------------\n\n\n\n@assert @isdefined Primes\n\n# ------------------------------------------------------------------------------------------\n# #### 7.2\n# Verify that you can now use the function `primes` to grab all prime numbers under\n# 1,000,000 and store it in variable `primes_list`\n# ------------------------------------------------------------------------------------------\n\n\n\n@assert primes_list == primes(1000000)\n\n# ------------------------------------------------------------------------------------------\n# Please click on `Validate` on the top, once you are done with the exercises.\n# ------------------------------------------------------------------------------------------\n", "meta": {"hexsha": "b844818af23861d4872053fc4a14c590c698fa0e", "size": 4201, "ext": "jl", "lang": "Julia", "max_stars_repo_path": ".nbexports/introductory-tutorials/intro-to-julia/long-version/060 Packages.jl", "max_stars_repo_name": "grenkoca/JuliaTutorials", "max_stars_repo_head_hexsha": "3968e0430db77856112521522e10f7da0d7610a0", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 535, "max_stars_repo_stars_event_min_datetime": "2020-07-15T14:56:11.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-25T12:50:32.000Z", "max_issues_repo_path": ".nbexports/introductory-tutorials/intro-to-julia/long-version/060 Packages.jl", "max_issues_repo_name": "grenkoca/JuliaTutorials", "max_issues_repo_head_hexsha": "3968e0430db77856112521522e10f7da0d7610a0", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 42, "max_issues_repo_issues_event_min_datetime": "2018-02-25T22:53:47.000Z", "max_issues_repo_issues_event_max_datetime": "2020-05-14T02:15:50.000Z", "max_forks_repo_path": ".nbexports/introductory-tutorials/intro-to-julia/long-version/060 Packages.jl", "max_forks_repo_name": "grenkoca/JuliaTutorials", "max_forks_repo_head_hexsha": "3968e0430db77856112521522e10f7da0d7610a0", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 394, "max_forks_repo_forks_event_min_datetime": "2020-07-14T23:22:24.000Z", "max_forks_repo_forks_event_max_datetime": "2022-03-28T20:12:57.000Z", "avg_line_length": 41.5940594059, "max_line_length": 92, "alphanum_fraction": 0.3741966199, "num_tokens": 627, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4035668680822513, "lm_q2_score": 0.13660839354130014, "lm_q1q2_score": 0.05513062153521014}}
{"text": "using StaticCompiler\nusing Test\nusing Libdl\nusing LinearAlgebra\nusing LoopVectorization\nusing ManualMemory\nusing StrideArraysCore\nusing Distributed\n\naddprocs(1)\n@everywhere using StaticCompiler\n\nremote_load_call(path, args...) = fetch(@spawnat 2 load_function(path)(args...))\n\n@testset \"Basics\" begin\n\n simple_sum(x) = x + one(typeof(x))\n\n # This probably needs a macro\n for T \u2208 (Int, Float64, Int32, Float32, Int16, Float16)\n _, path, = compile(simple_sum, (T,))\n @test remote_load_call(path, T(1)) == T(2)\n end\nend\n\n\nfib(n) = n <= 1 ? n : fib(n - 1) + fib(n - 2) # This needs to be defined globally due to https://github.com/JuliaLang/julia/issues/40990\n\n@testset \"Recursion\" begin\n _, path = compile(fib, (Int,))\n @test remote_load_call(path, 10) == fib(10)\n\n # Trick to work around #40990\n _fib2(_fib2, n) = n <= 1 ? n : _fib2(_fib2, n-1) + _fib2(_fib2, n-2)\n fib2(n) = _fib2(_fib2, n)\n \n _, path = compile(fib2, (Int,))\n @test remote_load_call(path, 20) == fib(20)\n #@test compile(fib2, (Int,))[1](20) == fib(20) \nend\n\n# Call binaries for testing\n# @testset \"Generate binary\" begin\n# fib(n) = n <= 1 ? n : fib(n - 1) + fib(n - 2)\n# libname = tempname() \n# generate_shlib(fib, (Int,), libname)\n# ptr = Libdl.dlopen(libname * \".\" * Libdl.dlext, Libdl.RTLD_LOCAL)\n# fptr = Libdl.dlsym(ptr, \"julia_fib\")\n# @assert fptr != C_NULL\n# # This works on REPL\n# @test_skip ccall(fptr, Int, (Int,), 10) == 55\n# end\n\n\n@testset \"Loops\" begin\n function sum_first_N_int(N)\n s = 0\n for a in 1:N\n s += a\n end\n s\n end\n _, path = compile(sum_first_N_int, (Int,))\n @test remote_load_call(path, 10) == 55\n \n function sum_first_N_float64(N)\n s = Float64(0)\n for a in 1:N\n s += Float64(a)\n end\n s\n end\n _, path = compile(sum_first_N_float64, (Int,))\n @test remote_load_call(path, 10) == 55.\n\n function sum_first_N_int_inbounds(N)\n s = 0\n @inbounds for a in 1:N\n s += a\n end\n s\n end\n _, path = compile(sum_first_N_int_inbounds, (Int,))\n @test remote_load_call(path, 10) == 55\n\n function sum_first_N_float64_inbounds(N)\n s = Float64(0)\n @inbounds for a in 1:N\n s += Float64(a)\n end\n s\n end\n _, path = compile(sum_first_N_float64_inbounds, (Int,))\n @test remote_load_call(path, 10) == 55.\nend\n\n# Arrays with different input types Int32, Int64, Float32, Float64, Complex?\n@testset \"Arrays\" begin\n function array_sum(n, A)\n s = zero(eltype(A))\n for i in 1:n\n s += A[i]\n end\n s\n end\n for T \u2208 (Int, Complex{Float32}, Complex{Float64})\n _, path = compile(array_sum, (Int, Vector{T}))\n @test remote_load_call(path, 10, T.(1:10)) == T(55)\n end\n\n # @test (10, Int.(1:10)) == 55\n # @test compile(array_sum, (Int, Vector{Complex{Float32}}))[1](10, Complex{Float32}.(1:10)) == 55f0 + 0f0im\n # @test compile(array_sum, (Int, Vector{Complex{Float64}}))[1](10, Complex{Float64}.(1:10)) == 55f0 + 0f0im\nend\n\n\n# Julia wants to treat Tuple (and other things like it) as plain bits, but LLVM wants to treat it as something with a pointer.\n# We need to be careful to not send, nor receive an unwrapped Tuple to a compiled function.\n# The interface made in `compile` should handle this fine. \n@testset \"Send and receive Tuple\" begin\n foo(u::Tuple) = 2 .* reverse(u) .- 1\n \n _, path = compile(foo, (NTuple{3, Int},))\n @test remote_load_call(path, (1, 2, 3)) == (5, 3, 1)\nend\n\n\n# Just to call external libraries\n@testset \"BLAS\" begin\n function mydot(a::Vector{Float64})\n N = length(a)\n BLAS.dot(N, a, 1, a, 1)\n end\n a = [1.0, 2.0]\n mydot_compiled, path = compile(mydot, (Vector{Float64},))\n @test_skip remote_load_call(path, a) == 5.0 # this needs a relocatable pointer to work\n @test mydot_compiled(a) == 5.0\nend\n\n\n@testset \"Hello World\" begin\n function hello(N)\n println(\"Hello World $N\")\n N\n end\n # How do I test this?\n # Also ... this segfaults\n @test_skip ccall(generate_shlib_fptr(hello, (Int,)), Int, (Int,), 1) == 1\nend\n\n# I can't beleive this works.\n@testset \"LoopVectorization\" begin\n function mul!(C, A, B)\n # note: @tturbo does NOT work\n @turbo for n \u2208 indices((C,B), 2), m \u2208 indices((C,A), 1)\n Cmn = zero(eltype(C))\n for k \u2208 indices((A,B), (2,1))\n Cmn += A[m,k] * B[k,n]\n end\n C[m,n] = Cmn\n end\n end\n\n C = Array{Float64}(undef, 10, 12)\n A = rand(10, 11)\n B = rand(11, 12)\n\n _, path = compile(mul!, (Matrix{Float64}, Matrix{Float64}, Matrix{Float64},))\n # remote_load_call(path, C, A, B) This won't work because @spawnat copies C\n C .= fetch(@spawnat 2 (load_function(path)(C, A, B); C))\n @test C \u2248 A*B\nend\n\n# This is a trick to get stack allocated arrays inside a function body (so long as they don't escape).\n# This lets us have intermediate, mutable stack allocated arrays inside our \n@testset \"Alloca\" begin\n function f(N)\n # this can hold at most 100 Int values, if you use it for more, you'll segfault\n buf = ManualMemory.MemoryBuffer{100, Int}(undef)\n GC.@preserve buf begin\n # wrap the first N values in a PtrArray\n arr = PtrArray(pointer(buf), (N,))\n arr .= 1 # mutate the array to be all 1s\n sum(arr) # compute the sum. It is very imporatant that no references to arr escape the function body\n end\n end\n _, path = compile(f, (Int,))\n @test remote_load_call(path, 20) == 20\nend \n\n\n# data structures, dictionaries, tuples, named tuples\n", "meta": {"hexsha": "9c1191babd84eba06c2ec7761fb54d4f332461c6", "size": 5744, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "test/runtests.jl", "max_stars_repo_name": "MasonProtter/StaticCompiler.jl", "max_stars_repo_head_hexsha": "4d348e52a430365a3e13aa8da6222a267e3e51d2", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2022-02-02T02:59:55.000Z", "max_stars_repo_stars_event_max_datetime": "2022-02-02T02:59:55.000Z", "max_issues_repo_path": "test/runtests.jl", "max_issues_repo_name": "MasonProtter/StaticCompiler.jl", "max_issues_repo_head_hexsha": "4d348e52a430365a3e13aa8da6222a267e3e51d2", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 1, "max_issues_repo_issues_event_min_datetime": "2022-02-04T04:17:41.000Z", "max_issues_repo_issues_event_max_datetime": "2022-02-04T04:49:10.000Z", "max_forks_repo_path": "test/runtests.jl", "max_forks_repo_name": "MasonProtter/StaticCompiler.jl", "max_forks_repo_head_hexsha": "4d348e52a430365a3e13aa8da6222a267e3e51d2", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 2, "max_forks_repo_forks_event_min_datetime": "2022-02-04T02:15:05.000Z", "max_forks_repo_forks_event_max_datetime": "2022-02-04T04:51:07.000Z", "avg_line_length": 29.7616580311, "max_line_length": 136, "alphanum_fraction": 0.5995821727, "num_tokens": 1811, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.46879062662624377, "lm_q2_score": 0.11757214127540647, "lm_q1q2_score": 0.05511671778228706}}
{"text": "module TestThis\n\n# package code goes here\nprintln(\"Sherlock! We are loading the module now...\")\nusing PyPlot\n\nfunction greet(name)\n\tprintln(\"Hello, $name\")\nend\n\nfunction addme(a,b)\n\treturn a + b\nend\n\nfunction plotStuff(savehere::String)\n\ta = [1,2,3,4]\n\tb = [5,4,5,3]\n\tplot(a,b)\n\tsavefig(savehere*\".png\")\nend\n\nend # module\n", "meta": {"hexsha": "a1386de03924ee30f24b1933ebf927ce48f56dba", "size": 322, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/TestThis.jl", "max_stars_repo_name": "rymanderson/TestThis.jl", "max_stars_repo_head_hexsha": "b6a236ddae2525a8c2c0abd8e94ed3135719c5ef", "max_stars_repo_licenses": ["MIT"], "max_stars_count": null, "max_stars_repo_stars_event_min_datetime": null, "max_stars_repo_stars_event_max_datetime": null, "max_issues_repo_path": "src/TestThis.jl", "max_issues_repo_name": "rymanderson/TestThis.jl", "max_issues_repo_head_hexsha": "b6a236ddae2525a8c2c0abd8e94ed3135719c5ef", "max_issues_repo_licenses": ["MIT"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "src/TestThis.jl", "max_forks_repo_name": "rymanderson/TestThis.jl", "max_forks_repo_head_hexsha": "b6a236ddae2525a8c2c0abd8e94ed3135719c5ef", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 14.0, "max_line_length": 53, "alphanum_fraction": 0.6894409938, "num_tokens": 101, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.48828339529583464, "lm_q2_score": 0.11279541523008142, "lm_q1q2_score": 0.05507612832234766}}
{"text": "module Grader\n\nexport Problem, rungolden!, runstudent!, grade!, pl_JSON\n\nusing Parameters\nimport Random\nimport JSON\n\n@with_kw mutable struct Image\n label::String = \"\"\n url::String = \"\"\nend\n\n@with_kw mutable struct TestResult\n name::String = \"\"\n description::String = \"\"\n points::Float64 = 0\n max_points::Float64 = 0\n message::String = \"\"\n output::String = \"\"\n images::Vector{Image} = []\nend\n\n@with_kw mutable struct Problem\n gradable::Bool = true\n score::Float64 = 0\n message::String = \"\"\n output::String = \"\"\n\n images::Vector{Image} = []\n\n tests::Vector{TestResult} = []\nend\n\n# Evaluate the provided code string inside a module,\n# and return the resulting module. May not \n# work if expected if the code string already is a module.\nfunction evalasmodule(code::AbstractString)\n mod = \"module \" * Random.randstring(Random.MersenneTwister(), \"abcdefghijklmnopqrstuvwxyz\", 20) * \"\\n\" * code * \"\\nend\"\n expr = Meta.parse(mod)\n eval(expr)\nend\n\nfunction rungolden!(p::Problem, goldencode::AbstractString)::Module\n goldenresult = Module()\n try\n goldenresult = evalasmodule(goldencode)\n catch err\n p.output = p.output * \"error running golden code:\\n\" * sprint(showerror, err, backtrace()) * \"\\n\"\n p.gradable = false\n p.message = \"Internal grading error, please notify instructor.\"\n end\n return goldenresult\nend\n\nfunction runstudent!(p::Problem, studentcode::AbstractString)::Module\n studentresult = Module()\n try\n studentresult = evalasmodule(studentcode)\n catch err\n p.output = p.output * \"error running student code:\\n\" * sprint(showerror, err, backtrace()) * \"\\n\"\n p.gradable = false\n p.message = \"There was an error running your code, please see information below.\"\n end\n return studentresult\nend\n\n\nfunction grade!(p::Problem, name::String, description::String, points::Real, expr::Expr, msg_if_incorrect::String)\n if !p.gradable \n return nothing\n end\n \n t = TestResult(max_points=points, name=name, description=description)\n\n correct = false\n try \n correct = eval(expr)\n catch err\n t.output = t.output * \"\\n\" * sprint(showerror, err)\n end\n\n if correct === missing\n t.message = \"your answer contains a 'missing' value\"\n else\n try\n correct ? t.points = points : t.message = msg_if_incorrect\n catch err\n t.message = msg_if_incorrect\n t.output = t.output * \"\\n\" * sprint(showerror, err)\n end\n end\n\n append!(p.tests, [t])\n\n pts = 0\n totalpts = 0\n for t in p.tests\n pts += t.points\n totalpts += t.max_points\n end\n p.score = float(pts) / float(totalpts)\n return nothing\nend\n\nfunction pl_JSON(io::IO, p::Problem)\n JSON.print(io, p)\nend\n\nend", "meta": {"hexsha": "eb5f189bc7b648803bba9b2ae6ae0c0112984e69", "size": 2818, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "src/Grader.jl", "max_stars_repo_name": "ctessum/Grader.jl", "max_stars_repo_head_hexsha": "26b39510625c578f3d7b1bc46f4d5db385810fb0", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 1, "max_stars_repo_stars_event_min_datetime": "2021-11-17T16:58:33.000Z", "max_stars_repo_stars_event_max_datetime": "2021-11-17T16:58:33.000Z", "max_issues_repo_path": "src/Grader.jl", "max_issues_repo_name": "ctessum/Grader.jl", "max_issues_repo_head_hexsha": "26b39510625c578f3d7b1bc46f4d5db385810fb0", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 1, "max_issues_repo_issues_event_min_datetime": "2021-06-15T00:47:33.000Z", "max_issues_repo_issues_event_max_datetime": "2021-06-15T00:47:33.000Z", "max_forks_repo_path": "src/Grader.jl", "max_forks_repo_name": "ctessum/Grader.jl", "max_forks_repo_head_hexsha": "26b39510625c578f3d7b1bc46f4d5db385810fb0", "max_forks_repo_licenses": ["MIT"], "max_forks_count": null, "max_forks_repo_forks_event_min_datetime": null, "max_forks_repo_forks_event_max_datetime": null, "avg_line_length": 25.6181818182, "max_line_length": 123, "alphanum_fraction": 0.64158978, "num_tokens": 720, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.4493926344647597, "lm_q2_score": 0.12252322533450248, "lm_q1q2_score": 0.05506103501619146}}
{"text": "# ------------------------------------------------------------------------------------------\n# # Introduction to DataFrames\n# **[Bogumi\u0142 Kami\u0144ski](http://bogumilkaminski.pl/about/), Apr 21, 2017**\n# ------------------------------------------------------------------------------------------\n\nusing DataFrames # load package\n\n# ------------------------------------------------------------------------------------------\n# ## Joining DataFrames\n# ------------------------------------------------------------------------------------------\n\n# ------------------------------------------------------------------------------------------\n# ### Preparing DataFrames for a join\n# ------------------------------------------------------------------------------------------\n\nx = DataFrame(ID=[1,2,3,4,missing], name = [\"Alice\", \"Bob\", \"Conor\", \"Dave\",\"Zed\"])\ny = DataFrame(id=[1,2,5,6,missing], age = [21,22,23,24,99])\nx,y\n\nrename!(x, :ID=>:id) # names of columns on which we want to join must be the same\n\n# ------------------------------------------------------------------------------------------\n# ### Standard joins: inner, left, right, outer, semi, anti\n# ------------------------------------------------------------------------------------------\n\njoin(x, y, on=:id) # :inner join by default, missing is joined\n\njoin(x, y, on=:id, kind=:left)\n\njoin(x, y, on=:id, kind=:right)\n\njoin(x, y, on=:id, kind=:outer)\n\njoin(x, y, on=:id, kind=:semi)\n\njoin(x, y, on=:id, kind=:anti)\n\n# ------------------------------------------------------------------------------------------\n# ### Cross join\n# ------------------------------------------------------------------------------------------\n\n# cross-join does not require on argument\n# it produces a Cartesian product or arguments\nfunction expand_grid(;xs...) # a simple replacement for expand.grid in R\n reduce((x,y) -> join(x, DataFrame(Pair(y...)), kind=:cross),\n DataFrame(Pair(xs[1]...)), xs[2:end])\nend\n\nexpand_grid(a=[1,2], b=[\"a\",\"b\",\"c\"], c=[true,false])\n\n# ------------------------------------------------------------------------------------------\n# ### Complex cases of joins\n# ------------------------------------------------------------------------------------------\n\nx = DataFrame(id1=[1,1,2,2,missing,missing],\n id2=[1,11,2,21,missing,99],\n name = [\"Alice\", \"Bob\", \"Conor\", \"Dave\",\"Zed\", \"Zoe\"])\ny = DataFrame(id1=[1,1,3,3,missing,missing],\n id2=[11,1,31,3,missing,999],\n age = [21,22,23,24,99, 100])\nx,y\n\njoin(x, y, on=[:id1, :id2]) # joining on two columns\n\njoin(x, y, on=[:id1], makeunique=true) # with duplicates all combinations are produced (here :inner join)\n\njoin(x, y, on=[:id1], kind=:semi) # but not by :semi join (as it would duplicate rows)\n", "meta": {"hexsha": "346e226953f6b20dea68c92780d7ef025aeb3316", "size": 2761, "ext": "jl", "lang": "Julia", "max_stars_repo_path": ".nbexports/introductory-tutorials/broader-topics-and-ecosystem/intro-to-julia-DataFrames/08_joins.jl", "max_stars_repo_name": "grenkoca/JuliaTutorials", "max_stars_repo_head_hexsha": "3968e0430db77856112521522e10f7da0d7610a0", "max_stars_repo_licenses": ["MIT"], "max_stars_count": 535, "max_stars_repo_stars_event_min_datetime": "2020-07-15T14:56:11.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-25T12:50:32.000Z", "max_issues_repo_path": ".nbexports/introductory-tutorials/broader-topics-and-ecosystem/intro-to-julia-DataFrames/08_joins.jl", "max_issues_repo_name": "grenkoca/JuliaTutorials", "max_issues_repo_head_hexsha": "3968e0430db77856112521522e10f7da0d7610a0", "max_issues_repo_licenses": ["MIT"], "max_issues_count": 42, "max_issues_repo_issues_event_min_datetime": "2018-02-25T22:53:47.000Z", "max_issues_repo_issues_event_max_datetime": "2020-05-14T02:15:50.000Z", "max_forks_repo_path": ".nbexports/introductory-tutorials/broader-topics-and-ecosystem/intro-to-julia-DataFrames/08_joins.jl", "max_forks_repo_name": "grenkoca/JuliaTutorials", "max_forks_repo_head_hexsha": "3968e0430db77856112521522e10f7da0d7610a0", "max_forks_repo_licenses": ["MIT"], "max_forks_count": 394, "max_forks_repo_forks_event_min_datetime": "2020-07-14T23:22:24.000Z", "max_forks_repo_forks_event_max_datetime": "2022-03-28T20:12:57.000Z", "avg_line_length": 40.6029411765, "max_line_length": 105, "alphanum_fraction": 0.367982615, "num_tokens": 583, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO", "lm_q1_score": 0.44939263446475963, "lm_q2_score": 0.12252321251815919, "lm_q1q2_score": 0.05506102925662117}}
{"text": "#!/usr/bin/env julia\n\nx = begin\n a = 5\n b = 6\n a + b\nend\n\nprintln(\"x = $x, a = $a, b = $b\")\n\nx = begin\n a = 9\n b = -9\n a + b\nend\n\nprintln(\"x = $x, a = $a, b = $b\")\n", "meta": {"hexsha": "37066ed48ea333ef1ba3891423fdb1e322d9b5ab", "size": 182, "ext": "jl", "lang": "Julia", "max_stars_repo_path": "source-code/semantics/compound_expressions.jl", "max_stars_repo_name": "gjbex/Julia_good_bad_ugly", "max_stars_repo_head_hexsha": "0d5d25bb6cc80aa36eae6758d2c86d3106e9bcee", "max_stars_repo_licenses": ["CC-BY-4.0"], "max_stars_count": 2, "max_stars_repo_stars_event_min_datetime": "2021-07-12T14:32:43.000Z", "max_stars_repo_stars_event_max_datetime": "2022-03-18T08:04:35.000Z", "max_issues_repo_path": "source-code/semantics/compound_expressions.jl", "max_issues_repo_name": "gjbex/Julia_good_bad_ugly", "max_issues_repo_head_hexsha": "0d5d25bb6cc80aa36eae6758d2c86d3106e9bcee", "max_issues_repo_licenses": ["CC-BY-4.0"], "max_issues_count": null, "max_issues_repo_issues_event_min_datetime": null, "max_issues_repo_issues_event_max_datetime": null, "max_forks_repo_path": "source-code/semantics/compound_expressions.jl", "max_forks_repo_name": "gjbex/Julia_good_bad_ugly", "max_forks_repo_head_hexsha": "0d5d25bb6cc80aa36eae6758d2c86d3106e9bcee", "max_forks_repo_licenses": ["CC-BY-4.0"], "max_forks_count": 1, "max_forks_repo_forks_event_min_datetime": "2022-03-18T08:03:54.000Z", "max_forks_repo_forks_event_max_datetime": "2022-03-18T08:03:54.000Z", "avg_line_length": 10.1111111111, "max_line_length": 33, "alphanum_fraction": 0.3846153846, "num_tokens": 87, "lm_name": "Qwen/Qwen-72B", "lm_label": "1. NO\n2. NO\n\n", "lm_q1_score": 0.4493926344647597, "lm_q2_score": 0.12252320931407355, "lm_q1q2_score": 0.0550610278167287}}
{"text": "### A Pluto.jl notebook ###\n# v0.14.0\n\nusing Markdown\nusing InteractiveUtils\n\n# This Pluto notebook uses @bind for interactivity. When running this notebook outside of Pluto, the following 'mock version' of @bind gives bound variables a default value (instead of an error).\nmacro bind(def, element)\n quote\n local el = $(esc(element))\n global $(esc(def)) = Core.applicable(Base.get, el) ? Base.get(el) : missing\n el\n end\nend\n\n# \u2554\u2550\u2561 db24490e-7eac-11ea-094e-9d3fc8f22784\nmd\"# Introducing _bound_ variables\n\nWith the new `@bind` macro, Pluto.jl can listen to real-time events from HTML objects!\"\n\n# \u2554\u2550\u2561 bd24d02c-7eac-11ea-14ab-95021678e71e\n@bind x html\"First Second